Note: Descriptions are shown in the official language in which they were submitted.
CA 02410688 2002-12-12
A SYS'fEllR Ft)lt (:Ii,LL-I~ASEI) SCREENING
Field of The Invention
This invention is in thc: held of fluorescence-based cell and molecular
biochemical assays for drug discovery.
Backt=_round of the Invention
Drug discovery, as currently pr~~rcticed in the art, is a long, multiple step
process
involving identification of specific riisease targets, development of an assay
based on a
specific target, validation of the a~:,s<~y, optimization and automation of
the assay to
produce a screen, hieh throughput ;screening of compound libraries using the
assay to
identify "lots", hit validation and hit compound optimization, The output of
tlvs
process is a lead compound that goes into pre-clinical and, if validated,
eventually into
clinical trials. In this process, the screening phase is distinct from the
assay
development phases, and involves;. ~Eesting compound efficacy in living
biological
systems.
CA 02410688 2002-12-12
Historically, drug discovery is a slow arld costly process, spanning numerous
years and consuming hundreds of millions of dollars per drug created.
Developments
in the areas of genomics and hi;h throughput screening have resulted in
increased
capacity and efficiency in the are,:l.s of target identification and volume of
compounds
screened. Significant advances in automated DNA sequencing, PCR application,
positional cloning, hybridizati<7n ~tr~ays, and b101Ilformatlcs have greatly
increased the
number of genes (and gene fra~rrnents) encoding potential drug screening
targets.
However, the basic scheme for dru; screening rernians the same.
Validation of genomic targets as points for therapeutic intervention using the
to existing methods and protocol , ha; hecoltre a bottlelaeck in the drug
discovery proce<.~s
due to the slow, manual metho;_ls employed, such as ~n uvo functional models,
functional analysis of recombinant proteins, and stable cell line expression
of candidate
genes. Primary DN.~1 sequence data acquired through automated sequencing does
not
permit identification of gene function, hut can provide; inforn~ation about
common
t 5 "motifs" and specific gene homology when compared to known sequence
databases.
Genomic methods such as subtrac~!ion hybridi-r,ation ;end It.,AL>1~ (rapid
amplification of
differential expression) can be used to identify genes that are up or down
regulated in a
disease state model. However, ider~cilication and validation still proceed
down the same
pathway. Some proteomic methods use protein ident5fication t~~lobal expression
arrays,
?0 2D electrophoresis, combinatorial libraries) in combination with reverse
genetics to
identify candidate genes of interest. Such putative "~.iisrase associated
sequences" or
DAS isolated as intact cDNA arc a ;;real advarotage to these methods. but they
are
identified by the hundreds without providing any intorrrlation regarding type,
activity,
and distribution of the encoded prc:~teirZ. t'hoosing a subset of DAS as drug
screening
CA 02410688 2002-12-12
targets is "random", and thus extremely inefficient, without functional data
to provide a
mechanistic link with disease. It is necessary, therefore, to pravide new
technologies to
rapidly screen DAS to establish biolagical function, thereby improving target
validation
and candidate optimization in dru,, discovery.
There are three major avenues for improving early drug discovery productivity.
First, there is a need for tools that l7rovide increased info>rmation handling
capability.
Bioinformatics has blossomed with the rapid development of DNA sequencing
systems
and the evolution of the genomic:s clatahase. Genomics is beginning to play a
critical
role in the identification of potenvial new targets. Proteomics has become
indispensible
1o in relating structure and function of Iv>rotein targets in order to predict
drug interactions.
However, the next level of bialoc;ical complexity is the cell. ~hherefr~re,
there is a need
to acquire, manage and search rvulti-dimensional infcrrmatian from cells.
Secondly,
there is a need for higher throu;_~hput tools. Automation is a key to
improving
productivity as has already been cicr77onstrated in DNA sequencing and high
throughput
n primary screening. The instant iia~ewtion provides for automated systems
that extract
multiple parameter information Irr>rr7 cells that meet the need for higher
throughput
tools. The instant invention else provides for miniaturizing the methods,
thereby
allowing increased throughput, w~~ile decreasing the volumes of reagents and
test
compounds requirecj in each assa~r.
2o Radioactivity has been tl~e uominant read-out in e<rriv drug discovery
assays.
However, the need for more inl~~rm~rtion, higher throughput and
miniaturization has
caused a shift towards using tluorescence detectior~_ Flr,wrc cence-based
reagents c:an
yield more powerful, multiple parameter assays that are higher rn throughput
and
- CA 02410688 2002-12-12
information content and require lower volumes of reagents and test compounds.
Fluorescence is also safer and less expensive than radioactivity-based
methods.
Screening of cells treated ~rith dyes and fluorescent reagents is well known
in
the art. There is a considerable body of literahtre related to genetic
engineering of cells
to produce fluorescent proteins, such as modified green fluorescent protein
(GFP), as a
reporter molecule. Some properties .of wild-type GF'P are disclosed by Morise
et al.
(Biochemistry 13 (1974), p. 2656-~2fa62), and Ward et al. (Photochem.
Photobiol. 31
(1980), p. 611-615). The GFP of.' the jellyfish .4eguorea victoria has an
excitation
maximum at 395 nm and an emission maximum at 510 nm, and does not require an
exogenous factor for fluorescence ~rctivity.. Uses for GFP disclosed in the
literature are
widespread and include the study of gene expression and protein localization
(Chalfie
et al., Science 263 (1994), p. 1501-12504)), as a tool for visualizing
subcellular
organelles (ltizzuto et al., (.,urr. Bi.;alc~y ~ (1995), p. ti35-fi42)),
visualization of protein
transport along the secretory pathv,~a;y (Kaether and Gerdes, FEES Letters 369
(1995),
p. 267-271)), expression in plant cells (I-lu and Cheng, FEBS Letters 369
(1995), p.
331-334)) and Drosophila embryos (Davis et al., f~ev. Bialo~ry 170 (1995), p.
726-
729)), and as a reporter molecule; hosed to another protein of" interest (U.
S. Patent
5,491,084 issued February 13,199t~). Similarly, W096/2:3898 published August
8,1996,
relates to methods of detecting l~ialogically active substances affecting
intracellular
processes by utilizing a GF'P constmct having a protein kinase activation
site.
Numerous references are :related to GFl' proteins in biological systems. For
example, WO 96/09598 published March 28,1996, describes a system for isolating
cells
of interest utilizing the expression caf a UFP like protein. WO 96/27675
published
September 12, 1996, describes tho expression of GI~F' in
4
CA 02410688 2002-12-12
plants. WO 95/21191 published August 10, 1995, describes modified GFP protein
expressed in transformed organisms to detect rnutagen Isis. U. S. Patents
5,401,629,
issued March 28, 1995, and 5,4:6,128, issued Juty 2'.i, 1995, describe assays
and
compositions for detecting and evaluating the intracellular transduction of an
extracellular
signal using recombinant cells that cyxl>ress cell surface receptors and
contain reporter
gene constructs that include transcripoional regulatory elements that are
responsive to the
activity of the cell surface receptors.
Performing a screen on many thousands of compounds requires parallel
handling and processing of many compounds and assay component reagents.
Standard
high throughput screens ("I-iTS") tisc~ mixtures of compounds and biological
reagents
along with some indicator compound loaded into arrays of wells in standard
microtiter
plates with 96 or 384 wells. The ~.:ignal measured from each well. either
fluorescence
emission, optical density, or radioactivity, integrates the signal from all
the material in
the well giving an overall population average of all the molecules in the
well.
Science Applications Intennational Corporation (SAIC.'j 130 Fifth Avenue,
Seattle, WA. 98109] describes arll inuaging plate reader. This system uses a
CCD
camera to image the whole area of a 96 well plate. 'The image is analyzed to
calculate
the total fluorescence per well for all the material in the well.
rM
Molecular Devices, lnc. (S:mnyvale, CA) describes a system (FLIPR) which
uses low angle laser scanning illumination and a mask to selectively excite
fluorescence
within approximately 200 microns of the bottoms of the wells in standard 96
well
plates in order to reduce backgroun.::l when imaging; cell monolayers. This
system uses
a CCD camera to image the whole: area of the plate bottom. Although this
system
measures signals originating from a cell monolayer at the bottom of the well,
the signal
measured is averaged over the area of the well and is therefore still
considered a
5
CA 02410688 2002-12-12
measurement of the average response of a population of cells. The image is
analyzed to
calculate the total fluorescence per- vrc;ll l~or cell-based assays. Fluid
delivery devices
have also been incorporated into cell based screening systems, such as the
FLIPRTM
system, in order to initiate a re~p<>rtse, which is then observed as a whole
well
population average response using .a macro-imaging system.
In contrast to high througtwput screens, various high-content screens ("HCS")
have been developed to address th<; need for more detailed information about
the
temporal-spatial dynamics of cell constituents and processes. High-content
screens
automate the extraction of multicculor fluorescence information derived from
specific
fluorescence-based reagents incorp;>rated into cells (Ciiuliana and Taylor
(1995), Curr.
Op. Cell Biol. 7:4; Criuliano et al. (1995) Ann. Rev. Biaphys. Biomol. Struct.
24:405).
Cells are analyzed using an optical system that can measure spatial, as well
as temporal
dynamics. (Farkas et al. {1993) Ar;~n. Rev. Physiol. :5:785; Ciiuliano et al.
(1990) In
Optical Microscopy ,for Biology. 13. :Herman and K:, Jacobson (eds.), pp. 543-
557.
Wiley-Liss, New York; Hahn et ,al ( 1992) Nature 359:73(i; Waggoner et al.
(1996)
Hum. Pathol. 27:494). The concept: is to treat each toll as a "well" that has
spatial and
temporal information on the activities of the labeled canstit:uents.
The types of biochemical ;:jlld molecular information now accessible through
fluorescence-based reagents applied to cells include ion concentrations,
membrane
potential, specific translocations, f~n~rytne activities, gene; expression, as
well as the
presence, amounts and patterns of o'etabolites, proteins, lipids,
carbohydrates, and
nucleic acid sequences (DeBiasio ~:~t ,:i1., ( 1996) Mol. Biol. (dell.
7:1259;Giuliano et al.,
(1995) Ann. Rev. Biaphys. Biomol. Stroct. 24:405:, Heirn and Tsien, ( 1996)
Curr. Biol.
6:178).
(a
CA 02410688 2002-12-12
High-content screens can be performed on either fixed cells, using
fluorescently
labeled antibodies, biological li~arus, and,%or nucleus acid hybridization
probes, or live
cells using multicolor fluorescent indicators and "biosensors." T'he choice of
fixed or
live cell screens depends on the :pec.ihc cell-based assay required.
Fixed cell assays are the: simplest, since an away of initially living cells
in a
microtiter plate for~tnat can be tr~f:ated with various corr~pounds and doses
being tested,
then the cells cart be fixed, lat:~eled with specific reagents, and measured.
No
environmental control of the cells is required after tixation. Spatial
information is
acquired, but only at one time point. The availability of thousands of
antibodies,
1ct ligands and nucleic: acid hybridisation probes that can ~~e applied to
cells makes this an
attractive approach for many types of cell-based screens. The fixation and
labeling
steps can be automated, allowing; efficient processing of assays.
Live cell assays are more: sophisticated and powerful, since an array of
living
cells containing the desired reagents can be screened over time, as well as
space.
1. Environmental control of the cells (temperature, humidity, and carbon
dioxide) is
required during measurement, since the physiological health of the cells must
be
maintained for multiple fluoresca~n~c~~ measurements over tune. There is a
growing list
of fluorescent physiological im'.lic;ators and "biosensors" that can report
changes in
biochemical and nuolecular acm,.~itit~s within calls (Ciiuliaruo et al_, (
L'~95) .~fttn. Rev.
?C~ Biophys. Biomol. 5'truct. '~4:~(.)s., Ilahn et al., ( I993p In
lvluur~esc~ent and Luminescent
Probes for Biologiccil Activity. ',%V'I'. Mason, (eed.), pp 349-359, .academic
Press, San
Diego).
The availability and use of tluorescence-based reagents has helped to advance
the development of both fixe~.l and live cell high-content screens. Advances
in
CA 02410688 2002-12-12
instrumentation to automatically extract multicolor, high-content information
has
recently made it possible to develc:~p I-1CS into an automated tool. An
article by Taylor.,
et al. (American Sci~>~trist ~;0 (199::'j, ~~. s~?-33pj desr.ribes many of
these methods and
their applications. For example, Proffitt et. al. (C:ytometry 24: 204-213
(1996)) describe
a semi-automated fluores~:ence digital imaging system for quantifying relative
cell
numbers in situ in a variety of tiss~.ie culture plate formats, especially 96-
well microtiter
plates. The system consists of an epifluorescence inverted microscope with a
motorized stage, video camera, im~~ge intensifier, and a microcomputer with a
PC-
TM
Vision digitizer. Turbo Pascal software controls thc~ stage and scans the
plate taking
multiple images per well. 'the software calculates total fluorescence per
well, provides
for daily calibration, and configur~;s easily for a variety of tissue culture
plate formats..
Thresholding of digital images arid reagents which fluoresce only when taken
up by
living cells are used to reduce background fluorescence without removing
excess
fluorescent reagent.
Scanning confocal microscope imaging (Go et al., (1997) Analytical
Biochemistry 247:210-215; Cioldman et al., ( 1995) Erperimental C.'ell
Research
221:311-319) and multiphoton minwrc>scope imaging (Denk et al., (1990)
Science'
248:73; Gratton et al., (1994) Pro~r, ojthe Microscohical ,,society of
America, pp. 154--
155) are also well established methods for acquiring; high resolution images
of
microscopic samples. The principle advantage of these optical systems is the
very
shallow depth of focus, which allows features of limited axial extent to be
resolved
against the background. F'or exarnl>le, it is possible to resolve internal
cytoplasmic
features of adherent cells from the features on the cell surface. Because
scanning
multiphoton imaging requires veryr short duration pulsed laser systems to
achieve the
8
CA 02410688 2002-12-12
high photon flux required, fluorescence lifetimes can also be measured in
these systems
(Lakowicz et al., ( 1992) Anal. urrachem. ?02:316-330; ~:ierrittsen et al. (
1997), J. of
Fluorescence 7:1 l -15)), providing, additional capability for different
detection med:s.
Small, reliable and relatively ins°xpensive laser systems, such as
laser diode pumped
lasers, are now available to allo~'N multiphoton confocal microscopy to be
applied in a
fairly routine fashion.
A combination of the biological heterogeneity of cells in populations (Bright,
et
al., (1989). J. Cell. Phvsiol. 141:41(1; CJiuliano, (1996) (,ell:'hfotil.
(:vtoskel. 35:237)} as
well as the high spatial and temporal frequency of cherrucal and molecular
information
iC~ present within cells, makes it impossible to extract high-content
information from
populations of cells using existin:whale microtiter plate readers. No existing
high-
content screening platform ha.; been designed for multicolor, fluorescence-
based
screens using cells that ,:rre ana:yrcci individually. Sirnilarly, no method
is currently
available that combines automatec.l fluid delivery t~:> arrays of cells for
the purpose of
15 systematically screening compot~ncls for the ability to induce a cellular
response that is
identified by HCS analysis, ,a~aE.Liallv fi~orn rolls ~;ro~n in microtiter
plates.
Furthermore, no method exists iru the art combining, high throughput well-by-
well
measurements to identify "hits" m one aissay tolowed by a socond high content
cell-by-
cell measurement on the same platc° of only those wells identified as
hits.
2D The instant invention prow: ides systems, methods, and screens that combine
high
throughput screening (>-TTSI arid high content screening (HC'S1 that
significantly
improve target validation and candidate optimization by combining many cell
screening
formats with fluorescence-basa:d molecular reagents and computer-based feature
extraction, data analysis, and automation, resulting in increased quantity and
speed of
c)
CA 02410688 2002-12-12
data collection, shortened cycle tithes, and, ultimately, faster evaluation of
promising
drug candidates. 'the instant inv~~rttion also provides for miniaturizing the
methods,
thereby allowing increased throi.~.g.l7put, while decreasing the volumes of
reagents and
test compounds required in each assay.
S
SUMMARY OF THE INVEN'h'ION
In one aspect, the present invention relates to a method for analyzing cells
comprising
~ providing cells containing fluorescent reporter molecules in an array of
locations,
~ treating the cells in thc: array of locations with one or more reagents,
~ imaging. numerous cells io each location with fluorescence optics,
~ converting the optical irofi~rnoation into digital data,
is ~ utilizing the digital data to determine thE: distribution, environment or
activity of the tluore~,cc:r~t?y labeled rcp~irter molecules in the cells and
the
distribution of the cel';s, and
~ interpreting that inforrn~:~ticm in ternis of at positive, negative or null
effect of
the compound being tested on the biological function
In this embodiment, the method rapidly determines the distribution,
environment, or activity of Iluarescently labeled reporter molecules in cells
for the
purpose of screening large numbers of compounds for those that specifically
affect
particular biological functions. fhe array of locations may be a microtiter
plate or a
2~ microchip which is a microplate leaving cells in art an;tv of locations. In
a preferred
embodiment, the method inclulc:., computcri.eed means for acquiring,
processing,
displaying and storing the data rc°ceived. In a preferred embodiment,
the method
further comprises automated fluid delivery to the array; of cells. In another
preferred
embodiment, the information oht;:~ined from high throughput measurements on
the
CA 02410688 2002-12-12
same plate are used to selectively perform high content screening on only a
subset of
the cell locations on the plate.
In another aspect of the present invention, a cell screening system is
provided
that comprises:
~ a high magnification fluorescence optical system having a microscope
objective,
~ an XY' stage adapted for holding a plate containing an array of cells and
having a means for proving the plate for proper alignment and focusing on
the cell arrays;
~ a digital camera;
~ a light source having optical means liar directing excitation light to cell
arrays and a means for directing flrtores;;ent light emitted from the cells to
the digital camera; and
~ a computer means for receiving and processing digital data from the digital
camera whercrn the c:c~rnputer mca,ns Includes a digital frame grabber for
receiving the irnage~ fr(rnr the camera, a ilisplay for user interaction and
display of assay resulta, digital storage media for data storage and
archiving,
and a means f~.~r control., :acduisition, pre>cessin::rnd display of results.
2o In a preferred embodiment, the cell screening system further comprises a
computer screen operatively associated with the computer for displaying data.
In
another preferred embodiment, f.te cc7mputer means for receiving and
processing digital
data from the digital camera store's tloe data in a bo>infonnatics data base.
In a further
preferred embodiment, the cell. ;screening systen; further comprises a reader
that
2.. measures a signal from many or all the walls in parallel. In another
preferred
embodiment, the cell screening system further comprises :i mechanical-optical
means
for changing the magnification ~:~f the system, to allow changing modes
between high
throughput and high content sr-reening. In ainottter preferred embodiment, the
cell
screening system further comprises a chamber and control system to maintain
the
3n temperature, CO~ concentration ,.rnd humidity surrounding the plate at
levels required to
CA 02410688 2002-12-12
keep cells alive. In a further prefe°rred embodiment, the cell
screening system utilizes a
confocal scanning illumination arid detection system,
In another aspect of the present invention, a machine readable storage
medium comprising a program ci>ntaining a set of instnrctions for causing a
cell
screening system to execute procedures for defining the distribution and
activity of
specific cellular constituents and processes is provided. In a preferred
embodiment, the
cell screening system comprises ~ high magnificatiot7 fluorescence optical
system with
a stage adapted for holding cells and a means far moving the stage, a digital
camera, a
light source for receiving and pro:=cessin~~ the digital Data from the digital
camera, an<i a
to computer means for receiving and processinri the di;~ital data from the
digital camera.
Preferred embodiments of the nnachine readable storage medium comprise
programs
consisting of a set of instnretiors fir causing a cell screening system to
execute the
procedures set forth in Figures 9, I 1, 12, 13, 14 or 1 ~. Another preferred
embodiment
compr7ses a program consisting o1' a set of in;atrur~tions for causing a cell
screening
is system to execute procedures fir clc:tecting the distribution and activity
of specific
cellular constituents and processes In mast preferred embodiments, the
cellular
processes include, but are not limited to. nuclear translocation i~f a
protein, cellular
hypertrophy, apoptosis, and protcasa...induced translocation of a protein.
?o BRIEF DESCRI1'TION,UF TIH_E t)RAWINGS
Figure 1 shows a diagram of the ct~nlponents of the cell-based scanning
system.
Figure 2 shows a schematic of the microscope subassembly.
Figure 3 shows the camera subasses~nbly.
?, Figure 4 illustrates cell scanning :~} stem process.
CA 02410688 2002-12-12
Figure 5 illustrates a user interfac:~ showing major functions to guide the
user.
Figure 6 is a block diagram of the two platforn~ architecture of the Dual Mode
System
for Cell Based Screening in which one platform uses a telescope lens to read
all wells
of a microtiter plate and a second platform that uses a higher magnification
lens to read
individual cells in a well.
Figure 7 is a detail of an optical system for a single platform architecture
of the Dual
Mode System for Cell Based Scre°.~er~ing that uses a moveable
'telescope' lens to read all
wells of a microtiter plate and a r~mveahle higher magnification lens to read
individual
cells in a well.
to Figure 8 is an illustration of the ~fluica delivery syster7o f«r acquiring
kinetic data on the
Cell Based Screening System.
Figure 9 is a flow chart of processinf:~, step for the cell-based scanning
system.
Figure 10 A-J illustrates the strategy of the Nuclear 'Cransloc:ation Assay.
Figure 11 is a flow chart detinirrg the processing steps in the Dual Mode
System for
Cell Based Screening combining high throughput and high content screening of
microtiter plates.
Figure 12 is a flow chart defining; tlac processing steps in the High
Throughput mode of
the System for Cell Based Screenin~~.
Figure 13 is a flow chart ~iefinin~~; tlr<v processing, steps in tire High
C"ontent mode of the
2o System for Cell Based Screening.
Figure 14 is a flow chart. defining the processing steps required for
acquiring kinetic
data in the High Content mode of the System for t'eil BasE;d Screening.
Figure 15 is a flow chart definin,; the processinstops performed within a well
during
the acquisition of kinetic data.
l
CA 02410688 2002-12-12
Figure 16 is an example of data from a known inhibitor oftranslocation.
Figure 17 is an example of data from a known stimulator oftrauslocation.
Figure 18 illustrates data presentation on a graphical display.
Figure 19 is an illustration of the data from the High Throughput mode of the
System
for Cell Based Screening. A) is an example of the data passed to the High
Content mode,
B) of the data acquired in the higl;~ content mode., and C.'} of the results
of the analysis of
the data.
Figure 20 shows the measuremerut <nf a drug-induced cytoplasm to nuclear
translocation.
The localization of GFP--hGR within the cell before and after stimulation with
dexamethasone is represented by (.~~.) and (B) respectivel~~.'1~he
translocation of GFP-hGR
from the cytoplasm to the nucleus of a cell is depicted in a cell nc>t treated
(C) and treated
(D) with dexamethasone.
Figure 21 illustrates a graphical user interface of the measurement shown in
Figure 20.
Figure 22 illustrates a graphical user interface, wil:h data presentation, of
the
measurement shown in Fig. 20.
Figure 23 is a graph representing; the kinetic data obtained from the
measurements
depicted in Fig. 20.
Figure 24 details a high-content screen of drug-induced apoptosrs.
DETAILED DESCRIPTION C>!F' ~l'HE INVENTION:
As used herein, the follcnwing terms have the specii'ic meaning:
Markers of cellrxlar dorr~a~~~ns. Luminescent probes that have high affinity
for
specific cellular constituents including specific organelles or molecules.
These probes
can either be small luminescent moln~<:ul~s or fluorescent 1y tagged
rnacromolecules used
as "labeling reagents", "environnnental indicators", or "Iiosensors".
14
CA 02410688 2002-12-12
Labeling r~ugen~.s. L.,;ribtvling reagents include, but are not limited to,
lumineseently labeled n~acrorrnvlccules incluifin~; fluoresec:nt protein
analogs and
biosensors, luminescent macromolecular chimera: including those formed with
'the
green fluorescent protein and mutants thereof, luntinescently labeled primary
or
secondary antibodies that react v~uth cellular antigens involved in a
physiological
response, luminescent stains, dyes, and other small molecules.
Markers of cellular tran:~loeatic>ns. Lumiruescently tagged macromolecules or
organelles that move from one ccrli oaomain to another during some cellular
process or
physiological response. Transl.oc:ation marke°rs can either simply
report location
to relative to the marl'ers of' eel lular olomains or they can also be
"biosensors" that report
some biochemical or molecular a~;~ti a ity as well.
Biosensors. Macromolecules consisting of a biological functional domain and a
luminescent probe or probes that report the environmental changes that occur
either
internally or on their surface. ~'~ class of luminescently labeled
macromolecules
designed to sense and report tl~eae changes lrav~: been termed "'fluorescent-
protein
biosensors". The protein comt7onent of the biosensor provides a highly evolved
molecular recognition moiety. .~~. fluorescent molecule attached to the
protein
component in the proximity of ,an active site transciuces environmental
changes into
fluorescence signals that are detcv~tcd using a system s~ ith an appropriate
temporal and
spatial resolution such as the coal scanning system of the present invention.
Because
the modulation of native protein ;~ctiviy within the living cell is
reversible, and because
fluorescent-protein biosensors can he designed to sense reversible changes in
protein
activity, these biosensors are esscwnt~ally reusable.
1
CA 02410688 2002-12-12
Disease associute~ seyuernce.s f"l~.~l~"j- This tf;rtr~ refers to nucleic acid
sequences
identified by standard techniqu~a, :,uch as primary UN.A sequence data,
genomic
methods such as subtracaion hvb~ndization and I~:ACtE, and proteomic methods
in
combination with reverse geneti~rs, as being of drug candidate compounds. The
term
does not mean that the sequence is only associated with a disease state.
High content screening (Iwi(_'S1 can be used to measure the effects of drugs
on
complex molecular events such as signal transduction pathways, as well as cell
functions including, but not limned to, apoptosis, cell division, cell
adhesion,
locomotion, exocytosis, and cell-~ccll communication. h9ulticolor fluorescence
permits
t0 multiple targets aril cell procc,sst:~s to be assayed in a single screen.
Cross-correlation
of cellular responses will yield 4~ mcalth of information required for target
validation
and lead optimization.
In one aspect of the present invention. a cell screening system is provided
comprising a high magnification fluorescence optical s;vstem having a
microscope
to objective, an XY stage adapted fc~r holding a plate with an array of
locations for
holding cells and having a mean's~ for moving the plKrte to align the
locations with the
microscope objective and a mean=; for movin' the plate in the direction to
effect
focusing; a digital camera; a light source having opmal means for directing
excitation
light to cells in the array of~loc:aticm~~ and a means far directing
i~luoreseent light emitted
'0 from the cells to the digital catnertt; and a computer means for receiving
and processing
digital data from tle digital canu~~ra wherein the c<>mputer means includes: a
digital
frame grabber for receiving the inrrges from the camera, a display for user
interaction
and display of assay results, cligitai storage media t:ur data storage and
archiving, and
means for control, acquisition, prixcmsing and display of results.
lb
CA 02410688 2002-12-12
Figure 1 is a schematic dia~~,ram of a preferred embodiment of the cell
scanning
system. An inverted fluorescence microscope is used l, such as a Zeiss
Axiovert
inverted fluorescence microscope w-hich uses standard objectives with
magnificatier, of
1-100x to the camera, and a white light source (e.g. 100W' mercury-arc lamp or
75W
xenon lamp) with power supply 2. 'lf'here is an XY stage ~ to move the plate 4
in the
XY direction over the microscolne objective. A ;Z-axis focus drive 5 moves the
objective in the Z direction for focusing. A joystick 6 provides for manual
movement
of the stage in the XYZ direction. A high resolution digital camera 7 acquires
images
from each well or location on the plate. There is a camera power supply 8 an
automation controller 9 and a central processing unit 10. The PC 11 provides a
display
12 and has associated software. 1i he printer 13 provides for printing of a
hard copy
record.
Figure 2 is a schematic of ~~nn embodiment ofathe microscope assembly 1_ of
the
invention, showing in more detail the XY stage 3, Z-axis focus drive 5,
joystick 6, light
source 2, and automation controller 9. Cables to the computer 1 S and
microscope 16,
respectively, are provided. In addition, Figure 2 shows a 9G well microtiter
plate 1 T
which is moved on the XY stage :3 in the XY direction. Light fiom the light
source
passes through the PC controlled shutter 18 to a. motorized filter wheel 19
with
excitation filters 20. The light pa;scs into filter cube 25 which has a
dichroic mirror 2fi
and an emission fitter 27. Excitation light reflects off the diehroic mirror
to the wells in
the microtiter plate 17 and fluorescent light 28 passes through the dichroic
mirror 26
and the emission filter 27 and to the digital camera 7.
Figure 3 shows a schematic drawing of a prefetTed camera assembly. The
digital camera 7, which contains ;gin automatic shuttc;r for Exposure control
and a power
17
CA 02410688 2002-12-12
supply 31, receives fluorescent light ?8 from the microscope assembly. A
digital cable
30 transports digital signals to the cup?~puter.
The standard optical configurations described above use microscope optics to
directly produce an enlarged image of the specimen on the camera sensor in
order to
capture a high resolution image of the specimen. 'this optical system is
commonly
referred to as 'wide field' rr~icr<nscopy. Those skilled in the art of
microscopy will
recognize that a high resolution image of the specimen can be created by a
variety of
other optical systems, includitzg, but not limited to, standard scanning
confocal
detection of a focused point or lure of illumination scanned over the specimen
(Go et al.
1U 1997, supra), and multi-photork scannin g confocal microscopy (Denk et al.,
1990,
supra), both of which can forn-~ images on a C'CIJ detector or by synchronous
digitization of the analog output o1 a photomultiplier tube.
In screening application's,, it is af~en necessary tea use a particular cell
tine:, or
primary cell culture, to take a~:lvantage of particular features of those
cells. Those
to skilled in the art of cell culture ~'vill recognize that sonic call lines
are contact inhibited,
meaning that they will stop ~ro~.vinY7 when they bei~ome surrounded by other
cells,
while other cell lines will continue: to grow under those conditions and the
cells will
literally pile up, forming many layers. An example c3f su;.h a cell fine is
the HEK. 293
(ATCC CRL.-15'?3) line. .An optical system that c:an ac~luire images of single
cell
2O layers in multilayer preparations is required for nse~ a itft cell lines
that tend to forni
layers. The large depth of field or wide field microscopes produces an image
that is a
projection through the many layers of cells, making analysis of subcellular
spatial
distributions extremely difficr:Lt in layer-fomriry, cells :alternatively, the
very shallow
depth of field that can be achieved on a confbca! rr~icroscope. (about one
micron),
1 ~;
CA 02410688 2002-12-12
allows discrimination of a single cell layer at high resolution, simplifying
the
determination of the subcellular spatial distribution. Similarly, confocal
imaging is
preferable when detection modes such as fluorescence lifetime imaging are
required.
The output of a standard eonfocal imaging attachment for a microscope is a
digital image that can be converted to the same format as the images produced
by the
other cell screening system err~bodiments described above, and can therefore
be
processed in exactly the same way as those images. The overall control,
acquisition
and analysis in this embodiment is essentially the same;. 'fhe optical roof
guration of
the confocal microscope system, is essentially the same as that described
above, except
for the illuminator and detectors. Illumination and detection systems required
far
confocal microscopy have been clesigned as accessories to be attached to
standard
microscope optical systems such as that of the present invention {Zeiss,
Germany).
These alternative optical systen-~s therefore can be easily integrated into
the system as
described above.
Figure 4 illustrates an alternative embodiment of the invention in which cell
arrays are in microwells 40 on a ~nicroplate 41. Typically the microplate is
20mm by
30mm as compared to a standard 9t~ well microtiter Plato which is $6mrn by
129mm. The
higher density array of cells on ,~ microplate allows the. mic:roplate to be
imaged at a low
resolution of few microns per laixel for high throlrghput and particuhu
locations on the
microplate to be imaged at a higher resolution of less than 0.5 microns per
pixel. These
two resolution modes help to improve the overall throughput of the systt'm.
lc
CA 02410688 2002-12-12
The microplate chamber 4::.' sewes as a microfluidic delivery system for the
addition of compounds to cells. fI"ltc microplate 4~1 in the microplate
chamber 42 is
placed in an XY microplate reaclea 43. Digital data is pracessed as described
above.
The small size of this micropl;ate system increases throughput, minimizes
reagent
volume and allows control of the distribution and placement of cells for fast
and precise
cell-based analysis. Processed data can be displayed c>n a I'C screen 1 l and
made part
of a bioinformatics data base 44. 'Plus data base not only permits storage and
retrieval
of data obtained through the methods of this in wention, but also permits
acquisition and
storage of external data relating: to cells. Figure ~ is a P(_' display which
illustrates the
to operation of the software.
In an alternative ernbr~diment, a high throughput system (HTS) is directly
coupled with the HGS either on the same platform or on tcvo separate platforms
connected electronically (e.g. via a Local area nerivark). This embodiment of
the
invention, referred to as a dual mole optical system, has the advantage of
increasing the
throughput of a HCS by coul7linft it with a HT'S and thereby requiring slower
high
resolution data acquisition and analysis only an the small subset of wells
that show a
response in the coupled HTS.
High throughput 'who'ie plate° reader systems are well known in the art
and are
commonly used as a <,ampor:ent <>f an H I~S system used to screen large
numbers of
Zo compounds (Bef;gs (l~)97),.1. cal~Biomolcc~. Sc~rer~nin,~ ~:?1-78;
_vlacaffrey et al., (1996)
J Btomolec. Scroerzing 1:187 -1 ~~()).
In one embodiment a9~ dual mode cell based screening, a two platform
architecture in which high throughput acquisition occurs on one platform and
high
content acquisition occurs on a second platform a prirsided (Figure b).
Processing
~cv
CA 02410688 2002-12-12
occurs on each platform independently, with results passed over a network
interface, or
a single controller is used to process the data from both platforms.
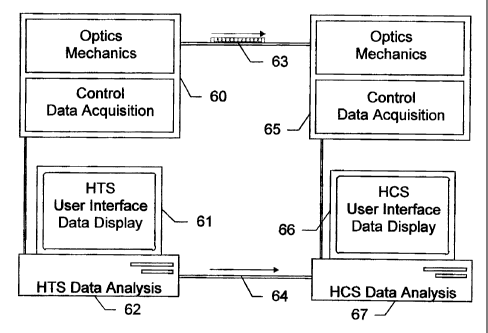
As illustrated in Figure 6, an exemplified two hlatforn~ dual mode optical
system consists of two light optical. instruments, a high throughput platform
60 and a
high content platform 6S which read fluorescent signals emitted from cells
cultwed in
microtiter plates or microwell arrays on a microplate, and communicate with
each other
via an electronic connection 64. The high throughput platform 60 analyzes all
the wells
in the whole plate either in parallel or rapid serial fashion. Those skilled
in the art of
screening will recognize that there are a many such commercially available
high
throughput reader systems that could be integrated into a dual mode cell based
TM
screening system (Topcount (hackard Instruments, Meriden, C'T); Spectramax,TM
Lumiskan (Molecular Devices, "~~unnyvale, CA); hluoroscan (Labsystems,
Beverly,
MA)). The high content platfornn fiS, as described above, scans from well to
well and
acquires and analyzes high resoluticm image data collected from individual
cells within
a well.
The HTS software, residing on the system's c:ornputer 62, controls the high
throughput instrument, and results ,are displayed on the monitor 61. The HCS
sofWane,
residing on it's computer system G7, controls the high content instrument
hardware G5,
optional devices (e.g. plate loader, environmental chamber, fluid dispenser),
analyzes
digital image data from the plan., displays results un the monitor 66 and
manages data
measured in an integrated database. The tvo systems can also share a single
computer,
in which case all data would be collected, processed and displayed on that
computer,
without the need for a local area network to transfer the. data. Microtiter
plates are
transferred from the high thr~:>ughput system tc~ the high content system 63
either
'? 1
CA 02410688 2002-12-12
manually or by a robotic plate ~:ransfer~ device, as is well known in the art
(Beggs
(1997), supra; Mcaffrey ( 1996), ,~~~:Prcr).
In a preferred embodirn~nt, the dual mode optical system utilizes a single
platform system (Figure 7). It consists of tw To separate optical modules, an
HCS
module 203 and an HTS module 209 that can be independently or collectively
moved
so that only one at a time is used to collect data tiom the microtiter plate
201. The
microtiter plate 201 is mounted irr a motorized X,Y stage so it can be
positioned for
imaging in either HTS or HC.'S mode. After collecting and analyzing the HTS
image
data as described below, the HTS optical module ?09 is moved out of the
optical path
to and the HCS optical module 20~f i<; moved into place.
The optical module for 1-l I S 209 consists of a projection lens 214,
excitation
wavelength fitter 213 and dichroic mirror 210 which are used to illuminate the
whole
bottom of the plate with a specific wavelength band from a conventional
microscope
lamp system (not illustrated). The fluorescence emission is collected through
the
dichroic mirror 210 and emissior~~ wavelength filter 21 1 by a lens 212 which
forms an
image on the camera 21 (~ with =~ertsor ,' 15.
The optical module far 1 1(.'S 203 consists oa' a projection lens 208,
excitation
wavelength filter 207 and dicnrcoic mirror 2t14 which are used to illuminate
the back
aperture of the microscope: objective 202, and thereby the. field of that
objective, from a
z0 standard microscope illuminatic>n system (not shown), he fluorescence
emission is
collected by the microscope of~j~:ctivc 202, pass;~s through the dichroic
mirror 204 and
emission wavelength filter 20°: ;rnd is focused by a tube lens 20G
which forms an image
on the same camera 21 (~ with :,~er~~or ? 1 S.
'1 7
CA 02410688 2002-12-12
In an alternative ernbodirnent of the present invention, the cell screening
system
further comprises a fluid delivery device for usi° with the live cell
embodiment of the
method of cell screening (see below). Figure ~ exemptities a fluid delivery
device for
use with the system of the invention. It consists of a bank of 12 syringe
pumps :701
.> driven by a single motor drive. fach syringe 702 is sized according to the
volume to be
delivered to each well, typically bi;tween 1 and IOf) 11L. Each syringe is
attached via
flexible tubing 703 to a similar bank of~ connectors which accept standard
pipette tips
705. The bank of pipette tips are ,rttached to a drive system so they can be
lowered and
raised relative to the microtiter Ivlate 706 to deliver fluid to each well.
The plate is
mounted on an X,Y stage, allc~wi~~r~~ nnovement relative to the optical system
707 for
data collection purposes. This set-up allows one set of pipette tips, or even
a single
pipette tip, to deliver reagent to all the wells on the plate. the bank of
syringe pumps
can be used to deliver fluid to I ~ wells simultaneously, or to fewer wells by
removing
some of the tips.
In another aspect, the prf~sc:nt invention tarovides a method for analyzing
cells
comprising providing an arra,,a ~~i locations which contain multiple cells
wherein the
cells contain one or more fluorescent reporter molecules; scanning multiple
cells in
each of the locations containi ng c<~lls to obtain ~luor~acent signals from
the fluorescent
reporter molecule in the cells; reconverting the fluorescent signals into
digital data; and
utilizing the digital data to c:~~twrZroine the distribution, environment or
activity of the
fluorescent reporter molecule v ithin the cells.
CA 02410688 2002-12-12
Cell Arrays
Screening large numbers of c:ompaunds far activity with respect to a
particular
biological function requires preparing arrays of cells for parallel handling
of cells and
reagents. Standard 96 well microtiter plates which arc 86 nun by 129 mm, with
6mm
diameter wells on a 9mm pitch, are used for compatibility with current
automated
loading and robotic handling systems. The microplate is typically 20 mm by 30
mm"
with cell locations that are 100-:.'00 microns in dimension on a pitch of
about 500
microns. Methods for making microplates are well known in the art. Microplates
may
consist of coplanar layers of matcri<;ls to which cells adhere, patterned with
materials to
which cells will not adhere, or tac~lued 3-dimensional surfaces of similarly
patterned
materials. For the purpose of the following discussion, the terms 'well" and
'microwell'
refer to a location in an array of any construction to which cells adhere and
within which
the cells are imaged. Microplate:; may also include fluid delivery channels in
the spaces
between the wells. The smaller for~~aat of a microplate increases the overall
efficiency of
the system by minimizing the quantities of the reagents, storage and handling
during
preparation and the overall movi:n~c:nt required for the scanning operation.
In addition,
the whole area of the microplate c:an be imaged more efficiently, allowing a
second mode
of operation for the microplate reader as described later in this document.
Fluorescence Reporter Molecarles
A major connponent of the new drug discovery paradigm is a continually
growing family of fluorescent and luminescent r~:agents that are used to
measure the
z~
CA 02410688 2002-12-12
temporal and spatia'I distribution, uc:~ntent, and activity of intracellular
ions, metabolites,
macromolecules, and organelles k.'lasses of these real;ents include labeling
reagents
that measure the distribution and amount of molecules in living and fi;;,:d
cells,
environmental indicators to repnrt srgrlal transduction events in time and
space, and
_°~ fluorescent protein biosensors to measure target, moiecular~
activities within living cells.
A multiparameter approach that corrtbines several reagents in a single cell is
a powerful
new tool for drug discovery.
The method of the present invention is baseii on the high affinity of
fluorescent
or luminescent molecules fe:~r specific cellular components. The affinity for
specific
to components is governed by phy:~iival forces such as ionic interactions,
covalent bonding
(which includes chimeric fusie;o v~ith protein-based chromophores,
fluorophores, and
lumiphores), as well as hydropluabic interactions, c~le~trical potential, and,
in some
cases, simple entrapment within a cellular component. The luminescent probes
ca.n be
small molecules, labeled nnacromolecules, or genetically engineered proteins,
1 ~ including, but not limited to gri:ern fluorescent protein chimeras.
Those skilled in this an mill recognize a wide variety of fluorescent reporter
molecules that can be used in the present lrlventlon, includir»:. but not
limited to,
fluorescently labeled biomcrlei:rzles such as proteins, phosphoiipids and DNA
hybridizing probes. Similarl~~, fluorescent reagents specifically synthesized
with
?o particular chemical properiie~ ~~t binding or ass~»iation have been used as
fluorescent
reporter molecules (Barak et u1.. ( I9971, J. Baol. ( 'hear. 272:27497-~7~00;
Southwick et
al., ( 1990), Cyto»rety 11:41 ~-43t); 'Tsien ( 1980) ire :Merhoais~ in Cell
Biology, Vol. 29
Taylor and Wang (eds.), pp. 12 .'-1507). Fluoresc-entlv labeled antibodies are
particularly
2>
CA 02410688 2002-12-12
useful reporter molecules due to their high degree of'specificity for
attaching to a single
molecular target in a mixture of rnol~c.cules as complex as a cell or tissue.
The lumirpsc~:r~t probes catu be synthesized within the living cell or can be
transported into the cell via several non-mechanical modes including
diffusion,
facilitated or active transport, signal-sequence-mediated transport, and
endocytotic or
pinocytotic uptake. Mechanical bulk loading methods, which are well known in
the art,
can also be used to load luminescent probes into living cells (Barber et al.
(1996),
Neuroscience Letters 207:17-20; lfiright et al. ( 1996), Cytometry 24:226-233;
McI~'eil
(1989) in Methods in Cell Biolo~v, Vol. 29, Taylor' and Wang (eds.), pp. 153-
173).
These methods include electroporation and other mechanical methods such as
scrape-
loading, bead-loading, irr~pact-loading, syringe-loading, hypersonic and
hypotonic
loading. Additionally, cells can be genetically engineered to express reporter
molecules, such as GFP, coupled to a protein of interest as previously
described
(Chalfie and Prasher U.S. Patent No. 5,491,084 issued February 13, 1996;
Cubitt et al.
(1995.), Trends in Biochemical Science 20:448-455.
Once in the cell, the lumine~>cent probes accumulate at their target domain as
a
result of specific and high affinity interactions with the target domain or
other modes of
molecular targeting such as signal-~~equence-mediated transport. Fluorescently
labeled
reporter molecules are useful for determining the: location, amount and
chemical
environment of the reporter. For example, whether the reporter is in a
lipophilic
membrane environment or in a more aqueous environment can be determined
(Giuliano
et al. (1995), Ann. Rev. of.Biophysi~~s and Biomolecular Structure 24:405-434;
Giuliano
and Taylor (1995), Methods in Neurorcience 27: ! -16). 'fhe pH environment of
the
reporter can be determined (BOght et al. (1989), J: (:'ell Biology 104:1019-
10:33;
2G
CA 02410688 2002-12-12
Giuliano et al. (1987), ,~nczl. NracIrE~m. 167:3b2-371; Thomas et al. (1979),
Biochernistrl' 18:2210-2218). It ~.ao be detennit+ed whcnher a reporter having
a
chelating group is bound to an ion, st.tch as (.'a-+-+, or not (Bright et al.
(1989), In
Methods in Cell Biology, Vol. 30, Taylor and Wang (eds.}, pp. 157-192;
Shimoura et al.
(i988), J. of Biochemistry ('Toky«) 251:405-410; Tsien (1989) In Methods in
Cell
Biology, Wol. 30, Taylor and War-tg (eds.), pp. 12'7-156).
Furthermore, certain cell types within an organism may contain components
that can be specifically labeled that may not occur in other cell types. For
example,
epithelial cells often contain p~.~larized membrane components. 'That is,
these cells
lei asymmetrically distribute macrrvnoolecules along their plasma membrane.
Connective
or supporting tissue cells often contain granules in ~y~hi~.h are trapped
molecules specific
to that cell type (e.g., heparin, histamine, serotonirt, etc.). V4ost muscular
tissue cells
contain a sarcoplasmic reticulurv, a specialized organelle whose function is
to regulate
the concentration of calcium forts ~~,~ithin the cell cytoplasm. Many nervous
tissue cells
contain secretocy granules arid vesicles in which are trapped neurohotmones or
neurotransmitters. Therefore, fluorescent molecules can be designed to label
not only
specific components within spG:~cific cells, but also specific cells w ithin a
population of
mixed cell types.
Those skilled in the ;art will recognize ;~ reide variety of ways to measure
?o fluorescence. For example, s~,~nnc: tluorescent rlorter molecules exhibit a
change in
excitation or emissiotl spectra. some exhibit resonance energy transfer where
one
fluorescent reporter loses fluorescence, while a second gains in fluorescence,
some
exhibit a loss (quenching) or al:3pearance of fluorescence, while some report
rotational
~ -.
CA 02410688 2002-12-12
movements (Giuliano et al. (1955), ,~lnn. Rev, o/~ Bio~ht:srcw and Biomol.
Structure
24:405-434; Giuliano et al. (19951, ~~lethods irr ~'~'ortru.scic:vc-a 2?:l-
16).
Scanning cell arrays
Referring to Figure 9, a preferred embodiment is provided to analyze cells
that
comprises operator-directed parameters being selected based on the assay being
conducted, data acquisition by tile cell screening system on the distribution
of
fluorescent signals within a sample. and interactive data review and analysis.
At the
start of an automated scan the operator enters information 100 that describes
the
tc) sample, specifies the filter settings and fluorescent channels to match
the biological
labels being used and the inforrllation sought, and then adjusts the camera
settings to
match the sample brightness. For flexibility to handle a range of samples, the
software
allows selection of various par~n.rrreter settings used to identify nuclei and
cytoplasm,
and selection of different tluorc~sc;erlt reagents, identification of cells of
interest based
is on morphology or brightness, anti cell numbers to be analyzed per well.
These
parameters are stored irr the systc:rn's database fir easy retrieval f'or each
automated
run. The system's int'ractive cell identification mode simplifies the
selection of
morphological parameter limits such as the range of size, shape. and intensity
of cells to
be analyzed. The user specitie,~~ which wells of the pl<rte the system will
scan and how
o many fields or how many cells to analyze in each well. Depending on the
setup mode
selected by the user at step 1 U I,_, the system either automatically pre-
focuses the region
of the plate to be scanned using an autofocus procedure to "find focus" of the
plate 102
or the user interactively pre-fiscuses 1U3 the scaruling region by selecting
three "tag"
points which define the rectan,~;ular area to beg sca~lncd. A least-squares
fit "focal plane
?8
CA 02410688 2002-12-12
model'" is then calculated from these tag points to estimate the focus of each
well
during an automated scat. The focus of each well is estimated by interpolating
from
the focal plane model during a sc:arr.
During an automated sc::ur, the software dynamically displays the scan status,
including the number of cells anahrzed, the current well being analyzed,
images of each
independent wavelength as they are acquired, and the result of the screen for
each well
as it is determined. The plate 4 (Figure 1 ) is scanned in a serpentine style
as the
software automatically moves tire motorized microscope X~r' stage 3 tTOm well
to well
and field to field within each will of a 96-well plate. (hose skilled in the
programming
to art will recognize how to adapt sc~flware for scanning of other microplate
formats such
as 24, 48, and 384 well plates. '~l he scan pattern of the entire plate as
well as the scan
pattern of fields within each v~~ell are programmed. The system adjusts sample
focus
with an autofocus procedure l>4 lFigure 9) through the Z axis focus drive S,
controls
filter selection via a motorized fl Iter w heel 19, and acquires and analyzes
images of up
l5 to four different colors ("channels"' or "wavelengths'")
The autofocus procedure i5 called at a user selected trequency, typically for
the
first field in each well and there c7nce every ~ =,o ~ tields within each
well. The autofocus
procedure calculates the starting. l-axis point b,~,~ interpolating from the
pre-calculated
plane focal model. Starting a t,r~»rar~lmable distance above or helwv this set
point, the
2o procedure moves the mechanical l,-axis through a number of different
positions,
acquires an image at each position, and finds the maximum of a calculated
focus score
that estimates the contrast ~:~f each image. Tho t.. position of the image
with the
maximum focus score deterntines the best focus for a particular field. Those
skilled in
the art will recognize this as ~:r variant of automatic focusing algorithms as
described in
CA 02410688 2002-12-12
Harms et al. in C~~torrzetn.~ S (198=1), 2 36-243, Groen et al. in C'vtometrv
6 (1985), 81-91,
and Firestone et al. in Cjuometn l a" 1.1991 ), 195-206
For image acquisition, tine camera's e~:posure time is separately adjusted for
each dye to ensure a high-quality image from each channel. Software procedures
can be
.. called, at the user's option, to correct for registration shifts between
wavelengths by
accounting for linear (X and Y) shifts between wavelengths before making any
further
measurements. The electronic shutter 18 is controlled so that sample photo-
bleaching is
kept to a minimum. Backgroun~:l shading and uneven hlumination can also be
corrected
by the software using methods; known in the art (Bright et al. ( 1987), J.
Cell i'3iol.
to 104:1019-1033).
In one channel, imaga~ are acquired of a primary marker 105 (Figure 9)
(typically cell nuclei counterstained with U.A.PI or PI fluorescent dyes)
which are
segmented ("identified") using an adaptive thresholding procedure. The
adaptive
thresholding procedure 106 is ~.ised to dynamically select the threshold of an
image for
t 5 separating cells from the background. The staining of cells with
fluorescent dyes can
vary to an unknown degree across calls in a microtiter plate sample as well as
within
images of a field of cells within each well of a microtiter plate. This
variation can occur
as a result of sample preparati~:in and-'or the dynamic nature of cells. A
global threshold
is calculated for the complete image to separate tire cells from background
and account
?.o for field to field variation. Clvese global ,rdaptive techniques are
variants of those
described in the art. (Kittler et al. in ( 'ornputer 1 'i.rion, Oruphics, and
image
Processing 30 ( 198 ), 125-1.47. Ridler et al in IF'E~' l fans ~Sp:stems, Man,
and
Cybernetics (1978), 630-632.1
3O
CA 02410688 2002-12-12
An alternative adaptive tlurcsholding method utilizes local region
thresholding
in contrast to global image threshol~ding. Image an alysi:~ of local regions
leads to better
overall segmentation since staining oaf cell nuclei (as well as other labeled
components)
can vary across an image. Using; this globalilocal procedure, a reduced
resolution
v> image (reduced in size by a factor of 2 to 4) is first globally segmented
(using adaptive
thresholding) to find regions of~ interest in the image. These regions then
serve as
guides to more fully analyze the same regions at full resolution. A more
localized
threshold is then calculated (oath using adaptive threshalding) for each
region of
interest.
I"he output of the segmentation proccdl.lre is a binary image wherein the
objects
are white and the background i~; black. 'This binary image, also called a mask
in the art,
is used to determine if the field contains objects 1i~7 T'he mask is labeled
with a blob
labeling algorithm whereby ea~:h object (or blab) has a unique number assigned
to it.
Morphological features, such u.s area and shape, c~f the blobs are used to
differentiate
i.s blobs likely to be cells froth tlaost. that are considered ;artifacts. The
user pre-sets the
morphological selection criteria by either typing in known cell morphological
features
or by using the interactive training utility. If objects of rnterest a.re
found in the field,
images are acquired far all ot'ncr active channel:, I(?~i, otherwise the stage
is advanced
to the next field 1()9 111 the current well. Earn oh~ect of interest is
located in the image
for further analysis 11,0. The s~:~ttware determll7es If the object meets the
criteria for a
valid cell nucleus 111 by measuring its morphological features (size and
shape). For
each valid cell, the XYZ staf;e location is recanted, a ;mall image of the
cell is stored,
and features are measured 1 l..?:.
31
CA 02410688 2002-12-12
The cell scanning method of the present invention can be used to perform many
different assays on cellular satnl~Ies 1y applying a n umber of analyrtical
methods
simultaneously to measure features at multiple wavelengths. An example of one
such
assay provides for the following measurements:
,;
1. The total fluorescent vint~.nsity within the cell
nucleus for colors 1-4
2. The area of the cell nuG:l~;us for color 1 (the primary
marker)
3. The shape of the ccIl nu<;leus for color 1 is described
by three shape
features:
1 o a) perimeter sqr:~.ared area
b) box area ration
c) height width r-atr0
4. The average tluoresc:errt intensity within the cell
nucleus for colors 1-4 (i.e.
#1 divided b~,-~ #? j
5. The total fluorescent intensity oi' a rind outside:
the nucleus (see Figure 10)
that represents tluorescc:nce of the cell's cytoplasm
(cytoplasmic mask;) for
colors 2-4
6. The area of the cytor~lasmic mask
7. The average fluoresct:nt intensity of the c:ytoplasmic
mask for colors 2-4
?0 (i.e. #~ divided by #ti)
8. 'The ratio of the average fluorescent iraterusity
of the cytoplasmic mask to
average fluorescent intensity within the: cell nucleus
for colors 2-4 (i.e. #7
divided by #4)
9. The difference of true average fluorescent intensity
of the cytoplasmic mask
2, and the average fluor~~,cent intensity ~~=itlir~
the cell nucleus for colors 2-4
(i.e. #7 mimes #4)
10. The number of fluc~resc~;nt domairrs (also call spots,
dots, or grains) within
the cell nucleus for ~:~c>lc>c~s '-4
;O Features 1 through 4 are general features of the
different cell screening
assays of the invention. Thescv ~t~ps art commonly caeci in a variety of image
analysis
applications and are we 11 knov.-n in art (Ross 1 l~)~i2) Tire lnra,~c~
Processing Handbook,
CRC' Press Inc.; Gonzales et ai ( 1957), Digitu! Imus,<<° f'f-ocessing.
Addison-Vv'esley
Publishing Co. pp. 391-448). i=eatures 5-9 have been developed specifically to
provide
3~ measurements of a cell's i7uorescent molecules within the local
c.y~toplasmic region of
the cell and the transloc:aticm ( i.e. movement) oC tluorcacent molecules from
the
cyrtoplasm to the nucleus. ~l hes~ featurca (step:; s-9) are used for
analyzing cells in
;1
CA 02410688 2002-12-12
microplates for the inhibition ol~ nuclear iranslocation. For example,
inhibition of
nuclear translocation of transcription factors provides a nc>vel approach to
screening
intact cells (detailed examples of other types of screens will be provided
bca>v,). A
specific algorithm measures the ~unount of probe in the nuclear region
(feature 4)
> versus the local cytoplasmic region (feature 'l) of each e;ell.
Quantification of the
difference between these two sub-cellular compartments provides a measure of
cytoplasm-nuclear translocation (fc°,ature 9).
Feature 10 describes a screen used for counting; of DN.A or RNA probes within
the nuclear region in colors 2-~. lvor example, probes ate commercially
available for
ti.i identifying chromosome-specific 9)N.~ sec~uerlce~ (1_ife hechnologies,
Gaithersburg,
MD; Genosys, Woodlands, ~T~; l:3iotechnologies. Inc.. Richmond, ('.4; Bio 101,
lnc.,
Vista, CA) Cells are three-d,mensicmal in nature and when examined at a high
magnification under a microscope one probe may be in-focus while another may
be
completely out-of focus. The deli screening method of the present invention
provides
for detecting three-dimension.il probes in nuclei by acquiring images from
multiple
focal planes. The software nuw~°,'. the Z-axis motor drive 5 (Figure 1
) in small steps
where the step distance is user selected to account for a wide range of
different nuclear
diameters. At each of the foc.~.l steps, an image is ai:quired. The maximum
gray-level
intensity from each pixel in e=ach image is touro.i arrd stored in a resulting
maximum
2o projection image. The maxim~.~.m projection imac;c i<< then used to count
the probes. 'The
above aigorithnn works well :n couratmg probes that: ;are not stacked directly
above or
below another one. 'I o acccurn for probes stacked on top of each other in the
Z-
direction, users can select an option to analyre probes in each of the focal
planes
acquired. In this mode, the ~:,c~~nning system perfor7ns the maximum plane
projection
CA 02410688 2002-12-12
algorithm as discussed above, detects probe regions of interest in this image,
then
further analyzes these regions in all the focal plane images.
After meu,urin~l cell features 112 (Figure ~)i, the, system checks if there
are any
unprocessed objects in the curr.vrG field 113. If there are any unprocessed
objects, it
locates the next object 110 and determines whether it meets the criteria for a
valid cell
nucleus 111, and measures its features. Once all the objects in the current
field are
processed, the system detern~inea whether analysis of the current plate is
complete 114;
if not, it determines the need to find more ce:Ils iii the current well I I5.
If the need
exists, the system advances the X fZ stage to tire next field rwithin the
current well 1U9
or advances the stage to the nest v>~;11 1 lh of~the pl;rte.
,After a plate scan is uornplete, images acrid data can be reviewed with the
system's image review, data r~:vlC'l1-, and summar~° renew facilities,
A11 images, data,
and settings from a scan aue archived in the system's database for later
review or for
interfacing with a network infornvation management system. Data can also be
exported
t:~ to other third-party statistical packages to tabulate results and generate
other reports.
Users can review the images alone of every rc:Il an.rlvred by the system with
an
interactive image review proec.dure 117. The user carr review data on a cell-
by-cell
basis using a combination cnf interactive f;raphs, a data spreadsheet of
measured
features, and images of all ilrt Ilur~rescence chanra els of a cell of
interest with the
interactive cell-by-cell. data r~.woev- procedure 1 I ~. t~.;~-apltical
platting capabilities are
provided in which data can h4 analyzed via interactive graphs such as
histograms and
scatter plots. Users can review ~ummary° data that are accumulated and
summarized for
all cells within each well of a plate with an interactive well-by-well data
review
3~
CA 02410688 2002-12-12
procedure 119. Hard copies of graphs and images can be printed on a wide range
of
standard printers.
As a final pha ;, c,f ~a co'op(ete scan, repon.s can be generated on one or
more
statistics of the measured feat~.irt;s. Users care generate a graphical report
of data
a summarized on a well-by-well basis for the scanned region of the plate using
an
interactive report generation procedure 12(?. This report includes a summary
of the
statistics by well in tabular and graphical format and identification
information on the
sample. The report window allows the operator to enter comments about the scan
for
later retrieval. Multiple reports ca.n be generated on many statistics and be
printed with
to the touch of one button. Reports can be previewed far placement and data
before being
printed.
The above-recited eminodiment of the metlroct operates in a single high
resolution mode referred to as the high content screening (HCS) mode. The HCS
mode
provides sufficient spatial resolution within a welt (on the order of 1 frm)
to define the
Is distribution of nnaterial withir7 thr: well, as well as within individual
cells in the well.
The high degree of information c~~ntent accessible in that made, comes at the
expense
of speed and complexity ofthe rc:qr.tired signal processing.
In an alternative embo<ifnnent, a high throughput system (HTS) is directly
coupled with the HCS either c>r7 the same platforni or on two separate
platforms
connected electronically (e.g. ~~ its a local ~irea net»~ork). This embodiment
of the
invention, referred to as a dual n~c.~cte optical system, has the advantage of
increasing the
throughput of an HCS by co~.r.pling it with an t-ITS .and thereby requiring
slower high
resolution data acquisition and analysis only on the small subset of wells
that show a
response in the coupled H rS.
CA 02410688 2002-12-12
High throughput 'whole l~~latr.' reader systems are well known in the art and
are
commonly used as a componenr of an HTS system used to screen large numbers of
compounds (Beggs et al. (1997), .sr~pra; Mc('affrey et a!. (1996), supra ).
The HTS of
the present invention is carried ol.rt on the microtiter plate or microwell
array by reading
many or all wells in the platen simultaneously with sufficient resolution to
make
determinations on a well-by-well basis. That is, calculations are made by
averaging the
total signal output of many or all the cells or the bulk of the material in
each well.
Wells that exhibit some defined response in the IiTS (the 'hits') are flagged
by the
system. Then on the same micrc~titer plate or rrucrawell array, each well
identified as a
1o hit is measured via HCS as described above.
Thus, the dual mode pro~.~e~ss involves:
1. Rapidly measuring numerous ~,~ ells of a microtirer plate or microwell
array,
2. Interpreting the data to determine the overall activity of fluorescently
labeled
reporter molecules in the calls on a well-byr-will basis to identify ''hits"
(wells that
1 s exhibit a defined response),
3. Imaging numerous cells in ~ arh "hit" well, and
4. Interpreting tire digital im<rgi: data to deterrorinc the distribution,
environment or
activity of the f7uorescentl~.~ labeled reporter n~ol~culcs in the individual
cells (i.e.
intracellular rneasurtments)~ ;rncl the distribution r~l~ the cells to test
for specific
biological functions
In a preferred embodiment of dual mode prowsslng (1~igure 1 I), at the start
of a
run 3U1, the operator enters irnfornnatron 302 drat de~serib~s the plate and
its contents,
specifies the filter settings and tltrorescent channels to match the
biological labels being
25 used, the information sought a:lnd the camera settings to match the sample
brightness.
These parameters are stored in the system's database for easy retrieval for
each
automated run. 'I'he microtiter plate car microwell array is loaded into the
cell screening
system 303 either manually c:~r automatically by controlling a robotic loading
device.
3 C:
CA 02410688 2002-12-12
An optional envircmmental chamber 3t>4 is controlled by the system to maintain
the
temperature, humidity and C(~~ '~ev-~:ls in the air surrounding live cello in
the microt.iter
plate or microwell array. Arn r,>lrtoonal fluid delivery ~~device 305 (see
Figure 8) is
controlled by the system to dispense fluids into the wells during the scan.
High throughput processing 30fi is first perf~nned on the microtiter plate or
mierowell array by acquiring and analyzing the srgnal from each of the wells
in the
plate. The processing performec:l in high throughput mode 30 i is illustrated
in Figure 12
and described below. Wells that exhibit some selected intensity response in
this high
throughput mode ("hits") ar~° identified by the system. The system
performs a
to conditional operation 308 that tests for hits. If hits ;ire found, those
specific hit wells are
further analyzed in high content (micro level ) mode ;409. The processing
performed in
high content mode 314 is iliuwtrated in Figure I ~. 'hhe system then updates
310 the
informatics database 311 witr~ results of the measurements on the plate. If
there are
more plates to he analyzed 3_1 ?~_ the system loads the next plate 303;
otherwise the
la analysis of the plates terminatt:s 314.
The following discus~~~ion describes the hi~~ll throughput mode illustrated in
Figure 12. The preferred emioodiruent of the system, the single platfonm dual
mode
screening system, will be d~~si:ribed. Those skilled in the art will recognize
that
operationally the dual platforrn system simply involves moving the plate
between two
?o optical systems rather than rn~:win~ the optics. C>nce the system has been
set up and the
plate loaded, the system begins tine Id 'CS acquisition and analysis 4U1. The
HTS optical
module is selected by controlling .~ motorized ~>ptical positioning device 402
on the
dual mode system. Irr one fluorescence channel, data from a primary marker on
the
plate is acquired 403 and wc,lls are isolated from the plate background using
a masking
t _.
CA 02410688 2002-12-12
procedure 404. Images are also ac~ltrired in other fluorescence channels being
used 405.
The region in each image come:>p~;~nding to each ~'vell 4i~(i is measured 407.
A feature
calculated from the measurements for a particular :veil is compared with a
predefined
threshold or intensity response ~~013, and based on the result the well is
either flagged as
a "hit" 409 or not. T he locatior7s of the wells flagged as hits are recorded
for
subsequent high content mode processing. If then. are wells remaining to be
processed
410 the program Ioops back 4~:aG until all the wells have been processed 411
and the
system exits high throughput mode.
Following HTS analysis, the system starts i~°~e high content mode
processing
to 501 defined in Figure 13. '1"he system selects the Hl"S optical module 50z
by
controlling the motorized posetic,ning system. l~or each "hit" well identified
in high
throughput mode, the ;~Y star;e location of the well is retrieved from memory
or disk
and the stage is then moved to the selected stage location ;503. The autofocus
procedure
5(~4 is called for the first hold in each hit well and then once every 5 to 8
fields within
each well. In one channel, in~a'.=.es are acquired of' the primary marker 505
(typically
cell nuclei counterstained witlo I:),4P1, Hoechst or P1 Iluorescent dye). The
images are
then segmented (separated irw;to regions of nuclei and non-nuclei) using an
adaptive
thresholding procedure 506. 'I he." output of the segmentation procedure is a
binary mask
wherein the objects are whit.:: aruii the background is black. This binary
image, also
2o called a mask in the art, is usccl to determine if tine field contains
objects 507. The mask
is labeled with a blob labelirng .~lgor-ithm whereby each object (or blob) has
a unique
number assigned to it. If objects are found in the field, images are acquired
for all other
active channels 508, otherwise the stage is advr~need to the: next field ~ 14
in the current
well. Each object is Iocatec:i ire the image for further analysis 509.
Morphological
CA 02410688 2002-12-12
features, such as area and shape oV~ the objects, are used to select objects
likely to be
cell nuclei 510, and discard (d<~ no further processing on 1 those that are
considered
artifacts. For each valid cell nucleus, the XYZ stage location is recorded, a
small image
of the cell is stored, and assay specific features are measured 5 i i . 'The
system then
performs multiple tests on the cc~lfs by applying several analytical methods
to measure
features at each of several wavelengths. After measuring the cell features,
the systems
checks if there are any unproc~:~ssed objects in the current Held 512. if
there are any
unprocessed objects, it locates the next object 50~? and determines whether it
meets the
criteria for a valid cell nucleus 51 ii, and measure~5 its features. After
processing all the
objects in the current field, the system det~rernmos whether it needs to find
more cells
or fields in the current well Sl..'s. if it needs to find t~nore cells or
fields in the current
well it advances the XYZ 4tagc: to the next field within the current well 515.
Otherwise, the system checks whither it has any remaining hit wells to measure
515. If
so, it advances to the next hit well 503 and pruceeds through another cycle of
I > acquisition and analysis, otherwise the hIC'S mode is finished 51 U.
In an alternative embo~.:lirnent of the present v.nwmtion, a method of kinetic
live
cell screening is provided. Tyne prc;viousiy described embodiments crf the
invention are
used to characterize the spatial distribution of cellular c:ornponents at a
specific point in
time, the time of chemical fi~at~r~n. .As such. these ern6odimerrts have
limited utility
for implementing kinetic based screens, due to the sequential nature of the
image
acquisition, and the amount of time required to read all the wells on a plate.
For
example, since a plate can reqr;ire 3t) -- (~U minutes to road through all the
wells, only
very slow kinetic processes can be treasured by simply preparing a plate of
live cells
and then reading through all the 4vells more than once Faster kinetic
processes can be
3y
CA 02410688 2002-12-12
measured by taking multiple reacl.ings ot: each well before proceeding to the
next well,
but the elapsed time between the first arid last well w oi.Ud be too long, and
fast kinetic
processes would likely be compie~te before reaching the last well.
The kinetic live cell extension of the invention enables the design and use of
screens in which a biological process is characterized by its kinetics instead
of; or in
addition to, its spatial characterstics. In many cases, a response in live
cells can be
measured by adding a reagent to a specific well and making multiple
measurements on
that well with the appropriate timing. 'This dynamic live cell embodiment of
the
invention therefore includes apparatus for fluid delivery to individual wells
of the
1~~ system in order to deliver reagents icE each well at a specific time in
advance of reading
the well. This embodiment trrereby allows kinetic measurements to be made with
temporal resolution of seconds to minutes on each well of the plate. 'to
improve the
overall efficiency of the dynamic live Cell system, the acquisition control
program is
modified to allow repetitive data collection front sub-regions of the plate,
allowing the
t 5 system to read other wells between the time points required for an
individual well.
Figure 8 describes an example of a fluid delivery device for use with the live
cell embodiment of the invewtian and is described abcwe. Mhis set-up allows
one set of
pipette tips 70~, or even a sinf~,le: pipette tip, to deliver reagent to all
the wells on the
plate. The bank of syringe pumps ~Ol can Ire: t.i:~e~d to deliver fluid to 12
wells
zo simultaneously, or to fewer wells by removing some of the tips iUS. The
temporal
resolution of the system can thcretore be adjusted, withocrt sacrificing data
collection
efficiency, by changing the nu,n~ber of tips and the scan pattern as follows.
Typically,
the data collectiim and analysis from a single well tab;es about S seconds.
'Moving, from
well to well and focusing in a moll requires about ~ seconds, so the overall
cycle time
:fit l
CA 02410688 2002-12-12
for a well is about IU seconds. '( h~refore, if a single pipette tip is used
to deliver fluid
to a single well, and data is collected repetitively from that well,
measurements can be
made with about 5 seconds temporal resolution. If 6 pipette tips are used to
ueiwer
fluids to 6 welts simultaneously, and the systerrr repetitively scans all 6
wells, each scan
will require 60 seconds, thereby establishing the temporal resolution. For
slower
processes which only require data collection every 8 minutes, fluids can be
delivered to
one half of the plate, by moving the plate during the fluid delivery phase,
and then
repetitively scanning that half of the plate. Theretc~re, try adjusting the
size of the sub-
region being scanned on the plate, the temporal resolution can be adjusted
without
to having to insert u-ait times bet;vc:en acquisitions. Because the system is
continuously
scanning and aeyuiring data, th;: cwGrall time to collect a kinetic data set
from the plate
is then simply the time to perfcnrnt a single scan c7f the plate, multiplied
by the number
of time points required. Typically, t tune point before addition of compounds
and 2 or
3 time points following additio:.n should be suf'ticient (or screening
purposes.
Figure 14 shows the ac~.a,uvsitic>n sequence used fir kinetic analysis. The
start of
processing 801 is configurati~:n~ of the system. much of which is identical to
the
standard HC.'S configuration. 1n addition, the operator must enter information
specific
to the kinetic analysis being p~:vrio.rmed 8U2, ,ucch as the sub-region size,
the number of
time points required, and the rwquirecl time increment. ~ sub-region is a
group of wells
z0 that will be scanned repetitiwlv in order to accumulate kinetic data. The
size of the
sub-region is adjusted so that tlic: system can scan a whole sub-region once
during a
single time increment. thus nnirrimizing wait tunes. fhe optimum sub-region
size is
calculated from the setup parrrneters, and adjusted if necessary by the
operator. The
system then moves the plate t::~ the first sub-region 8U3. and to the first
well in that sub-
41
CA 02410688 2002-12-12
region 804 to acquire the prestin~ulation (time - 0) time points. The
acquisition
sequence performed in each well is. exactly the same as that required for the
specific
HCS being run ;n kine'.ic mode. Figure 15 details a flow chart for that
processing. Alf
of the steps between the start 90_l and the return 9_U? are identical to those
described as
s steps 504 - 514 in Figure; l3.
After processing each well in a sub-region, the system checks to see if all
the
wells in the sub-region have been processed 8(76 (Figure 14j, and cycles
through all the
Wells until the whole region has been processed. ~l"he: systmn then moves the
plate into
position for fluid addition, and controls fluidic system delivery of fluids to
the entire
sub-region 807. this may r~~qiaire multiple: additions for sub-regions which
span
several rows on the plate, with the system moving the plate on the X,l' stage
between
additions. Once the fluids hav:~ ioeen added, the systenn moves to the first
well in the
sub-region 808 to begin acquisit.ican of~time points. The data is acquired
from each well
809 and as before the system cycles through all the ~~~ells in the sub-region
810. After
t s each pass through the sub-region, the system checks whether all the time
points have
been collected 811 and if not, f~atyses X13 if necess~uy 812 to stay
synchronized with the
requested time increment. Otlvervvise, the sy:,teni checks for additional sub-
regions on
the plate 814 and eith~:r movt,s t~-A the next sub-re~gi~c~n 5113 or tinishes
815. Thus, the
kinetic analysis mode comprises operator identifn-anon of sub-regions of the
mierotiter
2o plate or microwells to be scn::errecl, based on thc° kin~ai~:
response to be investigated,
with data acquisitions within ; scrh-region prior ro data acquisition in
subsequent sub-
regrons.
=t.'r
CA 02410688 2002-12-12
Specific Screens
In another aspect of the present invention, a rrrac:hine readable storage
medium
comprising a program containinl:~ a set of instructions for causing a cell
screening
system to execute procedures far defining the distribution and activity of
specific
S cellular constituents and processes is provided. In a preferred embodiment,
the cell
screening system comprises a high magnification fluorescence optical system
with a
stage adapted for holding cells and a means for rr~oving the stage, a digital
camera, a
light source for receiving and processing the digital data fiom the digital
camera, and a
computer means for receiving ,:rnd processing they digits( data from the
digital camera.
to This aspect of the invention comprises programs that instruct the cell
screening system
to define the distributian and ~zctivit~ of specitic cellular constituents and
processes,
using the luminescent probes.. th~~ optical imaging s~,~stem, and the pattern
recognition
software of the invention. Preferred embodimf~nts of the machine readable
storage
medium comprise programs consisting of ;a sees of instructions for causing a
cell
t5 screening system to execute the procedures set forth in Figures 9. 11, 12,
13, 1:1< ar 15.
Another preferred embodiment comprises a program consisting of a set of
instructions
for causing a cell screening systeru to execute procedures for detecting the
distribution
and activity of specific cellular constituents and processes. In most
preferred
embodiments, the collular ;~ro~:csscs include:. but. are not limited to,
nuclear
?0 translocation of a protein, ~<llular hypertrolrly, apc~plosis, and protease-
induced
translocation of a protein.
The following examlvles are intended for purposes of illustration only and
should not be construed to lin~rit the scope of the invention, as defined in
the claims
appended heretu.
4s
CA 02410688 2002-12-12
The various chemical con npounds, reagents.. dyes, and antibodies that are
referred to in the following frxanthlcs are commercially available from such
sources as
Sigma Chemical (St. Louis, M~J}, Molecular Probes (Eugene, OR}, Aldrich
Chemical
Company (Milwaukee, WI}, A.ecuratc Chemical C.'omparty (Westbury, NY), Jackson
Immunoresearch Laboratories ('Vest Cirove, P.4), and C.'lontech (Palo .Alto,
CA).
Example l Automated Serc:~err ,,for Compounds that Induce or Inhibit Nuclear
Translocation of a DNtf Transcvrr,ntio~r Factor
l0
Regulation of transcription of some genes involves activation of a
transcription
factor in the cytoplasm., resulting in that factor being transported into the
nucleus where
it can initiate transcription of a:c particular gene or genes. This change in
transcription
factor distribution is the basis of a screen for the cell-based screening
system to detect
compounds that inhibit or induce transcription cuf a particular gene or group
of genes.
A general description c>f the screen is given fc>Ilov~ed by a specific
example.
The distribution of the transcription factv.~r is detc;tmined by labeling the
nuclei
with a DNA specific fluoroph;>rc° like Hoechst i ~?3 and the
transcription factor with a
specific fluorescent antibody :otter autofocusing on the Hoechst labeled
nuclei, an
2o image of the nuclei is acquired in the cell-based scr~enir~g system at ?Ox
magnification
and used to create a mask by ~.~m of several optional thresholcling methods,
as described
supru. The mocpholo~~ical descriptors of the r~~;ions defined by the mask are
compared
with the user defined parameters and valid nuclear masks are identified and
used with
the following algorithm to extr;tct transcription factor distributions. Each
valid nuclear
?5 mask is eroded to define a sli;~,l~t9y smaller nuclear region. fhe original
nuclear mask is
then dilated in two steps tc; define a ring shaloed region around the nucleus,
which
,~a
CA 02410688 2002-12-12
represents a cytoplasmic region:. °fhe average antibody fluorescence in
each of these
two regions is determined, and the difference between these averages is
defined as the
NucCyt Difference. Twc:r exam~:rl~°,s of determining nuclear
translocation are discussed
below and illustrated in Figure lt)A-J. Figure 10A illustrates an unstimulated
cell with
its nucleus 200 labeled with a blue fluorophore and a transcription factor in
the
cytoplasm 201 labeled with a green fluorophore. Figure IUB illustrates the
nuclear
mask 202 derived by the cell-'based screening system. Figure IUC illustrates
the
cytoplasm 203 of the unstimr~latc:d cell imaged at a green wavelength. Figure
lUD
illustrates the nu~:fear mask 20? s eroded (reduced) once to define a nuclear
sampling
to region 204 with minimal cy~tolrlasrpic distribution. The nucleus boundary
202 is dilated
(expanded) several times to form a ring that is 2-> pixels wide that is used
to define the
cytoplasmic sampling region ~:O°} for the same cell. Figure lUE further
illustrates a side
view which shows the nuclear sartrpling region 204 and the cytoplasmic
sampling
region 205. Using these two sampling regions. data on nuclear translocation
can be
t ~ automatically analyzed by tlne c a ll-based screc:nin~; system on a cell
by cell basis.
Figure IOF-J illustrates the strategy for determining nuclear translocation in
a
stimulated cell. Figure l OF illi.istrates a stimulated cell vvnth its nucleus
206 labeled with
a blue fluorophore and a transcoption factor in the cytoplasm 207 labeled with
a green
tiuorophore. The nuclear mask ?~~$ ira i-igurc: 10(~ is derived by the cell
based
2o screening system. Figure IOFI illustrates the c;,noplasm 2U~) ofa
stimulated cell imaged
at a green wavelength. Fignru 101 illustrates the nuclear sampling region 211
and
cytoplasmic sampling region 21_2 of~ the stimulated cell. Figure 1 OJ further
illustrates a
side view w which shov~.~s the nuc: lean sampling region 21 l and the
c~~toplasmic sampling
region 212.
:)
CA 02410688 2002-12-12
A specific application of this cnethod has been used to validate this method
as a
screen. A human cell line was lrlated in 9b well n=icrotiter plates. Some rows
of wells
were titrated with agonist, a l:n~~wn inducer of a specific: :nuclear
transcription factor.
The cells were then fixed and stained by standard methods with a fluorescein
labeled
antibody to the transcription fi:~ctor, and Hoechst ~,4<'?3. The cell-based
screening
system was used to acquire anti, analyze images from this plate and the NucCyt
Difference was found to be stror~~glv correlated with the amount of agonist
added to the
wells as illustrated in Figure 1 (,. In a second c:xpc~riment, an antagonist
to the receptor
for the agonist was titrated in the presence of agonist. progressively
inhibiting agonist-
induced translocation of the transcription factor The NucCyc Difference was
found to
strongly correlate with this inhibition of translocation, as illustrated in
Figure 17.
Additional experiments have shown that the Nuc(.'vt Difference gives
consistent
results over a wide range of cel'I densities and reagent con centrattons, and
can therefore
be routinely used to screen compound libraries fc~r specific nuclear
translocation
t s activity. Furthermore, the ~~.arne method can be used writh antibodies to
other
transcription faca.ors, or GFP-transcription factor chimeras, in living and
fixed cells, to
screen for effects on the regulation of transcrilotion of this and other
genes.
Figure 18 is a representative display on a PC' <~crecn of data which was
obtained
in accordance with Example 1. Graph 1 18C! plots the dcfference between the
average
2o antibody fluorescence in the n~.rcl~;ar ~amplinle reunion and cytoplasmic
sampling region,
NucCyt Difference verses ~Vr:~il :~. (traph 2 18i_ plots the average
tluorescence of the
antibody in the nuclear sampling region, NPR average, vt.rsus the Well #.
Graph 3 182
plots the average antibody iluoresc:eoce in the ~ytoplasmic sampling region,
LIP1
average, versus Well f#. 'fh;v sofRware permits displaying data from each
cell. For
4~ >
CA 02410688 2002-12-12
example, Figure t 8 shows a s;:rc~i~n display t 83, the nuclear image 184, and
the
fluorescent antibody image 13~ f~~r c~:ll X26.
NucCyt Difference referred to in graph 1 180 of Figure l8 is the difference
between the average cytoplasmic probe (fluorescent reporter molecule)
intensity and
a the average nuclear probe (flut:~rt~scent reporter molecule) intensity. NPl
average
referred to in graph 2 t81 of Figure 18 is the average of cyloplasmic probe
(fluorescent
reporter molecule) intensity within the nuclear sampling region. 1.1 P 1
average referred
to in graph 3 1$2 of Figure la is the average prube (fluorescent reporter
molecule)
intensity within the cytoplasmic sampling region.
to
E.rample 2 Automated ScrE>cvn tf>r (~ompourrds that Irrcluce or Inhibit
Hypertrophy in
Cardiac Mvocvtes
Hypertrophy in cardiac: myocytes has been associated with a cascade of
t ~ alterations in gene expression artd can be characterir_ed in cell culture
by an alteration in
cell size, that is clearly visible irs adherent cells growing on a coverslip.
A screen is
implemented using the followin~.T strategy. My~~cvlc: cell line QM7 (Quail
muscle
clone 7; ATCC C'RL-19G?) c~~ltured in g6 well plates, can be treated with
various
compounds and then fixed and labeled with a Iluorescent antibody to a cell
surface
?o marker and a Dl'JA label like f-lcaLc.hst. After focnsin~; on the Hoechst
labeled nuclei,
two images are acquired, one ~:~t tl7e Hoechst labeled nuclei and one of the
fluorescent
antibody. The nuclei are identified by thresholding to create a mask and then
comparing
the morphological descriptors of the mask with a set of~ user defined
descriptor values.
Local regions containing cells are detined around the nuclei. The limits of
the cells in
a> those regions are them defined by a local dynamic threshold operation on
the same
region in the fluorescent antibody image. A sequence of erosions and dilations
is used
47
CA 02410688 2002-12-12
to separate slightly touching cell, and a second set ofmorphological
descriptors is used
to identify single cells. The areas crf thc: individual cell" os tabulated in
order to def ne
the distribution of cell sizes for comparison 4~ ittt size data Pram normal
and
hypertrophic cells. In addition, c:~ ~~~.~cond fluorescf;nt arntibody to a
particular cellular
protein, such as one of the major muscle proteins actin or myosin can be
included.
Images of this second antibody ~,. an be acquired and sicared with the above
images, for
later review, to identify anomalies in the distribution of these proteins in
hypertrophic
cells, or algorithms can be developed to automatically analyze the
distributions of the
labeled proteins in these images.
t0
Example 3 Dual Moele Higlr T~tr-r~iighput and flyh-Content Screen
The following example is a screen for activation of'a G-protein coupled
receptor
is (GPCR) as detected by the tran,~;location of the; <,;PCR from the plasma
membrane to a
proximal nuclear location. This e~:ample illustrates ho~~- a high throughput
screen can
be coupled with a high-content screen in the dual mode System for Cell Based
Screening.
G-protein coupled recep~tc~rs arc a large class of ? traps-membrane domain
cell
:!o surface receptors. Ligands for tltese receptors stimulate a cascade of
secondary signals
in the cell, which may includ;:v, I~ut are not limited to. C guy transients,
cyclic AMP
production, inositol triphosphate ( 1 P, ) produCa~on an<i phosphorylation.
Each of these
signals are rapid, occuring in a matter of second. to minutes, but are also
generic. For
example, many different CJPC'l~s produce a sea.ondat-v C'a" signal when
activated.
2~ Stimulation of a GPCR also re=suits in the transport of that tiPCR from the
cell surface
48
CA 02410688 2002-12-12
membrane to an internal, proxim,:rl nuclear compartment. ~fhis internalization
is a much
more receptor-specific indicator oi' activation of a particular receptor than
are the
secondary signals described above.
Figure 19 illustrates a dual mode screen for activation of a GPCR. Cells
carrying a stable chimera of the G:PC'R with a blue fluorescent protein (BFP)
would be
loaded with the acetoxymethylester form of Fluo-~, a :ell permeable calcium
indicator
(green fluorescence) that is trapped in living cells by the hvdroiysis of the
esters. They
would then be deposited into th~r ~4-ells of a microtiter plate 601. The wells
would then
be treated with an array of test compounds using a fluid delivery system, and
a short
to sequence of Fluo-3 images ou' the whole microtiter plate would be acquired
and
analyzed for wells exhibiting ..r. calcium response (t e., high throughput
mode). The
images would appear like the :,llusiration of the rrricrotit~r plate 601 in
Figure 1ST. A
small number of wells, such a.; wells C4 and Eti in the illustration, would
fluoresce
more brightly due to the Ca~+ ri:leased upon stimulation oiT the receptors.
The locations
of wells containing compound;~c tRiat induced ;:r reslaonse 6t)?, would then
be transferred
to the HCS program and the of~tica sv itched for detailed cell by cell
analysis of the blue
fluorescence for evidence of Gla(K translocation to the perinuclear region.
The bottom
of Figure 19 illustrates the twi;~ possible outcomes of the analysis of the
high resolution
cell data. The camera images a Sub-region 6(J4 of the well area 603, producing
images
of the fluorescent cells 60s. In well (:'4, the i.jnif~i,rm distribution of
the fluorescence in
the cells indicates that the receptor has not internalized. implying that the
Ca+' response
seen was the result of the stimulation of some other si'naliing system in the
cell. The
cells in well E9 606 on the other hand, clearly indicate a concentration of
the receptor
in the perinuclear region clearly indicating the full activation of the
receptor. Because
:1~3
CA 02410688 2002-12-12
only a few hit wells have to be analyzed with high resolution, the overall
throughput of
the dual mode system can be quoits; high, comparable to th-~ high Lhroughput
systE;m
alone.
Example 4 Kinetic High Content Screen
The following is an ~:::X~irIlplc; Of a screen to measure the kinetics of
internalization of a receptor. As described above, the stirrrulation of a
GPCR, results in
the internalization of the recelvtc:rr, with a time course of about I S min.
Simply
detecting the endpoint as internalir_ed or not, may nc5t he sufficient for
defining the
to potency of a compound as a (~F'CR agonist or antagonist. However, 3 time
points at ~
min intervals would provide inf~;:>n~~~ation not only about potency during the
time course
of measurement, but would also allow extrapolation of the data to much longer
time
periods. To perform this assay. the sub-regaon would be defined as two rows,
the
sampling interval as 5 minLtte~~, ~:um1 the total number of time points 3. The
system
t s would then start by scirnning tv,o> rows, and then adding reagent to the
two rows,
establishing the time=0 referenc::e <After reagent addition, the system would
again scan
the two row sub-region acquirving the first time point data. Since this
process would
take about 250 seconds. includm~~ scanning back to the beginning. of the sub-
region, the
system would wait 50 seconds to fyegin acquisition of the second time point.
Two more
2c~ cycles would praduce the thr~~ue rime points and thc: systen n would move
on to the
second 2 row sub-region. The final two 2-row sub-re~ior~s would be scanned to
finish
all the wells on the plate, resulting in four time points for each well over
the whole
plate. Although the time pi,~int~; for the wrens woul~3 be offset slightly
relative to
time=(), the spacing of the tune l~oirtts wcould be very elc5se to the
reguired S minutes,
5 (:'
CA 02410688 2002-12-12
and the actual acquisition times a.nd results recorded with much greater
precision than
in a fixed-cell screen.
Example S High-content sere~:~n of human ~IucocworW ~oicl receptor
trunslocation
One class of H~C'S involves the drug-induced dynamic redistribution of
intracellular constituents. The hunuan glucocorticoid receptor (hGR), a single
"sensor"
in the complex environmental response machinery ~.~f the cell, binds steroid
molecules
that have diffused into the cei 1. The ligand-re~:eptor complex transloeates
to the
nucleus where transcr-iptional o.etivation occurs 11-Itun et al., Proc. Natl.
Acacl. ,Sci.
to 93:4845, 1996).
In general, hormone receptors are excellent drug targets because their
activity
lies at the apex of key intracellular signaling pathways. 'therefore, a high-
content
screen of hGR translocation has distinct advantage over in vitro ligand-
receptor binding
assays. The availability of up to two more channels of~ fluorescence in the
cell
~ 5 screening system of the presen, invention permits the screen to contain
two additional
parameters in parallel, such as outer receptors. other distinct targets or
other cellular
processes.
Plas»iid construct. :~ ~:~ukaryotic expression plasmid containing a coding
sequence for a green fluorescent protein - human glucocorticoid receptor (GFP-
hGR)
20 chimera was prepared using C~FIa mutants (Palm ~a <r1., :Vat. .Strcco!.
Biol. 4:361 ( 1997).
The construct was used to t.ranv>f~:oa a human cewical carcinoma cell fine
(HeLa).
Cell preparation and transfection. Hel.a cells (.A'I'(.'(' C:t=I_-?) were
trypsinized
and plated using DM1M containing 5°io charcoal'dextran-treated fetal
bovine serum
(FBS) (HyGlone) and 1°,~o p~:nicillin-streptomycin ~C'-L~MEM) 12-24
hours prior to
>i
CA 02410688 2002-12-12
transfection and incubated at 37"(: and 5% (_'O~ Transfections were performed
by
calcium phosphate co-precipitat.~ur~ (Graham a~~d ~'an der Eb,, 6'iroloy
52:456, 1973;
Sambrook et al., ('1989). :~LTolerr~lc~r Cloning: ~1 Luhcrratow ,'t~unuul,
Second ed. Cold
Spring Harbor Laboratory Press, C'.~:~ld Spring Harbor, 1989) or with
Lipofectamine (Life
Technologies, Gaithersburg, MD)- For the calcium phosphate transfections, the
medium was replaced, prior to transfecaion, with DMEM containing 5%
charcoal/dextran-treated FBS. (;.'.ells were incubated with the calcium
phosphate-DNA
precipitate for 4-5 hours at 3~"°(' and 5% C'C),, w~uhed 3-4 times with
DMEM to
remove the precipitate, followed by the addition ol'C-DI~t>~.M.
:.0 Lipofectamine transfections were performed in serum-f=ree DMEM without
antibiotics according to the nrar~ufacturer'~. instructions (Life
Technologies,
Gaithersburg, MD). Followings a 2-a hour incubation with the DNA-liposome
complexes, the medium was removed and replaced with C -DMEM. All transfected
cells in 96-well microtiter plates were incubated at ~ 3"'(:'. and 5°,%
CO~ for 24-48 hours
15 prior to drug treatment. Experiments were performed with the receptor
expressed
transiently in HeLa cells.
Dexamethasone indur~tion of GFP-hGrR' translorution. 'fo obtain receptor-
ligand translocation kinetic dat,~, nuclei of transfected cells were tirst
labeled with 5
pg/ml Hoechst 63342 (Molecul;~r Probes) in C-~WII~M for ?0 minutes at 33"C and
5°,%
LO COz. Cells were w ~asl~ed oncw~ in Hzrnk's Balanced Salt Solution (HI3SS)
followed by
the addition of 100 nM dexa~°c~ethasone irr HBSS ~~~ith I%
charcoal/dextran-treated
FBS. To obtain fixed time point dcxamethasorle titration data, transfected I-
feLa cells
were first washed with DML~:!-1 and then incubated at 3v3°<~ and
5°'o CO, for 1 h in the
presence of 0 -- 1000 nM drxamethasone in ZrMEM containing f°,~o
charcoalidextran-
a ',
CA 02410688 2002-12-12
treated FBS. Cells were analyrew:l live or they were rinsed with HBSS, fixed
for 15 min
with 3.7°,% foonaldehyde in 1-ll3;p~, stained with 1-loec;hst 33342,
and washed before
analysis. The intracellular GFP-h(.iR fluorescence signal was not diminished
by this
fixation procedure.
Image acquisition and analysis. Kinetic. data were collected by acquiring
fluorescence image pairs (GFP-ilCUR and Hoechst 3332-labeled nuclei) from
fields of
living cells at 1 min interval,, for :30 min after the addition of
dexamethasone.
Likewise, image pairs were obt,:3.ir~~ed from each veil of the fixed time
point screening
plates 1 h after the addition of clc~aamethasone. In hoth cases, the image
pairs obtained
to at each time point were used tc:~~ t.tefine nuclear adld cyloplasnlic
regions in each cell.
Translocation of GFP-hGR w~a~s calculated by dividing the integrated
fluorescence
intensity of GFP-hGR in the nucleus by the integrated fluorescence intensity
of the
chimera in the cytoplasm or as a nuclear-cytoplasmic difference of GFP
fluorescence.
In the fixed time point screen this translocation ratio was calculated from
data obtained
t, from at least 200 cells at each ~-orlcentration of dexarnethasone tested.
Drug-induced
translocation of GFP-h(R from the cytoplasm to the nucleus was therefore
correlated
with an increase in the transloctrti~n ratio.
Results. Figure 20 schematically dispiay~, the drub:-induced cytoplasm 253 to
nucleus 252 translocation of tile human glucocorticoid receptor. The upper
pair of
U schematic diagrams depicts the localisation of GEP-h(~R wrthnl the cell
before 25(.1 (A)
and after 2~ 1 ~B) stimulation r~~ith dexamethasone. ~lnder these experimental
conditions, the drug induces a ',urge portion of the cytoplasmic CJFP-hGR to
translocate
into the nucleus. This redi~>tribution is quantitied by determining the
integrated
intensities ratio of the cvtopla~mic and nuclear fluorescence in treated 255
and
3
CA 02410688 2002-12-12
untreated 254 cells. The lower pair of' fluorescence micrographs show the
dynamic
redistribution of G:FP-hGR in a :,in~;le cell, before Z~4 arld after 2S5
treatment. The
HCS is performed on wells con~.~ainirrg hundreds to thousands of transfected
cells and
the translocation is quantified fc~r each cell in the tieLd exhibiting GFP
fluorescence.
Although the use of a stably tr~:rnsfected cell line would yield the most
consistently
labeled cells, the heterogeneous levels of GFP-hGR expression induced by
transient
transfection did not interfere with analysis by the cell screening system of
the present
invention.
To execute the screen, the cell screening system scans each well of the plate,
to images a population of cells in each, and anal.~ros cf~lls individually.
Here, two
channels of fluorescence are used to define the cyrtoplasmic and nuclear
distribution of
the GFP-hGR within each cell. Depicted in Fi;~ur~ 21 is the graphical user
interface of
the cell screening system near tine end of a GFF'-hGR sc:reert. The user
interface depicts
the parallel data collection and analysis capability i~f the system. The
windows labeled
t 5 "Nucleus" 261 and "GFP-hCaR" ?6? show the pair of fluorescetlce images
being
obtained and analyzed in a sin~_=fe freld. The window labeled "(dolor Overlay"
260 is
formed by pseudocotoring th~:° ,above images and mer:;ing them so the
user can
immediately identify cellular ch'4ryes. Within the "~~tc~red Object Regions"
window
265, an image containcng oacu ;tnalvred cell arrd its neighbors is presented
as it is
a0 archived. Furthermore, as the l-lt:'S data are being coIlectecf, they aro
analyzed, in this
case for GFP-hGR translocation, and translated into an mumediate "hit"
response. The
96 welt plate depicted in the livver window of the screen 2G l shows which
wells have
met a set of user-defined scrc:et7.in~~ c:nteria. For exan~ple~, a whito-
colored well 269
indicates that the drug-induce:cl translocation has exceeded a predetermined
threshold
j4
CA 02410688 2002-12-12
value of 50%. On the other hand, a black-colored well 2'70 indicates that the
drug being
tested induced less than 10°-r translcucation. Gray--colored walls 26$
indicate "hits"
where the translocation value fell between 10°~o and 50°,-0. low
"E" on the 96 well
plate being analyzed 266 show.<~ a titration with a drug known to activate GFP-
h(sR
translocation, dexamethasone. This examplf: screen used only two fluorescence
channels. Two additional chatrnels (Channels 3 263 and 4 264) are available
for
parallel analysis of other specific targets, cell processes, or cytotoxicity
to create
multiple parameter screens.
There is a link between the image database and the information database that
is
to a powerful tool during the valic9ation process of new screens At the
completion of a
screen, the user has total accvs~~ to image and calculated data (Figure 22).
The
comprehensive data analysis package of the cell screening system allows the
user to
examine HCS data at multiple levels. Images 276 and detailed data in a spread
sheet
279 for individual cells can be ~,eiewed separately, car summary data can be
plotted. For
t, example, the calculated results of a single parameter for each cell in a 96
well plate are
shown in the panel labeled Gr~xp1 1 275. By selecting a single point in the
graph, the
user can display the entire data sca for a particular cell that is recalled
from an existing
database. Shown here are the image pair 276 and detailed fluorescence and
morphometric data frorn a sin~ale cell (C'ell # l ~ 8, gray Line '~?7:9. hhe
large graphical
insert 278 shows the results of dcaamethasone concentration on the
translocation of
GFP-hGR. Each point is the average of data from at le~~st 200 cells. The
calculated
EC;~, for dexamethasone in thi~~ assay is 2 nM.
A powerful aspect of I~(-'S with the cell screening system is the capability
of
kinetic measurements using r7oulticolor fluorescence arid rnorphometric
parameters in
'i 5
CA 02410688 2002-12-12
living cells. Temporal and spatial measurements can be made on single cells
within a
population of cells in a field. Figure 2 3 shows kinetic data for the
dexamethasone-
induced translocation of GFP-hGEZ i,n several cells within a single field.
Human HeLa
cells transfected with GFP-hGR were treated with 100 nM dexamethasone and the
translocation of GFP-hGR was measured over time in a population of single
cells. The
graph shows the response of transfected cells 285, 286, 287, and 288 and non-
transfected cells 289. -Chese data also illustrate the ability to analyze
cells with
different expression levels.
1G E.rample 6 High-content scrce~r, ohdrug-inclucec! apo,~to.sis
Apoptosis is a complex c~ellawlar program that involves myriad molecular
events
and pathways. To understand th~~ mechanisms <af~ drug action on this process,
it is
essential to measure as many of vhese events within cells as possible with
temporal and
spatial resolution. Therefore, an apoptosis screen that requires little cell
sample
i 5 preparation yet provides an auto>nuated readout of several apoptosis-
related parameters
would be ideal. ,A cell-based as5av designed for the cell screening system has
been
used to simultaneously quantify sc:verai of ~he morphological, organellar, and
macromolecular hallmarks of p;:nclitaxel-induced apoptosis.
Cell preparation. The cells chosen for this study were mouse connective tissue
'o fibroblasts (L-92~>; ATf_'C CCI_-'t y and a highly mva~~i~re glioblastoma
cell line (~NB-
19; ATC'C C_'RL-2219] (Welclt et al., l~r Y~itro ('ell. DEw Hiol. 31:610,
1995). 'The day
before treatment with an apoptcasis inducing drug, 3500 cells were placed into
each well
of a 96-well plate and incubated ovemig,ht at ~i7"C' in a humidified 5% COs
atmosphere. The following day. the culture medium was removed fiom each well
and
56
CA 02410688 2002-12-12
replaced with fresh medium containing various concentrations of paclitaxel (0 -
SO
1M) from a 20 mM stock made in TaMiS(7. The maxin-ral concentration of DMSO
used
in these experiments was 0.25%. The cells were then incubated for 26 h as
abovE. At
the end of the paclitaxel treatment lner-iod, each well received flesh medium
containing
7S0 nM MitoTracker Red (Molecular Probes; Eugene, (UR) and 3 Irg/ml Hoechst
33342
DNA-binding dye (Molecular Probes) and was incubated as above for 20 min. Each
well on the plate was then washed with HBSS and fixed with 3.7% formaldehyde
in
HBSS for 1 S min at room temperature. The formaldehyde was washed out with
HBSS
and the cells were permeabilized for CIO s with 0.5% lulu) 'Triton X-1()~,
washed with
HBSS, incubated with 2 U ml-' Bwdipy FL phallacidin (Molecular Probes) for 30
min,
and washed with HBSS. °fhe wells on the plate were then filled with 200
ftl HBSS,
sealed, and the plate stored at 4°C.' ifi necessary. T'he fluorescence
signals from plates
stored this way were stable for at Yeast two weeks after preparation. As in
the nuclear'
translocation assay, fluorescence reagents can be designed to convert this
assay into a
live cell high-content screen.
Image acquisition and analysis on the .4rrayScan System. The fluorescence
intensity of intracellular Mito'I'ras~ker Red, Hoechst 33342, and Bodipy FL
phallacidin
was measured with the cell screening system as described supra. Morphometric
data
from each pair of images obtained from each well was also obtained to detect
each
object in the image field (e.g., ~ ells and nuclei), and to calculate its
size, shape, and
integrated intensity.
Calculations and outpua. A total of 50-2S0 cells were measured per image
field. For each field of cells, the following calculations were performed: (1
) The
average nuclear area (urnz) was calculated by dividing; the total nuclear area
in a field
S7
CA 02410688 2002-12-12
by the number of nuclei detected. ('?) The average nuclear perimeter (Irm) was
calculated by dividing the sum of thi; perimeters of all nuclei in a field by
the number
of nuclei detected it that field. Ijighly convoluted apoptotic nuclei. had the
largest
nuclear perimeter values. (~) The avt.rage nuclear brightness was calculated
by dividing
the integrated intensity of the entire field of nuclei by the number of nuclei
in that field,.
An increase in nuclear brightness 'was correlated with increased DNA content.
(4) The
average cellular brightness was calculated by dividing tire integrated
intensity of an
entire field of cells stained with Iv!lito"T racker dye by the number of
nuclei in that field.
Because the amount of MitoTrac:l~er dye that accumulates within the
mitochondria is
proportional to the mitochondria) potential, an increase in the average cell
brightness is
consistent with an increase in mitochondria) potential. (5) T'he average
cellular
brightness was also calculated bar dividing the integrated intensity of an
entire field of
cells stained with Bodipy FL phallacidin dye by the number of nuclei in that
field.
Because the phallotoxins bind with high affinity to the polymerized form of
actin, the
TM
amount of Bodipy FL phallacidirr dye that accumulates within the cell is
proportional to
actin polymerization state. An increarse in the average cell brightness is
consistent with
an increase in actin polymerization..
Results. Figure 24 (top panels) shows the changes paclitaxel induced in the
nuclear morphology of L-929 cells. Increasing arr~ounts of paclitaxel caused
nuclei to
enlarge and fragment 293; a hallmark oh apoptosis. (2uantitative analysis of
these and
other images obtained by the cell screening system is presented in the same
figure.
Each parameter measured showed that the L-929 cells 296 were less sensitive to
low
concentrations of paclitaxel tharu were SNB-19 cells 297. At higher
concentrations
though, the L-929 cells shovve:d a response for each parameter measured. The
S8
CA 02410688 2002-12-12
multiparameter approach of this assay is useful in dissecting the mechanisms
of drug
action. For example, the area, ~~nghtness, and fragmentation of the nucleus
298 and
actin polymerization vaiu~s 294 rx.<~ched a maximum value when SNB-19 cells
wore
treated with 10 nM paclitaxel (Figure 24; top and bottom graphs). However,
mitochondria) potential 295 was minimal at the same concentration of
paclitaxel
(Figure 24; middle graph). The fact that all the parameters measured
approached
control levels at increasing paclitaxel concentrations ( >10 nM) suggests that
SNB-19
cells have low affinity drug metabolic or clearance pathways that are
compensatory at
sufficiently high levels of the dnt,g. ( ontrasting the drug sensitivity of
SNB-19 cells
t0 297, L-929 showed a different response to paclitaxevl 296. These
fibroblastic cells
showed a maximal response in many parameters at S feM paclitaxel, a S00-fold
higher
dose than SNB-1~) cells. Furth;:rrnore, the L-929 cells did not show a sharp
decrease in
mitochondria) potential 295 at any of the paclitaxcf concentrations tested.
This result is
consistent with the presence of unique apoptosis pathways between a normal and
t5 cancer cell line. Therefore, tE~.ese results indicate that a relatively
simple fluorescence
labeling protocol can be coupl,~d with the cell scr~,enin~; system of the
present invention
to produce a high-content screen cri key events involved in programmed cell
death.
E.ranrple 7, Protease indu~rect trurrslococtior~ i~/~ rr si~rrraling cnt~me
contuin~ing a
2() disease-ussociclrc~d sequcjtoe fi~or~t <yuoBlusrn t<.> uuclcus.
Plasmid con.sr'ruct. ;1 eukaryotic a xpression plasmid containing a coding
sequence for a green fluorescent protein - caspase f C'ohen ( 1997 ),
Biochemical J.
326:1-15; Liang et al. (19t)7). J o;_,~folE~c. Biol. 2?4:~?~)1-3()2) chimera
is prepared using
2, GFP mutants. 'the construct is used to traps°ect eukan~otic cells.
5~)
CA 02410688 2002-12-12
Cell preparution and trans~''ectian. Cells are trysinized and plated 24 h
prior
to transfection and incubated at 3~" t' acrd ~~~~ C'O~' 'I'ransfe;ctions are
performed by
methods including, but not limited to calcium phosphate: coprecipitation or
lipofection.
Cells are incubated with the calcium phosphate-DNr'~ precipitate for 4-S hours
at 37°C
and 5% COz, washed 3-4 times ~nrith DMEM to remove the precipitate, followed
by the
addition of C-DMEM. Lipofectamine transfections are performed in serum-flee
DMEM without antibiotics according to the manufacturer's instructions.
Following a
2-3 hour incubation with the DN,~-liposome complexes, the medium is removed
and
replaced with C-DMEM.
t0 .4popototic induction a~ ('aspase-GFP trarrslacatiarr. To obtain Caspase-
GFP
translocation kinetic data, nuclei of transfected cells are first labeled with
5 Ilg/ml
Hoechst 33342 (Molecular Prows') in C-DMEM fiar ?() minutes at 37°C and
5% CO2.
Cells are washed once in Hank's Balanced Salt Solution (HBSS) followed by the
addition of compounds that insauce apoptosis. These compounds include, but are
not
limited to paclitaxel, staurospnrinc, ccramide, a~~d tumor necrosis factor. To
obtain
fixed time point titration data, tr;~nsfeeted cells are first washed with DMEM
and then
incubated at 37°C and 5°,% C(:~, for I h in the presenco of (-'~
-- l c>00 n~NI compound in
DMEM. Cells are analyzed lm~car thev are rinsed with HBSS. fixed for 1_S min
with
3.7% fotmaldelrvde in I-1BSS, ~~ta~ined with Hoechst 33s~:', and washed before
analysis.
?o Image acquisition arid analysis. Kinetic data are collected by acquiring
fluorescence image pairs (C.'a~sp;rse-CxFP and Hoechst 38342-labeled nuclei)
from tields
of living cells at 1 min interv;:~is for 3U min after the addition of
compound. Likewise,
image pairs are obtained fro.r each well of the fixed tune point screening
plates I h
after the addition of compound. In both cases, the image pairs obtained at
each time
6t)
CA 02410688 2002-12-12
point are used to define nuclear and cytoplasmic regions in each cell.
Translocation of
Caspase-GFP is calculated by dividing the integrated fluorescence intensity of
Caspase-
GFP in the nucleus by the int<f~rated fluorescence intensity of the chimera in
the
cytoplasm or as a nuclear-cytoplasmic difference of C:ifP fluorescence. In the
fixed
time point screen this translocatrorn ratio is calculated from data obtained
from at least
200 cells at each concentration of compound tested. Drug-induced translocation
of
Caspase-GFP from the cvtoplasrn to the nucleus is therefore correlated with an
increase
in the translocation ratio. Mole~Luiar interaction libraries including, but
not limited to
those comprising putative activators or inhibitors of apoptosis-activated
enzymes are
1~ use to screen the indicator cell ~i~ms and identify ~~ specafie ligand for
the DAS, and a
pathway activated by compound a<:tivity.
E.rumple 8. Identification of ~~u?~-ol steroid r47oeptors fi~orn D:4S
Two sources of materiw~l and/or information are required to make use of this
t ~ embodiment, which allows as;es~;ment of the iimction of an uncharacterized
gene.
First, disease associated segue-rvce banks) c:.antaining cDNA sequences
suitable for
transfection into mammalian c~eils can be used. Because every kZADE or
differential
expression experiment generntt~s up to several hundred sequences, it is
possible to
generate an ample supply caf 1=t.AS. S<<coi7d. information from primary
sequence
database searches can be use~.l. to r I<.ice DAS into broad categories,
including, but not
limited to, those that contain signal sequcr~ces, seven traps-membrane motifs,
conserved protease active site ciamains, or outer identifiable motifs. Based
on the
information acquired from these sources. al;~oritlm,: types and indicator cell
lines to be
transfected are selected. A largo number of motifs are already will
characterized and
CA 02410688 2002-12-12
encoded in the linear sequences ce~ntainod within the large number genes in
existing
genomic databases.
In one embodiment, the fn~ll-owing steps are taken:
l ) Information from the' I)AS identification experiment (including database
searches) is used as the basic: fttr selecting the relevant biological
processes. (for
example, look at the DAS fronn ,~ tumor line for cell c_,~cle modulation,
apoptosis,
metastatic proteases, etc. )
?) Sorting of DNA sequences or DAS by identifiable motifs (ie. signal
sequences, 7- transmembrane domarns, conser~~e~I protease active site domains,
etc.)
1~~ This initial grouping will dete~rroine fluorescent tagging strategies,
host cell lines,
indicator cell linca, and banks o1~ hioactive molecules tea be screened, as
described
supra.
3) Using well established molecular biology methods, ligate DAS into an
expression vector designed for this purpose. (.ieneralized expression vectors
contain
1, promoters, enhanccrs, and ternzinators for which td> deliver target
sequences to the cell
for transient expression. Such vectors may also contain antibody tagging
sequences,
direct association sequences, clorcvroophore fusiG~r~ ;;eq~ae;nces like
(:.iFP, etc. to facilitate
detection when expresse~.i k~sv the: host.
4) Transiently transfec:t cells with DAS c:~ntaining vectors using standard
transfeetion protocols ir~cluriin~;: calcium phosphate co-precipitation,
liposome
mediated, DEAF dexiran nrecfiated, polycatiunic mediated, viral mediated, or
electroporation, arid plate intiv :rrii:raciter plates or mrcrowell arrays.
Alternatively,
transfection can be done direct'~y is~ the microtiter ;>late. itself.
2, 5) Carry out the cell screening methods rrs described srsln"Cl.
In this embodiment, DAS shown to possess a motifs) suggestive of
transcr-iptional activation potential f for example, ONA binding domain, amino
terminal
modulating domain, hinge r..~gri~n, or carh~a::~; ternrit7al ligand binding
domain) are
utilized to identify novel steroid receptors.
Defining the fluorescent tags for this experiment involves identification of
the
nucleus through staining, and tagging the DAS by creatrng a (iFP chimera via
insertion
of DAS into an expressiorv vr:ctor, proximally fcrsecf to the gene encoding
GFP.
t l .,
CA 02410688 2002-12-12
Alternatively, a single chain antibody fragment with high affinity to some
portion of the
expressed DAS could be constru;~tc;~t using technolGyy available in the art
(Cambridge
Antibody Technologies) and linked to a iluorophore (Fl'fC) to tag the putative
transcriptional activator!receptor~ in the cells. This alternative would
provide an
external tag requiring no DNA tr,ansfection and therefore would be useful if
distribution
data were to be gathered from th~:~ ~:ariginal primary cultures used to
generate the DAS.
Plasmid construct. A eukaryotie expression plasmid containing a coding
sequence for a green fluorescent protein -- I~AS chimera is prepared using GFP
mutants. The construct is used t.o transfect HeLa cells ~('he plasmid, when
transfected
to into the host cell, produces a GFI" fused to the DAS pr~~tein product,
designated GFP-
DASpp.
Cell preparation and transfeetion. HcLa cells <~re trypsinized and plated
using
DMEM containing S°o charcoal dextrin-treated fetal bovine serum (FBS)
(Hyclone)
and 1°~° penicillin-streptomycin (('-l)MEM) 14"-:.'4 hours prior
to transfection and
t5 incubated at 37°C and 5°~o C.'('n . Transfections are
performed by calcium phosphate
coprecipitation or with Lipofe~.:t<rn~inc (Life 'I'ech~ol~.~'~ies). For the
calcium phosphate
transfections, the medium is rt:plai:ed, prior to trarjsfection, with DMEM
containing 5%
charcoal/dextran-treated FBS. ::'ells are incubated with the calcium phosphate-
DNA
precipitate for ~-5 hours at 3 ~'°~' and ~°,-o Ct:)~, ;md
wasi~ed 3-~ times with DMEM to
2o remove the precipitate, follm4~~.:d by the adciiticn~ of C-DMEM.
Lipofectamine
transfections are perfc~r~ned i~u senrm-free DMEM withc>ut antibiotics
according to the
manufacturer's instructions. Fcalluwing a 2-3 h«ur incubation with the DNA-
liposome
complexes, the rnediurn is rernc>ved and replaced with C'-DMEM. All
transfected cells
in 96-well microtiter plates a.re incubated at s3''(' end 5°it> C.'O~
Cor 24-48 hours prior to
(, _,
CA 02410688 2002-12-12
drug treatment. Experiments arc pexforrned with the receptor expressed
transiently in
HeLa cells.
Localization of cpxpress~~d GF'P-DASpp inside cells. To obtain cellular
distribution data, nuclei of transfected cells are first labeled with 5 llg/ml
Hoechst
33342 (Molecular Probes) in C-L?~"11M for 20 minutes at 33°C_' and 5%
CO~. Cells are
washed once in Hank's Balanced ;salt Solution (HBSS). I~he cells are analyzed
live or
they are rinsed with HBSS, fixed fc>r 15 min with 3_''°~ forrmldehyde
in HBSS, stained
with Hoechst 33342, and washed bef«re analysis.
In a prefewed embodiment, image acquisition and analysis are performed using
the cell screening syste~rn of tl.~e present inv~ertti~n. The intracellular
GFP-DA Spp
fluorescence signal is collected by acquiring fluorescence image pairs (GFP-
DASpp
and Hoechst 33342-labeled nucteil Iiom field cells. The image pairs obtained
at each
time point are used to define nui:lear and cy~toplasrr~ir regions in each
cell. Data
demonstrating dispersed signal rn the cvtoplasrn would be consistent with
known
steroid receptors that are DNA trans~,riptional ;~caivators.
Screening for inductiurt of GFP-D.9Spp translocation. t.Jsing the above
construct, confirnted for approlvriate expression ol~the (_iF~f-DASpp, as an
indicator cell
line, a screen of various ligands is performed using a series of steroid type
ligands
including, but riot limited to: estrogen, hro~~,est~rone. retinoids, growrth
factors,
2~ androgens, and many other stcu~oid and steroid ba,.sed »~c~lecules. Image
acquisition and
analysis are performed usini; the cell screening system of the invention. The
intracellular GFP-DASpp flu~::~rcvscTence signal Is CC~Ile:cted by acquiring
fluorescence
image pairs (CII~P-Dr'~Spp ancj llc:5echst 33342~1abeled nuclei) from fields
cells. The
image pairs obtained at each time: point aruae:d tc> c etine nuclear and
cytoplasmic
6~1
CA 02410688 2002-12-12
regions in each cell. Translocation of GFP-I)ASpp is calculated by dividing
the
integrated fluorescence intensity- 01 GFP-DASpp in the nucleus by the
integrated
fluorescence intensity of the chimera in the cynoplasm or as a nuclear-
cy~topiasmic
difference of GFP fluorescence. ~'~ translocation fTOrrr the cytoplasm into
the nucleus
indicates a ligand binding activ;anon of the DASpp thus identifying the
potential
receptor class and action. C'om'bining this data with other data obtained in a
similar
fashion using known inhibitors a.nci modifiers of steroid receptors, would
either validate
the DASpp as a target, or more iiata would be generated from various sources.
E.rample 9 .9dditivnal .fcf~ecet.s
Tran.slocation between the plamrtca mernbrano and the <~t.~t<ylc~.s~n~:
Profilactin complex dissociation and binding of profilin to the plasma
rs membrane. In one embodiment, a fluorescent prc7tein hiosensor of profilin
membrane
binding is prepared by Dabelin~~ Ivurified profilin (Federov et al.( 1 r)94),
J. Moles. Biol.
241:480-482; Lanbrechts et ai ; 1995), F.ur. .1. l~iaahenr. 2~i0:?81-?86} with
a probe
possessing a fluorescence lifetims~ iti the range of ?--aC)tl ns. The labeled
profilin is
introduced into living indicatoe~ cells using bulk leaading methodology and
the indicator
2o cells are treated with test cornp«unds. Fluorescence :rrcisotropy imaging
microscopy
(cough and Taylor ( L x)93), .l. c: a.7ll f3ioL 1 ~! 1 :1095-1 10 -') is used
to measure test-
compound dependent moveno.a~t of the fluorescent derivative of protilin
between the
cytoplasm and membrane for a taeriod of time after treatment ranging from O.I
s to 10
h.
zs Rho-RhoGDI comple:~ translocation to the membrane. In another
embodiment, indicator cells are treated with test compounds and then fixed,
washed,
CA 02410688 2002-12-12
and permeabilized. The indicator c~evll plasma rnembrant,, cytoplasm, and
nucleus are
ail labeled with distinctly colored rnarlcers followed by immunolocali2ation
of Rho
protein (Self ca al. ,1995), A~eti~u~is i» Enzymolo~v 256:3-lU; Tanaka et al.
(1995),
Methods in Enzymologv 25b:41-=19) with antibodies labeled with a fourth color.
Each
s of the four labels is imaged separately using the cell screening system, and
the images
used to calculate the amount of inhibition or activation o~ translocation
effected by the
test compound. To do this caculation, the imaes of the probes used to mark the
plasma membrane and cytoplasm are used to mask the image of the immunological
probe marking the location of intracellular Rho protean. 7 he integrated
brightness per
to unit area under each mask is .~s~~c:l tea form a tr~Lnsloeation quotient by
dividing the
plasma membrane integrated bric;htness~'area by tl7e cytoplasmic integrated
brightness/area. By comparing the. translocation quotient values from control
and
experimental wells, the percent translocation i_s calculated for each
potential lead
compound.
is ~3-Arrestin translocatio» to tire plciarno mernhrcu»e upon (r-protein
receptor activation.
In another embodiment of a cytoplasm to membrane translocation high-content
screen, the translocation of ~i-arrestin protein from the cytoplasm to the
plasma
membrane is measured in response to cell treatment. 'l ~r measure the
translocation,
20 living indicator cells containint, luminescent vlomain markers are treated
mith test
compounds and the movemer_t c>t~ the (3-arre~stin marker is measured in time
and space
using the cell screening s-vsteon c>r the present irtventitm. In a preferred
embodiment,
the indicator cells contain lunuinescent markers consisting of a green
fluorescent protein
(3-arrestin (GFl'-~3-arrestin) protein chimera (~3arak et al. 1,19 97), J.
Biol. Chern.
2s 272:27497-275!70; Daaka et 3.1. ) 1998), J. Btc~l. c'~yenn '73:685-d88)
that is expressed
(a c,
CA 02410688 2002-12-12
by the indicator cells through th<_ use o1~ transient oa- stable cell
transfection and other
reporters used to mark cytoplasn~ic ;u~d membrane dort7aios. Vfhen the
indicator cells
are in the resting state, tl.e dorr~ain marker molecules partition
predominately in the
plasma membrane or in the i;ytcrplasm. In the high-content screen, these
markers are
s used to delineate the cell cytolvlasm and plasma membrane in distinct
channels of
fluorescence. When the indicat.~r cells are treated with a test compound, the
dynamic
redistribution of the GFP-~i-arrestin is recorded as a series of images over a
time scale
ranging from 0.1 s to li> h. In a preferred embodiment, the time scale is 1 h.
Each
image is analyzed by a metho:l float quantifies the movement of the GFP-~3-
arrestin
to protein chimera between the plasma membrane and the cytoplasm. To do this
calculation, the images of the probes used to mark the plasma membrane and
cytoplasm
are used to mask the image of the GFP-~3-arrestin probe marking the location
of
intracellular GFP-~-arrestin prc~teir~. 'l~he integrated E>rightness per unit
area under each
mask is used to foml a translocation quotient by dmiding the plasma membrane
t5 integrated brightnessiarea by tl:.e cytoplasmic inte~.~;rat~~ct
brightnesslarea. By comparing
the translocation quotient values from control arnd experimental wells, the
percent
translocation is calculated for each potential lead compound. The output of
the high-
content screen relates quantita.tiw~ data describing ttte ma~;nit2~d~ of the
translocation
within a large number of individual cells that hay ~: bf:en treated with test
compounds of
2o interest.
Translocation tretrl~een the em,io,,nlasmic reticulunr arre~ the holgi
In one embodiment ~:~f an endoplasmic reticulurn to (Jolgi translocation high-
content screen, the translocation of a VW'G protein from the ts045 mutant
strain of
vesicular stomatitis virus (Ellenbc°rg et al. ( i ~19~; j, .,r. t oTl
Biol. 13f;:1193-1206; Presley
CA 02410688 2002-12-12
et al, (1997) Nature 389:81-85) frorrr the endoplasmic retuculum to the Golgi
domain is
measured in response to cell tre,rtrnent. To nn_asrjre ~:he translucation,
indicator cells
containing luminescent reporters are treated with test compounds and the
movement of
the reporters is measured in spare and time using the cell screening system of
the
present invention. The indicatc>r cells contain luminescent reporters
consisting of a
GFP-VSVG protein chimera that is expressed by the indicator cell through the
use of
transient or stable cell transfec:tiort and other domain markers used to
measure the
localization of the endoplasmic reticulum and Golg,i domains. When the
indicator cells
are in their resting state at 4U'(', the (~FP-VS~'t~; protein chimera
molecules are
partitioned predominately in ilne cndoplasmic reticulurn. In this high-content
screen,
domain markers of distinct color's used to delineate the endoplasmic reticulum
and the
Golgi domains in distinct channels c3f fluorescence V%hen the indicator cells
are treated
with a test compound and thc: temperature is .=~imultanoously lowered to
3~'''C, the
dynamic redistribution of the (:iFP-VSVG protein <;himera is recorded as a
series of
to images over a time scale ranching from 0,1 s to 10 h. Each image is
analysed by a
method that quantifies the mcw~c:nrent of the CaFP-V:~'v G protein chimera
between the
endoplasmic retrculum and the ~Tolgi domains. fc dc> this calculation, the
images of
the probes used to mark the enclaplasmic reticu'um and the Ciolgi domains are
used to
mask the image of the GFP-'~'f>~-G prohe rrrark~ng the location of
intracellular GFP-
2o VSVG protein. The integrated brightness her unit area under each mask is
used to form
a translocation quotient by dividing the: endoplasmic reticulum integrated
brightnessiarea by the Golgi integrated brightnessiarea By comparing the
translocation
quotient values from contr~::~i ~md c~xperirnental wells, the percent
translocation is
calculated for each potential land compound. The output of the high-content
screen
c,
CA 02410688 2002-12-12
relates quantitative data describirng tl~e magnitude c~f the translocation
within a large
number of individual cells that h,a~~r; been treated with test compounds of
interest at
final concentrations ranging fronu 10 '" M to 10 ; M for a period ranging from
1 min to
h.
Induction and inhibition of organellar function:
Intracellular microtubol~e stabilitw. In one embodiment of an organellar
function high-content screen, the assembly state of i~rtracellular
microtubules is
1o measured in response to cell treatment. T~> measure microtubule assembly
state,
indicator cells containing luminescent reporters are treated with test
compounds and the
distribution of the reporters is rrrc:asured in space and tin-re using the
cell screening
system of the present invention.
In a preferred embodin-cent, the reporter of inirtrcellular nricrotubule
assembly is
i> MAP 4 (Bulinski et al. (l'~)~~~')~ J. Cell Scimcn 110::3455-30<=Ij, a
ubiqtutous
microtubule associated protein tlaa.t is known to interact with microtubules
in interphase
and mitotic cells. Tho indic;itur cells contain luminescent reporters
consistinv~ of a
GFP-MAP 4 chimera that is e~,;pr~:ssed by the incficator cells through the use
of transient
or stable cell transfection and other reporters are used to measure the
localization of the
2o cytoplasmic and membrane ~~c:~noponents. :'~ t rFf~~tAP 4 construct is
prepared as
follows: PCR amplification of native or mutant (pFP molecules using primers to
introduce restriction enzyme sites is performed. The PCR product is ligated
into the
MAP 4 cDNA within a c:uf;arw>tic expression vector. Indicator cells are then
a~>
CA 02410688 2002-12-12
transfected with the expression v~;ec~or to produce esther transiently or
stably transfected
indicator cells.
Lndieator cells are treate~;i ~,~~ith test compounds at final concentrations
ranging
from IO~iz M to 10-; M for a loeriod ranging from 1 min to 10 h. Growth medium
a containing labeling reagent to rvark the nucleus and the cytoplasm are
added. After
incubation, the cells are washed with Hank's balani;ed salt solution (HBSS),
fixed with
3.7% formaldehyde for 10 min ,:rt ~~oom temperature, and washed and stored in
HBSS.
Image data are obtained fi-orn both fixed and living indicator cells. To
extract
morphometric data from each cyf the images obtained thc: following method of
analysis
i0 is used:
1. Threshold each nucleus acrd cytoplasmic image to product a mask that has
value
= 0 for each pixel outside a nucleus or cell boundary
2. Overlay the mask on h~: original image, detect each object in the field
(i.e.,
nucleus or cell), and calculate its size, shape, and integrated intensity.
1 a 3. Overlay the whole cell rrarsk obtained above on the corresponding GFP-
MAP 4
image and use an automa~.e~i measurement o(~ edge strength routine (Kolega et
al.
(1993). Biolrnagin~,y 1:13h-1 ~t)1 to calculate the total edge strength within
each cell.
To normalize for cell size, tlic total edge streo~;th is ~tivided by the cell
area to ~uive
a "fibrousnc:ss" v;rlue. I ar-~w i'ibrousness . aluf:~ arc associated with
strong edge
?o strength values and are tl~~~refor~ maximal ire cc°Lls f~ontaining
distinct microtubule
structures. Likewise, srn~ll fihrousness ~' clues are associated with weal';
edge
strength and are minimal in cells ~sitir depol~rnerized rnicrotubules. The
physiological range of fihorursness valu~:,s is set by treating cells with
either the
CA 02410688 2002-12-12
microtubule stabilizing drug pavlitarel (1U yM) or the microtubule
depolymerizing
drug nocodazole ( 1U pg,~ml).
High-content screens involving t~h~ functional localisation o~~mucromoleeules
Within this class of hif~h-content screen, the functional localization of
macromolecules in response to extor~~al Stimuli is measured within living
cells.
Glycolytic enzyme activity regulation, In a preferred embodiment of a
cellular enzyme activity high-c~ontc.nt screen, the activity of key glycolytic
regulatory
enzymes are measured in treated cells. To measure enzyme activity, indicator
cells
'. o containing luminescent labeling reagents are treated with test compounds
and the
activity of the reporters is measured in space and time using cell screening
system of
the present invention.
In one embodinnent, th~:weporter of intracellular enzyme activity is fructose-
6-
phosphate, 2-kinase/fn~ctose-2,6--bisphosphatase (I'FK-~'), a regulator's
enzyme whose
15 phosphorylation state indicates intracellular carhohydrate anabolism or
catabolism
(Deprez et al. ( 1997) J Bio~. l 'hcerrr. 27?: l ?2(~t~-l r 2~'S; Kealer et
al. ( 1996) hEBS
Letters 395:225-227; 1-ee et al. t; t 996), l3iochernist;ri' 3s:6U l U-6019).
The indicator
cells contain luminescent relyorters consistin~~ ~~f .a t~uorescerV protein
biosensor of
PFK-2 phosphorylation. 'l he fluorescent protein biosensor is constructed by
zo introducing an environmentall~~ _~cnsitme fli~oresc;ent dye near to the
known
phosphorylation site of the enzyX~~ l l:)eprez et al ( 19~)~), .supra;
~iutiano et al. (1995),
supra). The dye can be of the ketocyanine class ( Kessler and VVolfbeis ( 1991
),
Spectrochimica ..dicta 47A:18'T-1 ~)2 ) car any class that contains a protein
reactive moiety
and a tluorochrome whose excitation or emission spectrum is sensitive to
solution
CA 02410688 2002-12-12
polarity. The fluorescent protein biosensor is introduced into the indicator
cells using
bulk loading methodology.
Living indicator cells are treated with test canipounds, at final
concentrations
ranging from 10-'2 M to 10'' M fc?r times ranging from u.1 s to 10 h. In a
preferred
embodiment, ratio image data are obtained from licking treated indicator cells
by
collecting a spectral pair of f'.uorescence images at each time point. To
extract
morphometric data from each time point, a ratio is made between each pair of
images
by numerically dividing, the tuba spectral images at each time point, pixel by
pixel.
Each pixel value is then used to calculate the fractional phasphorylation of
PFK-2. At
tt~ small fractional values of~ phosl~horvlation, PFK-~ stimulates
carbohydrate catabolism.
At high fractional values c~f phosphorylatio~~, PFK-7 stimulates carbohydrate
anabolism.
Protein lcinase A ac ivity and Iocalrlation of subunits. In another
1 > embodiment of a high-content :,cretin, bath the datt2ain localization and
activity of
protein kinase A (PKA) within indicator cells are measured in response to
treatment
with test compounds.
The indicator eclls contain lun~inesceot rE°l>orters including a
fluorescent protein
biosensor of PKA activation. 1'he fluorcaeent protein biasensor is
constructf:d by
introducing an environmentally ~~nstive fluorescent ~3y~~ into the catalytic
subunit of
PKA near the site known to !ncertict with th. re4.~ulatory subunit of PKA
(Harootunian
et al. ( 1993), AToI. Biol. of thc- ('ell 4:993-100: Ji7hnson et al. ( 1996),
Cell 85:149-158;
Giuliano et al. ( 1995), supru). -1 he dye can be ~.nf the ketocyanine class
(Kessler, and
Wolfbeis (1991), Sp~~rtroeh:mic~a :lcta 47.~:1~7-1~)2) or any class that
contains a
,.
CA 02410688 2002-12-12
protein reactive moiety and a flu.orochrome whose excitation or emission
spectrum is
sensitive to solution polarity. Thc, tluoresccnt protein l~iosc;nsor of PKA
activation is
introduced into the indicator cell: using bulk loading methodology.
In one embodiment, livirug indicator cells are treated with test compounds, at
final concentrations ranging from l()'' M to 10' 1~9 for times ranging from
0.1 s to 10
h. In a preferred embodiment, ratio image data are obtained from living
treated
indicator cells. To extract bioseT~isc>r data from each time point, a ratio is
made between
each pair of images, and each l~i:~el value is then used to calculate the
fractional
activation of PKA (e.g., separation of the catalytic and regulatory subunits
after cAMP
1o binding). At high fractional valuca i>f activity, f'Fk-2 stimulates
biochemical cascades
within the living cell.
To measure the transloiaiion 4rf the catalytic. subunit of PKA, indicator
cells
containing luminescent reporters ~rre treated with test compounds and the
movement of
the reporters is measured in spare and time using the cell screening system.
The
indicator cells contain iumine:,cent reporters consisting of domain markers
used to
measure the localization of the cs.~toplasmic acrd nuclear domains. When the
indicator
cells are treated with a test ~.uympounds, tire dynamic redistribution of a
PKA
fluorescent protein biosensor i:s rvcoc-ded intracellularly as ~t series of
images over a
time scale ranging from 0.1 s to 1(t h. Eac-h irnat?;e is analyzed by a method
that
2o quantifies the movement of the PK.~~ between the cytoplasmic and nuclear
domains. To
do this calculation, the images o1~ the probes used to mark the cynoplasmic
and nuclear
domains are used to rr~ask the i~oag~, of the PKA fluorescent protein
biosensor. The
integrated brightness per unit area under each mask is used to torn a
translocation
quotient by dividing the c ~,~toplasmic integrated bri;~htness~=area by the
nuclear
.,
.,
CA 02410688 2002-12-12
integrated brightness/area. By comparing the translocation quotient values
from
control and experimental wells, the l:rercent translocation is G;rlculated for
each potential
lead compound. ~:'hc <output of the high-content screen relates yuantitative
data
describing the magnitude of the translocation within a large number of
individual cells
that have been treated with test compound in the concentratian range of l~-''
M to 103
M.
Nigh-content scrc.~ens involving !hE~ induction or inlubitron cy~gene
erpression
RNA-based flttorcascent hic~s~nsors
to Cytoskeletal protein trarrscriptiorr and message localization. Regulation
of
the general classes of cell physiological responses including cell-substrate
adhesion,
cell-cell adhesion, signal transd~r<:tion, cell-cyl~~ events, intermediary and
signaling
molecule metabolism, cell locornoticm, cell-fi~cll corumunication, and cell
death can
involve the alteration of gene e:~pression. High-cinrrteut Screens can also be
designed to
f , measure this class of physio(o~ictrl re>ponse.
In one embodiment, tl'ne reporter cat intracellular gene expression is an
oligonucleotide that cart hybridize with the tar~~tt rnl2NA and alter its
fluorescence
signal. In a preferred embodivment, the oiigunucleotide is a molecular beacon
(Tyagi
and Kramer ( 1906) ,'Vut. Brote~<~In~r~~:~l. 14:3()3- >t?~ 1, a
itirt~inescence-based reagent whose
?o fluorescence signal is depene?ent on intermolecular an,l intramolecular
interactions.
The fluorescent biosensor is c orastructed by introelucin~ a fluorescence
energy transfer
pair of fluorescent dyes such that there is one at each end (S' and 3') of the
reagent.
The dyes can be of-any class that cornains a protein reactive moiety and
fluorochromes
whose excitation and erniss~on ~sp4ctra ovcrlah sufficiently to provide
fluorescence
CA 02410688 2002-12-12
energy tra~tsfer between the dytas in the resting state. including, but not
limited to,
fluorescein and rhodamine (Molecular Probes, lnc ). In a preferred embodiment,
a
portion of the message :;oding for ~3-actin (Kislauskis et al.. (1994), J.
Cell Biol.
127:441-451; McC'ann et al, ( 1997), Proc. ~'Vcrti' Acu~. Sci. 94:5679-5684;
Sutoh
(1982), Biochemistry 21:3654-3f:.61 ) is inserted into the. loop region of a
hairpin-shaped
oligonucleotide with the ends tethered together due to intrarnolecular
hybridization. At
each end of the biosensor a flui>rescence donor (fluorescein) and a
fluorescence
acceptor (rhodamine) are covaletrtly bound. In the tethered state, the
fluorescence
energy transfer is maximal and therefore indicative of an unhybridized
molecule.
t~~ When hybridized with the r~rR1'~d~~ coding for ~-actin, the other is
broken and energy
transfer is lost. The complete fluorescent biosensor is introduced into the
indicator
cells using bulk loading methodolcyy.
In one embodiment, living indicator cells are treated with test compounds, at
ftnal concentrations ranging from 10~~' M to 117' "~~1 far tunes ranging from
0.1 s to 10
l 5 h. In a preferred embodiment, ratio image data are obtained from living
treated
indicator cells. ~I~o extract mcrrphometric data from each time point, a ratio
is made
between each pair of images, anif each pixel aaiue is then used to calculate
the
fractional hybridization of the: labeled nucleotide. .At small fractional
values of
hybridization little expression c:~f (3-actin is indicated. ~t hid=.h
fractional values of
?o hybridization, maximal expres~;sion of G3-actin ns iruclicated.
Furthermore, the distribution
of hybridized molecules within the cytoplasm c~f~the indicator cells is also a
measure of
the physiological response of the indicator cells.
Cell surface binding of a likancl
2>
7:
CA 02410688 2002-12-12
Labeled insulin binding to its cell surface receptor in living cells. Cells
whose plasma membrane dom;:~irr has been labeled wOh a labeling reagent of a
particular color are incubated with a solution contarning insulin molecules
(Lee et al.
(1997), Biochemistn~ 36:2701-2a'0~, Martinet-Zaguilan et al. (1996), Am. J.
Physlol.
s 270:C1438-C144fi) that are labeled. with a luminescent probe of a different
color for an
appropriate time under the appropriate conditions. After incubation, unbound
insulin
molecules are washed away, the ,.ells fixed and the distribution and
concentration of the
insulin on the plasma membranc.~ is measured. '1'o do this, the cell membrane
image is
used as a mask for the insulin in-age. 'The integrated intensity from the
masked insulin
tt) image is compared to a set of images containing known ,:amounts of labeled
insulin.
The amount of insulin bound tc~ the cell is deterzriined from the standards
and used in
conjunction with the tote! concentration of insulin incubated with the cell to
calculate a
dissociation constant or insulin to its cell surface rcc:eptor
1 s Labeling of cellukrr com~artmc~rrt,s
Whole cell labeling
Whole cell labeling is accomplished by labeling cellular components such that
dynamics of cell shape and molilit~ of the cell earl be measured over time by
analyzing
fluorescence images of cells.
>o In one embodiment, sunall resrctive: fluorLseent molecules are introduced
into
living cells. These membrane-pi>erroeant molecules both diffuse through and
react with
protein components in the pla:~noa membrane. 1.)ye molecules react with
intracellular
molecules to both increase the fluorescence signal emitted from each molecule
and to
entrap the fluorescent dve w ittnrn living cells 7,h~ae rnoleeules include
reactive
;6
CA 02410688 2002-12-12
chloromethyl derivatives of arninocoumarins, hydroxycoumarins, eosin
diacetate,
fluorescein diacetate, some l3odipy dye derivatives, and tetramethylrhodamine.
The
reactivity of these dyes toward macromolecules includes free primary amino
groups
and free sulfhydryl groups.
In another embodiment, the cell surface is labeled by allowing the cell to
interact with fluorescently labeled antibodies or lectins (Sigma Chemical
Company, St.
Louis, MO) that react specifically with molecules on the cell surface. Cell
surface
protein chimeras expressed by flue cell of intereat that contain a green
fluorescent
protein, or mutant thereof, component can also be used to tluorescently label
the entire
W cell surface. Once the entire cell is labeled, images of I:he entire cell or
cell array can
become a parameter in high content screens, involving the measurement of cell
shape,
motility, size, and growth and division.
Plasma membrane iabceling
to In one embodiment, labnlinthe whole plasma membrane employs some of the
same methodolo;y described albove for labclntg the entire cells. Luminescent
molecules that label the entire c:11 surface act 1o delineate tire plasnna
membrane.
In a second embodiment subdomains oi~ the plasma membrane, the extracellular
surface, the lipid bilayc:r, and ~:l~e sntracellular sr~rfac;e can bc; labeled
separately and
'0 used as components of high content screens. In floe first cmbodirnent, the
extracellular
surface is labeled using a brief treatment with a reactive fluorescent
molecule such as
the succinimidyl ester or iod~:~ac;etamde clcnvatives of fluorescent dyes such
as the
fluoresceins, rhodamines, cyanincs, and Bodipys.
~i
CA 02410688 2002-12-12
In a third embodiment, the: c:xtracellular surface is labeled using
fluorescently
labeled macromolecules with a hii;~h ailinity for cell surface molecules.
These include
fluorescently labeled Iectins such as the fluorcscein, rhodamine, and cyanine
derivatives of lectins derived fiom jack boar (Con A), red kidney bean
(erythroagglutinin PHA-E), or wheat germ.
In a fourth embodiment, fluarescently labeled antibodies with a high affinity
for
cell surface components are used to Label the extracellular region of the
plasma
membrane. Extracellular regions of cell surface receptors and ion channels are
examples of proteins that can be labeled with antibodies.
In a fifth embodiment, the lipid bilayer of the plasma membrane is labeled
with
fluorescent molecules. 'I'hese molecules include fluorescent dyes attached to
long chain
hydrophobic molecules that interact strongly with the: hydrophobic region in
the center
of the plasma membrane Lipid bilayer. Examples of these dyes include the PKH
series
of dyes (U.S. Patents Nos. 4,7;E3:i,4t~l issued November 8, 1988; 4,762,701
issued
August 9, 1988; and 4,8'.79,584 us~ued .August 22, 1989; available
commercially from
Sigma Chemical Company, SI. Louis, MO), fluorescent phospholipids such as
nitrobenzoxadiazole glycerol:>hosplioethanolarn in L and fluorscein-
derivatized
dihexadecanoylglycerophospha<;tha-nolamine, flui>resc:ent fatty acids such as
5-butyl-
4,4-difluoro-4-bora-3a,4a-diaza-s-indacene-3-nonamoic acid and 1-
pyrenedecanoic acid
(Molecular Probes, Inc.), fluorescent sterols including cholesteryl 4,4-
difluoro-5,7-
dimethyl-4-bora-3a,4a-diaza-s-indacene-3-dodecanoate and cholesteryl 1-
pyrenehexanoate, and fluorescently labeled proteins that interact specifically
with lipid
bilayer components such as the fluorescein derivative of annexin V (Caltag
Antibody
Co, Burlingame, f:A).
-rs
CA 02410688 2002-12-12
In another embodiment, tine intracellular component of the plasma membrane is
labeled with fluorescent molecul~a. Examples of these molecules are the
intracellular
components of the trimeric (r-protein receptor, adenylyl cyclase, and ionic
transport
proteins. These molecules can bc; Labeled as a result of tight binding to a
fluorescently
S labeled specific antibody or by thf: incorporation of a fluorescent protein
chimera that is
comprised of a membrane-associated protein and the green fluorescent protein,
and
mutants thereof.
Endosome fluorescence labeling
1o In one embodiment, ligands that are transported into cells by receptor-
mediated
endocytosis are used to trace the dynamics of endosornal organelles. Examples
of
labeled ligands include Bodily FL-labeled lo~wv density lipoprotein complexes,
tetramethylrhodamine transferrin analogs, and lluorescently labeled epidermal
growth
factor (Molecular Probes, lne.)
t~ In a second embodiment, Iluorescently labeled primary or secondary
antibodies
(Sigma Chemical C o. St. Lou:s, !~i0; Molecular Probes, Inc. Eugene, OR;
Caltag
Antibody Co.) that specifically lab~:l endosomal ligands are used to mark the
endosomal compartment in cells
In a third embodiment, ~;n~Iosomes are fluoresc:ently labeled in cells
expressing
2~a protein chimeras formed 'by fus~n~~ a green fluorescent protein, or
mutants thereof, with
a receptor whose internalization labels endosome~~ C'Iiimeras of the EGF,
transferrin,
and low density lipoprotein receptors are examples of these molecules.
c~
CA 02410688 2002-12-12
Lysosome labeling
In one embodirrrent, membrane permeant lysosome-specific luminescent
reagents are used to label the lysosonral compartment of living and fixed
cells. These
reagents include the luminescent molecules neutral red, N-(3-((2,4-
dinitrophenyl)amino)propyl)-N-{?~-trrninopropyl)methylamine, and the
LysoTracker
probes which report intralysos~on 7a1 pII as well as the dynamic distr7bution
of
lysosomes (Molecular Probes, Irn::. )
In a second embodimfvnt, antibodies against lysosomal antigens (Sigma
1o Chemical Co.; Molecular Prorr.es, lm.; C'altag "~ntil>ody t-'o.) are used
to label
lysosomal components drat are localized in specific ly:;osomal domains.
Examples of
these components are the degrad,:rtcve enzymes inv<Oved in cholesterol ester
hydrolysis,
membrane protein proteases, anti nucleases as well as the A fI'-driven
lysosomal proton
pump.
In a third embodiment, prcvtein chimer,:ts consisting of a lysosomal protein
genetically fused to an intrinsically luminescent protein such as the green
fluorescent
protein, or mutants thereof, are ns<.cl to label the lysoscan-ral domain.
Examples of these
components are the degradative enzv~mes in volvt°d in cholesterol ester
hydrolysis,
membrane protein prote,.rses. arn~;i ocizcleases as well as t'nc: !~-I-P-
driven iysosomal proton
2o pump.
Cytoplasmic fluarescer~~ck~ labeling
In one embodiment, cell pern~eant fluorescent dyes (Molecular Probes, Ine.)
with a reactive group are reacr.c~d with living cf:ll5. Reactrve dyes
including
'; CI
CA 02410688 2002-12-12
monobrornobimane, S-chloromethylfluorescein diacetate, carboxy fluorescein
diacetate
succinimidyl ester, and chloroi°n~~tln~~l tetramethylrhodarrnne are
examples of cell
permeant fluorescent dyes that art: used for long term labeling of the
cytoplasm of cells.
In a second embodiment, polar tracer molecules such as Lucifer yellow and
cascade blue-based fluorescent dyes (Molecular Probes, Inc.) are introduced
into cells
using bulk loading methods and Ure also used for cytoplrsmic labeling.
In a third embodiment, antibodies against cytoplasmic components (Sigma
Chemical Co.; Molecular Probes., lnc.; ~'altag Antibody C~o.) are used to
fluorescently
label the cytoplasm. Example<.: ~>f cytoplasmic antigens are rrrany of the
enzymes
~C~ involved in intermediary metabc;~lisrn. Enolase. phosphofi-uctokinase, and
acetyl-CoA
dehydrogenase are examples of uniformly distributed cytoplasmic antigens.
In a fourth embodiment, protein chimeras consisting of a cytoplasmic protein
genetically fused to an intrinsicaally luminescent protein such as the green
fluorescent
protein, or mutants thereof, are a sed to label the' .: ~,~toplasm.
Fluorescent chimeras of
t ~ uniformly distributed proteins arc: used to label the entire cytopiasmie
domain.
Examples of these proteins ~:rr roany of the proteins involved in intermediary
metabolism and include enolase, la:~<aat~ dehydr~7ge:nas~~, and hexokinase.
In a fifth emrrodimer~u, antibodies agarzns~ cytoplasmic antigens (Sigrrra
Chemical Co.; Molecular E'rev°.sc::s, lnc.; Calta'= .4ntihodw Co.) are
used to label
<.0 cytoplasmic components that are localized in specific cytoplasmic sub-
domains.
Examples of these componer7ts are the cytoskelei.al proteins actin, tubulin,
and
cytokeratin. A population of there proteins ~~ithin cells is assembled into
discrete
structures, which in this case, are fibrous. Fluorescence labeling of these
proteins with
antibody-based reagents therefc~r~ labels a specific: sub-domain of the
cytoplasm.
hl
CA 02410688 2002-12-12
In a sixth embodiment, non-,~ruibody-based iluorescently labeled molecules
that
interact strongly with cytoplasm~ic: proteins are toned to label specific
cytoplasmic
components. (Une example is a t'luorescent analag of the enzyme DNAse 1
(Molecular
Probes, Inc.) Fluc:rrescent analv:rgs of this en;~ynte bind tightly and
specifically to
s cytoplasmic actin, thus labeling a st b-domain of~ the cytoplasm. In another
example,
fluorescent analogs of the mushroom toxin phalloidin or the drug paclitaxel
(Molecular
Probes, Inc.) are used to label comp:>nents of the acain- and microtubule-
cytoskeletons,
respectively.
In a seventh embodiment, prc.,tein chimeras oansisting of a cytoplasmic
protein
to genetically fused to an ir"ttrirrsic:.~lly lurtzinESCerot protein such as
the green fluorescent
protein, or mutants thereof, ar. used to label specific domains of the
cytoplasm.
Fluorescent chimeras of highly localized proteins are used to label
cytoplasmic sub-
domains. Examples of these pr;~teiras sire many of the prcsteins involved in
regulating
the cytoskeleton. 'They include tha stnzctural protc;ins actin, tubulin, and
cytokeratin as
t vs well as the regulatory proteins nticrotubule associated l7rotein 4 and a-
actinin.
Nuclear labeling
In one embodiment, nnembrane pern~eant nucleic-acid-specific luminescent
reagents (Molecular Probes, Inc.) arv used to label ~hr nucleus of living and
fixed cells.
2o These reagents iroclude cyanint:-h~:tsed dyes (e g . TOTOr-. ~'C)YO~'. and
BOBOT"),
phenanthidines and acridines Ia:'.r-., ethidium hror-nide, pr~.~pidiurrt
iodide, and acridine
orange), indoles and irnidazole; (e_~., Hoechst y~t253. E-loechst 33342, and
4',6-
diamidino-2-phcnylindole), an~a ~,otlter similar re;.yents (t~.,sr., 7-
aminoactinomycin D,
hydroxystilbanudine, and the psaral~ns).
h-,
CA 02410688 2002-12-12
In a second embodiment, antibodies against nuclear antigens (Sigma Chemical
Co.; Molecular Probes, Inc.; ( 'a9tag Antibody ~:o.) are used to label nuclear
components that are localized in specific nuclear domains. Examples of these
components are the macromolecules involved in maintaining I)NA structure and
function. DNA, ItNA, histone;n, DNA polymor~ase, RNA polymerise, lamins, and
nuclear variants of cytoplasmic proteins such as actin are examples of nuclear
antigens.
In a third embodiment, protein chimeras consisting of a nuclear protein
genetically fused to an intrinsically luminescent protein such as the green
fluorescent
protein, or mutants thereof, are used to label the nuclear domain. Examples of
these
proteins are many of the proteins involved in maintaining DNA structure and
function.
Histones, DNA holymerase, I2.NA polymerise, lamins, and nuclear variants of
cytoplasmic proteins such as actin are examples of nuclear proteins.
Mitochondria) labeling
In one embodiment, mc;mbrane penneant mitochondria)-specif c luminescent
reagents (Molecular Probes, Inc.) .are used to label the mitochondria of
living and fixed
cells. These reagents include rl~adamine 123, tetralncthyl rosamine, JC-I, and
the
MitoTracker eactive dyes.
In a second embodiment, antibodies against mitochondria) antigens (Sigma
Chemical Co.; Molecular Prr>bes, Inc.; C'a'itag Antibody Co.) are used to
label
mitochondria) components tl~.at are localized in specific mitochondria)
domains.
Examples of these component:o are the macromolec:ulcs involved in maintaining
mitochondriat DNA structure and function. DN.A, ItNA, histones, I)NA
polymerise,
RNA polymerise, and mitochandrial variants of ~ytoplasmic macromolecules such
as
83
CA 02410688 2002-12-12
mitochondria) tRNA and rRNA vre examples mitochondria) antigens. Other
examples
of mitochondria) antigens are the cc>roponents o1 the oxiciativc:
phosphorylation system
found in the mitoehondr-ia (e.g., c~.nochxome c, cyt«chrorne c oxidise, and
succinate
dehydrogenase).
In a third embodiment, protein chimeras consisting of a mitochondria) protein
genetically fused to an intrinsically luminescent protein such as the green
fluorescent
protein, or mutants thereof, are usk:<i to label the rr~itoc;hondrial domain.
Examples of
these components are the macromolecules involved in maintaining mitochondria)
DNA
structure and function. Examples include histories, DNA polymerise, RNA
to polymerise, and the components oi~ the oxidati~;~ phosphorylation system
found in the
mitochondria (e.K., cvtochrorne c, cytochrorne ~ oxidise, and succinate
dehydrogenase).
Endoplasmic reticulum iaheling
t_> In one ernbodirnent, membrane penoeant c:ndoplasmic reticulum-specific
luminescent reagents (Molecular Probes, Inc.) are used to label the
endoplasmic
reticulum of living and fixed ec-lls. These reagents include short chain
carbocyanine
dyes (e.~., DiOC,, and DiOC~), loro~! chain carbocyanirn. dyes (e.y , DiIC'»,
and DiIC~s),
and luminescently labeled leetins such as concanav alin :~.
2o In a second embodiment, antibodies <rgatnst ~~ndoplasmic reticulum antigens
(Sigma Chemical <.'o.; ~Molecul~r (robes, Inc.; (.'all.ae, :Antibody ('o.) are
used to label
endoplasmic reticulum compom:nts that are localised xn specific endopiasmic
reticulum
domains. Examples of these components are the macromolc;cules involved in the
fatty
acid elongation systems, glucose-t:~-phc>sphitase, and HMCz C:'uA-reductase.
CA 02410688 2002-12-12
In a third embodiment, pr~;~tc::in chimeras consisting c~f a endaplasmic
reticulum
protein genetically fused to an intrinsically lurrliztescent protein such as
the green
fluorescent protein, or mutants i.hc~r~eof, are used to Iabel the endoplasmic
reticulum
domain. Examples of these comz~onents are the macromolecules involved in the
fatty
acid elongation systems, glucose ~6-~phosphatase. and 1-(MG C'oA-roductase.
Golgi labeling
to In one embodiment, membrane permeant Ciolgi-specific luminescent reagents
(Molecular Probes, Inc.) are used to lahel the CTolgi ~~f living and fixed
cells. These
reagents include luminescently labeled macromolecules such as wheat germ
agglutinin
and Brefeldin A as well as luminescently labeled c;~ramidt~.
In a second embodiment, antibodies against t_iolgr antigens (Sigma Chemical
Co.; Molecular Probes, lnc.; Caitag Antibody <'o.) are' used to label Golgi
components
that are localized in specific Ciol,~,i di>mains. Examples of these components
are N-
acetylglucosamine phc.~sphotr~~nsferase, G~:olgi-specific phosphodiesterase,
and
mannose-6-phosphate receptor l~r~ntein,
In a third embodirne~U, protein ch~meE-~~s c~~nyisting of a Golgi protein
genetically fused to an intrinsically lutninescc:nt protein such as the green
fluorescent
protein, or mutants thereof arc used to label thc~ Goigi domain. Examples of
these
components are N-acetylgfucosamine ph osphotransferase, Golgi-specific
phosphodiesterase, and manno:.~:-~~~-phosphate rccehtor protein.
~5
CA 02410688 2002-12-12
While many of the exarnpies presented involve the measurement of single
cellular processes, this is again i,~ intended for purposes . f illustration
only. Multiple
parameter high-content screens c,~.rt ho produced by combining several single
parameter
screens into a multlparameter hia;h-e:ontent screen or b,y adding cellular
parameters to
s any existing high-content screen. Furthermore, while each example is
described as
being based on either live or fixers ~~~,lls, each high-content screen can be
designed to be
used with both live and fixed cel3s.
Those skilled in the art w ill recognize a wide variety of distinct screens
that can
be developed based on the discla~siarc provided herein. 'Chore is a large and
growing list
to of known biochen a seal acid rool~:~c,ilar processes in :ells that involve
translocations or
reorganizations of specific com~:~oiients within cells. -hhe signaling pathway
from the
cell surface to target sites w,itloir~ the cell involves the translocation of
plasma
membrane-associated proteins tc> the cytoplasm. lvor r~xample, it is known
that one of
the src family of protein tyrosine: kinases, ppti0c-src ( Walker et al (
1993), J. Biol.
to C'hem. ?68:19552-19558) translo~.:ate;s from the plasma tt~embrane to the
cytoplasm
upon stimulation of fibrobl;tsts with platelet-derived growth factor (PDGF)
Additionally, the targets for screening can themselves be converted into
fluorescence-
based reagents that report mo)c~ctilar changes including ligand-binding and
post-
trans(ocational modifications.
:'o
RG