Note: Descriptions are shown in the official language in which they were submitted.
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
ENCAPSULATION OF PLASMID DNA (LIPOGENEST~ AND
THERAPEUTIC AGENTS WITH NUCLEAR LOCALIZATION
SIGNAL/FUSOGENIC PEPTIDE CONJUGATES INTO TARGETED
LIPOSOME COMPLEXES
CROSS-REFERENCE TO RELATED APPLICATIONS
This application claims priority under 35 U.S.C. ~ 119(e) to U.S. Provisional
Application Serial No. 60/210,925 filed June 9, 2000. The contents of this
application is hereby incorporated by reference into the present disclosure.
FIELD OF THE INVENTION
The present invention relates to the field of gene therapy and is specifically
directed toward methods fox producing peptide-lipid-polynucleotide complexes
suitable for delivery of polynucleotides to a subject. The peptide-lipid-
polynucleotide complexes so produced are useful in a subject for inhibiting
the
progression of neoplastic disease.
BACKGROUND OF THE INVENTION
Throughout this application various publications, patents and published
patent specifications are referenced by author and date or by an identifying
patent
number. Full bibliographical citations for the publications are provided
invnediately
preceding the claims. The disclosures of these publications, patents and
published
patent specifications are hereby incorporated by reference into the present
disclosure
to more fully describe the state of the art to which this invention pertains.
Gene therapy is a newly emerging field of biomedical research that holds
great promise for the treatment of both acute and chronic diseases and has the
potential to bring a revolutionary era to molecular medicine. However, despite
numerous preclinical and clinical studies, routine use of gene therapy for the
treatment of human disease has not yet been perfected. It remains an important
unmet need of gene therapy to create gene delivery systems that effectively
target
specific cells of interest in a subject while controlling harmful side
effects.
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
Gene therapy is aimed at introducing therapeutically important genes into
somatic cells of patients. Diseases already shown to be amenable to therapy
with
gene transfer in clinical trials include, cancer (melanoma, breast, lymphoma,
head
and neck, ovarian, colon, prostate, brain, chronic myelogenous leukemia, non-
small
cell lung, lung adenocarcinoma, colorectal, neuroblastoma, glioma,
glioblastoma,
astrocytoma, and others), AIDS, cystic fibrosis, adenosine deaminase
deficiency,
cardiovascular diseases (restenosis, familial hypercholesterolemia, peripheral
artery
disease), Gaucher disease, al-antitrypsin deficiency, rheumatoid arthritis and
others.
Human diseases expected to be the object of clinical trials include hemophilia
A and
B, Parkinson's disease, ocular diseases, xeroderma pigmentosum, high blood
pressure, obesity. ADA deficiency was the disease successfully treated by the
first
human "gene transfer" experiment conducted by Kenneth Culver in 1990. See,
Culver, K.W. (1996) in: Gene Therapy: A Primer for Physicians, Second Ed.,
Mary
Ann Liebert, Inc. Publ, New York, pp. 1-198.
The primary goals of gene therapy are to repair or replace mutated genes,
regulate gene expression and signal transduction, manipulate the immune
system, or
target malignant and other cells fox destruction. See, Anderson, W.F. (1992)
Science
256:808-813; Lasic, D. (1997) in: Liposomes in Gene Delivery, CRC Press, pp. 1-
295; Boulikas, T. (1998) Gene Ther. Mol. Biol. 1:l-172; Martin, F. and
Boulikas, T.
(1998) Gene Ther. Mol. Biol. 1:173-214; Ross, G. et al. (1996) Hum. Gene Ther.
7:1781-1790.
Human cancer presents a particular disease condition for which effective
gene therapy methods would provide a particularly useful clinical benefit.
Gene
therapy concepts for treatment of such diseases include stimulation of immune
responses as well as manipulation of a variety of alternative cellular
functions that
affect the malignant phenotype. Although many human tumors are non or weakly
immunogenic, the immune system can be reinforced and instructed to eliminate
cancer cells after transduction of a patient's cells ex vivo with the cytokine
genes
GM-CSF, IL-12, IL-2, IL-4, IL-7, IFN-y, and TNF-a, followed by cell
vaccination of
the patient (e.g. intradermally) to potentiate T-lymphocyte-mediated antitumor
effects (cancer immunotherapy). DNA vaccination with genes encoding tumor
antigens and immunotherapy with synthetic tumor peptide vaccines are further
2
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
developments that are currently being tested. The genes used for cancer gene
therapy in human clinical trials include a number of tumor suppressor genes
(p53,
RB, BRCA1, ElA), antisense oncogenes (antisense c fos, c-nayc, K ras), and
suicide
genes (HSV-tk, in combination with ganciclovir, cytosine deaminase in
combination
with 5-fluorocytosine). Other important genes that have been proposed for
cancer
gene therapy include bcl-2, MDR-l, p21, p16, bax, bcl-xs, E2F, IGF-I, VEGF,
angiostatin, CFTR, LDL-R, TGF-(3, and leptin. One major hurdle preventing
successful implementation of these gene therapies is the difficulty of
efficiently
delivering an effective dose of polynucleotides to the site of the tumor.
Thus, gene
delivery systems with enhanced transfection capabilities would be highly
advantageous.
A number of different vector technologies and gene delivery methods have
been proposed and tested for delivering genes in vivo, including viral vectors
and
various nucleic acid encapsulation techniques. Alternative viral delivery
vehicles for
genes include murine retroviruses, recombinant adenoviral vectors, adeno-
associated
virus, HSV, EBV, HIV vectors, and baculovirus. Nonviral gene delivery methods
use cationic or neutral liposomes, direct injection of plasmid DNA, and
polymers.
Various strategies to enhance efficiency of gene transfer have been tested
such as
fusogenic peptides in combination with liposomes or polymers to enhance the
release of plasmid DNA from endosomes.
Each of the various gene delivery techniques has been found to possess
different strengths and weaknesses. Recombinant retroviruses stably integrate
into
the chromosome but require host DNA synthesis to insert. Adenoviruses can
infect
non-dividing cells but cause immune reactions leading to the elimination of
therapeutically transduced cells. Adeno-associated virus (AAV) is not
pathogenic
and does not elicit immune responses but new production strategies are
required to
obtain high AAV titers for preclinical and clinical studies. Wild-type AAVs
integrate into chromosome 19, whereas recombinant AAVs are deprived of site-
specific integration and may also persist episomally.
Herpes Simplex Virus (HSV) vectors can infect non-replicating cells, such as
neuronal cells, and has a high payload capacity for foreign DNA but inflict
cytotoxic
effects. It seems that each delivery system will be developed independently of
the
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
others and that each will demonstrate strengths and weaknesses for certain
applications. At present, retroviruses are most commonly used in human
clinical
trials, followed by adenoviruses, cationic liposomes and AAV.
As the challenges of perfecting gene therapy techniques have become
apparent, a variety of additional delivery systems have been proposed to
circumvent
the difficulties observed with standard technologies. For example, cell-based
gene
delivery using polymer-encapsulated syngeneic or allogeneic cells implanted
into a
tissue of a patient can be used to secrete therapeutic proteins. This method
is being
tested in trials for amyotrophic lateral sclerosis using the ciliary
neurotrophic factor
gene, and may be extended to Factor VIII and IX for hemophilia, interleukin
genes,
dopamine-secreting cells to treat Parkinson's disease, nerve growth factor for
Alzheimer's disease and other diseases. Other techniques under development
include, vectors with the Cre-LoxP recombinase system to rid transfected cells
of
undesirable viral DNA sequences, use of tissue-specific promoters to express a
gene
in a particular cell type, or use of ligands recognizing cell surface
molecules to direct
gene vehicles to a particular cell type.
Additional methods that have been proposed for improving the efficacy of
gene therapy technologies include designing p53 "gene bombs" that explode into
tumor cells, exploiting the HIV-1 virus to engineer vectors for gene transfer,
combining viruses with polymers or cationic lipids to improve gene transfer,
the
attachment of nuclear localization signal peptides to oligonucleotides to
direct genes
to nuclei, and the development of molecular switch systems allowing genes to
be
turned on or off at will. Nevertheless, because of the wide range of disease
conditions for which gene therapies are required, and the complexities of
developing
treatments for such diseases, there remains a need for improved techniques for
performing gene therapy. The present invention provides methods and
compositions
for addressing these issues.
DISCLOSURE OF THE INVENTION
A method is disclosed for encapsulating DNA and negatively charged drugs
into liposomes having a different lipid composition between their inner and
outer
membrane bilayers. The liposomes are able to reach primary tumors and their
4
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
metastases after intravenous injection to animals and humans. The method
includes
micelle formation between DNA with a mixture of cationic lipid and peptide
molecules at molar ratios to nearly neutralization ratios in 10-90% ethanol;
the
cationic peptides specify nuclear localization and have a hydrophobic moiety
endowed with membrane fusion to improve entrance across the cell membrane of
the
complex. These peptides insert with their cationic portion directed toward
condensed DNA and their hydrophobic chain buried together with the hydrophobic
chains of the lipids in the micelle membrane monolayer. The DNA/lipidlpeptide
micelles are converted into liposomes by mixing with pre-made liposomes or
lipids
followed by dilution in aqueous solutions and dialysis to remove the ethanol
and
allow liposome formation and extrusion through membranes to a diameter below
160
nm entrapping and encapsulating DNA with a very high yield. The encapsulated
DNA has a high therapeutic efficacy in eradicating a variety of solid human
tumors
including, but not limited to, breast carcinoma and prostate carcinoma. A
plasmid is
constructed with DNA carrying anticancer genes including, but not limited to
p53,
RB, BRCA1, ElA, bcl-2, MDR-1, p21, p16, bax, bcl-xs, E2F, IGF-I VEGF,
angiostatin, oncostatin, endostatin, GIVI-CSF, IL-12, IL-2, IL-4, IL-7, IFN-y,
TNF-a,
HSV-tk (in combination with ganciclovir), E. coli cytosine deaminase (in
combination with 5-fluorocytosine) and is combined with encapsulated cisplatin
or
with other similarly systemically delivered antineoplastic drugs to suppress
cancer.
BRIEF DESCRIPTION OF THE DRAWINGS
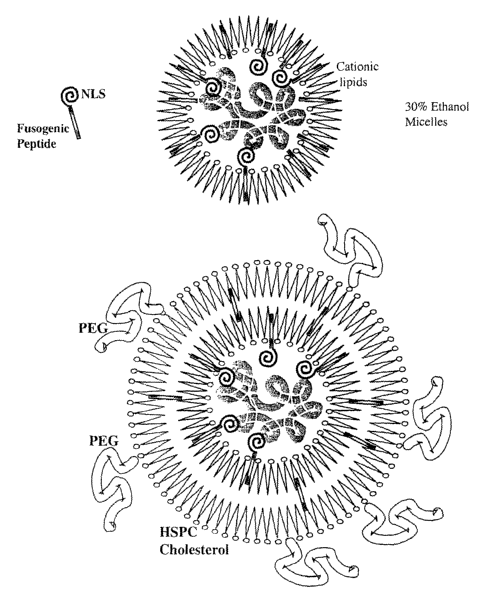
FIG. 1 illustrates the structure of the cancer targeted liposome complex.
FIG. 2 illustrates the results of plasmid DNA condensation with various
agents as well as various formulation of cationic liposomes in affecting the
level of
expression of the reporter beta-galactosidase gene after transfection of K562
human
erythroleukemia cell cultures.
FIG 3 illustrates tumor targeting in SCID mice. FIG 3A shows a SCID mouse
with a large and small human breast tumor before and after staining with X-Gal
to
test the expression of the transferred gene. Both tumors turn dark blue. The
intensity of the blue color is proportional to the expression of the beta-
galactosidase
gene.
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
FIG 3B shows that in the initial staining of the small tumor, the skin and the
intestines at the injection area are the first organs to turn blue. FIG 3C is
a view of
the back of the animal. The two tumors are clearly visible after removal of
the skin
(top). Dark staining of the small tumor and light blue staining of the large
tumor is
evident at an initial stage of staining (bottom). FIG 3D is a view of the
front side of
the animal. The two tumors are clearly visible after removal of the skin. On
the
figure to the bottom the dark staining of both tumors is evident at a later
stage during
staining.
FIG 3E shows the front (top) and rear (bottom) higher magnification view of
the dark staining of both tumors at a later stage during staining. Staining of
the
vascular system around the small tumor can also be seen (bottom).
BRIEF DESCRIPTION OF THE TABLES
Table 1 is a list of molecules able to form micelles.
Table 2 lists several fusogenic peptides and describes their properties, along
with a reference.
Table 3 lists simple Nuclear Localization Signal (NLS) peptides.
Table 4 shows a list of "bipartite" or "split" NLS peptides.
Table 5 lists "nonpositive NLS" peptides lacking clusters of
arginines/lysines.
Table 6 lists peptides with nucleolar localization signals (NoLS).
Table 7 lists peptides having karyophilic clusters on non-membrane protein
kinases.
Table 8 lists peptide nuclear localization signals on DNA repair proteins.
2S Table 9 lists NLS peptides in transcription factors.
Table 10 lists NLS peptides in other nuclear proteins.
MODES FOR CARRYING OUT THE INVENTION
Definitions
The practice of the present invention will employ, unless otherwise indicated,
conventional techniques of immunology, molecular biology, microbiology, cell
biology and recombinant DNA. These methods are described in the following
6
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
publications. See, e.g., Sambrook, et al. MOLECULAR CLONING: A LABORATORY
MANUAL, 2nd Edition (1989); CURRENT PROTOCOLS IN MOLECULAR BIOLOGY, F.M.
Ausubel, et al. eds., (1987); the series METHODS IN ENZYMOLOGY (Academic
Press, ...
Inc.); PCR: A PRACTICAL APPROACH, M. MacPherson, et al., IRL Press at Oxford
University Press (1991); PCR 2: A PRACTICAL APPROACH, MacPherson et al., eds.
(1995); ANTIBODIES, A LABORATORY MANUAL, Harlow and Lane, eds. (1988); and
ANIMAL CELL CULTURE, R.I. Freshney, ed. (1987).
As used in the specification and claims, the singular form "a," "an" and "the"
include plural references unless the context clearly dictates otherwise. For
example,
the term "a cell" includes a plurality of cells, including mixtures thereof.
The term "comprising" is intended to mean that the compositions and
methods include the recited elements, but not excluding others. "Consisting
essentially of when used to define compositions and methods, shall mean
excluding
other elements of any essential significance to the combination. Thus, a
composition
consisting essentially of the elements as defined herein would not exclude
trace
contaminants from the isolation and purification method and pharmaceutically
acceptable carriers, such as phosphate buffered saline, preservatives, and the
Like.
"Consisting of" shall mean excluding more than trace elements of other
ingredients
and substantial method steps for administering the compositions of this
invention.
Embodiments defined by each of these transition terms are within the scope of
this
invention.
The terms "polynucleotide" and "nucleic acid molecule" are used
interchangeably to refer to polymeric forms of nucleotides of any length. The
polynucleotides may contain deoxyribonucleotides, ribonucleotides, andlor
their
analogs. Nucleotides may have any three-dimensional structure, and may perform
any function, known or unknown. The term "polynucleotide" includes, for
example,
single-, double-stranded and triple helical molecules, a gene or gene
fragment,
exons, introns, mRNA, tRNA, rRNA, ribozymes, cDNA, recombinant
polynucleotides, branched polynucleotides, plasmids, vectors, isolated DNA of
any
sequence, isolated RNA of any sequence, nucleic acid probes, and primers. A
nucleic acid molecule may also comprise modified nucleic acid molecules.
7
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
A "gene" refers to a polynucleotide containing at least one open reading
frame that is capable of encoding a particular polypeptide or protein after
being
transcribed and translated.
A "gene product" refers to the amino acid (e.g., peptide or polypeptide)
generated when a gene is transcribed and translated.
The following abbreviations are used herein: DDAB: dimethyldioctadecyl
ammonium bromide (same as N,N-distearyl-N,N-dimethylammonium bromide);
DODAC: N,N-dioleyl-N,N-dimethylammonium chloride; DODAP: 1,2-dioleoyl-3-
dimethylammonium propane; DMRIE: N-[1-(2,3-dimyristyloxy)propyl]-N,N-
dimethyl-N-(2-hydroxyethyl) ammonium bromide; DMTAP: 1,2-dimyristoyl-3-
trimethylammonium propane; DOGS: Dioctadecylamidoglycylspermine; DOTAP
(same as DOTMA): N-(1-(2,3-dioleoyloxy)propyl)-N,N,N-trimethylammonium
chloride; DOSPA: N-(1-(2,3-dioleyloxy)propyl)-N-(2-
(sperminecarboxamido)ethyl)-N,N-dimethyl ammonium trifluoroacetate; DPTAP:
1,2- dipalmitoyl-3-trimethylammonium propane; DSTAP: 1,2-disteroyl-3-
trimethylammonium propane; DOPE, 1,2-sn-dioleoylphoshatidylethanolamine;
DC-Chol, 3(3-(N-(N',N'-dimethylaminoethane)carbamoyl)cholesterol. See, Gao et
al., Biochena. Biophys. Res. Comm. 179:280-285 (1991).
As used herein, the term "pharmaceutically acceptable anion" refers to
anions of organic and inorganic acids that provide non-toxic salts in
pharmaceutical
preparations. Examples of such anions include the halides anions, chloride,
bromide, and iodide, inorganic anions such as sulfate, phosphate, and nitrate,
and
organic anions. Organic anions may be derived from simple organic acids, such
as
acetic acid, propionic acid, glycolic acid, pyruvic acid, oxalic acid, malic
acid,
malonic acid, succinic acid, malefic, acid, fumaric acid, tartaric acid,
citric acid,
benzoic acid, cinnamic acid, mandelic acid, methane sulfonic acid, ethane
sulfonic
acid, p-toluenesulfonic acid, and the like. The preparation of
pharmaceutically
acceptable salts is described in Berge, et al., J. Pharm. Sci. 66:1-19 (1977),
incorporated herein by reference.
Physiologically acceptable carriers, excipients or stabilizers are nontoxic to
recipients at the dosages and concentrations employed, and include buffers
such as
phosphate, citrate, and other organic acids; antioxidants including ascorbic
acid; low
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
molecular weight (less than about 10 residues) polypeptides; proteins, such as
serum
albumin, gelatin, or immunoglobulins; hydrophilic polymers such as
polyvinylpyrrolidone; amino acids such as glycine, glutamine, asparagine,
arginine
or lysine; monosaccharides, disaccharides, and other carbohydrates including
glucose, mannose, or dextrins; chelating agents such as EDTA: sugar alcohols
such
as mannitol or sorbitol; salt-forming counter ions such as sodium; and/or
nonionic
surfactants such as Tween, Pluronics or polyethylene glycol (PEG). PEG
molecules
also contain a fusogenic peptide with an attached Nuclear Localization Signal
(NLS)
covalently Linked to the end of the PEG molecule.
The term "cationic lipid" refers to any of a number of lipid species that
carry
a net positive charge at physiological pH. Such lipids include, but are not
limited to,
DDAB, DMRIE, DODAC, DOGS, DOTAP, DOSPA and DC-Chol. Additionally, a
number of commercial preparations of cationic lipids are available that can be
used
in the present invention. These include, for example, LIPOFECTIN (commercially
available cationic liposomes comprising DOTMA and DOPE, from GIBCOBRL,
Grand Island, N.Y., USA); LIPOFECTAMINE (commercially available cationic
liposomes comprising DOSPA and DOPE, from GIBCOBRL); and
TRANSFECTAM (commercially available cationic lipids comprising DOGS in
ethanol from Promega Corp., Madison, Wis., USA).
This invention further provides a number of methods for producing micelles
with entrapped therapeutic drugs. The method is particularly useful to produce
micelles of drugs or compositions having a net overall negative charge, e.g.,
DNA,
RNA or negatively charged small molecules. For example, the DNA can be
comprised within a plasmid vector and encode for a therapeutic protein, e.g.,
wild-
type p53, HSV-tk, p21, Bax, Bad, IL-2, IL-12, GM-CSF, angiostatin, endostatin
and
oncostatin. In one embodiment, the method requires combining an effective
amount
of the therapeutic agent with an effective amount of cationic lipids. Cationic
lipids
useful in the methods of this invention include, but are not limited to, DDAB,
dimethyldioctadecyl ammonium bromide; DMRIE: N-[1-(2,3-
dimyristyloxy)propyl]-N,N-dimethyl-N-(2-hydroxyethyl) ammonium bromide;
DMTAP: 1,2-dimyristoyl-3-trimethylammonium propane; DOGS:
Dioctadecylamidoglycylspermine; DOTAP (same as DOTMA): N-(1-(2,3-
9
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
dioleoyloxy)propyl)-N,N,N-trimethylammonium chloride; DPTAP: 1,2-
dipalmitoyl-3-trimethylammonium propane; DSTAP: 1,2-disteroyl-3-
trimethylammonium propane.
In one aspect, a ratio of from about 30 to about 90% of phosphates contained
within the negatively charged therapeutic agent are neutralized by positive
charges
on lipid molecules (negative charges are in excess) to form an electrostatic
micelle
complex in an effective concentration of ethanol. In one aspect, the ethanol
solution
is from about 20% to about ~0% ethanol. In a further aspect, the ethanol
concentration is about 30%. The ethanol/cationic lipid/therapeutic agent
complex is
then combined with an effective amount of a fusogenic-karyophilic peptide
conjugate. In one aspect, an effective amount of the conjugate is a ratio
range from
about 0.0 to about 0.3 (positive charges on peptide to negative charges on
phosphate
groups) to neutralize the majority of the remaining negative charges on the
phosphate groups of the therapeutic agents thereby leading to an almost
complete
neutralization of the complex. The optimal conditions give to the complex a
slightly
negative charge. However, when the positive charges on cationic lipids exceed
the
negative charges on the DNA, the excess of positive charges are neutralized by
DPPG (dipalmitoyl phosphatidyl glycerol) and its derivatives, or by other
anionic
lipid molecules in the final micelle complex.
In an alternative embodiment, the above methods can be modified by
addition of DNA condensing agents selected from spermine, spermidine, and
magnesium or other divalent metal ions neutralizing a certain percentage (1-
20%) of
phosphate groups.
In a further embodiment, the cationic lipids are combined with an effective
amount of fusogenic lipid DOPE at various molar ratios for example, in a molar
ratio of from about 1:1 cationic lipid:DOPE. In an alternative embodiment, the
cationic lipids are combined with an effective amount of a fusogenic/NLS
peptide
conjugate. Examples of fusogenic/NLS peptide conjugates include, but are not
limited to (KAWLKAF)3 (SEQ ID NO:1), GLFKA.A.AKLLKSLWKLLLKA (SEQ
ID N0:2), LLLKAFAKLLKSLWKLLLKA (SEQ ID N0:3), as well as all
derivatives of the prototype (Hydrophobic3-Karyophilicl-Hydrophobic2-
Karyophilicl)2_3 where Hydrophobic is any of the A, I, L, V, P, G, W, F and
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
Karyophilic is any of the K, R, or H, containing a positively-charged residue
every
3rd or 4th amino acid, which form alpha helices and direct a net positive
charge to
the same direction of the helix. Additional examples include but are not
limited to
GLFKAIAGFIKNGWKGMIDGGGYC (SEQ ID N0:4) from influenza virus
hemagglutinin HA-2; YGRKKRRQRRR (SEQ ID NO:S) from TAT of HIV;
MSGTFGGILAGLIGLL(K/R/H)1_6 (SEQ ID N0:6), derived from the N-terminal
region of the S protein of duck hepatitis B virus, but with the addition of
one to six
positively-charged lysine, arginine or histidine residues, and combinations of
these,
able to interact directly with the phosphate groups of plasmid or
oligonucleotide
DNA, compensating for part of the positive charges provided by the cationic
lipids.
GAAIGLAWIPYFGPAA (SEQ ID N0:7) is derived from the fusogenic peptide of
the Ebola virus transmembrane protein; residues 53-70 (C-terminal helix) of
apolipoprotein (apo) All peptide; the 23-residue fusogenic N-terminal peptide
of
HIV-1 transmembrane glycoprotein gp4l; the 29-42-residue fragment from
Alzheimer's j3-amyloid peptide; the fusion peptide and N-terminal heptad
repeat of
Sendai virus; the 56-6~ helical segment of lecithin cholesterol
acyltransferase.
Included within these embodiments are shorter versions of these peptides, that
are
known to induce fusion of unilamellar lipid vesicles or all that are similarly
derivatized with the addition of one to six positively-charged lysine,
arginine or
histidine residues (K/R/H)1_6 able to interact directly with the phosphate
groups of
plasmid or oligonucleotide DNA, compensating for part of the positive charges
provided by the cationic lipids. The fusogenic peptides in the fusogenic/NLS
conjugates represent hydrophobic amino acid stretches, and smaller fragments
of
these peptide sequences, that include all signal peptide sequences used in
membrane
or secreted proteins that insert into the endoplasmic reticulum.
Alternatively, the
conjugates represent transmembrane domains and smaller fragments of these
peptide sequences.
In one aspect of the invention, the NLS peptide component in
fusogenic/NLS peptide conjugates is derived from the fusogenic hydrophobic
peptides. However, there is an addition of 5-6 amino acid karyophilic Nuclear
Localization Signals (NLS) derived from a number of known NLS peptides, as
well
as from searches of the nuclear protein databases, for stretches of five or
more
11
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
karyophilic amino acid stretches in proteins containing at least four
positively-
charged amino aids flanked by a proline (P) or glycine (G). Examples of NLS
peptides are shown in Tables 1-8. The NLS peptide component in fusogenic/NLS
peptide conjugates are synthetic peptides containing the above said NLS, but
further
modified by additional I~, R, H residues at the central part of the peptide or
with P
or G at the N- or C-terminus.
In a further aspect, the fusogenic/NLS peptide conjugates are derived from
the said fusogenic hydrophobic peptides but with the addition of a stretch of
H4_6
(four to six histidine residues) in the place of NIrS. Micelle formation takes
place at
pH 5-6 where histidyl residues are positively charged but lose their charge at
the
nearly neutral pH of the biological fluids, thus releasing the plasmid or
oligonucleotide DNA from their electrostatic interaction.
The fusogenic peptide/NLS peptide conjugates are linked to each other with
a short amino acid stretch representing an endogenous protease cleavage site.
In a preferred aspect of the invention, the structure of the preferred
prototype
fusogenic/NLS peptide conjugate used in this invention is: PKKRRGPSP(L/A/I)la-
ao (SEQ ID N0:8), where (L/A/I)12_ZO is a stretch of 12-20 hydrophobic amino
acids
containing A, L, I, Y, W, F and other hydrophobic amino acids.
The micelles made by the above methods are further provided by this
invention by conversion into liposomes. An effective amount of liposomes
(diameter from about 80 to about 160 nm), or of a lipid solution composed of
cholesterol (from about 10% to about 50%), neutral phospholipid such as
hydrogenated soy phosphatidylcholine (HSPC) (from about 40% to about 90%), and
the derivatized vesicle-forming lipid PEG-DSPE (distearoylphosphatidyl
ethanolamine) from about 1-to about 7 mole percent, is added to the micelle
solution.
In a specific embodiment, the liposomes are composed of vesicle-forming
lipids and between from about 1 to about 7 mole percent of
distearoylphosphatidyl
ethanolamine (DSPE) derivatized with a polyethyleneglycol. The compositiomof
claim 20, wherein the polyethyleneglycol has a molecular weight is between
about
1,000 to 5,000 daltons. Micelles are converted into liposomes with a
concomitant
decrease of the ethanol concentration which can be accomplished by removal of
the
12
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
ethanol by dialysis of the liposome complexes through permeable membranes or
reduced to a diameter of 80-160 nm by extrusion through membranes.
Liposome encapsulated therapeutic agents produced by the above methods
are further provided by this invention.
Also provided herein is a method for delivering a therapeutic agent such as
plasmid DNA or oligonucleotides to a tissue cell iri vivo by intravenous, or
other
type of inj ection of the micelles or liposomes. This method specifically
targets a
primary tumor and the metastases by the long circulating time of the micelle
or
liposome complex because of the exposure of PEG chains on its surface, its
small
size (80-160 nm) and the decrease in hydrostatic pressure in the solid tumor
from
the center to its periphery supporting a preferential extravasation through
the tumor
vasculature to the extracellular space in tumors. A method for delivering
plasmid or
oligonucleotide DNA across the cell membrane barrier of the tumors using the
micelle or liposome complexes described herein is capable because of the
presence
of the fusogenic peptides in the complex. In particular, a method for
delivering
plasmid or oligonucleotide DNA to the liver, spleen and bone marrow after
intravenous injection of the complexes is provided. Further provided is a
method
for delivering therapeutic genes to the liver, spleen and bone marrow of
cancer and
noncancer patients including but not limited to, factor VIII or IX for the
therapy of
hemophiliac, multidrug resistance, cytokine genes for cancer immunotherapy,
genes
for the alleviation of pain, genes for the alleviation of diabetes and genes
that can be
introduced to liver, spleen and bone marrow tissue, to produce a secreted form
of a
therapeutic protein.
The disclosed therapies also provide methods for reducing tumor size by
combining the encapsulated plasmid DNA carrying one or more anticancer genes
selected from the group consisting ofp53, RB, BRCA1, ElA, bcl-2, MDR-l, p21,
p16, bax, bcl-xs, E2F, IGF-I VEGF, angiostatin, oncostatin, endostatin, GM-
CSF,
IL-12, IL-2, IL-4, IL-7, IFN-y, TNF-a, HSV-tk (in combination with
ganciclovir),
E. coli cytosine deaminase (in combination with 5-fluorocytosine) with
encapsulated antisense oligonucleotides (antisense c-fos, c-myc, K-ras),
ribozymes
or triplex-forming oligonucleotides directed against genes that control the
cell cycle
or signaling pathways. These methods can be modified by combining the
13
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
encapsulated plasmid DNA carrying one or more anticancer genes of with
encapsulated or free antineoplastic drugs, consisting of the group of
adriamycin,
angiostatin, azathioprine, bleomycin, busulfane, camptothecin, carboplatin,
carmustine, chlorambucile, chlormethamine, chloroquinoxaline sulfonamide,
cisplatin, cyclophosphamide, cycloplatam, cytarabine, dacarbazine,
dactinomycin,
daunorubicin, didox, doxorubicin, endostatin, enloplatin, estramustine,
etoposide,
extramustinephosphat, flucytosine, fluorodeoxyuridine, fluorouracil, gallium
nitrate,
hydroxyurea, idoxuridine, interferons, interleukins, leuprolide, lobaplatin,
lomustine, mannomustine, mechlorethamine, mechlorethaminoxide, melphalan,
mercaptopurine, methotrexate, mithramycin, mitobronitole, mitomycin,
mycophenolic acid, nocodazole, oncostatin, oxaliplatin, paclitaxel,
pentamustine,
platinum-triamine complex, plicamycin, prednisolone, prednisone, procarbazine,
protein kinase C inhibitors, puromycine, semustine, signal transduction
inhibitors,
spiroplatin, streptozotocine, stromelysin inhibitors, taxol, tegafur,
telomerase
inhibitors, teniposide, thalidomide, thiamiprine, thioguanine, thiotepa,
tiamiprine,
tretamine, triaziquone, trifosfamide, tyrosine kinase inhibitors, uramustine,
vidarabine, vinblastine, vinca alcaloids, vincristine, vindesine, vorozole,
zeniplatin,
zeniplatin, and zinostatin.
The following examples are intended to illustrate, but not limit the
invention.
Liposome Composition
Liposomes are microscopic vesicles consisting of concentric lipid bilayers.
Structurally, liposomes range in size and shape from long tubes to spheres,
with
dimensions from a few hundred Angstroms to fractions of a millimeter. Vesicle-
forming lipids are selected to achieve a specified degree of fluidity or
rigidity of the
final complex providing the lipid composition of the outer layer. These are
neutral
(cholesterol) or bipolar and include phospholipids, such as
phosphatidylcholine (PC),
phosphatidylethanolamine (PE), phosphatidylinositol (PI), and sphingomyelin
(SM)
and other type of bipolar lipids including but not limited to
dioleoylphosphatidylethanolamine (DOPE), with a hydrocarbon chain length in
the
range of 14-22, and saturated or with one or more double C=C bonds. Examples
of
lipids capable of producing a stable liposome, alone, or in combination with
other
14
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
lipid components are phospholipids, such as hydrogenated soy
phosphatidylcholine
(HSPC), lecithin, phosphatidylethanolamine, lysolecithin,
lysophosphatidylethanolamine, phosphatidylserine, phosphatidylinositol,
sphingomyelin, cephalin, cardiolipin, phosphatidic acid, cerebrosides,
distearoylphosphatidylethanolamine (DSPE), dioleoylphosphatidylcholine (DOPC),
dipalmitoylphosphatidylcholine (DPPC), palmitoyloleoylphosphatidylcholine
(POPC), palmitoyloleoylphosphatidylethanolamine (POPE) and
dioleoylphosphatidylethanolamine 4-(N-maleimido-methyl)cyclohexane-1-
carboxylate (DOPE-mal). Additional non-phosphorous containing lipids that can
become incorporated into liposomes include stearylamine, dodecylamine,
hexadecylamine, isopropyl myristate, triethanolamine-lauryl sulfate, alkyl-
aryl
sulfate, acetyl palmitate, glycerol ricinoleate, hexadecyl stereate,
amphoteric acrylic
polymers, polyethyloxylated'fatty acid amides, and the cationic lipids
mentioned
above (DDAB, DODAC, DMRIE, DMTAP, DOGS, DOTAP (DOTMA), DOSPA,
DPTAP, DSTAP, DC-Chol). Negatively charged lipids include phosphatidic acid
(PA), dipalmitoylphosphatidylglycerol (DPPG), dioleoylphosphatidylglycerol and
(DOPG), dicetylphosphate that are able to form vesicles. Preferred lipids for
use in
the present invention are cholesterol, hydrogenated soy phosphatidylcholine
(HSPC)
and, the derivatized vesicle-forming lipid PEG-DSPE.
Typically, liposomes can be divided into three categories based on their
overall size and the nature of the lamellar structure. The three
classifications, as
developed by the New York Academy Sciences Meeting, "Liposomes and Their Use
in Biology and Medicine," December 1977, are mufti-lamellar vesicles (MLVs),
small uni-lamellar vesicles (SUVs) and large uni-lamellar vesicles (LUVs).
SUVs range in diameter from approximately 20 to 50 nm arid consist of a
single lipid bilayer surrounding an aqueous compartment. Unilamellar vesicles
can
also be prepared in sizes from about 50 nm to 600 nm in diameter. While
unilamellar are single compartmental vesicles of fairly uniform size, MLVs
vary
greatly in size up to 10,000 nm, or thereabouts, are mufti-compartmental in
their
structure and contain more than one bilayer. LUV liposomes are so named
because
of their large diameter that ranges from about 600 nm to 30,000 nm; they can
contain
more than one bilayer.
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
Liposomes may be prepared by a number of methods not all of which
produce the three different types of liposomes. For example, ultrasonic
dispersion
by means of immersing a metal probe directly into a suspension of MLVs is a
common way for preparing SUVs.
Preparing liposomes of the MLV class usually involves dissolving the lipids
in an appropriate organic solvent and then removing the solvent under a gas or
air
stream. This leaves behind a thin film of dry lipid on the surface of the
container.
An aqueous solution is then introduced into the container with shaking, in
order to
free lipid material from the sides of the container. This process disperses
the lipid,
causing it to form into lipid aggregates or liposomes. Liposomes of the LUV
variety
may be made by slow hydration of a thin layer of lipid with distilled water or
an
aqueous solution of some sort. Alternatively, liposomes may be prepared by
lyophilization. This process comprises drying a solution of lipids to a film
under a
stream of nitrogen. This film is then dissolved in a volatile solvent, frozen,
and
placed on a lyophilization apparatus to remove the solvent. To prepare a
pharmaceutical formulation containing a drug, a solution of the drug is added
to the
lyophilized lipids, whereupon Iiposomes are formed.
Preparing Cationic Liuosome/Cationic Peptide/Nucleic Acid Micelles
Cationic lipids, with the exception of sphingosine and some lipids in
primitive life forms, do not occur in nature. The present invention uses
single-chain
amphiphiles which are chloride and bromide salts of the alkyltrimethylammonium
surfactants including but not limited to C 12 and C 16 chains abbreviated DDAB
(same as DODAB) or CTAB. The molecular geometry of these molecules
determines the critical micelle concentration (ratio between free monomers in
solution and molecules in micelles). Lipid exchange between the two states is
a
highly dynamic process; phospholipids have critical micelle concentration
values
below I0-8 M and are more stable in Iiposomes; however, single chain
detergents,
such as stearylamine, may emerge from the liposome membrane upon dilution or
intravenous injection in milliseconds (Lasic, 1997).
Cationic lipids include, but are not limited to, DDAB: dimethyldioctadecyl
ammonium bromide (same as N,N-distearyl-N,N-dimethylammonium bromide);
16
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
DMRIE: N-[1-(2,3-dimyristyloxy)propyl]-N,N-dimethyl-N-(2-hydroxyethyl)
ammonium bromide; DODAC: N,N-dioleyl-N,N-dimethylammonium chloride;
DMTAP: 1,2-dimyristoyl-3-trimethylammonium propane; DODAP: 1,2-dioleoyl-3-
dimethylammonium propane; DOGS: Dioctadecylamidoglycylspermine; DOTAP
(same as DOTMA): N-(1-(2,3-dioleoyloxy)propyl)-N,N,N-trimethylammonium
chloride; DOSPA: N-(1-(2,3-dioleyloxy)propyl)-N-(2-(sperminecarboxamido)ethyl)-
N,N-dimethyl ammonium trifluoroacetate; DPTAP: 1,2- dipalmitoyl-3-
trimethylammonium propane; DSTAP: 1,2-disteroyl-3-trimethylammonium propane;
DC-Chol, 3(3-(N-(N',N'-dimethylaminoethane)carbamoyl)cholesterol.
Lipid-based vectors used in gene transfer have been formulated in one of two
ways. In one method, the nucleic acid is introduced into preformed liposomes
made
of mixtures of cationic lipids and neutral lipids. The complexes thus formed
have
undefined and complicated structures and the transfection efficiency is
severely
reduced by the presence of serum. Preformed liposomes are commercially
available
as LIPOFECTIN and LIPOFECTAMINE. The second method involves the
formation of DNA complexes with mono- or poly-cationic lipids without the
presence of a neutral lipid. These complexes are prepared in the presence of
ethanol
and are not stable in water. Additionally, these complexes are adversely
affected by
serum (see, Behr, Acc. Chem. Res. 26:274-78 (1993)). An example of a
commercially available poly-cationic lipid is TRANSFEGTAM. Other efforts to
encapsulate DNA in lipid-based formulations have not overcome these problems
(see, Szoka et al., Ann. Rev. Biophys. BioerZg. 9:467 (1980); and Deamer, U.S.
Patent
No. 4,515,736).
The nucleotide polymers can be single-stranded DNA or RNA, or double-
stranded DNA or DNA-RNA hybrids. Examples of double-stranded DNA include
structural genes, genes including control and termination regions, and self
replicating systems such as plasmid DNA. Particularly preferred nucleic acids
are
plasmids. Single-stranded nucleic acids include antisense oligonucleotides
(complementary to DNA and RNA), ribozymes and triplex-forming
oligonucleotides. In order to increase stability, some single-stranded nucleic
acids
will preferably have some or all of the nucleotide linkages substituted with
stable,
non-phosphodiester linleages, including, for example, phosphorothioate,
17
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
phosphorodithioate, phosphoroselenate, methylphosphonate, or O-alkyl
phosphotriester linkages.
Encapsulating Cationic Liposome/Cationic Peptide/Nucleic Acid
Micelles into Neutral Linosomes
Cationic lipids used with fusogenic peptide/NLS conjugates to provide the
inner layer of the particle can be any of a number of substances selected from
the
group of DDAB, DODAC, DMRIE, DMTAP, DOGS, DOTAP (DOTMA), DOSPA,
DPTAP, DSTAP, DC-Chol. The cationic lipid is combined with DOPE. In one
group of embodiments, the preferred cationic lipid is DDAB:DOPE 1:1.
Neutral lipids used herein to provide the outer layer of the particles can be
any of a number of lipid species that exist either in an uncharged or neutral
zwitterionic form at physiological pH. Such lipids are selected from a group
consisting of diacylphosphatidylcholine, diacylphosphatidylethanolamine,
ceramide,
sphingomyelin, cephalin, and cerebrosides. In one group of embodiments, lipids
containing saturated, mono-, or di-unsaturated fatty acids with carbon chain
lengths
in the range of C14 to C22 are preferred. In general, less saturated lipids
are more
easily sized, particularly when the liposomes must be sized below about 0.16
microns, for purposes of filter sterilization. Consideration of liposome size,
rigidity
and stability of the liposomes in the final preparation, its shelf life
without leakage of
the encapsulated DNA, and stability in the bloodstream generally guide the
selection
of neutral lipids for providing the outer coating of our gene vehicles. Lipids
having
a variety of acyl chain groups of varying chain length and degree of
saturation are
available or may be isolated or synthesized by well-known techniques. In
another
group of embodiments, lipids with carbon chain lengths in the range of C14 to
C22
are used. Preferably, the neutral lipids used in the present invention are
hydrogenated soy phosphatidylcholine (HSPC), cholesterol, and PEG-
distearoylphosphatidyl ethanolamine (DSPE) or PEG-ceramide.
Methods for preparing liposomes
A variety of methods for preparing various liposome forms have been
described in several issued patents, for example, U.S. Patent Nos. 4,229,360;
1~
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
4,224,179; 4,241,046; 4,737,323; 4,078,052; 4,235,871; 4,501,728; and
4,837,028,
as well as in the articles Szoka et al., Ann. Rev. Biophys. Bioeng. 9:467
(1980) and
Hope et al., Chem. Phys. Lip. 40:89 (1986). These methods do not produce all
three
different types of liposomes (MLVs, SUVs, LUVs). For example, ultrasonic
dispersion by means of immersing a metal probe directly into a suspension of
MLVs
is a common way for preparing SUVs.
Preparing liposomes of the MLV class usually involves dissolving the lipids
in an appropriate organic solvent and than removing the solvent under a gas or
air
stream. This leaves behind a thin film of dry lipid on the surface of the
container.
An aqueous solution is then introduced into the container with shaking, in
order to
free lipid material from the sides of the container. This process disperses
the lipid,
causing it to form into lipid aggregates or liposomes. Liposomes of the LW
variety
may be made by slow hydration of a thin layer of lipid with distilled water or
an
aqueous solution of some sort. Alternatively, liposomes may be prepared by
lyophilization. This process comprises drying a solution of lipids to a film
under a
stream of nitrogen. The film is then dissolved in a volatile solvent, frozen,
and
placed on a lyophilization apparatus to remove the solvent. To prepare a
pharmaceutical formulation containing a drug, a solution of the drug is added
to the
lyophilized lipids, whereupon liposomes are formed.
Following liposome preparation, the liposomes may be sized to achieve a
desired size range and relatively narrow distribution of liposome sizes.
Preferably,
the preformed liposomes are sized to a mean diameter of about 80 to 160 nm
(the
upper size limit for filter sterilization before in vivo administration).
Several
techniques are available for sizing liposomes to a desired size. Sonicating a
liposome suspension either by bath or probe sonication produces a progressive
size
reduction down to small unilamellar vesicles less than about 0.05 microns (50
nm) in
size. Extrusion of liposome through a small-pore polycarbonate is our
preferred
method for reducing liposome sizes to a relatively well-defined size
distribution.
The liposomes may be extruded through successively smaller-pore membranes, to
achieve a gradual reduction in liposome size.
One way used to coat DNA with lipid is by controlled detergent depletion
from a cationic lipid/DNA/detergent complex. This method can give complexes
19
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
with stability in plasma. Hofland et al. (1996), have prepared such complexes
by
dialysis of a mixture of DOSPA/DOPE/DNAloctylglucoside.
Pharmaceutical compositions comprising the cationic liposome/nucleic acid
complexes of the invention are prepared according to standard techniques and
further
comprise a pharmaceutically acceptable carrier. Generally, normal saline will
be
employed as the pharmaceutically acceptable Garner.
For in vivo administration, the pharmaceutical compositions are preferably
administered parenterally, i.e., intravenously, intraperitoneally,
subcutaneously,
intrathecally, inj ection to the spinal cord, intramuscularly,
intraarticularly, portal
vein injection, or intratumorally. More preferably, the pharmaceutical
compositions
are administered intravenously or intratumorally by a bolus injection. In
other
methods, the pharmaceutical preparations may be contacted with the target
tissue by
direct application of the preparation to the tissue. The application may be
made by
topical "open" or "closed" procedures. The term "topical" means the direct
application of the pharmaceutical preparation to a tissue exposed to the
environment,
such as the skin, to any surface of the body, nasopharynx, external auditory
canal,
ocular administration and administration to the surface of any body cavities,
inhalation to the lung, genital mucosa and the like.
"Open" procedures are those procedures that include incising the skin of a
patient and directly visualizing the underlying tissue to which the
pharmaceutical
preparations are applied. This is generally accomplished by a surgical
procedure,
such as a thoracotomy to access the lungs, abdominal laparotomy to access
abdominal viscera, or other direct surgical approach to the target tissue.
"Closed" procedures are invasive procedures in which the internal target
tissues are not directly visualized, but accessed via insertion of instruments
through
small wounds in the skin. For example, the preparations may be administered to
the
peritoneum by needle lavage. Likewise, the pharmaceutical preparations may be
administered to the meninges or spinal cord by infusion during a lumbar
puncture
followed by appropriate positioning of the patient as commonly practiced for
spinal
anesthesia or metrazamide imaging of the spinal cord. Alternatively, the
preparations may be administered through endoscopic devices.
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
EXAMPLES
Materials and Methods
DDAB, DOPE (dioleoylphosphatidylethanolamine) and most other lipids
used here were purchased from Avanti Polar Lipids; PEG-DSl'E was from Syngena.
Engineering of plasmid pLF
The pGL3-C (Promega) was cut with XbaI and blunt-end ligated using the
Klenow fragment of E. coli DNA polymerise. It was then cut with HindIII and
the
169-by fragment, carrying the luciferase gene, was gel-purified. The pGFP-Nl
plasmid (Clontech) was cut with SmaI and HindIII and the 4.7 kb fragment,
isolated
from an agarose gel, was ligated with the luciferase fragment. JM109 E. coli
cells
were transformed and 20 colonies were selected; about half of them showed the
presence of inserts; 8 clones with inserts were cut with BamHI and XhoI to
further
confirm the presence of the luciferase gene; seven of them were positive.
Radiolabeled plasmid pLF was generated by culturing Escherichia coli in
3H-thymidine-5'-triphosphate or 32P inorganic phosphate (5 mCi) (Dupont/NEN,
Boston, Mass.) and purified using standard techniques as described above.
DLS measurements
A Coulter N4M light scattering instrument was used, at a 90° angle,
set at a
run time of 200 sec, using 4 to 25 microsec sample time. The scan of the
particle
size distribution was obtained in 1 ml sample volume using plastic cuvettes,
at 20°C
and at 0.01 poise viscosity.
In one aspect, this invention provides a method for entrapping DNA into
lipids that enhances the content of plasmid per volume unit, and reduces the
toxicity
of the cationic lipids used to trap plasmid or oligonucleotide DNA. The DNA
becomes hidden in the inner membrane bilayer of the final complex.
Furthermore,
the gene transfer complex is endowed with long circulation time in body fluids
and
extravasates preferentially into solid tumors and their metastatic foci and
nodules.
The extravasation occurs through their vasculature at most sites of the human
or
animal body after intravenous inj ection of the gene-carrying vehicles. This
occurs
because of their small size (100-160 nm), their content in neutral to slightly
21
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
negatively-charged lipids in their outer membrane bilayers, and their coating
with
PEG. These gene delivery vehicles are able to cross the cell membrane barner
after
they reach the extracellular tumor space because of the presence of fusogenic
peptides conjugated with karyophilic peptides. The vehicles assume a certain
predefined orientation in the lipid membrane with their positive ends directed
toward
DNA and their hydrophobic tail buried inside the hydrophobic lipid bilayer.
The
labile NLS-fusogenic peptide linkage is cleaved after endocytosis and the
remaining
NLS peptide bound to plasmid DNA aids its nuclear uptake. This occurs
especially
when non-dividing cells are targeted, such as liver, spleen or bone marrow
cells that
represent the major sites for extravasation and concentration of these
vehicles other
than solid tumors.
Organic solvent
A suitable solvent for preparing a micelle from the desired lipid components
is ethanol, methanol, or other aliphatic alcohols such as propanol,
isopropanol,
butanol, tert-butanol, iso-butanol, pentanol and hexanol. Mixtures of two or
more
solvents may be used in the practice of the invention. It is also to be
understood that
any solvent that is miscible with an ethanol solution, even in small amounts,
can be
used to improve micelle formation and its subsequent conversion into
liposomes,
including chloroform, dichloromethane, diethylether, cyclohexane,
cyclopentane,
benzene, and toluene.
Cationic lipids
In a further embodiment, the Iiposome encapsulated DNA described herein
further comprises an effective amount of cationic lipids. Cationic lipids have
been
widely used for gene transfer; a number of clinical trials (34 out of 220
total RAC-
approved protocols as of December, 1997) use cationic lipids. Although many
cell
culture studies have been documented, systemic delivery of genes with cationic
lipids ifa ~iwo has been very limited. All clinical protocols use
subcutaneous,
intradermal, intratumoral, and intracranial injection as well as intranasal,
intrapleural, or aerosol administration but not LV. delivery, because of the
toxicity of
the cationic lipids and DOPE (see, Martin and Boulikas, 1990. Liposomes
22
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
formulated from DOPE and cationic lipids based on diacyltrimethylammonium
propane (dioleoyl-, dimyristoyl-, dipalmitoyl-, disteroyl-trimethylammonium
propane or DOTAP, DMTAP, DPTAP, DSTAP, respectively) or DDAB were highly
toxic when incubated in vitro with phagocytic cells (macrophages and U937
cells),
but not towards non-phagocytic T lymphocytes. The rank order of toxicity was
DOPE/DDAB > DOPE/DOTAP > DOPE/DMTAP > DOPE/DPTAP >
DOPE/DSTAP; and the toxicity was determined from the effect of the cationic
liposomes on the synthesis of nitric oxide (NO) and TNF-a produced by
activated
macrophages (Filion and Phillips, 1997).
Another aspect to be considered before LV. injection is undertaken, is thaf~
negatively charged serum proteins can interact and cause inactivation of
cationic
liposomes (Yang and Huang, 1997). Condensing agents used for plasmid delivery
including polylysine, transferrin-polylysine, a fifth-generation
poly(amidoamine)
(PAMAM) dendrimer, poly(ethyleneimine), and several cationic lipids (DOTAP,
DC-Chol/DOPE, DOGS/DOPE, and DOTMA/DOPE), were found to activate the
complement system to varying extents. Strong complement activation was seen
with
long-chain polylysines, the dendrimer, poly(ethyleneimine), and DOGS.
Modifying
the surface of preformed DNA complexes with polyethyleneglycol (Plank et al.,
1996) considerably reduced complement activation.
Cationic lipids increase the transfection efficiency by destabilizing the
biological membranes, including plasma, endosomal, and lysosomal membranes.
Incubation of isolated lysosomes with low concentrations of DOTAP caused a
striking increase in free activity of (3-galactosidase, and even a release of
the enzyme
into the medium. This demonstrates that the lysosomal membrane is deeply
destabilized by the lipid. The mechanism of destabilization was thought to
involve
an interaction between cationic liposomes and anionic lipids of the lysosomal
membrane, thus allowing a fusion between the lipid bilayers. The process was
less
pronounced at pH 5 than at pH 7.4, and anionic amphipathic lipids were able to
prevent partially this membrane destabilization (Wattiaux et al., 1997).
In contrast to DOTAP and DMRIE that were 100% charged at pH 7.4, DC-
CHOL was only about 50% charged as monitored by a pH-sensitive fluorophore.
This difference decreases the charge on the external surfaces of the
liposomes, and
23
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
was proposed to promote an easier dissociation of bilayers containing DC-CHOL
from the plasmid DNA, and an increase in release of the DNA-lipid complex into
the
cytosol from the endosomes (Zuidam and Barenholz, 1997).
Although cationic lipids have been used widely for the delivery of genes,
very few studies have used systemic LV. injection of cationic liposome-plasmid
complexes. This is because of the toxicity of the lipid component in animal
models,
not humans. Administration by LV. injection of two types of cationic lipids of
similar structure, DOTMA and DOTAP, shows that the transfection efficiency is
determined mainly by the structure of the cationic lipid and the ratio of
cationic lipid
to DNA; the luciferase and GFP gene expression in different organs was
transient,
with a peak level between 4 and 24 hr, dropping to less than 1 % of the peak
level by
day 4 (Song et al., 1997).
A number of different organs in vivo can be targeted after liposomal delivery
of genes or oligonucleotides. Intravenous injection of cationic liposome-
plasmid
complexes by tail vein in mice, targeted mainly the lung and to a smaller
extent the
liver, spleen, heart, kidney and other organs (Zhu et al., 1993).
Intraperitoneal
injection of a plasmid-liposome complex expressing antisense K-ras RNA in nude
mice inoculated i.p. with AsPC-1 pancreatic cancer cells harboring K-ras point
mutations and PCR analysis indicated that the injected DNA was delivered to
various organs except brain (Aoki et al., 1995).
A number of factors for DOTAP:cholesterollDNA complex preparation
including the DNA:liposome ratio, mild sonication, heating, and extrusion were
found to be crucial for improved systemic delivery; maximal gene expression
was
obtained when a homogeneous population of DNA:liposome complexes between
200 to 450 nm in size were used. Cryo-electron microscopy showed that the DNA
was condensed on the interior of invaginated liposomes between two lipid
bilayers in
these formulations, a factor that was thought to be responsible for the high
transfection efficiency in vivo and for the broad tissue distribution
(Templeton et al.,
1997).
Steps to improve liposome-mediated gene delivery to somatic cells include,
persistence of the-plasmid in blood circulation, port of entry and transport
across the
cell membrane, release from endosomal compartments into the cytoplasm, nuclear
24
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
import by docking through the pore complexes of the nuclear envelope,
expression
driven by the appropriate promoter/enhancer control elements, and persistence
of the
plasmid in the nucleus for long periods (Boulikas,y199~a).
Plasmid condensation with spermine
In a further embodiment, the liposome encapsulated DNA described herein is
condensed with spermine and/or spermidine. DNA can be presented to cells in
culture as a complex with polycations such as polylysine, or basic proteins
such as
protamine, total histones or specific histone fractions, protamine (Boulikas
and
Martin, 1997). The interaction of plasmid DNA with protamine sulfate, followed
by
the addition of DOTAP cationic liposomes, offered a better protection of
plasmid
DNA against enzymatic digestion. The method gave consistently higher gene
expression in mice via tail vein injection as compared with DOTAP/DNA
complexes. 50 ~g of luciferase-plasmid per mouse gave 20 ng luciferase protein
per
mg extracted tissue protein in the lung, that was detected as early as 1 h
after
injection, peaked at 6 h and declined thereafter. Intraportal injection of
protamine/DOTAP/DNA led to about a 100-fold decrease in gene expression in the
lung as compared with LV. injection. Endothelial cells were the primary locus
of
lacZ transgene expression (Li and Huang, 1997). Protamine sulfate enhanced
plasmid delivery into several different types of cells in vitro, using the
monovalent
cationic liposomal formulations (DC-Chol and lipofectin). This effect was less
pronounced with the multivalent cationic liposome formulation, lipofectamine
(Sorgi
et al., 1997).
Spermine is found to enhance the transfection efficiency of DNA-cationic
liposome complexes in cell culture and in animal studies. This biogenic
polyamine
at high concentrations caused liposome fusion most likely promoted by the
simultaneous interaction of one molecule of spermine (four positively charged
amino
groups) with the polar head groups of two or more molecules of lipids. At low
concentrations (0.03-0.1 mM) it promoted anchorage of the liposome-DNA complex
to the surface of cells and enhanced significantly transfection efficiency
(Boulikas,
unpublished).
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
The polycations polybrene, protamine, DEAE-dextran, and poly-L-lysine
significantly increased the efficiency of adenovirus-mediated gene transfer in
cell
culture. This was thought to act by neutralizing the negative charges
presented by
membrane glycoproteins that reduce the efficiency of adenovirus-mediated gene
transfer (Arcasoy et al., 1997).
Oligonucleotide transfer
In a further embodiment, the liposome encapsulates oligonucleotide DNA.
Encapsulation of oligonucleotides into liposomes increased their therapeutic
index,
prevented degradation in cultured cells, and in human serum and reduced
toxicity to
cells (Thierry and Dritschilo, 1992; Capaccioli et al., 1993; Lewis et al.,
1996).
However, most studies have been performed in cell culture, and very few in
animals
in vivo. There are still an important number of improvements needed before
these
approaches can move into clinical studies.
Zelphati and Szoka (1997), have found that complexes of fluorescently
labeled oligonucleotides with DOTAP liposomes, entered the cell using an
endocytic
pathway mainly involving uncoated vesicles. Oligonucleotides were
redistributed
from punctate cytoplasmic regions into the nucleus. This process was
independent
of acidification of the endosomal vesicles. The nuclear uptake of
oligonucleotides
depended on several factors, such as charge of the particle, where positively
charged
complexes were required for enhanced nuclear uptake. DOTAP increased over 100
fold the antisense activity of a specific anti-luciferase oligonucleotide.
Physicochemical studies of oligonucleotide-liposome complexes of different
cationic
lipid compositions indicated that either phosphatidylethanolamine or negative
charges on other lipids in the cell membrane are required for efficient fusion
with
cationic liposome-oligonucleotide complexes to promote entry to the cell
(Jaaskelainen et al., 1994).
Similar results were reported by Lappalainen et al. (1997). Digoxigenin
labeled oligodeoxynucleotides (ODNs) complexed with the polycationic DOSPA
and the monocationic DDAB (with DOPE as a helper lipid) were taken up by CaSki
cells in culture by endocytosis. The nuclear membrane was found to pose a
barrier
against nuclear import of ODNs that accumulated in the perinuclear area.
Although
26
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
DOSPA/DOPE liposomes could deliver ODNs into the cytosol, they were unable to
mediate nuclear import of ODNs. On the contrary, oligonucleotide-DDAB/DOPE
complexes with a net positive charge were released from vesicles into the
cytoplasm.
It was determined that DDAB/DOPE mediated nuclear import of the
oligonucleotides.
DOPE-heme (ferric protoporphyrin IX) conjugates, inserted in cationic lipid
particles with DOTAP, protected oligoribonucleotides from degradation in human
serum and increased oligoribonucleotide uptake into 2.2.15 human hepatoma
cells.
The enhancing effect of heme was evident only at a net negative charge in the
I O particles (Takle et al., 1997). Uptake of liposomes labeled with lIn and
composed
of DC-Chol and DOPE was primarily by liver, with some accumulation in spleen
and skin and very little in the lung after LV. tail injection. Preincubation
of cationic
liposomes with phosphorothioate oligonucleotide induced a dramatic, yet
transient,
accumulation of the lipid in lung that gradually redistributed to liver. The
mechanism of lung uptake involved entrapment of large aggregates of
oligonucleotides within pulmonary capillaries at 15 min post-injection via
embolism.
Labeled oligonucleotide was localized primarily to phagocytic vacuoles of
Kupffer
cells at 24 h post-injection. Nuclear uptake of oligonucleotides in vivo was
not
observed (Litzinger et al., 1996).
Polyethylene glycol (PEG)-coated liposomes
In a further embodiment, the liposome encapsulated DNA described herein,
further comprise coating of the final complex in step 2 (Fig. 1) with PEG. It
is often
desirable to conjugate a lipid to a polymer that confers extended half life,
such as
polyethylene glycol (PEG). Derivatized lipids that are employed, include PEG-
modified DSPE or PEG-ceramide. Addition of PEG components prevents complex
aggregation, increases circulation lifetime of particles (liposomes, proteins,
other
complexes, drugs) and increases the delivery of lipid-nucleic acid complexes
to the
target tissues. See, Maxfield et al., Polymer 16:505-509 (1975); Bailey, F.E.
et al.,
in: Nonionic Surfactants, Schick, M.J., ed., pp. 794-821 (1967); Abuchowski,
A. et
al., J. Biol. Chem. 252:3582-3586 (1977); Abuchowski, A. et al., Cancer
Biochem.
27
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
Biophys. 7:175-186 (I984); Katre, N.V. et aL, Proc. Natl. Acad. Sci. USA
84:1487-
1491 (1987); Goodson, R. et al. Bio Techfaology 8:343-346 (1990).
Conjugation to PEG is reported to have reduced immunogenicity and
toxicity. See, Abuchowski et al., J. Biol. Chem. 252:3578-3581 (1977). The
extent
S of enhancement of blood circulation time of liposomes, by coating with PEG
is
described in U.S. Patent No. 5,013,5S6. Typically, the concentration of the
PEG-
modified phospholipids, or PEG-ceramide in the complex will be about 1-7%. In
a
particularly preferred embodiment, the PEG-modified Lipid is a PEG-DSPE.
Coating the surface of liposomes with inert materials designed to camouflage
the liposome from the body's host defense systems was shown to increase
remarkably the plasma Longevity of liposomes. The biological paradigm for this
"surface modified" sub-branch was the erythrocyte, a cell that is coated with
a dense
layer of carbohydrate groups, and that manages to evade immune system
detection
and to circulate for several months (before being removed by the same type of
cell
1S responsible for removing Liposomes).
The first breakthrough came in 1987 when a glycolipid (the brain tissue-
derived ganglioside GM1), was identified that, when incorporated within the
lipid
matrix, allowed liposomes to circulate for many hours in the blood stream
(Allen and
Chonn, 1987). A second glycolipid, phosphatidylinositol, was also found to
impart
long plasma residence times to liposomes and, since it was extracted from
soybeans,
not brain tissue, was believed to be a more pharmaceutically acceptable
excipient
(Gabizon et al., 1989).
A major advance in the surface-modified sub-branch was the development of
polymer-coated liposomes (Allen et a1. 1991). Polyethylene glycol (PEG)
2S modification had been used for many years to prolong the half lives of
biological
proteins (such as enzymes and growth factors) and to reduce their
immunogenicity
(e.g. Beauchamp et al., 1983). It was reported in the early 1990s that PEG-
coated
Liposomes circulated for remarkably long times after intravenous
administration.
Half lives on the order of 24 h were seen in mice and rats, and over 30 hours
in dogs.
The term "stealth" was applied to these liposomes because of their ability of
evade
interception by the immune system. The PEG hydrophilic polymers form dense
"conformational clouds" to prevent other macromolecules from interaction with
the
28
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
surface, even at low concentrations of the protecting polymer (Gabizon and
Papahadjopoulos, 1988; Papahadjopoulos et al., 1991; reviewed by Torchilin,
1998).
The increased hydrophilicity of the liposomes after their coating with the
amphipathic PEG5000 leads to a reduction in nonspecific uptake by the
reticuloendothelial system.
Whereas the half life of antimyosin immunoliposomes was 40 min, by
coating with PEG, they increased their half life to 1000 min after intravenous
inj ection to rabbits (Torchilin et al., 1992). '
Micelles, surfactants and small unilamellar vesicles
In a further embodiment, the liposome encapsulated DNA described herein,
further comprise an initial step of micelle formation between cationic lipids
and
condensed plasmid or oligonucleotide DNA in ethanol solutions. Micelles are
small
amphiphilic colloidal particles formed by certain kinds of lipid molecules,
detergents
or surfactants under defined conditions of concentration, solvent and
temperature.
They are composed of a single lipid layer. Micelles can have their hydrophilic
head
groups assembled exposing their hydrophobic tails to the solvent (for example
in 30-
60% aqueous ethanol solution) or can reverse their structures exposing their
polar
heads toward the solvent such as by lowering the concentration of the ethanol
to
below 10% (reverse micelles). Micelle systems are in thermodynamic equilibrium
with the solvent molecules and environment. This results in constant phase
changes,
especially upon contact with biological materials, such as upon introduction
to cell
culture, injection to animals, dilution, contact with proteins or other
macromolecules.
These changes result in rapid micelle disassembly or flocculation. This is in
contrast
to the much higher stability of liposome bilayers.
Single-chain surfactants are able to form micelles (see Table 1, below).
These include the anionic (sodium dodecyl sulfate, cholate or oleate) or
cationic
(cetyl-trimethylammonium bromide, CTAB) surfactants. CTAB, CTAC, and DOIC
micelles yielded larger solubility gaps (lower concentration of colloidally
suspended
DNA) than corresponding SUV particles containing neutral lipid and CTAB (1:1)
(Lasic, 1997).
29
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
Table 1: Molecules able to form micelles
Molecule Reference
CTAB, CTAC, DOIC Lasic, 1997
Detergent/phospholipid micelles Lusa et a1.,1998
Dodecyl betaine (amphoteric surfactant) de la Maza et al.,
1998
Dodecylphosphocholine cholate Lasic, 1997
Glycine-conjugated bile salt (anionic Leonard and Cohen,
steroid detergent-like molecule) 1998
Lipid-dodecyl maltoside micelles Lambert et al.,
1998
mixed micelles (Triton X-100 & phosphatidylcholine)Lopez et al., 1998
Octylglucoside (non-ionic straight chainLeonard and Cohen,
detergent) 1998
Oleate Lasic, 1997
PEG- dialkylphosphatidic acid (dihexadecylphosphatidylTirosh et al., 1998
(DHP)-
PEG2000)
Phosphatidylcholine (neutral zwitterionic)Schroeder et al.,
1990
Polyethyleneglycol (MW 5000)-distearoyl Weissig et al.,
phosphatidyl ethanolamine 1998
(PEG-DSPE)
sodium dodecyl sulfate (anionic straightLeonard and Cohen,
chain detergent) 1998
Sodium taurofusidate (conjugated fungal Leonard and Cohen,
bile salt analog) 1998
Taurine- conjugated bile salts (anionic Leonard and Cohen,
steroid detergent-like 1998
molecule)
Triton X-100 surfactant Lasic, 1997
There is a critical detergentlphospholipid ratio at which lamellar-to-micellar
transition occurs. For example, the vesicle-micelle transition was observed
for
dodecyl maltoside with large unilamellar liposomes. A striking feature of the
solubilization process by dodecyl maltoside was the discovery of a new phase,
consisting of a very viscous "gel-like" structure composed of long filamentous
thread-like micelles, over 1 to 2 microns in length.
A long circulating complex needs to be slightly anionic. Therefore the
liposomes used for the conversion of the micelles into liposomes contain
bipolar
lipids (PC, PE) and 1-30% negatively charged lipids (DPPG). The cationic
lipids
which are toxic, are hidden in the inner liposome membrane bilayer. Those
reaching
the solid tumor will exert their toxic effects causing apoptosis. Apoptosis
will be
caused by the delivery of the toxic drug or anti-neoplastic gene or
oligonucleotide to
the cancer cell but also by the nuclear localization of the cationic lipids
(along with
plasmid DNA) to the nucleus. Indeed, a number of studies suggest that plasmid
DNA is imported to nuclei; its translocation docks cationic lipid molecules
electrostatically attached to the DNA. These cationic lipid molecules exert
their
toxicity by interfering with the nucleosome and domain structure of the
chromatin
causing local destabilization. This disturbance or aberrant chromatin
reorganization
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
could be exerted at the level of the nuclear matrix where plasmid DNA is
attached
for transcription, autonomous replication, or integration via recombination.
Surfactants have found wide application in formulations such as emulsions
(including microemulsions) and liposomes. The most common way of classifying
and ranking the properties of the many different types of surfactants, both
natural
and synthetic, is by the use of the hydrophile/lipophile balance (HLB). The
use of
surfactants in drug products, formulations and in emulsions has been reviewed
(Rieger, irz: Pharmaceutical Dosage Forms, Marcel Dekker, Inc., New York,
1988,
p. 285).
Nonionic surfactants find wide application in pharmaceutical and cosmetic
products and are usable over a wide range of pH values. In general, their HLB
values range from 2 to about 18, depending on their structure. Nonionic
surfactants
include, nonionic esters such as ethylene glycol esters, propylene glycol
esters,
glyceryl esters, polyglyceryl esters, sorbitan esters, sucrose esters, and
ethoxylated
esters. Nonionic alkanolamides and ethers, such as fatty alcohol ethoxylates,
propoxylated alcohols, and ethoxylated/propoxylated, block polymers are also
included in this class. The polyoxyethylene surfactants are the most popular
members of the nonionic surfactant class.
Anionic surfactants include carboxylates such as soaps, acyl lactylates, acyl
amides of amino acids, esters of sulfuric acid such as alkyl sulfates and
ethoxylated
alkyl sulfates, sulfonates such as alkyl benzene sulfonates, acyl
isethionates, acyl
taurates and sulfosuccinates, and phosphates. The most important members of
the
anionic surfactant class are the alkyl sulfates and the soaps.
Cationic surfactants include quaternary ammonium salts and ethoxylated
amines. The quaternary ammonium salts are the most used members of this class.
If
the surfactant molecule has the ability to carry either a positive or negative
charge,
the surfactant is classified as amphoteric. Amphoteric surfactants include
acrylic
acid derivatives, substituted alkylamides, N-alkylbetaines and phosphatides.
Classical micelles may not be effective as gene transfer vehicles, but
important intermediates in the formation of liposome complexes encapsulating
drugs
or nucleic acids. The stability of single chain surfactants-DNA-colloidal
systems is
lower than SUV particles containing neutral lipid and CTAB (1:1). However,
31
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
second generation micelles are able to target tumors in vivo. Weissig and co-
workers (1998) used the soybean trypsin inhibitor (STI) as a model protein to
target
tumors. STI was modified with a hydrophobic residue of N-glutaryl-phosphatidyl-
ethanolamine (NGPE) and incozporated into both polyethyleneglycol (MW 5000)-
distearoyl phosphatidyl ethanolamine (PEG-DSPE) micelles (< 20 nm) and PEG-
DSPE-modified long-circulating liposomes (ca. 100 nm). As determined from the
protein label by using mIn attached to soybean trypsin inhibitor via protein-
attached
diethylene triamine pentaacetic acid, DTPA, PEG-Iipid micelles accumulated
better
than the same protein anchored in long-circulating PEG-liposomes in
subcutaneously
established Lewis lung carcinoma in mice after tail vein injection.
Loading a liposomal dispersion with an amphiphilic drug may cause a phase
transformation into a micellar solution. The transition from high ratios of
phospholipid to drug (from 2:1 to 1:1 downwards) were accompanied by the
conversion of liposomal dispersions of milky-white appearance (particle size
200
nm) to nearly transparent micelles (particle size below 25 nm). See, Schutze
and
Muller-Goymann (1998).
Fusogenic peptides
In a further embodiment, the liposome encapsulated DNA described herein
further comprises an effective amount of a fusogenic peptide. Fusogenic
peptides
belong to a class of helical amphipathic peptides characterized by a
hydrophobicity
gradient along the Iong helical axis. This hydrophobicity gradient causes the
tilted
insertion of the peptides in membranes, thus destabilizing the lipid core and,
thereby,
enhancing membrane fusion (Decout et al., 1999).
Hemagglutinin (HA) is a homotrimeric surface glycoprotein of the influenza
virus. In infection, it induces membrane fusion between viral and endosomal
membranes at low pH. Each monomer consists of the receptor-binding HA1 domain
and the membrane-interacting HA2 domain. The NH2-terminal region of the HA2
domain (amino acids 1 to 127), the so-called "fusion peptide," inserts into
the target
membrane and plays a crucial role in triggering fusion between the viral and
endosomal membranes. Based on the substitution of eight amino acids in region
5-
14 with cysteines and spin-labeling electron paramagnetic resonance, it was
32
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
concluded that the peptide forms an alpha-helix tilted approximately 25
degrees from
the horizontal plane of the membrane with a maximum depth of 15 ~r from the
phosphate group (Macosko et al., 1997). Use of fusogenic peptides from
influenza
virus hemagglutinin HA-2 enhanced greatly the efficiency of transfernn-
polylysine-
DNA complex uptake by cells. The peptide was linked to polylysine and the
complex was delivered by the transferrin receptor-mediated endocytosis
(reviewed
by Boulikas, I998a). This peptide has the sequence: GLFEAIAGFI
ENGWEGMIDG GGYC (SEQ ID N0:9) and is able to induce the release of the
fluorescent dye calcein from liposomes prepared with egg yolk
phosphatidylcholine,
which was higher at acidic pH. This peptide was also able to increase up to 10-
fold
the anti-HIV potency of antisense oligonucleotides, at a concentration of 0.1-
I mM,
using CEM-SS lymphocytes in culture. This peptide changes conformation at the
slightly more acidic environment of the endosome, destabilizing and breaking
the
endosomal membrane (reviewed by Boulikas, 1998a).
The presence of negatively charged lipids in the membrane is important for
the manifestation of the fusogenic properties of some peptides, but not of
others.
Whereas the fusogenic action of a peptide, representing a putative fusion
domain of
fertilin, a sperm surface protein involved in sperm-egg fusion, was dependent
upon
the presence of negatively charged lipids, that of the HIV2 peptide was not
(Martin
and Ruysschaert, 1997).
For example, to analyze the two domains on the fusogenic peptides of
influenza virus hemagglutinin HA, HA-chimeras were designed in which the
cytoplasmic tail andlor transmembrane domain of HA was replaced with the
corresponding domains of the fusogenic glycoprotein F of Sendai virus.
Constructs
of HA were made in which the cytoplasmic tail was replaced by peptides of
human
neurofibromin type 1 (NFI) (residues 1441 to 1518) or c-Raf 1, (residues 51 to
131)
and were expressed in CV-1 cells by using the vaccinia virus-T7 polymerase
transient-expression system. Membrane fusion between CV-1 cells and bound
human erythrocytes (RBCs) mediated by parental or chimeric HA proteins showed
that, after the pH was lowered, a flow of the aqueous fluorophore calcein from
preloaded RBCs into the cytoplasm of the protein-expressing CV-l, cells took
place.
33
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
This indicated that membrane fusion involves both leaflets of the lipid
bilayers and
leads to formation of an aqueous fusion pore (Schroth-Diaz et al., 1998).
A remarkable discovery was that the TAT protein of HIV is able to cross cell
membranes (Green and Loewenstein, 1998) and that a 36-amino acid domain of
TAT, when chemically cross-linked to heterologous proteins, conferred the
ability to
transduce into cells. The 11-amino acid fusogenic peptide of TAT
(YGRI~KRRQRRR (SEQ ID NO:10)) is a nucleolar localization signal (see
Boulikas, 1998b).
Another protein of HIV, the glycoprotein gp4l, contains fusogenic peptides.
Linear peptides derived from the membrane proximal region of the gp41
ectodomain
have potential applications as anti-HIV agents and inhibit infectivity by
adopting a
helical conformation (Judice et al., 1997). The 23 amino acid residue, N-
terminal
peptide of HIV-1 gp41 has the capacity to destabilize negatively charged large
unilamellar vesicles. In the absence of cations, the main structure was a pore-
forming alpha-helix, whereas in the presence of Ca2+ the conformation switched
to a
fusogenic, predominantly extended beta-type structure. The fusion activity of
HIV(ala) (bearing the R22~A substitution) was reduced by 70%, whereas
fusogenicity was completely abolished when a second substitution (V2-~E) was
included, arguing that it is not an alpha-helical but an extended structure
adopted by
the HIV-1 fusion peptide that actively destabilizes cholesterol-containing,
electrically neutral membranes (Pereira et al., 1997).
The prion protein (PrP) is a glycoprotein of unknown function normally
found at the surface of neurons and of glial cells. It is involved in diseases
such as
bovine spongiform encephalopathy, and Creutzfeldt-Jakob disease in humans,
where
PrP is converted into an altered form (termed PrPSc). According to computer
modeling calculations, the 120 to 133 and 118 to 135 domains of PrP are tilted
lipid-
associating peptides inserting in a oblique way into a lipid bilayer and able
to
interact with liposomes to induce leakage of encapsulated calcein (Pillot et
al.,
1997b).
The C-terminal fragments of the Alzheimer amyloid peptide (amino acids 29-
and 29-42) have properties related to those of the fusion peptides of viral
proteins
inducing fusion of liposomes in vitro. These properties could mediate a direct
34
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
interaction of the amyloid peptide with cell membranes and account for part of
the
cytotoxicity of the amyloid peptide. In view of the epidemiologic and
biochemical
linkages between the pathology of Alzheimer's disease and apolipoprotein E
(apoE)
polymorphism, examination of the potential interaction between the three
common
apoE isoforms and the C-terminal fragments of the amyloid peptide showed that
only apoE2 and apoE3, not apoE4, are potent inhibitors of the amyloid peptide
fusogenic and aggregational properties. The protective effect of apoE against
the
formation of amyloid aggregates was thought to be mediated by the formation of
stable apoE/amyloid peptide complexes (Pillot et al., 1997a; Lins et al.,
1999).
The fusogenic properties of an amphipathic net-negative peptide (WAE 11),
consisting of 11 amino acid residues were strongly promoted when the peptide
was
anchored to a liposomal membrane. The fusion activity of the peptide appeared
to
be independent of pH and membrane merging, and the target membranes required a
positive charge that was provided by incorporating lysine-coupled
phosphatidylethanolamine (PE-I~). Whereas the coupled peptide could cause
vesicle
aggregation via nonspecific electrostatic interaction with PE-I~, the free
peptide
failed to induce aggregation of PE-K vesicles (Pecheur et al., 1997).
A number of studies suggest that stabilization of an alpha-helical secondary
structure of the peptide after insertion in lipid bilayers in membranes of
cells or
liposomes is responsible for the membrane fusion properties of peptides. Zn2~,
enhances the fusogenic activity of peptides because it stabilizes the alpha-
helical
structure. For example, the HEXXH (SEQ ID NO:11) domain of the salivary
antimicrobial peptide, located in the C-terminal functional domain of histatin-
5, a
recognized zinc-binding motif is in a helicoidal conformation (Martin et al.,
1999;
Melino et al., 1999; Curtain et al., 1999).
Fusion peptides have been formulated with DNA plasmids to create peptide-
based gene delivery systems. A combination of the YKAKnWK (SEQ ID NO:12)
peptide, used to condense plasmids into 40 to 200 nm nanoparticles, with the
GLFEALLELLESLWELLLEA (SEQ ID N0:13) amphipathic peptide, that is a pH-
sensitive lytic agent designed to facilitate release of the plasmid from
endosomes
enhanced expression systems containing the beta-galactosidase reporter gene
(Duguid et al., 1998). See Table 2, below.
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
Table 2. Fusogenic peptides
Fusogenic peptideSource ProteinProperties Reference
GLFEAIAGFIENGWEG Influenza Endowed with membraneBongartz et
virus al., 1994
MIDGGGYC (SEQ hemagglutininfusion properties
ID
N0:9) HA-2
YGRKKRRQRRR (SEQ TAT of HIV Endowed with membraneGreen and
ID NO:S) fusion properties Loewenstein,
1988
23-residue fusogenicHIV-1 trans-Was able to insertCurtain et
N- as an al., 1999
terminal peptide membrane alpha-helix into
neutral
glycoproteinphospholipid bilayers
gp41
70 residue peptideFusion peptideInduced lipid mixingGhosh and Shai,
(SV- of egg
117) and N-terminalphosphatidylcholine-1999
heptad repeatphosphatidyiglyceroI
of
Sendai virus(PC/PG) large unilamellar
vesicles (LUVs)
23 hydrophobic S protein A high degree of Rodriguez-Crespo
amino of similarity et
acids in the amino-terminalhepatitis with known fusogenical., 1994
B virus
region (HBV) peptides from other
viruses.
MSGTFGGILAGLIGLL N-terminal Was inserted into Rodriguez-Crespo
region the et
(SEQ ID N0:6) of the S hydrophobic core al., 1999
protein of the
of
duck hepatitislipid bilayer and
B induced
Virus (DHBV)leakage of internal
aqueous
contents from both
neutral
and negatively
charged
liposomes
MSPSSLLGLLAGLQVV S protein Was inserted into Rodriguez-Crespo
of the et~
(SEQ ID N0:14) woodchuck hydrophobic core al., 1999
of the
hepatitis lipid bilayer and
B virus induced
(WHV) leakage of internal
aqueous
contents from both
neutral
and negatively
charged
liposomes
N-terminus of Nef proteinMembrane-perturbingMacreadie et
Nef of and al.,
human fusogenic activities1997
in
immunodeficiencartificial membranes;
causes
y type 1 cell killing in
(HIV-1) E. coli and
yeast
Amino-terminal F1 polypeptideCan be used as Partidos et
sequence of a carrier al., 1996
F1 polypeptide measles system for CTL
virus epitopes
(MV)
19-27 amino acid GlycoproteinAdopts an amphiphilicVoneche et
segment al., 1992
gp51 of structure and plays
bovine a key
leukemia role in the fusion
virus events
induced by bovine
leukemia
virus
120 to 133 and Prion proteinTilted lipid-associatingPillot et al.,
118 to 135 1997b
domains peptide; interact
with
liposomes to induce
leakage
of encapsulated
calcein
29-42-residue Alzheimer'sEndowed with capacitiesLins et al.,
fragment beta- 1999
amyloid resembling those
peptide of the
tilted fragment
of viral
fusion proteins
Non-aggregated Alzheimer'sInduces apoptotic Pillot et al.,
amyloid beta- neuronal 1999
beta-peptide (1-40)amyloid cell death
peptide
36
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
Fusogenic peptide~ Source Properties Reference
Protein
LCAT 56-68 helicalLecithin Forms stable beta-sheetsPeelman et
in al., 1999;
segment cholesterollipids Decout et al.,
1999
acyltransferase
(LCAT)
Peptide sequence Membrane- Triggers fusion Ulrich et al.,
B 18 between 1999
associated lipid vesicles;
sea a histidine-
urchin spermrich motif for
binding zinc
protein is required for
binding the fusogenic
function
53-70 (C-terminalApolipoproteinInduces fusion Lambert et
helix) of al., 1998
(apo) All unilamellar lipid
vesicles
and displaces apo
AI from
HDL and r-HDL
Residues 90-111 PH-30 alphaMembrane-fusogenicNiidome et
(a al., 1997
protein activity to acidic
functioningphospholipid bilayers
in
sperm-egg
fusion)
Casein signal Alpha s2- Interact with Creuzenet et
peptides and al., 1997
beta-caseindimyristoylphosphatidyl-
glycerol and -choline
liposomes; show
both lytic
and fusogenic activities
Pardaxin AmphipathicForms voltage-gated,Lelkes and
polypeptide,cation-selective Lazarovici,
pores; 1988
purified mediated the aggregation
from the of
gland secretionliposomes composed
of of
the Red phosphatidylserine
Sea but not
Moses sole of phosphatidylcholine
flatfish
Pardachirus
marmoratus
Histatin-5 Salivary Aggregates and Melino et al.,
fuses 1999
antimicrobialnegatively charged
small
peptide unilamellar vesicles
in the
presence of Zn2+
Gramicidin (linearAntibiotic Induces aggregationMassari and
and Colonna,
hydrophobic polypeptide) fusion of vesicles1986; Tournois
et
al., 1990
Amphipathic negativelySynthetic Forms an alpha-helixMartin et al.,
1999
charged peptide inserted and anchored
consisting into
of 11 residues the membrane (favored
(WAE) at
37oC) oriented
almost
parallel to the
lipid aryl
chains; promotes
fusion of
large unilamellar
liposomes
(LUV)
A polymer of polylysineSynthetic Histidyl residues Midoux and
become
(average 190) cationic upon protonationMonsigny, 1999
partially of
substituted with the imidazole groups
histidyl at pH
residues below 6Ø; disrupt
endosomal membranes
GLFEALLELLESLWELLSynthetic Amphipathic peptide;Duguid et al.,
a pH- 1998
LEA (SEQ ID N0:4) sensitive lytic
agent to
facilitate release'
of the
plasmid from endosomes
37
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
Fusogenic peptideSource ProteinProperties Reference
(LKKL)4 (SEQ ID Synthetic Amphiphilic fusogenicGupta and Kothekar,
NO:15)
peptide, able to 1997
interact with
four molecules
of DMPC
Ac-(Leu-Ala-Arg-Leu)3-Synthetic; Caused a leakage Suenaga et
basic of al., 1989;
NHCH3 (SEQ ID amphipathiccontents from smallLee et al.,
N0:16) 1992
peptides unilamellar vesicles
composed of egg
yollc
phosphatidylcholine
and egg
yolk phosphatidic
acid (3:1)
Amphiphilic anionicSynthetic Can mimic the fusogenicMurata et al.,
1991
peptides ES and activity of influenza
ESL
hemagglutinin (HA)
30-amino acid Synthetic; Becomes an amphipathicParente et
peptide with al., 1988
the major repeat designed alpha-helix as
unit Glu- to mimic the pH is
Ala-Leu-Ala (GALA)the behaviorlowered to 5.0
of ; fusion of
(SEQ ID N0:17) the fusogenicphosphatidylcholine
small
sequences unilamellar vesicles
of viral induced
fusion proteinsby GALA requires
a peptide
length greater
than 16 amino
acids
Poly Glu-Aib-Leu-AibSynthetic Amphiphilic structureICono et al.,
upon 1993
(SEQ ID N0:18) the formation of
Aib alpha-
represents 2- helix; caused fusion
of
aminoisobutyric EYPC liposomes
acid and
dipalmitoylphosphatidylchol
ine liposomes more
strongly
with decreasing
pH
Fusogenic lipids
DOPE is a fusogenic lipid; elastase cleavage of N-methoxy-succinyl-Ala-
Ala-Pro-Val-DOPE (SEQ ID NO:19) converted this derivative to DOPE (overall
positive charge) to deliver an encapsulated fluorescent probe, calcein, into
the cell
cytoplasm (Pak et al., 1999). An oligodeoxynucleic sequence of 30 bases
complementary to a region of beta-endorphin mRNA elicited a concentration-
dependent inhibition of beta-endorphin production in cell culture after it was
encapsulated within small unilamellar vesicles (50 nm) containing dipalmitoyl-
DL-
alpha-phosphatidyl-L-serine endowed with fusogenic properties (Fresta et al.,
1998).
Nuclear localization signals (NLS)
In a further embodiment, the liposome encapsulated plasmid or
oligonucleotide DNA described herein further comprise an effective amount of
nuclear localization signal (NLS) peptides. Trafficking of nuclear proteins
from the
site of their synthesis in the cytoplasm to the sites of function in the
nucleus through
38
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
pore complexes is mediated by NLSs on proteins to be imported into nuclei
(Tables
3-10, below). Protein translocation from the cytoplasm to the nucleoplasm
involves:
(i) the formation of a complex of karyopherin a with NLS-protein; (ii)
subsequent
binding of karyopherin (3; (iii) binding of the complex to FXFG peptide
repeats on
nucleoporins; (iv) docking of Ran-GDP to nucleoporin and to karyopherin
heterodimer by p 10; (v) a number of association-dissociation reactions on
nucleoporins that dock the import substrate toward the nucleoplasmic side with
a
concomitant GDP-GTP exchange reaction transforming Ran-GDP into Ran-GTP and
catalyzed by karyopherin a; and (vi) dissociation from karyopherin (3 and
release of
the karyopherin a/NLS-protein by Ran-GTP to the nucleoplasm.
Karyophilic and acidic clusters were found in most non-membrane
serine/threonine protein kinases whose primary structure has been examined
(Table
6). These karyophilic clusters might mediate the anchoring of the kinase
molecules
to transporter proteins for their regulated nuclear import and might
constitute the
nuclear localization signals. In contrast to protein transcription factors
that are
exclusively nuclear possessing strong karyophilic peptides composed of at
least four
arginines, (R), and lysines, (K), within an hexapeptide flanked by proline and
glycine helix-breakers, protein kinases often contain one histidine and three
K+R
residues (Boulikas, 1996). This was proposed to specify a weak NLS structure
resulting in the nuclear import of a fraction of the total cytoplasmic kinase
molecules, as well as in their weak retention in the different ionic strength
nuclear
environment. Putative NLS peptides in protein kinases may also contain
hydrophobic or bulky aromatic amino acids proposed to further diminish their
capacity to act as strong NLS.
Most mammalian proteins that participate in DNA repair pathways seem to
possess strong karyophilic clusters containing at least four R+K over a
stretch of six
amino acids (Table 7).
Rules to predict nuclear localization of an unknown protein
Several simple rules have been proposed for the prediction of the nuclear
localization of a protein of an unknown function from its amino acid sequence:
39
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
(i) An NLS is defined as four arginines (R) plus lysines (K) within an
hexapeptide; the presence of one or more histidines (H) in the tetrad of the
karyophilic hexapeptide, often found in protein kinases that have a
cytoplasmic and a
nuclear function, may specify a weak NLS whose function might be regulated by
phosphorylation or may specify proteins that function in both the cytoplasm
and the
nucleus (Boulikas, 1996);
(ii) The K/R clusters are flanked by the a-helix breakers G and P thus placing
the NLS at a helix-turn-helix or end of a a-helix. Negatively-charged amino
acids
(D, E) are often found at the flank of the NLS and on some occasions may
interrupt
the positively-charged NLS cluster;
(iii) Bulky amino acids (W, F, Y) are not present within the NLS
hexapeptide;
(iv) NLS signals may not be flanked by long stretches of hydrophobic amino
acids (e.g. five); a mixture of charged and hydrophobic amino acids serves as
a
mitochondria) targeting signal;
(v) The higher the number of NLSs, the more readily a molecule is imported
to the nucleus (Dworetzky et al., 1988). Even small proteins, for example
histones
(10-22 kDa), need to be actively imported to increase their import rates
compared
with the slow rate of diffusion of small molecules through pores;
(vi) Signal peptides are stronger determinants than NLSs for protein
trafficking. Signal peptides direct proteins to the lumen of the endoplasmic
reticulum for their secretion or insertion into cellular membranes (presence
of
transmembrane domains) (Boulikas, 1994);
(vii) Signals for the mitochondria) import of proteins (a mixture of
hydrophobic and karyophilic amino acids) may antagonize nuclear import signals
and proteins possessing both type of signals may be translocated to both
mitochondria and nuclei;
(viii) Strong association of a protein with large cytoplasmic structures
(membrane proteins, intermediate filaments) make such proteins unavailable for
import even though they posses NLS-like peptides (Boulikas, 1994);
(ix). Transcription factors and other nuclear proteins posses a great
different
number of putative NLS stretches. Of the sixteen possible forms of putative
NLS
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
structures the most abundant types are the 90x60, A00x9, ease, and Sex~x0,
where
B is R or K, together accounting for about 70% of all karyophilic clusters on
transcription factors (Boulikas, 1994);
(x) A small number of nuclear proteins seem to be void of a typical
karyophilic NLS. Either non karyophilic peptides function for their nuclear
import,
as such molecules possess bipartite NLSs, or these NLS-less proteins depend
absolutely for import on their strong complexation in the cytoplasm with a
nuclear
protein partner able to be imported (Boulikas, 1994). This mechanism may
ensure a
certain stoichiometric ratio of the two molecules in the nucleus, and might be
of
physiological significance; and
(xi) A number of proteins may be imported via other mechanisms not
dependent on classical NLS.
A number of processes have been found to be regulated by nuclear import
including nuclear translocation of the transcription factors NF-xB, rNFIL-6,
ISGF3,
SRF, c-Fos, GR as well as human cyclins A and B1, casein kinase II, cAMP-
dependent protein kinase II, protein kinase C, ERI~1 and ERI~2. Failure of
cells to
import specific proteins into nuclei can lead to carcinogenesis. For example,
BRCAl is mainly localized in the cytoplasm in breast and ovarian cancer cells,
whereas in normal cells the protein is nuclear. mRNA is exported through the
same
route as a complex with nuclear proteins possessing nuclear export signals
(NES).
The majority of proteins with NES are RNA-binding proteins that bind to and
escort
RNAs to the cytoplasm. However, other proteins with NES function in the export
of
proteins; CRM1, that binds to the NES sequence on other proteins and interacts
with
the nuclear pore complex, is an essential mediator of the NES-dependent
nuclear
export of proteins in eukaryotic cells. Nuclear localization and export
signals (NLS
and NES) are found on a number of important molecules, including p53, v-Rel,
the
transcription factor NF-ATc, the c-Abl nonreceptor tyrosine kinase, and the
fragile X
syndrome mental retardation gene product. The deregulation of their normal
import/export trafficking has important implications for human disease. Both
nuclear import and export processes can be manipulated by conjugation of
proteins
with NLS or NES peptides. During gene therapy, the foreign DNA needs to enter
nuclei for its transcription. A pathway is proposed involving the complexation
of
41
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
plasmids and oligonucleotides with nascent nuclear proteins possessing NLSs as
a
prerequisite for their nuclear import. Covalent linkage of NLS peptides to
oligonucleotides and plasmids or formation of complexes of plasmids with
proteins
possessing multiple NLS peptides was proposed (Boulikas, 1998b) to increase
their
import rates and the efficiency of gene expression. Cancer cells were
predicted to
import more efficiently foreign DNA into nuclei, compared with terminally
differentiated cells because of their increased rates of proliferation and
protein
import.
Antineoplastic drubs
In a further embodiment, the liposome encapsulated plasmid or
oligonucleotide DNA described herein, further comprises its use for reducing
tumor
size or restricting its growth with combination with encapsulated or free
antineoplastic agents. Antineoplastic agents preferably are: (i) alkylating
agents
having the bis-(2-chloroethyl)-amine group such as chlormethine,
chlorambucile,
melphalan, uramustine, mannomustine, extramustinephosphat,
mechlorethaminoxide, cyclophosphamide, ifosfamide, or trifosfamide; (ii)
alkylating
agents having a substituted aziridine group, for example tretamine, thiotepa,
triaziquone, or mitomycine; (iii) alkylating agents of the methanesulfonic
ester type
such as busulfane; (iv) alkylating N-alkyl-N-nitrosourea derivatives, for
example
carmustine, lomustine, semustine, or streptozotocine; (v) alkylating agents of
the
mitobronitole, dacarbazine, or procarbazine type; (vi) complexing agents such
as cis-
platin; (vii) antimetabolites of the folic acid type, for example
methotrexate; (viii)
purine derivatives such as mercaptopurine, thioguanine, azathioprine,
tiamiprine,
vidarabine, or puromycine and purine nucleoside phosphorylase inhibitors; (ix)
pyrimidine derivatives, for example fluorouracil, floxuridine, tegafur,
cytarabine,
idoxuridine, flucytosine; (x) antibiotics such as dactinomycin, daunorubicin,
doxorubicin, mithramycin, bleomycin or etoposide; (xi) vinca alkaloids; (xii)
inhibitors of proteins overexpressed in cancer cells such as telomerase
inhibitors,
glutathione inhibitors, proteasome inhibitors; (xiii) modulators or inhibitors
of signal
transduction pathways such as phosphatase inhibitors, protein kinase C
inhibitors,
casein kinase inhibitors, insulin-like growth factor-1 receptor inhibitor, ras
42
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
inhibitors, ras-GAP inhibitor, protein tyrosine phosphatase inhibitors; (xiv)
tumor
angiogenesis inhibitors such as angiostatin, oncostatin, endostatin,
thalidomide; (xv)
modulators of the immune response and cytokines such as interferons,
interleukins,
TNF-alpha; (xvi) modulators of the extracellular matrix such as matrix
metalloproteinase inhibitors, stromelysin inhibitors, plasminogen activator
inhibitor;
(xvii) hormone modulators for hormone-dependent cancers (breast cancer,
prostate
cancer) such as antiandrogen, estrogens; (xviii) apoptosis regulators; (xix)
bFGF
inhibitor; (xx) multiple drug resistance gene inhibitor; (xxi) monoclonal
antibodies
or antibody fragments against antigenes overexpressed in cancer cells (anti-
Her2/neu
for breast cancer); (xxii) anticancer genes whose expression will cause
apoptosis,
arrest the cell cycle, induce an immune response against cancer cells, inhibit
tumor
angiogenesis i.e. formation of blood vessels, tumor suppressor genes (p53, RB,
BRCA1, ElA, bcl-2, MDR-1, p21, p16, bax, bcl-xs, E2F, IGF-I VEGF, angiostatin,
oncostatin, endostatin, GM-CSF, IL-12, IL-2, IL-4, IL-7, IFN-y, and TNF-a);
and
(xxiii) antisense oligonucleotides (antisense c-fos, c-myc, K-ras). Optionally
these
drugs are administered in combination with chlormethamine, prednisolone,
prednisone, or procarbazine or combined with radiation therapy. Future new
anticancer drugs added to the arsenal are expected to be ribozymes, triplex-
forming
oligonucleotides, gene inactivating oligonucleotides, a number of new genes
directed
against genes that control the cell proliferation or signaling pathways, and
compounds that block signal transduction.
Anti-cancer drugs include: acivicin, aclarubicin, acodazole hydrochloride,
acronine, adozelesin, adriamycin, aldesleukin, altretamine, ambomycin,
ametantrone
acetate, aminoglutethimide, amsacrine, anastrozole, anthramycin, asparaginase,
asperlin, azacitidine, azetepa, azotomycin, batimastat, benzodepa,
bicalutamide,
bisantrene hydrochloride, bisnafide dimesylate, bizelesin, bleomycin sulfate,
brequinar sodium, bropirimine, busulfan, cactinomycin, calusterone,
caracemide,
carbetimer, carboplatin, carmustine, carubicin hydrochloride, carzelesin,
cedefingol,
chlorambucil, cirolemycin, cisplatin, cladribine, crisnatol mesylate,
cyclophosphamide, cytarabine, dacarbazine, dactinomycin, daunorubicin
hydrochloride, decitabine, dexormaplatin, dezaguanine, dezaguanine mesylate,
diaziquone, docetaxel, doxorubicin, doxorubicin hydrochloride, droloxifene,
43
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
droloxifene citrate, dromostanolone propionate, duazomycin, edatrexate,
eflornithine
hydrochloride, elsamitrucin, enloplatin, enpromate, epipropidine, epirubicin
hydrochloride, erbulozole, esorubicin hydrochloride, estramustine,
estramustine
phosphate sodium, etanidazole, etoposide, etoposide phosphate, etoprine,
fadrozole
hydrochloride, fazarabine, fenretinide, floxuridine, fludarabine phosphate,
fluorouracil, flurocitabine, fosquidone, fostriecin sodium, gemcitabine,
gemcitabine
hydrochloride, hydroxyurea, idarubicin hydrochloride, ifosfamide, ilmofosine,
interferon alfa-2a, interferon a-2b, interferon a-nl, interferon a-n3,
interferon (3-i a,
interferon y-i b, iproplatin, irinotecan hydrochloride, lanreotide acetate,
letrozole,
leuprolide acetate, liarozole hydrochloride, lometrexol sodium, lomustine,
losoxantrone hydrochloride, masoprocol, maytansine, mechlorethamine
hydrochloride, megestrol acetate, melengestrol acetate, melphalan, menogaril,
mercaptopurine, methotrexate, methotrexate sodium, metoprine, meturedepa,
mitindomide, mitocarcin, mitocromin, mitogillin, mitomalcin, mitomycin,
mitosper,
mitotane, mitoxantrone hydrochloride, mycophenolic acid, nocodazole,
nogalamycin, ormaplatin, oxisuran, paclitaxel, pegaspargase, peliomycin,
pentamustine, peplomycin sulfate, perfosfamide, pipobroman, piposulfan,
piroxantrone hydrochloride, plicamycin, plomestane, porfimer sodium,
porfiromycin, prednimustine, prednisone, procarbazine hydrochloride,
puromycin,
puromycin hydrochloride, pyrazofurin, riboprine, rogletimide, safingol,
safingol
hydrochloride, semustine, simtrazene, sparfosate sodium, sparsomycin,
spirogermanium hydrochloride, spiromustine, spiroplatin, streptonigrin,
streptozocin,
sulofenur, talisomycin, taxol, tecogalan sodium, tegafur, teloxantrone
hydrochloride,
temoporfm, teniposide, teroxirone, testolactone, thiamiprine, thioguanine,
thiotepa,
tiazofurin, tirapazamine, topotecan hydrochloride, toremifene citrate,
trestolone
acetate, triciribine phosphate, trimetrexate, trimetrexate glucuronate,
triptorelin,
tubulozole hydrochloride, uracil mustard, uredepa, vapreotide, verteporfin,
vinblastine sulfate, vincristine sulfate, vindesine, vindesine sulfate,
vinepidine
sulfate, vinglycinate sulfate, vinleurosine sulfate, vinorelbine tartrate,
vinrosidine
sulfate, vinzolidine sulfate, vorozole, zeniplatin, zinostatin, zorubicin
hydrochloride.
Other anti-cancer drugs include: 20-epi-1,25 dihydroxyvitamin D3, 5-
ethynyluracil, abiraterone, aclarubicin, acylfulvene, adecypenol, adozelesin,
44
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
aldesleukin, ALL-TK antagonists, altretamine, ambamustine, amidox, amifostine,
aminolevulinic acid, amrubicin, amsacrine, anagrelide, anastrozole,
andrographolide,
angiogenesis inhibitors, antagonist D, antagonist G, antarelix, anti-
dorsalizing
morphogenetic protein-1, antiandrogen, antiestrogen, antineoplaston, antisense
oligonucleotides, aphidicolin glycinate, apoptosis gene modulators, apoptosis
regulators, apurinic acid, ara-CDP-DL-PTBA, arginine deaminase, asulacrine,
atamestane, atrimustine, axinastatin 1, axinastatin 2, axinastatin 3,
azasetron,
azatoxin, azatyrosine, baccatin III derivatives, balanol, batimastat, BCR/ABL
antagonists, benzochlorins, benzoylstaurosporine, beta lactam derivatives,
beta-
alethine, betaclamycin B, betulinic acid, bFGF inhibitor, bicalutamide,
bisantrene,
bisaziridinylspermine, bisnafide, bistratene A, bizelesin, breflate,
bropirimine,
budotitane, buthionine sulfoximine, calcipotriol, calphostin C, camptothecin
derivatives, canarypox IL-2, capecitabine, carboxamide-amino-triazole,
carboxyamidotriazole, CaRest M3, CARN 700, cartilage derived inhibitor,
carzelesin, casein kinase inhibitors (ICOS), castanospermine, cecropin B,
cetrorelix,
chlorlns, chloroquinoxaline sulfonamide, cicaprost, cis-porphyrin, cladribine,
clomifene analogues, clotrimazole, collismycin A, collismycin B,
combretastatin A4,
combretastatin analogue, conagenin, crambescidin 816, crisnatol, cryptophycin
8,
cryptophycin A derivatives, curacin A, cyclopentanthraquinones, cycloplatam,
cypemycin, cytarabine ocfosfate, cytolytic factor, cytostatin, dacliximab,
decitabine,
dehydrodidemnin B, deslorelin, dexifosfamide, dexrazoxane, dexverapamil,
diaziquone, didemnin B, didox, diethylnorspermine, dihydro-S-azacytidine,
dihydrotaxol, 9-dioxamycin, diphenyl spiromustine, docosanol, dolasetron,
doxifluridine, droloxifene, dronabinol, duocarmycin SA, ebselen, ecomustine,
edelfosine, edrecolomab, eflornithine, elemene, emitefur, epirubicin,
epristeride,
estramustine analogue, estrogen agonists, estiogen antagonists, etanidazole,
etoposide phosphate, exemestane, fadrozole, fazarabine, fenretinide,
filgrastim,
finasteride, flavopiridol, flezelastine, fluasterone, fludarabine,
fluorodaunorunicin
hydrochloride, forfenimex, formestane, fostriecin, fotemustine, gadolinium
gallium
nitrate texaphyrin, galocitabine, ganirelix, gelatinase inhibitors,
gemcitabine,
glutathione inhibitors, hepsulfam, heregulin, hexamethylene bisacetamide,
hypericin,
ibandronic acid, idarubicin, idoxifene, idramantone, ilmofosine, ilomastat,
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
imidazoacridones, imiquimod, immunostimulant peptides, insulin-like growth
factor-
1 receptor,inhibitor, interferon agonists, interferons, interleukins,
iobenguane,
iododoxorubicin, ipomeanol, 4-, irinotecan, iroplact, irsogladine,
isobengazole,
isohomohalicondrin B, itasetron, jasplakinolide, kahalalide F, lamellarin-N
triacetate, lanreotide, leinamycin, lenograstim, lentinan sulfate,
leptolstatin,
letrozole, leukemia inhibiting factor, leukocyte alpha interferon,
leuprolide+estrogen+progesterone, leuprorelin, levamisole, liarozole, linear
polyamine analogue, lipophilic disaccharide peptide, lipophilic platinum
compounds,
lissoclinamide 7, lobaplatin, lombricine, lometrexol, lonidamine,
losoxantrone,
lovastatin, loxoribine, lurtotecan, lutetium texaphyrin, lysofylline, lytic
peptides,
maitansine, mannostatin A, marimastat, masoprocol, maspin, matrilysin
inhibitors,
matrix metalloproteinase inhibitors, menogaril, merbarone, meterelin,
methioninase,
metoclopramide, MIF inhibitor, mifepristone, miltefosine, mirimostim,
mismatched
double stranded RNA, mitoguazone, mitolactol, mitomycin analogues, mitonafide,
mitotoxin fibroblast growth factor-saporin, mitoxantrone, mofarotene,
molgramostim, monoclonal antibody, human chorionic gonadotrophin,
monophosphoryl Iipid A+myobacterium cell wall sk, mopidamol, multiple drug
resistance gene inhibitor, multiple tumor suppressor 1-based therapy, mustard
anticancer agent, mycaperoxide B, mycobacterial cell wall extract,
myriaporone, N-
acetyldinaline, N-substituted benzamides, nafarelin, nagrestip, naloxone
+pentazocine, napavin, naphterpin, nartograstim, nedaplatin, nemorubicin,
neridronic acid, neutral endopeptidase, nilutamide, nisamycin, nitric oxide
modulators, nitroxide antioxidant, nitrullyn, 06-benzylguanine, octreotide,
okicenone, oligonucleotides, onapristone, ondansetron, ondansetron, oracin,
oral
cytokine inducer, ormaplatin, osaterone, oxaliplatin, oxaunomycin, paclitaxel
analogues, paclitaxel derivatives, palauamine, palmitoylrhizoxin, pamidronic
acid,
panaxytriol, panomifene, parabactin, pazelliptine, pegaspargase, peldesine,
pentosan
polysulfate sodium, pentostatin, pentrozole, perflubron, perfosfamide,
perillyl
alcohol, phenazinomycin, phenylacetate, phosphatase inhibitors, picibanil,
pilocarpine hydrochloride, pirarubicin, piritrexim, placetin A, placetin B,
plasminogen activator inhibitor, platinum complex, platinum compounds,
platinum-
triamine complex, porfimer sodium, porfiromycin, propyl bis-acridone,
46
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
prostaglandin J2, proteasome inhibitors, protein A-based immune modulator,
protein
kinase C inhibitor, protein kinase C inhibitors, microalgal., protein tyrosine
phosphatase inhibitors, purine nucleoside phosphorylase inhibitors, purpurins,
pyrazoloacridine, pyridoxylated hemoglobin polyoxyethylene conjugate, raf
antagonists, raltitrexed, ramosetron, ras farnesyl protein transferase
inhibitors, ras
inhibitors, ras-GAP inhibitor, retelliptine demethylated, rhenium Re 186
etidronate,
rhizoxin, ribozymes, RII retinamide, rogletimide, rohitukine, romurtide,
roquinimex,
rubiginone B 1, ruboxyl, safingol, saintopin, SaarCNU, sarcophytol A,
sargramostim,
Sdi 1 mimetics, semustine, senescence derived inhibitor l, sense
oligonucleotides,
signal transduction inhibitors, signal transduction modulators, single chain
antigen
binding protein, sizofiran, sobuzoxane, sodium borocaptate, sodium
phenylacetate,
solverol, somatomedin binding protein, sonermin, sparfosic acid, spicamycin D,
spiromustine, splenopentin, spongistatin 1, squalamine, stem cell inhibitor,
stem-cell
division inhibitors, stipiamide, stromelysin inhibitors, sulfmosine,
superactive
vasoactive intestinal peptide antagonist, suradista, suramin, swainsonine,
synthetic
glycosaminoglycans, tallimustine, tamoxifen methiodide, tauromustine,
tazarotene,
tecogalan sodium, tegafur, tellurapyrylium, telomerase inhibitors, temoporfin,
temozolomide, teniposide, tetrachlorodecaoxide, tetrazomine, thaliblastine,
thalidomide, thiocoraline, thrombopoietin, thrombopoietin mimetic,
thymalfasin,
thymopoietin receptor agonist, thymotrinan, thyroid stimulating hormone, tin
ethyl
etiopurpurin, tirapazamine, titanocene dichloride, topotecan, topsentin,
toremifene,
totipotent stem cell factor, translation inhibitors, tretinoin,
triacetyluridine,
triciribine, trimetrexate, triptorelin, tropisetron, turosteride, tyrosine
kinase
inhibitors, tyrphostins, UBC inhibitors, ubenimex, urogenital sinus-derived
growth
inhibitory factor, urokinase receptor antagonists, vapreotide, variolin B,
velaresol,
veramine, verdins, verteporfin, vinorelbine, vinxaltine, vitaxin, vorozole,
zanoterone,
zeniplatin, zilascorb, zinostatin stimalamer.
pH-sensitive peptide-DNA complexes
In a further embodiment of the invention, the genes in plasmid DNA are
brought in interaction with fusogenic peptide/NLS conjugates. In a further
embodiment the NLS moiety is a stretch of histidyl residues able to assume a
net
47
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
positive charge at a pH of about 5 to 6 and to show a reduction or loose
completely
this charge at pH above 7. The electrostatic interaction of these positively-
charged
peptides with the negatively-charged plasmid DNA molecules, established at pH
5-6
is weakened at physiological pH (pH-sensitive peptide-DNA complexes).
The first step of the present invention involves complex formation between
the plasmid or oligonucleotide DNA with the histidyl/fusogenic peptide
conjugate
and lipid components in 10-90% ethanol at pH 5.0 to 6Ø The conditions must
be
where the histidyl residues have a net positive charge and can establish
electrostatic
interactions with plasmids, oligonucleotides or negatively-charged drugs. At
the
same time, the presence of the positively-charged lipid molecules promotes
formation of micelles. At the second step, micelles are converted into
liposomes by
dilution with water and mixing with pre-made liposomes or lipids at pH 5-6.
This. is
followed by dialysis against pH 7 and extrusion through membranes, entrapping
and
encapsulating plasmids or oligonucleotides to with a very high yield.
Whereas the composition of peptides and cationic lipids in the first step
provides the lipids of the internal bilayer, the type of liposomes or lipids
added at
step 2 provide the external coating of the final liposome formulation (Figure
1 ).
Examples for the formulations of peptides include: HHHHHSPSL16 (SEQ ID
N0:623), and HHHHHSPS(LAI)5 (SEQ ID N0:624).
These are added at a 1:0.5:0.5 molar ratio (negative charge on DNA: cationic
liposome: histidine peptide). The peptide inserts in an alpha-helical
conformation
inside the lipid bilayer and not only carries out DNA condensation but also
endows
membrane fusion properties to the complex to improve entrance across the cell
membrane. The type of hydrophobic amino acids (for example, content in
aromatic
amino acids), in the peptide chain is very important as is the length of the
peptide
chain in ensuring integrity and rigidity of the complexes. Coating the outer
surface
of the complexes with polyethyleneglycol, hyaluronic acids and other polymers
conjugated to lipids gives the particles long circulation properties in body
fluids and
the ability to target solid tumors and their metastases after intravenous inj
ection, and
also the ability to cross the tumor cell membrane.
4~
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
Protease-sensitive linkages in peptides between the NLS and fusogenic
moieties
Conversion of micelles into liposomes
An important issue of the present invention is the conversion of micelles
formed between the DNA and the cationic lipids, in the presence of ethanol,
into
liposomes. This is done by the direct addition of the micelle complex into an
aqueous solution of preformed liposomes. The liposomes have an average size of
80-160 nm or vice versa, leading to a solution of a final ethanol
concentration below
10%. A formulation suitable for pharmaceutical use and for injection into
humans
and animals will require that the liposomes are of neutral composition (such
as
cholesterol, PE, PC) coated with PEG.
However, another important aspect is the research application of the present
invention, such as for transfection of cells in culture. The composition of
the
aqueous solution of liposomes is any type of liposomes containing cationic
lipids
and suitable therefore for transfection of cells in culture such as DDAB:DOPE
1:1.
These liposomes are pre-formed and downsized by sonication or extrusion
through
membranes to a diameter of 80-160 nm. The ethanolic micelle preparations are
then
added to the aqueous solution of liposomes with a concomitant dilution of the
ethanol solution to below 10%. This step will result in further condensation
of DNA
or interaction of the negatively-charged phosphate groups on DNA with
positively
charged groups on lipids. Care must be taken so as only part of the negative
charges
on DNA are neutralized by lipids in the micelle. The remaining charge
neutralization of the DNA is to be provided by the cationic component of the
preformed liposomes in the second step.
Regulatory DNA and nuclear matrix-attached DNA
In a further embodiment of the present invention, the genes in plasmid DNA
are driven by regulatory DNA sequences isolated from nuclear matrix-attached
DNA
using shotgun selection approaches.
' The compact structural organization of chromatin and the proper spatial
orientation of individual chromosomes within a cell are partially provided by
the
nuclear matrix. The nuclear matrix is composed of DNA, RNA and proteins and
49
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
serves as the site of DNA replication, gene transcription, DNA repair, and
chromosomal attachment in the nucleus. Diverse sets of DNA sequences have been
found associated with nuclear matrices and is referred to as matrix attachment
regions or MARS. The MARs serve many functions, acting as activators of gene
transcription, silencers of gene expression, insulators of transcriptional
activity,
nuclear retention signals and origins of DNA replication. Current studies
indicate
that different subsets of MARS are found in different tissue types and may
assist in
regulating the specific functions of cells. The presence of this complex
assortment
of structural and regulatory molecules in the matrix, as well as the in situ
localization
of DNA replication and transcription complexes to the matrix strongly suggest
that
the nuclear matrix plays a fundamental, unique role in nuclear processes. The
structuring of genomes into domains has a functional significance. The
inclusion of
specific MAR elements within gene transfer vectors could have utility in many
experimental and gene therapy applications. Many gene therapy applications
require
specific expression of one or more genes in targeted cell types for prolonged
time
periods. MARS within vectors could enhance transcription of the introduced
transgene, prolong the retention of that sequence within the nucleus or
insulate
expression of that transgene from the expression of a cotransduced gene
(reviewed
by Boulikas, 1995; Bode et al, 1996).
Various biochemical procedures have been used to identify regulatory
regions within genes. Traditionally, identification and selection of
regulatory DNA
sequences depend on tedious procedures such as transcription factor
footprinting in
vitro or in vivo, or subcloning of smaller fragments from larger genomic DNA
sequences upstream of reporter genes. These methods have been used primarily
to
identify regions proximal to the 5' end of genes. However, in many instances,
regulatory regions are found at considerable distances from the proximal 5'
end of
the gene, and confer cell type- or developmental stage- specificity. For
example,
studies from the groups of Grosveld and Engel (Lakshmanan et al., 1999) have
shown that over 625 kb of genomic sequences surrounding the GATA-3 locus are
required for the correct developmental expression of the gene in transgenic
mice.
Extensive DNA stretches at distances 5-20 kb upstream of the gene were found
to be
responsible for the central nervous system-specificity of expression. The
region
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
between 20 to 130 kb upstream of the gene harbored regulatory regions for
urogenital-specific expression of GATA-3, whereas sequences 90-180 kb
downstream of the gene conferred endocardial-specific expression.
The presently disclosed method has the potential of rapidly identifying
regulatory control regions. In cells, chromatin loops are formed and different
attachment regions are used in different cell types or stages of development
to
modulate the expression of a gene. The presently disclosed method for
isolating
regulatory regions based on their attachment to the nuclear matrix can
identify
regulatory regions irrespective of their distance from the gene. Although the
human
genome project is expected to be almost complete by the year 2000, information
on
the location and nature of the vast majority of the estimated 500,000
regulatory
regions will not be available.
Example 1
Plasmid DNA condenses with various agents, as well as various formulations
of cationic liposomes. The condensation affects the level of expression of the
reporter beta-galactosidase gene after transfection of K562 human
erythroleukemia
cell cultures. Liposome compositions are shown in the Table below and in FIG.
2.
All lipids were from Avanti Polar Lipids (700 Industrial Park Drive,
Alabaster, AL
35007). The optimal ratio of lipid to DNA was 7 nmoles total lipid/~,g DNA.
The
transfection reagent (10 ~g DNA mixed with 70 nmoles total lipid) was
transferred
to a small culture flask followed by the addition of 10 ml K562 cell culture
(about 2
million cells total); mixing of cells with the transfection reagent was at 5-
10 min
after mixing DNA with liposomes. Cells were assayed for beta-galactosidase
activity several times at 1-30 days post-transfection. The transfected cells
were
maintained in cell culture as normal cell cultures.
Best results were obtained when the cells used for transfection were at low
number, not near confluence. In all experiments the transfection material was
added
directly in the presence of serum and antibiotics without removal of the
transfection
reagent or washings of the cells. This simplifies the transfection procedure
and is
suitable for lymphoid and other type of cell cultures that do not attach to
the dish, but
grow in suspension. All DNA condensing agents were purchased from Sigma. They
51
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
were suspended at 0.1 mg/ml in water. Plasmid pCMV~3 was purchased from
Clontech and was purified using the Anaconda kit of Althea Technologies (San
Diego, CA). PolyK is polylysine, mw 9,400. PolyR is polyarginine. PolyH is
polyhistidine.
To 100 p.1 plasmid solution (10 ~,g total plasmid DNA) 20 p,1 or 50 p.1 of
polyK, polyR, polyH, were added; the volume was adjusted to 250 ~,1 with water
followed by addition of about 70 p,1 liposomes (7 nmoles /p.g DNA). After
incubation for 10 min to 1 h at 20°C the transfection mixture was
brought in contact
with the cell culture. The best DNA condensing reagent was polyhistidine
compared
with the popular polylysine. The best cationic lipid was DC-cholesterol (DC-
CHOL:
3(3 [N-(N',N'-dimethylaminoethane)carbamoyl]cholesterol). SFV is Semliki
Forest
virus expressing beta-galactosidase. The results are shown in FIG. 2.
LiposomeMolecular weight Composition Preparation
L2 DDAB mw 631 DDAB 4.2 ~moles/ml15 mg DDAB
DOPE mw 744 DOPE 4.2 ~moles/ml+ 0.88 ml 20
mg/ml
DOPE
L3 DOGS-NTA mw 1015.4DOGS-NTA 1 ~mole/ml5 mg DOGS
DOPE 1 pmole/ml 0.185 ml DOPE
L4 DC-Chol (mw 537) DC-Chol 1 ~mole/ml0.106 ml DC-Chol
(25
DOPE (mw 744) DOPE 1 ~molelml mg/ml)
+ 0.185 ml DOPE
(20
mg/ml)
LS DOTAP (mw 698) DOTAP 1.4 ltmole/ml0.5 ml 10 mg/ml
DOTAP
DOPE (mw 744) DOPE 1.3 ltmole/ml+ 0.25 ml DOPE
(20
mg/ml)
L6 DODAP (mw 648) DODAP 1.54 ~moles/ml0.5 ml 10 mg/ml
DOPE 1.3 ~mole/mlDODAP=5 mg=7.72
ltmoles
+ 0.25 ml DOPE
(20
mg/ml)
Example 2
Targeting Genes to Tumors Using Gene Vehicles (Lipogenes).
As shown in FIG. 3, tumor targeting in SLID (severe combined
immunodeficient) mice were implanted subcutaneously, at two sites, with human
MCF-7 breast cancer cells. The cells were allowed to develop into large,
measurable
solid tumors at about 30 days post-inoculation. Mice were injected
intraperitoneously with 0.2 mg plasmid pCMV(3 DNA (size of the plasmid is ~4
kb)
52
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
per animal carrying the bacterial beta-galactosidase reporter gene. Plasmid
DNA
(200~,g, 2.0 mg/ml, 0.1 ml ) was incubated for 5 min with 200p,1 neutral
liposomes of
the composition 40% cholesterol, 20% dioleoylphosphatidylethanolamine(DOPE),
12% palmitoyloleoylphosphatidylcholine (POPC), 10% hydrogenated soy
phosphatidylcholine (HSPC), 10% distearoylphosphatidylethanolamine (DSPE), 5%
sphingomyelin (SM), and 3% derivatized vesicle-forming lipid M-PEG-DSPE.
At this stage, weak complexation of plasmid DNA with neutral (zwitterionic)
liposomes takes place. This ensures homogeneous distribution of plasmid DNA to
liposomes at the subsequent step of addition of cationic liposomes. After
complexation of plasmid DNA with zwitterionic liposomes, 50 p,1 of cationic
liposomes (DC-Chol 1 ~,mole/mI:DOPE 1.4 ~,mole/ml) were added and incubated at
room temperature for 10 min. At this stage, a mixed liposome population is
present
and, most likely, formation of a type of liposome-DNA complexes containing
lipids
from the zwitterionic and cationic lipids takes place. The material was
injected (0.35
ml total volume) to the intraperitoneal cavity of the animal. At 5 days post-
injection
the animal was sacrificed, the skin was removed and the carcass was incubated
into
X-gal staining solution for about 30 min at 37°C. The animal was
incubated in
fixative in X-gal staining for about 30 min (addition of 100 ~,l concentrated
glutaraldehyde to 30 ml X-gal staining solution) and the incubation in
staining
solution continued. Photos were taken in a time course during the incubation
period
revealing the preferred organs where beta-galactosidase expression took place.
Because of the tumor vasculature targeting shown in FIG. 3E, the data imply
that transfer of the genes of angiostatin, endostatin, or oncostatin to the
tumors
(whose gene products restrict vascular growth and inhibit blood supply to the
tumor)
is expected to be a rational approach for cancer treatment. Also, a
combination
therapy using anticancer lipogenes with encapsulated drugs into tumor
targeting
liposomes appears as a rational cancer therapy.
It is to be understood that while the invention has been described in
conjunction with the above embodiments, that the foregoing description and the
following examples are intended to illustrate and not limit the scope of the
invention.
Other aspects, advantages and modifications within the scope of the invention
will
be apparent to those skilled in the art to which the invention pertains.
53
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
Table 3 Simple NLS
Signal oligopeptideProtein and features
PK_KICRKV (SEQ ID Wild-type SV40 large T protein
N0:20)
A point mutation converting lysine-128
(double underlined) to
threonine results in the retention of
large T in the cytoplasm. Transfer
of this peptide to the N-terminus of (3-galactosidase
or pyruvate kinase
at the gene level and microinjection of
plasmids into Vero cells
showed nuclear location of chimeric proteins.
PKKKRMV (SEQ ID SV40 large T with a K-~M change. Site-directed
N0:21) mutagenesis only
slightly impaired nuclear import of large
T.
PKKKRKVEDP (SEQ Synthetic NLS peptide from SV40 large
ID T antigen crosslinked to BSA
N0:22) or IgG mediated their nuclear localization
after microinjection in
~enopus oocytes. The PKKGSKKA from Xenopus
H2B was
ineffective and PKTKRKV was less effective.
CGYGPKKKRKVGG (SEQ Synthetic peptide from SV40 large T antigen
ID conjugated to various
N0:23) proteins and microinjected into the cytoplasm
of TC-7 cells. Specified
nuclear localization up to protein sizes
of 465 kD (ferritin). IgM of 970
kD and with an estimated radius of 25-40
nm was retained in the
cytoplasm.
CYDDEA_TADSQHSTPPKKKSV40 large T protein long NLS. The long
NLS but not the short NLS,
RKVEDPKDFESELLS was able to localize the bulky IgM (970
kD) into the nucleus.
(SEQ ID N0:24) Mutagenesis at the four possible sites
of phosphorylation (double
underlined) impaired nuclear import.
CGGPKKKRKVG SV40 large T protein. This synthetic peptide
crosslinked to chicken
(SEQ ID N0:25) serum albumin and microinjected into HeLa
cells caused nuclear
localization.
PKKKIKV (SEQ ID A mutated (R-~I) version of SV40 large
N0:26) T NLS. Effective NLS.
MKxIICRLKKLKCSKEKPKCYeast GAL4 (99 kD). Fusions of the GAL4
gene portion encoding the
AKCLKx5Rx3KTKR (SEQ74 N-terminal amino acid with E. coli
ID (3-galactosidase introduced into
N0:27) yeast cells specify nuclear localization.
74 N-terminal amino
acid
MKx 11 CRLKKLKCSKEKPKCYeast GAL4. Acted as an efficient nuclear
localization sequence when
A (SEQ ID N0:28) fused to invertase but not to ~i-galactosidase
introduced by
29 N-terminal aminotransformation into yeast cells.
acid
PKKARED (SEQ ID Polyoma large T protein. Identified by
N0:29) fusion with pyruvate kinase
VSRKRPR (SEQ ID cDNA and microinjection of Vero African
N0:30) green monkey cells.
Mutually independent NLS. Can exert cooperative
effects.
CGYGVSRKRPRPG Polyoma virus large T protein. This synthetic
peptide crosslinked to
(SEQ ID N0:31) chicken serum albumin and microinjected
into HeLa cells caused
nuclear localization.
APTKRKGS SV40 VP1 capsid polypeptide (46 kD). NLS
(N terminus) determined
(SEQ ID N0:32) by infection of monkey kidney cells with
a fusion construct containing
the 5' terminal portion of SV40 VP1 gene
and the complete cDNA
sequence ofpoliovirus capsid VP1 replacing
the VP1 gene of SV40.
APKRKSGVSKC (1-11) Polyoma virus major capsid protein VP1
(11 N-terminal amino acid).
(SEQ ID N0:33) Yeast expression vectors coding for 17
N-terminal amino acid of VP1
fused to (3-galactosidase gave a protein
that was transported to the
nucleus in yeast cells. Subtractive constructs
of VP1 lacking A1 to C11
were cytoplasmic. This, FITC-labeled,
synthetic peptide crosslinked to
BSA or IgG, caused nuclear import after
microinjection into 3T6 cells.
Replacement of K3 with T did not.
54
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
Signal oligopeptideProtein and features
PNKKKRK (SEQ ID SV40 VP2 capsid protein (39 kD). The 3'
N0:34) end of the SV40 VP2-VP3
(amino acid positiongenes containing this peptide when fused
317-323) to poliovirus VP1 capsid
protein at the gene level resulted in
nuclear import of the hybrid VP 1
in simian cells infected with the hybrid
SV40.
EEDGPQKKKRRL (307-318)Polyoma virus capsid protein Vl'2. A construct
having truncated VP2
(SEQ ID N0:35) lacking the 307-318 peptide transfected
into COS-7 cells showed
cytoplasmic retention of VP2. The 307-318
peptide crosslinked to
BSA or IgG specified nuclear import following
their microinjection
into NIH 3T6 cells.
GKKRSKA (SEQ ID Yeast histone H2B. This peptide specified
N0:36) nuclear import when fused
to (3-galactosidase.
KRPRP (SEQ ID N0:37)Adenovirus Ela. This pentapeptide, when
linked to the C-terminus of
E. coli galactokinase, was sufficient
to direct its nuclear accumulation
after microinjection in Vero monkey cells.
CGGLSSKRPRP (SEQ Adenovirus type 2/5 Ela. This synthetic
ID peptide crosslinked to chicken
N0:38) bovine albumin and microinjected into
HeLa cells caused nuclear
localization.
LVRKKRKTE3SP (NLS Xenopus N1 (590 amino acid). Abundant
1) in X. laevis oocytes, forming
(SEQ ID N0:39) complexes with histones H3, H4 via two
acidic domains each
LKDKDAKKSKQE (NLS2)containing 21 and 9 (D+E), respectively.
The NLS1 is required but not
(SEQ ID NO:40) sufficient for nuclear accumulation of
protein Nl. NLS 1 and 2 are
contiguous at the C-terminus.
GNKAKRQRST v-Rel or p59 v-rel the transforming protein,
product of the v-rel
(SEQ ID N0:41 ) oncogene of the avian reticuloendotheliosis
retrovirus strain T (Rev-T).
v-Rel NLS added to the normally cytoplasmic
~i-galactosidase directed
that protein to the nucleus.
PFLDRLRRDQK NS1 protein of influenza A virus, that
accumulates in nuclei of virus-
(SEQ ID N0:42) infected cells. Determined to be an NLS
by deletion mutagenesis of
PKQKRKMAR NS 1 in recombinant SV40. The 1 st NLS
is conserved among all NS 1
(SEQ ID N0:43) proteins of influenza A viruses.
SVTKKRKLE (SEQ ID Human lamin A. Dimerization of lamin A
N0:44) was proposed to give a
complex with two NLSs that was transported
more efficiently.
SASKRRRLE Xenopus lamin A. NLS inferred from its
similarity to human lamin A
(SEQ ID N0:45) NLS.
TKGKRKRID Xenopus lamin LI . NLS inferred from its
sequence similarity to
(SEQ ID N0:46) human lamin A NLS.
CVRTTKGKRKRIDV Xenopus lamin LI. This synthetic peptide
crosslinked to chicken
(SEQ ID N0:47) bovine albumin and microinjected into
HeLa cells caused nuclear
localization.
ACIDKRVKLD Human c-myc oncoprotein. This synthetic
peptide crosslinked to
(SEQ ID N0:48) chicken bovine albumin and microinjected
into HeLa cells caused
nuclear localization.
ACIDKRVKLD Human c-myc oncoprotein. Conjugation of
the M1 peptide to human
(SEQ ID N0:49) serum albumin and microinjection of Vero
cells gives complete
(M1, fully potent nuclear accumulation. M2 gave slower and
NLS) only partial nuclear
localization.
RQRRNELKRSP
(SEQ ID N0:50)
(M2, medium potency
NLS)
SALIKKKKKIvIAP Murine c-abl (IV) gene product. The p160gag/v-abl
has a cytoplasmic
(SEQ ID N0:51) and plasma membrane localization, whereas
the mouse type IV c-abl
protein is largely nuclear.
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
Signal oligopeptideProtein and features
~
PPKKRMRRRIE Adenovirus 5 DBP (DNA-binding protein)
found in nuclei of infected
(SEQ ID N0:52) cells and involved in virus replication
and early and late gene
PKKKKKRP (SEQ ID expression. Both NLS are needed, and disruption
N0:53) of either site
impaired nuclear localization of the 529
amino acid protein.
YRKCLQAGMNLEARKTKK Rat GR, glucocorticoid receptor (795 amino
acid) NLS 1 determined by
KIKGIQQATA (497-524fusion with (3-galactosidase (116 kD).
amino NLS1 is 100% conserved
acid) between human, mouse and rat GR. Whereas
the 407-615 amino acid
(SEQ ID N0:54) fragment of GR specifies nuclear location,
the 407-740 amino acid
fragment was cytoplasmic in the absence
of hormone, indicating that
sequence 615-740 may inhibit the nuclear
location activity. A second
(NLS2) is localized in an extensive 256
amino acid C-terminal
domain. NLS 2 requires hormone binding
for activity.
RKDRRGGRMLKHKR RDD Human ER (estrogen receptor, 595 amino
acid) NLS. NLS is between
GEGRGEVGSAGDMRAMIN the hormone-binding and DNA-binding regions;
ER, in contrast with
O ACIDNLWPSPLMIKRSKKGR, lacks a second NLS. Can direct a fusion
product with (3-
(amino acid 256-303)galactosidase to the nucleus.
(SEQ ID NO:55)
RKFKKFNK Rabbit PG (progesterone receptor). 100%
homology in humans; F~L
(SEQ ID N0:56) change in chickens. When this sequence
was deleted, the receptor
became cytoplasmic but could be shifted
into the nucleus by addition
of hormone; in this case the hormone mediated
the dimerization of a
mutant PG with a wild type PG molecule.
GKRKNKPK (SEQ ID Chicken Etsl core NLS. Within a 77 amino
N0:57) acid C-terminal segment
90% homologous to Ets2. When deleted by
deletion mutagenesis at the
gene level the mutant Etsl became cytoplasmic.
PLLKKIKQ (SEQ ID c-myb gene product; directs puruvate kinase
N0:58) to the nucleus.
PPQKKIKS (SEQ ID N-myc gene product; directs puruvate kinase
N0:59) to the nucleus.
PQPKKKP (SEQ ID p53; directs puruvate kinase to the nucleus.
N0:60)
SKRVAKRKL c-erb-A gene product; directs puruvate
kinase to the nucleus.
(SEQ ID N0:61)
CGGLSSKRPRP Adenovirus type2/5 Ela. This synthetic
peptide conjugated with a
(SEQ ID NO:62) bifunctional crosslinker to chicken serum
albumin (CSA) and
microinjected into HeLa cells directed
CSA to the nucleus.
MTGSKTRKHRGSGA Yeast ribosomal protein L29. Double-stranded
oligonucleotides
(SEQ ID NO:63) encoding the 7 amino acid peptides (underlined)
and inserted at the N-
MTGSKHRKHPGSGA terminus of the (i-galactosidase gene
resulted in nuclear import.
(SEQ ID N0:64)
RHRKHP (SEQ ID N0:65)Mutated peptides derived from yeast L29
ribosomal protein NLS,
KRRKIiP (SEQ ID found to be efficient NLS. 'The last two
N0:66) are less effective NLS,
KYRKHP (SEQ ID N0:67)resulting in both nuclear and cytoplasmic
location of ~i-galactosidase
KHRRHP (SEQ ID N0:68)fusion protein.
KHKKHP (SEQ ID N0:69)
RHLKHP (SEQ ID NO:70)
KHRKYP (SEQ ID N0:71)
KHRQHP (SEQ ID N0:72)
PETTVVRRRGRSPRRRTPSPDouble NLS of hepatitis B virus core antigen.
The two underlined
RRRRSPRRRRSQS (SEQ arginine clusters represent distinct and
ID independent NLS. Mutagenesis
N0:73) showed that the antigen fails to accumulate
in the nucleus only when
(One sequence, C-terminus)both NLS are simultaneously deleted or
mutated.
ASKSRKRKL Viral Jun, a transcription factor of the
AP-1 complex. Accumulates in
(SEQ ID N0:74) nuclei most rapidly during G2 and slowly
during G1 and S. The cell
cycle dependence of viral but not of cellular
Jun is due to a C-~S
mutation in NLS of viral Jun. This NLS
conjugated to rabbit IgG can
mediate cell cycle-dependent translocation.
56
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
Signal oligopeptideProtein and features
GGLCSARLHRHALLAT Human T-cell leukemia virus Tax trans-activator
protein. The most
(SEQ ID N0:75) basic region within the 48 N-terminal
segment. Missense mutations in
this domain result in its cytoplasmic
retention.
DTREKKKFLK Mouse nuclear Mxl protein (72 kD), Induced
RRLLRLDE by interferons (among
_ 20 other proteins) . Selectively inhibits
(604-620) influenza virus mRNA
(SEQ ID N0:76) synthesis in the nucleus and virus multiplication.
The cytoplasmic Mx2
has R->S and R-~E changes in this region.
CGYGPKKKRKV (SV40 Synthetic peptides crosslinked to bovine
large serum albumin (BSA) and
T) (SEQ ID N0:77) introduced into MCF 7 or HeLa S3 cells
with viral co-internalization
CGYGDRNKKKKE (humanmethod using adenovirus serotype 3B induced
nuclear import of BSA.
retinoic acid receptor)
(SEQ ID NO:78)
CGYGARKTKKKIK
(human glucocorticoid
receptor)
(SEQ ID N0:79)
CGYGIRKDRRGGR
(human estrogen
receptor)
(SEQ ID N0:80)
CGYGARKLKKLGN
(human androgen
receptor)
(SEQ ID N0:81)
RKRQRALMLRQAR Human XPAC (xeroderma pigmentosum group
A complementing
30-42 protein) involved in DNA excision repair.
By site-directed
(SEQ ID N0:82) mutagenesis and immunofluorescence. NLS
is encoded by exon 1
which is not essential for DNA repair
function.
EYLSRKGKLEL (SEQ T-DNA -linked VirD2 endonuclease of the
ID Agrobacterium
N0:83) ' tumefaciens tumor-inducing (Ti) plasmid.
A fusion protein with (3-
(at the N-terminus)galactosidase is targeted to the nucleus.
The T-plasmid integrates into
plant nuclear DNA; VirD2 produces a site-specific
nick for T
integration. VirD2 also contains a bipartite
NLS at its C-terminus (see
Table 2).
KKSKKKRC (SEQ ID Putative core NLS of yeast TRM1 (63 kD)
N0:84) that encodes the tRNA
(95-102) modification enzyme N2, N2-dimethylguanosine-specific
tRNA
methyltransferase. Localizes at the nuclear
periphery. The 70-213
amino acid segment of TRM1 causes nuclear
localization of (3-
galactosidase fusion protein in yeast
cells. Site-directed mutagenesis of
the 95-102 peptide resulted in its cytoplasmic
retention. TRM1 is both
nuclear and mitochondria). The 1-48 amino
acid segment specifies
mitochondria) import.
PQSRKKLR (SEQ ID Max protein; specifically interacts with
N0:85) c-Myc protein. Fusion of 126-
151 segment of Max to chicken pyruvate
kinase (PK) gene, including
this putative NLS, followed by transfection
of COS-1 cells and indirect
immunofluorescence with anti-PK showed
nuclear targeting.
QPQRYGGGRGRRW (SEQ Gag protein of human foamy retrovirus;
ID a mutant that completely lacks
N0:86) this box exhibits very little nuclear
localization; binds DNA and RNA
in vitro.
57
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
Table 4 "Bipartite" or "split" NLS
Signal Oligopeptide Protein and features
C-terminus Xenopus nucleoplasmin. Deletion analysis
demonstrated the
presence of a signal responsible for
nuclear location.
TKKAGQAKKK (SEQ ID Xenopus nucleoplasmin
N0:87)
TKKAGQAKKKKLD Xenopus nucleoplasmin. Whereas these
17 amino acids had NLS
(SEQ ID N0:88) activity, shorter versions of the 17
amino acid sequences were
unable to locate pyruvate kinase to
the nucleus.
TKKAGQAKKK(KLD) Xenopus nucleoplasmin. This 14 amino
acid segment was
(SEQ ID N0:89) identified as a minimal nuclear location
sequence but was unable
to locate puruvate kinase to the nucleus;
three more amino acids
at either end (shown in parenthesis)
were needed.
CGQAKKKKL,D Xenopus nucleoplasmin-derived synthetic
peptide; crosslinked to
(SEQ ID N0:90) chicken serum albumin and microinjected
to HeLa cells specified
nuclear localization. This suggests
that nucleoplasmin may
possess a simple NLS.
KRPAMINO ACID Xenopus nucleoplasmin bipartite NLS.
Two clusters of basic
TKKAGQAKKKK (SEQ ID amino acids (underlined) separated
N0:91) by 10 amino acid are half
NLS components.
HRKYEAPRHx6PRKR (SEQ Yeast L3 ribosomal protein (387 amino
ID acid) N-terminal 21
N0:92) amino acid. Possible bipartite NLS.
(Ribosomal proteins are
transported to the nucleus to assemble
with nascent rRNA).
Fusion genes with (3-galactosidase
were used to transform yeast
cells followed by fluorescence staining
with b-gal antibody. The
373 amino acid of L3 fused to (3-gal
failed to localize to the
nucleus, unless a 8 amino acid bridge
containing a proline was
inserted between L3 and (3-gal.
NKKKRKLSRGSSQKTKGTSASAKSV40 Vp3 structural protein. (35 amino
acid C-terminus). By
ARHKRRNRSSRS (one sequence)DEAE-dextran-mediated transfection
of TC7 cells with mutated
(SEQ ID N0:93) constructs.
RVTIRTVRVRRPPKGKHRK Simian sarcoma virus v-sis gene product
(p28sis). The cellular
(SEQ ID N0:94) counterpart c-sis gene encodes a precursor
of the PDGF B-chain
(platelet-derived growth factor). The
NLS is 100% conserved
between v-sis gene product and PDGF.
This protein is normally
transported across the ER; introduction
of a charged amino acid
within the hydrophobic signal peptide
results in a mutant protein
that is translocated into the nucleus.
Puruvate kinase-NLS fusion
product is transported less efficiently
than cytoplasmic v-sis
mutant proteins to the nucleus.
KRKIEEPEPEPKKAK Putative bipartite NLS ofXenopus laevis
protein factor xnf7.
(SEQ ID N0:95) Inferred by similarity to the bipartite
NLS of nucleoplasmin.
During oocyte maturation xnf7 is cytoplasmic
until mid-blastula-
gastrula stage due to high phosphorylation.
Partial
dephosphorylation results in nuclear
accumulation.
KKYENVVIKRSPRKRGRPRKD Yeast SWIS gene product, a transcription
factor. Underlined
(SEQ ID N0:96) basic amino acid show similarity to
bipartite NLS ofXenopus
nucleoplasmin. The SWIS gene is transcribed
during S, G2 and
M phases, during which the SWIS protein
remains cytoplasmic
due to phosphorylation by CDC28-dependent
histone H1 kinase
at three serine residues two near and
one (double underlined) in
the NLS. Translocated at the end of
anaphase/G1 due to
dephosphorylation of NLS. NLS confers
cell cycle-regulated
nuclear import of SWIS-(3-galactosidase
fusion protein.
5~
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
Signal Oligopeptide Protein and features
MKRKRNS 735-741 Bipartite NLS of influenza virus polymerase
basic protein 2
(SEQ ID N0:97) (PB2). Mutational analysis of PB2 and
transfection of BHK cells
GIESIDNVMGMIGILPDMTPSTEMshowed that both regions are involved
in nuclear import.
SMRGVRISKMGVDETSSAEKIVDeletion of 449-495 region gives perinuclear
localization to the
449-495 (SEQ ID N0:98)cytoplasmic side.
AHRARRLH (SEQ ID N0:99)"Tripartite" or "doubly bipartite"
NLS of adenovirus DNA
6-13 (BSI) polymerase (AdPol). BSI and II functioned
interdependently as
PPRRRVRQQPP (SEQ ID an NLS for the nuclear targeting of
NO:100) AdPol, for which BSIII was
23-33 (BSII) dispensable. BSII-III was more efficient
NLS than BSI-II.
PARARRRRAP (SEQ ID
NO:101)
39-48 (BSIII)
KRKxI IKK Human poly(ADP-ribose) polymerase (116
KSKK 207-226 kD). The linear
_ distance between the two basic clusters
(SEQ ID N0:102) is not crucial for NLS
activity in this bipartite NLS. Lysine
222 (double underlined) is
an essential NLS component. DNA binding
and poly(ADP-
ribosyl)ating active site are independent
of NLS.
GRKRAFHGDDPFGEGPPDKKGDHerpes simplex virus ICP8 protein (infected-cell
protein). This
(SEQ ID N0:103) C-terminal portion of ICP8 introduced
into pyruvate kinase (PK)
caused nuclear targeting in transfected
Vero cells. Inclusion of
additional ICP8 regions to PK led to
inhibition of nuclear
localization.
KRPREDDDGEPSERKRARDDR Bipartite NLS of VirD2 endonuclease
of rhizogenes strains of
(SEQ ID N0:104) Agrobacterium tumefaciens. Within the
C-terminal 34 amino
acid. Each region (underlined) independently
directs ~i-
glucuronidase to the nucleus, but both
motifs are necessary for
maximum efficiency. VirD2 is tightly
bound to the 5' end of the
single stranded DNA transfer intermediate
T-strand transferred
from Agrobacterium to the plant cell
genome.
59
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
Table 5. "Nonpositive NLS" lacking clusters of arginines/lysines
Signal oligopeptideProtein and features
OLVWMACNSAMIN Influenza virus nucleoprotein (NP). The
underlined region
O (327-345) when fused to chimpanzee al-globin
at the cDNA level and
ACIDFEDLRVLSFIRGTKVS~croinjected into Xenopus oocytes specifies
nuclear localization.
PRG 327-356
(SEQ ID N0:105)
MNKIPIKDLLNPQ Yeast MAT a2 repressor protein, containing
a homeodomain.
(NLS 1 at N-terminus)The two NLS are distinct, each capable
(SEQ ID of targeting (3-galactosidase to
N0:106) the nucleus. However, deletion of NLS2
results in a2 accumulation at
VRILESWFAKNIEN the pores. NLSl and 2 may act at different
steps in a localization
PYLDT (NLS2 at pathway. Part of the homeodomain mediates
amino acid nuclear localization in
141-159, part of addition to DNA binding. The core pentapeptide
the containing proline and
homeodomain) two other hydrophobic amino acids flanked
by lysines or arginines
(SEQ ID N0:107) (underlined) was suggested as one type
of NLS core.
~7~15~P~3HFY Drosophila HP1 (206 amino acids) that binds
to
EERLSWYSDNED (SEQ heterochromatin and is involved in gene
ID silencing. NLS identified by (3-
N0:108) galactosidase/HP1 fusion proteins introduced
by P-element mediated
152-206 (C-terminaltransformation into Drosophila embryos.
segment)
FVx7_ Adenovirus type 5 ElA internal, developmentally-regulated
20MxSLxYMx4MF NLS. This NLS functions in Xenopus oocytes
but not in somatic cells.
This NLS can be utilized up to the early
neurula stage.
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
Table 6. Nucleolar localization signals (NoLS)
Signal oligopeptide Protein and features
MPKTRRRPRRSQRKRPPTP Nucleolus localization signal in amino
terminus of human p27x-
(SEQ ID N0:109) III protein (also called Rex) of T
cell leukemia virus type I
(HTLV-I). When this peptide is fused
to N-terminus of (3-
galactosidase, directs it to the nucleolus.
Deletion of residues 2-
8 (underlined), 12-18 (double-underline)
or substitution of the
central RR (dotted-underlined) with
TT abolish nucleolar
localization. Other amino acids between
positions 20-80
increase nucleolar localization efficiency.
RLPVRRRRRRVP (SEQ ID Adenovirus pTPl and pTP2 (preterminal
NO:110) proteins, 80 kD)
between amino acid residues 362-373.
The 140 kD DNA
polymerase of adenovirus when it has
lost its own NLS can
enter the nucleus via its interaction
with pTP. The staining was
nuclear and nucleolar with some perinuclear
staining as well.
The NLS fused to the N-terminus of
E. coli (3-galactosidase was
functional in nuclear targeting.
GRKKRRQRRRP HIV (human immunodeficiency virus)
Tat protein; localizes
(SEQ ID NO:111) pyruvate kinase to the nucleolus.
Tat is constitutively nucleolar.
RKKRRQRRR(AHQ) Tat positive traps-activator protein
of HIV-1 (human
Nucleolar localization immunodeficiency virus type 1). The
signal 3 amino acids shown in
(SEQ ID N0:112) parenthesis are essential for the
localization of the ~3-
galactosidase to the nucleolus. The
9 amino acid basic region is
able to localize [3-gal to the nucleus
but not to the nucleolus.
KRVKLDQRRRP (SEQ ID Artificial sequence from c-Myc and
N0:113) HIV Tat NLSs that
effectively localizes pyruvate kinase
to the nucleolus.
FKRKHKKDISQNKRAVRR Human HSP70 (heat shock protein of
70 kD); localizes pyruvate
(SEQ ID N0:114) kinase to the nucleus and nucleolus.
HSP70 is physiologically
cytoplasmic but with heat-shock HSP70
redistributes to the
nucleoli, suggesting that the nucleolar
targeting sequence is
cryptic at physiological temperature
and is revealed under heat-
shock.
RQARRNRRRRWRERQR (35-50)HIV-1 Rev protein (116 amino acid,
nucleolar). Mutations in
(SEQ ID NO:115) either of the two regions of arginine
clusters severely impair
nuclear localization. (3-galactosidase
fused to R4W was targeted
to the nucleus, and fused to the entire
35-50 region, was targeted
to the nucleolus.
RQARRNRRRRWRERQRQ (35-51)HIV-1 Rev protein. A fusion of this
Rev peptide with (3-
(SEQ ID N0:116) galactosidase became nuclear but not
nucleolar. The 1-59 amino
acid segment of Rev fused to (3-galactosidase
localized entirely
within the nucleolus. Whereas the
NRRRRW (bold) is
responsible for nuclear targeting,
the RR and WRERQRQ
(double underlined) specify nucleolar
localization. Rev may
function to export HIV structural
mRNAs from the nucleus to
the cytoplasm.
61
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
Table 7. Karyophilic clusters on non-membrane protein kinases
Karyophilic Non-membrane Species Features
peptides
protein lcinase
73 FVVHKRCHE Protein kinaseBovine, Known to translocate
C (673 human to the
(SEQ ID N0:117)aa) (3 type nucleus following
treatment of
96 DDPRSKHKFKIH cells with mitogens.
(SEQ ID N0:118)
577 TKHPGKRLG
(SEQ ID N0:119)
71 FVVHRRCHEF Protein kinasebovine,
C (697 human y
(SEQ ID N0:120)aa) type
95 DDPRNKHKFRLH
(SEQ ID N0:121)
591 TKHPAKRLG
(SEQ ID N0:122)
72 FVVHKRCHE Protein kinaserabbit type
C (673 a and
(SEQ ID N0:123)aa) (3
96 DDPRSKHKFKIH
(SEQ ID N0:124)
577 TKHPGKRLG
(SEQ ID NO:125)
71 FVVHRRCHE PKC-I (701 rat brain
aa)
(SEQ ID N0:126)
95 DDPRNKHKFRLH
(SEQ ID N0:127)
594 TKHPGKRLG
(SEQ ID N0:128)
22 GENKMKSRLRKGProtein kinaseDrosophila 14 exons, 20 kb;
C 3 transcripts in
(not conserved)(639aa, 75 adult flies; not
kDa) expressed in 0-3h
(SEQ ID N0:129) Drosophila embryos;
the
80SYVVHKRCHEYVT VVHKRCHE (SEQ ID
(conserved) N0:133)motif (or
VVHRRCHE
(SEQ ID N0:130) (SEQ ID N0:134))
is conserved
211PDDKDQSKKKTR among all PKC known.
TIK (not conserved)
(SEQ ID N0:131)
614PPFKPKIKHRKMC
P (not conserved)
(SEQ ID N0:132)
148 KKVLQDKRFK Glycogen synthaserat brain Phosphorylates glycogen
synthase,
NRELQIMRKLD kinase 3 c-Jun, c-Myb; two
(SEQ isoforms
ID N0:135) GSK-3a encoded by discrete
genes; highly
(483 aa) expressed in brain;
both a and (3
forms are cytosolic
but also
GSK-3(3 associated with the
plasma
(420 aa) membrane consistent
with their
role in signal transduction
from the
cell surface.
LQDRRFKNRELQ Zw3 Drosophila Product of the segment
polarity
(SEQ ID N0:136)zeste-white gene zw3; the protein
3 encoded has
34% homology to cdc2;
mutations
in zw3 give embryos
that lack
most of the ventral
denticles,
differentiated structures
derived
from the most anterior
region of
each segment.
62
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
Karyophilic Non-membrane Species Features
peptides
protein kinase
289ECLKKFNARRKLCa'~/calinodulin-rat brain Composed of nine 50
kDa a-
KGAIL dependent protein subunits and three
60 kDa (3-
(SEQ ID N0:137)kinase II (CaM subunits; both are
kinase catalytic;
II) (3 subunit calmodulin- and ATP-binding
(542aa,
60.3 kDa) domains; highly expressed
in
forebrain neurons,
concentrated in
postsynaptic densities;
acts as a
z+
Ca -triggered switch
and could be
involved in long-lasting
changes in
synapses.
290LKKFNARRKL CaM kinase rat brain This particular isoform
II (478 is
KGAILTTM (SEQ aa, 54 kDa) exclusively expressed
ID in the brain;
N0:138) a-subunit high enzyme levels
in specific
450EETRVWHRRDGK brain areas; might
be involved in
(SEQ ID N0:139) short- and long-term
responses to
transient stimuli.
185 GFAKRVKGRT CADPK catalyticbovine By Edman degradation
(cardiac of protein
WTLCG subunit (349 muscle) fragments; mediates
aa, 40.6 the action of
(SEQ ID N0:140)kDa) and is activated by
cAMP; consists
of two regulatory
(R) and two
catalytic (C) subunits;
cAMP
releases the C subunit
from the
inactive R2C2 cADPK;
two
cDNAs were cloned
encoding two
isoforms of the catalytic
subunit of
cADPK in mouse.
186 GFAKRVKGRTWCADPK bovine cDNA was isolated
by screening a
TLCG (catalytic bovine pituitary cDNA
subunit) library;
(SEQ ID N0:141)(350 aa) 93% sequence similarity
to known
bovine cADPK; represents
the
second gene for the
catalytic
subunit of cADPK.
29 EEEIQELKRKLHCGDPK (SEQ bovine By protein sequencing;
ID lung composed
KCQSVLP (SEQ N0:144) of two identical subunits
ID activated
N0:142) (670 aa, 76.3 in an allosteric manner
kDa) by binding
389 KILKKRHIVDTR of cGMP and not by
dissociation
(SEQ ID N0:143) of catalytic subunit
as in cADPK;
sequence similar to
cADPK
117 KTLKKHTIVK TPK3 S. cerevisiaecAMP-DPK is a tetrameric
protein
(SEQ ID N0:145)(398 aa) with two catalytic
and two
cADPK regulatory subunits;
CAMP
activates the kinase
by dissociating
the catalytic subunits
from the
tetramer; all three
TPK 1, 2, 3 are
catalytic subunits.
16S2H13GHG2 SNF1 (633aa, S. cerevisiaeSer/Thr kinase;
72 kDa)
166 EYCHRHKIVHRD autophosphorylated;
plays a
LKP (SEQ ID central role is carbon
N0:146) catabolite
495 PLVTKKSKTRWH repression in yeast
required for
FG (SEQ ID N0:147) expression of glucose-repressible
genes; region 60-250
shows high
sequence similarity
to cAMP-
dependent protein
kinase
(cADPK).
63
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
Karyophilic Non-membrane Species Features
peptides
protein kinase
70 PVKKKICIKREIKCasein kinaseDrosophila CKII is composed of
II (a- a and (3
(SEQ ID N0:148)subunit, catalytic)melanogastersubunits in a a2~32
130-150 kDa
269 DILQRHSRKRW(336aa) protein; the a-subunit
is the
ERF (SEQ ID catalytic and the
N0:149) (3 is
146 PKSSRHHHTDGCKII ((3-subunit,Drosophila autophosphorylated.
(SEQ ID N0:150)regulatory) melanogaster
(215aa)
142 PKSSRHHHTDGCKII ((3-subunit,bovine (lung)
(SEQ ID N0:151)regulatory)
(209aa,
24.2 kDa)
108 PKQRHRKSLG KTN1 (1064 S. cerevisiae30% as similarity
aa, 117 to bovine
(SEQ ID N0:152)kDa) cADPK and 27% (K1N1)
or 25%
129 GSMCKVKLAK (KIN2) as similarity
to v-Src
HRYTNE within the kinase
domain; the
(SEQ ID N0:153) catalytic domains
of KINl and
506 DRKHAKIRNQ KIN2 are near the
N-terminus and
(SEQ ID N0:154) are structural mosaics
with features
638 GNIFRKLSQRR characteristic of
both Tyr and
KKTIEQ Ser/Thr kinases.
(SEQ ID NO:155)
773 PPLNVAKGRKL
HP (SEQ ID N0:156)
87 ELRQFHRRSLG KlN2 (1152 S. cerevisiae
aa, 126
(SEQ ID N0:157)kDa)
111 GKVKLVKHRQ
TKE (SEQ ID
N0:158)
217 GSLKEHHARKF
ARG (SEQ ID
N0:159)
807 LSVPKGRKLHP
(SEQ ID N0:160)
60FLRRGIKKKLTLDSTE7 (515 S. cerevisiaeImplicated in the
aa) control of the
(SEQ ID N0:161 three cell types in
) yeast: (a, a , and
472 PSKDDKFRHWC a/a) of which a and
a cells are
RKIKSKIKEDKRIKRE haploid and are specialized
for
(SEQ ID N0:162) mating whereas a/a
cells are
diploid and are specialized
for
meiosis and sporulation;
with the
exception of the mating
type locus,
MAT, all cells contain
the same
DNA sequences. STE7
gene
produces insensitivity
to cell-
division arrest induced
by the yeast
mating hormone, a-factor.
722 QRRVKKLPSTTLS6KIIa (733aa)Xenopus
(SEQ ID N0:163)
QRRVKKLPSITL S6KII (3 Xenopus
(SEQ ID N0:164)
742 QRRVKKLPSTTLS6KII (752 Chicken
aa)
(SEQ ID N0:165)
713QRRVRKLPSTTLS6KII (724aa)Mouse
(SEQ ID N0:166)
64
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
Karyophilic Non-membrane Species Features
peptides
protein ldnase
16GWYKGRHKTTG CDC2Hs Human Isolated by expressing
a human
(SEQ ID N0:167)(297aa) cDNA library in S.
pombe and
120 FCHSRRVLHRDp34cdc2 selecting for clones
that
LKP (SEQ ID complement a mutation
N0:168) in the cdc2
yeast gene; the human
CDC2 gene
can complement both
the
inviability of a null
allele of S.
cerevisiae CDC28 and
cdc2
mutants of S. pombe;
CDC2
mRNA appears after
that of
CDK2.
GVVYKARHKLSGR cdc2 (297aa) S. pombe High homology to S.
cerevisiae
(SEQ ID N0:169)
CDC28.
119HSHRVLHRDLKPCDK2 (cell Human The human CDK2 protein
division has 65%
(SEQ ID N0:170)kinase 2) sequence identity
(298 aa) to human
p34cdc2 and 89% sequence
identity to Xeuopus
Egl kinase;
human CDK2 was able
to
complement the inviability
of a
null allele of S.
cereuisiae CDC28
but not cdc2 mutants
in S. pombe.
CDK2 mRNA appears
in late
Gllearly S.
109 FCHSHRVLHRDEgl (297aa) Xenopus Cdk2-related
LKP (SEQ ID
N0:171)
125 GIAYCHSHRILHCDC28 (298a) S. cerevisiaeThe homolog of S.
pombe Cdc2
RDLKP
(SEQ ID N0:172)
119 HSHRVIHRDLKPcdk3 (305aa) Human
(SEQ ID N0:173)
56 KELKHKNIVR PSSALRE (291 Human cdc2 -related kinase.
aa)
(SEQ ID N0:174)(SEQ ID N0:175)
1 MDRMKKIKRQ PCTAIRE-1 Human cdc2-related kinase.
(N- (496 aa)
terminus) (SEQ
ID
N0:176)
141 DKPLSRRLRRV
(SEQ ID N0:177)
1 MKKFKRR PCTAIRE-2 Human cdc2 related kinase.
(523 aa)
(SEQ ID N0:178)
129 RNRIHRRIS
(SEQ ID N0:179)
172 SRRSRRAS
(SEQ ID N0:180)
304 HRRKVLHR
(SEQ ID N0:181)
512 GHGKNRRQSM
LF (SEQ ID N0:182)
163 HTRKILHR PCTAIRE-3 Human cdc2 related kinase.
(SEQ ID N0:183)(380 aa)
369 PGRGKNRRQSIF
(SEQ ID NO:184)
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
Karyophilic Non-membrane Species Features
peptides
protein kinase
69 EVFRRKRRLH KKIALRE (358 Human cdc2-related kinase.
aa)
(SEQ ID N0:185)(SEQ ID N0:187)
302 DKPTRKTLRKSR
KHH (SEQ ID
N0:186)
1 MVKRHKNT ~1+ gene productS. pombe
(SEQ ID N0:188)(new inducer
of
87 DGELFHYIRKHGP~tosis); protein
(SEQ ID N0:189)kinase (370
aa)
114 DAVAHCHRFRFR
HRD (SEQ ID
N0:190)
295 KKSSSKKVVRRL
QQRDD
(SEQ ID N0:191)
194 PAQKLRKKNNFDWeel+ gene S. pombe The Weel+ gene functions
product as a
(SEQ ID N0:192)(877aa) dose-dependent inhibitor
that
388 KQHRPRKNTNFT delays the initiation
of mitosis
PLPP (SEQ ID until the yeast cell
N0:193) has attained a
592 KYAVKKLKVKF certain size; Weel
has a protein
SGP (SEQ ID kinase consensus probably
N0:194)
regulating cdc2 kinase.
266 PNETRRIKRANCDC7 (497 aa) S. cerevisiaeRequired for mitotic
but not
RAG (SEQ ID meiotic DNA replication
N0:195)
presumably to phosphorylate
specific replication
protein factors;
implicated in DNA
repair and
meiotic recombination;
some
homology with CDC28
and
oncogene protein kinases
but
differs in a large
region within the
phosphorylation receptor
domain.
48YDHVRKTRVAIKKERK1 (MAP kinase)Rat Known to translocate
to the
(SEQ ID N0:196)(367 aa; 42 nucleus following
kDa) their activation
by phosphorylation
at T-190, and
Y-192 (T-183, Y-185
in ERK2).
59ILKHFKHE FUS3 (353aa) S. cerevisiaeMAP-(ERKl)-related.
(SEQ ID NO:197)
252 QIKSKRAKEY KSS1 (368 aa) S. cerevisiaeMAP-(ERKl)-related.
(SEQ ID N0:198)
ELVKHLVKHGSN SWI6 S. cerevisiaeActivator of CACGA-box
with
(SEQ ID N0:199)(803aa, 90kDa) sequence similarity
to cdcl0;
GKAKKIRSQLL required at START
of cell cycle.
(SEQ ID N0:200)
EQRLKRHRIDVSDEDcdcl0 S. pombe
(SEQ ID N0:201)
SNIKSKCRRW
(SEQ ID N0:202)
37 PPKRIRTD CTD kinase S. cerevisiaeConsists of 3 subunits
(528 aa) of 58, 38,
(suggested by 58 kDa subunit and 32 kDa; disruption
the of the 58
authors) (SEQ (catalytic) kDa gene gives cells
ID that lack CTD
N0:203) kinase, grow slowly,
are cold
492 KLARKQKRP sensitive, but have
different
(SEQ ID N0:204) phosphorylated forms
of RNA pol
II.
66
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
Karyophilic Non-membrane Species Features
peptides
protein kinase
29 GVSSVVRRCIHKPPhosphorylase Rabbit (skeletal
kinase
(SEQ ID N0:205)(catalytic muscle)
subunit)
(386aa)
489 KKYMARRKW Myosin light Chicken Ca2+/calmodulin-activated;
chain gizzard
QKTGHAV kinase (MLCK) phosphorylated by
(669 cADPK; first
(SEQ ID N0:206)aa) described as responsible
for the
phosphorylation of
a specific class
of myosin light chains;
required
for initiation of
contraction in
smooth muscle.
314 PWLNNLAEKAKMyosin light Rabbit (skeletalBy protein sequencing.
chain
RCNRRLKSQ kinase (partialmuscle)
368
(SEQ ID N0:207)carboxy-terminal
as
334ILLKKYLMKRR sequence)
WKKNFIAVS
(SEQ ID N0:208)
28 GVSSVVRRCIHKPPhosphorylase Mouse (muscle)Glycogenolytic regulatory
kinase enzyme;
(SEQ ID N0:209)(PhK) (catalytic undergoes complex
~y regulation;
subunit) (389 composed of 16 subunits
aa)
containing equimolar
ratios of a, [3,
y and 8 subunits;
high levels in
skeletal muscle; isoforms
in
cardiac muscle and
liver; cDNA
probe does not hybridize
to X
chromosome in mice
and is thus
distinct from the
mutant recessive
PhK deficiency that
results in
glycogen storage disease.
67
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
Table 8. Nuclear localization signals on DNA repair proteins
Putative NLS Gene Equivalent Features
protein
product in other species
HIGHER EUKARYOTES
None ERCC1 RAD10 297aa; DBD; interacts
(N-terminus)
strongly with ERCC4
(XPF)
MDPGKDKEGvpqpsgppaRKKF to form an excision
(bipartite NLS) endonuclease; unless
the
(SEQ ID N0:210) KDKxIIRKK is a
bipartite
NLS it may depend
upon its
binding with ERCC4
for its
nuclear import.
None ERCC2 RAD3 (S. cer) 760 aa; DNA helicase
681DKRFARGDKRGKLPR (XPD component of TFIIH,
)
(near the C-terminus) essential for cell
(four viability;
positive , one negative contains one nucleotide-
over a
heptapeptide stretch) binding, one DNA-binding,
(SEQ ID N0:211) and seven domains
characteristic
of helicases;
52% identity with
S. cer
RAD3 at the amino
acid
level.
8 DRDKKKSRKRHYEDEE ERCC3 SSL2 (S cer) 782 aa; helicase,
component
(SEQ ID N0:212) (XPB) Haywire(Dros) of TFIIH essential
for cell
522 YVAIKTKKRILLYTM viability; helix-turn-helix,
(SEQ ID N0:213) DNA-BD, and helicase
(weak NLS if at all, domains
hydrophobic
environment)
769 PSKHVHPLFKRFRK
(SEQ ID N0:214)
84 KKQTLVKRRQRKD ERCCS RAD2; 1186 as in human,
1196 in X.
(SEQ ID N0:215) (XPG) Radl3 laevis; 3' incision
210 EFTKRRRTL endonuclease; involved
in
(SEQ ID N0:216) homologous recombination;
390 DESMIKDRKDRLP strongly nuclear
(SEQ ID N0:217)
1170 GKI~RRKLRRARGRK
RKT (SEQ ID N0:218)
253PQKQEKKPRKIMLNEASGERCC6 RAD26 1493aa; involved
in the
(SEQ ID N0:219) CS-B preferential repair
of active
314 PNKKARVLSKKEERLKK genes; nonessential
for cell
HIKKLQKR (SEQ ID N0:220) 1 viability
406 PLPKGGKRQKKVP
(SEQ ID N0:221)
455 DGDEDYYK~WNK
LRLQDKEKRLKLEDDSEESD
(SEQ ID N0:222)
1028 DVQTPKCHLKRRIQP
XBPKRICKFP (SEQ ID
N0:223)
1180 KHKSKTKHHSVAEEETL
EKHLRPKQKPKX 1 SPHLVKK
RRY (SEQ ID N0:224)
1324 PAGKKSRFGKKRN
(SEQ ID N0:225)
68
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
Putative NLS Gene Equivalent Features
protein
product in other species
21 PASVRASIE~RALM XPA RAD14 273 aa; zinc finger
domain;
LRGAR (SEQ ID N0:226) involved in lesion
160 PPLKFIVKKNPHHSQW recognition
GD (weak) (SEQ ID
N0:227)
210 NREKMKQKKFDKKVKE
(weak because of
F)
(SEQ ID N0:228)
72 YLRRAMKRFN (weak)XPC RAD4 (23% identity,823 aas, 92.9
kDa; very
(SEQ ID N0:229) 44% similarity)hydrophilic protein;
might be
262 PSAKGKRNKGGRKKRSK involved in lesion
PSSSEEDEGPG (SEQ recognition since
ID XPC cells
N0:230) (40% of all XP
cases) can
297 QRRPHGRERR (weak) repair active
parts of the
(SEQ ID N0:231) genome whereas
inactive and
368 RTHRGSHRKDP (weak) the nontranscribed
strand of
(SEQ ID N0:232) active genes are
not repaired
384 SSSSSSSKRGKKMCSDG
(SEQ ID N0:233)
531 ALKRHLLKYE (weak)
(SEQ ID N0:234)
594 SNRARKARLAEP
(SEQ ID N0:235)
660 PNLHRVARKLD (weak)
(SEQ ID N0:236)
716 ERKEKEKKEKR
(SEQ ID N0:237)
740 IRERLKRRYG
(SEQ ID N0:238)
801 GGPKKTKRERK
(SEQ ID N0:239)
20 KSKAKSKARREEEEED XPC 940 aa; the first
117 as are
(SEQ ID NO:240) lacking in the
Legerski and
54 GKRKRG (SEQ ID Peterson, (1992)
N0:241) XPC
69 GPAKKKVAKVTVK sequence (see
above); the
(SEQ ID N0:242) following 823aa
are
103 PSDLKKAHHLKRG identical.
(SEQ ID N0:243)
82 EIDRRKKRPLENDGPVKKRep-3 Swi4 (S pom) 1137aa; mismatch
repair
KVKKVQQKE (SEQ ID (mouse) protein; Rep-3
is in the
N0:244) Duc-1 immediate 5' flanking
region
375 KENVRDKKKG (HeLa) of DHFR gene (89
bp) but
(SEQ ID N0:245) transcribed from
the opposite
57I FGRRKLKKWVT strand; a bidirectional
(SEQ ID N0:246) promoter is used
for both
710 PLIKI~RKDEIQG transcripts.
(SEQ ID N0:247)
1091 KELEGLINTKRKRLKYF
AKLW (SEQ ID N0:248)
422 EKHEGKHQKLL (weak)hMSH2 MSH2 (S cer) human mismatch
repair
(SEQ ID N0:249) protein; homologous
to S.
cerevisiae MSH2;
associated
with the hereditary
nonpolyposis colon
cancer
gene on chromosome
2p 16.
69
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
Putative NLS Gene Equivalent Features
protein
product in other species
397 PDIRRLTKKLNKRG MSH2
(SEQ ID N0:250) (S cer)
547 DAKELRKHKKYIE
(SEQ ID N0:251)
869 VKMAKRKANE
(SEQ ID N0:252)
95 GELAKRSERRAEAE Human Rad2 (S. porn)400 aa; required
Rad2 for fidelity
(SEQ ID N0:253) of chromosome separation
at
354 KRKEPEPKGSTKKKAK mitosis; limited
similarity to
TG (SEQ ID N0:254) RAD2 (ssDNA nuclease),
394 GKFKRGK (SEQ radl3, and XPG
ID (ERCCS).
N0:255)
None mouse 339 aa; recombination-repair
RAD51 protein; 83% homology
to S
cerevisiae RAD51
and 55%
homology to E.
coli RecA.
None HHR23B RAD23 Subunit of XPC
( 125 lcDa)
/p58
None ' HHR23A RAD23 Subunit of XPC
( 125 kDa)
32 PSQAEKKSRARAQ RPA (34 RPA (70, 34, and
kDa 14 lcDa
(SEQ ID N0:256) subunit) subunits) might
stabilize the
helicase-melted
DNA around
the lesion; antibodies
against
RPA 32 kDa subunit
inhibit
DNA replication.
GAKKRKIDDA ATPase RecQ (E. coli)649 aa; altered
Q1 in XPC cells;
(SEQ ID N0:257) undetermined role
in repair
PKKPRGKM (SEQ ID HMG-1 Calf thymus HMG
N0:258) 1
EHKICKIIP (SEQ ID (259 aa); involved
N0:259) in the
ETKKKFKDP (SEQ ID recognition of
N0:260) cisplatin
EKSKKKK(E/D)41 (SEQ lesions
ID
N0:261)
E3G2KKKKKFAK (SEQ
ID
N0:262)
512 RDEKKRKQLKKAKAK SSRP1 ABF (S cer) 709 aa, 81 kDa,
structure-
MAKDRKSRKKP (SEQ specific recognition
ID protein
N0:263) 1; involved in
recognition of
619 GESSKRDKSKKKKKVKV cisplatin-induced
lesions;
KMEKK (SEQ ID N0:264) also involved in
Ig gene
674 GENKSKKKRRRSEDSEE recombination;
one HMG-
EE (SEQ ID N0:265) box, similarity
to SRY,
MTFII, LEF-1, TCF-la,
and
ABF2.
1 MPKRGKKG (SEQ ID Ref 1 Redox factor 1
from HeLa
N0:266) (HAP1) cells; 37 kDa,
318 aa;
apurinic/apyrimidinic
(AP)
endonuclease for
DNA repair
but also of redox
activity
stimulating Jun/Fos
DNA
binding.
1 MPKRGKKG HAP1 ExoIII 323 aa; apurinic/apyrimidinic
(SEQ TD N0:267) (bovine)(E. coli) (AP)-endonuclease
ExoA (S.
pneumoniae)
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
Putative NLS Gene Equivalent Features
protein
product in other species
DROSOPHILA
1 MGPPKKSRKDRSGGDKF Haywire ERCC3 (XPB) helicase with 66%
identity to
GKKRRGQDE human ERCC3; flies
(SEQ ID N0:268) SSL2 (S cer) expressing marginal
levels of
EMSYSRKRQRFLVNQG Haywire display
motor
(weak) (SEQ ID N0:269) defects and reduced
life span
YYEHRKKNIGSVHPLFK
KFRG (bipartite)
(SEQ ID
N0:270)
77 ARGKKKQPK (SEQ Rrpl HAP1 Recombination repair
ID protein
N0:271) 1); 679 aa; the
252 as C-
98 KPKGRAKKA (SEQ terminal domain
ID is
N0:272) homologous to AP-
157 QAKGRKKKELP (SEQ endonucleases,
ID whereas the
N0:273) 1-426 as domain
is highly
179 EPPKQRARKE (SEQ charged, carries
ID all of the
NO:274) putative NLSs.
241 PPKAASKRAKKGK
(SEQ
ID N0:275)
282 PKKRAKKTT (SEQ
ID
N0:276)
317 EPAPGKKQKKSAD
(SEQ
ID N0:277)
336 EEEAKPSTETKPAKGR
KKAP (SEQ ID N0:278)
372 KPARGRKKA (SEQ
ID
N0:279)
394 GSKTTKKAKKAE
(SEQ ID N0:280)
S. CEREYISIAE
200 IEKRRKLYISGG RAD 1 ERCC4 1100 aa; 30% sequence
(SEQ ID N0:281) (XPF) identity to Radl6;
RAD1
515 NKKRGVRQVLLN Radl6 interacts strongly
(SEQ with
ID N0:282) RAD10
565 KEQVTTKRRRTRG
(conserved in Radl6)
(SEQ ID
N0:283)
1024 NLRKKIKSFNKLQ
(SEQ ID N0:284)
89 RQRKERRQGKRE RAD2 XPGC 1031 aa, 117.8
kDa; ssDNA
(SEQ ID NO:285) Radl3 endonuclease; rad
mutants
907 ENKFEKDLRKKLVNNE are defective in
incision
(SEQ ID N0:286)
984 RDVNKRKIKKGKQKRI
(SEQ ID N0:287)
1017 KRISTATGKLKKRKM
(SEQ ID N0:288)
672 GKDDYGVMVLADRRF RAD3 ERCC2 or XPD; 778 aa, 89,779
Da; 30%
SRKRSQLP (contains (S. cer)RadlS or Rhp3 sequence identity
the bulky to radl6;
F) (SEQ ID NO:289) ATP-dependent DNA
helicase; single-stranded
DNA-dependent ATPase.
71
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
Putative NLS Gene Equivalent Features
protein
product in other species
26 PLSRRRRVRRKNQPLPDRAD4 XPC 754 aa; mutations
in RAD4
AKKKFKTG (SEQ ID that that inactivate
N0:290) the
134 NEERKRRKYFHMLYL excision repair
function of
(SEQ ID N0:291) RAD4 result in
truncated
160 EWINSKRLSRKLSNL proteins missing
the C-
(weak) (SEQ ID N0:292) terminal one-third
of RAD4.
254 EMSANNKRKF'KTLKRSD
weak (SEQ ID N0:293)
382 WMNSKVRKRRITKDDF
GEK (SEQ ID N0:294)
403 RKVITALHHRKRTKID
DYED (SEQ ID N0:295) ,
504 KTGSRCKKVIKRTVGRP
(SEQ ID N0:296)
150 FHPKRRRIYGFR RADS 1169 aa; helicase
(SEQ ID involved in
NO:297) postreplication-repair
(RAD6
215 DSRGRKKASM (SEQ epistasis group);
ID binds DNA
N0:298) with the seven
helicase
297 DGESLMKRRRTEGGNK motifs and with
zinc forgers;
REK (SEQ ID N0:299) increases the instability
of
1152 DEDERRKRRIEE poly (GT) repeats
in the yeast
(SEQ ID N0:300) genome.
1 MSTPARRRLMRDFKRM RAD6 RAD6 mediates the
KEDAPP (SEQ ID N0:301) ubiquitination
of H2A and
H2B histones
15 GVAKLRKEKSGAD RAD10 ERCC1 210 aa; forms an
(SEQ ID N0:302) endonuclease with
RAD1;
76 DDYNRKRPFRSTRPGK the basic and tyrosine-rich
(SEQ ID N0:303) central domain
was
suggested to bind
DNA by
ionic interactions
and
tyrosine intercalation.
172 EGKAHRREKKYE RAD14 XPAC 247aa, 29.3 kDa;
two zinc
(SEQ ID N0:304) fingers; involved
in lesion
200 NRLREKKHGKAHIHH recognition; 27%
sequence
(SEQ ID N0:305) identity and 54%
sequence
similarity (if
conserved
residues are grouped
together) to human
XPA;
deletion of RAD
14 gene
generates high
UV
sensitivity.
345 ERRKQLKKQGPKRP Ixrl 591 aa; two consecutive
(SEQ ID N0:306) (S. cer) HMG boxes; involved
in
479 ETYKKRIKEWESCYPDE recognition of
1,2-intrastrand
(SEQ ID N0:307) d(GpG) and d(ApG)
cisplatin
crosslinks.
None RAD23 HHR23
72
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
Putative NLS Gene Equivalent Features
protein
product in other species
483 LTCKKLKTHNRIILSGRAD26 ERCC6 1075aa; disruption
of the
weak (SEQ ID N0:308)(yeast CS-B (hum) RAD26 gene gives
viable
934 NALRKSRKKITKQYEIGTERCC6) yeast cells unable
to
PXgGEIRKRDP preferentially
repair the
(SEQ ID N0:309) actively transcribed
strands;
surprisingly,
in contrast to
human CS-B cells,
disruption
of the RAD26 in
yeast does
not cause sensitivity
to UV,
Cisplatin, or
X-rays.
634 KPTSKPKRVRTATKKKIPMREl l Rad32 (S pom) meiotic recombination
(SEQ ID N0:310) protein; functions
in the
408 FYKKRSPVTRSKKSG same pathway with
RAD51
(SEQ ID N0:311 )
none; RAD51 RecA (E. coli)402 aa; essential
for repair of
361 GFKKGKGCQR DSBs and recombination;
(SEQ ID N0:312) ~ associates strongly
with
RAD52; self associates;
neither RAD51
nor RAD52
possess a typical
simple
NLS.
none; RAD51 364 as
(K.
328 GFKKGKGCQR lactis)
(SEQ ID N0:313)
none; RAD52 Rad22 504 aa; rad52
mutants are
155 ERAKKSAVTDALKRSLR defective in ionizing
GFGXBDKDFLAKIDKVKFDP radiation, mitotic
PD (tripartite) recombination,
mating-type
(SEQ ID N0:314) switching, and
repair of
DSDs.
1 MARRRLPDRPP RAD54 898 aa; recombination-repair
(SEQ ID N0:315) protein; ATP-binding
motif;
65 GGRSLRKRSA helicase domains;
in the
(SEQ ID N0:316) same subfamily
of helicases
99 QLTKRRKD with MOT1 and
SNF2.
(SEQ ID N0:317)
269 DETVFVKSKRVKASSSRAD55 Similarity to
RecA, and
(extremely weak if lower similarity
at all NLS) to RAD51,
(SEQ TD N0:318) RAD57, and DMC1
317 GEDRKREGRNLKR
(SEQ ID N0:319)
371 PISRQSKKRKFDYRVPRAD57 460 aa; nucleotide-binding
(SEQ TD N0:320) domain; limited
similarity to
RAD51
62 GLKKPRKKTKSSRH SSL2 ERCC3 (XPB) 843 aa; putative
helicase that
(SEQ ID N0:321) seems to function
in repair
688 GRILRAKRRNDEG but also in the
removal of
(SEQ ID N0:322) secondary structures
in the 5'
784 GRGSNGHKRFKS untranslated region
(weak) of mRNA
(SEQ ID N0:323) to allow ribosome
binding
and scanning.
73
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
Putative NLS Gene Equivalent Features
protein
product in other species
50 TRRHLCKIKGLSE DMC1 RecA 334 aa; yeast
(weak) homolog of
(SEQ ID N0:324) RecA, meiosis-specific;
277 DGRKPIGGHX12RKGRG dmcl mutants are
defective
DER (bipartite) (SEQ in reciprocal
ID recombination
N0:325) and accumulate
DSBs
11 ETEKRCKQKEQRY PMS1 904 aa, 103 kDa;
mismatch-
(SEQ ID N0:326) repair protein;
Mutt
(Salmonella) and
HexB
(Streptococcus)
homolog
None HRR25 Hhpl, Hhpl Mutations in HRR25
(S pom) Ser/Thr
1 MDLRVGRKFRIGRKIG CKI (mamm protein kinase
cause defects
(SEQ ID N0:327) in DNA repair
and
139 GRRGXBGLSKKYRDFNT retardation in
cell cycling
HRHIP (Bipartite
weak NLS)
(SEQ ID N0:328)
96 HELTKRSSRRVETEK YKL510 383 aa; structure-specific
(SEQ ID N0:329) endonuclease;
two domains
of about 100 as
with
sequence similarity
to N- and
C-terminal regions
~ of RAD2.
200 MLAMARRKKKIvISAKMOTl Modifier of transcription
1;
(SEQ ID N0:330) 1867 aa; DNA helicase
of S.
617 EHYKVKHTEK (weak cerevisiae required
for
NLS) (SEQ ID N0:331) viability; increases
gene
670 LHPEKKRSISE (weak expression of
several., but
NLS) (SEQ ID N0:332) not all, pheromone-
responsive genes
in the
absence of STE12;
the 1257
to 1825 as domain
(568 as
residues) has
homology to
SNF2 and RAD54
S. POMBE
60 SSIDExSSIK.RKRRI Swi4 Duc-1 113 kDa; KCII
(SEQ ID sites are
N0:333) Rep-3 upstream of NLS
like in
SV40 large T;
the
homologous prokaryotic
MutS and HexA
lack NLS
96 GELAKRVARHQKARE Rad2 380 as
(weak NLS) (SEQ ID
N0:334)
362 GSAKRKRDS
(SEQ ID N0:335)
372 KGGESKKKR
(SEQ ID N0:336)
None Rad9 427 aa; no homology
to other
DNA repair proteins;
rad9
fission yeast
mutants are
sensitive to both
UV and
ionizing radiation;
may be
involved in recombination-
repair.
None Rhp3 or ERCC2 772 aa; DNA helicase;
65%
681 DKRYGRSDKRTKLPK radl5 RAD3 identity to RAD3
and 55%
(SEQ ID N0:337) identity to ERCC2;
essential
for viability
74
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
Putative NLS Gene Equivalent Features
protein
roduct in other s
ecies
464 PPSKRRRVRGG Radl6 RAD1 Function in repair
of UV
(SEQ ID N0:338) damage for both
cyclobutane
dimer and (6-4)
photoproduct
lesions; Radl6
interacts with
SwI l O,
431 DFKQAILRKRKNESPERad21 628 aa, 67.8 kDa,
acidic
EVEP (SEQ ID N0:339) protein; a single
base
substitution in
mutant rad21-
45, changing an
Ile into a
Thr, is responsible
for the
low efficiency
in repair of
DSBs after g-radiation
although capable
of arresting
at G2.
490 DKKAKKG (SEQ Rad22 RAD52 496 aa; functions
ID in
N0:340) recombination-repair
and
matin - a switchin
.
394 DVVQFYLKKKYTRSKRNRad32 MRE11 (S cer)648 aa; meiotic
DC (weak because recombination protein;
of Y) (SEQ rad32
ID N0:341) mutants are sensitive
to b
575 PSPALLKKTNKRRELP and UV radiation;
functions
(SEQ ID N0:342) in the same pathway
with
Rll 51 RAD51 .
Rad51 recombination-re
air
GLAKKYRDHKTHLHIP Hhpl CKI (mammy Ser/Thr protein
(weak kinase;
NLS because of Y HRR25 (S cer)mutation in this
and H) (SEQ gene causes
ID N0:343 re air defects
None Hhp2 CKI (mammy Ser/Thr protein
kinase;
GLAKKYRD~"KTHVHIP HRR25 (S cer)mutation in this
(H in gene causes
Hhpl is replaced repair defects
by F in Hhp2)
SE ID N0:344
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
Table 9. NLS in Transcription factors
NLS and Flanks Protein factor and features
highly basic
HR4QRTRK7R (SEQ ID Human GCF (GC-factor)
N0:345)
LRRKSRP (SEQ ID N0:346)
SRRTKRRQ (SEQ ID
N0:347)
GRKRKKRT (SEQ ID Oct-6 protein transcription factor from
N0:348) mouse cells
GRRRKKRT (SEQ ID Mouse Oct-2 protein transcription factors
N0:349) (Oct-2.1 for Oct-2.6
isoforms)
ARKRKRT (SEQ ID N0:350)Oct-3 from mouse P19 embryonal carcinoma
cells
NRRQKGKRS (SEQ ID
N0:351)
ECRRKKKF Human ATF-1. In basic region/leucine zipper.
(SEQ ID N0:352)
ERKKRRRF (SEQ ID Human ATF-3 (in basic region that binds
N0:353) DNA)
AKCRNKKKEKT (SEQ
ID
NO:354)
SKKKIRL (SEQ ID N0:355)Mouse Pu.l (Friend erythroleukemia cells).
Related to ets oncogene
QKGNRKKM (SEQ ID
N0:356)
VKKVKKKI. (SEQ ID
N0:357)
VKRKKI (SEQ ID N0:358)Human PRDII-BFI that binds to IFN-/3 gene
promoter. (The largest
CRNRYRKLE (SEQ ID DNA-binding protein known, of 298 kD).
N0:359)
IRKRRKMK (SEQ ID
N0:360)
PKKKRLRL (SEQ ID
N0:361)
GKKKKRKREKL Murine LEF-1 (397 aa). Lymphoid-specific
with an HMG1-like box.
(within the HMG-box)NLS is identical to that of human TCF-la.
(SEQ ID N0:362)
GKKKKRKREKL Human TCF-la (399 aa)
(within the HMG-box)(T cell-specific transcription factor
that activates the T cell receptor
(SEQ ID N0:363) Ca). Contains an HMG box. NLS core is
identical to that of murine
LEF-1.
GKKKRRSREKH Human TCF-1
(within the HMG-box)(uniquely T cell-specific). HMG box containing.
(SEQ ID
N0:364)
PKKCRARF (SEQ ID
N0:365)
FKQRRIKL (SEQ ID Xenopus laevis Oct-I(within POU-domain)
N0:366)
NRRRKKRT (SEQ ID
N0:367)
NRRQKEKRI (SEQ ID
N0:368)
DKRSRKRKRSK (SEQ Drosophila Suvar (3) 7 gene product involved
ID in position-effect
N0:369) variegation (932 aas). Five widely spaced
zinc-forgers could help
RLRIDRKRN (SEQ ID condensation of the chromatin fiber.
N0:370)
AKRSRRS (SEQ ID N0:371)
76
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
NLS and Flanks Protein factor and features
IRKRRKMKSVGD2E2 (SEQHuman MBP-1 (class I MHC enhancer binding
ID protein 1) mw 200
N0:372) kD. Induced by phorbol esters and mitogens
in Jurkat T cells.
(not suggested as
NLS by the
authors; between
the 1st and 2nd
zinc forger)
PPKKKRLRLAE
(suggested as NLS
by the
authors; just before
2nd zinc
forger) (SEQ ID N0:373)
CRNRYRKLE
(within 1 st zinc
forger)
(SEQ ID N0:374)
PRRKRRV (SEQ ID N0:375)rat TTF-1 (thyroid nuclear factor that
binds to the promoter of
HRYKMKRQ (SEQ ID thyroid-specific genes). An homeodomain
N0:376) protein.
DGKRKRKN (SEQ ID Human thyroid hormone receptor a (c-erbA-1
N0:377) gene). Belongs to the
DDSKRVAKRKL (SEQ family of cytoplasmic proteins that are
ID receptors of hydrophobic
NO:378) ligands such as steroids, vitD, retinoic
acid, thyroid hormones. The
NRERRRKEE (SEQ ID ligand binding may expose the NLS for
N0:379) nuclear import of the
WKQRRKF (SEQ ID N0:380)receptor-ligand complex.
NRRKRKRS (SEQ ID Drosophila gcl (germ cell-less) gene product
N0:381) (569 aa, 65 kD), located
PKKKKL (SEQ ID N0:382)in nuclei, required for germ line formation.
ARRKRRRL (SEQ ID C. elegans Sdc-3 protein (sex-determining
N0:383) protein) (2,150 aas). A
LKFKKVRD (SEQ ID zinc finger protein.
N0:384)
FKKFRKF (SEQ ID N0:385)
GKQKRRF (SEQ ID N0:386)
ERLKRDKEKREKE (SEQ
ID
N0:387)
TRGRPKKVKE (SEQ ID
N0:388)
SKKRGRRRKKT (SEQ
ID
N0:389)
TRRQKRAKV (SEQ ID
N0:390)
SRKSKKRLRA (SEQ ID
N0:391 )
-
LKKIRRKIKNKI (SEQ Drosophila BBF-2 (related to CREB/ATF)
ID
N0:392) '
ESRRKKKE (SEQ ID
N0:393)
Group ease
DRNKKKKE (SEQ ID Xenopus RAR (retinoic acid receptor)
N0:394)
ARRRRP (SEQ ID N0:395)
GRRRRA (SEQ ID N0:396)Human ATF-2 (the 2nd and 3rd NLS are in
basic region that binds
DEKRRKV (SEQ ID NO:397)DNA)
CRQKRKV (SEQ ID N0:398).
ERKRRD (SEQ ID N0:399)Myn (murine homolog of Max). Forms a specific
DNA-binding
SRKKLRME (SEQ ID complex with c-Myc oncoprotein through
N0:400) a helix-loop-helix/leucine
zipper.
EEKRKRTYE (SEQ ID human NFKB p65 (550 aa).
N0:401)
Not binding DNA; complexed with p50 that
binds DNA. NFxB p50
also contains a NLS (Table 3b).
77
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
NLS and Flanks Protein factor and features
GRRRRA (SEQ ID N0:402)Human HB16, a cAMP response element-binding
protein
DEKRRKF (SEQ ID N0:403)
SRCRQKRKV (SEQ ID
N0:404)
SKKKKTKV (SEQ ID Human TFIIE-(3 (general transcription
N0:405) initiation protein factor; forms
NRPDKKKI (SEQ ID tetramer a2(32 with TFIIE-a)
N0:406)
QRRKKP (SEQ ID N0:407)
QKKRRFKT (SEQ ID
N0:408)
SRKRI~M (SEQ ID N0:409)Human kup transcriptional activator (433
aas). Two distantly spaced
zinc forgers. Expressed in hematopoietic
cells and testis.
ERKRLRNRLA (SEQ ID Mouse Jun-B homologue to avian sarcoma
virus 17 oncogene v-jun
N0:410) product. One region is similar to yeast
GCN4 and to Fos.
ATKCRKRKL (SEQ ID
N0:411 )
( 19 as stretch)
DKRx6ERKRRD (N-terminus)Max (specifically associates with c-Myc,
N-Myc, L-Myc). The Max-
(SEQ ID N0:412) Myc complex binds to DNA; neither Max
nor Myc alone exhibit
QSRKKLRME (C-terminus)appreciable DNA binding.
(SEQ ID N0:413)
DKEK_KIKLEEDE (withinChicken VBP (vitellogenin gene-binding
an protein). Leucine zipper.
acidic region) (SEQ Related to rat DBP.
ID N0:414)
IKKAKKV (SEQ ID N0:415)
TRRKKN (SEQ ID N0:416)
TRDDKRRA (SEQ ID Xenopus borealis B1 factor. Closely related
N0:417) to the mammalian USF.
EVERRRRDK (SEQ ID Binds to CACGTG in TFIIIA promoter to
developmentally regulate
N0:418) its expression.
TRDEKRRA (SEQ ID Human USF (upstream stimulatory factor)
N0:419) activating the major late
EVERRRRDK (SEQ ID adenovirus promoter
N0:420)
YRRYPRRRG (SEQ ID YB-1, a protein that binds to the MHC
class II Y box. YB-1 is a
NO:421) negative regulator.
QRRPYRRIE~RF (SEQ
ID
N0:422)
YRPRFRRG (SEQ ID
N0:423)
QRRYRRN (SEQ ID N0:424)
YRRRRP (SEQ ID N0:425)
AKERQKKD (SEQ ID Human TFEB Binds to IgH enhancer.
N0:426)
ERRRRF (SEQ ID N0:427)
LKERQKKD (SEQ ID Human TFE3 (536 aa). Binds to ~tE3 enhancer
N0:428) of IgH genes.
IERRRRFN (SEQ ID
N0:429)
YFRRRRLEKD (SEQ ID .
N0:430)
KTVALKRRKASSRL (SEQ Human Drl (176 aa, 19 kD). Interacts With
ID TBP (TATA-binding
N0:431) protein) thus inhibiting association of
TFIIA and/or TFIIB with TBP.
TBP-Drl association is affected by Drl
phosphorylation to repress
activated and basal transcription.
1 LRRRGRQTY (SEQ Drosophila ultrabithorax protein (from
ID the conserved 61 amino acid
N0:432) homeodomain segment only). Conserved in
the antenappedia
27 LTRRRRTEM (SEQ homeodomain protein.
ID
N0:433)
51 QNRRMKLKKEI (SEQ
ID
N0:434)
78
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
NLS and Flanks Protein factor and features
SNRRRPDHR (SEQ ID C. elegans sex-determining Tra-1 protein.
N0:435) Zinc forger. Peaks in the
VYRGRRRVRRE (SEQ second larval stage.
ID
N0:436)
P7AP2RRRRSADNKD2
(SEQ
ID N0:437)
PKKPRHQF (SEQ ID
N0:438)
EKRKKERN (SEQ ID Yeast NPS1 transcription protein factor
N0:439) (1359 aa) involved in cell
LLRRLKKEVE (SEQ ID growth control at G2 phase. Has a catalytic
domain of protein
N0:440) kinases.
EPLGRIRQKKRVY2D2
(SEQ
ID N0:441)
(EDAIKKRREARERRRLRQ)
(SEQ ID N0:442)
DKETTASRSKRRSSRKKRT
(SEQ ID N0:443)
ESKKKKPKL (SEQ ID
NO:444)
KKTAAKKTKTKS (SEQ
ID
N0:445)
QRKRQKL (SEQ ID N0:446)Human 243 transcriptional activator (968
aas), induced by mitogens
KAKKQK (SEQ ID N0:447)in T cells. N-terminal half is homologous
LRRKRQK (SEQ ID N0:448)to oncoprotein Rel and
Drosophila Dorsal protein involved in
development. The C-terminal
half contains repeats found in proteins
involved in cell-cycle control
of yeast and tissue differentiation in
Drosophila.
RDIRRRGKNKV (SEQ Mouse NF-E2 (45 kD), an erythroid transcription
ID factor from mouse
N0:449) erythroleukemia (MEL) cells. Involved
in globin gene regulation.
QNCRKRKLE (SEQ ID Binds to AP-I-like sites. Homology to
Jun B, GCN4, Fos, ATF1 and
N0:450) CREB in basic region/leucine zipper (see
Fig. 2).
Group eeexee
DKIRRKN (SEQ ID N0:451)Human glucocorticoid receptor
ARKTKKKI (SEQ ID
N0:452)
473 DKIRRKNCP (SEQ Mouse and human GR (glucocorticoid recptor)
ID
N0:453 )
EARKTKKKIKGIQ (SEQ
ID
N0:454)
Grouu A68xA
YRVRRERN (SEQ ID CIEBP (CCAAT/enhancer binding protein).
N0:455)
VRKSRDKA (SEQ ID Functions in Liver-specific gene expression.
N0:456)
DRLRKRVE (SEQ ID
N0:457)
DKIRRKN (SEQ ID N0:458)Human mineralocorticoid receptor
ARKSKKL (SEQ ID N0:459)
DKIRRKN (SEQ ID N0:460)Human PR (progesterone receptor)
GRKFKKF (SEQ ID N0:461)
EEVQRKRQKLMP (SEQ Human and mouse NFKB 105 kD precursor
ID of p50 (968 aas) (first R
N0:462) is at 361 position).
EEVQRKRQKL (SEQ ID Human NF-KB p50 (DNA-binding subunit).
Identical to protein
N0:463) KBF1, homologous to rel oncogene product.
NF-xB p65 also
contains a NLS (Table 3a).
GKTRTRKQ (SEQ ID Human TEF-1 (SV40 transcriptional enhances
N0:464) factor 1). 426 aa.
ARRKSRD (SEQ ID N0:465)
79
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
NLS and Flanks Protein factor and features
RKFRKSKS SE ID N0:466Rat, mouse, human IRF-1 (interferon regulatory
Q____________ ( Q factor-1). Induced in
)
TKSKTKRKL (SEQ ID lymphoma T cells by the pituitary peptide
N0:467) hormone prolactin.
Regulates the growth-inhibitory interferon
genes.
GKCKKKN (SEQ ID N0:468)Ehrlich ascites S-II transcription factor.
A general factor that acts at
the elongation step.
ERSKKRSRE (SEQ ID Tobacco TAF-1 transcriptional activator
N0:469)
ERELKREKRKQ (SEQ
ID
N0:470)
ARRSRLRKQ (SEQ ID
N0:471)
YKLDHMRRRIETDE (SEQ Drosophila TFIIEa, (433 aa), a general
ID transcription factor for RNA
N0:472) polymerase II. Composed of subunits a
and (3.
DKNRRKS (SEQ ID N0:473)Human ER (estrogen receptor); 595 aa.
IRKDRRG (SEQ ID N0:474)
IKRSKKN (SEQ ID N0:475)
EQRRHRIE (SEQ ID Yeast ADA2 (434 aa), a potential transcriptional
N0:476) adaptor required for
TTRAEKKRLL (SEQ ID the function of certain acidic activation
domains.
N0:477)
IDKKRSKEAKE (SEQ
ID
N0:478)
EAALRRKIRTISK Yeast GCNS gene product (439 aa)= reguired
for the function of
(SEQ ID N0:479) GCN4 transcriptional activator and for
the activity of the HAP2-3-4
complex.
Group sexes
NKKMRRNRF (SEQ ID Mouse LFB3
N0:480)
NRRKx4RQK (SEQ ID
N0:481)
TKKGRRNRF (SEQ ID Mouse LFB1
N0:482)
NRRKx4RHK (SEQ ID
N0:483)
NKKMRRNRFK (SEQ ID rat vHNFI-A
N0:484)
NKKMRRNR (SEQ ID murine HNF-1 (3
N0:485)
TKKGRRNRF (SEQ ID mouse HNF-I
N0:486)
NKKMRRNRF (SEQ ID human vHNFl
N0:487)
TKKGRILNRF (SEQ ID rat liver HNF1
N0:488)
LRRQKRFK (SEQ ID rat HNF-3(3
N0:489)
QQH3SH4Q (SEQ ID
N0:490)
LRRQKRFK (SEQ ID rat HNF-3y
N0:491)
LRRQKRFK (SEQ ID rat HNF-3a
N0:492)
LKEKERKA (SEQ ID rat DBP a protein factor that binds to
N0:493) the D site of the albumin gene
MKKARKV (SEQ ID N0:494)promoter
PR__R__E__R_ _R__Y rat AT-BP1. Highly acidic domain. Two
(SEQ ID N0:495) zinc fingers. Binds to the
B-domain of al-antitrypsin gene promoter
and to the NF-KB site in
the MHC gene enhancer.
DRRVRKGKV (SEQ ID A 19 kD Drosophila melanogaster nonhistone
associated with
N0:496) heterochromatin.
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
NLS and Flanks Protein factor and features
SKHGRRARRLDP (SEQ murine EBF (early B-cell factor) of 591
ID aa. Regulates the pre-B and
N0:497) B lymphocyte-specific mb-1 gene. Expxessed
in pre-B and B-cell
lines but not in plasmocytomas, T-cell
and nonlymphoid cell lines.
GRRTRRE (SEQ ID N0:498)human Spl
DEQKRAEKKAKE (SEQ yeast SNF2, a transcriptional regulator
ID of many genes.
N0:499)
IRRIHKVIRP (SEQ ID
N0:500)
LLRRLKKDVE (SEQ ID
N0:501)
Group Ox6Ax6
AKAKAKKA (SEQ ID mouse AGP/EBP (87% similarity to C/EBP),
N0:502) ubiquitously expressed
YKMRRERN (SEQ ID
N0:503)
VRKSRDKA (SEQ ID
N0:504)
AKAKAKKA (SEQ ID rat LAP, a 32-kD liver-enriched transcriptional
N0:505) activator, also present
YKMRRERN (SEQ ID in lung, with 71% sequence similarity
N0:506) to C/EBP. Leucine zipper.
VRKSRDKA (SEQ ID Accumulates to maximal levels around birth.
N0:507)
YRQRRER_(SEQ ID N0:508)Ig/EBP-1 (immunoglobulin gene enhancer-binding
protein). Forms
VKKSRLKSKQK (SEQ heterodimers with C/EBP.
ID
N0:509)
EDPEKE_K___R__IKELE mouse c-Myb
(SEQ ID
N0:510)
MRRKV (SEQ ID N0:511)
DYYKVKRPKTD (SEQ Drosophila eyes absent protein (760 aa),
ID a nuclear protein that
N0:512) functions in early development to prevent
programmed cell death and
GRARGRRHQ (SEQ ID to allow the event that generate the eye
to proceed. Mutations cause
N0:513) programmed cell death of eye progenitor
cells.
FRYRKIKDIY (SEQ ID
N0:514)
Group 6xAxAA
AKAKAKKA (SEQ ID rat IL-6DBP interacting with interleukin-6
N0:515) responsive elements. Has
a leucine zipper domain.
DKRQRNRC (SEQ ID mouse H-2RIIBP (MHC class I genes H-2
N0:516) region II binding protein).
FkrtirkD Member of the nuclear hormone receptor
superfamily.
FkrtirkD chicken RXR, related to RAR (retinoic
acid receptor), a nuclear
DKRQRNRC (SEQ ID protein factor from the thyroid/steroid
N0:517) hormone receptor family
VKSKAKKT (SEQ ID human NF-IL6 (345 aa). Specifically binds
N0:518) to IL1-responsive
YKIRRERN (SEQ ID element in the IL-6 gene. Leucine zipper.
N0:519) Homology to C/EBP.
VRKSRDKA (SEQ ID
N0:520)
QKKNRNKC (SEQ ID mouse PPAR (peroxisome proliferator activated
N0:521) receptor)
Group A6AxxAA
EQIRKLVKKHG (SEQ yeast RAP1
ID
N0:522) It binds regulatory sites at yeast mating
type silencers.
FRRSMKRKA (SEQ ID human vitamin D receptor (427 aa)
N0:523)
Group eexxee
~1
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
NLS and Flanks Protein factor and features
LKRHQRRH (SEQ ID mouse WTl (the murine homolog of human
N0:524) Wilms' tumor
predisposition gene WTl)
LKRHQRRH (SEQ ID human WT33 (Wilins' tumor predisposition)
N0:525)
Group A6AxxA
LKESKRK__ YDE (SEQ yeast SWI3 99 kD, highly acidic protein.
ID Global transcription
~
N0:526) activator.
EVLKVQKRRIYD (SEQ human RBAP-1 (retinoblastoma-associated
ID protein 1) factor (412 aa).
N0:527) A protein that binds to the pocket (functional
domain) of the
retinoblastoma (RB) protein involved in
suppression of cell growth
(tumor suppressor). The transcription
factor E2F, implicated in cell
growth, binds to the same pocket of RB.
82
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
Table 10. NLS in other nuclear proteins
Putative NLS Protein
YKSKKKA (SEQ ID N0:528) Yeast L3
TKKLPRKT (SEQ ID N0:529)
TRKKGGRRGRRL (SEQ ID N0:530) Yeast 59 ribosomal protein
C-terminus
ARATRRKRCKG (SEQ ID N0:531) Yeast L16 ribosomal protein
GKGKYRNRRW (SEQ ID N0:532) yeast L2 ribosomal protein
(homologous to
Xenopus L1). Encoded by intronless
genes.
GKGKMRNRRRIQRRG (SEQ ID N0:533)Xenopus laevis L1 ribosomal
protein (homologous
NKKVKRRELKKN (SEQ ID N0:534) to yeast L2) Encoded by intronless
genes.
AKTARRKA (SEQ ID N0:535)
IKAKE_KKP (SEQ ID N0:536)
GKPKAKKP (SEQ ID N0:537)
AKAKKRQ (SEQ ID N0:538)
ERKRKS (SEQ ID N0:539) human S6 ribosomal protein
(homologous to yeast
GKRPRTKA (SEQ ID N0:540) S 10)
HKRRRI (SEQ ID N0:541)
LKKQRTKKNKE (SEQ ID N0:542)
PKMRRRTYR (SEQ ID N0:543) Rat L17 ribosomal protein (184
aas)
KKKISQKKLKK (SEQ ID N0:544)
YMRRRTYRA (SEQ ID N0:545) Podocoryne carnea (hydrozoan,
Coelenteratum)
EVKKVSKKKL (SEQ ID N0:546) L17 ribosomal protein (184
aas) highly
homologous to rat L17.
ERNRKDKDAKFR (SEQ ID N0:547) human, rat ribosomal S 13 protein
ERKRKS (SEQ ID N0:548) yeast S10 ribosomal protein
(homologous to human
QRLQRKRH (SEQ ID N0:549) S6)
IRKRRA (SEQ ID NO:550)
GRRRKKIIRSRSRSRERRSRSRDRGRG12GRER35 kD subunit of U2 small nuclear
DRRRSRDRER (SEQ ID N0:551) ribonucleoprotein auxiliary
factor (U2AF), an
essential mammalian splicing
factor. U2AF35
interacts with the 65 kD subunit
(U2AF65). Both
proteins are concentrated in
a small number of
subnuclear organelles, the
coiled bodies.
EFEDPRD (SEQ ID N0:552) human UsnRNP-associated 70
k protein (437 aas)
ETREERME (SEQ ID N0:553) that is phosphorylated at Arg/Ser-rich
domains;
EAGDAPPDP (SEQ ID N0:554) involved in splicing
EERMERKRREK (SEQ ID N0:555)
HRDRDRDRERERRESRERDKERERRRSRSRD
RRRRSRSRDKEERRRSRERSKDKDRDRKRRS
SRSRERARRERERKEE (SEQ ID N0:556)
RDRDRERRRSHRSERERR___R__D___R_
DRDRDR__D___R__E__H__
~
KRGER (SEQ
ID NO:557)
QKRNNKKSKKKRCAE (SEQ ID N0:558)yeast TRM1 enzyme for the N2,N2-
EKLRKLKI (near C-terminus) dimethylguanosine modification
(SEQ ID N0:559) of both
mitochondrial and cytoplasmic
tRNAs. TRM1 is
both nuclear and mitochondrial.
The first motif is
within a region (70-213 as
segment) known to
cause nuclear localization
of j3-galactosidase.
NKRKRV (SEQ ID N0:560) Yeast nucleoporin NUP1 (1076
aa, 113 kD); an
SLKNRSNRKRE (SEQ ID N0:561) integral component of the pore
complex. Involved
EPKRKRRLP (SEQ ID N0:562) in both binding and translocation
steps of nuclear
ARMRHSKR (C-terminus) (SEQ import.
ID N0:563)
83
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
Putative NLS Protein
KAEKEx3KVD2E2 (SEQ ID N0:564) Chicken, ~enopus No 38 nucleolar
(38 kD);
~3~5~3R (SEQ ID N0:565) involved in intranuclear packaging
of preribosomal
particles. Shuttles between
nucleus and cytoplasm.
KTEREAEKALEEKx7R (SEQ ID N0:566)Chicken, hamster nucleolin (92
kD). Binds
Kx5Kx7Kx4RX3EDTTEETLR (SEQ preribosomal RNA. Shuttles between
ID N0:567) nucleus and
RG2RG2RG3RG2FG2RG3RGFG2RG3FRG2RG4cytoplasm.
DHKPQGKKIKFE (SEQ ID N0:568)
(C-terminus)
WYKHFKKTKD (SEQ ID N0:569) human SATB1 (763 aa) which binds
selectively to
AT-rich MARS with mixed
A, T, C on one strand excluding
G. Binds to minor
groove with little contact with
bases.
QKKKQMKAD (SEQ ID N0:570) yeast CBFSp, a centromere-binding
protein
(KKEKKE)5 (SEQ ID N0:571) (55kDa, 483aa). The KKE repeat
at its C-terminus
KKEKKRKSED (SEQ ID N0:572) occurs in microtubule-binding
domains; yeast cells
EEKKSKKSKK (SEQ ID N0:573) containing only three copies
of the KKE repeat of
CBFSp delay at G2/M; depletion
of CBESp arrests
cells at G1/S.
TKKKSFKL (SEQ ID N0:574) yeast CCEl, a cruciform cutting
endonuclease
KSERERMLRESLKEERRRF (SEQ ID rat nucleoporin 155 or Nup155
N0:575) (1390 aas, 155
kDa), a protein of the nuclear
pore complex;
contains 46 consensus sites
for various kinases;
associated with both the nucleoplasmic
and the
cytoplasmic region of pores.
PKKGSKKA (SEQ ID N0:576) human H2B variant differentially
expressed during
DGKKRKRSRKES (SEQ ID N0:577) the cell cycle
GAKRHRKVLRD (SEQ ID N0:578) Calf thymus histone H4
14-24 (102 aa)
PAIRRLARRG (SEQ ID N0:579)
32-41
EHARRKT (SEQ ID N0:580)
74-80
ARRIRGERA 127-135 (SEQ ID N0:581)Calf thymus H3
(135 aa)
GSHHKAKGK 121-129 (SEQ ID N0:582)Calf thymus H2A
(129 aa)
RGKSGKARTKAKSRSSR 3-19 (SEQ Sea urchin Psammechinus miliaris
ID H2A (123 aa)
N0:583)
PKKGSKKA 10-17 (SEQ ID N0:584)Calf thymus H2B
QKKDGKKRKRSRKES 22-36 (SEQ (125 aa)
ID N0:585)
GGKKRHRKRKGSY (SEQ ID N0:586) Sea urchin Psammechinus rniliaris
H2B (122 aa)
22-34
PRTDKKRRRKRKES 19-32 (SEQ ID Starfish H2B
N0:587)
(121 aa)
PAKAPKKKA 12-20 (SEQ ID N0:588)Trout testis H1
EAKKPAKKA 104-112 (SEQ ID N0:589)(194 aa)
AKKPKKV 128-134 (SEQ ID N0:590)
AKKSPKKAKKP 142-152 (SEQ ID
N0:591)
PKKVKKP 183-189 (SEQ ID N0:592)
~4
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
Putative NLS Protein
PRRKAKRA 30-37 (SEQ ID N0:593)Sea urchin Parechinus angulosus
sperm H1 (248
PKKAKKT 119-125 (SEQ ID N0:594)aa)
AKAKKAKA 129-136 (SEQ ID N0:595)
AKKARKAKA 139-147 (SEQ ID N0:596)
AKKAKKPKKKA 17I-181 (SEQ ID
N0:597)
AKKAKKPAKK 182-191 (SEQ ID
N0:598)
SPKKAKK:P 192-199 (SEQ ID N0:599)
AKKSPKKKKAKRS 200-212 (SEQ
ID N0:600)
PKKAKKA 213-219 (SEQ ID N0:601)
AKKAKKS 227-233 (SEQ ID N0:602)
PRKAGKRRSPKKARK 234-248 (SEQ
ID
N0:603)
ARRRKTA 1-7 (SEQ ID N0:604) Annelid sperm Hla
IRKFIRKA 55-61 (SEQ ID N0:605)(119 aa)
PKKKKP. 83-88 (SEQ ID N0:606)
AKKPKAKKVKKP 89-100 (SEQ ID
N0:607)
AKKKTNRARKPKTKKNR 104-120 (SEQ
ID
N0:608)
PKRKVSS 1-7 (SEQ ID N0:609) Calf thymus HMG14
EEPKRRSARLS 14-24 (SEQ ID N0:610)(100 aa)
PKRKAEGDAK 1-10 (SEQ ID N0:611)Calf thymus HMG17
PKGKKGKA 52-59 (SEQ ID N0:612)(89aa; 9,247 D)
PKKPRGKM (SEQ ID N0:613) Calf thymus HMG 1
EHKKKHP (SEQ ID N0:614) (2S9 aa)
ETKKKFKDP (SEQ ID N0:615)
EKSKKKK(E/D)41 (SEQ ID N0:616)
E3G2KKKKKFAK (SEQ ID N0:617)
EHKKKHP (SEQ ID N0:618) Calf thymus HMG 2
PKGDKKGKKKDP (SEQ ID N0:619) (256 aa)
E4G3KKKKKFAK (SEQ ID N0:620)
PKRKSATKGDEPARR 1-15 (SEQ ID Trout testis H6 (60 aa)
N0:621)
KPKKAAAPKKA 30-34 (SEQ ID N0:622)
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
References
U.S. Patent Documents
4,394,448 July, 1983 Szoka, Jr. et al.
4,598,051 July, 1986 Papahadjopoulos et al.
5,013,556 May, 1991 Woodle et al.
Journal Articles
Allen, T.M. and Chonn, A. (1987) "Large unilamellar liposomes with low uptake
into the reticuloendothelial system" FEBS Lett. 223:42-46.
Allen, T.M. et al. (1991) "Liposomes containing synthetic lipid derivatives of
polyethylene glycol show prolonged circulation half lives in vivo" Biochim.
Biophys. Acta 1066:29-36.
Anderson, W.F. (1992) "Human gene therapy" Science 256:808-813.
Aoki, K. et al. (1995) "Liposome-mediated in vivo gene transfer of antisense K-
ras
construct inhibits pancreatic tumor dissemination in the murine peritoneal
cavity" Cancer Res. 55:3810-3816.
Arcasoy, S.M. et al. (1997) "Polycations increase the efficiency of adenovirus-
mediated gene transfer to epithelial and endothelial cells in vitro" Gene
Ther.
4:32-38.
Beauchamp, C.O. et al. (1983) "A new procedure for the synthesis of
polyethylene
glycol-protein adducts; effects on function, receptor recognition, and
clearance of superoxide dismutase, lactoferrin, and alpha 2-macroglobulin"
Anal. Biochem. 131:25-33.
Bongartz, J.-P. et al. (1994) "Improved biological activity of antisense
oligonucleotides conjugated to a fusogenic peptide" Nucl. Acids Res.
22:4681-4688.
Boulikas, T. (1993) "Nuclear localization signals (NLS)" Crit. Rev. Eukar.
Gene
Expression 3:193-227.
Boulikas, T. (1994) "Putative nuclear localization signals (NLS) in protein
transcription factors" J. Cell. Biochem. 55:32-58.
Boulikas, T. (1996a) "Cancer gene therapy and immunotherapy" Intl. J. Oncol.
9:941-954.
Boulikas, T. (1996b) "Gene therapy to human diseases: ex vivo and in vivo
studies"
Intl. J. Oncol. 9:1239-1251.
Boulikas, T. (1996c) "Liposome DNA delivery and uptake by cells" Oncol. Rep.
3:989-995.
Boulikas, T. (1996d) "Nuclear import of protein kinases and cyclins" J. Cell.
Biochem. 60:61-82.
Boulikas, T. (1997a) "Gene therapy ofprostate cancer: p53, suicidal genes, and
other
targets" Anticancer Res. 17:1471-1506.
Boulikas, T. (1997b) "Nuclear import of DNA repair proteins" Anticancer Res.
17:843-864.
Boulikas, T. (1997c) "Nuclear localization signal peptides for the import of
plasmid
DNA in gene therapy" Int. .T. Oncol. 10:301-309.
86
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
Boulikas, T. (1998a) "Status of gene therapy in 1997: Molecular mechanisms,
disease targets, and clinical applications" Gene Ther. Mol. Biol. 1:l-172.
Boulikas, T. (1998b) "Nucleocytoplasmic trafficking: implications for the
nuclear
import of plasmid DNA during gene therapy" Gene Ther. Mol. Biol. 1:713-
740.
Boulikas, T. and Martin, F. (1997) "Histones, protamine, and polylysine but
not
poly(E:K) enhance transfection efficiency" Int. J. Oracol. 10:317-322.
Capaccioli, S. et al. (1993) "Cationic lipids improve antisense
oligonucleotide uptake
and prevent degradation in cultured cells and in human serum" Biochem.
BioplZys. Res. Comm. 197:818-825.
Creuzenet, C. et al. (1997) "Interaction of alpha s2- and beta-casein signal
peptides
with DMPC and DMPG liposomes" Peptides 18:463-472.
Culver, K.W. (1996) in: Gene Therapy: A primer for physicians, Second Edition.
Mary Ann Liebert, Inc. Publications, NY, pp. 1-198.
Curtain, C. et al. (1999) "The interactions of the N-terminal fusogenic
peptide of
HIV-1 gp41 with neutral phospholipids" Eur. Biophys. J. X8:427-436.
de la Maza, A. et al. (1998) "Solubilization of phosphatidylcholine liposomes
by the
amphoteric surfactant dodecyl betaine" Chem. Phys. Lipids 94:71-79.
Decout, A. et al. (1999) "Contribution of the hydrophobicity gradient to the
secondary structure and activity of fusogenic peptides" Mol. Membr. Biol.
16:237-246.
Duguid, J.G. et al. (1998) "A physicochemical approach for predicting the
effectiveness of peptide-based gene delivery systems for use in plasmid-
based gene therapy" Biophys. J. 74:2802-2814.
Filion, M.C. and Phillips, N.C. (1997) "Toxicity and immunomodulatory activity
of
liposomal vectors formulated with cationic lipids toward immune effector
cells" Biochim. Biophys. Acta 1329:345-356.
Fresta, M. et al. (1998) "Liposomal delivery of a 30-mer antisense
oligodeoxynucleotide to inhibit proopiomelanocortin expression" J. Pharm.
Sci.87:616-625.
Gabizon, A. and Papahadjopoulos, D. (1988) "Liposome formulations with
prolonged circulation time in blood and enhanced uptake by tumors" Proc.
Natl. Acad. Sci. ~ USA 85:6949-6953.
Gabizon, A. et al. (1989) "Pharmacokinetics and tissue localization of
doxorubicin
encapsulated in stable liposomes with long circulation times" J. Natl. Cancer
Inst. 81:1484-1488.
Ghosh, J.K. and Shai, Y. (1999) "Direct Evidence that the N-Terminal Heptad
Repeat of Sendai Virus Fusion Protein Participates in Membrane Fusion" J.
Mol. Biol. 292:531-546.
Green, M. and Loewenstein, P.M. (1988) "Autonomous functional domains of
chemically synthesized human immunodeficiency virus tat transactivator
protein" Cell 55:1179-1188.
Gupta, D. and Kothekar, V. (1997) "500 picosecond molecular dynamics
simulation
of amphiphilic polypeptide Ac(LKKL)4 NHEt with 1,2 di-mysristoyl-sn-
glycero-3-phosphorylcholine (DMPC) molecules" Indian J. Biochern.
Biophys. 34:501-511.
Hofland, H.E.J. et al. (1996) "Formation of stable cationic lipid/DNA
complexes for
gene transfer" Proc. Natl. Acad. Sci. USA 93:7305-7309.
87
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
Jaaskelainen, I. et al. (1994) "Oligonucleotide-cationic liposome
interactions. A
physicochemical study" Biochim. Biophys. Acta 1195:115-123.
Judice, J.K. et al. (1997) "Inhibition of HIV type 1 infectivity by
constrained alpha-
helical peptides: implications for the viral fusion mechanism" Proc. Natl.
Acad. Sci. USA 94:13426-13430.
Kono, K. et al. (1993) "Fusion activity of an amphiphilic polypeptide having
acidic
amino acid residues: generation of fusion activity by alpha-helix formation
and charge neutralization" Biochim. Biophys. Acta 1164:81-90.
Lambert, G. et al. (1998) "The C-terminal helix of human apolipoprotein All
promotes the fusion of unilamellar liposomes and displaces apolipoprotein AI
from high-density lipoproteins" Eur. J. Biochem. 253:328-338.
Lambert, O. et al. (1998) "A new "gel-Iike" phase in dodecyl maltoside-lipid
mixtures: implications in solubilization and reconstitution studies" Biophys.
J. 74:918-930.
Lappalainen, K. et al. (1997) "Intracellular distribution of oligonucleotides
delivered
by cationic liposomes: light and electron microscopic study" Histochem.
Cytochern. 45:265-274.
Lasic, D. (1997) in: Liposomes in Gene Delivery, CRC Press, pp. 1-295.
Lee, S. et al. (1992) "Effect of amphipathic peptides with different alpha-
helical
contents on liposome-fusion" Biochim. BiopTzys. Acta 1103:157-162.
Lelkes, P.I. and Lazarovici, P. (1988) "Pardaxin induces aggregation but not
fusion
of phosphatidylserine vesicles" FEBS Lett. 230:131-136.
Leonard, A.N. and Cohen, D.E. (1998) "Submicellar bile salts stimulate
phosphatidylcholine transfer activity of sterol carrier protein 2" J. Lipid
Res.
39:1981-1988.
Lewis, J.G. et al. (1996) "A serum-resistant cytofectin for cellular delivery
of
antisense oligodeoxynucleotides and plasmid DNA" Proc. Natl. Acad. Sci.
USA 93:3176-3181.
Li, S. and Huang, L. (1997) "In vivo gene transfer via intravenous
administration of
cationic lipid-protamine-DNA (LPD) complexes" Gene Ther. 4:891-900.
Lins, L. et al. (1999) "Molecular determinants of the interaction between the
C-
terminal domain of Alzheimer's beta-amyloid peptide and apolipoprotein E
alpha-helices" J. Neurochem. 73:758-769.
Litzinger, D.C. et al. (1996) "Fate of cationic Iiposomes and their complex
with
oligonucleotide in vivo" Biochim. Biophys. Acta 1281:139-149.
Lopez, O. et al. (1998) "Direct formation of mixed micelles in the
solubilization of
phospholipid liposomes by Triton X-100" FEBS Lett. 426:314-318.
Lusa, S. et al. (1998) "Direct observation of lipoprotein cholesterol ester
degradation
in lysosomes" Biochem. J. 332:451-457.
Macosko, J.C. et al. (1997) "The membrane topology of the fusion peptide
region of
influenza hemagglutinin determined by spin-labeling EPR" J. Mol. Biol.
267:1139-1148.
Macreadie, LG. et al. (1997) "Cytotoxicity resulting from addition of HIV-1
Nef N
terminal peptides to yeast and bacterial cells" Biochem. Biophys. Res.
Commun.232:707-711.
Martin, F. and Boulikas, T. (1998) "The challenge of liposomes in gene
therapy"
Gene Ther. Mol. Biol. 1:173-214.
88
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
Martin, I. et al. (1999) "Membrane fusion induced by a short fusogenic peptide
is
assessed by its insertion and orientation into target bilayers" Biochemistry
38:9337-9347.
Martin, I. and Ruysschaert, J.M. (1997) "Comparison of lipid vesicle fusion
induced
by the putative fusion peptide of fertilin (a protein active in sperm-egg
fusion) and the NH2-terminal domain of the HIV2 gp41" FEBS Lett.
405:351-355.
Massari, S. and Colonna, R. (1986) "Gramicidin induced aggregation and size
increase of phosphatidylcholine vesicles" Chem. Phys. Lipids 39:203-220.
Melino, S. et al. (1999) "Zn(2+) ions selectively induce antimicrobial
salivary
peptide histatin-5 to fuse negatively charged vesicles. Identification and
characterization of a zinc-binding motif present in the functional domain"
Biochemistry 38:9626-9633.
Midoux, P. and Monsigny, M. (1999) "Efficient gene transfer by histidylated
polylysine/pDNA complexes" Bioconjug. Chem. 10:406-411.
Murata, M. et al. (1991) "Modification of the N-terminus of membrane fusion-
active
peptides blocks the fusion activity" Biochem. Biophys. Res. Cornmun.
179:1050-1055.
Niidome, T. et al. (1997) "Membrane interaction of synthetic peptides related
to the
putative fusogenic region of PH-30 alpha, a protein in sperm-egg fusion'' J.
Peptide Res. 49:563-569.
Pak, C.C. et al. (1999) "Elastase activated liposomal delivery to nucleated
cells"
Biochim. Biophys. Acta 1419:111-126.
Papahadjopoulos, D. et al. (1991) "Sterically stabilized liposomes:
Improvements in
pharmacokinetics and antitumor therapeutic efficacy" Proc. Natl. Acad. Sci.
LISA 88:11460-11464.
Parente, R.A. et al. (1988) "pH-dependent fusion ofphosphatidylcholine small
vesicles. Induction by a synthetic amphipathic peptide" J. Biol. Chem.
263:4724-4730.
Partidos, C.D. et al. (1996) "Priming of measles virus-specific CTL responses
a$er
immunization with a CTL epitope linked to a fusogenic peptide" Virology
215:107-110.
Pecheur, E.I. et al. (1997) "Membrane anchorage brings about fusogenic
properties
in a short synthetic peptide" Biochemistry 36:3773-3781.
Peelman, F. et al. (1999) "Characterization of functional residues in the
interfacial
recognition domain of lecithin cholesterol acyltransferase (LCAT)" Protein
Eng. 12:71-78.
Pereira, F.B. et al. (1997) "Permeabilization and fusion of uncharged lipid
vesicles
induced by the HIV-1 fusion peptide adopting an extended conformation:
dose and sequence effects" Biophys. J. 73:1977-1986.
Pillot, T. et al. (1999) "The nonfibrillar amyloid beta-peptide induces
apoptotic
neuronal cell death: involvement of its C-terminal fusogenic domain" J.
Neurochem. 73:1626-1634.
Pillot, T. et al. (1997) "Specific modulation of the fusogenic properties of
the
Alzheimer beta-amyloid peptide by apolipoprotein E isoforms" Eur. J.
Biochem. 243:650-659.
Pillot, T. et al. (1997) "The 118-135 peptide of the human prion protein forms
amyloid fibrils and induces liposome fusion" J. Mol. Biol. 274:381-393.
89
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
Plank, C. et al. (1996) "Activation of the complement system by synthetic DNA
complexes: a potential barrier for intravenous gene delivery" Hunz. Gerze
Ther. 7:1437-1446.
Rodriguez-Crespo, I. et al. (1994) "Prediction of a putative fusion peptide in
the S
protein of hepatitis B virus" J. Gen. Yirol. 75:637-639.
Rodriguez-Crespo, I. et al. (1999) "Fusogenic activity of hepadenavirus
peptides
corresponding to sequences downstream of the putative cleavage site"
Virology 261:133-142.
Ross, G. et al. (1996) "Gene therapy in the United States: a five-year status
report"
Hum. Gene Ther. 7:1781-1790.
Schroeder, F. et al. (1990) "Intermembrane cholesterol transfer: role of
sterol carrier
proteins and phosphatidylserine" Lipids 25:669-674.
Schroth-Diez, B. et al. (1998) "Fusion activity of transmembrane and
cytoplasmic
domain chimeras of the influenza virus glycoprotein hemagglutinin" J. Yirol.
72:133-141.
Schutze, W. and Muller-Goymann, C.C. (1998) "Phase transformation of a
liposomal dispersion into a micellar solution induced by drug-loading"
Pharm. Res. 15:538-543.
Song, Y.I~. et al. (1997) "Characterization of cationic liposome-mediated gene
transfer in vivo by intravenous administration" Hum. Gene Ther. 8:1585-
1594.
Sorgi, F.L. et al. (1997) "Protamine sulfate enhances lipid-mediated gene
transfer"
Gerze Ther. 4:961-968.
Suenaga, M. et al. (1989) "Basic amphipathic helical peptides induce
destabilization
and fusion of acidic and neutral liposomes" Biochizzz. Biophys. Acta 981:143
150.
Takle, G.B. et al. (1997) "Delivery of oligoribonucleotides to human hepatoma
cells
using cationic lipid particles conjugated to fernc protoporphyrin IX (heme)"
Antisense Nucleic Aeid Drug Dev. 7:177-185.
Templeton, N.S. et al. (1997) "Improved DNA: liposome complexes for increased
systemic delivery and gene expression" Nature Biotechnol. 15:647-652.
Thierry, A.R. and Dritschilo, A. (1992) "Intracellular availability of
unmodified,
phosphorothioated and liposomally encapsulated oligodeoxynucleotides for
antisense activity" Nucl. Acids Res. 20:5691-5698.
Tirosh, O. et al. (1998) "Hydration of polyethylene glycol-grafted liposomes"
Biophys. J. 74:1371-1379.
Torchilin, V.P. (1998) "Polymer-coated long-circulating microparticulate
pharmaceuticals" J. Microencapsul. 15:1-19.
Torchilin, V.P. et al. (1992) "Targeted accumulation of polyethylene glycol-
coated
immunoliposomes in infarcted rabbit myocardium" FASEB J. 6:2716-2719.
Tournois, H. et al. (1990) "Gramicidin A induced fusion of large unilamellar
dioleoylphosphatidylcholine vesicles and its relation to the induction of type
II nonbilayer structures" Biochenzistry 29:8297-8307.
Ulrich, A.S. et al. (1999) "Ultrastructural characterization of peptide-
induced
membrane fusion and peptide self assembly in the lipid bilayer" Biophys. J.
77:829-841.
CA 02411542 2002-12-09
WO 01/93836 PCT/USO1/18657
Voneche, V. et al. (1992) "The 19-27 amino acid segment of gp51 adopts an
amphiphilic structure and plays a key role in the fusion events induced by
bovine leukemia virus" J. Biol. Chem. 267:15193-15197.
Wattiaux, R. et al. (1997) "Cationic lipids destabilize lysosomal membrane in
vitro"
FEBS Lett. 417:199-202.
Weissig, V. et al. (1998) "Accumulation of protein-loaded long-circulating
micelles
and liposomes in subcutaneous Lewis lung carcinoma in mice" Pharnz Res.
15:1552-1556.
Zelphati, O. and Szoka, Jr., F.C. (1997) "Intracellular distribution and
mechanism of
delivery of oligonucleotides mediated by cationic lipids" Pharm. Res.
13:1367-1372.
Zuidam, N.J. and Barenholz, Y. (1997) "Electrostatic parameters of cationic
liposomes commonly used for gene delivery as determined by 4-heptadecyl-
7-hydroxycoumarin" Biochim. Biophys. Acta 1329:211-222.
91