Note: Descriptions are shown in the official language in which they were submitted.
CA 02416603 2003-O1-20
WO 02/07761 PCT/USO1/22335
TITLE OF THE INVENTION
1NI-T1BITING HEPATITIS C VIRUS PROCESSING AND REPLICATION
CROSS-REFERENCE TO RELATED APPLICATIONS
The present application claims priority to provisional application U.S.
Serial No. 60/219,550, filed July 20, 2000, hereby incorporated by reference
herein.
BACKGROUND OF THE INVENTION
The references cited herein are not admitted to be prior art to the
claimed invention.
An estimated 170 million persons are infected with Hepatitis C virus
(HCV) worldwide. The infection is usually persistent, and following an
asymptomatic period often lasting years, many patients develop chronic liver
disease,
including cirrhosis and hepatocellular carcinoma.
HCV is a positive strand RNA virus. (Chop, et al., (1989) Science
244, 362-364; and Choo, et al., (1989) Science 244, 359-362.) The I3CV genome
encodes a single polyprotein of approximately 3000 amino acids, containing the
viral
proteins in the order: C-E1-E2-p7-NS2-NS3-NS4A-NS4B=NSSA-NSSB. The NS
proteins are thought to be non-structural and are involved with the enzymatic
functions of viral replication and processing of the viral polyprotein.
Release of the
individual proteins from the polyprotein precursor is mediated by both
cellular and
viral proteases. (Choo, et al., (1991) P.N.A.S. USA 88, 2451-2455; Takamizawa,
et
al., (1991) J. Virol. 65, 1105-1113; Neddermann, et al., (1997) Biol. Chem.
378, 469-
476; Lohmann, et al., (1996) J. Hepatol. 24, 11-19; and Houghton, et al.,
(1991)
Hepatology 14, 381-388.)
The proteolytic release of mature NS4A, NS4B, NSSA and NSSB is
catalyzed by the chymotrypsin-like serine protease contained within the N-
terminal
domain of NS3, while host cell proteases release C, El, E2, and p7, and create
the N-
terminus of NS2 at amino acid 810. (Mizushima, et al., (1994) J. Virol. 68,
2731-
2734, and Hijikata, et al., (1993) P.N.A.S. USA 90, 10773-10777.)
Cleavage between NS2 and NS3 is catalyzed by the NS2/3 protease.
The NS213 activity is likely to be essential for replication of HCV in humans.
(Kolykhalov, et al., (2000) Jounaal of virology 74, 2046-51.)
-1-
CA 02416603 2003-O1-20
WO 02/07761 PCT/USO1/22335
SUMMARY OF THE INVENTION
The present invention features methods for inhibiting HCV replication
and processing by targeting heat shock protein 90 (HSP90). HSP90 is a cellular
chaperone protein that was found to be an essential factor in NS2/3 self-
cleavage.
HSP90 can be targeted using compounds inhibiting the ability of HSP90 to
facilitate
NS2/3 cleavage.
Thus, a first aspect of the present invention describes a method of
inhibiting HCV replication or processing in a cell infected with HCV
comprising the
step of providing to the cell an effective amount of an HSP90 inhibitor. The
effective
amount is an amount sufficient to cause a detectable decrease in HCV
replication.
Preferably, replication is inhibited at Least about 20%, at least about 50%,
at Least
about 75%, or at least about 90%.
Another aspect of the present invention describes a method of
inhibiting NS2/3 cleavage in a polypeptide comprising NS2/3 activity. The
method
involves providing to the polypeptide an effective amount of an HSP90
inhibitor. The
effective amount is an amount sufficient to cause a detectable decrease in
NS2/3
cleavage. Preferably, NS2/3 cleavage is inhibited at least about 20%, at least
about
50%, at least about 75%, or at least about 90%.
A polypeptide "comprising NS2/3 activity" is made up in whole, or in
part, by an amino acid region that is derived from a naturally occurring NS2/3
region
and possesses NS2/3 autocatalytic activity. An amino acid region derived from
a
naturally occurring region contains the sequence of a naturally occurring
NS2/3 region
or is designed based on a naturally occurring region. The NS2/3 autocatalytic
activity
can be present on a longer length polypeptide.
Another aspect of the present invention features a method of inhibiting
HCV replication in a patient infected with HCV. The method involves the step
of
administering to the patent an effective amount of a HSP90 inhibitor. Patients
that
can be infected with HCV include humans and chimpanzees. Preferably, the
subject
is a human.
Another aspect of the present invention describes a method of
identifying a NS2/3 processing inhibitor comprising the steps of: (a)
measuring the
ability of a compound to inhibit HSP90 association to a polypeptide comprising
NS2/3 activity; and (b) measuring the ability of the compound to inhibit NS2/3
cleavage.
-2-
CA 02416603 2003-O1-20
WO 02/07761 PCT/USO1/22335
Another aspect of the present invention describes a method of
identifying an HCV replication inhibitor comprising the steps of: (a)
measuring the
ability of a compound to inhibit HSP90 activity; and (b) measuring the ability
of the
compound to inhibit HCV replication.
Another aspect of the present invention describes a method of
identifying an HCV replication inhibitor comprising the steps of: (a)
measuring the
ability of a compound to inhibit HSP90 association to a polypeptide comprising
NS2/3 activity; and (b) measuring the ability of the compound to inhibit HCV
replication.
Other features and advantages of the present invention are apparent
from the additional descriptions provided herein including the different
examples.
The provided examples illustrate different components and methodology useful
in
practicing the present invention. The examples do not limit the claimed
invention.
Based on the present disclosure the skilled artisan can identify and employ
other
components and methodology useful for practicing the present invention.
BRIEF DESCRIPTION OF THE DRAWINGS
Figure 1. Physical association of NS2/3 with HSP90. (A) Co-
immunoprecipitation of NS2/3 with HSP90-specific antibody. [35S] methionine-
labeled NS2/3 (810-16I5BI~), Ubi-849-1207J-BLA, or firefly Iuciferase
synthesized
in reticulocyte lysate were immunoprecipitated with anti-HSP90 mAb 3G3 or with
the
control IgM TEPC-183. (B) Geldanamycin interferes with the association of
NS2/3
and HSP90. [35S]-methionine-labeled NS2/3 synthesized in reticulocyte lysate
in the
absence (lane 1) or presence (lane 4) of 10 ~,M geldanamycin was
immunoprecipitated
with the anti-HSP90 mAb 3G3 (lanes 3 and 6) or the control IgM TEPC-183 (lanes
2
and 5).
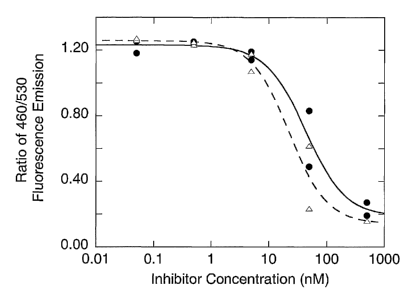
Figure 2. HSP90 inhibitors inhibit NS2/3 cleavage in a cell-based
assay. Cloned Jurkat cells expressing a fusion protein of NS2/3 in which (3-
lactamase
activity is the indicator of successful NS2/3 cleavage were treated with
either
geldanamycin or radicicol. Inhibitor treatment was for 5 hours, followed by
addition
of cycloheximide to stop protein synthesis (30 minutes) and subsequent
addition of
the (3-lactamase substrate, CCF2. After 2 hours, (3-lactamase activity was
quantified
by fluorescence readings (460nm/530nm ratio). Lower 460nm/530nm ratios
indicate
inhibition of NS2/3 cleavage. The geldanamycin (triangles) ICSO is 40 nM and
the
radicicol (circles) ICSp is 13 nM.
-3-
CA 02416603 2003-O1-20
WO 02/07761 PCT/USO1/22335
DETAILED DESCRIPTION OF THE INVENTION
HSP90 is a chaperone protein identified herein as a target for inhibiting
HCV replication or processing. Targeting of HSP90 can be achieved using
compounds that inhibit the ability of HSP90 to facilitate NS2/3 cleavage.
Chaperone proteins can prevent incorrect interactions within and
between non-native proteins and are thought to increase the yield but not rate
of
folding reactions of many newly synthesized proteins. (Hartl, (1996) Nature
381,
571.) In addition, by mechanisms related to their participation in protein-
folding,
chaperone proteins help modulate the activities of a variety of signaling
proteins,
including tyrosine kinases such as p60sr° (Whitesell, et al., (1994)
P.N.A.S. USA 91,
8324), steroid hormone receptors (for review see Pratt, et al., (1997)
E~edocr. Rev. I8,
306), and nitric oxide synthase (Garcia-Cardena, et al., (1998) Nature 392,
821,
Bender, et al., (1999) J. Biol. Chem. 274, 1472).
HSP90 is a chaperone protein involved in regulating the activity of
different proteins. (Scheibel, et al., (1998) Biochemical Pharmacology 56, 675-
682.)
In at least some instances, HSP90 functions as part of a chaperone complex
involving
partner proteins that assists cellular protein folding and preventing
irreversible side-
reactions. (Scheibel, et al., (1998) Bio,claemical Pharmacology 56, 675-682.)
HSP90 Inhibitors
HSP90 inhibitors physically associate with HSP90 and inhibit NS2/3
cleavage and/or HCV replication. The association between HSP90 may be a direct
association or may be mediated by other factors such as partner proteins.
Preferred
HSP90 inhibitors achieve a level of inhibition of at least about 20%, at least
about
50%, at least about 75%, or at least about 90%.
HSP90 activity can be inhibited using a variety of compounds that are
well known in the art. Such compounds may be used in methods for inhibiting
NS2/3
cleavage and/or HCV replication.
The Example section provided below illustrates the ability of
compounds well known in the art such as geldanamycin and radicicol to inhibit
NS2/3
cleavage. Geldanamycin and radicicol are compounds that have been described as
inhibitors of HSP90 activity. (Roe, et al., (1999) J. Med. Chefn. 42, 260-266;
and
Scheibel, et al., (1998) Biochemical Pharmacology 56, 675-682.) Numerous
different
derivatives of geldanamycin and radicicol are well known in the art. The
ability of
-4-
CA 02416603 2003-O1-20
WO 02/07761 PCT/USO1/22335
such derivatives to inhibit NS213 cleavage and HCV replication can be
determined
using standard techniques.
Geldanamycin is an ansamycin antibiotic having anti-tumor activity.
(Roe, et al., (1999) J. Med. Chem. 42, 260-266; and Scheibel, et al., (1998)
Biochemical Pharmacology 56, 675-682.) Geldanamycin has been described in
different references as exerting an anti-tumor drug effect by binding in the
ATP-
binding site present in the N-terminal domain of HSP90. (Roe, et al., (1999)
J. Med.
Chem. 42, 260-266; and Scheibel, et al., (1998) Biochemical Pharmacology 56,
675-
682, 1998.)
Derivatives of geldanamycin indicated to have anti-tumor activity are
described in different references. (See, for example, U.S. Patent No.
4,261,989 and
U.S. Patent No. 5,932,566, both of which are hereby incorporated by reference
herein.) These derivatives provide a class of compounds containing members
that are
expected to have activity in inhibiting NS2/3 cleavage and/or HCV replication.
The
ability of a particular compound to inhibit NS2/3 cleavage or HCV replication
can be
determined using techniques well known in the art.
Radicicol also exerts an anti-tumor drug effect by binding to HSP90 at
the N-terminal domain ATP-binding site. (Roe, et al., (1999) J. Med. Clzem.
42, 260-
266.) Derivatives of radicicol indicated to have anti-tumor activity are
described in
different references. (See, for example, U.S. Patent No. 5,597,846 and U.S.
Patent
No. 5,977,165, both of which are hereby incorporated by reference herein.)
These
derivatives provide a class of compounds containing members that are expected
to
have activity in inhibiting NS213 cleavage and/or HCV replication. The ability
of a
particular compound to inhibit NS2/3 cleavage or HCV replication can be
determined
using techniques well known in the art.
NS2/3 Cleavage
NS2/3 cleavage is part of HCV polyprotein processing leading to the
production of an active NS3 protease. In naturally occurring HCV polypeptide,
cleavage between amino acids1026 and 1027 separating NS2 from NS3 has been
found to be dependent upon protein regions of both NS2 and NS3 flanking the
cleaved site. (Grakoui, et al., (1993) P.N.A.S. USA 90, 10583-10587; and
Komoda, et
al., (1994) Gene 145, 221-226.) The cleavage is independent of the catalytic
activity
of the NS3 protease, as demonstrated with mutational studies. (Grakoui, et
al., (1993)
P.N.A.S. USA 90, 10583-10587; and Hijikata, et al., (1993) J. Virol. 67, 4665-
4675.)
-5-
CA 02416603 2003-O1-20
WO 02/07761 PCT/USO1/22335
NS2/3 cleavage can be measured in different systems using
polypeptides comprising NS2/3 catalytic activity. Examples of such
polypeptides
include naturally occurring NS2/3 regions having a sufficient amount of NS2/3
for
cleavage and derivatives thereof. The NS2/3 cleavage regions can be present
along
with other polypeptide regions such as those present in an HCV polypeptide or
those
not present in an HCV polypeptide. Fusion proteins containing NS2/3 cleavage
regions can be used to provide for NS2/3 activity and, for example, to provide
markers assisting in assaying for NS2/3 activity.
The NS2/3 cleavage reaction has been studied in bacterial, mammalian
and insect cells, and following in-vitro translation of the protein. (Grakoui,
et al.,
(1993) P.N.A.S. USA 90, 10583-10587; Selby, et al., (1993) J. GeyZ. Virol. 74,
1103-
1113; Hijikata, et al., (1993) J. Virol. 67, 4665-4675; Santolini, et al.,
(1995) J. Virol.
69, 7461-7471; D'Souza, et al., (1994) J. Ge~a. Virol. 75, 3469-3476; and
Pieroni, et
al., (1997) J. Virol. 71, 6373-6380.) The protein region essential for NS2/3
cleavage
activity has been approximately mapped to amino acids 898 to 1207 of the HCV
open
reading frame. (Grakoui, et al., (1993) P.N.A.S. USA 90, 10583-10587;
Hijikata, et
al., (1993) J. Virol. 67, 4665-4675; and Santolini, et al., (1995) J. Virol.
69, 7461-
7471.)
The catalytic mechanism of NS2/3 cleavage is unknown but is
speculated to be either a metalloprotease or cysteine protease (Wu, et al.,
(1998)
Trends Biochem. Sci. 23, 92-94; and Gorbalenya, et al., (1996) Perspect. Drug
Discovery Design, 64-86), and the NS2 N-terminus is believed to be a
transmembrane
polypeptide (Santolini, et al., (1995) J. Virol. 69, 7461.) Cleavage activity
of in-vitro
translated NS2/3 is inhibited by EDTA and activity is restored with metal ion
re-
addition. (Hijikata, et al., (1993) J. Virol. 67, 4665-4675; and Pieroni, et
al., (1997) J.
Virol. 71, 6373-6380.)
Identif~~ NS2/3 Processing and HCV Replication Inhibitors
Different assay formats can be employed to identify compounds
targeting HSP90 that inhibit NS2/3 processing or HCV replication. An example
of a
general assay format involves first identifying a compound that interacts with
HSP90
and then determining whether such a compound also inhibits NS2/3 cleavage or
HCV
replication.
Compounds interacting with HSP90 can directly bind to HSP90 or can
interact with an HSP90 partner protein such as that present in a chaperone
complex.
-6-
CA 02416603 2003-O1-20
WO 02/07761 PCT/USO1/22335
The ability of a compound to interact with HSP90 can be measured in a variety
of
ways such as carrying out binding assays and physical association assays.
Binding assays measure the ability of a compound to bind to HSP90 or
a HSP90 chaperone complex. Different binding assay formats can be performed to
measure the ability of a compound to bind to HSP90 or an HSP90 chaperone
complex. Examples of different assay formats include competitive and non-
competitive. A preferred target for a binding assay is the ATP binding site.
Compounds identified as binding to HSP90 or an HSP90 chaperone
complex can be labeled with a detectable moiety and used to evaluate the
ability of
other compounds to bind to HSP90 or an HSP90 chaperone complex. Suitable
detectable moieties include radioisotopes and fluorescent groups. A particular
detectable moiety is preferably selected and positioned on a compound such
that the
moiety will not substantially affect binding to HSP90 or a HSP90 chaperone
complex.
Physical association assays measure HSP90 association with a
polypeptide comprising NS2/3 fragments acted on by HSP90 or a HSP90 chaperone
complex. Techniques for measuring association of HSP90 and NS2/3 include those
using binding agents specific for HSP90 or NS2/3. Such binding agents can be
used,
for example, to indicate the presence of HSP90 and NS2/3 in a gel or column
fraction.
An example of specific binding agents are antibodies. Antibodies
specific for HSP90 and NS2/3 can be produced using standard immunological
techniques. General techniques for producing and using antibodies are
described in
Ausubel, Current Protocols in Molecular Biology, John Wiley, 1987-1998, and
Harlow, et al., Antibodies, A Laboratory Mahual, Cold Spring Harbor
Laboratory,
1988.
Compounds identified as interacting with HSP90 or an HSP90
chaperone complex can be further tested for an effect on NS2/3 processing or
HCV
replication. Inhibition of NS2/3 processing can be assayed for by measuring
NS2/3
autocleavage. Inhibition of HCV replication can be assayed for by measuring a
change in HCV levels in a subject infected with HCV. Subjects susceptible to
HCV
infection include chimpanzees. (Major, et al., (1999) Jounzal of Virology 73,
3317-
3325.)
A preferred target for inhibiting NS2/3 processing or HCV replication
is the HSP90 ATP binding site. Compounds binding to other sites, for example,
those
sites affecting the tertiary structure of HSP90, or other HSP90 activities,
can be
targeted and tested for their ability to inhibit NS213 processing or HCV
replication.
_7_
CA 02416603 2003-O1-20
WO 02/07761 PCT/USO1/22335
Assays can be performed using individual compounds or a preparation
containing different compounds. A preparation containing different compounds
wherein one or more compounds achieves a desired effect such as interacting
with
HSP90, inhibiting NS2/3 cleavage, or inhibiting HCV replication, can be
divided into
smaller groups to identify specific compounds) having a desired effect. In an
embodiment of the present invention a test preparation contains at least 10
different
compounds in an assay that measures interaction with HSP90, inhibition of
NS2/3
cleavage, or inhibition of HCV replication.
ADMllVISTRATION
Compounds targeting HSP90 can be formulated and administered to a
patient using the guidance provided herein along with techniques well known in
the
art. The preferred route of administration ensures that an effective amount of
compound reaches the target. Guidelines for pharmaceutical administration in
general
are provided in, for example, Remihgton's Pharmaceutical ScieyZCes 1 ~'j~
Edition, Ed.
Gennaro, Mack Publishing, 1990, and Modern Pharmaceutics 2'td Edition, Eds.
Banker and Rhodes, Marcel Dekker, Inc., 1990, both of which are hereby
incorporated
by reference herein.
Compounds having appropriate functional groups can be prepared as
acidic or base salts. Pharmaceutically acceptable salts (in the form of water-
or oil-
soluble or dispersible products) include conventional non-toxic salts or the
quaternary
ammonium salts that are formed, e.g., from inorganic or organic acids or
bases.
Examples of such salts include acid addition salts such as acetate, adipate,
alginate,
aspartate, benzoate, benzenesulfonate, bisulfate, butyrate, citrate,
camphorate,
camphorsulfonate, cyclopentanepropionate, digluconate, dodecylsulfate,
ethanesulfonate, fumarate, glucoheptanoate, glycerophosphate, hemisulfate,
heptanoate, hexanoate, hydrochloride, hydrobromide, hydroiodide, 2-
hydroxyethanesulfonate, lactate, maleate, methanesulfonate, 2-
naphthalenesulfonate,
nicotinate, oxalate, pamoate, pectinate, persulfate, 3-phenylpropionate,
picrate,
pivalate, propionate, succinate, tartrate, thiocyanate, tosylate, and
undecanoate; and
base salts such as ammonium salts, alkali metal salts such as sodium and
potassium
salts, alkaline earth metal salts such as calcium and magnesium salts, salts
with
organic bases such as dicyclohexylamine salts, N-methyl-D-glucamine, and salts
with
amino acids such as arginine and lysine.
_g_
CA 02416603 2003-O1-20
WO 02/07761 PCT/USO1/22335
Compounds can be administered using different routes including oral,
nasal, by injection, transdermal, and transmucosally. Active ingredients to be
administered orally as a suspension can be prepared according to techniques
well
known in the art of pharmaceutical formulation and may contain
microcrystalline
cellulose for imparting bulk, alginic acid or sodium alginate as a suspending
agent,
methylcellulose as a viscosity enhancer, and sweeteners/flavoring agents. As
immediate release tablets, these compositions may contain microcrystalline
cellulose,
dicalcium phosphate, starch, magnesium stearate and lactose and/or other
excipients,
binders, extenders, disintegrants, diluents and lubricants.
When administered by nasal aerosol or inhalation, compositions can be
prepared according to techniques well known in the art of pharmaceutical
formulation. Such compositions may be prepared, for example, as solutions in
saline,
employing benzyl alcohol or other suitable preservatives, absorption promoters
to
enhance bioavailability, fluorocarbons, and/or other solubilizing or
dispersing agents.
The compounds may also be administered in intravenous (both bolus
and infusion), intraperitoneal, subcutaneous, topical with or without
occlusion, or
intramuscular form, all using forms well known to those of ordinary skill in
the
pharmaceutical arts. When administered by injection, the injectable solutions
or
suspensions may be formulated using suitable non-toxic, parenterally-
acceptable
diluents or solvents, such as Ringer's solution or isotonic sodium chloride
solution, or
suitable dispersing or wetting and suspending agents, such as sterile, bland,
fixed oils,
including synthetic mono- or diglycerides, and fatty acids, including oleic
acid.
When rectally administered in the form of suppositories, these
compositions may be prepared by mixing the drug with a suitable non-irritating
excipient, such as cocoa butter, synthetic glyceride esters or polyethylene
glycols,
which are solid at ordinary temperatures, but liquidify and/or dissolve in the
rectal
cavity to release the drug.
Suitable dosing regimens for the therapeutic applications of the present
invention are selected taking into factors well known in the art including
age, weight,
sex and medical condition of the patient; the severity of the condition to be
treated;
the route of administration; the renal and hepatic function of the patient;
and the
particular compound employed.
Optimal precision in achieving concentrations of drug within the range
that yields efficacy without toxicity requires a regimen based on the kinetics
of the
drug's availability to target sites. This involves a consideration of the
distribution,
-9-
CA 02416603 2003-O1-20
WO 02/07761 PCT/USO1/22335
equilibrium, and elimination of a drug. The daily dose for a patient is
expected to be
between 0.01 and 1,000 mg per adult patient per day.
EXAMPLES
Examples are provided below to further illustrate different features of
the present invention. The examples also illustrate useful methodology for
practicing
the invention. These examples do not limit the claimed invention.
Example 1: Production of NS2/3
HCV residues 810-1615 of the BK strain, which includes all of NS2
and most of NS3 (termed 810-1615BK), was produced from the plasmid pCITE 810-
1615BK (a gift from Dr. Nicola La Monica) and has been described previously by
Pieroni, et al., (1997) J. Virol. 71, 6373. Following plasmid linearization
with BLPI,
RNA was transcribed with T7 RNA polymerase and purified. Protein translation
was
in rabbit reticulocyte lysate, 30°C for 40 minutes using [35S]-
methionine as a label.
Translation was then blocked by the addition of cycloheximide (250 ~,M final)
and the
samples were immediately frozen on dry ice.
Prior to processing experiments, an aliquot of translated NS2/3 was
thawed on ice. Processing of 810-1615BK was initiated by addition to room
temperature solutions containing Triton X-100 at I%, as described by Pieroni,
et al.,
(1997) J. Virol. 71, 6373. The distribution of [35S]-labeled proteins on dried
gels was
determined with a Phosphorimager (Molecular Dynamics). Product bands were
quantified and expressed as a proportion of total signal in the gel lane so
that
variations in gel lane loading were normalized. The product NS2 from 810-1615
BK
was used to generate data shown for inhibitor ICSO calculations, due to its
migration
on gels in a region with less background than the higher molecular weight NS3
fragment.
Example 2: Expression of NS2/3 Fusion Protein
The plasmid pM3A, derived from pCDNA (Invitrogen), encodes a
fusion protein termed Ubi-849-1207J-BLA that contains ubiquitin at its N-
terminus,
followed by NS2/3 residues 849-1207 (J strain) linked to bacterial TEM-1 [3-
lactamase at the C-terminus. The NS3 protease domain in this construct has the
inactivating mutation 51165A, which does not affect NS2/3 processing activity.
RNA
synthesis for this construct is driven by the T7 promoter, and RNA was
separately
-10-
CA 02416603 2003-O1-20
WO 02/07761 PCT/USO1/22335
prepared for translation as in Example 1. Upon translation in rabbit
reticulocyte
lysate, the ubiquitin is immediately cleaved from the protein by cellular
ubiquitin
hydrolases. (Hochstrasser, (1996) Annual Review of Genetics 30, 405.)
The cleavable linkage (SEQ. )D. NO. 1) is present as
ubiquitin.~ArgHisGlySerGluPhe-NS2/3. Translation of this construct inevitably
produced some processed NS2 and NS3 products, since NS2/3 processing for the J
strain does not require detergent or membranes as does NS2/3 from the BK
strain.
Translations were limited to 30 minutes for that reason. Quantification of
processing
at room temperature was by comparison of samples prepared immediately after
addition of cycloheximide with later time samples.
Example 3: Reconstitution Experiments
NS2/3 (810-1615BK) was synthesized with [35S]-Met-labeling in
rabbit reticulocyte lysate as described in Example 1. Lysate containing
translated,
[35S]-methionine labeled NS213 810-1615BK was centrifuged through a spin
column
containing P-6 polyacrylamide gel (Bio-Rad; exclusion limit 6000 Da)
equilibrated in
mM Tris-HCl buffer (pH 7.5). Rabbit reticulocyte lysate (Promega) was filtered
with Amicon Microcon-10 units to generate a 10 kDa filtrate.
Dilution of the labeled lysate fraction 10-fold into a rabbit reticulocyte
20 lysate filtrate containing solutes less than 10,000 Da (10 kDa filtrate)
supported the
NS2/3 cleavage to an extent similar to that observed in undiluted lysate as
previously
described (Darke, et al., (1999) J. Biol. Chem. 274, 34511). The same material
diluted into water or buffer failed to process NS2/3.
Commercial rabbit reticulocyte lysate, in addition to cellular
components, is supplemented with DTT, KOAc, GTP and creatine phosphate. The
filtered lysate was diluted 10-fold into the column buffer or into buffer
containing
combinations of 1 mM ATP, 1.5 mM Mg(OAc)2, 2 mM DTT and 79 mM KOAc. In
some samples, the amount of KOAc was varied (20 mM - 237 mM) or ATPyS was
substituted for ATP. Processing was initiated by the addition of Triton X-100
to 1%
and stopped after 30 minutes with an equal volume of SDS sample buffer. The
results
are shown in Table l, where activity is expressed as a percent of the amount
of
processing achieved in the presence of the 10 kDa filtrate control.
-11-
CA 02416603 2003-O1-20
WO 02/07761 PCT/USO1/22335
Table 1
Reconstitution of HCV NS2/3 Processing ifZ Reticulocyte Lysate
Addition HCV NS2/3 Processed a
lOkDa Filtrate 100
M ATP/DTT 7.5
M ATP S/DTT/79rnM KOAc 24
M ATP/DTT/79mM KOAc 68
M /ATPIDTT/20mM KOAc 20
M ATP/DTT/39.5mM KOAc 52
M ATP/DTT/158mM KOAc 102
M ATP/DTT/237mM KOAc 104
a Relative to the amount processed in the presence of lOkDa filtrate
The components of the 10 kDa filtrate that support processing are
stable to treatment with heat or trypsin (heat treatment was 100°C for
3 minutes, and
trypsin treatment was 0.1 mg/ml for 2 hours, followed by a 2-fold excess of
pancreatic
trypsin inhibitor), suggesting a non-proteinaceous nature. Small molecular
weight
compounds known to be present in rabbit reticulocyte lysate were examined
individually and in combination for their ability to support processing.
While no component alone or in pairs completely supported
processing, a solution containing ATP and a combination of salts enabled
processing
at a level comparable to that achieved with the 10 kDa filtrate, as shown in
Table 1.
Substitution of ATP with ATPyS in this combination caused a loss of
processing,
suggesting that the ATP contribution requires hydrolysis to ADP (Table 1).
Example 4: Inhibition of NS2/3 Processing_by Depleting ATP or Using ATP
Analogs
To compliment the reconstitution experiments indicating a role for
ATP in NS2/3 processing, rabbit reticulocyte lysate containing translated
[35S]-labeled
NS2/3 (810-1615BK) was depleted of ATP by treatment with glucose plus
hexokinase
before processing was initiated by the addition of Triton X-100 to 1%. While
neither
glucose nor hexokinase alone had a significant effect, the combination of the
two,
which consumes ATP in the phosphorylation of glucose, inhibited processing by
60%.
The results are shown in Table 2.
- 12-
CA 02416603 2003-O1-20
WO 02/07761 PCT/USO1/22335
Table 2
Effect of ATP Depletiooa a~ad Noh-Hydroly,zable ATP
Analogs orc HCV NS2/3 Processiy2g
Addition NS2/3 Processed (% of Control)
83 mM Glucose 101
0.5 units Hexokinase 88
Glucose + Hexokinase 38
5mM M ATP S 24
5mM M AMP-PNP 40
[ S]-methionine labeled NS2/3 810-1615BK was synthesized in rabbit
reticulocyte
lysate and 5 ~,l aliquots were directly combined with 1 x,1500 mM glucose, 0.5
unit
yeast hexokinase, or glucose plus hexokinase, and incubated for 30 minutes at
room
temperature. Similarly, lysate containing NS2/3 was incubated with Mg/ATPyS or
Mg/AMP-PNP, for final concentrations of the nucleoside analogs of 5 mM. A
stock
solution of Triton X-100 at 10% (w/v) was used to initiate autoprocessing of
the 810-
1615BK NS2/3. Following addition of SDS sample buffer, samples were heated to
100°C for 5 minutes and proteins separated on SDS/I4% polyacrylamide
gels.
Quantification of products was by phosphorimaging of the dried gels.
I5
Alternatively, inhibition was observed with the addition of either
ATP~yS or AMP-PNP, non-hydrolyzable analogs of ATP (Table 2). Titration of the
inhibition yielded ICso values of 2 mM and 4 mM for ATP~yS or AMP-PNP,
respectively, with inhibition at the maximum concentration tested (5 mM) of
77% and
60%, respectively (residual ATP is also present in these reactions, at a
concentration
of approximately 1 mM, so that complete inhibition is not expected).
Inhibition by
ATP~yS was also observed with NS2/3 from the J-strain of HCV, expressed as a
fusion
protein consisting of ubiquitin-NS2/3-~3-lactamase (Ubi-849-1207J-BLA).
Example 5: Inhibition of NS2/3 ActivitX
The involvement of ATP is consistent with the participation of ATP-
dependent cellular chaperones at a stage in the processing. Geldanamycin and
herbimycin A, two related benzoquinone ansamycins, and radicicol, a
macrocyclic
antibiotic, are examples of compounds that specifically inhibit HSP90 by
binding at
the ATP site. (Roe, et al., (1999) J. Med. Chern. 42, 260.)
-13-
CA 02416603 2003-O1-20
WO 02/07761 PCT/USO1/22335
Inhibition of NSZ/3 processing was observed with geldanamycin,
herbimycin A, and radicicol when added to izz vitro synthesized precursor 810-
1615BK (up to 50%). Somewhat greater inhibition of processing was observed, in
a
dose-dependent manner, if compounds were included during the synthesis phase
of
the experiment as well as in NS2/3 processing (Table 3). The compounds had no
effect on overall efficiency of protein synthesis and similar potencies of
inhibition
were observed using the NS2/3 fusion protein Ubi-849-1207J-BLA.
Table 3
Effect of I>zhibitors of HSP90 on HCV NS2/3 Processiyzg
Addition NS2/3 Processed (% of Control)
1 M Geldanam cin 53
10 M Geldanam cin 33
1 M Herbim cin 38
10 M Herbim cin 22
1 M Radicico1 58
10 ~M Radicicol I 46
[j5S]-methionine labeled NS2/3 8101615BK was synthesized in reticulocyte
lysate in
the presence of either I orl0 ~M geldanamycin, herbimycin A, or radicicol.
After
blocking further synthesis with cycloheximide, an aliquot was removed and
processing was initiated with the addition of Triton X-I00 to 1%. After 30
minutes
the reaction was terminated with SDS sample buffer and heated to 95°C.
NS2/3 processing reactions were performed with 810-1615BK. The
inhibitory effects of geldanamycin and radicicol were titrated using
techniques
described previously for peptide inhibition titrations. (Darke, et al., (1999)
J. Biol.
Chem. 274, 3451 l.) Inhibitors were dissolved in DMSO and protected from
light.
Dilutions were in DMSO, such that the final concentration of DMSO was 2% for
i>2
vitro experiments and I % for cell-based assays. Titration of geldanamycin and
radicicol yielded EC5o's in the low micromolar range (Table 4), similar to
what has
been observed in analogous in vitro studies of other proteins acted on by
HSP90. (Hu,
et al., (1996) P.N.A.S. USA 93, 1060; Thulasiraman, et al., (1996)
Biochemistry 35,
13443 (1996), Schneider, et al., (1996) P.N.A.S. USA 93, 14536.)
-14-
CA 02416603 2003-O1-20
WO 02/07761 PCT/USO1/22335
Table 4
Ifzhibitiozz of NS2/3 by HSP901>zlzibitors
Maximum ECso
Compound Name Structure Inhibition (~M)
(°Io)
Geldanamycin 91 1.8 ~ 0.5
OH O CH3
O
Radicicol ~ ~ '° ~ ~ 51 0.22 ~ 0.12
Ho
ci
The ICSO values were determined by first expressing the product level found as
a
fraction of the no-inhibitor control product level, then fitting the equation
Fractional Activity = a + b a
(1+xlc)
to the data, where a is the minimal level of fractional activity (tending to
0), a+b is the
maximal level (tending to 1), x is the concentration of inhibitor, a is the
ICso and d is a
slope coefficient. For both inhibitors, inhibition leveled out at the maximum
extent
indicated in the table, so that an effective concentration (ECso) is used to
describe the
relative potency. Values given are the average of two determinations.
-15-
CA 02416603 2003-O1-20
WO 02/07761 PCT/USO1/22335
Example 6: Physical Association Between NS2/3 and HSP90
Evidence for a physical association of ire vitro translated NS2/3 with
HSP90 was obtained by immunoprecipitation. The monoclonal IgM antibodies, 3G3
(anti-HSP90, Affinity Bioreagents) and TEPC-183 (control, Sigma), have been
previously described for use in immunoprecipitation of HSP90. (McGuire, et
al.,
(1994) Molecular and Cellular Biology 14, 2438.) Luciferase RNA was obtained
from Promega. Tinmunoaffinity beads were prepared by binding the primary
antibody
to a solid support by means of a bridging antibody.
Protein G-agarose (Boehringer) was used to immobilize goat anti-
mouse immunoglobulin M (IgM) (5 mg/ml gel) overnight at 4°C. The
monoclonal
anti-HSP90 antibody 3G3 or an equal concentration of control mouse IgM
antibody
TEPC-183 was then combined with the immobilized anti-mouse IgM. To
immunoprecipitate HSP90 and any associated proteins, Iysate containing
translated
[ssS]-labeled NS2/3 was incubated with the beads essentially as described.
(McGuire,
et al., (1994) Molecular and Cellular Biology 14, 2438.) Following binding for
2
hours at 4°C, the beads were washed, suspended in SDS sample buffer,
and heated to
95°C. Immunoprecipitates were resolved on SDSll4% polyacrylamide gels.
Immunoprecipitation of HSP90 with a monoclonal IgM antibody co-
immunoprecipitated NS2/3 derived from either BK or J strains, 810-1615BK and
Ubi-
849-1207J-BLA, as shown in Figure 1. A control IgM antibody, TEPC-183
immunoprecipitated only minimal amounts of the protein of interest.
Association
with HSP90 was not observed, however, with a control protein, translated
firefly
luciferase (Figure 1). The results indicate that de novo synthesized NS2/3
forms a
stable complex with HSP90 in solution.
Example 7: Inhibition of HSP90 Association With NS2/3
The ability of geldanamycin to interfere with the immunoprecipitation
of HSP90 with NS2/3 was examined. [35S]-labeled NS2/3 was synthesized in
reticulocyte Iysate in the absence or presence of 10 ~,M geldanamycin. In the
presence
of the geldanamycin the amount of NS2/3 co-immunoprecipitated with the anti-
HSP90 mAb was decreased by 60% (Figure 1B). Thus, some inhibition of NS2/3
processing by geldanamycin may be attributable to the prevention of HSP90
association with NS2/3 during or immediately following translation.
-16-
CA 02416603 2003-O1-20
WO 02/07761 PCT/USO1/22335
Example 8: Cell-Based Inhibition of NS2/3 Cleavage
Validation of the concept that HSP90 is essential for NS2/3 processing
in living cells was obtained by treating cells expressing NS2/3 with HSP90
inhibitors.
Through the use of a neomycin-selectable transfection vector, stable
expression of
NS2/3 in Jurkat cells was obtained. The plasmid pUbBla3X-NS2/3-3A conferring
neomycin (G418) resistance, expresses a fusion of the NS2/3 protein with 3
ubiquitin
domains appended to the N-terminus and (3-lactamase at the C-terminus. The
complete uncleaved protein has an in vivo half-life estimated to be 5-10
minutes.
The plasmid pUbBla3X NS2/3-3A was transfected into Jurkat cells.
The CMV promoter-driven ORF of the plasmid encodes a 91 kDa protein, ubiquitin-
ubiquitin-ubiquitin-NS2/3-(3-lactamase, with the C-termini of the 3 ubiquitin
domains
rendered non-cleaveable to ubiquitin-C-terminal hydrolases (ubiquitin C-
terminal
sequence ArgLeuArgGlyVal, SEQ. ID. NO. 2). The NS2/3 region includes HCV
residues 849-1207. The (3-lactamase domain is TEM-1 from E. coli. Expression
of
the ~i-lactamase moiety is readily detected with the fluorogenic, cell-
permeant
substrate, CCF2. (Zlokarnik, et al., (1998) Science 279, 84.)
Transfectants were sorted by FACS using treatment with CCF2 to
indicate (3-lactamase expression (Zlokarnik, et al., (1998) ScieyZCe 279, 84),
and
individual clones were grown under 6418 selection. The full fusion protein
expressed is highly unstable to ubiquitin-directed proteosomal degradation due
to its
ubiquitin N-terminal tag, while the C-terminal product of NS2/3 cleavage, NS3-
~3-
lactamase, is stable for hours. Thus, build-up in the cells of (3-lactamase
activity, as
indicated by CCF2 hydrolysis (high 460nm/530nm ratio) is indicative of
successful
NS2/3 cleavage, and suppression of ~i-lactarnase activity indicates NS2/3
inhibition.
Within this context, the NS2/3 mutation C993A, which is incapable of
processing, reduces (3-lactamase activity of the cells approximately 8-fold.
The
protein region essential for NS2/3 cleavage activity has been approximately
mapped
to amino acids 898 to 1207 of the HCV open reading frame. The conserved
cleavage
site sequence is ArgLeuLeu.~AlaProIle (SEQ. ID. NO. 3). Cys993 and His952 have
been identified as essential residues. (See, Grakoui, et al., (1993) P.N.A.S.
USA 90,
10583, Selby, et al., (1993) J. Gen. Virol. 74, 1103, Hijikata, et al., (1993)
J. Virol.
67, 4665, Santolini, et al., (1995) J. Virol. 69, 7461, D'Souza, et al.,
(1994) J. Gef2.
Virol. 75, 3469, and Pieroni, et al., (1997) J. Virol. 71, 6373.)
NS2/3 self-cleavage separates the destabilizing ubiquitin degradation
signal at the N-terminus from the NS3-~3-lactamase C-terminal product, thus
-17-
CA 02416603 2003-O1-20
WO 02/07761 PCT/USO1/22335
stabilizing the (3-lactamase activity within the cell. Using this system the
inhibitory
potency of HSP90 inhibitors toward NS2/3 processing in mammalian cells was
measured, as shown in Figure 2. Geldanamycin and radicicol are potent
inhibitors of
NS2/3 cleavage in this context, with ICSO values of 40 nM and 13 nM,
respectively.
In addition, inhibition is nearly complete at the highest concentrations
tested (Figure
2). The results are comparable to what others have noted for other HSP90
activities,
in that the concentration of geldanamycin required to inhibit HSP90 activity
in cells is
much lower than required in vitro. (Hu, et al., (1996) P.N.A.S. USA 93, 1060,
Holt, et
al., (1999) Genes Dev. 13, 817.)
Geldanamycin specifically interacts with HSP90 in cells, as
demonstrated by affinity labeling (Chavany, et al., (1996) J. Biol. Chem. 271,
4974),
and affinity chromatography (Schneider, et al., (1996) P.N.A.S. USA 93, 14536,
Whitesell, et al., (1994) P.N.A.S. USA 91, 8324, and Schulte, et al. (1998),
Cell Stress
and Clzaperohes 3, 100). Thus, the observed inhibition for NS2/3 cleavage is
HSP90-
mediated.
Other embodiments are within the following claims. While several
embodiments have been shown and described, various modifications may be made
without departing from the spirit and scope of the present invention.
-18-
CA 02416603 2003-O1-20
WO 02/07761 PCT/USO1/22335
SEQUENCE LISTING
<110> Merck & Co., Inc.
<120> INHIBITING HEPATITIS C VIRUS PROCESSING
AND REPLICATION
<130> 20695 PCT
<150> 60/219,550
<151> 2000-07-20
<160> 3
<170> FastSEQ for Windows Version 4.0
<210> 1
<211> 6
<212> PRT
<213> Artificial Sequence
<220>
<223> Cleavable linkage
<400> 1
Arg His Gly Ser Glu Phe
1 5
<210> 2
<211> 5
<212> PRT
<213> Artificial Sequence
<220>
<223> Ubiquitin C-terminal sequence
<400> 2
Arg Leu Arg Gly Val
1 5
<210> 3
<211> 6
<212> PRT
<213> Artificial Sequence
<220>
<223> Conserved cleavage site sequence
<400> 3
Arg Leu Leu Ala Pro Ile
1 5
-1-