Note: Descriptions are shown in the official language in which they were submitted.
CA 02444812 2003-10-28
3011-13CA
-1-
TITLE OF THE INVENTION: Genes and proteins involved in the biosynthesis of
enediyne ring structures.
CROSS-REFERENCE TO RELATED APPLICATIONS
This application claims benefit under 35 USC ~ 119 of provisional applications
USSN 601291,959 filed on May 21, 2001 and USSN 60/334,604 filed on December 3,
2001 which are hereby incorporated by reference in their entirety for all
purposes.
FIELD OF INVENTION
The present invention relates to the field of microbiology, and more
specifically
to genes and proteins involved in the production of enediynes.
BACKGROUND
Enediyne natural products are characterized by the presence of the enediyne
ring structure also referred to as the warhead. The labile enediyne ring
structure
undergoes a thermodynamically favorable Bergman cyclization resulting in
transient
formation of a biradical species. The biradical species is capable of inducing
irreversible DNA damage in the cell. This reactivity gives rise to potential
biological
activity against both bacterial and tumor cell lines. Enediynes have potential
as
anticancer agents because of their ability to cleave DNA. Calicheamicin is
currently in
clinical trials as an anticancer agent for acute myeloid leukemia (Nabhan C.
and
Tallman MS, Clin Lymphoma (2002) Mar;2 Suppl 1:S19-23). Enediynes also have
utility as anti-infective agents. Accordingly, processes for improving
production of
existing enediynes or producing novel modified enediynes are of great interest
to the
pharmaceutical industry.
Enediynes are a structurally diverse group of compounds. Chromoprotein
enediynes refer to enediynes associated with a protein conferring stability to
the
complex under physiological conditions. Non-chromoprotein enediynes refer to
enediynes that require no additional stabilization factors. The structure of
the
chromoprotein enediynes neocarzinostatin and C-1027, and the non-chromoprotein
enediynes calicheamicin and dynemicin are shown below with the dodecapolyene
backbone forming the warhead structure in each enediyne highlighted in bold.
CA 02444812 2003-10-28
3011-13CA
-2-
HC
HO~ ,
H3C O
Calicheamycin Neocarsinostatin
OOH
Dynemicin A C-1027 NH2
Efforts at discovering the genes responsible for synthesis of the warhead
20 structure that characterizes enediynes have been unsuccessful. Genes
encoding
biosynthetic enzymes for the aryltetrasaccharide of calicheamicin, and for
calicheamicin
resistance are described in WO 00137608. Additional genes involved in the
biosynthesis of the chromoprotein enediyne C-1027 have been isolated (Liu, et
al.
Antimicrobial Agents and Chemotherapy, vol. 44, pp 382-292 (2000); WO
00/40596).
Isotopic incorporation experiments have indicated that the enediyne backbones
of
esperamicin, dynemycin, and neocarzinostatin are acetate derived (Hansens,
O.D. et
al. J. Am. Chem Soc. 11, vol 111 pp. 3295-3299 (1989); Lam, K. et al. J. Am.
Chem.
Soc. vol. 115, pp 12340-12345 (1993); Tokiwa, Y et al. J. Am. Chem Soc. vol.
113 pp.
4107-4110). However, both PCR and DNA probes homologous to type I and type II
30 PKSs have failed to identify the presence of PKS genes associated with
biosynthesis of
enediynes in known enediyne producing microorganisms (WO 00/40596; W. Liu & B.
Shen, Antimicrobial Agents Chemotherapy, vol. 44 No. 2 pp.382-392 (2000)).
CA 02444812 2003-10-28
3011-13CA
-3-
Elucidation of the genes involved in biosynthesis of enediynes, particularly
the
warhead structure, would provide access to rational engineering of enediyne
biosynthesis for novel drug leads and makes it possible to construct
overproducing
strains by de-regulating the biosynthetic machinery. Elucidation of PKS genes
involved
in the biosynthesis of enediynes would contribute to the field of
combinatorial
biosynthesis by expanding the repertoire of PKS genes available for making
novel
enediynes via combinatorial biosynthesis.
Existing screening methods for identifying enediyne-producing microbes are
laborious, time-consuming and have not provided sufficient discrimination to
date to
detect organisms producing enediyne natural products at low levels. There is a
need
for improved tools to detect enediyne-producing organisms. There is also a
need for
tools capable of detecting organisms that produce enediynes at levels that are
not
detected by traditional culture tests.
SUMMARY OF THE INVENTION:
One embodiment of the present invention is an isolated, purified or enriched
nucleic acid comprising a sequence selected from the group consisting of: (a)
SEQ ID
NOS: 2, 14, 24, 34, 44, 54, 64, 74, 84, 94; sequences complementary to SEQ ID
NOS:
2, 14, 24, 34, 44, 54, 64, 74, 84, 94; fragments comprising 2000, preferably
3000, more
preferably 4000, still more preferably 5000, still more preferably 5600 and
most
preferably 5750 consecutive nucleotides of SEQ 1D NOS: 2, 14, 24, 34, 44, 54,
64, 74,
84, 94; and fragments comprising 2000, preferably 3000, more preferably 4000,
still
more preferably 5000, still more preferably 5600 and most preferably 5750
consecutive
nucleotides of the sequences complementary to SEQ ID NOS: 2, 14, 24, 34, 44,
54, 64,
74, 84, 94; (b) SEQ ID NOS: 4, 6, 16, 26, 36, 46, 56, 66, 76, 86, 96;
sequences
complementary to SEQ ID NOS: 4, 6, 16, 26, 36, 46, 56, 66, 76, 86, 96;
fragments
comprising 150, preferably 200, more preferably 250, still more preferably
300, still
more preferably 350 and most preferably 400 consecutive nucleotides of the
sequences complementary to SEQ ID NOS: 4, 6, 16, 26, 36, 46, 56, 66, 76, 86,
96; and
fragments comprising 150, preferably 200, more preferably 250, still more
preferably
300, still more preferably 350 and most preferably 400 consecutive nucleotides
of the
sequences complementary to SEQ ID NOS: 4, 6, 16, 26, 36, 46, 56, 66, 76, 86,
96; (c)
SEQ ID NOS: 8, 18, 28, 38, 48, 58, 68, 78, 88, 98; sequences complementary to
SEQ
CA 02444812 2003-10-28
3011-13CA
-4-
ID NOS: 8, 18, 28, 38, 48, 58, 68, 78, 88, 98; fragments comprising 700,
preferably
750, more preferably 800, still more preferably 850, still more preferably 900
and most
preferably 950 consecutive nucleotides of SEQ ID NOS: 8, 18, 28, 38, 48, 58,
68, 78,
88, 98; and fragments comprising 700, preferably 750, more preferably 800,
still more
preferably 850, still more preferably 900 and most preferably 950 consecutive
nucleotides of the sequences complementary to SEQ ID NOS: 8, 18, 28, 38, 48,
58, 68,
78, 88, 98; (d) SEO ID NOS: 10, 20, 30, 40, 50, 60, 70, 80, 90, 100; sequences
complementary to SEQ ID NOS: 10, 20, 30, 40, 50, 60, 70, 80, 90, 100;
fragments
comprising 600, preferably 700, more preferably 750, still more preferably
800, still
more preferably 850 and most preferably 900 consecutive nucleotides of SEQ ID
NOS:
10, 20, 30, 40, 50, 60, 70, 80, 90, 100; and fragments comprising 600,
preferably 700,
more preferably 750, still more preferably 800, still more preferably 850 and
most
preferably 900 consecutive nucleotides of SEQ ID NOS: 10, 20, 30, 40, 50, 60,
70, 80,
90, 100; and (e) SEQ ID NOS: 12, 22, 32, 42, 52, 62, 72, 82, 92, 102;
sequences
complementary to SEQ ID NOS: 12, 22, 32, 42, 52, 62, 72, 82, 92, 102;
fragments
comprising 700, preferably 750, more preferably 800, still more preferably
850, still
more preferably 900 and most preferably 950 consecutive nucleotides of the
sequences complementary to SEQ ID NOS: 12, 22, 32, 42, 52, 62, 72, 82, 92,
102; and
fragments comprising 700, preferably 750, more preferably 800, still more
preferably
850, still more preferably 900 and most preferably 950 consecutive nucleotides
of SEQ
ID NOS: 12, 22, 32, 42, 52, 62, 72, 82, 92, 102. One aspect of the present
invention is
an isolated, purified or enriched nucleic acid capable of hybridizing to the
nucleic acid
of this embodiment under conditions of high stringency. Another aspect of the
present
invention is an isolated, purified or enriched nucleic acid capable of
hybridizing to the
nucleic acid of this embodiment under conditions of moderate stringency.
Another
aspect of the present invention is an isolated, purified or enriched nucleic
acid capable
of hybridizing to the nucleic acid of this embodiment under low stringency.
Another
aspect of the present invention is an isolated, purified or enriched nucleic
acid having at
least 70% homology to the nucleic acid of this embodiment by analysis with
BLASTN
version 2.0 with the default parameters. Another aspect of the present
invention is an
isolated, purified or enriched nucleic acid having at least 99% homology to
the nucleic
acid of this embodiment as determined by analysis with BLASTN version 2.0 with
the
default parameters.
CA 02444812 2003-10-28
3011-13CA
-5-
Another embodiment is an isolated, purified or enriched nucleic acid that
encodes an enediyne polyketide synthase protein comprising a polypeptide
selected
from the group consisting of: (a) SEQ ID NOS: 1, 13, 23, 33, 43, 53, 63, 73,
83, 93; (b)
polypeptides having at least 75% homology to a polypeptide of SEQ ID NOS: 1,
13, 23,
33, 43, 53, 63, 73, 83, 93 as determined using the BLASTP algorithm with the
default
parameters and having the ability to substitute for a polypeptide of SEQ ID
NOS: 1, 13,
23, 33, 43, 53, 63, 73, 83 or 93 during synthesis a warhead structure in an
enediyne
compound; and (c) fragments of the polypeptides of (a) and (b), which
fragments have
the ability to substitute for a polypeptide of SEQ ID NOS: 1, 13, 23, 33, 43,
53, 63, 73,
83, 93 in the synthesis of the warhead structure in an enediyne compound. In
one
aspect of this embodiment, the nucleic acid encoding an enediyne polyketide
synthase
protein may be used in genetic engineering applications to synthesize the
warhead
structure of an enediyne compound.
Another embodiment is an isolated, purified or enriched nucleic acid that
encodes an enediyne polyketide synthase catalytic complex comprising (a) a
polypeptide selected from the group consisting of SEQ ID NOS: 1, 13, 23, 33,
43, 53,
63, 73, 83, 93; polypeptides having at least 75% homology to a polypeptide of
SEQ ID
NOS: 1, 13, 23, 33, 43, 53, 63, 73, 83, 93 as determined using the BLASTP
algorithm
with the default parameters and having the ability to substitute for a
polypeptide of SEQ
ID NOS: 1, 13, 23, 33, 43, 53, 63, 73, 83 or 93 during synthesis a warhead
structure in
an enediyne compound; and fragments thereof, which fragments have the ability
to
substitute for a polypeptide of SEQ ID NOS: 1, 13, 23, 33, 43, 53, 63, 73, 83,
93 in the
synthesis of the warhead structure in an enediyne compound; and (b) a
polypeptide
selected from the group consisting of SEQ ID NOS: 3, 5, 15, 25, 35, 45, 55,
65, 75, 85,
95; polypeptides having at least 75% homology to a polypeptide of SEQ ID NOS:
3, 5,
15, 25, 35, 45, 55, 65, 75, 85, 95 as determined using the BLASTP algorithm
with the
default parameters and having the ability to substitute for a polypeptide of
SEQ ID
NOS: 3, 5, 15, 25, 35, 45, 55, 65, 75, 85, 95 during synthesis of a warhead
structure in
an enediyne compound; and fragments thereof, which fragments have the ability
to
substitute for a polypeptide of SEQ ID NOS: 3, 5, 15, 25, 35, 45, 55, 65, 75,
85, 95 in
the synthesis of the warhead structure in an enediyne compound. In one aspect
of this
embodiment, the nucleic acid encoding an enediyne polyketide synthase
catalytic
CA 02444812 2003-10-28
3011-13CA
-6-
complex may be used in genetic engineering application to synthesize the
warhead
structure of an enediyne compound.
Another embodiment is an isolated, purified or enriched nucleic acid encoding
a
gene cassette comprising: (a) a nucleic acid encoding an enediyne polyketide
synthase
catalytic complex as described above; and (b) at least one nucleic acid
encoding a
polypeptide selected from the group consisting of (i) SEQ ID NOS: 7, 17, 27,
37, 47, 57,
67, 77, 87, 97; polypeptides having at least 75% homology to a polypeptide of
SEQ ID
NOS: 7, 17, 27, 37, 47, 57, 67, 77, 87, 97 as determined using the BLASTP
algorithm
with the default parameters and having the ability to substitute for a
polypeptide of SEQ
ID NOS: 7, 17, 27, 37, 47, 57, 67, 77, 87, 97 during synthesis of a warhead
structure in
an enediyne compound; and fragments thereof, which fragments have the ability
to
substitute for a polypeptide of SEQ ID NOS: 7, 17, 27, 37, 47, 57, 67, 77, 87,
97 in the
synthesis of the warhead structure in an enediyne compound; (ii) SEQ ID NOS:
9, 19,
29, 39, 49, 59, 69, 79, 89, 99; polypeptides having at least 75% homology to a
polypeptide of SEQ ID NOS: 9, 19, 29, 39, 49, 59, 69, 79, 89, 99 as determined
using
the BLASTP algorithm with the default parameters and having the ability to
substitute
for a polypeptide of SEQ ID NOS: 9, 19, 29, 39, 49, 59, 69, 79, 89, 99 during
synthesis
of a warhead structure in an enediyne compound; and fragments thereof, which
fragments have the ability to substitute for a polypeptide of SEQ ID NOS: 9,
19, 29, 39,
49, 59, 69, 79, 89, 99 in the synthesis of the warhead structure in an
enediyne
compound; and (iii) SEQ ID NOS: 11, 21, 31, 41, 51, 61, 71, 81, 91, 101;
polypeptides
having at least 75% homology to a polypeptide of SEQ ID NOS: 11, 21, 31, 41,
51, 61,
71, 81, 91, 101 as determined using the BLASTP algorithm with the default
parameters
and having the ability to substitute for a polypeptide of SEQ ID NOS: 11, 21,
31, 41, 51,
61, 71, 81, 91, 101 during synthesis of a warhead structure in an enediyne
compound;
and fragments thereof, which fragments have the ability to substitute for a
polypeptide
of SEQ ID NOS: 11, 21, 31, 41, 51, 61, 71, 81, 91, 101 in the synthesis of the
warhead
structure in an enediyne compound. In one aspect of this embodiment, the
nucleic acid
encoding the gene cassette may be used in genetic engineering application to
synthesize the warhead structure of an enediyne compound.
Another embodiment is an isolated, purified or enriched nucleic acid encoding
a
gene cassette comprising: (a) a nucleic acid encoding a polypeptide selected
from the
group consisting of SEQ ID NOS: 1, 13, 23, 33, 43, 53, 63, 73, 83, 93; a
polypeptide
CA 02444812 2003-10-28
3011-13CA
-7-
having at least 75% homology to a polypeptide of SEQ ID NOS: 1, 13, 23, 33,
43, 53,
63, 73, 83, 93 as determined using the BLASTP algorithm with the default
parameters
and having the ability to substitute for a polypeptide of SEQ ID NOS: 1, 13,
23, 33, 43,
53, 63, 73, 83 or 93 during synthesis a warhead structure in an enediyne
compound; or
a fragment thereof, which fragment has the ability to substitute for a
polypeptide of SEQ
ID NOS: 1, 13, 23, 33, 43, 53, 63, 73, 83, 93 in the synthesis of the warhead
structure
in an enediyne compound; (b) at least one nucleic acid encoding a polypeptide
selected from the group consisting of SEO ID NOS: 3, 5, 15, 25, 35, 45, 55,
65, 75, 85,
95; a polypeptide having at least 75% homology to a polypeptide of SEQ ID NOS:
3, 5,
15, 25, 35, 45, 55, 65, 75, 85, 95 as determined using the BLASTP algorithm
with the
default parameters and having the ability to substitute for a polypeptide of
SEQ ID
NOS: 3, 5, 15, 25, 35, 45, 55, 65, 75, 85, 95 during synthesis of a warhead
structure in
an enediyne compound; or a fragment thereof, which fragment has the ability to
substitute for a polypeptide of SEQ ID NOS: 3, 5, 15, 25, 35, 45, 55, 65, 75,
85, 95 in
the synthesis of the warhead structure in an enediyne compound; (c) at least
one
nucleic acid encoding a polypeptide selected from the group consisting of SEQ
ID
NOS: 7, 17, 27, 37, 47, 57, 67, 77, 87, 97; a polypeptide having at least 75%
homology
to a polypeptide of SEQ ID NOS: 7, 17, 27, 37, 47, 57, 67, 77, 87, 97 as
determined
using the BLASTP algorithm with the default parameters and having the ability
to
substitute for a polypeptide of SEQ ID NOS: 7, 17, 27, 37, 47, 57, 67, 77, 87,
97 during
synthesis of a warhead structure in an enediyne compound; and a fragment
thereof,
which fragment has the ability to substitute for a polypeptide of SEQ ID NOS:
7, 17, 27,
37, 47, 57, 67, 77, 87, 97 in the synthesis of the warhead structure in an
enediyne
compound; (d) at least one nucleic acid encoding a polypeptide selected from
SEQ ID
NOS: 9, 19, 29, 39, 49, 59, 69, 79, 89, 99; a polypeptide having at least 75%
homology
to a polypeptide of SEO ID NOS: 9, 19, 29, 39, 49, 59, 69, 79, 89, 99 as
determined
using the BLASTP algorithm with the default parameters and having the ability
to
substitute for a polypeptide of SEQ ID NOS: 9, 19, 29, 39, 49, 59, 69, 79, 89,
99 during
synthesis of a warhead structure in an enediyne compound; and a fragment
thereof,
which fragment has the ability to substitute for a polypeptide of SEQ ID NOS:
9, 19, 29,
39, 49, 59, 69, 79, 89, 99 in the synthesis of the warhead structure in an
enediyne
compound; and (e) at least one nucleic acid encoding a polypeptide selected
from SEQ
I D NOS: 11, 21, 31, 41, 51, 61, 71, 81, 91, 101; a polypeptide having at
least 75%
CA 02444812 2003-10-28
3011-13CA
_g_
homology to a polypeptide of SEO I D NOS: 11, 21, 31, 41, 51, 61, 71, 81, 91,
101 as
determined using the BLASTP algorithm with the default parameters and having
the
ability to substitute for a polypeptide of SEQ ID NOS: 11, 21, 31, 41, 51, 61,
71, 81, 91,
101 during synthesis of a warhead structure in an enediyne compound; and a
fragment
thereof, which fragment has the ability to substitute for a polypeptide of SEO
ID NOS:
11, 21, 31, 41, 51, 61, 71, 81, 91, 101 in the synthesis of the warhead
structure in an
enediyne compound. In one aspect of this embodiment, the nucleic acid encoding
the
gene cassette may be used in genetic engineering application to synthesize the
warhead structure of an enediyne compound.
Another embodiment of the present invention is an isolated or purified
polypeptides comprising a sequence selected from the group consisting of: (a)
SEQ ID
NOS: 1, 13, 23, 33, 43, 53, 63, 73, 83, 93 and fragments comprising 1300,
preferably
1450, more preferably 1550, still more preferably 1650, still more preferably
1750 and
most preferably 1850 consecutive amino acids of SEQ ID NOS: 1, 13, 23, 33, 43,
53,
63, 73, 83, 93; (b) SEQ ID NOS: 3, 5, 15, 25, 35, 45, 55, 65, 75, 85, 95; and
fragments
comprising 40, preferably 60, more preferably 80, still more preferably 100,
still more
preferably 120 and most preferably 130 consecutive amino acids of SEQ ID NOS:
3, 5,
15, 25, 35, 45, 55, 65, 75, 85, 95; (c) SEQ ID NOS: 7, 17, 27, 37, 47, 57, 67,
77, 87, 97;
and fragments comprising 220, preferably 240, more preferably 260, still more
preferably 280, still more preferably 300 and most preferably 310 consecutive
amino
acids of SEQ ID NOS: 7, 17, 27, 37, 47, 57, 67, 77, 87, 97; (d) SEQ ID NOS: 9,
19, 29,
39, 49, 59, 69, 79, 89, 99; and fragments comprising 520, preferably 540, more
preferably 560, still more preferably 580, still more preferably 600 and most
preferably
620 consecutive amino acids of SEQ ID NOS: 9, 19, 29, 39, 49, 59, 69, 79, 89,
99; and
(e) SEO I D NOS: 11, 21, 31, 41, 51, 61, 71, 81, 91, 101; and fragments
comprising
220, preferably 240, more preferably 260, still more preferably 280, still
more preferably
300 and most preferably 320 consecutive amino acids of SEO ID NOS: 11, 21, 31,
41,
51, 61, 71, 81, 91 and 101. One aspect of the present invention is an isolated
or
purified polypeptide having at least 70% homology to the polypeptide of this
embodiment by analysis with BLASTP algorithm with the default parameters.
Another
aspect of the present invention is an isolated or purified polypeptide having
at least
99% homology to the polypeptides of this embodiment as determined by analysis
with
BLASTP algorithm with the default parameters.
CA 02444812 2003-10-28
3011-13CA
_g_
Another embodiment is an isolated or purified enediyne polyketide synthase
comprising a polypeptide selected from the group consisting of (a) SEQ ID NOS:
1, 13,
23, 33, 43, 53, 63, 73, 83, 93; (b) polypeptides having at least 75% homology
to a
polypeptide of SEQ ID NOS: 1, 13, 23, 33, 43, 53, 63, 73, 83, 93 as determined
using
the BLASTP algorithm with the default parameters and having the ability to
substitute
for a polypeptide of SEQ ID NOS: 1, 13, 23, 33, 43, 53, 63, 73, 83 or 93
during
synthesis a warhead structure in an enediyne compound; and (c) fragments of
the
polypeptides of (a) and (b), which fragments have the ability to substitute
for a
polypeptide of SEQ ID NOS: 1, 13, 23, 33, 43, 53, 63, 73, 83, 93 in the
synthesis of the
warhead structure in an enediyne compound. In one aspect of this embodiment,
the
enediyne polyketide synthase protein may be used in genetic engineering
applications
to synthesize the warhead structure of an enediyne compound.
Another embodiment is an isolated, purified enediyne polyketide synthase
catalytic complex comprising (a) a polypeptide selected from the group
consisting of
SEQ ID NOS: 1, 13, 23, 33, 43, 53, 63, 73, 83, 93; polypeptides having at
least 75%
homology to a polypeptide of SEQ ID NOS: 1, 13, 23, 33, 43, 53, 63, 73, 83, 93
as
determined using the BLASTP algorithm with the default parameters and having
the
ability to substitute for a polypeptide of SEO ID NOS: 1, 13, 23, 33, 43, 53,
63, 73, 83 or
93 during synthesis a warhead structure in an enediyne compound; and fragments
thereof, which fragments have the ability to substitute for a polypeptide of
SEQ ID NOS:
1, 13, 23, 33, 43, 53, 63, 73, 83, 93 in the synthesis of the warhead
structure in an
enediyne compound; and (b) a polypeptide selected from the group consisting of
SEQ
ID NOS: 3, 5, 15, 25, 35, 45, 55, 65, 75, 85, 95; polypeptides having at least
75%
homology to a polypeptide of SEQ ID NOS: 3, 5, 15, 25, 35, 45, 55, 65, 75, 85,
95 as
determined using the BLASTP algorithm with the default parameters and having
the
ability to substitute for a polypeptide of SEQ ID NOS: 3, 5, 15, 25, 35, 45,
55, 65, 75,
85, 95 during synthesis of a warhead structure in an enediyne compound; and
fragments thereof, which fragments have the ability to substitute for a
polypeptide of
SEQ ID NOS: 3, 5, 15, 25, 35, 45, 55, 65, 75, 85, 95 in the synthesis of the
warhead
structure in an enediyne compound. In one aspect of this embodiment, the
enediyne
polyketide synthase catalytic complex may be used in genetic engineering
applications
to synthesize the warhead structure of an enediyne compound.
CA 02444812 2003-10-28
3011-13CA
-10-
In another embodiment, the invention is a polypeptide selected from the group
consisting of: (a) SEQ ID NOS: 7, 17, 27, 37, 47, 57, 67, 77, 87, 97; (b)
polypeptides
having at least 75% homology to a polypeptide of SEQ ID NOS: 7, 17, 27, 37,
47, 57,
67, 77, 87, 97 as determined using the BLASTP algorithm with the default
parameters
and having the ability to substitute for a polypeptide of SEQ ID NOS: 7, 17,
27, 37, 47,
57, 67, 77, 87, 97 during synthesis of a warhead structure in an enediyne
compound;
and (c) fragments of (a) or (b), which fragments have the ability to
substitute for a
polypeptide of SEQ ID NOS: 7, 17, 27, 37, 47, 57, 67, 77, 87, 97 in the
synthesis of the
warhead structure in an enediyne compound. In one aspect, the polypeptide of
this
embodiment may be used with an enediyne polyketide synthase catalytic complex
of
the invention in genetic engineering applications to synthesize the warhead
structure of
an enediyne compound.
In another embodiment, the invention is a polypeptide selected from the group
consisting of: (a) SEQ ID NOS: 9, 19, 29, 39, 49, 59, 69, 79, 89, 99; (b)
polypeptides
having at least 75% homology to a polypeptide of SEQ ID NOS: 9, 19, 29, 39,
49, 59,
69, 79, 89, 99 as determined using the BLASTP algorithm with the default
parameters
and having the ability to substitute for a polypeptide of SEO ID NOS: 9, 19,
29, 39, 49,
59, 69, 79, 89, 99 during synthesis of a warhead structure in an enediyne
compound;
and (c) fragments of (a) or (b), which fragments have the ability to
substitute for a
polypeptide of SEQ ID NOS: 9, 19, 29, 39, 49, 59, 69, 79, 89, 99 in the
synthesis of the
warhead structure in an enediyne compound. In one aspect, the polypeptide of
this
embodiment may be used with an enediyne polyketide synthase catalytic complex
of
the invention in genetic engineering applications to synthesize the warhead
structure of
an enediyne compound.
In another embodiment, the invention is a polypeptide selected from the group
consisting of (a) SEQ ID NOS: 11, 21, 31, 41, 51, 61, 71, 81, 91, 101; (b)
polypeptides
having at least 75% homology to a polypeptide of SEQ I D NOS: 11, 21, 31, 41,
51, 61,
71, 81, 91, 101 as determined using the BLASTP algorithm with the default
parameters
and having the ability to substitute for a polypeptide of SEQ ID NOS: 11, 21,
31, 41, 51,
61, 71, 81, 91, 101 during synthesis of a warhead structure in an enediyne
compound;
and (c) fragments of (a) or (b), which fragments have the ability to
substitute for a
polypeptide of SEQ ID NOS: 71, 21, 31, 41, 51, 61, 71, 81, 91, 101 in the
synthesis of
the warhead structure in an enediyne compound. fn one aspect of this
embodiment,
CA 02444812 2003-10-28
3011-13CA
-11-
the polypeptide of this embodiment may be used with an enediyne polyketide
synthase
catalytic complex of the invention in genetic engineering applications to
synthesize the
warhead structure of an enediyne compound.
An enediyne gene cluster may be identified using compositions of the invention
such as hybridization probes or PCR primers. Hybridization probes or PCR
primers
according to the invention are derived from protein families associated with
the
warhead structure characteristic of enediynes. To identify enediyne gene
clusters, the
hybridization probes or PCR primers are derived from any one or more nucleic
acid
sequences corresponding to the five protein families designated herein as
PKSE,
TEBC, UNBL, UNBV and UNBU. The compositions of the invention are used as
probes to identify enediyne biosynthetic genes, enediyne gene fragments,
enediyne
gene clusters, or enediyne producing organisms from samples including
potential
enediyne producing microorganisms. The samples may be in the form of
environmental biomass, pure or mixed microbial culture, isolated genomic DNA
from
pure or mixed microbial culture, genomic DNA libraries from pure or mixed
microbial
culture. The compositions are used in polymerase chain reaction, and nucleic
acid
hybridization techniques well known to those skilled in the art.
Environmental samples that harbour microorganisms with the potential to
produce enediynes are identified by PCR methods. Nucleic acids contained
within the
environmental sample are contacted with primers derived from the invention so
as to
amplify target orthosomycin biosynthetic gene sequences. Environmental samples
deemed to be positive by PCR are then pursued to identify and isolate the
enediyne
gene cluster and the microorganism that contains the target gene sequences.
The
enediyne gene cluster may be identified by generating genomic DNA libraries
(for
example, cosmid, BAC, etc.) representative of genomic DNA from the population
of
various microorganisms contained within the environmental sample, locating
genomic
DNA clones that contain the target sequences and possibly overlapping clones
(for
example, by hybridization techniques or PCR), determining the sequence of the
desired
genomic DNA clones and deducing the ORFs of the enediyne biosynthetic locus.
The
microorganism that contains the enediyne biosynthetic locus may be identified
and
isolated, for example, by colony hybridization using nucleic acid probes
derived from
either the invention or the newly identified enediyne biosynthetic locus. The
isolated
enediyne biosynthetic locus may be introduced into an appropriate surrogate
host to
CA 02444812 2003-10-28
3011-13CA
-12-
achieve heterologous production of the enediyne compound(s); alternatively, if
the
microorganism containing the enediyne biosynthetic locus is identified and
isolated it
may be subjected to fermentation to produce the enediyne compound(s).
A microorganism that harbours an enediyne gene cluster is first identified and
isolated as a pure culture, for example, by colony hybridization using nucleic
acid
probes derived from the invention. Beginning with a pure culture, a genomic
DNA
library (for example, cosmid, BAC, etc.) representative of genomic DNA from
this single
species is prepared, genomic DNA clones that contain the target sequences and
possibly overlapping clones are located using probes derived from the
invention (for
example, by hybridization techniques or PCR), the sequence of the desired
genomic
DNA clones is determined and the ORFs of the enediyne biosynthetic locus are
deduced. The microorganism containing the enediyne biosynthetic locus may be
subjected to fermentation to produce the enediyne compounds) or the enediyne
biosynthetic locus may be introduced into an appropriate surrogate host to
achieve
heterologous production of the enediyne compound(s).
An enediyne gene cluster may also be identified in silico using one or more
sequences selected from enediyne-specific nucleic acid code, and enediyne-
specific
polypeptide code as taught by the invention. A query from a set of query
sequences
stored on computer readable medium is read and compared to a subject selected
from
the reference sequences of the invention. The level of similarity between said
subject
and query is determined and queries sequences representing enediyne genes are
identified.
Thus another embodiment of the invention is a method of identifying an
enediyne biosynthetic gene or gene fragment comprising providing a sample
containing
genomic DNA, and detecting the presence of a nucleic acid sequence coding for
a
polypeptide from at least one or the groups consisting of: (a) SEQ ID NOS: 1,
13, 23,
33, 43, 53, 63, 73, 83, 93; and polypeptides having at least 75% homology to a
polypeptide of SEQ ID NOS: 1, 13, 23, 33, 43, 53, 63, 73, 83, 93 as determined
using
the BLASTP algorithm with the default parameters; (b) SEQ ID NOS: 3, 5, 15,
25, 35,
45, 55, 65, 75, 85, 95; and polypeptides having at least 75% homology to a
polypeptide
of SEQ ID NOS: 3, 5, 15, 25, 35, 45, 55, 65, 75, 85, 95 as determined using
the
BLASTP algorithm with the default parameters; (c) SEQ ID NOS: 7, 17, 27, 37,
47, 57,
67, 77, 87, 97; and polypeptides having at least 75% homology to a polypeptide
of SEQ
CA 02444812 2003-10-28
3011-13CA
- 13-
ID NOS: 7, 17, 27, 37, 47, 57, 67, 77, 87, 97as determined using the BLASTP
algorithm
with the default parameters; (d) SEQ ID NOS: 9, 19, 29, 39, 49, 59, 69, 79,
89, 99; and
polypeptides having at least 75% homology to a polypeptide of SEQ ID NOS: 9,
19, 29,
39, 49, 59, 69, 79, 89, 99 as determined using the BLASTP algorithm with the
default
parameters; and (e) SEQ I D NOS: 11, 21, 31, 41, 51, 61, 71, 81, 91, 101; and
polypeptides having at least 75% homology to a polypeptide of SEQ ID NOS: 11,
21,
31, 41, 51, 61, 71, 81, 91 and 101 as determined using the BLASTP algorithm
with the
default parameters. One aspect of this embodiment provides detecting a nucleic
acid
sequence coding a polypeptide from at least two of the above groups (a), (b),
(c), (d)
and (e). Another aspect of this embodiment provides detecting a nucleic acid
sequence coding a polypeptide from at least three of the groups (a), (b), (c),
(d) and (e).
Another aspect of this embodiment provides detecting a nucleic acid sequence
coding
a polypeptide from at least four of the groups (a), (b), (c), (d) and (e).
Another aspect of
this embodiment provides detecting a nucleic acid sequence coding a
polypeptide from
each of the groups (a), (b), (c), (d) and (e). Another aspect of this
embodiment of the
invention provide the further step of using the nucleic acid detected to
isolate an
enediyne gene cluster from the sample containing genomic DNA. Another aspect
of
this embodiment of the invention comprises identifying an organism containing
the
nucleic acid sequence detected from the genomic DNA in the sample.
It is understood that the invention, having provided, compositions and methods
to identify enediyne biosynthetic gene cluster, further provides enediynes
produced by
the biosynthetic gene clusters identified.
BRIEF DESCRIPTION OF THE DRAWINGS:
Figure 1 is a block diagram of a computer system which implements and
executes software tools for the purpose of comparing a query to a subject,
wherein the
subject is selected from the reference sequences of the invention
Figures 2A, 2B, 2C and 2D are flow diagrams of a sequence comparison
software that can be employed for the purpose of comparing a query to a
subject,
wherein the subject is selected from the reference sequences of the invention,
wherein
Figure 2A is the query initialization subprocess of the sequence comparison
software,
Figure 2B is the subject datasource initialization subprocess of the sequence
comparison software, Figure 2C illustrates the comparison subprocess and the
analysis
CA 02444812 2003-10-28
3011-13CA
-14-
subprocess of the sequence comparison software, and Figure 2D is the
Display/Report
subprocess of the sequence comparison software.
Figure 3 is a flow diagram of the comparator algorithm (238) of Figure 2C
which
is one embodiment of a comparator algorithm that can be used for pairwise
determination of similarity between a query/subject pair.
Figure 4 is a flow diagram of the analyzer algorithm (244) of Figure 2C which
is
one embodiment of an analyzer algorithm that can be used to assign identity to
a query
sequence, based on similarity to a subject sequence, where the subject
sequence is a
reference sequence of the invention.
1 o Figure 5 is a schematic representation comparing the calicheamicin
enediyne
biosynthetic locus from Micromonospora echinospora subsp. calichensis (CALI),
the
macromomycin (auromomycin) enediyne biosynthetic locus from Streptomyces
macromycetius (MACK), and a chromoprotein enediyne biosynthetic locus from
Streptomyces ghanaensis (009C). Open reading frames in each locus are
identified by
boxes; gray boxes indicate ORFs that are not common to the three enediyne
loci, black
boxes indicate ORFs that are common to the three enediyne loci and are labeled
using
a four-letter protein family designation. The scale is in kilobases.
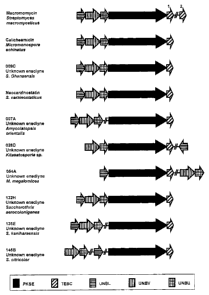
Figure 6 illustrates the 5 genes conserved throughout ten enediyne
biosynthetic
loci from diverse genera, including both chromoprotein and non-chromoprotein
20 enediyne loci.
Figure 7 is a graphical depiction of the domain architecture typical of
enediyne
polyketide synthases (PKSE).
Figure 8 is an amino acid clustal alignment of full length enediyne polyketide
synthase (PKSE) proteins from ten enediyne biosynthetic loci. Approximate
domain
boundaries are indicated above the alignment. Conserved residues or motifs
important
for the function of each domain are highlighted in black.
Figure 9A is an amino acid clustal alignment comparing the acyl carrier
protein
(ACP) domain of the PKSEs from three known enediynes, macromomycin (MACK),
calicheamicin (CALI), and neocarzinostatin (NEOC), and the ACP domain of the
30 actinorhodin Type !l PKS system (1AF8). Figure 9B depicts the space-filling
side-
chains of the conserved residues on the three dimensional structure of the ACP
of the
actinorhodin Type II PKS system (1AF8).
CA 02444812 2003-10-28
3011-13CA
-15-
Figure 1 OA is an amino acid clustal alignment comparing the 4'-
phosphopantetheinyl tranferase (PPTE) domain of the PKSEs from three known
enediynes, macromomycin (MACK), calicheamicin (CALI), and neocarzinostatin
(NEOC), and the 4'-phosphopantetheinyl transferase, Sfp, of Bacillus subtilis
(sfp).
Conserved residues are boxed. The known secondary structure of Sfp is shown
below
the aligned sequences and the predicted secondary struture of the PPTE domain
of the
PKSE is shown above the aligned sequences wherein the boxes indicate a-helices
and
the arrows indicate ~3-sheets. Figure 10B shows how the conserved residues of
the 4'-
phosphopantetheinyl transferase Sfp co-ordinate a magnesium ion and coenzyme
A;
corresponding residues in the neocarzinostatin PPTE domain are shown in bold.
Figure 11 is an amino acid clustal alignment of eleven TEBC proteins and 4-
hydroxybenzoyl-CoA thioesterase (1 BVQ) superimposed with the secondary
structure
of 1 BVQ. Alpha-helices (a) and beta-sheets (~3) are depicted by arrows.
Figure 12 is an amino acid clustal alignment of ten UNBL proteins.
Figure 13 is an amino acid clustal alignment of ten UNBV proteins highlighting
the putative N-terminal signal sequence that likely targets these proteins for
secretion.
Figure 14 is an amino acid clustal alignment of ten UNBU proteins highlighting
the putative transmembrane domains that likely anchor this family of proteins
within the
cell membrane.
Figure 15 shows restriction site and functional maps of plasmids pEC01202-
CALI-1 and pEC01202-CALI-4 of the invention. The open reading frames of the
genes
forming an expression cassette according to the invention are shown as arrows
pointing in the direction of transcription.
Figure 16 shows restriction site and functional maps of plasmids pEC01202-
CALI-5, pEC01202-CALI-2, pEC01202-CALI-3, pEC01202-CALI-6 and pEC01202-
CALI-7. The open reading frames of the genes forming the expression cassette
according to the invention are shown as arrows pointing in the direction of
transcription.
Figure 17 is an immunoblot analysis of His-tagged TEBC protein in total
protein
extracts from recombinant S. lividans TK24 clones harboring the pEC01202-CALI-
2 or
the pEC01202-CALL-4 expression vector.
Figure 18 is an immunoblot analysis of His-tagged TEBC protein in fractionated
extracts from recombinant S. lividans TK24 clones harboring the pEC01202-CALI-
2
expression vector.
CA 02444812 2003-10-28
3011-13CA
-16-
DETAILED DESCRIPTION OF THE INVENTION:
The invention provides enediyne related compositions. The compositions can
be used to produce enediyne-related compounds. The compositions can also be
used
to identify enediyne natural products, enediyne genes, enediyne gene clusters
and
enediyne producing organisms. The invention rests on the surprising discovery
that all
enediynes, including chromoprotein enediynes and non-chromoprotein enediynes,
use
a conserved set of genes for formation of the warhead structure.
To provide the compositions and methods of the invention, a sample of the
microorganism Streptomyces macromyceticus was obtained and the biosynthetic
locus
1 o for the chromoprotein enediyne macromomycin was identified. The gene
cluster was
identified as the biosynthetic locus for macromomycin from Streptomyces
macromyceticus NRRL B-5335 (sometimes referred to herein as MACR), firstly by
confirming the sequence encoding the apoprotein associated with the
chromoprotein,
which sequence is disclosed in Samy TS et a!., J. Biol. Chem (1983) Jan
10;258(1 )
pp.183-91, and secondly using the genome scanning procedure disclosed in co-
pending application USSN 09/910,813.
A sample of the microorganism Micromonospora echinospora subsp. calichensis
was then obtained and the full biosynthetic focus for the non-chromoprotein
enediyne
calicheamicin was identified. The gene cluster was identified as the
biosynthetic locus
20 for calicheamicin from Micromonospora echinospora subsp. calichensis NRRL
15839
(sometimes referred to herein as CALI) by comparing the sequence with the
partial
locus for CALI which was disclosed in WO 00/40596. We were able to overcome
the
problems encountered in prior attempts to isolate and clone the entire
biosynthetic
locus by using a shotgun-based approach as described in co-pending application
UUSN 09/910,813.
We identified two further enediyne natural products biosynthetic loci from
organisms not previously reported to produce enediyne compounds, namely a
chromoprotein enediyne from Streptomyces ghanaensis NRRL B-12104 (sometimes
referred to herein as 009C), and a chromoprotein enediyne from Amycolatopsis
30 orientalis ATCC 43491 (sometimes referred to herin as 007A). The presence
of an
apoprotein encoding gene in 009C and 007A confirms that 009C and 007A produce
chromoprotein enediyne compounds.
CA 02444812 2003-10-28
3011-13CA
-17-
Comparison of the MACR, CALI, 009C and 007A loci revealed that all loci
contain at least one a member of five (5) protein families. The five protein
families are
referred to throughout the description and figures by reference to a four-
letter
designation as indicated Table 1.
Table 1
Family
descriptions
FamiliesFunction
PKSE unusual polyketide synthase, found only in enediyne biosynthetic
loci and involved in
warhead formation; believed to act iteratively.
TEBC thioesterase unique to enediyne biosynthetic loci; significant
similarity to small (130-150 aa)
proteins of the 4-hydroxybenzoyl-CoA thioesterase family
in a number of bacteria.
UNBL unique to enediyne biosynthetic loci; these proteins are
rich in basic amino acids and
contain several conserved or invariant histidine residues.
UNBV unique to enediyne biosynthetic loci; secreted proteins;
contain putative cleavable N-
terminal signal sequence; believed to be associated with
stabilization and/or export of the
enediyne chromophore and/or late modifications in the
biosynthesis of enediyne
chromophores.
UNBU unique to enediyne biosynthetic loci; C-terminal domain
homology to bacterial putative ABC
transporters and permease transport systems; integral
membrane proteins with seven or
eight putative membrane-spanning alpha helices; believed
to be involved in transport of
enediynes and/or intermediates across the cell membrane.
A member of each of the five protein families was found in each of the more
than
ten biosynthetic loci for chromoprotein and non-chromoprotein enediynes
studied. Two
of the five protein families, PKSE and TEBC, form a polyketide synthase
catalytic
complex involved in formation of the warhead structure that distinguishes
enediyne
compounds. The other three protein families conserved throughout chromoprotein
and
non-chromoprotein enediyne biosynthetic loci are also associated with the
warhead
structure that characterizes enediyne compounds. Nucleic acid sequences and
polypeptide sequences related to these five protein families form the basis
for the
compositions and methods of the invention.
We have discovered at least one member of each of the protein families PKSE,
TEBC, UNBL, UNBV and UNBU in all of the 10 enediyne biosynthetic loci studied,
including MACK, CALI, 009C, 007A, an enediyne biosynthetic locus from
Kitasatosporia sp. (sometimes referred to herein as 028D), an enediyne
biosynthetic
locus from Micromonospora megalomicea (sometimes referred to herein as 054A),
an
CA 02444812 2003-10-28
3011-13CA
-18-
enediyne biosynthetic locus from Saccharothrix aerocolonigenes (sometimes
referred
to herein as 132H), an enediyne biosynthetic locus from Streptomyces
kaniharaensis
(sometimes referred to herein as 135E), an enediyne biosynthetic locus from
Streptomyces citricolor (sometimes referred to herein as 1458), and the
biosynthetic
locus for the chromoprotein enediyne neocarzinostatin from Streptomyces
carzinostaticus (sometimes referred to herein as NEOC).
The protein families PKSE, TEBC, UNBL, UNBV and UNBU of the present
invention are associated with warhead formation in enediyne compounds and are
found
in both chromoprotein and non-chromoprotein enediyne biosynthetic loci.
Members of
the protein families PKSE, TEBC, UNBL, UNBV and UNBU found within an enediyne
biosynthetic loci are necessarily present in a single operon and are therefore
not
necessarily transcriptionally linked to one another. However, the members of
the
protein families PKSE, TEBC, UNBL, UNBV and UNBU that are found within a
single
enediyne biosynthetic locus are functionally linked to one another in that
they act in a
concerted fashion in the production of an enediyne product. Although
expression of
functionally linked enediyne specific genes encoding members of the PKSE,
TEBC,
UNBL, UNBV and UNBU protein families may be under control of distinct
transcriptional
promoters, they may nonetheless be expressed in a concerted fashion.
Due to high overall sequence conservation between members of the PKSE,
TEBC, UNBL, UNBV and UNBU protein families, it is expected that members of the
PKSE, TEBC, UNBL, UNBV and UNBU protein families may be exchanged for another
member of the same protein family while retaining the ability of the new
enediyne
biosynthetic system to synthesize the warhead structure of an enediyne
compound.
Thus, it is contemplated that genes encoding a polypeptide from protein
families PKSE,
TEBC, UNBL, UNBV and UNBU from two or more different enediyne biosynthetic
systems may be combined so as to obtain a full complement of the five-gene
enediyne
cassette of the invention, wherein one or more genes in the enediyne cassette
has
inherent or engineered optimal properties.
Representative nucleic acid sequences and polypeptide sequences drawn from
each of the ten enediyne loci described herein are provided in the
accompanying
sequence listing as examples of the compositions of the invention. Referring
to the
sequence listing, a nucleic acid sequence encoding a member of the PKSE
protein
family of the invention from the biosynthetic locus for macromomycin from
CA 02444812 2003-10-28
3011-13CA
-19-
Streptomyces macromyceticus (MACR) is provided in SEO ID NO: 2, with the
corresponding deduced polypeptide sequence provided in SEQ ID NO: 1. Nucleic
acid
sequences encoding two members of the TEBC protein family from MACK are
provided
in SEQ ID NOS: 4 and 6 with the corresponding deduced polypeptide sequences
provided in SEO ID NOS: 3 and 5 respectively. A nucleic acid sequence encoding
a
member of the UNBL protein family from MACR is provided in SEO ID NO: 8 with
the
corresponding deduced polypeptide sequence provided in SEQ ID NO: 7. A nucleic
acid sequence encoding a member of the protein family UNBV from MACK is
provided
in SEQ ID NO: 10 with the corresponding deduced polypeptide provided in SEQ ID
NO:
9. A nucleic acid sequence encoding a member of the protein family UNBU from
MACK is provided in SEQ ID NO: 12 with the corresponding deduced polypeptide
provided in SEO ID NO: 11.
A nucleic acid sequence encoding a member of the PKSE protein family of the
invention from the biosynthetic locus for calicheamicin from Micromonospora
echinospora subsp. calichensis (CALI) is provided in SEQ ID NO: 14, with the
corresponding deduced polypeptide sequence provided in SEQ ID NO: 13. A
nucleic
acid sequence encoding a member of the TEBC protein family from CALI is
provided in
SEQ ID NO: 16, with the corresponding deduced polypeptide sequence provided in
SEQ ID NO: 15. A nucleic acid sequence encoding a member of the UNBL protein
2o family from CALI is provided in SEO ID NO: 18, with the corresponding
deduced
polypeptide sequence provided in SEQ ID NO: 17. A nucleic acid sequence
encoding
a member of the UNBV protein family from CALI is provided in SEQ ID NO: 20,
with the
corresponding deduced polypeptide sequence provided in SEQ ID NO: 19. A
nucleic
acid sequence encoding a member of the UNBU protein family from CALI is
provided in
SEQ ID NO: 22, with the corresponding deduced polypeptide sequence provided in
SEO ID NO: 21.
A nucleic acid sequence encoding a member of the PKSE protein family of the
invention from the enediyne biosynthetic locus from Streptomyces ghanaensis
(009C)
is provided in SEQ ID NO: 24, with the corresponding deduced polypeptide
sequence
3o provided in SEQ ID NO: 23. A nucleic acid sequence encoding a member of the
TEBC
protein family from 009C is provided in SEQ ID NO: 26, with the corresponding
deduced polypeptide sequence provided in SEQ ID NO: 25. A nucleic acid
sequence
encoding a member of the UNBL protein family from 009C is provided in SEQ ID
NO:
CA 02444812 2003-10-28
3011-13CA
-20-
28, with the corresponding deduced polypeptide sequence provided in SEQ ID NO:
27.
A nucleic acid sequence encoding a member of the UNBV protein family from 009C
is
provided in SEO ID NO: 30, with the corresponding deduced polypeptide sequence
provided in SEQ ID NO: 29. A nucleic acid sequence encoding a member of the
UNBU
protein family from 009C is provided in SEQ ID NO: 32, with the corresponding
deduced polypeptide sequence provided in SEQ ID NO: 31.
A nucleic acid sequence encoding a member of the PKSE protein family of the
invention from the biosynthetic locus for neocazinostatin from Streptomyces
carzinostaticus subsp. neocarzinostaticus (NEOC) is provided in SEQ ID NO: 34,
with
the corresponding deduced polypeptide sequence provided in SEQ ID NO: 33. A
nucleic acid sequence encoding a member of the TEBC protein family from NEOC
is
provided in SEQ ID NO: 36, with the corresponding deduced polypeptide sequence
provided in SEO ID NO: 35. A nucleic acid sequence encoding a member of the
UNBL
protein family from NEOC is provided in SEQ ID NO: 38, with the corresponding
deduced polypeptide sequence provided in SEQ ID NO: 37. A nucleic acid
sequence
encoding a member of the UNBV protein family from NEOC is provided in SEO ID
NO:
40, with the corresponding deduced polypeptide sequence provided in SEQ ID NO:
39.
A nucleic acid sequence encoding a member of the UNBU protein family from NEOC
is
provided in SEQ ID NO: 42, with the corresponding deduced polypeptide sequence
provided in SEQ ID NO: 41.
A nucleic acid sequence encoding a member of the PKSE protein family of the
invention from the enediyne biosynthetic locus from Amycolatopsis orientalis
(007A) is
provided in SEQ ID NO: 44, with the corresponding deduced polypeptide sequence
provided in SEQ ID NO: 43. A nucleic acid sequence encoding a member of the
TEBC
protein family from 007A is provided in SEQ ID NO: 46, with the corresponding
deduced polypeptide sequence provided in SEQ ID NO: 45. A nucleic acid
sequence
encoding a member of the UNBL protein family from 007A is provided in SEQ ID
NO:
48, with the corresponding deduced polypeptide sequence provided in SEQ 1D NO:
47.
A nucleic acid sequence encoding a member of the UNBV protein family from 007A
is
provided in SEQ ID NO: 50, with the corresponding deduced polypeptide sequence
provided in SEQ ID NO: 49. A nucleic acid sequence encoding a member of the
UNBU
protein family from 007A is provided in SEQ ID NO: 52, with the corresponding
deduced polypeptide sequence provided in SEO ID NO: 51.
CA 02444812 2003-10-28
3011-13CA
-21
A nucleic acid sequence encoding a member of the PKSE protein family of the
invention from the enediyne biosynthetic locus from Kitasatosporia sp. (028D)
is
provided in SEO ID NO. 54, with the corresponding deduced polypeptide sequence
provided in SEQ ID NO: 53. A nucleic acid sequence encoding a member of the
TEBC
protein family from 028D is provided in SEQ ID NO: 56, with the corresponding
deduced polypeptide sequence provided in SEQ ID NO: 55. A nucleic acid
sequence
encoding a member of the UNBL protein family from 028D is provided in SEO ID
NO:
58, with the corresponding deduced polypeptide sequence provided in SEO ID NO:
57.
A nucleic acid sequence encoding a member of the UNBV protein family from 028D
is
provided in SEQ ID NO: 60, with the corresponding deduced polypeptide sequence
provided in SEQ ID NO: 59. A nucleic acid sequence encoding a member of the
UNBU
protein family from 028D is provided in SEQ ID N0: 62, with the corresponding
deduced polypeptide sequence provided in SEQ ID NO: 61.
A nucleic acid sequence encoding a member of the PKSE protein family of the
invention from the enediyne biosynthetic locus from Micromonospora megalomicea
(054A) is provided in SEQ ID NO: 64, with the corresponding deduced
polypeptide
sequence provided in SEO ID NO: 63. A nucleic acid sequence encoding a member
of
the TEBC protein family from 054A is provided in SEQ ID NO: 66, with the
corresponding deduced polypeptide sequence provided in SEQ ID NO: 65. A
nucleic
acid sequence encoding a member of the UNBL protein family from 054A is
provided in
SEQ ID NO: 68, with the corresponding deduced polypeptide sequence provided in
SEQ ID NO: 67. A nucleic acid sequence encoding a member of the UNBV protein
family from 054A is provided in SEQ ID NO: 70, with the corresponding deduced
polypeptide sequence provided in SEQ ID NO: 69. A nucleic acid sequence
encoding
a member of the UNBU protein family from 054A is provided in SEQ ID NO: 72,
with
the corresponding deduced polypeptide sequence provided in SEQ ID NO: 71.
A nucleic acid sequence encoding a member of the PKSE protein family of the
invention from the enediyne biosynthetic locus from Saccharothrix
aerocolonigenes
(132H) is provided in SEQ ID NO: 74, with the corresponding deduced
polypeptide
sequence provided in SEQ ID NO: 73. A nucleic acid sequence encoding a member
of
the TEBC protein family from 132H is provided in SEQ ID NO: 76, with the
corresponding deduced polypeptide sequence provided in SEQ ID NO: 75. A
nucleic
acid sequence encoding a member of the UNBL protein family from 132H is
provided in
CA 02444812 2003-10-28
3011-13CA
-22-
SEO ID NO: 78, with the corresponding deduced polypeptide sequence provided in
SEO ID NO: 77. A nucleic acid sequence encoding a member of the UNBV protein
family from 132H is provided in SEQ ID NO: 80, with the corresponding deduced
polypeptide sequence provided in SEQ ID NO: 79. A nucleic acid sequence
encoding
a member of the UNBU protein family from 132H is provided in SEQ ID NO: 82,
with
the corresponding deduced polypeptide sequence provided in SEQ ID NO: 81.
A nucleic acid sequence encoding a member of the PKSE protein family of the
invention from the enediyne biosynthetic locus from Streptomyces kaniharaensis
(135E) is provided in SEQ ID NO: 84, with the corresponding deduced
polypeptide
sequence provided in SEQ ID NO: 83. A nucleic acid sequence encoding a member
of
the TEBC protein family from 135E is provided in SEQ ID NO: 86, with the
corresponding deduced polypeptide sequence provided in SEQ ID NO: 85. A
nucleic
acid sequence encoding a member of the UNBL protein family from 135E is
provided in
SEQ ID NO: 88, with the corresponding deduced polypeptide sequence provided in
SEO ID NO: 87. A nucleic acid sequence encoding a member of the UNBV protein
family from 135E is provided in SEQ ID NO: 90, with the corresponding deduced
polypeptide sequence provided in SEQ ID NO: 89. A nucleic acid sequence
encoding
a member of the UNBU protein family from 135E is provided in SEQ ID NO: 92,
with
the corresponding deduced polypeptide sequence provided in SEQ ID NO: 91.
A nucleic acid sequence encoding a member of the PKSE protein family of the
invention from the enediyne biosynthetic locus from Strepfomyces citricolor
(145B) is
provided in SEQ ID NO: 94, with the corresponding deduced polypeptide sequence
provided in SEQ ID NO: 93. A nucleic acid sequence encoding a member of the
TEBC
protein family from 145B is provided in SEQ ID NO: 96, with the corresponding
deduced polypeptide sequence provided in SEQ ID NO: 95. A nucleic acid
sequence
encoding a member of the UNBL protein family from 145B is provided in SEQ ID
NO:
98, with the corresponding deduced polypeptide sequence provided in SEQ ID NO:
97.
A nucleic acid sequence encoding a member of the UNBV protein family from 145B
is
provided in SEQ ID NO: 100, with the corresponding deduced polypeptide
sequence
provided in SEQ ID NO: 99. A nucleic acid sequence encoding a member of the
UNBU
protein family from 145B is provided in SEO ID NO: 102, with the corresponding
deduced polypeptide sequence provided in SEQ ID NO: 101.
CA 02444812 2003-10-28
3011-13CA
-23-
As used herein, PKSE refers to a family of polyketide synthase proteins that
are
uniquely associated with enediyne biosynthetic loci and that are involved in
synthesis of
the warhead structure that characterizes enediyne compounds. Representative
members of the protein family PKSE include the polypeptides of SEO ID NOS: 1,
13,
23, 33, 43, 53, 63, 73, 83, and 93. Other members of protein family PKSE
include
polypeptides having at least 75%, preferably 80%, more preferably, 85% still
more
preferably 90% and most preferably 95% or more homology to a polypeptide
having the
sequence of SEQ ID NOS: 1, 13, 23, 33, 43, 53, 63, 73, 83, 93 as determined
using the
BLASTP algorithm with the default parameters and having the ability to
substitute for
another PKSE protein and retaining the ability to act in a concerted fashion
with a
TEBC protein during synthesis of a warhead structure of an enediyne compound.
Other members of the protein family PKSE include fragments, analogs and
derivatives
of the above polypeptides, which fragments, analogs and derivatives have the
ability to
substitute for another PKSE protein and retain the ability to act in a
concerted fashion
with TEBC during synthesis of a warhead structure of an enediyne compound.
TEBC refers to a family of thioesterase proteins unique to enediyne
biosynthesis
which together with a protein from the protein family PKSE forms an enediyne
polyketide catalytic complex and is involved in synthesis of a warhead
structure that
characterizes enediyne compounds. Representative members of the protein family
TEBC include the polypeptides of SEO ID NOS: 3, 5, 15, 25, 35, 45, 55, 65, 75,
85, and
95. Other members of protein family TEBC include polypeptides having at least
75%,
preferably 80%, more preferably, 85% still more preferably 90% and most
preferably
95% or more homology to a polypeptide having the sequence of SEQ ID NOS: 3, 5,
15,
25, 35, 45, 55, 65, 75, 85, and 95 as determined using the BLASTP algorithm
with the
default parameters and retaining the ability to act in a concerted fashion
with a protein
from the protein family PKSE during synthesis of a warhead structure in an
enediyne
compound. Other members of the protein family TEBC include fragments, analogs
and
derivatives of the above polypeptides, which fragments, analogs and
derivatives have
the ability to substitute for another TEBC protein and retain the ability to
act in a
concerted fashion with a PKSE protein during formation of a warhead structure
in an
enediyne compound.
UNBL refers to a family of proteins indicative of enediyne biosynthetic loci
and
which are rich in basic amino acids and contain several conserved or invariant
histidine
CA 02444812 2003-10-28
3011-13CA
-24-
residues. Representative members of the protein family UNBL include the
polypeptides
of SEQ ID NOS: 7, 17, 27, 37, 47, 57, 67, 77, 87 and 97. Other members of
protein
family UNBL include polypeptides having at least 75%, preferably 80%, more
preferably, 85% still more preferably 90% and most preferably 95% or more
homology
to a polypeptide having the sequence of SEQ ID NOS: 7, 17, 27, 37, 47, 57, 67,
77, 87
and 97 as determined using the BLASTP algorithm with the default parameters
and that
are present in a gene cluster associated with the biosyntehsis of an enediyne
compound. Other members of the protein family UNBL include fragments, analogs
and
derivatives of the above polypeptides, which fragments, analogs and
derivatives have
the ability to substitute for another UNBL protein and retain the ability to
act in a
concerted fashion with genes in an enediyne biosynthetic locus to form a
warhead
structure of an enediyne compound.
UNBV refers to a family of proteins indicative of enediyne biosynthetic loci
and
which may contain a cleavable N-terminal signal sequence. Representative
members
of the protein family UNBV include the polypeptides of SEQ ID NOS: 9, 19, 29,
39, 49,
59, 69, 79, 89 and 99. Other members of protein family UNBV include
polypeptides
having at least 75%, preferably 80%, more preferably, 85% still more
preferably 90%
and most preferably 95% or more homology to a polypeptide having the sequence
of
SEQ ID NOS: 9, 19, 29, 39, 49, 59, 69, 79, 89 and 99 as determined using the
BLASTP
algorithm with the default parameters and that are present in a gene cluster
associated
with the biosynthesis of an enediyne compound. Other members of the protein
family
UNBV include fragments, analogs and derivatives of the above polypeptides,
which
fragments, analogs and derivatives have the ability to substitute for another
UNBV
protein and retain the ability to act in a concerted fashion with genes in an
enediyne
biosynthetic locus to form a warhead structure in an enediyne compound.
UNBU refers to a family of membrane proteins indicative of enediyne
biosynthetic loci. Representative members of the protein family UNBU include
the
polypeptides of SEO ID NOS: 11, 21, 31, 41, 51, 61, 71, 81, 91 and 101. Other
members of protein family UNBU include polypeptides having at least 75%,
preferably
80%, more preferably, 85% still more preferably 90% and most preferably 95% or
more
homology to a polypeptide having the sequence of SEQ ID NOS: 11, 21, 31, 41,
51, 61,
71, 81, 91 and 101 as determined using the BLASTP algorithm with the default
parameters and that are present in a gene cluster associated with the
biosynthesis of
CA 02444812 2003-10-28
3011-13CA
-25-
an enediyne compound. Other members of the protein family UNBU include
fragments,
analogs and derivatives of the above polypeptides, which fragments, analogs
and
derivatives have the ability to substitute for another UNBU protein and retain
the ability
to act in a concerted fashion with genes in an enediyne biosynthetic locus to
form the
warhead structure in an enediyne compound.
"Enediyne producer" or "enediyne-producing organism" refers to a
microorganism which carries the genetic information necessary to produce an
enediyne
compound, whether or not the organism is known to produce an enediyne product.
The
terms apply equally to organisms in which the genetic information to produce
an
enediyne compound is found in the organism as it exists in its natural
environment, and
to organisms in which the genetic information is introduced by recombinant
techniques.
For the sake of particularity, specific organisms contemplated herein include
organisms
of the family Micromonosporaceae, of which preferred genera include
Micromonospora,
Actinoplanes and Dactylosporangium; the family Streptomycetaceae, of which
preferred genera include Streptomyces and Kitasatospora; the family
Pseudonocardiaceae, of which preferred genera are Amycolatopsis and
Saccharopolyspora; and the family Actinosynnemataceae, of which preferred
genera
include Saccharothrix and Actinosynnema; however the terms are intended to
encompass all organisms containing genetic information necessary to produce an
2o enediyne compound.
"Enediyne biosynthetic gene product" refers to any enzyme involved in the
biosynthesis of an enediyne, whether a chromoprotein enediyne or a non-
chromoprotein enediyne. These gene products are located in any enediyne
biosynthetic locus in an organism of the family Micromonosporaceae, of which
preferred genera include Micromonospora, Actinoplanes and Dactylosporangium;
the
family Streptomycetaceae, of which preferred genera include Streptomyces and
Kitasatospora; the family Pseudonocardiaceae, of which preferred genera are
Amycolatopsis and Saccharopolyspora. For the sake of particularity, the
enediyne
biosynthetic loci described herein are associated with Streptomyces
macromyceticus,
30 Micromonospora echinospora subsp. calichensis, Streptomyces ghanaensis,
Streptomyces carzinostaticus subsp. neocarzinostaticus, Amycolatopsis
orientalis,
Kitasatosporia sp., Micromonospora megalomicea, Saccharothrix aerocolonigenes,
Streptomyces kaniharaensis, and Streptomyces citricolor; however, it should be
CA 02444812 2003-10-28
3011-13CA
-26-
understood that this term encompasses enediyne biosynthetic enzymes (and genes
encoding such enzymes) isolated from any microorganism of the genus
Streptomyces,
Micromonospora, Amycolatopsis, Kitesatosporia, or Saccharithrix and
furthermore that
these genes may have novel homologues in any microorganism, actinomycete or
non-
actinomycete, that falls within the scope of the claims stated herein.
Specific
embodiments include the polypeptides of SEO ID NOS: 1, 3, 5, 7, 9, 11, 13, 15,
17, 19,
21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45, 47, 49, 51, 53, 55, 57,
59, 61, 63, 65,
67, 69, 71, 73, 75, 77, 79, 81, 83, 85, 87, 89, 91, 93, 95, 97, 99, 101.
The term "isolated" means that the material is removed from its original
1o environment, e.g. the natural environment if it is naturally occurring. For
example, a
naturally-occurring polynucleotide or polypeptide present in a living organism
is not
isolated, but the same polynucleotide or polypeptide, separated from some or
all of the
coexisting materials in the natural system, is isolated. Such polynucleotides
could be
part of a vector and/or such polynucleotides or polypeptides could be part of
a
composition, and still be isolated in that such vector or composition is not
part of its
natural environment.
The term "purified" does not require absolute purity; rather, it is intended
as a
relative definition. Individual nucleic acids obtained from a library have
been
conventionally purified to electrophoretic homogeneity. The purified nucleic
acids of the
20 present invention have been purified from the remainder of the genomic DNA
in the
organism by at least 104 to 106 fold. However, the term "purified" also
includes nucleic
acids which have been purified from the remainder of the genomic DNA or from
other
sequences in a library or other environment by at least one order of
magnitude,
preferably two or three orders of magnitude, and more preferably four or five
orders of
magnitude.
"Recombinant" means that the nucleic acid is adjacent to "backbone" nucleic
acid to which it is not adjacent in its natural environment. "Enriched"
nucleic acids
represent 5% or more of the number of nucleic acid inserts in a population of
nucleic
acid backbone molecules. "Backbone" molecules include nucleic acids such as
30 expression vectors, self-replicating nucleic acids, viruses, integrating
nucleic acids, and
other vectors or nucleic acids used to maintain or manipulate a nucleic acid
of interest.
Preferably, the enriched nucleic acids represent 15% or more, more preferably
50% or
CA 02444812 2003-10-28
3011-13CA
-27-
more, and most preferably 90% or more, of the number of nucleic acid inserts
in the
population of recombinant backbone molecules.
"Recombinant polypeptides" or "recombinant proteins" refers to polypeptides or
proteins produced by recombinant DNA techniques, i.e. produced from cells
transformed by an exogenous DNA construct encoding the desired polypeptide or
protein. "Synthetic" polypeptides or proteins are those prepared by chemical
synthesis.
The term "gene" means the segment of DNA involved in producing a polypeptide
chain; it includes regions preceding and following the coding region (leader
and trailer)
as well as, where applicable, intervening regions (introns) between individual
coding
segments (exons).
The term "operon" means a transctional gene cassette under the control of a
single transcriptional promoter, which gene cassette encodes polypeptides that
may act
in a concerted fashion to carry out a biochemical pathway and/or cellular
process.
A DNA or nucleotide "coding sequence" or "sequence encoding" a particular
polypeptide or protein, is a DNA sequence which is transcribed and translated
into a
polypeptide or protein when placed under the control of appropriate regulatory
sequences.
"Oligonucleotide" refers to a nucleic acid, generally of at least 10,
preferably 15
and more preferably at least 20 nucleotides, preferably no more than 100
nucleotides,
that are hybridizable to a genomic DNA molecule, a cDNA molecule, or an mRNA
molecule encoding a gene, mRNA, cDNA or other nucleic acid of interest.
A promoter sequence is "operably linked to" a coding sequence recognized by
RNA polymerase which initiates transcription at the promoter and transcribes
the
coding sequence into mRNA.
"Plasmids" are designated herein by a lower case p followed by capital letters
and/or numbers. The starting plasmids herein are commercially available,
publicly
available on an unrestricted basis, or can be constructed from available
plasmids in
accord with published procedures. in addition, equivalent plasmids to those
described
herein are known in the art and will be apparent to the skilled artisan.
"Digestion" of DNA refers to enzymatic cleavage of the DNA with a restriction
enzyme that acts only at certain sequences in the DNA. The various restriction
enzymes used herein are commercially available and their reaction conditions,
cofactors and other requirements were used as would be known to the ordinary
skilled
CA 02444812 2003-10-28
3011-13CA
-28-
artisan. For analytical purposes, typically 1 Ng of plasmid or DNA fragment is
used with
about 2 units of enzyme in about 20 NI of buffer solution. For the purpose of
isolating
DNA fragments for plasmid construction, typically 5 to 50 Ng of DNA are
digested with
20 to 250 units of enzyme in a larger volume. Appropriate buffers and
substrate
amounts for particular enzymes are specified by the manufacturer. Incubation
times of
about 1 hour at 37°C are ordinarily used, but may vary in accordance
with the
supplier's instructions. After digestion, gel electrophoresis may be performed
to isolate
the desired fragment.
Two deposits have been made with the International Depositary Authority of
Canada, Bureau of Microbiology, Health Canada, 1015 Arlington Street,
Winnipeg,
Manitoba, Canada R3E 3R2 on April 3, 2002. The first deposit is an E, coli
DH10B
strain harbouring a cosmid clone (020CN) of a partial biosynthetic locus for
macromomycin from Streptomyces macromyceticus, including open reading frames
coding for the polypeptides of SEQ ID NOS: 1, 3, 5, 7, 9 and 11, which deposit
was
assigned deposit accession number IDAC030402-1. The second deposit is an E.
coli
DH10B strain harbouring a cosmid clone (061CR) of a partial biosynthetic locus
for
calicheamicin from Micromonospora echinospora subsp. calichensis, including
open
reading frames coding for the polypeptides of SEQ ID NOS: 13, 15, 17, 19, and
21,
which deposit was assigned accession number IDAC 030402-2. The E. coli strain
deposits are referred to herein as "the deposited strains".
The deposited strains comprise a member from each of the protein families
PKSE, TEBC, UNBL, UNBV and UNBU drawn from a chromoprotein enediyne
biosynthetic locus (macromomycin) and a member from each of the protein
families
PKSE, TEBC, UNBL, UNBV and UNBU drawn from a non-chromoprotein enediyne
biosynthetic locus (calicheamicin). The sequence of the polynucleotides
comprised in
the deposited strains, as well as the amino acid sequence of any polypeptide
encoded
thereby are controlling in the event of any conflict with any description of
sequences
herein.
The deposit of the deposited strains has been made under the terms of the
Budapest Treaty on the International Recognition of the Deposit of Micro-
organisms for
Purposes of Patent Procedure. The deposited strains will be irrevocably and
without
restriction or condition released to the public upon the issuance of a patent.
The
deposited strains are provided merely as convenience to those skilled in the
art and are
CA 02444812 2003-10-28
3011-13CA
-29-
not an admission that a deposit is required for enablement, such as that
required under
35 U.S.C. ~112. A license may be required to make, use or sell the deposited
strains
or nucleic acids therein, and compounds derived therefrom, and no such license
is
hereby granted.
Representative nucleic acid sequences encoding members of the five protein
families are provided in the accompanying sequence listing as SEQ ID NOS: 2,
4, 6, 8,
10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42, 44, 46,
48, 50, 52, 54,
56, 58, 60, 62, 64, 66, 68, 70, 72, 74, 76, 78, 80, 82, 84, 86, 88, 90, 92,
94, 96, 98, 100,
102. Representative polypeptides representing members of the five protein
families are
provided in the accompanying sequence listing as SEQ ID NOS: 1, 3, 5, 7, 9,
11, 13,
15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45, 47, 49, 51,
53, 55, 57, 59,
61, 63, 65, 67, 69, 71, 73, 75, 77, 79, 81, 83, 85, 87, 89, 91, 93, 95, 97,
99, 101.
One aspect of the present invention is an isolated, purified, or enriched
nucleic
acid comprising one of the sequences of SEQ ID NOS: 2, 4, 6, 8, 10, 12, 14,
16, 18, 20,
22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42, 44, 46, 48, 50, 52, 54, 56, 58,
60, 62, 64, 66,
68, 70, 72, 74, 76, 78, 80, 82, 84, 86, 88, 90, 92, 94, 96, 98, 100, 102, the
sequences
complementary thereto, or a fragment comprising at least 10, 15, 20, 25, 30,
35, 40, 50,
75, 100, 150, 200, 300, 400 or 500 consecutive bases of one of the sequences
of SEQ
ID NOS: 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36,
38, 40, 42, 44,
46, 48, 50, 52, 54, 56, 58, 60, 62, 64, 66, 68, 70, 72, 74, 76, 78, 80, 82,
84, 86, 88, 90,
92, 94, 96, 98, 100, 102 or the sequences complementary thereto. The isolated,
purified or enriched nucleic acids may comprise DNA, including cDNA, genomic
DNA,
and synthetic DNA. The DNA may be double stranded or single stranded, and if
single
stranded may be the coding or non-coding (anti-sense) strand. Alternatively,
the
isolated, purified or enriched nucleic acids may comprise RNA.
As discussed in more detail below, the isolated, purified or enriched nucleic
acids of one of SEQ ID NOS: 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26,
28, 30, 32,
34, 36, 38, 40, 42, 44, 46, 48, 50, 52, 54, 56, 58, 60, 62, 64, 66, 68, 70,
72, 74, 76, 78,
80, 82, 84, 86, 88, 90, 92, 94, 96, 98, 100, 102 may be used to prepare one of
the
polypeptides of SEQ ID NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27,
29, 31, 33,
35, 37, 39, 41, 43, 45, 47, 49, 51, 53, 55, 57, 59, 61, 63, 65, 67, 69, 71,
73, 75, 77, 79,
81, 83, 85, 87, 89, 91, 93, 95, 97, 99, 101 or fragments comprising at least
5, 10, 15,
20, 25, 30, 35, 40, 50, 75, 100 or 100 consecutive amino acids of one of the
CA 02444812 2003-10-28
3011-13CA
-30-
polypeptides of SEQ ID NO: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27,
29, 31, 33,
35, 37, 39, 41, 43, 45, 47, 49, 51, 53, 55, 57, 59, 61, 63, 65, 67, 69, 71,
73, 75, 77, 79,
81, 83, 85, 87, 89, 91, 93, 95, 97, 99, 101.
Accordingly, another aspect of the present invention is an isolated, purified
or
enriched nucleic acid which encodes one of the polypeptides of SEQ ID NOS: 1,
3, 5,
7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45,
47, 49, 51, 53,
55, 57, 59, 61, 63, 65, 67, 69, 71, 73, 75, 77, 79, 81, 83, 85, 87, 89, 91,
93, 95, 97, 99,
101, or fragments comprising at least 5, 10, 15, 20, 25, 30, 35, 40, 50, 75,
100 or 150
consecutive amino acids of one of the polypeptides of SEQ ID NOS: 1, 3, 5, 7,
9, 11,
13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45, 47, 49,
51, 53, 55, 57,
59, 61, 63, 65, 67, 69, 71, 73, 75, 77, 79, 81, 83, 85, 87, 89, 91, 93, 95,
97, 99, 101.
The coding sequences of these nucleic acids may be identical to one of the
coding
sequences of one of the nucleic acids of SEO ID NOS: 2, 4, 6, 8, 10, 12, 14,
16, 18, 20,
22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42, 44, 46, 48, 50, 52, 54, 56, 58,
60, 62, 64, 66,
68, 70, 72, 74, 76, 78, 80, 82, 84, 86, 88, 90, 92, 94, 96, 98, 100, 102, or a
fragment
thereof or may be different coding sequences which encode one of the
polypeptides of
SEQ ID NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35,
37, 39, 41,
43, 45, 47, 49, 51, 53, 55, 57, 59, 61, 63, 65, 67, 69, 71, 73, 75, 77, 79,
81, 83, 85, 87,
89, 91, 93, 95, 97, 99, 101, or fragments comprising at least 5, 10, 15, 20,
25, 30, 35,
40, 50, 75, 100 or 150 consecutive amino acids of one of the polypeptides of
SEQ ID
NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37,
39, 41, 43, 45,
47, 49, 51, 53, 55, 57, 59, 61, 63, 65, 67, 69, 71, 73, 75, 77, 79, 81, 83,
85, 87, 89, 91,
93, 95, 97, 99, 101 as a result of the redundancy or degeneracy of the genetic
code.
The genetic code is well known to those of skill in the art and can be
obtained, for
example, from Stryer, Biochemistry, 3~d edition, W. H. Freeman & Co., New
York.
The isolated, purified or enriched nucleic acid which encodes one of the
polypeptides of SEQ ID NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27,
29, 31, 33,
35, 37, 39, 41, 43, 45, 47, 49, 51, 53, 55, 57, 59, 61, 63, 65, 67, 69, 71,
73, 75, 77, 79,
81, 83, 85, 87, 89, 91, 93, 95, 97, 99, 101, may include, but is not limited
to: (1 ) only the
coding sequences of one of SEQ ID NOS: 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22,
24, 26,
28, 30, 32, 34, 36, 38, 40, 42, 44, 46, 48, 50, 52, 54, 56, 58, 60, 62, 64,
66, 68, 70, 72,
74, 76, 78, 80, 82, 84, 86, 88, 90, 92, 94, 96, 98, 100, 102; (2) the coding
sequences of
SEO ID NOS: 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34,
36, 38, 40,
CA 02444812 2003-10-28
3011-13CA
-31
42, 44, 46, 48, 50, 52, 54, 56, 58, 60, 62, 64, 66, 68, 70, 72, 74, 76, 78,
80, 82, 84, 86,
88, 90, 92, 94, 96, 98, 100, 102 and additional coding sequences, such as
leader
sequences or proprotein sequences; or (3) the coding sequences of SEQ ID NOS:
2, 4,
6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42, 44,
46, 48, 50, 52,
54, 56, 58, 60, 62, 64, 66, 68, 70, 72, 74, 76, 78, 80, 82, 84, 86, 88, 90,
92, 94, 96, 98,
100, 102 and non-coding sequences, such as introns or non-coding sequences 5'
and/or 3' of the coding sequence. Thus, as used herein, the term
"polynucleotide
encoding a polypeptide" encompasses a polynucleotide which includes only
coding
sequence for the polypeptide as well as a polynucleotide which includes
additional
coding and/or non-coding sequence.
The invention relates to polynucleotides based on SEQ ID NOS: 2, 4, 6, 8, 10,
12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42, 44, 46, 48,
50, 52, 54, 56,
58, 60, 62, 64, 66, 68, 70, 72, 74, 76, 78, 80, 82, 84, 86, 88, 90, 92, 94,
96, 98, 100,
102 but having polynucleotide changes that are "silent", for example changes
which do
not alter the amino acid sequence encoded by the polynucleotides of SEQ ID
NOS: 2,
4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42,
44, 46, 48, 50,
52, 54, 56, 58, 60, 62, 64, 66, 68, 70, 72, 74, 76, 78, 80, 82, 84, 86, 88,
90, 92, 94, 96,
98, 100, 102. The invention also relates to polynucleotides which have
nucleotide
changes which result in amino acid substitutions, additions, deletions,
fusions and
truncations of the polypeptides of SEQ ID NOS: 1, 3, 5, 7, 9, 1 i , 13, 15,
17, 19, 21, 23,
25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45, 47, 49, 51, 53, 55, 57, 59, 61,
63, 65, 67, 69,
71, 73, 75, 77, 79, 81, 83, 85, 87, 89, 91, 93, 95, 97, 99, 101. Such
nucleotide changes
may be introduced using techniques such as site directed mutagenesis, random
chemical mutagenesis, exonuclease III deletion, and other recombinant DNA
techniques.
The isolated, purified or enriched nucleic acids of SEQ ID NOS: 2, 4, 6, 8,
10,
12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42, 44, 46, 48,
50, 52, 54, 56,
58, 60, 62, 64, 66, 68, 70, 72, 74, 76, 78, 80, 82, 84, 86, 88, 90, 92, 94,
96, 98, i 00,
102, the sequences complementary thereto, or a fragment comprising at least
10, 15,
20, 25, 30, 35, 40, 50, 75, 100, 150, 200, 300, 400 or 500 consecutive bases
of one of
the sequence of SEQ ID NOS: 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26,
28, 30, 32,
34, 36, 38, 40, 42, 44, 46, 48, 50, 52, 54, 56, 58, 60, 62, 64, 66, 68, 70,
72, 74, 76, 78,
80, 82, 84, 86, 88, 90, 92, 94, 96, 98, 100, 102, or the sequences
complementary
CA 02444812 2003-10-28
3011-13CA
-32-
thereto may be used as probes to identify and isolate DNAs encoding the
polypeptides
of SEQ ID NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33,
35, 37, 39,
41, 43, 45, 47, 49, 51, 53, 55, 57, 59, 61, 63, 65, 67, 69, 71, 73, 75, 77,
79, 81, 83, 85,
87, 89, 91, 93, 95, 97, 99, 101 respectively.
For example, a genomic DNA library may be constructed from a sample
microorganism or a sample containing a microorganism capable of producing an
enediyne. The genomic DNA library is then contacted with a probe comprising a
coding sequence or a fragment of the coding sequence, encoding one of the
polypeptides of SEQ 1D NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27,
29, 31, 33,
35, 37, 39, 41, 43, 45, 47, 49, 51, 53, 55, 57, 59, 61, 63, 65, 67, 69, 71,
73, 75, 77, 79,
81, 83, 85, 87, 89, 91, 93, 95, 97, 99, 101, or a fragment thereof under
conditions which
permit the probe to specifically hybridize to sequences complementary thereto.
In one
embodiment, the probe is an oligonucleotide of about 10 to about 30
nucleotides in
length designed based on a nucleic acid of SEQ ID NOS: 2, 4, 6, 8, 10, 12, 14,
16, 18,
20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42, 44, 46, 48, 50, 52, 54, 56,
58, 60, 62, 64,
66, 68, 70, 72, 74, 76, 78, 80, 82, 84, 86, 88, 90, 92, 94, 96, 98, 100, 102.
Genomic
DNA clones which hybridize to the probe are then detected and isolated.
Procedures
for preparing and identifying DNA clones of interest are disclosed in Ausubel
et al.,
Current Protocols in Molecular Biology, John Wifey 503 Sons, Inc. 1997; and
Sambrook
et al., Molecular Cloning: A Laboratory Manual 2d Ed., Cold Spring Harbor
Laboratory
Press, 1989. In another embodiment, the probe is a restriction fragments or a
PCR
amplified nucleic acid derived from SEQ ID NOS: 2, 4, 6, 8, 10, 12, 14, 16,
18, 20, 22,
24, 26, 28, 30, 32, 34, 36, 38, 40, 42, 44, 46, 48, 50, 52, 54, 56, 58, 60,
62, 64, 66, 68,
70, 72, 74, 76, 78, 80, 82, 84, 86, 88, 90, 92, 94, 96, 98, 100, 102.
The isolated, purified or enriched nucleic acids of SEQ ID NOS: 2, 4, 6, 8,
10,
12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42, 44, 46, 48,
50, 52, 54, 56,
58, 60, 62, 64, 66, 68, 70, 72, 74, 76, 78, 80, 82, 84, 86, 88, 90, 92, 94,
96, 98, 100,
102, the sequences complementary thereto, or a fragment comprising at least
10, 15,
20, 25, 30, 35, 40, 50, 75, 100, 150, 200, 300, 400 or 500 consecutive bases
of one of
the sequences of SEQ ID NOS: 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26,
28, 30, 32,
34, 36, 38, 40, 42, 44, 46, 48, 50, 52, 54, 56, 58, 60, 62, 64, 66, 68, 70,
72, 74, 76, 78,
80, 82, 84, 86, 88, 90, 92, 94, 96, 98, 100, 102, or the sequences
complementary
thereto may be used as probes to identify and isolate related nucleic acids.
In some
CA 02444812 2003-10-28
3011-13CA
-33-
embodiments, the related nucleic acids may be genomic DNAs (or cDNAs) from
potential enediyne producers. In one embodiment, isolated, purified or
enriched
nucleic acids of SEO ID NOS: 2, 14, 24, 34, 44, 54, 64, 74, 84, 94 the
sequences
complementary thereto, or a fragment comprising at least 10, 15, 20, 25, 30,
35, 40, 50,
75, 100, 150, 200, 300, 400 or 500 consecutive bases of one of the sequences
of SEQ
ID NOS: 2, 14, 24, 34, 44, 54, 64, 74, 84, 94 or the sequences complementary
thereto
may be used as probes to identify and isolate related nucleic acids. In such
procedures, a nucleic acid sample containing nucleic acids from a potential
enediyne-
producer is contacted with the probe under conditions which permit the probe
to
specifically hybridize to related sequences. The nucleic acid sample may be a
genomic
DNA (or cDNA) library from the potential enediyne-producer. Hybridization of
the probe
to nucleic acids is then detected using any of the methods known in the art,
including
those referred to herein.
Hybridization may be carried out under conditions of low stringency, moderate
stringency or high stringency. As an example of nucleic acid hybridization, a
polymer
membrane containing immobilized denatured nucleic acids is first prehybridized
for 30
minutes at 45 °C in a solution consisting of 0.9 M NaCI, 50 mM NaH2P04,
pH 7.0, 5.0
mM Na2EDTA, 0.5% SDS, 10X Denhardt's, and 0.5 mg/ml polyriboadenylic acid.
Approximately 2 x 10' cpm (specific activity 4-9 x 10g cpm/ug) of 32P end-
labeled
oligonucleotide probe are then added to the solution. After 12-16 hours of
incubation,
the membrane is washed for 30 minutes at room temperature in 1X SET (150 mM
NaCI, 20 mM Tris hydrochloride, pH 7.8, 1 mM Na2EDTA) containing 0.5% SDS,
followed by a 30 minute wash in fresh 1 X SET at Tm-10 C for the
oligonucleotide probe
where Tm is the melting temperature. The membrane is then exposed to auto-
radiographic film for detection of hybridization signals.
By varying the stringency of the hybridization conditions used to identify
nucleic
acids, such as genomic DNAs or cDNAs, which hybridize to the detectable probe,
nucleic acids having different levels of homology to the probe can be
identified and
isolated. Stringency may be varied by conducting the hybridization at varying
3o temperatures below the melting temperatures of the probes. The melting
temperature
of the probe may be calculated using the following formulas:
For oligonucleotide probes between 14 and 70 nucleotides in length the melting
temperature (Tm) in degrees Celcius may be calculated using the formula:
CA 02444812 2003-10-28
3011-13CA
-34-
Tm=81.5+16.6(log [Na+]) + 0.41 (fraction G+C)-(6001N) where N is the length of
the
oligonucleotide.
If the hybridization is carried out in a solution containing formamide, the
melting
temperature may be calculated using the equation Tm=81.5+16.6(iog [Na +)) +
0.41 (fraction G + C)-(0.63% formamide)-(600/N) where N is the length of the
probe.
Prehybridization may be carried out in 6X SSC, 5X Denhardt's reagent,
0.5°~0
SDS, 0.1 mg/ml denatured fragmented salmon sperm DNA or 6X SSC, 5X Denhardt's
reagent, 0.5% SDS, 0.1 mg/ml denatured fragmented salmon sperm DNA, 50%
formamide. The composition of the SSC and Denhardt's solutions are listed in
Sambrook et al., supra.
Hybridization is conducted by adding the detectable probe to the hybridization
solutions listed above. Where the probe comprises double stranded DNA, it is
denatured by incubating at elevated temperatures and quickly cooling before
addition
to the hybridization solution. It may also be desirable to similarly denature
single
stranded probes to eliminate or diminish formation of secondary structures or
oligomerization. The filter is contacted with the hybridization solution for a
sufficient
period of time to allow the probe to hybridize to cDNAs or genomic DNAs
containing
sequences complementary thereto or homologous thereto. For probes over 200
nucleotides in length, the hybridization may be carried out at 15-25 °C
below the Tm.
For shorter probes, such as oligonucleotide probes, the hybridization may be
conducted at 5-10 °C below the Tm. Preferably, the hybridization is
conducted in 6X
SSC, for shorter probes. Preferably, the hybridization is conducted in 50%
formamide
containing solutions, for longer probes.
All the foregoing hybridizations would be considered to be examples of
hybridization performed under conditions of high stringency.
Following hybridization, the filter is washed for at least 15 minutes in 2X
SSC,
0.1 % SDS at room temperature or higher, depending on the desired stringency.
The
filter is then washed with 0.1 X SSC, 0.5% SDS at room temperature (again) for
30
minutes to 1 hour.
3o Nucleic acids which have hybridized to the probe are identified by
autoradiography or other conventional techniques.
The above procedure may be modified to identify nucleic acids having
decreasing levels of homology to the probe sequence. For example, to obtain
nucleic
CA 02444812 2003-10-28
3011-13CA
-35-
acids of decreasing homology to the detectable probe, less stringent
conditions may be
used. For example, the hybridization temperature may be decreased in
increments of 5
°C from 68 °C to 42 °C in a hybridization buffer having a
Na+ concentration of
approximately 1 M. Following hybridization, the filter may be washed with 2X
SSC,
0.5% SDS at the temperature of hybridization. These conditions are considered
to be
"moderate stringency" conditions above 50°C and "low stringency"
conditions below
50°C. A specific example of "moderate stringency" hybridization
conditions is when the
above hybridization is conducted at 55°C. A specific example of "low
stringency"
hybridization conditions is when the above hybridization is conducted at
45°C.
Alternatively, the hybridization may be carried out in buffers, such as 6X
SSC,
containing formamide at a temperature of 42 °C. In this case, the
concentration of
formamide in the hybridization buffer may be reduced in 5% increments from 50%
to
0% to identify clones having decreasing levels of homology to the probe.
Following
hybridization, the filter may be washed with 6X SSC, 0.5% SDS at 50 °C.
These
conditions are considered to be "moderate stringency" conditions above 25%
formamide and "low stringency" conditions below 25% formamide. A specific
example
of "moderate stringency" hybridization conditions is when the above
hybridization is
conducted at 30% formamide. A specific example of "low stringency"
hybridization
conditions is when the above hybridization is conducted at 10% formamide.
Nucleic acids which have hybridized to the probe are identified by
autoradiography or other conventional techniques.
For example, the preceding methods may be used to isolate nucleic acids
having a sequence with at least 97%, at least 95%, at least 90%, at least 85%,
at least
80%, or at least 70% homology to a nucleic acid sequence selected from the
group
consisting of the sequences of SEQ ID NOS: 2, 4, 6, 8, 10, 12, 14, 16, 18, 20,
22, 24,
26, 28, 30, 32, 34, 36, 38, 40, 42, 44, 46, 48, 50, 52, 54, 56, 58, 60, 62,
64, 66, 68, 70,
72, 74, 76, 78, 80, 82, 84, 86, 88, 90, 92, 94, 96, 98, 100, 102, fragments
comprising at
least 10, 15, 20, 25, 30, 35, 40, 50, 75, 100, 150, 200, 300, 400, or 500
consecutive
bases thereof, and the sequences complementary thereto. Homology may be
measured using BLASTN version 2.0 with the default parameters. For example,
the
homologous polynucleotides may have a coding sequence which is a naturally
occurring allelic variant of one of the coding sequences described herein.
Such allelic
variant may have a substitution, deletion or addition of one or more
nucleotides when
CA 02444812 2003-10-28
3011-13CA
-36-
compared to the nucleic acids of SEQ ID NOS: 2, 4, 6, 8, 10, 12, 14, 16, 18,
20, 22, 24,
26, 28, 30, 32, 34, 36, 38, 40, 42, 44, 46, 48, 50, 52, 54, 56, 58, 60, 62,
64, 66, 68, 70,
72, 74, 76, 78, 80, 82, 84, 86, 88, 90, 92, 94, 96, 98, 100, 102, or the
sequences
complementary thereto.
Additionally, the above procedures may be used to isolate nucleic acids which
encode polypeptides having at least 99%, 95%, at least 90%, at least 85%, at
least
80%, or at least 70% homology to a polypeptide having the sequence of one of
SEQ ID
NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37,
39, 41, 43, 45,
47, 49, 51, 53, 55, 57, 59, 61, 63, 65, 67, 69, 71, 73, 75, 77, 79, 81, 83,
85, 87, 89, 91,
93, 95, 97, 99, 101, or fragments comprising at least 5, 10, 15, 20, 25, 30,
35, 40, 50,
75, 100, or 150 consecutive amino acids thereof as determined using the BLASTP
version 2.2.2 algorithm with default parameters.
Structural features common to the biosynthesis of all enediyne compounds
require one or more proteins selected from a group of 5 specific protein
families,
namely PKSE, TEBC, UNBL, UNBV and UNBU. Thus, a polypeptide representing a
member of any one of these five protein families or a polynucleotide encoding
a
polypeptide representing a member of any one of these five protein families is
considered indicative of an enediyne gene cluster, a enediyne natural product
or an
enediyne producing organism. It is not necessary that a member of each of the
five
protein families considered indicative of an enediyne compound be detected to
identify
an enediyne biosynthetic locus and an enediyne-producing organism. Rather, the
presence of at least one, preferably two, more preferably three, still more
preferably
four, and most preferably five of the protein families PKSE, TEBC, UNBV and
UNBU
indicates the presence of an enediyne natural product, an enediyne
biosynthetic locus
or an enediyne producing organism.
To identify an enediyne natural product, an enediyne gene cluster or an
enediyne-producing organism, nucleic acids from cultivated microorganisms or
from an
environmental sample, e.g. soil, potentially harboring an organism having the
genetic
capacity to produce an enediyne compound may be contacted with a probe based
on
3o nucleotide sequences coding a member of the five protein families PKSE,
TEBC,
UNBL, UNBV and UNBU.
In such procedures, nucleic acids are obtained from cultivated microorganisms
or from an environmental sample potentially harboring an organism having the
genetic
CA 02444812 2003-10-28
3011-13CA
_37_
capacity to produce an enediyne compound. The nucleic acids are contacted with
probes designed based on the teachings and compositions of the invention under
conditions which permit the probe to specifically hybridize to any
complementary
sequences indicative of the presence of a member of the PKSE, TEBC, UNBL, UNBV
and UNBU protein families of the invention. The presence of at least one,
preferably
two, more preferably three, still more preferably 4 or 5 of the PKSE, TEBC,
UNBL,
UNBV and UNBU protein families indicates the presence of an enediyne gene
cluster
or an enediyne producing organism.
Diagnostic nucleic acid sequences encoding members of the PKSE, TEBC,
1 o UNBL, UNBV and UNBU protein families for identifying enediyne genes,
biosynthetic
loci, and microorganisms that harbor such genes or gene clusters may be
employed on
complex mixtures of microorganisms such as those from environmental samples
(e.g.,
soil). A mixture of microorganisms refers to a heterogeneous population of
microorganisms consisting of more than one species or strain. In the absence
of
amplification outside of its natural habitat, such a mixture of microorganisms
is said to
be uncultured. A cultured mixture of microorganisms may be obtained by
amplification
or propagation outside of its natural habitat by in vitro culture using
various growth
media that provide essential nutrients. However, depending on the growth
medium
used, the amplification may preferentially result in amplification of a sub-
population of
20 the mixture and hence may not be always desirable. If desired, a pure
culture
representing a single species or strain may obtained from either a cultured or
uncultured mixture of microorganisms by established microbiological techniques
such
as serial dilution followed by growth on solid media so as to isolate
individual colony
forming units.
Enediyne biosynthetic genes and/or enediyne biosynthetic gene clusters may be
identified from either a pure culture or cultured or uncultured mixtures of
microorganisms employing the diagnostic nucleic acid sequences disclosed in
this
invention by experimental techniques such as PCR, hybridization, or shotgun
sequencing followed by bioinformatic analysis of the sequence data. The
identification
30 of one or more members of the protein families PKSE, TEBC, UNBL, UNBV and
UNBU
or enediyne gene clusters including one or more members of the protein
families
PKSE, TEBC, UNBL, UNBV and UNBU in a pure culture of a single organism
directly
distinguishes such an enediyne-producer. The identification of one or more
members
CA 02444812 2003-10-28
3011-13CA
-38-
of the protein families PKSE, TEBC, UNBL, UNBV and UNBU or enediyne gene
clusters including one or more members of the protein families PKSE, TEBC,
UNBL,
UNBV and UNBU in a cultured or uncultured mixture of microorganisms requires
further
steps to identify and isolate the microorganisms) that harbors) them so as to
obtain
pure cultures of such microorganisms.
By way of example, the colony lift technique (Ausubel et al., Current
Protocols in
Molecular Biology, John Wiley 503 Sons, Inc. 1997; and Sambrook et al.,
Molecular
Cloning: A Laboratory Manual 2d Ed., Cold Spring Harbor Laboratory Press,
1989) may
be used to to identify microorganisms that harbour enediyne genes and/or
enediyne
biosynthetic loci from a cultured mixture of microorganisms. In such a
procedure, the
mixture of microorganisms is grown on an appropriate solid medium. The
resulting
colony forming units are replicated on a solid matrix such as a nylon
membrane. The
membrane is contacted with detectable diagnostic nucleic acid sequences, the
positive
colony forming units are identified, and the corresponding colony forming
units on the
original medium are identified, purified, and amplified.
Nucleic acids encoding a member of the protein families PKSE, TEBC, UNBL,
UNBV and UNBU may be used to survey a number of environmental samples for the
presence of organisms that have the potential to produce enediyne compounds,
i.e.,
those organisms that contain enediyne biosynthetic genes andlor an enediyne
biosynthetic locus. One protocol for use of a survey to identify polypeptides
encoded
by DNA isolated from uncultured mixtures of microorganisms is outlined in Seow
et al.
(1997) J. Bacteriol. Vol. 179 pp. 7360-7368.
Where necessary, conditions which permit the probe to specifically hybridize
to
complementary sequences from an enediyne-producer may be determined by placing
a
probe based on a member of the protein families PKSE , TEBC, UNBL, UNBV and
UNBU in contact with complementary sequences obtained from an enediyne-
producer
as well as control sequences which are not from an enediyne-producer, In some
analyses, the control sequences may be from organisms related to enediyne-
producers. Alternatively, the control sequences are not related to enediyne-
producers.
Hybridization conditions, such as the salt concentration of the hybridization
buffer, the
formamide concentration of the hybridization buffer, or the hybridization
temperature,
may be varied to identify conditions which allow the probe to hybridize
specifically to
nucleic acids from enediyne-producers.
CA 02444812 2003-10-28
3011-13CA
-39-
If the sample contains nucleic acids from enediyne-producers, specific
hybridization of the probe to the nucleic acids from the enediyne-producer is
then
detected. Hybridization may be detected by labeling the probe with a
detectable agent
such as a radioactive isotope, a fluorescent dye or an enzyme capable of
catalyzing the
formation of a detectable product. Many methods for using the labeled probes
to detect
the presenceof nucleic acids in a sample are familiar to those skilled in the
art. These
include Southern Blots, Northern Blots, colony hybridization procedures, and
dot blots.
Another aspect of the present invention is an isolated or purified polypeptide
comprising the sequence of one of SEQ ID NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17,
19, 21,
23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45, 47, 49, 51, 53, 55, 57, 59,
61, 63, 65, 67,
69, 71, 73, 75, 77, 79, 81, 83, 85, 87, 89, 91, 93, 95, 97, 99, 101 or
fragments
comprising at least 5, 10, 15, 20, 25, 30, 35, 40, 50, 75, 100, or 150
consecutive amino
acids thereof. As discussed above, such polypeptides may be obtained by
inserting a
nucleic acid encoding the polypeptide into a vector such that the coding
sequence is
operably linked to a sequence capable of driving the expression of the encoded
polypeptide in a suitable host cell. For example, the expression vector may
comprise a
promoter, a ribosome binding site for translation initiation and a
transcription terminator.
The vector may also include appropriate sequences for modulating expression
levels,
an origin of replication and a selectable marker.
Promoters suitable for expressing the polypeptide or fragment thereof in
bacteria
include the E.coli lac or trp promoters, the lacl promoter, the IacZ promoter,
the T3
promoter, the T7 promoter, the gpt promoter, the lambda PR promoter, the
lambda P~
promoter, promoters from operons encoding glycolytic enzymes such as 3-
phosphoglycerate kinase (PGK), and the acid phosphatase promoter. Fungal
promoters include the a factor promoter. Eukaryotic promoters include the CMV
immediate early promoter, the HSV thymidine kinase promoter, heat shock
promoters,
the early and late SV40 promoter, LTRs from retroviruses, and the mouse
metallothionein-I promoter. Other promoters known to control expression of
genes in
prokaryotic or eukaryotic cells or their viruses may also be used.
3o Mammalian expression vectors may also comprise an origin of replication,
any
necessary ribosome binding sites, a polyadenylation site, splice donors and
acceptor
sites, transcriptional termination sequences, and 5' flanking nontranscribed
sequences.
In some embodiments, DNA sequences derived from the SV40 splice and
CA 02444812 2003-10-28
3011-13CA
-40-
polyadenylation sites may be used to provide the required nontranscribed
genetic
elements.
Vectors for expressing the polypeptide or fragment thereof in eukaryotic cells
may also contain enhancers to increase expression levels. Enhancers are cis-
acting
elements of DNA, usually from about 10 to about 300 by in length that act on a
promoter to increase its transcription. Examples include the SV40 enhancer on
the late
side of the replication origin by 100 to 270, the cytomegalovirus early
promoter
enhancer, the polyoma enhancer on the late side of the replication origin, and
the
adenovirus enhancers.
In addition, the expression vectors preferably contain one or more selectable
marker genes to permit selection of host cells containing the vector. Examples
of
selectable markers that may be used include genes encoding dihydrofolate
reductase
or genes conferring neomycin resistance for eukaryotic cell culture, genes
conferring
tetracycline or ampicillin resistance in E. coli, and the S. cerevisiae TRP1
gene.
In some embodiments, the nucleic acid encoding one of the polypeptides of SEQ
I D NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35,
37, 39, 41, 43,
45, 47, 49, 51, 53, 55, 57, 59, 61, 63, 65, 67, 69, 71, 73, 75, 77, 79, 81,
83, 85, 87, 89,
91, 93, 95, 97, 99, 101, or fragments comprising at least 5, 10, 15, 20, 25,
30, 35, 40,
50, 75, 100, or 150 consecutive amino acids thereof is assembled in
appropriate phase
with a leader sequence capable of directing secretion of the translated
polypeptides or
fragments thereof. Optionally, the nucleic acid can encode a fusion
polypeptide in
which one of the polypeptide of SEQ ID NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19,
21, 23,
25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45, 47, 49, 51, 53, 55, 57, 59, 61,
63, 65, 67, 69,
71, 73, 75, 77, 79, 81, 83, 85, 87, 89, 91, 93, 95, 97, 99, 101 or fragments
comprising
at least 5, 10, 15, 20, 25, 30, 35, 40, 50, 75, 100, or 150 consecutive amino
acids
thereof is fused to heterologous peptides or polypeptides, such as N-terminal
identification peptides which impart desired characteristics such as increased
stability
or simplified purification or detection.
The appropriate DNA sequence may be inserted into the vector by a variety of
procedures. In general, the DNA sequence is ligated to the desired position in
the
vector following digestion of the insert and the vector with appropriate
restriction
endonucleases. Alternatively, appropriate restriction enzyme sites can be
engineered
into a DNA sequence by PCR. A variety of cloning techniques are disclosed in
Ausbel
CA 02444812 2003-10-28
3011-13CA
-41
et al. Current Protocols in Molecular Biology, John Wiley 503 Sons, Inc. 1997
and
Sambrook et al., Molecular Cloning: A Laboratory Manual 2d Ed., Cold Spring
Harbour
Laboratory Press, 1989. Such procedures and others are deemed to be within the
scope ofi those skilled in the art.
The vector may be, for example, in the form of a plasmid, a viral particle, or
a
phage. Other vectors include derivatives of chromosomal, nonchromosomal and
synthetic DNA sequences, viruses, bacterial plasmids, phage DNA, baculovirus,
yeast
plasmids, vectors derived from combinations of plasmids and phage DNA, viral
DNA
such as vaccinia, adenovirus, fowl pox virus, and pseudorabies. A variety of
cloning
and expression vectors for use with prokaryotic and eukaryotic hosts are
described by
Sambrook et al., Molecular Cloning: A Laboratory Manual, Second Edition, Cold
Spring
Harbor, N.Y., (1989).
Particular bacterial vectors which may be used include the commercially
available plasmids comprising genetic elements of the well known cloning
vector
pBR322 (ATCC 37017), pKK223-3 (Pharmacia Fine Chemicals, Uppsala, Sweden),
GEM1 (Promega Biotec, Madison, WI, USA) pQE70, pQE60, pQE-9 (Qiagen), pDlO,
psiX174 pBluescript II KS, pNHBA, pNHl6a, pNHl8A, pNH46A (Stratagene),
ptrc99a,
pKK223-3, pKK233-3, pDR540, pRITS (Pharmacia), pKK232-8 and pCM7. Particular
eukaryotic vectors include pSV2CAT, pOG44, pXT1, pSG (Stratagene) pSVK3, pBPV,
2o pMSG, and pSVL (Pharmacia). However, any other vector may be used as long
as it is
replicable and stable in the host cell.
The host cell may be any of the host cells familiar to those skilled in the
art,
including prokaryotic cells or eukaryotic cells. As representative examples of
appropriate hosts, there may be mentioned: bacteria cells, such as E, coli,
Streptomyces lividans, Bacillus subtilis, Salmonella typhimurium and various
species
within the genera Pseudomonas, Streptomyces, and Staphylococcus, fungal cells,
such
as yeast, insect cells such as Drosophila S2 and Spodoptera Sf9, animal cells
such as
CHO, COS or Bowes melanoma, and adenoviruses. The selection of an appropriate
host is within the abilities of those skilled in the art.
30 The vector may be introduced into the host cells using any of a variety of
techniques, including electroporation, transformation, transfection,
transduction, viral
infection, gene guns, or Ti-mediated gene transfer. Where appropriate, the
engineered
host cells can be cultured in conventional nutrient media modified as
appropriate for
CA 02444812 2003-10-28
3011-13CA
-42-
activating promoters, selecting transformants or amplifying the genes of the
present
invention. Following transformation of a suitable host strain and growth of
the host
strain to an appropriate cell density, the selected promoter may be induced by
appropriate means (e.g., temperature shift or chemical induction) and the
cells may be
cultured for an additional period to allow them to produce the desired
polypeptide or
fragment thereof.
Cells are typically harvested by centrifugation, disrupted by physical or
chemical
means, and the resulting crude extract is retained for further purification.
Microbial cells
employed for expression of proteins can be disrupted by any convenient method,
1 o including freeze-thaw cycling, sonication, mechanical disruption, or use
of cell lysing
agents. Such methods are well known to those skilled in the art. The expressed
polypeptide or fragment thereof can be recovered and purified from recombinant
cell
cultures by methods including ammonium sulfate or ethanol precipitation, acid
extraction, anion or cation exchange chromatography, phosphocellulose
chromatography, hydrophobic interaction chromatography, affinity
chromatography,
hydroxylapatite chromatography and lectin chromatography. Protein refolding
steps
can be used, as necessary, in completing configuration of the polypeptide. If
desired,
high performance liquid chromatography (HPLC) can be employed for final
purification
steps.
2o Various mammalian cell culture systems can also be employed to express
recombinant protein. Examples of mammalian expression systems include the COS-
7
lines of monkey kidney fibroblasts (described by Gluzman, Cell, 23:175(1981 ),
and
other cell lines capable of expressing proteins from a compatible vector, such
as the
C127, 3T3, CHO, HeLa and BHK cell lines.
The constructs in host cells can be used in a conventional manner to produce
the gene product encoded by the recombinant sequence. Depending upon the host
employed in a recombinant production procedure, the polypeptide produced by
host
cells containing the vector may be glycosylated or may be non-glycosylated.
Polypeptides of the invention may or may not also include an initial
methionine amino
30 acid residue.
Alternatively, the polypeptides of SEQ ID NOs: 1, 3, 5, 7, 9, 11, 13, 15, 17,
19,
21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45, 47, 49, 51, 53, 55, 57,
59, 61, 63, 65,
67, 69, 71, 73, 75, 77, 79, 81, 83, 85, 87, 89, 91, 93, 95, 97, 99, 101, or
fragments
CA 02444812 2003-10-28
3011-13CA
-43-
comprising at least 5, 10, 15, 20, 25, 30, 35, 40, 50, 75, 100, or 150
consecutive amino
acids thereof can be synthetically produced by conventional peptide
synthesizers. In
other embodiments, fragments or portions of the polynucleotides may be
employed for
producing the corresponding full-length polypeptide by peptide synthesis;
therefore, the
fragments may be employed as intermediates for producing the full-length
polypeptides.
Cell-free translation systems can also be employed to produce one of the
polypeptides of SEO ID NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27,
29, 31, 33,
35, 37, 39, 41, 43, 45, 47, 49, 51, 53, 55, 57, 59, 61, 63, 65, 67, 69, 71,
73, 75, 77, 79,
81, 83, 85, 87, 89, 91, 93, 95, 97, 99, 101, or fragments comprising at least
5, 10, 15,
20, 25, 30, 35, 40, 50, 75, 100, or 150 consecutive amino acids thereof using
mRNAs
transcribed form a DNA construct comprising a promoter operably linked to a
nucleic
acid encoding the polypeptide or fragment thereof. In some embodiments, the
DNA
construct may be linearized prior to conducting an in vitro transcription
reaction. The
transcribed mRNA is then incubated with an appropriate cell-free translation
extract,
such as a rabbit reticulocyte extract, to produce the desired polypeptide or
fragment
thereof.
The present invention also relates to variants of the polypeptides of SEQ ID
NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37,
39, 41, 43, 45,
47, 49, 51, 53, 55, 57, 59, 61, 63, 65, 67, 69, 71, 73, 75, 77, 79, 81, 83,
85, 87, 89, 91,
93, 95, 97, 99, 101, or fragments comprising at least 5, 10, 15, 20, 25, 30,
35, 40, 50,
75, 100, or 150 consecutive amino acids thereof. The term "variant" includes
derivatives or analogs of these polypeptides. In particular, the variants may
differ in
amino acid sequence from the polypeptides of SEQ lD NOS: 1, 3, 5, 7, 9, 11,
13, 15,
17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45, 47, 49, 51, 53,
55, 57, 59, 61,
63, 65, 67, 69, 71, 73, 75, 77, 79, 81, 83, 85, 87, 89, 91, 93, 95, 97, 99,
101, by one or
more substitutions, additions, deletions, fusions and truncations, which may
be present
in any combination.
The variants may be naturally occurring or created in vifro. In particular,
such
variants may be created using genetic engineering techniques such as site
directed
mutagenesis, random chemical mutagenesis, Exonuclease III deletion procedures,
and
standard cloning techniques. Alternatively, such variants, fragments, analogs,
or
derivatives may be created using chemical synthesis or modification
procedures.
CA 02444812 2003-10-28
3011-13CA
-44-
Other methods of making variants are also familiar to those skilled in the
art.
These include procedures in which nucleic acid sequences obtained from natural
isolates are modified to generate nucleic acids which encode polypeptides
having
characteristics which enhance their value in industrial or laboratory
applications. In
such procedures, a large number of variant sequences having one or more
nucleotide
differences with respect to the sequence obtained from the natural isolate are
generated and characterized. Preferably, these nucleotide differences result
in amino
acid changes with respect to the polypeptides encoded by the nucleic acids
from the
natural isolates.
For example, variants may be created using error prone PCR. In error prone
PCR, DNA amplification is performed under conditions where the fidelity of the
DNA
polymerase is low, such that a high rate of point mutation is obtained along
the entire
length of the PCR product. Error prone PCR is described in Leung, D.W., et
al.,
Technique, 1:11-15 (19 89) and Caldwell, R. C. & Joyce G.F., PCR Methods
Applic.,
2:28-33 (1992). Variants may also be created using site directed mutagenesis
to
generate site-specific mutations in any cloned DNA segment of interest.
Oligonucleotide mutagenesis is described in Reidhaar-Olson, J.F. and Sauer,
R.T.,
Science, 241:53-57 (1988). Variants may also be created using directed
evolution
strategies such as those described in US patents nos. 6,361,974 and 6,372,497.
The variants of the polypeptides of SEQ ID NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17,
19,
21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45, 47, 49, 51, 53, 55, 57,
59, 61, 63, 65,
67, 69, 71, 73, 75, 77, 79, 81, 83, 85, 87, 89, 91, 93, 95, 97, 99, 101, may
be (i)
variants in which one or more of the amino acid residues of the polypeptides
of SEQ ID
NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37,
39, 41, 43, 45,
47, 49, 51, 53, 55, 57, 59, 61, 63, 65, 67, 69, 71, 73, 75, 77, 79, 81, 83,
85, 87, 89, 91,
93, 95, 97, 99, 101, are substituted with a conserved or non-conserved amino
acid
residue (preferably a conserved amino acid residue) and such substituted amino
acid
residue may or may not be one encoded by the genetic code.
Conservative substitutions are those that substitute a given amino acid in a
3o polypeptide by another amino acid of like characteristics. Typically seen
as
conservative substitutions are the following replacements: replacements of an
aliphatic
amino acid such as Ala, Val, Leu and 11e with another aliphatic amino acid;
replacement
of a Ser with a Thr or vice versa; replacement of an acidic residue such as
Asp or Glu
CA 02444812 2003-10-28
3011-13CA
-45-
with another acidic residue; replacement of a residue bearing an amide group,
such as
Asn or Gln, with another residue bearing an amide group; exchange of a basic
residue
such as l_ys or Arg with another basic residue; and replacement of an aromatic
residue
such as Phe or Tyr with another aromatic residue.
Other variants are those in which one or more of the amino acid residues of
the
polypeptides of SEQ ID NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27,
29, 31, 33,
35, 37, 39, 41, 43, 45, 47, 49, 51, 53, 55, 57, 59, 61, 63, 65, 67, 69, 71,
73, 75, 77, 79,
81, 83, 85, 87, 89, 91, 93, 95, 97, 99, 101 includes a substituent group.
Still other variants are those in which the polypeptide is associated with
another
compound, such as a compound to increase the half-life of the polypeptide (for
example, polyethylene glycol).
Additional variants are those in which additional amino acids are fused to the
polypeptide, such as leader sequence, a secretory sequence, a proprotein
sequence or
a sequence which facilitates purification, enrichment, or stabilization of the
polypeptide.
In some embodiments, the fragments, derivatives and analogs retain the same
biological function or activity as the polypeptides of SEQ ID NOS: 1, 3, 5, 7,
9, 11, 13,
15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45, 47, 49, 51,
53, 55, 57, 59,
61, 63, 65, 67, 69, 71, 73, 75, 77, 79, 81, 83, 85, 87, 89, 91, 93, 95, 97,
99, 101. In
other embodiments, the fragment, derivative or analogue includes a fused
herterologous sequence which facilitates purification, enrichment, detection,
stabilization or secretion of the polypeptide that can be enzymatically
cleaved, in whole
or in part, away from the fragment, derivative or analogue.
Another aspect of the present invention are polypeptides or fragments thereof
which have at least 70%, at least 80%, at least 85%, at least 90%, or more
than 95°0
homology to one of the polypeptides of SEQ ID NOS: 1, 3, 5, 7, 9, 11, 13, 15,
17, 19,
21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45, 47, 49, 51, 53, 55, 57,
59, 61, 63, 65,
67, 69, 71, 73, 75, 77, 79, 81, 83, 85, 87, 89, 91, 93, 95, 97, 99, 101, or a
fragment
comprising at least 5, 10, 15, 20, 25, 30, 35, 40, 50, 75, 100, or 150
consecutive amino
acids thereof. Homology may be determined using a program, such as BLASTP
version 2.2.2 with the default parameters, which aligns the polypeptides or
fragments
being compared and determines the extent of amino acid identity or similarity
between
them. It will be appreciated that amino acid "homology" includes conservative
substitutions such as those described above.
CA 02444812 2003-10-28
3011-13CA
-46-
The polypeptides or fragments having homology to one of the polypeptides of
S EQ I D NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33,
35, 37, 39, 41,
43, 45, 47, 49, 51, 53, 55, 57, 59, 61, 63, 65, 67, 69, 71, 73, 75, 77, 79,
81, 83, 85, 87,
89, 91, 93, 95, 97, 99, 101, or a fragment comprising at least 5, 10, 15, 20,
25, 30, 35,
40, 50, 75, 100, or 150 consecutive amino acids thereof may be obtained by
isolating
the nucleic acids encoding them using the techniques described above.
Alternatively, the homologous polypeptides or fragments may be obtained
through biochemical enrichment or purification procedures. The sequence of
potentially homologous polypeptides or fragments may be determined by
proteolytic
digestion, gel electrophoresis and/or microsequencing. The sequence of the
prospective homologous polypeptide or fragment can be compared to one of the
polypeptides of SEQ ID NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27,
29, 31, 33,
35, 37, 39, 41, 43, 45, 47, 49, 51, 53, 55, 57, 59, 61, 63, 65, 67, 69, 71,
73, 75, 77, 79,
81, 83, 85, 87, 89, 91, 93, 95, 97, 99, 101, or a fragment comprising at least
5, 10, 15,
20, 25, 30, 35, 40, 50, 75, 100, or 150 consecutive amino acids thereof using
a
program such as BLASTP version 2.2.2 with the default parameters.
The polypeptides of SEQ ID NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25,
27,
29, 31, 33, 35, 37, 39, 41, 43, 45, 47, 49, 51, 53, 55, 57, 59, 61, 63, 65,
67, 69, 71, 73,
75, 77, 79, 81, 83, 85, 87, 89, 91, 93, 95, 97, 99, 101, or fragments,
derivatives or
analogs thereof comprising at least 5, 10, 15, 20, 25, 30, 35, 40, 50, 75,
100, or 150
consecutive amino acids thereof invention may be used in a variety of
application. For
example, the polypeptides or fragments, derivatives or analogs thereof may be
used to
biocatalyze biochemical reactions. In particular, the polypeptides of the PKSE
family,
namely SEQ ID NOS: 1, 13, 23, 33, 43, 53, 63, 73, 83, 93 fragments,
derivatives or
analogs thereof; the TEBC family, namely SEQ ID NOS: 3, 5, 15, 25, 35, 45, 55,
65, 75,
85, 95 or fragments, derivatives or analogs thereof, may be used in any
combination, in
vitro or in vivo, to direct the synthesis or modification of an enediyne
warhead or a
substructure thereof. Polypeptides of the UNBL family, namely SEQ ID NOS: 7,
17, 27,
37, 47, 57, 67, 77, 87, 97 or fragments, derivatives or analogs thereof; may
be used in
vitro or in vivo to direct or aid the synthesis or modification of an enediyne
warhead or a
substructure thereof. Polypeptides of the UNBV family, namely SEQ ID NOS: 9,
19,
29, 39, 49, 59, 69, 79, 89, 99 or fragments, derivatives or analogs thereof,
may be used
in vitro or in vivo to direct or aid the synthesis or modification of an
enediyne warhead
CA 02444812 2003-10-28
3011-13CA
-47-
or a substructure thereof. Polypeptides of the UNBU family, namely SEQ ID NOS:
11,
21, 31, 41, 51, 61, 71, 81, 91, 101 or fragments, derivatives or analogs
thereof may be
used in vitro or in vivo to direct or aid the synthesis or modification of an
enediyne
warhead or a substructure thereof.
The polypeptides of SEO I D NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23,
25, 27,
29, 31, 33, 35, 37, 39, 41, 43, 45, 47, 49, 51, 53, 55, 57, 59, 61, 63, 65,
67, 69, 71, 73,
75; 77, 79, 81, 83, 85, 87, 89, 91, 93, 95, 97, 99, 101, or fragments,
derivatives or
analogues thereof comprising at least 5, 10, 15, 20, 25, 30, 35, 40, 50, 75,
100, or 150
consecutive amino acids thereof, may also be used to generate antibodies which
bind
specifically to the polypeptides or fragments, derivatives or analogues. The
antibodies
generated from SEO ID NOS: 1, 3, 5, 7, 9, 11 may be used to determine whether
a
biological sample contains Streptomyces macromyceticus or a related
microorganism.
The antibodies generated from SEQ ID NOS: 13, 15, 17, 19, 21 may be used to
determine whether a biological sample contains Micromonospora echinospora
subsp.
calichensis or a related microorganism. The antibodies generated from SEO ID
NOS:
23, 25, 27, 29, 31 may be used to determine whether a biological sample
contains
Streptomyces ghanaensis or a related microorganism. The antibodies generated
from
SEO ID NOS: 33, 35, 37, 39, 41 may be used to determine whether a biological
sample
contains Streptomyces carzinostaticus subsp. neocarzinostaticus or a related
2o microorganism. The antibodies generated from 43, 45, 47, 49, 51 may be used
to
determine whether a biological sample contains Amycolatopsis orientalis or a
related
microorganism. The antibodies generated from 53, 55, 57, 59, 61 may be used to
determine whether a biological sample contains Kitasatosporia sp. or a related
microorganism. The antibodies generated from SEQ ID NOS: 63, 65, 67, 69, 71
may
be used to determine whether a biological sample contains Micromonospora
megalomicea or a related microorganism. The antibodies generated from SEQ ID
NOS: 73, 75, 77, 79, 81 may be used to determine whether a biological sample
contains Saccharothrix aerocolonigenes or a related microorganism. The
antibodies
generated from SEO ID NOS: 83, 85, 87, 89, 91 may be used to determine whether
a
30 biological sample contains Streptomyces kaniharaensis or a related
microorganism.
The antibodies generated from SEQ ID NOS: 93, 95, 97, 99, 101 may be used to
determine whether a biological sample contains Streptomyces citricoloror a
related
microorganism.
CA 02444812 2003-10-28
3011-13CA
-48-
In such procedures, a biological sample is contacted with an antibody capable
of
specifically binding to one of the polypeptides of SEQ ID NOS: 1, 3, 5, 7, 9,
11, 13, 15,
17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45, 47, 49, 51, 53,
55, 57, 59, 61,
63, 65, 67, 69, 71, 73, 75, 77, 79, 81, 83, 85, 87, 89, 91, 93, 95, 97, 99,
101, or
fragments comprising at least 5, 10, 15, 20, 25, 30, 35, 40, 50, 75, 100, or
150
consecutive amino acids thereof. The ability of the biological sample to bind
to the
antibody is then determined. For example, binding may be determined by
labeling the
antibody with a detectable label such as a fluorescent agent, an enzymatic
label, or a
radioisotope. Alternatively, binding of the antibody to the sample may be
detected
using a secondary antibody having such a detectable label thereon. A variety
of assay
protocols may be used to detect the presence of Micromonospora echinospora
subsp.
calichensis, Streptomyces ghanaensis, Streptomyces carzinostaticus subsp.
neocarzinostaticus, Amycolatopsis orientalis, Kitasatosporia sp.,
Micromonospora
megalomicea, Saccharothrix aerocolonigenes, Streptomyces kaniharaensis,
Streptomyces citricoloror the the present of polypeptides related to SEQ ID
NOS: 1, 3,
5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43,
45, 47, 49, 51,
53, 55, 57, 59, 61, 63, 65, 67, 69, 71, 73, 75, 77, 79, 81, 83, 85, 87, 89,
91, 93, 95, 97,
99, 101 in a sample. Particular assays include ELISA assays, sandwich assays,
radioimmunoassays, and Western Blots. Alternatively, antibodies generated from
SEQ
I D NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35,
37, 39, 41, 43,
45, 47, 49, 51, 53, 55, 57, 59, 61, 63, 65, 67, 69, 71, 73, 75, 77, 79, 81,
83, 85, 87, 89,
91, 93, 95, 97, 99, 101 may be used to determine whether a biological sample
contains
related polypeptides that may be involved in the biosynthesis of enediyne
natural
products or other enediyne-like compounds.
Pofyclonal antibodies generated against the polypeptides of SEQ ID NOS: 1, 3,
5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43,
45, 47, 49, 51,
53, 55, 57, 59, 61, 63, 65, 67, 69, 71, 73, 75, 77, 79, 81, 83, 85, 87, 89,
91, 93, 95, 97,
99, 101, or fragments comprising at least 5, 10, 15, 20, 25, 30, 35, 40, 50,
75, 100, or
150 consecutive amino acids thereof can be obtained by direct injection of the
3o polypeptides into an animal or by administering the polypeptides to an
animal. The
antibody so obtained will then bind the polypeptide itself. In this manner,
even a
sequence encoding only a fragment of the polypeptide can be used to generate
CA 02444812 2003-10-28
3011-13CA
-49-
antibodies which may bind to the whole native polypeptide. Such antibodies can
then
be used to isolate the polypeptide from cells expressing that polypeptide.
For preparation of monoclonal antibodies, any technique which provides
antibodies produced by continuous cell line cultures can be used. Examples
include
the hybridoma technique (Kholer and Milstein, 1975, Nature, 256:495-497), the
trioma
technique, the human B-cell hybridoma technique (Kozbor et al., 1983,
Immunology
Today 4:72), and the EBV-hybridoma technique (Cole, et al., 1985, in
Monoclonal
Antibodies and Cancer Therapy, Alan R. Liss, Inc., pp. 77-96).
Techniques described for the production of single chain antibodies (U.S.
Patent
4,946,778) can be adapted to produce single chain antibodies to the
polypeptides of
S EO I D NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33,
35, 37, 39, 41,
43, 45, 47, 49, 51, 53, 55, 57, 59, 61, 63, 65, 67, 69, 71, 73, 75, 77, 79,
81, 83, 85, 87,
89, 91, 93, 95, 97, 99, 101, or fragments comprising at least 5, 10, 15, 20,
25, 30, 35,
40, 50, 75, 100, or 150 consecutive amino acids thereof. Alternatively,
transgenic mice
may be used to express humanized antibodies to these polypeptides or fragments
thereof.
Antibodies generated against the polypeptides of SEQ ID NOS: 1, 3, 5, 7, 9,
11,
13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45, 47, 49,
51, 53, 55, 57,
59, 61, 63, 65, 67, 69, 71, 73, 75, 77, 79, 81, 83, 85, 87, 89, 91, 93, 95,
97, 99, 101, or
fragments comprising at least 5, 10, 15, 20, 25, 30, 35, 40, 50, 75, 100, or
150
consecutive amino acids thereof may be used in screening for similar
polypeptides from
a sample containing organisms or cell-free extracts thereof. In such
techniques,
polypeptides from the sample is contacted with the antibodies and those
polypeptides
which specifically bind the antibody are detected. Any of the procedures
described
above may be used to detect antibody binding. One such screening assay is
described
in "Methods for measuring Cellulase Activities", Methods in Enzymology, Vol
160, pp.
87-116.
As used herein, the term "enediyne-specific nucleic acid codes" encompass the
nucleotide sequences of SEQ ID NOS: 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22,
24, 26, 28,
30, 32, 34, 36, 38, 40, 42, 44, 46, 48, 50, 52, 54, 56, 58, 60, 62, 64, 66,
68, 70, 72, 74,
76, 78, 80, 82, 84, 86, 88, 90, 92, 94, 96, 98, 100, 102, fragments of SEQ ID
NOS: 2, 4,
6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42, 44,
46, 48, 50, 52,
54, 56, 58, 60, 62, 64, 66, 68, 70, 72, 74, 76, 78, 80, 82, 84, 86, 88, 90,
92, 94, 96, 98,
CA 02444812 2003-10-28
3011-13CA
-50-
100, 102, nucleotide sequences homologous to SEQ ID NOS: 2, 4, 6, 8, 10, 12,
14, 16,
18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42, 44, 46, 48, 50, 52, 54,
56, 58, 60, 62,
64, 66, 68, 70, 72, 74, 76, 78, 80, 82, 84, 86, 88, 90, 92, 94, 96, 98, 100,
102, or
homologous to fragments of SEQ ID NOS: 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22,
24, 26,
28, 30, 32, 34, 36, 38, 40, 42, 44, 46, 48, 50, 52, 54, 56, 58, 60, 62, 64,
66, 68, 70, 72,
74, 76, 78, 80, 82, 84, 86, 88, 90, 92, 94, 96, 98, 100, 102, and sequences
complementary to all of the preceding sequences. The fragments include
portions of
SEQ ID NOS: 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34,
36, 38, 40,
42, 44, 46, 48, 50, 52, 54, 56, 58, 60, 62, 64, 66, 68, 70, 72, 74, 76, 78,
80, 82, 84, 86,
88, 90, 92, 94, 96, 98, 100, 102 comprising at least 10, 15, 20, 25, 30, 35,
40, 50, 75,
100, 150, 200, 300, 400 or 500 consecutive nucleotides of SEQ ID NOS: 2, 4, 6,
8, 10,
12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42, 44, 46, 48,
50, 52, 54, 56,
58, 60, 62, 64, 66, 68, 70, 72, 74, 76, 78, 80, 82, 84, 86, 88, 90, 92, 94,
96, 98, 100,
102. Preferably, the fragments are novel fragments. Homologous sequences and
fragments of SEQ ID NOS: 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28,
30, 32, 34,
36, 38, 40, 42, 44, 46, 48, 50, 52, 54, 56, 58, 60, 62, 64, 66, 68, 70, 72,
74, 76, 78, 80,
82, 84, 86, 88, 90, 92, 94, 96, 98, 100, 102 refer to a sequence having at
least 99%,
98%, 97%, 96%, 95%, 90%, 80%, 75% or 70% homology to these sequences.
Homology may be determined using any of the computer programs and parameters
described herein, including BLASTN and TBLASTX with the default parameters.
Homologous sequences also include RNA sequences in which uridines replace the
thymines in the nucleic acid codes of SEQ ID NOS: 2, 4, 6, 8, 10, 12, 14, 16,
18, 20,
22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42, 44, 46, 48, 50, 52, 54, 56, 58,
60, 62, 64, 66,
68, 70, 72, 74, 76, 78, 80, 82, 84, 86, 88, 90, 92, 94, 96, 98, 100, 102. The
homologous
sequences may be obtained using any of the procedures described herein or may
result from the correction of a sequencing error. It will be appreciated that
the nucleic
acid codes of SEQ ID NOS: 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28,
30, 32, 34,
36, 38, 40, 42, 44, 46, 48, 50, 52, 54, 56, 58, 60, 62, 64, 66, 68, 70, 72,
74, 76, 78, 80,
82, 84, 86, 88, 90, 92, 94, 96, 98, 100, 102 can be represented in the
traditional single
character format in which G, A, T and C denote the guanine, adenine, thymine
and
cytosine bases of the deoxyribonucleic acid (DNA) sequence respectively, or in
which
G, A, U and C denote the guanine, adenine, uracil and cytosine bases of the
ribonucleic acid (RNA) sequence (see the inside back cover of Stryer,
Biochemistry, 3~d
CA 02444812 2003-10-28
3011-13CA
-51 -
edition, W. H. Freeman & Co., New York) or in any other format which records
the
identity of the nucleotides in a sequence.
"Enediyne-specific polypeptide codes" encompass the polypeptide sequences of
SEO ID NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35,
37, 39, 41,
43, 45, 47, 49, 51, 53, 55, 57, 59, 61, 63, 65, 67, 69, 71, 73, 75, 77, 79,
81, 83, 85, 87,
89, 91, 93, 95, 97, 99, 101 which are encoded by the cDNAs of SEQ ID NOS: 1,
3, 5, 7,
9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45, 47,
49, 51, 53,
55, 57, 59, 61, 63, 65, 67, 69, 71, 73, 75, 77, 79, 81, 83, 85, 87, 89, 91,
93, 95, 97, 99,
101; polypeptide sequences homologous to the polypeptides of SEQ ID NOS: 1, 3,
5,
7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45,
47, 49, 51, 53,
55, 57, 59, 61, 63, 65, 67, 69, 71, 73, 75, 77, 79, 81, 83, 85, 87, 89, 91,
93, 95, 97, 99,
101, or fragments of any of the preceding sequences. Homologous polypeptide
sequences refer to a polypeptide sequence having at least 99%, 98%, 97%, 96%,
95%,
90%, 85%, 80%, 75% or 70% homology to one of the polypeptide sequences of SEQ
I D NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35,
37, 39, 41, 43,
45, 47, 49, 51, 53, 55, 57, 59, 61, 63, 65, 67, 69, 71, 73, 75, 77, 79, 81,
83, 85, 87, 89,
91, 93, 95, 97, 99, 101. Polypeptide sequence homology may be determined using
any
of the computer programs and parameters described herein, including BLASTP
version
2.2.2 with the default parameters or with any user-specified parameters. The
homologous sequences may be obtained using any of the procedures described
herein
or may result from the correction of a sequencing error. The polypeptide
fragments
comprise at least 5, 10, 15, 20, 25, 30, 35, 40, 50, 75, 100 or 150
consecutive
polypeptides of the polypeptides of SEO ID NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17,
19, 21,
23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45, 47, 49, 51, 53, 55, 57, 59,
61, 63, 65, 67,
69, 71, 73, 75, 77, 79, 81, 83, 85, 87, 89, 91, 93, 95, 97, 99, 101.
Preferably the
fragments are novel fragments. It will be appreciated that the polypeptide
codes of the
SEQ I D NOS: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33,
35, 37, 39, 41,
43, 45, 47, 49, 51, 53, 55, 57, 59, 61, 63, 65, 67, 69, 71, 73, 75, 77, 79,
81, 83, 85, 87,
89, 91, 93, 95, 97, 99, 101 can be represented in the traditional single
character format
or three letter format (see the inside back cover of Stryer, Biochemistry, 3rd
edition,
W.H. Freeman & Co., New York) or in any other format which relates the
identity of the
polypeptides in a sequence.
CA 02444812 2003-10-28
3011-13CA
-52-
A single sequence selected from enediyne-specific nucleic acid codes and
enediyne-specific polypeptide codes is sometimes referred to herein as a
subject
sequence.
It will be readily appreciated by those skilled in the art that the enediyne-
specific
nucleic acid codes, a subset thereof, enediyne-specific polypeptide codes, a
subset
thereof, and a subject sequence can be stored, recorded and manipulated on any
medium which can be read and accessed by a computer. As used herein, the words
"recorded" and "stored" refer to a process for storing information on a
computer
medium. A skilled artisan can readily adopt any of the presently known methods
for
recording information on a computer readable medium to generate manufactures
comprising one or more of the enediyne-specific nucleic acid codes, a subset
thereof,
enediyne-specific polypeptide codes, a subset thereof, and a subject sequence.
Computer readable media include magnetically readable media, optically
readable media, electronically readable media and magnetic/optical media. For
example, the computer readable media may be a hard disk, a floppy disk, a
magnetic
tape, CD-ROM, Digital Versatile Disk (DVD), Random Access Memory (RAM), or
Read
Only Memory (ROM) as well as other types of media known to those skilled in
the art.
The enediyne-specific nucleic acid codes, a subset thereof and a subject
sequence may be stored and manipulated in a variety of data processor programs
in a
variety of formats. For example, the enediyne-specific nucleic acid codes, a
subset
thereof, enediyne-specific polypeptide codes, a subset thereof, and a subject
sequence
may be stored as ASCII or text in a word processing file, such as
MicrosoftWORD or
WORDPERFECT in a variety of database programs familiar to those of skill in
the art,
such as DB2 or ORACLE. In addition, many computer programs and databases may
be used as sequence comparers, identifiers or sources of query nucleotide
sequences
or query polypeptide sequences to be compared to the enediyne-specific nucleic
acid
codes, a subset thereof, the enediyne-specific polypeptide codes, a subset
thereof, and
a subject sequence.
The following list is intended not to limit the invention but to provide
guidance to
programs and databases useful with the enediyne-specific nucleic acid codes, a
subset
thereof, enediyne-specific polypeptide codes, a subset thereof, and a subject
sequence. The program and databases which may be used include, but are not
limited
to: MacPattern (EMBL), DiscoveryBase (Molecular Applications Group), GeneMine
CA 02444812 2003-10-28
3011-13CA
-53-
(Molecular Applications Group) Look (Molecular Applications Group), MacLook
(Molecular Applications Group), BLAST and BLAST2 (NCBI), BLASTN and BLASTX
(Altschul et al., J. Mol. Biol. 215:403 (1990)), FASTA (Person and Lipman,
Proc. Nalt.
Acad. Sci. USA, 85:2444 (1988)), FASTDB (Brutlag et al. Comp. App. Biosci. 6-
237-
245, 1990), Catalyst (Molecular Simulations Inc.), Catalyst/SHAPE (Molecular
Simulations Inc.), Cerius2.DBAccess (Molecular Simulations Inc.), HypoGen
(Molecular
Simulations Inc.), Insight II (Molecular Simulations Inc.), Discover
(Molecular
Simulations Inc.), CHARMm (Molecular Simulations Inc.), Felix (Molecular
Simulations
Inc.), DeIPhi (Molecular Simulations Inc.), QuanteMM (Molecular Simulations
Inc.),
Homology (Molecular Simulations Inc.), Modeler (Molecular Simulations Inc.),
ISIS
(Molecular Simulations Inc.), Quanta/Protein Design (Molecular Simulations
Inc.),
WetLab (Molecular Simulations Inc.), WetLab Diversity Explorer (Molecular
Simulations
Inc.), Gene Explorer (Molecular Simulations Inc.), SeqFold (Molecular
Simulations Inc.),
the MDL Available Chemicals Directory database, the MDL Drug Data Report data
base, the Comprehensive Medicinal Chemistry database, Derwents' World Drug
Index
database, the BioByteMasterFile database, the Genbank database, and the
Gensyqn
database. Many other programs and databases would be apparent to one of skill
in the
art given the present disclosure.
Embodiments of the present invention include systems, particularly computer
systems that store and manipulate the sequence information described herein.
As
used herein, "a computer system", refers to the hardware components, software
components, and data storage components used to analyze enediyne-specific
nucleic
acid codes, a subset thereof, enediyne-specific polypeptide codes, a subset
thereof, or
a subject sequence.
Preferably, the computer system is a general purpose system that comprises a
processor and one or more internal data storage components for storing data,
and one
or more data retrieving devices for retrieving the data stored on the data
storage
components. A skilled artisan can readily appreciate that any one of the
currently
available computer systems are suitable.
One example of a computer system is illustrated in Figure 1. The computer
system of Figure 4 will includes a number of components connected to a central
system
bus 116, including a central processing unit 118 with internal 118 and/or
external cache
memory 120, system memory 122, display adapter 102 connected to a monitor 100,
CA 02444812 2003-10-28
3011-13CA
-54-
network adapter 126 which may also be referred to as a network interface,
internal
modem 124, sound adapter 128, 10 controller 132 to which may be connected a
keyboard 140 and mouse 138, or other suitable input device such as a trackball
or
tablet, as well as external printer 134, and/or any number of external devices
such as
external modems, tape storage drives, or disk drives. One skilled in the art
will readily
appreciate that not all components illustrated in Figure 1 are required to
practice the
invention and, likewise, additional components not illustrated in Figure 1 may
be
present in a computer system contemplated for use with the invention.
One or more host bus adapters 114 may be connected to the system bus 116.
To host bus adapter 114 may optionally be connected one or more storage
devices
such as disk drives 112 (removable or fixed), floppy drives 110, tape drives
108, digital
versatile disk DVD drives 106, and compact disk CD ROM drives 104. The storage
devices may operate in read-only mode and / or in read-write mode. The
computer
system may optionally include multiple central processing units 118, or
multiple banks
of memory 122.
Arrows 142 in Figure 1 indicate the interconnection of internal components of
the
computer system. The arrows are illustrative only and do not specify exact
connection
architecture.
Software for accessing and processing the reference sequences (such as
sequence comparison software, analysis software as well as search tools,
annotation
tools, and modeling tools etc.) may reside in main memory 122 during
execution.
In one embodiment, the computer system further comprises a sequence
comparison software for comparing the nucleic acid codes of a query sequence
stored
on a computer readable medium to a subject sequence which is also stored on a
computer readable medium; or for comparing the polypeptide code of a query
sequence stored on a computer readable medium to a subject sequence which is
also
stored on computer readable medium. A "sequence comparison software" refers to
one or more programs that are implemented on the computer system to compare
nucleotide sequences with other nucleotide sequences stored within the data
storage
means. The design of one example of a sequence comparison software is provided
in
Figures 2A, 2B, 2C and 2D.
The sequence comparison software will typically employ one or more specialized
comparator algorithms. Protein and/or nucleic acid sequence similarities may
be
CA 02444812 2003-10-28
3011-13CA
-55-
evaluated using any of the variety of sequence comparator algorithms and
programs
known in the art. Such algorithms and programs include, but are no way limited
to,
TBLASTN, BLASTN, BLASTP, FASTA, TFASTA, CLUSTAL, HMMER, MAST, or other
suitable algorithm known to those skilled in the art. (Pearson and Lipman,
1988, Proc.
Natl. Acad. Sci USA 85(8): 2444-2448; Altschul et al, 1990, J. Mol. Biol.
215(3):403-
410; Thompson et al., 1994, Nucleic Acids Res. 22(2):4673-4680; Higgins et
al., 1996,
Methods Enzymol. 266:383-402; Altschul et al., 1990, J. Mol. Biol. 215(3):403-
410;
Altschul et al., 1993, Nature Genetics 3:266-272; Eddy S.R., Bioinformatics
14:755-
763, 1998; Bailey TL et aI,J Steroid Biochem Mol Biol 1997 May;62(1 ):29-44).
One
1o example of a comparator algorithm is illustrated in Figure 3. Sequence
comparator
algorithms identified in this specification are particularly contemplated for
use in this
aspect of the invention.
The sequence comparison software will typically employ one or more specialized
analyzer algorithms. One example of an analyzer algorithm is illustrated in
Figure 4.
Any appropriate analyzer algorithm can be used to evaluate similarities,
determined by
the comparator algorithm, between a query sequence and a subject sequence
(referred
to herein as a query/subject pair). Based on context specific rules, the
annotation of a
subject sequence may be assigned to the query sequence. A skilled artisan can
readily
determine the selection of an appropriate analyzer algorithm and appropriate
context
20 specific rules. Analyzer algorithms identified elsewhere in this
specification are
particularly contemplated for use in this aspect of the invention.
Figures 2A, 2B, 2C and 2D together provide a flowchart of one example of a
sequence comparison software for comparing query sequences to a subject
sequence.
The software determines if a gene or set of genes represented by their
nucleotide
sequence, polypeptide sequence or other representation (the query sequence) is
significantly similar to the enediyne-specific nucleic acid codes, a subset
thereof,
enediyne-specific polypeptide codes, a subset thereof, of the invention (the
subject
sequence). The software may be implemented in the C or C++ programming
language,
Java, Perl or other suitable programming language known to a person skilled in
the art.
30 Referring to Figure 2A, the query sequences) may be accessed by the program
by means of input from the user 210, accessing a database 208 or opening a
text file
206. The "query initialization process" allows a query sequence to be accessed
and
loaded into computer memory 122, or under control of the program stored on a
disk
CA 02444812 2003-10-28
3011-13CA
-56-
drive 112 or other storage device in the form of a query sequence array 216.
The
query array 216 is one or more query nucleotide or polypeptide sequences
accompanied by some appropriate identifiers.
A dataset is accessed by the program by means of input from the user 228,
accessing a database 226, or opening a text file 224. The "subject data source
initialization process" of Figure 2B refers to the method by which a reference
dataset
containing one or more sequence selected from the enediyne-specific nucleic
acid
codes, a subset thereof, enediyne-specific polypeptide codes, a subset
thereof, or a
subject sequence is loaded into computer memory 122, or under control of the
program
1 o stored on a disk drive 112 or other storage device in the form of a
subject array 234.
The subject array 234 comprises one or more subject nucleotide or polypeptide
sequences accompanied by some appropriate identifiers.
The "comparison subprocess" of Figure 2C is the process by which the
comparator algorithm 238 is invoked by the software for pairwise comparisons
between
query elements in the query sequence array 216, and subject elements in the
subject
array 234. The "comparator algorithm" of Figure 2C refers to the pairwise
comparisons
between a query sequence and subject sequence, i.e. a query/subject pair from
their
respective arrays 216, 234. Comparator algorithm 238 may be any algorithm that
acts
on a query/subject pair, including but not limited to homology algorithms such
as
2o BLAST, Smith Waterman, Fasta, or statistical representation/probabilistic
algorithms
such as Markov models exemplified by HMMER, or other suitable algorithm known
to
one skilled in the art. Suitable algorithms would generally require a
query/subject pair
as input and return a score (an indication of likeness between the query and
subject),
usually through the use of appropriate statistical methods such as Karlin
Altschul
statistics used in BLAST, Forward or Viterbi algorithms used in Markov models,
or other
suitable statistics known to those skilled in the art.
The sequence comparison software of Figure 2C also comprises a means of
analysis of the results of the pairwise comparisons performed by the
comparator
algorithm 238. The "analysis subprocess" of Figure 2C is a process by which
the
3o analyzer algorithm 244 is invoked by the software. The "analyzer algorithm"
refers to a
process by which annotation of a subject is assigned to the query based on
query/subject similarity as determined by the comparator algorithm 238
according to
context-specific rules coded into the program or dynamically loaded at
runtime.
CA 02444812 2003-10-28
3011-13CA
-57-
Context-specific rules are what the program uses to determine if the
annotation of the
subject can be assigned to the query given the context of the comparison.
These rules
allow the software to qualify the overall meaning of the results of the
comparator
algorithm 238.
In one embodiment, context-specific rules may state that for a set of query
sequences to be considered representative of an enediyne locus the comparator
algorithm 238 must determine that the set of query sequences contain at least
one
query sequence that shows a statistical similarity to reference sequences
corresponding to a nucleic acid sequence code for a polypeptide from two of
the
groups consisting of: (1) SEQ ID NOS: 1, 13, 23, 33, 43, 53, 63, 73, 83, 93
and
polypeptides having at least 75% homology to a polypeptide sequence of SEQ ID
NOS:
1, 13, 23, 33, 43, 53, 63, 73, 83, 93; (2) SEQ ID NOS: 3, 5, 15, 25, 35, 45,
55, 65, 75,
85, 95 and polypeptides having at least 75% homology to a polypeptide sequence
of
SEQ ID NOS: 3, 5, 15, 25, 35, 45, 55, 65, 75, 85, 95; (3) SEO ID NOS: 7, 17,
27, 37,
47, 57, 67, 77, 87, 97, and polypeptides having at least 75% homology to a
polypeptide
sequence of SEQ ID NOS: 7, 17, 27, 37, 47, 57, 67, 77, 87, 97; (4) SEQ ID NOS:
9,
19, 29, 39, 49, 59, 69, 79, 89, 99 and polypeptides having at least 75%
homology to a
polypeptide sequence of SEQ ID NOS: 9, 19, 29, 39, 49, 59, 69, 79, 89, 99; (5)
SEQ ID
NOS: 11, 21, 31, 41, 51, 61, 71, 81, 91, 101 and polypeptides having at least
75%
homology to a pofypeptide sequence of SEQ ID NOS: 11, 21, 31, 41, 51, 61, 71,
81,
91, 101. Of course preferred context specific rules may specify a wide variety
of
thresholds for identifying enediyne-biosynthetic genes or enediyne-producing
organisms without departing from the scope of the invention. Some thresholds
contemplate that at least one query sequence in the set of query sequences
show a
statistical similarity to the nucleic acid code corresponding to 2 or 3 or 4
or 5 of the
above 5 groups polypeptides diagnostic of enediyne biosynthetic genes. Other
context
specific rules set the level of homology required in each of the group may be
set at
70%, 80%, 85%, 90%, 95% or 98% in regards to any one or more of the subject
sequences.
3o In another embodiment context-specific rules may state that for a query
sequence to be considered an enediyne polyketide synthase, the comparator
algorithm
238 must determine that the query sequence shows a statistical similarity to
subject
sequences corresponding to a nucleic acid sequence code for a polypeptide of
SEO ID
CA 02444812 2003-10-28
3011-13CA
-58-
NOS: 1, 13, 23, 33, 43, 53, 63, 73, 83, 93, polypeptides having at least 75%
homology
to a polypeptide of SEQ iD NOS: 1, 13, 23, 33, 43, 53, 63, 73, 83, 93 and
fragment
comprising at least 500 consecutive amino acids of the polypeptides of SEQ ID
NOS: 1,
13, 23, 33, 43, 53, 63, 73, 83, 93. Of course preferred context specific rules
may
specify a wide variety of thresholds for identifying enediyne polyketide
synthase
proteins without departing from the scope of the invention. Some context
specific rules
set level of homology required of the query sequence at 70%, 80%, 85%, 90%,
95% or
98% in regards to the reference sequences.
Thus, the analysis subprocess may be employed in conjunction with any other
context specific rules and may be adapted to suit different embodiments. The
principal
function of the analyzer algorithm 244 is to assign meaning or a diagnosis to
a query or
set of queries based on context specific rules that are application specific
and may be
changed without altering the overall role of the analyzer algorithm 244.
Finally the sequence comparison software of Figure 2 comprises a means of
returning of the results of the comparisons by the comparator algorithm 238
and
analyzed by the analyzer algorithm 244 to the user or process that requested
the
comparison or comparisons. The "display / report subprocess" of Figure 2D is
the
process by which the results of the comparisons by the comparator algorithm
238 and
analyses by the analyzer algorithm 244 are returned to the user or process
that
requested the comparison or comparisons. The results 240, 246 may be written
to a file
252, displayed in some user interface such as a console, custom graphical
interface,
web interface, or other suitable implementation specific interface, or
uploaded to some
database such as a relational database, or other suitable implementation
specific
database.
Once the results have been returned to the user or process that requested the
comparison or comparisons the program exits.
The principle of the sequence comparison software of Figure 2 is to receive or
load a query or queries, receive or load a reference dataset, then run a
pairwise
comparison by means of the comparator algorithm 238, then evaluate the results
using
an analyzer algorithm 244 to arrive at a determination if the query or queries
bear
significant similarity to the reference sequences, and finally return the
results to the
user or calling program or process.
CA 02444812 2003-10-28
3011-13CA
-59-
Figure 3 is a flow diagram illustrating one embodiment of comparator algorithm
238 process in a computer for determining whether two sequences are
homologous.
The comparator algorithm receives a query/subject pair for comparison,
performs an
appropriate comparison, and returns the pair along with a calculated degree of
similarity.
Referring to Figure 3, the comparison is initiated at the beginning of
sequences
304. A match of (x) characters is attempted 306 where (x) is a user specified
number.
If a match is not found the query sequence is advanced 316 by one polypeptide
with
respect to the subject, and if the end of the query has not been reached 318
another
match of (x) characters is attempted 306. Thus if no match has been found the
query is
incrementally advanced in entirety past the initial position of the subject,
once the end
of the query is reached 318, the subject pointer is advanced by 1 polypeptide
and the
query pointer is set to the beginning of the query 318. If the end of the
subject has
been reached and still no matches have been found a null homology result score
is
assigned 324 and the algorithm returns the pair of sequences along with a null
score to
the calling process or program. The algorithm then exits 326. If instead a
match is
found 308, an extension of the matched region is attempted 310 and the match
is
analyzed statistically 312. The extension may be unidirectional or
bidirectional. The
algorithm continues in a loop extending the matched region and computing the
2o homology score, giving penalties for mismatches taking into consideration
that given
the chemical properties of the polypeptide side chains not all mismatches are
equal.
For example a mismatch of a lysine with an arginine both of which have basic
side
chains receive a lesser penalty than a mismatch between lysine and glutamate
which
has an acidic side chain. The extension loop stops once the accumulated
penalty
exceeds some user specified value, or of the end of either sequence is reached
312.
The maximal score is stored 314, and the query sequence is advanced 316 by one
polypeptide with respect to the subject, and if the end of the query has not
been
reached 318 another match of (x) characters is attempted 306. The process
continues
until the entire length of the subject has been evaluated for matches to the
entire length
30 of the query. All individual scores and alignments are stored 314 by the
algorithm and
an overall score is computed 324 and stored. The algorithm returns the pair of
sequences along with local and global scores to the calling process or
program. The
algorithm then exits 326.
CA 02444812 2003-10-28
3011-13CA
-60-
Comparator algorithm 238 algorithm may be represented in pseudocode as
follows:
INPUT: Q[m]: query, m is the length
S[n]: subject, n is the length
x: x is the size of a segment
START:
for each i in [1,n] do
for each j in [1,m] do
if ( j + x - 1 ) <= m and ( i + x -1 ) <= n then
if Q(j, j+x-1) - S(i, i+x-1) then
k=1;
while Q(j, j+x--1+k ) - S(i, i+x-1+ k) do
k++;
Store highest local homology
Compute overall homology score
Return local and overall homology scores
END.
The comparator algorithm 238 may be written for use on nucleotide sequences,
in which case the scoring scheme would be implemented so as to calculate
scores and
apply penalties based on the chemical nature of nucleotides. The comparator
algorithm 238 may also provide for the presence of gaps in the scoring method
for
nucleotide or polypeptide sequences.
BLAST is one implementation of the comparator algorithm 238. HMMER is
another implementation of the comparator algorithm 238 based on Markov model
analysis. in a HMMER implementation a query sequence would be compared to a
mathematical model representative of a subject sequence or sequences rather
than
using sequence homology.
3o Figure 4 is a flow diagram illustrating an analyzer algorithm 244 process
for
detecting the presence of an enediyne biosynthetic locus. The analyzer
algorithm of
Figure 4 may be used in the process by which the annotation of a subject is
assigned to
the query based on their similarity as determined by the comparator algorithm
238 and
according to context-specific rules coded into the program or dynamically
loaded at
runtime. Context sensitive rules are what determines if the annotation of the
subject
can be assigned to the query given the context of the comparison. Context
specific
rules set the thresholds for determining the level and quality of similarity
that would be
accepted in the process of evaluating matched pairs.
CA 02444812 2003-10-28
3011-13CA
-61 -
The analyzer algorithm 244 receives as its input an array of pairs that had
been
matched by the comparator algorithm 238. The array consists of at least a
query
identifier, a subject identifier and the associated value of the measure of
their similarity.
To determine if a group of query sequences includes sequences diagnostic of an
enediyne biosynthetic gene cluster, a reference or diagnostic array 406 is
generated by
accessing a data source and retrieving enediyne specific information 404
relating to
enediyne-specific nucleic acid codes and enediyne-specific polypeptide codes.
Diagnostic array 406 consists at least of subject identifiers and their
associated
annotation. Annotation may include reference to the five protein families
diagnostic of
enediyne biosynthetic genes clusters, i.e. PKSE, TEBC, UNBL, UNBV and UNBU.
Annotation may also include information regarding exclusive presence in loci
of a
specific structural class or may include previously computed matches to other
databases, for example databases of motifs.
Once the algorithm has successfully generated or received the two necessary
arrays 402, 406, and holds in memory any context specific rules, each matched
pair as
determined by the comparator algorithm 238 can be evaluated. The algorithm
will
perform an evaluation 408 of each matched pair and based on the context
specific
rules confirm or fail to confirm the match as valid 410. In cases of
successful
confirmation of the match 410 the annotation of the subject is assigned to the
query.
Results of each comparison are stored 412. The loop ends when the end of the
query /
subject array is reached. Once all query / subject pairs have been evaluated
against
enediyne-specific nucleic acid codes and enediyne-specific polypeptide codes,
a final
determination can be made if the query set of ORFs represents an enediyne
locus 416.
The algorithm then returns the overall diagnosis and an array of characterized
query / subject pairs along with supporting evidence to the calling program or
process
and then terminates 418.
The analyzer algorithm 244 may be configured to dynamically load different
diagnostic arrays and context specific rules. It may be used for example in
the
comparison of query/subject pairs with diagnostic subjects for other
biosynthetic
pathways, such as chromoprotein enediyne-specific nucleic acid codes or non-
chromoprotein enediyne-specific polypeptide codes, or other sets of annotated
subjects.
CA 02444812 2003-10-28
3011-13CA
-62-
The present invention will be further described with reference to the
following
examples; however, it is to be understood that the present invention is not
limited to
such examples.
EXAMPLES
Example 1: Identification and seguencing of the macromomycin (auromomycin)
biosynthetic locus
Macromomycin is a chromoprotein enediyne produced by Streptomyces
1o macromyceticus (NRRL B-5335). Macromomycin is believed to be a derivative
of a
larger chromoprotein enediyne compound referred to as auromomycin (Vandre and
Montgomery (1982) Biochemistry Vol 21 pp. 3343-3352; Yamashita et al. (1979)
J.
Antibiot. Vol. 32 pp. 330-339). Thus, throughout the specification, reference
to
macromomycin is intended to encompass the molecules referred to by some
authors as
auromomycin. Likewise, reference to the biosynthetic locus for macromomycin is
intended to encompass the biosynthetic locus that directs the synthesis of the
molecules some authors have referred to as macromomycin and auromomycin.
Streptomyces macromyceticus (NRRL B-5335) was obtained from the
Agricultural Research Service collection (National Center for Agricultural
Utilization
20 Research, 1815 N. University Street, Peoria, Illinois 61604) and cultured
using standard
microbiological techniques (Kieser et al., supra). The organism was propagated
on
oatmeal agar medium at 28 degrees Celsius for several days. For isolation of
high
molecular weight genomic DNA, cell mass from three freshly grown, near
confluent 100
mm petri dishes was used. The cell mass was collected by gentle scraping with
a
plastic spatula. Residual agar medium was removed by repeated washes with STE
buffer (75 mM NaCI; 20 mM Tris-HCI, pH 8.0; 25 mM EDTA). High molecular weight
DNA was isolated by established protocols (Kieser et al. supra) and its
integrity was
verified by field inversion gel electrophoresis (FIGE) using the preset
program number
6 of the FIGE MAPPERT"~ power supply (BIORAD). This high molecular weight
30 genomic DNA serves for the preparation of a small size fragment genomic
sampling
library (GSL), i.e., the small insert library, as well as a large size
fragment cluster
identification library (CIL), i.e., the large insert library. Both libraries
contained
CA 02444812 2003-10-28
3011-13CA
-63-
randomly generated S. macromyceticus genomic DNA fragments and, therefore, are
representative of the entire genome of this organism.
For the generation of the S. macromyceticus GSL library, genomic DNA was
randomly sheared by sonication. DNA fragments having a size range between 1.5
and
3 kb were fractionated on a agarose gel and isolated using standard molecular
biology
techniques (Sambrook et al., supra). The ends of the obtained DNA fragments
were
repaired using T4 DNA polymerase (Roche) as described by the supplier. This
enzyme
creates DNA fragments with blunt ends that can be subsequently cloned into an
appropriate vector. The repaired DNA fragments were subcloned into a
derivative of
1 o pBluescript SK+ vector (Stratagene) which does not allow transcription of
cloned DNA
fragments. This vector was selected as it contains a convenient polylinker
region
surrounded by sequences corresponding to universal sequencing primers such as
T3,
T7, SK, and KS (Stratagene). The unique EcoRV restriction site found in the
polylinker
region was used as it allows insertion of blunt-end DNA fragments. Ligation of
the
inserts, use of the ligation products to transform E. coli DH10B (Invitrogen)
host and
selection for recombinant clones were performed as previously described
(Sambrook et
al., supra). Plasmid DNA carrying the S. macromyceticus genomic DNA fragments
was
extracted by the alkaline lysis method (Sambrook et al., supra) and the insert
size of
1.5 to 3 kb was confirmed by electrophoresis on agarose gels. Using this
procedure, a
20 library of small size random genomic DNA fragments is generated that covers
the entire
genome of the studied microorganism. The number of individual clones that can
be
generated is infinite but only a small number is further analyzed to sample
the
microorganism's genome.
A CIL library was constructed from the S. macromyceticus high molecular weight
genomic DNA using the SuperCos-1 cosmid vector (StratageneT""). The cosmid
arms
were prepared as specified by the manufacturer. The high molecular weight DNA
was
subjected to partial digestion at 37 degrees Celsius with approximately one
unit of
Sau3Al restriction enzyme (New England Biolabs) per 100 micrograms of DNA in
the
buffer supplied by the manufacturer. This enzyme generates random fragments of
3o DNA ranging from the initial undigested size of the DNA to short fragments
of which the
length is dependent upon the frequency of the enzyme DNA recognition site in
the
genome and the extent of the DNA digestion. At various timepoints, aliquots of
the
digestion were transferred to new microfuge tubes and the enzyme was
inactivated by
CA 02444812 2003-10-28
3011-13CA
-64-
adding a final concentration of 10 mM EDTA and 0.1 % SDS. Aliquots judged by
FIGE
analysis to contain a significant fraction of DNA in the desired size range
(30-50kb)
were pooled, extracted with phenol/chloroform (1:1 vol:vol), and pelletted by
ethanol
precipitation.
The 5' ends of Sacf3Al DNA fragments were dephosphorylated using alkaline
phosphatase (Roche) according to the manufacturer's specifications at 37
degrees
Celcius for 30 min. The phosphatase was heat inactivated at 70 degrees Celcius
for 10
min and the DNA was extracted with phenol/chloroform (1:1 vol:vol), pelletted
by
ethanol precipitation, and resuspended in sterile water. The dephosphorylated
SatI3Al
1 o DNA fragments were then ligated overnight at room temperature to the
SuperCos-1
cosmid arms in a reaction containing approximately four-fold molar excess
SuperCos-1
cosmid arms.
The ligation products were packaged using Gigapack~ III XL packaging extracts
(StratageneT"") according to the manufacturer's specifications. The CIL
library
consisted of 864 isolated cosmid clones in E. coli DH10B (Invitrogen). These
clones
were picked and inoculated into nine 96-well microtiter plates containing LB
broth (per
liter of water: 10.0 g NaCI; 10.0 g tryptone; 5.0 g yeast extract) which were
grown
overnight and then adjusted to contain a final concentration of 25% glycerol.
These
microtiter plates were stored at -80 degrees Celcius and served as glycerol
stocks of
20 the CIL library. Duplicate microtiter plates were arrayed onto nylon
membranes as
follows. Cultures grown on microtiter plates were concentrated by pelleting
and
resuspending in a small volume of LB broth. A 3 X 3 96-pin-grid was spotted
onto
nylon membranes.
The membranes, representing the complete CIL library, were then layered onto
LB agar and incubated ovenight at 37 degrees Celcius to allow the colonies to
grow.
The membranes were layered onto filter paper pre-soaked with 0.5 N NaOH/1.5 M
NaCI for 10 min to denature the DNA and then neutralized by transferring onto
filter
paper pre-soaked with 0.5 M Tris (pH 8)/1.5 M NaCI for 10 min. Cell debris was
gently
scraped off with a plastic spatula and the DNA was crosslinked onto the
membranes by
30 UV irradiation using a GS GENE LINKERT"" UV Chamber (BIORAD). Considering
an
average size of 8 Mb for an actinomycete genome and an average size of 35 kb
of
genomic insert in the CIL library, this library represents roughly a 4-fold
coverage of the
microorganism's entire genome.
CA 02444812 2003-10-28
3011-13CA
-65-
The GSL library was analyzed by sequence determination of the cloned genomic
DNA inserts, The universal primers KS or T7, referred to as forward (F)
primers, were
used to initiate polymerization of labeled DNA. Extension of at least 700 by
from the
priming site can be routinely achieved using the TF, BDT v2.0 sequencing kit
as
specified by the supplier (Applied Biosystems). Sequence analysis of the small
genomic DNA fragments (Genomic Sequence Tags, GSTs) was performed using a
3700 ABI capillary electrophoresis DNA sequencer (Applied Biosystems). The
average
length of the DNA sequence reads was 700 bp. Further analysis of the obtained
GSTs was performed by sequence homology comparison to various protein sequence
databases. The DNA sequences of the obtained GSTs were translated into amino
acid
sequences and compared to the National Center for Biotechnology Information
(NCBI)
nonredundant protein database and the proprietary Ecopia natural product
biosynthetic
gene DecipherT"~ database using previously described algorithms (Altschul et
al.,
supra). Sequence similarity with known proteins of defined function in the
database
enables one to make predictions on the function of the partial protein that is
encoded
by the translated GST.
A total of 479 S. macromyceticus GSTs obtained with the forward sequencing
primer were analyzed by sequence comparison using the Blast algorithm
(Altschul et
al., supra). Sequence alignments displaying an E value of at least e-5 were
considered
2o as significantly homologous and retained for further evaluation. GSTs
showing
similarity to a gene of interest can be at this point selected and used to
identify larger
segments of genomic DNA from the CIL library that include the genes) of
interest.
Several S. macromyceticus GSTs that contained genes of interest were pursued.
One
of these GSTs encoded a portion of an oxidoreductase based on Blast analysis
of the
forward read and a portion of the macromomycin apoprotein based on Blast
analysis of
the reverse read. Oligonucleotide probes derived from such GSTs were used to
screen
the CIL library and the resulting positive cosmid clones were sequenced.
Overlapping
cosmid clones provided in excess of 125 kb of sequence information surrounding
the
macromomycin apoprotein gene (Figure 5).
30 Hybridization oligonucleotide probes were radiolabeled with P32 using T4
polynucleotide kinase (New England Biolabs) in 15 microliter reactions
containing 5
picomoles of oligonucleotide and 6.6 picomoles of [~y-P32]ATP in the kinase
reaction
buffer supplied by the manufacturer. After 1 hour at 37 degrees Celcius, the
kinase
CA 02444812 2003-10-28
3011-13CA
-66-
reaction was terminated by the addition of EDTA to a final concentration of 5
mM. The
specific activity of the radiolabeled oligonucleotide probes was estimated
using a Model
3 Geiger counter (Ludlum Measurements Inc., Sweetwater, Texas) with a built-in
integrator feature. The radiolabeled oligonucleotide probes were heat-
denatured by
incubation at 85 degrees Celcius for 10 minutes and quick-cooled in an ice
bath
immediately prior to use.
The S. macromyceticus CIL library membranes were pretreated by incubation for
at least 2 hours at 42 degrees Celcius in Prehyb Solution (6X SSC; 20mM
NaH2P04;
5X Denhardt's; 0.4% SDS; 0.1 mg/ml sonicated, denatured salmon sperm DNA)
using
a hybridization oven with gentle rotation. The membranes were then placed in
Hyb
Solution (6X SSC; 20mM NaH2P04; 0.4% SDS; 0. i mg/ml sonicated, denatured
salmon sperm DNA) containing 1 X106 cpm/ml of radiolabeled oligonucleotide
probe
and incubated overnight at 42 degrees Celcius using a hybridization oven with
gentle
rotation. The next day, the membranes were washed with Wash Buffer (6X SSC,
0.1
SDS) for 45 minutes each at 46, 48, and 50 degrees Celcius using a
hybridization oven
with gentle rotation. The S. macromyceticus CIL membranes were then exposed to
X-
ray film to visualize and identify the positive cosmid clones. Positive clones
were
identified, cosmid DNA was extracted from 30 ml cultures using the alkaline
lysis
method (Sambraok et ai., supra) and the inserts were entirely sequenced using
a
shotgun sequencing approach (Fleischmann et al., (1995) Science, 269:496-512).
Sequencing reads were assembled using the Phred-PhrapT"" algorithm
(University of Washington, Seattle, USA) recreating the entire DNA sequence of
the
cosmid insert. Reiterations of hybridizations of the CIL library with probes
derived from
the ends of the original cosmid allow indefinite extension of sequence
information on
both sides of the original cosmid sequence until the complete sought-after
gene cluster
is obtained. The structure of macromomycin (auromomycin) has not been
elucidated,
however the apoprotein component has been well characterized (Van Roey and
Beerman (1989) Proc Natl Acad Sci USA Vol. 86 pp. 6587-6591 ). An unusual
polyketide synthase (PKSE) was found approximately 40 kb upstream of the
3o macromomycin apoprotein gene (Figure 5). No other polyketide synthase or
fatty acid
synthase gene cluster was found in the vicinity of the macromomycin apoprotein
gene,
suggesting that the PKSE may be the only polyketide synthase involved in the
biosynthesis of macromomycin (auromomycin).
CA 02444812 2003-10-28
3011-13CA
-67-
Four other enediyne-specific genes clustered with or in close proximity to the
PKSE gene were found in the macromomycin biosynthetic locus. These genes and
the
polypeptides that they encode have been assigned the family designations TEBC,
UNBL, UNBV, and UNBU. The macromomycin locus contains two copies of the TEBC
gene (Figure 6, Table 2). Table 2 lists the results of sequence comparison
using the
Blast algorithm (Altschul et al., supra) for each of these enediyne-specific
polypeptides
from the macromomycin locus. Homology was determined using the BLASTP
algorithm with the default parameters.
Table 2
MACR
locus
Family#aa GenBank probabilityidentity similarity proposed function
homology of GenBank
Accession, match
#aa
PKSE1936T37056,2082aa6e-86 273/897 3721897 multi-domain beta
(30.43.6) (41.47%) keto-acyl
synthase, Streptomyces
coelicolor
NP_485686.1,1263aa5e-82 256/900 388/900 heterocyst glycolipid
(28.44%j (43.11%) synthase,
Nostoc sp.
AAL01060.1,2573aa6e-78 244/884 376/884 polyunsaturated
(27.6%) (42.53%) fatty acid
synthase, Photobacterium
profundum
TEBC1162 NP 249659.1,148aa4e-06 38/134 59/134 (44.03%)hypothetical
protein,
(28.36%)
Pseudomonas aeruginosa
CAB50777.1,150aa4e-06 39/145 65/145 (44.83%)hypothetical protein,
(26.9%)
Pseudomonas putida
NP_214031.1,128aa2e-04 33/129 55/129 (42.64%)hypothetical protein,
(25.58%) Aquifex
aeolicus
TEBC2157 NP 242865.1,138aa0.27 31/131 50!131 37%
(23%) ( ) 4-hydroxybenzoyl-CoA
thioesterase,
Bacillus hatodurans
UNBL327 NP 422192.1,423aa0.095 30/86 (34.88%)40/86 (46.51%)peptidase,
Caulobacter
crescentus
UNBV642 NO HOMOLOG
UNBU433 NP_486037.1,300aa1e-06 49/179 83/179 (46.37%)hypothetical protein,
(27.37%) Nostoc sp.
NP_107088.1,503aa2e-04 72/280 126/280 hypothetical protein,
(25.71%) (45%)
Mesorhizobium
loti
NP_440874.1,285aa4e-04 47!193 86/193 (44.56%)hypothetical protein,
(24.35%)
Synechocystis
sp.
The macromomycin genes listed in Table 2 are arranged as depicted in Figure 6.
The UNBL, UNBV, UNBU, PKSE, and TEBC1 genes span approximately 10.5 kb and
are tandemly arranged in the order listed. Thus these five genes may
constitute an
operon. A second TEBC gene (TEBC2) is found approximately 6.6 kb downstream of
the 5-gene enediyne-specific cassette. The macromomycin enediyne-specific
cassette
is composed of six functionally linked genes and polypeptides, five of which
may be
expressed as a single operon.
CA 02444812 2003-10-28
3011-13CA
-68-
Example 2: Identification and seguencing of the calicheamicin biosynthetic
locus
Calicheamicin is a non-chromoprotein enediyne produced by Micromonospora
echinospora subsp. calichensis NRRL 15839. Both GSL and CIL genomic DNA
libraries of M. echinospora genomic DNA were prepared as described in Example
1. A
total of 288 GSL clones were sequenced with the forward primer and analyzed by
sequence comparison using the Blast algorithm (Altschui et al., supra) to
identify those
clones that contained inserts related to the macromomycin (auromomycin)
biosynthetic
genes, particularly the PKSE. Such GST clones were identified and were used to
1 o isolate cosmid clones from the M. echinospora CIL library. Overlapping
cosmid clones
were sequenced and assembled as described in Example 1. The resulting DNA
sequence information was more than 125 kb in length and included the
calicheamicin
genes described in WO 00/37608. The calicheamicin biosynthetic genes disclosed
in
WO 00/37608 span only from 37140 by to 59774 by in Figure 5 and do not include
the
unusual PKS gene (PKSE) and four other flanking genes (UNBL, UNBV, UNBU, and
TEBC) that are homologuous to those in the macromomycin biosynthetic locus.
Table
3 lists the results of sequence comparison using the Blast algorithm (Altschul
et al.,
supra) for each of these enediyne-specific polypeptides from the calicheamicin
locus.
Homology was determined using the BLASTP algorithm with the default
parameters.
20 Table 3
CAL/
locus
Family#aaGenBank homologyprobabilityIdentity similarityproposed function
Accession, of GenBank
#aa match
PKSE1919AAF26923.1,2439aa1e-60 228/876 317/876 polyketide synthase,
(26.03%) (36.19%) Polyangium
cellulosum
NP 485686.1,1263aa5e-59 148/461 210/461 heterocyst glycolipid
(32.1 (45.55%) synthase,
%) Nostoc sp.
T37056,2082aa9e-58 161/466 213/466 multi-domain beta
(34.55%) (45.71%) keto-acyl
synthase, Streptomyces
coelicolor
TEBC148NP 249659.1,148aa8e-06 41/133 62/133 hypothetical protein,
(30.83%) (46.62%) Pseudomonas aeruginosa
AAD49752.1,148aa1e-05 41/138 63/138 orfl, Pseudomonas
(29.71%) (45.65%) aeruginosa
NP 242865.1,138aa2e-04 32/130 56/130 4-hydroxybenzoyl-CoA
(24.62%) (43.08%) thioesterase, Bacillus
~ halodurans
UNBL322NO HOMOLOG
UNBV651NO HOMOLOG
UNBU321NP 486037.1,300aa8e-09 61/210 99/210 hypothetical protein,
(29.05%) (47.14%) Nostoc sp.
NP_107088.1,503aa5e-05 58/208 96/208 hypothetical protein,
(27.88%) (46.15%) Mesorhizobium loll
CA 02444812 2003-10-28
3011-13CA
-69-
The calicheamicin genes listed in Table 3 are arranged as depicted in Figure
6.
The UNBL, UNBV, UNBU, PKSE, and TEBC genes span approximately 10.5 kb and
are tandemly arranged in the order listed. Thus these five genes may
constitute an
operon. Therefore, the calicheamicin enediyne-specific cassette is composed of
five
functionally linked genes and polypeptides that may be expressed as a single
operon.
Example 3: Identification and seauencing of the biosynthetic locus for an
unknown
chromo-protein enediyne in Stre~tomyces Qhanaensis
The genomic sampling method described in Example 1 was applied to genomic
DNA from Streptomyces ghanaensis NRRL B-12104. S. ghanaensis has not
previously
been described to produce enediyne compounds. Both GSL and CIL genomic DNA
libraries of S. ghanaensis genomic DNA were prepared as described in Example
1. A
total of 435 GSL clones were sequenced with the forward primer and analyzed by
sequence comparison using the Blast algorithm (Altschul et al., supra).
Surprisingly, two GSTs from S. ghanaensis were identified as encoding portions
of genes in the 5-gene cassette common to both the macromomycin and
calicheamicin
enediyne biosynthetic loci. One of these GSTs encoded a portion of a TEBC
homologue and the other encoded a portion of a UNBV homologue. These S.
ghanaensis GSTs were subsequently found in a genetic locus referred to herein
as
009C (Figure 5). As in the macromomycin and calicheamicin enediyne
biosynthetic
loci, the UNBV and TEBC genes in 009C were found to flank a PKSE gene and
adjacent to UNBL and UNBU genes. The 009C locus included a gene encoding a
homologue of the macromomycin apoprotein approximately 50 kb downstream of the
UNBV-UNBU-UNBL-PKSE-TEBC cassette. The presence of the 5-gene cassette in
the vicinity of an apoprotein suggests that 009C represents a biosynthetic
locus for an
unknown chromoprotein enediyne that was not previously described to be
produced by
S. ghanaensis NRRL B-12104.
Table 4 lists the results of sequence comparison using the Blast algorithm
(Altschul et al., supra) for each of these enediyne-specific poiypeptides from
the 009C
locus. Homology was determined using the BLASTP algorithm with the default
parameters.
CA 02444812 2003-10-28
3011-13CA
-70-
Table 4
009C
locus
Family#aaGenBank homologyprobabilityidentity similarityproposed function
of GenBank
Accession, match
#aa
PKSE1956T37056,2082aa1 e-101298/902 395/902 multi-domain beta
(33.04%) (43.79%) keto-acyl
synthase, Streptomyces
coelicolor
NP 485686.1,1263aa2e-99 274/900 407/900 heterocyst glycolipid
(30.44%j (45.22%) synthase,
N ostoc sp.
BAB69208.1,2365aa3e-89 282/880 366/880 polyketide synthase,
(32.05%) (41.59%) Streptomyces
avermitilis
TEBC152NP_249659.1,148aa5e-07 39/131 59/131 hypothetical protein,
(29.77%) (45.04%)
Pseudomonas aeruginosa
NP 231474.1,155aa2e-04 30/129 62/129 hypothetical protein,
(23.26%) (48.06%) Vibrio
cholerae
NP 214031.1,128aa2e-04 31/128 55/128 hypothetical protein,
(24.22%) (42.97%) Aquifex
aeolicus
UNBL329NO HOMOLOG
UNBV636NP 615809.1,2275aa6e-05 72/314 114/314 cell surface protein,
(22.93%) (36.31%)
M ethanosarcina acetivorans
UNBU382NP 486037.1,300aa4e-07 46/175 81/175 hypothetical protein,
(26.29%) (46.29%) Nostoc sp.
NP_107088.1,503aa6e-06 68/255 118/255 hypothetical protein,
(26.67%) (46.271)
Mesorhizobium loti
The 009C genes listed in Table 4 are arranged as depicted in Figure 6. The
UNBL, UNBV, UNBU, PKSE, and TEBC genes span approximately 10.5 kb and are
tandemly arranged in the order listed. These five genes may constitute an
operan.
Therefore, the 009C enediyne-specific cassette is composed of five
functionally linked
genes and polypeptides that may be expressed as a single operon.
Example 4: The 5-q_ene enediyne cassette is present in the neocarzinostatin
biosynthetic locus
Neocarzinostatin is a chromoprotein enediyne produced by Streptomyces
carzinostaticus subsp. neocarzinostaticus ATCC 15944. The neocarzinostatin
biosynthetic locus was sequenced and was shown to contain, in addition to the
neocarzinostatin apoprotein gene, the 5-gene cassette that is present in the
macromomycin and calicheamicin enediyne biosynthetic loci. The genes and
proteins
involved in the biosynthesis of neocarzinostatin are disclosed in co-pending
application
USSN 60/354,474. The presence of the 5-gene cassette in the neocarzinostatin
biosynthetic locus reconfirms that it is present in all enediyne biosynthetic
loci.
Table 5 lists the results of sequence comparison using the Blast algorithm
(Altschul et al., supra) for each of these enediyne-specific polypeptides from
the
CA 02444812 2003-10-28
3011-13CA
-71 -
neocarzinostatin locus. Homology was determined using the BLASTP algorithm
with
the default parameters.
Table 5
NEOC
locus
Family#aa GenBank probabilityidentity similarity proposed function
homology of GenBank
Accession, match
#aa
PKSE1977T37056,2082aa7e-93 285/891 384/891 mufti-domain beta
(31.99io) (43.1 %) keto-acyl
synthase, Streptomyces
coelicolor
NP_485686.1,1263aa8e-88 261!890 397/890 heterocyst glycolipid
(29.33%) (44.61 synthase,
%)
Nostoc sp.
BAB69208.1,2365aa2e-85 276/876 370/876 polyketide synthase,
(31.51o) (42.24%) Streptomyces
avermitilis
TEBC153 NP 249659.1,148aa3e-06 37/129 56/129 (43.41%)hypothetical protein,
(28.68%)
Pseudomonas aeruginosa
CAB50777.1,150aa1 e-04 32/114 53/114 (46.49%)hypothetical protein,
(28.07%)
Pseudomonas putida
NP_214031.1,i28aa2e-04 34/129 55/129 (42.64%)hypothetical protein,
(26.36ro) Aquifex
aeolicus
UNBL328
UNBV636 NP 618575.1,1881aa2e-05 77/317 f 17/317 cell surface protein,
(24.290) (36.91%)
Methanosarcina
acetivorans
UNBU364 NP_107088.1,503aa2e-05 49!158 79/158 (50%)hypothetical protein,
(31.01%)
Mesorhizobium
loti
NP 486037.1,300aa8e-05 33/126 60/126 (47.62%)hypothetical protein,
(26.19r) Nostoc sp.
The neocarzinostatin genes listed in Table 5 are arranged as depicted in
Figure
6. The UNBL, UNBV, UNBU, PKSE, and TEBC genes span approximately 10.5 kb and
are tandemly arranged in the order listed. Thus these five genes may
constitute an
operon. Therefore, the neocarzinostatin enediyne-specific cassette is composed
of five
functionally linked genes and polypeptides that may be expressed as a single
operon.
Example 5: The 5-Gene enediyne cassette is present in the bios~rnthetic locus
of an
unknown chromoprotein enediyne in Amycolafopsis orientalis
The genomic sampling method described in Example 1 was applied to genomic
DNA from Amycolatopsis orientalis ATCC 43491. A. orientalis has not previously
been
described to produce enediyne compounds. Both GSL and CIL genomic DNA
libraries
of A, orientalis genomic DNA were prepared as described in Example 1.
A total of 1025 GSL clones were sequenced with the forward primer and
analyzed by sequence comparison using the Blast algorithm (Altschul et al.,
supra).
Several secondary metabolism loci were identified and sequenced as described
in
Example 1. One of these loci (herein referred to as 007A) includes a 5-gene
cassette
CA 02444812 2003-10-28
3011-13CA
-72-
common to all enediyne biosynthetic loci. The arrangement of the five genes of
the
cassette in 007A is shown in Figure 6. Interestingly, the A. orientalis genome
also
contains an enediyne apoprotein gene that is similar to that from the
macromomycin
and 009C loci as well as other chromoprotein enediynes (data not shown).
Therefore,
A. orientalis, the producer of the well-known glycopeptide antibiotic
vancomycin, has
the genomic potential to produce a chromoprotein enediyne.
Table 6 lists the results of sequence comparison using the Blast algorithm
(Altschul et al., supra) for each of the enediyne-specific polypeptides from
the 007A
locus. Homology was determined using the BLASTP algorithm with the default
1 o parameters.
Table 6
007A
locus
Family#aaGenBank homologyprobabilityidentity similarityproposed function
of GenBank
Accession, match
#aa
PKSE 1939T37056,2082aa5e-96 291/906 399/906 multi-domain beta
(32.12%) (44.04%) keto-acyl
~ synthase, Streptomyces
coelicolor
NP 485686.1,1263aa9e-87 255/897 395/897 eterocyst glycolipid
(28.43%) (44.04%) synthase,
h
Nostoc sp.
BAB69208.1,2365aaSe-86 285/926 393/926 modular polyketide
(30.78%) (42.44%) synthase,
Streptomyces avermitilis
TEBC 146NP_ 214031.1,128aa0.052 28/124 511124 hypothetical protein,
(22.58%) (41.13%) Aquifex
aeolicus
UNBL 324NO HOMOLOG
UNBV 654NP 618575.1,1881aa0.001 80/332 1171332 cell surface protein,
(24.1%) (35.24%j
Methanosarcina
acetivorans
UNBU 329NP 486037.1,300aa0.005 56/245 96/245 hypothetical protein,
I I I , (22.86%) (39.18%) Nostoc
I ( p.
s
The 007A genes listed in Table 6 are arranged as depicted in Figure 6. The
UNBL, UNBV, and UNBU genes span approximately 4 kb and are tandemly arranged
in the order listed. The PKSE and TEBC genes span approximately 6.5 kb and are
tandemly arranged in the order listed. Thus these five genes may constitute
two
operons. The two putative operons are separated by approximately 5 kb.
Although
these two clusters of genes may not be transcriptionally linked to one
another, they are
still functionally linked. Therefore, the 007A enediyne-specific cassette is
composed of
20 five functionally linked genes and polypeptides, three of which may be
expressed as a
one operon and two of which may be expressed as a second operon.
Example 6: The 5-gene enediyne cassette is present in the biosynthetic locus
of an
unknown enediyne in Kitasatosporia sp. CECT 4991
CA 02444812 2003-10-28
3011-13CA
-73-
The genomic sampling method described in Example 1 was applied to genomic
DNA from Kitasatosporia sp. CECT 4991. This organism was not previously
described
to produce enediyne compounds. Both GSL and CIL genomic DNA libraries of
Kitasatosporia sp. genomic DNA were prepared as described in Example 1.
A total of 1390 GSL clones were sequenced with the forward primer and
analyzed by sequence comparison using the Blast algorithm (Altschul et al.,
supra).
Surprisingly, two GSTs from Kitasatosporia sp.were identified as encoding
portions of
genes in the 5-gene cassette common to enediyne biosynthetic loci. One of
these
GSTs encoded a portion of a PKSE homologue and the other encoded a portion of
a
UNBV homologue. These Kitasatosporia sp. GSTs were subsequently found in a
genetic locus referred to herein as 028D which includes a 5-gene cassette
common to
all enediyne biosynthetic loci. The arrangement of the five genes of the
cassette in
028D is shown in Figure 6. Therefore, Kitasatosporia sp. CECT 4991 has the
genomic
potential to produce enediyne compound(s).
Table 7 lists the results of sequence comparison using the Blast algorithm
(Altschul et al., supra) for each of the enediyne-specific polypeptides from
the 028D
locus. Homology was determined using the BLASTP algorithm with the default
parameters.
Table 7
028D
locus
Family#aa GenBank homologyprobabilityidentity similarity proposed function
Accession, of GenBank
#aa match
PKSE1958BAB69208.1,2365aa1e-81 273/926 354/926 polyketide synthase,
(29.48%) (38.23%) Streptomyces
avermitilis
T37056,2082aa3e-78 263!895 356/895 multi-domain beta
(29.39%) (39.78%) keto-acyl
synthase, Streptomyces
coelicolor
NP 485686.1,1263aa7e-71 231/875 345!875 heterocyst glycolipid
(26.4i) (39.43%) synthase,
Nostoc sp.
TEBC158 NP 249659.1,148aa1e-04 38/133 61/133 (45.86%)hypothetical protein,
(28.57%) Pseudomonas aeruginosa
AAD49752.1,148aa3e-04 38/138 62/7 38 orfi , Pseudomonas
(27.54%) (44.93%) aeruginosa
NP 231474.1,155aa7e-04 31!127 61/127 (48.03%)hypothetical protein,
(24.41%) Vibrio
cholerae
UNBL327 NO HOMOLOG
UNBV676 NO HOMOLOG
UNBU338 NP_486037.1,300aa5e-OS 66/240 105/240 hypothetical protein,
(27.5i) (43.75%) Nostoc sp.
NP__440874.1,285aa2e-04 51/190 98/190 (51.58%)hypothetical protein,
(26.84%) Synechocystis
sp.
The 028D genes listed in Table 7 are arranged as depicted in Figure 6. The
UNBV, UNBU, PKSE, and TEBC genes span approximately 9.5 kb and are tandemly
CA 02444812 2003-10-28
3011-13CA
-74-
arranged in the order listed. Thus these four genes may constitute an operon.
This
putative operon is separated from the UNBL gene, which is oriented in the
opposite
direction relative to the putative operon, by approximately 10.5 kb. Although
the UNBL
gene cannot be transcriptionally linked to the other genes, it is still
functionally linked to
the former. Therefore, the 028D enediyne-specific cassette is composed of five
functionally linked genes and polypeptides, four of which may be expressed as
a single
operon. Although expression of functionally linked enediyne-specific genes may
be
under control of distinct transcriptional promoters they may, nonetheless, be
expressed
in a concerted fashion. As depicted in Figure 6, the 028D biosynthetic locus
is unique
1 o in that it is the only example whose enediyne-specific genes are not all
oriented in the
same direction.
Example 7: The 5-giene enediyne cassette is present in the biosynthetic locus
of an
unknown enedi ny a in Micromonospara megalomicea
The genomic sampling method described in Example 1 was applied to genomic
DNA from Micromonospora megalomicea NRRL 3275. This organism was not
previously described to produce enediyne compounds. Both GSL and CIL genomic
DNA libraries of M. megalomicea genomic DNA were prepared as described in
Example 1.
20 A total of 1390 GSL clones were sequenced with the forward primer and
analyzed by sequence comparison using the Blast algorithm (Altschul et al.,
supra).
Surprisingly, one GST from M. megalomicea was identified as encoding a portion
of the
PKSE gene present in the 5-gene cassette common to biosynthetic loci. The
forward
read of this GST encoded the C-terminal portion of the KS domain and the N-
terminal
portion of the AT domain of a PKSE gene. The complement of the reverse read of
this
GST encoded the C-terminal portion of the AT domain of a PKSE gene. This M.
megalomicea GST was subsequently found in a genetic locus referred to herein
as
054A which includes a 5-gene cassette common to all enediyne biosynthetic
loci. The
arrangement of the five genes of the cassette in 054A is shown in Figure 6.
Therefore,
3o M. megalomicea has the genomic potential to produce enediyne compound(s).
Table 8 lists the results of sequence comparison using the Blast algorithm
(Altschul et al., supra) for each of the enediyne-specific polypeptides from
the 054A
CA 02444812 2003-10-28
3011-13CA
-75-
locus. Homology was determined using the BLASTP algorithm with the default
parameters.
Table 8
054A
locus
Family#aaGenBank homologyprobabilityidentity similarityproposed function
Accession, of GenBank
#aa match
PKSE 1927NP_485686.1,1263aa3e-76 247/886 365/886 heterocyst glycolipid
(27.88%) (41.2%) synthase,
Nostoc sp.
T37056,2082aa3e-75 269/903 354/903 multi-domain beta
(29.790) (39.2%) keto-acyl
synthase, Streptomyces
coelicolor
BAB69208.1,2365aa9e-74 277/923 359/923 polyketide synthase,
(30.01%) (38,89%) Streptomyces
avermitilis
TEBC 154NP 249659.1,148aa2e-O6 43/147 66/147 hypothetical protein,
(29.25,%) (44.9%) Pseudomonas aeruginosa
AAD49752.1,148aa2e-05 42/t47 65/147 orfi, Pseudomonas
(28.57%) (44.22%) aeruginosa
CAB50777.1,150aa1 e-04 40/139 61 /139 hypothetical protein,
(28.78%) (43.88%) Pseudomonas putida
UNBL 322NO HOMOLOG
UNBV 659CAC44518.1,706aa0.048 50/166 67/166 putative secreted
(30.12%) (40.36%) esterase,
Streptomyces coelicolor
UNBU 354NP_486037.1,300aa5e-06 66/268 118!268 hypothetical protein,
(24.63%) (44.03%) Nostoc sp.
The 054A genes listed in Table 8 are arranged as depicted in Figure 6. The
UNBL, PKSE, and TEBC genes span approximately 7.5 kb and are tandemly arranged
in the order listed. The UNBV and UNBU genes span approximately 3 kb and are
tandemly arranged in the order listed. Thus these five genes may constitute
two
operons. The two putative operons are separated by approximately 2 kb.
Therefore,
the 054A enediyne-specific cassette is composed of five functionally linked
genes and
polypeptides, three of which may be expressed as a one operon and two of which
may
be expressed as another operon.
Example 8: The 5-gene enediyne cassette is present in the bios~rnthetic locus
of an
unknown enediyne in Saccharothrix aerocoloni
The genomic sampling method described in Example 1 was applied to genomic
DNA from Saccharothrix aerocolonigenes ATCC 39243. This organism was not
previously described to produce enediyne compounds. Both GSL and CIL genomic
DNA libraries of Saccharothrix aerocolonigenes genomic DNA were prepared as
described in Example 1.
A total of 513 GSL clones were sequenced with the forward primer and analyzed
by sequence comparison using the Blast algorithm (Altschul et al., supra).
Several
CA 02444812 2003-10-28
3011-13CA
-76-
secondary metabolism loci were identified and sequenced as described in
Example 1.
One of these loci (herein referred to as 132H) includes a 5-gene cassette
common to
all enediyne biosynthetic loci. The arrangement of the five genes of the
cassette in
132H is shown in Figure 6. Therefore, Saccharothrix aerocolonigenes has the
genomic
potential to produce enediyne compound(s).
Table 9 lists the results of sequence comparison using the Blast algorithm
(Altschul et al., supra) for each of these enediyne-specific polypeptides from
the 132H
locus. Homology was determined using the BLASTP algorithm with the default
parameters.
Table 9
132H
locus
Family
#aa
GenBank
homology
probability
identity
similarity
proposed
function
of
GenBank
Accession,
#aa
match
PKSE1892BAB69208.1,2365aa1e-108 312/872 404/872 polyketide synthase,
(35.78%) (46.33%) Streptomyces
avermitilis
T37056,2082aa1e-101 290!886 407/886 multi-domain beta
(32.73%) (45.94%) keto-acyl
synthase, Streptomyces
coelicolor
T30183,2756aa4e-94 271/886 398/886 hypothetical protein,
(30.59%) (44.92%) Shewanella
sp.
TEBC143 NP 442358.1,138aa0.001 32/127 48/127 hypothetical protein,
(25.2%) (37.8%)
Synechocystis sp.
UNBL313 NO HOMOLOG
UNBV647 AAD34550.1,1529aa0.012 76/304 1051304 esterase, Aspergillus
(25i) (34.54%) terreus
UNBU336 NP_486037.1,300aa1e-04 42!172 79/172 hypothetical protein,
(24.42%) (45.93%) Nostoc sp.
NP 440874.1,285aaie-04 48/181 90/181 hypothetical protein,
(26.52%) (49.72%)
Synechocystis sp.
The 132H genes listed in Table 9 are arranged as depicted in Figure 6. The
UNBL, UNBV, UNBU, PKSE, and TEBC genes span approximately 10.5 kb and are
tandemly arranged in the order fisted. Thus, these five genes may constitute
an
operon. Therefore, the 132H enediyne-specific cassette is composed of five
functionally linked genes and polypeptides that may be expressed as a single
operon.
Example 9: The 5-gene enediyne cassette is present in the biosynthetic locus
of an
unknown enediyne in Streptom~ces kaniharaensis
2o The genomic sampling method described in Example 1 was applied to genomic
DNA from Streptomyces kaniharaensis ATCC 21070. This organism was not
previously described to produce enediyne compounds. Both GSL and CIL genomic
CA 02444812 2003-10-28
3011-13CA
_77_
DNA libraries of S. kaniharaensis genomic DNA were prepared as described in
Example 1.
A total of 1020 GSL clones were sequenced with the forward primer and
analyzed by sequence comparison using the Blast algorithm (Altschul et al.,
supra).
Surprisingly, one GST from S. kaniharaensis was identified as encoding a
portion of the
PKSE gene present in the 5-gene cassette common to biosynthetic loci. The
forward
read of this GST encoded the N-terminal portion of the KS domain of a PKSE
gene.
The complement of the reverse read of this GST encoded the C-terminal portion
of the
AT domain of a PKSE gene. This S. kaniharaensis GST was subsequently found in
a
genetic locus referred to herein as 135E which includes a 5-gene cassette
common to
all enediyne biosynthetic loci. The arrangement of the five genes of the
cassette in
135E is shown in Figure 6. Therefore, S. kaniharaensis has the genomic
potential to
produce enediyne compound(s).
Table 10 lists the results of sequence comparison using the Blast algorithm
(Altschul et al., supra) for each of the enediyne-specific polypeptides from
the 135E
locus. Homology was determined using the BLASTP algorithm with the default
parameters.
Table 10
135E
locus
Family#aaGenBank homologyprobabilityidentity similarityproposed function
of GenBank
Accession, match
#aa
PKSE 1933T37056,2082aa1 e-85 282/909 365/909 multi-domain beta
(31.02%) (40.15%) keto-acyl
synthase, Streptomyces
coelicolor
BAB69208.1,2365aa3e-84 285/925 366/925 polyketide synthase,
(30.81 (39.57%) Streptomyces
%)
avermitilis
T30937,1053aa2e-69 246/907 356/907 glycolipid synthase,
(27.12%) (39.25%) Nostoc
punctiforme
TEBC 154NP_249659.1,148aa2e-07 41/132 63/132 hypothetical protein,
(31.06%) (47.73%)
Pseudomonas aeruginosa
AAD49752.1,148aa2e-06 40/132 62/132 orf1, Pseudomonas
(30.3%) (46.97%) aeruginosa
NP 214031.1,128aa5e-04 35/127 60/127 hypothetical protein,
(27.56%) (47.24%) Aquifex
aeolicus
UNBL 323NO HOMOLOG
UNBV 655CAC44518.i,706aa9e-04 41/135 59/135 putative secreted
(30.37%) (43.7%) esterase,
Streptomyces coelicolor
UNBU 346NP_486037.1,300aa4e-09 521191 87/191 hypothetical protein,
(27.23%) (45.55%) Nostoc sp.
NP 440874.1,285aa9e-06 47/197 89/197 hypothetical protein,
(23.86%) (45.18%)
Synechocystis sp.
2o The 135E genes listed in Table 10 are arranged as depicted in Figure 6. The
UNBL, UNBV, and UNBU genes span approximately 4 kb and are tandemly arranged
CA 02444812 2003-10-28
3011-13CA
-78-
in the order listed. The PKSE and TEBC genes span approximately 6.5 kb and are
tandemly arranged in the order listed. Thus these five genes may constitute
two
operons. The two putative operons are separated by approximately 6 kb.
Although
these two clusters of genes may not be transcriptionally linked to one
another, they are
still functionally linked. Therefore, the 135E enediyne-specific cassette is
composed of
five functionally linked genes and palypeptides, three of which may be
expressed as a
one operon and two of which may be expressed as another operon.
Example 10: The 5-Gene enediyne cassette is present in the biosynthetic locus
of an
unknown enedi~me in Streptomyces citricolor
The genomic sampling method described in Example 1 was applied to genomic
DNA from Streptomyces citricoior IFO 13005. This organism was not previously
described to produce enediyne compounds. Both GSL and CIL genomic DNA
libraries
of S. citricolor genomic DNA were prepared as described in Example 1.
A total of 1245 GSL clones were sequenced with the forward primer and
analyzed by sequence comparison using the Blast algorithm (Altschul et al.,
supra).
Several secondary metabolism loci were identified and sequenced as described
in
Example 1. One of these loci (herein referred to as 145B) includes a 5-gene
cassette
common to all enediyne biosynthetic loci. The arrangement of the five genes of
the
cassette in 145B is shown in Figure 6. Therefore, S. citricolor has the
genomic
potential to produce enediyne compound(s).
Table 11 lists the results of sequence comparison using the Blast algorithm
(Altschul et al., supra) for each of the enediyne-specific polypeptides from
the 145B
locus. Homology was determined using the BLASTP algorithm with the default
parameters.
CA 02444812 2003-10-28
3011-13CA
_79_
Table 11
1458
locus
Family#aaGenBank homologyprobabilityidentity similarityproposed function
of GenBank
Accession, match
#aa
PKSE 1958T37056,2082aa4e-88 285/929 378/929 multi-domain beta
(30.68%) (40.69%) keto-acyl
synthase, Streptomyces
coelicolor
BAB69208.1,2365aa3e-82 284/923 375/923 polyketide synthase,
(30.77%) (40.63%) Streptomyces
avermitilis
AAL01060.1,2573aa5e-78 240/855 354/855 polyunsaturated
(28.07%) (41.4%) fatty acid
synthase, Photobacterium
profundum
TEBC 165NP 249659.1,148aa2e-07 39/133 60/133 hypothetical protein,
(29.32%) (45.11%)
P seudomonas aeruginosa
NP__231474.1,155aa3e-04 30/127 60/127 hypothetical protein,
(23.62~) (47.24%) Vit>rio
cholerae
CAB50777.1,150aa4e-04 37/135 58/135 hypothetical protein,
(27.41%) (42.96%)
Pseudomonas putida
UNBL 324NO HOMOLOG
UNBV 659NP 618575.1,1881aa0.003 57/245 85/245 cell surface protein,
(23.27i) (34.69%)
M ethanosarcina acetivorans
UNBU 337NP 486037.1,300aa0.002 62/267 109/267 hypothetical protein,
(23.22%) (40.82%) Nostoc sp.
The 1458 genes listed in Table 11 are arranged as depicted in Figure 6. The
UNBV, and UNBU genes span approximately 3 kb and are tandemly arranged in the
order listed. The PKSE and TEBC genes span approximately 6.5 kb and are
tandemly
arranged in the order listed. Thus these four genes may constitute two
operons. The
two putative operons are separated by approximately 9.5 kb that includes the
UNBL
gene. Although these genes may not be transcriptionally linked to one another,
they
are still functionally linked. Therefore, the 1458 enediyne-specific cassette
is
composed of five functionally linked genes and polypeptides, four of which may
be
expressed as two operons each containing two genes.
Example 11: Analysis of the~olyheptides encoded b ty he 5-gene enediyne-
specific
cassette
The amino acid sequences of the PKSE, TEBC, UNBL, UNBV, and UNBU
protein families from the ten enediyne biosynthetic loci described above were
compared to one another by multiple sequence alignment using the Clustal
algorithm
(Thompson et al., 1994, Nucleic Acids Res. 22(2):4673-4680; Higgins et al.,
1996,
Methods Enzymol. 266:383-402; Higgins and Sharp (1988) Gene Vol. 73 pp.237-
244).
The alignments are shown in Figures 8, 11, 12, 13, and 14, respectively. Where
CA 02444812 2003-10-28
3011-13CA
-80-
applicable, conserved residues or motifs important for the function are
highlighted in
black and additional features are indicated.
The PKSE family is a family of polyketide synthases that are involved in
formation of enediyne warhead structures. Figure 7 summarizes schematically
the
domain organization of a typical PKSE, showing the position and relative size
of the
putative domains based on Markov modeling of PKS domains: ketosynthase (KS),
acyltransferase (AT), acyl carrier protein (ACP), ketoreductase (KR),
dehydratase (DH),
and 4'-phosphopantetheinyl transferase (PPTE) activities. Using the
calicheamicin
PKSE as an example, the full-length PKSE protein is 1919 amino acids in
length. As
indicated in Figure 8 for the calicheamicin PKSE, the KS domain spans
positions 3 to
467 of the PKSE; the AT domain spans positions 482 to 905 of the PKSE; the ACP
domain spans positions 939 to 1009 of the PKSE; a small domain of unknown
function
of approximately 130 amino acids (spanning positions 1025 to 1144 of the PKSE)
is
present between the ACP and the KR domains; the KR domain spans positions 1153
to
1414 of the PKSE; the DH domain spans positions 1421 to 1563 of the PKSE; a C-
terminal 4'-phosphopantetheinyl transferase (PPTE) domain spans positions 1708
to
1914 of the PKSE; a small domain of about 110 amino acids (spanning positions
1591
to 1701 of the PKSE) is present between the DH and the PPTE domains.
The PKSE contains a conserved unusual ACP domain (Figure 9A). This ACP
domain contains several conserved residues that are also present in the well-
characterized ACP of the actinorhodin type II PKS (PDBid:IAF8 in Figure 9B).
The
most important conserved resudue is the serine residue to which a 4'-
phosphopantetheine prosthetic group is covalently attached (corresponding to
Ser-42
of 1AF8). In addition to Ser-42, several surface-exposed charged residues are
conserved, namely Glu-20, Asp-37, and Glu-84 (highlighted in the alignment of
Figure
9A and highlighted and labeled in the three dimensional structure shown in
Figure 9B).
Several buried uncharged or non-polar residues that may be important in
stabilizing the
overall fold of the ACP domain are also conserved, namely Leu-14, Val-15, Gly-
57,
Pro-71, Ala-83, and Ala-85 (highlighted in the alignment and three dimensional
3o structure shown in Figure 9). Interestingly, the conserved serine (Ser-42)
is almost
always immediately preceeded by another serine in the ACP domains of PKSEs. As
shown in Figure 8, nine of the ten PKSE members contain this double serine
arrangement, the only exception being that from the 132H locus in which the
first of the
CA 02444812 2003-10-28
3011-13CA
_81 _
serine is replaced by a threonine. Therefore, PKSEs contain ACP domains with
two
potential hydroxyl-containing residues in close proximity to one another.
These ACPs
may carry two 4'-phosphopantetheine prosthetic groups. The positioning of the
KR and
DH domains after the ACP is unusual among PKSs, but is described in one of the
three
PKS-like components of the eicosapentaenoic acid (EPA) and docosahexaenoic
acid
(DHA) biosynthetic machinery (Metz et al. (2001 ) Science Vol. 293 pp. 290-
293). The
unusual domain organization shared by the PKSE genes of the invention and the
PKS-
like synthetase involved in synthesis of polyunsaturated fatty acids suggests
that
enediyne warhead formation involves intermediates similar to those generated
during
assembly of polyunsaturated fatty acids.
The presence of an unusual ACP domain in the PKSE, and the absence of any
obvious 4'-phosphopantetheinyl transferase or holo-ACP synthase (involved in
phosphopantetheinyl transfer onto the conserved serine of the ACP) common to
enediyne biosynthetic loci led us to search for the presence of a 4'-
phosphopantetheinyl transferase. We examined the conserved domains of the PKSE
whose functions were unaccounted for as well as the UNBL, UNBV, and UNBU
polypeptides in more detail and determined that the PPTE domain was a 4'-
phosphopantetheinyl transferase.
The C-terminal domains of the PKSEs from the biosynthetic loci of three known
enediynes, namely neocarzinostatin (NEOC, as 1620-1977), calicheamicin (CALI,
as
1562-1919) and macromomycin (MACR, as 1582-1936), were analyzed for their
folding
using secondary structure predictions and solvation potential information
(Kelley et al.
(2000) J. Mol. Biol. Vol. 299 pp. 499-520). Comparison searches using a
database of
known 3-D structures of proteins revealed similarities between the C-terminal
domains
of the PKSEs and Sfp, the 4'-phosphopantetheinyl transferase from the Bacillus
subtilis
surfactin biosynthetic locus (Reuter et al. (1999) EMBO Vol. 18 pp. 6823-6831
).. The
alignment shown in Figure 10A indicates the predicted secondary structures of
all three
C-terminal PKSE domains (PPTE domains) along with the X-ray crystallography-
determined secondary structure of Sfp (PDB id: 1 QRO). Alpha-helices are
indicated by
rectangles and (3-sheets by arrows.
An overall conservation of secondary structure over the entire length of the
proteins is evident. Ali major structural constituents of Sfp, namely a-
helices a1-a5 and
CA 02444812 2003-10-28
3011-13CA
-82-
~-sheets a2- X34 and ~8 are also present in PPTE domains. Similar to Sfp, the
PPTE
domains are predicted to have an intramolecular 2-fold pseudosymmetry.
The loop formed between a5 and ~7 in Sfp is not present in the PPTE domains.
It is believed that this region of Sfp is in part responsible for ACP
recognition and
contributes to the broad substrate specificity observed for this enzyme. The
size of this
loop appears to vary among phosphopantetheinyl transferases, as the EntD
enzyme,
which exhibits a greater ACP substrate specificity than Sfp, has a region
between a5
and ~7 structures shorter than that of Sfp but longer than that found in the
PPTE
domains. The short a5/~7 loop region found in the PPTE domains may reflect the
need
for a specific interaction with the rather unusual ACP domain found in the
PKSE
enzymes. Residues conserved in all phosphopantetheinyl transferases and shown
in
Sfp to make contacts with the CoA substrate and Mg++ cofactor are also
conserved in
the PPTE domains (highlighted in Figure 10A).
Referring to Figure 10B, Sfp residues Lys-28 and Lys-31 make salt bridges with
the 3'-phosphate of CoA and are not found in the PPTE domains; however, a
similar
interaction could be provided by the corresponding conserved residue Arg-26.
Sfp Thr-
44 makes a hydrogen bond and His-90 a salt bridge with the 3'-phosphate of
CoA;
similar hydrogen bonding potential is provided by the conserved serine found
at the
corresponding position 44 of the PPTE domains, while the histidine 90 residue
is
absolutely conserved in all three PPTE domains.
Sfp amino acid residues 73-76 hold in place the adenine base of CoA. The main
chain carbonyl of Tyr-73 forms a hydrogen bond with the adenine amino group
and
residues Gly-74, Lys-75 and Pro-76 hold firmly in place the adenine ring. In
the PPTE
domains, a conserved aspartic acid that may form a salt bridge with the
adenine amino
group is substituted for Tyr-73 and a conserved arginine residue is
substituted for Lys-
75. The remaining two residues, Gly-74 and Pro-76, are also found in the PPTE
domains.
Sfp residues Ser-89 and His-90 interact via hydrogen bonding and salt bridging
with the a-phosphate of the CoA substrate. Similarly, Lys-155 in helix a5
interacts with
the CoA a-phosphate. The His-90 and Lys-155 residues are highly conserved in
the
PPTE domains whereas Ser-89 is found only in the neocarzinostatin PPTE domain.
CA 02444812 2003-10-28
3011-13CA
-83-
Sfp residues Asp-107, Glu-109 in the a4 sheet and Glu-151 in the a5 helix
participate in the complexation of a metal ion (presumably Mg++) together with
the a
and (3 phosphates of the CoA pyrophosphate and a water molecule. All three
residues
are also conserved in PPTE domains. Importantly, Asp-107 was altered by
mutagenesis in Sfp and shown to be critical for catalytic activity but not for
CoA binding
of the protein suggesting the Mg++ ion is important for catalysis (Quadri et
al., 1998,
Biochemistry, Vol. 37, 1585-1595).
In the Sfp protein, residue Glu-127 salt-bridges the amino group of Lys-150.
In
the PPTE domains, a Glu/Asp residue is found at the corresponding position
127,
whereas Lys-150 is not conserved. Since Glu-127 is highly conserved in the
PPTE
domains, it is conceivable that the role of Lys-150 is served by other basic
residues in
the vicinity, namely the conserved arginine at the corresponding position 145.
Residue
Trp-147, conserved in all phosphopantetheinyl transferases and shown to be
critical for
catalytic activity, is also present in all three PPTE domains (Quadri et al.,
1998,
Biochemistry, Vol. 37, 1585-1595).
The presence of a phosphopantetheinyl domain (PPTE) in the C-terminal part of
the PKSE enediyne warhead PKS is reminiscent of the 4'-phosphopantetheinyl
domain
found in the yeast fatty acid synthase (FAS) complex, where it resides in the
C-terminal
region of the FAS a subunit. FAS is capable of auto-pantetheinylation
resulting in a
post-translational autoactivation of this enzyme (Fichtlscherer et al., 2000,
Eur. ,J.
Biochem., Vol. 267, 2666-2671 ). In a similar manner, the PKSE warhead PKSs
are
likely to be capable of auto-pantetheinylation and activation of their ACP
domains
before proceeding to the iterative synthesis of the polyunsaturated polyketide
intermediate forming the enediyne core.
The ACP and KR domains of the PKSEs are separated by approximately 130
amino acids. The presence of a considerable number of invariable residues
within this
stretch of amino acids suggests that the putative domain formed by these 130
amino
acids has a functional role. The putative domain may serve a structural role,
for
example as a protein-protein interaction domain or it may form a cleft
adjacent to the
ACP that acts as a "chain length factor" for the growing polyketide chain. A
search of
NCBI's Conserved Domain Database with Reverse Position Specific BLAST revealed
several short stretches of homology to proteins that bind substrates such as
ATP, AMP,
NAD(P), as well as folates and double stranded RNA (adenosine deaminase).
Thus,
CA 02444812 2003-10-28
3011-13CA
-84-
the putative domain may adopt a structure accommodating an adenosine or
adenosine-
like structure and serve as a cofactor-binding site. Alternatively, the domain
might
interact with the adenosine moiety of coenzyme A (CoA). As such, the physical
proximity of the CoA to the ACP domain may facilitate the
phosphopantetheinylation of
the ACP. Yet another possibility is that a molecule of CoA is noncovalently-
bound to
the putative domain downstream of the ACP via its adenosine moiety and its
phosphopantetheinyl tail protrudes out from the enzyme, as would the
phosphopantetheinyl tail on the holo-ACP. Alternatively, the PPTE domain can
carry a
molecule of noncovalently-bound CoA. Thus, it is expected that KS carries out
several
iterations of condensation reactions involving the transfer of an acetyl group
from an
acetyl-ACP-thioester to a growing acyl-CoA chain that is non-covalently bound
to the
enzyme. The proposed scenario explains the presence of the TEBC, an acyl-CoA
thioesterase rather than a "conventional" PKS-type thioesterase: the full-
length
polyketide chain generated by the PKSE is not tethered to the holo-ACP, but
rather to a
non-covalently bound CoA and the TEBC hydrolyzes the thioester bond of a
polyketide-
CoA to release the full-length polyketide and CoA. A CoA-activated thioester
may
render the polyketide more accessible to auxiliary enzymes involved in
cyclization and
acetylenation prior to or concomitant to hydrolytic release by TEBC.
Figure 11 is a Clustal amino acid alignment showing the relationship between
the TEBC family of proteins and the enzyme 4-hydroxybenzoyl-CoA thioesterase
(1 BVQ) of Pseudomonas sp. Strain CBS-3 for which the crystal structure has
been
previously determined (Benning et al. (1998) J. Biol. Chem. Vol. 273 pp. 33572-
33579).
The black bars highlight the three regions of conservation believed to play
important
roles in the catalysis for 4-hydroxybenzoyl-CoA thioesterase. Homology between
the
TEBC family of proteins and 1 BVQ is concentrated in these three highlighted
regions.
Figure 12 is a Clustal amino acid alignment of the UNBL family of proteins.
The
UNBL family of proteins represents a novel group of conserved proteins that
are unique
to enediyne biosynthetic loci. The UNBL proteins are rich in basic residues
and contain
several conserved or invariant histidine residues. Besides the PKSE and TEBC
proteins, the UNBL proteins are the only other proteins predicted by the PSORT
program (Nakai et al. (1999) Trends Biochem. Sci. Vol. 24 pp. 34-36) to be
cytosolic
that are encoded by the enediyne warhead gene cassette and thus represent the
best
CA 02444812 2003-10-28
3011-13CA
-85-
candidates for the acetylenase activity that is required to introduce triple
bonds into the
warhead structure.
Figure 13 is a Clustal amino acid alignment of the UNBV family of proteins.
PSORT analysis of the UNBV family of proteins predicts that they are secreted
proteins. The approximate position of the putative cleavable N-terminal signal
sequence is indicated above the alignment. The UNBV proteins display
considerable
amino acid conservation but do not have any known homologue. Thus, the UNBV
family of proteins represents a novel group of conserved proteins of unknown
function
that are unique to enediyne biosynthetic loci.
Figure 14 is a Clustal amino acid alignment of the UNBU family of proteins.
PSORT analysis of the UNBU family of proteins predicts that they are integral
membrane proteins with seven or eight putative membrane-spanning alpha helices
(indicated by dashes in Figure 14). The UNBU proteins display considerable
amino
acid conservation but do not have any known homologue. The UNBU family of
proteins
represents a novel group of conserved proteins that are unique to enediyne
biosynthetic loci.
UNBU is likely involved in transport of the enediynes across the cell
membrane.
UNBU may also contribute, in part, to the biochemistry involved in the
completion of the
warhead. In the case of chromoprotein enediynes, the apoprotein carries its
own
2o cleavable N-terminal signal sequence and is probably exported independently
of the
chromoprotein by the general protein secretion machinery. Formation of the
bioactive
warhead, export, and binding of the chromophore and protein component must
occur in
and around the cell membrane to minimize damage to the producer and to
maximize
the stability of the natural product. UNBV is predicted to be an extracellular
protein.
UNBV may finalize or stabilize the warhead structure. UNBV may act in close
association with the extracellularly exposed portions) of UNBU.
To date, we have sequenced over ten enediyne biosynthetic loci that contain
the
5-gene cassette made up of PKSE, TEBC, UNBL, UNBV, and UNBU genes. In all
cases, the PKSE and TEBC genes are adjacent to one another and the TEBC gene
is
30 always downstream of the PKSE gene. Moreover, these two genes are usually,
if not
always, translationally coupled. These observations suggest that the
expression of the
PKSE and TEBC genes is tightly coordinated and that their gene products, i.e.,
polypeptides, act together. Likewise, the UNBV and UNBU genes are always
adjacent
CA 02444812 2003-10-28
3011-13CA
-86-
to one another and the UNBU gene is always downstream of the UNBV gene.
Moreover, these two genes are usually, if not always, translationally coupled.
These
observations suggest that the expression of the UNBV and UNBU genes is tightly
coordinated and that their gene products, i.e., polypeptides, act together.
Example 12: Common mechanism for the biosynthesis of enediyne warheads
Without intending to be limited to any particular biosynthetic scheme or
mechanism of action, the genes and proteins of the present invention can
explain
formation of enediyne warheads in both chromoprotein enediynes and non-
chromoprotein enediynes.
The PKSE is proposed to generate a highly conjugated polyunsaturated
hepta/octaketide intermediate in a manner analogous to the action of
polyunsaturated
fatty acid synthases (PUFAs). The polyunsaturated fatty acyl intermediate is
then
modified by tailoring enzymes involving one or more of UNBL, UNBU and UNBV to
introduce the acetylene bonds and form the ring structure(s), The conserved
auxiliary
proteins UNBL, UNBU and UNBV are expected to be involved in modulating
iterations
performed by the PKSE, or in subsequent transformations to produce the
enediyne
core in a manner analogous to action of lovastatin monaketide synthase, a
fungal
iterative type I polyketide synthase that is able to perform different
oxidative/reductive
chemistry at each iteration with the aid of at least one auxiliary protein
(Kennedy et al.,
1999, Science Vol. 284 pp. 1368-1372).
The acetate enrichment pattern of the enediyne moiety of esperamicin and
dynemicin suggest that both are derived from an intact heptaketide/octaketide.
There
has been suggestion that esperamicin and dynemicin may share a common
precursor
(Lam et, al., J. Am. Chem. Soc. 1993, Vol. 115 pp. 12340). However, in the
case of
neocarzinostatin, representative of other chromoprotein enediynes,
incorporation
studies investigating carbon-carbon connectivities revealing that the final
enediyne core
contains uncoupled acetate atoms (Hensens et al., 1989 JACS, Vol. 111, pp.
3295-
3299), and other studies regarding polyacetylene biosynthesis (Hensens et.
al., supra),
suggest that the chromoprotein enediyne precursors are distinct from those of
the non-
chromoprotein enediynes. Thus, prior art studies regarding formation of the
enediyne
core teach away from the present invention that genes and proteins common to
both
CA 02444812 2003-10-28
3011-13CA
-87-
chromoprotein enediynes and non-chromoprotein enediynes are responsible for
formation of the warhead in both classes of enediynes.
We propose that skeletal rearrangements may account for the distinct
chromoprotein/nonchromoprotein enediyne labeling patterns. For instance,
thermal
electrocyclic rearrangement of an intermediate cyclobutene to a 1,3 diene
could result
in an isotopic labeling pattern consistent with that which has been reported.
___ _ ___ '°n40R'
'-.~ ________ .___ '_______ ORZ
OR,
OR,
R3C~COOH
Accordingly, the warhead precursor in the formation of neocarzinostatin could
be
a heptaketide, similar to that proposed for the other classes of enediynes.
Since
calicheamicin and esperimicin do not contain any uncoupled acetates, the
common
unsaturated polyketidic precursor must rearrange differently from the
chromoprotein
class. However, the proposed biosynthetic scheme is consistent with one aspect
of the
present invention, namely that warhead formation in all enediynes involves
common
genes, proteins and common precursors.
Example 13: Heterologous expression of Genes and loroteins of the
calicheamicin
enediyne cassette
Escherichia coli was used as a general host for routine subcloning.
2o Streptomyces lividans TK24 was used as a heterologous expression host. The
plasmid
pEC01202 was derived from plasmid pANT1202 (Desanti, C. L. 2000. The molecular
biology of the Streptomyces snp Locus, 262 pp., Ph.D dissertation, Ohio State
Univ.,
Columbus, OH) by deleting the Kpnl site in the multi-cloning site (MCS).
pEC01202RBS contains a DNA sequence encoding a putative ribosome-binding site
(AGGAG) introduced just upstream of the Clal site located in the MCS of
pEC01202.
E. coli strains carrying plasmids were grown in Luria-Bertani (LB) medium and
were selected with appropriate antibiotics. S. lividans TK24 strains were
grown on
R2YE medium. (Kieser, T. et al., Practical Streptomyces Genetics, The John
Innes
Foundation, Norwich, United Kingdom, 2000).
CA 02444812 2003-10-28
3011-13CA
_88_
Preparation of S. lividans TK24 protoplasts was carried out using the standard
protocols. (Kieser et al., supra). Polyethylene glycol-induced protoplast
transformation
was carried out with 1 pg DNA per transformation. After protoplast
regeneration on R5
agar medium for 16 h at 30 °C, transformants were selected by
overlaying each R5
plate with 50 p,g/ml apramycin solutions. Transformants were grown in 50 ml
flasks
containing R2YE medium plus apramycin for seven days.
SDS-PAGE and Western-blotting were carried out by standard procedures
(Sambrook, J. et al. 1989. Molecular cloning: a laboratory manual, 2"d ed.
Cold Spring
Harbor Laboratory, Cold Spring Harbor, N.Y.). Penta-His antibody was obtained
from
Qiagen. Western blots were performed using the ECL detection kit from Amersham
Pharmacia biotech using the manufacturer's suggested protocols. One milliliter
of
seven-day S. lividans culture was centrifuged and mycelium resuspended in cold
extraction buffer (0.1 M Tris-HCI, pH 7.6, 10 mM MgCl2 and 1 mM PMSF). The
mycelium was sonicated 4 x 20 sec on ice with 1 min intervals to release
soluble
protein. After 10 min centrifugation at 20,000g, the supernatant and pellet
fractions
were diluted with sample buffer and subjected to SDS-PAGE and Western-blotting
analysis.
DNA manipulations used in construction of expression plamsids were carried out
using standard methods (Sambrook, J. et al., supra). The plasmid pEC01202 was
2o used as the parent plasmid. Cosmid 061 CR, carrying the calicheamicin
biosynthetic
gene locus was digested with Mfel, and the restriction fragments were made
blunt
ended by treatment with the Klenow fragment of DNA polymerase 1. Upon
additional
digestion with Bglll after phenol extraction and ethanol precipitation, the
resulting 11.5
kb blunt-ended, Bglll fragment was gel purified and cloned into pEC01202
(previously
digested with EcoRl, made blunt ended by treatment with Klenow fragment of
polymerase I, then digested with BamHl ), to yield pEC01202-CALI-1, as shown
in
Figure 15.
PCR was carried out on a PTC-100 programmable thermal controller (MJ
research) with Pfu polymerase and buffer from Stratagene. A typical PCR
mixture
30 consisted of 10 ng of template DNA, 20 ~.M dNTPs, 5% dimethyl sulfoxide, 2U
of Pfu
polymerase, 1 ~.M primers, and 1 X buffer in a final volume of 50 ~I. The PCR
temperature program was the following: initial denaturation at 94 °C
for 2 min, 30 cycles
of 45 sec at 94 °C, 1 min at 55 °C, and 2 min at 72 °C,
followed by an additional 7 min
CA 02444812 2003-10-28
3011-13CA
_89_
at 72 °C. A PCR product amplified by primer 1402, 5' -
GAGTTGTATCGATGAGCAGGATCGCCGTCGTCGGC -3' [containing Cla I site (italic)
and the start codon of PKSE gene (bold)], and primer 1420,
5'GTAGCCGGCCGCCTCCGGCC (corresponding to the nucleotide sequence 940 to
959 by of PKSE), was digested with Clal and Nhel and gel purified. This
fragment was
then cloned into Clal, Nhel digested pEC01202-CALI-1 to yield pEC01202-CALI-5
(Figure 16).
PCR products were amplified by primer 1421, 5'-
GACCTGCCGTACACCGTCTCC -3' (corresponding to the nucleotide sequence 5367
to 5387 by of PKSE), and primer 1403, 5'-
CCCAAGCTTCAGTGGTGGTGGTGGTGGTGCCCCTGCCCCACCGTGGCCGAC-
3'[containing a His Tag (underlined), Hindlll site (italic) and stop codon of
TEBC (bold)],
or primer 1500, 5'- CCCAAGCT'TCACCCCTGCCCCACCGTGGCCGAC- 3' (containing
Hindlll site (italic) and stop codon (bold) of TEBC). These PCR products were
digested
with Hindlll and Psii, gel purified, and then cloned into Hindlll, Psfl
digested pEC01205
to yield pEC01202-CALI-2 (with HisTag) and pEC01202-CALI-3 (without HisTag),
respectively (Figure 16).
The Clal and Hind III fragments from pEC01202-CALI-2 and pEC01202-CALI-3
were cloned into pEC01202RBS to yield pEC01202-CALI-6 (with HisTag) and
pEC01202-CALI-7 (without HisTag), respectively, as shown in Figure 16.
Six transformants of S. lividans TK24 harboring pEC01202-CALI-2 were
analyzed for expression of the His-tagged TEBC protein. Referring to Figure
17, lane
M provides molecular weight markers; lanes 1 to 6 represent crude extracts of
independent transformants of S. lividans TK24 harboring pEC01202-CALI-2; lane
7
represents a crude extract of S. lividans TK24 harboring pEC01202-CALI-4; and
lane 8
represents a crude extract of S. lividans TK24 harboring pEC01202 (control).
TEBC
protein expression was detected in four pEC01202-CALI-2 transformants by
Western
blotting using an antibody that recognizes the His-tag (lanes 2, 3, 5, 6).
TEBC protein
expression was also observed in the transformant of S. lividans TK24 harboring
pEC01202-CALI-4 (lane 7).
As shown in Figure 12, the TEBC protein was expressed as a soluble protein in
S. lividans although the pellet fraction also contains TEBC protein, perhaps
reflecting
insoluble protein or incomplete lysis of S. lividans by the sonication
procedure used.
CA 02444812 2003-10-28
3011-13CA
-90-
Figure 12 provides an analysis of His-tagged TEBC protein derived from
recombinant
S. lividans TK24 by immunoblotting. The soluble and insoluble protein
fractions of S.
lividans transformants were separated by 12% SDS-polyacrylamide gel
electrophoresis, blotted to PVDF membrane, and detected detection with the
Penta-His
antibody. Referring to Figure 12, lane M provides molecular weight markers;
lane 1 to 6
represent soluble (S) and pellet (P) protein fractions of independent
transformants of S.
lividans TK24 harboring pEC01202-CALi-2; lane C represents protein fractions
of S.
lividans TK24 harboring pEC01202 (control).
Example 14: Disruption of the PKSE giene abolishes production of enediyne
To confirm that the PKSE is critical to the biosynthesis of enediynes, the
PKSE
gene of the calicheamicin producer, M. echinospora, was disrupted by
introduction of
an apramycin selectable marker as follows. M, echinospora was grown with a
1:100
fresh inoculum in 50 mL MS medium (Kieser et al., supra) supplemented with 5 %
PEG
8000 and 5 mM MgCl2 for 24 - 36 h and 6 h prior to harvest, 0.5 % glycine was
added.
The digest of the cell wall was accomplished via published procedures with the
exception that 5 mg mL~' lysozyme and 2000 U mutanolysin were used. Under
these
conditions, protoplast formation was complete within 30-60 min after which the
mixture
was filtered twice through cotton wool. Transformation was accomplished via
typical
methodology (Kieser et al., supra) with a 1:1 mixture of T-buffer and PEG 2000
containing up to 10 Ng of alkaline denatured DNA per transformation. The
protoplasts
were then plated on R2YE plates supplemented with 10 mg L-' CoCl2 and
submitted to
antibiotic pressure (70 Ng mL-' apramycin) after 3 - 4 days. To date, all
attempts to
use methods other than protoplast chemical transformation (e.g. phage
transduction,
conjugation and electroporation) have failed to introduce DNA into M.
echinospora.
Low transformation efficiencies were observed in all calicheamicin-producing
Micromonospora strains tested, including those developed from strain
improvement
efforts. In comparison to other actinomycetes, M. echinospora protoplast
regeneration
was found to be slow (~ 4 weeks). Moreover, integration into the locus
requires
homologous fragments exceeding 3 kb in size as constructs containing PKSE
fragments (or other calicheamicin gene fragments) smaller than 3 kb all failed
to
integrate into the chromosome (data not shown).
CA 02444812 2003-10-28
3011-13CA
-91 -
Nine independent apramycin-resistant PKSE disruption clones were obtained.
All nine isolates mapped consistently with the expected PKSE gene disruption
both by
PCR fragment amplification and by Southern hybridization (data not shown). All
nine
PKSE disruption mutants and two parental controls were subsequently tested in
parallel
for calicheamicin production. Extracts from these strains were prepared as
follows.
Fresh M. echinospora cells grown in R2YE were inoculated 1:100 in 10 mL medium
E
(Kieser et al., supra) in stoppered 25 ml glass tubes containing a 4 cm
stainless coil
spring for better aeration and incubated on an orbital shaker with 230 rpm at
28 °C for
one to three weeks. A 600 NI aliquot was removed at various time points,
extracted
with an equal volume of EtOAc and centrifuged at 10000 xg for 5 min in a
benchtop
centrifuge. The supernatant was concentrated to dryness, the pellet
redissolved in 200
NI acetonitrile, centrifuged again and the supernatant removed, concentrated
to dryness
and the residual material finally dissolved in 10 p1 acetonitrile. One NI of
this solution
was utilized for the bioassays and the remaining 8 NI aliquot was utilized for
analysis by
HPLC (Ultrasphere-ODS chromatography, 5 gym, 4.6 mm x 250 mm, 55:45 CH3CN- 0.2
NH40Ac, pH 6.0, 1.0 mL min-', 280 nm detection). A typical M. echinospora
fermentation contains a mixture of calicheamicins that are resolved by HPLC -
~~,~
(retention time - 7 min, ~60%), s,~ (retention time - 5.7 min, ~30%), and a3~
(retention
time - 3.8 min, ~10%) - and all of these calicheamicin components contribute
to
bioassay activities. The best production was found to occur during late log or
early
stationary phase growth. The estimate of calicheamicin production by parental
M.
echinospora is 0.78-0.85 mg mL-'. Extracts were analyzed by i) the biological
induction
assay, a modified prophage induction assay used in the original discovery of
the
calicheamicins (Greenstein et al. (1986) Antimicrob. Agents Chemotherap. Vol.
29,
861 ); ii) the molecular break light assay, a DNA-cleavage assay based upon
intramolecular fluorescence quenching optimized for DNA-cleavage by enediynes
(in
which fM calicheamicin concentrations are detectable) (Biggins et al. (2000)
Proc. Natl.
Acad. Sci. USA Vol. 97, 13537); and iii) high-performance liquid
chromatography
(HPLC) (described above). As expected, all three methods revealed that the
parental
3o M. echinospora fermentations produced 0.5-0.8 mg L-'. In contrast, the PKSE
gene
disruption mutant strains were both devoid of any calicheamicin, known
calicheamicin
derivatives and/or enediyne activity by all three methods of detection. The
elimination
of calicheamicin production brought about by disruption of the PKSE gene
indicates
CA 02444812 2003-10-28
3011-13CA
-92-
that it provides an essential activity for biosynthesis of calicheamicin.
Based on the
presence of the PKSE in all enediyne biosynthetic loci sequenced to date and
on their
overall conservation, it is expected that PKSEs fulfill the same, essential
function in the
biosynthesis of all enediyne structures.
The present invention is not to be limited in scope by the specific
embodiments
described herein. Indeed, various modifications of the invention in addition
to those
described herein will become apparent to those skilled in the art from the
foregoing
description and the accompanying figures. Such modifications are intended to
fall
within the scope of the appended claims.
It is further to be understood that all sizes and all molecular weight or mass
values are approximate, and are provided for description.
Some open reading frames listed herein initiate with non-standard initiation
codons (e.g. GTG - Valine or TTG - Leucine) rather than the standard
initiation codon
ATG, namely SEQ ID NOS: 2, 8, 16, 28, 30, 32, 38, 40, 42, 48, 54, 56, 70, 74,
76, 78,
80, 82, 84, 86, 88, 92, 98, 100. All ORFs are listed with M, V or L amino
acids at the
amino-terminal position to indicate the specificity of the first codon of the
ORF. It is
expected, however, that in all cases the biosynthesized protein will contain a
methionine residue, and more specifically a formylmethionine residue, at the
amino
terminal position, in keeping with the widely accepted principle that protein
synthesis in
bacteria initiates with methionine (formylmethionine) even when the encoding
gene
specifies a non-standard initiation codon (e.g. Stryer, Biochemistry 3'd
edition, 1998,
W.H. Freeman and Co., New York, pp. 752-754).
Patents, patent publications, procedures and publications cited throughout
this
application are incorporated herein in their entirety for afl purposes.