Note: Descriptions are shown in the official language in which they were submitted.
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
ANTHER-SPECIFIC TAA1 GENES ENCODING FATTY ACYL CO-A
REDUCTASES, AND USES THEREOF
1. FILED OF THE INVENTION
The present invention relates to genes that are specifically expressed in the
anthers of plants. More particularly, the present invention relates to genes
encoding fatty acyl Co-A reductase enzymes that are required for pollen grain
maturation.
2. BACKGROUND TO THE INVENTION
There is a significant degree of commercial interest in the development of
transgenic plants with altered lipid metabolism, which generate altered or
increased yields of lipid products. The development of such modified plants
and
crops may facilitate the manufacture of nutritional and medicinal products in
crops. Therefore, the possibility of successfully generating lipid-modified
plants
has implications for both the agricultural and pharmaceutical industries.
The metabolic pathways that regulate lipid metabolism in plants are not fully
understood. Different regions and organs of a plant generate alternative
profiles
of lipid products, with certain regions of a plant comprising a greater
concentration of lipid products than others. For this reason, the genes
involved in
lipid metabolism must undergo differential regulation for specific lipid
products
to be concentrated in particular regions of the plant. Delineation of plant
lipid
metabolic pathways, and the generation ~ of modified transgenic plants with
beneficial characteristics, represents a considerable challenge to those of
skill in
the art.
The outer surface of pollen grains represents one region of a plant known to
harbor higher concentrations of lipid products. The anthers of plants have
1
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
evolved to coat pollen grains with an oily substance to preserve and increase
the
viability of the pollen. For this purpose, male gametophyte development, and
in
particular the interplay between the sporophytic tapetum and gametophytic
microspore, is a well-orchestrated process in plants (Goldberg et al., 1993).
To
date, a number of genes underlying this process have been isolated and
characterized. These genes may be grouped into pollen-specific and
antherltapetum-specific genes. The former are usually predominantly expressed
during advanced stages of pollen development. The examples include genes
encoding cytoskeletal proteins (I~andasamy et al., 1999; Lopez et al., 1996),
cell
wall-degrading enzymes (Brown and Crouch, 1990; Futamura et al., 2000),
pollen allergens (Rafnar et al., 1991) and other genes with unknown functions
(Zou et al., 1994). The other group includes genes preferentially expressed in
the
tapetum at relatively early stages of microsporogenesis. These include genes
associated with programmed cell death (Walden et al., 1999), pollen excine
formation (Aarts et al., 1997; Fuerstenberg et al., 2000; I~oltunow et al.,
1990),
lipid transfer (Aguirre and Smith, 1993), cell wall-degradation (Bih et al.,
1999;
Hird et al., 1993; Rubinelli et al., 1998) and unknown functions (Jeon et al.,
1999).
The anther tapetum plays a pivotal role in plant gametophyte development
(Piffanelli et al., 1998; Shivanna et al., 1997). In addition to breakdown of
callus
wall around microspore tetrads and supply of nutrients to developing pollens,
the
essential function of the tapetum is thought to form two extracellular lipid-
derived structures (pollen exine and pollen coating) of pollen grains. This
assumption is established on the base of the earlier cytological observations,
and
recent ultrastructural and molecular studies (for recent reviews see Furness
and
Rudall, 2001; Huysmans et al., 1998; Piffanelli et al., 1998). For example, it
has
been shown that during the development of the extracellular lipidic
structures, the
tapetum and not the microspore is the major site of fatty acid biosynthesis
(Piffanelli et al., 1997). Mutation of a tapetum-specific gene encoding a
putative
2
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
fatty lipid reductase leads to formation of exine-free pollen and male
sterility
(Aarts et al., 1997). Recent progress also includes the fording that two
tapetum-
unique lipidic organelles whose major constituents are neutral esters and
polar
lipids, upon lysis of the tapetal cells, are discharged into the anther locule
and
their components contribute to the formation of the lipidic coating of mature
pollen grains (Hernandez-Pinzon et al., 1999; Piffanelli~and Murphy, 1998;
Ting
et al., 1998; Wu et al., 1997).
These findings substantially facilitate our understanding of the intrinsic
link
between the tapetal lipid biosynthesis and microspore development. However,
the
enzymes that catalyze and regulate the biochemical production of these tapetal
lipidic compounds have remained unclear. Isolation and characterization of
these
lipid-specific enzymes would permit an improved understanding of the
mechanisms of plant lipid metabolism.
3. SUMMARY OF THE INVENTION
It is an object of the present invention to isolate and characterize anther-
specific
genes involved in lipid metabolism, for the commercial development of useful
transgenic plants and plant products.
It is a further object of the present invention to provide a means for
modifying
lipid metabolism in plants, preferably by increasing or altering the yield of
useful
lipid-based products in the plants. The present invention further aims to
provide a
transgenic plant with increased levels of fatty alcohols, which can be
harvested
for use in the production of, for example, nutritional and pharmaceutical
products.
It is another object of the present invention to provide a means of increasing
the
levels of fatty alcohols in designated regions or organs of a plant, for
specific
3
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
commercial purposes. These commercial purposes may include, but are not
limited to, the production of crops with increased pesticide resistance, crops
with
altered cross-breeding activity, plants with increased levels of lipid
products
concentrated in regions that permit facile harvesting and extraction.
It is further an object of the present invention to isolate and characterize
genes
expressed in the tapetum, and their corresponding proteins, that are required
for
the formation of the outer cell wall of pollen grains during microspore
development. In this way, the present invention aims to alter anther-specific
properties of a plant to induce, for example, male sterility, and
developmental or
reproductive modifications that are commercially useful properties. It is
still
further an obj ect of the present invention to provide a transgenic plant,
comprising a construct wherein anther-specific promoters are utilized to
generate
useful products in the anthers and pollen cells of the transgenic plants.
It is a further object of the present invention to provide isolated
recombinant
proteins involved in plant lipid metabolism, which can be used in the
commercial
ex vivo production of fatty alcohols.
The peptides of the present invention, and their corresponding nucleotide
sequences, have significant potential for use in the generation of genetically
modified plants with altered profiles, or increased or otherwise altered
levels of
lipid compounds, as well as plant having desirable anther-specific and whole-
plant phenotypic modifications.
The inventors of the present application have succeeded in isolating and
purifying
both the genomic and cDNA sequences of a family of three closely related genes
that are predominantly expressed in the tapetum of anthers. The genes were
isolated from the bread wheat species 'Triticuna aestivum', and are designated
TAAI a, TAAl b, and TAAl c. The cDNA sequences of these genes have
4
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
permitted the characterization of the corresponding protein products, which
can
function as fatty acyl Co-A reductases. Transgenic plants overexpressing a
TAAI
gene can comprise higher than normal levels of fatty alcohols. It is
considered
that similar transgenic plants will have strong potential for the generation
of
crops capable of producing fatty acid products for agricultural, nutritional
and
pharmaceutical purposes.
Moreover, isolation of the corresponding genomic DNA sequences for the TAAl
genes has permitted the characterization of the anther-specific TAAl gene
promoters. These promoters are tapetum specific and have significant potential
for the generation of constructs for use in transgenic plants, wherein the
constructs comprise a gene of choice under the control of the anther-specific
promoter. In this way, numerous properties of the plant can be modified to
alter,
for example, the developmental, reproductive, and aesthetic properties of the
plant.
In accordance with a first embodiment of the present invention, there is
provided
an isolated and purified nucleotide sequence, characterized in that the
nucleotide
sequence is endogenously expressed in wheat anthers, and encodes a peptide
having fatty acyl Co-A reductase (FAR) activity. The present invention
therefore
provides characterization of a novel family of genes that are involved in
lipid
metabolism in anthers of plants and encompasses all such corresponding
homologous genes.
In an alternative embodiment the present invention provides an isolated and
purified nucleotide sequence, characterized in that the nucleotide sequence is
selected from:
(a) a TAAl gene, or a part thereof, or a complement thereof; and
(b) a nucleotide sequence having at least 50% identity to a peptide
encoded by a TAAl gene, or a part thereof, or a complement thereof;
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
the nucleotide sequence encoding a protein or a part thereof, that alters
lipid
metabolism in a transgenic plant exogenously expressing said nucleotide
sequence.
Preferably, the isolated and purified nucleotide sequence is selected from:
(a) SEQ ID NO: 1, 3, or 5, or a part thereof, or a complement thereof;
and
(b) a nucleotide sequence having at least 50% identity to a peptide
encoded by a SEQ ID NO: l, 3, or 5, or a part thereof, or a complement
thereof;
the nucleotide sequence encoding a protein or a part thereof, that alters
lipid
metabolism in a transgenic plant exogenously expressing said nucleotide
sequence. More preferably, the nucleotide sequence has at least 70%, more
preferably 90%, more preferably 95% and most preferably 99% identity to a
peptide encoded by a SEQ ID NO: 1, 3, or 5, or a part thereof, or a complement
thereof. In this way, the nucleotide sequences of the present invention
include
TAAl homologous genes from species of plants other than wheat, as well as
closely related wheat homologues, polymorphisms and mutated variants of the
genes. The invention further encompasses nucleotide sequences that will bind
to
SEQ ID NOS: 1, 3, or, 5 under stringent hybridization conditions, including
nucleotide sequences suitable for use as hybridization probes, PCR primers and
DNA sequencing primers.
In further embodiments, the present invention also encompasses isolated and
purified peptides, or parts thereof, encoded by TAAl genes, or possible
variants
of the TAAl genes disclosed herein. Such peptides may be used in the
production
of pharmaceutical or nutritional agents as appropriate.
The present invention further encompasses expression cassettes and constructs
comprising TAAI gene sequences and variants, complements, or parts thereof.
Preferably, the expression cassettes and constructs include a TAAI gene
sequence
6
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
open reading frame operably linked to a promoter for expression of the THAI
gene product, or part or variant thereof. Preferably, the expression cassettes
and
constructs of the present invention are suitable for transformation into
plants. In
this way, transgenic plants having altered lipid metabolism or altered lipid
content can be generated. More preferably, the altered lipid metabolism or
altered
lipid content at least partly occurs within the anthers and / or pollen of the
transgenic plant.
The transgenic plants of the present invention therefore include plants
expressing
the nucleotide sequences disclosed herein, and homologues and variants
thereof,
thereby increasing, decreasing or changing the lipid content of the plant
compared to an unmodified plant. More preferably, the change in lipid content
may specifically relate to the fatty alcohol content of the plant, and more
preferably the fatty alcohol content of the anthers and / or pollen of the
plant.
The transgenic plants of the present invention include species of a woody
plants,
non-woody plants, and grasses, as well as plants selected from the group
consisting of crucifer crops, tobacco, wheat, corn, sugar cane, and apple.
In an alternative embodiment, the transgenic plants of the present invention
may
include constructs wherein the TAA1 gene or part or variant thereof is under
the
control of an organ-specific promoter. In this way, the promoter can direct
the
expression of the nucleotide sequence to affect a particular organ or organs
of the
plant. The transgenic plants of the present invention may exhibit one or more
modified characteristics compared to an unmodified plant including, but not
limited to: increased pest resistance; male sterility; reduced height; reduced
internode spacing; increased resistance to wind damage; reduced growth rate;
altered cross-pollination specification ; increased fruit or nut aesthetic
appeal;
delayed vegetative development; and delayed propagative development.
7
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
The transgenic plants of the present invention may contain constructs
characterized in that the nucleotide sequence expressed is oriented fox
antisense
expression from the construct, thereby causing a reduction in the levels of
fatty
acyl Co-A reductase compared to an unmodified plant, and a corresponding
decrease in the levels of fatty alcohols present in the plant.
The present invention further encompasses an isolated and purified nucleotide
sequence, characterized in that the nucleotide sequence is selected from:
(a) SEQ ID NO: 7, 8, or 9 or a complement thereof; and
(b) a nucleotide sequence that can hybridize to SEQ ID NO: 7, 8, or 9
or a complement thereof under stringent hybridization conditions. Therefore,
the
invention encompasses the corresponding genomic DNA sequences for the TAAI
family of genes, including promoter sequence disclosed in SEQ ID NOS: 7, 8,
and 9, or TAA1 promoter sequence obtained by chromosome walking a genomic
DNA library for 5' (and 3') untranslated regions of the TAAI genomic DNAs.
Furthermore, in alternative embodiments the invention includes nucleotide
sequences for use as hybridization probes, PCR primers or DNA sequencing
primers, that bind to the TAA1 sequences under stringent hybridization
conditions. Preferably, the promoters of the present invention can be used to
direct the expression of a gene unrelated to fatty aryl Co-A reductases in the
anthers and pollen grains of transgenic plants. Most preferably, the promoter
of
the present invention may comprise of genomic DNA sequence of about 1.6kb
upstream from the start codon of SEQ ID NO: 8.
In additional embodiments, the present invention includes constructs
comprising
TAAl promoter sequences in operative association with an open reading frame,
or
a part thereof or a complement thereof, for use in modifying anther, tapetum
or
pollen metabolism. The constructs may be transformed into plants to generate
transgenic plants with altered characteristics. For example, the invention
encompasses transgenic plants transformed with a construct having a Tf4Al
8
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
promoter or part thereof in operative association with an anther or pollen
inactivating gene, wherein expression of the open-reading frame induces male
sterility of the transgenic plant. Alternatively, the open-reading frame may
encode a transposase, and expression of the open-reading frame may induce an
increased rate of genomic DNA rearrangement in anther or pollen cells of the
transgenic plant. Alternatively, the open-reading frame may encode a peptide
suitable for use as a nutritional or pharmaceutical agent, the peptide being
expressed in anthers or pollen of the transgenic plant. Alternatively, the
open-
reading frame may encode a peptide required for the production of a
nutritional
or pharmaceutical agent, or a protein that inhibits the production and / or
accumulation of an unwanted substance selected from the group consisting of a
toxin, and an allergen, or a peptide for altering the cross-pollination
specification
of the transgenic plant. Alternatively, the open reading frame may be oriented
for
antisense expression within the construct, thereby inducing antisense
repression
of endogenous gene expression within the anthers, tapetum or pollen of the
transgenic plant.
The present invention further provides, in alternative embodiments, for a
means
for generating fatty alcohols that may be used as nutritional or
pharmaceutical
agents. The fatty alcohols may be purified from extracts of the transgenic
plants
using techniques that are well known in the art. Preferably, the fatty
alcohols
generated by the transgenic plants of the present invention include
Octacosanol; a
fatty alcohol known to produce health benefits including enhances physical
endurance and reproductive health. Moreover, in another preferred embodiment,
the transgenic plants of the present invention may be used to generate fatty
alcohols for the washing and cleaning industry. In alternative embodiments,
the
transgenic plants of the present invention may bear fruit with increased
levels of
fatty alcohols, wherein the fruit include wax derived from the fatty alcohols
to
help preserve the fruit and improve the aesthetic appeal of the fruit, thereby
improving shelf life. The increased levels of wax production in the plants of
the
9
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
present invention are further predicted to confer enhanced properties such as
reduced rates of moisture loss, and increased resistance to pests.
The invention further encompasses the fatty alcohols derived or extracted from
the transgenic plants or other transformed organisms (e.g. bacteria) of the
present
invention, and their use, for example as a wax, as a cleaning agent, as a
cosmetic
agent, as a dermatological agent, as a pharmaceutical agent, or as a
nutritional
agent.
The invention further encompasses pharmaceutical and nutritional compositions
and agents comprising the plant extracts and fatty alcohols obtained from the
transgenic plants of the present invention, as well as methods for treating or
preventing a medical condition, or for providing a dietary supplement, by the
administration of the plant extracts or fatty alcohols of the present
invention.
The invention further encompasses method for the production and isolation of
fatty alcohols, characterized in that the method comprises the steps of:
transforming an organism with a construct comprising a TAAI gene sequence, or
part thereof, or complement thereof in accordance with the present invention;
growing or propagating said organism containing said construct; and
extracting said fatty alcohols from said organism. Preferably, the organism is
an
E. toll bacterium, such that recombinant E. eoli comprising increased or
altered
levels of fatty alcohols may be cultured and harvested. In an alternative
embodiment, the organism may comprise a plant or a plant embryo, preferably a
tobacco plant or tobacco plant embryo, that is induced to express the
construct
and generate increased or altered levels of fatty alcohols. Similarly, such
transgenic plants may be grown and / or propagated thereby allowing plant
extracts to be harvested and fatty alcohols to be purified by standard
techniques.
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
The invention further encompasses a method of inducing dwarfism in a plant,
characterized in that the method comprises the steps of:
transforming a plant cell, plant embryo or plant with a construct according
to the present invention; and
growing or propagating said plant cell, plant embryo, or plant, thereby
generating a plant expressing a DNA sequence encoded by said construct, said
plant having a reduced size compared to an unmodified plant.
4. BRIEF DESCRIPTION OF THE DRAWINGS
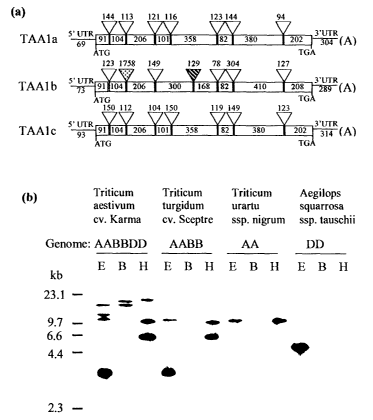
Figure 1 (a) schematically illustrates the genomic organization of TAAI a,
TAAI b,
and TAAl c. Triangles and rectangles represent introns and exons,
respectively.
The length (bp: base pair) of each intron and exon is shown above and in the
corresponding triangle and rectangle. The stippled and hatched triangles
indicate
a very long intron and an intron with alternative insertion position in TAAI
b,
respectively. The putative translation start (AUG) and stop (TGA) codons, and
5'
and 3' UTR (untranslational region) are given. (A)n represents a poly(A) tail.
For
clarity, cDNA sequences and introns are not drawn to scale.
Figure 1 (b) provides a genomic DNA blot analysis of different wheat species.
Molecular size makers are indicated at left in kilobases. Sources of DNA are
shown. Total DNA (10 ~,g) was digested with EcoRI (E), BanzHI (B) and Hiha'III
(H), separated on a 1 % agarose gel, blotted onto a nylon membrane, probed
with
the coding region of TAAIa, and visualized by exposure to an x-film.
Figure 2 (a) demonstrates anther-specific expression of TAA1. R, roots; S,
stems;
L, leaves; A, anthers; O, ovary; G, glume and pilea. Northern blot analysis of
T~4A1 expression in wheat. Total RNA (about 5 ~,g) purified from root, stem,
leaf,
anther, ovary, and pilea and glume was loaded. A 0.7 kb fragment of the TAAla
11
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
cDNA resulting from 5' RACE was used as a probe. The estimated size of
hybridizing RNA species is shown to the left side. Underneath is the same blot
hybridized with a 28S rRNA probe.
Figure 2 (b) demonstrates anther-specific expression of TAAl. R, roots; S,
stems;
L, leaves; A, anthers; O, ovary; G, glume and pilea. RT-PCR amplification of
cDNA derived from different wheat tissues. Underneath is the same cDNA
amplified with a pair of primers to a glyceraldehyde-3-phosphate dehydrogenase
gene (GPD).
Figure 3 In situ RNA hybridization. The cross-sections of wheat flower buds
were hybridized with TAAl anti-sense and sense transcript. Hybridization was
shown by the formation of a dark bluish precipitate. Solid arrow head:
tapetum;
unfilled arrow head: micropore; m, microspores; ov, ovary; and ps, pollen sac.
Scale bars=100~m.
(a) probed with a TAAl a sense transcript.
(b) probed with a TAAI antisense transcript.
(c) sectioned at stage pre-meiosis, probed with a TAAl antisense transcript.
(d) sectioned at stage young microspore, probed with a TAAI antisense
transcript.
(e) sectioned at stage vacuolated microspore, probed with a TAAI antisense
transcripts.
Figure 4 Immunocytochemical detection of the TAA1 protein on the wheat
anthers. The sections were immunoblotted with either pre-immune serum or
TAAla antiserum. Positive antibody recognition was shown by the formation of
bluish deposits. M, microspores; ps, pollen sac, arrow heads, tapetum. Scale
bars=100~,m.
(a) immunoreacted with pre-immune serum (control).
(b) with TAAla antiserum.
(c) an enlarged pollen sac of (a) (control).
12
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
(d) sectioned at the young microspore stage and immunoreacted with TAAla
antiserum.
(e) sectioned at the vacuolated microspore stage.
Figure 5 (a). Amino acid sequence comparison and phylogenetic analysis of
TAAl. Pair-wise alignment of the amino acid sequence of TAAla with that of
FAR according to Pearson and Lipman (1998). D represents gaps which are
introduced to allow the best matches. The dashes in FAR indicate the identical
residues to TAAla. Two potential transmembrane helixes predicted by Metz et
al. (2000) are underlined.
Figure 5 (b). Phylogenetic analysis of TAAl and its related genes. The
sequences
of all related genes were obtained from public databases and refer to the
following: FAR, the jojoba acyl coenzyme A reductase (accession no.
AF149917); MS2-like; a predicted gene from Arabidopsis (accession no.
AB012244); MS2, the Arabidopsis male sterility 2 gene (accession no. 533804);
B-MS2, the Brassica MS2 gene (accession no. T08096).
Figure 6 (a) Fatty alcohols in transgenic seeds and E. coli. Fatty alcohol
content
in the tobacco seeds transformed with the Napin-TAAla chimeric gene. The
amounts of fatty alcohols obtained from GC analysis were normalized against
the
internal standard beta-sitosterol. The y-axis of the graph illustrates
percentage
'FA' of the relative amounts of fatty alcohols to beta-sitosterol (%). Line
723-0-D
was transformed with the control vector. All the remaining (477-0-4, 477-0-18,
477-0-2, and 477-0-10) were the Napin-TAAI a transgenic lines.
Figure 6 (b) Gas chromatography (GC) analysis of fatty alcohol amounts and
compositions in bacterial cells without ((i) upper graph), or with ((ii) lower
graph) expression of TAAI a. t = retention time in minutes, and CL = chain
length of fatty alcohol standards.
13
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
Figure 7. Over-expression of TAAI results in significant dwarfism in
transgenic tobacco. Vector: transgenic plants containing NPTII resistant
gene only; 35S::TAA1: transgenic containing both NPTII resistant gene and
35S::TAAla chimeric gene. In this example, TAAl was over-expressed
constitutively. (a) Three-week old seedlings in MS medium. (b) Plants three
weeks after transplanting in a greenhouse. (c) Plants two months after
transplanting in greenhouse.
Figure 8 Transient expression assay of TAAI promoter specificity. Hand cross-
section of Daylily flower buds were bombarded with microprojectiles coated
with
either the CaMV35S-uidA (a) or TAAl-uidA chimeric genes (b). 35S-GUS
transient expression was observed in anther walls, filaments, and petals (a).
In
contrast, TAA1-GUS transient expression limited in microspores and tapetum
(arrow) (b). an: Anther, f Filament, m: microspores, pe: petal, ps: pollen
sac.
Scale bars= 1 mm.
Figure 9 GUS expression pattern in transgenic tobacco anthers. (a) and (b)
show
GUS assays on hand cross-sections of anthers at different developmental stages
of a transgenic plant containing a TAAl -uidA chimeric gene. (a) at the tetrad
stage
and (b) at the microspore separation stage. (c) and (d) show paraffin cross-
sections of anthers of transgenic plants containing a 35S-uidA chimeric gene
and
a TAAI-uidA chimeric gene, respectively. aw: anther wall; cn: connective
tissue;
ep: epidermis; m: microspores; t: tapetum. Scale bars=200~.m.
6. GLOSSARY OF TERMS
Amplification of DNA / amplified DNA: "amplified DNA" refers to the product
of nucleic-acid amplification of a target nucleic-acid sequence. Nucleic-acid
amplification can be accomplished by any of the various nucleic-acid
14
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
amplification methods known in the art, including the polymerase chain
reaction
(PCR). A variety of amplification methods are known in the art and are
described, inter alia, in U.S. Pat. Nos. 4,683,195 and 4,683,202, and in Innis
et al.
(eds.), PCR Protocols: A Guide to Methods and Applications, Academic Press,
San Diego, 1990.
Construct: A construct comprises a vector and a DNA molecule operatively
linked to the vector, such that the vector and operatively linked DNA molecule
can be replicated and transformed as required.
Expression: The generation of a protein product derived from a DNA sequence
encoding the protein, comprising a combination of transcription and
translation.
Homologous: DNA or peptide sequences exhibiting similarity to another DNA or
peptide sequences in terms of the chemical nature, order and position of the
individual residues relative to one another in the sequence. For the purposes
of
this application, unless stated otherwise homology is characterized according
to
BLAST search results, wherein a best-fit sequence alignment is obtained. In
this
way, sequences comprising residues that are similar or identical may be
aligned,
and gaps provided as necessary. Homology is therefore expressed as a
percentage of similarity or identity, wherein similarity encompasses both
similar
and identical residues. Unless stated otherwise, all BLAST searches were
carried
out using default parameters: e.g. gaps permitted, E-value =1, organism
selected
as required, filter for low complexity, standard genetic code, BLOSUM62
general
purpose matrix; for more information see
http://www.ncbi.nlm.nih.gov/Education/BLASTinfo/tutl .html.
Identity: Comparison of homologous DNA or peptide sequences provides
identification of residues that are identical in the same relative position of
the
sequence, following best fit alignment. For the purposes of this application,
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
unless stated otherwise, homology, best fit alignment and identity are
calculated
according to BLAST search results (BLAST searching is available, for example,
from the following website: http://www.ncbi.nlm.nih.gov/BLAST/). Identity is
provided as a percentage, indicating the percentage of residues that are
identical
along the sequences under comparison, excluding regions of gaps between the
aligned sequences. BLAST searching permits a standard alignment configuration
to automatically take into account regions of gaps or truncations between
sequences, thereby providing a 'best fit' alignment.
Isolated: A nucleotide or peptide is "isolated" if it has been separated from
other
cellular components (nucleic acids, liquids, carbohydrates, and other
nucleotides
or peptides) that naturally accompany it. Such a nucleotide or peptide can
also be
referred to as "pure" or "homogeneous" or "substantially" pure or homogeneous.
Thus, a nucleotide or peptide which is chemically synthesized or recombinant
is
considered to be isolate. A nucleotide or peptide is isolated when at least 60-
90%
by weight of a sample is composed of the nucleotide or peptide, preferably
95°/~
or more, and more preferably more than 99%. Protein purity or homogeneity is
indicated, for example, by polyacrylamide gel electrophoresis of a protein
sample, followed by visualization of a single peptide band upon staining the
polyacrylamide gel; high-performance liquid chromatography; or other
conventional methods. The peptides of the present invention can be purified by
any of the means known in the art. Various methods of protein purification are
described, e.g., in Guide to Protein Purification, in Deutscher (ed.), Meth.
Enzymol. 185, Academic Press, San Diego, 1990; and Scopes, Protein
Purification: Principles and Practice, Springer Verlag, New York, 1982.
Operably linked: two ~~acleotide sequences are operable linked if the linkage
allows the two sequences to carry out their normal functions relative to each
other. For instance, a promoter region would be operably linked to a coding
sequence if the promoter were capable of effecting transcription of that
coding
16
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
sequence, and the coding sequence encoded a product intended to be expressed
in
response to the activity of the promoter.
Organ: A specific region of a plant defined in terms of structure and
function, for
example, a stem, a leaf, an anther, a pollen grain, or a root.
Promoter: A recognition site on a DNA sequence or group of DNA sequences
that provides at least one expression control element for a gene encoding a
polypeptide, and to which RNA polymerase specifically binds and initiates RNA
synthesis (transcription) of the gene.
Stringent conditions, or stringent hybridization conditions: includes
reference to
conditions under which a probe will hybridize to its target sequence, to a
detectably greater degree than other sequences (e.g. at least 2-fold over
background). Stringent conditions are sequence-dependent and will be different
in different circumstances. Longer sequences hybridize specifically at higher
temperatures. Generally, stringent conditions are selected to be about 5
° C lower
than the thermal melting point (Tin) for the specific sequence at a defined
ionic
strength and pH. The Tm is the temperature (under defined ionic strength and
pH
at which 50% of a complementary target sequence hybridizes to a perfectly
matched probe. Typically, stringent conditions will be those in which the salt
concentration is less than about 1.0 M Na ion, typically about 0.01 to 1.0 M
Na
ion concentration (or other salts) at pH 7.0 to 8.3 and the temperature is at
least
about 30°C for short probes (e.g. 10 to 50 nucleotides) and at least
about 60°C for
long probes (e.g. greater than 50 nucleotides). Stringent conditions may also
be
achieved with the addition of destabilizing agents such as formamide.
Exemplary
low stringency conditions include hybridization with a buffer solution of 30%
formamide, 1 M NaCI, 1% SDS at 37°C, and a wash in 2X SSC at
50°C.
Exemplary high stringency conditions include hybridization in 50% formamide, 1
M NaCI, 1% SDS at 37°C, and a wash in O.1X SSC at 60°C.
Hybridization
17
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
procedures are well-known in the art and are described in Ausubel et
al.,(Ausubel
F.M., et a1.,1994, Current Protocols in Molecular Biology, John Wiley & Sons
Inc.).
Transformation: Modification of a cell by the introduction of exogeneous DNA
sequence (e.g. a vector or recombinant DNA molecule).
Transgenic: A cell or organism derived from a process of cellular
transformation,
wherein the cell or organism comprises the introduced exogenous DNA molecule
not originally present in a non-transgenic cell or organism.
Transgenic plant: A plant or progeny thereof derived from a transformed plant
cell or protoplast, wherein the plant DNA contains an introduced exogenous
DNA molecule not originally present in a native, non-transgenic plant of the
same strain. The terms "transgenic plant" and "transformed plant" have
sometimes been used in the art as synonymous terms to define a plant whose
DNA contains an exogenous DNA molecule. However, it is thought more
scientifically correct to refer to a regenerated plant or callus obtained from
a
transformed plant cell or protoplast as being a transgenic plant.
Vector: A DNA molecule capable of replication in a host cell and/or to which
another DNA segment (or insert) can be operatively linked so as to bring about
replication of the attached insert. A plasmid is an exemplary vector.
Moreover, a
vector may include promoter sequence to facilitate expressi9n of an open
reading
frame present in the DNA insert. All vectors used for the present application
were generated by the inventors, with the exception of: T/A vectors
(Invitrogen),
pRSET A (Invitrogen), phagemids (Stratagene), pRD400 and pRD410 (Datla et
al. 1992), pHS724 (Huang et al., 2000), pJOY43 (Hair et al., 2000).
18
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
7. DETAILED DESCRIPTION OF THE PREFERRED EMBODIMENTS
To support this application, two deposits of biological material have been
made
under the Budapest Treaty regarding Deposits of Biological Material. The
deposits were made at the International Depository Authority of Canada, Bureau
of Microbiology, Health Canada, Winnipeg, Manitoba, Canada, on June 7, 2001,
under accession numbers IDAC 070601-2 and IDAC 070601-1. The deposits
both comprise E.coli bacterial cells, strain DHSoc, transformed with
constructs
comprising DNA sequence of the present invention. Deposit number IDAC
070601-2 consists of DHSoc cells transformed with pAMW133 comprising the
full length coding region of TAAla cDNA. Deposit number IDAC 070601-1
consists of DHSoc cells transformed with pAMW170 comprising the promoter
region of the TAAI b gene.
Nucleotide sequences encofnpassed by tlae present invention
The present invention provides a polynucleotide molecule comprising nucleotide
sequences derived from the TAAl family. The genetic sequences encompassed
by the present invention include, but are not limited to, TAAl a cDNA (SEQ ID
NO: 1 ), TAAI b cDNA (SEQ ID NO: 3), TAAI c cDNA (SEQ ID NO: 5), TAAl a
genomic DNA (SEQ ID NO: 7), TAAl b genomic DNA (SEQ ID NO: 8), and
TAAI c genomic DNA (SEQ ID NO: 9).
Whilst the present invention discloses polynucleotide sequences for three
closely
related genes, homologous nucleotide sequences encoding peptides with
significant amino acid sequence identity to those encoded by SEQ ID NOS: 1, 3,
5, 7, 8, and 9, can be readily obtained in accordance with the teachings of
the
present application (and references disclosed herein), and are encompassed
within
the scope of the present invention. In this regard, nucleotide sequences of
the
present invention can be used to produce (degenerate) nucleotide probes, for
the
19
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
purposes of screening cDNA and genomic DNA libraries of various plant
species. Related techniques are well understood in the art, for example as
provided in Sambrook et al., Molecular Cloning: A Laboratory Manual, Cold
Spring Harbor Press, Cold Spring Harbor, N.Y.(1989). In this way, sequences
homologous to those of the present application are readily obtainable. For
this
reason, it is the intention of the present invention to encompass
polynucleotide
molecules comprising DNA sequences that encode peptides with significant
sequence identity to those disclosed in the present application, wherein SEQ
ID
NOS: 1, 3, 5, 7, 8, or 9, or parts thereof, may be utilized as polynucleotide
probes
to search for and isolate homologous polynucleotide molecules. Moreover,
polynucleotides encoding proteins with significant sequence identity to those
of
the present application are expected give rise to similar protein products
with
similar biochemical characteristics, to those described in the present
application.
Indeed, such techniques were used by the inventors to isolate the various TAAI
cDNA and genomic DNA homologous sequences disclosed herein. More details
in this regard are provided in the examples.
The present invention therefore encompasses DNA sequences obtained by
techniques known in the art for isolating homologous DNA sequences, wherein
the techniques utilize degenerate oligonucleotide probes derived from a
sequence
selected from SEQ ID NO:1, 3, 5, 7, 8, and 9, or parts thereof. The degree of
amino acid sequence identity will vary for each identified sequence. It is the
intention of the present invention to encompass polynucleotide sequences
comprising at least 50% sequence identity with regard to the peptide sequences
encoded by the corresponding polynucleotides. Without wishing to be bound by
theory, it is generally expected in the art that enzymes with at least 50%
identity
can have enzymatic activities that are similar in scope. In this regard, the
essential structural features of the enzyme are preserved to scaffold the
conformation of the catalytic site of the enzyme. Therefore, the present
invention
encompasses polynucleotide molecules derived by screening genomic and cDNA
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
libraries of plant types including wheat and other species, using degenerate
DNA
probes derived from the sequences disclosed in the present application. Such
species include, but are not restricted to: rye, barley, rice and other
grasses, and
monocots such as maize, and lily.
The present invention also encompasses polynucleotide sequences obtained by
screening DNA libraries using degenerate oligonucleotide probes derived from
the polynucleotides of the present invention, wherein the sequences encode
peptides comprising at least 70% amino acid sequence identity to peptides
encoded by SEQ ID NOS: l, 3, 5, 7, ~, and 9. In this regard, homologous
proteins with at least 70% predicted amino acid sequence identity are expected
to
encompass proteins with similar fatty acyl Co-A reductase activity as those
defined by the present invention, but possibly with altered substrate
specificity.
Such proteins may be derived from related species of plant.
The present invention also encompasses polynucleotide sequences encoding
peptides comprising at least 90%, 95% or 99% sequence identity to the peptides
encoded by SEQ ID NOS: 1, 3, 5, 7, 8, and 9. This class of related proteins is
intended to include close gene family members with very similar or identical
catalytic activity. In addition, peptides with 90%, 95% or 99% amino acid
sequence identity may be derived from functional homologues of similar species
of plant, or from directed mutations to the sequences disclosed in the present
application.
Isolation of TAA1 cDNA and genomic DNA laonaologues
With the provision of several TAAI cDNA and genomic DNAs, the polymerase
chain reaction (PCR) may now be utilized in a preferred method for
isolating further TAAl homologous nucleotide sequences from wheat and other
species of plant. PCR amplification of the TAAI cDNA sequence may be
accomplished either by direct PCR from a plant cDNA library or by
21
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
Reverse-Transcription PCR (RT-PCR) using RNA extracted from plant cells as a
template. Methods and conditions for both direct PCR and RT-PCR are known
in the art and are described in numerous standard textbooks. Similarly, the
TAA1 genomic sequences may be amplified directly from genomic DNA
extracted from plants, or from plant genomic DNA libraries. Amplification may
be used to obtain the full length cDNA or genomic sequence, or may be used to
amplify selected portions of these molecules (for example for use in antisense
constructs).
Moreover, the well known technique of chromosome walking can be readily used
to isolate regions of genomic DNA that are 5' or 3' to the coding region of
the
gene. The technique of chromosome walking is described, for example, in
Sambrook et al., Moleculao Cloning: A Laboratory Manual, Cold Spring Harbor
Press, Cold Spring Harbor, N.Y.(1989). The present disclosure includes
analysis
of a region upstream of to the TAAI b genomic DNA start codon, that is
suitable
for use as an anther specific promoter.
The selection of PCR primers will be made according to the portions of the
TAAI
nucleic acids which are to be amplified, including full-length TAAI clones.
Variations in amplification conditions may be required to accommodate primers
of differing lengths; such considerations are well known in the art.
Oligonucleotides which are derived from the TAAI nucleic acid sequences
described herein, and which are suitable for use as PCR primers to amplify
additional TAAl nucleic acid sequences are encompassed within the scope of the
present invention. Preferably, such oligonucleotide primers will comprise a
sequence of 15-20 consecutive nucleotides of the TAAl nucleic acid sequences.
To enhance amplification specificity, primers comprising at least 20-30
consecutive nucleotides of these sequences may also be used.
22
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
With the provision herein of the TAAl nucleic acid sequences, the cloning by
standard methodologies of corresponding cDNAs and genes from other ecotypes
and plant species, as well as polymorphic forms of the disclosed sequences is
now enabled. Thus, the present invention includes methods of isolating a
nucleotide sequence encoding a TAA1 enzyme from a plant. Both conventional
hybridization and PCR amplification procedures may be utilized to clone such
sequences. Common to both of these techniques is the hybridization of probes
or
primers derived from the disclosed TAAI nucleic acid sequences to a target
nucleotide preparation, which may be, in the case of conventional
hybridization
approaches, a cDNA or genomic library or, in the in the case of PCR
amplification, extracted genomic DNA, mRNA, a cDNA library or a genomic
library.
Direct PCR amplification may be performed on cDNA libraries prepared from
the plant species in question, or RT-PCR may be performed using mRNA
extracted from the plant cells using standard methods. PCR primers will
comprise
at least 15 consecutive nucleotides of the TAAl nucleic acid sequences. One of
skill in the art will appreciate that sequence differences between the
disclosed
TAAI nucleic acid sequences and the target gene to be amplified may result in
lower amplification efficiencies. To compensate for this, longer PCR primers
or
lower annealing temperatures may be used during the amplification cycle. Where
lower annealing temperatures are used, sequential rounds of amplification
using
nested primer pairs may be necessary to enhance specificity.
Genef~ation of TAAl variants and mutants
For conventional hybridization techniques, the hybridization probe is
preferably
labeled with a detectable label such as a radioactive label, and the probe is
of at
least 20 nucleotides in length. As is well known in the art, increasing length
of
hybridization probes tends to give enhanced specificity. The labeled probe
23
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
derived from, for example, the TAA1 cDNA sequence may be hybridized to a
plant cDNA or genomic library and the hybridization signal detected using
means
known in the art. The hybridizing colony or plaque (depending on the type of
library used) is then purified and the cloned sequence contained in that
colony or
plaque isolated and characterized.
It will also be understood to a person of skill in the art that site-directed
mutagenesis techniques are readily applicable to the polynucleotide sequences
of
the present invention. Related techniques are well understood in the art, for
example as provided in Sambrook et al., Molecular Clof2ing: A Laboratory
Mafzual, Cold Spring Harbor Press, Cold Spring Harbor, N.Y.(1989). In this
regard, the present invention teaches the isolation and characterization of
the
DNA sequences as provided as SEQ ID NOS: 1, 3, 5, 7, 8, and 9. However, the
present invention is not intended to be limited to these specific sequences.
Numerous directed mutagenesis techniques would permit the non-informed
technician to alter one or more residues in the nucleotide, thus changing the
subsequently expressed polypeptide sequences. Moreover, commercial 'kits' are
available from numerous companies that permit directed mutagenesis to be
carried out (available for example from Promega and Biorad). These include the
use of plasmids with altered antibiotic resistance, uracil incorporation and
PCR
techniques to generate the desired mutations. The mutations generated may
include point mutations, deletions and truncations as required. The present
invention is therefore intended to encompass corresponding mutants of the TAA1
cDNA and genomic DNA sequences disclosed in the present application.
The mutated variants of the sequences of the present application are predicted
to
include enzymes with reduced or increased fatty acyl Co-A reductase activity,
as
well as altered substrate specificity. Such mutants may confer advantageous
properties to subsequently transformed transgenic cell lines and plants. For
example, a transgenic plant comprising a construct overexpressing an inactive
24
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
mutant of the enzymes of the present invention can be expected to have a
significantly altered profile of lipid constituents, including a possible
reduction in
fatty alcohol content. In contrast, the expression of mutant fatty acyl Co-A
reductase enzymes with increased catalytic turnover are expected to give rise
to
transgenic plants with an high level of fatty alcohols. Mutant fatty acyl Co-A
reductase enzymes with altered substrate specificity will likely be useful in
altering the relative quantities of lipid metabolism products generated in a
correspondingly transformed plant, or altering the distribution of the lipid
metabolism products within the organs of the plant.
Genenatiota of eofastructs comprising TAA1 sequence
The polynucleotide sequences of the present invention can be ligated into
suitable
vectors before transfer of the genetic material into plants. For this purpose,
standard ligation techniques that are well known in the art may be used. Such
techniques are readily obtainable from any standard textbook relating to
protocols
in molecular biology, and suitable ligase enzymes are readily available from
commercial sources. A number of recombinant vectors suitable for stable
transfection of plant cells or for the establishment of transgenic plants have
been
described, which are also readily available from commercial sources.
Typically,
plant transformation vectors include one or more cloned plant genes (or cDNAs)
under the transcriptional control of 5' and 3' regulatory sequences and a
dominant
selectable marker. Such plant transformation vectors typically also contain a
promoter regulatory region (e.g., a regulatory region controlling inducible or
constitutive, environmentally- or developmentally-regulated, or cell- or
tissue-
specific expression), a transcription initiation start site, a ribosome
binding site,
an RNA processing signal, a transcription termination site, andlor a
polyadenylation signal.
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
As noted above, the particular arrangement of the TAAI nucleic acid in the
transformation vector will be selected according to the expression of the
nucleic
acid desired.
Where enhanced fatty alcohol synthesis is desired, the TAAI nucleic acid may
be
operably linked to a constitutive high-level promoter such as the CaMV 35S
promoter. Modification of fatty alcohols synthesis may also be achieved by
introducing into a plant a transformation vector containing a variant form of
the
TAAI nucleic acid, for example a form which varies from the exact nucleotide
sequence of the TAAI nucleic acid, but which encodes a protein that retains
the
functional characteristic of the TAA1 protein, i.e., fatty acyl Co-A reductase
activity.
In contrast, a reduction of fatty alcohol synthesis may be obtained by
introducing
antisense constructs based on the TAAI nucleic acid sequence into plants. For
antisense suppression, the TAAI nucleic acid is arranged in reverse
orientation
relative to the promoter sequence in the transformation vector. The introduced
sequence need not be the full length TAAI nucleic acid, and need not be
exactly
homologous to the TAAl nucleic acid. Generally, however, where the introduced
sequence is of shorter length, a higher degree of homology to the native TAAI
sequence will be needed for effective antisense suppression. Preferably, the
introduced antisense sequence in the vector will be at least 30 nucleotides in
length, and improved antisense suppression will typically be observed as the
length of the antisense sequence increases. Preferably, the length of the
antisense
sequence in the vector will be greater than 100 nucleotides. Transcription of
an
antisense construct as described results in the production of RNA molecules
that
are the reverse complement of mRNA molecules transcribed iom the
endogenous TAAI gene in the plant cell. Although the exact mechanism by which
antisense RNA molecules interfere with gene expression has not been
elucidated,
it is believed that antisense RNA molecules bind to the endogenous mRNA
26
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
molecules and thereby inhibit translation of the endogenous mRNA or trigger
the
degradation of mRNA, or inhibit transcription by causing methylation of the
gene. A variation of the antisense suppression includes RNAi strategy as
published in the literature under various names such as double stranded
(dsRNA)
RNA suppression.
Suppression of endogenous TAAI gene expression can also be achieved using
ribozymes. Ribozymes are synthetic RNA molecules that possess highly specific
endoribonuclease activity. The production and use of ribozymes are disclosed
in
U.S. Pat. Nos. 4,987,071 to Cech and 5,543,508 to Haselhoff, which are hereby
incorporated by reference. The inclusion of ribozyme sequences within
antisense
RNAs may be used to confer RNA cleaving activity on the antisense RNA, such
that endogenous mRNA molecules that bind to the antisense RNA are cleaved,
which in turn leads to an enhanced antisense inhibition of endogenous gene
expression.
Constructs in which the TAAI nucleic acids (or variants thereon) are over-
expressed may also be used to obtain co-suppression of the endogenous TAAI
gene in the manner described in U.S. Pat. No. 5,231,021 to Jorgensen. Such co-
suppression (also termed sense suppression) does not require that the entire
TAAI
nucleic acid be introduced into the plant cells, nor does it require that the
introduced sequence be exactly identical to the TAA1 nucleic acid. However, as
with antisense suppression, the suppressive efficiency will be enhanced as (1)
the
introduced sequence is lengthened and (2) the sequence similarity between the
introduced sequence and the endogenous TAAl gene is increased.
Transfornaatiou of TAA1 constructs
The present invention also encompasses a plant cell transformed with a
nucleotide sequence of the present invention, and as well as plants derived
from
27
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
propagation of the transformed plant cells. Numerous methods for plant
transformation have been developed, including biological and physical, plant
transformation protocols. See, for example, Miki et al., "Procedures for
Introducing Foreign DNA into Plants" in Methods in Plant Molecular Biology
and Biotechnology, Glick, B. R. and Thompson, J. E. Eds. (CRC Press, Inc.,
Boca Raton, 1993) pages 67-88. In addition, expression vectors and in vitro
culture methods for plant cell or tissue transformation and regeneration of
plants
are available. See, for example, Gruber et al., "Vectors for Plant
Transformation"
in Methods in Plant Molecular Biology and Biotechnology, Glick, B. R. and
Thompson, J. E. Eds. (CRC Press, Inc., Boca Raton, 1993) pages 89-119.
The following are examples, and are not limiting:
A. Agrobacterium-mediated Transformation: One method for introducing an
expression vector into plants is based on the natural transformation system of
Agrobacterium. See, for example, Horsch et al., Science 227: 1229 (1985). A.
tumefaciens and A. rhizogenes are plant pathogenic soil bacteria which
genetically transform plantcells. The Ti and Ri plasmids of A. tumefaciens and
A. rhizogenes, respectively, carry genes responsible for genetic
transformation of
the plant. See, for example, Kado, C. L, Crit~ Rev. Plant. Sci.lO: 1 (1991).
Descriptions of Agrobacterium vector systems and methods for Agrobacterium-
mediated gene transfer are provided by Gruber et al., supra, Miki et al.,
supra,
and Moloney et al., Plant Cell Reports 8: 238 (1989). Bechtold et al., C. R.
Acad.
Sci. Paris Life Sciences, 316:1194-9 (1993).
B. Direct Gene Transfer: Several methods of plant transformation, collectively
referred to as direct gene transfer, have been developed as an alternative to
Agrobacterium-mediated transformation. A generally applicable method of plant
transformation is microprojectile-mediated transformation wherein DNA is
carried on the surface of microprojectiles measuring 1 to 4 µm. The
expression
28
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
vector is introduced into plant tissues with a biolistic device that
accelerates the
microprojectiles to speeds of 300 to 600 m/s which is sufficient to penetrate
plant
cell walls and membranes. Sanford et al., Part. Sci. Technol. 5: 27 (1987),
Sanford, J. C., Trends Biotech. 6: 299 (1988), Klein et al., Bio/Technology 6:
559-563 (1988), Sanford, J. C., Physiol Plant 79: 206 (1990), Klein et al.,
Biotechnology 10: 268 (1992). See also U.S. Pat. No. 5,015,580 (Christou, et
al),
issued May 14, 1991; U.S. Pat. No. 5,322,783 (Tomes, et al.), issued Jun. 21,
1994.
C. Other methods: Another method for physical delivery of DNA to plants is
sonication of target cells. Zhang et al., Bio/Technology 9: 996 (1991).
Alternatively, liposome or spheroplast fusion have been used to introduce
expression vectors into plants. Deshayes et al., EMBO J., 4: 2731 (1985),
Christou et al., Proc Natl. Acad. Sci. U.S.A. 84: 3962 (1987). Direct uptake
of
DNA into protoplasts using CaCl2 precipitation, polyvinyl alcohol or poly-L-
ornithine have also been reported. Hain et al., Mol. Gen. Genet.199: 161
(1985)
and Draper et al., Plant Cell Physiol.23: 451 (1982). Electroporation of
protoplasts and whole cells and tissues have also been described. Donn et al.,
In
Abstracts of VIIth International Congress on Plant Cell and Tissue Culture
IAPTC, A2-38, p 53 (1990); D'Halluin et al., Plant Cell 4: 1495-1505 (1992)
and
Spencer et al., Plant Mol. Biol. 24: 51-61 (1994).
Following transformation of target cells) or tissues, expression of the above-
described selectable marker genes allows for preferential selection of
transformed
cells, tissues and/or plants, using regeneration and selection methods now
well
known in the art.
The foregoing methods for transformation would typically be used for producing
a transgenic variety. The transgenic variety could then be crossed, with
another
29
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
(non-transformed or transformed) variety, in order to produce a new transgenic
variety.
Alternatively, a genetic trait which has been engineered into a particular
line
using the foregoing transformation techniques could be moved into another line
using traditional backcrossing techniques that are well known in the plant
breeding arts. For example, a backcrossing approach could be used to move an
engineered trait from a public, non-elite variety into an elite variety, or
from a
variety containing a foreign gene in its genome into a variety or varieties
which
do not contain that gene. As used herein, "crossing" can refer to a simple X
by Y
cross, or the process of backcrossing, depending on the context. Once a
transgenic plant has been established, it is important to determine the
phenotype
of the seeds of the plant.
Accordingly, in a preferred embodiment of the invention a method is provided
for
modifying the seed of a plant comprising the steps of
(a) introducing into a plant cell capable of being transformed and
regenerated into a whole plant a construct comprising, in addition to the DNA
sequences required for transformation and selection in plants, a nucleotide
. sequence in accordance with the nucleotide sequences encompassed by the
present invention, operably linked to a promoter; and
(b) recovery of a plant which contains the nucleotide sequence.
The present invention therefore encompasses the transformation of a variety of
plant species, including woody, non-woody, fruit bearing and grass species,
with
the DNA sequences disclosed. Particularly preferred varieties include
crucifier
crops, tobacco, wheat, corn, sugar cane, and apple. The present invention is
particularly considered to be useful in the generation of modified fruits such
as
apples, since increased expression of fatty acyl Co-A reductase enzymes of the
present invention is expected to increase the fatty alcohol concentration in
the
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
fruits, thus providing the fruits with a more waxy texture, and a more
aesthetically pleasing coating.
Fatty acid analysis and purification
Once a transgenic plant has been established, it is important to determine the
fatty
alcohol content of the plant, or various plant organs. For this purpose,
several
techniques are known in the art to for the analysis of the chemical content of
plant material, and in particular, the lipid and fatty alcohol content of the
plant.
These techniques include Gas Chromatography (GC), high performance liquid
chromatography, and MS-GC, as well as other techniques that are familiar to
those of skill in the art. Moreover, the fatty alcohol products may be
extracted
from the plant by any one of a range of techniques that are well known in the
art
for the purposes of lipid extraction.
One example of fatty alcohol purification from plant materials is outlined in
the
Experimental Procedures where fatty alcohols. were purified for Gas
Chromatography analysis. An alternative method was published by Miwa, T.K.
1971. Journal of The American Oil Chemists' Society. 48:259-264, in relation
to
jojoba oil analysis.
Pf-oductiofa of ~ecofrzbinant TAAI protein usifag heter~ologous expression
systerras
Many different expression systems are available for expressing cloned nucleic
acid molecules. Examples of prokaryotic and eukaryotic expression systems that
are routinely used in laboratories are described in Chapters 16-17 of Sambrook
et
al. (1989), which are herein incorporated by reference. Such systems may be
used
to express a TAAl protein or derivatives thereof at high levels to facilitate
purification and functional analysis of the enzyme. Apart from permitting the
activity of the enzyme to be determined (which is particularly useful to
assess the
31
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
activity of homologous and derivative proteins), heterologous expression
facilitates other uses of the purified enzyme. For example the purified enzyme
produced by recombinant means may be used to synthesize fatty alcohols and
other fatty acid metabolites in vitro, particularly radio- or fluorescent-
labeled
forms of fatty alcohols and metabolites. These molecules may be used as
tracers
to determine the location in plant tissues and cells of fatty alcohols and
their
metabolites. The purified recombinant enzyme may also be used as an
immunogen to raise enzyme-specific antibodies. Such antibodies are useful as
both research reagents (such as in the study of fatty alcohol regulation in
plants)
as well as diagnostically to determine expression levels of the enzyme in
agricultural products, including pollen.
By way of example only, high level expression of the TAAl protein may be
achieved by cloning and expressing the cDNA in yeast cells using the pYES2
yeast expression vector (Invitrogen, San Diego, Calif.). Secretion of the
recombinant TAA1 from the yeast cells may be achieved by placing a yeast
signal sequence adjacent to the TAAI coding region. A number of yeast signal
sequences have been characterized, including the signal sequence for yeast
invertase. This sequence has been successfully used to direct the secretion of
heterologous proteins from yeast cells. Alternatively, the enzyme may be
expressed at high level in standard prokaryotic expression systems, such as E.
coli.
Homology of TAAI nucleic acids to other plant genes
It is predicted that the TAAl genes are the first anther-specific genes in
wheat to
be reported. TAA1 appears to be related to the jojoba FAR (reported by Metz et
al., 2000). Both have an Mr of ~ 58,000, and share ~44% as identity and ~63%
similarity. This FAR is the only biochemically characterized enzyme for which
a
deduced structure is available. It belongs to the category of alcohol-forming
32
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
FARs that produces fatty alcohols directly from fatty acyl CoA, one of the two
penultimate substrates in wax biosynthesis (I~olattukudy, 1970). The other
category of fatty acyl CoA reductases is smaller and produces aldehydes (Mr ~
30
kDa) (Vioque and I~olattukudy, 1997), and thus TAA1 is further distinct from
these. In jojoba, FAR was isolated from developing seeds. It is not clear if
it is
also expressed in the anther and other tissues.
The predicted TAA1 gene products share a lesser, yet significant homology with
two known anther-specific genes, the Arabidopsis MS2 gene (Aarts et al., 1997)
and the Brassica MS2 gene (Aarts et al., 1997; Hodge et al., 1992). Both of
them
also share significant homology with FAR (~40% identity and ~59% similarity).
Any functional implication of TAA1 and MS2 relationship must be considered in
light of the observation that TAA1 shows a greater relationship to the jojoba
FAR. The A~abidopsis MS2 gene is required for pollen development (Aarts et
al.,
1993; Aarts et al., 1997). The post-meiotic, tapetal expression of MS2 in
Arabidopsis and that of TAAl in wheat are almost identical. Thus, despite the
deviation in the deduced primary structures, both appear to be functionally
similar: While a partial redundancy of MS2 function has not been ruled out
(Aarts
et al., 1993; Aarts et al., 1995), the Ai°abidopsis genome database
(Arabidopsis
thaliana geneome CD) was searched using the MS2 as sequence. Excluding the
MS2 itself, seven hypothetical proteins including two named male sterility 2 -
like, four called acyl coA reductase-like, and one putative protein were
found.
These proteins are 37-42% identical to and 59-65% similar with the MS2 as
sequence. These proteins range from 409 to 527 as in length and also share as
homology to TAAl at similar levels. Moreover, BLASTN searches using the
MS2 cDNA did not retrieve any more sequences than itself, indicating that at
the
nt level, the MS2 does not share significant homology with these predictive
genes. In contrast, these genes seem to be more closely related to each other
as
they conserve significant similarities both at the as level and the nt level
(data not
shown). However, it is not know if these genes function in pollen development.
33
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
Aarts et al. (1993) identified a short segment of homology between a wheat
mitochondria) sequence (Spencer et al., 1992) and the Arabidopsis MS2 at the
deduced as sequence level. This (93/153 aa) occurred over two stretches of the
mitochondria) sequence with an unrelated sequence flanked by hallmarks of
nuclear splice junctions that connected the two parts. Since splicing in
mitochondria) transcripts follows a different scheme, Aarts et al. (1997)
proposed
that the mitochondria) sequence is of nuclear origin that had recently
migrated to
the organellar genome. Were this the case, there should be a greater homology
between the nt sequence of TAA1, a wheat genomic sequence, and the wheat
mitochondria) sequence. Instead, there was no significant homology even though
the deduced TAAla protein did show a similar rate of homology to a
discontiguously translated polypeptide of the mitochondria) sequence at the
same
region as MS2 (93/153 aa). Therefore, the corresponding mitochondria) sequence
is more likely due to the biochemical convergence in evolution than a
genealogical relationship.
This application -provides convincing evidence that TAAl is an FAR. FAR
converts fatty acyl coA to fatty alcohol (Kolattukudy, 1970; Kolattukudy and
Rogers, 1986; Lardizabal et al., 2000; Metz et al., 2000). This is the first
plant
tapetum-specific gene which is enzymatically identified to be associated with
lipid and wax biosynthesis. Pollen grains are coated with two layers of
lipidic
structures, i.e., the pollen outer wall (exine) and the pollen outmost coating
(tryphine) that is overlaid on exine. Sporopollenin is the major constituent
of
exine and contains metabolites derived from long chain fatty acids and
phenylpropanoids (reviewed by Scott, 1994). Although long chain fatty lipids
seem to be definitely required for the synthesis of sporopollenin, how
sporopollenin is polymerized and what precursors participate in the
polymerization still remain unclear. In crucifer plants such as Arabidopsis
and
Brassica, the deposition of the exine takes place from the completion of
meiosis
34
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
II, through the tetrad and ring-vacuolate stages, to the time of the first
pollen
mitosis (Piffanelli et al., 1998). During this process, the tapetum is
performing
very active lipid biosynthesis (Piffanelli et al., 1998). So, it is logically
assumed
that the tapetum plays a major role in exine formation. Prior to this report,
the
only functionally characterized anther tapetum-specific gene was the
Arabidopsis
MS2 gene, whose expression pattern is concomitant with the formation of the
pollen outer cell wall. Disruption of the MS2 gene with a transposon results
in
male sterility. Pollen development in the nzs2 mutant shows most dramatic
defect
upon release from tetrads (Aarts et al., 1997). These pollens lack exine.
Though
the enzymatic nature of its encoded polypetide has yet to be identified, it
shares
40% as sequence identity with FAR. The present application includes evidence
that TAAl has the MS2 expression pattern during formation of microspore exine
and its encoded polypeptide has fatty alcohol forming capacity. Thus, as
proposed by Scott (1994) and Aarts et al. (1997), fatty alcohols, the TAA1 or
MS2 enzymatic products are likely to be the precursors for sporopollenin
polymerization.
The outmost layer of the pollen grain is the pollen coating or tryphine
derived
from two tapetum-specific lipid-rich organelles, elaioplasts and tapetosomes
(Hernandez-Pinzon et al., 1999; Piffanelli et al., 1998; Ting et al:, 1998; Wu
et
al., 1997). The former is a plastid with triacylgycerol (TAG) and neutral
esters,
and the later is a lipid body containing neutral lipids including TAG and wax
esters, and also oleosin-like proteins. The main functions of tryphine include
pollen-stigma recognition and subsequent pollen hydration (Piffanelli et al.,
1998). Analysis of tryphine lipid fractions shows that the tryphine lipids
contain
TAG, triterpene esters, sterol esters and very long-chain wax esters (Bianchi
et
al., 1990; Preuss et al., 1993). Of these compounds, long-chain lipids and
linear
waxes are thought to be essential for the functions of the pollen coating
(Lemieux, 1996; Mariani and Wolters-Arts, 2000; Negruk et al., 1996; Preuss et
al., 1993). This is in agreement with the previous observations that wax
defective
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
mutants such as the cerl, ce~2, cer3, cer6, cef~~ and cerl0 mutants
inArabidopsis
exhibit conditional male sterility (Hannoufa et al., 1993; Koornneef et al.,
1989).
Of these mutants, some have tryphine with smaller lipid droplets than wild-
type
pollen and some have tryphine without lipids. Recently, a complementation
experiment of the cer6 mutants by transgenically expressing the CER6 gene has
shown that the two phenotypic effects, i.e. wax defection and male sterility
cannot be rescued equally (Fiebig et al., 2000). CER6 is identical to CUTI
encoding an enzyme responsible for elongation of fatty acyl CoA, and silencing
CUTI induces waxless stem and male sterility (Millar et al., 1999).
Interestingly,
some fertility-restored lines (CER6 transformants) of the cey~6 mutnats still
show
wax-defective stem. Analysis on very long fatty lipids reveals that low
amounts
of long fatty lipids are sufficient for pollen hydration and germination,
suggesting
that this remarkable difference results from the different requirements for
CER6
activity on stems and the pollen coating (Fiebig et al., 2000). Apart from the
ingenious very long chain lipids, a class of exogenous TAGS which is absent in
the pollen coating of Arabdopsis also can rescue the fertility of an Af-
abidopsis
wax-defective mutant (Wolters-Arts et al., 1998). These findings raise a
possibility that the tryphine function is dependent on the nature of the
mixture of
lipids including TAG, very long fatty acids and waxes in the tryphine. The
composition of and the relative amounts of each species of the lipid pool
rather
than a single macromolecule determine a functional pollen coating.
Apart from their. presence in pollen lipidic structures (Bianchi et al.,
1990), linear
wax esters are also ubiquitously present in the plant cuticle (Piffanelli et
al.,
1998; Post-Beittenmiller, 1996), indicating plant encoded FAR genes have
evolved divergent expression mechanisms. Indeed, an alcohol-forming FAR was
previously purified from pca leaves (Vioque and Kolattukudy, 1997). Since the
lipid composition of tryine is significantly different from that of the
cuticle and
even that of the intracellular contents of the pollen grains in the same
plants
(Bianchi et al., 1989; Bianchi et al., 1990; Piffanelli et al., 1997),.there
must be a
36
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
mechanism by which the wax biosynthesis is spatially and temporally regulated.
Searches of the Arabidopsis genome database identify 7 FAR-like hypothetical
proteins excluding the MS2. It would be possible to investigate if these FAR-
like
genes contribute to divergent lipid biosynthesis in plants. Alternatively,
isolation
and characterization of more FAR genes will definitely assist in understanding
this complicate regulation mechanism.
The present application provides convincing evidence that TAAl can reduce long
chain acyl CoA to fatty alcohols. Fatty alcohols can be further esterified
with
fatty acids to generate linear wax esters. Transgenic plants that overexpress
TAAl proteins via their natural (tapetum specific) promoters are predicted to
have an increased consumption of fatty acyl CoA by TAA1 in the tapetum, which
in turn may impact upon lipid-related biosynthesis in the anthers. In this
regard,
alteration of the lipid composition in the tapetum by TAA1 may be also
required
for the tryphine development to assure its recognition and hydration function.
Corresponding effects upon lipid metabolism are predicted to occur if the TAA1
protein is overexpressed in a plant organ other than the tapetum. For this
reason,
the --present invention- encompasses DNA constructs, and the corresponding
transgenic plants transformed with the constructs, wherein the over- or under-
expression of TAA1-like proteins gives rise to altered lipid metabolism by
virtue
of an abnormal level of fatty acyl Co-A. Such modifications to lipid
metabolism
can have profound effects upon phenotype, developmental, reproductive, growth
and structural characteristics of the plant. Moreover, the nature and impact
of
these effects are expected to depend upon the extent of TAA1 expression, and
the
localisation of TAAl expression to specific plant organs. Both of these
factors
are regulated in part the strength and specificity of the promoter.
Specific embodiments of the present invention are illustrated by way of the
following examples:
37
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
Example 1 - The TAAl group of genes in wheat - identification and structural
characteristics of the cDNA clones
RT-PCR experiments were conducted using an anther-specific cDNA library with
primers specific for the rice PSI gene (Zou et al., 1994). At a moderate
annealing
temperature of 43°C a 0.7-kb amplicon was obtained from mRNA isolated
from
anther but not from root, stem, leaf, glume and pilea tissues. DNA sequencing
of
the amplicon en masse gave an unambiguous result indicating that the PCR
product was composed of a homogeneous sequence within the detection limits of
sequencing reaction. Since this amplicon was specific to anther mRNA, a full-
length cDNA clone encompassing the amplicon sequence was obtained by 5'-
and 3'- RACE. On probing an anther cDNA library with the full-length cDNA,
12 clones were identified and these were grouped into three groups according
to
their restriction pattern (data not shown). The longest clones from each group
were studied further. 5'-RACE experiments did not produce further extensions,
suggesting that the longest clones were full-length cDNA clones and hereafter
referred as TAAla, TAAIb and TAAlc (SEQ ID NOS: 1, 3, and 5 respectively) .
Both TAAI a cDNA (GenBank accession number AJ459249) and TAA1 c cDNA
(GenBank accession number AJ459253) clones have a predicted open reading
frame (ORF) of 1524 nucleotides (nt) encoding 507 amino acids (aa) but with
different lengths of 5' UTRs (TAAI a: 69 nt and TAAI c: 93 nt) (Figure 1 a).
The
TAA1 b cDNA (GenBank accession number AJ459251) ORF has a slightly larger
ORF (1569 nt) encoding 522 amino acids and a 5' UTR of 73 nt. The putative
polyadenylation signals, AATAA or TATAA, were found in the 3' UTR of all
three TAAl cDNAs. Database analysis on the deduced as sequences encoded by
these three genes revealed that they all shared similarity to the fatty acyl-
coenzymeA reductase (FAR) gene of jojoba (Metz et al., 2000) and the A.
thaliatza MSS gene (Aarts et al., 1993) (see later re.: Figure 5). On further
examination, the primers initially used contained at their 3' end a high level
of
3S
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
identity to two portions of the cDNA clone encompassing the 0.7 kb cDNA
fragment.
Example 2 - The TAAI group of genes in wheat - identification and structural
characteristics of the geraomic DNA clones
The genomic counterparts of the entire coding region of all three TAAl cDNAs
were obtained by PCR amplification of the genomic DNA with primers based on
the cDNAs. The genomic DNA sequences for TAAI a, TAAl b, and TAAl c are
shown in SEQ ID NOS. 7, 8, and 9 respectively. The TAAla genomic DNA has
been assigned Genbank accession number AJ459250, the TAAl b genomic DNA
has been assigned GenBank accession number AJ459252, and the TAAl c
genomic DNA has been assigned GenBank Accession number AJ459254
(genomic sequences submitted to GenBank include only the genomic DNA
regions encompassing coding sequence).
Nucleotide sequence analysis of the amplicons showed 7 introns interrupting
the
coding -regions in all three genes. The length and composition varied among
the
three genes. The most significant difference was in the length of the second
intron (1758 nt in TAAl b, but only 113 in TAAI a and 112 in TAAI c). The
position of the 4th intron in TAAl b also deviated substantially (Figure 1 a).
Example 3 - TAA1 genes are likely to exist as single copy per haploid genonze
Southern blot analysis of the wheats of different genetic constitution --
namely,
AABBDD, AABB, AA and DD-- revealed that the TAAI genes are likely to exist
as single copy per haploid genome. This interpretation was possible because of
the choice of restriction enzymes that either did not cut within the coding
sequence or cut only rarely and the use of the entire coding sequence of TAAI
a
cDNA as the probe. Despite this probe coverage, only one hybridization band
39
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
was observed in the diploids, two in the tetraploid and no more than 4 in the
hexaploid (Figure 1 b). Assuming that introns of any paralogs would have
caused a restriction polymorphism at this level, these results are consistent
with a
single copy gene per haploid genome equivalent. The presence of four bands in
the hexaploid blot is due to restriction within a TAAI gene (data not shown).
Example 4 - TAA1 expf°essiou is confined pf°inaaYily to the
anther tapetuna and
associated with microsporogenesis - molecular biology studies
The expression pattern of TAAI in wheat was determined by probing RNA blots
with the 0.7 kb amplicon of TAAla. The TAAla probe strongly hybridized only to
the anther mRNA, and did not show any hybridization with the root, stem, leaf,
ovary or glume transcripts (Figure 2 a). For enhanced detection, RT-PCR of
these RNA preparations was done with a primer pair designed to cover parts of
two exons with an intron in-between so as to discriminate amplicons of mRNA (~
0.4 kb) and genomic DNA origin (~ 0.8 kb). A strong band of ~ 0.4 kb was
detected in the anther sample, and there was a weak signal in the stem but not
in
other samples (Figure 2 b). The identity of the amplicon was confirmed by nt
sequencing. Thus, TAAI gene expression is specific to the anther tissue.
Example 5 - TAA1 expression is eonfined primarily to the ajzther tapetum and
associated with microsporogeTZesis - In situ hybridisatiofz studies
In situ RNA hybridization of transverse sections of flowers at various stages
of
development with an antisense probe (Figure 3 c and d) showed expression in
the
tapetum but not in ovary, epidermis, connective tissues and the filament at
various ages. With a sense probe, no significant signal was detected (Figure 3
a).
Generally, wheat anther development follows seven stages (Saini, 1984; Dorion
et al., 1996; Lalonde et al., 1997). These stages include pre-meiosis,
meiosis,
young microspore, vacuolated microspore (microspores irregularly shaped and in
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
contact with the tapetum, and microspore wall and pore formation in progress),
PGM1 (microspore nucleus divides to form vegetative and generative nuclei),
PGM2 (tapetal cell walls break down), and mature pollen grain. The onset of
TAAI transcription was not evident until the microspore separation occurred at
a
stage corresponding to the presence of a young microspore. From then on, TAA1
mRNA was predominantly distributed in tapetum cells and to a lesser extent in
some microspores (Figure 3 b and c). TAAI was strongly expressed at the
vacuolated microspore stage when microspore cell walls were evident. The
disappearance of TAA1 transcripts coincided with tapetmn degeneration (PGM2
stage). Thus, TAAl transcription is confined (with the exception of weak
expression in stem) to anthers, and within anthers it is localized in the
tapetum
from the point of the formation of young microspores to the degeneration of
the
tapetum (PGM2 stage).
To further test whether the TAAl gene product is also dominantly localized in
the
tapetum, the TAA1 specific poly-clonal antibodies were generated and used in
in
situ immunochemical analysis (Figure 4) of floral tissues. The results were
consistent with RNA in situ-hybridization, suggesting the TAA1 protein was
indeed produced in the tapetum as young microspores developed to the PGM2
. stage. There was also evidence of its production, albeit at a much lower
level, in
microspore cells at this stage. Other floral tissues did not show any
distinctive
signals. Taken together these data indicate that the TAAI gene products are
associated with microspore development.
Example 6 - Tlae TAAl gene pr°oduct shares hofnolog~~ with the
jojoba seed-
borne FAR and the Arabidopsis antherspecific MS2-encoded protein
Database analysis of TAAl was performed to search homologous genes and
explore its potential function. While BLAST searches (conducted in accordance
with Altschul et al., 1997) of GenBank with the ORF of TAAI a, TAAI b and
41
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
TAAI c did not identify any significant homologs, the deduced as sequence
(>84% similarity, >74% identity among the three TAAl peptide sequences) had
homologs from A. tlaaliana (MS2 and MS2-like; Aarts et al., 1993), Brassica
napus (B-MS2; Hodge et al., 1992) and Simnaondsia clZinensis (fatty acyl CoA
reductase gene (FAR); Metz et al., 2000). TAAI gene products were found to be
most similar to the jojoba (S. clzinensis) FAR and the Arabidopsis putative
MS2-
like protein (61-65% similarity and 42-46% identity), and to a lesser extent
to the
MS2 and the Brassica MS2 (54-57% similarity and 35 to 39% identity). Notably,
MS2 and its functional ortholog from B. napus (89% identical to MS2; Aarts et
al., 1997) have an additional stretch of 117-as at their amino-terminal region
in
comparison with TAA1, MS2-like and FAR. Of all these related gene products,
only FAR has been biochemically characterized (Metz et al., 2000). Thus, even
though the vt~heat TAAl gene products share a developmental connotation with
the Arabidopsis MS2 gene, TAAl bears a greater relationship to the
characterized
jojoba FAR (Figure 5 a and b). The latter is associated with accumulation of
storage lipids in seeds and thus presumably inconsequential to anther
development. The pair-wise alignment of the as sequence of the TAA1 a-encoded
polypeptides to that of the jojoba FAR was carried out to explore conserved
domains. There are two consensus regions containing more than 12 consecutive
amino acids (Fig 5 a). Interestingly, these two regions are located at the two
predicted transmembrane helices (Figure 5 a) (Metz et al., 2000). Further
examination of the corresponding regions on the other related gene products
revealed that the first putative transmembrane helix of FAR is globally
conserved
while the second one is not (data not shown).
Example 7 - Accufnulation of fatty alcohols in the TAA1 tnansgenic tobacco
seeds, and influence of TAAl expy~ession on plant phenotype
To initiate functional characterization of the TAAI gene in planta, the TAAI a
cDNA was cloned into a binary vector under the control of a napin promoter.
The
42
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
napin-TAA1 a chimeric gene was transformed into tobacco. Tobacco seeds
contain 30-43% oil and are rich in fatty acids. The potential TAA1 substrates,
fatty acyl coA, are actively synthesized in the developing seeds (Frega et
al.,
1991). Total fatty alcohols were extracted from transgenic seeds. GC analysis
on
fatty alcohol contents and compositions showed that the TAAl-encoded enzyme
significantly modified the pathway of fatty alcohol synthesis in the napin-
TAAl
transgenic seeds. The amounts of the eve major fatty alcohols, i.e., C18:1,
C20:1,
C22:1, C24:1 and C24:1 increased by 8.75%-357.47%, 57.78%-426.8.95%,
130.96%-307.72%; 145.00%-361.49% and 99.89%-5929.47%, respectively
(Figure 6 a).
Unexpectedly, the overexpression of the TAAI a gene in tobacco under the
control
of a 35S promoter results in significant changes to the phenotype of the
corresponding transgenic plants (Figure 7). In this regard, the modified
transgenic plants are significantly smaller, with shorter internodes, and
delayed
flowering. The expression of fatty acyl Co-A reductase in these plants
therefore
gives rise to considerable developmental alterations in the plant. This
provides
evidence that changes in lipid metabolism via altered expression of TAAl genes
can generate desirable changes to plant phenotype. Specifically, the reduction
in
internode length and reduction in overall size of the plants will render the
plants
less susceptible to wind damage. Moreover, the reduction in size of the plants
may permit the generation of dwarf plant specifies for horticultural purposes,
and
plants with increased wind resistance. The delay in flowering may also be a
desirable attribute for certain horticultural situations.
Example 8 - Accumulation of fatty alcohols frofn TAA1 expression in E. coli
To further verify TAA1's alcohol-forming ability, a bacterial expression
system
was employed. Previously this approach had been originally used to determine
the enzymatic activity of the FAR isolated from jojoba (Metz et al., 2000).
The
43
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
neutral components from bacterial cells with the control plasmid or with the
plasmid containing the coding region of the TAAI a cDNA under the control of
the T7 promoter were subjected to GC analysis. In the control bacterial cells,
there was one major peak observed. The nature of this compound was not clear.
In the bacterial cells expressing TAAla, two additional major peaks and one
additional minor peak were detected (Figure 6 b). Detention times of these
three
peaks were found to be identical to those of three authentic fatty alcohol
standards (C 14, myristi alcohol; C 16:0, hexadecyl alcohol; and C 18 :1,
oleyl
alcohol). The identity of the two major peaks (C16 and C18:1) was confirmed by
MS-GC while the C14 fatty alcohol was not due to its very low concentration.
These results suggest that TAA1 is a wheat FAR.
Example 9 - PROMOTER STUDIES - Tlae wheat TAAl b promoter retains its
spatial and temporal expy~ession specificity in a distant In0320COt and also
in a
dicot
A 1.6-I~b genomic segment upstream of the predicted start codon of the TAAI b
was isolated by genome walking (and given GenBank accession no: AJ488930).
Particle-bombardment of daylily, a distant monocot in the phylogeny of wheat,
with a construct containing GUS ORF immediately 3' to this fragment elaborated
~3 glucuronidase, as shown by histochemical staining, only in the anther
tapetum
and microspores (Figure 8); none of the other parts such as leaves, stems, and
the
anther epidermis and connective tissues showed GUS expression. Furthermore,
when anthers of various stages of development were bombarded, only those past
the tetrad formation showed GUS expression, and this result was consistent
with
the observations in wheat. In contrast, a CaMV 35S-GUS construct gave
expression in all tissues examined. Thus, the wheat promoter is likely to be
useful
to manipulate male gametophyte development in other important monocot crops.
44
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
The wheat TAAI promoter was also found to be functional in the anthers of
stably
transformed tobacco (Figure 9). GUS expression was absent in other floral
parts,
roots and stems. In contrast, and as expected, the CaMV 35S-GUS plants showed
expression in all these tissues, and an example of floral tissue is shown for
comparison (Figure 9 d). While GUS activity was evident in the epidermis,
connective tissues, the tapetum, microspores, and other tissues, the
expression in
the tapetum was not as strong as in the case of the TAAl b-GUS plant.
Interestingly, the heterologous promoter maintained its developmental
specificity
in transgenic tobacco. GUS staining was not detectable in very young anthers
at
the time of tetrad formation (Figure 9 a). According to Koltunow et al.
(1990),
this would be flower buds <12 mm. In flower buds of 14 to 24 mm (Stages 2 to
6), GUS activity was detected in the tapetum, and in buds >15 mm (Stage 4), a
strong GUS staining was also seen in microspores (Figure 9 b and c).
Collectively, these results show cross-functionality of any cis elements of
the
TAA1 promoter with tratzs-acting factors in daylily and tobacco.
Experimental procedures
Plafzt materials, DNA and RNA isolatioya
Hexaploid spring wheat (Triticuna aestivum L. cv. Karma, genomic complement
AABBDD), tetraploid wheat (Triticum turgidum L. cv. Sceptre, genomic
complement AABB), and two diploid wheat species (Triticum urartu, ssp.
Nigrum, genomic complement AA; Aegilops squarrosa, ssp. Tauschii, genomic
complement DD) were grown in an open filed or in a greenhouse under standard
conditions. Five-week-old seedlings were used for DNA isolation and for leaf,
root and stem RNA isolation. Anther, ovary, glume and pilea tissues were
collected 1-3 days prior to anthesis for RNA purification. DNA extraction was
carried out following the published protocol (Wang et al., 1998). Total RNA
was
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
extracted using TrizolTM reagent (Life Technologies/Gibco-BRL, Burlington,
Ontario) following the supplier's recommendations.
RT PCR, 5'- and 3'-RACE, ahd otlze~ RNA and DNA techfzologies
The nucleotide acid-related enzymes used were from Life Technologies
(Burlington, Canada) except otherwise stated. Five ~,g of total RNA derived
from
various tissues was used to synthesize the first strand cDNA by reverse
transcriptase using primers OL2707 (5'GACTACGTCGTCCAAGGCCG3'
SEQ ID NO: 10) and OL2708 (5'GTCGAACTGCTTGAGCAG CGC3' - SEQ
ID NO: 11). The PCR reactions were carried out with a Techne Genius DNA
thernlal cycler (Duxford, Cambridge, UI~) under the following conditions:
94°C
for 1 min, 43°C for 1 min, 72°C for 2 min, 35 cycles, followed
by 10 min's
incubation at 72°C. The amplified products were subjected to DNA
sequencing.
Based on obtained sequences, primers OL2881
(5'GCAGAACCTGACATACTTC3' - SEQ ID NO: 12) and OL2885
(5'GAGGCGGTACCT GAGCAT3' - SEQ ID NO: 13) were designed for RT-
PCR detection and genomic DNA amplification.
Antisense primers OL2884 (5'TTCGCATAGCCGATCACG3' - SEQ ID NO:
14) and OL2883 (AATGCCGGCCCTGGTAAG3' - SEQ ID NO: 15) and sense
primers OL2880 (5'CAGGTGGCCAAA CACATA SEQ ID NO: 16 and OL2881
(5'GCAGAACCTGACATACTTC - SEQ ID NO: 12) were designed for 5'- and
3'-RACE which were conducted using 5' and 3' RACE kits (Life Technologies,
Burlington, Ontario) following the manufacturer's protocol. The resulting cDNA
fragments were either directly subjected to DNA sequencing or cloned into a
T/A
vector using the Original TA Cloning KitTM (Invitrogen, Carlsbad, CA) for
sequencing.
46
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
RNA and DNA gel blot analyses were performed essentially as described (Hair et
al., 2000). The predicted coding region of TAAl a was used as a probe.
cDNA library constt-uction and screetzing
Wheat anther poly(A)+ RNA was isolated using an mRNA kit (Clontech, Palo
Alto, CA). Approximately 5 ~,g poly(A)+ RNA was used for cDNA library
construction using a ZAP cDNA Gigapack III Gold Cloning KitTM according to
the supplier's instructions (Stratagene, La Jolla, CA). The ligated vector was
packaged into phage particles using Gigapack III gold packaging extracts
(Stratagene, La Jolla, CA). A total of 2.2 x 10' primary pfu were obtained.
Library screening was conducted using the 5' RACE PCR cDNA fragment as a
probe to hybridize phage'plaques containing approximately 250, 000 recombinant
clones. The positive plaques were isolated and the phagemids were excised in
vivo from the Uni-ZAP XRTM vector using the ExAssist/SOLRTM system
(Stratagene, La Jolla, CA). The inserted cDNA sequences in the purified
phagemids were determined by DNA sequencing.
Production of polyclonal antibodies against TAAI
The entire coding region of TAA1 a was directionally cloned in-frame into the
BarnHl-EcoRI sites of plasmid pRSET A (Invitrogen, Carlsbad, California) to
make plasmid pTAA238. Fusion protein was expressed in E. coli strain
BL21(DE3)pLysS (Invitrogen). The TAA1 fusion protein was purified and
injected into rabbits following the procedures (Wang et al., 1999). Polyclonal
antibodies were harvested and purified as described (Wang et al., 1999).
In situ RNA hybridization and inamurao-cytolocalization
47
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
Plant materials were infiltrated overnight at 4°C in 4%
paraformaldehyde (PFA)
with a 100mM phosphate buffer pH 7.2. The fixed material were dehydrated in a
graded ethanol series and then embedded in paraffin (Paraplast plus x-tray.
Sections were cut at 8 ~.l.m thickness and mounted on glass slides
(SuperfrostTM
plus, Fisher Scientific, Nepean, Ontario). For in situ RNA hybridization,
probes
were prepared using MAXIscript T"" in vitro transcription kit and BrightStarTM
P
soralen-Biotin nonisotopic labeling kit (Ambion, Austin, Texas) according to
manufacture's protocols. A DNA fragment of 550 by of the TAAla cDNA
starting from the predicted start codon was directionally cloned into
pBluescriptTM II KS+ phagemid vector (Stratagene, La Jolla, CA) at the BarnHI-
.Jr'hoI sites to produce plasmid pTAA253. The antisense transcripts
synthesized in
vita°o by T3 polymerase using XbaI-linearlized plasmid pTAA253 as a
template
were used to detect the TAAl naRNA. The partial TAAI sense transcripts
generated by T7 polymerase using plasmid Jr'TZOI-linearized plasmid pTAA253 as
a terriplate were served as a control. In situ hybridization was carried out
essentially following the instructions of the mRNA locator-HybTM kit (Ambion).
For ifz situ immunological detection, slides mounted with fixed sections of
wheat
florets are incubated in the blocking solution containing 1:1000 TAAla immune
or pre-immune serum for overnight at 4 °C. Visualization of immuno-
reaction
was as described (Cho and Kende, 1998).
Tlector~ coizst~~uctioh and genetic transformation
Plasmid pRD400 (Dada et al., 1992) was modified by flipping-over the region
containing the polylinker and the NPT II gene cassette to generate a binary
transformation vector pAMW281. Two pieces of DNA fragments including a 2.4
kb fragment containing a CaMV 35S promoter and a uidA gene from plasmid
pRD410 (Datla et al., 1992) digested with HindIII and EcoRI, and a 0.7 kb
fragment containing a CaMV 35S terminator from plasmid pHS724 restricted
48
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
with EcoRI and KpfzI (Huang et al., 2000) were co-ligated into the backbone of
pAMW281 digested with Hif2dIII and Kp~I to produce plasmid pAMW287. A 1.4
kb napin promoter obtained from digestion of plasmid pJOY43 with HiudIII-
BamHI (Hair et al., 2000) and the 1.4 kb TAAl a entire coding region resulting
from plasmid pTAA238 restricted with BafnHI and EcoRI were co-ligated into
the HindIII-EcoRI sites of plasmid pAMW287. The resulting plasmid pAMW
458 consisted of the Napin promoter, the TAAla coding region and the 35S
transcription terminator.
Agf°obacteriurra-mediated transformation was employed for production of
tobacco
(Nicotiafaa tabacus~2 cv Xanthi) transgenic plants using published protocols
(Huang et al., 2000). The presence of foreign genes in independently derived
kanamycin-resistant cell lines was confirmed by PCR and Southern blot
analyses,
according to standard techniques.
Gas chrosnatog~aphy (GC) analysis
For, plant. GC, analysis, mature , seeds_were _ harvested from TAAl transgenic
plants, non-transgenic wild-type control plants and control transgenic plants.
Seed samples were ground and saponified with 10% potassium hydroxide
dissolved in methanol with 1 % water, incubated at 80 °C for 2 hours.
The mixture
was then extracted twice with hexane and subjected to GS analyses for total
fatty
alcohol and fatty acid contents and compositions as described by I~atavic et
al.
(1995). Analysis was done on a 30 M DB-5 column starting at 250 °C to
300 °C at
5 °C/min. The identity of the fatty alcohol peaks was based on
retention times of
authentic fatty alcohol standards and confirmed by GC-MS. The relative amount
of fatty alcohols was calculated on the basis of fresh weight of the seeds and
normalized according to internal contents of (3-sitosterol extracted in the
same
procedure.
49
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
For E. coli GC analyses, 200 ml bacterial cells with appropriate plasmids were
grown to OD value of 0.5 at 30 °C. After addition of IPTG (0.2mM), the
culture
was allowed to grow for 3 hr. The bacterial cells were harvested. Subsequent
extraction and GC analysis were essentially as above. Qualification of fatty
alcohols was based on flame ionization detector peak areas, which were
converted to mass units by comparison with an internal standard which was
added before the extraction.
TAAI promotes' isolation and analysis
The upstream regulation region of TAAl b was isolated from the hexaploid
spring
wheat cultivar Karma (genetic complements: AABBDD) using a Universal
GenomeWalkerTM I~it (Clontech, Palo Alto, CA). The resulting 1.7 kb DNA
fragment was cloned into a T/A vector (Original TA Cloning I~it, Invitrogen,
Carlsbad, CA) for further analysis.
Plasmid pRD400 (Datla et al., 1992) was modified by flipping-over the region
containing the polylinker and the NPT II gene cassette to generate a binary
transformation vector pAMW281. Two pieces of DNA fragments including a 2.4
kb fragment containing a CaMV 35S promoter and a uidA gene from plasmid
pRD410 (Dada et al., 1992) digested with HindIII and EcoRI, and a 0.7 kb
fragment containing a CaMV 35S terminator from plasmid pHS724 restricted
with EcoRI and KpnI (Huang et al., 2000) were co-ligated into the backbone of
pAMW281 digested with HindIII and KpnI to produce plasmid pAMW287,
consisting of the 35S-GUS-PolyA cassette. Plasmid pAMW445 containing
TAAl b promoter-GUS-PolyA was obtained by cloning the isolated 1.5 kb TAAI b
promoter into the HindIII-BamHI sites of plasmid pAMW287. Agr~obacteYiurn-
mediated transformation was employed for production of tobacco (Nicotiana
tabacurn cv Xanthi) transgenic plants using published protocols (Huang et al.,
2000). Presence of foreign genes in independently derived kanamycin-resistant
cell lines was confirmed by PCR and Southern blot analyses.
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
For transient expression analysis, microprojectiles coated with 35SlGUS or
TAAI
promoter/GUS chimeric genes were bombarded into the transverse sections of
flowers of a monocotyledonous plant species, daylily (Hemet~ocallis
lilioasplZOdelus) essentially as described (Chen et al., 1998).
To analyze GUS expression in transgenic plants, the flower buds were collected
from the primary transgenic tobacco plants (F0). Anthers Were cut transversely
and incubated in a GUS-assay buffer (0.1 M phosphate buffer pH7.0, 2 mM
I~3 [Fe(CN)6], 2 mM I~[Fe(CN)G], 1 mM EDTA. 0.1 % Triton) with 1 mM X-
Gluc (5-bromo-4-chloro-3-indoyl-(3-D-glucuronide) overnight at
37°C. After
incubation, the anthers were observed under a microscope. Typical anthers were
embedded in paraffin and then sectioned in 6 ~,m thickness for the further
observation. The histochemical assay on the daylily flowers was performed 24 h
post bombardment essentially as described (Wang et al., 1998).
Whilst the present invention has been described with particular reference to
specific examples and techniques, the invention is not intended to be limited
in
this regard, and numerous peptide and nucleotide sequences, constructs,
transformed organisms, methods, and products that are not directly described
herein are intended to be encompassed within the scope of the present
invention.
5l
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
Seduence listing
SEQ ID NO:1 (TAAla cDNA sequence)
(Putative start and stop codons are in bold/underlined)
CTCCTCTCTCTCTCTCTCTCTCTCTCTCTCTCGCACGCACGCACGCACGCACACAAGAAAAAAATCAAG_ATGGTTGAC
AC
ACTGAGTGAAGAGAACATCATTGGATACTTCAAGAACAAGAGCATCCTCATCACTGGATCAACAGGCTTTCTTGGAAAG
A
TACTGGTGGAGAAGATACTGAGAGTTCAACCTGATGTGAAGAAGATCTACCTCCCGGTGCGAGCGGTGGATGCCGCGGC
G
1O
GCAAAGCATCGGGTGGAGACTGAGGTGGTAGGGAAGGAGTTGTTCGGGCTTCTGAGGGAGAAGCACGGGGGCAGGTTTC
A
ATCTTTCATCTGGGAAAAGATCGTCCCATTGGCCGGAGACGTGATGCGCGAGGACTTCGGCGTCGACAGCGAGACCCTG
A
GGGAGCTCCGGGTGACCCAGGAGCTCGATGTCATCGTTAATGGCGCCGCCACCACCAACTTCTACGAAAGGTATGATGT
G
GCTCTAGACGTGAACGTGATGGGAGTGAAGCATATGTGCAACTTCGCCAAGAAGTGCCCCAATCTCAAGGTGCTCCTCC
A
TGTCTCCACGGCTTACGTGGCGGGTGAGAAGCAAGGGCTGGTGCAAGAGAGACCATTCAAGAATGGCGAGACGCTGCTC
G
IS
AGGGGACCCGCCTCGACATCGACACTGAGCTGAAACTGGCCAAGGACCTGAAAAAGCAGCTTGAGGCCGACGTTGATTC
G
TCGCCCAAGGCCGAAAGGAAGGCCATGAAGGATCTTGGCCTTACCAGGGCCCGGCACTTCAGGTGGCCAAACACATACG
T
GTTCACCAAGTCGATGGGGGAGATGGTGCTAAGCCAGTTGCAGTGTGATGTCCCCGTTGTCATCGTCCGTCCCAGCATC
A
TCACAAGTGTCCAGAACGACCCACTGCCCGGATGGATCGAAGGCACCAGGACGATCGACACGATCGTGATCGGCTATGC
G
AAGCAGAACCTGACATACTTCTTGGCCGACCTCAACCTCACCATGGATGTGATGCCGGGCGACATGGTGGTGAATGCGA
T
2O
GATGGCGGCAATAGTGGCACACAGCTCGTCCTCATTGGAGAAGACAAAGTCACATCCCAAGCAACATGCACCGGCGGTG
T
ACCACGTGAGCTCGTCGCTGCGTAATCCGGCACCATACAATGTGCTTCATGAGGCTGGGTTTCGGTACTTCACGGAGCA
C
CCTCGCGTGGGCCCTGACGGTCGCACCGTGCGTACCCATAAGATGACATTCCTCAGCAGCATGGCTTCCTTCCACCTAT
T
TATGATGCTCAGGTACCGCCTCCTCCTCGAGCTCCTCCACCTGCTCTCCATCCTCTGCTGCGGCCTCTTCGGCCTCGAC
A
CCCTCTACCACGACCAAGCACGCAAGTACAGGTTCGTGATGCACCTGGTGGATCTGTACGGGCCCTTTGCGCTGTTCAA
G
ZS
GGGTGCTTCGATGACGTCAACCTAAACAAGCTCAGGCTCGCCATGACCAGCAACCATGGTAGCCTCTTCAATTTCGACC
C
GAAGACCATTGATTGGGACGAGTACTTCTACAGGGTCCACATCCCCGGGGTCATAAAGTACATGCTCAAG_TGAAATAT
CC
GTGCACCGAAATTCAGGCGTTGCCTTAATAATTAAATAATCGTACGATTGTAAGAAATGTCCTCCCAAATTGGGTTCTA
A
CCGTTAAATAATCTATATGAAAGAGGAAAATGTGTCTGTCGCTTGATAGAGCAGGCATCAACGTTGCATAAGTTTCTTG
A
AGACAAGGAATGCTACATTATGTAGTCCCATTCTGGTTCATGTATTGTATTGCTAATTATGACTAGTATTGATTGTTGT
T
3 O ACCAAGCCGAATTCCAGCACACTGGCGGCCGTTACTAGTGGATCCGAGCTCGGTACC
3S
4S
SO
SS
6S
7S
S2
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
SEQ ID N0:3 (TAAlb cDNA sequence)
(Putative start and stop codons are in bold/underlined)
CTCTCTTCTTCCTCCGTCTCTCATTCTCTCCTCCCGAGCTTCATAGCTCACACGCACACAAAACCAAGGCAAGATGGTG
G
S
GCACGCTGGATGAGGGGAAGATCGTCGACTACTTCAGGAACAAGAGCGTGCTCATCACCGGAGCCACGGGATTCCTTGG
C
AAGATAATGGTGGAGAAGATCCTGCGGGTCCAGCCGGACGTGAAGAGGATCTACCTGCCGGTGCGGGCGGCCGACGCCG
C
GGCGGCGAGGCGCCGGGTGGAGACCGAGGTGGTGGGGAAGGAGCTGTTCTGCGTGCTGCGGGAGCGCCACGGCGCCGGG
T
TCGACGCCTTCGTCGCCGACAAGGTGGTGGGGCTGGCCGGCGACGTCATGCGCGAGGGCTTCGGCGTCGACCCCGCCAC
G
CTGCGGGACCTCCGGCTCGCCGACGAGCTCAACGTCATCGTCAACGGCGCCGCCACCACCAACTTCTACGAAAGGTACG
A
1O
CGTGGCCCTGGACGTGAACGTGGTGGGGGTGAAGCACATGTGCGACTTCGCCCGGAGGTGCCCCAACCTCGAGGTGCTC
A
TGCACGTCTCCACGGCCTACGTCGCCGGCGAGAAGCAGGGGCTGGTTCCGGAGAGGCCGTTCAGGGACGGCGAGACGCT
G
CGCGACGACGGCACCCAACTCGACATCGACGCCGAGATGAGGCTGGCCAAGGACCTCAGGAAGCAGATGGAGGCCGACG
A
CGATGTGGACCCCAAGGCCCAGAGGAAGGCCATGAAGGACCTCGGCCTCACCAGGGCCAGGCACTTTGGGTGGCCCAAC
A
CGTACGTGTTCACCAAATCCATGGGGGAGATGATGCTGGCCCAGATGATGCGCGGGGGCGACGTGCCCGTCGTCATCGT
C
1 S
CGGCCCAGCATCATCACCAGCGTCCAGAACGACCCACTGCCAGGATGGATCGAAGGCACCAGGACGATCGACGCAATCC
T
GATCGGGTACGCGAAGCAGAGCCTGTCGTGCTTCCTCGCCGACCTCGACCTAACCATGGACGTGATGCCCGGCGACATG
G
TGGTGAACGCGATGATGGCGGCCACGGTGGCACATGCCTCCTCCACTCAGACATCAGAGCCAGAGAAGAAGCCGCCTCC
G
CAGCAGCAACACCCTCACTCGGTGCCGGCAGCGCCAACGGTGTACCACGTGAGCTCGTCGCTGCGGCACCCGGCTCCGT
A
CGCGGTGTTGTACCGAACGGGGATCCGGTACTTCGAGGAGCACCCACGGGTGGGGCCTGATGGCCGCCCCGTGCGCACC
C
O
GTAAGGTGCGGTTCCTCGGCAGCATCGCGGCGTTCCACCTATTCATGGTGCTCAAGTACCGTGTCCCCCTTGAACTCCT
C
CGCCTGCTCTCCATCCTCTGTTGCGGCCTCTTTGGCCTTGCCGCCCTCTACCACGACCTCGCCCGCAAGTACAGGTTCG
T
GATGCAGCTGGTGGACCTGTACGGGCCCTTCTCGCTCTTCAAGGGTTGCTTCGACGATGTAAACCTCAACAAGCTCAGG
C
TCGCCATGGCCGACGGTGACCATGCCGATTCCGCATTCAACTTTGACCCCAAGACCATTGACTGGGACGACTACTTCTT
C
AAGGTCCACATCCCTGGTGTCATGAAGTACGTCCACAAGTGATGTTCTGTGTGCGATCTGCTTCTGCGTGCTGAGAAGG
A
ZS -
ATGGAGGAAATCAAATTAATGGTAGCGCTAGTGTGCCTTGCTTGTGTTGTGTAACCTCCTTCTTCGTTCATCGAGTATT
A
TTGGTTGAGTATTGATTGTATTGTCATTGGAAGTTAAATTAACCAGTGACTATGAGTATAACTAAGATGAAATTACTTG
C
ATCATGGCGGGTCTCTAAAACTAAGATAGTACAAGGATCCTATGAAGTACATTGAAATTACTTAGTACTTTTCATGGTA
C
TATCATAATAC
3S
4S
SS
6S
7S
S3
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
SEQ ID NO:S (TAAlc cDNA sequence)
(Putative start and stop codons are in bold/underlined)
CTCCTTCTTCCTCTCTCTCTCTCTCTCTCCCTGCTCTCCCTGCTCTCTCTCTCTCTCTCTCTCTCTCTTGCGCACACAA
G
S
P.AAAAAAATCAAGATGGTTGACACACTGAGTGAAGAGAAGATCATTGGATACTTCAAGAACAAGAGCATCCTCATCAC
TG
GATCAACAGGCTTTCTTGGAAAGATACTAGTGGAGAAGATACTGAGAGTTCAACCTGATGTAAAGAAGATCTATCTCCC
G
GTGCGAGCGGTGGATGCCGCGGCGGCGAAGGATCGGGTGGAGACTGAGGTGGTAGGGAAGGAGTTGTTCGGGCTTCTGA
G
GGAGAAGCACGGGGACTGGTTTCAATCTTTCATCTGTGAAAAGATCGTCCCATTGGCCGGAGATGTGATGCGTGAGGAC
T
TTGGCGTCGACAGCGAGACCCTGAGGGAGCTCCGGGTGACCCAGGAGCTCGATGTCATCGTTAATGGCGCCGCCACCAC
C
IO
AACTTCTACGAAAGGTATGATGTGGCCCTGGACGTGAACGTGATGGGAGTGAAGCATATGTGCAACTTCGCCAAGAAGT
G
CCCCAATCTCAAGGTGCTCCTCCATGTCTCCACGGCTTATGTTGCGGGTGAGAAGCAAGGACTCGTGCAAGAGAGACCA
T
TCAAGAATGGCGAGACGCTGCTCGAGGGGACCCACCTCGATATCGACACCGAGCTGAAACTGGCCAAGGACCTGAAAAA
G
CAGCTTGAGGCCGACGCCGACTCGTCGCCCAAGTCCCAAAGGAAGGCCATGAAGGACCTTGGCATCACCAGGGCCCGGC
A
CTTCGGGTGGCCGAACACATACGTGTTCACCAAGTCGATGGGGGAGATGGTGCTGGGCCAGTTGAAGTGTGATCTCCCT
G
IS
TTGTCATCGTCCGTCCCAGCATCATCACCAGTGTCCAGAACGACCCACTGCCCGGATGGATCGAAGGCACCAGGACGAT
C
GACACGATCGTGATCGGCTATGCGAAGCAGAACCTGACATACTTCTTGGCGGACCTCAACCTCACCATGGATGTGATGC
C
GGGCGACATGGTGGTGAATGCGATGATGGCTGCCATCGTGGCGCACAGCTCGTCCTTATTGGAGAAGACACAGTCACAT
C
CCGAGCCACACGCACCGGCGGTGTACCACGTGAGCTCGTCGCGGCGTAACCCGGCGCCGTACAATGTGCTGCACGAGGC
T
GGGTTTCGGTACTTCACGGAGCACCCTCGGGTGGGCCCTGACGGCCGCACGGTGCGCACCCATAAGATGACATTCCTCA
G
ZO
CAGCATGGCTTCCTTCCACCTCTTTATGATGCTCAGGTACCGCCTCCTCCTTGAGCTCCTCCACCTGCTCTCCGTCCTC
T
GTTGTGGCCTCTTCGGCCTCGACACCCTCTACCACGACCAAGCACGCAAGTACAGGTTCGTGATGCACCTGGTGGATCT
G
TATGGGCCCTTCGCGCTGTTCAAGGGCTGCTTCGATGACGTCAACCTAAACAAGCTCAGGCTCGCCATGACCAGCAACC
A
TGGAAGCCTCTTTAATTTCGACCCCAAGACCATTGACTGGGACGATTACTTCTACAGCGTCCACATCCCCGGGGTCCTA
A
AGCACATGCTCAACTGAAATATCCATGCACCGAAAATTTAGGCGTTGCCTTAATAATTAAATAATCGTACGTTGTAAGA
A
ZS
ATATCCTCCCAAATTGGGTTCTAACCGTTAAATAATCTGTGTGAAAGAGGGAAATGTGTCTGTCGCTTGATAGTGCAGG
C
ATCAACTTGCATATGCTTCTTGAAGAACAAGGAATGCTACATCATGTAGTTTCGTTCTGGCTCGTGCTGTATTGCCAAT
T
ATGACTAGTATTGGTTGATAACAATTTCTTGTTCAGTCACATATTGTACCTTGACTAGTATTGATTGTTAATTATAATT
T
CTTTTCAAGAG
35
45
54
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
SEQ ID NO: 7 (TAAla genomic sequence)
(introns are underlined, transcription start site and start and stop codons
are
indicated in bold/underlined)
CCTCTTTAAGTCGGTCAGGTACGTAGCAAATAAACCAAACAAGCAGGACATGTTTGAAAATCACTGTTCTAACTTTAAT
A
ATGCACACAAGGAATTGAAGGCTTGATAAATCCTGTGCTAGTACCACCCATCCGACTGTCGGTTTTTAAGACATGGTAG
A
GTAGAGTACTAACATAACGATCTTCAAGATATTGTGTCCAAGTTCTTTGCCACAGGTTTGTCGTATTGAGCCAAAATAT
A
CCACTGAATTATGTGCTAATAGTGTAATTATTGGTCAATTTTTTGTTCTAACGAGAGAAAAATACGCTATGCGTTTTGA
A
1O
GGGATGGAGTACGATGATACTACTCTATTGTACTCCACGGATCAATTGTAGGAGTACCCTCTAATTTCTAATGGTATAA
C
TTTGATGACATACGATTTGTATTACGTTGGTTGAATTGTACATGGGCCTTCTATAAAGCGAAAAAGAAAGGAATACTGA
G
CTTCTCTAGTTTTCTATAAMCCAGAGTTAGACGTCCTACTGGACAACTCATTCCTAAGTGTGTAGCAATAATACTGAGT
A
CTACCGGTTGTCCACTTGAGCTATTAATTTGATGGCTAGCAACGTACTGCAACCCCAATAATAACCCTGTTTGAAAATC
T
AGAGTACCTCTACGCGTTATTTTGACTGCATGTGATTCCTCTCATGATCACCAGGTCCCTTGICTTTGGGTTGGACAGC
GC
IS
ATAAATGTTGCGCTCAAGTGCCTCAAATTAAGACTCCCCTCTCCTACCTACAACACAAAAGACCTTTTTTTCAGCAAAA
G
AGGGCGCCACCCCCTCCGATTTCATATAATGAAACCAACTGGTTCAACTGCCAAACAAACAACAAAAACAAAACGAAAC
A
CCTTCATGGCCCGTGTATAAATATTGTGTACATGACCCTCTTTCCACTCACCCACACAAATCAT_CTCCTCTCTCTCTC
TC
TCTCTCTCTCTCTCTCGCACGCACGCACGCACGCACACAAGAAAAAAATCAAG_ATGGTTGACACACTGAGTGAAGAGA
AC
ATCATTGGATACTTCAAGAACAAGAGCATCCTCATCACTGGATCAACAGGCTTTCTTGGAAAGAGTATGCATGCATGCA
T
ZO
AATGTTTATGCACACATATTACGCATGCATATACTTTTTTTTGGCAAATTCTAGAAGTTTTTTGCGTCAAGTTCATATG
T
GCTGTCCTCATCTGTTAATTAGTTCACTGTTGGTGTGCGTATATGCAGTACTGGTGGAGAAGATACTGAGAGTTCAACC
T
GATGTGAAGAAGATCTACCTCCCGGTGCGAGCGGTGGATGCCGCGGCGGCAAAGCATCGGGTGGAGACTGAGGTAGTGT
T
ACTAATTTCTTTTCTATATGCGCAGGTGGTAGGGAAGGAGTTGTTCGGGCTTCTGAGGGAGAAGCACGGGGGCAGGTTT
C
ZS
AATCTTTCATCTGGGAAAAGATCGTCCCATTGGCCGGAGACGTGATGCGCGAGGACTTCGGCGTCGACAGCGAGACCCT
G
AGGGAGCTCCGGGTGACCCAGGAGCTCGATGTCATCGTTAATGGCGCCGCCACCACCAACTTCTACGAAAGGTGCGTCA
T
TGTCAACTGATTGTTTTGTCCAAGAAAGGAAAAATCAGCAGAGAACTTGAGTAGGTGCAGCAAGAGTACGTACGCAGTG
A
ATGGTCTCAGAAGAACACTTGTGTGCGTGCAGGTATGATGTGGCTCTAGACGTGAACGTGATGGGAGTGAAGCATATGT
G
CAACTTCGCCAAGAAGTGCCCCAATCTCAAGGTGCTCCTCCATGTCTCCACGGGTACGTACACCAACTCTACAGGTAAA
A
3O
ACAAAAATAAAGTTCTTGGATGTTAATTATAATACACCTAGATTTGATTTACAAATGAAGTTAATAAATTCATATATGA
G
TTGGTGCAGCTTACGTGGCGGGTGAGAAGCAAGGGCTGGTGCAAGAGAGACCATTCAAGAATGGCGAGACGCTGCTCGA
G
GGGACCCGCCTCGACATCGACACTGAGCTGAAACTGGCCAAGGACCTGAAAAAGCAGCTTGAGGCCGACGTTGATTCGT
C
GCCCAAGGCCGAAAGGAAGGCCATGAAGGATCTTGGCCTTACCAGGGCCCGGCACTTCAGGTGGCCAAACACATACGTG
T
TCACCAAGTCGATGGGGGAGATGGTGCTAAGCCAGTTGCAGTGTGATGTCCCCGTTGTCATCGTCCGTCCCAGCATCAT
C
3S
ACAAGTGTCCAGAACGACCCACTGCCCGGATGGATCGAAGGCACCAGGTTCATTATATGTTTCTTGTCCTTTCTCTGCC
T
CCAAATTTAGAGTGCAATCGTCTTACTCTGTTGCAAATGCCAAAAGAAGTAAAATATGATATTTGTTCAATGTAAAAAT
G
TAAATTGCAGGACGATCGACACGATCGTGATCGGCTATGCGAAGCAGAACCTGACATACTTCTTGGCCGACCTCAACCT
C
ACCATGGATGTGGTAAGCAACGTTGTACTATGCATGCAGTTAAGATATATTCCAGGCAATGGTTGGTTGTCAGTCCAGT
C
CAGGAATCCGTACAGTAAGATGAATTTCGACGACGATGGTGAAAGCAATCGTTTGGTTGGGTATATGTTGCTGTAGATG
C
4O
CGGGCGACATGGTGGTGAATGCGATGATGGCGGCAATAGTGGCACACAGCTCGTCCTCATTGGAGAAGACAAAGTCACA
T
CCCAAGCAACATGCACCGGCGGTGTACCACGTGAGCTCGTCGCTGCGTAATCCGGCACCATACAATGTGCTTCATGAGG
C
TGGGTTTCGGTACTTCACGGAGCACCCTCGCGTGGGCCCTGACGGTCGCACCGTGCGTACCCATAAGATGACATTCCTC
A
GCAGCATGGCTTCCTTCCACCTATTTATGATGCTCAGGTACCGCCTCCTCCTCGAGCTCCTCCACCTGCTCTCCATCCT
C
TGCTGCGGCCTCTTCGGCCTCGACACCCTCTACCACGACCAAGCACGCAAGTACAGGTTAGTTAGTTGGTTGAAATCTT
G
4S
TGCGGTTGTATCTTCTTGATGGCTCCCACATAATTAAGATGACACGACTTTTATTGTTGTATTGTTATAGGTTCGTGAT
G
CACCTGGTGGATCTGTACGGGCCCTTTGCGCTGTTCAAGGGGTGCTTCGATGACGTCAACCTAAACAAGCTCAGGCTCG
C
CATGACCAGCAACCATGGTAGCCTCTTCAATTTCGACCCGAAGACCATTGATTGGGACGAGTACTTCTACAGGGTCCAC
A
TCCCCGGGGTCATAAAGTACATGCTCAAGTGA
SS
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
SEQ ID NO: 8 (TAAlb genomic sequence)
(introns are underlined, transcription start site and start and stop colons
are
indicated in boldlunderlined)
(tlie region for promoter analysis is italicized)
CTGACCAAACATCTCCCATCAGGAACCATGTTACTATTTAATTTTGCATGTTCTTTGCTTCGTGCTGTTTGTCAAGGAC
A
CGCTAGCTTCTGCATTATACAGACAGGCTGGAAATCTGTAAATAATGTGTTGGTGAATCTTTGGAGTATTTTTCTAATG
T
TGATCAGTACTTGTTGACCTAGGTGGCGACTAACTTTGGACAGGGGCAAGCAAGCAAAAGCAGAGACAGCTAGCTTCTG
C
ATTATCAGAAAATAGCCTGTGTTGGTGAATCTTCAGAGTGCTCCTACTCCCCCGTCCCCCCAAATGTTCAGTACTTGTT
G
IO
ACCTACTGGAGCTGGACGACCCGCTCTGAAAGTTTTACTTGTCCACACTTGTCTGGCCAAAATCCCCAATGAATCAGAA
T
ATAGGCTGAAAAATGAGTGGGTCGTTTTTGGAGCTCGGCCTCCACGGAGGCCGAAAAAAAACTCAAAATTCTAATTTCT
C
AGTTTAAAAAAAAATGAAAAAAATTGTACAAGTAAACAAGGTTGTGATGTGTATGTGTGTAAAATTTCAGAACAAAATA
C
TTTGAAATGCGATCTGAGCAAAAAAAAACAAATTCATAGACTTTTGAGAATGAATAGTGCAAGCTACTAAAACGCATGA
G
ATTCTTTTTGCGTGTAGCACTCATTTTAATGTATTTCGTCCTGAAAATTACAAACTTGTGCATTATGTCTATAAGTATA
T
IS
GGTGGATTTAAAAAAAAACTGAAACATGCAAATTTGAATTTTGATCATTTCAAAATCGGCCTCCATGGAGCATTTTTGC
T
GAAAAATCTAAAAGAAAACGCAGTCCAACCTTAGTAATTACCCATGTTGGTGAATCCAGAGTGTTTACGATAAATATAG
G
ACCTCTTTAGCTAGCGAACGATGGCATTTGCGAATGTTATTTATGGCAAATCCTTCAGAACACACATGCCAATTTCTTC
A
TAAACACATGGCAATTCTGGCAAATATTTTGCTGTGAAAATTGTTGTGCTTGCTTCAAGGAATTGCCATGTTCCCGCAA
T
ATAAAATGCCATGAAAAACGTTCATTTTGTCATATCCCCAGCGAACGTCCGCCGAGTAGCATTACCGTAAATATAGGGG
G
2O
TGTTTGGTTCAGAAGTTCTAGGACTTTTTCTAGTCCCAGGGACTAATCAAAAAAGACTCTCTAATAGAGTCTTTTTCTA
G
TCCCTATAGAAAAAGTCCCTCCCGTTTGGTTCCTAGGGACTTTTTTTAGTCTCTAGGACTAAAAAGTCCCTGAACCAAA
C
ACCCCATATTAGTAGTACCGCAAAAAGTAGTAAGTACTACAGTATTGGTGGATCTTTAATGAGTTAATTAACCTAGAAT
G
GCTCCTGCTCCTCCGGTTTCACTGGAAAAGCAGGTTGTGGTTGGGAAGCACAAGCGTGTTGCCGTTGCTAATTACGTGC
A
CTCAATAGCCAGCCAGCCACCCTCAACACCTCGCTGGCCGGCCCTGCGCGTGTATAAATACTGGGTGCCGGAGCACACA
A
?,S
GCCTCACACACCAAGCACAC_CTCTCTTCTTCCTCCGTCTCTCATTCTCTCCTCCCGAGCTTCATAGCTCACACGCACA
CA
AAACCAAGGCAAGA_TGGTGGGCACGCTGGATGAGGGGAAGATCGTCGACTACTTCAGGAACAAGAGCGTGCTCATCAC
CG
GAGCCACGGGATTCCTTGGCAAGAGTAAGTGAGCATGATGAGGATATATCATATATGCATGCCTTGCTATTTTCTGAAG
A
ACAGTTCATGCATCCATGCTGAGAGTCCTTGGTGTTGCATTGTGTGCTCACTCTTCGACCTGTGCAGTAATGGTGGAGA
A
GATCCTGCGGGTCCAGCCGGACGTGAAGAGGATCTACCTGCCGGTGCGGGCGGCCGACGCCGCGGCGGCGAGGCGCCGG
G
3 O
TGGAGACCGAGGTGCGTGCCTCTCTGCCCGTCCATCATGCTCCAGCATGATGGTGATCCTTGCACCCCCTCCATCAATC
T
AAGGAACACATGCGAGCTTTCTCTCAGACAGAGAAGCCCCCAAGCTGCTCCCACTGTGCCCCTTGTGTCTCTTCATTCC
G
TTTCATATTCACTACTCTTTTTTTCATTGATAATCAATACTGTAATACTAGTAATATTATGTGGCGCATCGGAACAAAA
C
ATGCATAGATATCACAGAACCATAAATAGTCCAGCTGCCCGCTGCTATATCAGTGATGCAAACATGAACATGGGTAGAA
T
3S
AGTTATCTTATAGGCATAAACCGGCATTAGTCAGGACTAGGTGCACACTGTTGAGTCTCATCCCTGCCGAACATTCAGG
C
TACATACAGAATAAAATGAATGAATCTATATTCTAAAATATGTCTACATACATCCATATGTTATAGTCCATTTAAAATG
T
TGGAAACCGGGAACCTTGGGAAAACCGGCCGCAGGATGACAATTAAACCGGCGGCGAAGTAGCGCAGCTGTGTGTGTAT
C
CCTAGGAGTCGTATAGGAGTTATGGTGTGGGTAGGCCGGTAGGTTTGGTCAGGTCGACTCGATTCACAAGTTTTCAATG
G
CATGCATGCATGGCTGGCTGGCTGCTGCTGGGAAATGACGAGCGCAGGGCACCATCTGGCACTGGCACCGGCACCACTA
A
4S
CTACAGCTTCATCAACTTGAGAATTCAGAACCCAGAGTTTCTTGTCATATGTGTATAGAGTGGACCGCAAGTCTGCAAT
G
AAAAGAAAAAAAGTGTAGTTATTGAGTGGTGCCAAGGAGATCCAGAGCAGAGACAATGGGCAGTATGCTTGGTCCTCCA
C
SO
GCAATTTATAGTTTTTACCAAAAGTTGCCTGAGACACTCACCATGTTGGGTGGTGGATCCACAGTGCGTGCATGCATGT
A
GTTCGCAGGTGGTGGGGAAGGAGCTGTTCTGCGTGCTGCGGGAGCGCCACGGCGCCGGGTTCGACGCCTTCGTCGCCGA
C
AAGGTGGTGGGGCTGGCCGGCGACGTCATGCGCGAGGGCTTCGGCGTCGACCCCGCCACGCTGCGGGACCTCCGGCTCG
C
CGACGAGCTCAACGTCATCGTCAACGGCGCCGCCACCACCAACTTCTACGAAAGGTGCTGTGTACTATTGACTGTTTTA
A
SS
TTAACTTCGTCAGAGAGAAAATTCAGCGGGAGAGAACGTGCATGGCTGCACCTTCTTCTCCAGAGTAGTGAGCTCATCA
A
TGGCCTCTGCAACTAACTTTGGAATGGACGTGTGCGTGCTCAGGTACGACGTGGCCCTGGACGTGAACGTGGTGGGGGT
G
AAGCACATGTGCGACTTCGCCCGGAGGTGCCCCAACCTCGAGGTGCTCATGCACGTCTCCACGGCCTACGTCGCCGGCG
A
GAAGCAGGGGCTGGTTCCGGAGAGGCCGTTCAGGGACGGCGAGACGCTGCGCGACGACGGCACCCAACTCGACATCGAC
G
CCGAGATGAGGCTGGCCAAGGACCTCAGGAAGCAGATGGAGGCCGACGACGATGTGGACCCCAAGGCCCAGAGGAAGGC
C
C7O
ATGAAGGACCTCGGCCTCACCAGGTTAGCAATCCTATATTTTTCCAAAGTTTCTCCTCTTTCCTGTGAAATTCAAGTTC
A
AAAAACAGAGGGTTTTATTTTGTGAAACTTGGAACTGAAATTCAATATATTTTTTTGTAATTGTGTGTGCAGGGCCAGG
C
ACTTTGGGTGGCCCAACACGTACGTGTTCACCAAATCCATGGGGGAGATGATGCTGGCCCAGATGATGCGCGGGGGCGA
C
GTGCCCGTCGTCATCGTCCGGCCCAGCATCATCACCAGCGTCCAGAACGACCCACTGCCAGGATGGATCGAAGGCACCA
G
GTCGGTTCAAACTCTTTGGGTCTAAATATGAATGCCAAGTTTCAAATTCAAATTCTAACACCGAAATGAAAAATGCAGG
A
C)S
CGATCGACGCAATCCTGATCGGGTACGCGAAGCAGAGCCTGTCGTGCTTCCTCGCCGACCTCGACCTAACCATGGACGT
G
GAGTAATAAGTATACTAGTACAGTGTGTGACCATGCTGCAGTAAGTTGCATCTGAAGCGTCTGTGCCAACCAGGCCAAG
T
GTTACATATGCTGCTACTGGATTATCAAGTTCTAACCCAAATGCGCTACCATGGTGTCTGCAGATGCCCGGCGACATGG
T
7O
GGTGAACGCGATGATGGCGGCCACGGTGGCACATGCCTCCTCCACTCAGACATCAGAGCCAGAGAAGAAGCCGCCTCCG
C
AGCAGCAACACCCTCACTCGGTGCCGGCAGCGCCAACGGTGTACCACGTGAGCTCGTCGCTGCGGCACCCGGCTCCGTA
C
GCGGTGTTGTACCGAACGGGGATCCGGTACTTCGAGGAGCACCCACGGGTGGGGCCTGATGGCCGCCCCGTGCGCACCC
G
TAAGGTGCGGTTCCTCGGCAGCATCGCGGCGTTCCACCTATTCATGGTGCTCAAGTACCGTGTCCCCCTTGAACTCCTC
C
GCCTGCTCTCCATCCTCTGTTGCGGCCTCTTTGGCCTTGCCGCCCTCTACCACGACCTCGCCCGCAAGTACAGGTACAT
T
7S
CTTCTTCCGGAATTAGTTGACGCTCAAACGAATTATCTAGAGGGAGCACAGTTTAAACACTTTTGCGGTTGTACCGTGT
T
GATGTTATACCATTGGCTTAAGAACATGCATTTTGTGTAGGTTCGTGATGCAGCTGGTGGACCTGTACGGGCCCTTCTC
G
S6
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
CTCTTCAAGGGTTGCTTCGACGATGTAAACCTCAACAAGCTCAGGCTCGCCATGGCCGACGGTGACCATGCCGATTCCG
C
ATTCAACTTTGACCCCAAGACCATTGACTGGGACGACTACTTCTTCAAGGTCCACATCCCTGGTGTCATGAAGTACGTC
C
ACAAGTGA
57
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
SEQ ID NO: 9 (TAAlc genomic sequence)
(introns are underlined, transcription start site and start and stop codons
are
indicated in bold/underlined)
J
ACTCCGACACAATATTTTGACTGCATGTGATTCCTCTTATGATCACTGAGTCTTTGTTTCGGTTGGACAGCGCATAAAT
G
TTGCGCTCAAGTGCCTCAAATTAAGACACCCCTCTTCTACCTACAACACGAAAGACCCTTTTTTCTTAGCAAAAATTTC
A
TACAATGAAACCAACTGGTTCAACTGCCAAAACAACAGACTCCAAGACAAAACACCTTGATGGCCCGTGTATAAATATT
G
TGTACAGGAGCCTCTTTCCACTCACCTACACAAATCATCTCCTTCTTCCTCTCTCTCTCTCTCTCCCTGCTCTCCCTGC
T
CTCTCTCTCTCTCTCTCTCTCTCTTGCGCACACAAGAAAAAAAATCAAGATGGTTGACACACTGAGTGAAGAGAAGATC
A
1O -
TTGGATACTTCAAGAACAAGAGCATCCTCATCACTGGATCAACAGGCTTTCTTGGAAAGAGTATGCATGCATGCACGCA
T
AATGTTTATACAAACATATTACGCATGCATATACTATTTGTTGCAAATTCTAGAAGTTTTTTTTTTGCGTCAAGTTGAT
A
TGTGTTGTGCTCATCTGTTAATTAGTTCTACTGTTGGTGCATATGTGCAGTACTAGTGGAGAAGATACTGAGAGTTCAA
C
CTGATGTAAAGAAGATCTATCTCCCGGTGCGAGCGGTGGATGCCGCGGCGGCGAAGGATCGGGTGGAGACTGAGGTAGT
G
TTGTCCATCATGGTTCTTCATGATCTAGACCTCTACTGCTCTCATGATTCTTTTGATCCCTTTGCTTGAGTGTTGGTAC
T
IS
GAATAATTTATTTTCTATGTGCCCAGGTGGTAGGGAAGGAGTTGTTCGGGCTTCTGAGGGAGAAGCACGGGGACTGGTT
T
CAATCTTTCATCTGTGAAAAGATCGTCCCATTGGCCGGAGATGTGATGCGTGAGGACTTTGGCGTCGACAGCGAGACCC
T
GAGGGAGCTCCGGGTGACCCAGGAGCTCGATGTCATCGTTAATGGCGCCGCCACCACCAACTTCTACGAAAGGTGCGTC
G
TCGTCAACTGATTGTTTTGTCCAAGAAAGGAAAAAAAATCAGCAGAGAACTTGAGTAGGTGCAGCAAGAGTACGGAGTG
G
AATTGTGTGCGTGCAGGTATGATGTGGCCCTGGACGTGAACGTGATGGGAGTGAAGCATATGTGCAACTTCGCCAAGAA
G
,O
TGCCCCAATCTCAAGGTGCTCCTCCATGTCTCCACGGGTACGTACACCAACTCTACAAATTACCATCATGGACTATGAA
C
TTGGATGCTTCTGGGGAAAACAAAAATGAAGTTCTTGGATGTAATTAAAGTACACCTAGATTTGATTTACAAATCAAGT
T
AATGAATTCATACATGAGTTGGTGCAGCTTATGTTGCGGGTGAGAAGCAAGGACTCGTGCAAGAGAGACCATTCAAGAA
T
GGCGAGACGCTGCTCGAGGGGACCCACCTCGATATCGACACCGAGCTGAAACTGGCCAAGGACCTGAAAAAGCAGCTTG
A
GGCCGACGCCGACTCGTCGCCCAAGTCCCAAAGGAAGGCCATGAAGGACCTTGGCATCACCAGGGCCCGGCACTTCGGG
T
~S
GGCCGAACACATACGTGTTCACCAAGTCGATGGGGGAGATGGTGCTGGGCCAGTTGAAGTGTGATCTCCCTGTTGTCAT
C
GTCCGTCCCAGCATCATCACCAGTGTCCAGAACGACCCACTGCCCGGATGGATCGAAGGCACCAGGTTCGTTATATGCT
T
CTCTTTCCTTTCCCTGCCTCTAAATTTAAAGTGCAATCGTTTTAATCTGTTGCAAATGCAAGTAAATAAAGGATGTTTG
T
TCAATGTAAAAATGTAAATTGCAGGACGATCGACACGATCGTGATCGGCTATGCGAAGCAGAACCTGACATACTTCTTG
G
CGGACCTCAACCTCACCATGGATGTGGTAAGCAACGTTGCACTATGCATGCAGTTAATTAACATATATTCCAGGCATGC
A
3 O
ATGGTTGGTTGTCAGTCCAGGAATCCATACAGTAAGATATGGATTTCAACGATGGCGGTGAATGCAATCGTGTGGTTGG
G
TATATGTTGGTGCAGATGCCGGGCGACATGGTGGTGAATGCGATGATGGCTGCCATCGTGGCGCACAGCTCGTCCTTAT
T
GGAGAAGACACAGTCACATCCCGAGCCACACGCACCGGCGGTGTACCACGTGAGCTCGTCGCGGCGTAACCCGGCGCCG
T
ACAATGTGCTGCACGAGGCTGGGTTTCGGTACTTCACGGAGCACCCTCGGGTGGGCCCTGACGGCCGCACGGTGCGCAC
C
CATAAGATGACATTCCTCAGCAGCATGGCTTCCTTCCACCTCTTTATGATGCTCAGGTACCGCCTCCTCCTTGAGCTCC
T
3 S
CCACCTGCTCTCCGTCCTCTGTTGTGGCCTCTTCGGCCTCGACACCCTCTACCACGACCAAGCACGCAAGTACAGGTTA
G
TCGGTTTAAATCTTTTGCGGATGGCATTTTTGATAACAAGTATTTCCGGACGGAGGGAGTATCTTCTTGATGGCGCGGC
A
TATATGATGACACGGCCTTTATTGTTATATTGTTGTAGGTTCGTGATGCACCTGGTGGATCTGTATGGGCCCTTCGCGC
T
GTTCAAGGGCTGCTTCGATGACGTCAACCTAAACAAGCTCAGGCTCGCCATGACCAGCAACCATGGAAGCCTCTTTAAT
T
4O -
TCGACCCCAAGACCATTGACTGGGACGATTACTTCTACAGCGTCCACATCCCCGGGGTCCTAAAGCACATGCTCAACTG
A
50
5~
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
SEQ ID NO: 2
TAAla amino acid sequence
S
MVDTLSEENIIGYFKNKSILITGSTGFLGKILVEKILRVQPDVKKIYLPVRAVDAAAAKHRVETEWGKELFGLLREKHG
GRFQSFTWEKIVPLAGDVMREDFGVDSETLRELRVTQELDVIVNGAATTNFYERYDVALDVNVMGVKHMCNFAKKCPNL
K
VLLHVSTAYVAGEKQGLVQERPFKNGETLLEGTRLDIDTELKLAKDLKKQLEADVDSSPKAERKAMKDLGLTRARHFRW
P
NTYVFTKSMGEMVLSQLQCDVPWIVRPSIITSVQNDPLPGWIEGTRTIDTIVIGYAKQNLTYFLADLNLTMDVMPGDMV
VNAMMAAIVAHSSSSLEKTKSHPKQHAPAWHVSSSLRNPAPYNVLHEAGFRYFTEHPRVGPDGRTVRTHKMTFLSSMAS
IO
FHLFMMLRYRLLLELLHLLSILCCGLFGLDTLYHDQARKYRFVMHLVDLYGPFALFKGCFDDVNLNKLRLAMTSNHGSL
F
NFDPKTIDWDEYFYRVHIPGVIKYMLK
20
30
40
50
60
70
59
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
SEQ ID NO: 4
TAAlb amino acid sequence
S
MVGTLDEGKIVDYFRNKSVLITGATGFLGKIMVEKILRVQPDVKRIYLPVRAADAAAARRRVETEWGKELFCVLRERHG
AGFDAFVADKWGLAGDVMREGFGVDPATLRDLRLADELNVIVNGAATTNFYERYDVALDVNWGVKHMCDFARRCPNLE
VLMHVSTAYVAGEKQGLVPERPFRDGETLRDDGTQLDIDAEMRLAKDLRKQMEADDDVDPKAQRKAMKDLGLTRARHFG
W
PNTWFTKSMGEMMLAQMMRGGDVPWIVRPSIITSVQNDPLPGWIEGTRTIDAILIGYAKQSLSCFLADLDLTMDVMPG
DMVVNAMMAATVAHASSTQTSEPEKKPPPQQQHPHSVPAAPTWHVSSSLRHPAPYAVLYRTGIRYFEEHPRVGPDGRPV
1O
RTRKVRFLGSIAAFHLFMVLKYRVPLELLRLLSILCCGLFGLAALYHDLARKYRFVMQLVDLYGPFSLFKGCFDDVNLN
K
LRLAMADGDHADSAFNFDPKTIDWDDYFFKVHIPGVMKYVHK
20
30
40
50
60
70
60
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
SEQ ID NO: 6
TAAlc amino acid sequence
J
MVDTLSEEKIIGYFKNKSILITGSTGFLGKILVEKILRVQPDVKKIYLPVRAWAAAAKDRVETEWGKELFGLLREKHG
DWFQSFICEKIVPLAGDVMREDFGVDSETLRELRVTQELDVIVNGAATTNFYERYDVALDVNVMGVKHMCNFAKKCPNL
K
VLLHVSTA'YVAGEKQGLVQERPFKNGETLLEGTHLDIDTELKLAKDLKKQLEADADSSPKSQRKAMKDLGITRARHFG
WP
NTYVFTKSMGEMVLGQLKCDLPWIVRPSIITSVQNDPLPGWIEGTRTIDTIVIGYAKQNLTYFLADLNLTMDVMPGDMV
VNAMMAAIVAHSSSLLEKTQSHPEPHAPAVYHVSSSRRNPAPYNVLHEAGFRYFTEHPRVGPDGRTVRTHKMTFLSSMA
S
1O
FHLFMMLRYRLLLELLHLLSVLCCGLFGLDTLYHDQARKYRFVMHLVDLYGPFALFKGCFDDVNLNKLRLAMTSNHGSL
F
NFDPKTIDWDDYFYSVHIPGVLKHMLN
61
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
SEQUENCE LISTING FREE TEXT:
SEQ ID NO: 10 OL2707 primer
SEQ ID NO: 11 OL2708 primer
SEQ ID NO: OL2881 primer
12
SEQ ID NO: 13 OL2885 primer
SEQ ID NO: 14 OL2884 primer
SEQ ID NO: 15 OL2883 primer
SEQ ID NO: 16 OL2880 primer
62
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
International Depositary Authority of Canada
Bureau of Microbiology, T~ealth Canada
1015 Arlington Street Tel: (204) 789-2002
Winnipeg, Ndan9toba, Canada R3E 3R2 Pax: (204) 789-203f
Interaatioual Form IDAC/BP/4
RECEIPT IN THE CASE OF AN ORIGINAL DEPOSIT
(issued pursuant to Rule 7.1 of the Budapest 2"reary Rcgularions)
A1TACH COPIES OF'IliE ORIGINAL DF~061T CONTRACT Aid VIpBILrtY S'I'ATE1~E:N'f
This Intenea~ional Depository Ar~hority accepts the d'eposft e, f the
tnicroorganrsm spet'~c'ed
8elox~, which was receirred by it on Jun 7. 2 I
To (l~Iame of I<??eposlLor) Dr. Gopalan Selvaraj~
Address NRC/PBX
110 GNnnasi~ Place, Saskatoon, SK, S7N OW9
1DENTIhICA'IZON OF DEPOSIT
Reference sssi,g~ned by depositor pAMW170
Accession Nwa~bcr assigned by this IDA IDAC 070501-1
The deposit identified aborre was accompanied by;
C1 a scientific description (specify)
d a proposed taxonomic designation (specify)
Signature of person(s)authorized to represent IDAC:
ce
Date Jun 7, 2D01
R~dpc ~ ~ caa~ or an o,~~ D~c
63
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
International De~os~tary Authority of Canada
Bureau of lVlic~robiology, Health Canada
1015 Arlingwc Street
Tei: (204) 789-2070
Wionl-beg, Manitnb~a Canada R3E 3R2 flax: (Z04) 789-2097
Interuabonal Form IDACJDP/9
STATEA~NT OF vIABILIT'Y
(Issued pursuant to Rule 10.2 of the Budapest T't~eQty RegulaGods)
P.~R7oY TO W~IOM T~:'~IABa,rt'3r STATF~.SNr IS ISSUED
Name r. Wa ne Anderson
Address~tadonal Rr$earch ConnciI of Canada
,_.....,r...._~_...
M 58, Mornreal Rd., Ottawa, ON, K1A OR6
D»osTrott
Name Dr. rxoualan Selvaraj-, ~ ..
...~,
IDEN'~FZCATtON OF THE DEPOSIT
Accession Number given by the International Depository Authority iD.A,C 070601-
1
Date of the orig3ual deposit (or most zecent relevant date) Jwa 7 2001
'YrABU.rrY TEST
The viability of the deposit identified above was tested on (most recent test
date) June 8. 2001
On the date indicated above, the culture was:
~ viable
no longer viable
Conditions under which the Viability Test were performed (to be filled in if
the information
has been requested and the results of the test were negative)
SSgnatiu (s) authorized to represent mAC
Date June 8, 2001
6mt of V9abitil~
64
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
IBtecnationoi I~positary Aufhoritp of Canada
Bureau of Microbiology, Health Canada
1015 Arlington Street ~ Tel: (Z04) 789-20pZ
'Wi>'mipeg, Manitoba, Canada R3E 3R2 Fax: (204.) 789-2036
Jnternatyonal Form IDAC/BP/4
RIEICEIPT 1N THE CASE OF AN O~tIGrNAL DEPOSIT
(issued pursuant to Rale 7_ 10~ the Budapest Treasy Regulations)
ATTACH COPIES OF TII'E ORIGINAL DEPOSIT CONTRACT AN'p VIABILrIy $'j'A
This Intet~onal Depository Authority accepts !he delrasit of the microorganism
specified
be~otv, whi'cb was received by it on Jun 7, 2001
To (Name of l7epositor)_ _ Dr. Copalan Selvarai
Address
Place, Saslsatoon, SK, S7N OW9
IDIrNT~'ICAT1,ON OF DEPOSIT
Reference assig~aed by depositor pAMW133
Accession Number assigned by Ibis ~7A IDAC 070601-2
The deposit ideutif~d above was accou~panied by:
a scientific description (specify)
a proposed raxononuc desig~acion (specifyj-. _
Signa~ure of person(s)autho~ed to represent IDAC:
Date Jun 7, 2001
'R~oeipt is the case d an O~gmal Deposit
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
Internations~ Depositary Authoritp o~ Canada
Bureau of Microbiology, ~3ealth Canada
1015 Arlington Strett Tel: (204) x'89-2070
Wianipeg, llr~anitol~ Canada R31: 3R2 Pax: (204) 78~-20f7
International Form IDAClBPl9
STATEMENT OF ~1~,~'Y
(Issued pursuant to Rule 10.2 of the Budapest Tree~y Reg~,a~tions)
PwB~rY To WHOM T.E~ V~gnrry Strw~c Its ISSUED
Dame Mr. W~ra ~e Anderson
Address,_ National Research Council of Canada
M-58, Montreal Rd., Ottawa, ON, l~lA OR6
D~POSrroR
Name Dr. Gopallan Selvarai
Address NRC/PBI
IbEN~IFtGITION OF T.~ DEPOSIT
Accesaiou Number given by the lnt~ernational Depository Authority IDAC 070601
2
Date of the original deposit (or most recent relevant date) Jun 7, 2001
~tA~iTY T~
The viability of the deposit identified above wag tested on (most recent test
date) June Ss 2001
On the dale indicated above, the culture was:
~ viable
no longer viable
Conditions under which the Viabiliry Tcsc were gerformsd (to be filled in if
the inf~ormaiion
has i~een requested and. the results of tlxe test were negative)
Signature s s) authori2ed to represent 1DAC
f
Date >~ne 8, 2001
sca~me~e at vt
66
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
References
Aarts MGM, Dirkse WG, Stiekma WJ, Pereira A (1993) Transposon tagging
of a male sterility gene in Arabidopsis. Nature 353: 715-717
Aarts MGM, Hodge R, Kalantidis K, Florack D, Wilson ZA, Mulligan B,
Stiekema WJ, Scott R, Pereira A (1997) The A~abidopsis MALE
STERILITY 2 protein shares similarity with reductases in elongation
condensation complexes. Plant Journal 12: 615-623
Aarts MGM, Keijzer CJ, Stiekema WJ, Pereira A (1995) Molecular
characterization of the CER1 gene of Arabidopsis involved in epicuticular
wax biosynthesis and pollen fertility. Plant Cell 7: 2115-2127
Aguirre PJ, Smith AG (1993) Molecular characterization of a gene encoding a
cycteine-rich protein preferentially expressed in anthers of Lycopersicon
esculefatuna. Plant Molecular Biology 23: 477-487
Altschul SF, Madden TL, Schaffer A, Zhang J, Zhang Z, Miller W, Lipman
DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein
database search programs. Nucleic Acids Research 25: 3389-3402
Bih FY, Wu SSH, Ratnayake C, Walling LL, Nothnagel EA, Huang AHC
( 1999) The predominant protein on the surface of maize pollen is an
endoxylanase synthsized by a tapetum mRNA with a long 5' leader. The
Journal of Biological Chemistry 274: 22884-22894
Brown SM, Crouch ML (1990) Characterization of a gene family abundantly
expressed in Oenothera organensis pollen that shows sequence similarity
to polygalacturonase. Plant Cell 2: 263-274
Chen L, Zhang S, Beachy RN, Fauquet CM (1998) A protocol for consistent,
large-scale production of fertile transgenic rice plants. Plant Cell Report
18: 25-31. Plant Cell Report 18: 25-31
Cho H-T, Kende H (1998) Tissue localization of expression in deep water rice.
Plant Journal 15: 805-812
67
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
Datla RSS, Hammerlindl JK, Panchuk B, Pelcher LE, Keller W (1992)
Modified binary plant transformation vectors with the wild-type gene
encoding NPTII. Gene 122: 383-384
Fuerstenberg S, Bucciaglia PA, Smith AG (2000) Molecular characterization
of an anther-specific Been from tobacco shows sequence similarity to a
tapetum-specific gene from tomato. Sex Plan Reprod 12: 250-252
Futamura N, Mori H, Kouchi H, Shinohara K (2000) Male flower-specific
expression of gene for polygalacturonase, pectin methylesterase and (3-
1,3-glucanase in a dioecious willow (Salix gilgiana Seemen). Plant Cell
Physiology 41: 16-26
Goldberg RB, Beals TP, Sanders PM (1993) Anther development: basic
principles and practical applications. Plant Cell 5: 1217-1229
Hernandez-Pinzon I, Ross JHE, Barnes KA, Damant AP, Murphy DJ (1999)
Composition and role of tapetal lipid bodies in the biogenesis of the
pollen coat of Brassica napus. Planta 208: 588-598
Hird DL, Worral D, Hodge R, Smartt S, Wyatt P, Scott R (1993) The anther-
specific protein encoded by the Brassica napus and Arabidopsis thaliana
a6 gene displays similarity to (3-13-y~,vxava6~a. Plant Journal 4: 1023-
1033
Hodge R, Paul W, Draper J, Scott R (1992) Cold-plaque screening: a simple
technique for the isolation of low abundance, differentially expressed
transcripts from conventional cDNA libraries. Plant Journal 2: 257-267
Huang J, Hirji R, Adam L, Rozwadowski KL, Hammerlindl JK, Keller WA,
Selvaraj G (2000) Genetic engineering of glycinebetaine production
toward enhancing stress tolerance in plants: metabolic limitations. Plant
Physiology 122: 747-756
Jeon JS, Chung YY, Lee S, Yi GH, Uh BH, An G (1999) Isolation and
characterization of an anther-specific gene, RA8, from rice (Oryza sativa
L.). Plant Molecular Biology 39: 35-44
68
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
Kandasamy MK, McKinney EC, Meagher RB (1999) The late pollen-specific
actins in angiosperms. The Plant Journal 18: 681-691
Katavic, V., Reed, D. W., Taylor, D. C., Giblin, E. M., Barton, D. L., Zou, J:
T., MacKenzie, S. L., Covello, P. S. & Kunst, L. (1995). Alternation of
seed fatty acid composition by an ethyl methanesulfonate-induced
mutation in Arabidopsis thaliana affecting diacylglycerol acyltransferase
activity. Plant Physiology 108, 399-409.
Kolattukudy PE (1970) Reduction of fatty acids to alcohols by cell-free
preparations of Euglena gracilis. Biochemistry 9: 1095-1102
Koltunow AM, Truettner J, Cox KH, Wallroth M, Goldberg RB (1990)
Different temporal and spatial gene expression patterns occur during
anther development. Plant Cell 2: 1201-1224
Lopez I, Anthony RG, Maciver SK, Jiang C-J, Khan S, Weeds AG, Hussey
PJ (1996) Pollen specific expression of maize genes encoding actin
deploymerizing factor-like proteins. Proc Natl Acad Sci USA 93: 7415
7420
Mariani C, de Beuckeleer M, Truettner J, Leemans J, Goldberg RB (1990)
Induction of male sterility in plants by a chimaeric ribonuclease gene.
Nature 347: 737-741
Mascarenhas JP (1975) The biochemistry of angiosperm pollen development.
Bot Rev 41: 259-314
May CE, Appels R (1987) Wheat worldwide in "Wheat and Wheat
Improvement" (Ed. E.G. Heyne) pp 165-198. Agronomy No. 13.
American Society of Agronomy, Inc Madison, Wisconsin, USA
Metz JG, Pollard MR, Anderson L, Hayes TR, Lassner MW (2000)
Purification of a jojoba embryo fatty acyl-coenzyme A reductase and
expression of its cDNA in high erucic acid rapeseed. Plant Physiology
122:635-644
Millar AA, Clemens S, Zachgo S, Giblin EM, Taylor DC, Kunst L (1999)
CUTl, an Arabidopsis gene required for cuticular wax biosynthesis and
69
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
pollen fertility, encodes a very-long-chain fatty acid condensing enzyme.
Plant Cell 11: 825-838
Nakamura T, Yamamori M, Hirano H, Hidaka S, Nagamine T (1995)
Production of waxy (amylose-free) wheats. Molecular General Genetics
248:253-259
Nair, R. B., Joy IV, R. W., Kurylo, E., Shi, X., Schnaider, J., Datla, R. S.
S.,
Keller, W. A. & Selvaraj, G. (2000). Identification of a CYP84 family of
cytochrome P450-depdendent mono-oxygenase genes in Brassica napus
and perturbation of their expression for engineering sinapine reduction in
the seeds. Plant Physiology 123, 1623-1634.
Pearson WR, Lipman DJ (1988) Improved tools for ' biological sequence
comparison. Proc Natl Acad Sci USA 85: 2444-2448
Pickett AA, Galwey NW (1997) A further evaluation of hybrid wheat. Plant
Varieties and Seeds 10: 15-32
Piffanelli P, Ross JHE, Murphy DJ (1998) Biogenesis and function of the
lipidic structures of pollen grains. Sex Plan Reprod 11: 65-80
Preuss D, Lemieux B, Yen G, Davis RW (1993) A conditional sterile mutation
_eliminates_surface components from_Arabidopsis pollen and disrupts cell
signaling during fertilization. Genes & Development 7: 974-985
Rafnar T, Griffith IJ, Kuo M, Bond JF, Rogers BL, Klapper DG (1991)
Cloning of Afnb a I (antigen E), the major allergen family of short
ragweed pollen. Journal of Biological Chemistry 266: 1229-1236
Rubinelli P, Hu Y, Ma H (1998) Identification, sequencing analysis and
expression studies of novel anther-specific genes of Arabidopsis thaliafza.
Plant Molecular Biology 37: 607-619
Scott RJ Pollen exine - the sporopollenin enigma and the physics of pattern.
In:
Scott RJ, Stead AD (eds) Molecular and cellular aspects of plant
reproduction, pp. 49-81. Cambridge University Press, Cambridge (1994).
Shivanna KR, Cresti M, Ciampolini F (1997) Pollen development and pollen-
pistil interaction. In: Shivanna KR, Sawhney VK (eds) Pollen
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
biotechnology for crop production and improvement. Cambridge
University Press, Cambridge, United Kingdom.
Spencer DF, Schnare MN, Coulthart MB, Gray MW (1992) Sequence and
organization of a 7.2 kb region of wheat mitochondria) DNA containing
the large subunit (26S) rRNA gene. Plant Molecular Biology 20: 347-352
Ting JTL, Wu SSH, Ratnayake C, Huang AHC (1998) Constituents of the
tapetosomes and elaioplasts in Brassica campestris tapetum and their
degradation and retention during microsporogenesis. Plant Journal 16:
541-551
Vioque J, Kolattukudy PE (1997) Resolution and purification of an aldehyde-
generating and an alcohol-generating fatty acyl-CoA reductase from pea
leaves (Pisuna sativutn L.). Arch Biochem Biophys 340: 64-72
Walden AR, Walter C, Gardner RC (1999) Genes expressed in Pihus radiata
male cones include homologs to anther-specific and pathogenesis
response genes. Plant Physiology 121: 1103-1116
Wang A, Carrier K, Chisholm J, Wieczorek A, Huguenot C, Sanfacon H
(1999) Porteolytic processing of tomato ringspot nepovirus 3C-like
protease pr-ecursors: definition _of the domains for the VPg, protease and
putative RNA-depedent RNA polymerase. J Gen Virol 80: 799-809
Wang A, Fan H, Singsit C, Ozias-Akins P (1998) Transformation of peanut
with a soybean vspB promoter-uidA chimeric gene. I. Optimization of a
transformation system and analysis of GUS expression in primary
transgenic tissues and plants. Physiologia Plantarum 102: 38-48
Zou J-T, Zhan X-Y, Wu H-M, Wang H, Cheung AY (1994) Characterization
of a rice pollen-specific gene and its expression. American Journal of
Botany 81: 552-561
71
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
1 / 27
SEQUENCE LISTING
<110> National Research Council of Canada
<120> Anther-specific TAA1 genes encoding fatty acyl Co-A reductases,
and uses thereof
<130> 46450-PT
<150> US 60/296,159
<151> 2001-06-07
<160> 16
<170> PatentIn version 3.1
<210> 1
<211> 1919
<212> DNA
<213> Triticum aestivum
<220>
<221> CDS
<222> (70)..(1593)
<223>
<400> 1
ctcctctctc tctctctctc tctctctctc tcgcacgcac gcacgcacgc acacaagaaa 60
aaaatcaag atg gtt gac aca ctg agt gaa gag aac atc att gga tac ttc 111
Met
Val
Asp
Thr
Leu
Ser
Glu
Glu
Asn
Ile
Ile
Gly
Tyr
Phe
1 5 10
aagaac aagagcatc ctcatc actggatcaaca ggctttctt ggaaag 159
LysAsn LysSerIle LeuIle ThrGlySerThr GlyPheLeu GlyLys
15 20 25 30
atactg gtggagaag atactg agagttcaacct gatgtgaag aagatc 207
IleLeu ValGluLys IleLeu ArgValGlnPro AspValLys LysIle
35 40 45
tacctc ccggtgcga gcggtg gatgccgcggcg gcaaagcat cgggtg 255
TyrLeu ProValArg AlaVal AspAlaAlaAla AlaLysHis ArgVal
50 55 60
gagact gaggtggta gggaag gagttgttcggg cttctgagg gagaag 303
GluThr GluValVal GlyLys GluLeuPheGly LeuLeuArg GluLys
65 70 75
cacggg ggcaggttt caatct ttcatctgggaa aagatcgtc ccattg 351
HisGly GlyArgPhe GlnSer PheIleTrpGlu LysIleVal ProLeu
80 85 90
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
2/27
gcc gga gac gtg atg cgc gag gac ttc ggc gtc gac agc gag acc ctg 399
Ala Gly Asp Val Met Arg Glu Asp Phe Gly Val Asp Ser Glu Thr Leu
95 100 105 110
agg gag ctc cgg gtg acc cag gag ctc gat gtc atc gtt aat ggc gcc 447
Arg Glu Leu Arg Val Thr Gln Glu Leu Asp VaI Ile Val Asn Gly Ala
115 120 125
gcc acc ace aac ttc tac gaa agg tat gat gtg get cta gac gtg aac 495
Ala Thr Thr Asn Phe Tyr Glu Arg Tyr Asp Val Ala Leu Asp Val Asn
130 135 140
gtgatgggagtg aagcatatg tgcaacttc gccaagaag tgccccaat 543
ValMetGlyVal LysHisMet CysAsnPhe AlaLysLys CysProAsn
145 150 155
ctcaaggtgcte ctecatgtc tccacgget tacgtggcg ggtgagaag 591
LeuLysValLeu LeuHisVal SerThrAla TyrValAla GlyGluLys
160 165 170
caagggctggtg caagagaga ccattcaag aatggcgag acgctgctc 639
GlnGlyLeuVal GlnGluArg ProPheLys AsnGlyGlu ThrLeuLeu
175 180 185 190
gaggggacccgc ctcgacatc gacactgag ctgaaactg gccaaggac 687
GluGlyThrArg LeuAspIle AspThrGlu LeuLysLeu AlaLysAsp
195 200 205
ctgaaaaagcag cttgaggcc gacgttgat tcgtcgccc aaggccgaa 735
LeuLysLysGln LeuGluAla AspValAsp SerSerPro LysAlaGlu
210 215 220
aggaaggccatg aaggatctt ggccttacc agggcccgg cacttcagg 783
ArgLysAlaMet LysAspLeu GlyLeuThr ArgAlaArg HisPheArg
225 230 235
tggccaaacaca tacgtgttc accaagtcg atgggggag atggtgcta 831
TrpProAsnThr TyrValPhe ThrLysSer MetGlyGlu MetValLeu
240 245 250
agccagttgcag tgtgatgtc cccgttgtc atcgtccgt cccagcatc 879
SerGlnLeuGln CysAspVal ProValVal IleValArg ProSerIle
255 260 265 270
atcacaagtgtc cagaacgac ccactgccc ggatggatc gaaggcacc 927
IleThrSerVal GlnAsnAsp ProLeuPro GlyTrpIle GluGlyThr
275 280 285
aggacgatcgac acgatcgtg atcggctat gcgaagcag aacctgaca 975
ArgThrIleAsp ThrIleVal IleGlyTyr AlaLysGln AsnLeuThr
290 295 300
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
3 / 27
tac ttc ttg gcc gac ctc aac ctc acc atg gat gtg atg ccg ggc gac 1023
Tyr Phe Leu Ala Asp Leu Asn Leu Thr Met Asp Val Met Pro Gly Asp
305 310 315
atg gtg gtg aat gcg atg atg gcg gca ata gtg gca cac agc tcg tcc 1071
Met Val Val Asn Ala Met Met Ala Ala Ile Val Ala His Ser Ser Ser
320 325 330
tca ttg gag aag aca aag tea cat ccc aag caa cat gca ccg gcg gtg 1119
Ser Leu Glu Lys Thr Lys Ser His Pro Lys Gln His Ala Pro Ala Val
335 340 345 350
tac cac gtg agc tcg tcg ctg cgt aat ccg gca cca tac aat gtg ctt 1167
Tyr His Val Ser Ser Ser Leu Arg Asn Pro Ala Pro Tyr Asn Val Leu
355 360 365
cat gag get ggg ttt cgg tac ttc acg gag cac cct cgc gtg ggc cct 1215
His Glu Ala Gly Phe Arg Tyr Phe Thr Glu His Pro Arg Val Gly Pro
370 375 380
gac ggt cgc acc gtg cgt acc cat aag atg aca ttc ctc agc agc atg 1263
Asp Gly Arg Thr Val Arg Thr His Lys Met Thr Phe Leu Ser Ser Met
385 390 395
get tcc ttc cac cta ttt atg atg ctc agg tac cgc ctc ctc ctc gag 1311
Ala Ser Phe His Leu Phe Met Met Leu Arg Tyr Arg Leu Leu Leu Glu
400 405 410
ctc ctc cac ctg ctc tcc atc ctc tgc tgc ggc ctc ttc ggc ctc gac 1359
Leu Leu His Leu Leu Ser Ile Leu Cys Cys Gly Leu Phe Gly Leu Asp
415 420 425 430
acc ctc tac cac gac caa gca cgc aag tac agg ttc gtg atg cac ctg 1407
Thr Leu Tyr His Asp Gln Ala Arg Lys Tyr Arg Phe Val Met His Leu
435 440 445
gtg gat ctg tac ggg ccc ttt gcg ctg ttc aag ggg tgc ttc gat gac 1455
Val Asp Leu Tyr Gly Pro Phe Ala Leu Phe Lys Gly Cys Phe Asp Asp
450 455 460
gtc aac cta aac aag ctc agg ctc gcc atg acc agc aac cat ggt agc 1503
Val Asn Leu Asn Lys Leu Arg Leu Ala Met Thr Ser Asn His Gly Ser
465 470 475
ctc ttc aat ttc gac ccg aag acc att gat tgg gac gag tac ttc tac 1551
Leu Phe Asn Phe Asp Pro Lys Thr Ile Asp Trp Asp Glu Tyr Phe Tyr
480 485 490
agg gtc cac atc ccc ggg gtc ata aag tac atg ctc aag tga 1593
Arg Val His Ile Pro Gly Val Ile Lys Tyr Met Leu Lys
495 500 505
aatatccgtg caccgaaatt caggcgttgc cttaataatt aaataatcgt acgattgtaa 1&53
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
4/27
gaaatgtcct cccaaattgg gttctaaccg ttaaataatc tatatgaaag aggaaaatgt 1713
gtctgtcgct tgatagagca ggcatcaacg ttgcataagt ttcttgaaga caaggaatgc 1773
tacattatgt agtcccattc tggttcatgt attgtattgc taattatgac tagtattgat 1833
tgttgttacc aagccgaatt ccagcacact ggcggccgtt actagtggat ccgagctcgg 1893
taccaaaaaa aaaaaaaaaa aaaaaa 1919
<210> 2
<211> 507
<212> PRT
<213> Triticum aestivum
<400> 2
Met Val Asp Thr Leu Ser Glu Glu Asn Ile Ile Gly Tyr Phe Lys Asn
1 5 10 15
Lys Ser Ile Leu Ile Thr Gly Ser Thr Gly Phe Leu Gly Lys Ile Leu
20 25 30
Val Glu Lys Ile Leu Arg Val Gln Pro Asp Val Lys Lys Ile Tyr Leu
35 40 45
Pro Val Arg Ala Val Asp Ala Ala Ala Ala Lys His Arg Val Glu Thr
50 55 60
Glu Val Val Gly Lys Glu Leu Phe Gly Leu Leu Arg Glu Lys His Gly
65 70 75 80
Gly Arg Phe Gln Ser Phe Ile Trp Glu Lys Ile Val Pro Leu Ala Gly
85 90 95
Asp Val Met Arg Glu Asp Phe Gly Val Asp Ser Glu Thr Leu Arg Glu
100 105 110
Leu Arg Val Thr Gln Glu Leu Asp Val Ile Val Asn Gly Ala Ala Thr
115 120 125
Thr Asn Phe Tyr Glu Arg Tyr Asp Val Ala Leu Asp Val Asn Val Met
130 135 140
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
/ 27
Gly Val Lys His Met Cys Asn Phe Ala Lys Lys Cys Pro Asn Leu Lys
145 150 155 160
Val Leu Leu His Val Ser Thr Ala Tyr Val Ala Gly Glu Lys Gln Gly
165 170 175
Leu Val Gln Glu Arg Pro Phe Lys Asn Gly Glu Thr Leu Leu Glu Gly
180 185 190
Thr Arg Leu Asp Ile Asp Thr Glu Leu Lys Leu Ala Lys Asp Leu Lys
195 200 205
Lys Gln Leu Glu Ala Asp Val Asp Ser Ser Pro Lys Ala Glu Arg Lys
210 215 220
Ala Met Lys Asp Leu Gly Leu Thr Arg Ala Arg His Phe Arg Trp Pro
225 230 235 240
Asn Thr Tyr Val Phe Thr Lys Ser Met Gly Glu Met Val Leu Ser Gln
245 250 255
Leu Gln Cys Asp Val Pro Val Val Ile Val Arg Pro Ser Ile Ile Thr
260 265 270
Ser Val Gln Asn Asp Pro Leu Pro Gly Trp Ile Glu Gly Thr Arg Thr
275 280 285
Ile Asp Thr Ile Val Ile Gly Tyr Ala Lys Gln Asn Leu Thr Tyr Phe
290 295 300
Leu Ala Asp Leu Asn Leu Thr Met Asp Val Met Pro Gly Asp Met Val
305 310 315 320
Val Asn Ala Met Met Ala Ala Ile Val Ala His Ser Ser Ser Ser Leu
325 330 335
Glu Lys Thr Lys Ser His Pro Lys Gln His Ala Pro Ala Val Tyr His
340 345 350
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
6/27
Val Ser Ser Ser Leu Arg Asn Pro Ala Pro Tyr Asn Val Leu His Glu
355 360 365
Ala Gly Phe Arg Tyr Phe Thr Glu His Pro Arg Val Gly Pro Asp Gly
370 375 380
Arg Thr Val Arg Thr His Lys Met Thr Phe Leu Ser Ser Met Ala Ser
385 390 395 400
Phe His Leu Phe Met Met Leu Arg Tyr Arg Leu Leu Leu Glu Leu Leu
405 410 415
His Leu Leu Ser Ile Leu Cys Cys Gly Leu Phe Gly Leu Asp Thr Leu
420 425 430
Tyr His Asp Gln Ala Arg Lys Tyr Arg Phe Val Met His Leu Val Asp
435 440 445
Leu Tyr Gly Pro Phe Ala Leu Phe Lys Gly Cys Phe Asp Asp Val Asn
450 455 460
Leu Asn Lys Leu Arg Leu Ala Met Thr Ser Asn His Gly Ser Leu Phe
465 470 475 480
Asn Phe Asp Pro Lys Thr I1e Asp Trp Asp Glu Tyr Phe Tyr Arg Val
485 490 495
His Ile Pro Gly Val Ile Lys Tyr Met Leu Lys
500 505
<210> 3
<211> 1949
<212> DNA
<213> Triticum aestivum
<220>
<221> CDS
<222> (74)..(1642)
<223>
<400> 3
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
7/27
ctctcttctt tcatagctca 60
cctccgtctc cacgcacaca
tcattctctc
ctcccgagct
r
aaaccaaggc 109
aag
atg
gtg
ggc
acg
ctg
gat
gag
ggg
aag
atc
gtc
gac
Met
Val
Gly
Thr
Leu
Asp
Glu
Gly
Lys
Ile
Val
Asp
1 5 10
tacttcaggaac aagagcgtgctc atcaccgga gccacg ggattcctt 157
TyrPheArgAsn LysSerValLeu IleThrGly AlaThr GlyPheLeu
15 20 25
ggcaagataatg gtggagaagatc ctgcgggtc cagccg gacgtgaag 205
GlyLysIleMet ValGluLysIle LeuArgVal GlnPro AspValLys
30 35 40
aggatctacctg ccggtgcgggcg gccgacgcc gcggcg gcgaggcgc 253
ArgIleTyrLeu ProValArgAla AlaAspAla AlaAla AlaArgArg
45 50 55 60
cgggtggagacc gaggtggtgggg aaggagctg ttctgc gtgctgcgg 301
ArgValGluThr GluValValG1y LysGluLeu PheCys ValLeuArg
65 70 75
gagcgccacggc gccgggttcgac gccttcgtc gccgac aaggtggtg 349
GluArgHisGly AlaGlyPheAsp AlaPheVal AlaAsp LysValVal
80 85 90
gggctggccggc gacgtcatgcgc gagggcttc ggcgtc gaccccgcc 397
GlyLeuAlaGly AspValMetArg GluGlyPhe GlyVal AspProAla
95 100 105
acgctgcgggac ctccggctcgcc gacgagctc aacgtc atcgtcaac 445
ThrLeuArgAsp LeuArgLeuAla AspGluLeu AsnVal IleValAsn
110 115 120
ggcgccgccacc accaacttctac gaaaggtac gacgtg gccctggac 493
GlyAlaAlaThr ThrAsnPheTyr GluArgTyr AspVa1 AlaLeuAsp
125 130 135 140
gtgaacgtggtg ggggtgaagcac atgtgcgac ttcgcc cggaggtgc 541
ValAsnValVal GlyValLysHis MetCysAsp PheAla ArgArgCys
145 150 155
cccaacctcgag gtgctcatgcac gtctccacg gcctac gtcgccggc 589
ProAsnLeuGlu ValLeuMetHis ValSerThr AlaTyr ValA1aGly
160 165 170
gagaagcagggg ctggttccggag aggccgttc agggac ggcgagacg 637
GluLysGlnGly LeuValProGlu ArgProPhe ArgAsp GlyGluThr
175 180 185
ctgcgcgacgac ggcacccaactc gacatcgac gccgag atgaggctg 685
LeuArgAspAsp GlyThrGlnLeu AspIleAsp AlaGlu MetArgLeu
190 195 200
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
8/27
gccaaggacctc aggaagcag atggaggcc gacgacgat gtggacccc 733
AlaLysAspLeu ArgLysGln MetGluAla AspAspAsp ValAspPro
205 210 215 220
aaggcccagagg aaggccatg aaggacctc ggcctcacc agggccagg 781
LysAlaGlnArg LysAlaMet LysAspLeu GlyLeuThr ArgAlaArg
225 230 235
cactttgggtgg cccaacacg tacgtgttc accaaatcc atgggggag 829
HisPheGlyTrp ProAsnThr TyrValPhe ThrLysSer MetGlyGlu
240 245 250
atgatgctggcc cagatgatg cgcgggggc gacgtgccc gtcgtcatc 877
MetMetLeuAla GlnMetMet ArgGlyGly AspValPro ValValIle
255 260 265
gtccggcccagc atcatcacc agcgtccag aacgaccca ctgccagga 925
ValArgProSer IleIleThr SerValGln AsnAspPro LeuProGly
270 275 280
tggatcgaaggc accaggacg atcgacgca atcctgatc gggtacgcg 973
TrpIleGluGly ThrArgThr IleAspAla IleLeuIle GlyTyrAla
285 290 295 300
aagcagagcctg tcgtgcttc ctcgccgac ctcgaccta accatggac 1021
LysGlnSerLeu SerCysPhe LeuAlaAsp LeuAspLeu ThrMetAsp
305 310 315
gtgatgcccggc gacatggtg gtgaacgcg atgatggcg gccacggtg 1069
ValMetProGly AspMetVal ValAsnAla MetMetAla AlaThrVal
320 325 330
gcacatgcctcc tccactcag acatcagag ccagagaag aagccgcct 1117
AlaHisAlaSer SerThrGln ThrSerGlu ProGluLys LysProPro
335 340 345
ccgcagcagcaa caccctcac tcggtgccg gcagcgcca acggtgtac 1165
ProGlnGlnGln HisProHis SerValPro AlaAlaPro ThrValTyr
350 355 360
cacgtgagctcg tcgctgcgg cacccgget ccgtacgcg gtgttgtac 1213
HisValSerSer SerLeuArg HisProAla ProTyrAla ValLeuTyr
365 370 375 380
cgaacggggatc cggtacttc gaggagcac ccacgggtg gggcctgat 1261
ArgThrGlyIle ArgTyrPhe GluGluHis ProArgVal GlyProAsp
385 390 395
ggccgccccgtg cgcacccgt aaggtgcgg ttcctcggc agcatcgcg 1309
GlyArgProVal ArgThrArg LysValArg PheLeuGly SerIleAla
400 "405 410
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
9/27
gcg caccta ttc gtg ctcaagtaccgt gtccccctt gaactc 1357
ttc atg
Ala HisLeu Phe Val LeuLysTyrArg ValProLeu GluLeu
Phe Met
415 420 425
ctc ctgctc tcc ctc tgttgcggcctc tttggcctt gccgcc 1405
cgc atc
Leu LeuLeu Ser Leu CysCysGlyLeu PheGlyLeu AlaAla
Arg Ile
430 435 440
ctc cacgac ctc cgc aagtacaggttc gtgatgcag ctggtg 1453
tac gcc
Leu HisAsp Leu Arg LysTyrArgPhe ValMetGln LeuVal
Tyr Ala
445 450 455 460
gac tacggg ccc tcg ctcttcaagggt tgcttcgac gatgta 1501
ctg ttc
Asp TyrGly Pro Ser LeuPheLysGly CysPheAsp AspVal
Leu Phe
465 470 475
aac aacaag ctc ctc gccatggccgac ggtgaccat gccgat 1549
ctc agg
Asn AsnLys Leu Leu AlaMetAlaAsp GlyAspHis AlaAsp
Leu Arg
480 485 490
tcc ttcaac ttt ccc aagaccattgac tgggacgac tacttc 1597
gca gac
Ser PheAsn Phe Pro LysThrIleAsp TrpAspAsp TyrPhe
Ala Asp
495 500 505
ttc gtccac atc ggt gtcatgaagtac gtccacaag tga 1642
aag cct
Phe ValHis Ile Gly ValMetLysTyr ValHisLys
Lys Pro
510 515 520
tgttctgtgtgcgatctgct ggaggaaatc aaattaatgg
1702
tctgcgtgct
gagaaggaat
tagcgctagtgtgccttgct ttcgttcatc gagtattatt
1762
tgtgttgtgt
aacctccttc
ggttgagtattgattgtatt ccagtgacta tgagtataac
1822
gtcattggaa
gttaaattaa
taagatgaaattacttgcat aagatagtac aaggatccta
1882
catggcgggt
ctctaaaact
tgaagtacattgaaattact tcataataca aaaaaaaaaa
1942
tagtactttt
catggtacta
aaaaaaa 1949
<210> 4
<211> 522
<212> PRT
<213> Triticum aestivum
<400> 4
Met Val Gly Thr Leu Asp Glu Gly Lys Ile Val Asp Tyr Phe Arg Asn
1 5 10 15
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
10/27
Lys Ser Val Leu Ile Thr Gly Ala Thr Gly Phe Leu Gly Lys Ile Met
20 25 30
Val Glu Lys Ile Leu Arg Val Gln Pro Asp Val Lys Arg Ile Tyr Leu
35 40 45
Pro Val Arg Ala Ala Asp Ala Ala Ala Ala Arg Arg Arg Val Glu Thr
50 55 60
Glu Val Val Gly Lys Glu Leu Phe Cys Val Leu Arg Glu Arg His Gly
65 70 75 80
Ala Gly Phe Asp Ala Phe Val Ala Asp Lys Val Val Gly Leu Ala Gly
85 90 95
Asp Val Met Arg Glu Gly Phe Gly Val Asp Pro Ala Thr Leu Arg Asp
100 105 110
Leu Arg Leu Ala Asp Glu Leu Asn Val Ile Val Asn Gly Ala Ala Thr
115 120 125
Thr Asn Phe Tyr Glu Arg Tyr Asp Val Ala Leu Asp Val Asn Val Val
130 135 140
Gly Val Lys His Met Cys Asp Phe Ala Arg Arg Cys Pro Asn Leu Glu
145 150 155 160
Val Leu Met His Val Ser Thr Ala Tyr Val Ala Gly Glu Lys Gln Gly
165 170 175
Leu Val Pro Glu Arg Pro Phe Arg Asp Gly Glu Thr Leu Arg Asp Asp
180 185 190
Gly Thr Gln Leu Asp Ile Asp Ala Glu Met Arg Leu Ala Lys Asp Leu
195 200 205
Arg Lys Gln Met Glu Ala Asp Asp Asp Val Asp Pro Lys Ala Gln Arg
210 215 220
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
11 l27
Lys Ala Met Lys Asp Leu Gly Leu Thr Arg Ala Arg His Phe Gly Trp
225 230 235 240
Pro Asn Thr Tyr Val Phe Thr Lys Ser Met Gly Glu Met Met Leu Ala
245 250 255
Gln Met Met Arg Gly Gly Asp Val Pro Val Val Ile Val Arg Pro Ser
260 265 270
Ile Ile Thr Ser Val Gln Asn Asp Pro Leu Pro Gly Trp Ile Glu Gly
275 280 285
Thr Arg Thr Ile Asp Ala Ile Leu Ile Gly Tyr Ala Lys Gln Ser Leu
290 295 300
Ser Cys Phe Leu Ala Asp Leu Asp Leu Thr Met Asp Val Met Pro Gly
305 310 315 320
Asp Met Val Val Asn Ala Met Met Ala Ala Thr Val Ala His Ala Ser
325 330 335
Ser Thr Gln Thr Ser Glu Pro Glu Lys Lys Pro Pro Pro Gln Gln Gln
340 345 350
His Pro His Ser Val Pro Ala Ala Pro Thr Val Tyr His Val Ser Ser
355 360 365
Ser Leu Arg His Pro Ala Pro Tyr Ala Val Leu Tyr Arg Thr Gly Ile
370 375 380
Arg Tyr Phe Glu Glu His Pro Arg Val Gly Pro Asp Gly Arg Pro Val
385 390 395 400
Arg Thr Arg Lys Val Arg Phe Leu Gly Ser Ile Ala Ala Phe His Leu
405 410 415
Phe Met Val Leu Lys Tyr Arg Val Pro Leu Glu Leu Leu Arg Leu Leu
420 425 430
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
12/27
Ser Ile Leu Cys Cys Gly Leu Phe Gly Leu Ala Ala Leu Tyr His Asp
435 440 445
Leu Ala Arg Lys Tyr Arg Phe Val Met Gln Leu Val Asp Leu Tyr Gly
450 455 460
Pro Phe Ser Leu Phe Lys Gly Cys Phe Asp Asp Val Asn Leu Asn Lys
465 470 475 480
Leu Arg Leu Ala Met Ala Asp Gly Asp His Ala Asp Ser Ala Phe Asn
485 490 495
Phe Asp Pro Lys Thr Ile Asp Trp Asp Asp Tyr Phe Phe Lys Val His
500 505 510
Ile Pro Gly Val Met Lys Tyr Val His Lys
515 520
<210> 5
<211> 1949
<212> DNA
<213> Triticum aestivum
<220>
<221> CDS
<222> (94)..(1617)
<223>
<400> 5
C'tCCttCttC CtC'tCtCtCt CtCtCtCtCC CtgCtC'tCCC tgCtCtCtCt CtCtCtCtCt 6O
ctctctcttg cgcacacaag aaaaaaaatc aag atg gtt gac aca ctg agt gaa 114
Met Val Asp Thr Leu Ser Glu
1 5
gag aag atc att gga tac ttc aag aac aag agc atc ctc atc act gga 162
Glu Lys Ile Ile Gly Tyr Phe Lys Asn Lys Ser Ile Leu Ile Thr Gly
15 20
tca aca ggc ttt ctt gga aag ata cta gtg gag aag ata ctg aga gtt 210
Ser Thr Gly Phe Leu Gly Lys Ile Leu Val Glu Lys Ile Leu Arg Val
25 30 35
caa cct gat gta aag aag atc tat ctc ccg gtg cga gcg gtg gat gcc 258
Gln Pro Asp Val Lys Lys Ile Tyr Leu Pro Val Arg Ala Val Asp Ala
40 45 50 55
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
13!27
gcg gcg gcg aag gat cgg gtg gag act gag gtg gta ggg aag gag ttg 306
Ala Ala Ala Lys Asp Arg Val Glu Thr Glu Val Val Gly Lys Glu Leu
60 65 70
ttc ggg ctt ctg agg gag aag cac ggg gac tgg ttt caa tct ttc atc 354
Phe Gly Leu Leu Arg Glu Lys His Gly Asp Trp Phe Gln Ser Phe Ile
75 80 85
tgt gaa aag atc gtc cca ttg gcc gga gat gtg atg cgt gag gac ttt 402
Cys Glu Lys Ile Val Pro Leu Ala Gly Asp Val Met Arg Glu Asp Phe
90 95 100
ggc gtc gac agc gag acc ctg agg gag ctc cgg gtg acc cag gag ctc 450
Gly Val Asp Ser Glu Thr Leu Arg Glu Leu Arg Val Thr Gln Glu Leu
105 110 115
gat gtc atc gtt aat ggc gcc gcc acc acc aac ttc tac gaa agg tat 498
Asp Val Ile Val Asn Gly Ala Ala Thr Thr Asn Phe Tyr Glu Arg Tyr
120 125 130 135
gat gtg gcc ctg gac gtg aac gtg atg gga gtg aag cat atg tgc aac 546
Asp Val Ala Leu Asp Val Asn Val Met Gly Val Lys His Met Cys Asn
140 145 150
ttc gcc aag aag tgc ccc aat ctc aag gtg ctc ctc cat gtc tcc acg 594
Phe Ala Lys Lys Cys Pro Asn Leu Lys Val Leu Leu His Val Ser Thr
155 160 165
get tat gtt gcg ggt gag aag caa gga ctc gtg caa gag aga cca ttc 642
Ala Tyr Val Ala Gly Glu Lys Gln Gly Leu Val Gln Glu Arg Pro Phe
170 175 180
aag aat ggc gag acg ctg ctc gag ggg acc cac ctc gat atc gac acc 690
Lys Asn Gly Glu Thr Leu Leu Glu Gly Thr His Leu Asp Ile Asp Thr
185 190 195
gag ctg aaa ctg gcc aag gac ctg aaa aag cag ctt gag gcc gac gcc 738
Glu Leu Lys Leu Ala Lys Asp Leu Lys Lys Gln Leu Glu Ala Asp Ala
200 205 210 2I5
gac tcg tcg ccc aag tcc caa agg aag gcc atg aag gac ctt ggc atc 786
Asp Ser Ser Pro Lys Ser Gln Arg Lys Ala Met Lys Asp Leu Gly Ile
220 225 230
acc agg gcc cgg cac ttc ggg tgg ccg aac aca tac gtg ttc acc aag 834
Thr Arg Ala Arg His Phe Gly Trp Pro Asn Thr Tyr Val Phe Thr Lys
235 240 245
tcg atg ggg gag atg gtg ctg ggc cag ttg aag tgt gat ctc cot gtt 882
Ser Met Gly Glu Met Val Leu Gly Gln Leu Lys Cys Asp Leu Pro Val
250 255 260
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
14/27
gtcatcgtccgtccc agcatcatc accagtgtc cagaac gacccactg 930
ValIleValArgPro SerIleIle ThrSerVal GlnAsn AspProLeu
265 270 275
cccggatggatcgaa ggcaccagg acgatcgac acgatc gtgatcggc 978
ProGlyTrpIleGlu GlyThrArg ThrIleAsp ThrIle ValIleGly
280 285 290 295
tatgcgaagcagaac ctgacatac ttcttggcg gacctc aacctcacc 1026
TyrAlaLysGlnAsn LeuThrTyr PheLeuAla AspLeu AsnLeuThr
300 305 310
atggatgtgatgccg ggcgacatg gtggtgaat gcgatg atggetgcc 1074
MetAspValMetPro GlyAspMet ValValAsn AlaMet MetAlaAla
315 320 325
atcgtggcgcacagc tcgtcctta ttggagaag acacag tcacatccc 1122
IleV'alAlaHisSer SerSerLeu LeuGluLys ThrGln SerHisPro
330 335 340
gagccacacgcaccg gcggtgtac cacgtg-agctcgtcg cggcgtaac 1170
GluProHisAlaPro AlaValTyr HisVal5er SerSer ArgArgAsn
345 350 355
ccggcgccgtac aatgtgctgcac gaggetggg tttcggtac ttcacg 1218
ProAlaProTyr AsnValLeuHis GluAlaGly PheArgTyr PheThr
360 365 370 375
gagcaccctcgg gtgggccctgac ggccgcacg gtgcgcacc cataag 1266
GluHisProArg ValGlyProAsp GlyArgThr ValArgThr HisLys
380 385 390
atgacattcctc agcagcatgget tccttccac ctctttatg atgctc 1314
MetThrPheLeu SerSerMetAla SerPheHis LeuPheMet MetLeu
395 400 405
aggtaccgcctc ctccttgagctc ctccacctg ctctccgtc ctctgt 1362
ArgTyrArgLeu LeuLeuGluLeu LeuHisLeu LeuSerVal LeuCys
410 415 420
tgtggcctcttc ggcctcgacacc ctctaccac gaccaagca cgcaag 1410
CysGlyLeuPhe GlyLeuAspThr LeuTyrHis AspGlnAla ArgLys
425 430 435
tacaggttcgtg atgcacctggtg gatctgtat gggcccttc gcgctg 1458
TyrArgPheVal MetHisLeuVal AspLeuTyr GlyProPhe AlaLeu
440 445 450 455
ttcaagggctgc ttcgatgacgtc aacctaaac aagctcagg ctcgcc 1506
PheLysGlyCys PheAspAspVal AsnLeuAsn LysLeuArg LeuAla
460 465 470
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
15 / 27
atg acc aac cat gga agc ctc ttt gac ccc acc att 1554
agc aat ttc aag
Met Thr Asn His Gly Ser Leu Phe Asp Pro Thr Ile
Ser Asn Phe Lys
475 480 485
gac tgg gat tac ttc tac agc gtc ccc ggg cta aag 1602
gac cac atc gtc
Asp Trp Asp Tyr Phe Tyr Ser Val Pro Gly Leu Lys
Asp His Ile Val
490 495 500
cac atg aac tga aatatccatg caccgaaaat ttaataat 1657
ctc ttaggcgttg cc
His Met Asn
Leu
505
taaataatcgtacgttgtaa gaaatatcct cccaaattgggttctaaccgttaaataatc1717
tgtgtgaaagagggaaatgt gtctgtcgct tgatagtgcaggcatcaacttgcatatgct1777
tcttgaagaacaaggaatgc tacatcatgt agtttcgttctggctcgtgctgtattgcca1837
attatgactagtattggttg ataacaattt cttgttcagtcacatattgtaccttgacta1897
gtattgattgttaattataa tttcttttca agagaaaaaaaaaaaaaaaaas 19.49
<210>
6
<211>
507
<212>
PRT
<213> icum aestivum
Trit
<400> 6
Met Val Asp Thr Leu Ser Glu Glu Lys Ile Ile Gly Tyr Phe Lys Asn
1 5 10 15
Lys Ser Ile Leu Ile Thr Gly Ser Thr Gly Phe Leu Gly Lys Ile Leu
20 25 30
Val Glu Lys Ile Leu Arg Val Gln Pro Asp Val Lys Lys Ile Tyr Leu
35 40 45
Pro Val Arg Ala Val Asp Ala Ala Ala Ala Lys Asp Arg Val Glu Thr
50 55 60
Glu Val Val Gly Lys Glu Leu Phe Gly Leu Leu Arg Glu Lys His Gly
65 70 75 80
Asp Trp Phe Gln Ser Phe Ile Cys Glu Lys Ile Val Pro Leu Ala Gly
85 90 95
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
16/27
Asp Val Met Arg Glu Asp Phe Gly Val Asp Ser Glu Thr Leu Arg Glu
100 105 110
Leu Arg Val Thr Gln Glu Leu Asp Val Ile Val Asn Gly Ala Ala Thr
115 120 125
Thr Asn Phe Tyr Glu Arg Tyr Asp Val Ala Leu Asp Val Asn Val Met
130 135 140
Gly Val Lys His Met Cys Asn Phe Ala Lys Lys Cys Pro Asn Leu Lys
145 150 155 160
Val Leu Leu His Val Ser Thr Ala Tyr Val Ala Gly Glu Lys Gln Gly
165 170 175
Leu Val Gln Glu Arg Pro Phe Lys Asn Gly Glu Thr Leu Leu Glu Gly
180 185 190
Thr His Leu Asp Ile Asp Thr Glu Leu Lys Leu Ala Lys Asp Leu Lys
195 200 205
Lys Gln Leu Glu Ala Asp Ala Asp Ser Ser Pro Lys Ser Gln Arg Lys
210 215 220
Ala Met Lys Asp Leu Gly Ile Thr Arg Ala Arg His Phe Gly Trp Pro
225 230 235 240
Asn Thr Tyr Val Phe Thr Lys Ser Met Gly Glu Met Val Leu Gly Gln
245 250 255
Leu Lys Cys Asp Leu Pro Val Val Ile Val Arg Pro Ser Ile Ile Thr
260 265 270
Ser Val Gln Asn Asp Pro Leu Pro Gly Trp Ile Glu Gly Thr Arg Thr
275 280 285
Ile Asp Thr Ile Val Ile Gly Tyr Ala Lys Gln Asn Leu Thr Tyr Phe
290 295 300
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
17127
Leu Ala Asp Leu Asn Leu Thr Met Asp Val Met Pro Gly Asp Met Val
305 310 315 320
Val Asn Ala Met Met Ala Ala Ile Val Ala His Ser Ser Ser Leu Leu
325 330 335
Glu Lys Thr Gln Ser His Pro Glu Pro His Ala Pro Ala Val Tyr His
340 345 350
Val Ser Ser Ser Arg Arg Asn Pro Ala Pro Tyr Asn Val Leu His Glu
355 360 365
Ala Gly Phe Arg Tyr Phe Thr Glu His Pro Arg Val Gly Pro Asp Gly
370 375 380
Arg Thr Val Arg Thr His Lys Met Thr Phe Leu Ser Ser Met Ala Ser
385 390 395 400
Phe His Leu Phe Met Met Leu Arg Tyr Arg Leu Leu Leu Glu Leu Leu
405 410 415
His Leu Leu Ser Val Leu Cys Cys Gly Leu Phe Gly Leu Asp Thr Leu
420 425 430
Tyr His Asp Gln Ala Arg Lys Tyr Arg Phe Val Met His Leu Val Asp
435 440 445
Leu Tyr Gly Pro Phe Ala Leu Phe Lys Gly Cys Phe Asp Asp Val Asn
450 455 460
Leu Asn Lys Leu Arg Leu Ala Met Thr Ser Asn His Gly Ser Leu Phe
465 470 475 480
Asn Phe Asp Pro Lys Thr Ile Asp Trp Asp Asp Tyr Phe Tyr Ser Val
485 490 495
His Ile Pro Gly Val Leu Lys His Met Leu Asn
500 505
<210> 7
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
18/27
<211> 3392
<212> DNA
<213> Triticum aestivum
<400>
7
cctctttaagtcggtcaggtacgtagcaaataaaccaaacaagcaggacatgtttgaaaa 60
tcactgttctaactttaataatgcacacaaggaattgaaggcttgataaatcctgtgcta 120
gtaccacccatccgactgtcggtttttaagacatggtagagtagagtactaacataacga 180
tcttcaagatattgtgtccaagttctttgccacaggtttgtcgtattgagccaaaatata 240
ccactgaattatgtgctaatagtgtaattattggtcaattttttgttctaacgagagaaa 300
aatacgctatgcgttttgaagggatggagtacgatgatactactctattgtactccacgg 360
atcaattgtaggagtaccctctaatttctaatggtataactttgatgacatacgatttgt 420
attacgttggttgaattgtacatgggccttctataaagcgaaaaagaaaggaatactgag 480
cttctctagttttctataamccagagttagacgtcctactggacaactcattcctaagtg 540
tgtagcaataatactgagtactaccggttgtccacttgagctattaatttgatggctagc 600
aacgtactgcaaccccaataataaccctgtttgaaaatctagagtacctctacgcgttat 660
tttgactgcatgtgattcctctcatgatcaccaggtcccttgktttgggttggacagcgc 720
ataaatgttgcgctcaagtgcctcaaattaagactcccctctcctacctacaacacaaaa 780
gacctttttt'tcagcaaaagagggcgccaccccctccgatttcatataatgaaaccaact 840
ggttcaactg ccaaacaaac aacaaaaaca aaacgaaaca ccttcatggc ccgtgtataa 900
atattgtgta catgaccctc tttccactca cccacacaaa tcatctcctc tctctctctc 960
tctctctctctctctcgcacgcacgcacgcacgcacacaagaaaaaaatcaagatggttg1020
acacactgagtgaagagaacatcattggatacttcaagaacaagagcatcctcatcactg1080
gatcaacaggctttcttggaaagagtatgcatgcatgcataatgtttatgcacacatatt1140
acgcatgcatatacttttttttggcaaattctagaagttttttgcgtcaagttcatatgt1200
gctgtcctcatctgttaattagttcactgttggtgtgcgtatatgcagtactggtggaga1260
agatactgagagttcaacctgatgtgaagaagatctacctcccggtgcgagcggtggatg1320
ccgcggcggcaaagcatcgggtggagactgaggtagtgttgtccatcatggttcttcatg1380
atctataccctttacccctctcatgattcttttgatcccttttcttcagagttagcgctg1440
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
19/27
actaatttcttttctatatgcgcaggtggtagggaaggagttgttcgggcttctgaggga1500
gaagcacgggggcaggtttcaatctttcatctgggaaaagatcgtcccattggccggaga1560
cgtgatgcgcgaggacttcggcgtcgacagcgagaccctgagggagctccgggtgaccca1620
ggagctcgatgtcatcgttaatggcgccgccaccaccaacttctacgaaaggtgcgtcat1680
tgtcaactgattgttttgtccaagaaaggaaaaatcagcagagaacttgagtaggtgcag1740
caagagtacgtacgcagtgaatggtctcagaagaacacttgtgtgcgtgcaggtatgatg1800
tggctctagacgtgaacgtgatgggagtgaagcatatgtgcaacttcgccaagaagtgcc1860
ccaatctcaaggtgctcctccatgtctccacgggtacgtacaccaactctacaggtaaaa1920
acaaaaataaagttcttggatgttaattataatacacctagatttgatttacaaatgaag1980
ttaataaattcatatatgagttggtgcagcttacgtggcgggtgagaagcaagggctggt2040
gcaagagagaccattcaagaatggcgagacgctgctcgaggggacccgcctcgacatcga2100
cactgagctgaaactggccaaggacctgaaaaagcagcttgaggccgacgttgattcgtc2160
gcccaaggccgaaaggaaggccatgaaggatcttggccttaccagggcccggcacttcag2220
gtggccaaacacatacgtgttcaccaagtcgatgggggagatggtgctaagccagttgca2280
gtgtgatgtccccgttgtcatcgtccgtcccagcatcatcacaagtgtccagaacgaccc2340
actgcccggatggatcgaaggcaccaggttcattatatgtttcttgtcctttctctgcct2400
ccaaatttagagtgcaatcgtcttactctgttgcaaatgccaaaagaagtaaaatatgat2460
atttgttcaatgtaaaaatgtaaattgcaggacgatcgacacgatcgtgatcggctatgc2520
gaagcagaacctgacatacttcttggccgacctcaacctcaccatggatgtggtaagcaa2580
cgttgtactatgcatgcagttaagatatattccaggcaatggttggttgtcagtccagtc2640
caggaatccgtacagtaagatgaatttcgacgacgatggtgaaagcaatcgtttggttgg2700
gtatatgttgctgtagatgccgggcgacatggtggtgaatgcgatgatggcggcaatagt2760
ggcacacagctcgtcctcattggagaagacaaagtcacatcccaagcaacatgcaccggc2820
ggtgtaccacgtgagctcgtcgctgcgtaatccggcaccatacaatgtgcttcatgaggc2880
tgggtttcggtacttcacggagcaccctcgcgtgggccctgacggtcgcaccgtgcgtac2940
ccataagatgacattcctcagcagcatggcttccttccacctatttatgatgctcaggta3000
CCCJCCtCCtCCtCgagCtCCtCC3CCtgCtCtCCatCCtCtgctgcggcctcttcggcct3060
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
20 / 27
cgacaccctctaccacgaccaagcacgcaagtacaggttagttagttggttgaaatcttg3120
tgcggttgtatcttcttgatggctcccacataattaagatgacacgacttttattgttgt3180
attgttataggttcgtgatgcacctggtggatctgtacgggccctttgcgctgttcaagg3240
ggtgcttcgatgacgtcaacctaaacaagctcaggctcgccatgaccagcaaccatggta3300
gcctcttcaatttcgacccgaagaccattgattgggacgagtacttctacagggtccaca3360
tccccggggtcataaagtacatgctcaagtga 3392
<210>
8
<211>
5848
<212>
DNA
<213> icum aestivum
Trit
<400>
8
ctgaccaaacatctcccatcaggaaccatgttactatttaattttgcatgttctttgctt60
cgtgctgtttgtcaaggacacgctagcttctgcattatacagacaggctggaaatctgta120
aataatgtgttggtgaatctttggagtatttttctaatgttgatcagtacttgttgacct180
aggtggcgactaactttggacaggggcaagcaagcaaaagcagagacagctagcttctgc240
attatcagaaaatagcctgtgttggtgaatcttcagagtgctcctactcccccgtccccc300
caaatgttcagtacttgttgacctactggagctggacgacccgctctgaaagttttactt360
gtccacacttgtctggccaaaatccccaatgaatcagaatataggctgaaaaatgagtgg420
gtcgtttttggagctcggcctccacggaggccgaaaaaaaactcaaaattctaatttctc480
agtttaaaaaaaaatgaaaaaaattgtacaagtaaacaaggttgtgatgtgtatgtgtgt540
aaaatttcagaacaaaatactttgaaatgcgatctgagcaaaaaaaaacaaattcataga600
cttttgagaatgaatagtgcaagctactaaaacgcatgagattctttttgcgtgtagcac660
tcattttaatgtatttcgtcctgaaaattacaaacttgtgcattatgtctataagtatat720
ggtggatttaaaaaaaaactgaaacatgcaaatttgaattttgatcatttcaaaatcggc780
ctccatggagcatttttgctgaaaaatctaaaagaaaacgcagtccaaccttagtaatta840
cccatgttggtgaatccagagtgtttacgataaatataggacctctttagctagcgaacg900
atggcatttgcgaatgttatttatggcaaatccttcagaacacacatgccaatttcttca960
taaacacatggcaattctggcaaatattttgctgtgaaaattgttgtgcttgcttcaagg1020
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
21 / 27
aattgccatgttcccgcaatataaaatgccatgaaaaacgttcattttgtcatatcccca1080
gcgaacgtccgccgagtagcattaccgtaaatatagggggtgtttggttcagaagttcta1140
ggactttttctagtcccagggactaatcaaaaaagactctctaatagagtctttttctag1200
tccctatagaaaaagtccctcccgtttggttcctagggactttttttagtctctaggact1260
aaaaagtccctgaaccaaacaccccatattagtagtaccgcaaaaagtagtaagtactac1320
agtattggtggatctttaatgagttaattaacctagaatggctcctgctcctccggtttc1380
actggaaaagcaggttgtggttgggaagcacaagcgtgttgccgttgctaattacgtgca1440
ctcaatagccagccagccaccctcaacacctcgctggccggccctgcgcgtgtataaata1500
ctgggtgccggagcacacaagcctcacacaccaagcacacctctcttcttcctccgtctc1560
tcattctctcctcccgagcttcatagctcacacgcacacaaaaccaaggcaagatggtgg1620
gcacgctggatgaggggaagatcgtcgactacttcaggaacaagagcgtgctcatcaccg1680
gagccacgggattccttggcaagagtaagtgagcatgatgaggatatatcatatatgcat1740
gccttgctattttctgaagaacagttcatgcatccatgctgagagtccttggtgttgcat1800
tgtgtgctcactcttcgacctgtgcagtaatggtggagaagatcctgcgggtccagccgg1860
acgtgaagaggatctacctgccggtgcgggcggccgacgccgcggcggcgaggcgccggg1920
tggagaccgaggtgcgtgcctctctgcccgtccatcatgctccagcatgatggtgatcct1980
tgcaccccctccatcaatctcgctctgaacactcctctgttctagagtaatactagtagt2040
agacaaaggatgcatgccggcagcatggttctcatgctgcaaggaacacatgcgagcttt2100
ctctcagacagagaagcccccaagctgctcccactgtgccccttgtgtctcttcattccg2160
tttcatattcactactctttttttcattgataatcaatactgtaatactagtaatattat2220
gtggcgcatcggaacaaaacatgcatagatatcacagaaccataaatagtccagctgccc2280
gctgctatatcagtgatgcaaacatgaacatgggtagaatagttatcttataggcataaa2340
ccggcattagtcaggactaggtgcacactgttgagtctcatccctgccgaacattcaggc2400
aggtcacatgccattttttcttctctttctgaggttgagagtcacatgccatactctgca2460
tgagaggtggccatgaataatctcagctgtacaccgctcattcttctctactactccctc2520
cgttcctaaatatgtctttttaaagattccaacaagtgactacatacagaataaaatgaa2580
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
22 / 27
tgaatctata ttctaaaata tgtctacata catccatatg ttatagtcca tttaaaatgt 2640
ctaaaaaaac ttatatttag gaacggaggg agtagatact agttaaatgc acaaatattc 2700
tgccagcttt tggtatgcca ggattaattt gaacgagaaa ccagatacta gtaggcgtat 2760
ggttgacgaa attttcaaca ttttcattta cgcccgatcc tggaaaccgg gaaccttggg 2820
aaaaccggcc gcaggatgac aattaaaccg gcggcgaagt agcgcagctg tgtgtgtatc 2880
cctaggagtc gtataggagt tatggtgtgg gtaggccggt aggtttggtc aggtcgactc 2940
gattcacaagttttcaatggcctctgccgcatgcagtgcacaactggtactgcaccacca3000
cctagctagctagccggccagtgtgcagcctgcatgcatgcatgcatgcatggctggctg3060
gctgctgctgggaaatgacgagcgcagggcac,catctggcactggcaccggcaccactaa3120
ctacagcttcatcaacttgagaattcagaacccagagtttcttgtcatatgtgtatagag3180
tggaccgcaagtctgcaatgatgtgttggtacctgattgcatcaacaatgaacacatata3240
actgcaattatgttttggttagtagattctagtatattgcaaaagaaaaaaagtgtagtt3300
attgagtggtgccaaggagatccagagcagagacaatgggcagtatgcttggtcctccac3360
cctactcctactccatctagtaaaagccgggaaagaaacccttccagcacgcctggcccc3420
tcttcatttcttcatatgcattcactgatccttcattgttcggcatgggcaatttacagc3480
tgcagacagttgcacgggtcggttgtcgtgcgctgcagaagcaatttatagtttttacca3540
aaagttgcctgagacactcaccatgttgggtggtggatccacagtgcgtgcatgcatgta3600
tcaaacgcacattgcaggttaagaattggtgtaaatcaagcacgtctattgatcaaacta3660
attttatcgagaccggcgtggttcgcaggtggtggggaaggagctgttctgcgtgctgcg3720
ggagcgccacggcgccgggttcgacgccttcgtcgccgacaaggtggtggggctggccgg3780
cgacgtcatgcgcgagggcttcggcgtcgaccccgccacgctgcgggacctccggctcgc3840
cgacgagctcaacgtcatcgtcaacggcgccgccaccaccaacttctacgaaaggtgctg3900
tgtactattgactgttttaattaacttcgtcagagagaaaattcagcgggagagaacgtg3960
catggctgcaccttcttctccagagtagtgagctcatcaatggcctctgcaactaacttt4020
ggaatggacgtgtgcgtgctcaggtacgacgtggccctggacgtgaacgtggtgggggtg4080
aagcacatgtgcgacttcgcccggaggtgccccaacctcgaggtgctcatgcacgtctcc4140
acggcctacgtcgccggcgagaagcaggggctggttccggagaggccgttcagggacggc4200
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
23 / 27
gagacgctgc gcgacgacgg cacccaactc gacatcgacg ccgagatgag gctggccaag 4260
gacctcaggaagcagatggaggccgacgacgatgtggaccccaaggcccagaggaaggcc4320
atgaaggacctcggcctcaccaggttagcaatcctatatttttccaaagtttctcctctt4380
tcctgtgaaattcaagttcaaaaaacagagggttttattttgtgaaacttggaactgaaa4440
ttcaatatatttttttgtaattgtgtgtgcagggccaggcactttgggtggcccaacacg4500
tacgtgttcaccaaatccatgggggagatgatgctggcccagatgatgcgcgggggcgac4560
gtgcccgtcgtcatcgtccggcccagcatcatcaccagcgtccagaacgacccactgcca4620
ggatggatcgaaggcaccaggtcggttcaaactctttgggtctaaatatgaatgccaagt4680
ttcaaattcaaattctaacaccgaaatgaaaaatgcaggacgatcgacgcaatcctgatc4740
gggtacgcga agcagagcct gtcgtgcttc ctcgccgacc tcgacctaac catggacgtg 4800
gtaattgctt cactcgttat tagttcagca aatgcagagc tgctagtgct actgtcttct 4860
tgcgaacgtg ctgcagtaat gagtaataag tatactagta cagtgtgtga ccatgctgca 4920
gtaagttgca tctgaagcgt ctgtgccaac caggccaagt ggctgctgca gtgcatcagc 4980
caaccttaga tgaacttggc tgcgtgcggt tactatctga atcttgcctg ttacagcaag 5040
gttacatatg ctgctactgg attatcaagt tctaacccaa atgcgctacc atggtgtctg 5100
cagatgcccg gcgacatggt ggtgaacgcg atgatggcgg ccacggtggc acatgcctcc 5160
tccactcaga catcagagcc agagaagaag ccgcctccgc agcagcaaca ccctcactcg 5220
gtgccggcag cgccaacggt gtaccacgtg agctcgtcgc tgcggcaccc ggctccgtac 5280
gcggtgttgt accgaacggg gatccggtac ttcgaggagc acccacgggt ggggcctgat 5340
ggccgccccg tgcgcacccg taaggtgcgg ttcctcggca gcatcgcggc gttccaccta 5400
ttcatggtgc tcaagtaccg tgtccccctt gaactcctcc gcctgctctc catcctctgt 5460
tgcggcctct ttggccttgc cgccctctac cacgacctcg cccgcaagta caggtacatt 5520
cttcttccgg aattagttga cgctcaaacg aattatctag agggagcaca gtttaaacac 5580
ttttgcggtt gtaccgtgtt gatgttatac cattggctta agaacatgca ttttgtgtag 5640
gttcgtgatg cagctggtgg acctgtacgg gcccttctcg ctcttcaagg gttgcttcga 5700
cgatgtaaac ctcaacaagc tcaggctcgc catggccgac ggtgaccatg ccgattccgc 5760
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
24127
attcaacttt gaccccaaga ccattgactg ggacgactac ttcttcaagg tccacatccc 5820
tggtgtcatg aagtacgtcc acaagtga 5848
<210> 9
<211> 2800
<212> DNA
<213> Triticum aestivum
<400> 9
actccgacac aatattttga ctgcatgtga ttcctcttat gatcactgag tctttgtttc 60
ggttggacag cgcataaatg ttgcgctcaa gtgcctcaaa ttaagacacc cctcttctac 120
ctacaacacg aaagaccctt ttttcttagc aaaaatttca tacaatgaaa ccaactggtt 180
caactgccaaaacaacagactccaagacaaaacaccttgatggcccgtgtataaatattg240
tgtacaggagcctctttccactcacctacacaaatcatctccttcttcctctctctctct300
C'tCtCCCtgCtCtCCCtgCtCtCtC'tCtCtctCtCtCtCtctcttgcgcacacaagaaaa360
aaaatcaagatggttgacacactgagtgaagagaagatcattggatacttcaagaacaag420
agcatcctcatcactggatcaacaggctttcttggaaagagtatgcatgcatgcacgcat480
aatgtttatacaaacatattacgcatgcatatacta'tttgttgcaaattctagaagtttt540
ttttttgcgtcaagttgatatgtgttgtgctcatctgttaattagttctactgttggtgc600
atatgtgcagtactagtggagaagatactgagagttcaacctgatgtaaagaagatctat660
ctcccggtgcgagcggtggatgccgcggcggcgaaggatcgggtggagactgaggtagtg720
ttgtccatcatggttcttcatgatctagacctctactgctctcatgattcttttgatccc780
tttgcttgagtgttggtactgaataatttattttctatgtgcccaggtggtagggaagga840
gttgttcgggcttctgagggagaagcacggggactggtttcaatctttcatctgtgaaaa900
gatcgtcccattggccggagatgtgatgcgtgaggactttggcgtcgacagcgagaccct960
gagggagctccgggtgacccaggagctcgatgtcatcgttaatggcgccgccaccaccaa1020
cttctacgaaaggtgcgtcgtcgtcaactgattgttttgtccaagaaaggaaaaaaaatc1080
agcagagaacttgagtaggtgcagcaagagtacggagtggaattgtgtgcgtgcaggtat1140
gatgtggccctggacgtgaacgtgatgggagtgaagcatatgtgcaacttcgccaagaag1200
tgccccaatctcaaggtgctcctccatgtctccacgggtacgtacaccaactctacaaat1260
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
25 / 27
taccatcatg gactatgaac ttggatgctt ctggggaaaa caaaaatgaa gttcttggat 1320
gtaattaaag tacacctaga tttgatttac aaatcaagtt aatgaattca tacatgagtt 1380
ggtgcagcttatgttgcgggtgagaagcaaggactcgtgcaagagagaccattcaagaat1440
ggcgagacgctgctcgaggggacccacctcgatatcgacaccgagctgaaactggccaag1500
gacctgaaaaagcagcttgaggccgacgccgactcgtcgcccaagtcccaaaggaaggcc1560
atgaaggaccttggcatcaccagggcccggcacttcgggtggccgaacacatacgtgttc1620
accaagtcgatgggggagatggtgctgggccagttgaagtgtgatctccctgttgtcatc1680
gtccgtcccagcatcatcaccagtgtccagaacgacccactgcccggatggatcgaaggc1740
accaggttcgttatatgcttctctttcctttccctgcctctaaatttaaagtgcaatcgt1800
tttaatctgttgcaaatgcaagtaaataaaggatgtttgttcaatgtaaaaatgtaaatt1860
gcaggacgatcgacacgatcgtgatcggctatgcgaagcagaacctgacatacttcttgg1920
cggacctcaacctcaccatggatgtggtaagcaacgttgcactatgcatgcagttaatta1980
acatatattccaggcatgcaatggttggttgtcagtccaggaatccatacagtaagatat2040
ggatttcaacgatggcggtgaatgcaatcgtgtggttgggtatatgttggtgcagatgcc2100
gggcgacatggtggtgaatgcgatgatggctgccatcgtggcgcacagctcgtccttatt2160
ggagaagacacagtcacatcccgagccacacgcaccggcggtgtaccacgtgagctcgtc2220
gcggcgtaacccggcgccgtacaatgtgctgcacgaggctgggtttcggtacttcacgga2280
gcaccctcgggtgggccctgacggccgcacggtgcgcacccataagatgacattcctcag2340
cagcatggcttccttccacctctttatgatgctcaggtaccgcctcctccttgagctcct2400
CC3CCtgCtCtCCgtCCtCtgttgtggCCtCttCggCCtCgaC3CCCtCtaCCaCgaCCa2460
agcacgcaagtacaggttagtcggtttaaatcttttgcggatggcatttttgataacaag2520
tatttccggacggagggagtatcttcttgatggcgcggcatatatgatgacacggccttt2580
attgttatattgttgtaggttcgtgatgcacctggtggatctgtatgggcccttcgcgct2640
gttcaagggctgcttcgatgacgtcaacctaaacaagctcaggctcgccatgaccagcaa2700
ccatggaagcctctttaatttcgaccccaagaccattgactgggacgattacttctacag2760
cgtccacatccccggggtcctaaagcacatgctcaactga 2800
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
26 / 27
<210> 10
<211> 20
<212> DNA
<213> Artificial sequence
<220>
<223> OL2707 primer
<400> 10
gactacgtcg tccaaggccg 20
<210> 11
<211> 21
<212> DNA
<213> Artificial sequence
<220>
<223> OL2708 primer
<400> 11
gtcgaactgc ttgagcagcg c 21
<210> 12
<211> 19
<212> DNA
<213> OL2881 primer
<400> 12
gcagaacctg acatacttc 19
<210> 13
<211> 18
<212> DNA
<213> Artificial sequence
<220>
<223> OL2885 primer
<400> 13
gaggcggtac ctgagcat 18
<210> 14
<211> 18
<212> DNA
<213> OL2884 primer
<400> 14
ttcgcatagc cgatcacg 18
CA 02449810 2003-12-04
WO 02/099111 PCT/CA02/00834
27 / 27
<210> 15
<211> 18
<212> DNA
<213> Artificial sequence
<220>
<223> 0L2883 primer
<400> 15
aatgccggcc ctggtaag 18
<210> 16
<211> 18
<212> DNA
<213> Artificial sequence
<220>
<223> 0L2880 primer
<400> 16
caggtggcca aacacata 18