Note: Descriptions are shown in the official language in which they were submitted.
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
METHODS FOR GENERATING ANTIBIOTIC RESISTANT MICROBES
AND NOVEL ANTIBIOTICS
FIELD OF THE INVENTION
This invention relates to the field of antimicrobial treatments and gene
targets for the
discovery of antimicrobial agents. In particular, it relates to the discovery
of genes essential
for growth and virulence of bacteria.
BACKGROUND OF THE INVENTION
Despite the development of new classes of antimicrobial agents over the past
decade
(reviewed in http://vet.purdue.edu/bms), microbial infections remain a serious
health problem.
While antibiotics treatment has been effective in controlling infectious
diseases, an increase
in the number of antibiotic-resistant (AR) microbes have emerged and are now
posing a major
therapeutic problem. In today's industrialized societies, infectious strains
can be found that are
resistant to all classes of antimicrobial agents used in the clinic.
Infections due to resistant
strains include higher morbidity and mortality, longer patient
hospitalization, and an increase
in treatment costs (Murray (1994) New Ehgl. J. Med. 330:1229-1230). In light
of these
findings, an unmet need exists for the development of new therapeutic agents
that can worlc by
inhibiting the ever-increasing number of novel antibiotic resistance
mechanisms.
One approach for generating new therapies and/or therapeutic strategies
against AR
microbes is to develop methods that can generate a wide array of genomic
alterations in a
microbe's genome that can yield maximal number altered target genes that are
capable of
eliciting antibiotic resistance. Once an AR strain is developed, it can be
used for rapid
genome analysis to identify mutant genes) that are capable of rendering a
microbe resistant to
an antibiotic for target identification. Such analysis can involve any of a
variety of methods
used by those skilled in the art for identifying mutations and/or differential
gene expression,
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
-2-
including but not limited to differential gene expression using microarrays,
cDNA subtraction,
differential protein analysis, complementation assays, single nucleotide
polymorphosm (SNP)
analysis or whole genome sequencing to identify altered loci.
A bottleneck to generating genetically diverse microbes is the inability to
generate
nonbiased genome-wide mutations. Many mutagenesis methods are available
whereby the use
of chemical and radiation exposure has been successful in generating genomic
mutations. A
limitation of this approach is that these various methods are usually DNA site
specific or are
extremely toxic, therefore limiting the mutation spectra and the opportunity
to identify a
maximal number of genes, when mutated, that are able to confer resistance to
an antibiotic.
Recently, work done by Nicolaides, et al. (Nicolaides et al. (1998) Mol. Cell.
Biol. 18:1635-
1641; U.S. patent 6,146,894) has demonstrated the utility of introducing
dominant negative
mismatch repair mutants into cells to confer global DNA hypermutability. These
mutations
are in the form of point mutations or small insertion-deletions that are
distributed equally
throughout the genome. The ability to manipulate the mismatch repair (MMR)
process of a
target host organism can lead to an increase in the mutability of the target
host genome,
leading to the generation of innovative cell subtypes with varying phenotypes
from the
original wild-type cells. Variants can be placed under a specified, desired
selective process
the result of which is the capacity to select for a novel organism that
expresses an altered
biological molecules) and has a new phenotype. The concept of creating and
introducing
dominant negative allele of a gene, including the MMR alleles, in bacterial
cells has been
documented to result in genetically altered prokaryotic mismatch repair genes
(Aronshtam and
Marinus (1996) Nucl. Acids Res. 24:2498-2504; Wu and Marinus (1994) J.
Bacte~iol.
176:5393-400; Brosh and Matson (1995) J. Bacteriol. 177:5612-5621).
Furthermore, altered
MMR activity has been demonstrated when MMR genes from different species
including
yeast and mammalian cells are over-expressed (Fishel et al. (1993) Cell 7:1027-
1038; Lipkin
et al. (2000) Nat. Genet. 24:27-35). The ability to create hypermutable
organisms by blocking
MMR has great commercial value for the generation of AR bacterial strains for
drug screening
and target discovery.
There is an urgent need in the art to elucidate the mechanisms of
antimicrobial
resistance, and to identify novel antimicrobial agents.
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
-3-
SUMMARY
The invention provides new uses of MMR deficiency in bacteria to identify
antibiotic
resistance (AR) genes and pathways that can lead to the generation of novel
therapeutic
strategies for alternative clinical strategies.
Antibiotic resistant (AR) microbes express a number of genes that are
essential for
growth of the organism in an infection, and serve as useful reagents for
target discovery and/or
screening lines for the discovery of novel antimicrobial agents. This
invention provides an
approach to the identification of genes that confer anti-microbial resistance,
and the use of
those genes, and bacterial strains expressing mutant forms of genes, in the
identification,
characterization, and evaluation of targets for therapeutic development. In
addition, this
application teaches of the use of employing structural information of the
gene, gene product
and mutant strains in screening for antimicrobial agents active against the
genes and their
corresponding products and pathways. Positive compounds can then be used as
final products
or precursors to be further developed into antibacterial agents. This
invention also provides
methods of treating microbial infections in mammals by administering an
antimicrobial agent
active against such an identified target gene or product, and the
pharmaceutical compositions
effective for such treatment.
To identify genes capable of rendering bacteria antibiotic-resistant, the
invention
provides methods of decreasing MMR activity of a microbial host to produce AR
strains.
Using this process, commercially viable microbes that are resistant to a wide
range of
antibiotics can be directly selected for the resistance to an anti-microbial
agent of interest. AR
microbes may be genetically screened to identify novel therapeutic targets for
drug
develoment. Once a bacterium with a specified resistance is isolated, the MMR
activity may
be restored by several methods well known to those skilled in the art as a
means to gentically
"fix" the new mutations in the host genome, thereby making the AR microbe
suitable for
comparative molecular analysis to the wild-type strain as well as for drug
screening to identify
novel antimicrobial compounds. For example, if MMR is decreased by the use of
a dominant-
negative allele or antisense vector directed to an internal MMR gene, the
endogenous repair
activity can be restored if the gene is expressed by an inducible promoter
system, including
but not limited to promoters such as: TAC-LACI, tryp (Brosius et al. (1984)
Gene 27:161-
172), araBAD (Guzman et al. (1995) J. Bact. 177:4121-4130) pLex (La Vallie et
al. (1992)
Bio.Tecla~ology 11:187-193), pRSET (Schoepfer, R. (1993) Gene 124:83-85), pT7
(Studier
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
-4-
(1991) J. Mol. Biol. 219(1):37-44) etc., by removing the inducer and,
therefore, reducing the
the promoter activity. In the case that the expression vector employs a Cre-
lox system, MMR
can be restored by disrupting the cDNA gene insert from the host cell
harboring the expression
vector (Hasan, N. et al. (1994) Gene 2:51-56). Yet other methods include
homologous
knockout of the expression vector that can turn off the actively expressed
gene used to inhibit
MMR activity. In addition to the recombinant methods outlined above that have
the capacity
to eliminate the MMR activity from the microbe, it has been demonstrated that
many
chemicals have the ability to "cure" microbial cells of plasmids. For example,
chemical
treatment of cells with drugs including bleomycin (Attfield et al. (1985)
Arctimicrob. Agents
Chemothe~. 27:985-988) or novobiocin, coumercycin, and quinolones (Fu et al.
(1988)
Chemotherapy 34:415-418) have been shown to result in microbial cells that
laclc endogenous
plasmid as evidenced by Southern analysis of cured cells as well as
sensitivity to the
appropriate antibiotic (Attfield et al. (1985) Antimic~ob. Agefats
Claemother°. 27(6):985-988, Fu
et al. (1988) Cherra. Abstracts 34(5):415-418; BiWang et al. (1999) J. of
Fujian Agricultural
Universiy 28(1):43-46; Brosius, J. (1988) Biotech~zology 10:205-225). Whether
by use of
recombinant means or treatment of cells with chemicals, removal of the MMR-
expression
plasmid results in the reestablislnnent of a genetically stable microbial cell
line. Therefore,
the restoration of MMR allows host bacteria to function normally to repair
DNA. The newly
generated mutant bacterial strain that exhibits a novel anti-microbial
resistance is now suitable
for gene/protein discovery to identify new biomolecules that are involved in
generating
resistance as well as a model system to screen for novel anti-microbial agents
targeted against
certain antibiotic resistant strains.
In certain embodiments, the invention provides methods for generating
antibiotic
resistant bacteria comprising the steps of:
blocking mismatch repair in the bacterium whereby the bacterium becomes
hypermutable;
contacting the bacterium with at least one antibiotic
determining whether the bacterium is resistant to the antibiotic, thereby
generating
antibiotic resistant bacteria.
In the methods of the invention, mismatch repair may be blocked in some
embodiments by introducing a polynucleotide encoding a wild-type allele of a
mismatch repair
gene into a cell, whereby the wild-type allele inactivates the endogenous MMR
activity by
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
-5-
binding to and interfering with the resident activity. The cell becomes
hypermutable as a
result of the introduction of the gene.
In other embodiments of the invention, a polynucleotide encoding a dominant
negative
allele of a mismatch repair gene is introduced into a cell, where the dominant
negative gene is
derived from a mismatch repair gene from a different organism. The cell
becomes
hypermutable as a result of the introduction of the gene. In particular
embodiments of this
method, MMR activity is inhibited for ten rounds of cell division and then the
MMR activity
is restored therefore restoring the genetic stability. An example of a
dominant negative MMR
gene is the PMS2-134 gene.
In other embodiments of the invention, a polynucleotide encoding an allele of
a
mismatch repair gene is introduced into a bacterial cell, where the mismatch
repair gene is
derived from a wild-type or altered mammalian, yeast, fungal, amphibian,
insect, plant or
bacterial mismatch repair gene. The cell becomes hypermutable as a result of
the introduction
of the gene.
In another embodiment, mismatch repair may be blocked by introducing an
antisense
nucleic acid molecule into the bacterium wherein the antisense nucleic acid
molecule
specifically binds to a mismatch repair gene and inhibits mismatch repair in
the bacterium.
In other embodiments of the invention, methods are provided for generating a
genetic
alteration of a bacterial host genome to produce variant strains expressing
new output traits.
Transgenic bacterium comprising a polynucleotide encoding a wild-type allele
of a mismatch
repair gene is grown. The bacteria are comprised of a set of altered genes for
a desired
biological phenotype.
In other embodiments of the invention, methods are provided for generating a
genetic
alteration of a bacterial host genome to produce variant strains that are
resistant to
antimicrobial agents. Bacteria with decreased mismatch repair are grown. The
bacteria are
comprised of a set of altered genes for a desired antibiotic-resistance
phenotype.
In further embodiments of the invention, methods are provided for creating a
hypermutable bacterium using a wild-type MMR allele for antibiotic-resistance
selection, and
restoring genomic stability of a selected host by inactivating or decreasing
the expression of
the wild-type MMR allele.
In another embodiment of the invention, a method is provided for creating a
hypermutable bacteria using a dominant negative MMR allele for antibiotic-
resistance
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
-6-
selection, and restoring genomic stability of a selected host by inactivating
or decreasing the
expression of the dominant negative MMR gene allele.
In another embodiment of the invention, a method is provided for creating a
hypermutable bacteria expressing an antisense gene to a MMR gene for
antibiotic-resistance
selection, and restoring genomic stability of a selected host by inactivating
or decreasing the
expression of the dominant negative MMR gene allele.
In another embodiment of the invention, a method is provided for creating a
hypermutable bacteria using chemical inhibitors of MMR for antibiotic-
resistance selection,
and restoring genomic stability of a selected host by removing the chemical
inhibitor.
by introducing a dominant negative allele of a mismatch repair gene into the
bacterium. The
dominant negative allele may be, for example, a PMS2-134 gene.
In another embodiment, mismatch repair may be blocked by exposing the
bacterium a
to a compound that inhibits mismatch repair whereby cells are grown in the
presence of the
compound and undergo multiple rounds of cell divison in the absence of MMR,
yielding sibs
that are genetically diverse. Sibs are then selected for antibiotic
resistance. AR strains are
removed from chemical inhibitor and the endogenous MMR activity is restored
leaving
genetically stable strains that are now suitable for gene discovery and/or
therapeutic agent
development. For example, the compound that blocks mismatch repair may be an
anthracene
derivative, including, but not limited 1,2-dimethylanthracene, 9,10-dimethyl
anthracene, 7,8-
dimethylanthracene, 9,10-diphenylanthracene, 9,10-dihydroxymethylanthracene, 9-
hydroxymethyl-10-methylanthracene, dimethylanthracene-1,2-diol, 9-
hydroxymethyl-10-
methylanthracene-1,2-diol, 9-hydroxymethyl-10-methylanthracene-3,4-diol, 9, 10-
di-m-
tolyanthracene. In other embodiments, the compound that blocks MMR activity is
an ATP
analog. In other embodiments, the compound that blocks MMR activity is a
nuclease inhibitor.
In other embodiments, the compound that blocks MMR activity is a DNA
polymerase
inhibitor.
The methods of the invention may further comprise exposing the bacteria to
chemical
mutagens. While it has been documented that MMR deficiency can lead to as much
as a 1000-
fold increase in the endogenous DNA mutation rate of a host, there is no
assurance that MMR
deficiency alone will be sufficient to alter every gene within the DNA of the
host bacterium to
create altered biochemicals with new activity(s). Therefore, the use of
chemical agents and
their respective analogues such as methane sulfonate, dimethyl sulfonate, O-6-
methyl
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
_7_
benzadine, ethylnitrosourea (ENU), ethidium bromide, ethyl methanesulfonate
(EMS), N-
methyl-N'-nitro-N-nitrosoguanidine (MNNG), methylnitrosourea (MNU), Tamoxifen,
8-
hydroxyguanine, as well as others listed but not limited to in publications
by: I~hromov-
Borisov et al. (1999) Mutat. Res. 430:55-74; Ohe et al. (1999) Mutat. Res.
429:189-199; Hour
et al. (1999) Food Clzem. Toxicol. 37:569-579; Hrelia et al. (1999) Chem.
Biol. Interact.
118:99-111; Garganta et al. (1999) Environ. Mol. Mutagen. 33:75-85; Ukawa-
Ishikawa et al
(1998) Mutat. Res. 412:99-107; www.ehs.utah.edu/ohh/mutagens, etc. can be used
in the
methods of the invention to further enhance the spectrum of mutations and
increase the
likelihood of obtaining alterations in one or more genes that can in turn
generate host bacteria
with a complex antibiotic resistant phenotype (Fu et al. (1988) Chemotherapy
34(5):415-418;
Lee et al. (1994) Mutagenesis 9:401-405; Vidal et al. (1995) Carcinogenesis
16:817-821).
Prior art teaches us that mismatch repair deficiency leads to hosts with an
increased resistance
to toxicity by chemicals with DNA damaging activity. This feature allows for
the creation of
additional genetically diverse hosts when MMR defective bacteria are exposed
to such agents,
which would be otherwise impossible due to the toxic effects of such chemical
mutagens
(Colella et al. (1999) Br. J. Cancer 80:338-343; Moreland et al. (1999) Cancer
Res. 59:2102-
2106; Humbert et al. (1999) Carcinogeyaesis 20:205-214; Glaab et al. (1998)
Mutat. Res.
398:197-207). Moreover, prior art teaches us that MMR is responsible for
repairing chemical-
induced DNA adducts, so therefore blocking this process could theoretically
increase the
number, types, mutation rate and genomic alterations of a bacterial host
[Rasmussen et al.
(1996) Carcinogenesis 17:2085-2088; Sledziewska-Gojska et al. (1997) Mutat.
Res. 383:31-
37; Janion et al. (1989) Mutat. Res. 210:15-22). In addition to the chemicals
listed above,
other types of DNA mutagens include ionizing radiation and UV-irradiation,
which are known
to cause DNA mutagenesis in bacteria can also be used to potentially enhance
this process.
These agents, which are extremely toxic to host cells and, therefore, result
in a decrease in the
actual pool size of altered bacterial cells, are more tolerated in MMR
defective hosts and in
turn allow for a enriched spectrum and degree of genomic mutation ( Drummond
et al. (1996)
J. Biol. Chem. 271:9645-19648). such as, but not limited to methane sulfonate,
dimethyl
sulfonate, O-6-methyl benzadine, ethylnitrosourea, ethidium bromide, ethyl
methanesulfonate,
N-methyl-N'-nitro-N-nitrosoguanidine, methylnitrosourea, Tamoxifen, and 8-
hydroxyguanine.
The methods of the invention may be used to generate AR bacteria which are
resistant
to such antibiotic compounds as, for example, quinilones, aminoglycosides,
magainins,
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
_$_
defensins, tetracyclines, beta-lactams, macrolides, lincosamide, sulfonamides,
chloramphenicols, nitrofurantoins, and isoniazids.
In the methods of the invention, the step of determining whether the bacterium
is
resistant to an antibiotic may comprise analyzing the bacterium for
multiantiboitic resistance.
Further, the methods of the invention may comprise making antibiotic resistant
bacteria
genetically stable, such as by removing the MMR inhibitory molecule, for
example.
In the methods of the invention, the genome of the antibiotic resistant
bacterium and
the genome of a wild-type strain of the bacterium may be compared by sequence
analysis of
the entire genomes, or compared by microarray analysis, for example. In
another embodiment,
the genome of said antibiotic resistant bacterium and the genome of said wild-
type strain of
said bacterium are compared by:
introducing gene fragments from the antibiotic resistant bacterium into the
wild-type
bacterium, thereby producing mutant bacteria;
selecting a mutant bacterium with antibiotic resistance; and sequencing the
gene
fragment from the mutant bacterium with antibiotic resistance, thereby
identifying the
antibiotic resistant gene.
The invention also provides methods of using microbial strains that are
naturally
defective for MMR due to defects in genes encoding for MMR proteins. Strains
in which
mutS, mutt, mutes, o~ mutY genes are defective have been reported to be
defective in MMR
activity (Modrich (1994) Science 266:1959-1960). The methods of the invention
may employ
bacterial strains with mutant endogenous MMR genes for selecting for variants
that are AR.
Once an AR variant strain is identified, the genetic stability of the microbe
can be restored by
expressing a functional gene that can complement the defective MMR gene
activity.
Mutant strains can be used for gene identification by isolating DNA fragments
derived
from the MMR defective antibiotic-resistant strains. These bacteria contain
DNA fragments
with altered sequences that can be introduced into wild-type counterparts
(antibiotic
susceptible) and screened for fragments that confer antibiotic resistance.
Conversely, DNA
fragments derived from the wild-type bacteria can be introduced into mutant
bacterial strains
to screen for genes effective via loss-of function mutated genes. The fact
that a clone is
complemented suggests the introduced fragment contains a gene encoding for an
antibiotic-
resistant gene product. Other methods can also be used to identify AR genes
including but not
limited to microarray analysis of gene expression, differential expression
andlor differential
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
-9-
protein analysis know by those skilled in the art.
The microbial strains described herein have either been generated and
characterized in
a manner which essentially provides a process by which the manipulation of MMR
can confer
AR against a wide range of anti-microbial compounds and that these strains are
now useful for
target discovery and/or therapeutic agent discovery as screening lines.
In other embodiments of the invention, methods of producing a stable bacterium
expressing a new phenotype is provided. Turning off the expression of the MMR-
wild-type
alleles, MMR-dominant negative alleles, or MMR-antisense alleles, results in
genetically
stable bacteria expressing a new output trait(s).
The invention also provides antibiotic resistant strains of bacteria produced
by the
methods of the invention.
These and other aspects of the invention provide the art with methods that can
generate
enhanced mutability in bacteria as well as providing prokaryotic organisms
harboring
potentially useful mutations to generate novel output traits for commercial
applications, and
are set forth in greater detail below.
BRIEF DESCRIPTION OF THE FIGURE
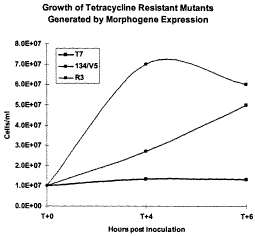
Figure 1 shows growth of tetracyclin-resistant mutant bacteria carrying a
dominant
negative allele of PMS2 in the pT7Ea plasmid (134/V5), tetracyclin-resistant
mutant bacteria
carrying a the PMSR3 gene in the pT7Ea plasmid (R3), and wild-type bacteria
carrying the
empty pT7Ea plasmid (T7), on medium containing tetracyclin at 0, 4 and 6 hours
after
tetracycline addition.
DETAILED DESCRIPTION OF THE INVENTION
The inventors present a method for developing hypermutable bacteria by
altering the
activity of endogenous mismatch repair (MMR) activity of hosts to generate
antibiotic
resistant (AR) microbes for target discovery and the development of novel anti-
microbial
agent by screening for new compounds. Wild-type and some dominant negative
alleles of
mismatch repair genes, when introduced and expressed in bacteria, increase the
rate of
spontaneous mutations by reducing the effectiveness of the endogenous MMR-
mediated DNA
repair activity, thereby rendering the bacteria highly susceptible to genetic
alterations due to
hypermutability. Hypermutable bacteria can then be utilized to screen for
novel mutations in a
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
-10-
gene or a set of genes that produce variant siblings exhibiting new output
traits not found in
the wild-type cells such as antibiotic resistance.
The process of mismatch repair, also called mismatch proofreading, is an
evolutionarily highly conserved process that is carried out by protein
complexes described in
cells as disparate as prokaryotic cells such as bacteria to more complex
mammalian cells
(Modrich (1994) Science 266:1959-1960; Strand et al. (1993) Nature 365:274-
276; Su et al.
(1988) J. Biol. Chem. 263:6829-6835; Aronshtam.and Marinus (1996) Nucl. Acids
Res.
24:2498-2504; Wu and Marinus (1994) J. Bacte~iol. 176:5393-400). A mismatch
repair gene
is a gene that encodes one of the proteins of such a mismatch repair complex.
Although not
wanting to be bound by any particular theory of mechanism of action, a
mismatch repair
complex is believed to detect distoutions of the DNA helix resulting from non-
complementary
pairing of nucleotide bases. The non-complementary base on the newer DNA
strand is
excised, and the excised base is replaced with the appropriate base that is
complementary to
the older DNA strand. In this way, cells eliminate many mutations that occur
as a result of
mistakes in DNA replication, resulting in genetic stability of the sibling
cells derived from the
parental cell.
Some wild-type MMR gene alleles as well as dominant negative alleles cause a
mismatch repair defective phenotype even in the presence of a wild-type MMR
gene allele in
the same cell. An example of a dominant negative allele of a MMR gene is the
human gene
hPMS2-134, which carries a truncation mutation at codon 134 (Nicolaides et al.
(1998) Mol.
Cell. Biol. 18:1635-1641). The mutation causes the product of this gene to
abnormally
terminate at the position of the 134th amino acid, resulting in a shortened
polypeptide
containing the N-terminal 133 amino acids. Such a mutation causes an increase
in the rate of
mutations, which accumulate in cells after DNA replication. Expression of a
dominant
negative allele of a mismatch repair gene results in impairment of mismatch
repair activity,
even in the presence of the wild-type allele. Any mismatch repair allele,
which produces such
effect, can be used in this invention. In addition, the use of over-expressed
wild-type MMR
gene alleles from human, mouse, plants, and yeast in bacteria has been shown
to cause a
dominant negative effect on the bacterial hosts MMR activity (Fishel et al.
(1993) Cell
7:1027-1038; Aronshtam.and Marinus (1996) Nucl. Acids Res. 24:2498-2504; Wu
and
Marinus (1994) J. Bacteriol. 176:5393-400; Lipkin et al. (2000) Nat. Ger2et.
24:27-35).
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
-11-
Dominant negative alleles of a mismatch repair gene can be obtained from the
cells of
humans, animals, yeast, bacteria, plants or other organisms. Screening cells
for defective
mismatch repair activity can identify such alleles. Mismatch repair genes may
be mutant or
wild-type. Bacterial host MMR may be mutated or not. The term bacteria used in
this
application include any organism from the prokaryotic kingdom. These organisms
include
genera such as but not limited to Agrobactey°ium, Anae~obacte~,
Aquabactef°ium,
Azof°hizobiuna, Bacillus, Bs°ady~hizobium, Cnyobacteriuna,
Eschericlaia, Enterococcus,
Heliobacterium, Klebsiella, Lactobacillus, Methanococcus,
Methanothernaobacten,
Micr~ococcus, Mycobacterium, Oceanomonas, Pseudoynonas, RlZizobium,
Staphylococcus,
Streptococcus, Stneptomyces, The~musaquaticus, Thermaerobacter,
The~mobacillus, etc.
Other procaryotes that can be used for this application are listed at
(www.bacterio.cict.frlvalid~enericnames). Bacteria exposed to chemical
mutagens or radiation
exposure can be screened for defective mismatch repair. Genomic DNA, cDNA, or
mRNA
from any cell encoding a mismatch repair protein can be analyzed for
variations from the
wild-type sequence. Dominant negative alleles of a mismatch repair gene can
also be created
artificially, for example, by producing variants of the hPMS2-134 allele or
other mismatch
repair genes (Nicolaides et al. (1998) Mol. Cell. Biol. 18:1635-1641). Various
techniques of
site-directed mutagenesis can be used. The suitability of such alleles,
whether natural or
artificial, for use in generating hypermutable bacteria can be evaluated by
testing the mismatch
repair activity (using methods described in Nicolaides et al. (1998) Mol.
Cell. Biol. 18:1635-
1641) caused by the allele in the presence of one or more wild-type alleles,
to determine if it is
a dominant negative allele.
A bacterium that over-expresses a wild-type mismatch repair allele or a
dominant
negative allele of a mismatch repair gene will become hypermutable. This means
that the
spontaneous mutation rate of such bacteria is elevated compared to bacteria
without such
alleles. The degree of elevation of the spontaneous mutation rate can be at
least 2-fold, 5-fold,
10-fold, 20-fold, 50-fold, 100-fold, 200-fold, 500-fold, or 1000-fold that of
the normal bacteria
as measured as a function of bacterial doubling/minute.
According to one aspect of the invention, a polynucleotide encoding either a
wild-type
or a dominant negative form of a mismatch repair protein is introduced into
bacteria. The
gene can be any dominant negative allele encoding a protein which is part of a
mismatch
repair complex, for example, nzutS, mutt, mutes, or mutYhomologs of the
bacterial, yeast,
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
-12-
plant or mammalian genes (Modrich (1994) Science 266:1959-1960; Prolla et al.
(1994)
Science 264:1091-1093). The dominant negative allele can be naturally
occurring or made in
the laboratory. The polynucleotide can be in the form of genomic DNA, cDNA,
RNA, or a
chemically synthesized polynucleotide or polypeptide. The molecule can be
introduced into
the cell by transfection or other methods well described in the literature.
Transfection is any process whereby a polynucleotide or polypeptide is
introduced into
a cell. The process of transfection can be carried out in a bacterial culture
using a suspension
culture. The bacteria can be any type classified under the prokaryotes.
In general, transfection will be carried out using a suspension of cells but
other
methods can also be employed as long as a sufficient fraction of the treated
cells incorporate
the polynucleotide or polypeptide so as to allow transfected cells to be grown
and utilized.
The protein product of the polynucleotide may be transiently or stably
expressed in the cell.
Techniques for transfection are well known to those skilled in the art.
Available techniques to
introduce a polynucleotide or polypeptide into a prokaryote include but are
not limited to
electroporation, transduction, cell fusion, the use of chemically competent
cells (e.g., calcium
chloride), and packaging of the polynucleotide together with lipid for fusion
with the cells of
interest. Once a cell has been transformed with the inhibitory mismatch repair
gene or protein,
the cell can be propagated and manipulated in either liquid culture or on a
solid agar matrix,
such as a petri dish. If the transfected cell is stable, the gene will be
retained and expressed at
a consistent level when the promoter is constitutively active, or when in the
presence of
appropriate inducer molecules when the promoter is inducible, for many cell
generations, and
a stable, hypermutable bacterial strain results.
An isolated bacterial cell is a clone obtained from a pool of a bacterial
culture by
chemically selecting out strains using antibiotic selection of an expression
vector. If the
bacterial cell is derived from a single cell, it is defined as a clone.
Bacterial cultures may be screened for antibiotic resistance against a wide
array of
antibiotic compounds. For example, but not by way of limitation, bacteria
produced by the
methods of the invention may be screened for resistance to quinilones,
aminoglycosides,
magainins, defensins, tetracyclines, beta-lactams, macrolides, lincosamide,
sulfonamides,
chloramphenicols, nitrofurantoins, and isoniazids. The antibiotics may be
incorporated into
solid or liquid growth medium, for example.
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
-13-
A polynucleotide encoding an inhibitory form of a mismatch repair protein can
be
introduced into the genome of a bacterium or propagated on an extra-
chromosomal plasmid.
Selection of clones harboring the mismatch repair gene expression vector can
be accomplished
by addition of any of several different antibiotics, including but not limited
to ampicillin,
kanamycin, chloramphenicol, zeocin, and tetracycline. The microbe can be any
species for
which suitable techniques are available to produce transgenic microorganisms,
such as but not
limited to geheva including Bacillus, Pseudomohas, Staphylococcus, Esche~ichia
and others.
Any method for making transgenic bacteria known in the art can be used.
According
to one process of producing a transgenic microorganism, the polynucleotide is
transfected into
the microbe by one of the methods well known to those in the art. Next, the
microbial culture
is grown under conditions that select for cells in which the polynucleotide
encoding the
mismatch repair gene is either incorporated into the host genome as a stable
entity or
propagated on a self replicating extra-chromosomal plasmid, and the protein
encoded by the
polynucleotide fragment transcribed and subsequently translated into a
functional protein
within the cell. Once a transgenic microbe is engineered to harbor the
expression construct, it
is then propagated to generate and sustain a culture of transgenic microbes
indefinitely.
Once a stable, transgenic microorganism has been engineered to express a
functional
MMR protein, the microbe can be exploited to create novel mutations in one or
more target
genes) of interest harbored within the same microorganism. A gene of interest
can be any
gene naturally possessed by the bacterium or one introduced into the bacterial
host by standard
recombinant DNA techniques. The target genes) may be known prior to the
selection or
unknown. One advantage of employing such transgenic microbes to induce
mutations in
resident or extra-chromosomal genes within the microbe is that it is
unnecessary to expose the
microorganism to mutagenic insult, whether it be chemical or radiation in
nature, to produce a
series of random gene alterations in the target gene(s). This is due to the
highly efficient
nature and the spectrum of naturally occurring mutations that result as a
consequence of the
altered mismatch repair process. However, it is possible to increase the
spectrum and
frequency of mutations by the concomitant use of either chemical and/or
radiation together
with MMR defective cells. The net effect of the combination treatment is the
increase in
altered gene pool in the genetically altered microbe that result in an
increased alteration of an
alleles) that are useful for producing new output traits. Other benefits of
using MMR-
defective microbes that are taught in this application are genetic screens for
the DIRECT
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
-14-
selection of variant sub-clones that exhibit new output traits with
commercially important
applications such as antibiotic resistance, which allows the bypassing of the
tedious and time
consuming gene identification, isolation and characterization stages.
Mutations can be detected by analyzing the recombinant microbe for alterations
in the
genotype and/or phenotype post-activation of the decreased mismatch repair
activity of the
transgenic microorganism. Novel genes that produce altered phenotypes in MMR-
defective
microbial cells can be discerned by any variety of molecular techniques well
known to those
in the art. For example, the microbial genome can be isolated and a library of
restriction
fragments cloned into a plasmid vector. The library can be introduced into a
"normal" cell
and the cells exhibiting the novel phenotype screened. Transformed cells are
then screened
for the new phenotype (e.g., antibiotic resistance). A plasmid is isolated
from those normal,
transformed cells that exhibit the novel phenotype and the inserted genes)
characterized by
DNA sequence analysis.
Alternatively, differential messenger RNA screen can be employed utilizing
driver and
tester RNA (derived from wild-type and novel mutant respectively) followed by
cloning the
differential transcripts and characterizing them by standard molecular biology
methods well
known to those skilled in the art. Furthermore, if the mutant sought is on
encoded by an
extrachromosmal plasmid, then following co-expression of the dominant negative
MMR gene
and the gene of interest to be altered and phenotypic selection, the plasmid
is isolated from
mutant clones and analyzed by DNA sequence analysis by methods well known to
those in the
art.
In another embodiment, the screening of cells may be performed by microarray
analysis. In microarray analysis, microchips containing all or a subset of all
expressed
bacterial genes may be screened using RNA molecules derived from the wild-type
or
antibiotic resistant strain whereby RNA derived from one strain is reverse
transcribed using
FluoroLink Cy3 and the other RNA sample is reverse transcribe-labelled using
Cy5 dUTP.
Labelled cDNAs from each organism are used to probe the microchip whereby
unique
message from one source will .generate a distinct signal while message
expressed from both
sources will generate a cormnon fluorescence. Alternatively, microchips
containing
olignucleotide derived from the wild-type strain can be used to hybridize
genomic fragments
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
-15-
from the antibiotic resistant strain to identify fragments containing a
mutated gene by loss of
hybridization.
Phenotypic screening for output traits in MMR-defective mutants can be by
biochemical activity and/or a physical phenotype of the altered gene product.
A mutant
phenotype can also be detected by identifying alterations in electrophoretic
mobility, DNA
binding in the case of transcription factors, spectroscopic properties such as
IR, CD, X-ray
crystallography or high field NMR analysis, or other physical or structural
characteristics of a
protein encoded by a mutant gene. It is also possible to screen for altered
novel function of a
protein izz situ, in isolated form, or in model systems. One can screen for
alteration of any
property of the microorganism associated with the function of the gene of
interest, whether the
gene is known prior to the selection or unknown. The aforementioned screening
and selection
discussion is meant to illustrate the potential means of obtaining novel
mutants with
commercially valuable output traits.
Plasmid expression vectors that harbor the mismatch repair (MMR) gene inserts
can be
used in combination with a number of commercially available regulatory
sequences to control
both the temporal and quantitative biochemical expression level of the
dominant negative
MMR protein. The regulatory sequences can be comprised of a promoter, enhancer
or
promoter/enhancer combination and can be inserted either upstream or
downstream of the
MMR gene to control the expression level. The regulatory promoter sequence can
be any of
those well known to those in the art, including but not limited to the lacI,
tetracycline,
tryptophan-inducible, phosphate inducible, T7-polymerase-inducible (Studier et
al. (1991) J.
Mol. Biol. 219(1):37-44), and steroid inducible constructs as well as
sequences which can
result in the excision of the dominant negative mismatch repair gene such as
those of the Cre-
Lox system. These types of regulatory systems have been listed in scientific
publications and
are familiar to those skilled in the art.
Once a microorganism with a novel, desired output trait of interest is
created, the
activity of the aberrant MMR activity is attenuated or eliminated by any of a
variety of
methods, including removal of the inducer from the culture medium that is
responsible for
promoter activation, gene disruption of the aberrant MMR gene constructs,
electroporation
and or chemical curing of the expression plasmids (Brosius(1988)
Biotechjzology 10:205-225;
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
- 16-
Wang et al. (1999) J. of Fujian Agricultural University 28:43-46; Fu et. al.
(1988) Chern.
Abstracts 34:415-418). The expression of the dominant negative MMR gene will
be turned on
to select for hypermutable microbes with new output traits. Next, the
expression of the
dominant negative dominant negative MMR allele is rapidly turned off to
reconstitute a
genetically stable strain that produces a new output trait of commercial
interest. The resulting
microbe is now useful as a stable strain that can be applied to various
commercial applications,
depending upon the selection process placed upon it.
In cases where genetically deficient mismatch repair bacteria (strains such as
but not
limited to: M1 (mutS) and in EC2416 (mutS delta umuDC), and mutt or mutt
strains) are
used to derive new output traits, transgenic constructs will be used that
express wild-type
mismatch repair genes sufficient to complement the genetic defect and
therefore restore
mismatch repair activity of the host after trait selection (Grzesiuk et al.
(1988) Mutagenesis
13:127-132; Bridges et al. (1997) EMBO J. 16:3349-3356; LeClerc (1996) Science
15:1208-
1211; Jaworski, A. et al. (1995) Proc. Natl. Acad. Sci USA 92:11019-11023).
The resulting
microbe is genetically stable and can be applied to various commercial
practices.
The use of over expressing foreign mismatch repair genes from human and yeast
such
as human PMSl (SEQ ID N0:7), human PMS2 (SEQ ID N0:5), mouse PMSZ (SEQ ID
N0:3), human MSH2 (SEQ ID N0:9), human MLHI (SEQ ID NO:11), yeast MLHl (SEQ ID
NO:1), human MLH3 (SEQ ID N0:28), as well as the other homologs identified in
other
species for these encoded polypeptides etc.have been previously demonstrated
to produce a
dominant negative mutator phenotype in bacterial hosts (Brosh and Matson
(1995) J.
Bacteriol. 177:5612-5621; Studamire et al. (1998) Mol. Cell. Biol. 18:7590-
7601; Alani et al.
(1997) Mol. Cell. Biol.17:2436-2447). In addition, the use of bacterial
strains expressing
prokaryotic dominant negative MMR genes as well as hosts that have genomic
defects in
endogenous MMR proteins have also been previously shown to result in a
dominant negative
mutator phenotype (Strand et al. (1993) Nature 365:274-276; Nicolaides et al.
(1998) Mol.
Cell. Biol. 18:1635-1641). However, the findings disclosed here teach the use
of MMR
genes, including the human PMSR2 and PMSR3 gene (Nicolaides et al. (1995)
Genojnics
30:195-206); the related PMS134 truncated MMR gene (Nicolaides et a1.(1998)
Mol. Cell.
Biol. 18:1635-1641); the plant mismatch repair genes (derived from Arabidopsis
thaliana),
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
- 17-
ATPMS2 (SEQ ID N0:30), At PMSI (SEQ ID N0:32), and MutS homolog (SEQ ID N0:34)
and those genes that are homologous to the 134 N-terminal amino acids of the
PMS2 gene
which include the Mutt family of MMR proteins and including the PMSR and PMS2L
homologs described by Hori et al. (PMS2L8 (SEQ ID N0:36) and PMS2L9 (SEQ ID
N0:38)) and Nicolaides (Nicolaides et al. (1995) Genomics 30:195-206) to
create
hypermutable microbes. The corresponding polypeptide sequences for the above-
referenced
nucleic acid sequences are as follows: yeast MLHI (SEQ ID N0:2); mouse PMS2
(SEQ ID
N0:4); human PMS2 (SEQ ID N0:6); human PMS1 (SEQ ID N0:8); human MSH2 (SEQ ID
NO:10); human MLH1 (SEQ ID N0:12); PMS2-134 (SEQ ID N0:14); human MSH6 (SEQ
ID N0:16); human PMSR2 (SEQ ID N0:18); human PMSR3 (SEQ ID N0:20); human
PMSL9 (SEQ ID N0:22); human MLH3 (SEQ ID N0:29); ATPMS2 (SEQ ID NO:31);
ATPMSl (SEQ ID N0:33); At MutS homolog (SEQ ID NO:35); PMS2L8 (SEQ ID N0:37);
and PMS2L9 (SEQ ID N0:39).
In addition, the invention provides the use of DNA mutagens in combination
with
MMR defective microbial hosts to enhance the hypermutable production of
genetic alterations.
This has not been demonstrated in the art previously as a means to accentuate
MMR activity
for generation of microorganisms with clinically relevant output traits such
as antibiotic
resistance.
In some embodiments of the invention, the bacteria cells are rendered
hypermutable by
introducing a chemical inhibitor of mismatch repair into the growth medium.
Chemical
inhibitors of mismatch repair that may be used to generate hypermutable
bacterial cells
include anthracene-derived compounds comprising the formula:
8 ~9 ~1
R~ / / / 2
R6 RS R10 R4 R3
In certain preferred embodiments of the invention, the anthracene has the
formula:
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
-18-
8 ~9 ~1
Rg RS R10 R4 R3
wherein Rl-Rlo are independently hydrogen, hydroxyl, amino, alkyl, substituted
alkyl, alkenyl,
substituted alkenyl, alkynyl, substituted alkynyl, O-alkyl, S-alkyl, N-alkyl,
O-alkenyl, S-
alkenyl, N-alkenyl,0-allcynyl, S-alkynyl, N-alkynyl, aryl, substituted aryl,
aryloxy, substituted
aryloxy, heteroaryl, substituted heteroaryl, aralkyloxy, arylalkyl, alkylaryl,
alkylaryloxy,
arylsulfonyl, alkylsulfonyl, alkoxycarbonyl, aryloxycarbonyl, guanidine,
carboxy, an alcohol,
an amino acid, sulfonate, alkyl sulfonate, CN, NOZ, an aldehyde group, an
ester, an ether, a
crown ether, a ketone, an organosulfur compound, an organometallic group, a
carboxylic acid,
an organosilicon or a carbohydrate that optionally contains one or more
alkylated hydroxyl
groups;
wherein said heteroalkyl, heteroaryl, and substituted heteroaryl contain at
least one
heteroatom that is oxygen, sulfur, a metal atom, phosphorus, silicon or
nitrogen; and
wherein said substituents of said substituted alkyl, substituted alkenyl,
substituted
allcynyl,
substituted aryl, and substituted heteroaryl are halogen, CN, NOZ, lower
alkyl, aryl, heteroaryl,
aralkyl, aralkyloxy, guanidine, alkoxycarbonyl, alkoxy, hydroxy, carboxy and
amino;
and wherein said amino groups optionally substituted with an acyl group, or 1
to 3 aryl
or lower alkyl groups;
or wherein any two of Rl-Rlo can together form a polyether;
or wherein any two of Rl-Rlo can, together with the intervening carbon atoms
of the
anthracene core, form a crown ether.
As used herein, "alkyl" refers to a hydrocarbon containing from 1 to about 20
carbon
atoms. Alkyl groups may straight, branched, cyclic, or combinations thereof.
Alkyl groups
thus include, by way of illustration only, methyl, ethyl, propyl, isopropyl,
butyl, isobutyl,
cyclopentyl, cyclopentylmethyl, cyclohexyl, cyclohexylmethyl, and the like.
Also included
within the definition of "alkyl" are fused and/or polycyclic aliphatic cyclic
ring systems such
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
-19-
as, for example, adamantane. As used herein the term "alkenyl" denotes an
alkyl group
having at least one carbon-carbon double bond. As used herein the term
"alkynyl" denotes an
alkyl group having at least one carbon-carbon triple bond. In certain
preferred embodiments of
the invention, the anthracene has the formula:
8 ~9 ~1
R~ ~ ~ ~ z
R6 RS R10 R4 R3
wherein Rl-Rlo are independently hydrogen, hydroxyl, amino, alkyl, substituted
alkyl, alkenyl,
substituted alkenyl, alkynyl, substituted allcynyl, O-alkyl, S-alkyl, N-alkyl,
O-alkenyl, S-
alkenyl, N-alkenyl,0-alkynyl, S-alkynyl, N-alkynyl, aryl, substituted aryl,
aryloxy, substituted
aryloxy, heteroaryl, substituted heteroaryl, aralkyloxy, arylalkyl, alkylaryl,
alkylaryloxy,
arylsulfonyl, alkylsulfonyl, alkoxycarbonyl, aryloxycarbonyl, guanidino,
carboxy, an alcohol,
an amino acid, sulfonate, alkyl sulfonate, CN, NOz, an aldehyde group, an
ester, an ether, a
crown ether, a ketone, an organosulfur compound, an organometallic group, a
carboxylic acid,
an organosilicon or a carbohydrate that optionally contains one or more
alkylated hydroxyl
groups;
wherein said heteroalkyl, heteroaryl, and substituted heteroaryl contain at
least one
heteroatom that is oxygen, sulfur, a metal atom, phosphorus, silicon or
nitrogen; and
wherein said substituents of said substituted alkyl, substituted alkenyl,
substituted
alkynyl,
substituted aryl, and substituted heteroaryl are halogen, CN, NOz, lower
alkyl, aryl, heteroaryl,
aralkyl, aralkyloxy, guanidino, alkoxycarbonyl, alkoxy, hydroxy, carboxy and
amino;
and wherein said amino groups optionally substituted with an acyl group, or 1
to 3 aryl
or lower alkyl groups;
or wherein any two of Rl-Rlo can together form a polyether;
or wherein any two of Rl-Rlo can, together with the intervening carbon atoms
of the
anthracene core, form a crown ether.
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
-20-
As used herein, "alkyl" refers to a hydrocarbon containing from 1 to about 20
carbon
atoms. Alkyl groups may straight, branched, cyclic, or combinations thereof.
Alkyl groups
thus include, by way of illustration only, methyl, ethyl, propyl, isopropyl,
butyl, isobutyl,
cyclopentyl, cyclopentylmethyl, cyclohexyl, cyclohexylmethyl, and the like.
Also included
within the definition of "alkyl" are fused and/or polycyclic aliphatic cyclic
ring systems such
as, for example, adamantane. As used herein the teen "alkenyl" denotes an
alkyl group
having at least one carbon-carbon double bond. As used herein the term
"alkynyl" denotes an
alkyl group having at least one carbon-carbon triple bond.
In some embodiments, the anthracene has the formula:
8 ~9 ~1
R3
wherein R1-RIO are independently hydrogen, hydroxyl, amino, alkyl, substituted
alkyl, alkenyl,
substituted alkenyl, alkynyl, substituted alkynyl, O-alkyl, S-alkyl, N-alkyl,
O-alkenyl, S-
alkenyl, N-allcenyl,0-alkynyl, S-alkynyl, N-alkynyl, aryl, substituted aryl,
aryloxy, substituted
aryloxy, heteroaryl, substituted heteroaryl, aralkyloxy, arylalkyl, alkylaryl,
alkylaryloxy,
arylsulfonyl, alkylsulfonyl, alkoxycarbonyl, aryloxycarbonyl, guanidino,
carboxy, an alcohol,
an amino acid, sulfonate, alkyl sulfonate, CN, NO2, an aldehyde group, an
ester, an ether, a
crown ether, a ketone, an organosulfur compound, an organometallic group, a
carboxylic acid,
an organosilicon or a carbohydrate that optionally contains one or more
alkylated hydroxyl
groups;
wherein said heteroalkyl, heteroaryl, and substituted heteroaryl contain at
least one
heteroatom that is oxygen, sulfur, a metal atom, phosphorus, silicon or
nitrogen; and
wherein said substituents of said substituted alkyl, substituted alkenyl,
substituted
alkynyl,
substituted aryl, and substituted heteroaryl are halogen, CN, N02, lower
alkyl, aryl, heteroaryl,
aralkyl, aralkyloxy, guanidino, alkoxycarbonyl, alkoxy, hydroxy, carboxy and
amino;
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
-21 -
and wherein said amino groups optionally substituted with an acyl group, or 1
to 3 aryl
or lower alkyl groups.
Examples of such anthracenes include, but are not limited to 1,2-
dimethylanthracene,
9,10-dimethyl anthracene, 7, ~-dimethylanthracene, 9,10-diphenylanthracene,
9,10-
dihydroxymethylanthracene, 9-hydroxymethyl-10-methylanthracene,
dimethylanthracene-1,2-
diol, 9-hydroxymethyl-10-methylanthracene-1,2-diol, 9-hydroxymethyl-10-
methylanthracene-
3,4-diol, and 9, 10-di-m-tolyanthracene.
As used herein the term "anthracene" refers to the compound anthracene.
However,
when referred to in the general sense, such as "anthracenes," "an anthracene"
or "the
anthracene," such terms denote any compound that contains the fused triphenyl
core structure
of anthracene, i.e.,
" ,
regardless of extent of substitution.
In some embodiments, the alkyl, alkenyl, alkynyl, aryl, aryloxy, and
heteroaryl
substituent groups described above may bear one or more further substituent
groups; that is,
they may be "substituted". In some preferred embodiments these substituent
groups can
include~halogens (for example fluorine, chlorine, bromine and iodine), CN,
NOZ, lower alkyl
groups, aryl groups, heteroaryl groups, aralkyl groups, aralkyloxy groups,
guanidino,
alkoxycarbonyl, alkoxy, hydroxy, carboxy and amino groups. In addition, the
alkyl and aryl
portions of aralkyloxy, arylalkyl, arylsulfonyl, alkylsulfonyl,
alkoxycarbonyl, and
aryloxycarbonyl groups also can bear such substituent groups. Thus, by way of
example only,
substituted alkyl groups include, for example, alkyl groups fluoro-, chloro-,
bromo- and
iodoalkyl groups, aminoallcyl groups, and hydroxyalkyl groups, such as
hydroxymethyl,
hydroxyethyl, hydroxypropyl, hydroxybutyl, and the like. In some preferred
embodiments
such hydroxyalkyl groups contain from 1 to about 20 carbons.
As used herein the term "aryl" means a group having 5 to about 20 carbon atoms
and
which contains at least one aromatic ring, such as phenyl, biphenyl and
naphthyl. Preferred
aryl groups include unsubstituted or substituted phenyl and naphthyl groups.
The term
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
"aryloxy" denotes an aryl group that is bound through an oxygen atom, for
example a phenoxy
group.
In general, the prefix "hetero" denotes the presence of at least one hetero
(i.e., non-
carbon) atom, which is in some preferred embodiments independently one to
three O, N, S, P,
Si or metal atoms. Thus, the term "heteroaryl" denotes an aryl group in which
one or more
ring carbon atom is replaced by such a heteroatom. Preferred heteroaryl groups
include
pyridyl, pyrimidyl, pyrrolyl, furyl, thienyl, and imidazolyl groups.
The term "aralkyl" (or "arylalkyl") is intended to denote a group having from
6 to 15
carbons, consisting of an alkyl group that bears an aryl group. Examples of
aralkyl groups
include benzyl, phenethyl, benzhydryl and naphthylmethyl groups.
The term "alkylaryl" (or "alkaryl") is intended to denote a group having from
6 to 15
carbons, consisting of an aryl group that bears an allcyl group. Examples of
aralkyl groups
include methylphenyl, ethylphenyl and methylnaphthyl groups.
The term "arylsulfonyl" denotes an aryl group attached through a sulfonyl
group, for
example phenylsulfonyl. The term "alkylsulfonyl" denotes an alkyl group
attached through a
sulfonyl group, for example methylsulfonyl.
The teen "alkoxycarbonyl" denotes a group of formula -C(=O)-O-R where R is
alkyl,
alkenyl, or alkynyl, where the alkyl, alkenyl, or allcynyl portions thereof
can be optionally
substituted as described herein.
The term "aryloxycarbonyl" denotes a group of formula -C(=O)-O-R where R is
aryl,
where the aryl portion thereof can be optionally substituted as described
herein.
The terms "arylalkyloxy" or "aralkyloxy" are equivalent, and denote a group of
formula -O-R'-R~~, where R~ is R is alkyl, alkenyl, or alkynyl which can be
optionally
substituted as described herein, and wherein R~~ denotes a aryl or substituted
aryl group.
The terms "alkylaryloxy" or "alkaryloxy" are equivalent, and denote a group of
formula -O-R~-R~~, where R~ is an aryl or substituted aryl group, and R~~ is
alkyl, alkenyl, or
alkynyl which can be optionally substituted as described herein.
As used herein, the term "aldehyde group" denotes a group that bears a moiety
of
formula -C(=O)-H. The term "ketone" denotes a moiety containing a group of
formula -R-
C(=O)-R=, where R and R= are independently alkyl, alkenyl, alkynyl, aryl,
heteroaryl, aralkyl,
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
- 23 -
or alkaryl, each of which may be substituted as described herein.
As used herein, the term "ester" denotes a moiety having a group of formula -R-
C(=O)-O-R= or -R-O-C(=O)-R= where R and R= are independently alkyl, alkenyl,
alkynyl,
aryl, heteroaryl, aralkyl, or alkaryl, each of which may be substituted as
described herein.
The term "ether" denotes a moiety having a group of formula -R-O-R= or where R
and
R= are independently alkyl, alkenyl, alkynyl, aryl, heteroaryl, aralkyl, or
alkaryl, each of
which may be substituted as described herein.
The term "crown ether" has its usual meaning of a cyclic ether containing
several
oxygen atoms. As used herein the term "organosulfur compound" denotes
aliphatic or
aromatic sulfur containing compounds, for example thiols and disulfides. The
term
"organometallic group" denotes an organic molecule containing at least one
metal atom.
The term "organosilicon compound" denotes aliphatic or aromatic silicon
containing
compounds, for example alkyl and aryl silanes.
The teen "carboxylic acid" denotes a moiety having a carboxyl group, other
than an
amino acid.
As used herein, the term "amino acid" denotes a molecule containing both an
amino
group and a carboxyl group. In some preferred embodiments, the amino acids are
a , ~3-, ~y or
S-amino acids, including their stereoisomers and racemates. As used herein the
term "L-
amino acid" denotes an tx amino acid having the L configuration around the a
carbon, that is,
a carboxylic acid of general formula CH(COOH)(NHZ)-(side chain), having the L-
configuration. The term "D-amino acid" similarly denotes a carboxylic acid of
general
formula CH(COOH)(NHZ)-(side chain), having the D-configuration around the cx
carbon.
Side chains of L-amino acids include naturally occurring and non-naturally
occurring
moieties. Non-naturally occurring (i.e., unnatural) amino acid side chains are
moieties that are
used in place of naturally occurring amino acid side chains in, for example,
amino acid
analogs. See, for example, Lehninger, Bioclaemist~y, Second Edition, Worth
Publishers, Inc,
1975, pages 72-77, incorporated herein by reference. Amino acid substituents
may be
attached through their carbonyl groups through the oxygen or carbonyl carbon
thereof, or
through their amino groups, or through functionalities residing on their side
chain portions.
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
-24-
In some embodiments of the methods of the invention, the cells are made
hypermutable using ATP analogs capable of blocking ATPase activity required
for MMR.
MMR reporter cells are screened with ATP compound libraries to identify those
compounds
capable of blocking MMR in vivo. Examples of ATP analogs that are useful in
blocking MMR
activity include, but are not limited to, nonhydrolyzable fot~ns of ATP such
as AMP-PNP and
ATP[gamma]S block the MMR activity (Galio, L. et al. (1999) Nucl. Acia's Res.
27:2325-
2331; Allen, D.J. et al. (1997) EMBO J. 16:4467-4476; Bjornson K.P. et al.
(2000) Biochem.
39:3176-3183). The ATPase inhibitors inhibit MMR and the cells become
hypermutable as a
result.
In other embodiments of the methods of the invention, the cells are made
hypermutable
using nuclease inhibitors that are able to block the exonuclease activity of
the MMR
biochemical pathway. MMR reporter cells are screened with nuclease inhibitor
compound
libraries to identify compounds capable of blocking MMR in vivo. Examples of
nuclease
inhibitors that are useful in blocking MMR activity include, but are not
limited to analogs of
N-Ethylmaleimide, an endonuclease inhibitor (Huang, Y.C., et.al. (1995)
Ay°ch. Biochem.
Biophys. 316:485), heterodimeric adenine-chain-acridine compounds, exonulcease
III
inhibitors (Belmont P, et.al., Bioorg Med Chem Lett (2000) 10:293-295), as
well as antibiotic
compounds such as Heliquinomycin, which have helicase inhibitory activity
(Chino, M, et.al.
J. Antibiot. (Tokyo) (1998) 51:480-486). The nuclease inhibitors inhibit MMR
and the cells
become hypennutable as a result.
In other embodiments of the methods of the invention, the cells are made
hypermutable
using DNA polymerase inhibitors that are able to block the polymerization
required for
mismatch-mediated repair. MMR reporter cells are screened with DNA polymerase
inhibitor
compound libraries to identify those compounds capable of blocking MMR in
vivo. Examples
of DNA polymerase inhibitors that are useful in blocking MMR activity include,
but are not
limited to, analogs of actinomycin D (Martin, S.J., et.al. (1990) J. Irnmunol.
145:1859),
Aphidicolin (Kuwakado, K. et.al. (1993) Bioclzem. Pha~macol. 46:1909) 1-(2'-
Deoxy-2'-
fluoro-beta-L-arabinofuranosyl)-5-methyluracil (L-FMAU) (Kukhanova M, et.al.,
Biochen2
Pha~rnacol (1998) 55:1181-1187), and 2',3'-dideoxyribonucleoside 5'-
triphosphates (ddNTPs)
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
- 25 -
(Ono, K., et.al., Biomed Plaarmacotlze~ (1984) 38:382-389). The polymerase
inhibitors inhibit
MMR and the cells become hypermutable as a result.
Bacterial cells rendered hypermutable using chemical inhibitors of MMR may be
made
genetically stable when the desired phenotype is obtained by removing the MMR
inhibitory
molecule.
In certain embodiments, the bacterial cells are made hypermutable by
introducing
plamids that generate antisense messages wherein the antisense RNA
specifically bind to
MMR genes and prevent efficient expression of MMR proteins. Preferably, the
antisense
transcripts are at least 12 nucleotides in length and, more preferably are at
least 20, 30, 40, 50
nucleotides or more in length. The antisense transcripts specifically bind to
regions of the
MMR gene to inhibit expression. Preferably, the antisense transcripts
specifically bind to
regulatory regions of the MMR gene such as to the MMR promoter region, Kozak
consensus
sequences, and the like. As used herein, "specifically bind" refers to
association of nucleic
acid strands forming complementary base pairing in Watson-Crick arrangement,
allowing for
mismatches such that 100% complementarity is not required. In general, the
complementarity
will be about 85%, 90%, 95% or more. Plasmids that may be used to express an
antisense
MMR transcript include any vector generally known in the art to express
antisense transcripts,
such as for example, those found in Qian Y. et al. (1998) Mutat. Res. 418(2-
3):61-71.
The above disclosure generally describes the present invention. A more
complete
understanding can be obtained by reference to the following specific examples
that will be
provided herein for purposes of illustration only, and are not intended to
limit the scope of the
invention.
Example 1: Generation of MMR defective bacteria.
Bacterial expression constructs were prepared using the human PMS2 related
gene
(hPMSR3) (Nicolaides et al. (1995) GefZOmics 30:195-206) and the human PMS134
cDNA
(Nicolaides et al. (1998) Mol. Cell. Biol. 18:1635-1641), both of which are
capable of
inactivating MMR activity and thereby increase the overall frequency of
genomic
hypermutation. Moreover, the use of regulatable expression vectors will allow
for
suppression of dominant negative MMR alleles and restoration of the MMR
pathway and
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
-26-
genetic stability in hosts cells (Brosius, J. (1988) Biotechnology 10:205-
225). For these
studies, a plasmid encoding the hPMS 134 cDNA was altered by polymerase chain
reaction
(PCR). The 5' oligonucleotide has the following sequence: 5'-acg cat atg gag
cga get gag agc
tcg agt-3' (SEQ ID NO:23)that includes the NdeI restriction site (cat atg).
The 3'-
oligonucleotide has the following sequence: 5'-gaa ttc tta tca cgt aga atc gag
acc gag gag agg
gtt agg gat agg ctt acc agt tcc aac ctt cgc cga tgc-3' (SEQ ID N0:24) that
includes an EcoRI
site (gaattc) and the 14 amino acid epitope for the VS antibody. The
oligonucleotides were
used for PCR under standard conditions that included 25 cycles of PCR
(95°C for 1 minute,
55°C for 1 minute, 72°C for 1.5 minutes for 25 cycles followed
by 3 minutes at 72°C). The
PCR fragment was purified by gel electrophoresis and cloned into pTA2.1
(InVitrogen) by
standard cloning methods (Sambrook et al., MOLECULAR CLONING: A LABORATORY
MANUAL,
THIRD EDITION, 2001), creating the plasmid pTA2.1-hPMS134. The pTA2.1-hPMS134
plasmid was digested with the restriction enzyme EcoRI to release the insert
(there are two
EcoRI restriction sites in the multiple cloning site of pTA2.1 that flank the
insert) and the
fragment was end-filled using Klenow fragment and dNTPs. Next, the fragment
was gel
purified, digested with NdeI, and inserted in pT7-Ea (which had been digested
with NdeI and
BarnHI, end-filledusing Klenow, and phosphatase treated). The new plasmid was
designated
pT7-Ea-hPMS 134.
The following strategy, similar to that described above to clone human PMS
134, was
used to construct an expression vector for the human related gene PMSR3.
First, the hPMSR3
fragment was amplified by PCR to introduce two restriction sites: an NdeI
restriction site at
the 5'- end, and an Eco RI site at the 3'-end of the fragment. The 5'-
oligonucleotide that was
used for PCR has the following sequence: 5'-acg cat atg tgt cct tgg cgg cct
aga-3' (SEQ ID
NO:25) that includes the NdeI restriction site (CAT ATG). The 3'-
oligonucleotide used for
PCR has the following sequence: 5'-gaa ttc tta tta cgt aga atc gag acc gag gag
agg gtt agg gat
agg ctt acc cat gtg tga tgt ttc aga gct-3' (SEQ ID N0:26) that includes an
EcoRI site (gaattc)
and the VS epitope to allow for antibody detection. The plasmid that contained
human
PMSR3 in pBluescript SK (Nicolaides et al. (1995) Gefaomics 30:195-206) was
used as the
PCR target with the hPMS2-specific oligonucleotides above. Following 25 cycles
of PCR
(95°C for 1 minute, 55°C for 1 minute, 72°C for 1.5
minutes for 25 cycles followed by 3
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
_27_
minutes at 72°C). The PCR fragment was purified by gel electrophoresis
and cloned into
pTA2.1 (InVitrogen) by standard cloning methods (Sambrook et al., MOLECULAR
CLONING: A
LABORATORY MANUAL, THIRD EDITION, 2001), creating the plasmid pTA2.l-hR3. The
pTA2.l-hR3 plasmid was next digested with the restriction enzyme EcoRI to
release the insert
(there are two EcoRI restriction sites in the multiple cloning site of pTA2.1
that flame the
insert) and the fragment was end-filled using Klenow fragment and dNTPs. Then,
the
fragment was gel purified, digested with NdeI, and inserted in pT7-Ea (which
had been
digested with NdeI and BamHI, end-filled using Klenow, and phosphatase
treated). The new
plasmid was designated pT7-Ea-hR3.
BL21 cells harbor an additional expression vector for the lysozyme protein,
which has
been demonstrated to bind to the T7 polymerase in situ; this results in a
bacterial strain that
has very low levels of T7 polymerase expression. However, upon addition of the
inducer
isopropyl-beta-D-thiogalactopyranoside (IPTG), the cells express high-levels
of T7
polymerase due to the IPTG-inducible element that drives expression of the
polymerase that is
resident within the genome of the BL21 cells (Studier et al. (1991) J. Mol.
Biol. 219(1):37-
44). The BL21 cells are chloramphenicol resistant due to the plasmid that
expresses lysozyme
within the cell. To introduce the pT7-hPMS 134 or the pT7-hPMSR3 genes into
BL21 cells,
the cells were made competent by incubating the cells in ice cold SOmM CaCl2
for 20 minutes,
followed by concentrating the cells and adding super-coiled plasmid DNA as
describe
Sambrook et al., MOLECULAR CLONING: A LABORATORY MANUAL, THIRD EDITION, Cold
Spring Harbor Laboratory Press, 2001). Ampicillin resistant BL21 were selected
on LB-agar
plates [5% yeast extract, 10% bactotryptone, 5% NaCI, 1.5% bactoagar, pH 7.0
(Difco)] plates
containing 25 ,ug/ml chloramphenicol and 100,ug/ml ampicillin. The next day,
bacterial
colonies were selected and analyzed by restriction endonuclease digestion and
sequence
analysis for plasmids containing an intact pTACPMS 134 or pTAC empty plasmid .
In addition to constructing a VS-epitope tagged PMS134 construct, we also
constructed and tested a non-epitope tagged version. This was prepared to
demonstrate that
the epitope tag did not cause the alteration of the dominant-negative
phenotype that PMS 134
has on mismatch repair activity. For these studies, a BamHI restriction
fragment containing
the hPMS 134 cDNA was filled-in using Klenow fragment and then sub-cloned into
a
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
_28_
Klenow-filled, blunt-ended NdeI-XhoI site of the pTACLAC expression vector
(which
contains the IPTG-inducible bacterial TAC promoter and ampicillin resistance
gene as
selectable marker). The NdeI-XhoI cloning site is flanked by the TACLAC
promoter that
contains the LacI repressor site followed by a Shine-Dalgarno ribosome-binding
site at the 5'
flanking region and the T1T2 ribosomal RNA terminator in the 3' flanking
region. The
TACLAC vector also contains the LacI gene, which is constitutively expressed
by the TAC
promoter.
DH10B bacterial cells containing the pBCSK vector (Stratagene), which
constitutively
expresses the (3-galactosidase gene and contains the chloramphenicol
resistance marker for
selection, were made competent via the CaCl2 method (Sambrook, et al.
MOLECULAR
CLONING: A LABORATORY MANUAL, Cold Spring Harbor Laboratory Press, 1982). This
vector turns bacterial cells blue when grown in the presence of IPTG and X-gal
that aids in the
detection of bacterial colonies. Competent cells were transfected with the
pTAC empty vector
or the pTACPMSl34 vector following the heat-shock protocol. Transfected
cultures were
plated onto LB-agar [5% yeast extract, 10% bactotryptone, 5% NaCI, 1.5%
bactoagar, pH 7.0
(Difco)] plates containing 25 ,ug/ml chloramphenicol and 100 ~.g/ml
ampicillin. The next day,
bacterial colonies were selected and analyzed by restriction endonuclease
digestion and
sequence analysis for plasmids containing an intact pTACPMS 134 or pTAC empty
plasmid.
Ten clones of each bacteria containing correct empty or PMS 134 inserts were
then grown to
confluence overnight in LB media (5% yeast extract, 10% bactotryptone, 5%
NaCI, pH 7.0)
containing 10 ~,g/ml chloramphenicol and 50 ~,g/ml ampicillin. The next day
TAC empty or
pTACPMS134 cultures were diluted 1:4 in LB medium plus 50 ~,M IPTG (Gold
Biotechnology) and cultures were grown for 12 and 24 hours at 37°C.
After incubation, 50 ~,1
aliquots were taken from each culture and added to 150 ~,ls of 2X SDS buffer
and cultures
were analyzed for PMS 134 protein expression by western blot.
Western blots were carried out as follows: 50 ,uls of each PMS 134 or empty
plasmid
culture was directly lysed in 2X lysis buffer (60 mM Tris, pH 6.8, 2% SDS, 10%
glycerol, 0.1
M 2-mercaptoethanol, 0.001% bromophenol blue) and samples were boiled for 5
minutes.
Lysate proteins were separated by electrophoresis on 4-20% Tris glycine gels
(Novex). Gels
were electroblotted onto Immobilon-P (Millipore) in 48 mM Tris base, 40 mM
glycine,
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
-29-
0.0375% SDS, 20% methanol and blocked overnight at 4°C in Tris-buffered
saline plus 0.05%
Tween-20 and 5% condensed milk. Filters were probed with a rabbit polyclonal
antibody
generated against the N-terminus of the human PMS2 polypeptide (Santa Cruz),
which is able
to recognize the PMS134 polypeptide (Su et al. (1988) J. Biol. Chem. 263:6829-
6835),
followed by a secondary goat anti-rabbit horseradish peroxidase-conjugated
antibody.
Alternatively, blots were probed with an anti-VS monoclonal antibody followed
by a
secondary goat anti-mouse horseradish peroxidase-conjugated antibody. After
incubation
with the secondary antibody, blots are developed using chemilmninescence
(Pierce) and
exposed to film to measure PMS 134 expression.
For induction of PMS gene product, 5 ml cultures of Luria Broth (LB) plus 50
~.g/ml
ampicillin were inoculated from glycerol stocks of the transformants pT7Ea
(BL21),
pT7PMS 134/V5 (BL21 ), or pT7PMSR3 (BL21 ) and grown overnight at 37°C
with shaking.
200 ~1 of each overnight culture was inoculated in 20 ml (1:100) fresh LB
broth plus
ampicillin and grown to an OD6oo of 0.6. 20 ~1 of 100 mM IPTG (final
concentration O.lmM)
was added and cultures were grown overnight. Western analysis confirmed the
presence of
inducible PMS expression in the presence of inducer molecule (not shown).
Example 2: Generation of antibiotic resistant bacteria
To demonstrate the ability to produce antibiotic resistant bacterial strains
by inhibiting
MMR, 10~ bacterial cells expressing either the vector (pT7Ea) or pT7PMS 134/V5
were
inoculated into 5 ml LB broth plus the appropriate antibiotic concentrations
as shown below
(Table 1) and grown overnight at 37°C with shaking. Antibiotic
concentrations were based on
O.SX the minimum inhibitory concentrations (MIC) observed to inhibit the
growth of bacteria
constitutively expressing the mar operon (Goldman et al. (1996) Antimicrobial
Agents
Chemother. 40: 1266-1269). Titration analysis found the following amounts to
be effective in
inhibiting bacterial growth in the presence of various compounds.
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
-30-
Table 1. Half minimum inhibitory concentrations (MIC) on BL21 cells.
DRUG O.SX MIC (u,~/ml)
Tetracycline 4.70
Nalidixic Acid 7.10
Ofloxacin 0.13
Norfloxacin 0.13
Vancomycin 250.0
The next day, cultures were analyzed for cell growth in the presence or
absence of antibiotics.
Table 2 summarizes typical data from these studies. No growth was observed in
bacterial
control cells (pT7Ea), which had OD levels similar to blank culture. In
contrast, significant
culture growth was observed in pT7PMS 134V5 and pT7PMSR3 (not shown) cultures
grown
in all antibiotics tested (Table 2)
Table 2. Overnight Growth of Drug Resistant Mutants Expressing the PMS2-134.
Drug pT7Ea pT7PMS134V5
growth Cell # growth Cell # (X109)
Tetracycline - 0 + 1.10
Nalidixic - 0 + 0.97
Acid
Ofloxacin - 0 + 1.20
Norfloxacin - 0 + 1.40
Vancomycin - 0 + ND
To test the stability of antibiotic resistance, cells were replated and
followed for
growth in the presence of 1X MIC concentration of antibiotic. Table 3 shows an
example in
which bacterial cells were inoculated at 1 x 10~ cells/ml and grown for 6
hours in the presence
of tetracycline (Tet). As shown in Figure 1, pT7Ea control culture did not
grow in the
presence of Tet while pT7PMS 134 and pT7PMSR3 cultures resistant to Tet grew
to
confluence at time 4 hours after inoculation. These data demonstrate the
ability to generate
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
-31-
antibiotic resistant cultures by blocking MMR and reestablishing genetically
stable cultures
that can be used for gene discovery.
EXAMPLE 3: Genomic analysis of antibiotic resistant bacteria and target
discovery.
The ability to generate a wide degree of genomic mutation in MMR defective
bacteria
allow for the rapid analysis of the AR host's genome in comparison to the wild-
type strain.
While many methods for mutation analysis exist that are know by those skilled
in the art,
several approaches exist that allow for the screening of unknown genes as well
as those that
exist which are capable of screening for mutants within "candidate" genes that
are capable of
conferring an antiobiotic resistant phenotype. One such method includes the
use of in vit~o-
coupled-translation strategies, which is a rapid method that is used to screen
for mutations that
result in truncated polypeptides (Liu et al. (1996) Nat. Med. 2:169-174;
Nicolaides et al.
(1994) Nature 371: 75-80; Papadopoulos et al. (1994) Seience 263:1625-1629;
Nicolaides et
al. (1998) Mol. Cell. Biol. 18:1635-1641; Alelcshun, M.N. and S.B. Levy (1999)
J. Bacte~iol.
181:3303-3306).
Ih vitro transcription-coupled-translation.
Linear DNA fragments containing candidate gene sequences were prepared by PCR,
incorporating sequences for ih vitro transcription and translation in the
sense primer. The sense
primer contains the leader sequence 5'-tttaatacgactcactatagggagaccaccatggnnn
nnn nnn nnn
nnn-3' (SEQ ID N0:27) where the series of "n" nuclsotides indicates sequence
corresponding
to the first 5 codons. The antisense primer consists of nucleotide sequences
surrounding and
including the natural stop codon of the gene. DNA fragments are PCR amplified
using buffers
and condions as described (Nicolaides et al. (1995) Gehomics 30:195-206). Two
to five
microliters of whole bacteria are added to the PCR reaction mix and reactions
are carried out
at 95°C for 1 minute for one cycle followed by thirty cycles at
95°C for 30 sec, 52°C for 2
minute and 72°C for 2 minutes. PCR products are then directly added to
a rabbiti reticulolysate
mixture to carry out transcription-coupled-translation (Promega). The reaction
mixtures were
supplemented with [35S]-methionine for detection purposes. Translation
reactions are
incubated for 2 hours. After the reaction is complete, an equal volume of 2X
SDS lysis buffer
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
-32-
is added to the samples, and the samples are boiled and then loaded onto 12%
NuPAGE gels
(Novex). Gels are run at 150V, dried and exposed to autoradiography. Products
that are
smaller than the expected molecular weight of the wild-type protein (as
compared to the
control samples) are then determined to be mutant and DNA, fragments are
sequenced to
confirm the presence of a frame-shift/nonsense mutation. This approach has
been used to
identify mutations in bacterial genes that have been previously been reported
to produce
antibiotic resistance in bacteria.
Discussion
The results described above lead to several conclusions. The inhibition of MMR
results in an increase in hypermutability in bacteria. This activity is due to
the inhibition of
MMR biochemical activity in these hosts. This invention provides a novel
method of
producing antibiotic resistant strains for target discovery and the rational
design of novel anti-
microbial agents to each target identified by generating AR bacteria through
the inhibition of
mismatch repair.
The disclosures of the following references, as well as the references cited
herein, are
hereby incorporated by reference in their entirety.
References
1. Attfield, P.V. and Pinney, R.J. 1985. Elimination of multicopy plasmid R6K
by
bleomycin. Antimicrob. Agents Chemother. 27(6):985-988.
2. Baker S.M., Bronner, C.E., Zhang, L., Plug, A.W., Robatez, M., Warren, G.,
Elliott,
E.A., Yu, J., Ashley, T., Arnheim, N., Bradley, N., Flavell, R.A., and Liskay,
R.M. 1995.
Male defective in the DNA mismatch repair gene PMS2 exhibit abnormal
chromosome
synapsis in meiosis. Cell 82:309-319.
3. Berry, A. 1996. Improving production of aromatic compounds in Escherichia
coli by
metabolic engineering. Trends Biotechnol. 14(7):250-256.
4. Bronner, C.E., Baker, S.M., Morrison, P.T., Warren, G., Smith, L.G.,
Lescoe, M.K.,
Kane, M., Earabino, C., Lipford, J., Lindblom, A., Tannergard, P.,
Bollag,R.J., Godwin, A., R.,
Ward, D.C., Nordenskjold, M., Fishel, R., I~olodner, R., and Liskay, R.M.
1994. Mutation in
the DNA mismatch repair gene homologue hMLHl is associated with hereditary
non-polyposis colon cancer. Nature 368:258-261.
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
- 33 -
5. de Wind N., Dekker, M., Berns, A., Radman; M., and Riele, H.T. 1995.
Inactivation of
the mouse Msh2 gene results in mismatch repair deficiency, methylation
tolerance,
hyperrecombination, and predisposition to cancer. Cell 82:321-300.
6. Drummond, J.T., Li, G.M., Longley, M.J., and Modrich, P. 1995. Isolation of
an
hMSH2-p160 heterodimer that restores mismatch repair to tumor cells. Science
268:1909-1912.
7. Drummond, J.T., Anthoney, A., Brown, R., and Modrich, P. 1996. Cisplatin
and
adriamycin resistance are associated with MutLa and mismatch repair deficiency
in an ovarian
tumor cell line. J.Biol.Chem. 271:9645-19648.
8. Edelmann, W., Cohen, P.E., Kane, M., Lau, K., Morrow, B., Bennett, S.,
Umar, A.,
Ktu~lcel, T., Cattoretti, G., Chagnatti, R., Pollard, J.W., Kolodner, R.D.,
and Kucherlapati, R.
1996. Meiotic pachytene arrest in MLH1-deficient mice. Cell 85:1125-1134.
9. Fishel, R., Lescoe, M., Rao, M.R.S., Copeland, N.J., Jenlcins, N.A.,
Garber, J., Kane,
M., and Kolodner, R. 1993. The human mutator gene homolog MSH~ and its
association with
hereditary nonpolyposis colon cancer. Cell 7:1027-1038.
10. Fu, K.P., Grace, M.E., Hsiao, C.L., and Hung, P.P. 1988. Elinination
ofantibiotic-
resistant plasmids by quinolone antibiotics. Chemotherapy 34(5):415-418.
11. Leach, F.S., Nicolaides, N.C, Papadopoulos, N., Liu, B., Jen, J., Parsons,
R., Peltomaki,
P., Sistonen, P., Aaltonen, L.A., Nystrom-Lahti, M., Guan, X.Y., Zhang, J.,
Meltzer, P.S., Yu,
J.W., Kao, F.T., Chen, D.J., Cerosaletti, K.M., Fournier, R.E.K., Todd, S.,
Lewis, T., Leach
R.J., Naylor, S.L., Weissenbach, J., Mecklin, J.P., Jarvinen, J.A., Petersen,
G.M., Hamilton,
S.R., Green, J., Jass, J., Watson, P., Lynch, H.T., Trent, J.M., de la
Chapelle, A., Kinzler,
K.W., and Vogelstein, B. 1993. Mutations of a mutS homolog in hereditary non-
polyposis
colorectal cancer. Cell 75:1215-1225.
12. Li, G.-M., and Modrich, P. 1995. Restoration of mismatch repair to nuclear
extracts of
H6 colorectal tumor cells by a heterodimer of human mutt homologs. Proc. Natl.
Acad. Sci.
USA 92:1950-1954.
13. Liu, B., Parsons, R., Papadopoulos, N., Nicolaides, N.C., Lynch, H.T.,
Watson, P., Jass,
J.R., Dunlop, M., Wyllie, A., Peltomaki, P., de la Chapelle, A., Hamilton,
S.R., Vogelstein, B.,
and Kinzler, K.W. 1996. Analysis of mismatch repair genes in hereditary non-
polyposis
colorectal cancer patients. Nat. Med. 2:169-174.
14. Modrich, P. 1994. Mismatch repair, genetic stability, and cancer. Science
266:1959-1960.
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
-34-
15. Nicolaides, N.C., Gualdi, R., Casadevall, C., Manzella, L., and
Calabretta, B. 1991.
Positive autoregulation of c-nayb expression via MYB binding in the 5'
flanking region of the
human c-myb gene. Mol. Cell. Biol. 11:6166-6176.
16. Nicolaides, N.C., Correa, L, Casadevall, C., Travali, S., Soprano, K.J.,
and Calabretta,
B. 1992. The Jun family members , c-JUN and JUND, transactivate the human c-
myb
promoter via an Apl like element. J. Biol. Chem. 267, 19665-19672.
17. Nicolaides, N.C., Papadopoulos, N., Liu, B., Wei, Y.F., Carter, K.C.,
Ruben, S.M.,
Rosen, C.A., Haseltine, W.A., Fleischmann, R.D., Fraser, C.M., Adams, M.D.,
Venter, C.J.,
Dunlop, M.G., Hamilton, S.R., Petersen, G.M., de la Chapelle, A., Vogelstein,
B., and kinzler,
K.W. 1994. Mutations of two PMS homologs in hereditary nonpolyposis colon
cancer.
Nature 371: 75-80.
18. Nicolaides N.C., Kinzler, K.W., and Vogelstein, B. 1995. Analysis of the
5' region of
PMS2 reveals heterogenous transcripts and a novel overlapping gene. Genomics
29:329-334.
19. Nicolaides, N.C., Carter, K.C., Shell, B.K., Papadopoulos, N., Vogelstein,
B., and
Kinzler, K.W. 1995. Genomic organization of the human PMS2 gene family.
Genomics
30:195-206.
20. Nicolaides, N.C., Palombo, F., Kinzler, K.W., Vogelstein, B., and Jiricny,
J. 1996.
Molecular cloning of the N-terminus of GTBP. Genomics 31:395-397.
21. Palombo, F., Hughes, M., Jiricny, J., Truong, O., Hsuan, J. 1994. Mismatch
repair and
cancer. Nature 36:417.
22. Palombo, F., Gallinari, P., Iaccarino, L, Lettieri, T., Hughes, M.A.,
Truong, O., Hsuan,
J.J., and Jiricny, J. 1995. GTBP, a 160-kilodalton protein essential for
mismatch-binding
activity in human cells. Science 268:1912-1914.
23. Pang, Q., Prolla, T.A., and Liskay, R.M. 1997. Functional domains of the
Sacchaf omyces cerevisiae Mlhlp and Pmslp DNA mismatch repair proteins and
their
relevance to human hereditary nonpolyposis colorectal cancer-associated
mutations. Mol.
Cell. Biol. 17:4465-4473.
24. Papadopoulos, N., Nicolaides, N.C., Wei, Y.F., Carter, K.C., Ruben, S.M.,
Rosen,
C.A., Haseltine, W.A., Fleischmann, R.D., Fraser, C.M., Adams, M.D., Venter,
C.J., Dunlop,
M.G., Hamilton, S.R., Petersen, G.M., de la Chapelle, A.,Vogelstein, B., and
kinzler, K.W.
1994. Mutation of a mutt homolog is associated with hereditary colon cancer.
Science
263:1625-1629.
25. Parsons, R., Li, G.M., Longley, M.J., Fang, W.H., Papadopolous, N., Jen,
J., de la
Chapelle, A., Kinzler, K.W., Vogelstein, B., and Modrich, P. 1993.
Hypermutability and
mismatch repair deficiency in RER+ tumor cells. Cell 75:1227-1236.
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
-35-
26. Parsons, R., Li, G.M., Longley, M., Modrich, P., Liu, B., Berk, T.,
Hamilton, S.R.,
Kinzler, K.W., and Vogelstein, B. 1995. Mismatch repair deficiency in
phenotypically
normal human cells. Science 268:738-740.
27. Perucho, M. 1996. Cancer of the microsattelite mutator phenotype. Biol
Chem.
377:675-684.
28. Prolla, T.A, Pang, Q., Alani, E., Kolodner, R.A., and Liskay, R.M. 1994.
MLH1,
PMS l, and MSH2 Interaction during the initiation of DNA mismatch repair in
yeast. Science
264:1091-1093.
29. Strand, M., Prolla, T.A., Liskay, R.M., and Petes, T.D. Destabilization of
tracts of
simple repetitive DNA in yeast by mutations affecting DNA mismatch repair.
1993. Nature
3 65:274-276.
30. Studier, F.W. Use of bacteriophage T7 lysozyme to improve an inducible T7
expression
system. 1991. J. Mol. Biol. 219(1):37-44.
31. Su, S.S., Lahue, R.S., Au, K.G., and Modrich, P. 1988. Mispair specificity
of methyl
directed DNA mismatch corrections in vitro. J. Biol. Chem. 263:6829-6835.
32. Nicolaides NC, Littman SJ, Modrich P, Kinzler KW, Vogelstein B 1998. A
naturally
occurring hPMS2 mutation can confer a dominant negative mutator phenotype. Mol
Cell Biol
18:1635-1641.
33. Aronshtam A, Marinus MG. 1996. Dominant negative mutator mutations in the
mutt
gene of Escherichia coli. Nucleic Acids Res 24:2498-2504.
34. Wu TH, Marinus MG. 1994. Dominant negative mutator mutations in the mutS
gene
of Escherichia coli. J Bacteriol 176:5393-400.
35. Brosh RM Jr, Matson SW. 1995. Mutations in motif II of Escherichia coli
DNA
helicase II render the enzyme nonfunctional in both mismatch repair and
excision repair with
differential effects on the unwinding reaction. J Bacteriol 177:5612-5621.
36. Studamire B, Quach T, Alani, E. 1998. Saccharomyces cerevisiae Msh2p and
Msh6p
ATPase activities are both required during mismatch repair. Mol Cell Biol
18:7590-7601.
37. Alani E, Solcolsky T, Studamire B, Miret JJ, Lahue RS. 1997. Genetic and
biochemical
analysis of Msh2p-Msh6p: role of ATP hydrolysis and Msh2p-Msh6p subunit
interactions in
mismatch base pair recognition.
Mol Cell Biol 17:2436-2447.
38. Lipkin SM, Wang V, Jacoby R, Banerjee-Basu S, Baxevanis AD, Lynch HT,
Elliott
RM, and Collins FS. 2000. MLH3: a DNA mismatch repair gene associated with
mammalian
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
-36-
microsatellite instability. Nat Genet 24:27-35.
39. Lee CC, Lin HK, Lin JK. 1994. A reverse mutagenicity assay for alkylating
agents
based on a point mutation in the beta-lactamase gene at the active site serine
codon.
Mutagenesis 9:401-405.
40. Vidal A, Abril N, Pueyo C. 1995. DNA repair by Ogt alkyltransferase
influences EMS
mutational specificity. Carcinogenesis 16:817-821.
41. Fu, K.P., Grace, M.E., Hsiao, C.L., and Hung, P.P. 1988. Elimination of
antibiotic-
resistant plasmids by quinolone antibiotics. Chem. Abstracts 34(5):415-418.
42. BiWang, H., ZhiPeng, H., and Xiong, G. 1999. Transformation of Escherichia
coli
and Bacillus thuringiensis and their plasmid curing by electroporation. J. of
Fujian
Agricultural University 28(1):43-46.
43. Brosius, J. 1988. Expression vectors employing lambda-, trp-, lac-, and
lpp-derived
promoters. Biotechnology 10:205-225.
44. Alekshun, M.N. and Levy, S.B. 1999. Characterization of MarR
Superrepressor
Mutants. J. Bacteriol. 181:3303-3306.
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
SEQUENCE LISTING
<110> Nicolaides, Nicholas C
Sass, Philip M
Grasso, Luigi M
Kline, J Bradford
<120> METHODS FOR GENERATING ANTIBIOTIC RESISTANT MICROBES AND NOVEL
ANTIBIOTICS
<130> MOR-0039
<140> 00/000,000
<141> 2001-07-25
<160> 39
<170> PatentIn version 3.1
<210> 1
<211> 3218
<212> DNA
<213> Saccharomyces cerevisiae
<400>
1
aaataggaatgtgataccttctattgcatgcaaagatagtgtaggaggcgctgctattgc 60
caaagacttttgagaccgcttgctgtttcattatagttgaggagttctcgaagacgagaa 120
attagcagttttcggtgtttagtaatcgcgctagcatgctaggacaatttaactgcaaaa 180
ttttgatacgatagtgatagtaaatggaaggtaaaaataacatagacctatcaataagca 240
atgtctctcagaataaaagcacttgatgcatcagtggttaacaaaattgctgcaggtgag 300
atcataatatcccccgtaaatgctctcaaagaaatgatggagaattccatcgatgcgaat 360
gctacaatgattgatattctagtcaaggaaggaggaattaaggtacttcaaataacagat 420
aacggatctggaattaataaagcagacctgccaatcttatgtgagcgattcacgacgtcc 480
aaattacaaaaattcgaagatttgagtcagattcaaacgtatggattccgaggagaagct 540
ttagccagtatctcacatgtggcaagagtcacagtaacgacaaaagttaaagaagacaga 600
tgtgcatggagagtttcatatgcagaaggtaagatgttggaaagccccaaacctgttgct 660
ggaaaagacggtaccacgatcctagttgaagacctttttttcaatattccttctagatta 720
agggccttgaggtcccataatgatgaatactctaaaatattagatgttgtcgggcgatac 780
gccattcattccaaggacattggcttttcttgtaaaaagttcggagactctaattattct 840
ttatcagttaaaccttcatatacagtccaggataggattaggactgtgttcaataaatct 900
gtggcttcgaatttaattacttttcatatcagcaaagtagaagatttaaacctggaaagc 960
gttgatggaaaggtgtgtaatttgaatttcatatccaaaaagtccatttcattaattttt 1020
ttcattaataatagactagtgacatgtgatcttctaagaagagctttgaacagcgtttac 1080
tccaattatctgccaaagggcttcagaccttttatttatttgggaattgttatagatccg 1140
gcggctgttgatgttaacgttcacccgacaaagagagaggttcgtttcctgagccaagat 1200
Page 1
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
gagatcatagagaaaatcgccaatcaattgcacgccgaattatctgccattgatacttca1260
cgtactttcaaggcttcttcaatttcaacaaacaagccagagtcattgataccatttaat1320
gacaccatagaaagtgataggaataggaagagtctccgacaagcccaagtggtagagaat1380
tcatatacgacagccaatagtcaactaaggaaagcgaaaagacaagagaataaactagtc1440
agaatagatgcttcacaagctaaaattacgtcatttttatcctcaagtcaacagttcaac1500
tttgaaggatcgtctacaaagcgacaactgagtgaacccaaggtaacaaatgtaagccac1560
tcccaagaggcagaaaagctgacactaaatgaaagcgaacaaccgcgtgatgccaataca1620
atcaatgataatgacttgaaggatcaacctaagaagaaacaaaagttgggggattataaa1680
gttccaagcattgccgatgacgaaaagaatgcactcccgatttcaaaagacgggtatatt1740
agagtacctaaggagcgagttaatgttaatcttacgagtatcaagaaattgcgtgaaaaa1800
gtagatgattcgatacatcgagaactaacagacatttttgcaaatttgaattacgttggg1860
gttgtagatgaggaaagaagattagccgctattcagcatgacttaaagctttttttaata1920
gattacggatctgtgtgctatgagctattctatcagattggtttgacagacttcgcaaac1980
tttggtaagataaacctacagagtacaaatgtgtcagatgatatagttttgtataatctc2040
ctatcagaatttgacgagttaaatgacgatgcttccaaagaaaaaataattagtaaaata2100
tgggacatgagcagtatgctaaatgagtactattccatagaattggtgaatgatggtcta2160
gataatgacttaaagtctgtgaagctaaaatctctaccactacttttaaaaggctacatt2220
ccatctctggtcaagttaccattttttatatatcgcctgggtaaagaagttgattgggag2280
gatgaacaagagtgtctagatggtattttaagagagattgcattactctatatacctgat2340
atggttccgaaagtcgatacactcgatgcatcgttgtcagaagacgaaaaagcccagttt2400
ataaatagaaaggaacacatatcctcattactagaacacgttctcttcccttgtatcaaa2460
cgaaggttcctggcccctagacacattctcaaggatgtcgtggaaatagccaaccttcca2520
gatctatacaaagtttttgagaggtgttaactttaaaacgttttggctgtaataccaaag2580
tttttgtttatttcctgagtgtgattgtgtttcatttgaaagtgtatgccctttccttta2640
acgattcatccgcgagatttcaaaggatatgaaatatggttgcagttaggaaagtatgtc2700
agaaatgtatattcggattgaaactcttctaatagttctgaagtcacttggttccgtatt2760
gttttcgtcctcttcctcaagcaacgattcttgtctaagcttattcaacggtaccaaaga2820
cccgagtccttttatgagagaaaacatttcatcatttttcaactcaattatcttaatatc2880
attttgtagtattttgaaaacaggatggtaaaacgaatcacctgaatctagaagctgtac2940
cttgtcccataaaagttttaatttactgagcctttcggtcaagtaaactagtttatctag3000
ttttgaaccgaatattgtgggcagatttgcagtaagttcagttagatctactaaaagttg3060
tttgacagcagccgattccacaaaaatttggtaaaaggagatgaaagagacctcgcgcgt3120
Page 2
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
aatggtttgc atcaccatcg gatgtctgtt gaaaaactca ctttttgcat ggaagttatt 3180
aacaataaga ctaatgatta ccttagaata atgtataa 3218
<210> 2
<211> 769
<212> PRT
<213> Saccharomyces cerevisiae
<400> 2
Met Ser Leu Arg Ile Lys Ala Leu Asp Ala Ser Val Val Asn Lys Ile
1 5 10 15
Ala Ala Gly Glu Ile Ile Ile Ser Pro Val Asn Ala Leu Lys Glu Met
20 25 30
Met Glu Asn Ser Ile Asp Ala Asn Ala Thr Met Ile Asp Ile Leu Val
35 40 45
Lys Glu Gly Gly Ile Lys Val Leu Gln Ile Thr Asp Asn Gly Ser Gly
50 55 60
Ile Asn Lys Ala Asp Leu Pro Ile Leu Cys Glu Arg Phe Thr Thr Ser
65 70 75 80
Lys Leu Gln Lys Phe Glu Asp Leu Ser Gln Ile Gln Thr Tyr Gly Phe
85 90 95
Arg Gly Glu Ala Leu Ala Ser Ile Ser His Val Ala Arg Val Thr Val
100 105 110
Thr Thr Lys Val Lys Glu Asp Arg Cys Ala Trp Arg Val Ser Tyr Ala
115 120 125
Glu Gly Lys Met Leu Glu Ser Pro Lys Pro Val Ala Gly Lys Asp Gly
130 135 140
Thr Thr Ile Leu Val Glu Asp Leu Phe Phe Asn Tle Pro Ser Arg Leu
145 150 155 160
Arg Ala Leu Arg Ser His Asn Asp Glu Tyr Ser Lys Ile Leu Asp Val
165 170 175
Val Gly Arg Tyr Ala Ile His Ser Lys Asp Ile Gly Phe Ser Cys Lys
180 185 190
Lys Phe Gly Asp Ser Asn Tyr Ser Leu Ser Val Lys Pro Ser Tyr Thr
195 200 205
Page 3
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
Val Gl.n Asp Arg Ile Arg Thr Val Phe Asn Lys Ser Val Ala Ser Asn
210 215 220
Leu Ile Thr Phe His Ile Ser Lys Val Glu Asp Leu Asn Leu Glu Ser
225 230 235 240
Val Asp Gly Lys Val Cys Asn Leu Asn Phe Ile Ser Lys Lys Ser Ile
245 250 255
Ser Leu Ile Phe Phe Ile Asn Asn Arg Leu Val Thr Cys Asp Leu Leu
260 265 270
Arg Arg Ala Leu Asn Ser Val Tyr Ser Asn Tyr Leu Pro Lys Gly Phe
275 280 285
Arg Pro Phe Ile Tyr Leu Gly Ile Val Ile Asp Pro Ala Ala Val Asp
290 295 300
Val Asn Val His Pro Thr Lys Arg Glu Val Arg Phe Leu Ser Gln Asp
305 310 315 320
Glu Ile Ile Glu Lys Ile Ala Asn Gln Leu His Ala Glu Leu Ser Ala
325 330, 335
Ile Asp Thr Ser Arg Thr Phe Lys Ala Ser Ser Ile Ser Thr Asn Lys
340 345 350
Pro Glu Ser Leu Ile Pro Phe Asn Asp Thr Ile Glu Ser Asp Arg Asn
355 360 365
Arg Lys Ser Leu Arg Gln Ala Gln Val Val Glu Asn Ser Tyr Thr Thr
370 375 380
Ala Asn Ser Gln Leu Arg Lys Ala Lys Arg Gln Glu Asn Lys Leu Val
385 390 395 400
Arg Ile Asp Ala Ser Gln Ala Lys Ile Thr Ser Phe Leu Ser Ser Ser
405 410 415
Gln Gln Phe Asn Phe Glu Gly Ser Ser Thr Lys Arg Gln Leu Ser Glu
420 425 430
Pro Lys Val Thr Asn Val Ser His Ser Gln Glu Ala Glu Lys Leu Thr
435 440 445
Leu Asn Glu Ser Glu Gln Pro Arg Asp Ala Asn Thr Ile Asn Asp Asn
450 455 460
Page 4
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
Asp Leu Lys Asp Gln Pro Lys Lys Lys Gln Lys Leu Gly Asp Tyr Lys
465 470 475 480
Val Pro Ser Ile Ala Asp Asp Glu Lys Asn Ala Leu Pro Ile Ser Lys
485 490 495
Asp Gly Tyr Ile Arg Val Pro Lys Glu Arg Val Asn Val Asn Leu Thr
500 505 510
Ser Ile Lys Lys Leu Arg Glu Lys Val Asp Asp Ser Ile His Arg Glu
515 520 525
Leu Thr Asp Ile Phe Ala Asn Leu Asn Tyr Val Gly Val Val Asp Glu
530 535 540
Glu Arg Arg Leu Ala Ala Ile Gln His Asp Leu Lys Leu Phe Leu Ile
545 550 555 560
Asp Tyr Gly Ser Val Cys Tyr Glu Leu Phe Tyr Gln Ile Gly Leu Thr
565 570 575
Asp Phe Ala Asn Phe Gly Lys Ile Asn Leu Gln Ser Thr Asn Val Ser
580 585 590
Asp Asp Ile Val Leu Tyr Asn Leu Leu Ser Glu Phe Asp Glu Leu Asn
595 600 605
Asp Asp Ala Ser Lys Glu Lys Ile Ile Ser Lys Ile Trp Asp Met Ser
610 615 620
Ser Met Leu Asn Glu Tyr Tyr Ser Ile Glu Leu Val Asn Asp Gly Leu
625 630 635 640
Asp Asn Asp Leu Lys Ser Val Lys Leu Lys Ser Leu Pro Leu Leu Leu
645 650 655
Lys Gly Tyr Ile Pro Ser Leu Val Lys Leu Pro Phe Phe Ile Tyr Arg
660 665 670
Leu Gly Lys Glu Val Asp Trp Glu Asp Glu Gln Glu Cys Leu Asp Gly
675 680 685
Ile Leu Arg Glu Ile Ala Leu Leu Tyr Ile Pro Asp Met Val Pro Lys
690 695 700
Val Asp Thr Leu Asp Ala Ser Leu Ser Glu Asp Glu Lys Ala Gln Phe
705 710 715 720
Page 5
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
Ile Asn Arg Lys Glu His Ile Ser Ser Leu Leu Glu His Val Leu Phe
725 730 735
Pro Cys Ile Lys Arg Arg Phe Leu Ala Pro Arg His Ile Leu Lys Asp
740 745 750
Val Val Glu Ile Ala Asn Leu Pro Asp Leu Tyr Lys Val Phe Glu Arg
755 760 765
Cys
<210>
3
<211>
3056
<212>
DNA
<213> musculus
Mus
<400>
3
gaattccggtgaaggtcctgaagaatttccagattcctgagtatcattggaggagacaga 60
taacctgtcgtcaggtaacgatggtgtatatgcaacagaaatgggtgttcctggagacgc 120
gtcttttcccgagagcggcaccgcaactctcccgcggtgactgtgactggaggagtcctg 180
catccatggagcaaaccgaaggcgtgagtacagaatgtgctaaggccatcaagcctattg 240
atgggaagtcagtccatcaaatttgttctgggcaggtgatactcagtttaagcaccgctg 300
tgaaggagttgatagaaaatagtgtagatgctggtgctactactattgatctaaggctta 360
aagactatggggtggacctcattgaagtttcagacaatggatgtggggtagaagaagaaa 420
actttgaaggtctagctctgaaacatcacacatctaagattcaagagtttgccgacctca 480
cgcaggttgaaactttcggctttcggggggaagetctgagctctctgtgtgcactaagtg 540
atgtcactatatctacctgccacgggtctgcaagcgttgggactcgactggtgtttgacc 600
ataatgggaaaatcacccagaaaactccctacccccgacctaaaggaaccacagtcagtg 660
tgcagcacttattttatacactacccgtgcgttacaaagagtttcagaggaacattaaaa 720
aggagtattccaaaatggtgcaggtcttacaggcgtactgtatcatctcagcaggcgtcc 780
gtgtaagctgcactaatcagctcggacaggggaagcggcacgctgtggtgtgcacaagcg 840
gcacgtctggcatgaaggaaaatatcgggtctgtgtttggccagaagcagttgcaaagcc 900
tcattccttttgttcagctgccccctagtgacgctgtgtgtgaagagtacggcctgagca 960
cttcaggacgccacaaaaccttttctacgtttcgggcttcatttcacagtgcacgcacgg 1020
cgccgggaggagtgcaacagacaggcagtttttcttcatcaatcagaggccctgtgaccc 1080
agcaaaggtctctaagcttgtcaatgaggttttatcacatgtataaccggcatcagtacc 1140
catttgtcgtccttaacgtttccgttgactcagaatgtgtggatattaatgtaactccag 1200
ataaaaggcaaattctactacaagaagagaagctattgctggccgttttaaagacctcct 1260
Page 6
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
tgataggaat gtttgacagt gatgcaaaca agcttaatgt caaccagcag ccactgctag 1320
atgttgaagg taacttagta aagctgcata ctgcagaact agaaaagcct gtgccaggaa 1380
agcaagataa ctctccttca ctgaagagca cagcagacga gaaaagggta gcatccatct 1440
ccaggctgag agaggccttt tctcttcatc ctactaaaga gatcaagtct aggggtccag 1500
agactgctga actgacacgg agttttccaa gtgagaaaag gggcgtgtta tcctcttatc 1560
cttcagacgt catctcttac agaggcctcc gtggctcgca ggacaaattg gtgagtccca 1620
cggacagccc tggtgactgt atggacagag agaaaataga aaaagactca gggctcagca 1680
gcacctcagc tggctctgag gaagagttca gcaccccaga agtggccagt agctttagca 1740
gtgactataa cgtgagctcc ctagaagaca gaccttctca ggaaaccata aactgtggtg 1800
acctggactg ccgtcctcca ggtacaggac agtccttgaa gccagaagac catggatatc 1860
aatgcaaagc tctacctcta gctcgtctgt cacccacaaa tgccaagcgc ttcaagacag 1920
aggaaagacc ctcaaatgtc aacatttctc aaagattgcc tggtcctcag agcacctcag 1980
cagctgaggt cgatgtagcc ataaaaatga ataagagaat cgtgctcctc gagttctctc 2040
tgagttctct agctaagcga atgaagcagt tacagcacct aaaggcgcag aacaaacatg 2100
aactgagtta cagaaaattt agggccaaga tttgccctgg agaaaaccaa gcagcagaag 2160
atgaactcag aaaagagatt agtaaatcga tgtttgcaga gatggagatc ttgggtcagt 2220
ttaacctggg atttatagta accaaactga aagaggacct cttcctggtg gaccagcatg 2280
ctgcggatga gaagtacaac tttgagatgc tgcagcagca cacggtgctc caggcgcaga 2340
ggctcatcac accccagact ctgaacttaa ctgctgtcaa tgaagctgta ctgatagaaa 2400
atctggaaat attcagaaag aatggctttg actttgtcat tgatgaggat gctccagtca 2460
ctgaaagggc taaattgatt tccttaccaa ctagtaaaaa ctggaccttt ggaccccaag 2520
atatagatga actgatcttt atgttaagtg acagccctgg ggtcatgtgc cggccctcac 2580
gagtcagaca gatgtttgct tccagagcct gtcggaagtc agtgatgatt ggaacggcgc 2640
tcaatgcgag cgagatgaag aagctcatca cccacatggg tgagatggac cacccctgga 2700
actgccccca cggcaggcca accatgaggc acgttgccaa tctggatgtc atctctcaga 2760
actgacacac cccttgtagc atagagttta ttacagattg ttcggtttgc aaagagaagg 2820
ttttaagtaa tctgattatc gttgtacaaa aattagcatg ctgctttaat gtactggatc 2880
catttaaaag cagtgttaag gcaggcatga tggagtgttc ctctagctca gctacttggg 2940
tgatccggtg ggagctcatg tgagcccagg actttgagac cactccgagc cacattcatg 3000
agactcaatt caaggacaaa aaaaaaaaga tatttttgaa gccttttaaa aaaaaa 3056
<210> 4
<2l1> 859
Page 7
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
<212> PRT
<213> Mus musculus
<400> 4
Met Glu Gln Thr Glu Gly Val Ser Thr Glu Cys Ala Lys Ala Ile Lys
1 5 10 15
Pro Ile Asp Gly Lys Ser Val His Gln Ile Cys Ser G1y Gln Val Ile
20 25 30
Leu Ser Leu Ser Thr Ala Val Lys Glu Leu Ile Glu Asn Ser Val Asp
35 40 45
Ala Gly Ala Thr Thr Ile Asp Leu Arg Leu Lys Asp Tyr Gly Val Asp
50 55 60
Leu Ile Glu Val Ser Asp Asn Gly Cys Gly Val Glu Glu Glu Asn Phe
65 70 75 80
Glu Gly Leu Ala Leu Lys His His Thr Ser Lys Ile Gln Glu Phe Ala
85 90 95
Asp Leu Thr Gln Val Glu Thr Phe Gly Phe Arg Gly Glu Ala Leu Ser
100 105 110
Ser Leu Cys Ala Leu Ser Asp Val Thr Ile Ser Thr Cys His Gly Ser
115 120 125
Ala Ser Val Gly Thr Arg Leu Val Phe Asp His Asn Gly Lys Ile Thr
130 135 140
Gln Lys Thr Pro Tyr Pro Arg Pro Lys Gly Thr Thr Val Ser Val Gln
145 150 155 160
His Leu Phe Tyr Thr Leu Pro Val Arg Tyr Lys Glu Phe Gln Arg Asn
165 170 175
Ile Lys Lys Glu Tyr Ser Lys Met Val Gln Val Leu Gln Ala Tyr Cys
180 185 190
Ile Ile Ser Ala Gly Val Arg Val Ser Cys Thr Asn Gln Leu Gly Gln
195 200 205
Gly Lys Arg His Ala Val Val Cys Thr Ser Gly Thr Ser Gly Met Lys
210 215 220
Glu Asn Ile Gly Ser Val Phe Gly Gln Lys Gln Leu Gln Ser Leu Ile
225 230 235 240
Page 8
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
Pro Phe Val Gln Leu Pro Pro Ser Asp Ala Val Cys Glu Glu Tyr Gly
245 250 255
Leu Ser Thr Ser Gly Arg His Lys Thr Phe Ser Thr Phe Arg Ala Ser
260 265 270
Phe His Ser Ala Arg Thr Ala Pro Gly Gly Val Gln Gln Thr Gly Ser
275 280 285
Phe Ser Ser Ser Ile Arg Gly Pro Val Thr Gln Gln Arg Ser Leu Ser
290 295 300
Leu Ser Met Arg Phe Tyr His Met Tyr Asn Arg His Gln Tyr Pro Phe
305 310 315 320
Val Val Leu Asn Val Ser Val Asp Ser Glu Cys Val Asp Ile Asn Val
325 330 335
Thr Pro Asp Lys Arg Gln I1e Leu Leu Gln Glu Glu Lys Leu Leu Leu
340 345 350
Ala Val Leu Lys Thr Ser Leu Ile Gly Met Phe Asp Ser Asp Ala Asn
355 360 365
Lys Leu Asn Val Asn Gln Gln Pro Leu Leu Asp Val Glu Gly Asn Leu
370 375 380
Val Lys Leu His Thr Ala Glu Leu Glu Lys Pro Val Pro Gly Lys Gln
385 390 395 400
Asp Asn Ser Pro Ser Leu Lys Ser Thr Ala Asp Glu Lys Arg Val Ala
405 410 4l5
Ser Ile Ser Arg Leu Arg Glu Ala Phe Ser Leu His Pro Thr Lys Glu
420 425 430
Ile Lys Ser Arg Gly Pro Glu Thr Ala Glu Leu Thr Arg Ser Phe Pro
435 440 445
Ser Glu Lys Arg Gly Val Leu Ser Ser Tyr Pro Ser Asp Val Ile Ser
450 455 460
Tyr Arg Gly Leu Arg Gly Ser Gln Asp Lys Leu Val Ser Pro Thr Asp
465 470 475 480
Ser Pro Gly Asp Cys Met Asp Arg Glu Lys Ile Glu Lys Asp Ser Gly
485 490 495
Page 9
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
Leu Ser Ser Thr Ser Ala Gly Ser Glu Glu Glu Phe Ser Thr Pro Glu
500 505 510
Val Ala Ser Ser Phe Ser Ser Asp Tyr Asn Val Ser Ser Leu Glu Asp
515 520 525
Arg Pro Ser Gln Glu Thr Ile Asn Cys Gly Asp Leu Asp Cys Arg Pro
530 535 540
Pro Gly Thr Gly Gln Ser Leu Lys Pro Glu Asp His Gly Tyr Gln Cys
545 550 555 560
Lys Ala Leu Pro Leu Ala Arg Leu Ser Pro Thr Asn Ala Lys Arg Phe
565 570 575
Lys Thr Glu Glu Arg Pro Ser Asn Val Asn Ile Ser Gln Arg Leu Pro
580 585 590
Gly Pro Gln Ser Thr Ser Ala Ala Glu Val Asp Val Ala Ile Lys Met
595 600 605
Asn Lys Arg Ile Val Leu Leu Glu Phe Ser Leu Ser Ser Leu Ala Lys
610 615 620
Arg Met Lys Gln Leu Gln His Leu Lys Ala Gln Asn Lys His Glu Leu
625 630 635 640
Ser Tyr Arg Lys Phe Arg Ala Lys Ile Cys Pro Gly Glu Asn Gln Ala
645 650 655
Ala Glu Asp Glu Leu Arg Lys Glu Ile Ser Lys Ser Met Phe Ala Glu
660 665 670
Met Glu Ile Leu Gly Gln Phe Asn Leu Gly Phe Ile Val Thr Lys Leu
675 680 685
Lys Glu Asp Leu Phe Leu Val Asp Gln His Ala Ala Asp Glu Lys Tyr
690 695 700
Asn Phe Glu Met Leu Gln Gln His Thr Val Leu Gln Ala Gln Arg Leu
705 710 715 720
Ile Thr Pro Gln Thr Leu Asn Leu Thr Ala Val Asn Glu Ala Val Leu
725 730 735
Ile Glu Asn Leu Glu Ile Phe Arg Lys Asn Gly Phe Asp Phe Val Ile
740 745 750
Page 10
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
Asp Glu Asp Ala Pro Val Thr Glu Arg Ala Lys Leu Ile Ser Leu Pro
755 760 765
Thr Ser Lys Asn Trp Thr Phe Gly Pro Gln Asp Ile Asp Glu Leu Ile
770 775 780
Phe Met Leu Ser Asp Ser Pro Gly Val Met Cys Arg Pro Ser Arg Val
785 790 795 800
Arg Gln Met Phe Ala Ser Arg Ala Cys Arg Lys Ser Val Met Ile Gly
805 810 815
Thr Ala Leu Asn Ala Ser Glu Met Lys Lys Leu Ile Thr His Met Gly
820 825 830
Glu Met Asp His Pro Trp Asn Cys Pro His Gly Arg Pro Thr Met Arg
835 840 845
His Val Ala Asn Leu Asp Val Ile Ser Gln Asn
850 855
<210>
<211>
2771
<212>
DNA
<213>
Homo
Sapiens
<400>
5
cgaggcggatcgggtgttgcatccatggagcgagctgagagctcgagtacagaacctgct60
aaggccatcaaacctattgatcggaagtcagtccatcagatttgctctgggcaggtggta120
ctgagtctaagcactgcggtaaaggagttagtagaaaacagtctggatgctggtgccact180
aatattgatctaaagcttaaggactatggagtggatcttattgaagtttcagacaatgga240
tgtggggtagaagaagaaaacttcgaaggcttaactctgaaacatcacacatctaagatt300
caagagtttgccgacctaactcaggttgaaacttttggctttcggggggaagctctgagc360
tcactttgtgcactgagcgatgtcaccatttctacctgccacgcatcggcgaaggttgga420
actcgactgatgtttgatcacaatgggaaaattatccagaaaaccccctacccccgcccc480
agagggaccacagtcagcgtgcagcagttattttccacactacctgtgcgccataaggaa540
tttcaaaggaatattaagaaggagtatgccaaaatggtccaggtcttacatgcatactgt600
atcatttcagcaggcatccgtgtaagttgcaccaatcagcttggacaaggaaaacgacag660
cctgtggtatgcacaggtggaagccccagcataaaggaaaatatcggctctgtgtttggg720
cagaagcagttgcaaagcctcattccttttgttcagctgccccctagtgactccgtgtgt780
gaagagtacggtttgagctgttcggatgctctgcataatcttttttacatctcaggtttc840
atttcacaatgcacgcatggagttggaaggagttcaacagacagacagtttttctttatc900
Page 11
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
aaccggcggccttgtgacccagcaaaggtctgcagactcgtgaatgaggtctaccacatg960
tataatcgacaccagtatccatttgttgttcttaacatttctgttgattcagaatgcgtt1020
gatatcaatgttactccagataaaaggcaaattttgctacaagaggaaaagcttttgttg1080
gcagttttaaagacctctttgataggaatgtttgatagtgatgtcaacaagctaaatgtc1140
agtcagcagccactgctggatgttgaaggtaacttaataaaaatgcatgcagcggatttg1200
gaaaagcccatggtagaaaagcaggatcaatccccttcattaaggactggagaagaaaaa1260
aaagacgtgtccatttccagactgcgagaggccttttctcttcgtcacacaacagagaac1320
aagcctcacagcccaaagactccagaaccaagaaggagccctctaggacagaaaaggggt1380
atgctgtcttctagcacttcaggtgccatctctgacaaaggcgtcctgagacctcagaaa1440
gaggcagtgagttccagtcacggacccagtgaccctacggacagagcggaggtggagaag1500
gactcggggcacggcagcacttccgtggattctgaggggttcagcatcccagacacgggc1560
agtcactgcagcagcgagtatgcggccagctccccaggggacaggggctcgcaggaacat1620
gtggactctcaggagaaagcgcctgaaactgacgactctttttcagatgtggactgccat1680
tcaaaccaggaagataccggatgtaaatttcgagttttgcctcagccaactaatctcgca1740
accccaaacacaaagcgttttaaaaaagaagaaattctttccagttctgacatttgtcaa1800
aagttagtaaatactcaggacatgtcagcctctcaggttgatgtagctgtgaaaattaat1860
aagaaagttgtgcccctggacttttctatgagttctttagctaaacgaataaagcagtta1920
catcatgaagcacagcaaagtgaaggggaacagaattacaggaagtttagggcaaagatt1980
tgtcctggagaaaatcaagcagccgaagatgaactaagaaaagagataagtaaaacgatg2040
tttgcagaaatggaaatcattggtcagtttaacctgggatttataataaccaaactgaat2100
gaggatatcttcatagtggaccagcatgccacggacgagaagtataacttcgagatgctg2160
cagcagcacaccgtgctccaggggcagaggctcatagcacctcagactctcaacttaact2220
gctgttaatgaagctgttctgatagaaaatctggaaatatttagaaagaatggctttgat2280
tttgttatcgatgaaaatgctccagtcactgaaagggctaaactgatttccttgccaact2340
agtaaaaactggaccttcggaccccaggacgtcgatgaactgatcttcatgctgagcgac2400
agccctggggtcatgtgccggccttcccgagtcaagcagatgtttgcctccagagcctgc2460
cggaagtcggtgatgattgggactgctcttaacacaagcgagatgaagaaactgatcacc2520
cacatgggggagatggaccacccctggaactgtccccatggaaggccaaccatgagacac2580
atcgccaacctgggtgtcatttctcagaactgaccgtagtcactgtatggaataattggt2640
tttatcgcagatttttatgttttgaaagacagagtcttcactaaccttttttgttttaaa2700
atgaaacctgctacttaaaaaaaatacacatcacacccatttaaaagtgatcttgagaac2760
cttttcaaacc 2771
Page 12
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
<210> 6
<211> 932
<212> PRT
<213> Homo Sapiens
<400> 6
Met Lys Gln Leu Pro Ala Ala Thr Val Arg Leu Leu Ser Ser Ser Gln
1 5 l0 15
Ile Ile Thr Ser Val Val Ser Val Val Lys Glu Leu Ile Glu Asn Ser
20 25 30
Leu Asp Ala Gly Ala Thr Ser Val Asp Val Lys Leu Glu Asn Tyr Gly
35 40 45
Phe Asp Lys Ile Glu Val Arg Asp Asn Gly Glu Gly Ile Lys Ala Val
50 55 60
Asp Ala Pro Val Met Ala Met Lys Tyr Tyr Thr Ser Lys Ile Asn Ser
65 70 75 80
His Glu Asp Leu Glu Asn Leu Thr Thr Tyr Gly Phe Arg Gly Glu Ala
85 90 95
Leu Gly Ser Ile Cys Cys Ile Ala Glu Val Leu Ile Thr Thr Arg Thr
100 105 110
Ala Ala Asp Asn Phe Ser Thr Gln Tyr Val Leu Asp Gly Ser Gly His
115 120 125
Ile Leu Ser Gln Lys Pro Ser His Leu Gly Gln Gly Thr Thr Val Thr
130 135 140
Ala Leu Arg Leu Phe Lys Asn Leu Pro Val Arg Lys Gln Phe Tyr Ser
145 150 155 160
Thr Ala Lys Lys Cys Lys Asp Glu Ile Lys Lys Ile Gln Asp Leu Leu
165 170 175
Met Ser Phe Gly Ile Leu Lys Pro Asp Leu Arg Ile Val Phe Val His
180 185 190
Asn Lys Ala Val Ile Trp Gln Lys Ser Arg Val Ser Asp His Lys Met
195 200 205
Ala Leu Met Ser Val Leu Gly Thr Ala Val Met Asn Asn Met Glu Ser
210 215 220
Page 13
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
Phe Gln Tyr His Ser Glu Glu Ser Gln Ile Tyr Leu Ser Gly Phe Leu
225 230 235 240
Pro Lys Cys Asp Ala Asp His Ser Phe Thr Ser Leu Ser Thr Pro Glu
245 250 255
Arg Ser Phe Ile Phe Ile Asn Ser Arg Pro Val His Gln Lys Asp Ile
260 265 270
Leu Lys Leu Ile Arg His His Tyr Asn Leu Lys Cys Leu Lys Glu Ser
275 280 285
Thr Arg Leu Tyr Pro Val Phe Phe Leu Lys Ile Asp Val Pro Thr Ala
290 295 300
Asp Val Asp Val Asn Leu Thr Pro Asp Lys Ser Gln Val Leu Leu Gln
305 310 315 320
Asn Lys Glu Ser Val Leu Ile Ala Leu Glu Asn Leu Met Thr Thr Cys
325 330 335
Tyr Gly Pro Leu Pro Ser Thr Asn Ser Tyr Glu Asn Asn Lys Thr Asp
340 345 350
Val Ser Ala Ala Asp Ile Val Leu Ser Lys Thr Ala Glu Thr Asp Val
355 360 365
Leu Phe Asn Lys Val Glu 5er Ser Gly Lys Asn Tyr Ser Asn Val Asp
370 375 380
Thr Ser Val Ile Pro Phe Gln Asn Asp Met His Asn Asp Glu Ser Gly
385 390 395 400
Lys Asn Thr Asp Asp Cys Leu Asn His Gln Ile Ser Ile Gly Asp Phe
405 410 415
Gly Tyr Gly His Cys Ser Ser Glu Ile Ser Asn Ile Asp Lys Asn Thr
420 425 430
Lys Asn Ala Phe Gln Asp Ile Ser Met Ser Asn Val Ser Trp Glu Asn
435 440 445
Ser Gln Thr Glu Tyr Ser Lys Thr Cys Phe Ile Ser Ser Val Lys His
450 455 460
Thr Gln Ser Glu Asn Gly Asn Lys Asp His Ile Asp Glu Ser Gly Glu
465 470 475 480
Page 14
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
Asn Glu Glu Glu Ala Gly Leu Glu Asn Ser Ser Glu Ile Ser Ala Asp
485 490 495
Glu Trp Ser Arg Gly Asn Ile Leu Lys Asn Ser Val Gly Glu Asn Ile
500 505 510
Glu Pro Val Lys Ile Leu Val Pro Glu Lys Ser Leu Pro Cys Lys Val
515 520 525
Ser Asn Asn Asn Tyr Pro Ile Pro Glu Gln Met Asn Leu Asn Glu Asp
530 535 540
Ser Cys Asn Lys Lys Ser Asn Val Ile Asp Asn Lys Ser Gly Lys Val
545 550 555 560
Thr Ala Tyr Asp Leu Leu Ser Asn Arg Val Ile Lys Lys Pro Met Ser
565 570 575
Ala Ser Ala Leu Phe Val Gln Asp His Arg Pro Gln Phe Leu Ile Glu
580 585 590
Asn Pro Lys Thr Ser Leu Glu Asp Ala Thr Leu Gln Ile Glu Glu Leu
595 600 605
Trp Lys Thr Leu Ser Glu Glu Glu Lys Leu Lys Tyr Glu'Glu Lys Ala
610 615 620
Thr Lys Asp Leu Glu Arg Tyr Asn Ser Gln Met Lys Arg Ala Ile Glu
625 630 635 640
Gln Glu Ser Gln Met Ser Leu Lys Asp Gly Arg Lys Lys Ile Lys Pro
645 650 655
Thr Ser Ala Trp Asn Leu Ala Gln Lys His Lys Leu Lys Thr Ser Leu
660 665 670
Ser Asn Gln Pro Lys Leu Asp Glu Leu Leu Gln Ser Gln Ile Glu Lys
675 680 685
Arg Arg Ser Gln Asn Ile Lys Met Val Gln Ile Pro Phe Ser Met Lys
690 695 700
Asn Leu Lys Ile Asn Phe Lys Lys Gln Asn Lys Val Asp Leu Glu Glu
705 710 715 720
Lys Asp Glu Pro Cys Leu Ile His Asn Leu Arg Phe Pro Asp Ala Trp
725 730 735
Page 15
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
Leu Met Thr Ser Lys Thr Glu Val Met Leu Leu Asn Pro Tyr Arg Val
740 745 750
Glu Glu Ala Leu Leu Phe Lys Arg Leu Leu,Glu Asn His Lys Leu Pro
755 760 765
Ala Glu Pro Leu Glu Lys Pro Ile Met Leu Thr Glu Ser Leu Phe Asn
770 775 780
Gly Ser His Tyr Leu Asp Val Leu Tyr Lys Met Thr Ala Asp Asp Gln
785 790 795 800
Arg Tyr Ser Gly Ser Thr Tyr Leu Ser Asp Pro Arg Leu Thr Ala Asn
805 810 815
Gly Phe Lys Ile Lys Leu Ile Pro Gly Val Ser Ile Thr Glu Asn Tyr
820 825 830
Leu Glu Ile Glu Gly Met Ala Asn Cys Leu Pro Phe Tyr Gly Val Ala
835 840 845
Asp Leu Lys Glu Ile Leu Asn Ala Ile Leu Asn Arg Asn Ala Lys Glu
850 855 860
Val Tyr Glu Cys Arg Pro Arg Lys Val Ile Ser Tyr Leu Glu Gly Glu
865 870 875 880
Ala Val Arg Leu Ser Arg Gln Leu Pro Met Tyr Leu Ser Lys Glu Asp
885 890 895
Ile Gln Asp Ile Ile Tyr Arg Met Lys His Gln Phe Gly Asn Glu Ile
900 905 910
Lys Glu Cys Val His Gly Arg Pro Phe Phe His His Leu Thr Tyr Leu
915 920 925
Pro Glu Thr Thr
930
<210> 7
<211> 3063
<212> DNA
<213> Homo Sapiens
<400> 7
ggcacgagtg gctgcttgcg gctagtggat ggtaattgcc tgcctcgcgc tagcagcaag 60
ctgctctgtt aaaagcgaaa atgaaacaat tgcctgcggc aacagttcga ctcctttcaa 120
gttctcagat catcacttcg gtggtcagtg ttgtaaaaga gcttattgaa aactccttgg 180
Page 16
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
atgctggtgccacaagcgtagatgttaaactggagaactatggatttgataaaattgagg240
tgcgagataacggggagggtatcaaggctgttgatgcacctgtaatggcaatgaagtact300
acacctcaaaaataaatagtcatgaagatcttgaaaatttgacaacttacggttttcgtg360
gagaagccttggggtcaatttgttgtatagctgaggttttaattacaacaagaacggctg420
ctgataattttagcacccagtatgttttagatggcagtggccacatactttctcagaaac480
cttcacatcttggtcaaggtacaactgtaactgctttaagattatttaagaatctacctg540
taagaaagcagttttactcaactgcaaaaaaatgtaaagatgaaataaaaaagatccaag600
atctcctcatgagctttggtatccttaaacctgacttaaggattgtctttgtacataaca660
aggcagttatttggcagaaaagcagagtatcagatcacaagatggctctcatgtcagttc720
tggggactgctgttatgaacaatatggaatcctttcagtaccactctgaagaatctcaga780
tttatctcagtggatttcttccaaagtgtgatgcagaccactctttcactagtctttcaa840
caccagaaagaagtttcatcttcataaacagtcgaccagtacatcaaaaagatatcttaa900
agttaatccgacatcattacaatctgaaatgcctaaaggaatctactcgtttgtatcctg960
ttttctttctgaaaatcgatgttcctacagctgatgttgatgtaaatttaacaccagata1020
aaagccaagtattattacaaaataaggaatctgttttaattgctcttgaaaatctgatga1080
cgacttgttatggaccattacctagtacaaattcttatgaaaataataaaacagatgttt1140
ccgcagctgacatcgttcttagtaaaacagcagaaacagatgtgctttttaataaagtgg1200
aatcatctggaaagaattattcaaatgttgatacttcagtcattccattccaaaatgata1260
tgcataatgatgaatctggaaaaaacactgatgattgtttaaatcaccagataagtattg1320
gtgactttggttatggtcattgtagtagtgaaatttctaacattgataaaaacactaaga1380
atgcatttcaggacatttcaatgagtaatgtatcatgggagaactctcagacggaatata1440
gtaaaacttgttttataagttccgttaagcacacccagtcagaaaatggcaataaagacc1500
atatagatgagagtggggaaaatgaggaagaagcaggtcttgaaaactcttcggaaattt1560
ctgcagatgagtggagcaggggaaatatacttaaaaattcagtgggagagaatattgaac1620
ctgtgaaaattttagtgcctgaaaaaagtttaccatgtaaagtaagtaataataattatc1680
caatccctgaacaaatgaatcttaatgaagattcatgtaacaaaaaatcaaatgtaatag1740
ataataaatctggaaaagttacagcttatgatttacttagcaatcgagtaatcaagaaac1800
ccatgtcagcaagtgctctttttgttcaagatcatcgtcctcagtttctcatagaaaatc1860
ctaagactagtttagaggatgcaacactacaaattgaagaactgtggaagacattgagtg1920
aagaggaaaaactgaaatatgaagagaaggctactaaagacttggaacgatacaatagtc1980
aaatgaagagagccattgaacaggagtcacaaatgtcactaaaagatggcagaaaaaaga2040
taaaacccaccagcgcatggaatttggcccagaagcacaagttaaaaacctcattatcta2100
Page 17
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
atcaaccaaaacttgatgaactccttcagtcccaaattgaaaaaagaaggagtcaaaata2160
ttaaaatggtacagatccccttttctatgaaaaacttaaaaataaattttaagaaacaaa2220
acaaagttgacttagaagagaaggatgaaccttgcttgatccacaatctcaggtttcctg2280
atgcatggctaatgacatccaaaacagaggtaatgttattaaatccatatagagtagaag2340
aagccctgctatttaaaagacttcttgagaatcataaacttcctgcagagccactggaaa2400
agccaattatgttaacagagagtctttttaatggatctcattatttagacgttttatata2460
aaatgacagcagatgaccaaagatacagtggatcaacttacctgtctgatcctcgtctta2520
cagcgaatggtttcaagataaaattgataccaggagtttcaattactgaaaattacttgg2580
aaatagaaggaatggctaattgtctcccattctatggagtagcagatttaaaagaaattc2640
ttaatgctatattaaacagaaatgcaaaggaagtttatgaatgtagacctcgcaaagtga2700
taagttatttagagggagaagcagtgcgtctatccagacaattacccatgtacttatcaa2760
aagaggacatccaagacattatctacagaatgaagcaccagtttggaaatgaaattaaag2820
agtgtgttcatggtcgcccattttttcatcatttaacctatcttccagaaactacatgat2880
taaatatgtttaagaagattagttaccattgaaattggttctgtcataaaacagcatgag2940
tctggttttaaattatctttgtattatgtgtcacatggttattttttaaatgaggattca3000
ctgacttgtttttatattgaaaaaagttccacgtattgtagaaaacgtaaataaactaat3060
aac 3063
<210> 8
<211> 932
<212> PRT
<213> Homo sapiens
<400> 8
Met Lys Gln Leu Pro Ala Ala Thr Val Arg Leu Leu Ser Ser Ser Gln
1 5 10 15
Ile Ile Thr Ser Val Val Ser Val Val Lys Glu Leu Ile Glu Asn Ser
20 25 30
Leu Asp Ala Gly Ala Thr Ser Val Asp Val Lys Leu Glu Asn Tyr Gly
35 40 45
Phe Asp Lys Ile Glu Val Arg~Asp Asn Gly Glu Gly Ile Lys Ala Val
50 55 60
Asp Ala Pro Val Met Ala Met Lys Tyr Tyr Thr Ser Lys Ile Asn Ser
65 70 75 80
His Glu Asp Leu Glu Asn Leu Thr Thr Tyr Gly Phe Arg Gly Glu Ala
Page 18
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
85 90 95
Leu Gly Ser Ile Cys Cys Ile Ala Glu Val Leu Ile Thr Thr Arg Thr
100 105 110
Ala Ala Asp Asn Phe Ser Thr Gln Tyr Val Leu Asp Gly Ser Gly His
115 120 125
Ile Leu Ser Gln Lys Pro Ser His Leu Gly Gln Gly Thr Thr Val Thr
130 135 140
Ala Leu Arg Leu Phe Lys Asn Leu Pro Val Arg Lys Gln Phe Tyr Ser
145 150 155 160
Thr Ala Lys Lys Cys Lys Asp Glu Ile Lys Lys Ile Gln Asp Leu Leu
165 170 175
Met Ser Phe Gly Ile Leu Lys Pro Asp Leu Arg Ile Val Phe Val His
180 185 190
Asn Lys. Ala Val Ile Trp Gln Lys Ser Arg Val Ser Asp His Lys Met
195 200 205
Ala Leu Met Ser Val Leu Gly Thr Ala Val Met Asn Asn Met Glu Ser
210 215 220
Phe Gln Tyr His Ser Glu Glu Ser Gln Ile Tyr Leu Ser Gly Phe Leu
225 230 235 240
Pro Lys Cys Asp Ala Asp His Ser Phe Thr Ser Leu Ser Thr Pro Glu
245 250 255
Arg Ser Phe Ile Phe Ile Asn Ser Arg Pro Val His Gln Lys Asp Ile
260 265 270
Leu Lys Leu Ile Arg His His Tyr Asn Leu Lys Cys Leu Lys Glu Ser
275 280 285
Thr Arg Leu Tyr Pro Val Phe Phe Leu Lys Ile Asp Val Pro Thr Ala
290 295 300
Asp Val Asp Val Asn Leu Thr Pro Asp Lys Ser Gln Val Leu Leu G1n
305 310 315 320
Asn Lys Glu Ser Val Leu Ile Ala Leu Glu Asn Leu Met Thr Thr Cys
325 330 335
Tyr Gly Pro Leu Pro Ser Thr Asn Ser Tyr Glu Asn Asn Lys Thr Asp
Page 19
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
340 345 350
Val Ser Ala Ala Asp Ile Val Leu Ser Lys Thr Ala Glu Thr Asp Val
355 360 365
Leu Phe Asn Lys Val Glu Ser Ser Gly Lys Asn Tyr Ser Asn Val Asp
370 375 380
Thr Ser Val Ile Pro Phe Gln Asn Asp Met His Asn Asp Glu Ser Gly
385 390 395 400
Lys Asn Thr Asp Asp Cys Leu Asn His Gln Ile Ser Ile Gly Asp Phe
405 410 415
Gly Tyr Gly His Cys Ser Ser Glu Ile Ser Asn Ile Asp Lys Asn Thr
420 425 430
Lys Asn Ala Phe Gln Asp Ile Ser Met Ser Asn Val Ser Trp Glu Asn
435 440 445
Ser Gln Thr Glu Tyr Ser Lys Thr Cys Phe Ile Ser Ser Val Lys His
450 455 460
Thr Gln Ser Glu Asn Gly Asn Lys Asp His Tle Asp Glu Ser Gly Glu
465 470 475 480
Asn Glu Glu Glu Ala Gly Leu Glu Asn Ser Ser Glu Ile Ser Ala Asp
485 490 495
Glu Trp Ser Arg Gly Asn Ile Leu Lys Asn Ser Val Gly Glu Asn Ile
500 505 510
Glu Pro Val Lys Ile Leu Val Pro Glu Lys Ser Leu Pro Cys Lys Val
5l5 520 525
Ser Asn Asn Asn Tyr Pro Ile Pro Glu Gln Met Asn Leu Asn Glu Asp
530 535 540
Ser Cys Asn Lys Lys Ser Asn Val Ile Asp Asn Lys Ser Gly Lys Val
545 550 555 560
Thr Ala Tyr Asp Leu Leu Ser Asn Arg Val Ile Lys Lys Pro Met Ser
565 570 575
Ala Ser Ala Leu Phe Val Gln Asp His Arg Pro Gln Phe Leu Ile Glu
580 585 590
Asn Pro Lys Thr Ser Leu Glu Asp Ala Thr Leu Gln Ile Glu Glu Leu
Page 20
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
595 600 605
Trp Lys Thr Leu Ser Glu Glu Glu Lys Leu Lys Tyr Glu Glu Lys Ala
610 615 620
Thr Lys Asp Leu Glu Arg Tyr Asn Ser Gln Met Lys Arg Ala Ile Glu
625 630 635 640
Gln Glu Ser Gln Met Ser Leu Lys Asp Gly Arg Lys Lys Ile Lys Pro
645 650 655
Thr Ser Ala Trp Asn Leu Ala Gln Lys His Lys Leu Lys Thr Ser Leu
660 665 670
Ser Asn Gln Pro Lys Leu Asp Glu Leu Leu Gln Ser Gln Ile Glu Lys
675 680 685
Arg Arg Ser Gln Asn Ile Lys Met Val Gln Ile Pro Phe Ser Met Lys
690 695 700
Asn Leu Lys Ile Asn Phe Lys Lys Gln Asn Lys Val Asp Leu Glu Glu
705 710 715 720
Lys Asp Glu Pro Cys Leu Ile His Asn Leu Arg Phe Pro Asp Ala Trp
725 730 735
Leu Met Thr Ser Lys Thr Glu Val Met Leu Leu Asn Pro Tyr Arg Val
740 745 750
Glu Glu Ala Leu Leu Phe Lys Arg Leu Leu Glu Asn His Lys Leu Pro
755 760 765
Ala Glu Pro Leu Glu Lys Pro Ile Met Leu Thr Glu Ser Leu Phe Asn
770 775 780
Gly Ser His Tyr Leu Asp Val Leu Tyr Lys Met Thr Ala Asp Asp Gln
785 790 795 800
Arg Tyr Ser Gly Ser Thr Tyr Leu Ser Asp Pro Arg Leu Thr Ala Asn
805 810 815
Gly Phe Lys Ile Lys Leu Ile Pro Gly Val Ser Ile Thr Glu Asn Tyr
820 825 830
Leu Glu Ile Glu Gly Met Ala Asn Cys Leu Pro Phe Tyr Gly Val Ala
835 840 845
Asp Leu Lys Glu Ile Leu Asn Ala Ile Leu Asn Arg Asn Ala Lys Glu
Page 21
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
850 855 ' 860
Val Tyr Glu Cys Arg Pro Arg Lys Val Ile Ser Tyr Leu Glu Gly Glu
865 870 875 880
Ala Val Arg Leu Ser Arg Gln Leu Pro Met Tyr Leu Ser Lys Glu Asp
885 890 895
Ile Gln Asp Ile Ile Tyr Arg Met Lys His Gln Phe Gly Asn Glu Ile
900 905 910
Lys Glu Cys Val His Gly Arg Pro Phe Phe His His Leu Thr Tyr Leu
915 920 925
Pro Glu Thr Thr
930
<210>
9
<211>
3145
<212>
DNA
<213>
Homo
Sapiens
<400>
9
ggcgggaaacagcttagtgggtgtggggtcgcgcattttcttcaaccaggaggtgaggag 60
gtttcgacatggcggtgcagccgaaggagacgctgcagttggagagcgcggccgaggtcg 120
gcttcgtgcgcttctttcagggcatgccggagaagccgaocaccacagtgcgccttttcg 180
accggggcgacttctatacggcgcacggcgaggacgcgctgctggccgcccgggaggtgt 240
tcaagacccagggggtgatcaagtacatggggccggcaggagcaaagaatctgcagagtg 300
ttgtgcttagtaaaatgaattttgaatcttttgtaaaagatcttcttctggttcgtcagt 360
atagagttgaagtttataagaatagagctggaaataaggcatccaaggagaatgattggt 420
atttggcatataaggcttctcctggcaatctctctcagtttgaagacattctotttggta 480
acaatgatatgtcagcttccattggtgttgtgggtgttaaaatgtccgcagttgatggcc 540
agagacaggttggagttgggtatgtggattccatacagaggaaactaggactgtgtgaat 600
tccctgataatgatcagttctccaatcttgaggctctcctcatccagattggaccaaagg 660
aatgtgttttacccggaggagagactgctggagacatggggaaactgagacagataattc 720
aaagaggaggaattctgatcacagaaagaaaaaaagctgacttttccacaaaagacattt 780
atcaggacctcaaccggttgttgaaaggcaaaaagggagagcagatgaatagtgctgtat 840
tgccagaaatggagaatcaggttgcagtttcatcactgtctgcggtaatcaagtttttag 900
aactcttatcagatgattocaactttggacagtttgaactgactacttttgacttcagcc 960
agtatatgaaattggatattgcagcagtcagagcccttaacctttttcagggttctgttg 1020
aagataccactggctctcagtctctggctgccttgctgaataagtgtaaaacccctcaag 1080
Page 22
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
gacaaagacttgttaaccagtggattaagcagcctctcatggataagaacagaatagagg1140
agagattgaatttagtggaagcttttgtagaagatgcagaattgaggcagactttacaag1200
aagatttacttcgtcgattcccagatcttaaccgacttgccaagaagtttcaaagacaag1260
cagcaaacttacaagattgttaccgactctatcagggtataaatcaactacctaatgtta1320
tacaggctctggaaaaacatgaaggaaaacaccagaaattattgttggcagtttttgtga1380
ctcctcttactgatcttcgttctgacttctccaagtttcaggaaatgatagaaacaactt1440
tagatatggatcaggtggaaaaccatgaattccttgtaaaaccttcatttgatcctaatc1500
tcagtgaattaagagaaataatgaatgacttggaaaagaagatgcagtcaacattaataa1560
gtgcagccagagatcttggcttggaccctggcaaacagattaaactggattccagtgcac1620
agtttggatattactttcgtgtaacctgtaaggaagaaaaagtccttcgtaacaataaaa1680
actttagtactgtagatatccagaagaatggtgttaaatttaccaacagcaaattgactt1740
ctttaaatgaagagtataccaaaaataaaacagaatatgaagaagcccaggatgccattg1800
ttaaagaaattgtcaatatttcttcaggctatgtagaaccaatgcagacactcaatgatg1860
tgttagctcagctagatgctgttgtcagctttgctcacgtgtcaaatggagcacctgttc1920
catatgtacgaccagccattttggagaaaggacaaggaagaattatattaaaagcatcca1980
ggcatgcttgtgttgaagttcaagatgaaattgcatttattcctaatgacgtatactttg2040
aaaaagataaacagatgttccacatcattactggccccaatatgggaggtaaatcaacat2100
atattcgacaaactggggtgatagtactcatggcccaaattgggtgttttgtgccatgtg2160
agtcagcagaagtgtccattgtggactgcatcttagcccgagtaggggctggtgacagtc2220
aattgaaaggagtctccacgttcatggctgaaatgttggaaactgcttctatcctcaggt2280
ctgcaaccaaagattcattaataatcatagatgaattgggaagaggaacttctacctacg2340
atggatttgggttagcatgggctatatcagaatacattgcaacaaagattggtgcttttt2400
gcatgtttgcaacccattttcatgaacttactgccttggccaatcagataccaactgtta2460
ataatctacatgtcacagcactcaccactgaagagaccttaactatgctttatcaggtga2520
agaaaggtgtctgtgatcaaagttttgggattcatgttgcagagcttgctaatttcccta2580
agcatgtaatagagtgtgctaaacagaaagccctggaacttgaggagtttcagtatattg2640
gagaatcgcaaggatatgatatcatggaaccagcagcaaagaagtgctatctggaaagag2700
agcaaggtgaaaaaattattcaggagttcctgtccaaggtgaaacaaatgccctttactg2760
aaatgtcagaagaaaacatcacaataaagttaaaacagctaaaagctgaagtaatagcaa2820
agaataatagctttgtaaatgaaatcatttcacgaataaaagttactacgtgaaaaatcc2880
cagtaatggaatgaaggtaatattgataagctattgtctgtaatag~ttttatattgtttt2940
atattaaccctttttccatagtgttaactgtcagtgcccatgggctatcaacttaataag3000
Page 23
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
atatttagta atattttact ttgaggacat tttcaaagat ttttattttg aaaaatgaga 3060
gctgtaactg aggactgttt gcaattgaca taggcaataa taagtgatgt gctgaatttt 3120
ataaataaaa tcatgtagtt tgtgg 3145
<210> 10
<211> 934
<212> PRT
<213> Homo sapiens
<400> 10
Met Ala Val Gln Pro Lys Glu Thr Leu Gln Leu Glu Ser Ala Ala Glu
1 ' 5 10 15
Val Gly Phe Val Arg Phe Phe Gln Gly Met Pro Glu Lys Pro Thr Thr
20 25 30
Thr Val Arg Leu Phe Asp Arg Gly Asp Phe Tyr Thr Ala His Gly Glu
35 40 45
Asp Ala Leu Leu Ala Ala Arg Glu Val Phe Lys Thr Gln Gly Val Ile
50 55 60
Lys Tyr Met Gly Pro Ala Gly Ala Lys Asn Leu Gln Ser Val Val Leu
65 70 75 80
Ser Lys Met Asn Phe Glu Ser Phe Val Lys Asp Leu Leu Leu Val Arg
85 90 95
Gln Tyr Arg Val Glu Val Tyr Lys Asn Arg Ala Gly Asn Lys Ala Ser
100 105 110
Lys Glu Asn Asp Trp Tyr Leu Ala Tyr Lys Ala Ser Pro Gly Asn Leu
115 120 125
Ser Gln Phe Glu Asp Ile Leu Phe Gly Asn Asn Asp Met Ser Ala Ser
130 135 140
Ile Gly Val Val Gly Val Lys Met Ser Ala Val Asp Gly Gln Arg Gln
145 150 155 160
Val Gly Val Gly Tyr Val Asp Ser Ile Gln Arg Lys Leu Gly Leu Cys
165 170 175
Glu Phe Pro Asp Asn Asp Gln Phe Ser Asn Leu Glu Ala Leu Leu Ile
180 185 190
Gln Ile Gly Pro Lys Glu Cys Val Leu Pro Gly Gly Glu Thr Ala Gly
Page 24
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
195 200 205
Asp Met Gly Lys Leu Arg Gln Ile Ile Gln Arg Gly Gly Ile Leu Ile
210 215 220
Thr Glu Arg Lys Lys Ala Asp Phe Ser Thr Lys Asp Ile Tyr Gln Asp
225 230 235 240
Leu Asn Arg Leu Leu Lys Gly Lys Lys Gly Glu Gln Met Asn Ser Ala
245 250 255
Val Leu Pro Glu Met Glu Asn Gln Val Ala Val Ser Ser Leu Ser Ala
260 265 270
Val Ile Lys Phe Leu Glu Leu Leu Ser Asp Asp Ser Asn Phe Gly Gln
275 280 285
Phe Glu Leu Thr Thr Phe Asp Phe Ser Gln Tyr Met Lys Leu Asp Ile
290 295 300
Ala Ala Val Arg Ala Leu Asn Leu Phe Gln Gly Ser Val Glu Asp Thr
305 310 315 320
Thr Gly Ser Gln Ser Leu Ala Ala Leu Leu Asn Lys Cys Lys Thr Pro
325 330 335
Gln Gly Gln Arg Leu Val Asn Gln Trp Ile Lys Gln Pro Leu Met Asp
340 345 350
Lys Asn Arg Ile Glu Glu Arg Leu Asn Leu Val Glu Ala Phe Val Glu
355 360 365
Asp Ala Glu Leu Arg Gln Thr Leu Gln Glu Asp Leu Leu Arg Arg Phe
370 375 380
Pro Asp Leu Asn Arg Leu Ala Lys Lys Phe Gln Arg Gln Ala Ala Asn
385 390 395 400
Leu Gln Asp Cys Tyr Arg Leu Tyr Gln Gly Ile Asn Gln Leu Pro Asn
405 410 415
Val Ile Gln Ala Leu Glu Lys His Glu Gly Lys His Gln Lys Leu Leu
420 425 430
Leu Ala Val Phe Val Thr Pro Leu Thr Asp Leu Arg Ser Asp Phe Ser
435 440 445
Lys Phe Gln Glu Met Ile Glu Thr Thr Leu Asp Met Asp Gln Val Glu
Page 25
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
450 455 460
Asn His Glu Phe Leu Val Lys Pro Ser Phe Asp Pro Asn Leu Ser Glu
465 470 475 480
Leu Arg Glu Ile Met Asn Asp Leu Glu Lys Lys Met Gln Ser Thr Leu
485 490 495
Ile Ser Ala Ala Arg Asp Leu Gly Leu Asp Pro Gly Lys Gln Ile Lys
500 505 510
Leu Asp Ser Ser Ala Gln Phe Gly Tyr Tyr Phe Arg Val Thr Cys Lys
515 520 525
Glu Glu Lys Val Leu Arg Asn Asn Lys Asn Phe Ser Thr Val Asp Ile
530 535 540
Gln Lys Asn Gly Val Lys Phe Thr Asn Ser Lys Leu Thr Ser Leu Asn
545 550 555 560
Glu Glu Tyr Thr Lys Asn Lys Thr Glu Tyr Glu Glu Ala Gln Asp Ala
565 570 575
Ile Val Lys Glu Ile Val Asn Ile Ser Ser Gly Tyr Val Glu Pro Met
580 585 590
Gln Thr Leu Asn Asp Val Leu Ala Gln Leu Asp Ala Val Val Ser Phe
595 600 605
Ala His Val Ser Asn Gly Ala Pro Val Pro Tyr Val Arg Pro Ala Ile
610 615 620
Leu Glu Lys Gly Gln Gly Arg Ile Ile Leu Lys Ala Ser Arg His Ala
625 630 635 640
Cys Val Glu Val Gln Asp Glu Ile Ala Phe Ile Pro Asn Asp Val Tyr
645 650 655
Phe Glu Lys Asp Lys Gln Met Phe His Ile Ile Thr Gly Pro Asn Met
660 665 670
Gly Gly Lys Ser Thr Tyr Ile Arg Gln Thr Gly Val Ile Val Leu Met
675 680 685
Ala Gln Ile Gly Cys Phe Val Pro Cys Glu Ser Ala Glu Val Ser Ile
690 695 700
Val Asp Cys Ile Leu Ala Arg Val Gly Ala Gly Asp Ser Gln Leu Lys
Page 26
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
705 710 715 720
Gly Val Ser Thr Phe Met Ala Glu Met Leu Glu Thr Ala Ser Ile Leu
725 730 735
Arg Ser Ala Thr Lys Asp Ser Leu Ile Ile Ile Asp Glu Leu Gly Arg
740 745 750
Gly Thr Ser Thr Tyr Asp Gly Phe Gly Leu Ala Trp Ala Ile Ser Glu
755 760 765
Tyr Ile Ala Thr Lys Ile Gly Ala Phe Cys Met Phe Ala Thr His Phe
770 775 780
His Glu Leu Thr Ala Leu Ala Asn Gln Ile Pro Thr Val Asn Asn Leu
785 790 795 800
His Val Thr Ala Leu Thr Thr Glu Glu Thr Leu Thr Met Leu Tyr Gln
805 8l0 815
Val Lys Lys Gly Val Cys Asp Gln Ser Phe Gly Ile His Val Ala Glu
820 825 830
Leu Ala Asn Phe Pro Lys His Va1 Ile Glu Cys Ala Lys Gln Lys Ala
835 840 845
Leu Glu Leu Glu Glu Phe Gln Tyr Ile Gly Glu Ser Gln Gly Tyr Asp
850 855 860
Ile Met Glu Pro Ala~Ala Lys Lys Cys Tyr Leu Glu Arg Glu Gln Gly
865 870 875 880
Glu Lys Ile Ile Gln Glu Phe Leu Ser Lys Val Lys Gln Met Pro Phe
885 890 895
Thr Glu Met Ser Glu Glu Asn Ile Thr Ile Lys Leu Lys Gln Leu Lys
900 905 910
Ala Glu Val Ile Ala Lys Asn Asn Ser Phe Val Asn Glu Ile Ile Ser
915 920 925
Arg Ile Lys Val Thr Thr
930
<210> 11
<211> 2484
<212> DNA
<213> Homo Sapiens
Page 27
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
<400>
11
cttggctcttctggcgccaaaatgtcgttcgtggcaggggttattcggcggctggacgag60
acagtggtgaaccgcatcgcggcgggggaagttatccagcggccagctaatgctatcaaa120
gagatgattgagaactgtttagatgcaaaatccacaagtattcaagtgattgttaaagag180
ggaggcctgaagttgattcagatccaagacaatggcaccgggatcaggaaagaagatctg240
gatattgtatgtgaaaggttcactactagtaaactgcagtcctttgaggatttagccagt300
atttctacctatggctttcgaggtgaggctttggccagcataagccatgtggctcatgtt360
actattacaacgaaaacagctgatggaaagtgtgcatacagagcaagttactcagatgga420
aaactgaaagcccctcctaaaccatgtgctggcaatcaagggacccagatcacggtggag480
gaccttttttacaacatagccacgaggagaaaagctttaaaaaatccaagtgaagaatat540
gggaaaattttggaagttgttggcaggtattcagtacacaatgcaggcattagtttctca600
gttaaaaaacaaggagagacagtagctgatgttaggacactacccaatgcctcaaccgtg660
gacaatattcgctccatctttggaaatgctgttagtcgagaactgatagaaattggatgt720
gaggataaaaccctagccttcaaaatgaatggttacatatccaatgcaaactactcagtg780
aagaagtgcatcttcttactcttcatcaaccatcgtctggtagaatcaacttccttgaga840
aaagccatagaaacagtgtatgcagcctatttgcccaaaaacacacacccattcctgtac900
ctcagtttagaaatcagtccccagaatgtggatgttaatgtgcaccccacaaagcatgaa960
gttcacttcctgcacgaggagagcatcctggagcgggtgcagcagcacatcgagagcaag1020
ctcctgggctccaattcctccaggatgtacttcacccagactttgctaccaggacttgct1080
ggcccctctggggagatggttaaatccacaacaagtctgacctcgtcttctacttctgga1140
agtagtgataaggtctatgcccaccagatggttcgtacagattcccgggaacagaagctt1200
gatgcatttctgcagcctctgagcaaacccctgtccagtcagccccaggccattgtcaca1260
gaggataagacagatatttctagtggcagggctaggcagcaagatgaggagatgcttgaa1320
ctcccagcccctgctgaagtggctgccaaaaatcagagcttggagggggatacaacaaag1380
gggacttcagaaatgtcagagaagagaggacctacttccagcaaccccagaaagagacat1440
cgggaagattctgatgtggaaatggtggaagatgattcccgaaaggaaatgactgcagct1500
tgtaccccccggagaaggatcattaacctcactagtgttttgagtctccaggaagaaatt1560
aatgagcagggacatgaggt.tctccgggagatgttgcataaccactccttcgtgggctgt1620
gtgaatcctcagtgggccttggcacagcatcaaaccaagttataccttctcaacaccacc1680
aagcttagtgaagaactgttctaccagatactcatttatgattttgccaattttggtgtt1740
ctcaggttatcggagccagcaccgctctttgaccttgccatgcttgccttagatagtcca1800
gagagtggctggacagaggaagatggtcccaaagaaggacttgctgaatacattgttgag1860
tttctgaagaagaaggctgagatgcttgcagactatttctctttggaaattgatgaggaa1920
Page 28
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
gggaacctgattggattaccccttctgattgacaactatgtgccccctttggagggactg1980
cctatcttcattcttcgactagccactgaggtgaattgggacgaagaaaaggaatgtttt2040
gaaagcctcagtaaagaatgcgctatgttctattccatccggaagcagtacatatctgag2100
gagtcgaccctctcaggccagcagagtgaagtgcctggctccattccaaactcctggaag2160
tggactgtggaacacattgtctataaagccttgcgctcacacattctgcctcctaaacat2220
ttcacagaagatggaaatatcctgcagcttgctaacctgcctgatctatacaaagtcttt2280
gagaggtgttaaatatggttatttatgcactgtgggatgtgttcttctttctctgtattc2340
cgatacaaagtgttgtatcaaagtgtgatatacaaagtgtaccaacataagtgttggtag2400
cacttaagacttatacttgccttctgatagtattcctttatacacagtggattgattata2460
aataaatagatgtgtcttaarata 2484
<210> 12
<211> 756
<212> PRT
<213> Homo sapiens
<400> 12
Met Ser Phe Val Ala Gly Val Ile Arg Arg Leu Asp Glu Thr Val Val
1 5 10 15
Asn Arg Ile Ala Ala Gly Glu Val Ile Gln Arg Pro Ala Asn Ala Ile
20 25 30
Lys Glu Met Ile Glu Asn Cys Leu Asp Ala Lys Ser Thr Ser Ile Gln
35 40 45
Val Ile Val Lys Glu Gly Gly Leu Lys Leu Ile Gln Ile Gln Asp Asn
50 55 60
Gly Thr Gly Ile Arg Lys Glu Asp Leu Asp Ile Val Cys Glu Arg Phe
65 70 75 80
Thr Thr Ser Lys Leu Gln Ser Phe Glu Asp Leu Ala Ser Ile Ser Thr
85 90 95
Tyr Gly Phe Arg Gly Glu Ala Leu Ala Ser Ile Ser His Val Ala His
100 105 110
Val Thr Ile Thr Thr Lys Thr Ala Asp Gly Lys Cys Ala Tyr Arg Ala
115 120 125
Ser Tyr Ser Asp Gly Lys Leu Lys Ala Pro Pro Lys Pro Cys Ala Gly
130 135 140
Page 29
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
Asn Gln Gly Thr Gln Ile Thr Val Glu Asp Leu Phe Tyr Asn Ile Ala
145 150 155 160
Thr Arg Arg Lys Ala Leu Lys Asn Pro Ser Glu Glu Tyr Gly Lys Ile
165 170 175
Leu Glu Val Val Gly Arg Tyr Ser Val His Asn Ala Gly Ile Ser Phe
180 185 190
Ser Val Lys Lys Gln Gly Glu Thr Val Ala Asp Val Arg Thr Leu Pro
195 200 205
Asn Ala Ser Thr Val Asp Asn Ile Arg Ser Ile Phe Gly Asn Ala Val
210 215 220
Ser Arg Glu Leu Ile Glu Ile Gly Cys Glu Asp Lys Thr Leu Ala Phe
225 230 235 240
Lys Met Asn Gly Tyr Ile Ser Asn Ala Asn Tyr Ser Val Lys Lys Cys
245 250 255
Ile Phe Leu Leu Phe Ile Asn His Arg Leu Val Glu Ser Thr Ser Leu
260 265 270
Arg Lys Ala Ile Glu Thr Val Tyr Ala Ala Tyr Leu Pro Lys Asn Thr
275 280 285
His Pro Phe Leu Tyr Leu Ser Leu Glu Ile Ser Pro Gln Asn Val Asp
290 295 300
Val Asn Val His Pro Thr Lys His Glu Val His Phe Leu His Glu Glu
305 310 315 320
Ser Ile Leu Glu Arg Val Gln Gln His Ile Glu Ser Lys Leu Leu Gly
325 330 335
Ser Asn Ser Ser Arg Met Tyr Phe Thr Gln Thr Leu Leu Pro Gly Leu
340 345 350
Ala Gly Pro Ser Gly Glu Met Val Lys Ser Thr Thr Ser Leu Thr Ser
355 360 365
Ser Ser Thr Ser Gly Ser Ser Asp Lys Val Tyr Ala His Gln Met Val
370 375 380
Arg Thr Asp Ser Arg Glu Gln Lys Leu Asp Ala Phe Leu Gln Pro Leu
385 390 395 400
Page 30
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
Ser Lys Pro Leu Ser Ser Gln Pro Gln Ala Ile Val Thr Glu Asp Lys
405 410 415
Thr Asp Ile Ser Ser Gly Arg Ala Arg Gln Gln Asp Glu Glu Met Leu
420 425 430
Glu Leu Pro Ala Pro Ala Glu Val Ala Ala Lys Asn Gln Ser Leu Glu
435 440 445
Gly Asp Thr Thr Lys Gly Thr Ser Glu Met Ser Glu Lys Arg Gly Pro
450 455 460
Thr Ser Ser Asn Pro Arg Lys Arg His Arg Glu Asp Ser Asp Val Glu
465 470 475 480
Met Val Glu Asp Asp Ser Arg Lys Glu Met Thr Ala Ala Cys Thr Pro
485 490 495
Arg Arg Arg Ile Ile Asn Leu Thr Ser Val Leu Ser Leu Gln Glu Glu
500 505 510
Ile Asn Glu Gln Gly His Glu Val Leu Arg Glu Met Leu His Asn His
515 520 525
Ser Phe Val Gly Cys Val Asn Pro Gln Trp Ala Leu Ala Gln His Gln
530 535 540
Thr Lys Leu Tyr Leu Leu Asn Thr Thr Lys Leu Ser Glu Glu Leu Phe
545 550 555 560
Tyr Gln Ile Leu Ile Tyr Asp Phe Ala Asn Phe Gly Val Leu Arg Leu
565 570 575
Ser Glu Pro Ala Pro Leu Phe Asp Leu Ala Met Leu Ala Leu Asp Ser
580 585 590
Pro Glu Ser Gly Trp Thr Glu Glu Asp Gly Pro Lys Glu Gly Leu Ala
595 600 605
Glu Tyr Ile Val Glu Phe Leu Lys Lys Lys Ala Glu Met Leu Ala Asp
610 615 620
Tyr Phe Ser Leu Glu Ile Asp Glu Glu Gly Asn Leu Ile Gly Leu Pro
625 630 635 640
Leu Leu Ile Asp Asn Tyr Val Pro Pro Leu Glu Gly Leu Pro Ile Phe
645 650 655
Page 31
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
Ile Leu Arg Leu Ala Thr Glu Val Asn Trp Asp Glu Glu Lys Glu Cys
660 665 670
Phe Glu Ser Leu Ser Lys Glu Cys Ala Met Phe Tyr Ser Ile Arg Lys
675 680 685
Gln Tyr Ile Ser Glu Glu Ser Thr Leu Ser Gly Gln Gln Ser Glu Val
690 695 700
Pro Gly Ser Ile Pro Asn Ser Trp Lys Trp Thr Val Glu His Ile Val
705 710 715 720
Tyr Lys Ala Leu Arg Ser His Ile Leu Pro Pro Lys His Phe Thr Glu
725 730 735
Asp Gly Asn Ile Leu Gln Leu Ala Asn Leu Pro Asp Leu Tyr Lys Val
740 745 750
Phe Glu Arg Cys
755
<210> 13
<211> 426
<212> DNA
<213> Homo Sapiens
<400>
13
cgaggcggatcgggtgttgcatccatggagcgagctgagagctcgagtacagaacctgct 60
aaggccatcaaacctattgatcggaagtcagtccatcagatttgctctgggcaggtggta 120
ctgagtctaagcactgcggtaaaggagttagtagaaaacagtctggatgctggtgccact 180
aatattgatctaaagcttaaggactatggagtggatcttattgaagtttcagacaatgga 240
tgtggggtagaagaagaaaacttcgaaggcttaactotgaaacatcacacatctaagatt 300
caagagtttgccgacctaactcaggttgaaacttttggctttcggggggaagctctgagc 360
tcactttgtgcactgagcgatgtcaccatttctacctgccacgcatcggcgaaggttgga 420
acttga 426
<210> 14
<211> 133
<212> .PRT
<213> Homo Sapiens
<400> 14
Met Lys Gln Leu Pro Ala Ala Thr Val Arg Leu Leu Ser Ser Ser Gln
1 5 10 15
Ile Ile Thr Ser Val Val Ser Val Val Lys Glu Leu Ile Glu Asn Ser
Page 32
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
20 25 30
Leu Asp Ala Gly Ala Thr Ser Val Asp Val Lys Leu Glu Asn Tyr Gly
35 40 45
Phe Asp Lys Ile Glu Val Arg Asp Asn Gly Glu Gly Ile Lys Ala Val
50 55 60
Asp Ala Pro Val Met Ala Met Lys Tyr Tyr Thr Ser Lys Ile Asn Ser
65 70 75 80
His Glu Asp Leu Glu Asn Leu Thr Thr Tyr Gly Phe Arg Gly Glu Ala
85 90 95
Leu Gly Ser Ile Cys Cys Ile Ala Glu Val Leu Ile Thr Thr Arg Thr
100 105 110
Ala Ala Asp Asn Phe Ser Thr Gln Tyr Val Leu Asp Gly Ser Gly His
115 120 125
Ile Leu Ser Gln Lys
130
<210>
15
<21l>
4264
<212>
DNA
<213>
Homo
Sapiens
<400>
15
atttcccgccagcaggagccgcgcggtagatgcggtgcttttaggagctccgtccgacag60
aacggttgggccttgccggctgtcggtatgtcgcgacagagcaccctgtacagcttcttc120
cccaagtctccggcgctgagtgatgccaacaaggcctcggccagggcctcacgcgaaggc180
ggccgtgccgccgctgcccccggggcctctccttccccaggcggggatgcggcctggagc240
gaggctgggcctgggcccaggcccttggcgcgatccgcgtcaccgcccaaggcgaagaac300
ctcaacggagggctgcggagatcggtagcgcctgctgcccccaccagttgtgacttctca360
ccaggagatttggtttgggccaagatggagggttacccctggtggccttgtctggtttac420
aaccacccctttgatggaacattcatccgcgagaaagggaaatcagtccgtgttcatgta480
cagttttttgatgacagcccaacaaggggctgggttagcaaaaggcttttaaagccatat540
acaggttcaaaatcaaaggaagcccagaagggaggtcatttttacagtgcaaagcctgaa600
atactgagagcaatgcaacgtgcagatgaagccttaaataaagacaagattaagaggctt660
gaattggcagtttgtgatgagccctcagagccagaagaggaagaagagatggaggtaggc720
acaacttacgtaacagataagagtgaagaagataatgaaattgagagtgaagaggaagta780
cagcctaagacacaaggatctaggcgaagtagccgccaaataaaaaaacgaagggtcata840
Page 33
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
tcagattctg agagtgacat tggtggctct gatgtggaat ttaagccaga cactaaggag 900
gaaggaagca gtgatgaaat aagcagtgga gtgggggata gtgagagtga aggcctgaac 960
agccctgtca aagttgctcg aaagcggaag agaatggtga ctggaaatgg ctctcttaaa 1020
aggaaaagct ctaggaagga aacgccctca gccaccaaac aagcaactag catttcatca 1080
gaaaccaaga atactttgag agctttctct gcccctcaaa attctgaatc ccaagcccac 1140
gttagtggag gtggtgatga cagtagtcgc cctactgttt ggtatcatga aactttagaa 1200
tggcttaagg aggaaaagag aagagatgag cacaggagga ggcctgatca ccccgatttt 1260
gatgcatcta cactctatgt gcctgaggat ttcctcaatt cttgtactcc tgggatgagg 1320
aagtggtggc agattaagtc tcagaacttt gatcttgtca tctgttacaa ggtggggaaa 1380
ttttatgagc tgtaccacat ggatgctctt attggagtca gtgaactggg gctggtattc 1440
atgaaaggca actgggccca ttctggcttt cctgaaattg catttggccg ttattcagat 1500
tccctggtgc agaagggcta taaagtagca cgagtggaac agactgagac tccagaaatg 1560
atggaggcac gatgtagaaa gatggcacat atatccaagt atgatagagt ggtgaggagg 1620
gagatctgta ggatcattac caagggtaca cagacttaca gtgtgctgga aggtgatccc 1680
tctgagaact acagtaagta tcttcttagc ctcaaagaaa aagaggaaga ttcttctggc 1740
catactcgtg catatggtgt gtgctttgtt gatacttcac tgggaaagtt tttcataggt 1800
cagttttcag atgatcgcca ttgttcgaga tttaggactc tagtggcaca ctatccccca 1860
gtacaagttt tatttgaaaa aggaaatctc tcaaaggaaa ctaaaacaat tctaaagagt 1920
tcattgtcct gttctcttca ggaaggtctg atacccggct cccagttttg ggatgcatcc 1980
aaaactttga gaactctcct tgaggaagaa tattttaggg aaaagctaag tgatggcatt . 2040
ggggtgatgt taccccaggt gcttaaaggt atgacttcag agtctgattc cattgggttg 2100
acaccaggag agaaaagtga attggccctc tctgctctag gtggttgtgt cttctacctc 2160
aaaaaatgcc ttattgatca ggagctttta tcaatggcta attttgaaga atatattccc 2220
ttggattctg acacagtcag cactacaaga tctggtgcta tcttcaccaa agcctatcaa 2280
cgaatggtgc tagatgcagt gacattaaac aacttggaga tttttctgaa tggaacaaat 2340
ggttctactg aaggaaccct actagagagg gttgatactt gccatactcc ttttggtaag 2400
cggctcctaa agcaatggct ttgtgcccca ctctgtaacc attatgctat taatgatcgt 2460
ctagatgcca tagaagacct catggttgtg cctgacaaaa tctccgaagt tgtagagctt 2520
ctaaagaagc ttccagatct tgagaggcta ctcagtaaaa ttcataatgt tgggtctccc 2580
ctgaagagtc agaaccaccc agacagcagg gctataatgt atgaagaaac tacatacagc 2640
aagaagaaga ttattgattt tctttctgct ctggaaggat tcaaagtaat gtgtaaaatt 2700
atagggatca tggaagaagt tgctgatggt tttaagtcta aaatccttaa gcaggtcatc 2760
Page 34
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
tctctgcagacaaaaaatcctgaaggtcgttttcctgatttgactgtagaattgaaccga2820
tgggatacagcctttgaccatgaaaaggctcgaaagactggacttattactcccaaagca2880
ggctttgactctgattatgaccaagctcttgctgacataagagaaaatgaacagagcctc2940
ctggaatacctagagaaacagcgcaacagaattggctgtaggaccatagtctattggggg3000
attggtaggaaccgttaccagctggaaattcctgagaatttcaccactcgcaatttgcca3060
gaagaatacgagttgaaatctaccaagaagggctgtaaacgatactggaccaaaactatt3120
gaaaagaagttggctaatctcataaatgctgaagaacggagggatgtatcattgaaggac3180
tgcatgcggcgactgttctataactttgataaaaattacaaggactggcagtctgctgta3240
gagtgtatcgcagtgttggatgttttactgtgcctggctaactatagtcgagggggtgat3300
ggtcctatgtgtcgcccagtaattctgttgccggaagataccccccccttcttagagctt3360
aaaggatcacgccatccttgcattacgaagactttttttggagatgattttattcctaat3420
gacattctaataggctgtgaggaagaggagcaggaaaatggcaaagcctattgtgtgctt3480
gttactggaccaaatatggggggcaagtctacgcttatgagacaggctggcttattagct3540
gtaatggcccagatgggttgttacgtccctgctgaagtgtgcaggctcacaccaattgat3600
agagtgtttactagacttggtgcctcagacagaataatgtcaggtgaaagtacatttttt3660
gttgaattaagtgaaactgccagcatactcatgcatgcaacagcacattctctggtgctt3720
gtggatgaattaggaagaggtactgcaacatttgatgggacggcaatagcaaatgcagtt3780
gttaaagaacttgctgagactataaaatgtcgtacattattttcaactcactaccattca3840
ttagtagaagattattctcaaaatgttgctgtgcgcctaggacatatggcatgcatggta3900
gaaaatgaatgtgaagaccccagccaggagactattacgttcctctataaattcattaag3960
ggagcttgtcctaaaagctatggctttaatgcagcaaggcttgctaatctcccagaggaa4020
gttattcaaaagggacatagaaaagcaagagaatttgagaagatgaatcagtcactacga4080
ttatttcgggaagtttgcctggctagtgaaaggtcaactgtagatgctgaagctgtccat4140
aaattgctgactttgattaaggaattatagactgactacattggaagctttgagttgact4200
tctgaccaaaggtggtaaattcagacaacattatgatctaataaactttattttttaaaa4260
atga 4264
<210>
16
<211>
1360
<212>
PRT
<213>
Homo
Sapiens
<400> 16
Met Ser Arg Gln Ser Thr Leu Tyr Ser Phe Phe Pro Lys Ser Pro Ala
1 5 10 15
Page 35
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
Leu Ser Asp Ala Asn Lys Ala Ser Ala Arg Ala Ser Arg Glu Gly Gly
20 25 30
Arg Ala Ala Ala Ala Pro Gly Ala Ser Pro Ser Pro Gly Gly Asp Ala
35 40 45
Ala Trp Ser Glu Ala Gly Pro Gly Pro Arg Pro Leu Ala Arg Ser Ala
50 55 60
Ser Pro Pro Lys Ala Lys Asn Leu Asn Gly Gly Leu Arg Arg Ser Val
65 70 75 80
Ala Pro Ala Ala Pro Thr Ser Cys Asp Phe Ser Pro Gly Asp Leu Val
85 90 95
Trp Ala Lys Met Glu Gly Tyr Pro Trp Trp Pro Cys Leu Val Tyr Asn
100 105 110
His Pro Phe Asp Gly Thr Phe Ile Arg Glu Lys Gly Lys Ser Val Arg
115 120 125
Val His Val Gln Phe Phe Asp Asp Ser Pro Thr Arg Gly Trp Val Ser
130 135 140
Lys Arg Leu Leu Lys Pro Tyr Thr Gly Ser Lys Ser Lys Glu Ala Gln
145 150 155 160
Lys Gly Gly His Phe Tyr Ser Ala Lys Pro Glu Ile Leu Arg Ala Met
165 170 175
Gln Arg Ala Asp Glu Ala Leu Asn Lys Asp Lys Ile Lys Arg Leu Glu
180 185 190
Leu Ala Val Cys Asp Glu Pro Ser Glu Pro Glu Glu Glu Glu Glu Met
195 200 205
Glu Val Gly Thr Thr Tyr Val Thr Asp Lys Ser Glu Glu Asp Asn Glu
210 215 220
Ile Glu Ser Glu Glu Glu Val Gln Pro Lys Thr Gln Gly Ser Arg Arg
225 , 230 235 240
Ser Ser Arg Gln Ile Lys Lys Arg Arg Val Ile Ser Asp Ser Glu Ser
245 250 255
Asp Ile Gly Gly Ser Asp Val Glu Phe Lys Pro Asp Thr Lys Glu Glu
260 265 270
Page 36
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
Gly Ser Ser Asp Glu Ile Ser Ser Gly Val Gly Asp Ser Glu Ser Glu
275 280 285
Gly Leu Asn Ser Pro Val Lys Val Ala Arg Lys Arg Lys Arg Met Val
290 295 300
Thr Gly Asn Gly Ser Leu Lys Arg Lys Ser Ser Arg Lys Glu Thr Pro
305 310 315 320
Ser Ala Thr Lys Gln Ala Thr Ser Ile Ser Ser Glu Thr Lys Asn Thr
325 ' 330 335
Leu Arg Ala Phe Ser Ala Pro Gln Asn Ser Glu Ser Gln Ala His Val
340 345 350
Ser Gly Gly Gly Asp Asp Ser Ser Arg Pro Thr Val Trp Tyr His Glu
355 360 365
Thr Leu Glu Trp Leu Lys Glu Glu Lys Arg Arg Asp Glu His Arg Arg
370 375 380
Arg Pro Asp His Pro Asp Phe Asp Ala Ser Thr Leu Tyr Val Pro Glu
385 390 395 400
Asp Phe Leu Asn Ser Cys Thr Pro Gly Met Arg Lys Trp Trp Gln Ile
405 410 415
Lys Ser Gln Asn Phe Asp Leu Val Ile Cys Tyr Lys Val Gly Lys Phe
420 425 430
Tyr Glu Leu Tyr His Met Asp Ala Leu Ile Gly Val Ser Glu Leu Gly
435 440 445
Leu Val Phe Met Lys Gly Asn Trp Ala His Ser Gly Phe Pro Glu Ile
450 455 460
Ala Phe Gly Arg Tyr Ser Asp Ser Leu Val Gln Lys Gly Tyr Lys Val
465 470 475 480
Ala Arg Val Glu Gln Thr Glu Thr Pro Glu Met Met Glu Ala Arg Cys
485 490 495
Arg Lys Met Ala His Ile Ser Lys Tyr Asp Arg Val Val Arg Arg Glu
500 505 510
Ile Cys Arg Ile Ile Thr Lys Gly Thr Gln Thr Tyr Ser Val Leu Glu
515 520 525
Page 37
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
Gly Asp Pro Ser Glu Asn Tyr Ser Lys Tyr Leu Leu Ser Leu Lys Glu
530 535 540
Lys Glu Glu Asp Ser Ser Gly His Thr Arg Ala Tyr Gly Val Cys Phe
545 550 555 560
Val Asp Thr Ser Leu Gly Lys Phe Phe Ile Gly Gln Phe Ser Asp Asp
565 570 575
Arg His Cys Ser Arg Phe Arg Thr Leu Val Ala His Tyr Pro Pro Val
580 585 590
Gln Val Leu Phe Glu Lys Gly Asn Leu Ser Lys Glu Thr Lys Thr Ile
595 600 605
Leu Lys Ser Ser Leu Ser Cys Ser Leu Gln Glu Gly Leu Ile Pro Gly
610 615 620
Ser Gln Phe Trp Asp Ala Ser Lys Thr Leu Arg Thr Leu Leu Glu Glu
625 630 635 640
Glu Tyr Phe Arg Glu Lys Leu Ser Asp Gly Ile Gly Val Met Leu Pro
645 650 655
Gln Val Leu Lys Gly Met Thr Ser Glu Ser Asp Ser Ile Gly Leu Thr
660 665 670
Pro Gly Glu Lys Ser Glu Leu Ala Leu Ser Ala Leu Gly Gly Cys Val
675 680 685
Phe Tyr Leu Lys Lys Cys Leu Ile Asp Gln Glu Leu Leu Ser Met Ala
690, 695 700
Asn Phe Glu Glu Tyr Ile Pro Leu Asp Ser Asp Thr Val Ser Thr Thr
705 710 715 720
Arg Ser Gly Ala Ile Phe Thr Lys Ala Tyr Gln Arg Met Val Leu Asp
725 730 735
Ala Val Thr Leu Asn Asn Leu Glu Ile Phe Leu Asn Gly Thr Asn Gly
740 745 750
Ser Thr Glu Gly Thr Leu Leu Glu Arg Val Asp Thr Cys His Thr Pro
755 760 765
Phe Gly Lys Arg Leu Leu Lys Gln Trp Leu Cys Ala Pro Leu Cys Asn
770 775 780
Page 38
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
His Tyr Ala Ile Asn Asp Arg Leu Asp Ala Ile Glu Asp Leu Met Val
785 790 795 800
Val Pro Asp Lys Ile Ser Glu Val Val Glu Leu Leu Lys Lys Leu Pro
805 810 815
Asp Leu Glu Arg Leu Leu Ser Lys Ile His Asn Val Gly Ser Pro Leu
820 825 830
Lys Ser Gln Asn His Pro Asp Ser Arg Ala Ile Met Tyr Glu Glu Thr
835 840 845
Thr Tyr Ser Lys Lys Lys Ile Ile Asp Phe Leu Ser Ala Leu Glu Gly
850 855 860
Phe Lys Val Met Cys Lys Ile Ile Gly Ile Met Glu Glu Val Ala Asp
865 870 875 880
Gly Phe Lys Ser Lys Ile Leu Lys Gln Val Ile Ser Leu Gln Thr Lys
885 890 895
Asn Pro Glu Gly Arg Phe Pro Asp Leu Thr Val Glu Leu Asn Arg Trp
900 905 910
Asp Thr Ala Phe Asp His Glu Lys Ala Arg Lys Thr Gly Leu Ile Thr
915 920 925
Pro Lys Ala Gly Phe Asp Ser Asp Tyr Asp Gln Ala Leu Ala Asp Ile
930 935 940
Arg Glu Asn Glu Gln Ser Leu Leu Glu Tyr Leu Glu Lys Gln Arg Asn
945 950 955 960
Arg Ile Gly Cys Arg Thr Ile Val Tyr Trp Gly Ile Gly Arg Asn Arg
965 970 975
Tyr Gln Leu Glu Ile Pro Glu Asn Phe Thr Thr Arg Asn Leu Pro Glu
980 985 990
Glu Tyr Glu Leu Lys Ser Thr Lys Lys Gly Cys Lys Arg Tyr Trp Thr
995 1000 1005
Lys Thr Ile Glu Lys Lys Leu Ala Asn Leu Ile Asn Ala Glu Glu
1010 1015 1020
Arg Arg Asp Val Ser Leu Lys Asp Cys Met Arg Arg Leu Phe Tyr
1025 1030 1035
Page 39
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
Asn Phe Asp Lys Asn Tyr Lys Asp Trp Gln Ser Ala Val Glu Cys
1040 1045 1050
Ile Ala Val Leu Asp Val Leu Leu Cys Leu Ala Asn Tyr Ser Arg
1055 1060 1065
Gly Gly Asp Gly Pro Met Cys Arg Pro Val Ile Leu Leu Pro Glu
1070 1075 1080
Asp Thr Pro Pro Phe Leu Glu Leu Lys Gly Ser Arg His Pro Cys
1085 1090 1095
Ile Thr Lys Thr Phe Phe Gly Asp Asp Phe Ile Pro Asn Asp Ile
1100 1105 1110
Leu Ile Gly Cys Glu Glu Glu Glu Gln Glu Asn Gly Lys Ala Tyr
1115 1120 1125
Cys Val Leu Val Thr Gly Pro Asn Met Gly Gly Lys Ser Thr Leu
1130 1135 1140
Met Arg Gln Ala Gly Leu Leu Ala Val Met Ala Gln Met Gly Cys
1145 1150 1155
Tyr Val Pro Ala Glu Val Cys Arg Leu Thr Pro Ile Asp Arg Val
1160 1165 1170
Phe Thr Arg Leu Gly Ala Ser Asp Arg Ile Met Ser Gly Glu Ser
1175 1180 ' 1185
Thr Phe Phe Val Glu Leu Ser Glu Thr Ala Ser Ile Leu Met His
1190 1195 1200
Ala Thr Ala His Ser Leu Val Leu Val Asp Glu Leu Gly Arg Gly
1205 1210 1215
Thr Ala Thr Phe Asp Gly Thr Ala Ile Ala Asn Ala Val Val Lys
1220 1225 1230
Glu Leu Ala Glu Thr Ile Lys Cys Arg Thr Leu Phe Ser Thr His
1235 1240 1245
Tyr His Ser Leu Val Glu Asp Tyr Ser Gln Asn Val Ala Val Arg
1250 1255 1260
Leu Gly His Met Ala Cys Met Val Glu Asn Glu Cys Glu Asp Pro
1265 1270 1275
Page 40
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
Ser Gln Glu Thr Ile Thr Phe Leu Tyr Lys Phe Ile Lys Gly Ala
1280 1285 1290
Cys Pro Lys Ser Tyr Gly Phe Asn Ala Ala Arg Leu Ala Asn Leu
1295 1300 1305
Pro Glu Glu Val Ile Gln Lys Gly His Arg Lys Ala Arg Glu Phe
1310 .1315 1320
Glu Lys Met Asn Gln Ser Leu Arg Leu Phe Arg Glu Val Cys Leu
1325 1330 1335
Ala Ser Glu Arg Ser Thr Val Asp Ala Glu Ala Val His Lys Leu
1340 1345 1350
Leu Thr Leu Ile Lys Glu Leu
1355 1360
<210>
17
<211>
1408
<212>
DNA
<213> sapiens
Homo
<400>
17
ggcgctcctacctgcaagtggctagtgccaagtgctgggccgccgctcctgccgtgcatg60
ttggggagccagtacatgcaggtgggctccacacggagaggggcgcagacccggtgacag120
ggctttacctggtacatcggcatggcgcaaccaaagcaagagagggtggcgcgtgccaga180
caccaacggtcggaaaccgccagacaccaacggtcggaaaccgccaagacaccaacgctc240
ggaaaccgccagacaccaacgctcggaaaccgccagacaccaaggctcggaatccacgcc300
aggccacgacggagggcgactacctcccttctgaccctgctgctggcgttcggaaaaaac360
gcagtccggtgtgctctgattggtccaggctctttgacgtcacggactcgacctttgaca420
gagccactaggcgaaaaggagagacgggaagtattttttccgccccgcccggaaagggtg480
gagcacaacgtcgaaagcagccgttgggagcccaggaggcggggcgcctgtgggagccgt540
ggagggaactttcccagtccccgaggcggatccggtgttgcatccttggagcgagctgag600
aactcgagtacagaacctgctaaggccatcaaacctattgatcggaagtcagtccatcag660
atttgctctgggccggtggtaccgagtctaaggccgaatgcggtgaaggagttagtagaa720
aacagtctggatgctggtgccactaatgttgatctaaagcttaaggactatggagtggat780
ctcattgaagtttcaggcaatggatgtggggtagaagaagaaaacttcgaaggctttact840
ctgaaacatcacacatgtaagattcaagagtttgccgacctaactcaggtggaaactttt900
ggctttcggggggaagctctgagctcactttgtgcactgagtgatgtcaccatttctacc960
tgccgtgtatcagcgaaggttgggactcgactggtgtttgatcactatgggaaaatcatc1020
Page 41
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
cagaaaaccccctacccccgccccagagggatgacagtcagcgtgaagcagttattttct1080
acgctacctgtgcaccataaagaatttcaaaggaatattaagaagaaacgtgcctgcttc1140
cccttcgccttctgccgtgattgtcagtttcctgaggcctccccagccatgcttcctgta1200
cagcctgtagaactgactcctagaagtaccccaccccacccctgctccttggaggacaac1260
gtgatcactgtattcagctctgtcaagaatggtccaggttcttctagatgatctgcacaa1320
atggttcctctcctccttcctgatgtctgccattagcattggaataaagttcctgctgaa1380
aatccaaaaaaaaaaaaaaaaaaaaaaa 1408
<2l0> 18
<211> 389
<212> PRT °
<2l3> Homo sapiens
<400> 18
Met Ala Gln Pro Lys Gln Glu Arg Val Ala Arg Ala Arg His Gln Arg
1 5 10 15
Ser Glu Thr Ala Arg His Gln Arg Ser Glu Thr Ala Lys Thr Pro Thr
20 25 30
Leu Gly Asn Arg Gln Thr Pro Thr Leu Gly Asn Arg Gln Thr Pro Arg
35 40 45
Leu Gly Ile His Ala Arg Pro Arg Arg Arg Ala Thr Thr Ser Leu Leu
50 55 60
Thr Leu Leu Leu Ala Phe Gly Lys Asn Ala Val Arg Cys Ala Leu Ile
65 70 75 80
Gly Pro Gly Ser Leu Thr Ser Arg Thr Arg Pro Leu Thr Glu Pro Leu
85 90 95
Gly Glu Lys Glu Arg Arg Glu Val Phe Phe Pro Pro Arg Pro Glu Arg
100 105 110
Val Glu His Asn Val Glu Ser Ser Arg Trp Glu Pro Arg Arg Arg Gly
115 120 125
Ala Cys Gly Ser Arg Gly Gly Asn Phe Pro Ser Pro Arg Gly Gly Ser
130 135 140
Gly Val Ala Ser Leu Glu Arg Ala Glu Asn Ser Ser Thr Glu Pro Ala
145 150 155 160
Lys Ala Ile Lys Pro Ile Asp Arg Lys Ser Val His Gln Ile Cys Ser
Page 42
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
165 170 175
Gly Pro Val Val Pro Ser Leu Arg Pro Asn Ala Val Lys Glu Leu Val
180 185 190
Glu Asn Ser Leu Asp Ala Gly Ala Thr Asn Val Asp Leu Lys Leu Lys
195 200 205
Asp Tyr Gly Val Asp Leu Ile Glu Val Ser Gly Asn Gly Cys Gly Val
210 215 220
Glu Glu Glu Asn Phe Glu Gly Phe Thr Leu Lys His His Thr Cys Lys
225 230 235 240
Ile Gln Glu Phe Ala Asp Leu Thr Gln Val Glu Thr Phe Gly Phe Arg
245 250 255
Gly Glu Ala Leu Ser Ser Leu Cys Ala Leu Ser Asp Val Thr Ile Ser
260 265 270
Thr Cys Arg Val Ser Ala Lys Val Gly Thr Arg Leu Val Phe Asp His
275 280 285
Tyr Gly Lys Ile Ile Gln Lys Thr Pro Tyr Pro Arg Pro Arg Gly Met
290 295 f 300
Thr Val Ser Val Lys Gln Leu Phe Ser Thr Leu Pro Val His His Lys
305 310 315 320
Glu Phe Gln Arg Asn Ile Lys Lys Lys Arg Ala Cys Phe Pro Phe Ala
325 330 335
Phe Cys Arg Asp Cys Gln Phe Pro Glu Ala Ser Pro Ala Met Leu Pro
340 345 350
Val Gln Pro Val Glu Leu Thr Pro Arg Ser Thr Pro Pro His Pro Cys
355 360 365
Ser Leu Glu Asp Asn Val Ile Thr Val Phe Ser Ser Val Lys Asn Gly
370 375 380
Pro Gly Ser Ser Arg
385
<210> 19
<211> 1785
<212> DNA
<213> Homo sapiens
Page 43
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
<400>
19
tttttagaaactgatgtttattttccatcaaccatttttccatgctgcttaagagaatat60
gcaagaacagcttaagaccagtcagtggttgctcctacccattcagtggcctgagcagtg120
gggagctgcagaccagtcttccgtggcaggctgagcgctccagtcttcagtagggaattg180
ctgaataggcacagagggcacctgtacaccttcagaccagtctgcaacctcaggctgagt240
agcagtgaactcaggagcgggagcagtccattcaccctgaaattcctccttggtcactgc300
cttctcagcagcagcctgctcttctttttcaatctcttcaggatctctgtagaagtacag360
atcaggcatgacctcccatgggtgttcacgggaaatggtgccacgcatgcgcagaacttc420
ccgagccagcatccaccacattaaacccactgagtgagctcccttgttgttgcatgggat480
ggcaatgtccacatagcgcagaggagaatctgtgttacacagcgcaatggtaggtaggtt540
aacataagatgcctccgtgagaggcgaaggggcggcgggacccgggcctggcccgtatgt600
gtccttggcggcctagactaggccgtcgctgtatggtgagccccagggaggcggatctgg660
gcccccagaaggaCa.CCCgCCtggatttgCCCCgtagCCCggcccgggcccctcgggagc720
agaacagccttggtgaggtggacaggaggggacctcgcgagcagacgcgcgcgccagcga780
cagcagccccgccccggcctctcgggagccggggggcagaggctgcggagccccaggagg840
gtctatcagccacagtctctgcatgtttccaagagcaacaggaaatgaacacattgcagg900
ggccagtgtcattcaaagatgtggctgtggatttcacccaggaggagtggcggcaactgg960
accctgatgagaagatagcatacggggatgtgatgttggagaactacagccatctagttt1020
ctgtggggtatgattatcaccaagccaaacatcatcatggagtggaggtgaaggaagtgg1080
agcagggagaggagccgtggataatggaaggtgaatttccatgtcaacatagtccagaac1140
ctgctaaggccatcaaacctattgatcggaagtcagtccatcagatttgctctgggccag1200
tggtactgagtctaagcactgcagtgaaggagttagtagaaaacagtctggatgctggtg1260
ccactaatattgatctaaagcttaaggactatggagtggatctcattgaagtttcagaca1320
atggatgtggggtagaagaagaaaactttgaaggcttaatctctttcagctctgaaacat1380
cacacatgtaagattcaagagtttgccgacctaactgaagttgaaactttcggttttcag1440
ggggaagctctgagctcactgtgtgcactgagcgatgtcaccatttctacctgccacgcg1500
ttggtgaaggttgggactcgactggtgtttgatcacgatgggaaaatcatccaggaaacc1560
ccctacccccaccccagagggaccacagtcagcgtgaagcagttattttctacgctacct1620
gtgcgccataaggaatttcaaaggaatattaagaagacgtgcctgcttccccttcgcctt1680
ctgccgtgattgtcagtttcctgaggcctccccagccatgcttcctgtacagcctgcaga1740
actgtgagtcaattaaacctcttttcttcataaattaaaaaaaaa 1785
<210> 20
<211> 264
Page 44
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
<212> PRT
<213> Homo Sapiens
<400> 20 ,
Met Cys Pro Trp Arg Pro Arg Leu Gly Arg Arg Cys Met Val Ser Pro
1 5 10 15
Arg Glu Ala Asp Leu Gly Pro Gln Lys Asp Thr Arg Leu Asp Leu Pro
20 25 30
Arg Ser Pro Ala Arg Ala Pro Arg Glu Gln Asn Ser Leu Gly Glu Val
35 40 45
Asp Arg Arg Gly Pro Arg Glu Gln Thr Arg Ala Pro Ala Thr Ala Ala
50 55 60
Pro Pro Arg Pro Leu Gly Ser Arg Gly Ala Glu Ala Ala Glu Pro Gln
65 70 75 80
Glu Gly Leu Ser Ala Thr Val Ser Ala Cys Phe Gln Glu Gln Gln Glu
85 90 95
Met Asn Thr Leu Gln Gly Pro Val Ser Phe Lys Asp Val Ala Val Asp
100 105 110
Phe Thr Gln Glu Glu Trp Arg Gln Leu Asp Pro Asp Glu Lys Ile Ala
115 120 125
Tyr Gly Asp Val Met Leu Glu Asn Tyr Ser His Leu Val Ser Val Gly
130 135 140 .
Tyr Asp Tyr His Gln Ala Lys His His His Gly Val Glu Val Lys Glu
145 150 155 160
Val Glu Gln Gly Glu Glu Pro Trp Ile Met Glu Gly Glu Phe Pro Cys
165 170 175
Gln His Ser Pro Glu Pro Ala Lys Ala Ile Lys Pro Ile Asp Arg Lys
180 185 190
Ser Val His Gln Ile Cys Ser Gly Pro Val Val Leu Ser Leu Ser Thr
195 200 205
Ala Val Lys Glu Leu Val Glu Asn Ser Leu Asp Ala Gly Ala Thr Asn
210 215 220
Ile Asp Leu Lys Leu Lys Asp Tyr Gly Val Asp Leu Ile Glu Val Ser
225 230 235 240
Page 45
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
Asp Asn Gly Cys Gly Val Glu Glu Glu Asn Phe Glu Gly Leu Ile Ser
245 250 255
Phe Ser Ser Glu Thr Ser His Met
260
<210> 21
<211> 795
<212> DNA
<213> Homo Sapiens
<400>
21
atgtgtccttggcggcctagactaggccgtcgctgtatggtgagccccagggaggcggat60
ctgggcccccagaaggacacccgcctggatttgccccgtagcccggcccgggcccctcgg120
gagcagaacagccttggtgaggtggacaggaggggacctcgcgagcagacgcgcgcgcca180
gcgacagcagCCCCgCCCCggCCtCtCgggagccggggggcagaggctgcggagccccag240
gagggtctatcagccacagtctctgcatgtttccaagagcaacaggaaatgaacacattg300
caggggccagtgtcattcaaagatgtggctgtggatttcacccaggaggagtggcggcaa360
ctggaccctgatgagaagatagcatacggggatgtgatgttggagaactacagccatcta420
gtttctgtggggtatgattatcaccaagccaaacatcatcatggagtggaggtgaaggaa480
gtggagcagggagaggagccgtggataatggaaggtgaatttccatgtcaacatagtcca540
gaacctgctaaggccatcaaacctattgatcggaagtcagtccatcagatttgctctggg600
ccagtggtactgagtctaagcactgcagtgaaggagttagtagaaaacagtctggatgct660
ggtgccactaatattgatctaaagcttaaggactatggagtggatctcattgaagtttca720
gacaatggatgtggggtagaagaagaaaactttgaaggcttaatetctttcagctctgaa780
acatcacacatgtaa 795
<210> 22
<211> 264
<212> PRT
<213> Homo Sapiens
<400> 22
Met Cys Pro Trp Arg Pro Arg Leu Gly Arg Arg Cys Met Val Ser Pro
1 5 10 15
Arg Glu Ala Asp Leu Gly Pro Gln Lys Asp Thr Arg Leu Asp Leu Pro
20 25 30
Arg Ser Pro Ala Arg Ala Pro Arg Glu Gln Asn Ser Leu Gly Glu Val
35 40 45
Asp Arg Arg Gly Pro Arg Glu Gln Thr Arg Ala Pro Ala Thr Ala Ala
Page 46
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
50 55 60
Pro Pro Arg Pro Leu Gly Ser Arg Gly Ala Glu Ala Ala Glu Pro Gln
65 70 75 80
Glu Gly Leu Ser Ala Thr Val Ser Ala Cys Phe Gln Glu Gln Gln Glu
85 90 95
Met Asn Thr Leu Gln Gly Pro Val Ser Phe Lys Asp Val Ala Val Asp
100 105 110
Phe Thr Gln Glu Glu Trp Arg Gln Leu Asp Pro Asp Glu Lys Ile Ala
115 120 125
Tyr Gly Asp Val Met Leu Glu Asn Tyr Ser His Leu Val Ser Val Gly
130 135 140
Tyr Asp Tyr His Gln Ala Lys His His His Gly Val Glu Val Lys Glu
145 150 155 160
Val Glu Gln Gly Glu Glu Pro Trp Ile Met Glu Gly Glu Phe Pro Cys
165 170 175
Gln His Ser Pro Glu Pro Ala Lys Ala Ile Lys Pro Ile Asp Arg Lys
l80 185 190
Ser Val His Gln Ile Cys Ser Gly Pro Val Val Leu Ser Leu Ser Thr
195 200 205
Ala Val Lys Glu Leu Val Glu Asn Ser Leu Asp Ala Gly Ala Thr Asn
210 215 220
Ile Asp Leu Lys Leu Lys Asp Tyr Gly Val Asp Leu Ile Glu Val Ser
225 230 235 240
Asp Asn Gly Cys Gly Val Glu Glu Glu Asn Phe Glu Gly Leu Ile Ser
245 250 255
Phe Ser Ser Glu Thr Ser His Met
260
<210> 23
<211> 30
<212> DNA
<213> Artificial Sequence
<220>
<223> synthetic oligonucleotide primer
<400> 23
Page 47
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
acgcatatgg agcgagctga gagctcgagt 30
<210> 24
<211> 75
<212> DNA
<213> Artificial Sequence
<220>
<223> synthetic oligonucleotide primer
<400> 24
gaattcttat cacgtagaat cgagaccgag gagagggtta gggataggct taccagttcc 60
aaccttcgcc gatgc 75
<210> 25
<211> 27
<212> DNA
<213> Artificial Sequence
<220>
<223> synthetic oligonucleotide primer
<400> 25
acgcatatgt gtccttggcg gcctaga 27
<210> 26
<211> 75
<212> DNA
<213> Artificial Sequence
<220>
<223> synthetic oligonucleotide primer
<400> 26
gaattcttat tacgtagaat cgagaccgag gagagggtta gggataggct tacccatgtg 60
tgatgtttca gagct 75
<210> 27
<2l1> 49
<212> DNA
<213> Artificial Sequence
<220>
<223> synthetic oligonucleotide primer
<220>
<221> misc_feature
<222> (35) . (35)
<223> corresponds to the first nucleotide of the first codon of the tar
get mismatch repair gene
<220>
<221> misc_feature
<222> (36) . (36) .
<223> corresponds to the second nucleotide of the first codon of the to
rget mismatch repair gene
Page 48
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
<220>
<221> misC_feature
<222> (37) . (37)
<223> corresponds to the third nucleotide of the first codon of the tar
get mismatch repair gene
<220>
<221> misc_feature
<222> (38) . (38)
<223> corresponds to the first nucleotide of the second codon of the to
rget mismatch repair gene
<220>
<221> misc_feature
<222> (39) . (39)
<223> corresponds to the second nucleotide of the second codon of the t
arget mismatch repair gene
<220>
<221> misC_feature
<222> (40) . (40)
<223> corresponds to the third nucleotide of the second codon of the to
rget mismatch repair gene
<220>
<221> misc_feature
<222> (41) . (41)
<223> corresponds to the first nucleotide of the third codon of the tar
get mismatch repair gene
<220>
<221> misc_feature
<222> (42) . (42)
<223> corresponds to the second nucleotide of the third codon of the to
rget mismatch repair gene
<220>
<221> misc_feature
<222> (43) . (43)
<223> corresponds to the third nucleotide of the third codon of the tar
get mismatch repair gene
<220>
<221> misc_feature
<222> (44) . (44)
<223> corresponds to the first nucleotide of the fourth codon of the to
rget mismatch repair gene
<220>
<221> misc_feature
<222> (45) . (45)
<223> corresponds to the second nucleotide of the fourth codon of the t
arget mismatch repair gene
Page 49
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
<220>
<221> misC_feature
<222> (46) . (46)
<223> corresponds to the third nucleotide of the fourth codon of the to
rget mismatch repair gene
<220>
<221> misc_feature
<222> (47) . (47)
<223> corresponds to the first nucleotide of the fifth codon of the tar
get mismatch repair gene
<220>
<221> misc_feature
<222> (48) . (48)
<223> corresponds to the second nucleotide of the fifth codon of the to
rget mismatch repair gene
<220>
<221> misc_feature
<222> (49) . (49)
<223> corresponds to the third nucleotide of the fifth codon of the tar
get mismatch repair gene
<400> 27
tttaatacga ctcactatag ggagaccacc atggnnnnnn nnnnnnnnn 49
<210>
28
<211>
4290
<212>
DNA
<213> Sapiens
Homo
<400>
28
atgatcaagtgcttgtcagttgaagtacaagccaaattgcgttctggtttggccataagc60
tccttgggccaatgtgttgaggaacttgccctcaacagtattgatgctgaagcaaaatgt120
gtggctgtcagggtgaatatggaaaccttccaagttcaagtgatagacaatggatttggg180
atggggagtgatgatgtagagaaagtgggaaatcgttatttcaccagtaaatgccactcg240
gtacaggacttggagaatccaaggttttatggtttccgaggagaggccttggcaaatatt300
gctgacatggccagtgctgtggaaatttcgtccaagaaaaacaggacaatgaaaactttt360
gtgaaactgtttcagagtggaaaagccctgaaagcttgtgaagctgatgtgactagagca420
agcgctgggactactgtaacagtgtataacctattttaccagcttcctgtaaggaggaaa480
tgcatggaccctagactggagtttgagaaggttaggcagagaatagaagctCtCtCaCtC540
atgcacccttccatttctttctctttgagaaatgatgtttctggttccatggttcttcag600
ctccctaaaaccaaagacgtatgttcccgattttgtcaaatttatggattgggaaagtcc660
caaaagctaagagaaataagttttaaatataaagagtttgagcttagtggctatatcagc720
tctgaagcacattacaacaagaatatgcagtttttgtttgtgaacaaaagactagtttta780
Page 50
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
aggacaaagctacataaactcattgactttttattaaggaaagaaagtattatatgcaag 840
ccaaagaatggtcccaccagtaggcaaatgaattcaagtcttcggcaccggtctacccca 900
gaactctatggcatatatgtaattaatgtgcagtgccaattctgtgagtatgatgtgtgc 960
atggagccagccaaaactctgattgaatttcagaactgggacactctcttgttttgcatt 1020
caggaaggagtgaaaatgtttttaaagcaagaaaaattatttgtggaattatcaggtgag 1080
gatattaaggaatttagtgaagataatggttttagtttatttgatgctactcttcagaag 1140
cgtgtgacttccgatgagaggagcaatttccaggaagcatgtaataatattttagattcc 1200
tatgagatgtttaatttgcagtcaaaagctgtgaaaagaaaaactactgcagaaaacgta 1260
aacacacagagttctagggattcagaagctaccagaaaaaatacaaatgatgcatttttg 1320
tacatttatgaatcaggtggtccaggccatagcaaaatgacagagccatctttacaaaac 1380
aaagacagctcttgctcagaatcaaagatgttagaacaagagacaattgtagcatcagaa 1440
gctggagaaaatgagaaacataaaaaatctttcctggaacatagctctttagaaaatccg 1500
tgtggaaccagtttagaaatgtttttaagcccttttcagacaccatgtcactttgaggag 1560
agtgggcaggatctagaaatatggaaagaaagtactactgttaatggcatggctgccaac 1620
atcttgaaaaataatagaattcagaatcaaccaaagagatttaaagatgctactgaagtg 1680
ggatgccagcctctgccttttgcaacaacattatggggagtacatagtgctcagacagag 1740
aaagagaaaaaaaaagaatctagcaattgtggaagaagaaatgtttttagttatgggcga 1800
gttaaattatgttccactggctttataactcatgtagtacaaaatgaaaaaactaaatca 1860
actgaaacagaacattcatttaaaaattatgttagacctggtcccacacgtgcccaagaa 1920
acatttggaaatagaacacgtcattcagttgaaactccagacatcaaagatttagccagc 1980
actttaagtaaagaatctggtcaattgcccaacaaaaaaaattgcagaacgaatataagt 2040
tatgggctagagaatgaacctacagcaacttatacaatgttttctgcttttcaggaaggt 2100
agcaaaaaatcacaaacagattgcatattatctgatacatCCCCCtCtttcccctggtat 2160
agacacgtttccaatgatagtaggaaaacagataaattaattggtttctccaaaccaatc 2220
gtccgtaagaagctaagcttgagttcacagctaggatctttagagaagtttaagaggcaa 2280
tatgggaaggttgaaaatcctctggatacagaagtagaggaaagtaatggagtcactacc 2340
aatctcagtcttcaagttgaacctgacattctgctgaaggacaagaaccgcttagagaac 2400
tctgatgtttgtaaaatcactactatggagcatagtgattcagatagtagttgtcaacca 2460
gcaagccacatccttgactcagagaagtttccattctccaaggatgaagattgtttagaa 2520
caacagatgcttagtttgagagaaagtcctatgaccctgaaggagttatctctctttaat 2580
agaaaacctttggaccttgagaagtcatctgaatcactagcctctaaattatccagactg 2640
aagggttccgaaagagaaactcaaacaatggggatgatgagtcgttttaatgaacttcca 2700
Page 51
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
aattcagattccagtaggaaagacagcaagttgtgcagtgtgttaacacaagatttttgt 2760
atgttatttaacaacaagcatgaaaaaacagagaatggtgtcatcccaacatcagattct 2820
gccacacaggataattcctttaataaaaatagtaaaacacattctaacagcaatacaaca 2880
gagaactgtgtgatatcagaaactcctttggtattgccctataataattctaaagttacc 2940
ggtaaagattcagatgttcttatcagagcctcagaacaacagataggaagtcttgactct 3000
cccagtggaatgttaatgaatccggtagaagatgccacaggtgaccaaaatggaatttgt 3060
tttcagagtgaggaatctaaagcaagagcttgttctgaaactgaagagtcaaacacgtgt 3120
tgttcagattggcagcggcatttcgatgtagccctgggaagaatggtttatgtcaacaaa 3180
atgactggactcagcacattcattgccccaactgaggacattcaggctgcttgtactaaa 3240
gacctgacaactgtggctgtggatgttgtacttgagaatgggtctcagtacaggtgtcaa 3300
ccttttagaagcgaccttgttcttcctttccttccgagagctcgagcagagaggactgtg 3360
atgagacaggataacagagatactgtggatgatactgttagtagcgaatcgcttcagtct 3420
ttgttctcagaatgggacaatccagtatttgcccgttatccagaggttgctgttgatgta 3480
agcagtggccaggctgagagcttagcagttaaaattcacaacatcttgtatccctatcgt 3540
ttcaccaaaggaatgattcattcaatgcaggttctccagcaagtagataacaagtttatt 3600
gcctgtttgatgagcactaagactgaagagaatggcgaggcagattcctacgagaagcaa 3660
caggcacaaggctctggtcggaaaaaattactgtcttctactctaattcctccgctagag 3720
ataacagtgacagaggaacaaaggagactcttatggtgttaccacaaaaatctggaagat 3780
ctgggccttgaatttgtatttccagacactagtgattctctggtccttgtgggaaaagta 3840
ccactatgttttgtggaaagagaagccaatgaacttcggagaggaagatctactgtgacc 3900
aagagtattgtggaggaatttatccgagaacaactggagctactccagaccaccggaggc 3960
atccaagggacattgccactgactgtccagaaggtgttggcatcccaagcctgccatggg 4020
gccattaagtttaatgatggcctgagcttacaggaaagttgccgccttattgaagctctg 4080
tcctcatgccagctgccattccagtgtgctcacgggagaccttctatgctgccgttagct 4140
gacatagaccacttggaacaggaaaaacagattaaacccaacctcactaaacttcgcaaa 4200
atggcccaggcctggcgtctctttggaaaagcagagtgtgatacaaggcagagcctgcag 4260
caatccatgcctccctgtgagccaccatga 4290
<210>
29
<211>
1429
<212>
PRT
<213>
Homo
Sapiens
<400> 29
Met Ile Lys Cys Leu Ser Val Glu Val Gln Ala Lys Leu Arg Ser Gly
1 5 10 15
Page 52
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
Leu Ala Ile Ser Ser Leu Gly Gln Cys Val Glu Glu Leu Ala Leu Asn
20 25 30
Ser Ile Asp Ala Glu Ala Lys Cys Val Ala Val Arg Val Asn Met Glu
35 40 45
Thr Phe Gln Val Gln Val Ile Asp Asn Gly Phe Gly Met Gly Ser Asp
50 55 60
Asp Val Glu Lys Val Gly Asn Arg Tyr Phe Thr Ser Lys Cys His Ser
65 70 75 80
Val Gln Asp Leu Glu Asn Pro Arg Phe Tyr Gly Phe Arg Gly Glu Ala
85 90 95
Leu Ala Asn Ile Ala Asp Met Ala Ser Ala Val Glu Ile Ser Ser Lys
l00 105 110
Lys Asn Arg Thr Met Lys Thr Phe Val Lys Leu Phe Gln Ser Gly Lys
l15 120 125
Ala Leu Lys Ala Cys Glu Ala Asp Val Thr Arg Ala Ser Ala Gly Thr
130 135 140
Thr Val Thr Val Tyr Asn Leu Phe Tyr Gln Leu Pro Val Arg Arg Lys
145 150 155 160
Cys Met Asp Pro Arg Leu Glu Phe Glu Lys Val Arg Gln Arg Ile Glu
165 170 175
Ala Leu Ser Leu Met His Pro Ser Ile Ser Phe Ser Leu Arg Asn Asp
180 185 190
Val Ser Gly Ser Met Val Leu Gln Leu Pro Lys Thr Lys Asp Val Cys
195 200 205
Ser Arg Phe Cys Gln Ile Tyr Gly Leu Gly Lys Ser Gln Lys Leu Arg
210 215 220
Glu Ile Ser Phe Lys Tyr Lys Glu Phe Glu Leu Ser Gly Tyr Ile Ser
225 230 235 240
Ser Glu Ala His Tyr Asn Lys Asn Met Gln Phe Leu Phe Val Asn Lys
245 250 255
Arg Leu Val Leu Arg Thr Lys Leu His Lys Leu Ile Asp Phe Leu Leu
260 265 270
Page 53
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
Arg Lys Glu Ser Ile Ile Cys Lys Pro Lys Asn Gly Pro Thr Ser Arg
275 280 285
Gln Met Asn Ser Ser Leu Arg His Arg Ser Thr Pro Glu Leu Tyr Gly
290 295 300
Ile Tyr Val Ile Asn Val Gln Cys Gln Phe Cys Glu Tyr Asp Val Cys
305 310 315 320
Met Glu Pro Ala Lys Thr Leu Ile Glu Phe Gln Asn Trp Asp Thr Leu
325 330 335
Leu Phe Cys Ile Gln Glu Gly Val Lys Met Phe Leu Lys Gln Glu Lys
340 345 350
Leu Phe Val Glu Leu Ser Gly Glu Asp Ile Lys Glu Phe Ser Glu Asp
355 360 365
Asn Gly Phe Ser Leu Phe Asp Ala Thr Leu Gln Lys Arg Val Thr Ser
370 375 380
Asp Glu Arg Ser Asn Phe Gln Glu Ala Cys Asn Asn Ile Leu Asp Ser
385 390 395 400
Tyr Glu Met Phe Asn Leu Gln Ser Lys Ala Val Lys Arg Lys Thr Thr
405 410 415
Ala Glu~Asn Val Asn Thr Gln Ser Ser Arg Asp Ser Glu Ala Thr Arg
420 425 430
Lys Asn Thr Asn Asp Ala Phe Leu Tyr Ile Tyr Glu Ser Gly Gly Pro
435 440 445
Gly His Ser Lys Met Thr Glu Pro Ser Leu Gln Asn Lys Asp Ser Ser
450 455 460
Cys Ser Glu Ser Lys Met Leu Glu Gln Glu Thr Ile Val Ala Ser Glu
465 470 475 480
Ala Gly Glu Asn Glu Lys His Lys Lys Ser Phe Leu Glu His Ser Ser
485 490 495
Leu Glu Asn Pro Cys Gly Thr Ser Leu Glu Met Phe Leu Ser Pro Phe
500 505 510
Gln Thr Pro Cys His Phe Glu Glu Ser Gly Gln Asp Leu Glu Ile Trp
515 520 525
Page 54
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
Lys Glu Ser Thr Thr Val Asn Gly Met Ala Ala Asn Ile Leu Lys Asn
530 535 540
Asn Arg Ile Gln Asn Gln Pro Lys Arg Phe Lys Asp Ala Thr Glu Val
545 550 555 560
Gly Cys Gln Pro Leu Pro Phe Ala Thr Thr Leu Trp Gly Val His Ser
565 570 575
Ala Gln Thr Glu Lys Glu Lys Lys Lys Glu Ser Ser Asn Cys Gly Arg
580 585 590
Arg Asn Val Phe Ser Tyr Gly Arg Val Lys Leu Cys Ser Thr Gly Phe
595 600 605
Ile Thr His Val Val Gln Asn Glu Lys Thr Lys Ser Thr Glu Thr Glu
610 615 620
His Ser Phe Lys Asn Tyr Val Arg Pro Gly Pro Thr Arg Ala Gln Glu
625 630 635 640
Thr Phe Gly Asn Arg Thr Arg His Ser Val Glu Thr Pro Asp Ile Lys
645 650 655
Asp Leu Ala Ser Thr Leu Ser Lys Glu Ser Gly Gln Leu Pro Asn Lys
660 665 670
Lys Asn Cys Arg Thr Asn Ile Ser Tyr Gly Leu Glu Asn Glu Pro Thr
675 680 685
Ala Thr Tyr Thr Met Phe Ser Ala Phe Gln Glu Gly Ser Lys Lys Ser
690 695 700
Gln Thr Asp Cys Ile Leu Ser Asp Thr Ser Pro Ser Phe Pro Trp Tyr
705 710 715 720
Arg His Val Ser Asn Asp Ser Arg Lys Thr Asp Lys Leu Ile Gly Phe
725 730 735
Ser Lys Pro Ile Val Arg Lys Lys Leu Ser Leu Ser Ser Gln Leu Gly
740 745 750
Ser Leu Glu Lys Phe Lys Arg Gln Tyr Gly Lys Val Glu Asn Pro Leu
755 760 ' 765
Asp Thr Glu Val Glu Glu Ser Asn Gly Val Thr Thr Asn Leu Ser Leu
770 775 780
Page 55
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
Gln Val Glu Pro Asp Ile Leu Leu Lys Asp Lys Asn Arg Leu Glu Asn
785 790 795 800
Ser Asp Val Cys Lys Ile Thr Thr Met Glu His Ser Asp Ser Asp Ser
805 810 815
Ser Cys Gln Pro Ala Ser His Ile Leu Asp Ser Glu Lys Phe Pro Phe
820 825 830
Ser Lys Asp Glu Asp Cys Leu Glu Gln Gln Met Leu Ser Leu Arg Glu
835 840 845
Ser Pro Met Thr Leu Lys Glu Leu Ser Leu Phe Asn Arg Lys Pro Leu
850 855 860
Asp Leu Glu Lys Ser Ser Glu Ser Leu Ala Ser Lys Leu Ser Arg Leu
865 870 875 880
Lys Gly Ser Glu Arg Glu Thr Gln Thr Met Gly Met Met Ser Arg Phe
885 890 895
Asn Glu Leu Pro Asn Ser Asp Ser Ser Arg Lys Asp Ser Lys Leu Cys
900 905 910
Ser Val Leu Thr Gln Asp Phe Cys Met Leu Phe Asn Asn Lys His Glu
915 920 925
Lys Thr Glu Asn Gly Val Ile Pro Thr Ser Asp Ser Ala Thr Gln Asp
930 935 940
Asn Ser Phe Asn Lys Asn Ser Lys Thr His Ser Asn Ser Asn Thr Thr
945 950 955 960
Glu Asn Cys Val Ile Ser Glu Thr Pro Leu Val Leu Pro Tyr Asn Asn
965 970 975
Ser Lys Val Thr Gly Lys Asp Ser Asp Val Leu Ile Arg Ala Ser Glu
980 985 990
Gln Gln Ile Gly Ser Leu Asp Ser Pro Ser Gly Met Leu Met Asn Pro
995 1000 1005
Val Glu Asp Ala Thr Gly Asp Gln Asn Gly Ile Cys Phe Gln Ser
1010 1015 1020
Glu Glu Ser Lys Ala Arg Ala Cys Ser Glu Thr Glu Glu Ser Asn
1025 1030 1035
Page 56
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
Thr Cys Cys Ser Asp Trp Gln Arg His Phe Asp Val Ala Leu Gly
1040 1045 1050
Arg Met Val Tyr Val Asn Lys Met Thr Gly Leu Ser Thr Phe Ile
1055 1060 1065
Ala Pro Thr Glu Asp Ile Gln Ala Ala Cys Thr Lys Asp Leu Thr
1070 1075 1080
Thr Val Ala Val Asp Val Val Leu Glu Asn Gly Ser Gln Tyr Arg
1085 1090 1095
Cys Gln Pro Phe Arg Ser Asp Leu Val Leu Pro Phe Leu Pro Arg
1100 1105 1110
Ala Arg Ala Glu Arg Thr Val Met Arg Gln Asp Asn Arg Asp Thr
1115 1120 1125
Val Asp Asp Thr Val Ser Ser Glu Ser Leu Gln Ser Leu Phe Ser
1130 1135 1140
Glu Trp Asp Asn Pro Val Phe Ala Arg Tyr Pro Glu Val Ala Val
1145 1150 1155
Asp Val Ser Ser Gly Gln Ala Glu Ser Leu Ala Val Lys Ile His
1160 1165 1170
Asn Ile Leu Tyr Pro Tyr Arg Phe Thr Lys Gly Met Ile His Ser
1175 1180 1185
Met Gln Val Leu Gln Gln Val Asp Asn Lys Phe Ile Ala Cys Leu
1190 1195 1200
Met Ser Thr Lys Thr Glu Glu Asn Gly Glu Ala Asp Ser Tyr Glu
1205 1210 1215
Lys Gln Gln Ala Gln Gly Ser Gly Arg Lys Lys Leu Leu Ser Ser
1220 1225 1230
Thr Leu Ile Pro Pro Leu Glu Ile Thr Val Thr Glu Glu Gln Arg
1235 1240 1245
Arg Leu Leu Trp Cys Tyr His Lys Asn Leu Glu Asp Leu Gly Leu
1250 1255 1260
Glu Phe Val Phe Pro Asp Thr Ser Asp Ser Leu Val Leu Val Gly
1265 1270 1275
Page 57
Page 56
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
Lys Val Pro Leu Cys Phe Val Glu Arg Glu Ala Asn Glu Leu Arg
1280 1285 1290
Arg Gly Arg Ser Thr Val Thr Lys Ser Ile Val Glu Glu Phe Ile
1295 1300 1305
Arg Glu Gln Leu Glu Leu Leu Gln Thr Thr Gly Gly Ile Gln Gly
1310 1315 1320
Thr Leu Pro Leu Thr Val Gln Lys Val Leu Ala Ser Gln Ala Cys
1325 1330 1335
His Gly Ala Ile Lys Phe Asn Asp Gly Leu Ser Leu Gln Glu Ser
1340 1345 1350
Cys Arg Leu Ile Glu Ala Leu Ser Ser Cys Gln Leu Pro Phe Gln
1355 1360 1365
Cys Ala His Gly Arg Pro Ser Met Leu Pro Leu Ala Asp Ile Asp
1370 1375 1380
His Leu Glu Gln Glu Lys Gln Ile Lys Pro Asn Leu \Thr Lys Leu
1385 1390 1395
Arg Lys Met Ala Gln Ala Trp Arg Leu Phe Gly Lys Ala Glu Cys
1400 1405 1410
Asp Thr Arg Gln Ser Leu Gln Gln Ser Met Pro Pro Cys Glu Pro
1415 1420 1425
Pro
<210>
30
<211>
2340
<212>
DNA
<213> idopsis
Arab thaliana
<400>
30
atgcaaggagattcttctccgtctccgacgactactagctctcctttgataagacctata60
aacagaaacgtaattcacagaatctgttccggtcaagtcatcttagacctctcttcggcc120
gtcaaggagcttgtcgagaatagtctcgacgccggcgccaccagtatagagattaacctc180
cgagactacggcgaagactattttcaggtcattgacaatggttgtggcatttccccaacc240
aatttcaaggtttgtgtccaaattctccgaagaacttttgatgttcttgcacttaagcat300
catacttctaaattagaggatttcacagatcttttgaatttgactacttatggttttaga360
Page 58
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
ggagaagcct tgagctctct ctgtgcattg ggaaatctca ctgtggaaac aagaacaaag 420
aatgagccag ttgctacgct cttgacgttt gatcattctg gtttgcttac tgctgaaaag 480
aagactgctc gccaaattgg taccactgtc actgttagga agttgttctc taatttacct 540
gtacgaagca aagagtttaa gcggaatata cgcaaagaat atgggaagct tgtatcttta 600
ttgaacgcat atgcgcttat tgcgaaagga gtgcggtttg tctgctctaa cacgactggg 660
aaaaacccaa agtctgttgt gctgaacaca caagggaggg gttcacttaa agataatatc 720
ataacagttt tcggcattag tacctttaca agtctacagc ctggtactgg acgcaattta 780
gcagatcgac agtatttctt tataaatggt cggcctgtag atatgccaaa agtcagcaag 840
ttggtgaatg agttatataa agatacaagt tctcggaaat atccagttac cattctggat 900
tttattgtgc ctggtggagc atgtgatttg aatgtcacgc ccgataaaag aaaggtgttc 960
ttttctgacg agacttctgt tatcggttct ttgagggaag gtctgaacga gatatattcc 1020
tccagtaatg cgtcttatat tgttaatagg ttcgaggaga attcggagca accagataag 1080
gctggagttt cgtcgtttca gaagaaatca aatcttttgt cagaagggat agttctggat 1140
gtcagttcta aaacaagact aggggaagct attgagaaag aaaatccatc cttaagggag 1200
gttgaaattg ataatagttc gccaatggag aagtttaagt ttgagatcaa ggcatgtggg 1260
acgaagaaag gggaaggttc tttatcagtc catgatgtaa ctcaccttga caagacacct 1320
agcaaaggtt tgcctcagtt aaatgtgact gagaaagtta ctgatgcaag taaagacttg 1380
agcagccgct ctagctttgc ccagtcaact ttgaatactt ttgttaccat gggaaaaaga 1440
aaacatgaaa acataagcac catcctctct gaaacacctg tcctcagaaa ccaaacttct 1500
agttatcgtg tggagaaaag caaatttgaa gttcgtgcct tagcttcaag gtgtctcgtg 1560
gaaggcgatc aacttgatga tatggtcatc tcaaaggaag atatgacacc aagcgaaaga 1620
gattctgaac taggcaatcg gatttctcct ggaacacaag ctgataatgt tgaaagacat 1680
gagagagtac tcgggcaatt caatcttggg ttcatcattg caaaattgga gcgagatctg 1740
ttcattgtgg atcagcatgc agctgatgag aaattcaact tcgaacattt agcaaggtca 1800
actgtcctga accagcaacc cttactccag cctttgaact tggaactctc tccagaagaa 1860
gaagtaactg tgttaatgca catggatatt atcagggaaa atggctttct tctagaggag 1920
aatccaagtg CtCCtCCCgg aaaacacttt agactacgag ccattcctta tagcaagaat 1980
atcacctttg gagtcgaaga tcttaaagac ctgatctcaa ctctaggaga taaccatggg 2040
gaatgttcgg ttgctagtag ctacaaaacc agcaaaacag attcgatttg tccatcacga 2100
gtccgtgcaa tgctagcatc ccgagcatgc agatcatctg tgatgatcgg agatccactc 2160
agaaaaaacg aaatgcagaa gatagtagaa cacttggcag atctcgaatc tccttggaat 2220
tgcccacacg gacgaccaac aatgcgtcat cttgtggact tgacaacttt actcacatta 2280
Page 59
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
cctgatgacg acaatgtcaa tgatgatgat gatgatgatg caaccatctc attggcatga 2340
<210> 31
<211> 779
<212> PRT
<213> Arabidopsis thaliana
<400> 31
Met Gln Gly Asp Ser Ser Pro Ser Pro Thr Thr Thr Ser Ser Pro Leu
1 5 10 15
Ile Arg Pro Ile Asn Arg Asn Val Ile His Arg Ile Cys Ser Gly Gln
20 25 30
Val Ile Leu Asp Leu Ser Ser Ala Val Lys Glu Leu Val Glu Asn Ser
35 40 45
Leu Asp Ala Gly Ala Thr Ser Ile Glu Ile Asn Leu Arg Asp Tyr Gly
50 55 60
Glu Asp Tyr Phe Gln Val Ile Asp Asn Gly Cys Gly Ile Ser Pro Thr
65 70 75 80
Asn Phe Lys Val Cys Val Gln Ile Leu Arg Arg Thr Phe Asp Val Leu
85 90 95
Ala Leu Lys His His Thr Ser Lys Leu Glu Asp Phe Thr Asp Leu Leu
100 105 110
Asn Leu Thr Thr Tyr Gly Phe Arg Gly Glu Ala Leu Ser Ser Leu Cys
115 120 125
Ala Leu Gly Asn Leu Thr Val Glu Thr Arg Thr Lys Asn Glu Pro Val
130 135 140
.J
Ala Thr Leu Leu Thr Phe Asp His Ser Gly Leu Leu Thr Ala Glu Lys
145 150 155 160
Lys Thr Ala Arg Gln Ile Gly Thr Thr Val Thr Val Arg Lys Leu Phe
165 170 175
Ser Asn Leu Pro Val Arg Ser Lys Glu Phe Lys Arg Asn Ile Arg Lys
180 185 190
Glu Tyr Gly Lys Leu Val Ser Leu Leu Asn Ala Tyr Ala Leu Ile Ala
195 200 205
Lys Gly Val Arg Phe Val Cys Ser Asn Thr Thr Gly Lys Asn Pro Lys
210 215 220
Page 60
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
Ser Val Val Leu Asn Thr Gln Gly Arg Gly Ser Leu Lys Asp Asn Ile
225 230 235 240
Ile Thr Val Phe Gly Ile Ser Thr Phe Thr Ser Leu Gln Pro Gly Thr
245 250 255
Gly Arg Asn Leu Ala Asp Arg Gln Tyr Phe Phe Ile Asn Gly Arg Pro
260 265 270
Val Asp Met Pro Lys Val Ser Lys Leu Val Asn Glu Leu Tyr Lys Asp
275 280 285
Thr Ser Ser Arg Lys Tyr Pro Val Thr Ile Leu Asp Phe Ile Val Pro
290 295 300
Gly Gly Ala Cys Asp Leu Asn Val Thr Pro Asp Lys Arg Lys Val Phe
305 310 315 320
Phe.Ser Asp Glu Thr Ser Val Ile Gly Ser Leu Arg Glu Gly Leu Asn
325 330 335
Glu Ile Tyr Ser Ser Ser Asn Ala Ser Tyr Ile Val Asn Arg Phe Glu
340 345 350
Glu Asn Ser Glu Gln Pro Asp Lys Ala Gly Val Ser Ser Phe Gln Lys
355 360 365
Lys Ser Asn Leu Leu Ser Glu Gly Ile Val Leu Asp Val Ser Ser Lys
370 375 380
Thr Arg Leu Gly Glu Ala Ile Glu Lys Glu Asn Pro Ser Leu Arg Glu
385 390 395 400
Val Glu Ile Asp Asn Ser Ser Pro Met Glu Lys Phe Lys Phe Glu Ile
405 410 415
Lys Ala Cys Gly Thr Lys Lys Gly Glu Gly Ser Leu Ser Val His Asp
420 425 430
Val Thr His Leu Asp Lys Thr Pro Ser Lys Gly Leu Pro Gln Leu Asn
435 440 445
Val Thr Glu Lys Val Thr Asp Ala Ser Lys Asp Leu Ser Ser Arg Ser
450 455 460
Ser Phe Ala Gln Ser Thr Leu Asn Thr Phe Val Thr Met Gly Lys Arg
465 470 475 480
Page 61
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
Lys His Glu Asn Ile Ser Thr Ile Leu Ser Glu Thr Pro Val Leu Arg
485 490 495
Asn Gln Thr Ser Ser Tyr Arg Val Glu Lys Ser Lys Phe Glu Val Arg
500 505 510
Ala Leu Ala Ser Arg Cys Leu Val Glu Gly Asp Gln Leu Asp Asp Met
515 520 525
Val Ile Ser Lys Glu Asp Met Thr Pro Ser Glu Arg Asp Ser Glu Leu
530 535 540
Gly Asn Arg Ile Ser Pro Gly Thr Gln Ala Asp Asn Val Glu Arg His
545 550 555 560
Glu Arg Val Leu Gly Gln Phe Asn Leu Gly Phe Ile Ile Ala Lys Leu
565 570 575
Glu Arg Asp Leu Phe Ile Val Asp Gln His Ala Ala Asp Glu Lys Phe
580 585 590
Asn Phe Glu His Leu Ala Arg Ser Thr Val Leu Asn Gln Gln Pro Leu
595 600 605
Leu Gln Pro Leu Asn Leu Glu Leu Ser Pro Glu Glu Glu Val Thr Val
610 615 620
Leu Met His Met Asp Ile Ile Arg Glu Asn Gly Phe Leu Leu Glu Glu
625 630 635 640
Asn Pro Ser Ala Pro Pro Gly Lys His Phe Arg Leu Arg Ala Ile Pro
645 650 655
Tyr Ser Lys Asn Ile Thr Phe Gly Val Glu Asp Leu Lys Asp Leu Ile
660 665 670
Ser Thr Leu Gly Asp Asn His Gly Glu Cys Ser Val Ala Ser Ser Tyr
675 680 685
Lys Thr Ser Lys Thr Asp Ser Ile Cys Pro Ser Arg Val Arg Ala Met
690 695 700
Leu Ala Ser Arg Ala Cys Arg Ser Ser Val Met Ile Gly Asp Pro Leu
705 710 715 720
Arg Lys Asn Glu Met Gln Lys Ile Val Glu His Leu Ala Asp Leu Glu
725 730 735
Page 62
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
Ser Pro Trp Asn Cys Pro His Gly Arg Pro Thr Met Arg His Leu Val
740 745 750
Asp Leu Thr Thr Leu Leu Thr Leu Pro Asp Asp Asp Asn Val Asn Asp
755 760 765
Asp Asp Asp Asp Asp Ala Thr Ile Ser Leu Ala
770 775
<210>
32
<211>
3456
<212>
DNA
<213>
Arabidopsis
thaliana
<400>
32
atgaagacgatcaagcccttgccggaaggagttcgtcactccatgcgttctggaattatc60
atgttcgacatggcgagggtcgtggaagaactcgtcttcaacagtctcgatgctggggcg120
accaaggtgtctatcttcgtgggtgttgtttcatgctctgtgaaagttgtggatgatgga180
tcaggcgtttcaagagatgatttggttttgttgggagaaagatatgctacttcaaagttt240
cacgacttcaccaacgtggagacagctagtgaaacttttggatttcgtggagaggcctta300
gcttcaatatcagatatctcgttactggaggttaggacaaaagctattgggaggcctaat360
ggttatcgaaaggttatgaagggatccaagtgtctacatcttggaattgatgatgataga420
aaagactctggcacgacggtaactgtccgagatctattttacagtcagccagtgagacga480
aaatatatgcaatccagccccaagaaagttttggaatctatcaaaaagtgtgtgttccgg540
attgcccttgtgcactccaatgtttccttcagtgttcttgatatcgaaagtgatgaagag600
cttttccaaaccaatccttcttcttcagcattctcactactgatgagagatgcagggacc660
gaagctgtaaattcgctttgtaaagtaaacgttacagatggcatgctgaatgtctctggt720
tttgagtgtgcggatgactggaagcctacggatgggcaacaaacaggaagacgcaataga780
cttcaatccaaccctggttacattctgtgcatagcatgtccacgccgtctttatgaattc840
tcgtttgaaccatcaaagacgcacgttgagttcaagaagtggggacctgtacttgccttt900
atagaaagaatcactctagccaactggaagaaagatagaattcttgaactttttgatggg960
ggagctgatatactggcaaaaggtgatagacaagacctgattgatgacaaaattagactt1020
caaaacggcagccttttctcaattcttcattttctggatgcagattggccagaagctatg1080
gaacctgcaaaaaagaagctgaagagaagtaatgatcatgcaccttgtagttctctcttg1140
tttccgtctgctgactttaaacaagatggtgattatttttctccacgaaaggatgtatgg1200
tctccagaatgtgaagtcgaactgaaaattcagaatcccaaagagcaaggtactgtagct1260
ggatttgaaagccggactgattctcttctacagtcacgtgacatagaaatgcaaacgaat1320
Page 63
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
gaagacttcccacaagttactgacctccttgaaacaagcttggttgctgactctaagtgc 1380
cgtaaacagtttctaacaagatgtcagattaccacacctgtcaatatcaaccatgatttt 1440
atgaaagattcagacgtgttaaattttcagtttcaaggattgaaagatgagttggatgtc 1500
agcaattgcattggaaagcatctcttgcgtggttgctcttcaagagtaagcctaaccttt 1560
catgagcctaaactatctcatgttgaagggtatgaatccgtcgtgcctatgatacctaat 1620
gaaaaacaaagtagtccgcgggtcctagagaccagagaaggtggttcgtactgtgatgtt 1680
tattctgataagactcctgattgttccctagggagttcatggcaggatactgattggttt 1740
actccacagtgttcctcagataggggatgtgttggaattggagaagattttaacattacc 1800
cccatagatactgcggaatttgattcttatgatgaaaaagttggtagtaaaaagtatctt 1860
tcttctgtcaatgtggggagctctgttactggtagtttctgtttaagttctgagtggtct 1920
ccaatgtactccacaccttctgcgaccaagtgggagtctgagtaccagaaaggttgtcga 1980
attcttgaacagagtttgagactgggaaggatgcctgaccctgaattttgtttcagtgca 2040
gctaacaacatcaaatttgaccacgaggtcatacctgaaatggattgctgtgaaaccggt 2100
acagactctttcacagctattcagaactgcactcagttagctgataaaatttgcaagtct 2160
tcgtgggggcatgcagatgatgtgcgtattgaccaatatagtatcaggaaggaaaagttc 2220
agttatatggatggcacacagaacaatgctggtaaacaaaggtcaaaaagaagtcgatct 2280
gctcctccattttatcgagagaagaagagatttatcagcttaagttgtaaatcagacaca 2340
aaaccaaagaactctgatccatcagaacctgatgatctggagtgtttgacacaaccttgt 2400
aatgcatctcaaatgcatcttaagtgcagcatccttgatgatg~.gtcgtatgaccacata 2460
caagaaacagaaaaaagattgagttctgcctcagacttgaaagcatctgctggttgcagg 2520
actgtgcactcagagacccaagatgaggatgtgcacgaagacttcagctcagaggaattt 2580
ctggatccaattaaatccacaacaaaatggcgccataactgtgcggtctctcaggttccc 2640
aaggaatcacacgagcttcatggtcaagatggtgtatttgatatatcttcgggacttctg 2700
cacttacgatccgatgaatccttggttcctgaatctatcaacagacactcccttgaagat 2760
gccaaggttctacaacaggttgataaaaaatatatcccaatcgttgcttgtggaacagtt 2820
gccatcgttgatcagcatgctgccgatgaaagaattcgtttggaagagctgcgtacaaag 2880
tttattaatgatgcattattaatttttgtgttgacattaaaggtactgccggagatgggt 2940
tatcagttactccagagttattcagagcagataagagactggggttggatctgcaacatt 3000
actgtagaagggtcaacgtcctttaagaaaaacatgagcatcatccagcggaaaccaaca 3060
ccaatcacacttaatgcggttccatgcattctgggtgtaaatctatcagatgttgatcta 3120
ttagagtttcttcagcagcttgctgatactgacggatcatcaactattcctccatctgtt 3180
cttcgagtcctaaattccaaagcctgtagaggtgcaattatgtttggagatagtctgtta 3240
Page 64
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
ccgtcagaat gctctttaat cattgatgga ctgaagcaga cctcactttg tttccagtgt 3300
gctcatgggc gacctacaac agttcctctt gtcgatttga aggcattgca caaacagata 3360
gcaaagctca gtggaagaca agtgtggcat ggcttacaac gcagagaaat tacacttgat 3420
cgtgcaaaat cacgcttaga caacgctaaa agttaa 3456
<210> 33
<211> 1151
<212> PRT
<213> Arabidopsis thaliana
<400> 33
Met Lys Thr Ile Lys Pro Leu Pro Glu Gly Val Arg His Ser Met Arg
1 5 10 15
Ser Gly Ile Ile Met Phe Asp Met Ala Arg Val Val Glu Glu Leu Val
20 25 30
Phe Asn Ser Leu Asp Ala Gly Ala Thr Lys Val Ser Ile Phe Val Gly
35 40 45
Val Val Ser Cys Ser Val Lys Val Val Asp Asp Gly Ser Gly Val Ser
50 55 60
Arg Asp Asp Leu Val Leu Leu Gly Glu Arg Tyr Ala Thr Ser Lys Phe
65 70 ' 75 80
His Asp Phe Thr Asn Val Glu Thr Ala Ser Glu Thr Phe Gly Phe Arg
85 90 95
Gly Glu Ala Leu Ala Ser Ile Ser Asp Ile Ser Leu Leu Glu Val Arg
100 105 110
Thr Lys Ala Ile Gly Arg Pro Asn Gly Tyr Arg Lys Val Met Lys Gly
115 120 125
Ser Lys Cys Leu His Leu Gly Ile Asp Asp Asp Arg Lys Asp Ser Gly
130 135 140
Thr Thr Val Thr Val Arg Asp Leu Phe Tyr Ser Gln Pro Val Arg Arg
145 150 155 160
Lys Tyr Met Gln Ser Ser Pro Lys Lys Val Leu Glu Ser Ile Lys Lys
165 170 175
Cys Val Phe Arg Ile Ala Leu Val His Ser Asn Val Ser Phe Ser Val
180 185 190
Page 65
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
Leu Asp Ile Glu Ser Asp Glu Glu Leu Phe Gln Thr Asn Pro Ser Ser
195 200 205
Ser Ala Phe Ser Leu Leu Met Arg Asp Ala Gly Thr Glu Ala Val Asn
210 215 220
Ser Leu Cys Lys Val Asn Val Thr Asp Gly Met Leu Asn Val Ser Gly
225 230 235 240
Phe Glu Cys Ala Asp Asp Trp Lys Pro Thr Asp Gly Gln Gln Thr Gly
245 250 255
Arg Arg Asn Arg Leu Gln Ser Asn Pro Gly Tyr Ile Leu Cys Ile Ala
260 265 270
Cys Pro Arg Arg Leu Tyr Glu Phe Ser Phe Glu Pro Ser Lys Thr His
275 280 285
Val Glu Phe Lys Lys Trp Gly Pro Val Leu Ala Phe Ile Glu Arg Ile
290 295 300
Thr Leu Ala Asn Trp Lys Lys Asp Arg Ile Leu Glu Leu Phe Asp Gly
305 310 315 320
Gly Ala Asp Ile Leu Ala Lys Gly Asp Arg Gln Asp Leu Ile Asp Asp
325 330 335
Lys Ile Arg Leu Gln Asn Gly Ser Leu Phe Ser Ile Leu His Phe Leu
340 345 350
Asp Ala Asp Trp Pro Glu Ala Met Glu Pro Ala Lys Lys Lys Leu Lys
355 360 365
Arg Ser Asn Asp His Ala Pro Cys Ser Ser Leu Leu Phe Pro Ser Ala
370 375 380
Asp Phe Lys Gln Asp Gly Asp Tyr Phe Ser Pro Arg Lys Asp Val Trp
385 390 395 400
Ser Pro Glu Cys Glu Val Glu Leu Lys Ile Gln Asn Pro Lys Glu Gln
405 410 415
Gly Thr Val Ala Gly Phe Glu Ser Arg Thr Asp Ser Leu Leu Gln Ser
420 425 430
Arg Asp Ile Glu Met Gln Thr Asn Glu Asp Phe Pro Gln Val Thr Asp
435 440 445
Page 66
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
Leu Leu Glu Thr Ser Leu Val Ala Asp Ser Lys Cys Arg Lys Gln Phe
450 455 460
Leu Thr Arg Cys Gln Ile Thr Thr Pro Val Asn Ile Asn His Asp Phe
465 470 475 480
Met Lys Asp Ser Asp Val Leu Asn Phe Gln Phe Gln Gly Leu Lys Asp
485 490 495
Glu Leu Asp Val Ser Asn Cys Ile Gly Lys His Leu Leu Arg Gly Cys
500 505 510
Ser Ser Arg Val Ser Leu Thr Phe His Glu Pro Lys Leu Ser His Val
515 520 525
Glu Gly Tyr Glu Ser Val Val Pro Met Ile Pro Asn Glu Lys Gln Ser
530 535 540
Ser Pro Arg Val Leu Glu Thr Arg Glu Gly Gly Ser Tyr Cys Asp Val
545 550 555 560
Tyr Ser Asp Lys Thr Pro Asp Cys Ser Leu Gly Ser Ser Trp Gln Asp
565 570 575
Thr Asp Trp Phe Thr Pro Gln Cys Ser Ser Asp Arg Gly Cys Val Gly
580 585 590
Ile Gly~Glu Asp Phe Asn Ile Thr Pro Ile Asp Thr Ala Glu Phe Asp
595 600 605
Ser Tyr Asp Glu Lys Val Gly Ser Lys Lys Tyr Leu Ser Ser Val Asn
610 615 620
Val Gly Ser Ser Val Thr Gly Ser Phe Cys Leu Ser Ser Glu Trp Ser
625 630 635 640
Pro Met Tyr Ser Thr Pro Ser Ala Thr Lys Trp Glu Ser Glu Tyr Gln
645 650 655
Lys Gly Cys Arg Ile Leu Glu Gln Ser Leu Arg Leu Gly Arg Met Pro
660 665 670
Asp Pro Glu Phe Cys Phe Ser Ala Ala Asn Asn Ile Lys Phe Asp His
675 680 685
Glu Val Ile Pro Glu Met Asp Cys Cys Glu Thr Gly Thr Asp Ser Phe
690 695 700
Page 67
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
Thr Ala Ile Gln Asn Cys Thr Gln Leu Ala Asp Lys Ile Cys Lys Ser
705 710 715 720
Ser Trp Gly His Ala Asp Asp Val Arg Ile Asp Gln Tyr Ser Ile Arg
725 730 735
Lys Glu Lys Phe Ser Tyr Met Asp Gly Thr Gln Asn Asn Ala Gly Lys
740 745 750
Gln Arg Ser Lys Arg Ser Arg Ser Ala Pro Pro Phe Tyr Arg Glu Lys
755 760 765
Lys Arg Phe Ile Ser Leu Ser Cys Lys Ser Asp Thr Lys Pro Lys Asn
770 775 780
Ser Asp Pro Ser Glu Pro Asp Asp Leu Glu Cys Leu Thr Gln Pro Cys
785 790 795 800
Asn Ala Ser Gln Met His Leu Lys Cys Ser Ile Leu Asp Asp Val Ser
805 810 815
Tyr Asp His Ile Gln Glu Thr Glu Lys Arg Leu Ser Ser Ala Ser Asp
820 825 830
Leu Lys Ala Ser Ala Gly Cys Arg Thr Val His Ser Glu Thr Gln Asp
835 840 845
Glu Asp Val His Glu Asp Phe Ser Ser Glu Glu Phe Leu Asp Pro Ile
850 855 860
Lys Ser Thr Thr Lys Trp Arg His Asn Cys Ala Val Ser Gln Val Pro
865 870 875 880
Lys Glu Ser His Glu Leu His Gly Gln Asp Gly Val Phe Asp Ile Ser
885 890 895
Ser Gly Leu Leu His Leu Arg Ser Asp Glu Ser Leu Val Pro Glu Ser
900 905 910
Ile Asn Arg His Ser Leu Glu Asp Ala Lys Val Leu Gln Gln Val Asp
915 920 925
Lys Lys Tyr Ile Pro Ile Val Ala Cys Gly Thr Val Ala Ile Val Asp
930 935 940
Gln His Ala Ala Asp Glu Arg Ile Arg Leu Glu Glu Leu Arg Thr Lys
945 950 955 960
Page 68
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
Phe Ile Asn Asp Ala Leu Leu Ile Phe Val Leu Thr Leu Lys Val Leu
965 970 975
Pro Glu Met Gly Tyr Gln Leu Leu Gln Ser Tyr Ser Glu Gln Ile Arg
980 985 990
Asp Trp Gly Trp Ile Cys Asn Ile Thr Val Glu Gly Ser Thr Ser Phe
995 1000 1005
Lys Lys Asn Met Ser Ile Ile Gln Arg Lys Pro Thr Pro Ile Thr
1010 1015 1020
Leu Asn Ala Val Pro Cys Ile Leu Gly Val Asn Leu Ser Asp Val
1025 1030 1035
Asp Leu Leu Glu Phe Leu Gln Gln Leu Ala Asp Thr Asp Gly Ser
1040 1045 1050
Ser Thr Ile Pro Pro Ser Val Leu Arg Val Leu Asn Ser Lys Ala
1055 1060 1065
Cys Arg Gly Ala Ile Met Phe Gly Asp Ser Leu Leu Pro Ser Glu
1070 1075 1080
Cys Ser Leu Ile Ile Asp Gly Leu Lys Gln Thr Ser Leu Cys Phe
1085 1090 1095
Gln Cys Ala His Gly Arg Pro Thr Thr Val Pro Leu Val Asp Leu
1100 1105 1110
Lys Ala Leu His Lys Gln Ile Ala Lys Leu Ser Gly Arg Gln Val
1115 1120 1125
Trp His Gly Leu Gln Arg Arg Glu Ile Thr Leu Asp Arg Ala Lys
1130 1135 1140
Ser Arg Leu Asp Asn Ala Lys Ser
1145 1150
<210> 34
<211> 3330
<2l2> DNA
<213> Arabidopsis thaliana
<400> 34
atgcagcgcc agagatcgat tttgtctttc ttccaaaaac ccacggcggc gactacgaag 60
ggtttggttt ccggcgatgc tgctagcggc gggggcggca gcggaggacc acgatttaat 120
gtgaaggaag gggatgctaa aggcgacgct tctgtacgtt ttgctgtttc gaaatctgtc 180
Page 69
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
gatgaggttagaggaacggatactecaccggagaaggttccgcgtcgtgtcctgccgtct240
ggatttaagccggctgaatccgccggtgatgcttcgtccctgttctccaatattatgcat300
aagtttgtaaaagtcgatgatcgagattgttetggagagaggagccgagaagatgttgtt360
ccgctgaatgattcatctctatgtatgaaggctaatgatgttattcctcaatttcgttcc420
aataatggtaaaactcaagaaagaaaccatgcttttagtttcagtgggagagctgaactt480
agatcagtagaagatataggagtagatggcgatgttcctggtccagaaacaccagggatg540
cgtccacgtgcttctcgcttgaagcgagttctggaggatgaaatgacttttaaggaggat600
aaggttcctgtattggactctaacaaaaggctgaaaatgctccaggatecggtttgtgga660
gagaagaaagaagtaaacgaaggaaccaaatttgaatggcttgagtettctcgaatcagg720
gatgccaatagaagacgtcctgatgatcccctttacgatagaaagaccttacacatacca780
ectgatgttttcaagaaaatgtctgcatcacaaaagcaatattggagtgttaagagtgaa840
tatatggacattgtgcttttctttaaagtggggaaattttatgagctgtatgagctagat900
gcggaattaggtcacaaggagcttgactggaagatgaccatgagtggtgtgggaaaatgc960
agacaggttggtatetctgaaagtgggatagatgaggcagtgcaaaagctattagctegt1020
ggatataaagttggacgaatcgagcagctagaaacatctgaccaagcaaaagccagaggt1080
gctaatactataattccaaggaagctagttcaggtattaactccatcaacagcaagcgag1140
ggaaacategggcctgatgccgtccatcttcttgctataaaagagatcaaaatggagcta1200
caaaagtgttcaactgtgtatggatttgcttttgttgactgtgctgccttgaggttttgg1260
gttgggtccatcagcgatgatgcatcatgtgctgctcttggagcgttattgatgcaggtt1320
tctccaaaggaagtgttatatgacagtaaagggctatcaagagaagcacaaaaggctcta1380
aggaaatatacgttgacagggtctacggcggtacagttggctccagtaccacaagtaatg1440
ggggatacagatgctgctggagttagaaatataatagaatctaacggatactttaaaggt1500
tcttctgaatcatggaactgtgctgttgatggtctaaatgaatgtgatgttgcecttagt1560
gctcttggagagctaattaatcatctgtetaggctaaagctagaagatgtacttaagcat1620
ggggatatttttccataccaagtttacaggggttgtctcagaattgatggccagacgatg1680
gtaaatcttgagatatttaacaatagctgtgatggtggtccttcagggaccttgtacaaa1740
tatcttgataactgtgttagtccaactggtaagcgactcttaaggaattggatctgccat1800
ccactcaaagatgtagaaagcatcaataaacggcttgatgtagttgaagaattcacggca1860
aactcagaaagtatgcaaatcactggccagtatctccacaaacttccagacttagaaaga1920
ctgctcggacgcatcaagtctagcgttcgatcatcagcctctgtgttgcctgetcttctg1980
gggaaaaaagtgctgaaacaacgagttaaagcatttgggcaaattgtgaaagggttcaga2040
agtggaattgatctgttgttggctctacagaaggaatcaaatatgatgagtttgetttat2100
Page 70
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
aaactctgtaaacttcctatattagtaggaaaaagcgggctagagttatttctttctcaa 2160
ttcgaagcagccatagatagcgactttcca,aattatcagaaccaagatgtgacagatgaa 2220
aacgctgaaactctcacaatacttatcgaactttttatcgaaagagcaactcaatggtct 2280
gaggtcattcacaccataagctgcctagatgtcctgagatcttttgcaatcgcagcaagt 2340
ctctctgctggaagcatggccaggcctgttatttttcccgaatcagaagctacagatcag 2400
aatcagaaaacaaaagggccaatacttaaaatccaaggactatggcatccatttgcagtt 2460
gcagccgatggtcaattgcctgttccgaatgatatactccttggcgaggctagaagaagc 2520
agtggcagcattcatcctcggtcattgttactgacgggaccaaacatgggcggaaaatca 2580
actcttcttcgtgcaacatgtctggccgttatctttgcccaacttggctgctacgtgccg 2640
tgtgagtcttgcgaaatctccctcgtggatactatcttcacaaggcttggcgcatctgat 2700
agaatcatgacaggagagagtacctttttggtagaatgcactgagacagcgtcagttctt 2760
cagaatgcaactcaggattcactagtaatccttgacgaactgggcagaggaactagtact 2820
ttcgatggatacgccattgcatactcggtttttcgtcacctggtagagaaagttcaatgt 2880
cggatgctctttgcaacacattaccaccctctcaccaaggaattcgcgtctcacccacgt 2940
gtcacctcgaaacacatggcttgcgcattcaaatcaagatctgattatcaaccacgtggt 3000
tgtgatcaagacctagtgttcttgtaccgtttaaccgagggagcttgtcctgagagctac 3060
ggacttcaagtggcactcatggctggaataccaaaccaagtggttgaaacagcatcaggt 3120
gctgctcaagccatgaagagatcaattggggaaaacttcaagtcaagtgagctaagatct 3180
gagttctcaagtctgcatgaagactggctcaagtcattggtgggtatttctcgagtcgcc 3240
cacaacaatgcccccattggcgaagatgactacgacactttgttttgcttatggcatgag 3300
atcaaatcctcttactgtgttcccaaataa 3330
<210> 35
<211> 1109
<212> PRT
<213> Arabidopsis thaliana
<400> 35
Met Gln Arg Gln Arg Ser Ile Leu Ser~Phe Phe Gln Lys Pro Thr Ala
1 5 10 15
Ala Thr Thr Lys Gly Leu Val Ser Gly Asp Ala Ala Ser Gly Gly Gly
20 25 30
Gly Ser Gly Gly Pro Arg Phe Asn Val Lys Glu Gly Asp Ala Lys Gly
35 40 45
Asp Ala Ser Val Arg Phe Ala Val Ser Lys Ser Val Asp Glu Val Arg
50 55 60
Page 7l
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
Gly Thr Asp Thr Pro Pro Glu Lys Val Pro Arg Arg Val Leu Pro Ser
65 70 75 80
Gly Phe Lys Pro Ala Glu Ser Ala Gly Asp Ala Ser Ser Leu Phe Ser
85 90 95
Asn Ile Met His Lys Phe Val Lys Val Asp Asp Arg Asp Cys Ser Gly
100 105 110
Glu Arg Ser Arg Glu Asp Val Val Pro Leu Asn Asp Ser Ser Leu Cys
115 120 125
Met Lys Ala Asn Asp Val Ile Pro Gln Phe Arg Ser Asn Asn Gly Lys
130 135 140
Thr Gln Glu Arg Asn His Ala Phe Ser Phe Ser Gly Arg Ala Glu Leu
145 150 155 160
Arg Ser Val Glu Asp Ile Gly Val Asp Gly Asp Val Pro Gly Pro Glu
165 170 175
Thr Pro Gly Met Arg Pro Arg Ala Ser Arg Leu Lys Arg Val Leu Glu
180 185 190
Asp Glu Met Thr Phe Lys Glu Asp Lys Val Pro Val Leu Asp Ser Asn
195 200 205
Lys Arg Leu Lys Met Leu Gln Asp Pro Val Cys Gly Glu Lys Lys Glu
210 215 220
Val Asn Glu Gly Thr Lys Phe Glu Trp Leu Glu Ser Ser Arg Ile Arg
225 230 235 240
Asp Ala Asn Arg Arg Arg Pro Asp Asp Pro Leu Tyr Asp Arg Lys Thr
245 250 255
Leu His Ile Pro Pro Asp Val Phe Lys Lys Met Ser Ala Ser Gln Lys
260 265 270
Gln Tyr Trp Ser Val Lys Ser Glu Tyr Met Asp Ile Val Leu Phe Phe
275 280 285
Lys Val Gly Lys Phe Tyr Glu Leu Tyr Glu Leu Asp Ala Glu Leu Gly
290 295 300
His Lys Glu Leu Asp Trp Lys Met Thr Met Ser Gly Val Gly Lys Cys
305 310 315 320
Page 72
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
Arg Gln Val Gly Ile Ser Glu Ser Gly Ile Asp Glu Ala Val Gln Lys
325 330 335
Leu Leu Ala Arg Gly Tyr Lys Val Gly Arg Ile Glu Gln Leu Glu Thr
340 345 350
Ser Asp Gln Ala Lys Ala Arg Gly Ala Asn Thr Ile Ile Pro Arg Lys
355 360 365
Leu Val Gln Val Leu Thr Pro Ser Thr Ala Ser Glu Gly Asn Ile Gly
370 375 380
Pro Asp Ala Val His Leu Leu Ala Ile Lys Glu Ile Lys Met Glu Leu I' ;
385 390 395 400
Gln Lys Cys Ser Thr Val Tyr Gly Phe Ala Phe Val Asp Cys Ala Ala
405 410 415
Leu Arg Phe Trp Val Gly Ser Ile Ser Asp Asp Ala Ser Cys Ala Ala
420 425 430
Leu Gly Ala Leu Leu Met Gln Val Ser Pro Lys Glu Val Leu Tyr Asp
435 440 445
Ser Lys Gly Leu Ser Arg Glu Ala Gln Lys Ala Leu Arg Lys Tyr Thr
450 455 460
Leu Thr Gly Ser Thr Ala Val Gln Leu Ala Pro Val Pro Gln Val Met
465 470 475 480
Gly Asp Thr Asp Ala Ala Gly Val Arg Asn Ile Ile Glu Ser Asn Gly
485 490 495
Tyr Phe Lys Gly Ser Ser Glu Ser Trp Asn Cys Ala Val Asp Gly Leu
500 505 510
Asn Glu Cys Asp Val Ala Leu Ser Ala Leu Gly Glu Leu Ile Asri His
515 520 525
Leu Ser Arg Leu Lys Leu Glu Asp Val Leu Lys His Gly Asp Ile Phe
530 535 540
Pro Tyr Gln Val Tyr Arg Gly Cys Leu Arg Ile Asp Gly Gln Thr Met
545 550 555 560
Val Asn Leu Glu Ile Phe Asn Asn Ser Cys Asp Gly Gly Pro Ser Gly
565 570 575
Page 73
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
Thr Leu Tyr Lys Tyr Leu Asp Asn Cys Val Ser Pro Thr Gly Lys Arg
580 585 590
Leu Leu Arg Asn Trp Ile Cys His Pro Leu Lys Asp Val Glu Ser Ile
595 600 605
Asn Lys Arg Leu Asp Val Val Glu Glu Phe Thr Ala Asn Ser Glu Ser
610 615 620
Met Gln Ile Thr Gly Gln Tyr Leu His Lys Leu Pro Asp Leu Glu Arg
625 630 635 640
Leu Leu Gly Arg Ile Lys Ser Ser Val Arg Ser Ser Ala Ser Val Leu
645 650 655
Pro Ala Leu Leu Gly Lys Lys Val Leu Lys Gln Arg Val Lys Ala Phe
660 665 670
Gly Gln Ile Val Lys Gly Phe Arg Ser Gly Ile Asp Leu Leu Leu Ala
675 680 685
Leu Gln Lys Glu Ser Asn Met Met Ser Leu Leu Tyr Lys Leu Cys Lys
690 695 700
Leu Pro Ile Leu Val Gly Lys Ser Gly Leu Glu Leu Phe Leu Ser Gln
705 710 715 720
Phe Glu Ala Ala Ile Asp Ser Asp Phe Pro Asn Tyr Gln Asn Gln Asp
725 730 735
Val Thr Asp Glu Asn Ala Glu Thr Leu Thr Ile Leu Ile Glu Leu Phe
740 745 750
Ile Glu Arg Ala Thr Gln Trp Ser~Glu Val Ile His Thr Ile Ser Cys
755 760 765
Leu Asp Val Leu Arg Ser Phe Ala Ile Ala Ala Ser Leu Ser Ala Gly
770 775 780
Ser Met Ala Arg Pro Val Ile Phe Pro Glu Ser Glu Ala Thr Asp Gln
785 790 795 800
Asn Gln Lys Thr Lys Gly Pro Ile Leu Lys Ile Gln Gly Leu Trp His
805 810 815
Pro Phe Ala Val Ala Ala Asp Gly Gln Leu Pro Val Pro Asn Asp Ile
820 825 830
Page 74
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
Leu Leu Gly Glu Ala Arg Arg Ser Ser Gly Ser Ile His Pro Arg Ser
835 840 845
Leu Leu Leu Thr Gly Pro Asn Met Gly Gly Lys Ser Thr Leu Leu Arg
850 855 860
Ala Thr Cys Leu Ala Val Ile Phe Ala Gln Leu Gly Cys Tyr Val Pro
865 870 875 880
Cys Glu Ser Cys Glu Ile Ser Leu Val Asp Thr Ile Phe Thr Arg Leu
885 890 895
Gly Ala Ser Asp Arg Ile Met Thr Gly Glu Ser Thr Phe Leu Val Glu
900 905 910
Cys Thr Glu Thr Ala Ser Val Leu Gln Asn Ala Thr Gln Asp Ser Leu
915 920 925
Val Ile Leu Asp Glu Leu Gly Arg Gly Thr Ser Thr Phe Asp Gly Tyr
930 935 940
Ala Ile Ala Tyr Ser Val Phe Arg His Leu Val Glu Lys Val Gln Cys
945 950 955 960
Arg Met Leu Phe Ala Thr His Tyr His Pro Leu Thr Lys Glu Phe Ala
965 970 975
Ser His Pro Arg Val Thr Ser Lys His Met Ala Cys Ala Phe Lys Ser
980 985 990
Arg Ser Asp Tyr Gln Pro Arg Gly Cys Asp Gln Asp Leu Val Phe Leu
995 1000 1005
Tyr Arg Leu Thr Glu Gly Ala Cys Pro Glu Ser Tyr Gly Leu Gln
1010 ~ 1015 1020
Val Ala Leu Met Ala Gly Ile Pro Asn Gln Val Val Glu Thr Ala
1025 1030 1035
Ser Gly Ala Ala Gln Ala Met Lys Arg Ser Ile Gly Glu Asn Phe
1040 1045 1050
Lys Ser Ser Glu Leu Arg Ser Glu Phe Ser Ser Leu His Glu Asp
1055 1060 1065
Trp Leu Lys Ser Leu Val Gly Ile Ser Arg Val Ala His Asn Asn
1070 1075 1080
Page 75
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
Ala Pro Ile Gly Glu Asp Asp Tyr Asp Thr Leu Phe Cys Leu Trp
1085 1090 1095
His Glu Ile Lys Ser Ser Tyr Cys Val Pro Lys
1100 1105
<210> 36
<211> 1170
<212> DNA
<213> Homo sapiens
<400>
36
atggcgcaaccaaagcaagagagggtggcgcgtgccagacaccaacggtcggaaaccgcc60
agacaccaacggtcggaaaccgccaagacaccaacgctcggaaaccgccagacaccaacg120
ctcggaaaccgccagacaccaaggctcggaatccacgccaggccacgacggagggcgact180
aCCtCCCttCtgaccctgctgctggcgttcggaaaaaacgcagtccggtgtgctctgatt240
ggtccaggctctttgacgtcacggactcgacctttgacagagccactaggcgaaaaggag300
agacgggaagtattttttccgccccgcccggaaagggtggagcacaacgtcgaaagcagc360
cgttgggagcccaggaggcggggcgcctgtgggagccgtggagggaactttcccagtccc420
cgaggeggatccggtgttgcatccttggagcgagctgagaactcgagtacagaacctgct480
aaggccatcaaacctattgatcggaagtcagtccatcagatttgctctgggccggtggta540
ccgagtctaaggccgaatgcggtgaaggagttagtagaaaacagtctggatgctggtgcc600
actaatgttgatctaaagcttaaggactatggagtggatctcattgaagtttcaggcaat660
ggatgtggggtagaagaagaaaacttcgaaggctttactctgaaacatcacacatgtaag720
attcaagagtttgccgacctaactcaggtggaaacttttggctttcggggggaagctctg780
agctcactttgtgcactgagtgatgtcaccatttctacctgccgtgtatcagcgaaggtt840
gggactcgactggtgtttgatcactatgggaaaatcatccagaaaaccccctacccccgc900
cccagagggatgacagtcagcgtgaagcagttattttctacgctacctgtgcaccataaa960
gaatttcaaaggaatattaagaagaaacgtgcctgcttccccttcgccttctgccgtgat1020
tgtcagtttcctgaggcctccccagccatgcttcctgtacagcctgtagaactgactcct1080
agaagtaccccaccccacccctgctccttggaggacaacgtgatcactgtattcagctct1140
gtcaagaatggtccaggttcttctagatga 1170
<210> 37
<211> 389
<212> PRT
<213> Homo Sapiens
<400> 37
Page 76
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
Met Ala Gln Pro Lys Gln Glu Arg Val Ala Arg Ala Arg His Gln Arg
1 5 10 15
Ser Glu Thr Ala Arg His Gln Arg Ser Glu Thr Ala Lys Thr Pro Thr
20 25 30
Leu Gly Asn Arg Gln Thr Pro Thr Leu Gly Asn Arg Gln Thr Pro Arg
35 40 45
Leu Gly Ile His Ala Arg Pro Arg Arg Arg Ala Thr Thr Ser Leu Leu
50 55 60
Thr Leu Leu Leu Ala Phe Gly Lys Asn Ala Val Arg Cys Ala Leu Ile
65 70 75 80
Gly Pro Gly Ser Leu Thr Ser Arg Thr Arg Pro Leu Thr Glu Pro Leu
85 90 95
Gly Glu Lys Glu Arg Arg Glu Val Phe Phe Pro Pro Arg Pro Glu Arg
100 105 l10
Val Glu His Asn Val Glu Ser Ser Arg Trp Glu Pro Arg Arg Arg Gly
115 120 125
Ala Cys Gly Ser Arg Gly Gly Asn Phe Pro Ser Pro Arg Gly Gly Ser
130 135 140
Gly Val Ala Ser Leu Glu Arg Ala Glu Asn Ser Ser Thr Glu Pro Ala
145 150 155 160
Lys Ala Ile Lys Pro Ile Asp Arg Lys Ser Val His Gln Ile Cys Ser
165 170 175
Gly Pro Val Val Pro Ser Leu Arg Pro Asn Ala Val Lys Glu Leu Val
180 185 190
Glu Asn Ser Leu Asp Ala Gly Ala Thr Asn Val Asp Leu Lys Leu Lys
195 200 205
Asp Tyr Gly Val Asp Leu Ile Glu Val Ser Gly Asn Gly Cys Gly Val
210 215 220
Glu Glu Glu Asn Phe Glu Gly Phe Thr Leu Lys His His Thr Cys Lys
225 230 235 240
Ile Gln Glu Phe Ala Asp Leu Thr Gln Val Glu Thr Phe Gly Phe Arg
245 250 255
Page 77
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
Gly Glu Ala Leu Ser Ser Leu Cys Ala Leu Ser Asp Val Thr Ile Ser
260 265 270
Thr Cys Arg Val Ser Ala Lys Val Gly Thr Arg Leu Val Phe Asp His
275 280 285
Tyr Gly Lys Ile Ile Gln Lys Thr Pro Tyr Pro Arg Pro Arg Gly Met
290 295 300
Thr Val Ser Val Lys Gln Leu Phe Ser Thr Leu Pro Val His His Lys
305 310 315 320
Glu Phe Gln Arg Asn Ile Lys Lys Lys Arg Ala Cys Phe Pro Phe Ala
325 330 335
Phe Cys Arg Asp Cys Gln Phe Pro Glu Ala Ser Pro Ala Met Leu Pro
340 345 350
Val Gln Pro Val Glu Leu Thr Pro Arg Ser Thr Pro Pro His Pro Cys
355 360 365
Ser Leu Glu Asp Asn Val Ile Thr Val Phe Ser Ser Val Lys Asn Gly
370 375 380
Pro Gly Ser Ser'Arg
385
<210>
' 38
<211>
795
<212>
DNA
<213> Sapiens
Homo
<400>
38
atgtgtccttggcggcctagactaggccgtcgctgtatggtgagccccagggaggcggat 60
CtgggCCCCCagaaggaCaCCCgCCtggatttgCCCCgtagCCCggCCCgggCCCCtCgg 120
gagcagaacagccttggtgaggtggacaggaggggacctcgcgagcagacgcgcgcgcca 180
gcgacagcagccccgccccggcctctcgggagccggggggcagaggctgcggagccccag 240
gagggtctatcagccacagtctctgcatgtttccaagagcaacaggaaatgaacacattg 300
caggggccagtgtcattcaaagatgtggctgtggatttcacccaggaggagtggcggcaa 360
ctggaccctgatgagaagatagcatacggggatgtgatgttggagaactacagccatcta 420
gtttctgtggggtatgattatcaccaagccaaacatcatcatggagtggaggtgaaggaa 480
gtggagcagggagaggagccgtggataatggaaggtgaatttccatgtcaacatagtcca 540
gaacctgctaaggccatcaaacctattgatcggaagtcagtccatcagatttgctctggg 600
ccagtggtactgagtctaagcactgcagtgaaggagttagtagaaaacagtctggatgct 660
Page 78
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
ggtgccacta atattgatct aaagcttaag gactatggag tggatctcat tgaagtttca 720
gacaatggat gtggggtaga agaagaaaac tttgaaggct taatctcttt cagctctgaa 780
acatcacaca tgtaa 795
<210> 39
<211> 264
<212> PRT
<2l3> Homo sapiens
<400> 39
Met Cys Pro Trp Arg Pro Arg Leu Gly Arg Arg Cys Met Val Ser Pro
1 5 10 15
Arg Glu Ala Asp Leu Gly Pro Gln Lys Asp Thr Arg Leu Asp Leu Pro
20 25 30
Arg Ser Pro Ala Arg Ala Pro Arg Glu Gln Asn Ser Leu Gly Glu Val
35 40 45
Asp Arg Arg Gly Pro Arg Glu Gln Thr Arg Ala Pro Ala Thr Ala Ala
50 55 60
Pro Pro Arg Pro Leu Gly Ser Arg Gly Ala Glu Ala Ala Glu Pro Gln
65 70 75 80
Glu Gly Leu Ser Ala Thr Val Ser Ala Cys Phe Gln Glu Gln Gln Glu
85 90 95
Met Asn Thr Leu Gln Gly Pro Val Ser Phe Lys Asp Val Ala Val Asp
100 105 110
Phe Thr Gln Glu Glu Trp Arg Gln Leu Asp Pro Asp Glu Lys Ile Ala
115 120 125
Tyr Gly Asp Val Met Leu Glu Asn Tyr Ser His Leu Val Ser Val Gly
130 135 140
Tyr Asp Tyr His Gln Ala Lys His His His Gly Val Glu Val Lys Glu
145 150 155 160
Val Glu Gln Gly Glu Glu Pro Trp Ile Met Glu Gly Glu Phe Pro Cys
165 170 175
Gln His Ser Pro Glu Pro Ala Lys Ala Ile Lys Pro Ile Asp Arg Lys
180 185 190
Ser Val His Gln Ile Cys Ser Gly Pro Val Val Leu Ser Leu Ser Thr
195 200 205
Page 79
CA 02455686 2004-O1-26
WO 03/012130 PCT/USO1/23888
Ala Val Lys Glu Leu Val Glu Asn Ser Leu Asp Ala Gly Ala Thr Asn
210 215 220
Ile Asp Leu Lys Leu Lys Asp Tyr Gly Val Asp Leu Ile Glu Val Ser
225 230 235 240
Asp Asn Gly Cys Gly Val Glu Glu Glu Asn Phe Glu Gly Leu Ile Ser
245 250 255
Phe Ser Ser Glu Thr Ser His Met
260
Page 80