Note: Descriptions are shown in the official language in which they were submitted.
CA 02536698 2006-02-15
SYSTEM AND METHOD OF DETECTING PATHOGENS
Field of the Invention
[00011 The present invention relates to the field of detecting pathogens. In
particular,
it relates to a system and method for detecting, identifying, characterizing
and surveilling
pathogen and host markers, collecting and disseminating information concerning
those
pathogens and their hosts to and from an instant location, providing treatment
recommendation to and from an instant location, and providing educational
information
to and from an instant location.
Background of the Invention
[00021 Detection and characterization of an infectious disease is a complex
process
that ideally begins with the identification of the causative agent (pathogen).
This has
traditionally been accomplished by direct examination and culture of an
appropriate
clinical specimen. However, direct examination is limited by the number of
organisms
present and by the observer's ability to successfully recognize the pathogen.
Similarly, in
vitro culture of the etiologic agent depends on selection of appropriate
culture media as
well as on the microbe's fastidiousness. The utility of pathogen culture is
further
restricted by lengthy incubation periods and limited sensitivity and
specificity.
[00031 When in vitro culture remains a feasible option, the identification and
differentiation of microorganisms has principally relied on microbial
morphology and
growth variables which, in some cases, are sufficient for strain
characterization (i.e.
isoenzyme profiles, antibiotic susceptibility profiles, and chematographic
analysis of fatty
acids).
[00041 If culture is difficult, or specimens are not collected at the
appropriate time,
the detection of infection is often made retrospectively, if at all, by
demonstrating a
serum antibody response in the infected host. Antigen and antibody detection
methods
have relied on developments in direct (DFA) and indirect (IFA)
immunofluorescence
analysis and enzyme immunoassay (EIA)-based techniques, but these methods can
also
possess limited sensitivity.
-1-
CA 02536698 2006-02-15
[0005] These existing methods have several drawbacks. First, the process can
take
several days to return results. In the case of highly communicable and/or
dangerous
pathogens, confirmation of pathogen type may not be received until the host
has already
exposed others or has passed beyond treatment. Second, the transportation of
samples to
laboratories for culture growth increases the risk of errors, such as
misidentifying the
sample, or exposure of unprotected personnel to a sample containing a highly
communicable pathogen. Lastly, the pathogen tests are limited based on the
suspected
pathogen list provided by the observer (i.e. doctor), meaning that additional
unsuspected
pathogens are not tested for but may be present.
100061 Related to this method of diagnosis is the response to an outbreak of
infectious disease. If an outbreak is suspected or detected, the existing
response is the
hundreds of years old method of quarantine. One of the problems with
quarantine is the
delaying of correct diagnosis and treatment for individuals in quarantine.
Another is that
healthy, unexposed individuals may be quarantined with infected individuals
and contract
a disease they would not have otherwise. Resorting to quarantine is seen as
necessary
due to the delays involved in detecting pathogens through blood sampling as
discussed
previously.
[00071 A final problem lies in identifying unknown and unsuspected pathogens.
The
blood sampling tests are limited to those pathogens which are listed as
suspected by the
observing doctor. As many doctors may not be familiar with rare or recently
identified
pathogens, or may not recognize the relevant identifying symptoms, they may
not list
them as suspected on the sample. As a result, these pathogens remain
undetected and
may be unknowingly spread by the host.
[00081 In contrast to reliance on morphological characteristics, pathogen
genotypic
and proteomic traits generally provide reliable and quantifiable information
for the
detection and characterization of infectious agents. Moreover, microbial
DNA/RNA can
be extracted directly from clinical specimens without the need for
purification or isolation
of the agent.
[00091 On a global scale, molecular techniques can be applied in a high
throughput
manner in screening and surveillance studies monitoring disease prevalence and
distribution, evaluation of control measures, and identification of outbreaks.
-2-
CA 02536698 2006-02-15
[0010] Point-of-care diagnostic devices (PDDs) have been developed for a
number of
individual infectious diseases. In most cases these assays are
immunochromatographic
single colorimetric strip tests designed to detect a single infectious agent
(either a
pathogen-specific antigen or an antibody response to one) in a small volume of
blood or
serum.
[0011] None of these current assays has the capability to detect multiple
pathogens or
simultaneously detect genomes and proteomes of multiple pathogens. Similar
limitations
exist for other rapid diagnostic assays. Since almost all these tests rely on
a single visual
colorimetric change for their readout, the opportunities to detect multiple
pathogens are
severely impeded and the majority of current PDDs are restricted to the
detection of a
single pathogen. Consequently, evaluating patients for potential infectious
agents or
testing a unit of blood for common transmissible agents requires multiple
consecutive
point-of-care tests to be performed, complicating clinical management, slowing
results
and significantly escalating costs.
[0012] Many PDDs do not meet what are considered essential requirements
including: ease of performance, a requirement for minimal training, the
generation of
unambiguous results, high sensitivity and specificity, the generation of same
day results
(preferably within minutes), relative low cost, and no requirement for
refrigeration or
specialized additional equipment.
[0013] In summary, despite current availability of excellent diagnostic
reagents (e.g.
antibody and nucleic acid probes) that recognize specific targets for many
microbial
pathogens, the current strategies have inadequate performance characteristics.
Contributing to this is the fact that these reagents are conjugated to organic
dyes, gold-
labelled particles or enzymes that lack sufficient sensitivity to be detected
at the single
molecule level. Furthermore, the current PDD platforms and detection schemes
typically
rely on single macroscopic colorimetric changes and are not well suited to the
simultaneous detection of multiple pathogens.
100141 More recent advances in molecular diagnostics, including real-time PCR
combined with automated specimen processing, have addressed a number of the
limitations of earlier "in-house" and non-standardized gene amplification
assays. These
assays represent a significant advance in detecting, quantifying, and
characterizing many
-3-
CA 02536698 2006-02-15
microbes and currently represent the "gold" or reference standard for
infectious disease
diagnostics for a number of pathogens. However, these assays are still
complex,
expensive, and require specialized equipment, creating a number of barriers to
their
potential application at point-of-care.
[0015] Finally, current genomic or proteomic detection strategies require a
sample
processing and technical commitment to one strategy or the other. There is no
current
capacity to simultaneously detect both antigenic targets for some pathogens
and genetic
targets for others. This limits the simultaneous detection of preferred
pathogen-specific
targets and presents a barrier to fully exploiting the complementary power of
both
strategies.
[0016] A system is needed which enables pathogen detection, identification and
characterization, as well as host characterization in a much more timely
manner than
existing methods. Preferably, such a system would also enable detection,
identification
and characterization of many or all pathogens in a sample based on an existing
database
and not be limited to a suspected list provided by a observing doctor.
Summary of the Invention
[0017] According to an aspect of the invention there is provided a method of
performing one or more of: detecting, identifying and characterizing pathogens
and
characterizing pathogen hosts using markers for pathogens and hosts,
comprising the
steps of: a) preparing a marker-detection medium containing signatures of the
identity
and characteristics of pathogens and optionally of hosts; b) collecting a
sample from a
host; c) combining the sample with the marker-detection medium and d)
analyzing the
signatures to detect, identify and characterize the pathogens, and optionally,
characterize
the host.
[0018] Preferably, the sample collected is a blood sample, although plasma,
serum
and other types of samples can also be used, and the pathogen-detection medium
preferably consists of nanobeads conjugated to biorecognition molecules (BRMs)
and the
nanobeads are injected with quantum dots. Also preferably, each of the
nanobeads
contains a unique combination of quantum dots to provide a unique optical
barcode
-4-
CA 02536698 2006-02-15
associated with each nanobead for detecting unique molecular signatures of
each
pathogen and for characteristics of each pathogen and host.
[00191 Preferably, the analyzing step consists of illuminating the combined
sample
with a laser and obtaining resulting spectra with a spectrometer/CCD camera.
These
spectra correlate with identified individual molecular markers of the pathogen
and host.
[00201 Optionally, the method may include producing a list of host
characterization
markers associated with said host sample as part of analysis step d). .
[00211 Optionally, the method may include an additional step e) of providing a
list of
treatment options based on the list of pathogen and host markers generated in
analysis
step d).
100221 Optionally, the method may include step f) of providing to an instant
location
educational information based on the list of pathogen and host markers
generated in
analysis step d).
[00231 Preferably, the method further includes an additional step g) of
transmitting
said list of pathogen markers and said list of host identifier markers to a
remote database.
[00241 According to another aspect of the invention a system of components is
provided which is capable of executing any of the above methods.
[00251 The advantages of the present invention include a vast reduction in the
amount
of time necessary to identify pathogens in a patient sample, as well as the
ability to
provide immediate on-site information concerning treatment and quarantine
measures for
any identified pathogens. Another advantage is the ability to collect patient
and pathogen
data in a global database and mine the information contained in this database
to produce
trends and tracking measures for various pathogens and their hosts, which
information
may be used for surveillance, research, therapeutic design, and other
purposes.
100261 Other and further advantages and features of the invention will be
apparent to
those skilled in the art from the following detailed description thereof,
taken in
conjunction with the accompanying drawings.
-5-
CA 02536698 2006-02-15
Brief Description of the Drawings
[00271 The invention will now be described in more detail, by way of example
only,
with reference to the accompanying drawings, in which like numbers refer to
like
elements, wherein:
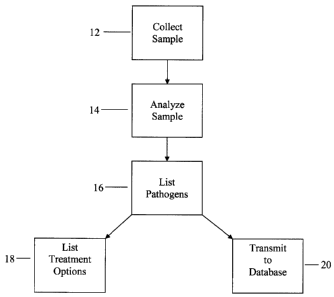
Figure 1 is a flow chart detailing the series of steps in the inventive method
disclosed herein;
Figure 2 is a block diagram for a pathogen detection device; and
Figure 3 is a block diagram of multiple devices communicating with a central
database.
Detailed Description of the Preferred Embodiments
[00281 Referring now to Figure 1, the present inventive method is described by
a
series of steps set out in a flowchart.
100291 The first step 12 is to collect a sample from a host (e.g. a human,
animal or
environmental sample), preferably a blood sample, although plasma samples,
serum
samples and other types of physical samples can be used, as appropriate. This
sample is
then analyzed 14 and a list of pathogens contained in the sample is generated
16.
[00301 The analysis 14 is performed by a pathogen detection device 30 as shown
in
Figure 2. This device 30 is portable, preferably hand-held, and has an outlet
32 for
receiving a sample and a display 36 to show the list of detected pathogens
within the
sample. An input device 38, such as a keyboard, is also provided to enable
scrolling and
viewing of the display and input of additional information (field notes,
etc.). The list of
pathogens is generated from a database of known pathogens, which may be an
internal
database on the device 30 (kept in flash memory or similar storage to allow
for updating)
or retrieved by communicating with an external database. The pathogen
detection device
30 is ideally capable of detecting multiple pathogen and host markers within a
single
sample, and preferably markers of different types, such as protein-based
markers and
gene-based markers.
-6-
CA 02536698 2006-02-15
[00311 The method of detection used can be varied among suitable available
methods, however, a preferred method is the use of quantum dots bonded to
biorecognition molecules (BRMs). Quantum dots, also known as semiconductor
nanocrystals, are electromagnetically active nanotechnologic particles,
ranging in size
from 2 nanometers (nm) to 8 nm. A particularly useful property of quantum dots
is that
they are fluorescent, that is they emit light after brief illumination by a
laser. In addition,
quantum dots of different sizes will fluoresce in different colors and the
fluorescing color
can be modified by the particle's shape, size and composition. BRMs are
biological
molecules that bind only to a single other biological molecule. For example,
"antibodies"
are BRMs that bind to proteins and "oligonucleotide probes" are BRMs that bind
to genes
(e.g. DNA or RNA). Pathogens and hosts have both unique and shared genetic and
protein markers, and each marker can be bonded to by a specific BRM.
[00321 A nanobead, which is a plastic bead that can be 1-10 microns in
diameter and
injected with a collection of quantum dots, is physically conjugated to a BRM.
By
injecting unique combinations of quantum dots into the nanobeads, thousands of
nanobeads with distinctive combinations of quantum dots can be created. When a
laser
illuminates the nanobeads, the quantum dots fluoresce to produce a distinctive
combination of colors. These color combinations are an example of a barcode,
in this
case an optical bar code, analogous to a UPC symbol, and similar known types
of
imprinted barcodes. Since each BRM recognizes a distinct pathogen or host
marker and
each nanobead has a unique barcode, each BRM-conjugated nanobead provides a
barcode
for a specific pathogen or host marker. Thousands of these BRM-conjugated
nanobeads
may be dried into a powder and provided in sample vials.
[0033] The biological (e.g. blood) sample is added to a vial, and the various
nanobeads become attached to different pathogens based on the BRM conjugated
to the
nanobeads. The vial is then illuminated by a laser, causing the quantum dots
to fluoresce.
The resulting spectrum is analyzed by the marker detection device and, by
comparing the
spectrum data with a database of known spectra corresponding to different
pathogens and
host characteristics, a list of detected pathogens and pathogen and host
characteristics is
produced. The response time from the taking of the original biological sample
to the
production of the pathogen list can be measured in minutes.
-7-
CA 02536698 2006-02-15
[0034] Ideally, the pathogen detection device 30 is a portable, hand-held
device with
an integrated laser and spectrometer and includes a supply of sample vials.
The device
30 may store a pathogen identity database on board, or access a remote
database,
preferably via the Internet, preferably wirelessly, and identify the pathogen
from a
remote, central database. If an on-board database is used, a communications
system 34
for contacting and receiving updates from a larger, central database should be
provided.
[00351 Once the pathogen list is produced, the pathogen detection device 30
may
additionally provide further information of value to the diagnosing doctor.
Ideally, a
treatment protocol is provided (step 18), including any special measures
necessary to
avoid communication of the pathogen. Other information, such as
pathophysiology,
disease history and bibliographic references can be provided, enabling the
pathogen
detection device 30 to also be used as an educational tool in the appropriate
scenarios.
[00361 An outbreak scenario for use of the device in a standard pathogen
detection
setting follows. An airport is a point of entry representing a major pathogen
travel
vector, as well as presenting problems with implementing traditional detection
and
quarantine methods. By equipping medical staff with a number of pathogen
detection
devices as described herein, and a supply of nanobead sample vials, incoming
passengers
can be processed on-site by taking a blood sample and injecting it into a
sample vial. The
analysis is performed by the pathogen detection device within minutes and the
sampled
passenger can be quickly released or redirected for treatment and observation,
as
necessary. While a single device is limited in processing capability, the
ability to provide
multiples of identical devices can enable processing of passengers in a matter
of hours,
not days. Faster processing allows appropriate treatment and quarantine
measures to be
taken earlier, and be more effective, reducing the probability of the pathogen
spreading
unchecked.
[00371 Detecting and providing a treatment protocol for a pathogen represents
merely
the first step in a potentially much larger process for tracking and
controlling pathogens
as shown in Figure 3. By providing a sufficient number of BRM-conjugated
nanobeads,
the pathogen and the human or animal host for the pathogen can be
characterized by
thousands of genetic and protein markers contained in a biological sample,
such as blood.
This information, which does not include any other information about the
patient (e.g.
-8-
CA 02536698 2006-02-15
name, address and other privacy-protected data), but which does include GPS
locator
information to identify the location of the sample, is then sent to a central
database 40.
The information is preferably sent wirelessly, and immediately upon generation
of the
pathogen list (step 20). The central database 40 is in contact with a
substantial number of
pathogen detection devices 30 at any given time.
[0038] The central database 40 can be local, national or global, or a
combination of
different databases of these types. Ideally, one top-level central database 40
is provided
which receives information constantly from all devices 30 worldwide. Over
time, the
database becomes a collection of information concerning every known pathogen
and
probable or possible mutations, the locations where outbreaks have occurred
and the
types of humans or animals (based on gene and protein code data) who have been
exposed to certain pathogens. The information contained in this database can
then be
mined to detect patterns.
[0039] Some patterns developed from the collected data include outbreak
tracking, by
identifying geographic regions where outbreaks of specific pathogens occur
regularly.
Other patterns can be developed from the human or animal host data, including
susceptibility of certain segments of the population to certain pathogens, and
resistance of
other segments of the population to other pathogens.
[0040] This concludes the description of a presently preferred embodiment of
the
invention. The foregoing description has been presented for the purpose of
illustration
and is not intended to be exhaustive or to limit the invention to the precise
form
disclosed. Many modifications and variations are possible in light of the
above teaching
and will be apparent to those skilled in the art. It is intended the scope of
the invention be
limited not by this description but by the claims that follow.
-9-