Note: Descriptions are shown in the official language in which they were submitted.
CA 02538147 2004-O1-21
3005-7PCT-9CA
-1-
TITLE OF INVENTION; FARNESYL DIBENZODIAZEPINONE, PROCESSES FOR
ITS PRODUCTION AND ITS USE AS A PHARMACEUTICAL
RELATED APPLICATIONS
This application claims priority to U.S. Provisional Application 60/441,126,
filed January 21, 2003; U.S. Provisional Application 60/492,997, filed August
7,
2003; and U.S. Provisional Application 60/518,286, filed November 10, 2003.
The
entire teachings of the above provisional applications are incorporated herein
by
reference.
FIELD OF THE INVENTION
This invention relates to a novel farnesylated dibenzodiazepinone, named
ECO-04601, its pharmaceutically acceptable salts and derivatives, and to
methods
for obtaining the compound. One method of obtaining the compound is by
cultivation of a novel strain of Micromonospora sp., i.e., 046-EC011 or
[S01]046;
another method involves expression of biosynthetic pathway genes in
transformed
host cells. The present invention further relates to Micromonospora sp.
strains 046-
EC011 and [S01]046, to the use of ECO-04601 and its pharmaceutically
acceptable
salts and derivatives as pharmaceuticals, in particular to their use as
inhibitors of
cancer cell growth, bacterial cell growth, mammalian lipoxygenase, and for
treating
acute and chronic inflammation, and to pharmaceutical compositions comprising
ECO-04601 or a pharmaceutically acceptable salt or derivative thereof.
Finally, the
invention relates to novel polynucleotide sequences and their encoded
proteins,
which are involved in the biosynthesis of ECO-04601.
BACKGROUND OF THE INVENTION
The euactinomycetes are a subset of a large and complex group of Gram-
positive bacteria known as actinomycetes. Over the past few decades these
organisms, which are abundant in soil, have generated significant commercial
and
scientific interest as a result of the large number of therapeutically useful
compounds, particularly antibiotics, produced as secondary metabolites. The
intensive search for strains able to produce new antibiotics has led to the
identification of hundreds of new species.
Many of the euactinomycetes, particularly Streptomyces and the closely
CA 02538147 2004-O1-21
3005-7PCT-9CA
-2-
related Saccharopolyspora genera, have been extensively studied. Both of these
genera produce a notable diversity of biologically active metabolites. Because
of the
commercial significance of these compounds, much is known about the genetics
and physiology of these organisms.
Another representative genus of euactinomycetes, Micromonospora, has also
generated commercial interest. For example, U.S. Patent No. 5,541,181 (Ohkuma
et al.) discloses a dibenzodiazepinone compound, specifically 5-farnesyl-4,7,9-
trihydroxy-dibenzodiazepin-11-one (named "BU-4664L"), produced by a known
euactinomycetes strain, Micromonospora sp. M990-6 (ATCC 55378). The Ohkurma
et al. patent reports that BU-4664L and its chemically synthesized di- and tri-
alkoxy
and acyloxy derivatives possess anti-inflammatory and anti-tumor cell
activities.
Although many biologically active compounds have been identified from
bacteria, there remains the need to obtain novel naturally occurring compounds
with
enhanced properties. Current methods of obtaining such compounds include
screening of natural isolates and chemical modification of existing compounds,
both
of which are costly and time consuming. Current screening methods are based on
general biological properties of the compound, which require prior knowledge
of the
structure of the molecules. Methods for chemically modifying known active
compounds exist, but still suffer from practical limitations as to the type of
compounds obtainable.
Thus, there exists a considerable need to obtain pharmaceutically active
compounds in a cost-effective manner and with high yield. The present
invention
solves these problems by providing a novel strain of Micromonospora capable of
producing a potent new therapeutic compound, as well as reagents (e.g.,
polynucleotides, vectors comprising the polynucleotides and host cells
comprising
the vectors) and methods to generate novel compounds by de novo biosynthesis
rather than by chemical synthesis.
SUMMARY OF THE INVENTION
In one aspect, the invention relates to a compound of the formula
CA 02538147 2004-O1-21
3005-7PCT-9CA
-3-
I
i
n
(Formula II) or a pharmaceutically acceptable salt thereof.
In another aspect, the invention relates to a pharmaceutical composition
comprising a compound of the formula
I
i
HO
or a pharmaceutically acceptable salt thereof, together with a
pharmaceutically
acceptable carrier.
In a further aspect, the invention relates to a class of compounds represented
by Formula I:
0
i ~ ,~W~ 3~CHg
w
Rz0 ORa
Formula I
wherein,
W 1, W2 and W3 is each independently selected from
CH3 CH3 CH3 H CH3
or
HZ I I . H I ,
-C-H-~_ ; _~-H-C-~_ _ - \ / - _
O OH OH
the chain from the tricycle may terminate at W3, W2 or W1 with W3, W2 or
W 1 respectively being either -CH=O or -CH20H;
CA 02538147 2004-O1-21
3005-7PCT-9CA
-4-
A is selected from -NH--, -NCH2R1, -NC(O)R1;
R1 is selected from C1-6 alkyl, C2-6 alkene, aryl or heteroaryl;
R2, R3, and R4 is each independently selected from H, R5, -C(O)RE
R5 is each independently selected from C1_6 alkyl, C2_7 alkalene, aryl or
heteroaryl;
R6 is each independently selected from H, C1 _g alkyl, C2_7 alkalene, aryl or
heteroaryl; or a pharmaceutically acceptable salt thereof.
in one embodiment, A is NH.
In another embodiment, A is -NCH2R'.
In another embodiment, A is -NC(O)R'.
In another embodiment, R2 is H.
In another embodiment, R3 is H.
In another embodiment, R4 is H.
In another embodiment, R2, R3 and R4 are each H.
In another embodiment, R2, R3 and R4 are each H, and W' is -CH =CH-.
In another embodiment, R2, R3 and R4 are each H, and W2 is -CH =CH-.
In another embodiment, R2, R3 and R4 are each H, and W3 is -CH =CH-.
In another embodiment, A is NH and R2, R3 and R4 are each H.
In another embodiment, A is NH, each of W', W2, and W3 is -CH =CH-.
The invention further encompasses a compound selected from the group
consisting of:
O CH- CH, CH-
N~~I~~CH,
N
HO _ OH
O=C
H3C HO
CA 02538147 2004-O1-21
~00~-PCT-9CA
o cH, cH, cH,
N / / / CHj
N /
HO
_ OH
H'CJ HO .
-5-
0 CH, CH, CH, 0 CH, CH, CH,
N I / / CH ~ N / / CH
0 ~~/ 0
N / ~ i N (\
HO H OH HO H ~~~~~OH
HO ~ HO
0 CH, CH, CH, O CH, CH, CH,
~N / / CH, / N / CH,
0 \ ~ O 0
/ ~ % /
HO H _ 'OH HO H OH
HO ~ HO
0 CH, CH, CH, O CH, CH, CH,
N / CH, / N / CH,
0 O ~ ~ 0 0
HO H OH HO H OH
HO ~ HO
O CH, CH, CH, 0 CH, CH, CH,
i ~ i
N CH, N / CH,
O O O \ ~ O
% / ~ % /
HO H _ OH HO H ' OH
HO ~ HO
CA 02538147 2004-O1-21
~OU5-7PCT-9CA
-6-
0 CH, CH, CH, 0 CH, CH, CH,
N / CH, / ~ ~N / CH,
\ p \ ~ O
1~ ~N / ~ i /
HO H OH HO H ~ OH
HO ~ HO
Q OH, CH, CH, 0 CH, CH, CH,
i i
N CH, ~ N~ CH,
\ / O 0 \ ~ 0 O
j /
HO H OH HO H OH
HO ~ HO
O ~ H, CH, CH, O CH, CH, CH,
i
~~N CH, N / CH,
O O \ ~ O
HO H / ~ OH HO H / ~ OH
HO ~ HO
O CH, CH, CH, O CH, CH, CH,
\ ~ N / CH, \ ~ N / CH,
0 0
N /
HO H ~~ OH HO ~ _ ~ OH
HG ~ HO
O CH, CH, CH, O CH, CH, CH,
i i
N ~ CH, ~N CH,
\ ~ o \ ~ o
i / \ i /
HO H OH HO H OH
HO ~ HO
CA 02538147 2004-O1-21
3005-7PCT-9CA
-7-
O CH, CH, CH, O CH, CH, CH,
/ / /
C ~ 1 CH ~ / CH
\ 0
O ~ N
HO ~ OH O ~ _ OH
H H
HO ~ H'C HO
O CH, CH, CH,
i N~ / / CH;
HC ~ ~~ v
r-~N--~~ 0
Ho ; ~ p
~ H ~ ~CH3
HO
O CH, CH, CH,
N~ / / CH
0~ N ~ ~ O
O IIII
H O~CH,
HsC HO
O CH3 CH3 CH, 0 CH, CH3 CHI
i
N/ / / C N / / / CH
v
N
N ~ \ HO
H,C'0 ~ _ pH H _ OH
H
HO ~ H3C~0
CA 02538147 2004-O1-21
3005-7PCT-9CA
_g_
O CH, CH, CH,
N / / / CH,
N ~ ~ H C,
HO ~ O-CH,
H
HO
0 CH, CH, CH,
I
0 CH, CH, CH=
N / / / CH,
N / / / CH
N
HO H ~ ~ O~CH,
H C~ 0 H _ ~ O~CH,
CH; ~ HO
O CH3 CH3 CH;
i
N / / CH3
HC 0
N
HO ~ OH
H
HO
O CHI CHI CH, 0 CH3 CH3 CH,
N / / CH ~ N / / CH
HO N ' ~ HO ~ ~ ~ H
~OH H _ ~O
HO~ ~ HO
0 CH3 CH3 CHa 0 CH3 CH3 CH3
N ~ / CH3 ~ ~ N / CH3
i N---~ \\ N
HO H/' ~OH HO H ' OH
HC/;~''/ ~ HO
CA 02538147 2004-O1-21
3005-7PCT-9CA
_g_
0 CHI CH3 CH3 O CH3 CHI CH3
i / i \ ~
N CH3 ~ ' /r 'N CH3
i /
HO H _ OH HO H ~ OH
HO , HO
0 CH, CH, O CH, CH,
N / / /O ~ ~ N i / OH
N / ~ N /
HO H _ OH HO H ~ OH
HO , HO
O CH, O CH3
/ /O ~ ~N / OH
/ ~ % /
HO H OH HO H _ OH
HO , HO
O O
N~O ~ ~ N~OH
N ~ \ ~ N /
HC /~OH HO ~ OH
H ~ H
HO , HO
O CH, CH, CH O CH, CH, CH,
i
w
(( N/ /. /~CH3 O ~ N / / CH3
v off ~ off
~N / OH N OH
HO H ~OH HO H ' ~ OH
HO ~ HO
CA 02538147 2004-O1-21
3005-7PCT-9CA
-10-
0 CH, CH; CH; 0 CH, CH, CH;
N / / CH; /N / CH,
OH \ ~ OH OH
N / OH N / OH OH
HO ; _ OH HO ~ _ ~ OH
H H
HC/ ; HO
0 CH; CH; CH, O CH; CH, CH;
'\ N / CH; / N / CH3
OH OH \ ~ OH OH
OH OH OH OH
/ ~ % /
HO H _ OH HO H _ OH
HC' ; H~~ ; and
O CH; CH, CH,
i
~N CH,
OH OH OH
/ OH OH OH
N
HO
H _ ~OH
HO
In one embodiment, the invention relates to compositions of the compounds
of Formula I together with a pharmaceutically acceptable carrier.
The invention further encompasses a farnesyl dibenzodiazepinone obtained
by a method comprising: a) cultivating Micromonospora sp. strain [S01]046,
wherein
the cultivation is performed under aerobic conditions in a nutrient medium
comprising at least one source of carbon atoms and at least one source of
nitrogen
atoms; and b) isolating a farnesyl dibenzodiazepinone from the bacteria
cultivated in
step (a). In one embodiment the farnesyl dibenzodiazapinone is the compound of
Formula II.
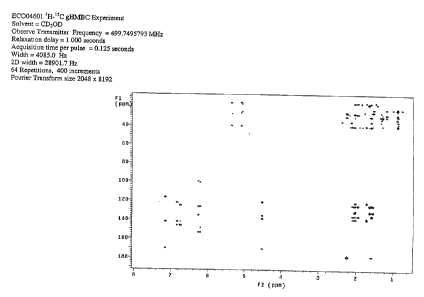
In one embodiment, the farnesyl dibenzodiazepinone generates NMR spectra
essentially as shown in Figure 3, 4, 5, 6 and 7. In another embodiment, the
farnesyl
dibenzodiazepinone generates an'H NMR spectrum of Figure 3.
The invention further encompasses a process for making a farnesyl
dibenzodiazapinone compound, comprising cultivation of Micromonospora sp.
strain
046-EC011, in a nutrient medium comprising at least one source of carbon atoms
and at least one source of nitrogen atoms, and isolation and purification of
the
CA 02538147 2004-O1-21
3005-7PCT-9CA
compound.
-11-
The invention further encompasses a process for making a farnesyl
dibenzodiazepinone compound comprising cultivation of Micromonospora sp.
strain
[S01]046 in a nutrient medium comprising at least one source of carbon atoms
and
at least one source of nitrogen atoms, and isolation and purification of the
compound.
In one embodiment, the cultivation occurs under aerobic conditions.
In another embodiment, the carbon atom and nitrogen atom sources are
chosen from the components shown in Table 16.
In another embodiment, the cultivation is carried out at a temperature ranging
from 18°C to 40°C. In a further embodiment, the temperature
range is 18°C to 29°C.
In another embodiment, the cultivation is carried out at a pH ranging from 6
to
9.
The invention further encompasses the Micromonospora sp. having IDAC
Accession No. 231203-01.
The invention further encompasses a method of inhibiting the growth of a
cancer cell, the method comprising contacting the cancer cell with a compound
of
Formula I, such that growth of the cancer cell is inhibited.
In one embodiment, the compound is ECO-04601.
The invention further encompasses a method of inhibiting the growth of a
cancer cell in a mammal, the method comprising administering a compound of
Formula I to a mammal comprising a cancer cell, such that growth of the cancer
cell
is inhibited in the mammal.
In one embodiment, the compound is ECO-04601.
The invention further encompasses a method of treating a pre-cancerous or
cancerous condition in a mammal, comprising the step of administering to the
mammal a therapeutically effective amount of a compound of Formula I, such
treat a
pre-cancerous or cancerous condition is treated.
In one embodiment, the compound is ECO-04601.
The invention further encompasses a method of treating a bacterial infection
in a mammal, comprising administering a therapeutically effective amount of a
compound of Formula I to a mammal having a bacterial infection, such that the
bacterial infection is treated.
CA 02538147 2004-O1-21
3005-7PCT-9CA
-12-
In one embodiment, the compound is ECO-04601.
The invention further encompasses a method of reducing inflammation in a
mammal, comprising administering to a mammal having inflammation a
therapeutically effective amount of a compound of Formula I, such that the
inflammation is reduced.
In one embodiment, the compound is ECO-04601.
The invention further encompasses an isolated polynucleotide comprising one
or more of SEQ ID NOs. 1, 64 and 73, wherein the polynucleotide encodes a
polypeptide that participates in a biosynthetic pathway for a farnesyl
dibenzodiazepinone.
The invention further encompasses an isolated polynucleotide comprising
SEQ ID NOs. 1, 64 and 73, wherein the polynucleotide encodes a polypeptide
that
participates in a biosynthetic pathway for a farnesyl dibenzodiazepinone.
The invention further encompasses an isolated polynucleotide that encodes a
polypeptide selected from the group consisting of SEQ ID NOs. 3, 5, 7, 9, 11,
13, 15,
17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45, 47, 49, 51, 53,
55, 57, 59,
61, 63, 66, 68, 70, 72, 75, 77, 79, 81, 83, 85, 87 and 89.
In one embodiment, the isolated polynucleotide comprising SEQ ID No. 1
encodes a polypeptide selected from the group consisting of SEQ ID Nos. 3, 5,
7, 9,
11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45, 47,
49, 51, 53,
55, 57, 59, 61 and 63.
In another embodiment, the isolated polynucleotide comprising SEQ ID No.
64 encodes a polypeptide selected from the group consisting of SEQ ID NOS: 66,
68, 70 and 72.
In another embodiment, the isolated polynucleotide comprising SEQ ID No.
73, encodes a polypeptide selected from the group consisting of SEQ ID NOS:
75,
77, 79, 81, 83, 85, 87 and 89.
The invention further encompasses an isolated polypeptide of SEQ ID NO. 3,
5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43,
45, 47, 49,
51, 53, 55, 57, 59, 61, 63, 66, 68, 70, 72, 75, 77, 79, 81, 83, 85, 87 or 89.
In one embodiment, the polypeptide participates in a biosynthetic pathway for
a farnesyl dibenzodiazepinone.
The invention further encompasses an expression vector comprising one or
CA 02538147 2004-O1-21
3005-7PCT-9CA
-13-
more of the polynucleotides described herein.
The invention further encompasses a recombinant prokaryotic organism
comprising one or more such expression vectors.
In one embodiment, the organism is an actinomycete.
In another embodiment, the organism requires the expression vector to
synthesize a farnesyl dibenzodiazepinone. That is, the organism is deficient
in the
ability to synthesize a farnesyl dibenzodiazepinone before transformation with
a
polynucleotide as described herein.
The invention further encompasses a method of making a farnesyl
dibenzodiazepinone de novo in a prokaryote, comprising the steps of: (a)
providing a
prokaryote that is incapable of synthesizing a farnesyl dibenzodiazepinone;
(b)
transforming the prokaryote with an expression vector as described herein; and
(c)
culturing the prokaryote; wherein the culturing results in the synthesis of a
farnesyl
dibenzodiazepinone in the prokaryote.
In one embodiment, the prokaryote is an actinomycete.
In another embodiment, the vector expresses a polypeptide of SEQ ID NO: 3,
5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43,
45, 47, 49,
51, 53, 55, 57, 59, 61, 63, 66, 68, 70, 72, 75, 77, 79, 81, 83, 85, 87 or 89.
Brief Description of the Figures
FIGURE 1 shows the mass of ECO-04601 determined by electrospray mass
spectrometry to be 462.6.
FIGURE 2 shows the absorption spectrum of purified ECO-04601 with a UVmax
at 230 nm and a shoulder at 290nm.
FIGURE 3 shows proton NMR data for the compound dissolved in MeOH-d4 .
FIGURE 4 shows multidimensional pulse sequences gDQCOSY.
FIGURE 5 shows multidimensional pulse sequences gHSQC.
FIGURE 6 shows multidimensional pulse sequences gHMBC.
FIGURE 7 shows multidimensional pulse sequences NOESY.
FIGURE 8 shows the in vitro anti-inflammatory activity of ECO-04601. Graph
shows percent inhibition of 5-lipoxygenase activity plotted against the
Log pM concentration of ECO-04601 and NDGA. Graph shows the
EC5o of ECO-04601 to be 0.93pM.
CA 02538147 2004-O1-21
3005-7PCT-9CA
-14-
FIGURE 9 shows inhibition of tumor growth resulting from administration of 1 U
to
30 mg/kg of ECO-04601 to glioblastoma-bearing mice beginning one
day after tumor cell inoculation.
FIGURE 10 shows inhibition of tumor growth resulting from administration of 20-
30
mg/kg of ECO-04601 to glioblastoma-bearing mice beginning ten days
after tumor cell inoculation.
FIGURE 11 shows micrographs of tumor sections from mice bearing glioblastoma
tumors and treated with saline or ECO-04601. The cell density of
tumor treated with ECO-04601 appears decreased and nuclei from
ECO-04601-treated tumor cells are larger and pynotic suggesting a
cytotoxic effect.
FIGURE 12 shows the biosynthetic locus of ECO-04601, isolated from
Micromonospora sp. strain 046-EC011, including the positions of
cosmids 046KM and 046KQ.
FIGURE 13 shows a schematic diagram of the biosynthetic pathway for the
production of the farnesyl-diphosphate group of ECO-04601 with
biosynthetic enzymes indicated by their ORF number and family
designation.
FIGURE 14 shows a schematic diagram of the biosynthetic pathway for the
production of (a) 3-hydroxy-anthranilate-adenylate, and (b) 2-amino-6-
hydroxy-[1,4]benzoquinone components as specified by ORFs present
in the locus encoding ECO-04601. Biosynthetic enzymes are indicated
by their ORF number and family designation.
FIGURE 15 shows a schematic diagram of the biosynthetic pathway for the
assembly_of the ECO-04601 precursors, farnesyl-diphosphate, 3-
hydroxy-anthranilate-adenylate and 2-amino-6-hydroxy-
[1,4]benzoquinone. Biosynthetic enzymes are indicated by their ORF
number and family designation.
FIGURE 16 shows a sequence listing table indicating the SEQ ID NO. and
function
for each of the open reading frames (ORFs) of the 046D biosynthetic
locus and the corresponding gene product.
FIGURE 17 shows results of the fatty acid analysis of Micromonospora sp.
strain
046EC011 (Accession No. IDAC 070303-01 ). Analysis was conducted
CA 02538147 2004-O1-21
3005-7PCT-9CA
-15-
using gas chromatography on fatty acid methyl esters (FAME).
FIGURE 18 illustrates the 16S ribosomal RNA analysis of Micromonospora sp.
strain 046EC011 (Accession No. IDAC 070303-01). Alignment of 16S
ribosomal RNA sequences demonstrates the phylogenetic relatedness
of Micromonospora sp. strain 046EC011 (indicated as MID352
ECOPIA#1 con) to Micromonospora chalcea.
FIGURE 19 shows the complete'H and'3C NMR assignments for ECO-04601
when measured in MeOH-d4.
DETAILED DESCRIPTION OF THE INVENTION
The present invention relates to a novel farnesyl dibenzodiazepinone,
referred to herein as "ECO-04601," which was isolated from novel strains of
actinomycetes, Micromonospora sp. strain 046-EC011 and strain [S01]046. These
microorganisms were analysed using gas chromatography as Fatty acid methyl
esters (FAME) (Figure 17) 6S ribosomal RNA determination (Figure 18) and were
found to belong to the genus of Micromonospora. These organisms were deposited
on March 7, 2003, and December 23, 2003, respectively, with the International
Depository Authority of Canada (IDAC), Bureau of Microbiology, Health Canada,
1015 Arlington Street, Winnipeg, Manitoba, Canada R3E 3R2, under Accession
Nos. I DAC 070303-01 and I DAC 231203-01, respectively.
The invention further relates to pharmaceutically acceptable salts and
derivatives of ECO-04601, and to methods for obtaining such compounds. One
method of obtaining the compound is by cultivating Micromonospora sp. strain
046-
EC011, or a mutant or a variant thereof, under suitable Micromonospora culture
conditions, preferably using the fermentation protocol described hereinbelow.
The invention also relates to a method for producing novel polyketide
compounds, namely farnesyl dibenzodiazepinones, by selectively altering the
genetic information of an organism. The present invention further provides
isolated
and purified polynucleotides that encode farnesyl dibenzodiazepinone domains,
i.e.,
polypeptides from farnesyl dibenzodiazepinone-producing microorganisms,
fragments thereof, vectors containing those polynucleotides, and host cells
transformed with those vectors. These polynucleotides, fragments thereof, and
CA 02538147 2004-O1-21
3005-7PCT-9CA
-16-
vectors comprising the polynucleotides can be used as reagents in the above
described method. Portions of the polynucleotide sequences disclosed herein
are
also useful as primers for the amplification of DNA or as probes to identify
related
domains from other farnesyl dibenzodiazepinone producing microorganisms.
The present invention also relates to pharmaceutical compositions comprising
ECO-04601 and its pharmaceutically acceptable salts and derivatives. ECO-04601
is useful as a pharmaceutical, in particular for use as an inhibitor of cancer
cell
growth, bacterial cell growth, and mammalian lipoxygenase. The invention also
relates to novel polynucleotide sequences and their encoded proteins, which
are
involved in the biosynthesis of ECO-04601.
The following detailed description discloses how to make and use ECO-
04601 and compositions containing this compound to inhibit microbial growth
and/or
specific disease pathways.
Accordingly, certain aspects of the present invention relate to pharmaceutical
compositions comprising the farnesylated dibenzodiazepinone compounds of the
present invention together with a pharmaceutically acceptable carrier, methods
of
using the compositions to inhibit bacterial growth, and methods of using the
pharmaceutical compositions to treat diseases, including cancer, and chronic
and
acute inflammation.
I. Definitions
For convenience, the meaning of certain terms and phrases used in the
specification, examples, and appended claims, are provided below.
As used herein, the term "farnesyl dibenzodiazepinone" refers to a class of
dibenzodiazepinone compounds containing a farnesyl moiety. The term includes,
but is not limited to, the exemplified compound of the present invention, 10-
farnesyl-
4,6,3-trihydroxy-dibenzodiazepin-11-one, which is referred to herein as "ECO-
04601." As used herein, the term "farnesyl dibenzodiazepinone" includes
compounds of this class that can be used as intermediates in chemical
syntheses.
As used herein, the term "alkyl" refers to linear or branched hydrocarbon
groups.
Examples of alkyl groups include, without limitation, methyl, ethyl, n-propyl,
isopropyl, n-butyl, pentyl, hexyl, heptyl, cyclopentyl, cyclohexyl,
cyclohexymethyl,
and the like. Alkyl may optionally be substituted with substituents selected
from
CA 02538147 2004-O1-21
3005-7PCT-9CA
-17-
acyl, amino, acylamino, acyloxy, carboalkoxy, carboxy, carboxyamido, cyano,
halo,
hydroxyl, nitro, thio, alkyl, alkenyl, alkynyl, cycloalkyl, heterocyclyl,
aryl, heteroaryl,
alkoxy, aryloxy, sulfinyl, sulfonyl, oxo, guanidino and formyl.
The term "alkenyl" refers to linear, branched or cyclic hydrocarbon groups
containing at least one carbon-carbon double bond. Examples of alkenyl groups
include, without limitation, vinyl, 1-propen-2-yl, 1-buten-4-yl, 2-buten-4-yl,
1-penten-
5-yl and the like. Alkenyl may optionally be substituted with substituents
selected
from acyl, amino, acylamino, acyloxy, carboalkoxy, carboxy, carboxyamido,
cyano,
halo, hydroxyl, nitro, thio, alkyl, alkenyl, alkynyl, cycloalkyl,
heterocyclyl, aryl,
heteroaryl, alkoxy, aryloxy, sulfinyl, sulfonyl, formyl, oxo and guanidino.
The double
bond portions) of the unsaturated hydrocarbon chain may be either in the cis
or
traps configuration.
The terms "cycloalkyl" and "cycloalkyl ring" refer to a saturated or partially
unsaturated carbocyclic ring in a single or fused carbocyclic ring system
having from
three to fifteen ring members. Examples of cycloalkyl groups include, without
limitation, cyclopropyl, cyclobutyl, cyclohexyl, and cycloheptyl. Cycloalkyl
may
optionally be substituted with substituents selected from acyl, amino,
acylamino,
acyloxy, carboalkoxy, carboxy, carboxyamido, cyano, halo, hydroxyl, nitro,
thio, alkyl,
alkenyl, alkynyl, cycloalkyl, heterocyclyl, aryl, heteroaryl, alkoxy, aryloxy,
sulfinyl,
sulfonyl and formyl.
The terms "heterocyclyl" and "heterocyclic" refer to a saturated or partially
unsaturated ring containing one to four hetero atoms or hetero groups selected
from
O, N, NH, NRx, P02, S, SO or S02 in a single or fused heterocyclic ring system
having from three to fifteen ring members. Examples of a heterocyclyl or
heterocyclic ring include, without limitation, morpholinyl, piperidinyl, and
pyrrolidinyl.
~leterocyclyl, heterocyclic or heterocyclyl ring may optionally be substituted
with
substituents selected from acyl, amino, acylamino, acyloxy, oxo, thiocarbonyl,
imino,
carboalkoxy, carboxy, carboxyamido, cyano, halo, hydroxyl, nitro, thio, alkyl,
alkenyl,
alkynyl, cycloalkyl, heterocyclyl, aryl, heteroaryl, alkoxy, aryloxy,
sulfinyl, sulfonyl and
formyl.
The term "amino acid" refers to any natural amino acid, all natural amino
acids are well known to a person skilled in the art.
CA 02538147 2004-O1-21
X005-7PCT-9CA
-18-
The term "halo" refers to a halogen atom, e.g., bromine, chlorine, fluorine
and
iodine.
The terms "aryl" and "aryl ring" refer to aromatic groups in a single or fused
ring system, having from five to fifteen ring members. Examples of aryl
include,
without limitation, phenyl, naphthyl, biphenyl, terphenyl. Aryl may optionally
be
substituted with one or more substituent group selected from acyl, amino,
acylamino,
acyloxy, azido, alkythio, carboalkoxy, carboxy, carboxyamido, cyano, halo,
hydroxyl,
nitro, thio, alkyl, alkenyl, alkynyl, cycloalkyl, heterocyclyl, aryl,
heteroaryl, alkoxy,
aryloxy, sulfinyl, sulfonyl and formyl.
The terms "heteroaryl" and "heteroaryl ring" refer to aromatic groups in a
single or fused ring system, having from five to fifteen ring members and
containing
at least one hetero atom such as O, N, S, SO and S02. Examples of heteroaryl
groups include, without limitation, pyridinyl, thiazolyl, thiadiazoyl,
isoquinolinyl,
pyrazolyl, oxazolyl, oxadiazoyl, triazolyl, and pyrrolyl groups. Heteroaryl
groups may
opitionally be substituted with one or more substituent group selected from
acyl,
amino, acylamino, acyloxy, carboalkoxy, carboxy, carboxyamido, cyano, halo,
hydroxyl, nitro, thio, thiocarbonyl, alkyl, alkenyl, alkynyl, cycloalkyl,
heterocyclyl, aryl,
heteroaryl, alkoxy, aryloxy, sulfinyl, sulfonyl, and formyl.
The terms "aralkyl" and "heteroaralkyl" refer to an aryl group or a heteroaryl
group, respectively bonded directly through an alkyl group, such as benzyl.
Aralkyl
and heteroaralkyl may be optionally substituted as the aryl and heteroaryl
groups.
Similarly, the terms "aralkenyl" and "heteroaralkenyl" refer to an aryl group
or
a heteroaryl group, respectively bonded directly through an alkene group, such
as
benzyl. Aralkenyl and heteroaralkenyl may be optionally substituted as the
aryl and
heteroaryl groups.
The compounds of the present invention can possess one or more
asymmetric carbon atoms and can exist as optical isomers forming mixtures of
racemic or non-racemic compounds. The compounds of the present invention are
useful as single isomers or as a mixture of stereochemical isomeric forms.
~iastereoisomers, i.e., nonsuperimposable stereochemical isomers, can be
separated by conventional means such as chromatography, distillation,
crystallization or sublimation. The optical isomers can be obtained by
resolution of
the racemic mixtures according to conventional processes.
CA 02538147 2004-O1-21
3005-7PCT-9CA
-19-
The invention encompasses isolated or purified compounds. An "isolated" or
"purified" compound refers to a compound which represents at least 10%, 20%,
50%, 80% or 90% of the compound of the present invention present in a mixture,
provided that the mixture comprising the compound of the invention has
demonstrable (i.e. statistically significant) biological activity including
antibacterial,
cytostatic, cytotoxic, antiinflammatory or enzyme inhibitory action when
tested in
conventional biological assays known to a person skilled in the art.
The terms "farnesyl dibenzodiazepinone-producing microorganism" and
"producer of farnesyl dibenzodiazepinone," as used herein, refer to a
microorganism
that carries genetic information necessary to produce a farnesyl
dibenzodiazepinone
compound, whether or not the organism naturally produces the compound. The
terms apply equally to organisms in which the genetic information to produce
the
farnesyl dibenzodiazepinone compound is found in the organism as it exists in
its
natural environment, and to organisms in which the genetic information is
introduced
by recombinant techniques.
Specific organisms contemplated herein include, without limitation, organisms
of the family Micromonosporaceae, of which preferred genera include
Micromonospora, Actinoplanes and Dactylosporangium; the family
Streptomycetaceae, of which preferred genera include Streptomyces and
Kitasatospora; the family Pseudonocardiaceae, of which preferred genera are
Amycolatopsis and Saccharopolyspora; and the family Actinosynnemataceae, of
which preferred genera include Saccharothrix and Actinosynnema; however the
terms are intended to encompass all organisms containing genetic information
necessary to produce a farnesyl dibenzodiazepinone compound. A preferred
producer of a farnesyl dibenzodiazepinone compound includes microbial strain
046-
EC011, a deposit of which was made on March 7, 2003, with the International
Depository Authority of Canada (IDAC), Bureau of Microbiology, Health Canada,
1015 Arlington Street, Winnipeg, Manitoba, Canada R3E 3R2, under Accession No.
I DAC 070303-01.
The term "gene" means the segment of DNA involved in producing a
polypeptide chain; it includes regions preceding and following the coding
region
(leader and trailer) as well as, where applicable, intervening regions
(introns)
between individual coding segments (exons).
CA 02538147 2004-O1-21
3005-7PCT-9CA
-20-
The terms "gene locus, "gene cluster," and "biosynthetic locus" refer to a
group of genes or variants thereof involved in the biosynthesis of a farnesyl
benzodiazepinone compound. The biosynthetic locus in strain 046-EC011 that
directs the production of ECO-04601 is often referred to herein, in both the
written
description and Figures, as "046D." Genetic modification of gene locus, gene
cluster
or biosynthetic locus refers to any genetic recombinant techniques known in
the art
including mutagenesis, inactivation, or replacement of nucleic acids that can
be
applied to generate variants of ECO-04601.
A DNA or nucleotide "coding sequence" or "sequence encoding" a particular
polypeptide or protein, is a DNA sequence which is transcribed and translated
into a
polypeptide or protein when placed under the control of an appropriate
regulatory
sequence.
"Oligonucleotide" refers to a nucleic acid, generally of at least 10,
preferably
15 and more preferably at least 20 nucleotides in length, preferably no more
than
100 nucleotides in length, that are hybridizable to a genomic DNA molecule, a
cDNA
molecule, or an mRNA molecule encoding a gene, mRNA, cDNA or other nucleic
acid of interest.
A promoter sequence is "operably linked to" a coding sequence recognized by
RNA polymerise which initiates transcription at the promoter and transcribes
the
coding sequence into mRNA.
The term "replicon" as used herein means any genetic element, such as a
plasmid, cosmid, chromosome or virus, that behaves as an autonomous unit of
polynucleotide replication within a cell. A "expression vector" or "vector" is
a replicon
in which another polynucleotide fragment is attached, such as to bring about
the
replication and/or expression of the attached fragment. "Plasmids" are
designated
herein by a lower case "p" preceded or followed by capital letters and/or
numbers.
The starting plasmids disclosed herein are commercially available, publicly
available
on an unrestricted basis, or can be constructed from available plasmids in
accordance with published procedures. In addition, equivalent plasmids to
those
described herein are known in the art and will be apparent to the skilled
artisan.
The terms "express" and "expression" means allowing or causing the
information in a gene or DNA sequence to become manifest, for example
producing
a protein by activating the cellular functions involved in transcription and
translation
CA 02538147 2004-O1-21
0005-7~CT-9CA
-21-
of a corresponding gene or DNA sequence. A DNA sequence is expressed in or by
a
cell to form an "expression product" such as a protein. The expression product
itself,
e.g. the resulting protein, may also be said to be "expressed" by the cell. An
expression product can be characterized as intracellular, extracellular or
secreted.
"Digestion" of DNA refers to enzymatic cleavage of the DNA with a restriction
enzyme that acts only at certain sequences in the DNA. The various restriction
enzymes used herein are commercially available and their reaction conditions,
cofactors and other requirements were used as would be known to the ordinary
skilled artisan. For analytical purposes, typically 1 pg of plasmid or DNA
fragment is
used with about 2 units of enzyme in about 20 pl of buffer solution. For the
purpose
of isolating DNA fragments for plasmid construction, typically 5 to 50 pg of
DNA are
digested with 20 to 250 units of enzyme in a larger volume. Appropriate
buffers and
substrate amounts for particular enzymes are specified by the manufacturer.
Incubation times of about 1 hour at 37°C are ordinarily used, but may
vary in
accordance with the supplier's instructions. After digestion the gel
electrophoresis
may be performed to isolate the desired fragment.
The term "isolated" as used herein means that the material is removed from
its original environment (e.g. the natural environment where the material is
naturally
occurring). For example, a naturally occurring polynucleotide or polypeptide
present
in a living animal is not isolated, but the same polynucleotide or
polypeptide, which is
separated from some or all of the coexisting materials in the natural system,
is
isolated. Such polynucleotides could be part of a vector and/or such
polynucleotides
or polypeptides could be part of a composition, and still be isolated in that
the vector
or composition is not part of the natural environment.
The term "restriction fragment" as used herein refers to any linear DNA
generated by the action of one or more restriction enzymes.
The term "transformation" means the introduction of a foreign gene; foreign
nucleic acid, DNA or RNA sequence to a host cell, so that the host cell will
express
the introduced gene or sequence to produce a desired substance, typically a
protein
or enzyme coded by the introduced gene or sequence. The introduced gene or
sequence may also be called a "cloned" or "foreign" gene or sequence, may
include
regulatory or control sequences, such as start, stop, promoter, signal,
secretion, or
other sequences used by a cell's genetic machinery. The gene or sequence may
CA 02538147 2004-O1-21
3005-7PCT-9CA
-22-
include nonfunctional sequences or sequences with no known function. A host
cell
that receives and expresses introduced DNA or RNA has been "transformed" and
is
a "transformant" or a "clone" or "recombinant". The DNA or RNA introduced to a
host cell can come from any source, including cells of the same genus or
species as
the host cell, or cells of a different genus or species.
The terms "recombinant polynucleotide" and "recombinant polypeptide" as
used herein mean a polynucleotide or polypeptide which by virtue of its origin
or
manipulation is not associated with all or a portion of the polynucleotide or
polypeptide with which it is associated in nature and/or is linked to a
polynucleotide
or polypeptide other than that to which it is linked in nature.
The term "host cell" as used herein, refer to both prokaryotic and eukaryotic
cells which are used as recipients of the recombinant polynucleotides and
vectors
provided herein. In one embodiment, the host cell is a prokaryote.
The terms "open reading frame" and "ORF" as used herein refers to a region
of a polynucleotide sequence which encodes a polypeptide; this region may
represent a portion of a coding sequence or a total coding sequence.
As used herein and as known in the art, the term "identity" is the
relationship
between two or more polynucleotide sequences, as determined by comparing the
sequences. Identity also means the degree of sequence relatedness between
polynucleotide sequences, as determined by the match between strings of such
sequences. Identity can be readily calculated (see, e.g., Computation
Molecular
Biology, Lesk, A.M., eds., Oxford University Press, New York (1998), and
Biocomputing: Informatics and Genome Projects, Smith, D.W., ed., Academic
Press,
New York (1993), both of which are incorporated by reference herein). While
there
exist a number of methods to measure identity between two polynucleotide
sequences, the term is well known to skilled artisans (see, e.g., Sequence
Analysis
in Molecular Biology, von Heinje, G., Academic Press (1987); and Sequence
Analysis Primer, Gribskov., M. and Devereux, J., eds., M. Stockton Press, New
York
(1991 )). Methods commonly employed to determine identity between sequences
include, for example, those disclosed in Carillo, H., and Lipman, D., SIAM J.
Applied
Math. (1988) 48:1073. "Substantially identical," as used herein, means there
is a
very high degree of homology (preferably 100% sequence identity) between
subject
polynucleotide sequences. However, polynucleotides having greater than 90%, or
CA 02538147 2004-O1-21
3005-7PCT-9CA
-23-
95% sequence identity may be used in the present invention, and thus sequence
variations that might be expected due to genetic mutation, strain
polymorphism, or
evolutionary divergence can be tolerated.
As used herein, the term "treatment" refers to the application or
administration
of a therapeutic agent to a patient, or application or administration of a
therapeutic
agent to an isolated tissue or cell line from a patient, who has a disorder,
e.g., a
disease or condition, a symptom of disease, or a predisposition toward a
disease,
with the purpose to cure, heal, alleviate, relieve, alter, remedy, ameliorate,
improve,
or affect the disease, the symptoms of disease, or the predisposition toward
disease.
As used herein, a "pharmaceutical composition" comprises a
pharmacologically effective amount of a farnesyl dibenzodiazepinone and a
pharmaceutically acceptable carrier. As used herein, "pharmacologically
effective
amount," "therapeutically effective amount" or simply "effective amount"
refers to that
amount of a farnesyl dibenzodiazepinone effective to produce the intended
pharmacological, therapeutic or preventive result. For example, if a given
clinical
treatment is considered effective when there is at least a 25% reduction in a
measurable parameter associated with a disease or disorder, a therapeutically
effective amount of a drug for the treatment of that disease or disorder is
the amount
necessary to effect at least a 25% reduction in that parameter.
The term "pharmaceutically acceptable carrier" refers to a carrier for
administration of a therapeutic agent. Such carriers include, but are not
limited to,
saline, buffered saline, dextrose, water, glycerol, ethanol, and combinations
thereof.
The term specifically excludes cell culture medium. For drugs administered
orally,
pharmaceutically acceptable carriers include, but are not limited to
pharmaceutically
acceptable excipients such as inert diluents, disintegrating agents, binding
agents,
lubricating agents, sweetening agents, flavoring agents, coloring agents and
preservatives. Suitable inert diluents include sodium and calcium carbonate,
sodium
and calcium phosphate, and lactose, while corn starch and alginic acid are
suitable
disintegrating agents. Binding agents may include starch and gelatin, while
the
lubricating agent, if present, will generally be magnesium stearate, stearic
acid or
talc. If desired, the tablets may be coated with a material such as glyceryl
monostearate or glyceryl distearate, to delay absorption in the
gastrointestinal tract.
CA 02538147 2004-O1-21
~00~-7PCT-9CA
-24-
The term "pharmaceutically acceptable salt" refers to both acid addition salts
and base addition salts. The nature of the salt is not critical, provided that
it is
pharmaceutically acceptable. Exemplary acid addition salts include, without
limitation, hydrochloric, hydrobromic, hydroiodic, nitric, carbonic,
sulphuric,
phosphoric, formic, acetic, citric, tartaric, succinic, oxalic, malic,
glutamic, propionic,
glycolic, gluconic, malefic, embonic (pamoic), methanesulfonic,
ethanesulfonic, 2-
hydroxyethanesulfonic, pantothenic, benzenesulfonic, toluenesulfonic,
sulfanilic,
mesylic, cyclohexylaminosulfonic, stearic, algenic, /3-hydroxybutyric,
malonic,
galactaric, galacturonic acid and the like. Suitable pharmaceutically
acceptable
base addition salts include, without limitation, metallic salts made from
aluminium,
calcium, lithium, magnesium, potassium, sodium and zinc or organic salts made
from N,N'-dibenzylethylenediamine, chloroprocaine, choline, diethanolamine,
ethylenediamine, N-methylglucamine, lysine, procaine and the like. Additional
examples of pharmaceutically acceptable salts are listed in Journal of
Pharmaceutical Sciences (1977) 66:2. All of these salts may be prepared by
conventional means from a farnesyi dibenzodiazepinone by treating the compound
with the appropriate acid or base.
II. Farnesylated Dibenzodiazepinone Compounds
In one aspect, the invention relates to a novel farnesyl dibenzodiazepinone,
referred to herein as "ECO-04601" and having the chemical structure
represented by
the following formula:
0
N
UH
N
OH
HO
ECO-04601 may be described as a new dibenzodiazepinone having a 10-
farnesyl substituent located on the nitrogen atom in the 10 position of the
dibenzodiazepine ring (i.e., the amide nitrogen in the diazepinone ring), and
three
CA 02538147 2004-O1-21
3005-7PCT-9CA
-25-
phenolic hydroxy substituents in the 4,6 and 8 positions of the
dibenzodiazepinone
ring. ECO-04601 may be characterized by any one or more of its physicochemical
and spectral properties given below, such as its mass, UV, and NMR
spectroscopic
data. Mass was determined by electrospray mass spectrometry to be 462.6
(FIGURE 1 ); UV = 230 nm with a shoulder at 290nm (FIGURE 2). NMR data were
collected using MeOH- d4, including proton (FIGURE 3), and multidimensional
pulse
sequences gDQCOSY (FIGURE 4), gHSOC (FIGURE 5), gHMBC (FIGURE 6), and
NOESY (FIGURE 7).
In another aspect, the invention relates to a novel class of farnesyl
dibenzodiazepinone compounds represented by Formula I:
~W~ 3~CH3
W
Formula I.
wherein,
R4
W1, W2 and W3 is each independently selected from
H CH3
CH3 CH3 CH3
~_C? H-~- , _~-H-C-~_ ; _~-C-C-~_ ; ; Or
\O OH OH
the chain from the tricycle may terminate at W3, W2 or W1 with W3, W2 or
W 1 respectively being either -CH=O or -CH20H;
A is selected from -NH--, -NCH2R1, -NC(O)R1;
R1 is selected from C1-6 alkyl, C2-6 alkene, aryl or heteroaryl;
R2, R3, and R4 is each independently selected from H, R5, -C(O)RE
R5 is each independently selected from C1_6 alkyl, C2_7 alkalene, aryl or
CA 02538147 2004-O1-21
3Q0~-7PCT-9CA
heteroaryl;
-26-
R~ is each independently selected from H, C~ _g alkyl, C2_7 alkalene, aryl or
heteroaryl; or a pharmaceutically acceptable salt thereof.
In other embodiments, the invention provides compounds of Formula I,
wherein A is selected from the group cansisting of NH, NCH2R1, and NC(O)R~ ;
wherein R2 is H; R3 is H; and R4 is H. In another embodiment, R2, R3 and R4
are
each H; and all other groups are as previously defined. In a further
embodiment,
R2, R3 and R4 are each H; ar7d W1 is -CH =CH- and all other groups are as
previously defined. In a further embodiment, R2, R3 and R4 are each H, and W2
is
-CH =CH- and all other groups are as previously defined. In a further
embodiment,
R2, R3 and R4 are each H; and V'J3 is -CH =CH- ; and all other groups are as
previously defined. In a further embodiment, A is NH; R2, R3 and R4 are each
H;
and all other groups are as previously defined. In a further embodiment, A is
NHn
each of W ~ , W2, and W3 is -CH =CH-; and all other groups are as previously
defined. The invention encompasses ail pharmaceutically acceptable salts of
the
foregoing compounds.
The following are exemplary compounds of the invention:
CA 02538147 2004-O1-21
X005-7~CT-9~A
-27-
O N
Oli
NY
OH
HO
Formula II
O CH, CH, CH,
0 CH, CH, CH, i N / / / CH,
/ N / / / CH ' I N
N ~ ~ HO ~H ~ OH
HO ~ OH
0-C ~ HO
H3C HO ~ /
Formula lii Formula IV
o cH, cH, cH,
N / / / CH
N
HO J/ _ OH
H'C/ HO .
Formula V Formula VI
CH, CH, CH, 0 CH, CH, CH,
N / / CH, /N / / CH,
/ o ~ / o
HO H ~ ~OH HO H ~OH
HO ~ ~ HO
Formula VII Formula VIII
O CH, CH, CH, O CH, CH, CH,
~~N/ / / ~ CH, / N / CH,
O \ / O O
HO ~ ~_~OH HO H ~ ~ OH
HO ~ HO
Formula VIX Formula X
CA 02538147 2004-O1-21
3005-7PCT-9CA
-28-
O CH; CH; CH, O CH; CH; CH,
I
~i v
~~~ N 0 O CH' ~ / N/ / O O CH
I N / N /
HO H ~ OH HO H _ ~ OH
HO ~ HO
Formula XI Formula XII
O CH, CH, CH; O CH; CH; CHI
i
i i
N ~ CH; N / CH;
O 0 O \ ~ O
/ ~ % /
HO H _ OH HO H OH
HO ~ HO
Formula XIII Formula XIV
o cH; cH; cH; o cH; cH, cH;
N CH; ~ i ~CH;
0 \ 0
------(( \ N~ /
HO H ~OH HO H ~OH
HO/~~/ ~ HO~
Formula XV Formula XVI
0 CH; CH; H 0 i H; CH, CH;
N I I OH; N ~ CH;
O 0 O O
/ ~ % /
HO H OH HO H _ OH
HO ~ HO
Formula XVII Formula XVIII
0 CH, CH; i H; O CH, CH; CH,
i
N CH; N / CH;
O O ~ ~ 0
N /
HO N /' OH HO H OH
HO ~ HO
Formula XIX Formula XX
CA 02538147 2004-O1-21
3005-7PCT-9CA
-29-
O CH, CH, CH, O CH, CH, CH,
N / CH, ~ ~ N / CH,
0 O
N / ~ N /
HO H _ OH HO H _ OH
HO ~ HO
Formula XXI Formula XXII
0 CH, CH, CH, O CH, CH, CH,
i i
~N CH, ~ N CH,
0 ~ 0
/ ~ % /
HO H ' OH HO H _ OH
HO ~ HO
Formula XXIII Formula XXIV
0 CH, CH, CH, O CH, CH, CH,
i i
N CH, ~ N / / / CH,
0
°\ ~ /
NO H OH ~~0 H _ OH
HO ~ H'C HO
Formula XXV Formula XXVI
0 CH, CH, CH,
N / / / CH,
N ~' ~~Of
HO ~ \\ / \O \
H ~ CH
HO
Formula XXVII Formula XXVIII
0
H,y.
Formula XXIX Formula XXX
CA 02538147 2004-O1-21
X005-71'CT-9CA
O CH, CH, CH,
N / / / CH
I
O ~ ~ O
N IIII
O /
H O~CH,
HO
-30-
Formula XXXI Formula XXXII
0 CH, CH, CH, O CH, CH, CH,
N / / / CH, / / N / / / CH,
N ~ \ HO N
H,C'O ~ OH H -_-~~OH
H
H3C~0
HU
Formula XXXIII Formula XXXIV
cH, cH,
o cH, cH, cH, / /
CH,
N / / / CH
N ~
Ho i ~ o-cH,
r;
Ho ;
Formula XXXV Formula XXXVI
O CH, CH, CH,
0 CH, CH, CH,
N / / / CH,
N / / / CH
/~~ (, /
N \
l //~~
HO H ~~O~CH, ~ \N~~~\
O / H,C~O ~ ~O~CH,
/~ /H
CH, 9 HO ?
Formula XXXVII Formula XXXVIII
CA 02538147 2004-O1-21
3005-7PCT-9CA
-31-
O ~ H3 CH3 CH3
i
N / / CH3
O N
HO j OH
H
;, HO
Formula XXXIX Formula XL
O CH3 "H3 CH3 0 CH3 CH3 CH3
II I ~ i
~'~N /" " CH3 ~ / N / / "' CH3
/ N ~ \ ~ N
HO H _ OH HO H _ OH
HO , HO
Formula XLI Formula XLII
O CH3 CHj CH3 O CH3 CH3 CHI
I
N / CH3 ~ ~ N / CH3
HO H ~ ~ OH HO H ~ ~ OH
HO . HO
Formula XLIII Formula XLIV
O CH3 CH~ CH3 O CHa CH3 CH3
I I
'/ ~N /CHI N CH3
I N ~ \ N
HO H _ OH 0 H ~ OH
HO , HO
Formula XLV Formula XLVI
O CH, i.H, O CH, CH,
~/~\ i
I N ~ ~ N
HO H _ ~ OH HO H ~ OH
HO ~ HO
Formula XLVII Formula XLVIII
CA 02538147 2004-O1-21
X005-7PCT-9CA
-32-
O CH, O CH,
i ~ /O ~ ~ OH
\ ~ ~N \ ~ N
/_ \ % /_ \
HO H OH HO H OH
HO , H / 7
Formula XLIX Formula L
0 0
\ ,o ~ ~oH
\ ~ N~ \ ~ N
/ \ -~/ \
HO HN _ OH HO H _ OH
HO ~ , HO
Formula LI Formula LII
0 CH, CH, CH, 0 CH, CH, CH,
N /~ / CH, / N / / CH,
\ / H \ / ~ OH
~/ \ OH OH
He HN / \ OH HO H / \ OH
HO y HO
Formula LIII Formula LIV
CH, CH, CH, O CH, CH CH
i ~ /~ ~ i ~ ~/ ~~I
N~~~CH, N ~ " " CH,
\ OH \ ~ OH OH
OH ~ OH OH
/ ~ N
HO H ~ _ \ OH HO H _ OH
H~ ~ HO
Formula LV Formula LVI
O CH, CH, CH, O CH, CH, CH,
N / CH, ~ N / CH,
\ ~ OH OH OH ~ OH
OH OH \ OH OH
I N--~ I N \
HG H ~~OH O H _ OH
HO ~/ ~ HO ~ c'LIZ
Formula LVII Formula LVIII
CA 02538147 2004-O1-21
3005-7PCT-9CA
-33-
OH3 CH3 CH3
/ ~~ I~ \~ ~ \~ CHI
OH ~ OH OH
~/~\ OH OH OH
/ N
HO /
H 'OH
HO
Formula LIX
Certain embodiments expressly exclude one or more of the compounds of
Formula I. In one embodiment, the compound of Formula II is excluded.
The compounds of this invention may be formulated into pharmaceutical
compositions comprised of compounds of Formula I in combination with a
pharmaceutical acceptable carrier, as discussed in Section V below.
III. Method of MakincLa Farnesyl Dibenzodiazepinone by Fermentation
In one embodiment, ECO-04601 is obtained by cultivating a novel strain of
Micromonospora, namely Micromonospora sp. strain 046-EC011. Strain 046-
EC011 was deposited on March 7, 2003, with the International Depositary
Authority
of Canada (IDAC), Bureau of Microbiology, Health Canada, 1015 Arlington
Street,
Winnipeg, Manitoba, Canada R3E 3R2, under Accession No. 070303-01. The
deposit of the strain was made under the terms of the Budapest Treaty on the
International Recognition of the Deposit of Microorganisms for Purposes of
Patent
Procedure. The deposited strains will be irrevocably and without restriction
or
condition released to the public upon the issuance of a patent. The deposited
strains are provided merely as convenience to those skilled in the art and are
not an
admission that a deposit is required for enablement, such as that required
under 35
U.S.C. ~112.
It is to be understood that the present invention is not limited to use of the
particular strain 046-EC011. Rather, the present invention contemplates the
use of
other ECO-04601 producing organisms, such as mutants or variants of 046-ECO11
that can be derived from this organism by known means such as X-ray
irradiation,
ultraviolet irradiation, treatment with nitrogen mustard, phage exposure,
antibiotic
selection and the like; or through the use of recombinant genetic engineering
techniques, as described in Section IV below.
The farnesyl dibenzodiazepinone compounds of the present invention may be
CA 02538147 2004-O1-21
3005-7PCT-9CA
-34-
biosynthesized by various microorganisms. Microorganisms that may synthesize
the
compounds of the present invention include but are not limited to bacteria of
the
order Actinomycetales, also referred to as actinomycetes. Non-limiting
examples of
members belonging to the genera of Actinomycetes include Nocardia,
Geodermatophilus, Actinoplanes, Micromonospora, Nocardioides, Saccharothrix,
Amycolatopsis, Kutzneria, Saccharomonospora, Saccharopolyspora, Kitasatospora,
Streptomyces, Microbispora, Streptosporangium, and Actinomadura. The taxonomy
of actinomycetes is complex and reference is made to Goodfellow, Suprageneric
Classification ofActinomycetes (1989); 8ergey's Manual of Systematic
bacteriology,
Voi. 4 (Williams and Wilkins, Baltimore, pp. 2322-2339); and to Embley and
Stackebrandt, ''The molecular phylogeny and systematics of the
actinomycetes,"'
Annu. Rev. Microbiol. (1994) 48:257-289, each of which is hereby incorporated
by
reference in its entirety, for genera that may synthesize the compounds of the
invention.
Farnesyl dibenzodiazepinone-producing microorganisms are cultivated in
culture medium containing known nutritional sources for actinomycetes. Such
media
having assimilable sources of carbon, nitrogen, plus optional inorganic salts
and
other known growth factors at a pH of about 6 to about 9. Suitable media
include,
without limitation, the growth media provided in Table 16. Microorganisms are
cultivated at incubation temperatures of about 18 °C to about 40
°C for about 3 to
about 40 days.
The culture media inoculated with the farnesyl dibenzodiazepinone-producing
microorganisms may be aerated by incubating the inoculated culture media with
agitation, for example, shaking on a rotary shaker, or a shaking water bath.
Aeration
may also be achieved by the injection of air, oxygen or an appropriate gaseous
mixture to the inoculated culture media during incubation. Following
cultivation, the
farnesyl dibenzodiazepinone compounds can be extracted and isolated from the
cultivated culture media by techniques known to a skilled person in the art
and/or
disclosed herein, including for example centrifugation, chromatography,
adsorption,
filtration. For example, the cultivated culture media can be mixed with a
suitable
organic solvent such as n-butanol, n-butyl acetate or 4-methyl-2-pentanone,
the
organic layer can be separated for example, by centrifugation followed by the
removal of the solvent, by evaporation to dryness or by evaporation to dryness
CA 02538147 2004-O1-21
~00~-71'CT-9CA
-35-
under vacuum. The resulting residue can optionally be reconstituted with for
example water, ethanol, ethyl acetate, methanol or a mixture thereof, and re-
extracted with a suitable organic solvent such as hexane, carbon
tetrachloride,
methylene chloride or a mixture thereof. Following removal of the solvent, the
compounds may be further purified by the use of standard techniques, such as
chromatography.
The farnesyl dibenzodiapezinones biosynthesized by microorganisms may
optionally be subjected to random and/or directed chemical modifications to
form
compounds that are derivatives or structural analogs. Such derivatives or
structural
analogs having similar functional activities are within the scope of the
present
invention. Farnesyl dibenzodiapezinone compounds may optionally be modified
using methods known in the art and described herein.
IV. Method of Making a Farnesyl Dibenzodiazepinone by Recombinant
Technology
In another embodiment, the present invention relates to nucleic acid
molecules that encode proteins useful in the production of farnesyl
benzodiazepinones. Specifically, the present invention provides recombinant
DNA
vectors and nucleic acid molecules that encode all or part of the biosynthetic
locus in
strain 046-EC011, which directs the production of ECO-04601, and is referred
to
herein as "046D." The invention further includes genetic modification of 046D
using
conventional genetic recombinant techniques, such as mutagenesis,
inactivation, or
replacement of nucleic acids, to produce chemical variants of ECO-04601.
The invention thus provides a method for making a farnesyl
benzodiazepinone compound using a transformed host cell comprising a
recombinant DNA vector that encodes one or more of the polypeptides of the
present invention, and culturing the host cell under conditions such that
farnesyl
benzodiazepinone is produced. The host cell is a prokaryote. In one
embodiment,
the host cell is an actinomycete. In another embodiment, the host cell is a
Streptomyces host cell.
The invention provides recombinant nucleic acids that produce a variety of
farnesyl dibenzodiazepinone compounds that cannot be readily synthesized by
CA 02538147 2004-O1-21
3005-7PCT-9CA
-36-
chemical methodology alone. The invention allows direct manipulation of 046D
biosynthetic focus via genetic engineering of the enzymes involved in the
biosynthesis of a farnesyl benzodiazepinone according to the invention. The
046A
biosynthetic locus is described in Example 11.
Recombinant DNA Vectors
Vectors of the invention typically comprise the DNA of a transmissible agent,
into which foreign DNA is inserted. A common way to insert one segment of DNA
into another segment of DNA involves the use of specific enzymes called
restriction
enzymes that cleave DNA at specific sites (specific groups of nucleotides)
called
restriction sites. A "cassette" refers to a DNA coding sequence or segment of
DNA
that codes for an expression product that can be inserted into a vector at
defined
restriction sites. The cassette restriction sites are designed to ensure
insertion of the
cassette in the proper reading frame. Generally, a nucleic acid molecule that
encodes a protein useful in the production of a farnesyl benzodiazepinone is
inserted at one or more restriction sites of the vector DNA, and then is
carried by the
vector into a prokaryote e.g. actinomycte, by transformation (see below). A
segment
or sequence of DNA having inserted or added DNA, such as an expression vector,
can also be called a "DNA construct". A common type of vector is a "plasmid"
which
generally is a self-contained molecule of double-stranded DNA, usually of
bacterial
origin, that can readily accept additional (foreign) DNA and which can be
readily
introduced into a suitable host cell. A plasmid vector often contains coding
DNA and
promoter DNA and has one or more restriction sites suitable for inserting
foreign
DNA. Coding DNA is a DNA sequence that encodes a particular amino acid
sequence for a particular protein or enzyme. In one embodiment of the
invention, the
coding DNA encodes for polypeptides of SEQ ID NOs. 3, 5, 7, 9, 11, 13, 15, 17,
19,
21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45, 47, 49, 51, 53, 55, 57,
59, 61, 63,
66, 68, 70, 72, 75, 77, 79, 81, 83, 85, 87 or 89 that are required for the
biosynthesis
of a farnesyl benzodiazepinone.
Promoter DNA of a recombinant vector is a DNA sequence that initiates,
regulates, or otherwise mediates or controls the expression of the coding DNA.
Promoter DNA and coding may be from the same or different organisms.
Recombinant cloning vectors will often include one or more replication systems
for
CA 02538147 2004-O1-21
3005-7PCT-9CA
-37-
cloning or expression, one or more markers for selection in the host, e.g.
antibiotic
resistance, and one or more expression cassettes. Vector constructs may be
produced using conventional molecular biology and recombinant DNA techniques
within the skill of the art. Such techniques are explained fully in the
literature. See,
e.g., Sambrook, Fritsch & Maniatis, Molecular Cloning: A Laboratory Manual,
Second Edition (1989) Cold Spring Harbor Laboratory Press, Cold Spring Harbor,
New York (herein "Sambrook et al., 1989"); DNA Cloning: A Practical Approach,
Volumes I and II (D. N. Glover ed. 1985); F. M. Ausubel et al. (eds.), Current
Protocols in Molecular Biology, John Wiley & Sons, Inc. (1994).
Examples of promoters that function in actinomycetes, e.g. Streptomyces, are
taught in US Patent Nos. 5,830,695 and 5,466,590. Another example of a
transcription promoter useful in Actinomycetes expression vectors is tipA, a
promoter inducible by the antibiotic thiostrepton [c.f. Murakami, T., et al.,
(1989), J.
Bacteriol, 171, 1459].
Transformation of Actinomycetes
A suitable transformation method for use with an actinomycete comprises
forming the actinomycete culture into spheroplasts using lysozyme. A buffer
solution
containing recombinant DNA vectors and polyethylene glycol is then added, in
order
to introduce the vector into the host cells, by using either of the methods of
Thompson or Keiser [c. f. Thompson, C. J., et al., (1982), J. Bacteriol., 151,
668-677
or Keiser, T. et al. (2000), "Practical Streptomyces Genetics", The John lnnes
Foundation, Norwich], for example. A thiostrepton-resistance gene is
frequently used
as a selective marker in the transformation plasrnid [c.f. Hopwood, D. A., et
al.,
(1987), "Methods in Enzymology" 153, 116, Academic Press, New York], but the
present invention is not limited thereto. Additional methods for the
transformation of
actinomycetes are taught in US 5,393,665.
Assay for farnesyl dibenzodiazepinone or biosynthetic intermediates
Actinomycetes defective in farnesyl dibenzodiazepinone biosynthesis are
transformed with one or more expression vectors encoding one or more proteins
in
the farnesyl benzodiazepinone biosynthetic pathway, thus restoring farnesyl
benzodiazepinone biosynthesis by genetic complementation of the specific
defect.
CA 02538147 2004-O1-21
3~05-7PCT-9CA
-38-
The presence or absence of farnesyl dibenzodiazepinone or intermediates in
the biosynthetic pathway (see Figures 13, 14 and 15) in a recombinant
actinomycete
can be determined using methodologies that are well known to persons of skill
in the
art. For example, ethyl acetate extracts of fermentation media used for the
culture of
a recombinant actinomycete are processed as described in Example 2 and
fractions
containing farnesyl dibenzodiazepinone or intermediates detected by TLC on
commercial Kieselgel 60F25a plates. Farnesyl dibenzodiazepinone and
intermediate
compounds are visualized by inspection of dried plates under UV light or by
spraying
the plates with a spray containing vanillin (0.75%) and concentrated sulfuric
acid
(1.5%, v/v) in ethanol and subsequently heating the plate. The exact identity
of the
compounds separated by TLC is then determined using gas chromatography-mass
spectroscopy. Methods of mass spectroscopy are taught in the published U.S.
Patent Application No. US2003/0052268.
Mutac~enesis
The invention allows direct manipulation of 046D biosynthetic locus via
genetic engineering of the enzymes involved in the biosynthesis of a farnesyl
benzodiazepinone according to the invention.
A number of methods are known in the art that permit the random as well as
targeted mutation of the DNA sequences of the invention (see for example,
Ausubel
et. al. Short Protocols in Molecular Biology (1995) 3rd Ed. John Wiley & Sons,
Inc.).
In addition, there are a number of of commercially available kits for site-
directed
mutagenesis, including both conventional and PCR-based methods. Examples
include the E~SITET"' PCR-Based Site-directed Mutagenesis Kit available from
Stratagene (Catalog No. 200502) and the QUIKCHANGETM Site-directed
mutagenesis Kit from Stratagene (Catalog No. 200518), and the CHAMELEON~
double-stranded Site-directed mutagenesis kit, also from Stratagene (Catalog
No.
200509).
In addition the nucleotides of the invention may be generated by insertional
mutation or truncation (N-terminal, internal or C-terminal) according to
methodology
known to a person skilled in the art.
Older methods of site-directed mutagenesis known in the art rely on sub-
cloning of the sequence to be mutated into a vector, such as an M13
bacteriophage
CA 02538147 2004-O1-21
3005-7PCT-9CA
-39-
vector, that allows the isolation of single-stranded DNA template. In these
methods,
one anneals a mutagenic primer (i.e., a primer capable of annealing to the
site to be
mutated but bearing one or more mismatched nucleotides at the site to be
mutated)
to the single-stranded template and then polymerizes the complement of the
template starting from the 3' end of the mutagenic primer. The resulting
duplexes
are then transformed into host bacteria and plaques are screened for the
desired
mutation.
More recently, site-directed mutagenesis has employed PCR methodologies,
which have the advantage of not requiring a single-stranded template. In
addition,
methods have been developed that do not require sub-cloning. Several issues
must
be considered when PCR-based site-directed mutagenesis is performed. First, in
these methods it is desirable to reduce the number of PCR cycles to prevent
expansion of undesired mutations introduced by the polymerise. Second, a
selection must be employed in order to reduce the number of non-mutated
parental
molecules persisting in the reaction. Third, an extended-length PCR method is
preferred in order to allow the use of a single PCR primer set. And fourth,
because
of the non-template-dependent terminal extension activity of some thermostable
polymerises it is often necessary to incorporate an end-polishing step into
the
procedure prior to blunt-end ligation of the PCR-generated mutant product.
The protocol described below accommodates these considerations through
the following steps. First, the template concentration used is approximately
1000-
fold higher than that used in conventional PCR reactions, allowing a reduction
in the
number of cycles from 25-30 down to 5-10 without dramatically reducing product
yield. Second, the restriction endonuclease Dpn I (recognition target
sequence: 5-
Gm6ATC-3, where the A residue is methylated) is used to select against
parental
DNA, since most common strains of E. coli Dam methylate their DNA at the
sequence 5-GATC-3. Third, Taq Extender is used in the PCR mix in order to
increase the proportion of long (i.e., full plasmid length) PCR products.
Finally, Pfu
DNA polymerise is used to polish the ends of the PCR product prior to
intramolecular ligation using T4 DNA ligase.
A non-limiting example for the isolation of mutant polynucleotides is
described
in detail as follows:
Plasmid template DNA (approximately 0.5 pmole) is added to a PCR cocktail
CA 02538147 2004-O1-21
3005-7PCT-9CA
-40-
containing: 1x mutagenesis buffer (20 mM Tris HCI, pH 7.5; 8 mM MgCl2;
40~gi'ml
BSA); 12-20 pmole of each primer (one of skill in the art may design a
mutagenic
primer as necessary, giving consideration to those factors such as base
composition, primer length and intended buffer salt concentrations that affect
the
annealing characteristics of oligonucleotide primers; one primer must contain
the
desired mutation, and one (the same or the other) must contain a 5' phosphate
to
facilitate later ligation), 250 ~M each dNTP, 2.5 U Taq DNA polymerise, and
2.5 U
of Taq Extender (Available from Stratagene; See Nielson et al. (1994)
Strategies 7:
27, and U.S. Patent No. 5,556,772). Primers can be prepared using the triester
method of Matteucci et al., 1981, J. Am. Chem. Soc. 103:3185-3191,
incorporated
herein by reference. Alternatively automated synthesis may be preferred, for
example, on a Biosearch 8700 DNA Synthesizer using cyanoethyl phosphoramidite
chemistry.
The PCR cycling is performed as follows: 1 cycle of 4 min at 94oC, 2 min at
50oC and 2 min at 72oC; followed by 5-10 cycles of 1 min at 94oC, 2 min at
54oC
and 1 min at 72oC. The parental template DNA and the linear, PCR-generated DNA
incorporating the mutagenic primer are treated with Dpnl (10 U) and Pfu DNA
polymerise (2.5U). This results in the Dpnl digestion of the in vivo
methylated
parental template and hybrid DNA and the removal, by Pfu DNA polymerise, of
the
non-template-directed Taq DNA polymerise-extended bases) on the linear PCR
product. The reaction is incubated at 37oC for 30 min and then transferred to
72oC
for an additional 30 min. Mutagenesis buffer (115 ul of 1x) containing 0.5 mM
ATP
is added to the Dpnl-digested, Pfu DNA polymerise-polished PCR products. The
solution is mixed and 10 ul are removed to a new microfuge tube and T4 DNA
ligase
(2-4 U) is added. The ligation is incubated for greater than 60 min at 37oC.
Finally,
the treated solution is transformed into competent E. coli according to
standard
methods.
Methods of random mutagenesis, which will result in a panel of mutants
bearing one or more randomly situated mutations, exist in the art. Such a
panel of
mutants may then be screened for those exhibiting reduced uracil detection
activity
relative to the wild-type polymerise (e.g., by measuring the incorporation of
10nmoles of dNTPs into polymeric form in 30 minutes in the presence of 200~M
dUTP and at the optimal temperature for a given DNA polymerise). An example of
CA 02538147 2004-O1-21
3005-7PCT-9CA
-41-
a method for random mutagenesis is the so-called "error-prone PCR method". As
the name implies, the method amplifies a given sequence under conditions in
which
the DNA polymerise does not support high fidelity incorporation. The
conditions
encouraging error-prone incorporation for different DNA polymerises vary,
however
one skilled in the art may determine such conditions for a given enzyme. A key
variable for many DNA polymerises in the fidelity of amplification is, for
example,
the type and concentration of divalent metal ion in the buffer. The use of
manganese ion and/or variation of the magnesium or manganese ion concentration
may therefore be applied to influence the error rate of the polymerise.
Genes for desired mutant polypeptides generated by mutagenesis may be
sequenced to identify the sites and number of mutations. For those mutants
comprising more than one mutation, the effect of a given mutation may be
evaluated
by introduction of the identified mutation to the wild-type gene by site-
directed
mutagenesis in isolation from the other mutations borne by the particular
mutant.
Screening assays of the single mutant thus produced will then allow the
determination of the effect of that mutation alone.
V. Genes and proteins for the production of ECO-04601
As discussed in more detail below, the isolated, purified or enriched nucleic
acids of one of SEQ I D NOS: 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 2 7,
29, 31,
33, 35, 37, 39, 41, 43, 45, 47, 49, 51, 53, 55, 57, 59, 61, 63, 66, 68, 70,
72, 75, 77,
79, 81, 83, 85, 87 and 89 may be used to prepare one of the polypeptides of
SEQ ID
NOS: 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38,
41, 42, 44,
4C, 48, 50, 52, 54, 56, 58, 60, 62, 65, 67, 69, 70, 71, 74, 76, 78, 80,' 82,
84, 86 and
88, respectively, or fragments comprising at least 50, 75, 100, 200, 300, 500
or more
consecutive amino acids of one of the polypeptides of SEQ ID NO: 2, 4, 6, 8,
10, 12,
14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 41, 42, 44, 46, 48, 50,
52, 54, 56,
58, 60, 62, 65, 67, 69, 70, 71, 74, 76, 78, 80, 82, 84, 86 and 88.
Accordingly, another aspect of the present invention is an isolated, purified
or
enriched nucleic acid which encodes one of the polypeptides of SEQ ID NOS: 2,
4,
5, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 41, 42, 44,
46, 48, 50,
52, 54, 56, 58, 60, 62, 65, 67, 69, 70, 71, 74, 76, 78, 80, 82, 84, 86 and 88
or
CA 02538147 2004-O1-21
3005-7PCT-9CA
-42-
fragments comprising at least 50, 75, 100, 150, 200, 300 or more consecutive
amino
acids of one of the polypeptides of SEQ ID NOS: 2, 4, 6, 8, 10, 12, 14, 16,
18, 20,
22, 24, 26, 28, 30, 32, 34, 36, 38, 41, 42, 44, 46, 48, 50, 52, 54, 56, 58,
60, 62, 65,
67, 69, 70, 71, 74, 76, 78, 80, 82, 84, 86 and 88. The coding sequences of
these
nucleic acids may be identical to one of the coding sequences of one of the
nucleic
acids of SEQ I D NOS: 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31,
33, 35,
37, 39, 41, 43, 45, 47, 49, 51, 53, 55, 57, 59, 61, 63, 66, 68, 70, 72, 75,
77, 79, 81,
83, 85, 87 and 89 or a fragment thereof, or may be different coding sequences
which encode one of the polypeptides of SEQ ID NOS: 2, 4, 6, 8, 10, 12, 14,
16, 18,
20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 41, 42, 44, 46, 48, 50, 52, 54, 56,
58, 60, 62,
65, 67, 69, 70, 71, 74, 76, 78, 80, 82, 84, 86 and 88 or fragments comprising
at least
50, 75, 100, 150, 200, 300 consecutive amino acids of one of the polypeptides
of
SEQ ID NOS: 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34,
36, 38, 41,
42, 44, 46, 48, 50, 52, 54, 56, 58, 60, 62, 65, 67, 69, 70, 71, 74, 76, 78,
80, 82, ~'34,
86 and 88 as a result of the redundancy or degeneracy of the genetic code. The
genetic code is well known to those of skill in the art and can be obtained,
for
example, from Stryer, Biochemistry, 3rd edition, W. H. Freeman & Co., New
York.
The isolated, purified or enriched nucleic acid which encodes one of the
polypeptides of SEQ ID NOS: 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26,
28, 30,
32, 34, 36. 38, 41, 42, 44, 46, 48, 50, 52, 54, 56, 58, 60, 62, 65, 67, 69,
70, 71, 74,
76, 78, 80, 82, 84, 86 and 88 may include, but is not limited to: (1 ) only
the coding
sequences of one of SEQ ID NOS: 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25,
27, 29,
31, 33, 35, 37, 39, 41, 43, 45, 47, 49, 51, 53, 55, 57, 59, 61, 63, 66, 68,
70, 72, 75,
77, 79, 81, 83, 85, 87 and 89; (2) the coding sequences of SEQ ID NOS: 3, 5,
7, 9,
11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45, 47,
49, 51, 53,
55, 57, 59, 61, 63, 66, 68, 70, 72, 75, 77, 79, 81, 83, 85, 87 and 89 and
additional
coding sequences, such as leader sequences or proprotein; and (3) the coding
sequences of SEQ ID NOS: 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29,
31, 33,
35, 37, 39, 41, 43, 45, 47, 49, 51, 53, 55, 57, 59, 61, 63, 66, 68, 70, 72,
75, 77, 79,
81, 83, 85, 87 and 89 and non-coding sequences, such as non-coding sequences
5'
and/or 3' of the coding sequence. Thus, as used herein, the term
"polynucleotide
encoding a polypeptide" encompasses a polynucleotide that includes only coding
sequence for the polypeptide as well as a polynucleotide that includes
additional
CA 02538147 2004-O1-21
3005-7PCT-9CA
-43-
coding and/or non-coding sequence.
The invention relates to polynucleotides based on SEQ ID NOS: 3, 5, 7, 9,
11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45, 47,
49, 51, 53,
55, 57, 59, 61, 63, 66, 68, 70, 72, 75, 77, 79, 81, 83, 85, 87 and 89 but
having
polynucleotide changes that are "silent", for example changes which do not
alter the
amino acid sequence encoded by the polynucleotides of SEQ ID NOS: 3, 5, 7, 9,
11,
13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45, 47, 49,
51, 53, 55,
57, 59, 61, 63, 66, 68, 70, 72, 75, 77, 79, 81, 83, 85, 87 and 89. The
invention also
relates to polynucleotides which have nucleotide changes which result in amino
acid
substitutions, additions, deletions, fusions and truncations of the
polypeptides of
SEQ ID NOS: 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34,
36, 38, 41,
42, 44, 46, 48, 50, 52, 54, 56, 58, 60, 62, 65, 67, 69, 70, 71, 74, 76, 78,
80, 82, 84,
86 and 88. Such nucleotide changes may be introduced using techniques such as
site directed mutagenesis, random chemical mutagenesis, exonuclease III
deletion,
and other recombinant DNA techniques.
The isolated, purified or enriched nucleic acids of SEQ ID NOS: 3, 5, 7, 9,
11,
13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45, 47, 49,
51, 53, 55,
57, 59, 61, 63, 66, 68, 70, 72, 75, 77, 79, 81, 83, 85, 87 and 89, the
sequences
complementary thereto, or a fragment comprising at least 100, 150, 200, 300,
400 or
more consecutive bases of one of the sequence of SEQ ID NOS: 3, 5, 7, 9, 11,
13,
15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45, 47, 49, 51,
53, 55, 57,
59, 51, 63, 66, 68, 70, 72, 75, 77, 79, 81, 83, 85, 87 and 89, or the
sequences
complementary thereto may be used as probes to identify and isolate DNAs
encoding the polypeptides of SEQ ID NOS: 2, 4, 6, 8, 10, 12, 14, 16, 18, 20,
22, 24,
26, 28, 30, 32, 34, 36, 38, 41, 42, 44, 46, 48, 50, 52, 54, 56, 58, 60, 62,
65, 67, 69,
70, 71, 74, 76, 78, 80, 82, 84, 86 and 88 espectively. In such procedures, a
genomic DNA library is constructed from a sample microorganism or a sample
containing a microorganism capable of producing a farnesyl dibenzodiazepinone.
The genomic DNA library is then contacted with a probe comprising a coding
sequence or a fragment of the coding sequence, encoding one of the
polypeptides
of SEO ID NOS: 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34,
36, 38,
41, 42, 44, 46, 48, 50, 52, 54, 56, 58, 60, 62, 65, 67, 69, 70, 71, 74, 76,
78, 80, 82,
84, 86 and 88, or a fragment thereof under conditions which permit the probe
to
CA 02538147 2004-O1-21
3005-7PCT-9CA
-44-
specifically hybridize to sequences complementary thereto. In a preferred
embodiment, the probe is an oligonucleotide of about 10 to about 30
nucleotides in
length designed based on a nucleic acid of SEQ ID NOS: 3, 5, 7, 9, 11, 13, 15,
17,
19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45, 47, 49, 51, 53, 55,
57, 59, 61,
63, 66, 68, 70, 72, 75, 77, 79, 81, 83, 85, 87 and 89. Genomic DNA clones
which
hybridize to the probe are then detected and isolated. Procedures for
preparing and
identifying DNA clones of interest are disclosed in Ausubel et al., Current
Protocols
in Molecular Biology, John Wiley 503 Sons, Inc. 1997; and Sambrook et al.,
Molecular Cloning: A Laboratory Manual 2d Ed., Cold Spring Harbor Laboratory
Press, 1989. In another embodiment, the probe is a restriction fragment or a
PCR
amplified nucleic acid derived from SEQ I D NOS: 3, 5, 7, 9, 11, 13, 15, 17,
19, 21,
23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45, 47, 49, 51, 53, 55, 57, 59,
61, 63, 66,
68, 70, 72, 75, 77, 79, 81, 83, 85, 87 and 89.
The isolated, purified or enriched nucleic acids of SEQ ID NOS: 3, 5, 7, 9,
11,
13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45, 47, 49,
51, 53, 55,
57, 59, 61, 63, 66, 68, 70, 72, 75, 77, 79, 81, 83, 85, 87 and 89, the
sequences
complementary thereto, or a fragment comprising at least 10, 15, 20, 25, 30,
35, 40,
50, 75, 100, 150, 200, 300, 400 or 500 consecutive bases of one of the
sequences
of SEQ I D NOS: 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33,
35, 37, 39,
41, 43, 45, 47, 49, 51, 53, 55, 57, 59, 61, 63, 66, 68, 70, 72, 75, 77, 79,
81, 83, 85,
87 and 89 or the sequences complementary thereto may be used as probes to
identify and isolate related nucleic acids. In some embodiments, the related
nucleic
acids may be genomic DNAs (or cDNAs) from potential farnesyl
dibenzodiazepinone
producers. In such procedures, a nucleic acid sample containing nucleic acids
from
a potential farnesyl dibenzodiazepinone producer is contacted with the probe
under
conditions that permit the probe to specifically hybridize to related
sequences. The
nucleic acid sample may be a genomic DNA (or cDNA) library from the potential
farnesyl dibenzodiazepinone-producer. Hybridization of the probe to nucleic
acids is
then detected using any of the methods described above.
Hybridization may be carried out under conditions of low stringency, moderate
stringency or high stringency. As an example of nucleic acid hybridization, a
polymer membrane containing immobilized denatured nucleic acids is first
prehybridized for 30 minutes at 45 °C in a solution consisting of 0.9 M
NaCI, 50 mM
CA 02538147 2004-O1-21
3005-7PCT-9CA
-45-
NaH2P04, pH 7.0, 5.0 mM Na2EDTA, 0.5% SDS, 10X Denhardt's, and 0.5 mg/ml
polyriboadenylic acid. Approximately 2 x 10' cpm (specific activity 4-9 x 10$
cpm/ug)
of 32P end-labeled oligonucleotide probe are then added to the solution. After
12-16
hours of incubation, the membrane is washed for 30 minutes at room temperature
in
1X SET (150 mM NaCI, 20 mM Tris hydrochloride, pH 7.8, 1 mM Na2EDTA)
containing 0.5% SDS, followed by a 30 minute wash in fresh 1X SET at Tm-
10°C for
the oligonucleotide probe where Tm is the melting temperature. The membrane is
then exposed to autoradiographic film for detection of hybridization signals.
By varying the stringency of the hybridization conditions used to identify
nucleic acids, such as genomic DNAs or cDNAs, which hybridize to the
detectable
probe, nucleic acids having different levels of homology to the probe can be
identified and isolated. Stringency may be varied by conducting the
hybridization at
varying temperatures below the melting temperatures of the probes. The melting
temperature of the probe may be calculated using the following formulas:
For oligonucleotide probes between 14 and 70 nucleotides in length the
melting temperature (Tm) in degrees Celcius may be calculated using the
formula:
Tm=81.5+16.6(log [Na+]) + 0.41 (fraction G+C)-(600/N) where N is the length of
the
oligonucleotide.
If the hybridization is carried out in a solution containing formamide, the
melting temperature may be calculated using the equation Tm=81.5+16.6(log [Na
+])
+ 0.41 (fraction G + C)-(0.63% formamide)-(600/N) where N is the length of the
probe.
Prehybridization may be carried out in 6X SSC, 5X Denhardt's reagent, 0.5%
SDS, 0.1 mg/ml denatured fragmented salmon sperm DNA or 6X SSC, 5X
Denhardt's reagent, 0.5% SDS, 0.1 mg/ml denatured fragmented salmon sperm
DNA, 50% formamide. The composition of the SSC and Denhardt's solutions are
listed in Sambrook et al., supra.
Hybridization is conducted by adding the detectable probe to the hybridization
solutions listed above. Where the probe comprises double stranded DNA, it is
denatured by incubating at elevated temperatures and quickly cooling before
addition to the hybridization solution. It may also be desirable to similarly
denature
single stranded probes to eliminate or diminish formation of secondary
structures or
oligomerization~ The filter is contacted with the hybridization solution for a
sufficient
CA 02538147 2004-O1-21
3005-7PCT-9CA
-46-
period of time to allow the probe to hybridize to cDNAs or genomic DNAs
containing
sequences complementary thereto or homologous thereto. For probes over 200
nucleotides in length, the hybridization may be carried out at 15-25 °C
below the Tm.
For shorter probes, such as oligonucleotide probes, the hybridization may be
conducted at 5-10 °C below the Tm. Preferably, the hybridization is
conducted in 6X
SSC, for shorter probes. Preferably, the hybridization is conducted in 50%
formamide containing solutions, for longer probes. All the foregoing
hybridizations
would be considered to be examples of hybridization performed under conditions
of
high stringency.
Following hybridization, the filter is washed for at least 15 minutes in 2X
SSC,
0.1 % SDS at room temperature or higher, depending on the desired stringency.
The
filter is then washed with 0.1X SSC, 0.5% SDS at room temperature (again) for
30
minutes to 1 hour. Nucleic acids which have hybridized to the probe are
identified
by conventional autoradiography and non-radioactive detection methods.
The above procedure may be modified to identify nucleic acids having
decreasing levels of homology to the probe sequence. For example, to obtain
nucleic acids of decreasing homology to the detectable probe, less stringent
conditions may be used. For example, the hybridization temperature may be
decreased in increments of 5 °C from 68 °C to 42 °C in a
hybridization buffer having
a Na+ concentration of approximately 1 M. Following hybridization, the filter
may be
washed with 2X SSC, 0.5% SDS at the temperature of hybridization. These
conditions are considered to be "moderate stringency" conditions above
50°C and
"low stringency" conditions below 50°C. A specific example of "moderate
stringency"
hybridization conditions is when the above hybridization is conducted at
55°C. A
specific example of "low stringency" hybridization conditions is when the
above
hybridization is conducted at 45°C.
Alternatively, the hybridization may be carried out in buffers, such as 6X
SSC,
containing formamide at a temperature of 42 °C. In this case, the
concentration of
formamide in the hybridization buffer may be reduced in 5% increments from 50%
to
0% to identify clones having decreasing levels of homology to the probe.
Following
hybridization, the filter may be washed with 6X SSC, 0.5% SDS at 50 °C.
These
conditions are considered to be "moderate stringency" conditions above 25%
formamide and "low stringency" conditions below 25% formamide. A specific
CA 02538147 2004-O1-21
3005-7PCT-9CA
-47-
example of "moderate stringency" hybridization conditions is when the above
hybridization is conducted at 30% formamide. A specific example of "low
stringency"
hybridization conditions is when the above hybridization is conducted at 10%
formamide. Nucleic acids which have hybridized to the probe are identified by
conventional autoradiography and non-radioactive detection methods.
The preceding methods may be used to isolate nucleic acids having at least
97%, at least 95%, at least 90%, at least 85%, at least 80%, or at least 70%
sequence identity to a nucleic acid sequence selected from the group
consisting of
the sequences of SEQ I D NOS: 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27,
29, 31,
33, 35, 37, 39, 41, 43, 45, 47, 49, 51, 53, 55, 57, 59, 61, 63, 66, 68, 70,
72, 75, 77,
79, 81, 83, 85, 87 and 89. The isolated nucleic acid may have a coding
sequence
that is a naturally occurring allelic variant of one of the coding sequences
described
herein. Such allelic variant may have a substitution, deletion or addition of
one or
more nucleotides when compared to the nucleic acids of SEQ ID NOS: 3, 5, 7, 9,
11,
13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 37, 39, 41, 43, 45, 47, 49,
51, 53, 55,
57, 59, 61, 63, 66, 68, 70, 72, 75, 77, 79, 81, 83, 85, 87 and 89, or the
sequences
complementary thereto.
Additionally, the above procedures may be used to isolate nucleic acids which
encode polypeptides having at least 99%, at least 95%, at least 90%, at least
85%,
at least 80°f0, or at least 70% identity to a polypeptide having the
sequence of one of
SEQ ID NOS: 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34,
36, 38, 41,
42, 44, 46, 48, 50, 52, 54, 56, 58, 60, 62, 65, 67, 69, 70, 71, 74, 76, 78,
80, 82, 84,
86 and 88 or fragments comprising at least 50, 75, 100, 150, 200, 300
consecutive
amino acids thereof.
Another aspect of the present invention is an isolated or purified polypeptide
comprising the sequence of one of SEQ ID NOS: 2, 4, 6, 8, 10, 12, 14, 16, 18,
20,
22, 24, 26, 28, 30, 32, 34, 36, 38, 41, 42, 44, 46, 48, 50, 52, 54, 56, 58,
60, 62, 65,
67, 69, 70, 71, 74, 76, 78, 80, 82, 84, 86 and 88 or fragments comprising at
least 50,
75, 100, 150, 200 or 300 consecutive amino acids thereof. As discussed herein,
such polypeptides may be obtained by inserting a nucleic acid encoding the
polypeptide into a vector such that the coding sequence is operably linked to
a
sequence capable of driving the expression of the encoded polypeptide in a
suitable
host cell. For example, the expression vector may comprise a promoter, a
ribosome
CA 02538147 2004-O1-21
3005-7PCT-9CA
-48-
binding site for translation initiation and a transcription terminator. The
vector may
also include appropriate sequences for modulating expression levels, an origin
of
replication and a selectable marker.
Promoters suitable for expressing the polypeptide or fragment thereof in
bacteria include the E.coli lac or trp promoters, the lacl promoter, the iacZ
promoter,
the T3 promoter, the T7 promoter, the gpt promoter, the lambda PR promoter,
the
lambda P~ promoter, promoters from operons encoding glycolytic enzymes such as
3-phosphoglycerate kinase (PGK), and the acid phosphatase promoter. Fungal
promoters include the a factor promoter. Eukaryotic promoters include the CMV
immediate early promoter, the HSV thymidine kinase promoter, heat shock
promoters, the early and late SV40 promoter, LTRs from retroviruses, and the
mouse metallothionein-I promoter. Other promoters known to control expression
of
genes in prokaryotic or eukaryotic cells or their viruses may also be used.
Mammalian expression vectors may also comprise an origin of replication,
any necessary ribosome binding sites, a polyadenylation site, splice donors
and
acceptor sites, transcriptional termination sequences, and 5' flanking
nontranscribed
sequences. In some embodiments, DNA sequences derived from the SV40 splice
and polyadenylation sites may be used to provide the required nontranscribed
genetic elements.
Vectors for expressing the polypeptide or fragment thereof in eukaryotic cells
may also contain enhancers to increase expression levels. Enhancers are cis-
acting
elements of DNA, usually from about 10 to about 300 by in length that act on a
promoter to increase its transcription. Examples include the SV40 enhancer on
the
late side of the replication origin by 100 to 270, the cytomegalovirus early
promoter
enhancer, the polyoma enhancer on the late side of the replication origin, and
the
adenovirus enhancers.
In addition, the expression vectors preferably contain one or more selectable
marker genes to permit selection of host cells containing the vector. Examples
of
selectable markers that may be used include genes encoding dihydrofolate
reductase or genes conferring neomycin resistance for eukaryotic cell culture,
genes
conferring tetracycline or ampicillin resistance in E. coli, and the S.
cerevisiae TRP1
gene.
The appropriate DNA sequence may be inserted into the vector by a variety
CA 02538147 2004-O1-21
3005-7PCT-9CA
-49-
of procedures. In general, the DNA sequence is ligated to the desired position
in the
vector following digestion of the insert and the vector with appropriate
restriction
endonucleases. Alternatively, appropriate restriction enzyme sites can be
engineered into a DNA sequence by PCR. A variety of cloning techniques are
disclosed in Ausbel et al. Current Protocols in Molecular Biology, John Wiley
503
Sons, Inc. 1997 and Sambrook et al., Molecular Cloning: A Laboratory Manual 2d
Ed., Cold Spring Harbour Laboratory Press, 1989. Such procedures and others
are
deemed to be within the scope of those skilled in the art.
The vector may be, for example, in the form of a plasmid, a viral particle, or
a
phage. Other vectors include derivatives of chromosomal, nonchromosomal and
synthetic DNA sequences, viruses, bacterial plasmids, phage DNA, baculovirus,
yeast plasmids, vectors derived from combinations of plasmids and phage DNA,
viral DNA such as vaccinia, adenovirus, fowl pox virus, and pseudorabies. A
variety
of cloning and expression vectors for use with prokaryotic and eukaryotic
hosts are
described by Sambrook et al., Molecular Cloning: A Laboratory Manual, Second
Edition, Cold Spring Harbor, N.Y., (1989).
Particular bacterial vectors which may be used include the commercially
available plasmids comprising genetic elements of the well known cloning
vector
pBR322 (ATCC 37017), pKK223-3 (Pharmacia Fine Chemicals, Uppsala, Sweden),
pGEM1 (Promega Biotec, Madison, WI, USA) pQE70, pQE60, pQE-9 (Qiagen),
pD10, phiX174, pBluescriptT"' II KS, pNHBA, pNH16a, pNH18A, pNH46A
(Stratagene), ptrc99a, pKK223-3, pKK233-3, pDR540, pRIT5 (Pharmacia), pKK232-
8 and pCM7. Particular eukaryotic vectors include pSV2CAT, pOG44, pXT1, pSG
(Stratagene) pSVK3, pBPV, pMSG, and pSVL (Pharmacia). However, any other
vector may be used as long as it is replicable and stable in the host cell.
The host cell may be any of the host cells familiar to those skilled in the
art,
including prokaryotic cells or eukaryotic cell s. As representative examples
of
appropriate hosts, there may be mentioned: bacteria cells, such as E. coli,
Streptomyces lividans, Streptomyces griseofuscus, Streptomyces ambofaciens,
Bacillus subtilis, Salmonella typhimurium and various species within the
genera
Pseudomonas, Streptomyces, Bacillus, and Staphylococcus, fungal cells, such as
yeast, insect cells such as Drosophila S2 and Spodoptera Sf9, animal cells
such as
CHO, COS or Bowes melanoma, and adenoviruses. The selection of an appropriate
CA 02538147 2004-O1-21
3005-7PCT-9CA
-50-
host is within the abilities of those skilled in the art.
The vector may be introduced into the host cells using any of a variety of
techniques, including electroporation transformation, transfection,
transduction, viral
infection, gene guns, or Ti-mediated gene transfer. Where appropriate, the
engineered host cells can be cultured in conventional nutrient media modified
as
appropriate for activating promoters, selecting transformants or amplifying
the genes
of the present invention. Following transformation of a suitable host strain
and
growth of the host strain to an appropriate cell density, the selected
promoter may
be induced by appropriate means (e.g., temperature shift or chemical
induction) and
the cells may be cultured for an additional period to allow them to produce
the
desired polypeptide or fragment thereof.
Cells are typically harvested by centrifugation, disrupted by physical or
chemical means, and the resulting crude extract is retained for further
purification.
Microbial cells employed for expression of proteins can be disrupted by any
convenient method, including freeze-thaw cycling, sonication, mechanical
disruption,
or use of cell lysing agents. Such methods are well known to those skilled in
the art.
The expressed polypeptide or fragment thereof can be recovered and purified
from
recombinant cell cultures by methods including ammonium sulfate or ethanol
precipitation, acid extraction, anion or cation exchange chromatography,
phosphocellulose chromatography, hydrophobic interaction chromatography,
affinity
chromatography, hydroxylapatite chromatography and lectin chromatography.
Protein refolding steps can be used, as necessary, in completing configuration
of the
polypeptide. If desired, high performance liquid chromatography (HPLC) can be
employed for final purification steps.
Various mammalian cell culture systems can also be employed to express
recombinant protein. Examples of mammalian expression systems include the
COS-7 lines of monkey kidney fibroblasts (described by Gluzman, Cell,
23:175(1981 )), and other cell lines capable of expressing proteins from a
compatible
vector, such as the C127, 3T3, CHO, HeLa and BHK cell lines. The constructs in
host cells can be used in a conventional manner to produce the gene product
encoded by the recombinant sequence. Polypeptides of the invention may or may
not also include an initial methionine amino acid residue.
Alternatively, the polypeptides of SEQ ID NOS: 2, 4, 6, 8, 10, 12, 14, 16, 18,
CA 02538147 2004-O1-21
3005-7PCT-9CA
-51-
20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 41, 42, 44, 46, 48, 50, 52, 54, 56,
58, 60, h2,
65, 67, 69, 70, 71, 74, 76, 78, 80, 82, 84, 86 and 88 or fragments comprising
at least
50, 75, 100, 150, 200 or 300 consecutive amino acids thereof can be
synthetically
produced by conventional peptide synthesizers. In other embodiments, fragments
or
portions of the polynucleotides may be employed for producing the
corresponding
full-length polypeptide by peptide synthesis; therefore, the fragments may be
employed as intermediates for producing the full-length polypeptides.
Cell-free translation systems can also be employed to produce one of the
polypeptides of SEQ ID NOS: 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26,
28, 30,
32, 34, 36, 38, 41, 42, 44, 46, 48, 50, 52, 54, 56, 58, 60, 62, 65, 67, 69,
70, 71, 74,
76, 78, 80, 82, 84, 86 and 88 or fragments comprising at least 50, 75, 100,
150, 200
or 300 consecutive amino acids thereof using mRNAs transcribed from a DNA
construct comprising a promoter operably linked to a nucleic acid encoding the
polypeptide or fragment thereof. In some embodiments, the DNA construct may be
linearized prior to conducting an in vitro transcription reaction. The
transcribed
mRNA is then incubated with an appropriate cell-free translation extract, such
as a
rabbit reticulocyte extract, to produce the desired polypeptide or fragment
thereof.
The present invention also relates to variants of the polypeptides of SEQ ID
NOS: 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38,
41, 42, 44,
46, 48, 50, 52, 54, 56, 58, 60, 62, 65, 67, 69, 70, 71, 74, 76, 78, 80, 82,
84, 86 and
88 or fragments comprising at least 50, 75, 100, 150, 200 or 300 consecutive
amino
acids thereof. The term "variant" includes derivatives or analogs of these
polypeptides. In particular, the variants may differ in amino acid sequence
from the
polypeptides of SEQ ID NOS: 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26,
28, 30,
32, 34, 36, 38, 41, 42, 44, 46, 48, 50, 52, 54, 56, 58, 60, 62, 65, 67, 69,
70, 71, 74,
76, 78, 80, 82, 84, 86 and 88 by one or more substitutions, additions,
deletions,
fusions and truncations, which may be present in any combination.
The variants may be naturally occurring or created in vitro. In particular,
such
variants may be created using genetic engineering techniques such as site
directed
mutagenesis, random chemical mutagenesis, exonuclease III deletion procedures,
and standard cloning techniques. Alternatively, such variants, fragments,
analogs,
or derivatives may be created using chemical synthesis or modification
procedures.
Other methods of making variants are also familiar to those skilled in the
art.
CA 02538147 2004-O1-21
3005-7PCT-9CA
-52-
These include procedures in which nucleic acid sequences obtained from natural
isolates are modified to generate nucleic acids that encode polypeptides
having
characteristics which enhance their value in industrial or laboratory
applications. In
such procedures, a large number of variant sequences having one or more
nucleotide differences with respect to the sequence obtained from the natural
isolate
are generated and characterized. Preferably, these nucleotide differences
result in
amino acid changes with respect to the polypeptides encoded by the nucleic
acids
from the natural isolates.
The variants of the polypeptides of SEQ ID NOS: 2, 4, 6, 8, 10, 12, 14, 15,
18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 41, 42, 44, 46, 48, 50, 52, 54,
56, 58, b0,
62, 65, 679 69, 70, 71, 74, 76, 78, 80, 82, 84, 86 and 88 may be variants in
which
one or more of the amino acid residues of the polypeptides of SEQ ID NOS: 2,
4, 6,
8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 41, 42, 44, 46,
48, 50,
52, 54, 56, 58, 60, 62, 65, 67, 69, 70, 71, 74, 76, 78, 80, 82, 84, 86 and 88
are
substituted with a conserved or non-conserved amino acid residue (preferably a
conserved amino acid residue) and such substituted amino acid residue may or
may
not be one encoded by the genetic code.
Conservative substitutions are those that substitute a given amino acid in a
polypeptide by another amino acid of like characteristics. Typically seen as
conservative substitutions are the following replacements: replacements of an
aliphatic amino acid such as Ala, Val, Leu and Ile with another aliphatic
amino acid;
replacement of a Ser with a Thr or vice versa; replacement of an acidic
residue such
as Asp or Glu with another acidic residue; replacement of a residue bearing an
amide group, such as Asn or Gln, with another residue bearing an amide group;
exchange of a basic residue such as Lys or Arg with another basic residue; and
replacement of an aromatic residue such as Phe or Tyr with another aromatic
residue.
Other variants are those in which one or more of the amino acid residues of
the polypeptides of SEQ ID NOS: 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24,
26, 28,
30, 32, 34, 36, 38, 41, 42, 44, 46, 48, 50, 52, 54, 56, 58, 60, 62, 65, 67,
69, 70, 71,
74, 76, 78, 80, 82, 84, 86 and 88 include a substituent group. Still other
variants are
those in which the polypeptide is associated with another compound, such as a
compound to increase the half-life of the polypeptide (for example,
polyethylene
CA 02538147 2004-O1-21
3005-7PCT-9CA
-53-
glycol). Additional variants are those in which additional amino acids are
fused to
the polypeptide, such as leader sequence, a secretory sequence, a proprotein
sequence or a sequence that facilitates purification, enrichment, or
stabilization of
the polypeptide.
In some embodiments, the fragments, derivatives and analogs retain the
same biological function or activity as the polypeptides of SEQ ID NOS: 2, 4,
6, 8,
10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 41, 42, 44, 46,
48, 50, 52,
54, 56, 58, 60, 62, 65, 67, 69, 70, 71, 74, 76, 78, 80, 82, 84, 86 and 88. In
other
embodiments, the fragment, derivative or analogue includes a fused
heterologous
sequence that facilitates purification, enrichment, detection, stabilization
or
secretion of the polypeptide that can be enzymatically cleaved, in whole or in
part,
away from the fragment, derivative or analogue.
Another aspect of the present invention are polypeptides or fragments thereof
which have at least 70%, at least 80%, at least 85%, at least 90%, or more
than
95% identity to one of the polypeptides of SEQ ID NOS: 2, 4, 6, 8, 10, 12, 14,
16,
18, 20, 22, 24, 26, 28, 30, 32, 34, 36, 38, 41, 42, 44, 46, 48, 50, 52, 54,
56, 58, 50,
62, 65, 67, 69, 70, 71, 74, 76, 78, 80, 82, 84, 86 and 88 or a fragment
comprising at
least 50, 75, 100, 150, 200 or 300 consecutive amino acids thereof. It will be
appreciated that amino acid "substantially identity" includes conservative
substitutions such as those described above.
The polypeptides or fragments having homology to one of the polypeptides of
SEQ ID NOS: 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30, 32, 34,
36, 38, 41,
42, 44, 46, 48, 50, 52, 54, 56, 58, 60, 62, 65, 67, 69, 70, 71, 74, 76, 78,
80, 82, 84,
86 and 88 or a fragment comprising at least 50, 75, 100, 150, 200 or 300
consecutive amino acids thereof may be obtained by isolating the nucleic acids
encoding them using the techniques described above.
Alternatively, the homologous polypeptides or fragments may be obtained
through biochemical enrichment or purification procedures. The sequence of
potentially homologous polypeptides or fragments may be determined by
proteolytic
digestion, gel electrophoresis and/or microsequencing. The sequence of the
prospective homologous polypeptide or fragment can be compared to one of the
polypeptides of SEQ ID NOS: 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26,
28, 30,
32, 34, 36, 38, 41, 42, 44, 46, 48, 50, 52, 54, 56, 58, 60, 62, 65, 67, 69,
70, 71, 74,
CA 02538147 2004-O1-21
3005-7PCT-9CA
-54-
76, 78, 80, 82, 84, 86 and 88 or a fragment comprising at least 5, 10, 15, 20,
25, 30,
35, 40, 50, 75, 100, or 150 consecutive amino acids thereof.
The polypeptides of SEQ ID NOS: 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24,
26, 28, 30, 32, 34, 36, 38, 41, 42, 44, 46, 48, 50, 52, 54, 56, 58, 60, 62,
65, 67, 59,
70, 71, 74, 76, 78, 80, 82, 84, 86 and 88 or fragments, derivatives or analogs
thereof
comprising at least 40, 50, 75, 100, 150, 200 or 300 consecutive amino acids
thereof
invention may be used in a variety of applications. For example, the
polypeptides or
fragments, derivatives or analogs thereof may be used to catalyze biochemical
reactions as described elsewhere in the specification.
VI. Pharmaceutical compositions comprising farnesyl dibenzodiazepinones
In another embodiment, the invention relates to a pharmaceutical composition
comprising a farnesyl dibenzodiazepinone, as described in the preceding
section,
and a pharmaceutically acceptable carrier, as described below. The
pharmaceutical
composition comprising the farnesyl dibenzodiazepinone is useful for treating
a
variety of diseases and disorders, including cancer, inflammation and
bacterial
infections.
The compounds of the present invention, or pharmaceutically acceptable
salts thereof, can be formulated for oral, intravenous, intramuscular,
subcutaneous,
topical or parenteral administration for the therapeutic or prophylactic
treatment of
diseases, particularly bacterial infections, acute and chronic inflammation
and
cancer. For oral or parental administration, compounds of the present
invention can
be mixed with conventional pharmaceutics! carriers and excipients and used in
the
form of tablets, capsules, elixirs, suspensions, syrups, wafers and the like.
The
compositions comprising a compound of this present invention will contain from
about 0.1 % to about 99.9%, about 1 % to about 98%, about 5% to about 95%,
about
10% to about 80% or about 15% to about 60% by weight of the active compound.
The pharmaceutical preparations disclosed herein are prepared in
accordance with standard procedures and are administered at dosages that are
selected to reduce, prevent, or eliminate bacterial infection, cancer or
inflammation.
(See, e.g., Remington's Pharmaceutical Sciences, Mack Publishing Company,
Easton, PA; and Goodman and Gilman, Pharmaceutical Basis of Therapeutics,
CA 02538147 2004-O1-21
3005-7PCT-9CA
-55-
Pergamon Press, New York, NY, the contents of which are incorporated herein by
reference, for a general description of the methods for administering various
antimicrobial agents for human therapy). The compositions of the present
invention
can be delivered using controlled (e.g., capsules) or sustained release
delivery
systems (e.g., bioerodable matrices). Exemplary delayed release delivery
systems
for drug delivery that are suitable for administration of the compositions of
the
invention (preferably of Formula I) are described in U.S. Patent Nos 4,452,775
(issued to Kent), 5,239,660 (issued to Leonard), 3,854,480 (issued to
Zaffaroni),
The pharmaceutically acceptable compositions of the present invention
comprise one or more compounds of the present invention in association with
one or
more non-toxic, pharmaceutically acceptable carriers and/or diluents and/or
adjuvants and/or excipients, collectively referred to herein as "carrier"
materials, and
if desired other active ingredients. The compositions may contain common
carriers
and excipients, such as corn starch or gelatin, lactose, sucrose,
microcrystalline
cellulose, kaolin, mannitol, dicalcium phosphate, sodium chloride and alginic
acid.
The compositions may contain crosarmellose sodium, microcrystalline
cellulose;,
sodium starch glycolate and alginic acid.
Tablet binders that can be included are acacia, methylcellulose, sodium
carboxymethylcellulose, polyvinylpyrrolidone (Providone), hydroxypropyl
methylcellulose, sucrose, starch and ethylcellulose.
Lubricants that can be used include magnesium stearate or other metallic
stearates, stearic acid, silicon fluid, talc, waxes, oils and colloidal
silica.
Flavouring agents such as peppermint, oil of wintergreen, cherry flavouring or
the like can also be used. It may also be desirable to add a coloring agent to
make
the dosage form more aesthetic in appearance or to help identify the product
comprising a compound of the present invention.
For oral use, solid formulations such as tablets and capsules are particularly
useful. Sustained released or enterically coated preparations may also be
devised.
For pediatric and geriatric applications, suspension, syrups and chewable
tablets are
especially suitable. For oral administration, the pharmaceutical compositions
are in
the form of, for example, a tablet, capsule, suspension or liquid. The
pharmaceutical
composition is preferably made in the form of a dosage unit containing a
therapeutically-effective amount of the active ingredient. Examples of such
dosage
CA 02538147 2004-O1-21
3005-7PCT-9CA
-56-
units are tablets and capsules. For therapeutic purposes, the tablets and
capsules
which can contain, in addition to the active ingredient, conventional carriers
such as
binding agents, for example, acacia gum, gelatin, polyvinylpyrrolidone,
sorbitol, or
tragacanth; fillers, for example, calcium phosphate, glycine, lactose, maize-
starch,
sorbitol, or sucrose; lubricants, for example, magnesium stearate,
polyethylene
glycol, silica or talc: disintegrants, for example, potato starch, flavoring
or coloring
agents, or acceptable wetting agents. Oral liquid preparations generally are
in the
form of aqueous or oily solutions, suspensions, emulsions, syrups or elixirs
and may
contain conventional additives such as suspending agents, emulsifying agents,
non-
aqueous agents, preservatives, coloring agents and flavoring agents. Examples
of
additives for liquid preparations include acacia, almond oil, ethyl alcohol,
fractionated
coconut oil, gelatin, glucose syrup, glycerin, hydrogenated edible fats,
lecithin,
methyl cellulose, methyl or propyl para-hydroxybenzoate, propylene glycol,
sorbitol,
or sorbic acid.
For intravenous (iv) use, compounds of the present invention can be
dissolved or suspended in any of the commonly used intravenous fluids and
administered by infusion. Intravenous fluids include, without limitation,
physiological
saline or Ringer's solution.
Formulations for parental administration can be in the form of aqueous or
non-aqueous isotonic sterile injection solutions or suspensions. These
solutions or
suspensions can be prepared from sterile powders or granules having one or
more
of the carriers mentioned for use in the formulations for oral administration.
The
compounds can be dissolved in polyethylene glycol, propylene glycol, ethanol,
corn
oil, benzyl alcohol, sodium chloride, and/or various buffers.
For intramuscular preparations, a sterile formulation of compounds of the
present invention or suitable soluble salts forming the compound, can be
dissolved
and administered in a pharmaceutical diluent such as Water-for-Injection
(WFI),
physiological saline or 5% glucose. A suitable insoluble form of the compound
may
be prepared and administered as a suspension in an aqueous base or a
pharmaceutically acceptable oil base, e.g. an ester of a long chain fatty acid
such as
ethyl oleate.
For topical use the compounds of present invention can also be prepared in
suitable forms to be applied to the skin, or mucus membranes of the nose and
CA 02538147 2004-O1-21
3005-7PCT-9CA
-57-
throat, and can take the form of creams, ointments, liquid sprays or
inhalants,
lozenges, or throat paints. Such topical formulations further can include
chemical
compounds such as dimethylsulfoxide (DMSO) to facilitate surface penetration
of
the active ingredient.
For application to the eyes or ears, the compounds of the present invention
can be presented in liquid or semi-liquid form formulated in hydrophobic or
hydrophilic bases as ointments, creams, lotions, paints or powders.
For rectal administration the compounds of the present invention can be
administered in the form of suppositories admixed with conventional carriers
such as
cocoa butter, wax or other glyceride.
Alternatively, the compound of the present invention can be in powder form
for reconstitution in the appropriate pharmaceutically acceptable carrier at
the time
of delivery. In another embodiment, the unit dosage form of the compound can
be a
solution of the compound or a salt thereof in a suitable diluent in sterile,
hermetically
sealed ampoules.
The amount of the compound of the present invention in a unit dosage
comprises a therapeutically-effective amount of at least one active compound
of the
present invention which may vary depending on the recipient subject, route and
frequency of administration. A recipient subject refers to a plant, a cell
culture or an
animal such as an ovine or a mammal including a human.
According to this aspect of the present invention, the novel compositions
disclosed herein are placed in a pharmaceutically acceptable carrier and are
delivered to a recipient subject (including a human subject) in accordance
with
known methods of drug delivery. In general, the methods of the invention for
delivering the compositions of the invention in vivo utilize art-recognized
protocols for
delivering the agent with the only substantial procedural modification being
the
substitution of the compounds of the present invention for the drugs in the
art-
recognized protocols.
Likewise, the methods for using the claimed composition for treating cells in
culture, for example, to eliminate or reduce the level of bacterial
contamination of a
cell culture, utilize art-recognized protocols for treating cell cultures with
antibacterial
agents) with the only substantial procedural modification being the
substitution of
the compounds of the present invention for the agents used in the art-
recognized
CA 02538147 2004-O1-21
3005-7PCT-9CA
-58-
protocols.
The compounds of the present invention provide a method for treating
bacterial infections, pre-cancerous or cancerous conditions, and acute or
chronic
inflammatory disease. As used herein, the term "unit dosage" refers to a
quantity of
a therapeutically effective amount of a compound of the present invention that
elicits
a desired therapeutic response. As used herein, the phrase "therapeutically
effective amount" means an amount of a compound of the present invention that
prevents the onset, alleviates the symptoms, or stops the progression of a
bacterial
infection, inflammatory condition, or pre-cancerous or cancerous condition.
The
term "treating" is defined as administering, to a subject, a therapeutically
effective
amount of at least one compound of the present invention, both to prevent the
occurrence of a bacterial infection, inflammation or pre-cancer or cancer
condition,
or to control or eliminate a bacterial infection, inflammation or pre-cancer
or cancer
condition. The term "desired therapeutic response" refers to treating a
recipient
subject with a compound of the present invention such that a bacterial or
inflammatory condition or pre-cancer or cancer condition is reversed, arrested
or
prevented in a recipient subject.
The compounds of the present invention can be administered as a single
daily dose or in multiple doses per day. The treatment regime may require
administration over extended periods of time, e.g., for several days or for
from two to
four weeks. The amount per administered dose or the total amount administered
will depend on such factors as the nature and severity of the disease
condition, the
age and general health of the recipient subject, the tolerance of the
recipient subject
to the compound and the type of the bacterial infection, inflammatory
disorder, or
type of cancer.
A compound according to this invention may also be administered in the diet
or feed of a patient or animal. The diet for animals can be normal foodstuffs
to
which the compound can be added or it can be added to a premix.
The compounds of the present invention may be taken in combination,
together or separately with any known clinically approved antibiotic,
inflammation or
anti-cancer agent to treat a recipient subject in need of such treatment.
VII. Method of Inhibiting Tumor Growth
CA 02538147 2004-O1-21
3005-7PCT-9CA
-59-
In another embodiment, the present invention relates to a method of inhibiting
tumor growth. Compounds as described herein can possess antitumor activity.
The
compounds are effective against mammalian tumor cells such as leukemia cells,
melanoma cells, breast carcinoma cells, lung carcinoma cells, pancreatic
carcinoma
cells, ovarian carcinoma cells, renal carcinoma cells, colon carcinoma cells
prostate
carcinoma cells and glioma cells. The antitumor method of the invention
results in
inhibition of tumor cells. The term "inhibition", when used in conjunction
with the
antitumor method refers to suppression, killing, stasis, or destruction of
tumor cells.
The antitumor method preferably results in prevention, reduction or
elimination of
invasive activity and related metastasis of tumor cells. The term "effective
amount"
when used in conjunction with the antitumor cell method refers to the amount
of the
compound sufficient to result in the inhibition of mammalian tumor cells.
The inhibition of mammalian tumor growth according to this method can be
monitored in several ways. First, tumor cells grown in vitro can be treated
with the
compound and monitored for growth or death relative to the same cells cultured
in
the absence of the compound. A cessation of growth or a slowing of the growth
rate
(i.e., the doubling rate), e.g., by 10% or more, is indicative of tumor cell
inhibition.
Alternatively, tumor cell inhibition can be monitored by administering the
compound
to an animal model of the tumor of interest. Examples of experimental animal
tumor
models are known in the art and described in the examples herein. A cessation
of
tumor growth (i.e., no further increase in size) or a reduction in tumor size
(i.e.,
tumor volume) or cell number (e.g., at least a 10% decrease in either) in
animals
treated with a compound as described herein relative to tumors in control
animals
not treated with the compound is indicative of tumor growth inhibition.
To monitor the efficacy of tumor treatment in a human, tumor size or tumor
cell titer is measured before and after initiation of the treatment, and
treatment is
considered effective if either the tumor size or titer ceases further growth,
or if the
tumor is reduced in size or titer, e.g., by at least 10% or more (e.g., 20%,
30%, 40%,
50%, 60%, 70%, 80%, 90% or even 100%, that is, the absence of the tumor).
Methods of determining the size or cell titer of a tumor in vivo vary with the
type of
tumor, and include, for example, various imaging techniques well known to
those in
the medical imaging or oncology fields (MRI, CAT, PET, etc.), as well as
histological
CA 02538147 2004-O1-21
3005-7PCT-9CA
-60-
techniques and flow cytometry.
For the antitumor method of the invention, a typical effective dose of the
compounds given orally or parenterally would be from about 5 to about 100
mg/kg of
body weight of the subject with a daily dose ranging from about 15 to about
300
mg/kg of body weight of the subject.
VIII. Method of Inhibiting Lipoxyaenase
In another embodiment, the present invention also provides for a method of
treating diseased states, in particular inflammation, caused by the 5-
lipoxygenase
system and/or by the synthesis of the Leukotrienes C4, D4, E4 and F4 as well
as
Leukotriene B4 in mammals, especially in human subjects. This method comprises
administering to a subject an effective amount of ECO-04601. Compound ECO-
04601 may be used alone or in combination with other anti-inflammatory
compounds
to treat or prevent disease states related to inflammation including pulmonary
conditions, inflammation, cardiovascular conditions, central nervous system
conditions or skin conditions. More specific diseases include gastritis;
erosive
esophagitis; inflammatory bowel disease; ethanol-induced hemorrhagic erosions;
hepatic ischemia; ischemic neuronal injury; noxious agent induced damage or
necrosis of hepatic, pancreatic, renal, neuronal or myocardial tissue; liver
parenchymal damage caused by hepatoxic agents such as CC14 and D-
galactosamine; ischemic renal failure; disease-induced hepatic damage; trauma-
or
stress-induced cell damage; asthma; multiple sclerosis; ischemic reperfusion;
edema; rheumatoid arthritis; viral encephalitis; bacterial pneumonia;
neurodegeneration; Alzheimer's disease and glycerol-induced renal failure.
For the method of the invention related to the 5-lipoxygenase system and/or
the biosynthesis of Leukotrienes, a typical effective unit dose of ECO-04601
given
orally or parenterally would be from about 5 to about 100 mg/kg of body weight
of
the subject with a daily dose ranging from about 15 to about 300 mg/kg of body
weight of the subject.
The inhibition of lipoxygenase enzymes is monitored using methods well
known in the art and as described in the examples herein. A decrease in enzyme
activity by at least 10%, relative to the activity in the absence of a
compound as
CA 02538147 2004-O1-21
3005-7PCT-9CA
-61-
described herein is indicative of effective inhibition of lipoxygenase
activity.
Farnesyl dibenzodiazepinone compounds useful according to the invention
can be used to reduce or prevent inflammation. Among the hallmarks of local
acute
inflammation are heat, redness, swelling, pain and loss of function. These
changes
are induced largely by changes in vascular flow and caliber, changes in
vascular
permeability and leukocyte exudation (Bobbins et al., "Pathologic Basis of
Disease",
6'" Ed., W.B. Saunders Co., Philadelphia, PA). Anti-inflammatory therapy
performed
using compounds useful according to the invention can be monitored for success
by
tracking any of these changes. For example, a decrease in swelling (e.g., at
least
10% decrease following treatment) or reported pain (e.g., a sustained decrease
of 1
point or more on a 1-10 scale reported by the patient, with 10 being the worst
pain
experienced in association with this disorder prior to treatment, and 0 being
no pain)
can be used to indicate successful treatment.
Other measurable hallmarks of inflammation include leukocyte infiltration and
inflammatory cytokine levels. These hallmarks can be monitored by biopsy of
the
affected tissue. A decrease of 10% or more in leukocyte infiltration in fixed,
stained
tissue relative to infiltration in similar tissue prior to treatment can be
used to indicate
successful treatment, as can a decrease of 10% or more in the level of any
given
inflammatory cytokine, relative to the level before treatment. Those skilled
in the art
can readily assay for inflammatory cytokine levels in tissue, blood, or other
fluid
samples. Alternatively, the level of systemic indicators of inflammation such
as C
reactive protein levels and erythrocyte sedimentation rate can be monitored.
Each
of these has established normal ranges in medicine, and treatment is
considered
successful if one or more of such indicators goes from outside the normal
range to
inside the normal range after the initiation of treatment.
IX. Method of Inhibitina Bacterial Growth
In another embodiment, the present invention relates to a method for treating
bacterial infection in a mammalian subject in need thereof, comprising the
step of
administering to the mammal a therapeutically effective amount of compound ECO-
0~601, a compound as described herein, or a pharmaceutically acceptable
derivative or prod rug thereof.
CA 02538147 2004-O1-21
3005-7PCT-9CA
-62-
According to another embodiment, the invention provides a method of
decreasing bacterial quantity in a biological sample. This method comprises
the step
of contacting the biological sample with a compound ECO-04601, a compound as
described herein, or a pharmaceutically acceptable derivative or prodrug
thereof.
This method is effective if the number of bacteria decreases by at least 10%,
and
preferably more, e.g., 25%, 50%, 75% or even 100% after contacting the
biological
sample with compound ECO-04601, a compound as described herein, or a
pharmaceutically acceptable derivative or prod rug thereof.
These pharmaceutical compositions effective to treat or prevent a bacterial
infection which comprise ECO-04601, a compound as described herein, or a
pharmaceutically acceptable derivative or prodrug thereof in an amount
sufficient to
measurably decrease bacterial quantity, and a pharmaceutically acceptable
carrier,
are another embodiment of the present invention. The term "measurably decrease
bacterial quantity", as used herein means a measurable change in the number of
bacteria between a sample containing the inhibitor and a sample not containing
the
inhibitor.
Agents which increase the susceptibility of bacterial organisms to antibiotics
are known. For example, U.S. Pat. No. 5,523,288, U.S. Pat. No. 5,783,561 and
U.S.
Pat. No. 6,140,306 describe methods of using bactericidal/permeability-
increasing
protein (BPI) for increasing antibiotic susceptibility of gram-positive and
gram-
negative bacteria. Agents that increase the permeability of the outer membrane
of
bacterial organisms have been described by Vaara, M. in Microbiological
Reviews
(1992) pp. 395-411, and the sensitization of gram-negative bacteria has been
described by Tsubery, H., et al, in J. Med. Chem . (2000) pp. 3085-3092.
For the method of the invention related to treatment of subjects with a
bacterial infection, a typical effective unit dose of ECO-04601, a compound
described herein or a pharmaceutically acceptable derivative or prod rug
thereof
given orally or parenterally would be from about 5 to about 100 mg/kg of body
weight
of the subject with a daily dose ranging from about 15 to about 300 mg/kg of
body
weight of the subject.
Another preferred embodiment of this invention relates to a method, as
described above, of treating a bacterial infection in a mammal in need
thereof, but
further comprising the step of administering to the mammal an agent which
CA 02538147 2004-O1-21
3005-7PCT-9CA
-63-
increases the susceptibility of bacterial organisms to antibiotics.
According to another preferred embodiment, the invention provides a method,
as described above, of decreasing bacterial quantity in a biological sample,
but
further comprising the step of contacting the biological sample with an agent
which
increases the susceptibility of bacterial organisms to antibiotics.
Methods of decreasing bacterial quantity are effective if the number of
bacteria decreases at least 10%, and preferably more, e.g., 25%, 50%, 75% or
even
100% after contacting the biological sample with compound ECO-04601, a
compound as described herein, or a pharmaceutically acceptable derivative or
prod rug thereof.
The pharmaceutical compositions and methods of this invention will be useful
generally for controlling bacterial infections in vivo. Examples of bacterial
organisms
that may be controlled by the compositions and methods of this invention
include,
but are not limited to the following organisms: Streptococcus pneumoniae,
Streptococcus pyrogenes, Enterococcus fecalis, Enterococcus faecium,
Klebsiella
pneumoniae, Enterobacter spp., Proteus spp., Pseudomonas aeruginosa, E. coli,
Serratia marcesens, Staphylococcus aureus, Coagulase negative Staphylococcus,
Haemophilus infuenzae, Bacillus anthracis, Mycoplasma pneumoniae, and
Staphylococcus epidermidis. The compositions and methods will therefore be
useful
for controlling, treating or reducing the advancement, severity or effects of
nosocomial or non-nosocomial infections. Examples of nosocomial uses include,
but
are not limited to, urinary tract infections, pneumonia, surgical wound
infections,
bacteremia and therapy for febrile neutropenic patients. Examples of non-
nosocomial uses include but are not limited to urinary tract infections,
pneumonia,
prostatitis, skin and soft tissue infections and intra-abdominal infections.
In addition to the compounds of this invention, pharmaceutically acceptable
derivatives or prod rugs of the compounds of this invention may also be
employed in
compositions to treat or prevent the above-identified disorders.
A "pharmaceutically acceptable derivative or prod rug" means any
pharmaceutically acceptable salt, ester, salt of an ester or other derivative
of a
compound of this invention which, upon administration to a recipient, is
capable of
providing, either directly or indirectly, a compound of this invention or an
inhibitorily
active metabolite or residue thereof. Particularly favored derivatives or prod
rugs are
CA 02538147 2004-O1-21
3005-7PCT-9CA
-64-
those that increase the bioavailability of the compounds of this invention
when such
compounds are administered to a mammal (e.g., by allowing an orally
administered
compound to be more readily absorbed into the blood) or which enhance delivery
of
the parent compound to a biological compartment (e.g., the brain or lymphatic
system) relative to the parent species.
Pharmaceutically acceptable prodrugs of the compounds of this invention
include, without limitation, esters, amino acid esters, phosphate esters,
metal salts
and sulfonate esters.
Unless otherwise indicated, all numbers expressing quantities of ingredients,
properties such as molecular weight, reaction conditions, IC5o and so forth
used in
the specification and claims are to be understood as being modified in all
instances
by the term "about". Accordingly, unless indicated to the contrary, the
numerical
parameters set forth in the present specification and attached claims are
approximations. At the very least, and not as an attempt to limit the
application of
the doctrine of equivalents to the scope of the claims, each numerical
parameter
should at least be construed in light of the number of significant figures and
by
applying ordinary rounding techniques. Notwithstanding that the numerical
ranges
and parameters setting forth the broad scope of the invention are
approximations,
the numerical values set in the examples, Tables and Figures are reported as
precisely as possible. Any numerical values may inherently contain certain
errors
resulting from variations in experiments, testing measurements, statistical
analyses
and such.
Unless otherwise defined, all technical and scientific terms used herein have
the same meaning as commonly understood by one of ordinary skill in the art to
which this invention belongs. Although methods and materials similar or
equivalent
to those described herein can be used in the practice or testing of the
present
invention, suitable methods and materials are described below. All
publications,
patent applications, patents, and other references mentioned herein are
incorporated by reference in their entirety. In case of conflict, the present
specification, including definitions, will control. In addition, the
materials, methods,
and examples are illustrative only and not intended to be limiting.
FX~4MP1 F
CA 02538147 2004-O1-21
3005-7PCT-9CA
-65-
EXAMPLE 1: PREPARATION OF PRODUCTION CULTURE
lJnless otherwise noted, all reagents were purchased from Sigma Chemical
Co. (St. Louis, MO), (Aldrich). Micromonospora spp. (deposit accession number
IDAC 070303-01 ) was maintained on agar plates of ISP2 agar (Difco
Laboratories,
Detroit, MI). An inoculum for the production phase was prepared by
transferring the
surface growth of the Micromonospora spp. from the agar plates to 125-mL
flasks
containing 25 mL of sterile medium comprised of 24 g potato dextrin, 3 g beef
extract, 5 g Bacto-casitone, 5 g glucose, 5 g yeast extract, and 4 g CaC03
made up
to one liter with distilled water (pH 7.0). The culture was incubated at about
28°C for
approximately 60 hours on a rotary shaker set at 250 rpm. Following
incubation, 10
mL of culture was transferred to a 2L baffled flask containing 500 mL of
sterile
production medium containing 20 g/L potato dextrin, 20 g/L glycerol, 10 g/L
Fish
meal, 5 g/L Bacto-peptone, 2 g/L CaC03, and 2 g/L (NH4)2S04, pH 7Ø
Fermentation broth was prepared by incubating the production culture at
28°C in a
rotary shaker set at 250 rpm for one week.
EXAMPLE 2: ISOLATION
500 mL ethyl acetate was added to 500 mL of fermentation broth prepared as
described in Example 1 above. The mixture was agitated for 30 minutes on an
orbital shaker at 200 rpm to create an emulsion. The phases were separated by
centrifugation and decantation. Between 4 and 5 g of anhydrous MgS04 was added
to the organic phase, which was then filtered and the solvents removed in
vacuo.
An ethyl acetate extract from 2 L fermentation was mixed with HP-20 resin
(100 mL; Mitsubishi Casei Corp., Tokyo, Japan) in water (300 mL). Ethyl
acetate
was removed in vacuo, the resin was filtered on a Buchner funnel and the
filtrate
was discarded. The adsorbed HP-20 resin was then washed successively with 2 x
125 mL of 50% acetonitrile in water, 2X125 mL of 75% acetonitrile in water and
2 X
125 mL of acetonitrile.
Fractions containing the compound of Formula II were evaporated to dryness
and 100 mg was digested in the 5 mL of the upper phase of a mixture prepared
from
chloroform, cyclohexane, methanol, and water in the ratios, by volume, of
5:2:10:5.
The sample was subjected to centrifugal partition chromatography using a High
CA 02538147 2004-O1-21
3005-7PCT-9CA
-66-
Speed Countercurrent (HSCC) system (Kromaton Technologies, Angers, France)
fitted with a 200 mL cartridge and prepacked with the upper phase of this two-
phase
system. The HSCC was run with the lower phase mobile and the compound of
Formula II was eluted at approximately one-half column volume. Fractions were
collected and the compound of Formula II was detected by TLC of aliquots of
the
fractions on commercial Kieselgel 60F25a plates. Compound could be visualized
by
inspection of dried plates under UV light or by spraying the plates with a
spray
containing vanillin (0.75%) and concentrated sulfuric acid (1.5%, v/v) in
ethanol and
subsequently heating the plate. Fractions contained substantially pure
compound of
Formula II, although highly colored. A buff-colored sample could be obtained
by
chromatography on HPLC as follows.
6 mg of sample was dissolved in acetonitrile and injected onto a preparative
HPLC column (XTerra ODS (10ym), 19x150mm, Waters Co., Milford, MA), with a 9
mL/min flow rate and UV peak detection at 300 nm. The column was eluted with
acetonitrile/buffer (20 mM of NH4HC03) according to the following gradient
shown in
Table 1.
Table 1
Time min Water % Acetonit_rile
0 70 30
5 95 i
5 95
70 30
Fractions containing the compound of Formula II eluted at approximately 11:0
min and were combined, concentrated and lyophilized to give a yield of 3.8 mg
compound.
Alternative Protocol 1
The compound of Formula II was also isolated using the following alternative
protocol. At the end of the incubation period, the fermentation broth from the
baffled
flasks of Example 1 was centrifuged and the supernatant decanted from the
pellet
containing the bacterial mycelia. 100 mL of 100% MeOH was added to the
mycelial
pellet and the sample was stirred for 10 minutes and centrifuged for 15
minutes.
CA 02538147 2004-O1-21
3005-7PCT-9CA
-67-
The methanolic supernatant was decanted and saved. 100 mL of acetone was then
added to the mycelial pellet and stirred for 10 minutes then centrifuged for
15
minutes. The acetonic supernatant was decanted and combined with the
methanolic supernatant. Finally, 100 mL of 20% MeOH/H20 was added to the
mycelial pellet, stirred for 10 minutes and centrifuged for 15 minutes. The
supernatant was combined with the acetonic and methanolic supernatants.
The combined supernatant was added to 400 ml of HP-20 resin in 1000 rnL of
water and the organics were removed in vacuo. The resulting slurry was
filtered on
a Buchner funnel and the filtrate was discarded. Adsorbed HP-20 resin was
washed
successively with 2x500mL of 50% MeOH/H20, 2x500mL of 75% MeOH/H20 and
2x500mL of MeOH.
The individual washes were collected separately and analyzed by TLC as
described above. Those fractions containing the compound of Formula II were
evaporated to near dryness and lyophilized. The lyophilizate was dissolved in
methanol and injected onto a preparative HPLC column (Xterra ODS (10~m),
19x150mm, Waters Co., Milford, MA) with a flow rate of 9 mL/min and peak
detection at 300 nm.
The column was eluted with acetonitrile/buffer (5 mM of NH4HC03) according
to gradient shown in Table 2.
Table 2
i Time min Buffer % Acetonitrile
0 95 5
15 45 55
20 5 95
30 5 95
35 95 5
Fractions containing the compound of Formula II were combined,
concentrated and lyophilized to yield about 33.7 mg of compound.
Alternative Protocol 2
liters of the whole broth from Example 1 are extracted twice with equal
volumes of ethyl acetate and the two extracts are combined and concentrated to
dryness. The dried extract is weighed, and for every gram of dry extract, 100
mL of
MeOH-H20 (2:1 v/v) and 100 mL of hexane is added. The mixture is swirled
gently
CA 02538147 2004-O1-21
3005-7PCT-9CA
-68-
but well to achieve dissolution. The two layers are separated and the aqueous
layer
is washed with 100 mL of hexane. The two hexane layers are combined and the
combined hexane solution is washed with 100 mL methanol:water (2:1, v/v). The
two methanol:water layers are combined and treated with 200 mL of EtOAc and
400
mL of water. The layers are separated and the aqueous layer is extracted twice
more with 200 mL portions of EtOAc. The EtOAc layers are combined and
concentrated. The residue obtained will be suitable for final purification,
either by
HSCC or by HPLC as described above. This extraction process achieves a ten-
fold
purification when compared with the extraction protocol used above.
EXAMPLE 3: ELUCIDATION OF THE STRUCTURE OF COMPOUND OF
FORMULA II.
The structure of the compound of Formula II was derived from spectroscopic
data, including mass, UV, and NMR spectroscopy. Mass was determined by
electrospray mass spectrometry to be 462.6 (FIGURE 1 ), UVmax 230nm with a
shoulder at 290 nm (FIGURE 2). NMR data were collected dissolved in MeOH-d4
including proton (FIGURE 3), and multidimensional pulse sequences gDQCOSY
(FIGURE 4), gHSQC (FIGURE 5), gHMBC (FIGURE 6), and NOESY (FIGURE 7).
A number of cross peaks in the 2D spectra of ECO-04601 are key in the
structural determination. For example, the farnesyl chain is placed on the
amide
nitrogen by a strong cross peak between the proton signal of the terminal
methylene
of that chain at 4.52 ppm and the amide carbonyl carbon at 170 ppm in the
gHMBC
experiment. This conclusion is confirmed by a cross peak in the NOESY spectrum
between the same methylene signals at 4.52 ppm and the aromatic proton signal
at
6.25 ppm from one of the two protons of the tetra substituted benzenoid ring.
Based on the mass, UV and NMR spectroscopy data, the structure of the
compound was determined to be the structure of Formula II.
EXAMPLE 4: ANTIBACTERIAL ACTIVITY (MINIMAL INHIBITORY
CONCENTRATION DETERMINATION)
CA 02538147 2004-O1-21
3005-7 PCT-9CA
-69-
Minimal Inhibitory Concentration (MIC) is defined as the lowest concentration
of drug
that inhibits more than 99% of the bacterial population. The MIC determination
of
ECO-04601 against bacteria strains (Bacillus subtilis - ATCC 23857;
Micrococcus
luteus - ATCC 9341 ) was performed using broth microdilution assay (Methods
for
Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow
Aerobically;
Approved Standard-Fifth Edition. NCCLS document M7-A5 (ISBN 1-56238-394-9).
NCCLS, 940 West Valley Road, Suite 1400, Wayne, Pennsylvania 19087-1898
USA.).
Test compound preparation: The test article ECO-04601 is prepared as 100X
stock
solutions in DMSO, with concentrations ranging from 3.2 mg/ml to 0.0625 mg/ml
(a
two-fold dilution series over 10 points). The first dilution (3.2mg/ml) was
prepared by
resuspending 0.5 mg of each test article in 156.25,u1 of DMSO. The stock is
then
serially diluted by two-fold decrement to obtain the desired concentration
range.
Inoculum preparation: From an overnight culture in Mueller Hinton (MH) broth,
cell
density for each indicator strain (Bacillus subtilis; Micrococcus luteus) was
adjusted
to 0.5 Mc Farland units in 0.85% saline, then further diluted 1/100 in
appropriate
assay medium (~ 1 X 106 cells/ml).
MIC determination: The 100X ECO-04601 solutions was diluted 50 times in MH
broth and dispensed in a 96 well plate, one test concentration per column of
wells,
columns in total. The 11t" column of wells contained MH broth with 1% DMSO,
the 12'" column of wells contained 100,u1 of broth alone. 50 ~I of the final
cell dilution
of each indicator strain was added to each corresponding well of the
microplate
containing 50 ~I of diluted drug or media alone. Assay plates were incubated
at 35°C
for 24 hrs.
The results of the MIC for the compound of ECO-04601, shown in Table 3,
demonstrate a range of antibacterial effects:
CA 02538147 2004-O1-21
3005-7PCT-9CA
-70
Table 3
Indicator strain MIC (~g/mL)
Bacillus subtilis ATCC 23857 12.5
Micrococcus luteus ATCC 9341 6.25
EXAMPLE 5. ANTICANCER ACTIVITY IN VITRO AGAINST HUMAN AND ANIMAL
TUMOR CELL LINES FROM VARIOUS TISSUES
Culture conditions: The cell lines listed in Table 4 were used to characterize
the
cytotoxicity of ECO-04601 against human and animal tumor cell lines. These
cell
lines were shown to be free of mycoplasma infection and were maintained on the
appropriate media (Table 4) supplemented with 10% heat-inactivated fetal
bovine
serum and 1 % penicillin-streptomycin, under 5% C02 at 37°C. Cells were
passaged
twice to three times per week. Viability was examined by staining with 0.25%
trypan
blue and only flasks where cell viability was >95% were used for this study.
Cell lines amplification and plating: Tumor cells were seeded (1-3 x 103 cells
per 100
pL) in 96-wells flat bottom microtiter plates and incubated at 37°C and
5% COZ for
16 hrs before treatment in drug-free medium supplemented with 10% serum.
Evaluation of inhibitory activity on cell proliferation: Cells were incubated
for 96 hrs
with 6 loglo-fold concentrations of the test substance starting at 10pg/ml (20
pM;P.
The test substance stock solution (5 mg/mL) was initially diluted at 1/70 fold
in
medium supplemented with serum. Other concentrations were then obtained from
1/10fold successive dilutions in the same supplemented medium. Cell survival
vvas
evaluated 96 h later by replacing the culture media with 150 pL fresh medium
containing 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid buffer, pH
7.4.
Next, 50 pL of 2.5 mg/mL of 3-(4,5-dimethylthiazo-2-yl)-2,5-
diphenyltetrazolium
bromide (MTT) in phosphate buffer solution, pH 7.4, was added. After 3-4h of
incubation at 37°C, the medium and soluble MTT was removed, and 200 pL
of
dimethylsulfoxide was added to dissolve the precipitate of reduced MTT
followed by
addition of 25 pL glycine buffer (0.1 M glycine plus 0.1 M NaCI, pH 10.5). The
absorbance was determined at 570 nm with a microplate reader. Results were
CA 02538147 2004-O1-21
3005-7PCT-9CA
-71-
expressed as the concentration of drug which inhibits 50% of the cell growth
(IC~o).
The IC~o values shown in Table 4 demonstrated a pharmacologically relevant
cytotoxic activity of ECO-04601 against a variety of tumor types such as
leukemias,
melanomas, pancreatic and breast carcinomas.
Table 4
~II lines Type Origin Source Culture medium IC5o
x10'6M
K562 Leukemia Human ATCC RPMI 1640 8.6
I ~
m elo eneous
P388 Leukemia Mouse ATCC RPM11640 10.9
183 Leukemia Human ATCC RPM11640 2.7
B16 F10 Melanoma Mouse ATCC RPMI 1640 11.4
SK-MEL 28 Melanoma Human ATCC RPMI 1640 14.0
SK-MEL Melanoma Human ATCC RPM11640 14.3
28VEGF (expressing
VEGF
SK-MEL-1 Melanoma ~ Human ATCC EMEM 1 % non- 14.1
~ i
i ~ essential amino
i I acid 1 % Sodium
i
ur vate ~ i
Panc 96 Pancreatic Human ATCC RPMI 1% Sodium 12.5
carcinoma ur vate
Panc 10.05 Pancreatic Human ATCC RPMI 1% Sodium 14.2
carcinoma ur vate Insulin
i MCF-7 Breast Human ATCC RPMI 1640 9.7
adenocarcinoma
EXAMPLE 6. ANTICANCER ACTIVITY IN VITRO AGAINST VARIOUS HUMAN
TUMOR CELL LINES FROM THE U.S. NATIONAL CANCER INSTITUTE PANEL
A study measuring the in vitro antitumor activity of ECO-04601 was
performed by the National Cancer Institute (National Institutes of Health,
Bethesda,
Maryland, USA) against panel of human cancer cell lines in order to determine
the
ECO-04601 concentrations needed to obtain a 50% inhibition of cell
proliferation
(Gl5o). The operation of this unique screen utilizes 50 different human tumor
cell
lines, representing leukemia, melanoma and cancers of the lung, colon, brain,
ovary,
breast, prostate, and kidney.
CA 02538147 2004-O1-21
3005-7PCT-9CA
Culture conditions and plating:
-72-
The human tumor cell lines of the cancer-screening panel were grown in
RPMI 1640 medium containing 5% fetal bovine serum and 2 mM L-glutamine. For a
typical screening experiment, cells were inoculated into 96 well microtiter
plates in
100 ~L at plating densities ranging from 5,000 to 40,000 cells/well depending
on the
doubling time of individual cell lines (Table 5). After cell inoculation, the
microtiter
plates were incubated at 37°C, 5% C02, 95% air and 100% relative
humidity for 24 h
prior to addition of experimental drugs. After 24 h, two plates of each cell
line were
fixed in situ with TCA, to represent a measurement of the cell population for
each
cell line at the time of drug addition (Tz).
Evaluation of inhibitory activity on cell proliferation:
ECO-04601 was provided as a lyophilized powder with an estimated purity of
90+%. The compound was stored at -20°C until day of use. ECO-04601 was
solubilized in dimethyl sulfoxide at 400-fold the desired final maximum test
concentration. At the time of drug addition, an aliquot of frozen concentrate
was
thawed and diluted to twice the desired final maximum test concentration with
complete medium containing 50pg/mL gentamicin. Additional four, 10-fold or'/2
log
serial dilutions were made to provide a total of five drug concentrations plus
control.
Aliquots of 100 pl of these different drug dilutions were added to the
appropriate
microtiter wells already containing 100 ~I of medium, resulting in the
required final
drug concentrations (8.0 x 10-5 M to 8.0 x 10-9 M).
Following drug addition, the plates were incubated for an additional 48 h at
37°C, 5°/~ COz, 95% air, and 100% relative humidity. For
adherent cells, the assay
was terminated by the addition of cold TCA. Cells were fixed in situ by the
gentle
addition of 50 pl of cold 50% (w/v) TCA (final concentration, 10% TCA) and
incubated for 60 minutes at 4°C. Supernatants were discarded, and the
plates were
washed five times with tap water and air-dried. Sulforhodamine B (SRB)
solution
(100 pl) at 0.4% (w/v) in 1 % acetic acid was added to each well, and plates
were
incubated for 10 minutes at room temperature. After staining, unbound dye was
removed by washing five times with 1 % acetic acid and the plates were air-
dried.
Bound stain was subsequently solubilized with 10 mM trizma base, and the
absorbance was read on an automated plate reader at a wavelength of 515 nm.
For
CA 02538147 2004-O1-21
3005-7PCT-9CA
-73-
suspension cells, the methodology was the same except that the assay was
terminated by fixing settled cells at the bottom of the wells by gently adding
50 pl of
80% TCA (final concentration, 16% TCA).
The growth inhibitory activity of ECO-04601 was measured by NCI utilizing
the Gl5o value, rather than the classical IC5o value. The GlSO value
emphasizes the
correction for the cell count at time zero and, using the seven absorbance
measurements [time zero, (Tz), control growth, (C), and test growth in the
presence
of drug at the five concentration levels (Ti)], Gl5o is calculated as [(Ti-
Tz)/(C-Tz)] x
100 = -50, which is the drug concentration resulting in a 50% reduction in the
net
protein 'increase (as measured by SRB staining) in control cells during the
drug
incubation.
Result:
ECO-04601 shows a significant antitumor activity against several types of
tumor as revealed by the NCI screening. Results of the screen are shown in
Table 5,
and more detailed results of activity against gliomas are shown in Example 7
(Table
6).
Table 5
Inoculation GlSo
Cell Line Name Type Origin Density (x10-6M)
(number of
cells/well)
CCRF-CEM Leukemia Human 40,000 1.08
K-562 Leukemia Human 5,000 1.43
RPMI-8226 Leukemia Human 20,000 3.15
A549/ATCC Non-Small Cell Human 7,500 9.10
Lung
EKVX Non-Small Cell Human 20,000 0.23
Lung
HOP-62 Non-Small Cell Human 10,000 8.29
Lung
NCI-H226 Non-Small Cell Human 20,000 2.00
Lung
NCI-H23 Non-Small Cell Human 20,000 2.02
Lung
NCI-H460 Non-Small Cell Human 7,500 13.60
Lung
NCI-H522 Non-Small Cell Human 20,000 3.44
Lung
COLD 205 Colon Human 15,000 12.70
HCT-116 Colon Human 5,000 2.92
HCT-15 Colon Human 10,000 9.73
HT29 Colon Human 5,000 20.70
~SW-620 Colon Human 10,000 2.72
CA 02538147 2004-O1-21
3005-7PCT-9CA
-74-
SF-268 CNS Human 15,000 4.94
SF-295 CNS Human 10,000 12.70
SF-539 CNS Human 15,000 0.0075
SNB-19 CNS Human 15,000 2.90
SNB-75 CNS Human 20,000 7.71
U251 CNS Human 7,500 2.19
LOX IMVI Melanoma Human 7,500 4.53
M14 Melanoma Human 15,000 4.57
SK-MEL-2 Melanoma Human 20,000 25.0
SK-MEL-28 Melanoma Human 10,000 11.6
SK-MEL-5 Melanoma Human 10,000 7.80
UACC-257 Melanoma Human 20,000 2.31
UACC-62 Melanoma Human 10,000 1.55
IGR-OV1 Ovarian Human 10,000 3.11
OVCAR-3 Ovarian Human 10,000 13.50
OVCAR-4 Ovarian Human 15,000 9.67
OVCAR-5 Ovarian Human 20,000 2.81
OVCAR-8 Ovarian Human 10,000 ~ 2.65 i
SK-OV-3 Ovarian Human 20,000 4_.00
786-0 Renal Human 10,000 6.99
A498 Renal Human 25,000 22.30
ACHN Renal Human 10,000 3.10
CAKI-1 Renal Human 10,000 15.20
RXF 393 Renal Human 15,000 7.71
SN12C Renal Human 15,000 3.85
UO-31 Renal Human 15,000 19.70
DU-145 Prostate Human 10,000 3.56
MCF7 Breast Human 10,000 10.10
NCI/ADR-RES Breast Human 15,000 18.30
MDA-MB- Breast Human 20,000 2.72
231/ATCC
HS 578T Breast Human 20,000 2.76
MDA-MB-435 Breast Human 15,000 15.30
BT-549 Breast Human 20,000 U.11
T-47D Breast Human 20,000 U.77
The results indicate that ECO-04601 was effective against most of the human
tumor
cell lines that have been assayed in the NCI screening panel suggesting a
broad
anticancer activity against several types of human cancer.
EXAMPLE 7: IN VITRO ANTIPROLIFERATIVE STUDY AGAINST A PANEL OF
GLIOMA CELL LINES
CA 02538147 2004-O1-21
3005-7PCT-9CA
-75-
The anticancer activity of ECO-04601 was evaluated using a panel of glioma
cancer cell lines shown in Table 6, and the 50% inhibition of cell
proliferation (IC5o)
was determined.
Culture conditions:
The cell lines listed in Table 6 were shown to be free of mycoplasma infection
and were maintained on DMEM medium supplemented with 10% heat-inactivated
fetal bovine serum and 1 % penicillin-streptomycin, under 5% C02 at
37°C. Cells
were passaged once a week. Prior to use the cells were detached from the
culture
flask by treating with trypsin for five to ten minutes. The cells were counted
with a
Neubauer glass slide and viability assessed by 0.25% trypan blue exclusion.
Only
flasks with >95% cell viability, were used in the study.
Cell lines amplification and plating:
Cells, 5 x 103 cells per well in 100 pL drug-free medium supplemented with
10% serum, were plated in 96-well flat bottom microtiter plates and incubated
at
37°C for 48 hrs before treatment.
Evaluation of inhibitory activity on cell proliferation:
Cells (in triplicate wells) were incubated 96 hrs with medium containing
different concentrations of ECO-04601, starting at 5.0 pg/ml (10 pM). The
compound
was used in a solution of 1 % DMSO in D-MEM or RPMI media (or other equivalent
media). The concentrations of ECO-04601 were as follows: 10 pM (5.0 pg/ml), 1
pM
(0.50 pg/ml), 0.5 pM (0.25 pg/ml), 0.1 pM (0.050 pg/ml), 0.5 pM (0.025 pg/ml),
0.01
pM (0.0050 pg/ml), 0.001 pM (0.00050 pg/ml). Negative controls were cells
treated
with vehicle alone (1 % DMSO in culture medium). Positive controls were cells
treated with 4 to 6 increasing concentrations of cisplatin (CDDP) (data not
shown).
The optical density was measured before incubation (time 0) and following 96
hrs of
incubation with test compound in order to measure the growth rate of each cell
line.
At the end of the cell treatment, cell culture media was replaced with 150 pl
of
fresh medium containing 10 mM of 4-(2-hydroxyethyl)-1-piperazineethanesulfonic
acid buffer, pH 7.4. Then 50 pl of 2.5 mg/ml of 3-(4,5-dimethylthiazo-2-yl)-
2,5-
diphenyltetrazolium bromide in PBS pH 7.4, were added to each well and the
culture
plates incubated for 4 hrs at 37°C. The resulting supernatant was
removed and
CA 02538147 2004-O1-21
3005-7PCT-9CA
-76-
formazan crystals were dissolved with 200 pl of DMSO followed by 25 pl of
glycine
buffer (0.1 M glycine plus 0.1 M NaCI, pH 10.5). The optical density was read
in
each well using a single wavelength spectrophotometer plate reader at 570 nm.
Results were expressed as the concentration of drug, which inhibits 50% of the
cell
growth (IC5o). Each of the cell lines was tested in at least 3 independent
experiments.
Results shown in Table 6 confirmed the activity of ECO-04601 against
different brain cancer cell lines including gliosarcoma, which is the most
malignant
form of type IV glioblastoma multiform. Gliosarcomas are a mixture of glial
and
endothelial cells and are resistant to any chemotherapy.
Table 6
Cell linesType Origin Source ICso
(x 10-6
M)
9L Gliosarcoma Rat ATCC 6.82 2.90
GHD Astrocytoma Human ATCC 6.29 2.98
U 373 Astrocytoma Human ATCC 3.83 1.37
GL26 Glioblastoma Human ATCC 8.93 1.10
C6 Glioblastoma Rat ATCC 4.28 2.82
Oligodendrogliom ATCC
DN a Human 3.26 0.93
OligodendrogliomI ATCC
GHA a Hurnan 1.78 0.84
'... ~
EXAMPLE 8: EFFECT ON THE ENZYMATIC ACTIVITY OF HUMAN
LIPOXYGENASE (5-LO)
5-Lipoxygenase catalyzes the oxidative metabolism of arachidonic acid to 5-
hydroxyeicosatetraenoic acid (5-HETE), the initial reaction leading to
formation of
leukotrienes. Eicosanoids derived from arachidonic acid by the action of
lipoxygenases or cycloxygenases have been found to be involved in acute and
chronic inflammatory diseases (i.e. asthma, multiple sclerosis, rheumatoid
arthritis,
ischemia, edema) as well in neurodegeneration (Alzheimer disease), aging and
CA 02538147 2004-O1-21
3005-7PCT-9CA
-77-
various steps of carcinogenesis, including tumor promotion, progression and
metastasis.
The aim of this study was to determine whether ECO-04601, is able to block
the formation of leukotrienes by inhibiting the enzymatic activity of human 5-
LO.
Methods employed are based on Carter et al (1991 ) J. Pharmacol. Exp. Ther.
256(3):929-937, and Safayhi (2000), Planta Medica 66:110-113 which are
incorporated herein in their entirety by reference.
Experimental Design:
Human peripheral blood mononuclear cells (PMNs) were isolated through a
Ficoll-Paque density gradient. PMNs were stimulated by addition A23187 (30 pM
final concentration). Stimulated PMNs were adjusted to a density of 5 x106
cells/mL
in HBBS medium and incubated with the vehicle control (DMSO), ECO-04601 (at
final concentrations of 0.1, 0.5, 1, 2.5, 5 and 10 pM) and NDGA as positive
control
(at final concentrations of 3, 1, 0.3, 0.1 and 0.03 pM) for 15 minutes at
37°C.
Following incubation, samples were neutralized with NaOH and centrifuged.
Leukotriene B4 content was measured in the supernatant using an Enzyme
Immunosorbant Assay (EIA) assay.
Results:
Results shown in Figure 8 demonstrated that ECO-04601 inhibited the activity
of human 5-LO with an apparent ICSO = 0.93 pM (versus 0.1 NM for the positive
control NDGA) and therefore displays anti-inflammatory properties.
EXAMPLE 9: IN VIVD EFFICACY IN A GLIOMA MODEL
The aim of this study was to test whether ECO-04601 administered by i.p.
route prevents or delays tumor growth in C6 glioblastoma cell-bearing mice,
and to
determine an effective dosage regimen.
Animalc~
A total of 60 six-week-old female mice (Mus musculus nude mice), ranging
between 18 to 25 g in weight, were observed for 7 days before treatment.
Animal
experiments were performed according to ethical guidelines of animal
CA 02538147 2004-O1-21
3005-7PCT-9CA
-78-
experimentation (Charte du comite d'ethique du CNRS, juillet 2003) and the
English
guidelines for the welfare of animals in experimental neoplasia (WORKMAN, P.,
TWENTYMAN, P., BALKWILL, F., et al. (1998). United Kingdom Coordinating
Committee on Cancer Research (UKCCCR) Guidelines for the welfare of animals in
experimental neoplasia (Second Edition, July 1997; British Journal of Cancer
77:1-
10). Any dead or apparently sick mice were promptly removed and replaced with
healthy mice. Sick mice were euthanized upon removal from the cage. Animals
were
maintained in rooms under controlled conditions of temperature
(23~2°C), humidity
(45~5%), photoperiodicity (12 hrs light / 12 hrs dark) and air exchange.
Animals were
housed in polycarbonate cages (5/single cage) that were equipped to provide
food
and water. Animal bedding consisted of sterile wood shavings that were
replaced
every other day. Food was provided ad libitum, being placed in the metal lid
on the
top of the cage. Autoclaved tap water was provided ad libitum. Water bottles
were
equipped with rubber stoppers and sipper tubes. Water bottles were cleaned,
sterilized and replaced once a week. Two different numbers engraved on two
earrings identified the animals. Each cage was labelled with a specific code.
Tumor Cell Line:
The C6 cell line was cloned from a rat glial tumor induced by N-
nitrosomethyurea (NMU) by Premont et al. (Premont J, Benda P, Jard S., [3H]
norepinephrine binding by rat glial cells in culture. Lack of correlation
between
binding and adenylate cyclase activation. Biochim Biophys Acta. 1975 Feb
13;381 (2):368-76.) after series of alternate culture and animal passages.
Cells were grown as adherent monolayers at 37°C in a humidified
atmosphere (5% COz, 95% air). The culture medium was DMEM supplemented with
2 mM L-glutamine and 10% fetal bovine serum. For experimental use, tumor cells
were detached from the culture flask by a 10 min treatment with trypsin-
versen. The
cells were counted in a hemocytometer and their viability assessed by 0.25%
trypan
blue exclusion.
Preparation of the Test Article:
For the test article, the following procedure was followed for reconstitution
(performed immediately preceding injection). The vehicle consisted of a
mixture of
benzyl alcohol (1.5%), ethanol (8.5%), propylene glycol (27%), PEG 400 (27%),
CA 02538147 2004-O1-21
3005-7PCT-9CA
_79_
dimethylacetamide (6%) and water (30%). The vehicle solution was first
vortexed in
order to obtain a homogeneous liquid. 0.6 mL of the vortexed vehicle solution
was
added to each vial containing the test article (ECO-04601 ). Vials were mixed
thoroughly by vortexing for 1 minute and inverted and shaken vigorously. Vials
were
mixed again prior to injection into each animal.
Animal Inoculation with tumor cells:
Experiment started at day 0 (Do). On Do, mice received a superficial
intramuscular injection of C6 tumor cells (5 x 105 cells) in 0.1 mL of DMEM
complete
medium into the upper right posterior leg.
Treatment regimen and Results:
In a first series of experiments, treatment started 24 hrs following
inoculation
of C6 cells. On the day of the treatment, each mouse was slowly injected with
100
pL of test or control articles by i.p. route. For all groups, treatment was
performed
until the tumor volume of the saline-treated mice (group 1 ) reached
approximately 3
crra3 (around day 16). Mice of group 1 were treated daily with a saline
isosmotic
solution for 16 days. Mice of group 2 were treated daily with the vehicle
solution for
16 days. Mice of group 3 were treated daily with 10 mg/kg of ECO-04601 for 16
days. Mice of group 3 were treated every two days with 30 mg/kg of ECO-04601
and
received 8 treatments. Mice of group 5 were treated every three days with 30
mg/kg
of ECO-04601 and received 6 treatments. Measurement of tumor volume started as
soon as tumors became palpable (>100 mm3; around day 11 post-inoculation) and
was evaluated every second day until the end of the treatment using callipers.
As
shown in Table 7 and Figure 9, the mean value of the tumor volume of all ECO-
04601 treated groups (6 mice/group) was significantly reduced as demonstrated
by
the one-way analysis of variance (Anova) test followed by the non-parametric
Dunnett's multiple comparison test comparing treated groups to the saline
group. An
asterisk in the P value column of Table '7 indicates a statistically
significant valuE:,
while "ns'° signifies not significant.
Table 7
CA 02538147 2004-O1-21
3005-7PCT-9CA
-80-
Treatment Treatment Tumor volume % P
regimen (mm3) Inhibitiovalue
I
(mean SEM) n
Saline Q1 x 16 3,004.1 - -
I 249.64
Vehicle Q1 x 16 2,162.0 350.028.0% >0.05
solution ns
ECO-04601 Q1 x 16 1,220.4 59.4% <0.01
(10 mg/kg) 283:46
ECO-04601 Q2 x 8 1,236.9 58.8% <0.01
(30 mg/kg) 233.99
I
ECO-04601 Q3 x 6 1,184.1 60.6% <0.01
I
(30 mg/kg) 221.45
In a second series of experiments, treatment started at day 10 following
inoculation of C6 cells when tumors became palpable (around 100 to 200 mm3)
Treatment was repeated daily for 5 consecutive days. On the day of the
treatment,
each mouse was slowly injected with 100 pL of ECO-04601 by i.p. route. Mice of
group 1 were treated daily with saline isosmotic solution. Mice of group 2
were
treated daily with the vehicle solution. Mice of group 3 were treated daily
with 20
mg/kg of ECO-04601. Mice of group 4 were treated daily with 30 mg/kg of ECO-
04601. Mice were treated until the tumor volume of the saline-treated control
mice
(group 1 ) reached around 4 cm3. Tumor volume was measured every second day
until the end of the treatment using callipers. As shown in Table 8 and Figure
70, the
mean value of the tumor volume of all ECO-04601 treated groups (6 mice/group)
was significantly reduced as demonstrated by the one-way analysis of variance
(Anova) test followed by the non-parametric Dunnett's multiple comparison test
comparing treated groups to the saline group. An asterisk in the P value
column of
Table 8 indicates a statistically significant value, while "ns" signifies not
statistically
significant.
Histological analysis of tumor sections showed pronounced morphological
CA 02538147 2004-O1-21
3005-7PCT-9CA
-81-
changes between ECO-04601-treated tumors and control groups. In tumors treated
with ECO-04601 (20 - 30 mg/kg), cell density was decreased and the nuclei of
remaining tumor cells appeared larger and pycnotic while no such changes were
observed for vehicle-treated mice (Figure 11 ).
Table 8
Treatment Treatment ~ Tumor volume% P ~I,
i
regimen (mm3) Inhibitiovalue
I
(mean SEM) n
Saline Q1 x 5 4,363.1 - -
614.31
Vehicle solutionQ1 x 5 3,205.0 26.5% >0.05
i
632.37 ns
ECO-04601 Q1 x 5 1,721.5 60.5% <0.01
!, (20 mg/kg) 374.79
ECO-04601 Q1 x 5 1,131.6 74.1% <0.01
(30 mglkg) 525.21
EXAMPLE 10: GENERATION OF VARIANTS OF ECO-04601 ACCORDING TO
THE INVENTION
Variants of the ECO-04601 molecule, for example those identified herein as
Formulae III-LIX, can be generated by standard organic chemistry approaches.
General principles of organic chemistry required for making and manipulating
the
compounds described herein, including functional moieties, reactivity and
common
protocols are described, for example, in "Advanced Organic Chemistry," 3~d
Edition
by Jerry March (1985) which is incorporated herein by reference in its
entirety. In
addition, it will be appreciated by one of ordinary skill in the art that the
synthetic
methods described herein may use a variety of protecting groups, whether or
not
they are explicitly described. A "protecting group" as used herein means a
moiety
used to block one or more functional moieties such as reactive groups
including
oxygen, sulfur or nitrogen, so that a reaction can be carried out selectively
at another
CA 02538147 2004-O1-21
3005-7PCT-9CA
-82-
reactive site in a polyfunctional compound. General principles for the use of
protective groups, their applicability to specific functional groups and their
uses are
described for example in T. H. Greene and P. G. M. Wuts, Protective Groups in
Organic Synthesis, 3rd Edition, John Wiley & Sons, lVew York (1999).
Scheme 1: Epoxide variants
The epoxide compounds of the present invention (e.g., compounds according
to exemplary Formulae VII-XIV) are made from the compound of Formula II (ECO-
04601 ) by treatment with any of a number of epoxidizing reagents such as
perbenzoic acid, monoperphthalic acid or more preferably by m-chloroperbenzoic
acid in an inert solvent such as tetrahydrofuran (THF) dichloromethane or 1,2-
dichloroethane. It will be appreciated by one of ordinary skill in the art
that slightly
greater than one molecule equivalent of epoxidizing agent will result in the
maximal
yield of mono-epoxides, and that the reagent, solvent, concentration and
temperature of the reaction will dictate the ratio of specific mono-epoxides
formed. It
will also be appreciated that the mono-epoxides will be enantiorneric
mixtures, and
that the di-epoxides and the tri-epoxide can be prepared as diastereomers and
that
the conditions of the reaction will determine the ratios of the products. One
skilled in
the art will appreciate that under most conditions of reactions the product
will be a
mixture of all possible epoxides and that these may be separated by standard
methods of chromatography. Exemplary approaches to the generation of mono-, di-
and tri-epoxides are provided below.
A) Mono-epoxides of the Formulae VII, VIII, and IX by epoxidation of the
compound of Formula II:
Formula VII Formula VIII
O CH, CH3 CH, 0 CH, CH, CH,
i
N / / CH, N / / CHI
O ~ 0
N
~N ~ \ HO
HO / ~ OH H ~OH
H
H HO
0 CH, CH, CH,
i
N / / CH,
O
HO H ~ ~ OH Formula IX
HO
CA 02538147 2004-O1-21
;005-PCT-OCR
-g~_
To a solution of the compound of Formula II dissolved in tetrahydrofuran
~TI~F) is added 1.1 equivalents of meta-chloroperbenzoic acid. The reaction is
cooled in an 'ice bath and stirred at 0 °C for 1-2 hours. The reaction
mixture is then
evaporated to dryness, re-dissolved in methanol and subjected to liquid
chromatography on a column of Sephadex LH-20 to isolate a mixture of
predominantly the compounds of Formulae VII, VIII and IX, contaminated with
some
unchanged starting material and some di- and tri- epoxides. The compounds of
Formulae VII, Vill and XIX are separated and purified by HPLC using the system
described in Example 2 for the purification of the compound of Formulae II. In
a
typical experiment yields of 15% to 25% are obtained for each of the compounds
of
Formulae VII, VIII and IX.
B) Synthesis of Compounds of Formulae X, XI, and XII by di-epoxidation of
Compound of Formula II~
~orrnu~a ~ Forrnu~a ~9
o cH, cH, !~H,
;,~cH, cH; ;1
~~i w ' ~ ~cH, ,
' ~, ~ '
,, c i~~'j /%~
r,_~'~ HO '
~~oH
"_cH
HG
c CH, CH3 CH,
~~i\/~N~ CH,
/~ ~Y ~ ~.
HO H ~ ~~OH Formula X~~
HO
To a solution of the compound of Formula II dissolved in tetrahydrofuran
~THF) is added 2.3 equivalents of meta-chloroperbenzoic acid. The reaction is
cooled in an ice bath and stirred at 0 °C for 1-2 hours. The reaction
mixture is then
evaporated to dryness, re-dissolved in methanol and subjected to liquid
chromatography on a column of Sephadex LH-20 to isolate a mixture of
predominantly the compounds of Formulae X, XI and XII, contaminated with
traces
of unchanged starting material and some mono- and tri- epoxides. The Compounds
of Formulae X, XI and XII are separated and purified by HPLC using the system
CA 02538147 2004-O1-21
3005-7PCT-9CA
-84-
described in Example 2 for the purification of the compound of Formulae II. In
a
typical experiment, yields of 15% to 20% are obtained for each of the
compounds of
Formulae X, XI and XII.
C) Synthesis of Compound of Formula XIII by tri-epoxidation of Compound of
Formula II:
O CH; CHj i H3
N~ CHj
O O O
HO ~N~~ ~ OH Formula XI ll
H
HO
To a solution of the compound of Formula II, dissolved in tetrahydrofuran
(THF), is added 3.5 equivalents of meta-chloroperbenzoic acid. The reaction is
cooled in an ice bath and stirred at 0 °C for 1-2 hours. The reaction
mixture is then
evaporated to dryness, re-dissolved in methanol and subjected to liquid
chromatography on a column of Sephadex LH-20 to isolate the compound of
Formula XIII as a mixture of diasteriomers in a yield of 80+%.
Scheme 2: Synthesis of Compound of Formula III by N-acetylation of Compound of
Formula II.
UII CH CH, CH,
~N~ /I / CHo
Formula III
N
Ho ,
o=c ~ off
H,C HO
To a solution of Compound of Formula II dissolved in tetrahydrofuran (THF) is
added 1.2 equivalents of acetic anhydride and a few drops of triethylamine.
The
reaction mixture allowed to stand at room temperature for 1-2 hours and then
evaporated to dryness under reduced pressure to obtain the Compound of Formula
III in an essentially pure form in an almost quantitative yield.
Scheme 3: Syntheses of Compounds of Formulae IV and V by N-alkylation of
Compound of
Formula II.
CA 02538147 2004-O1-21
3005-7PCT-9CA
-85-
CH, CH, CH,
i' ~ IAN ~ ~~ ~~H Formula IV R = benzyl
Formula V R = ethyl
off
Ho
To a solution of Compound of Formula II dissolved in terachloroethylene is
added 1.2 equivalents of the appropriate alkyl bromide (benzyl bromide for the
compound of formula IV or ethyl bromide for the Compound of Formula V). The
reaction mixture the reaction mixture is heated under reflux for 1-2 hours and
then
evaporated to dryness under reduced pressure to obtain the Compound of Formula
IV or the Compound of Formula V respectively, in an essentially pure form in
an
almost quantitative yield.
Scheme 4: Syntheses of Compounds of Formulae XL, XLI and XLII by catalytic
reduction of Compound of Formula II.
O CH3 CH3 CH3 ".,3 .."3 '
N / / CH3 ~ ~ N / / CH3
N ~ ~ Formula XL ~ ~ ~ Formula XLI
HO HO
OH H ' OH
HO HO
0 CH3 CH, CH3
i
N / / CH3
H° ; ~ ~ off Formula XLII
H ~
HO
A solution of the Compound of Formula II (462 mg) in ethanol (200 ml) with
palladium on charcoal (25 mg of 5%) is shaken in an hydrogenation apparatus in
an
atmosphere of hydrogen. The uptake of hydrogen by the reaction is measured
carefully and at the point where one millimole of hydrogen has been consumed,
shaking is stopped, the vessel is rapidly evacuated and the atmosphere is
replaced
with nitrogen. The catalyst is removed by filtration and the filtrate is
concentrated to
obtain a crude mixture of the Compounds of Formulae XL, XLI and XLII
contaminated by unreacted starting material and minor amounts of over reduced
products. The desired products may be separated and purified by HPLC or HSCC
chromatography using the systems as described in Example 2 above, to obtain
approximately 100 mg of each of the Compounds of Formulae XL, XLI and XLII.
CA 02538147 2004-O1-21
3005-7 PCT-9CA
-86-
Scheme 5: Syntheses of Compounds of Formulae XLIII, XLIV and XLV by catalytic
reduction of
Compound of Formula II.
O CHI CH3 CH3
i \ ~
i '~N / CHa
N / CHj
Formula XLIV
Formula XLIII H
HO OH
H _ OH
HO
HO
0 CH, CH3 CHI
i
N / CH3
Formula XLV
N
HO / OH
H
HO
A solution of the Compound of Formula II (462 mg) in ethanol (200 ml) with
palladium on charcoal (25 mg of 5%) is shaken in an hydrogenation apparatus in
an
atmosphere of hydrogen. The uptake of hydrogen by the reaction is measured
carefully and at the point where two millimoles of hydrogen has been consumed,
shaking is stopped, the vessel is rapidly evacuated and the atmosphere is
replaced
with nitrogen. The catalyst is removed by filtration and the filtrate is
concentrated to
obtain a crude mixture of the Compounds of Formulae XLIII, XLIV and XLV
contaminated by trace amounts unreacted starting material and minor amounts of
under and over reduced products. The desired products may be separated and
purified by HPLC or HSCC chromatography using the systems as described in
Example 2 above, to obtain approximately 100 mg of each of the Compounds of
Formulae XLIII, XLIV and XLV.
Scheme 6: Syntheses of Compound of Formula XLVI by catalytic reduction of
Compound of Formula ll.
O CH3 CH3 ~ H3
i ~ ~
N~ CH3
HO ; / ~ off Formula XLVI
H
HO
A solution of the Compound of Formula II (462 mg) in ethanol (200 ml) with
palladium on charcoal (25 mg of 5%) is shaken in an hydrogenation apparatus in
an
CA 02538147 2004-O1-21
3005-7PCT-9CA
_87_
atmosphere of hydrogen. The uptake of hydrogen by the reaction is measured
carefully and at the point where three millimoles of hydrogen has been
consumed,
shaking is stopped, the vessel is rapidly evacuated and the atmosphere is
replaced
with nitrogen. The catalyst is removed by filtration and the filtrate is
concentrated to
obtain an essentially pure sample of the Compound of Formula XLVI.
Scheme 7: Syntheses of Compound of Formula VI by peracetylation of Compound
of Formula II.
H3~
A solution of the Compound of Formula II (100 mg) in acetic anhydride (5 ml)
is treated with pyridine (250 ul). The reaction mixture is allowed to stand
overnight
at room temperature and is then diluted with toluene (100 ml). The toluene
solution
is washed well with aqueous 5% sodium bicarbonate solutions, then with water
and
is finally concentrated under reduced pressure to give an essentially pure
sample of
the Compound of Formula VI in almost quantitative yield.
Scheme 8: Syntheses of Compound of Formula LI by opening the epoxide of
Compound of Formula VII.
O i H~ CH, CHI
N / / CHI
OH OH
N i ~ Formula LI
Ho ~ off
H ~
HO
A solution of the Compound of Formula VII (100 mg) in tetrahydrofuran (50
ml) is treated with 1 N aqueous hydrochloric acid (5 ml). The reaction mixture
is
stirred overnight at room temperature and is then diluted with toluene (100
ml) and
water (200 ml). The toluene layer is separated and the aqueous layer is
extracted
with a further 100 ml of toluene. The combined toluene layers are washed once
more with water (50 ml) and the separated and dried under vacuum to give the
CA 02538147 2004-O1-21
3005-7PCT-9CA
_88_
vicinal glycol Compound of Formula LI.
Scheme 9: Syntheses of Compounds of Formulae XLVII, XLIX and LI by ozonolysis
of Compound of Formula II.
0 CFi,
O CH, CH, i ~~0
/ NN
/ N Ho N i~ Formula XLIX
Ho ; ~ ~ Formula XLVII H OH
H ~ ~OH HO
HO
0
i ~ ~O
N
Formula LI
N ~
HO / OH
H
HO
A solution of the Compound of Formula II (462 mg) in dry ethyl acetate (200
ml) in an ozonolysis apparatus is cooled to below -20°C. A stream of
ozone-
containing oxygen is passed into the solution from an ozone generator, which
has
been precalibrated such that the rate of ozone generation is known. To obtain
predominantly the compound of Formula XLVII the passage of ozone is halted
after
0.9 millimole have been generated. To obtain predominantly the compound of
Formula XLIX the ozone passage is halted after 2 millimoles have been
generated
and to obtain the compound of Formula LI as the predominant product 3.3
millimoles
of ozone are generated.
At the completion of the ozonolysis, the reaction mixture is transferred to an
hydrogenation apparatus, 5% palladium on calcium carbonate catalyst (0.2 g) is
added to the reaction mixture which is maintained at less than -20°C
and is
hydrogenated. When hydrogen uptake is complete the hydrogen atmosphere is
r eplaced with nitrogen and the reaction mixture is allowed to come to room
temperature, filtered to remove catalyst and the filtrate is concentrated. The
crude
product may be purified by chromatography using either HPLC or HSCC with the
systems as described in Example 2 to give, dependent on the amount of ozone
used, Compounds of Formulae XLVII, XLIX and LI.
Scheme 10: Synthesis of Compound of Formulae XLVIII by reduction of the
3005-7PCT-9CA
CA 02538147 2004-O1-21
_89_
aldehyde of Compound of Formula XLVII.
O CH CH,
i
i N ~~,~~.OH
y y
Ho N ~ \ off Formula XLVIII
H
HO
A solution of the Compound of Formula XLVIII (50 mg) in isopropanol (5 ml)
is cooled in an ice-salt bath and sodium borohydride (10 mg) is added and the
mixture is stirred for 20 minutes. It is then diluted with water (20 ml) and
extracted
twice with toluene (10 ml portions) at ambient temperature. The combined
toluene
extracts are filtered and the filtrate is concentrated to give the Compound of
Formula XLVII.
Scheme 11: Syntheses of Compounds of Formulae XIV and XV by epoxidation of
the Compound of Formula XLII.
0 CH, !'H, CH 0 ~ ,, ~H..
l~ . v
":~% ~ ,r
~N~~ / ~CH3 ~ ~~ .i ~ w,
~0 ~
\ Formula XIV Ho ;N ~ \ Formul;
HO / OH H ,~OH
H
HO HO
To a solution of Compound of Formula XLII dissolved in tetrahydrofuran
(THF) is added 1.1 equivalents of meta-chloroperbenzoic acid. The reaction is
cooled in an ice bath and stirred at 0 °C for 1-2 hours. The reaction
mixture is then
evaporated to dryness, re-dissolved in methanol and subjected to liquid
chromatography on a column of Sephadex LH-20 to isolate a mixture of
predominantly the Compounds of Formulae XIV, and XV, contaminated with some
unchanged starting material and some diepoxide. The Compounds of Formulae XIV
and XV are separated and purified by HPLC or HSCC using one of the systems
described in Example 2 for the purification of the Compound of Formulae II. In
a
typical experiment yields of 35% to 40% are obtained for each of the Compounds
of
Formulae XIV and XV.
Scheme 12: Synthesis of Compound of Formulae XIX by epoxidation of the
CA 02538147 2004-O1-21
3005-7PCT-9CA
Compound of Formula XL.
-90-
O CH, CH, CH,
i
~N CHI
0 O
Ho ; ~ \ off Formula XIX
H
HO
To a solution of Compound of Formula XL dissolved in tetrahydrofuran (THF)
is added 2.2 equivalents of meta-chloroperbenzoic acid. The reaction is cooled
in
an ice bath and stirred at 0 °C for 1-2 hours. The reaction mixture is
then
evaporated to dryness, re-dissolved in methanol and subjected to liquid
chromatography on a column of Sephadex LH-20 to isolate essentially pure
Compound of Formulae XIX in good yield.
Scheme 13: Syntheses of Compounds of Formulae XXVI, XXVII and XXVIII by
esterification of the Compound of Formula II.
O CH, CH, CH,
i~
N~' / / CH,
o , N ~ \ Formula XXVI
H ~~OH
H,C HO
O CH, CH3 CH,
i
~N ~ ~ ~ C;H,
HO ; ~ \ o~ Formula XXVIII
" ~ cH,
HO
To a solution of Compound of Formula II dissolved in toluene (9 parts)
tetrahydrofuran (1 part), cooled in an ice-bath is added 1.1 equivalents of
acetic
anhydride and two drops of boron trifluoride etherate. The reaction is
maintained
cool in an ice bath and stirred at 0 °C for 1-2 hours. The reaction
mixture is then
poured into aqueous 5% sodium bicarbonate solution shaken and the toluene
layer
is removed. The aqueous layer is re-extracted with toluene and the combined
toluene layers are concentrated to a mixture of predominantly the Compounds of
Formulae XXVI, XXVII and XXVIII, contaminated with some unchanged starting
material and some diacetates. The Compounds of Formulae XXVI, XXVII and
XXVIII are separated and purified by HPLC or HSCC using one of the systems
described in Example 2 for the purification of the Compound of Formulae II. In
a
CA 02538147 2004-O1-21
3005-7PCT-9CA
-91-
typical experiment yields of 25% to 30% are obtained for each of the Compounds
of
Formulae XXVI, XXVII and XXVIII.
Scheme 14: Syntheses of Compounds of Formulae XXXIII, XXXIV and XXXV by
methylation of the Compound of Formula II.
CH, CHI CH3 CH CHI CHI
I ~ ~~ ~
N~~~~ ~ CH
N~ / / CH,
Formula XXXIV
N ~ \ Formula XXXIII N ~ \
HO
H,C'O H _ OH H OH
HO H C'0
3
O CH, CH, CH,
~N / /~ / CHa
Formula XXXV
N
HO O-CH,
H
HO
A solution of the Compound of Formula II (1 g) in tetrahydrofuran 50 (ml) is
titrated with exactly one equivalent of sodium methoxide, allowed to stand for
30
minutes at room temperature and then treated with 1.2 equivalents of
dimethylsulphate. Heat the mixture under reflux for one hour, cool to room
temperature and pour into a mixture of toluene (200 ml) and water (200 ml).
The
layers are separated and the aqueous layer is extracted once more with an
equal
portion of toluene. The combined toluene layers are washed once with 1 N
aqueous
acetic acid and then concentrated to s crude product, which is predominantly a
mixture of the Compounds of Formulae XXXIII, XXXIV and XXXV with some
unchanged starting material and traces of over-methylated derivatives. The
desired
products may be separated and purified by HPLC or HSCC chromatography using
the systems as described in Example 2 above, to obtain approximately 200 mg of
each of the Compounds of Formulae XXXII1, XXXIV and XXXV.
EXAMPLE 11: GENES AND PROTEINS FOR THE PRODUCTION OF
COMPOUNDS OF FORMULA
Micromonospora sp. strain 046-EC011 is a representative microorganism
useful in the production of the compound of the invention. Strain 046-EC011
has
been deposited with the International Depositary Authority of Canada (IDAC),
CA 02538147 2004-O1-21
3005-7PCT-9CA
-92-
Bureau of Microbiology, Health Canada, 1015 Arlington Street, Winnipeg,
Manitoba,
Canada R3E 3R2 on March 7, 2003 and was assigned IDAC accession no. 070303-
01. The biosynthetic locus for the production of the compound of Formula ll wa
s
identified in the genome of Micromonospora sp. strain 046-EC011 using the
genome scanning method described in USSN 10/232,370, CA 2,352,451 and
Zazopoulos et. al., Nature 8iotechnol., 21, 187-190 (2003).
The biosynthetic locus spans approximately 52,400 base pairs of DNA and
encodes 43 proteins. More than 10 kilobases of DNA sequence were analyzed on
each side of the locus and these regions were deemed to contain primary genes
or
genes unrelated to the synthesis of the compound of Formula II. As illustrated
in
FIGURE 12, the locus is contained within three sequences of contiguous base
pairs,
namely Contig 1 having the 36,602 contiguous base pairs of SEQ ID NO: 1 and
comprising ORFs 1 to 31 (SEQ ID NOS: 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23,
25, 27,
29, 31, 33, 35, 37, 39, 41, 43, 45, 47, 49, 51, 53, 55, 57, 59, 61 and 63),
Contig 2
having the 5,960 contiguous base pairs of SEQ ID NO: 64 and comprising ORFs 32
to 35 (SEQ ID NOS: 66, 68, 70 and 72), and Contig 3 having the 9,762 base
pairs of
SEO ID NO: 73 and comprising ORFs 36 to 43 (SEQ ID NOS: 75, 77, 79, 81, 83,
85n
87 and 89). The order, relative position and orientation of the 43 open
reading
frames representing the proteins of the biosynthetic locus are illustrated
schematically in FIGURE 12. The top line in FIGURE 12 provides a scale in base
pairs. The gray bars depict the three DNA contigs (SEQ ID NOS: 1, 64 and 73)
that
cover the locus. The empty arrows represent the 43 open reading frames of this
biosynthetic locus. The black arrows represent the two deposited cosmid clones
covering the locus.
The biosynthetic locus will be further understood with reference to the
sequence listing which provides contiguous nucleotide sequences and deduced
amino acid sequences of the locus from Micromonospora sp. strain 046-EC011.
The contiguous nucleotide sequences are arranged such that, as found within
the
biosynthetic locus, Contig 1 (SEQ ID NO: 1 ) is adjacent to the 5' end of
Contig 2
(SEQ ID NO: 64), which in turn is adjacent to Contig 3 (SEQ ID NO: 73). The
ORFs
illustrated in FIGURE 12 and provided in the sequence listing represent open
reading frames deduced from the nucleotide sequences of Contigs 1, 2 and 3
(SEQ
ID NOS: 1, 64 and 73). Referring to the Sequence Listing, ORF 1 (SEQ ID NO: 3)
is
CA 02538147 2004-O1-21
3005-7PCT-9CA
-93-
the polynucleotide drawn from residues 2139 to 424 of SEQ ID NO: 1, and SEQ ID
NO: 2 represents that polypeptide deduced from SEQ ID NO: 3. ORF 2 (SEQ ID
NO: 5) is the polynucleotide drawn from residues 2890 to 4959 of SEQ ID NO: 1,
and SEQ ID NO: 4 represents the polypeptide deduced from SEQ ID NO: 5. ORF 3
(SEQ ID NO: 7) is the polynucleotide drawn from residues 7701 to 5014 of SEQ
ID
NO: 1, and SEQ ID NO: 6 represents the polypeptide deduced from SEQ ID NO: 7.
ORF 4 (SEQ ID NO: 9) is the polynucleotide drawn from residues 8104 to 9192 of
SEQ ID NO: 1, and SEQ ID NO: 8 represents the polypeptide deduced from SEQ ID
N0: 9. ORF 5 (SEQ ID .NO: 11 ) is the polynucleotide drawn from residues 9192
to
10256 of SEQ ID NO: 1, and SEQ ID NO: 10 represents the polypeptide deduced
from SEQ ID NO: 11. ORF 6 (SEQ ID NO: 13) is the poiynucleotide drawn from
residues 10246 to 11286 of SEQ ID NO: 1, and SEQ ID NO: 12 represents the
polypeptide deduced from SEQ ID NO: 13. ORF 7 (SEQ ID NO: 15) is the
polynucleotide drawn from residues 11283 to 12392 of SEQ ID NO: 1, and SEQ ID
NO: 14 represents the polypeptide deduced from SEQ ID NO: 15. ORF 8 (SEQ ID
NO: 17) is the polynucleotide drawn from residues 12389 to 13471 of SEQ ID N0:
1,
and SEQ ID NO: 16 represents the polypeptide deduced from SEQ ID NO: 17. ORF
9 (SEQ ID NO: 19) is the polynucleotide drawn from residues 13468 to 14523 of
SEQ ID NO: 1, and SEO ID NO: 18 represents the polypeptide deduced from SEQ
ID NO: 19. ORF 10 (SEQ ID NO: 21 ) is the polynucleotide drawn from residues
14526 to 15701 of SEQ ID NO: 1, and SEQ ID NO: 20 represents the poiypeptide
deduced from SEQ JD NO: 21. ORF 11 (SEQ ID NO: 23) is the polynucleotide
drawn from residues 15770 to 16642 of SEQ ID NO: 1, and SEQ ID NO: 22
represents the polypeptide deduced from SEQ ID NO: 23. ORF 12 (SEQ ID NO: 25)
is the polynucleotide drawn from residues 16756 to 17868 of SEQ ID NO: 1, and
SEQ ID NO: 24 represents the polypeptide deduced from SEQ ID NO: 25. ORF 13
(SEQ ID NO: 27) is the polynucleotide drawn from residues 17865 to 18527 of
SEQ
ID NO: 1, and SEQ ID NO: 26 represents the polypeptide deduced from SEQ ID NO:
27. ORF 14 (SEQ ID NO: 29) is the polynucleotide drawn from residues 18724 to
19119 of SEQ ID NO: 1, and SEQ ID NO: 28 represents the polypeptide deduced
from SEQ ID NO: 29. ORF 15 (SEQ ID NO: 31 ) is the polynucleotide drawn from
residues 19175 to 19639 of SEQ ID N0: 1, and SEQ ID NO: 30 represents the
polypeptide deduced from SEQ ID NO: 31. ORF 16 (SEQ ID NO: 33) is the
CA 02538147 2004-O1-21
3005-7PCT-9CA
-94-
polynucieotide drawn from residues 19636 to 21621 of SEQ JD NO: 1, and SEQ ID
NO: 32 represents the polypeptide deduced from SEQ ID NO: 33. ORF 17 (SEQ ID
NO: 35) is the polynucleotide drawn from residues 21632 to 22021 of SEQ ID NO:
1,
and SEQ ID NO: 34 represents the polypeptide deduced from SEQ ID NO: 35. ORF
18 (SEQ ID NO: 37) is the poiynucleotide drawn from residues 22658 to 22122 of
SEQ ID NO: 1, and SEQ ID NO: 36 represents the polypeptide deduced from SEQ
ID NO: 37. ORF 19 (SEQ ID NO: 39) is the polynucleotide drawn from residues
24665 to 22680 of SEQ ID NO: 1, and SEQ ID NO: 38 represents the polypeptide
deduced from SEQ ID NO: 39. ORF 20 (SEQ ID NO: 41 ) is the polynucleotide
drawn from residues 24880 to 26163 of SEQ ID NO: 1, and SEQ ID NO: 40
represents the polypeptide deduced from SEQ ID NO: 41. ORF 21 (SEQ ID NO: 43)
is the polynucleotide drawn from residues 26179 to 27003 of SEQ ID NO: 1, and
SEQ ID NO: 42 represents the polypeptide deduced from SEQ ID NO: 43. ORF 22
(SEQ ID NO: 45) is the polynucleotide drawn from residues 27035 to 28138 of
SEQ
ID NO: 1, and SEQ ID NO: 44 represents the polypeptide deduced from SEQ ID NO:
45. ORF 23 (SEQ ID NO: 47) is the polynucleotide drawn from residues 28164 to
28925 of SEQ iD NO: 1, and SEQ ID NO: 46 represents the polypeptide deduced
from SEQ ID NO: 47. ORF 24 (SEQ ID NO: 49) is the polynucleotide drawn from
residues 28922 to 30238 of SEQ ID NO: 1, and SEQ ID NO: 48 represents the
polypeptide deduced from SEQ ID NO: 49. ORF 25 (SEQ ID NO: 51 ) is the
polynucleotide drawn from residues 30249 to 31439 of SEQ ID NO: 1, and SEQ ID
NO: 50 represents the poiypeptide deduced from SEQ ID NO: 51. ORF 26 (SEQ ID
NO: 53) is the polynucleotide drawn from residues 31439 to 32224 of SEQ ID NO:
1,
and SEQ ID NO: 52 represents the polypeptide deduced from SEQ ID NO: 53. ORF
27 (SEQ ID NO: 55) is the polynucleotide drawn from residues 32257 to 32931 of
SEQ ID NO: 1, and SEQ ID NO: 54 represents the polypeptide deduced from SEQ
ID NO: 55. ORF 28 (SEQ ID NO: 57) is the polynucleotide drawn from residues
32943 to 33644 of SEQ ID NO: 1, and SEQ ID NO: 56 represents the poiypeptide
deduced from SEQ ID NO: 57. ORF 29 (SEQ ID NO: 59) is the polynucleotide
drawn from residues 34377 to 33637 of SEQ ID NO: 1, and SEQ ID NO: 58
represents the polypeptide deduced from SEQ ID NO: 59. ORF 30 (SEQ ID NO: 61)
is the polynucleotide drawn from residues 34572 to 34907 of SEQ ID NO: 1, and
SEQ ID NO: 60 represents the polypeptide deduced from SEQ ID NO: 61. ORF 31
CA 02538147 2004-O1-21
3005-7PCT-9CA
-95-
(SEQ ID NO: 63) is the polynucleotide drawn from residues 34904 to 36583 of
SEQ
ID NO: 1, and SEQ ID NO: 62 represents the polypeptide deduced from SEQ ID NO:
63. ORF 32 (SEQ ID NO: 66) is the polynucleotide drawn from residues 23 to
1621
of SEQ ID NO: 64, and SEQ ID NO: 65 represents the polypeptide deduced from
SEQ ID NO: 66. ORF 33 (SEQ ID NO: 68) is the polynucleotide drawn from
residues 1702 to 2973 of SEQ ID NO: 64, and SEQ ID NO: 67 represents the
polypeptide deduced from SEQ ID NO: 68. ORF 34 (SEQ ID NO: 70) is the
polynucleotide drawn from residues 3248 to 4270 of SEQ ID NO: 64, and SEQ ID
NO: 69 represents the polypeptide deduced from SEQ ID NO: 70. ORF 35 (SEQ ID
NO: 72) is the polynucleotide drawn from residues 4452 to 5933 of SEQ ID NO:
64,
and SEQ ID NO: 71 represents the polypeptide deduced from SEQ ID NO: 72. ORF
36 (SEQ ID NO: 75) is the polynucleotide drawn from residues 30 to 398 of SEQ
ID
NO: 73, and SEQ ID NO: 74 represents the polypeptide deduced from SEQ ID NO:
75. ORF 37 (SEQ ID NO: 77) is the polynucleotide drawn from residues 395 to
1372
of SEQ ID NO: 73, and SEQ ID NO: 76 represents the. polypeptide deduced from
SEQ ID NO: 77. ORF 38 (SEO ID NO: 79) is the polynucleotide drawn from
residues
3388 to 1397 of SEQ ID NO: 73, and SEQ ID NO: 78 represents the polypeptide
deduced from SEQ ID N0: 79. ORF 39 (SEQ ID NO: 81) is the polynucleotide drawn
from residues 3565 to 5286 of SEQ ID NO: 73, and SEQ ID NO: 80 represents the
polypeptide deduced from SEQ ID NO: 81. ORF 40 (SEQ ID NO: 83) is the
polynucleotide drawn from residues 5283 to 7073 of SEQ ID NO: 73, and SEQ ID
NO: 82 represents the polypeptide deduced from SEQ ID NO: 83. ORF 41 (SEQ ID
NO: 85) is the polynucleotide drawn from residues 7108 to 8631 of SEQ ID NO:
73,
and SEQ ID NO: 84 represents the polypeptide deduced from SEQ ID NO: 85. ORF
42 (SEQ ID NO: 87) is the polynucleotide drawn from residues 9371 to 8673 of
SEQ
ID NO: 73, and SEQ ID NO: 86 represents the polypeptide deduced from SEQ ID
NO: 87. ORF 43 (SEQ ID NO: 89) is the polynucleotide drawn from residues 97Ei2
to
9364 of SEQ ID NO: 73, and SEQ ID NO: 88 represents the polypeptide deduced
from SEQ ID NO: 89.
Some open reading frames provided in the Sequence Listing, namely ORF 2
(SEQ ID NO: 5), ORF 5 (SEQ ID NO: 11), ORF 12 (SEQ ID NO: 25), ORF 13 (SEQ
ID NO: 27), ORF 15 (SEQ ID NO: 31 ), ORF 17 (SEQ ID NO: 35), ORF 19 (SEQ ID
NO: 39), ORF 20 (SEQ ID NO: 41 ), ORF 22 (SEQ ID NO: 45), ORF 24 (SEQ ID N0:
CA 02538147 2004-O1-21
3005-7PCT-9CA
-96-
49), ORF 26 (SEQ ID NO: 53) and ORF 27 (SEQ ID NO: 55) initiate with non-
standard initiation codons (eg. GTG - Valine, or CTG - Leucine) rather than
standard initiation codon ATG methionine. All ORFs are listed with the
appropriate
M, V or L amino acids at the amino-terminal position to indicate the
specificity of the
first codon of the ORF. It is expected, however, that in all cases the
biosynthesized
protein will contain a methionine residue, and more specifically a
formylmethionine
residue, at the amino terminal position, in keeping with the widely accepted
principle
that protein synthesis in bacteria initiate with methionine (formylmethionine)
even
when the encoding gene specifies a non-standard initiation codon (e.g. Stryer
Biochemistry 3~d edition, 1998, W.H. Freeman and Co., New York, pp. 752-754).
ORF 32 (SEQ ID NO: 65) is incomplete and contains a truncation of 10 to 20
amino acids from its carboxy terminus. This is due to incomplete sequence
information between Contigs 2 and 3 (SEQ ID NOS: 64 and 73, respectively).
Deposits of E. coli DH10B vectors, each harbouring a cosmid clone
(designated in FIGURE 12 as 046KM and 046KQ respectively) of a partial
biosynthetic locus for the compound of Formula II from Micromonospora sp.
strain
046-EC011 and together spanning the full biosynthetic locus for production of
the
compound of Formula II have been deposited with the International Depositary
Authority of Canada, Bureau of Microbiology, Health Canada, 1015 Arlington
Street,
Winnipeg, Manitoba, Canada R3E 3R2 on February 25, 2003. The cosmid clone
designated 046KM was assigned deposit accession numbers IDAC 250203-06, and
the cosmid clone designated 046KQ was assigned deposit accession numbers IDAC
250203-07. Cosmid 046KM covers residue 1 to residue 32,250 of Contig 1 (SEQ ID
NO: 1 ). Cosmid 046KQ covers residue 21,700 of Contig 1 (SEQ ID NO: 1 ) to
residue 9,762 of Contig 3 (SEQ ID NO: 73). The sequence of the polynucfeotides
comprised in the deposited strains, as well as the amino acid sequence of any
polypeptide encoded thereby are controlling in the event of any conflict with
any
description of sequences herein.
The deposit of the deposited strains has been made under the terms of the
Budapest Treaty on the International Recognition of the Deposit of Micro-
organisms
for Purposes of Patent Procedure. The deposited strains will be irrevocably
and
without restriction or condition released to the public upon the issuance of a
patent.
The deposited strains are provided merely as convenience to those skilled in
the art
CA 02538147 2004-O1-21
3005-7PCT-9CA
-97-
and are not an admission that a deposit is required for enablement, such as
that:
required under 35 U.S.C. ~112. A license may be required to make, use or sell
the
deposited strains, and compounds derived therefrom, and no such license is
hereby
granted.
In order to identify the function of the proteins coded by the genes forming
the
biosynthetic locus for the production of the compound of Formula II the gene
products of ORFs 1 to 43, namely SEQ ID NOS: 2, 4, 6, 8, 10, 12, 14, 16, 18,
20,
22, 24, 26, 28, 30, 32, 34, 36, 38, 40, 42, 44, 46, 48, 50, 52, 54, 56, 58,
60, 62, 65,
67, 69, 71, 74, 76, 78, 80, 82, 84, 86 and 88 were compared, using the BLASTP
version 2.2.10 algorithm with the default parameters, to sequences in the
National
Center for Biotechnology Information (NCBI) nonredundant protein database and
the
DECIPHER~ database of microbial genes, pathways and natural products (Ecopia
BioSciences Inc. St.-Laurent, QC, Canada).
The accession numbers of the top GenBankTM hits of this BLAST analysis are
presented in Table 14 along with the corresponding E values. The E value
relates
the expected number of chance alignments with an alignment score at least
equal to
the observed alignment score. An E value of 0.00 indicates a perfect homolog.
The
E values are calculated as described in Altschul et al. J. Mol. Biol., 215,
403-410
(1990). The E value assists in the determination of whether two sequences
display
sufficient similarity to justify an inference of homology.
CA 02538147 2004-O1-21
3005-7PCT-9CA
_98_
a
T
Q
L N
N~
O
f0p .(0O
C _
tn
~/JN f1O N
U N m o N ,mcnY
N i
E ~'coy U y U vto
t
n
N U
C ~ N O p ~ d ' w U
U
V O) O _ O N O Ip '
'
.N _ ~ OO
tlJf O
C C C C d ~~ U~ J O ~ ~ L VN N N~
N
N _
~
U U U U p / O N~ pM U ~ J N ON ~ C
_ t M fn7 ~ O
7 N
p tG ~ ~ N U QU ~ Mp E= ~ m E N Y ~ O
O N Q7p7>, p~ N Q) 7N ~ f4'Jc6 c0 E E N~
E
O E E E ~ ~ O ~~ O 7 ~O
~ ~ OO Q 7 UO O U X N X _~w C N ~C
~ C
c ' ' ' ~ _ ~~ ~ ~ ~ n U)Y a a ~0
~ g c 0
a a a ~ a~c ~ . ~' ~ >.. ~, ,
~ ~ ~ X ~,
w O OO NU ~ (0p N a7~ ~ OU
~ ~ ~ U
c0 c0c0p f~UN Q UO C~ ~ ~ U ViV7c0~ 'O
N p Q7O Op N UN 2'.,.~..N (4~ ~ ~ ~ C G y~ w p~0
N
p ~ ~ ~ _ U Y Y C 7
7
Q ?, >.~ ~ U~' U~ ~ Y ~ ~ ~ Ot . =
O O O ~ ~ ~~ ~O U ? " (dO N ~ ~ OO. d d~
~ ~7
d U U U ~ ~ ~ oU ~ ~ N ~ o = o ~ c op o o~
s
~ ~
a = ' ~= a QN ~~ c c c ~ n ~ o o ~n. a aa~
_ a~
r p L y Y Y p _ O j ~ -E
'O E
O O O U C (nU d _~(U ~V N ~ p E N E N 7 N, ~ OM
N N ~ N N ~> ~ -M
c0(Of0O (0O E E O, . C>
C C >
,
C C C L C = O O L p.
X
X
c9 N c0~ 'U~O 'pON ~~ O O O N'O d Q-~ ~ G~
C C ~
p
N p (0a. N N (Of4c4 O O d d N p O-O
N 'O
O
U U U m C Q~ ?~a ~N ' ' ' n ' Q O O co 0 0~r
m m m ~ , . a a ~ ~ .
~ v ~ > a a
~ a~ ~~ Q:oE E E a E ~ a a ~~n N Ncb
c:~
w
c o ~~ cc ~ ~ o c ~ ~ c o 0o c c~
o ~ o v o ~o c yo 00 0 0 o c o 0 0 0 00 0 0o
0
0
W o M O ~ M t0.~O NM tnO M h N WN CON07
O N O O (DOO ~ r ~M N r O N O O)N ~ O~ M 07O
M
M ~ d'M CD . u O ~ WO (D O
c0 M ~ h ~CC CJ O NV h O
V ~~ v ~..~. CDO v CvCO~ CoV V'v h ~
v .
r
CO O ~ O O "O h ~N O N N h tn~ h ~ 07V O)O~00
h
U)O ~ MM h MM N O h O COtnt()CO 'd'VV
M M
o ~ O ~ ~ ~ OM N ~~ MM M M N c\M M MM M MM
M
00 O ~,r ~ O(~ ~0 \VM ~ ~ n ~ ~ O N O)~ O Nh
O
h N QO00 ~'d'O OO ~ O cDV N O ~(O h
-
N 00 .- r rN NN r ' r r N N rN N NM
M
R
o ~ ~ ~ ao ~ 00 00 0 o 0 0 0 ~ 0 0 00 0 00
~ . 0
O o 0 0 o o N p~N ~ M M T h M ~h
M O
f~ r O M~ M O(V OJN O h ~ O h N h OJ07 NCO
O ~ N
~ M O CO h 07M hCOCflV ~ 07N ~ O h O~
p " N ~ ~ ~ N~ ~
O . ~ ~ O (OV ~ ~ 0 t0d0V O GO00
h
O ~ QO O h O h ~
v 0 N M MC\~ M M M (V~ M M MM M M~
c'
~
~ OM M~ a0 O if N~ N
N
a 0~is~ NN O ~~ ~M s ~ h t0~ M o ~ ~V
0 ~
N r t0~ m 0~m ~ ~ O h ~ r 07h ON N ~
w h O N h N hO ~ NO O7O~r (p(pp~p~~ M h rM O M~
O
,-O a0r _ _ ON r a0 h1~I~(OM 00(O~ ~ 1~MN N h(O
(O
Oj lJJW IJJW lJJlJJUJWt1JL1JLLW W tJJUJLLll1JUJW Wj j W
~ O ~.f
O O O O O OO O OO OO O O O O O O O O O OW
O O O
p O O O O O OO O OO OO O O O O O O O O OO O OO
O
M M M MM r MN MN N ~ M N N M ~ CONr r ~O
r
r
(0 N N p O (9(Q f0c9O NN O p t9N N p f0N N pN
N
N N N ~p~pOc9 f0O~ OO m m O N cD~ N N cb mc0
O O N
O ~ M M MM M 0 ~ ~ M O O N ~ O 00~ m hM
M
lC7~O~ON ~ N V h (O~ ~ M ~ O ,.h COO M
O OM MM M ~ ~ ~ M
O O) O O M M M M ~ M M M M
O N r _ _ _ _ _ _M M r
~
L h ofo7dj~jV07 tnWc0 h07Lj~ O ofh ~j07NcD N m
y N M M O O Nh hOJO ~f7~ O O V11 (~Mh
CO
C CO COCO~ N rh N h~ MO h O~O h ofM h 00V~ ~ V
CO O O M ofC O ~
O
O
m M o o M ~ MM ~ Mo M_ h h N h h ~ h h NY Y _V
m ~ m ,n o o p o o O o
h
h
y '~'~U U ~U U h~'~ yN m m '~m m him m C7o a~~Q
n a o Q Q dQ Q aa oo Q Q ~ Q Q a Q Q U c~iQ
a a
_ z _ U m zU m zz __ m m z m m z m m ~ zm
z z zz m
CO M ('~ M M M M M
m w w > ~ > a
p
Q ~ ~ - ~ o ~ - z
r N M V u~ CD h a0 O
CA 02538147 2004-O1-21
OU05~7F'~~~9C~
_99_
a
0
U J ~ m
>,U '
c a
. ~
o a ~ m
a~
Q o c m
v~ u~~ m o a~ C
O
~ Q O O
_~O C U O
U
r1J~ ~m ~ _ d E O E
~ -o ~
C ~_
O Q UQ C N E ~
tn N 0
Ov f0~_, ( 0
U ~ O O
UM ~ N M O ~ N 7
N O C N
U ~i--0Q D_ ~ Q C C_ L ~ O
~C~ ~O Np Q ~ ~ ~ ~ d
(n c0 _ ~ _
~
N p .L-..O ~N ME O O -OO O L (~ c/7
U
U O CU NO QN Q (0.U O U N 0 c0 N
' N
Q f/7N (0~ U7U L U U ~ ,-,,-, ~ N N C ~
d
O ~ ~O ~~ pH ~~~ O ~ ~ '-'L ~ ~ ~ -
U U ~ _ ~. >,
(6Ntn 07j ~V~ ~ f0cO N N tn7 O O 7, ~ N N U
=
N , ~ ~
U ~ OC N U O O m ~UQ O ~ E ~ ~ N
a
C Q 7 ~
O O O UE ~a UY 7 O ~ ~ ~ O a7N ~ '~7 L O O (0c0
U U m ~O U~ NN . E O O O m t0/7N C ~ O C C C
E ~
UC O C_ ~ N O U y
E E ~~ ~
~ ON >'-a~ 47N E N a d U L 9 L U O
Q
cp(0 N Q L N N O ~p O O
O. O
7 ~ Y ~(O op in~ H ~ t ~ ~ ~ O c ~-L ~ C C w,0
i7 y
_ _ U dL " flQ L ~ ~ O ~ O ~ ~ (OO
O m U
O OC (OC OC ~ O O d 'pO U U ~ ~ (O N O_L
> > (~tO N O Q L
, , N O ~ N N O N 7 C _ tnH
U C
L ~'~ ~ O O O O N ~ Q -O~ w C ~
j,
OJN ~ C j ~ ~ _ D D ~ O U U v77 '~ f0 (O
O fn
E E d O U O O p7 ~ a 0 N ~ 'p~UU Q. Q Q7
O
M w~ ?-T O N~ O N ~ OJQ C 7 "ON ~ ~ O O (~6
Q L
> Q QU NQ ~~ N ~ ~ p O ~ X ~ ~ f4 C C
~
O
. >. O O~ ~.~ N O C C C C ' 1'
~ ~
X X U U= ~o Y" m U O O V C U o ~ ~ 3 ~ c ,~
~ ~ ~
.N
0 o o 0 0 o 0 3 .> , c
M ~ n
-
CJC7o V ~~ ~ c-o- c io~ E c ~ L ' co
V a~ '
c L
O- ~O CO)~ ~ N = j N O Y 1' C O ~ a
7 C
O
L L ~ ~t 3 ~tn~ ~ C~C~ Y E ~ ~ n a ~'Q
chc~ Z = v ~ U U C~= ~ ~ . Q
Q~
0 0 0 00 "" "'~0 0 0 ~ o o ~ ~ ~ '1 0 0 0
0 0 0 00 00 00 0 0 0 0 0 0 0 0 0 0 0 0
0
0
f~N M Mh OO~ 0000COI~I~ .-..-..-. M I~0 0 o O O ~ M
O)
h r ~m NO 07~ M i~o~ 0 0 o ~ O CO07~ M ~ ~ O
~
COf~ OJ~M ON Nf~~ ~ M O O O ~ . . C7N ~ ~ O N
' ~
O_JIv iv~_~ ~CO cD~_o~IvI_~ COCOCO~ ~ V_Ivv CO (O (O ~
C_D
f~c0 M MN MO~ 00O~O 00N ~ ~ ~ M c0of 1~N N N O ~I7
N
V M d'o~M I~O Op N N ~ ~ M ~ N O N r N N N N
N
M M M MN NN Nr N N N C7M M V V _ r CO CO V V
CO
67O ~ OV N V 6)M ~ M M M ~ ~ O I~COCO of M ~ o~
M
N ~ N (VN rM Mr I~c0(O r I~f~00OJf~ ~ V N N
~
0 0 0 oa o0 0~ 0 0 0 ~ ~ ~ 0 0 0 o a o 0 0 0 0
0
M CO N N ~ ~~ f~I~(D o 0 o O f~O 07 O M r
O
M M COr~ MO~ N~ N r M ofo~OJI~O~r M r M N N M r
N
O I~ O)M rM M f~07O r ~-r OjO~O~ 07P
~
I_~CO CO~_~ Nv ~y 0 y _O N ofofN N N_~ ~ . tn ~ M M
v ~.. _~
I~CO M ~ M ~v O N N M M M M COo~ v N N O
N M ~ " " " N N
V OJ V N O O N _ N ~ ~ ~ M N O ~ N N N
O N N
M M M MN NN N N N N ~ ~ ~7V V V ~ r CO CO ~
r .. cD
I~O ~ CO N N OJ07V ~ M ~ O~ O~ V
O O~
N MN ~ ~ ~ M N N N N ~ r ~ cD~ w ~ r
~
N r r 7 M
M
M ON COO~ NV O)t~CO N N N N N r M M
f~N OV VM I~(OCO O O O ~ M M M M N CO(O
W W LIJ~ WW 11JW W IJJW W W 11JliJW W W W W W W LiJW
W W
O O O OO OO OO O O O O O O O O O O O O O O O O
O
O O O OO OO OO O O O cDcDCOO O O O O O O O O O
O
r r M~f1~~ ~07M ~ CO 1~I~I~r M r I~r r r r N
N c~ (i3 cB N (6
N ciFN NO (6N Oc9N N GO (~O O N (6N N ~ N N N (Q
N
caca sncu~ mc~ ~~ ca co cBca~ cao m co~ ca a~ ca m n
M O CflM~ V'~t NO V N r ofM M O o0M I~M _ Cp f~ ~p~
O~
O N~ MV N N N N t~N O ~yCON N M I~ N ~
CO
M M M MN MM M N _ N COV ~ ~ _ V r r CD
r r r r
_ _ _ __- __ _ _ _ _ _ ~ _ _ ~ _ N p
L ~
COtn N O~ O1(7~O)o~O V O O djc0 I~O 6jI~ V C
O O M 0
COO N N~ MM Om O O)CO ~ r V c0~ c0~ M (O O r
I~ O
OJVN NV ~M tnO~M Z ~ N O O V M p~N t~ ~ f~
f~ ~
~ O O ~ r ' N M ~ M O
M
O O NM CO O O N n V ~ O 7 O
~f
Q CO 07D~I Z~ ofO ofO I ~I~I~ NI NI~IO I O NI d O
O
Q Q Q Q~ Q~ o_I a I a a a Q n_O a.o_Q a I a I
I I
a7m 07UZ Q ZN Z N Z Z Z Q Z N Z Z Q Z N Z
N
N
0 0 o v o~ oo r~
O O I~ N M tn CO N f~ (O N
M N M N r (p r Cp
Z m
u7 Y W W p
Y ~ a ~ >
c > O > > U Q
n
O N M ~Y~ cD I~ M ~ O
r r r r r r r r N
CA 02538147 2004-O1-21
3005-7PCT-9CA
-100-
L
'
N - O O~ O
Q ~
N N N ~ O) 01N
~
U ~ U ~ ._~ .-. ~j j
~ ~
W (0 (0
O
O. O_ y UM C O y
Y O ~ ~ ~ O O
V ~ C O O N -m 3
O (~=~ '
O ,-,
m ~ ', N
(07 ~ p d ~ ~ N L M LN ~ N
N
. Q _ O M
E Q ~ ~ ~ f0~ 7 7C ~ N
C '
. 3N Y
t ~ L ~ U~~O N G C m mO j ~ ~
N N . O O O
E r O ~.~ O ~ O V O~ :p m~ p :7
N
N ~ N ~ O j O ' c9N _ ~ O
O ~ p
~tnU N U ON U ~ ~ ~ N c O c0L N ~ U N (0
~ ~ I~N 6 j' O
(0 T
a t U C ~O N U
O . O L ~ .~ ~ ~ ~ O N OO'O (0 p N
O ~ O . ~ f9 N U L ~ 7
N
o E N E L ~U N v w ~ ~ ; ~~ ~~ ~ ',~ ~
~
~
O ~ '-'o ~ O L , L ~ , E U
-
~ ~ a ~ ~
N L ON' U ~ O O OC ~ .L1O CLO ~ ~ Q
E O N O d M O O C O.CN ~ 7N aU Q O .
O (0 ~ L
N
O
N ~ (CO~~ p Upfn~A~ t N =.~ ~ ~f0 N~ Q C
- p
p
L L .7dc6O . w N fl.(nQX O C 7U f6O fO~
O -
a p . - c~O Z 7, = L X x~ ~ ~ w E t
..
~ - U o a _v ~ c~U
a~ O V'O~ O afO' ~ p M
N
c ~ ~ ~ .7. C C ~ L N LD O O O~ O~O ~'
O >,
C C ~ _ L OL C C C
L L '~NL V m~ ' '
'pU N N
~ Q C _ ~ N ~ N N U T N~ I,-,O . C
, d 'N
'
N ~ 'U ~ E ~ _ O
-
~ p a~ O O ~ ~ ~ ~,jN ~dj N Np Lp O O O
p ' O
O O ~,-Od d M M M C CON O (9cD ~9c ON d O_
0
m a N Q c~s~ ~ li~ > ~ > c Em m ~,a -aco
c~
d ~ ~ N N O , t , ~E E EN ~ ~ U U
U
~a~. '~~ E E ~ ~ N'O O v~ ~ny- ~-o C
p '
. a U ~ ~
O N
N O U C N fJN ~ '' t ~ U~ L L~ ~ __
O -
V
U N N~ O O C C p O U O~ L UU U U ~'pO O E
O C Q C C O
. ~ ~ p d d N N Q . p_~ >. O. d Q 2
N O ~ O O ~ > O O XO O O U
N . . O O U LX >,>,>,
c0U LL U (0UE L L U U U N NN D ~~ ~ ~ E
.~
U N (p rnO L L L
a o o 0 0 00 0 0 0 0 0 0 0 00 0 00 0 00 00
0 ~
M M N u7NO O ~.,~o~f~f~~ M MO MN 1~ fW~ ~ ~ o
V'
of ~ N 00ppCOM N ~ ~ 07~ COO O OI~ I~d'~ (Dc0 ~
O
O
~ M vv ~ ~r ~ ~~ V~ O
_ ~ ~ V
N O O~ ~ ~ h In~ 07O t() O f~O~ (bMO O7O
M O
M V O ~ ~~ lyC7'~V V tf7N OCO N ~I7O) O O NN O f~
M
~ N N M N Nr ~ r M M M M V MN N.- r NN NN ~ ~ Q)
N
CO!~cD M O~ 00(O ~ r r (OOW ~ atN O WN..OO h
N V V (O~ppt~f~Ivt~I~ OV r ~~ ~ Mr OO ~ ~ V
~ r.
e-c- r r-~ N N Nr r r r ~-
0 0 0 0 0 00 \ a o 0 0 0 0 00 0 00 0 0~ ~o ~ o o
0 'v
67N M (O aOOo ~.._ (OQ)1~(OM (OO m M(O (ON 0 0 0
0
00O M CD O ('~O ~ ~ r V O ~ M OIO I~MIn ~ N~ MN ~
~ M
c0tnM f~ ofOO tfjy (7N o~c0tIjV tIj~ tnN ~ ~
~ ~ ~ ~~ ~ M ~ ~ ~ ~~ ~ ~~ ~ ~ N O
d'
V N .- v N N N tv
v ~ '-'.-
N O 00 ~ LC7N f~1IWC7O~O ~ O f~OJ ~7MO O~m CO(O
M V O O ~I~ V d'~ ~ N O(D 1(Wf)6) O O r
' ~
V N N c~ N Nr ~ ~ C\M M M d~MN N N _ NN NN ~ Or
N 7
N ~ V c0 tnM~ N ~ N c0W c0hO O c0 tn
O7O O O ~ V ~ N r ~ ~ On ' M
O
r r r r r r r COcD~hV M
V M N CO ~ M~ N r COr opop ~f7MO opop ~~ N N O
O O~
cD~ ~ V~ ~ ~r ~ r V'~ ~ h.I~c01n ~ ~(O M ~N ~-.-O O ~
u~ O
W W LL W lLLLJW W W t1JL11LIJW LLJWW W LLJW W WW LLW W W LLJ
W W
O O O O O OO O O O O O O O OO O OO O OO OO O O O
O O
O O O O O OO O O O O O O O OO O OO O OO OO lC707O
O O
c0N N V N (VdW- N ~OI~O~r-NCO r QiIs .-NOi ~Is' ti~
N dW r
p f9N c6 c6NN c9N c0N N (gypN ~N (m0 ON cB N N
N s0
m f4N cb (0(6N N N c9f4c6N f0Mf0 O ~p (O(B O(4 N O
c0 ~p N
COO O~ N _1~f~o~M ~ I~~ O O_ CO~~p ~pNN ~~ _ M
N (OI~ O O O
~
1 f~ O (D~ ~ ~ M M M ~ N ct(O N ~~p ~p V M of~-
N c0 N
N
N M ~ MN N N t V V _ ~ N _ N~ ~ MN NN ~ N -
r r r r rr r r r r - 'N ~ O -
r
~ r ~ ~~ N rr r CV ~
' ,
N N M ~ N m d'O N O ~p l Np pj~
CO ~ 0 O ~
N O N O)~tf O 7 Q7N O ~ ~ ofof ~ 0
O
~ ~ ~~ _ O ~ MO ~ ~ _ OM ON ~ ~
tfJ~t ~ N ~ d0N N ~ l~
~ (~
I~ I~ ~1~~ N I~ M O N OQ) O M_ _ WO O O N O
M
NI~I NI ~INI I I ~I~I~I ~IOM ~ ~ M ~ ~ o
~I ~ t~
o_ a a~ n.a ~.o_a O a ~I O ~a a lzl ~. n I
~ ~ ,
z z z z zz z z z z z N z Nz N z z
z
N
~ M ~
C tn CD N M
O
N M N d' M N N N N r
o ~ o ~ x ~ m ~ a
a ~ z >
Q o
N N N N
N N N N N M
Image
CA 02538147 2004-O1-21
3005-7PCT-9CA
-102-
N
N
U
L c9
C
roU ~ N
N E tl5, N
U
Q N
w
,U r..7 _'
.
roi 7 . C
/~ U
N N ro~
T N U m 7 m
N C =
.
7 ~ L N . .U
U U
c O _
T
ro
ro~ ' OJD
"
U O
U O_ ~
ro7 m X O ~
O
_ ~ Q
- ~ ro7
ro~'N O
p]~ m E N N ~
ro
. ~ O
N .,.C (0C_
~
N W ~ U
O 7
~ N
~ E ~ J L Q d
N
~ _
ro
N ~n;~~, m ~
a
a ~ s N
O a~o a~~ ~
3
~,U L - N .~
O
.cW N p
~ ~ Y
d N d
E ~ L H L J L
7
0 0 0 0 0
0
I~ I~ 0
I~ 0
N N N CO
N
N ~
~ M M M_
V
0
~ 0 0
N N N N N
N
M CO~.' n
CO
~ t ~
V
n r
0 0 ~ ~ 0 0
0 0
M N O)O y f
c 7
D
N
M M N ~
M
N N N
~N..v '-'~.N_v
N
O O
N N O I~
N
N CNON N N O
N
M M ~ ~ V ~
M N
O O O O O O O O
W W W W W W W W
O O O O O O O O)
~ N NI ~ N ~1 ~
ro rororororo
ro m
m rom m m m
m m
ca .-o~o ~ r-
cc ~ O ~ O N
due.. N
~
CO c0N N N ~
c0 r
O O m f~N N
~ r
f~ m O M tn1~
M 07
u7 N o~I~1~N
f~ V
V O V t(7I~
CO
M <tM ~ t!7N
~ V
0~ ~ ~ OJCO00
CO I I I I N
I
I
I
a d d a ~ a
a a
z z z z z z
z z
I~ N N
O M M
N
a
z
rt
CA 02538147 2004-O1-21
3005-7PCT-9CA
- 103 -
The ORFs encoding proteins involved in the biosynthesis of compounds of
Formula II are assigned a putative function and grouped together in families
based
on sequence similarity to known proteins. To correlate structure and function,
the
protein families are given a four-letter designation used throughout the
description
and figures as indicated in Table 15. The meaning of the four letter
designations is
as follows: AAKD designates an amino acid kinase; ABCA and ABCC designate
ABC transporters; ADSA designates an amide synthetase; ALDB designates an
aidolase function; CSMB designates a chorismate transaminase; DAHP designates
a 3,4-dideoxy-4-amino-D-arabino-heptulosonic acid 7-phosphate synthase
activity;
DHBS designates a 2,3-dihydro-2,3-dihydroxybenzoate synthase activity; DMDA
designates a diphosphomevalonate decarboxylase; EFFT designates an efflux
protein; HMGA designates a 3-hydroxy-3-methylglutaryl-CoA reductase; HOXV
designates a monooxygenase activity; HOYH designates a hydroxylase/
decarboxylase activity; HYDK designates a hydrolase activity; IDSA designates
an
isopentenyl diphosphate synthase; IPPI designates an isopentenyl diphosphate
isomerase; IPTN designates an isoprenyltransferase; KASH designates 3-hydrcsxy-
3-
methylglutaryl-CoA synthase; MVKA designates a mevalonate kinase; MVPK
designates a phosphomevalonate kinase; OXAH designates an acyICoA oxidase;
OXDS designates an oxidoreductase; RECH, RECI, RECD, REGG and RREB
designate regulators; SDRA designates a dehydrogenase/ketoreductase, SPKG
designates a sensory protein kinase; UNES, UNEZ, UNFA, UNFC, UNFD, UNFE,
UNFJ and UNIQ designate proteins of unknown function.
Table 15
FAMILY FUNCTION
AAKD amino acid kinase; strong homology to primary aspartate
kinases, converting L-
aspartate to 4-phospho-L-aspartate
ABCA ABC transporter
ABCC ABC transporter
ADSA adenylating amide synthetase
ALDB aldolase; similarity to fructose-1,6-biphosphate aldolase
that generates D-
i glyceraldehyde-3Ph, precursor of D-erythrose-4Ph involved
in the shikimate
pathway
~SMB chorismate transaminase, similarity to anthranilate synthase
DAHP DAHP synthase, class II; involved in formation of aminoDAHP
from PEP and
j
a erythrose-4-phosphate
CA 02538147 2004-O1-21
3005-7PCT-9CA
- 104 -
i DHBS 2,3-dihydro-2,3-dihydroxybenzoate synthase (isochorismatase)
DMDA diphosphomevalonate decarboxylase (mevalonate pyrophosphate
decarboxylase)
EFFT ~ efflux protein
HMGA HMG-CoA reductase; converts 3-hydroxy-3-methylglutaryl-CoA
to mevalonate
plus CoA in isoprenoid biosynthesis
HOXV FAD monooxygenase; shows homology to a variety of monooxygenases
including
salicylate hydroxylases, zeaxanthin epoxidases
HOYH hydroxylase/decarboxylase; FAD-dependent monooxygenase
i
HYDK hydrolase ~
IDSA isoprenyl diphosphate synthase, catalyzes the addition
of 2 molecules of l
isopentenyl pyrophosphate to dimethylallyl pyrophosphate
to generate GGPP
IPPI I isopentenyl diphosphate isomerase, catalyzes the isomerization
i j of IPP to produce
dimethylallyl diphosphate
IPTN ~ isoprenyltransferase; catalyzes covalent N-terminal attachment
of isoprenyl units i
to amide groups of nitrogen-containing heterocycle rings
i
KASH HMG-CoA synthase; condenses acetyl-CoA with acetoacetyl-CoA
to form 3-
hydroxy-3-methylglutaryl-CoA
MVKA mevalonate kinase; converts mevalonate to 5-phosphomevalonate
in the
mevalonate pathway of isoprenoid biosynthesis
MVKP phosphomevalonate kinase; converts 5-phosphomevalonate
to 5-
diphosphomevalonate in the mevalonate pathway of isoprenoid
biosynyhesis
OXAH acyl CoA oxidase
OXDS oxidoreductase
RECH regulator
RECI regulator; similarity to PadR transcriptional regulators
involved in repression of
phenolic acid metabolism
)
RECD transcriptional regulator; relatively large regulators
with an N-terminal ATP-binding
domain containing Walker A and B motifs and a C-terminal
LuxR type DNA-
binding domain
REGG regulator
RREB response regulator; similar to response regulators that
I are known to bind DNA
i and act as
transcriptional activators
SDRA I dehydrogenase/ketoreductase, NAD-dependent
SPKG sensory protein kinase, two component system
UNES unknown function
UNEZ unknown function
UNFA unknown function
I UNFC unknown function
i UNFD unknown function
UNFE putative membrane protein
UNFJ unknown function
UNIQ unknown function
CA 02538147 2004-O1-21
3005-7PCT-9CA
- 105 -
Biosynthesis of the compound of Formula II involves the action of various
enzymes that synthesize the three building blocks of the compound, namely the
farnesyl-diphosphate component (FIGURE 13), the 3-hydroxy-anthranilate-
adenylate
component (FIGURE 14a) and the 2-amino-6-hydroxy-benzoquinone component
(FIGURE 14b) that are subsequently condensed to form the final compound
(FIGURE 15).
The farnesyl-diphosphate biosynthesis involves the concerted action of seven
enzymes (FIGURE 13). ORF 10 (KASH) (SEQ ID NO: 20) encodes a
hydroxymethylglutaryl-CoA synthase that catalyzes an aldol addition of acetyl-
CoA
onto acetoacyl-CoA to yield 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA). This
product is subsequently reduced through the action of ORF 9 (HMGA) (SEQ ID NO:
18) to form mevalonic acid (MVA). ORF 5 (MVKA) (SEQ ID NO: 10) phosphorylates
mevalonate to 5'-phosphomevalonate using ATP as the phosphate donor. The next
step in the farnesyl-diphosphate biosynthesis is the phosphorylation reaction
of the
5'-phosphomevalonate to 5'-pyrophosphomevalonate (DPMVA) that is catalyzed by
ORF 7 (MVKP) (SEQ ID NO: 14). Subsequent decarboxylation of 5'-
pyrophosphomevalonate catalyzed by ORF 6 (DMDA) (SEQ ID NO: 12) yields
isopentenyl diphosphate (IPP) which is then converted to
dimethylallyldiphosphate
(DMADP) through the action of ORF 8 (IPPI) (SEQ ID NO: 16) that has isomerase
enzymatic activity. The final step in the biosynthesis of farnesyl-diphosphate
is the
condensation of one molecule of dimethylallyldiphosphate with two molecules of
isopentenyl diphosphate catalyzed by the isoprenyl diphosphate synthase ORF 4
(IDSA) (SEQ ID NO: 8). The described pathway involved in synthesis of farnesyl-
diphosphate is entirely consistent with related mevalonate pathways described
in
other actinomycete species (Takagi et al., J. Bacteriol. 182, 4153-4157,
(2000)).
Biosynthesis of the 3-hydroxy-anthranilate component involves the use of
precursors derived from the shikimate pathway (FIGURE 14a). Chorismic acid is
transaminated through the action of ORF 19 (CSMB) (SEQ ID NO: 38) to form
aminodeoxyisochorismic acid. This enzyme resembles anthranilate syntheses and
is
likely to catalyze specifically the transfer of the amino group using
glutamine as the
amino donor. The next step involves isochorismatase activity and is mediated
by
ORF 27 (DHBS) (SEQ ID NO: 54). This reaction consists in the removal of the
pyruvate side chain from aminodeoxyisochorismic acid to form 6-amino-5-hydroxy-
CA 02538147 2004-O1-21
3005-7PCT-9CA
- 106 -
cyclohexa-1,3-dienecarboxylic acid. This compound is subsequently oxidized
through the action of ORF 26 (SDRA) (SEQ ID NO: 52) yielding 3-hydroxy-
anthranilic acid. ORF 24 (ADSA) (SEQ ID NO: 48) catalyzes the activation of 3-
hydroxy-anthranilic acid through adenylation generating the 3-hydroxy-
anthranilate-
adenylate component (FIGURE 14a).
Biosynthesis of the 2-amino-6-hydroxy-benzoquinone component of the
compound of Formula II, requires components derived from the aminoshikimate
pathway. FIGURE 14b depicts the series of enzymatic reactions involved in the
biosynthesis of this constituent. ORF 21 (ALDB) (SEQ ID NO: 42) resembles
aldolases involved in the generation of precursors of D-erythrose-4-phosphate
which
is part of the aminoshikimate pathway used for the generation of 2-amino-6-
hydroxy-
[1,4]-benzoquinone. ORF 33 (DAHP) (SEQ ID NO: 67) catalyzes the initial step
in
the aminoshikimate pathway that corresponds to the formation of 3,4-dideoxy-4~-
amino-D-arabino-heptulosonic acid 7-phosphate (amino DAHP) from
phosphoenolpyruvate (PEP) and erythrose 4-phosphate (E-4Ph). Subsequent
reactions leading to 3-amino-5-hydroxy-benzoic acid are catalyzed by enzymes
provided by primary metabolism biosynthetic pathways present in Micromonospora
sp. strain 046-EC011. ORF 25 (HOXV) (SEQ ID NO: 50) hydroxylates 3-amino-5-
hydroxy-benzoic acid at position 2, generating 3-amino-2,5-dihydroxy-benzoic
acid.
This intermediate is further modified by ORF 32 (HOYH) (SEQ ID NO: 65) that
catalyzes a decarboxylative oxidation reaction yielding 6-amino-benzene-1,2,4-
trios.
A final oxidation reaction is performed by ORF 16 (OXDS) (SEQ ID NO: 32)
yielding
2-amino-6-hydroxy-[1,4]-benzoquinone (FIGURE 14b).
Assembly of the three components resulting in the compound of Formula II is
catalyzed by ORFs 24 and 11 (FIGURE 15). ORF 24 (ADSA) (SEQ ID NO: 48)
catalyzes the condensation of the adenylated 3-hydroxy-anthranilate with the 2-
amino-6-hydroxy-[1,4]-benzoquinone component. A spontaneous condensation
between the free amino group of the 3-hydroxy-anthranilate and one of the
carbonyl
groups present on the 2-amino-6-hydroxy-[1,4]-benzoquinone component occurs
yielding a dibenzodiazepinone intermediate. This compound is further modified
through transfer of the farnesyl group of the farnesyl-diphosphate
intermediate onto
the nitrogen of the amide of the dibenzodiazepinone catalyzed by ORF 11 (IPTN)
(SEQ ID NO: 22) and resulting in the formation of the compound of Formula II
CA 02538147 2004-O1-21
3005-7PCT-9CA
- 107 -
(FIGURE 15).
Additional ORFs, namely ORF 2 (RECH) (SEQ ID NO: 4), ORF 3 (REGD)
(SEQ ID NO: 6), ORF 12 (SPKG) (SEQ ID NO: 24), ORF 13 (RREB) (SEQ ID NO:
26), ORF 34 (REGG) (SEQ ID NO: 69) and ORF 36 (RECI) (SEQ ID NO: 74) are
involved in the regulation of the biosynthetic locus encoding the compound of
Formula II. Other ORFs, namely ORF 1 (ABCC) (SEQ ID NO: 2), ORF 31 (EFFT)
(SEQ ID NO: 62), ORFs 39 and 40 (ABCA) (SEQ ID NOS: 80 and 82, respectively)
and ORF 42 (SEQ ID NO: 86) are involved in transport. Other ORFs involved in
the
biosynthesis of the compound of Formula II include ORF 20 (AAKD) (SEQ ID NO:
40), ORF 23 (HYDK) (SEQ ID NO: 46), ORF 38 (OXAH) (SEQ ID NO: 78) as well as
ORFs 14, 15, 17, 18, 22, 29, 30, 35, 37, 41 and 43 (SEQ ID NOS: 28, 30, 34,
34,
44, 58, 60, 71, 76, 84 and 88, respectively) of unknown function.
CA 02538147 2004-O1-21
3005-7PCT-9CA
- 108 -
TABLE 16: PREFERRED MEDIA COMPOSITION FOR PRODUCTION OF ECO-
04601
Component QB MA KH RM JA FA
.
pH* 7.27.5 7 6.857.3 7.0
Glucose 12 10 10 10
I
Sucrose I I I 100 i
I Lactose
i
Cane molassesi 15
Corn starch 30
Soluble starch10 25
Potato dextrin 20 40
Corn steep
solid
Corn steep 5 15
liquor
Dried yeast 2
Yeast extract 5
Malt extract 35
- 10 ~ 15
~harmamediaTMI
~
~ Glycerol
NZ-Amine 5 10
Soybean powder 15
Soybean flour
Meat extract
Bacto-peptone
MgS04.7H20 1
MgCl2. 6H20 I
CaC03 f I 4 1 2 2
I
~ NaCi 5
(NHa)2 SOa 2
KZ S04 0.25
MnClz.4H20
MgC12.6H20 10
FeC12.4H20
Z
Na2HP04 j ~ ~ 3
i
Thiamine
Gasamino ' 0.1
acid
Proflo oil 4
MOPS 21
I Trace element 2
I solution
*3 ml/L
Unless otherwise indicated all the ingredients are in gm/L.
*~ Trace elements solution contains: ZnCl2 40 mg; Fe C13 6H20 (200 mg); CuCl2
2H20 (10 mg);
MnC12.4H20; Na2B40~.10H20 (10mg); (NH4)6 M0,0z4.4H20 (10 mg) per litre.
*5 The pH is to adjusted as marked prior to the addition of CaC03.
CA 02538147 2004-O1-21
3005-7PCT-9CA
- 109
All patents, patent applications, and published references cited herein are
hereby incorporated by reference in their entirety. While this invention has
been
particularly shown and described with references to preferred embodiments
thereof,
it will be understood by those skilled in the art that various changes in form
and
details may be made therein without departing from the scope of the invention
encompassed by the appended claims.
CA 02538147 2004-O1-21
SEQUENCE LISTING
APPLICANT: ECOPIA BIOSCIENCES INC.
Zazopoulos, Emmanuel
Farnet, Chris M.
TITLE OF INVENTION: FARNESYL DIBENZODIAZEPINONE, PROCESSES FOR ITS
PRODUCTION AND ITS USE AS A PHARMACEUTICAL
NUMBER OF SEQUENCES: 89
CORRESPONDANCE ADDRESS: 7290 Frederick-Banting
Saint-Laurent, Quebec, Canada, H4S 2A1
COMPUTER-READABLE FORM
SOFTWARE: PatentIn version 3.0
FILE REFERENCE: 3005-7PCT-9CA
PRTOR APPLICATION DATA
APPLICATION NUMBER: USSN 60/441,126
FILING DATE: 2003-O1-21
APPLICATION NUMBER: USSN 60/492,997
FILING DATE: 2003-08-07
APPLICATION NUMBER: USSN 60/518,286
FILING DATE: 2003-11-10
PATENT AGENT INFORMATION
NAME: Ywe J. Looper
REFERENCE NUMBER: 10961
SEQ ID 1
NO:
LENGTH:
36602
TYPE:
DNA
ORGANISM:Micromonospora
sp. strain
046-ECO11
SEQUENCE:1
ccggtgcaccgggttctccaggatcgccgtcgcgcccaccggccccgacaggtagacgac60
gttcagggacttgccgcgcccttcgtagttggcccgcaccacctgcgcgtcgccgatccg120
gcccctggtctccagcgtgcggttctcccacacctgccatccgacgaaggtcaggaacag180
cgcggtgaacagggacgtgacgagcagccagaggccagctgtcagcacggtcgccccctc240
gccccgtagcaggccgaggacgacctcctcgtagcgcgaggggcggccgacggggccggt300
gcccgctccgtcgacagccatcccgccgctccttcgccgactgccccggacatccacggt360
agccagcgagtccagtccggtgaggaaggggtggcgagaagtcgatatgactgagaggca420
tatttatgactcccagtcatatcgctcggaagtgaccgaacgacctgacgccgccggggc480
tgtgagcggcagcgtgggccaggccgcgaggtcctggagcatctgccggtcgtgggtggc540
gacgaccaccgccgcccgggtcgtcagcagggcggcggtgaggtcgtcgaccagcggcgc600
cgacaggtggttcgtcggttcgtcgaggatcagcaggtcgggacgttcggccaggcgcag660
1
CA 02538147 2004-O1-21
cgccaggttcagccgccgttgctgtccgtgcgacatccggccgacgggggtacgccgggc720
ctcggcgtcgagcaggttcgtcgcgctcagcggcagggccgtgccggagccgacgcgccc780
gctggagcggagccggcccacgtgctgctcgtacaggtcgtgcgcgagcagcgccggcgg840
ccagtcgggcacctcctgaccgaggtacgcgacgcgcgcgccggacaggtgccggacctc900
cccggtcgacggcgtgaggtcgccggccagcacggagagcagcgtcgacttgcccgcgcc960
gttgggtccggtcaccagcaggcggtccccgccgtcgagcgtgagcgtgacccgggtacg1020
caggcgcccggccaccgtgacgtcgtggcatcgcaggatgggcagtccggcacgggtgtc1080
cagcggcggccagcgcagcggctgcggtggctccggcacggtgacgcggtgcgcgtcgag1140
cgcctcctgccggcggcgcagcgcctggaccagtccgggcgcgcgggactggcgctggtg1200
cttgccgtgccccttctccggccgccagccggtgctgagccggtcccgcgcctcccgtac1260
cccgtcggccagccgctggtgctcggcctgctgcgcctcgtggtcgcgtacccagtgcgc1320
gaagtcgcggcggcgcccgtcctgccaggcgacgtagtccccggcgtagcggcgcgggcg1380
cccgtccgcgctggggtcgaggtccaggaactccgtggcgacgtcccgcagcagggcgcg1440
gtcgtgggtcaccagcacgacgccgcccgggtggtcgcgtagccgggcggtgaggaaggc1500
caggctgtcggcgtcgaggtggttcgtcggctcgtccagcatcagcaggtcgaccctcgc1560
tcccagcaggcacgccagccgtacccggtagcgctggccgacggacaacgtggccagctg1620
ccggtcccggtccgggcacgcgtcgaggccggccagcgccacgtcgacgcgccgctgcgc1680
gtcccaggcgtccagccgggtcgccgcgtcgagcgcggccgcgtacgcgtcgtccgcgcc1740
cgcccggccctcggtgagcgcgatcgtcgcctcgtcgagcgcccgcagcgcgcgttcgga1800
ctcccggatcgcctcccggacgagcgtgcccactgtctcgccgtggcgcgactccaggtt1860
ctgccgggcgacgccgatcgtgcccagccgttccaccacgccctggtcgggcgcgatgag1920
gccggccagcacgtgcagcagggtggtcttgccgcggccgttctcgccgacgactgcgag1980
gcgggaagcggcggagacggtcacgctgacgtcggacagcacgacccggccgccgcgtac2040
gacgcggacgccgtcggcccgcacgtgcgcccggtgcccggcgggcagcgaaccgcccga2100
ggtggatggggaggaaggaatgttgtcgaggttgtgcacagtccgctcttcggctcgtcg2160
tggagccgggcagcgcgaggacaccgcccggcgggaacgccgggacggcggagcagagct2220
ggtacgtcagaagaagccggtcaccctgccgccgtcagcggagggaccagggcttcatga2280
cagcggcgtagaacctcatgcggtcaacactacccggggccgggccggagatcgccgcag2340
ttatcggcggcggcgggcgtcggcctcggcgtcgagcaggtcgttcaccgccagcgccga2400
gttgatcagagcgaggtggctgaacgcctgcggatccacggttgcggtactccatttgca2460
2
CA 02538147 2004-O1-21
gtacacctgtcggtatccggtcagcgccgtatcctgcgctttctctgtcggcagagcggc2520
gcggtcgcccgccgcccgccgacgtggctgcggggccggtcgggctcggaccgctcggtg2580
cggcgtcgcggcccggccgtagcatgtttcacctgttcagagcggcttccgggcgctcgg2640
gccgtcggccgcggtggttaccggcgagggctatttcggtcatgcgagagggttctgcca2700
atcgtggcattgtttagttaagtccgatatcagcgggatgctgcctgatatatgacggct2760
gcgcccgggcctgccggatagctatgatgagcgacgacggtgatcgatggcaaatgttgt2820
tgctgtggggtagcgtcaccgccgagtccaggcttttcttgagctgtgtgcgcatattcc2880
ggggggattatgacaacgggacggccgggggagaaccgggcgacagacgcggcacgaaat2940
ccggggtgggccgccggggggccggcgtcccagccatggggcggggggaacgacgagcag3000
gtcctgcgcgagatcctcggggtcgacgtgcaccgcgagctgattgacttcgcgggtggt3060
gccggcggaaatccgcacctggtcgccgaactcgcgcgcgggctcgccgaagagggattg3120
attcgggagacaaacggtcgggcggaattggtgtcccggcgaattccccggcgcgtgctg3180
agttttgtcatgcgtcgattgaatgatgtcagcgccggctgccagcagttcttgaaggtt3240
gccgcggcattgggcagatccttcatgctggaggacgtttcgagaatgctgggccgatcg3300
tcggcggccctgctcccgccggtggacgaggcgatcgcatcgggcttcgtcgtcgccgcc3360
gagcatcaactcgcctttcagagcgacttcctgctgcgcggcatcatcgagtccattccc3420
gggcccgcccgcgacgccttacgacgtgaggcgatgagcctttccgggcgacggcgcccg3480
gcggccgaccagaatcgccggttggacgcggcgcctaccgcgccggtgagcgcgaccggg3540
gaggacgccaccggatcctgttcccgggcgcaccgcctgataatgaacgggaacgcgaag3600
gccggcattcgcgtcgccgaggcggttctcgccggcccggccgcgtcgctcgctgcccgg3660
cgtgacgcggaggcgtgtctggtgctggccgatctgctgctcggcggggagggcggcggc3720
ccgatgaccgaggcgatcctgcgcgaacgcgacgccgagtccggtgacgccgcactggcg3780
atggcgctgaccgcccggtccaccgggctgtggtcggcgggaaagctggcggagggcctg3840
aagctgggacgggcggcggtgcgggcgggcgcggaggccgaaccggtgtggcgtctgcac3900
gcccagctcgcgctcgccgggaaactcgcgaacctccgcgagttcgacgaggccgaggcg3960
ttgatcaacgaggcggaagcgggcctgcgcggactgcccgcgccgatctggacggccgcg4020
acggcggtgatgcggtcccggttgctgctccaggcggggcggatcggggaggcgcgtcgg4080
gaggcggcgctggccaccaccgccgtggagggggacgcggtgccgatgctgcggcctctc4140
gcctacgcggtgctcagcaccgcctccttctacatgggggacctgcccgccgcgatcgag4200
tacctcaggcgggggcagcgggacgcggaccgccacgtggtcctcgactcggtgcagtac4260
tcgtgggcggaagtgctgatcacggtcaagcaggaaggcccgcgggccgccgcccagctg4320
3
CA 02538147 2004-O1-21
ctcgcgggcaagcaccaccgcctgcccacgcagcgccgcctctacgtcgaggtgccgagc4380
gccgccgccttcctggtcctgctcgcccgcgacgtggacgaccgtgacctcgaacgccgc4440
gtcctcgacacggtcaacgggctcgccgcggacaaccccaggatccaggtcgtcagcctc4500
accgccatgcacgcccacgcgctggcgaacagcgctccggccgccctggcgctcatcatc4560
gtgcagtcacgggacccgatctcggtggcgctggccaccgaggaactcgccaagctctac4620
gccgcgcaggcccaggcggggggacggccggcgacgccggcccgcgccgaggaggccgcc4680
accccgccggcgagctgctggtcgaccctgtccgacatggagcagcggatcgcctacctg4740
gtgagcg~gggtctgacgaaccggcagatcgccaagcaggtccacctgtccgcgcacacc4800
gtcaactaccacctgcggaagatctaccggaaactgggtttcaacacccgggccgagctg4860
gcgcacgccgcggccacgtactccggccgggcggcgatctactccatgagcggcgaccag4920
gactggggcgccggatccatgaccggcaaggccagctgaaccgcattcccggcgtccgcc4980
ggctgaaccgcgccccggcgtacgccggccggttcagccggcggacgccggctggcgtgt5040
ggtggccagcgccggccggaccgcctcgtgcgcgatgaagcagcgggtcagttccacccg5100
gctgttgatgtcgagcttggagaagacgcgccgcaggtgactgtcgacggtgtgcgggga5160
caggaacagcgaactcgccgcctcgcggttggtcatcccgtccacgatggcccgcacgac5220
ccgcagctccgcgctggtcaggctctcccaccccgaccggggccggtcggggaccagcgg5280
gcggacgttgtgagccggcaggccacgcagctcggcctccacgcgctccaggtcgcgtcg5340
cgcgccgcactcccggtagccgtccgtcgcggcctcgagcagacgggtggcctcggcccg5400
gtcccgggtgctgcgggccgcgtcctccaccgcgccggccgccgcgagcgtacggccggc5460
gagccggtgcagatccgcggcccgcagcagcgccgccggatcgtcgcgcaggagacccgc5520
ggcgtgttccgccgccgccgccagcgactggacgaacgggttgccgcgggcgacgcgccg5580
ggcgacctccacggcgcgctcggcctccgcgtcgagccccgcccggcgggcctggcgtac5640
gagcgtcgccgcagcggccggcgcctcggtgaacagcagcggatcgggtgcgacctgtcc5700
ggcgacgttgatcagcgtctgcaccatcatcgccggacggccgctggcagcgtggaaccg5760
ggccagcgcccagtccatccgcgccgagtcgtcggcggaggccagccgctcggccgcccg5820
caactggtcgctggccgtggcgaggtcaccgtggtgcacgccgaggtgggccaggaccag5880
gcgcgccggcacgcagtcgcccggccgggagtggtcggcggctcgcagcgccgcctccgc5940
ctcggcgcgtgcctcgtccagccgtccggccgctgccagcagctcggcccggtggccgcg6000
ccagagcgactccgagccggtgtgactgggctcctgcgccagcggtcgtacggtgtccag6060
caccgcctgcgcctcgtcgagctgatcggccgcgcccagcgcccggaccagccaggtcca6120
4
CA 02538147 2004-O1-21
cagcggccgccggccgggcgcgcagcccggggactggtgccggggctccagctcggcgga6180
ggaggcaccgcccaggtgcttcgtggtgtccgcgagcgcccggtccagcttggcgcggtc6240
cagctcgcacacgtcgtggcgggcctgcgtccggcgcaggaagccggccgccaggcggtg6300
gctgccggcggcccgcatcccgtgtcccagttcgagcacgagctgcgcctcgacgtccgc6360
cgcgaggtcgcggcggagcatcacctccgcgaggcggccggcctcggcggcccgccccgc6420
cccggccagcaggcgcagcgcacgggccagtgctcgtggcgcctcggcggatccgttctc6480
caggtgggacacggcggctgccgccacgtcgtcgcacccgcaccggcccgagcgcggccc6540
cgccgtcgcggcgggcgtggccgcgtccggcgcggagcgcgtgacgcgtacgccggcggg6600
ggagtggggcgtcccgggccgcggatcgggccgcccgcgccggaccgggtcgcccgccgc6660
cggtgecggcgcggatccgggctcggcacgctccggttccgggtacgcggcgtggcgaag6720
cgcctctccgagcaccgggtgggcgaaggtcagctccgcgccgtcgcgtcgtatcagccc6780
gacccgcaccgcctcgtcgatcgcggcggacacgtcggcggccgagccgtccagcaggcc6840
cgtcacccggtcgacgggaaacgtgtggccgagccggccgccggccgcgagcaggcgccg6900
cagcgggggcggcagctcctccagcagcccgcgaacggcggcgaggacaccgtcgggcag6960
ctcgtcggacaccaccgacgccgccccgtccacgatgatcatctggccggccttgatgaa7020
cgcgctgaagacgatctccatcaccttcgggttgccgccgcagcgggccgcccagcgcag7080
gacggaggcgtccggccgggcgccgaggatgccggcgcacaggtcggccaccgcctcctc7140
gcccggctcgcgcagccgtacccgtaccgcgacgtgctcggccagccagtcgacggcgtg7200
ctgagcgatcgacccggcggcgaccggccggcgggccagcagccagagcaccggcgagga7260
cgccaggcgcggcacgagcccgcgcagggccagggcgctgacgtcgtcgatgcgctgggc7320
gtcgtccagggcgaccacgagcgggcgccggcgcgccgcgacctcgaccagatcgccgac7380
ccggtcgatcagccagaacgggttggcgcccggcagggcgagctgctcgaccgccgcttc7440
gccgggcatcgcgtggcgcaggaagttgacgagcaggtgtacgggcaccggctgatccgt7500
gacgcttgcccgcccggccaccactgtcagcccgcgggccgccgcctccaggccggtgac7560
cttcagcaggtgggtcttgccgatgccgaacggcccgtcgacgacgacgcagcccccgga7620
tccccgcatggtggcgtcgagcagttcccccaatgaggacaattcctgcccgcgccccgc7680
catgcgattcatgatgaccatcccgttttcctctgctgaatcgtccgacgtgcgccgcga7740
gccgatgtcccaccgcgttcgaccgtccgttctggacagttgaacgccggatcggggcgg7800
gctactcagttatacgggatctgcggccgttcgtcggcgacgtcgctggcagcgcgcact7860
actcgcgtgagtagtgggcagggtgtcaggccgcgattactgtcaggccatgccgggctc7920
ggcgtgccggcgcggacgaaatggcgacgccgatggggagatcggcgtcgtttccgcgcc7980
CA 02538147 2004-O1-21
ggcgcaaaacgtccggaacggaatcgactaatcgccgctcgacgcgactggtccagcgaa8040
tccaggggagtccgagatgcgtgagtgtaatggtgaccgccgtcttgatcgggagacgcg8100
ggcatgaccgtcggatatctcgggacggtcaccgactcggcgcccgtcgacgccgcgctg8160
cgcgacttcttcgccgagcgccgcgccgaggcacgcgagctcggcgacgacttcgcggcc8220
ctggtcgccgagctggagagctacgtcctgcggggcggcaagcgcatccggcccgccttc8280
gcctggctgggctggatcggcgccggcggcgacccggaggacccggtggcgaccgcggtg8340
ctgaacgcctgcgccgggttcgagctgctgcacgcgtccggcctcatccacgacgacatc8400
atcgacgcgtcgcagacccgccgcggccatcccgccgcgcacgtcgcgtacgccgaacgg8460
catcgggcgcggcgcttctccggtgacccgggaacgttcggcaccggcaccgccatcctg8520
atcggagacctcgtcctgatctgggccgacgtcctggtccgcgcctccggcctgccggcc8580
gacgcgcacgtgcgggtctcgccggtgtggtcggcggtgcgctccgaggtcatgtacggc8640
cagctgctcgatctgatcagccaggtgagccggagcgaggacgtcgacgcggcgctgcgc8700
atcaaccagtacaagaccgcgtcgtacacggtggagcggccactgcagttcggcgcggcg8760
atcgccggcgcggacgacgacctcttcgcggcctaccgcgccttcggcgccgacgtgggt8820
attgccttccagctgcgcgacgacctgctcggcgtgttcggcgacccggtggtgacgggc8880
aagccgtccggcgacgacctgcgggagggcaagcggacggtcctgctcgccacggcgctc8940
aagcgcgccgacgaacgggacccggacgcggcggcctacctgcgggcgaaggtcggcacg9000
gacctcgcggacgaggagatcgcccgcatccgcgccatcttccgcgacgtcggcgcggtc9060
gaggagatcgagcggcagatctcgcagcgcaccgaccgggcgctggccgcgctggaggcg9120
agcagcgccaccgcccccgcgaagcatcagctcgccgacatggcgatcaaggccacccag9180
cgggcccagtgatgtccacggaaccggtgaccgtcgtcgcccgcggcgttctcgacggcc9240
ggggtgacgggccgggccgcctcggcaccggccgcgcccacggcaaggccatcctgctgg9300
gcgaacacgccgtcgtgtacggcgctccggcgctcgccgtcccggtgccgcaactgaccg9360
ccgtggccaaggcgcggcgggccggcggcgacggcggcgacgaggtctccttcgccatcg9420
ccgggctggagagcccggaggtgacgtcgcttccgaccgacggcctgcaacatctggtga9480
cggagttccggcagcgggccgccgtcaccgagccgatgcgcgtcgacgtgctcgtggact9540
gcgccatcccgcagggccgggggctcgggtcgagcgccgcctgcgcccgcgccgcggtgc9600
tggccctcgcggacgcgttcgaccgccgcctcgacgccgccacggtgttcgatctggtgc9660
agacctcggagaacgtggcgcacggccgggccagcggcatcgacgccctggccaccggtg9720
cgaccgcgccgctgatcttccgcaacggcgtgggccgggaactgccggtcgccatggcgg9780
6
CA 02538147 2004-O1-21
gcgccgcgcg tgccgcgcga gggtcgggcc cggccggctt cgacgcggtg ctcgtcatcg 9840
ccgacagcgg cgtcagcggc agcacccggg acgcggtgga gctgctgcgg ggtgccttcg 9900
agcgctcccc gcgcacgcgc gacgagttcg tcagccgggt gaccagcctg accgaggcgg 9960
cggcgcacga cctgctccag ggccgggtcg ccgacttcgg cgcgcggctg accgagaacc 10020
accggctgtt gcgcgaggtc ggcatcagca ccgaacggat cgaccggatg gtcgacgccg 10080
cgctcgcggc gggcagcccg ggcgccaaga tcagcggcgg tggcctgggc ggctgcatga 10140
tcgcactggc ccgggaccgc caggaatccg cggcggtggt gcggagcgtc cagcaggccg 10200
gcgccgtccg cacctggacc gtcccgatgg ggaggttcac cggccatgac gactgaccac 10260
cgggcggagc cgtccgagcc ggcgctcgac cggcccgcga ccgccgtggc ccatccgaac 10320
atcgcgctga tcaagtactg gggcaagcgc gacgagcagc tgatgatccc gtacgccgac 10380
agcctgtcga tgacgctcga cgtcttcccg accaccacca ccgtccggat cgacagcggc 10440
gcggcggccg acgaggtcgt cctcgacggc tcgcccgccg acggcgaacg gcgacagcgc 10500
gtcgtcacct tcctggacct ggtacgcaag ctggccgggc gcacggaacg ggcctgcgtc 10560
gacacccgca actccgtgcc caccggcgcc ggcctggcgt cctcggcgag cggattcgcc 10620
gCCCtCgCCC tCgCCggCgC CgCCgCgtaC ggCCtCgaCC tggaCaCCaC CgCgCtgtCC 10680
cgcctggccc ggcggggatc cgtgtcggcc tcccggtcgg tcttcggcgg cttcgcgatg 10740
tgccacgcag gccccggcgc cgggaccgcc gcggacctcg gctcctacgc cgagccggtg 10800
cccgtcgcgc ccctcgacgt cgcgctggtg atcgcgatcg tcgacgccgg gccgaaggcg 10860
gtgtcgagcc gcgaggggat gcggcgaacc gtccggacct ccccgctcta tcagtcgtgg 10920
gtcgcctccg gccgcgccga cctggccgag atgcgggccg cgctgctcca gggagacctg 10980
gacgcggtcg gcgagatcgc cgaacgcaac gccctcggca tgcacgccac catgctggcc 11040
gcccggccgg cggtgcgcta cctggcgccg gtcactgtcg ccgtgctcga cagcgtgctg 11100
cgcctgcgcg ccgacggcgt ctccgcctac gccacgatgg acgcgggacc gaacgtcaag 1116c)
gtgctctgcc gccgcgcgga cgccgaccgg gtcgccgaca ccctgcgcga cgccgcgccg 11220
agctgcgccg tggtcgtcgc cggaccgggg ccggcggccc ggccggaccc gggcagccgg 11280
ccgtgaccgg cccgggcgcc gtgcgccgcc acgcgccggg caagctgttc gtcgccggtg 11340
agtacgcggt gctggagccg ggccacccgg cgctgctggt ggcggtcgac aggggagtgg 1140()
acgtcaccgt ctccggcgcc gacgcccacc tcgttgtcga ctccgacctc tgcccggagc 11460
aggcgtgcct gcggtggcag gacggccggc tcgtcggcgc gggcgacggg cagccggcgc 11520
ccgacgccct cggcgccgtg gtctcggcga tcgaggtggt cggcgaactc ctgaccggac 11580
gagggctgcg cccgctgccc atgcgggtgg cgatcaccag ccggctgcac cgcgacggca 11640
7
CA 02538147 2004-O1-21
cgaagttcgg cctcgggtcg agcggggcgg tgacagtcgc cacggtgacc gcagtggccg 11700
cgtaccacgg ggtggagctg tcgctcgaat cgcggttccg gctggcgatg ctggcgacgg 11760
tgcgtgacgg cgccgacgcc tccggcggtg atctggccgc gagcgtctgg ggcggctgga 11820
tcgcctacca ggcgcccgac cgcgcggccg tgcgcgagat ggcgcggcgg cgcggcgtcg 11880
aggagacgat gcgcgcgccc tggccgggcc tgcgggtccg gcggctgcca ccaccgcgtg 11940
gcctcgcgct ggaggtgggc tggaccggcg agccggcgag cagcagctcg ttgaccgggc 12000
ggctggccgc ctcccggtgg cggggcagcc cggcgcggtg gagcttcacc agccgtagcc 12060
aggagtgtgt gcgtaccgcc atcgacgcgc tggagcgggg cgacgaccag gaactgctgc 12120
accaggtccg gcgggcccgg cacgtgcttg ccgagctgga cgacgaggtc cggctcggga 12180
tcttcacccc ccggctgacg gcgctgtgcg acgccgccga gaccgtcggc ggcgcggcca 12240
aaccgtccgg cgccggtggc ggggactgcg gcatcgcgtt gctggacgcc accgccgcga 12300
cgcggaccgc gcggctgcgc gagcagtggg ccgccgccgg ggtgctcccc atgccgatcc 12360
aggtccatca gacgaacggg agcgcgcgat gatcgccaac cgcaaggacg accacgtccg 12420
gctcgccgcc gagcagcagg gccggctcgg cggtcaccac gagttcgacg acgtgtcctt 12480
cgtgcaccac gccctggccg gcatcgaccg gtccgacgtc tcgctggcca cgtcgttcgg 12540
cggcatcgac tggccggtgc cgctgtgcat caacgcgatg accggcggca gcaccaagac 12600
cggcctgatc aaccgggacc tggcgatcgc ggcccgggag accggcgtac cgatcgccac 12660
cgggtcgatg agcgcctact tcgccgacga gtcggtggcc gagagtttca gcgtgatgcg 12720
ccgggagaac cccgacgggt tcatcatggc caacgtcaac gccaccgcct ccgtcgaacg 12780
ggcccggcgg gctgtcgacc tgatgcgggc cgacgcgctg cagatccacc tgaacaccat 12840
ccaggagacg gtgatgccgg agggggaccg gtcgttcgcc gcctgggggc cgcggatcga 12900
acagatcgtc gccggcgtcg gtgtgccggt gatcgtcaag gaggtcggct tcgggctcag 12960
ccgcgaaacg ctgctgcggc tgcgggacat gggcgtccgg gtggccgacg tcgccggccg 13020
cggcggcacg aacttcgcgc gcatcgagaa cgaccggcgg gacgccgccg actactcctt 13080
cctcgacggg tggggacagt cgacacccgc ctgcctgctg gacgcccagg gcgtggacct 13140
gcccgtgctg gcctccggcg gcatccgcaa cccgctcgac gtggtccgcg ggctggcgct 13200
cggcgccggc gcggccgggg tgtccggact gttcctgcgc acgctcctgg acggcggcgt 13260
gccggcgctg ctgtcgctgc tgtccacctg gctcgaccag atcgaagccc tgatgaccgc 13320
cctgggcgcg cggaccccgg ccgacctgac ccgctgcgac ctgctgatcc agggtcggct 13380
gagcgcgttc tgcgcggccc ggggcatcga cacccaccgc ctcgccaccc gttccggcgc 13440
8
CA 02538147 2004-O1-21
cacccacgag atgatcggag gcattcgatg aacgacgcga tcgccggtgt gcccatgaaa 13500
tgggtaggtc ccgtgcggat ctcgggaaac gtggcgcaga tcgagacgga ggttccgctc 13560
gccacgtacg agtcgccgct ctggccgtcc gtcggccggg gcgcgaagat ctcccggatg 13620
gtcgaggcgg gcatcgtcgc cacgctcgtc gacgagcgca tgacccgctc ggtgttcgtg 13680
cgcgccaagg acgcgcagac cgcctacctg gcctcgcttg aggtcgacgc gcggttcgac 13740
gaactgcgtg acatcgtgcg cacctgcggc aggttcgtcg agctgatcgg gttccaccac 13800
gagatcaccg cgaacctgct gttcctgcgg ttcagtttca ccaccggcga cgcgtccggg 13860
cacaacatgg cgacgctggc cgccgacgcg ctgctgaagc acatcctgga caccattccg 13920
ggcatctcgt acggctcgat ctcgggcaac tactgcaccg acaagaaggc caccgcgata 13980
aacggcattc tcggccgggg caagaacgtg gtcaccgagc tggtcgtgcc gcgggagatc 14040
gtccacgaca gcctgcacac gacggcggcg gcgatcgccc agctgaacgt gcacaagaac 14100
atgatcggca cgttgctcgc cggcggtatc cgctcggcca acgcccacta cgcgaacatg 14160
ctgctcgggt tctacctggc cacgggtcag gacgccgcga acatcgtcga gggctcccag 14220
ggcgtgacgg tcgccgagga ccgcgacggc gacctctact tctcctgcac gctgcccaac 14280
ctgatcgtgg gcaccgtcgg caacggcaag gggctcggct tcgtcgagga gaacctggag 14340
cggctcggct gccgcgcctc gcgtgatccg ggcgagaacg cccggcggct cgcggtcatc 14400
gcggccgcga cggtgctctg cggcgagctg tccctgctcg ccgcgcagac caacccgggc 14460
gagctgatgc gggcgcacgt ccggctcgaa cgcccgaccg agaccacgaa gatcggagcc 14520
tgacgatggc cgagagaccc gccgtcggca tccacgacct gtccgccgcg acggcgcatc 14580
acgtgctgac acacgagacc ctggccgcga gcaacggcgc cgacgtggcc aagtaccacc 14640
gtggcatcgg gctgcgggcg atgagcgtgc ccgccccgga cgaggacatc gtgacgatgg 14700
ctgctgccgc cgccgcgccg gtggtcgccc gccacggcac cgaccggatc cggaccgtcg 14760
tgttcgccac ggagtcgtcg gtcgaccagg cgaaggcggc cgggatacac gtccactccc 14820
tgctcggcct cccctcggcc acccgggtgg tcgagctgaa gcaggcctgc tacggcggta 14880
cggcgggact gcagttcgcc atcggcctgg tgcaccgtga cccgtcgcag caggtcctgg 14940
tgatcgccag cgacgtgtcg aagtacgcgc tgggtgagcc cggcgaggcg acccagggcg 15000
ccgcggcggt cgccatgctc gtcggcgcgg acccggcgct ggtacgcgtc gaggacccgt 15060
cgggcatgtt caccgccgac gtcatggact tctggcggcc gaactaccgc accaccgccc 15120
tggtcgacgg gcacgagtcc atctccgcct acctgcaggc gctggagggc tcgtggaagg 15180
actacaccga gcgcggcggt cgcaccctgg acgagttcgg cgcgttctgc taccaccagc 15240
cgttcccgag gatggccgac aaggcgcacc ggcacctgct caactactgc gggcgcgacg 15300
9
CA 02538147 2004-O1-21
tcgacgacgc gctggtggcc ggggccatcg ggcacaccac cgcgtacaac gccgagatcg 15360
gcaacagcta cacggcgtcg atgtatctcg ggctcgcggc actgctcgac accgccgacg 15420
acctgaccgg ccggaccgtc ggcttcctca gctacgggtc cggcagcgtc gccgagttct 15480
tcgccggcac tgtcgtgccc gggtaccgcg cgcacacgcg acccgaccag caccgcgcgg 15540
cgatcgaccg gcggcaggag atcgactacg cgacgtaccg ggagttgcac gagcacgcct 15600
tcccggtcga cggcggcgac tatccggcgc cggaggtgac caccgggccg taccggctgg 15660
ccgggctctc cggtcacaag cgcgtctacg agccgcgata ggaccggcca cgccggccgc 15720
cctgaccgaa cgaaccatgc ttggaggatc gatgtccgga actcccgagg tggccgagct 15780
ctactcgacc atcgaggaat cggcccggca actggacgtg ccgtgttcgc gcgaccgggt 15840
ctggcccatc ctgtccgcgt acggcgacgc gttcgcccat cccgaggcgg tggtcgcctt 15900
ccgggtggcg accgcgctgc gtcacgcggg cgagctggac tgccggttcc ggacgcatcc 15960
ggacgaccgg gacccgtacg cctcggcgct cgcccggggc ctcaccccgc gcacggacca 16020
ccccgtcggc gcgctgctct ccgaggtcca ccggcgctgc ccggtggaga gccacggcat 16080
cgacttcggg gtggtcggcg gcttcaagaa gatctacgcg gccttcgccc cggacgagct 16140
gcaggtggcc acgtcgctcg ccggcattcc ggcgatgccc cgcagcctcg ccgcgaacgc 16200
cgacttcttc acccggcacg gcctcgacga ccgggtcggc gtgctgggat tcgactaccc 16260
ggcccggacc gtgaacgtct acttcaacga cgtgccgcgt gagtgcttcg agccggagac 16320
catccggtcg acgctgcgcc ggaccgggat ggccgagccg agcgagcaga tgctccggct 16380
cggcaccggg gcgttcgggc tctacgtcac gctgggctgg gactccccgg agatcgagcg 16440
gatctgctac gccgcggcga ccacggacct gaccacgctt ccggtacccg tggaaccgga 16500
gatcgagaag ttcgtgaaaa gcgttccgta cggcggcggg gaccggaagt tcgtctacgg 16560
cgtggcgctg acccccaagg gggagtacta caaactcgag tcgcactaca aatggaagcc 16620
gggcgcggtg aacttcattt gaacagcggc cggttccgcc gcccgggcgg cggaaccggg 16680
atcaatgcct gttcgctcgg gttcaacact ggcgcgctcc gctaaagtgc gaacatgacg 16740
actggactgt ccagtgtgtg ggcccgggtg aagaactggg tcgtcgcgtt ggctgtggcg 16800
gcggtgctga tgatcagcgc gctggccggt gaccatcctg cccccgaggg cctcggtctg 16860
ctcggcttcg cgctggtggc ggcgagcggc ctggcgctgg ccgccagtcg tcgggccccg 16920
atcgccgtgc tggtcgccac cgggctgtgc gtggtgggct acaacgcgat cggcttcggg 16980
gtgcccgcca tcgcgtacct gttcgcggtc tacgcggcgg tccgggccgg gcaccggctc 17040
gtcacgctcg gggcgagcgc cgccctgctc gtcgtcctgc cgctggcgat catggtctcg 17100
CA 02538147 2004-O1-21
cccgcggacg gcgccctcaa ggaggcgctc gcgcagtcgc ggggcgtgct ggaactggcc 17160
tggctgatcg ccgcggcggc ggccggtgag gcgctgcggc aggccgaacg gcgagcggac 17220
gaggcggaac ggacccgcga ggagaccgcc cggctgcgcg ccacccagga gcggctgcac 17280
atcgcacggg agctgcacga ctcgctcacc caccagatct cgatcatcaa ggtgcaggcg 17340
gaggtggcgg tccacctggc ccgcaagcgg ggcgagcagg tgccggagtc gctgctggcg 17400
atccaggagg ccggccgggc ggcgactcgc gagctgcgcg cgaccctgga gacgctgcgt 17460
gacctgacca agtccccgtc gcacgggctc gaccacctcc cggagctgct ggccggggcc 17520
gagaagatcg gcctggccac cacgctgacc atcgagggcg acCagcggga cgtgccggag 17580
gcggtgggcc gcaccgcgta ccggatcgtg caggagtcgc tcaccaacac cgcccggcac 17640
gcctccgccg cggccgccgc ggtccggatc gactaccgcc cggacgcgct gagcatccgg 17700
atcgacgacg acgggacggc ccggccgggc gccgccccgg tgcccggcgt cgggctgctg 17760
gggatgcacg agcgcgtcct cgcgctgggc ggccggctgc gggcggaacc ccgcaccggc 17820
ggaggcttca ccgtccaggc cgaactcccg gtggtgcgcg tcccatgatc aggatcatgc 17880
tgctcgacga ccagccgctg ctgcgcagcg ggttccgcgc gctcctcgac gccgaggacg 17940
acatcgaggt ggtggccgag ggcgggaacg gccgggaggg cctggcgctg gcccggcagc 18000
acctgcccga tctcgccctg atcgacatcc agatgccggt catggacggc gtcgagacga 18060
cccggcagat cgtcgcggat ccggcgctgg ccggggtacg cgtcgtcatc ctcaccaact 18120
acggcctcga cgagtacgtc ttccacgcgc tgcgcgccgg cgccaccggc ttcctggtca 18180
aggacatcga gccggacgac ctgctgcacg ccgtgcgggt cgccgcgcgc ggtgacgcgc 18240
tgctcgcgcc gtcgatcacc cggatgctga tcaacaggta cgtgtcggag ccgctctgcg 18300
cggacgtcac gcccggcatg gaggagctga ccaaccggga acgcgaggcg gtcgccctgg 18360
ccgcccgggg cctgtccaac gacgagatcg ccgatcgcat ggtgatcagc ccgctgaccg 18420
cgaagaccca cgtcaaccgc gccatgacca agctgcaggc ccgcgaccgc gcccagctgg 18480
tggtgttcgc ctacgagtcc ggcctggtgt cacccggcaa tcgctgaccg ggcagcccgc 18540
ccggtctgtc gcctcggcag tgctgcggct gcggtatgcg gctgctcccg gcgcagacgc 18600
cggagcccgt ggataccgtc accgcagtag atcgatcgat tgtctccttc ggcatgacga 18660
cccgtagcgg ggtcgttacc tacgctggcg cagatgcctg ttcccgcagc cgaaggggct 18720
tccatgttca tccgtcgttt gctcaccgcc gccgcagccg gcgtcctcgg tgggctcgca 18780
ctcgtcgcac cggcggccgc gcaggtgacg gccgccgacg gtgacggtgg ttccggccgc 18840
gccggatccg tgctggcgct cgcgctcgcg ttgctcggcc tcgtcctggg cgggtgggcg 18900
ttgcgctccg cggggcgcgg cggcggtcgt ggcaacgcga tcgccgcgct ggtgctcgcg 18960
~l
CA 02538147 2004-O1-21
gtggccggcc tgatcgccgg cgtggtcgcc ctggccggct ccgacggtgg tgtcggcagc 19020
ggcaacggcc gtggtggcgc catcgtggcc gtcgtgctgg cgctgatcgg gatcgccgtc 19080
ggcggcctgg cattcacccg ctcccggcgc gccgcctgac cggcgctgcc gaccgaacac 19140
cccggtgacc caaccgaacc cgaaggggag tcccatgcgc aaagtgttcg ccggactggc 19200
agcgttcctg ctgctcgtgc tcgtggtgca gttcttcctg gccgccagcg gcgcgttcag 19260
caacgaggcc aacgaggagg cgttccgccc tcaccggatc ctgggcctgg ggagcatcct 19320
cgtcgccgtg gtgctgacgg tggccgccgc ggtgatgcgg atgcccggcc ggatcatcgg 19380
cctgtccggc ctggtcgccg ggctgggcat cctgcaggcc ctgatcgcgg tcatcgccaa 19440
ggcgttcggc gactcggccg gtgactcggc cgtcggccgg tacgtgttcg gcctgcacgc 19500
ggtcaacgga ctggtgatgg tggccgtcgc ccgcgtcatc ctgcgcagcg tccgggcggc 19560
gccggacacg accaccacgc ccggcgtgga cacgacggtc accggtccgg cggccgactc 19620
ggcgcgaacg gcgtcatgag cacgctccaa tggatcctcg tggaccacgt cgtggcgctg 19680
ctcggtgtcg cgacgtggtt cgcaacgggt gtcacggcag ctctcggccg ccaccggatc 19740
gcgttggcgc tcctcggcgc cgcggtgctg gtgacagtcg cccgcctggg caccgtggcg 19800
ctgctggccg accgcggctg gtggttcgtc caggagaagg ttctgctggg gctgccgatg 19860
ctcggcgccg cggggctcgt cgcggtgctc ctggccggcc cgcgcctgct cgcggcccgg 19920
cagtcaccgg cggcggacct gccggccggc gcgctggtcg cggtgctgac cgccggcttc 19980
gccgcgctgg ccggcctggt ggtgacgttc accgccgggt acccgctgac gtggagcacc 20040
gcgctgatcg ccgtcgccct cgtctgcgcc gccgcgctgc tcaccgcgcg ggtggtcgga 20100
cgacccgccg ccccggccgc ggaggccggc tccccggagc acacgccggc ggcggccggg 20160
cccacggcgc tgtcccgccg ccggttcctc ggcgtggccg ggggagtggt cgcggcgggc 20220
gccggcgcca ccggcgtcgg cctgctcttc cgcgacccgg aggcgatggt caccggaggc 20280
ggccccggac acgccggtgg cgcccgcccc aaggtctccg tggcggacct gcgcggcccc 20340
ggcgctccgg cggcgggcgg cacggcgcga cgccacgtgc tcaccgcccg gacgggcacc 20400
gtcacgattc cgtccggacg tccgatcgac gcctggagct acgagggccg cctgcccggg 20460
ccggccatca ccgcgaccga gggcgacctg atcgaggtga cgctccgcaa cgccgacatc 20520
gaggacggcg tcaccgtgca ctggcacggg tacgacgtgc cgtgcggcga ggacggcgcg 20580
ccgggcgcca cgcagcacgc ggtgcagccc ggcggcgagt tcgtctaccg gttccaggcg 20640
gaccaggtgg ggacgtactg gtaccacacc caccaggcgt cgcaccccgc cgtgcgcaaa 20700
gggctgtacg ggacgctcgt cgtgacgccg cgcgaggacc ggccggaagc ggagcgcggg 20760
12
CA 02538147 2004-O1-21
ctggacctga cgctgccggt gcacacgttc gacgacgtca cgatcctcgg cgaccaggag 20820
ggacgcgccg tccacgacgt ccgccccggc cagccggtgc gactgcgtct gatcaacacc 20880
gactccaacc cgcactggtt cgccgtcgtc ggctcgccct tccgcgtggt ggccgtcgac 20940
ggccgcgacc tcaaccagcc gggcgaggta cgcgaggtcg ggctccgcct gcccgccgga 21000
ggccggtacg acctgaccct ggccatgccg gacgccaagg tcacgctgct gctcgacaac 21060
gactccgacc agggcgtcct gctgcgcccg ccgggcgtcg gcggtggtga ccgcccgctg 21120
ccggacaccg ccgactggcc cgagttcgac ctgctgggct acggcgagcc ggcgcccgtg 21180
ccgttcgacg ccgacgacgc cgaccgccac ttcaccatcg tcctcgaccg ggccctggcc 21240
atggtcgacg gcaagcccgc gtacgcccag accgtcgacg gtcgcgcaca tccctccgtc 21300
cccgaccagc tcgtccggga gggggacgtc gtgcgcttca cggtggtcaa ccggagcctc 21360
gaaacccacc cgtggcacct gcacggccat ccggtgctga tcctgtcccg cgacggccgg 21420
ccgtactccg gcagcccgct gtggatggac accttcgacg tgcggccggg agaggtgtgg 21480
gaggtggcgt tccgggcgga caatccgggt gtctggatga accactgcca caacctgccg 21540
caccaggagc agggcatgat gctgcggctc gtctacgacg gtgtcaccac gcccttcgcc 21600
agcacgagcc acgcacactg aggggactcg catgaccgca gacctgcacg gcctggccag 21660
cgtccgctac atcgtcgacg acgtgtcggc ggcgatcgag ttctacacca cccacctggg 21720
tttcacggtg tcgaccgcgt tcccgccggc cttcgccgac gtggtgcgcg ggccgctgcg 21780
gctcctgctg tccgggccga ccagctcggg cgcccgggtc accccggcgg acgcggccgg 21840
gtgcgggcgc aaccgcatcc acctgatcgt cgacgatctc gacgccgaac gggagcggct 21900
ggagcgcgcc ggggtgacgt tgcgcagcga cgtcgtggcc gggccgggcg gccgtcagtt 21960
cctgatcgcc gacccggcgg gcaacctggt cgaggtgttc gagccggcag cccgcggctg 22020
aaccgccgac ggacgccctc ccacctcgcg acgcccgaag cccgacacct ggccgcgtcg 22080
cggccacgat caccgtggcc gcgacgcggt gacggggtgc cttaccgggg cggggtgggc 22140
gcggcgagcc gcgcggccag gatggagatg atcacggcgc cggcgatcac gtgggtgccg 22200
gcgaggacga gctgcgtcga caccggggtg tcccgggcga aggcgggcgc ggcgagggac 22260
agcacggtga acgcgacggt gccggccacg aaggcacgca cgggccgccg ggcccgccgc 22320
gccacgacca ccgccaggac gattccgccg atcgaccaga gcacgacgct gcgggcgatg 22380
gcccccaccg ggatcgcctg cgcctgctcc tcccagacgc cggccgcctc catcggtacg 22440
ccgaagcccc gggcggcgag cgtgaacgcc tccgcggcca cggccccggc gagggtggcc 22500
agcacgccga ccagccacac cggagcggtg gccggcgacc aggtgggccg tgccgcgacg 22560
ggagttcggg gagtggcctc atccacggcg tcgcctccgg tcgggtgcct cgatgtgttc 22620
13
CA 02538147 2004-O1-21
tcgggagaat gcggggacgc cacgacggca gtcaacatgg acagttgaac gccctggcgt 22680
cacgggcggt tcccgcgccg gcccgccgcc tcggccgcgg cggcggccgt gccgtcggcg 22740
agcagggaga ccagcaggtc gcccaggatc cgtgggccgt gctgggtgag gacggactcc 22800
aggtggaact ggacggaacg gaatcccggg ccgcgcagcg cgtgcacgtc cccgctgtcc 22860
gggctgcggc tgatctcgat cgggccccgc cggccaccgg ccaccacgtc gtgcgcggag 22920
cgggcggtgt aggtgttgta gaaccccacg agttccggcc ggccgaacag gtcgatccgc 22980
ttctgcacac cctggttggg caccgcgcgc cgggcgaggg ggaaccccag ttcggcggcg 23040
agcacctggt ggcccaggca gatggacagg aacggcaccg ttccggcgag caggtcgcgg 23100
gtgagcccgc gcagggtccg catacgcggg tcggtcaggt cgcccgggtc gccggggccg 23160
ggaccgacga cgacgaggtc gtgtccgtcc ggccgcagcc ggctgtcgaa ccgggcgatg 23220
ctcgaccgca gcccgagggc ccgcaactgg tggtcgagca tggccatgaa cgtgtcctcg 23280
ttgtcgacga cgagcacgcg gcgtccggtc agcgccgggt tcggggtgcg ccgctccgcg 23340
ccgtcgagcc agaacctcga cagtgtggtg ttgcgctcgc gcaacgcccg ccgtacccgg 23400
gggtcggtgg ccagggacga acgagcccgc gcggccgtgg tccgcccgcc gtccgggccg 23460
tccgggtcga cgccgaggcc gagcgccgcg cgcatggcgc ccgccttggc ccgcgtctcg 23520
gccacctccg actccggctt ggagtcccgc acgagggtgg cgccgacgcc caggcgcagc 23580
gtgcccgcgt cgtcgatctc ggcggtgcgg atcatgatgg ccgagtcgag cgtacggctg 23640
ccggccgagt cacggcccat caacgcgagc acgccgccgt agtagccgcg gccggtcgtc 23700
tcgtggcggg tgatgacccg gaacgcgttc tcgatcgggc tgccggtgac cgtcggcgcg 23760
agcagggtct cccgcagcac gtcgcgcacg tccaggtcgc tgcggccggt caggatgtac 23820
tcggagtgcg tcacccgcgc catttccttg aggaacgggc cgtgcacctg gccgccggag 23880
gcgcacatcc gcgccatcat tttcagttcc tcgtcgacga ccatgtagag ttcgttagcc 23940
tctttcgggt cgttcaggaa ttccagcaga ccggaaacgg ccgggccgtt cggggggtgc 24000
cggtaggtcc cgctgatggg attcatcgag acggttccgt cgatcatgct gacgtgtcgt 24060
tccggtgacg cgccgatgaa cgtgccggcg ccggagtgga acagaaacgt ccagtaggaa 24120
cccagttcgc cggtcagcaa ccggcggaag agcgccagtt ccgtggcgat cgagtagtcg 24180
gccagccgcg cggtgaaggt gcgccggatg acgaagttgg atccggcgcc cagcccgatc 24240
tcgtcaccca ccacccgctt gacgatcgcg gcgtagtcct cgtcgctgag gtcgaagtcg 24300
gcgtcggtca ccggcacacc gcgttcgggc aggcccgcca gcgcctgtcc gcggtcgagc 24360
ccgaactgct cgtggacgcg catcgcgagc agcggcgcgc cgtcgtcgtg gcagtcgaac 24420
14
CA 02538147 2004-O1-21
ccccgttcgg tgacctgccg gtacggcacc gccacgagca ggtcgtgccg cgcgccggtc 24480
gccggctcgg tgggcagggg cagctcgccg agagtgtcca cgtcgcacac ctcgccggtc 24540
agaacctcca cgtacgcgca cccggccgcg ccgggccggt gcagcagggc gaaggcgcgc 24600
ccgtcgccgc cgagaccgga cagcagatcg gggaatccgg tcacgttcga ttccgtcccg 24660
tccatgtcgc tccctttgcc tgagagatcg cctgtcgata ctgcgtccgg caaaaggcgt 24720
cgcacatgac gtgaagtcgc cgacggcatc acgtgtttcc ggtaacgcgc cgacgttatg 24780
gcgtgaacga ctgaatcggc gggctactac tcgggcgagt agtgcccacg cagatcgacc 24840
gcgattactg tcgaccgcaa tgccgatacg acgagggcgg tgaagacgac tgtggacgtg 24900
ctggtccaga aatacggggg cacctcgctg cagaccctcg accgcgttcg gcacgccgcg 24960
ctgcggatcg ccgaggcgcg gcggcacggc tccgccgtga cagtggtcgt gtcggcgcgc 25020
ggcagccgga ccgacgacct gctgcggctg gcggccgacg tcggcgccgc gggtccgtcc 25080
cgggaactcg accagttgct cgcagtcggc gagtccgagt cggcggcgct gatggcgctg 25140
gcgttgaccg ggctgggagt gccggccgtc tcgctgaccg ggcaccaggc ggagatccac 25200
accaccgacc ggcacggcga cgcgctgatc tcgcggatcg gggcggcgcg ggtggaagcg 25260
gcgctgggcc gtggcgaggt cgccgtggtc accggattcc agggcatcga ccgggccggt 25320
gacgtcgcca cgctggggcg cggcggctcc gacacgacag cggtggcgct cgcggcccgg 25380
ctccgcgcgt cggcgtgcga gatctacacc gacgtggacg gcgtcttcag cgccgacccc 25440
cgcatccttc cggcggcgcg ttgcctgccg tgggtggagc ccggcgtcat ggcggagatg 25500
gcgttcgccg gcgcgcgggt cctgcacacc cgatgcatcg agctggccgc catggaaggg 25560
gtcgaagtgc gcgtgcgcaa cgcgtcgtcg caggcgcccg gaacgatagt cgtggaccgg 25620
cccgacgacc ggccgctgga gacccggcgg gccgtggtgg cggtcaccca cgacaccgat 25680
gtcgtccgcg tgctggtgca ctgccgcgac ggccgccggg acatggcacc cgacgtgttc 25740
gaggtgctgg ccgcccatgg ggcggtggcg gacctggtgg cccggtccgg gccctacgag 25800
agcgagttcc ggatggggtt caccatccgc cgcagccagg ccgaagcggt gcggaccgcg 25860
ctgcacgacc tcaccgcgtc cttcgacggc ggggtccact tcgacgagaa cgtcggcaag 25920
gtgtccgtgg tcggcatggg cctgctcagc cgccccgagc acacggcccg gctgatggcg 25980
gcgctggccg cggcggggat ctcgacgagc tggatctcca cctcccagat gcggctgtcg 26040
gtgatcgtgt cgcgggaccg caccgtcgac gccgtcgaag ccctgcaccg cgcgttccgc 26100
ctggaccggt ccgagccggc ggacgccacg tccctgacct cccgccgttc cgccaccgcc 26160
tgagagaggt aggaaaccgt ggccgtactc aacgcttcgt tcgctcgtgg cctgcgtctg 26220
cgccgactgt tccgacgcgg cgacggacgc ctgctcgtcg tcccgctcga ccactccgtc 26280
CA 02538147 2004-O1-21
accgacgggc cgctgcgccg cggcgacctg aactcgctgc tcggtgagct cgccggcacc 26340
ggcgtggacg ccgtggtgct gcacaagggc agcctgcggc acgtcgacca cggctggttc 26400
ggcgacatgt cgctgatcgt gcatctgagc gtgagcaccc ggcacgcccc ggacccggac 26460
gcgaagtacc tggtcgcgca cgtggaggag gcgctgcggc tgggcgccga cgcggtcagc 26520
gtgcacgtca acctcggctc accgcaggag gcgcggcaga tcgccgacct ggcggcggtg 26580
gcgggggagt gcgaccgctg gaacgtcccg ctgctggcca tggtgtacgc ccgcgggccg 26640
cagatcaccg actcccgggc accggagctg gtggcgcacg ccgcgacgct cgccgcggac 26700
ctcggcgccg acatcgtcaa gaccgactac gtgggcacgc ccgagcagat ggccgaggtg 26760
gtgcgcggct gcccgatccc gctgatcgtg gccggcggcc cgcgctcggc cgacactccg 26820
acggtgctcg cctacgtctc ggacgcgctg cgcggcggcg tggccgggat ggccatgggc 26880
cgcaacgtgt tccaggccga gcagcccggc ctgatggccg ccgccgtggc acggctggtg 26940
cacgagccac ggcacgtgcc ggaccggtac gacgtcgacg accggctcgc ccttacgtcc 27000
tgagactccc tgaccgtcca ccgaggagaa acccgtgaag ctgtgctggc tggacatccg 27060
taacgtcaac ggcgccaagg aggcaatcgt cgaggaggcg gtccaccagc gggtggacgc 27120
cgtcgtggcg gccgatccgg ccgacctgga gacgcttccc ccgacggtga agaaggtgct 27180
gttcccgcag ggcgggccgc tgccggagaa gctggaaccg gccgacctgg tgatcgtcga 27240
gccggcccgg cacggcgagc ccgccgagct ggcggcccgg tacccggagg tggagttcgg 27300
ccggttcgtc gagatcgtcg acgcggacag cctggaggac gcctgccggt ccgcgcgcca 27360
cgaccggtgg agcctgctgt acttccgcga ccccaccaag atcccgctgg agatcgtgct 27420
ggcggccgcg gcgggcgcgg agggcagcat catcacccag gtcgccgacg tcgaggaggc 27480
ggagatcgtc ttcggcgtcc tggagcacgg ctcggacgga gtgatgctgg cgccccgcgc 27540
cgtgggggag gccaccgagc tgcggaccgc cgcggtgagc acggcggcgg acctgtcgct 27600
cgtggagctg gaggtcaccg gcatccggcg ggtgggcatg ggcgagcgcg cctgcgtcga 27660
cacgtgcacg aacttccgtc tggacgaggg catcctggtc ggctcgcact ccaccggcat 27720
gatcctgtgc tgcagcgaga cgcatccgct gccgtacatg ccgacccggc cgttccgggt 27780
caacgccggc gcgctgcact cgtacacgct ctccgccggc gggcggacca actacctcag 27840
cgagctggtc tccggcggcc gggtgctcgc cgtggactcg caggggaagt cccgcgtcgt 27900
cacagtggga cgggtcaaga tcgagacgcg tccgctgctg gcgatcgacg cggtctcccc 27960
ctccgggaca cgcgtcaacc tcatcgtcca ggacgactgg cacgtgcgcg tgctcgggcc 28020
gggcggcacc gtgctcaacg tgaccgagct gaccgccggc acgaaggtgc tcggttacct 28080
16
CA 02538147 2004-O1-21
gccggtggag aagcggcacg tcggctaccc gatcgacgag ttctgcatcg agaagtgaca 2814()
ggcggcggga aggggagcgg gcgatgaccg cgcagccggt gctggacttc cacgtacgcc 28200
tggcgccccg gcccggggcg cgggagcggc tgctcgccgc gctgcgcgag tgcgggctgg 28260
cgcgggcggt ggtgtgcgcg ggcggcacca tcgacctgga ccggctgtcc cgccagctcg 28320
tcaccggcgg ccacgtcgag accgacgccg acaacgacgc ggtggcggcg gcctgcgccg 28380
gcaccgacgg ccggctggtg ccgttcttct tcgccaaccc gcaccggccg gccgaggcgt 28440
accgggcccg cgccgccgag ttccgcggcc tggagatctc acccgccgtc cacggcgtcg 28500
ccctgaccga cccgcgggtc gccgacctcg tggccgtggc ggcggagttc gaccatccgg 28560
tgtacgtggt ctgcctggac cgacccggcg cgggcgtggc cgacctggtc ggcctgagcc 28620
gccggttccc gcaggtgagc ttcgtgctcg ggcacagcgg cgtcggcaac atcgacctct 28680
acgccctgac cctgatccag gacgagccga acatctcgct ggagacctcc ggcggctaca 28740
cctgcgtggc cgaggcggcg ctacgccgcc tcggcgacga ccgggtggtg ttcggctccg 28800
agtacccgct gcagcacccg gccgtggaac tggccaagtt ccaggcgttg cgactgccgc 28860
cggagcggtg gcggcggatc gcctgggaca acgcgcatcg actgctagga gaggagaagc 28920
ggtgagcgag ccaagttcga gcctgccccg gctcggccag tggcacggcc tcgaggacct 28980
gcggcgcctc caggagaagc aactggcgga gacgttcacc tgggcggccc ggtcgccgtt 29040
ctaccgggcg cggctggcct ccggcgcgcc gccggtgacg cccgccgacc tggccgacct 29100
gccgctgacc accaagcagg acctgcggga caactacccc ttcggcatgc tcgccgtgcc 29160
ccgcgaacgg ctggcgacct accacgagtc gagcgggacc gccgggaagc ((a(((cct( 29220
ctactacacc gcggaggact ggaccgacct ggcggagcgc ttcgcccgca agtggatcgg 29280
catgtccgcc gacgacgtct tcctggtccg cacgccgtac gcgctgctgc tgaccgggca 2934()
tctcgcccac gccgcagccc ggctgcgtgg ggccacggtg gtacctggcg acaaccggtc 29400
gctggcgatg ccgtacgccc gggtggtccg ggtgatgcac gacctggacg tcacgctcac 2946C
ctggtcggtg ccgacggagt gcctgatctg ggccgccgcg gcgatcgcgg ccgggcaccg 29520
gcccgacatc gacttcccgg cgctgcgcgc gctgttcgtc ggcggcgagc cgatgaccga 29580
cgcccgccgg cggcggatca gccgcctgtg gggggtgccg gtcatcgagg agtacggctc 29640
gacggagacc ggcagcctgg ccggggagtg ccccgaggga cgcctgcacc tgtgggccga 29700
ccgggcgctg ttcgaggtgt acgacccgga caccggcgcc gtccgcgcgg acggcgacgg 29760
ccagctcgtg gtcacgccgc tgttccggga ggcgatgccg ctgctgcggt acaacctgga 29820
ggacaacgtg tcggtctcct acgacgactg cggatgcggc tggaagctgc ccaccgtgcg 29880
ggtgctcggc cggtcggcgt tcggctaccg ggtcggcggc accaccatca cccagcacca 29940
17
CA 02538147 2004-O1-21
gctggaggaa ctggtcttct ccctgccgga ggcgcaccgg gtgatgttct ggcgggccaa 30000
ggcggagccg gcgctgttgc gggtcgagat cgaggtggcc gccgcgcacc gggtcgccgc 3006()
cgaggcggag ctgaccgccg cgatccgggc cgccttcggc gtggacagcg aggtcaccgg 30120
cctggcgccg ggaaccctga tcccgctcga cgcgctgacc agcatgccgg acgtggtgaa 30180
gccacgcagc ctgttcggtc cggacgagga ctggagcaaa gcgctcctct actactgagg 30240
gaaccgacat gccgcagatg agggtcgccg tggccggcgc cggcatcgcc gggctcgcct 30300
tcgccgccgc cctgcgccgg accgggatcg actgccacgt gtacgaacag gccgaccagc 30360
tcatggaggt gggcgcgggc gtgcaggtcg cgccgaacgc cacccggctg ctgcaccggc 30420
tgggcctgcg tgaccgcctg cgtacggtgg ctgtcgcgcc gcaggcgatc gagatgcgcc 30480
gctgggacga cggcacgctg ctgcaacgca cccagctggg cagcgtgtgc ggacgccgct 30540
tcggcgcgcc gtactacgtg gtgcaccgcg cggacctgca cagcagcctg ctgtcgctgg 30600
tgccgccgga ccgggtgcac ctgggcgccc gcctcaccgc cgtgacgcag accgccgacg 3066()
aggcgtacct gcacctgtcc aacggcacca cggtcgcggc ggatctcgtc gtgggcgccg 30720
acggcatcca ctcggtcgcg cgggagcaga tcgtggcgga ccggccgcgc ttctccggac 30780
agtccatcta ccgcgggctg gtgccggccg agcgggtgcc gttcctgctc accgaacccc 30840
gggtgcagtt gtggttcggg ccggaccagc actgcgtctg ctacccggtg tccgccggcc 30900
ggcaggtgag cttcggcgcg acggtgcccg ccaccgactg gcggcaggag tcgtggtcgg 30960
gccggggcga cgtgacgcaa ctcgcggccg cgtacgcggg ctggcacccg gacgtcaccc 31020
ggctgatcgc cgcggccgac cgggtcggca ggtgggcgct gcacgaccgg gacagcatcg 31080
accggctcag cgcgggacgg gtgaccctga tcggcgacgc cgcgcacccg atgctgccgt 31140
tccaggcgca gggcgcgaac caggccgtcg aggacgcggt ggtgctcgcg gtctgcctgg 31200
ccggcgtgga accggcgggc ctgggcgccg cgctgcgccg ctacgaacgg atccgcctgc 31260
cccggaccac ccggatccag cggcagtccc gggccaacgc cgagatgttc cacctggccg 31320
acggcgccga ccagcgccgc cgggacgtcg ccgcacaatc ctcgtccggc ctggaccgcc 31380
acgaatggct cttcgggtac gacgccgaga aagccaccac gaccagcggg agcgcctgat 31440
ggaactgacc ggaatcgagt cgaaggtcgc cctggtcacg ggcgcggggc agggcatcgg 31500
cgccgccgtg gccggtgtcc tggcgagggc gggcgcgcag gtggcggcgg tggaccgcaa 31560
cgccgaggcg ctgaccaccg tcgtgacgaa gctcgccgcc gagggcgact cggcgcgcgc 31620
ctactgcgtc gacgtgtgcg acagcgaggc ggtggacgcg ctggtgcgcc gggtcgagga 31680
cgagatgggg ccggtcgcca tcctggtcaa cgccgccggc gtgctgcaca ccggacgggt 31740
18
CA 02538147 2004-O1-21
cgtcgagctg tcggaccggc agtggcgccg gaccttctcg gtgaacgccg acggcgtgtt 31800
ccacgtgtcc cgggcggtgg cgcggcggat ggtgggccgc cgtcgtggcg cgatcgtcac 31860
cgtggcgtcg aacgccgccg gggtgccgcg taccgagatg gccgcgtacg ccgcctccaa 31920
ggccgcgtcc gcgcagttca cccgctgcct ggggcttgag ctgtccggct acggcatccg 31980
gtgcaacgtg gtctcgcccg gctccaccga cacccccatg ctgcgggcca tgctcggcga 32040
gggcgccgac ccgagcgcgg tgatcgaggg cacgccgggc gcgtaccgcg tcggcatccc 32100
gctgcgcaag ctggcccagc cgcgcgacgt ggccgaggcg gtcgcctatc tggtgtccga 32160
ccaggcgggc cacgtgacca tgcacgacct gtacgtcgac ggcggcgcgg ccctgcacgt 32220
gtgacgccct cgcacggaaa ccggaggcga gaaccgatgg ccatgacccc gatcgcgccg 32280
taccgcatgc ccggcgacgg cgacctgccc ggcaccgcgc tgccctggcg tccgcacccg 32340
gaccgggccg ccgtgctggt gcacgacctg caacgctact tcctgcgccc gttcgaggcc 32400
ggggagtccc cgatggccga actgctcccc aacgtcgcga agctgctcgc cacggcgcgg 32460
gcggccggcg tgccggtgct gtacaccgcg cagcccggcg gcatgagccg gcaggaccgc 32520
gggttgctgc acgacctgtg gggccccggc atgagcagcg ccgaggacga ccggggcatc 32580
gtcgacgacg tcgccccgca gccgggcgac acggtgctga ccaagtggcg ctacagcgcg 32640
ttcttccgca gcgacctgga ggagcgactg cgcggtgcgg gacgggacca gctcgtggtc 32700
tgcggcgtgt acgcgcacat ggggtgcctg atcaccgcct gcgacgcgtt cagccgcgac 32760
atcgaggcgt tcctggtggc ggacgcgctg gccgacctat cgcgcgagga ccacctgatg 32820
gcgctgcgct acgccgcgga ccgctgcgcg gtgccgttgt ggacggcgga tgtgctggac 32880
gggctggcgg acgccgccgg gcgtccggat cagagcagca cccaacgatg aggagaacat 32940
cgatgtcgga tcggacccgg gtcgtggtcg tcggcggaac ctcggggatc gggcggcact 33000
tcgcccgatt ctgcgccgaa cgcggagacg acgtggtgat caccggccgt tcggcggccc 33060
ggaccaagac cgtggcggac gagatcggcg ggcggacccg tgggctcgct ctcgacctgg 33120
ccgagccgga gacgatcgcg gacgcgctcg ccgacgl=gcc gcacgtcgac cggctcgtgg 33180
tcgcggcgct ggaccgcgac tacaacaccg tccgcgcgta ccggccgggc gacgcggcgc 33240
ggctgctgac cgtcaagctg gtcggctaca cggcggtcct gcacgccctc gccccgcgga 33300
tgaccgacga gagcgcagtc gtgctgctcg gcggcctggc cagccaccgg ccgtatcccg 33360
gctccacctc cgtcacgacc gccaacggcg ggatcagcgc gctggtgcgg accctggctg 33420
tggaactctc gccggtccgg gtcaacgccc tgcacccgag catcgtctcc gacacgccgt 33480
tctggagcga caagcccgcc gcgcgggagg ccgccgcgac ccgcgcgctc agccgacggc 33540
cggtcaccat gcaggactgc gccgaggcga tcgacttcct gctgacgaac cgctcgataa 33600
19
CA 02538147 2004-O1-21
acggggtcaa cctgaacatc gacggcgggg acgtgctcat ctgacgccgg aggcgatccg 33660
ccacggcccc caccacccgg tcgcgccctg cccgtgctcc cgctgctcgc gggggtaccg 33720
ggccaggtcg cgggcggaga agagcgccat gccggcgtgg aatccggtca ccggcaccgg 33780
gacccgcgcc cagtaggcga gccggccgtc gacgtggaac tccacctccg acgtcggcgc 33840
ccggtaggtg atggcgtatc cgtgcgcccg gcccggctcc gtcggcacgt ccaggaccac 33900
ccggtggatg tagtgctcgt gcggctgggt cacgccgggc agcaccaggc gctcgaccgt 33960
cgcgtacacg gtgtcgttcg tggcggcggc gttgaacacg acgccggtct ccaggtcgaa 34020
caggttcacc gtgccgaacg cgtccagcag gtcgtgcggg atctgccggt acgtccgcac 34080
gcccatctcc acctcgacgg tcagcgagcc ctccgccggc acggcgaagc gccgcaccga 34140
ccggtacatc tgcttggcgt tgttctgccg gggatcggtg tcgtggaagc gggtgaacgg 34200
gtcgacggtc agctccagcc gcccgtcgcc ggtgcggacc tgggcgttgc ggtcctggta 34260
cctgtgggtc tgcccgtccg cgccggcgat cgacatgatc gcccagcggg cggggtccag 34320
ctcgcggctg gtgaagtcgt cgtacgtcca cgcgctggtt ctcagtgccg acgtcatgca 34380
gtcaccatcg gacgccggcc gggcgcgggc atcacccgtt cacgcggttc ggccggaccc 34440
ggcacgccaa tgcgccggcc acgccccgga aatcccgtga ttaagccatg ccggagcgtg 34500
aacggtcgcc gagactgacg ccgcacccat ctccgcatcg tctgcgacgt tctcaccagg 34560
gggagagagc aatggacacg gcagctccgg caacggacgg cggtcgctac ctcgccgtcc 34620
atcacagcgc agagttcagg gaactacggc gacgatcgag cacgttcacg ctctgggcca 34680
gcgtcgcctt cttcggctgg tggttcctcg gcagcctgct cgccacctac gcgccggact 34740
tcttccggga gaaggtggcc ggcccggtca acgtgggtct gctcttcgtc ttcctgtcgt 34800
tcgccttcgt ggtgacgctc gccgccttct acctgcgtta cgcccgcacg catctcgatc 34860
cgctcagcga gaagatccgt gccgacctgg aaggagcgtc ccgatgagcg tcatcctcgc 34920
cgacccgcca cccccggtcg acaacacgtg ggcgacgccc gcgatcgccg tgccggtcac 34980
catcgtcctc gcgctcgcgg tgctctacct ggtccggtcg gcgcgcgcca gcaccaccac 35040
cgcggacggc ttcctgctgg ccgaccggcg gatcgggccg gtgcagaacg cgctggcggt 35100
ggcctccgcg ccgctgatgt actcgacgat gtacatcatc accggccaca tcgcgctcag 35160
cggctacgac gccatcctgc tgatgaccgc cttcaccatg ggcaccatgc tcgcgctgtt 35220
cctcttcgcc gggccggtgc gcaacgtggg cggctar_acg ctcggtgacc tgctcgcggt 35280
ccgtacccgg gagcggccgg cgcggatcgc gtcggcggtg ctcacgctgc tgacgtacgt 35340
catgctgacg gtgatcatga tggccgccat cgcgttcatc ttcaaccgct ggttcggcgt 35400
CA 02538147 2004-O1-21
cgacgccctc gtcggcctgg tcctcccggt gttcgtcgtc ggtctgatca cggtggggta 35460
cgtgtacctc ggcgggatgc tcggggtcac ccgcatcctg gtgttcaagc tggtgctgtc 3552()
ggtggtcgtc gtgggcgtgc tgaccgcctg ggtgctggcc cgcttcgacc tgaacctctt 35580
cagcctgctg gagcgggccg aggcgaacgc ggcgccggtg cccagcggca gcgacctgct 35640
gggcccgggc cggctgttcg gcgagggcgc gaccacgctc gtgcacctgt cgaagctgtt 35700
cgccatcgcc gtcggagtgg cggccattcc gttcctgttc atgcgcaact tcgcggtgac 3576()
cagcgggcgg gacgcgcgcc ggtcgaccgg gtgggcgtcg atgatcatcg tcgggttcta 35820
cctgtgcctg tccgtcgtcg ggctcggtgc cgtcgcgatc ctcggccggg acaacatcgg 35880
cgtcatcaag gcccaccgcg acatcagctt ccccaagctc gccgacgagc tcggcggtcc 35940
ggtgatggtc ggctccctgg ccggcgtcgc ggtcctgacg atcgtcggcg tcttcgcgcc 3600()
gctgctgcac agcgccgtga cgacggtgac caaggacctg aacgtgatcc gcggccggcg 3606()
gctggatccg gccgccgagc tgcgggacat caagcgcaac accctgatca tcggcgtcgg 36120
ctccgtgctg ctggcggtcg tgatgctgcc ggtacggacc cacatcttca tcccgacctc 36180
gatcgacatt gccggcgcgg tggtcctgcc gatcgtc:gtc tacgcgttgt tctggcggcg 3624()
tttcaacacc cgcggactgc agtggacggt ctacggcggc ctcgcgctca ccgcgttcct 36300
ggtgctgttc tccaacggtg tctcgggcga gccggacgcc atcttcccgg accgcaactt 36360
caagttcgtg gacgtcgagc ccgcgctgat cacggtgccg gtcggcttcc tgctcggcta 36420
cctcggctcg atcaccagcc gggagcgcga cgacgccgcg ttcgccgaga tgcaggtccg 36480
gtccctcacc ggagctgtcg tcacgggacc gccgcggccg gccgccgtgg acgacgagga 36540
ccgcgacggc cgccaggacc gggcgcccag cccggtgagc tgaacatccg caacggtgtg 36600
gg 36602
SEQ ID NO: 2
LENGTH: 571
TYPE: PRT
ORGANISM: Micromonospora sp. strain 046-ECO11
SEQUENCE: 2
Val His Asn Leu Asp Asn Ile Pro Ser Ser Pro Ser Thr Ser Gly Gly
1 5 10 15
Ser Leu Pro Ala Gly His Arg Ala His Val Arg Ala Asp Gly Val Arg
2C 25 30
Val Val Arg GIy Gly Arg Val Val Leu Ser Asp Val Ser Val Thr Val
35 40 45
Ser Ala Ala Ser Arg Leu Ala Val Val Gly Glu Asn Gly Arg Gly Lys
50 55 60
21
CA 02538147 2004-O1-21
Thr Thr Leu Leu His Val Leu Ala Gly Leu Ile Ala Pro Asp Gln Gly
65 70 75 80
Val Val Glu Arg Leu Gly Thr Ile Gly Val Ala Arg Gln Asn Leu Glu
85 90 95
Ser Arg His Gly Glu Thr Val Gly Thr Leu Val Arg Glu Ala Ile Arg
100 105 110
Glu Ser Glu Arg Ala Leu Arg Ala Leu Asp Glu Ala Thr Ile Ala Leu
115 120 125
Thr Glu Gly Arg Ala Gly Ala Asp Asp Ala Tyr Ala Ala Ala Leu Asp
130 135 140
Ala Ala Thr Arg Leu Asp Ala Trp Asp Ala Gln Arg Arg Val Asp Val
145 150 155 160
Ala Leu Ala Gly Leu Asp Ala Cys Pro Asp Arg Asp Arg Gln Leu Ala
165 170 175
Thr Leu Ser Val Gly Gln Arg Tyr Arg Val Arg Leu Ala Cys Leu Leu
180 185 190
Gly Ala Arg Val Asp Leu Leu Met Leu Asp Glu Pro Thr Asn His Leu
195 200 205
Asp Ala Asp Ser Leu Ala Phe Leu Thr Ala Arg Leu Arg Asp His Pro
210 215 220
Gly Gly VaI Val Leu VaI Thr His Asp Arg Ala Leu Leu Arg Asp Val
225 23C 235 240
Ala Thr Glu Phe Leu Asp Leu Asp Pro Ser Ala Asp Gly Arg Pro Arg
245 250 255
Arg Tyr Ala Gly Asp Tyr Val Ala Trp Gln Asp Gly Arg Arg Arg Asp
260 265 270
Phe Ala His Trp Val Arg Asp His Glu Ala Gln Gln Ala Glu His Gln
275 280 285
Arg Leu Ala Asp Gly Val Arg Glu Ala Arg Asp Arg Leu Ser Thr Gly
290 295 300
Trp Arg Prc Glu Lys Gly His Gly Lys His Gln Arg Gln Ser Arg Ala
305 310 315 320
Pro Gly Leu Val Gln A1a Leu Arg Arg Arg Gln Glu Ala Leu Asp Ala
325 330 335
His Arg Val Thr Val Pro Glu Pro Pro Gln Pro Leu Arg Trp Pro Pro
340 345 350
Leu Asp Thr Arg Ala Gly Leu Pro Ile Leu Arg Cys His Asp Val Thr
355 360 365
Val Ala Gly Arg Leu Arg Thr Arg Val Thr Leu Thr Leu Asp Gly Gly
370 375 380
22
CA 02538147 2004-O1-21
Asp Arg Leu Leu Val Thr Gly Pro Asn Gly Ala Gly Lys Ser Thr Leu
385 390 395 400
Leu Ser Val Leu Ala Gly Asp Leu Thr Pro Ser Thr Gly Glu Val Arg
405 410 415
His Leu Ser Gly Ala Arg Val Ala Tyr Leu Gly Gln Glu Val Pro Asp
420 425 430
Trp Pro Pro Ala Leu Leu Ala His Asp Leu Tyr Glu Gln His Val Gly
435 440 445
Arg Leu Arg Ser Ser Gly Arg Val Gly Ser Gly Thr Ala Leu Pro Leu
450 455 460
Ser Ala Thr Asn Leu Leu Asp Ala Glu Ala Arg Arg Thr Pro Val Gly
465 470 475 480
Arg Met Ser His Gly Gln Gln Arg Arg Leu Asn Leu Ala Leu Arg Leu
485 490 495
Ala Glu Arg Pro Asp Leu Leu Ile Leu Asp Glu Pro Thr Asn His Leu
500 505 510
Ser Ala Fro Leu Val Asp Asp Leu Thr Ala Ala Leu Leu Thr Thr Arg
515 520 525
Ala Ala Val Val Val Ala Thr His Asp Arg Gln Met Leu Gln Asp Leu
530 535 540
AIa Ala Trp Pro Thr Leu Pro Leu Thr Ala Pro Ala Ala Ser Gly Arg
545 550 555 560
Ser Val Thr Ser Glu Arg Tyr Asp Trp Glu Ser
565 570
SEQ ID 3
N0:
LENGTH: 716
1
TYPE:
DNA
ORGANISM:Micromonospora
sp. strain
046-ECO11
SEQUENCE:3
gtgcacaacctcgacaacattccttcctccccatccacctcgggcggttcgctgcccgcc60
gggcaccgggcgcacgtgcgggccgacggcgtccgcgtcgtacgcggcggccgggtcgtg120
ctgtccgacgtcagcgtgaccgtctccgccgcttcccgcctcgcagtcgtcggcgagaac180
ggccgcggcaagaccaccctgctgcacgtgctggccggcctcatcgcgcccgaccagggc240
gtggtggaacggctgggcacgatcggcgtcgcccggcagaacctggagtcgcgccacggc300
gagacagtgggcacgctcgtccgggaggcgatccgggagtccgaacgcgcgctgcgggcg360
ctcgacgaggcgacgatcgcgctcaccgagggccgggcgggcgcggacgacgcgtacgcg420
gccgcgctcgacgcggcgacccggctggacgcctgggacgcgcagcggcgcgtcgacgtg480
gcgctggccggcctcgacgcgtgcccggaccgggaccggcagctggccacgttgtccgtc540
ggccagcgctaccgggtacggctggcgtgcctgctgggagcgagggtcgacctgctgatg600
23
CA 02538147 2004-O1-21
ctggacgagccgacgaaccacctcgacgccgacagcctggccttcctcaccgcccggcta 660
cgcgaccacccgggcggcgtcgtgctggtgacccacgaccgcgccctgctgcgggacgtc 720
gccacggagttcctggacctcgaccccagcgcggacgggcgcccgcgccgctacgccggg 780
gactacgtcgcctggcaggacgggcgccgccgcgacttcgcgcactgggtacgcgaccac 840
gaggcgcagcaggccgagcaccagcggctggccgacggggtacgggaggcgcgggaccgg 900
ctcagcaccggctggcggccggagaaggggcacggcaagcaccagcgccagtcccgcgcg 960
cccggactggtccaggcgctgcgccgccggcaggaggcgctcgacgcgcaccgcgtcacc 1020
gtgccggagccaccgcagccgctgcgctggccgccgctggacacccgtgccggactgccc 1080
atcctgcgatgccacgacgtcacggtggccgggcgcctgcgtacccgggtcacgctcacg 1140
ctcgacggcggggaccgcctgctggtgaccggacccaacggcgcgggcaagtcgacgctg 1200
ctctccgtgctggccggcgacctcacgccgtcgaccggggaggtccggcacctgtccggc 1260
gcgcgcgtcgcgtacctcggtcaggaggtgcccgactggccgccggcgctgctcgcgcac 1320
gacctgtacgagcagcacgtgggccggctccgctccagcgggcgcgtcggctccggcacg 1380
gccctgccgctgagcgcgacgaacctgctcgacgccgaggcccggcgtacccccgtcggc 1440
cggatgtcgcacggacagcaacggcggctgaacctggcgctgcgcctggccgaacgtccc 1500
gacctgctgatcctcgacgaaccgacgaaccacctgtcggcgccgctggtcgacgacctc 1560
accgccgccctgctgacgacccgggcggcggtggtcgtcgccacccacgaccggcagatg 1620
ctccaggacctcgcggcctggcccacgctgccgctcacagccccggcggcgtcaggtcgt 1680
tcggtcacttccgagcgatatgactgggagtcataa 171c;
SEQ ID NO: 4
LENGTH: 689
TYPE: PRT
ORGANISM: Micromonospora sp. strain 046-EC011
SEQUENCE: 4
Met Thr Thr Gly Arg Pro Gly Glu Asn Arg Ala Thr Asp Ala Ala Arg
1 5 10 15
Asn Pro Gly Trp Ala Ala Gly Gly Pro Ala Ser Gln Pro Trp Gly Gly
20 25 30
Gly Asn Asp Glu Gln Val Leu Arg Glu I1e Leu Gly Val Asp Val His
35 40 45
Arg Glu Leu Ile Asp Phe Ala Gly Gly Ala Gly Gly Asn Pro His Leu
50 55 60
Val Ala Glu Leu Ala Arg GIy Leu Ala Glu Glu Gly Leu Ile Arg GIu
65 70 75 80
24
CA 02538147 2004-O1-21
Thr Asn Gly Arg Ala Glu Leu Val Ser Arg Arg Ile Pro Arg Arg Val
85 90 95
Leu Ser Phe Val Met Arg Arg Leu Asn Asp Val Ser Ala Gly Cys Gln
100 105 110
Gln Phe Leu Lys Val Ala Ala Ala Leu Gly Arg Ser Phe Met Leu Glu
115 120 125
Asp Val Ser Arg Met Leu Gly Arg Ser Ser Ala Ala Leu Leu Pro Pro
130 135 140
Val Asp Glu Ala IIe Ala Ser Gly Phe Val Val Ala Ala Glu His Gln
145 150 155 160
Leu Ala Phe Gln Ser Asp Phe Leu Leu Arg Gly Ile Ile Glu Ser Ile
165 170 175
Pro Gly Pro Ala Arg Asp Ala Leu Arg Arg Glu Ala Met Ser Leu Ser
180 185 190
GIy Arg Arg Arg Pro Ala Ala Asp Gln Asn Arg Arg Leu Asp Ala Ala
195 200 205
Pro Thr Ala Pro Val Ser Ala Thr Gly Glu Asp Ala Thr Gly Ser Cys
210 215 220
Ser Arg Ala His Arg Leu Ile Met Asn Gly Asn Ala Lys Ala Gly Ile
225 230 235 240
Arg Val Ala Glu Ala Val Leu Ala Gly Pro Ala Ala Ser Leu Ala Ala
245 250 255
Arg Arg Asp Ala Glu Ala Cys Leu Val Leu Ala Asp Leu Leu Leu Gly
260 265 270
G1y Glu Gly Gly GIy Pro Met Thr Glu Ala Ile Leu Arg Glu Arg Asp
275 280 285
Ala Glu Ser Gly Asp Ala Ala Leu Ala Met Ala Leu Thr Ala Arg Ser
290 295 300
Thr Gly Leu Trp Ser Ala Gly Lys Leu Ala Glu Gly Leu Lys Leu Gly
305 310 315 320
Arg AIa Ala Val Arg Ala Gly Ala Glu Ala Glu Pro Val Trp Arg Leu
325 330 335
His AIa Gln Leu Ala Leu Ala Gly Lys Leu Ala Asn Leu Arg Glu Phe
340 345 350
Asp Glu Ala Glu Ala Leu Ile Asn Glu Ala Glu Ala Gly Leu Arg Gly
355 360 365
Leu Pro Ala Pro Ile Trp Thr Ala Ala Thr Ala Val Met Arg Ser Arg
370 375 380
Leu Leu Leu Gln Ala Gly Arg Ile Gly Glu Ala Arg Arg Glu Ala Ala
385 390 395 400
CA 02538147 2004-O1-21
Leu Ala Thr Thr Ala Val Glu Gly Asp Ala Val Pro Met Leu Arg Pro
405 410 415
Leu Ala Tyr Ala Val Leu Ser Thr Ala Ser Phe Tyr Met Gly Asp Leu
420 425 430
Pro Ala Ala Ile Glu Tyr Leu Arg Arg Gly GIn Arg Asp AIa Asp Arg
435 440 445
His Val Val Leu Asp Ser Val Gln Tyr Ser Trp Ala Glu Val Leu Ile
450 455 460
Thr Val Lys Gln Glu Gly Pro Arg Ala Ala Ala Gln Leu Leu Ala Gly
465 470 475 480
Lys His His Arg Leu Pro Thr Gln Arg Arg Leu Tyr Val Glu Val Pro
485 490 495
Ser Ala Ala Ala Phe Leu Val Leu Leu Ala Arg Asp Val Asp Asp Arg
500 505 510
Asp Leu Glu Arg Arg Val Leu Asp Thr Val Asn Gly Leu Ala Ala Asp
515 520 525
Asn Pro Arg Ile Gln Val Val Ser Leu Thr Ala Met His Ala His Ala
530 535 540
Leu Ala Asn Ser Ala Pro Ala Ala Leu Ala Leu Ile Ile Val Gln Ser
545 550 555 560
Arg Asp Pro Ile Ser Val Ala Leu Ala Thr Glu Glu Leu Ala Lys Leu
565 570 575
Tyr Ala Ala Gln Ala Gln Ala Gly Gly Arg Pro Ala Thr Pro Ala Arg
580 585 590
Ala Glu Glu Ala Ala Thr Pro Pro Ala Ser Cys Trp Ser Thr Leu Ser
595 600 605
Asp Met Glu Gln Arg Ile Ala Tyr Leu Val Ser Val Gly Leu Thr Asn
610 615 620
Arg GIn Ile Ala Lys Gln Val His Leu Ser Ala His Thr Val Asn Tyr
625 630 635 640
His Leu Arg Lys Ile Tyr Arg Lys Leu Gly Phe Asn Thr Arg Ala Glu
645 650 655
Leu Ala His Ala Ala Ala Thr Tyr Ser Gly Arg Ala Ala Ile Tyr Ser
660 665 670
Met Ser Giy Asp Gln Asp Trp Gly Ala Gly Ser Met Thr Gly Lys Ala
675 680 685
Ser
SEQ ID N0: 5
LENGTH: 2070
TYPE: DNA
ORGANISM: Micromonospora sp. strain 046-ECO11
2G
CA 02538147 2004-O1-21
SEQUENCE:5
atgacaacgggacggccgggggagaaccgggcgacagacgcggcacgaaatccggggtgg60
gccgccggggggccggcgtcccagccatggggcggggggaacgacgagcaggtcctgcgc120
gagatcctcggggtcgacgtgcaccgcgagctgattgacttcgcgggtggtgccggcgga180
aatccgcacctggtcgccgaactcgcgcgcgggctcgccgaagagggattgattcgggag240
acaaacggtcgggcggaattggtgtcccggcgaattccccggcgcgtgctgagttttgtc300
atgcgtcgattgaatgatgtcagcgccggctgccagcagttcttgaaggttgccgcggca360
ttgggcagatccttcatgctggaggacgtttcgagaatgctgggccgatcgtcggcggcc420
ctgctcccgccggtggacgaggcgatcgcatcgggcttcgtcgtcgccgccgagcatcaa480
ctcgcctttcagagcgacttcctgctgcgcggcatcatcgagtccattcccgggcccgcc540
cgcgacgccttacgacgtgaggcgatgagcctttccgggcgacggcgcccggcggccgac600
cagaatcgccggttggacgcggcgcctaccgcgccggtgagcgcgaccggggaggacgcc660
accggatcctgttcccgggcgcaccgcctgataatgaacgggaacgcgaaggccggcatt720
cgcgtcgccgaggcggttctcgccggcccggccgcgtcgctcgctgcccggcgtgacgcg780
gaggcgtgtctggtgctggccgatctgctgctcggcggggagggcggcggcccgatgacc840
gaggcgatcctgcgcgaacgcgacgccgagtccggtgacgccgcactggcgatggcgctg900
accgcccggtccaccgggctgtggtcggcgggaaagctggcggagggcctgaagctggga960
cgggcggeggtgcgggcgggcgcggaggccgaaccggtgtggcgtctgcacgcccagctc1020
gcgctcgccgggaaactcgcgaacctccgcgagttcgacgaggccgaggcgttgatcaac1080
gaggcggaagCgggCCtgCgcggactgcccgcgccgatctggacggccgcgacggcggtg1140
atgcggtcccggttgctgctccaggcggggcggatcggggaggcgcgtcgggaggcggcg1200
ctggccaccaccgccgtggagggggacgcggtgccgatgctgcggcctctcgcctacgcg1260
gtgctcagcaccgcctccttctacatgggggacctgcccgccgcgatcgagtacctcagg1320
cgggggcagcgggacgcggaccgccacgtggtcctcgactcggtgcagtactcgtgggcg1380
gaagtgctgatcacggtcaagcaggaaggcccgcgggccgccgcccagctgctcgcgggc1440
aagcaccaccgcctgcccacgcagcgccgcctctacgtcgaggtgccgagcgccgccgcc1500
ttcctggtcctgctcgcccgcgacgtggacgaccgtgacctcgaacgccgcgtcctcgac1560
acggtcaacgggctcgccgcggacaaccccaggatccaggtcgtcagcctcaccgccatg1620
cacgcccacgcgctggcgaacagcgctccggccgccctggcgctcatcatcgtgcagtca1680
cgggacccgatctcggtggcgctggccaccgaggaactcgccaagctctacgccgcgcag1740
gcccaggcgg ggggacggcc ggcgacgccg gcccgcgccg aggaggccgc caccccgccg 1800
CA 02538147 2004-O1-21
gcgagctgctggtcgaccctgtccgacatggagcagcgga tcgcctacctggtgagcgtg1860
ggtctgacgaaccggcagatcgccaagcaggtccacctgt ccgcgcacaccgtcaactac1920
cacctgcggaagatctaccggaaactgggtttcaacaccc gggccgagctggcgcacgcc1980
gcggccacgtactccggccgggcggcgatctactccatga gcggcgaccaggactggggc2040
gccggatccatgaccggcaaggccagctga 2070
SEQ ID N0: 6
LENGTH: 895
TYPE: PRT
ORGANISM: Micromonospora sp. strain 046-ECO11
SEQUENCE: 6
Met Vai Ile Met Asn Arg Met Ala Gly Arg Gly Gln Glu Leu Ser Ser
1 5 10 15
Leu Gly Glu Leu Leu Asp Ala Thr Met Arg Gly Ser Gly Gly Cys Val
20 25 30
Val Val Asp Gly Pro Phe Gly Ile Gly Lys Thr His Leu Leu Lys Val
35 40 45
Thr Gly Leu Glu Ala Ala Ala Arg Gly Leu Thr Val Val Ala Gly Arg
50 55 60
Ala Ser Val Thr Asp Gln Pro Val Pro Val His Leu Leu Val Asn Phe
65 70 75 80
Leu Arg His Ala Met Pro Gly Glu Ala Ala Val Glu Gln Leu Ala Leu
85 90 95
Pro Gly AIa Asn Pro Phe Trp Leu Ile Asp Arg Val Gly Asp Leu Val
100 105 110
Glu Val Ala Ala Arg Arg Arg Pro Leu Val Val Ala Leu Asp Asp Ala
115 120 125
Gin Arg Ile Asp Asp Val Ser Ala Leu Ala Leu Arg Gly Leu Val Pro
130 135 140
Arg Leu Ala Ser Ser Pro Val Leu Trp Leu Leu Ala Arg Arg Pro Val
145 150 155 160
Ala Ala Gly Ser Ile Ala Gln His Ala Val Asp Trp Leu Ala Glu His
165 170 175
Val Ala Va1 Arg Val Arg Leu Arg Glu Pro Gly Glu Glu Ala Val Ala
180 185 190
Asp Leu Cys Ala Gly Ile Leu Gly Ala Arg Pro Asp Ala Ser Val Leu
195 200 205
Arg Trp Ala Ala Arg Cys Gly Gly Asn Pro Lys Val Met Glu Ile Val
210 215 220
28
CA 02538147 2004-O1-21
Phe Ser Ala Phe Ile Lys Ala Gly Gln Met Ile Ile Val Asp Gly Ala
225 230 235 240
Ala Ser Val Val Ser Asp Glu Leu Pro Asp Gly Val Leu Ala Ala Val
245 250 255
Arg Gly Leu Leu Glu Glu Leu Pro Pro Pro Leu Arg Arg Leu Leu Ala
260 265 270
Ala Gly Gly Arg Leu Gly His Thr Phe Pro Val Asp Arg Val Thr Gly
275 280 285
Leu Leu Asp Gly Ser Ala Ala Asp Val Ser Ala Ala Ile Asp Glu Ala
290 295 300
Val Arg Val Gly Leu Ile Arg Arg Asp Gly Ala Glu Leu Thr Phe Ala
305 310 315 320
His Pro Val Leu Gly Glu Ala Leu Arg His Ala Ala Tyr Pro Glu Pro
325 330 335
Glu Arg Ala Glu Pro Gly Ser Ala Pro Ala Pro Ala Ala Gly Asp Pro
340 345 350
Val Arg Arg Gly Arg Pro Asp Pro Arg Pro Gly Thr Pro His Ser Pro
355 360 365
Ala Gly Val Arg Val Thr Arg Ser Ala Pro Asp Ala Ala Thr Pro Ala
370 375 380
Ala Thr Ala Gly Pro Arg Ser Gly Arg Cys Gly Cys Asp Asp Val Ala
385 390 395 400
Ala Ala Ala Val Ser His Leu Glu Asn Gly Ser Ala Glu Ala Pro Arg
405 410 415
Ala Leu Aia Arg Ala Leu Arg Leu Leu Ala Gly Ala Gly Arg Ala Ala
420 425 430
Glu Ala Gly Arg Leu Ala Glu Val Met Leu Arg Arg Asp Leu Ala Ala
435 440 445
Asp Val Glu Ala Gln Leu Val Leu Glu Leu Gly His Gly Met Arg Ala
450 455 460
Ala Gly Ser His Arg Leu Ala Ala Gly Phe Leu Arg Arg Thr Gln Ala
465 470 475 480
Arg His Asp Val Cys Glu Leu Asp Arg Ala Lys Leu Asp Arg Ala Leu
485 490 495
Ala Asp Thr Thr Lys His Leu Gly Gly Ala Ser Ser Ala Glu Leu Glu
500 505 510
Pro Arg His Gln Ser Pro Gly Cys Ala Pro Gly Arg Arg Pro Leu Trp
515 520 525
Thr Trp Leu Val Arg Ala Leu Gly Ala Ala Asp Gln Leu Asp Glu Ala
530 535 540
Glra Ala Val Leu Asp Thr Val Arg Pro Leu Ala Gln Glu Pro Ser His
CA 02538147 2004-O1-21
545 550 555 560
Thr Gly Ser Glu Ser Leu Trp Arg Gly His Arg Ala Glu Leu Leu Ala
565 570 575
Ala Ala Gly Arg Leu Asp Glu Ala Arg Ala Glu Ala Glu Ala Ala Leu
580 585 590
Arg AIa Ala Asp His Ser Arg Pro Gly Asp Cys Val Pro Ala Arg Leu
595 600 605
Val Leu Ala His Leu Gly Val His His Gly Asp Leu Ala Thr Ala Ser
610 615 620
Asp Gln Leu Arg Ala Ala Glu Arg Leu Ala Ser Ala Asp Asp Ser Ala
625 630 635 640
Arg Met Asp Trp Ala Leu Ala Arg Phe His Ala Ala Ser Gly Arg Pro
645 650 655
Ala Met Met Val Gln Thr Leu Ile Asn Val Ala Gly Gln Val Ala Pro
660 665 670
Asp Pro Leu Leu Phe Thr Glu Ala Pro Ala Ala Ala Ala Thr Leu Val
675 680 685
Arg Gln Ala Arg Arg Ala Gly Leu Asp Ala Glu Ala Glu Arg Ala Val
690 695 700
Glu Val Ala Arg Arg Val Ala Arg Gly Asn Pro Phe Val Gln Ser Leu
705 710 715 720
Ala Ala Ala Ala Glu His Ala Ala Gly Leu Leu Arg Asp Asp Pro Ala
725 730 735
Ala Leu Leu Arg Ala Ala Asp Leu His Arg Leu Ala Gly Arg Thr Leu
740 745 750
Ala Ala Ala Gly Ala Val Glu Asp Ala Ala Arg Ser Thr Arg Asp Arg
755 760 765
Ala Glu Ala Thr Arg Leu Leu Glu Ala Ala Thr Asp Gly Tyr Arg Glu
770 775 780
Cys Gly Ala Arg Arg Asp Leu Glu Arg Val Glu Ala Glu Leu Arg Gly
785 790 795 800
Leu Pro Ala His Asn Val Arg Pro Leu Val Pro Asp Arg Pro Arg Ser
805 810 815
Gly Trp Glu Ser Leu Thr Ser Ala Glu Leu Arg Val Val Arg Ala Ile
820 825 830
Val Asp Gly Met Thr Asn Arg Glu Ala Ala Ser Ser~Leu Phe Leu Ser
835 840 845
Pro His Thr Val Asp Ser His Leu Arg Arg Val Phe Ser Lys Leu Asp
850 855 860
Ile Asn Ser Arg Val Glu Leu Thr Arg Cys Phe Ile Ala His Glu Ala
865 870 875 880
CA 02538147 2004-O1-21
Val Arg Pro Ala Leu Ala Thr Thr Arg Gln Pro Ala Ser Ala Gly
885 890 895
SEQ ID NO: 7
LENGTH: 2688
TYPE: DNA
ORGANISM: Micromonospora sp. strain 046-EC011
SEQUENCE:7
atggtcatcatgaatcgcatggcggggcgcgggcaggaattgtcctcattgggggaactg60
ctcgacgccaccatgcggggatccgggggctgcgtcgtcgtcgacgggccgttcggcatc120
ggcaagacccacctgctgaaggtcaccggcctggaggcggcggcccgcgggctgacagtg180
gtggccgggcgggcaagcgtcacggatcagccggtgcccgtacacctgctcgtcaacttc240
ctgcgccacgcgatgcccggcgaagcggcggtcgagcagctcgccctgccgggcgccaac300
ccgttctggctgatcgaccgggtcggcgatctggtcgaggtcgcggcgcgccggcgcccg360
ctcgtggtcgccctggacgacgcccagcgcatcgacgacgtcagcgccctggccctgcgc420
gggctcgtgccgcgcctggcgtcctcgccggtgctctggctgctggcccgccggccggtc480
gccgccgggtcgatcgctcagcacgccgtcgactggctggccgagcacgtcgcggtacgg540
gtacggctgcgcgagccgggcgaggaggcggtggccgacctgtgcgccggcatcctcggc600
gcccggccggacgcctccgtcctgcgctgggcggcccgctgcggcggcaacccgaaggtg660
atggagatcgtcttcagcgcgttcatcaaggccggccagatgatcatcgtggacggggcg720
gcgtcggtggtgtccgacgagctgcccgacggtgtcctcgccgccgttcgcgggctgctg780
gaggagctgccgcccccgctgcggcgcctgctcgcggccggcggccggctcggccacacg840
tttcccgtcgaccgggtgacgggcctgctggacggctcggccgccgacgtgtccgccgcg900
atcgacgaggcggtgcgggtcgggctgatacgacgcgacggcgcggagctgaccttcgcc960
cacccggtgctcggagaggcgcttcgccacgccgcgtacccggaaccggagcgtgccgag1020
cccggatccgcgccggcaccggcggcgggcgacccggtccggcgcgggcggcccgatccg1080
cggcccgggacgccccactcccccgccggcgtacgcgtcacgcgctccgcgccggacgcg1140
gccacgcccgccgcgacggcggggccgcgctcgggccggtgcgggtgcgacgacgtggcg1200
gcagccgccgtgtcccacctggagaacggatccgccgaggcgccacgagcactggcccgt1260
gcgctgcgcctgctggccggggcggggcgggccgccgaggccggccgcctcgcggaggtg1320
atgctccgccgcgacctcgcggcggacgtcgaggcgcagctcgtgctcgaactgggacac1380
gggatgcgggccgccggcagccaccgcctggcggccggcttcctgcgccggacgcaggcc1440
cgccacgacgtgtgcgagctggaccgcgccaagctggaccgggcgctcgcggacaccacg1500
aagcacctgggcggtgcctcctccgccgagctggagccccggcaccagtccccgggctgc1560
31
CA 02538147 2004-O1-21
gcgcccggccggcggccgctgtggacctggctggtccgggcgctgggcgcggccgatcag 1620
ctcgacgaggcgcaggcggtgctggacaccgtacgaccgctggcgcaggagcccagtcac 1680
accggctcggagtcgctctggcgcggccaccgggccgagctgctggcagcggccggacgg 1740
~ctggacgaggcacgcgccgaggcggaggcggcgctgcgagccgccgaccactcccggccg 1800
ggcgactgcgtgccggcgcgcctggtcctggcccacctcggcgtgcaccacggtgacctc 1860
gccacggccagcgaccagttgcgggcggccgagcggctggcctccgccgacgactcggcg 1920
cggatggactgggcgctggcccggttccacgctgccagcggccgtccggcgatgatggtg 1980
cagacgctgatcaacgtcgccggacaggtcgcacccgatccgctgctgttcaccgaggcg 2040
ccggccgctgcggcgacgctcgtacgccaggcccgccgggcggggctcgacgcggaggcc 2100
gagcgcgccgtggaggtcgcccggcgcgtcgcccgcggcaacccgttcgtccagtcgctg 2160
gcggcggcggcggaacacgccgcgggtctcctgcgcgacgatccggcggcgctgctgcgg 2220
gccgcggatctgcaccggctcgccggccgtacgctcgcggcggccggcgcggtggaggac 2280
gcggcccgcagcacccgggaccgggccgaggccacccgtctgctcgaggccgcgacggac 2340
ggctaccgggagtgcggcgcgcgacgcgacctggagcgcgtggaggccgagctgcgtggc 2400
ctgccggctcacaacgtccgcccgctggtccccgaccggccccggtcggggtgggagagc 2460
ctgaccagcgcggagctgcgggtcgtgcgggccatcgtggacgggatgaccaaccgcgag 2520
gcggcgagttcgctgttcctgtccccgcacaccgtcgacagtcacctgcggcgcgtcttc 2580
tccaagctcgacatcaacagccgggtggaactgacccgctgcttcatcgcgcacgaggcg 2640
gtccggccggcgctggccaccacacgccagccggcgtccgccggctga 2688
SEQ ID NO: 8
LENGTH: 362
TYPE: PRT
ORGANISM: Micromonospora sp. strain 046-EC011
SEQUENCE: 8
Met Thr Val Gly Tyr Leu Gly Thr Val Thr Asp Ser Ala Pro Val Asp
1 5 10 15
Ala Ala Leu Arg Asp Phe Phe Ala Glu Arg Arg Ala Glu Ala Arg Glu
20 25 30
Lei. Gly Asp Asp Phe Ala AIa Leu Va1 Ala Glu Leu Glu Ser Tyr Val
35 40 45
Leu Arg Gli~ Gly Lys Arg Ile Arg Pro Ala Phe Ala Trp Leu Gly Trp
50 55 60
Ile Gly Ala Gly Gly Asp Pro Glu Asp Pro Val Ala Thr Ala Val Leu
65 70 75 80
32
CA 02538147 2004-O1-21
Asn AIa Cys Ala Gly Phe Glu Leu Leu His AIa Ser Gly Leu Ile His
85 90 95
Asp Asp Ile Ile Asp Ala Ser Gln Thr Arg Arg Gly His Pro Ala Ala
100 105 110
His Val Ala Tyr Ala Glu Arg His Arg Ala Arg Arg Phe Ser Gly Asp
115 120 125
Pro Gly Thr Phe Gly Thr Gly Thr Ala Ile Leu Ile Gly Asp Leu Val
I30 135 140
Leu IIe Trp Ala Asp Val Leu Val Arg Ala Ser Gly Leu Pro Ala Asp
145 150 155 160
AIa His Val Arg Val Ser Pro Val Trp Ser Ala Val Arg Ser Glu Val
165 170 175
Met Tyr Gly Gln Leu Leu Asp Leu Ile Ser GIn Val Ser Arg Ser Glu
180 185 190
Asp Val Asp Ala Ala Leu Arg Ile Asn Gln Tyr Lys Thr Ala Ser Tyr
195 200 205
Thr Val Glu Arg Pro Leu Gln Phe Gly Ala Ala Ile Ala Gly Ala Asp
210 215 220
Asp Asp Leu Phe Ala Ala Tyr Arg Ala Phe Gly Ala Asp Val Gly Ile
225 230 235 240
Ala Phe Gln Leu Arg Asp Asp Leu Leu Gly Val Phe Gly Asp Pro Val
245 250 255
Val Thr Gly Lys Pro Ser Gly Asp Asp Leu Arg Glu Gly Lys Arg Thr
260 265 270
Val Leu Leu Ala Thr Ala Leu Lys Arg Ala Asp Glu Arg Asp Pro Asp
275 280 285
Ala Ala Ala Tyr Leu Arg Ala Lys Val Gly Thr Asp Leu Ala Asp Glu
290 295 300
Glu Ile Ala Arg Ile Arg Ala Ile Phe Arg Asp Val Gly Ala Val Glu
305 310 315 320
Glu Ile Glu Arg Gln Ile Ser Gln Arg Thr Asp Arg Ala Leu Ala Ala
325 330 335
Leu Glu Ala Ser Ser Ala Thr Ala Pro Ala Lys His Gln Leu Ala Asp
340 345 350
Met Ala Ile Lys Ala Thr Gln Arg Ala Gln
355 360
SEQ ID N0: 9
LENGTH: 1089
TYPE: DNA
ORGANISM: Micromonospora sp. strain 046-ECO11
SEQUENCE: 9
3s
CA 02538147 2004-O1-21
atgaccgtcggatatctcgggacggtcaccgactcggcgcccgtcgacgccgcgctgcgc60
gacttcttcgccgagcgccgcgccgaggcacgcgagctcggcgacgacttcgcggccctg120
gtcgccgagctggagagctacgtcctgcggggcggcaagcgcatccggcccgccttcgcc180
tggctgggctggatcggcgccggcggcgacccggaggacccggtggcgaccgcggtgctg240
aacgcctgcgccgggttcgagctgctgcacgcgtccggcctcatccacgacgacatcatc300
gacgcgtcgcagacccgccgcggccatcccgccgcgcacgtcgcgtacgccgaacggcat360
cgggcgcggcgcttctccggtgacccgggaacgttcggcaccggcaccgccatcctgatc420
ggagacctcgtcctgatctgggccgacgtcctggtccgcgcctccggcctgccggccgac480
gcgcacgtgcgggtctcgccggtgtggtcggcggtgcgctccgaggtcatgtacggccag540
ctgctcgatctgatcagccaggtgagccggagcgaggacgtcgacgcggcgctgcgcatc600
aaccagtacaagaccgcgtcgtacacggtggagcggccactgcagttcggcgcggcgatc660
gccggcgcggacgacgacctcttcgcggcctaccgcgccttcggcgccgacgtgggtatt720
gccttccagctgcgcgacgacctgctcggcgtgttcggcgacccggtggtgacgggcaag780
ccgtccggcgacgacctgcgggagggcaagcggacggtcctgctcgccacggcgctcaag840
cgcgccgacgaacgggacccggacgcggcggcctacctgcgggcgaaggtcggcacggac900
ctcgcggacgaggagatcgcccgcatccgcgccatcttccgcgacgtcggcgcggtcgag960
gagatcgagcggcagatctcgcagcgcaccgaccgggcgctggccgcgctggaggcgagc1020
agcgccaccgcccccgcgaagcatcagctcgccgacatggcgatcaaggccacccagcgg1080
gcccagtga 1089
SEQ ID NO: 10
LENGTH: 354
TYPE: PRT
ORGANISM: Micromonospora sp. strain 046-ECO11
SEQUENCE: 10
Met Ser Thr Glu Pro Val Thr Val Val Ala Arg Gly Val Leu Asp Gly
1 5 10 15
Arg Gly Asp Gly Pro Gly Arg Leu Gly Thr Gly Arg Ala His Gly Lys
20 25 30
Ala Ile Leu Leu Gly Glu His Ala Val Val Tyr Gly Ala Pro Ala Leu
35 40 45
Ala Val Pro Val Pro Gln Leu Thr Ala Val Ala Lys Ala Arg Arg Ala
50 55 60
Gly Gly Asp Gly Gly Asp Glu Val Ser Phe Ala Ile Ala Gly Leu Glu
65 70 75 80
34
CA 02538147 2004-O1-21
Ser Pro Glu Val Thr Ser Leu Pro Thr Asp Gly Leu Gln His Leu Val
85 90 95
Thr Glu Phe Arg Gln Arg Ala Ala Val Thr Glu Pro Met Arg Val Asp
100 105 110
Val Leu Val Asp Cys Ala Ile Pro Gln Gly Arg Gly Leu Gly Ser Ser
115 120 125
Ala Ala Cys Ala Arg Ala Ala Val Leu Ala Leu Ala Asp Ala Phe Asp
130 135 140
Arg Arg Leu Asp Ala Ala Thr Val Phe Asp Leu Val Gln Thr Ser Glu
145 150 155 160
Asn Val Ala His Gly Arg Ala Ser Gly Ile Asp Ala Leu Ala Thr Gly
165 170 175
Ala Thr Ala Pro Leu Ile Phe Arg Asn Gly Val Gly Arg Glu Leu Pro
180 185 190
Val Ala Met Ala Gly Ala Ala Arg Ala Ala Arg Gly Ser Gly Pro Ala
195 200 205
Gly Phe Asp Ala Val Leu Val Ile Ala Asp Ser Gly Val Ser Gly Ser
210 215 220
Thr Arg Asp Ala Val Glu Leu Leu Arg Gly Ala Phe Glu Arg Ser Pro
225 230 23S 240
Arg Thr Arg Asp Glu Phe Val Ser Arg Val Thr Ser Leu Thr Glu Ala
245 250 255
Ala Ala His Asp Leu Leu Gln Gly Arg Val Ala Asp Phe Gly Ala Arg
260 265 270
Leu Thr Glu Asn His Arg Leu Leu Arg Glu Val Gly Ile Ser Thr Glu
275 280 285
Arg Ile Asp Arg Met Val Asp Ala Ala Leu Ala Ala Gly Ser Pro Gly
290 295 300
Ala Lys Ile Ser Gly Gly Gly Leu Gly Gly Cys Met Ile Ala Leu Ala
305 310 315 320
Arg Asp Arg Gln Glu Ser Ala Ala val Val Arg Ser Val Gln Gln Ala
325 330 335
Gly Ala Val Arg Thr Trp Thr Val Pro Met Gly Arg Phe Thr Gly His
340 345 350
Asp Asp
SEQ ID NO: 11
LENGTH: 1065
'TYPE : DNA
ORGANISM: Micromonospora sp. strain 046-ECO11
SEQUENCE: 11
atgtccacgg aaccggtgac cgtcgtcgcc cgcggcgttc tcgacggccg gggtgacggg 60
CA 02538147 2004-O1-21
ccgggccgcctcggcaccggccgcgcccacggcaaggccatcctgctgggcgaacacgcc120
gtcgtgtacggcgctccggcgctcgccgtcccggtgccgcaactgaccgccgtggccaag180
gcgcggcgggccggcggcgacggcggcgacgaggtctccttcgccatcgccgggctggag240
agcccggaggtgacgtcgcttccgaccgacggcctgcaacatctggtgacggagttccgg300
cagcgggccgccgtcaccgagccgatgcgcgtcgacgtgctcgtggactgcgccatcccg360
cagggccgggggctcgggtcgagcgccgcctgcgcccgcgccgcggtgctggccctcgcg420
gacgcgttcgaccgccgcctcgacgccgccacggtgttcgatctggtgcagacctcggag480
aacgtggcgcacggccgggccagcggcatcgacgccctggccaccggtgcgaccgcgccg540
ctgatcttccgcaacggcgtgggccgggaactgccggtcgccatggcgggcgccgcgcgt600
gccgcgcgagggtcgggcccggccggcttcgacgcggtgctcgtcatogccgacagcggc660
gtcagcggcagcacccgggacgcggtggagctgctgcggggtgccttcgagcgctccccg720
cgcacgcgcgacgagttcgtcagccgggtgaccagcctgaccgaggcggcggcgcacgac780
ctgctccagggccgggtcgccgacttcggcgcgcggctgaccgagaaccaccggctgttg840
cgcgaggtcggcatcagcaccgaacggatcgaccggatggtcgacgccgcgctcgcggcg900
ggcagc~~cgggcgccaagatcagcggcggtggcctgggcggctgcatgatcgcactggcc960
cgggaccgccaggaatccgcggcggtggtgcggagcgtccagcaggccggcgccgtccgc1020
acctggaccgtcccgatggggaggttcaccggccatgacgactga 1065
SEQ ID N0: 12
LENGTH: 346
TYPE: PRT
ORGANISM: Micromonospora sp. strain 046-ECO11
SEQUENCE: 12
Met Thr Thr Asp His Arg Ala Glu Pro Ser Glu Pro Ala Leu Asp Arg
1 5 10 15
Pro Ala Thr Ala Val Ala His Pro Asn Ile Ala Leu Ile Lys Tyr Trp
20 25 30
Gly Lys Arg Asp Glu Gln Leu Met Ile Pro Tyr Ala Asp Ser Leu Ser
35 40 45
Met Thr Leu Asp Val Phe Pro Thr Thr Thr Thr Val Arg Ile Asp Ser
50 55 60
Gly Ala Ala Ala Asp Glu Val Val Leu Asp Gly Ser Pro Ala Asp Gly
65 70 75 g0
Glu Arg Arg Gln Arg Val Val Thr Phe Leu Asp Leu Val Arg Lys Leu
85 90 95
36
CA 02538147 2004-O1-21
Aia Gly Arg Thr Glu Arg Ala Cys Val Asp Thr Arg Asn Ser Val Pro
100 105 110
Thr Gly Ala Gly Leu Ala Ser Ser Ala Ser Gly Phe Ala Ala Leu Ala
115 120 125
Leu Ala Gly Ala Ala Ala Tyr Gly Leu Asp Leu Asp Thr Thr Ala Leu
130 135 140
Ser Arg Leu Ala Arg Arg Gly Ser Val Ser Ala Ser Arg Ser Val Phe
145 150 155 160
Gly Gly Phe Ala Met Cys His Ala Gly Pro Gly Ala Gly Thr Ala Ala
165 170 175
Asp Leu Gly Ser Tyr Ala Glu Pro Val Pro Val Ala Pro Leu Asp Val
180 185 190
Ala Leu Val Ile Ala Ile VaI Asp AIa Gly Pro Lys Ala Val Ser Ser
195 200 205
Arg Glu Gly Met Arg Arg Thr Val Arg Thr Ser Pro Leu Tyr Gln Ser
210 215 220
Trp Val. Ala Ser Gly Arg Ala Asp Leu Ala Glu Met Arg Ala Ala Leu
225 230 235 240
Leu Gln Gly Asp Leu Asp Ala Val Gly Glu Ile Ala Glu Arg Asn Ala
245 250 255
Leu Gly Met His Ala Thr Met Leu Ala Ala Arg Pro Ala Val Arg Tyr
260 265 270
Leu Ala Pro Val Thr Val Ala Val Leu Asp Ser Val Leu Arg Leu Arg
275 280 285
Ala Asp Gly Val Ser Ala Tyr Ala Thr Met Asp Ala Gly Pro Asn Val
290 295 300
Lys Val Leu Cys Arg Arg Ala Asp Ala Asp Arg Val Ala Asp Thr Leu
305 310 315 320
Arg Asp Ala Ala Pro Ser Cys Ala Val Val Val Ala Gly Pro Gly Pro
325 330 335
Ala Ala Arg Pro Asp Pro Gly Ser Arg Pro
340 345
SEQ ID N0: 13
LENGTH: 1041
TYPE: DNA
ORGANISM: Micromonospora sp. strain 045-ECO11
SEQUENCE: 13
atgacgactg accaccgggc ggagccgtcc gagccggcgc tcgaccggcc cgcgaccgcc 60
gtggcccatc cgaacatcgc gctgatcaag tactggggca agcgcgacga gcagctgatg 120
atcccgtacg ccgacagcct gtcgatgacg ctcgacgtct tcccgaccac caccaccgtc 180
cggatcgaca gcggcgcggc ggccgacgag gtcgtcctcg acggctcgcc cgccgacggc 240
37
CA 02538147 2004-O1-21
gaacggcgacagcgcgtcgtcaccttcctggacctggtacgcaagctggccgggcgcacg300
gaacgggcctgcgtcgacacccgcaactccgtgcccaccggcgccggcctggcgtcctcg360
gcgagcggattcgccgccctcgccctcgccggcgccgccgcgtacggcctcgacctggac420
accaccgcgctgtcccgcctggcccggcggggatccgtgtcggcctcccggtcggtcttc480
ggcggcttcgcgatgtgccacgcaggccccggcgccgggaccgccgcggacctcggctcc540
tacgccgagccggtgcccgtcgcgcccctcgacgtcgcgctggtgatcgcgatcgtcgac600
gccgggccgaaggcggtgtcgagccgcgaggggatgcggcgaaccgtccggacctccccg660
ctctatcagtcgtgggtcgcctccggccgcgccgacctggccgagatgcgggccgcgctg720
ctccagggagacctggacgcggtcggcgagatcgccgaacgcaacgccctcggcatgcac780
gccaccatgctggccgcccggccggcggtgcgctacctggcgccggtcactgtcgccgtg840
ctcgacagcgtgctgcgcctgcgcgccgacggcgtctccgcctacgccacgatggacgcg900
ggaccgaacgtcaaggtgctctgccgccgcgcggacgccgaccgggtcgccgacaccctg960
cgcgacgccgcgccgagctgcgccgtggtcgtcgccggaccggggccggcggcccggccg1020
gacccgggcagccggccgtga 1041
SEQ ID 14
N0:
LENGTH:
369
TYPE:
PRT
ORGANISM:Micromonospora
sp. strain
046-ECO11
SEQUENCE: 14
Val Thr Gly Pro Gly Ala Val Arg Arg His Ala Pro Gly Lys Leu Phe
1 5 10 15
Val Ala Gly Glu Tyr Ala VaI Leu Glu Pro Gly His Pro Ala Leu Leu
20 25 30
Val Ala Val Asp Arg Gly Val Asp Val Thr Val Ser Gly Ala Asp Ala
35 40 45
His Leu Val Val Asp Ser Asp Leu Cys Pro Glu Gln Ala Cys Leu Arg
50 55 60
Trp Gln Asp Gly Arg Leu Val Gly Ala Gly Asp Gly Gln Pro Ala Pro
65 70 75 80
Asp Ala Leu Gly Ala Val Val Ser Ala Ile Glu Val Val Gly Glu Leu
85 90 95
Leu Thr Gly Arg Gly Leu Arg Pro Leu Pro Met Arg Val Ala Ile Thr
100 105 110
Ser Arg Leu His Arg Asp Gly Thr Lys Phe Gly Leu Gly Ser Ser Gly
115 120 125
38
CA 02538147 2004-O1-21
Ala Val Thr Val Ala Thr Val Thr Ala Val Ala Ala Tyr His Gly Val
130 135 140
Glu Leu Ser Leu Glu Ser Arg Phe Arg Leu Ala Met Leu Ala Thr Val
145 150 155 160
Arg Asp Gly Ala Asp Ala Ser Gly Gly Asp Leu Ala Ala Ser Val Trp
165 170 175
Gly Gly Trp Ile Ala Tyr Gln Ala Pro Asp Arg Ala Ala Val Arg Glu
180 185 190
Met Ala Arg Arg Arg Gly Val Glu Glu Thr Met Arg Ala Pro Trp Pro
195 200 205
Gly Leu Arg Val Arg Arg Leu Pro Pro Pro Arg Gly Leu Ala Leu Glu
210 215 220
Val Gly Trp Thr Gly Glu Pro Ala Ser Ser Ser Ser Leu Thr Gly Arg
225 230 235 240
Leu Ala Ala Ser Arg Trp Arg Gly Ser Pro Ala Arg Trp Ser Phe Thr
245 250 255
Ser Arg Ser Gln Glu Cys Val Arg Thr Ala Ile Asp Ala Leu Glu Arg
260 265 270
Gly Asp Asp Gln Glu Leu Leu His Gln Val Arg Arg Ala Arg His Val
275 280 285
Leu Ala Glu Leu Asp Asp Glu Val Arg Leu Gly Ile Phe Thr Pro Arg
290 295 300
Leu Thr Aia Leu Cys Asp Ala Ala Glu Thr Val Gly Gly Ala Ala Lys
305 310 315 320
Pro Ser Gly Ala Gly Gly Gly Asp Cys Gly Ile Ala Leu Leu Asp Ala
325 330 335
Thr Ala Ala Thr Arg Thr Ala Arg Leu Arg Glu Gln Trp Ala Ala Ala
340 345 350
GIy Val Leu Pro Met Pro Ile Gln Val His Gln Thr Asn Gly Ser Ala
355 360 365
Ar g
SEQ ID NO: 15
LENGTH: 1110
TYPE: DNA
ORGANISM: Micromonospora sp. strain 046-EC011
SEQUENCE: 15
gtgaccggcc cgggcgccgt gcgccgccac gcgccgggca agctgttcgt cgccggtgag 60
tacgcggtgc tggagccggg ccacccggcg ctgctggtgg cggtcgacag gggagtggac 120
gtcaccgtct ccggcgccga cgcccacctc gttgtcgact ccgacctctg cccggagcag 180
gcgtgcctgc ggtggcagga cggccggctc gtcggcgcgg gcgacgggca gccggcgccc 240
39
CA 02538147 2004-O1-21
gacgccctcggcgccgtggtctcggcgatcgaggtggtcggcgaactcctgaccggacga300
gggctgcgcccgctgcccatgcgggtggcgatcaccagccggctgcaccgcgacggcacg360
aagttcggcctcgggtcgagcggggcggtgacagtcgccacggtgaccgcagtggccgcg420
taccacggggtggagctgtcgctcgaatcgcggttccggctggcgatgctggcgacggtg480
cgtgacggcgccgacgcctccggcggtgatctggccgcgagcgtctggggcggctggatc540
gcctaccaggcgcccgaccgcgcggccgtgcgcgagatggcgcggcggcgcggcgtcgag600
gagacgatgcgcgcgccctggccgggcctgcgggtccggcggctgccaccaccgcgtggc660
ctcgcgctggaggtgggctggaccggcgagccggcgagcagcagctcgttgaccgggcgg720
ctggccgcctcccggtggcggggcagcccggcgcggtggagcttcaccagccgtagccag780
gagtgtgtgcgtaccgccatcgacgcgctggagcggggcgacgaccaggaactgctgcac840
caggtccggcgggcccggcacgtgcttgccgagctggacgacgaggtccggctcgggatc900
ttcaccccccggctgacggcgctgtgcgacgccgccgagaccgtcggcggcgcggccaaa960
ccgtccggcgccggtggcggggactgcggcatcgcgttgctggacgccaccgccgcgacg1020
cggaccgcgcggctgcgcgagcagtgggccgccgccggggtgctccccatgccgatccag1080
gtccatcagacgaacgggagcgcgcgatga 1110
SEQ ID N0: 16
LENGTH: 360
TYPE: PRT
ORGANISM: Micromonospora sp. strain 046-EC011
SEQUENCE: 16
MetIleAla Asn Lys AspAspHis ValArgLeu AlaAlaGlu Gln
Arg
1 5 10 15
GlnGlyArg LeuGlyGly HisHisGlu PheAspAsp ValSerPhe Val
20 25 30
HisHisAla LeuAlaGly IleAspArg SerAspVal SerLeuAla Thr
35 40 45
SerPheGly GlyIleAsp TrpProVai ProLeuCys IleAsnAla Met
50 55 60
ThrGlyG1y SerThrLys ThrGlyLeu IIeAsnArg AspLeuAla Ile
65 70 75 80
AlaAlaArg GluThrGly ValProIle AlaThrGly SerMetSer Ala
85 90 95
Tyr Phe Ala Asp Glu Ser Val Ala Glu Ser Phe Ser Val Met Arg Arg
100 105 110
Glu Asn Pro Asp Gly Phe Ile Met Ala Asn Val Asn Ala Thr Ala Ser
CA 02538147 2004-O1-21
115 120 125
Val Glu Arg Ala Arg Arg Ala Val Asp Leu Met Arg Ala Asp Ala Leu
130 135 140
Gln Ile His Leu Asn Thr Ile Gln Glu Thr Val Met Pro Glu Gly Asp
145 150 155 160
Arg Ser Phe Ala Ala Trp Gly Pro Arg Ile Glu Gln Ile Val Ala Gly
165 170 175
Val Gly Vai Pro Val Ile Val Lys Glu Val Gly Phe Gly Leu Ser Arg
180 185 190
Glu Thr Leu Leu Arg Leu Arg Asp Met Gly Val Arg Val Ala Asp Val
195 200 205
Ala Gly Arg Gly Gly Thr Asn Phe Ala Arg Ile Glu Asn Asp Arg Arg
210 215 220
Asp Ala Ala Asp Tyr Ser Phe Leu Asp Gly Trp Gly Gln Ser Thr Pro
225 230 235 240
Aia Cys Leu Leu Asp Ala Gln Gly Val Asp Leu Pro Val Leu Ala Ser
245 250 255
Gly Gly Ile Arg Asn Pro Leu Asp Val Val Arg Gly Leu Ala Leu Gly
260 265 270
Ala Gly Ala Ala Gly Val Ser Gly Leu Phe Leu Arg Thr Leu Leu Asp
275 280 285
Gly Gly° Val Pro Ala Leu Leu Ser Leu Leu Ser Thr Trp Leu Asp Gln
290 295 300
Ile Glu Ala Leu Met Thr Ala Leu Gly Ala Arg Thr Pro Ala Asp Leu
305 310 315 320
Thr Arg Cys Asp Leu Leu Ile Gln Gly Arg Leu Ser Ala Phe Cys Ala
325 330 335
Ala Arg Gly Ile Asp Thr His Arg Leu Ala Thr Arg Ser Gly Ala Thr
340 345 350
His Glu Met Ile Gly Gly Ile Arg
355 360
SEQ ID NO: 17
LENGTH: 1083
TYPE: DNA
ORGANISM: Micromonospora sp. strain 046-ECOll
SEQLENCE: 17
atgatcgcca accgcaagga cgaccacgtc cggctcgccg ccgagcagca gggccggctc 60
ggcggtcacc acgagttcga cgacgtgtcc ttcgtgcacc acgccctggc cggcatcgac 120
cggtccgacg tctcgctggc cacgtcgttc ggcggcatcg actggccggt gccgctgtgc 180
atcaacgcga tgaccggcgg cagcaccaag accggcctga tcaaccggga cctggcgatc 240
41
CA 02538147 2004-O1-21
gcggcccgggagaccggcgtaccgatcgccaccgggtcgatgagcgcctacttcgccgac300
gagtcggtggccgagagtttcagcgtgatgcgccgggagaaccccgacgggttcatcatg360
gccaacgtcaacgccaccgcctccgtcgaacgggcccggcgggctgtcgacctgatgcgg420
gccgacgcgctgcagatccacctgaacaccatccaggagacggtgatgccggagggggac480
cggtcgttcgccgcctgggggccgcggatcgaacagatcgtcgccggcgtcggtgtgccg540
gtgatcgtcaaggaggtcggcttcgggctcagccgcgaaacgctgctgcggctgcgggac600
atgggcgtccgggtggccgacgtcgccggccgcggcggcacgaacttcgcgcgcatcgag660
aacgaccggcgggacgccgccgactactccttcctcgacgggtggggacagtcgacaccc720
gcctgcctgctggacgcccagggcgtggacctgcccgtgctggcctccggcggcatccgc780
aacccgctcgacgtggtccgcgggctggcgctcggcgccggcgcggccggggtgtccgga840
ctgttcctgcgcacgctcctggacggcggcgtgccggcgctgctgtcgctgctgtccacc900
tggctcgaccagatcgaagccctgatgaccgccctgggcgcgcggaccccggccgacctg960
acccgctgcgacctgctgatccagggtcggctgagcgcgttctgcgcggcccggggcatc1020
gacacccaccgcctcgccacccgttccggcgccacccacgagatgatcggaggcattcga1080
tga 108:3
SEQ ID N0: 18
LENGTH: 351
TYPE: PRT
ORGANISM: Micromonospora sp. strain 046-ECO11
SEQUENCE: 18
Met Asn Asp Ala Ile Ala Gly Val Pro Met Lys Trp Val Gly Pro Val
1 5 10 15
Arg Ile Ser Gly Asn Val Ala Gln Ile Glu Thr Glu Val Pro Leu Ala
20 25 30
Thr Tyr Glu Ser Pro Leu Trp Pro Ser Val Gly Arg Gly Ala Lys Ile
35 40 45
Ser Arg Met Val Glu Ala Gly Ile Val Ala Thr Leu Val Asp Glu Arg
50 55 60
Met Thr Arg Ser Val Phe Val Arg Ala Lys Asp Ala Gln Thr Ala Tyr
65 70 75 80
Leu Ala Ser Leu Glu Val Asp Ala Arg Phe Asp Glu Leu Arg Asp Ile
85 90 95
Val Arg Thr Cys Gly Arg Phe Val Glu Leu Ile Gly Phe His His Glu
100 105 110
Ile Thr A:la Asn Leu Leu Phe Leu Arg Phe Ser Phe Thr Thr Gly Asp
115 120 125
42
CA 02538147 2004-O1-21
Ala Ser Gly His Asn Met Ala Thr Leu Ala Ala Asp Ala Leu Leu Lys
130 135 140
His Ile Leu Asp Thr Ile Pro Gly Ile Ser Tyr Gly Ser Ile Ser Gly
145 150 155 160
Asn Tyr Cys Thr Asp Lys Lys Ala Thr Ala Ile Asn Gly Ile Leu Gly
165 170 175
Arg Gly Lys Asn Val Val Thr Glu Leu Val Val Pro Arg Glu Ile Val
180 185 190
His Asp Ser Leu His Thr Thr Ala Ala Ala Ile Ala Gln Leu Asn Val
195 200 205
His Lys Asn Met Ile Gly Thr Leu Leu Ala Gly Gly Ile Arg Ser Ala
210 215 220
Asn AIa His Tyr Ala Asn Met Leu Leu Gly Phe Tyr Leu Ala Thr Gly
225 230 235 240
Gln Asp Ala Ala Asn Ile Val Glu Gly Ser Gln Gly Val Thr Val Ala
245 250 255
Glu Asp Arg Asp Gly Asp Leu Tyr Phe Ser Cys Thr Leu Pro Asn Leu
260 265 270
Ile Val Gly Thr Val Gly Asn Gly Lys Gly Leu Gly Phe Val Glu Glu
275 280 285
Asn Leu Glu Arg Leu Gly Cys Arg Ala Ser Arg Asp Pro Gly Glu Asn
290 295 300
Ala Arg Arg Leu Ala Val Ile Ala Ala Ala Thr Val Leu Cys Gly Glu
305 310 315 320
Leu Ser Leu Leu Ala Ala Gln Thr Asn Pro Gly Glu Leu Met Arg Ala
325 330 335
His VaI Arg Leu Glu Arg Pro Thr Glu Thr Thr Lys IIe Gly Ala
340 345 350
SEQ ID 19
N0:
LENGTH:
1056
TYPE:
DNA
ORGANISM:Micromonospora
sp. strain
046-ECO11
SEQUENCE:19
atgaacgacgcgatcgccggtgtgcccatgaaatgggtaggtcccgtgcggatctcggga60
aacgtggcgcagatcgagacggaggttccgctcgccacgtacgagtcgccgctctggccg120
tccgtcggccggggcgcgaagatctcccggatggtcgaggcgggcatcgtcgccacgctc180
gtcgacgagcgcatgacccgctcggtgttcgtgcgcgccaaggacgcgcagaccgcctac240
ctggcctcgcttgaggtcgacgcgcggttcgacgaactgcgtgacatcgtgcgcacctgc300
ggcaggttcgtcgagctgatcgggttccaccacgagatcaccgcgaacctgctgttcctg360
43
CA 02538147 2004-O1-21
cggttcagtttcaccaccggcgacgcgtccgggcacaacatggcgacgctggccgccgac420
gcgctgctgaagcacatcctggacaccattccgggcatctcgtacggctcgatctcgggc480
aactactgcaccgacaagaaggccaccgcgataaacggcattctcggccggggcaagaac540
gtggtcaccgagctggtcgtgccgcgggagatcgtccacgacagcctgcacacgacggcg600
gcggcgatcgcccagctgaacgtgcacaagaacatgatcggcacgttgctcgccggcggt660
atccgctcggccaacgcccactacgcgaacatgctgctcgggttctacctggccacgggt720
caggacgccgcgaacatcgtcgagggctcccagggcgtgacggtcgccgaggaccgcgac780
ggcgacctctacttctcctgcacgctgcccaacctgatcgtgggcaccgtcggcaacggc840
aaggggctcggcttcgtcgaggagaacctggagcggctcggctgccgcgcctcgcgtgat900
ccgggcgagaacgcccggcggctcgcggtcatcgcggccgcgacggtgctctgcggcgag960
ctgtccctgctcgccgcgcagaccaacccgggcgagctgatgcgggcgcacgtccggctc1020
gaacgcccgaccgagaccacgaagatcggagcctga 1056
SEQ ID N0: 20
LENGTH: 391
TYPE: PRT
ORGANISM: Micromonospora sp. strain 046-EC011
SEQUENCE: 20
Met Ala Glu Arg Pro Ala Val Gly Ile His Asp Leu Ser Ala Ala Thr
1 5 10 15
Ala His His Val Leu Thr His Glu Thr Leu Ala Ala Ser Asn Gly Ala
20 25 30
Asp Vai Ala Lys Tyr His Arg Gly Ile Gly Leu Arg Ala Met Ser Val
35 40 45
Pro Ala Pro Asp Glu Asp Ile Val Thr Met Ala Ala Ala Ala Ala Ala
50 55 60
Pro Val Val Ala Arg His Gly Thr Asp Arg Ile Arg Thr Val Val Phe
65 70 75 80
Ala Thr Glu Ser Ser Val Asp Gln Ala Lys Ala Ala Gly Ile His Val
85 90 95
His Ser Leu Leu Gly Leu Pro Ser Ala Thr Arg Val Val Glu Leu Lys
100 105 110
Gln Ala Cys Tyr GIy Gly Thr Ala Gly Leu Gln Phe Ala Ile Gly Leu
115 120 125
VaI His Arg Asp Pro Ser Gln Gln Val Leu Val Ile Ala Ser Asp Val
130 135 140
Ser Lys Tyr Ala Leu Gly Glu Pro Gly Glu Ala Thr Gln Gly Ala Ala
145 150 155 160
44
CA 02538147 2004-O1-21
Ala Val Ala Met Leu Val Gly Ala Asp Pro Ala Leu Val Arg Val Glu
165 170 175
Asp Pro Ser Gly Met Phe Thr Ala Asp Val Met Asp Phe Trp Arg Pro
180 185 190
Asn Tyr Arg Thr Thr Ala Leu Val Asp Gly His Glu Ser Ile Ser Ala
195 200 205
Tyr Leu Gln Ala Leu Glu Gly Ser Trp Lys Asp Tyr Thr Glu Arg Gly
210 215 220
Gly Arg Thr Leu Asp Glu Phe Gly Ala Phe Cys Tyr His Gln Pro Phe
225 230 235 240
Pro Arg Met Ala Asp Lys Ala His Arg His Leu Leu Asn Tyr Cys Gly
245 250 255
Arg Asp Val Asp Asp Ala Leu Val Ala Gly Ala Ile Gly His Thr Thr
260 265 270
Ala Tyr Asn Ala Glu Ile Gly Asn Ser Tyr Thr Ala Ser Met Tyr Leu
275 280 285
Gly Leu Ala Ala Leu Leu Asp Thr Ala Asp Asp Leu Thr Gly Arg Thr
290 295 300
Val Gly Phe Leu Ser Tyr Gly Ser Gly Ser Val Ala Glu Phe Phe Ala
305 310 315 320
Gly Thr Val Val Pro Gly Tyr Arg Ala His Thr Arg Pro Asp Gln His
325 330 335
Arg Ala Ala Ile Asp Arg Arg Gln Glu Ile Asp Tyr Ala Thr Tyr Arg
340 345 350
Glu Leu His Glu His Ala Phe Pro Val Asp Gly Gly Asp Tyr Pro Ala
355 360 365
Pro Glu Val Thr Thr Gly Pro Tyr Arg Leu Ala Gly Leu Ser Gly His
370 375 380
Lys Ara Val Tyr Glu Pro Arg
385 390
SEQ ID NO: 2I
LENGTH: 1176
TYPE: DNA
ORGANISM: Micromonospora sp. strain 046-ECO11
SEQUENCE: 21
atggccgaga gacccgccgt cggcatccac gacctgtccg ccgcgacggc gcatcacgtg 60
ctgacacacg agaccctggc cgcgagcaac ggcgccgacg tggccaagta ccaccgtggc 120
atcgggctgc gggcgatgag cgtgcccgcc ccggacgagg acatcgtgac gatggctgct 180
gccgccgccg cgccggtggt cgcccgccac ggcaccgacc ggatccggac cgtcgtgttc 240
gccacggagt cgtcggtcga ccaggcgaag gcggccggga tacacgtcca ctccctgctc 300
CA 02538147 2004-O1-21
ggcctcccctcggccacccgggtggtcgagctgaagcaggcctgctacggcggtacggcg360
ggactgcagttcgccatcggcctggtgcaccgtgacccgtcgcagcaggtcctggtgatc420
gccagcgacgtgtcgaagtacgcgctgggtgagcccggcgaggcgacccagggcgccgcg480
gcggtcgccatgctcgtcggcgcggacccggcgctggtacgcgtcgaggacccgtcgggc540
atgttcaccgccgacgtcatggacttctggcggccgaactaccgcaccaccgccctggtc600
gacgggcacgagtccatctccgcctacctgcaggcgctggagggctcgtggaaggactac660
accgagcgcggcggtcgcaccctggacgagttcggcgcgttctgctaccaccagccgttc720
ccgaggatggccgacaaggcgcaccggcacctgctcaactactgcgggcgcgacgtcgac780
gacgcgctggtggccggggccatcgggcacaccaccgcgtacaacgccgagatcggcaac840
agctacacggcgtcgatgtatctcgggctcgcggcactgctcgacaccgccgacgacctg900
accggccggaccgtcggcttcctcagctacgggtccggcagcgtcgccgagttcttcgcc960
ggcactgtcgtgcccgggtaccgcgcgcacacgcgacccgaccagcaccgcgcggcgatc1020
gaccggcggcaggagatcgactacgcgacgtaccgggagttgcacgagcacgccttcccg1080
gtcgacggcggcgactatccggcgccggaggtgaccaccgggccgtaccggctggccggg1140
ctctccggtcacaagcgcgtctacgagccgcgatag 1176
SEQ ID NO: 22
LENGTH: 290
TYPE: PRT
ORGANISM: Micromonospora sp. strain 046-ECO11
SEQUENCE: 22
Val Ala Giu Leu Tyr Ser Thr Ile Glu Glu Ser Ala Arg Gln Leu Asp
1 5 10 15
Val Pro Cys Ser Arg Asp Arg Val Trp Pro Ile Leu Ser Ala Tyr Gly
20 25 30
Asp Ala Phe Ala His Pro Glu Ala Val Val Ala Phe Arg Val Ala Thr
35 40 45
Ala Leu Arg His Ala Gly Glu Leu Asp Cys Arg Phe Arg Thr His Pro
50 55 60
Asp Asp Arg Asp Pro Tyr Ala Ser Ala Leu Ala Arg Gly Leu Thr Pro
55 70 75 80
Arg Thr Asp His Pro Val Gly Ala Leu Leu Ser Glu Val His Arg Arg
85 90 95
Cys Pro Val Glu Ser His Gly Ile Asp Phe Gly Val Val Gly Gly Phe
100 105 110
Lys Lys Ile Tyr Ala Ala Phe Ala Pro Asp Glu Leu Gln Val Ala Thr
4G
CA 02538147 2004-O1-21
115 120 125
Ser Leu Ala Gly Ile Pro Ala Met Pro Arg Ser Leu Ala Ala Asn Ala
130 135 140
Asp Phe Phe Thr Arg His Gly Leu Asp Asp Arg Val Gly Val Leu Gly
i45 150 155 160
Phe Asp Tyr Pro Ala Arg Thr Val Asn Val Tyr Phe Asn Asp Val Pro
165 170 175
Arg Glu Cys Phe Glu Pro Glu Thr Ile Arg Ser Thr Leu Arg Arg Thr
180 185 190
Gly Met Ala Glu Pro Ser Glu Gln Met Leu Arg Leu Gly Thr Gly Ala
195 200 205
Phe Gly Leu Tyr Val Thr Leu Gly Trp Asp Ser Pro Glu Ile Glu Arg
210 215 220
Ile Cys Tyr Ala Ala Ala Thr Thr Asp Leu Thr Thr Leu Pro Val Pro
225 230 235 240
Vai Glu Pro Glu Ile Glu Lys Phe Val Lys Ser Val Pro Tyr Gly Gly
245 250 255
Gly Asp Arg Lys Phe Val Tyr Gly Val Ala Leu Thr Pro Lys Gly Glu
260 265 270
Tyr Tyr Lys Leu Glu Ser His Tyr Lys Trp Lys Pro Gly Ala Val Asn
275 280 285
Phe Ile
29C
SEQ ID 23
NO:
LENGTH:
873
TYPE:
DNA
ORGANISM:Micromonospora
sp. strain
046-ECO11
SEQUENCE:23
gtggccgagctctactcgaccatcgaggaatcggcccggcaactggacgtgccgtgttcg60
cgcgaccgggtctggcccatcctgtccgcgtacggcgacgcgttcgcccatcccgaggcg12()
gtggtcgccttccgggtggcgaccgcgctgcgtcacgcgggcgagctggactgccggttc180
cggacgcatccggacgaccgggacccgtacgcctcggcgctcgcccggggcctcaccccg240
cgcacggaccaccccgtcggcgcgctgctctccgaggtccaccggcgctgcccggtggag300
agccacggcatcgacttcggggtggtcggcggcttcaagaagatctacgcggccttcgcc360
ccggacgagctgcaggtggccacgtcgctcgccggcattccggcgatgccccgcagcctc42()
gccgcgaacgccgacttcttcacccggcacggcctcgacgaccgggtcggcgtgctggga480
ttcgactacccggcccggaccgtgaacgtctacttcaacgacgtgccgcgtgagtgcttc540
gagccggagaccatccggtcgacgctgcgccggaccgggatggccgagccgagcgagcag600
47
CA 02538147 2004-O1-21
atgctccggctcggcaccggggcgttcgggctctacgtcacgctgggctgggactccccg660
gagatcgagcggatctgctacgccgcggcgaccacggacctgaccacgcttccggtaccc720
gtggaaccggagatcgagaagttcgtgaaaagcgttccgtacggcggcggggaccggaag780
ttcgtctacggcgtggcgctgacccccaagggggagtactacaaactcgagtcgcactac840
aaatggaagccgggcgcggtgaacttcatttga 87:3
SEQ ID N0: 24
LENGTH: 370
TYPE: PRT
ORGANISM: Micromonospora sp. strain 046-ECO11
SEQUENCE: 24
Val Trp Ala Arg Val Lys Asn Trp Val Val Ala Leu Ala Val Ala Ala
1 5 10 15
Val Leu Met Ile Ser Ala Leu Ala Gly Asp His Pro Ala Pro Glu Gly
20 25 30
Leu Gly Leu Leu Gly Phe Ala Leu Val Ala Ala Ser Gly Leu Ala Leu
35 40 45
Ala Ala Ser Arg Arg Ala Pro Ile Ala Val Leu Val Ala Thr Gly Leu
50 55 60
Cys Val Val Gly Tyr Asn Ala Ile Gly Phe Gly Val Pro Ala Ile Ala
65 70 75 80
Tyr Leu Phe Ala Val Tyr Ala Ala Val Arg Ala Gly His Arg Leu Val
85 90 95
Thr Leu G1V Ala Ser Ala Ala Leu Leu Val Val Leu Pro Leu Ala Ile
y 100 105 110
Met '~'al Ser Pro Ala Asp Gly Ala Leu Lys Glu Ala Leu Ala Gln Ser
~15 i20 125
Arg Gly Val Leu Glu Leu Ala Trp Leu Ile Ala Ala Ala Ala Ala Cllr
130 135 140
Glu Ala Leu Arg Gln Ala Glu Arg Arg Ala Asp Glu Ala Glu Arg Thr
145 150 155 160
Arg Glu Glu Thr Ala Arg Leu Arg Ala Thr Gln Glu Arg Leu His Ile
165 170 175
Ala Arg Glu Leu His Asp Ser Leu Thr His Gln Ile Ser Ile Ile Lys
180 185 190
Val Gln Ala GIu Val Ala Val His Leu Ala Arg Lys Arg Gly Glu Gln
195 200 205
Val Pro Glu Ser Leu Leu Ala Ile Gln Glu Ala Gly Arg Ala Ala Thr
210 215 220
Arg Glu Leu Arg Ala Thr Leu Glu Thr Leu Arg Asp Leu Thr Lys Ser
48
CA 02538147 2004-O1-21
225 230 235 240
Pro Ser His Gly Leu Asp His Leu Pro Glu Leu Leu Ala Gly Ala Glu
245 250 255
Lys Ile Gly Leu Ala Thr Thr Leu Thr Ile Glu Gly Asp Gln Arg Asp
260 265 270
Val Pro Glu Ala Val Gly Arg Thr Ala Tyr Arg Ile Val Gln Glu Ser
275 280 285
Leu Thr Asn Thr Ala Arg His Ala Ser Ala Ala Ala Ala Ala Val Arg
290 295 300
Ile Asp Tyr Arg Pro Asp Ala Leu Ser Ile Arg Ile Asp Asp Asp Gly
305 310 315 320
Thr Ala Arg Pro Gly Ala Ala Pro Val Pro Gly Val Gly Leu Leu Gly
325 330 335
Met His Glu Arg Val Leu Ala Leu Gly Gly Arg Leu Arg Ala Glu Pro
340 345 350
Arg Thr Gly Gly Gly Phe Thr Val Gln Ala Glu Leu Pro Val Val Arg
355 360 365
Val Pro
370
SEQ ID N0: 25
LENGTH: 1113
TYPE: DNA
ORGANISM: Micromonospora sp. strain 046-ECOll
SEQUENCE:25
gtgtgggcccgggtgaagaactgggtcgtcgcgttggctgtggcggcggtgctgatgatc60
agcgcgctggccggtgaccatcctgcccccgagggcctcggtctgctcggcttcgcgctg120
gtggcggcgagcggcctggcgctggccgccagtcgtcgggccccgatcgccgtgctggtc180
gccaccgggctgtgcgtggtgggctacaacgcgatcggcttcggggtgcccgccatcgcg240
tacctgttcgcggtctacgcggcggtccgggccgggcaccggctcgtcacgctcggggcg300
agcgecgccctgctcgtcgtcctgccgctggcgatcatggtctcgcccgcggacggcgcc360
ctcaaggaggcgctcgcgcagtcgcggggcgtgctggaactggcctggctgatcgccgcg420
gcggcggccggtgaggcgctgcggcaggccgaacggcgagcggacgaggcggaacggacc480
cgcgaggagaccgcccggctgcgcgccacccaggagcggctgcacatcgcacgggagctg540
cacgactcgctcacccaccagatctcgatcatcaaggtgcaggcggaggtggcggtccac600
ctggcccgcaagcggggcgagcaggtgccggagtcgctgctggcgatccaggaggccggc660
cgggcggcgactcgcgagctgcgcgcgaccctggagacgctgcgtgacctgaccaagtcc720
ccgtcgcacgggctcgaccacctcccggagctgctggccggggccgagaagatcggcctg780
49
CA 02538147 2004-O1-21
gccaccacgctgaccatcgagggcgaccagcgggacgtgccggaggcggtgggccgcacc840
gcgtaccggatcgtgcaggagtcgctcaccaacaccgcccggcacgcctccgccgcggcc900
gccgcggtccggatcgactaccgcccggacgcgctgagcatccggatcgacgacgacggg960
acggcccggccgggcgccgccccggtgcccggcgtcgggctgctggggatgcacgagcgc1020
gtcctcgcgctgggcggccggctgcgggcggaaccccgcaccggcggaggcttcaccgtc1080
caggccgaactcccggtggtgcgcgtcccatga 1113
SEQ ID NO: 26
LENGTH: 220
TYPE: PRT
ORGANISM: Micromonospora sp. strain 046-ECO11
SEQUENCE: 26
Met Ile Arg Ile Met Leu Leu Asp Asp Gln Pro Leu Leu Arg Ser Gly
1 5 10 15
Phe Arg Ala Leu Leu Asp Ala Glu Asp Asp Ile Glu Val Val Ala Glu
20 25 30
GIy Gly Asn Gly Arg Glu GIy Leu Ala Leu Ala Arg Gln His Leu Pro
35 40 45
Asp Leu Ala Leu Ile Asp Ile Gln Met Pro Val Met Asp Gly Val Glu
50 55 60
Thr Thr Arg Gln Ile Val Ala Asp Pro Ala Leu Ala Gly Val Arg Val
65 70 75 80
Val Ile Leu Thr Asn Tyr Gly Leu Asp Glu Tyr Val Phe His Ala Leu
85 90 95
Arg Ala Giy Ala Thr Gly Phe Leu Val Lys Asp Ile Glu Pro Asp Asp
100 105 110
Leu Leu His Ala Val Arg Val Ala Ala Arg Gly Asp Ala Leu Leu Ala
115 120 125
Pro Ser Ile Thr Arg Met Leu Ile Asn Arg Tyr Val Ser Glu Pro Leu
130 135 140
Cys Ala Asp Val Thr Pro Gly Met Glu Glu Leu Thr Asn Arg Glu Arg
145 150 155 160
GIu Ala Val Ala Leu Ala Ala Arg Gly Leu Ser Asn Asp Glu Ile Ala
165 170 175
Asp Arg Met Va1 Ile Ser Pro Leu Thr Ala Lys Thr His Val Asn Arg
180 185 190
Ala Met Thr Lys Leu Gln Ala Arg Asp Arg Ala Gln Leu Val Val Phe
195 200 205
Ala Tyr Glu Ser Gly Leu Val Ser Pro Gly Asn Arg
2I0 215 220
CA 02538147 2004-O1-21
SEQ ID NO: 27
LENGTH: 663
TYPE: DNA
ORGANISM: Micromonospora sp. strain 046-ECO11
SEQUENCE:27
atgatcaggatcatgctgctcgacgaccagccgctgctgcgcagcgggttccgcgcgctc60
ctcgacgccgaggacgacatcgaggtggtggccgagggcgggaacggccgggagggcctg120
gcgctggcccggcagcacctgcccgatctcgccctgatcgacatccagatgccggtcatg180
gacggcgtcgagacgacccggcagatcgtcgcggatccggcgctggccggggtacgcgtc240
gtcatcctcaccaactacggcctcgacgagtacgtcttccacgcgctgcgcgccggcgcc300
accggcttcctggtcaaggacatcgagccggacgacctgctgcacgccgtgcgggtcgcc360
gcgcgcggtgacgcgctgctcgcgccgtcgatcacccggatgctgatcaacaggtacgtg420
tcggagccgctctgcgcggacgtcacgcccggcatggaggagctgaccaaccgggaacgc480
gaggcggtcgccctggccgcccggggcctgtccaacgacgagatcgccgatcgcatggtg540
atcagcccgctgaccgcgaagacccacgtcaaccgcgccatgaccaagctgcaggcccgc600
gaccgcgcccagctggtggtgttcgcctacgagtccggcctggtgtcacccggcaatcgc660
tga 663
SEQ ID N0: 28
LENGTH: 131
TYPE: PRT
ORGANISM: Micromonospora sp. strain 046-ECO11
SEQUENCE: 28
Met Phe Ile Arg Arg Leu Leu Thr Ala Ala Ala Ala Gly Val Leu Gly
1 5 10 15
Gly Leu Ala Leu Val Ala Pro Ala Ala Ala Gln Val Thr Ala Ala Asp
20 25 30
Gly Asp Gly Gly Ser Gly Arg Ala Gly Ser Val Leu Ala Leu Ala Leu
35 40 45
Ala Leu Leu Gly Leu Val Leu Gly Gly Trp Ala Leu Arg Ser Ala Gly
50 55 60
Arg Gly Gly Gly Arg Gly Asn Ala Ile Ala Ala Leu Val Leu Ala Val
65 70 75 80
Ala Gly Leu Ile Ala Gly Val Val Ala Leu Ala Gly Ser Asp Gly Gly
85 90 95
Val Gly Ser Gly Asn Gly Arg Gly Gly Ala Ile Val Ala Val Val Leu
100 105 110
Ala Leu Ile Gly Ile Ala Val Gly Gly Leu Ala Phe Thr Arg Ser Arg
SI
CA 02538147 2004-O1-21
115 120 125
Arg Ala Ala
130
SEQ ID N0: 29
LENGTH: 396
TYPE: DNA
ORGANISM: Micromonospora sp. strain 046-EC011
SEQUENCE: 29
atgttcatcc gtcgtttgct caccgccgcc gcagccggcg tcctcggtgg gctcgcactc 60
gtcgcaccgg cggccgcgca ggtgacggcc gccgacggtg acggtggttc cggccgcgcc 120
ggatccgtgc tggcgctcgc gctcgcgttg ctcggcctcg tcctgggcgg gtgggcgttg 180
cgctccgcgg ggcgcggcgg cggtcgtggc aacgcgatcg ccgcgctggt gctcgcggtg 240
gccggcctga tcgccggcgt ggtcgccctg gccggctccg acggtggtgt cggcagcggc 300
aacggccgtg gtggcgccat cgtggccgtc gtgctggcgc tgatcgggat cgccgtcggc 360
ggcctggcat tcacccgctc ccggcgcgcc gcctga 395
SEQ ID NO: 30
LENGTH: 154
TYPE: PRT
ORGANISM: Micromonospora sp. strain 046-ECO11
SEQUENCE: 30
Met Arg Lys Val Phe Ala Gly Leu Ala Ala Phe Leu Leu Leu Val Leu
1 5 10 15
Val Val Gln Phe Phe Leu Ala Ala Ser Gly Ala Phe Ser Asn Glu Ala
20 25 30
Asn Glu Glu Ala Phe Arg Pro His Arg Ile Leu Gly Leu Gly Ser Ile
35 40 45
Leu Val Ala Val Val Leu Thr Val Ala Ala Ala Val Met Arg Met Pro
50 55 60
Gly Arg Ile Ile Gly Leu Ser Gly Leu Val Ala Gly Leu Gly Ile Leu
65 70 75 80
Gln Ala Leu 31e Ala Val Ile Ala Lys Ala Phe Gly Asp Ser Ala Gly
85 90 95
Asp .Ser Ala Val Gly Arg Tyr Va1 Phe Gly Leu His Ala Val Asn Gly
100 105 110
Leu Val Met Val Ala Val Ala Arg Val Ile Leu Arg Ser Val Arg Ala
115 120 125
Ala Pro Asp Thr Thr Thr Thr Pro Gly Val Asp Thr Thr Val Thr Gly
130 135 140
Pro Ala Ala Asp Ser Ala Arg Thr Ala Ser
52
CA 02538147 2004-O1-21
145 150
SEQ ID N0: 31
LENGTH: 465
TYPE: DNA
ORGANISM: Micromonospora sp. strain 046-ECO11
SEQUENCE:31
atgcgcaaagtgttcgccggactggcagcgttcctgctgctcgtgctcgtggtgcagttc60
ttcctggccgccagcggcgcgttcagcaacgaggccaacgaggaggcgttccgccctcac120
cggatcctgggcctggggagcatcctcgtcgccgtggtgctgacggtggccgccgcggtg180
atgcggatgcccggccggatcatcggcctgtccggcctggtcgccgggctgggcatcctg240
caggccctgatcgcggtcatcgccaaggcgttcggcgactcggccggtgactcggccgtc300
ggccggtacgtgttcggcctgcacgcggtcaacggactggtgatggtggccgtcgcccgc360
gtcatcctgcgcagcgtccgggcggcgccggacacgaccaccacgcccggcgtggacacg420
acggtcaccggtccggcggccgactcggcgcgaacggcgtcatga 465
SEQ ID N0: 32
LENGTH: 661
TYPE: PRT
ORGANISM: Micromonospora sp. strain 046-ECO11
SEQUENCE: 32
Met Ser Thr Leu Gln Trp Ile Leu Val Asp His Val Val Ala Leu Leu
1 5 10 15
Gly Val Ala Thr Trp Phe Ala Thr Gly Val Thr Ala Ala Leu Gly Arg
20 25 30
His Arg Ile Ala Leu Ala Leu Leu GIy Ala Ala Val Leu Val Thr Val
35 40 45
Ala Arg Leu Gly Thr Val Ala Leu Leu Ala Asp Arg Gly Trp Trp Phe
50 55 60
Val Gln Glu Lys Val Leu Leu Gly Leu Pro Met Leu Gly Ala Ala Gly
65 70 75 80
Leu Val Ala Val Leu Leu Ala Gly Pro Arg Leu Leu Ala Ala Arg Gln
85 90 95
Ser Pro Ala Ala Asp Leu Pro Ala Gly Ala Leu Val Ala Val Leu Thr
100 105 110
Ala Gly Phe Ala AIa Leu Ala Gly Leu Val Val Thr Phe Thr Ala Gly
115 120 125
Tyr Pro Leu Thr Trp Ser Thr AIa Leu Ile Ala Val Ala Leu Val Cys
130 135 140
Ala Ala Ala Leu Leu Thr Ala Arg Val Val Gly Arg Pro Ala Ala Pro
145 150 155 160
Si
CA 02538147 2004-O1-21
Ala Ala Glu Ala Gly Ser Pro Glu His Thr Pro Ala Ala Ala Gly Pro
165 170 175
Thr Ala Leu Ser Arg Arg Arg Phe Leu Gly Val Ala Gly Gly Val Val
180 185 190
Ala Ala Gly Ala Gly Ala Thr Gly Val Gly Leu Leu Phe Arg Asp Pro
195 200 205
Glu Ala Met Val Thr Gly Gly Gly Pro Gly His Ala Gly Gly Ala Arg
210 215 220
Pro Lys Val Ser Val Ala Asp Leu Arg Gly Pro Gly Ala Pro Ala Ala
225 230 235 240
Gly Gly Thr Ala Arg Arg His Val Leu Thr Ala Arg Thr Gly Thr Val
245 250 255
Thr Ile Pro Ser Gly Arg Pro Ile Asp Ala Trp Ser Tyr Glu Gly Arg
260 265 270
Leu Pro Gly Pro Ala Ile Thr Ala Thr Glu Gly Asp Leu Ile Glu Val
275 280 285
Thr Leu Arg Asn Ala Asp Ile Glu Asp Gly Val Thr Val His Trp His
290 295 300
Gly Tyr Asp Val Pro Cys Gly Glu Asp Gly Ala Pro Gly Ala Thr Gln
305 310 315 320
His Ala Val Gln Pro Gly Gly Glu Phe Val Tyr Arg Phe Gln Ala Asp
325 330 335
Gln Val Gly Thr Tyr Trp Tyr His Thr His Gln Ala Ser His Pro Ala
340 345 350
Val Arg Lys Gly Leu Tyr Gly Thr Leu Val Val Thr Pro Arg Glu Asp
355 360 365
Arg Pro Glu Ala Glu Arg Gly Leu Asp Leu Thr Leu Pro Val His Thr
370 375 380
Phe Asp Asp Val Thr Ile Leu Gly Asp Gln Glu Gly Arg Ala Val His
385 390 395 400
Asp Val Arg Pro Gly Gln Pro Val Arg Leu Arg Leu Ile Asn Thr Asp
405 410 415
Ser Asn Pro His Trp Phe Ala Val Val Gly Ser Pro Phe Arg Val Val
420 425 430
Ala Val Asp Gly Arg Asp Leu Asn Gln Pro Gly Glu Val Arg Glu Val
435 440 445
Gly Leu Arg Leu Pro Ala Gly Gly Arg Tyr Asp Leu Thr Leu Ala Met
450 455 460
Pro Asp Ala Lys Val Thr Leu Leu Leu Asp Asn Asp Ser Asp Gln Gly
465 470 475 480
54
CA 02538147 2004-O1-21
Vai Leu Leu Arg Pro Pro Gly Val Gly Gly Gly Asp Arg Pro Leu Pro
485 490 495
Asp Thr Ala Asp Trp Pro Glu Phe Asp Leu Leu Gly Tyr Gly Glu Pro
500 505 510
Ala Pro Val Pro Phe Asp Ala Asp Asp Ala Asp Arg His Phe Thr Ile
515 520 525
Val Leu Asp Arg Ala Leu Ala Met Val Asp Gly Lys Pro Ala Tyr Ala
530 535 540
Gln Thr Val Asp Gly Arg Ala His Pro Ser Val Pro Asp Gln Leu Val
545 550 555 560
Arg Glu Gly Asp Val Val Arg Phe Thr Val Val Asn Arg Ser Leu Glu
565 570 575
Thr His Pro Trp His Leu His Gly His Pro Val Leu Ile Leu Ser Arg
580 585 590
Asp Gly Arg Pro Tyr Ser Gly Ser Pro Leu Trp Met Asp Thr Phe Asp
595 600 605
Val Arg Pro Gly Glu Val Trp Glu Val Ala Phe Arg Ala Asp Asn Pro
610 615 620
Gly Val Trp Met Asn His Cys His Asn Leu Pro His Gln Glu Gln Gly
625 630 635 640
Met Met Leu Arg Leu Val Tyr Asp Gly Val Thr Thr Pro Phe Ala Ser
645 650 655
Thr Ser His Ala His
660
SEQ ID 33
NO:
LENGTH:
1986
TYPE:
DNA
ORGANISM:Micromonospora
sp. strain
046-ECO11
SEQUENCE:33
atgagcacgctccaatggatcctcgtggaccacgtcgtggcgctgctcggtgtcgcgacg60
tggttcgcaacgggtgtcacggcagctctcggccgccaccggatcgcgttggcgctcctc120
ggcgccgcggtgctggtgacagtcgcccgcctgggcaccgtggcgctgctggccgaccgc180
ggctggtggttcgtccaggagaaggttctgctggggr_tgccgatgctcggcgccgcgggg240
ctcgtcgcggtgctcctggccggcccgcgcctgctcgcggcccggcagtcaccggcggcg300
gacctgccggccggcgcgctggtcgcggtgctgaccgccggcttcgccgcgctggccggc360
ctggtggtgacgttcaccgccgggtacccgctgacgtggagcaccgcgctgatcgccgtc420
gccctcgtctgcgccgccgcgctgctcaccgcgcgggtggtcggacgacccgccgccccg480
gccgcggaggccggctccccggagcacacgccggcggcggccgggcccacggcgctgtcc540
cgccgccggttcctcggcgtggccgggggagtggtcgcggcgggcgccggcgccaccggc600
S$
CA 02538147 2004-O1-21
gtcggcctgctcttccgcgacccggaggcgatggtcaccggaggcggccccggacacgcc660
ggtggcgcccgccccaaggtctccgtggcggacctgcgcggccccggcgctccggcggcg720
ggcggcacggcgcgacgccacgtgctcaccgcccggacgggcaccgtcacgattccgtcc780
ggacgtccgatcgacgcctggagctacgagggccgcctgcccgggccggccatcaccgcg8417
accgagggcgacctgatcgaggtgacgctccgcaacgccgacatcgaggacggcgtcacc900
gtgcactggcacgggtacgacgtgccgtgcggcgaggacggcgcgccgggcgccacgcag960
oacgcggtgcagcccggcggcgagttcgtctaccggttccaggcggaccaggtggggacg1020
tactggtaccacacccaccaggcgtcgcaccccgccgtgcgcaaagggctgtacgggacg1080
ctcgtcgtgacgccgcgcgaggaccggccggaagcggagcgcgggctggacctgacgctg1140
ccggtgcacacgttcgacgacgtcacgatcctcggcgaccaggagggacgcgccgtccac1200
gacgtccgccccggccagccggtgcgactgcgtctgatcaacaccgactccaacccgcac1260
tggttcgccgtcgtcggctcgcccttccgcgtggtggccgtcgacggccgcgacctcaac1320
cagccgggcgaggtacgcgaggtcgggctccgcctgcccgccggaggccggtacgacctg1380
accctggccatgccggacgccaaggtcacgctgctgctcgacaacgactccgaocagggc1440
gtcctgctgcgcccgccgggcgtcggcggtggtgaccgcccgctgccggacaccgccgac1500
tggcccgagttcgacctgctgggctacggcgagccggcgcccgtgccgttcgacgccgac1560
gacgccgaccgccacttcaccatcgtcctcgaccgggccctggccatggtcgacggcaag1620
cccgcgtacgcccagaccgtcgacggtcgcgcacatccctccgtccccgaccagctcgtc1680
cgggagggggacgtcgtgcgcttcacggtggtcaaccggagcctcgaaacccacccgtgg1740
cacctgcacggccatccggtgctgatcctgtcccgcgacggccggccgtactccggcagc1800
ccgctgtggatggacaccttcgacgtgcggccgggagaggtgtgggaggtggcgttccgg1860
gcggacaatccgggtgtctggatgaaccactgccacaacctgccgcaccaggagcagggc1920
atgatgctgcggctcgtctacgacggtgtcaccacgr_ccttcgccagcacgagccacgca1980
cactga 1986
SEQ ID N0: 34
LENGTH: 129
TYPE : PRT
OP.GANISM: Micromonospora sp. strain 046-ECO11
SEQUENCE: 34
Met Thr Ala Asp Leu His Gly Leu Ala Ser Val Arg Tyr Ile Val Asp
1 5 10 15
Asp Val Ser Ala Ala Ile Glu Phe Tyr Thr Thr His Leu Gly Phe Thr
CA 02538147 2004-O1-21
20 25 30
Val Ser Thr Ala Phe Pro Pro Ala Phe Ala Asp Val Val Arg Gly Pro
35 40 45
Leu Arg Leu Leu Leu Ser Gly Pro Thr Ser Ser Gly Ala Arg Val Thr
50 55 60
Pro Ala Asp Ala Ala Gly Cys Gly Arg Asn Arg Ile His Leu Ile Val
65 70 75 80
Asp Asp Leu Asp Ala Glu Arg Glu Arg Leu Glu Arg Ala Gly Val Thr
85 90 95
Leu Arg Ser Asp Val Val Ala Gly Pro Gly Gly Arg Gln Phe Leu Ile
100 105 110
Ala Asp Pro Ala Gly Asn Leu Val Glu Val Phe Glu Pro Ala Ala Arg
115 120 125
Giy
SEQ ID N0: 35
LENGTH: 390
TYPE: DNA
ORGANISM: Micromonospora sp. strain 046-ECOll
SEQUENCE: 35
atgaccgcagacctgcacggcctggccagcgtccgctacatcgtcgacgacgtgtcggcg60
gcgatcgagttctacaccacccacctgggtttcacggtgtcgaccgcgttcccgccggcc120
ttcgccgacgtggtgcgcgggccgctgcggctcctgctgtccgggccgaccagctcgggc180
gcccgggtcaccccggcggacgcggccgggtgcgggcgcaaccgcatccacctgatcgtc240
gacgatctcgacgccgaacgggagcggctggagcgcgccggggtgacgttgcgcagcgac300
gtcgtggccgggccgggcggccgtcagttcctgatcgccgacccggcgggcaacctggtc360
gaggtgttcgagccggcagcccgcggctga 390
SEQ ID NO: 36
LENGTH: 178
TYPE: PRT
ORGANISM: Micromonospora sp. strain 046-ECO11
SEQUENCE: 36
Met Leu Thr Ala Val Val Ala Ser Pro His Ser Pro Glu Asn Thr Ser
1 5 10 15
Arg His Pro Thr Gly Gly Asp Ala Val Asp Glu Ala Thr Pro Arg Thr
20 25 30
Pro Val Ala Ala Arg Pro Thr Trp Ser Pro Ala Thr Ala Pro Val Trp
35 40 45
Leu Val Gly Val Leu Ala Thr Leu Ala Gly Ala Val Ala Ala Glu Ala
57
CA 02538147 2004-O1-21
50 55 60
Phe Thr Leu Ala Ala Arg Gly Phe Gly Val Pro Met Glu Ala Ala Gly
65 70 75 80
Vai Trp GIu Glu Gln Ala Gln Ala Ile Pro Val Gly Ala Ile Ala Arg
85 90 95
Ser Val Val Leu Trp Ser Ile Gly Gly Ile Val Leu Ala Val Val Val
100 105 110
Ala Arg Arg Ala Arg Arg Pro Val Arg Ala Phe Val Ala Gly Thr Val
115 120 125
Ala Phe Thr Val Leu Ser Leu Ala Ala Pro Ala Phe Ala Arg Asp Thr
130 135 140
Pro Val Ser Thr Gln Leu Val Leu Ala Gly Thr His Val Ile Ala Gly
145 150 155 160
Ala Val Ile Ile Ser Ile Leu Ala Ala Arg Leu Ala Ala Pro Thr Pro
165 170 175
Pro Arg
SEQ ID 37
N0:
LENGTH:
537
TYPE:
DNA
ORGANISM:Micromonospora
sp. strain
046-ECO11
SEQUENCE:37
atgttgactgccgtcgtggcgtccccgcattctcccgagaacacatcgaggcacccgacc60
ggaggcgacgccgtggatgaggccactccccgaactcccgtcgcggcacggcccacctgg120
tcgccggccaccgctccggtgtggctggtcggcgtgctggccaccctcgccggggccgtg180
gccgcggaggcgttcacgctcgccgcccggggcttcggcgtaccgatggaggcggccggc240
gtctgggaggagcaggcgcaggcgatcccggtgggggccatcgcccgcagcgtcgtgctc300
tggtcgatcggcggaatcgtcctggcggtggtcgtggcgcggcgggcccggcggcccgtg360
cgtgccttcgtggccggcaccgtcgcgttcaccgtgctgtccctcgccgcgcccgccttc420
goccgggacaccccggtgtcgacgcagctcgtcctcgccggcacccacgtgatcgccggc480
gccgtgatcatctccatcctggccgcgcggctcgccgcgcccaccccgccccggtaa 53'7
SEQ ID NO: 38
LENGTH: 66i
TYPE: PRT
ORGANISM: Micromonospora sp. strain 046-ECO11
SEQUENCE: 38
Met Asp Gly Thr Glu Ser Asn Val Thr Gly Phe Pro Asp Leu Leu Ser
1 5 10 15
5g
CA 02538147 2004-O1-21
Gly Leu Gly Gly Asp Gly Arg Ala Phe Ala Leu Leu His Arg Pro Gly
20 25 30
Ala Ala Gly Cys Ala Tyr Val Glu Val Leu Thr Gly Glu Val Cys Asp
35 40 45
Val Asp Thr Leu Gly Glu Leu Pro Leu Pro Thr Glu Pro Ala Thr Gly
50 55 60
Ala Arg His Asp Leu Leu Val Ala Val Pro Tyr Arg Gln Val Thr Glu
65 70 75 80
Arg Gly Phe Asp Cys His Asp Asp Gly Ala Pro Leu Leu Ala Met Arg
85 90 95
Val His Glu Gln Phe Gly Leu Asp Arg Gly Gln Ala Leu Ala Gly Leu
100 105 110
Pro Glu Arg Gly Val Pro Val Thr Asp Ala Asp Phe Asp Leu Ser Asp
115 120 125
Glu Asp Tyr Ala Ala Ile Val Lys Arg Val Val Gly Asp Glu Ile Gly
130 135 140
Leu Gly Ala Gly Ser Asn Phe Val Ile Arg Arg Thr Phe Thr Ala Arg
145 150 155 160
Leu Ala Asp Tyr Ser Ile Ala Thr Glu Leu Ala Leu Phe Arg Arg Leu
165 170 175
Leu Thr Gly Glu Leu Gly Ser Tyr Trp Thr Phe Leu Phe His Ser Gly
180 185 190
Ala Gly Thr Phe Ile Gly Ala Ser Pro Glu Arg His Val Ser Met Ile
195 200 205
Asp Gly Thr Val Ser Met Asn Pro Ile Ser Gly Thr Tyr Arg His Pro
210 215 220
Pro Asn Gly Pro Ala Val Ser Gly Leu Leu Glu Phe Leu Asn Asp Pro
225 230 235 240
Lys Glu Ala Asn Glu Leu Tyr Met Val Val Asp Glu Glu Leu Lys Met
245 250 255
Met Aia Arg Met Cys Ala Ser Gly Gly GIn Val His GIy Pro Phe Leu
260 265 270
Lys Glu Met Ala Arg Val Thr His Ser Glu Tyr Ile Leu Thr Gly Arg
275 280 285
Ser Asp Leu Asp Val Arg Asp Val Leu Arg Glu Thr Leu Leu Ala Pro
290 295 300
Thr Val Thr Gly Ser Pro Ile Glu Asn Ala Phe Arg Val Ile Thr Arg
305 310 315 320
His Glu Thr Thr Gly Arg Gly Tyr Tyr Gly Gly Val Leu Ala Leu Met
325 330 335
Gly Arg Asp Ser Ala Gly Ser Arg Thr Leu Asp Ser Ala Ile Met Ile
$~
CA 02538147 2004-O1-21
340 345 350
Arg Thr Ala Glu Ile Asp Asp Ala Gly Thr Leu Arg Leu Gly Val Gly
355 360 365
Ala Thr Leu Val Arg Asp Ser Lys Pro Glu Ser Glu Val Ala Glu Thr
370 375 380
Arg AIa Lys Ala Gly Ala Met Arg Ala Ala Leu Gly Leu GIy Val Asp
385 390 395 400
Pro Asp Gly Pro Asp Gly Gly Arg Thr Thr Ala Ala Arg Ala Arg Ser
405 410 415
Ser Leu Ala Thr Asp Pro Arg Val Arg Arg Ala Leu Arg Glu Arg Asn
420 425 430
Thr Thr Leu Ser Arg Phe Trp Leu Asp Gly Ala Glu Arg Arg Thr Pro
435 440 445
Asn Pro Ala Leu Thr Gly Arg Arg Val Leu Val Val Asp Asn Glu Asp
450 455 460
Thr Phe Met Ala Met Leu Asp His Gln Leu Arg Ala Leu Gly Leu Arg
465 470 475 480
Ser Ser Ile Ala Arg Phe Asp Ser Arg Leu Arg Pro Asp Gly His Asp
485 490 495
Leu Val Val Val Gly Pro Gly Pro Gly Asp Pro Gly Asp Leu Thr Asp
500 505 510
Pro Arg Met Arg Thr Leu Arg Gly Leu Thr Arg Asp Leu Leu Ala Gly
515 520 525
Thr Val Pro Phe Leu Ser Ile Cys Leu Gly His Gln Val Leu Ala Ala
530 535 540
Glu Leu Gly Phe Pro Leu Ala Arg Arg Ala Val Pro Asn Gln Gly Val
545 550 555 560
Gln Lys Arg Ile Asp Leu Phe Gly Arg Pro Glu Leu Val Gly Phe Tyr
565 570 575
Asn Thr Tyr Thr Ala Arg Ser Ala His Asp Val Val Ala Gly Gly Arg
580 585 590
Arg Gly Pro Ile Glu Ile Ser Arg Ser Pro Asp Ser Gly Asp Val His
595 600 605
Ala Leu Arg Gly Pro Gly Phe Arg Ser Val Gln Phe His Leu Glu Ser
610 615 620
Val Leu Thr Gln His Gly Pro Arg Ile Leu Gly Asp Leu Leu Val Ser
625 630 635 640
Leu Leu Ala Asp Gly Thr Ala Ala Ala Ala Ala Glu Ala Ala Gly Arg
645 650 655
Arg Gly Asn Arg Pro
660
CA 02538147 2004-O1-21
SEQ ID 39
NO:
LENGTH:
1986
TYPE:
DNA
ORGANISM:Micromonospora
sp. strain
046-ECOll
SEQUENCE:39
atggacgggacggaatcgaacgtgaccggattccccgatctgctgtccggtctcggcggc60
gacgggcgcgccttcgccctgctgcaccggcccggcgcggccgggtgcgcgtacgtggag120
gttctgaccggcgaggtgtgcgacgtggacactctcggcgagctgcccctgcccaccgag180
ccggcgaccggcgcgcggcacgacctgctcgtggcggtgccgtaccggcaggtcaccgaa240
cgggggttcgactgccacgacgacggcgcgccgctgctcgcgatgcgcgtccacgagcag300
ttcgggctcgaccgcggacaggcgctggcgggcctgcccgaacgcggtgtgccggtgacc360
gacgccgacttcgacctcagcgacgaggactacgccgcgatcgtcaagcgggtggtgggt420
gacgagatcgggctgggcgccggatccaacttcgtcatccggcgcaccttcaccgcgcgg480
ctggccgactactcgatcgccacggaactggcgctcttccgccggttgctgaccggcgaa540
ctgggttcctactggacgtttctgttccactccggcgccggcacgttcatcggcgcgtca600
ccggaacgacacgtcagcatgatcgacggaaccgtctcgatgaatcccatcagcgggacc660
taccggcaccccccgaacggcccggccgtttccggtctgctggaattcctgaacgacccg720
aaagaggctaacgaactctacatggtcgtcgacgaggaactgaaaatgatggcgcggatg780
tgcgcctccggcggccaggtgcacggcccgttcctcaaggaaatggcgcgggtgacgcac840
tccgagtacatcctgaccggccgcagcgacctggacgtgcgcgacgtgctgcgggagacc900
ctgctcgcgccgacggtcaccggcagcccgatcgagaacgcgttccgggtcatcacccgc960
cacgagacgaccggccgcggctactacggcggcgtgctcgcgttgatgggccgtgactcg102()
gccggcagccgtacgctcgactcggccatcatgatccgcaccgccgagatcgacgacgcg1080
ggcacgctgcgcctgggcgtcggcgccaccctcgtgcgggactccaagccggagtcggag114()
gtggccgagacgcgggccaaggcgggcgccatgcgcgcggcgctcggcctcggcgtcgac1200
ccggacggcccggacggcgggcggaccacggccgcgcgggctcgttcgtccctggccacc1260
gacccccgggtacggcgggcgttgcgcgagcgcaacaccacactgtcgaggttctggctc132()
gacggcgcggagcggcgcaccccgaacccggcgctgaccggacgccgcgtgctcgtcgtc1380
gacaacgaggacacgttcatggccatgctcgaccaccagttgcgggccctcgggctgcgg1440
tcgagcatcgcccggttcgacagccggctgcggccggacggacacgacctcgtcgtcgtc1500
ggtcccggceccggcgacccgggcgacctgaccgacccgcgtatgcggaccctgcgcggg1560
ctcacccgcgacctgctcgccggaacggtgccgttcctgtccatctgcctgggccaccag162()
~1
CA 02538147 2004-O1-21
gtgctcgccgccgaactggggttccccctcgcccggcgcgcggtgcccaaccagggtgtg 1680
cagaagcggatcgacctgttcggccggccggaactcgtggggttctacaacacctacacc 1740
gcccgctccgcgcacgacgtggtggccggtggccggcggggcccgatcgagatcagccgc 1800
agcccggacagcggggacgtgcacgcgctgcgcggcccgggattccgttccgtccagttc 1860
cacctggagtccgtcctcacccagcacggcccacggatcctgggcgacctgctggtctcc 1920
ctgctcgccgacggcacggccgccgccgcggccgaggcggcgggccggcgcgggaaccgc 1980
ccgtga 1986
SEQ ID NO: 40
LENGTH: 427
TYPE: PRT
ORGANISM: Micromonospora sp. strain 046-ECOll
SEQUENCE: 40
ValLysThr ThrValAsp ValLeuVal GinLysTyr GlyGly ThrSer
1 5 10 15
LeuGlnThr LeuAspArg ValArgHis AlaAlaLeu ArgIle AlaGlu
20 25 30
AlaArgArg HisGlySer AlaValThr ValValVal SerAla ArgGly
35 40 45
SerArgThr AspAspLeu LeuArgLeu AlaAlaAsp ValGly AlaAla
50 55 60
GIyProSer ArgGluLeu AspGlnLeu LeuAlaVal GlyGlu SerGlu
65 70 75 80
Ser Ala Ala Leu Met Ala Leu Ala Leu Thr Gly Leu Gly Val Pro Ala
85 90 95
Val Ser Leu Thr Gly His Gln Ala Glu Ile His Thr Thr Asp Arg His
100 105 110
Gly Asp Ala Leu Ile Ser Arg Ile Gly Ala Ala Arg Val Glu Ala Ala
115 120 125
Leu Gly Arg Gly Glu Val Ala Val Val Thr Gly Phe Gln Gly Ile Asp
130 135 140
Arg Ala Gly Asp Val Ala Thr Leu Gly Arg Gly Gly Ser Asp Thr Thr
145 150 155 160
Ala Val Ala Leu Ala Ala Arg Leu Arg Ala Ser Ala Cys Glu Ile Tyr
165 170 175
Thr Asp Val Asp Gly Val Phe Ser Ala Asp Pro Arg Ile Leu Pro Ala
180 185 190
Ala Arg Cys Leu Pro Trp Val Glu Pro Gly Val Met Ala Glu Met Ala
195 200 205
CA 02538147 2004-O1-21
Phe Ala Gly Ala Arg Val Leu His Thr Arg Cys Ile Glu Leu Ala AIa
210 215 220
Met Glu Gly Val Glu Val Arg Val Arg Asn Ala Ser Ser Gln Ala Pro
225 230 235 240
Gly Thr Ile Val Val Asp Arg Pro Asp Asp Arg Pro Leu Glu Thr Arg
245 250 255
Arg Ala Val Val Ala Val Thr His Asp Thr Asp Val Val Arg Val Leu
260 265 270
Val His Cys Arg Asp Gly Arg Arg Asp Met Ala Pro Asp Val Phe Glu
275 280 285
Val Leu Ala Ala His Gly Ala Val Ala Asp Leu Val Ala Arg Ser Gly
290 295 300
Pro Tyr Glu Ser Glu Phe Arg Met Gly Phe Thr Ile Arg Arg Ser Gln
305 310 315 320
Ala Glu Ala Val Arg Thr Ala Leu His Asp Leu Thr Ala Ser Phe Asp
325 330 335
Gly Gly Val His Phe Asp Glu Asn Val Gly Lys Val Ser Val Val Gly
340 345 350
Met Gly Leu Leu Ser Arg Pro Glu His Thr Ala Arg Leu Met Ala Ala
355 360 365
Leu Ala Ala Ala Gly Ile Ser Thr Ser Trp Ile Ser Thr Ser Gln Met
370 375 380
Arg Leu Ser val Ile Val Ser Arg Asp Arg Thr Val Asp Ala Val Glu
385 390 395 400
Ala Leu His Arg Ala Phe Arg Leu Asp Arg Ser Glu Pro Ala Asp Ala
405 410 415
Thr Ser Leu Thr Ser Arg Arg Ser Ala Thr Ala
420 425
SEQ ID 41
NO:
LENGTH:
1284
TYPE:
DNA
ORGANISM:Micromonospora
sp. strain
046-EC011
SEQUENCE:41
gtgaagacgactgtggacgtgctggtccagaaatacgggggcacctcgctgcagaccctc60
gaccgcgttcggcacgccgcgctgcggatcgccgaggcgcggcggcacggctccgccgtg120
acagtggtcgtgtcggcgcgcggcagccggaccgacgacctgctgcggctggcggccgac180
gtcggcgccgcgggtccgtcccgggaactcgaccagttgctcgcagtcggcgagtccgag240
tcggcggcgctgatggcgctggcgttgaccgggctgggagtgccggccgtctcgctgacc300
gggcaccaggcggagatccacaccaccgaccggcacggcgacgcgctgatctcgcggatc360
ggggcggcgcgggtggaagcggcgctgggccgtggcgaggtcgccgtggtcaccggattc420
63
CA 02538147 2004-O1-21
cagggcatcgaccgggccggtgacgtcgccacgctggggcgcggcggctccgacacgaca 480
gcggtggcgctcgcggcccggctccgcgcgtcggcgtgcgagatctacaccgacgtggac 540
ggcgtcttcagcgccgacccccgcatccttccggcggcgcgttgcctgccgtgggtggag 600
cccggcgtcatggcggagatggcgttcgccggcgcgcgggtcctgcacacccgatgcatc 660
gagctggccgccatggaaggggtcgaagtgcgcgtgcgcaacgcgtcgtcgcaggcgccc 720
ggaacgatagtcgtggaccggcccgacgaccggccgctggagacccggcgggccgtggtg 780
gcggtcacccacgacaccgatgtcgtccgcgtgctggtgcactgccgcgacggccgccgg 840
gacatggcacccgacgtgttcgaggtgctggccgcccatggggcggtggcggacctggtg 900
gcccggtccgggccctacgagagcgagttccggatggggttcaccatccgccgcagccag 960
gccgaagcggtgcggaccgcgctgcacgacctcaccgcgtccttcgacggcggggtccac 1020
ttcgacgagaacgtcggcaaggtgtccgtggtcggcatgggcctgctcagccgccccgag 1080
cacacggcccggctgatggcggcgctggccgcggcggggatctcgacgagctggatctcc 1140
acctcccagatgcggctgtcggtgatcgtgtcgcgggaccgcaccgtcgacgccgtcgaa 1200
gccctgcaccgcgcgttccgcctggaccggtccgagccggcggacgccacgtccctgacc 1267
tcccgccgttccgccaccgcctga 1284
SEQ ID NO: 42
LENGTH: 274
TYPE: PRT
ORGANISM: Micromonospora sp. strain 046-ECO11
SEQUENCE: 42
Val Ala Val Leu Asn Ala Ser Phe Ala Arg Gly Leu Arg Leu Arg Arg
1 5 10 15
Leu Phe Arg Arg Gly Asp Gly Arg Leu Leu Val Val Pro Leu Asp His
20 25 30
Ser Val Thr Asp Gly Pro Leu Arg Arg Gly Asp Leu Asn Ser Leu Leu
35 40 45
Gly G1u Leu Ala Gly Thr Gly Val Asp Ala Val Val Leu His Lys Gly
50 55 60
Ser Leu Arg His Val Asp His Gly Trp Phe Gly Asp Met Ser Leu Ile
65 70 75 80
Val His Leu Ser Val Ser Thr Arg His Ala Pro Asp Pro Asp Ala Lys
85 9C 95
Tyr Leu Vai Ala His Val Glu Glu Ala Leu Arg Leu Gly Ala Asp Ala
100 105 110
VaI Ser Val His Val Asn Leu Gly Ser Pro Gln Glu Ala Arg Gln Ile
G4
CA 02538147 2004-O1-21
115 120 125
Ala Asp Leu Ala Ala Val Ala Gly Glu Cys Asp Arg Trp Asn Val Pro
130 135 140
Leu Leu Ala Met Val Tyr Ala Arg Gly Pro Gln Ile Thr Asp Ser Arg
145 150 155 160
Ala Pro Glu Leu Val Ala His Ala Ala Thr Leu Ala Ala Asp Leu Gly
165 170 175
Ala Asp Ile Val Lys Thr Asp Tyr Val Gly Thr Pro Glu Gln Met Ala
180 185 190
Glu Val Val Arg Gly Cys Pro Ile Pro Leu Ile Val Ala Gly Gly Pro
195 200 205
Arg Ser Ala Asp Thr Pro Thr Val Leu AIa Tyr Val Ser Asp Ala Leu
210 215 220
Arg Gly Gly Val Ala Gly Met Ala Met Gly Arg Asn Val Phe Gln Ala
225 230 235 240
Glu Gln Pro Gly Leu Met Ala Ala Ala Val Ala Arg Leu Val His Glu
245 250 255
Pro Arg His Val Pro Asp Arg Tyr Asp Val Asp Asp Arg Leu Ala Leu
260 265 270
Thr Ser
SEQ ID 43
NO:
LENGTH:
825
TYPE:
DNA
ORGANISM:Micromonospora
sp. strain
046-ECOll
SEQUENCE:43
gtggccgtactcaacgcttcgttcgctcgtggcctgcgtctgcgccgactgttccgacgc60
ggcgacggacgcctgctcgtcgtcccgctcgaccactccgtcaccgacgggccgctgcgc120
cgcggcgacctgaactcgctgctcggtgagctcgccggcaccggcgtggacgccgtggtg180
ctgcacaagggcagcctgcggcacgtcgaccacggctggttcggcgacatgtcgctgatc240
gtgcatctgagcgtgagcacccggcacgccccggacccggacgcgaagtacctggtcgcg300
cacgtggaggaggcgctgcggctgggcgccgacgcggtcagcgtgcacgtcaacctcggc360
tcaccgcaggaggcgcggcagatcgccgacctggcggcggtggcgggggagtgcgaccgc420
tggaacgtcccgctgctggccatggtgtacgcccgcgggccgcagatcaccgactcccgg480
gcaccggagctggtggcgcacgccgcgacgCtCgCCgCggacctcggcgccgacatcgtc540
aagaccgactacgtgggcacgcccgagcagatggccgaggtggtgcgcggctgcccgatc600
ccgctgatcgtggccggcggcccgcgctcggccgacactccgacggtgctcgcctacgtc660
tcggacgcgctgcgcggcggcgtggccgggatggccatgggccgcaacgtgttccaggcc720
CA 02538147 2004-O1-21
gagcagcccg gcctgatggc cgccgccgtg gcacggctgg tgcacgagcc acggcacgtg 780
ccggaccggt acgacgtcga cgaccggctc gcccttacgt cctga 82.5
SEQ ID N0: 44
LENGTH: 367
TYPE: PRT
ORGANISM: Micromonospora sp. strain 046-ECO11
SEQUENCE: 44
Val Lys Leu Cys Trp Leu Asp Ile Arg Asn Val Asn Gly Ala Lys Glu
1 5 10 15
Aia Ile Val Glu Glu Ala Val His Gln Arg Val Asp Ala Val Val Ala
20 25 30
Ala Asp Pro Ala Asp Leu Glu Thr Leu Pro Pro Thr Val Lys Lys Val
35 40 45
Leu Phe Pro GIn Gly Gly Pro Leu Pro Glu Lys Leu Glu Pro Ala Asp
50 55 60
Leu Val Ile Val Glu Pro Ala Arg His Gly Glu Pro Ala Glu Leu Ala
65 70 75 80
Ala Arg Tyr Pro Glu Val Glu Phe Gly Arg Phe Val Glu Ile Val Asp
85 90 95
Ala Asp Ser Leu Glu Asp Ala Cys Arg Ser Ala Arg His Asp Arg Trp
100 105 110
Ser Leu Leu Tyr Phe Arg Asp Pro Thr Lys Ile Pro Leu Glu Ile Val
115 120 125
Leu Ala Ala Ala Ala Gly Ala Glu Gly Ser Ile Ile Thr Gln Val Ala
130 135 140
Asp Val Glu Glu Ala Glu Ile Val Phe Gly Val Leu Glu His Gly Ser
145 150 155 160
Asp Gly Val Met Leu Ala Pro Arg Ala Val Gly Glu Ala Thr Glu Leu
165 170 175
Arg Thr Ala Ala Val Ser Thr Ala Ala Asp Leu Ser Leu VaI Glu Leu
180 185 190
Glu Val Thr Gly Ile Arg Arg Val Gly Met Gly Glu Arg Ala Cys Val
195 200 205
Asp Thr Cys Thr Asn Phe Arg Leu Asp Glu Gly Ile Leu Val Gly Ser
210 215 220
His Ser Thr Gly Met Ile Leu Cys Cys Ser Glu Thr His Pro Leu Pro
225 230 235 240
Tyr Met Pro Thr Arg Pro Phe Arg Val Asn Ala Gly Ala Leu His Ser
245 250 255
66
CA 02538147 2004-O1-21
Tyr Thr Leu Ser Ala Gly Gly Arg Thr Asn Tyr Leu Ser Glu Leu Val
260 265 270
Ser Gly Gly Arg Val Leu Ala Val Asp Ser Gln Gly Lys Ser Arg Val
275 280 285
Val Thr Val Gly Arg Val Lys Ile Glu Thr Arg Pro Leu Leu Ala Ile
290 295 300
Asp Ala Val Ser Pro Ser Gly Thr Arg Val Asn Leu Ile Val Gln Asp
305 310 315 320
Asp Trp His Val Arg Val Leu Gly Pro Gly Gly Thr Val Leu Asn Val
325 330 335
Thr Glu Leu Thr Ala Gly Thr Lys Val Leu Gly Tyr Leu Pro Val Glu
340 345 350
Lys Arg His Val Gly Tyr Pro Ile Asp Glu Phe Cys Ile Glu Lys
355 360 365
SEQ ID 45
N0:
LENGTH:
1104
TYPE:
DNA
ORGANISM:Micromonospora
sp. strain
046-ECO11
SEQUENCE:45
gtgaagctgtgctggctggacatccgtaacgtcaacggcgccaaggaggcaatcgtcgag60
gaggcggtccaccagcgggtggacgccgtcgtggcggccgatccggccgacctggagacg120
cttcccccgacggtgaagaaggtgctgttcccgcagggcgggccgctgccggagaagctg180
gaaccggccgacctggtgatcgtcgagccggcccggcacggcgagcccgccgagctggcg240
gcccggtacccggaggtggagttcggccggttcgtcgagatcgtcgacgcggacagcctg300
gaggacgcctgccggtccgcgcgccacgaccggtggagcctgctgtacttccgcgacccc360
accaagatcccgctggagatcgtgctggcggccgcggcgggcgcggagggcagcatcatc420
acccaggtcgccgacgtcgaggaggcggagatcgtcttcggcgtcctggagcacggctcg480
gacggagtgatgctggcgccccgcgccgtgggggaggccaccgagctgcggaccgccgcg540
gtgagcacggcggcggacctgtcgctcgtggagctggaggtcaccggcatccggcgggtg600
ggcatgggcgagcgcgcctgcgtcgacacgtgcacgaacttccgtctggacgagggcatc660
ctggtcggctcgcactccaccggcatgatcctgtgcr_gcagcgagacgcatccgctgccg720
tacatgccgacccggccgttccgggtcaacgccggcgcgctgcactcgtacacgctctcc780
gccggcgggcggaccaactacctcagcgagctggtctccggcggccgggtgctcgccgtg840
gactcgcaggggaagtcccgcgtcgtcacagtgggacgggtcaagatcgagacgcgtccg900
ctgctggcgatcgacgcggtctccccctccgggacacgcgtcaacctcatcgtccaggac960
gactggcacgtgcgcgtgctcgggccgggcggcaccgtgctcaacgtgaccgagctgacc1020
G7
CA 02538147 2004-O1-21
gccggcacga aggtgctcgg ttacctgccg gtggagaagc ggcacgtcgg ctacccgatc 1080
gacgagttct gcatcgagaa gtga 1104
SEQ ID NO: 46
LENGTH: 253
TYPE: PRT
ORGANISM: Micromonospora sp. strain 046-EC011
SEQUENCE. 46
Met Thr Ala Gln Pro Val Leu Asp Phe His Val Arg Leu Ala Pro Arg
1 5 10 15
Pro Gly Ala Arg Glu Arg Leu Leu Ala Ala Leu Arg Glu Cys Gly Leu
20 25 30
Ala Arg Ala Val Val Cys Ala Gly Gly Thr Ile Asp Leu Asp Arg Leu
35 40 45
Ser Arg Gln Leu Val Thr Gly Gly His Val Glu Thr Asp Ala Asp Asn
50 55 60
Asp Ala Val Ala Ala Ala Cys Ala Gly Thr Asp Gly Arg Leu Val Pro
65 70 75 80
Phe Phe Phe Ala Asn Pro His Arg Pro Ala Glu Ala Tyr Arg Ala Arg
85 90 95
Ala A1a Glu Phe Arg Gly Leu Glu Ile Ser Pro Ala Val His Gly Val
100 105 110
Ala Leu Thr Asp Pro Arg Val Ala Asp Leu val Ala Val Ala Ala Glu
115 120 125
Phe Asp His Pro Val Tyr Val Val Cys Leu Asp Arg Pro Gly Ala Gly
130 135 140
Val Ala Asp Leu Val Gly Leu Ser Arg Arg Phe Pro Gln Val Ser Phe
145 150 155 160
Val Leu Gly His Ser Gly Val Gly Asn Ile Asp Leu Tyr Ala Leu Thr
165 170 175
Leu Ile Gln Asp Glu Pro Asn Ile Ser Leu Glu Thr Ser Gly Gly Tyr
180 185 190
Thr Cys Val Ala Glu Ala Ala Leu Arg Arg Leu Gly Asp Asp Arg Val
195 200 205
Val Phe Gly Ser Glu Tyr Pro Leu Gln His Pro Ala Val Glu Leu Ala
210 215 220
Lys Phe Gln Ala Leu Arg Leu Pro Pro Glu Arg Trp Arg Arg Ile Ala
225 230 235 240
Trp Asp Asn Ala His Arg Leu Leu Gly Glu Glu Lys Arg
245 250
SEQ ID NO: 47
CA 02538147 2004-O1-21
LENGTH:
762
TYPE:
DNA
ORGANISM:Micromonospora
sp. strain
046-ECO11
SEQUENCE:47
atgaccgcgcagccggtgctggacttccacgtacgcctggcgccccggcccggggcgcgg60
gagcggctgctcgccgcgctgcgcgagtgcgggctggcgcgggcggtggtgtgcgcgggc120
ggcaccatcgacctggaccggctgtcccgccagctcgtcaccggcggccacgtcgagacc180
gacgccgacaacgacgcggtggcggcggcctgcgccggcaccgacggccggctggtgccg240
ttcttcttcgCCaaCCCgCaCCggCCggCCgaggcgtaccgggcccgcgccgccgagttc300
cgcggcctggagatctcacccgccgtccacggcgtcgccctgaccgacccgcgggtcgcc360
gacctcgtggccgtggcggcggagttcgaccatccggtgtacgtggtctgcctggaccga420
cccggcgcgggcgtggccgacctggtcggcctgagccgccggttcccgcaggtgagcttc480
gtgctcgggcacagcggcgtcggcaacatcgacctctacgccctgaccctgatccaggac540
gagccgaacatctcgctggagacctccggcggctacacctgcgtggccgaggcggcgcta600
cgccgcctcggcgacgaccgggtggtgttcggctccgagtacccgctgcagcacccggcc660
gtggaactggccaagttccaggcgttgcgactgccgccggagcggtggcggcggatcgcc720
tgggacaacgcgcatcgactgctaggagaggagaagcggtga 762
SEQ ID N0: 48
LENGTH: 438
TYPE: PRT
ORGANISM: Micromonospora sp. strain 045-ECO11
SEQUENCE: 48
v'al Ser ProSer Ser Pro ArgLeu Gly TrpHis
Glu Ser Leu Gln Gly
1 5 10 15
Leu. Glu LeuArg Leu Glu LysGln Leu GluThr
Asp Arg Gln Ala Phe
20 25 30
Thr Trp AlaArg Pro Tyr ArgAla Arg AlaSer
Ala Ser Phe Leu Gly
35 40 45
Ala Pro Pro Val Thr Pro Ala Asp Leu Ala Asp Leu Pro Leu Thr Thr
50 55 60
Lys Gln Asp Leu Arg Asp Asn Tyr Pro Phe Gly Met Leu Ala Val Pro
65 7C 75 80
Arg GIu Arg Leu Ala Thr Tyr His Glu Ser Ser Gly Thr Ala Gly Lys
85 90 95
Pro Thr Pro Ser Tyr Tyr Thr Ala Glu Asp Trp Thr Asp Leu Ala Glu
100 105 110
Arg Phe Ala Arg Lys Trp Ile Gly Met Ser Ala Asp Asp Val Phe Leu
CA 02538147 2004-O1-21
115 120 125
Val Arg Thr Pro Tyr Ala Leu Leu Leu Thr Gly His Leu Ala His Ala
130 135 140
Aia Ala Arg Leu Arg Gly Ala Thr Val Val Pro Gly Asp Asn Arg Ser
145 150 155 160
Leu Ala Met Pro Tyr Ala Arg Val Val Arg Val Met His Asp Leu Asp
165 170 175
Vai Thr Leu Thr Trp Ser Val Pro Thr Glu Cys Leu Ile Trp Ala Ala
180 185 190
Ala Ala Ile Ala Ala Gly His Arg Pro Asp Ile Asp Phe Pro Ala Leu
195 200 205
Arg Ala Leu Phe Val Gly Gly Glu Pro Met Thr Asp Ala Arg Arg Arg
210 215 220
Arg Ile Ser Arg Leu Trp Gly Val Pro Val Ile Glu Glu Tyr Gly Ser
225 230 235 240
Thr Glu Thr Gly Ser Leu Ala Gly Glu Cys Pro Glu Gly Arg Leu His
245 250 255
Leu Trp Ala Asp Arg Ala Leu Phe Glu vaI Tyr Asp Pro Asp Thr Gly
260 265 270
Ala Val Arg Ala Asp Gly Asp Gly Gln Leu Val Val Thr Pro Leu Phe
275 280 285
Arg Glu Ala Met Pro Leu Leu Arg Tyr Asn Leu Glu Asp Asn Val Ser
290 295 300
Val Ser Tyr Asp Asp Cys Gly Cys Gly Trp Lys Leu Pro Thr Vai Arg
3C5 310 315 320
Val Leu Gly Arg Ser Ala Phe Gly Tyr Arg Val Gly Gly Thr Thr Ile
325 330 335
Thr Gln His Gln Leu Glu Glu Leu Val Phe Ser Leu Pro Glu Ala His
340 345 350
Arg Val Met Phe Trp Arg Ala Lys Ala Glu Pro Ala Leu Leu Arg Val
355 360 365
Giu ile Glu Val Ala Ala Ala His Arg Val Ala Ala Glu Ala Glu Leu
370 375 380
Thr Ala Ala I1e Arg Ala Ala Phe Gly Val Asp Ser Glu Val Thr Gly
385 390 395 400
Leu Ala Pro Gly Thr Leu Ile Pro Leu Asp Ala Leu Thr Ser Met Pro
405 410 415
Asp Val Vai Lys Pro Arg Ser Leu Phe Gly Pro Asp Glu Asp Trp Ser
420 425 430
Lys Ala Leu Leu Tyr Tyr
435
CA 02538147 2004-O1-21
SEQ ID 49
N0:
LENGTH:
1317
TYPE:
DNA
ORGANISM:Micromonospora
sp. strain
046-ECO11
SEQUENCE:49
gtgagcgagccaagttcgagcctgccccggctcggccagtggcacggcctcgaggacctg60
cggcgcctccaggagaagcaactggcggagacgttcacctgggcggcccggtcgccgttc12c)
taccgggcgcggctggcctccggcgcgccgccggtgacgcccgccgacctggccgacctg180
ccgctgaccaccaagcaggacctgcgggacaactaccccttcggcatgctcgccgtgccc240
cgcgaacggctggcgacctaccacgagtcgagcgggaccgccgggaagcccaccccctcc300
tactacaccgcggaggactggaccgacctggcggagcgcttcgcccgcaagtggatcggc360
atgtccgccgacgacgtcttcctggtccgcacgccgtacgcgctgctgctgaccgggcat420
ctcgcccacgccgcagcccggctgcgtggggccacggtggtacctggcgacaaccggtcg480
ctggcgatgccgtacgcccgggtggtccgggtgatgcacgacctggacgtcacgctcacc540
tggtcggtgccgacggagtgcctgatctgggccgccgcggcgatcgcggccgggcaccgg600
cccgacatcgacttcccggcgctgcgcgcgctgttcgtcggcggcgagccgatgaccgac660
gcccgccggcggcggatcagccgcctgtggggggtgccggtcatcgaggagtacggctcg720
acggagaccggcagcctggccggggagtgccccgagggacgcctgcacctgtgggccgac780
cgggcgctgttcgaggtgtacgacccggacaccggcgccgtccgcgcggacggcgacggc840
cagctcgtggtcacgccgctgttccgggaggcgatgccgctgctgcggtacaacctggag90()
gacaacgtgtcggtctcctacgacgactgcggatgcggctggaagctgcccaccgtgcgg960
gtgctcggccggtcggcgttcggctaccgggtcggcggcaccaccatcacccagcaccag1020
ctggaggaactggtcttctccctgccggaggcgcaccgggtgatgttctggcgggccaag1080
gcggagccggcgctgttgcgggtcgagatcgaggtggccgccgcgcaccgggtcgccgcc1140
gaggcggagctgaccgccgcgatccgggccgccttcggcgtggacagcgaggtcaccggc1200
ctggcgccgggaaccctgatcccgctcgacgcgctgaccagcatgccggacgtggtgaag1260
ccacgcagcctgttcggtccggacgaggactggagcaaagcgctcctctactactga 1317
SEQ ID NO: 50
LENGTH: 396
TYPE: PRT
ORGANISM: Micromonospora sp. strain 046-ECO11
SEQUENCE: 50
Met Pro Gln Met Arg Val Ala Val Ala Gly Ala Gly Ile Ala Gly Leu
1 5 10 15
71
CA 02538147 2004-O1-21
Ala Phe Ala Ala Ala Leu Arg Arg Thr Gly Ile Asp Cys His Val Tyr
20 25 30
Glu Gln Ala Asp Gln Leu Met Glu Val Gly Ala Gly Val Gln Val Ala
35 40 45
Pro Asn Ala Thr Arg Leu Leu His Arg Leu Gly Leu Arg Asp Arg Leu
50 55 60
Arg Thr Val Ala Val Ala Pro Gln Ala Ile Glu Met Arg Arg Trp Asp
65 70 75 80
Asp Gly Thr Leu Leu Gln Arg Thr Gln Leu Gly Ser Val Cys Gly Arg
85 90 95
Arg Phe Gly Ala Pro Tyr Tyr Val Val His Arg Ala Asp Leu His Ser
100 105 110
Ser Leu Leu Ser Leu Val Pro Pro Asp Arg Val His Leu Gly Ala Arg
115 120 125
Leu Thr Ala Val Thr Gln Thr Ala Asp Glu Ala Tyr Leu His Leu Ser
130 135 140
Asn Gly Thr Thr Val Ala Ala Asp Leu Val Val Gly Ala Asp Gly Ile
145 150 155 160
His Ser Val Ala Arg Glu Gln Ile Val Ala Asp Arg Pro Arg Phe Ser
165 170 175
Gly Gln Ser Ile Tyr Arg Gly Leu Val Pro Ala Glu Arg Val Pro Phe
180 185 190
Leu Leu Thr Glu Pro Arg Val Gln Leu Trp Phe Gly Pro Asp Gln His
195 200 205
Cys Val Cys Tyr Pro Val Ser Ala Gly Arg Gln Val Ser Phe Gly Ala
210 215 220
Thr Val Pro Ala Thr Asp Trp Arg Gln Glu Ser Trp Ser Gly Arg Gly
225 230 235 240
Asp Val Thr Gln Leu Ala Ala Ala Tyr Ala Gly Trp His Pro Asp Val
245 250 255
Thr Arg Leu Ile Ala Ala Ala Asp Arg Val Gly Arg Trp Ala Leu His
260 265 270
Asp A_rg Asp Ser Ile Asp Arg Leu Ser Ala Gly Arg Val Thr Leu Ile
275 280 285
Gly Asp Ala Ala His Pro Met Leu Pro Phe Gln Ala Gln Gly Ala Asn
290 295 300
Gln Ala Val Glu Asp Ala Val Val Leu Ala Val Cys Leu Ala Gly Val
305 310 315 320
Glu Pro Ala Gly Leu Gly Ala Ala Leu Arg Arg Tyr Glu Arg Ile Arg
325 330 335
CA 02538147 2004-O1-21
Leu Pro Arg Thr Thr Arg Ile Gln Arg Gln Ser Arg Ala Asn Ala Glu
340 345 350
Met Phe His Leu Ala Asp Gly Ala Asp Gln Arg Arg Arg Asp Val Ala
355 360 365
Ala. Gln Ser Ser Ser Gly Leu Asp Arg His Glu Trp Leu Phe Gly Tyr
370 375 380
Asp Ala Glu Lys Ala Thr Thr Thr Ser Gly Ser Ala
385 390 395
SEQ ID N0: 51
LENGTH: 1191
TYPE: DNA
ORGANISM: Micromonospora sp. strain 046-EC011
SEQUENCE:51
atgccgcagatgagggtcgccgtggccggcgccggcatcgccgggctcgccttcgccgcc60
gccctgcgccggaccgggatcgactgccacgtgtacgaacaggccgaccagctcatggag120
gtgggcgcgggcgtgcaggtcgcgccgaacgccacccggctgctgcaccggctgggcctg180
cgtgaccgcctgcgtacggtggctgtcgcgccgcaggcgatcgagatgcgccgctgggac240
gacggcacgctgctgcaacgcacccagctgggcagcgtgtgcggacgccgcttcggcgcg300
ccgtactacgtggtgcaccgcgcggacctgcacagcagcctgctgtcgctggtgccgccg360
gaccgggtgcacctgggcgcccgcctcaccgccgtgacgcagaccgccgacgaggcgtac42()
ctgcacctgtccaacggcaccacggtcgcggcggatctcgtcgtgggcgccgacggcatc480
cactcggtcgcgcgggagcagatcgtggcggaccggccgcgcttctccggacagtccatc540
taccgcgggctggtgccggccgagcgggtgccgttcctgctcaccgaaccccgggtgcag600
ttgtggttcgggccggaccagcactgcgtctgctacccggtgtccgccggccggcaggtg660
agcttcggcgcgacggtgcccgccaccgactggcggcaggagtcgtggtcgggccggggc720
gacgtgacgcaactcgcggccgcgtacgcgggctggcacccggacgtcacccggctgatc78()
gccgcggccgaccgggtcggcaggtgggcgctgcacgaccgggacagcatcgaccggctc840
agcgcgggacgggtgaccctgatcggcgacgccgcgcacccgatgctgccgttccaggcg900
cagggcgcgaaccaggccgtcgaggacgcggtggtgctcgcggtctgcctggccggcgtg960
gaaccggcgggcctgggcgccgcgctgcgccgctacgaacggatccgcctgccccggacc1020
acccggatccagcggcagtcccgggccaacgccgagatgttccacctggccgacggcgcc1080
gaccagcgccgccgggacgtcgccgcacaatcctcgt:ccggcctggaccgccacgaatgg114()
ctcttcgggtacgacgccgagaaagccaccacgaccagcgggagcgcctga 119=
SEQ ID NO: 52
LENGTH: 261
CA 02538147 2004-O1-21
TYPE: PRT
ORGANISM: Micromonospora sp. strain 046-ECO11
SEQUENCE: 52
Met Glu Leu Thr Gly Tle Glu Ser Lys Val Ala Leu Val Thr Gly Ala
1 5 10 15
Gly Gln Gly Ile Gly Ala Ala Val Ala Gly Val Leu Ala Arg Ala Gly
20 25 30
A1a Gln Val Ala Ala Val Asp Arg Asn Ala Glu Ala Leu Thr Thr Val
35 40 45
Val Thr Lys Leu Ala Ala Glu Gly Asp Ser Ala Arg Ala Tyr Cys Val
50 55 60
Asp Val Cys Asp Ser Glu Ala Val Asp Ala Leu Val Arg Arg Val Glu
65 70 75 80
Asp Glu Met Gly Pro Val Ala Ile Leu Val Asn Ala Ala Gly Val Leu
85 90 95
His Thr Gly Arg Val Val Glu Leu Ser Asp Arg Gln Trp Arg Arg Thr
100 105 110
Phe Ser Val Asn Ala Asp Gly Val Phe His Val Ser Arg Ala Val Ala
115 120 125
Arg Arg Met Val Gly Arg Arg Arg Gly Ala Ile Val Thr Val Ala Ser
130 135 140
Asn Ala Ala Gly Val Pro Arg Thr Glu Met Ala Ala Tyr Ala Ala Ser
145 150 155 160
Lys Ala Ala Ser Ala Gln Phe Thr Arg Cys Leu Gly Leu Glu Leu Ser
165 170 175
Gly Tyr Gly Ile Arg Cys Asn Val Val Ser Pro Gly Ser Thr Asp Thr
180 185 190
Pro Met Leu Arg Ala Met Leu Gly Glu Gly Ala Asp Pro Ser Ala Val
195 200 205
Ile Glu Gly Thr Pro Gly Ala Tyr Arg Val Gly Ile Pro Leu Arg Lys
210 215 220
Leu Ala Gln Pro Arg Asp Val Ala Glu Ala Val Ala Tyr Leu Val Ser
225 230 235 240
Asp Gln Ala Gly His Val Thr Met His Asp Leu Tyr Val Asp Gly Gly
245 250 255
Ala Ala Leu His Val
260
SEQ ID N0: 53
LENGTH: 786
TYPE: DNA
ORGANISM: Micromonospora sp. strain 046-ECO11
74
CA 02538147 2004-O1-21
SEQUENCE:53
atggaactgaccggaatcgagtcgaaggtcgccctggtcacgggcgcggggcagggcatc 6U
ggcgccgccgtggccggtgtcctggcgagggcgggcgcgcaggtggcggcggtggaccgc 120
aacgccgaggcgctgaccaccgtcgtgacgaagctcgccgccgagggcgactcggcgcgc 180
gcctactgcgtcgacgtgtgcgacagcgaggcggtggacgcgctggtgcgccgggtcgag 240
gacgagatggggccggtcgccatcctggtcaacgccgccggcgtgctgcacaccggacgg 30U
gtcgtcgagctgtcggaccggcagtggcgccggaccttctcggtgaacgccgacggcgtg 360
ttccacgtgtcccgggcggtggcgcggcggatggtgggccgccgtcgtggcgcgatcgtc 420
accgtggcgtcgaacgccgccggggtgccgcgtaccgagatggccgcgtacgccgcctcc 48U
aaggccgcgtccgcgcagttcacccgctgcctggggcttgagctgtccggctacggcatc 540
cggtgcaacgtggtctcgcccggctccaccgacacccccatgctgcgggccatgctcggc 60()
gagggcgccgacccgagcgcggtgatcgagggcacgccgggcgcgtaccgcgtcggcatc 660
ccgctgcgcaagctggcccagccgcgcgacgtggccgaggcggtcgcctatctggtgtcc 720
gaccaggcgggccacgtgaccatgcacgacctgtacgtcgacggcggcgcggccctgcac 780
gtgtga 786
SEQ ID N0: 54
LENGTH: 224
TYPE: PRT
ORGANISM: Micromonospora sp. strain 046-ECO11
SEQUENCE: 54
Met Ala Met Thr Pro Ile Ala Pro Tyr Arg Met Pro Gly Asp Gly Asp
1 5 10 15
Leu Pro Gly Thr Ala Leu Pro Trp Arg Pro His Pro Asp Arg Ala Ala
2G 25 30
Val Leu Val His Asp Leu Gln Arg Tyr Phe Leu Arg Pro Phe Glu Ala
35 40 45
Gly Glu Ser Pro Met Ala Glu Leu Leu Pro Asn Val Ala Lys Leu Leu
50 55 60
Ala Thr Ala Arg Ala Ala Gly Val Pro Val Leu Tyr Thr Ala Gln Pro
65 70 75 80
Gly Gly Met Ser Arg Gln Asp Arg Gly Leu Leu His Asp Leu Trp Gly
85 90 95
Pro Gly Met Ser Ser Ala Glu Asp Asp Arg Gly Ile Val Asp Asp Val
100 105 110
Ala Pro Gln Pro Gly Asp Thr Val Leu Thr Lys Trp Arg Tyr Ser Ala
115 120 125
7$
CA 02538147 2004-O1-21
Phe Phe Arg Ser Asp Leu Glu Glu Arg Leu Arg Gly Ala Gly Arg Asp
130 135 140
Gln Leu Val Val Cys Gly Val Tyr Ala His Met Gly Cys Leu Ile Thr
145 150 155 160
Ala Cys Asp Ala Phe Ser Arg Asp Ile Glu Ala Phe Leu Val Ala Asp
165 170 175
Ala Leu Ala Asp Leu Ser Arg Glu Asp His Leu Met Ala Leu Arg Tyr
180 185 190
Ala Ala Asp Arg Cys Ala Val Pro Leu Trp Thr Ala Asp Val Leu Asp
195 200 205
Gly Leu Asp Ala Ser Ser Gln Arg
Ala Ala Gly Thr
Arg Pro
Asp Gln
210 215 220
SEQ ID 55
N0:
LENGTH: 5
67
TYPE:
DNA
ORGANISM:Micromonospora
sp. strain
046-ECO11
SEQUENCE:55
atggccatgaccccgatcgcgccgtaccgcatgcccggcgacggcgacctgcccggcacc60
gcgctgccctggcgtccgcacccggaccgggccgccgtgctggtgcacgacctgcaacgc120
tacttcctgcgcccgttcgaggccggggagtccccgatggccgaactgctccccaacgtc180
gcgaagctgctcgccacggcgcgggcggccggcgtgccggtgctgtacaccgcgcagccc240
ggcggcatgagccggcaggaccgcgggttgctgcacgacctgtggggccccggcatgagc300
agcgccgaggacgaccggggcatcgtcgacgacgtcgccccgcagccgggcgacacggtg360
etgaccaagtggcgctacagcgcgttcttccgcagcgacctggaggagcgactgcgcggt420
gcgggacgggaccagctcgtggtctgcggcgtgtacgcgcacatggggtgcctgatcacc480
gcctgcgacgcgttcagccgcgacatcgaggcgttcctggtggcggacgcgctggccgac540
ctatcgcgcgaggaccacctgatggcgctgcgctacgccgcggaccgctgcgcggtgccg600
ttgtggacggcggatgtgctggacgggctggcggacgccgccgggcgtccggatcagagc660
agcacccaacgatga 675
SEQ ID NO: 56
LENGTH: 233
TYPE: PRT
ORGANISM: Micromonospora sp. strain 046-ECO11
SEQUENCE: 56
Met Ser Asp Arg Thr Arg Val Val Val Val Gly Gly Thr Ser Gly Ile
1 5 10 15
Gly Arg His Phe Ala Arg Phe Cys Ala Glu Arg Gly Asp Asp Val Val
20 25 30
7G
CA 02538147 2004-O1-21
Ile Thr Gly Arg Ser Ala Ala Arg Thr Lys Thr Val Ala Asp Glu Ile
35 40 45
Gly Gly Arg Thr Arg Gly Leu Ala Leu Asp Leu Ala Glu Pro Glu Thr
50 55 60
Ile Ala Asp Ala Leu Ala Asp Val Pro His Val Asp Arg Leu Val Val
65 70 75 80
Ala Ala Leu Asp Arg Asp Tyr Asn Thr Val Arg Ala Tyr Arg Pro Gly
85 90 95
Asp Ala Ala Arg Leu Leu Thr Val Lys Leu Val Gly Tyr Thr Ala Vai
100 I05 110
Leu His Ala Leu Ala Pro Arg Met Thr Asp Glu Ser Ala Val Val Leu
315 120 125
Leu Gly GIy Leu Ala Ser His Arg Pro Tyr Pro Gly Ser Thr Ser Val
130 135 140
Thr Thr Ala Asn Gly Gly Ile Ser Ala Leu Val Arg Thr Leu Ala Val
145 150 155 160
Glu Leu Ser Pro Val Arg Val Asn Ala Leu His Pro Ser Ile Val Ser
165 170 175
Asp Thr Pro Phe Trp Ser Asp Lys Pro Ala Ala Arg Glu Ala Ala Ala
180 185 190
Thr Arg Ala Leu Ser Arg Arg Pro Val Thr Met Gln Asp Cys Ala GIu
195 200 205
Ala Ile Asp Phe Leu Leu Thr Asn Arg Ser Ile Asn Gly Val Asn Leu
210 215 220
Asn Ile Asp Gly Gly Asp Val Leu Ile
225 230
SEQ ID 57
NO:
LENGTH:
702
TYPE:
DNA
ORGANISM:MiCromonospora
sp. strain
046-ECO11
SEQUENCE:57
atgtcggatcggacccgggtcgtggtcgtcggcggaacctcggggatcgggcggcacttc60
gcccgattctgcgccgaacgcggagacgacgtggtgatcaccggccgttcggcggcccgg120
accaagaccgtggcggacgagatcggcgggcggacccgtgggctcgctctcgacctggcc180
gagccggagacgatcgcggacgcgctcgccgacgtgccgcacgtcgaccggctcgtggtc240
gcggcgctggaccgcgactacaacaccgtccgcgcgtaccggccgggcgacgcggcgcgg300
ctgctgaccgtcaagctggtcggctacacggcggtcctgcacgccctcgccccgcggatg360
accgacgagagcgcagtcgtgctgctcggcggcctggccagccaccggccgtatcccggc420
tccacctccgtcacgaccgccaacggcgggatcagcgcgctggtgcggaccctggctgtg480
77
CA 02538147 2004-O1-21
gaactctcgc cggtccgggt caacgccctg cacccgagca tcgtctccga cacgccgttc 540
tggagcgaca agcccgccgc gcgggaggcc gccgcgaccc gcgcgctcag ccgacggccg 600
gtcaccatgc aggactgcgc cgaggcgatc gacttcctgc tgacgaaccg ctcgataaac 660
ggggtcaacc tgaacatcga cggcggggac gtgctcatct ga 702
SEQ ID NO: 58
LENGTH: 246
TYPE: PRT
ORGANISM: Micromonospora sp. strain 046-ECO11
SEQUENCE: 58
Met Thr Ser Ala Leu Arg Thr Ser Ala Trp Thr Tyr Asp Asp Phe Thr
1 5 10 15
Ser Arg Glu Leu Asp Pro Ala Arg Trp Ala Ile Met Ser Ile Ala Gly
20 25 30
Ala Asp Gly GIn Thr His Arg Tyr Gln Asp Arg Asn Ala Gln VaI Arg
35 40 45
Thr Gly Asp Gly Arg Leu Glu Leu Thr Val Asp Pro Phe Thr Arg Phe
50 55 60
His Asp Thr Asp Pro Arg Gln Asn Asn Ala Lys Gln Met Tyr Arg Ser
65 70 75 80
Val Arg Arg Phe Ala Val Pro Ala Glu Gly Ser Leu Thr Val Glu Val
85 90 95
Glu Met Gly Val Arg Thr Tyr Arg Gln Ile Pro His Asp Leu Leu Asp
100 105 110
Ala Phe Gly Thr Val Asn Leu Phe Asp Leu Glu Thr Gly Val Val Phe
115 120 125
Asn Ala Ala Ala Thr Asn Asp Thr Val Tyr Ala Thr Val Glu Arg Leu
130 135 140
Val Leu Pro Gly Val Thr Gln Pro His Glu His Tyr Ile His Arg Val
145 150 155 160
Val Leu Asp Val Pro Thr Glu Pro GIy Arg Ala His Gly Tyr AIa Ile
165 170 175
Thr Tyr Arg Ala Pro Thr Ser Glu Val Glu Phe His Val Asp Gly Arg
180 185 190
Leu Ala Tyr Trp Ala Arg VaI Pro Val Pro Va1 Thr Gly Phe His Ala
195 200 205
Gly Met Ala Leu Phe Ser AIa Arg Asp Leu Ala Arg Tyr Pro Arg Glu
210 215 220
Gln Arg Glu His Gly Gln Gly Ala Thr Gly Trp Trp Gly Pro Trp Arg
225 230 235 240
7g
CA 02538147 2004-O1-21
Ile Ala Ser Gly Val Arg
245
SEQ ID N0: 59
LENGTH: 74i
TYPE: DNA
ORGANISM: Micromonospora sp. strain 046-ECO11
SEQUENCE:59
atgacgtcggcactgagaaccagcgcgtggacgtacgacgacttcaccagccgcgagctg60
gaccccgcccgctgggcgatcatgtcgatcgccggcgcggacgggcagacccacaggtac120
caggaccgcaacgcccaggtccgcaccggcgacgggcggctggagctgaccgtcgacccg180
ttcacccgcttccacgacaccgatccccggcagaacaacgccaagcagatgtaccggtcg240
gtgcggcgcttcgccgtgccggcggagggctcgctgaccgtcgaggtggagatgggcgtg300
cggacgtaccggcagatcccgcacgacctgctggacgcgttcggcacggtgaacctgttc360
gacctggagaccggcgtcgtgttcaacgccgccgccacgaacgacaccgtgtacgcgacg420
gtcgagcgcctggtgctgcccggcgtgacccagccgcacgagcactacatccaccgggtg480
gtcctggacgtgccgacggagccgggccgggcgcacggatacgccatcacctaccgggcg540
ccgacgtcggaggtggagttccacgtcgacggccggctcgcctactgggcgcgggtcccg60()
gtgccggtgaccggattccacgccggcatggcgctcttctccgcccgcgacctggcccgg66()
tacccccgcgagcagcgggagcacgggcagggcgcgaccgggtggtgggggccgtggcgg720
atcgcctccggcgtcagatga 74:L
SEQ ID NO: 60
LENGTH: 111
TYPE: PRT
ORGANISM: Micromonospora sp. strain 046-ECO11
SEQUENCE: 60
Met Asp Thr Ala Ala Pro Ala Thr Asp Gly Gly Arg Tyr Leu Ala Val
1 5 10 15
His His Ser Ala Glu Phe Arg Glu Leu Arg Arg Arg Ser Ser Thr Phe
20 25 30
Thr Leu Trp Ala Ser Val Ala Phe Phe Gly Trp Trp Phe Leu Gly Ser
35 40 45
Leu Leu Ala Thr Tyr Ala Pro Asp Phe Phe Arg Glu Lys Val Ala Gly
50 55 60
Pro Val Asn Val Gly Leu Leu Phe Val Phe Leu Ser Phe Ala Phe Val
65 70 75 80
Val Thr Leu Ala Ala Phe Tyr Leu Arg Tyr Ala Arg Thr His Leu Asp
85 90 95
CA 02538147 2004-O1-21
Pro Leu Ser Glu Lys Ile Arg Ala Asp Leu Glu Gly Ala Ser Arg
100 105 110
SEQ ID 61
N0:
LENGTH:
336
TYPE:
DNA
ORGANISM:Micromonospora
sp. strain
046-ECO11
SEQUENCE:61
atggacacggcagctccggcaacggacggcggtcgctacctcgccgtccatcacagcgca6t)
gagttcagggaactacggcgacgatcgagcacgttcacgctctgggccagcgtcgccttc120
ttcggctggtggttcctcggcagcctgctcgccacctacgcgccggacttcttccgggag180
aaggtggccggcccggtcaacgtgggtctgctcttcgtcttcctgtcgttcgccttcgtg24()
gtgacgctcgccgccttctacctgcgttacgcccgcacgcatctcgatccgctcagcgag300
aagatccgtgccgacctggaaggagcgtcccgatga 336
SEQ ID 62
N0:
LENGTH:
559
TYPE:
PRT
ORGANISM:Micromonospora
sp. strain
046-ECO11
SEQUENCE: 62
Met Ser Val Ile Leu Ala Asp Pro Pro Pro Pro Val Asp Asn Thr Trp
1 5 10 15
Ala Thr Pro Ala Ile Ala Val Pro Val Thr Ile Val Leu Ala Leu Ala
20 25 30
Val Leu Tyr Leu Val Arg Ser Ala Arg Ala Ser Thr Thr Thr Ala Asp
35 40 45
Gly Phe Leu Leu Ala Asp Arg Arg Ile Gly Pro Val Gln Asn Ala Leu
50 55 60
Ala Val Ala Ser Ala Pro Leu Met Tyr Ser Thr Met Tyr Ile Ile Thr
65 70 75 80
Gly His Ile Aia Leu Ser Gly Tyr Asp Ala Ile Leu Leu Met Thr Ala
85 90 95
Phe Thr Met Gly Thr Met Leu Ala Leu Phe Leu Phe Ala Gly Pro Val
100 105 110
Arg Asn Val Gly Gly Tyr Thr Leu Gly Asp Leu Leu Ala Val Arg Thr
115 120 125
Arg Glu Arg Pro Ala Arg Ile Ala Ser Ala Val Leu Thr Leu Leu Thr
130 135 140
Tyr Val Met Leu Thr Val Ile Met Met Ala Ala Ile Ala Phe Ile Phe
145 150 155 160
Asn Arg Trp Phe Gly Val Asp Ala Leu Val Gly Leu Val Leu Pro Val
CA 02538147 2004-O1-21
165 170 175
Phe Val Val Gly Leu Ile Thr Val Gly Tyr Val Tyr Leu Gly Gly Met
180 185 190
Leu Gly Val Thr Arg Ile Leu Val Phe Lys Leu Val Leu Ser Val Val
195 200 205
Val Val Gly Val Leu Thr Ala Trp Val Leu Ala Arg Phe Asp Leu Asn
210 215 220
Leu Phe Ser Leu Leu Glu Arg Ala Glu Ala Asn Ala Ala Pro Val Pro
225 230 235 240
Ser Gly Ser Asp Leu Leu Gly Pro Gly Arg Leu Phe Gly Glu Gly Ala
245 250 255
Thr Thr Leu Val His Leu Ser Lys Leu Phe Ala Ile Ala Val Gly Val
260 265 270
Ala Ala Ile Pro Phe Leu Phe Met Arg Asn Phe Ala Val Thr Ser Gly
275 280 285
Arg Asp Ala Arg Arg Ser Thr Gly Trp Ala Ser Met Ile Ile Val Gly
290 295 300
Phe Tyr Leu Cys Leu Ser Val Val Gly Leu Gly Ala Val Ala Ile Leu
305 310 315 320
G1y Arg Asp Asn Ile Gly Val Ile Lys Ala His Arg Asp Ile Ser Phe
325 330 335
Pro Lys Leu Aia Asp Glu Leu Gly Gly Pro Val Met Val Gly Ser Leu
340 345 350
Ala Gly Val Ala Val Leu Thr Ile Val Gly Val Phe Ala Pro Leu Leu
355 360 365
His Ser Ala Val Thr Thr Val Thr Lys Asp Leu Asn Val Ile Arg Gly
370 375 380
Arg Arg Leu Asp Pro Ala Ala Glu Leu Arg Asp Ile Lys Arg Asn Thr
385 390 395 400
Leu Ile Ile Gly Val Gly Ser Val Leu Leu Ala Val Val Met Leu Pro
405 410 415
Val Arg Thr His Ile Phe Ile Pro Thr Ser Ile Asp Ile Ala Gly Ala
420 425 430
Val Val Leu Pro Ile Val Val Tyr Ala Leu Phe Trp Arg Arg Phe Asn
435 440 445
Thr Arg Gly Leu Gln Trp Thr Val Tyr Gly Gly Leu Ala Leu Thr Ala
450 455 460
Phe Leu Val Leu Phe Ser Asn Gly Val Ser Gly Glu Pro Asp Ala Ile
465 470 475 480
Phe Pro Asp Arg Asn Phe Lys Phe Val Asp Val Glu Pro Ala Leu Ile
485 490 495
81
CA 02538147 2004-O1-21
Thr Val Pro Val Gly Phe Leu Leu Gly Tyr Leu Gly Ser Ile Thr Ser
500 505 510
Arg Glu Arg Asp Asp Ala Ala Phe Ala Glu Met Gln Val Arg Ser Leu
515 520 525
Thr Gly Ala Val Val Thr Gly Pro Pro Arg Pro Ala Ala Val Asp Asp
530 535 540
Glu Asp Arg Asp Gly Arg Gln Asp Arg Ala Pro Ser Pro Val Ser
545 550 555
SEQ ID 63
NO:
LENGTH:
1680
TYPE:
DNA
ORGANISM:Micromonospora
sp. strain
046-ECO11
SEQUENCE:63
atgagcgtcatcctcgccgacccgccacccccggtcgacaacacgtgggcgacgcccgcg60
atcgccgtgccggtcaccatcgtcctcgcgctcgcggtgctctacctggtccggtcggcg120
cgcgccagcaccaccaccgcggacggcttcctgctggccgaccggcggatcgggccggtg180
cagaacgcgctggcggtggcctccgcgccgctgatgtactcgacgatgtacatcatcacc240
ggccacatcgcgctcagcggctacgacgccatcctgctgatgaccgccttcaccatgggc300
accatgctcgcgctgttcctcttcgccgggccggtgcgcaacgtgggcggctacacgctc360
ggtgacctgctcgcggtccgtacccgggagcggccggcgcggatcgcgtcggcggtgctc420
acgctgctgacgtacgtcatgctgacggtgatcatgatggccgccatcgcgttcatcttc480
aaccgctggttcggcgtcgacgccctcgtcggcctggtcctcccggtgttcgtcgtcggt540
ctgatcacggtggggtacgtgtacctcggcgggatgctcggggtcacccgcatcctggtg600
ttcaagctggtgctgtcggtggtcgtcgtgggcgtgctgaccgcctgggtgctggcccgc660
ttcgacctgaacctcttcagcctgctggagcgggccgaggcgaacgcggcgccggtgccc720
agcggcagcgacctgctgggcccgggccggctgttcggcgagggcgcgaccacgctcgtg780
cacctgtcgaagctgttcgccatcgccgtcggagtggcggccattccgttcctgttcatg840
cgcaacttcgcggtgaccagcgggcgggacgcgcgccggtcgaccgggtgggcgtcgatg900
atcatcgtcgggttctacctgtgcctgtccgtcgtcgggctcggtgccgtcgcgatcctc960
ggccgggacaacatcggcgtcatcaaggcccaccgcgacatcagcttccccaagctcgcc1020
gacgagctcggcggtccggtgatggtcggetccctggccggcgtcgcggtcctgacgatc1080
gtcggcgtcttcgcgccgctgctgcacagcgccgtgacgacggtgaccaaggacctgaac1140
gtgatccgcggccggcggctggatccggccgccgagctgcgggacatcaagcgcaacacc1200
ctgatcatcggcgtcggctccgtgctgctggcggtcgtgatgctgccggtacggacccac1260
CA 02538147 2004-O1-21
atcttcatcccgacctcgatcgacattgccggcgcggtggtcctgccgatcgtcgtctac 1320
gcgttgttctggcggcgtttcaacacccgcggactgcagtggacggtctacggcggcctc 138()
gcgctcaccgcgttcctggtgctgttctccaacggtgtctcgggcgagccggacgccatc 1440
ttcccggaccgcaacttcaagttcgtggacgtcgagcccgcgctgatcacggtgccggtc 1500
ggcttcctgctcggctacctcggctcgatcaccagccgggagcgcgacgacgccgcgttc 1560
gccgagatgcaggtccggtccctcaccggagctgtcgtcacgggaccgccgcggccggcc 1620
gccgtggacgacgaggaccgcgacggccgccaggaccgggcgcccagcccggtgagctga 168()
SEQ ID 64
NO:
LENGTH:
5960
TYPE:
DNA
ORGANISM:Micromonospora rain 046-ECO11
sp. st
SEQUENCE:64
ccacacccctcgggaggcaactgtggatccggtaccggttctggtcgtgggcgcgggccc50
ggtcggcatggtcaccgcgctggcgctcgcccgtcacggcgtcgcctgcgtcctcgtcga120
ccagggcttcgagacgtcggtccatcccaagctggactacgtcaacgcccgcagcatgga180
gttcctccgccagttcggcctcgccgacgacgtccgtgccgccggcgtcgcgcccgagca240
ccgggccgacgtcatctggtcgaccggcctggccggtgagccgatcaccaggtgggggct300
gccctcggtgacgcaggagtggcgccgcatcgccgagcacaacgacggcacccagccggc360
cgagcccggccagcggatctcccagatcgacctggaaccggtcctgcgggcccgctgccg420
gcgggagccccttgtcgacctgcgcctcggcgtacggttcgactcgctgacccaggacga480
cgcgggggtcaccagcgtcctcgccgacgacaccggcggcgaggtccgggtgcggtcgga540
gtacgtggtcgggtgcgacggcgcgtcgagccaggtccgccgggccgtgggcatcggtga60()
ggaggggttegacgtgcccggcctgccgggcgccttcatggtgcacttcaccagccggga66()
cctggacagcctgcaccggcacggccggttctggcactacttcgcgttccggtacgtgat720
catcgcccaggacgaggtcgacacctggaccgcgcacgtcaacggcgtcgacccgaacga780
gttcgacgagccgccggccgacccggaggcgttcctgctcgacacgatccgcaccgagct840
gcggatcgacaaggtgctgctcacctcgcgctggcgtcccggcttcatgctcgccgacag900
gtaccgcgccggccgggtgctgctcgccggtgactcggcccaccggatgttccccaccgg960
cgcgtacggcatgaacaccggcatcggcgacgccgtcgacgtggcctggaagctggccgc1020
tgtcgtccggggcttcggcggccccgggctgctcgacagctacgacgccgaacgccgccc1080
ggtggggcggcgcaacatgcgcacctcgcaccggcacctgggcgtgcacctgcgggcggg1140
cgagctcctgcgcggcggcgccccgctgccgtccgtcgcggccttcctcgacgccgagcg1200
83
CA 02538147 2004-O1-21
gggcgagaac gagtaccggg ggatcgagct cggctaccgc tactccggct cgccggtgct 1260
ctggccggag ggcccggggg agccctcgga cgacccgcgg gcgtacgccc cgacgacctg 1320
gcccggcgcc cgtccgccca gcctcctgct gagcgacggg cagcagatct tcgaccggtt 1380
cgacccggcc tcgttcaccc tcgtggactt caccggtgac ggcgccgccg gtccgctgct 1440
ggcggcggcg gccgcgcggg ggctcccggt cacccacacc gtggtgaccg acccccgggc 1500
tcgtgagctg tgggaacgcg acctcgtcct gctgcggccg gaccaccacg tcgcctggcg 1560
gggaaacacc gtgccgccgg accccgacgc cgtggtccag cgcgtgcggg gtggcggata 1620
ggcgcgacgt gccgtcaccg gcggcccggg tcacgcgcac acgcgaccgg ccggtccggc 1680
tgactctcga ctggaggaca gatgcagcaa tccggttcaa cggcggaacg cagcccactc 1740
gggccgtggg agggcatgcc ggcggtccag caaccggact ggcaggacca cccggcgtac 1800
gcggagacct gtcaggcgtt ggcgtcggcc ccgccgctgg tcccacccgg ggaggtacgg 1860
gggttccggc agctgttgtc ggagctggcg tcgaccgacg ggctcctgct gcagttgggc 1920
gactgcgccg agagcctcta cgagtgcacc ccccggcaca cctcggacaa gatcgaggtc 1980
atcgaccggc tgggggaccg gctcagcgag ctcaccgggc gcaacgtgct gcgggtgggc 2040
cggatggccg ggcagttcgc caagccccgg tcgcaggcga cggagtggca cgacgcgctg 2100
agcatcccct ccttccgcgg ccacatgatc aattccgagc tggccgcgcc cggtacgcgc 2160
aaggccgacc ctcgccgcat gtggtgggcg tacgaggcga gcgaccgggt gcagcgggtc 2220
ctgcgcgccc accgggaggg caaccggcgt gccgcgcgga ccgaggggcc gtggtcgagc 2280
cacgaggccc tggtcgtcga ctacgagtcc cgcctgatcc gccgggaccc ggacacgggc 2340
gagcactacc tggcgtcgac ccacctgccg tgggtggggg agcggacccg ccggtccgcc 2400
gaggcgcacg tggccatgct gtccacggtg gtgaacccgg tcggctgcaa gatcgggccg 2460
gacgccgacc cggacgacgt cctgcgggtg tgcgaggcgc tcgaccCgcg gcgcgatccg 2520
ggccgtctcg tcctgatccc gcggatgggc cgggaccgga tccgggagtc cctgccgccg 2580
atcgtccgcg cggtggtgaa cgcggggcac cccgtgctct ggctgagcga tcccatgcac 2640
ggcaacaccg tcaaggcctc ggtcggcctg aagacgc~gcc acctctccga cgtggtcacc 2700
gaggcgctgt ggttccgcga catcctcgac cagcagcggc agcacgccgc cgggctgcac 2760
atcgaggtcg ccgccaccga cgtgaccgag tgcgtcggcg gttcggtggc cggcgaggag 2820
gacctggcgc ggcactacac ctcgctgtgc gacccgcggc tcaacccggg tcaggccacc 2880
gagctgatcg aagcgtgggc caaggacacc gcgacggtcg gcccgggacc gcggcgctcc 2940
ggcccttcgg cgcggccgga ggtcgccgcc tgacgtcgcc ggtctttgcg ccggccgttt 3000
ccgaactgcg ggaaaattga cagaaggaga cctgccggag caaattcggc caggctagcc 3060
84
CA 02538147 2004-O1-21
gcgccgtagttcgtcgtccactacttgcgtgggtagtgtcaactacccgtgccgggaccg3120
tcggtggtgttgctcagcaggaatcccatcgcaatgatgtgtgagaaggcgtaatccttc318c)
gatcggtgacgcgcgtacctcatcctatccgcactgaatcctgtctcagctgaagcgagt3240
gtttccaatgtggggcagctcaaacacgctggaagtgaagggcaacgacgagagattccc3300
cctgcccgatgcagctacggaggatcggtctgtgcttggcgagacggttccggtttccgc336c)
gctgctgcccggtgactccccgcggctggcgggcgagaacgtcgagcacatccggctgct3420
ggccgcgatgcacgacctcccgccgatcctggtgcaacgcggcacgatgcgggtgatcga3480
cggcatgcaccggctgcgggccgccaagctgcgcggcgacgagaccgtgcgggtgacgtt3540
cttcgacggggacgacgccgcggcgttcctgctctcggtcgacgccaacatcaaacacgg3600
gctgccgttgtcccgcgccgaccgggaggccgccgcc~acccgcatcctgcggttgtatcc3660
gcagtggtcggaccgcgccgtcgccgcggcggccgggctgtcaccgaccacggcgagcgg3720
catccggcgccgcctgctgcaaccggcggcgcgggagggcagccgggtgggacgggacgg3780
gcgggtgcgcccgctggacggctcggcgggccgacggcgggccagcgcggtcatcgcgct3840
ccggccggacgcgcccctgcgtgccatcgcgcaggaggccggggtgtcggtgggcacggc3900
gcgggacgtgcgcgcccggttgcaggcgggccgggaccccgtcctgacctcgcagcgacc3960
ggcggccgagcccgagccggccgccgacgacgggccggaggcgcgcagacgccggctcgg4020
ccagccctccgtgccgcctgtcgactggccggcggtacggggcaacctgatccgggaccc4080
cgcggtgaagtacgccgagctgggccgggccttcgtccgctgggccgacgggcacgtggt4140
ggatccggcggcctggcgcgagttcgtcgacgccgtgccgccgtactggcgcaaatcggt4200
ggccgagctggcccgttcgtgcgccagcgcctggctggcgttcgcccaggaactggagga4260
ccgggcgtgaaaatggcggccggcatatttacggtggttgccgacagcgcgtcgcattcc4320
actgtcgcggccactacccgatcgagtagtggaccggcttgaataacgcgcgttaatgtt4380
ccttcgatccgctgccctcatttttcggtgagcacatttttgcggcggtccaatggagag4440
gagaattcccggtgaacattctgaggcggccgcggaaacggcatctcgggggtgtcgcgg4500
ccgtcgccgcggcgatcgccctggtggcgtcgctgacaaacggtgtggcggctgccccgc4560
aggcgccgaccttcgacctcgacaacgggaacgccctgaccgacgtcatctacccggccc4620
tcaacaccgagccgcgggtcgagtacagcggccggcccgggtcctgggccgcggaccgcg4680
ccatgctcatcgaactgccgtggttcgacgccctggcggcgtaccaccccaccgcggtcg4740
gcatcttctccaccatcggccgccgtcccgccgaggagcacacgacgcgcaacaagaaca4800
tcgccgtcatctactcggcctacacctcgctcagcaagctctacccccagcacgaggcga4860
g5
CA 02538147 2004-O1-21
cctggcagcggatgatggccaccgcgggcctggacc<:ggccgtcaccgcggaggaccgga 4920
ccaccgccagcggcatcggcatcctcgcctcgaagaacgcgatggcggcgcgccggaacg 4980
acggcacgaaccgcgacggcgacgcgggcggccgtcgctacaaccgtgagccgtacgccg 5040
accacaccggctaccggccggtcaacagcccgtacgagctgcgcttcccgtcgcgctggc 5100
agccgaacaccatctccaagcgcgaggtcgtcctgacgcaggagttcgcgacgccccagt 5160
tcggccgggtcaagccgatcaccttcgagcggcccgagcagttccggctcaccccgccgc 5220
cgaaccaccacctgttgaacccgaagggctaccggaagcaggccgacgaggtgctgcgcg 528()
cctcggcgggcctggacgaccgcaagaagatgagcgcggagatcttcagcgacaacatca 5340
cgccgtacggcgccatcgcgcacacgctcctgcggggccggtacaacaccgaggactccg 5400
tccggttcatcgtgatgactgacgtcgccgggttcgacgtggcgatcgcgtcctggtact 5460
acatgcgcaagtacgactcggtgcagccgttcagcgcgatccgccacctgtacccgaaca 5520
agaagctgaccgcgtggggcggcccgggccggggcaccgtcaacgacatcaccggcaccc 5580
agtggcgcagctacctcagctcggtcgccatcgcggctccggattacccgtcggtcaacg 5640
cggcggtctgcgtcgcctacgcccaggtcgcgcgccggttcaccggcacggacaagctga 5700
ccgtcgtgatcccggtccgcaagggctcctcgatcgtggaaccgggcgtgaccccggccg 5760
ccgacatgatgctcacctggaacagctactcggagtgggccgccgagtgcgggcagagcc 5820
gggtctgggccggcgagaacttccccgcctcggtcgcggccgccgaccagtacgcgccgc 5880
agatcggcgaccgtgccttcgacttcgtccagagcaagctgaacgggcgctgacgcccgc 5940
gtaccggtccgtgctgccgg
5960
SEQ ID N0: 65
LENGTH: 532
TYPE: PRT
ORGANISM: Micromonospora 5p. strain 046-ECOll
SEQUENCE: 65
Val Asp Pro Va1 Pro Val Leu Val Val Gly Ala Gly Pro Val Gly Met
I 5 10 15
Val Thr Ala Leu Ala Leu Ala Arg His Gly Val Ala Cys Val Leu Val
20 25 30
Asp Gln Gly Phe Glu Thr Ser Val His Pro Lys Leu Asp Tyr Val Asn
35 40 45
Ala Arg Ser Met Glu Phe Leu Arg Gln Phe Gly Leu Ala Asp Asp Val
50 55 60
Arg Ala Aia Gly Val Ala Pro Glu His Arg Ala Asp Val Iie Trp Ser
65 70 75 80
g6
CA 02538147 2004-O1-21
Thr Gly Leu Ala GIy Glu Pro Ile Thr Arg Trp Gly Leu Pro Ser Val
85 90 95
Th-r G1n Glu Trp Arg Arg Ile Ala Glu His Asn Asp Gly Thr Gln Pro
100 105 110
Ala Glu Pro Gly Gln Arg Ile Ser Gln Ile Asp Leu Glu Pro Val Leu
115 120 125
Arg Ala Arg Cys Arg Arg Glu Pro Leu Val Asp Leu Arg Leu Gly Val
130 135 140
Arg Phe Asp Ser Leu Thr Gln Asp Asp Ala Gly Val Thr Ser Val Leu
145 150 155 160
Ala Asp Asp Thr Gly Gly Glu Val Arg Val Arg Ser Glu Tyr Val Val
165 170 175
Gly Cys Asp Gly Ala Ser Ser Gln Val Arg Arg Ala Val Gly Ile Gly
180 185 190
Glu Glu Gly Phe Asp Val Pro Gly Leu Pro Gly Ala Phe Met Val His
195 200 205
Phe Thr Ser Arg Asp Leu Asp Ser Leu His Arg His Gly Arg Phe Trp
210 215 220
His Tyr Phe Aia Phe Arg Tyr Val Ile Ile Ala Gln Asp Glu Val Asp
225 230 235 240
Thr Trp Thr Aia His VaI Asn Gly Val Asp Pro Asn Glu Phe Asp Glu
245 250 255
Pro Pro Ala Asp Pro Glu Ala Phe Leu Leu Asp Thr Ile Arg Thr Glu
260 265 270
Leu Arg Ile Asp Lys Val Leu Leu Thr Ser Arg Trp Arg Pro Gly Phe
275 280 285
Met Leu Ala Asp Arg Tyr Arg Ala Gly Arg Val Leu Leu Ala Gly Asp
290 295 300
Ser Ala His Arg Met Phe Pro Thr Gly Ala Tyr Gly Met Asn Thr Gly
305 310 315 320
Ile Gly Asp Ala Val Asp Val Ala Trp Lys Leu Ala Ala Val Val Arg
325 330 335
Gly Phe Gly Gly Pro Gly Leu Leu Asp Ser Tyr Asp Ala Glu Arg Arg
340 345 350
Pro Val Gly Arg Arg Asn Met Arg Thr Ser His Arg His Leu Gly Val
355 360 365
His Leu Arg Ala GIy Glu Leu Leu Arg Gly Gly Ala Pro Leu Pro Ser
370 375 380
Val AIa Ala Phe Leu Asp Ala Glu Arg Gly Glu Asn Glu Tyr Arg Gly
385 390 395 400
Ile Glu Leu Gly Tyr Arg Tyr Ser Gly Ser Pro Val Leu Trp Pro Glu
CA 02538147 2004-O1-21
405 410 415
Gly Pro Gly Glu Pro Ser Asp Asp Pro Arg Ala Tyr Ala Pro Thr Thr
420 425 430
Trp Pro Gly Ala Arg Pro Pro Ser Leu Leu Leu Ser Asp Gly Gln Gln
435 440 445
Ile Phe Asp Arg Phe Asp Pro Ala Ser Phe Thr Leu Val Asp Phe Thr
450 455 460
Gl~° Asp Gly Ala Ala Gly Pro Leu Leu Ala Ala Ala Ala Ala Arg Gly
465 470 475 480
Leu Pro Val Thr His Thr Val Val Thr Asp Pro Arg Ala Arg Glu Leu
485 490 495
Trp Glu Arg Asp Leu Val Leu Leu Arg Pro Asp His His Val Ala Trp
500 505 510
Arg Gly Asn Thr Val Pro Pro Asp Pro Asp Ala Val Val Gln Arg Val
515 520 525
Arg Gly Gly Gly
530
SEQ ID 66
N0:
LENGTH:
1599
TYPE:
DNA
ORGANISM:Micromonospora
sp. strain
046-ECO11
SEQUENCE:56
gtggatccggtaccggttctggtcgtgggcgcgggcccggtcggcatggtcaccgcgctg60
gCgCtCgCCCgtcacggcgtcgcctgcgtcctcgtcgaccagggcttcgagacgtcggtc12c)
catcccaagctggactacgtcaacgcccgcagcatggagttcctccgccagttcggcctc18t)
gccgacgacgtccgtgccgccggcgtcgcgcccgagcaccgggccgacgtcatctggtcg24c)
accggcctggccggtgagccgatcaccaggtgggggctgccctcggtgacgcaggagtgg300
cgccgcatcgccgagcacaacgacggcacccagccggccgagcccggccagcggatctcc360
cagatcgacctggaaccggtcctgcgggcccgctgccggcgggagccccttgtcgacctg420
cgcctcggcgtacggttcgactcgctgacccaggacgacgcgggggtcaccagcgtcctc480
gccgacgacaccggcggcgaggtccgggtgcggtcggagtacgtggtcgggtgcgacggc540
gcgtcgagccaggtccgccgggccgtgggcatcggtgaggaggggttcgacgtgcccggc600
ctgccgggcgccttcatggtgcacttcaccagccgggacctggacagcctgcaccggcac660
ggccggttctggcactacttcgcgttccggtacgtgatcatcgcccaggacgaggtcgac720
acctggaccgcgcacgtcaacggcgtcgacccgaacgagttcgacgagccgccggccgac78U
ccggaggcgttcctgctcgacacgatccgcaccgagctgcggatcgacaaggtgctgctc840
acctcgcgctggcgtcccggcttcatgctcgccgacaggtaccgcgccggccgggtgctg900
88
CA 02538147 2004-O1-21
ctcgccggtgactcggcccaccggatgttccccaccggcgcgtacggcatgaacaccggc 960
atcggcgacgccgtcgacgtggcctggaagctggccgctgtcgtccggggcttcggcggc 1020
cccgggctgctcgacagctacgacgccgaacgccgcccggtggggcggcgcaacatgcgc 1080
acctcgcaccggcacctgggcgtgcacctgcgggcgc)gcgagctcctgcgcggcggcgcc 1140
ccgctgccgtccgtcgcggccttcctcgacgccgagcggggcgagaacgagtaccggggg 1200
atcgagctcggctaccgctactccggctcgccggtgctctggccggagggcccgggggag 126()
ccctcggacgacccgcgggcgtacgccccgacgacctggcccggcgcccgtccgcccagc 1320
ctcctgctgagcgacgggcagcagatcttcgaccggttcgacccggcctcgttcaccctc 1380
gtggacttcaccggtgacggcgccgccggtccgctgctggcggcggcggccgcgcggggg 1440
ctcccggtcacccacaccgtggtgaccgacccccgggctcgtgagctgtgggaacgcgac 1500
ctcgtcctgctgcggccggaccaccacgtcgcctggcggggaaacaccgtgccgccggac 1560
cccgacgccgtggtccagcgcgtgcggggtggcggatag 1599
SEQ ID NO: 67
LENGTH: 423
TYPE: PRT
ORGANISM: Micromonospora sp. strain 046-ECO11
SEQUENCE: 67
Met Gln Gln Ser Gly Ser Thr Ala Glu Arg Ser Pro Leu Gly Pro Trp
1 5 10 15
Glu Gly Met Pro Ala Val Gln Gln Pro Asp Trp Gln Asp His Pro Ala
20 25 30
Tyr Ala Glu Thr Cys Gln Ala Leu Ala Ser AIa Pro Pro Leu Val Pro
35 40 45
Pro Gly Glu Val Arg Gly Phe Arg Gln Leu Leu Ser Glu Leu Ala Ser
50 55 60
Thr Asp Giy Leu Leu Leu Gln Leu Gly Asp Cys Ala Glu Ser Leu Tyr
65 70 75 80
Giu Cys Thr Pro Arg His Thr Ser Asp Lys Ile Glu Val Ile Asp Arg
85 90 95
Leu Gly Asp Arg Leu Ser Glu Leu Thr Gly Arg Asn Val Leu Arg Val
100 105 110
G1y Arg Met Ala Gly Gln Phe Ala Lys Pro Arg Ser Gln Ala Thr Glu
115 120 125
Trp His Asp Ala Leu Ser Ile Pro Ser Phe Arg Gly His Met Ile Asn
130 135 140
Ser Glu Leu Ala Ala Pro Gly Thr Arg Lys Ala Asp Pro Arg Arg Met
g9
CA 02538147 2004-O1-21
145 150 155 160
Trp Trp Ala Tyr Glu Ala Ser Asp Arg Val Gln Arg Val Leu Arg Ala
165 170 175
His Arg Glu Gly Asn Arg Arg Ala Ala Arg Thr Glu Gly Pro Trp Ser
180 185 190
Ser His Glu Ala Leu Val Val Asp Tyr Glu Ser Arg Leu Ile Arg Arg
195 200 205
Asp Pro Asp Thr Gly Glu His Tyr Leu Ala Ser Thr His Leu Pro Trp
210 215 220
Val Gly Glu Arg Thr Arg Arg Ser Ala Glu Ala His Val Ala Met Leu
225 230 235 240
Ser Thr Val Val Asn Pro Val Gly Cys Lys Ile Gly Pro Asp Ala Asp
245 250 255
Pro Asp Asp Val Leu Arg Val Cys Glu Ala Leu Asp Pro Arg Arg Asp
260 265 270
Pro Gly Arg Leu Val Leu Ile Pro Arg Met Gly Arg Asp Arg Ile Arg
275 280 285
Glu Ser Leu Pro Pro Ile Val Arg Ala Val Val Asn Ala Gly His Pro
290 295 300
Val Leu Trp Leu Ser Asp Pro Met His Gly Asn Thr Val Lys Ala Ser
305 310 315 320
Val Gly Leu Lys Thr Arg His Leu Ser Asp Val Val Thr Glu Ala Leu
325 330 335
Trp Phe Arg Asp Ile Leu Asp Gln Gln Arg Gln His Ala Ala Gly Leu
340 345 350
His Ile Glu Val Ala Ala Thr Asp Val Thr Glu Cys Val Gly Gly Ser
355 360 365
Val Ala Gly Glu Glu Asp Leu Ala Arg His Tyr Thr Ser Leu Cys Asp
370 375 380
Pro Arg Leu Asn Pro Gly Gln Ala Thr Glu Leu Ile Glu Ala Trp Ala
385 390 395 400
Lys Asp Thr Ala Thr Val Gly Pro Gly Pro Arg Arg Ser Gly Pro Ser
405 410 415
Ala Arg Pro Glu Val Ala Ala
420
SEQ ID NO: 68
LENGTH: 1272
TYPE: DNA
ORGANISM: Micromonospora sp. strain 046-ECO11
SEQUENCE: 68
atgcagcaat ccggttcaac ggcggaacgc agcccactcg ggccgtggga gggcatgccg 60
9 C)
CA 02538147 2004-O1-21
gcggtccagcaaccggactggcaggaccacccggcgtacgcggagacctgtcaggcgttg 120
gcgtcggccccgccgctggtcccacccggggaggtacgggggttccggcagctgttgtcg 180
gagctggcgtcgaccgacgggctcctgctgcagttgggcgactgcgccgagagcctctac 240
gagtgcaccccccggcacacctcggacaagatcgagc~tcatcgaccggctgggggaccgg 300
ctcagcgagctcaccgggcgcaacgtgctgcgggtgggccggatggccgggcagttcgcc 360
aagccccggtcgcaggcgacggagtggcacgacgcgctgagcatcccctccttccgcggc 420
cacatgatcaattccgagctggccgcgcccggtacgcgcaaggccgaccctcgccgcatg 480
tggtgggcgtacgaggcgagcgaccgggtgcagcgggtcctgcgcgcccaccgggagggc 540
aaccggcgtgccgcgcggaccgaggggccgtggtcgagccacgaggccctggtcgtcgac 600
tacgagtcccgcctgatccgccgggacccggacacgggcgagcactacctggcgtcgacc 660
cacctgccgtgggtgggggagcggacccgccggtccgccgaggcgcacgtggccatgctg 720
tccacggtggtgaacccggtcggctgcaagatcgggccggacgccgacccggacgacgtc 780
ctgcgggtgtgcgaggcgctcgacccgcggcgcgatccgggccgtctcgtcctgatcccg 840
cggatgggccgggaccggatccgggagtccctgccgccgatcgtccgcgcggtggtgaac 900
gcggggcaccccgtgctctggctgagcgatcccatgcacggcaacaccgtcaaggcctcg 960
gtcggcctgaagacgcgccacctctccgacgtggtcaccgaggcgctgtggttccgcgac 1020
atcctcgaccagcagcggcagcacgccgccgggctgcacatcgaggtcgccgccaccgac lOBU
gtgaccgagtgcgtcggcggttcggtggccggcgaggaggacctggcgcggcactacacc 1140
tcgctgtgcgacccgcggctcaacccgggtcaggccaccgagctgatcgaagcgtgggcc 1200
aaggacaccgcgacggtcggcccgggaccgcggcgctccggcccttcggcgcggccggag 1260
gtcgccgcctga 127'?
SEQ ID NO: 69
LENGTH: 340
TYPE: PRT
ORGANISM: Micromonospora sp. strain 046-ECOll
SEQUENCE: 69
Met Trp Gly Ser Ser Asn Thr Leu Glu Val Lys GIy Asn Asp Glu Arg
1 5 10 15
Phe Pro Leu Pro Asp Ala Ala Thr Glu Asp Arg Ser Val Leu Gly Glu
20 25 30
Thr Val Pro Val Ser Ala Leu Leu Pro Gly Asp Ser Pro Arg Leu Ala
35 40 45
Gly Glu Asn Val Glu His Ile Arg Leu Leu Ala Ala Met His Asp Leu
50 55 60
91
CA 02538147 2004-O1-21
Pro Pro Ile Leu Val Gln Arg Gly Thr Met Arg Val Ile Asp Gly Met
65 70 75 80
His Arg Leu Arg Ala Ala Lys Leu Arg Gly Asp Glu Thr Val Arg Val
85 90 95
Thr Phe Phe Asp Gly Asp Asp Ala Ala Ala Phe Leu Leu Ser Val Asp
100 105 110
Ala Asn Ile Lys His Gly Leu Pro Leu Ser Arg Ala Asp Arg Glu Ala
115 120 125
Ala Ala Thr Arg Ile Leu Arg Leu Tyr Pro Gln Trp Ser Asp Arg Ala
130 135 140
Val Ala Ala Ala Ala Gly Leu Ser Pro Thr Thr Ala Ser Gly Ile Arg
145 150 155 160
Arg Arg Leu Leu Gln Pro Ala Ala Arg Glu Gly Ser Arg Val Gly Arg
165 170 175
Asp Gly Arg Val Arg Pro Leu Asp Gly Ser Ala Gly Arg Arg Arg Ala
180 185 190
Ser Ala Val Ile Ala Leu Arg Pro Asp Ala Pro Leu Arg Ala Ile Ala
195 200 205
Uln Glu Ala Gly Val Ser Val Gly Thr Ala Arg Asp Val Arg Ala Arg
210 215 220
Leu Gln Ala Gly Arg Asp Pro Val Leu Thr Ser Gln Arg Pro Ala Ala
225 230 235 240
Glu Pro Glu Pro Ala Ala Asp Asp Gly Pro Glu Ala Arg Arg Arg Arg
245 250 255
Leu Gly Gln Pro Ser Val Pro Pro Val Asp Trp Pro Ala Val Arg Gly
260 265 270
Asn Leu IIe Arg Asp Pro Ala Val Lys Tyr Ala Glu Leu Gly Arg Ala
275 280 285
Phe Val Arg Trp Ala Asp Gly His Val Val Asp Pro Ala Ala Trp Arg
290 295 300
Glu Phe Vai Asp Ala Val Pro Pro Tyr Trp Arg Lys Ser Val Ala Glu
305 310 315 320
Leu Ala Arg Ser Cys Ala Ser Ala Trp Leu Ala Phe Ala Gln Glu Leu
325 330 335
Glu Asp Arg Ala
340
SEQ ID NO: 70
LENGTH: 1023
TYPE: DNA
ORGANISM: Micromonospora sp. strain 046-ECO11
SEQUENCE: 70
92
CA 02538147 2004-O1-21
atgtggggcagctcaaacacgctggaagtgaagggcaacgacgagagattccccctgccc5c)
gatgcagctacggaggatcggtctgtgcttggcgagacggttccggtttccgcgctgctg120
cccggtgactccccgcggctggcgggcgagaacgtcgagcacatccggctgctggccgcg180
atgcacgacctcccgccgatcctggtgcaacgcggcacgatgcgggtgatcgacggcatg240
caccggctgcgggccgccaagctgcgcggcgacgagaccgtgcgggtgacgttcttcgac300
ggggacgacgccgcggcgttcctgctctcggtcgacgccaacatcaaacacgggctgccg360
ttgtcccgcgccgaccgggaggccgccgccacccgcatcctgcggttgtatccgcagtgg420
tcggaccgcgccgtcgccgcggcggccgggctgtcaccgaccacggcgagcggcatccgg480
cgccgcctgctgcaaccggcggcgcgggagggcagccgggtgggacgggacgggcgggtg540
cgcccgctggacggctcggcgggccgacggcgggccagcgcggtcatcgcgctccggccg600
gacgcgcccctgcgtgccatcgcgcaggaggccggggtgtcggtgggcacggcgcgggac660
gtgcgcgcccggttgcaggcgggccgggaccccgtcctgacctcgcagcgaccggcggcc720
gagcccgagccggccgccgacgacgggccggaggcgcgcagacgccggctcggccagccc780
tccgtgecgcctgtcgactggccggcggtacggggcaacctgatccgggaccccgcggtg840
aagtacgccgagctgggccgggccttcgtccgctgggccgacgggcacgtggtggatccg900
gcggcctggcgcgagttcgtcgacgccgtgccgccgtactggcgcaaatcggtggccgag960
ctggcccgttcgtgcgccagcgcctggctggcgttcgcccaggaactggaggaccgggcg1020
tga 1023
SEQ ID NO: 71
LENGTH: 493
TYPE: PRT
ORGANISM: Micromonospora sp. strain 046-ECOll
SEQUENCE: 71
Val Asn Ile Leu Arg Arg Pro Arg Lys Arg His Leu Gly Gly Val Ala
1 5 10 15
Ala Val Ala Ala Ala Ile Ala Leu Val Ala Ser Leu Thr Asn Gly Val
20 25 30
Ala Ala Ala Pro G1n Ala Pro Thr Phe Asp Leu Asp Asn Gly Asn Ala
35 40 45
Leu Thr Asp Val Ile Tyr Pro Ala Leu Asn Thr Glu Pro Arg Val Glu
50 55 60
Tyr Ser Gly Arg Pro Gly Ser Trp Ala Ala Asp Arg Ala Met Leu Ile
65 70 75 80
G1u Leu Pro Trp Phe Asp Ala Leu Ala Ala Tyr His Pro Thr Ala Val
85 90 95
93
CA 02538147 2004-O1-21
Gly Ile Phe Ser Thr Ile Gly Arg Arg Pro Ala Glu Glu His Thr Thr
100 105 110
Arg Asn Lys Asn Ile Ala Val Ile Tyr Ser Ala Tyr Thr Ser Leu Ser
115 120 125
Lys Leu Tyr Pro Gln His Glu Ala Thr Trp Gln Arg Met Met Ala Thr
130 135 140
Ala Giy Leu Asp Pro Ala Val Thr Ala Glu Asp Arg Thr Thr Ala Ser
145 150 155 160
Gly Ile Gly Ile Leu Ala Ser Lys Asn Ala Met Ala Ala Arg Arg Asn
165 170 175
Asp Gly Thr Asn Arg Asp Gly Asp Ala Gly Gly Arg Arg Tyr Asn Arg
180 185 190
Glu Pro Tyr Ala Asp His Thr Gly Tyr Arg Pro Val Asn Ser Pro Tyr
195 200 205
Glu Leu Arg Phe Pro Ser Arg Trp Gln Pro Asn Thr Ile Ser Lys Arg
210 215 220
Glu Val Val Leu Thr Gln Glu Phe Ala Thr Pro Gln Phe Gly Arg Val
225 230 235 240
Lys Pro Ile Thr Phe Glu Arg Pro Glu Gln Phe Arg Leu Thr Pro Pro
245 250 255
Pro Asn His His Leu Leu Asn Pro Lys Gly Tyr Arg Lys Gln Ala Asp
260 265 270
Glu Val Leu Arg Ala Ser Ala Gly Leu Asp Asp Arg Lys Lys Met Ser
275 280 285
Ala Giu Ile Phe Ser Asp Asn Ile Thr Pro Tyr Gly Ala Ile Ala His
290 295 300
Thr Leu Leu Arg Gly Arg Tyr Asn Thr Glu Asp Ser Val Arg Phe Ile
305 310 315 320
Val Met Thr Asp Val Ala Gly Phe Asp Val Ala Ile Ala Ser Trp Tyr
325 330 335
Tyr Met Arg Lys Tyr Asp Ser Val Gln Pro Phe Ser Ala Ile Arg His
340 345 350
Leu Tyr Pro Asn Lys Lys Leu Thr Ala Trp Gly Gly Pro Gly Arg Gly
355 360 365
Thr Val Asn Asp Ile Thr Gly Thr Gln Trp Ar_g Ser Tyr Leu Ser Ser
370 375 380
Val Ala Ile Ala Ala Pro Asp Tyr Pro Ser VaI Asn Ala Ala Val Cys
385 390 395 400
Val Ala Tyr Ala Gln Val Ala Arg Arg Phe Thr Gly Thr Asp Lys Leu
405 410 415
94
CA 02538147 2004-O1-21
Thr Val Val Ile Pro Val Arg Lys Gly Ser Ser Ile Val Glu Pro Gly
420 425 430
Val Thr Pro Ala Ala Asp Met Met Leu Thr Trp Asn Ser Tyr Ser Glu
435 440 445
Trp Ala Ala Glu Cys Gly Gln Ser Arg Val Trp Ala Gly Glu Asn Phe
450 455 460
Pro Ala Ser Val Ala Ala Ala Asp Gln Tyr Ala Pro Gln Ile Gly Asp
465 470 475 480
Arg Ala Phe Asp Phe Val Gln Ser Lys Leu Asn Gly Arg
485 490
SEQ ID NO: 72
LENGTH: 1482
TYPE: DNA
ORGANISM: Micromonospora sp. strain 046-EC011
SEQUENCE:72
gtgaacattctgaggcggccgcggaaacggcatctcgggggtgtcgcggccgtcgccgcg60
gcgatcgccctggtggcgtcgctgacaaacggtgtggcggctgccccgcaggcgccgacc120
ttcgacctcgacaacgggaacgccctgaccgacgtc<~tctacccggccctcaacaccgag180
ccgegggtcgagtacagcggccggcccgggtcctgggccgcggaccgcgccatgctcatc240
gaactgccgtggttcgacgccctggcggcgtaccaccccaccgcggtcggcatcttctcc300
accatcggccgccgtcccgccgaggagcacacgacgr_gcaacaagaacatcgccgtcatc360
tactcggcctacacctcgctcagcaagctctacccccagcacgaggcgacctggcagcgg420
atgatggccaccgcgggcctggacccggccgtcaccgcggaggaccggaccaccgccagc480
ggcatcggcatcctcgcctcgaagaacgcgatggcggcgcgccggaacgacggcacgaac54c)
cgcgacggcgacgcgggcggccgtcgctacaaccgtgagccgtacgccgaccacaccggc600
taccggccggtcaacagcccgtacgagctgcgcttcccgtcgcgctggcagccgaacacc660
atctccaagcgcgaggtcgtcctgacgcaggagttcgcgacgccccagttcggccgggtc720
aagccgatcaccttcgagcggcccgagcagttccggctcaccccgccgccgaaccaccac780
ctgttgaacccgaagggctaccggaagcaggccgacgaggtgctgcgcgcctcggcgggc840
ctggacgaccgcaagaagatgagcgcggagatcttcagcgacaacatcacgccgtacggc900
gccatcgcgcacacgctcctgcggggccggtacaacaccgaggactccgtccggttcatc960
gtgatgactgacgtcgccgggttcgacgtggcgatcgcgtcctggtactacatgcgcaag1020
tacgactcggtgcagccgttcagcgcgatccgccacctgtacccgaacaagaagctgacc1080
gcgtggggcggcccgggccggggcaccgtcaacgacatcaccggcacccagtggcgcagc1140
tacctcagctcggtcgccatcgcggctccggattacccgtcggtcaacgcggcggtctgc1200
CA 02538147 2004-O1-21
gtcgcctacgcccaggtcgcgcgccggttcaccggcacggacaagctgaccgtcgtgatc1260
ccggtccgcaagggctcctcgatcgtggaaccgggcgtgaccccggccgccgacatgatg1320
ctcacctggaacagctactcggagtgggccgccgagtgcgggcagagccgggtctgggcc1380
ggcgagaacttccccgcctcggtcgcggccgccgaccagtacgcgccgcagatcggcgac1440
cgtgccttcgacttcgtccagagcaagctgaacgggcgctga 148:2
SEQ ID N0: 73
LENGTH: 9762
TYPE: DNA
ORGANISM: Micromonospora sp. strain 046-ECOll
SEQUENCE:73
cagccacggcgttccgaccccccgcaagatggcttgtatagcaaggtatcttgcgatgca60
tggacggggcacgtgagcggatcactacgaacatccgcaagggcgtgctggagtactgcg120
tgctcgccctgctctcgcggcgcgacatgtacggcctggaactggccgactggctcgccg180
tccgcggtctgaccgcgagcgagggcagcctgtatccgctgctcgcccgcatgcggcagg240
ccggctccgtgcagacccggtgggtggcccccgagcaggggcacgcccggcggtactacg300
cgatcaccgaccaggggcgggcgcacctgcgggtgttcgcggcggtgtggcaggagatcc360
agccgcacgtggacgacctgatgggggaggaagcatgagcgacgacggcctcccggaggc42i)
ggcgtggacctatctgcgcgcgctcgacgcggagttgtccgacgtcccgtccggcacggc480
ggaggagatcgtcgcggatgtccgcgcgcacatcgccgacgccctcgacagcggacggag540
cgcccacgagatcctcgccggcctcggcgccgcgcgggacgtggcccggcaggcgcgcga600
ggagctggggctgccggcccaggaccgcccggcccgggccggccggaccctgtccctggc660
cgcggtggcggtcggcgtgctgatcgccgtgtgcgtgagcttcctgctgccgtccgcagt720
gccggtggagccgatccaggccggccccggcgagcagggcgtcctccgccggctcggccc780
cggaatcgcgctgctcacgctgctgccggcgctcgtcgcggccgcgccgctcgtggcgcc840
cgcccgggcacgtgccggggtacggttcgccggcgcggcggtcctgacgatgttcgcctg900
cgcggccggcgagacgggcctgtactacttcccgctcgcgctgatggcctgggcggcggc960
gatcgtgccgtgggccctgcggcgcggagccggtggacggtggtggcgctatctgaccgg1020
tggattcgtggcgatgcccggcgtgctggtggcggtcgcgtcggccggtggctcggtcgg1080
cgtcggctgggtcggcgcggcgctgtggatcgccgggccgctcgcggccggcgcgctgtg1140
cgcctacgggatccgggccggctacgccgtgaccgcc;ctggccggcgcgctggccatagc1200
gctctcgatggccgagcgcggcttcctgttcgccgccttctggctgttcggcgggctgta126()
cctggcgctcggcgccgctgcgtacaccgcctcgcgggccgtcgacggcgacgccgccgc132()
96
CA 02538147 2004-O1-21
gacgcccggcccgccggcccggccggaacccgcgccggcccccggaggctgacccggggg1380
ccgtggcgccggccggctaggcggggacggcctgcgggtcgccggcggcgtcgtgcgcgg1440
ccatcgtctcctgccggacgggctcctcgcgcaggatcgccgcgtgcagccacgcgtccg1500
ggatggcgaagccgtccacgagcgtgcgcatgtccgggcgcagctccttgagcagcccgt1560
tcaccacgctggtgatggtcttcgagcgggccggggtgagccggccgtgctcgagcagcc1620
agcccttgttcgcctcgatcacggtgagcgcgtacaggtcgcagacccgggacagcagtt1680
ccttgaccgccgggtcggcgatggcgtcgatcccggcgacgaacgcctccagcgtcaccc1740
ggtcgatgtgcgccgcggcgacggcgaggacgtggtcctggacgtcgttgaagatgtcga1800
aggggcggtccttcttggtggacgcgccaccgcgcaggcggcggaccgcgctgtcgagca1860
ggtgctcctcgcggtcctcgaagagcttgagctgccagccccggtcggtgacggcgacct1920
cgtcgtcgcgcccgggcacggcgctgaccagacgtgcgatcagcgcccgcgcggcggtgc1980
gttccagcaccatctcgcgtacctgctcggccacgaaggaggcgcgtccccagccgtcga2040
gcgagccgaactcgtcccggtagccggtcagcagccccttggcgaccagttgcagcagca2100
ccgtgttgtcgccctcgaaggtggtgaagacatcggtgtcggccttgaggctgggcaggc2160
ggttctcggacaggtagccggcgccgccacacgcctcccggcagatctggatggtgcggg2220
tggcgtgccaggtctgcgccgccttcagaccggcggcccgggactccagctcccgctgcc2280
ggtgctcgtcgaccggcccgtcgccgccctggatgtcgtcgagcgccgcgaccagctccg2340
cctgggcgaaggtcagcgcgtacgtggtggccagcgcgggcagcagcttgcgctggtgcg2400
ccaggtagtcgttgagcagcacctcgcggtcgccgtcggcgtcggcgaactgccggcgga2460
tgtcgccgtagcgcaccgcgatggccagcgccgacttggtggccgccgacgcggcgccgc2520
CCaCaCtcaCcCggCCCCggaccagggtgcccagcatggtgaagaagcgccgggagtcgt2580
tctcgatcgggctggagtacgtgccgtcctcggcgacctgcgcgtactggtccagcagca2640
tctcccgcggcacccgcacgtggtcgaagctgagccgcccgttgtccacgccgagcaggc2700
cggccttgggcccggcgtcgccgatggtcacgccgggcatcggcttgccgtgctcgtcgc2760
ggatcggcaccagccaggcgtgcaccccgtggcggcgcccgccggtgacgagctgggcga2820
acaccacagccatccgcccgtcccgggccgcgttgccgatgtagtccttgcgcgcggcct2880
cgtgcggggtgtgcaggtcgaaggtctgcgtctgcgggtcgtagacgcaggtggtgcgca2940
gttgctgcacgtccgagccgtggccggtctcggtcatcgcgaagcagccgaagagccggc3000
ccgcgacgatgtcccgcaggtaggcgtcgtggtgccgcttcgtgccgagggcggcgaccg3060
cgccgccgaacaggccccactgcacgccggccttcaccatcagtgacaggtccacctggg3120
ccagcatctcggtggcgacgatcgaggcgcccacgtcgccgcggccgccgtactcggcgg3180
97
CA 02538147 2004-O1-21
ggaaaccgga ggcgatgccc agctcgacgg ggagttcgga cagcagccgg gtgatgcgct 3240
cgcgggcctg gtcaccggtc tcgccgtaca ccgggaggaa gcgttcgtcg aggtgttcgc 3300
ggtgcgcccg gcggacctcg gcccaccggc cgtcgagcgc ttcccgcagg cgtgtgacgt 3360
cgatgcggcc ggatgcgtga tcgagcattg tcactcctcg gggcagcgga catttgcgta 3420
tactctcggc ctgatcaaca ttaccggcgg tgatcgcacc ccgctggcgg agcgcgtggt 3480
gagcccggcc acccccggcg gttcggccac ccgtgaagct gaggttaggc tgtcctcact 3540
tcacagcact ggaggcatcc cctcgtgtcc ccgcttcccc ccggcagcgc cgtcaccgcc 3600
cggcacgtgc tccgccaggc gctgcgccgc cagcgccgcc cggtgctgat cggcgtgacc 3660
ctgctcgggc tgcaccaggt caccgaggcg ctcgtgccgg tggcgatcgg cgtcatcatc 3720
gaccgggccg tggtgaccgg cgacccgtgg gcgctcgcgt actccgtcgc cggcctcgcc 3780
gccctgttca ccgtgctggc gttcgcctac cgcaacggcg cccgccaggc gttcgcggcg 3840
gtggaacggg aggcgcacct gctgcgggtc gagctggccg agcgcgcgct cgacccgcgc 3900
gggcaccgct ccggcctgcg cgacggcgag ctgctctcgg tcgccgcctc cgacgccgaa 3960
ctctccgcgt acgtggtccg ggtggccggc ttcggcgtcg ccgcggtgag cgcgctgacc 4020
gtcgcggcgg tcgcgctgct ggtcatcgac gtcccgctcg gactcggcgt gctcatcggc 4080
gtaccggtgc tggtcctggc gctgcaacgg atggcgccgc tgctgtcccg gcgcagcgcc 4140
tcccagcagg aggccctcgc ggagaccacg gcgctcgccg tggacctcgt ctccggcctg 4200
cgcgtgctgc gcggcatcgg cgcccagcac cacgccgccg gccggtacgc cgaggccagc 4260
cgacgcgccc tcgccgtgac gctgcgcgcc gccaacacca agggcctgca cctcgggctc 4320
accaccgccg cgaacggcct cttcctcgcc gccgtcgccg gggtcgccgg ctggctcgcg 4380
ctgcgcggcc ggctcaccat cggcgagctg gtcaccgtgg tcgggctcgc gcagttcgtc 4440
gccgagccgg tgcagacgct gggctactgc gtgcagctgt tcgcgatggc ccgcgcctcc 4500
gccgcccggg tcgggcgcgt gctcggcgcc gagccgctga cccggccggg cagcgcgccc 4560
cggccggacc gcacggacgg gccgcggctc gtcctcgacc acgtcggcca cgccgcgctg 4620
gacggggtgt gcctgcgcgt cgacccggga gagatcgtcg gcgtcctggc gtacgacccg 4680
gccgacgcgg acgcgctggt ggcgctgctg tccgggr_ggg tgcccgcgga ccggcgccgg 4740
ggcacggtac gcgtcgacgg ggtacccgcc gacgacctgg acgtcgacgc gctgcgcggc 4800
gccgtcctgg tcgagccgca cgacgtgacg ctgttcgagg gaaccgtggc cgccaacctc 4860
gccgccggga gcaggaccga ggaggggcgc ctgcgcgccg cggtccgggc ggccgcggcg 4920
gacgacgtgg tggacgcgca ccccggcggc ctcggccacc ggctcgtcga gcggggcgcc 4980
98
CA 02538147 2004-O1-21
aacctctccg gcgggcagcg ccagcggctc gggctggcgc gggcgctgca cgccgacccg 5040
ccggtgctgg tgctgcacga ccccaccacc gccgtggacg cggccaccga ggcccaactc 5100
gccgacggac tggccggcgc gcgccgcgaa gcgccccggg gcacgctgct ggtcaccagc 5160
agccccgccc tgctgcggat caccgaccgg gtggtggtga tcgccgacgg ccgggtgacc 5220
gccgagggga cgcacgagca cctgctggcc accgacgccc gctaccgcga ggagacactg 5280
cggtgaccgc tgacccgcgt accgccgaac ccacccgggt gttgctgccc accgcgaccg 5340
cccggcggac ctggacgacg ctcggcgcgg agttccgccg gcggcccggc ctcagcgccg 5400
ccgcgaccgc cgtgctcgtc gccgccgcca ccggcgggct ggtcgcgccc tgggtgctcg 5460
gccgcctcgt cgacgacgtc atcgccgacg ccccggtctc ccggatcgcc ggccgggtgg 5520
cggtgatcgc cggcgcggca gtgctcaccg gactgctcac cgccgccggg gccgcgctcg 5580
cgtcccgcct gggggagacg gtgctggccc ggctgcgcga gcgggtcctc gaccgggcgc 5640
tgcacctgcc ctcggcgacg ctggaacggg ccggcaccgg cgacctgctg gcccgggtcg 5700
gcgacgacgt ggcggtggtg acgaacgtga tcgcggtcag cggcccggcg ttcgtcggcg 5760
cgctgctgtc cgtggtgctg accgtgttcg ggctggtcgc gctcgactgg cggctcggcc 5820
tcgccgggct ggtcgccgcg cccgcctacg cgctggcgct gcgctggtac ctgcgccggt 5880
cggcgccgta ctacgcccgc gagcgcgtcg ccaccggcga gcggacgcag gcgatggccg 5940
gcgcgctgcg tggcgcggcc accgtgcgcg cgtaccggac cgaggacgcg cacgtcgcgg 6000
cgatcgccga gcgctccggc gtggcgcgcg acctgtcgct ggagatcttc aacctgcaca 6060
cccggttcgg gctgcggatc aacaggtcgg agttcctcgg cctggccgcg gtgctcgtcg 6120
ccgggttctt cctggtccgc gccgacctgg tcacagtggg cgcggcgacc accgccgcgc 6180
tctacttcca ccggctgttc aacccgatcg gcctgctgct gatggagtcc gactcggtgc 6240
tgcaggccgg cgcgagcctc gcccggctgg tcggcgtggc cacgctgccc gacaccgccc 6300
cgtccgggcc cgcgccgtcg gcggccgggc ggcgcggccc ggcggcgctg gacgtcacgg 6360
tccgccggca ccgctacgac gacgacggcc ctctggtcct ggccgacgtc gacctgcgcc 6420
tggccccggg cgagcgggtc gcgctcgtgg gcgccagcgg cgcgggcaag agcacgctcg 6480
ccggcatcgc cgccgggatc atcgcgccca ccgacgggtc ggtacgcctg ggcggcgtgc 6540
cgctgaccga gcggggcgag cacgccgtgc ggcgcgacgt cgcgctggtc agccaggagg 6600
tgcacgtctt cgctggaccg ctcgccgagg atctgcgcct ggctgccccg gacgccaccg 6660
acgccgaact gctcgacgcg ctggaccggg tcggcgccac cacctggctg cgcgcgctgc 6720
cggacgggct ggccacagcg gtcggcgagg gcggccaccg gctcaccgcc gcgcaggccc 6780
agcaggtcgc cctggcccgg ctggtgctgg ccgcgcccgc cgtcgccgtg ctggacgagg 6840
99
CA 02538147 2004-O1-21
ccaccgccga ggccggcagc gccggagcgc gtgacctgga ccgggcggcg ctggccgcca 6900
ccgagggacg gaccacgctg atcgtggcgc accggci~cag ccaggcggtc gccgccgacc 6960
ggatcgtcct gctcgaccac gggcggatcg tggagcaggg cacgcactcg gaactgctcg 7020
ccgccgacgg ccggtacggg catctgtggc gctcctggag cgtcccggta tgatcgcgca 7080
ccgcccatcg gcccaggtga ggggaacatg accgacgcgc cggcccgctt cgtgctcttc 7140
ccggggcggc accacctgct gacccggttc caggccgact acctgcggcg gctggccggg 7200
gacgacgcca cagtggtctg ggcggtgacg tcggccaacc acgagaacac caggcgcaac 7260
ccggtgccct accaccggcg ggaggccgcg atcgaacgat tcagcgtgct gagcgggctg 7320
cgctcggtgg tggtgccgat cttcgacacc gcgtacaccg acgcgttcgc cgaggtgacg 7380
ctgaagtcca tcgcggtggc caccgggctc gaactcaccc ccgccgacac cgtgctggcc 7440
tgctccacgc cggaggtcgc gaagctgtac gagcagctcg gcttttcgat cgcgccggtc 7500
gaggcggacc cggacctgcc cgagccgccc gaacggccgt gggacgtgct gctgcgcctg 7560
gccgccgggg acgagacctg gcgcgcgctc acccacccgg ccaccatcga cgtgttcgag 7620
cgctaccgcc tggtcgagtc gatccggtcg gtggtgaacg acccgctcgt cggcgacgag 7680
ggcggtctca cagtgacccg cgactaccgg acctacgtcg aggcgttcgc cacggccgcg 7740
cagcgcaagt gggactcggt acgccggtac gtgcagcccg gccgcatcgt ggacatcggc 7800
tgcggcgcgg gcgccgtcct ggaactcgcc gaccgggagg ccgcgctgcg tgagagcgac 7860
ctgatcggcg tggaggtcgc ccgccacctc taccaggagt gcctgcacaa gaaggcgcag 7920
ggcgtgttcc gcaacgccaa cgtctacttc ttccaccgca acgtcctcgg cggcgcggtg 7980
ttcaaggacc gctcggtcga caccacgctc acgttcgcgc tgacccacga gatctggtcg 8040
tacgggcggc ggcgggagtc gctgctgcag ttcgcccgcc gcatccacga ccacacggtg 8100
cccggcggcg tctggatcaa cagcgacgtg tgcggtccgg acgacccccg gcggcaggtg 8160
ctcctgcgac tgtccaccga cgacggcgac aacccggccg cgccccgccc cgacctcgcc 8220
gagctgacct cggcggaggt ccggcgttac gtcggcgggc tgtcgacgcg ggcgcggctg 8280
gaccagttcg ccgtcgactt cgcgttcgac ttcgactacg agccgctccc cgacggcgcg 8340
gtacgcctga cgctgggcgc cgcgatggac tacctgaccc gcaaggacta cacggacaac 8400
tggctgtcgg agacgcagga gcagttctgc ggcctgagct tcgccgactg gacggacctg 8460
ctcaccgagg cggggttcga gatcggcccg gcgtcggcgc cggtgcgcaa cgagtgggtg 8520
atcgacaacc ggatcgcgcc agtcgcgtcc ctcaccgacc tcgacggccg gccgctggac 8580
tggccgacca cccacgtcct caccgtcgcc caccgccccc gcaaccagtg agaccgacgg 8640
100
CA 02538147 2004-O1-21
cgcccgccgcgttcggcgggcgccgtcgtcgctcaccggctcagcgcgatccggatcgcc 8700
aggacgatcaggatgagcccggtcagccgttcgatcaccagcagcacggacggccgggtc 8760
agccagggctgcaacctgtcgatgagcatgatgtagcaggcccaccagagcaccgcgagg 8820
ccgatgaacgtggcggcgagcaccgccgtacgggccgccgccccctcgccgggcttgacg 8880
aactgcggcacgaacgagacgtagaagacgaccaccttgacgttcagcagctggctggtg 8940
acgcccatgacgaacgagcggcgggccacgtgcggctcgtcggcggccggggtgtccggc 900()
accggcgcggggccggtgtccgtgtccggcccggcgccgcccgcgccgacagtgaccggc 9060
tgcgccgccgggaccgtccggcgcggccgggtcgcccagaggatcgtgccgcccaggtag 9120
agcaggtacagcgcgccggcgacgcgcagcaccgtgtagagcgtcggcgaggagaccagc 9180
agggcggacaggccggcggtcgcgaacgacgcgtgcaccagcgcggcgacgaacagcccg 924()
gccagcaccacgaacccggcccgccggccgtacctgacggtctgccgggtgacgagcgcg 9300
aagtcgacgcccggcacgatgatgatgagcaggctggcggcgacgaaactgatgatctgg 9360
atgt.cagacacgacgccggctctcctgtcctccggcgagcgccggcactgcctcctcgat 9420
gacggagacgccgctgtcctggcgtggtccgtgccggcgccactgttcccgcagccggat 948()
ccggccgtccggcagccgttcgggccgggactcgcactcgccgatgactatggtgccgtc 954()
ggtgagcacctccaggtaggcgaagcgcacgacgccctgcgcgtcgcaggtgccggccag 9600
ccggccgtgccggaccgggccgccggtgatctccgcccagaccaggtcgccacgctggtg 9660
gtagtgcccccgcagcggctcggcgccgtcaccggcgtcgtggtccaccgagacgaagac 972()
gcggccgtcgtagtcgaatgtcgtcatcgcgctcacgcccac 976:?
SEQ ID N0: 74
LENGTH: 112
TYPE: PRT
ORGANISM: Micromonospora sp. strain 046-ECO11
SEQUENCE: 74
Met Asp Gly Ala Arg Glu Arg Ile Thr Thr Asn Ile Arg Lys Gly Val
1 5 10 15
Leu Glu Tyr Cys Val Leu Ala Leu Leu Ser Arg Arg Asp Met Tyr Gly
20 25 30
Leu Glu Leu Ala Asp Trp Leu Ala Vai Arg Gly Leu Thr Ala Ser Glu
35 40 45
G1y Ser Leu Tyr Pro Leu Leu Ala Arg Met Arg Gln Ala Gly Ser Val
50 55 60
Gln Thr Arg Trp Val Ala Pro Glu Gln Gly His Ala Arg Arg Tyr Tyr
65 70 75 80
101
CA 02538147 2004-O1-21
Ala Ile His Leu Val Phe
Thr Asp Arg Ala Ala
Gln Gly Val
Arg Ala
85 90 95
Trp Gln u Ile Asp Asp Met Gly
Gl Gln Pro Leu Glu Glu
His Val Ala
100 105 110
SEQ ID 75
NO:
LENGTH:
339
TYPE:
DNA
ORGANISM:Micromonospora rain 046-ECO11
sp. st
SEQUENCE:75
atggacggggcacgtgagcggatcactacgaacatccgcaagggcgtgctggagtactgc60
gtgctcgccctgctctcgcggcgcgacatgtacggcctggaactggccgactggctcgcc120
gtccgcggtctgaccgcgagcgagggcagcctgtatccgctgctcgcccgcatgcggcag180
gccggctccgtgcagacccggtgggtggcccccgagcaggggcacgcccggcggtactac240
gcgatcaccgaccaggggcgggcgcacctgcgggtgttcgcggcggtgtggcaggagatc300
cagccgcacgtggacgacctgatgggggaggaagcatga 339
SEQ ID N0: 76
LENGTH: 325
TYPE: PRT
ORGANISM: Micromonospora sp. strain 046-EC011
SEQUENCE: 76
Met Ser Asp Asp Gly Leu Pro Glu Ala Ala Trp Thr Tyr Leu Arg Ala
1 5 10 15
Leu Asp Ala Glu Leu Ser Asp Val Pro Ser Gly Thr Ala Glu Glu Ile
20 25 30
Val Ala Asp Val Arg Ala His Ile Ala Asp Ala Leu Asp Ser Gly Arg
35 40 45
her Ala His Glu Ile Leu Ala Gly Leu Gly Ala Ala Arg Asp Val Ala
50 55 60
Arg Gln Ala Arg Glu Glu Leu Gly Leu Pro Ala Gln Asp Arg Pro Ala
65 70 75 $0
Arg Ala Gly Arg Thr Leu Ser Leu Ala Ala Val Ala Val Gly Val Leu
85 90 95
Ile Ala Val Cys Val Ser Phe Leu Leu Pro Ser Ala Val Pro Val Glu
100 105 110
Pro Ile Gln Ala Gly Pro Gly Glu Gln Gly VaI Leu Arg Arg Leu Gly
115 120 125
Pro Gly Ile Ala Leu Leu Thr Leu Leu Pro Ala Leu Val Ala Ala Ala
130 135 140
Pro Leu Val Ala Pro Ala Arg Ala Arg Ala Gly Val Arg Phe Ala Gly
145 150 155 160
1~~
CA 02538147 2004-O1-21
Ala Ala Val Leu Thr Met Phe Ala Cys Ala Ala Gly Glu Thr Gly Leu
165 170 175
Tyr Tyr Phe Pro Leu Ala Leu Met Ala Trp Ala Ala Ala Ile Val Pro
180 185 190
Trp Ala Leu Arg Arg Gly Ala Gly Gly Arg Trp Trp Arg Tyr Leu Thr
195 200 205
Gly Gly Phe Val Ala Met Pro Gly Val Leu Val Ala Val Ala Ser Ala
210 215 220
Gly Gly Ser Val Gly Val Gly Trp Val Gly Ala Ala Leu Trp Ile Ala
225 230 235 240
Gly Pro Leu Ala Ala Gly Ala Leu Cys Ala Tyr Gly Ile Arg Ala Gly
245 250 255
Tyr Ala Val Thr Ala Leu Ala Gly Ala Leu Ala Ile Ala Leu Ser Met
260 265 270
Ala Glu Arg Gly Phe Leu Phe AIa Ala Phe Trp Leu Phe Gly Gly Leu
275 280 285
Tyr Leu Ala Leu Gly Ala Ala Ala Tyr Thr Ala Ser Arg Ala Val Asp
290 295 300
Gly Asp Ala Ala Ala Thr Pro Gly Pro Pro Ala Arg Pro Glu Pro Ala
305 310 315 320
Pro Ala Pro Gly Gly
325
SEQ ID 77
N0:
LENGTH.
978
TYPE,
DNA
ORGANISM:Micromonospora
sp. strain
046-ECO11
SEQUENCE:77
atgagcgacgacggcctcccggaggcggcgtggacctatctgcgcgcgctcgacgcggag 60
ttgtccgacgtcccgtccggcacggcggaggagatcgtcgcggatgtccgcgcgcacatc 120
gccgacgccctcgacagcggacggagcgcccacgagatcctcgccggcctcggcgccgcg 180
cgggacgtggcccggcaggcgcgcgaggagctggggctgccggcccaggaccgcccggcc 240
cgggccggccggaccctgtccctggccgcggtggcggtcggcgtgctgatcgccgtgtgc 300
gtgagcttcctgctgccgtccgcagtgccggtggagccgatccaggccggccccggcgag 360
cagggcgtcctccgccggctcggccccggaatcgcgctgctcacgctgctgccggcgctc 420
gtcgcggccgcgccgctcgtggcgcccgcccgggcacgtgccggggtacggttcgccggc 480
gcggcggtcctgacgatgttcgcctgcgcggccggcgagacgggcctgtactacttcccg 540
ctcgcgctgatggcctgggcggcggcgatcgtgccgtgggccctgcggcgcggagccggt 600
ggacggtggtggcgctatctgaccggtggattcgtggcgatgcccggcgtgctggtggcg 66U
1 ~;3
CA 02538147 2004-O1-21
gtcgcgtcggccggtggctcggtcggcgtcggctgggtcggcgcggcgctgtggatcgcc 72()
gggccgctcgcggccggcgcgctgtgcgcctacgggatccgggccggctacgccgtgacc 780
gcgctggccggcgcgctggccatagcgctctcgatggccgagcgcggcttcctgttcgcc 840
gccttctggctgttcggcgggctgtacctggcgctcggcgccgctgcgtacaccgcctcg 900
cgggccgtcgacggcgacgccgccgcgacgcccggcccgccggcccggccggaacccgcg 960
ccggcccccggaggctga 978
SEQ ID N0: 78
LENGTH: 663
TYPE: PRT
ORGANISM: Micromonospora sp. strain 046-EC011
SEQUENCE: 78
Met Leu Asp His Ala Ser Gly Arg Ile Asp Val Thr Arg Leu Arg Glu
1 5 10 15
Ala Leu Asp Gly Arg Trp Ala Glu Val Arg Arg Ala His Arg Glu His
20 25 30
Leu Asp Glu Arg Phe Leu Pro Val Tyr Gly Glu Thr Gly Asp Gln Ala
35 40 45
Arg Glu Arg Ile Thr Arg Leu Leu Ser Glu Leu Pro Val Glu Leu Gly
50 55 60
Ile Ala Ser Gly Phe Pro Ala Glu Tyr Gly Gly Arg Gly Asp Val Gly
65 70 75 80
Ala Ser Ile Val Ala Thr Glu Met Leu Ala Gln Val Asp Leu Ser Leu
85 90 95
Met Val Lys Ala Gly Val Gln Trp Gly Leu Phe Gly Gly Ala Val Ala
100 105 110
Ala Leu Gly Thr Lys Arg His His Asp Ala Tyr Leu Arg Asp Ile Val
115 120 125
Ala Gly Arg Leu Phe Gly Cys Phe Ala Met Thr Glu Thr Gly His Gly
130 135 140
Ser Asp Va1 Gln Gln Leu Arg Thr Thr Cys Val Tyr Asp Pro Gln Thr
145 150 155 160
Gln Thr Phe Asp Leu His Thr Pro His Glu Ala Ala Arg Lys Asp Tyr
165 170 175
Ile Gly Asn AIa Ala Arg Asp Gly Arg Met Ala Val Val Phe Ala Gln
i80 185 190
Leu VaI Thr Gly Gly Arg Arg His Gly Val His Ala Trp Leu Val Pro
195 200 205
Ile Arg Asp Glu His Gly Lys Pro Met Pro Gly Val Thr Ile Gly Asp
1~~
CA 02538147 2004-O1-21
210 215 220
Ala Gly Pro Lys Ala Gly Leu Leu Gly Val Asp Asn Gly Arg Leu Ser
225 230 235 240
Phe Asp His Val Arg Val Pro Arg Glu Met Leu Leu Asp Gln Tyr Ala
245 250 255
Gln Val Ala Glu Asp Gly Thr Tyr Ser Ser Pro Ile Glu Asn Asp Ser
260 265 270
Arg Arg Phe Phe Thr Met Leu Gly Thr Leu Vai Arg Gly Arg Val Ser
275 280 285
Val Gly Gly Ala Ala Ser Ala Ala Thr Lys Ser Ala Leu Ala Ile Ala
290 295 300
Val Arg Tyr Gly Asp Ile Arg Arg Gln Phe Ala Asp Ala Asp Gly Asp
305 310 315 320
Arg Glu Val Leu Leu Asn Asp Tyr Leu Ala His Gln Arg Lys Leu Leu
325 330 335
Pro Ala Leu Ala Thr Thr Tyr Ala Leu Thr Phe Ala Gln Ala Glu Leu
340 345 350
Val Ala Ala Leu Asp Asp Ile Gln Gly Gly Asp Gly Pro Val Asp Glu
355 360 365
His Arg Gln Arg Glu Leu Glu Ser Arg Ala Ala Gly Leu Lys Ala Ala
370 375 380
Gln Thr Trp His Ala Thr Arg Thr Ile Gln Ile Cys Arg Glu Ala Cys
385 390 395 400
Gly GIy Ala GIy Tyr Leu Ser Glu Asn Arg Leu Pro Ser Leu Lys Ala
405 410 415
Asp Thr Asp Val Phe Thr Thr Phe Glu Gly Asp Asn Thr Val Leu Leu
420 425 430
Gln Leu Val Ala Lys Gly Leu Leu Thr Gly Tyr Arg Asp Glu Phe Gly
435 440 445
Ser Leu Asp Gly Trp Gly Arg Ala Ser Phe Val Ala Glu Gln Val Arg
450 455 460
Glu Met Val Leu Glu Arg Thr Ala Aia Arg Ala Leu Ile Ala Arg Leu
465 470 475 480
Val Ser Ala Val Pro Gly Arg Asp Asp Glu Val Ala Val Thr Asp Arg
485 490 495
Gly Trp Gln Leu Lys Leu Phe Glu Asp Arg Glu Glu His Leu Leu Asp
500 505 510
Ser Ala Val Arg Arg Leu Arg Gly Gly Ala Ser Thr Lys Lys Asp Arg
515 520 525
Pro Phe Asp Ile Phe Asn Asp Val GIn Asp His Val Leu Ala Val Ala
530 535 540
105
CA 02538147 2004-O1-21
Ala Ala His Ile Asp Arg Val Thr Leu Glu Ala Phe Val Ala Gly Ile
545 550 555 560
Asp Ala Ile Ala Asp Pro Ala Val Lys Glu Leu Leu Ser Arg Val Cys
565 570 575
Asp Leu Tyr Ala Leu Thr Val Ile G1u Ala Asn Lys Gly Trp Leu Leu
580 585 590
Glu His Gly Arg Leu Thr Pro Ala Arg Ser Lys Thr Ile Thr Ser Val
595 600 605
Val Asn Gly Leu Leu Lys Glu Leu Arg Pro Asp Met Arg Thr Leu Val
610 615 620
Asp Gly Phe Ala Ile Pro Asp Ala Trp Leu His Ala Ala Ile Leu Arg
625 630 635 640
Glu Glu Prc Val Arg Gln Glu Thr Met Ala Ala His Asp Ala Ala Gly
645 650 655
Asp Pro Gln Ala Val Pro Ala
660
SEQ ID 79
N0:
LENGTH:
1992
TYPE:
DNA
ORGANISM:Micromonospora
sp. strain
046-EC011
SEQUENCE:79
atgctcgatcacgcatccggccgcatcgacgtcacacgcctgcgggaagcgctcgacggc60
cggtgggccgaggtccgccgggcgcaccgcgaacacctcgacgaacgcttcctcccggtg120
tacggcgagaccggtgaccaggcccgcgagcgcatcacccggctgctgtccgaactcccc180
gtcgagctgggcatcgcctccggtttccccgccgagtacggcggccgcggcgacgtgggc240
gcctcgatcgtcgccaccgagatgctggcccaggtggacctgtcactgatggtgaaggcc300
ggcgtgeagtggggcctgttcggcggcgcggtcgccgccctcggcacgaagcggcaccac360
gacgcctaectgcgggacatcgtcgcgggccggctct:tcggctgcttcgcgatgaccgag420
accggccacggctcggacgtgcagcaactgcgcaccacctgcgtctacgacccgcagacg480
cagaccttcgacctgcacaccccgcacgaggccgcgcgcaaggactacatcggcaacgcg540
gcccgggacgggcggatggctgtggtgttcgcccagctcgtcaccggcgggcgccgccac600
ggggtgcacgcctggctggtgccgatccgcgacgagcacggcaagccgatgcccggcgtg660
accatcggcgacgccgggcccaaggccggcctgctcggcgtggacaacgggcggctcagc720
ttcgaccacgtgcgggtgccgcgggagatgctgctggaccagtacgcgcaggtcgccgag780
gacggcacgtactccagcccgatcgagaacgactcccggcgcttcttcaccatgctgggc840
accctggtccggggccgggtgagcgtgggcggcgccgcgtcggcggccaccaagtcggcg900
10~
CA 02538147 2004-O1-21
ctggccatcgcggtgcgctacggcgacatccgccggcagttcgccgacgccgacggcgac960
cgcgaggtgctgctcaacgactacctggcgcaccagcgcaagctgctgcccgcgctggcc1020
accacgtacgcgctgaccttcgcccaggcggagctggtcgcggcgctcgacgacatccag1080
ggcggcgacgggccggtcgacgagcaccggcagcgggagctggagtcccgggccgccggt1140
ctgaaggcggcgcagacctggcacgccacccgcaccatccagatctgccgggaggcgtgt1200
ggcggcgccggctacctgtccgagaaccgcctgccc<~gcctcaaggccgacaccgatgtc1260
ttcaccaccttcgagggcgacaacacggtgctgctgcaactggtcgccaaggggctgctg1320
accggctaccgggacgagttcggctcgctcgacggctggggacgcgcctccttcgtggcc1380
gagcaggtacgcgagatggtgctggaacgcaccgccgcgcgggcgctgatcgcacgtctg1440
gtcagcgccgtgcccgggcgcgacgacgaggtcgccgtcaccgaccggggctggcagctc1500
aagctcttcgaggaccgcgaggagcacctgctcgacagcgcggtccgccgcctgcgcggt1560
ggcgcgtccaccaagaaggaccgccccttcgacatcttcaacgacgtccaggaccacgtc1620
ctcgccgtcgccgcggcgcacatcgaccgggtgacgctggaggcgttcgtcgccgggatc1680
gacgccatcgccgacccggcggtcaaggaactgctgtcccgggtctgcgacctgtacgcg1740
ctcaccgtgatcgaggcgaacaagggctggctgctcgagcacggccggctcaccccggcc1800
cgctcgaagaccatcaccagcgtggtgaacgggctgctcaaggagctgcgcccggacatg1860
cgcacgctcgtggacggcttcgccatcccggacgcgtggctgcacgcggcgatcctgcgc1920
gaggagcccgtccggcaggagacgatggccgcgcacgacgccgccggcgacccgcaggcc1980
gtccccgcctag 1992
SEQ ID NO: 80
LENGTH: 573
TYPE: PRT
ORGANISM: Micromonospora sp. strain 046-ECO11
SEQUENCE: 80
'~'al Ser Pro Leu Pro Pro Gly Ser Ala Val Thr Ala Arg His Val Leu
1 5 10 15
Arg Gln Ala Leu Arg Arg Gln Arg Arg Pro Val Leu Ile Gly Val Thr
20 25 30
Leu Leu Gly Leu His Gln Val Thr Glu Ala Leu Val Pro Val Ala Ile
35 40 45
Gly Val Ile Ile Asp Arg Ala Val Val Thr Gly Asp Pro Trp Ala Leu
50 55 60
Ala Tyr Ser Val Ala Gly Leu Ala Ala Leu Phe Thr Val Leu Ala Phe
65 70 75 80
1~~
CA 02538147 2004-O1-21
Ala Tyr Arg Asn Gly Ala Arg Gln Ala Phe Ala Ala Val Glu Arg Glu
85 90 95
Ala His Leu Leu Arg Val Glu Leu Ala Glu Arg Ala Leu Asp Pro Arg
100 105 110
Gly His Arg Ser G1y Leu Arg Asp Gly Glu Leu Leu Ser Val Ala Ala
115 120 125
Ser Asp Ala Glu Leu Ser Ala Tyr Val Val Arg Val Ala Gly Phe Gly
130 135 140
Val Ala Ala Val Ser Ala Leu Thr Val Ala Ala Val Ala Leu Leu Val
145 150 155 160
Ile Asp Val Pro Leu Gly Leu Gly Val Leu Ile Gly Val Pro Val Leu
165 170 175
Val Leu Ala Leu Gln Arg Met Ala Pro Leu Leu Ser Arg Arg Ser Ala
180 185 190
Ser Gln Gln Glu Ala Leu Ala Glu Thr Thr Ala Leu Ala Val Asp Leu
195 200 205
Val Ser Gly Leu Arg Val Leu Arg Gly Ile Gly Ala Gln His His Ala
210 215 220
Ala Gly Arg Tyr Ala Glu Ala Ser Arg Arg Ala Leu Ala Val Thr Leu
225 230 235 240
Arg Ala Ala Asn Thr Lys Gly Leu His Leu Gly Leu Thr Thr Ala Ala
245 250 255
Asn G1y Leu Phe Leu Ala Ala Val Ala Gly Val Ala Gly Trp Leu Ala
260 265 270
Leu Arg Gly Arg Leu Thr Ile Gly Glu Leu Vai Thr Val Val Gly Leu
275 280 285
Ala Gln Phe Val Ala Glu Pro Val Gln Thr Leu Gly Tyr Cys Val Gln
290 295 300
Leu Phe Ala Met Ala Arg Ala Ser Ala Ala Arg Val Gly Arg Val Leu
305 310 315 320
Gly Ala Glu Pro Leu Thr Arg Pro Gly Ser Ala Pro Arg Pro Asp Arg
325 330 335
Thr Asp Gly Pro Arg Leu Val Leu Asp His Val Gly His Ala Ala Leu
340 345 350
Asp Gly Val Cys Leu Arg Val Asp Pro Gly Glu Ile Val Gly Val Leu
355 360 365
Ala Tyr Asp Pre Ala Asp A1a Asp Ala Leu VaI Ala Leu Leu Ser Gly
370 375 380
Arg Val Pro Ala Asp Arg Arg Arg Gly Thr Val Arg Val Asp Gly Val
385 390 395 400
Pro Ala Asp Asp Leu Asp Val Asp Ala Leu Arg Gly Ala Val Leu Val
1~g
CA 02538147 2004-O1-21
405 410 415
Glu Pro His Asp Val Thr Leu Phe Glu Gly Thr Val Ala Ala Asn Leu
420 425 430
Ala Ala Gly Ser Arg Thr Glu Glu Gly Arg Leu Arg Ala Ala Val Arg
435 440 445
Ala Ala Ala Ala Asp Asp Val Val Asp Ala His Pro Gly Gly Leu Gly
450 455 460
His Arg Leu Val Glu Arg Gly Ala Asn Leu Ser Gly Gly Gln Arg Gln
465 470 475 480
Arg Leu Gly Leu Ala Arg AIa Leu His Ala Asp Pro Pro Val Leu Val
485 490 495
Leu His Asp Pro Thr Thr Ala Val Asp Ala Ala Thr Glu Ala Gln Leu
500 505 510
Ala Asp Gly Leu Ala Gly Ala Arg Arg Glu Ala Pro Arg Gly Thr Leu
515 520 525
Leu Val Thr Ser Ser Pro Ala Leu Leu Arg Ile Thr Asp Arg Val Val
530 535 540
Val Ile Ala Asp Gly Arg Val Thr Ala Glu Gly Thr His Glu His Leu
545 550 555 560
Leu Ala Thr Asp Ala Arg Tyr Arg Glu Glu Thr Leu Arg
565 570
SEQ ID 81
NO:
LENGTH:
1722
TYPE:
DNA
ORGANISM:Micromonospora
sp. strain
046-ECO11
SEQUENCE:81
gtgtccccgcttccccccggcagcgccgtcaccgcccggcacgtgctccgccaggcgctg60
CgCCgCCagCgccgcccggtgctgatcggcgtgaccctgctcgggctgcaccaggtcacc120
gaggcgctcgtgccggtggcgatcggcgtcatcatcgaccgggccgtggtgaccggcgac180
ccgtgggcgctcgcgtactccgtcgccggcCtCgCCgCCCtgttcaccgtgctggcgttc240
gcctaccgcaa.cggcgcccgccaggcgttcgcggcggtggaacgggaggcgcacctgctg300
cgggtcgagctggccgagcgcgcgctcgacccgcgcgggcaccgctccggcctgcgcgac360
ggcgagctgctctcggtcgccgcctccgacgccgaactctccgcgtacgtggtccgggtg420
gccggcttcggcgtcgccgcggtgagcgcgctgaccgtcgcggcggtcgcgctgctggtc480
atcgacgtcccgctcggactcggcgtgctcatcggcgtaccggtgctggtcctggcgctg540
caacggatggcgccgctgctgtcccggcgcagcgcctcccagcaggaggccctcgcggag600
accacggcgctcgccgtggacctcgtctccggcctgcgcgtgctgcgcggcatcggcgcc660
cagcaccacgccgccggccggtacgccgaggccagccgacgcgccctcgccgtgacgctg720
1~9
CA 02538147 2004-O1-21
Lgcgccgccaacaccaagggcctgcacctcgggctcaccaccgccgcgaacggcctcttc 780
ctcgccgccgtcgccggggtcgccggctggctcgcgctgcgcggccggctcaccatcggc 840
gagctggtcaccgtggtcgggctcgcgcagttcgtcgccgagccggtgcagacgctgggc 900
tactgcgtgcagctgttcgcgatggcccgcgcctccgccgcccgggtcgggcgcgtgctc 960
ggcgccgagccgctgacccggccgggcagcgcgccccggccggaccgcacggacgggccg 1020
cggctcgtcctcgaccacgtcggccacgccgcgctggacggggtgtgcctgcgcgtcgac 1080
ccgggagagatcgtcggcgtcctggcgtacgacccggccgacgcggacgcgctggtggcg 1140
ctgctgtccgggcgggtgcccgcggaccggcgccggggcacggtacgcgtcgacggggta 1200
cccgccgacgacctggacgtcgacgcgctgcgcggcgccgtcctggtcgagccgcacgac 1260
gtgacgctgttcgagggaaccgtggccgccaacctcgccgccgggagcaggaccgaggag 1320
gggcgcctgcgcgccgcggtccgggcggccgcggcggacgacgtggtggacgcgcacccc 1380
ggcggcctcggccaccggctcgtcgagcggggcgccaacctctccggcgggcagcgccag 1440
cggctcgggctggcgcgggcgctgcacgccgacccgccggtgctggtgctgcacgacccc 1500
accaccgccgtggacgcggccaccgaggcccaactcgccgacggactggccggcgcgcgc 1560
cgcgaagcgccccggggcacgctgctggtcaccagcagccccgccctgctgcggatcacc 1620
gaccgggtggtggtgatcgccgacggccgggtgaccgccgaggggacgcacgagcacctg 1680
ctggccaccgacgcccgctaccgcgaggagacactgcggtga 1722
SEQ ID NO: 82
LENGTH: 596
TYPE: PRT
ORGANISM: Micromonospora sp. strain 046-ECOll
SEQUENCE: 82
Val Thr Ala Asp Pro Arg Thr Ala GIu Pro Thr Arg Val Leu Leu Pro
1 5 10 15
Thr Ala Thr Ala Arg Arg Thr Trp Thr Thr Leu Gly Ala Glu Phe Arg
20 25 30
Arg Arg Pro Gly Leu Ser Ala Ala AIa Thr Ala Val Leu Val Ala Ala
35 40 45
Ala 'Ihr Gly Giy Leu Val Ala Pro Trp Val Leu Gly Arg Leu Val Asp
50 55 60
Asp Val Ile Ala Asp Ala Pro Val Ser Arg Ile Ala Gly Arg Val Ala
65 70 75 80
Val IIe Ala Gly Ala Ala Val Leu Thr Gly Leu Leu Thr Ala Ala Gly
85 90 95
110
CA 02538147 2004-O1-21
Aia Ala Leu Ala Ser Arg Leu Gly Glu Thr Val Leu Ala Arg Leu Arg
100 105 110
Glu Arg Val Leu Asp Arg Ala Leu His Leu Pro Ser Ala Thr Leu Glu
115 120 125
Arg Ala Gly Thr Gly Asp Leu Leu Ala Arg Val Gly Asp Asp Val Ala
130 135 140
Val Val Thr Asn Val Ile Ala Val Ser Gly Pro Ala Phe Val Gly Ala
145 150 155 160
Leu Leu Ser Val Val Leu Thr Val Phe Gly Leu Val Ala Leu Asp Trp
165 170 175
Arg Leu Gly Leu Ala Gly Leu Val Ala Ala Pro Ala Tyr Ala Leu Ala
180 185 190
Leu Arg Trp Tyr Leu Arg Arg Ser Ala Pro Tyr Tyr Ala Arg Glu Arg
195 200 205
Val Ala Thr Gly Glu Arg Thr Gln Ala Met Ala Gly Ala Leu Arg Gly
210 215 220
Ala Ala Thr Val Arg Ala Tyr Arg Thr Glu Asp Ala His Val Ala Ala
225 230 235 240
Ile Ala Glu Arg Ser Gly Val Ala Arg Asp Leu Ser Leu Glu Ile Phe
245 250 255
Asn Leu His Thr Arg Phe Gly Leu Arg Ile Asn Arg Ser Glu Phe Leu
260 265 270
Gly Leu Ala Ala Val Leu Val Ala Gly Phe Phe Leu Val Arg Ala Asp
275 280 285
Leu Vai Thr Vai Gly Ala Ala Thr Thr Ala Ala Leu Tyr Phe His Arg
290 295 300
Leu Phe Asn Pro Ile Gly Leu Leu Leu Met Glu Ser Asp Ser Val Leu
305 310 315 320
Gln Ala Gly Ala Ser Leu Ala Arg Leu Val Gly Val Ala Thr Leu Pro
325 330 335
Asp Thr Ala Pro Ser Gly Pro Ala Pro Ser Ala Ala Gly Arg Arg Gly
340 345 350
Fro Ala Ala Leu Asp Val Thr Val Arg Arg His Arg Tyr Asp Asp Asp
355 360 365
Gly Pro Leu Val Leu Ala Asp Val Asp Leu Arg Leu Ala Pro Gly Glu
370 375 380
Arg Val Ala Leu Val Gly Ala Ser Gly Ala Gly Lys Ser Thr Leu Ala
385 390 395 400
Gly Ile Ala Aia Gly Ile Ile Ala Pro Thr Asp Gly Ser Val Arg Leu
405 410 415
Gly Gly Val Pro Leu Thr Glu Arg Gly Glu His Ala Val Arg Arg Asp
111
CA 02538147 2004-O1-21
420 425 430
Val Ala Leu Val Ser Gln Glu Val His Val Phe Ala Gly Pro Leu Ala
435 440 445
Glu Asp Leu Arg Leu Ala Ala Pro Asp Ala Thr Asp Ala Glu Leu Leu
450 455 460
Asp Ala Leu Asp Arg Val Gly Ala Thr Thr Trp Leu Arg Ala Leu Pro
465 470 475 480
Asp Gly Leu Ala Thr Ala Val Gly Glu Gly Gly His Arg Leu Thr Ala
485 490 495
Ala Gln Ala Gln Gln Val Ala Leu Ala Arg Leu Val Leu Ala Ala Pro
500 505 510
Ala Val Aia Val Leu Asp Glu Ala Thr Ala Glu Ala Gly Ser Ala Gly
515 520 525
Ala Arg Asp Leu Asp Arg Ala Ala Leu Ala Ala Thr Glu Gly Arg Thr
530 535 540
Thr Leu Ile Val Ala His Arg Leu Ser Gln Ala Val Ala Ala Asp Arg
545 550 555 560
Ile Val Leu Leu Asp His Gly Arg Ile Val Glu Gln Gly Thr His Ser
565 570 575
Glu Leu Leu Ala Ala Asp Gly Arg Tyr Gly His Leu Trp Arg Ser Trp
580 585 590
Ser Val Pro Val
595
SEQ ID 83
N0:
LENGTH:
1791
TYPE:
DNA
ORGANISM:Micromonospora
sp. strain
046-ECO11
SEQUENCE:83
gtgaccgctgacccgcgtaccgccgaacccacccgggtgttgctgcccaccgcgaccgcc60
cggcggacctggacgacgctcggcgcggagttccgccggcggcccggcctcagcgccgcc120
gcgaccgccgtgctcgtcgccgccgccaccggcgggctggtcgcgccctgggtgctcggc180
cgcctcgtcgacgacgtcatcgccgacgccccggtctcccggatcgccggccgggtggcg240
gtgatcgccggcgcggcagtgctcaccggactgctcaccgccgccggggccgcgctcgcg300
tcccgcctgggggagacggtgctggcccggctgcgcgagcgggtcctcgaccgggcgctg360
cacctgccctcggcgacgctggaacgggccggcaccggcgacctgctggcccgggtcggc420
gacgacgtggcggtggtgacgaacgtgatcgcggtcagcggcccggcgttcgtcggcgcg480
ctgctgtccgtggtgctgaccgtgttcgggctggtcgcgctcgactggcggctcggcctc540
gccgggctggtcgccgcgcccgcctacgcgctggcgctgcgctggtacctgcgccggtcg600
mz
CA 02538147 2004-O1-21
gcgccgtactacgcccgcgagcgcgtcgccaccggcgagcggacgcaggcgatggccggc660
gcgctgcgtggcgcggccaccgtgcgcgcgtaccggaccgaggacgcgcacgtcgcggcg720
atcgccgagcgctccggcgtggcgcgcgacctgtcgctggagatcttcaacctgcacacc780
cggttcgggctgcggatcaacaggtcggagttcctcggcctggccgcggtgctcgtcgcc840
gggttcttcctggtccgcgccgacctggtcacagtgggcgcggcgaccaccgccgcgctc900
tacttccaccggctgttcaacccgatcggcctgctgctgatggagtccgactcggtgctg960
caggccggcgcgagcctcgcccggctggtcggcgtggccacgctgcccgacaccgccccg1020
tccgggcccgcgccgtcggcggccgggcggcgcggcr_cggcggcgctggacgtcacggtc1080
cgccggcaccgctacgacgacgacggccctctggtcctggccgacgtcgacctgcgcctg1140
gccccgggcgagcgggtcgcgctcgtgggcgccagcggcgcgggcaagagcacgctcgcc1200
ggcatcgccgccgggatcatcgcgcccaccgacgggtcggtacgcctgggcggcgtgccg1260
ctgaccgagcggggcgagcacgccgtgcggcgcgacgtcgcgctggtcagccaggaggtg1320
cacgtcttcgctggaccgctcgccgaggatctgcgcctggctgccccggacgccaccgac1x80
gccgaactgctcgacgcgctggaccgggtcggcgccaccacctggctgcgcgcgctgccg1440
gacgggctggccacagcggtcggcgagggcggccaccggctcaccgccgcgcaggcccag1500
caggtcgccctggcccggctggtgctggccgcgcccgccgtcgccgtgctggacgaggcc1560
accgccgaggccggcagcgccggagcgcgtgacctggaccgggcggcgctggccgccacc1620
gagggacggaccacgctgatcgtggcgcaccggctcagccaggcggtcgccgccgaccgg1680
atcgtcctgctcgaccacgggcggatcgtggagcagggcacgcactcggaactgctcgcc1740
gccgacggccggtacgggcatctgtggcgctcctggagcgtcccggtatga 1791
SEQ ID N0: 84
LENGTH: 507
TYPE: PRT
ORGANISM: Micromonospora sp. strain 046-ECO11
SEQUENCE: 84
Met Thr Asp Ala Pro Ala Arg Phe Val Leu Phe Pro Gly Arg His His
1 5 10 15
Leu Leu Thr Arg Phe Gln Ala Asp Tyr Leu Arg Arg Leu Ala Gly Asp
20 25 30
Asp Ala Thr Val Val Trp Ala Val Thr Ser Ala Asn His Glu Asn Thr
35 40 45
Arg Arg Asn Pro Val Pro Tyr His Arg Arg Glu Ala Ala Ile Glu Arg
50 55 60
Phe Ser Val Leu Ser Gly Leu Arg Ser Val Val Val Pro Ile Phe Asp
113
CA 02538147 2004-O1-21
65 70 75 80
Thr Ala Tyr Thr Asp Ala Phe Ala Glu Val Thr Leu Lys Ser Ile Ala
85 90 95
Val Ala Thr Gly Leu Glu Leu Thr Pro Ala Asp Thr Val Leu Ala Cys
100 105 110
Ser Thr Pro Glu Val Ala Lys Leu Tyr GIu Gln Leu Gly Phe Ser Ile
115 120 125
Ala Pro Val Glu Ala Asp Pro Asp Leu Pro Glu Pro Pro Glu Arg Pro
130 135 i40
Trp Asp Val Leu Leu Arg Leu Ala Ala Gly Asp Glu Thr Trp Arg Ala
145 150 155 160
Leu Thr His Pro Ala Thr Ile Asp Val Phe Glu Arg Tyr Arg Leu Val
165 170 175
Glu Ser Ile Arg Ser Val Val Asn Asp Pro Leu Val Gly Asp Glu Gly
180 185 190
Gly Leu Thr Val Thr Arg Asp Tyr Arg Thr Tyr Val Glu Ala Phe Ala
195 200 205
Thr Aia Ala Gln Arg Lys Trp Asp Ser Val Arg Arg Tyr Val Gln Pro
210 215 220
Gly Arg Ile Val Asp Ile Gly Cys Gly Ala Gly Ala Val Leu GIu Leu
225 230 235 240
Ala Asp Arg Glu Ala Ala Leu Arg Glu Ser Asp Leu Ile Gly Val Glu
245 250 255
Val Ala Arg His Leu Tyr Gln Glu Cys Leu His Lys Lys Ala Gln Gly
260 265 270
'~'al Phe Arg Asn Ala Asn Val Tyr Phe Phe His Arg Asn Val Leu Gly
275 280 285
Gly Ala Vai Phe Lys Asp Arg Ser Val Asp Thr Thr Leu Thr Phe Ala
290 295 300
Leu Thr His Glu Ile Trp Ser Tyr Gly Arg Arg Arg Glu Ser Leu Leu
305 310 315 320
Gln Phe Ala Arg Arg Ile His Asp His Thr Val Pro Gly Gly Val Trp
325 330 335
Ile Asn Ser Asp Val Cys Gly Pro Asp Asp Pro Arg Arg Gln Val Leu
340 345 350
Leu Arg Leu Ser Thr Asp Asp Gly Asp Asn Pro Ala Ala Pro Arg Pro
355 360 365
Asp Leu Ala Glu Leu Thr Ser Ala Glu Val Arg Arg Tyr Val Gly Gly
370 375 380
Leu Ser Thr Arg Ala Arg Leu Asp Gln Phe Ala Val Asp Phe Ala Phe
385 390 395 400
ll~
CA 02538147 2004-O1-21
Asp Phe Asp Tyr Glu Pro Leu Pro Asp Gly Ala Val Arg Leu Thr Leu
405 410 415
Gly Ala Ala Met Asp Tyr Leu Thr Arg Lys Asp Tyr Thr Asp Asn Trp
420 425 430
Leu Ser Glu Thr Gln Glu Gln Phe Cys Gly Leu Ser Phe Ala Asp Trp
435 440 445
Thr Asp Leu Leu Thr Glu Ala Gly Phe Glu Ile Gly Pro Ala Ser Ala
450 455 460
Pro Val Arg Asn Glu Trp Val Ile Asp Asn Arg Ile Ala Pro Val Ala
465 470 475 480
Ser Leu Thr Asp Leu Asp Gly Arg Pro Leu Asp Trp Pro Thr Thr His
485 490 495
Val Leu Thr Val Ala His Arg Pro Arg Asn Gln
500 505
SEQ ID 85
N0:
LENGTH:
1524
TYPE:
DNA
ORGANISM:Micromonospora
sp. strain
046-EC011
SEQUENCE:85
atgaccgacgcgccggcccgcttcgtgctcttcccggggcggcaccacctgctgacccgg60
ttccaggccgactacctgcggcggctggccggggacgacgccacagtggtctgggcggtg120
acgtcggccaaccacgagaacaccaggcgcaacccggtgccctaccaccggcgggaggcc180
gcgatcgaacgattcagcgtgctgagcgggctgcgct=cggtggtggtgccgatcttcgac240
accgcgtacaccgacgcgttcgccgaggtgacgctgaagtccatcgcggtggccaccggg30()
ctcgaactcacccccgccgacaccgtgctggcctgctccacgccggaggtcgcgaagctg360
tacgagcagctcggcttttcgatcgcgccggtcgaggcggacccggacctgcccgagccg420
cccgaacggccgtgggacgtgctgctgcgcctggccgccggggacgagacctggcgcgcg480
ctcacccacccggccaccatcgacgtgttcgagcgctaccgcctggtcgagtcgatccgg540
tcggtggtgaacgacccgctcgtcggcgacgagggcggtctcacagtgacccgcgactac600
cggacctacgtcgaggcgttcgccacggccgcgcagcgcaagtgggactcggtacgccgg660
tacgtgcagcccggccgcatcgtggacatcggctgcggcgcgggcgccgtcctggaactc720
gccgaccgggaggccgcgctgcgtgagagcgacctgatcggcgtggaggtcgcccgccac780
ctctaccaggagtgcctgcacaagaaggcgcagggcgtgttccgcaacgccaacgtctac840
ttcttccaccgcaacgtcctcggcggcgcggtgttcaaggaccgctcggtcgacaccacg900
ctcacgttcgcgctgacccacgagatctggtcgtacgggcggcggcgggagtcgctgctg960
cagttcgcccgccgcatccacgaccacacggtgcccggcggcgtctggatcaacagcgac1020
115
CA 02538147 2004-O1-21
gtgtgcggtccggacgacccccggcggcaggtgctcctgcgactgtccaccgacgacggc1080
gacaacccggccgcgccccgccccgacctcgccgagctgacctcggcggaggtccggcgt1140
tacgtcggcgggctgtcgacgcgggcgcggctggaccagttcgccgtcgacttcgcgttc1200
gacttcgactacgagccgctccccgacggcgcggtacgcctgacgctgggcgccgcgatg1260
gactacctgacccgcaaggactacacggacaactggctgtcggagacgcaggagcagttc132()
tgcggcctgagcttcgccgactggacggacctgctcaccgaggcggggttcgagatcggc1380
ccggcgtcggcgccggtgcgcaacgagtgggtgatcgacaaccggatcgcgccagtcgcg1440
tccctcaccgacctcgacggccggccgctggactggccgaccacccacgtcctcaccgtc150()
gcccaccgcccccgcaaccagtga 1524
SEQ ID N0: 86
LENGTH: 232
TYPE: PRT
ORGANISM: Micromonospora sp. strain 046-ECO11
SEQUENCE: 86
Val Ser Asp Ile Gln Ile Ile Ser Phe Val Ala Ala Ser Leu Leu Ile
1 5 10 15
Ile Ile VaI Pro Gly Val Asp Phe Ala Leu Val Thr Arg Gln Thr Val
20 25 30
Arg Tyr Gly Arg Arg Ala Gly Phe Val Val Leu Ala Gly Leu Phe Val
35 40 45
Ala Ala Leu Val His Ala Ser Phe Ala Thr Ala Gly Leu Ser Ala Leu
50 55 60
Leu Val Ser Ser Pro Thr Leu Tyr Thr Val Leu Arg Val Ala Gly Ala
65 70 75 80
Leu Tyr Leu Leu Tyr Leu Gly Gly Thr Ile Leu Trp Ala Thr Arg Pro
85 90 95
Arg Arg Thr Val Pro Ala Ala Gln Pro Val Thr Val Gly Ala Gly Gly
100 105 110
Ala Gly Pro Asp Thr Asp Thr Gly Pro Ala Pro Val Pro Asp Thr Pro
115 120 125
Ala Ala Asp Glu Pro His Val Aia Arg Arg Ser Phe Val Met Gly Val
130 135 140
Thr Ser Gln Leu Leu Asn Val Lys Val Val Val Phe Tyr Val Ser Phe
145 150 155 160
Val Pro Gln Phe Val Lys Pro Gly Glu Gly Ala Ala Ala Arg Thr Ala
165 170 175
Val Leu Ala Ala Thr Phe Ile Gly Leu Ala Val Leu Trp Trp Ala Cys
116
CA 02538147 2004-O1-21
180 185 190
Tyr Ile Met Leu Ile Asp Arg Leu Gln Pro Trp Leu Thr Arg Pro Ser
195 200 205
Val Leu Leu Val Ile Glu Arg Leu Thr Gly Leu Ile Leu Ile Val Leu
210 215 220
Ala Ile .Arg Ile Ala Leu Ser Arg
225 230
SEQ ID 87
NO:
LENGTH.
699
TYPE:
DNA
ORGANISM:Micromonospora
sp. strain
046-ECO11
SEQUENCE:87
gtgtctgacatccagatcatcagtttcgtcgccgccagcctgctcatcatcatcgtgccg 60
ggcgtcgacttcgcgctcgtcacccggcagaccgtcaggtacggccggcgggccgggttc 120
gtggtgctggccgggctgttcgtcgccgcgctggtgcacgcgtcgttcgcgaccgccggc 180
ctgtccgccctgctggtctcctcgccgacgctctacacggtgctgcgcgtcgccggcgcg 240
ctgtacctgctctacctgggcggcacgatcctctgggcgacccggccgcgccggacggtc 300
ccggcggcgcagccggtcactgtcggcgcgggcggcgccgggccggacacggacaccggc 360
cccgcgccggtgccggacaccccggccgccgacgagccgcacgtggcccgccgctcgttc 420
gtcatgggcgtcaccagccagctgctgaacgtcaaggtggtcgtcttctacgtctcgttc 480
gtgccgcagttcgtcaagcccggcgagggggcggcggcccgtacggcggtgctcgccgcc 540
acgttcatcggcctcgcggtgctctggtgggcctgctacatcatgctcatcgacaggttg 600
cagccctggctgacccggccgtccgtgctgctggtgatcgaacggctgaccgggctcatc 660
ctgatcgtcctggcgatccggatcgcgctgagccggtga 699
SEQ ID N0: 88
LENGTH: 132
TYPE: PRT
ORGANISM: Micromonospora sp. strain 046-ECO11
SEQUENCE: 88
Val Gly Val Ser Ala Met Thr Thr Phe Asp Tyr Asp Gly Arg Val Phe
1 5 10 15
Val Ser Val Asp His Asp Ala Gly Asp Gly Ala Glu Pro Leu Arg Gly
20 25 30
His Tyr His Gln Arg Gly Asp Leu VaI Trp Aia Glu Ile Thr Gly Gly
35 40 45
Pro Val Arg His Gly Arg Leu Ala Gly Thr Cys Asp Ala Gln Gly Val
50 55 60
117
CA 02538147 2004-O1-21
Val Arg Phe Ala Tyr Leu Glu Val Leu Thr Asp Gly Thr Ile Val Ile
65 70 75 80
Gly Glu Cys Glu Ser Arg Pro Glu Arg Leu Pro Asp Gly Arg Ile Arg
85 90 95
Leu Arg Glu Gln Trp Arg Arg His Gly Pro Arg Gln Asp Ser Gly Val
100 105 110
Ser Val Ile Glu Glu Ala Val Pro Ala Leu Ala Gly Gly Gln Glu Ser
115 120 125
Arg Arg Arg Val
130
SEQ ID 89
NO:
LENGTH:
399
TYPE:
DNA
ORGANISM:Micromonospora
sp. strain
046-EC011
SEQUENCE:89
gtgggcgtgagcgcgatgacgacattcgactacgacggccgcgtcttcgtctcggtggac60
cacgacgccggtgacggcgccgagccgctgcgggggcactaccaccagcgtggcgacctg120
gtctgggcggagatcaccggcggcccggtccggcacggccggctggccggcacctgcgac180
gcgcagggcgtcgtgcgcttcgcctacctggaggtgctcaccgacggcaccatagtcatc240
ggcgagtgcgagtcccggcccgaacggctgccggacggccggatccggctgcgggaacag300
tggcgccggcacggaccacgccaggacagcggcgtctccgtcatcgaggaggcagtgccg360
gcgctcgccggaggacaggagagccggcgtcgtgtctga 399
118