Note: Descriptions are shown in the official language in which they were submitted.
DEMANDES OU BREVETS VOLUMINEUX
LA PRESENTE PARTIE I)E CETTE DEMANDE OU CE BREVETS
COMPRI~:ND PLUS D'UN TOME.
CECI EST ~.E TOME 1 DE 2
NOTE: Pour les tomes additionels, veillez contacter le Bureau Canadien des
Brevets.
JUMBO APPLICATIONS / PATENTS
THIS SECTION OF THE APPLICATION / PATENT CONTAINS MORE
THAN ONE VOLUME.
THIS IS VOLUME 1 OF 2
NOTE: For additional vohxmes please contact the Canadian Patent Oi~ice.
CA 02553468 2006-07-13
WO 2005/071079 PCT/EP2005/000559
INHIBTTORS OF MAMMALIAN HDAC 11 USEFUL FOR TREATING HDAC 11 MEDIATED
DISORDERS
BACKGROUND OF THE INVENTION
This invention relates to histone deacetylases, in particular HDAC 11, its
role in cellular
proliferative diseases such as cancer and methods of treating said disorders
via the use of newly
identified HDAC 11 inhibitors.
In eukaryotic cells, the orderly packaging of DNA in the nucleus plays an
important role
in the regulation of gene transcription. In the resting cell, DNA is tightly
compacted to prevent
transcription factor accessibility.
The compact complex termed chromatic results from the tight association of
nuclear
DNA with histories. The basic repeating unit in chromatin is the nucleosome,
which consists of 146
bases of DNA wrapped around a complex of eight histone proteins, two molecules
each of the core
histories, H2A, H2B, H3, and H4. Each core histone octamer is comprised of
several highly conserved
structural motifs including a globular domain and an N-terminal tail domain
that extends outside of the
nucleosome. Importantly, the packaging of DNA into nucleosomes acts as a
barrier to the initiation of
transcription by preventing the access of transcription factors, and RNA
polymerase II, to their cognate
recognition sequences (Workman, J. L. & A. R. Buchman (1993) Trends. Biochem.
Sci. 18:90-95).
During activation of the cell this compact, inaccessible DNA is made available
to DNA-
binding proteins, thus allowing the induction of gene transcription (Beato, M.
(1996) J. Mol. Med.
74:711-724; Wolffe, A. P. (1997) Nature 387:16-17). The physical interaction
between the core histone
particle and DNA principally occurs through the negatively charged phosphate
groups of the DNA and
the basic amino acid moieties of the histone proteins. These histone N-
terminal tails are enriched in
basic amino acids that are sites for post-transcriptional modifications, and
are thought to mediate histone-
DNA contacts through electrostatic interactions with DNA's negatively charged
phosphate backbone.
(Beato, M (1996) J. Mol. Med. 74:711-724; Beato, M. & K. Eisfeld (1997)
Nucleic. Acids. Res. 25:3559-
3563). Core histories may be modified by acetylation, phosphorylation,
methylation, ADP ribosylation or
ubiquitinylation of specific amino acid residues (Wu, R. S et al (1986) CRC
Crit. Rev. Biochem. 20:201-
263).
There is a vast body of evidence suggesting that increased gene transcription
is
associated with an increase in histone acetylation, whereas hypo-acetylation
is correlated with reduced
transcription or gene silencing (Ura, K et al (1997) EMBO J. 16:2096-2107;
Wolffe, A. P (1997) Nature
387:16-17).
CA 02553468 2006-07-13
WO 2005/071079 PCT/EP2005/000559
Histone acetylation, a reversible modification, occurs on actively transcribed
chromatin
via the action of histone acetyl transferases (HATS) (Perry, M. & R. Chalkley
(1982) J. Biol. Chem.
257:7336-7347), while histone deacetylation is catalyzed by a family of
enzymes termed histone
deacetylases (I3DACs), which serve to repress gene expression. See, for
example, Grunstein, Nature
389, 349-352 (1997); Pazin et al_, Cell 89, 325-328 (1997); Wade et al.,
Trends Biochem. Sci. 22, 128-
132 (1997); and Wolffe, Science 272, 371-372 (1996). Essentially, acetylation
of histones reduces their
positive charge, thereby relaxing the structure of the nucleosome and
facilitating the interaction of
transcription factors to the DNA. This, in turn, effectively enhances gene
transcription. On the other
hand, deacetylation or removal of the acetyl group restores the positive
charge condensing the structure
of the nucleosome, thereby repressing gene expression. Thus, the balance
between activities of histone
acetylases, determines the level of histone acetylation.
Eighteen different HDACs have been cloned from vertebrate organisms. These
proteins
were grouped into three different classes based on sequence homologies and
catalytic mechanism. Class
I proteins share sequence homology with the yeast protein RPD3 and comprise
HDACs 1, 2, 3 and 8.
Class II proteins are homologous to the yeast HDA1 protein. HDACs 4, 5, 6, 7,
9, and 10 belong to this
class. HDAC11 shares homology to both class I and class II enzymes. All class
I and class II proteins, as
well as HDAC11, are likely to make use of the same catalytic mechanism that
involves the hydrolysis of
an amide bond via a catalytic zinc ion. Class III enzymes, in contrast, which
are homologous to the yeast
Sir protein, differ from class I and class II proteins because they transfer
an acetyl group from an
acetylated lysine residue to NAD, simultaneously cleaving the dinucleotide.
The molecular cloning of
gene sequences encoding proteins with HDAC activity has established the
existence of a set of discrete
HDAC enzyme isoforms.
Recent research efforts have highlighted the important role of HDACs in cancer
biology.
Leukemic fusion proteins such as PML-RAR, PLZF-RAR or AML-ETO were shown to
recruit HDACs
inappropriately (Minucci et al. Molecular Cell 5, 811, 2000; Amann et al.,
Mol. Cell. Biol. 21, 6470,
2001). In B-cell lymphoma, aberrant expression of BCL6 was shown to lead to an
anomalous
recruitment of HDACs (Bereshchenko et al. Nat Genet. 2002 Dec;32(4):606-13.)
In prostate and breast
cancer, the severity of the disease was found to correlate with high
expression levels of the polycomb
protein EZH2, that recruits histone deacetylases and shows histone methyl
transferase activities
(Varambally et al., Nature 419, 624, 2002; Kleer et al., Proc. Natl. Acad. Sci
USA 100, 11606, 2003).
Cancer is the second leading cause of human death next to coronary disease.
Worldwide, millions of
people die from cancer every year. In the United States alone, as reported by
the American Cancer
Society, cancer causes the death of well over a half-million people annually,
with over 1.2 million new
cases diagnosed per year.
-2-
CA 02553468 2006-07-13
WO 2005/071079 PCT/EP2005/000559
Representative cancer types include carcinoma (e.g., adenocarcinoma), sarcoma,
myeloma, leukemia, and lymphoma, and mixed types of cancers, such as
adenosquarnous carcinoma,
mixed mesodermal tumor, carcinosarcoma, and teratocarcinoma. Exemplary cancers
include, lung
cancer, breast cancer, colon cancer, rectal cancer, endometrial cancer, and
ovarian cancer as well as
AIDS-related cancers (e.g., Kaposi's Sarcoma, Aff)S-related lymphoma). Also
included are brain cancers
(e.g., adult brain tumor, childhood brain stem glioma,
digestive/gastrointestinal cancers (e.g., colon
cancer, esophageal cancer, gallbladder cancer, anal cancer, extrahepatic bile
duct cancer, gastrointestinal
carcinoid tumor). Also included are neurologic cancers (e.g., neuroblastoma,
pituitary tumor, and
primary central nervous system lymphoma), respiratory/thoracic cancers (e.g.,
non-small cell lung cancer,
small cell lung cancer, malignant mesothelioma, and malignant thymoma), and
gynecologic cancers (e.g.,
cervical cancer, endometrial cancer, gestational trophoblastic tumor, ovarian
epithelial cancer, ovarian
germ cell tumor, ovarian low malignant potential tumor, uterine sarcoma,
vaginal cancer, and vulvar
cancer), and unknown primary cancers.
HDAC inhibitors (HDACi) have been shown to inhibit tumor growth in animal
models
of breast, prostate, lung and stomach cancer, neuroblastoma and leukemia.
These inhibitors have been
found to have anti- proliferative effects, including induction of Gl/S and
G2lM cell cycle arrest,
differentiation and apoptosis of transformed and normal cells and reversal of
transformation. Arrest at
Gl/S is believed to be meditated by induction of the CDK-inhibitor p21.
Support for this observation
becomes evident when considering that a g21 defective colon cancer cell line
failed undergo Gl arrest in
response to HDACi. Several inhibitors are presently being evaluated as single
agents and in combination
regimens with cytotoxic agents for the treatment of advanced malignancies
(reviewed in P. A. Marks et
al., Curr. Opin. Oncol. 2001 Nov.;l3(6):477-83).
Indeed, several small molecule inhibitors of HDAC have shown anti-
proliferative
activities on a number of tumor cell lines and potent anti-tumor activity in
pre-clinical tumor xenograft
models. Refer to CBHA (D. C. Coffey et al., 2001, Cancer Res. 61 (9):3591-4),
pyroxamide, (L. M.
Butler et al, 2001, Clin. Cancer Res. 7(4): 962-70), and CHAP31 (Y. Komatsu et
al., 2001, Cancer Res.
61 ( 11):4459-66). For example, histone daacetylase inhibitors, phenylbutyrate
and valproic acid have
shown promise in the treatment of promyelocytic leukemia and several other
IiDAC inhibitors are being
studied as treatments for cancers. Other potential HDAC inhibitors effective
in arresting cell-
proliferation include suberoylanilide hydroxamic acid (SARA) (Butler et al.,
2000; Marks et al., 2001);
m-carboxycinnamic acid bis-hydroxanude (Coffey et al., 2001); and pyroxamide
(Butler et al., 2001) as
well as LAQ824 (Remiszewski, SW Curr_ Med. Chem. 10, 2393, 2003.), PND101
(Plumb et al., Mol.
Cancer Ther. 2, 721, 2003), FK228 (Yoshida et al., Curr. Med. Chem. 10, 2351,
2003), MS27-275 (Saito
et al., Proc. Natl. Acad. Sci. USA 96, 4592, 1999) and CI994 (Kraker et al.,
Mol. Cancer Ther. 2, 401,
-3-
CA 02553468 2006-07-13
WO 2005/071079 PCT/EP2005/000559
2003). The data suggest that inhibition of the action of HDACs causes a
variety of cellular responses
including the accumulation of hyperacetylatad histones, altered gene
expression, and cell cycle arrest.
Consequently, regulation of gene expression through the inhibition of the
nuclear
enzyme histone deacetylase (HDAC) thus represents one of several possible
regulatory mechanisms
whereby chromatin activity can be affected. As such, the inhibition of H1~AC
activity represents a novel
approach for the intervention of cell cycle regulation and that HDAC
inhibitors have great therapeutic
potential in the treatment of cell proliferativa diseases or conditions.
However, a distinct drawback attending the use of conventional HI~AC
inhibitors is the
fording that all of the known histone deacetylase inhibitors are non-specific
for a particular histone
deacetylase isoform, and more or less inhibit all members of both the histone
deacetylase families
equally. See Grozinger, C. M., et al., Proc. Natl. Acad. Sci. U.S.A.
96:48684873, 1999. See also Marks
et al., J. National Cancer Inst. 92:1210-1216 (2000), who review histone
deacetylase inhibitors and their
role in studying differentiation and apoptosis.
As well, there is a paucity of understanding of the pharmacology of HDACi
molecules
and their interaction with a target HDAC. Also, the identity of specific HDAC
proteins and their
association with specific anti-proliferative diseases is lacking. In addition,
all known HDAC inhibitors
have been shown to possess adverse effects such as mylosuppession and GI-
toxicity, which, in turn, leads
to dose-limitation, thereby adversely effecting their inhibitory action. This
paucity of understanding,
together with the limited knowledge in the art of the identification of
specific HDAC subtypes and
diseases associated therewith, has hampered the rational design, testing and
screening of potent, selective
HDAC inhibitors that interact with specific human HDAC subtypes. Thus, while a
number of natural
product and synthetic HDAC inhibitors have been reported (J. Med. Chem. 1999,
42, 3001; and PNAS,
1998, 95, 3003), there still exists a need for inhibitors with improved
profiles of activity.
The instant application meets an unmet need by specifically identifying a
specific HDAC
protein as a potential target for inhibition as a means of treating specified
cell proliferative disorders. As
well, the invention provides for the development of methods for identifying
HDAC modulators. An
important feature of the invention is that not only does HDAC 11 knockdown
strongly inhibit cell
growth, it does so with less toxicity relative to other HDACi molecules.
SUMMARY OF THE INVENTION
The present invention relates on the unexpected discovery of the role of HDAC
11 in
certain cell proliferative disorders. The present inventors have established a
nexus between HDAC 11
and certain cancers. The findings show that inhibitors of human HDAC 11 are
useful in the treatment of
certain cell proliferative disorders l cancers in that specific inhibition of
HDAC 11 is sufficient to induce
-4-
CA 02553468 2006-07-13
WO 2005/071079 PCT/EP2005/000559
cell cycle arrest in said cell proliferative disorders notwithstanding that
the expression of or activity of
HDAC gene or its gene product in said cell proliferative disorders is normal
or near normal.
Importantly, the surprising discovery forms the basis of the invention in that
specific
HDAC 11 inhibitors (HDAC 1 li /HDAC Therapeutics) will aid in the treatment of
specific cell
proliferative disorders, i.e., colon cancer, lung cancer and cervical cancer.
Consequently, in a broad aspect, the invention relates to inhibition of human
HDAC 11
as a means of treating certain cell proliferation disorders. The HDAC 11
inhibiting molecules may be
biological entities such as dsRNA, antisense, or antibodies, etc or may be
chemical moieties (small
molecules) specific for HDAC 11.
In a first aspect, the invention provides a method for the treatment of a cell
proliferative
disorder, e.g., cancer, the method comprising administering to a subject in
need thereof an agent which
inhibits the bioactivity of human HDAC 11, or an agent which decreases
expression of HDAC 11.
In a second aspect, the present invention provides a method for inhibiting the
growth or
proliferation of a tumor cell, the method comprising contacting a tumor cell
with an agent which inhibits
the activity of I~AC 11 or an agent which decreases expression of HI~AC 11 in
an amount sufficient in
induce cell-cycle arrest.
In another aspect, the present invention provides a tumor/cancer cell into
which a nucleic
acid molecule has been introduced, the nucleic acid molecule comprising or
encoding (i) an agent which
decreases expression of HDAC l for (ii) an agent which inhibits HDAC 11
mediated activity.
In another aspect, the present invention provides a method of screening for an
agent
which inhibits cell proliferative disorders, the method comprising testing a
putative agent for the ability
to inhibit HI~AC 11 bioactivity, or decrease expression of HI~AC 11.
In a preferred embodiment of the present invention the agent is selected from
the group
consisting of a RNAi construct targeted for silencing HDAC 11 gene expression,
an HDAC 11 antisense
oligonu<cleotide, a ribozyme targeted against HDAC 11, an antibody specific
for HDAC 11, a ssDNA
targeted against HDAC lldsDNA such that the ssDNA forms a triplex with the
HDAC 11 dsDNA, an
enzyme targeted against HDAC 11 and a chemical moiety (small molecule) that
inhibits HDAC 11
function or activity.
The method of the first aspect may involve indirect inhibition of cancerous
cell growth
by inhibiting HDAC 11 mediated bioactivity or andlor direct inhibition by
blocking HDAC 11 expression
or activity in said cancer cells. As will be recognized by those skilled in
this field there are a number of
means by which the method of the present invention may be achieved.
In one embodiment, the method according to the first aspect proposes
specifically
inducing gene silencing , i.e., silencing the gene encoding HDAC 11 by a
phenomenon commonly
referred to as RNA interference.
-5-
CA 02553468 2006-07-13
WO 2005/071079 PCT/EP2005/000559
In another embodiment, the method is achieved by targeting the HDAC 11 gene
directly
for example by using triple helix (triplex) methods in which a ssDNA molecule
can bind to the dsDNA
and prevent transcription.
In another embodiment, the method is achieved by inhibiting translation of the
HDAC 11
mRNA using dsRNA constructs specific for a region of the HDAC 11 gene wherein
the HDAC 11 gene
expression is silenced via RNA interference.
In another embodiment, the method is achieved by inhibiting translation of the
HDAC 11
mRNA using synthetic antisense DNA molecules that do not act as a substrate
for RNase H and act by
sterically blocking gene expression.
In another embodiment, the method is achieved by inhibiting translation of the
HDAC 11
mRNA by destabilizing the mRNA using synthetic antisense DNA molecules that
act by directing the
RNase H-mediated degradation of the HDAC 11 mRNA present in the heteroduplex
formed between the
antisense DNA and mRNA.
In another embodiment, the method is achieved by inhibition of the function of
a HDAC
11 gene product by drugs that effectively inhibit or block the deacetylation
activity attendant a native
HDAC 11 protein.
Within the context of the present invention, the HDAC l linhibitory agents may
be
administered either alone or in combination with one or more additional anti-
cancer agents which will be
known to a person skilled in the art.
Thus, an embodiment of the invention proposes using an RNAi construct that
specifically downregulate expression of human HDAC 11, thereby effectively
inducing cell-cycle arrest
of cancerous cells. Thus, an aspect of the invention features small
interfering nucleic acid molecule for
modulating HDAC gene expression.
In accordance with the above, the invention provides methods of silencing a
specific
gene, via the use of a novel mechanism referred to herein as RNA interference
(RNAi), which uses a
specific RNAi construct effective to silence a specified gene. The method
involves introducing a dsRNA
specific for a region within a human HDAC 11 nucleotide sequence into a cell,
and maintaining the cell
under conditions and for a time sufficient to obtain degradation of mRNA of
the target gene. The
double-stranded structure comprises a nucleotide sequence which is
substantially similar or identical to at
least a part of a target gene in a mammalian cell. The RNA comprises a first
complementary RNA strand
and a second RNA strand, wherein the first complementary RNA strand comprises
the nucleotide
sequence corresponding to the target gene. Each of the first two RNA strands
of the dsRNA have a 3'-
terminus and a 5'-terminus. The siRNA comprises between about 20 and about 24
nucleotides in length,
preferably 22 nucleotides (siRNA construct), and the target gene comprises a
contiguous sequence of
nucleotides within the target gene, e.g., human IIDAC gene.
-6-
CA 02553468 2006-07-13
WO 2005/071079 PCT/EP2005/000559
At least one of the two RNA strands of the dsRNA (siRNA construct) may have a
nucleotide overhang of between one and four nucleotides, preferably one or two
nucleotides, in length.
The dsRNA may have only one nucleotide overhang, preferably on the 3'-terminus
of the complementary
RNA strand. At least one of the ends of the dsRNA may further comprise a
chemical linker, such as a
hexaethylene glycol linker. The linker may connect the 5'-terminus of the
first complementary RNA
strand and the 3'-terminus of the second RNA strand.
The first RNA strand of the dsRNA may have the nucleotide sequence of any one
of
SEQ ID NOS: 3-6; and the second RNA strand may have a complementary nucleotide
sequence. Each of
the first strand has a sequence of nucleotides substantially
identical/complementary to a portion of the
sequence of nucleotides set forth in SEQ ID NO:1. The dsRNA may comprise a
single-self
complementary RNA strand, wherein one end comprises a loop structure and the
other end comprises the
two termini. The dsRNA may hare a nucleotide overhang of between about one and
about four
nucleotides, preferably one or two nucleotides, in length. The small
interfering nucleic acid molecule of
the invention can be unmodified or chemically-modified.
In a further aspect, the invention relates to a pharmaceutical composition
comprising the
dsRNA and a pharmaceutically acceptable carrier. The mammalian cell may be a
neoplastic cell or other
cancerous cell. The target gene is human HDAC 11.
In another embodiment, the invention features dsRNA constructs comprising a
sequence
selected from the group consisting of SEQ ID NOS: 3-10.
An alternative embodiment of the present invention is drawn to a purified or
isolated
antisense nucleic acid comprising a nucleic acid sequence complementary to at
least a portion of a target
gene effective to silence said gene. The portion may be an intragenic
sequence, intergenic sequence,
sequences spanning at least a portion of two or more genes, 5' noncoding
region, or 3' noncoding region
within an operon comprising a proliferation-required gene whose activity or
expression is inhibited by an
antisense nucleic acid. The target gene is human HDAC 11 of SEQ ID NO: l
including variants and
biologically active fragments thereof.
Another embodiment of the present invention is a vector comprising a promoter
operably
linked to a nucleic acid encoding a polypeptide whose expression is inhibited
by an antisense nucleic
acid comprising a nucleic acid sequence complementary to at least a portion of
a target gene effective to
silence said gene. Preferably, the target gene is HDAC 11. Another embodiment
of the present
invention is a method of inhibiting cell proliferative disorders comprising
inhibiting the activity or
reducing the amount of a gene product whose expression is inhibited by an
antisense nucleic acid as
noted above. The gene product may comprise a polypeptide comprising a sequence
as set forth in SEQ
11? N0:2. Variants and fragments of said polypeptide are included.
CA 02553468 2006-07-13
WO 2005/071079 PCT/EP2005/000559
In an exemplary embodiment, an antisense method is used to treat tumor cells
by
antagonizing HDAC activity and blocking cell cycle progression. The method
includes, but is not limited
to, the treatment of lung tissue, cervical tissue or colon cell so as to
modulate HDAC 11 expression or
activity with a biological or chemical moiety effective to inhibit said HDAC
11 activity or expression.
In certain embodiments, the biological moiety may be an antibody specific for
the gene
product of SEQ m NO:1 effective to antagonize HDAC 11 mediated activity. In
other embodiments the
HDACi moiety may be a chemical entity capable of antagonizing the histone
deacetylase activity of
I~AC 11 effective to induce cell-cycle arrest in said cells contacted with
said chemical entity.
Another aspect of the invention is directed to methods for the identification
of molecules
that can bind to the gene product of SEQ ll~ NO: l or variants there such as
to inhibit activity of said gene
product, e.g., deacetylase activity attendant a native HDAC enzyme.
In accordance with the above, the invention provides methods for the
identification of
molecules that can bind to a polypeptide comprising an amino acid sequence set
forth in SEQ m NO: 2
and or modulate the activity of a polypeptide comprising an amino acid
sequence set forth in SEQ m
NO: 2, or molecules that can bind to nucleic acid sequences that modulate the
transcription or translation
of a polynucleotide encoding a polypeptide comprising an amino acid sequence
set forth in SEQ ~ NO:
2.
Another aspect of the present invention involves a method for modulating HDAC
bioactivity, e.g., by inhibiting the deacetylase activity of HDAC proteins, or
disrupting certain protein-
protein interactions. In general, whether carried out in vivo, in vitro, ex
vivo, or in situ, the method
comprises treating a cell with an effective amount of an HDAC therapeutic so
as to alter, relative to an
effect in the absence of treatment, one or more of (i) rate of growth or
proliferation, (ii) differentiation, or
(iii) survival of the cell. Accordingly, the method can be carried out with
HI?AC therapeutics, such as
peptide and peptidomimetics, or other molecules identified in the drug
screening methods as described
herein which antagonize the effects of a naturally-occurring HI?AC protein on
a cell.
Yet another aspect of the invention relates to compounds and compositions
useful for
modulating mammalian HDAC l lfunction and/or gene expression in a cell.
Molecules identified by such methods also fall within the scope of the present
invention.
The proposed nucleic acid molecules of the instant invention, e.g., siRNA and
antisense
constructs, antibodies, peptides etc. together with chemical moieties , e.g.,
small molecules identified
using the methods of the invention provide useful reagents and methods for a
variety of therapeutic
applications including in particular the treatment of cell proliferative
disorders such as cancers of the
cervix, lung and colon.
_g_
CA 02553468 2006-07-13
WO 2005/071079 PCT/EP2005/000559
BRIEF DESCRIPTION OF THE DRAWINGS
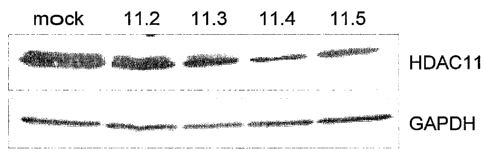
Figure 1 details the gene knock down of HDAC11 in human cells. The upper panel
details the results of a western blot analysis of HCT116 cells transfected
with 4 different siRNA
constructs , e.g., 11.2, 11.3, 11.4, 11.5 each of which is directed against a
specified region within the
HDAC11 mRNA 48 hours post-transfection.
Figure 2a and 2b detail the cell growth curves of HCT116 or A549 cells
transfected with
siRNA constructs of the invention directed against HDAC11 or GL2 (Control).
Figure 3 represents the nucleotide sequence of Human HDAC 11 (SEQ ID NO: l)
according to an embodiment of the invention.
Figure 4 shows the deduced amino acid sequence of the HDAC 11 polypeptide of
the
invention (SEQ ll~ NO: 2).
DETAILED DESCRIPTION OF THE INVENTION
The description below of the various aspects and embodiments is provided with
reference to the exemplary human HDAC 11 protein. However, the various aspects
and embodiments are
also directed to other genes that are members of the HDAC family, particularly
those that exhibit similar
biological functions.
Before the present proteins, nucleotide sequences, and methods are described,
it is to be
understood that the present invention is not limited to the particular
methodologies, protocols, cell lines,
vectors, and reagents described, as these may vary. It is also understood that
the terminology used herein
is for the purpose of describing particular embodiments only, and is not to
limit the scope of the present
invention.
Glossary:
The singular forms "a," "an," and "the" include plural reference unless the
context
clearly dictates otherwise.
All technical and scientific terms used herein have the same meanings as
commonly
understood by one of ordinary skill in the art to which this invention
pertains. The practice of the present
invention will employ, unless otherwise indicated, conventional techniques of
protein chemistry and
biochemistry, molecular biology, microbiology and recombinant DNA technology,
which are within the
skill of the art. Such techniques are explained fully in the literature. All
patents, patent applications, and
publications mentioned herein, whether supra or infra, are each incorporated
by reference in its entirety.
For convenience, the meaning of certain terms and phrases employed in the
specification, examples, and appended claims are provided below. It is also to
be understood that the
-9-
CA 02553468 2006-07-13
WO 2005/071079 PCT/EP2005/000559
terminology used herein is for the purpose of describing particular
embodiments only and is not intended
to be limiting.
8y "gene" oligonucleotide nucleic acid as used herein, refers to an
oligonucleotide,
nucleotide, or polynucleotide (e.g., DNA, cDNA, RNA), and fragments or
portions thereof, and to DNA
or RNA of genomic or synthetic origin which may be single- or double-stranded,
and represent the sense
(coding) or antise:nse (non-coding) strand. The exact size will depend on many
factors, which in turn
depends on the ultimate function or use of the oligonucleotide. The
oligonucleotide may be generated in
any manner, including chemical synthesis, DNA replication, reverse
transcription. The term should also
be understood to include, as equivalents, analogs of either RNA or DNA made
from nucleotide analogs,
and, as applicable to the embodiment being described, single (sense or
antisense) and double-stranded
polynucleotides. As a non-limiting example, fragments include nucleic acid
sequences that can be about
10 to 60 contiguous nucleotides in length, preferably, at least 15-60
contiguous nucleotides in length, and
also preferably include fragments that are at least 70-100 contiguous
nucleotides, or which are at least
1000 contiguous nucleotides or greater in length. Nucleic acids for use as
probes or primers may differ
in length as described herein.
A.s used herein, "target gene" refers to a section of a DNA strand of a double-
stranded
DNA that is complementary to a section of a DNA strand, including all
transcribed regions, that serves as
a matrix for transcription. The target gene is therefore usually the sense
strand. The target gene can be a
gene derived from a cell, an endogenous gene, a transgene, or exogenous genes
such as genes of a
pathogen, for example a virus, which is present in the cell after infection
thereof. The cell containing the
target gene can ba derived from or contained in any organism, for example a
plant, animal, protozoan,
virus, bacterium, or fungus. Non-limiting examples of animals include
vertebrates or invertebrates.
Non-limiting examples of fungi include molds or yeasts. Preferably, the target
gene is human HDAC 11
including variants and biologically equivalent sequences thereof.
When refernng to a sequence that "consists of about" a certain number of
nucleotides,
this is intended to refer to a sequence that consists of the certain number of
nucleotides plus or minus
20% or 10% of the number of nucleotides. For example, a sequence consisting of
about 10 nucleotides
refers to a sequence of from 8 -12 nucleotides.
A "delivery complex" or "siRNA vehicle" shall mean a targeting means for
delivering an
siRNA complex to a target gene. Examples of targeting means include: sterols
(e.g. cholesterol), lipids
(e.g. a cationic lipid, virosome or liposome), viruses (e.g_ adenovirus, adeno-
associated virus, and
retrovirus) or target cell specific binding agents (e.g. ligands recognized by
target cell specific receptors).
T'he term "equivalent" is understood to include nucleotide sequences encoding
functionally equivalent polypeptides. Equivalent nucleotide sequences will
include sequences that differ
by one or more nucleotide substitutions, additions or deletions, such as
allelic variants; and will,
-10-
CA 02553468 2006-07-13
WO 2005/071079 PCT/EP2005/000559
therefore, include sequences that differ from the nucleotide sequence of a
nucleic acid of interest due to
the degeneracy of the genetic code. Thus "equivalent" nucleotide sequences of
human HDAC of SEQ ID
NO:l include degenerate sequences.
"Identity," as known in the art, is the relationship between two or more
polynucleotide
sequences, as determined by comparing the sequences. Identity also means the
degree of sequence
relatedness between polynucleotide sequences, as determined by the match
between strings of such
sequences. Identity can be readily calculated (see, .e.g, Computation
Molecular Biology, Lesk, A. M.,
eds., Oxford University Press, New York (1998), and Biocornputing: Informatics
and Genome Projects,
Smith, D. W., ed., Academic Press, New York (1993), both of which are
incorporated by reference
herein). While there exist a number of methods to measure identity between two
polynucleotide
sequences, the term is well known to skilled artisans (see, e.g., Sequence
Analysis in Molecular Biology,
von Heinje, G., Academic Press (1987); and Sequence Analysis Primer,
Gribskov., M. and Devereux, J.,
eds., M Stockton Press, New York (1991)). Methods commonly employed to
determine identity between
sequences include, for example, those disclosed in Carillo, H., and Lipman,
D., SIAM J. Applied Math.
(1988) 48:1073. "Substantially identical," as used herein, means there is a
very high degree of homology
(preferably 100% sequence identity) between the respective target sequence and
the reference sequence.
Thus, where an embodiment is drawn to silencing a particular gene via t RNA
interference, "substantially
identical" refers to the nucleotide sequence comprising the strand of the
dsRNA which is complementary
to an mRNA of the target gene or a region contained therein_ The percentage of
identity between the
substantially similar nucleotide sequences is at least 80%, more desirably at
least 85%. However,
dsRNA having greater than 90% , or 95% sequence identity may be used in the
present invention, and
thus .sequence variations that might be expected due to genetic mutation,
strain polymorphism, or
evolutionary divergence can be tolerated. Although 100% identity is preferred,
the dsRNA may contain
single or multiple base-pair random mismatches between the RNA and the target
gene.
The term "complementary RNA strand" refers to the strand of the dsRNA which is
complementary to an mRNA transcript that is formed during expression of the
target gene, or its
processing products. "dsRNA" refers to a ribonucleic acid molecule having a
duplex structure
comprising two complementary and anti-parallel nucleic acid strands. Not all
nucleotides of a dsRNA
must exhibit Watson-Crick base pairs. The maximum number of base pairs is the
number of nucleotides
in the shortest strand of the dsRNA.
As an example, a nucleotide sequence when referring to an anti-sense construct
(reference sequence) is "substantially similar" to the target nucleotide
sequence when said sequence
hybridizes to the reference nucleotide sequence in 7% sodium dodecyl sulfate
(SDS), 0.5 M NaP04, 1
mM EDTA at 50°C with washing in 2X SSC, 0.1% SDS at SO°C, more
desirably in 7% sodium dodecyl
sulfate (SDS), 0.5 M NaP04, 1 mM EDTA at 50°C with washing in 1X SSC,
0.1% SDS at 50°C, more
-11-
CA 02553468 2006-07-13
WO 2005/071079 PCT/EP2005/000559
desirably still in 7% sodium dodecyl sulfate (SDS), 0.5 M NaP04, 1 mM EDTA at
50°C with washing
in 0.5X SSC, 0.1% SDS at 50°C, preferably in 7% sodium dodecyl sulfate
(SDS), 0.5 M NaP04, 1 mM
EDTA at 50°C with washing in 0.1X SSC, 0.1% SDS at 50°C, more
preferably in 7% sodium dodecyl
sulfate (SDS), 0.5 M NaP04, 1 mM EDTA at 50°C with washing in O. 1X
SSC, 0.1% SDS at 65°C, yet
still encodes a functionally equivalent gene product.
An allele or allelic sequence is an alternative form of an HDAC nucleic acid
sequence.
Alleles may result from at least one mutation in the nucleic acid sequence and
may yield altered mRNAs
or polypeptides whose structure or function may or may not be altered. Any
given gene, whether natural
or recombinant, may have none, one, or many allelic forms. Common mutational
changes that give rise
to alleles are generally ascribed to natural deletions, additions, or
substitutions of nucleotides. Each of
these types of changes may occur alone, or in combination with the others, one
or more times in a given
sequence.
"RNAi" stands for RNA-mediated interference.
The term "short interfering RNA" or "siRNA" as used herein refers to a double
stranded
nucleic acid molecule capable of RNA interference "RNAi", see for example
Bass, 2001, Nature, 411,
428-429; Elbashir et al., 2001, Nature, 411, 494-498; and Kreutzer et al.,
International PCT Publication
No. WO 00/44895; Zernicka-Goetz et al., International PCT Publication No. WO
01/36646; Fire,
International PCT Publication No. WO 99/32619; Plaetinck et al., International
PCT Publication No. WO
00/01846; Mello and Fire, International PCT Publication No. WO 01129058;
Deschamps-Depaillette,
International PCT Publication No. WO 99/07409; and Li et al., International
PCT Publication No. WO
00/44914. As used herein, siRNA molecules need not be lixW ted to those
molecules containing only
RNA, but further encompasses chemically modified nucleotides and non-
nucleotides.
"A therapeutically effective amount" of a compound is an amount which results
in a
therapeutic effect in the subject to whom it was administered.
"Inhibiting gene expression" "gene silencing" refers to a phenomenon whereby a
function of a gene is completely or partially inhibited. For example, the
action may result in decreased
production of a polypeptide encoded by the gene or decreased levels of an RNA
encoded by the target
gene. Inhibiting gene expression includes inhibiting transcription,
translation or degrading the DNA
template or RNA encoded thereby. As such, HDACi molecules may be characterized
as having the
ability to interfere with the function of a gene or gene product in such a way
as to decrease expression of
the gene or to reduce the level or activity of a product of the gene. Agents
which inhibit the activity of a
gene include agents that inhibit transcription of the gene, agents that
inhibit processing of the transcript
of the gene, agents that reduce the stability of the transcript of the gene,
and agents that inhibit translation
of the mRNA transcribed from the gene. Of particular utility to the present
invention are dsRNA
molecules, antisense RNAs that have activities against the oparons or genes to
which they specifically
-12-
CA 02553468 2006-07-13
WO 2005/071079 PCT/EP2005/000559
hybridze. For example, inhibiting histone deacetylation causes cells to arrest
in the G1 and G2 phases of
the cell cycle. In general, whether the HDAC l li molecule is an antisense or
an siRNA molecule,
inhibition of gene expression with the siRNA molecule or an anti-sense
molecule is greater in the
presence of the siRN.A or anti-sense molecule than in its absence.
According to an aspect of the invention, there is provided a HDAC therapeutic
which
causes an inhibition of cell proliferation of the contacted cells. The phrase
"inhibiting cell proliferation"
is used to denote an ability of an inhibitor of histone deacetylase to retard
the growth of cells contacted
with the inhibitor as compared to cells not contacted. An assessment of cell
proliferation can be made by
counting contacted and non-contacted cells using a Coulter Cell Counter
(Coulter, Miami, Fla.) or a
hemocytometer. Where the cells are in a solid growth (e.g., a solid tumor or
organ), such an assessment
of cell proliferation can be made by measuring the growth with calipers and
comparing the size of the
growth of contacted cells with non-contacted cells.
Preferably, growth of cells contacted with the inhibitor is retarded by at
least 50% as
compared to growth of non-contacted cells. More preferably, cell proliferation
is inhibited by 100°Io (i.e.,
the contacted cells do not increase in number). Most preferably, the phrase
"inhibiting cell proliferation"
includes a reduction in the number or size of contacted cells, as compared to
non-contacted cells. Thus,
an inhibitor of histone deacetylase according to the invention that inhibits
cell proliferation in a contacted
cell may induce the c ontacted cell to undergo growth retardation, to undergo
growth arrest, to undergo
programmed cell death (i.e., to apoptosis), or to undergo necrotic cell death.
In some preferred embodiments, the contacted cell is a neoplastic cell. The
term
"neoplastic cell" is used to denote a cell that shows aberrant cell growth.
Preferably, the aberrant cell
growth of a neoplastic cell is increased cell growth. A neoplastic cell may be
a hyperplastic cell, a cell
that shows a lack of contact inhibition of growth in vitro, a benign tumor
cell that is incapable of
metastasis in vivo, or a cancer cell that is capable of metastasis in vivo and
that may recur after attempted
removal. The term "tumorigenesis" is used to denote the induction of cell
proliferation that leads to the
development of a neoplastic growth.
In some preferred embodiments, the contacted cell is in an animal. Thus, the
invention
provides a method for treating a cell proliferative disease or condition in an
animal, comprising
administering to an animal in need of such treatment a therapeutically
effective amount of a histone
deacetylase inhibitor of the invention. Preferably, the animal is a mammal,
more preferably a
domesticated mammal. Most preferably, the animal is a human. Preferably, the
histone deacetylase is
I~AC 11.
The term "cell proliferative disease or condition" is meant to refer to any
condition
characterized by aberrant cell growth, preferably abnormally increased
cellular proliferation. In
particularly preferred embodiments, the invention provides a method for
inhibiting neoplastic cell
-13-
CA 02553468 2006-07-13
WO 2005/071079 PCT/EP2005/000559
proliferation in an animal comprising administering to an animal having at
least one neoplastic cell
present in its body a therapeutically effective amount of a histone
deacetylase inhibitor of the invention.
As used herein, the terms "histone deacetylase" and "HDAC" are intended to
refer to any
one of a family of enzymes that remove acetyl groups from the ~-amino groups
of lysine residues at the
N-terminus of a histone. Unless otherwise indicated by context, the term
"histone" is meant to refer to
any histone protein, including Hl, H2A, H2B, H3, H4, and H5, from any species.
Preferably the histone
deacetylase is a human HDAC, and most preferably the human histone deacetylase
is human HDAC 11.
The HDAC 11 of the invention is characterized as proteins capable of removing
acetyl groups from
primary amines or amino acids, either free or in the context of a polypeptide
chain.
The term "IiDACi" (histone deacetylase inhibitor) refers to any agent that
inhibits
IIDAC 11 activity or expression of the HDAC 11 gene. Where the IiDACi is a
small molecule, it is
capable of inhibiting HDAC activity, by for example, interacting with a
histone deacetylase and
inhibiting its enzymatic activity. Inhibiting histone deacetylase enzymatic
activity means reducing the
ability of a histone deacetylase to remove an acetyl group from a histone. In
some preferred
embodiments, such reduction of histone deacetylase activity is at least about
50%, more preferably at
least about 75%, and still more preferably at least about 90%. In other
preferred embodiments, histone
deacetylase activity is reduced by at least 95% and more preferably by at
least 99%.
An "HDAC therapeutic," whether inhibitory or potentiating with respect to
modulating
histone deacetylation, can be, as appropriate, any of the preparations
described herein, including isolated
polypeptides, gene therapy constructs, antisense molecules, or agents
identified in the drug and bioactive
screening assays and methods well known to one skilled in the art as well as
those described herein.
The term "sample" or "biological sample", is meant to be interpreted in its
broadest
sense. A biological sample suspected of containing nucleic acid encoding an
FiDAC protein, or
fragments thereof, or an HDAC protein itself, may comprise a body fluid, an
extract from cells or tissue,
chromosomes isolated from a cell (e.g., a spread of metaphase chromosomes),
organelle, or membrane
isolated from a cell, a cell, nucleic acid such as genomic DNA (in solution or
bound to a solid support
such as for Southern analysis), RNA (in solution or bound to a solid support
such as for Northern
analysis), cDNA (in solution or bound to a solid support), a tissue, a tissue
print and the like. Methods
for detecting HDAC 11 expression are well known, e.g., Northern Blot, Southern
Blot, Western Blot etc.
An HDAC-11 protein refers to the HDAC 11 proteins or polypeptides described
herein,
as well as other human homologs of these HDAC sequences, in addition to
orthologs and paralogs
(homologs) of the HDAC sequences in other species, ranging from yeast to other
mammals, e.g.,
homologous histone deacetylase. The full-length cDNA for HDAC 11 is as set
forth in SEQ ID NO:1
and it predicts a protein comprising the amino acid sequence as set forth in
SEQ ID N0:2. The term
ortholog refers to genes or proteins that are homologs via speciation, e.g.,
closely related and assumed to
-14-
CA 02553468 2006-07-13
WO 2005/071079 PCT/EP2005/000559
have common descent based on structural and functional considerations.
Orthologous proteins function
as recognizably the same activity in different species. The term paralog
refers to genes or proteins that
are homologs via gene duplication, e.g., duplicated variants of a gene within
a genome. (See, W. M.
Fritch, 1970, Syst. Zool., 19:99-113.
It will be appreciated that, under certain circumstances, it may be
advantageous to
provide homologs of the preferred HDAC polypeptides which function in a
limited capacity as one of
either an HDAC agonist (i.e., mimetic), or an HDAC antagonist, in order to
promote or inhibit only a
subset of the biological activities of the naturally-occurring form of the
protein. Thus, specific biological
effects can be elicited by treatment with a homolog of limited function, and
with fewer side effects,
relative to treatment with agonists or antagonists which are directed to all
of the biological activities of
naturally-occurring forms of HDAC proteins. Homologs (i.e., isofonns or
variants) of a target HDAC
polypeptides can be generated by mutagenesis, such as by discrete point
znutation(s), or by truncation.
For example, mutation can yield homologs that retain substantially the same,
or merely a subset of, the
biological activity of the HDAC polypeptide from which it was derived.
Alternatively, antagonistic
forms of the protein can be generated which are able to inhibit the function
of the naturally-occurring
form of the protein, such as by competitively binding to an IiDAC substrate,
or HDAC-associated
protein. Non-limiting examples of such situations include competing with wild-
type HIIAC in the
binding of p53 or a histone. Also, agonistic forms of the protein can be
generated which are
constitutively active, or have an altered Kcat or Km for deacylation
reactions. Thus, the HDAC protein
and homologs thereof may be either positive or negative regulators of
transcription and/or replication.
As used herein, "HDAC activity", including refers to the ability of an HDAC
polypeptide
to deacetylate hi stone proteins.
The term "biologically active", i.e., functional, refers to a protein or
polypeptide or
peptide fragment thereof having structural, regulatory, or biochenucal
functions of a naturally occurring
molecule. Likewise, "immunologically active" refers to the capability of the
natural, recombinant, or
synthetic HDAC, or any oligopeptide thereof, to induce a specific immune
response in appropriate
animals or cells, for example, to generate antibodies, and to bind with
specific antibodies.
In specific embodiments, a polynucleotide of the invention can comprise a
sequence of
nucleotides that specifically target a region of an HI~AC 11 nucleotide
sequence via RNA interference.
Said regions may include nucleotides positions 511-531, 580-600, 1030-1050 and
1342-1362 of the
native IiDAC gene sequence, represented herein as SEQ ID NO:1.
Methods of the Invention
Several human cancers have been associated with malfunctions in HAT and HDAC
activity. Indeed, it is well documented that multiple classical features of
cancer cells can be manifested
-15-
CA 02553468 2006-07-13
WO 2005/071079 PCT/EP2005/000559
by improper histone deacetylation. For review see Wade, P. A. Hum Mol Genet;
10(7):693-698 (2001.
In addition, perturbations of gene expression have long been acknowledged to
account for a vast number
of diseases including, numerous forms of cancer, vascular diseases, neuronal
and endocrine diseases.
Consequently, one of the major challenges of medicine has been to regulate the
expression of targeted
genes that are implicated in a wide diversity of physiological responses.
However, conventional treatments for cancer, are marred by major drawbacks.
For
example, current treatment protocols for cell proliferative disorders, such as
cancer require empirically
derived cytotoxic chemotherapy that is marginally more toxic for the hyper
proliferative cell than normal
cells.
The therapeutic effects of HDAC inhibition are believed to occur through the
induction
of differentiation and/or apoptosis through the up-regulation of genes such as
the cyclin dependent kinase
inhibitors, p21 and p27 (see, e.g., W. Wharton et al., 2000, J. Biol. Chem.
275(43):33981-7; L. Huang et
al., 2000, Mol. Med. 6(10):849-66). However, although known HDAC inhibitors
are efficacious as anti-
tumor agents, they are also associated with toxicity (see, e.g., V. Sandor et
al., 2002, Clin. Cancer Res.
8(3):718-28). Such toxicity is believed to be caused by a non-selective
mechanism of targeting multiple
HDACs. Thus, despite the potent anti-tumor activity of HDAC inhibitors, it is
still unclear which
HDACs are necessary to produce an anti-proliferative response. Furthermore,
little progress has been
made in comparing the HDAC gene expression profiles in tumor versus normal
cells. Differential
HDAC expression may underlie the tumor-selective responses of HDAC inhibition.
In addition, a
cellular growth advantage may be conferred by the expression of particular
I3DAC's.
As regards gene inhibition as an alternative strategy for treating cancer,
targeted
inhibition of specific genes has been very difficult to achieve. As well,
current approaches for
suppressing gene expression, including site-directed gene disruption etc
require complex genetic
manipulations or heavy dosages of suppressors that often exceeds the toxicity
tolerance level of the host
cell. Therefore, there is a need for further insight into the consequences of
selective HDAC inhibition, or
activation.
Thus, while conventional methods for either the prevention or the alleviation
of
symptoms and/or the slowing down of disease progression may improve quality of
life in cancer patients,
there exists an unmet need in the comprehensive treatment and prevention of
said cancerous disorders,
especially those treatments where specific enzymes are targeted, thereby
improving the overall treatment
protocol. As such, there exists the need for therapeutics effective not only
in reversing the physiological
changes associated with hyper cell proliferative disorders but also being more
selective and potent with
fewer toxic side effects. The use of compounds to modulate the expression of
selective HDAC subtypes,
e.g., HDAC 11 is of therapeutic significance.
-16-
CA 02553468 2006-07-13
WO 2005/071079 PCT/EP2005/000559
With respect to therapeutic intervention via agents which target expression of
genes,
these generally require acute specificity and/or are time consuming. For
example, zinc-finger proteins
(Choo, Y., et al., Nature (1994) 372:642), act at the DNA level, interacting
with the target sequence and
blocking transcription. However, gene fusions occur randomly and within
introns, hence requiring a
unique or "custom" zinc-forger for each patient. Hammerhead ribozymes, (James,
H. A, and I. Gibson,
Blood (1998) 91:371) in turn, require specific nucleotide sequences in the
target gene, which are not
always present.
More recently, double-stranded RNA molecules (dsRNA) have been shown to block
gene expression in a highly conserved regulatory mechanism known as RNA
interference (RNAi). The
presence of long dsRNAs in cells is believed to stimulate the activity of a
ribonuclease III enzyme
referred to as dicer. Dicer is involved in the processing of the dsRNA into
short pieces of dsRNA known
as short interfering RNAs (siRNA) (Berstein et al., 2001, Nature, 409, 363).
Short interfering RNAs
derived from dicer activity are typically about 21-23 nucleotides in length
and comprise about 19 base
pair duplexes. Dicer has also been implicated in the excision of 21 and 22
nucleotide small temporal
RNAs (stRNA) from precursor RNA of conserved structure that are implicated in
translational control
(Hutvagner et al., 2001, Science, 293, 834). The RNAi response also features
an endonuclease complex
containing a siRNA, commonly referred to as an RNA-induced silencing complex
(RISC), which
mediates cleavage of single stranded RNA having sequence complementary to the
antisense strand of the
siRNA duplex. Cleavage of the target RNA takes place in the middle of the
region complementary to the
antisense strand of the siRNA duplex (Elbashir et al., 2001, Genes Dev., 15,
188). In other words, RNAi
involves a catalytic-type reaction whereby new siRNAs are generated through
successive cleavage of
long dsRNA. Thus, unlike antisense, RNAi degrades target RNA in a non-
stoichiornetric manner.
Initial attempts to harness this phenomenon for experimental manipulation of
mammalian cells were foiled by a robust and nonspecific antiviral defense
mechanism activated in
response to long dsRNA molecules. Gil et al. Apoptosis 2000, 5:107-114. The
field was significantly
advanced upon the demonstration that synthetic duplexes of 21 nucleotide RNAs
when administered to a
cell or organism, provoke degradation of endogenous messenger RNA (mRNA)
through RNAi., without
invoking a generic antiviral defense mechanisms. Elbashir et al. Nature 2001,
411:494-498; Caplen et al.
Proc Natl Acad Sci 2001, 98:9742-9747. As a result, small-interfering RNAs
(siRNAs) have become
powerful tools to dissect gene function. Numerous groups have sought the
development of DNA-based
vectors capable of generating such siRNA within cells.
Several groups have recently attained this goal and published similar
strategies that, in
general, involve transcription of short hairpin (sh)RNAs that are efficiently
processed to form siRNAs
within cells. Paddison et al. PNAS 2002, 99:1443-1448; Paddison et al. Genes &
Dev 2002, 16:948-958;
Sui et al. PNAS 2002, 8:5515-5520; and Brummelkamp et al. Science 2002,
296:550-553. These reports
-17-
CA 02553468 2006-07-13
WO 2005/071079 PCT/EP2005/000559
describe methods to generate siRNAs capable of specifically targeting numerous
endogenously and
exogenously expressed genes. In addition, dsRNA has been reported to have anti-
proliferative
properties, which makes it possible also to envisage therapeutic applications
(Aubel et al_, Proc. Natl.
Aced. Sci., USA 88:906 (1991)). For example, synthetic dsRNA has been shown to
inhibit tumor growth
in mice (Levy et al. Proc. Nat. Aced. Sci. USA, 62:357-361 (1969)), is active
in the treatment of
leukemic mice (Zeleznick et al., Proc. Soc. Exp. Biol. Med. 130:126-128
(1969)); and inhibits
chemically-induced tumorigenesis in mouse skin (Gelboin et al., Science
167:205-207 (1970)).
Consequently, the invention provides a reliable method for inhibiting the
proliferation
and migration of tumor cells in human patients, and for inhibiting metastatic
cancer development. It is
believed that such a method would have implications for the therapeutic
treatment of cell proliferative
disorder sin general, and in particular those disorders that are mediated by
HDAC 11. In the main, the
invention relies on the discovery that various hyper proliferative disorders
such as some forms of cancer
may be amenable to treatment by specifically inhibiting the expression or
biological activity of HDAC
11. The details of these experiments are outlined in the Examples herebelow.
In its broadest aspect, the invention provides I~AC modulatory (i.e.,
preferably
inhibitory) compounds e.g., HDACi molecules that are likely to play an
important role in treating or
reducing cellular proliferation. Preferred histone deacetylase inhibitors are
those that interact with and
reduce the enzymatic activity of a histone deacetylase that is involved in
tumorigenesis. Specifically, the
HDAC modulating moieties can be chemical moieties capable of inhibiting 131~AC
11 activity in vivo or
ex vivo or biological moieties capable of inhibiting HDAC 11 activity or
expression levels in vivo.
Exemplary HDAC modulating moieties that are biological in nature include siRNA
duplexes capable of
specifically silencing HI~AC expression i~a vivo or anti-sense constructs
capable of specifically inhibiting
HDAC 11 gene transcription or translation in vivo or antibodies or peptides
that inhibit I3T)AC 11 protein
function, some of which are exemplified in greater detail here after.
In certain aspects of the inventions, the disclosed HDAC 11 inhibitors,
antisense
molecules, anti-HDAC antibodies, or antibody fragments can be used as
treatments for colon, cervical or
lung cancer.
Inhibition of HDAC 11 Expression
(I) RNAi based Embodiments:
In one approach, the present invention relates to the specific inhibition of
expression of a
histone deacetylase gene in a mammal using a short double stranded RNA
(dsRNA). As noted, supra,
dsRNA directs the sequence-specific degradation of mRNA through a process
known as RNA
interference (RNAi). AS detailed in the Examples section, the present
inventors have demonstrated that
dsRNA of approximately 19-24 nucleotides, preferably 20-23 nucleotides, and
most preferably 22
-18-
CA 02553468 2006-07-13
WO 2005/071079 PCT/EP2005/000559
nucleotides in length, which have a nucleotide sequence complementary to the
target gene or to a region
contained therein, can specifically and efficiently mediate RNAi. The present
invention encompasses
these short dsRNAs and their use for specifically inactivating gene function.
The use of these dsRNAs
enables the targeting of mRNAs of mammalian gene involved in cell
proliferative disorders. Thus, the
dsRNAs of the present invention are useful for treating diseases caused by a
specified HDAC gene,
particularly malignant diseases such as lung cancer, cervical cancer and
cancer of the colon.
The dsRNAs of the present invention comprises a double stranded structure, and
have a
nucleotide sequence which is substantially identical to at least a part or
portion of the target gene.
Preferably, the target gene is HDAC 11 including fragments and biologically
equivalent variants thereof.
Consequently, the discovery of a nexus between HDAC 11 and certain cancerous
conditions, the siRNA molecules of the invention together with the methods
detailed herein represent a
novel therapeutic approach to treat a variety of pathologic indications,
particularly cancer and any other
diseases or conditions that are related to the levels of human HDAC level of
expression or actW ity in a
cell or tissue, alone or in combination with other therapies. The reduction of
HDAC activity or
expression and thus reduction in the level of the respective protein relieves,
to some extent, the
symptoms of the disease or condition.
Preferably, each sequence of a siRNA molecule of the invention is
independently about
11 to about 24 nucleotides in length, more preferably of from about 17 to
about 23, e.g, about 17, 18, 19,
20, 21, 22, or 23 base pairs. In some instances, the length of the nucleotide
sequence is such as to be
effective in silencing the target gene.
In accordance with an embodiment of the invention, the invention relates to an
RNA
having a double-stranded structure and a nucleotide sequence which is
substantially identical to at least a
part of the target gene. The RNA is between about 19 and about 24 nucleotides
in length. The dsRNA
comprises two complementary RNA strands, one of which comprises a nucleotide
sequence which is
substantially identical to a portion of the target gene. In a preferred
embodiment, at least one end of
the dsRNA has a single-stranded nucleotide overhang of between one and four,
preferably one or two
nucleotides. As used herein, a "nucleotide overhang" refers to the unpaired
nucleotide or nucleotides that
protrude from the duplex structure when the 5'-terminal end of one RNA strand
extends beyond the 3'
terminus end of the other strand, or vice versa dsRNAs having at least one
nucleotide overhang have
unexpectedly superior inhibitory properties than their blunt-ended
counterparts. It is well accepted that
the presence of one or two nucleotide overhang appears to strengthen the
interference activity of the
respective dsRNA, without diminishing the overall stability of the structure,
as typically happens with
other dsRNA having no overhang. Preferably, the single-stranded overhang is
located at the 3'-terminal
end of the complementary RNA strand (also referred to herein as the "S 1"
strand). It is believed that
such a configuration produces a further increase in efficiency.
-19-
CA 02553468 2006-07-13
WO 2005/071079 PCT/EP2005/000559
The nucleotide sequence on the complementary RNA strand (S 1 strand)
preferably has
between 18 and 24 nucleotides, most preferably 19-21 nucleotide: s. The
complementary RNA strand of
the dsRNA preferably has fewer that 23 nucleotides because as noted by
researchers in the field of RNAi
such dsRNA molecules exhibit superior intracellular stability and avoid an
interferon response.
At least one end of the dsRNA may be modified to improve resistance to
degradation
and/or dissociation of the two strands of the duplex. As well, the
cohesiveness of the double-stranded
structure formed by base pairing between the complementary RIA strands can be
further improved by
the presence of one, and preferably two, chemical linkages. Chemical linking
may be achieved by a_ny of
a variety of well-known techniques, including through covalent, ionic or
hydrogen bonds; hydrophobic
interactions, preferably van der Waals or stacking interactions; or by means
of metal-ion coordination.
The purines of the dsRNA may also be replaced with purine analogues. Most
preferably, the chemical
linkage is achieved using a hexa-ethylene glycol linker on one end of the
dsRNA. In a preferred
embodiment, the linkage is formed between the 5'-terminus of the complementary
RNA strand and the 3'-
terminus of the second RNA strand.
In another embodiment, the present invention relates to a method for
inhibiting the
expression of a target gene comprising a fusion site using a dsRhTA. The
method comprises introducing a
dsRNA having a nucleotide sequence which is substantially identical to at
least a part of a target gene
into a mammalian cell. The RNA is preferably between 20 and 23 nucleotides in
length, most prefe:~rably
22 nucleotides. The resulting cell is maintained under conditions and for a
time sufficient to achieve
degradation of mRNA of the target gene, thereby silencing expression of the
target gene.
In still another embodiment, the invention relates to a method for treating a
mammal
having a disease caused by the expression of a mammalian HDAC, preferably HDAC
11. The method
comprises administering the dsRNA of the invention to the animal, such that
expression of the targets
gene is silenced.
In an exemplified embodiment, the target gene comprises a HDAC gene, i.e.,
HDAC 11
gene. In this example, the complementary RNA (S1) strand of the dsRNA has the
sequence selected
from the groups consisting of SEQ m NOS: 3-6, and the second f S2) strand has
the sequence of SEQ )D
N0:7-10. Such a construct is useful for treating either colon, cervical or
lung cancer. The dsRNA
(siRNA molecule) may be administered using any acceptable method known to one
skilled in the art,
including liposomes etc. Where the HI?AC l li is a siRNA molecule and its use
is envisioned as a
therapeutic, such therapeutic agents can be administered by a varaety of well
known techniques,
including inhalation, oral ingestion, and injection, particularly intravenous
or intraperitoneal injection, or
injection directly into the affected bone marrow.
The siRNA molecules of the invention are added directly, or can be complexed
with
cationic lipids, packaged within liposomes, or otherwise delivered to target
cells or tissues. The nucleic
- 20 _
CA 02553468 2006-07-13
WO 2005/071079 PCT/EP2005/000559
acid or nucleic acid complexes can be locally administered to relevant tissues
ex vivo, or in vivo through
injection, infusion pump or stmt, with or without their incorporation in
biopolymers. In particular
embodiments, the nucleic acid molecules of the invention comprise sequences
shown in Table I and/or
FIGS. 4 and 5. Examples of such nucleic acid molecules consist essentially of
sequences defined in this
table.
In another aspect, the invention provides mammalian cells containing one or
more
siRNA molecules of this invention. The one or more siRNA molecules can
independently be targeted to
the same or different sites.
The nucleic acid molecules of the instant invention, individually, or in
combination or in
conjunction with other drugs, can be used to treat diseases or conditions
discussed herein. For example,
to treat a particular disease or condition, the siRNA molecules can be
administered to a subject or can be
administered to other appropriate cells evident to those skilled in the art,
individually or in combination
with one or more drugs under conditions suitable for the treatment.
In a further embodiment, the siRNA molecules can be used in combination with
other
known treatments to treat conditions or diseases discussed above. For example,
the described molecules
could be used in combination with one or more known therapeutic agents to
treat a disease or condition.
Non-limiting examples of other therapeutic agents that can be readily combined
with a siRNA molecule
of the invention are enzymatic nucleic acid molecules, allosteric nucleic acid
molecules, antisense,
decoy, or aptamer nucleic acid molecules, antibodies such as monoclonal
antibodies, small molecules,
and other organic and/or inorganic compounds including metals, salts and ions.
In one embodiment, the invention features an expression vector comprising a
nucleic
acid sequence encoding at least one siRNA molecule of the invention, in a
manner which allows
expression of the siRNA molecule. For example, the vector can contain
sequences) encoding both
strands of a siRNA molecule comprising a duplex. The vector can also contain
sequences) encoding a
single nucleic acid molecule that is self complementary and thus forms a siRNA
molecule. Non-limiting
examples of such expression vectors are described in Paul et al., 2002, Nature
Biotechnology, 19, 505;
Miyagishi and Taira, 2002, Nature Biotechnology, 19, 497; Lee et al., 2002,
Nature Biotechnology, 19,
500; and Novina et al., 2002, Nature Medicine, advance online publication
doi:10.1038/nm725.
In another embodiment, the invention features a mammalian cell, for example, a
human
cell, including an expression vector of the invention.
In yet another embodiment, the expression vector of the invention comprises a
sequence
for a siRNA molecule having complementarity to a RNA molecule referred to by a
Genbank Accession
numbers, for example BALE genes such as Genbank Accession Nos. NM__p12104
(RACE), NM__
006222 (PIN-1), L76517 (PS-1) and/or L43964 (PS-2).
-21-
CA 02553468 2006-07-13
WO 2005/071079 PCT/EP2005/000559
In one embodiment, an expression vector of the invention comprises a nucleic
acid
sequence encoding two or more siRNA molecules, which can be the same or
different.
In another aspect of the invention, siRNA molecules that interact with target
RNA
molecules and down-regulate gene encoding target RNA molecules for example
target RNA molecules
referred to by Genbank Accession numbers herein) are expressed from
transcription units inserted into
DNA or RNA vectors. The recombinant vectors can be DNA plasmids or viral
vectors. siRNA
expressing viral vectors can be constructed based on, but not limited to,
adeno-associated virus,
retrovirus, adenovirus, or alphavirus. The recombinant vectors capable of
expressing the siRNA
molecules can be delivered as described herein, and persist in target cells.
Alternatively, viral vectors
can be used that provide for transient expression of siRNA molecules. Such
vectors can be repeatedly
administered as necessary. Once expressed, the siRNA molecules bind and down-
regulate gene function
or expression via RNA interference (RNAi). Delivery of siRNA expressing
vectors can be systemic,
such as by intravenous or intramuscular administration, by administration to
target cells ex-planted from
a subject followed by reintroduction into the subject, or by any other- means
that would allow for
introduction into the desired target cell.
Pharmaceutical composition comprising formulations containing a dsRNA of the
invention are also encompassed by the invention. The pharmaceutical
composition may be administered
in a dosage sufficient to inhibit expression of the target gene.
As used herein, a "pharmaceutical composition" comprises a pharmacologically
effective
amount of a dsRNA and a pharmaceutically acceptable carrier. As a sed herein,
"pharmacologically
effective amount," "therapeutically effective amount" or simply "effective
amount" refers to that amount
of an siRNA effective to produce the intended pharmacological, therapeutic or
preventive result. For
example, if a given clinical treatment is considered effective when there is
at least a 50% reduction in a
measurable parameter associated with a disease or disorder, a therapeutically
effective amount of a drug
for the treatment of that disease of disorder is the amount necessary to
effect that at least 50% reduction.
The term "pharmaceutically acceptable carrier" refers to a carrier for
administration of a
therapeutic agent. Such carriers are well known to one skilled in the art.
Therapeutic kits are also envisioned by the inventors. The kits comprise the
reagents,
agents, and materials that may be required to practice the above methods of
the invention, including, but
not limited to those reagents necessary for transfection or transformation of
cells with siRNA. Such kits
may also comprise siRNA made by the methods of the present invention. The kits
will generally contain,
in suitable container means, a pharmaceutically acceptable formulate on of
siRNA. The kit may have a
single container means, and/or it may have distinct container means for each
compound or each reaction
mixture or step.
(II) Antisense Embodiments
_22_
CA 02553468 2006-07-13
WO 2005/071079 PCT/EP2005/000559
The present invention also encompasses various methods and compositions for
inhibiting
the transcription of the HDAC 11 gene. Similarly, the invention also provides
methods and compositions
for inhibiting the translation of HDAC 11 mRNA into protein.
For example, in one approach, a method of inhibiting the transcription of the
HDAC 11
gene comprises contacting the HDAC 11 gene with a I~AC 11 antisense
polynucleotide. In yet another
embodiment, there is provided a method of inhibiting I3DAC 11 mRNA translation
comprising
contacting the HDAC 11 mRNA with an antisense polynucleotide. An alternative
approach proposes a
BAC 11 specific ribozyme to cleave the HI~AC 11 rriessage, thereby inhibiting
translation. Such
antisense and ribozyme based methods can also be directed to the regulatory
regions of the HDAC 11
gene, such as the HDAC 11 promoter and/or enhancer elements. The use of
antisense and ribozyme
molecules to inhibit transcription and translation is well known in the art.
Other factors that inhibit the transcription of BAC 11 are also useful to
treat cancers
expressing HDAC 11. Similarly, factorslagent that specifically interfere with
HDAC 11 mediated
processes, e.g. deacetylation etc. are also useful to treat cancers. Thus,
cancer treatment methods
utilizing such factorslagents also fall within the scope of the invention.
Representative, nucleic acid related en-sbodiments of the invention disclosed
herein are
genomic DNA, cDNAs, ribozymes, and antisense molecules, as well as nucleic
acid molecules based on
an alternative backbone, or including alternative bases, whether derived from
natural sources or
synthesized, and include molecules capable of inhibitirig the RNA or protein
expression of HDAC 11.
Antisense RNA technology has been developed as an approach to inhibiting gene
expression. An "antisense" RNA molecule is one which contains the complement
of, and can therefore
hybridize with, messenger RNAs of the cell. It is widely believed that the
hybridization of antisense
RNA to its cellular RNA complement effectively prevents expression of the
cellular RNA, perhaps by
limiting its translatability. While various studies have involved the
processing of RNA or direct
introduction of antisense RNA oligonucleotides to cells for the inhibition of
gene expression (Brown, et
al., 1989; Wickstrom, et al., 1988; Smith, et al., 1986; Buvoli, et al.,
1987), the more common means of
cellular introduction of antisense RNAs has been through the construction of
recombinant vectors which
will express antisense RNA once the vector is introduced into the cell.
A principle application of antisense RNA technology has been in connection
with
attempts to affect the expression of specific genes. Antisense technology has
also been applied in
attempts to inhibit the expression of various oncogenes _ For example, Kasid,
et al., 1989, report the
preparation of recombinant vector construct employing Craf 1 cDNA fragments in
an antisense
orientation, brought under the control of an adenovirus 2 late promoter.
According to the report, the
introduction of the recombinant construct into a human squamous carcinoma
resulted in a greatly
reduced tumorigenic potential relative to cells transfected with control sense
transfectants. Similar
- 23 -
CA 02553468 2006-07-13
WO 2005/071079 PCT/EP2005/000559
results have been reported in the use of Cmyc antisense constructs which were
effective to differentiate
and inhibit G-1 progression in Friend Murine Erythroleukemia cells. See
Prochownik, et al., 1988.
Therefore, it appears that antisense technology shows potential promise as a
means of external control of
gene expression.
A representative method of the invention encompasses antisense molecules that
may be
RNAs or other molecules, including peptide nucleic acids (PNAs) or non-nucleic
acid molecules such as
phosphorothioate derivatives, that specifically bind DNA or RNA in a base pair-
dependent manner.
Applicants note that a skilled artisan may readily obtain these classes of
nucleic acid molecules using the
nucleic acid sequence of human HDAC 11 )(SEQ ID NO:1) polynucleotides
sequences disclosed herein.
In general, antisense technology entails the administration of exogenous
oligonucleotides
that bind to a target polynucleotide located within the cells. The term
"antisense" refers to the fact that
such oligonucleotides are complementary to their intracellular targets
sequences, specifically human
HDAC 11 of SEQ ID NO:1 including variants and fragments thereof. Refer to Jack
Cohen,
Oligodeoxynucleotides, Antisense Inhibitors of Gene Expression, CRC Press,
1989; and Synthesis 1:1-5
(1988). The HI)AC 11 specific antisense oligonucleotides of the present
invention include derivatives
detailed in Jack Cohen, supra, such as phosphorothioate derivatives or S-
oligonucleotides which are
believed to exhibit enhanced cancer cell growth inhibitory action. As noted is
said reference, S-oligos
(nucleoside phosphorothioates) are isoelectronic analogs of an oligonucleotide
(O-oligo) in which a
nonbridging oxygen atom of the phosphate group is replaced by a sulfur atom.
The S-oligos of the
present invention can be prepared by treatment of the corresponding O-oligos
with 3H-1,2-benzodithiol-
3-one-1,1-dioxide, which is a sulfur transfer reagent. See Iyer, R. P. et al,
J. Org. Chem. 55:4693-4698
(1990); and Iyer, R. P. et al., J. Am. Chem. Soc. 112:1253-1254 (1990).
Other HDAC 11 specific antisense oligonucleotides of the present invention may
include
morpholino antisense oligonucleotides known in the art (see, e.g., Partridge
et al., 1996, Antisense &
Nucleic Acid Drug Development 6: 169-175).
The HDAC 11 specific antisense oligonucleotides of the present invention
typically can
be RNA or DNA that is complementary to and stably hybridizes with the first
100 5' codons or last 100 3'
codons of the HDAC 11 specific genomic sequence or the corresponding mRNA.
Absolute
complementarity is not required, although high degrees of complementarity are
preferred. Use of an
oligonucleotide complementary to this region allows for the selective
hybridization to HDAC 11 specific
mRNA and not to mRNA specifying other proteins. In one embodiment, HDAC 11
specific antisense
oligonucleotides of the present invention are 15 to 30-mer fragments of the
antisense DNA molecule that
have a sequence that hybridizes to HDAC 11 specific mRNA. Optionally, HDAC 11
specific antisense
oligonucleotide is a 30-mer oligonucleotide that is complementary to a region
in the first 10 5' codons or
last 10 3' codons of HDAC 11 specific. Alternatively, the antisense molecules
are modified to employ
-24-
CA 02553468 2006-07-13
WO 2005/071079 PCT/EP2005/000559
ribozymes in the inhibition of HDAC 11 specific expression, see, e.g., L. A.
Cou.-ture & D. T.
Stinchcomb; Trends Genet 12: 510-515 (1996).
(III) Inhibition of HDAC 11 specific Protein Function
The invention also provides various methods and compositions for inhibiting
the
biological, enzymatic functions of specific HDAC molecules or its association
with other proteins) as
well as methods for inhibiting HDAC 11 specific function.
(A) Inhibition of HDAC 11 specific with Intracellular Antibodies
Cancer is known to be a multistep process where cellular growth becomes
progressively
dysregulated and cells progress from a normal physiological state to
precancerous and then cancerous
states (see, e.g., Alers et al., Lab Invest. 77(5): 437-438 (1997) and Isaacs
et al., Cancer Surv. 23: 19-32
(1995)). In this context, upon presentation of clinical signs of colon,
cervical or lung cancer,
examination of a biological sample for evidence of dysregulated cell growth,
will allow for early
detection of such aberrant physiology, before a pathologic state such as
cancer has progressed to a stage
that therapeutic options are more limited and or the prognosis is worse.
In such examinations, the status of various markers for one of lung, colon or
cervical
cancer in a biological sample of interest can be compared, for example, to the
status of the same markers
in a corresponding normal sample (e.g. a sample from that individual or
alternatively another individual
that is not affected by a pathology). An alteration in the status of said
markers in the biological sample
(as compared to the normal sample) provides evidence of dysregulated cellular
g-zowth. In addition to
using a biological sample that is not affected by a pathology as a normal
sample, one can also use a
predetermined normative value such as a predetermined normal level of mRNA
expression to compare
the marker status in a sample.
The term "status" in this context is used according to its art accepted
meaning and refers
to the condition or state of a gene and its products. Typically, skilled
artisans use a number of
parameters to evaluate the condition or state of a gene and its products.
These include, but are not
limited to the location of expressed gene products (including the location of
the marker expressing cells
etc) as well as the level, and biological activity of expressed gene products
including HDAC 11.
Consequently, HDAC 11 proteins of SEQ ID NO: 2 or fragments or immunologically
active fragments there have a number of different uses including generating
antibodies. Antibodies
raised against an HDAC protein or fragment thereof are useful in the
management of human cancers
characterized by aberrant cell growth. Thus, for patients having or presenting
symptoms consistent with
any one of more of the above mentioned cancers, early intervention by way of
inhibiting I~AC 11
protein function or expression level may present an amicable means of early
intervention notwithstanding
the observations of the inventors that in said cell proliferative disorders
the expression levels of HDAC
11 is normal or near normal. Thus, an aspect of the invention provides
antibodies that bind to HDAC 11
- 25 -
CA 02553468 2006-07-13
WO 2005/071079 PCT/EP2005/000559
proteins or related proteins in a manner specific to abrogate the biological
proteins attendant said
proteins, e.g., enzymatic activities such as deacetylation etc. Preferred
antibodies specifically bind to
HDAC 11 and do not bind (or bind weakly) to peptides or proteins that are not
HDAC 11.
In another approach, a recombinant vector that encodes single chain antibodies
that
specifically bind to HDAC l late introduced into HDAC 11 expressing cells
previously correlated to a
specific cancer, via gene transfer technologies. Accordingly, the encoded
single chain ant-HDAC
l lantibody is expressed intracellularly, binds to HDAC 11 protein, and
thereby inhibits its function.
Methods for engineering such intracellular single chain antibodies are well
known. Such intracellular
antibodies, also known as "intrabodies", are specifically targeted to a
particular compartment within the
cell, providing control over where the inhibitory activity of the treatment is
focused. This technology has
been successfully applied in the art (for review, see Richardson and Marasco,
1995, T1BTECH vol. 13).
Various methods for the preparation of antibodies are well known in the art.
For
example, antibodies can be prepared by immunizing a suitable mammalian host
using a HDAC 11
protein, peptide, or fragment, in isolated or immunoconjugated form
(Antibodies: A Laboratory Manual,
CSH Press, Eds., Harlow, and Lane (1988); Harlow, Antibodies, Cold Spring
Harbor Press, NY (1989)).
(B) Inhibition of HDAC 11 with Recombinant Proteins
Recombinant molecules that specifically bind to HDAC 11 and thereby inhibit
HDAC 11
specific function are also within the scope of the invention. Fir example,
these recombinant molecules
specifically prevent or inhibit HDAC 11 mediated activity within a cell.
Screening Methods
Since HDAC 11 and its polymorphism variant play a role in transcription,
chromosome
stability, cell cycle progression, aging, regulation of neuronal phenotype,
DNA replication and the
response to DNA damage, and in view of the nexus found by the present
inventors vis-a-vis normal
expression levels of the protein of SEQ ID NO: 2 and certain cell
proliferation disorders, the HDAC 11
protein, and respective nucleic acids can be used in screening assays to
identify candidate agents or drugs
that modulate IiDAC bioactivity, for potential use to treat neoplastic
disorders, for example, to kill
cancer cells and tumor cells exhibiting uncontrolled cell growth for numerous
reasons, e.g., the lack of a
suppressor molecule such as p53. As well, HDAC 11 proteins and encoding
nucleic acids, as well as the
agents that modulate HDAC 11 activity or function, can be used as effectors in
methods to regulate cell
growth, e.g., to kill neoplastic cells.
Antagonists or inhibitors of HDAC 11 may be antibodies specific for the HDAC
11
protein that can be used directly as an antagonist, or indirectly as a
targeting or delivery mechanism for
bringing a pharmaceutical agent to cells or tissue which express the HDAC 11
protein. Other methods to
inhibit the expression of the HDAC 11 protein include antisense and triple
helix strategies as described
herein.
-26-
CA 02553468 2006-07-13
WO 2005/071079 PCT/EP2005/000559
Other antagonists or inhibitors of the HDAC 11 protein may be produced using
methods
which are generally known in the art, including the screening of libraries of
pharmaceutical agents to
identify those which specifically bind the HDAC 11 protein. The HDAC 11
protein, or fragment thereof,
preferably its functional or immunogenic fragments, or oligopeptides related
thcreto, can be used for
screening libraries of compounds in any of a variety of drug screening
techniques. The fragment
employed in such screening may be free in solution, affixed to a solid
support, borne on a cell surface, or
located intracellularly. The formation of binding complexes, between the HDA_C
11 protein, or fragment
thereof, or derivative thereof, and the agent being tested, may be measured
using methods known in the
art.
An alternative technique for drug screening provides for high throughput
screening of
compounds having suitable binding affinity to the HDAC 11 protein. See publi
shed PCT application
W084/03564.
Since the protein of SEQ )D N0:2 may deacetylate substrates, preferably
acetylated
histones, either directly or indirectly as enzymes cofactors, this property
may be exploited to identify
HDAC 11 inhibitors. The deacetylation activity of the protein of SEQ >D N0:2:
or fragment thereof may
be assayed using any of a number of methods known to those skilled in the art.
The following techniques may be used to detect or assay acetylation or
deacetylation.
Such methods, when applicable, may be used for in i~itro assays. A
representatiEVe assay proposes
treating a specific cell line with different concentrations of a candidate
substance. Methods for
measuring hyperacetylation of histones have been described in detail (Verdel
and Khochbin, 1999;
Fischle et al., 1999; Grozinger et al., 1999) and are known in the art. For
example, the appearance of
hyperacetylated H3histone can be monitored using antibody raised against
hyperacetylated histone H3
and detected by cytofluorimetric measurement of immunofluorescence (see, Vam
Lint et al., 1996).
Alternatively, cells may be lysed, the histones purified and analyzed on a
Tritor~lacid/urea gel.
Analytical ultracentrifugation is often used also to detect histone
acetylation.
Acetylated substrates used in such methods are preferably acetylated histones
and
acetyltransferases. Measuring the rate of deacetylation of [3H]-labeled
acetylated histones also is a
useful assay in the present invention. For example, the HDAC 11 protein or a
fragment thereof is added
to a sample containing a substrate under conditions favoring deacetylation,
and allowed to catalyze the
deacetylation of the substrate. In a preferred embodiment, the deacetylation
is carried out using a
standard assay such as those described in Landry and collaborators. Refer to
La~dry et al., Proc. Natl.
Acad. Sci. 97:5807-5811 (2000), the disclosure of which is incorporated by
reference in its entirety.
Deacetylated histones obtained by this method may be mixed with purified naked
DNA (plasmid
preparations for example) in order to reconstitute chromatine-like structures
in vitro. Such structures are
of great interest in the study of enzymatic factors involved in transcription
and replication. Natural
_27_
CA 02553468 2006-07-13
WO 2005/071079 PCT/EP2005/000559
transcription factors are unable to enter the condensed chromatin and the gene
function is effectively
switched-off.
A representative method according to the inventi on is directed to a method
for
determining the ability of a candidate substance to inhibit HDAG 11 activity;
generally including the
steps of:
(a) providing a source of HDAC 11 enzyme,
(b) contacting the enzyme with a candidate substance;
(c) determining the enzyme function in step (b); and
(d) comparing the enzyme function in step (c) with the enzyme function of the
enzyme in the absence of the candidate substance, wherein increased enzyme
function in the presence of
the candidate substance, as compared to enzyme function in the absence of the
candidate substance,
identifies the candidate substance as an inhibitor of cell proliferation.
An exemplary embodiment proposes a method for screening test agents to
identify
modulating agents which inhibit or antagonize deacetylation activity of a
histone deacetylase, proposes
(i) combining an isolated polypeptide of SEQ ID N0:2 or polymorphic or
biologically active variants thereof having a histone deacetylase activity
with a histone deacetylase
substrate and a test agent in a reaction mixture; and
(ii) determining the conversion of the substrate to product; wherein a
statistically significant decrease in the conversion of the substrata in the
presence of the test agent
indicates identification of a modulating agent which inhibits or antagonizes
the deacetylation activity of
histone deacetylase.
Also contemplated are methods for screening test agents to identify modulating
agents
that inhibit or antagonize interaction of histone deacetylase with a histone
deacetylase binding protein.
This method proposes : (i) combining a HDAC 11 polypeptide having a histone
deacetylase activity with
the histone deacetylase binding protein and a test agent in a reaction
mixture; and (ii) detecting the
interaction of the polypeptide with the histone deacetylase bindir~g protein
to form a complex; wherein a
statistically significant decrease in the interaction of the polypeptide and
protein in the presence of the
test agent indicates identification of a modulating agent which inJ~ibits or
antagonizes interaction of the
HDAC 11 polypeptide with the histone deacetylase binding protein.
High Throughput Screening - Test compounds can be screened for the ability to
bind to
HIaAC 11 polypeptides or polynucleotides or to affect HDAC 11 activity or HDAC
11 gene expression
using high throughput screening. Using high throughput screening, many
discrete compounds can be
tested in parallel so that large numbers of test compounds can be quickly
screened. The most widely
established techniques utilize microtiter plates. The wells of the inicrotiter
plates typically require assay
_ 28 _
CA 02553468 2006-07-13
WO 2005/071079 PCT/EP2005/000559
volumes that range from 2 to 500 ~,1. In addition to the plates, many
instruments, materials, pipettors,
robotics, plate washers, and plate readers are commercially available to fit
the microtiter format.
Yet another example is described by Salmon et al., Molecular Diversity 2, 57-
63 (1996).
In this example, combinatorial libraries were screened for compounds that had
cytotoxic effects on
cancer cells growing in agar.
Another high throughput screening method is described in Beutel et al., U.S.
Pat. No.
5,976,813, which proposes placing test samples in a porous matrix. One or more
assay components are
then placed within, on top of, or at the bottom of a matrix such as a gel, a
plastic sheet, a filter, or other
form of easily manipulated solid support. When samples are introduced to the
porous matrix they diffuse
sufficiently slowly, such that the assays can be performed without the test
samples running together.
Other assays include binding assays, functional assays etc which are well
known to one
skilled in the art.
It is recognized that the HDAC 11 therapeutics, e.g., HDAC 11 inhibitor
molecules
(HDAC l li) contemplated by the present invention, e.g., antisense molecules,
anti-HDAC antibodies, or
antibody fragments, small molecules, (chemical moieties, or dsRNA constructs
(siRNA molecules) of the
invention can be used in combination with other HDAC inhibitory agents, e.g.,
trichostatin A (D. M.
Vigushin et al., 2001, Clin. Cancer Res. 7(4):971-6); trapoxin A (Itazaki et
al., 1990, J. Antibiot.
43:1524-1532), MS-275 (T. Suzuki et al., 1999, J. Med. Chem. 42(15):3001-3),
CHAP (Y. Komatsu et
al., 2001, Cancer Res. 61(11):4459-66), Cl-994 (see, e.g., P. M. LoRusso et
al., 1996, New Drugs
14(4):349-56), SARA (V. M. Richon et al., 2001, Blood Cells Mol. Dis.
27(1):260-4), depsipeptide
(FR901228; FK228; V. Sandor et al., 2002, Clin. Cancer Res. 8(3):718-28), CBHA
(D. C. Coffey et al.,
2001, Cancer Res. 61 (9):3591-4), pyroxamide, (L. M. Butler et al, 2001, Clin.
Cancer Res. 7(4):962-70),
CHAP31 (Y. Komatsu et al., 2001, Cancer Res. 61(11):4459-66), HC-toxin (Liesch
et al., 1982,
Tetrahedron 38:45-48), chlamydocin (Closse et al., 1974, Helv. Chim. Acta
57:533-545), Cly-2 (Hirota et
al., 1973, Agri. Biol. Chem. 37:955-56), WF-3161 (Umehana et al., 1983, J.
Antibiot. 36, 478-483; M.
Kawai et al., 1986, J. Med. Chem. 29(11):2409-11), Tan-1746 (Japanese Pat. No.
7196686 to Takeda
Yakuhin Kogyo KK), apicidin (S. H. Kwon et al., 2002, J. Biol. Chem.
277(3):2073-80), as LAQ824
(Remiszewski, SW Curr. Med. Chem 10, 2393, 2003.), PKD101 (Plumb et al., Mol.
Cancer Ther. 2, 721,
2003), FK228 (Yoshida et al., Curr. Med. Chem 10, 2351, 2003), MS27-275 (Saito
et al., Proc. Natl.
Acad. Sci. USA 96, 4592, 1999) and CI994 (Kraker et al., Mol. Cancer Ther. 2,
401, 2003) and analogs
thereof.
Kits
For use in the therapeutic applications contemplated herein, kits are also
within the scope
of the invention. Such kits can comprise a carrier, package or container that
is compartmentalized to
receive one or more containers such as vials, tubes, and the like, each of the
containers) comprising one
-29-
CA 02553468 2006-07-13
WO 2005/071079 PCT/EP2005/000559
of the separate elements to be used in the method. For example, the
containers) can comprise an
antisense construct with detectable label. Alternatively, the cori-tainer may
comprise a probe that is or
can be detectably labeled. Such probes can be an antibody or polynucleotide
specific for an HDAC 11
specific-related protein or a HDAC 11 specific gene or message ~ respectively.
The kit can include all or
part of the amino acid sequence of SEQ m NO: 2 or analogs thereof, or a
nucleic acid molecules that
encodes such amino acid sequences.
A label can be present on the container to indicate that the composition is
used for a
specific therapy or non-therapeutic application, and can also incZicate
directions for either in vivo or- in
vitro use, such as those described above. Directions and or othe=r information
can also be included on a
label or on an insert which is included with the kit.
General Considerations for Therapeutic Strategies
Gene transfer and gene therapy technologies can be used to deliver therapeutic
polynucleotide molecules i.e., antisense or dsRNA i molecules to cancer cells
or tissue. Synthesizing
HDAC 11 specific (i.e., antisense, ribozyme, RNAi or polynuclcotides encoding
intrabodies and other
I~AC 11 specific inhibitory molecules). A number of gene therapy approaches
are known in the axt.
For example, recombinant vectors encoding HDAC 11 specific antisense
polynucleotides, ribozyrnes,
factors capable of interfering with HDAC 11 specific transcripti on, and so
forth, can be delivered to
target tumor cells using such gene therapy approaches. Other techniques
including those detailed in the
Examples below may also be used to deliver the HDAC therapeutics to the target
cell or tissue.
Importantly, the aforementioned therapeutic approaches can be combined with
any one
of a wide variety of surgical, chemotherapy or radiation therapy regimens. The
therapeutic approaches of
the invention can enable the use of reduced dosages of chemotherapy (or other
therapies) and/or les s
frequent administration, an advantage for all patients and particularly for
those that do not tolerate the
toxicity of the chemotherapeutic agent well.
The anti-tumor activity of a particular composition (e.g., antisense,
ribozyme, intrabody,
small molecule inhibitor), or a combination of such compositions, can be
evaluated using various in vitro
and in vivo assay systems. In vitro assays that evaluate therape~.tic activity
include cell growth assays,
soft agar assays and other assays indicative of tumor promoting activity,
binding assays capable of
determining the extent to which a therapeutic composition will inhibit the
binding of HDAC 11 specific
to a binding partner, etc.
In vivo, the effect of a HDAC 11 specific therapeutic composition may be
evaluated in a
suitable animal model. For example, xenogenic lung cancer or colon cancer
models can be used, wherein
human lung or colon cancer explants or passaged xenograft tissues are
introduced into immune
compromised animals, such as nude or SCll~ mice (Shibayama et al., 1991, J
Urol., 146(4):1136-7;
-30-
CA 02553468 2006-07-13
WO 2005/071079 PCT/EP2005/000559
Beecken et al., 2000, Urology, 56(3):521-526). Indeed, the overall efficacy
can be predicted using assays
that measure inhibition of tumor formation, tumor regression or metastasis,
and the like.
In vivo assays that evaluate progression of apoptosis are useful in evaluating
therapeutic
compositions. In an exemplary embodiment, xenografts from tumor bearing mice
treated with an HDAC
11 therapeutic can be examined for the presence of apoptotic foci and compared
to untreated control
xenograft-bearing mice. The extent to which apoptotic foci are found in the
tumors of tha treated mice,
in turn, may provide an indication of the therapeutic efficacy of the
composition.
The HI~AC 11 therapeutic compositions identified herein can be formulated into
pharmaceutical compositions comprising a carrier suitable for the desired
delivery method. Suitable
carriers include any material that when combined with the therapeutic
composition retains the anti-tumor
function of the therapeutic composition and is generally non-reactive with the
patient's immune system.
See, Remington's Pharmaceutical Sciences l6th Edition, A. Osal., Ed.,
1980).
Dosages and administration protocols for the treatment of cancers using the
foregoing
methods will vary with the method and the target cancer and will generally
depend on a number of other
factors appreciated in the art.
EXAMPLE 1
Materials and Methods.
siRNA design.
Sequences of the type AA(N19)dTdT (N, any nucleotide) from the targeted mRNA
were
designed based on the rules suggested by Elbashir et al. (Genes Dev, 2001) and
purchased from
Dharmacon Research or Oligo Engine as annealed, deprotected, double-stranded
2lmers. The N1g
sequences targeting HDAC11 mRNA corresponded to the following nucleotide
positions relative to the
Genbank accession number NM 024827: HDAC11.2 nt. 513-531; HDAC11.3 nt. 582-
600; HDAC11.4
nt. 1032-1050; HDAC11.5 nt. 1344-1362.
Representative dsRNA constructs for specifically silencing human HDAC 11 via
RNA
interference include
11.2
1 AAGUUUCUGU UUGAGCGUGU G (SEQ ID NO: 3)
CAAAGACA AACUCGCACA CAA (SEQ ~ NO: 7)
11.3
1 AAUGGGCAUG AGCGAGACUU AAC (SEQ ~ NO: 4)
ACCCGUAC UCGCUCUGAA UUGAA (SEQ ID NO: 8)
-31 -
CA 02553468 2006-07-13
WO 2005/071079 PCT/EP2005/000559
11.4
1 AACUCt~GACA CACCGCUGCU U (SEQ ~ NO: 5)
GAGLJCUGU GUGGCGACGA AAA (SEQ ID NO: 9)
11.5
1 AACUG~1GAAU UGGAGAGGAC A (SEQ m NO: 6)
GACLJCUUA ACCUCUCCUG UAA (SEQ m NO: 10)
Construct 11.2 targets nucleotides 511-531 of human HDAC gene, i.e. SEQ )D NO:
1,
comprising the nucleotide sequence aagtttctgtttgagcgtgtg.
Construct 11.3 targets nucleotides 580-600 of human HDAC gene, i.e. SEQ m NO:
1,
comprising the nucleotide sequenc a aatgggcatgagcgagacttc.
Construct 11.4 targets nucleotides 1030-1050 of human HDAC gene, i.e. SEQ ~
NO: 1,
comprising the nucleotide sequence aactcagacacaccgctgctt.
Construct 11.5 targets nucleotides 1342-1362 of human HDAC gene, i.e. SEQ m
NO: 1,
comprising the nucleotide sequence aactgagaattggagaggaca..
Cell culture and siRNA transfection.
HeLa human cervical carcinoma cell line was grown in Dulbecco modified Eagle
medium (DMEM, Gibco); HCT-116 colorectal carcinoma cells were grown in McCoy's
5A medium
(Gibco); A549 human lung carcinoma cells were cultured in F12K Nutrient
Mixture (Gibco). All media
were supplemented with 10% fetal bovine serum (FBS, Gibco), 2 mM L-glutamine
(Gibco), 100 U of
penicillin per ml and 100 p,g of streptomycin per ml (Gibco).
The day before transfection cells were trypsinized and transferred to 6-well
plates at 70-
80% confluency, in a final volume of 2 ml. Transfection was performed by using
Lipofectamine 2000
(Invitrogen) according to manufacturer's instructions with minor
modifications. Briefly, a few hours
before transfection, culture medium was replaced with 1 ml of fresh medium
supplemented with 12%
FBS without antibiotics. Lipofectamine 2000 (4 p,l per well) was incubated
with 100 ~l of Optimem 1
medium (Gibco) for 5 minutes at room temperature; in the meantime 3 p,l of 20
p,l~ siRNA were diluted
in another 100 ~1 of Optimem 1. T'he two mixtures were then mixed and
incubated for 25 minutes at
room temperature. The resulting final mixture was added to the cells of a 6-
well plate to have a final
siRNA concentration of 50 nM. Transfection was carried out for 4 hours, then
transfection mixture was
-32-
CA 02553468 2006-07-13
WO 2005/071079 PCT/EP2005/000559
removed, cells were washed, trypsinized, and replated at different
concentrations for the different assays.
Silencing was assayed 48 hours post transfection by western blot and Taqman
analysis.
Immunoblotting.
A rabbit polyclonal antibody was raised against a peptide corresponding to the
first 15
amino acids of the open reading frame of HDAC11 (MLHTTQLYQHVPETR) (SEQ ID NO:
14). For
Western blot analysis, transfected cells were washed once with phosphate-
buffered saline (PBS) and
lysed directly in the culture dish with 150 ~l per well of 1 % SDS in PB S.
Protein extracts were sheared
with Qiashredder columns (Qiagen). Samples were diluted in sample buffer,
heated for 5 min at 95°C,
electrophoresed on 10% SDS-polyacrylamide gels and blotted onto a
nitrocellulose membrane. Primary
antibody against HDAC11 was diluted 1:5000 in blocking buffer and incubated at
4°C overnight. A
peroxidase-conjugated secondary antibody (Pierce) diluted 1:5000 in blocking
buffer was incubated 1
hour at room temperature and detection was carried out using the SuperSignal
West Pico
Chemiluminescent Substrate (Pierce). To confirm equal loading, the filter was
then reprobed with an
anti-GAPDH as the primary antibody, diluted 1:5000 in blocking buffer and
incubated at 4°C overnight.
An alkaline phosphatase-conjugated secondary antibody was used at a 1:2000
dilution in blocking buffer,
and revealed with NBT and BC1P substrates.
The results are disclosed in Figure 1 which details the gene knock down of
HDAC11 in
human cells. The upper panel details the results of a western blot analysis of
HCT116 cells transfected
with 4 different siRNA constructs , e.g., 11.2, 11.3, 11.4, 11.5 each of which
is directed against a
specified region within the HDAC11 mRNA 48 hours post-transfection.
Cell growth assay.
After 4 hours of transfection, cells were washed, trypsinized and replated in
96-well
Cytostar-T scintillating microplates at 2500 or 5000 cells per well in
triplicates. 80 nCi of methyl-14C-
thymidine (Amersham Pharmacia) were added to each well. Labeled thymidine
incorporation was
measured with a TOP Count NXT Microplate Scintillation and Luminescence
Counter (Packard) every
8-12 hours up to 72 hours post transfection.
Figure 2a and 2b, detail the cell growth curves of HCT 116 or A549 cells
transfected with
siRNA constructs of the invention directed against HDAC11 or GL2 (Control).
RNA extraction and Taqman analysis.
At the indicated time points transfected cells were quickly washed once in PBS
and total
RNA was extracted using the Rneasy kit (Qiagen), according to manufacturer's
instructions. A Dnase I
-33-
CA 02553468 2006-07-13
WO 2005/071079 PCT/EP2005/000559
treatment was added to the basic extraction protocol, as suggested by the
manufacturer. RNAs were
quantified by spectrophotometric absorbance and their quality was analyzed on
1 % denaturing gel.
Quantitative RT-PCR of HDAC11 mRNA was performed in triplicate on 100 ng of
total
RNA per well, by using the One Step RT-PCR Master Mix (Applied Biosystem) with
the following set of
primers and probe: primer sense: 5'-CCTCAGGCGGGTACCAGAA-3' (SEQ ID NO: 11),
200 nM;
primer antisense: 5'-CAGGCCAAACAGATTAAGTATGGA-3', (SEQ )D N0:12 ) 400 nM;
probe
(FAM-TAMRA): 5'-CGCACAGCCCGCATCATTGCT-3' (SEQ ID NO: 13), 150 nM.
Normalization
was done on the same amount of template by amplification of human GAPDH or
human (3-actin using
the corresponding Pre-Developed Taqman Assay Reagents (Applied Biosystem), in
triplicate, in the same
plate as HDAC11. Detection was performed with an ABI Prism 7900HT Sequence
Detection System.
HDAC11 NM 024827
HDAC11.1
AA GUGGUCCUUUGCUGUUGCU (SEQ 11? NO: 15) nt. 274-294 %GC=47.6 Tm=52.9
HI~AC11.2
AA GUUUCUGUUUGAGCGUGUG (SEQ >D NO: 16) nt_ 511-531 %GC=42.9 Tm=47
HDAC11.3
AA UGGGCAUGAGCGAGACUUC (SEQ 1D NO: 17) nt_ 580-600 %GC=52.4 Tm=53.6 DG dimer=-
2.1
HDAC 11.4
AA CUCAGACACACCGCUGCUU (SEQ ID NO: 18) nt. 1030-1050 %GC=52.4 Tm=54.7 DG loop=
-
1.
HDAC11.5
AA CUGAGAAUUGGAGAGGACA (SEQ ID NO: 19) nt. 1342-1362 %GC=42.9 Tm= 49.2
-34-
DEMANDES OU BREVETS VOLUMINEUX
LA PRESENTE PARTIE DE CETTE DEMANDE OU CE BREVETS
COMPRI~:ND PLUS D'UN TOME.
CECI EST L,E TOME 1 DE 2
NOTE: Pour les tomes additionels, veillez contacter le Bureau Canadien des
Brevets.
JUMBO APPLICATIONS / PATENTS
THIS SECTION OF THE APPLICATION / PATENT CONTAINS MORE
THAN ONE VOLUME.
THIS IS VOLUME 1 OF 2
NOTE: For additional valumes please contact the Canadian Patent Office.