Note: Descriptions are shown in the official language in which they were submitted.
DEMANDE OU BREVET VOLUMINEUX
LA PRESENTE PARTIE DE CETTE DEMANDE OU CE BREVET COMPREND
PLUS D'UN TOME.
CECI EST LE TOME 1 DE 3
CONTENANT LES PAGES 1 A 189
NOTE : Pour les tomes additionels, veuillez contacter 1e Bureau canadien des
brevets
JUMBO APPLICATIONS/PATENTS
THIS SECTION OF THE APPLICATION/PATENT CONTAINS MORE THAN ONE
VOLUME
THIS IS VOLUME 1 OF 3
CONTAINING PAGES 1 TO 189
NOTE: For additional volumes, please contact the Canadian Patent Office
NOM DU FICHIER / FILE NAME
NOTE POUR LE TOME / VOLUME NOTE:
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
METHODS FOR IDENTIFYING RISK OF OSTEOARTHRITIS AND
TREATMENTS THEREOF
Field of the Invention
[0001] The invention relates to genetic methods for identifying risk of
osteoarthritis and treatments
that specifically target such diseases.
Back rg ound
[0002] Osteoarthritis (OA) is a chronic disease usually affecting weight-
bearing synovial joints.
There are approximately 20 million Americans affected by OA and it is the
leading cause of disability in
the United States. In addition to extensive human suffering, OA also accounts
for nearly all knee
replacements and more than half of all hip replacements in the United States.
Despite its prevalence,
OA is poorly understood and there are few treatments available besides anti-
inflammatory drugs and
joint replacement.
[0003] Osteoarthritis (OA) is a disease caused by degeneration of articular
cartilage and subsequent
joint deformation. In addition to risk factors like body weight, joint injury
and age, there is a strong
hereditary component to OA, reflected by high heritability estimates from twin
studies. So far, few of
the genes responsible for this genetic component have been identified.
Summary
[0004] It has been discovered that certain polymorphic variations in human
genomic DNA are
associated with osteoarthritis. In particular, polymorphic variants in loci
containing KIAA0296, Chr~om
4, PSMBI, TBP, PDCD2, ELP3, LRCHl, SNWl and ERG regions and other regions in
Table A of
human genomic DNA have been associated with risk of osteoarthritis. Some of
the associated
polymorphic variants fall in an intergenic region on chromosome 4 that does
not include a known gene;
therefore, the region is referred to herein as the Chronz 4 region. Also, the
PSMBl, TBP and PDCD2
regions are located in a larger region referred to herein as the Chrom 6
region.
[0005] Thus, featured herein are methods for identifying a subject at risk of
osteoarthritis and/or a
risk of osteoarthritis in a subject, which comprise detecting the presence or
absence of one or more
polymorphic variations associated with osteoarthritis in or around the loci
described herein in a human
nucleic acid sample. In an embodiment, two or more polymorphic variations are
detected in two or
more regions of which one is the KIAA0296, Cht~om 4, Chrona 6, ELP3, LRCHl,
SNWl or ERG region
or other region in Table A. In certain embodiments, 3 or more, or 4, 5, 6, 7,
8, 9, 10, 11, 12, 13, 14, 15,
16, 17, 18, 19 or 20 or more polymorphic variants are detected.
[0006] Also featured are nucleic acids that include one or more polymorphic
variations associated
with occurrence of osteoarthritis, as well as polypeptides encoded by these
nucleic acids. In addition,
provided are methods for identifying candidate therapeutic molecules for
treating osteoarthritis, as well
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
as methods for treating osteoarthritis in a subject by identifying a subject
at risk of osteoarthritis and
treating the subject with a suitable prophylactic, treatment or therapeutic
molecule.
[0007] Also provided are compositions comprising a cell from a subject having
osteoarthritis or at
risk of osteoarthritis and/or a KIAA0296, Chrona 4, Chrom 6, ELP3, LRCHI, SNWI
or ERG nucleic acid
or other nucleic acid referenced in Table A, with a RNAi, siRNA, antisense DNA
or RNA, or ribozyme
nucleic acid designed from a KIAA0296, Chrona 4, Chr~ona 6, ELP3, LRCHI, SNWI
or ERG nucleotide
sequence or other nucleotide sequence referenced in Table A. In an embodiment,
the RNAi, siRNA,
antisense DNA or RNA, or ribozyme nucleic acid is designed from a KIAA0296,
Chrom 4, Ch~om 6,
ELP3, LRCHI, SNWI or ERG nucleotide sequence or other nucleotide sequence
referenced in Table A
that includes one or more polymorphic variations associated with
osteoarthritis, and in some instances,
specifically interacts with such a nucleotide sequence. Further, provided are
arrays of nucleic acids
bound to a solid surface, in which one or more nucleic acid molecules of the
array have a KIAA0296,
Ch~om 4, Cla~~om 6, ELP3, LRCHl, SNWI or ERG nucleotide sequence or other
nucleotide sequence
referenced in Table A, or a fragment or substantially identical nucleic acid
thereof, or a complementary
nucleic acid of the foregoing. Featured also are compositions comprising a
cell from a subject having
osteoarthritis or at risk of osteoarthritis and/or a KIAA0296, Chrorn 4, Chrom
6, ELP3, LRCHl, SNWI
or ERG polypeptide or other polypeptide referenced in Table A, with an
antibody that specifically binds
to the polypeptide. In an embodiment, the antibody specifically binds to an
epitope in the polypeptide
that includes a non-synonymous amino acid modification associated with
osteoarthritis (e.g., results in
an amino acid substitution in the encoded polypeptide associated with
osteoarthritis). In certain
embodiments, the antibody selectively binds to an epitope in the KIAA0296,
Chf°ona 4, Clarom 6, ELP3,
LRCHl, SNWI or ERG polypeptide, or other polypeptide referenced in Table A,
having an amino acid
associated with osteoarthritis. Thus, featured is an antibody that binds an
epitope having an amino acid
encoded by rs734784, rs1042164, rs749670, rs955592, rs241448 and/or rs1040461,
such as a valine or
isoleucine encoded by rs734784 (e.g., a valine at position 489 in a KCNSI
polypeptide), a valine or
alanine encoded by rs1042164 (e.g., a valine at position 133 in a IER2
polypeptide), a glutamate or
glycine encoded by rs749670 (e.g., a glutamate at position 327 in a KIAA0296
polypeptide), a threonine
or isoleucine encoded by rs955592 (e.g., a threonine at position 70 in a RBEDl
polypeptide), a
glutamine or termination encoded by rs241448 (e.g., a glutamine at position
687 in a TAP2 polypeptide)
or a glycine or serine encoded by rs1040461 (e.g., a glycine at position 207
in a RAB23 polypeptide) at
the corresponding position in the polypeptide.
Brief Description of the Drawinus
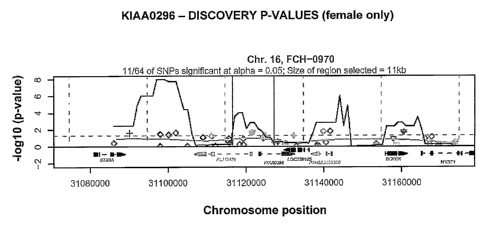
[0008] Figures lA-1G show proximal SNPs in a 100-kb window in KIAA0296, Chr~om
4, Chrofn 6,
ELP3, LRCHl, SNWI and ERG regions of genomic DNA, respectively, that were
compared between
pools of cases and controls. The x-axis corresponds to their chromosomal
position and the y-axis to the
test P-values (shown on the -logo scale). The continuous dark line presents
the results of a goodness-of
2
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
fit test for an excess of significance (compared to 0.05) in a 10 kb sliding
window assessed at 1 kb
increments.
Detailed Description
(0009] It has been discovered that polymorphic variants in a locus containing
a KIAA0296, Chrorn
4, Ch~om 6, ELP3, LRCHl, SNWI or ERG region are associated with occurrence of
osteoarthritis in
subjects. Thus, detecting genetic determinants associated with an increased
risk of osteoarthritis
occurrence can lead to early identification of a predisposition to
osteoarthritis and early prescription of
preventative measures. Also, associating a KIAA0296, Chrom 4, Chr om 6, ELP3,
LRCHI, SNWI or
ERG polymorphic variant and other variants referenced in Table A with
osteoarthritis has provided new
targets for screening molecules useful in treatments of osteoarthritis.
Osteoarthritis and Sample Selection
[0010] Osteoarthritis (OA), or degenerative joint disease, is one of the
oldest and most common
types of arthritis. It is characterized by the breakdown of the joint's
cartilage. Cartilage is the part of the
joint that cushions the ends of bones, and its breakdown causes bones to rub
against each other, causing
pain and loss of movement. Type II collagen is the main component of
cartilage, comprising 15-25% of
the wet weight, approximately half the dry weight, and representing 90-95% of
the total collagen
content in the tissue. It forms fibrils that endow cartilage with tensile
strength (Mayne, R. Arthritis
Rhuem. 32:241-246 (1989)).
[0011] Most commonly affecting middle-aged and older people, OA can range from
very mild to
very severe. It affects hands and weight-bearing joints such as knees, hips,
feet and the back. Knee OA
can be as disabling as any cardiovascular disease except stroke.
[0012] Osteoarthritis affects an estimated 20.7 million Americans, mostly
after age 45, with
women more commonly affected than men. Physicians make a diagnosis of OA based
on a physical
exam and history of symptoms. X-rays are used to confirm diagnosis. Most
people over 60 reflect the
disease on X-ray, and about one-third have actual symptoms.
[0013] There are many factors that can cause OA. Obesity may lead to
osteoarthritis of the knees.
In addition, people with joint injuries due to sports, work-related activity
or accidents may be at
increased risk of developing OA.
[0014] Genetics has a role in the development of OA too. Some people may be
born with defective
cartilage or with slight defects in the way that joints fit together. As a
person ages, these defects may
cause early cartilage breakdown in the joint or the inability to repair
damaged or deteriorated cartilage
in the joint.
[0015] Inclusion or exclusion of samples for an osteoarthritis pool may be
based upon the
following criteria: ethnicity (e.g., samples derived from an individual
characterized as Caucasian);
parental ethnicity (e.g., samples derived from an individual of British
paternal and maternal descent);
relevant phenotype information for the individual (e.g., case samples derived
from individuals
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
diagnosed with specific knee, hand or hip osteoarthritis (OA); case samples
recruited from an OA knee
replacement clinic). Control samples may be selected based on relevant
phenotype information for the
individual (e.g., derived from individuals free of OA at several sites (knee,
hand, hip etc)); and no
family history of OA and/or rheumatoid arthritis. Additional phenotype
information collected for both
cases and controls may include age of the individual, gender, family history
of OA, diagnosis with
osteoarthritis (joint location of OA (e.g., knee, hips, hands and spine), date
of primary diagnosis, age of
individual as of primary diagnosis), knee history (current symptoms, any major
knee injury,
menisectomy, knee replacement surgery, age of surgery), HRT history,
osteoporosis diagnosis.
[0016] Based in part upon selection criteria set forth above, individuals
having osteoarthritis can be
selected for genetic studies. Also, individuals having no history of
osteoarthritis often are selected for
genetic studies, as described hereafter.
Pol,~o~hic Variants Associated with Osteoarthritis
[0017] A genetic analysis provided herein linked osteoarthritis with
polymorphic variant nucleic
acid sequences in the human genome. As used herein, the term "polymorphic
site" refers to a region in
a nucleic acid at which two or more alternative nucleotide sequences are
observed in a significant
number of nucleic acid samples from a population of individuals. A polymorphic
site may be a
nucleotide sequence of two or more nucleotides, an inserted nucleotide or
nucleotide sequence, a
deleted nucleotide or nucleotide sequence, or a microsatellite, for example. A
polymorphic site that is
two or more nucleotides in length may be 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13,
14, 15 or more, 20 or more,
30 or more, 50 or more, 75 or more, 100 or more, 500 or more, or about 1000
nucleotides in length,
where all or some of the nucleotide sequences differ within the region. A
polymorphic site is often one
nucleotide in length, which is referred to herein as a "single nucleotide
polymorphism" or a "SNP."
[0018] Where there are two, three, or four alternative nucleotide sequences at
a polymorphic site,
each nucleotide sequence is referred to as a "polymorphic variant" or "nucleic
acid variant." Where two
polymorphic variants exist, for example, the polymorphic variant represented
in a minority of samples
from a population is sometimes referred to as a "minor allele" and the
polymorphic variant that is more
prevalently represented is sometimes referred to as a "major allele." Many
organisms possess a copy of
each chromosome (e.g., humans), and those individuals who possess two major
alleles or two minor
alleles are often referred to as being "homozygous" with respect to the
polymorphism, and those
individuals who possess one major allele and one minor allele are normally
referred to as being
"heterozygous" with respect to the polymorphism. Individuals who are
homozygous with respect to one
allele are sometimes predisposed to a different phenotype as compared to
individuals who are
heterozygous or homozygous with respect to another allele.
[0019] In genetic analysis that associate polymorphic variants with
osteoarthritis, samples from
individuals having osteoarthritis and individuals not having osteoarthritis
often are allelotyped and/or
genotyped. The term "allelotype" as used herein refers to a process for
determining the allele frequency
for a polymorphic variant in pooled DNA samples from cases and controls. By
pooling DNA from each
4
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
group, an allele frequency for each SNP in each group is calculated. These
allele frequencies are then
compared to one another. The term "genotyped" as used herein refers to a
process for determining a
genotype of one or more individuals, where a "genotype" is a representation of
one or more
polymorphic variants in a population.
[0020] A genotype or polymorphic variant may be expressed in terms of a
"haplotype," which as
used herein refers to two or more polymorphic variants occurring within
genomic DNA in a group of
individuals within a population. For example, two SNPs may exist within a gene
where each SNP
position includes a cytosine variation and an adenine variation. Certain
individuals in a population may
carry one allele (heterozygous) or two alleles (homozygous) having the gene
with a cytosine at each
SNP position. As the two cytosines corresponding to each SNP in the gene
travel together on one or
both alleles in these individuals, the individuals can be characterized as
having a cytosine/cytosine
haplotype with respect to the two SNPs in the gene.
[0021] As used herein, the term "phenotype" refers to a trait which can be
compared between
individuals, such as presence or absence of a condition, a visually observable
difference in appearance
between individuals, metabolic variations, physiological variations,
variations in the function of
biological molecules, and the like. An example of a phenotype is occurrence of
osteoarthritis.
[0022] Researchers sometimes report a polymorphic variant in a database
without determining
whether the variant is represented in a significant fraction of a population.
Because a subset of these
reported polymorphic variants are not represented in a statistically
significant portion of the population,
some of them are sequencing errors and/or not biologically relevant. Thus, it
is often not known
whether a reported polymorphic variant is statistically significant or
biologically relevant until the
presence of the variant is detected in a population of individuals and the
frequency of the variant is
determined. Methods for detecting a polymorphic variant in a population are
described herein,
specifically in Example 2. A polymorphic variant is statistically significant
and often biologically
relevant if it is represented in 5% or more of a population, sometimes 10% or
more, 15% or more, or
20% or more of a population, and often 25% or more, 30% or more, 35% or more,
40% or more, 45% or
more, or 50% or more of a population.
[0023] A polymorphic variant may be detected on either or both strands of a
double-stranded
nucleic acid. Also, a polymorphic variant may be located within an intron or
exon of a gene or within a
portion of a regulatory region such as a promoter, a 5' untranslated region
(UTR), a 3' UTR, and in
DNA (e.g., genomic DNA (gDNA) and complementary DNA (cDNA)), RNA (e.g., mRNA,
tRNA, and
rRNA), or a polypeptide. Polymorphic variations may or may not result in
detectable differences in
gene expression, polypeptide structure, or polypeptide function.
[0024] It was determined that polymorphic variations associated with an
increased risk of
osteoarthritis existed in SEQ ID NO: 1-7 or a nucleotide sequence referenced
in Table A. In certain
embodiments, polymorphic variants at positions rs552, rs12904, rs2282146,
rs734784, rs1042164,
rs749670, rs955592, rs1143016, rs755248, rs1055055, rs835409, rs927663,
rs8162, rs831038, rs33079,
rs1710880, rs1078153, rs799570, rs1282730, rs1518875, rs1568694, rs905042,
rs1957723, rs794018,
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
rs707723, rs893861, rs1914903, rs2062232, rs26609, rs1370987, rs1012414,
rs435903, rs1248,
rs703508, rs226465, rs241448, rs763155, rs1040461, rs462832, rs804194,
rs1022646, rs756519,
rs1042327, rs8770, rs1569112, rs1563055, rs805623, rs1019850, rs1599931, AA,
rs912428, rs279941,
rs1062230, rs1859911, rs1477261, rs1191119, rs657780, rs1393890, rs1478714,
rs868213, rs690115,
rs1465501, rs899173, rs10477, rs926393, rs465271, rs1888475, rs13847 and/or
rs738658 in the human
genome were associated with an increased risk of osteoarthritis, and in
specific embodiments, the
corresponding allele in the right-most column in Table A for each position is
associated with an
increased risk of osteoarthritis. In other embodiments polymorphic variants at
positions rs734784,
rs1042164, rs749670, rs955592, rs241448 and rs1040461 were associated with an
increased risk of
osteoarthritis, and in specific embodiments, a valine encoded by rs734784, a
valine encoded by
rs1042164, a glutamate encoded by rs749670, a threonine encoded by rs955592, a
glutamine encoded
by rs241448, and a glycine encoded by rs1040461 were associated with an
increased risk of
osteoarthritis.
[0025] Polymorphic variants in and around the KIAA0296 locus were tested for
association with
osteoarthritis. These include polymorphic variants at positions in SEQ ID NO:
1 selected from the
group consisting of 247, 1535, 2386, 6440, 9133, 9143, 9471, 13150, 13717,
14466, 15769, 16870,
18545, 18749, 19123, 20736, 21038, 21046, 21050, 21056, 21706, 23170, 25028,
27871, 28070, 31717,
32019, 32318, 33080, 33101, 34236, 34285, 34818, 35168, 37981, 38113, 38117,
38481, 38615, 38944,
39288, 41385, 42136, 42185, 42353, 42434, 44580, 44675, 45739, 46439, 47457,
47735, 50319, 50708,
51185, 53002, 53064, 53637, 55274, 55825, 55986, 56684, 57653, 57659, 57692,
57775, 61313, 61431,
61699, 62906, 63619, 64664, 68452, 69665, 69681, 70091, 74637, 74760, 76523,
78559, 79549, 79882,
81339, 81681, 81696, 83517, 85431, 86332, 87358, 87725, 89052, 90020, 90231,
90284, 90447, 90601,
90724, 92559, 95176, 95195 and 96822. Polymorphic variants at the following
positions in SEQ 117
NO: 1 in particular were associated with an increased risk of osteoarthritis:
13150, 21046, 23170,
25028, 44580, 62906, 64664 and 83517. In particular, the following polymorphic
variants in SEQ ID
NO: 1 were associated with risk of osteoarthritis: a guanine at position
13150, a thymine at position
21046, an adenine at position 23170, an adenine at position 25028, a guanine
at position 44580, a
guanine at position 62906, a cytosine at position 64664 and a cytosine at
position 83517. A
polymorphic variant in a KIAA0296 polypeptide encoded by rs749670 (e.g., a
glutamate at position 327
in the polypeptide) also was associated with increased risk of osteoarthritis.
[0026] Polymorphic variants in and around the clzrom 4 locus were tested for
association with
osteoarthritis. These include polymorphic variants at positions in SEQ ID NO:
2 selected from the
group consisting of 211, 7217, 7895, 13308, 14279, 17026, 18271, 20417, 21843,
22069, 22145, 22519,
22539, 23236, 23256, 23402, 23499, 23620, 23871, 24136, 25427, 25866, 26541,
26576, 26689, 26720,
27113, 27164, 27186, 28341, 29160, 29844, 30665, 30830, 31061, 31523, 32326,
32346, 32358, 34909,
34975, 35066, 35096, 35375, 36304, 36712, 36770, 37342, 37412, 37884, 38077,
38300, 38301, 41189,
44408, 44493, 44571, 44670, 45219, 45258, 47261, 48473, 48771, 55292, 56479,
56747, 60620, 60688,
61058, 61129, 61577, 61961, 63351, 63926, 65798, 66043, 66044, 66246, 66318,
66547, 71238, 71283,
6
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
71492, 72274, 73762, 74209, 75284, 77347, 77589, 78096, 78606, 78862, 79135,
79146, 79456, 79609,
80086, 80119, 80766, 81110, 81269, 81668, 82433, 82559, 83298, 83821, 84121,
84147, 84543, 84554,
84691, 84727, 85678, 86699, 86700, 86792, 86832, 87045, 87140, 87365, 88342,
88498, 88589, 95502,
96968, 97448, 97568 and 98724. Polymorphic variants at the following positions
in SEQ ID NO: 2 in
particular were associated with an increased risk of osteoarthritis: 23236,
32358, 47261, 48771, 55292,
60688, 72274, 74209, 77589, 79135, 79456, 79609, 80119, 80766, 81110, 82433,
84121, 84147, 85678,
86699, 86832, 87140 and 88589, where specific embodiments are directed to a
polymorphic variant at
position 32358, 47261, 74209 and/or 79456. In particular, the following
polymorphic variants in SEQ
ID NO: 2 were associated with risk of osteoarthritis: an adenine at position
23236, a cytosine at
position 32358, a guanine at position 47261, a guanine at position 48771, a
cytosine at position 55292,
an adenine at position 60688, a guanine at position 72274, a guanine at
position 74209, a cytosine at
position 77589, an adenine at position 79135, a thymine at position 79456, an
adenine at position
79609, an adenine at position 80119, a cytosine at position 80766, an adenine
at position 81110, a
cytosine at position 82433, a cytosine at position 84121, a thymine at
position 84147, a cytosine at
position 85678, a thymine at position 86699, an adenine at position 86832, a
guanine at position 87140
and an adenine at position 88589.
[0027] Polymorphic variants in and around the ch~om 6 region were tested for
association with
osteoarthritis. These include polymorphic variants at positions in SEQ ID NO:
3 selected from the
group consisting of 229, 6310, 11840, 11870, 12064, 13392, 16354, 16559,
16935, 17616, 17737,
18321, 18453, 18811, 20020, 21662, 23197, 23446, 24339, 25504, 27174, 28008,
29294, 29759, 30832,
44512, 44850, 45884, 46345, 48589, 53371, 53911, 53990, 55152, 55667, 58952,
59315, 60029, 61477,
62988, 63090, 64021, 65685, 70220, 70323, 70959, 73436, 82945, 82958, 82961,
82964, 82965, 83006,
83025, 83034, 83074 ,83132, 83155, 83172, 83174, 83206, 83216, 83234, 83252,
83260, 83263, 83296,
83319, 83322, 83324, 83357, 83375, 83381, 83389, 83443, 83499, 83545, 83566,
83591, 83619, 83698,
83780, 83784, 83826, 83832, 83852, 86297, 86315, 86420, 86460, 86714, 86718,
86736, 86753, 86766,
88162, 88218, 88246, 88255, 88309, 88310, 88471, 88619, 88904, 89044, 90531,
90534, 90613 and
46252. Polymorphic variants at the following positions in SEQ ID NO: 3 in
particular were associated
with an increased risle of osteoarthritis: 229, 6310, 16559, 18453, 25504,
27174, 30832, 44850, 45884,
48589, 61477, 82961 and 46252, with specific embodiments directed to variants
at positions 229,
16559, 44850 and/or 46252. In particular, the following polymorphic variants
in SEQ ID NO: 3 were
associated with risk of osteoarthritis: a thymine at position 229, a guanine
at position 6310, a thymine
at position 16559, an adenine at position 18453, an adenine at position 25504,
an adenine at position
27174, an adenine at position 30832, a guanine at position 44850, an adenine
at position 45884, an
adenine at position 48589, a cytosine at position 61477, a cytosine at
position 82961 and a thymine at
position 46252.
[0028] Polymorphic variants in and around the ELP3 region were tested for
association with
osteoarthritis. These include polymorphic variants at positions in SEQ ID NO:
4 selected from the
group consisting of 211, 473, 1536, 5639, 17186, 17335, 25029, 25111, 28811,
28863, 30809, 40985,
7
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
45147, 45282, 46168, 46328, 49077, 51925, 52141, 52168, 60852, 62468, 65572,
79089, 79541, 79790,
90843, 90978, 91052, 91131, 91132, 94439 and 94621. Polymorphic variants at
the following positions
in SEQ ID NO: 4 in particular were associated with an increased risk of
osteoarthritis: 40985, 46168,
51925 and 52168. In particular, the following polymorphic variants in SEQ ID
NO: 4 were associated
with risk of osteoarthritis: a cytosine at position 40985, a guanine at
position 46168, a thymine at
position 51925 and a cytosine at position 52168.
[0029] Polymorphic variants in and around the LRCHI region were tested for
association with
osteoarthritis. These include polymorphic variants at positions in SEQ ID NO:
5 selected from the
group consisting of 243, 10208, 15049, 15111, 15272, 15287, 15326, 15327,
17038, 19391, 21702,
22431, 22881, 27744, 32564, 32698, 33104, 33181, 33256, 33543, 35567, 40085,
40482, 45641, 46059,
48504, 48919, 49693, 49874, 50020, 50616, 50719, 55511, 65533, 70529, 75591,
77266, 80368, 82475,
92462, 92480, 95819 and 96275. Polymorphic variants at the following positions
in SEQ ID NO: 5 in
particular were associated with an increased risk of osteoarthritis: 15111,
45641, 46059, 49693, 49874,
50020, 50719, 70529, 82475, 92462, 92480 and 96275, with specific embodiments
directed to variants
at positions 82475 and/or 92462. In particular, the following polymorphie
variants in SEQ ID NO: 5
were associated with risk of osteoarthritis: a guanine at position 15111, a
thymine at position 45641, an
adenine at position 46059, a cytosine at position 49693, an adenine at
position 49874, an adenine at
position 50020, a guanine at position 50719, an adenine at position 70529, an
adenine at position 82475,
a thymine at position 92462, a thymine at position 92480 and a cytosine at
position 96275.
[0030] Polymorphic variants in and around the SNWI locus were tested for
association with
osteoarthritis. These include polymorphic variants at positions in SEQ ID NO:
6 selected from the
group consisting of 218, 1440, 1442, 2611, 4317, 4724, 4788, 5202, 5780, 5974,
6644, 7430, 7938,
8095, 8183, 8312, 8352, 9348, 9378, 9617, 9727, 9834, 9899, 10211, 10377,
10695, 10729, 10730,
11433, 11951, 12697, 12982, 14419, 14501, 14983, 15280, 15475, 15888, 15976,
16307, 16442, 17255,
18948, 19435, 19753, 20021, 20022, 20503, 20590, 21804, 21919, 21990, 22412,
22536, 23432, 23468,
23772, 24325, 24773, 26274, 27440, 28561, 30071, 31764, 33008, 35310, 35460,
37112, 37285, 37747,
38057, 38859, 38860, 39525, 40216, 40281, 41453, 42091, 42513, 42935, 42985,
43003, 43281, 43716,
43866, 44234, 44596, 44871, 45005, 45282, 47178, 47816, 47887, 48134, 48135,
48276, 48400, 48798,
48803, 49146, 49969, 51059, 51064, 53285, 54560, 54748, 54785, 55102, 55644,
55705, 55841, 56623,
56825, 56827, 56892, 59150, 59958, 60231, 60524, 61871, 62226, 63230, 63468,
63787, 65732, 65989,
68832, 69904, 70365, 70886, 73088, 73103, 75934, 75966, 76273, 77943, 78466,
78861, 78872, 79836,
80908, 81509, 83576, 83662, 83782, 84282, 84444, 85129, 85151, 85296, 85809,
86387, 86494, 89786,
89894, 90122, 92067, 92187, 92312, 92824, 93733, 96553 and 96941. Polymorphic
variants at the
following positions in SEQ ID NO: 6 in particular were associated with an
increased risk of
osteoarthritis: 4788, 8312, 9378, 9727, 9899, 10211, 27440, 40216, 40281,
42091, 43866, 48803,
51059, 55644, 56623, 73103, 78872, 79836, 85129, 92824 and 96941. In
particular, the following
polymorphic variants in SEQ ID NO: 6 were associated with risk of
osteoarthritis: a guanine at position
4788, a thymine at position 8312, a deletion at position 9378, a cytosine at
position 9727, a guanine at
8
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
position 9899, a cytosine at position 10211, a guanine at position 27440, a
guanine at position 40216, a
cytosine at position 40281, an adenine at position 42091, a guanine at
position 43866, an adenine at
position 48803, an adenine at position 51059, an adenine at position 55644, a
cytosine at position
56623, a cytosine at position 73103, an adenine at position 78872, a guanine
at position 79836, a
cytosine at position 85129, a guanine at position 92824 and an adenine at
position 96941.
[0031] Polymorphic variants in and around the ERG region were tested for
association with
osteoarthritis. These include polymorphic variants at positions in SEQ ID NO:
7 selected from the
group consisting of 231, 882, 960, 1194, 1530, 1673, 2096, 2285, 5873, 7256,
7988, 8222, 8381, 8814,
8915, 9642, 9902, 10619, 10927, 11032, 14377, 15608, 15928, 16296, 17598,
19272, 20084, 20577,
28051, 29466, 29530, 29987, 30012, 30322, 32216, 32516, 32544, 32746, 33137,
33538, 33798, 33802,
33964, 34132, 34210, 34317, 34499, 34753, 34845, 35335, 36423, 36450, 36481,
38447, 38784, 39387,
39458, 39822, 40305, 40869, 40926, 41010, 41134, 41984, 42172, 42753, 43011,
43176, 43320, 43381,
44142, 44383, 44726, 45087, 45141, 45359, 45421, 45456, 45467, 45486, 45709,
45716, 47626, 49413,
49796, 49962, 50075, 50093, 50571, 50615, 50780, 50851, 51459, 53193, 53702,
53736, 53795, 54109,
54126, 54230, 54894, 55455, 55499, 56522, 56662, 56954, 57267, 58282, 58916,
59544, 59666, 59913,
66846, 67245, 67652, 67955, 67966, 68420, 70226, 70810, 72246, 73330, 73457,
74389, 74638, 74640,
75358, 75952, 76098, 77836, 78449, 78507, 80031, 81695, 82775, 82795, 84611,
84657, 84693, 85020,
85048, 85100, 85325, 85452, 85868, 85936, 85990, 86139, 86497, 87236, 87248,
87533, 87912, 88108,
88494, 89598, 90235, 91287, 91359, 92384, 92410, 92900, 94495, 94512, 97777
and 98333.
Polymorphic variants at the following positions in SEQ ID NO: 7 in particular
were associated with an
increased risk of osteoarthritis: 1673, 20577, 33137, 39822, 45716, 49962,
51459, 54894, 55455,
55499, 58282, 68420 and 80031, with specific embodiments directed to variants
at positions 33137,
55499 and/or 58282. In particular, the following polymorphic variants in SEQ
ID NO: 7 were
associated with risk of osteoarthritis: a guanine at position 1673, a thymine
at position 20577, a guanine
at position 33137, a guanine at position 39822, an adenine at position 45716,
a guanine at position
49962, an adenine at position 51459, a cytosine at position 54894, an adenine
at position 55455, an
adenine at position 55499, a guanine at position 58282, an adenine at position
68420 and a thymine at
position 80031.
[0032] Based in part upon analyses summarized in Figures lA-1G, regions with
significant
association have been identified in regions associated with osteoarthritis.
Any polymorphic variants
associated with osteoarthritis in a region of significant association can be
utilized for embodiments
described herein. For example, polymorphic variants in a region spanning
chromosome positions
31118000 to 31129000 (approximately 11,000 nucleotides in length) in a
KIAA0296 locus, a region
spanning chromosome positions 36914000 to 36931000 (approximately 17,000
nucleotides in length) in
a chrona 4 region, a region spanning chromosome positions 170719500 to
170766500 (approximately
47,000 nucleotides in length) in a ch~om 6 region, a region spanning
chromosome positions 27963000
to 27983000 (approximately 20,000 nucleotides in length) in an ELP3 locus, a
region spanning
chromosome positions 44962000 to 45013000 (approximately 51,000 nucleotides in
length) in a LRCHI
9
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
locus, a region spanning chromosome positions 76196500 to 76221500
(approximately 25,000
nucleotides in length) in a SNWI locus, and a region spanning chromosome
positions 38830000 to
38844000 (approximately 14,000 nucleotides in length) in an ERG locus have
significant association
(chromosome positions are within NCBI's Genome build 34).
Additional Polyphic Variants Associated with Osteoarthritis
[0033] Also provided is a method for identifying polymorphic variants proximal
to an incident,
founder polymorphic variant associated with osteoarthritis. Thus, featured
herein are methods for
identifying a polymorphic variation associated with osteoarthritis that is
proximal to an incident
polymorphic variation associated with osteoarthritis, which comprises
identifying a polymorphic variant
proximal to the incident polymorphic variant associated with osteoarthritis,
where the incident
polymorphic variant is in a KIAA0296, Ch~om 4, Clz~onz 6, ELP3, LRCHl, SNWI or
ERG nucleotide
sequence or other nucleotide sequence referenced in Table A. The nucleotide
sequence often comprises
a polynucleotide sequence selected from the group consisting of (a) a
polynucleotide sequence of SEQ
ID NO: 1-7 or referenced in Table A; (b) a polynucleotide sequence that
encodes a polypeptide having
an amino acid sequence encoded by a polynucleotide sequence of SEQ ID NO: 1-7
or referenced in
Table A; and (c) a polynucleotide sequence that encodes a polypeptide having
an amino acid sequence
that is 90% or more identical to an amino acid sequence encoded by a
nucleotide sequence of SEQ ID
NO: 1-7 or referenced in Table A or a polynucleotide sequence 90% or more
identical to the
polynucleotide sequence of SEQ ID NO: 1-7 or referenced in Table A. The
presence or absence of an
association of the proximal polymorphic variant with osteoarthritis then is
determined using a known
association method, such as a method described in the Examples hereafter. In
an embodiment, the
incident polymorphic variant is a polymorphic variant associated with
osteoarthritis described herein.
In another embodiment, the proximal polymorphic variant identified sometimes
is a publicly disclosed
polymorphic variant, which for example, sometimes is published in a publicly
available database. In
other embodiments, the polymorphic variant identified is not publicly
disclosed and is discovered using
a known method, including, but not limited to, sequencing a region surrounding
the incident
polymorphic variant in a group of nucleic samples. Thus, multiple polymorphic
variants proximal to an
incident polymorphic variant are associated with osteoarthritis using this
method.
[0034] The proximal polymorphic variant often is identified in a region
surrounding the incident
polymorphic variant. In certain embodiments, this surrounding region is about
50 kb flanking the first
polymorphic variant (e.g. about 50 kb 5' of the first polymorphic variant and
about 50 kb 3' of the first
polymorphic variant), and the region sometimes is composed of shorter flanking
sequences, such as
flanking sequences of about 40 kb, about 30 kb, about 25 kb, about 20 kb,
about 15 kb, about 10 kb,
about 7 kb, about 5 kb, or about 2 kb 5' and 3' of the incident polymorphic
variant. In other
embodiments, the region is composed of longer flanking sequences, such as
flanking sequences of about
55 kb, about 60 kb, about 65 kb, about 70 kb, about 75 kb, about 80 kb, about
85 kb, about 90 kb, about
95 kb, or about 100 kb 5' and 3' of the incident polymorphic variant.
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
[0035] In certain embodiments, polymorphic variants associated with
osteoarthritis are identified
iteratively. For example, a first proximal polymorphic variant is associated
with osteoarthritis using the
methods described above and then another polymorphic variant proximal to the
first proximal
polymorphic variant is identified (e.g., publicly disclosed or discovered) and
the presence or absence of
an association of one or more other polymorphic variants proximal to the first
proximal polymorphic
variant with osteoarthritis is determined.
[0036] The methods described herein are useful for identifying or discovering
additional
polymorphic variants that may be used to further characterize a gene, region
or loci associated with a
condition, a disease (e.g., osteoarthritis), or a disorder. For example,
allelotyping or genotyping data
from the additional polymorphic variants may be used to identify a functional
mutation or a region of
linkage disequilibrium. In certain embodiments, polymorphic variants
identified or discovered within a
region comprising the first polymorphic variant associated with osteoarthritis
are genotyped using the
genetic methods and sample selection techniques described herein, and it can
be determined whether
those polymorphic variants are in linkage disequilibrium with the first
polymorphic variant. The size of
the region in linkage disequilibrium with the first polymorphic variant also
can be assessed using these
genotyping methods. Thus, provided herein are methods for determining whether
a polymorphic variant
is in linkage disequilibrium with a first polymorphic variant associated with
osteoarthritis, and such
information can be used in prognosis/diagnosis methods described herein.
Isolated Nucleic Acids
[0037] Featured herein are isolated KIAA0296, Chrom 4, Cht~om 6, ELP3, LRCHl,
SNWI or ERG
nucleic acid variants depicted in SEQ 117 NO: 1-7 or referenced in Table A,
and substantially identical
nucleic acids thereof. A nucleic acid variant may be represented on one or
both strands in a double-
stranded nucleic acid or on one chromosomal complement (heterozygous) or both
chromosomal
complements (homozygous).
[0038] As used herein, the term "nucleic acid" includes DNA molecules (e.g., a
complementary
DNA (cDNA) and genomic DNA (gDNA)) and RNA molecules (e.g., mRNA, rRNA, siRNA
and
tRNA) and analogs of DNA or RNA, for example, by use of nucleotide analogs.
The nucleic acid
molecule can be single-stranded and it is often double-stranded. The term
"isolated or purified nucleic
acid" refers to nucleic acids that are separated from other nucleic acids
present in the natural source of
the nucleic acid. For example, with regard to genomic DNA, the term "isolated"
includes nucleic acids
which are separated from the chromosome with which the genomic DNA is
naturally associated. An
"isolated" nucleic acid is often free of sequences which naturally flank the
nucleic acid (i.e., sequences
located at the 5' and/or 3' ends of the nucleic acid) in the genomic DNA of
the organism from which the
nucleic acid is derived. For example, in various embodiments, the isolated
nucleic acid molecule can
contain less than about 5 kb, 4 kb, 3 kb, 2 kb, 1 kb, 0.5 kb or 0.1 kb of 5'
and/or 3' nucleotide sequences
which flank the nucleic acid molecule in genomic DNA of the cell from which
the nucleic acid is
derived. Moreover, an "isolated" nucleic acid molecule, such as a cDNA
molecule, can be substantially
11
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
free of other cellular material, or culture medium when produced by
recombinant techniques, or
substantially free of chemical precursors or other chemicals when chemically
synthesized. As used
herein, the term "gene" refers to a nucleotide sequence that encodes a
polypeptide.
[0039] Also included herein are nucleic acid fragments. These fragments often
have a nucleotide
sequence identical to a nucleotide sequence of SEQ ID NO: 1-7 or referenced in
Table A, a nucleotide
sequence substantially identical to a nucleotide sequence of SEQ ID NO: 1-7 or
referenced in Table A,
or a nucleotide sequence that is complementary to the foregoing. The nucleic
acid fragment may be
identical, substantially identical or homologous to a nucleotide sequence in
an exon or an intron in a
nucleotide sequence of SEQ ID NO: 1-7 or referenced in Table A, and may encode
a domain or part of a
domain of a polypeptide. Sometimes, the fragment will comprises one or more of
the polymorphic
variations described herein as being associated with osteoarthritis. The
nucleic acid fragment is often
50, 100, or 200 or fewer base pairs in length, and is sometimes about 300,
400, 500, 600, 700, 800, 900,
1000, 1100, 1200, 1300, 1400, 1500, 2000, 3000, 4000, 5000, 10000, 15000, or
20000 base pairs in length.
A nucleic acid fragment that is complementary to a nucleotide sequence
identical or substantially identical
to a nucleotide sequence in SEQ 117 NO: 1-7 or referenced in Table A and
hybridizes to such a nucleotide
sequence under stringent conditions is often referred to as a "probe." Nucleic
acid fragments often include
one or more polymorphic sites, or sometimes have an end that is adjacent to a
polymorphic site as described
hereafter.
[0040] An example of a nucleic acid fragment is an oligonucleotide. As used
herein, the term
"oligonucleotide" refers to a nucleic acid comprising about 8 to about 50
covalently linked nucleotides,
often comprising from about 8 to about 35 nucleotides, and more often from
about 10 to about 25
nucleotides. The backbone and nucleotides within an oligonucleotide may be the
same as those of
naturally occurring nucleic acids, or analogs or derivatives of naturally
occurring nucleic acids,
provided that oligonucleotides having such analogs or derivatives retain the
ability to hybridize
specifically to a nucleic acid comprising a targeted polymorphism.
Oligonucleotides described herein
may be used as hybridization probes or as components of prognostic or
diagnostic assays, for example,
as described herein.
[0041] Oligonucleotides are typically synthesized using standard methods and
equipment, such as
the ABITM3900 High Throughput DNA Synthesizer and the EXPEDITETM 8909 Nucleic
Acid
Synthesizer, both of which are available from Applied Biosystems (Foster City,
CA). Analogs and
derivatives are exemplified in U.S. Pat. Nos. 4,469,863; 5,536,821; 5,541,306;
5,637,683; 5,637,684;
5,700,922; 5,717,083; 5,719,262; 5,739,308; 5,773,601; 5,886,165; 5,929,226;
5,977,296; 6,140,482;
WO 00/56746; WO 01114398, and related publications. Methods for synthesizing
oligonucleotides
comprising such analogs or derivatives are disclosed, for example, in the
patent publications cited above
and in U.S. Pat. Nos. 5,614,622; 5,739,314; 5,955,599; 5,962,674; 6,117,992;
in WO 00/75372; and in
related publications.
[0042] Oligonucleotides may also be linked to a second moiety. The second
moiety may be an
additional nucleotide sequence such as a tail sequence (e.g., a polyadenosine
tail), an adapter sequence
12
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
(e.g., phage M13 universal tail sequence), and others. Alternatively, the
second moiety may be a non-
nucleotide moiety such as a moiety which facilitates linkage to a solid
support or a label to facilitate
detection of the oligonucleotide. Such labels include, without limitation, a
radioactive label, a
fluorescent label, a chemiluminescent label, a paramagnetic label, and the
like. The second moiety may
be attached to any position of the oligonucleotide, provided the
oligonucleotide can hybridize to the
nucleic acid comprising the polymorphism.
Uses for Nucleic Acid Sequence
[0043] Nucleic acid coding sequences may be used for diagnostic purposes for
detection and
control of polypeptide expression. Also, included herein are oligonucleotide
sequences such as
antisense RNA, small-interfering RNA (siRNA) and DNA molecules and ribozymes
that function to
inhibit translation of a polypeptide. Antisense techniques and RNA
interference techniques are known
in the art and are described herein.
[0044] Ribozymes are enzymatic RNA molecules capable of catalyzing the
specific cleavage of
RNA. The mechanism of ribozyme action involves sequence specific hybridization
of the ribozyme
molecule to complementary target RNA, followed by endonucleolytic cleavage.
For example,
hammerhead motif ribozyme molecules may be engineered that specifically and
efficiently catalyze
endonucleolytic cleavage of RNA sequences corresponding to or complementary to
KIAA0296, Chrom
4, ChronZ 6, ELP3, LRCHl, SNWI or ERG nucleotide sequences or other nucleotide
sequences
referenced in Table A. Specific ribozyme cleavage sites within any potential
RNA target are initially
identified by scanning the target molecule for ribozyme cleavage sites which
include the following
sequences, GUA, GUU and GUC. Once identified, short RNA sequences of between
fifteen (15) and
twenty (20) ribonucleotides corresponding to the region of the target gene
containing the cleavage site
may be evaluated for predicted structural features such as secondary structure
that may render the
oligonucleotide sequence unsuitable. The suitability of candidate targets may
also be evaluated by
testing their accessibility to hybridization with complementary
oligonucleotides, using ribonuclease
protection assays.
[0045] Antisense RNA and DNA molecules, siRNA and ribozymes may be prepared by
any
method known in the art for the synthesis of RNA molecules. These include
techniques for chemically
synthesizing oligodeoxyribonucleotides well known in the art such as solid
phase phosphoramidite
chemical synthesis. Alternatively, RNA molecules may be generated by in vitro
and ira vivo
transcription of DNA sequences encoding the antisense RNA molecule. Such DNA
sequences may be
incorporated into a wide variety of vectors which incorporate suitable RNA
polymerase promoters such
as the T7 or SP6 polymerase promoters. Alternatively, antisense cDNA
constructs that synthesize
antisense RNA constitutively or inducibly, depending on the promoter used, can
be introduced stably
into cell lines.
[0046] DNA encoding a polypeptide also may have a number of uses for the
diagnosis of diseases,
including osteoarthritis, resulting from aberrant expression of a target gene
described herein. For
13
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
example, the nucleic acid sequence may be used in hybridization assays of
biopsies or autopsies to
diagnose abnormalities of expression or function (e.g., Southern or Northern
blot analysis, in situ
hybridization assays).
[0047] In addition, the expression of a polypeptide during embryonic
development may also be
determined using nucleic acid encoding the polypeptide. As addressed, infra,
production of functionally
impaired polypeptide is the cause of various disease states, such as
osteoarthritis. In situ hybridizations
using polypeptide as a probe may be employed to predict problems related to
osteoarthritis. Further, as
indicated, infra, administration of human active polypeptide, recombinantly
produced as described
herein, may be used to treat disease states related to functionally impaired
polypeptide. Alternatively,
gene therapy approaches may be employed to remedy deficiencies of functional
polypeptide or to
replace or compete with dysfunctional polypeptide.
Expression Vectors Host Cells and Geneticall~gineered Cells
[0048] Provided herein are nucleic acid vectors, often expression vectors,
which contain a
KIAA0296, Chrom 4, Chrom 6, ELP3, LRCHl, SNWI or ERG nucleotide sequence or
other nucleotide
sequence referenced in Table A, or a substantially identical sequence thereof.
As used herein, the term
"vector" refers to a nucleic acid molecule capable of transporting another
nucleic acid to which it has
been linked and can include a plasmid, cosmid, or viral vector. The vector can
be capable of
autonomous replication or it can integrate into a host DNA. Viral vectors may
include replication
defective retroviruses, adenoviruses and adeno-associated viruses for example.
[0049] A vector can include a KIAA0296, Chrom 4, Chrom 6, ELP3, LRCHl, SNWl or
ERG
nucleotide sequence or other nucleotide sequence referenced in Table A in a
form suitable for
expression of an encoded target polypeptide or target nucleic acid in a host
cell. A "target polypeptide"
is a polypeptide encoded by a KIAA0296, Chrom 4, Clarom 6, ELP3, LRCHl, SNWI
or ERG nucleotide
sequence or other nucleotide sequence referenced in Table A, or a
substantially identical nucleotide
sequence thereof. The recombinant expression vector typically includes one or
more regulatory
sequences operatively linked to the nucleic acid sequence to be expressed. The
term "regulatory
sequence" includes promoters, enhancers and other expression control elements
(e.g., polyadenylation
signals). Regulatory sequences include those that direct constitutive
expression of a nucleotide
sequence, as well as tissue-specific regulatory and/or inducible sequences.
The design of the expression
vector can depend on such factors as the choice of the host cell to be
transformed, the level of
expression of polypeptide desired, and the like. Expression vectors can be
introduced into host cells to
produce target polypeptides, including fusion polypeptides.
[0050] Recombinant expression vectors can be designed for expression of target
polypeptides in
prokaryotic or eukaryotic cells. For example, target polypeptides can be
expressed in E, coli, insect
cells (e.g., using baculovirus expression vectors), yeast cells, or mammalian
cells. Suitable host cells
are discussed further in Goeddel, Gene Expression Technology.' Methods in
Er~zy»aology 185,
Academic Press, San Diego, CA (1990). Alternatively, the recombinant
expression vector can be
14
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
transcribed and translated in vitro, for example using T7 promoter regulatory
sequences and T7
polymerase.
[0051] Expression of polypeptides in prokaryotes is most often carried out in
E. coli with vectors
containing constitutive or inducible promoters directing the expression of
either fusion or non-fusion
polypeptides. Fusion vectors add a number of amino acids to a polypeptide
encoded therein, usually to
the amino terminus of the recombinant polypeptide. Such fusion vectors
typically serve three purposes:
1) to increase expression of recombinant polypeptide; 2) to increase the
solubility of the recombinant
polypeptide; and 3) to aid in the purification of the recombinant polypeptide
by acting as a ligand in
affinity purification. Often, a proteolytic cleavage site is introduced at the
junction of the fusion moiety
and the recombinant polypeptide to enable separation of the recombinant
polypeptide from the fusion
moiety subsequent to purification of the fusion polypeptide. Such enzymes, and
their cognate
recognition sequences, include Factor Xa, thrombin and enterokinase. Typical
fusion expression
vectors include pGEX (Pharmacia Biotech Inc; Smith & Johnson, Gene 67: 31-40
(1988)), pMAL (New
England Biolabs, Beverly, MA) and pRITS (Pharmacia, Piscataway, NJ) which fuse
glutathione S-
transferase (GST), maltose E binding polypeptide, or polypeptide A,
respectively, to the target
recombinant polypeptide.
[0052] Purified fusion polypeptides can be used in screening assays and to
generate antibodies
specific for target polypeptides. In a therapeutic embodiment, fusion
polypeptide expressed in a
retroviral expression vector is used to infect bone marrow cells that are
subsequently transplanted into
irradiated recipients. The pathology of the subject recipient is then examined
after sufficient time has
passed (e.g., six (6) weeks).
[0053] Expressing the polypeptide in host bacteria with an impaired capacity
to proteolytically
cleave the recombinant polypeptide is often used to maximize recombinant
polypeptide expression
(Gottesman, S., Gefze Expressioh Technology: Methods in EnzynZOlogy, Academic
Press, San Diego,
California 185: 119-128 (1990)). Another strategy is to alter the nucleotide
sequence of the nucleic
acid to be inserted into an expression vector so that the individual codons
for each amino acid are those
preferentially utilized in E. coli (Wada et al., Nucleic Acids Res. 20: 2111-
2118 (1992)). Such
alteration of nucleotide sequences can be carried out by standard DNA
synthesis techniques.
[0054] When used in mammalian cells, the expression vector's control functions
are often provided
by viral regulatory elements. For example, commonly used promoters are derived
from polyoma,
Adenovirus 2, cytomegalovirus and Simian Virus 40. Recombinant mammalian
expression vectors are
often capable of directing expression of the nucleic acid in a particular cell
type (e.g., tissue-specific
regulatory elements are used to express the nucleic acid). Non-limiting
examples of suitable tissue-
specific promoters include an albumin promoter (liver-specific; Pinkert et
al., Genes Dev. 1: 268-277
(1987)), lymphoid-specific promoters (Calame ~ Eaton, Adv. Immunol. 43: 235-
275 (1988)),
promoters of T cell receptors (Winoto & Baltimore, EMBO J. 8: 729-733 (1989))
promoters of
immunoglobulins (Banerji et al., Cell 33: 729-740 (1983); Queen & Baltimore,
Cell 33: 741-748
(1983)), neuron-specific promoters (e.g., the neurofilament promoter; Byrne &
Ruddle, Proc. Natl.
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
Acad. Sci. USA 86: 5473-5477 (1989)), pancreas-specific promoters (Edlund et
al., Science 230: 912-
916 (1985)), and mammary gland-specific promoters (e.g., milk whey promoter;
U.S. Patent No.
4,873,316 and European Application Publication No. 264,166). Developmentally-
regulated promoters
are sometimes utilized, for example, the murine hox promoters (I~essel &
Gruss, Science 249: 374-379
(1990)) and the oc-fetopolypeptide promoter (Campes & Tilghman, Genes Dev. 3:
537-546 (1989)).
[0055] A KIAA0296, Ch~om 4, Chrom 6, ELP3, LRCHl, SNWI or ERG nucleic acid or
other
nucleic acid referenced in Table A also may be cloned into an expression
vector in an antisense
orientation. Regulatory sequences (e.g., viral promoters andlor enhancers)
operatively linked to a
KIAA0296, Ch~om 4, Chrom 6, ELP3, LRCHl, SNWl or ERG nucleic acid or other
nucleic acid
referenced in Table A cloned in the antisense orientation can be chosen for
directing constitutive, tissue
specific or cell type specific expression of antisense RNA in a variety of
cell types. Antisense
expression vectors can be in the form of a recombinant plasmid, phagemid or
attenuated virus. For a
discussion of the regulation of gene expression using antisense genes see,
e.g., Weintraub et al.,
Antisense RNA as a molecular tool for genetic analysis, Reviews - Trends in
Genetics, Vol. 1(1) (1986).
[0056] Also provided herein are host cells that include a KIAA0296, Ch~om 4,
Chrona 6, ELP3,
LRCHl, SNWI or ERG nucleotide sequence or other nucleotide sequence referenced
in Table A within
a recombinant expression vector or a fragment of such a nucleotide sequence
which facilitate
homologous recombination into a specific site of the host cell genome. The
terms "host~cell" and
"recombinant host cell" are used interchangeably herein. Such terms refer not
only to the particular
subject cell but rather also to the progeny or potential progeny of such a
cell. Because certain
,:
modifications may occur in succeeding generations due to either mutation or
environmental influences,
such progeny may not, in fact, be identical to the parent cell, but are still
included within the scope of
the term as used herein. A host cell can be any prokaryotic or eukaryotic
cell. For example, a target
polypeptide can be expressed in bacterial cells such as E. coli, insect cells,
yeast or mammalian cells
(such as Chinese hamster ovary cells (CHO) or COS cells). Other suitable host
cells are known to those
skilled in the art.
[0057] Vectors can be introduced into host cells via conventional
transformation or transfection
techniques. As used herein, the terms "transformation" and "transfection" are
intended to refer to a
variety of art-recognized techniques for introducing foreign nucleic acid
(e.g., DNA) into a host cell,
including calcium phosphate or calcium chloride co-precipitation,
transductiouinfection, DEAE-
dextran-mediated transfection, lipofection, or electroporation.
[0058] A host cell provided herein can be used to produce (i. e., express) a
target polypeptide or a
substantially identical polypeptide thereof. Accordingly, further provided are
methods for producing a
target polypeptide using host cells described herein. In one embodiment, the
method includes culturing
host cells into which a recombinant expression vector encoding a target
polypeptide has been introduced
in a suitable medium such that a target polypeptide is produced. In another
embodiment, the method
further includes isolating a target polypeptide from the medium or the host
cell.
16
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
[0059] Also provided are cells or purified preparations of cells which include
a KIAA0296, Chrom
4, Chrona 6, ELP3, LRCHl, SNWI or ERG transgene, or other transgene in Table
A, or which otherwise
misexpress target polypeptide. Cell preparations can consist of human or non-
human cells, e.g., rodent
cells, e.g., mouse or rat cells, rabbit cells, or pig cells. In preferred
embodiments, the cell or cells
include a I~IIAA0296, Chrorn 4, Chrom 6, ELP3, LRCHl, SNWI or ERG transgene or
other transgene
referenced in Table A (e.g., a heterologous form of a KIAA0296, Chrom 4, Chrom
6, ELP3, LRCHl,
SNWI or ERG gene or other gene referenced in Table A, such as a human gene
expressed in non-human
cells). The transgene can be misexpressed, e.g., overexpressed or
underexpressed. In other preferred
embodiments, the cell or cells include a gene which misexpress an endogenous
target polypeptide (e.g.,
expression of a gene is disrupted, also known as a knockout). Such cells can
serve as a model for
studying disorders which are related to mutated or mis-expressed alleles or
for use in drug screening.
Also provided are human cells (e.g., a hematopoietic stem cells) transfected
with a KI~1A0296, Chrona 4,
Chrom 6, ELP3, LRCHl, SNWI or ERG nucleic acid or other nucleic acid
referenced in Table A.
[0060] Also provided are cells or a purified preparation thereof (e.g., human
cells) in which an
endogenous KIAA0296, Chronz 4, Chrom 6, ELP3, LRCHl, SNWI or ERG nucleic acid
or other nucleic
acid referenced in Table A is under the control of a regulatory sequence that
does not normally control
the expression of the endogenous gene. The expression characteristics of an
endogenous gene within a
cell (e.g., a cell line or microorganism) can be modified by inserting a
heterologous DNA regulatory
element into the genome of the cell such that the inserted regulatory element
is operably linked to the
corresponding endogenous gene. For example, an endogenous corresponding gene
(e.g., a gene which
is "transcriptionally silent," not normally expressed, or expressed only at
very low levels) may be
activated by inserting a regulatory element which is capable of promoting the
expression of a normally
expressed gene product in that cell. Techniques such as targeted homologous
recombinations, can be
used to insert the heterologous DNA as described in, e.g., Chappel, US
5,272,071; WO 91/06667,
published on May 16, 1991.
Transgenic Animals
[0061] Non-human transgenic animals that express a heterologous target
polypeptide (e.g.,
expressed from a KIAA0296, Chrorrr 4, Chrorn 6, ELP3, LRCHl, SNWI or ERG
nucleic acid or other
nucleic acid referenced in Table A, or substantially identical sequence
thereof) can be generated. Such
animals are useful for studying the function and/or activity of a target
polypeptide and for identifying
and/or evaluating modulators of the activity of KI~4A0296, ChronZ 4, Chrom 6,
ELP3, LRCHl, SNWI or
ERG nucleic acids, other nucleic acids referenced in Table A, and encoded
polypeptides. As used
herein, a "transgenic animal" is a non-human animal such as a mammal (e.g., a
non-human primate such
as chimpanzee, baboon, or macaque; an ungulate such as an equine, bovine, or
caprine; or a rodent such
as a rat, a mouse, or an Israeli sand rat), a bird (e.g., a chicken or a
turkey), an amphibian (e.g., a frog,
salamander, or newt), or an insect (e.g., Drosophila rraelarcogaster), in
which one or more of the cells of
the animal includes a transgene. A transgene is exogenous DNA or a
rearrangement (e.g., a deletion of
17
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
endogenous chromosomal DNA) that is often integrated into or occurs in the
genome of cells in a
transgenic animal. A transgene can direct expression of an encoded gene
product in one or more cell
types or tissues of the transgenic animal, and other transgenes can reduce
expression (e.g., a knockout).
Thus, a transgenic animal can be one in which an endogenous nucleic acid
homologous to a KIAA0296,
Chrorn 4, Chf~om 6, ELP3, LRCHl, SNWI or ERG nucleic acid or other nucleic
acid referenced in Table
A has been altered by homologous recombination between the endogenous gene and
an exogenous
DNA molecule introduced into a cell of the animal (e.g., an embryonic cell of
the animal) prior to
development of the animal.
[0062] Intronic sequences and polyadenylation signals can also be included in
the transgene to
increase expression efficiency of the transgene. One or more tissue-specific
regulatory sequences can
be operably linked to a KI~1A0296, Chf°ona 4, Chf°om 6, ELP3,
LRCHI, SNWl or ERG nucleotide
sequence or other nucleotide sequence referenced in Table A to direct
expression of an encoded
polypeptide to particular cells. A transgenic founder animal can be identified
based upon the presence
of a KIAA0296, Chrom 4, Clzf~ona 6, ELP3, LRCHl, SNWI or ERG nucleotide
sequence or other
nucleotide sequence referenced in Table A in its genome and/or expression of
encoded mRNA in tissues
or cells of the animals. A transgenic founder animal can then be used to breed
additional animals
carrying the transgene. Moreover, transgenic animals carrying a KIAA0296,
Chron2 4, Chrom 6, ELP3,
LRCHl, SNWI or ERG nucleotide sequence or other nucleotide sequence referenced
in Table A can
further be bred to other transgenic animals carrying other transgenes.
[0063] Target polypeptides can be expressed in transgenic animals or plants by
introducing, for
example, a KIAA0296, Chrom 4, Chrom 6, ELP3, LRCHl, SNWI or ERG nucleic acid
or other nucleic
acid referenced in Table A into the genome of an animal that encodes the
target polypeptide. In
preferred embodiments the nucleic acid is placed under the control of a tissue
specific promoter, e.g., a ,
milk or egg specific promoter, and recovered from the milk or eggs produced by
the animal. Also
included is a population of cells from a transgenic animal.
Tar-e~ t PolYpeptides
[0064] Also featured herein are isolated target polypeptides, which are
encoded by a KIAA0296,
Chf°om 4, Ch~om 6, ELP3, LRCHl, SNWI or ERG nucleotide sequence or a
nucleotide sequence
referenced in Table A (e.g., SEQ ID NO: 8-17 or a sequence referenced in Table
A), or a substantially
identical nucleotide sequence thereof. Examples of KIf1A0296, Chrom 4, Cht~om
6, ELP3, LRCHl,
SNWI or ERG polypeptides are set forth in SEQ ID NO: 18-27. The term
"polypeptide" as used herein
includes proteins and peptides. An "isolated" or "purified" polypeptide or
protein is substantially free
of cellular material or other contaminating proteins from the cell or tissue
source from which the protein
is derived, or substantially free from chemical precursors or other chemicals
when chemically
synthesized. In one embodiment, the language "substantially free" means
preparation of a target
polypeptide having less than about 30%, 20%, 10% and more preferably 5% (by
dry weight), of non-
target polypeptide (also referred to herein as a "contaminating protein"), or
of chemical precursors or
18
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
non-target chemicals. When the target polypeptide or a biologically active
portion thereof is
recombinantly produced, it is also preferably substantially free of culture
medium, specifically, where
culture medium represents less than about 20%, sometimes less than about 10%,
and often less than
about 5% of the volume of the polypeptide preparation. Isolated or purified
target polypeptide
preparations are sometimes 0.01 milligrams or more or 0.1 milligrams or more,
and often 1.0 milligrams
or more and 10 milligrams or more in dry weight.
[0065] Further included herein are target polypeptide fragments. The
polypeptide fragment may be
a domain or part of a domain of a target polypeptide. The polypeptide fragment
may have increased,
decreased or unexpected biological activity. The polypeptide fragment is often
50 or fewer, 100 or fewer,
or 200 or fewer amino acids in length, and is sometimes 300, 400, 500, 600,
700, or 900 or fewer amino
acids in length. Specific embodiments are directed to a PTPNI polypeptide
fragment (e.g., rs2282146 in
Table A), such as a catalytic domain starting at about amino acid 3 and ending
at about amino acid 279.
Other embodiments are directed to a KCNSl polypeptide fragment (e.g., rs734784
in Table A), such as a
voltage gated postassium ion channel domain (e.g., starting at about amino
acid 21 and ending at about
amino acid 509), a postassium channel tetramerization domain (e.g., starting
at about amino acid 52 and
ending at about amino acid 155) or an ion transport protein domain (e.g.,
starting at about amino acid 271
and ending at about amino acid 456), for example. Certain embodiments are
directed to PSMBI
polypeptide fragments (e.g., sequence accessed by NP_002784; rs756519 in Table
A), such as a
proteasome protease domain (e.g., starting at about amino acid 34 and ending
at about amino acid 226) or a
proteasome B domain (e.g., starting at about amino acid 41 and ending at about
amino acid 88). Certain
embodiments are directed to a ANXA6 polypeptide fragment (e.g., rs1012414 in
Table A), such as an
annexin domain starting at about amino acid 5 and ending at about amino acid
325, an annexin domain
startuig at about amino acid 179 and ending at about amino acid 507, or an
annexin domain starting at about .
amino acid 355 and ending at about amino acid 673 in isoform 1 or isoform 2
(e.g., an isoform 1 sequence
can be accessed using accession number NP_001146 and an isoform 2 sequence can
be accessed using
accession number NP 004024; isoform 2 lacks exon 21 and encodes a protein
isoform lacking the six
amino acids VAAEIL,). Amino acid sequences can be accessed using information
in Table A and in SEQ
117 NO: 18-27.
[0066] Substantially identical target polypeptides may depart from the amino
acid sequences of
target polypeptides in different manners. For example, conservative amino acid
modifications may be
introduced at one or more positions in the amino acid sequences of target
polypeptides. A "conservative
amino acid substitution" is one in which the amino acid is replaced by another
amino acid having a
similar structure and/or chemical function. Families of amino acid residues
having similar structures
and functions are well known. These families include amino acids with basic
side chains (e.g., lysine,
arginine, histidine), acidic side chains (e.g., aspartic acid, glutamic acid),
uncharged polar side chains
(e.g., glycine, asparagine, glutamine, serine, threonine, tyrosine, cysteine),
nonpolar side chains (e.g.,
alanine, valine, leucine, isoleucine, proline, phenylalanine, methionine,
tryptophan), beta-branched side
chains (e.g., threonine, valine, isoleucine) and aromatic side chains (e.g.,
tyrosine, phenylalanine,
19
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
tryptophan, histidine). Also, essential and non-essential amino acids may be
replaced. A "non-
essential" amino acid is one that can be altered without abolishing or
substantially altering the biological
function of a target polypeptide, whereas altering an "essential" amino acid
abolishes or substantially
alters the biological function of a target polypeptide. Amino acids that are
conserved among target
polypeptides are typically essential amino acids. In certain embodiments, the
polypeptide includes one
or more non-synonymous polymorphic variants associated with osteoarthritis, as
described above (e.g.,
a valine encoded by rs734784, a valine encoded by rs1042164, a glutamate
encoded by rs749670, a
threonine encoded by rs955592, a glutamine encoded by rs241448, and a glycine
encoded by
rs 1040461 ).
[0067] Also, target polypeptides may exist as chimeric or fusion polypeptides.
As used herein, a
target "chimeric polypeptide" or target "fusion polypeptide" includes a target
polypeptide linked to a
non-target polypeptide. A "non-target polypeptide" refers to a polypeptide
having an amino acid
sequence corresponding to a polypeptide which is not substantially identical
to the target polypeptide,
which includes, for example, a polypeptide that is different from the target
polypeptide and derived
from the same or a different organism. The target polypeptide in the fusion
polypeptide can correspond
to an entire or nearly entire target polypeptide or a fragment thereof. The
non-target polypeptide can be
fused to the N-terminus or C-terminus of the target polypeptide.
[0068] Fusion polypeptides can include a moiety having high affinity for a
ligand. For example,
the fusion polypeptide can be a GST-target fusion polypeptide in which the
target sequences are fused
to the C-terminus of the GST sequences, or a polyhistidine-target fusion
polypeptide in which the target
polypeptide is fused at the N- or C-terminus to a string of histidine
residues. Such fusion polypeptides
can facilitate purification of recombinant target polypeptide. Expression
vectors are commercially
available that already encode a fusion moiety (e.g., a GST polypeptide), and a
nucleotide sequence in
SEQ ID NO: 1-7 or referenced in Table A, or a substantially identical
nucleotide sequence thereof, can
be cloned into an expression vector such that the fusion moiety is linked in-
frame to the target
polypeptide. Further, the fusion polypeptide can be a target polypeptide
containing a heterologous
signal sequence at its N-terminus. In certain host cells (e.g., mammalian host
cells), expression,
secretion, cellular internalization, and cellular localization of a target
polypeptide can be increased
through use of a heterologous signal sequence. Fusion polypeptides can also
include all or a part of a
serum polypeptide (e.g., an IgG constant region or human serum albumin).
[0069] Target polypeptides can be incorporated into pharmaceutical
compositions and
administered to a subject in vivo. Administration of these target polypeptides
can be used to affect the
bioavailability of a substrate of the target polypeptide and may effectively
increase target polypeptide
biological activity in a cell. Target fusion polypeptides may be useful
therapeutically for the treatment
of disorders caused by, for example, (i) aberrant modification or mutation of
a gene encoding a target
polypeptide; (ii) mis-regulation of the gene encoding the target polypeptide;
and (iii) aberrant post-
translational modification of a target polypeptide. Also, target p0lypeptides
can be used as immunogens
to produce anti-target antibodies in a subject, to purify target polypeptide
ligands or binding partners,
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
and in screening assays to identify molecules which inhibit or enhance the
interaction of a target
polypeptide with a substrate.
[0070] In addition, polypeptides can be chemically synthesized using
techniques known in the art
(See, e.g., Creighton, 1983 Proteins. New York, N.Y.: W. H. Freeman and
Company; and Hunkapiller
et al., (1984) Nature July 12 -18;310(5973):105-11). For example, a relative
short fragment can be
synthesized by use of a peptide synthesizer. Furthermore, if desired, non-
classical amino acids or
chemical amino acid analogs can be introduced as a substitution or addition
into the fragment sequence.
Non-classical amino acids include, but are not limited to, to the D-isomers of
the common amino acids,
2,4-diaminobutyric acid, a-amino isobutyric acid, 4-aminobutyric acid, Abu, 2-
amino butyric acid, g-
Abu, e-Ahx, 6-amino hexanoic acid, Aib, 2-amino isobutyric acid, 3-amino
propionic acid, ornithine,
norleucine, norvaline, hydroxyproline, sarcosine, citrulline, homocitrulline,
cysteic acid, t-butylglycine,
t-butylalanine, phenylglycine, cyclohexylalanine, b-alanine, fluoroamino
acids, designer amino acids
such as b-methyl amino acids, Ca-methyl amino acids, Na-methyl amino acids,
and amino acid analogs
in general. Furthermore, the amino acid can be D (dextrorotary) or L
(levorotary).
[0071] Polypeptides and polypeptide fragments sometimes are differentially
modified during or
after translation, e.g., by glycosylation, acetylation, phosphorylation,
amidation, derivatization by
known protecting/blocking groups, proteolytic cleavage, linkage to an antibody
molecule or other
cellular ligand, etc. Any of numerous chemical modifications may be carried
out by known techniques,
including but not limited, to specific chemical cleavage by cyanogen bromide,
trypsin, chymotrypsin,
papain, V8 protease, NaBH4; acetylation, formylation, oxidation, reduction;
metabolic synthesis in the
presence of tunicamycin; and the like. Additional post-translational
modifications include, for example,
N-linked or O-linked carbohydrate chains, processing of N-terminal or C-
terminal ends), attachment of
chemical moieties to the amino acid backbone, chemical modifications of N-
linked or O-linked
carbohydrate chains, and addition or deletion of an N-terminal methionine
residue as a result of
prokaryotic host cell expression. The polypeptide fragments may also be
modified with a detectable
label, such as an enzymatic, fluorescent, isotopic or affinity label to allow
for detection and isolation of
the polypeptide.
[0072] Also provided are chemically modified derivatives of polypeptides that
can provide
additional advantages such as increased solubility, stability and circulating
time of the polypeptide, or
decreased immunogenicity (see e.g., U.S. Pat. No: 4,179,337. The chemical
moieties for derivitization
may be selected from water soluble polymers such as polyethylene glycol,
ethylene glycollpropylene
glycol copolymers, carboxymethylcellulose, dextran, polyvinyl alcohol and the
like. The polypeptides
may be modified at random positions within the molecule, or at predetermined
positions within the
molecule and may include one, two, three or more attached chemical moieties.
[0073] The polymer may be of any molecular weight, and may be branched or
unbranched. For
polyethylene glycol, the preferred molecular weight is between about 1 kDa and
about 100 kDa (the
term "about" indicating that in preparations of polyethylene glycol, some
molecules will weigh more,
some less, than the stated molecular weight) for ease in handling and
manufacturing. Other sizes may be
21
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
used, depending on the desired therapeutic profile (e.g., the duration of
sustained release desired, the
effects, if any on biological activity, the ease in handling, the degree or
lack of antigenicity and other
known effects of the polyethylene glycol to a therapeutic protein or analog).
[0074] The polymers should be attached to the polypeptide with consideration
of effects on
functional or antigenic domains of the polypeptide. There are a number of
attachment methods available
to those skilled in the art (e.g., EP 0 401 384 (coupling PEG to G-CSF) and
Malik et al. (1992) Exp
Hematol. September;20(8):1028-35 (pegylation of GM-CSF using tresyl
chloride)). For example,
polyethylene glycol may be covalently bound through amino acid residues via a
reactive group, such as
a free amino or carboxyl group. Reactive groups are those to which an
activated polyethylene glycol
molecule may be bound. The amino acid residues having a free amino group may
include lysine
residues and the N-terminal amino acid residues; those having a free carboxyl
group may include
aspartic acid residues, glutamic acid residues and the C-terminal amino acid
residue. Sulfhydryl groups
may also be used as a reactive group for attaching the polyethylene glycol
molecules. For therapeutic
purposes, the attaclnnent sometimes is at an amino group, such as attachment
at the N-terminus or
lysine group.
[0075] Proteins can be chemically modified at the N-terminus. Using
polyethylene glycol as an
illustration of such a composition, one may select from a variety of
polyethylene glycol molecules (by
molecular weight, branching, and the like), the proportion of polyethylene
glycol molecules to protein
(polypeptide) molecules in the reaction mix, the type of pegylation reaction
to be performed, and the
method of obtaining the selected N-terminally pegylated protein. The method of
obtaining the N-
terminally pegylated preparation (i.e., separating this moiety from other
monopegylated moieties if
necessary) may be by purification of the N-terminally pegylated material from
a population of pegylated
protein molecules. Selective proteins chemically modified at the N-terminus
may be accomplished by
reductive alkylation, which exploits differential reactivity of different
types of primary amino groups
(lysine versus the N-terminal) available for derivatization in a particular
protein. Under the appropriate
reaction conditions, substantially selective derivatization of the protein at
the N-terminus with a
carbonyl group containing polymer is achieved.
Substantially Identical Nucleic Acids and Polypeptides
[0076] Nucleotide sequences and polypeptide sequences that are substantially
identical to a
KIAA0296, Chrom 4, CI2f'o3rZ 6, ELP3, LRCHl, SNWI or ERG nucleotide sequence
or other nucleotide
sequence referenced in Table A and the target polypeptide sequences encoded by
those nucleotide
sequences, respectively, are included herein. The term "substantially
identical" as used herein refers to
two or more nucleic acids or polypeptides sharing one or more identical
nucleotide sequences or
polypeptide sequences, respectively. Included are nucleotide sequences or
polypeptide sequences that
are 55% or more, 60% or more, 65% or more, 70% or more, 75% or more, 80% or
more, 85% or more,
90% or more, 95% or more (each often within a 1%, 2%, 3% or 4% variability)
identical to a
KIAA0296, Chrom 4, Chrom 6, ELP3, LRCHl, SNWI or ERG nucleotide sequence, or
other nucleotide
22
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
sequence referenced in Table A, or the encoded target polypeptide amino acid
sequences. One test for
determining whether two nucleic acids are substantially identical is to
determine the percent of identical
nucleotide sequences or polypeptide sequences shared between the nucleic acids
or polypeptides.
[0077] Calculations of sequence identity are often performed as follows.
Sequences are aligned for
optimal comparison purposes (e.g., gaps can be introduced in one or both of a
first and a second amino
acid or nucleic acid sequence for optimal alignment and non-homologous
sequences can be disregarded
for comparison purposes). The length of a reference sequence aligned for
comparison purposes is
sometimes 30% or more, 40% or more, 50% or more, often 60% or more, and more
often 70% or more,
80% or more, 90% or more, or 100% of the length of the reference sequence. The
nucleotides or amino
acids at corresponding nucleotide or polypeptide positions, respectively, are
then compared among the
two sequences. When a position in the first sequence is occupied by the same
nucleotide or amino acid
as the corresponding position in the second sequence, the nucleotides or amino
acids are deemed to be
identical at that position. The percent identity between the two sequences is
a function of the number of
identical positions shared by the sequences, taking into account the number of
gaps, and the length of
each gap, introduced for optimal alignment of the two sequences.
[0078] Comparison of sequences and determination of percent identity between
two sequences can
be accomplished using a mathematical algorithm. Percent identity between two
amino acid or
nucleotide sequences can be determined using the algorithm of Meyers & Miller,
CABIOS 4: 11-17
(1989), which has been incorporated into the ALIGN program (version 2.0),
using a PAM120 weight
residue table, a gap length penalty of 12 and a gap penalty of 4. Also,
percent identity between two
amino acid sequences can be determined using the Needleman ~ Wunsch, J. Mol.
Biol. 4~: 444-453
(1970) algorithm which has been incorporated into the GAP program in the GCG
software package
(available at the http address www.gcg.com), using either a Blossum 62 matrix
or a PAM250 matrix,
and a gap weight of 16, 14, 12, 10, 8, 6, or 4 and a length weight of 1, 2, 3,
4, 5, or 6. Percent identity
between two nucleotide sequences can be determined using the GAP program in
the GCG software
package (available at http address www.gcg.com), using a NWSgapdna.CMP matrix
and a gap weight
of 40, 50, 60, 70, or 80 and a length weight of 1, 2, 3, 4, 5, or 6. A set of
parameters often used is a
Blossum 62 scoring matrix with a gap open penalty of 12, a gap extend penalty
of 4, and a frameshift
gap penalty of 5.
[0079] Another manner for determining if two nucleic acids are substantially
identical is to assess
whether a polynucleotide homologous to one nucleic acid will hybridize to the
other nucleic acid under
stringent conditions. As use herein, the term "stringent conditions" refers to
conditions for
hybridization and washing. Stringent conditions are known to those skilled in
the art and can be found
in Cu~f~ent Protocols in Moleeular Biology, John Wiley & Sons, N.Y. , 6.3.1-
6.3.6 (1989). Aqueous
and non-aqueous methods are described in that reference and either can be
used. An example of
stringent hybridization conditions is hybridization in 6X sodium
chloride/sodium citrate (SSC) at about
45°C, followed by one or more washes in 0.2X SSC, 0.1% SDS at
50°C. Another example of stringent
hybridization conditions are hybridization in 6X sodium chloridelsodium
citrate (SSC) at about 45°C,
23
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
followed by one or more washes in 0.2X SSC, 0.1% SDS at 55°C. A further
example of stringent
hybridization conditions is hybridization in 6X sodium chloride/sodium citrate
(SSC) at about 45°C,
followed by one or more washes in 0.2X SSC, 0.1% SDS at 60°C. Often,
stringent hybridization
conditions are hybridization in 6X sodium chloride/sodium citrate (SSC) at
about 45°C, followed by
one or more washes in 0.2X SSC, 0.1% SDS at 65°C. More often,
stringency conditions are 0.5M
sodium phosphate, 7% SDS at 65°C, followed by one or more washes at
0.2X SSC, 1% SDS at 65°C.
[0080] An example of a substantially identical nucleotide sequence to a
nucleotide sequence in
SEQ ID NO: 1-7 or referenced in Table A is one that has a different nucleotide
sequence but still
encodes the same polypeptide sequence encoded by the nucleotide sequence in
SEQ ID NO: 1-7 or
referenced in Table A. Another example is a nucleotide sequence that encodes a
polypeptide having a
polypeptide sequence that is more than 70% or more identical to, sometimes
more than 75% or more,
80% or more, or 85% or more identical to, and often more than 90% or more and
95% or more identical
to a polypeptide sequence encoded by a nucleotide sequence in SEQ ID NO: 1-7
or referenced in Table
A.
[0081] Nucleotide sequences in SEQ ID NO: 1-7 or referenced in Table A and
amino acid
sequences of encoded polypeptides can be used as "query sequences" to perform
a search against public
databases to identify other family members or related sequences, for example.
Such searches can be
performed using the NBLAST and XBLAST programs (version 2.0) of Altschul et
al., J. Mol. Biol.
215: 403-10 (1990). BLAST nucleotide searches can be performed with the NBLAST
program, score =
100, wordlength = 12 to obtain nucleotide sequences homologous to nucleotide
sequences in SEQ ID
NO: 1-7, SEQ ID NO: 8-17 or referenced in Table A. BLAST polypeptide searches
can be performed
with the XBLAST program, score = 50, wordlength = 3 to obtain amino acid
sequences homologous to
polypeptides encoded by the nucleotide sequences of SEQ ID NO: 8-17 or
referenced in Table A. To
obtain gapped alignments for comparison purposes, Gapped BLAST can be utilized
as described in
Altschul et al., Nucleic Acids Res. 25(17): 3389-3402 (1997). When utilizing
BLAST and Gapped
BLAST programs, default parameters of the respective programs (e.g., XBLAST
and NBLAST) can be
used (see the http address www.ncbi.nlm.nih.gov).
[0082] A nucleic acid that is substantially identical to a nucleotide sequence
in SEQ ID NO: 1-7 or
referenced in Table A may include polymorphic sites at positions equivalent to
those described herein
when the sequences are aligned. For example, using the alignment procedures
described herein, SNPs
in a sequence substantially identical to a sequence in SEQ ID NO: 1-7 or
referenced in Table A can be
identified at nucleotide positions that match (i.e., align) with nucleotides
at SNP positions in each
nucleotide sequence in SEQ ID NO: 1-7 or referenced in Table A. Also, where a
polymorphic variation
results in an insertion or deletion, insertion or deletion of a nucleotide
sequence from a reference
sequence can change the relative positions of other polymorphic sites in the
nucleotide sequence.
[0083] Substantially identical nucleotide and polypeptide sequences include
those that are naturally
occurring, such as allelic variants (same locus), splice variants, homologs
(different locus), and
24
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
orthologs (different organism) or can be non-naturally occurring. Non-
naturally occurring variants can
be generated by mutagenesis techniques, including those applied to
polynucleotides, cells, or organisms.
The variants can contain nucleotide substitutions, deletions, inversions and
insertions. Variation can
occur in either or both the coding and non-coding regions. The variations can
produce both
conservative and non-conservative amino acid substitutions (as compared in the
encoded product).
Orthologs, homologs, allelic variants, and splice variants can be identified
using methods known in the
art. These variants normally comprise a nucleotide sequence encoding a
polypeptide that is 50% or
more, about 55% or more, often about 70-75% or more or about 80-85% or more,
and sometimes about
90-95% or more identical to the amino acid sequences of target polypeptides or
a fragment thereof.
Such nucleic acid molecules can readily be identified as being able to
hybridize under stringent
conditions to a nucleotide sequence in SEQ ID NO: 1-7 or referenced in Table A
or a fragment of this
sequence. Nucleic acid molecules corresponding to orthologs, homologs, and
allelic variants of a
nucleotide sequence in SEQ ID NO: 1-7 or referenced in Table A can further be
identified by mapping
the sequence to the same chromosome or locus as the nucleotide sequence in SEQ
ID NO: 1-7 or
referenced in Table A.
[0084] Also, substantially identical nucleotide sequences may include codons
that are altered with
respect to the naturally occurring sequence for enhancing expression of a
target polypeptide in a
particular expression system. For example, the nucleic acid can be one in
which one or more codons are
altered, and often 10% or more or 20% or more of the codons are altered for
optimized expression in
bacteria (e.g., E. coli.), yeast (e.g., S. cervesiae), human (e.g., 293
cells), insect, or rodent (e.g., hamster)
cells.
Methods for Identif5ring Risk of Osteoarthritis
[0085] Methods for prognosing and diagnosing osteoarthritis are included
herein. These methods
include detecting the presence or absence of one or more polymorphic
variations in a nucleotide
sequence associated with osteoarthritis, such as variants in or around the
loci set forth herein, or a
substantially identical sequence thereof, in a sample from a subject, where
the presence of a
polymorphic variant described herein is indicative of a risk of
osteoarthritis. Determining a risk of
osteoarthritis sometimes refers to determining whether an individual is at an
increased risk of
osteoarthritis (e.g., intermediate risk or higher risk).
[0086] Thus, featured herein is a method for identifying a subject who is at
risk of osteoarthritis,
which comprises detecting an aberration associated with osteoarthritis in a
nucleic acid sample from the
subject. An embodiment is a method for detecting a risk of osteoarthritis in a
subject, which comprises
detecting the presence or absence of a polymorphic variation associated with
osteoarthritis at a
polymorphic site in a nucleotide sequence in a nucleic acid sample from a
subject, where the nucleotide
sequence comprises a polynucleotide sequence selected from the group
consisting of: (a) a nucleotide
sequence of SEQ D7 NO: 1-7 or referenced in Table A; (b) a nucleotide sequence
which encodes a
polypeptide consisting of an amino acid sequence encoded by a nucleotide
sequence of SEQ ID NO: 1-7
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
or referenced in Table A; (c) a nucleotide sequence which encodes a
polypeptide that is 90% or more
identical to an amino acid sequence encoded by a nucleotide sequence of SEQ ID
NO: 1-7 or referenced
in Table A, or a nucleotide sequence about 90% or more identical to a
nucleotide sequence of SEQ ID
NO: 1-7 or referenced in Table A; and (d) a fragment of a nucleotide sequence
of (a), (b), or (c)
comprising the polymorphic site; whereby the presence of the polymorphic
variation is indicative of a
predisposition to osteoarthritis in the subject. In certain embodiments,
polymorphic variants at the
positions described herein are detected for determining a risk of
osteoarthritis, and polymorphic variants
at positions in linkage disequilibrium with these positions are detected for
determining a risk of
osteoarthritis. As used herein, the terms "SEQ ID NO: 1-7" and other
nucleotide sequences "referenced
in Table A" refers to individual sequences in SEQ ID NO: l, 2, 3, 4, 5, 6 or
7, or any individual
sequence referenced in Table A, each sequence being separately applicable to
embodiments described
herein.
[0087] Risk of osteoarthritis sometimes is expressed as a probability, such as
an odds ratio,
percentage, or risk factor. Risk often is based upon the presence or absence
of one or more polymorphic
variants described herein, and also may be based in part upon phenotypic
traits of the individual being
tested. Methods for calculating risk based upon patient data are well known
(see, e.g., Agresti,
Categorical Data Analysis, 2nd Ed. 2002. Wiley). Allelotyping and genotyping
analyses may be
carried out in populations other than those exemplified herein to enhance the
predictive power of the
prognostic method. These further analyses are executed in view of the
exemplified procedures
described herein, and may be based upon the same polymorphic variations or
additional polymorphic
variations.
[0088] In certain embodiments, determining the presence of a combination of
two or more
polymorphic variants associated with osteoarthritis in one or more genetic
loci (e.g., one or more genes)
of the sample is determined to identify, quantify and/or estimate, risk of
osteoarthritis. The risk often is
the probability of having or developing osteoarthritis. The risk sometimes is
expressed as a relative risk
with respect to a population average risk of osteoarthritis, and sometimes is
expressed as a relative risk
with respect to the lowest risk group. Such relative risk assessments often
are based upon penetrance
values determined by statistical methods, and are particularly useful to
clinicians and insurance
companies for assessing risk of osteoarthritis (e.g., a clinician can target
appropriate detection,
prevention and therapeutic regimens to a patient after determining the
patient's risk of osteoarthritis, and
an insurance company can fme tune actuarial tables based upon population
genotype assessments of
osteoarthritis risk). Risk of osteoarthritis sometimes is expressed as an odds
ratio, which is the odds of a
particular person having a genotype has or will develop osteoarthritis with
respect to another genotype
group (e.g., the most disease protective genotype or population average). In
related embodiments, the
determination is utilized to identify a subject at risk of osteoarthritis. In
an embodiment, two or more
polymorphic variations are detected in two or more regions in human genomic
DNA associated with
increased risk of osteoarthritis, such as a locus containing a KIAA0296,
Chr~om 4, Chrom 6, ELP3,
LRCHl, SNWI or ERG or other locus referenced in Table A, for example. In
certain embodiments, 3 or
26
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
more, or 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 25, 30,
35, 40, 45, 50, 55, 60, 65, 70,
80, 90, 100 or more polymorphic variants are detected in the sample. In
specific embodiments,
polymorphic variants are detected in a KIAA0296, Chrom 4, Chrom 6, ELP3,
LRCHl, SNWI or ERG
region or other region referenced in Table A, for example. In another
embodiment, polymorphic
variants are detected at two or three positions in a nucleotide sequence of
SEQ ID NO: 1-7 or referenced
in Table A. In certain embodiments, polymorphic variants are detected at other
genetic loci (e.g., the
polymorphic variants can be detected in a KIAA0296, Chrom 4, Chrom 6, ELP3,
LRCHI, SlVWl or ERG
nucleotide sequence or other nucleotide sequence referenced in Table A in
addition to other loci or only
in other loci), where the other loci include but are not limited to those
described in patent applications
60/559,011; 60/559,202; 60/559,203; 60/559,042; 60/559,275; 60/559,040 and
60/559,225, each of
which is entitled "Methods for Identifying Risk of Osteoarthritis and
Treatments Thereof," each of
which was filed on 1 April 2004 and each of which is incorporated herein by
reference in its entirety in
jurisdictions allowing incorporation by reference.
[0089] Results from prognostic tests may be combined with other test results
to diagnose
osteoarthritis. For example, prognostic results may be gathered, a patient
sample may be ordered based
on a determined predisposition to osteoarthritis, the patient sample is
analyzed, and the results of the
analysis may be utilized to diagnose osteoarthritis. Also osteoaxthritis
diagnostic method can be
developed from studies used to generate prognostic methods in which
populations are stratified into
subpopulations having different progressions of osteoarthritis. In another
embodiment, prognostic
results may be gathered, a patient's risk factors for developing
osteoarthritis (e.g., age, weight,
occupational history, race, diet) analyzed, and a patient sample may be
ordered based on a determined
predisposition to osteoarthritis.
[0090] The nucleic acid sample typically is isolated from a biological sample
obtained from a
subject. For example, nucleic acid can be isolated from blood, saliva, sputum,
urine, cell scrapings, and
biopsy tissue. The nucleic acid sample can be isolated from a biological
sample using standard
techniques, such as the technique described in Example 2. As used herein, the
term "subject" refers
primarily to humans but also refers to other mammals such as dogs, cats, and
ungulates (e.g., cattle,
sheep, and swine). Subjects also include avians (e.g., chickens and turkeys),
reptiles, and fish (e.g.,
salmon), as embodiments described herein can be adapted to nucleic acid
samples isolated from any of
these organisms. The nucleic acid sample may be isolated from the subject and
then directly utilized in
a method for determining the presence of a polymorphic variant, or
alternatively, the sample may be
isolated and then stored (e.g., frozen) for a period of time before being
subjected to analysis.
[0091] The presence or absence of a polymorphic variant is determined using
one or both
chromosomal complements represented in the nucleic acid sample. Determining
the presence or
absence of a polymorphic variant in both chromosomal complements represented
in a nucleic acid
sample from a subject having a copy of each chromosome is useful for
determining the zygosity of an
individual for the polymorphic variant (i.e., whether the individual is
homozygous or heterozygous for
the polymorphic variant). Any oligonucleotide-based diagnostic may be utilized
to determine whether a
27
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
sample includes the presence or absence of a polymorphic variant in a sample.
For example, primer
extension methods, ligase sequence determination methods (e.g., U.S. Pat. Nos.
5,679,524 and
5,952,174, and WO 01/27326), mismatch sequence determination methods (e.g.,
U.S. Pat. Nos.
5,851,770; 5,958,692; 6,110,684; and 6,183,958), microarray sequence
determination methods,
restriction fragment length polymorphism (RFLP), single strand conformation
polymorphism detection
(SSCP) (e.g., U.S. Pat. Nos. 5,891,625 and 6,013,499), PCR-based assays (e.g.,
TAQMAN~ PCR
System (Applied Biosystems)), and nucleotide sequencing methods may be used.
[0092] Oligonucleotide extension methods typically involve providing a pair of
oligonucleotide
primers in a polymerase chain reaction (PCR) or in other nucleic acid
amplification methods for the
purpose of amplifying a region from the nucleic acid sample that comprises the
polymorphic variation.
One oligonucleotide primer is complementary to a region 3' of the polymorphism
and the other is
complementary to a region 5' of the polymorphism. A PCR primer pair may be
used in methods
disclosed in U.S. Pat. Nos. 4,683,195; 4,683,202, 4,965,188; 5,656,493;
5,998,143; 6,140,054; WO
01127327; and WO 01/27329 for example. PCR primer pairs may also be used in
any commercially
available machines that perform PCR, such as any of the GENEAMP~ Systems
available from Applied
Biosystems. Also, those of ordinary skill in the art will be able to design
oligonucleotide primers based
upon a KIAA0296, Chrom 4, Chr~om 6, ELP3, LRCHl, SNWI or ERG nucleotide
sequence or other
nucleotide sequence referenced in Table A using knowledge available in the
art.
[0093] Also provided is an extension oligonucleotide that hybridizes to the
amplified fragment
adjacent to the polymorphic variation. As used herein, the term "adjacent"
refers to the 3' end of the
extension oligonucleotide being often 1 nucleotide from the 5' end of the
polymorphic site, and
sometimes 2, 3, 4, 5, 6, 7, 8, 9, or 10 nucleotides from the 5' end of the
polymorphic site, in the nucleic
acid when the extension oligonucleotide is hybridized to the nucleic acid. The
extension
oligonucleotide then is extended by one or more nucleotides, and the number
and/or type of nucleotides
that are added to the extension oligonucleotide determine whether the
polymorphic variant is present.
Oligonucleotide extension methods are disclosed, for example, in U.S. Pat.
Nos. 4,656,127; 4,851,331;
5,679,524; 5,834,189; 5,876,934; 5,908,755; 5,912,118; 5,976,802; 5,981,186;
6,004,744; 6,013,431;
6,017,702; 6,046,005; 6,087,095; 6,210,891; and WO 01/20039. Oligonucleotide
extension methods
using mass spectrometry are described, for example, in U.S. Pat. Nos.
5,547,835; 5,605,798; 5,691,141;
5,849,542; 5,869,242; 5,928,906; 6,043,031; and 6,194,144, and a method often
utilized is described
herein in Example 2.
[0094] A microarray can be utilized for determining whether a polymorphic
variant is present or
absent in a nucleic acid sample. A microarray may include any oligonucleotides
described herein, and
methods for making and using oligonucleotide microarrays suitable for
diagnostic use are disclosed in
U.S. Pat. Nos. 5,492,806; 5,525,464; 5,589,330; 5,695,940; 5,849,483;
6,018,041; 6,045,996;
6,136,541; 6,142,681; 6,156,501; 6,197,506; 6,223,127; 6,225,625; 6,229,911;
6,239,273; WO
00/52625; WO 01/25485; and WO 01/29259. The microarray typically comprises a
solid support and
the oligonucleotides may be linked to this solid support by covalent bonds or
by non-covalent
28
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
interactions. The oligonucleotides may also be linked to the solid support
directly or by a spacer
molecule. A microarray may comprise one or more oligonucleotides complementary
to a polymorphic
site set forth herein.
[0095] A kit also may be utilized for determining whether a polymorphic
variant is present or
absent in a nucleic acid sample. A kit often comprises one or more pairs of
oligonucleotide primers
useful for amplifying a fragment of a nucleotide sequence of SEQ ID NO: 1-7 or
referenced in Table A
or a substantially identical sequence thereof, where the fragment includes a
polymorphic site. The kit
sometimes comprises a polymerizing agent, for example, a thermostable nucleic
acid polymerase such
as one disclosed in U.S. Pat. Nos. 4,889,818 or 6,077,664. Also, the kit often
comprises an elongation
oligonucleotide that hybridizes to a KIAA0296, Chrom 4, Chr~om 6, ELP3, LRCHI,
SlVYT~l or ERG
nucleotide sequence or other nucleotide sequence referenced in Table A in a
nucleic acid sample
adjacent to the polymorphic site. Where the kit includes an elongation
oligonucleotide, it also often
comprises chain elongating nucleotides, such as dATP, dTTP, dGTP, dCTP, and
dITP, including
analogs of dATP, dTTP, dGTP, dCTP and dITP, provided that such analogs are
substrates for a
thermostable nucleic acid polymerase and can be incorporated into a nucleic
acid chain elongated from
the extension oligonucleotide. Along with chain elongating nucleotides would
be one or more chain
terminating nucleotides such as ddATP, ddTTP, ddGTP, ddCTP, and the like. In
an embodiment, the
kit comprises one or more oligonucleotide primer pairs, a polymerizing agent,
chain elongating
nucleotides, at least one elongation oligonucleotide, and one or more chain
terminating nucleotides.
Fits optionally include buffers, vials, microtiter plates, and instructions
for use.
[0096] An individual identified as being at risk of osteoarthritis may be
heterozygous or
homozygous with respect to the allele associated with a higher risk of
osteoarthritis. A subject
homozygous for an allele associated with an increased risk of osteoarthritis
is at a comparatively high
risk of osteoarthritis, a subject heterozygous for an allele associated with
an increased risk of
osteoarthritis is at a comparatively intermediate risk of osteoarthritis, and
a subject homozygous for an
allele associated with a decreased risk of osteoarthritis is at a
comparatively low risk of osteoarthritis. A
genotype may be assessed for a complementary strand, such that the
complementary nucleotide at a
particular position is detected.
[0097] Also featured are methods for determining risk of osteoarthritis and/or
identifying a subject
at risk of osteoarthritis by contacting a polypeptide or protein encoded by a
KIAA0296, Chf~om 4, Chrorra
6, ELP3, LRCHl, SNWI or ERG nucleotide sequence or other nucleotide sequence
referenced in Table
A from a subject with an antibody that specifically binds to an epitope
associated with increased risk of
osteoarthritis in the polypeptide.
Applications of Prognostic and Diagnostic Results to Pharmaco~;enomic Methods
[0098] Pharmacogenomics is a discipline that involves tailoring a treatment
for a subject according
to the subject's genotype as a particular treatment regimen may exert a
differential effect depending
upon the subject's genotype. For example, based upon the outcome of a
prognostic test described
29
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
herein, a clinician or physician may target pertinent information and
preventative or therapeutic
treatments to a subject who would be benefited by the information or treatment
and avoid directing such
information and treatments to a subject who would not be benefited (e.g., the
treatment has no
therapeutic effect and/or the subject experiences adverse side effects).
[0099] The following is an example of a pharmacogenomic embodiment. A
particular treatment
regimen can exert a differential effect depending upon the subject's genotype.
Where a candidate
therapeutic exhibits a significant interaction with a major allele and a
comparatively weak interaction
with a minor allele (e.g., an order of magnitude or greater difference in the
interaction), such a
therapeutic typically would not be administered to a subject genotyped as
being homozygous for the
minor allele, and sometimes not administered to a subject genotyped as being
heterozygous for the
minor allele. In another example, where a candidate therapeutic is not
significantly toxic when
administered to subjects who are homozygous for a major allele but is
comparatively toxic when
administered to subjects heterozygous or homozygous for a minor allele, the
candidate therapeutic is not
typically administered to subjects who are genotyped as being heterozygous or
homozygous with
respect to the minor allele.
(0100] The methods described herein are applicable to pharmacogenomic methods
for preventing,
alleviating or treating osteoarthritis. For example, a nucleic acid sample
from an individual may be
subjected to a prognostic test described herein. Where one or more polymorphic
variations associated
with increased risk of osteoarthritis are identified in a subject, information
for preventing or treating
osteoarthritis and/or one or more osteoarthritis treatment regimens then may
be prescribed to that
subj ect.
[0101] In certain embodiments, a treatment or preventative regimen is
specifically prescribed
and/or administered to individuals who will most benefit from it based upon
their risk of developing
osteoarthritis assessed by the methods described herein. Thus, provided are
methods for identifying a
subject predisposed to osteoarthritis and then prescribing a therapeutic or
preventative regimen to
individuals identified as having a predisposition. Thus, certain embodiments
are directed to a method
for reducing osteoarthritis in a subject, which comprises: detecting the
presence or absence of a
polymorphic variant associated with osteoarthritis in a nucleotide sequence in
a nucleic acid sample
from a subject, where the nucleotide sequence comprises a polynucleotide
sequence selected from the
group consisting of (a) a nucleotide sequence of SEQ ID NO: 1-7 or referenced
in Table A; (b) a
nucleotide sequence which encodes a polypeptide consisting of an amino acid
sequence encoded by a
nucleotide sequence of SEQ ID NO: 1-7 or referenced in Table A; (c) a
nucleotide sequence which
encodes a polypeptide that is 90% or more identical to an amino acid sequence
encoded by a nucleotide
sequence of SEQ ID NO: 1-7 or referenced in Table A, or a nucleotide sequence
about 90% or more
identical to a nucleotide sequence of SEQ 117 NO: 1-7 or referenced in Table
A; and (d) a fragment of a
polynucleotide sequence of (a), (b), or (c); and prescribing or administering
a treatment regimen to a
subject from whom the sample originated where the presence of a polymorphic
variation associated
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
with osteoarthritis is detected in the nucleotide sequence. In these methods,
predisposition results may
be utilized in combination with other test results to diagnose osteoarthritis.
[0102] Certain preventative treatments often are prescribed to subjects having
a predisposition to
osteoarthritis and where the subject is diagnosed with osteoarthritis or is
diagnosed as having symptoms
indicative of an early stage of osteoarthritis. The treatment sometimes is
preventative (e.g., is
prescribed or administered to reduce the probability that osteoarthritis
arises or progresses), sometimes
is therapeutic, and sometimes delays, alleviates or halts the progression of
osteoarthritis. Any known
preventative or therapeutic treatment for alleviating or preventing the
occurrence of osteoarthritis is
prescribed and/or administered. For example, the treatment often is directed
to decreasing pain and
improving joint movement. Examples of OA treatments include exercises to keep
joints flexible and
improve muscle strength. Different medications to control pain, including
corticosteroids and
nonsteroidal anti-inflammatory drugs (NSAIDs, e.g., Voltaren); cyclooxygenase-
2 (COX-2) inhibitors
(e.g., Celebrex, Vioxx, Mobic, and Bextra); monoclonal antibodies (e.g.,
Remicade); tumor necrosis
factor inhibitors (e.g., Enbrel); or injections of glucocorticoids, hyaluronic
acid or chondrotin sulfate
into joints that are inflamed and not responsive to NSAIDS. Orally
administered chondroitin sulfate
also may be used as a therapeutic, as it may increase hyaluronic acid levels
and viscosity of synovial
fluid, and decrease collagenase levels in synovial fluid. Also, glucosamine
can serve as an OA
therapeutic as delivering it into joints may inhibit enzymes involved in
cartilage degradation and
enhance the production of hyaluronic acid. For mild pain without inflammation,
acetaminophen may be
used. Other treatments include: heat/cold therapy for temporary pain relief;
joint protection to prevent
strain or stress on painful joints; surgery to relieve chronic pain in damaged
joints; and weight control to
prevent extra stress on weight-bearing joints.
[0103] As therapeutic approaches for treating osteoarthritis continue to
evolve and improve, the
goal of treatments for osteoarthritis related disorders is to intervene even
before clinical signs first
manifest. Thus, genetic markers associated with susceptibility to
osteoarthritis prove useful for early
diagnosis, prevention and treatment of osteoarthritis.
[0104] As osteoarthritis preventative and treatment information can be
specifically targeted to
subjects in need thereof (e.g., those at risk of developing osteoarthritis or
those in an early stage of
osteoarthritis), provided herein is a method for preventing or reducing the
risk of developing
osteoarthritis in a subject, which comprises: (a) detecting the presence or
absence of a polymorphic
variation associated with osteoarthritis at a polymorphic site in a nucleotide
sequence in a nucleic acid
sample from a subject; (b) identifying a subject with a predisposition to
osteoarthritis, whereby the
presence of the polymorphic variation is indicative of a predisposition to
osteoarthritis in the subject;
and (c) if such a predisposition is identified, providing the subject with
information about methods or
products to prevent or reduce osteoarthritis or to delay the onset of
osteoarthritis. Also provided is a
method of targeting information or advertising to a subpopulation of a human
population based on the
subpopulation being genetically predisposed to a disease or condition, which
comprises: (a) detecting
the presence or absence of a polymorphic variation associated with
osteoarthritis at a polymorphic site
31
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
in a nucleotide sequence in a nucleic acid sample from a subject; (b)
identifying the subpopulation of
subjects in which the polymorphic variation is associated with osteoarthritis;
and (c) providing
information only to the subpopulation of subjects about a particular product
which may be obtained and
consumed or applied by the subject to help prevent or delay onset of the
disease or condition.
[0105] Pharmacogenomics methods also may be used to analyze and predict a
response to
osteoarthritis treatment or a drug. For example, if pharmacogenomics analysis
indicates a likelihood
that an individual will respond positively to osteoarthritis treatment with a
particular drug, the drug may
be administered to the individual. Conversely, if the analysis indicates that
an individual is likely to
respond negatively to treatment with a particular drug, an alternative course
of treatment may be
prescribed. A negative response may be defined as either the absence of an
efficacious response or the
presence of toxic side effects. The response to a therapeutic treatment can be
predicted in a background
study in which subjects in any of the following populations are genotyped: a
population that responds
favorably to a treatment regimen, a population that does not respond
significantly to a treatment
regimen, and a population that responds adversely to a treatment regimen
(e.g., exhibits one or more
side effects). These populations are provided as examples and other
populations and subpopulations
may be analyzed. Based upon the results of these analyses, a subject is
genotyped to predict whether he
or she will respond favorably to a treatment regimen, not respond
significantly to a treatment regimen,
or respond adversely to a treatment regimen.
[0106] The tests described herein also are applicable to clinical drug trials.
One or more
polymorphic variants indicative of response to an agent for treating
osteoarthritis or to side effects to an
agent for treating osteoarthritis may be identified using the methods
described herein. Thereafter,
potential participants in clinical trials of such an agent may be screened to
identify those individuals
most likely to respond favorably to the drug and exclude those likely to
experience side effects. In that
way, the effectiveness of drug treatment may be measured in individuals who
respond positively to the
drug, without lowering the measurement as a result of the inclusion of
individuals who are unlikely to
respond positively in the study and without risking undesirable safety
problems.
[0107] Thus, another embodiment is a method of selecting an individual for
inclusion in a clinical
trial of a treatment or drug comprising the steps of: (a) obtaining a nucleic
acid sample from an
individual; (b) determining the identity of a polymorphic variation which is
associated with a positive
response to the treatment or the drug, or at least one polymorphic variation
which is associated with a
negative response to the treatment or the drug in the nucleic acid sample, and
(c) including the
individual in the clinical trial if the nucleic acid sample contains said
polymorphic variation associated
with a positive response to the treatment or the drug or if the nucleic acid
sample lacks said
polymorphic variation associated with a negative response to the treatment or
the drug. In addition, the
methods described herein for selecting an individual for inclusion in a
clinical trial of a treatment or
drug encompass methods with any further limitation described in this
disclosure, or those following,
specified alone or in any combination. The polymorphic variation may be in a
sequence selected
individually or in any combination from the group consisting of (i) a
nucleotide sequence of SEQ )D
32
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
NO: 1[-7ror referenced in Table A; (ii) a nucleotide sequence which encodes a
polypeptide consisting of
an amino acid sequence encoded by a nucleotide sequence of SEQ B7 NO: 1-7 or
referenced in Table A;
(iii) a nucleotide sequence which encodes a polypeptide that is 90% or more
identical to an amino acid
sequence encoded by a nucleotide sequence of SEQ >D NO: 1-7 or referenced in
Table A, or a
nucleotide sequence about 90% or more identical to a nucleotide sequence of
SEQ ID NO: 1-7 or
referenced in Table A; and (iv) a fragment of a polynucleotide sequence of
(i), (ii), or (iii) comprising
the polymorphic site. The including step (c) optionally comprises
administering the drug or the
treatment to the individual if the nucleic acid sample contains the
polymorphic variation associated with
a positive response to the treatment or the drug and the nucleic acid sample
lacks said biallelic marker
associated with a negative response to the treatment or the drug.
[0108] Also provided herein is a method of partnering between a
diagnostic/prognostic testing
provider and a provider of a consumable product, which comprises: (a) the
diagnostic/prognostic
testing provider detects the presence or absence of a polymorphic variation
associated with osteoarthritis
at a polymorphic site in a nucleotide sequence in a nucleic acid sample from a
subject; (b) the
diagnostic/prognostic testing provider identifies the subpopulation of
subjects in which the polymorphic
variation is associated with osteoarthritis; (c) the diagnostic/prognostic
testing provider forwards
information to the subpopulation of subjects about a particular product which
may be obtained and
consumed or applied by the subject to help prevent or delay onset of the
disease or condition; and (d)
the provider of a consumable product forwards to the diagnostic test provider
a fee every time the
diagnostic/prognostic test provider forwards information to the subject as set
forth in step (c) above.
Compositions Comprising Osteoarthritis-Directed Molecules
[0109] Featured herein is a composition comprising a cell from a subject
having osteoarthritis or at
risk of osteoarthritis and one or more molecules specifically directed and
targeted to a nucleic acid
comprising a KIAA0296, Ch~om 4, Chrom 6, ELP3, LRCHl, SNWI or ERG nucleotide
sequence, other
nucleotide sequence referenced in Table A, or an encoded amino acid sequence.
Such directed
molecules include, but are not limited to, a compound that binds to a
KIAA0296, Chrom 4, Chrom 6,
ELP3, LRCHl, SNWI or ERG nucleotide sequence, or other nucleotide sequence
referenced in Table A,
or encoded amino acid sequence; a RNAi or siRNA molecule having a strand
complementary or
substantially complementary to a KIAA0296, Chrom 4, Chrom 6, ELP3, LRCHl, SNWI
or ERG
nucleotide sequence or other nucleotide sequence referenced in Table A (e.g.,
hybridizes to a
KIAA0296, Chrona 4, Chr°om 6, ELP3, LRCHI, SNWl or ERG nucleotide
sequence or other nucleotide
sequence referenced in Table A under conditions of high stringency); an
antisense nucleic acid
complementary or substantially complementary to an RNA encoded by a KIAA0296,
Cht~om 4, Chr~ona
6, ELP3, LRCHl, SNWl or ERG nucleotide sequence or other nucleotide sequence
referenced in Table
A (e.g., hybridizes to a KIAA0296, Ch~om 4, Chrom 6, ELP3, LRCHI, SNWI or ERG
nucleotide
sequence or other nucleotide sequence referenced in Table A under conditions
of high stringency); a
ribozyme that hybridizes to a KIAA0296, ClZrom 4, Clar~om 6, ELP3, LRCHl, SNWI
or ERG nucleotide
33
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
sequence or other nucleotide sequence referenced in Table A (e.g., hybridizes
to a KIAA0296, Ch~om 4,
Chrom 6, ELP3, LRCHl, SNWI or ERG nucleotide sequence or other nucleotide
sequence referenced
in Table A under conditions of high stringency); a nucleic acid aptamer that
specifically binds a
polypeptide encoded by a KIAA0296, Ch~om 4, Chrom 6, ELP3, LRCHl, SNWl or ERG
nucleotide
sequence or other nucleotide sequence referenced in Table A; and an antibody
that specifically binds to
a polypeptide encoded by a KIAA0296, Ch~om 4, ChronZ 6, ELP3, LRCHl, SNWI or
ERG nucleotide
sequence or other nucleotide sequence referenced in Table A or binds to a
nucleic acid having such a
nucleotide sequence. In an embodiment, the antibody selectively binds to an
epitope comprising an
amino acid encoded by rs734784, rs1042164, rs749670, rs955592, rs241448 and
rs1040461. In specific
embodiments, the osteoarthritis directed molecule interacts with a nucleic
acid or polypeptide variant
associated with osteoarthritis, such as variants referenced herein. In other
embodiments, the
osteoarthritis directed molecule interacts with a polypeptide involved in a
signal pathway of a
polypeptide encoded by a KIAA0296, Ch~om 4, Chrof~a 6, ELP3, LRCHI, SNWI or
ERG nucleotide
sequence or other nucleotide sequence referenced in Table A, or a nucleic acid
comprising such a
nucleotide sequence.
[0110] Compositions sometimes include an adjuvant known to stimulate an immune
response, and
in certain embodiments, an adjuvant that stimulates a T-cell lymphocyte
response. Adjuvants are
known, including but not limited to an aluminum adjuvant (e.g., aluminum
hydroxide); a cytokine
adjuvant or adjuvant that stimulates a cytokine response (e.g., interleukin
(IL)-12 and/or gamma-
interferon cytokines); a Freund-type mineral oil adjuvant emulsion (e.g.,
Freund's complete or
incomplete adjuvant); a synthetic lipoid compound; a copolymer adjuvant (e.g.,
TitreMax); a saponin;
Quil A; a liposome; an oil-in-water emulsion (e.g., an emulsion stabilized by
Tween 80 and pluronic
polyoxyethlene/polyoxypropylene block copolymer (Syntex Adjuvant Formulation);
TitreMax;
detoxified endotoxin (MPL) and mycobacterial cell wall components (T1~W, CWS)
in 2% squalene
(Ribi Adjuvant System)); a muramyl dipeptide; an immune-stimulating complex
(ISCOM, e.g., an Ag-
modified saponin/cholesterol micelle that forms stable cage-like structure);
an aqueous phase adjuvant
that does not have a depot effect (e.g., Gerbu adjuvant); a carbohydrate
polymer (e.g., AdjuPrime); L-
tyrosine; a manide-oleate compound (e.g., Montanide); an ethylene-vinyl
acetate copolymer (e.g., Elvax
40W1,2); or lipid A, for example. Such compositions are useful for generating
an immune response
against osteoarthritis directed molecule (e.g., an HLA-binding subsequence
within a polypeptide
encoded by a KIAA0296, Ch~orn 4, Clarom 6, ELP3, LRCHI, SNWI or ERG nucleotide
sequence). In
such methods, a peptide having an amino acid subsequence of a polypeptide
encoded by a KIAA0296,
Chrom 4, Chf~om 6, ELP3, LRCHI, SNWI or ERG nucleotide sequence is delivered
to a subject, where
the subsequence binds to an HLA molecule and induces a CTL lymphocyte
response. The peptide
sometimes is delivered to the subject as an isolated peptide or as a minigene
in a plasmid that encodes
the peptide. Methods for identifying HLA-binding subsequences in such
polypeptides are known (see
e.g., publication W002/20616 and PCT application US98/01373 for methods of
identifying such
sequences).
34
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
[0111] The cell may be in a group of cells cultured in vitro or in a tissue
maintained in vitt°o or
present in an animal in vivo (e.g., a rat, mouse, ape or human). In certain
embodiments, a composition
comprises a component from a cell such as a nucleic acid molecule (e.g.,
genomic DNA), a protein
mixture or isolated protein, for example. The aforementioned compositions have
utility in diagnostic,
prognostic and pharmacogenomic methods described previously and in
therapeutics described hereafter.
Certain osteoarthritis directed molecules are described in greater detail
below.
Compounds
[0112] Compounds can be obtained using any of the numerous approaches in
combinatorial library
methods known in the art, including: biological libraries; peptoid libraries
(libraries of molecules having
the functionalities of peptides, but with a novel, non-peptide backbone which
are resistant to enzymatic
degradation but which nevertheless remain bioactive (see, e.g., Zuckermann et
al., J. Med. Chem.37:
2678-85 (1994)); spatially addressable parallel solid phase or solution phase
libraries; synthetic library
methods requiring deconvolution; "one-bead one-compound" library methods; and
synthetic library
methods using affinity chromatography selection. Biological library and
peptoid library approaches are
typically limited to peptide libraries, while the other approaches are
applicable to peptide, non-peptide
oligomer or small molecule libraries of compounds (Lam, Anticancer Drug Des.
12: 145, (1997)).
Examples of methods for synthesizing molecular libraries are described, for
example, in DeWitt et al.,
Proc. Natl. Acad. Sci. U.S.A. 90: 6909 (1993); Erb et al., Proc. Natl. Acad.
Sci. USA 91: 11422 (1994);
Zuckermann et al., J. Med. Chem. 37: 2678 (1994); Cho et al., Science 261:
1303 (1993); Carrell et al.,
Angew. Chem. Int. Ed. Engl. 33: 2059 (1994); Carell et al., Angew. Chem. Int.
Ed. Engl. 33: 2061
(1994); and in Gallop et al., J. Med. Chem. 37: 1233 (1994).
[0113] Libraries of compounds may be presented in solution (e.g., Houghten,
Biotechniques 13:
412-421 (1992)), or on beads (Lam, Nature 354: 82-84 (1991)), chips (Fodor,
Nature 364: 555-556
(1993)), bacteria or spores (Ladner, United States Patent No. 5,223,409),
plasmids (Cull et al., Proc.
Natl. Acad. Sci. USA 89: 1865-1869 (1992)) or on phage (Scott and Smith,
Science 249: 386-390
(1990); Devlin, Science 249: 404-406 (1990); Cwirla et al., Proc. Natl. Acad.
Sci. 87: 6378-6382
(1990); Felici, J. Mol. Biol. 222: 301-310 (1991); Ladner supra.).
[0114] A compound sometimes alters expression and sometimes alters activity of
a polypeptide
target and may be a small molecule. Small molecules include, but are not
limited to, peptides,
peptidomimetics (e.g., peptoids), amino acids, amino acid analogs,
polynucleotides, polynucleotide
analogs, nucleotides, nucleotide analogs, organic or inorganic compounds
(i.e., including heteroorganic
and organometallic compounds) having a molecular weight less than about 10,000
grams per mole,
organic or inorganic compounds having a molecular weight less than about 5,000
grams per mole,
organic or inorganic compounds having a molecular weight less than about 1,000
grams per mole,
organic or inorganic compounds having a molecular weight less than about 500
grams per mole, and
salts, esters, and other pharmaceutically acceptable forms of such compounds.
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
Antisense Nucleic Acid Molecules, Riboz~nes. RNAi, siRNA and Modified Nucleic
Acid
Molecules
[0115] An "antisense" nucleic acid refers to a nucleotide sequence
complementary to a "sense"
nucleic acid encoding a polypeptide, e.g., complementary to the coding strand
of a double-stranded
cDNA molecule or complementary to an mRNA sequence. The antisense nucleic acid
can be
complementary to an entire coding strand, or to a portion thereof or a
substantially identical sequence
thereof. In another embodiment, the antisense nucleic acid molecule is
antisense to a "noncoding
region" of the coding strand of a nucleotide sequence (e.g., 5' and 3'
untranslated regions in SEQ ID
NO: 1-7 or a nucleotide sequence referenced in Table A).
[0116] An antisense nucleic acid can be designed such that it is complementary
to the entire coding
region of an mRNA encoded by a nucleotide sequence (e.g., SEQ ID NO: 1-7, SEQ
ID NO: 8-17 or a
nucleotide sequence referenced in Table A), and often the antisense nucleic
acid is an oligonucleotide
antisense to only a portion of a coding or noncoding region of the mRNA. For
example, the antisense
oligonucleotide can be complementary to the region surrounding the translation
start site of the mRNA,
e.g., between the -10 and +10 regions of the target gene nucleotide sequence
of interest. An antisense
oligonucleotide can be, for example, about 7, 10, 15, 20, 25, 30, 35, 40, 45,
50, 55, 60, 65, 70, 75, 80, or
more nucleotides in length. The antisense nucleic acids, which include the
ribozymes described
hereafter, can be designed to target a KIAA0296, Ch~om 4, Chr~om 6, ELP3,
LRCHl, SNWI or ERG
nucleotide sequence, often a variant associated with osteoarthritis, or a
substantially identical sequence
thereof. Among the variants, minor alleles and major alleles can be targeted,
and those associated with
a higher risk of osteoarthritis are often designed, tested, and administered
to subjects.
[0117] An antisense nucleic acid can be constructed using chemical synthesis
and enzymatic
ligation reactions using standard procedures. For example, an antisense
nucleic acid (e.g., an antisense
oligonucleotide) can be chemically synthesized using naturally occurring
nucleotides or variously
modified nucleotides designed to increase the biological stability of the
molecules or to increase the
physical stability of the duplex formed between the antisense and sense
nucleic acids, e.g.,
phosphorothioate derivatives and acridine substituted nucleotides can be used.
Antisense nucleic acid
also can be produced biologically using an expression vector into which a
nucleic acid has been
subcloned in an antisense orientation (i.e., RNA transcribed from the inserted
nucleic acid will be of an
antisense orientation to a target nucleic acid of interest, described further
in the following subsection).
[0118] When utilized as therapeutics, antisense nucleic acids typically are
administered to a subject
(e.g., by direct injection at a tissue site) or generated in situ such that
they hybridize with or bind to
cellular mRNA and/or genomic DNA encoding a polypeptide and thereby inhibit
expression of the
polypeptide, for example, by inhibiting transcription and/or translation.
Alternatively, antisense nucleic
acid molecules can be modified to target selected cells and then are
administered systemically. For
systemic administration, antisense molecules can be modified such that they
specifically bind to
receptors or antigens expressed on a selected cell surface, for example, by
linking antisense nucleic acid
molecules to peptides or antibodies which bind to cell surface receptors or
antigens. Antisense nucleic
36
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
acid molecules can also be delivered to cells using the vectors described
herein. Sufficient intracellular
concentrations of antisense molecules are achieved by incorporating a strong
promoter, such as a pol II
or pol III promoter, in the vector construct.
[0119] Antisense nucleic acid molecules sometimes are alpha-anomeric nucleic
acid molecules.
An alpha-anomeric nucleic acid molecule forms specific double-stranded hybrids
with complementary
RNA in which, contrary to the usual beta-units, the strands run parallel to
each other (Gaultier et al.,
Nucleic Acids. Res. 15: 6625-6641 (1987)). Antisense nucleic acid molecules
can also comprise a 2'-0-
methylribonucleotide (moue et al., Nucleic Acids Res. 15: 6131-6148 (1987)) or
a chimeric RNA-DNA
analogue (moue et al., FEBS Lett. 215: 327-330 (1987)). Antisense nucleic
acids sometimes are
composed of DNA or PNA or any other nucleic acid derivatives described
previously.
[0120] In another embodiment, an antisense nucleic acid is a ribozyme. A
ribozyme having
specificity for a KIAA0296, Ch~om 4, Ch~om 6, ELP3, LRCHI, SNWI or ERG
nucleotide sequence or
other nucleotide sequence referenced in Table A can include one or more
sequences complementary to
such a nucleotide sequence, and a sequence having a known catalytic region
responsible for mRNA
cleavage (see e.g., U.S. Pat. No. 5,093,246 or Haselhoff and Gerlach, Nature
334: 585-591 (1988)). For
example, a derivative of a Tetrahymena L-19 IVS RNA is sometimes utilized in
which the nucleotide
sequence of the active site is complementary to the nucleotide sequence to be
cleaved in a mRNA (see
e.g., Cech et al. U.S. Patent No. 4,987,071; and Cech et al. U.S. Patent No.
5,116,742). Also, target
mRNA sequences can be used to select a catalytic RNA having a specific
ribonuclease activity from a
pool of RNA molecules (see e.g., Bartel & Szostak, Science 261: 1411-1418
(1993)).
[0121] Osteoarthritis directed molecules include in certain embodiments
nucleic acids that can
form triple helix structures with a KIAA0296, Ch~om 4, Chrom 6, ELP3, LRCHI,
SNWI or ERG
nucleotide sequence or other nucleotide sequence referenced in Table A, or a
substantially identical
sequence thereof, especially one that includes a regulatory region that
controls expression of a
polypeptide. Gene expression can be inhibited by targeting nucleotide
sequences complementary to the
regulatory region of a nucleotide sequence referenced herein or a
substantially identical sequence (e.g.,
promoter and/or enhancers) to form triple helical structures that prevent
transcription of a gene in target
cells (see e.g., Helene, Anticancer Drug Des. 6(6): 569-84 (1991); Helene et
al., Ann. N.Y. Acad. Sci.
660: 27-36 (1992); and Maher, Bioassays 14(12): 807-15 (1992). Potential
sequences that can be
targeted for triple helix formation can be increased by creating a so-called
"switchback" nucleic acid
molecule. Switchback molecules are synthesized in an alternating 5'-3', 3'-S'
manner, such that they
base pair with first one strand of a duplex and then the other, eliminating
the necessity for a sizeable
stretch of either purines or pyrimidines to be present on one strand of a
duplex.
[0122] Osteoarthritis directed molecules include RNAi and siRNA nucleic acids.
Gene expression
may be inhibited by the introduction of double-stranded RNA (dsRNA), which
induces potent and
specific gene silencing, a phenomenon called RNA interference or RNAi. See,
e.g., Fire et al., US
Patent Number 6,506,559; Tuschl et al. PCT International Publication No. WO
01/75164; Kay et al.
PCT International Publication No. WO 03/010180A1; or Bosher JM, Labouesse, Nat
Cell Biol 2000
37
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
Feb;2(2):E31-6. This process has been improved by decreasing the size of the
double-stranded RNA to
20-24 base pairs (to create small-interfering RNAs or siRNAs) that "switched
off' genes in mammalian
cells without initiating an acute phase response, i.e., a host defense
mechanism that often results in cell
death (see, e.g., Caplen et al. Proc Natl Acad Sci U S A. 2001 Aug
14;98(17):9742-7 and Elbashir et al.
Methods 2002 Feb;26(2):199-213). There is increasing evidence of post-
transcriptional gene silencing
by RNA interference (RNAi) for inhibiting targeted expression in mammalian
cells at the mRNA level,
in human cells. There is additional evidence of effective methods for
inhibiting the proliferation and
migration of tumor cells in human patients, and for inhibiting metastatic
cancer development (see, e.g.,
U. S. Patent Application No. US2001000993183; Caplen et al. Proc Natl Acad Sci
U S A; and
Abderrahmani et al. Mol Cell Biol 2001 Nov21(21):7256-67).
[0123] An "siRNA" or "RNAi" refers to a nucleic acid that forms a double
stranded RNA and has
the ability to reduce or inhibit expression of a gene or target gene when the
siRNA is delivered to or
expressed in the same cell as the gene or target gene. "siRNA" refers to short
double-stranded RNA
formed by the complementary strands. Complementary portions of the siRNA that
hybridize to form
the double stranded molecule often have substantial or complete identity to
the target molecule
sequence. In one embodiment, an siRNA refers to a nucleic acid that has
substantial or complete
identity to a target gene and forms a double stranded siRNA.
[0124] When designing the siRNA molecules, the targeted region often is
selected from a given
DNA sequence beginning 50 to 100 nucleotides downstream of the start codon.
See, e.g., Elbashir et
al,. Methods 26:199-213 (2002). Initially, 5' or 3' UTrs and regions nearby
the start codon were
avoided assuming that UTR-binding proteins and/or translation initiation
complexes may interfere with
binding of the siRNP or RISC endonuclease complex. Sometimes regions of the
target 23 nucleotides
in length conforming to the sequence motif AA(N19)TT (N, an nucleotide), and
regions with
approximately 30% to 70% G/C-content (often about 50% G/C-content) often are
selected. If no
suitable sequences are found, the search often is extended using the
motifNA(N21). The sequence of
the sense siRNA sometimes corresponds to (N19) TT or N21 (position 3 to 23 of
the 23-nt motif),
respectively. In the latter case, the 3' end of the sense siRNA often is
converted to TT. The rationale
for this sequence conversion is to generate a symmetric duplex with respect to
the sequence composition
of the sense and antisense 3' overhangs. The antisense siRNA is synthesized as
the complement to
position 1 to 21 of the 23-nt motif. Because position 1 of the 23-nt motif is
not recognized sequence-
specifically by the antisense siRNA, the 3'-most nucleotide residue of the
antisense siRNA can be
chosen deliberately. However, the penultimate nucleotide of the antisense
siRNA (complementary to
position 2 of the 23-nt motif) often is complementary to the targeted
sequence. For simplifying
chemical synthesis, TT often is utilized. siRNAs corresponding to the target
motifNAR(N17)YNN,
where R is purine (A,G) and Y is pyrimidine (C,U), often are selected.
Respective 21 nucleotide sense
and antisense siRNAs often begin with a purine nucleotide and can also be
expressed from pol III
expression vectors without a change in targeting site. Expression of RNAs from
pol III promoters often
is efficient when the first transcribed nucleotide is a purine.
38
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
[0125] The sequence of the siRNA can correspond to the full length target
gene, or a subsequence
thereof. Often, the siRNA is about 15 to about 50 nucleotides in length (e.g.,
each complementary
sequence of the double stranded siRNA is 15-50 nucleotides in length, and the
double stranded siRNA
is about 15-50 base pairs in length, sometimes about 20-30 nucleotides in
length or about 20-25
nucleotides in length, e.g., 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, or 30
nucleotides in length. The siRNA
sometimes is about 21 nucleotides in length. Methods of using siRNA are well
known in the art, and
specific siRNA molecules may be purchased from a number of companies including
Dharmacon
Research, Inc.
[0126] Antisense, ribozyme, RNAi and siRNA nucleic acids can be altered to
form modified
nucleic acid molecules. The nucleic acids can be altered at base moieties,
sugar moieties or phosphate
backbone moieties to improve stability, hybridization, or solubility of the
molecule. For example, the
deoxyribose phosphate backbone of nucleic acid molecules can be modified to
generate peptide nucleic
acids (see Hyrup et al., Bioorganic & Medicinal Chemistry 4 (1): 5-23 (1996)).
As used herein, the
terms "peptide nucleic acid" or "PNA" refers to a nucleic acid mimic such as a
DNA mimic, in which
the deoxyribose phosphate backbone is replaced by a pseudopeptide backbone and
only the four natural
nucleobases are retained. The neutral backbone of a PNA can allow for specific
hybridization to DNA
and RNA under conditions of low ionic strength. Synthesis of PNA oligomers can
be performed using
standard solid phase peptide synthesis protocols as described, for example, in
Hyrup et al., (1996) supra
and Perry-O'Keefe et al., Proc. Natl. Acad. Sci. 93: 14670-675 (1996).
[0127] PNA nucleic acids can be used in prognostic, diagnostic, and
therapeutic applications. For
example, PNAs can be used as antisense or antigene agents for sequence-
specific modulation of gene
expression by, for example, inducing transcription or translation arrest or
inhibiting replication. PNA
nucleic acid molecules can also be used in the analysis of single base pair
mutations in a gene, (e.g., by
PNA-directed PCR clamping); as "artificial restriction enzymes" when used in
combination with other
enzymes, (e.g., S1 nucleases (Hyrup (1996) supra)); or as probes or primers
for DNA sequencing or
hybridization (Hyrup et al., (1996) supra; Perry-O'Keefe supra).
[0128] In other embodiments, oligonucleotides may include other appended
groups such as
peptides (e.g., for targeting host cell receptors in vivo), or agents
facilitating transport across cell
membranes (see e.g., Letsinger et al., Proc. Natl. Acad. Sci. USA 86: 6553-
6556 (1989); Lemaitre et al.,
Proc. Natl. Acad. Sci. USA 84: 648-652 (1987); PCT Publication No. W088/09810)
or the blood-brain
barrier (see, e.g., PCT Publication No. W089/10134). In addition,
oligonucleotides can be modified
with hybridization-triggered cleavage agents (See, e.g., Krol et al., Bio-
Techniques 6: 958-976 (1988))
or intercalating agents. (See, e.g., Zon, Pharm. Res. 5: 539-549 (1988) ).
To.this end, the
oligonucleotide may be conjugated to another molecule, (e.g., a peptide,
hybridization triggered cross-
linking agent, transport agent, or hybridization-triggered cleavage agent).
[0129] Also included herein are molecular beacon oligonucleotide primer and
probe molecules
having one or more regions complementary to a KIAA0296, Clarom 4, Chrorra 6,
ELP3, LRCHl, SNWI
or ERG nucleotide sequence or other nucleotide sequence referenced in Table A,
or a substantially
39
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
identical sequence thereof, two complementary regions one having a fluorophore
and one a quencher
such that the molecular beacon is useful for quantifying the presence of the
nucleic acid in a sample.
Molecular beacon nucleic acids are described, for example, in Lizardi et al.,
U.S. Patent No. 5,854,033;
Nazarenko et al., U.S. Patent No. 5,866,336, and Livak et al., U.S. Patent
5,876,930.
Antibodies
[0130] The term "antibody" as used herein refers to an immunoglobulin molecule
or
immunologically active portion thereof, i.e., an antigen-binding portion.
Examples of immunologically
active portions of immunoglobulin molecules include Flab) and F(ab')z
fragments which can be
generated by treating the antibody with an enzyme such as pepsin. An antibody
sometimes is a
polyclonal, monoclonal, recombinant (e.g., a chimeric or humanized), fully
human, non-human (e.g.,
murine), or a single chain antibody. An antibody may have effector function
and can fix complement,
and is sometimes coupled to a toxin or imaging agent.
[0131] A full-length polypeptide or antigenic peptide fragment encoded by a
nucleotide sequence
referenced herein can be used as an immunogen or can be used to identify
antibodies made with other
immunogens, e.g., cells, membrane preparations, and the like. An antigenic
peptide often includes at
least 8 amino acid residues of the amino acid sequences encoded by a
nucleotide sequence referenced
herein, or substantially identical sequence thereof, and encompasses an
epitope. Antigenic peptides
sometimes include 10 or more amino acids, 15 or more amino acids, 20 or more
amino acids, or 30 or
more amino acids. Hydrophilic and hydrophobic fragments of polypeptides
sometimes are used as
immunogens.
[0132] Epitopes encompassed by the antigenic peptide are regions located on
the surface of the
polypeptide (e.g., hydrophilic regions) as well as regions with high
antigenicity. For example, an Emini
surface probability analysis of the human polypeptide sequence can be used to
indicate the regions that
have a particularly high probability of being localized to the surface of the
polypeptide and are thus
likely to constitute surface residues useful for targeting antibody
production. The antibody may bind an
epitope on any domain or region on polypeptides described herein.
[0133] Also, chimeric, humanized, and completely human antibodies are useful
for applications
which include repeated administration to subjects. Chimeric and humanized
monoclonal antibodies,
comprising both human and non-human portions, can be made using standard
recombinant DNA
techniques. Such chimeric and humanized monoclonal antibodies can be produced
by recombinant
DNA techniques known in the art, for example using methods described in
Robinson et al International
Application No. PCT/LJS86/02269; Akira, et al European Patent Application
184,187; Taniguchi, M.,
European Patent Application 171,496; Morrison et al European Patent
Application 173,494; Neuberger
et al PCT International Publication No. WO 86/01533; Cabilly et al U.S. Patent
No. 4,816,567; Cabilly
et al European Patent Application 125,023; Better et al., Science 240: 1041-
1043 (1988); Liu et al.,
Proc. Natl. Acad. Sci. USA 84: 3439-3443 (1987); Liu et al., J. Immunol. 139:
3521-3526 (1987); Sun
et al., Proc. Natl. Acad. Sci. USA 84: 214-218 (1987); Nishimura et al., Canc.
Res. 47: 999-1005
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
(1987); Wood et al., Nature 314: 446-449 (1985); and Shaw et al., J. Natl.
Cancer Inst. 80: 1553-1559
(1988); Morrison, S. L., Science 229: 1202-1207 (1985); Oi et al.,
BioTechniques 4: 214 (1986);
Winter U.S. Patent 5,225,539; Jones et al., Nature 321: 552-525 (1986);
Verhoeyan et al., Science 239:
1534; and Beidler et al., J. Immunol. 141: 4053-4060 (1988).
[0134] Completely human antibodies are particularly desirable for therapeutic
treatment of human
patients. Such antibodies can be produced using transgenic mice that are
incapable of expressing
endogenous immunoglobulin heavy and light chains genes, but which can express
human heavy and
light chain genes. See, for example, Lonberg and Huszar, Int. Rev. Immunol.
13: 65-93 (1995); and
U.S. Patent Nos. 5,625,126; 5,633,425; 5,569,825; 5,661,016; and 5,545,806. In
addition, companies
such as Abgenix, Inc. (Fremont, CA) and Medarex, Inc. (Princeton, NJ), can be
engaged to provide
human antibodies directed against a selected antigen using technology similar
to that described above.
Completely human antibodies that recognize a selected epitope also can be
generated using a technique
referred to as "guided selection." In this approach a selected non-human
monoclonal antibody (e.g., a
murine antibody) is used to guide the selection of a completely human antibody
recognizing the same
epitope. This technology is described for example by Jespers et al.,
Bio/Technology 12: 899-903
( 1994).
[0135] An antibody can be a single chain antibody. A single chain antibody
(scFV) can be
engineered (see, e.g., Colcher et al., Ann. N Y Acad. Sci. 880: 263-80 (1999);
and Reiter, Clin. Cancer
Res. 2: 245-52 (1996)). Single chain antibodies can be dimerized or
multimerized to generate
multivalent antibodies having specificities for different epitopes of the same
target polypeptide.
[0136] Antibodies also may be selected or modified so that they exhibit
reduced or no ability to
bind an Fc receptor. For example, an antibody may be an isotype or subtype,
fragment or other mutant,
which does not support binding to an Fc receptor (e.g., it has a mutagenized
or deleted Fc receptor
binding region).
[0137] Also, an antibody (or fragment thereof) may be conjugated to a
therapeutic moiety such as a
cytotoxin, a therapeutic agent or a radioactive metal ion. A cytotoxin or
cytotoxic agent includes any
agent that is detrimental to cells. Examples include taxol, cytochalasin B,
gramicidin D, ethidium
bromide, emetine, mitomycin, etoposide, tenoposide, vincristine, vinblastine,
colchicin, doxorubicin,
daunorubicin, dihydroxy anthracin dione, mitoxantrone, mithramycin,
actinomycin D, 1
dehydrotestosterone, glucocorticoids, procaine, tetracaine, lidocaine,
propranolol, and puromycin and
analogs or homologs thereof. Therapeutic agents include, but are not limited
to, antimetabolites (e.g.,
methotrexate, 6-mercaptopurine, 6-thioguanine, cytarabine, 5-fluorouracil
decarbazine), alkylating
agents (e.g., mechlorethamine, thiotepa chlorambucil, melphalan, carmustine
(BCNU) and lomustine
(CCNU), cyclophosphamide, busulfan, dibromomannitol, streptozotocin, mitomycin
C, and cis-
dichlorodiamine platinum (II) (DDP) cisplatin), anthracyclines (e.g.,
daunorubicin (formerly
daunomycin) and doxorubicin), antibiotics (e.g., dactinomycin (formerly
actinomycin), bleomycin,
mithramycin, and anthramycin (AMC)), and anti-mitotic agents (e.g.,
vincristine and vinblastine).
41
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
[0138] Antibody conjugates can be used for modifying a given biological
response. For example,
the drug moiety may be a protein or polypeptide possessing a desired
biological activity. Such proteins
may include, for example, a toxin such as abrin, ricin A, pseudomonas
exotoxin, or diphtheria toxin; a
polypeptide such as tumor necrosis factor, gamma-interferon, alpha-interferon,
nerve growth factor,
platelet derived growth factor, tissue plasminogen activator; or, biological
response modifiers such as,
for example, lymphokines, interleukin-1 ("IL-1"), interleukin-2 ("IL-2"),
interleukin-6 ("IL-6"),
granulocyte macrophage colony stimulating factor ("GM-CSF"), granulocyte
colony stimulating factor
("G-CSF"), or other growth factors. Also, an antibody can be conjugated to a
second antibody to form
an antibody heteroconjugate as described by Segal in U.S. Patent No.
4,676,980, for example.
[0139] An antibody (e.g., monoclonal antibody) can be used to isolate target
polypeptides by
standard techniques, such as affinity chromatography or immunoprecipitation.
Moreover, an antibody
can be used to detect a target polypeptide (e.g., in a cellular lysate or cell
supernatant) in order to
evaluate the abundance and pattern of expression of the polypeptide.
Antibodies can be used
diagnostically to monitor polypeptide levels in tissue as part of a clinical
testing procedure, e.g., to
determine the efficacy of a given treatment regimen. Detection can be
facilitated by coupling (i.e.,
physically linking) the antibody to a detectable substance (i.e., antibody
labeling). Examples of
detectable substances include various enzymes, prosthetic groups, fluorescent
materials, luminescent
materials, bioluminescent materials, and radioactive materials. Examples of
suitable enzymes include
horseradish peroxidase, allealine phosphatase, (3-galactosidase, or
acetylcholinesterase; examples of
suitable prosthetic group complexes include streptavidin/biotin and
avidin/biotin; examples of suitable
fluorescent materials include umbelliferone, fluorescein, fluorescein
isothiocyanate, rhodamine,
dichlorotriazinylamine fluorescein, dansyl chloride or phycoerythrin; an
example of a luminescent
material includes luminol; examples of bioluminescent materials include
luciferase, luciferin, and
aequorin, and examples of suitable radioactive material include lzsh i3ih 3sS
or 3H. Also, an antibody
can be utilized as a test molecule for determining whether it can treat
osteoarthritis, and as a therapeutic
for administration to a subject for treating osteoarthritis.
[0140] An antibody can be made by immunizing with a purified antigen, or a
fragment thereof,
e.g., a fragment described herein, a membrane associated antigen, tissues,
e.g., crude tissue preparations,
whole cells, preferably living cells, lysed cells, or cell fractions.
[0141] Included herein are antibodies which bind only a native polypeptide,
only denatured or
otherwise non-native polypeptide, or which bind both, as well as those having
linear or conformational
epitopes. Conformational epitopes sometimes can be identified by selecting
antibodies that bind to
native but not denatured polypeptide. Also featured are antibodies that
specifically bind to a
polypeptide variant associated with osteoarthritis.
Methods for Identi , ink Candidate Therapeutics for Treating Osteoarthritis
[0142] Current therapies for the treatment of osteoarthritis have limited
efficacy, limited
tolerability and significant mechanism-based side effects, and few of the
available therapies adequately
42
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
address underlying defects. Current therapeutic approaches were largely
developed in the absence of
defined molecular targets or even a solid understanding of disease
pathogenesis. Therefore, provided
are methods of identifying candidate therapeutics that target biochemical
pathways related to the
development of osteoarthritis.
[0143] Thus, featured herein are methods for identifying a candidate
therapeutic for treating
osteoarthritis. The methods comprise contacting a test molecule with a target
molecule in a system. A
"target molecule" as used herein refers to a KIAA0296, Cla~om 4, Chrom 6,
ELP3, LRCHI, SNWI or
ERG nucleic acid or other nucleotide sequence referenced in Table A, a
substantially identical nucleic
acid thereof, or a fragment thereof, and an encoded polypeptide of the
foregoing. The methods also
comprise determining the presence or absence of an interaction between the
test molecule and the target
molecule, where the presence of an interaction between the test molecule and
the nucleic acid or
polypeptide identifies the test molecule as a candidate osteoarthritis
therapeutic. The interaction
between the test molecule and the target molecule may be quantified.
[0144] Test molecules and candidate therapeutics include, but are not limited
to, compounds,
antisense nucleic acids, siRNA molecules, ribozymes, polypeptides or proteins
encoded by a KIAA0296,
Chrom 4, Chrom 6, ELP3, LRCHl, SNWI or ERG nucleotide sequence or other
nucleotide sequence
referenced in Table A , or a substantially identical sequence or fragment
thereof, and
immunotherapeutics (e.g., antibodies and HLA-presented polypeptide fragments).
A test molecule or
candidate therapeutic may act as a modulator of target molecule concentration
or target molecule
function in a system. A "modulator" may agonize (i.e., up-regulates) or
antagonize (i.e., down-
regulates) a target molecule concentration partially or completely in a system
by affecting such cellular
functions as DNA replication and/or DNA processing (e.g., DNA methylation or
DNA repair), RNA
transcription and/or RNA processing (e.g., removal of intronic sequences
and/or translocation of spliced
mRNA from the nucleus), polypeptide production (e.g., translation of the
polypeptide from mRNA),
and/or polypeptide post-translational modification (e.g., glycosylation,
phosphorylation, and proteolysis
of pro-polypeptides). A modulator may also agonize or antagonize a biological
function of a target
molecule partially or completely, where the function may include adopting a
certain structural
conformation, interacting with one or more binding partners, ligand binding,
catalysis (e.g.,
phosphorylation, dephosphorylation, hydrolysis, methylation, and
isomerization), and an effect upon a
cellular event (e.g., effecting progression of osteoarthritis).
[0145] As used herein, the term "system" refers to a cell free in vitro
environment and a cell-based
environment such as a collection of cells, a tissue, an organ, or an organism.
A system is "contacted"
with a test molecule in a variety of manners, including adding molecules in
solution and allowing them
to interact with one another by diffusion, cell injection, and any
administration routes in an animal. As
used herein, the term "interaction" refers to an effect of a test molecule on
test molecule, where the
effect sometimes is binding between the test molecule and the target molecule,
and sometimes is an
observable change in cells, tissue, or organism.
43
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
[0146] There are many standard methods for detecting the presence or absence
of interaction
between a test molecule and a target molecule. For example, titrametric,
acidimetric, radiometric,
NMR, monolayer, polarographic, spectrophotometric, fluorescent, and ESR assays
probative of a target
molecule interaction may be utilized. Any modulator can be tested in such
methods and modulators for
certain targets described in Table A are known. For example, modulators of
protein tyrosine
phosphatases (e.g., PTPNI includes a protein phosphatase domain) are described
in WO-03072537,
WO-03020688, WO-00218321, WO-00218323, WO-03055883, WO-03041729, WO-00226707,
WO-
00226743 and WO-03037328; modulators of potassium channels (e.g., KCNSI
includes a potassium
channel domain) are described in WO-09962891, WO-09716437, WO-09521813, WO-
09521823, WO-
09521824, WO-09521825 and WO-03088908; modulators of annexin (e.g., AN~YA6
includes an annexin
domain) are described in WO-2004018670, WO-02067857, WO-2004013303 and WO-
00147510;
proteasome modulators (e.g., PSMBI includes a proteasome domain) are described
in WO-2004014882
and Roesel et al. Proceedings of the American Association of Cancer Research
2003, 44:1 st Ed (Abs
1769), and bortezomib (Velcade, MLN-341, LDP-341 and PS-341), a ubiquitin
proteosome inhibitor, is
used for the treatment of multiple myeloma; and modulators of protein kinases
(e.g., FYN is a protein
kinase) are described in WO-03081210, WO-02080926, WO-02076986, WO-03077921,
W003026666,
W003026665 and W003026664.
[0147] Test molecule/target molecule interactions can be detected and/or
quantified using assays
known in the art. For example, an interaction can be determined by labeling
the test molecule and/or
the target molecule, where the label is covalently or non-covalently attached
to the test molecule or
target molecule. The label is sometimes a radioactive molecule such as
lzslysih ssS or 3H, which can be
detected by direct counting of radioemission or by scintillation counting.
Also, enzymatic labels such as
horseradish peroxidase, alkaline phosphatase, or luciferase may be utilized
where the enzymatic label
can be detected by determining conversion of an appropriate substrate to
product. In addition, presence
or absence of an interaction can be determined without labeling. For example,
a microphysiometer
(e.g., Cytosensor) is an analytical instrument that measures the rate at which
a cell acidifies its
environment using a light-addressable potentiometric sensor (LAPS). Changes in
this acidification rate
can be used as an indication of an interaction between a test molecule and
target molecule (McConnell,
H. M. et al., Science 257: 1906-1912 (1992)).
[0148] In cell-based systems, cells typically include a KIAA0296, Chrona 4,
Chrom 6, ELP3,
LRCHl, SNWI or ERG nucleic acid or other nucleotide sequence referenced in
Table A, an encoded
polypeptide, or substantially identical nucleic acid or polypeptide thereof,
and are often of mammalian
origin, although the cell can be of any origin. Whole cells, cell homogenates,
and cell fractions (e.g.,
cell membrane fractions) can be subjected to analysis. Where interactions
between a test molecule with
a target polypeptide are monitored, soluble and/or membrane bound forms of the
polypeptide may be
utilized. Where membrane-bound forms of the polypeptide are used, it may be
desirable to utilize a
solubilizing agent. Examples of such solubilizing agents include non-ionic
detergents such as n-
octylglucoside, n-dodecylglucoside, n-dodecylmaltoside, octanoyl-N-
methylglucamide, decanoyl-N-
44
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
methylglucamide, Triton~ X-100, Triton~ X-114, Thesit~,
Isotridecypoly(ethylene glycol ether)n, 3-
[(3-cholamidopropyl)dimethylamminio]-1-propane sulfonate (CHAPS), 3-[(3-
cholamidopropyl)dimethylamminio]-2-hydroxy-1-propane sulfonate (CHAPSO), or N-
dodecyl-N,N-
dimethyl-3-ammonio-1-propane sulfonate.
[0149] An interaction between a test molecule and target molecule also can be
detected by
monitoring fluorescence energy transfer (FET) (see, e.g., Lakowicz et al.,
U.S. Patent No. 5,631,169;
Stavrianopoulos et al. U.S. Patent No. 4,868,103). A fluorophore label on a
first, "donor" molecule is
selected such that its emitted fluorescent energy will be absorbed by a
fluorescent label on a second,
"acceptor" molecule, which in turn is able to fluoresce due to the absorbed
energy. Alternately, the
"donor" polypeptide molecule may simply utilize the natural fluorescent energy
of tryptophan residues.
Labels are chosen that emit different wavelengths of light, such that the
"acceptor" molecule label may
be differentiated from that of the "donor". Since the efficiency of energy
transfer between the labels is
related to the distance separating the molecules, the spatial relationship
between the molecules can be
assessed. In a situation in which binding occurs between the molecules, the
fluorescent emission of the
"acceptor" molecule label in the assay should be maximal. An FET binding event
can be conveniently
measured through standard fluorometric detection means well known in the art
(e.g., using a
fluorimeter).
[0150] In another embodiment, determining the presence or absence of an
interaction between a
test molecule and a target molecule can be effected by monitoring surface
plasmon resonance (see, e.g.,
Sjolander & Urbaniczk, Afzal. Chem. 63: 2338-2345 (1991) and Szabo et al.,
Cur. Opin. Struct. Biol. S:
699-705 (1995)). "Surface plasmon resonance" or "biomolecular interaction
analysis (BIA)" can be
utilized to detect biospecific interactions in real time, without labeling any
of the interactants (e.g.,
BIAcore). Changes in the mass at the binding surface (indicative of a binding
event) result in
alterations of the refractive index of light near the surface (the optical
phenomenon of surface plasmon
resonance (SPR)), resulting in a detectable signal which can be used as an
indication of real-time
reactions between biological molecules.
[0151] In another embodiment, the target molecule or test molecules are
anchored to a solid phase,
facilitating the detection of target molecule/test molecule complexes and
separation of the complexes
from free, uncomplexed molecules. The target molecule or test molecule is
immobilized to the solid
support. In an embodiment, the target molecule is anchored to a solid surface,
and the test molecule,
which is not anchored, can be labeled, either directly or indirectly, with
detectable labels discussed
herein.
[0152] It may be desirable to immobilize a target molecule, an anti-target
molecule antibody,
and/or test molecules to facilitate separation of target molecule/test
molecule complexes from
uncomplexed forms, as well as to accommodate automation of the assay. The
attachment between a test
molecule and/or target molecule and the solid support may be covalent or non-
covalent (see, e.g., U.S.
Patent No. 6,022,688 for non-covalent attachments). The solid support may be
one or more surfaces of
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
the system, such as one or more surfaces in each well of a microtiter plate, a
surface of a silicon wafer, a
surface of a bead (see, e.g., Lam, Nature 354: 82-84 (1991)) that is
optionally linked to another solid
support, or a channel in a microfluidic device, for example. Types of solid
supports, linker molecules
for covalent and non-covalent attaclunents to solid supports, and methods for
immobilizing nucleic
acids and other molecules to solid supports are well known (see, e.g., U.S.
Patent Nos. 6,261,776;
5,900,481; 6,133,436; and 6,022,688; and WIPO publication WO 01/18234).
[0153] In an embodiment, target molecule may be immobilized to surfaces via
biotin and
streptavidin. For example, biotinylated target polypeptide can be prepared
from biotin-NHS (N-
hydroxy-succinimide) using techniques known in the art (e.g., biotinylation
kit, Pierce Chemicals,
Rockford, IL), and immobilized in the wells of streptavidin-coated 96 well
plates (Pierce Chemical). In
another embodiment, a target polypeptide can be prepared as a fusion
polypeptide. For example,
glutathione-S-transferase/target polypeptide fusion can be adsorbed onto
glutathione sepharose beads
(Sigma Chemical, St. Louis, MO) or glutathione derivitized microtiter plates,
which are then combined
with a test molecule under conditions conducive to complex formation (e.g., at
physiological conditions
for salt and pH). Following incubation, the beads or microtiter plate wells
are washed to remove any
unbound components, or the matrix is immobilized in the case of beads, and
complex formation is
determined directly or indirectly as described above. Alternatively, the
complexes can be dissociated
from the matrix, and the level of target molecule binding or activity is
determined using standard
techniques.
[0154] In an embodiment, the non-immobilized component is added to the coated
surface
containing the anchored component. After the reaction is complete, unreacted
components are removed
(e.g., by washing) under conditions such that a significant percentage of
complexes formed will remain
immobilized to the solid surface. The detection of complexes anchored on the
solid surface can be
accomplished in a number of manners. Where the previously non-immobilized
component is pre-
labeled, the detection of label immobilized on the surface indicates that
complexes were formed. Where
the previously non-immobilized component is not pre-labeled, an indirect label
can be used to detect
complexes anchored on the surface, e.g., by adding a labeled antibody specific
for the immobilized
component, where the antibody, in turn, can be directly labeled or indirectly
labeled with, e.g., a labeled
anti-Ig antibody.
[0155] In another embodiment, an assay is performed utilizing antibodies that
specifically bind
target molecule or test molecule but do not interfere with binding of the
target molecule to the test
molecule. Such antibodies can be derivitized to a solid support, and unbound
target molecule may be
immobilized by antibody conjugation. Methods for detecting such complexes, in
addition to those
described above for the GST-immobilized complexes, include immunodetection of
complexes using
antibodies reactive with the target molecule, as well as enzyme-linked assays
which rely on detecting an
enzymatic activity associated with the target molecule.
[0156] Cell free assays also can be conducted in a liquid phase. In such an
assay, reaction products
are separated from unreacted components, by any of a number of standard
techniques, including but not
46
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
limited to: differential centrifugation (see, e.g., Rivas, G., and Minton,
Trends Biochem Sci Aug;18(8):
284-7 (1993)); chromatography (gel filtration chromatography, ion-exchange
chromatography);
electrophoresis (see, e.g., Ausubel et al., eds. Current Protocols in
Molecular Biology , J. Wiley: New
York (1999)); and immunoprecipitation (see, e.g., Ausubel et al., eds.,
supra). Media and
chromatographic techniques are known to one skilled in the art (see, e.g.,
Heegaard, JMoI. Recognit.
Winter; 1l (I-6): 141-8 (1998); Hage & Tweed, J. Chromatogr. B Biomed. Sci.
Appl. Oct 10; 699 (1-2):
499-525 (1997)). Further, fluorescence energy transfer may also be
conveniently utilized, as described
herein, to detect binding without further purification of the complex from
solution.
[0157] In another embodiment, modulators of target molecule expression are
identified. For
example, a cell or cell free mixture is contacted with a candidate compound
and the expression of target
mRNA or target polypeptide is evaluated relative to the level of expression of
target mRNA or target
polypeptide in the absence of the candidate compound. When expression of
target mRNA or target
polypeptide is greater in the presence of the candidate compound than in its
absence, the candidate
compound is identified as an agonist of target mRNA or target polypeptide
expression. Alternatively,
when expression of target mRNA or target polypeptide is less (e.g., less with
statistical significance) in
the presence of the candidate compound than in its absence, the candidate
compound is identified as an
antagonist or inhibitor of target mRNA or target polypeptide expression. The
level of target mRNA or
target polypeptide expression can be determined by methods described herein.
[0158] In another embodiment, binding partners that interact with a target
molecule are detected.
The target molecules can interact with one or more cellular or extracellular
macromolecules, such as
polypeptides in vivo, and these interacting molecules are referred to herein
as "binding partners."
Binding partners can agonize or antagonize target molecule biological
activity. Also, test molecules
that agonize or antagonize interactions between target molecules and binding
partners can be ,useful as
therapeutic molecules as they can up-regulate or down-regulated target
molecule activity in vivo and
thereby treat osteoarthritis.
[0159] Binding partners of target molecules can be identified by methods known
in the art. For
example, binding partners may be identified by lysing cells and analyzing cell
lysates by electrophoretic
techniques. Alternatively, a two-hybrid assay or three-hybrid assay can be
utilized (see, e.g., U.S.
Patent No. 5,283,317; Zervos et al., Cell 72:223-232 (1993); Madura et al., J.
Biol. Chem. 268: 12046-
12054 (1993); Bartel et al., BiotechrZiques 14: 920-924 (1993); Iwabuchi et
al., Oncogerae 8: 1693-1696
(1993); and Brent W094/10300). A two-hybrid system is based on the modular
nature of most
transcription factors, which consist of separable DNA-binding and activation
domains. The assay often
utilizes two different DNA constructs. In one construct, a KIAA0296, Chrom 4,
Chrom 6, ELP3,
LRCHI, SNWI or ERG nucleic acid or other nucleic acid referenced in Table A
(sometimes referred to
as the "bait") is fused to a gene encoding the DNA binding domain of a known
transcription factor (e.g.,
GAL-4). In another construct, a DNA sequence from a library of DNA sequences
that encodes a
potential binding partner (sometimes referred to as the "prey") is fused to a
gene that encodes an
activation domain of the known transcription factor. Sometimes, a KIAA0296,
Chrom 4, Chrona 6,
47
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
ELP3, LRCHl, SNWI or ERG nucleic acid or other nucleic acid referenced in
Table A can be fused to
the activation domain. If the "bait" and the "prey" molecules interact in
vivo, the DNA-binding and
activation domains of the transcription factor are brought into close
proximity. This proximity allows
transcription of a reporter gene (e.g., LacZ) which is operably linked to a
transcriptional regulatory site
responsive to the transcription factor. Expression of the reporter gene can be
detected and cell colonies
containing the functional transcription factor can be isolated and used to
identify the potential binding
partner.
[0160] In an embodiment for identifying test molecules that antagonize or
agonize complex
formation between target molecules and binding partners, a reaction mixture
containing the target
molecule and the binding partner is prepared, under conditions and for a time
sufficient to allow
complex formation. The reaction mixture often is provided in the presence or
absence of the test
molecule. The test molecule can be included initially in the reaction mixture,
or can be added at a time
subsequent to the addition of the target molecule and its binding partner.
Control reaction mixtures are
incubated without the test molecule or with a placebo. Formation of any
complexes between the target
molecule and the binding partner then is detected. Decreased formation of a
complex in the reaction
mixture containing test molecule as compared to in a control reaction mixture
indicates that the
molecule antagonizes target molecule/binding partner complex formation.
Alternatively, increased
formation of a complex in the reaction mixture containing test molecule as
compared to in a control
reaction mixture indicates that the molecule agonizes target molecule/binding
partner complex
formation. In another embodiment, complex formation of target molecule/binding
partner can be
compared to complex formation of mutant target molecule/binding partner (e.g.,
amino acid
modifications in a target polypeptide). Such a comparison can be important in
those cases where it is
desirable to identify test molecules that modulate interactions of mutant but
not non-mutated.target gene
products.
[0161] The assays can be conducted in a heterogeneous or homogeneous format.
In heterogeneous
assays, target molecule and/or the binding partner are immobilized to a solid
phase, and complexes are
detected on the solid phase at the end of the reaction. In homogeneous assays,
the entire reaction is
carried out in a liquid phase. In either approach, the order of addition of
reactants can be varied to
obtain different information about the molecules being tested. For example,
test compounds that
agonize target molecule/binding partner interactions can be identified by
conducting the reaction in the
presence of the test molecule in a competition format. Alternatively, test
molecules that agonize
preformed complexes, e.g., molecules with higher binding constants that
displace one of the
components from the complex, can be tested by adding the test compound to the
reaction mixture after
complexes have been formed.
(0162] In a heterogeneous assay embodiment, the target molecule or the binding
partner is
anchored onto a solid surface (e.g., a microtiter plate), while the non-
anchored species is labeled, either
directly or indirectly. The anchored molecule can be immobilized by non-
covalent or covalent
attachments. Alternatively, an immobilized antibody specific for the molecule
to be anchored can be
4~
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
used to anchor the molecule to the solid surface. The partner of the
immobilized species is exposed to
the coated surface with or without the test molecule. After the reaction is
complete, unreacted
components are removed (e.g., by washing) such that a significant portion of
any complexes formed
will remain immobilized on the solid surface. Where the non-immobilized
species is pre-labeled, the
detection of label immobilized on the surface is indicative of complex. Where
the non-immobilized
species is not pre-labeled, an indirect label can be used to detect complexes
anchored to the surface;
e.g., by using a labeled antibody specific for the initially non-immobilized
species. Depending upon the
order of addition of reaction components, test compounds that inhibit complex
formation or that disrupt
preformed complexes can be detected.
[0163] In another embodiment, the reaction can be conducted in a liquid phase
in the presence or
absence of test molecule, where the reaction products are separated from
unreacted components, and the
complexes are detected (e.g., using an immobilized antibody specific for one
of the binding components
to anchor any complexes formed in solution, and a labeled antibody specific
for the other partner to
detect anchored complexes). Again, depending upon the order of addition of
reactants to the liquid
phase, test compounds that inhibit complex or that disrupt preformed complexes
can be identified.
[0164] In an alternate embodiment, a homogeneous assay can be utilized. For
example, a
preformed complex of the target gene product and the interactive cellular or
extracellular binding
partner product is prepared. One or both of the target molecule or binding
partner is labeled, and the
signal generated by the labels) is quenched upon complex formation (e.g., U.S.
Patent No. 4,109,496
that utilizes this approach for immunoassays). Addition of a test molecule
that competes with and
displaces one of the species from the preformed complex will result in the
generation of a signal above
background. In this way, test substances that disrupt target molecule/binding
partner complexes can be
identified.
[0165] Candidate therapeutics for treating osteoarthritis are identified from
a group of test
molecules that interact with a target molecule. Test molecules are normally
ranked according to the
degree with which they modulate (e.g., agonize or antagonize) a function
associated with the target
molecule (e.g., DNA replication and/or processing, RNA transcription and/or
processing, polypeptide
production and/or processing, and/or biological function/activity), and then
top ranleing modulators are
selected. Also, pharmacogenomic information described herein can determine the
rank of a modulator.
The top 10% of ranked test molecules often are selected for further testing as
candidate therapeutics,
and sometimes the top 1 S%, 20%, or 25% of ranked test molecules are selected
for further testing as
candidate therapeutics. Candidate therapeutics typically are formulated for
administration to a subject.
Therapeutic Formulations
[0166] Formulations and pharmaceutical compositions typically include in
combination with a
pharmaceutically acceptable carrier one or more target molecule modulators.
The modulator often is a
test molecule identified as having an interaction with a target molecule by a
screening method described
above. The modulator may be a compound, an antisense nucleic acid, a ribozyme,
an antibody, or a
49
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
binding partner. Also, formulations may comprise a target polypeptide or
fragment thereof in
combination with a pharmaceutically acceptable carrier.
[0167] As used herein, the term "pharmaceutically acceptable carrier" includes
solvents,
dispersion media, coatings, antibacterial and antifungal agents, isotonic and
absorption delaying agents,
and the like, compatible with pharmaceutical administration. Supplementary
active compounds can also
be incorporated into the compositions. Pharmaceutical compositions can be
included in a container,
pack, or dispenser together with instructions for administration.
[0168] A pharmaceutical composition typically is formulated to be compatible
with its intended
route of administration. Examples of routes of administration include
parenteral, e.g., intravenous,
intradermal, subcutaneous, oral (e.g., inhalation), transdermal (topical),
transmucosal, and rectal
administration. Solutions or suspensions used for parenteral, intradermal, or
subcutaneous application
can include the following components: a sterile diluent such as water for
injection, saline solution, fixed
oils, polyethylene glycols, glycerin, propylene glycol or other synthetic
solvents; antibacterial agents
such as benzyl alcohol or methyl parabens; antioxidants such as ascorbic acid
or sodium bisulfate;
chelating agents such as ethylenediaminetetraacetic acid; buffers such as
acetates, citrates or phosphates
and agents for the adjustment of tonicity such as sodium chloride or dextrose.
pH can be adjusted with
acids or bases, such as hydrochloric acid or sodium hydroxide. The parenteral
preparation can be
enclosed in ampoules, disposable syringes or multiple dose vials made of glass
or plastic.
[0169] Oral compositions generally include an inert diluent or an edible
carrier. For the purpose of
oral therapeutic administration, the active compound can be incorporated with
excipients and used in the
form of tablets, troches, or capsules, e.g., gelatin capsules. Oral
compositions can also be prepared
using a fluid carrier for use as a mouthwash. Pharmaceutically compatible
binding agents, and/or
adjuvant materials can be included as part of the composition. The tablets,
pills, capsules, troches and
the like can contain any of the following ingredients, or compounds of a
similar nature: a binder such as
microcrystalline cellulose, gum tragacanth or gelatin; an excipient such as
starch or lactose, a
disintegrating agent such as alginic acid, Primogel, or corn starch; a
lubricant such as magnesium
stearate or Sterotes; a glidant such as colloidal silicon dioxide; a
sweetening agent such as sucrose or
saccharin; or a flavoring agent such as peppermint, methyl salicylate, or
orange flavoring.
/[0170] Pharmaceutical compositions suitable for injectable use include
sterile aqueous solutions
(where water soluble) or dispersions and sterile powders for the
extemporaneous preparation of sterile
injectable solutions or dispersion. For intravenous administration, suitable
carriers include
physiological saline, bacteriostatic water, Cremophor ELTM (BASF, Parsippany,
NJ) or phosphate
buffered saline (PBS). In all cases, the composition must be sterile and
should be fluid to the extent that
easy syringability exists. It should be stable under the conditions of
manufacture and storage and must
be preserved against the contaminating action of microorganisms such as
bacteria and fungi. The
carrier can be a solvent or dispersion medium containing, for example, water,
ethanol, polyol (for
example, glycerol, propylene glycol, and liquid polyethylene glycol, and the
like), and suitable mixtures
thereof. The proper fluidity can be maintained, for example, by the use of a
coating such as lecithin, by
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
the maintenance of the required particle size in the case of dispersion and by
the use of surfactants.
Prevention of the action of microorganisms can be achieved by various
antibacterial and antifungal
agents, for example, parabens, chlorobutanol, phenol, ascorbic acid,
thimerosal, and the like. In many
cases, it will be preferable to include isotonic agents, for example, sugars,
polyalcohols such as
mannitol, sorbitol, sodium chloride in the composition. Prolonged absorption
of the injectable
compositions can be brought about by including in the composition an agent
which delays absorption,
for example, aluminum monostearate and gelatin.
[0171] Sterile injectable solutions can be prepared by incorporating the
active compound in the
required amount in an appropriate solvent with one or a combination of
ingredients enumerated above,
as required, followed by filtered sterilization. Generally, dispersions are
prepared by incorporating the
active compound into a sterile vehicle which contains a basic dispersion
medium and the required other
ingredients from those enumerated above. In the case of sterile powders for
the preparation of sterile
injectable solutions, the preferred methods of preparation are vacuum drying
and freeze-drying which
yields a powder of the active ingredient plus any additional desired
ingredient from a previously sterile-
filtered solution thereof.
[0172] For administration by inhalation, the compounds are delivered in the
form of an aerosol
spray from pressured container or dispenser which contains a suitable
propellant, e.g., a gas such as
carbon dioxide, or a nebulizer.
[0173] Systemic administration can also be by transmucosal or transdermal
means. For
transmucosal or transdermal administration, penetrants appropriate to the
barrier to be permeated are
used in the formulation. Such penetrants are generally known in the art, and
include, for example, for
transmucosal administration, detergents, bile salts, and fusidic acid
derivatives. Transmucosal
administration can be accomplished through the use of nasal sprays or
suppositories. For transdermal
administration, the active compounds are formulated into ointments, salves,
gels, or creams as generally
known in the art. Molecules can also be prepared in the form of suppositories
(e.g., with conventional
suppository bases such as cocoa butter and other glycerides) or retention
enemas for rectal delivery.
[0174] In one embodiment, active molecules are prepared with carriers that
will protect the
compound against rapid elimination from the body, such as a controlled release
formulation, including
implants and microencapsulated delivery systems. Biodegradable, biocompatible
polymers can be used,
such as ethylene vinyl acetate, polyanhydrides, polyglycolic acid, collagen,
polyorthoesters, and
polylactic acid. Methods for preparation of such formulations will be apparent
to those skilled in the
art. Materials can also be obtained commercially from Alza Corporation and
Nova Pharmaceuticals,
Inc. Liposomal suspensions (including liposomes targeted to infected cells
with monoclonal antibodies
to viral antigens) can also be used as pharmaceutically acceptable carriers.
These can be prepared
according to methods known to those skilled in the art, for example, as
described in U.S. Patent
No. 4, 522, 811.
[0175] It is advantageous to formulate oral or parenteral compositions in
dosage unit form for ease
of administration and uniformity of dosage. Dosage unit form as used herein
refers to physically
51
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
discrete units suited as unitary dosages for the subject to be treated; each
unit containing a
predetermined quantity of active compound calculated to produce the desired
therapeutic effect in
association with the required pharmaceutical carrier.
[0176] Toxicity and therapeutic efficacy of such compounds can be determined
by standard
pharmaceutical procedures in cell cultures or experimental animals, e.g., for
determining the LDso (the
dose lethal to 50% of the population) and the EDso (the dose therapeutically
effective in 50% of the
population). The dose ratio between toxic and therapeutic effects is the
therapeutic index and it can be
expressed as the ratio LDso/EDso. Molecules which exhibit high therapeutic
indices are preferred.
While molecules that exhibit toxic side effects may be used, care should be
taken to design a delivery
system that targets such compounds to the site of affected tissue in order to
minimize potential damage
to uninfected cells and, thereby, reduce side effects.
[0177] The data obtained from the cell culture assays and animal studies can
be used in
formulating a range of dosage for use in humans. The dosage of such molecules
lies preferably within a
range of circulating concentrations that include the EDSO with little or no
toxicity. The dosage may vary
within this range depending upon the dosage form employed and the route of
administration utilized.
For any molecules used in the methods described herein, the therapeutically
effective dose can be
estimated initially from cell culture assays. A dose may be formulated in
animal models to achieve a
circulating plasma concentration range that includes the ICSO (i.e., the
concentration of the test
compound which achieves a half maximal inhibition of symptoms) as determined
in cell culture. Such
information can be used to more accurately determine useful doses in humans.
Levels in plasma may be
measured, for example, by high performance liquid chromatography.
[0178] As defined herein, a therapeutically effective amount of protein or
polypeptide (i.e., an
effective dosage) ranges from about 0.001 to 30 mg/kg body weight, sometimes
about 0.01 to 25 mg/kg
body weight, often about 0.1 to 20 mg/kg body weight, and more often about 1
to 10 mg/kg, 2 to 9
mg/kg, 3 to 8 mg/kg, 4 to 7 mg/kg, or 5 to 6 mg/kg body weight. The protein or
polypeptide can be
administered one time per week for between about 1 to 10 weeks, sometimes
between 2 to 8 weeks,
often between about 3 to 7 weeks, and more often for about 4, 5, or 6 weeks.
The skilled artisan will
appreciate that certain factors may influence the dosage and timing required
to effectively treat a
subject, including but not limited to the severity of the disease or disorder,
previous treatments, the
general health and/or age of the subject, and other diseases present.
Moreover, treatment of a subject
with a therapeutically effective amount of a protein, polypeptide, or antibody
can include a single
treatment or, preferably, can include a series of treatments.
[0179] With regard to polypeptide formulations, featured herein is a method
for treating
osteoarthritis in a subject, which comprises contacting one or more cells in
the subject with a first
polypeptide, where the subject comprises a second polypeptide having one or
more polymorphic
variations associated with cancer, and where the first polypeptide comprises
fewer polymorphic
variations associated with cancer than the second polypeptide. The first and
second polypeptides are
encoded by a nucleic acid which comprises a nucleotide sequence in SEQ ID NO:
1-7 or referenced in
52
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
Table A; a nucleotide sequence which encodes a polypeptide consisting of an
amino acid sequence
encoded by a nucleotide sequence referenced in SEQ ID NO: 1-7 or referenced in
Table A; a nucleotide
sequence which encodes a polypeptide that is 90% or more identical to an amino
acid sequence encoded
by a nucleotide sequence of SEQ ID NO: 1-7 or referenced in Table A and a
nucleotide sequence 90%
or more identical to a nucleotide sequence in SEQ ID NO: 1-7 or referenced in
Table A. The subject
often is a human.
[0180] For antibodies, a dosage of 0.1 mg/kg of body weight (generally 10
mg/kg to 20 mg/kg) is
often utilized. If the antibody is to act in the brain, a dosage of 50 mg/kg
to 100 mg/kg is often
appropriate. Generally, partially human antibodies and fully human antibodies
have a longer half life
within the human body than other antibodies. Accordingly, lower dosages and
less frequent
administration is often possible. Modifications such as lipidation can be used
to stabilize antibodies and
to enhance uptake and tissue penetration (e.g., into the brain). A method for
lipidation of antibodies is
described by Cruikshank et al., J. Acquired Immune Deficiency Syndromes and
Human Retrovirology
14:193 (1997).
[0181] Antibody conjugates can be used for modifying a given biological
response, the drug
moiety is not to be construed as limited to classical chemical therapeutic
agents. For example, the drug
moiety may be a protein or polypeptide possessing a desired biological
activity. Such proteins may
include, for example, a toxin such as abrin, ricin A, pseudomonas exotoxin, or
diphtheria toxin; a
polypeptide such as tumor necrosis factor, alpha-interferon, beta-interferon,
nerve growth factor, platelet
derived growth factor, tissue plasminogen activator; or, biological response
modifiers such as, for
example, lymphokines, interleukin-1 ("IL,-1"), interleukin-2 ("IL-2"),
interleukin-6 ("IL-6"),
granulocyte macrophage colony stimulating factor ("GM-CSF"), granulocyte
colony stimulating factor
("G-CSF"), or other growth factors. Alternatively, an antibody can be
conjugated to a second antibody
to form an antibody heteroconjugate as described by Segal in U.S. Patent No.
4,676,980.
[0182] For compounds, exemplary doses include milligram or microgram amounts
of the
compound per kilogram of subject or sample weight, for example, about 1
microgram per kilogram to
about 500 milligrams per kilogram, about 100 micrograms per kilogram to about
5 milligrams per
kilogram, or about 1 microgram per kilogram to about 50 micrograms per
kilogram. It is understood
that appropriate doses of a small molecule depend upon the potency of the
small molecule with respect
to the expression or activity to be modulated. When one or more of these small
molecules is to be
administered to an animal (e.g., a human) in order to modulate expression or
activity of a polypeptide or
nucleic acid described herein, a physician, veterinarian, or researcher may,
for example, prescribe a
relatively low dose at first, subsequently increasing the dose until an
appropriate response is obtained.
In addition, it is understood that the specific dose level for any particular
animal subject will depend
upon a variety of factors including the activity of the specific compound
employed, the age, body
weight, general health, gender, and diet of the subject, the time of
administration, the route of
administration, the rate of excretion, any drug combination, and the degree of
expression or activity to
be modulated.
53
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
[0183] With regard to nucleic acid formulations, gene therapy vectors can be
delivered to a subject
by, for example, intravenous injection, local administration (see, e.g., U.S.
Patent 5,328,470) or by
stereotactic injection (see e.g., Chen et al., (1994) Proc. Natl. Acad. Sei.
USA 91:3054-3057).
Pharmaceutical preparations of gene therapy vectors can include a gene therapy
vector in an acceptable
diluent, or can comprise a slow release matrix in which the gene delivery
vehicle is imbedded.
Alternatively, where the complete gene delivery vector can be produced intact
from recombinant cells
(e.g., retroviral vectors) the pharmaceutical preparation can include one or
more cells which produce the
gene delivery system. Examples of gene delivery vectors are described herein.
Therapeutic Methods
[0184] A therapeutic formulation described above can be administered to a
subject in need of a
therapeutic for inducing a desired biological response. Therapeutic
formulations can be administered by
any of the paths described herein. With regard to both prophylactic and
therapeutic methods of
treatment, such treatments may be specifically tailored or modified, based on
knowledge obtained from
pharmacogenomic analyses described herein.
[0185] As used herein, the term "treatment" is defined as the application or
administration of a
therapeutic formulation to a subject, or application or administration of a
therapeutic agent to an isolated
tissue or cell line from a subject with the purpose to cure, heal, alleviate,
relieve, alter, remedy,
ameliorate, improve or affect osteoarthritis, symptoms of osteoarthritis or a
predisposition towards
osteoarthritis. A therapeutic formulation includes, but is not limited to,
small molecules, peptides,
antibodies, ribozymes and antisense oligonucleotides. Administration of a
therapeutic formulation can
occur prior to the manifestation of symptoms characteristic of osteoarthritis,
such that osteoarthritis is
prevented or delayed in its progression. The appropriate therapeutic
composition can be determined
based on screening assays described herein.
[0186] As discussed, successful treatment of osteoarthritis can be brought
about by techniques that
serve to agonize target molecule expression or function, or alternatively,
antagonize target molecule
expression or function. These techniques include administration of modulators
that include, but are not
limited to, small organic or inorganic molecules; antibodies (including, for
example, polyclonal,
monoclonal, humanized, anti-idiotypic, chimeric or single chain antibodies,
and Fab, F(ab')Z and Fab
expression library fragments, scFV molecules, and epitope-binding fragments
thereof); and peptides,
phosphopeptides, or polypeptides.
[0187] Further, antisense and ribozyme molecules that inhibit expression of
the target gene can
also be used to reduce the level of target gene expression, thus effectively
reducing the level of target
gene activity. Still further, triple helix molecules can be utilized in
reducing the level of target gene
activity. Antisense, ribozyme and triple helix molecules are discussed above.
It is possible that the use
of antisense, ribozyme, and/or triple helix molecules to reduce or inhibit
mutant gene expression can
also reduce or inhibit the transcription (triple helix) and/or translation
(antisense, ribozyme) of mRNA
produced by normal target gene alleles, such that the concentration of normal
target gene product
54
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
present can be lower than is necessary for a normal phenotype. In such cases,
nucleic acid molecules
that encode and express target gene polypeptides exhibiting normal target gene
activity can be
introduced into cells via gene therapy method. Alternatively, in instances in
that the target gene
encodes an extracellular polypeptide, it can be preferable to co-administer
normal target gene
polypeptide into the cell or tissue in order to maintain the requisite level
of cellular or tissue target gene
activity.
[0188] Another method by which nucleic acid molecules may be utilized in
treating or preventing
osteoarthritis is use of aptamer molecules specific for target molecules.
Aptamers are nucleic acid
molecules having a tertiary structure which permits them to specifically bind
to ligands (see, e.g.,
Osborne, et al., Curr. Opin. Chem. Biol. l (1): 5-9 (1997); and Patel, D. J.,
Curr. Opira. Claem. Biol.
Jun; I (1): 32-46 (1997)).
[0189] Yet another method of utilizing nucleic acid molecules for
osteoarthritis treatment is gene
therapy, which can also be referred to as allele therapy. Provided herein is a
gene therapy method for
treating osteoarthritis in a subject, which comprises contacting one or more
cells in the subject or from
the subject with a nucleic acid having a first nucleotide sequence (e.g., the
first nucleotide sequence is
identical to or substantially identical to a nucleotide sequence of SEQ ID NO:
1-7 or other nucleotide
sequence referenced in Table A). Genomic DNA in the subject comprises a second
nucleotide sequence
having one or more polymorphic variations associated with osteoarthritis
(e.g., the second nucleotide
sequence is identical to or substantially identical to a nucleotide sequence
of SEQ ID NO: 1-7 or other
nucleotide sequence referenced in Table A). The first and second nucleotide
sequences typically are
substantially identical to one another, and the first nucleotide sequence
comprises fewer polymorphic
variations associated with osteoarthritis than the second nucleotide sequence.
The first nucleotide
sequence may comprise a gene sequence that encodes a full-length polypeptide
or a fragment thereof.
The subject is often a human. Allele therapy methods often are utilized in
conjunction with a method of
first determining whether a subject has genomic DNA that includes polymorphic
variants associated
with osteoarthritis.
[0190] In another allele therapy embodiment, provided herein is a method which
comprises
contacting one or more cells in the subject or from the subject with a
polypeptide encoded by a nucleic
acid having a first nucleotide sequence (e.g., the first nucleotide sequence
is identical to or substantially
identical to the nucleotide sequence of SEQ ID NO: 1-7 or other nucleotide
sequence referenced in
Table A). Genomic DNA in the subject comprises a second nucleotide sequence
having one or more
polymorphic variations associated with osteoarthritis (e.g., the second
nucleotide sequence is identical
to or substantially identical to a nucleotide sequence of SEQ ID NO: 1-7 or
other nucleotide sequence
referenced in Table A). The first and second nucleotide sequences typically
are substantially identical
to one another, and the first nucleotide sequence comprises fewer polymorphic
variations associated
with osteoarthritis than the second nucleotide sequence. The first nucleotide
sequence may comprise a
gene sequence that encodes a full-length polypeptide or a fragment thereof.
The subject is often a
human.
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
[0191] For antibody-based therapies, antibodies can be generated that are both
specific for target
molecules and that reduce target molecule activity. Such antibodies may be
administered in instances
where antagonizing a target molecule function is appropriate for the treatment
of osteoarthritis.
[0192] In circumstances where stimulating antibody production in an animal or
a human subject by
injection with a target molecule is harmful to the subject, it is possible to
generate an immune response
against the target molecule by use of anti-idiotypic antibodies (see, e.g.,
Herlyn, Anfa. Med.; 31 (1): 66-78
(1999); and Bhattacharya-Chatterjee & Foon, Cancer Treat. Res.; 94: 51-68
(1998)). Introducing an
anti-idiotypic antibody to a mammal or human subject often stimulates
production of anti-anti-idiotypic
antibodies, which typically are specific to the target molecule. Vaccines
directed to osteoarthritis also
may be generated in this fashion.
[0193] In instances where the target molecule is intracellular and whole
antibodies are used,
internalizing antibodies may be preferred. Lipofectin or liposomes can be used
to deliver the antibody
or a fragment of the Fab region that binds to the target antigen into cells.
Where fragments of the
antibody are used, the smallest inhibitory fragment that binds to the target
antigen is preferred. For
example, peptides having an amino acid sequence corresponding to the Fv region
of the antibody can be
used. Alternatively, single chain neutralizing antibodies that bind to
intracellular target antigens can
also be administered. Such single chain antibodies can be administered, for
example, by expressing
nucleotide sequences encoding single-chain antibodies within the target cell
population (see, e.g.,
Marasco et al., Proc. Natl. Acad. Sci. USA 90: 7889-7893 (1993)).
[0194] Modulators can be administered to a patient at therapeutically
effective doses to treat
osteoarthritis. A therapeutically effective dose refers to an amount of the
modulator sufficient to result
in amelioration of symptoms of osteoarthritis. Toxicity and therapeutic
efficacy of modulators can be
determined by standard pharmaceutical procedures in cell cultures or
experimental animals, e.g., for
determining the LDSO (the dose lethal to 50% of the population) and the EDso
(the dose therapeutically
effective in 50% of the population). The dose ratio between toxic and
therapeutic effects is the
therapeutic index and it can be expressed as the ratio LDSO/EDso. Modulators
that exhibit large
therapeutic indices are preferred. While modulators that exhibit toxic side
effects can be used, care
should be taken to design a delivery system that targets such molecules to the
site of affected tissue in
order to minimize potential damage to uninfected cells, thereby reducing side
effects.
[0195] Data obtained from cell culture assays and animal studies can be used
in formulating a
range of dosages for use in humans. The dosage of such compounds lies
preferably within a range of
circulating concentrations that include the EDSO with little or no toxicity.
The dosage can vary within
this range depending upon the dosage form employed and the route of
administration utilized. For any
compound used in the methods described herein, the therapeutically effective
dose can be estimated
initially from cell culture assays. A dose can be formulated in animal models
to achieve a circulating
plasma concentration range that includes the ICSO (i. e., the concentration of
the test compound that
achieves a half maximal inhibition of symptoms) as determined in cell culture.
Such information can be
56
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
used to more accurately determine useful doses in humans. Levels in plasma can
be measured, for
example, by high performance liquid chromatography.
[0196] Another example of effective dose determination for an individual is
the ability to directly
assay levels of "free" and "bound" compound in the serum of the test subject.
Such assays may utilize
antibody mimics and/or "biosensors" that have been created through molecular
imprinting techniques.
Molecules that modulate target molecule activity are used as a template, or
"imprinting molecule", to
spatially organize polymerizable monomers prior to their polymerization with
catalytic reagents. The
subsequent removal of the imprinted molecule leaves a polymer matrix which
contains a repeated
"negative image" of the compound and is able to selectively rebind the
molecule under biological assay
conditions. A detailed review of this technique can be seen in Ansell et al.,
Cu~~ent Opinion in
Biotechnology 7: 89-94 (1996) and in Shea, Trends in Polymer Science 2: 166-
173 (1994). Such
"imprinted" affinity matrixes are amenable to ligand-binding assays, whereby
the immobilized
monoclonal antibody component is replaced by an appropriately imprinted
matrix. An example of the
use of such matrixes in this way can be seen in Vlatakis, et al., Nature 361:
645-647 (1993). Through
the use of isotope-labeling, the "free" concentration of compound which
modulates target molecule
expression or activity readily can be monitored and used in calculations of
ICSO. Such "imprinted"
affinity matrixes can also be designed to include fluorescent groups whose
photon-emitting properties
measurably change upon local and selective binding of target compound. These
changes readily can be
assayed in real time using appropriate fiberoptic devices, in turn allowing
the dose in a test subject to be
quickly optimized based on its individual ICSO. An example of such a
"biosensor" is discussed in Kriz
et al., Analytical Chemistry 67.' 2142-2144 (1995).
[0197] The examples set forth below illustrate but not limit the invention.
Examples
[0198] In the following studies a group of subjects was selected according to
specific parameters
pertaining to osteoarthritis. Nucleic acid samples obtained from individuals
in the study group were
subjected to genetic analyses that identified associations between
osteoarthritis and certain polymorphic
variants in human genomic DNA. The polymorphisms were genotyped again in two
replication cohorts
consisting of individuals selected for OA. In addition, SNPs proximal to the
incident polymorphism in
the KIAA0296 region, the Cla~~om 4 region, the Ch~om 6 region, the ELP3
region, the LRCHI region, the
SNWI region and in the ERG region were identified and allelotyped in OA case
and control pools.
Methods are described for producing target polypeptides encoded by the nucleic
acids of Table A in
vitf~o or in vivo, which can be utilized in methods that screen test molecules
for those that interact with
target polypeptides. Test molecules identified as being interactors with
target polypeptides can be
screened further as osteoarthritis therapeutics.
57
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
Example 1
Samples and Poolin S~ trategies
Sample Selection
[0199] Blood samples were collected from individuals diagnosed with knee
osteoarthritis, which
were referred to as case samples. Also, blood samples were collected from
individuals not diagnosed
with knee osteoarthritis as gender and age-matched controls. A database was
created that listed all
phenotypic trait information gathered from individuals for each case and
control sample. Genomic
DNA was extracted from each of the blood samples for genetic analyses.
DNA Extraction from Blood Samples
[0200] Six to ten milliliters of whole blood was transferred to a 50 ml tube
containing 27 ml of red
cell lysis solution (RCL). The tube was inverted until the contents were
mixed. Each tube was
incubated for 10 minutes at room temperature and inverted once during the
incubation. The tubes were
then centrifuged for 20 minutes at 3000 x g and the supernatant was carefully
poured off. 100-200 w1 of
residual liquid was left in the tube and was pipetted repeatedly to resuspend
the pellet in the residual
supernatant. White cell lysis solution (WCL) was added to the tube and
pipetted repeatedly until
completely mixed. While no incubation was normally required, the solution was
incubated at 37°C or
room temperature if cell clumps were visible after mixing until the solution
was homogeneous. 2 ml of
protein precipitation was added to the cell lysate. The mixtures were vortexed
vigorously at high speed
for 20 sec to mix the protein precipitation solution uniformly with the cell
lysate, and then centrifuged
for 10 minutes at 3000 x g. The supernatant containing the DNA was then poured
into a clean 15 ml
tube, which contained 7 ml of 100% isopropanol. The samples were mixed by
inverting the tubes gently
until white threads of DNA were visible. Samples were centrifuged for 3
minutes at 2000 x g and the
DNA was visible as a small white pellet. The supernatant was decanted and 5 ml
of 70% ethanol was
added to each tube. Each tube was inverted several times to wash the DNA
pellet, and then centrifuged
for 1 minute at 2000 x g. The ethanol was decanted and each tube was drained
on clean absorbent
paper. The DNA was dried in the tube by inversion for 10 minutes, and then
1000 ~,1 of 1X TE was
added. The size of each sample was estimated, and less TE buffer was added
during the following DNA
hydration step if the sample was smaller. The DNA was allowed to rehydrate
overnight at room
temperature, and DNA samples were stored at 2-8°C.
[0201] DNA was quantified by placing samples on a hematology mixer for at
least 1 hour. DNA
was serially diluted (typically 1:80, 1:160, 1:320, and 1:640 dilutions) so
that it would be within the
measurable range of standards. 125 p,1 of diluted DNA was transferred to a
clear U-bottom microtitre
plate, and 125 ~,1 of 1X TE buffer was transferred into each well using a
multichannel pipette. The
DNA and 1X TE were mixed by repeated pipetting at least 15 times, and then the
plates were sealed. 50
p1 of diluted DNA was added to wells AS-H12 of a black flat bottom microtitre
plate. Standards were
58
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
inverted six times to mix them, and then 50 ~,1 of 1X TE buffer was pipetted
into well A1, 1000 ng/ml
of standard was pipetted into well A2, 500 ng/ml of standard was pipetted into
well A3, and 250 ng/ml
of standard was pipetted into well A4. PicoGreen (Molecular Probes, Eugene,
Oregon) was thawed and
freshly diluted 1:200 according to the number of plates that were being
measured. PicoGreen was
vortexed and then 501 was pipetted into all wells of the black plate with the
diluted DNA. DNA and
PicoGreen were mixed by pipetting repeatedly at least 10 times with the
multichannel pipette. The plate
was placed into a Fluoroskan Ascent Machine (microplate fluorometer produced
by Labsystems) and
the samples were allowed to incubate for 3 minutes before the machine was run
using filter pairs 485
nm excitation and 538 nm emission wavelengths. Samples having measured DNA
concentrations of
greater than 450 ng/~,1 were re-measured for conformation. Samples having
measured DNA
concentrations of 20 ng/~,1 or less were re-measured for confirmation.
Poolin S~trate-dies - Discovery Cohort
[0202] Samples were derived from the Nottingham knee OA family study (UK)
where index cases
were identified through a knee replacement registry. Siblings were approached
and assessed with knee
x-rays and assigned status as affected or unaffected. In all 1,157 individuals
were available. In order to
create same-sex pools of appropriate sizes, 335 unrelated female individuals
with OA from the
Nottingham OA sample were selected for the case pool. The control pool was
made up of unrelated
female individuals from the St. Thomas twin study (England) with normal knee x-
rays and without
other indications of OA, regardless of anatomical location, as well as lacking
family history of OA. The
St. Thomas twin study consists of Caucasian, female participants from the St.
Thomas' Hospital,
London, adult-twin registry, which is a voluntary registry of >4,000 twin
pairs ranging from 18 to
76 years of age. The female case samples and female control samples are
described further in Table 1
below.
[0203] A select set of samples from each group were utilized to generate
pools, and one pool was
created for each group. Each individual sample in a pool was represented by an
equal amount of
genomic DNA. For example, where 25 ng of genomic DNA was utilized in each PCR
reaction and
there were 200 individuals in each pool, each individual would provide 125 pg
of genomic DNA.
Inclusion or exclusion of samples for a pool was based upon the following
criteria: the sample was
derived from an individual characterized as Caucasian; the sample was derived
from an individual of
British paternal and maternal descent; case samples were derived from
individuals diagnosed with
specific knee osteoarthritis (OA) and were recruited from an OA knee
replacement clinic. Control
samples were derived from individuals free of OA, family history of OA, and
rheumatoid arthritis.
Also, sufficient genomic DNA was extracted from each blood sample for all
allelotyping and
genotyping reactions performed during the study. Phenotype information from
each individual was
collected and included age of the individual, gender, family history of OA,
general medical information
(e.g., height, weight, thyroid disease, diabetes, psoriasis, hysterectomy),
joint history (previous and
current symptoms, joint-related operations, age at onset of symptoms, date of
primary diagnosis, age of
59
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
individual as of primary diagnosis and order of involvement), and knee-related
findings (crepitus,
restricted passive movement, bony swelling/deformity). Additional knee
information included knee
history, current symptoms, any major knee injury, menisectomy, knee
replacement surgery, age of
surgery, and treatment history (including hormone replace therapy (HRT)).
Samples that met these
criteria were added to appropriate pools based on disease status.
[0204] The selection process yielded the pools set forth in Table 1, which
were used in the studies
that follow:
TABLE 1
Female case Female control
Pool size 335 335
umber)
Pool Criteria
ntr case
l
(ex: case/control)co
o
Mean Age 5'7,21 69.95
(ex: years)
Example 2
Association of Polymorphic Variants with Osteoarthritis
[0205] A whole-genome screen was performed to identify particular SNPs
associated with
occurrence of osteoarthritis. As described in Example 1, two sets of samples
were utilized, which
included samples from female individuals having knee osteoarthritis
(osteoarthritis cases), and samples
from female individuals not having knee osteoarthritis (female controls). The
initial screen of each pool
was performed in an allelotyping study, in which certain samples in each group
were pooled. By
pooling DNA from each group, an allele frequency for each SNP in each group
was calculated. These
allele frequencies were then compared to one another. Particular SNPs were
considered as being
associated with osteoarthritis when allele frequency differences calculated
between case and control
pools were statistically significant. SNP disease association results obtained
from the allelotyping study
were then validated by genotyping each associated SNP across all samples from
each pool. The results
of the genotyping then were analyzed, allele frequencies for each group were
calculated from the
individual genotyping results, and a p-value was calculated to determine
whether the case and control
groups had statistically significant differences in allele frequencies for a
particular SNP. When the
genotyping results agreed with the original allelotyping results, the SNP
disease association was
considered validated at the genetic level.
SNP Panel Used for Genetic Anal'tses
[0206] A whole-genome SNP screen began with an initial screen of approximately
25,000 SNPs
over each set of disease and control samples using a pooling approach. The
pools studied in the screen
are described in Example 1. The SNPs analyzed in this study were part of a set
of 25,488 SNPs
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
confirmed as being statistically polymorphic as each is characterized as
having a minor allele frequency
of greater than 10%. The SNPs in the set reside in genes or in close proximity
to genes, and many
reside in gene exons. Specifically, SNPs in the set are located in exons,
introns, and within 5,000 base-
pairs upstream of a transcription start site of a gene. In addition, SNPs were
selected according to the
following criteria: they are located in ESTs; they are located in Locuslink or
Ensembl genes; and they
are located in Genomatix promoter predictions. SNPs in the set were also
selected on the basis of even
spacing across the genome, as depicted in Table 2.
[0207] A case-control study design using a whole genome association strategy
involving
approximately 28,000 single nucleotide polymorphisms (SNPs) was employed.
Approximately 25,000
SNPs were evenly spaced in gene-based regions of the human genome with a
median inter-marker
distance of about 40,000 base pairs. Additionally, approximately 3,000 SNPs
causing amino acid
substitutions in genes described in the literature as candidates for various
diseases were used. The case-
control study samples were of female Caucasian origin (British paternal and
maternal descent) 670
individuals were equally distributed in two groups: female controls and female
cases. The whole
genome association approach was first conducted on 2 DNA pools representing
the 2 groups.
Significant markers were confirmed by individual genotyping.
TABLE 2
General Statistics Spacing Statistics
Total # of SNPs 25,488 Median 37,058 by
# of Exonic SNPs>4,335 (17%) Minimum* 1,000 by
# SNPs with refSNP20,776 (81%) Maximum* 3,000,000
ID by
Gene Coverage >10,000 Mean 122,412
by
Chromosome CoverageAll Std Deviation 373,325
by
*Excludes outliers
Alleloty~ing and Genoty~ing Results
[0208] The genetic studies summarized above and described in more detail below
identified allelic
variants associated with osteoarthritis, which are summarized in Table A.
Assay for Verifying, Alleloty~ing, and Genotypin~ SNPs
[0209] A MassARRAYTM system (Sequenom, Inc.) was utilized to perform SNP
genotyping in a
high-throughput fashion. This genotyping platform was complemented by a
homogeneous, single-tube
assay method (hMETM or homogeneous MassEXTENDTM (Sequenom, Inc.)) in which two
genotyping
primers anneal to and amplify a genomic target surrounding a polymorphic site
of interest. A third
primer (the MassEXTENDTM primer), which is complementary to the amplified
target up to but not
including the polymorphism, was then enzymatically extended one or a few bases
through the
polymorphic site and then terminated.
61
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
[0210] For each polymorphism, SpectroDESIGNERTM software (Sequenom, Inc.) was
used to
generate a set of PCR primers and a MassEXTENDTM primer which where used to
genotype the
polymorphism. Other primer design software could be used or one of ordinary
skill in the art could
manually design primers based on his or her knowledge of the relevant factors
and considerations in
designing such primers. Table 3 shows PCR primers and Table 4 shows extension
primers used for
analyzing polymorphisms. The initial PCR amplification reaction was performed
in a 5 ~,l total volume
containing 1X PCR buffer with 1.5 mM MgCl2 (Qiagen), 200 p,M each of dATP,
dGTP, dCTP, dTTP
(Gibco-BRL), 2.5 ng of genomic DNA, 0.1 units of HotStar DNA polymerase
(Qiagen), and 200 nM
each of forward and reverse PCR primers specific for the polymorphic region of
interest.
TABLE 3: PCR Primers
SNP
Forward PCR primer Reverse PCR primer
Reference
rs552 ACGTTGGATGGACTGAGGTAGATGATGCACGTTGGATGGCTTTCTTTCCCTTGGTTTC
rs12904 ACGTTGGATGGAACCACTCCCACCACAGACGTTGGATGGGTGGGGATGGCACTGTC
rs2282146ACGTTGGATGTCCCACGAGGACCTGGAGACGTTGGATGTTCGTTTGGGTGGCCGGG
rs734784ACGTTGGATGTCGGGATGTCTCCAGAGATGACGTTGGATGGCAACCACCAAGAGTTTGAG
rs1042164ACGTTGGATGTTTCTTCCAGACGGGCTTTCACGTTGGATGCAAAGTCAGCCGCAAACGAC
rs749670ACGTTGGATGTCTCATCTGTGTGCCCATTGACGTTGGATGATGAGGGTGAAAGGCAGGAG
rs955592ACGTTGGATGTTCCCATTCTTCTTGGGCTCACGTTGGATGTCTCAGAGGGTCTCCTTTTC
rs1143016ACGTTGGATGTTGTCCAGCAGGTAGGGCAGACGTTGGATGACCCATCGCGGATACATGTG
rs755248ACGTTGGATGGGTCTCTGCTGAGGAAGTGGACGTTGGATGACACTCACTACGGGGCCAG
rs1055055ACGTTGGATGTTGTGCTTGCTGAGGAATCCACGTTGGATGGTTGCAGAGAGCGTCTATAC
rs835409ACGTTGGATGTCCTGTTGGCTTTTGCAGACACGTTGGATGACTGCTCATGGTGGTTGAAC
rs927663ACGTTGGATGTTTGACTGGTTGCCCCAAACACGTTGGATGAAGAATCTTCAGTGCCAGCC
rs8162 ACGTTGGATGCTTCATCCAGAACCTCCAGGACGTTGGATGTGCATATGGCTTGTCAGAGC
rs831038ACGTTGGATGTGAAAGAGCTGCCTTCTTTCACGTTGGATGAAATGACACTCACGGTAAGC
rs33079 ACGTTGGATGTTATTTCATTGGCCAAGCCCACGTTGGATGGTGTTCACTTGTTCATGCAC
rs1710880ACGTTGGATGCGAAGGCAGAGAATAAACTGACGTTGGATGAACTCTGTGGTTTAAGAAAG
rs1078153ACGTTGGATGTCCTGCGTGTAACTGAGAGGACGTTGGATGAACATACACACAGTGCGAGC
rs799570ACGTTGGATGATGCATATGGGCAGGTTGCCACGTTGGATGCCAGGAAAGCATCCTCAGAC
rs1282730ACGTTGGATGTCCTTTGACTTACTGTGCTGACGTTGGATGAGAAAAGAGGTTGTGTACAG
rs1518875ACGTTGGATGAGAATGCGTTCAATGCCTGCACGTTGGATGAGCGAAAAGCTCTGCCATTG
rs1568694ACGTTGGATGGTTCATTCAGTTATGGACGGACGTTGGATGTGATAGGAGGGAGCCATCTC
rs905042ACGTTGGATGTAACAATGGTAAGGGCCAAGACGTTGGATGGGTCCATAATGGTCATTGTG
rs1957723ACGTTGGATGTACTCACTTGTGTACTGCTCACGTTGGATGGCTGCAGCGTCACATTAATC
rs794018ACGTTGGATGGGATGATGATGAAATGACTGACGTTGGATGGCTCTAGTTAGATGAGTCTC
rs707723ACGTTGGATGTGTGGCTGAAGTTTGCTCTGACGTTGGATGCACACACAAACCTTGAAGAG
rs893861ACGTTGGATGGAGGCATGTACACAAAACTGACGTTGGATGGCTCACGACTGTAATAGTTG
rs1914903ACGTTGGATGTGCGTCAAGT1'GAAGTCCTCACGTTGGATGAGGGTAGTGAGTTCACATGC
rs2062232ACGTTGGATGTCCTGCTCAGATAACTGCTGACGTTGGATGGCGGTAGTTTTCCCTAAACC
rs26609 ACGTTGGATGCAAGGGAGATCAGAAACATCACGTTGGATGAATTCATTGTTCTTGATGGC
rs1370987ACGTTGGATGATACTTTGGATGTCTGGTGGACGTTGGATGGGTCTTTGGTCACAACTATC
rs1012414ACGTTGGATGACTTGGAAAGTCAGTCTGGGACGTTGGATGGAAACCGAGAAATGGCTATG
rs435903ACGTTGGATGGGCATAAGTTAGAGACAACCACGTTGGATGGGCTATGTTATGCTGCTGTG
rs1248 ACGTTGGATGGAGATTGTGCATTTTGGCAAGACGTTGGATGCAGACACCATCTTAACCAAG
rs703508ACGTTGGATGAGCTCTGTGGCCTCTTTTGGACGTTGGATGTACTCACAGTCTTCCCGGCG
rs226465ACGTTGGATGAATTTTGACCCCTGCCAACCACGTTGGATGTATGTGAAAGAGGCGTGAAG
rs241448ACGTTGGATGCAAGCTGCAGAAGCTTGCCACGTTGGATGTGAGAAGAGGGCCCAGTATC
rs763155ACGTTGGATGGGGAAACCCAAAATAGTGTCACGTTGGATGTCACAGGAGAGTAATGCCTC
rs1040461ACGTTGGATGACATCTGGTGGAAGTCACTCACGTTGGATGGGTCCTTTGTTTGTTGGGTC
rs462832~ ACGTTGGATGCACTTTCTCTGTAATATTTGACGTTGGATGTGAGACAACAAAAATTTGCC
~
62
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
SNP Forward PCR primer Reverse PCR primer
Reference
rs804194ACGTTGGATGTAATCCGGTGGCAGATCAAGACGTTGGATGGAAATTCATGTGCTGACGGG
rs1022646ACGTTGGATGACTGTCACCTAATCATCCTGACGTTGGATGGACTATGTTGGAGTTCAGAG
rs1569112ACGTTGGATGTCATGATCTGCCTGTGGAGAACGTTGGATGACCATCCTCACACCCATCCA
rs805623ACGTTGGATGATCAACCACTCATACACTGCACGTTGGATGCACAGAAACAGCTGGATTGC
rs1019850ACGTTGGATGTTTTACTCCAGGAAGCCACCACGTTGGATGAGCAGGGAGAATTGTTCCAG
rs1599931ACGTTGGATGTCAAACCCTTCCTGTAGACGACGTTGGATGTGAACATAGTAGGCGCTCAG
AA ACGTTGGATGTAGGAGTGCTCGTATTTTGGACGTTGGATGCTGGGAACAGCTTTTGATCC
rs279941ACGTTGGATGCATAGGGAACACCGAGAATGACGTTGGATGGGTTGTCATCTATGGGCTAG
rs1062230ACGTTGGATGAAACTCCTTTCCCTCTCAAGACGTTGGATGGGCCCATCAGTCTATAGTTT
rs1859911ACGTTGGATGCTGTITfTCCGAGCATCTACACGTTGGATGCCTCTTGCATATGAGATAGG
rs1477261ACGTTGGATGCAGGGTTATGTGGTATTATCACGTTGGATGGGGAAAGTAAAAGATAAGAG
rs1191119ACGTTGGATGACTCTCAGGGTGATTATCTGACGTTGGATGTGTAAGATTCTGGCACTGTC
rs657780ACGTTGGATGTTTAAGAAGCCGCCAAGGAGACGTTGGATGCCCATTTTCAGACCACTTGG
rs1393890ACGTTGGATGGTCTGATTATCTTTCTGCCGACGTTGGATGGGTACCTTTATCCTTGCTTC
rs1478714ACGTTGGATGAATAATTTGCTGACACCCCCACGTTGGATGGGAGTCCAGAGGTTAAACAG
rs868213ACGTTGGATGTGTCAGAACTGGGCACATTCACGTTGGATGAGGGATAGGGATCAGGAATG
rs690115ACGTTGGATGAATAGCCAAGGCCGTGTGGGACGTTGGATGCACCTGGGAGATAGCAGGG
rs1465501ACGTTGGATGTCAGGAATTGTTACCTGGACACGTTGGATGCCCTCATCTAGACACTTTTG
rs899173ACGTTGGATGAGTGCCACATCACTCTTGTGACGTTGGATGTTCTGCTCCACTACAGTCTC
rs10477 ACGTTGGATGGGGGCTACGTGGAAGTTACCACGTTGGATGATGGCAATCAAGAGAGTCTAA
rs926393ACGTTGGATGAGATCAGCCCAGGAAATGTGACGTTGGATGTGTTGGAGAAGGTTTCCACC
rs465271ACGTTGGATGAATCACAGCTCATGGCTCACACGTTGGATGATGGTAGTGTGCACCTATGG
rs13847 ACGTTGGATGCGCCCGTAGTGATAAGCACACGTTGGATGCAGGACAGGGCAGAGTGAG
rs738658ACGTTGGATGGATGGTATGTGTGCATCAGGACGTTGGATGCTTTCCAAGAGATGGCGTTC
rs756519ACGTTGGATGTCTAGAGACACCTGAGGTTGACGTTGGATGTGTTTCACTTCAGAGCCCTG
rs1042327ACGTTGGATGAACTTCACATCACAGCTCCCACGTTGGATGCAGAAGTTGGGTTTTCCAGC
rs8770 ACGTTGGATGCTGTCACTGGACACTTTTGACGTTGGATGAAAATAGAGGTGCAGAGATG
rs1563055ACGTTGGATGAGTTCTTTCTCCTCACATTGACGTTGGATGCCCTTTAGAAGCACATACTC
rs912428ACGTTGGATGACTACATCCATTCCAGGGAGACGTTGGATGTCAGATCAGAGTGAGTTTAG
rs1888475ACGTTGGATGACCCCTGGCAAGTGAATTACACGTTGGATGGGGAGGTGGATGTTCTTATC
[0211] Samples were incubated at 95°C for 15 minutes, followed by 45
cycles of 95°C for 20
seconds, 56°C for 30 seconds, and 72°C for 1 minute, finishing
with a 3 minute final extension at 72°C.
Following amplification, shrimp alkaline phosphatase (SAP) (0.3 units in a 2
~l volume) (Amersham
Pharmacia) was added to each reaction (total reaction volume was 7 ~1) to
remove any residual dNTPs
that were not consumed in the PCR step. Samples were incubated for 20 minutes
at 37°C, followed by
minutes at 85°C to denature the SAP.
[0212] Once the SAP reaction was complete, a primer extension reaction was
initiated by adding a
polymorphism-specific MassEXTENDTM primer cocktail to each sample. Each
MassEXTENDTM
cocktail included a specific combination of dideoxynucleotides (ddNTPs) and
deoxynucleotides
(dNTPs) used to distinguish polymorphic alleles from one another. Methods for
verifying, allelotyping
and genotyping SNPs are disclosed, for example, in U.S. Pat. No. 6,258,538,
the content of which is
hereby incorporated by reference. In Table 4, ddNTPs are shown and the fourth
nucleotide not shown is
the dNTP.
63
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
TABLE 4: Extension Primers
SNP Termination
ReferenceExtend Probe Mix
rs552 TGATGCTGTTGTCAGATACC ACT
rs12904 AGCCTCAAAACGGGTCA ACT
rs2282146GGACCTGGAGCCCCCACC ACG
rs734784GCCTCCGACACCCCATCAA ACG
rs1042164CTTGCTCGGGACCAGTCCA ACT
rs749670GGTGGTGGGCATCCCTTTC ACG
rs955592TTGGGCTCTGACCACCTCT ACT
rs1143016ATGCAGCGTCACCAGCAC ACT
rs755248TGAGGAAGTGGCAGGTGTG ACG
rs1055055CCCAGTTCAGGCTCACTTTC ACT
rs835409TTGCAGACCAGCCAATTAAGAA ACT
rs927663GGTTGCCCCAAACTCCCTT ACT
rs8162 AACACAGAGCAAAGCACC ACT
rs831038CGTTATAGTAAAGGAAAGGCAG ACG
rs33079 CCCATCACCTGGAGCTTTG ACG
rs1710880CTGTATTATGTTTCCCCTTGG CGT
rs1078153GCCGGCACCGTCAGAAAC CGT
rs799570GCAGTTCCTAGAAGACAGCT ACT
rs1282730TGCTGGCCCAACTTTTGTCT ACG
rs1518875CTGCAATGTTTCCAAACCCC ACT
rs1568694AGTTATGGACGGAAGAAGGG ACG
rs905042GGTAAGGGCCAAGTGAGTG CGT
rs1957723AGCATGGCATAGGCACTGG ACG
rs794018AAATGACTGAAAATGTGTACTATA ACG
rs707723CCTGAGGTATATTCAATA ACG
rs893861CATGTACACAAAACTGTTAAGTAA ACG
rs1914903TCCCCATAGATGGACCTGC ACG
rs2062232GCTGAAGACAAGGATTAGGTT ACG
rs26609 GAGATCAGAAACATCACCTTG CGT
rs1370987TTTGGGAGTTACTGCCTTAGAA ACT
rs1012414ACTAGGAACCAGAATATGAGCATC ACG
rs435903AAGCTAACAATGGAATAATGGC ACG
rs1248 GTGCATTTTGGCAAGAATATATG CGT
rs703508GGGGTCCAGGCAGAAAGAAAC ACG
rs226465CCTCTTCCCCTCCTCCCT ACT
rs241448GCAGAAGCTTGCCCAGCTC ACG
rs763155GCAGCCTGCAAGTGAGTGA ACT
rs 1040461AAGTCACTCCGGTCAGAATTCA ACT
rs462832ATAAGAATCTTTTAGATCCCAACA CGT
rs804194GATCAAGGCTGATCTCGCC ACT
rs1022646CCTAATCATCCTGCCACCC ACT
rs1569112ACCAGGCCGCATGGGCTG ACG
rs805623CTGTGTTCAAATAAGGCAACC ACT
rs1019850AGGAAGCCACCAGCTAATAC CGT
rs1599931CTGAGGCCGGGAGGGATT ACT
AA TAGTTTTAAATTCTGCACA ACT
rs279941AACACCGAGAATGAAAACATC ACT
rs1062230ATGCTGGTTCTGTCCAA ACG
rs1859911TCCGAGCATCTACATGCTCA ACT
rs1477261AGGAGGAGCCCAAATATGAAA CGT
rs1191119GTCTTTTTGTTAACTGGGGAACCC ACG
rs657780CGCCAAGGAGTTTCCCACA ACT
rs1393890CTGCCGTACCTGGCAAGC ACT
rs147871~CCCCGAGGGGACAGTCCA ~ ACG
64
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
SNP Extend Probe Termination
Reference Mix
rs868213GGGCACATTCTTGAGGAGGT ACG
rs690115AGCCGAGGGAGCTGACCCTG ACG
rs1465501TCCAGGAGCCCTCAGAATG ACT
rs899173CCTCTGGCAAAGTGTGGAGC ACG
rs10477 AGTACGATATCAAAGATC ACG
rs926393CAGGAAATGTGCTTTCGAGTTCC ACG
rs465271GGCTCAAGGGATCCTCCCA ACG
rs13847 AAGCACACCGGCACGAAC ACT
rs738658GAGGCATTTTCATTAATGCATG CGT
rs756519CAGAGCCCTGTTCTTTGATTT ACG
rs1042327CATCACAGCTCCCCACCAT ACT
rs8770 TAGACACTGTGTAAGCAATC ACG
rs 1563055TTCTCCTCACATTGTTTCTACT ACG
rs912428CCATTCCAGGGAGACTCCCA ACT
rs1888475GACATCAAATGATTCCCCTGT ~ ACT
[0213] The MassEXTENDTM reaction was performed in a total volume of 9 ~,1,
with the addition of
1X ThermoSequenase buffer, 0.576 units of ThermoSequenase (Amersham
Pharmacia), 600 nM
MassEXTENDTM primer, 2 mM of ddATP and/or ddCTP and/or ddGTP and/or ddTTP, and
2 mM of
dATP or dCTP or dGTP or dTTP. The deoxy nucleotide (dNTP) used in the assay
normally was
complementary to the nucleotide at the polymorphic site in the amplicon.
Samples were incubated at
94°C for 2 minutes, followed by 55 cycles of 5 seconds at 94°C,
5 seconds at 52°C, and 5 seconds
at 72°C.
[0214] Following incubation, samples were desalted by adding 16 ~1 of water
(total reaction
volume was 25 ~l), 3 mg of SpectroCLEANTM sample cleaning beads (Sequenom,
Inc.) and allowed to
incubate for 3 minutes with rotation. Samples were then robotically dispensed
using a piezoelectric
dispensing device (SpectroJETTM (Sequenom, Inc.)) onto either 96-spot or 384-
spot silicon chips
containing a matrix that crystallized each sample (SpectroCHIPTM (Sequenom,
Inc.)). Subsequently,
MALDI-TOF mass spectrometry (Biflex and Autoflex MALDI-TOF mass spectrometers
(Bruker
Daltonics) can be used) and SpectroTYPER RTTM software (Sequenom, Inc.) were
used to analyze and
interpret the SNP genotype for each sample.
Genetic Anal
[0215] Minor allelic frequencies for the polymorphisms set forth in Table A
were verified as being
10% or greater using the extension assay described above in a group of samples
isolated from 92
individuals originating from the state of Utah in the United States, Venezuela
and France (Coriell cell
repositories).
[0216] Genotyping results are shown for female pools in Table 5. In Table 5,
"AF" refers to allelic
frequency; and "F case" and "F control" refer to female case and female
control groups, respectively.
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
TABLE 5: Genotyping Results
SNP Reference p-~alue
F c se F control
A=0.190 A=0.123
rs552 G = 0.810 G = 0.877 0.0011
A = 0.455 A = 0.375
rs12904 G = 0,545 G = 0.625 0.0012
C = 0.906 C = 0.939
rs2282146 T = 0.094 T = 0.061 0.0105
G=0.483 G=0.416
rs734784 A = 0.517 A = 0.584 0.0052
T=0.233 T=0.159
rs1042164 C = 0.767 C = 0.841 0.0002
C=0.342 C=0.419
rs749670 T = 0.658 T = 0.581 0.0038
T = 0.045 T = 0.076
rs955592 C = 0.955 ' C = 0.924 0.0177
T = 0.093 T = 0.054
rs1143016 C = 0.907 C = 0.946 0.0071
G=0.146 G=0.069
rs755248 A = 0.854 A = 0.931 0.0000
A = 0.432 A = 0.355
rs 1055055 G = 0.568 G = 0.645 0.0046
T=0.620 T=0.681
rs835409 G = 0.380 G = 0.319 0.0222
T=0.301 T=0.358
rs927663 G = 0.699 G = 0.642 0.0289
A=0.591 A=0.657
rs8162 G = 0.409 G = 0.343 0.0149
C=0.617 C=0.666
rs831038 T = 0.383 T = 0.334 0.0359
G=0.823 G=0.881
rs33079 A = 0.177 A = 0.119 0.0013
C=0.303 C=0.371
rs1710880 A = 0.697 A = 0.629 0.0129
T=0.818 T=0.875
rs1078153 A = 0.182 A = 0.125 0.0039
A=0.675 A=0.740
rs799570 G = 0.325 G = 0.260 0.0100
G=0.086 G=0.127
rs1282730 A = 0.914 A = 0.873 0.0150
T = 0.033 T = 0.055
rs1518875 C = 0.967 C = 0.945 0.0508
G=0.045 G=0.081
rs1568694 A = 0.955 A = 0.919 0.0064
A = 0.832 A = 0.769
rs905042 T = 0.168 T = 0.231 0.0047
G = 0.778 G = 0.839
rs1957723 A = 0.222 A = 0.161 0.0048
G = 0.273 G = 0.220
rs794018 A = 0.727 A = 0.780 0.0034
C=0.759 C=0.811
rs707723 T = 0.241 T = 0.189 0.0195
G=0.246 G=0.196
rs893861 A = 0.754 A = 0.804 0.0251
G=0.861 G=0.910
rs1914903 A = 0.139 A = 0.090 0.0055
C=0.064 C=0.117
rs2062232 T = 0.936 T = 0.883 0.0012
A = 0.777 A = 0.840
rs26609 T = 0.223 T = 0.160 0.0039
A=0.422 A=0.341
rs1370987 G = 0.578 G = 0.659 0.0007
G=0.876 G=0.833
rs1012414 A = 0.124 A = 0.167 0.0289
rs435903 G = 0.766 G = 0.685 0.0013
66
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
SNP ReferenceF case F co trol p-value
A=0.234 A=0.315
T = 0.668 T = 0.593
rs1248 A = 0.332 A = 0.407 0.0014
G=0.875 G=0.910
rs703508 A = 0.125 A = 0.090 0.0375
G=0.094 G=0.129
rs226465 C = 0.906 C = 0.871 0.0454
C=0.294 C=0.212
rs241448 T = 0.706 T = 0.788 0.0010
A=0.160 A=0.114
rs763155 C = 0.840 C = 0.886 0.0140
T = 0.069 T = 0.098
rs1040461 C = 0.931 C = 0.902 0.0281
A=0.218 A=0.145
rs462832 T = 0.782 T = 0.855 0.0008
T=0.583 T=0.679
rs804194 C = 0.417 C = 0.321 0.0004
A=0.169 A=0.103
rs1022646 G = 0.831 G = 0.897 0.0007
G=0.853 G=0.812
rs1569112 A = 0.147 A = 0.188 0.0468
A=0.097 A=0.140
rs805623 G = 0.903 G = 0.860 0.0143
A = 0.330 A = 0.240
rs 1019850 T = 0.670 T = 0.760 0.0005
A = 0.581 A = 0.659
rs1599931 G = 0.419 G = 0.341 0.0037
A = 0.506 A = 0.577
AA G = 0.494 G = 0.423 0.0102
T=0.100 T=0.138
rs279941 G = 0.900 G = 0.862 0.0324
C=0.778 C=0.717
rs1062230 T = 0.222 T = 0.283 0.0109
T = 0.295 T = 0.243
rs1859911 C = 0.705 C = 0.757 0.0328
T=0.861 T=0.809
rs1477261 A = 0.139 A = 0.191 0.0105
G=0.121 G=0.078
rs1191119 A = 0.879 A = 0.922 0.0079
A = 0.674 A = 0.583
rs657780 G = 0.326 G = 0.417 0.0009
G = 0.639 G = 0.724
rs1393890 C = 0.361 C = 0.276 0.0014
G=0.331 G=0.269
rs 1478714 A = 0.669 A = 0.731 0.0136
C=0.078 C=0.044
rs868213 T = 0.922 T = 0.956 0.0083
G = 0.839 G = 0.784
rs690115 A = 0.161 A = 0.216 0.0111
A = 0.846 A = 0.903
rs1465501 G = 0.154 G = 0.097 0.0020
C = 0.895 C = 0.858
rs899173 T = 0.105 T = 0.142 0.0408
C=0.087 C=0.146
rs 10477 T = 0.913 T = 0.854 0.0010
C=0.715 C=0.647
rs926393 T = 0.285 T = 0.353 0.0082
C=0.194 C=0.130
rs465271 T = 0.806 T = 0.870 0.0019
A=0.111 A=0.163
rs13847 G = 0.889 G = 0.837 0.0056
C = 0.898 C = 0.855
rs738658 A = 0.102 A = 0.145 0.0183
rs756519 C = 0.581 C = 0.656 0.0055
T=0.419 T=0.344
67
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
SNP Reference p-~alue
F c se F control
rs1042327 T = 0.472 T = 0.563 0
0012
C=0.528 C=0.437 .
rs8770 C = 0.529 C = 0.432 0
0001
T = 0.471 T = 0.568 .
rs1563055 C = 0.653 C = 0.736 0
0013
T = 0.347 T = 0.264 .
rs912428 T = 0.228 T = 0.170 0076
0
C=0.772 C=0.830 .
rs1888475 A = 0.188 A = 0.135 0
0087
G=0.812 G=0.865 .
[0217] All of the single marker alleles set forth in Table A were considered
validated, since the
genotyping data agreed with the allelotyping data and each SNP significantly
associated with
osteoarthritis. Particularly significant associations with osteoarthritis are
indicated by a calculated p-
value of less than 0.05 for genotype results.
Example 3
Association of Pol,~phic Variants with Osteoarthritis in Replication Cohorts
[0218] The single marker polymorphisms set forth in Table A were genotyped
again in two
replication cohorts consisting of individuals selected for OA.
Sample Selection and Poolin S~gies - Replication Sample 1
[0219] A second case control sample (replication sample #1) was created by
using 100 Caucasian
female cases from Chingford, UK, and 148 unrelated female cases from the St.
Thomas twin study.
Cases were defined as having I~ellgren-Lawrence (KL) scores of at least 2 in
at least one knee x-ray. In
addition, 199 male knee replacement cases from Nottingham were included. (For
a cohort description,
see the Nottingham description provided in Example 1). The control pool was
made up of unrelated
female individuals from the St. Thomas twin study (England) with normal knee x-
rays and without
other indications of OA, regardless of anatomical location, as well as lacking
family history of OA. The
St. Thomas twin study consists of Caucasian, female participants from the St.
Thomas' Hospital,
London, adult-twin registry, which is a voluntary registry of >4,000 twin
pairs ranging from 18 to
76 years of age. The replication sample 1 cohort was used to replicate the
initial results. Table 6 below
summarizes the selected phenotype data collected from the case and conh~ol
individuals.
TABLE 6
Phenotype Female cases (n=248):Male cases (n=199):Female controls
median (range)! median (range)/ (n=313):
(n,%) (n,%) mean (range)/ (n,%)
Age 59 (39- 73) 66 (45- 73) 55 (50- 72)
Height (cm) 162 (141- 178) 175 (152- 198) 162 (141- 176)
Weight (kg) 68 (51- 123) 86 (62- 127) 64 (40- 111)
Body mass 26 (18- 44) 29 (21- 41 ) 24 (18- 46)
index
68
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
Phenotype Female cases (n=248):Male cases (n=199):Female controls
(n=313):
median (range)/ median (range)/mean (range)/ (n,%)
(n,%) (n,%)
(kg/m')
Kellgren- 0 (63, 26%), 1
(20, 8%), 2
Lawrence* (105, 43%), 3 (58,NA NA
left 23%), 4
knee (1, 0%)
Kellgren- 0 (43, 7%), 1 (18,
7%), 2
Lawrence* (127, 52%), 3 (57,NA NA
right 23%), 4
knee (1, 0%)
KL* >2 both No (145, 59%), NA NA
Yes (101,
knees 41 %)
KL* >2 eitherNo (0, 0%), Yes NA NA
(248, 100%)
knee
* 0: normal, 1: doubtful, 2: definite osteophyte (bony protuberance), 3: joint
space narrowing
(with or without osteophyte), 4: joint deformity
Sample Selection and Pooling Strategies - Replication Sample 2
[0220] A third case control sample (replication sample #2) was created by
using individuals with
symptoms of OA from Newfoundland, Canada. These individuals were recruited and
examined by
rheumatologists. Affected joints were x-rayed and a final diagnosis of
definite or probable OA was
made according to American College of Rheumatology criteria by a single
rheumatologist to avoid any
inter-examiner diagnosis variability. Controls were recruited from volunteers
without any symptoms
from the musculoskeletal system based on a normal joint exam performed by a
rheumatologist. Only
cases with a diagnosis of definite OA were included in the study. Only
individuals of Caucasian origin
were included. The cases consisted of 228 individuals with definite knee OA,
106 individuals with
definite hip OA, and 74 individuals with hip OA.
TABLE 7
Phenotype Case Control
Age at Visit 62.7 52.5
Sex (Female/Male) 227/119 174/101
Knee OA Xray: No 35% (120) 80% (16)
Unknown 1 % (4) 0% (0)
Yes 64% (221 ) 20% (4)
Hip OA Xray: No 63% (215) 80% (16)
2 /0 7
Unknown ( ) 0 / 0
( )
Yes 35% (121) 20% (4)
Assay for Veri ins, Allelotypin~, and Genotyping SNPs
[0221] Genotyping of the replication cohorts described in Tables 6 and 7 was
performed using the
same methods used for the original genotyping, as described herein. A
MassARRAYTM system
69
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
(Sequenom, Inc.) was utilized to perform SNP genotyping in a high-throughput
fashion. This
genotyping platform was complemented by a homogeneous, single-tube assay
method (hMETM or
homogeneous MassEXTENDTM (Sequenom, Inc.)) in which two genotyping primers
anneal to and
amplify a genomic target surrounding a polymorphic site of interest. A third
primer (the
MassEXTENDTM primer), which is complementary to the amplified target up to but
not including the
polymorphism, was then enzymatically extended one or a few bases through the
polymorphic site and
then terminated.
[0222] For each polymorphism, SpectroDESIGNERTM software (Sequenom, Inc.) was
used to
generate a set of PCR primers and a MassEXTENDTM primer which where used to
genotype the
polymorphism. Other primer design software could be used or one of ordinary
skill in the art could
manually design primers based on his or her knowledge of the relevant factors
and considerations in
designing such primers. Table 3 shows PCR primers and Table 4 shows extension
probes used for
analyzing (e.g., genotyping) polymorphisms in the replication cohorts. The
initial PCR amplification
reaction was performed in a 5 p1 total volume containing 1X PCR buffer with
1.5 mM MgCl2 (Qiagen),
200 pM each of dATP, dGTP, dCTP, dTTP (Gibco-BRL), 2.5 ng of genomic DNA, 0.1
units of HotStar
DNA polymerase (Qiagen), and 200 nM each of forward and reverse PCR primers
specific for the
polymorphic region of interest.
[0223] Samples were incubated at 95°C for 15 minutes, followed by 45
cycles of 95°C for 20
seconds, 56°C for 30 seconds, and 72°C for 1 minute, finishing
with a 3 minute final extension at 72°C.
Following amplification, shrimp alkaline phosphatase (SAP) (0.3 units in a 2
p.1 volume) (Amersham
Pharmacia) was added to each reaction (total reaction volume was 7 p,1) to
remove any residual dNTPs
that were not consumed in the PCR step. Samples were incubated for 20 minutes
at 37°C, followed by
minutes at 85°C to denature the SAP.
[0224] Once the SAP reaction was complete, a primer extension reaction was
initiated by adding a
polymorphism-specific MassEXTENDTM primer cocktail to each sample. Each
MassEXTENDTM
cocktail included a specific combination of dideoxynucleotides (ddNTPs) and
deoxynucleotides
(dNTPs) used to distinguish polymorphic alleles from one another. Methods for
verifying, allelotyping
and genotyping SNPs are disclosed, for example, in U.S. Pat. No. 6,258,538,
the content of which is
hereby incorporated by reference. In Table 7, ddNTPs are shown and the fourth
nucleotide not shown is
the dNTP.
[0225] The MassEXTENDTM reaction was performed in a total volume of 9 p,1,
with the addition of
1X ThermoSequenase buffer, 0.576 units of ThermoSequenase (Amersham
Pharmacia), 600 nM
MassEXTENDTM primer, 2 mM of ddATP and/or ddCTP and/or ddGTP and/or ddTTP, and
2 mM of
dATP or dCTP or dGTP or dTTP. The deoxy nucleotide (dNTP) used in the assay
normally was
complementary to the nucleotide at the polymorphic site in the amplicon.
Samples were incubated at
94°C for 2 minutes, followed by 55 cycles of 5 seconds at 94°C,
5 seconds at 52°C, and 5 seconds
at 72°C.
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
[0226] Following incubation, samples were desalted by adding 16 p,1 of water
(total reaction
volume was 25 p.1), 3 mg of SpectroCLEANTM sample cleaning beads (Sequenom,
Inc.) and allowed to
incubate for 3 minutes with rotation. Samples were then robotically dispensed
using a piezoelectric
dispensing device (SpectroJETTM (Sequenom, Inc.)) onto either 96-spot or 384-
spot silicon chips
containing a matrix that crystallized each sample (SpectroCHl1'TM (Sequenom,
Inc.)). Subsequently,
MALDI-TOF mass spectrometry (Biflex and Autoflex MALDI-TOF mass spectrometers
(Bruker
Daltonics) can be used) and SpectroTYPER RTTM software (Sequenom, Inc.) were
used to analyze and
interpret the SNP genotype for each sample.
Genetic Analysis
[0227] Genotyping results for replication cohorts #1 and #2 are provided in
Tables 8 and 9,
respectively.
TABLE 8
Replication Meta-analysis
rslD #1 Disc. + Rep
(Mixed #1
MaleIFemale P-value
cases
and Female
controls)
AF OA Con
AF OA
Cas Delta
P-value
rs552 0.87 0.85 0.02 0.344 0.0300
rs12904 0.57 0.57 0.00 0.936 0.2700
rs22821460.08 0.1 -0.020.342 0.0190
rs734784 0.52 0.54 -0.020.451 0.7200
rs10421640.79 0.82 -0.030.161 0.9100
rs749670 0.62 0.66 -0.040.173 0.0019
rs955592 0.93 0.94 -0.010.521 0.0600
rs11430160.93 0.93 0.00 0.869 NA
rs755248 0.9 0.89 0.01 0.544 0.1600
rs10550550.64 0.64 0.00 0.947 0.3300
rs835409 0.34 0.35 -0.010.715 0.1300
rs927663 0.64 0.65 -0.010.611 0.0690
rs831038 0.35 0.37 -0.020.399 NA
rs33079 0.14 0.14 0.00 0.995 0.3100
rs17108800.66 0.62 0.04 0.087 0.9000
rs799570 0.29 0.29 0.00 0.903 0.2500
rs12827300.88 0.87 0.01 0.751 0.4800
rs15686940.93 0.94 0.00 0.928 0.2600
rs905042 0.21 0.2 0.01 0.829 0.2200
rs19577230.13 0.16 -0.030.124 0.0009
rs794018 0.74 0.72 0.02 0.518 0.0710
rs707723 0.18 0.19 -0.010.658 0.0650
rs19149030.15 0.14 0.01 0.605 0.5500
rs20622320.91 0.91 0.00 0.788 0.2100
rs26609 0.16 0.19 -0.020.226 0.0032
rs13709870.63 0.63 -0.010.857 0.3900
rs10124140.12 0.13 -0.010.669 0.5600
rs435903 0.27 0.27 0.00 0.950 0.2800
rs 1248 0.36 0.36 0.00 0.917 0.2400
~
71
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
Replication Meta-analysis
rslD #1 Disc. + Rep
(Mixed #1
MaleIFemale P-value
cases
and Female
controls)
AF OA
Con AF
OA Cas
Delta
P-value
rs703508 0.11 0.12 -0.010.558 0.0660
rs226465 0.87 0.88 -0.010.436 0.0500
rs241448 0.74 0.75 -0.010.805 0.4100
rs763155 0.86 0.88 -0.020.273 0.8800
rs1040461 0.92 0.92 0.00 0.826 NA
rs1022646 0.85 0.87 -0.020.219 0.8200
rs1569112 0.16 0.18 -0.020.402 0.8800
rs805623 0.87 0.88 -0.010.460 0.0370
rs1019850 0.69 0.7 0.00 0.890 0.3700
AA 0.47 0.48 -0.010.681 0.1200
rs279941 0.87 0.89 -0.010.400 0.0340
rs1062230 0.26 0.26 0.00 0.896 0.4200
rs1859911 0.71 0.75 -0.040.128 0.9000
rs1477261 0.16 0.16 0.00 0.986 0.3000
rs 11911190.89 0.88 0.01 0.569 0.1200
rs1393890 0.29 0.31 -0.020.527 0.1400
rs1478714 0.69 0.67 0.03 0.300 0.0140
rs868213 0.92 0.93 -0.010.455 0.7000
rs690115 0.2 0.21 -0.010.729 0.4900
rs1465501 0.11 0.1 0.01 0.718 0.5600
rs899173 0.1 0.11 0.00 0.924 0.3300
rs10477 0.89 0.88 0.01 0.691 0.4700
rs926393 0.3 0.31 -0.010.830 0.4200
rs465271 0.86 0.85 0.01 0.516 0.0660
rs13847 0.86 0.85 0.01 0.547 0.5900
rs738658 0.14 0.15 -0.010.536 0.6700
rs756519 0.4 0.43 -0.040.140 0.0098
rs1042327 0.49 0.52 -0.030.234 0.0430
rs8770 0.51 0.48 0.03 0.303 0.0480
rs1563055 0.31 0.35 -0.040.083 0.0002
rs912428 0.86 0.8 0.06 0.004 0.00001
rs1888475 0.86 0.81 _ 0.032 0.0002
I
0.04
TABLE 9
Replication Meta-analysis
rslD #2 (Newfoundland) Disc. + Rep
(MaleIFemale #2
cases Not Done
and controls)
AF OA Con
AF OA
Cas Delta
P-value
rs552 0.85 0.86 -0.0140.496
rs12904 0.58 0.57 0.011 0.719
rs22821460.08 0.08 0.002 0.876
rs734784 0.53 0.54 -0.0030.907
rs10421640.83 0.80 0.026 0.248
rs749670 0.66 0.62 0.036 0.208
rs955592 0.95 0.92 0.033 0.027
rs 11430160.96 0.94 0.015 0.236
rs755248 0.89 0.90 -0.0090.608
72
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
Replication Meta-analysis
rslD #2 (Newfoundland) Disc. + Rep
(MaleIFemale #2
cases Not Done
and controls)
AF OA
Con AF
OA Cas
Delta
P-value
rs 10550550.64 0.61 0.034 0.249
rs835409 0.36 0.31 0.047 0.101
rs927663 0.67 0.68 -0.013Ø631
rs831038 0.34 0.35 -0.0140.612
rs33079 0.17 0.19 -0.0190.417
rs1710880 0.64 0.62 0.029 0.309
rs799570 0.35 0.30 0.058 0.033
rs1282730 0.89 0.89 -0.0010.982
rs1568694 0.95 0.94 0.009 0.518
rs905042 0.19 0.20 -0.0020.933
rs1957723 0.18 0.20 -0.0170.454
rs794018 0.73 0.72 0.015 0.586
rs707723 0.20 0.21 -0.0070.759
rs1914903 0.14 0.16 -0.0220.285
rs2062232 0.92 0.91 0.008 0.632
rs26609 0.19 0.18 0.005 0.827
rs1370987 0.59 0.61 -0.0230.423
rs 10124140.15 0.14 0.008 0.679
rs435903 0.24 0.26 -0.0260.316
rs1248 0.33 0.38 -0.0510.078
rs703508 0.10 0.11 -0.0020.916
rs226465 0.89 0.89 -0.0070.699
rs241448 0.76 0.77 -0.0070.778
rs763155 0.89 0.84 0.049 0.016
rs 10404610.91 0.91 0.001 0.948
rs1022646 0.86 0.86 -0.0010.974
rs1569112 0.16 0.17 -0.0160.446
rs805623 0.89 0.87 0.022 0.256
rs1019850 0.71 0.69 0.026 0.341
AA 0.48 0.44 0.035 0.234
rs279941 0.91 0.87 0.037 0.047
rs1062230 0.23 0.22 0.011 0.653
rs1859911 0.72 0.71 0.015 0.560
rs1477261 0.17 0.14 0.031 0.143
rs1191119 0.86 0.88 -0.0170.377
rs1393890 0.30 0.28 0.017 0.516
rs1478714 0.68 0.70 -0.0250.358
rs868213 0.91 0.93 -0.0190.260
rs690115 0.19 0.18 0.005 0.811
rs1465501 0.10 0.12 -0.0200.282
rs899173 0.14 0.12 0.020 0.319
rs10477 0.86 0.88 -0.0160.442
rs926393 0.37 0.32 0.042 0.137
rs465271 0.87 0.85 0.023 0.263
rs13847 0.84 0.85 -0.0120.582
rs738658 0.18 0.15 0.021 0.340
rs756519 0.39 0.40 -0.0070.816
rs1042327 0.49 0.51 -0.0240.405
rs8770 0.53 I 0.49 0.039 0.195
73
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
Replication Meta-analysis
rslD #2 (Newfoundland) Disc. + Rep
(MaleIFemale #2
cases Not Done
and controls)
AF OA Con
AF OA
Cas Delta
P-value
rs15630550.34 0.34 -0.0050.864
rs912428 0._82 0.76 0.058 0.016
- -.
~ ~ _ -0.0250.280
rs1888475x.80 0.82 I
[0228] To combine the evidence for association from multiple sample
collections, a meta-analysis
procedure was employed. The allele frequencies were compared between cases and
controls within the
discovery sample, as well as within the replication cohort #1 using the
DerSimian-Laird approach
(DerSimonian, R. and N. Laird. 1986. Meta-analysis in clinical trials. Control
Clin Trials 7: 177-188.)
[0229] The absence of a statistically significant association in one or more
of the replication
cohorts should not be interpreted as minimizing the value of the original
fording. There are many
reasons why a biologically derived association identified in a sample from one
population would not
replicate in a sample from another population. The most important reason is
differences in population
history. Due to bottlenecks and founder effects, there may be common disease
predisposing alleles
present in one population that are relatively rare in another, leading to a
lack of association in the
candidate region. Also, because common diseases such as arthritis-related
disorders are the result of
susceptibilities in many genes and many environmental risk factors,
differences in population-specific
genetic and environmental backgrounds could mask the effects of a biologically
relevant allele. For
these and other reasons, statistically strong results in the original,
discovery sample that did not replicate
in one or more of the replication samples may be further evaluated in
additional replication cohorts and
experimental systems.
Example 4
KIAA0296 Reeion Proximal SNPs
[0230] SNP rs749670 is associated with osteoarthritis and is described in
Table A. It lies within
the KIAA0296 gene and codes for a G327E amino acid change. The thymine allele
of SNP rs749670 is
associated with osteoarthritis (see Table 5) and codes for glutamic acid.
KIAA0296 shares homology
with C2H2-type Zn-finger protein and is likely a novel transcription factor.
One-hundred one additional
allelic variants proximal to rs749670 were identified and subsequently
allelotyped in osteoarthritis case
and control sample sets as described in Examples 1 and 2. The polymorphic
variants are set forth in
Table 10. The chromosome positions provided in column four of Table 10 are
based on Genome
"Build 34" of NCBI's GenBank.
TABLE 10
dbSNP Position ChromosomeAllele
Chromosomein SEQ
rs# ID NO: Position Variants
1
rs750017616 247 31077197 a/
rs656521216 1535 31078485 c/t
74
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Position ChromosomeAllele
rs# Chromosomein SEQ Position Variants
1D NO:
1
rs805404616 2386 3107933_6 c/t
rs805684216 6440 3108339 c/t
0
rs73217316 9133 _ It
31086083
rs73217216 9143 31086093 a/
rs718855716 9471 3108642 alt
1
rs228800416 13150 _ c/
31090100
rs433731016 13717 31090667 c/t
rs201655416 14466 31091416 al
rs656521316 15769 31092719 a/c
rs720476216 16870 31093820 a/
rs488952916 18545 31095495 clt
rs656521416 18749 31095699 c/t
rs749967416 19123 31096073 /t
rs656521516 20736 31097686 al
rs102362316 21038 31097988 c/t
rs102362416 21046 31097996 c/t
rs102362516 21050 31098000 c/t
rs154929716 21056 31098006 alt
rs308489416 21706 31098656 -/acc
rs804822816 23170 31100120 a/
rs740543216 25028 31101978 a/t
rs805424916 27871 31104821 a/
rs806104716 28070 31105020 c/t
rs718722016 31717 31108667 a/
rs804697816 32019 31108969 a/
rs228800316 32318 31109268 a/
rs719642116 33080 31110030 a/
rs719643116 33101 31110051 a/
rs720315816 34236 31111186 a/
rs230322316 34285 31111235 c/t
rs203291716 34818 31111768 c/
rs804413416 35168 31112118 c/
rs488953116 37981 31114931 c/t
rs488953216 38113 31115063 c/
rs488953316 38117 31115067 c/t
rs88192916 38481 31115431 /t
rs804710416 38615 31115565 c/
rs804780316 38944 31115894 a/c
rs464487416 39288 31116238 a/c
rs235967316 41385 31118335 c/t
rs443527116 42136 31119086 a/t
rs719771716 42185 31119135 a/c
rs235967416 42353 31119303 a/
rs656521716 42434 31119384 a/
rs230322216 44580 31121530 a/
rs488961516 44675 31121625 alt
rs462419716 45739 31122689 /t
rs375185316 46439 31123389 c/t
rs74967116 47457 31124407 c/t
rs74967016 47735 31124685 c/t
rs375185516 50319 31127269 clt
rs375185616 50708 31127658 a/
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP ChromosomePosition ChromosomeAllele
rs# in SEQ Position Variants
ID NO:
1
rs719672616 51185 31128135 a/
rs88955016 53002 31129952 a/
rs75095216 53064 31130014 c/t
rs207763316 53637 31130587 a/
rs719994916 55274 31132224 c/
rs203291616 55825 31132775 c/t
rs446864116 55986 31132936 a/c
rs488953516 56684 31133634 c/
rs431677516 57653 31134603 c/t
rs431381916 57659 31134609 c/
rs656521816 57692 31134642 /t
rs431822416 57775 31134725 c/t
rs104603016 61313 31138263 clt
rs7294 16 61431 31138381 a/
rs720074916 61699 31138649 al
rs235961216 62906 31139856 a/
rs805089416 63619 31140569 c/
rs288473716 64664 31141614 a/c
rs189551416 68452 31145402 /t
rs806020916 69665 31146615 c/t
rs806021716 69681 31146631 c/t
rs719616116 70091 31147041 a/
rs806233616 74637 31151587 a/
rs804377816 74760 31151710 a/
rs203291516 76523 31153473 a/
rs488961616 78559 31155509 c/
rs104556416 79549 31156499 a/c
rs230322116 79882 31156832 c/t
rs154929616 81339 31158289 a/
rs88955516 81681 31158631 c/t
rs581652116 81696 31158646 -/
rs74976716 83517 31160467 c/t
rs288473816 85431 31162381 a/c
rs205258116 86332 31163282 c/t
rs488961716 87358 31164308 a/
rs488961916 87725 31164675 c/t
rs197848716 89052 31166002 a/
rs197848616 90020 31166970 a/
rs197848516 90231 31167181 al
rs488962016 90284 31167234 a/
rs488962116 90447 31167397 c/t
rs321447716 90601 31167551 -!
rs452703416 90724 31167674 a/
rs106050616 92559 31169509 c/t
rs720012516 95176 31172126 a/
rs656521916 95195 31172145 c/t
rs88954816 96822 31173772 a/
76
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
Assay for Verifying and Allelotypin~NPs
[0231] The methods used to verify and allelotype the 101 proximal SNPs of
Table 10 are the same
methods described in Examples 1 and 2 herein. The primers and probes used in
these assays are
provided in Table 11 and Table 12, respectively.
TABLE 11
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs7500176ACGTTGGATGACAGTGGCTCATGCCTGTAAACGTTGGATGTTTCACCATATTGGCCAGGC
rs6565212ACGTTGGATGTTAGGAAGGATGTGGAAGGGACGTTGGATGGACCTGACCTCAAAGAGAAG
rs8054046ACGTTGGATGCACTGAAGTTTAGAGCAGCCACGTTGGATGTGCACAGTGGGTAACTGTAG
rs8056842ACGTTGGATGATGAGGTTTCACCTTGTTGGACGTTGGATGATCATAGCACTTTGCGAGGC
rs732173ACGTTGGATGAGACCAGGCTCAGTCCAAACACGTTGGATGTGGCCAAACCTGGAAGACAC
rs732172ACGTTGGATGCTCAGTCCAAACTGCCAGACACGTTGGATGCATGGCCAAACCTGGAAGAC
rs7188557ACGTTGGATGAACATCTGTACAAGGCTGGGACGTTGGATGATTGGCTGTAGCATGACTGA
rs2288004ACGTTGGATGAAAGACACTGGAAGGCTGTGACGTTGGATGAGAGAAGGTGGAGCTCTTTC
rs4337310ACGTTGGATGAGGGAAGAGATGTACACAGGACGTTGGATGTTTGGAGCAGATCTGGTAGG
rs2016554ACGTTGGATGAAGCAATCCTCCCACCTCAGACGTTGGATGCAAGAGCAAAACTCCCTCTC
rs6565213ACGTTGGATGAGATGGAGTCTCACTCCATCACGTTGGATGTGAGGCAGGAGAATCGCTTG
rs7204762ACGTTGGATGAGTGGCTCACACCTGTAATCACGTTGGATGGCTGGTCTTGAACTTCTGAC
rs4889529ACGTTGGATGCAAGCAATCCTTGCCTCAAGACGTTGGATGGGTGGTTCACATCTGCAATC
rs6565214ACGTTGGATGTGATCTCGGCTCACTGCAAGACGTTGGATGAAAATTAGCCGGGCATGGTG
rs7499674ACGTTGGATGAACTAGGGAACTCTTCCCACACGTTGGATGTGGGCCCCACTAAGTCTAAA
rs6565215ACGTTGGATGAGACGGAAAGTTCCAGCTTGACGTTGGATGTGGGACCACTCTGTTCTATG
rs1023623ACGTTGGATGACAGAGCAAGACTCCATCTCACGTTGGATGTCCTCTTCAGAGCTGTTCAC
rs1023624ACGTTGGATGTGACAGAGCAAGACTCCATCACGTTGGATGGTCCTAACCAGTGAGCCTAT
rs1023625ACGTTGGATGTGGTGACAGAGCAAGACTCCACGTTGGATGTCAGGTCCTAACCAGTGAGC
rs1549297ACGTTGGATGTTGCATTGATCCGAGATCGCACGTTGGATGTCAGGTCCTAACCAGTGAGC
rs3084894ACGTTGGATGTCCCAGGTTCAAGCGATTCTACGTTGGATGCCATGAAACCCCATCTCTAC
rs8048228ACGTTGGATGAATTGCTTGAACCTGGGAGGACGTTGGATGTTCGACAGTCTCCCTCTATC
rs7405432ACGTTGGATGAGATCATGCCACTGCACTACACGTTGGATGCACTGCACTTGGCCTAATTG
rs8054249ACGTTGGATGATCTCCTGACCTCATGATCTACGTTGGATGTAATCAAACACCAGGCTGGG
rs8061047ACGTTGGATGATGATCACAGCTCACTGCAGACGTTGGATGCTCCCTGCCTCTACAAAAAG
rs7187220ACGTTGGATGAAGGAGACCTTCTCCACAATACGTTGGATGCCGGTCAGAGAAGCTCTTGC
rs8046978ACGTTGGATGTGCACAGGAGCTGGTGGTGACGTTGGATGATCACACCACCTGACTCCGG
rs2288003ACGTTGGATGACCGGCCGTTCAAGTGCCTGACGTTGGATGAGAGTGCACCAGCGCGTGC
rs7196421ACGTTGGATGTTCACGCCATTCTCCTGCCTACGTTGGATGAAATTAGCCAGGCGTGGTGG
rs7196431ACGTTGGATGAGATCTCGGCTCACTGCAAGACGTTGGATGATGTAGTCCCAGCTACTCGG
rs7203158ACGTTGGATGAAGCCTATGCGGAGCTCAAGACGTTGGATGATTGGCTGCAGCAACGCTGT
rs2303223ACGTTGGATGACCCTCACCGCTCATGGTTGACGTTGGATGTGCGGCCCTACAGCTGTGA
rs2032917ACGTTGGATGCCTGGGCGCGTTTGGAAATGACGTTGGATGAGCCCCCGGCTACAAGCGCT
rs8044134ACGTTGGATGACTAAGAAAGGAGGCTGAGGACGTTGGATGACAGTGTTTGGAAAAGCCCG
rs4889531ACGTTGGATGATTCCTCACCCAACTCTGTCACGTTGGATGGACCGTGTGTAATGTACTGC
rs4889532ACGTTGGATGGGGACAAGAATCCCTATCTCACGTTGGATGTAGAGCCAGACACATTGCTG
rs4889533ACGTTGGATGCTCTGTAAAGTAGGGACAAGACGTTGGATGTAGAGCCAGACACATTGCTG
rs881929ACGTTGGATGTTGACCCAGTGGTTCTGAGCACGTTGGATGCCAGCTACCTGGTGTCTAAC
rs8047104ACGTTGGATGGTGGGATGTTAGACAGAGACACGTTGGATGTGCCAGGTTGGTCTCAGCAT
rs8047803ACGTTGGATGAAAGTGCTGGGATTACAGGCACGTTGGATGAAATACAGATTCCTGAGGCC
rs4644874ACGTTGGATGAGTCTTGCTATGTTGCCTGGACGTTGGATGTAATCCCAGCACTTTGGGAG
rs2359673ACGTTGGATGGTGTGATGTCAGTTCACTGCACGTTGGATGATCCCAAATACTTGGGAGGC
rs4435271ACGTTGGATGACAGTGGTCTCAAGAACTCCACGTTGGATGTGGCTCATGCCTGTAATCAC
77
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs7197717ACGTTGGATGTGTGATTACAGGCATGAGCCACGTTGGATGGCTTGCAAGGAGTATTGTCC
rs2359674ACGTTGGATGGCCTAGCAGTTCATTATGAGACGTTGGATGCCTTGTCTCCAAATACAGTC
rs6565217ACGTTGGATGAAGAACGCTAATCCTACTGGACGTTGGATGTGGAGACAAGGCCTTTATGG
rs2303222ACGTTGGATGTTGGGAAAAGTCCTCCAGAGACGTTGGATGGCGCAGAAAGGGAGAAAAAG
rs4889615ACGTTGGATGTAAGTTCTAGGTCTGCACGGACGTTGGATGATGCACCGGAACGATTCTAG
rs4624197ACGTTGGATGCGCTAAGAGAGTCTTTTGGGACGTTGGATGCAGAGCGAGACTCCATCTCA
rs3751853ACGTTGGATGTCCCCTAGGCTTAAGTCATCACGTTGGATGGGTCTGTGATCAGAAGTAGG
rs749671ACGTTGGATGTGACTACATTTGTACCGCCGACGTTGGATGTCAGTAGTGAACTTCACAGG
rs749670ACGTTGGATGTCTCATCTGTGTGCCCATTGACGTTGGATGATGAGGGTGAAAGGCAGGAG
rs3751855ACGTTGGATGAAGAAGAGGTGTGGGAGGAGACGTTGGATGTCAGAGCTGGCTTCAGTCTG
rs3751856ACGTTGGATGAGCTGTACTGGCCCGTCTCGACGTTGGATGCAGTGCGGGCGGACCTATC
rs7196726ACGTTGGATGGACCTAGTTAGGAACTGAGGACGTTGGATGTCAGGGCAGCAAGCTCAGAAG
rs889550ACGTTGGATGTCCACCCAGCACTGCTGGAACGTTGGATGCAGGTCCTGCTGAGGGAAC
rs750952ACGTTGGATGTTCCCTCAGCAGGACCTGGACGTTGGATGGGTGGCCACTAGATGGAATG
rs2077633ACGTTGGATGTTTCTCAGGAGTAGTTCGGGACGTTGGATGAAAGAAGCCAGATCTGGGTC
rs7199949ACGTTGGATGTCCCCATCAGGCAGGTGGTACGTTGGATGCAGCCTGTGACACTGGGAG
rs2032916ACGTTGGATGGTTCCCCTCATTACTGAAGGACGTTGGATGTGCCACTTGCCTGTAGTTAC
rs4468641ACGTTGGATGATGAGTCAGGAATACGGGAGACGTTGGATGAATGCCCCTACTTGTCACTC
rs4889535ACGTTGGATGCTATGGCAGACACCCTCTGAACGTTGGATGGAAGAGAAGGAGCAGAAGGG
rs4316775ACGTTGGATGAGTAGCTCACGCTTGTAATCACGTTGGATGCTATGTTGCACAGGCTAGTC
rs4313819ACGTTGGATGTGCACAGGCTAGTCTTGAACACGTTGGATGAGTAGCTCACGCTTGTAATC
rs6565218ACGTTGGATGTTAAAGTCACAGACTGAGGCACGTTGGATGTTGAACTCTTGGGCTCAAGC
rs4318224ACGTTGGATGTCAGTCTGTGACTTTAAGCGACGTTGGATGACCACCTTTCATGGTAGAAG
rs1046030ACGTTGGATGGTCTCCAAAGCTCTTTCCATTACGTTGGATGGATTGATCTAAGAAACTTTA
rs7294 ACGTTGGATGGCACTGGGTGTAAAAAAGAGACGTTGGATGTTCTAGATTACCCCCTCCTC
rs7200749ACGTTGGATGGAGCACGAAGAACAGGATCCACGTTGGATGTCTGTCCTGATGCTGCTGAG
rs2359612ACGTTGGATGAAATCGGCCAAGTCTGAACCACGTTGGATGTCCAGAGAAGGCATCACTGA
rs8050894ACGTTGGATGAATCTTGGTGATCCACACAGACGTTGGATGTAGTTACCTCCCCACATCCC
rs2884737ACGTTGGATGTCATTATGCTAACGCCTGGCACGTTGGATGTTGACGATGGTCTCAAGGAC
rs1895514ACGTTGGATGCAATCTCAGCTCACTGCAACACGTTGGATGTAATCCCAGCTACTTGGGAG
rs8060209ACGTTGGATGGGTCAGGAGTTTAAGACAAGACGTTGGATGCCATGCCCGGCTAATTTTTG
rs8060217ACGTTGGATGTGAGTAGCTGGGATTACAGGACGTTGGATGAGACAAGCTTGGCCAACATG
rs7196161ACGTTGGATGGTGTTTTTAGTAGAGACGGGACGTTGGATGATCCCAGCACTTTAGGAAGC
rs8062336ACGTTGGATGTGCTCCCCACATCTCAGACGACGTTGGATGAAGCGAGGAGCGCCTCTTC
rs8043778ACGTTGGATGTTCCTCACTTCTCAGACGGGACGTTGGATGATCGTCTGAGATGTGGGGAG
rs2032915ACGTTGGATGATTCCCACCCGTTCTTTCCCACGTTGGATGTTCCCGCTCCCTTTTACCAC
rs4889616ACGTTGGATGGAACCAAGAACTGGAAGGAGACGTTGGATGTGTAAAGCGCACAGATCACG
rs1045564ACGTTGGATGTGTCAGCATCCTCGACGCACACGTTGGATGACCCAGGCGACCCAAAATGG
rs2303221ACGTTGGATGAGAACCCCCAACACTCTCCCACGTTGGATGAGCGGAGAAGGTGCGCAAG
rs1549296ACGTTGGATGATGCTGCTGAACTTCCTAACACGTTGGATGAGCAGGGTTTCTCAACCATG
rs889555ACGTTGGATGAGACCAGTAGGTACAAGCACACGTTGGATGTCAAGAATGCCATGAGGTGG
rs5816521ACGTTGGATGATTGTGGCTCTATGCAGAGGACGTTGGATGTCAAGAATGCCATGAGGTGG
rs749767ACGTTGGATGCTGATAGAAAGGACCAAGGAACGTTGGATGCTGGAGTTCTGATTCAGGTC
rs2884738ACGTTGGATGAGAACTGCTTGAACCCAGGAACGTTGGATGATGGAGTCTTGTTGTGTCGG
rs2052581ACGTTGGATGTGGGACATGCGGATATGGAGACGTTGGATGGAGGGTTCTGTGAGAGTCAG
rs4889617ACGTTGGATGCAGAGCGAGACTCCATCTCAACGTTGGATGACACTCGCGCTGGCCTAATG
rs4889619ACGTTGGATGAAAATTAGATGGGCGTGGTGACGTTGGATGATCTCGGCTCACTGCAACCT
rs1978487ACGTTGGATGTCCCTTCTCTATGTTCCTGCACGTTGGATGATGGAGGAAGACAGAGAGAG
rs1978486ACGTTGGATGTACCTAGGGTCACAGATTTGACGTTGGATGGGGTATGTGGTAAAATGAGC
rs1978485ACGTTGGATGTCAAGCAATTTTCCTGCCTCACGTTGGATGCCATCTGTACCAAAAAGACG
rs4889620ACGTTGGATGTGGCAAAACCCCATCTGTACACGTTGGATGAGTAGTTGGGATTACAGGTG
rs4889621ACGTTGGATGTACTCAATCACTGCCACAACACGTTGGATGGCCAGTTATTTTCTCATTCG
78
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs3214477ACGTTGGATGACTCGAGACTGGATCACTTCACGTTGGATGCCTTTTGTTCCAGCCTTACC
rs4527034ACGTTGGATGAAGTATGGGCCATAAGAGTGACGTTGGATGTATGTACACTACGTGGGCTG
rs1060506ACGTTGGATGATCAGGAGTGCAAACCAGAGACGTTGGATGGGATGAAGCTGCAATAGCTG
rs7200125ACGTTGGATGATTTTGCCATTGCACTCCAGACGTTGGATGTACAGGCATGAGCCATAGCC
rs6565219ACGTTGGATGCTTGGCCTCTCAAAGTGCTGACGTTGGATGAGGGCGAGGCTCCATTTCAA
rs889548ACGTTGGATGCTGGCCAAGTCCTAATACAGACGTTGGATGCCCAATTCCAGAGATGTCAG
'TABLE 12
dbSNP Extend Term
rs# Primer Mix
rs7500176 GATCACGAGGTCAGGAGTTC ACT
rs6565212 GCTGGAAAACTGTTGAGGGT ACT
rs8054046 TTTAGAGCAGCCGATACCCA ACG
rs8056842 GCTGGTCTCGAACTCCTGA ACG
rs732173 GCTCAGTCCAAACTGCCAG CGT
rs732172 ACTGCCAGACTCCCGCCA ACG
rs7188557 CCTGGCCCTGGTTGTGAGT CGT
rs2288004 CGGCAGATCCAGTGTGTC ACT
rs4337310 CACGGAATCTCCAGTGCAC ACT
rs2016554 GGCACGTACCACTGACATG ACG
rs6565213 GCAGTGGCGCAATCTTGAC ACT
rs7204762 CCCAGCACTTTGGGAGGC ACG
rs4889529 CTCAAGTGATCCTCCTGCCT ACG
rs6565214 GAGTAGCTGGGACTACAGG ACG
rs7499674 GTTCTTCTCAACATCTGCCCA ACT
rs6565215 TTTCCTTCAGACAGGGCTCT ACT
rs1023623 GACTCCATCTCAAAAAAAAAAAAAACT
rs1023624 GAGCAAGACTCCATCTCAAAAA ACT
rs1023625 CAGAGCAAGACTCCATCTCA ACT
rs1549297 GGTGACAGAGCAAGACTCC CGT
rs3084894 CGAGTAGGTGGGACTACAG ACT
rs8048228 TGAGCCGAGATGGCAACAC ACG
rs7405432 CTACAGGCTAGGAGACAGAG CGT
rs8054249 AAAGTGCTGGGATTACAGGC ACT
rs8061047 CCTCCTGAGGAGCTGGTCT ACT
rs7187220 GGCCCTTCCCCTGCACC ACG
rs8046978 AGAGTTCAGCCGCCCCGG ACG
rs2288003 GTGACAAGACGTTCGTGGC ACT
rs7196421 CTCAGCCTCCCGAGTAGC ACG
rs7196431 CGGGTTCACGCCATTCTCC ACG
rs7203158 CAACCATGAGCGGTGAGGG ACG
rs2303223 TTGAGCTCCGCATAGGCTTT ACT
rs2032917 TGGAAATGTCTTGGTACAGGACA ACT
rs8044134 CCTACACGTCCCCCCCC ACT
rs4889531 CAACTCTGTCAGGTAAGTACT ACT
rs4889532 CAAGAATCCCTATCTCAGAAAG ACT
rs4889533 GGACAAGAATCCCTATCTCAG I ACT
79
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Extend Term
rs# Primer Mix
rs881929 CTGCCTCTTGCCAGCTCTG ACT
rs8047104 CAGAGACCTAGCCTACCTG ACT
rs8047803 TTACAGGCGAGAGCCACCA CGT
rs4644874 GGGCTCAAGTGATCCTCCC CGT
rs2359673 ACTGCGACCTCTGCCTCC ACG
rs4435271 GCTTCAGATGCTCCTCCACT CGT
rs7197717 GCATGAGCCGTGACCAGC CGT
rs2359674 GAATGTTTGTGTTCCCTGTCC ACT
rs6565217 CCAGGGCCATACCCTTATGA ACG
rs2303222 AAAGTGTCACCAAAGTAC ACG
rs4889615 GCGGCGTCTTTGCACGCTA CGT
rs4624197 AGAGAGTCTTTTGGGGTTTTTT ACT
rs3751853 CCTACAGGTATAGCTAAGGAA ACT
rs749671 ATTTGTACCGCCGCTCCTC ACG
rs749670 GGTGGTGGGCATCCCTTTC ACG
rs3751855 AGAGCCCAGGCTGGAGAC ACG
rs3751856 CCGTCTCGTGGCTGCGC ACG
rs7196726 GTTAGGAACTGAGGAACCCAG ACG
rs889550 AGCACTGCTGGAAGCCGC ACT
rs750952 GCTGGCCTCTCCACCTCC ACG
rs2077633 CCATATCTTCTCCTCTCCCC ACG
rs7199949 CAGGCAGGTGGTGGTCAG ACT
rs2032916 CCAAAGTTCCAGAGAGGTTAA ACT
rs4468641 ATACGGGAGGCAGGCCCA ACT
rs4889535 CAGACACCCTCTGATTGCAG ACT
rs4316775 GAGGATCGCTTGAGCCCAA ACT
rs4313819 GCTAGTCTTGAACTCTTGGG ACT
rs6565218 CTCACGCTTGTAATCCCAGC CGT
rs4318224 TTCCCTTGCAACCTGAGTTTT ACG
rs1046030 GCCCAGGGAGGGAAGGTT ACG
rs7294 TTGGTCCATTGTCATGTG ACG
rs7200749 GAAGAACAGGATCCAGGCCA ACT
rs2359612 CCATGTGTCAGCCAGGACC ACT
rs8050894 CCAGCTAGCTGCTCATCAC ACT
rs2884737 TCGCCAACACCCCCCTTC CGT
rs1895514 CCCCTCTCGGGTTCAAGC CGT
rs8060209 TGGCCAACATGGCGAAACC ACG
rs8060217 CCATGCCCGGCTAATTTTTGT ACT
rs7196161 AACTCCTGACCTCATGATCC ACT
rs8062336 TCACTTCCTAGATGGGAAGG ACG
rs8043778 CGCTCCTCACCTCCCAGA ACG
rs2032915 TTCTTTCCCAACGTCCTGGA ACT
rs4889616 GAACTGGAAGGAGGACAAGA ACT
rs1045564 GTCCCTGAAGTCGGAGAAG CGT
rs2303221 CTCTCCCTCCCGCCTACAT ACG
rs1549296 TGCACGGGGCAGCCCCT ACT
rs889555 AGCACCCCGGTTCCTGTCC ACT
rs5816521 CCAGTA ACT
GGTACAAGCACCC
rs749767 _ ~ ACT
I GACCAAGGATTTGGGCAAAG
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Extend Term
rs# Primer Mix
rs2884738 CCAGGAGGTGGAGGTTGCA ACT
rs2052581 GGATATGGAGGGCCGATTGT ACT
rs4889617 GAGACTCCATCTCAAAAAAAAAA ACT
rs4889619 GCAGAGGAATCGCTTGAACC ACG
rs1978487 GTTCCTGCAACATTTTTTTCCTA ACG
rs1978486 GGGTCACAGATTTGAAAAGTG ACT
rs1978485 TTTTCCTGCCTCAGCCTCC ACG
rs4889620 ACCCCATCTGTACCAAAAAGA ACG
rs4889621 CTGTGAGGTGGATCAGGTTG ACT
rs3214477 GCAGAATCTGTGATGGAAAAAG ACT
rs4527034 CCAGGGCAGCCAACTCCC ACG
rs1060506 AAGTCTCCAGACACCCAGA ACG
rs7200125 AGGCTCCATTTCAAAAAAAAAAAAACT
rs6565219 AAAGTGCTGGGATTACAGGC ACT
rs889548 AGTCCTAATACAGTGGATGTC ACT
I
Genetic Analysis
[0232] Allelotyping results from the discovery cohort are shown for cases and
controls in
Table 13. The allele frequency for the A2 allele is noted in the fifth and
sixth columns for osteoarthritis
case pools and control pools, respectively, where "AF" is allele frequency.
The allele frequency for the
A1 allele can be easily calculated by subtracting the A2 allele frequency from
1 (Al AF = 1-A2 AF).
For example, the SNP rs732173 has the following case and control allele
frequencies: case A1 (G) _
0.55; case A2 (T) = 0.45; control A1 (G) = 0.58; and control A2 (T) = 0.42,
where the nucleotide is
provided in paranthesis. Some SNPs are labeled "untyped" because of failed
assays.
'TABLE 13
dbSNP PositionChromosomeAl/A2 F A2 F A2 F p-
rs# in position Allele Case Control Value
SEQ iD AF AF
NO:
rs7500176247 31077197 A/G
rs65652121535 31078485 C/T
rs80540462386 31079336 C/T
rs80568426440 31083390 C/T
rs732173 9133 31086083 G/T 0.45 0.42 0.382
rs732172 9143 31086093 A/G
rs71885579471 31086421 A/T
rs228800413150 31090100 C/G 0.52 0.45 0.026
rs433731013717 31090667 C/T 0.18 unt ed
rs201655414466 31091416 A/G
rs656521315769 31092719 A/C
rs720476216870 31093820 A/G
rs488952918545 31095495 C/T
rs656521418749 31095699 C/T
rs749967419123 31096073 GlT
rs656521520736 31097686 A/G
rs102362321038 31097988 C/T 0.02 unt ed
rs102362421046 31097996 C/T 0.16 0.11 0.035
rs102362521050 31098000 C/T 0.32 NA
rs154929721056 31098006 A/T
81
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP PositionCpromosomeAl/A2 F A2 F A2 F p-
rs# in position Allele Case Control Value
SEA iD AF AF
NO:
rs308489421706 31098656 -/ACC
rs804822823170 31100120 A/G 0.54 0.61 0.040
rs740543225028 31101978 A/T 0.35 0.43 0.025
rs805424927871 31104821 A/G
rs806104728070 31105020 C/T 0.21 0.21 0.903
rs718722031717 31108667 A/G
rs804697832019 31108969 A/G 0.34 0.28 0.083
rs228800332318 31109268 A/G
rs719642133080 31110030 A/G
rs719643133101 31110051 A/G
rs720315834236 31111186 A/G
rs230322334285 31111235 C/T 0.52 0.45 0.060
rs203291734818 31111768 C/G
rs804413435168 31112118 C/G 0.97 0.97 0.856
rs488953137981 31114931 ClT
rs488953238113 31115063 CIG
rs488953338117 31115067 C/T
rs881929 38481 31115431 G/T 0.38 0.34 0.228
rs804710438615 31115565 C/G 0.60 0.65 0.117
rs804780338944 31115894 A/C 0.35 0.33 0.437
rs464487439288 31116238 A/C
rs235967341385 31118335 C/T 0.18 0.20 0.563
rs443527142136 31119086 A/T
rs719771742185 31119135 A/C
rs235967442353 31119303 A/G 0.22 0.18 0.122
rs656521742434 31119384 A/G 0.35 0.33 0.608
rs230322244580 31121530 A/G 0.60 0.52 0.022
rs488961544675 31121625 A/T
rs462419745739 31122689 G/T
rs375185346439 31123389 C/T
rs749671 47457 31124407 C/T 0.32 0.37 0.095
rs'T4967047735 31124685 C/T
rs375185550319 31127269 C/T 0.53 0.57 0.287
rs375185650708 31127658 A/G
rs719672651185 31128135 A/G 0.41 0.37 0.258
rs889550 53002 31129952 A/G
rs750952 53064 31130014 C/T 0.43 0.41 0.535
rs207763353637 31130587 A/G
rs719994955274 31132224 C/G 0.46 0.53 0.051
rs203291655825 31132775 C/T
rs446864155986 31132936 A/C 0.26 0.25 0.902
rs488953556684 31133634 C/G
rs431677557653 31134603 C/T
rs431381957659 31134609 C/G
rs656521857692 31134642 G/T
rs431822457775 31134725 C/T
rs104603061313 31138263 C/T
rs7294 61431 31138381 A/G 0.38 0.37 0.669
rs720074961699 31138649 A/G
rs235961262906 31139856 A/G 0.56 0.48 0.017
rs805089463619 31140569 C/G 0.48 0.45 0.320
rs288473764664 31141614 AIC 0.68 0.60 0.016
rs189551468452 31145402 G/T
rs806020969665 31146615 C/T
rs806021769681 31146631 C/T
rs719616170091 31147041 A/G
rs806233674637 31151587 A/G
rs804377874760 31151710 A/G
rs203291576523 31153473 A/G 0.43 0.41 0.505
rs488961678559 31155509 C/G
rs104556479549 31156499 A/C
rs230322179882 31156832 C/T
rs154929681339 31158289 A/G
82
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP PositionChromosomeAl/A2 F A2 F A2 F p-
rs# in position Allele Case Control Value
SEQ 1D AF AF
NO:
rs889555 81681 31158631 C/T 0.49 0.50 0.740
rs581652181696 31158646 -/G
rs749767 83517 31160467 C/T 0.28 0.36 0.020
rs288473885431 31162381 AlC
rs205258186332 31163282 C/T
rs488961787358 31164308 A/G
rs488961987725 31164675 ClT
rs197848789052 31166002 A/G 0.62 0.57 0.124
rs197848690020 31166970 A/G
rs197848590231 31167181 A/G 0.90 0.88 0.513
rs488962090284 31167234 A/G
rs488962190447 31167397 C/T
rs321447790601 31167551 -/G
rs452703490724 31167674 A/G 0.37 0.43 0.079
rs106050692559 31169509 C/T 0.29 0.28 0.720
rs720012595176 31172126 AlG
rs656521995195 31172145 C/T
rs889548 96822 31173772 A/G 0.54 0.51 0.320
rs6145813Not mappedNot mapped~TTTTT p,33 0.32 0.909
TTTTTT
[0233] Allelotyping results were considered particularly significant with a
calculated p-value of
less than or equal to 0.05 for allelotype results. These values are indicated
in bold. The allelotyping p-
values were plotted in Figure 1A for the discovery cohort. The position of
each SNP on the
chromosome is presented on the x-axis. The y-axis gives the negative logarithm
(base 10) of the p-
value comparing the estimated allele in the case group to that of the control
group. The minor allele
frequency of the control group for each SNP designated by an X or other symbol
on the graphs in
Figure 1 C can be determined by consulting Table 13. For example, the left-
most X on the left graph is
at position 31077197. By proceeding down the Table from top to bottom and
across the graphs from
left to right the allele frequency associated with each symbol shown can be
determined.
[0234] To aid the interpretation, multiple lines have been added to the graph.
The broken
horizontal lines are drawn at two common significance levels, 0.05 and 0.01.
The vertical broken lines
are drawn every 20kb to assist in the interpretation of distances between
SNPs. Two other lines are
drawn to expose linear trends in the association of SNPs to the disease. The
generally bottom-most
curve is a nonlinear smoother through the data points on the graph using a
local polynomial regression
method (W.S. Cleveland, E. Grosse and W.M. Shyu (1992) Local regression
models. Chapter 8 of
Statistical Models in S eds J.M. Chambers and T.J. Hastie, Wadsworth &
Brooks/Cole.). The black line
provides a local test for excess statistical significance to identify regions
of association. This was
created by use of a l Okb sliding window with lkb step sizes. Within each
window, a chi-square
goodness of fit test was applied to compare the proportion of SNPs that were
significant at a test wise
level of 0.01, to the proportion that would be expected by chance alone (0.05
for the methods used
here). Resulting p-values that were less than 10-8 were truncated at that
value.
[0235] Finally, the exons and introns of the genes in the covered region are
plotted below each
graph at the appropriate chromosomal positions. The gene boundary is indicated
by the broken
83
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
horizontal line. The exon positions are shown as thick, unbroken bars. An
arrow is place at the 3' end
of each gene to show the direction of transcription.
Example 5
Chromosome 4 Region Proximal SNPs
[0236] SNP rs1957723 is associated with osteoarthritis and is described in
Table A. SNP
rs 1957723 falls in an intergenic region on chromosome 4 that does not include
a known gene, therefore,
the region is referred to herein as the Ch~om 4 region. One hundred-thirty
additional allelic variants
proximal to rs1957723 were identified and subsequently allelotyped in
osteoarthritis case and control
sample sets as described in Examples 1 and 2. The polymorphic variants are set
forth in Table 14. The
chromosome positions provided in column four of Table 14 are based on Genome
"Build 34" of NCBI's
GenBank.
TABLE 14
dbSNP Position ChromosomeAllele
rs# Chromosomein SEQ Position Variants
ID NO:
2
rs38490234 211 36870611 /t
rs14443114 7217 36877617 a/
rs20442954 7895 36878295 alC
rs21660934 13308 36883708 C/t
rs23763344 14279 36884679 /t
rs14443204 17026 36887426 c/t
rs20442944 18271 36888671 a/
rs18998644 20417 36890817 C/t
rs15620944 21843 36892243 a/
rs15620984 22069 36892469 a/
rs15620974 22145 36892545 al
rs15620964 22519 36892919 a/
rs15620954 22539 36892939 a/
rs14443194 23236 36893636 alC
rs14443184 23256 36893656 a/c
rs10259384 23402 36893802 c/t
rs10259374 23499 36893899 a/c
rs10259364 23620 36894020 C/t
rs10203334 23871 36894271 a/t
rs21206544 24136 36894536 c/
rs25885474 25427 36895827 a/
rs20442934 25866 36896266 /t
rs27603244 26541 36896941 a/
rs25885464 26576 36896976 /t
rs25885454 26689 36897089 a/
rs27603284 26720 36897120 a/c
rs25885444 27113 36897513 c/t
rs27603314 27164 36897564 c/t
rs25885434 27186 36897586 a/
rs25885424 28341 36898741 alt
rs25885414 29160 36899560 C/t
rs25885404 29844 36900244 a/g
~
84
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Position ChromosomeAllele
rs# Chromosomein SEQ Position Variants
ID NO:
2
rs27603364 30665 36901065 /t
rs27603374 30830 36901230 a/
rs20287324 31061 36901461 a/c
rs25885384 31523 36901923 c/t
rs19926174 32326 36902726 c/t
rs19984694 32346 36902746 al
rs19984704 32358 36902758 c/t
rs19754984 34909 36905309 c/t
rs15620934 34975 36905375 a/
rs19754974 35066 36905466 c/t
rs15620924 35096 36905496 /t
rs22487884 35375 36905775 c/t
rs18998624 36304 36906704 a/
rs25885324 36712 36907112 a/t
rs18858784 36770 36907170 c/t
rs9866484 37342 36907742 c/t
rs9866474 37412 36907812 c/t
rs10100104 37884 36908284 al
rs10100094 38077 36908477 a/c
rs27603254 38300 36908700 c/t
rs25885314 38301 36908701 c/t
rs18383884 41189 36911589 c/t
rs19754954 44408 36914808 c/t
rs21814914 44493 36914893 a/c
rs19754964 44571 36914971 a/
rs21814924 44670 36915070 a/
rs22247194 45219 36915619 a/
rs22247204 45258 36915658 C/t
rs19517704 47261 36917661 a/
rs22960404 48473 36918873 a/c
rs19577234 48771 36919171 a/
rs19577254 55292 36925692 c/t
rs28893464 56479 36926879 a/
rs18858794 56747 36927147 a/c
rs19577264 60620 36931020 /t
rs19577274 60688 36931088 a/c
rs18858804 61058 36931458 a/c
rs18858814 61129 36931529 clt
rs9421084 61577 36931977 c/t
rs19517714 61961 36932361 a/
rs23763234 63351 36933751 /t
rs20133584 63926 36934326 a/
rs21814944 65798 36936198 a/
rs19577284 66043 36936443 a/c
rs19577294 66044 36936444 a/
rs19577304 66246 36936646 c/t
rs19577314 66318 36936718 c/t
rs19984684 66547 36936947 /t
rs19577324 71238 36941638 c/t
rs19577334 71283 36941683 a/
rs23763224 71492 36941892 a/
_ ~ 4 72274 36942674 alg
rs2889345
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Position ChromosomeAllele
rs# Chromosomein SEQ Position Variants
ID NO:
2
rs18152674 73762 36944162 alt
rs19577344 74209 36944609 /t
rs19577354 75284 36945684 a/t
rs19577364 77347 36947747 a/c
rs19577374 77589 36947989 c/t
rs19577384 78096 36948496 a/
rs19577394 78606 36949006 a/
rs19577404 78862 36949262 /t
rs19577414 79135 36949535 a/
rs19577424 79146 36949546 a/
rs19577434 79456 36949856 c/t
rs19577444 79609 36950009 a/
rs19577454 80086 36950486 a/
rs19577464 80119 36950519 a/
rs19577474 80766 36951166 c/t
rs21466704 81110 36951510 a/
rs21466714 81269 36951669 a/t
rs19577484 81668 36952068 c/t
rs21623074 82433 36952833 clt
rs19628394 82559 36952959 c/
rs23763154 83298 36953698 c/t
rs14264104 83821 36954221 a/
rs18959214 84121 36954521 c/t
rs18959224 84147 36954547 c/t
rs10357794 84543 36954943 a/
rs10357804 84554 36954954 a/
rs10357814 84691 36955091 a/
rs10357824 84727 36955127 a/
rs14264114 85678 36956078 c/t
rs18346024 86699 36957099 clt
rs18346034 86700 36957100 a/
rs18346044 86792 36957192 a/
rs18346054 86832 36957232 a/
rs21623084 87045 36957445 a/
rs13653414 87140 36957540 a/
rs18204584 87365 36957765 a/c
rs14693104 88342 36958742 c/t
rs30578794 88498 36958898 -/tca
rs14693114 88589 36958989 a/
rs7683264 95502 36965902 a/
rs18635234 96968 36967368 c/t
rs14693124 97448 36967848 c/t
rs14693134 97568 36967968 c/t
rs19517734 98724 36969124 c/t
rs21206554 Not ma Not ma t/
ed ed
rs21814954 Not ma Not ma /a
ed ed
86
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
Assay for Verifyin~ and AllelotYpin~ SNPs
[0237] The methods used to verify and allelotype the 130 proximal SNPs of
Table 14 are the same
methods described in Examples 1 and 2 herein. The primers and probes used in
these assays are
provided in Table 15 and Table 16, respectively.
TABLE 15
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs3849023ACGTTGGATGGGTAATTGCTAACCATGTTCACGTTGGATGGACCCAGTCAAGTCAATAAAC
rs1444311ACGTTGGATGGCCTATTGGTTTAACTAGGCACGTTGGATGTCTGGCTTCTTCAGGAGTTC
rs2044295ACGTTGGATGCCACACCACTACTATTCAAGACGTTGGATGGTGGTGTGTTAGAAGGTTAC
rs2166093ACGTTGGATGAAAATCCTGGAGATGGATGGACGTTGGATGTAGGTGTACAGTTCAGTGTC
rs2376334ACGTTGGATGTCTCAGAGAACCAGCTTTTGACGTTGGATGGGGAATATTAAACATTGGGG
rs1444320ACGTTGGATGTAATTCTCTCCTCCAAATGCACGTTGGATGCTAGAAACAAAAGACTACATG
rs2044294ACGTTGGATGAACCTAAATCTCCTCAAGCCACGTTGGATGTTCTGACCACTTCTCTATGG
rs1899864ACGTTGGATGTTTATAGGCGTGGGCAATCGACGTTGGATGTTGTCAGAAAGTGTCGTGCC
rs1562094ACGTTGGATGTGGATTCCTTTCTTGAAGACACGTTGGATGGCAACAAAGAAACTTAATGC
rs1562098ACGTTGGATGTCTGAGTCCGAGTGATCATCACGTTGGATGAAACAATTAGCAGGGCACAG
rs1562097ACGTTGGATGCACAGGATCTTACTCTGTTGACGTTGGATGCGGACTCAGAAATTCAAGTC
rs1562096ACGTTGGATGACCCAGGGCATGTTATATAGACGTTGGATGTTTCTCTCTGGTACCCTCTC
rs1562095ACGTTGGATGTGTTAGTAACCCAGGGCATGACGTTGGATGTGACAGATGCCACCAGTTAC
rs1444319ACGTTGGATGTTCAACTTTAGCCTCTGGGCACGTTGGATGCCCTGCAAAGTCAAAGGAAC
rs1444318ACGTTGGATGCTCTGGGCAATTATCAAGCCACGTTGGATGAGTTCGCTGATGTGTTTGGG
rs1025938ACGTTGGATGCAGGTAAGAAAAGCTTTTTGGACGTTGGATGCCCTGCTAATGACTGAATTTC
rs1025937ACGTTGGATGGAATAGGAAAGGTAGTATACCACGTTGGATGAAATTCAGTCATTAGCAGGG
rs1025936ACGTTGGATGTCTCCAGGTAGATGAGTCAGACGTTGGATGCCACACACCAAAGCAATCAC
rs1020333ACGTTGGATGGCATCTCTTCAATCTGGACGACGTTGGATGGTGGATCACAGAAGTCAGAG
rs2120654ACGTTGGATGACCAGAAAGACCAGGGCATGACGTTGGATGAACCTTTAGCTCTTCTCCCC
rs2588547ACGTTGGATGTCACAAATGTAATATAAATCACGTTGGATGGATAGCTACGTTTAAAAATG
rs2044293ACGTTGGATGTGTCAACAATACAAGACTAAACGTTGGATGTGCACTGGACTTTTTTTTT
rs2760324ACGTTGGATGACAAACCAGTGGTTGAGGAGACGTTGGATGCCTCACGAATCCAACAGAAC
rs2588546ACGTTGGATGCTTAGAGGATGGAGTCAGTCACGTTGGATGTACTACCAGAGATGCTGGTG
rs2588545ACGTTGGATGCAACACAGCTACAGTGCATCACGTTGGATGTGGGTAAAGGGAAAAGAAGG
rs2760328ACGTTGGATGGCCATAAAATTGGGTAAAGGGACGTTGGATGGCATCTATTTGACACCAACG
rs2588544ACGTTGGATGTAAGAATTAGCATGTGAAAGACGTTGGATGTTTGTGCACAAAGAATTTGG
rs2760331ACGTTGGATGAAACAGTATGCCTTTTGTGCACGTTGGATGCTTCTCGTAATTTTACATGAC
rs2588543ACGTTGGATGGTGCCAAATTCTTTGTGCACACGTTGGATGCTAAGATAGGTAGATACCAG
rs2588542ACGTTGGATGTGGCAGCAAAGCTTAAGCTCACGTTGGATGTCCACAGTCACCTCTCATTC
rs2588541ACGTTGGATGTGACAAGGTCTATGTCAGGGACGTTGGATGGGCATTGTCATGGTGATGAG
rs2588540ACGTTGGATG.TGCTGTATGATCCAGCAATCACGTTGGATGGGTGCAAATACTGTCTCTTC
rs2760336ACGTTGGATGAAGCTGAGGCAGGAGAATGGACGTTGGATGTGTTTTGAGACGGAGTCTCG
rs2760337ACGTTGGATGGGTGTTCGAACTGATACAAGACGTTGGATGACTACCATTCTACTCTCTGC
rs2028732ACGTTGGATGTTCCTGGACAGCTAAATAGGACGTTGGATGGCCATTGTCGTTTTCTTGTT
rs2588538ACGTTGGATGTATCTTCTGGGAAGCCTTTCACGTTGGATGGACTTGAAATCACTCCATGC
rs1992617ACGTTGGATGGGAGGACATTGCCTTCAAAGACGTTGGATGCTGACCTTCTGTCTAGTCAC
rs1998469ACGTTGGATGTATATGCCAAGGACCAACGGACGTTGGATGCTGACCTTCTGTCTAGTCAC
rs1998470ACGTTGGATGATTTCCCCCATTAAGCTTTGACGTTGGATGGAAAAGTATTATATGCCAAGG
rs1975498ACGTTGGATGAGCTCTCTTTTTGCCTGCTGACGTTGGATGAGGAGGCTTCACAATCATGG
rs1562093ACGTTGGATGTGATTGTGAAGCCTCCTCTGACGTTGGATGAAAGACATACCCAAGACTGG
rs1975497ACGTTGGATGTCAGCAGCATGAAAACTGACACGTTGGATGCATTTAGACTTTTTCTGGGG
Irs1562092ACGTTGGATGTTCCAGTGACTGGACCATAGACGTTGGATGTCAGCAGCATGAAAACTGAC
87
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs2248788ACGTTGGATGGGGAAAAGAAAAAAGACTTCCACGTTGGATGGTAGTAGCTGCTTCTAAAAG
rs1899862ACGTTGGATGTAATCTCCCATAATAAGTGCACGTTGGATGGCTACAAAAGAAAATGAATAC
rs2588532ACGTTGGATGCAAACAATAGTGGCTGAGAGACGTTGGATGTTTGTAGCACAGGCGCATAG
rs1885878ACGTTGGATGTGACTCAGCGAGTTTGTAGCACGTTGGATGAGCCAGATTGGGTGCTTTTC
rs986648ACGTTGGATGTGAGAAAGCTTTCTGAGGACACGTTGGATGGGTTTTCTGTTGTGAATGGG
rs986647ACGTTGGATGACACACTCTTTCTCAAGCAGACGTTGGATGCTTATTTGTCCTCAGAAAGC
rs1010010ACGTTGGATGACTGTAGCTAAGTTGGCATACGTTGGATGTTCACCAACACCAATAAGGC
rs1010009ACGTTGGATGTCTCATCAGCTCTTTCCTGGACGTTGGATGAAAGGGATGAGGAAGTGAGG
rs2760325ACGTTGGATGATCCCCAGCATGTAGCATAGACGTTGGATGCTGCCCATAAGTCTCTTCTG
rs2588531ACGTTGGATGATCCCCAGCATGTAGCATAGACGTTGGATGCTGCCCATAAGTCTCTTCTG
rs1838388ACGTTGGATGGTACCTCATGGATATTTACACACGTTGGATGTTGGTGTTGTTATAAATGAC
rs1975495ACGTTGGATGCAGGTCAGGAGTTTAAGACCACGTTGGATGAGCTGGGATTACAGTCATGC
rs2181491ACGTTGGATGGTACCTAATATATGCTTCTGGACGTTGGATGTTATTCCCGTCTTACTTTCC
rs1975496ACGTTGGATGTATATTAGGTACAGTGTGGCACGTTGGATGCAACCAACTTCACTGAAAGC
rs2181492ACGTTGGATGCTTGCAGGAAGAGGAAGAAGACGTTGGATGACAATCACCTTTGGAGGCAG
rs2224719ACGTTGGATGTCAAGGGTGTAGATGTGTAGACGTTGGATGCCAGAGAGGAGTAATGGTAT
rs2224720ACGTTGGATGCCAATTACTCAAGGGTGTAGACGTTGGATGAATTCAGTACAGACAGAGGG
rs1951770ACGTTGGATGCCTGGGAACTTCAGCTTTTCACGTTGGATGTGGCACAGCAGGAATATCAG
rs2296040ACGTTGGATGGGGCATCATGAAATGCAGACACGTTGGATGGCATGTACAGGAAAGCAGTG
rs1957723ACGTTGGATGTACTCACTTGTGTACTGCTCACGTTGGATGGCTGCAGCGTCACATTAATC
rs1957725ACGTTGGATGTTATTGGAATTCTCCAGGTCACGTTGGATGAAGATGATTAGTCCAGCCTG
rs2889346ACGTTGGATGTGACTGACTTCCTAGGTCAGACGTTGGATGTGACAGTGTTTGAGTGGCAG
rs1885879ACGTTGGATGTTCACCCCTTCACATCTGATACGTTGGATGCTACAAGGAAGATAACAGAG
rs1957726ACGTTGGATGAAATTCAGCCACTCAACCAGACGTTGGATGAAGTGGTTGGGATTTGTGAG
rs1957727ACGTTGGATGGCCAACGTATCTTTAAAACCCACGTTGGATGGTTTTGTCTTGGTTCTCATC
rs1885880ACGTTGGATGTGGAATGCCCCAAGATTTCAACGTTGGATGCTGGAATCCCAAGGTTCCTG
rs1885881ACGTTGGATGTAGACGTGTTCTGCATCATGACGTTGGATGATGAAATCTTGGGGCATTCC
rs942108ACGTTGGATGGAGCTGTTAGGGTAGAAATGACGTTGGATGGTCCTTGGACTAATTTTGACC
rs1951771ACGTTGGATGGGCATTCCCTTTTGTCTAAGACGTTGGATGAGTAAACAAGGACTAGAGCC
rs2376323ACGTTGGATGTCCTTACTTGCTAGCACTGCACGTTGGATGGCATCCCTTGGTGACTGATA
rs2013358ACGTTGGATGGGAATTTTAGGAGTACTGTAGACGTTGGATGGCCAACCATAGAACCTAAATC
rs2181494ACGTTGGATGATTCAATTACCTCCCACTGGACGTTGGATGTATCCCCACCCAAATGTCAC
rs1957728ACGTTGGATGAAATAGATCCCAACCAAGGGACGTTGGATGGTAACATTTACCTAAGCGGG
rs1957729ACGTTGGATGAAATAGATCCCAACCAAGGGACGTTGGATGGTAACATTTACCTAAGCGGG
rs1957730ACGTTGGATGGGTCTAAACATGAGAGACTCACGTTGGATGTCTTTATGGATATAGGGTCC
rs1957731ACGTTGGATGTATTGGAACCTGGTACCTGGACGTTGGATGGACCTGAATCATGTCTCCAG
rs1998468ACGTTGGATGTATAAAGCCTCAAAAGTGGGACGTTGGATGACCTTATTCCAGAATGAAAC
rs1957732ACGTTGGATGAAGAGAGGAGTTTATTGGCCACGTTGGATGCGGCCTGATCTTTATTTTCG
rs1957733ACGTTGGATGCTATCAAGACTCTGATTGCCACGTTGGATGTGTTTGCAGGTAAACTTGGC
rs2376322ACGTTGGATGTCGTTCTCTCTCTGTGCATGACGTTGGATGTTAGTCAGATGCTTGGTGAG
rs2889345ACGTTGGATGTGGAATCCCAAACCTTTCAGACGTTGGATGTTCTTGCTAAATGTAGGCC
rs1815267ACGTTGGATGCAGGAAAGGGCTACTATCAGACGTTGGATGGTAGGCCAAACTAGCTTTGG
rs1957734ACGTTGGATGCTACCCCTGCCTTATAATTCACGTTGGATGCAAGTGGTAAAAGGATGTGG
rs1957735ACGTTGGATGAGCTTCCCATGGTTATAGAGACGTTGGATGCTGAAAACAATACCGGTCTC
rs1957736ACGTTGGATGCTGAAGCAAAGATTTCTCTCACGTTGGATGAGCATCTTTTGCTGTCACTG
rs1957737ACGTTGGATGACATGGAAGCTGAAGCCAAGACGTTGGATGCAGAGCTTTGACCTTACTCC
rs1957738ACGTTGGATGATGTCCCTTAAAAGGCTGCCACGTTGGATGCAGATGATCTTGCTTCCCAG
rs1957739ACGTTGGATGTCACTGCCTGAGTGCTTTAGACGTTGGATGCTGATGGCCTGAGAACTAAG
rs1957740ACGTTGGATGGCCCAGTCAAGTTGACATACACGTTGGATGCACCTGCTCCAGTTATATAC
rs1957741ACGTTGGATGAGGAGCATTATCCCTATTAGACGTTGGATGCCTCTTAGTAAAATATGGATG
rs1957742ACGTTGGATGGGATGATATCTACTTTGTACGACGTTGGATGGACTCCATCTGAGATGTTAG
rs1957743ACGTTGGATGCAACTGTCTTGTATTTGAAGACGTTGGATGGACAGACTTTCATTGTTTTC
88
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs1957744ACGTTGGATGTCAGTGTACCCTGTAATGCCACGTTGGATGTGCCCAGCAGTGAGTAATTG
rs1957745ACGTTGGATGGTTTAGAAAGTGTTGGGTTCCACGTTGGATGAGCAAATGCAGCTTATTACC
rs1957746ACGTTGGATGTCTCATACAACATAGTTAGCACGTTGGATGGGTTTAGGTTTGGTTTGATG
rs1957747ACGTTGGATGGTCACTCAAGATAACAGTTCCACGTTGGATGTTACCTAACGTGAAGGTAGC
rs2146670ACGTTGGATGCCTAACACATCTTTATGAGCACGTTGGATGCTCATAAGATATGCTAAGCAC
rs2146671ACGTTGGATGATGAGGAGCAACTAGAAGGCACGTTGGATGAAAGGGCTGGAAGAAACAGG
rs1957748ACGTTGGATGTGAAGTTTGTAGTAGGGAGCACGTTGGATGTTCTGTCACACAAACACTCC
rs2162307ACGTTGGATGACATGCGGTGCCTGGCCCTTTACGTTGGATGCCTTTGTAGGGACATGGATG
rs1962839ACGTTGGATGGGCTGCATAGTATTCCATGGACGTTGGATGAGGGAATCCTTTCCCCATTG
rs2376315ACGTTGGATGTGGCCTTGGATTTCTTCCACACGTTGGATGAGAATTGGACAGAGTGGCAG
rs1426410ACGTTGGATGGAGAAAGTTGCATCTTGCCCACGTTGGATGGGGAAGTTTTACCTTGGCTC
rs1895921ACGTTGGATGGGTGATGGTGTTTGAGGTACACGTTGGATGATTAGGCTTCTCCCACCATC
rs1895922ACGTTGGATGCAATGCATTAGGCTTCTCCCACGTTGGATGGAGGTACATTTCTCAGGCAG
rs1035779ACGTTGGATGGAGAATCACTTGAACCCGGGACGTTGGATGTGGAGTGCAGTGGCATGATC
rs1035780ACGTTGGATGTTTGAGATGGAGTCTCGCTCACGTTGGATGAATCACTTGAACCCGGGAGG
rs1035781ACGTTGGATGGGAAGATGCTGACTCTGAACACGTTGGATGCCTTGACTGTTTAGGGATCC
rs1035782ACGTTGGATGGGATCCCTAAACAGTCAAGGACGTTGGATGAGTTGGCTAGACTTGCGTTC
rs1426411ACGTTGGATGCAAGAGTGCTACACAAGTCGACGTTGGATGTGTACCTTGGTCAGGTGATC
rs1834602ACGTTGGATGGATGGGCCCTATTTTTCTTGACGTTGGATGCTTTTCCAACCCAGTAATGTC
rs1834603ACGTTGGATGGATGGGCCCTATTTTTCTTGACGTTGGATGTCTTTTCCAACCCAGTAATG
rs1834604ACGTTGGATGGAAAGACATTACTGGGTTGGACGTTGGATGAGAATTCTTCCTGACTGTGG
rs1834605ACGTTGGATGGCCCACAGTCAGGAAGAATTACGTTGGATGTTGTGGAGACTGGCCAAAAG
rs2162308ACGTTGGATGTAAAGAAACAGAGGGACACCACGTTGGATGTATGATCAGAGTCATCAGGG
rs1365341ACGTTGGATGTCCCTCTGTTTCTTTAGGCAACGTTGGATGCATCTCCCCTGGTAGCATTT
rs1820458ACGTTGGATGCACCCTCAGACTTGGAAATGACGTTGGATGGTCAGGTGACTCTATTCAGC
rs1469310ACGTTGGATGTACTACAGCGTGTTTAGCAGACGTTGGATGTGTCAAAGGGAGAGTTAGAG
rs3057879ACGTTGGATGGGCACATTGGAAAATAAAGCCACGTTGGATGACGGCATGAACAATTCTCAG
rs1469311ACGTTGGATGCCTGAGAATTGTTCATGCCGACGTTGGATGTTTTCAGTGTTCTCTCCAGG
rs768326ACGTTGGATGAATTAGCCAGGCATGGTGTCACGTTGGATGACATCCTAGGCTCAAGTGAC
rs1863523ACGTTGGATGGGCAGACACATTCCTATTCGACGTTGGATGGGGAAAGGTGTGCTGAGTAA
rs1469312ACGTTGGATGCATTTCGTCAGCATTCTAGCACGTTGGATGGGACTCATGTCATCTCTTGG
rs1469313ACGTTGGATGAGTGAGGGAGAAAAGTGAACACGTTGGATGCCTAACTTCTCTCCAATCTC
rs1951773ACGTTGGATGAAGGTTCAAGTTACCGCATGACGTTGGATGCACTGTGGTCCATGAAAAA
rs2120655ACGTTGGATGACAGGGTTTCTGCATGTTGCACGTTGGATGACGCCTGTAATCCCAGCACT
rs2181495ACGTTGGATGGAATTGTGGGAGTTACAATTC~ACGTTGGATGGAATCAAGCTAATTAACATGTG~
TABLE 16
dbSNP Extend Term
rs# Primer Mix
rs3849023 CTCATAACATAAGAAGTTGATGC CGT
rs1444311 CTAGGCATGCTAGCTTGGC ACT
rs2044295 CACTACTATTCAAGATTACCCTTTACT
rs2166093 GGTGGTGATGGCTGCACAA ACG
rs2376334 TCAGAGAACCAGCTTTTGATTTCAACT
rs1444320 GCCTAGACCCCGTGCAAC ACG
rs2044294 CTCCTCAAGCCAATAGGTCTTA ACG
rs1899864 CGCACCTGGCCGAAAATAAC ACT
rs1562094 AACCTGCAAAAGATTTACACTTGCACT
rs1562098 TCCTGCCTCAGCCTTCCTAGA ACT
~9
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Extend Term
rs# Primer Mix
rs1562097 ACTCTGTTGTTCAGGCTGGGGT ACT
rs1562096 TAAGCTAGCTAGTAACTGGTG ACT
rs1562095 ATGTTATATAGAACATCCCTTTTTACT
rs1444319 TCTGGGCAATTATCAAGCCTTT ACT
rs1444318 CTTTGCATTTTCCTGAGTTCCTTTACT
rs1025938 AAGAAAAGCTTTTTGGTTTGGG ACT
rs1025937 GGTAGTATACCTAAAAAAACAGC CGT
rs1025936 TCAAAGGACACCCAGCATTCA ACG
rs1020333 ACGTTTATCTGTAACCTTTCCA CGT
rs2120654 GAAAGACCAGGGCATGATTAGA ACT
rs2588547 ACAAATGTAATATAAATCAAGCTCACG
rs2044293 ACCAGCCTGGGTAACATAGCCA ACT
rs2760324 GGTTGAGGAGAAGCACCAGCA ACG
rs2588546 TACAATTTCTAGCCTTAATAAGATACT
rs2588545 TACAGTGCATCTATTTGACACCAAACG
rs2760328 AAATTGGGTAAAGGGAAAAGAAG ACT
rs2588544 ATTAGCATGTGAAAGACTTCTC ACT
rs2760331 AGTATGCCTTTTGTGCACAAAGA ACT
rs2588543 ATTCTTTGTGCACAAAAGGCATA ACG
rs2588542 GCTTAAGCTCTTACAGGCAG CGT
rs2588541 AGGTCTATGTCAGGGAAAACCTTAACG
rs2588540 GATCCAGCAATCCCACTGAT ACG
rs2760336 AGGCGGAGCTTGCAGTGAG ~ ACT
rs2760337 CACCAATACTGTATGATTCTTTT ACT
rs2028732 CAGCTAAATAGGGCTTGAGTCAAT~ CGT
rs2588538 AATTTGTACAAATTTATGGGGTATACT
rs1992617 ATTGCCTTCAAAGAACATCAAAGCACG
rs1998469 GACCAACGGGAGGACATTG ACG
rs1998470 CTTTGAAGGCAATGTCCTCC ACG
rs1975498 TTTTGCCTGCTGCTATCCAC ACT
rs1562093 CTCCTCTGCCATGTGGAAC ACG
rs1975497 AAAACTGACTAATACACACTGTT ACT
rs1562092 TTTGGTTAATGGACATTTAGACT ACT
rs2248788 TGTGGGATTTTATTATTTTCATCAACT
rs1899862 TAAGTGCATAACTTGTCTTTGAGGACT
rs2588532 ATAGTGGCTGAGAGCCAGAT CGT
rs1885878 GCGAGTTTGTAGCACAGGC ACT
rs986648 GTACATGTAATGCTAGTAAAGAAAACG
rs986647 CTCTTTCTCAAGCAGGAGTTA ACG
rs1010010 AGCTAAGTTGGCATGTGGGA ACT
rs1010009 CCTGGCTACCTTCCAAAAAG ACT
rs2760325 TCTCAGGAAGTATGAAATAAATAGACG
rs2588531 CTCAGGAAGTATGAAATAAATAGTACT
rs1838388 TCATGGATATTTACACCTACTAC ACT
rs1975495 AGGAGTTTAAGACCAGCCTG ACT
rs2181491 TGCTTCTGGATTTTTAATGATCACACT
rs1975496 ATGATCAAATCATTTTGAGGGC ACT
rs2181492 GTTGCATTGCTATGGTCTGC ACT
rs2224719 CATATATCCCTCTGTCTGTAC ~ ACG
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Extend Term
rs# Primer Mix
rs2224720 GGGTGTAGATGTGTAGATTTATA ACT
rs1951770 ACAAGCATTAGAGACTTGATTG ACG
rs2296040 CTTTGTTTCTAAAATCTGATAGTCACT
rs1957723 AGCATGGCATAGGCACTGG ACG
rs1957725 GCGAGGAAAGACCTGTTCTA ACG
rs2889346 GGTCAGCTCAGCTGGTTTTT ACG
rs1885879 CACATCTGATGCTCTCCTAAA ACT
rs1957726 GCCACTCAACCAGTAGGAAA ACT
rs1957727 GTATCTTTAAAACCCTCACAAAT CGT
rs1885880 TTACGTTAGTCTGCCTACTTCCA ACT
rs1885881 TGGGCTATCAATGATGGAAAC ACT
rs942108 AAATGAAATAGAATTGTGTACTTCACT
rs1951771 TCCCTTTTGTCTAAGAATATTAG ACG
rs2376323 CTAGCACTGCCAAGTGCAAC ACT
rs2013358 TTTTAGGAGTACTGTAGAACACA ACG
rs2181494 TGGGTCCCTCCCATAACAC ACT
rs1957728 AGAAGCATGTGCTTATAACAATAACGT
rs1957729 GAAGCATGTGCTTATAACAATAAAACT
rs1957730 ACATGAGAGACTCTGAAGACT ACT
rs1957731 GGGTGAGCTTTGGGATCAC ACT
rs1998468 GGGCATAATTAATCCATGTTAG ACT
rs1957732 GGCCAAGTTTACCTGCAAAC ACT
rs1957733 TCTAATGTTAAAGAGAGGAGTTTAACG
rs2376322 GCGCCAAGGAAAGGCCAC ACT
rs2889345 TCATTTCTCACCCTTGATATCCA ACT
rs1815267 AAAGGGCTACTATCAGTTTTGT CGT
rs1957734 CTGCCTTATAATTCTAAAAAGGT ACT
rs1957735 CTAAAACTAAGAAATGTTTCCAC CGT
rs1957736 TAATACTAAGGAGAGGGCTCCT ACT
rs1957737 AGCCAAGGGTGTGGATGAG ACT
rs1957738 CCTTAAAAGGCTGCCTACAAAATAACT
rs1957739 CTGAGTGCTTTAGCTGGATTA ACG
rs1957740 TTAAGCATCACACTGAGTTTGAG ACT
rs1957741 AGCTGAATTAAGCGCGACAGCTA ACG
rs1957742 TCTACTTTGTACGTAGCTGTCGC ACT
rs1957743 GAAAATATTACTAAAAAAGACCTCACG
rs1957744 TGTACCCTGTAATGCCTAAAGC ACG
rs1957745 TTTTCAAAGGTTTAGGTTTGGTTTACT
rs1957746 ACAACATAGTTAGCAAATGCAG ACG
rs1957747 GATAACAGTTCCAATTACAACAA ACG
rs2146670 ATCTTTATGAGCTTTTCCTTTCTTACG
rs2146671 TACAACCCTTTCAGGACTTCA CGT
rs1957748 TTGTAGTAGGGAGCCATGGT ACT
rs2162307 CCTGGCCCTTTGTCCCTG ACG
rs1962839 CCACATCTTTGACAAACCTGA ACT
rs2376315 CCCCCTTCCTTTTCCAGGC ACT
rs1426410 CATCTTGCCCTAAAATCACTC ACG
rs1895921 GTACATTTCTCAGGCAGCTC ACG
rs1895922 ATTAGGCTTCTCCCACCATC I ACT
91
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Extend Term
rs# Primer Mix
rs1035779 ACCCGGGAGGGTTGCAGT ACT
rs1035780 GGCTGGAGTGCAGTGGCA ACG
rs1035781 ACCTAGACTAAGAGAGTGATTGCAACT
rs1035782 CCTAAACAGTCAAGGCAAAGG ACT
rs1426411 TTTATGGTCTTCTTAGGATATCAACG
rs1834602 AGGAAGGTGCCCAGATCCT ACG
rs1834603 AGGAAGGTGCCCAGATCCTT ACT
rs1834604 AGTTTTCTAGTAACTTCTCTAAAAACT
rs1834605 ACAGTCAGGAAGAATTCTGTCT ACT
rs2162308 CACCTACAGAGTTTAAGTAAATTTACG
rs1365341 AAATCTCCTGGAGGGCTTCATAAACT
rs1820458 TGGAAATGGCAACTGAATCCT ACT
rs1469310 ACCCACACAATGCCAATAGCAC ACT
rs3057879 TGGAAAATAAAGCCTTTTGAGGTTACT
rs1469311 TGCCGTTAAAGAGGAAAAGCT ACT
rs768326 CAGCTACTCTGTAAAGCTGAA ACT
rs1863523 ATATTCTTGCTCATCTTTCTCTATACT
rs1469312 TAGTCCAGCAAACGCCAGC ACT
rs1469313 GTGAACAAATAATGCAAGTTCAGACT
rs1951773 CCCTTTGGGAGAGAAGGGC ACT
rs2120655 AGCAATCCTCCCACTTTGGC CGT
rs2181495 GGTGACATTTGGGTGGGGATACAACT
Genetic Analysis
[0238] Allelotyping results from the discovery cohort are shown for cases and
controls in Table 17.
The allele frequency for the A2 allele is noted in the fifth and sixth columns
for osteoarthritis case pools
and control pools, respectively, where "AF" is allele frequency. The allele
frequency for the A1 allele
can be easily calculated by subtracting the A2 allele frequency from 1 (A1 AF
= 1-A2 AF). For
example, the SNP rs1444311 has the following case and control allele
frequencies: case A1 (A) = 0.74;
case A2 (G) = 0.26; control A1 (A) = 0.75; and control A2 (G) = 0.25, where
the nucleotide is provided
in paranthesis. Some SNPs are labeled "untyped" because of failed assays.
TABLE 17
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in Position AlleleCase Control Value
SEQ ID AF AF
NO: 2
rs3849023211 36870611 G/T
rs14443117217 36877617 A/G 0.26 0.25 0.566
rs20442957895 36878295 A/C
rs216609313308 36883708 C/T
rs237633414279 36884679 G/T 0.15 0.16 0.734
rs144432017026 36887426 C/T
rs204429418271 36888671 A/G 0.16 0.14 0.412
rs189986420417 36890817 C/T
rs156209421843 36892243 A/G 0.22 0.23 0.586
rs156209822069 36892469 A/G
rs156209722145 36892545 A/G NA 0.97 NA
rs156209622519 36892919 A/G 0.20 0.21 0.773
rs156209522539 36892939 A/G 0.53 0.51 0.407
rs144431923236 36893636 A/C 0.74 0.79 0.023
~
92
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in Position Allele Case Control Value
SEQ ID AF AF
NO: 2
rs144431823256 36893656 A/C 0.12 0.13 0.559
rs102593823402 36893802 C/T 0.18 0.19 0.633
rs102593723499 36893899 A/C
rs102593623620 36894020 C/T 0.84 0.84 0.907
rs102033323871 36894271 A/T
rs212065424136 36894536 CIG 0.15 0.16 0.718
rs258854725427 36895827 A/G 0.39 0.40 0.603
rs204429325866 36896266 G/T
rs276032426541 36896941 A/G 0.59 0.61 0.395
rs258854626576 36896976 G/T 0.07 0.05 0.352
rs258854526689 36897089 A/G
rs276032826720 36897120 A/C 0.25 0.26 0.791
rs258854427113 36897513 C/T
rs276033127164 36897564 C/T 0.91 0.94 0.184
rs258854327186 36897586 A/G 0.59 0.59 0.828
rs258854228341 36898741 A/T
rs258854129160 36899560 ClT 0.61 0.59 0.313
rs258854029844 36900244 A/G 0.62 0.62 0.999
rs276033630665 36901065 G/T
rs276033730830 36901230 A/G 0.16 0.16 0.826
rs202873231061 36901461 A/C 0.60 0.58 0.432
rs258853831523 36901923 C/T 0.62 0.61 0.853
rs 199261732326 36902726 C/T 0.61 0.59 0.282
rs199846932346 36902746 A/G
rs199847032358 36902758 C/T 0.81 0.86 0.018
rs197549834909 36905309 C/T
rs156209334975 36905375 A/G 0.89 0.87 0.529
rs197549735066 36905466 C/T 0.13 0.13 0.691
rs156209235096 36905496 G/T
rs224878835375 36905775 C/T 0.29 0.31 0.368
rs189986236304 36906704 A/G 0.18 0.16 0.274
rs258853236712 36907112 A/T 0.30 0.32 0.443
rs188587836770 36907170 C/T 0.35 0.35 0.866
rs986648 37342 36907742 C/T 0.74 0.73 0.679
rs986647 37412 36907812 C/T 0.78 0.76 0.263
rs101001037884 36908284 A/G 0.25 0.26 0.649
rs101000938077 36908477 A/C 0.26 0.25 0.781
rs276032538300 36908700 C/T
rs258853138301 36908701 C/T
rs183838841189 36911589 C/T 0.75 0.74 0.650
rs197549544408 36914808 C/T
rs218149144493 36914893 A/C 0.14 _0.12 0.235
rs197549644571 36914971 A/G 0.26 0.26 0.944
rs218149244670 36915070 A/G 0.11 0.09 0.311
rs222471945219 36915619 A/G 0.78 0.78 0.866
rs222472045258 36915658 C/T 0.20 0.21 0.641
rs 195177047261 36917661 A/G 0.22 0.18 0.029
rs229604048473 36918873 A/C 0.41 0.43 0.459
rs995772348771 36919171 A/G 0.42 0.38 0.113
rs195772555292 36925692 CIT 0.75 0.78 0.196
rs288934656479 36926879 A/G 0.54 0.55 0.677
rs188587956747 36927147 A/C 0,44 0.48 0.123
rs195772660620 36931020 G/T 0.14 0.14 0.741
rs195772760688 36931088 A/C 0.73 0.76 0.271
rs188588061058 36931458 A/C 0.43 0.43 0.935
rs188588161129 36931529 C/T 0.12 0.11 0.681
rs942108 61577 36931977 C/T 0.49 0.52 0.317
rs195177161961 36932361 A/G 0.93 NA NA
rs237632363351 36933751 G/T
rs201335863926 36934326 A/G 0.13 0.13 0.821
rs218149465798 36936198 A/G 0.42 0.43 0.512
rs195772866043 36936443 A/C
rs 195772966044 36936444 A/G 0.79 0.77 0.405
rs195773066246 36936646 C/T 0.15 0.16 0.719
93
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in Position Allele Case Control Value
SEQ ID AF AF
N~: 2
rs195773166318 36936718 C/T 0.14 0.16 0.413
rs199846866547 36936947 G/T 0.13 0.12 0.468
rs195773271238 36941638 C/T 0.10 0.10 0.841
rs195773371283 36941683 A/G 0.63 0.61 0.632
rs237632271492 36941892 A/G 0.26 0.28 0.509
rs288934572274 36942674 A/G 0.20 0.18 0.234
rs181526773762 36944162 A/T 0.46 0.45 0.674
rs195773474209 36944609 G/T 0.55 0.64 0.003
rs195773575284 36945684 A/T 0.63 0.61 0.430
rs195773677347 36947747 A/C 0.05 0.05 0.903
rs195773777589 36947989 C/T 0,71 0.75 0.164
rs195773878096 36948496 A/G
rs195773978606 36949006 A/G
rs195774078862 36949262 G/T
rs195774179135 36949535 A/G 0.76 0.80 0.077
rs195774279146 36949546 A/G 0.95 0.96 0.500
rs195774379456 36949856 C/T 0.21 0.16 0.039
rs195774479609 36950009 A/G 0.66 0.70 0.088
rs195774580086 36950486 A/G 0.88 0.90 0.354
rs195774680119 36950519 A/G 0.40 0.44 0.120
rs195774780766 36951166 C/T 0.72 0.76 0.093
rs214667081110 36951510 A/G 0.73 0.77 0.072
rs214667181269 36951669 A/T 0.17 0.15 0.250
rs195774881668 36952068 C/T 0.16 0.14 0.407
rs216230782433 36952833 C/T 0.73 0.76 0.170
rs196283982559 36952959 C/G
rs237631583298 36953698 C/T 0.62 0.66 0.179
rs142641083821 36954221 A/G 0.75 0.77 0.307
rs189592184121 36954521 C/T 0.75 0.78 0.175
rs189592284147 36954547 C/T 0.15 0.12 0.095
rs103577984543 36954943 A/G 0.66 0.64 0.649
rs103578084554 36954954 A/G
rs103578184691 36955091 A/G 0.73 0.77 0.100
rs103578284727 36955127 A/G
rs142641185678 36956078 C/T 0.76 0.80 0.084
rs183460286699 36957099 C/T 0.20 0.16 0.072
rs183460386700 36957100 A/G 0.94 0.92 0.326
rs183460486792 36957192 A/G 0.70 0.73 0.287
rs183460586832 36957232 A/G 0.72 0.76 0.057
rs216230887045 36957445 A/G
rs136534187140 36957540 A/G 0.18 0.15 0.086
rs182045887365 36957765 A/C 0.23 0.21 0.298
rs146931088342 36958742 C/T 0.20 0.18 0.265
rs305787988498 36958898 -ITCA 0.70 0.71 0.649
rs146931188589 36958989 A/G 0.70 0.74 0.065
rs768326 95502 36965902 A/G
rs186352396968 36967368 C/T 0.21 0.18 0.247
rs146931297448 36967848 C/T 0.78 0.76 0.312
rs146931397568 36967968 C/T 0.81 0.80 0.617
rs195177398724 36969124 C/T
rs2120655Not ma Not ma T/G
ed ed
rs2181495Not ma Not ma G/A 0.78 0.76 0.617
ed ed
[0239] The chrorn 4 proximal SNPs were also allelotyped in the replication
cohorts using the
methods described herein and the primers provided in Tables 15 and 16. The
replication allelotyping
results for replication cohort #1 and replication cohort #2 are provided in
Tables 18 and 19,
respectively.
94
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
TABLE 18
Position
dbSNP in ChromosomeAl/A2 A2 CaseA2 Control
rs# position Allele AF AF -Value
SEQ D
NO:
rs3849023211 36870611 G/T
rs14443117217 36877617 A/G 0.27 0.25 0.441
rs20442957895 36878295 A/C
rs216609313308 36883708 C/T
rs237633414279 36884679 G/T 0.14 0.14 0.970
rs144432017026 36887426 C/T
rs204429418271 36888671 A/G 0.16 0.14 0.423
rs189986420417 36890817 C/T
rs156209421843 36892243 A/G 0.22 0.21 0.725
rs156209822069 36892469 A/G
rs156209722145 36892545 A/G
rs156209622519 36892919 A/G 0.20 0.19 0.795
rs156209522539 36892939 A/G 0.55 0.53 0.453
rs144431923236 36893636 A/C 0.70 0.80 0.003
rs144431823256 36893656 A/C 0.12 0.13 0.645
rs102593823402 36893802 C/T 0.18 0.18 0.824
rs102593723499 36893899 A/C
rs102593623620 36894020 C/T 0.85 0.83 0.622
rs102033323871 36894271 A/T
rs212065424136 36894536 C/G 0.16 0.16 0.914
rs258854725427 36895827 A/G 0.40 0.40 0.980
rs204429325866 36896266 G/T
rs276032426541 36896941 A/G 0.57 0.61 0.287
rs258854626576 36896976 G/T 0.08 0.05 0.265
rs258854526689 36897089 A/G
rs276032826720 36897120 A/C 0.25 unt ed NA
rs258854427113 36897513 C/T
rs276033127164 36897564 C/T 0.88 0.92 0.193
rs258854327186 36897586 A/G 0.57 0.58 0.869
rs258854228341 36898741 A/T
rs258854129160 36899560 C/T 0.61 0.57 0.230
rs258854029844 36900244 A/G 0.64 0.64 0.926
rs276033630665 36901065 G/T
rs276033730830 36901230 A/G 0.16 0.16 0.956
rs202873231061 36901461 A/C 0.60 0.57 0.330
rs258853831523 36901923 C/T 0.62 0.61 0.747
rs199261732326 36902726 C/T 0.62 0.59 0.341
rs199846932346 36902746 A/G
rs199847032358 36902758 C/T 0.78 0.88 0.0001
rs197549834909 36905309 CIT
rs156209334975 36905375 A/G 0.89 0.90 0.905
rs197549735066 36905466 C/T 0.12 0.12 0.873
rs156209235096 36905496 G/T
rs224878835375 36905775 C/T 0.28 0.31 0.308
rs189986236304 36906704 A/G 0.19 0.14 0.088
rs258853236712 36907112 A/T 0.30 0.33 0.347
rs188587836770 36907170 C/T 0.36 0.34 0.362
rs98664837342 36907742 C/T 0.74 0.75 0.773
rs98664737412 36907812 C/T 0.78 0.77 0.693
rs101001037884 36908284 A/G 0.25 0.26 0.690
rs101000938077 36908477 A/C 0.27 0.26 0.870
rs276032538300 36908700 C/T
rs258853138301 36908701 C/T
rs183838841189 36911589 C/T 0.74 0.75 0.826
rs197549544408 36914808 C/T
rs218149144493 36914893 A/C 0.16 0.10 0.057
rs 197549644571 36914971 A/G 0.25 0.26 0.596
rs218149244670 36915070 A/G 0.11 0.08 0.167
rs222471945219 36915619 A/G 0.78 0.79 0.705
rs222472045258 36915658 C/T 0.19 0.21 0.478
rs195177047261 36917661 A/G 0.25 0.16 0.003
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
Position
dbSNP in ChromosomeAl/A2 A2 CaseA2 Control
rs# position Allele AF AF -Value
SEQ D
NO:
rs229604048473 36918873 A/C 0.39 0.43 0
.241
rs195772348771 36919171 A/G 0.44 0.36 _
0.027
rs195772555292 36925692 C/T 0.73 0.80 0.024
rs288934656479 36926879 A/G 0.53 0_.55 0.552
rs188587956747 36927147 A/C 0.43 0.50 0.057
rs195772660620 36931020 G/T 0.14 0.14 0.918
rs19577276_0688 36931088 A/C 0.71 0_.78 0.038
rs188588061058 36931458 A/C 0.44 0.42 0.627
rs188588161129 36931529 C/T 0.12 0.12 0.833
rs94210861577 36931977 C/T 0.42 0.49 0.096
rs195177161961 36932361 A/G 0.93 NA NA
rs237632363351 36933751 G/T
rs201335863926 36934326 A/G 0.13 0.12 0.795
rs218149465798 36936198 A/G 0.38 0.41 0.424
rs195772866043 36936443 A/C
rs195772966044 36936444 A/G 0.78 0.77 0.672
rs195773066246 36936646 C/T 0.15 0.15 0.885
rs 195773166318 36936718 CIT 0.15 0.16 0.719
rs199846866547 36936947 G/T 0.14 0.10 0.243
rs195773271238 36941638 C/T 0.10 0.09 0.817
rs195773371283 36941683 A/G 0.62 NA 0.628
rs237632271492 36941892 A/G 0.26 0.27 0.660
rs288934572274 36942674 A/G 0.22 0.16 0.020
rs181526773762 36944162 A/T 0.46 0.48 0.626
rs195773474209 36944609 G/T 0.44 0.61 0.0001
rs195773575284 36945684 A/T 0.63 0.63 0.792
rs195773677347 36947747 A/C 0.03 0.03 0.987
rs195773777589 36947989 C/T 0.69 0.77 0.024
rs195773878096 36948496 A/G
rs195773978606 36949006 A/G
rs195774078862 36949262 G/T
rs195774179135 36949535 A/G 0.75 0.83 0.008
rs195774279146 36949546 A/G 0.94 0.96 0.459
rs195774379456 36949856 C/T 0.24 0.14 0.009
rs195774479609 36950009 A/G 0.63 0.72 0.006
rs195774580086 36950486 A/G 0.86 0.90 0.229
rs195774680119 36950519 A/G 0.42 0.50 0.019
rs195774780766 36951166 C/T 0.71 0.79 0.009
rs214667081110 36951510 A/G 0.72 0.81 0.004
rs214667181269 36951669 A/T 0.17 0.13 0.106
rs195774881668 36952068 C/T 0.17 0.13 0.133
rs216230782433 36952833 C/T 0.72 0.78 0.020
rs196283982559 36952959 C/G
rs237631583298 36953698 C/T 0.61 0.67 0.074
rs142641083821 36954221 A/G 0.73 0.79 0.058
rs189592184121 36954521 C/T 0.72 0.80 0.013
rs189592284147 36954547 C/T 0.17 0.11 0.014
rs103577984543 36954943 A/G 0.66 0.64 0.613
rs103578084554 36954954 A/G
rs103578184691 36955091 A/G 0.71 0.78 0.059
rs103578284727 36955127 A/G
rs142641185678 36956078 C/T 0.75 0.82 0.008
rs183460286699 36957099 C/T 0.22 0.15 0.020
rs183460386700 36957100 A/G 0.94 0.92 0.483
rs183460486792 36957192 A/G 0.69 0.75 0.056
rs183460586832 36957232 A/G 0.71 0.79 0.007
rs216230887045 36957445 A/G
rs136534187140 36957540 A/G 0.19 0.13 0.017
rs182045887365 36957765 A/C 0.24 0.19 0.141
rs146931088342 36958742 C/T 0.22 0.17 0.061
rs305787988498 36958898 -/TCA 0.67 NA NA
rs146931188589 36958989 A/G 0.67 0.76 0.006
rs76832695502 36965902 A/G
96
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
Position
dbSNP in ChromosomeAl/A2 A2 CaseA2 Control
rs# position Allele AF AF -Value
SEQ D
NO:
rs186352396968 36967368 C/T 0.22 0.17 0.103
rs146931297448 36967848 C/T 0.80 0.77 0.236
rs146931397568 36967968 C/T 0.83 0.80 0.422
rs195177398724 36969124 C/T
rs2120655Not ma Not ma T/G
ed ed
rs2181495Not ma Not ma G/A 0.78 0.76 0.617
ed ed
TABLE 19
Position
dbSNP in ChromosomeAllA2 A2 CaseA2 Control
rs# position Allele AF AF -Value
SEQ D
NO:
rs3849023211 36870611 G/T
rs14443117217 36877617 A/G 0.25 0.26 0.876
rs20442957895 36878295 A/C
rs216609313308 36883708 C/T
rs237633414279 36884679 G/T 0.16 0.18 0.532
rs144432017026 36887426 C/T
rs204429418271 36888671 A/G NA 0.15 NA
rs189986420417 36890817 C/T
rs156209421843 36892243 A/G NA 0.28 NA
rs156209822069 36892469 A/G
rs156209722145 36892545 A/G
rs156209622519 36892919 A/G 0.20 0.23 0.364
rs156209522539 36892939 A/G 0.50 0.48 0.588
rs144431923236 36893636 A/C 0.79 0.79 0.923
rs144431823256 36893656 A/C 0.12 0.13 0.711
rs102593823402 36893802 ClT 0.18 0.22 0.247
rs102593723499 36893899 A/C
rs102593623620 36894020 C/T 0.84 0.86 0.403
rs102033323871 36894271 A/T
rs212065424136 36894536 ClG 0.14 0.16 0.682
rs258854725427 36895827 A/G 0.37 NA
rs204429325866 36896266 G/T
rs276032426541 36896941 A/G 0.60 0.60 0.965
rs258854626576 36896976 G/T 0.05 0.06 0.797
rs258854526689 36897089 A/G
rs276032826720 36897120 A/C 0.25 0.26 0.816
rs258854427113 36897513 C/T
rs276033127164 36897564 C/T 0.95 0.96 0.597
rs258854327186 36897586 A/G 0.60 0.61 0.801
rs258854228341 36898741 A/T
rs258854129160 36899560 C/T 0.62 0.61 0.972
rs258854029844 36900244 A/G 0.60 0.59 0.810
rs276033630665 36901065 G/T
rs276033730830 36901230 A/G 0.16 0.17 0.659
rs202873231061 36901461 A/C 0.60 0.60 0.976
rs258853831523 36901923 ClT 0.61 0.61 0.912
rs199261732326 36902726 C/T 0.61 0.59 0.583
rs199846932346 36902746 A/G
rs199847032358 36902758 C/T 0.84 0.81 0.338
rs197549834909 36905309 C/T
rs156209334975 36905375 A/G 0.88 0.84 0.199
rs197549735066 36905466 C/T 0.13 0.15 0.613
rs156209235096 36905496 G/T
rs224878835375 36905775 C/T 0.30 0.31 0.884
rs189986236304 36906704 A/G 0.17 0.18 0.563
rs258853236712 36907112 A/T 0.29 0.29 0.952
rs188587836770 36907170 C/T 0.33 0.36 0.405
rs98664837342 36907742 C/T 0.75 0.72 0.283
rs98664737412 36907812 C/T 0.79 0.74 0.186
97
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
Position
dbSNP in ChromosomeAllA2 A2 CaseA2 Control
rs# position Allele AF AF -Value
SEQ D
NO:
rs101001037884 36908284 A/G _0.25 0.26 0.843_
rs101000938077 36908477 A/C 0.25 0.24 0.764
rs276032538300 36908700 C/T
rs258853138301 36908701 C/T
rs183838841189 36911589 C/T 0.76 0.72 0.284
rs197549544408 36914808 ClT
rs218149144493 36914893 A/C 0.12 0.15 0.464
rs197549644571 36914971 A/G 0.27 0.25 0.577
rs218149244670 36915070 A/G 0.10 0.11 0.844
rs222471945219 36915619 A/G 0.78 0.75 0.426
rs222472045258 36915658 C/T 0.21 0.20 0.790
rs195177047261 36917661 A/G 0.19 0.20 0.796
rs229604048473 36918873 A/C 0.43 0.42 0.804
rs195772348771 36919171 A/G 0.41 0.42 0.653
rs195772555292 36925692 C/T 0.77 0.75 0.439
rs288934656479 36926879 A/G 0.56 0.56 0.948
rs188587956747 36927147 A/C 0.46 0.45 0.959
rs195772660620 36931020 G/T 0.14 0.15 0.673
rs195772760688 36931088 A/C 0.76 0.73 0.255
rs188588061058 36931458 A/C 0.43 0.45 0.614
rs188588161129 36931529 C/T 0.13 0.10 0.346
rs94210861577 36931977 C/T 0.58 0.56 0.730
rs195177161961 36932361 A/G
rs237632363351 36933751 GlT
rs201335863926 36934326 A/G 0.13 0.15 0.469
rs218149465798 36936198 A/G 0.46 0.47 0.820
rs195772866043 36936443 A/C
rs195772966044 36936444 A/G 0.80 0.78 0.440
rs195773066246 36936646 C/T 0.15 0.17 0.668
rs195773166318 36936718 C/T 0.14 0.16 0.387
rs199846866547 36936947 G/T 0.12 0.13 0.615
rs195773271238 36941638 C/T 0.09 0.11 0.469
rs195773371283 36941683 A/G 0.60 0.02
rs237632271492 36941892 A/G 0.27 0.29 0.582
rs288934572274 36942674 A/G 0.17 0.21 0.308
rs181526773762 36944162 A/T 0.46 0.41 0.151
rs195773474209 36944609 G/T 0.68 0.69 0.766
rs195773575284 36945684 A/T 0.62 0.58 0.311
rs195773677347 36947747 A/C 0.07 0.08 0.688
rs195773777589 36947989 C/T 0.75 0.71 0.305
rs195773878096 36948496 A/G
rs195773978606 36949006 A/G
rs195774078862 36949262 G/T
rs195774179135 36949535 A/G 0.78 0.76 0.446
rs195774279146 36949546 A/G 0.96 0.96 0.938
rs 195774379456 36949856 C/T 0.17 0.19 0.667
rs195774479609 36950009 A/G 0.69 0.66 0.423
rs195774580086 36950486 A/G 0.90 0.89 0.738
rs195774680119 36950519 A/G 0.37 0.35 0.708
rs195774780766 36951166 C/T 0.72 0.70 0.639
rs214667081110 36951510 A/G 0.75 0.72 0.306
rs214667181269 36951669 A/T 0.16 0.17 0.806
rs195774881668 36952068 C/T 0.14 0.17 0.453
rs216230782433 36952833 C/T 0.76 0.73 0.465
rs196283982559 36952959 CIG
rs237631583298 36953698 C/T 0.64 0.63 0.767
rs142641083821 36954221 A/G 0.77 0.74 0.465
rs189592184121 36954521 C/T 0.78 0.75 0.320
rs189592284147 36954547 C/T 0.12 0.13 0.586
rs103577984543 36954943 A/G NA 0.65 NA
rs103578084554 36954954 A/G
rs103578184691 36955091 A/G 0.75 0.76 0.830
rs103578284727 36955127 A/G
98
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
Position
dbSNP in ChromosomeAllA2 A2 CaseA2 Control
rs# position Allele AF AF -Value
SEQ ~D
NO:
rs142641185678 36956078 C/T 0.78 0.76 0.488
rs183460286699 36957099 C/T 0.18 0.18 0.945
rs183460386700 36957100 A/G 0.95 NA
rs183460486792 36957192 A/G 0.72 0.69 0.427
rs183460586832 36957232 A/G 0.73 0.72 0.647
rs216230887045 36957445 A/G
rs136534187140 36957540 A/G 0.16 0.17 0.667
rs182045887365 36957765 A/C 0.23 0.24 0.670
rs146931088342 36958742 C/T 0.19 0.21 0.592
rs305787988498 36958898 -/TCA 0.74 0.71 0.478
rs146931188589 36958989 A/G 0.74 0.72 0.582
rs76832695502 36965902 A/G
rs186352396968 36967368 C/T 0.20 0.21 0.687
rs146931297448 36967848 C/T 0.76 0.75 0.807
rs146931397568 36967968 C/T 0.78 0.79 0.824
rs195177398724 36969124 C/T
rs2120655Not ma Not ma T/G
ed ed
rs2181495Not mapped~ Not G/A
~ mapped
[0240] Allelotyping results were considered particularly significant with a
calculated p-value of
less than or equal to 0.05 for allelotype results. These values are indicated
in bold. The allelotyping p-
values were plotted in Figure 1B for the discovery cohort. The position of
each SNP on the
chromosome is presented on the x-axis. The y-axis gives the negative logarithm
(base 10) of the p-
value comparing the estimated allele in the case group to that of the control
group. The minor allele
frequency of the control group for each SNP designated by an X or other symbol
on the graphs in
Figure 1B can be determined by consulting Table 17. For example, the left-most
X on the left graph is
at position 36870611. By proceeding down the Table from top to bottom and
across the graphs from
left to right the allele frequency associated with each symbol shown can be
determined.
[0241] To aid the interpretation, multiple lines have been added to the graph.
The broken
horizontal lines are drawn at two common significance levels, 0.05 and 0.01.
The vertical broken lines
are drawn every 20kb to assist in the interpretation of distances between
SNPs. Two other lines are
drawn to expose linear trends in the association of SNPs to the disease. The
generally bottom-most
curve is a nonlinear smoother through the data points on the graph using a
local polynomial regression
method (W.S. Cleveland, E. Grosse and W.M. Shyu (1992) Local regression
models. Chapter 8 of
Statistical Models in S eds J.M. Chambers and T.J. Hastie, Wadsworth &
Brooks/Cole). The black line
provides a local test for excess statistical significance to identify regions
of association. This was
created by use of a l Okb sliding window with lkb step sizes. Within each
window, a chi-square
goodness of fit test was applied to compare the proportion of SNPs that were
significant at a test wise
level of 0.01, to the proportion that would be expected by chance alone (0.05
for the methods used
here). Resulting p-values that were less than 10-8 were truncated at that
value.
[0242] Finally, the exons and introns of the genes in the covered region are
plotted below each
graph at the appropriate chromosomal positions. The gene boundary is indicated
by the broken
99
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
horizontal line. The exon positions are shown as thick, unbroken bars. An
arrow is place at the 3' end
of each gene to show the direction of transcription.
Example 6
Chrom 6 Region Proximal SNPs
[0243] It has been discovered that SNPs rs756519, rs1042327 and rs8770 on
chromosome 6 (6q27)
are associated with occurrence of osteoarthritis in subjects. This region
contains genes that encode
proteasome (prosome, macropain) subunit, beta type, 1 (PSMBI), TATA box
binding protein (TBP),
and programmed cell death 2 (PDCD2).
[0244] One hundred-nine additional allelic variants proximal to rs756519,
rs1042327 and rs8770
were identified and subsequently allelot0yped in osteoarthritis case and
control sample sets as described
in Examples 1 and 2. The polymorphic variants are set forth in Table 20. The
chromosome positions
provided in column four of Table 20 are based on Genome "Build 34" of NCBI's
GenBank.
TABLE 20
dbSNP Position Chromosome Allele
rs# Chromosomein SEQ Position Variants
ID NO:
3
rs14745556 229 170689279 c/t
rs14745546 6310 170695360 a/
rs10334 6 11840 170700890 /t
rs10541 6 11870 170700920 a/t
rs38232996 12064 170701114 a/
rs742348 6 13392 170702442 c/
rs14746446 16354 170705404 a/
rs14746436 16559 170705609 c/t
rs20569706 16935 170705985 a/
rs22234746 17616 170706666 c/t
rs22062846 17737 170706787 c/t
rs756519 6 18321 170707371 c/t
rs756518 6 18453 170707503 a/
rs756517 6 18811 170707861 c/t
rs14746426 20020 170709070 c/t
rs20380936 21662 170710712 c/
rs20380926 23197 170712247 cl
rs22234736 23446 170712496 /t
rs760909 6 24339 170713389 /t
rs20763196 25504 170714554 a/
rs37785896 27174 170716224 a/
rs38002366 28008 170717058 alt
rs22062866 29294 170718344 c/t
rs12717 6 29759 170718809 c/
rs21793736 30832 170719882 a/
rs38002356 44512 170733562 a/c
rs38232986 44850 170733900 c/
rs20763186 45884 170734934 a/
rs22355066 46345 170735395 clt
rs20729166 48589 170737639 a/
rs37347ti36 53371 170742421 a/
100
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP ChromosomePosition ChromosomeAllele
rs# in SEQ Position Variants
ID NO:
3
rs31775716 53911 170742961 /t
rs8770 6 53990 170743040 al
rs31732196 55152 170744202 c!
rs9607446 55667 170744717 c/t
rs20669546 58952 170748002 a/c
rs20729176 59315 170748365 /t
rs31732206 60029 170749079 a/
rs7342496 61477 170750527 a/c
rs20923106 62988 170752038 c/t
rs20923096 63090 170752140 c/
rs10165366 64021 170753071 alc
rs22355066 65685 170754735 c/t
rs20769986 70220 170759270 a/
rs20769976 70323 170759373 a/c
rs23454786 70959 170760009 a/c
rs20218996 73436 170762486 c/
rs20218986 82945 170771995 a/
rs23456826 82958 170772008 /t
rs23456836 82961 170772011 c/
rs28811956 82964 170772014 c/t
rs23456846 82965 170772015 It
rs30462616 83006 170772056 -/cttt
rs40834136 83025 170772075 c/t
rs40834126 83034 170772084 a/
rs23456856 83074 170772124 /t
rs20218976 83132 170772182 /t
rs40362116 83155 170772205 c/t
rs40362126 83172 170772222 alt
rs40362136 83174 170772224 /t
rs23456866 83206 170772256 c/t
rs40362146 83216 170772266 /t
rs40362156 83234 170772284 /t
rs23456876 83252 170772302 a/
rs23456886 83260 170772310 a/c
rs28811966 83263 170772313 a/c
rs30462886 83296 170772346 -/at
rs40362166 83319 170772369 a/
rs40362056 83322 170772372 c/
rs20923076 83324 170772374 alc
rs40362066 83357 170772407 c/
rs23456896 83375 170772425 c/t
rs23456906 83381 170772431 c/t
rs23456916 83389 170772439 a/t
rs23456926 83443 170772493 a/
rs30463066 83499 170772549 -/ t
rs40362076 83545 170772595 clt
rs23456936 83566 170772616 clt
rs23456946 83591 170772641 c/t
rs23456956 83619 170772669 /t
rs23456966 83698 170772748 a/
rs40362096 83780 170772830 /t
rs2345697~ 83784 170772834 g/t
101
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Position Chromosome Allele
rs# Chromosomein SEQ Position Variants
ID NO:
3
rs28811976 83826 170772876 /t
rs23456986 83832 170772882 c/t
rs23456996 83852 170772902 c/t
rs27446406 86297 170775347 c/t
rs27446396 86315 170775365 /t
rs27446386 86420 170775470 c!
rs27446376 86460 170775510 c/
rs27446366 86714 170775764 c/t
rs27446356 86718 170775768 clt
rs27446346 86736 170775786 c/
rs27446336 86753 170775803 c/t
rs27446326 86766 170775816 It
rs27446306 88162 170777212 c/
rs27446296 88218 170777268 a/
rs27446286 88246 170777296 al
rs27446276 88255 170777305 c/t
rs29776166 88309 170777359 /t
rs29776176 88310 170777360 a/t
rs27446266 88471 170777521 a/
rs27446256 88619 170777669 c/t
rs31158476 88904 170777954 c/t
rs27446236 89044 170778094 c/
rs40361936 90531 170779581 -/aaaaa
rs40361946 90534 170779584 a/
rs40361966 90613 170779663 c/
rs10423276 46252 170735302 c/t
Assay for Verifying and Allelotypin~ SNPs
[0245] The methods used to verify and allelotype the 109 proximal SNPs of
Table 20 are the same
methods described in Examples 1 and 2 herein. The primers and probes used in
these assays are
provided in Table 21 and Table 22, respectively.
TABLE 21
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs1474555ACGTTGGATGACATCAACTGAAGCCGACAGACGTTGGATGAATGGTGGAATGTGATGAGA
rs1474554ACGTTGGATGATACACCTAGGACACCTCCAACGTTGGATGCAGAAGGAGATAAACCCAGC
rs10334ACGTTGGATGAACAGTTTCCTCCCTGATGCACGTTGGATGCGGCTGGTGAAAGATGTCTT
rs10541ACGTTGGATGACTATGCAGATCCGGAGTGCACGTTGGATGGTCCTTGGACAGAGCCATG
rs3823299ACGTTGGATGCTCATGTGTACGAGGATTTGACGTTGGATGGTCTGGAAGGGTCTTTATTC
rs742348ACGTTGGATGTGTGGATTTTCCAGTGCTCGACGTTGGATGCTGTACTTGAACTCCCAAGC
rs1474644ACGTTGGATGGCAAGACAAGCATAATTGGGACGTTGGATGTAAAGGGCATTTTGGCTTCC
rs1474643ACGTTGGATGTCTCCCAAATTAAAAGTGGCACGTTGGATGGATACCAAAGTCCTACTTAC
rs2056970ACGTTGGATGTGGGACTACAGGAAGAGAAGACGTTGGATGCAAAACACAGACCTTCAGCC
rs2223474ACGTTGGATGCCAGGGTAAAGAAAAGATCCACGTTGGATGAGAGGCTTACCTCCTAAAAG
rs2206284ACGTTGGATGTCACATACTAGGTGGATCCCACGTTGGATGAAAGAGGAGAACACAGGATG
rs756519ACGTTGGATGTCTAGAGACACCTGAGGTTGACGTTGGATGTGTTTCACTTCAGAGCCCTG
rs756518ACGTTGGATGCCCAGATTAGACTCTCTAACACGTTGGATGAAATAGCTGAGCTGCCATTG
102
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs756517ACGTTGGATGCTCGGTTGTTGACTCCTATCACGTTGGATGGCGGATGTTAAGAGTCAGAG
rs1474642ACGTTGGATGGGAGGTCATACATTAGCTTCACGTTGGATGTACCATCTGACACAATTCTC
rs2038093ACGTTGGATGGAGACAGAGTTTCACTCTTGACGTTGGATGTAATCACTTGAACCCAGGAG
rs2038092ACGTTGGATGTTACCTGAGGTCAGGAGTTTACGTTGGATGCCACACCCAGCTGATTTTTG
rs2223473ACGTTGGATGCCTTTATGTTATTGCTTTCCACGTTGGATGCAGGGAAATTTAAGAATAGC
rs760909ACGTTGGATGGGAAGAGGCAAGCTTAGTTCACGTTGGATGGCAGCATTAACGAATGCCTG
rs2076319ACGTTGGATGGACATTTCACAATGCCTTTGACGTTGGATGCCAACAGCAACTTAAAAACTC
rs3778589ACGTTGGATGGCAAGAGAGAGAAAAGTTCCACGTTGGATGGTGTTTCTGTCCCATTTCAC
rs3800236ACGTTGGATGAGAGAATGAGGCCTCATTTTACGTTGGATGCTCAGTCATTGTTCTTTTTC
rs2206286ACGTTGGATGTTCAGACGCTAACCCTCTACACGTTGGATGAACATAGCCTCTGCTCTGTG
rs12717ACGTTGGATGAAAATCGCAGCTGCAAAGGGACGTTGGATGAGACAGCAAGTGTCGGATCC
rs2179373ACGTTGGATGGAAGTGACCTATGCTCACACACGTTGGATGAATGTCACTTCCGCCAGTTC
rs3800235ACGTTGGATGCTATGTGTTGATACCTCCAAGACGTTGGATGGCTTCATAAATGAACTGAAC
rs3823298ACGTTGGATGGGTGGTTTCTTGTCTTGATGACGTTGGATGTTTTTGTCCCAGAGCATCTG
rs2076318ACGTTGGATGTCCGCCAAATTATTGTAGCCACGTTGGATGCTCAGTAGAAATGCATGGGC
rs2235506ACGTTGGATGTAACCATGTCAACTGTTCTCACGTTGGATGCCCACCAACAATTTAGTAGG
rs2072916ACGTTGGATGACGCTGGAGTCACTAAGATGACGTTGGATGCAGATTAAGGCACAGGCATG
rs3734763ACGTTGGATGGCCTTTTGCCTTTCAGTGTCACGTTGGATGTAAAGAGGCTGGACCTTCAG
rs3177571ACGTTGGATGGTCTGTTGTCAATATAGGTGACGTTGGATGACAAAAGTGTCCAGTGACAG
rs8770 ACGTTGGATGAATTCCCTGTCACTGGACACACGTTGGATGCCAAAAATAGAGGTGCAGAG
rs3173219ACGTTGGATGACATAACCACACTGGAGGTGACGTTGGATGCCTAGTTTTCAGACACGGTC
rs960744ACGTTGGATGAAAGGCATGTCACAGTTCCCACGTTGGATGGCCCTCTGAGTCAGATAAAC
rs2066954ACGTTGGATGGAGGTTCTGGGTATAACTTTCACGTTGGATGCTACAAACCAGTAAGCTGATG
rs2072917ACGTTGGATGTGCTAGGCACTCACACTATCACGTTGGATGAGGCTTGGTAAGTTCCTCTG
rs3173220ACGTTGGATGTATCTGGGTTGACAAAGGCGACGTTGGATGACATAAGCAGGCTTGTGCAC
rs734249ACGTTGGATGAGGTGGACACCAGCAGGGAAACGTTGGATGTCACCTCTGCACATGTCTTG
rs2092310ACGTTGGATGTTAGTCAGGTAAAGCGGGACACGTTGGATGTCAGTGGAAGGCTGATCAAG
rs2092309ACGTTGGATGATCTAATTGCTTCCCCTCCCACGTTGGATGCAGCCTTCCACTGAATACAC
rs1016536ACGTTGGATGCCCCAAAAATTGGAGACAGGACGTTGGATGGGCTGTCATAATCGTGTGTC
rs2235506ACGTTGGATGAAGTGATTCTCCTGCCTCAGACGTTGGATGTGGTGAAACCCTGTCTCTAC
rs2076998ACGTTGGATGGCTCTGTGATTTCGATGATGACGTTGGATGAGCTACTTCTTGCAGGAGTC
rs2076997ACGTTGGATGCAGAGCTTCCAAGTGTTTTCACGTTGGATGAAAGGAGTGCTTAAAGGAGC
rs2345478ACGTTGGATGCCTTCAACAAGTGCTGACACACGTTGGATGATCCAGGCATTATTGCCAGC
rs2021899ACGTTGGATGGTTTTGTGGTGGATGATGGGACGTTGGATGAGAGTGCCCATAATGGACAG
rs2021898ACGTTGGATGCGCAAGAAACTCCTTGGATGACGTTGGATGCCAATTAAAGCCAAGGTCAC
rs2345682ACGTTGGATGATTCGCAAGAAACTCCTTGGACGTTGGATGGGAAGAAATCTTACCAGAAC
rs2345683ACGTTGGATGATTCGCAAGAAACTCCTTGGACGTTGGATGGGAAGAAATCTTACCAGAAC
rs2881195ACGTTGGATGATTCGCAAGAAACTCCTTGGACGTTGGATGGGAAGAAATCTTACCAGAAC
rs2345684ACGTTGGATGATTCGCAAGAAACTCCTTGGACGTTGGATGGGAAGAAATCTTACCAGAAC
rs3046261ACGTTGGATGCTCCACTCAGACATCAAAAGACGTTGGATGGTGACCTTGGCTTTAATTGG
rs4083413ACGTTGGATGGTGACCTTGGCTTTAATTGGACGTTGGATGCTCCACTCAGACATCAAAAG
rs4083412ACGTTGGATGGTGACCTTGGCTTTAATTGGACGTTGGATGCTCCACTCAGACATCAAAAG
rs2345685ACGTTGGATGGTTCTGGTAAGATTTCTTCCACGTTGGATGAGTCTTACAATAGATGACTG
rs2021897ACGTTGGATGGCAATTATTTACAGAAGCCCACGTTGGATGTCCCACACAGTCATCTATTG
rs4036211ACGTTGGATGCCCATTACAAGTTGGGCAGTTACGTTGGATGCTTTCTGATTCCTTTTTTTTCC
rs4036212ACGTTGGATGCTTTCTGATTCCTTTTTTTTCCACGTTGGATGCCCATTACAAGTTGGGCAGTT
rs4036213ACGTTGGATGCCCATTACAAGTTGGGCAGTTACGTTGGATGCTTTCTGATTCCTTTTTTTTCC
rs2345686ACGTTGGATGCCCATTACAAGTTGGGCAGTTACGTTGGATGCTTTCTGATTCCTTTTTTTTCC
rs4036214ACGTTGGATGCCCATTACAAGTTGGGCAGTTACGTTGGATGCTTTCTGATTCCTTTTTTTTCC
rs4036215ACGTTGGATGCTTTCTGATTCCTTTTTTTTCCACGTTGGATGCCCATTACAAGTTGGGCAGTT
rs2345687ACGTTGGATGGGATTGTAAGGTGAGACTTGACGTTGGATGTTCCTCCCCATTACAAGTTG
rs2345688ACGTTGGATGAGGGTCCCATCTAAGAATTC~ ACGTTGGATGGGATTGTAAGGTGAGACTTG
103
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs2881196ACGTTGGATGAGGGTCCCATCTAAGAATTCACGTTGGATGGGATTGTAAGGTGAGACTTG
rs3046288ACGTTGGATGCCAACTTGTAATGGGGAGGAACGTTGGATGCAGTTTTTACAGAGGGTCCC
rs4036216ACGTTGGATGCTTGTAATGGGGAGGAAAAAAACGTTGGATGTTCTCATTTTAATCTGTCAG
rs4036205ACGTTGGATGCTTGTAATGGGGAGGAAAAAAACGTTGGATGTTCTCATTTTAATCTGTCAG
rs2092307ACGTTGGATGCTTGTAATGGGGAGGAAAAAAACGTTGGATGTTCTCATTTTAATCTGTCAG
rs4036206ACGTTGGATGGACCCTCTGTAAAAACTGACACGTTGGATGCCACTGCACCTCAAATCTTC
rs2345689ACGTTGGATGTTCCCTGAGTATCTCCCATGACGTTGGATGGGGACCCTCTGTAAAAACTG
rs2345690ACGTTGGATGTTCCCTGAGTATCTCCCATGACGTTGGATGGGGACCCTCTGTAAAAACTG
rs2345691ACGTTGGATGGCCACCTGTTGGAGATTTACACGTTGGATGGGGACCCTCTGTAAAAACTG
rs2345692ACGTTGGATGTACATGGGAGATACTCAGGGACGTTGGATGCCACTGCACCTCAAATCTTC
rs3046306ACGTTGGATGGTATAACAAACCTTACCCTTGACGTTGGATGTAAAGAAAGAAGATTTGAGG
rs4036207ACGTTGGATGTATCAATGGAGAATGCGTGGACGTTGGATGGGGAGTTAACCAGCAAAAGC
rs2345693ACGTTGGATGTCGACAACAAGAAGAGAAGGACGTTGGATGCACATTAGACAAGGGTAAGG
rs2345694ACGTTGGATGCTACCTCTCTCGACAACAAGACGTTGGATGCTTAAGTCCACGCATTCTCC
rs2345695ACGTTGGATGCGCATTCTCCATTGATAAGACACGTTGGATGCCATTTAAAAGCTACCTCTC
rs2345696ACGTTGGATGCCTTACACAAGTGTAACTTCACGTTGGATGCCCCAAAATATAATGGTAGG
rs4036209ACGTTGGATGGGAACACAGTGTATAAGACCACGTTGGATGGTTTTCACAACTTCGTTAGC
rs2345697ACGTTGGATGGTTTTCACAACTTCGTTAGCACGTTGGATGGCCACCCCAAAATATAATGG
rs2881197ACGTTGGATGGCTGGAGGAAAAACAAGAACACGTTGGATGCCTACCATTATATTTTGGGG
rs2345698ACGTTGGATGCTGGAGGAAAAACAAGAACTCACGTTGGATGCATTATATTTTGGGGTGGCAT
rs2345699ACGTTGGATGGCTGGAGGAAAAACAAGAACACGTTGGATGGGGTGGCATATTTTGGTCTT
rs2744640ACGTTGGATGGCAACAGCACTTAGTATGCCACGTTGGATGTGTGAAGCTGCAAATCTGGC
rs2744639ACGTTGGATGGCAACAGCACTTAGTATGCCACGTTGGATGTGTGAAGCTGCAAATCTGGC
rs2744638ACGTTGGATGAACCGTGGCAATACCACGTCACGTTGGATGTGGGTTTGGGCTGGATTTGG
rs2744637ACGTTGGATGTGAGTTGACAGCCTCTGCTGGACGTTGGATGCACGTCAGTAAGGCAGAGAC
rs2744636ACGTTGGATGTCGGAGATGACATTGTCACCACGTTGGATGTTCCAGGGGTTACGTGTGTG
rs2744635ACGTTGGATGTGAGTCTGACTGTGTCACGGACGTTGGATGTCGGAGATGACATTGTCACC
rs2744634ACGTTGGATGCGTGTTCCAGGGATTATATGACGTTGGATGGCACATAACGCTTGGAACTC
rs2744633ACGTTGGATGTATGAGTGTGACGGGTGTAGACGTTGGATGGCACATAACGCTTGGAACTC
rs2744632ACGTTGGATGTAGCTGCCTTCCACATCCAAACGTTGGATGTGTGACGGGTGTAGCGTTAG
rs2744630ACGTTGGATGGGGTTCAAATGCCTCTGATAGACGTTGGATGGGTCTAGGACAAGACCCATT
rs2744629ACGTTGGATGAACTTTCCCTTAGCCAGTGGACGTTGGATGATCAGAGGCATTTGAACCCC
rs2744628ACGTTGGATGTTGACCTCAAATCATGTCACACGTTGGATGTATCAGAGGCATTTGAACCC
rs2744627ACGTTGGATGGGGTGGTTTATGTTCCACTGACGTTGGATGCCAGAACTAATGCTAGCTTC
rs2977616ACGTTGGATGTTCCACTGGCTAAGAGAAAGACGTTGGATGCCAGAACTAATGCTAGCTTC
rs2977617ACGTTGGATGCCAGAACTAATGCTAGCTTCACGTTGGATGTTCCACTGGCTAAGAGAAAG
rs2744626ACGTTGGATGACAGTGAAATTGTATTTCCGACGTTGGATGGCACAAACTTAAGAATCTCC
rs2744625ACGTTGGATGAGCAAAATCCACCTATGTCCACGTTGGATGCTGAATTTTGTCTCCAGTAC
rs3115847ACGTTGGATGTCGAGGCAGAGGCGTAGTAACGTTGGATGATAGGAATGACATGAACCCG
rs2744623ACGTTGGATGACGCGAGTCCGTAGGTGCTGACGTTGGATGAAGAGGCTGCTACCCAGAG
rs4036193ACGTTGGATGAGAGCAAGACTCCGTCTCAAACGTTGGATGACATGTCGCTTGATGTGTGC
rs4036194ACGTTGGATGACATGTCGCTTGATGTGTGCACGTTGGATGAGAGCAAGACTCCGTCTCAA
rs4036196ACGTTGGATGCCCCAGCGTTCATATTTGTCACGTTGGATGTCTGGCCAAATGGTCATACC
rs1042327ACGTTGGATGAACTTCACATCACAGCTCCCACGTTGGATGCAGAAGTTGGGTTTTCCAGC
TABLE 22
dbSNP Extend 'Perm
rs# Primer Mix
rs1474555 TGAAGCCGACAGTGACACC ACT
rs1474554 CCAATTTTGCACACCTCCAGCA ( ACG
104
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Extend Term
rs# Primer Mix
rs10334 CAGATCCGGAGTGCGTCC CGT
rs10541 TCTCTCTCAGCCGCAGAA CGT
rs3823299 GAGGATTTGTGATGAAAATACTA ACG
rs742348 AATCCCCGTGTTGTTCAAGG ACT
rs1474644 AAGGATGTTCATCATAGTGTTTA ACG
rs1474643 ACATGTTTATACATACACTCATG ACG
rs2056970 TTGGCAGCTTTTTAGGCCTC ACT
rs2223474 AAGTCTCAAAAAGGTCCC ACT
rs2206284 TAGGTGGATCCCTTTTCCC ACG
rs756519 CAGAGCCCTGTTCTTTGATTT ACG
rs756518 CAAAGGATGCTGTCTGGCC ACG
rs756517 GTTCCATGAGCGTTTTCTTTG ACG
rs1474642 CTTCAGTTTCTTCATCACTTTC ACT
rs2038093 TTTCACTCTTGTTGCCCAGG ACT
rs2038092 CCAACATGGTGAAACCCCATCT ACT
rs2223473 TAGAATTAAAATTAGACTTTGGGGACT
rs760909 GCAAGCTTAGTTCTAGGTCAG CGT
rs2076319 TCACAATGCCTTTGTAATGATTT ACT
rs3778589 GTTTTAGGAAGACTGCTCTGACAAACG
rs3800236 CTGAGAGCCAGCTGCAGTAA CGT
rs2206286 CCTCGCCGGCTGGCATAA ACT
rs12717 CCATCCCCAAGTCTCTGCCAG ACT
rs2179373 TGACCTATGCTCACACTTCTCA ACG
rs3800235 GTGTTGATACCTCCAAGTACATTTCGT
rs3823298 CTTGATGAAATAGTCATCCAACTAACT
rs2076318 TGAATTATCACCATCATCA ACT
rs2235506 TGTTGCCAATAACAATCA ACG
rs2072916 TGTGACAAGGGATTCCAC ACG
rs3734763 CATCTGTAAGCAGGGCCGC ACG
rs3177571 AAGACTGTGTAGCCTTCCTCTG ACT
rs8770 GTAGACACTGTGTAAGCAATC ACG
rs3173219 CACTGGAGGTGGAGAGCA ACT
rs960744 CCCCATCAGACCTGGCTGT ACT
rs2066954 TTACAATTTGAGCCTTGAGC CGT
rs2072917 CTATCCCGACCCGAGAAAC CGT
rs3173220 GCGATGAAACTGAACTGA ACT
rs734249 CACCAGCAGGGAAGGTTTG CGT
rs2092310 TTGAGGTGAGGGCTTCCAG ACT
rs2092309 TCCCCTCCCCTATTGTTTAC ACT
rs1016536 AAATTGGAGACAGGTCTCAGT ACT
rs2235506 CTGGGAGTACAGGTGCGC ACT
rs2076998 GTTTTTGTATAGTCTGCAGATGC ACT
rs2076997 ATCCATTTTAATGGGTTGCTAGCTACT
rs2345478 ACAACTGTACTTATTGGGCATA ACT
rs2021899 CTTTCTTGGAAACTCTTCCCA ACT
rs2021898 TTGGATGGGGTTAATGGCAG ACG
rs2345682 GTTAATGGCAGCTGTATTTTTCTGACT
rs2345683 GGCAGCTGTATTTTTCTGTGA ACT
rs2881195 CAGCTGTATTTTTCTGTGACCT ACG
105
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Extend Term
rs# Primer Mix
rs2345684 GCAGCTGTATTTTTCTGTGACCTTACT
rs3046261 GAAAACATTTGAGATACTGAAGATACT
rs4083413 TTCCTTTATCTTCAGTATCTCAA ACT
rs4083412 TCTTCAGTATCTCAAATGTTTTCAACG
rs2345685 CAACTTTTGATGTCTGAGTGGA ACT
rs2021897 ATTATTTACAGAAGCCCTATTCA ACT
rs4036211 TTTCCAAACAAAAGCTACCATGCAACT
rs4036212 AAATAATTGCATGGTAGCTTTTG CGT
rs4036213 ACAACTACTTTGATGTTATTTCC CGT
rs2345686 ACAATCCAAAAATCACATTCCTA ACT
rs4036214 GTCTCACCTTACAATCCAAAAAT CGT
rs4036215 AATGTGATTTTTGGATTGTAAGG ACT
rs2345687 AAGGTGAGACTTGTTTAGCTTT ACT
rs2345688 TCCTCCCCATTACAAGTTGGGCA ACT
rs2881196 TTTTCCTCCCCATTACAAGTTGG ACT
rs3046288 TAATGGGGAGGAAAAAAATTTTCTACT
rs4036216 ATGTTTTTGGAATTCTTAGATGG ACT
rs4036205 GTTTTTGGAATTCTTAGATGGGACACT
rs2092307 TGGAATTCTTAGATGGGACCC ACT
rs4036206 ACTGACAGATTAAAATGAGAAAAAACT
rs2345689 TCCCATGTATCCATAAGGTATAC ACT
rs2345690 GTATCTCCCATGTATCCATAAG ACT
rs2345691 CCCTGAGTATCTCCCATGTA CGT
rs2345692 TCTCCAACAGGTGGCTTTCA ACT
rs3046306 TTGCTGGTTAACTCCCCACT CGT
rs4036207 GCGTGGACTTAAGTCTGTATAAC ACT
rs2345693 AGAGTCTTATCAATGGAGAATGC ACT
rs2345694 GAAGAGAAGGATAACTAAATCACTACT
rs2345695 ATTTAGTTATCCTTCTCTTCTTG ACT
rs2345696 ACACAAGTGTAACTTCTACTCT ACT
rs4036209 GGAAACCAGAATATGCCACC CGT
rs2345697 AGCCAAAGGGACATATTTTGTGGTACT
rs2881197 GGAACACAGTGTATAAGACCAAA CGT
rs2345698 CGGTGGAACACAGTGTATAAG ACT
rs2345699 AAAACAAGAACTCTTTTCATTGCCACT
rs2744640 TTTATCTCCAGTTCCCCAGC ACG
rs2744639 AGCACTTAGTATGCCTTCTCCTT ACT
rs2744638 TGGCAATACCACGTCAGTAAG ACT
rs2744637 GCTGGGCTGGGTTTGGGCTG ACT
rs2744636 ACCCGTCACACTCATATAATCCC ACG
rs2744635 ACACATGCGTGTTCCAGGG ACT
rs2744634 GGGATTATATGAGTGTGACGG ACT
rs2744633 GGGTGTAGCGTTAGGTGAC ACT
rs2744632 GCGCACATAACGCTTGGAAC ACT
rs2744630 CGTGTTAAAACTCATGGCCAAAC ACT
rs2744629 ATAAACCACCCTGGAGTTCAT ACT
rs2744628 TTGAAGAAAACTTTCCCTTAGCCAACT
rs2744627 GTTTATGTTCCACTGGCTAAG ACT
~rs2977616 TTGAGGTCAAACATTAATATCAAGI ACT
106
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Extend Term
rs# Primer Mix
rs2977617 CTAGCTTCTCAATCTTTTGAGTT CGT
rs2744626 GTGAAATTGTATTTCCGGATTTC ACT
rs2744625 TCCTGAACACTTATCCACTTTAC ACT
rs3115847 CCAGGGCTGGAGGGGCC ACT
rs2744623 GGTGCTGGCGGGAGCGAGAGT ACT
rs4036193 GACTCCGTCTCAAAAAAAAAAAAAACT
rs4036194 CTTGATGTGTGCTTCAGGGTA ACG
rs4036196 CAGTGCAAGTAAAGAGCCTTA ACT
rs1042327 CATCACAGCTCCCCACCAT ACT
Genetic Anal,
[0246] Allelotyping results from the discovery cohort are shown for cases and
controls in Table 23.
The allele frequency for the A2 allele is noted in the fifth and sixth columns
for osteoarthritis case pools
and control pools, respectively, where "AF" is allele frequency. The allele
frequency for the A1 allele
can be easily calculated by subtracting the A2 allele frequency from 1 (A1 AF
= 1-A2 AF). For
example, the SNP rs1474555 has the following case and control allele
frequencies: case Al (C) = 0.64;
case A2 (T) = 0.36; control A1 (C) = 0.70; and control A2 (T) = 0.30, where
the nucleotide is provided
in paranthesis. Some SNPs are labeled "untyped" because of failed assays.
TABLE 23
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in Position AlleleCase Control Value
SEQ ID AF AF
NO: 3
rs1474555 229 170689279 C/T 0.36 0.30 0.024
rs1474554 6310 170695360 A/G 0.48 0.43 0.058
rs10334 11840 170700890 G/T
rs10541 11870 170700920 A/T
rs3823299 12064 170701114 A/G 0.45 0.41 0.125
rs742348 13392 170702442 C/G 0.46 0.44 0.275
rs1474644 16354 170705404 A/G 0.75 0.77 0.270
rs1474643 16559 170705609 C/T 0.45 0.40 0.042
rs2056970 16935 170705985 A/G 0.36 0.33 0.242
rs2223474 17616 170706666 C/T 0.42 0.46 0.140
rs2206284 17737 170706787 C/T 0.37 0.35 0.493
rs756519 18321 170707371 C/T
rs756518 18453 170707503 A/G 0.49 0.53 0.133
rs756517 18811 170707861 C/T
rs1474642 20020 170709070 ClT 0.12 0.12 0.904
rs2038093 21662 170710712 C/G
rs2038092 23197 170712247 C/G
rs2223473 23446 170712496 G/T 0.42 0.45 0.296
rs760909 24339 170713389 G/T 0.49 0.52 0.255
rs2076319 25504 170714554 A/G 0.43 0.46 0.219
rs3778589 27174 170716224 A/G 0.49 0.54 0.081
rs3800236 28008 170717058 A/T 0.47 0.50 0.319
rs2206286 29294 170718344 C/T 0.81 0.82 0.831
rs12717 29759 170718809 C/G 0.52 0.57 0.081
rs2179373 30832 170719882 A/G 0.58 0.62 0.089
rs3800235 44512 170733562 A/C 0.60 0.64 0.077
rs3823298 44850 170733900 C/G 0.44 0.38 0.022
rs2076318 45884 170734934 A/G 0.41 0.45 0.109
rs2235506 46345 170735395 C/T 0.68 0.66 0.320
rs2072916 48589 170737639 A/G 0.48 0.51 0.192
107
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in Position Allele . Control Value
SEQ ID Case AF
NO: 3 AF
rs373476353371 170742421A/G 0.50 0.54 0.142
rs317757153911 170742961G/T
rs8770 53990 170743040A/G
rs317321955152 170744202C/G 0.49 0.53 0.056
rs960744 55667 170744717C/T 0.39 0.35 0.179
rs206695458952 170748002A/C 0.37 0.32 0.057
rs207291759315 170748365G/T 0.46 0.42 0.153
rs317322060029 170749079A/G
rs734249 61477 170750527A/C 0.48 0.40 0.022
rs209231062988 170752038C/T
rs209230963090 170752140C/G 0.43 0.47 0.165
rs101653664021 170753071A/C 0.10 0.10 0.985
rs223550665685 170754735C/T
rs207699870220 170759270A/G
rs207699770323 170759373AlC 0.90 0.90 0.814
rs234547870959 170760009A/C 0.09 0.09 0.947
rs202189973436 170762486C/G 0.46 0.43 0.218
rs202189882945 170771995A/G
rs234568282958 170772008G/T
rs234568382961 170772011C/G 0.28 0.34 0.019
rs288119582964 170772014C/T
rs234568482965 170772015G/T
rs304626183006 170772056-/CTTT
rs408341383025 170772075C/T
rs408341283034 170772084A/G
rs234568583074 170772124G/T 0.71 0.71 0.835
rs202189783132 170772182G/T
rs403621183155 170772205C/T
rs403621283172 170772222A/T
rs403621383174 170772224G/T
rs234568683206 170772256C/T
rs403621483216 170772266G/T
rs403621583234 170772284G/T
rs234568783252 170772302A/G 0.55 0.50 0.085
rs234568883260 170772310AlC 0.53 0.52 0.958
rs288119683263 170772313A/C
rs304628883296 170772346-/AT
rs403621683319 170772369A/G
rs403620583322 170772372C/G
rs209230783324 170772374A/C
rs403620683357 170772407C/G
rs234568983375 170772425C/T
rs234569083381 170772431C/T
rs234569183389 170772439A/T
rs234569283443 170772493A/G
rs304630683499 170772549-/GGTG 0.42 0.43 0.761
rs403620783545 170772595C/T
rs234569383566 170772616C/T
rs234569483591 170772641C/T
rs234569583619 170772669G/T
rs234569683698 170772748A/G
rs403620983780 170772830G/T 0.79 0.73 0.156
rs234569783784 170772834G/T
rs288119783826 170772876G/T
rs234569883832 170772882C/T
rs234569983852 170772902C/T
rs274464086297 170775347C/T 0.53 0.53 0.973
rs274463986315 170775365G/T 0.40 0.40 0.789
rs274463886420 170775470C/G 0.39 0.39 0.941
rs274463786460 170775510C/G 0.40 0.42 0.497
rs274463686714 170775764C/T 0.76 0.73 0.271
rs274463586718 170775768C/T 0.03 0.02 0.425
rs274463486736 170775786C/G 0.96 0.94 0.436
rs274463386753 170775803C/T 0.14 0.16 0.409
108
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in Position AlleleCase Control Value
SEQ ID AF AF
NO: 3
rs274463286766 170775816G/T 0.80 0.83 0.217
rs274463088162 170777212CIG
rs274462988218 170777268A/G 0.80 0.80 0.978
rs274462888246 170777296AlG 0.71 0.67 0.206
rs274462788255 170777305C/T 0.32 0.30 0.335
rs297761688309 170777359G/T
rs297761788310 170777360A/T
rs274462688471 170777521A/G
rs274462588619 170777669C/T
rs311584788904 170777954C/T
rs274462389044 170778094ClG
rs403619390531 170779581-/AAAAA
rs403619490534 170779584AlG
rs403619690613 170779663C/G
rs 104232746252 170735302ClT 0.45 0.39 0.028
[0247] The Chrom 6 proximal SNPs were also allelotyped in the replication
cohorts using the
methods described herein and the primers provided in Tables 11 and 12. The
replication allelotyping
results for replication cohort # 1 and replication cohort #2 are provided in
Tables 24 and 25,
respectively.
TABLE 24
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in Position Allele Case Control Value
SEQ ID AF AF
NO: 3
rs1474555 229 170689279C/T 0.37 0.27 0.004
rs1474554 6310 170695360A/G 0.50 0.42 0.020
rs10334 11840 170700890G/T
rs10541 11870 170700920A/T
rs3823299 12064 170701114A/G 0.45 0.40 0.080
rs742348 13392 170702442C/G 0.47 0.41 0.075
rs1474644 16354 170705404A/G 0.75 0.79 0.231
rs1474643 16559 170705609C/T 0.46 0.39 0.028
rs2056970 16935 170705985A/G 0.38 0.33 0.129
rs2223474 17616 170706666C/T 0.41 0.48 0.052
rs2206284 17737 170706787C/T 0.37 0.34 0.342
rs756519 18321 170707371C/T
rs756518 18453 170707503A/G 0.48 0.56 0.013
rs756517 18811 170707861C/T
rs1474642 20020 170709070C/T 0.10 0.13 0.27_7_
rs2038093 21662 170710712C/G
rs2038092 23197 170712247C/G
rs2223473 23446 170712496G/T 0.42 0.48 0.070
rs760909 24339 170713389G/T 0.47 0.54 0.077
rs2076319 25504 170714554A/G 0.41 0.49 0.017
rs3778589 27174 170716224A/G 0.50 0.57 0.035
rs3800236 28008 170717058A/T 0.47 0.52 0.126
rs2206286 29294 170718344ClT 0.80 0.80 0.952
rs12717 29759 170718809C/G 0.53 0.59 0.059
rs2179373 30832 170719882AlG 0.57 0.64 0.025
rs3800235 44512 170733562A/C 0.59 0.65 0.065
rs3823298 44850 170733900C/G 0.46 0.36 0.003
rs2076318 45884 170734934A/G 0.40 0.47 0.017
rs2235506 46345 170735395C/T 0.68 0.65 0.434
rs2072916 48589 170737639A/G 0.47 0.54 0.026
rs3734763 53371 170742421A/G 0.49 0.56 0.052
rs3177571 53911 170742961G/T
rs8770 53990 170743040A/G
rs3173219 55152 170744202C/G 0.49 0.55 0.069
rs960744 55667 170744717C/T 0.39 0.34 0.131
109
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in Position AlleleCase Control Value
SEQ ID AF AF
NO: 3
rs2066954 58952 170748002 A/C 0.36 0.31 0.096
rs2072917 59315 170748365 G/T 0.46 0.41 0.070
rs3173220 60029 170749079 A/G
rs734249 61477 170750527 A/C 0.37 NA 0.484
rs2092310 62988 170752038 C/T
rs2092309 63090 170752140 C/G 0.43 0.49 0.102
rs1016536 64021 170753071 A/C 0.08 0.11 0.277
rs2235506 65685 170754735 C/T
rs2076998 70220 170759270 A/G
rs2076997 70323 170759373 A/C 0.89 0.91 0.655
rs2345478 70959 170760009 A/C 0.08 0.09 0.660
rs2021899 73436 170762486 C/G 0.48 0.42 0.081
rs2021898 82945 170771995 A/G
rs2345682 82958 170772008 G/T
rs2345683 82961 170772011 C/G 0.32 0.39 0.046
rs2881195 82964 170772014 C/T
rs2345684 82965 170772015 G/T
rs3046261 83006 170772056 -/CTTT
rs4083413 83025 170772075 C/T
rs4083412 83034 170772084 A/G
rs2345685 83074 170772124 G/T 0.69 0.70 0.772
rs2021897 83132 170772182 G/T
rs4036211 83155 170772205 C/T
rs4036212 83172 170772222 A/T
rs4036213 83174 170772224 G/T
rs2345686 83206 170772256 C/T
rs4036214 83216 170772266 G/T
rs4036215 83234 170772284 G/T
rs2345687 83252 170772302 A/G 0.62 NA NA
rs2345688 83260 170772310 A/C 0.46 0.49 0.383
rs2881196 83263 170772313 A/C
rs3046288 83296 170772346 -/AT
rs4036216 83319 170772369 AIG
rs4036205 83322 170772372 C/G
rs2092307 83324 170772374 A/C
rs4036206 83357 170772407 C/G
rs2345689 83375 170772425 ClT
rs2345690 83381 170772431 C/T
rs2345691 83389 170772439 A/T
rs2345692 83443 170772493 A/G
rs3046306 83499 170772549 -/GGTG0.39 0.40 0.729
rs4036207 83545 170772595 C/T
rs2345693 83566 170772616 C/T
rs2345694 83591 170772641 C/T
rs2345695 83619 170772669 G/T
rs2345696 83698 170772748 A/G
rs4036209 83780 170772830 G/T 0.79 0.73 0.156
rs2345697 83784 170772834 G/T
rs2881197 83826 170772876 G/T
rs2345698 83832 170772882 C/T
rs2345699 83852 170772902 C/T
rs2744640 86297 170775347 C/T 0.49 0.51 0.583
rs2744639 86315 170775365 G/T 0.45 0.43 0.745
rs2744638 86420 170775470 C/G 0.38 0.38 0.852
rs2744637 86460 170775510 C/G 0.35 0.40 0.216
rs2744636 86714 170775764 C/T 0.71 0.73 0.482
rs2744635 86718 170775768 CIT 0.05 0.03 0.195
rs2744634 86736 170775786 C/G 0.93 0.92 0.601
rs2744633 86753 170775803 C/T 0.19 0.20 0.681
rs2744632 86766 170775816 G/T 0.85 0.90 0.070
rs2744630 88162 170777212 C/G
rs2744629 88218 170777268 A/G 0.78 0.79 0.891
rs2744628 88246 170777296 A/G 0.68 0.67 0.766
rs2744627 88255 170777305 C/T 0.32 0.30 0.636
110
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in Position Allele Case Control Value
SEQ ID AF AF
NO: 3
rs2977616 88309 170777359G/T
rs2977617 88310 170777360A/T
rs2744626 88471 170777521A/G
rs2744625 88619 170777669C/T
rs3115847 88904 170777954C/T
rs2744623 89044 170778094C/G
rs4036193 90531 170779581-/AAAAA
rs4036194 90534 170779584A/G
rs4036196 90613 170779663C/G
rs1042327 46252 170735302C/T 0.46 0.37 0.004
TABLE 25
dbSNP Position ChromosomAl/A2 F A2 F A2 F p-
rs# in a PositionAllele Case Control Value
SEQ ID AF AF
NO: 3
rs1474555 229 170689279C/T 0.35 0.36 0.770
rs1474554 6310 170695360A/G 0.45 0.44 0.873
rs10334 11840 170700890G/T
rs10541 11870 170700920A/T
rs3823299 12064 170701114A/G unt 0.43 NA
ed
rs742348 13392 170702442C/G 0.45 0.47 0.600
rs1474644 16354 170705404A/G 0.74 0.75 0.775
rs 147464316559 170705609C/T 0.43 0.41 0.614
rs2056970 16935 170705985A/G 0.33 0.33 0.978
rs2223474 17616 170706666C/T 0.44 0.43 0.944
rs2206284 17737 170706787C/T 0.36 0.37 0.901
rs756519 18321 170707371C/T
rs756518 18453 170707503A/G 0.50 0.47 0.453
rs756517 18811 170707861C/T
rs1474642 20020 170709070C/T 0.15 0.11 0.147
rs2038093 21662 170710712C/G
rs2038092 23197 170712247ClG
rs2223473 23446 170712496G/T 0.43 0.40 0.408
rs760909 24339 170713389G/T 0.51 0.48 0.506
rs2076319 25504 170714554A/G 0.44 0.40 0.264
rs3778589 27174 170716224A/G 0.49 0.48 0.910
rs3800236 28008 170717058A/T 0.48 0.46 0.670
rs2206286 29294 170718344C/T 0.83 0.84 0.685
rs12717 29759 170718809C/G 0.51 0.53 0.726
rs2179373 30832 170719882A/G 0.59 0.58 0.880
rs3800235 44512 170733562A/C 0.60 0.62 0.632
rs3823298 44850 170733900C/G 0.41 0.41 0.945
rs2076318 45884 170734934A/G 0.43 0.42 0.636
rs2235506 46345 170735395C/T 0.69 0.67 0.594
rs2072916 48589 170737639A/G 0.49 0.46 0.399
rs3734763 53371 170742421A/G 0.51 0.51 0.888
rs3177571 53911 170742961G/T
rs8770 53990 170743040A/G
rs3173219 55152 170744202C/G 0.48 0.51 0.493
rs960744 55667 170744717C/T 0.38 0.37 0.738
rs2066954 58952 170748002A/C 0.37 0.34 0.378
rs2072917 59315 170748365G/T 0.45 0.45 0.982
rs3173220 60029 170749079A/G
rs734249 61477 170750527A/C 0.46 0.02
rs2092310 62988 170752038C/T
rs2092309 63090 170752140C/G 0.43 0.44 0.891
rs1016536 64021 170753071A/C 0.13 0.09 0.173
rs2235506 65685 170754735C/T
rs2076998 70220 170759270A/G
rs2076997 70323 170759373A/C 0.92 0.89 0.256
rs2345478 70959 170760009A/C 0.11 0.10 0.545
rs2021899 73436 170762486C/G 0.44 0.45 0.797
111
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Position Chromosom~ Al/A2F A2 F A2 F p-
rs# in a PositionAllele Case Control Value
SEQ ID AF AF
NO: 3
rs202189882945 170771995A/G
rs234568282958 170772008G/T
rs234568382961 170772011C/G 0.23 0.26 0.407
rs288119582964 170772014C/T
rs234568482965 170772015G/T
rs304626183006 170772056-/CTTT
rs408341383025 170772075C/T
rs408341283034 170772084A/G
rs234568583074 170772124G/T 0.74 0.71 0.533
rs202189783132 170772182GlT
rs403621183155 170772205C/T
rs403621283172 170772222A/T
rs403621383174 170772224G/T
rs234568683206 170772256C/T
rs403621483216 170772266G/T
rs403621583234 170772284G/T
rs234568783252 170772302A/G 0.47 0.50 0.457
rs234568883260 170772310A/C 0.61 0.58 0.434
rs288119683263 170772313A/C
rs304628883296 170772346-/AT
rs403621683319 170772369A/G
rs403620583322 170772372C/G
rs209230783324 170772374A/C
rs403620683357 170772407C/G
rs234568983375 170772425C/T
rs234569083381 170772431C/T
rs234569183389 170772439A/T
rs234569283443 170772493A/G
rs304630683499 170772549-/GGTG
rs403620783545 170772595C/T
rs234569383566 170772616C/T
rs234569483591 170772641C/T
rs234569583619 170772669G/T
rs234569683698 170772748A/G
rs403620983780 170772830G/T
rs234569783784 170772834G/T
rs288119783826 170772876G/T
rs234569883832 170772882C/T
rs234569983852 170772902C/T
rs274464086297 170775347C/T 0.57 0.55 0.595
rs274463986315 170775365G/T 0.35 0.34 0.752
rs274463886420 170775470C/G 0.41 0.40 0.793
rs274463786460 170775510C/G 0.47 0.46 0.836
rs274463686714 170775764C/T 0.83 NA
rs274463586718 170775768C/T
rs274463486736 170775786C/G unt 0.97 NA
ed
rs274463386753 170775803C/T 0.09 0.10 0.691
rs274463286766 170775816G/T 0.74 0.72 0.529
rs274463088162 170777212C/G
rs274462988218 170777268A/G 0.81 0.81 0.959
rs274462888246 170777296A/G 0.74 NA
rs274462788255 170777305C/T 0.33 0.29 0.341
rs297761688309 170777359G/T
rs297761788310 170777360AIT
rs274462688471 170777521A/G
rs274462588619 170777669C/T
rs311584788904 170777954C/T
rs274462389044 170778094C/G
rs403619390531 170779581-lAAAAA
rs403619490534 170779584A/G
rs403619690613 170779663C/G
rs104232746252 170735302C/T 0.42 0.43 0.880
112
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
[0248] Allelotyping results were considered particularly significant with a
calculated p-value of
less than or equal to 0.05 for allelotype results. These values are indicated
in bold. The allelotyping p-
values were plotted in Figure 1 C for the discovery cohort. The position of
each SNP on the
chromosome is presented on the x-axis. The y-axis gives the negative logarithm
(base 10) of the p-
value comparing the estimated allele in the case group to that of the control
group. The minor allele
frequency of the control group for each SNP designated by an X or other symbol
on the graphs in Figure
1 C can be determined by consulting Table 23. For example, the left-most X on
the left graph is at
position 170689279. By proceeding down the Table from top to bottom and across
the graphs from left
to right the allele frequency associated with each symbol shown can be
determined.
[0249] To aid the interpretation, multiple lines have been added to the graph.
The broken
horizontal lines are drawn at two common significance levels, 0.05 and 0.01.
The vertical broken lines
are drawn every 20kb to assist in the interpretation of distances between
SNPs. Two other lines are
drawn to expose linear trends in the association of SNPs to the disease. The
generally bottom-most
curve is a nonlinear smoother through the data points on the graph using a
local polynomial regression
method (W.S. Cleveland, E. Grosse and W.M. Shyu (1992) Local regression
models. Chapter 8 of
Statistical Models in S eds J.M. Chambers and T.J. Hastie, Wadsworth &
Brooks/Cole.). The black line
provides a local test for excess statistical significance to identify regions
of association. This was
created by use of a l Okb sliding window with lkb step sizes. Within each
window, a chi-square
goodness of fit test was applied to compare the proportion of SNPs that were
significant at a test wise
level of 0.01, to the proportion that would be expected by chance alone (0.05
for the methods used
here). Resulting p-values that were less than 10-8 were truncated at that
value.
[0250] Finally, the exons and introns of the genes in the covered region are
plotted below each
graph at the appropriate chromosomal positions. The gene boundary is indicated
by the broken
horizontal line. The exon positions are shown as thick, unbroken bars. An
arrow is place at the 3' end
of each gene to show the direction of transcription.
Example 7
ELP3 Region Proximal SNPs
[0251] It has been discovered that SNP rs1563055 in elongation protein 3
homolog (ELP3) is
associated with occurrence of osteoarthritis in subjects.
[0252] Thirty-three additional allelic variants proximal to rs1563055 were
identified and
subsequently allelotyped in osteoarthritis case and control sample sets as
described in Examples 1 and 2.
The polymorphic variants are set forth in Table 26. The chromosome positions
provided in column
four of Table 26 are based on Genome "Build 34" of NCBI's GenBank.
TABLE 26
dbSNP Position Chromosome Allele
in SEQ
rs# ChromosomeID NO: Position Variants
4
rs10006588 211 27927511 c/t
113
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Position ChromosomeAllele
rs# Chromosomein SEQ Position Variants
ID NO:
4
rs19848808 473 27927773 c/t
rs999112 8 1536 27928836 c/t
rs735880 8 5639 279'32939 c/t
rs20450298 17186 27944486 a/
rs20450288 17335 27944635 c/t
rs19473848 25029 27952329 c/
rs19473858 25111 27952411 c/t
rs19017448 28811 27956111 a/
rs19017458 28863 27956163 a/t
rs971882 8 30809 27958109 alc
rs13773388 40985 27968285 a/c
rs23054528 45147 27972447 c/t
rs23054518 45282 27972582 a/
rs21234728 46168 27973468 /t
rs21677688 46328 27973628 al
rs15630558 49077 27976377 a/
rs22903718 51925 27979225 c/t
rs22903708 52141 27979441 a/
rs22903698 52168 27979468 c/t
rs28749048 60852 27988152 c/t
rs32139978 62468 27989768 a/
rs32139988 65572 27992872 /t
rs15309298 79089 28006389 a/c
rs10002758 79541 28006841 c/t
rs10002748 79790 28007090 c/t
rs37578968 90843 28018143 a/
rs37578958 90978 28018278 c/t
rs37578948 91052 28018352 c/
rs37578938 91131 28018431 a/
rs37578928 91132 28018432 c/t
rs37578918 94439 28021739 a/
rs37578908 94621 28021921 a/t
Assay for Verifying and Allelotypin~ SNPs
[0253] The methods used to verify and allelotype the 33 proximal SNPs of Table
26 are the same
methods described in Examples 1 and 2 herein. The primers and probes used in
these assays are
provided in Table 27 and Table 28, respectively.
TABLE 27
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs1000658ACGTTGGATGTTCTCAAAAAAGAAACACATACGTTGGATGGGGTTATCAGTTTGAGATTC
rs1984880ACGTTGGATGCCATTTGCCAATTCCTGTGGACGTTGGATGATGGGCTGAAATGTATCCCC
rs999112ACGTTGGATGCTAAGCACATGCCTTTCTTGACGTTGGATGCTATTTTCTACTGGGAGATG
rs735880ACGTTGGATGTGCCTTCATTCTCCAACCACACGTTGGATGAACAGAGTGAGACCCATCTG
rs2045029ACGTTGGATGAGTCATTGCTAGCTTTCTGGACGTTGGATGGGGACTTTAGGGAAGTTATAG
rs2045028ACGTTGGATGAGCTTGTAGTGAGCCGAGATACGTTGGATGTGAGACAGAGTCTTGCTCTG
rs1947384ACGTTGGATGATTCTCCACCGAGAAACCAGACGTTGGATGTTGTGGCAGCAAGAAGGAAC
~
114
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs1947385ACGTTGGATGAAATTTCAACAGTCAACAATACGTTGGATGGTCAGTTTTGAAAACTGATC
rs1901744ACGTTGGATGCCTTGATTGAAGAGTAAAGCACGTTGGATGATCAAATATTCCTCATCCCC
rs1901745ACGTTGGATGCTTCTGCCTTTACCTGTGTCACGTTGGATGAAATGAAGCAGCACTCACAG
rs971882ACGTTGGATGAAGCCCTAATCATTGGTACGACGTTGGATGGATGGGTGCTAAAAAGACAC
rs1377338ACGTTGGATGCCCACATATCTACACATCAAGACGTTGGATGAGGGAGATAGGTGGTTAAAG
rs2305452ACGTTGGATGCCGTGTTGCAACTAACAGGGACGTTGGATGAGACGTTCCCATCCTCCATC
rs2305451ACGTTGGATGGCAGAGCCACCAGAGATAAAACGTTGGATGTTTTACGACAGGCGGGATTG
rs2123472ACGTTGGATGCACTTAGAATTGTTGCTTGGACGTTGGATGGCTGTATCTGTGACCTCAAA
rs2167768ACGTTGGATGGAATCAACATGACTTGGTGACACGTTGGATGATCTCACTCTAACTTGCTCC
rs1563055ACGTTGGATGAGTTCTTTCTCCTCACATTGACGTTGGATGCCCTTTAGAAGCACATACTC
rs2290371ACGTTGGATGATCCTCTTGGTAGCTTGTCCACGTTGGATGCTGTCTTGGTTTTCACCCTG
rs2290370ACGTTGGATGCAACCTCTACCTCACTACACACGTTGGATGATGAGGTATCGACACACTGG
rs2290369ACGTTGGATGACACACTGGGTATCTGTTCTACGTTGGATGTCAGAATCCCCAACCTCTAC
rs2874904ACGTTGGATGAAATTCCAGGCTGGGTACAGACGTTGGATGTGCTGACCTTAAGTGATCCG
rs3213997ACGTTGGATGGGTTGGCTAGAAGAGAAAAAACGTTGGATGTACAGTCCTTTTGAAACTAC
rs3213998ACGTTGGATGACAGTTTGTTGACATAGTAGACGTTGGATGAGGCTGAAAAGACATTCATG
rs1530929ACGTTGGATGGGCTTTCACTATATTTCCTCACGTTGGATGGAATACAGTAAGCCTATGGG
rs1000275ACGTTGGATGAACCCCAGAAAGCAAAAAGCACGTTGGATGCACGCTTGCTAACTTAATGG
rs1000274ACGTTGGATGGCCTAAGACAGGATCCAAACACGTTGGATGTTACTGCGTGCCTTAGTACC
rs3757896ACGTTGGATGCCTTCAAGCAAGTCAGTTACACGTTGGATGCAGAAACTGTGTGACTGATC
rs3757895ACGTTGGATGAAAATCATTGGCCAAACTGCACGTTGGATGCTCCTTAGTATTCTTAGGTG
rs3757894ACGTTGGATGAGAAGGGTTGAACAACAAGGACGTTGGATGCACCTAAGAATACTAAGGAG
rs3757893ACGTTGGATGCCCTTGTTGTTCAACCCTTCACGTTGGATGCTGCATGTGGATACCTACAC
rs3757892ACGTTGGATGTCCTGCATGTGGATACCTACACGTTGGATGCCCTTGTTGTTCAACCCTTC
rs3757891ACGTTGGATGATGGGCCAATTCTCCATAGGACGTTGGATGAGGCCTGTTAAGGAAACCTG
rs3757890ACGTTGGATGCAGGTGGATGTAGGCTTAAGACGTTGGATGGCACCACTGCCTCTTGTTTT
TABLE 28
dbSNP Extend Term
rs# Primer Mix
rs1000658 AATTGACAATGTTGGGACTGTT ACG
rs1984880 TGTGGTGTAAATAGGAGTTAGTGGACT
rs999112 GCACATGCCTTTCTTGGAACTG ACG
rs735880 AACCTTTACTTGTACTACATGC ACG
rs2045029 GCTAGCTTTCTGGTAATGAAAAT ACT
rs2045028 GATCGCACCACTGCACTCCAG ACG
rs1947384 ATAGCGGCAGTCCAAAAAGC ACT
rs1947385 TTCAACAGTCAACAATGAAACC ACT
rs1901744 ATAGTCAAGTATGCAAATGAAGC ACT
rs1901745 CCTTTACCTGTGTCTTCCCT CGT
rs971882 CCTAATCATTGGTACGGTCTCA ACT
rs1377338 AGTATTAGCTCAAATATCACATTGACT
rs2305452 CAGGGTAGCAGGCGGCC ACG
rs2305451 CCACAAACTCAGACCACGG ACT
rs2123472 CAGTTAATGTCAAGAAGCATAG ACT
rs2167768 ACATGACTTGGTGACAGAAGAA ACT
rs1563055 TTCTCCTCACATTGTTTCTACT ACG
rs2290371 GGTAGCTTGTCCTTAAATAACCGTACT
rs2290370 GGAGCAGGGACTTCTGCCA ACT
115
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Extend Term
rs# Primer Mix
rs2290369 AGTCCCTGCTCCATGTGAC ACT
rs2874904 GGCTAACGCCTGTAATCCCA ACT
rs3213997 AGAAAAATATTGTTATGCCCACA ACG
rs3213998 TAGTATTCTCAAATAGAGAGATTCACT
rs1530929 TTTCCTCTTTCCAGAATTGTATTTACT
rs1000275 ATGAGAATATCCTAGAATGAGGCAACG
rs1000274 GAATCATCAGGTCCTGTGCC ACG
rs3757896 TAATTCTCCTTAAGTAGTTAATTCACT
rs3757895 TTGGCCAAACTGCAGGATCT ACT
rs3757894 AAGGGCCACACAAGCAATTTCAA ACT
rs3757893 CCAAAGGACATTAGGTGGTG ACG
rs3757892 TGTGGATACCTACACTGCTC ACG
rs3757891 AGGATAAGTGTAACGGGGTC ACT
rs3757890 AGTGACACTCTTACTTCACAC I
CGT
Genetic Anal, sis
[0254] Allelotyping results from the discovery cohort are shown for cases and
controls in Table 29.
The allele frequency for the A2 allele is noted in the fifth and sixth columns
for osteoarthritis case pools
and control pools, respectively, where "AF" is allele frequency. The allele
frequency for the A1 allele
can be easily calculated by subtracting the A2 allele frequency from 1 (A1 AF
= 1-A2 AF). For
example, the SNP rs1000658 has the following case and control allele
frequencies: case A1 (C) = 0.36;
case A2 (T) = 0.64; control A1 (C) = 0.37; and control A2 (T) = 0.63, where
the nucleotide is provided
in paranthesis. Some SNPs are labeled "untyped" because of failed assays.
TABLE 29
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in Position Allele Case Control Value
SEQ ID AF AF
NO: 4
rs1000658211 27927511 C/T 0.79 0.80 0.591
rs1984880473 27927773 C/T 0.47 0.48 0.735
rs999112 1536 27928836 C/T 0.72 0.72 0.775
rs735880 5639 27932939 C/T 0.20 0.19 0.561
rs204502917186 27944486 A/G 0.54 0.56 0.361
rs204502817335 27944635 C/T
rs194738425029 27952329 C/G 0.63 0.60 0.122
rs194738525111 27952411 C/T
rs190174428811 27956111 A/G 0.18 0.18 0.796
rs190174528863 27956163 A/T 0.14 0.18 0.117
rs971882 30809 27958109 A/C
rs137733840985 27968285 A/C 0.28 0.24 0.085
rs230545245147 27972447 C/T 0.31 0.27 0.078
rs230545145282 27972582 A/G 0.48 0.52 0.130
rs212347246168 27973468 G/T 0.42 0.45 0.239
rs216776846328 27973628 A/G 0.38 0.35 0.350
rs156305549077 27976377 A/G
rs229037151925 27979225 C/T 0.28 0.24 0.039
rs229037052141 27979441 A/G 0.85 0.84 0.551
rs229036952168 27979468 C/T 0.43 0.47 0.138
rs287490460852 27988152 C/T 0.26 0.23 0.132
rs321399762468 27989768 A/G 0.44 0.47 0.201
rs321399865572 27992872 G/T 0.83 0.80 0.223
116
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in Position AlleleCase C_on_trolValue
SEQ ID AF AF
NO: 4
rs153092979089 28006389 A/C 0.47 0.49 0.556
rs100027579541 28006841 C/T 0.86 0.87 0.771
rs100027479790 28007090 C/T 0.54 0.56 0.510
rs375789690843 28018143 AlG
rs375789590978 28018278 C/T 0.46 0.47 0.874
rs375789491052 28018352 C/G 0.08 0.09 0.709
rs375789391131 28018431 A/G 0.16 0.15 0.590
rs375789291132 28018432 C/T 0.09 0.08 0.595
rs375789194439 28021739 A/G
rs375789094621 28021921 A/T 0.98 0.96 0.167
[0255] The ELP3 proximal SNPs were also allelotyped in the replication cohorts
using the methods
described herein and the primers provided in Tables 27 and 28. The replication
allelotyping results for
replication cohort #1 and replication cohort #2 are provided in Tables 30 and
31, respectively.
TABLE 30
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in Position Allele Case Control Value
SEQ ID AF AF
NO: 4
rs1000658211 27927511 C/T 0.78 0.79 0.863
rs1984880473 27927773 C/T 0.46 0.48 0.594
rs999112 1536 27928836 C/T 0.71 0.70 0.759
rs735880 5639 27932939 C/T 0.20 0.17 0.255
rs204502917186 27944486 A/G 0.55 0.57 0.526
rs204502817335 27944635 C/T
rs194738425029 27952329 C/G 0.65 0.61 0.198
rs194738525111 27952411 C/T
rs190174428811 27956111 A/G 0.19 0.18 0.674
rs190174528863 27956163 A/T 0.15 0.18 0.448
rs971882 30809 27958109 A/C
rs137733840985 27968285 A/C 0.29 0.22 0.039
rs230545245147 27972447 C/T 0.31 0.26 0.067
rs230545145282 27972582 A/G 0.49 0.56 0.063
rs212347246168 27973468 G/T 0.42 0.49 0.039
rs216776846328 27973628 A/G 0.36 0.34 0.396
rs156305549077 27976377 AlG
rs229037151925 27979225 C/T 0.28 0.23 0.054
rs229037052141 27979441 A/G 0.85 0.83 0.488
rs229036952168 27979468 C/T 0.41 0.49 0.036
rs287490460852 27988152 C/T 0.29 0.22 0.062
rs321399762468 27989768 A/G 0.44 0.50 0.064
rs321399865572 27992872 G/T 0.84 0.82 0.336
rs153092979089 28006389 A/C 0.48 0.52 0.311
rs100027579541 28006841 C/T 0.86 0.87 0.566
rs100027479790 28007090 C/T 0.54 0.59 0.159
rs375789690843 28018143 A/G
rs375789590978 28018278 C/T 0.45 0.49 0.308
rs375789491052 28018352 C/G 0.09 0.09 0.914
rs375789391131 28018431 A/G 0.15 0.14 0.803
rs375789291132 28018432 C/T 0.09 0.08 0.798
rs375789194439 28021739 A/G
rs375789094621 28021921 A/T 0.98 0.95 0.159
TABLE 31
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
in
rs# SEQ ID Position AlleleCase Control Value
NO: 4 AF AF
rs1000658211 27927511 C/T 0.80 0.82 0.443
rs1984880473 27927773 C/T 0.48 0.47 0.898
117
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in Position AlleleCase Control Value
SEQ ID AF AF
NO: 4
rs9991121536 27928836 C/T 0.72 0.76 0.319
rs7358805639 27932939 C/T 0.20 0.22 0.598
rs204502917186 27944486 AIG 0.52 0.54 0.581
rs204502817335 27944635 C/T
rs194738425029 27952329 C/G 0.62 0.59 0.348
rs194738525111 27952411 C/T
rs190174428811 27956111 A/G 0.18 0.18 0.928
rs190174528863 27956163 A/T 0.13 0.17 0.113
rs97188230809 27958109 A/C
rs137733840985 27968285 A/C 0.27 0.27 0.961
rs230545245147 27972447 C/T 0.32 0.30 0.673
rs230545145282 27972582 AlG 0.47 0.47 0.911
rs212347246168 27973468 G/T 0.41 0.38 0.348
rs216776846328 27973628 A/G 0.39 0.37 0.664
rs956305549077 27976377 A/G
rs229037151925 27979225 C/T 0.28 0.25 0.403
rs229037052141 27979441 A/G 0.85 0.84 0.939
rs229036952168 27979468 C/T 0.46 0.44 0.712
rs287490460852 27988152 C/T 0.24 0.24 0.888
rs321399762468 27989768 A/G 0.45 0.43 0.752
rs321399865572 27992872 G/T 0.81 0.78 0.373
rs153092979089 28006389 A/C 0.46 0.43 0.445
rs100027579541 28006841 C/T 0.87 0.86 0.767
rs100027479790 28007090 C/T 0.54 0.51 0.394
rs375789690843 28018143 A/G
rs375789590978 28018278 C/T 0.47 0.42 0.202
rs375789491052 28018352 C/G 0.07 0.09 0.478
rs375789391131 28018431 A/G 0.17 0.16 0.653
rs375789291132 28018432 C/T 0.09 0.07 0.567
rs375789194439 28021739 A/G
rs375789094621 28021921 A/T 0.97 0.97 0.728
[0256] Allelotyping results were considered particularly significant with a
calculated p-value of
less than or equal to 0.05 for allelotype results. These values are indicated
in bold. The allelotyping p
values were plotted in Figure 1D for the discovery cohort. The position of
each SNP on the
chromosome is presented on the x-axis. The y-axis gives the negative logarithm
(base 10) of the p-
value comparing the estimated allele in the case group to that of the control
group. The minor allele
frequency of the control group for each SNP designated by an X or other symbol
on the graphs in Figure
1D can be determined by consulting Table 29. For example, the left-most X on
the left graph is at
position 27927511. By proceeding down the Table from top to bottom and across
the graphs from left
to right the allele frequency associated with each symbol shown can be
determined.
[0257] To aid the interpretation, multiple lines have been added to the graph.
The broken
horizontal lines are drawn at two common significance levels, 0.05 and 0.01.
The vertical broken lines
are drawn every 201cb to assist in the interpretation of distances between
SNPs. Two other lines are
drawn to expose linear trends in the association of SNPs to the disease. The
generally bottom-most
curve is a nonlinear smoother through the data points on the graph using a
local polynomial regression
method (W.S. Cleveland, E. Grosse and W.M. Shyu (1992) Local regression
models. Chapter 8 of
Statistical Models in S eds J.M. Chambers and T.J. Hastie, Wadsworth &
BrookslCole.). The black line
provides a local test for excess statistical significance to identify regions
of association. This was
created by use of a lOkb sliding window with lkb step sizes. Within each
window, a chi-square
118
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
goodness of fit test was applied to compare the proportion of SNPs that were
significant at a test wise
level of 0.01, to the proportion that would be expected by chance alone (0.05
for the methods used
here). Resulting p-values that were less than 10-8 were truncated at that
value.
[0258] Finally, the exons and introns of the genes in the covered region are
plotted below each
graph at the appropriate chromosomal positions. The gene boundary is indicated
by the broken
horizontal line. The exon positions are shown as thick, unbroken bars. An
arrow is place at the 3' end
of each gene to show the direction of transcription.
Example 8
LRCHI Reason Proximal SNPs
[0259] It has been discovered that SNP rs912428 in leucine-rich repeats and
calponin homology
(CIT) domain containing 1 (LRCHI) is associated with occurrence of
osteoarthritis in subjects.
[0260] Forty-three additional allelic variants proximal to rs912428 were
identified and
subsequently allelotyped in osteoarthritis case and control sample sets as
described in Examples 1 and 2.
The polymorphic variants are set forth in Table 32. The chromosome positions
provided in column
four of Table 32 are based on Genome "Build 34" of NCBI's GenBank.
TABLE 32
dbSNP Position ChromosomeAllele
rs# Chromosomein SEQ Position Variants
ID NO:
5
rs101262813 243 44917643 c/t
rs157097613 10208 44927608 c/t
rs912436 13 15049 44932449 c/t
rs912435 13 15111 44932511 a/
rs912433 13 15272 44932672 c/t
rs912432 13 15287 44932687 a/
rs912431 13 15326 44932726 a/
rs912430 13 15327 44932727 c/t
rs140822513 17038 44934438 c/t
rs998657 13 19391 44936791 a/
rs132400613 21702 44939102 c/t
rs192441713 22431 44939831 c/
rs203872813 22881 44940281 a/
rs912429 13 27744 44945144 a/t
rs374226913 32564 44949964 a/
rs374227013 32698 44950098 a/c
rs380319213 33104 44950504 /t
rs380319113 33181 44950581 c/t
rs754106 13 33256 44950656 c/t
rs200505313 33543 44950943 c/t
rs153579313 35567 44952967 c/t
rs188622013 40085 44957485 c/t
rs188621913 40482 44957882 alt
rs153579213 45641 44963041 a/t
rs153579113 46059 44963459 a/
rs912428 13 48504 44965904 c/t
rs188621813 48919 44966319 a/c
119
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP ChromosomePosition ChromosomeAllele
rs# in SEQ Position Variants
ID NO:
5
rs157062213 49693 44967093 c/t
rs912427 13 49874 44967274 a/
rs912426 13 50020 44967420 al
rs306869313 50616 44968016 -/ttt
rs157062113 50719 44968119 a/
rs188696513 55511 44972911 c/t
rs100884913 65533 44982933 a/
rs912434 13 70529 44987929 a/c
rs388909513 75591 44992991 c/t
rs716223 13 77266 44994666 /t
rs289720713 80368 44997768 /t
rs157062013 82475 44999875 a/
rs146760513 92462 45009862 /t
rs146760413 92480 45009880 c/t
rs140822413 95819 45013219 c/t
rs140822313 96275 45013675 c/t
Assax for Veri in and Allelotyping SNPs
[0261] The methods used to verify and allelotype the 43 proximal SNPs of Table
32 are the same
methods described in Examples 1 and 2 herein. The primers and probes used in
these assays are
provided in Table 33 and Table 34, respectively.
TABLE 33
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs1012628ACGTTGGATGGATTTTCTGTGTCCCCCAAGACGTTGGATGTTGCAACAGAGAGAGCTCTG
rs1570976ACGTTGGATGTGATGTGTCTGCTGTGTTGGACGTTGGATGTTCACATGGCGAGGTCTTAG
rs912436ACGTTGGATGCCATATAAGGTGGTTATGGGACGTTGGATGCAAACAGGTTTTTCTGAGGC
rs912435ACGTTGGATGCAAGCCAATATCCAAGACAGACGTTGGATGAAAAACCTGTTTGTGAGGCC
rs912433ACGTTGGATGTGCCTTCCATCCTTAACACGACGTTGGATGGGCTTGAGCTTAGATATGGC
rs912432ACGTTGGATGAAATAGTTGGGTTTTGTGCCACGTTGGATGATTTGGTGTTAATTGCAGTG
rs912431ACGTTGGATGTGGAAGGCACAAAACCCAACACGTTGGATGCAGAAGCTAGGCTTCCTATG
rs912430ACGTTGGATGTGGAAGGCACAAAACCCAACACGTTGGATGCAGAAGCTAGGCTTCCTATG
rs1408225ACGTTGGATGGGGCACCATGACAATATTCCACGTTGGATGACACCTTGATCTTGGACTTC
rs998657ACGTTGGATGACTGGGCCAGGGAGGAATAGACGTTGGATGGTTGGGGAGATAATACAGAAG
rs1324006ACGTTGGATGGCTGAAAACCCAAATGTGTGACGTTGGATGCCAGCTATCAGCTCCATTTC
rs1924417ACGTTGGATGACAAAAGCAAGCCTTCACAGACGTTGGATGGTACTGTAAAAGGTACTGTG
rs2038728ACGTTGGATGAAGGCTTTTGGACACAAGTCACGTTGGATGGCACCTCTTATGATGTTCCC
rs912429ACGTTGGATGTTCAATTCCCCAAAGCCCTCACGTTGGATGGGCAAGTTCCATAACCTCTC
rs3742269ACGTTGGATGGAGAAAAGAGAACGAGAAGGACGTTGGATGTAAATGACAGCAGTCTGGAG
rs3742270ACGTTGGATGCTAAAACCAAAGCTGACGGGACGTTGGATGTTCTGCTCCTGTGGCATAGC
rs3803192ACGTTGGATGTCCTTTTGCTTCTGCGATGCACGTTGGATGTGCTTCCCCATCAGTTCTTG
rs3803191ACGTTGGATGCTGTCTGTACATTACCAGGCACGTTGGATGAATAGCAGCTGGAGGATCTC
rs754106ACGTTGGATGTTCTTACCATCCAGCAAGGCACGTTGGATGGCCTGGTAATGTACAGACAG
rs2005053ACGTTGGATGCTGTTGCTAGCTTGGATTTGACGTTGGATGTTCCCTGTCCTTTCTGGCAT
rs1535793ACGTTGGATGAACAAAGAGGAACAGAGCCCACGTTGGATGGCATAAGCCCCTTTTCCTAG
rs1886220ACGTTGGATGTCACCGTGTTAGCGAGAATGACGTTGGATGTAATCCCAGCACTTTGGGAG
rs1886219ACGTTGGATGTGTAACTGGATTTGCTGGAGACGTTGGATGTACATCAATAGCCGAGGAAG
120
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs1535792ACGTTGGATGCTGTATATCAGTGACTGTCCACGTTGGATGCAGAGAAGAAACATCTCAGC
rs1535791ACGTTGGATGGAGGGTTTATCCTTACAATTGACGTTGGATGTTTTAGGGTCCCTTGATAAG
rs912428ACGTTGGATGACTACATCCATTCCAGGGAGACGTTGGATGTCAGATCAGAGTGAGTTTAG
rs1886218ACGTTGGATGTCCCGAAAACAAGTCAAGACACGTTGGATGAGTCCAGGCAAAACAGTAAG
rs1570622ACGTTGGATGATAGCTGCCACACTCTTTAGACGTTGGATGGCGCAGTTTAGAAAAACCTG
rs912427ACGTTGGATGTAGGGTTCTCGATGGGTATGACGTTGGATGTTTGCCCTGGTCACTTTAGG
rs912426ACGTTGGATGTTAGAGGATGCATAGGCCAGACGTTGGATGAAGTCACTTACTGCATGGTC
rs3068693ACGTTGGATGAAATTGGCCACATGGAATCCACGTTGGATGCTACCTTTAACATCCCTGTC
rs1570621ACGTTGGATGAATTAAGAATGGCAGCTATGACGTTGGATGGTTTAAAACTAAAAACAC
rs1886965ACGTTGGATGCTGCTAAGGATATGTGTTTCCACGTTGGATGACACCAGTGCTCAGTATTTG
rs1008849ACGTTGGATGGCAGTTGTGAATTGTGCAGCACGTTGGATGTGGTGCAGAACATGTCAGAC
rs912434ACGTTGGATGTTCTGACATGTACAGACGTGACGTTGGATGTCCTGGGAAATCTTTCCATC
rs3889095ACGTTGGATGAAGGTAATGATATGTCCCCCACGTTGGATGCGCATTTTACAGAGACATTG
rs716223ACGTTGGATGACACTGTCTCTAGAAGCAGGACGTTGGATGGAAGCAGGAAAAGAGTGAGG
rs2897207ACGTTGGATGTCAGCCTCCAGAACTATGAGACGTTGGATGAACAGAGAGAGACCCTGTCT
rs1570620ACGTTGGATGCTGTTCCTGCCTTGATATGGACGTTGGATGGAAGGAAGTCTATTCAGCCC
rs1467605ACGTTGGATGATGTTACAGGGTGGTAAGCGACGTTGGATGTAAAGTTGCCACGCTTCTCC
rs1467604ACGTTGGATGATATACGGCATGTTACAGGGACGTTGGATGTTAAAGTTGCCACGCTTCTC
rs1408224ACGTTGGATGACTTCCCACTCCTCTAGACAACGTTGGATGTATTGGCTGGGTAGCACTCC
rs1408223ACGTTGGATGTCATTACCAGTTCCACAGAGACGTTGGATGTTGAGACATCATGAGGAGTG
TABLE 34
dbSNP Extend Term
rs# Primer Mix
rs1012628 CTGTGTCCCCCAAGTCTTTG ACG
rs1570976 TTGGCATTTCTTTGAGAA ACT
rs912436 AGGTGGTTATGGGTTTGTCACTCAACT
rs912435 TCCAAAAAGCCCAAGAAATTCT ACT
rs912433 CCTTAACACGTTTATAATAGATTAACG
rs912432 GTGCCTTCCATCCTTAACAC ACT
rs912431 GGCACAAAACCCAACTATTTTTC ACG
rs912430 GCACAAAACCCAACTATTTTTCC ACT
rs1408225 CCTCAGACTGGGTGGCTTA ACT
rs998657 CACCCACCTGAGGGAGGC ACT
rs1324006 GATACCTTGAAGAATTTTTAAAACACG
rs1924417 TTTAGGCACATTTGTACTTATAAAACT
rs2038728 TGGACACAAGTCCATGCAACA ACG
rs912429 CTGTGACAGGTGCTATTATCA CGT
rs3742269 TTTTGGACCGATTTCCGGTG ACT
rs3742270 GCTGACGGGGATTCCCTTTA ACT
rs3803192 GATGCACTAAAAGCAGCAATGT ACT
rs3803191 TCCAGCCTTCATATTTTCCTC ACG
rs754106 ATCCAGCAAGGCACTTAGAAT ACT
rs2005053 TGTGGCCTTCAGATGCTTACAT ACG
rs1535793 GAGGAACAGAGCCCAAAGGACA ACT
rs1886220 CTGACCTCGTGATCCGCC ACG
rs1886219 ACTGGATTTGCTGGAGTTAAGAA CGT
rs1535792 TATCAGTGACTGTCCTTTTCTTTTCGT
rs1535791 TTATCCTTACAATTGAAGAAAGGAI ACT
121
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Extend Term
rs# Primer Mix
rs912428 CCATTCCAGGGAGACTCCCA ACT
rs1886218 GAAAACAAGTCAAGACATTTATTGACT
rs1570622 CTGCCACACTCTTTAGATGAAGTTACG
rs912427 GGGAGATGACAGAACAAAACT ACT
rs912426 AGGTGCCAAGTGTTAGAAGAAACACG
rs3068693 GCCTCACATTGTTTTTTTTTTTTTACT
rs1570621 TCGGTCATAACTTTAATGAAGG ACG
rs1886965 TGATTTTATGACTCACATTATTTCACT
rs1008849 GTGAATTGTGCAGCTATAAACATGACG
rs912434 AGACGTGCCCAGCTATGATA ACT
rs3889095 TCCCCCATAACATTTCAGCAT ACT
rs716223 GTGGTTTGTATTTCCAGTGTCA ACT
rs2897207 AACTATGAGAAATAAATGTGTGGGACT
rs1570620 TTGATATGGTTCTTGGTTGTTGGACG
rs1467605 GTAAGCGCTAGAAAGAAAAATAAACT
rs1467604 ACGGCATGTTACAGGGTGGTAAGACG
rs1408224 GGGCACACATTCAGAACTGCCC ACG
rs1408223 ACAGAGGAAGACCAAATGACA I ACG
Genetic Anal,
[0262] Allelotyping results from the discovery cohort are shown for cases and
controls in Table 35.
The allele frequency for the A2 allele is noted in the fifth and sixth columns
for osteoarthritis case pools
and control pools, respectively, where "AF" is allele frequency. The allele
frequency for the Al allele
can be easily calculated by subtracting the A2 allele frequency from 1 (Al AF
= 1-A2 AF). For
example, the SNP rs1570976 has the following case and control allele
frequencies: case A1 (C) = 0.49;
case A2 (T) = 0.51; control A1 (C) = 0.53; and control A2 (T) = 0.47, where
the nucleotide is provided
in paranthesis. Some SNPs are labeled "untyped" because of failed assays.
TABLE 35
dbSNP Position ChromosomeAl/A2 F A2 F A2
rs# in Position Allele Case C AF Value
SEQ ID AF ~l
NO: 5
rs1012628243 44917643 C/T 0.70 0.70 0.768
rs157097610208 44927608 C/T 0.51 0.47 0.125
rs912436 15049 44932449 C/T 0.98 unt ed
rs912435 15111 44932511 A/G 0.64 0.36 0.0001
.
rs912433 15272 44932672 C/T 0.22 0.23 0.581
rs912432 15287 44932687 AIG 0.46 0.44 0.282
rs912431 15326 44932726 A/G 0.46 0.46 0.969
rs912430 15327 44932727 C/T 0.20 0.19 0.584
rs140822517038 44934438 C/T
rs998657 19391 44936791 A/G 0.47 0.44 0.254
rs132400621702 44939102 C/T 0.55 0.53 0.419
rs192441722431 44939831 C/G 0.53 0.49 0.108
rs203872822881 44940281 A/G 0.34 0.38 0.082
rs912429 27744 44945144 A/T
rs374226932564 44949964 A/G 0.83 0.83 0.967
rs374227032698 44950098 A/C 0.53 0.50 0.170
rs380319233104 44950504 G/T
rs380319133181 44950581 C/T
122
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Position ChromosomeAl/A2 F A2 Co t of F p-
rs# in Position Allele Case AF Value
SEQ ID AF
NO: 5
rs754106 33256 44950656 C/T 0.40 0.41 0.714
rs200505333543 44950943 C/T 0.40 0.40 0.877
rs153579335567 44952967 C/T 0.26 0.26 0.910
rs188622040085 44957485 C/T
rs188621940482 44957882 A/T 0.21 0.22 0.867
rs153579245641 44963041 A/T 0.73 0.71 0.550
rs153579146059 44963459 A/G 0.08 0.15 0.009
rs912428 48504 44965904 C/T
rs188621848919 44966319 A/C
rs157062249693 44967093 C/T 0.73 0.75 0.451
rs912427 49874 44967274 A/G 0.68 0.70 0.352
rs912426 50020 44967420 A/G 0.76 0.77 0.680
rs306869350616 44968016 -/TTT 0.22 0.21 0.597
rs157062150719 44968119 A/G 0.19 0.18 0.569
rs188696555511 44972911 C/T
rs100884965533 44982933 AIG 0.48 0.43 0.160
rs912434 70529 44987929 A/C 0.23 0.23 0.988
rs388909575591 44992991 C/T 0.90 0.90 0.880
rs716223 77266 44994666 G/T 0.91 0.90 0.981
rs289720780368 44997768 G/T 0.46 0.46 0.921
rs157062082475 44999875 A/G 0.67 0.68 0.738
rs146760592462 45009862 G/T 0.29 0.22 0.044
rs146760492480 45009880 C/T 0.68 0.67 0.537
rs140822495819 45013219 ClT 0.66 0.65 0.683
rs140822396275 45013675 C/T 0.29 0.28 0.587
[0263] The LRCHI proximal SNPs were also allelotyped in the replication
cohorts using the
methods described herein and the primers provided in Tables 33 and 34. The
replication allelotyping
results for replication cohort #1 and replication cohort #2 are provided in
Tables 36 and 37,
respectively.
TABLE 36
dbSNP Position ChromosomeAl/A2 F A2 Co t F p
rs# in Position Allele Case of Value
SEQ ID AF AF
NO: 5
rs1012628243 44917643 C/T 0.69 0.72 0.337
rs157097610208 44927608 C/T 0.48 0.46 0.490
rs912436 15049 44932449 C/T
rs912435 15111 44932511 A/G 0.16 unt 0.637
ed
rs912433 15272 44932672 C/T 0.28 0.28 0.984
rs912432 15287 44932687 AIG 0.46 0.42 0.260
rs912431 15326 44932726 A/G 0.46 0.48 0.602
rs912430 15327 44932727 C/T 0.18 0.20 0.476
rs140822517038 44934438 C/T
rs998657 19391 44936791 A/G 0.46 0.43 0.380
rs132400621702 44939102 C/T 0.54 0.53 0.811
rs192441722431 44939831 C/G 0.51 0.49 0.440
rs203872822881 44940281 A/G 0.35 0.39 0.181
rs912429 27744 44945144 A/T
rs374226932564 44949964 A/G 0.84 0.85 0.911
rs374227032698 44950098 A/C 0.56 0.50 0.090
rs380319233104 44950504 G/T
rs380319133181 44950581 C/T
rs754106 33256 44950656 C/T 0.40 0.40 0.827
rs200505333543 44950943 C/T 0.40 0.37 0.328
rs153579335567 44952967 C/T 0.27 0.24 0.259
rs188622040085 44957485 C/T
rs188621940482 44957882 A/T 0.22 0.19 0.302
123
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in Position Allele Case C AF Value
SEQ ID AF of
NO: 5
rs 153579245641 44963041 A/T 0.73 0.76 0.435
rs 153579146059 44963459 A/G 0.08 0.08 0.958
rs912428 48504 44965904 Cn- See replication
genotyping
results
in
Tables
8 &
9.
rs188621848919 44966319 A/C
rs157062249693 44967093 C/T 0.71 0.79 0.007
rs912427 49874 44967274 A/G 0.65 0.73 0.007
rs912426 50020 44967420 A/G 0.74 0.80 0.047
rs306869350616 44968016 -/TTT 0.25 0.21 0.236
rs157062150719 44968119 A/G 0.22 0.15 0.028
rs188696555511 44972911 C/T
rs100884965533 44982933 A/G 0.47 unt NA
ed
rs912434 70529 44987929 AIC 0.24 0.19 0.083
rs388909575591 44992991 C/T 0.91 0.91 0.867
rs716223 77266 44994666 G/T 0.91 0.93 0.598
rs289720780368 44997768 G/T 0.48 0.45 0.321
rs157062082475 44999875 A/G 0.66 0.72 0.034
rs146760592462 45009862 G/T 0.29 0.22 0.044
rs146760492480 45009880 C/T 0.66 0.70 0.307
rs140822495819 45013219 C/T 0.64 0.67 0.312
rs140822396275 45013675 C/T 0.31 0.23 0.028
TABLE 37
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in Position Allele Case Control Value
SEQ ID AF AF
NO: 5
rs1012628243 44917643 C/T 0.71 0.68 0.438
rs157097610208 44927608 C/T 0.55 0.50 0.159
rs912436 15049 44932449 C/T
rs912435 15111 44932511 A/G 0.66 unt ed
rs912433 15272 44932672 C/T 0.14 0.17 0.479
rs912432 15287 44932687 A/G 0.47 0.46 0.806
rs912431 15326 44932726 A/G 0.46 0.44 0.513
rs912430.15327 44932727 C/T 0.23 0.17 0.084
rs140822517038 44934438 C/T
rs998657 19391 44936791 A/G 0.48 0.45 0.518
rs132400621702 44939102 C/T 0.55 0.52 0.324
rs192441722431 44939831 C/G 0.54 0.49 0.123
rs203872822881 44940281 A/G 0.34 0.37 0.295
rs912429 27744 44945144 A/T
rs374226932564 44949964 A/G 0.82 0.82 0.861
rs374227032698 44950098 A/C 0.50 0.49 0.873
rs380319233104 44950504 G/T
rs380319133181 44950581 C/T
rs754106 33256 44950656 C/T 0.41 0.44 0.346
rs200505333543 44950943 C/T 0.40 0.44 0.302
rs 153579335567 44952967 C/T 0.25 0.31 0.096
rs188622040085 44957485 C/T
rs188621940482 44957882 A/T 0.20 0.27 0.053
rs153579245641 44963041 A/T 0.73 0.63 0.007
rs153579146059 44963459 A/G NA 0.27 NA
rs912428 48504 44965904 Cn- See
replication
genotyping
results
in
Tables
8 &
9.
rs188621848919 44966319 A/C
rs 157062249693 44967093 C/T 0.75 0.67 0.040
rs912427 49874 44967274 A/G 0.71 0.64 0.059
rs912426 50020 44967420 A/G 0.78 0.72 0.065
rs306869350616 44968016 -/TTT 0.19 0.21 0.520
rs157062150719 44968119 A/G 0.15 0.21 0.077
rs188696555511 44972911 C/T
rs100884965533 44982933 A/G 0.49 0.43 0.138
124
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in Position Allele Case Control Value
SEQ ID AF AF
NO: 5
rs912434 70529 44987929 A/C 0.21 0.28 0.027
rs388909575591 44992991 C/T 0.89 0.88 0.583
rs716223 77266 44994666 G/T 0.90 0.87 0.368
rs289720780368 44997768 G/T 0.44 0.48 0.276
rs157062082475 44999875 A/G 0.70 0.62 0.026
rs146760592462 45009862 G/T
rs146760492480 45009880 C/T 0.71 0.62 0.018
rs140822495819 45013219 C/T 0.68 0.61 0.060
rs140822396275 45013675 C/T 0.27 0.34 0.023
[0264] Allelotyping results were considered particularly significant with a
calculated p-value of
less than or equal to 0.05 for allelotype results. These values are indicated
in bold. The allelotyping p-
values were plotted in Figure 1E for the discovery cohort. The position of
each SNP on the
chromosome is presented on the x-axis. The y-axis gives the negative logarithm
(base 10) of the p-
value comparing the estimated allele in the case group to that of the control
group. The minor allele
frequency of the control group for each SNP designated by an X or other symbol
on the graphs in Figure
1E can be determined by consulting Table 35. For example, the left-most X on
the left graph is at
position 44917643. By proceeding down the Table from top to bottom and across
the graphs from left
to right the allele frequency associated with each symbol shown can be
determined.
[0265] To aid the interpretation, multiple lines have been added to the graph.
The broken
horizontal lines are drawn at two common significance levels, 0.05 and 0.01.
The vertical broken lines
are drawn every 20kb to assist in the interpretation of distances between
SNPs. Two other lines are
drawn to expose linear trends in the association of SNPs to the disease. The
generally bottom-most
curve is a nonlinear smoother through the data points on the graph using a
local polynomial regression
method (W.S. Cleveland, E. Grosse and W.M. Shyu (1992) Local regression
models. Chapter 8 of
Statistical Models in S eds J.M. Chambers and T.J. Hastie, Wadsworth &
Brooks/Cole.). The black line
provides a local test for excess statistical significance to identify regions
of association. This was
created by use of a lOkb sliding window with lkb step sizes. Within each
window, a chi-square
goodness of fit test was applied to compare the proportion of SNPs that were
significant at a test wise
level of 0.01, to the proportion that would be expected by chance alone (0.05
for the methods used
here). Resulting p-values that were less than 10-8 were truncated at that
value.
[0266] Finally, the exons and introns of the genes in the covered region are
plotted below each
graph at the appropriate chromosomal positions. The gene boundary is indicated
by the broken
horizontal line. The exon positions are shown as thick, unbroken bars. An
arrow is place at the 3' end
of each gene to show the direction of transcription.
Example 9
SNWI Region Proximal SNPs
[0267] SNP rs 1477261 is associated with osteoarthritis and is described in
Table A. It lies within
an intron of the SKI-interacting protein gene (SNWI ). This gene, a member of
the SNW gene family,
125
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
encodes a coactivator that enhances transcription from some Pol II promoters.
This coactivator can bind
to the ligand-binding domain of the vitamin D receptor and to retinoid
receptors to enhance vitamin D-,
retinoic acid-, estrogen-, and glucocorticoid-mediated gene expression. It
also can interact with
poly(A)-binding protein 2 to directly control the expression of muscle-
specific genes at the
transcriptional level. One hundred sixty-three additional allelic variants
proximal to rs1477261 were
identii ed and subsequently allelotyped in osteoarthritis case and control
sample sets as described in
Examples 1 and 2. The polymorphic variants are set forth in Table 38. The
chromosome position
provided in column four of Table 38 is based on Genome "Build 34" of NCBI's
GenBank.
TABLE 38
dbSNP Position ChromosomeAllele
rs# Chromosomein SEQ Position Variants
ID NO: 6
rs714392614 218 76161268 a/t
rs154907114 1440 76162490 c/t
rs801285814 1442 76162492 c/t
rs715561114 2611 76163661 c/t
rs176941 14 4317 76165367 a/c
rs 17694214 4724 76165774 alg
rs176943 14 4788 76165838 g/t
rs176944 14 5202 76166252 g/t
rs436522114 5780 76166830 c/t
rs316895214 5974 76167024 c/t
rs176945 14 6644 76167694 c/g
rs176946 14 7430 76168480 a/g
rs176947 14 7938 76168988 .~ c/t
rs176948 14 8095 76169145 c/t
rs176949 14 8183 76169233 a/c
rs176950 14 8312 76169362 c/t
rs176951 14 8352 76169402 a/c
rs715690514 9348 76170398 c/t
rs321719714 9378 76170428 -/tctc
rs227044314 9617 76170667 a/g
rs176952 14 9727 76170777 c/t
rs176953 14 9834 76170884 c/t
rs176954 14 9899 76170949 g/t
rs176955 14 10211 76171261 c/t
rs321441614 10377 76171427 -/t
rs176956 14 10695 76171745 c/t
rs254456614 10729 76171779 c/g
rs254456714 10730 76171780 c/t
rs176957 14 11433 76172483 a/g
rs176958 14 11951 76173001 c/g
rs176959 14 12697 76173747 c/t
rs180222714 12982 76174032 a/c
rs176961 14 14419 76175469 c/t
rs176962 14 14501 76175551 c/t
rs740128514 14983 76176033 a/c
rs176963 14 15280 76176330 c/t
rs176964 14 15475 76176525 a/g
rs490363114 15888 76176938 a/g
126
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Position Chromosome Allele
rs# Chromosomein SEQ Position Variants
ID NO:
6
rs490363214 15976 76177026 a/t
rs176965 14 16307 76177357 a/c
rs490363314 16442 76177492 a/c
rs176966 14 17255 76178305 clt
rs176968 14 18948 76179998 g/t
rs176969 14 19435 76180485 a/t
rs176970 14 19753 76180803 clt
rs714919814 20021 76181071 c/t
rs714791814 20022 76181072 a/c
rs714868514 20503 76181553 a/g
rs118423214 20590 76181640 g/t
rs118423314 21804 76182854 g/t
rs118423414 21919 76182969 c/t
rs740199814 21990 76183040 a/t
rs176974 14 22412 76183462 a/g
rs657439014 22536 76183586 c/t
rs176975 14 23432 76184482 a/g
rs176976 14 23468 76184518 g/t
rs176977 14 23772 76184822 c/t
rs801372714 24325 76185375 c/t
rs176978 14 24773 76185823 c/t
rs211182914 26274 76187324 clt
rs176980 14 27440 76188490 c/g
rs580984814 28561 76189611 -/acag
rs580984914 30071 76191121 -la
rs438307014 31764 76192814 a/t
rs749365214 33008 76194058 c/t
rs211213314 35310 76196360 a/t
rs196383314 35460 76196510 a/c
rs657439114 37112 76198162 a/g
rs715506214 37285 76198335 a/g
rs489967414 37747 76198797 c/t
rs802251614 38057 76199107 c/t
rs714083814 38859 76199909 a/c
rs714112714 38860 76199910 a/g
rs657439214 39525 76200575 a/g
rs800369114 40216 76201266 a/g
rs800397914 40281 76201331 c/t
rs801054114 41453 76202503 c/g
rs801641614 42091 76203141 a/t
rs801617514 42513 76203563 a/g
rs715457114 42935 76203985 c/t
rs715882614 42985 76204035 a/g
rs715931014 43003 76204053 a/g
rs740190014 43281 76204331 a/g
rs716035514 43716 76204766 c/t
rs203278114 43866 76204916 a/g
rs657439414 44234 76205284 g/t
rs800759814 44596 76205646 a/g
rs226776714 44871 76205921 c/t
rs657439514 45005 76206055 a/g
rs715006614 45282 76206332 a/c
127
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP . Position Chromosome Allele
rs# Chromosomein SEQ Position Variants
I~ NO:
6
rs749233414 47178 76208228 a/c
rs435936114 47816 76208866 g/t
rs460508914 47887 76208937 a/g
rs714644614 48134 76209184 c/t
rs434614414 48135 76209185 alg
rs714807814 48276 76209326 g/t
rs714828614 48400 76209450 c/t
rs378398014 48798 76209848 a/g
rs154911914 48803 76209853 a/t
rs198492514 49146 76210196 c/t
rs147726114 49969 76211019 alt
rs801644714 51059 76212109 a/g
rs749404414 51064 76212114 c/t
rs202328814 53285 76214335 alt
rs715168514 54560 76215610 c/t
rs211213514 54748 76215798 a/g
rs216108814 54785 76215835 c/g
rs490363814 55102 76216152 c/g
rs147726214 55644 76216694 alg
rs147726314 55705 76216755 g/t
rs147726414 55841 76216891 a/g
rs227791714 56623 76217673 c/g
rs227791814 56825 76217875 ~~ ~a/c
rs227791914 56827 76217877 a/g
rs197841614 56892 76217942 c/t
rs375972814 59150 76220200 a/t
rs657439914 59958 76221008 ea/t
rs715533614 60231 76221281 c/t
rs715618614 60524 76221574 a/g
rs714239014 61871 76222921 c/t
rs714587514 62226 76223276 c/t
rs801463514 63230 76224280 g/t
rs801593814 63468 76224518 g/t
rs801531314 63787 76224837 c/t
rs800631514 65732 76226782 a/c
rs657440014 65989 76227039 a/g
rs714081614 68832 76229882 g/t
rs456607814 69904 76230954 c/t
rs714105014 70365 76231415 a/g
rs304935614 70886 76231936 -/tatc
rs490363914 73088 76234138 a/t
rs490364114 73103 76234153 c/t
rs236483814 75934 76236984 c/t
rs236483914 75966 76237016 c/t
rs463206614 76273 76237323 c/t
rs211213614 77943 76238993 c/t
rs464165514 78466 76239516 c/t
rs463526914 78861 76239911 c/t
rs457076414 78872 76239922 a/g
rs759808 14 79836 76240886 g/t
rs715053114 80908 76241958 c/t
rs715496814 81509 76242559 c/g
128
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Position Chromosome Allele
rs# Chromosomein SEQ Position Variants
ID NO:
6
rs714665714 83576 76244626 c/t
rs714585914 83662 76244712 c/g
rs490364314 83782 76244832 c/t
rs71768214 84282 76245332 g/t
rs71768314 84444 76245494 a/g
rs147725914 85129 76246179 clg
rs801906414 85151 76246201 a/g
rs801897114 85296 76246346 a/c
rs147726014 85809 76246859 c/g
rs580985114 86387 76247437 -/t
rs198514914 86494 76247544 a/g
rs100898814 89786 76250836 a/g
rs100898914 89894 76250944 a/t
rs801822214 90122 76251172 glt
rs100604014 92067 76253117 a/g
rs100603914 92187 76253237 c/t
rs100603814 92312 76253362 a/g
rs800978414 92824 76253874 g/t
rs490364414 93733 76254783 c/t
rs714949614 96553 76257603 c/g
rs657440214 96941 76257991 a/c
Assay for Verifying and Allelotypin~ SNPs
[0268] The methods used to verify and allelotype the 101 proximal SNPs of
Table 38 are the same
methods described in Examples 1 and 2 herein. The primers and probes used in
these assays are
provided in Table 39 and Table 40, respectively.
TABLE 39
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs7143926ACGTTGGATGGAGTCACCCAAAATTAAGGCACGTTGGATGGAAAGCCAAAATTAGCCTGC
rs1549071ACGTTGGATGGTGAGACGCTGTCTCAGTAAACGTTGGATGCTCCACACTTGGAGAAGTTG
rs8012858ACGTTGGATGGTGAGACGCTGTCTCAGTAAACGTTGGATGCTCCACACTTGGAGAAGTTG
rs7155611ACGTTGGATGATGGAATACAGGCACCGTTCACGTTGGATGCCCCTTCTTAATCTCCATGG
rs176941ACGTTGGATGTTAGTATGGGAAAAGGGCTCACGTTGGATGCAACAATCCTATGAGTTGGG
rs176942ACGTTGGATGAGTGGCTCAGATGTGAGTAGACGTTGGATGTGGTCTTCACCAACCACATG
rs176943ACGTTGGATGACCAAGCCCAGTAAAGTCTCACGTTGGATGGCATCCGCAAGATGCTAATG
rs176944CGTTGGATGGGCCTCAATATTGGCTAAATGACGTTGGATGCTTAACCATTAGAGCCCTTC
rs4365221ACGTTGGATGAAATAAGGCAGGAAGGGTAGACGTTGGATGTCCCAACTTACTGGTCTTTC
rs3168952ACGTTGGATGATGTACCAGACTTGGTGGTGACGTTGGATGTTTGCTGAGGATGGAGACTG
rs176945ACGTTGGATGCCTACTATACACTCACAAAAACGTTGGATGTTTTTTAAAACACTTTAAGC
rs176946ACGTTGGATGGCTTTATCATAGGTATTTGTGACGTTGGATGGAGAGATGTGTTGTTTTTGAG
rs176947ACGTTGGATGTGAGTAGCTGGGACTACAGGACGTTGGATGGGCCAACATAGCGAAACTCC
rs176948ACGTTGGATGCAGAGCCAAAGGTCAACAAGACGTTGGATGTACAGGTGTGAGCCTTCATG
rs176949ACGTTGGATGTAGGAACTCCCTGCAGTTCCACGTTGGATGCCTTGCTGGCTTTAAAGAAG
rs176950ACGTTGGATGAATCACAGGAGTGACATCCCACGTTGGATGTGGAGGAGAAACCTGACTTG
rs176951ACGTTGGATGCCCTATATAATCTCCTCCCCACGTTGGATGCAGGAGTGACATCCCATTAC
129
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs7156905ACGTTGGATGTGAGAGAGAGAACCTGTCTCACGTTGGATGAAAGGCGGCTTTGATGTTGG
rs3217197ACGTTGGATGTTGATTGTGCCACTGCACTCACGTTGGATGACTCTAGTTGGAAATCCTGG
rs2270443ACGTTGGATGATAACTCAGTCCAGGTGTGGACGTTGGATGCACTCAAGCAGTCTACTCAC
rs176952ACGTTGGATGGATCTCAGCTCACTGCAATCACGTTGGATGTATCTGGGTGACTGAGGAAG
ys176953ACGTTGGATGTTGAGGTCAGGAGTTTGGGAACGTTGGATGGCCACCACACCCAGCTAATT
rs176954ACGTTGGATGAAAACATAGGCCAGGTGCAGACGTTGGATGAAACTCCTGACCTCAAGCCA
rs176955ACGTTGGATGCTAGAGTGCTTGGATGTACCACGTTGGATGGTCATCTACAGGGACTAGAC
rs3214416ACGTTGGATGACGACTATCATCACGTGTTCACGTTGGATGACCAGAAGTCTGTAACTAGG
rs176956ACGTTGGATGTACAGGCATAAGCCACCATGACGTTGGATGAGGAAGGGTGTAAAGCAAGG
rs2544566ACGTTGGATGCAAGCAATCTTCCCATCTGGACGTTGGATGTGATCCGATTTTTGGCTGGG
rs2544567ACGTTGGATGCAAGCAATCTTCCCATCTGGACGTTGGATGTGATCCGATTTTTGGCTGGG
rs176957ACGTTGGATGTTTCACCGTGTTAGCCAGGAACGTTGGATGTAATCCCAGCACTTTGGGAG
rs176958ACGTTGGATGAAAACTGGGCACTCTACCACACGTTGGATGAAAATCGCGCCATTGCACTC
rs176959ACGTTGGATGCAGGCAGTTTTTATTTGTCCCACGTTGGATGGGTTAGGGAGTCATAATACC
rs1802227ACGTTGGATGAACAAATAGTTGCACCAAGACGTTGGATGTTTTAATTTGGAGTGGGCA
rs176961ACGTTGGATGAACCCAGTTTAAGACCGGCCACGTTGGATGTACAGGTGTGTGCCACCATG
rs176962ACGTTGGATGATATTTCTGGCTGGGCACTGACGTTGGATGACTGGGTTCAAGCAATCTGC
rs7401285ACGTTGGATGACAGAGTGGGACTCCATATCACGTTGGATGGATTCAAACTGGGTGTCTTG
rs176963ACGTTGGATGTAAGCCTGGGAAAACACACGACGTTGGATGCCCACTCTACTTTCCAGTAG
rs176964ACGTTGGATGAGAGTCAGTGTCCTACAAAAACGTTGGATGTAATCCCGTTTTACAGCTTC
rs4903631ACGTTGGATGGTAAATGCCAGCATGATGACACGTTGGATGTCTCAGCCCACTATAAGAAG
rs4903632ACGTTGGATGTGTGAATACCTATCCTCAGGACGTTGGATGGTCATCATGCTGGCATTTAC
rs176965ACGTTGGATGAATGCTTTATAAGGGCTGCCACGTTGGATGTCTCAGAAACAAAGGATGTG
rs4903633ACGTTGGATGCAACCCCCAAACCATCATATACGTTGGATGCTAACAGATTCGTTGACATGG
rs176966ACGTTGGATGCTCTCGAGTAGCTGGGACTAACGTTGGATGTGGCCAACATGGTGAAACCC
rs176968ACGTTGGATGGCGAAACTCCGTCTCAAAACACGTTGGATGTAGTGATCTTCCCACCTAGG
rs176969ACGTTGGATGCTGTCTGTCCGATTTACTGCACGTTGGATGTCTAGAATCAAGCATGCGGC
rs176970ACGTTGGATGCTAATGTTCCTAGTACAGTGGACGTTGGATGCTTCTCTTCTAGCTATTTTGC
rs7149198ACGTTGGATGCAATGGGATATTACTCAGCCACGTTGGATGTTTCTGTGCCGGGCTTATTC
rs7147918ACGTTGGATGCAATGGGATATTACTCAGCCACGTTGGATGTTTCTGTGCCGGGCTTATTC
rs7148685ACGTTGGATGTGTCTTCTTTTGAGACCGTCACGTTGGATGCTCAATCGCAAAGAAACGAG
rs1184232ACGTTGGATGAAGAGGCCACCTACAGAATGACGTTGGATGCTCGTTTCTTTGCGATTGAG
rs1184233ACGTTGGATGAAGTGTTGGGATTACAGGTGACGTTGGATGAGTGAAAGATCGCCACAAAG
rs1184234ACGTTGGATGGCTATGTGCAGTGACTCATGACGTTGGATGTCTCAGACCTCAGGTGATCT
rs7401998ACGTTGGATGTGAGTAGCTAGGACAACAGGACGTTGGATGAACGTGGTGAAACCCCATCT
rs176974ACGTTGGATGTTACAGCGAGCTGAGATCATACGTTGGATGAGGATCATACTGTCTCTGAC
rs6574390ACGTTGGATGTGATGAAACCCCGTCTGTACACGTTGGATGTCCTGAGTAGCTGGGATTAC
rs176975ACGTTGGATGTGTAGAATCTAGGTGGTAGGACGTTGGATGCCAGCCTTTCCTGACATTTT
rs176976ACGTTGGATGGGTAGGAGATACAGGTGTTCACGTTGGATGCCCAGCCTTTCCTGACATTT
rs176977ACGTTGGATGTTGCATCATTACACTTCAGCACGTTGGATGGGGAAACATTATGCATAATTCC
rs8013727ACGTTGGATGTGCCTGGTTGTATACCTAACACGTTGGATGCTTGAGAACGATTCTGTTGTC
rs176978ACGTTGGATGGGGACCATGTTTTTGTTACCACGTTGGATGAATACTGTGGAATGGGCATG
rs2111829ACGTTGGATGCATGTGGAAAAAGGTATGACACGTTGGATGCCTACTTTATATGCAGTAGG
rs176980ACGTTGGATGATGGCCAATGCTATGAACGCACGTTGGATGAAGGGCAGTTGCAGGAAAAG
rs5809848ACGTTGGATGTCTATTTTTCCAGAGCTTGGGACGTTGGATGCCATTTCACTGATGCTTTGG
rs5809849ACGTTGGATGGTGAATACCGTGTCAGTTCCACGTTGGATGTGCAGTGAGCTGAGATCATG
rs4383070ACGTTGGATGAGCGATTCTCTTGTCTCAGCACGTTGGATGAACTTAGCTGGGCATTGTGG
rs7493652ACGTTGGATGGGTCATATACCACAAGTAACACGTTGGATGCTGGCCCTATGCTATTTTCA
rs2112133ACGTTGGATGGCCACCACAACTGGCTAATTACGTTGGATGTGTGGTCAGGAGATCGAAAC
rs1963833ACGTTGGATGTAAGCCAAGATTGCGTCACTACGTTGGATGAGCATTAAAGGTAGAATGCC
rs6574391ACGTTGGATGTAACCGTTGCTATGGAGAAGACGTTGGATGACCTATACAACCCTAAGCTG
rs7155062ACGTTGGATGGCTCCTTATTTGGGCATTCCACGTTGGATGCACTCAGCCTTGTGAGATAC
130
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs4899674ACGTTGGATGAATGTGCTGAGGAAACTGAGACGTTGGATGGCTTCTGATACTTTCAAGAG
rs8022516ACGTTGGATGGTTGAAGGCATTCTTTTGGGACGTTGGATGCTAGCCTGGGCAATATAATG
rs7140838ACGTTGGATGCCTCGTTTCTGAAGAATACCACGTTGGATGGAGACTGAACAGGTTATTGG
rs7141127ACGTTGGATGCCTCGTTTCTGAAGAATACCACGTTGGATGGAGACTGAACAGGTTATTGG
rs6574392ACGTTGGATGAGAAAATAGCATAGGCTGGGACGTTGGATGAAATGATCCATCCTCCTCAG
rs8003691ACGTTGGATGACTGAAGTCAAGTGAAGGCCACGTTGGATGTTAGGCCCCTATACATGGAG
rs8003979ACGTTGGATGCACAAAACCACTTCTGAAGCACGTTGGATGGGGCCTAATTTTCCTTTTGC
rs8010541ACGTTGGATGCACTTTTCTTGGCTAGCTTCACGTTGGATGCAGAATGGCTAAAACTGAAC
rs8016416ACGTTGGATGTGCCCATAACTTCCTTTGACACGTTGGATGGCCACGGAATCCTATATAGA
rs8016175ACGTTGGATGTTGAGCACTGAGTGAGTGAGACGTTGGATGTCCTAACCGTGAGTGATCTG
rs7154571ACGTTGGATGATGTGAGGAGCACCTCTGCCACGTTGGATGCTCTTCCCTTCTCAGACGG
rs7158826ACGTTGGATGCACCTCCCTCCTGGACGGGACGTTGGATGGCCACCCCGTCTGAGAAGG
rs7159310ACGTTGGATGACCCCGTCTGAGAAGGGAAGACGTTGGATGCACCTCCCTCCTGGACGGG
rs7401900ACGTTGGATGCCCAACAGCTCATTGAGAACACGTTGGATGTCTTTTCCCCACATTTCCCC
rs7160355ACGTTGGATGTCACTTGTTTATCTGCTGACACGTTGGATGTTATTGATCATTCTTGGGTG
rs2032781ACGTTGGATGTATATCACTGTAGTAACAGCACGTTGGATGACCATAAGTATATATCACAAG
rs6574394ACGTTGGATGACCACACCCAGCCTATTTGTACGTTGGATGTTATGCTGAAAGCCTGGGAG
rs8007598ACGTTGGATGCTGGCAAAAGTCTCTTAACACACGTTGGATGTTGGTTAAAGTCACAGAATG
rs2267767ACGTTGGATGGTTTCACCATGTTAGCCAGGACGTTGGATGTAATCCCAGCACTTTGGGAG
rs6574395ACGTTGGATGAACCTTGAACTCTTGGGCTCACGTTGGATGAAAAAATTCACCGGGCATGG
rs7150066ACGTTGGATGAAGCAATCCTCCTGCTTCTGACGTTGGATGAGATCAGGTGTAGATCCAGG
rs7492334ACGTTGGATGGCCTTTGCATTGGCTATTTGACGTTGGATGTAGAAAGCAGTCATGGGAAG
rs4359361ACGTTGGATGGTAGTATTTGCTTAGTACACACGTTGGATGTTCTAAGCCTGAATGTTTCC
rs4605089ACGTTGGATGAATACCTATGAGATCTCAGGACGTTGGATGCCTTGTAACTCTTTAACATC
rs7146446ACGTTGGATGATTCACTTTTACAAGACCTCACGTTGGATGGCATATTGTACTTAGGAACTC
rs4346144ACGTTGGATGATTCACTTTTACAAGACCTCACGTTGGATGGCATATTGTACTTAGGAACTC
rs7148078ACGTTGGATGTGTGTCAGATTGATGGCTTGACGTTGGATGCCAAGAGAATAAAGCTGAGAG
rs7148286ACGTTGGATGGTGGTCATTAAGCTTGCCAGACGTTGGATGTGCTATGGATGCTGCTTGAG
rs3783980ACGTTGGATGTTTTTTGCCCCAGGTAAGACACGTTGGATGTGGTGCTTTTGTTCTCTCTG
rs1549119ACGTTGGATGTTTCATCTTCCTCTGCCTCCACGTTGGATGGTGAAGGCCAGTCATATTGC
rs1984925ACGTTGGATGAAGTAGCCAGGATTACAGGCACGTTGGATGCCAGCCTAGCAAACATGGTG
rs1477261ACGTTGGATGCAGGGTTATGTGGTATTATCACGTTGGATGGGGAAAGTAAAAGATAAGAG
rs8016447ACGTTGGATGAATTACAGACGTGTGCCACCACGTTGGATGTGACACAGAGAGACTCTGTC
rs7494044ACGTTGGATGAATTACAGACGTGTGCCACCACGTTGGATGTGACACAGAGAGACTCTGTC
rs2023288ACGTTGGATGGAGAAAAATTGTGATTGATTGACGTTGGATGGCCATCAAATCAATCTAATC
rs7151685ACGTTGGATGACAGTGCTGGCATTACTGGCACGTTGGATGTAAAGATCGTCTGCCACTGC
rs2112135ACGTTGGATGAGTGCAGTGGCCCAATCACAACGTTGGATGGTCTAGAGTCCCAGCTACTC
rs2161088ACGTTGGATGTATAGGGTCTCACTCTTGCCACGTTGGATGAGGAGGATCACCTGAGCCTT
rs4903638ACGTTGGATGATAGGGTGTTACTGCGTTGGACGTTGGATGAGGCCTAGGTGAGAAGATTG
rs1477262ACGTTGGATGATGCGTGAGGAGAATGAAGGACGTTGGATGAAGGCTAGTGTTCAGGAAGG
rs1477263ACGTTGGATGAACCTTCCTGAACACTAGCCACGTTGGATGCCTTGCTGCCCCATTTTAAG
rs1477264ACGTTGGATGCGTAGATAGAACCACCTCAGACGTTGGATGAAAGGCGGAGAGCACTTTAC
rs2277917ACGTTGGATGGCATTTGTTGCTAGCTGAAGACGTTGGATGTTGAACAGGAGTACCGTTTG
rs2277918ACGTTGGATGTTACGTTCCTTACTCAGTCCACGTTGGATGACCTGTCGTTTTAAACGCCC
rs2277919ACGTTGGATGTTACGTTCCTTACTCAGTCCACGTTGGATGACCTGTCGTTTTAAACGCCC
rs1978416ACGTTGGATGAGGGCGTTTAAAACGACAGGACGTTGGATGCGGGTGAGAGGATATGGTTT
rs3759728ACGTTGGATGATAGTCCCTCGCTGTTTTGGACGTTGGATGAGAAAGCACTAGGCCTTTGG
rs6574399ACGTTGGATGATGCTCTGATGCCATTATGCACGTTGGATGAGGGCACGTAAAACACATCC
rs7155336ACGTTGGATGGAGGAAGACTCGGTCTAAAAACGTTGGATGAACAATCTGACACTAGGTGC
rs7156186ACGTTGGATGATTACGGGTATGAGCCACTGACGTTGGATGGAACTGGACATTAGGTCTGG
rs7142390ACGTTGGATGTAATCAAGACAGTGTGGTACACGTTGGATGGGGTTTATTTCAGGACTCTC
rs7145875ACGTTGGATGGTCCTTTGAAGCACAAAACCACGTTGGATGCTTCATGATCTTGGATTTGGC
131
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs8014635ACGTTGGATGGTCTTCTCACTCAAGAACACACGTTGGATGCAACAGAGCAAGACTCCAAC
rs8015938ACGTTGGATGCCCTGAACTCAAGTGATCTGACGTTGGATGATCTAAACAGTGTTCCTGGC
rs8015313ACGTTGGATGAAAACTGATTCTGTACCTGGACGTTGGATGGTCAGTCATTTTATAGGCAG
rs8006315ACGTTGGATGTCACTTGAGGTCAGGAGTTCACGTTGGATGCCATGCCTGGCTAAGTTTTG
rs6574400ACGTTGGATGTTATCCTTCCTCTGCCAGTGACGTTGGATGCCTTTGAACTTCCTACCCAG
rs7140816ACGTTGGATGCAATAGAGTGAGACTCTGTCACGTTGGATGATTCATGAGCCTCTCTTTAC
rs4566078ACGTTGGATGTAGAGTCTTGTTCTGTCACCACGTTGGATGAGGAGAATCGCTTGAACCCA
rs7141050ACGTTGGATGATATGTTATACATATTAGTCACGTTGGATGCAAGTTAACCATTATCAACTC
rs3049356ACGTTGGATGTACCACTGGCAGAGTAGAAGACGTTGGATGCACATGGTTTGGGTACTGAG
rs4903639ACGTTGGATGAGCGAGACTCCGTCTCAAAAACGTTGGATGTCAAAGGTAGCCTTGACTGG
rs4903641ACGTTGGATGACTCCAACCTGGGCAACAGAACGTTGGATGCTGGCTCCAGCACACTTATC
rs2364838ACGTTGGATGTGTAGTCCCAGCTACTTGTGACGTTGGATGTGATCATAGCTCACTGCAGC
rs2364839ACGTTGGATGTGGGCAACATAGCAAGATCCACGTTGGATGCTCACAAGTAGCTGGGACTA
rs4632066ACGTTGGATGGAGAAAAAAGAGATGGAGGGACGTTGGATGGCCCTGACTGTGTTTTTATG
rs2112136ACGTTGGATGTTTCTTGGGGACTAAGGCTCACGTTGGATGTAACAGGCCCTGAAGGAATG
rs4641655ACGTTGGATGCGATAGAGCAACCCTGTCTCACGTTGGATGGCCCTACACCCAGATTCAAG
rs4635269ACGTTGGATGAAAGTGCTGGGATTACAGGCACGTTGGATGCTTGCAGCATATTTCTGAGG
rs4570764ACGTTGGATGCTTGCAGCATATTTCTGAGGACGTTGGATGAAAGTGCTGGGATTACAGGC
rs759808ACGTTGGATGATGAGCTGTGATCATGCCACACGTTGGATGCCTGAACTTCATTGTGCTCC
rs7150531ACGTTGGATGATGTGCGGTGTGAAGCAAAGACGTTGGATGTTGTTTGGCCTGGTCTGATG
rs7154968ACGTTGGATGTACCCAGGTAACAAACCTGCACGTTGGATGTCCCCTATAAGGCTTTCAGG
rs7146657ACGTTGGATGTGAGTAGCTGGGACTACAGGACGTTGGATGTAACACGGTGAAACCCCGTC
rs7145859ACGTTGGATGAGGCAGGAGAATGGCGTGAAACGTTGGATGTTTTTGAGACGGAGTCTTGC
rs4903643ACGTTGGATGTATTCCATGCTGTCTGCCTCACGTTGGATGAGTTGACCTTAAAGGCTGGG
rs717682ACGTTGGATGTTTAGGGACAGAGGCTGAGGACGTTGGATGAAGTGCAGTGGCCTGATCTC
rs717683ACGTTGGATGTTGGCAAAAAAGGTGGAGGCACGTTGGATGTGATGATGGCACAGGGAATG
rs1477259ACGTTGGATGTGACTGAGACTACCTTCACCACGTTGGATGAAGTGCTCACGTAGGTTGTC
rs8019064ACGTTGGATGCCTTGCAGCAAACTTCAGAGACGTTGGATGTGACTGAGACTACCTTCACC
rs8018971ACGTTGGATGATGGTCTCACTCTGTCACTCACGTTGGATGAATTGTTTGAGCCCAGGAGG
rs1477260ACGTTGGATGAGTGTCATGGTAGCAAGGACACGTTGGATGTGCCATCTGTTTCCCATAGG
rs5809851ACGTTGGATGACAGAGAGTGTTCAGCACAGACGTTGGATGTTGGGCAACAGAGAGAGACT
rs1985149ACGTTGGATGACTGAAATCTTTGCCTCCCGACGTTGGATGGTGGTGCACTTATGTAGTCC
rs1008988ACGTTGGATGAGTGTGTCTCAGGGAATGTGACGTTGGATGCCTGGCAATTTGTTCTCTGC
rs1008989ACGTTGGATGGGAATAGCAAGTGTAACGGCACGTTGGATGACTCCAACCGCATCAGCTTC
rs8018222ACGTTGGATGATCCTCCATATGCTGAACGCACGTTGGATGAAGGTGGAACGAGAGACTTG
rs1006040ACGTTGGATGTTTAGCTCTCTCTCTGTTGCACGTTGGATGTCTTGAGCCCAGGAGTTCAA
rs1006039ACGTTGGATGTGAAGCTGGGAGTTAGAGACACGTTGGATGCCACCATGCCCAGCTAATTT
rs1006038ACGTTGGATGATAAGCCACTGTGCTCAGTCACGTTGGATGGGTAGGGTTTATTAAGTGCC
rs8009784ACGTTGGATGTGTTTTGGCTATGCTTTGCCACGTTGGATGTGACAGAGCGAGACTTTGTC
rs4903644ACGTTGGATGTTGCAGTGAGCTGAGATTGGACGTTGGATGGTGAATGAATGAATAAGGGCC
rs7149496ACGTTGGATGACAACACACAGTACTGGACCACGTTGGATGTGGGTGCATGTTAGAAACGC
rs6574402ACGTTGGATGCAGGTCCTTTGTCTGACAAGACGTTGGATGGGGATGTGCGATTTGATCTG
TABLE 40
dbSNP Extend Term
rs# Primer Mix
rs7143926 ACCCAAAATTAAGGCAAAATGG CGT
rs1549071 CACACACATATATACACACACA ACG
rs8012858 CACACATATATACACACACACA ACG
132
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Extend Term
rs# Primer Mix
rs7155611 GGCACCGTTCTCTCTCTCA ACT
rs176941 CTGGGCCTCAGTTTACTCAT CGT
rs176942 AATAGGTTGGTTTGTGCCCC ACT
rs176943 CCCGTAGTCCCTGTGAAAC ACT
rs176944 AAAAGTCCACTAATCCTTCCAA CGT
rs4365221 GAGGGCAACTCAACACATTTTA ACG
rs3168952 TTGGTGGTGAGATGGACAGA ACT
rs176945 TACTATACACTCACAAAAATTGTTACT
rs176946 TTGTATAACAAAATACCACAAGCACT
rs176947 GGCGCCCGCCACTACGC ACG
rs176948 AAAACAGACCTCAGTCCTACA ACT
rs176949 CTCCCTGCAGTTCCTTGTTA CGT
rs176950 GGAGTGACATCCCATTACTTT ACG
rs176951 TCCTCCCCTCCTTGGGTG ACT
rs7156905 CTGTCTCAAAAAAGGAACCAG ACT
rs3217197 CTCCAGCCTGAGTGAGAGA ACT
rs2270443 CAGGTGTGGTGGCTCATGC ACG
rs176952 CACTGCAATCGCTGCCTCC ACG
rs176953 GGACCAGCCTGGCCAACAT ACT
rs176954 GCAGTGGCTCAATCCCAGC CGT
rs176955 CTGCCCCTCCAGCCCTTC ACT
rs3214416 CATCACGTGTTCCTAATGAAAA CGT
rs176956 AAGCCACCATGCCCAGCC ACT
rs2544566 CATCTGGGCCTCCCAAAGTA ACT
rs2544567 CATCTGGGCCTCCCAAAGT ACT,
rs176957 GGTCTCGATCTCCTGACCT ACG
rs176958 GGAGTTTTGCTCTTGTTGCC ACT
rs176959 TTTTATTTGTCCCTTGTTCTTTCACT
rs1802227 AATAGTTGCACCAAGCAAGAG ACT
rs176961 TATGGCAAAACCCTGTCTACA ACT
rs176962 GGCTCACGCCTGTAATCCTA ACT
rs7401285 GGGACTCCATATCAGAAAACA CGT
rs176963 GAAAACACACGCGGGCGC ACT
rs176964 CAGTGTCCTACAAAAGTGCCT ACG
rs4903631 CTTGAGACAAGATGAAACAGTT ACG
rs4903632 ATCCTCAGGGAAACGAAAATTA CGT
rs176965 ATAAGGGCTGCCAGCTTGAT ACT
rs4903633 TAGCAATTTTATATCTCAGCATGACT
rs176966 ACCACACCCAGCTAATTTTTG ACG
rs176968 TCACACCTGTGACTCCAGC CGT
rs176969 CCGATTTACTGCATTGCATTTC CGT
rs176970 GTACAGTGGGGTGAATAGTTA ACT
rs7149198 GATATTACTCAGCCATAAAAAAGACT
rs7147918 GGATATTACTCAGCCATAAAAAAACT
rs7148685 TTGAGACCGTCTATTCAGATC ACT
rs1184232 GAATGGAAGAAAATGGTTGCAAACGT
rs1184233 TGCCCAGCCTCTTCAATTAC ACT
rs1184234 TACCAGCACTTTGGGAGGC ACG
rs7401998 ~ CCACGCCTGGCTAATTTTTTTT~ CGT
133
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Extend Term
rs# Primer Mix
rs176974 CTGGGCAACAAAGCAAGACT ACG
rs6574390 AAATTAGCTGGGTATGATGGC ACT
rs176975 AATCTAGGTGGTAGGAGATAC ACT
rs176976 TATAATTCTTTCAGCTTTTCTGTAACT
rs176977 TCAGCCTGGGCAACAAGAG ACG
rs8013727 CCTAACCATAGAAGATAATTAGAAACT
rs176978 ATGTTTTTGTTACCTCTTGTTAC ACG
rs2111829 GAATTTTGCTTGGTGAACAAAAT ACT
rs176980 GCTATGAACGCCATTTTATGTA ACT
rs5809848 TGGGTTCTGAAATCCTGCTG CGT
rs5809849 CGTGTCAGTTCCTTTTTTTTTTT ACT
rs4383070 GCCTCCTGAGTAGCTGGG CGT
rs7493652 TATACCACAAGTAACTGTTAATTTACG
rs2112133 CCACAACTGGCTAATTTTTTGT CGT
rs1963833 CTGGGTGACAGAGCAAGAC CGT
rs6574391 TGGAGAAGTGATAAACTC ACG
rs7155062 ATAACCCTTCAAATGAGCATCA ACT
rs4899674 GGCAAATGGGCTGGGGAG ACG
rs8022516 AGGCATTCTTTTGGGTATAGTA ACG
rs7140838 TCTGAAGAATACCAGACCTCT CGT
rs7141127 CTGAAGAATACCAGACCTCTC ACT
rs6574392 CTGGGCACAGCGACTCAC ACT
rs8003691 TGAAGGCCTCCATGGTATAG ACT
rs8003979 TCTGAAGCCAGTGAGGAAGT ACT
rs8010541 GCTAGCTTCAACTCTCCTGAT ACT
rs8016416 TAACTTCCTTTGACTTGCTTTTT CGT
rs8016175 GTCTGCAATCCCGGCACCT ACG
rs7154571 GTGAGGAGCGTCTCTGCC ACG
rs7158826 TCGCTCCTCACTTCCCAGA ACG
rs7159310 CATCTGGGAAGTGAGGAGC ACT
rs7401900 TGAGAACAGGCCATGATGAC ACT
rs7160355 CCTGCCAAATCCCCCTCTC ACG
rs2032781 GAGAAAAGCGGGCAGGACT ACT
rs6574394 CACCCAGCCTATTTGTATAATT ACT
rs8007598 CTCTTAACACATTTTTTTACAGCAACG
rs2267767 CTGACCTCGTGATCTGCCC ACT
rs6574395 GGCTCAGGCGATCATCGTA ACG
rs7150066 CTGCCACCCAAAGTGCTGG ACT
rs7492334 TTGTGTGTGTGTGTGTGTGG ACT
rs4359361 GCTTAGTACACTTTAAACATGAT ACT
rs4605089 TCAGGAACACCGCTTAATTTTT ACG
rs7146446 CAAGACCTCTTTAAGTAATACTC ACG
rs4346144 AGACCTCTTTAAGTAATACTCC ACT
rs7148078 GGCTTGGGTACGGGAAGC CGT
rs7148286 CATTAAGCTTGCCAGAAAATCA ACG
rs3783980 CATCTTCCTCTGCCTCCCA ACG
rs1549119 CTTCCTCTGCCTCCCATAAAT CGT
rs1984925 CAGGCACGTGCCACCACA ACG
rs1477261 AGGAGGAGCCCAAATATGAAA I
CGT
134
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Extend Term
rs# Primer Mix
rs8016447 CACACCTGGCCATGCTTCC ACT
rs7494044 CCTGGCCATGCTTCCGTATT ACG
rs2023288 AATTGTGATTGATTGATTGCGAT CGT
rs7151685 GTGAGCCACCACATCATCTG ACT
rs2112135 TCAGGTGATCCTCCTGCCT ACG
rs2161088 CCAATCACAGCCCACTGCA ACT
rs4903638 GCCAGAGTGGTCTCCAACT ACT
rs1477262 AGAGCTCAAGCTGATGTCCT ACT
rs1477263 TTTTCCTGTTGAGTTCGCATG ACT
rs1477264 ACCACCTCAGTTTTGCTGTTT ACG
rs2277917 CCTTGATAACCGCTTGGTCT ACT
rs2277918 AAAAGCTTCCCGGGGACAG CGT
rs2277919 AGCTTCCCGGGGACAGCT ACT
rs1978416 TGAGACTAGCTAATGGAGAGT ACG
rs3759728 AGCAAATCTACTGCAAACGTG CGT
rs6574399 AAGTAGAGCTGCTCCACC CGT
rs7155336 GAAGACTCGGTCTAAAAAAAAAA ACT
rs7156186 GCCACTGCACCTGGCCG ACT
rs7142390 TGGTACTGGCATAAGGATAGA ACG
rs7145875 CACAAAACCTTAACTTTTGATTTAACT
rs8014635 CAAGAACACTGGTTTTGGTTTT ACT
rs8015938 CTCAAGTGATCTGCCTGCC ACT
rs8015313 GATTCTGTACCTGGTTGATCAT ACT
rs8006315 AACATGGTGAAGCCCCATCT CGT
rs6574400 GAGATCGCCAGAGACACCA ACG
rs7140816 TGAGACTCTGTCTCAAATACTA CGT
rs4566078 CTCAGCTCACTGCAACCTC ACG
rs7141050 AGCACATAGTAAGTGCCCTAT ACT
rs3049356 GAATAGTGGAAGGTATTGAAATA ACT
rs4903639 GAGACTCCGTCTCAAAAAAAAAA CGT
rs4903641 GGCAACAGAGCGAGACTCC ACT
rs2364838 CCAGCTACTTGTGAGGCCAA ACT
rs2364839 AGCCAGACGTGGTGGCAC ACT
rs4632066 GAGATGGAGGGGGAGCCT ACT
rs2112136 GGGACTAAGGCTCGCATCC ACT
rs4641655 GGATTTCTGGGTCCCACTC ACG
rs4635269 AGCCACCGCGCCCGGCC ACT
rs4570764 GTGATTATTGGCCGGGCGC ACT
rs759808 TGCACCACACAGCCTGGG CGT
rs7150531 AGCAAAGTTAATGGGAGGCC ACT
rs7154968 AACAAACCTGCATATGTACCC ACT
rs7146657 CACCCACCACCCCGCCC ACT
rs7145859 CGGGAGGTGGAGCTTGCA ACT
rs4903643 GCTCCCTTCTGTCTACTGC ACT
rs717682 AGGCTGAGGCAGGAGAATC ACT
rs717683 AAAAGGTGGAGGCCAAAGAC ACT
rs1477259 CGGAATAATTATATCTGCCTCT ACT
rs8019064 CAGAGGCAGATATAATTATTCC ACT
rs8018971 GTCACTCAGGCTGGAGTGC CGT
135
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Extend Term
rs# Primer Mix
rs1477260 GACGAGGAGGAAAGCCATC ACT
rs5809851 CACAGCAGTGTCTTTTTTTTTTT ACT
rs1985149 TTCTTCTCCCTCAGCCTCC ACG
rs1008988 GGGGATGACCTCTCTGGAG ACT
rs1008989 GCCAGCTTGGCAGATTGAG CGT
rs8018222 TGCTGAACGCTGGTCCCC CGT
rs1006040 TGGAGTGCAGTGGCAAGAC ACG
rs1006039 CATAGCCAGACCCTATGAGA ACG
rs1006038 ACTGTGCTCAGTCTATGCTG ACG
rs8009784 ATGCTTTGCCTTAAAGTGGTG ACT
rs4903644 GCCTGGGCAACAGAGCAAG ACT
rs7149496 GATTCTGTAAGTCTGGTATGAG ACT
rs6574402 CTGACAAGAAAATGACTGCATA ACT
Genetic Analysis
[0269] Allelotyping results are shown for cases and controls in Table 41. The
allele frequency for
the A2 allele is noted in the fifth and sixth columns for osteoarthritis case
pools and control pools,
respectively, where "AF" is allele frequency. The allele frequency for the A1
allele can be easily
calculated by subtracting the A2 allele frequency from 1 (A1 AF = 1-A2 AF).
For example, the SNP
rs7143926 has the following case and control allele frequencies: case Al (A) =
0.75; case A2 (T) _
0.25; control A1 (A) = 0.71; and control A2 (T) = 0.29, where the nucleotide
is provided in parenthesis.
Some SNPs are labeled "untyped" because of failed assays.
TABLE 41
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in Position Allele Case Control Value
SEQ ID AF AF
NO: 6
rs7143926218 76161268 A/T 0.25 0.29 0.216
rs15490711440 76162490 C/T 0.15 0.20 0.098
rs80128581442 76162492 C/T 0.93 0.95 0.335
rs71556112611 76163661 C/T 0.02 0.02 0.949
rs176941 4317 76165367 A/C 0.31 0.35 0.271
rs176942 4724 76165774 A/G 0.02 0.02 0.911
rs176943 4788 76165838 G/T 0.13 0.18 0.037
rs 1769445202 76166252 G/T 0.09 0.14 0.107
rs43652215780 76166830 C/T
rs31689525974 76167024 C/T
rs176945 6644 76167694 C/G 0.95 0.96 0.801
rs176946 7430 76168480 A/G 0.10 0.15 0.054
rs176947 7938 76168988 C/T 0.10 0.08 0.473
rs176948 8095 76169145 C/T 0.31 0.35 0.132
rs176949 8183 76169233 A/C 0.03 0.02 0.887
rs176950 8312 76169362 C/T 0.78 0.70 0.008
rs176951 8352 76169402 A/C
rs71569059348 76170398 C/T 0.89 0.90 0.794
rs32171979378 76170428 -/TCTC 0.29 0.35 0.036
rs22704439617 76170667 A/G 0.39 0.34 0.176
rs176952 9727 76170777 C/T 0.17 0.24 0.018
rs176953 9834 76170884 C/T
rs176954 9899 76170949 G/T 0.43 0.52 0.010
rs176955 10211 76171261 C/T 0.12 0.18 0.028
136
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in Position Allele Case Control Value
SEQ ID AF AF
NO: 6
rs321441610377 76171427 -/T 0.91 0.89 0.544
rs 17695610695 76171745 C/T 0.51 0.49 0.492
rs254456610729 76171779 C/G
rs254456710730 76171780 C/T
rs17695711433 76172483 A/G
rs17695811951 76173001 C/G 0.02 NA NA
rs17695912697 76173747 C/T 0.30 0.34 0.147
rs180222712982 76174032 A/C 0.92 0.95 0.332
rs17696114419 76175469 C/T 0.51 0.47 0.158
rs 17696214501 76175551 C/T 0.82 0.79 0.192
rs740128514983 76176033 A/C
rs17696315280 76176330 C/T 0.51 0.46 0.155
rs 17696415475 76176525 A/G 0.53 0.49 0.197
rs490363115888 76176938 A/G
rs490363215976 76177026 A/T
rs17696516307 76177357 A/C 0.55 0.52 0.368
rs490363316442 76177492 A/C 0.83 0.83 0.970
rs17696617255 76178305 C/T
rs17696818948 76179998 G/T 0.23 0.27 0.246
rs17696919435 76180485 A/T 0.14 0.20 0.052
rs17697019753 76180803 C/T 0.35 0.38 0.328
rs714919820021 76181071 C/T
rs714791820022 76181072 A/C
rs714868520503 76181553 A/G 0.19 0.18 0.669
rs 118423220590 76181640 G/T 0.16 0.19 0.316
rs118423321804 76182854 G/T 0.36 0.36 0.895
rs118423421919 76182969 C/T 0.36 0.35 0.797
rs740199821990 76183040 A/T
rs17697422412 76183462 A/G
rs657439022536 76183586 C/T
rs17697523432 76184482 A/G 0.18 0.23 0.147
rs17697623468 76184518 G/T 0.86 0.80 0.087
rs17697723772 76184822 C/T 0.42 0.41 0.794
rs801372724325 76185375 C/T
rs17697824773 76185823 C/T 0.10 0.12 0.512
rs211182926274 76187324 C/T 0.02 NA
rs17698027440 76188490 C/G 0.79 0.73 0.018
rs580984828561 76189611 -IACAG 0.11 0.16 0.091
rs580984930071 76191121 -/A 0.60 0.57 0.355
rs438307031764 76192814 A/T
rs749365233008 76194058 C/T
rs211213335310 76196360 A/T
rs196383335460 76196510 A/C
rs657439137112 76198162 A/G 0.69 0.63 0.064
rs715506237285 76198335 A/G 0.17 0.18 0.878
rs489967437747 76198797 C/T 0.57 0.52 0.201
rs802251638057 76199107 C/T 0.57 0.51 0.135
rs714083838859 76199909 A/C 0.17 0.17 0.957
rs714112738860 76199910 A/G
rs657439239525 76200575 A/G 0.27 0.32 0.099
rs800369140216 76201266 A/G 0.70 0.63 0.029
rs800397940281 76201331 C/T 0.10 0.15 0.024
rs801054141453 76202503 C/G 0.38 0.38 0.993
rs801641642091 76203141 A/T 0.09 0.14 0.035
rs801617542513 76203563 A/G
rs715457142935 76203985 C/T
rs715882642985 76204035 AlG
rs715931043003 76204053 A/G 0.62 NA
rs740190043281 76204331 A/G
rs716035543716 76204766 C/T
rs203278143866 76204916 A/G 0.80 0.74 0.047
rs657439444234 76205284 G/T 0.61 0.54 0.091
rs800759844596 76205646 A/G 0.09 0.10 0.734
rs226776744871 76205921 C/T
137
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in Position Allele Case Control Value
SEQ ID AF AF
NO: 6
rs657439545005 76206055 A/G 0.10 0.14 0.203
rs715006645282 76206332 A/C 0.91 NA
rs749233447178 76208228 A/C
rs435936147816 76208866 G/T
rs460508947887 76208937 A/G
rs714644648134 76209184 C/T 0.09 0.09 0.981
rs434614448135 76209185 A/G 0.83 0.85 0.368
rs714807848276 76209326 G/T 0.44 0.50 0.098
rs714828648400 76209450 C/T 0.96 0.96 0.893
rs378398048798 76209848 A/G 0.15 0.20 0.073
rs154911948803 76209853 A/T 0.18 0.25 0.027
rs198492549146 76210196 C/T 0.04 0.04 0.882
rs147726149969 76211019 A/T
rs801644751059 76212109 A/G 0.10 0.15 0.049
rs749404451064 76212114 C/T
rs202328853285 76214335 A/T 0.97 0.98 0.774
rs715168554560 76215610 C/T
rs211213554748 76215798 A/G 0.05 NA
rs216108854785 76215835 C/G
rs490363855102 76216152 C/G 0.59 0.59 0.975
rs 147726255644 76216694 A/G 0.12 0.17 0.040
rs 147726355705 76216755 G/T 0.18 0.23 0.057
rs147726455841 76216891 A/G 0.45 0.42 0.271
rs227791756623 76217673 C/G 0.30 0.36 0.039
rs227791856825 76217875 A/C 0.49 0.45 0.232
rs227791956827 76217877 A/G 0.20 _ 0.310
0.17
rs197841656892 76217942 C/T 0.79 0.73 0.074
rs375972859150 76220200 A/T 0.13 0.18 0.083
rs657439959958 76221008 A/T 0.33 0.36 0.396
rs715533660231 76221281 C/T 0.25 _ 0.250
0.28
rs715618660524 76221574 A/G 0.85 0.85 0.965
rs714239061871 76222921 C/T
rs714587562226 76223276 C/T
rs801463563230 76224280 G/T 0.07 0.11 0.062
rs801593863468 76224518 G/T 0.08 0.07 0.693
rs801531363787 76224837 C/T 0.67 0.71 0.135
rs800631565732 76226782 A/C
rs657440065989 76227039 A/G 0.75 0.70 0.099
rs714081668832 76229882 G/T 0.54 0.48 0.095
rs456607869904 76230954 C/T
rs714105070365 76231415 A/G
rs304935670886 76231936 -/TATC 0.64 0.69 0.091
rs490363973088 76234138 A/T
rs490364173103 76234153 C/T 0.54 0.66 ~0.0001
rs236483875934 76236984 C/T
rs236483975966 76237016 C/T 0.18 0.18 0.988
rs463206676273 76237323 C/T 0.66 0.66 0.961
rs211213677943 76238993 C/T 0.70 0.64 0.064
rs464165578466 76239516 C/T 0.52 0.48 0.174
rs463526978861 76239911 C/T
rs457076478872 76239922 A/G 0.55 0.68 ~0.0001
rs75980879836 76240886 G/T 0.12 0.18 0.043
rs715053180908 76241958 C/T 0.33 0.31 0.491
rs715496881509 76242559 C/G 0.03 NA
rs714665783576 76244626 C/T 0.57 NA NA
rs714585983662 76244712 C/G
rs490364383782 76244832 C/T 0.10 0.14 0.074
rs71768284282 76245332 G/T 0.11 0.13 0.624
rs71768384444 76245494 A/G 0.79 0.75 0.121
rs147725985129 76246179 C/G 0.11 0.16 0.022
rs801906485151 76246201 A/G 0.90 0.93 0.192
rs801897185296 76246346 A/C
rs147726085809 76246859 C/G 0.12 0.16 0.085
rs580985186387 ~ 76247437 -/T 0.30 0.30 0 993
~ ~ ~ ~
138
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Position ChromosomeAllA2 F A2 F A2 F p-
rs# in Position Allele Case Control Value
SEQ ID AF AF
NO: 6
rs198514986494 76247544 A/G 0.22 0.23 0.892
rs100898889786 76250836 A/G 0.61 0.58 0.380
rs100898989894 76250944 A/T 0.14 0.18 0.172
rs801822290122 76251172 G/T
rs100604092067 76253117 A/G 0.13 0.18 0.092
rs100603992187 76253237 C/T 0.06 0.10 0.133
rs 100603892312 76253362 AIG 0.19 0.24 0.114
rs800978492824 76253874 G/T 0.13 0.18 0.037
rs490364493733 76254783 C/T 0.41 0.38 0.383
rs714949696553 76257603 ClG
rs657440296941 76257991 A/C 0.12 0.17 0.037
[0270] Allelotyping results were considered particularly significant with a
calculated p-value of
less than or equal to 0.05 for allelotype results. These values are indicated
in bold. The allelotyping p-
values were plotted in Figure 1F for the discovery cohort. The position of
each SNP on the
chromosome is presented on the x-axis. The y-axis gives the negative logarithm
(base 10) of the p-
value comparing the estimated allele in the case group to that of the control
group. The minor allele
frequency of the control group for each SNP designated by an X or other symbol
on the graphs in
Figure 1F can be determined by consulting Table 41. For example, the left-most
X on the left graph is
at position 76161268. By proceeding down the Table from top to bottom and
across the graphs from
left to right the allele frequency associated with each symbol shown can be
determined.
[0271] To aid the interpretation, multiple lines have been added to the graph.
The broken
horizontal lines are drawn at two common significance levels, 0.05 and 0.01.
The'vertical broken lines
are drawn every 20kb to assist in the interpretation of distances between
SNPs. Two other lines are
drawn to expose linear trends in the association of SNPs to the disease. The
generally bottom-most
curve is a nonlinear smoother through the data points on the graph using a
local polynomial regression
method (W.S. Cleveland, E. Grosse and W.M. Shyu (1992) Local regression
models. Chapter 8 of
Statistical Models in S eds J.M. Chambers and T.J. Hastie, Wadsworth &
Brooks/Cole.). The black line
provides a local test for excess statistical significance to identify regions
of association. This was
created by use of a lOkb sliding window with lkb step sizes. Within each
window, a chi-square
goodness of fit test was applied to compare the proportion of SNPs that were
significant at a test wise
level of 0.01, to the proportion that would be expected by chance alone (0.05
for the methods used
here). Resulting p-values that were less than 10-$ were truncated at that
value.
[0272] Finally, the exons and introns of the genes in the covered region are
plotted below each
graph at the appropriate chromosomal positions. The gene boundary is indicated
by the broken
horizontal line. The exon positions are shown as thick, unbroken bars. An
arrow is place at the 3' end
of each gene to show the direction of transcription.
139
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
Example 10
ERG Region Proximal SNPs
[0273] It has been discovered that SNP rs1888475 in v-ets erythroblastosis
virus E26 oncogene
like (ERG) is associated with occurrence of osteoarthritis in subjects. One
hundred sixty-six additional
allelic variants proximal to rs1888475 were identified and subsequently
allelotyped in osteoarthritis case
and control sample sets as described in Examples 1 and 2. The polymorphic
variants are set forth in
Table 42. The chromosome positions provided in column four of Table 42 are
based on Genome "Build
34" of NCBI's GenBank.
TABLE 42
dbSNP Position Chromosome Allele
rs# Chromosomein SEQ Position Variants
ID NO:
7
rs289835321 231 38783681 alt
rs960818 21 882 38784332 al
rs960819 21 960 38784410 a/c
rs241003421 1194 38784644 a/c
rs283643721 1530 38784980 al
rs283643821 1673 38785123 a/
rs283643921 2096 38785546 c/t
rs283644021 2285 38785735 a/
rs222668321 5873 38789323 c/t
rs283644121 7256 38790706 a/
rs283644221 7988 38791438 a/
rs283644321 8222 38791672 /t
rs283644421 8381 38791831 c/t
rs378790621 8814 38792264 c/t
rs383810821 8915 38792365 -/c
rs283644521 9642 38793092 a/
rs283644621 9902 38793352 a/t
rs378790821 10619 38794069 a/
rs283644721 10927 38794377 c/t
rs283644821 11032 38794482 clt
rs283645021 14377 38797827 c/t
rs283645121 15608 38799058 c/t
rs101502221 15928 38799378 c!
rs283645221 16296 38799746 a/
rs283645321 17598 38801048 a/t
rs378790921 19272 38802722 a/
rs283645421 20084 38803534 a/
rs283645521 20577 38804027 a/t
rs215571821 28051 38811501 a/
rs283645621 29466 38812916 a/
rs283645721 29530 38812980 c/t
rs283645821 29987 38813437 a/
rs203232321 30012 38813462 clt
rs205140021 30322 38813772 /t
rs283645921 32216 38815666 c/t
rs283646021 32516 38815966 clt
rs283646121 32544 38815994 a/
rs283646221 32746 38816196 a/
140
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Position ChromosomeAllele
rs# Chromosomein SEQ Position Variants
ID NO:
7
rs283646321 33137 38816587 /t
rs283646421 33538 38816988 a/
rs283646521 33798 38817248 c/t
rs283646621 33802 38817252 a/c
rs283646721 33964 38817414 c/t
rs382720421 34132 38817582 a/
rs283646821 34210 38817660 c/t
rs378791121 34317 38817767 a/
rs283646921 34499 38817949 c/t
rs283647021 34753 38818203 a/c
rs221259921 34845 38818295 c/t
rs283647221 35335 38818785 c/t
rs283647321 36423 38819873 clt
rs188846921 36450 38819900 a/
rs188847021 36481 38819931 /t
rs203232221 38447 38821897 c!
rs241003521 38784 38822234 c/t
rs157333221 39387 38822837 alt
rs283647421 39458 38822908 c/t
rs283647521 39822 38823272 c/
rs378791421 40305 38823755 c/
rs188847121 40869 38824319 c/t
rs188847221 40926 38824376 c/t
rs188847321 41010 38824460 c/t
rs188847421 41134 38824584 clt
rs283647621 41984 38825434 a/
rs378791621 42172 38825622 a/t
rs283647721 42753 38826203 /t
rs970043 21 43011 38826461 c/t
rs221260021 43176 38826626 a/
rs283647821 43320 38826770 /t
rs283647921 43381 38826831 alt
rs147587721 44142 38827592 a/
rs283648021 44383 38827833 a/
rs283648121 44726 38828176 c/t
rs283648321 45087 38828537 a/
rs283648421 45141 38828591 clt
rs283648521 45359 38828809 c/
rs283648621 45421 38828871 c/t
rs283648721 45456 38828906 c/t
rs189319921 45467 38828917 c/t
rs283648821 45486 38828936 c/t
rs189320021 45709 38829159 a/
rs189320121 45716 38829166 al
rs283648921 47626 38831076 c/t
rs188847521 49413 38832863 a/
rs283649021 49796 38833246 c/t
rs283649121 49962 38833412 a/
rs283649221 50075 38833525 c/t
rs283649321 50093 38833543 a/
rs283649421 50571 38834021 c/t
rs283649521 50615 38834065 a/
141
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Position Chromosome Allele
rs# Chromosomein SEQ Position Variants
ID NO:
7
rs289835421 50780 38834230 a/
rs306539021 50851 38834301 -/ta
rs283649621 51459 38834909 alc
rs283649721 53193 38836643 c/t
rs283649821 53702 38837152 c/t
rs283649921 53736 38837186 a/c
rs283650021 53795 38837245 c/t
rs283650121 54109 38837559 a/t
rs283650221 54126 38837576 c/t
rs283650321 54230 38837680 a/c
rs283650421 54894 38838344 c/t
rs378791721 55455 38838905 a/
rs283650521 55499 38838949 a/
rs283650621 56522 38839972 c/t
rs283650721 56662 38840112 c/t
rs283650821 56954 38840404 a/
rs283650921 57267 38840717 a/
rs283651021 58282 38841732 a/
rs283651121 58916 38842366 a/c
rs221260121 59544 38842994 c/
rs221260221 59666 38843116 c/t
rs222668221 59913 38843363 a/t
rs283651221 66846 38850296 a/
rs283651321 67245 38850695 /t
rs199932821 67652 38851102 a/c
rs221260321 67955 38851405 a/
rs378791921 67966 38851416 a/c
rs283651421 68420 38851870 a/
rs102315321 70226 38853676 a/
rs102337221 70810 38854260 c/t
rs221260421 72246 38855696 a/
rs222668421 73330 38856780 /t
rs221260521 73457 38856907 c/t
rs218730721 74389 38857839 a/
rs306541221 74638 38858088 -/aa
rs289835521 74640 38858090 a/c
rs283651821 75358 38858808 a/c
rs383811021 75952 38859402 -/
rs283651921 76098 38859548 al
rs382720721 77836 38861286 a/
rs283652021 78449 38861899 alc
rs283652121 78507 38861957 /t
rs283652221 80031 38863481 /t
rs283652321 81695 38865145 clt
rs283652421 82775 38866225 al
rs283652521 82795 38866245 a/
rs383335021 84611 38868061 -!c
rs283652621 84657 38868107 c/t
rs283652721 84693 38868143 a/c
rs383467621 85020 38868470 -/t
rs283652821 85048 38868498 clt
rs376136421 85100 38868550 c/t
142
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Position Chromosome Allele
rs# Chromosomein SEQ Position Variants
ID NO:
7
rs2836529 21 85325 38868775 a/c
rs2836530 21 85452 38868902 c/t
rs3761366 21 85868 38869318 a/
rs2836531 21 85936 38869386 a/
rs2836532 21 85990 38869440 a/t
rs2836533 21 86139 38869589 c/t
rs2836534 21 86497 38869947 c/t
rs2836535 21 87236 38870686 a/
rs2836536 21 87248 38870698 c/t
rs3827208 21 87533 38870983 c/
rs715860 21 87912 38871362 a/
rs717231 21 88108 38871558 /t
rs2836537 21 88494 38871944 a/c
rs2836538 21 89598 38873048 a/c
rs2836539 21 90235 38873685 a/t
rs2836540 21 91287 38874737 /t
rs2836541 21 91359 38874809 c/t
rs2836542 21 92384 38875834 a/c
rs2836543 21 92410 38875860 c/t
rs881837 21 92900 38876350 c/t
rs3949052 21 94495 38877945 a/
rs2065307 21 94512 38877962 a/
rs3216105 21 97777 38881227 -/a
rs2073427 21 98333 38881783 c/t
Assay for Verif~n~ and Allelotypin~ SNPs
[0274] The methods used to verify and allelotype the 166 proximal SNPs of
Table 42 are the same
methods described in Examples 1 and 2 herein. The primers and probes used in
these assays are
provided in Table 43 and Table 44, respectively.
TABLE 43
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs2898353ACGTTGGATGAATGTGAATGTGGAGGTAGCACGTTGGATGCTCCCTTGCTGGTTTTTTTG
rs960818ACGTTGGATGTGGGATTTTTCCCAGAAGAGACGTTGGATGCTGTGCAGAGAAACATGATG
rs960819ACGTTGGATGCTGTCTCCCTTCTCTTTATCACGTTGGATGCATCATGTTTCTCTGCACAG
rs2410034ACGTTGGATGTTTAGAGACATTTCTCCTAGACGTTGGATGTTAGGATGATGTTAGTTTGG
rs2836437ACGTTGGATGAGCTTCTGCGATATCAGTGGACGTTGGATGTTCCTGTCAGCACATTCTCC
rs2836438ACGTTGGATGAACATGTCTTGGCCAAGCTCACGTTGGATGCCACTGTGACCTCTGGATTT
rs2836439ACGTTGGATGCCTAGTGTATAAAGTGATGCACGTTGGATGTCCTTTCTAGGCACCAATAC
rs2836440ACGTTGGATGAGATCCTAACCAACCACAGCACGTTGGATGAGGTAGGTAGATACAAGGCC
rs2226683ACGTTGGATGAATATGGCTCCTATAGACAGACGTTGGATGTTTTGGGTCACAAAATCAAG
rs2836441ACGTTGGATGTTACCTTAATAGTGCTGGCCACGTTGGATGACTTTCTGGTCAGAGAGAAG
rs2836442ACGTTGGATGCAAGGACTCTAGGCTTACAGACGTTGGATGGGGACATTTGTAGTCACTTC
rs2836443ACGTTGGATGGGGCCCCATTACATGTCTAAACGTTGGATGTTCGCTGTACTTCCTTCGAG
rs2836444ACGTTGGATGCTGCAACCAGGAATTGTCAGACGTTGGATGAGGACCCATAAAGAGGTGTG
rs3787906ACGTTGGATGTGAAAAGAGCGGAAATCAACACGTTGGATGGTAAGAAAATCATTCTGTGG
rs3838108ACGTTGGATGATGAATAAGATGGCAGGCTGACGTTGGATGAAGCTGCCCAGATAAAACAG
143
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs2836445ACGTTGGATGCATTTCCAAAATTAGACGCAGACGTTGGATGAAAAAGAGAAAAACAGATGC
rs2836446ACGTTGGATGGTGCCTTGTCCTATCAAGAGACGTTGGATGAGCATCCAAGCCTGGTAATC
rs3787908ACGTTGGATGAATCACCACACTAGACCAGCACGTTGGATGCATGCAAGGGAAATGTGTGC
rs2836447ACGTTGGATGATCTCCTCTCTTTGCTCTGCACGTTGGATGGAGGAAGGTTAGGAGCTAAG
rs2836448ACGTTGGATGTGTAGGGATGTATAGGGCAGACGTTGGATGAAAGAGAGGAGATCCGTCTG
rs2836450ACGTTGGATGTGTGGGCATCAGATGACAACACGTTGGATGATCCCGTTAAATGCACCGAC
rs2836451ACGTTGGATGCAGACAAACAACTGTCACCCACGTTGGATGGTATTTCCTTTTCTCGCCGC
rs1015022ACGTTGGATGTCGAGCCAGCGTCTTTTATCACGTTGGATGGTAACAGTCGTACATTCCGG
rs2836452ACGTTGGATGATCACTGACACAGTCATGAGACGTTGGATGCCAGTAACTTTGCAGGTTTG
rs2836453ACGTTGGATGTGTATTTCCCAAGATGGCCCACGTTGGATGCCTCACTTTCTGATGGAAGC
rs3787909ACGTTGGATGACTTCTCAGTGTTCTGGCTGACGTTGGATGCGTCACTCTCTGTTTCATGG
rs2836454ACGTTGGATGAGGAATGATTCACAACCTCCACGTTGGATGGAATGTTCAAATGTAGGGTGG
rs2836455ACGTTGGATGGGTCTATTGCTGTGACATTTACGTTGGATGCATCCCAATTTTTAAGCAAG
rs2155718ACGTTGGATGAGAACTCTCACACACAGCTGACGTTGGATGTGCCTCTTATTACAGCCCTG
rs2836456ACGTTGGATGGGGATTGTCTGATCTCCTTGACGTTGGATGCCAGCTTTCCTTTGTGCATG
rs2836457ACGTTGGATGAACTCCTGGAATGAGTCACCACGTTGGATGATGCACAAAGGAAAGCTGGG
rs2836458ACGTTGGATGATCACTTAGAAGCCCAGCAGACGTTGGATGTGATGCACACTCACTGAAGC
rs2032323ACGTTGGATGGTAGCCGCACTTTGAGATGCACGTTGGATGAGCACAGAGTCGAGGAGGAG
rs2051400ACGTTGGATGACAGACCTCAGACCAAAGTCACGTTGGATGTTTGTCCTAGAGTAACCCCC
rs2836459ACGTTGGATGGCAAGAATGTTACTTTCTGGACGTTGGATGCCATCAAATAGTTGGTTGTC
rs2836460ACGTTGGATGCAATATCTGAGTTTCACCCCACGTTGGATGGTAGATGAGAATTCCGTGTG
rs2836461ACGTTGGATGGTTACCCACACGGAATTCTCACGTTGGATGCCAGATCCAGGTTCTTTCTG
rs2836462ACGTTGGATGTCTCCTCCGTATGTCTCCATACGTTGGATGATCCCGGAACTCTCTGTTTC
rs2836463ACGTTGGATGGCACTATTTGACTTGAGCTCACGTTGGATGAATTCAAGCCAGAAAGGCTC
rs2836464ACGTTGGATGGTCTTTTTCACCCCAGTAAAGACGTTGGATGATAAGCAAAAGGACCTTTGG
rs2836465ACGTTGGATGTGAGCTCTTGTGTTTTGCCCACGTTGGATGGAGAATTCTCCAGCCTTCTC
rs2836466ACGTTGGATGTGAGCTCTTGTGTTTTGCCCACGTTGGATGGAGAATTCTCCAGCCTTCTC
rs2836467ACGTTGGATGGACTCTGCTCATTTCCTTGGACGTTGGATGAAGAGTAGGGGTAGATGCAG
rs3827204ACGTTGGATGTGAAGATCACACGTGGTGTAACGTTGGATGGGGTGAATGCCAAAAAGAGG
rs2836468ACGTTGGATGTAGAGGCAGGAAAGAGCATGACGTTGGATGTTTTTGGCATTCACCCTCTC
rs3787911ACGTTGGATGTAACCCTCTTCTGGATTCGGACGTTGGATGTCATGTGCTCTGAGAGCATC
rs2836469ACGTTGGATGATTTCTCTACCTCATCCCCCACGTTGGATGGGTTGAAGTCACGTAACAGC
rs2836470ACGTTGGATGCCACTGTTAATCGTATTGCCACGTTGGATGACGGACTGAAAGCCAAATGG
rs2212599ACGTTGGATGAGGAGTTATTCTTCCCCAACACGTTGGATGCAGTGGTCCATTAAGAATCC
rs2836472ACGTTGGATGGAGTATCGTTCTCTATCATGACGTTGGATGTAAAAGAGTCAGAGCAGGAC
rs2836473ACGTTGGATGTCTCAGCCAGAGTTTTGACCACGTTGGATGAATCAACGCCTCCTCTTCAG
rs1888469ACGTTGGATGACCACCAGGAAGGGTCTGAAACGTTGGATGGAGGATCAGAGGCAGAAAAC
rs1888470ACGTTGGATGGCGTTGATTGCAGTTTTCTGACGTTGGATGTTCTTTGGCCTCCGTGTAAG
rs2032322ACGTTGGATGTGATACTCTGTTGAGCCTCCACGTTGGATGGGGGAGCAGTGATGAGTTAT
rs2410035ACGTTGGATGAATCACTTGAACCCAGGAGGACGTTGGATGTTTTTGAGACGGAGTTTCGC
rs1573332ACGTTGGATGGGGTGAACTTTACAGAGAGGACGTTGGATGCTGCCAGACAGTTTTGAGAC
rs2836474ACGTTGGATGAATTCTGCACAGGAGAGTCCACGTTGGATGCAGGAAATGAAGATGTCGCC
rs2836475ACGTTGGATGAGTTCTACATGGGAAGCTGCACGTTGGATGATATCTGTGTCTACAGGCCC
rs3787914ACGTTGGATGGGCTGAAGGCTAAAATCACCACGTTGGATGGTCTGAGAAGTAGGAATGGC
rs1888471ACGTTGGATGACTGAGGCAATTGTGTAGACACGTTGGATGTTGACTTTGTTTTGAGAGGC
rs1888472ACGTTGGATGTTGCCTCTCAAAACAAAGTCACGTTGGATGCTATTATTCTGGAAGCAGCC
rs1888473ACGTTGGATGAGAAAGTTCAGTTCTCAGCCACGTTGGATGTGTTTGCTCCTGTGAGTAAC
rs1888474ACGTTGGATGTGTTATGTGAGTCCAGGGTGACGTTGGATGTCTTGTTATGTGGGTGGGTG
rs2836476ACGTTGGATGTTACCTGTGACCTCATTTGGACGTTGGATGGAACACACAACATACGGTAC
rs3787916ACGTTGGATGAAGGCATCTCAGTCATTCTCACGTTGGATGTGAGTTTGACACAAAGAAGC
rs2836477ACGTTGGATGTTTAGCTCTCCTGGATGATGACGTTGGATGCCATGATTAGTGCATGAAGG
rs970043ACGTTGGATGTATAACTCCCCTCTCTCCTG~ ACGTTGGATGAGAGCAGACCCTTATCAGAG
144
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs2212600ACGTTGGATGGAAACAGGTGTTCATTTGGCACGTTGGATGTCTGCATGAACCAGTAAGTC
rs2836478ACGTTGGATGAGCTATTGAGTGTCACTTGCACGTTGGATGCAGAAGCTTCTGACTTCAAC
rs2836479ACGTTGGATGAGTAGCCATCCTAATAGGTGACGTTGGATGAGCAAGTGACACTCAATAGC
rs1475877ACGTTGGATGAATCAACACTCCCCGTGTTCACGTTGGATGGGTACCTAGAGTAGTCCAAG
rs2836480ACGTTGGATGTACCAAACCCACTGTACATCACGTTGGATGCATAACCTAACACATTGTGGG
rs2836481ACGTTGGATGTAAGAAGTTCTTTCTCCCCCACGTTGGATGGCTGCTTCTTTCATAAGAGG
rs2836483ACGTTGGATGCACTGAGGTAATCTCCAACCACGTTGGATGGGTGGAGATATGGCTTGATG
rs2836484ACGTTGGATGAAGCCCACCAGAGTCATCAAACGTTGGATGACTACTGACCAGCTTTCCAG
rs2836485ACGTTGGATGTTCTAAGTGAAGCCCTCCTCACGTTGGATGTACAGCTGTGCAAACAGTTG
rs2836486ACGTTGGATGCATGGTCTGTTGCCTCTAAGACGTTGGATGCCCTAGCATTTTATGCATCC
rs2836487ACGTTGGATGTGAATACCCACTAGGTCTCGACGTTGGATGCCACCACTAAACTTAGAGGC
rs1893199ACGTTGGATGGGCAACAGACCATGGTTTTGACGTTGGATGCTTCCCTTCAACATGCACTG
rs2836488ACGTTGGATGGGCAACAGACCATGGTTTTGACGTTGGATGCTTCCCTTCAACATGCACTG
rs1893200ACGTTGGATGAGTTAAGTCTTCGCATAACCACGTTGGATGCCTCTCACACACTAAATCTTG
rs1893201ACGTTGGATGGTCTTCGCATAACCAAAACAGACGTTGGATGCCTCTCACACACTAAATCTTG
rs2836489ACGTTGGATGGTCAACCATGGAGCTTGAACACGTTGGATGAGAAGACATGTGGGCTTGTG
rs1888475ACGTTGGATGACCCCTGGCAAGTGAATTACACGTTGGATGGGGAGGTGGATGTTCTTATC
rs2836490ACGTTGGATGAAAGGCAGAGCTAAAGCAAGACGTTGGATGAGCACAACCCAGCAATGCAG
rs2836491ACGTTGGATGACAACTTGGAGTGGAAAGGGACGTTGGATGATCCAGATGGATTCCACAGC
rs2836492ACGTTGGATGACATATGGGCATGGAAGAGCACGTTGGATGAATCCATCTGGATGGAAGAC
rs2836493ACGTTGGATGTTAAGAGTTCCGATGCTTGCACGTTGGATGGTAATCTGGACTTCTCTTCC
rs2836494ACGTTGGATGGTGCATTCATTTGAATTGCTGACGTTGGATGCAGTCTTACTTAAAACTGAC
rs2836495ACGTTGGATGGAATTTAACGAAACTTCAGCACGTTGGATGGGATATTTTCAGGATATCTG
rs2898354ACGTTGGATGTGTAACAAACCTGCACATCCACGTTGGATGGGTACTTTCCAAATATCTGC
rs3065390ACGTTGGATGCGAGACTCCATCTCAAAAAAGACGTTGGATGTGGAAAGTACCAATAGCTTC
rs2836496ACGTTGGATGTGGAGCTTAATGTGTTCCTGACGTTGGATGGTTAGCCATGCATAAGACAG
rs2836497ACGTTGGATGAGCCGGGATGACTGCTAGACACGTTGGATGAGATGAGGCTGAAGAAGTAA
rs2836498ACGTTGGATGGGTCCTGGGAAAATAGGATGACGTTGGATGCACCCTTGCTCTTTCTGAAG
rs2836499ACGTTGGATGACTAGTCAGAGCACAGTGAGACGTTGGATGGCTCTCTCCTTCTTTGACTC
rs2836500ACGTTGGATGGCTTCCTGGTTAGTAAGAGGACGTTGGATGATCAACTCAGGGCTCTTCTC
rs2836501ACGTTGGATGACTCACAAAGGTTGACCTTGACGTTGGATGGAGGTCCAGGTTGAAAGAAC
rs2836502ACGTTGGATGGAGGTCCAGGTTGAAAGAACACGTTGGATGACTCACAAAGGTTGACCTTG
rs2836503ACGTTGGATGGAGCAATTATCAACCCTACGACGTTGGATGATTCTCCCCCTTCACTCTTG
rs2836504ACGTTGGATGGAGTCTGGGTATGGAAAGAGACGTTGGATGTTCCTAGAAATGGTG1'CTGC
rs3787917ACGTTGGATGTTTGGAGGAGGAATGCCTTGACGTTGGATGCGCCCACAAACCTAAGAGAA
rs2836505ACGTTGGATGTTTTCGACTGCTCCACTCTGACGTTGGATGGCTCTCCCTCATTGTTCTTC
rs2836506ACGTTGGATGGGCTAAGGGCATCATTTTATCACGTTGGATGGTTTGCTGATTCATGGATGC
rs2836507ACGTTGGATGAGCAAAGGTTCTGGTGTTGGACGTTGGATGAAATGATGCCCTTAGCCCAG
rs2836508ACGTTGGATGGTGTGATGATATTTTTCTCCACGTTGGATGTTTCAGGTATTCCTCTTTGC
rs2836509ACGTTGGATGTAAAGCTTTCTAAGTCAATGACGTTGGATGTCATATGATAATGGTCTCTG
rs2836510ACGTTGGATGCAGGGAGAGATCTAAACAGCACGTTGGATGGCCAAAGCTATAACACGTGG
rs2836511ACGTTGGATGAGAACCTGACTTTTGGAGTGACGTTGGATGCTTCCTCATTGGTCAGAGTC
rs2212601ACGTTGGATGCCAGCCTTTAGAACTGTGAGACGTTGGATGTGGGCTGCTGTAACAAAGTG
rs2212602ACGTTGGATGACTACAACCAGCCAGAGATGACGTTGGATGCACAAACCTTGTGTGAACCC
rs2226682ACGTTGGATGCCAAGATTGAACCAGGAAAGACGTTGGATGCACAAAAGAATTCAGGAGGTG
rs2836512ACGTTGGATGCCCCAAAACTTAGCATCCTGACGTTGGATGTGTTCTCCCTGCACTTCAAC
rs2836513ACGTTGGATGCACTGGGGTTAGCAAGAAACACGTTGGATGGACTGTGATTCACCCTGTCT
rs1999328ACGTTGGATGAGTTACAGCGCAAATTGAGGACGTTGGATGGCCTTTATGACTCCATTTCTC
rs2212603ACGTTGGATGTGGAGGGTGTCTGTGAGTACACGTTGGATGTCATGGAGCAAGGTCTGTGG
rs3787919ACGTTGGATGCCATCAGCTAGGATTCATGGACGTTGGATGTCTGTGAGTACCCCACAATG
rs2836514ACGTTGGATGCAGGTCTAACTAACTGATGACACGTTGGATGGCCTCTACTGTTATTTAAGG
rs1023153ACGTTGGATGTACAAAAGTGACCTAGAGCC~ ACGTTGGATGTTCTTGCAGGACATTGTGCC
145
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Forward Reverse
rs# PCR primer PCR primer
rs1023372ACGTTGGATGCAAATTCCAAAATTCTGGTTGACGTTGGATGCTCAGAAGTAACATGTACTC
rs2212604ACGTTGGATGCAGACTTGAGCATATACCACACGTTGGATGACCCATGTGGGAAAATGTTG
rs2226684ACGTTGGATGGGTGTTGGAAAAGGAACATCACGTTGGATGTTAATGATAGTTCCCCTCAG
rs2212605ACGTTGGATGATATGAGTGATTTGCATGGGACGTTGGATGTGCATATAAGCTGTCTGCAC
rs2187307ACGTTGGATGCACATCCTGCAGCTTTAACCACGTTGGATGCCTGGCACTTTCAAGTAACG
rs3065412ACGTTGGATGGGCTGAGATAGAATGTGCTCACGTTGGATGTCTCCTGCTTTGTTCTGGAG
rs2898355ACGTTGGATGGGCTGAGATAGAATGTGCTCACGTTGGATGTCTCCTGCTTTGTTCTGGAG
rs2836518ACGTTGGATGCACTTGTTGCTTCTTCCACCACGTTGGATGATGCCAACCTTGCTGATGTC
rs3838110ACGTTGGATGGAAGTAGTGAAGTGTTCCCCACGTTGGATGAGCCTCACTGAATCTTAACG
rs2836519ACGTTGGATGTGTTTCTCCTTCTCACTGGGACGTTGGATGAAAGGCTACAGGAACTGAGC
rs3827207ACGTTGGATGTGTAGTCTGCACCTTCACCTACGTTGGATGAGCGGCTGCTGAACATAGAT
rs2836520ACGTTGGATGCCTGCAAAGGTGTTTGCTTCACGTTGGATGGCCACCTAATTTTTCCTCTC
rs2836521ACGTTGGATGAAGAATAAGAAGCAAACACCACGTTGGATGGTTTTAGGGGAAAGGCATAAG
rs2836522ACGTTGGATGTGCATCTTTGGTTGTGACAGACGTTGGATGGCACATCTACTCTTAGCATG
rs2836523ACGTTGGATGTCTCTCTTTCTTTTCCCTACACGTTGGATGACTCTCAGTTATGATTTCTC
rs2836524ACGTTGGATGGTGTGTTGGTAGAAACGTTCACGTTGGATGGTCACCCCTTCAGATAATAAG
rs2836525ACGTTGGATGCAGAGCCGAAAACATAGTTCACGTTGGATGGTGTGTTGGTAGAAACGTTC
rs3833350ACGTTGGATGGTTGTTCCTTTTGTCTTCTAGACGTTGGATGGAATCATGTCCTTCAGTAAGC
rs2836526ACGTTGGATGATTGTGTCCTGTCCTGCTAGACGTTGGATGGACGGCTAGAAGACAAAAGG
rs2836527ACGTTGGATGGTGTTTTATGTTCTAGCAGGACGTTGGATGGATGCCTTTAGGCAAACATG
rs3834676ACGTTGGATGAAGCTGAAAAGGATGTGCAGACGTTGGATGACAGGGCATACTTCTCTATC
rs2836528ACGTTGGATGCCAAAACTCATGCGATCTGCACGTTGGATGTGGCGCTGAAGTACTCAATG
rs3761364ACGTTGGATGAAACAGCACAGCTACCATTCACGTTGGATGATGAGAAAATGTGTGTGGAG
rs2836529ACGTTGGATGAGCGGTGTTTTAAAATGTCCACGTTGGATGCAGAGCCCAAAAAAAATTTGG
rs2836530ACGTTGGATGACAGACAGTGGTCAGAACATACGTTGGATGAAAGATGCCTATAATCCAGG
rs3761366ACGTTGGATGCAGGTGATAAAAAGCAAGTGACGTTGGATGGCCATCAGTTCTTTTTTGGC
rs2836531ACGTTGGATGGCCTTCGAAAATGTCTCAAGACGTTGGATGCACTTGCTTTTTATCACCTG
rs2836532ACGTTGGATGGAAAGACAGCCTTCGAAAATGACGTTGGATGCAATGGCTCTTTGCAGTAAC
rs2836533ACGTTGGATGTTTCTGACCTCTCACGGTACACGTTGGATGTGCAGATCTGGAGGTAGATG
rs2836534ACGTTGGATGAGAAGAGGCTGGGAGAGGATACGTTGGATGTGCTGCTCTTAGGATAAGGG
rs2836535ACGTTGGATGACAGGAGGAGTTGAGTGTTGACGTTGGATGTAGAGGCACGGAGAAGATAG
rs2836536ACGTTGGATGAAAAGCATGGGTACAGGAGGACGTTGGATGTAGAGGCACGGAGAAGATAG
rs3827208ACGTTGGATGGAGGATGAGAGGTACCTGAGACGTTGGATGGGGATGATCAAACGTAGT
rs715860ACGTTGGATGTTCTGGTGGAGGTTTCTTGGACGTTGGATGCGAGACATGATCTCAAACCC
rs717231ACGTTGGATGCAAGAGACTCAAACAGTTGCACGTTGGATGTCATAGAAGTTACAGCAGCC
rs2836537ACGTTGGATGTTGGTGTGTGATCACTCTGGACGTTGGATGGAACCTAAGTTTCTCCCAGC
rs2836538ACGTTGGATGGGTTAGAGCTTACGTAATTCACGTTGGATGCTACTTGTGTCACTTCTTTG
rs2836539ACGTTGGATGTTATCCTCCAAGAGCCTTAGACGTTGGATGGGGCAAATGGAGTTCTTATT
rs2836540ACGTTGGATGCCCAGTTGGTATCAGTGTTGACGTTGGATGTGCTGAACATCGTTTGGAGG
rs2836541ACGTTGGATGCTTGCACTGACACCTTTGTGACGTTGGATGGTACTGGCGAAGACATGATG
rs2836542ACGTTGGATGAGATGAGCCATTTCCTACTGACGTTGGATGCAGCATGAGAAACTGAATGC
rs2836543ACGTTGGATGAAATGGACTTCTTCAGTAGGACGTTGGATGGATACAATTCAACCCATAGC
rs881837ACGTTGGATGAATGGATGTGGCTCTTGAGGACGTTGGATGTATGGAGGGACTTACGAAAG
rs3949052ACGTTGGATGTTTTCAACGGAAACAGATGCACGTTGGATGCCAAGTAAAATATTCAATCCCC
rs2065307ACGTTGGATGTTTTCAACGGAAACAGATGCACGTTGGATGCCAAGTAAAATATTCAATCCCC
rs3216105ACGTTGGATGACCACCATGCCTGGCTAATTACGTTGGATGGGCCTGGACAAAATAGTGAG
I
rs2073427ACGTTGGATGTTTTGCTTGGGTGTTCTGCCACGTTGGATGGGATTTACACTGGTGTTGGG
~'
146
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
TABLE 44
dbSNP Extend Term
rs# Primer Mix
Rs2898353 TCCTGTCTTCAGTGCTTGATTCTGCGT
rs960818 AGTAGATAACATAAAGTAACCAGCACT
rs960819 GCTATTCACCCTAGCTGTACATAGACT
Rs2410034 AAATGTAGCTGTAGTATCTTGAA ACT
Rs2836437 TTCACACTCAACAACAAACACA ACT
Rs2836438 TGGAAAGTAAGCTAGACCAAACAGACT
Rs2836439 GTATAAAGTGATGCTGCTTGC ACT
Rs2836440 AACAATTGGGATATGTCTCTCCACACG
Rs2226683 GAGAGTTAATGTGCCCTACTT ACT
Rs2836441 TAATAGTGCTGGCCATAATGC ACT
Rs2836442 CTCTAGGCTTACAGTAAACAC ACT
Rs2836443 TATAAGTTCAGGGTCACAGGTC ACT
Rs2836444 TGTGTTCTTGGGGTCGCCT ACT
Rs3787906 TAATGTAGGTGCTGAGAACTTAG ACT
Rs3838108 GGCTGATTAAAATTCTGTTTCCCCACT
Rs2836445 AGACGCAGTAAAACTTATGGAT ACG
Rs2836446 GCCTTGTCCTATCAAGAGCCAAAGCGT
Rs3787908 CATACAGTAGCTGTGGACAGC ACT
Rs2836447 ATGTATTACATTGAGAACCATGTGACT
Rs2836448 TGTATAGGGCAGGGATAAAGAC ACT
Rs2836450 AACAACAAATTTACTGATATCATCACT
Rs2836451 CTGTCACCCATTGACCTCAC ACT
Rs1015022 CTTTTATCTGCAGTTGCACCC ACT
Rs2836452 CGGGAAGATGGCTGCCTTC ACG
Rs2836453 CCAAGATGGCCCAGTAGGA CGT
Rs3787909 AAATAGTAAAATAAAAAGAGCTCCACG
Rs2836454 CACAACCTCCCAAATGAATAAATCACT
Rs2836455 TGCTGTGACATTTTAGTGCTTCTGCGT
Rs2155718 CTCACACACAGCTGGAGTTTA ACT
Rs2836456 CGTTCTGAAGGTTTTGTGTACA ACT
Rs2836457 GAGTCACCCGTCCCCTAGA ACT
Rs2836458 ACAGAAGAGCCAGCCGACA ACT
Rs2032323 TGCACACTCACTGAAGCCC ACT
Rs2051400 AAACACTATGTGACGCCACC ACT
Rs2836459 AGAATGTTACTTTCTGGATTCTACACT
Rs2836460 ATTGTAATTCTCCGTAAAACCC ACG
Rs2836461 TACCCACACGGAATTCTCATCTACACT
Rs2836462 TCCGTATGTCTCCATCCATCTCA ACT
Rs2836463 AAACTTAAATTGCTTTAATCAGCTACT
Rs2836464 AATATCTTATCACTGCTCCTGTCTACG
Rs2836465 GCCCACTTTTGTGTTTGCTTTAG ACT
Rs2836466 TTTGCCCACTTTTGTGTTTGCT ACT
Rs2836467 TTAATTTTCTTGTCTCTTTCTGTAACT
Rs3827204 CCCTCACATCTTCCCCGC ACT
Rs2836468 GCAGGAAAGAGCATGGGCATTAACACT
Rs3787911 TACATCCAAAAGCCTGCCAG ACT
Rs2836469 TCCTGCGAGATCCTGCTCA ACG
147
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
;~",- ,~"".. ....., ....
.~ . ...., , ..,m.
....~ ~.,. .,... ...
.......
..
dbSNP Extend Term
rs# Primer Mix
Rs2836470 ACAAGCTTAATGTTTTGTTCAGA ACT
Rs2212599 TTCCCCAACAATAGTCAGAAAA ACT
Rs2836472 TTCTCTATCATGATGCAGTCC ACT
Rs2836473 GATGATGAACAGGGCTGTGA ACG
Rs1888469 AAGGGTCTGAAGAGGAGGC ACT
Rs1888470 GTTTTCTGCCTCTGATCCTCA ACT
Rs2032322 CCTATAGGTAACGTGGCTTCT ACT
Rs2410035 AGGCAGAAGTTGCAGTGAAC ACG
Rs1573332 GAGAGGCCAGAAAGCCTTC CGT
Rs2836474 GCACAGGAGAGTCCTCAATT ACG
Rs2836475 CATGGGAAGCTGCTGAACTA ACT
Rs3787914 ACAGTGTTTGAGCCCTCCTT ACT
Rs1888471 AACTGACAGAAGAAAGAAAAATATACG
Rs1888472 TGTGTTGGTGTATAAATCAAGATTACG
Rs1888473 CAGTTCTCAGCCAGACGATC ACG
Rs1888474 GAGTCCAGGGTGCTAATTTC ACG
Rs2836476 GGTGTTAGCCCTGGGTTCTAATAAACG
Rs3787916 TCTCTTATGTAAATACAAAGACG CGT
Rs2836477 CCTCTTAAAATAGCCTGCCTTCA ACT
rs970043 GCTCCTTGACTCAAGTATTTC ACG
Rs2212600 AAAACAACTTTCTCTCCCAAAC ACG
Rs2836478 CTTGCTTATCTTCAAGCAGTC CGT
Rs2836479 CCTAATAGGTGTGAAGTGTAAAA CGT
Rs1475877 CTCCCCGTGTTCTGCATGC ACG
Rs2836480 CCCACTGTACATCTTACACTC ACT
Rs2836481 TCCCCCTGAAATCCCATAGC ACT
Rs2836483 AGGTAATCTCCAACCAAACCT ACT
Rs2836484 AGTCATCAAGCCATATCTCCA ACG
Rs2836485 CTCCTCTGGGACGTCAGC ACT
Rs2836486 CCTCTAAGTTTAGTGGTGGAT ACT
Rs2836487 TGTTGGGTTCTACACATTCAAA ACT
Rs1893199 CAGACCATGGTTTTGAATGTG ACG
Rs2836488 GTAGAACCCAACACAGAGCC ACG
Rs1893200 AGTCTTCGCATAACCAAAACAGA ACT
Rs1893201 CGCATAACCAAAACAGAAAAGAACACT
Rs2836489 CAAGAGCTCTTTTCAATTCCAG ACT
Rs1888475 GACATCAAATGATTCCCCTGT ACT
Rs2836490 GAGCCAAAGCTTTCCTGATG ACT
Rs2836491 GTGGAAAGGGCACTGTGGT ACT
Rs2836492 GGCATGGAAGAGCAAGCATC ACT
Rs2836493 TCCGATGCTTGCTCTTCCAT ACT
Rs2836494 TGAAGTTTCGTTAAATTCACTACAACT
Rs2836495 CTTCAGCAATTCAAATGAATGCACACT
Rs2898354 TCCGGCACATATATCCTGGAAC ACT
Rs3065390 AAACAAACAAACAAAAACAGTGTAACT
Rs2836496 GTGTTCCTGATGTTTCTGGAGT CGT
Rs2836497 CTGCTAGACATTGTCAGTCC ACT
Rs2836498 AATAGGATGAGTCAAAGAAGGAG ACT
Rs283649~ GAGAAGAGCCCTGAGTTGATAAA I ACT
148
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Extend Term
rs# Primer Mix
Rs2836500 AGAGGATGAGCAATTTCAGGGA ACT
Rs2836501 CAAAGGTTGACCTTGTTTTCTAT CGT
Rs2836502 AAGAACTTACATTTTATGGCTTC ACT
Rs2836503 GATTTGGGAGCAAGGGAGC ACT
Rs2836504 AGAGTTAAAGATGACTCTAGGCTCACT
Rs3787917 GCAGCCAGAGTGGAGCAGT ACG
Rs2836505 AAGGCATTCCTCCTCCAAATCAC ACT
Rs2836506 GAAAATCAAATCAGTTTCTACAACACT
Rs2836507 GTGTTGGAATATTGTTGGCCT ACT
Rs2836508 ATTCTCTACCATTTCATTCTCTTTACT
Rs2836509 TTTCTAAGTCAATGTAGGCAAC ACT
Rs2836510 CAGCTAGTTATCTTACTTCACC ACT
Rs2836511 AGCAGGTGACAACCCAGACAT ACT
Rs2212601 TAAGTTTCTGTTGTTTATATGCCAACT
Rs2212602 CCAGCCAGAGATGGGATCA ACG
Rs2226682 GATTGAACCAGGAAAGAAATAGTTCGT
Rs2836512 AATGCCAGTTGCCATAGGATA ACG
Rs2836513 ATAAGAAGATGAGTACTATTATTGACT
Rs1999328 ATTGAGGGAAGAGTAAATGATTTCCGT
Rs2212603 TGTCTGTGAGTACCCCACAATGAAACT
Rs3787919 TCTGTGGCTTCAATGCTGGG ACT
Rs2836514 ACAGACTTTAACAAAATCACTGA ACT
Rs1023153 GGGTCATCTCCTTACCTGTCCAA ACG
Rs1023372 TTCCAAAATTCTGGTTGTGTTTT ACT
Rs2212604 CTGCCCCTATACATACATAGCTTCACG
Rs2226684 AAAAACAATCTGCACAACAAATATACT
Rs2212605 GCAGTGAATATGAACAAAAAAAAAACT
Rs2187307 CAGCTTTAACCTCACTCCAC ACT
Rs3065412 AGTTACAAATCAGGTGGTGCTGG ACT
Rs2898355 GTTACAAATCAGGTGGTGCTG ACT
Rs2836518 TAGGAATCGGAGTCAATAATTTT ACT
Rs3838110 GCTGCACAATCCCCCCCC CGT
Rs2836519 CCTTCTCACTGGGTTCCTG ACG
Rs3827207 TATCACCCCTGTGTCCTGC ACG
Rs2836520 CACAAATAGATTATATATCCTGTTACT
Rs2836521 AATAAGAAGCAAACACCTTTGCA ACT
Rs2836522 CCACCCCTTCAGAGAGTTG ACT
Rs2836523 TCATATTGGTTGATCGTATTGGTTACT
Rs2836524 GATTTCAGGAATGAACTATGTTTTACG
Rs2836525 AGCCGAAAACATAGTTCATTCCTGACT
Rs3833350 CTTTTGTCTTCTAGCCGTCAG ACT
Rs2836526 AGAACATAAAACACAGAAATGCA ACT
Rs2836527 TTATGTTCTAGCAGGACAGGA CGT
Rs3834676 AAAAGGATGTGCAGATCGCAT ACT
Rs2836528 ATCTGCACATCCTTTTCAGCTT ACG
Rs3761364 CTACCATTCATTGAGTACTTCAG ACG
Rs2836529 CTTCAAAATGTGGGTTGATACC ACT
Rs2836530 GGTCAGAACATGCTGCTTTAT ACT
Rs3761366 GTGATGGCTTCTAAAAATGTAAA I ACG
149
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Extend Term
rs# Primer Mix
Rs2836531 GCATTTGTTACTGCAAAGAGCCATACG
Rs2836532 AGCCTTCGAAAATGTCTCAAG CGT
Rs2836533 CACACCCATTCCAACCCAAT ACG
Rs2836534 GCTGAAGGTTTCTGGGAGCA ACG
Rs2836535 GAGGAGTTGAGTGTTGGAACCA ACG
Rs2836536 ATGGGTACAGGAGGAGTTGA ACT
Rs3827208 CACCCACCCCAATCACCC ACT
rs715860 CTTGGTTATCCTTCAGTTTCCA ACT
rs717231 CTCATTTAGTTTATGTCTTGGTTGACT
Rs2836537 GCTCATACGCCCTTGGTCTCTAATACT
Rs2836538 AGCTTACGTAATTCAAATCAAGT ACT
Rs2836539 TTACACATTTGCACAATGAGGATACGT
Rs2836540 GTATCAGTGTTGAATGACTGGT ACT
Rs2836541 TGACACCTTTGTGAATTGCTGAACACT
Rs2836542 CCATTTCCTACTGAAGAAGTCCA ACT
Rs2836543 CTTCTTCAGTAGGAAATGGCT ACG
rs881837 GGCTCTTGAGGCCATGCC ACG
Rs3949052 ACAATTTCTCATGTTGTAAGGATTACG
Rs2065307 GGAAACAGATGCCATTTACAATTTACG
Rs3216105 GCCTGGCTAATTTTTAAAAAAAAACGT
Rs2073427 CTGCCCCCACATGACCCA I
ACG
Genetic Anal. skis
[0275] Allelotyping results from the discovery cohort are shown for cases and
controls in Table 45.
The allele frequency for the A2 allele is noted in the fifth and sixth columns
for osteoarthritis case pools
and control pools, respectively, where "AF" is allele frequency. The allele
frequency for the A1 allele
can be easily calculated by subtracting the A2 allele frequency from 1 (A1 AF
= 1-A2 AF). For
example, the SNP rs2898353 has the following case and control allele
frequencies: case A1 (A) = 0.79;
case A2 (T) = 0.21; control A1 (A) = 0.81; and control A2 (T) = 0.19, where
the nucleotide is provided
in paranthesis. Some SNPs are labeled "untyped" because of failed assays.
TABLE 45
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in Position Allele Case Control Value
SEQ ID AF AF
NO: 7
rs2898353231 38783681 A/T 0.21 0.19 0.560
rs960818 882 38784332 A/G 0.59 0.57 0.330
rs960819 960 38784410 A/C 0.13 0.09 0.101
rs24100341194 38784644 A/C
rs28364371530 38784980 AlG 0.14 0.14 0.956
rs28364381673 38785123 A/G 0.79 0.75 0.077
rs28364392096 38785546 C/T 0.70 0.71 0.508
rs28364402285 38785735 A/G 0.19 0.18 0.623
rs22266835873 38789323 C/T 0.79 0.76 0.312
rs28364417256 38790706 A/G 0.12 0.12 0.765
rs28364427988 38791438 A/G 0.31 0.30 0.746
rs28364438222 38791672 G/T 0.22 0.23 0.728
rs28364448381 38791831 C/T 0.19 0.20 0.807
rs37879068814 38792264 C/T 0.97 unt ed NA
150
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
in
rs# SEQ ID Position AlleleCase Control Value
NO: 7 AF AF
rs3838108 8915 38792365 -/C 0.58 0.56 0.425
rs2836445 9642 38793092 A/G 0.32 0.35 0.190
rs2836446 9902 38793352 A/T 0.12 0.14 0.274
rs3787908 10619 38794069 A/G
rs2836447 10927 38794377 C/T 0.68 0.67 0.816
rs2836448 11032 38794482 C/T 0.12 0.14 0.235
rs2836450 14377 38797827 C/T 0.70 0.68 0.460
rs2836451 15608 38799058 C/T 0.92 0.95 0.157
rs1015022 15928 38799378 C/G 0.31 0.36 0.072
rs2836452 16296 38799746 A/G 0.18 0.18 0.822
rs2836453 17598 38801048 A/T 0.02 0.02 0.836
rs3787909 19272 38802722 A/G 0.06 0.03 0.091
rs2836454 20084 38803534 A/G 0.04 0.03 0.397
rs2836455 20577 38804027 A/T 0.17 0.13 0.050
rs2155718 28051 38811501 A/G 0.78 0.78 0.950
rs2836456 29466 38812916 A/G 0.94 0.92 0.569
rs2836457 29530 38812980 C/T
rs2836458 29987 38813437 A/G 0.48 0.46 0.455
rs2032323 30012 38813462 C/T
rs2051400 30322 38813772 GlT 0.03 NA NA
rs2836459 32216 38815666 C/T 0.19 0.17 0.319
rs2836460 32516 38815966 C/T
rs2836461 32544 38815994 AlG
rs2836462 32746 38816196 A/G
rs2836463 33137 38816587 G/T 0.67 0.72 0.032
rs2836464 33538 38816988 A/G 0.67 0.67 0.991
rs2836465 33798 38817248 C/T
rs2836466 33802 38817252 A/C 0.39 0.40 0.627
rs2836467 33964 38817414 C/T
rs3827204 34132 38817582 A/G 0.45 0.42 0.213
rs2836468 34210 38817660 C/T 0.13 0.14 0.678
rs3787911 34317 38817767 A/G 0.13 0.12 0.862
rs2836469 34499 38817949 C/T 0.38 0.40 0.250
rs2836470 34753 38818203 A/C 0.73 0.74 0.939
rs2212599 34845 38818295 C/T 0.66 0.64 0.474
rs2836472 35335 38818785 C/T 0.40 0.35 0.071
rs2836473 36423 38819873 C/T 0.53 0.54 0.755
rs1888469 36450 38819900 A/G 0.45 0.49 0.175
rs1888470 36481 38819931 G/T 0.17 0.18 0.623
rs2032322 38447 38821897 C/G 0.50 0.50 0.879
rs2410035 38784 38822234 C/T
rs1573332 39387 38822837 A/T 0.57 0.58 0.609
rs2836474 39458 38822908 C/T 0.33 0.35 0.564
rs2836475 39822 38823272 C/G 0.17 0,14 0.113
rs3787914 40305 38823755 ClG 0.73 0.73 0.987
rs1888471 40869 38824319 C/T 0.29 0.26 0.175
rs1888472 40926 38824376 C/T 0.62 0.63 0.818
rs1888473 41010 38824460 C/T 0.63 0.65 0.435
rs1888474 41134 38824584 C/T 0.28 0.23 0.099
rs2836476 41984 38825434 A/G 0.46 0.44 0.379
rs3787916 42172 38825622 A/T 0.45 0.43 0.314
rs2836477 42753 38826203 G/T 0.94 0.96 0.196
rs970043 43011 38826461 C/T 0.04 0.04 0.549
rs2212600 43176 38826626 A/G
rs2836478 43320 38826770 G/T 0.76 0.75 0.914
rs2836479 43381 38826831 AIT 0.44 0.43 0.670
rs1475877 44142 38827592 A/G 0.35 0.32 0.110
rs2836480 44383 38827833 A/G 0.46 0.43 0.153
rs2836481 44726 38828176 C/T 0.42 0.40 0.434
rs2836483 45087 38828537 A/G 0.47 0.45 0.393
rs2836484 45141 38828591 C/T 0.46 0.47 0.671
rs2836485 45359 38828809 C/G 0.16 0.17 0.643
rs2836486 45421 38828871 C/T
rs2836487 45456 38828906 C/T 0.02 0.03 0.758
151
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in Position Allele Case Control Value
SEQ ID AF AF
NO: 7
rs189319945467 38828917 C/T 0.62 0.65 0.220
rs283648845486 38828936 C/T 0.25 0.23 0.360
rs189320045709 38829159 A/G 0.16 0.14 0.177
rs189320145716 38829166 A/G 0.84 0.87 0.060
rs283648947626 38831076 C/T 0.29 0.31 0.502
rs1S8847549413 38832863 AlG
rs283649049796 38833246 C/T 0.94 0.93 0.731
rs283649149962 38833412 AIG 0.10 0.08 0.219
rs283649250075 38833525 C/T 0.20 0.22 0.518
rs283649350093 38833543 A/G 0.95 0.94 0.850
rs283649450571 38834021 C/T 0.72 0.70 0.536
rs283649550615 38834065 A/G 0.82 0.78 0.142
rs289835450780 38834230 A/G 0.25 0.25 0.728
rs306539050851 38834301 -/TA 0.10 0.11 0.845
rs283649651459 38834909 A/C 0.80 0.84 0.064
rs283649753193 38836643 C/T 0.65 0.65 0.935
rs283649853702 38837152 C/T 0.43 0.44 0.682
rs283649953736 38837186 A/C 0.33 0.30 0.169
rs283650053795 38837245 C/T
rs283650154109 38837559 A/T 0.36 0.34 0.234
rs283650254126 38837576 C/T 0.31 0.29 0.427
rs283650354230 38837680 A/C 0.32 0.29 0.194
rs283650454894 38838344 C/T 0.51 0.54 0.170
rs378791755455 38838905 A/G 0.56 0.60 0.137
rs283650555499 38838949 A/G 0.73 0.78 0.022
rs283650656522 38839972 C/T 0.52 0.56 0.145
rs283650756662 38840112 C/T 0.51 0.54 0.173
rs283650856954 38840404 A/G 0.53 0.56 0.376
rs283650957267 38840717 A/G 0.35 0.31 0.089
rs283651058282 38841732 A/G 0.65 0.59 0.034
rs283651158916 38842366 A/C 0.32 0.30 0.315
rs221260159544 38842994 ClG 0.45 0.46 0.568
rs221260259666 38843116 C/T 0.30 0.28 0.644
rs222668259913 38843363 A/T 0.38 0.35 0.164
rs283651266846 38850296 A/G 0.94 0.94 0.896
rs283651367245 38850695 G/T 0.23 0.22 0.713
rs199932867652 38851102 A/C 0.79 0.79 0.973
rs221260367955 38851405 A/G 0.73 0.72 0.776
rs378791967966 38851416 A/C
rs283651468420 38851870 A/G 0.52 0.54 0.319
rs102315370226 38853676 A/G 0.09 0.09 0.985
rs102337270810 38854260 C/T 0.83 0.81 0.518
rs221260472246 38855696 A/G 0.68 0.71 0.237
rs222668473330 38856780 G/T 0.83 0.81 0.462
rs221260573457 38856907 C/T 0.82 0,85 0.255
rs218730774389 38857839 A/G 0.13 0.13 0.869
rs306541274638 38858088 -/AA
rs289835574640 38858090 A/C 0.96 0.94 0.413
rs283651875358 38858808 A/C 0.10 0.12 0.261
rs383811075952 38859402 -/G 0.66 0.67 0.790
rs283651976098 38859548 AIG 0.60 0.61 0.509
rs382720777836 38861286 A/G 0.62 0.63 0.575
rs283652078449 38861899 A/C
rs283652178507 38861957 G/T 0.07 0.08 0.551
rs283652280031 38863481 GIT 0.11 0.08 0.155
rs283652381695 38865145 C/T
rs283652482775 38866225 A/G 0.05 0.04 0.321
rs283652582795 38866245 A/G 0.11 0.11 0.875
rs383335084611 38868061 -/C
rs283652684657 38868107 C/T 0.83 0.86 0.292
rs283652784693 38868143 A/C 0.08 0.08 0.936
rs383467685020 38868470 -/T 0.80 0.83 0.191
rs283652885048 38868498 C/T 0.84 0.87 0.089
rs376136485100 38868550 C/T 0.06 0.04 0.159
152
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in Position AlleleCase Control Value
SEQ ID AF AF
NO: 7
rs2836529 85325 38868775 A/C 0.09 0.06 0.100
rs2836530 85452 38868902 C/T
rs3761366 85868 38869318 A/G 0.06 0.04 0.179
rs2836531 85936 38869386 A/G 0.49 0.50 0.729
rs2836532 85990 38869440 A/T 0.30 0.29 0.766
rs2836533 86139 38869589 C/T 0.47 0.48 0.751
rs2836534 86497 38869947 C/T 0.87 0.87 0.874
rs2836535 87236 38870686 A/G 0.93 0.92 0.628
rs2836536 87248 38870698 C/T 0.86 0.84 0.474
rs3827208 87533 38870983 C/G 0.51 0.53 0.459
rs715860 87912 38871362 A/G 0.08 0.09 0.627
rs717231 88108 38871558 G/T 0.65 0.67 0.382
rs2836537 88494 38871944 A/C 0.43 0.40 0.239
rs2836538 89598 38873048 A/C
rs2836539 90235 38873685 A/T 0.98 0.97 0.796
rs2836540 91287 38874737 G/T
rs2836541 91359 38874809 C/T 0.07 0.06 0.403
rs2836542 92384 38875834 A/C 0.36 0.38 0.418
rs2836543 92410 38875860 C/T 0.54 0.50 0.202
rs881837 92900 38876350 C/T 0.29 0.28 0.639
rs3949052 94495 38877945 A/G
rs2065307 94512 38877962 A/G
rs3216105 97777 38881227 -/A 0.32 0.28 0.265
rs2073427 98333 38881783 C/T 0.09 0.07 0.242
[0276] The ERG proximal SNPs were also allelotyped in the replication cohorts
using the methods
described herein and the primers provided in Tables 43 and 44. The replication
allelotyping results for
replication cohort #1 and replication cohort #2 are provided in Tables 46 and
47, respectively.
TABLE 46
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in Position Allele Case Control Value
SEQ ID AF AF
NO: 7
rs2898353231 38783681 A/T 0.19 0.19 0.773
rs960818 882 38784332 A/G 0.59 0.57 0.600
rs960819 960 38784410 A/C 0.07 NA 0.132
rs24100341194 38784644 A/C
rs28364371530 38784980 A/G 0.14 0.14 0.957
rs28364381673 38785123 A/G 0.80 0.77 0.402
rs28364392096 38785546 C/T 0.68 0.73 0.089
rs28364402285 38785735 A/G 0.20 0.18 0.421
rs22266835873 38789323 C/T 0.78 0.76 0.622
rs28364417256 38790706 A/G 0.12 0.12 0.946
rs28364427988 38791438 A/G 0.30 __ 0.674
0.32
rs28364438222 38791672 G/T 0.22 0.25 0.332
rs28364448381 38791831 C/T 0.20 0.20 0.908
rs37879068814 38792264 C/T 0.97 unt ed NA
rs38381088915 38792365 -lC 0.58 0.56 0.604
rs28364459642 38793092 A/G 0.33 0.37 0.211
rs28364469902 38793352 A/T 0.13 0.15 0.481
rs378790810619 38794069 A/G
rs283644710927 38794377 C/T 0.67 0.67 0.843
rs283644811032 38794482 C/T 0.13 0.15 _
0.521
rs283645014377 38797827 C/T 0.67 0.67 0.989
rs283645115608 38799058 C/T 0.92 0.95 0.214
rs101502215928 38799378 C/G 0.30 0.36 0.076
rs283645216296 38799746 A/G 0.18 0.18 0.982
rs283645317598 38801048 A/T 0.02 unt ed NA
rs378790919272 38802722 A/G 0.06 0.03 0.110
rs283645420084 38803534 A/G 0.03 0.03 0.746
rs283645520577 ~ 38804027 A/T 0.17 0 12 0 080
~ ~ ~
153
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Position Chromosome~ Al/A2F A2 F A2 F p-
rs# in Position Allele Case Control Value
SEQ ID AF AF
NO: 7
rs215571828051 38811501 A/G 0.78 0.79 0.747
rs283645629466 38812916 A/G 0.91 0.91 0.915
rs283645729530 38812980 C/T
rs283645829987 38813437 A/G 0.48 0.47 0.626
rs203232330012 38813462 C/T
rs205140030322 38813772 G/T 0.02 un ed NA
rs283645932216 38815666 C/T 0.20 0.16 0.278
rs283646032516 38815966 C/T
rs283646132544 38815994 A/G
rs283646232746 38816196 A/G
rs283646333137 38816587 G/T 0.67 0.75 0.011
rs283646433538 38816988 A/G 0.66 0.68 0.586
rs283646533798 38817248 C/T
rs283646633802 38817252 A/C 0.39 0.41 0.507
rs283646733964 38817414 C/T
rs382720434132 38817582 A/G 0.45 0.41 0.229
rs283646834210 38817660 C/T 0.13 0.14 0.736
rs378791134317 38817767 A/G 0.14 0.13 0.856
rs283646934499 38817949 C/T 0.37 0.41 0.168
rs283647034753 38818203 A/C 0.72 0.73 0.854
rs221259934845 38818295 C/T 0.63 0.65 0.636
rs283647235335 38818785 C/T 0.41 0.35 0.145
rs283647336423 38819873 C/T 0.51 0.54 0.291
rs188846936450 38819900 A/G 0.45 0.49 0.281
rs188847036481 38819931 G/T 0.17 0.17 0.949
rs203232238447 38821897 C/G 0.51 0.53 0.476
rs241003538784 38822234 C/T
rs157333239387 38822837 A/T 0.56 0.60 0.279
rs283647439458 38822908 C/T 0.33 0.36 0.330
rs283647539822 38823272 ClG 0.18 0.13 0.049
rs378791440305 38823755 C/G 0.73 0.74 0.977
rs188847140869 38824319 C/T 0.31 0.26 0.134
rs188847240926 38824376 C/T 0.62 0.65 0.247
rs188847341010 38824460 C/T 0.63 0.67 0.210
rs188847441134 38824584 C/T 0.28 0.21 0.091
rs283647641984 38825434 A/G 0.47 0.44 0.346
rs378791642172 38825622 A/T 0.46 0.41 0.171
rs283647742753 38826203 G/T 0.94 0.97 0.294
rs970043 43011 38826461 C/T 0.05 0.03 0.331
rs221260043176 38826626 A/G
rs283647843320 38826770 G/T 0.75 0.75 0.983
rs283647943381 38826831 A/T 0.44 0.43 0.752
rs147587744142 38827592 A/G 0.35 0.31 0.166
rs283648044383 38827833 A/G 0.45 0.41 0.254
rs283648144726 38828176 ClT 0.42 0.39 0.330
rs283648345087 38828537 A/G 0.46 0.46 0.797
rs283648445141 38828591 C/T 0.45 0.47 0.553
rs283648545359 38828809 CIG 0.18 0.18 0.993
rs283648645421 38828871 ClT
rs283648745456 38828906 C/T 0.03 0.03 0.955
rs189319945467 38828917 C/T 0.61 0.67 0.071
rs283648845486 38828936 C/T 0.27 0.23 0.246
rs189320045709 38829159 A/G 0.16 0.13 0.203
rs189320145716 38829166 A/G 0.83 0.89 0.021
rs283648947626 38831076 C/T 0.30 0.31 0.702
rs188847549413 38832863 A/G
rs283649049796 38833246 C/T 0.94 0.95 0.662
rs283649149962 38833412 A/G 0.10 0.06 0.038
rs283649250075 38833525 C/T 0.20 0.22 0.651
rs283649350093 38833543 A/G 0.93 0.95 0.397
rs283649450571 38834021 C/T 0.73 0.71 0.592
rs283649550615 38834065 A/G 0.81 0.77 0.212
rs289835450780 38834230 A/G 0.24 0.24 0.827
rs306539050851 38834301 -/TA 0.10 0.11 0.743
154
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in Position Allele Case Control Value
SEQ ID AF AF
NO: 7
rs283649651459 38834909 A/C 0.78 0.86 0.022
rs283649753193 38836643 C/T 0.65 0.66 0.733
rs283649853702 38837152 C/T 0.44 0.46 0.576
rs283649953736 38837186 A/C 0.33 0.29 0.200
rs283650053795 38837245 C/T
rs283650154109 38837559 A/T 0.36 0.32 0.167
rs283650254126 38837576 C/T 0.31 0.27 0.206
rs283650354230 38837680 A/C 0.32 0.28 0.173
rs283650454894 38838344 C/T 0.50 0.57 0.033
rs378791755455 38838905 A/G 0.56 0.62 0.033
rs283650555499 38838949 A/G 0.72 0.81 0.004
rs283650656522 38839972 C/T 0.52 0.58 0.093
rs283650756662 38840112 C/T 0.51 0.56 0.134
rs283650856954 38840404 A/G 0.53 0.58 0.170
rs283650957267 38840717 A/G 0.35 0.30 0.136
rs283651058282 38841732 A/G 0.62 0.56 0.035
rs283651158916 38842366 A/C 0.33 0.30 0.273
rs221260159544 38842994 C/G 0.44 0.46 0.675
rs221260259666 38843116 C/T 0.29 0.27 0.571
rs222668259913 38843363 A/T 0.38 0.33 0.127
rs283651266846 38850296 A/G 0.93 0.96 0.261
rs283651367245 38850695 G/T 0.23 0.22 0.692
rs199932867652 38851102 A/C 0.79 0.80 0.618
rs221260367955 38851405 A/G 0.73 0.74 0.676
rs378791967966 38851416 A/C
rs283651468420 38851870 A/G 0.51 0.57 0.044
rs102315370226 38853676 A/G 0.09 0.09 0.699
rs102337270810 38854260 C/T 0.82 unt ed NA
rs221260472246 38855696 A/G 0.67 0.73 0.063
rs222668473330 38856780 G/T 0.82 0.82 0.992
rs221260573457 38856907 C/T 0.83 0.86 0.180
rs218730774389 38857839 A/G 0.14 0.13 0.901
rs306541274638 38858088 -/AA
rs289835574640 38858090 A/C 0.95 0.93 0.442
rs283651875358 38858808 A/C 0.11 0.14 0.248
rs383811075952 38859402 -/G 0.65 0.68 0.399
rs283651976098 38859548 A/G 0.59 0.64 0.134
rs382720777836 38861286 A/G 0.60 0.64 0.205
rs283652078449 38861899 A/C
rs283652178507 38861957 G/T 0.08 0.09 0.765
rs283652280031 38863481 GlT 0.12 0.07 0.033
rs283652381695 38865145 C/T
rs283652482775 38866225 A/G 0.05 0.04 0.539
rs283652582795 38866245 A/G 0.12 0.09 0.179
rs383335084611 38868061 -/C
rs283652684657 38868107 C/T 0.83 0.85 0.536
rs283652784693 38868143 A/C 0.08 0.07 0.444
rs383467685020 38868470 -/T 0.79 0.82 0.270
rs283652885048 38868498 C/T 0.82 0.86 0.130
rs376136485100 38868550 C/T 0.08 0.05 0.132
rs283652985325 38868775 A/C 0.09 0.07 0.214
rs283653085452 38868902 C/T
rs376136685868 38869318 A/G 0.07 0.04 0.259
rs283653185936 38869386 A/G 0.49 0.50 0.741
rs283653285990 38869440 A/T 0.30 0.30 0.921
rs283653386139 38869589 C/T 0.48 0.48 0.843
rs283653486497 38869947 C/T 0.86 0.89 0.374
rs283653587236 38870686 A/G 0.91 0.91 0.933
rs283653687248 38870698 C/T 0.86 0.86 0.945
rs382720887533 38870983 C/G 0.51 0.55 0.183
rs715860 87912 38871362 A/G 0.07 0.07 0.893
rs717231 88108 38871558 G/T 0.65 0.68 0.506
rs283653788494 38871944 A/C 0.43 0.39 0.251
rs283653889598 38873048 A/C
155
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Position ChromosomeAllA2 F A2 F A2 F p-
rs# in Position Allele Case Control Value
SEQ ID AF AF
NO: 7
rs283653990235 38873685 A/T 0.98 0.98 0.910
rs283654091287 38874737 G/T
rs283654191359 38874809 C/T 0.09 0.06 0.324
rs283654292384 38875834 A/C 0.37 0.41 0.365
rs283654392410 38875860 C/T 0.54 0.55 0.863
rs881837 92900 38876350 C/T 0.30 0.28 0.673
rs394905294495 38877945 A/G
rs206530794512 38877962 A/G
rs321610597777 38881227 -/A 0.31 0.29 0.603
rs207342798333 38881783 C/T 0.09 0.06 0.249
TABLE 47
dbSNP Position ChromosomeAllA2 F A2 F A2 F p-
rs# in Position AlleleCase Control Value
SEQ ID AF AF
NO: 7
rs2898353231 38783681 A/T 0.22 0.21 0.629
rs960818882 38784332 A/G 0.59 0.55 0.351
rs960819960 38784410 A/C 0.12 0.01
rs24100341194 38784644 A/C
rs28364371530 38784980 A/G 0.14 0.14 0.989
rs28364381673 38785123 A/G 0.78 0.71 0.047
rs28364392096 38785546 C/T 0.72 0.68 0.265
rs28364402285 38785735 A/G 0.18 0.19 0.789
rs22266835873 38789323 C/T 0.80 0.77 0.342
rs28364417256 38790706 A/G 0.11 0.12 0.559
rs28364427988 38791438 A/G 0.32 0.28 0.269
rs28364438222 38791672 G/T 0.23 0.21 0.504
rs28364448381 38791831 C/T 0.19 0.19 0.829
rs37879068814 38792264 C/T 0.97 unt ed
rs38381088915 38792365 -/C 0.58 0.55 0.526
rs28364459642 38793092 A/G 0.30 0.32 0.722
rs28364469902 38793352 A/T 0.11 0.14 0.425
rs378790810619 38794069 A/G
rs283644710927 38794377 C/T 0.68 0.68 0.908
rs283644811032 38794482 C/T 0.11 0.14 0.302
rs283645014377 38797827 C/T 0.73 0.70 0.314
rs283645115608 38799058 C/T 0.93 0.94 0.499
rs101502215928 38799378 C/G 0.33 0.35 0.527
rs283645216296 38799746 A/G 0.17 0.18 0.750
rs283645317598 38801048 A/T 0.02 0.02 0.934
rs378790919272 38802722 A/G 0.05 0.04 0.546
rs283645420084 38803534 A/G 0.05 0.03 0.379
rs283645520577 38804027 A/T 0.17 0.15 0.472
rs215571828051 38811501 A/G 0.79 0.78 0.704
rs283645629466 38812916 A/G 0.97 0.94 0.174
rs283645729530 38812980 C/T
rs283645829987 38813437 A/G 0.48 0.45 0.532
rs203232330012 38813462 C/T
rs205140030322 38813772 G/T 0.04 0.02 0.476
rs283645932216 38815666 C/T 0.19 0.18 0.921
rs283646032516 38815966 C/T
rs283646132544 38815994 A/G
rs283646232746 38816196 A/G
rs283646333137 38816587 G/T 0.68 0.68 0.988
rs283646433538 38816988 A/G 0.69 0.66 0.430
rs283646533798 38817248 C/T
rs283646633802 38817252 A/C 0.39 0.39 0.948
rs283646733964 38817414 C/T
rs382720434132 38817582 A/G 0.45 0.43 0.614
rs283646834210 38817660 C/T 0.12 0.12 0.879
rs378791134317 38817767 A/G 0.12 0.11 0.901
rs283646934499 38817949 C/T 0.38 0.39 0.914
156
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in PositionAllele Case Control Value
SEQ ID AF AF
NO: 7
rs283647034753 38818203A/C 0.75 0.74 0.960
rs221259934845 38818295C/T 0.71 0.64 0.095
rs283647235335 38818785C/T 0.40 0.36 0.321
rs283647336423 38819873ClT 0.56 0.53 0.433
rs188846936450 38819900A/G 0.45 0.49 0.399
rs188847036481 38819931G/T 0.16 0.19 0.356
rs203232238447 38821897ClG 0.50 0.45 0.190
rs241003538784 38822234C/T
rs157333239387 38822837A/T 0.58 0.56 0.554
rs283647439458 38822908C/T 0.34 0.33 0.762
rs283647539822 38823272C/G 0.15 0.14 0.817
rs378791440305 38823755C/G 0.73 0.73 0.934
rs188847140869 38824319C/T 0.28 0.27 0.760
rs188847240926 38824376C/T 0.63 0.58 0.302
rs188847341010 38824460C/T 0.63 0.62 0.683
rs188847441134 38824584C/T 0.27 0.26 0.853
rs283647641984 38825434A/G 0.46 0.45 0.838
rs378791642172 38825622A/T 0.44 0.45 0.827
rs283647742753 38826203GlT 0.94 0.95 0.505
rs97004343011 38826461C/T 0.04 0.04 0.848
rs221260043176 38826626A/G
rs283647843320 38826770G/T 0.76 0.75 0.893
rs283647943381 38826831A/T 0.44 0.43 0.801
rs147587744142 38827592A/G 0.35 0.33 0.450
rs283648044383 38827833A/G 0.47 0.44 0.444
rs283648144726 38828176C/T 0.41 0.41 0.999
rs283648345087 38828537A/G 0.48 0.44 0.306
rs283648445141 38828591C/T 0.46 0.46 0.939
rs283648545359 38828809C/G 0.15 0.17 0.483
rs283648645421 38828871C/T
rs283648745456 38828906C/T NA 0.03 NA
rs189319945467 38828917C/T 0.63 0.62 0.868
rs283648845486 38828936C/T 0.23 0.22 0.913
rs189320045709 38829159A/G 0.17 0.16 0.653
rs189320145716 38829166A/G 0.85 0.85 0.947
rs283648947626 38831076C/T 0.27 0.30 0.597
rs188847549413 38832863A/G
rs283649049796 38833246CIT 0.94 0.91 0.196
rs283649149962 38833412A/G 0.09 0.11 0.493
rs283649250075 38833525C/T 0.20 0.21 0.669
rs283649350093 38833543A/G 0.96 0.93 0.211
rs283649450571 38834021C/T 0.70 0.69 0.697
rs283649550615 38834065A/G 0.82 0.80 0.510
rs289835450780 38834230A/G 0.27 0.26 0.846
rs306539050851 38834301-/TA 0.11 0.10 0.936
rs283649651459 38834909A/C 0.81 0.80 0.746
rs283649753193 38836643C/T 0.66 0.64 0.756
rs283649853702 38837152C/T 0.41 0.40 0.844
rs283649953736 38837186A/C 0.32 0.30 0.567
rs283650053795 38837245C/T
rs283650154109 38837559A/T 0.36 0.36 0.917
rs283650254126 38837576C/T 0.31 0.32 0.738
rs283650354230 38837680A/C 0.32 0.31 0.730
rs283650454894 38838344C/T 0.52 0.50 0.620
rs378791755455 38838905A/G 0.57 0.56 0.759
rs283650555499 38838949A/G 0.74 0.74 0.982
rs283650656522 38839972C/T 0.52 0.53 0.907
rs283650756662 38840112C/T 0.51 0.52 0.785
rs283650856954 38840404A/G 0.53 0.52 0.709
rs283650957267 38840717A/G 0.35 0.33 0.453
rs283651058282 38841732A/G 0.68 0.65 0.457
rs283651158916 38842366A/C 0.32 0.31 0.832
rs221260159544 38842994C/G 0.45 0.47 0.717
rs221260259666 38843116C/T 0.30 0.30 0.994
157
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
dbSNP Position ChromosomeAl/A2 F A2 F A2 F p-
rs# in PositionAllele Case Control Value
SEQ ID AF AF
NO: 7
rs222668259913 38843363A/T 0.39 0.38 0.801
rs283651266846 38850296A/G 0.94 0.91 0.184
rs283651367245 38850695G/T 0.23 0.23 0.949
rs199932867652 38851102A/C 0.80 0.77 0.487
rs221260367955 38851405A/G 0.74 0.70 0.289
rs378791967966 38851416A/C
rs283651468420 38851870A/G 0.53 0.49 0.363
rs102315370226 38853676A/G 0.08 0.09 0.611
rs102337270810 38854260C/T 0.84 0.81 0.315
rs221260472246 38855696A/G 0.69 0.68 0.641
rs222668473330 38856780G/T 0.85 0.81 0.216
rs221260573457 38856907C/T 0.82 0.82 0.927
rs218730774389 38857839A/G 0.12 0.13 0.685
rs306541274638 38858088-/AA
rs289835574640 38858090A/C 0.96 0.96 0.893
rs283651875358 38858808A/C 0.10 0.11 0.823
rs383811075952 38859402-/G 0.68 0.65 0.457
rs283651976098 38859548A/G 0.60 0.57 0.357
rs382720777836 38861286AIG 0.64 0.61 0.449
rs283652078449 38861899A/C
rs283652178507 38861957G/T 0.06 0.07 0.625
rs283652280031 38863481G/T 0.09 0.10 0.810
rs283652381695 38865145C/T _
rs283652482775 38866225A/G 0.05 0.04 0.419
rs283652582795 38866245A/G 0.10 0.14 0.132
rs383335084611 38868061-/C
rs283652684657 38868107C/T 0.83 0.86 0.342
rs283652784693 38868143A/C 0.08 0.11 0.209
rs383467685020 38868470-/T 0.81 0.84 0.442
rs283652885048 38868498C/T 0.86 0.88 0.350
rs376136485100 38868550C/T 0.04 0.03 0.643
rs283652985325 38868775A/C 0.08 0.06 0.271
rs283653085452 38868902C/T
rs376136685868 38869318A/G 0.06 0.04 0.473
rs283653185936 38869386A/G 0.49 0.49 0.915
rs283653285990 38869440A/T 0.31 0.28 0.446
rs283653386139 38869589C/T 0.47 0.48 0.810
rs283653486497 38869947C/T 0.88 0.84 0.149
rs283653587236 38870686A/G 0.94 0.92 0.378
rs283653687248 38870698C/T 0.86 0.82 0.311
rs382720887533 38870983C/G 0.51 0.49 0.598
rs715860 87912 38871362A/G 0.09 0.11 0.463
rs717231 88108 38871558G/T 0.65 0.67 0.588
rs283653788494 38871944A/C 0.42 0.41 0.694
rs283653889598 38873048A/C
rs283653990235 38873685A/T 0.97 0.97 0.749
rs283654091287 38874737G/T
rs283654191359 38874809C/T 0.05 0.05 0.895
rs283654292384 38875834A/C 0.34 0.34 0.998
rs283654392410 38875860C/T unt 0.43 NA
ed
rs881837 92900 38876350C/T 0.29 0.28 0.811
rs394905294495 38877945A/G
rs206530794512 38877962A/G
rs321610597777 38881227-/A 0.32 0.28 0.273
rs207342798333 38881783C/T 0.08 0.07 0.700
[0277] Allelotyping results were considered particularly significant with a
calculated p-value of
less than or equal to 0.05 for allelotype results. These values are indicated
in bold. The allelotyping p-
values were plotted in Figure 1G for the discovery cohort. The position of
each SNP on the
chromosome is presented on the x-axis. The y-axis gives the negative logarithm
(base 10) of the p-
158
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
value comparing the estimated allele in the case group to that of the control
group. The minor allele
frequency of the control group for each SNP designated by an X or other symbol
on the graphs in Figure
1 G can be determined by consulting Table 45. For example, the left-most X on
the left graph is at
position 38783681. By proceeding down the Table from top to bottom and across
the graphs from left
to right the allele frequency associated with each symbol shown can be
determined.
[0278] To aid the interpretation, multiple lines have been added to the graph.
The broken
horizontal lines are drawn at two common significance levels, 0.05 and 0.01.
The vertical broken lines
are drawn every 20kb to assist in the interpretation of distances between
SNPs. Two other lines are
drawn to expose linear trends in the association of SNPs to the disease. The
generally bottom-most
curve is a nonlinear smoother through the data points on the graph using a
local polynomial regression
method (W.S. Cleveland, E. Grosse and W.M. Shyu (1992) Local regression
models. Chapter 8 of
Statistical Models in S eds J.M. Chambers and T.J. Hastie, Wadsworth &
Brooks/Cole.). The black line
provides a local test for excess statistical significance to identify regions
of association. This was
created by use of a lOkb sliding window with lkb step sizes. Within each
window, a chi-square
goodness of fit test was applied to compare the proportion of SNPs that were
significant at a test wise
level of 0.01, to the proportion that would be expected by chance alone (0.05
for the methods used
here). Resulting p-values that were less than 10-8 were truncated at that
value.
[0279] Finally, the exons and introns of the genes in the covered region are
plotted below each
graph at the appropriate chromosomal positions. The gene boundary is indicated
by the broken
horizontal line. The exon positions are shown as thick, unbroken bars. An
arrow is place at the 3' end
of each gene to show the direction of transcription.
Example 11
Expression of LRCHI in Human Chondroblastoma Cells
[0280] Human chondrosarcoma cells were cultured either in monolayers or in a
solid alginate
matrix to address the possibilty that chondrocytes would dedifferentiate in
monolayer culture but would
retain a chondrocytic phenotype in matrix environments (Lee, D.A., T. Reisler,
and D.L. Bader,
Expansion of chondrocytes for tissue engineering in alginate beads enhances
chondrocytic phenotype
compared to conventional monolayer techniques. Acta Orthop Scand, 2003. 74(1):
p. 6-15).
Methods
[0281] SW1353 chondrosarcoma cells (ATCC, HTB-94 ) were propagated in
Leibovitz's L-15
medium supplemented with 2 mM L-glutamine,l0% fetal calf serum and
penicillin/streptomycin
(100U/ml) as per ATCC protocol. Confluent SW1353 cells were made into single
cell suspensions by
treatment with trypsin-EDTA and were resuspended in 1.2% alginate (Keltone
LVCR, Kelco, Chicago,
USA) in 0.9%NaCI at a density of 4x106 cells/ml (10 million cells per
stimuli). Alginate beads of
uniform diameter were prepared by dispensing the cell-alginate suspension
dropwise through a 22 gauge
needle into 100mm CaCl2 from a height of approximately 2cm. After
polymerization (10 minutes),
159
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
beads were washed 3 times in PBS and then once with medium. The encapsulated
cells were
differentiated in a 24 well plate (10 beads/we11;25-SOK cells/bead) for 2
weeks under standard
conditions with medium changes every 3 days. At the end of 14 days, a few
randomly selected beads
were stained for the presence of glycosaminoglycans by alcian blue staining
suggesting a chondrocytic
phenotype [46J. After 14 days, the alginate cultured cells were stimulated
with either recombinant
human IL1-beta (R&D Systems) or phorbol 12-myristate 15 - acetate (PMA, Sigma)
alongside serum-
starved controls for 3 hours (PMA) and 24 hours (no serum and IL1-beta).
Similar experimental
conditions were applied on confluent plates of undifferentiated SW1353 cells
to compare the effects of
monolayer culture to alginate culture on gene expression. Encapsulated cells
were released from the
alginate beads by sodium citrate (SSmM in O.15M NaCI) treatment and the
expression of target genes
plus control genes (matrix metalloproteinases 8 and 13) was determined by mRNA
isolation
(Dynabeads oligo dT(25), Dynal Biotech), followed by cDNA synthesis
(Superscript II, Invitrogen) and
semi-quantitative PCR using standard molecular biology techniques and
manufacturer's protocols. PCR
was performed using a standard protocol of 30 cycles. LRCHI forward primer: 5'-
CCAAAGATCAGGACATGGATA-3'; LRCHI reverse primer: 5'-TGCTGTTTGTGGTAGGAGAG-
3'; MMP8 forward primer: 5'-CAATACTGGGCTCTGAGTGG-3'; MMP8 reverse primer: 5'-
GGAAAGGCACCTGATATGC-3'; MMP13 forward primer: 5'-ATATCTGAACTGGGTCTTCC-3';
MMPl3 reverse primer: 5'-GACAGCATCTACTTTATCACC-3'; GAPDH forward primer: 5'-
ATCATCTCTGCCCCCTCTG-3'; GAPDHreverse primer: 5'-GAGGCATTGCTGATGATCTTG-3';
Single band PCR products were resolved on 2% agarose gels and visualized by
ethidium bromide
staining. cDNA levels were normalized for cell number differences by the
housekeeping gene, GAPDH.
Control cDNA is composed of an equimolar mixture of 56 cDNA preparations from
various human cell
lines and was used to verify that the selected primers only amplified a single
predicted product.
Results
[0282] Analysis of LRCHI expression in alginate cultured human chondrosarcoma
cells treated
with inflammatory stimuli, IL1-beta and PMA revealed substantial increases in
the expression of the
known IL1-beta responsive gene, MMP13 [52], in both IL1-beta and PMA
stimulated cells.
Interestingly, MMP8 was strongly upregulated by IL1-beta but weakly
upregulated by PMA, suggesting
that MMP8 may be regulated by different inflammatory stimuli and pathways than
MMP13. LRCHl
expression after IL1-beta and PMA stimulation was unchanged from controls.
This suggests that the
effect that LRCH1 has on the etiology of osteoarthritis may be via an
inflammatory independent
mechanism, possibly involving compressive stress. There were no differences in
expression of LRCH1
or control genes in monolayer cultured SW1353 cells compared to alginate
cultured cells suggesting
that SW1353 cells retain a chondrocytic phenotype even in monolayer culture
conditions (data not
shown).
160
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
Example 12
Ire Vitro Production of Target Polyp~tides
[0283] cDNA is cloned into a pIVEX 2.3-MCS vector (Roche Biochem) using a
directional
cloning method. A cDNA insert is prepared using PCR with forward and reverse
primers having 5'
restriction site tags (in frame) and 5-6 additional nucleotides in addition to
3' gene-specific portions, the
latter of which is typically about twenty to about twenty-five base pairs in
length. A Sal I restriction site
is introduced by the forward primer and a Sma I restriction site is introduced
by the reverse primer. The
ends ~f PCR products are cut with the corresponding restriction enzymes (i.
e., Sal I and Sma I) and the
products are gel-purified. The pIVEX 2.3-MCS vector is linearized using the
same restriction enzymes,
and the fragment with the correct sized fragment is isolated by gel-
purification. Purified PCR product is
ligated into the linearized pIVEX 2.3-MCS vector and E. coli cells transformed
for plasmid
amplification. The newly constructed expression vector is verified by
restriction mapping and used for
protein production.
[0284] E. coli lysate is reconstituted with 0.25 ml of Reconstitution Buffer,
the Reaction Mix is
reconstituted with 0.8 ml of Reconstitution Buffer; the Feeding Mix is
reconstituted with 10.5 ml of
Reconstitution Buffer; and the Energy Mix is reconstituted with 0.6 ml of
Reconstitution Buffer. 0.5 ml
of the Energy Mix was added to the Feeding Mix to obtain the Feeding Solution.
0.75 ml of Reaction
Mix, 50 ~1 of Energy Mix, and 10 ~g of the template DNA is added to the E.
coli lysate.
[0285] Using the reaction device (Roche Biochem), 1 ml of the Reaction
Solution is loaded into the
reaction compartment. The reaction device is turned upside-down and 10 ml of
the Feeding Solution is
loaded into the feeding compartment. All lids are closed and the reaction
device is loaded into the
RTS500 instrument. The instrument is run at 30°C for 24 hours with a
stir bar speed of 150 rpm. The
pIVEX 2.3 MCS vector includes a nucleotide sequence that encodes six
consecutive histidine amino
acids on the C-terminal end of the target polypeptide for the purpose of
protein purification. Target
polypeptide is purified by contacting the contents of reaction device with
resin modified with Niz+ ions.
Target polypeptide is eluted from the resin with a solution containing free
Ni2+ ions.
Example 13
Cellular Production of Tar e_g t PolY~eptides
[0286] Nucleic acids are cloned into DNA plasmids having phage recombination
cites and target
polypeptides are expressed therefrom in a variety of host cells. Alpha phage
genomic DNA contains
short sequences known as attP sites, and E. coli genomic DNA contains unique,
short sequences known
as attB sites. These regions share homology, allowing for integration of phage
DNA into E. coli via
directional, site-specific recombination using the phage protein Int and the
E. coli protein IHF.
Integration produces two new att sites, L and R, which flank the inserted
prophage DNA. Phage
excision from E. coli genomic DNA can also be accomplished using these two
proteins with the
addition of a second phage protein, Xis. DNA vectors have been produced where
the
161
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
integration/excision process is modified to allow for the directional
integration or excision of a target
DNA fragment into a backbone vector in a rapid in vitt~o reaction (GatewayT"'
Technology (Invitrogen,
Inc.)).
[0287] A first step is to transfer the nucleic acid insert into a shuttle
vector that contains attL sites
surrounding the negative selection gene, ccdB (e.g. pENTER vector, Invitrogen,
Inc.). This transfer
process is accomplished by digesting the nucleic acid from a DNA vector used
for sequencing, and to
ligate it into the multicloning site of the shuttle vector, which will place
it between the two attL sites
while removing the negative selection gene ccdB. A second method is to amplify
the nucleic acid by
the polymerise chain reaction (PCR) with primers containing attB sites. The
amplified fragment then is
integrated into the shuttle vector using Int and IHF. A third method is to
utilize a topoisomerase-
mediated process, in which the nucleic acid is amplified via PCR using gene-
specific primers with the
5' upstream primer containing an additional CACC sequence (e.g., TOPO~
expression kit (Invitrogen,
Inc.)). In conjunction with Topoisomerase I, the PCR amplified fragment can be
cloned into the shuttle
vector via the attL sites in the correct orientation.
[0288] Once the nucleic acid is transferred into the shuttle vector, it can be
cloned into an
expression vector having attR sites. Several vectors containing attR sites for
expression of target
polypeptide as a native polypeptide, IvT-fusion polypeptide, and C-fusion
polypeptides are commercially
available (e.g., pDEST (Invitrogen, Inc.)), and any vector can be converted
into an expression vector for
receiving a nucleic acid from the shuttle vector by introducing an insert
having an attR site flanked by
an antibiotic resistant gene for selection using the standard methods
described above. Transfer of the
nucleic acid from the shuttle vector is accomplished by directional
recombination using Int, IHF, and
Xis (LR clonase). Then the desired sequence can be transferred to an
expression vector by carrying out
a one hour incubation at room temperature with Int, IHF, and Xis, a ten minute
incubation at 37°C with
proteinase I~, transforming bacteria and allowing expression for one hour, and
then plating on selective
media. Generally, 90% cloning efficiency is achieved by this method. Examples
of expression vectors
are pDEST 14 bacterial expression vector with att7 promoter, pDEST 15
bacterial expression vector
with a T7 promoter and a N-terminal GST tag, pDEST 17 bacterial vector with a
T7 promoter and a N-
terminal polyhistidine affinity tag, and pDEST 12.2 mammalian expression
vector with a CMV
promoter and neo resistance gene. These expression vectors or others like them
are transformed or
transfected into cells for expression of the target polypeptide or polypeptide
variants. These expression
vectors are often transfected, for example, into murine-transformed a
adipocyte cell line 3T3-L1,
(ATCC), human embryonic kidney cell line 293, and rat cardiomyocyte cell line
H9C2.
[0289] Modifications may be made to the foregoing without departing from the
basic aspects of the
invention. Although the invention has been described in substantial detail
with reference to one or more
specific embodiments, those of skill in the art will recognize that changes
may be made to the
embodiments specifically disclosed in this application, yet these
modifications and improvements are
within the scope and spirit of the invention, as set forth in the claims which
follow. All publications or
162
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
patent documents cited in this specification are incorporated herein by
reference as if each such
publication or document was specifically and individually indicated to be
incorporated herein by
reference.
[0290] Citation of the above publications or documents is not intended as an
admission that any of
the foregoing is pertinent prior art, nor does it constitute any admission as
to the contents or date of
these publications or documents. U.S. patents and other publications
referenced herein are hereby
incorporated by reference.
Nucleotide and Amino Acid Sequence Examples
[0291] Table A includes information pertaining to the incident polymorphic
variant associated with
osteoarthritis identified herein. Public information pertaining to the
polymorphism and the genomic
sequence that includes the polymorphism are indicated. The genomic sequences
identified in Table A
may be accessed at the http address
http:l/www.ncbi.nlm.nih.gov/entrez/query.fcgi?CMD=search&DB=snp, for example,
by using the
publicly available SNP reference number (e.g., rs552). The chromosome position
refers to the position
of the SNP within NCBI's Genome Build 34, which may be accessed at the
following http address:
www.ncbi.nlm.nih.gov/mapview/map search.cgi?chr=hum chr.inf~query=. The
"Contig Position"
provided in Table A corresponds to a nucleotide position set forth in the
contig sequence (see "Contig
Accession No."), and designates the polymorphic site corresponding to the SNP
reference number. The
sequence containing the polymorphisms also may be referenced by the
"Nucleotide Accession No." set
forth in Table A. The "Sequence Identification" corresponds to cDNA sequence
that encodes associated
target polypeptides (e.g., Q96FX2). The position of the SNP within the cDNA
sequence is provided in
the "Sequence Position" column of Table A. If the SNP falls within an exon,
the corresponding amino
acid position (and amino acid change, if applicable) is provided as well. The
amino acid found to be
associated with OA is in bold. Also, the allelic variation at the polymorphic
site and the allelic variant
identified as associated with osteoarthritis is specified in Table A. All
nucleotide and polypeptide
sequences referenced and accessed by the parameters set forth in Table A are
incorporated herein by
reference. Genomic nucleotide sequences for KLAA0296, Ch~ona 4, Chnom 6, ELP3,
LRCHI, SNWl and
ERG regions are set forth in SEQ ID NO: 1-7, respectively. A polymorphism in
Table A designated by
"AA" is present in the genomic nucleotide sequence of SEQ ID NO: 28, which
follows Table A.
163
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
ti
O
a a 1-e~I-~-U I-c~a c~c~c~I-a a a c~a U aa a e~I-c~a 1-
aa
c~c~ E'Qv E e~v Q c~c~c~c~E'Q Q Q c~Q v Q Q a ~ Q Q E'
a a ~ ~~,~ ~ ~,~ a ~ ~ a~ ~ ~ ~,a ~ ~ ~~ C~C~U CJC7U
, , ,
LL ~ ~~ I~~ ~ ~ tLtiu..u.u.~ L m.Ii~ ~ ~~ ~ ~ u.~ ~ t~
a
o r
00N ~ O I~N CO~ N ~ ~ ~ M I~(ON 07~ O d N ON N O ~ O M
~ ~ ~ M~ ~ ~ O ~O ~ ~ _O O O p N ~ c0
V ~ .d ~ st~ V C ~ O ~ O M
' O
r ~ InMO O M lf~~ N ~ N Wd"(OC4~ O)LnC COC C C ~ r 'd'
'
'
d
d' CO r O O ~ J N O
~ r N
fn N ~- r r ~ r O G(7 00O N p
z U
r J
v ~ z ~ Z~ ~ L X W d ~ ~ ~a ~ z D ~ c Xc c c ~ Q
U
O U u o U~ m uJ; r N X~ d U U J r o Uo 0 0 ~ ~ ~ ~'
_.
I-
N . U ~ _ ~ ' ~ U J V OJ (nJ N = J C V~ C C a Z U ~
LL '
O Y ~ J ( - L
j O cn n a U L
J Y t~~ ~ U . ~
O
~
O M OM ~ p
~ ~ W
;=
a 0 Q> f'> c
O >
a ..
Cc c C
OO O O O = U
,
VO U L O ~ TT >,>,~ ~ ~ L U U U U ~ O
C ~ .~7Ln>,~C C C ~'7 >,~ ~ ~C C ~ C C C '~C~ C C ~ ~ p)
O ~ ~ ~
i C
d ~ ~ OO O O N ' ~ ' M 'O O ' O O O O OO O O ~
= A (0 N ~ f9 (B CO ~
_L~ _L_L_LO)_L~ CDO)y N L _
~ r L
L ~ C ~~ ~ ~ s_C L r LC C L C C C ~ C~ ~ ~ ~ ' N C
E '- ._ ._
c o o a ' ~ o ~ ~ ~ ~ c c
C_
y a - o
o
UU U U
ODlf~h r1~O)M I~N N r OptnM N I~M ~1' r [v[vpp(O
ODN 'V'N ~O O M ' O
'
~ r r d O rN r ~ Lnlf7O O 0000~ r
~ tt7d'0000NO CON COf~f0N O I~Inr O M I~M Ln I
n O O r
p O ~'M N N'V'd'N N 'd'M a0~ 'd'd'M M O ltdOON MO N N CflO 00O
N
a
N N O O O OO O O O O r O M OO O O O ~-O O OO O O O O ~ O
p
c~ I I I I II I I I I I I r"II I I 1 t I ~ I~ ~ O I I I I
O I
Z
3
V a
Z
a
x Z Z Z ZZ Z Z Z Z Z Z ZZ Z Z Z Z Z Z Z Z X Z
C M O) N N~ON O I~M O N N~ O p r d'00CO ~ I~
(O O ~ d' N o1~ O d'I~~ h
0 M M 00(~c0a0 _ O t~
r ~N N ~'~ ~ ~ ~ ~ ON O M ~ 00r COO~ M O O
w ~ ~ l C N O O
(O O r M O ~1'r ~ N r r ~i'I~ p O CO
O N u7 N COh 00 ~ r (p;jr M_ ~ ~ ~ 0 ~ d ~
N I~N o0 O o0 ~
.d C ~t ~ N c O~ ~ ~ d
. I~.~ d . CO'V' lO ~ 0 r r M InCO
p ~ d ~.M r rO r O 0000M LI~ O ,d.
r GOti'N r 'V' d' ~ ~ 'd'I~h t~
r p N NN N r r r O) ~.N
COr 0000r r T (pC4C4(Or ~tf'~i'O d'd'~Yd'Cf7Ct7~ ~ O O O p
'd''~d'~t r r r r r r r r rr r r r r r r
~N ~Y~ M MM M ~t ~ ~ ~ ~ ~1' d' d'd~ ~ if
M ~ M M M M .~ d~ vt~te!-d'tYd' ~ ~
I II I M ,~ M M M M M
V I I o~o~N ooI M I 1 I I ~M M I M M M M MM I I M M 1
~' M O r rG(7COO I ~ M M 0 II I I I I I II I I
O
, . ~ t L d O O M O O O O d'M M M ln~ O N
-, 1~a. Inlf7'~I'O d' ' ~ ~ ~ lI~
V
~ O r r r In (OC4 CflC4COCflCflI~ 07O)r CO
a p r rr 'ctM M O ~ ~ ~ r
O
N Cp r rr N N ~ M M M W NtnInN I~f~1~I~c1'd'Cp(pM M (p(p
~ '
'
N I II I N ~ M M M h N~ lf~N ~ ~ ~ InCO(Or r COCOr r
I I I I I I I I I I I I II I I
C MI~ N N~ ~ NIN ~ N N N NI~ ~ I I I . I
I
p tn ~nvim u~vu ~ M N ~ ~ ~ ~N ~ ~ ~ ~ d.~.
N N ~
= = Z Z Z Z _ = Z = Z Z ZZ Z Z ~ ~n
V 2 Z ZZ 2 = Z Z Z Z Z Z
C M ~ COCOOOtn0~~ r10 1~O d'O ~jO CODM M t~ r '~i'r M ~
CO O LOO 00In O r (O Op M N n1~[~O O M
~ te O M O p
p ~ f~OM COM ~ V CON ~ CO.d~ M a.~ O O Or r COf~CO ~
CO r OInh O 0 I~ . C O
~ 0 M N I~ 07O CO1~N
I~N 00ON N I~p O O tn'Wt~O ~ CVr O N M d' ~ 0
0
V N C . M M ~ ~N r CO~ 1~~
N O O I~Op
a c ~ ~ ~ccr l N ~ N O ~ NO r O N 0 C
O i t7 0 (OC4O h i~
r r 'd'~r M 00N M 'd'~ ~ 'd'~ N r ~ ~ ~ ~ ~r M '~f''~I'~l'~ Cfl
O
O
O M r O OW CO
N Nr r N r r r r r NN N M M M M M d''d'd'd'd'~1'd'~f'
y"
O
t
~
U
O ~ 0 (j M ~ W N O M
O N 00 O M 00 O N 00M r
N O r Mr r'~ 'd'~ O CONM O)0 t f~ d' r N Cfl
O O h 0 r h 00O 1~
M
I~ COIn N ~i'COCOO ~ CO O ~ M O N
tnO N d.N M ~ O O a0 N o000 I~ d'N
N N O O ~ ~ ~ ~ r,rr p ~ d.I~M
M MM
s ~ ~ ~ ~ O O ~ O M I~O ~ N l(7l~~ ~ ~ ~ ~ 0 0
- N O O
r r r r r r r r r r N
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
i-Q C~C~I-Q U U QU Q U Q f-U U C~I-C~Q C~C~I- CJU I-I-C~QU C7U C~U
C7Q Q Q Q U E'UU U C~~ U E'Q ~ C7 C7C7U C'7F U Q Q C7U Q ~ Q C~
C7CO~ C7C U a~ ~ ~ a U ~ U ~ U a~ a ~ ~ ~ ~ ~ C C ~
, 7 C a U ( aO O U C
~ m ~ ~ ~ ~ ~ ~ ~~ ~ ~ ~ ~ ~ ~ ~ ~ ~~ m ~ ~ rl~ ~ m ~ rL~ ~
N ~ ~ ~ O N r O~ O d'~ O o0'd'M O NO N M M ~ r M 'd~ V ~
O o0 07O o N
f~ 1~COd)CO 00M M 00O M M ~ C O ~ ~ M r 0
M N M ~ N N c00 _ 0 Cr ~ M M
N C N~ 7 ( ~ ~ C ~ N C N ~ N N C'd~ a a G
O 0 O 0 0
r N N
(pN N m ~ MM J r N r m ~ NaCO Z ~ r O~O~
tt7 d ~ N 11 m Q M ON ~ r ~ CO r
r
. p 0 N M Z Z a p I-a cC7c ~ = ~ m (n J c~ M M
. U
. ~ . m o o
tiu.z ~ Q C7m > ~-7-~ m U z J oJ o U '- d. O O
~ ~ p _
Q U U Y ~ ~ U~ ~ d ~ V ~ cp c Cn~ ~ ~ ~ N.LucU ~ U U c
V
J Z cn ~ O O O
x
n
N N
O
M
cn Z
c c
O O
c
c c o L U U ~ L U L U
C C C C C ~ C C C~ C C C C 3 C C CC = V C 3 7 C C C ~ C C
~
~ O
O O O O O ' O p OO O O O O U ' O O OO O C O ' ' O O O O~ O O
L L L L L N L L L L L ~ N ~ N
_ _ _ _ _ _ _L L _L~_L~ O L _L_L_L~I-_L_LN
~1-~~1-~ C C .1uC rtu O C .1-n L L ~1-iE L L L
C C C C C L C C C C C C L C C C O C C G C ~ C C L
~ .~ . _ ~. _ ~ _ _O _O
C C E ~ C C E E ~ C E
O C
O O U
U U
f~r LOCO(O1~O V'MI~N I~I~M 'd'COO r d' d'f0 N 00(O~ ~h 00M (Oa0
00h ~ I~1~M O ~i't!7h '
r M M O O O COO CO <fr LOIwO cf'M O N r N
O O r M M r ~ ~ IwN O O O I~r tn00O M (O ~
-
r / r N N V O 1~lf7r
O)M r ~ ~ ~ r O NCON N N N M N CO00Or O (Ol(7O d'V'N V OCO000000N
r ~ O r c-O O O Lnr O O O O O O O r CO C O r O r O r O CQ7r [~[~C
O N O O O O O O rO O O O O O O O O OO O O O O O O O O OO O r r O
I I I I I I I I II I I I I I I I cI c I I I c
~ ~ ~ ~ ~ ~ ~ ~ ~~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~I~I~I~I ~I~I~I~Ic
Z X Z Z Z Z Z Z ZZ Z Z Z Z Z Z Z Z Z Z Z Z Z Z Z Z X Z Z Z
O O d'~ ~ 00N Lf~ON O 'd'O MInCO d' 'V' COO 00ODr O
QO In M OCON (pO O ~ N ' ~' ' O O M
'
M O M ~ ~ 00O O Wd'W.r I~W r ) I~~ N00tn O d N N r N Od ~ r
'd' N ~ ~r r O) CO ~f'O M OO O
O O ~ r M N
COM O N ~ O ~ ON ~OI'000000~I7LO O00N I~ I~ r r N CON N
~ r ~~ ~ ~ ~ N M L~~ N ~d 0 ue N O ~ ~ ' O
~ C O I~~ O ~ O o a c V M ' d c ~ c N ~N ~ ~ COM
O O O - ' ' - - ~ ~
M 00r r r ~ r M ~1~InCOCOInLnInN O re/'O 00 CO~ N 00O MN '~lf~~Or
r r N Ntfr r r
rr N N r Intn00Mr ~-
M M O r r r ~,,~ MM M M M r r d,COLn(Or r r r r r r rr r r r r '
r r r r r r r
r r rr r r r r
~t'd'~h'~h~ N ~I'd''d' M d'd'M M M M M M M M MM M M M M
M M M M M M MIO Mc~M M M M op~ M M MI I I I I I I I II I I I I
I I I I I I N ~ II I I I I ~ I I I IN o0 O o~d''d'~t'V'rr O tc>opM
00O ~ O O O 'd'~ O)O ~ ~ ~ O ~'O CON N00I~ 00 O O O O O ~M O t0daN
'
00I~~ M 0000O p ,d.~ O)O O d.O .~.~ N COO lf~ CO r (OCOCO(OM~l'I~(OI~r
'
Ino0d N N N ~ O ~~ 000000~ O ~ O 0000~l'O) d' O COCOCOCOOO O O O r
O M M M ~ tnIn
M o0M N N r N N N N rr r r r r
ml~IN N N N MII-~I~IN N N ~II-~I~'IN II I I I I I I I II I I I I
I I I I ~pZ OO I I I Z I Or N M M 'V'etd"d'tnInc0CO!wop
U O N N ~ N O UU ~ ~ ~ O O ~ ~ ~~ ~ ~ ~ N ~
Z Z = ZZ ~ U U
= = Z Z = 2 Z = Z Z = =Z Z Z Z Z Z Z Z ZZ Z Z Z Z
r I~~ 0~0~ W N M ODO ~ l~jO I~O ~ ~ d'~hf~(Od' ~ I~(OO)00I~N ttN M
CONO M r 1'OCOr (pO I~N r N Oo0~ O O Op
O)00Lnr ~ 00L(]Q)M O r
~(7M N 1~00.0d.ON r M tI'I~.M M M CMOONO~ t~ ~ O ~ ~-o N N~ O ~ r ~
s C
O (~r'ODCYr CO O 1~M r M COM O f~O
CO
N N O ~ ~pO O ~tO~tf~I~O M 'd'tnN M o0O ~ O)V'O
M
M Mj r O N NM t~N ~ N ~ ~ ~ O~ ~ O ~
r r r r ~ r r r ~ ~ O V ~ C C
O ' D O
InLL~Intn~ ~f2COCOCOCOCO(OCO(O(OC4f~00Or N M M M 'd''d'd''d'~t0(OCO1~00
rr r r r r r r r r rr r r r r
~'M OOIn0p~Or N d'O O f~ N ~ MO r M r O r r O OO d' r
O M '- O ~ N
O 00O (OV'~ M O r O r ~ N~ M N .~.M r (pr MO r ~ ~ O
O ~ d' ' ' ' ~ O M '-O 00O N O)N I~
O '~1InWitV r 00r Lt~ 1~ ~p r 00 tt~
~
N M M cN-O N _ CMOQ ~ O ~ ~ ~ ~ ~ ~
'd'
O a ~ ~ 00 ~ '- O O ~ ~ ~M ~ 0 0
~ ~ N N ~ ~'M 0 ~ O O
r r ~- ~-y - r r r r N r r r r Or r M O r
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
UII-IUIUIQIC71U
E' ~ U C~ Q
U U U Ula a U
u-I~IL~I~I~Iu.lu..
N~~ N c
~ N ~ O N ~
r
m O ~ ~ 11
C7 Z
C~
Q ~' D O ~ a 1~1
a W i.u
U ~ c~
~
C7 C C ~ C
N
O O
O O O O O
cE ~ c
~ E ~ U
rInW Ctr Op
00tt~r N ChO O)
rr N ~ O N r ~O
OO O O O O O
~I~I~I~ ~I~I~I
zz z z z z
M ~ ~
MO ~ '-0~0000~ ~'
t!7O W O ~
M
~
'dCOD~ N ~7r N I~ ~,
"i
~N N ~
b
rDO00W O r O U
~t' ' '
~ '
d l d'd V'~t~
pp ~
bn
W
II I I I I l
N~ a~o~OBN t~~d
l(7r r COCOI~I~
~
Wit'~c7~ (flCOCOCOO ~ V ~ (.t.~
rr r r r r r
rr r r r r ~
r _
II I I I I U
I ~
OO O r r r N ~ t~
rN N N N N N ' L
U N O O
Z= Z 2 Z Z Z
y U y
~(flr 1~N O OO
OM M O M ~ CO~ ;~ ~ ;~ U
O
ON M tt~00N r .~,
'd'00a0COa0d'O ~ ~
.~ p ~-~ C
rM M N M d'M
f~
~
: N
~
WO O r r r N ~ '
rN N N N N N ~
a~
U
U
tf7 bl7 O ~" U O
N N f~ '
:
U
y
,
N E ~ M M U ~ C%~ ~
00r ~ C O r 1~~
O D ~
d'
r n
n
n
a ,.~, a ,.T' a
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
AA ~enomic sequence (SEQ ID NO: 28)
TCATTAGCTTTTTCAGTTTTTCACATTCCTGATACAGACGTAGGAGTGCTCGTATTTTGGATTTTGCATCCAACTTGTA
CTTAGTT
TTAAATTCTGCACA[A/G]AAATGTTCCACTAACTTTTCATCGAAGTTTTTTCCTCCTAAGAAAGGATCAAAAGCTGTT
CCCAGTA
CCTAATTTGGTTGAAACAACAAATAGTCTGGTT
[0292] Following are genomic nucleotide sequences for a KIAA0296 region (SEQ
ID NO: 1), a
ch~om 4 region (SEQ ID NO: 2), a ch~~om 6 region (SEQ ID NO: 3), a ELP3 region
(SEQ ID NO: 4), a
LRCHI region (SEQ ID NO: 5), a SNWI region (SEQ ID NO: 6), and a ERG region
(SEQ ID NO: 7).
The following nucleotide representations are used throughout: "A" or "a" is
adenosine, adenine, or
adenylic acid; "C" or "c" is cytidine, cytosine, or cytidylic acid; "G" or "g"
is guanosine, guanine, or
guanylic acid; "T" or "t" is thymidine, thymine, or thymidylic acid; and "I"
or "i" is inosine,
hypoxanthine, or inosinic acid. Exons are indicated in italicized lower case
type, introns are depicted in
normal text lower case type, and polymorphic sites are depicted in bold upper
case type. SNPs are
designated by the following convention: "R" represents A or G, "M" represents
A or C; "W" represents A
or T; "Y" represents C or T; "S" represents C or G; "K" represents G or T; "V"
represents A, C or G; "H"
represents A, C, or T; "D" represents A, G, or T; "B" represents C, G, or T;
and "N" represents A, G, C,
or T.
KIAA0296 ~enomic sequence~S~ ID NO' 1)
>16:31076951-31174000
1 ccccaccccccaacagctgcacagtctggagcgaatatacacgcccaccacccacacacc
61 caagacccaatacacttttttaaactttatttttacttctatttatttatttttaattat
121 tttttaaaaatctaattagagatgaggtcttaggctgggcacagtggctcatgcctgtaa
181 ccccagcacttcgggaggccgaggcaggcagatcacgaggcgggaggatcacgaggtcag
241 gagttcRagaccagcctggccaatatggtgaaaccccatctctgctaaaaatacaaaaat
301 gagctgggcgcggtggtgtgcacctgtaatctcagctacttgggaggctgaggcagaatt
361 gtttgaactcaggaggcggatgctgcagtgagctgagatcgtgccactgcactccagtct
421 gggagacagagcgagactacgtctcaaaacaaacaaacaaacaaacaacaacaacaaaaa
481 cagagataaggtcttggcatgttgcccaggctggtctcaagtcctgggctcaaaggattc
541 tcctgcctcagcctcccaaagtgctaggattacaggcgtgaaccactgcacccaccctac
601 ttttttttttttttttttttatacaggatctcactctgtcacccgggctggagtgcagtg
661 gcaagatcactgctgactgtacccttgacctcagggactcaagtgatcctcctgcctcag
721 cctcctgagtagctgggactacaggagagcgccagcacacctgggtaattaagatttttt
781 ttgtagagacagacgctatgttgcccaggctgctctcgaactcctggcttcaagtgatac
841 acccttggcctcctaaagtgbtgggatcacaggcatgagccactgcacctagcctaatat
901 agttaatatccccgtcaaggctgctcagagggcctgagaggaacaaagggctcagctctg
961 gagagctccacccccagcgccaatctctctaaatggcctctttcctctccatattccacc
1021acaaggcttggagtccagcttcctgtgaccttaagtcaccattccaaagccctgcgatct
1081cacccagagaccacaagtgaaataatattataatcctgagaagtttagtggaccaagatg
1141gcatgccatcaagacgctgagaaacaaagaggaagatgggaccagggggcccagaagacg
1201ctggaacccacagtattaaaagctcagagaggctgggcacagtggctcacacctgtaatc
1261ccagcactttgggaggccaaggtgggtggatcacttgagcccaggggtttgagaacagcc
1321tgggcaacatggcgaaacccagtctctaccaaaaaatatacaaaaattagccaggcatgg
1381tggtgcgtgccttagtaccagctacttgggaggctgaggcaggaggattgactgaacctg
1441agagcacaccactgcactccagcctggatgacagaaccagacctgacctcaaagagaaga
1501aaaaaaaaaaaaaaaaaaagcccagaggggagggYaccctcaacagttttccagcccctt
1561ccacatccttcctaacctcacttgatagtgttcaagtcctaccttaggcaaggcagaaat
1621tataggaccaagccgccaaatggggaaattgagtcccagagagaagtaatgcattattta
1681agatcccatgcaggactatgagtcaggggtccaagagcccttccaccgtgtgccactcag
1741agacacagagtaggagggggaagggggtcgggtggcaggggacaaaagatgcaggaggca
1801agcagcagtgactgaagaggcagaggctgacatgaaagacccaggagcagagaatctttc
1861cttatcatctccaggggacaccactgggcagggcttggcctccggaaaaaccctgcattc
1921cctctgtgggttcatcagggcaccactctcctactagctgggttttttttttttgttttg
1981ttttgtttttgagacagagtcttactctgtcacctaggctggagtgcaatggcgtgatct
2041cagctcactgtaacctccacctcccatgttcaagcaatcctcctgtctcagcctcccaag
2101tagctgggattacaggcacctgccatcatgcctggctaatttttgtatgtttgtagagac
167
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
2161agggtttcgccatgttggccaggctggtctccaactcctggcctcaggtgatctgcctgc
2221ctcagcctcccaaagtgctgggattacaggcatgagccaccacaccctgcctgagctggg
2281ttttaacaggaagaggagaagagccaaaactcctcacatagaatcacacagcacttgaca
2341gtttccaacctcatcatcactgaagtttagagcagccgatacccaYaaagatgatctccc
2401catccccctacagttacccactgtgcagagggagatccacacttagagacaggaagcgat
2461ttccagaagtccatagcaactcagtcccaggaatctaggtttcctgaccagggcatagca
2521gaaagggtccattcctttccttgcttgtaccttcacagaagcttcctggacagagccctg
2581gggtccaggagacctgttattcattcccggctatgctgagacttgctgagtgaccttggg
2641gactccttctagagaatataagttccacgagtgcaggaatttttgtctattagtccttga
2701tgtatctccagccctagaacagtgtttggcccatactatgtgcccaaaaaatatccatta
2761aatgactgaatgttgctgtgcatggtggtgcatgcctgtaatcccagcactttgggaagc
2821tgaggcagaaggattgcttaagcccaggagttagagaccagcttggacaacatagtgaga
2881ccgcatctcgtaaaaatttttaaaaataaaaaatgagtgaatatctagatagccaggatt
2941agagaagtgtcacagtcagaaagcctgaagcctaaagaagaccaaggaaccaggggcttt
3001atcctcagatacatgaaagcctgaaattctgtccacaagtatttatagagggcccgtaat
3061gttcttggtactgggctaggaactccccagattcagttaagaacaaagtcattacctggc
3121ctcagatgcaaggcaggggctggggggtgtgagtggcagggaggcagcgtgatcaataca
3181aacacttttcttagcctgagctgccctgacatggtctgacggctcacaaggtggtgagtg
3241cagccgggctgcagtgttcaaggagggcgccggctggccgcccacctgtcagaggctgcg
3301ccagaaggatgcggaagaagagatttctgccttggctgaggtcacttcccacccccagat
3361tccctgcccacacaaccctgcaattttctgacgctgacgactcggatcctattatttccc
3421gattttcaaggtcccatgatgctgacagccccaaatgctaagtcgtcagtccgcccacgc
3481cctggacccgaaagcaataaaggcgaggtcagcaagggtcctaccacccactgcctcgaa
3541aggcctctgggggtggtcggcgcgcccctccccacctcgcgggggccgtgtgggcgtcgc
3601tcggtcgttggggtgccggggacgtcgtgatgagaacggcgtcccagagacggcggtgac
3661agagccgggacacgtgacagtcacagggtcacattctgcggtccacgagtttgggaccgg
3721gctggtcacgtgacgcggtgggggcaccatggggtgatgtgagatgcgggtgtctcggat
3781tacgtacaaatgacgtattcctaccccttttggcaaccagatttccgttggaagatgcaa
3841cggttccggtgacggtagcaagttctcgcgtccaggcatctccgcttccgctcggggcgc
3901aacaacttccgactccaccttcccagcctcgggcaaggaagagacgcgaccatgtgcgca
3961tgccccgaatttatcacggaggggcggggctgaggctgcgggagctggagcggggaagaa
4021aagggaattccaacctgtggaaccttggggggtccccggggtcggcgccttcccattgac
4081tgtgggcggtgcaagggacggagcctctggcggctcgtgggggtgttggggtccgcaggg
4141ggagggaggggagtgtcagagtgtgagcggggtacgggaattccaaatttgagggcctcc
4201cggctctggcgccggggagggagagctcaggccgccatgcgggacaggacccacgagctg
4261agacaggtgagacgccagggcagcggggatggggacgggcggacgaactggaacgcagga
4321cttctggtcttcgggatagggaggggtggctgatggccaggaaggaaagtcccggaagcc
4381tgtgggtcctgcggggtaagagccgcagcgaaacggtggtgccaatgactccgggcctgg
4441cagggggatgacagctcggacgaagaggacaaggagcgggtcgcgctggtggtgcacccg
4501ggcacggcacggctggggagcccggacgaggagttcttccacaaggtaaggggctggggt
4561ctccgcctggattcgcgagggtgtaggaggacccgaggagtagcgtggtctggagtaccc
4621catatctctttcagccctctcggtcaccctccccaggtccggacaattcggcagactatt
4681gtcaaactggggaataaagtccaggagttggagaaacagcaggtcaccatcctggccacg
4741ccccttcccgaggagagtgagtgaaaccccggctgcagggcgcatgctccgccccaggga
4801ttgtgggggttgtagttccacgcaggtggtggccagagtggtttgttgaggtgggggctg
4861ctgtttgggagtcttggccttctcttattcaggcatgaagcaggagctgcagaacctgcg
4921cgatgagatcaaacagctggggagggagatccgcctgcagctgaagggtgagctcctggg
4981acctcagacagatccttccctctgatcctgccctgttgttggtatatctggggagtgtgt
5041ggcccagagaagccagtgatatatccaggtcacacagcaggcctgggtctagcatctgtc
5101tcctggcctccaggccattgtactctccacagcacaagtccgcctctcaggttcttttat
5161ttacaatgaaaccatttacttacacagttatcgctgcccactgggcattctttgggcagg
5221gagatggagttttgttaggtggcctctgcatacctatgggaactcagtgatgtaatgcaa
5281agaaaaataaacttactttctcctcttagaggctcagccttagtcattttatgataaatt
5341atatttccctaaaaatcctatggagacaagtacccccaatacccctgtgtcttcccacag
5401ccatagagccccagaaggaggaagctgatgagaactataactccgtcaacacaagaatga
5461gaaaaacccaggtgggttttttttctcagaaatgaggacatttcagcaaatgtttcatga
5521agtattagatgacaggtgtatgaaggaagggcctgcagagatcatggagtccaattggat
5581gacttttccaaatggggaaactgagctcagagagagaaagaacttgctcaaggtcaggaa
5641gccaggtctcctgatgctcagtccggttataacaccctgctttattttcttccattcaat
5701aggaagttactgtgaccccagacaagacctagtcttggctgtgggacacatgttttcttt
5761tctttttttgcctcagcctcctgaatagctgggattacaggcggacacccccatgcccag
5821ctaatttttgtagttttagtagagactgggcttcaccatgttggccaggctggtttcgaa
5881ctcctgacctcaggtgaccctcctgcctcggcctcccaaagtgctgaattacaggcgtga
5941gccaccatgcccagctgggacacatgttttctgggagtcaagatgaggagttagggttca
6001ataggggataaagacattactcacgtgggacctggtggctaacggcgctgcccagggaag
6061gagagtgagaagtcataaatgactggcaggtttcctatctatgtgacagggacatcctta
6121gtcccacaggtggaattcaagaagtcaggaagaggaacttccttggggcaacactgaaga
6181ggaactcccctggtgtgatatcttatttttttaattattattatttttttgagatggagt
6241ctcactctgtccctcaggctggagtacagtggcacaatcttggctcactgcaacctccac
6301ctccttcaagcgattctcctgcctcagcctcacgagtagctgggattacaggtgtgcacc
6361accacacctggctaattttttatatttttggtagaaatgaggtttcaccttgttggccag
1fR
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
6421 gctggtctcg aactcctgaY ctcaactgat ccacctgctt tggcctcgca aagtgctatg
6481 attataggca tgagccaccg cgcgcggccc ctggggtgat atcttagtaa ggagatttgc
6541 agtgatctga ctggccctct ctgggtcccc agtgaggagg ataccaggag gtcagggttg
6601 gagtagttgg gcccagggct cagcagggac cccagattga agatggagca gcttgggcat
6661 cttggaaggg tgaagctgga accaggaaag cagatgtatc tctggaaaag gaactccaag
6721 gaatgagcat atttaaggcc tcagaagaag gggcaaggca gagcagatgc cccagaacca
6781 gtgtttctgg ggaagcctgt ggtggtgatt ggcatgagtg gttgagggtc catgtgggcc
6841 tgttgcacct gtttcgccca ggcaacatgt tcatctctag gcgtaggagc tgtggtgtag
6901 gcagcgaggt tggcattcag caagcattca gcagttacat attgggtgcc tactgtgtgc
6961 cagacctttt tggaactgtt taggatacag cagtgaacca gtgatccctg tcctcatgga
7021 acttcccttc tggtgtagac aatcaccata ataaataagt gaattattta gaacataata
7081 agcattaagg aaaaaagagc aggggaagag ggactaagca tgctggagga ggtagagttg
7141 cagttgaaag caggtggagg aagcttcatt cagaaggtaa catctgaaca agagacttaa
7201 aggtgtttgc tgggaatgag cattctaggt agaaggaaaa gtgaatgcaa aggcttaagc
7261 tgagagtgtg ctttgtctag ggaggggtaa ggagaccagt gtggatgggc agaggaaggg
7321 aacagtaaga ggaagtaaga tcagagaggt catgggagaa ggagagatca tagagggcta
7381 gccaggcacc gtggctcacg cctgtaatcc cagcactttg gaggctgagg tgggaggatt
7441 ggttgagccc aggagtttga gaccagcctg ggcaatatag tgagaccccc cccccttttt
7501 tttttttcct ttgagacagg gtctcactct gttgcccagg ctggagtaca gtggtgccat
7561 ctctgctcac tgcaacctcc gcctcctggg ttcaagccat tctcttgctt cagcctccca
7621 agtagctggg actacaggcg cccaccactg caccaagcta atttctgtac ttttagtaga
7681 gatggggttt caccacgttg gccaggctgg tcttgacctc ctgacctcag gtgatccacc
7741 tgcctcagcc tcccaaagtg ctgggatcac aggcatgagc caccgtgccc ggccaaccct
7801 gtctctatta aaaataaaaa taggccaggt gcagtggctc acgcctgtaa tggaggccga
7861 ggcaggtgga tcacaaggtc aagagatcaa gaccatcctg gccaacatgg tgaaacccca
7921 tctctactaa aaatacaaaa attagccgtg cgtggtggcg cgtgcctgta gtcccagcta
7981 ctcgggaggc tgaggcaaga gaattgcttg aacccgggag gccaaggttg cagtgagccg
8041 agattgtgcc actgcactcc agcctgggca acaagagtga aactctgtct caaaaaacaa
8101 ataataaata aataaataaa taaataaata aataaataaa taaaaaagat catggaggac
8161 cacataggcc tgataagggc tttggctttt agtctaagag aaatggggga gcctgtcaag
8221 gtcatcacaa ggtggttaag gtggcagatc ccgcataaga gctcatgcta tttgctcact
8281 gtactatggg gttgccgagg caccgaccgg gcagggatcc tcccaggggc actcagccta
8341 tattcttcat ctttagcatg gggtcctgtc ccagcaattc gtggagctca tcaacaagtg
8401 caattcaatg cagtccgaat accgggagaa gaacgtggag cggattcgga ggcagctgaa
8461 gatcagtgag ttgtgcatgc ccagcctggc ccgcaggggc aggtaatccc aacccaaccc
8521 tgagcctggc cttttccttc acagccaatg ctgggatggt gtctgatgag gagttggagc
8581 agatgctgga cagtgggcaa agcgaggtgt ttgtgtccaa tgtgagtggc cacagccagc
8641 ccctctctgc tgtgcctccc atcccctctg agtcctgtcc gtttctcgac ctcctgggct
8701 caggtgatcc tcctgcctca gcctcccgag tagctgggac tataggtgca agccactgca
8761 ccccgcttgc tgtggccctt tctgattaag ggcaccctga ggcctctaag ggaattaatt
8821 agcctgcctg gagtcaccca tcagattcca ggctgagggc tccccagaag ctcaacagga
8881 gtttctgacc tgctgtcggt ctccctgtga acagttgccc cactcctgtc caccccccag
8941 atcctgaagg acacgcaggt gactcgacag gccttaaatg agatctcggc ccggcacagt
9001 gagatccagc agcttgaacg cagtattcgt gagctgcacg acatattcac ttttctggct
9061 accgaagtgg agatgcaggt gggtgccccg cgcagcccca gacgtgagac caggctcagt
9121 ccaaactgcc agMctcccgc caYccttaga ttctctccct gaggcttttg tgtcttccag
9181 gtttggccat gcccccagat tggtgcttat tcctatcctt agctgtaccc cgagaatggc
9241 acctgcctct gctgctacac agatgcccac tcccttctgc atagcaccct gccccctctc
9301 caaaacttga gcctgcccag gtctggcccc agccctcact ccccctccac taacagcatc
9361 cacccttata cctctcagag gtccagtcag agttgcccta gaggggctgc ctcctaacat
9421 ctgtacaagg ctggggtggg ggcggcgttc ccctggccct ggttgtgagt Wgagttgagc
9481 ttccagccct gtcctggagg agctggcctc agtcatgcta cagccaatgc ccttttgcag
9541 ctgagactta caggaaagag atctcattca gtaggagtac tgagacctga ggctggtggt
9601 gccaggagga ggcagggata gggagggctt tgcagcagct gtagataggc ctggaagaat
9661 gggtaaattc agacagattt gtgaaggcac agttcaccat ctgtgaaagg tatgagccat
9721 ttgaggccct tagctccaag ctaccactgc agatagaggt tgtatgggat aagtgagcag
9781 gggacaaggg actacatgat agaaggggcc tggaagccat ccccaaggag tctgaacttt
9841 tgtcagatca agtcttgccc ttgtctttgt tagtgcaatt tttttttcct gccaggaatg
9901 ttcttcagtc atctggggtg gggtgggcaa aggcatcctt acctccctga accaccccat
9961 cctctgagca gggggagatg atcaatcgga ttgagaagaa catcctgagc tcagcggact
10021 acgtggaacg tgggcaggag cacgtcaaga cggccctgga gaaccagaag aaggcgagga
10081 aggtgagcct cccaggcccg gccactgccc caggcaccct gtgtgacttc cctgaccccc
10141 tcctctccca cagaagaaag tcttgattgc catctgtgtg tccatcaccg tcgtcctcct
10201 agcagtcatc attggcgtca cagtggttgg ataatgtcgc acattgttgg tgagatgttg
10261 tgggctgccc cctggcctgc cccagccctg gccccagccc tccctcctcc ctcagaccct
10321 gttctccctc ctttccttac aggcactagg agcaccagga acccagggcc tggccttctc
10381 tcccagcagc ctggggggca gggcagagcc tccagtcgga ccccttcctc acactggccc
10441 ctatgcagaa gggcagacag ttcttctggg gttggcagct gctcattcat gatggcctcc
10501 tccttcaggc ctcaatgcct gggggaggcc tgcactgtcc tgattggccg ggacacacgg
10561 ttttgtaaaa aattaaaaaa caaaaaaaga gcatagaaag ccctgtgcac gtgtgttcct
10621 ggaagggctg gcccaaggct tccgggcatc caacctcctt acctcctgga cgtccccagg
1H9
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
10681 gccaggtctg gccctggctg ctcaggtcaa actgccaggg gtgctgtgcc cacagcaggc
10741 tggttctgcc tttctgcacc cccataggaa tgggtgggca gggaggggta acaccggcat
10801 ctagctcctg gctcagtact gtccccggga aaggaccact gtgagtatct gtcttggaaa
10861 tgatgaggct gaccaggcca ggctgggacg caggtgagat gggggtttgg gtggcatcag
10921 tgggccttct tgtggcccag aggaagaggc accatgaaaa aatgcctaat tgaggctgtc
10981 actttggatg cagtggatag ggatggtctg gtttcagcag ggatgacatt ggagtgggat
11041 gttaagctgg ggaagaggtt gccagtcaga aagcacagga ggccgggccc tgtgaccaac
11101 aaaagcatca tcttttacat aagcgtttag gcagggtgtg gtggctcgca ccagtaatcc
11161 cagcactttg ggaggcccag gcaggaggat ctcttgagcc caggagcttg agaccagcct
11221 aggcaggatg gggcaacctc ttctctttag agaataataa ttttacaaat tagccaggcg
11281 tgatggcaag tgtctgtcgt cccagctact ccagaggctg aggtgggagg atcgcttgag
11341 cccaggagat taaggctgca gtgagccatg gtcatcccac tgcactccat cctgggtgac
11401 agagcgagac cctgtctcaa aaataatagc aatcatcatc agtagcagca gcagcagcag
11461 cagcagcata gagagccagt gatcctggat cagtgcacct ggttgctgag ggttacctgg
11521 ctgaagcagg tggtggcagc agaaaagcct gacctctgat ttcttccata aggtacctga
11581 aatccaagcc ctgactaaat ttcttttttt cttttttttt gagacagagt cttgttcttt
11641 tgcceaggct ggagtgcagt ggcactatct cagctcactg caagctccgt ctcccaggtt
11701 cacgccattc tcctgcctca gcctcccgag tagctgggac tgcaggcacc cgccaccaca
11761 cccggctaat tttttgtatt tttagtagag acggggtttc accgtgttag gatggtctcg
11821 atctactgac atcgtgatct gccctcctcg gcctcccaaa gtgttgggat tacaggcgtg
11881 agccaccgcc taaatttcta agggctccta gtcctgatgc ctaatttctg gagtggacgt
11941 ggctcctgtt ccccgacacc tagagttttt gtttgtttgt ttgtttgttt tgagacagag
12001 tctcgctctg tcgcccagcc tggggtgcag tggcgcaatc tcggctcact gcaagctccg
12061 cctcccgggt tcacgccatt ctcctgcctc agcctccaga gtagctggga ctacaggcgc
12121 ccgccaccat gcccggctaa tttttttttt ttttttgaga cggagtcttg ctctatcgcc
12181 cagactggag tgcagtggtg cgatctccgc tcactgcaaa cttcgcctcc cgggttcacg
12241 ccattctcct gcctcaggct cctgagtagc tgggactaca ggcacccgcc accgcgcccg
12301 gctaattttt tgtattttta gtagagacgg ggtttcatcg tgttagccag gatggtctcg
12361 atctcctgac ctcgtgatcc gcccgcctca gcttcccaaa gtgctgggat tacaggcatg
12421 agccaccgcg cccggccccc gacacctagt tttaaagggt aagccggctc ctggcacctg
12481 cctacttgca gtagggcggc gcctagctct gacctccaag gtctggggac tgcgtcgcag
12541 ccgcccagtc catcccactt tcaatcttac aggcccctgc tgttgctgcc gctgccgccg
12601 ctcccagctg cccagtctgg cgggctcagt cccgcgttgc catgtgtggg agaccgcgtc
12661 gcgtaagcgc tggatgtggc ttcgctgatg cacattggac cgggctctgg actgggctag
12721 gggaagggca ggagggcgga attgggcccg agggccaggc ctcgccgacc cccgactgcg
12781 cctcccggtg gccccgcagc gcctcccggt ggccctggag tgcaggtctt accgtccgag
12841 atcgtccgca actgggcgag ctgtgcatgg ggcgtggcta aggccgtggt ttggttacga
12901 ttggccagcg ggacttaagt gttgtctctg aagagcatgg acattagtct ggagggtcct
12961 ggaagagtga tccccgcccc accatcaaat ggcgcttagg tctaggaagc gggtgtgggt
13021 ggggccttag ggcgaggcgc agacataccc cgaagtggtt ggattgtata ccgcaagggg
13081 ctggatcgaa ccccccaaag acactggaag gctgtgtggc tgaggagggc ccggcagatc
13141 cagtgtgtcS tgggctttac aggaaagagc tccaccttct ctggagtgtg cagatgcgat
13201 ctaggtgtgt ccacccgatg ggagctgcgg gccgggcaga tgctgcccca gtacaaagct
13261 gatttggacc tggggcctct ggacttccct gattctctgc ttgcatctcc agcaaagtcc
13321 tgtcccgttg gctgccttca tccactctct cacttctctg ccttcagagt aaaattgcaa
13381 gatctgtggt gcttactggg atctgataga gtctctcggc atccactgtc tatgcagcgg
13441 gtgtccacct gcagcggggg ccatgtgcag cggggggcca cgtgcagtgt gtgcctcttc
13501 ttagccatgc tggacagcgc cgcccctgaa aagcagctcc ccggtttcac ccagaaagcc
13561 atccagaacc tcctggaaaa ggtggcctga tggccaagtg gcctcggatg ccaggctcaa
13621 tcctttgaac ttttcctgtg ggctgtcagg acccatagaa ggtctttgag caggtgagtt
13681 tggagcagat ctggtaggca agcgaacaga tggatgYgtg cactggagat tccgtgggtt
13741 cccctgtgta catctcttcc ctttgggaaa ctgccctgag tgaggggcta agggcaggat
13801 ttgcattgaa atcctagctt tgctgctgtc agcccaactt ttaggcaaca gggtcttggt
13861 ttgatgtgac atttccaagt ccatcttgta tcacaacctg tcagctgcag ctcacttatt
13921 caatctattg tggttcaagt tcccaagaaa atgaatcagt ctggtctgct ctccagatca
13981 gattacgttt acttgcctag gaattgtctg ccctttaact caagactttg cactgttgtt
14041 cacatttgta atcccagcac tttgggaggc caaagcagga gtattgcttg agcccaggag
14101 ttcaagacca gccagggaaa tataacaaga ccctatctct acaaaaatta aaattaggtt
14161 gggcactgtg gctcatgcct gtaatcccag cactttggga ggccctggca ggtggatcac
14221 ctgatgtcag gcgttcgaga ccagcctgac caacatggtg aaaccccgtc tctactaaat
14281 acaaaaagtt agctggatat ggtggtgcag gcctgtaatc ctacttggga ggctgaggca
14341 gaagaatcac ttgaacccgg gaggtggagg ttgcagtgag ccgagattgt cccattgcac
14401 tccaacttgg gcaacaagag caaaactccc tctcaaaaaa aaaaaaaaaa aaaaaaaagc
14461 caggtRcatg tcagtggtac gtgcctgtgg tcccagctac ttgggaggct gaggtgggag
14521 gattgcttgg gcctggggtt gagaccacag tgagccaata ttgcaccact gcactccagc
14581 ctggacaaca gaataatacc ctgtctcaaa aaaaaaaaaa aaaaaaaaga aaaaaaagaa
14641 aagaaaaaga ctttgccctt gagtcaagac tttacccttt tacccttggc taagatggat
14701 gtaggaagtg acatggtaca aaatgctgca gcagagcgtg tgtatgtgct ggaagaggag
14761 ttgactaggg cagtgattga catctctgtt ccagatattt gcttaccttc cctgctgggc
14821 ccctccctat aggagcatta tatgctcatt ccctacttac aataggtttg gctataggac
14881 ttgctttggc cagtggaata tgggtaggaa ggcaaaatat cggccgggcg caatggctca
~~n
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
14941cacctgtaatcccagcactttgggaggatgaggcgggtggatcagctgaggtcaagagtt
15001cgagaccagtctggccaacgtggtgaaaccctgtctctactaaaagtacaaaattagcca
15061ggcatggtggcacgggcctgtaatcccagctgctaggaaggctgaggcaggagaatcact
15121tgaacccgggaaggaggaggttgcaatgagcccagatcatgccattgcactccagcctgg
15181acaaaaagtgaaactccgtctcaaaaaaaaaaaaaaaaggtaaagtatcacttctgcata
15241gaagctttagggcaccattgagtactctagcagcttccagtctcttctccctctgctcag
15301gctcataaacctggcagtttccagatctagacttctctttcagcctgcaacccagaatga
15361caatgacatgaagctgggctacagcctacctataaaatgatgcagaatttaagaaataaa
15421tctctcttgctgtgagccattgatatatggaggttgtttgttagcacatccaaatgttta
15481aacaaactgttacagaattaatacccagaagtggtgtgctgcaacaataaaaattgagcc
15541tcagccgggcacggtggctcacacctgtaatcccagcattttgggaggccaaggtaagtg
15601ggtcacctaaggttaggagtttgaaaccagcctggccaacatgacaaaaccctgtctcta
15661ctaaaaatacaaaaaaaattagccaggcatggtggtaggtgcctgtaatcccagctagag
15721gctgaggcaggagaatcgcttggacccaggaggcagaggttggcagtgMgtcaagattgc
15781gccactgcactccagcctgggcgatggagtgagactccatctcaaaaaattaaaaaataa
15841aaataaaaatattattaaaaattagccaggtgtgatggcatgtgcctgtagtcccagcta
15901cttgggaggctgagatgggaggatcacttgagcccaggaagcagaggttgcagtgagcca
15961agattgcaccactgcactccagcctgggtccaaaaaaaaaaaaatceccagccaggcatg
16021gtggctcatgcctgtaatcccagcactttgggaggctgaggtgggtgctgaggtcaggag
16081tttgatactagcctggcaaacatggtgaaaccctgtctccactaaaaatacaaaaattat
16141ccaggcatggtggtgggcacctgtaatcccagctactcaggaggctgaggctggagaatc
16201gcttgaaccttggatgcggaggttgcagtgagccaagatcaagccactgcactccagcct
16261gggcgacagagcaagactatctcaaaaaaaaaaaaaaaagcctaaactatgtaaactata
16321tgacattgacgttgagctggacagtggctggtaagggaactgtcattggaagttggaaag
16381atggtgacgtgtgttatgcaatggtgaatcgtttggttaaactgtaagcttatgaccaaa
16441tgagctttaggctttaggtaaagaactggggaaagggagtattggtagcatgctgtcact
16501actattgcatgcatttgaggagttactagaagaaagagatgactcagaaattaaatggtc
16561agtttataagcagaaatggaagagaatatagaaattcgaggcaagtgatccacattttca
16621gtaaaagatacaactgagaaagtccttgagccacaaggttttcgtttttgtttttgagac
16681agtcttgctcttgtttccaaggccaccttctgggttcaagcctttctcctgactcagcct
16741cccaagtagctgggattacaggcgtgcaccaccacgctcagctaatttttgtattttcag
16801tagagacaggtttcaccatgttggccaagctggtcttgaacttctgacctcaaatgatcc
16861tcccacctcRgcctcccaaagtgctgggattacaggtgtgagccactgcgaccggctgag
16921ctacaagttttgattaaaagtcatctttgtggcaagggccatatcaagtatatggctatt
16981atgccctttgtaaaaatctccaaactgatcaaagtggttcctaataaatcctctcagcta
17041gtcaagatgattcaaaggaaagaggttaagagtgtaactcaccttggctgggcgtggtgg
17101ctcacgcctgtaattccagcactttgggaggctgaggtgggcggatcacctgaggtcagg
17161agtttgagactagcctaaccaacatggagaaaccccgtctctactaaaaatacaaaatta
17221gccaggcatggtggtgcatgcctgtaatcccaactacttgggaggctgaggcaggagaat
17281tgcttgaacctgggaggcggaggttgcagtgagccaagatcacccatggcactccagcct
17341gggcaacaagagtgaaactccatctcaaaaaaaaaaaaaaatgtagcttacctgagggag
17401tcagtaggctcaactacagttaagtctaacgtcatggttatgtctgaaaagaattatggg
17461tatgctgttgacccatggatctgaatggagtaaaatacgtaagttcagttttggagggaa
17521ttgccctgcttcccctgcctaacaccccctcaccctgacaaaaagccaccaggttaaatc
17581ttgaccatgagtgttcaatacttagtatgatttttaggtccccaagtttctttctttttt
17641tttatttcggagaccgggtctcactctgtcacccagcctggagtgcagtgatgcaaccac
17701agctctctataacctcgaacttctgggctcacacgatcctcctgcctcagcctcccaagt
17761agctgggactacaggcccatgccaccccagcaggctaatttttgtttttcaaattttttt
17821gaaacaaaatctcactctgccacccaggctgaagtgcagtggcacgatcttggctcactg
17881caacctccgcttcctgggcttgagtgatccacttacctcagcctcccaagtagctgggac
17941tacaggtgtgcgctaccatgcccggctaatttttgtatttttttggtagagacagggtct
18001tgctatattgcccaggctggtctcgaactcctgaactcaagcgattcacctgtcttggcc
18061tcccaaagtgctggcattataggcgtgcagtgtaccaccatgcccagcctatttttgttt
18121tgttttgctttgttttgttttgagatgaagtcttgctctgtcactccagctggagtgcag
18181tggcacaatcaagcctcactgcagcctctacctctagggctccagtgatccccccacctc
18241agccttctgagtagctgggactacaggcatgcgccaccacacctggctaatttttctatt
18301tttttctggagaggatttcagcctgttgcccaagctggtcttgaacttctggtcttaagg
18361agttctccctcgttggcttcccaaagtgatgggattacaggtgtgagccaccatgcccag
18421cctaatttttgtatttcaggtttttttttgttttgttttgttttgtttttagtagagatg
18481ggggtctctgtatgttgcccaggctggcctcaagcaatccttgcctcaagtgatcctcct
18541gcctYagcctctcaaaatactgtgattgcagatgtgaaccaccatgcccggcctgggtct
18601ccaaatttcttttttttttttagagacggagtctcgttctgtcacccaggctggagtgca
18661gtggtgtgatctcggctcactgcaagctctgcctcccaggttcacgccgttctcccgcct
18721cagcctcccgagtagctgggactacaggYgcccgccaccatgcccggctaattttttttt
18781gtatttttagtagagacagggtttcactgtgttcgccaggatggtttcgatctcctgacc
18841tcgtgatctgcccgcctcggcctcccaaagtgttggggttataggcgtgagccaccgcac
18901ctggccatgggtctccaaatttctatgggcatgaaggagactgagaaagctactctactt
18961cagaaagacataaccaccagtgtcctctcaattgtggccaaggagaataagtggaaaagg
19021gtggtttactctaagggcagagccaagaacatggtgaagaatgaactagggaactcttcc
19081cactcccagggaaaagtgggggttcttctcaacatctgcccaKcagcactttagacttag
19141tggggcccagagcctgctgtgtgtctcctgtccttccttccttttttttttttttttttt
171
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
19201 tttttgagac agagtttcac ttttgtcacc catgttgtag tgcaatggca ctatctcggc
19261 tcactgcaac ctctgcctcc tgggttcaag cgattctctt gcctcagcct cccgagtagc
19321 tgggactaca ggtgcatgcc accacgcctg gctaattttt ggttttgggg ttttttgttt
19381 ttgtttttga gacggagtct tgcactgtcg cccaggctgg agtggaatgg cacgatctcg
19441 gctcactgca acctctgcct cctgggttca agcgattctc ctgtctcagc ctcctgagta
19501 gctgggacta caggggcccg ccaccacgcc cggctaactt tttgtatttt tagtagagac
19561 cgggtttcac tatgttggcc tggctggtct tgaactcctg accttgtgat ctgcccccct
19621 cggcctccca aagtgctgga attacagacg tgagccactg cgcctggcta atttttgtat
19681 ttttagtaga gacaggtttt caccgtgttg gccagggtgg tctcaaactc ctgacctcag
19741 gtgatccacc ggcctcggcc tcccaaagtg ctgggattac aggcatgagc caccgcaccg
19801 ggccctgtcc ttccttctga acgggagtgt gctctgctgt tctcctgtcc ttgttctgct
19861 ttatatgttg gatgtgttcg tgtgtgtgtg tgtgtagaaa tggggcacag gtaacttgtc
19921 tctgtctctc tcttattttg tagctcatag gtctctgaat caagagaagc cacatctgga
19981 cctgatatag aagagactat tagagatcct gggcttgagg ctgattccat gtcagatggg
20041 tcacttaggt ggtctccctt gggaagggga tgcatttatt ttgcatatgg aagaaaatgc
20101 aaaggcagta tttgtaagga agagggcaga cgggggaaga ttttataatt gttcaaaaac
20161 attcactggg atgtgtgtgg tggctcacgc ctataatccc agtgctttgg gagggtgaag
20221 caggaggatc acttgaggcc aggagtttga gaccagcatg ggcaacatag tgagacccta
20281 tctctacaaa aaataaaaca ataaaaaaaa attagctggg cgtggtggtg cttgcctgta
20341 gtcctagcta cttaggaggc tgaggtggga ggatcactta agctcaggag gtagaggctg
20401 cagtgagtta tgattgcacc atgcacctat gcactccagc ctgggcaaca caacaaaaca
20461 ctgactctaa aaaaacaacc aacaaaaaaa aatcacatgt attcactggc cctctctttg
20521 gggacctgct acatagaatg gttttttgtc cccagttcac tgacatcagg tatggctatg
20581 tggcttgctt tagaccatgg actttgagtg gaaatgacat gtgccacttc cacgaggaag
20641 ctttaaaagc cgtcatgggg tctgccacct ttcctctctt cggtgtctgg agacggaaag
20701 ttccagcttg agacttttcc ttcagacagg gctctRgaat gaagatagca tagaacagag
20761 tggtcccatg gaggacatgg atatgagtga gaaatcaaca tggtgttgtg agcccctaag
20821 atttgggggc tgctattact gcagcgtaac tggatcccag ctgatagatg cagcctccct
20881 gtgggatacc ctgctcaggt atcctttccc atcaccatga caactgacac accataatga
20941 gctatgctga tgttaggaag tctccgcctt tgctcctctt cagagctgtt caccctcagg
21001 tcctaaccag tgagcctatt tctttttttc tttctttYtt tttttYtttY tgagaWggag
21061 tcttgctctg tcaccaggct ggagtgcagt ggtgcgatct cggatcaatg caacctctgc
21121 cttctggatt taagcaaata ttgtgcttca gcctcctgag taggtctgga actcctgacc
21181 tcaggccatc cgccagcttt ggccttctaa agtgctggga ttacaggcat gaaccaccgt
21241 gcccagccaa gccgagtctt cttgattctt gctggcattt ggcaactagt agcagctgct
21301 cacaggaact gtaaaaacat ctggtggggc ccagaccttc tagcatcaac atggtgccta
21361 gtaaatatca atctcacatg catcctgaga tgcattaaaa agaagctgtc caggccgggc
21421 acgggggctc acgcctgtaa tcccagcact ttgggaggca gaggcgggtg gattgcttga
21481 gcccaggagt ttgagaccag tctaggaaac atggcaaaat cctagctcta tttttaaaaa
21541 gggggggaaa aagaaataaa aaagctgggc atggtggttc acacctgtaa tcccaacact
21601 ttgggtggct gaggcaggtg gatcacttga gagaccagcc tggtcaacac catgaaaccc
21661 catctctact aaaaatacaa aaattagcta cacctcatgg tgcacNcctg tagtcccacc
21721 tactcgggag gctgaggcag gagaatcgct tgaacctggg aggtggaggt tgcagtgagc
21781 ccagatcacg ccactgcact ctaacctggg ctagagagtg agactctgaa aaaaaaaaaa
21841 aaaaaaaaaa gagaaaagaa cataatgttt ggccaggcat ggtgccttac acctgtaatc
21901 ccagcagttt gggaagccga gggggcggat cacctgaggt tagttcaaga ccaacctaat
21961 caacatggtg aaacccatat ctactaaaaa aaaaaaaaaa attagccagg cgtggtggtg
22021 gatgcctgaa atcccagcta cttgggaggc tgaggcagaa gaattgtttg aaccctggag
22081 gcagaggttg cagtgaaccg agattgtgtc actgcactcc agcctgggcg acaagagtaa
22141 aactccgtct caaaaacaaa acaaaacaaa aaagaatcat aatggttagt aagtgaaaat
22201 tctgaattag tttgtgtatg tgtattgttg catataatag agacccaaat taactgtggc
22261 ttaaataaga tagaagttta tttctctctt ctataaaagt ccaagttagt atgatggatc
22321 tttccatgaa atcattagga gccagatttt ttgtatcatt cattcattca ttgattcatt
22381 actaccatta atagagacaa ttttctgcac cattcaggct ggagtgcagt ggtgcaatca
22441 taattcactg taacctcaaa atcctgggct ccagcgattc tcctgcctta gccccaacaa
22501 agtagcaggg actacaagca catgccacca cgcctggcta atttttcttt ttcttttttg
22561 tagaggtggg gtgttactat gttgcccagg ctggtctcaa actcctggcc tcaagtgatc
22621 ctcctgcttc accctcccaa agctctggga tgacaggcat gagccactct gcccctccag
22681 gtctttttta tcttgttgct gttccatccc tagggcgttg ccctcaccca catgatccaa
22741 tatgattcac caccacttcc acagtctggc ccttctgagg ggtgatggtt tgccctttgc
22801 cctaaagagc atgattcaga agtacagatc atttttgctc taatccccat agccaggatg
22861 tagtcatatg gctacatccc gatgaaagtg ttgctgagaa atagaatctc taccctgagc
22921 agctttttgc ccagataaaa gttcagttac tctgggagaa gggtagaatg gatactgggg
22981 gaccataagc tgttgccacc acacacattg aatgttaacc catcccaact gtatcaattt
23041 ttCCttCCtt tCCttCCttC CtCCCtCCCt CCCtCCCtCC CtCCCtCCtt CCttCCttCC
23101 ttCCttCatt CCttCCttCC tttgttCCtt tCtttCgaCa gtCtCCCtCt atCCCCtagg
23161 ctggagtgcR gtgttgccat ctcggctcac tgcaacctct gcctcccagg ttcaagcaat
23221 tctcctgcct cagcctcctg agtagctggg attacaggcg tgctccacca tgcccagcta
23281 atttttgtat ttttagtaga gacaggattt ccccatgttg gccaggctgg tcttgaactc
23341 ctgccctcag gtgatccacc cacctcagcc tccaaaagtg ctgggattat aggcgtgagc
23401 cactgccttg gcctcaaacg gtatcaattt tctgttactg atttaaccaa ttatcataca
177
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
23461 ctcagtggtt taaaaccaca cacatttact ttcttacagt tctggaagtc agaagttcaa
23521 aatcagtttc attgagccaa tgtctggtgt cagcagggct ggtttttgtt ggtggctctg
23581 gtggacaatg tttccttgcc ttcttcagct cttttttttt tttttttttt tgagacaggg
23641 tctcgctctg ttacccaggc tggggtgcag tggtgcaatc atagctcact gcagcctcca
23701 tctcccaggc tcaggcgatc ctcccgtgtt agccttctca gtagctggga ccacaggctc
23761 acgccaccac gccctgctaa ttttgtttat tttttgtaga gatgaggtct cactccattg
23821 cccagactgg tctcaaagtc ctggattcag gagatcctcc tgcctcagcc tcccaaaggt
23881 ctgggattac aggtgtgagc cgttgcaccc caccctcttt cagtttagaa aggctacctg
23941 tattccttgg ctggtgggtc catcctccat tggaaagcac atgaatccat ctctgccttc
24001 atcatcactc cactttctcc tctgagactt attcctcctg tgtgcctctt aggaggatgt
24061 tcatgattac ataccgccct cttggataat cctgaataat ctctccatct caggatcctt
24121 cacattttca aaatcccttt caccatataa cgtgacattc acagattcca ggaataggac
24181 gtagacatat ttaggggggt tctctattca gcctactgta ccatgccatt ccacacttaa
24241 ctccttcact catttattca taaaatatgt attgagcaag acctgtgtgc caggcattgt
24301 gttaggtgct agagaaatag aggtgaaaat acagacaagg cctctgcttt catggagttt
24361 atattctagt gaagaggaca agtaaatagc taagctattc tttttttttt tttttttttt
24421 tttgagacgg agtctccctc tgtcgcccag gctggagtgc agtggcgcaa tctcggttca
24481 ctgcaagccc cacctcctgg gttcacgcca ttctcctgcc tcagcctcct gagtagctgg
24541 gactacaggc gcccgccacc acgcccagct aattttttgt atttttagta gagacggggt
24601 ttcaccgtgt tagccaggat ggtctcgatc tcctgacctc atgatccacc cgcctcggcc
24661 tcccaaagtg ctgggattat aggcgtgagc caccatgccc ggccaagagc taagctattc
24721 taagctataa cgtgtattat caaaacaatt aaggccaggc acagttgctc acacctgtaa
24781 tcacaacact ttgggaggct gaggcgggtg gatcatttga ggtcaggagt ttgagaccag
24841 cctggccaac atggtaaaac cctgtctcta ctaaaaatac aaaaaaatta tccaggtgtg
24901 gtggtgcatg cctgcagtcc cggctactcg ggaggctgag gcacaagaat aagaattgct
24961 tgagtgggga ggtggaggtt gcagtgagcc aagatcatgc cactgcacta caggctagga
25021 gacagagWga gaccctgtct taaaaaaaaa gcaattaggc caagtgcagt ggctcatgcc
25081 tgtaatccca gcactttggg aggccaagga gggcagatca cgaggtcaag aaatcgagac
25141 cagcctggcc aacatggtga aaccctgtct ctactaaaaa tacgaaaatt agctgggtgt
25201 ggtggcgcgt gcctgtagtc ccagctactc gggaggctga ggcaggagaa tgccttgaac
25261 ccgggaggtg gaggttgcag tgagccgaga tcacgccact gcactccagc ctgacgacag
25321 agtgggaatc catctaaaaa aagaaagaaa gaaattggct ggagaatcgc ttggacccag
25381 gggtggaggt tgccatgagc tgagattgtg ccactgcact ccagcctagg caacaagagc
25441 aaaactccgt ctcaaaaaaa aaaaaaaaaa tcccagcact ttgggaggcc aaggagggca
25501 gatcacgagg tcaagaaatc gagaccagcc tggccaacat ggtgaaaccc tgtctctact
25561 aaaaatacaa aaaattagct gggtgtggtg gcgggtgcct gtagtcccag ctacttggga
25621 ggctgaggca ggagaatggc atgaacctgg gaggcggagc ttgcagtgag ccgagatcac
25681 accactgcac tccagcctgg gcaacagagc aagactctgt ctcaaaaaaa aaaaaaaaaa
25741 gaaaagaaaa gaaattaaac agtgtgatgt gacaaaaagt gatagggggt tggagacagc
25801 ttttctgttg gatggttagg aatggcttct tagaaaagat gactgacaca tgggaggctg
25861 atgtggcaga tcacgaggtc aggagatcaa gaccatcctg gctaacacgg tgaaaccccg
25921 tctctactaa aaaatagaaa aaattagccg ggtgtggtgg cgggcgcctg cagtcccagc
25981 tactaaggag gctgaggcag gagaatggcg tgaacccggg aggcagagct tgcagtgagc
26041 tgagatcacg ccactgcact ccagcctgga cgacagagcg agactccatc tcaaaaaaaa
26101 aaaaagaaag aaaagatggc tgacacagag ggcagagctg agagccaaga gggcagaaaa
26161 gagccataga aaaccatttc caggcctgga agcctaaagg aatttcccag ctggatttgc
26221 agttgctttg gattggtgac tcctttttac ctttcattgt taggggacct gcaggttcct
26281 ttgcctgctg tgcagctaca gctccattac accaagacaa tagggatgca gcagagagag
26341 ttactggtgc agggcaccta gtgcagagat gggaagaggc cctcaaatct atctccccga
26401 gcaattctgg gagagggttt ctaaggggac tgtggagggt aggggattgt ggagggtaag
26461 gttttgggca actgggtcat tgattgattg ggggaaggat gtagaagctg cgtttttggg
26521 ggaattagct ccttgtgggg tccttcaggt cagctgagtc agtagttcca tgaggacctg
26581 aaggaatctc ttttcttttc ttcttcttct tctttttttt tttttttttt gagatggagt
26641 ctctctctgt cgccaggcta gaggtgcagg gggtcgcagg ctagaggtgc agtggcatga
26701 tcttggctca ctgcaacctc cacctcccgg gttcaagcaa ttctcctgcc tcagcctccc
26761 aagtagctgg gactagaggt gcgtgccacc acacccagct aatttttgta tttttagtag
26821 agacagggtt tcaccatgtt ggccagggtg gtctcgatct cttgacttcg tgatcggccc
26881 ccgccccacc ctcggcctcc caaagtgctg ggatcacagg agtgagccac ggtgcccagc
26941 cttaattttt gtattttcag tggagacggg gtttcaccat gttgatcagg ctggagtgca
27001 atggtgcaat cttggctcac tgcaacattc gcctcctgga tttgaatgat tctcctgcct
27061 cagcctccca agtaactggg attacaggaa tgcgtcacca cgcccggcta attttgtatt
27121 tttttagtag agacggggtt tcaccatgtt ggtcaggctg tcttgaactt ctgacctcaa
27181 gtgatccacc tgctttggcc tcccagagtc tgaaggaata tctcaaaggg aacacttaat
27241 gttgtgtaat gtccaggttg tgatccatag agcagttaaa ggtaaaggta actataattt
27301 tttttttttt tttttagaca gagtctccct ctctgtcacc caggctggag tgcagtttca
27361 cgatctcggc gcactgcaac ctccgcctcc ctggttcaac caattctcct gcctcagcct
27421 ctcaagtgtg tgctgccatg ccaggctaat tttttttttt agacggagtc ttgctctgtc
27481 acccaggctg gagtgcagtg gcacaatctc ggctcactgc aacctccggc tcctgggctc
27541 aaacaatgtg ttttttcccc tagtactttg gtgtttgatt atcttttttt tttttttttt
27601 tttttttttg agatagagtc tcgctctgtc acccaggctg gagtgcagtg gtgcaatctt
27661 agctcactgc aagctctgcc tcctgggttc atcccattct cctgcctcag cctcccaagt
17'~
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
27721 agctgggact acaggcaccc accaccacgc ccggctaatt ttttgtattt ttagtagaga
27781 cggggtttca ccatgttagc caggatggtc tcgatctcct gacctcatga tctgcccgcc
27841 tcagcctccc aaagtgctgg gattacaggc Rtgagccact gcacccagcc tggtgtttga
27901 ttatctatta tgtcaaacag gctgggttta gtggctcacg cctgtaatcc cagcactttg
27961 ggagactgag gtgggaggat cacttgagcc caggagctga agaccagcgt agcaatgtag
28021 caactccctg cctctacaaa aagttaaaaa atttagctgg gtgcaccagY agaccagctc
28081 ctcaggaggc tgaggaggga ggatcactcg agcccaggag ttcaaggctg cagtgagctg
28141 tgatcatgcc actgtactcc agcccaggca atggagcgag accctgtctc aaaataaata
28201 aaacatgaag aatgtcgaac acattatctg gtttttgttt ttgttttctt tttttgagat
28261 gttgtctcgc tctgtcaccc tggctggagt gtagtggtgc gatctcggct cactgcaacc
28321 tctgcctccc gggttcaagc gattctcccg cctcagcctc ccgagtagct gggactacag
28381 gtacgtgcca ccatgcctgg ctaatttttg ttattttttt tttttcagta gggacagggt
28441 ttcgccatgt tggccaggct gttctgaaac tcctgacctc agatgatcca cccacctcgg
28501 cctcccaaag tgctgggatt acaggtgtga gccatcgtgc ccggcctgtt ttaaaaaacc
28561 atattggccc aactcggtgg ctcatgcctg taatcccagc actttgggaa gccaaagcag
28621 gaggattggt tgagcttagg agtttgagac cattctgggc aacatggtga aaccctgtct
28681 ctgcacaaaa atagaaaaat ttgccacctg tgctggtgtg tgcctgtagt cccagctact
28741 ctcaaggctg agggaggagg attgcttgta gagcctggga agtcggagct gcagtgagcc
28801 atgatcacac caccacactc tagcctgaca gaatgagacc ttatcccaaa agaaaaaata
28861 aatgatattg tattatatgt gaactttgaa ttatattgtg ttgtatctga agtttgaatt
28921 ttcacgttat gtttaaaaat cttggctggg cgtggtgggt cacgcctgta atcccagcac
28981 tttcggaggc caaggcgggt ggatcacctg aggtcaggag ttcgagacaa gcctggccaa
29041 catggtgaaa ccccgtctct actaaaaata caaaacttag ccgggcatag tgacatgcac
29101 ctgtagttcc agctactcgg gaggctgagg caggagaatc gcttgaaccc aggaggcaga
2916T ggttgcagtg agctgagatc gtggcattgt actccagtct gggcaacaag agtgaaactc
29221 catctaaaaa ataaaaaaga aaaagaaaaa ataatacaag aaattagccg ggcgtggtga
29281 caggcacttg tagtccctcc cagctactca ggaggctgac gcaagagaat tgcttgaact
29341 tgggaggtgg aggttgcagt gagctgagat cgtgccattg cactctagcc tgggaaacaa
29401 gagcaaaact cagtctcaaa aataaatagc ttgaacccgg gaggcagagg ttgcagtgag
29461 ctgagattgc accacttcat tccagcctgg gtgatagagc aagactctat ctctaaataa
29521 ataaataaat aatcctttag gatggcaatg aatttaagga ctaaactagg gagaatcgac
29581 tttttttttt aaaatggagt cttggtctgt cgcccagact agggtgcagt gggcgccatc
29641 tcggctcact gcaacctcca ccttccaggt tcaagggatt cttgtccctc agcctcccaa
29701 gtagctggga ttacaggcac ccgccaccat gcctggctaa tttttgtatt tttagtagag
29761 atggggtttc accatgttgg ctatggttgg ccaggctggt cttgaactcc tgacctgagg
29821 tgatctgcct gcctcggcct cccaaagtgc tgggattaca ggcatgagcc actgtaccca
29881 gcccattcga cattatttat ttatttattt atttatttat tttttgaggt ggagtctcac
29941 tctgtcgccc aggctggagt gcagtggcac aatctcggct cactgaaacc tccgcctccc
30001 gggttcaagc cgattctcct gcctcagtct cccgagtagc tgagattaga ggcaaccacc
30061 actatacccg actaattttt gtatttttca gtagagatag ggcttcacca tgttggccag
30121 gctggtctcg aactcctgac gtcagttgat cctcccacct cagcctccca aaatgctgga
30181 attaaagctg taagccagcg ggcctggtgg acatctttta ataatcagtc tttccattca
30241 ggtatatggt atatgtctcc atttacttag gtcttatttc atatccttca ggttggagct
30301 atcatttctt ttcatacagg ttttgcacat ttcttgtgag gtttattcct tcatggtcca
30361 tggattttgt tgtgaattgg gaatcctttt tccaccaagt atattttcta atttgttact
30421 ttagtataca ggaaagataa ctaattttta tctgcagttt attatctatg aaaggataaa
30481 agtagaacta ctcagtaaaa ggtttccata atcaaataag tatgggctaa acaaagctaa
30541 acagatgtgt tcactgctgg acttatcaat gcttgtgata attttttttt ttttttttga
30601 ggcagagttt tgctctgtag cccaggctgg agtgcagtgg cgggatcttg gctcactgca
30661 acctccacct cccgggttca agtgattctc ctgcctcagc ctcccgagta gctgggacta
30721 tggcatgcac caccacatct ggctaatttt tgtaatttta atagagacag ggtttcacca
30781 tgttgactag gctggtctca gaactgttga cctcaggtga tctgcctgcc tcagcctccc
30841 aaagtgctgg gattacaggt gtgagccacc accaccaggc aattgaagac gtatattcta
30901 tgaagaaatg ggtagatttt aatgaacaat accccttttg tgggcagatt cctaagtccc
30961 aggccctcac aacaaagggg cagtgggcct ggagatgcca gcttcagctg ccagagggac
31021 tgctcctcca gggccacccc agcccacttt tgatcaccaa gttttgatca ccaagaatcc
31081 caagaagggc acagggaatt tcctttctta cctgcccatg aaaccttttg tcactagaca
31141 tcctgaaaca tactttggga aactgcatcc aaagaccctt ctagtttcaa atctgtggat
31201 ccaggggtct ccactgaacc ttacctgatg cccaaactcc cacccattca ctcccaacca
31261 gaacacagaa gatgacctgg tgccaaaatg aaagctttaa tgagtgttac tcctagacag
31321 tcacgtctca gcttctgcca gcctccactg tcccagctct cttagctggc cgacagggga
31381 gctagttgct gaggggtagg gatctggagt ctaaagagca gagccaggca aaaggaggta
31441 caggaagccc ccgatggggg ctgggctccc ggagtgtggt gctggggggt catgggcttc
31501 aggccggccc ctcttcaggc attcctagca aagccaccag gggctccagg ggtgtggggg
31561 tccccatggg cacagggtgg gtgcgttcat gcttgcgcaa gtcgctggca ctcaagaagg
31621 ccttgggaca atgggggcag gtgtaggggc gcactgagct gtgagtgcgg ctgtgtttgc
31681 gcagcccagc ccggtcagag aagctcttgc cgcactRggt gcaggggaag ggccggagct
31741 ccgggtgtga gcgctcgtgc cgacgcagca gcgtcattgt ggagaaggtc tccttgcact
31801 ctcggcacac aaactggggg ggcttctcgt cagcctcctc accccccgcc tcgcctgccc
31861 cttccaaggg accaggagcc tcccggacac cagcatcttg gcattccaca tgctccaccg
31921 tcatgcccac cacctgccac tgtgtggcca tcacaccacc tgactccggg ggcagcccta
174
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
31981 gcagccctgc tggagggtcc cccagccccg cccctgctRc cggggcggct gaactctcac
32041 ctgccacgcc cacaggcagc gccaacccca ccaccagctc ctgtgcaggg ggcacacccg
32101 cggcctcact gcttcgatgg gtccgctcgt gcttcctcag gctcgacgac accacaaagg
32161 atttcccaca tgcgttacag tggaaggggc gctcccccga gtgcacccgg ctatgcttcg
32221 tgaggctggc acgctcggcg aaggctcgcc cgcactcctc acagcggaag ggccgctggc
32281 cagagtgcac cagcgcgtgc cgcttgaggt cccaggaYgc cacgaacgtc ttgtcacatt
32341 gcaggcactt gaacggccgg tcgcctgtgt gcacacgccg gtgcatggcc aggtccgccg
32401 gctgccggaa gtccttgccg cacttctcgc agtggtatgg cttcacccct tcatgggcgc
32461 gctggtggcg acggaagctc gaggggtcgg agaacatgcg gccgcagcgc gggcacagga
32521 agggcttctc ccccgagtgc gtgcgctcgt ggctctggta ggaactgagc tgcgtgaagc
32581 ccttgccgca ggccgggcag cggtagggct tctgtgccgc gtggatgcgc tggtggcacg
32641 tgagcgagga tgagcgggag aagctcttcc cgcactcgga gcagaggaag gggcgctcgc
32701 cggtgtggga cctgcggggg tgtggaggac ttggcatgaa ggcgacagac ccataacgtg
32761 accccactgc ctgtctgggc tgtactttag gggctcccca aacgttcgtg ggggcctagg
32821 cttaatcccc taagagccac atggctgcac cccagaggaa gaagccttca ggctggctgg
32881 gtgtctctat tccaaagacc tgtctctgca cattaaagac caagatatgg gccgggcgcg
32941 gtggetcacg cctgtaattc cagcactttg ggaggccgag gtcaggagat cgagaccatc
33001 ccggctaaca cggtgaaacc ccgtctctac taaaactata aaaaattagc caggcgtggt
33061 ggtgggtgca tgtagtcccR gctactcggg aggctgaggc Rggagaatgg cgtgaacccg
33121 ggaggcggag cttgcagtga gccgagatct tgccactgca ctccagcctg ggcggcagag
33181 cgagactccg tctcaaaaaa aaaaaaaaaa aaagaaaaag aaaaaaaaaa aagaccaaga
33241 catggccagg cgcggtggct cacgcctgta atcccagcac tttgggaggc cgaggcgggc
33301 ggatcacctg aggtcaggag ttcaagacca gtgtgaccaa aacggagaaa ccccgtctct
33361 actaaaaata caaaattagc cgggcatggt ggcgcatgcc tgtaatccca gctactcgga
33421 aggctgaggc aggagaatcg cttgaacccg ggaggcggag gttgcggtga gccgagatcg
33481 cgccattgca ctctagcctg ggcaacaaga tcaaaactcc gtctcaaaaa acaaacaaac
33541 aaaaacaaaa caaaacaaaa cagaaaacca agatacgtgt cctccgcctt ttttttcctg
33601 ttccccaggc tggaatgcag tggcctgacc atagctcact gcagcctcga cctcccaggc
33661 tcaggccatc ctcccacctt atcctcccaa gtacccggga ctagaagtgt acatccccac
33721 gctcgggtaa tttttttatt tttatagaga cgaggcttgc tgtgttgccc aggctggtct
33781 tgaactcttg ggctcaagca atcctcctgc ctcagcctcc caaagtgctg gaattatagg
33841 cgtgagctat tgtgcccagc ctagaaacat gtcattaatg tagaggctga gaaaaagaaa
33901 aaaaaaaatg acctagacaa accaggcccc actcacacct cctggtctcc acaaaagacc
33961 ctcagaactg cccaactcca aaccccgccc cctttccagc tggcctacaa cggaggccaa
34021 tctgacccaa tcccattctc agagatcaac ctcaaggtgg ttgccacctc tgcccaatca
34081 ggggcaccaa tttctcccac atgcctagcc cctccccttg gatctgccat gcccaccttc
34141 ccattggctc actttaccct gagactcaaa cccaggcccc attggctgca gcaacgctgt
34201 cgccctgccc cggaaggcgc cctgccccgg aaggcRCCCt caccgctcat ggttgcggag
34261 gtccttgagc tccgcatagg ctttRccgca acgctcacag ctgtagggcc gcaggccagc
34321 gtgagtacgc cggtgcttgc ggaacactga agggtcagca aagctcttgc cgcagtcggc
34381 gcaggcgtaa ggccgctcgc ctgtgtggcc acgctggtgg atcttgagct tggagagcgc
34441 gccataggcc ttcgggcagt gcgcacagcg gaagggcagt tcgccagcgt gcgaggccag
34501 gtgcacgcgc aggcacacgg gctgcatgaa gcggcggccg cactcggggc acggaaaggg
34561 cttctccccc gtgtggctgc gcccgtggct gcgcagctcg ggtgccgtct tgtaggcctt
34621 ggggcatagc ggacacgcat agggcctagg cttggccgcg gagcctgaca ccttctcccc
34681 actggcttcc tctgccttag cttctgtctc tggctttggc ttcacctcgg CCaCCtCttC
34741 agagcagtct gccggcccat gtgtggcagc gtggcgcgct gccctgggcg cgtttggaaa
34801 tgtcttggta caggacaSgc acttgtagcg gcggcccgag cgcttgtagc cgggggctgg
34861 ggaccgggcc tctgcagcct ccacttccat ggccttggtg aacggggttt ctctgcaaga
34921 gaagcaaagt tagaccaaag ccacatacct tcgccactcc tgaaagcctc agagagaacc
34981 ctatctcatc tgcatttctc acctcggaac ccacacatcc ttcctgccca gcattcctgg
35041 ctctgacatc ctgcgttcgt ttcctccctg atctgctcat tgaagaaagg agttggacca
35101 agtgtccgca gagccactaa gaaaggaggc tgagggtcac aaaagattca cctacacgtc
35161 cccccccScc cccaacgggc ttttccaaac actgtggcat tcccagaggc ccaggttcca
35221 tctgtctcac catcttcctt cttcagctca gtgtccaaga actatgccag gataaagagt
35281 gtacccagac gtgggcctgg cctgaaggtc tccaagcgcc cagaaaagac agacctggga
35341 ccagaaaagg gctgagccaa tgggctaaat ctggtagctg gcactgtctg gaagtgacag
35401 gtccccagca tttgtgtttt ctttcctcct ctggatggtt agtcctcaga gacagcaact
35461 gttcacacag aattctggcc ttgcacagct gtacgggcct ccgccccaga ctggaatctg
35521 tccactctct gctctggaat cttgttggcc tgttcccaca actctggtaa tggagaatca
35581 ctcaaggcag cctgagccat tgctagcagc tggaagcctc tttctgagtc ataactgatg
35641 tatctgatct aacatggcct cctgggatac cagctctagc tgagatccct actttctggt
35701 ccagaaccca gacgccctcc acccagctgc tcctggggat catggttggg aggaaacagg
35761 attaatggct gtattagtct taacaccagc tcatcctccc tgggggatga agggaagagg
35821 attatggcag atccacttaa ggagtgctca gcagctgctg ctgggggaag gggtctgagg
35881 agtgggggct gcagggagcc aggtgtgccc agaggctagg gggcctacgt tctacttgca
35941 gccctgtgga ttactatgag acctcagtga aatgagtgtt gtttataaga ctatttccgc
36001 ccggctgggc gtggtggctc acgcctgtaa tcccagcact ttgggaggcc gaggggggcg
36061 gatcacgagg tcaggagacc gagaccatcc tggctaacac agtgaaaccc cgtctctact
36121 gaaaatacaa aaaaattagc cgggcgtggt ggcgggcgcc tgtagtccca gctactcggg
36181 aggctgaggc aggagaatgg tgtgaacctg ggaggcggag cttgcagtga gccaagatcg
178
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
36241 tgccaccgca ctccagcctg ggcgacagag caagactccg tctcaaaaaa aaaaaaataa
36301 aaagactatt tcctcatctg gacacgttag gggttggtgg cttctaagtt atgacactgg
36361 ggttcaggag gaggaaactg agctgagcca cgagggtgct aggggaaata cagagactca
36421 gggcccctta tgccaggaaa gggcgggaga agcagcttac ctggttgacc cagggaggac
36481 acaggagccc ctgacgcgtg gcaagggcca catccttaaa gtcagcaccc ttctgcctga
36541 aatcccggcc tcaggcctgc acgctgccct ccctccccaa ccccatgcag ccggtcttct
36601 cccaaaagta atgaatgatg tttcctgttc cctgctcaag aactttccat ggcttccacc
36661 acaccatgca aatccccagg tccttcctgc ttgttccaaa tctgacaagt ccttcactgg
36721 agccccactt ccaccaggag gatcctcaag catctcccct ttgggtttgg ccccaatccc
36781 acaaaactcc atcttattct cacttggtta tttcactttt cccactaaat cctagggctt
36841 agccaggtgt ggtggcatgc gccttaagtc ccagctacgt gggaggctga ggcgggagga
36901 tcctgtgaac ccaggggttc gaggctgcag ggaactatga tcgtaccact gcgctccagc
36961 ctgggcaacc tggtgagacc ccatctctac taaaaataaa aattagcaga tgtggtggca
37021 tgtgcctgta gtactgccta cttgagaggc tggggtggga ggactgcttg agcccaggag
37081 ttcaaggctg taccactgca ctccagcctg ggagacaggg caagacactg tctcaatcaa
37141 tcaatcaatc aataatcaat cctggggctt gaagataagt taaagggact gaattctaac
37201 ctttctgatg acttgaattc ttcctacagt ttccaaggga tccctcccta tttctggatg
37261 aggtactcac tacctcttcc agacggtttc tggagagtct gcctgataat gttcctcctt
37321 aataaaaatg atagcttggg ccaggttcag tggctcacac tagcactttg ggagaccaag
37381 gcgggtggat cgcttgagcc caggagttca agacaaggcg ggtggatccc ttgagcccag
37447. gagttcaaga ccagcctggg cgatatagca aaatcccatt tctacaaaaa atacaaaaat
37501 tagccaggca tggtggagca tgcctgtact cccagctact ccagaaggct gaggtaggag
37561 aaatgcttga gcctaaggag actgaggttg cagtaagcca agatggtgcc actgcactcc
37621 agcctggcaa cagagtgaga ccccatctca aaaataaata aataaataaa tgataaaaaa
37681 gatagttcac atttactgag cactcgccaa ataccaggca gtatcctaaa ctccttatgt
37741 gtattagctc agttaccctt catggcaacc ccatgaggaa agttctatta ttccattttc
37801 acagataagg gaaccaaggt ccagagaaat ggttcagtat tttgttaagt gcccagtccc
37861 tgaagccaaa ctgtctggct tcagattttg cctccatcac ttcccagctg atgtgaccgt
37921 gtgtaatgta ctgcatgtct tagaacctca gttttctaat ctgaaaaatg gagataatga
37981 Yagtacttac ctgacagagt tgggtgagga atgaatgagt caaaaataat tactgtcctc
38041 aattatcaaa gcgtcttctt agagccagac acattgctgg gtgtgctggt ttatttaatt
38101 taatgtctta taScttYctg agatagggat tcttgtccct actttacaga ggaataaatc
38161 gaggttccaa gagttaagtg acttgcccat ggtcccacaa tgggtaacct aagcagctgg
38221 gacttcagtc caggtgttta atttgcctta agttgctggg gtcttgctca gtggtctggg
38281 gcctcttacg cttgtctgct gcctccgcca gccccacagt gaccagaacc ctgagctcag
38341 gtcatacctg tgtcttctct catccttgca gaactgccct gagaccctgg ccggcaccct
38401 ttatgtctct gcttccctct cagagggctt gacccagtgg ttctgagctc tggcccttct
38461 actgcctctt gccagctctg ICgtctcagcc ttcctatctg tgagttagac accaggtagc
38521 tggaggggaa atccctcctc ccatggcact tcccagggga aaaggtaggg gagtgccagg
38581 ttggtctcag catgcgccca gctacacaaa gaggScaggt aggctaggtc tctgtctaac
38641 atcccaccat taaaaaaaaa aaaaaaaaaa atatatatat atatatatat atatataatt
38701 tttgagatgg agtcttactc tgttgcccag gctggagtgc agtggcgcga tcttggttca
38761 ctgcaacctc cgcctcctga gtagctggga ctacaggcac ctgccaccac acctggctag
38821 ttttttgtat ttttagtaga gacggggttt cactgtgtta gccaggatgg tctcaatctc
38881 ctgacctcgt gatccgaccg cctcggcctc ccaaagtgct gggattacag gcgagagcca
38941 ccaMgcccag actttttttt ttttttttat taaagagctt gaggtaggcc tcaggaatct
39001 gtattttaaa tacactctgg atcattccag ccaatacttt tgtttgtttg gttttgaaac
39061 aagatctcac tctgtcgctc aggctggagt gcagtggtgc agtcatggct tactgcagcc
39121 ttgacttccc agactcaagc aatcctccca cttcagcctc ccaagtagct gggactacag
39181 gcatgcacca tcatgcctgg ctaattttta ttttattttt agtagagatg aagtcttgct
39241 atgttgcctg ggctggtcta gaactcctgg gctcaagtga tcctcccMcc gcagcctccc
39301 aaagtgctgg gattacaggc gtgagccacc gtgcctgccc aaccaatact taagaaccaa
39361 acacacatcc ttaggtctcc acgagctctc aggagaggag cattttaagt gttcactaca
39421 cctctttttc agatattgag attaaggtcc ccacaaagga aaaactgtac acaaggacac
39481 acagctggtc aaggagccag actcgaaccc aagtctccat tctctccccc aggttgaatc
39541 atgagacttc ccactgctcc caggaaaaag accaatatct tttccatggc cagcatagcc
39601 ccaaaccatc taaatcctgc ctacctgggc agatcacttg aggccaggag tttgagacca
39661 gcctggccaa catggtgaaa ccccatctct actaaaaata cacacacaca cacacacaca
39721 cacacacaca cacacacctg cctacctcac ctcccactcc tctccctggc ccactgggct
39781 ctacccacag aggcctcctt tcttctcctc aaagagctaa attccttccc acctcagggc
39841 agtggcacta gcagttccct ctgtctgagc cactcttctc ccacgatctt tgtgtagctg
39901 tcttttttgg tgttatttgg atctcagctc ccagtcacct cctcaaaaag agctttcttg
39961 accacctttc cttttcttcc ccccttttaa tattccaaat tttttccttt tttaaccaac
40021 caaggagcac tgaatgacta cctttctcaa tgctatcttt acccctgata atcattctct
40081 atctactctt tattattatt attatttttt gacggaatct catgattatc tatcaagcag
40141 ttctcctgcc tcagcctcct aagtagctgg gactacaggt gcccgccacc acgcccggct
40201 attttttttt tttttttttt tttgagacgg agtctcactc cgtcacccag gctggagtgc
40261 agtggcacaa tcctggctca ctgcaagctc cgcctcccgg gttcatgcca ttctcctgcc
40321 ttagcctcct gagtagctgg gactacaggt gcccgccacc acgcccggct aattttttgt
40381 atttttagta gagacagggt ctcactgtgt tagccaggat ggtctcaatc tcctgacctc
40441 gtgatccacc tgcctcggcc tcccagagtg ctgggattac aggcgtgagc cactgcaccc
17~
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
40501 ggcccaatcc cggctaattt ttgtattttt agtagagatg gggtttcacc atgttggcca
40561 ggatggtctc gatctattga cctcgtgatc cgcccgcctc ggcctcccag agtgctggga
40621 ttacaggtgt gagccaccgc gcccggccct ttttttgaga cggagtctta ctctgtcccc
40681 caggctggag tgcaatggca caatatctgc tcactgcaac ctccgcctcc cgggttcaag
40741 cggttctcct gcctcagctt cccgagtagc tggaattata ggcgcccgcc actacatctg
40801 gctcattttt gtatttttag tagagagagg atttcaccat gttggccagg ctggtcttga
40861 actcctgacc tcaagtgatc cacccacctt ggcttcccaa agtgcttgga ttacaggcat
40921 gagccaccgc acccagccct ctttactttt taaaaaatgt ttttattttt atttatatat
40981 ttatttttga gacagagttt cactcttgtt gccaaggctg gagtgcaatg gcaccatctc
41041 tgctcactcc aacctccgcc tccccggttc acgagatttt cctgcctcag cctcccgagt
41101 agctggaatt acaggcatcc accaccacgc ctggttaatt ttttgtattt ttagtagaga
41161 tggagtttca ccatgttggc caggctggtc tcgaactcct gacctcagat gatccactgc
41221 ctcggcttcc cagagtgctg ggattacagg catgagccac cgtgcctggc ttatttttat
41281 ttattttatt atttattgtt attattattt gagacagagt ctccctctgt tgcccaggct
41341 ggagtgcaat ggtgtgatgt cagttcactg cgacctctgc ctccYgggtt caagcaattc
41401 ttctgcctca gcctcccaag tatttgggat tacaggtgcc tgccaccaca gccagctaat
41461 tttttgtatt tttagtagag atggccatgt tggctaggct ggtctggaac tcctgacctc
41521 aggtgatcca cccaccttgg cctcccaaag tgctgggatt acaggcttga gccaccatgc
41581 ccggcctatt tatttcattt ttatttattt attttctttg agagaaagtc tttgttgccc
41641 aggctggagt gcagtggctg catctcagct cactgcagcc tccacctccc ggattcaagt
41701 gattctccag cctcagcctc ccgagtagct gggactacag gcgaaagcca tcacacctgg
41761 caaacttttg tatttttagt agagacaggg tttcaccaca ttggccacgc tggtctcgaa
41821 ctcctgacct caagtgatcc gaccgcctca gcctcccaaa gtgctgggat tacaggcgtg
41881 agccaccgtg cctggccttt atttttattt agagattggg tctcactctg tcaccctgga
41941 gtgcagtggc tcaatcatag ctcacttcag cctcaaactc ctgcactcaa gcaatcctcc
42001 tgagtagcta ggactatagg cacccaccac cacacctcgc taatttatta aaaattcttt
42061 gtatatagag atggaggtct cactacgctg ccgacagtgg tctcaagaac tcctggcttc
42121 agatgctcct ccactWtggc ttcctaaagt gctgtgatta caggcatgag ccacagtgac
42181 cagcMcccct cctctctaat ttcctttatg gtgtcatctg gacaatactc cttgcaagct
42241 taccatgggc aaggtatcat tctaagcatt ttgtgcataa tactcaacta ctcaagccaa
42301 ctgcacagct gcctagcagt tcattatgag tgaatgtttg tgttccctgt ccRttcatat
42361 attgaaattc taactctcat tgtgactgta tttggagaca aggcctttat ggaagtaatt
42421 aaggctgaat gacRtcataa gggtatggcc ctggtccagt aggattagcg ttcttatgag
42481 aagtgacacc acaaagaaac aagcttacta gtcacggtcc cagccagtgt tcaaatccca
42541 aacacctgct ctctgagccc tgactattgc tttgctagca ttacacatct tatggtttgg
42601 ctgttaattc ttcatcacca gcaccagagt ccaggctggc aaagggctag gaaaccgatc
42661 atctgcctcc tctacaccca gaaccctgtg tggtgaccca aaacaaatgg aaagaaccaa
42721 cctcagatga aatttgaacc caggtctgta gcctctgcct cctccacagt aggagtttgt
42781 gagaatgtcc accaataact gtttattaac taaattcctc cacctttcca actccacaag
42841 gctcaactat tgccccaata tcccacagtg ggctccctgt cgtggcgatg aagccttgct
42901 ttgcctcact actggcttag gaaggggatg aagttctgct gtctcactag gcaagcaaag
42961 attcaatttc caaaaatcct aggctaggac cctggggcag ggatgaggag aaaaaggagg
43021 cacctcaatc ttcccatctc taaacaagca gtcaccacac aggcctcacg gccaggagtg
43081 acgtagtgag cggatgaacc ctgcagaagt gagcgctaat catatcctag gcctcctacc
43141 gggccagttc acaacctgat cccaacctac ttttccagcc tcagctccag cgacagagtc
43201 ctcctcccgg cagcgaccga acctccagcc acaattctca atcctccacc tctagatgca
43261 gtttcctggg ctcggggagc ccttgcctgc cttctccaac gggaaagccc ctttacatcc
43321 ttcaagaacc cccttcccta gtgccctcag gaatacttta tggcagtaga gaggtaaggg
43381 ccccacgccg ctctggacct cagtttcctc atcagtaaaa tggggtccct taagtttgtg
43441 ggagtttgaa gggcggtggg gcctacgaag cgcgccgagg agccgagagt tgcagaaacc
43501 cggagctcct cgctcctcgc aaccgtctgt acgcggcgcc cccgccagcc aggcagcccc
43561 tggagggcag gaccccggtt ataagcctca gaaaacgtgg cttcggagga cgtggcaagg
43621 aggactgaac gaaggatgag gagatgaaca aatgaatgga cggaaggccg aatgagggac
43681 aaaggcttgc aagatggcgt tctctaggac cgcgggagtg gtggccgggc ctcagcaggg
43741 gagggggccg ccggcgcctg ggatcttgca gcgcgggcca cgcgaccggg acaaaaaccc
43801 aaaacatggc gggctctagg acccccggga ccacaccgcg ccgggccagc agccgcgcgg
43861 gccgagcctg ggtgtccgca gcccagaacc gcggagacag ccggcgggtt ctaggacctg
43921 ctgggcccgc aatgcgcccg gggccgctca cagcccgccc cgcccgcgct gctcgcgctg
43981 ggccaacccg gcccgccctc ggcgcccgca gggaaactga ggccagctcg gatcgtggcc
44041 gcgtgggagc tgcccggggc cctcgcggct tcccgccgac gtttcctacc tgatgagact
44101 tgtgctgact ccgtggcgtc ggcgtcggct cctcgcaccg acggagcccg gaccctgcca
44161 aacaggggcc ggcgctagga cccagcgggg ggcggggagg tgggaccggt ggcgcggcga
44221 gcggaagtga gggatcttcc tcagctagga aggaagggaa agttcccggg gaacctccag
44281 cctatggcgg cagagaagca tcttgcaaga ggtctctgtg tgtgctgagg caaggggacg
44341 ccaggcaggc tgacggtata cgcccgcctt gtgttagtct ggggccaggt caccggcaat
44401 gtcttcaaga accagaaggt gggaggacaa aaaggccatg ctcaagctct gcaaaaccta
44461 agtccaatta tattccacaa catttccttt aatagcaagg agtgggtttc acgaagtttt
44521 ctccatctgc cttgggaaaa gtcctccaga gagccccacg aaaagtgtca ccaaagtacY
44581 gtggccaggg ctgtagaatc tttttctccc tttctgcgcc tattcaatat gcaccggaac
44641 gattctagct gctcttgccg gaagtgtggc tagcWtagcg tgcaaagacg ccgcgttgtg
44701 acctacgtgg gcggagtcag ccgtgcagac ctagaactta gcgccggaag tgtgtctgcg
177
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
44761 taaaactgcg gctcggaggc gggacaggca gtgctcccga agcggaagtt tcgcggggaa
44821 gcttttgcac ctaaggacac ttcctgtttc ctagtaacaa taggaagtgt ccgtagagct
44881 gggagtagac tcctggctca tgccagctgc gccctctttt ctctacctcc ttcctcttcc
44941 ccccttctcc tttccctctt tcccttccct cccacctccc gggaaccctg gctgagtgtg
45001 cgtgtgtggg agcgcgagag ccccccgaca gccacccctt ggggcgcgcg gctgcagtta
45061 gggtaagggt cccagtgcgc aggcgcgctc ttgttcgcgg tccccaactg acgcgcccgc
45121 ggcgggaagg agagggggcc gccggtgcga ggtaggcgtc ctgagaggaa attggcagac
45181 gagatgagtg aggtcagagg acttcattgg tggttgacta ggggagggga cttttgatca
45241 aaggggcttt gtgaggtgag aattccgggg gcaggttcag tgaaggggtt tacggtcgga
45301 gacagttgta ctggggtccc atgggaagaa gagtcagagt tggagaagga caagacgatg
45361 tagggggcat gagagagcaa gggctttggg atgaagaccg cctcagtcag ggggtttgct
45421 gttacgtgaa ctagaataac aggcattgcc gtgtgttctg aggattgagt aacacaatga
45481 atatagttag cacagtgcct ggcacatggt aatataatac tcttcatgag ttgctgtcat
45541 catcagtatt aagagaggag gcagaagaaa aaaatgagaa agactggtga gggtaagatc
45601 aggaggcatg ggagagaaaa ggtaggcaat gaagtgacat tacaacctgg tattgatgtt
45661 attcccaaga tggaagatag tttgagttca agggaagtag ataaaagaag tcgctaagag
45721 agtcttttgg ggttttttKt ttttttgaga tggagtctcg ctctgtcacc caggctggag
45781 tgctgtggtg ggatctcagc tcactgcaac ctccacctcc caggttcaag cagttcttca
45841 gcctcagcct cctgagtagc tagaattaca ggtgcccacc accacgcccg gctaattttt
45901 gtatttttag tagagacagg gtttcaccat gttgtccagg ctggtcttga actcctgaac
45961 tcaggtgatc caccagcctc ggcctcccaa agtgctggga ttacaggcgt gagccaccac
46021 acctggccca gttaagagag tcttaactct cttaactctc ttgtcacaag gaaaagagac
46081 cttgtgacac tgaaatgact cggggtggtg ggggaacaag ccagcccttt ccctgaagga
46141 ggcctctaac ctctcctctc aggtcctcag ctattaactg gaggaaacag ctgctttttc
46201 agtgcttctc agctactctg tttagctgag agatgaagta ggaagatttg gacttctctt
46261 attgaaaggc ctagagaagg ttttggtgtc cttttaagat gtcacagaaa atttttgttt
46321 caggattgta gggagcagat tcctactgtt cttaaaggac agtaatgcct tttgagtctg
46381 gtctgaagaa cataacaggt ctgtgatcag aagtaggttg catctctctc aactttaaYt
46441 tccttagcta tacctgtagg gatgacttaa gcctagggga gctcctatat ttgggaagct
46501 tgtgcacagg gaagccttaa atgatggtgc ctgcagattg gatctagtag aaattaggtc
46561 cttgggcatg gatgcttggg gaacctctca gtgacctcag gtgaacttgt tgctcgtaga
46621 gccaagaggc gaagttaatt caggccttcc ttttgaccac tgccccctct tcctaggcct
46681 tggcccctcc accagaggaa ggtgctgcca cgtgtctgct ccttctgaac ctccaggttt
46741 ctgctacgtt gccccatgga ggacacaccc ccctcactca gctgctccga ctgtcagcgc
46801 cactttccca gcctcccaga gctctctcgg caccgagaac tgctccatcc atctcccaac
46861 caggacagtg aggaggctga cagcatccct cggccctacc gttgtcagca gtgtgggcgg
46921 ggctaccgtc accccgggag cctggttaac catcgtcgga cccacgagac tggccttttc
46981 ccctgtacca cctgtggcaa ggacttctcc aatcccatgg ctctcaagag ccatatgagg
47041 acacatgctc ctgagggccg ccgcaggcac aggcccccac gccccaagga agccactcca
47101 cacctccagg gtgagacggt gtccactgac tcctggggcc aaaggcttgg ctctagtgaa
47161 ggctgggaaa accagacaaa acatacagaa gagacacctg actgtgaatc tgtacctgac
47221 cccagggcag cttcgggtac gtgggaagat ctgcccacca gacaaagaga aggcttggca
47281 agccacccag gtcctgagga tggtgcagac ggctggggac cctccactaa ctctgccaga
47341 gcccctcctc tccccatccc agccagcagc cttcttagca acttggaaca gtatctggct
47401 gaatcagtag tgaacttcac agggggccag gagcccaccc agtcccctcc tgctgaRgag
47461 gagcggcggt acaaatgtag tcagtgtggc aagacctaca agcacgccgg gagcctcacc
47521 aaccaccgcc agagccacac gctgggcatc tacccctgtg ccatctgttt caaggagttc
47581 tctaacctca tggctctgaa gaaccactct cgactgcatg cccagtatcg gccttaccac
47641 tgtccccact gcccccgtgt cttccggctc ccccgggagc tgctggaaca ccagcagtcc
47701 catgagggtg aaaggcagga gccacgctgg gaggRgaaag ggatgcccac caccaatggg
47761 cacacagatg agagcagcca ggaccagctc cccagtgcac agatgctgaa tggctctgcg
47821 gagctcagca cctctgggga gctggaggac agtggcctgg aggaataccg gcctttccgc
47881 tgtggggact gtggccgtac ttaccgccat gctgggagcc tcatcaacca tcgaaagagc
47941 caccagacag gtgtctaccc ctgctcactc tgttctaagc agctgttcaa tgcggctgcc
48001 ctcaaaaacc atgtgcgggc tcatcacagg cccaggcaag gagttgggga aaatgggcag
48061 ccatcagtcc caccagctcc cctgctgctg gctgagacca cccacaaaga ggaagaggac
48121 cccaccacca ccctggacca tcggccctat aagtgcagtg agtgtggtcg tgcttaccgc
48181 caccggggga gcctggtgaa ccatcgccac agccatcgga ctggagagta ccagtgctca
48241 ctctgtcccc gcaagtaccc caatctcatg gccctgcgca accacgtgcg ggtacattgc
48301 aaggctgctc gccgaagtgc agacatcggg gctgagggtg cccccagcca cctcaaggta
48361 gaactcccgc ctgacccagt ggaggcagag gcagccccgc acacagatca ggaccatgtg
48421 tgcaaacatg aagaagaggc cacggacatc accccagcag cagacaagac agcagcacat
48481 atctgtagca tctgtgggct gctctttgaa gacgctgaga gccttgaacg tcatggcctg
48541 actcatgggg caggggaaaa ggaaaatagc agaacagaga ccacaatgtc acctcctagg
48601 gcctttgcct gccgagactg tggaaagagc tatcgccact caggcagcct tatcaaccac
48661 aggcagaccc accagacagg agacttcagt tgtggggcct gtgccaagca cttccacacc
48721 atggctgcca tgaagaacca cttgcgccgg cacagtcggc ggcggagcag gcggcatcgg
48781 aagcgggctg gcggtgccag cggtgggaga gaagccaaac tcctggcagc ggagagctgg
48841 acccgggagc tagaagacaa tgaaggcctg gagtctcccc aagacccttc aggggaaagt
48901 cctcatgggg ctgaaggcaa cctggaaagt gatggggact gtttgcaggc tgaatctgaa
48961 ggggacaaat gtgggcttga gagggatgag acccatttcc agggtgataa agagagcgga
17R
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
49021 ggcactgggg aaggactgga aaggaaggat gccagtttac ttgacaactt ggacatccca
49081 ggtgaggaag gtggtggcac tcacttctgc gatagcctca ctggggtgga tgaagaccag
49141 aagccagcca ctggccaacc caactcctct tcccactctg ccaatgctgt cactggctgg
49201 caggctgggg ccgctcacac atgctctgac tgtgggcatt ctttccccca tgccactggc
49261 ctgctgagcc acaggccctg ccacccacca ggcatctatc agtgctccct ctgcccgaag
49321 gagtttgact ctctgcctgc cctccgcagc cacttccaga accataggcc tggggaggcg
49381 acctcagcac agcctttcct ctgctgcctc tgtggcatga tcttccctgg gcgggctggc
49441 tacaggcttc accggcgcca ggcccacagc tcctctggca tgactgaggg ctcagaggag
49501 gagggggaag aggaaggagt ggcagaggca gcccctgcac gcagtccacc actgcagctc
49561 tcggaagcag agctgctgaa tcagctgcag cgggaggtgg aagcgctgga cagtgcaggg
49621 tatgggcaca tctgtggctg ctgtggtcag acctacgatg acctggggag cctggagcgt
49681 caccaccaaa gtcagagttc tgggactact gcagacaagg ctcccagccc cttgggagtg
49741 gcaggtgatg ccatggagat ggtcgtggac agtgtcttgg aggacatagt gaattctgtc
49801 tctggagagg gtggagatgc caagtctcaa gagggagcag gcaccccctt gggagacagc
49861 ctctgcatcc agggtgggga aagtttgttg gaggctcagc cccgcccctt ccgctgcaac
49921 cagtgtggca agacctatcg ccatgggggc agcctggtga accaccgcaa gatccaccag
49981 actggagact ttctctgccc tgtctgctcc cgctgctacc ccaacctggc tgcctaccgt
50041 aatcatctgc ggaaccaccc tcgctgcaaa ggctctgagc cccaggttgg gcccatccca
50101 gaggcagcag gtagcagtga gctgcaggtt gggcccatcc cagaaggagg cagcaacaag
50161 ccccagcaca tggcagagga ggggccgggg caagcagaag tcgagaagct ccaggaagaa
50221 cttaaagtgg agcccctgga ggaagtggcc agggtgaaag aagaggtgtg ggaggagacc
50281 actgtgaagg gggaggagat agagcccagg ctggagacYg ccgagaaggg ctgccagact
50341 gaagccagct ctgagcggcc cttcagctgc gaggtgtgtg gccgatccta caagcacgcc
50401 ggcagcctca tcaaccaccg gcagagccac cagaccggcc actttggctg tcaggcctgc
50461 tccaagggct tctcaaacct catgtccctc aagaaccacc ggcgcatcca tgcagatccc
50521 cgacgtttcc gctgcagcga gtgtgggaag gccttccgcc tgcggaaaca gctggccagc
50581 caccagcggg tccacatgga acggcgtggg ggtgggggca cccgaaaggc gactcgggaa
50641 gatcggccct tccgctgtgg gcagtgcggg cggacctatc gccacgccgg cagcctcctg
50701 aaccaccRgc gcagccacga gacgggccag tacagctgcc ccacctgccc caagacctac
50761 tccaaccgca tggccctgaa ggaccaccag aggctgcact cagagaatcg gcggcgacgg
50821 gctggacggt ccaggcgcac agctgtgcgt tgcgccctct gtggccgcag cttccctggc
50881 cggggatctt tggagcggca cctgcgggag catgaggaga cagaaaggga gccagccaat
50941 ggccagggag gcctggatgg cacagcggcc agtgaggcga acctgactgg cagccaggga
51001 ctagagaccc aattgggtgg tgctgagcca gtaccccact tggaggatgg agtcecaagg
51061 ccaggggagc gcagtcagag ccccatcagg gcagcaagct cagaagcccc agagccactg
51121 tcctggggtg cagggaaggc aggtgggtgg ccggtaggtg ggggactggg gaatcatagt
51181 ggagRctggg ttcctcagtt cctaactagg tcagaggagc cagaggacag tgtccacagg
51241 agtccttgcc acgctggtga ctgccagctc aatggaccta ctctgagtca catggatagc
51301 tgggacaaca gagacaacag ctctcagctg cagccaggga gccactcctc ttgcagccag
51361 tgtggcaaga cttactgcca gtcaggcagc ctcttgaacc acaacaccaa caagacagac
51421 cgacactatt gcctgctctg ctccaaggag ttcttaaatc ctgtggccac aaagagccac
51481 agccacaacc acatagacgc ccagaccttt gcctgtcctg actgtggcaa agcctttgag
51541 tcccaccagg aactggccag ccacctgcag gctcatgccc ggggccacag ccaggtgcca
51601 gcccagatgg aggaggccag agatcccaaa gccgggactg gggaggacca ggtggttctc
51661 cctggtcaag ggaaagccca ggaggcccca tcagaaaccc ccagaggccc aggagagagt
51721 gtggagagag ccaggggagg acaagcggtg acgtccatgg cggctgagga caaggagcgg
51781 cccttccgct gcacccagtg cgggcgctcc taccgccatg ctggcagcct gctgaaccac
51841 cagaaggccc acaccacagg gttgtacccg tgctccctct gtcccaaact tctccctaac
51901 ctgctgtctc ttaagaacca cagcaggacc cacacggacc ccaagcgcca ctgctgcagc
51961 atctgtggca aggcctttcg gacagctgcc cggctggagg gccacgggcg ggtccatgca
52021 ccccgggagg ggcctttcac ctgcccccat tgtccccgcc acttccgccg ccgaatcagc
52081 ttcgtgcagc accagcagca gcaccaggag gagtggacgg tggccggctc cggtaggggg
52141 catgaagggt cccaggagga ggtgggcaca cagtggaggg ggaagtccag ccccaaagtc
52201 ggtgggggag caaggagtga gaggagagag ccccggggat tctaagaggt gggtgggggc
52261 ttggctatgg ggtgagagaa gtagcttgag gatgtgctga gctgagcacc cgcaagtcag
52321 gtataacaaa tagcagggtg ggttgggcag cacgtggggg cgtggtcagg ccgaggctgc
52381 tacctgggct cctccattac actgtagcca gaatggaatg gtctttctgt tcaggggaag
52441 gtcactgggt accccctggc tgctgtgtct ggaaaccctc ctgagtcagc cagtaaagta
52501 atgacttcca gagaaaaaga ggaagccatt ggtttggtct aggttccatt ctttcctgga
52561 gcaggccggg tgccagggaa caagggatgg ggcatgggct ccacggcttc cctgctgact
52621 tggccacgga aactggttca ctggttggca ccctactccc tgtccctctt tccctgcgcc
52681 ttgtctctgc tgctcctctc cttggaaact agacctctgg tccttccctg tcagtgttgc
52741 tcccatctct tctctaacct ttattcagcc ccttttccct ctgctgccaa cggccttttt
52801 aggatccaac caaaccaccc tttctacctg cgcaccctgc caccctctgc acacctttaa
52861 ctggaggact gagtcacaga taattgtttc cttgaagtcc aggcccagct gcagcaacaa
52921 cagtcattag cccgtgtcac atccctgatc agagggcatc tccgtgggga atcgcctcca
52981 cccagcactg ctggaagccg cRgctgccag ggagtggggc ggccggttcc ctcagcagga
53041 cctgggctgg cctctccacc tccYctagta gaggcggacc cattccatct agtggccacc
53101 gagggtgggt ggccctgaga tggtgggccc ttgacaggcc ttgtcagagc agagggcagg
53161 tgggagtcac ctgaaagctg aaggaatggc tttaaggata gaagatttct catgacctca
53221 agggatatga gggaggagcc agtttgccag ggctgggaaa ataattagga ggcctagaat
179
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
53281 ccctgttctc atctgggcct ccggggccag gggcagggga atggcctgca gggctgggag
53341 ggggtacacg ctgtgcgggg tctgcccctc agttggtgac ctcctctctc tctcccccca
53401 ggagccccag tggcaccagt gacgggcaga ggggacttgc cattgccccc tccacccacc
53461 cccacgaccc cactcctgga tccttcaccc cagtggcctg cagacctcag cttctccctc
53521 tgaacttcaa gtctccaaag atcagaatct gggggaggga gcgcgtgcag ggaggggctt
53581 gatctccaca ttttctcagg agtagttcgg gcatccccat atcttctcct ctccccYtgt
53641 gaagaggacc cagatctggc ttctttccca aggagggggt ggggtgttcc tcgcgtccct
53701 gtccttgaag gacctccttc ccccagcctc atcaccgtgc tcttctcagc gccaccctca
53761 gcagccagat tgcaacacca gggagaggcg gatgcagagc cccaccggtg ggaaagttgc
53821 ctgtggaagg gagccttttg ctacaatttg taacttattt tctaaagtct attttgtaac
53881 aatttattta agtttaaaaa aaggaaaact gctgcccccc aaaaaaagaa attttcaaaa
53941 caacgtggct ggcgtgattg tatctgaaag ggtaaaggag gaggaaagct gagacgcctg
54001 cttggtagca gagttgggtg tgggagtgtc cacagacacc cctgtcctgc agggtgggga
54061 gtgggcacct gtggccccag gcaggttcct tcccacagct gctgggcttc tgggcctgcc
54121 ctggtgcctg gaatcacaca tgacagggtg gggaggacag gggcagtaat gccatttgcc
54181 tgcctgcatt ctcttgtcct gagaatggcc aggtcccctg tcagcagctg gttggttggc
54241 ctgtggggaa ggaaggaggg tggagttgtc ctcatcctca cggctttggt ccctccctcc
54301 ctccccattc ctcgaaggaa cagggtctgt cttggccgcc atgacagatg agaatactga
54361 ggctcaaagc ggttgagcag cctgctccaa gtcacacgat gacaaagaac cagaatctga
54421 atcaaatggg tctgcctgtt gctccaccct acccaaggca gctggagtgg gttagaacgg
54481 cacgttctca ctggagagag aagggtcctg gagaggcagg gtttggcagg aggccccggg
54541 gccacatact tatgttggcc aggcagcttc caggctcagc ctcgggctct ggttcctcgg
54601 cgaagtagac ctgccagtcc aaactgctga cccagtcctc ataggcaggg agcgcggtga
54661 agaccgccgg cctggcgggg ccttggcaag catctccgaa gctgtgcagc ccggccagga
54721 accatgtgcc cctcacctca tgcaccagtg gtgccccaga caggccctgt caggggtcag
54781 gtgacactgg gtgacttttt ataggcagct gtgcaggacg gtagagcagg ctagggaacc
54841 tctggctgtt tgggggctaa ttggcaaaaa ggcttagttt caggtgggag gtgggtccag
54901 aggccaagcc tggaaggtcc gttcttgagc tattgggtga ggggatgcca ggggcctgtt
54961 aggtaaggtg tgggggcctg gggctcacct cacagctggg cagctcaccc acagcactgg
55021 tacacaccat ccccggcaga atagggctgc catcaccccc aggagctgca tgcagccggc
55081 tgcaggccct aggccccagg agggtcacgg gcactgtctg gagggagctg atgcctgtgg
55141 agcaagggaa agctggctgc cccggcctgc aggttggatg gacagcagcc ctggccctgt
55201 gcccacctac ctgctcctgg gcgggcccgt cccagaaccc agccacgctc cccatcaggc
55261 aggtggtggt cagSataggg caggcagagg ggccgcaggc tggctcccag tgtcacaggc
55321 tgggccagca gcaggagggc catgtcgtag cccccctcag ggtgggtgta ggctccatgc
55381 aggatgagct gcttcaggcc ccactcctcc ggtctggtcc ccagccctac gctccattcc
55441 tctggggcct ggcgcctgtg cagaggcagt gtggacggca ccaggtgaca gggctgccgg
55501 caggggcagg gggcacagca gagagggatc caagactcac ccaatgaagc agtgggcagc
55561 agttagcacc gcctcctctg acaccagggc tccgccacag gccagctgtc cctggtgcat
55621 cagcctggcc tcccagggcc atggggaggg tgctcctgcc tggggacctg ctgtcctcaa
55681 ggatccacag gctgaaacag ataagtgcct catctgactt cttgctaagc acttcccact
55741 tcccatcaaa tcctcacagt agcttttgaa attgaatttt gttcccctca ttactgaagg
55801 taaccaaagt tccagagagg ttaaRagact tccttgctct gtaactacag gcaagtggca
55861 gacttgcgat tcgcaagcca gggcctgagc ttttagccct gttggtctga tgggacccct
55921 gtcccagcct cgtgggaatg cccctacttg tcactcccct gctccacccg tctgcaccct
55981 ccaggMtggg cctgcctccc gtattcctga ctcattagtg catctcttat catcaagaca
56041 taatatccgt cccaggaatt actttgcatt caatttgcat gccagtcact taatgccgga
56101 attctgactg ggagccctac cctgtgcagg ctcgctgggt ccctgctgga agcctgccac
56161 ttccccagaa acccaagtca ggtctcagag attcctcttc tcacctaaac tccaaacctg
56221 tagagttcca aagtgcctgt gcctctcagc cctaacaggg ctgttcccat cccagggggt
56281 gaaagagccc cctaattagg ctcggcggtg tggatgccta tgccagttct ctagtcctaa
56341 ctgaggtttg cttcacagtg gcttctgccc actcccagca tgccccactc gtacctacac
56401 agctgtcctc atcactcatc tccggggtct ctgggctctg ggccaggaaa gctgccccct
56461 gaactcgagc ctgcagccag gaactgtgag cagctgtgtt ggtcagcagc acaggagcgt
56521 cctcctgggc acagcttgat gcaaagctga tgatgccagc ctgaacccag tgtccgtcag
56581 gctcgaggca cagcacaggg cccccggaat ctccctggag ccaggcaaca aagccaagga
56641 cagatgcctg agcccagcta tggcagacac cctctgattg cagStctttc ccccagtcct
56701 gtgagtccaa cccccagccc agggttaggc ccctcccctt ctgctccttc tcttctccct
56761 atcagacctg acaggggccc tgcaccccag gctgggggcc cccacatagc atcccaggcc
56821 gggccgggtt ggacaggtgt cgctggtgca gctggttgta gatacagtta catgtggggc
56881 gactgatgag acgcaggcgc agattgcgta gggtcccagg agctggtgaa agagacgggg
56941 ctggggctag agtctgggat ctgggaagca agggagactg gaagccagga cagttgggag
57001 ctgagactgt gacgctgggc aagcagagtg cctctccgag cttcagtctg ggccagcact
57061 taccatcact ggtgtcctga tcccagccag tggcccagca ggaggctcca aaggggaagc
57121 gatgggcggg ctggggcagg cagaggggtg tgtgggtcgt ggggtgggcg agctgcagca
57181 gggccaggtc tgagccctgg ctgtagtggt tataggccct gggcaactgc agggcagcca
57241 cccccacctc ttcggcccca gggctgagtc cctcacgctg cagagaaccc aggaccactg
57301 accaggaatt cagttctgtt gctgctgccc tggaagggga aggacaaggt ccaggctgct
57361 gagtgcaagg gtagacagtg acaccatggg agaccccaca ggtaggtgga agatagcaga
57421 gagcactcca cggctttttc tttttgaggt agggtctcgt cctgtcgccc aggctgcagt
57481 gcagtggcat gaccatgact cactacaagc cttgacctcc caggctcaag cgatcctccc
~Rn
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
57541 acctcagcct cgtgagtagc tgggactaca ggcgtgcacc accatgcctg gctaatttta
57601 cagtttttgt agagacatgg tcttgctatg ttgcacaggc tagtcttgaa ctYttgggst
57661 caagcgatcc tccctccgtg gcctcccaaa gKgctgggat tacaagcgtg agctactgtg
57721 cccggcctca gtctgtgact ttaagcgagg tcattccctt gcaacctgag ttttYtgatt
57781 tgtaaatctt ctaccatgaa aggtggttgt tacaattaac ttgacaatat gtacccagta
57841 catggcagat tctcagtaaa tggttcttta ttattactct agaggcaggg actagccttg
57901 tggggcttga gacaaattgt ccactcccca gagccggaga ccctgcctga ccccagccat
57961 gacttacttt tcaaagcagt gggcagcagt gaggacccag gtgtctgcca ccagggagcc
58021 gctgcagatg tgggctcctt gcctcctcac actggcctgc cagggccact cgccagggac
58081 tgtgttgccc tcctgaggct tgggggggcc ggggccacgc tgtccacagg ctgtggaaag
58141 gagttagtca cactgacagc agatacctgt cctgacctca ccccagtccc aacccagacc
58201 caagtaagac ctaggccccg tgtccccaga ccttaccacg ctgagcggct tgaagacctg
58261 ggaagagaga gacacaggta agatgcaggg actccaggcc tgcctagctt tggggaggag
58321 aggggatggg ctggggggcg ggcccagggt ccttgcctgt gactctgatt accttaccag
58381 cccctcccag aatgaaaata tttatatgag gccgagacag cttttattgg aacctattca
58441 gtgtgcacac tcagtaatta attctccttc agctgtgctc agcactctgg ggcttggggt
58501 tcagcttggg agtaggccag ccctcctcca ggcttcagaa cccccaactc ctgcccccgc
58561 cactgagtca gccaggcggc ctgtgtgtgt agagagcatt agcttaattg tcctcttagc
58621 agcagaggcc taagaggaag gattagaggc ctgcatcatt tccaagtggg gagggcccca
58681 agaaatggag acttacctca ccctggtcta gagactcagt cttccccacc ttcccagaaa
58741 ctgtctgaga gcccgccaga gagagggccc ctgcccaccg cccctcacag gcacacaggc
58801 accccatgag acagctgagc caggctgccc agaggatgga tgaaaagaaa gggaaactga
58861 ggccagagga gcccagagtt tgcctgacgt cattgtggaa gtcgaggggg aggcaggcac
58921 aggacacacg caggcagcac ctcacacaca cacaagacca caggccccgc caacgcaaac
58981 tgcagctggc ccgagaaaat ctcatccatg ttgacacagg tggccacata taccacccca
59041 cagagtccta cggagttaca tccccactgg tggactgtcg cccacaggcg gtccccccac
59101 caagaagcag ggactgctgg ggcagagatg gcccctgagc ccccaccagg ccacacccat
59161 accccagcac atggcggctt accctccatg aggactgtgg cacccgcgat gagcagcact
59221 gggccccagc accacttcat gctgccccgg gccactctgc cacctgtgct ccactctgag
59281 agaggccacc tgggtctccc tggctccacc tctgctccac ctccagttgg ctaggattca
59341 gctgtctgct tgccctagcc agcagttcct agctctgggg ctgtggctgt gtgaccctag
59401 gcttgtcacc acccctctct gggccttagc cgtggagcca aattgtctgg gtttaaatct
59461 gtttccttac tacttgagtg gcctgggaga gctaacctgc ctgtgccttg tctgagaaac
59521 agttgttgaa tggaaaacgt ctcaatgaca gcctggcaca gggcaagctc actcccagca
59581 tgaaagtgcc atcctggaac catggagggc acgtgggcag ctcctcccag tccccacttt
59641 tcatttccac tcccccactt ctcttccagc cagggctagg tgggtttcac atgccttctc
59701 ccaggtctgc tcagagacag tgtgtgccag gctgagtggt gaggagccag cacccactgc
59761 tgcttgctcg ctgtgagcct ttggcaaatc ccagtatctc cctgggcctg ctggcttcct
59821 gggaaacagg gtgacagcgg tgcccacctt cctcataggc gcagaggact cggtcaggcg
59881 ggaagaacac tttggcgcgt gcctcctctt cctcttccca ctcctaactc aggctggccc
59941 agaccattca agaaccctga ccccaagaca gaggcagctg tgggtaaggt ttagcatata
60001 ttatagatgc tggccgggca tggtggctca cgcctgtaat cccagcactt tgggaggcca
60061 aagcaggcag atcacctgag gtcgggagtg cgagaccagc ctgaccaaca tagagaaaca
60121 ctgtctctac taaaaataca aaattagcca ggcctggtgg cgcatgcctg taatcccagc
60181 tactcgggag gctgaggcag gagaatcact tgaacctggg aggtggaggt tgtggtgagc
60241 cgaaattgtg ccattgcact ccagcctgga caacaagagc gaaagtccgt ctcaaaaaca
60301 aaccaacaaa aaaaaaagca tatagatgct ggagccagat ggaccaggtg gattcaaatc
60361 ctgagagcct cctgcagtag ctgtgtgatc tcaggcatac taattagcca ctctgtccct
60421 atcatcaaga tagggataat aatagtaccc acctcataaa tcactgtaaa gattaaacga
60481 gtttaacacg ttaacacaag tttttttttg ttttttgttt ttttttggag acggtcttgc
60541 tctgttgctc aggctggagt gcagtgatca tggctcactg cagcctcaac cttctgggct
60601 caagcagtct tcccgccagc ttcccgatta gttgggacta taggtacata caccaccatg
60661 cctggctaat tttttttttt tttttttttt gtagagacag gggtctcact attttgccca
60721 ggctggtctg gaactcctgg cctcaagtga tcctcctgcc ttggcctccc aaagtacagg
60781 gattacaggt gtgagccact gcaccctgcc aacacaagtt cttaaacagt gcttggcatg
60841 taggtaagtg gtcagggcat aataggcaaa acaaaaacct tcacaacctg gccctgaccc
60901 tccaaggcta ccaatactgg cttggaatgg tcgataaggc aactggaggg gttaaagtta
60961 aactcaagga agaacttccc agcaagcatt tacagaacca gaagggcagc ctgcccttcc
61021 aggtgtgtgc tcagccttcc caggagagga ggcctggctc ctgtgggcag caggagcgag
61081 ctgccagcct gtttcctggg ggtgggggcc aatggtgccc caggccttgc tgactccaca
61141 cactggagat gagactaccc ataaccaccc ttcccagcag gccctccact ctccctctga
61201 ctcacccttc ccagctccag agaaggcaac accgagggag gcccagcacc acagtccatg
61261 gcagacacat ggttcagact tggctgattg atctaagaaa ctttattgct caRaaccttc
61321 cctccctggg caatggaaag agctttggag accagcccat ggggacagag tcagaggcac
61381 tgggtgtaaa aaagagcgag cgtgtggcac atttggtcca ttgtcatgtg Ygggtatggc
61441 aggaggaggg ggtaatctag aagccccaca tctagggcct tctagggacc cagatatgcc
61501 cccttaggca aggctcacat gccaaagcaa agcagatgag gtcagcctgg cttgggttga
61561 gggctcagtg cctcttagcc ttgccctggg gttcttggac cttccggaaa ctgagccaca
61621 tcaggctcac gttgatagca taggtggtga tacaaacaat gcagaaatca tagagcacga
61681 agaacaggat ccaggccaRg tagacagaac cagcgagaga caccagggag ctcagcagca
61741 tcaggacaga ggcccagcgt gtccgcaggc aacctgcaag gcagaagagg gtccggtgtg
1R1
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
61801 ggcttcaggc actggccacc tcccgaacac tccatgatgt cactgcacta cctagatgcc
61861 aggagctgct gggaagggtc tctaaaacaa gaggctccag ggcagatgtg gtggcttacg
61921 cacgtattcc aaacactctg ggaggccgag gtgggaagac tgctttaggc taggagttca
61981 agaccagctt gggcaacata gaaagacccg atctctatca aaaatttaga aattcagctg
62041 ggcacggtgg ctcatgcctg taatcccagt attttgggag gccaatgggg gtggatcacc
62101 tgaggttagg agttegagag cagcctggcc aacatggcaa aaccccatct ctaataaaaa
62161 tacaaaaatt agccgggtgt gttgatgggc acctgtaatc ccagctgctg gggaggctaa
62221 ggtggagaat cgcttgaacc taggaggtgg aggttgcagt gagccgagat catgccacca
62281 cattccagct tgggcagcaa gagcaagact tcgtctcaaa aaaaaaatgc ccggtgaggt
62341 tgactcacgc ctgtaatcct agcactttgg gaggccaagg cggatggatc accaggtcaa
62401 gagattgaga ccatcctagc caacatggtg aaaccctgtc tccactaaaa atacaaaaat
62461 tagctgggcg tggtgacacg cgcctgtagt cccagctact caggaggctg aggcaggaaa
62521 attgcttgaa cccgggaggc agaggttgca gtgagccaag atcgtgccac tgcactccag
62581 cctggtgaca gagcgagact atgtcgcaaa aaaaaaaaaa aaaaaaaaaa aaaaattagc
62641 tgggtgcagt gacatgccta tagtcctagc tactcaggag gatcgcttga gccaggttac
62701 agtgagctat gattgtgcca ctgcactcca gcctgggcaa cagagcaaga ttatttctta
62761 aaaaaaaaaa aaaagaaggc ttcaacaggt cccctccaag ggactggtct ctgaagctct
62821 tgccattgcc cagggaggga aagttctgag caataaaatt tcttaaataa atcggccaag
62881 tctgaaccat gtgtcagcca ggaccRtggt gctgggcctc cctcagtgat gccttctctg
62941 gagctgggaa gggtgactca aagggagcgt gggagcctgc tgggaagggt ggtaatggat
63001 agtctcatct ccggcatatg gcatcagcaa ggcctggggc gccatcgtct tccactccct
63061 tggttcctct ctctgttctt atgggactag atacaaattt tcctgctgag cactaaatga
63121 gacaaaagat agctcatgct cagcttctcc ttaaaaagga atttcggcat cttttccaca
63181 aaactggggt gttggtgggg catggtagct cacgcctgta atccccccag cactttggga
63241 ggctgaggca gacagattgc ttgagaccag cctgggcaac atggcgagac accatctcta
63301 ccaaaaaaaa acaaaaacaa aaattagctg ggcatagtgg tgcacgcctg tgattccagc
63361 tgcttgggag gctaaggtgg gaggatccct tgggcaggga ggcagaggtt gccatgaact
63421 gagatcacgc cagtgcacac taagggcatc ctagacctca ctttgggcaa cagagccaga
63481 ccctgtctca aaacaacaac aaacaaaaaa cctggggacc taggatgtct ttaagggccc
63541 ttcagcctct aacagtactt aaaccaatta aaagactcct gttagttacc tccccacatc
63601 cccacccgca ggacgctcSg tgatgagcag ctagctggct gtcagctgtg tggatcacca
63661 agattgcatg gagtggggct gagctgacca agggggatga ggggcggggc ggggcgggca
63721 gggagggggc ggagccactc acctaacaat agctgtagtg tgtagaagat gcaaccgaat
63781 atgctgttgg attgattgag gatgctgtcc tgtcccagca catgctccac cagcccgaaa
63841 cccctgcccc acctggcaga ggggtggggt ggggtggaac caggttagga ctgtcaaccc
63901 agtgccttgg accctgcccg agaaaggtga tttccaagaa gccacctggg ctatcctctg
63961 ttccccgacc tcccatccta gtccaagggt cgatgatctc ctggcaccgg gcacctttgg
64021 ccacgtcagg attccatgtc actgacccta tcctcccctc tccccagacc aggcccggac
64081 gtggctactc cgtaggccct gcttttcatc ttagacctta agtaagtctc tttttttttt
64141 tttttttttt tttttttgag acggagtctc actctggccc aggctggagt gcagtggcgc
64201 gatctcggct cactgaaacc tccgcctccc gggttcaagc gattctcctg ccttagcccc
64261 cccgagtagc tgggattaag gcacccgcca tagcgcccga ttaatttttc tatttttggt
64321 agagacaggc tttcaccatg ttggccaggc tggtcaactc ctgacttcaa gtgatccatc
64381 cgcctcggcc tcccaaagtg ctgggattac aggagtgggc caccgcgccc ggcccttaag
64441 taattcttaa aatggcaagg ctggtataac ggttcactcg gttttgcatc agagactggg
64501 agtcgggggc agattatctt tgccctggac cccagaatct ccagctccct ggccactcac
64561 tcgcctcctc tgtattccgt cattatgcta acgcctggcc atcacgcaca gccagaccgg
64621 gccaccttgt tcctgggcgc agccatcgcc aacacccccc ttcMcctgcg cgccgtcctt
64681 gagaccatcg tcaatctcta ccgccatcct gcctccccgc ctttcctggt caccgttatt
64741 ccttggcatc caattcacgt gcgagtcccc ggaataatcc cagtccccag cactgtctgg
64801 tcccttgcct cgcactctta tttcgaacac cagtatcgct gggtagctca gcccctgtgc
64861 aacgaccccg cgagcagtcc agccccgtgt ccgttccccg ggcacaccga tcccagactc
64921 cagaataatc atctggcatc ctggccgccc tgctccgagg ccccacgcct cccactcccg
64981 tgcacacctg gaggagaaga cgcgcgaaca gctgatggcg gtgcccacgt cgcagagcgc
65041 gcggtaatcc cggtcccggg cgcgcgccgc cttcacgtgc agcgcgtaga gcgagagcac
65101 taagcccgtc aggcaaagag cgagccgcac ccagccaggg ctcccccagg tgctgcccat
65161 tatctccagg ttccgcccga ggcgcccgcg gagaaaacca gccacggagc aggggccggg
65221 cggcgaatgg ccgcgcccct cctggccctc tgactcggcg attggccggc cgtgctcgca
65281 ctccacgacc caaatggctg ttccagggcg ctagtcaagc gggcgagtta ggaaaacagc
65341 gaagaatgcc gggactagtg aagcgggtaa gggacgtgcg gaatcgcggc cccagcggct
65401 gccaggcatg atgggagttg tagtcggcgc ggctgcaagg catcaaggga aatgaagtct
65461 ccacagattt aaaaactgtt ggccgggcac ggtggctcac gcttgtaatc ccagcacttt
65521 gggaggccga ggctggcgga ttacctgagg tcaggagttc aagaccagcc tcgccaacat
65581 ggtgaaaccc catctcaact aaaaatacaa aaattagccg ggcgtggtgg cacatgcctg
65641 taatcccagc tactcgggag gctgaggcag gagaattgct tgagccgggg agccggaggt
65701 tgcagtgagc cgagatcgtg ccactgcact ccagccaggc cgacagagtg agactctgtc
65761 tcaaaaaaaa aaaagaaaaa aaaaagtgtg ttagtgtggt taacagcatt tgcgcttacc
65821 ctatgccaag tcctgttgta agaattgcag catccgggac ctagagacca gcggatcagg
65881 ggatccagcg aatacggcga tccgattcgg gaaccaagca tttcccctga aactatttca
65941 ggcaccattc gggctgcagc ctcccatcct cccgggtcct gcctcaccag tgcttcctgg
66001 tggtcggtct ccctttctcc catacattca cagaaccact cctttggcca cacacaccct
1R7
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
1261gaaggaaaagctggaatactgtgagagacttaccagtggcctgtctgcgtgtaactaact
1321cctcatcccgacttggtcacgcaaaggacaggtgaccatacctccaggaaggtggaaagg
1381ggccctacatgagtgagaggctcaagtcctgcccaagtggaatctgtgattctgtgatct
1441aaagaaagtttgtggctatttttagaaactaaagtttatctcatattgacacaaactcaa
1501aaatcaatgatactttgagatatgtcatatcacaaaaaagatttctcctctagtacatta
1561tcatttaaccaatacaaacaggtctgggccaggtgctgtggttcatgcctataaacccag
1621cactttgagaggctgaggggggtggatccctagagcccaggagtttcaggccagcctggg
1681caacatggtgaaactctgtctctaccaaaagtacaaaaattagccaggtatagtggcaca
1741cacctgtagttccagctactaaggaggctaaggtgagaggaccactgagcccagggacgt
1801agagactacggtgagacatgattataccactgcactccagcctgggcaacagagggagac
1861cttctctcaaaaagaaaagaaaagaaaaagaaacaggtctaatttgttcatctaagcaat
1921gataagatttatatgaacataagttgctttattgatgaaaaattgaacataatctaatca
1981agcctctaaatttaactgccaatttataggaaaagacaggacagaacctacaggaatgca
2041atcatttatatctagaatgtggaagattctgcatgacaaacgacttggattcttcaacat
2101gtaaatttcaaggaaagagagagagagagagataaaaaggcttgtttccagatttgaatg
2161acatgttaatagacatttatgagacaatcaaggaaatttgaacatggactgcatattgaa
2221tgttgagggattatagttaatttttaaaggtacaattgtgatactgtgattatattttta
2281aatgcgatcattatcttttaggagcactaaaatatttactaataaaattataggatttac
2341ttcaaaataaacaacgataataagattaccatgaatagtggtgggtgaaatatacaaggg
2401gcttatttgacatttccataataaaaaacacgaatgaataacaaagcattatagaaattc
2461aagtaaaaaatagctcgtgcttagtaataaatcaaagcgtgctggacactttaaataata
2521aacaaagataagatcttgccctctagaaaacagctctaattcaggacagacttgctcaca
2581tcagatggaagagtcagaatgagatgtgctgagtagaaggtagaacggtcgctagcagag
2641ggtgggaaggtggtatgtgtgggggagaggagaaagagaggttgattaatgggtacaaac
2701agacagttagatggaaggagtgagatctgttgttcgacagtagagtagggtgactacagt
2761taacaacaatatttgcatatttcaaaatagctagaagacaggacttggaatgttcccaac
2821acatagaaaggtgatgaactcaaggtgctgaacaccccaaataccctgacttgatcatta
2881tgcaataacaaaatatcacatgtaccctgtaaatatgtacaaatactacatatcaattta
2941aaaattttcacacaagaatgagatgtgctgatgcatgtggaggccctagctccagactgg
3001gtgtaaacttcaagccactgaagatcttatttccaaggtttttctactttgaaattccaa
3061acttatttttctagcagatttataagggacacgggacaacataaacttgttaaagtgaca
3121agagaaagtaaatatgcccttaaatttataccaaatcttctgacaagtcttgactgataa
3181ttgtttccttcaaatttgtgaaaaacatgagagaaaacgtgtttgtat~ctcatttttaag
.
3241tgtggacacttggcattgctcacggcttccaacggaataaaatagggcttagttgttttc
3301cattagctttttcctttgtgtgtgtgtgtgtgtgtgtgtgtgtgtgtgtgtgttgtgtgt
3361acatattggctctaagcatttattagcccaataatttttagtgaaactctccacttctca
3421atatcttttgccgtattagatttttgtaacatgtgctaaaggttaaaacaccttttcccc
,
3481ttcatgaagcctgagagaagtcggttttctgggttttctcaagacaacccagaggttttg
3541tatacgtctgtctaaaaagtctcagatttttcttgctaattgtgcaccttcataatcaag
3601cagacaaatcagaatattattttggtgaggccatcatctaaactaacagtctttatgtac
3661agaagcagcactgaccgggttcatactcctcagttagcaagtcaacatcttcctcctgcc
3721agcaacccatcccagaatatctgtggcttattaatacttatgaaaacaacagtcttcatt
3781atttactaattaggagatgatcagatgtatctattgatagcaacaggctatttaaaagtg
3841aaataatctatcaaacagattttttatcaactcaaagtttccagttagatatttttcatt
3901aaattgattgctagattgcagccacaatcaaacttaagtattataagaagtttggttggt
3961cttttaaaatcatgcaaaaattcaaggggtgctattaaatatagaattccaaatgtataa
4021agtcttgtcctaaaatggtcaataaaatgaaccagtccactggttcatttatagggggcc
4081agtccactacattaatttggatgttcttcgtctgcattttcatgttttcacacacctgca
4141gtcattgtgtacaattctggactcttgattttcatttagcattctgcgttaatattctga
4201catgttgctgccaggtcttcatggccaccacttttaatggccagaaaatatttcatgtag
4261tgaacatttcataaattacttaaccatgtcccaattattggttatttaaggaatttaaaa
4321ctaaaacaccactcttgtgacaaagttctgatgtatccagatgtactcatgccaagtcgt
4381ccatcaatagttctgctaaaccctgtcagagcccttttctgaagggaaccaggaaacatc
4441tcacaacaagaagcttaaggcctctacaaaatgacttcagggttaggatttcaatttcac
4501tctgaggcacatacaggaaccactaggttatttggcttagaatggagggaagtgtcattt
4561tgtttctgttcggcctgcaggaagctccttcccaggccctgcattgccaattgaacaatt
4621gaacaattctcattgttcaattcccacctatgagtgagaacatgcattgtttggtttcct
4681gtccttgcgatagtttactgagaatgatggtttccagcttcatccatgtccctacgaagg
4741acatgaactcatcattttttatggctgcatagtattccatggtgtatatgtgccacattt
4801tcttaatccagtctatcactgatggacatttgggttggttccaagtctttgctattgtga
4861atagtaccgcaataaacatacgtgtgcatgtgtctttatagtgcctagaacataatagga
4921gctccagaaatactgttgaaaaaatgaataaattgagcacactaagtgtctgaataaaat
4981accctgaccatacccctaaataaacaacataaataagcaaatttcaatttctcggaaaag
5041ttatattttagtgtccaatgctcttgttatgcagtaatggcatctttgattattcatatt
5101cgttagagcttccagaaaggagtattgcaaatcacacgggcctctgactctcatgaccaa
5161atccccctgtcactgtccttgttctctgatccttcctctgggccctcggaaatgctggtc
5221tccatcaaattgctgtaaacagttttcagaaaaagttctctttgggaagtttcaagagaa
5281aaccacaaatttcctggaaatgcttcatgcttcatgacatttaaggctttcagagccttg
5341aacttcaggagcaagatagctggtaggtcttctggaggtcttgcctaacctgagaaggtc
5401ccagagaatttgtgcatagaacctcccaggaagcagtaagacaggctggtgcagtcccac
5461aggatgagaaaggtaagagcttaccaatgtctccagttttgtaaatgaatgttcttagca
?.14.
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
5521 tttttggtgaggagaaaaaaatatcaaacccatcacagacagatcagcagtctcttgacc
5581 taggtcactcaaggggttctcaacaatcaattctaaaatgcaagtgaaaaatgtcaatat
5641 aaggctagacacaggggctcatgcctgtaattccaacactttggaaggccaaggcagacc
5701 tagaactttgtaaatctgagattttgctccatgactttgtggatactttctactaatacc
5761 acctacacaaaatcttcaaccaagaatcttaattgcatttatcaacttgtgtccttagat
5821 cataacatttagattcatttgaaagaaatttattgaagtaaaagaaataagaagaacttg
5881 atagcacaaaacaaagaaaacccagaaagagagagagagagagaagcagaaaaactaaac
5941 actgaaacccagagagagagagagaagtagaaaaactaaacactgagtcttaatctgttc
6001 tgccactagtcagcagcctgcacagagcactttaatgtattctctgtccattagtttcct
6061 tgtttatgaaagtatatagctccaaaaaaattctgtgttctgatttttgtctctccaaac
6121 cacaaccagtcccctgctcccttctctcctcctcctcctccttcttcttctttctctctc
6181 tctccctgtctetctctctctctctctctctccccctttccctctcccccacccacccac
6241 ctcccaacccctgcatacacctaggacacctccagcataggttactaccaattttgcaca
6301 cctccagcaYaggttactaccaattttgctgggtttatctccttctgtcctttccgcttt
6361 gatctgaggaatagctgagatttaggacagcaacaaggtgtacctccttccaggttataa
6421 aacaggattaatgattaggctcaaggccccttcctagtcactcagtaaagtctgtgcact
6481 ggaaaactgtggtagcagttttctgagcattagaaaactgtggttctcacagaggctgga
6541 tgagtcaacactgccatctggcggcctcttggagggtgatgtggagcctggctttcattg
6601 aagaatgaaggtcccttgatttcctgaccctcagccaactctccagcagcttctcactgc
6661 agaagaggtcaggccattggtcagcttgaggacaaagtgggaggatcacactttcgatca
6721 cctatactttctaacaatcagccctgtggacatctgctcacgccccatgctgtttttaaa
6781 atattttcccattatagaaatatttttcaaagatgattcacaagctccaggagccattca
6841 gacaagggagagcaaattggcagctaaactcattcaagagtggagcagatgcacatgaag
6901 ctctgtctggcggaggaggcacaagacaccgagcctggctgggagggtgctcatgacaag
6961 agtggggccacaggcctctcctttcattggacatagtggccatacaaagtggctgccttg
7021 aaatgcacccacatacactatcggtcctcccggtctagggagaaaactagcacagtatgt
7081 caacagataaatcagcgcaatcctgtgggtgaagcagtgcacccatactcttcattctgc
7141 tgagcggaactcaagggtgagacacttgtattcctaactccacttgccttcaggctcctc
7201 caacattccaaattgcccatggacagagctactacccctccatagaagtgacaacttgaa
7261 ataaaagatttgaagctccttcccacgtttagacccaggcctgctgctaggaactccaga
7321 ggagggatggaaagaagtttgcactgctcacagatttaatgtttctctcacaaccagaag
7381 taggcagcaggatggattttcaatcaacactaacaaacggatcactcctgggtcctttaa
7441 acaagttccatgtctctttataggttttaggtgcctcctatgtgttcagacttCgtataa
7501 tggatatataagaaaagtaaatgggaggggcaatatttaagaaataacagcaaacaattg
7561 cattcatatttaatataaactacaagtatcaagttaatagacgctcagaagaaattcata
7621 aaatctagaaaagttgggaaaatctcaatagagaaaataaaacttgatctggactttaag
7681 agtatgatttataaagttcaaggggacagtacacaaagtcagacaatgttagctgaatca
7741 ctgaagaatgagaatatcacacccacttaacaaagcagcccagctagttaaagaagtatg
7801 ttgaagaggtagggagttgggtgggggaggcgaggatggcgtggagggatctgaaaacta
7861 tcaaccttaaataatgagatttagaaaatatgattaagtagagggttaattcgagtgcag
7921 agcttgagaatggccacctggaaacactgactctaaaccagtaaggttaatgtttcaaag
7981 tggagaagttaaggtttcacttagaaattttagcaggatcacattttccatacaagacca
8041 gtgcattcgccacagcaatttgattggttatagattgctgctcattccaagattacttta
8101 ttactctgtgcggaggagtagtgatttgaggggtcttatgtctggtgcctttttgtcttg
8161 tttacaggggaaaaggcagaagttgcgcctgcatgccgcataactcaggctctgcatagc
8221 cacatgtctttcaaggctcagaataatttgaagttccaacagctttaagtttgaattaat
8281 ttcacaaagcggagaaagacttggacttcgtgcagtaatgaagaccaccgatgggtactg
8341 aacagctctggtgggtttgcactcagagaaaggagcctaagatgcagaaggtgcagtgag
8401 gcagcagccatctgctcttgtgctgctgagcagagcatgatgggctgtcaccgctcacgc
8461 gtgttctttacctgctgactcacctggcggcatgggctgcatatggggtcccagctcctt
8521 aagctcctgtcaggcccttctgcaacttctcccaagctcttgggccaggtgcatgtctag
8581 ccatgaaaaaggaggccagtacctgatcactaagtgaaagttctaaggtagtgggactgc
8641 cacaggtgtcccccatggtcccaggtcacaatccagtctgttgaccctcctccttttgca
8701 ccatcgcccctttgacagcctgtgctatgggttttaggctcctagcaccaaacagaaaca
8761 ggcttatatagatcatctaatcagctcttcttatgtgaggccgaactctgtaataaatct
8821 ttttatgtctcctagggcttctctgattgaacctgtctgatgaggagttaaagaagttct
8881 caaattgtgtatgcataagaatcacctgggatttctagctcagagactgggacaataggt
8941 ctattgaacctaggagttcattttgaacaagtgctctacgtaattctgatacagggaatc
9001 ttccagttacagtttgaaattcacaaatacaaggaatgagagacctagaatcagaaagca
9061 tgttaatacacttttgggccattaagtgctcacccaggtaacaggtacagaaaggcagaa
9121 aaaggaaacctatcagggtaataattatgcctttttttttttttttttttgagacagggt
9181 cttgctctatcacccaggctggagtgcagtggcacaaccacagctcactgcagccttgac
9241 ctcctgggctcaagtgattcttccagttcagcctcttgagtaactacgactacaggcatg
9301 tgccaccatccctggctcattttttgtagagagggggtttggccatgctgcacaggctgg
9361 tctcaaattcctgggctcatgtgatttccccacctcagcctcctaaagtattaggattac
9421 aggcgtaagccactgtgcttggccaagacattttggtaagaaatattattttcctactaa
9481 attgtctacattccccttgggtaggcttgcaaagtcactgtgactacagcaggagctatt
9541 gctgcatgggaaatatggagacacgagtggtacctggcagtcacgggctcagtttgtttc
9601 taacctcccaagtcagcacagccccactgagcagactgccggaaagtatttatgccatct
9661 gtcggataattaagacaaatccaaacatctacgtgcattctgtgtgtataaatggagtca
9721 tggccaacctctcaagcagttttccatcaatcacttgtaatattaccagatacttccaat
?.1 S
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
66061 tgacagtatc ctaacctctc tactatcatg gcgcccgcct ggcaacacct gagcttgcat
66121 caactcaaca gtcagtctct cctttccaag ctgtgggcca gatgaggtct ctccttcaca
66181 gacgtcccat ctgatggtga cccatctctc ctacaccttc aacctctcca tcctccccat
66241 ctacactgca acttgtttct ctttcccatc tcagaaacag caacaaatct cccttcacta
66301 agaggtcctt caccagcctc ctctcccggc attatcccat ctacccctcc acattcaagt
66361 ttttggaaag attctacact cccagtctct acttcctcac ttcttccttg ctgcccacgc
66421 cataaactag ctgctgcctc cagcattgcc ctgacaccta gtggctggtg tcaccaagac
66481 gctagaccca atggttattt atttatttat ttacttattt tgagacggag tctcactctg
66541 tcgcccaggc tggagtgcag cggtgccatc tcggctcact gcaacttccg cctccagggt
66601 tcaagtggtt ctcgtgcctc agcctcccaa gtagtttgga ctacaggtgc ctgccaccat
66661 gtctggctaa tttttgtatt tttagtagag acagggtttc accatgttgg ccaggcttgt
66721 cttaaactcc tgacctcaag tgatccaccc acctcggcct cccaaaatgc taggattata
66781 ggcgtgagcc accgcacccg gccaatggtt gtttttcagg tcttctcttg cttgacttcc
66841 cagagggatc ccttactgtt gcacctaccc ttctgggaac tctcttcctc tggcgtctgt
66901 gatatttccc tctcctgctg gctcctccct ctccagatgc tgtttctcac atctactctc
66961 ttctagagag tgtggtagac agaataatgg tcaccaaaga tgtccctgca tgaatccctg
67021 gaacttgtga atatgatagg ttaaatggcc aaaagggaat taaggttgca gatggaatta
67081 agctgaccaa tctcctgatt ttattttatt ttattttgtt tttgaggtgg agtttcgctc
67141 ttgttgccca actggagtgc aatggtgtga tctcggctca ctgcaacctc cgcctgccag
67201 gttcgagaga ttctcctgcc tcagcctccc gagtagctgg gattacaggc acccgccatc
67261 atgcctggct aattttttaa atttttagta gagacagggt ttcgctatat tggccaggct
67321 ggtcttgaac tcctgacctc aggtgatccg cccacctcgg cctcccaaag tgctgggatt
67381 acaggcgtga gccaccgtgc ccagcctatc tatctattta tttatttatt tttgagatgg
67441 agttttgctc ttgttgccca ggctagaatg caatggtgct atctcgactc accgcaacct
67501 ccacctcccc ggttaaagcg attctcctgc ctcaggctcc tgagtagctg ggattatagg
67561 catgtgccac cacgcctggc taattttttg tatttttagt agagatgggg tttctccatg
67621 ttggtcaggc tggtctcaaa ctcctgacct cagatgatcc acccacctgg gcctcccaaa
67681 gtgctgggat tataggcgtg agccatcata ccaggctcta ttgatttatt tttattttta
67741 tttttgagac ggagtctcgc tctgttgcct aggctggagt gcagtggcac aatcttggct
67801 cattataact tccgcccccc cccaggttca agccattctc ctgcctcagc ctcccgagca
67861 gttgggacta caggcgcgtg caaccatgcc tggctaattt ttgtattttt agtagagacg
67921 gggtttcact gtgttggcca ggctggtctc gaactcctga ctttgtgatc tgcctgcctc
67981 agcctcccaa agtgctggga ttacaagtgt aagccaccac gcccagccta ttttgtttat
68041 tttttcaaag acccttgaca cccaggctgg agtgtagtgg cactgtcata actcactgca
68101 acctccgtct cccaggttca agcgattctt gcacctcagc ctccctagta gctaggagta
68161 caagtacgtc ccaccacacc tggctaattt atttttattt ttgtagagat ggggtctcac
68221 tttgtttccc aggctggtct aaacttctgg tttcaagcaa cctteccacc tcaaagtgct
68281 gggagtacag gcatgagcca ccaccacacc tggcctaatt tgctgatttt tatttatttt
68341 ttattattta tcttaatttt tattttgaga cagagtcttg ctctgtcatc caggctggag
68401 tacggtggtg caatctcagc tcactgcaac ctccccctct cgggttcaag cMattcttgt
68461 gcctcaacct cccaagtagc tgggattata ggtgctggcc accacgcctg actaattatt
68521 gtaatttttt ttttttttag tagagacggg atttcaccat gttggccagg ctggtcttga
68581 actcctgacc tcaagtgatc cacctgcctc agcctctcaa agtatgggga ttacgggtgt
68641 gagccgccgt gcctggccca atttttgtat tttcagtgga gatggggttt tgccatgttg
68701 gccaggctgg tctggaactc ctgacctcag gtgacccgcc tgcctccgcc tctcaaagtg
68761 ctgggattac aggcataagc caccatgcct ggcccacagg ggtccttaaa aaatgaagga
68821 ggatggcaga agaaagtcag agggagatgt gagtaaagaa aaaagacaca gagagctgca
68881 atgtttctgg tttgaagatg gaggaagggg attgtgagct aataaatacg ggtggcctct
68941 aaaggcaaga aagggtaaag aactggattc tcactctaga gtcaccggga aggaactatc
69001 aacatcttga tttcagccca gtgagactct gtcagacttc taagctacag aactgtaaga
69061 taaatttgtg ttgttttaca tcattaaatg tgcagtaact tgttacagca gcaattagaa
69121 atgaatacag aggactgggc attaggcctg tatctcagct ttctctgatc tcctggtgtg
69181 ttcctgttat ttattgttgg tttcccccag aatgagtgat ctaagaggaa gcaaaataga
69241 agccgcaatc tctttatgac ttagcctcag aagacacaca ctggggccag gtgcagtggc
69301 ttatgcctat aatcccagca ctttgggagg ctgaggcagg aggaccactt gagcccagga
69361 gtttgagaca ccctggacaa cacagggaga ccctcactct ataaaaaata aacaaaatta
69421 gccaggtgtg gtggtgcaca cctgtagtcc cagaactttg ggaggctgag acaagacaat
69481 gacttgagcc caggagtttg agacaggtct ggacaacgtg gtaagactct gtctttataa
69541 acatttttaa aattaggcgg ggcatggtgg ctcatgcctg taatcccagc aatttgggag
69601 gctgaggtgg gtggatcacc tgaggtcagg agtttaagac aagcttggcc aacatggcga
69661 aaccYcgtct ctactaaaaa Yacaaaaatt agccgggcat ggtggtgggt gcctgtaatc
69721 ccagctactc aggaggctga ggcaggagaa tcatttgaac ccgggaggtg gaagttgtag
69781 tgagccgaga ttgccttcct cactccaaga gttataaaag attttgacca tattttcttc
69841 tagcatttaa tcaattaatt aattaatgtg agacagtccc actctgctgc ccaggctgaa
69901 atgcagtggt gcaatctcgg ctcactgcaa cctctgcctc ccggattcaa gtgattctcc
69961 tgccttagcc tcctgagtag ctgtgattac aggcaccagc cactatgcgt ggctgatttt
70021 tgtgttttta gtagagacgg ggtttcacca tattggccag gctggtctca aactcctgac
70081 ctcatgatcc Rccctccttg gcttcctaaa gtgctgggat tacaggcgtg agccactgtg
70141 cctggccttt ttctttcttt tttttttttt ttcattagag atgagttgtt gttatgttgc
70201 ctctaactcc tgggctcaag cagttctccc accctggctt cccaaagtgc tgctgggatt
70261 acaggagtga gccactgccc ccagcctctg acagtttttg tgcactagga atttgggaag
1R~
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
70321 acaattttac ctggctattt ctggctcata caattgcagt cagatggtgg ctaaagctgg
70381 aacaataagc agctaaaaca gctgaaagat aacctagcat tctctctccc tttctctgag
70441 tagtctccga acctatctat gttgtcctct gcatgggcta gcttgggctt cctcacagca
70501 tggcagcctt aaggctttaa tagtcagctt ccaaaatggt cctcagtgat tcctgcttcc
70561 tggtattgat accattgtga agtctcttct cacattgaaa ggggctgaac tggcccattg
70621 ggataatgca gaaatgacag tgtgtgactt tagaggctaa atcatgaaga tattgtggct
70681 tccatcttgc tcctttgtag atcactcatt ctagacaaag ccagctacca tgatatgaaa
70741 gcactcaagt aaccctaggg agagaggtct ccttagtgag gaactgaggc cctgtaagaa
70801 acgtgggtgt gctgcagtca agtgggcata ggccaaagta aacatccaga gtgactcagt
70861 gagtttagag tgcaggcata tagctccact tgttatcaca gccgtgtagc cataacatgg
70921 gaaggctcat cacttggctc tgagccactg ttgtctgtaa aaggtataat tgccctgctg
70981 acactgtgca cagggctcgg cccaacatgg cttgacatgg gacatggctc ttgtgcaggt
71041 gcttgtaccc agagaaagag agaaagccag agctgtccat ctcggggaag ccaagacaca
71101 gctcagctag ctcatgccca gagggagaaa gagtaaggct gtggggtgtg gtggctcatg
71161 cccataatcc cagcactttg ggaggccaag gcaggtggat cacaaggtta ggagtttgag
71221 accagcctgg tcaacatggt gaaacccctt ctcaactaaa aatacaaaaa ttagctggcc
71281 atggtggtgc atgcctgtaa tcccagctac tcaggaggct gaggcaggag aattgcttga
71341 acccaggagg cagaggttgc agtgagccga gatcacacca ctgcactcca gcctgggcaa
71401 cagcgcgaga ctccatctca ggaaaaaaag aaaaaagaaa agaaaagaaa gagtgaagct
71461 gctgaccctg aagggagagc tggccacaca gctgtgtgtg tgtgggagct gccggagtaa
71521 gcagctgaga cagagcagac agtgcgagag taagatgttg atgatgagag agctgctgaa
71581 taaagccatg tctcatttac ctgctgtctc tcgagtgttc ttctagctcc ctgcctcacg
71641 tccactgctt cctctcacac ctcagctggg gctggacccc aaccctgagc atgacgggcc
71701 ttctgtcaac aaccagcagt aacctgctgg gcatgtgagg gagctacctt ggaatcagat
71761 tctgtaaaac agtcacgcct tcagatgacg gtagcattgg ccaacatttt gactgcactt
71821 catgagagac cctgagccag aaccccctag attectaacc caaggaaact gtgtgtgata
71881 agtgtttatt gttttttttt tttttttttt tttttgagaa agagtctcgc tctattgccc
71941 aggctggagc acagtggcac aatcttggct cactgcaagc tccgcctctc aggttcacac
72001 cattctcctg aatcagcctc ctgagtagct gggactacag gcacccacca ccacgcctag
72061 ttaatttttt tgtattttca gtagagacag ggtttcaccg tgttagccag gatggtctca
72121 atctcctgac ctcgtgatcc gCCCgCCtCC gcctcccaaa atgctgggat tacaggcgtg
72181 agtcaccaca cccggccagt gtttattgtt ttaagatatt ggctaggcgc agtggttcac
72241 acctgtaatc ccagcacttt ggaaggccga agtgggagga tcacttgagc tcaggagttc
72301 aagttcaaga acagcctggg caacatagtg agaccttgtc tctatttaaa aaaatgtttt
72361 taagatgtta tgtttgagct gggtatggtg tggctcacgc ctgtaatccc agcactttgg
72421 gaggctgagg tgggtggatc acctgtggtc aggagatgga gaccagcctg gccaacatag
72481 tgaaaccccg tctctactaa aaatacaaaa aattagctgg gcatggtggt gggcgcctgt
72541 aatcccagct actagggagg ctgaggcagg agaatcgctt gaacccggga ggcagaggtt
72601 gcagtgagcc aagatcgtgc cattgcactc cagcatggtg ctatgttttg gaggtaattt
72661 gttacacagc aataaataat tcgtacaggg caccagcctg gccaacatgg agaaaccctt
72721 ctctagtaaa aattatccgg gtatggtggt gcatgcctgt aatcccagct acttgggagg
72781 ctgaagcagg agaatccctt gaacctggga ggtggaggct gcagtgagcc aagatcgcac
72841 cactgcactc cagcctgggt cacagagcaa gactctgtct caatttaaaa ataaaataat
72901 aataatatag ggcagtcaga ctgcccacct ggcagctcag gactagcaca tgtgctccag
72961 aaagccaggt ggaagctaca tattttatga tctaaactca gaagtcatat agcatctgtt
73021 ccactgtaat cacaagcctt cccagttcca aggggaggga acatagactc cctcacctct
73081 tgatacaaga agtgtcaaag ttatatggta agaagttggc caggccctgc ttgtctctgt
73141 tgttcatgcc tgtaatccca gcactttggg aagacgaggc agatggatca cctgaggtca
73201 ggagtttgaa gccagcctgg ccaacatggt gaaaccctat ctctacaaaa atacaaaaat
73261 tagctgggca tggtggtatg cacctgtaat cccagctact tgggaggcca aggcacgaga
73321 attgcttgaa gctgggaggc agaagttgca gtgagccgag attgtgccac tacactctgg
73381 cctgggttac agtgcaagac tctgtctcaa aaaaaaaaaa aaaaaaaaag agagaagttg
73441 gttgggccca gtggctcacg cctgtaatcc caacactttg ggaagctgag atgggaggat
73501 cgcttcaggc cagaagatcc atcgttacca gcctgagcaa cacaaggaga tcccgtcctt
73561 acaaaatttt tttaaaaatc agctgggtgt ggtggcaggc acctgtggtc acagctactc
73621 gggatgctga ggtaggagga tcgcttgagt cagggaggtt gtggctgccg taagccatga
73681 acatgccatt gcattctagc ctgggtaaca gagtgagaca ctgtttcaga aaaaataata
73741 aaataaaata aataatgttg taggacaggc gtggggctca cgcttggaat ttcagtgctt
73801 tgggagactg aggcaggagg attgcttgag aacaggagtt cgaggctgca gtgagctgtg
73861 atcgcaccac tgcactccag ccttggtgac atgagcgata tcttgtctca ataaataaat
73921 acatacagtt ctcttttaca tcgagtatat gtaaattttt aaaaatacat tgaaagcgct
73981 tagaaagccg cctgactctc cctctccctc tccctctccc tctccctctc cgtctccgtc
74041 tccgtctccg tctccgtctc cgtctccgtc tccctccacg gtctccttcc acggtctccc
74101 tctgatgccg agccaaggct ggacggtgct gctgccatct cggctcactg cagcctccct
74161 gcctgattct cctgcctcag cctgctgagt gcctgcgatt gcaggcgcac gccgccacgc
74221 ctcactggtt ttcgtttttt tttttggtgg agacggggtt ttgctgtgtt ggccgggctg
74281 gtctccagct cctagccgcg agtgatccgc cagcctcggc ctcccggggt gccgggattg
74341 cggacggagt ctcgttcact cagtgctctg tggtgcccag gctggagtgc agtggcgtga
74401 tctcggctcg ctacagcctc cacctcccag ccgcctgcct tggcccccca aagtgccgag
74461 attgcagcct ctgcccagcc gccaccccgt ctgggaagtg aggagcgtct ctgcttggcc
74521 acccatcgtc tgggatatga ggagcctctc tgcctggctg cccagtctgg aaagtgagga
1R4
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
74581 gcgtctctgc ccggccgcca tcccatctag gaagcgagga gcgcctcttc cccgccRcct
74641 tcccatctag gaagtgagga gcgtctctgc ccggccgccc atcgtctgag atgtggggag
74701 cacctctgcc ccgccgccct gtctgggatg tgaggagcgc ctctgctggc cgcaaccctR
74761 tctgggaggt gaggagcgtc tctgcccggc cgccccgtct gagaagtgag gaaaccctct
74821 gcctggcaac cgccccgtct gagaagtgag gagcccctcc gtccggcagc caccccgtct
74881 gggaagtgag gagcgtctcc gcccggcagc caccccgtcc gggagggagg tggggggggt
74941 cagccccccg cccggccagc cgccccatcc gggaggtgag gggctcctct gcccggccgc
75001 ccctactggg aagtgaggag cccctctgcc tggccagtcg ccccgtccag gagggaggtg
75061 ggggggtcag ccccccgccc ggccagccgc ccagtccggg aggtgagggg cgcctctgcc
75121 cggccgcccc tactgggaag tgaggagccc ctctgcccgg ccagccgccc cgtccgggag
75181 gggggagggg gggtcagccc cctgcccggc cagccgcccc gtccgggagg gaggtggtgg
75241 gggtcagccc cccgcccggc cagccgcccc gtccgggagg tgaggggtgc ctctgcccgg
75301 ccgcccctac tgggaagtga ggagcccctc tgcccggcca gccgccccgt ccgggaggga
75361 ggtggggggg tcagcccccc gcccggccgg ccgccccgtc cgggaggtga ggggcgcctc
75421 tgccccgccg cccctactgg gaagtgagga cccctctgcc cagccagccg ccccgtccgg
75481 gagggaggtg ggggggtcag ccccccgccc ggccagccgc ccagtccggg agggaggtgg
75541 ggggatcagc cccccgcccg gccagccgcc cagtccggga gggaggtggg gggatcagcc
75601 ccccgcctgg ccagccgccc cgtccgggag gtgaggggcg cctctgcccg gccgccccta
75661 ctgggaagtg aggagcccct ctgcccggcc agccgccccg tccgggaggg aggtgggggg
75721 gtcagccccc cgcccggcca gccgccccgt ccgggaggga agtggggggg gtcagccccc
75781 cgcccgacca gccgccccgt ccgggaggga ggtgggggga tcagcccccc gcctggccag
75841 ccgccccgtc cgggaggtga ggggcgcctc tgcccggccg cccctactgg gaagtgagga
75901 gcccctctgc cctgcttgaa ggcagcatgc tcgttaagag tcatcaccac tccctaatct
75961 taagtaccca gggacacaaa cactgcggaa ggccgcaggg tcctctgcct aggaaaacca
76021 gagacctttg ttcacttgtt tatctgctga ccttccctcc actattgtcc tatgaccctg
76081 ccaaatcccc ctctgcgaga aacacccaag aatgatcaat aaaaaataaa aataaaaaaa
76141 aaaaaataaa aaaataaaaa aaaaaaaaaa gaaagccgcc tgacctgtat acagtattct
76201 gaaaaggggg tcgcgaggtg catgtccaac ctccgccgcc gggggcagca gcgagtccag
76261 gccgagccgg ggcctagcga gcggggtcaa atggggtgag gcctgtgcca gacctctcca
76321 cctcggtggc agccgcagcc tcctccgcct gcggctcctg tccacgccgc ggccacgtga
76381 gcgccagatt ctggcgcaca gaccactgcc agtcctttgc tgctttgcgc agcctgtcct
76441 ccccgccagg agcacccttc ccgctccctt ttaccacggg ctccagccgt ggctgccttg
76501 gggctgccgc cgcctggctg taYtccagga cgttgggaaa gaacgggtgg gaatggtgtg
76561 ggtgggggtc aaagaggaaa cccagagatg cagggcgccc ctttcccgtg gtctgccccc
76621 aattgctcag gcaggccagt cacggtgagg cgtcctccct ccaagtttat atttattatt
76681 atttattatt tatttttttc accttcaagt ttattattta ttatttattt atttattttt
76741 gagacggagc ctctctctgt cgcccaggct ggagtgcatt ggcacgatct tggctcactg
76801 caacctccgc ctcccgggtt caagcgattc ttctgcctca gcctcccgag tagcagggat
76861 tacaggtgca tgccaccaca tccggctaat ttttgtattt ttagtaaaga cagagtttca
76921 ccatattggc caggctggtc tcgaactcct gacctcaggt catctgcccg ccttggcctc
76981 ccaaagtgct gggattacaa gcatgagcca ctgcacctgg ctaacctcca agtttaaaga
77041 cagccgccag gcccagtggc tcactcctgt aaccccaaca actcaggagg ctgaggccag
77101 gaatttgaga ccagcctggg caacatagcg agaccccggc tctaagaaaa ataggccagg
77161 cacggtgggt tacgtctgta atcccagcac tttgggaggc tgtggcaaaa ggattgcctg
77221 agggggaaaa aatcaccctg ggggtagtgg tgcacactta cagtctcggc tacttgagag
77281 gctgaggtgg gaggatcatt taagtcggag gctgcagtga gctactatgg agcgactgta
77341 ttacagcttg agcaacagag cgagacccca tctccaaata aataaataaa gatagcctcc
77401 aaagatgtca cttgcttcac ttagcacttt ttattgaaca tatttaggaa atatataacc
77461 atggtaaaga aaattgcaat tagtacaaat acacactaca cacatatgca cgtgcacaca
77521 ctgaaacgtg tcccttccag cgcctcctgc ctggataatt tttttttttt ttgcaacgga
77581 gttttgctct tgttgcccag gatggagagc agtggcggga tatcggctca ctgcaacctc
77641 ctcctcccgg gttcgagcga ttctcctgcc tcagcctcgc gagtagctgg gattacaggc
77701 gccagccaca acacccggct gatttttgta tttttagtag agacggggtt ttgccaagtt
77761 ggccaggctg gtctggaact cctgagatcc gcccacctcg tcctctcaaa gtgctgggat
77821 tacaggcctg agccactgcg cctggcctaa agtaattgtc ttcttattgg tttctctgcc
77881 tttggtctca ccaaacgccc cactctaaat cactgcagac aaggggttct gtctaaggag
77941 gagagcccac cagttaaaac ccttcagtag ttcctaacta ccctaggaca aatgcagact
78001 tatccttcgc tctctgctgc cgcccctctc cttcccagat cccatatggc tccagccata
78061 tcctcagacc atctgggctg ttgtctgtcc tgccacacac ccttcccacc ccgctccctt
78121 gcaagtccta ctcaggctgc acctacacag ctgtctcagt ttctatctga ggctcccgca
78181 gcctcctctg aaagtcctca tcacagccaa tgatactgaa actgttttct tatgcagggc
78241 acggaaagat ctattcaact cactgcccaa tctcctctcc tggtgcagaa aagacaccca
78301 gtcagcgact tgcattatct gcaggcatga gtgaataata ataatgccta acccttatat
78361 agtgctaatt ccacgcctgg cactgttcta ggcactcata taaattcatg taatccacac
78421 aaccccaaaa ctgatgatat ctcttcatct taccaagcac tgagcagtta aataacttgc
78481 tccagatatt aaggggtcga gctggggttt gaagcctggt tacccataaa tgaaccaaga
78541 actggaagga ggacaagaSc tccgagaagg agtcaggtag ggcgtgatct gtgcgcttta
78601 catctaagat cttccagctc ccagggagcc cgtttcatag agcaggagat agaggctggg
78661 agggacacgg agagcctcga gagccgtgtg gaggaagcgg tgctgtttgg ggtccgggag
78721 caagggcgtg gcctggatgc gcgggcgccc gggacggcac gtcctcagac caaactacaa
78781 ctcccaggac ccagcgggcg ctgccgccca cgcgacgtca cggcggcgga gggcgcaggc
1R5
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
78841 ggctgggcgc ctggcgagtg gactgttcga gcccttccgc tgggacccgg gccctggctc
78901 cggccccgcg gtaagtgggg cgaccccagc ctactcagtc cgcggaggcc ccgcggcgca
78961 cgtccgcagc ctccatcaca gcgcgggcgc gcagacgggg ctggcatcta ccatatgggg
79021 ggcatccggg ccgaaccaag tgacccgcgt ggggggtccc gctggggact ccgtgccgca
79081 ccctcccaag ccggccccag gggcccaggg ctggtgtcgc acgttcgctg gccgcgctcc
79141 cagggcccgg gtttgaaggc gctgggcagg caggggcagc cccgccccct gagaagggta
79201 cccgggaccc cggggcgctg gggcgaggtt ttcgggctgg aagggtctga ggggctcctc
79261 ccccgacagc cctcccaccg ccagtagagc ctcgggttgg ggaatagaag cccccgggag
79321 gctaggtcct ttgggcgcgg cctgtgtgca tctggggaga cggtgggagt ggtggggaga
79381 ggtcgcccgg gtctggggag accgatgcac aggtggagag atggtgcggg ttctgtggat
79441 tcggatcctt acaacttcct cttccccgcc ccggtagatg ggagctgctc tccgcgggct
79501 gagcctgtca gcatcctcga cgcaccctgg tccctgaagt cggagaagMg cccctaccca
79561 cccacacccc cttgccccat tttgggtcgc ctgggtcctc agtcctagcg gatcctcagt
79621 cctagcggcc accgggtctg aaaggagcaa gacgatgatc ctggcgtcgg tgctgaggag
79681 cggtcccggg ggcgggcttc cgctccggcc cctcctggga cccgcactcg cgctccgggc
79741 ccgctcgacg tcggccaccg acacacacca cgtggagatg gctcgggagc gctccaagac
79801 cgtcacctcc ttttacaacc agtcggccat cgacgcggca gcggagaagg tgcgcaaggg
79861 ggcagccagc ccagggtccg gRatgtaggc gggagggaga gtgttggggg ttctctgctc
79921 aaggcctctc tccctctcta gccctcagtc cgcctaacgc ccaccatgat gctctacgct
79981 ggccgctctc aggacggcag ccaccttctg gtaagattca cgccctctat tttcctcgtg
80041 gatcctggag ctctcccaga cactcaggct ccagccccgc cttcccttct cattttctcc
80101 cagaaaagtg ctcggtacct gcagcaagaa cttccagtga ggattgctca ccgcatcaag
80161 ggcttccgct gccttccttt catcattggc tgcaacccca ccatactgca cgtggtaagg
80221 tagagaggac cttaggtcag cgggccaccc tgccccgggg gcaagtgggg agtctggggc
80281 ccagagtggc agacgattgc ttgcctaaag gtgtcagggc cacacaggat tcaaccccag
80341 gccttcagaa gccaaaggtg tgtattcacg gagcctggaa gggtcgaagt gggggtttga
80401 tcacgtggtc gaccagctgg gtggtgatcc ccatgggtag gtgggggtgg ctgttctctg
80461 ctcagtgccc atgcggcttt gtgaattccc acacctcttc cttgcagcat gagctatata
80521 tccgtgcctt ccagaagctg acagacttcc ctccggtgag tgctgggcca gagcagggtg
80581 aggggctgag aggttgggct tggaccaccc ttcctcatga ctctgtgacc tgcagatcaa
80641 ggaccaggcg gacgaggccc agtactgcca gctggtgcga cagctgctgg atgaccacaa
80701 ggatgtggtg accctcttgg cagagggcct acgtgagagc cggaagcaca tagaggttgg
80761 ggcagcaaag gagaggccgg gcctgctggg ggtgggaagg gcacgggatt ctgagacctc
80821 actctttaca ggatgaaaag ctcgtccgct acttcttgga caagacgctg acttcgaggc
80881 ttggaatccg catgttggcc acgcatcacc tggcgctgca tgaggacaag gtggggctct
80941 gggacctgag acccacctgg gaacattaag tgagacagag gagactgggc tggggatccg
81001 ggtcaagggc ctgggggctg aggctgtggg gctggtgctt tggggcagtt ccgaagttgc
81061 cagcatcttg gggtggggct aggggcgtgg gtagtcctga cctcctttct ccggccagcc
81121 tgactttgtc ggcatcatct gtactcgtct ctcaccaaag aagattattg agaagtgggt
81181 ggactttgcc aggtgaggca agaatggctc agggggtggg cagacatctg gggcagggaa
81241 ggcttgggtc tgagcccttg cccggggcat gatctgcggg gagcagggtt tctcaaccat
81301 ggcactattg acatttccag ccagataatt ctttgtcaYa ggggctgccc cgtgcacgtt
81361 aggaagttca gcagcatccc tggcgccagc agtactgcct agttgtgaca aacaaaaatg
81421 tctctgcaca ttgccatatg ttacttaggg gggcagaatt gtttccagtt gcaaaccact
81481 ggtggagggg cccctgactg aaccctcgct cctatccgca gacgcctgtg tgagcacaag
81541 tatggcaatg cgccccgtgt ccgcatcaat ggccatgtgg ctgcccggtt ccccttcatc
81601 cctatgccac tggactacat cctgccggag ctgctcaaga atgccatgag gtggggtggc
81661 ttgatgtgct ggcttggggg Yggacaggaa ccgggNtgct tgtacctact ggtctttccc
81721 ctctgcatag agccacaatg gagagtcacc tagacactcc ctacaatgtc ccagatgtgg
81781 tcatcaccat cgccaacaat gatgtcgatc tgatcatcag gtttgccctg agtgggagtt
81841 gagctgaggt ggatgggatg ggggtctagg cactgtttct gacttgattt aggaccttga
81901 gccccttcct gccccattct gggacttggt ccctgaccag acaaactatt ctctgaatcc
81961 tgagatggcc atgagctgct tattaatgga tctggggcca gctgcaggcc taggtatcct
82021 gcctctgtca gcagctgagg agcttgaaat tgagaaatag tcaggagtcg gtctaggatg
82081 ctgggccgag gataaatgtc acatcctgtg agaaggtata agcagtcagt ggccctggca
82141 ggggtgagga tgatataaac aaggcccaag ggtctaggtg gaccacattc cagctctggg
82201 tggaaggaac aggaaggcag actttgcact gtctgcttgg ggggtggtga gtaccccatc
82261 aaagctgagc caagcccatt gttgttgcca tcttgctagg atctcagacc gtggtggagg
82321 aatcgctcac aaagatctgg accgggtcat ggactaccac ttcactactg ctgaggccag
82381 cacacaggac ccccggatca gccccctctt tggccatctg gacatgcata gtggcgccca
82441 gtcaggaccc atgcacgggt gagaccctgc caggccagga tggaggggtg ggggacccca
82501 ggagactcaa gCCtCtgaag CCtCCtgtCC tgtCCCCCtg CCCaCCCCCa gCtttggCtt
82561 cgggttgccc acgtcacggg cctacgcgga gtacctcggt gggtctctgc agctgcagtc
82621 cctgcagggc attggcacgg acgtctacct gcggctccgc cacatcgatg gccgggagga
82681 aagcttccgg atctgacccc acagcctttg gcctgctcac ccgaccagcc tgggccgcat
82741 tccctgcagg acctcccggg tcaggcaggg cggccccctg ctccacacac tgctgcatct
82801 tgggtctcag ggacccagac agatggactt acatggagct gggcactgcc ctgcctcaac
82861 agggtccatt gcctcctcgc ctccagaact tggagcaggg aagtgggcac cctgaggcct
82921 ccagcaccag ttccgtcatt ctcgttcctg gggaaccccc actctgacct gttattaaag
82981 ttcacatttt gaatgccctc tcgggccccg tgtgtgggga gggcaggtga acttttgttt
83041 ctgcccccat tcaggttcac tgagcccttg ggttgaactg gttcgtgtcc cagtctctta
1 St H
CA 02561669 2006-09-27
WO 2005/097421 PCT/US2005/010913
83101 cctgccctga gagcctggca ggccaggagt agaatgggtc ccaagtctgt tgcatgtttg
83161 atttggtggg agtgggatga ctgcagcacc ttatacaaag agctttcatt catcttgttg
83221 aacaaatgtt tccgggtccc agataatatt gaaggcccag actgacccag cttcgggcat
83281 cagttttgac tcttcctttc ctggcagtca cagtttctag aggtgaaggt caccagactg
83341 ggcaaactcc tgagccaact gcttcccaag cctgagtagg ttaaaaatac tgtgtctgct
83401 gctgccaagg aaaagaacat acaaggttgt gccttggcag gccctagcag ggactgggtg
83461 ccccactgca aggaaaggtg gggccctgat agaaaggacc aaggatttgg gcaaagRtat
83521 caggtaggct caaggttaga cctgaatcag aactccagat gacatcttag gtaggaacac
83581 cctacccacc ttgccaggga agaaaggcct aagggcggcc tggtggggct gggaggagaa
83641 ctggaaagtt ctcttgcctt cacatgtgag ctcccacagc aaacttcctg aggctggctc
83701 taggcctgta ccatctccta cccttcacgg ggatggaggg gaagttgtat gtggaagcca
83761 aatggcaggg gctaggaaac cacagtgact tgctagactg aaaaatcccg ccagctgcaa
83821 ggcagggtgc tgaggctgga gaggcaggca gcagtcagag gccagggccc tgaaacatgg
83881 gatttatctt gagccatagg gatccatggg tgagttttta tttatttaga aatggggtct
83941 tgctctgttg cccaggctgg aatatggtgg ctgcagagtt cactgcagcc ttgaactcct
84001 gggatcaaga gattctccca cctcagcctt ctgagtagct tggaccatca tgccaggcta
84061 aattttaaaa ttttttgtag aaacagggtt tctacaaagc cctatgttgc cccgggctgg
84121 acttgaactt ctgggctcaa atgatccttc caccccagcc tcccaaagtg gtggggttac
84181 aggcatgagc cactgcagct ggcccatgag tgggttttga gctgggaagg gatgtttctg
84241 gttggagtcc ctgagaggat tcatgtccac gtgatttctt aagaaagtgc tcccagaaca
84301 gagtagggga agtaggaagg ggaaggggag gaagccaagc aaggatgtga cctcaggcaa
84361 aagcccagaa ccagtcaatt atgcctcagg gttgaaggta agagagctaa acctcagagt
84421 tactgattaa tttctccact tggcagtcac tggttaaagt cagttgggaa agtgaacagc
84481 tctattaacc taaggatggt tttttaagaa gagcctcagg tgctggtgtg ggtctttgaa
84541 agcacatcaa aggtaatctg ggcacacaga aacagcaaga actcccagag gatctgggtg
84601 gagcacctac attgtttttt tgtttgtttt gtttcgtttt gtttttttaa acggagtctc
84661 aatctgttgc tcaggctgga gtgcagtggc tggatcttcg ctcactgcaa cctccgcccc
84721 accccccccc aaccccaggt tcaagcgatt ctcctgcctc agcctcccga gtagctggga
84781 ttacaggcgc gtgccaccac acccagctaa tttttctatt tttagtagag atggggtttc
84841 accacgttgg ccaggctggt ctcgaactcc caacctcgtg atccatccac ctcagcctcc
84901 caaagtgcca ggattacagg catgagccac catgcctgtc ggatgtttct tgatttgtaa
84961 cctctgagag acccatccgc aggccctgag cattccactc ctctcagaat tgtttccaag
85021 cccaataacc acattataaa tcaaacaaga ttcagagaat agccaaaggg aatgtttact
85081 gagtacctac ccggtctggc actttgcaat acacttgtat attgctaaga cggatagttc
85141 aaccgttaca tagttatatg attgatagtt atacatgctt aactgctggg gattggttcc
85201 aggaccgcct gtgaataccg aaatctgcag gcgctcaagt cctacagttg gccctgccaa
85261 acagcagata tgaagtcagc tcttcagatc tgtgggttct gcatccttac aatatttcct
85321 ttcctttcct tttcttttcc tcccttcctc cctctttttt ctttttcttt tttgagatgg
85381 agtcttgttg tgtcggccag gttggagtgc agtggcgcga tctcggctca Mtgcaacctc
85441 cacctcctgg gttcaagcag ttctcctgcc tcagcctccc aagtagctgg gattacaggc
85501 acacgccacc acccctgact gttttgtatt ttcagtagag acggggtttc acaatgtggg
85561 ccaagctggt tttgaactcc tgacctcaag taatccacct gcttcggcct cccaaagtgc
85621 tgggattaca ggtgtgagcc accgcgccca gtcttttttt tttttttttt gaggcagagt
85681 ttcactcttg ttgcccaggc tggagtgcaa tggcacaatc tcagctcacc acaacctctg
85741 cctcccaggt tcaagcggtt ctcctgcctc agcctcccga gtagctggga ttacaggcat
85801 gcggccacca cgcctggcta attttgtatt tttagtagag atggggtttc tccatgttgg
85861 tcaggctggt ctcgaactcc cgacctcagg tgatctgcct gcctcggcct cccaaagtgg
85921 tgggattaca ggagtgagcc actgcgccca gcctcctttt ctttcccccc tttttttttg
85981 agacagggtc tctgtcaccc aagctggagt gcagtggagg gattatagct cactcagcct
86041 cgacctcctg ggtttaagcg atccctctgc ctcagcctcc tgagtaggtg ggactacagg
86101 tgcgggcccc gaggcccagc taattttttt tttcccccaa atttttagta gaaaggaggt
86161 ctctatgctg cccaggctgg tcttgaactc ctggcctgaa gcgatcctcc tgcttggatt
86221 cctgaagtgc gagattacag gtgtgagcca ccatacctca acactgtatt ttcaacccgc
86281 tcttcgttca atctccaaag gtgggacatg cggatatgga gggccgattg tRtatggttg
86341 gaccatacac atataaatgg ctttaacctt tactgactct cacagaaccc tcagtgcagt
86401 ggcgtgatct cagctcactg caagctccac ctcccgggtt cacaccattc tcctgcctca
86461 gcctcccgag tagctgggac tacaggggcc cgccaccacg cccggctaat tgttttgtat
86521 ttttttttag tagagacgga gtttcatcgt gttagccaga atagtctcga tcttctgacc
86581 tcgtgatcca cccgcctagg cctcccaaag tgctgggatt acaggcgtga accaccgcac
86641 ccggcctttt tatttttttt gagatggagt ctggctcttg gtccccaggc tggagtgcaa
86701 tggcgggatc tcggctcact gcaacctccg cctcccgggt tcaagcgatt ctcctgcctc
86761 agcctcccga gtagctggga ctacaggtgc gtgccaccac gcccggctaa attttgtatt
86821 tttagtagag acggagtttc acggtgttag ccaggatggt ctcgatctcc gcccgcctcg
86881 gcctctcaaa gtgctgagat tacaggcgtg agccaccacg ccccgcccaa ctcgtccttt
86941 ctttagactt tatcctgtga gggtgaatta tggcctgtcc ctggacacac ccgttctgct
87001 ttccccgcac caactgtatc ccaaataggg gaagtagtct cttcaacctt caaaaatggg
87061 gcactggctg ggcacggtgg ctcacgcctg taaccctagc actttgggag gccgaggcgg
87121 gcggatcacc tgaggtcagg agttcgagac cagcctggcc aacagggtga agccctctct
87181 ctactaaaaa tacaaaaaat agccgggcgt ggtggcgcgc gattgtaatc ccagctattc
87241 aggaggctga ggcaggagaa tcgcttgaac ccgggaggcg gaggctgcag tgagccgaga
87301 tcgcgtcact gcactccagc ctgggcgaca gagcgagact ccatctcaaa aaaaaaaRag
1R7
DEMANDE OU BREVET VOLUMINEUX
LA PRESENTE PARTIE DE CETTE DEMANDE OU CE BREVET COMPREND
PLUS D'UN TOME.
CECI EST LE TOME 1 DE 3
CONTENANT LES PAGES 1 A 189
NOTE : Pour les tomes additionels, veuillez contacter 1e Bureau canadien des
brevets
JUMBO APPLICATIONS/PATENTS
THIS SECTION OF THE APPLICATION/PATENT CONTAINS MORE THAN ONE
VOLUME
THIS IS VOLUME 1 OF 3
CONTAINING PAGES 1 TO 189
NOTE: For additional volumes, please contact the Canadian Patent Office
NOM DU FICHIER / FILE NAME
NOTE POUR LE TOME / VOLUME NOTE: