Note: Descriptions are shown in the official language in which they were submitted.
CA 02567937 2006-11-23
WO 2005/116250 PCT/IB2005/002352
VIRAL AND VIRAL ASSOCIATED MIRNAS AND USES THEREOF
CROSS REFERENCE TO RELATED APPLICATIONS
[0001] The present application is a continuation-in-part of U.S. Patent
Application
No. 10/709,739, filed May 26, 2004, and claims the benefit of U.S. Provisional
Patent
Application No. 60/522,459, filed October 4, 2004 and U.S. Provisional Patent
Application
No. 60/665,094, filed March 25, 2005, each of which is incorporated herein by
reference.
FIELD OF THE INVENTION
[0002] The invention relates in general to viral microRNA molecules and to a
group of human
microRNA molecules associated with viral infections, as well as various
nucleic acid molecules
relating thereto or derived therefrom.
BACKGROUND OF THE INVENTION
[0003] MicroRNAs (miRNAs) are short RNA oligonucleotides of approximately 22
nucleotides
that are involved in gene regulation. MicroRNAs regulate gene expression by
targeting mRNAs
for cleavage or translational repression. Although miRNAs are present in a
wide range of
species including C. elegans, Drosophila and humans, they have only recently
been identified.
More importantly, the role of miRNAs in the development and progression of
disease has only
recently become appreciated.
[0004] As a result of their small size, miRNAs have been difficult to identify
using standard
methodologies. A limited number of miRNAs have been identified by extracting
large quantities
of RNA. MiRNAs have also been identified that contribute to the presentation
of visibly
discemable phenotypes. Expression array data shows that miRNAs are expressed
in different
developmental stages or in different tissues. The restriction of miRNAs to
certain tissues or at
limited developmental stages indicates that the miRNAs identified to date are
likely only a small
fraction of the total miRNAs.
[0005] Computational approaches have recently been developed to identify the
remainder of
miRNAs in the genome. Tools such as MiRscan and MiRseeker have identified
miRNAs that
were later experimentally confirmed. Based on these computational tools, it
has been estimated
that the human genome contains 200-255 miRNA genes. These estimates are based
on an
CA 02567937 2006-11-23
WO 2005/116250 PCT/IB2005/002352
assumption, however, that the miRNAs remaining to be identified will have the
same properties
as those miRNAs already identified. Based on the fundamental importance of
miRNAs in
mammalian biology and disease, the art needs to identify unknown miRNAs. The
present
invention satisfies this need and provides a significant number of miRNAs and
uses therefore.
To date, no viral miRNAs have been detected.
SUMMARY OF THE INVENTION
[0006] The present invention is related to an isolated nucleic acid comprising
a sequence of a
pri-miRNA, pre-miRNA, miRNA, miRNA*, anti-miRNA, or a miRNA binding site, or a
variant
thereof. The nucleic acid may comprise SEQ ID NOS: 4097721-4204913; the
sequence of a
precursor referred to in Table 1, 11-12 or 21-23; SEQ ID NOS: 1-1142416 or
4204914-4204915;
the sequence of a miRNA referred to in Table 1, 13-14 or 21-23; SEQ ID NOS:
1142417-
4097720; the sequence of a target gene binding site referred to in Tables 4,
10 or 15-16; a
complement thereof; or a sequence comprising at least 12 contiguous
nucleotides at least 70%
identical thereto. The isolated nucleic acid may be from 5-250 nucleotides in
length.
[0007] The present invention is also related to a probe comprising the nucleic
acid. The probe
may comprise at least 8-22 contiguous nucleotides complementary to SEQ ID NOS:
1-1142416
or 4204914-4204915, a miRNA referred to in Table 1, 13-14 or 21-23, or a
variant thereof. The
probe may also comprise at least 8-22 contiguous nucleotides complementary to
a human
miRNA differentially expressed in viral infection, or variant thereof.
[0008] The present invention is also related to a plurality of the probes. The
plurality of probes
may comprise at least ten of the probes. The plurality of probes may also
comprise at least 100
nf th~pr Pc 1_..0Z1lt i~yant~~n-is_a1s$4e1at.ed Ao a rmmpneitinn cmmprieiner a
nrohc or
plurality of probes. The present invention is also related to a biochip
comprising a solid
substrate, said substrate comprising a plurality of the probes. Each of the
probes may be attached
to the substrate at a spatially defined address. The biochip may comprise
probes that are
complementary to a viral miRNA. The biochip may also comprise probes that are
complementary to a human miRNA characterized by expression during viral
infection.
[0009] The present invention is also related to a method of detecting
differential expression of a
disease-associated miRNA. A biological sample may be provided and the level of
a nucleic acid
measured that is at least 70% identical to SEQ ID NOS: 1-1142416 or 4204914-
4204915; the
-2-
CA 02567937 2006-11-23
WO 2005/116250 PCT/IB2005/002352
--- - -- -- -
sequence of a miRNA referred to in Table 1, 13-14 and 21-23; or a variant
thereof. A difference
in the level of the nucleic acid compared to a control is indicative of
differential expression.
[0010] The present invention is also related to a method of identifying a
compound that
modulates a pathological condition. A cell may be provided that is capable of
expressing a
nucleic acid at least 70% identical to SEQ ID NOS: 1-1142416; the sequence of
a miRNA
referred to in Table 1, 13-14 and 21-23; or a variant thereof. The cell may be
contacted with a
candidate modulator and then measuring the level of expression of the nucleic
acid. A difference
in the level of the nucleic acid compared to a control identifies the compound
as a modulator of a
pathological condition associated with the nucleic acid.
[0011] The present invention is also related to a method of inhibiting
expression of a target gene
in a cell. Into the cell, a nucleic acid may be introduced in an amount
sufficient to inhibit
expression of the target gene. The target gene may comprise a binding site
substantially identical
to a binding site referred to in Tables 4, 10 or 15-16, or a variant thereof.
The nucleic acid may
comprise a portion of SEQ ID NOS: 1-1142416 or 4204914-4204915; the sequence
of a miRNA
referred to in Table 1, 13-14 or 21-23; or a variant thereof. Expression of
the target gene may be
inhibited in vitro or in vivo.
[0012] The present invention is also related to a method of increasing
expression of a target gene
in a cell. Into the cell, a nucleic acid may be introduced in an amount
sufficient to increase
expression of the target gene. The target gene may comprise a binding site
substantially identical
to a binding site referred to in Tables 4, 10 or 15-16, or a variant thereof.
A portion of the
nucleic acid may be substantially complementary to SEQ ID NOS: 1-1142416 or
4204914-
4204915; the sequence of a miRNA referred to in Table 1, 13-14 or 21-23; or a
variant thereof.
Expre~s
may be increased in vitro or in vivo.
[0013] The present invention is also related to a method of treating a patient
with a disorder set
forth on Table 6 comprising administering to a patient in need thereof a
nucleic acid comprising
a sequence of SEQ ID NOS: 1-760616; a sequence set forth on Table 10; a
sequence set forth on
Table 17; or a variant thereof.
[0014] The present invention is also related to a method of treating a patient
with a viral
infection or a condition associated with a viral infection comprising
administering to a patient in
need thereof a nucleic acid, wherein a portion of the nucleic acid is
substantially complementary
-3-
CA 02567937 2006-11-23
WO 2005/116250 PCT/IB2005/002352
to SEQ ID NOS: 1-1142416 or 4204914-4204915; the sequence of a miRNA referred
to in Table
1, 13-14 or 21-23; or a variant thereof.
BRIEF DESCRIPTION OF THE DRAWINGS
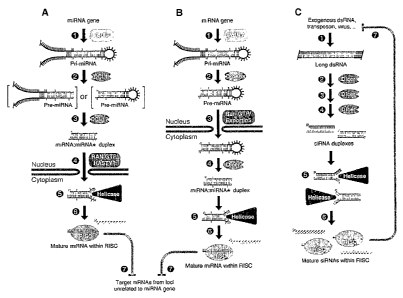
[0015] Figure 1 demonstrates a model of maturation for miRNAs.
[0016] Figure 2A shows the 5'UTR of HIV 1(U5R) containing two predicted miRNAs
in bold.
The mature miRNAs are underlined, one closer to the 5' end (Fig. 2B) and the
second closer to
the 3' end (Fig. 2C). The 5'-most miRNA matches the known HIV 1 RNA structure
named TAR
to which the TAT protein binds (Nature 1987. 330:489-93). A similar miRNA (GAM
NAME
506033) was also lit on the chip. This miRNA probe was designed based on the
sequence of T-
tropic HIV-1 (LAV-1), Subtype B, which is one nucleotide different from the
miRNA presented
in Fig. 2B. Figs. 2B and 2C depict Northern blot analysis of miRNA
oligonucleotides that are
present in U5R, hybridized with predicted mature miRNA probes. The upper arrow
indicates the
molecular size of the entire 355 nt U5R transcript. The predicted molecular
sizes of the two
GAM RNAs are 22 nt and 17 nt, respectively. The lower arrow indicates the 22
nt molecular
marker. Lanes: 1- Hela lysate; 2 -U5R transcript in HeLa Lysate without
incubation; and 3 -
U5R transcript incubated for 24 hours with Hela lysate. Figs. 2D and 2E
present partial
transcripts of HIV 1 RNA reacted with predicted mature HIV 1 miRNA probes. In
each figure, the
experimental transcript sequence is shown, and the predicted mature miRNA is
underlined.
Northern blot analyses of miRNA precursors are presented. It is demonstrated
that one miRNA
precursor transcript is 163 nt and the other miRNA precursor transcript is 200
nt. The predicted
molecular sizes of mature miRNA are both 24 nt. The 22 nt molecular marker is
indicated.
La:Tl=-1=Tran$cript in HaT a-Lysa:t~~kt4l-lC-utJa:tloi-l-cuau c~Ti-ran~~sv~'1-
pt-i-nC~b&ti~-fvi-24-
hours with HeLa lysate.
[0017] Figure 3 shows the results of a Northern Blot. The expression profile
of GAM506333 and
GAM506336 in EBV-infected (B95/8 EBV) and non-infected (pBMC) cells are
presented. The
expression of these miRNAs was demonstrated on a miRNA microarray hybridized
with RNA
from B-95/8 cell lines infected with EBV. Probes against these validated miRNA
predictions
were hybridized with total RNA on a Northern blot. Northern blots confirmed
high expression of
these two miRNAs in the infected cells on the microarray.
[0018] Figure 4 shows validation of miRNAs expressed by EBV.
-4-
CA 02567937 2006-11-23
WO 2005/116250 PCT/IB2005/002352
[0019] Figure 5 shows the knockout of EBV miRNAs.
DETAILED DESCRIPTION
[0020] The present invention provides nucleotide sequences of viral and viral-
associated
miRNAs, precursors thereto, targets thereof and related sequences. Such
nucleic acids are useful
for diagnostic purposes, and also for modifying target gene expression. Other
aspects of the
invention will become apparent to the skilled artisan by the following
description of the
invention.
1. Definitions
[0021] Before the present compounds, products and compositions and methods are
disclosed and
described, it is to be understood that the terminology used herein is for the
purpose of describing
particular embodiments only and is not intended to be limiting. It must be
noted that, as used in
the specification and the appended claims, the singular forms "a," "an" and
"the" include plural
referents unless the context clearly dictates otherwise. It must further be
noted that the terms
"and" and "or" may encompass both conjunctive and disjunctive meaning unless
the context
clearly dictates otherwise.
[0022] "Animal" as used herein may mean fish, amphibians, reptiles, birds, and
mammals, such
as mice, rats, rabbits, goats, cats, dogs, cows, apes and humans.
[0023] "Attached" or "immobilized" as used herein to refer to a probe and a
solid support may
mean that the binding between the probe and the solid support is sufficient to
be stable under
conditions of binding, washing, analysis, and removal. The binding may be
covalent or non-
covalent. Covalent bonds may be formed directly between the probe and the
solid support or
may he fnrmed by a cross linkPr or by inr111sinn of aspPcifiIrl, rPa~ti P
arnnn nn Pither tje-sp.lid-
support or the probe or both molecules. Non-covalent binding may be one or
more of
electrostatic, hydrophilic, and hydrophobic interactions. Included in non-
covalent binding is the
covalent attachment of a molecule, such as streptavidin, to the support and
the non-covalent
binding of a biotinylated probe to the streptavidin. Immobilization may also
involve a
combination of covalent and non-covalent interactions.
[0024] "Biological sample" as used herein may mean a sample of biological
tissue or fluid that
comprises nucleic acids. Such samples include, but are not limited to, tissue
isolated from
animals. Biological samples may also include sections of tissues such as
biopsy and autopsy
-5-
CA 02567937 2006-11-23
WO 2005/116250 PCT/IB2005/002352
samples, frozen sections taken for histologic purposes, blood, plasma, serum,
sputum, stool,
tears, mucus, hair, and skin. Biological samples also include explants and
primary and/or
transformed cell cultures derived from patient tissues. A biological sample
may be provided by
removing a sample of cells from an animal, but can also be accomplished by
using previously
isolated cells (e.g., isolated by another person, at another time, and/or for
another purpose), or by
performing the methods of the invention in vivo. Archival tissues, such as
those having treatment
or outcome history, may also be used.
[0025] "Complement" or "complementary" as used herein may mean Watson-Crick or
Hoogsteen base pairing between nucleotides or nucleotide analogs of nucleic
acid molecules.
[0026] "Differential expression" may mean qualitative or quantitative
differences in the temporal
and/or cellular gene expression patterns within and among cells and tissue.
Thus, a differentially
expressed gene can qualitatively have its expression altered, including an
activation or
inactivation, in, e.g., normal versus disease tissue. Genes may be turned on
or turned off in a
particular state, relative to another state thus permitting comparison of two
or more states. A
qualitatively regulated gene will exhibit an expression pattern within a state
or cell type which
may be detectable by standard techniques. Some genes will be expressed in one
state or cell type,
but not in both. Alternatively, the difference in expression may be
quantitative, e.g., in that
expression is modulated, either up-regulated, resulting in an increased amount
of transcript, or
down-regulated, resulting in a decreased amount of transcript. The degree to
which expression
differs need only be large enough to quantify via standard characterization
techniques such as
expression arrays, quantitative reverse transcriptase PCR, northern analysis,
and RNase
protection.
f 0027j-11 n rnay be ~genomri a~rscript~dtor
translational regulatory sequences and/or a coding region and/or non-
translated sequences (e.g.,
introns, 5'- and 3'-untranslated sequences). The coding region of a gene may
be a nucleotide
sequence coding for an amino acid sequence or a functional RNA, such as tRNA,
rRNA,
catalytic RNA, siRNA, miRNA and antisense RNA. A gene may also be an mRNA or
eDNA
corresponding to the coding regions (e.g., exons and miRNA) optionally
comprising 5'- or 3'-
untranslated sequences linked thereto. A gene may also be an amplified nucleic
acid molecule
produced in vitro comprising all or a part of the coding region and/or 5'- or
3'-untranslated
sequences linked thereto.
-6-
CA 02567937 2006-11-23
WO 2005/116250 PCT/IB2005/002352
[0025] "Host cell" used herein may be a naturally occurring cell or a
transformed cell that
contains a vector and supports the replication of the vector. Host cells may
be cultured cells,
explants, cells in vivo, and the like. Host cells may be prokaryotic cells
such as E. coli, or
eukaryotic cells such as yeast, insect, amphibian, or mammalian cells, such as
CHO, HeLa.
[0029] "Identical" or "identity" as used herein in the context of two or more
nucleic acids or
polypeptide sequences, may mean that the sequences have a specified percentage
of nucleotides
or amino acids that are the same over a specified region. The percentage may
be calculated by
comparing optimally aligning the two sequences, comparing the two sequences
over the
specified region, determining the number of positions at which the identical
residue occurs in
both sequences to yield the number of matched positions, dividing the number
of matched
positions by the total number of positions in the specified region, and
multiplying the result by
100 to yield the percentage of sequence identity. In cases where the two
sequences are of
different lengths or the alignment produces staggered end and the specified
region of comparison
includes only a single sequence, the residues of single sequence are included
in the denominator
but not the numerator of the calculation. When comparing DNA and RNA, thymine
(T) and
uracil (U) are considered equivalent. Identity may be performed manually or by
using computer
sequence algorithm such as BLAST or BLAST 2Ø
[0030] "Inhibit" as used herein may mean prevent, suppress, repress, reduce or
eliminate.
[0031] "Label" as used herein may mean a composition detectable by
spectroscopic,
photochemical, biochemical, immunochemical, chemical, or other physical means.
For example,
useful labels include 32P, fluorescent dyes, electron-dense reagents, enzymes
(e.g., as commonly
used in an ELISA), biotin, digoxigenin, or haptens and other entities which
can be made
dEteet s~nd .
[0032] "Nucleic acid" or "oligonucleotide" or "polynucleotide" used herein may
mean at least
two nucleotides covalently linked together. As will be appreciated by those in
the art, the
depiction of a single strand also defines the sequence of the complementary
strand. Thus, a
nucleic acid also encompasses the complementary strand of a depicted single
strand. As will
also be appreciated by those in the art, many variants of a nucleic acid may
be used for the same
purpose as a given nucleic acid. Thus, a nucleic acid also encompasses
substantially identical
nucleic acids and complements thereof. As will also be appreciated by those in
the art, a single
strand provides a probe for a probe that may hybridize to the target sequence
under stringent
-7-
CA 02567937 2006-11-23
WO 2005/116250 PCT/IB2005/002352
hybridization conditions. Thus, a nucleic acid also encompasses a probe that
hybridizes under
stringent hybridization conditions.
[0033] Nucleic acids may be single stranded or double stranded, or may contain
portions of both
double stranded and single stranded sequence. The nucleic acid may be DNA,
both genomic and
cDNA, RNA, or a hybrid, where the nucleic acid may contain combinations of
deoxyribo- and
ribo-nucleotides, and combinations of bases including uracil, adenine,
thymine, cytosine,
guanine, inosine, xanthine hypoxanthine, isocytosine and isoguanine. Nucleic
acids may be
obtained by chemical synthesis methods or by recombinant methods.
[0034] A nucleic acid will generally contain phosphodiester bonds, although
nucleic acid
analogs may be included that may have at least one different linkage, e.g.,
phosphoramidate,
phosphorothioate, phosphorodithioate, or O-methylphosphoroamidite linkages and
peptide
nucleic acid backbones and linkages. Other analog nucleic acids include those
with positive
backbones; non-ionic backbones, and non-ribose backbones, including those
described in U.S.
Pat. Nos. 5,235,033 and 5,034,506, which are incorporated by reference.
Nucleic acids
containing one or more non-naturally occurring or modified nucleotides are
also included within
one definition of nucleic acids. The modified nucleotide analog may be located
for example at
the 5'-end and/or the 3'-end of the nucleic acid molecule. Representative
examples of nucleotide
analogs may be selected from sugar- or backbone-modified ribonucleotides. It
should be noted,
however, that also nucleobase-modified ribonucleotides, i.e. ribonucleotides,
containing a non-
naturally occurring nucleobase instead of a naturally occurring nucleobase
such as uridines or
cytidines modified at the 5-position, e.g. 5-(2-amino)propyl uridine, 5-bromo
uridine; adenosines
and guanosines modified at the 8-position, e.g. 8-bromo guanosine; deaza
nucleotides, e.g. 7-
lea~u=ade , - - , - -
OH-group may be replaced by a group selected from H, OR, R, halo, SH, SR, NH2,
NHR, NR2
or CN, wherein R is C1-C6 alkyl, alkenyl or alkynyl and halo is F, Cl, Br or
I. Modifications of
the ribose-phosphate backbone may be done for a variety of reasons, e.g., to
increase the stability
and half-life of such molecules in physiological environments or as probes on
a biochip.
Mixtures of naturally occurring nucleic acids and analogs may be made;
alternatively, mixtures
of different nucleic acid analogs, and mixtures of naturally occurring nucleic
acids and analogs
may be made.
-8-
CA 02567937 2006-11-23
WO 2005/116250 PCT/IB2005/002352
[0035] "Operably linked" used herein may mean that expression of a gene is
under the control of
a promoter with which it is spatially connected. A promoter may be positioned
5' (upstream) or
3' (downstream) of the gene under its control. The distance between the
promoter and the gene
may be approximately the same as the distance between that promoter and the
gene it controls in
the gene from which the promoter is derived. As is known in the art, variation
in this distance
can be accommodated without loss of promoter function.
[0036] "Probe" as used herein may mean an oligonucleotide capable of binding
to a target
nucleic acid of complementary sequence through one or more types of chemical
bonds, usually
through complementary base pairing, usually through hydrogen bond formation.
Probes may
bind target sequences lacking complete complementarity with the probe sequence
depending
upon the stringency of the hybridization conditions. There may be any number
of base pair
mismatches which will interfere witli hybridization between the target
sequence and the single
stranded nucleic acids of the present invention. However, if the number of
mutations is so great
that no hybridization can occur under even the least stringent of
hybridization conditions, the
sequence is not a complementary target sequence. A probe may be single
stranded or partially
single and partially double stranded. The strandedness of the probe is
dictated by the structure,
composition, and properties of the target sequence. Probes may be directly
labeled or indirectly
labeled such as with biotin to which a streptavidin complex may later bind.
[0037] "Promoter" as used herein may mean a synthetic or naturally-derived
molecule which is
capable of conferring, activating or enhancing expression of a nucleic acid in
a cell. A promoter
may comprise one or more specific regulatory elements to further enhance
expression and/or to
alter the spatial expression and/or temporal expression of same. A promoter
may also comprise
dYSII~;:11 , severa ousan ase
pairs from the start site of transcription. A promoter may be derived from
sources including viral,
bacterial, fungal, plants, insects, and animals. A promoter may regulate the
expression of a gene
component constitutively, or differentially with respect to cell, the tissue
or organ in which
expression occurs or, with respect to the developmental stage at which
expression occurs, or in
response to external stimuli such as physiological stresses, pathogens, metal
ions, or inducing
agents. Representative examples of promoters include the bacteriophage T7
promoter,
bacteriophage T3 promoter, SP6 promoter, lac operator-promoter, tac promoter,
SV40 late
-9-
CA 02567937 2006-11-23
WO 2005/116250 PCT/IB2005/002352
promoter, SV40 early promoter, RSV-LTR promoter, CMV IE promoter, SV40 early
promoter
or SV40 late promoter and the CMV IE promoter.
[0038] "Selectable marker" used herein may mean any gene which confers a
phenotype on a cell
in which it is expressed to facilitate the identification and/or selection of
cells which are
transfected or transformed with a genetic construct. Representative examples
of selectable
markers include the ampicillin-resistance gene (Ampr), tetracycline-resistance
gene (Tcr),
bacterial kanamycin-resistance gene (Kan'), zeocin resistance gene, the AURI-C
gene which
confers resistance to the antibiotic aureobasidin A, phosphinothricin-
resistance gene, neomycin
phosphotransferase gene (nptll), hygromycin-resistance gene, beta-
glucuronidase (GUS) gene,
chloramphenicol acetyltransferase (CAT) gene, green fluorescent protein-
encoding gene and
luciferase gene.
[0039] "Stringent hybridization conditions" used herein may mean conditions
under which a first
nucleic acid sequence (e.g., probe) will hybridize to a second nucleic acid
sequence (e.g., target),
such as in a complex mixture of nucleic acids, but to no other sequences.
Stringent conditions
are sequence-dependent and will be different in different circumstances.
Generally, stringent
conditions are selected to be about 5-10 C lower than the thermal melting
point (Tm) for the
specific sequence at a defined ionic strength pH. The Tm may be the
temperature (under defined
ionic strength, pH, and nucleic concentration) at which 50% of the probes
complementary to the
target hybridize to the target sequence at equilibrium (as the target
sequences are present in
excess, at Tm, 50% of the probes are occupied at equilibrium). Stringent
conditions may be
those in which the salt concentration is less than about 1.0 M sodium ion,
typically about 0.01-
1.0 M sodium ion concentration (or other salts) at pH 7.0 to 8.3 and the
temperature is at least
about 30 C for short probes (e.g., about 10-50 nucleotides) and at least about
60 C for long
probes (e.g., greater than about 50 nucleotides). Stringent conditions may
also be achieved with
the addition of destabilizing agents such as formamide. For selective or
specific hybridization, a
positive signal may be at least 2 to 10 times background hybridization.
Exemplary stringent
hybridization conditions include the following: 50% formamide, 5x SSC, and 1%
SDS,
incubating at 42 C, or, 5x SSC, 1% SDS, incubating at 65 C, with wash in 0.2x
SSC, and 0.1%
SDS at 65 C.
[0040] "Substantially complementary" used herein may mean that a first
sequence is at least
60%, 65%, 70%, 75%, 80%, 85%, 90%, 95%, 97%, 98% or 99% identical to the
complement of
-10-
CA 02567937 2006-11-23
WO 2005/116250 PCT/IB2005/002352
a second sequence over a region of 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18,
19, 20, 21, 22, 23, 24,
25, 30, 35, 40, 45, 50 or more nucleotides, or that the two sequences
hybridize under stringent
hybridization conditions.
[0041] "Substantially identical" used herein may mean that a first and second
sequence are at
least 60%, 65%, 70%, 75%, 80%, 85%, 90%, 95%, 97%, 98% or 99% identical over a
region of
8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 30, 35,
40, 45, 50 or more
nucleotides or amino acids, or with respect to nucleic acids, if the first
sequence is substantially
complementary to the complement of the second sequence.
[0042] "Target" as used herein may mean a polynucleotide that may be bound by
one or more
probes under stringent hybridization conditions.
[0043] "Terminator" used herein may mean a sequence at the end of a
transcriptional unit which
signals termination of transcription. A terminator may be a 3'-non-translated
DNA sequence
containing a polyadenylation signal, which may facilitate the addition of
polyadenylate
sequences to the 3'-end of a primary transcript. A terminator may be derived
from sources
including viral, bacterial, fungal, plants, insects, and animals.
Representative examples of
terminators include the SV40 polyadenylation signal, HSV TK polyadenylation
signal, CYC1
terminator, ADH terminator, SPA terminator, nopaline synthase (NOS) gene
terminator of
Agrobacterium tumefaciens, the terminator of the Cauliflower mosaic virus
(CaMV) 35S gene,
the zein gene terminator from Zea mays, the Rubisco small subunit gene (SSU)
gene terminator
sequences, subclover stunt virus (SCSV) gene sequence terminators, rho-
independent E. coli
terminators, and the lacZ alpha terminator.
[0044] "Treat" or "treating" used herein when referring to protection of an
animal from a
condit' , , uppressiT, repx , 'tirnrPreventing
the condition involves administering a composition of the present invention to
an animal prior to
onset of the condition. Suppressing the condition involves administering a
composition of the
present invention to an animal after induction of the condition but before its
clinical appearance.
Repressing the condition involves administering a composition of the present
invention to an
animal after clinical appearance of the condition such that the condition is
reduced or prevented
from worsening. Elimination of the condition involves administering a
composition of the
present invention to an animal after clinical appearance of the condition such
that the animal no
longer suffers from the condition.
-11-
CA 02567937 2006-11-23
WO 2005/116250 PCT/IB2005/002352
[0045] "Vector" used herein may mean a nucleic acid sequence containing an
origin of
replication. A vector may be a plasmid, bacteriophage, bacterial artificial
chromosome or yeast
artificial chromosome. A vector may be a DNA or RNA vector. A vector may be
either a self-
replicating extrachromosomal vector or a vector which integrates into a host
genome.
2. MicroRNA
[0046] While not being bound by theory, the current model for the maturation
of mammalian
miRNAs is shown in Figure 1. A gene coding for a miRNA may be transcribed
leading to
production of an miRNA precursor known as the pri-miRNA. The pri-miRNA may be
part of a
polycistronic RNA comprising multiple pri-miRNAs. The pri-miRNA may form a
hairpin with a
stem and loop. As indicated on Figure 1, the stem may comprise mismatched
bases.
[0047] The hairpin structure of the pri-miRNA may be recognized by Drosha,
which is an RNase
III endonuclease. Drosha may recognize terminal loops in the pri-miRNA and
cleave
approximately two helical turns into the stem to produce a 60-70 nt precursor
known as the pre-
miRNA. Drosha may cleave the pri-miRNA with a staggered cut typical of RNase
III
endonucleases yielding a pre-miRNA stem loop with a 5' phosphate and -2
nucleotide 3'
overhang. Approximately one helical turn of stem (-10 nucleotides) extending
beyond the
Drosha cleavage site may be essential for efficient processing. The pre-miRNA
may then be
actively transported from the nucleus to the cytoplasm by Ran-GTP and the
export receptor Ex-
portin-5.
[0048] The pre-miRNA may be recognized by Dicer, which is also an RNase III
endonuclease.
Dicer may recognize the double-stranded stem of the pre-miRNA. Dicer may also
recognize the
5' phosphate and 3' overhang at the base of the stem loop. Dicer may cleave
off the terminal
loep t~ve 13e1ic~1-t~trns ~wa~fr eaving~rradd~t~nal-5'-phosphate
and -2 nucleotide 3' overhang. The resulting siRNA-like duplex, which may
comprise
mismatches, comprises the mature miRNA and a similar-sized fragment known as
the miRNA*.
The miRNA and miRNA* may be derived from opposing arms of the pri-miRNA and
pre-
miRNA. MiRNA* sequences may be found in libraries of cloned miRNAs but
typically at lower
frequency than the miRNAs.
[0049] Although initially present as a double-stranded species with miRNA*,
the miRNA may
eventually become incorporated as single-stranded RNAs into a
ribonucleoprotein complex
known as the RNA-induced silencing complex (RISC). Various proteins can form
the RISC,
-12-
CA 02567937 2006-11-23
WO 2005/116250 PCT/IB2005/002352
which can lead to variability in specifity for miRNA/miRNA* duplexes, binding
site of the target
gene, activity of miRNA (repress or activate), which strand of the
miRNA/miRNA* duplex is
loaded in to the RISC.
[0050] When the miRNA strand of the miRNA:miRNA* duplex is loaded into the
RISC, the
miRNA* may be removed and degraded. The strand of the miRNA:miRNA* duplex that
is
loaded into the RISC may be the strand whose 5' end is less tightly paired. In
cases where both
ends of the miRNA:miRNA* have roughly equivalent 5' pairing, both miRNA and
miRNA* may
have gene silencing activity.
[0051] The RISC may identify target nucleic acids based on high levels of
complementarity
between the miRNA and the mRNA, especially by nucleotides 2-8 of the miRNA.
Only one
case has been reported in animals where the interaction between the miRNA and
its target was
along the entire length of the miRNA. This was shown for mir-196 and Hox B8
and it was
further shown that mir-196 mediates the cleavage of the Hox B8 mRNA (Yekta et
al 2004,
Science 304-594). Otherwise, such interactions are known only in plants
(Bartel & Bartel 2003,
Plant Physiol 132-709).
[0052] A number of studies have looked at the base-pairing requirement between
miRNA and its
mRNA target for achieving efficient inhibition of translation (reviewed by
Bartel 2004, Cell 116-
281). In mammalian cells, the first 8 nucleotides of the miRNA may be
important (Doench &
Sharp 2004 GenesDev 2004-504). However, other parts of the microRNA may also
participate
in mRNA binding. Moreover, sufficient base pairing at the 3' can compensate
for insufficient
pairing at the 5' (Brennecke at al, 2005 PLoS 3-e85). Computation studies,
analyzing miRNA
binding on whole genomes have suggested a specific role for bases 2-7 at the
5' of the miRNA in
z-uf-t1e first'nucl~t'~d~; f~n-d -u-sually t be was a so recognize
(Lewis et at 2005 Cell 120-15). Similarly, nucleotides 1-7 or 2-8 were used to
identify and
validate targets by Krek et al (2005, Nat Genet 37-495).
[0053] The target sites in the mRNA may be in the 5' UTR, the 3' UTR or in the
coding region.
Interestingly, multiple miRNAs may regulate the same mRNA target by
recognizing the same or
multiple sites. The presence of multiple miRNA complementarity sites in most
genetically
identified targets may indicate that the cooperative action of multiple RISCs
provides the most
efficient translational inhibition.
-13-
CA 02567937 2006-11-23
WO 2005/116250 PCT/IB2005/002352
[0054] MiRNAs may direct the RISC to downregulate gene expression by either of
two
mechanisms: mRNA cleavage or translational repression. The miRNA may specify
cleavage of
the mRNA if the mRNA has a certain degree of complementarity to the miRNA.
When a
miRNA guides cleavage, the cut may be between the nucleotides pairing to
residues 10 and 11 of
the miRNA. Alternatively, the miRNA may repress translation if the miRNA does
not have the
requisite degree of complementarity to the miRNA. Translational repression may
be more
prevalent in animals since animals may have a lower degree of complementarity.
[0055] It should be notes that there may be variability in the 5' and 3' ends
of any pair of
miRNA and miRNA*. This variability may be due to variability in the enzymatic
processing of
Drosha and Dicer with respect to the site of cleavage. Variability at the 5'
and 3' ends of
miRNA and miRNA* may also be due to mismatches in the stem structures of the
pri-miRNA
and pre-miRNA. The mismatches of the stem strands may lead to a population of
different
hairpin structures. Variability in the stem structures may also lead to
variability in the products
of cleavage by Drosha and Dicer.
3. Nucleic Acid
[0056] The present invention relates to an isolated nucleic acid comprising a
nucleotide sequence
referred to in SEQ ID NOS: 1-4204915, the sequences referred to in Tables 1,
4, 10-14 and 21-
23, and variants thereof. The variant may be a complement of the referenced
nucleotide
sequence. The variant may also be a nucleotide sequence that is substantially
identical to the
referenced nucleotide sequence or the complement thereof. The variant may also
be a nucleotide
sequence which hybridizes under stringent conditions to the referenced
nucleotide sequence,
complements thereof, or nucleotide sequences substantially identical thereto.
[0e omcil
have a length of at least 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22,
23, 24, 25, 26, 27, 28,
29, 30, 35, 40, 45, 50, 60, 70, 80 or 90 nucleotides. The nucleic acid may be
synthesized or
expressed in a cell (in vitro or in vivo) using a synthetic gene described
below. The nucleic acid
may be synthesized as a single strand molecule and hybridized to a
substantially complementary
nucleic acid to form a duplex, which is considered a nucleic acid of the
invention. The nucleic
acid may be introduced to a cell, tissue or organ in a single- or double-
stranded form or capable
of being expressed by a synthetic gene using methods well known to those
skilled in the art,
including as described in U.S. Patent No. 6,506,559 which is incorporated by
reference.
-14-
CA 02567937 2006-11-23
WO 2005/116250 PCT/IB2005/002352
a. Pri-miRNA
[0058] The nucleic acid of the invention may comprise a sequence of a pri-
miRNA or a variant
thereof. The pri-miRNA sequence may comprise from 45-250, 55-200, 70-150 or 80-
100
nucleotides. The sequence of the pri-miRNA may comprise a pre-miRNA, miRNA and
miRNA* as set forth below. The pri-miRNA may also comprise a miRNA or miRNA*
and the
complement thereof, and variants thereof. The pri-miRNA may comprise at least
19% adenosine
nucleotides, at least 16% cytosine nucleotides, at least 23% thymine
nucleotides and at least 19%
guanine nucleotides.
[0059]. The pri-miRNA may form a hairpin structure. The hairpin may comprise a
first and
second nucleic acid sequence that are substantially complementary. The first
and second nucleic
acid sequence may be from 37-50 nucleotides. The first and second nucleic acid
sequence may
be separated by a third sequence of from 8-12 nucleotides. The hairpin
structure may have a free
energy less than -25 Kcal/mole as calculated by the Vienna algorithm with
default parameters, as
described in Hofacker et al., Monatshefte f. Chemie 125: 167-188 (1994), the
contents of which
are incorporated herein. The hairpin may comprise a terminal loop of 4-20, 8-
12 or 10
nucleotides.
[0060] The sequence of the pri-miRNA may comprise SEQ ID NOS: 4097721-4204913,
a
precursor referred to in Table 1, the sequence of a sequence referred to in
Tables 11-12 and 21-
23, or a variant thereof.
b. Pre-miRNA
[0061] The nucleic acid of the invention may also comprise a sequence of a pre-
miRNA or a
variant thereof. The pre-miRNA sequence may comprise from 45-90, 60-80 or 60-
70
z=ReleQtides. T~ze seeluenee ~r~riRl'd~ Tnay~anrp ' t
forth below. The pre-miRNA may also comprise a miRNA or miRNA* and the
complement
thereof, and variants thereof. The sequence of the pre-miRNA may also be that
of a pri-miRNA
excluding from 0-160 nucleotides from the 5' and 3' ends of the pri-miRNA.
[0062] The sequence of the pre-miRNA may comprise SEQ ID NOS: 4097721-4204913,
a
precursor referred to in Table 1, the sequence of a sequence referred to in
Tables 11-12 and 21-
23, or a variant thereof.
-15-
CA 02567937 2006-11-23
WO 2005/116250 PCT/IB2005/002352
c. MiRNA
[0063] The nucleic acid of the invention may also comprise a sequence of a
miRNA, miRNA* or
a variant thereof. The miRNA sequence may comprise from 13-33, 18-24 or 21-23
nucleotides.
The sequence of the miRNA may be the first 13-33 nucleotides of the pre-miRNA.
The
sequence of the miRNA may be the last 13-33 nucleotides of the pre-miRNA.
[0064] The sequence of the miRNA may comprise SEQ ID NOS: 1-1142416 or 4204914-
4204915, a miRNA referred to in Table 1, the sequence of a sequence referred
to in Tables 11-12
and 21-23, or a variant thereof.
d. Anti-miRNA
[0065] The nucleic acid of the invention may also comprise a sequence of an
anti-miRNA that is
capable of blocking the activity of a miRNA or miRNA*. The anti-miRNA may
comprise a total
of 5-100 or 10-60 nucleotides. The anti-miRNA may also comprise a total of at
least 5, 6, 7, 8,
9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25 or 26
nucleotides. The sequence of
the anti-miRNA may comprise (a) at least 5 nucleotides that are substantially
identical to the 5'
of a miRNA and at least 5-12 nucleotide that are substantially complementary
to the flanking
regions of the target site from the 5' end of said miRNA, or (b) at least 5-12
nucleotides that are
substantially identical to the 3' of a miRNA and at least 5 nucleotide that
are substantially
complementary to the flanking region of the target site from the 3' end of
said miRNA.
[0066] The sequence of the anti-miRNA may comprise the complement of SEQ ID
NOS: 1-
1142416 or 4204914-4204915, a sequence of a miRNA referred to in Tables 1, 13-
14 or 21-23,
or a variant thereof.
e. Binding Site of Target
[Owl~ 11~11biG1Ui~ a6~d (~f th~ 1"z~~ vr+'=~vi~ ~'' ~~.a'~' a1S~ ~.~:::Y ~$~
a S~qklgn6g 8f a ta~ggt~lllPd Vt~
binding site, or a variant thereof. The target site sequence may comprise a
total of 5-100 or 10-
60 nucleotides. The target site sequence may comprise at least 5 nucleotides
of SEQ ID NOS:
1142417-4097720, the sequence of a target gene binding site referred to in
Tables 4, 10 or 15-16,
or a variant thereof.
4. Synthetic Gene
[0068] The present invention also relates to a synthetic gene comprising a
nucleic acid of the
invention operably linked to a transcriptional and/or translational regulatory
sequences. The
synthetic gene may be capable of modifying the expression of a target gene
with a binding site
-16-
CA 02567937 2006-11-23
WO 2005/116250 PCT/IB2005/002352
for the nucleic acid of the invention. Expression of the target gene may be
modified in a cell,
tissue or organ. The synthetic gene may be synthesized or derived from
naturally-occurring
genes by standard recombinant techniques. The synthetic gene may also comprise
terminators at
the 3'-end of the transcriptional unit of the synthetic gene sequence. The
synthetic gene may also
comprise a selectable marker.
5. Vector
[0069] The present invention also relates to a vector comprising a synthetic
gene of the
invention. The vector may be an expression vector. An expression vector may
comprise
additional elements. For example, the expression vector may have two
replication systems
allowing it to be maintained in two organisms, e.g., in mammalian or insect
cells for expression
and in a prokaryotic host for cloning and amplification. For integrating
expression vectors, the
expression vector may contain at least one sequence homologous to the host
cell genome, and
preferably two homologous sequences which flank the expression construct. The
integrating
vector may be directed to a specific locus in the host cell by selecting the
appropriate
homologous sequence for inclusion in the vector. The vector may also comprise
a selectable
marker gene to allow the selection of transformed host cells.
6. Host Cell
[0070] The present invention also relates to a host cell comprising a vector
of the invention. The
cell may be a bacterial, fungal, plant, insect or animal cell.
7. Probes
[0071] The present invention also relates to a probe comprising a nucleic acid
of the invention.
Probes may be used for screening and diagnostic methods, as outlined below.
The probe may be
attached-ar-inunobilia , ohip -
[0072] The probe may have a length of from 8 to 500, 10 to 100 or 20 to 60
nucleotides. The
probe may also have a length of at least 8, 9, 10, 11, 12, 13, 14, 15, 16, 17,
18, 19, 20, 21, 22, 23,
24, 25, 26, 27, 28, 29, 30, 35, 40, 45, 50, 60, 70, 80, 90, 100, 120, 140,
160, 180, 200, 220, 240,
260, 280 or 300 nucleotides. The probe may further comprise a linker sequence
of from 10-60
nucleotides.
8. Biochip
[0073] The present invention also relates to a biochip. The biochip may
comprise a solid
substrate comprising an attached probe or plurality of probes of the
invention. The probes may
-17-
CA 02567937 2006-11-23
WO 2005/116250 PCT/IB2005/002352
be capable of hybridizing to a target sequence under stringent hybridization
conditions. The
probes may be attached at spatially defined address on the substrate. More
than one probe per
target sequence may be used, with either overlapping probes or probes to
different sections of a
particular target sequence. The probes may be capable of hybridizing to target
sequences
associated with a single disorder.
[0074] The probes may be attached to the biochip in a wide variety of ways, as
will be
appreciated by those in the art. The probes may either be synthesized first,
with subsequent
attachment to the biochip, or may be directly synthesized on the biochip.
[0075] The solid substrate may be a material that may be modified to contain
discrete individual
sites appropriate for the attachment or association of the probes and is
amenable to at least one
detection method. Representative examples of substrates include glass and
modified or
functionalized glass, plastics (including acrylics, polystyrene and copolymers
of styrene and
other materials, polypropylene, polyethylene, polybutylene, polyurethanes,
TeflonJ, etc.),
polysaccharides, nylon or nitrocellulose, resins, silica or silica-based
materials including silicon
and modified silicon, carbon, metals, inorganic glasses and plastics. The
substrates may allow
optical detection without appreciably fluorescing.
[0076] The substrate may be planar, although other configurations of
substrates may be used as
well. For example, probes may be placed on the inside surface of a tube, for
flow-through
sample analysis to minimize sample volume. Similarly, the substrate may be
flexible, such as a
flexible foam, including closed cell foams made of particular plastics.
[0077] The biochip and the probe may be derivatized with chemical functional
groups for
subsequent attachment of the two. For example, the biochip may be derivatized
with a chemical
, > > cuiboxyl >
thiol groups. Using these functional groups, the probes may be attached using
functional groups
on the probes either directly or indirectly using a linker. The probes may be
attached to the solid
support by either the 5' terminus, 3' terminus, or via an internal nucleotide.
[0078] The probe may also be attached to the solid support non-covalently. For
example,
biotinylated oligonucleotides can be made, which may bind to surfaces
covalently coated with
streptavidin, resulting in attachment. Alternatively, probes may be
synthesized on the surface
using techniques such as photopolymerization and photolithography.
-18-
CA 02567937 2006-11-23
WO 2005/116250 PCT/IB2005/002352
9. miRNA expression analysis
[0079] The present invention also relates to a method of identifying miRNAs
that are associated
with disease or a pathological condition, such as viral infection, comprising
contacting a
biological sample with a probe or biochip of the invention and detecting the
amount of
hybridization. PCR may be used to amplify nucleic acids in the sample, which
may provide
higher sensitivity.
[0080] The ability to identify miRNAs that are overexpressed or underexpressed
in pathological
cells compared to a control can provide high-resolution, high-sensitivity
datasets which may be
used in the areas of diagnostics, therapeutics, drug development,
pharmacogenetics, biosensor
development, and other related areas. An expression profile generated by the
current methods
may be a "fingerprint" of the state of the sample with respect to a number of
miRNAs. While
two states may have any particular miRNA similarly expressed, the evaluation
of a number of
miRNAs simultaneously allows the generation of a gene expression profile that
is characteristic
of the state of the cell. That is, normal tissue may be distinguished from
diseased tissue. By
comparing expression profiles of tissue in known different disease states,
information regarding
which miRNAs are associated in each of these states may be obtained. Then,
diagnosis may be
performed or confirmed to determine whether a tissue sample has the expression
profile of
normal or disease tissue. This may provide for molecular diagnosis of related
conditions.
10. Determining Expression Levels
[0081] The present invention also relates to a method of determining the
expression level of a
disease-associated miRNA comprising contacting a biological sample with a
probe or biochip of
the invention and measuring the amount of hybridization. The expression level
of a disease-
-as-seeiated rxiFNAis infex33atien-in a nt~r~fi , -ential-expr-e~n-aÃ
a disease-associated miRNA compared to a control may be used as a diagnostic
that a patient
suffers from the disease. Expression levels of a disease-associated miRNA may
also be used to
monitor the treatment and disease state of a patient. Furthermore, expression
levels of a disease-
associated miRNA may allow the screening of drug candidates for altering a
particular
expression profile or suppressing an expression profile associated with
disease.
[0082] A target nucleic acid may be detected by contacting a sample comprising
the target
nucleic acid with a biochip comprising an attached probe sufficiently
complementary to the
target nucleic acid and detecting hybridization to the probe above control
levels.
-19-
CA 02567937 2006-11-23
WO 2005/116250 PCT/IB2005/002352
[0083] The target nucleic acid may also be detected by immobilizing the
nucleic acid to be
examined on a solid support such as nylon membranes and hybridizing a labeled
probe with the
sample. Similarly, the target nucleic may also be detected by immobilizing the
labeled probe to
the solid support and hybridizing a sample comprising a labeled target nucleic
acid. Following
washing to remove the non-specific hybridization, the label may be detected.
[0084] The target nucleic acid may also be detected in situ by contacting
permeabilized cells or
tissue samples with a labeled probe to allow hybridization with the target
nucleic acid. Following
washing to remove the non-specifically bound probe, the label may be detected.
[0085] These assays can be direct hybridization assays or can comprise
sandwich assays, which
include the use of multiple probes, as generally outlined in U.S. Pat. Nos.
5,681,702; 5,597,909;
5,545,730; 5,594,117; 5,591,584; 5,571,670; 5,580,731; 5,571,670; 5,591,584;
5,624,802;
5,635,352; 5,594,118; 5,359,100; 5,124,246; and 5,681,697, each of which is
hereby
incorporated by reference.
[0086] A variety of hybridization conditions may be used, including high,
moderate and low
stringency conditions as outlined above. The assays may be performed under
stringency
conditions which allow hybridization of the probe only to the target.
Stringency can be
controlled by altering a step parameter that is a thermodynamic variable,
including, but not
limited to, temperature, formamide concentration, salt concentration,
chaotropic salt
concentration pH, or organic solvent concentration.
[0087] Hybridization reactions may be accomplished in a variety of ways.
Components of the
reaction may be added simultaneously, or sequentially, in different orders. In
addition, the
reaction may include a variety of other reagents. These include salts,
buffers, neutral proteins,
"~.aZii~ a~~er-gen~ , ,
ti-.g~ "oti r,~atiroirarrd
detection, and/or reduce non-specific or background interactions. Reagents
that otherwise
improve the efficiency of the assay, such as protease inhibitors, nuclease
inhibitors and anti-
microbial agents may also be used as appropriate, depending on the sample
preparation methods
and purity of the target.
a. Diagnostic
[0088] The present invention also relates to a method of diagnosis comprising
detecting a
differential expression level of a disease- or infection-associated miRNA in a
biological sample.
The miRNA may be a viral miRNA, which may be expressed in the infected
subject. The
-20-
CA 02567937 2006-11-23
WO 2005/116250 PCT/IB2005/002352
miRNA may also be from the subject, the expression level of which is modified
due to a viral
infection. The sample may be derived from a patient. Diagnosis of a disease
state in a patient
allows for prognosis and selection of therapeutic strategy. Further, the
developmental stage of
cells may be classified by determining temporarily expressed miRNA-molecules.
[0089] In situ hybridization of labeled probes to tissue arrays may be
performed. When
comparing the fingerprints between an individual and a standard, the skilled
artisan can make a
diagnosis, a prognosis, or a prediction based on the findings. It is further
understood that the
genes which indicate the diagnosis may differ from those which indicate the
prognosis and
molecular profiling of the condition of the cells may lead to distinctions
between responsive or
refractory conditions or may be predictive of outcomes.
b. Drug Screening
[0090] The present invention also relates to a method of screening
therapeutics comprising
contacting a pathological cell capable of expressing a disease related miRNA
with a candidate
therapeutic and evaluating the effect of a drug candidate on the expression
profile of the disease
associated miRNA. Having identified the differentially expressed miRNAs, a
variety of assays
may be executed. Test compounds may be screened for the ability to modulate
gene expression
of the disease associated miRNA. Modulation includes both an increase and a
decrease in gene
expression.
[0091] The test compound or drug candidate may be any molecule, e.g., protein,
oligopeptide,
small organic molecule, polysaccharide, polynucleotide, etc., to be tested for
the capacity to
directly or indirectly alter the disease phenotype or the expression of the
disease associated
miRNA. Drug candidates encompass numerous chemical classes, such as small
organic
~eeules having ~maleetrlar weiglit af~re tiran 160 arre~ {i~800; 1~-00;
2,000 or 2,500 daltons. Candidate compounds may comprise functional groups
necessary for
structural interaction with proteins, particularly hydrogen bonding, and
typically include at least
an amine, carbonyl, hydroxyl or carboxyl group, preferably at least two of the
functional
chemical groups. The candidate agents may comprise cyclical carbon or
heterocyclic structures
and/or aromatic or polyaromatic structures substituted with one or more of the
above functional
groups. Candidate agents are also found among biomolecules including peptides,
saccharides,
fatty acids, steroids, purines, pyrimidines, derivatives, structural analogs
or combinations thereof.
-21-
CA 02567937 2006-11-23
WO 2005/116250 PCT/IB2005/002352
[0092] Combinatorial libraries of potential modulators may be screened for the
ability to bind to
the disease associated miRNA or to modulate the activity thereof. The
combinatorial library
may be a collection of diverse chemical compounds generated by either chemical
synthesis or
biological synthesis by combining a number of chemical building blocks such as
reagents.
Preparation and screening of combinatorial chemical libraries is well known to
those of skill in
the art. Such combinatorial chemical.libraries include, but are not limited
to, peptide libraries
encoded peptides, benzodiazepines, diversomers such as hydantoins,
benzodiazepines and
dipeptide, vinylogous polypeptides, analogous organic syntheses of small
compound libraries,
oligocarbamates, and/or peptidyl phosphonates, nucleic acid libraries, peptide
nucleic acid
libraries, antibody libraries, carbohydrate libraries, and small organic
molecule libraries.
11. Gene Silencing
[0093] The present invention also relates to a method of using the nucleic
acids of the invention
to reduce expression of a target gene in a cell, tissue or organ. Expression
of the target gene
may be reduced by expressing a nucleic acid of the invention that comprises a
sequence
substantially complementary to one or more binding sites of the target mRNA.
The nucleic acid
may be a miRNA or a variant thereof. The nucleic acid may also be pri-miRNA,
pre-miRNA,
or a variant thereof, which may be processed to yield a miRNA. The expressed
miRNA may
hybridize to a substantially complementary binding site on the target mRNA,
which may lead to
activation of RISC-mediated gene silencing. An example for a study employing
over-expression
of miRNA is Yekta et al 2004, Science 304-594, which is incorporated herein by
reference. One
of ordinary skill in the art will recognize that the nucleic acids of the
present invention may be
used to inhibit expression of target genes using antisense methods well known
in the art, as well
~ v ~n
as D'lk,Ai rneth0ds-describe~i~t T-o:- .-Pator-~~ s: 6;5063-59 aria ~~a 6,-
5r~~~9-vvhiel~aie-
incorporated by reference.
[0094] The target gene may be a viral gene, which may be reduced by expressing
a viral or
human miRNA. The target gene may also be a human gene that is expressed upon
viral
infection, which may be reduced by expressing a viral or human miRNA. The
target of gene
silencing may be a protein that causes the silencing of a second protein. By
repressing
expression of the target gene, expression of the second protein may be
increased. Examples for
efficient suppression of miRNA expression are the studies by Esau et al 2004
JBC 275-52361;
and Cheng et al 2005 Nucleic Acids Res. 33-1290, which is incorporated herein
by reference.
-22-
CA 02567937 2006-11-23
WO 2005/116250 PCT/IB2005/002352
12. Gene Enhancement
[0095] The present invention also relates to a method of using the nucleic
acids of the invention
to increase expression of a target gene in a cell, tissue or organ. Expression
of the target gene
may be increased by expressing a nucleic acid of the invention that comprises
a sequence
substantially complementary to a pri-miRNA, pre-miRNA, miRNA or a variant
thereof. The
nucleic acid may be an anti-miRNA. The anti-miRNA may hybridize with a pri-
miRNA, pre-
miRNA or miRNA, thereby reducing its gene repression activity. Expression of
the target gene
may also be increased by expressing a nucleic acid of the invention that is
substantially
complementary to a portion of the binding site in the target gene, such that
binding of the nucleic
acid to the binding site may prevent miRNA binding.
[0096] The target gene may be a viral gene, expression of which may reduce
infectivity of the
virus. The target gene may also be a human gene, expression of which may
reduce infectivity of
the virus or increase resistance or immunity to the viral infection.
13. Therapeutic
[0097] The present invention also relates to a method of using the nucleic
acids of the invention
as modulators or targets of disease or disorders, such as those associated
with viral infection. In
general, the claimed nucleic acid molecules may be used as a modulator of the
expression of
genes which are at least partially complementary to said nucleic acid.
Further, miRNA molecules
may act as target for therapeutic screening procedures, e.g. inhibition or
activation of miRNA
molecules might modulate a cellular differentiation process, e.g. apoptosis.
[0098] Furthermore, existing miRNA molecules may be used as starting materials
for the
manufacture of sequence-modified miRNA molecules, in order to modify the
target-specificity
#~~ f a tr an nn~g.ggna~ a mnltirarii~~~6~ggn~ er~llgthar~""tin target rran-e-
.
Further, miRNA molecules can be modified, in order that they are processed and
then generated
as double-stranded siRNAs which are again directed against therapeutically
relevant targets.
Furthermore, miRNA molecules may be used for tissue reprogramming procedures,
e.g. a
differentiated cell line might be transformed by expression of miRNA molecules
into a different
cell type or a stem cell.
14. Compositions
[0099] The present invention also relates to a pharmaceutical composition
comprising the
nucleic acids of the invention and optionally a pharmaceutically acceptable
carrier. The
-23-
CA 02567937 2006-11-23
WO 2005/116250 PCT/IB2005/002352
compositions may be used for diagnostic or therapeutic applications. The
administration of the
pharmaceutical composition may be carried out by known methods, wherein a
nucleic acid is
introduced into a desired target cell in vitro or in vivo. Commonly used gene
transfer techniques
include calcium phosphate, DEAE-dextran, electroporation, microinjection,
viral methods and
cationic liposomes.
15. Kits
[0100] The present invention also relates to kits comprising a nucleic acid of
the invention
together with any or all of the following: assay reagents, buffers, probes
and/or primers, and
sterile saline or another pharmaceutically acceptable emulsion and suspension
base. In addition,
the kits may include instructional materials containing directions (e.g.,
protocols) for the practice
of the methods of this invention.
EXAMPLE 1
Prediction Of MiRNAs
[00100] We surveyed a number of viral genomes for potential miRNA coding genes
using
three computational approaches similar to those described in U.S. Patent
Application Nos.
60/522,459, 10/709,577 and 10/709,572, the contents of which are incorporated
herein by
reference, for predicting miRNAs. The predicted hairpins and potential miRNAs
were scored by
thermodynamic stability, as well as structural and contextual features. The
algorithm was
calibrated by using miRNAs in the Sanger Database which had been validated.
1. First and Second Screen
[0100] Tables 11 and 12 show the sequence ("PRECURSOR SEQUENCE"), sequence
identifier
"-gRE~ _ " rr
from the first computational screen, together with the predicted miRNAs ("GAM
NAME").
Tables 13 and 14 show the sequence ("GAM RNA SEQUENCE") and sequence
identifier
("GAM SEQ-ID") for each miRNA ("GAM NAME"), along with the organism of origin
("GAM
ORGANISM") and Dicer cut location ("GAM POS").
2. Third Screen
[0101] Table 1 lists the SEQ ID NO for each predicted hairpin ("HID") of the
third
computational screen of a particular viral genome ("V"; See also Table 10).
Table 1 also lists the
genomic location for each hairpin ("Hairpin Location"). The format for the
genomic location is
-24-
CA 02567937 2006-11-23
WO 2005/116250 PCT/IB2005/002352
a concatenation of <strand><start position>. The genetic location is based on
the NCBI - Entrez
Nucleotides database. The Entrez Nucleotides database is a collection of
sequences from several
sources, including GenBank, RefSeq, and PDB. Table 10 shows the accession
number and the
build (version) are presented for each of the genomes used in this screen.
[0102] Table 1 also lists the SEQ ID NO ("MID") for each predicted miRNA and
miRNA*.
Table 1 also lists the prediction score grade for each hairpin ("P") on a
scale of 0-1 (1 the hairpin
is the most reliable), as described in Hofacker et al., Monatshefte f. Chemie
125: 167-188, 1994.
Table 1 also lists the p-value ("Pval") calculated out of background hairpins
for the values of
each P scores. All the p-values are significant - lower than 0.05. As shown in
Table 1, there are
few instances where the Pval is 0Ø In each of these cases, the value is less
than 0.0001. The p-
values were calculated by comparing the palgrade of the tested hairpin to the
palgrade of other
sequences without pre-selection of hairpins..
[0103] Table 1 also lists whether the miRNAs were validated by expression
analysis ("E")
(Y=Yes, N=No), as detailed in Table 2. Table 1 also lists whether the miRNAs
were validated
by sequencing ("S") (Y=Yes, N=No), as detailed in Table 3. If there was a
difference in
sequences between the predicted and sequenced miRNAs, the sequenced sequence
is presented.
It should be noted that failure to sequence or detect expression of a miRNA
does not necessarily
mean that a miRNA does not exist. Such undetected miRNAs may be expressed in
tissues other
than those tested. In addition, such undetected miRNAs may be expressed in the
test tissues, but
at a difference stage or under different condition than those of the
experimental cells.
[0104] Table 1 also listed whether the miRNAs were shown to be differentially
expressed ("D")
(Y=Yes, N=No) in at least one disease, as detailed in Table 2). Table 1 also
whether the
mi s were presen - es, N- =A-pril2005)
(http://nar.oupjournals.orgl) as being detected in humans or mice or predicted
in humans. As
discussed above, the miRNAs'listed in the Sanger database are a component of
the prediction
algorithm and a control for the output.
[0105] Table 1 also lists a genetic location cluster ("LC") for those hairpins
that are within 1,000
nucleotides of each other of a particular virus. Each miRNA that has the same
LC share the
same genetic cluster. Those hairpins that overlap are not clustered. Table 1
also lists a seed
cluster ("SC") to group miRNAs by their seed of 2-7 by an exact match,
regardless of the source
-25-
CA 02567937 2006-11-23
WO 2005/116250 PCT/IB2005/002352
virus. Each miRNA that has the same SC have the same seed. For a discussion of
seed lengths
of 6 nucleotides, see Lewis et al., Cell, 120;15-20 (2005).
EXAMPLE 2
Prediction of Target Genes
[0106] The predicted miRNAs from the three computational screens of Example 1
were then
used to predict human and viral target genes and their binding sites using two
computational
approaches similar to those described in U.S. Patent Application Nos.
60/522,459, 10/709,577
and 10/709,572, the contents of which are incorporated herein by reference,
for predicting
miRNAs.
1. First and Second Screen
[0107] Tables 15 and 16 list the predicted target genes ("TARGET") and binding
site sequence
("TARGET BINDING SITE SEQUENCE") and binding site sequence identifier ("TARGET
BINDING SITE SEQ-ID") from the first computational screen, as well as the
organism of origin
for the target ("TARGET ORGANISM").
2. Third Screen
a. Human Target Genes
[0108] Table 4 lists the predicted human target gene for each miRNA (MID) from
a particular
virus (V) and its hairpin (HID) from the third computational screen. The names
of the target
genes were taken from NCBI Reference Sequence release
9(http://www.ncbi.nlm.nih.gov; Pruitt
et al., Nucleic Acids Res, 33(1):D501-D504, 2005; Pruitt et al., Trends
Genet., 16(1):44-47,
2000; and Tatusova et al., Bioinformatics, 15(7-8):536-43, 1999). Target genes
were identified
b~~ilig r~pe~feE~ c e,""iide riri-IZNA-seel (poSitio lS-28)-and-a3Y
A on the UTR (total=8 nucleotides). For a discussion on identifying target
genes, see Lewis et
al., Cell, 120: 15-20, (2005). For a discussion of the seed being sufficient
for binding of a
miRNA to a UTR, see Lim Lau et al., (Nature 2005) and Brenneck et al, (PLoS
Bio12005).
[0109] The binding site screen only considered the first 4000 nucleotides per
UTR and
considered the longest transcript when there were several transcripts per
gene. The filtering
reduced the total number of transcripts from 23626 to 14239. Table 4 lists the
SEQ ID NO for
the predicted binding sites for each target gene. The sequence of the binding
site includes the 20
nucleotides 5' and 3' of the binding site as they are located on the spliced
mRNA. In cases that
-26-
CA 02567937 2006-11-23
WO 2005/116250 PCT/IB2005/002352
the binding site is comprised from 2 exons, 20 nucleotides are included from
both 5' and 3' ends
of both exons.
[0110] Table 5 shows the relationship between the miRNAs ("MID")/hairpins
("HID") of a
particular virus ("V") and diseases by their human target genes. The name of
the diseases are
taken from OMIM. For a discussion of the rationale for connecting the host
gene the hairpin is
located upon to disease, see Baskerville and Bartel, RNA, 11: 241-247 (2005)
and Rodriguez et
al., Genome Res., 14: 1902-1910 (2004). Table 5 shows the number of miRNA
target genes
("N") that are related to the disease. Table 5 also shows the total number of
genes that are
related to the disease ("T"), which is taken from the genes that were
predicted to have binding
sites for miRNAs. Table 5 also shows the percentage of N out of T and the p-
value of
hypergeometric analysis ("Pval"). In cases that the pval is listed as 0.0, it
means that the value is
less than 0.0001. For a reference of hypergeometric analysis, see Schaum's
Outline of Elements
of Statistics II: Inferential Statistics. Table 7 shows the disease codes for
Tables 5 and 6.
b. Viral Target Genes
[0111] Similar to the date described above in Table 4 for human target genes,
Table 10 lists the
predicted viral target gene for each miRNA (MID) from the same particular
virus (V) and its
hairpin (HID) from the third computational screen. The prediction of viral
binding sites used
complete genes not UTRs as in the Table 4 in the method described above for
human target
genes Table 10. Candidate target genes were included in the screen if they
were known to have a
role in the virus life cycle. Those miRNAs that have binding sites on a viral
gene that takes part
in the virus life cycle they may affect the diseases that may be related to
the virus
[0112] Human Herpes virus 1 and 2 are related to any of several inflammatory
diseases caused
by-a-hei~esvirus ~~~narked~ri ene-ease b~gr~ps uf~atery-b''~ste~o~s'~___
orm=asoas
membranes (as of the mouth and lips) above the waist and in the other by such
blisters on the
genitals. Human herpesvirus 4 (Epstein-Barr virus) causes infectious
mononucleosis and is
associated with Burkitt's lymphoma and nasopharyngeal carcinoma. HIV strains
are related to
Acquired Immune Deficiency Syndrome (AIDS). Hepatitis B and C viruses cause
inflammation
of the liver.
-27-
CA 02567937 2006-11-23
WO 2005/116250 PCT/IB2005/002352
EXAMPLE 3
Validation of miRNAs
[0113] To confirm the hairpins and miRNAs predicted in Example 1, we detected
expression in
various tissues using the high-throughput microarrays similar to those
described in U.S. Patent
Application Nos. 60/522,459, 10/709,577 and 10/709,572, the contents of which
are incorporated
herein by reference. For each predicted precursor miRNA, mature miRNAs derived
from both
stems of the hairpin were tested.
1. Expression Analysis - Set 1 and Set 2
[0114] Tables 17-19 list the results of microarray expression analysis to
detch miRNA sequence
("GAM RNA SEQUENCE").
2. Expression Analysis - Set 3
[0115] Table 2 shows the hairpins ("HID") of the third prediction set that
were validated by
detecting expression of related miRNAs ("MID") from a particular virus ("V"),
as well as a code
for the tissue ("Tissue") that expression was detected. In cases where there
is more than one
score from the same miRNA in the same tissue, only the one with the higher
score is presented.
[0116] The tissue and diseases codes are listed in Table 6 and Table 7,
respectively. Table 8
shows the relationship between gene and disease. This enables the connection
of all miRNAs to
disease. Table 4 assign at least one target gene to each miRNA. Table 5
presents the outcome of
statistical analysis of table 4 and OMIM to depict significant relations of
miRNAs and disease.
Table 8 is basically a condensed version of OMIM. It lists for each gene all
the numeric codes of
the diseases that are related to it.
[0117] All the tissues disclosed give an indication of a viral disease. The
fact that significant
expression of the virus was measured implies that in this tissue it may be
involve in a viral
disease(s). E.g. when a mir from HIV was expressed in T cell line it may have
an effect on
AIDS. Of course cell lines represent only subset of the features of a tissue
as it function in an
organ however we can deduce from the expression as it is measured in the cell
line.
[0118] Table 2 also shows the chip expression score grade (range of 500-
65000)("S"). A
threshold of 500 was used to eliminate non-significant signals and the score
was normalized by
MirChip probe signals from different experiments. Variations in the
intensities of fluorescence
material between experiments may be due to variability in RNA preparation or
labeling
efficiency. We normalized based on the assumption that the total amount of
miRNAs in each
-28-
CA 02567937 2006-11-23
WO 2005/116250 PCT/IB2005/002352
sample is relatively constant. First we subtracted the background signal from
the raw signal of
each probe, where the background signal is defined as 400. Next, we divided
each miRNA
probe signal by the average signal of all miRNAs, multiplied the result by
10000 and added back
the background signal of 400. Thus, by definition, the sum of all miRNA probe
signals in each
experiment is 10400.
[0119] Table 2 also shows a statistical analysis of the normalized signal
("Spval") calculated on
the normalized score. For each miRNA, we used a relevant control group out of
the full
predicted miRNA list. Each miRNA has an internal control of probes with
mismatches. The
relevant control group contained probes with similar C and G percentage (abs
diff < 5%) in order
to have similar Tm. The probe signal P value is the ratio over the relevant
control group probes
with the same or higher signals. The results are p-value <0.05 and score is
above 500. In those
cases that the SPVaI is listed as 0.0, the value is less than 0.0001.
3. Sequencing - Set 3
[0120] To further validate the hairpins ("HID") of the second prediction, a
number of miRNAs
were validated by sequencing methods similar to those described in U.S. Patent
Application Nos.
60/522,459, 10/709,577 and 10/709,572, the contents of which are incorporated
herein by
reference. Table 3 shows the hairpins ("HID") that were validated by
sequencing a miRNA
(MID) from a virus ("V") in the indicated tissue ("Tissue").
4. Northern Analysis
[0121] A group of miRNA were validated by Northern analysis, as shown in
Figures 2 and 3.
EXAMPLE 4
Differential Expression of miRNAs
1. Viral miRNAs
[0122] Table 20 provides validated viral miRNAs that were demonstrated to be
differentially
expressed in diseased compared to healthy human tissue or human-derived cell
lines. All miRNA
sequences were validated using a miRNA microarray as described hereinabove.
For Alzheimer
Disease, GAM RNA expression was studied in a mixture of tissue from diseased
and healthy
human amygdala, cingulate cortex, caudate nucleus, globus pallidus, posterior
parietal cortex,
and superior parietal cortex, all brain regions that were shown to be affected
mildly, moderately,
or severely by Alzheimer pathology. For Parkinson Disease, GAM RNA expression
was studied
-29-
CA 02567937 2006-11-23
WO 2005/116250 PCT/IB2005/002352
in substantia nigra tissue from diseased and healthy human tissue. MT2 cell
lines were infected
with a T-tropic clinical isolation of Clade A Human Immunodeficiency Virus
(HIV), while
healthy controls were not infected. cMagi cell lines were infected with a M-
tropic Clade B HIV,
while healthy controls were not infected. Human fibroblast cells (TC) were
infected with HSV 1
or HSV2 or were not infected and served as controls. GAM RNA SEQUENCE: the
sequence (5'
to 3') of the mature, "diced" GAM RNA. CHIP SEQUENCE is the sequence of the
oligonucleotide including the predicted GAM RNA that was placed on the
microarray (not
including the non-genomic sequence used as a separator from the microarray
surface).
DISEASE: the disease in which the GAM RNA was differentially expressed - BAL
refers to M-
tropic HIV 1 Subtype B, lab strain, and BLAI refers to T-tropic HIV- 1 (LAV-
1), Subtype B;
SIGNAL (HEALTHY): the signal on the microarray for the GAM RNA in samples
comprised of
human tissue or human-derived cell lines that are not afflicted with the
specified disease;
SIGNAL (DISEASE): the signal on the microarray for the GAM RNA in samples
comprised of
human tissue or human-derived cell lines that are afflicted with the specified
disease.
2. Human miRNAs
[0123] Table 21 lists expression data of miRNAs by the following: HID: hairpin
SEQ ID NO;
MID: MiRNA SEQ ID NO; Tissue: tested tissue; S: chip expression score grade
(range=100-
65000); Dis. Diff. Exp.: disease related differential expression and the
tissue it was tested in; R:
ratio of disease related expression (range=0.01-99.99); and abbreviations:
Brain Mix A - a
mixture of brain tissue that are affected in Alzheimer; Brain Mix B - a
mixture of all brain
tissues; and Brain SN - Substantia Nigra. Tables 22 and 23 provide the details
regarding the
differentially expressed miRNAs by the following: HID: hairpin SEQ ID NO;
Hairpin_Loc:
=)
19+135460000 means chrl9 +strand, start position 135460000); C: conservation
in evolution
(Yes/No and "-" when data is not available; Yes-conservation level above
threshold of 0.7); T:
genomic type, InterGenic (G), Intron (I), Exon (E); MID: MiRNA SEQ ID NO;
Target Gene,
Disease: target gene (HUGO database) and related disease (OMIM database); P:
prediction score
grade, on range 0-9; E: chip expression information - Yes/No (Y/N); S:
validation by eequencing
- Yes/No (Y/N); HID: hairpin SEQ ID NO. Table 24 provides the sequence for the
sequences
referred to in Tables 21-23.
-30-
CA 02567937 2006-11-23
WO 2005/116250 PCT/IB2005/002352
EXAMPLE 4
Analysis of EBV miRNAs
1. Validation of Expression
[0124] Figure 4 shows the validation of expression of miRNAs predicted in EBV
(Epstein's Barr
Virus) miRNAs; expression validation. Three cell line were tested. Two were
freshly infected
normal B-cells (PBMC-1/2-EBV), and one EBV-transformed cell line (B-95-8). The
3 cell lines
exhibit the same extent of EBV infection (Figure 4A). However, in contrast to
the freshly
infected B cells, EBV-miR-RG-1 and -2 are highly expressed in the B-95-8 cell-
line (Figure 4B).
2. Knockout of Expression
[0125] Figure 5 shows the knockout of EBV miRNAs. Addition of 2-0-Methyl
against EBV-
miR-RG-1 to B-95-8 cell line resulted is dramatic reduction of cells
expressing EBV antigens.
Addition of 2-0-Methyl against EBV-miR-RG-2 to B-95-8, had a moderate effect,
slightly
increasing EBV expression.
-31-