Note: Descriptions are shown in the official language in which they were submitted.
CA 02590286 2007-06-15
- 1 -
DESCRIPTION
GENES CODING FOR FLAVONE SYNTHASES
This application is a division of Application Serial No.
2,324,505 filed January 28, 2000.
Technical Field
The present invention relates to the control and
utilization of biosynthesis of flavones, which have effects on
flower color, protection from ultraviolet ray, symbiosis with
microorganisms, etc. in plants, by a genetic engineering
technique. More specifically, it relates to genes encoding
proteins with activity of synthesizing flavones from
flavanones, and to their utilization.
Background Art
The abundance of different flower colors is one of the
pleasant aspects of life that enriches human minds and hearts.
It is expected to increase food production to meet future
population increase by the means of accelerating the growth of
plants through symbiosis with microorganisms, or by increasing
the number of nitrogen-fixing leguminous bacteria, thus
improving the plant productivity as a result of increasing the
content of nitrogen in the soil. Elimination or reduction of
the use of agricultural chemicals is also desirable to achieve
more environmentally friendly agriculture, and this requires
improvement of the soil by the above-mentioned biological
means, as well as higher resistance of plants against
microbial infection. Another desired goal is to obtain plants
with high protective functions against ultraviolet rays as a
means of protecting the plants from the destruction of the
ozone layer.
"Flavonoid" is a general term for a group of compounds
with a C6-C3-C6 carbon skeleton, and they are widely
distributed throughout plant cells. Flavonoids are known to
have such functions as attracting insects and other
pollinators, protecting plant from ultraviolet rays, and
participating in interaction with soil microorganisms
(BioEssays, 16 (1994), Koes at al p.123;
CA 02590286 2007-06-15
2 -
Trends in Plant Science, 1 (1997), Shirley, B.W., p.377).
Of flavonoids, flavone plays an important role in
interaction of plants with microorganisms, especially in
legumes, where they participate in the initial steps of
the symbiosis with leguminous bacteria (Plant Cell, 7
(1995), Dixon and Paiva, p.1085; Annu. Rev. Phytopathol.,
33 (1995), Spaink, p.345). Flavones in petals play a
role in recognition by insects and act as copigments
which form complexes with anthocyanins. (Gendai Kagaku,
(May, 1998), Honda and Saito, p.25; Prog. Chem. Org.
Natl. Prod., 52 (1987), Goto, T., p.114). It is known
that when flavone forms a complex with anthocyanin, the
absorption maximum of the anthocyanin shifts toward the
longer wavelength, i.e. toward blue.
The biosynthesis pathways for flavonoids have been
widely studied (Plant Cell, 7 (1995), Holton and Cornish,
p.1071), and the genes for all of the enzymes involved in
the biosynthesis of anthocyanidin 3-glucoside and
flavonol, for example, have been isolated. However, the
genes involved in the biosynthesis of flavones have not
yet been isolated. The enzymes'that synthesize flavones
include those belonging to the dioxygenase family that
depends on 2-oxoglutaric acid (flavone synthase I) and
monooxygenase enzymes belonging to the cytochrome P450
family (flavone synthase II). These groups of enzymes
are completely different enzymes with no structural
homology.
It has been reported that in parsley, 2-oxoglutaric
acid-dependent dioxygenase catalyzes a reaction which
- produces apigenin, a flavone, from naringenin, a
flavanone (Z. Naturforsch., 36c (1981), Britsch et al.,
p.742; Arch. Biochem. Biophys., 282 (1990), Britsch,
p.152). The other type, flavone synthase II, is known to
exist in snapdragon (Z. Naturforsch., 36c (1981), Stotz
and Forkmann, p.737) and soybean (Z. Naturforsch., 42c
(1987), Kochs and Grisebach, p.343; Planta, 171 (1987),
Kochs et al., p.519). A correlation has been recently
CA 02590286 2007-06-15
3 -
reported between a gene locus and flavone synthase II
activity in the petals of gerbera (Phytochemi-stry, 49
(1998), Martens and Forkmann, p.1953). However, there
are no reports that the genes for these flavone synthases
I and II were isolated or that flavone synthase II was
highly purified.
The properties of a cytochrome P450 protein, which
had licodione-synthesizing activity that was induced when
cultured cells of licorice (Glycyrrhiza echinata) were
treated with an elicitor, were investigated. The protein
is believed to catalyze the hydroxylation of 2-position
of liquiritigenin which is a 5-deoxyflavanone, followed
by non-enzymatic hemiacetal ring opening to produce
licodione (Plant Physiol., 105 (1994), Otani et al.,
p.1427). For cloning of licodione synthase, a cDNA
library was prepared from elicitor-treated Glycyrrhiza
cultured cells, and 8 gene fragments encoding cytochrome
P450 were cloned (Plant Science, 126 (1997), Akashi et
al., p.39).
From these fragments there.were obtained two
different full-length cDNA sequences, each encoding a
cytochrome P450, which had been unknown until that time.
Specifically, they were CYPGe-3 (cytochrome P450
No.CYP81E1) and CYPGe-5 (cytochrome P450 No.CYP93B1,
hereinafter indicated as CYP93B1) (Plant Physiol., 115
(1997), Akashi et al., p.1288). By further expressing
the CYP93B1 cDNA in a system using cultured insect cells,
the protein derived from the gene was shown to catalyze
the reaction synthesizing licodione from liquiritigenin,
a flavanone, and 2-hydroxynaringenin from naringenin,
also a flavanone.
2-Hydroxynaringenin was converted to apigenin, a
flavone, by acid treatment with 10% hydrochloric acid
(room temperature, 2 hours). Also, eriodictyol was
converted to luteolin, a flavone, by reacting eriodictyol
with microsomes of CYP93B1-expressing yeast followed by
acid treatment. It was therefore demonstrated that the
CA 02590286 2007-06-15
4 -
cytochrome P450 gene encodes the function of flavanone 2-
hydroxylase activity (FEBS Lett., 431 (1998),- Akashi et
al., p.287). Here, production of apigenin from
naringenin required CYP93B1 as well as another unknown
enzyme, so that it was concluded that a total of two
enzymes were necessary.
However, no genes have yet been identified for
enzymes with activity of synthesizing flavones (such as
apigenin) directly from flavanones (such as naringenin)
without acid treatment. Thus, despite the fact that
flavones have numerous functions in plants, no techniques
have yet been reported for controlling their biosynthesis
in plants, and improving the biofunctions in which
flavones are involved, such as flower color. The
discovery of an enzyme which by itself can accomplish
synthesis of flavones from flavanones and acquisition of
its gene, and introduction of such a gene into plants,
would be more practical and industrially applicable than
the introduction into a plant of genes for two enzymes
involved in the synthesis of flavones from flavanones.
Disclosure of the Invention
It is an aim of the present invention to provide
flavone synthase genes, preferably flavone synthase II
genes, and more preferably genes for flavone synthases
with activity of synthesizing flavones directly from
flavanones. The obtained flavone synthase genes may be
introduced into plants and over-expressed to alter flower
colors.
Moreover, in the petals of flowers that naturally
contain large amounts of flavones, it is expected that
controlling expression of the flavone synthase genes by
an antisense method or a cosuppression method can also
alter flower colors. Also, expression of the flavone
synthase genes in the appropriate organs, in light of the
antibacterial activity of flavones and their interaction
with soil microorganisms, will result in an increase in
the antibacterial properties of plants and improvement in
CA 02590286 2010-02-17
-
the nitrogen fixing ability of legumes due to promoted
symbiosis with rhizosphere microorganisms, as-well as a
protective effect against ultraviolet rays and light.
The present invention therefore provides genes
5 encoding proteins that can synthesize flavones directly
from flavanones. The genes are, specifically, genes
encoding flavone synthase II that can synthesize flavones
from flavanones by a single-enzyme reaction (hereinafter
referred to as "flavone synthase II").
More specifically, the present invention provides
genes encoding P450 proteins having the amino acid
sequences listed as SEQ.ID..No. 2, 4 or 7 of the Sequence
Listing and possessing activity of synthesizing flavones
from flavanones, or genes encoding proteins having amino
acid sequences modified by additions or deletions of one
or more amino acids and/or a substitution with different
amino acids in said amino acid sequence,.-and possessing
activity of synthesizing flavones from flavanones.
The invention further provides a gene encoding
proteins having amino acid sequences with at least 55%
identity with the amino acid sequences listed as SEQ.ID.
No. 2, 4 or 7 of the Sequence Listing and possessing
activity of synthesizing flavones from flavanones.
The invention still further provides genes encoding
proteins possessing activity of synthesizing flavones
from flavanones, and hybridizing with all or a part of
the nucleotide sequences listed as SEQ.ID. No. 1 or 3
of the Sequence List under the conditions of 5 x SSC,
50 C.
The invention still further provides a vector,
particularly an expression vector, containing any one of
the aforementioned genes.
The invention still further provides a host
transformed with the aforementioned vector.
The invention still further provides a protein
encoded by any of the aforementioned genes.
The invention still further provides a process for
CA 02590286 2007-06-15
6 -
producing the aforementioned protein which is
characterized by culturing or growing the aforementioned
host, and collecting the protein with flavone-
synthesizing activity from the host.
The invention still further provides a plant into
which any one of the aforementioned genes has been
introduced, or progenies of the plant or a tissue
thereof, such as cut flowers, which exhibit the same
properties.
The invention still further provides a method of
altering amounts and compositions of flavonoid using the
aforementioned genes; a method of altering amounts of
flavones using the aforementioned genes; a method of'
altering flower colors using the aforementioned genes; a
method of bluing the color of flowers using the
aforementioned genes; a method of reddening the color of
flowers using the aforementioned genes; a method of
modifying the photosensitivity of plants using the
aforementioned genes; and a method of controlling the
interaction between plants and microbes using the
aforementioned genes.
Brief Description of the Drawings
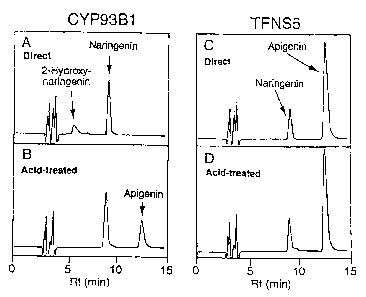
Fig. 1 is a chromatogram showing the results of HPLC
analysis of products obtained from a substrate,
naringenin, using proteins encoded by CYP93B1 and TFNS5.
A and B: Obtained by adding a crude enzyme fraction
of CYP93B1-expressing yeast.
C and D: Obtained by adding a crude enzyme fraction
of TFNS5-expressing yeast.
A and C: Direct products obtained by addition of
enzyme fraction.
B: Products obtained by acid treatment after
reaction of A.
D: Products obtained by acid treatment after
reaction of C.
Embodiments for Carrying out the Invention
Flavanone 2-hydroxylase encoded by the Glycyrrhiza
CA 02590286 2007-06-15
7 -
CYP93B1 gene produces 2-hydroxyflavanones from flavanones
as the substrates, and the products are converted to
flavones by acid treatment. The present inventors viewed
that it would be possible to obtain a gene encoding a
flavone synthase II, which was an object of the
invention, by using the Glycyrrhiza-derived cDNA, CYP93B1
for screening of a cDNA library of, for example, a flower
containing a large amount of flavones, to thus obtain
cDNA encoding proteins with activity of synthesizing
flavones directly from flavanones as substrates.
According to the invention, a cDNA library of
snapdragon which contains a large amount of flavones is
screened using the Glycyrrhiza-derived cDNA, CYP93B1 as a
probe, to obtain cDNA encoding a novel cytochrome P450
(see Example 1). The snapdragon cDNA, ANFNS2, obtained
in this manner and the Glycyrrhiza CYP93B1 cDNA were then
used as a mixed probe to obtain TFNS5, a cDNA encoding a
novel cytochrome P450, from a cDNA library of torenia
flower petals (see Example 2).
The torenia-derived cDNA was expressed in yeast and
reacted with naringenin, a flavanone, as a substrate
which resulted in production not of 2-hydroxynaringenin
but rather of the flavone apigenin, without acid
treatment (see Example 3). In other words, this enzyme
directly produced flavones from flavanones without acid
treatment, and its gene was confirmed to be a flavone
synthase II which had never been cloned. The amino acid
sequence encoded by the snapdragon-derived ANFNS2 of
Example 1 exhibited high identity of 77% with the flavone
synthase II encoded by TFNS5, and it exhibited the enzyme
activity of flavone synthase II (Example 4). In
addition, since an amino acid sequence encoded by
perilla-derived cDNA also exhibited high identity'of 76%
and 75% with TFNS5 and ANFNS2, respectively (Example 8),
it is speculated that the protein encoded by this cDNA
also possesses the same enzymatic activity as the flavone
synthases encoded by TFNS5 and ANFNS2.
CA 02590286 2010-02-17
8 -
The genes of the present invention may be, for
example, one encoding the amino acid sequences listed as
SEQ.ID. No. 2, 4 or 7 of the Sequence Listing. However,
it is known that proteins whose amino acid sequences are
modified by additions or deletions of multiple amino
acids and/or substitutions with different amino acids can
maintain the same enzyme activity as the original
protein. Consequently, proteins having the amino acid
sequences listed as SEQ.ID. No. 2, 4 or 7 of the Sequence
Listing wherein the amino acid sequence is modified by
additions or deletions of one or more amino acids and/or
substitutions with different amino acids, and genes
encoding those proteins, are also encompassed by the
present invention so long as they maintain the activity
of producing flavones directly from flavanones.
The present invention also relates to genes that
have the nucleotide sequences listed as SEQ.ID.No. 1 or 3
and nucleotide sequences encoding the amino acid
sequences listed therein, or that hybridize with portions
of their nucleotide sequences under conditions of 5 x
SSC, 50 C, for example, providing they encode proteins
possessing activity of producing f lavones from
flavanones. The suitable hybridization temperature will
differ depending on nucleotide sequences and the length
of nucleotide sequences, and for example, when the probe
used is a DNA fragment comprising 18 bases coding for 6
amino acids, the temperature is preferably not higher
than 50 C.
A gene selected by such hybridization may be a
naturally derived one, such as a plant-derived gene, for
example, a gene derived from snapdragon, torenia or
perilla; it may also be a gene from another plant, such
as gentian, verbena, chrysanthemum, iris, or the like. A
gene selected by hybridization may be cDNA or genomic
DNA.
The invention also relates to genes encoding
proteins that have amino acid sequences with identity of
CA 02590286 2010-02-17
9
at least 55%, preferably at least 70%, such as 80% or
greater and even 90% or greater, with any one-of the
amino acid sequences listed as SEQ.ID. Nos. 2, 4 or 7 of
the Sequence Listing, and that possess activity of
synthesizing flavones from flavanones.
A gene with the natural nucleotide sequence can be
obtained by screening of a cDNA library, for example, as
demonstrated in detail in the examples. DNA encoding
enzymes with modified amino acid sequences can be
synthesized using common site-directed mutagenesis or a
PCR method, using DNA with a natural nucleotide sequence
as a starting material. For example, a DNA fragment into
which a modification is to be introduced may be obtained
by restriction enzyme treatments of natural cDNA or
genomic DNA and then used as a template for site-directed
mutagenesis or PCR using a primer having the desired
mutation introduced therein, to obtain a DNA fragment
having the desired modification introduced therein.
Mutation-introduced DNA fragments may then be linked to a
DNA fragment encoding another portion of a target enzyme.
Alternatively, in order to obtain DNA encoding an
enzyme consisting of a shortened amino acid sequence, for
example, DNA encoding an amino acid sequence which is
longer than the aimed amino acid sequence, such as the
full length amino acid sequence, may be cut with desired
restriction endonucleases, and if the DNA fragment
obtained thereby does not encode the entire target amino
acid sequence, it may be linked with synthesized DNA
comprising the rest of the sequence.
Thus obtained genes may be expressed in an
expression system using-E. coli or yeast and its enzyme
activity measured to confirm that the obtained gene
encodes flavone synthase. By expressing the gene, it is
also possible to obtain the flavone synthase protein as
the gene product. Alternatively, it is also possible to
obtain a flavone synthase protein even using antibodies
for a full or a.partial amino acid sequence' listed as
CA 02590286 2010-02-17
- 10 -
SEQ.ID. No. 2, 4 or 7, and such antibodies may be used
for cloning of a flavone synthase gene in another
organism.
Consequently, the invention also relates to
recombinant vectors, and especially expression vectors,
containing the aforementioned genes, and to-hosts
transformed by these vectors. The hosts used.may be
prokaryotic or eukaryotic organisms. Examples of
prokaryotic organisms that may commonly be used as hosts
include bacteria belonging to the genus Escherichia, such
as Escherichia coli, and microorganisms belonging to the
genus Bacillus, such as Bacillus subtilis.
Examples of eukaryotic hosts that may be used
include lower eukaryotic organisms, for example,
eukaryotic microorganisms, for example, Eumycota such as
yeast and filamentous fungi. As yeast there may be
mentioned microorganisms belonging to the genus
Saccharomyces, such as Saccharomyces cerevisiae., and as
filamentous fungi there may be mentioned microorganisms
belonging to the genus Aspergillus, such as Aspergillus
oryzae and Aspergillus niger and microorganisms belonging
to the genus Penicillium. Animal cells and plant cells
may also be used, the animal cells being cell lines from
mice, hamsters, monkeys or humans. Insect cells, such as
silkworm cells, or the adult silkworms themselves, may
also be used.
The expression vectors of the invention will include
expression regulating regions such as a promoter and a
terminator, a replication origin, etc., depending on the
type of hosts into which they are to be introduced.
Examples of promoters for bacterial expression vectors
which may be used include conventional promoters such as
trc promoter, tac promoter, lac promoter, etc., examples
of yeast promoters that may be used include
glyceraldehyde-3-phosphate dehydrogenase promoter, PH05
promoter, etc., and examples of filamentous fungi
promoters that may be used include amylase promoter,
CA 02590286 2007-06-15
- 11 -
trpC, etc. Examples of animal cell host promoters that
may be used include viral promoters such as SV40 early
promoter, SV40 late promoter, etc.
The expression vector may be prepared according to a
conventional method using restriction endonucleases,
ligases and the like. The transformation of a host with
an expression vector may also be carried out according to
conventional methods.
The hosts transformed by the expression vector may
be cultured, cultivated or raised, and the target protein
may be recovered and purified from the cultured product,
etc. according to conventional methods such as
filtration, centrifugal separation, cell crushing, gel
filtration chromatography, ion-exchange chromatography
and the like.
The present specification throughout discusses
flavone synthases II derived from snapdragon, torenia and
perilla that are capable of synthesizing flavones
directly from flavanones, and it is also known that the
cytochrome P450 genes constitute a superfamily (DNA and
Cell Biology, 12 (1993), Nelson et al., p.1) and that
cytochrome P450 proteins within the same family have 40%
or greater identity in their amino acid sequences while
cytochrome P450 proteins within a subfamily have 55% or
greater identity in their amino acid sequences, and their
genes hybridize to each other (Pharmacogenetics, 6
(1996), Nelson et al., p.1).
For example, a gene for flavonoid 3',51-hydroxylase,
which was a type of cytochrome P450 and participated in
the pathway of flavonoid synthesis, was first isolated
from petunias (Nature, 366 (1993), Holton et al., p.276),
and the petunia flavonoid 3',5'-hydroxylase gene was used
as a probe to easily isolate a flavonoid 3',5'-
hydroxylase gene from gentian (Plant Cell Physiol., 37
(1996), Tanaka et al., p.711), prairie-gentian,
bellflower (W093/18155 (1993), Kikuchi et al.), lavender,
torenia and verbena (Shokubutsu no Kagaku Chosetsu, 33
CA 02590286 2007-06-15
- 12 -
(1998), Tanaka et al., p.55).
Thus, a part or all of any of the flavone synthase
II genes of the invention derived from snapdragon,
torenia or perilla, which are capable of synthesizing
flavones directly from flavanones, can be used as a
probe, in order to obtain flavone synthase II genes
capable of synthesizing flavones directly from
flavanones, from different species of plants.
Furthermore, by purifying the snapdragon-, torenia- or
perilla-derived flavone synthase II enzymes described in
this specification which can synthesize flavones directly
from flavanones, and obtaining antibodies against the
enzymes by conventional methods, it is possible to obtain
different flavone synthase II proteins that react with
the antibodies, and obtain genes coding for those
proteins.
Consequently, the present invention is not limited
merely to snapdragon-, torenia- or perilla-derived genes
for flavone synthases II capable of synthesizing flavones
directly from flavanones, but further relates to flavone
synthases II derived from numerous other plants, which
are capable of synthesizing flavones directly from
flavanones. The sources for such flavone synthase II
genes may be, in addition to snapdragon, torenia and
perilla described here, also gentian, verbena,
chrysanthemum, iris, commelina, centaurea, salvia,
nemophila and the like, although the scope of the
invention is not limited to these plants.
The invention still further relates to plants whose
colors are modified by introducing a gene or genes for
flavone synthases II that can synthesize flavones
directly from flavanones, and to progenies of the plants
or their tissues, which may also be in the form of cut
flowers. By using the flavone synthases II or their
genes which have been cloned according to the invention,
it is possible to produce flavones in plant species or
varieties that otherwise produce little or absolutely no
CA 02590286 2007-06-15
- 13 -
flavones. By expressing the flavone synthase II gene or
the genes in flower petals, it is possible to-increase
the amount of flavones in the flower petals, thus
allowing the colors of the flowers to be modified toward
the blue, for example.
Conversely, by repressing synthesis of flavones in
flower petals, it is possible to modify the colors of the
flowers toward the red, for example. However, flavones
have myriad effects on flower colors, and the changes in
flower colors are therefore not limited to those
mentioned here. With the current level of technology, it
is possible to introduce a gene into a plant and express
the gene in a constitutive or tissue-specific manner,
while it is also possible to repress the expression of a
target gene by an antisense method or a cosuppression
method.
As examples of transformable plants there may be
mentioned rose, chrysanthemum, carnation, snapdragon,
cyclamen, orchid, prairie-gentian, freesia, gerbera,
gladiolus, baby's breath, kalanchoe, lily, pelargonium,
geranium, petunia, torenia, tulip, rice, barley, wheat,
rapeseed, potato, tomato, poplar, banana, eucalyptus,
sweet potato, soybean, alfalfa, lupin, corn, etc., but
there is no limitation to these.
Because flavones have various physiological
activities as explained above, they can impart new
physiological activity or economic value to plants. For
example, by expressing the gene to produce flavones in
roots, it is possible to promote growth of microorganisms
that are beneficial for the plant, and thus promote
growth of the plant. It is also possible to synthesize
flavones that exhibit physiological activity in humans,
animals or insects.
Examples
The invention will now be explained in further
detail by way of the following examples. Unless
CA 02590286 2007-06-15
- 14 -
otherwise specified, the molecular biological methods
were carried out according to Molecular Cloning (Sambrook
et al., 1989).
Example 1. Cloning of snapdragon flavone synthase II
gene
RNA was extracted from about 5 g of young buds of a
Yellow Butterfly snapdragon (commercial name by Sakata-
no-lane, KK.), and polyA+ RNA was obtained by an
Oligotex. This polyA+ RNA was used as a template to
prepare a cDNA library using a Lambda ZAPII cDNA Library
Synthesis Kit (Stratagene) by the method recommended by
Stratagene (Stratagene Instruction Manual, Revision
#065001). The cDNA library was screened using the full
length cDNA CYP93B1 as the probe. The screening and
detection of positive clones were carried out using a
DIG-DNA-labeling and detection kit (Boehringer) based on
the method recommended by the same company, under a low
stringent condition.
Specifically, a hybridization buffer (5 x SSC, 30%
formamide, 50 mM sodium phosphate buffer (pH 7.0), 1%
SDS, 2% blocking reagent (Boehringer), 0.1%
lauroylsarcosine, 80 pg/ml salmon sperm DNA) was used for
prehybridization at 42 C for 2 hours, after which the
DIG-labeled probe was added and the mixture was kept
overnight. The membrane was rinsed in 5 x SSC rinsing
solution containing 1% SDS at 65 C for 1.5 hours. One
positive clone was obtained, and it was designated as
ANFNS1. Upon determining the nucleotide sequence at the
5' end of ANFNS1 it was expected that ANFNS1 encodes a
sequence with high identity with the flavanone 2-
hydroxylase encoded by licorice CYP93B1, and it was
assumed that it encoded a P450 with a function similar to
that of flavanone 2-hydroxylase.
However, a comparison with the amino acid sequence
of flavanone 2-hydroxylase encoded by CYP93B1 suggested
that the cDNA of ANFNS1 is not a full-length cDNA,
lacking the portion corresponding to approximately 65
CA 02590286 2007-06-15
- 15 -
amino acid residues from the initiating methionine. The
ANFNS1 cDNA was therefore used as a probe for- rescreening
of the snapdragon cDNA library, to obtain cDNA (ANFNS2)
which was believed to include the full-length amino acid
sequence. The protein encoded by ANFNS2 obtained here
exhibited 53% identity on the amino acid level with
flavanone 2-hydroxylase encoded by licorice CYP93B1. The
nucleotide sequence of ANFNS2 is listed as SEQ.ID. No.1, and
the amino acid sequence deduced therefrom is listed as
SEQ.ID. No.2.
Example 2. Cloning of torenia flavone synthase II gene
RNA was extracted from approximately 2 g of buds of a
torenia variety (variety name: Sunrenive, Variety
Registration Application No.: 7433 according to the Seeds
and Seedlings Law, by Suntory Ltd.) and the polyA+ RNA was
obtained with an Oligotex. The polyA+ RNA was used as a
template to prepare a cDNA library using a Lambda ZAPII cDNA
Library Synthesis Kit (Stratagene) by the method recommended
by Stratagene as mentioned in Example 1. The cDNA library
was screened using a mixture of the aforementioned CYP93B1
cDNA and ANFNS1 cDNA as the probes. The screening and
detection of positive clones were carried out under the low
stringent conditions as described in Example 1.
One positive clone was obtained, and was designated as
TFNSS. Upon determining the full nucleotide sequence of
TFNS5 cDNA, it was found that the protein encoded by TFNS5
cDNA exhibited 52% identity on the amino acid level with
flavanone-2-hydroxylase encoded by licorice CYP93B1. This
TFNS5 cDNA also had high identity of 77% with the protein
encoded by ANFNS2, the snapdragon-derived cDNA obtained in
Example 1. The determined nucleotide sequence is listed as
SEQ.ID. No.3, and the amino acid sequence deduced therefrom
is listed as SEQ.ID. No.4.
CA 02590286 2007-06-15
- 16 -
Example 3. Expression of torenia flavone synthase II
gene in yeast -
The following experiment was conducted in order to
detect the enzyme activity of the protein encoded by
TFNS5, the torenia cDNA obtained in Example 2. Parts of
the outside of the translated region of the gene were
modified to introduce restriction enzyme sites therein to
prepare a sense primer (5'-
AAATAGGATCCAAGCatgGACACAGTCTTAA-3'; underline = BamHI
site; lowercase letters: initiation codon) (SEQ.ID. No.5)
and an antisense primer (5'-
CCCTTCTAGAtcaAGCACCCGATATTGTGGCCGGG-3'; underline = XbaI
site; lowercase letters: termination codon) (SEQ.ID.
No.6) were used with KOD polymerase (Toyobo) for PCR
reaction. The PCR conditions were 98 C for one minute,
cycles of (98 C for 15 seconds, 55 C for 10 seconds,
74 C for 30 seconds), followed by 74 C for 10 minutes.
After introducing the resultant PCR product into the
EcoRV site of pBluescriptll SK(-) (Stratagene), it was
20 digested with restriction enzymes BamHI and XbaI and
introduced at the BamHI-XbaI sites of the yeast
expression vector pYES2 (Invitrogen). The resultant
plasmid was then introduced into BJ2168 yeast (Nihon
Gene). The enzyme activity was measured by the method
described by Akashi et al. (FEBS Lett., 431 (1998),
Akashi et al., p.287). The transformed yeast cells were
cultured in 20 ml of selective medium (6.7 mg/ml amino
acid-free yeast nitrogen base (Difco), 20 mg/ml glucose,
pg/ml leucine, 20 pg/ml tryptophan and 5 mg/ml
30 casamino acid), at 30 C for 24 hours.
After harvesting the yeast cells with
centrifugation, the harvested yeast cells were cultured
at 30 C for 48 hours in an expressing medium (10 mg/ml
yeast extract, 10 mg/ml peptone, 2 pg/ml hemin, 20 mg/ml
galactose). After collecting the yeast cells, they were
washed by suspending in water and collecting them. Glass
beads were used for 10 minutes of disrupting, after which
CA 02590286 2007-06-15
- 17 -
the cells were centrifuged at 8000 x g for 10 minutes.
The supernatant was further centrifuged at 15-,000 x g for
minutes to obtain a crude enzyme fraction.
A mixture of 15 pg of (R,S)-naringenin (dissolved in
5 30 p1 of 2-methoxyethanol), 1 ml of crude enzyme solution
and 1 mM NADPH (total reaction mixture volume: 1.05 ml)
was reacted at 30 C for 2 hours. After terminating the
reaction by addition of 30 pl of acetic acid, 1 ml of
ethyl acetate was added and mixed therewith. After
10 centrifugation, the ethyl acetate layer was dried with an
evaporator. The residue was dissolved in 100 pl of
methanol and analyzed by HPLC. The analysis was carried
out according to the method described by Akashi et al.
The acid treatment involved dissolution of the
evaporator-dried sample in 150 p1 of ethanol containing
10% hydrochloric acid, and stirring for 30 minutes. This
was diluted with 1.3 ml of water, 800 pl of ethyl acetate
was further added and mixed therewith, and after
centrifugation, the ethyl acetate layer was recovered.
This was then dried, dissolved in 200 pl of methanol, and
analyzed by HPLC.
The yeast expressing licorice CYP93B1 produced 2-
hydroxynaringenin from naringenin, but yielded no
apigenin (Fig. 1, A). Only upon acid treatment of the
reaction mixture, apigenin was yielded from 2-
hydroxynaringenin (Fig. 1, B). In contrast, the yeast
expressing torenia TFNS5 yielded apigenin from naringenin
without acid treatment of the reaction mixture (Fig. 1,
C). This demonstrated that TFNS5 encodes a flavone
synthase II.
Example 4. Expression of snapdragon flavone synthase II
gene in yeast
An approximately 1400 bp DNA fragment obtained by
digesting ANFNS2 cDNA with BamHI and SphI, an
approximately 350 bp DNA fragment obtained by digesting
the same with SphI and BamHI, and pYES2 digested with
BamHI and XhoI were ligated to obtain a plasmid, which
CA 02590286 2007-06-15
- 18 -
was then introduced into yeast by the same method as
described in Example 3. The resultant recomb-inant yeast
cells were used to measure the flavone synthesis activity
by the same method as in Example 3. The yeast expressing
the snapdragon-derived ANFNS2 produced apigenin without
acid treatment, thus demonstrating that ANFNS2 encodes a
flavone synthase II.
Example 5. Construction of expression vector in plants
A plant expression vector was constructed to
introduce TFNS5, the torenia cDNA obtained in Example 2,
into plants. After digesting pBE2113-GUS (Plant Cell
Physiol., 37 (1996), Mitsuhara et al., p.49) with Sacl, a
blunting kit (Takara) was used to blunt the ends, after
which a XhoI linker (Toyobo) was inserted. The resulting
plasmid was then digested with Hindlll and EcoRI, and an
approximately 3 kb DNA fragment was recovered. The DNA
fragment was linked to the Hindlll/EcoRI site of the
binary vector pBINPLUS to prepare pBE2113'. vector
pBINPLUS used here was obtained by modifying the binary
vector Bin19 (Nucl. Acids Res., 12 (1984), Bevan,
p.8711), which is widely used for gene introduction into
plants using Agrobacterium cells, in the manner reported*
by van Engelen et al. (Transgenic Research, 4 (1995), van
Engelen et al., p.288).
The TFNS5 cDNA was cut out of SK(-) vector by
cleavage with BamHI/XhoI, and an approximately 1.7 kb
fragment thus obtained was ligated to the BamHI/XhoI
sites of the aforementioned binary vector pBE2113'. The
construct thus obtained, pSPB441, expresses TFNS5 cDNA in
the sense direction under the control of 35S cauliflower
mosaic virus promoter having a double repeat of the
enhancer sequence (Plant cell Physiol., 37 (1996),
Mitsuhara et al., p.49).
Example 6. Alteration of torenia flower color
A torenia variety (variety name: Sunrenive, variety
Registration Application No.: 7433 according to the Seeds
and Seedlings Law, by Suntory Ltd.) was transformed with
CA 02590286 2007-06-15
- 19 -
pSPB441 constructed in Example 5 above, according to the
method of Aida et al. (Breeding Science, 45 (1995), Aida
et al., p.71). Over 95% of the obtained transformants
showed alteration of the flower color from the dark
purple of the parent strain to a light purple. The left
and the right flower petal colors of four flower petals
were measured. While the flower petal color of the
parent strain was Number 89A according to the Royal
Horticultural Society Color Chart, the typical flower
petal colors of the transformants were 82C, 87D, 87C,
88D, 91A, etc. These results indicated that introduction
of TFNS5 into plants can alter flower colors.
In the transformed individuals, the amount of
flavones ranged 1/5 to 1/10 that of the host, while the
amount of anthocyanins was reduced to about 1/3 that of
the host. Also detected were flavanones (naringenin,
eriodictyol and pentahydroxyflavanone) which are flavone
biosynthesis precursors that were not detected in the
host.
Example 7. Expression of flavone synthase in petunias
Plasmid pSPB441 was introduced into a petunia
variety (variety name: Revolution Violet Mini, Variety
Registration Application No.: 9217, according to the
Seeds and Seedlings Law, by Suntory Ltd.) according to
the method of Napoli et al. (Plant Cell, 2, (1990),
Napoli et al., p.279). Changes in flower colors occurred
in two of the resultant transformants, where the flower
colors were lighter than the parent strain. The flower
color of the, parent strain was Number 88A according to
the Royal Horticultural Society Color Chart, whereas 87A
in the transformants. Also, while no flavones were
detected in the parent strain, a flavone, luteolin, was
detected in the transformed strains.
Example 8. Cloning of perilla flavone synthase II gene
The method described in Example 1 was used to screen
a cDNA library prepared from leaves of red perilla
(Perilla frutescens) according to the method of Gong et
CA 02590286 2010-02-17
- 20 -
al. (Plant Mol. Biol., 35, (1997), Gong et al., p.915)
using Xgt10 (Stratagene) as the vector. Culturing, DNA
preparation and subcloning of the resulting phage clones
#3 were carried out according to the method of Gong et
al. (Plant Mol. Biol., 35, (1997), Gong et al., p.915),
and the nucleotide sequence was determined.
The deduced amino acid sequence encoded by
this nucleotide sequence was listed as SEQ.ID. No.7.
This amino acid sequence showed 76% and 75% identity with
TFNS5 and ANFNS2, respectively. It also showed 52%
identity with CYP93B1.
Example 9. Expression of perilla flavone synthase II
gene in yeast
The phage clone #3 obtained in Example 8 was used as
a template for PCR by the method described in Example 3,
using Lambda Arm Primer (Stratagene). The amplified DNA
fragment was subcloned at the EcoRV site of pBluescript
KS(-). A clone with the initiation codon of perilla
flavone synthase II cDNA on the Sall site side of
pBluescript KS(-) was selected, and was designated as
pFS3. The nucleotide sequence of the cDNA insert of pFS3
was determined, and PCR was conducted to confirm the
absence of errors.
An approximately 1.8 kb DNA fragment obtained by
25. digesting pFS3 with Sall and XbaI was ligated with pYES2
digested with XhoI and XbaI (Example 3) to obtain a
plasmid which was designated as pYFS3, and this was
-introduced into yeast BJ2168 by the method described in
Example 3. When the flavone synthase activity of this
recombinant yeast was measured by the method described in
Example 3, production of apigenin from naringenin was
confirmed, indicating that the perilla phage clone #3
cDNA encodes a protein with flavone synthase II activity.
Industrial Applicability
It is possible to alter flower colors by linking
cDNA of the invention to an appropriate plant expression
vector and introducing it into plants to express or
CA 02590286 2007-06-15
- 21 -
inhibit expression of flavone synthases. Furthermore, by
expressing the flavone synthase genes not only in petals
but also in entire plants or their appropriate organs, it
is possible to increase the resistance agasint
microorganisms of plants or to improve the nitrogen
fixing ability of legumes by promoting association with
rhizosphere microorganisms, as well as to improve the
protective effects of plants against ultraviolet rays and
light.
CA 02590286 2010-02-17
SEQUENCE LISTING
<110> INTERNATIONAL FLOWER DEVELOPMENTS PROPRIETARY LIMITED
<120> GENE ENCODING FLAVONE SYNTHASE
<130> 993009
<160> 7
<210> 1
<211> 1724
<212> DNA
<213> Antirrhinum majus
<220>
<223> Nucleotide sequence encoding a protein having an
activity to directly convert flavanone to flavone
<400> 1
gctttacaca cacacacaca cacacacaca caaacaaaa atg tct aca ctt gtc 54
Met Ser Thr Leu Val
1 5
tac agc aca ctc ttc atc ctc tca acc ctc ctc ctc acc ctc cta acc 102
Tyr Ser Thr Leu Phe Ile Leu Ser Thr Leu Leu Leu Thr Leu Leu Thr
15 20
cgc acc cgc cgc aag acc cgc ccg ccc ggc cca tta gcc ctc ccc tta 150
Arg Thr Arg Arg Lys Thr Arg Pro Pro Gly Pro Leu Ala Leu Pro Leu
25 30 35
ata ggc cac tta cac ctc ctc ggc cca aag ctc cac cac acc ttc cac 198
Ile Gly His Leu His Leu Leu Gly Pro Lys Leu His His Thr Phe His
40 45 50
caa ttc tcc caa cgc tac ggc ccg ctc atc cag ctc tac ctc ggc tcc 246
Gln Phe Ser Gln Arg Tyr Gly Pro Leu Ile Gin Leu Tyr Leu Gly Ser
55 60 65
gtc cca tgc gtc gtc get tcc acg ccc gaa ctc gcc cgc gaa ttc ctc 294
Val Pro Cys Val Val Ala Ser Thr Pro Glu Leu Ala Arg Glu Phe Leu
70 75 80 85
aag acg cac gaa ctc gac ttc tcg tcc cgc aag cac tcc acc gcc atc 342
Lys Thr His Glu Leu Asp Phe Ser Ser Arg Lys His Ser Thr Ala Ile
21a
CA 02590286 2010-02-17
90 95 100
gac atc gtc acg tac gac tcc tcg ttc gcc ttc gcg ccg tac ggg ccg 390
Asp Ile Val Thr Tyr Asp Ser Ser Phe Ala Phe Ala Pro Tyr Gly Pro
105 110 115
tac tgg aaa ttc atc aag aaa tta tgt act tac gag cta ctg ggt gcc 438
Tyr Trp Lys Phe Ile Lys Lys Leu Cys Thr Tyr Glu Leu Leu Gly Ala
120 125 130
cgg aac ttg agc cat ttc cag ccc att aga get ttg gag gtc aac agt 486
Arg Asn Leu Ser His Phe Gln Pro Ile Arg Ala Leu Glu Val Asn Ser
135 140 145
ttc ttg aga att ttg tac gag aaa aca gag cag aaa cag agt gtt aat 534
Phe Leu Arg Ile Leu Tyr Glu Lys Thr Glu Gln Lys Gln Ser Val Asn
150 155 160 165
gtg act gag gag ctt gtg aag ctg acg agt aat gtg atc agt aac atg 582
Val Thr Glu Glu Leu Val Lys Leu Thr Ser Asn Val Ile Ser Asn Met
170 175 180
atg ttg ggg atc agg tgt tcg ggg acg gaa ggg gag gcg gag gtg gcg 630
Met Leu Gly Ile Arg Cys Ser Gly Thr Glu Gly Glu Ala Glu Val Ala
185 190 195
agg acg gtg ata agg gag gtg acg cag ata ttt ggg gag ttt gat gtg 678
Arg Thr Val Ile Arg Glu Val Thr Gln Ile Phe Gly Glu Phe Asp Val
200 205 210
tcg gag att gtt tgg ttt tgt aag aat ttg gat ctg cag ggg att agg 726
Ser Glu Ile Val Trp Phe Cys Lys Asn Leu Asp Leu Gln Gly Ile Arg
215 220 225
aag agg tcg gag gat att agg agg agg tat gat get ttg ttg gag aag 774
Lys Arg Ser Glu Asp Ile Arg Arg Arg Tyr Asp Ala Leu Leu Glu Lys
230 235 240 245
att att agt gat agg gag agg ttg agg ttg agg ggg ggt ggt ggt gga 822
Ile Ile Ser Asp Arg Glu Arg Leu Arg Leu Arg Gly Gly Gly Gly Gly
250 255 260
ggg ggt gga gag gtg aag gat ttt ttg gat atg ttg ttg gat gtg atg 870
Gly Gly Gly Glu Val Lys Asp Phe Leu Asp Met Leu Leu Asp Val Met
265 270 275
gag agt gag aaa tcg gag gtg gag ttt acg agg gag cat ctc aaa get 918
Glu Ser Glu Lys Ser Glu Val Glu Phe Thr Arg Glu His Leu Lys Ala
280 285 290
ttg att ctg gat ttc ttc act gcc ggt aca gac aca aca gca atc aca 966
Leu Ile Leu Asp Phe Phe Thr Ala Gly Thr Asp Thr Thr Ala Ile Thr
295 300 305
2lb
CA 02590286 2010-02-17
aca gaa tgg gca ata gca gaa ctc att agc aat cca aat gta ctc aaa 1014
Thr Glu Trp Ala Ile Ala Glu Leu Ile Ser Asn Pro Asn Val Leu Lys
310 315 320 325
aaa get caa gaa gag atg gac aaa gtc ata gga tca caa agg ttg ttg 1062
Lys Ala Gln Glu Glu Met Asp Lys Val Ile Gly Ser Gln Arg Leu Leu
330 335 340
caa gaa tcc gac gcc cct aac ttg cct tac ctc aac gcg atc ata aaa 1110
Gln Glu Ser Asp Ala Pro Asn Leu Pro Tyr Leu Asn Ala Ile Ile Lys
345 350 355
gaa acg ttc cgt ctc cac cct cca atc ccc atg ctc act aga aaa tca 1158
Glu Thr Phe Arg Leu His Pro Pro Ile Pro Met Leu Thr Arg Lys Ser
360 365 370
att tct gac gtt gtg gtc aac ggg tac acg atc cct gcc aaa acg cta 1206
Ile Ser Asp Val Val Val Asn Gly Tyr Thr Ile Pro Ala Lys Thr Leu
375 380 385
ttg ttt gtc aac ctt tgg tcc atg gga agg aat cct aac tac tgg gaa 1254
Leu Phe Val Asn Leu Trp Ser Met Gly Arg Asn Pro Asn Tyr Trp Glu
390 395 400 405
aat ccg atg gag ttc cga ccc gag agg ttt ctc gag aaa ggg acc ggg 1302
Asn Pro Met Glu Phe Arg Pro Glu Arg Phe Leu Glu Lys Gly Thr Gly
410 415 420
tcg ata gac gtt aaa ggg cag cat ttc gag ttg ctg ccg ttt ggc acg 1350
Ser Ile Asp Val Lys Gly Gln His Phe Glu Leu Leu Pro Phe Gly Thr
425 430 435
ggc agg cgg ggc tgc ccg ggg atg ttg tta ggc atg cag gag ttg ttt 1398
Gly Arg Arg Gly Cys Pro Gly Met Leu Leu Gly Met Gln Glu Leu Phe
440 445 450
agt att atc ggg get atg gtg cag tgc ttc gat tgg aaa ctg ccc gat 1446
Ser Ile Ile Gly Ala Met Val Gln Cys Phe Asp Trp Lys Leu Pro Asp
455 460 465
ggt gtg aag tcg gtc gac atg acc gag cgg ccc ggg ttg acg get cca 1494
Gly Val Lys Ser Val Asp Met Thr Glu Arg Pro Gly Leu Thr Ala Pro
470 475 480 485
cgt gcc aat gat ttg gtg tgc caa ttg gtg cca cgg att gac ccg gtc 1542
Arg Ala Asn Asp Leu Val Cys Gln Leu Val Pro Arg Ile Asp Pro Val
490 495 500
gtt gtc tcc gga ccg tgaaccttaa ggtagtatcg ataatctgtt taatt 1592
Val Val Ser Gly Pro
505
aaattgttat ttgttgtgag gatttgattt ttgttatgta tgattatgcg tggattaaga 1652
21c
CA 02590286 2010-02-17
taagcctgca aggacaaatt ccctttcttt gattgatgtc aatgagtttg tgtcaaaaaa 1712
aaaaaaaaaa as 1724
<210> 2
<211> 506
<212> PRT
<213> Antirrhinum majus
<220>
<223> Amino acid sequence of a protein having an activity to
directly convert flavanone to flavone
<400> 2
Met Ser Thr Leu Val Tyr Ser Thr Leu Phe Ile Leu Ser Thr Leu Leu
1 5 10 15
Leu Thr Leu Leu Thr Arg Thr Arg Arg Lys Thr Arg Pro Pro Gly Pro
20 25 30
Leu Ala Leu Pro Leu Ile Gly His Leu His Leu Leu Gly Pro Lys Leu
35 40 45
His His Thr Phe His Gln Phe Ser Gln Arg Tyr Gly Pro Leu Ile Gln
50 55 60
Leu Tyr Leu Gly Ser Val Pro Cys Val Val Ala Ser Thr Pro Glu Leu
65 70 75 80
Ala Arg Glu Phe Leu Lys Thr His Glu Leu Asp Phe Ser Ser Arg Lys
85 90 95
His Ser Thr Ala Ile Asp Ile Val Thr Tyr Asp Ser Ser Phe Ala Phe
100 105 110
Ala Pro Tyr Gly Pro Tyr Trp Lys Phe Ile Lys Lys Leu Cys Thr Tyr
115 120 125
Glu Leu Leu Gly Ala Arg Asn Leu Ser His Phe Gln Pro Ile Arg Ala
130 135 140
Leu Glu Val Asn Ser Phe Leu Arg Ile Leu Tyr Glu Lys Thr Glu Gln
145 150 155 160
Lys Gln Ser Val Asn Val Thr Glu Glu Leu Val Lys Leu Thr Ser Asn
165 170 175
Val Ile Ser Asn Met Met Leu Gly Ile Arg Cys Ser Gly Thr Glu Gly
180 185 190
Glu Ala Glu Val Ala Arg Thr Val Ile Arg Glu Val Thr Gln Ile Phe
195 200 205
Gly Glu Phe Asp Val Ser Glu Ile Val Trp Phe Cys Lys Asn Leu Asp
21d
CA 02590286 2010-02-17
210 215 220
Leu Gln Gly Ile Arg Lys Arg Ser Glu Asp Ile Arg Arg Arg Tyr Asp
225 230 235 240
Ala Leu Leu Glu Lys Ile Ile Ser Asp Arg Glu Arg Leu Arg Leu Arg
245 250 255
Gly Gly Gly Gly Gly Gly Gly Gly Glu Val Lys Asp Phe Leu Asp Met
260 265 270
Leu Leu Asp Val Met Glu Ser Glu Lys Ser Glu Val Glu Phe Thr Arg
275 280 285
Glu His Leu Lys Ala Leu Ile Leu Asp Phe Phe Thr Ala Gly Thr Asp
290 295 300
Thr Thr Ala Ile Thr Thr Glu Trp Ala Ile Ala Glu Leu Ile Ser Asn
305 310 315 320
Pro Asn Val Leu Lys Lys Ala Gln Glu Glu Met Asp Lys Val Ile Gly
325 330 335
Ser Gln Arg Leu Leu Gln Glu Ser Asp Ala Pro Asn Leu Pro Tyr Leu
340 345 350
Asn Ala Ile Ile Lys Glu Thr Phe Arg Leu His Pro Pro Ile Pro Met
355 360 365
Leu Thr Arg Lys Ser Ile Ser Asp Val Val Val Asn Gly Tyr Thr Ile
370 375 380
Pro Ala Lys Thr Leu Leu Phe Val Asn Leu Trp Ser Met Gly Arg Asn
385 390 395 400
Pro Asn Tyr Trp Glu Asn Pro Met Glu Phe Arg Pro Glu Arg Phe Leu
405 410 415
Glu Lys Gly Thr Gly Ser Ile Asp Val Lys Gly Gln His Phe Glu Leu
420 425 430
Leu Pro Phe Gly Thr Gly Arg Arg Gly Cys Pro Gly Met Leu Leu Gly
435 440 445
Met Gln Glu Leu Phe Ser Ile Ile Gly Ala Met Val Gln Cys Phe Asp
450 455 460
Trp Lys Leu Pro Asp Gly Val Lys Ser Val Asp Met Thr Glu Arg Pro
465 470 475 480
Gly Leu Thr Ala Pro Arg Ala Asn Asp Leu Val Cys Gln Leu Val Pro
485 490 495
Arg Ile Asp Pro Val Val Val Ser Gly Pro
500 505
<210> 3
<211> 1730
21e
CA 02590286 2010-02-17
<212> DNA
<213> Torenia hybrida
<220>
<223> Nucleotide sequence encoding a protein having an
activity to directly convert flavanone to flavone
<400> 3
atcgaaaccg ctatatcatt acatttacaa cagcgctaaa aaaatatata taaagc 56
atg gac aca gtc tta atc aca ctc tac acc gcc ctg ttc gtc atc acc 104
Met Asp Thr Val Leu Ile Thr Leu Tyr Thr Ala Leu Phe Val Ile Thr
1 5 10 15
acc acc ttc ctc ctc ctc ctc cgc cga agg gga cca ccg tct ccg ccc 152
Thr Thr Phe Leu Leu Leu Leu Arg Arg Arg Gly Pro Pro Ser Pro Pro
20 25 30
ggt cct ctc tcc cta ccc ata att ggc cac ctc cac ctc ctc ggc cca 200
Gly Pro Leu Ser Leu Pro Ile Ile Gly His Leu His Leu Leu Gly Pro
35 40 45
aga ctc cac cac acg ttc cat gaa ttc tca ctc aaa tac ggc cca ttg 248
Arg Leu His His Thr Phe His Glu Phe Ser Leu Lys Tyr Gly Pro Leu
50 55 60
atc cag ctc aag ctc ggc tcg atc ccg tgc gtc gtg gcc tcg acg ccc 296
Ile Gln Leu Lys Leu Gly Ser Ile Pro Cys Val Val Ala Ser Thr Pro
65 70 75 80
gag ctc gcg aga gag ttt ctt aag acg aac gag ctc gcg ttc tcc tct 344
Glu Leu Ala Arg Glu Phe Leu Lys Thr Asn Glu Leu Ala Phe Ser Ser
85 90 95
cgc aag cac tct acg gcc ata gac atc gtc acc tac gac tcg tcc ttt 392
Arg Lys His Ser Thr Ala Ile Asp Ile Val Thr Tyr Asp Ser Ser Phe
100 105 110
get ttc tct ccg tac gga ccc tac tgg aag tac atc aag aaa ctg tgt 440
Ala Phe Ser Pro Tyr Gly Pro Tyr Trp Lys Tyr Ile Lys Lys Leu Cys
115 120 125
acc tac gag ctg ctc gga gcg agg aac ctc gga cac ttt cag ccc att 488
Thr Tyr Glu Leu Leu Gly Ala Arg Asn Leu Gly His Phe Gln Pro Ile
130 135 140
agg aat ctc gag gtc agg tcc ttt ctg cag ctt ctg atg cac aag agc 536
Arg Asn Leu Glu Val Arg Ser Phe Leu Gln Leu Leu Met His Lys Ser
145 150 155 160
ttt aag ggc gag agt gtg aat gtg aca gac gag ctg gtg agg ctg acg 584
21f
CA 02590286 2010-02-17
Phe Lys Gly Glu Ser Val Asn Val Thr Asp Glu Leu Val Arg Leu Thr
165 170 175
agc aat gtg ata tcc cac atg atg ctg agc ata agg tgc tcg gaa gat 632
Ser Asn Val Ile Ser His Met Met Leu Ser Ile Arg Cys Ser Glu Asp
180 185 190
gaa ggc gat get gag gcg gcg aga aca gtg ata cgc gag gtg acg cag 680
Glu Gly Asp Ala Glu Ala Ala Arg Thr Val Ile Arg Glu Val Thr Gln
195 200 205
ata ttt ggg gaa ttc gat gtt acg gac ata ata tgg ttt tgc aag aaa 728
Ile Phe Gly Glu Phe Asp Val Thr Asp Ile Ile Trp Phe Cys Lys Lys
210 215 220
ttc gat ctg cag ggg ata aag aag agg tca gag gat att cag agg agg 776
Phe Asp Leu Gln Gly Ile Lys Lys Arg Ser Glu Asp Ile Gln Arg Arg
225 230 235 240
tat gat get ttg ctc gag aag att att agt gat aga gag aga tcg agg 824
Tyr Asp Ala Leu Leu Glu Lys Ile Ile Ser Asp Arg Glu Arg Ser Arg
245 250 255
agg caa aat cgt gat aag cat ggt ggc ggt aac aat gag gag gcc aag 872
Arg Gln Asn Arg Asp Lys His Gly Gly Gly Asn Asn Glu Glu Ala Lys
260 265 270
gat ttt ctt gat atg ttg ctt gat gtg atg gag agt ggg gac acg gag 920
Asp Phe Leu Asp Met Leu Leu Asp Val Met Glu Ser Gly Asp Thr Glu
275 280 285
gtc aaa ttc act aga gag cat ctc aag get ttg att ctg gat ttc ttc 968
Val Lys Phe Thr Arg Glu His Leu Lys Ala Leu Ile Leu Asp Phe Phe
290 295 300
acg gcc ggt acg gac aca aca gcc ata gcc acc gag tgg gcc atc gcc 1016
Thr Ala Gly Thr Asp Thr Thr Ala Ile Ala Thr Glu Trp Ala Ile Ala
305 310 315 320
gag ctc atc aac aac ccg aac gtc ttg aag aag gcc caa gaa gaa ata 1064
Glu Leu Ile Asn Asn Pro Asn Val Leu Lys Lys Ala Gln Glu Glu Ile
325 330 335
tcc cgg atc atc gga acc aag cgg atc gta caa gaa tcc gac gcc cca 1112
Ser Arg Ile Ile Gly Thr Lys Arg Ile Val Gln Glu Ser Asp Ala Pro
340 345 350
gac cta ccc tac ctc cag gcc atc atc aag gag acg ttc cgg ctc cac 1160
Asp Leu Pro Tyr Leu Gln Ala Ile Ile Lys Glu Thr Phe Arg Leu His
355 360 365
cca ccg atc ccg atg ctc tcg cgt aag tcc acc tcc gat tgc acg gtc 1208
Pro Pro Ile Pro Met Leu Ser Arg Lys Ser Thr Ser Asp Cys Thr Val
21g
CA 02590286 2010-02-17
370 375 380
aac ggc tac aaa atc caa gcc aag agc ctc ttg ttc gtg aac ata tgg 1256
Asn Gly Tyr Lys Ile Gln Ala Lys Ser Leu Leu Phe Val Asn Ile Trp
385 390 395 400
tcc atc ggt cga aac cct aat tac tgg gaa agc cct atg gag ttc agg 1304
Ser Ile Gly Arg Asn Pro Asn Tyr Trp Glu Ser Pro Met Glu Phe Arg
405 410 415
ccc gag cgg ttc ttg gag aag gga cgc gag tcc atc gac gtc aag ggc 1352
Pro Glu Arg Phe Leu Glu Lys Gly Arg Glu Ser Ile Asp Val Lys Gly
420 425 430
cag cac ttt gag ctc ttg cct ttt ggg acg ggc cgc agg ggc tgt ccc 1400
Gln His Phe Glu Leu Leu Pro Phe Gly Thr Gly Arg Arg Gly Cys Pro
435 440 445
ggt atg ttg ctg get ata caa gag gtg gtc agc atc att ggg acc atg 1448
Gly Met Leu Leu Ala Ile Gln Glu Val Val Ser Ile Ile Gly Thr Met
450 455 460
gtt cag tgc ttc gac tgg aaa ttg gca gat ggt tcg ggc aat aat gtg 1496
Val Gln Cys Phe Asp Trp Lys Leu Ala Asp Gly Ser Gly Asn Asn Val
465 470 475 480
gac atg acc gaa cgg tct gga ttg acc get ccg aga gcg ttc gat ctg 1544
Asp Met Thr Glu Arg Ser Gly Leu Thr Ala Pro Arg Ala Phe Asp Leu
485 490 495
gtt tgc cgg ttg tat cca cgg gtt gac ccg gcc aca ata tcg ggt get 1592
Val Cys Arg Leu Tyr Pro Arg Val Asp Pro Ala Thr Ile Ser Gly Ala
500 505 510 512
tgatgtagta gggtgaggcg cgtgttggtg ttttatcttt cggttttgtt ctgttagtat 1652
tattatggtc tgtgttgaag cctcaaggat tttaaaaaaa aaaaaaaaaa aaaaaaaaaa 1712
aaaaaaaaaa aaaaaaaa 1730
<210> 4
<211> 512
<212> PRT
<213> Torenia hybrida
<220>
<223> Amino acid sequence of a protein having an activity to
directly convert flavanone to flavone
<400> 4
Met Asp Thr Val Leu Ile Thr Leu Tyr Thr Ala Leu Phe Val Ile Thr
21h
CA 02590286 2010-02-17
1 5 10 15
Thr Thr Phe Leu Leu Leu Leu Arg Arg Arg Gly Pro Pro Ser Pro Pro
20 25 30
Gly Pro Leu Ser Leu Pro Ile Ile Gly His Leu His Leu Leu Gly Pro
35 40 45
Arg Leu His His Thr Phe His Glu Phe Ser Leu Lys Tyr Gly Pro Leu
50 55 60
Ile Gln Leu Lys Leu Gly Ser Ile Pro Cys Val Val Ala Ser Thr Pro
65 70 75 80
Glu Leu Ala Arg Glu Phe Leu Lys Thr Asn Glu Leu Ala Phe Ser Ser
85 90 95
Arg Lys His Ser Thr Ala Ile Asp Ile Val Thr Tyr Asp Ser Ser Phe
100 105 110
Ala Phe Ser Pro Tyr Gly Pro Tyr Trp Lys Tyr Ile Lys Lys Leu Cys
115 120 125
Thr Tyr Glu Leu Leu Gly Ala Arg Asn Leu Gly His Phe Gln Pro Ile
130 135 140
Arg Asn Leu Glu Val Arg Ser Phe Leu Gin Leu Leu Met His Lys Ser
145 150 155 160
Phe Lys Gly Glu Ser Val Asn Val Thr Asp Glu Leu Val Arg Leu Thr
165 170 175
Ser Asn Val Ile Ser His Met Met Leu Ser Ile Arg Cys Ser Glu Asp
180 185 190
Glu Gly Asp Ala Glu Ala Ala Arg Thr Val Ile Arg Glu Val Thr Gln
195 200 205
Ile Phe Gly Glu Phe Asp Val Thr Asp Ile Ile Trp Phe Cys Lys Lys
210 215 220
Phe Asp Leu Gln Gly Ile Lys Lys Arg Ser Glu Asp Ile Gln Arg Arg
225 230 235 240
Tyr Asp Ala Leu Leu Glu Lys Ile Ile Ser Asp Arg Glu Arg Ser Arg
245 250 255
Arg Gln Asn Arg Asp Lys His Gly Gly Gly Asn Asn Glu Glu Ala Lys
260 265 270
Asp Phe Leu Asp Met Leu Leu Asp Val Met Glu Ser Gly Asp Thr Glu
275 280 285
Val Lys Phe Thr Arg Glu His Leu Lys Ala Leu Ile Leu Asp Phe Phe
290 295 300
Thr Ala Gly Thr Asp Thr Thr Ala Ile Ala Thr Glu Trp Ala Ile Ala
305 310 315 320
Glu Leu Ile Asn Asn Pro Asn Val Leu Lys Lys Ala Gln Glu Glu Ile
21i
CA 02590286 2010-02-17
325 330 335
Ser Arg Ile Ile Gly Thr Lys Arg Ile Val Gln Glu Ser Asp Ala Pro
340 345 350
Asp Leu Pro Tyr Leu Gln Ala Ile Ile Lys Glu Thr Phe Arg Leu His
355 360 365
Pro Pro Ile Pro Met Leu Ser Arg Lys Ser Thr Ser Asp Cys Thr Val
370 375 380
Asn Gly Tyr Lys Ile Gln Ala Lys Ser Leu Leu Phe Val Asn Ile Trp
385 390 395 400
Ser Ile Gly Arg Asn Pro Asn Tyr Trp Glu Ser Pro Met Glu Phe Arg
405 410 415
Pro Glu Arg Phe Leu Glu Lys Gly Arg Glu Ser Ile Asp Val Lys Gly
420 425 430
Gln His Phe Glu Leu Leu Pro Phe Gly Thr Gly Arg Arg Gly Cys Pro
435 440 445
Gly Met Leu Leu Ala Ile Gln Glu Val Val Ser Ile Ile Gly Thr Met
450 455 460
Val Gln Cys Phe Asp Trp Lys Leu Ala Asp Gly Ser Gly Asn Asn Val
465 470 475 480
Asp Met Thr Glu Arg Ser Gly Leu Thr Ala Pro Arg Ala Phe Asp Leu
485 490 495
Val Cys Arg Leu Tyr Pro Arg Val Asp Pro Ala Thr Ile Ser Gly Ala
500 505 510
<210> 5
<211> 31
<212> DNA
<213> Artificial Sequence
<220>
<223> Primer
<400> 5
aaataggatc caagcatgga cacagtctta a 31
<210> 6
<211> 35
<212> DNA
<213> Artificial Sequence
21j
CA 02590286 2010-02-17
<220>
<223> Primer
<400> 6
cccttctaga tcaagcaccc gatattgtgg ccggg 35
<210> 7
<211> 506
<212> PRT
<213> Perilla frutescens
<220>
<223> Amino acid sequence of a protein having an activity to
directly convert flavanone to flavone
<400> 7
Met Ala Leu Tyr Ala Ala Leu Phe Leu Leu Ser Ala Ala Val Val Arg
1 5 10 15
Ser Val Leu Asp Arg Lys Arg Gly Arg Pro Pro Tyr Pro Pro Gly Pro
20 25 30
Phe Pro Leu Pro Ile Ile Gly His Leu His Leu Leu Gly Pro Arg Leu
35 40 45
His Gln Thr Phe His Asp Leu Ser Gln Arg Tyr Gly Pro Leu Met Gln
50 55 60
Leu Arg Leu Gly Ser Ile Arg Cys Val Ile Ala Ala Ser Pro Glu Leu
65 70 75 80
Ala Lys Glu Cys Leu Lys Thr His Glu Leu Val Phe Ser Ser Arg Lys
85 90 95
His Ser Thr Ala Ile Asp Ile Val Thr Tyr Asp Ser Ser Phe Ala Phe
100 105 110
Ser Pro Tyr Gly Pro Tyr Trp Lys Phe Ile Lys Lys Leu Cys Thr Tyr
115 120 125
Glu Leu Leu Gly Ala Arg Asn Leu Ala His Phe Gln Pro Ile Arg Thr
130 135 140
Leu Glu Val Lys Ser Phe Leu Gln Ile Leu Met Arg Lys Gly Glu Ser
145 150 155 160
Gly Glu Ser Phe Asn Val Thr Glu Glu Leu Val Lys Leu Thr Ser Asn
165 170 175
Val Ile Ser His Met Met Leu Ser Ile Arg Cys Ser Glu Thr Glu Ser
180 185 190
21k
CA 02590286 2010-02-17
Glu Ala Glu Ala Ala Arg Thr Val Ile Arg Glu Val Thr Gln Ile Phe
195 200 205
Gly Glu Phe Asp Val Ser Asp Ile Ile Trp Leu Cys Lys Asn Phe Asp
210 215 220
Phe Gln Gly Ile Arg Lys Arg Ser Glu Asp Ile Gln Arg Arg Tyr Asp
225 230 235 240
Ala Leu Leu Glu Lys Ile Ile Thr Asp Arg Glu Lys Gln Arg Arg Thr
245 250 255
His Gly Gly Gly Gly Gly Gly Gly Glu Ala Lys Asp Phe Leu Asp Met
260 265 270
Phe Leu Asp Ile Met Glu Ser Gly Lys Ala Glu Val Lys Phe Thr Arg
275 280 285
Glu His Leu Lys Ala Leu Ile Leu Asp Phe Phe Thr Ala Gly Thr Asp
290 295 300
Thr Thr Ala Ile Val Cys Glu Trp Ala Ile Ala Glu Val Ile Asn Asn
305 310 315 320
Pro Asn Val Leu Lys Lys Ala Gln Glu Glu Ile Ala Asn Ile Val Gly
325 330 335
Phe Asp Arg Ile Leu Gln Glu Ser Asp Ala Pro Asn Leu Pro Tyr Leu
340 345 350
Gln Ala Leu Ile Lys Glu Thr Phe Arg Leu His Pro Pro Ile Pro Met
355 360 365
Leu Ala Arg Lys Ser Ile Ser Asp Cys Val Ile Asp Gly Tyr Met Ile
370 375 380
Pro Ala Asn Thr Leu Leu Phe Val Asn Leu Trp Ser Met Gly Arg Asn
385 390 395 400
Pro Lys Ile Trp Asp Tyr Pro Thr Ala Phe Gln Pro Glu Arg Phe Leu
405 410 415
Glu Lys Glu Lys Ala Ala Ile Asp Val Lys Gly Gln His Phe Glu Leu
420 425 430
Leu Pro Phe Gly Thr Gly Arg Arg Gly Cys Pro Gly Met Leu Leu Ala
435 440 445
Ile Gln Glu Val Val Ile Ile Ile Gly Thr Met Ile Gln Cys Phe Asp
450 455 460
Trp Lys Leu Pro Asp Gly Ser Gly His Val Asp Met Ala Glu Arg Pro
465 470 475 480
Gly Leu Thr Ala Pro Arg Glu Thr Asp Leu Phe Cys Arg Val Val Pro
485 490 495
Arg Val Asp Pro Leu Val Val Ser Thr Gln
500 505
211