Note: Descriptions are shown in the official language in which they were submitted.
CA 02594790 2007-07-12
WO 2006/075013 PCT/EP2006/050169
1
Genes encoding the synthetic pathway for the production of disorazole
The present invention relates to nucleic acid sequences and proteins derivable
therefrom
which are catalytically active or participate in the biosynthetic pathway of
disorazoles. The
catalytically active proteins, i.e. enzymes, are also known as polyketide
synthases and
nonribosomal peptide synthetases.
It is known that myxobacteria produce a large variety of biologically active
compounds, also
known as secondary metabolites. Among these secondary metabolites, the group
of
disorazoles has attracted attention as inhibitors for the polymerisation of
tubulin, for the
induction of apoptosis and for the arrest of the cell cycle or inhibition of
cell proliferation at
concentrations as low as e.g. 3 pM. The present invention provides nucleic
acid sequences
and proteins which can be translated from the nucleic acid sequences into
catalytically active
proteins or proteins participating in the biosynthesis of disorazoles. In
cooperation, these
translated proteins in vivo and/or in vitro catalyze the formation of
disorazoles. Accordingly,
the present invention also provides a production process using the nucleic
acid sequences
and/or proteins derivable therefrom for the production of disorazoles, for
example using
CA 02594790 2007-07-12
WO 2006/075013 PCT/EP2006/050169
2
homologous or heterologous expression of proteins derivable from these nucleic
acid
sequences in microorganisms for fermentation or the peptides in an immobilized
state to
produce disorazoles from precursor compounds.
State of the art
WO 2004/053065 A2 describes nucleic acid sequences encoding disorazole
polyketide
synthases DszA, DszB, DszC and DszD obtained from Sorangium cellulosum So ce
12 using
transposon generated cosmids. In very general terms, synthetic synthases are
described which
can be obtained by rearrangement of domains that can be identified in the
wildtype disorazole
synthase enzymes, namely a ketoreductase domain, a dehydratase domain, an
enoylreductase
domain, a ketosynthase domain, a nonribosomal protein synthetase domain, a
methyltransferase domain, an acyl carrier protein domain, a serine cyclization
domain, a
serine condensation domain, an adenylation domain, a peptidyl carrier protein
domain, a
thiolation domain, an oxidase domain, a thioesterase domain, and an acyl
transferase domain
from a total number of 8 domains in the disorazole synthetase. These domains
are predicted
from the DNA sequence obtained. However, specific synthetic rearrangements of
these
domains are not identified. The nucleotide sequence disclosed for the
disorazole polyketide
synthase and/or nonribosomal peptide synthetase comprises 77294 bp and
allegedly includes
the coding sequences for DszA, DszB, DszC, DszD and several other open reading
frames
which are located adjacent one another.
The present invention relates to the group of disorazoles, namely disorazole
Al and
derivatives thereof, for example dizorazoles according to the following
formulae 1-8 and
specific embodiments of these as detailed below:
Formula 1 Formula 2
X CH3 x CH3
X H3C CH3 x H C CH3
0 0 3
N R2 N
x O O
x R2
R3 00 X N ~0 R3 00 X N 0
H3c 0 H3c ~ 0
H3C CH3 H3C CH3
X. R1 R1
CA 02594790 2007-07-12
WO 2006/075013 PCT/EP2006/050169
3
Formula 3 Formula 4
X CH3 X CH3
X 11 H C CH3 X H3C CH3
O 3
N 0 N R2
O X O
X R2
0 R3 N 0 0 00 X N ~0
H3C 0 H3C 0
H3C CH3 H 3 C CH3
R1 R1
X X
Formula 5 Formula 6
R4 CH3 CH3
X H 3C CH3 H 3 C CH3
O O \
N R2 N R2
X 0 R4 O
R3 O O X N~O R3 O 0 X N O
H3c o H3c o
H 3 C CH3 H3C CH3
R1 R1
X X
Formula 7 Formula 8
X CH3 R4 CH3
X H3C CH3 X H3C CH3
O ~ R2 0 ~
N N R2
X O ~ X O
0 R3 X N~O R3 00 X =0
H3C O H3C N
H3C CH3 R1 H 3 C CH3 O
X
wherein
X represents an 0, two vicinal OH, or a single bond and
Rl, R2, R3, R4 each represent independently H, OH, OCH3.
Specific embodiments of general formulae 1-8 are:
Disorazole Al - A7 Disorazole F1 - F3
Disorazole B1 - B4 Disorazole G1 - G3
Disorazole Cl - C2 Disorazole H
Disorazole D1 - D5 Disorazole I
Disorazole El - E3
(R. Janssen et al., Liebigs Ann. Chem. 1994, 759-773).
CA 02594790 2007-07-12
WO 2006/075013 PCT/EP2006/050169
4
General description of the invention
The present invention provides the complete nucleic acid sequences encoding
not only a gene
cluster but further additional genetic elements which are necessary for
correct biosynthesis of
disorazoles. The entire biosynthetic gene cluster is disclosed, having high
homology to the
DszA - D disclosed in WO 2004/053065 A2 including its functional analysis.
The core biosynthetic gene cluster for the biosynthetic pathway for
disorazoles comprises
genes disA through disD. The gene disA is preceded by a putative ribosomal
binding site
located 11 base pairs upstream from the designated start codon (GTG). DisB
presumably
starts with an ATG and a putative ribosomal binding site could be localized 7
base pairs
upstream from the start codon. Arranged with disA and disB, which are
polyketide synthases,
in one transcriptional unit is disC, the latter encoding a mixed polyketide
synthase /
nonribosomal peptide synthetase. DisC most likely starts with an ATG, preceded
by a
putative ribosomal binding site located 8 base pairs upstream. An alternative
start codon of
disC could be found 36 base pairs downstream of the putative start codon.
Downstream this
transcriptional unit of disA, disB and disC, a probable transcription
terminator is located.
Following orf 9, located downstream of the transcriptional unit disA through
disC, disD was
identified having its putative ribosomal binding site 7 base pairs upstream
its start codon. The
gene disD shows significant similarities to the bifunctional proteins LnmG
from the
leinamycin biosynthetic gene cluster and to MmpIII from the mupirocin
biosynthetic gene
cluster. The C-terminus of DisD has close sequence similarity to the
oxidoreductase
superfamily. From a total of four transposon mutants, listed in Table 3 below,
plasmids were
recovered, harbouring the hygromycin resistance gene and the kpir dependent
origin of
replication (ori) R6K together with parts of chromosomal DNA of Sorangium
cellulosum So
ce 12 which originally flanked the transposition site. A computer assisted
analysis of the
chromosomal DNA portions using BLAST searches identified two of the proteins
predicted
from the recovered DNA portions as putative fragments of a polyketide synthase
and a
nonribosomal peptide synthetase. Using these two chromosomal DNA portions as
probes for
hybridization with a BAC library, previously established for Sorangium
cellulosum So ce 12,
sequencing of hybridizing BAC clones yielded orfs encoding proteins
participating in the
biosynthesis of disorazoles, which are summarised in Table 1 below.
CA 02594790 2007-07-12
WO 2006/075013 PCT/EP2006/050169
Detailed description of the invention
When analysing the biosynthetic pathway for the production of disorazoles, the
genomic
DNA of Sorangium cellulosum So ce 12 has been analyzed to identify the genes
whose
translation products are necessary components of the synthetic pathway, fmally
producing
disorazoles including known variants or derivatives of disorazole A, e. g.
according to
formulae 1- 8 above. The gene cluster encoding the enzymes catalyzing the
biosynthesis of
disorazoles comprises the translation products of disA, disB, disC, disD. It
is possible that
translation products from open reading frame (orf) orf 9, arranged between
disC and disD,
may participate in or be beneficial to the biosynthesis of disorazoles.
In the following, reference is made to the figures, wherein
= Figure 1 is a schematic representation of the synthetic pathway for
disorazoles,
= Figure 2 schematically shows the arrangement of genes adjacent to the
insertion site of
the transposon in the transposon mutant So ce 12_EXI_IE-2 and sequenced from
its
plasmid pTn-Rec_IE-2, and
= Figure 3 lists nucleic acid and amino acid sequences relevant to the
invention, namely
the nucleic acid sequence ofpTn-Rec_IE-2 (Seq.-ID No. 1), the amino acid
sequences
of orf 1-pTn-Rec_IE-2 (Seq.-ID No. 2), orf 2-pTn-Rec_IE-2 (Seq.-ID No. 3), orf
3-
pTn-Rec_IE-2 (Seq.-ID No. 4), orf 4-pTn-Rec_IE-2 (Seq.-ID No. 5), orf 5-pTn-
Rec IE-2 (Seq.-ID No. 6), the nucleic acid sequence disA-disD (Seq.-ID No. 7)
comprising genes disA, disB, disC, orf 9 and disD, and amino acid sequences of
DisA
(Seq.-ID No. 8), DisB (Seq.-ID No. 9), DisC (Seq.-ID No. 10), orf 9 (Seq.-ID
No. 11)
and DisD (Seq.-ID No. 12).
The functions proposed in Table 1 above have been identified by a similarity
search on
known sequences, however, the gene products from the orfs of Table 1 can
differ according to
their function in the biosynthetic gene cluster for disorazoles.
An analysis of the genomic DNA region encoding disA through disD has revealed
several
orfs in the vicinity of disA through disD, summarised in Table 1.
CA 02594790 2007-07-12
WO 2006/075013 PCT/EP2006/050169
6
Table 1: Orfs identified in the biosynthetic gene cluster for disorazoles
Acc.
Orienta- Proposed Function No. of
Size tion of the Similar Similarity to Similarity/ similar
Gene (Da/bp) (strand) Protein Source Identity protein
orfl 49316/ - b1r4832 Bradyrhizobium 49%/67%, NC 00
1374 Sugar (and other) japonicum USDA 4463.1
transporter 110
orf2 51696/ - probable two- Pseudomonas 49%/66% NC 00
1449 component aeruginosa PAO1 2516.1
response regulator,
signal receiver
domain
orf3 45545/ + hypothetical Leptospira 27%/40% NC 00
1293 protein interrogans 4342.1
serovar Lai str.
56601
orf4 56119/ - no prediction
1641
orf5 48994/ - probable two- Pseudomonas 51%/69% NC 00
1371 component aeruginosa PAO1 2516.1
response regulator,
signal receiver
domain
orf6 105961 - sensory box Pseudomonas 39%/55% NC 00
/3021 histidine kinase putida KT2440 2947.3
orfl 34954/ - phosphotransferase Escherichia coli 29%/40% 4~5
975
orf$ 37435/ + putative Streptomyces 33%/48% NC 00
1053 serine/threonine avermitilis MA- 3155.2
protein kinase 4680
disA +
-C
orf9 30717/ + no functional
822 prediction
disD +
orf 23476/ - phosphotransferase Bacillus subtilis 38%/56% NC 00
642 subsp. subtilis str. 0964.2
168
orf 46773/ + putative sugar Streptomyces 27%/41% NC 00
11 1287 transporter avermitilis MA- 3155.2
4680
orf 32992/ + ABC membrane Brevibacterium 36%/53% 093RD
12 912 transporter fuscum var. 7
homologue dextranlyticum
orf 31993/ + ABC membrane Brevibacterium 51o/a/72o/a 093RD
13 882 transporter fuscum var. 6
homologue dextranlyticum
CA 02594790 2007-07-12
WO 2006/075013 PCT/EP2006/050169
7
Table 1 continued:
orf 86590/ + putative sugar Streptomyces 61 %/72% NP_73
14 2355 hydrolase coelicolor A3(2) 3521
orf 105005 + putative sugar Streptomyces 46%/59% NP_62
15 /2892 hydrolase coelicolor A3(2) 9813
orf 121293 + serine-threonine Mycobacterium 36%/53% NP_30
16 /3273 protein kinase leprae TN 1681
orf 23384/ - no prediction
17 642
orf 35402/ - no prediction
18 999
orf 25075/ - no prediction
19 657
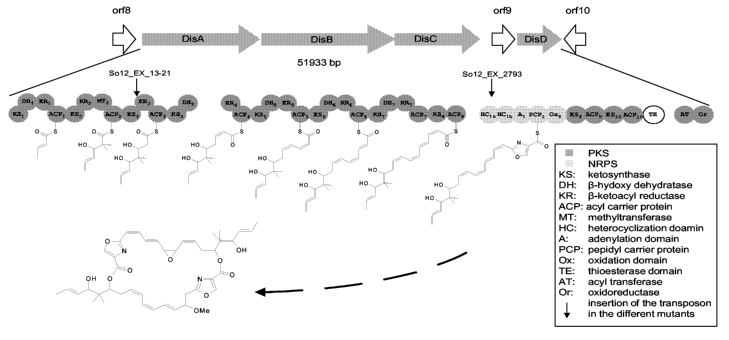
Figure 1 schematically depicts the arrangement of genes disA, disB, disC,
orf9, and disD,
wherein the abbreviations refer to catalytic centers and domains as follows:
Dark shade : polyketide synthase (PKS), Light shade nonribosomal protein
synthetase (NRPS), KS: ketosynthase, DH: 0-hydroxydehydratase, KR: 0-ketoacyl
reductase, ACP: acyl carrier protein, MT: methyltransferase, HC:
heterocyclization
domain, A: adenylation domain, PCP: peptidyl carrier protein, Ox: oxidation
domain,
TE: thioesterase domain,
AT: acyl transferase, Or: oxidoreductase, and 1: site of insertion of
transposon in
different mutants.
The sites indicated by the arrows (1) are designated as So12_EX_13-21 and
So12_EX 2793,
which are So ce 12 mutants from which the plasmids pTn-Rec13-21 and pTn-
Rec2793,
respectively, were recovered.
The arrangement of genes adjacent to the insertion site of the transposon
mutant So ce
12_EXI IE-2 is schematically depicted in Figure 2.
For the gene products of disA through disD, functions can be proposed for
individual protein
domains by homology search. These proposed functions, including their relative
positions in
the individual nucleic acid sequences are listed in Table 2 below.
CA 02594790 2007-07-12
WO 2006/075013 PCT/EP2006/050169
8
Table 2: Disorazole biosynthetic genes disA, disB, disC and disD
Proposed Function
Protein Size (Protein domains with their positions in the amino acid sequences
of
(Gene) (Da/bp) Figure 3)
DisA 647772/ PKS Domains: KS1 (3-428), DH1 (953-1144), KR1(1528-1779),
(disA) 18036 ACP1 (1821-1889), KS2 (1971-2395); KR2 (2856-3105), MT2
(3225-3463), ACP2 (3537-3606), ACP2b (3672-3741), KS3 (3779-
4201), KR3 (4642-4898), ACP3 (4918-4987), KS4 (5059-5490),
DH4 (5649-5878)
DisB 672408/ PKS Domains: KR4 (238-492), ACP4 (547-615), KS5 (676-1114),
(disB) 18771 DH5 (1274-1476), KR5 (1836-2093), ACP5 (2108-2176), KS6
(2255-2686), DH6 (2944-3149), KR6 (3490-3738), ACP6 (3776-
3824), KS7 (3876-4304), DH7 (4472-4679), KR7 (5049-
5302),ACP7 (5316-5398), KS8 (5500-5926), ACP8 (6123-6192)
DisC 409960/ NRPS Domains:HC1a (58-506), HC1b (532-955), Al (1035-1551),
(disC) 11379 PCP1 (1580-1647), OX (1649-1836),
PKS Domains: KS9 (1882-2309), ACP9 (2542-2609), KS10 (2668-
3098), ACP10 (3399-3468), TE (3521-3701)
DisD 90953/ PKS-Domains: AT (1-280), OX (393-839)
(disD) 2526
Abbreviations are according to Figure 1.
However, when analysing the synthesis of disorazoles in microorganisms
expressing the
biosynthetic gene cluster consisting of the sequences encoding DisA, DisB,
DisC and DisD
only, homologous sequences of which have been described in WO 2004/053065 A2,
it is
considered impossible that the full range of derivative disorazoles could be
produced with the
translation products DisA, DisB, DisC and DisD only. The reason is that
comparative analysis
showed that DisA, DisB, DisC and DisD lack at least some functions, e.g.
necessary for
hydroxylation, epoxidation and methoxylation, that are assumed necessary for
synthesis of at
least some known derivatives of disorazole.
Further analysis of the genomic region adjacent the genes disA through disD,
for example the
gene products of those orfs listed in Table 2 above, did not identify coding
sequences for
accessory functions to complement the biosynthetic pathway of DisA through
DisD to allow
production of disorazole or the range of known disorazole derivatives.
Analysis of the two additional disorazole negative mutants revealed further
sequences
obtainable from Sorangium cellulosum So ce 12, at least one of which encodes a
translation
CA 02594790 2007-07-12
WO 2006/075013 PCT/EP2006/050169
9
product that is necessary for synthesis of disorazoles in combination with the
translation
products of disA, disB, disC and disD, preferably in combination with the
translation product
of orf 9. These additional nucleic acid sequences have been identified on
recovered plasmids
of disorazole negative So ce 12 mutants and are summarised in Table 3 below.
Table 3: Recovered plasmids and proposed function of the encoded proteins
Identity/
Plasmid Proposed function of the similar Source of the Similarity
protein similar protein (DNA/
protein)
pTn-Rec_2793 BarG (PKS) Lyngbya majuscula
barbamide biosynthetic gene cluster 39%/57%
pTn-Rec_13-3 5' to transposition site: no prediction
Rhodopirellula
3' to transposition site: baltica SH 1 28%/45%
carbamoyltransferase B1mD
pTn-Rec_13-21 LnmJ (PKS) Streptomyces
leinamycin biosynthetic gene cluster atroolivaceus 29%/40%
pTn-Rec_IE-2 beta-lactamase Oceanobacillus
iheyensis 38%/53%
putative esterase
Rhodopirellula 30%/48%
baltica SH 1
The proposed functions have been identified by similarity searches with known
proteins but
may be different from the proposed functions indicated here according to their
functions
within the biosynthetic gene pathway.
Sequencing of pTn-Rec_IE-2 identified a total of 5 orfs and their putative
functions, which
are summarized in Table 4 below:
CA 02594790 2007-07-12
WO 2006/075013 PCT/EP2006/050169
Table 4: Proteins encoded on the plasmid pTn-RecIE-2 and their putative
function
Position Size Proposed Source Similarity/
orf on DNA (Da/bp) Function Identity
sequence of the Similar (DNA/
of pTn- Protein protein)
RecIE-2
orf 1- 58 - 579 18008/522 arylesterase- Caulobacter 29%/43%
pTn- related protein crescentus
RecIE-2
orf 2 - 1665 - 20979/591 SAM-dependent Gloeobacter 48%/58%
pTn- 2255 methyl- violaceus
RecIE-2 transferase
orf 3 - 3159 - 46369/128 putative esterase Rhodopirellula 35%/51%
pTn- 4442 4 beta-lactamase baltica SH 1
RecIE-2 Oceanobacillus
iheyensis
orf 4 - 4459 - 62063/178 adenylate cyclase Stigmatella 31 %/51 %
pTn- 6240 2 2 aurantiaca
RecIE-2
orf 5 - 6328 - 29564/854 outer membrane Myxococcus 36%/46
pTn- 7181 protein xanthus
RecIE-2 (incomplete)
In a first embodiment of the present invention, at least one of the
translation products of Table
4 is used in combination with the translation products of disA through disD to
provide the
biosynthetic pathway for disorazoles, in a preferred embodiment, at least 2,
more preferred
three or four translation products of the sequences identified in Table 4
participate in the
biosynthetic pathway for disorazoles in combination with disA through disD,
preferably
including the translation product of orf 9.
The DNA sequences of disA, disB, disC, disD and orf 1-pTn-Rec_IE-2, orf 2-pTn-
Rec_IE-2,
orf 3-pTn-Rec_IE-2, orf 4-pTn-Rec_IE-2, and orf 5-pTn-Rec_IE-2 as well as
their translation
products obtained from Sorangium cellulosum So ce 12 are listed in Figure 3.
These specific
sequences are preferred for performing the present invention, but other coding
sequences and
peptides derivable therefrom providing the respective activity necessary in
the disorazole
synthetic pathway are also applicable in the present invention and can replace
the sequences
of Figure 3.
The present invention will now be described in greater detail by way of
examples, which are
not intended to limit the scope of the invention.
CA 02594790 2007-07-12
WO 2006/075013 PCT/EP2006/050169
11
Example 1: Cloning and sequencing of nucleic acid sequences complementing the
biosynthetic pathway enzymes for disorazoles
Nucleic acid sequences, the translation products of which participate in the
biosynthetic
pathway for disorazoles have been identified using a transposon recovery
procedure from
disorazole negative transposon mutants of Sorangium cellulosum strain So ce
12. Strain So ce
12 is available at NCIMB Aberdeen, UK, under accession No. NCIB 12134.
For transposon mutagenesis, transposon termed pMiniHimarHyg which is
applicable to
myxobacteria was used, comprising the hygromycin resistance, but lacking the
genes for
conjugational DNA transfer. The transformation of Sorangium cellulosum was
obtained by
electroporation as described in European patent application EP 04 103 546.0,
filed on 23 July
2004 with the European patent office.
Disorazole negative mutants were detected in a bioassay using an overlay with
the disorazole
sensitive yeast R. glutinis. In this bioassay, transposon mutants were plated
on PM 12 agar
plates without hygromycin at 32 C until colonies became visible, then
overlayed with R.
glutinis, incubated overnight at 30 C and growth inhibition zones were
compared to a wild
type Sorangium cellulosum So ce 12.
Transposon recovery from disorazole negative transposon mutant colonies was
essentially
carried out as described in Kopp et al. (J. Biotech 107, 29 (2004))
Example 2: Heterolo opression of biosynthetic pathway enzymes for the
production of
disorazole
The core biosynthetic gene cluster and their respective translation products
sufficient for the
biosynthesis of disorazoles was determined by heterologous gene expression
experiments. As
expected, the core enzymes comprising disA, disB, disC as well as disD are
regarded as
necessary components for the biosynthetic pathway. An optional and preferably
included
component is orf 9.
The core cluster comprising disA, disB, disC as well as disD needs
complementation with at
least an expression cassette encoding orf 3-pTn-Rec_IE-2, optionally in
combination with orf
1-pTn-Rec_IE-2, optionally in combination with orf 2-pTn-Rec_IE-2, optionally
in
CA 02594790 2007-07-12
WO 2006/075013 PCT/EP2006/050169
12
combination with orf 4-pTn-Rec_IE-2, and optionally in combination with orf 5-
pTn-Rec_IE-
2.
When expressing sequences encoding at least one, preferably two, more
preferably three or
four and most preferably all of the group comprising orf 1-pTn-Rec_IE-2, orf 2-
pTn-Rec_IE-
2, orf 4-pTn-Rec_IE-2, and orf 5-pTn-Rec_IE-2, in combination with orf 3 - pTn-
Rec_IE-2 to
supplement the expression cassettes encoding disA - disD, optionally orf 9,
respectively,
production of disorazoles was found.
The number of derivative disorazoles varied according to the sequences
selected among orf 1-
pTn-Rec_IE-2, orf 2-pTn-Rec_IE-2, orf 4-pTn-Rec_IE-2, and orf 5-pTn-Rec_IE-2
for
expression in combination with orf 3 - pTn-Rec_IE-2 and disA - disD,
optionally orf 9. It is
preferred that the coding sequences are contained intra-chromosomally in their
natural
arrangement.
For production of disorazoles, the identification of the set of genes or gene
cluster according
to the invention allows to modify producer strains, for example by
specifically targeted
modification of regulatory elements, e.g. the introduction of stronger
promoters for disA,
disB, disC, orf 9, and/or disD, and/or for the complementing genes orf 1-pTn-
Rec_IE-2, orf 2-
pTn-Rec_IE-2, orf 3 - pTn-Rec_IE-2, orf 4-pTn-Rec_IE-2, and/or orf 5-pTn-
Rec_IE-2.
Alternatively, heterologous expression can be employed using microorganisms
which are no
natural producers of disorazole. For heterologous expression, Myxococcales,
preferably
Myxococcus xanthus, or Polyangium, also termed Sorangium, e. g. Sorangium
cellulosum
accessible as ATCC 25531, ATCC 29479 (DSMZ 2044), Stigmatella aurantiaca,
Angiococcus disciforrnis and strains of the genus Pseudomonas, e.g.
Pseudomonasputida,
Pseudomonas stutzeri, and Pseudomonas syringae can be used.
Alternatively, the expression products, i. e. proteins derivable from the
aforementioned sets of
genes for the synthetic pathway, can be used in an extracellular synthesis
system, e. g. as
catalysts like an immobilized enzyme system for synthesis of disorazoles.