Note: Descriptions are shown in the official language in which they were submitted.
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
HUMAN CANCER SUPPRESSOR GENE, PROTEIN ENCODED THEREIN
TECHNICAL FIELD
The present invention relates to a human cancer suppressor gene, a protein
encoded therein, an expression vector containing the same and a cell
transformed with
the vector.
BACKGROUND ART
Tumor suppressor gene products function to suppress normal cells from being
transformed into certain cancer cells, and therefore loss of this function of
the tumor
suppressor gene products allows the normal cells to become malignant
transformants
(Klein, G., FASEB J, 7, 821-825 (1993)). In order to allow cancer cells to
grow into a
cancer, the cells should lose a function to control the normal copy number of
a tumor
suppressor gene. It was found that modification in a coding sequence of a p53
tumor
suppressor gene is one of the most general genetic changes in the human
cancers
(Bishop, J.M., Cell, 64, 235-248 (1991); and Weinberg, R.A., Science, 254,
1138-1146
(1991)).
However, it was estimated that only some of breast cancer tissues exhibit a
p53
mutation because the p53 mutation reported in the breast cancer amounts to a
range of
30 % of the total breast cancer (Keen, J.C. & Davidson, N. E., Cancer, 97, 825-
833
(2003)) and Borresen-Dale, A-L., Human Mutation, 21, 292-300 (2003)). Also, it
was
estimated that only some of leukemia tissues exhibit a p53 mutation because
the p53
mutation reported in the leukemia amounts to a range of 20 % of the total
leukemia
1
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
(Boyapati, A., et al., Acta Haematol., 111(1-2), 100-106 (2004)).
The p53 mutation accounts to at least 50 % of the liver cancer especially in
the
region exposed to aflatoxin B 1 or having a high frequency of infection by
hepatitis B
virus, and it is mainly characterized by a missense mutation at a codon 249 in
the p53
tumor gene (Montesano, R. et al., J. Natl. Cancer Inst., 89, 1844-1851 (1997);
Szymanska, K. & Hainaut, P. Acta Biochimica Polonica, 50, 231-238 (2003)).
However, it was reported that the p53 mutation amounts to nothing but a range
of 30 %
of the liver cancer in U.S. and Western Europe, and there is no hot spot in
which such
mutation occurs frequently (Szymanska, K. & Hainaut, P. Acta Biochimica
Polonica, 50,
231-238 (2003)).
Meanwhile, it was estimated that only some of the cervical cancer tissues
exhibit
a p53 mutation because the p53 mutation reported in the cervical cancer
amounts to only
a range of 2 to 11 % of the toatal cervical cancer (Crook, T. et al., Lancet,
339,
1070-1073 (1992); and Busby-Earle, R.M.C. et al., Br. j. Cancer, 69, 732-737
(1994)).
Also, it was reported that the mutation frequency of a p53 tumor suppressor
gene
in the non-small-cell lung cancer and the small-cell lung cancer amounts to
approximately 50 % and 70 % of the total lung cancer, respectively (Takahashi,
T. et al.,
Science, 246, 491-494 1989; Bodner, S.M. et al., Oncogene, 7, 743-749 (1992);
Mao, L.
Lung Cancer, 34, S27-S34 (2001)). Smoking is one of the most critical factors
in
development and progress of the lung cancer, and other various tumor
suppressor genes
and cancer genes in addition to the p53 are concurrently involved in the
development
and the progress of the lung cancer (Osada, H. & Takahashi, T. Oncogene, 21,
7421-7434 (2002)).
2
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
Accordingly, the present inventors have ardently attempted to separate a novel
tumor suppressor gene from normal tissues such as breast, liver, cervix,
lungs, etc. using
an mRNA differential display (DD) method for more effectively displaying genes
differentially expressed between the normal tissues such as the breast, the
liver, the
cervix and the lungs and the cancer tissues such as the breast cancer, the
liver cancer,
the cervical cancer and the lung cancer (Liang, P. and Pardee, A. B., Science,
257,
967-971 (1992); and Liang, P. et al., Cancer Res., 52, 6966-6968 (1993)).
DISCLOSURE OF INVENTION
Accordingly, the present invention is designed to solve the problems of the
prior
art, and therefore it is an object of the present invention to provide a novel
human
cancer suppressor gene.
It is another object of the present invention to provide a cancer suppressor
protein encoded by the cancer suppressor gene.
It is still another object of the present invention to provide an expression
vector
including the cancer suppressor gene.
In order to accomplish one of the above objects, the present invention
provides a
human cancer suppressor gene having a DNA sequence selected from the group
consisting of SEQ ID NO: 1; SEQ ID NO: 5; SEQ ID NO: 9; SEQ ID NO: 13; SEQ ID
NO: 17; SEQ ID NO: 21; SEQ ID NO: 25; SEQ ID NO: 29; SEQ ID NO: 33; SEQ ID
NO: 37; SEQ ID NO: 41; SEQ ID NO: 45; SEQ ID NO: 49; SEQ ID NO: 53; SEQ ID
NO: 57; SEQ ID NO: 61; SEQ ID NO: 65; SEQ ID NO: 69; SEQ ID NO: 73; SEQ ID
3
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
NO: 77; SEQ ID NO: 81; SEQ ID NO: 85; SEQ ID NO: 89; SEQ ID NO: 93; SEQ ID
NO: 97; SEQ ID NO: 101; SEQ ID NO: 105; SEQ ID NO: 109 and SEQ ID NO: 113.
According to another of the above objects, the present invention provides a
human cancer suppressor protein having an amino acid sequence selected from
the
group consisting of SEQ ID NO: 2; SEQ ID NO: 6; SEQ ID NO: 10; SEQ ID NO: 14;
SEQ ID NO: 18; SEQ ID NO: 22; SEQ ID NO: 26; SEQ ID NO: 30; SEQ ID NO: 34;
SEQ ID NO: 38; SEQ ID NO: 42; SEQ ID NO: 46; SEQ ID NO: 50; SEQ ID NO: 54;
SEQ ID NO: 58; SEQ ID NO: 62; SEQ ID NO: 66; SEQ ID NO: 70; SEQ ID NO: 74;
SEQ ID NO: 78; SEQ ID NO: 82; SEQ ID NO: 86; SEQ ID NO: 90; SEQ ID NO: 94;
SEQ ID NO: 98; SEQ ID NO: 102; SEQ ID NO: 106; SEQ ID NO: 110; and SEQ ID
NO: 114.
According to still another of the above objects, the present invention
provides an
expression vector including the cancer suppressor gene.
According to yet another of the above objects, the present invention provides
a
cell transformed with the expression vector.
BRIEF DESCRIPTION OF THE DRAWINGS
These and other features, aspects, and advantages of preferred embodiments of
the present invention will be more fully described in the following detailed
description,
taken accompanying drawings. In the drawings:
FIG. 1 is a gel diagram showing a PCR result using a random 5'-13-mer primer
H-AP33 of SEQ ID NO: 3 and an anchored oligo-dT primer of SEQ ID NO: 4;
FIG. 2 is a gel diagram showing a PCR result using a random 5'-13-mer primer
4
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
H-AP 10 of SEQ ID NO: 7 and an anchored oligo-dT primer of SEQ ID NO: 8;
FIG. 3 is a gel diagram showing a PCR result using a random 5'-13-mer primer
H-AP5 of SEQ ID NO: 11 and an anchored oligo-dT primer of SEQ ID NO: 12;
FIG. 4 is a gel diagram showing a PCR result using a random 5'-13-mer primer
H-AP2 of SEQ ID NO: 15 and an anchored oligo-dT primer of SEQ ID NO: 16;
FIG. 5 is a gel diagram showing a PCR result using a random 5'-13-mer primer
H-AP8 of SEQ ID NO: 19 and an anchored oligo-dT primer of SEQ ID NO: 20;
FIG. 6 is a gel diagram showing a PCR result using a random 5'-13-mer primer
H-AP7 of SEQ ID NO: 23 and an anchored oligo-dT primer of SEQ ID NO: 24;
FIG. 7 is a gel diagram showing a PCR result using a random 5'-13-mer primer
H-AP11 of SEQ ID NO: 27 and an anchored oligo-dT primer of SEQ ID NO: 28;
FIG. 8 is a gel diagram showing a PCR result using a random 5'-13-mer primer
H-AP3 of SEQ ID NO: 31 and an anchored oligo-dT primer of SEQ ID NO: 32;
FIG. 9 is a gel diagram showing a PCR result using a random 5'-13-mer primer
H-AP4 of SEQ ID NO: 35 and an anchored oligo-dT primer of SEQ ID NO: 36;
FIG. 10 is a gel diagram showing a PCR result using a random 5'-13-mer primer
H-AP8 of SEQ ID NO: 39 and an anchored oligo-dT primer of SEQ ID NO: 40;
FIG. 11 is a gel diagram showing a PCR result using a random 5'-13-mer primer
H-AP33 of SEQ ID NO: 43 and an anchored oligo-dT primer of SEQ ID NO: 44;
FIG. 12 is a gel diagram showing a PCR result using a random 5'-13-mer primer
H-AP35 of SEQ ID NO: 47 and an anchored oligo-dT primer of SEQ ID NO: 48;
FIG. 13 is a gel diagram showing a PCR result using a random 5'-13-mer primer
H-AP3 of SEQ ID NO: 51 and an anchored oligo-dT primer of SEQ ID NO: 52;
5
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
FIG. 14 is a gel diagram showing a PCR result using a random 5'-13-mer primer
H-AP12 of SEQ ID NO: 55 and an anchored oligo-dT primer of SEQ ID NO: 56;
FIG. 15 is a gel diagram showing a PCR result using a random 5'-13-mer primer
H-AP12 of SEQ ID NO: 59 and an anchored oligo-dT primer of SEQ ID NO: 60;
FIG. 16 is a gel diagram showing a PCR result using a random 5'-13-mer primer
H-AP7 of SEQ ID NO: 63 and an anchored oligo-dT primer of SEQ ID NO: 64;
FIG. 17 is a gel diagram showing a PCR result using a random 5'-13-mer primer
H-AP8 of SEQ ID NO: 67 and an anchored oligo-dT primer of SEQ ID NO: 68;
FIG. 18 is a gel diagram showing a PCR result using a random 5'-13-mer primer
H-AP 10 of SEQ ID NO: 71 and an anchored oligo-dT primer of SEQ ID NO: 72;
FIG. 19 is a gel diagram showing a PCR result using a random 5'-13-mer primer
H-AP16 of SEQ ID NO: 75 and an anchored oligo-dT primer of SEQ ID NO: 76;
FIG. 20 is a gel diagram showing a PCR result using a random 5'-13-mer primer
H-AP2 of SEQ ID NO: 79 and an anchored oligo-dT primer of SEQ ID NO: 80;
FIG. 21 is a gel diagram showing a PCR result using a random 5'-13-mer primer
H-AP9 of SEQ ID NO: 83 and an anchored oligo-dT primer of SEQ ID NO: 84;
FIG. 22 is a gel diagram showing a PCR result using a random 5'-13-mer primer
H-AP9 of SEQ ID NO: 87 and an anchored oligo-dT primer of SEQ ID NO: 88;
FIG. 23 is a gel diagram showing a PCR result using a random 5'-13-mer primer
H-AP32 of SEQ ID NO: 91 and an anchored oligo-dT primer of SEQ ID NO: 92;
FIG. 24 is a gel diagram showing a PCR result using a random 5'-13-mer primer
H-AP 10 of SEQ ID NO: 95 and an anchored oligo-dT primer of SEQ ID NO: 96;
FIG. 25 is a gel diagram showing a PCR result using a random 5'-13-mer primer
6
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
H-AP22 of SEQ ID NO: 99 and an anchored oligo-dT primer of SEQ ID NO: 100;
FIG. 26 is a gel diagram showing a PCR result using a random 5'-13-mer primer
H-AP12 of SEQ ID NO: 103 and an anchored oligo-dT primer of SEQ ID NO: 104;
FIG. 27 is a gel diagram showing a PCR result using a random 5'-13-mer primer
H-AP10 of SEQ ID NO: 107 and an anchored oligo-dT primer of SEQ ID NO: 108;
FIG. 28 is a gel diagram showing a PCR result using a random 5'-13-mer primer
H-AP 12 of SEQ ID NO: 111 and an anchored oligo-dT primer of SEQ ID NO: 112;
and
FIG. 29 is a gel diagram showing a PCR result using a random 5'-13-mer primer
H-AP4 of SEQ ID NO: 115 and an anchored oligo-dT primer of SEQ ID NO: 116.
FIGs. 30 to FIG. 58 are diagrams showing SDS-PAGE analysis results of a GIG8
gene product (FIG. 30); a GIG10 gene product (FIG. 31); a GIG13 gene product
(FIG.
32); a GIG15 gene product (FIG. 33); a GIG16 gene product (FIG. 34); a GIG24
gene
product (FIG. 35); a GIG26 gene product (FIG. 36); a GIG29 gene product (FIG.
37); a
GIG30 gene product (FIG. 38); a GIG32 gene product (FIG. 39); a GIG33 gene
product
(FIG. 40); a GIG34 gene product (FIG. 41); a GIG35 gene product (FIG. 42); a
GIG38
gene product (FIG. 43); a GIG39 gene product (FIG. 44); a GIG40 gene product
(FIG.
45); a GIG42 gene product (FIG. 46); a GIG43 gene product (FIG. 47); a GIG46
gene
product (FIG. 48); a PIG33 gene product (FIG. 49); a PIG35 gene product (FIG.
50); a
PIG36 gene product (FIG. 51); an MIG20 gene product (FIG. 52); a PIG49 gene
product
(FIG. 53); a PIG51 gene product (FIG. 54); an MIG12 gene product (FIG. 55); a
PIG37
gene product (FIG. 56); a GIG44 gene product (FIG. 57); and a GIG31 gene
product
(FIG. 58) of the present invention, respectively.
FIG. 59, FIG. 60, FIG. 61, FIG. 67, FIG. 68, FIG. 69, FIG. 70, FIG. 71, FIG.
72,
7
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
FIG. 73, FIG. 76, FIG. 82, FIG. 83, FIG. 86 and FIG. 87 are diagrams showing
northern
blotting results that the GIG8 gene; the GIG10 gene; the GIG13 gene; the GIG30
gene;
the GIG32 gene; the GIG33 gene; the GIG34 gene; the GIG35 gene; the GIG38
gene;
the GIG39 gene; the GIG43 gene; the PIG49 gene; the PIG51 gene; the GIG44
gene;
and the GIG31 gene are differentially expressed in a normal breast tissue, a
primary
breast cancer tissue and a breast cancer cell line, respectively;
FIG. 62 is a diagram showing a northern blotting result that the GIG15 gene of
the present invention is differentially expressed in a normal bone marrow
tissue, a
leukemic bone marrow tissue, and a K562 leukemia cell;
FIG. 63, FIG. 64, FIG. 65, FIG. 66, FIG. 74, FIG. 75, FIG. 78, FIG. 79, FIG.
80
and FIG. 85 are diagrams showing northern blotting results that the GIG16
gene; the
GIG24 gene; the GIG26 gene; the GIG29 gene; the GIG40 gene; the GIG42 gene;
the
PIG33 gene; the PIG35 gene; the PIG36 gene; and the PIG37 gene are
differentially
expressed in a normal liver tissue, a primary liver cancer tissue and a liver
cancer cell
line, respectively;
FIG. 77 and FIG. 81 are diagrams showing northern blotting results that the
GIG46 gene; and the MIG20 gene are differentially expressed in a normal
exocervical
tissue, a primary uterine cancer tissue and a uterine cancer cell line,
respectively;
FIG. 84 is a diagram showing a northern blotting result that the MIG12 gene of
the present invention is differentially expressed in a normal lung tissue, a
primary lung
cancer tissue, a metastatic lung cancer tissue and a lung cancer cell line,
and bottoms of
FIGs. 59 to 87 are diagrams showing northern blotting results obtained by
hybridizing
the same blots with j3 -actin probe, respectively.
8
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
FIGs. 88 to 116 are diagrams showing northern blotting results that the GIG8
gene (FIG. 88); the GIG10 gene (FIG. 89); the GIG13 gene (FIG. 90); the GIG15
gene
(FIG. 91); the GIG16 gene (FIG. 92); the GIG24 gene (FIG. 93); the GIG26 gene
(FIG.
94); the GIG29 gene (FIG. 95); the GIG30 gene (FIG. 96); the GIG32 gene (FIG.
97);
the GIG33 gene (FIG. 98); the GIG34 gene (FIG. 99); the GIG35 gene (FIG. 100);
the
GIG38 gene (FIG. 101); the GIG39 gene (FIG. 102); the GIG40 gene (FIG. 103);
the
GIG42 gene (FIG. 104); the GIG43 gene (FIG. 105); the GIG46 gene (FIG. 106);
the
PIG33 gene (FIG. 107); the PIG35 gene (FIG. 108); the PIG36 gene (FIG. 109);
the
MIG20 gene (FIG. 110); the PIG49 gene (FIG. 111); the PIG51 gene (FIG. 112);
the
MIG12 gene (FIG. 113); the PIG37 gene (FIG. 114); the GIG44 gene (FIG. 115);
and
the GIG31 gene (FIG. 116) are differentially expressed in various normal
tissues,
respectively, and bottoms of FIGs. 88 to 116 are diagrams showing northern
blotting
results obtained by hybridizing the same blots with j3 -actin probe,
respectively.
FIGs. 117 to 145 are diagrams showing northern blotting results that the GIG8
gene (FIG. 117); the GIG10 gene (FIG. 118); the GIG13 gene (FIG. 119); the
GIG15
gene (FIG. 120); the GIG16 gene (FIG. 121); the GIG24 gene (FIG. 122); the
GIG26
gene (FIG. 123); the GIG29 gene (FIG. 124); the GIG30 gene (FIG. 125); the
GIG32
gene (FIG. 126); the GIG33 gene (FIG. 127); the GIG34 gene (FIG. 128); the
GIG35
gene (FIG. 129); the GIG38 gene (FIG. 130); the GIG39 gene (FIG. 131); the
GIG40
gene (FIG. 132); the GIG42 gene (FIG. 133); the GIG43 gene (FIG. 134); the
GIG46
gene (FIG. 135); the PIG33 gene (FIG. 136); the PIG35 gene (FIG. 137); the
PIG36
gene (FIG. 138); the MIG20 gene (FIG. 139); the PIG49 gene (FIG. 140); the
PIG51
gene (FIG. 141); the MIG12 gene (FIG. 142); the PIG37 gene (FIG. 143); the
GIG44
9
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
gene (FIG. 144); and the GIG31 gene (FIG. 145) are differentially expressed in
various
cancer cell lines, respectively, and bottoms of FIGs. 117 to 145 are diagrams
showing
northern blotting results obtained by hybridizing the same blots with Ji -
actin probe,
respectively.
FIG. 146, FIG. 147, FIG. 148, FIG. 154, FIG. 155, FIG. 156, FIG. 157, FIG.
158,
FIG. 159, FIG. 160, FIG. 169, FIG. 170, FIG. 173 and FIG. 174 are diagrams
showing
growth curves of a wild-type MCF-7 cell; MCF-7 breast cancer cells transfected
with
the GIG8 gene; the GIG10 gene; the GIG13 gene; the GIG30 gene; GIG32 gene; the
GIG33 gene; the GIG34 gene; the GIG35 gene; the GIG38 gene; the GIG39 gene;
the
PIG49 gene; the PIG33 gene; the GIG44 gene; and the GIG31 gene, respectively;
and a
MCF-7 cell transfected with the expression vector pcDNA3.1, respectively;
FIG. 149 is a diagram showing growth curves of a wild-type K562 cell line; a
K562 leukemia cell transfected with the GIG15 gene; and a K562 cell
transfected with
the expression vector pcDNA3.1;
FIG. 150, FIG. 151, FIG. 152, FIG. 153, FIG. 161, FIG. 162, FIG. 163, FIG.
165,
FIG. 166, FIG. 167 and FIG. 172 are diagrams showing growth curves of a wild-
type
HepG2 liver cancer cell line; HepG2 liver cancer cells transfected with the
GIG16 gene;
the GIG24 gene; the GIG26 gene; the GIG29 gene; the GIG40 gene; the GIG42
gene;
the GIG43 gene; the PIG33 gene; the PIG35 gene; the PIG36 gene; and the PIG37
gene,
respectively; and a HepG2 cell transfected with the expression vector
pcDNA3.1;
FIG. 164 and FIG. 168 are diagrams showing growth curves of a wild-type HeLa
cell; HeLa uterine cancer cells transfected with the GIG46 gene; and the MIG20
gene,
respectively; and a HeLa cell transfected with the expression vector pcDNA3.1,
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
respectively; and
FIG. 171 is a diagram showing growth curves of a wild-type A549 lung cancer
cell line; an A549 lung cancer cell transfected with the MIG12 gene; and an
A549 cell
transfected with the expression vector pcDNA3.1.
BEST MODES FOR CARRYING OUT THE INVENTION
Hereinafter, preferred embodiments of the present invention will be described
in
detail with reference to the accompanying drawings.
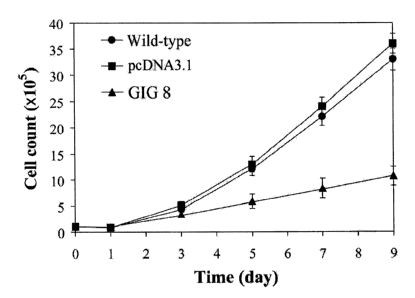
1. GIG8
The gene of the present invention is a human cancer suppressor gene 8 (GIG8)
having a DNA sequence of SEQ ID NO: 1, which was deposited with Accession No.
AY634687 into the GenBank database of U.S. National Institutes of Health (NIH)
(Scheduled Release Date: December 31, 2005), and a DNA sequence of the
deposited
gene is similar to that of the Homo sapiens inhibitor of DNA binding 2,
dominant
negative helix-loop-helix protein (ID2) gene deposited with Accession No.
NM002166
into the database. From this study result, it was however found that the GIG8
gene
was closely related to various human carcinogenesis. From the study result, it
was
found that the GIG8 tumor suppressor gene was rarely expressed or not
expressed in
various human tumors including the breast cancer, while its expression was
significantly
increased in various normal tissues.
The DNA sequence of SEQ ID NO: 1 has one open reading frame (ORF)
corresponding to base positions from 120 to 524 of the DNA sequence (base
positions
from 522 to 524 represent a stop codon).
11
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
A protein expressed from the gene of the present invention consists of 134
amino acid residues, and has an amino acid sequence of SEQ ID NO: 2 and a
molecular
weight of approximately 15 kDa.
The gene and the protein of the present invention may be separated from human
tissues, or also be synthesized according to the known methods for
synthesizing DNA or
peptide. For example, the gene of the present invention may be screened and
cloned
according to the conventional methods on the basis of the information on the
DNA
sequence set forth in SEQ ID NO: 1. As another example, a 163-bp cDNA
fragment,
which is not expressed or rarely expressed in the cancer tissue or the cancer
cell line but
differentially expressed in the normal tissue, may be obtained by carrying out
a reverse
transcription-polymerase chain reaction (RT-PCR) on the total RNA extracted
from a
normal tissue, and a cancer tissue or a cancer cell line using a random primer
H-AP33 of
SEQ ID NO: 3 (5'-AAGCTTGCTGCTC-3') and an anchored oligo-dT primer of SEQ
ID NO: 4 (5'-AAGCTTTTTTTTTTTA-3'), and the resultant fragment, which is used
as
the probe, may be plaque-hybridized with a cDNA library to obtain a full-
length cDNA
clone.
It is regarded that the gene of the present invention is overexpressed in the
normal tissues, preferably the breast, the brain, the heart, the muscles, the
large intestine,
the thymus, the spleen, the kidney, the liver, the small intestine, the
placenta and the
lungs to suppress the carcinogenesis. The gene of the present invention is
mainly
overexpressed in these tissues as an mRNA transcript having a size of
approximately 1.3
kb.
2. GIG10
12
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
The gene of the present invention is a human cancer suppressor gene 10 (GIG10)
having a DNA sequence of SEQ ID NO: 5, which was deposited with Accession No.
AY542305 into the GenBank database of U.S. National Institutes of Health (NIH)
(Scheduled Release Date: December 31, 2005), and a DNA sequence of the
deposited
gene is similar to that of the Homo sapiens c-Cbl-interacting protein (CIN85)
mRNA
gene deposited with Accession No. AF230904 into the database.
From this study result, it was however found that the GIG10 gene was closely
related to various human carcinogenesis. From the study result, it was found
that the
GIG10 tumor suppressor gene was rarely expressed or not expressed in various
human
tumors including the breast cancer, while its expression was significantly
increased in
various normal tissues.
The DNA sequence of SEQ ID NO: 5 has one open reading frame (ORF)
corresponding to base positions from 52 to 2,049 of the DNA sequence (base
positions
from 2,047 to 2,049 represent a stop codon).
A protein expressed from the gene of the present invention consists of 665
amino acid residues, and has an amino acid sequence of SEQ ID NO: 6 and a
molecular
weight of approximately 73 kDa.
The gene and the protein of the present invention may be separated from human
tissues, or also be synthesized according to the known methods for
synthesizing DNA or
peptide. For example, the gene of the present invention may be screened and
cloned
according to the conventional methods on the basis of the information on the
DNA
sequence set forth in SEQ ID NO: 5. As another example, a 321-bp cDNA
fragment,
which is not expressed or rarely expressed in the cancer tissue or the cancer
cell line but
13
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
differentially expressed in the normal tissue, may be obtained by carrying out
a reverse
transcription-polymerase chain reaction (RT-PCR) on the total RNA extracted
from a
normal tissue, and a cancer tissue or a cancer cell line using a random primer
H-AP 10 of
SEQ ID NO: 7 (5'-AAGCTTCCACGTA-3') and an anchored oligo-dT primer of SEQ
ID NO: 8 (5'-AAGCTTTTTTTTTTTC-3'), and the resultant fragment, which is used
as
the probe, may be plaque-hybridized with a cDNA library to obtain a full-
length cDNA
clone.
It is regarded that the gene of the present invention is overexpressed in the
normal tissues, preferably the breast, the brain, the heart, the muscles, the
thymus, the
spleen, the kidney, the liver, the small intestine, the placenta and the lungs
to suppress
the carcinogenesis. The gene of the present invention is mainly overexpressed
in these
tissues as an mRNA transcript having a size of approximately 3.5 kb.
3. GIG13
The gene of the present invention is a human cancer suppressor gene 13 (GIG13)
having a DNA sequence of SEQ ID NO: 9, which was deposited with Accession No.
AY493418 into the GenBank database of U.S. National Institutes of Health (NIH)
(Scheduled Release Date: December 31, 2005), and a DNA sequence of the
deposited
gene is similar to that of the Homo sapiens period homolog 3 (Drosophila)
(PER3) gene
deposited with Accession No. NM_016831 into the database.
From this study result, it was however found that the GIG13 gene was closely
related to various human carcinogenesis. From the study result, it was found
that the
GIG13 tumor suppressor gene was rarely expressed or not expressed in various
human
tumors including the breast cancer, while its expression was significantly
increased in
14
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
various normal tissues. The DNA sequence of SEQ ID NO: 9 has one open reading
frame (ORF) corresponding to base positions from 72 to 3,677 of the DNA
sequence
(base positions from 3,675 to 3,677 represent a stop codon).
A protein expressed from the gene of the present invention consists of 1,201
amino acid residues, and has an amino acid sequence of SEQ ID NO: 10 and a
molecular weight of approximately 132 kDa.
The gene and the protein of the present invention may be separated from human
tissues, or also be synthesized according to the known methods for
synthesizing DNA or
peptide. For example, the gene of the present invention may be screened and
cloned
according to the conventional methods on the basis of the information on the
DNA
sequence set forth in SEQ ID NO: 1. As another example, a 347-bp cDNA
fragment,
which is not expressed or rarely expressed in the cancer tissue or the cancer
cell line but
differentially expressed in the normal tissue, may be obtained by carrying out
a reverse
transcription-polymerase chain reaction (RT-PCR) on the total RNA extracted
from a
normal tissue, and a cancer tissue or a cancer cell line using a random primer
H-AP5 of
SEQ ID NO: 11 (5'-AAGCTTAGTAGGC-3') and an anchored oligo-dT primer of SEQ
ID NO: 12 (5'-AAGCTTTTTTTTTTTC-3'), and the resultant fragment, which is used
as
the probe, may be plaque-hybridized with a cDNA library to obtain a full-
length cDNA
clone.
It is regarded that the gene of the present invention is overexpressed in the
normal tissues, preferably the breast and the liver to suppress the
carcinogenesis. The
gene of the present invention is mainly overexpressed in these tissues as an
mRNA
transcript having a size of approximately 1.3 kb. Especially, the gene of the
present
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
invention is differentially expressed only in the normal tissues. For example,
the gene
of the present invention is rarely expressed or not expressed in the cancer
tissues and the
cancer cells such as the breast cancer tissue, and the breast cancer cell line
MCF-7, but
differentially increasingly expressed only in the normal breast tissues.
4. GIG15
The gene of the present invention is a human cancer suppressor gene 15 (GIG15)
having a DNA sequence of SEQ ID NO: 13, which was deposited with Accession No.
AY927233 into the GenBank database of U.S. National Institutes of Health (NIH)
(Scheduled Release Date: October 1, 2006), and a DNA sequence of the deposited
gene
is similar to that of the Homo sapiens chromosome 11, clone RP11-466H18 gene
deposited with Accession No. AC116533 into the database. From this study
result, it
was however found that the GIG15 gene was closely related to various human
carcinogenesis. From the study result, it was found that the GIG15 tumor
suppressor
gene was rarely expressed or not expressed in various human tumors including
the
leukemia, while its expression was significantly increased in various normal
tissues.
The DNA sequence of SEQ ID NO: 13 has one open reading frame (ORF)
corresponding to base positions from 18 to 338 of the DNA sequence (base
positions
from 336 to 338 represent a stop codon).
A protein expressed from the gene of the present invention consists of 106
amino acid residues, and has an amino acid sequence of SEQ ID NO: 14 and a
molecular weight of approximately 12 kDa.
The gene and the protein of the present invention may be separated from human
tissues, or also be synthesized according to the known methods for
synthesizing DNA or
16
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
peptide. For example, the gene of the present invention may be screened and
cloned
according to the conventional methods on the basis of the information on the
DNA
sequence set forth in SEQ ID NO: 13. As another example, a 133-bp cDNA
fragment,
which is rarely expressed in the cancer tissue or the cancer cell line but
differentially
expressed in the normal tissue, may be obtained by carrying out a reverse
transcription-polymerase chain reaction (RT-PCR) on the total RNA extracted
from a
normal tissue, and a cancer tissue or a cancer cell line using a random primer
H-AP2 of
SEQ ID NO: 15 (5'-AAGCTTCGACTGT-3') and an anchored oligo-dT primer of SEQ
ID NO: 16 (5'-AAGCTTTTTTTTTTTC-3'), and the resultant fragment, which is used
as
the probe, may be plaque-hybridized with a cDNA library to obtain a full-
length cDNA
clone.
It is regarded that the gene of the present invention is overexpressed in the
normal tissues, preferably the breast, the brain, the heart, the muscles, the
large intestine,
the thymus, the spleen, the kidney, the liver, the small intestine, the
placenta and the
lungs to suppress the carcinogenesis. The gene of the present invention is
mainly
overexpressed in these tissues as an mRNA transcript having a size of
approximately 0.5
kb. Especially, the gene of the present invention is differentially expressed
only in the
normal tissues. For example, the gene of the present invention is rarely
expressed in
the cancer tissues and the cancer cells such as the leukemia cell and the
leukemia cell
line K562, but differentially increasingly expressed only in the normal breast
tissues.
5. GIG16
The gene of the present invention is a human cancer suppressor gene 16 (GIG16)
having a DNA sequence of SEQ ID NO: 17, which was deposited with Accession No.
17
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
AY513277 into the GenBank database of U.S. National Institutes of Health (NIH)
(Scheduled Release Date: December 31, 2005), and some DNA sequence of the
deposited gene is different from that of the Homo sapiens hydroxyacid oxidase
2 (long
chain) (HAO2) gene deposited with Accession No. NM_016527 into the database,
the
HAO2 gene being known to be one of three genes having 2-hydroxyacid oxidase
activity (Jones, J.M., et al., J. Biol. Chem. 275(17), 12590-12597 (2000)).
From this
study result, it was however found that the GIG16 tumor suppressor gene was
not
expressed at all in various human tumors including the liver cancer, while its
expression
was significantly increased in various normal tissues.
The DNA sequence of SEQ ID NO: 17 has one open reading frame (ORF)
corresponding to base positions from 41 to 1,096 of the DNA sequence (base
positions
from 1,094 to 1,096 represent a stop codon).
A protein expressed from the gene of the present invention consists of 351
amino acid residues, and has an amino acid sequence of SEQ ID NO: 18 and a
molecular weight of approximately 39 kDa.
The gene and the protein of the present invention may be separated from human
tissues, or also be synthesized according to the known methods for
synthesizing DNA or
peptide. For example, the gene of the present invention may be screened and
cloned
according to the conventional methods on the basis of the information on the
DNA
sequence set forth in SEQ ID NO: 17. As another example, a 213-bp cDNA
fragment,
which is not expressed in the cancer tissue or the cancer cell line but
differentially
expressed in the normal tissue, may be obtained by carrying out a reverse
transcription-polymerase chain reaction (RT-PCR) on the total RNA extracted
from a
18
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
normal tissue, and a cancer tissue or a cancer cell line using a random primer
H-AP8 of
SEQ ID NO: 19 (5'-AAGCTTTTACCGC-3') and an anchored oligo-dT primer of SEQ
ID NO: 20 (5'-AAGCTTTTTTTTTTTC-3'), and the resultant fragment, which is used
as
the probe, may be plaque-hybridized with a cDNA library to obtain a full-
length cDNA
clone. The present inventors inserted the full-length GIG16 cDNA into the
expression
vector pBAD/Thio-Topo (Invitrogen, U.A.), and then transformed E. coli DH5 a
with
the resultant expression vector to obtain a transformant, which was designated
E. coli
DH5 a /GIG 16/pBAD/Thio-Topo.
It is regarded that the gene of the present invention is overexpressed in the
normal tissues, preferably the liver and the kidney to suppress the
carcinogenesis. Also,
it is regarded that its gene expression is suppressed in the leukemia, the
uterine cancer,
the malignant lymphoma, the colon cancer, the lung cancer and the skin cancer
to induce
the carcinogenesis. The gene of the present invention is mainly overexpressed
in these
tissues as an mRNA transcript having a size of approximately 2.0 kb.
Especially, the
gene of the present invention is differentially expressed only in the normal
tissues. For
example, the gene of the present invention is not expressed in the cancer
tissues and the
cancer cells such as the liver cancer tissue, and the liver cancer cell line
HepG2, but
differentially expressed only in the normal breast tissues.
6. GIG24
The gene of the present invention is a human cancer suppressor gene 24 (GIG24)
having a DNA sequence of SEQ ID NO: 21, which was deposited with Accession No.
AY513275 into the GenBank database of U.S. National Institutes of Health (NIH)
(Scheduled Release Date: December 31, 2005), and a DNA sequence of the
deposited
19
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
gene is similar to that of the Homo sapiens cDNA FLJ35730 fis, clone
TESTI2003131,
highly similar to ALPHA- I -ANTICHYMOTRYPSIN PRECURSOR gene deposited
with Accession No. AK093049 into the database. From this study result, it was
however found that the GIG24 tumor suppressor gene was not expressed at all in
various human tumors including the liver cancer, while its expression was
significantly
increased in the normal liver tissue.
The DNA sequence of SEQ ID NO: 21 has one open reading frame (ORF)
corresponding to base positions from 34 to 1,305 of the DNA sequence (base
positions
from 1,303 to 1,305 represent a stop codon).
A protein expressed from the gene of the present invention consists of 423
amino acid residues, and has an amino acid sequence of SEQ ID NO: 22 and a
molecular weight of approximately 47 kDa.
The gene and the protein of the present invention may be separated from human
tissues, or also be synthesized according to the known methods for
synthesizing DNA or
peptide. For example, the gene of the present invention may be screened and
cloned
according to the conventional methods on the basis of the information on the
DNA
sequence set forth in SEQ ID NO: 1. As another example, a 221-bp cDNA
fragment,
which is not expressed in the cancer tissue or the cancer cell line but
differentially
expressed in the normal tissue, may be obtained by carrying out a reverse
transcription-polymerase chain reaction (RT-PCR) on the total RNA extracted
from a
normal tissue, and a cancer tissue or a cancer cell line using a random primer
H-AP7 of
SEQ ID NO: 23 (5'-AAGCTTAACGAGG-3') and an anchored oligo-dT primer of SEQ
ID NO: 24 (5'-AAGCTTTTTTTTTTTC-3'), and the resultant fragment, which is used
as
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
the probe, may be plaque-hybridized with a cDNA library to obtain a full-
length cDNA
clone. The present inventors inserted the full-length GIG24 cDNA into the
expression
vector pBAD/Thio-Topo (Invitrogen, U.A.), and then transformed E. coli DH5 a
with
the resultant expression vector to obtain a transformant, which was designated
E. coli
DH5 a /GIG24/pBAD/Thio-Topo.
It is regarded that the gene of the present invention is overexpressed in the
normal tissues, preferably the liver, the heart and the muscles to suppress
the
carcinogenesis. Also, it is regarded that its gene expression is suppressed in
the
leukemia, the uterine cancer, the malignant lymphoma, the colon cancer, the
lung cancer
and the skin cancer to induce the carcinogenesis. The gene of the present
invention is
mainly overexpressed in these tissues as an mRNA transcript having a size of
approximately 2.4 kb.
7. GIG26
The gene of the present invention is a human cancer suppressor gene 26 (GIG26)
having a DNA sequence of SEQ ID NO: 25, which was deposited with Accession No.
AY544126 into the GenBank database of U.S. National Institutes of Health (NIH)
(Scheduled Release Date: December 31, 2005), and a DNA sequence of the
deposited
gene is similar to those of the Homo sapiens prostate-specific membrane
antigen-like
(PSMAL) gene and the Homo sapiens prostate-specific membrane antigen-like
protein
(PSMAL/GCP 111) mRNA gene, deposited with Accession No. NM_153696 and
AF261715 into the database, respectively, the prostate-specific membrane
antigen being
known to be a marker protein that is mainly expressed in a prostate epithelium
((Lee,
S.J., et al., J. Mol. Biol. 330(4), 749-760 (2003)). From this study result,
it was
21
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
however found that the GIG26 tumor suppressor gene was not expressed at all in
various human tumors including the liver cancer, while its expression was
significantly
increased in various normal tissues.
The DNA sequence of SEQ ID NO: 25 has one open reading frame (ORF)
corresponding to base positions from 26 to 1,354 of the DNA sequence (base
positions
from 1,352 to 1,354 represent a stop codon).
A protein expressed from the gene of the present invention consists of 442
amino acid residues, and has an amino acid sequence of SEQ ID NO: 26 and a
molecular weight of approximately 50 kDa.
The gene and the protein of the present invention may be separated from human
tissues, or also be synthesized according to the known methods for
synthesizing DNA or
peptide. For example, the gene of the present invention may be screened and
cloned
according to the conventional methods on the basis of the information on the
DNA
sequence set forth in SEQ ID NO: 25. As another example, a 204-bp cDNA
fragment,
which is not expressed in the cancer tissue or the cancer cell line but
differentially
expressed in the normal tissue, may be obtained by carrying out a reverse
transcription-polymerase chain reaction (RT-PCR) on the total RNA extracted
from a
normal tissue, and a cancer tissue or a cancer cell line using a random primer
H-AP 11 of
SEQ ID NO: 27 (5'-AAGCTTCGGGTAA-3') and an anchored oligo-dT primer of SEQ
ID NO: 28 (5'-AAGCTTTTTTTTTTTG-3'), and the resultant fragment, which is used
as
the probe, may be plaque-hybridized with a cDNA library to obtain a full-
length cDNA
clone.
The present inventors inserted the full-length GIG26 cDNA into the expression
22
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
vector pBAD/Thio-Topo (Invitrogen, U.A.), and then transformed E. coli DH5 a
with
the resultant expression vector to obtain a transformant, which was designated
E. coli
DH5 a /GIG26/pBAD/Thio-Topo.
It is regarded that the gene of the present invention is overexpressed in the
normal tissues, preferably the liver, the kidney, the brain and the heart to
suppress the
carcinogenesis. Also, it is regarded that its gene expression is suppressed in
leukemia,
uterine cancer, malignant lymphoma, colon cancer, lung cancer and skin cancer
to
induce the carcinogenesis. The gene of the present invention is mainly
overexpressed
in these tissues as an mRNA transcript having a size of approximately 2.0 kb.
8. GIG29
The gene of the present invention is a human cancer suppressor gene 29 (GIG29)
having a DNA sequence of SEQ ID NO: 29, which was deposited with Accession No.
AY544127 into the GenBank database of U.S. National Institutes of Health (NIH)
(Scheduled Release Date: December 31, 2005), and a DNA sequence of the
deposited
gene is similar to that of the Homo sapiens solute carrier family 10
(sodium/bile acid
cotransporter family) gene deposited with Accession No. NM_003049 into the
database.
From this study result, it was however found that the GIG29 tumor suppressor
gene
was not expressed at all in various human tumors including the liver cancer,
while its
expression was significantly increased in the normal liver tissue.
The DNA sequence of SEQ ID NO: 29 has one open reading frame (ORF)
corresponding to base positions from 62 to 1,111 of the DNA sequence (base
positions
from 1,109 to 1,111 represent a stop codon).
A protein expressed from the gene of the present invention consists of 349
23
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
amino acid residues, and has an amino acid sequence of SEQ ID NO: 30 and a
molecular weight of approximately 38 kDa.
The gene and the protein of the present invention may be separated from human
tissues, or also be synthesized according to the known methods for
synthesizing DNA or
peptide. For example, the gene of the present invention may be screened and
cloned
according to the conventional methods on the basis of the information on the
DNA
sequence set forth in SEQ ID NO: 29. As another example, a 277-bp cDNA
fragment,
which is not expressed in the cancer tissue or the cancer cell line but
differentially
expressed in the normal tissue, may be obtained by carrying out a reverse
transcription-polymerase chain reaction (RT-PCR) on the total RNA extracted
from a
normal tissue, and a cancer tissue or a cancer cell line using a random primer
H-AP3 of
SEQ ID NO: 31 (5'-AAGCTTTGGTCAG-3') and an anchored oligo-dT primer of SEQ
ID NO: 32 (5'-AAGCTTTTTTTTTTTA-3'), and the resultant fragment, which is used
as
the probe, may be plaque-hybridized with a cDNA library to obtain a full-
length cDNA
clone. The present inventors inserted the full-length GIG29 cDNA into the
expression
vector pBAD/Thio-Topo (Invitrogen, U.A.), and then transformed E. coli DH5 a
with
the resultant expression vector to obtain a transformant, which was designated
E. coli
DH5 a /GIG29/pBAD/Thio-Topo.
It is regarded that the gene of the present invention is overexpressed in the
normal tissues, preferably the liver to suppress the carcinogenesis. Also, it
is regarded
that its gene expression is suppressed in the leukemia, the uterine cancer,
the malignant
lymphoma, the colon cancer, the lung cancer and the skin cancer to induce the
carcinogenesis. The gene of the present invention is mainly overexpressed in
these
24
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
tissues as an mRNA transcript having a size of approximately 1.4 kb.
9. GIG30
The gene of the present invention is a human cancer suppressor gene 30 (GIG30)
having a DNA sequence of SEQ ID NO: 33, which was deposited with Accession No.
AY524045 into the GenBank database of U.S. National Institutes of Health (NIH)
(Scheduled Release Date: December 31,, 2005), and a DNA sequence of the
deposited
gene is similar to that of the Homo sapiens clone DNA43305 RIPK2 (UNQ277) gene
deposited with Accession No. AY358814 into the database. From this study
result, it
was however found that the GIG30 gene was closely related to various human
carcinogenesis. From the study result, it was found that the GIG30 tumor
suppressor
gene was rarely expressed or not expressed in various human tumors including
the
breast cancer, while its expression was significantly increased in various
normal tissues.
The DNA sequence of SEQ ID NO: 33 has one open reading frame (ORF)
corresponding to base positions from 88 to 1,710 of the DNA sequence (base
positions
from 1,708 to 1,710 represent a stop codon).
A protein expressed from the gene of the present invention consists of 540
amino acid residues, and has an amino acid sequence of SEQ ID NO: 34 and a
molecular weight of approximately 61 kDa.
The gene and the protein of the present invention may be separated from human
tissues, or also be synthesized according to the known methods for
synthesizing DNA or
peptide. For example, the gene of the present invention may be screened and
cloned
according to the conventional methods on the basis of the information on the
DNA
sequence set forth in SEQ ID NO: 35. As another example, a 278-bp cDNA
fragment,
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
which is not expressed or rarely expressed in the cancer tissue or the cancer
cell line but
differentially expressed in the normal tissue, may be obtained by carrying out
a reverse
transcription-polymerase chain reaction (RT-PCR) on the total RNA extracted
from a
normal tissue, and a cancer tissue or a cancer cell line using a random primer
H-AP4 of
SEQ ID NO: 35 (5'-AAGCTTCTCAACG-3') and an anchored oligo-dT primer of SEQ
ID NO: 36 (5'-AAGCTTTTTTTTTTTG-3'), and the resultant fragment, which is used
as
the probe, may be plaque-hybridized with a cDNA library to obtain a full-
length cDNA
clone.
It is regarded that the gene of the present invention is overexpressed in the
normal tissues, preferably the breast, the heart, the muscles and the liver to
suppress the
carcinogenesis. The gene of the present invention is mainly overexpressed in
these
tissues as an mRNA transcript having a size of approximately 1.9 kb.
10. GIG32
The gene of the present invention is a human cancer suppressor gene 32 (GIG32)
having a DNA sequence of SEQ ID NO: 37, which was deposited with Accession No.
AY762103 into the GenBank database of U.S. National Institutes of Health (NIH)
(Scheduled Release Date: December 31, 2005), and a DNA sequence of the
deposited
gene is similar to that of the Homo sapiens caveolin 1, caveolae protein, 22
kDa
(CAV1) gene deposited with Accession No. NM001753 into the database. From this
study result, it was however found that the GIG32 gene was closely related to
various
human carcinogenesis. From the study result, it was found that the GIG32 tumor
suppressor gene was rarely expressed or not expressed in various human tumors
including the breast cancer, while its expression was significantly increased
in various
26
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
normal tissues.
The DNA sequence of SEQ ID NO: 37 has one open reading frame (ORF)
corresponding to base positions from 43 to 579 of the DNA sequence (base
positions
from 577 to 579 represent a stop codon).
A protein expressed from the gene of the present invention consists of 178
amino acid residues, and has an amino acid sequence of SEQ ID NO: 38 and a
molecular weight of approximately 20 kDa.
The gene and the protein of the present invention may be separated from human
tissues, or also be synthesized according to the known methods for
synthesizing DNA or
peptide. For example, the gene of the present invention may be screened and
cloned
according to the conventional methods on the basis of the information on the
DNA
sequence set forth in SEQ ID NO: 37. As another example, a 172-bp cDNA
fragment,
which is not expressed or rarely expressed in the cancer tissue or the cancer
cell line but
differentially expressed in the normal tissue, may be obtained by carrying out
a reverse
transcription-polymerase chain reaction (RT-PCR) on the total RNA extracted
from a
normal tissue, and a cancer tissue or a cancer cell line using a random primer
H-AP8 of
SEQ ID NO: 39 (5'-AAGCTTTTACCGC-3') and an anchored oligo-dT primer of SEQ
ID NO: 40 (5'-AAGCTTTTTTTTTTTG-3'), and the resultant fragment, which is used
as
the probe, may be plaque-hybridized with a cDNA library to obtain a full-
length cDNA
clone.
It is regarded that the gene of the present invention is overexpressed in the
normal tissues, preferably the breast, the brain, the heart, the muscles, the
large intestine,
the thymus, the spleen, the kidney, the liver, the small intestine, the
placenta and the
27
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
lungs to suppress the carcinogenesis. The gene of the present invention is
mainly
overexpressed in these tissues as an mRNA transcript having a size of
approximately 4.0
kb.
11. GIG3 3
The gene of the present invention is a human cancer suppressor gene 33 (GIG33)
having a DNA sequence of SEQ ID NO: 41, which was deposited with Accession No.
AY871273 into the GenBank database of U.S. National Institutes of Health (NIH)
(Scheduled Release Date: October 1, 2006), and a DNA sequence of the deposited
gene
is similar to that of the Homo sapiens ribosomal protein L35a (RPL35A) gene
deposited
with Accession No. NM 000996 into the database. A ribosomal gene is an
intracellular organelle that catalyzes the protein synthesis and consists of a
small 40S
subunit and a large 60S subunit. However, a function of the gene remains to be
specified (Herzog, H., et al., Nucleic Acids Res., 18(15), 4600 (1990);
Kenmochi, N., et
al., Genome Res., 8(5), 509-523 (1998); Lopez, C.D., et al., Cancer Lett.
180(2),
195-202 (2002)). From this study result, it was however found that the GIG33
gene
was closely related to various human carcinogenesis. From the study result, it
was
found that the GIG33 tumor suppressor gene was rarely expressed or not
expressed in
various human tumors including the breast cancer, while its expression was
significantly
increased in various normal tissues.
The DNA sequence of SEQ ID NO: 41 has one open reading frame (ORF)
corresponding to base positions from 74 to 406 of the DNA sequence (base
positions
from 404 to 406 represent a stop codon).
A protein expressed from the gene of the present invention consists of 110
28
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
amino acid residues, and has an amino acid sequence of SEQ ID NO: 42 and a
molecular weight of approximately 12 kDa.
The gene and the protein of the present invention may be separated from human
tissues, or also be synthesized according to the known methods for
synthesizing DNA or
peptide. For example, the gene of the present invention may be screened and
cloned
according to the conventional methods on the basis of the information on the
DNA
sequence set forth in SEQ ID NO: 41. As another example, a 182-bp cDNA
fragment,
which is not expressed or rarely expressed in the cancer tissue or the cancer
cell line but
differentially expressed in the normal tissue, may be obtained by carrying out
a reverse
transcription-polymerase chain reaction (RT-PCR) on the total RNA extracted
from a
normal tissue, and a cancer tissue or a cancer cell line using a random primer
H-AP33 of
SEQ ID NO: 43 (5'-AAGCTTGCTGCTC-3') and an anchored oligo-dT primer of SEQ
ID NO: 44 (5'-AAGCTTTTTTTTTTTG-3'), and the resultant fragment, which is used
as
the probe, may be plaque-hybridized with a cDNA library to obtain a full-
length cDNA
clone.
It is regarded that the gene of the present invention is overexpressed in the
normal tissues, preferably the breast, the brain, the heart, the muscles, the
large intestine,
the thymus, the spleen, the kidney, the liver, the small intestine, the
placenta, the lungs
and the peripheral blood leukocyte to suppress the carcinogenesis. The gene of
the
present invention is mainly overexpressed in these tissues as an mRNA
transcript having
a size of approximately 0.6 kb.
12. GIG34
The gene of the present invention is a human cancer suppressor gene 34 (GIG34)
29
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
having a DNA sequence of SEQ ID NO: 45, which was deposited with Accession No.
AY871274 into the GenBank database of U.S. National Institutes of Health (NIH)
(Scheduled Release Date: October 1, 2006), and a DNA sequence of the deposited
gene
is similar to that of the Homo sapiens ribosomal protein L11 gene deposited
with
Accession No. BC018970 into the database. From this study result, it was
however
found that the GIG34 gene was closely related to various human carcinogenesis.
From
the study result, it was found that the GIG34 tumor suppressor gene was rarely
expressed or not expressed in various human tumors including the breast
cancer, while
its expression was significantly increased in various normal tissues.
The DNA sequence of SEQ ID NO: 45 has one open reading frame (ORF)
corresponding to base positions from 5 to 538 of the DNA sequence (base
positions
from 536 to 538 represent a stop codon).
A protein expressed from the gene of the present invention consists of 177
amino acid residues, and has an amino acid sequence of SEQ ID NO: 46 and a
molecular weight of approximately 20 kDa.
The gene and the protein of the present invention may be separated from human
tissues, or also be synthesized according to the known methods for
synthesizing DNA or
peptide. For example, the gene of the present invention may be screened and
cloned
according to the conventional methods on the basis of the information on the
DNA
sequence set forth in SEQ ID NO: 45. As another example, a 205-bp cDNA
fragment,
which is not expressed or rarely expressed in the cancer tissue or the cancer
cell line but
differentially expressed in the normal tissue, may be obtained by carrying out
a reverse
transcription-polymerase chain reaction (RT-PCR) on the total RNA extracted
from a
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
normal tissue, and a cancer tissue or a cancer cell line using a random primer
H-AP35 of
SEQ ID NO: 47 (5'-AAGCTTCAGGGCA-3') and an anchored oligo-dT primer of SEQ
ID NO: 48 (5'-AAGCTTTTTTTTTTTC-3'), and the resultant fragment, which is used
as
the probe, may be plaque-hybridized with a cDNA library to obtain a full-
length cDNA
clone.
It is regarded that the gene of the present invention is overexpressed in the
normal tissues, preferably the breast, the brain, the heart, the muscles, the
large intestine,
the thymus, the spleen, the kidney, the liver, the small intestine, the
placenta, the lungs
and the peripheral blood leukocyte to suppress the carcinogenesis. The gene of
the
present invention is mainly overexpressed in these tissues as an mRNA
transcript having
a size of approximately 0.6 kb.
13. GIG35
The gene of the present invention is a human cancer suppressor gene 35 (GIG35)
having a DNA sequence of SEQ ID NO: 49, which was deposited with Accession No.
AY542307 into the GenBank database of U.S. National Institutes of Health (NIH)
(Scheduled Release Date: December 31, 2005), and a DNA sequence of the
deposited
gene is similar to that of the Homo sapiens eukaryotic translation elongation
factor 1
gamma (EEF 1 G) gene deposited with Accession No. NM_001404 into the database.
The gene is a subunit of the elongation factor 1 that takes an important role
in
transferring aminoacyl tRNAs to ribosome ((Kumabe, T., et al., Nucleic Acids
Res.,
20(10), 2598 (1992)). From this study result, it was however found that the
GIG35
gene was closely related to various human carcinogenesis. From the study
result, it
was found that the GIG35 tumor suppressor gene was rarely expressed in various
human
31
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
tumors including the breast cancer, while its expression was significantly
increased in
various normal tissues.
The DNA sequence of SEQ ID NO: 49 has one open reading frame (ORF)
corresponding to base positions from 19 to 1,332 of the DNA sequence (base
positions
from 1,330 to 1,332 represent a stop codon).
A protein expressed from the gene of the present invention consists of 437
amino acid residues, and has an amino acid sequence of SEQ ID NO: 50 and a
molecular weight of approximately 50 kDa.
The gene and the protein of the present invention may be separated from human
tissues, or also be synthesized according to the known methods for
synthesizing DNA or
peptide. For example, the gene of the present invention may be screened and
cloned
according to the conventional methods on the basis of the information on the
DNA
sequence set forth in SEQ ID NO: 49. As another example, a 212-bp cDNA
fragment,
which is not expressed or rarely expressed in the cancer tissue or the cancer
cell line but
differentially expressed in the normal tissue, may be obtained by carrying out
a reverse
transcription-polymerase chain reaction (RT-PCR) on the total RNA extracted
from a
normal tissue, and a cancer tissue or a cancer cell line using a random primer
H-AP3 of
SEQ ID NO: 51 (5'-AAGCTTTGGTCAG-3') and an anchored oligo-dT primer of SEQ
ID NO: 52 (5'-AAGCTTTTTTTTTTTC-3'), and the resultant fragment, which is used
as
the probe, may be plaque-hybridized with a cDNA library to obtain a full-
length cDNA
clone.
It is regarded that the gene of the present invention is overexpressed in the
normal tissues, preferably the breast, the brain, the heart, the muscles, the
large intestine,
32
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
the thymus, the spleen, the kidney, the liver, the small intestine, the
placenta and the
lungs to suppress the carcinogenesis. The gene of the present invention is
mainly
overexpressed in these tissues as an mRNA transcript having a size of
approximately 1.3
kb.
14. GIG38
The gene of the present invention is a human cancer suppressor gene 38 (GIG38)
having a DNA sequence of SEQ ID NO: 53, which was deposited with Accession No.
AY550970 into the GenBank database of U.S. National Institutes of Health (NIH)
(Scheduled Release Date: December 31, 2005), and a DNA sequence of the
deposited
gene is similar to that of the Homo sapiens sin3-associated polypeptide, 18
kDa
(SAP18) gene deposited with Accession No. NM_005870 into the database. From
this
study result, it was however found that the GIG38 gene was closely related to
various
human carcinogenesis. From the study result, it was found that the GIG38 tumor
suppressor gene was rarely expressed or not expressed in various human tumors
including the breast cancer, while its expression was significantly increased
in various
normal tissues.
The DNA sequence of SEQ ID NO: 53 has one open reading frame (ORF)
corresponding to base positions from 17 to 478 of the DNA sequence (base
positions
from 476 to 478 represent a stop codon).
A protein expressed from the gene of the present invention consists of 153
amino acid residues, and has an amino acid sequence of SEQ ID NO: 54 and a
molecular weight of approximately 17 kDa.
The gene and the protein of the present invention may be separated from human
33
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
tissues, or also be synthesized according to the known methods for
synthesizing DNA or
peptide. For example, the gene of the present invention may be screened and
cloned
according to the conventional methods on the basis of the information on the
DNA
sequence set forth in SEQ ID NO: 53. As another example, a 172-bp cDNA
fragment,
which is not expressed or rarely expressed in the cancer tissue or the cancer
cell line but
differentially expressed in the normal tissue, may be obtained by carrying out
a reverse
transcription-polymerase chain reaction (RT-PCR) on the total RNA extracted
from a
normal tissue, and a cancer tissue or a cancer cell line using a random primer
H-AP12 of
SEQ ID NO: 55 (5'-AAGCTTGAGTGCT-3') and an anchored oligo-dT primer of SEQ
ID NO: 56 (5'-AAGCTTTTTTTTTTTC-3'), and the resultant fragment, which is used
as
the probe, may be plaque-hybridized with a cDNA library to obtain a full-
length cDNA
clone.
It is regarded that the gene of the present invention is overexpressed in the
normal tissues, preferably the breast, the heart, the muscles, the kidney, the
liver and the
placenta to suppress the carcinogenesis. The gene of the present invention is
mainly
overexpressed in these tissues as an mRNA transcript having a size of
approximately 0.7
kb.
15. GIG39
The gene of the present invention is a human cancer suppressor gene 39 (GIG39)
having a DNA sequence of SEQ ID NO: 57, which was deposited with Accession No.
AY550972 into the GenBank database of U.S. National Institutes of Health (NIH)
(Scheduled Release Date: December 31, 2005), and a DNA sequence of the
deposited
gene is similar to that of the Homo sapiens chromosome 1 open reading frame 24
gene
34
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
deposited with Accession No. BC030531 into the database. From this study
result, it
was however found that the GIG39 gene was closely related to various human
carcinogenesis. From the study result, it was found that the GIG39 tumor
suppressor
gene was rarely expressed or not expressed in various human tumors including
the
breast cancer, while its expression was significantly increased in various
normal tissues.
The DNA sequence of SEQ ID NO: 57 has one open reading frame (ORF)
corresponding to base positions from 70 to 2,856 of the DNA sequence (base
positions
from 2,854 to 2,856 represent a stop codon).
A protein expressed from the gene of the present invention consists of 928
amino acid residues, and has an amino acid sequence of SEQ ID NO: 58 and a
molecular weight of approximately 103 kDa.
The gene and the protein of the present invention may be separated from human
tissues, or also be synthesized according to the known methods for
synthesizing DNA or
peptide. For example, the gene of the present invention may be screened and
cloned
according to the conventional methods on the basis of the information on the
DNA
sequence set forth in SEQ ID NO: 57. As another example, a 327-bp cDNA
fragment,
which is not expressed or rarely expressed in the cancer tissue or the cancer
cell line but
differentially expressed in the normal tissue, may be obtained by carrying out
a reverse
transcription-polymerase chain reaction (RT-PCR) on the total RNA extracted
from a
normal tissue, and a cancer tissue or a cancer cell line using a random primer
H-AP 12 of
SEQ ID NO: 59 (5'-AAGCTTGAGTGCT-3') and an anchored oligo-dT primer of SEQ
ID NO: 60 (5'-AAGCTTTTTTTTTTTA-3'), and the resultant fragment, which is used
as
the probe, may be plaque-hybridized with a cDNA library to obtain a full-
length cDNA
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
clone.
It is regarded that the gene of the present invention is overexpressed in the
normal tissues, preferably the breast and the liver to suppress the
carcinogenesis. The
gene of the present invention is mainly overexpressed in these tissues as an
mRNA
transcript having a size of approximately 2.4 kb.
16. GIG40
The gene of the present invention is a human cancer suppressor gene 40 (GIG40)
having a DNA sequence of SEQ ID NO: 61, which was deposited with Accession No.
AY550966 into the GenBank database of U.S. National Institutes of Health (NIH)
(Scheduled Release Date: December 31, 2005), and a DNA sequence of the
deposited
gene is similar to that of the Homo sapiens epidermal growth factor receptor
(erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) (EGFR)
deposited
with Accession No. NM005228 into the database. From this study result, it was
however found that the GIG40 tumor suppressor gene was rarely expressed in
various
human tumors including the liver cancer, while its expression was
significantly
increased in the normal liver tissue.
The DNA sequence of SEQ ID NO: 61 has one open reading frame (ORF)
corresponding to base positions from 47 to 3,679 of the DNA sequence (base
positions
from 3,677 to 3,679 represent a stop codon).
A protein expressed from the gene of the present invention consists of 1,210
amino acid residues, and has an amino acid sequence of SEQ ID NO: 62 and a
molecular weight of approximately 134 kDa.
The gene and the protein of the present invention may be separated from human
36
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
tissues, or also be synthesized according to the known methods for
synthesizing DNA or
peptide. For example, the gene of the present invention may be screened and
cloned
according to the conventional methods on the basis of the information on the
DNA
sequence set forth in SEQ ID NO: 61. As another example, a 275-bp cDNA
fragment,
which is not expressed in the cancer tissue or the cancer cell line but
differentially
expressed in the normal tissue, may be obtained by carrying out a reverse
transcription-polymerase chain reaction (RT-PCR) on the total RNA extracted
from a
normal tissue, and a cancer tissue or a cancer cell line using a random primer
H-AP7 of
SEQ ID NO: 63 (5'-AAGCTTAACGAGG-3') and an anchored oligo-dT primer of SEQ
ID NO: 64 (5'-AAGCTTTTTTTTTTTG-3'), and the resultant fragment, which is used
as
the probe, may be plaque-hybridized with a cDNA library to obtain a full-
length cDNA
clone. The present inventors inserted the full-length GIG40 cDNA into the
expression
vector pBAD/Thio-Topo (Invitrogen, U.A.), and then transformed E. coli DH5 a
with
the resultant expression vector to obtain a transformant, which was designated
E. coli
DH5 a /GIG40/pBAD/Thio-Topo.
It is regarded that the gene of the present invention is overexpressed in the
normal tissues, preferably the liver, the heart and the muscles to suppress
the
carcinogenesis. Also, it is regarded that its gene expression is suppressed in
the
leukemia, the uterine cancer, the malignant lymphoma, the colon cancer, the
lung cancer
and the skin cancer to induce the carcinogenesis. The gene of the present
invention is
mainly overexpressed in these tissues as an mRNA transcript having a size of
approximately 1.5 kb.
17. GIG42
37
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
The gene of the present invention is a human cancer suppressor gene 42 (GIG42)
having a DNA sequence of SEQ ID NO: 65, which was deposited with Accession No.
AY550967 into the GenBank database of U.S. National Institutes of Health (NIH)
(Scheduled Release Date: December 31, 2005), and a DNA sequence of the
deposited
gene is similar to that of the Homo sapiens albumin gene deposited with
Accession No.
BC034023 into the database. From this study result, it was however found that
the
GIG42 tumor suppressor gene was not expressed at all in various human tumors
including the liver cancer, while its expression was significantly increased
in various
normal tissues.
The DNA sequence of SEQ ID NO: 65 has one open reading frame (ORF)
corresponding to base positions from 8 to 1,837 of the DNA sequence (base
positions
from 1,835 to 1,837 represent a stop codon).
A protein expressed from the gene of the present invention consists of 609
amino acid residues, and has an amino acid sequence of SEQ ID NO: 66 and a
molecular weight of approximately 69 kDa.
The gene and the protein of the present invention may be separated from human
tissues, or also be synthesized according to the known methods for
synthesizing DNA or
peptide. For example, the gene of the present invention may be screened and
cloned
according to the conventional methods on the basis of the information on the
DNA
sequence set forth in SEQ ID NO: 65. As another example, a 327-bp cDNA
fragment,
which is not expressed in the cancer tissue or the cancer cell line but
differentially
expressed in the normal tissue, may be obtained by carrying out a reverse
transcription-polymerase chain reaction (RT-PCR) on the total RNA extracted
from a
38
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
normal tissue, and a cancer tissue or a cancer cell line using a random primer
H-AP8 of
SEQ ID NO: 67 (5'-AAGCTTTTACCGC-3') and an anchored oligo-dT primer of SEQ
ID NO: 68 (5'-AAGCTTTTTTTTTTTG-3'), and the resultant fragment, which is used
as
the probe, may be plaque-hybridized with a cDNA library to obtain a full-
length cDNA
clone. The present inventors inserted the full-length GIG42 cDNA into the
expression
vector pBAD/Thio-Topo (Invitrogen, U.A.), and then transformed E. coli DH5 a
with
the resultant expression vector to obtain a transformant, which was designated
E. coli
DH5 a /GIG42/pBAD/Thio-Topo.
It is regarded that the gene of the present invention is overexpressed in the
normal tissues, preferably the liver to suppress the carcinogenesis. Also, it
is regarded
that its gene expression is suppressed in the leukemia, the uterine cancer,
the malignant
lymphoma, the colon cancer, the lung cancer and the skin cancer to induce the
carcinogenesis. The gene of the present invention is mainly overexpressed in
these
tissues as an mRNA transcript having a size of approximately 2.5 kb.
18. GIG43
The gene of the present invention is a human cancer suppressor gene 43 (GIG43)
having a DNA sequence of SEQ ID NO: 69, which was deposited with Accession No.
AY550971 into the GenBank database of U.S. National Institutes of Health (NIH)
(Scheduled Release Date: December 31, 2005), and a DNA sequence of the
deposited
gene is similar to that of the Homo sapiens phospholipid scramblase 4 gene
deposited
with Accession No. BC028354 into the database. From this study result, it was
however found that the GIG43 gene was closely related to various human
carcinogenesis. From the study result, it was found that the GIG43 tumor
suppressor
39
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
gene was rarely expressed or not expressed in various human tumors including
the
breast cancer, while its expression was significantly increased in various
normal tissues.
The DNA sequence of SEQ ID NO: 69 has one open reading frame (ORF)
corresponding to base positions from 96 to 1,085 of the DNA sequence (base
positions
from 1,083 to 1,085 represent a stop codon).
A protein expressed from the gene of the present invention consists of 329
amino acid residues, and has an amino acid sequence of SEQ ID NO: 70 and a
molecular weight of approximately 37 kDa.
The gene and the protein of the present invention may be separated from human
tissues, or also be synthesized according to the known methods for
synthesizing DNA or
peptide. For example, the gene of the present invention may be screened and
cloned
according to the conventional methods on the basis of the information on the
DNA
sequence set forth in SEQ ID NO: 69. As another example, a 273-bp cDNA
fragment,
which is not expressed or rarely expressed in the cancer tissue or the cancer
cell line but
differentially expressed in the normal tissue, may be obtained by carrying out
a reverse
transcription-polymerase chain reaction (RT-PCR) on the total RNA extracted
from a
normal tissue, and a cancer tissue or a cancer cell line using a random primer
H-AP10 of
SEQ ID NO: 71 (5'-AAGCTTCCACGTA-3') and an anchored oligo-dT primer of SEQ
ID NO: 72 (5'-AAGCTTTTTTTTTTTG-3'), and the resultant fragment, which is used
as
the probe, may be plaque-hybridized with a cDNA library to obtain a full-
length cDNA
clone.
It is regarded that the gene of the present invention is overexpressed in the
normal tissues, preferably the breast, the heart, the kidney, the liver, the
placenta and the
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
lungs to suppress the carcinogenesis. The gene of the present invention is
mainly
overexpressed in these tissues as an mRNA transcript having a size of
approximately 3.5
kb.
19. GIG46
The gene of the present invention is a human cancer suppressor gene 1(GIG46)
having a DNA sequence of SEQ ID NO: 73, which was deposited with Accession No.
AY692464 into the GenBank database of U.S. National Institutes of Health (NIH)
(Scheduled Release Date: May 1, 2006), and a DNA sequence of the deposited
gene is
similar to that of the Homo sapiens actin, alpha 2, smooth muscle, aorta
(ACTA2) gene
deposited with Accession No. NM_001613 into the database. Contrary to its
functions
as reported previously, it was however found from this study result that the
GIG46 gene
was closely related to various human carcinogenesis. From the study result, it
was
found that the GIG8 tumor suppressor gene was very rarely expressed in various
human
tumors including the uterine cancer, while its expression was significantly
increased in
various normal tissues.
The DNA sequence of SEQ ID NO: 73 has one open reading frame (ORF)
corresponding to base positions from 393 to 1,526 of the DNA sequence (base
positions
from 1,524 to 1,526 represent a stop codon).
A protein expressed from the gene of the present invention consists of 377
amino acid residues, and has an amino acid sequence of SEQ ID NO: 74 and a
molecular weight of approximately 42 kDa.
The gene and the protein of the present invention may be separated from human
tissues, or also be synthesized according to the known methods for
synthesizing DNA or
41
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
peptide. For example, the gene of the present invention may be screened and
cloned
according to the conventional methods on the basis of the information on the
DNA
sequence set forth in SEQ ID NO: 73. As another example, a 255-bp cDNA
fragment
(corresponding to base positions from 1,323 to 1,577), which is not expressed
in the
cancer tissue or the cancer cell line but differentially expressed in the
normal tissue, may
be obtained by carrying out a reverse transcription-polymerase chain reaction
(RT-PCR)
on the total RNA extracted from a normal tissue, and a cancer tissue or a
cancer cell line
using a random primer H-AP16 of SEQ ID NO: 75 (5'-AAGCTTTAGAGCG-3') and an
anchored oligo-dT primer of SEQ ID NO: 76 (5'-AAGCTTTTTTTTTTTA-3'), and the
resultant fragment, which is used as the probe, may be plaque-hybridized with
a cDNA
library to obtain a full-length cDNA clone.
It is regarded that the gene of the present invention is overexpressed in the
normal tissues, preferably the uterus, the brain, the heart, the skeletal
muscles, the large
intestine, the thymus, the spleen, the kidney, the liver, the small intestine,
the placenta,
the lungs and the peripheral blood leukocyte to suppress the carcinogenesis.
The gene
of the present invention is mainly overexpressed in these tissues as an mRNA
transcript
having a size of approximately 1.5 kb, and an mRNA transcript having a size of
approximately 2.0 kb is also expressed in addition to the 1.5-kb mRNA
transcript.
20. PIG33
The gene of the present invention is a human cancer suppressor gene (PIG33)
having a DNA sequence of SEQ ID NO: 77, which was deposited with Accession No.
AY513278 into the GenBank database of U.S. National Institutes of Health (NIH)
(Scheduled Release Date: December 31, 2005), and a DNA sequence of the
deposited
42
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
gene is similar to that of the Homo sapiens SPARC-like 1(mast9, hevin) gene
deposited
with Accession No. BC033721 into the database. From this study result, it was
however found that the PIC'735 tumor suppressor gene was rarely expressed in
various
human tumors including the liver cancer, while its expression was
significantly
increased in the normal liver tissue.
The DNA sequence of SEQ ID NO: 77 has one open reading frame (ORF)
corresponding to base positions from 81 to 2,075 of the DNA sequence (base
positions
from 2,073 to 2,075 represent a stop codon).
A protein expressed from the gene of the present invention consists of 664
amino acid residues, and has an amino acid sequence of SEQ ID NO: 78 and a
molecular weight of approximately 75 kDa.
The gene and the protein of the present invention may be separated from human
tissues, or also be synthesized according to the known methods for
synthesizing DNA or
peptide. For example, the gene of the present invention may be screened and
cloned
according to the conventional methods on the basis of the information on the
DNA
sequence set forth in SEQ ID NO: 77. As another example, a 256-bp cDNA
fragment,
which is not expressed or rarely expressed in the cancer tissue or the cancer
cell line but
differentially expressed in the normal tissue, may be obtained by carrying out
a reverse
transcription-polymerase chain reaction (RT-PCR) on the total RNA extracted
from a
normal tissue, and a cancer tissue or a cancer cell line using a random primer
H-AP2 of
SEQ ID NO: 79 (5'-AAGCTTCGACTGT-3') and an anchored oligo-dT primer of SEQ
ID NO: 80 (5'-AAGCTTTTTTTTTTTA-3'), and the resultant fragment, which is used
as
the probe, may be plaque-hybridized with a cDNA library to obtain a full-
length cDNA
43
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
clone.
The present inventors inserted the full-length PIG33 cDNA into the expression
vector pBAD/Thio-Topo (Invitrogen, U.A.), and then transformed E. coli DH5 a
with
the resultant expression vector to obtain a transformant, which was designated
E. coli
DH5 a / PIG33/pBAD/Thio-Topo.
It is regarded that the gene of the present invention is overexpressed in the
normal tissues, preferably the liver, the brain, the heart, the muscles, the
large intestine,
the thymus, the spleen, the kidney, the small intestine, the placenta and the
lungs to
suppress the carcinogenesis. Also, it is regarded that its gene expression is
suppressed
in the leukemia, the uterine cancer, the colon cancer, the lung cancer and the
skin cancer
to induce the carcinogenesis. The gene of the present invention is mainly
overexpressed in these tissues as an mRNA transcript having a size of
approximately 3.0
kb.
21. PIG35
The gene of the present invention is a human cancer suppressor gene (PIG35)
having a DNA sequence of SEQ ID NO: 81, which was deposited with Accession No.
AY513280 into the GenBank database of U.S. National Institutes of Health (NIH)
(Scheduled Release Date: December 31, 2005), and a DNA sequence of the
deposited
gene is similar to that of the Homo sapiens serine (or cysteine) proteinase
inhibitor
deposited with Accession No. NM002615 into the database. From this study
result, it
was however found that the PIG35 tumor suppressor gene was rarely expressed in
various human tumors including the breast cancer, while its expression was
significantly
increased in the normal liver tissue.
44
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
The DNA sequence of SEQ ID NO: 81 has one open reading frame (ORF)
corresponding to base positions from 50 to 1,306 of the DNA sequence (base
positions
from 1,304 to 1,306 represent a stop codon).
A protein expressed from the gene of the present invention consists of 418
amino acid residues, and has an amino acid sequence of SEQ ID NO: 82 and a
molecular weight of approximately 46 kDa.
The gene and the protein of the present invention may be separated from human
tissues, or also be synthesized according to the known methods for
synthesizing DNA or
peptide. For example, the gene of the present invention may be screened and
cloned
according to the conventional methods on the basis of the information on the
DNA
sequence set forth in SEQ ID NO: 81. As another example, a 312-bp cDNA
fragment,
which is not expressed or rarely expressed in the cancer tissue or the cancer
cell line but
differentially expressed in the normal tissue, may be obtained by carrying out
a reverse
transcription-polymerase chain reaction (RT-PCR) on the total RNA extracted
from a
normal tissue, and a cancer tissue or a cancer cell line using a random primer
H-AP9 of
SEQ ID NO: 83 (5'-AAGCTTCATTCCG-3') and an anchored oligo-dT primer of SEQ
ID NO: 84 (5'-AAGCTTTTTTTTTTTC-3'), and the resultant fragment, which is used
as
the probe, may be plaque-hybridized with a cDNA library to obtain a full-
length cDNA
clone.
The present inventors inserted the full-length PIG35 cDNA into the expression
vector pBAD/Thio-Topo (Invitrogen, U.A.), and then transformed E. coli DH5 a
with
the resultant expression vector to obtain a transformant, which was designated
E. coli
DH5 a /PIG35/pBAD/Thio-Topo.
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
It is regarded that the gene of the present invention is overexpressed in the
normal tissues, preferably the liver, the heart, the muscles, the brain, the
small intestine,
the lungs and the placenta to suppress the carcinogenesis. Also, it is
regarded that its
gene expression is suppressed in the leukemia, the uterine cancer, the
malignant
lymphoma, the colon cancer, the lung cancer and the skin cancer to induce the
carcinogenesis. The gene of the present invention is mainly overexpressed in
these
tissues as an mRNA transcript having a size of approximately 1.7 kb.
22. PIG36
The gene of the present invention is a human cancer suppressor gene (PIG36)
having a DNA sequence of SEQ ID NO: 85, which was deposited with Accession No.
AY544129 into the GenBank database of U.S. National Institutes of Health (NIH)
(Scheduled Release Date: December 31, 2005), and a DNA sequence of the
deposited
gene is similar to that of the Homo sapiens ATP synthase, H+ transporting,
mitochondrial FO complex, subunit F6 (ATP5J) deposited with Accession No.
NM001685 into the database. From this study result, it was however found that
the
PIG36 tumor suppressor gene was rarely expressed in various human tumors
including
the liver cancer, while its expression was significantly increased in various
normal
tissues.
The DNA sequence of SEQ ID NO: 85 has one open reading frame (ORF)
corresponding to base positions from 102 to 428 of the DNA sequence (base
positions
from 426 to 428 represent a stop codon).
A protein expressed from the gene of the present invention consists of 108
amino acid residues, and has an amino acid sequence of SEQ ID NO: 86 and a
46
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
molecular weight of approximately 13 kDa.
The gene and the protein of the present invention may be separated from human
tissues, or also be synthesized according to the known methods for
synthesizing DNA or
peptide. For example, the gene of the present invention may be screened and
cloned
according to the conventional methods on the basis of the information on the
DNA
sequence set forth in SEQ ID NO: 85. As another example, a 162-bp cDNA
fragment,
which is not expressed or rarely expressed in the cancer tissue or the cancer
cell line but
differentially expressed in the normal tissue, may be obtained by carrying out
a reverse
transcription-polymerase chain reaction (RT-PCR) on the total RNA extracted
from a
normal tissue, and a cancer tissue or a cancer cell line using a random primer
H-AP9 of
SEQ ID NO: 87 (5'-AAGCTTCATTCCG-3') and an anchored oligo-dT primer of SEQ
ID NO: 88 (5'-AAGCTTTTTTTTTTTG-3'), and the resultant fragment, which is used
as
the probe, may be plaque-hybridized with a cDNA library to obtain a full-
length cDNA
clone. The present inventors inserted the full-length PIG36 cDNA into the
expression
vector pBAD/Thio-Topo (Invitrogen, U.A.), and then transformed E. coli DH5 a
with
the resultant expression vector to obtain a transformant, which was designated
E. coli
DH5 a / PIG36/pBAD/Thio-Topo.
It is regarded that the gene of the present invention is overexpressed in the
normal tissues, preferably the liver, the heart, the muscles, kidney and the
placenta to
suppress the carcinogenesis. Also, it is regarded that the gene of the present
invention
was suppressed in the leukemia, the uterine cancer, the malignant lymphoma,
the colon
cancer, the lung cancer and the skin cancer to induce the carcinogenesis. The
gene of
the present invention is mainly overexpressed in these tissues as an mRNA
transcript
47
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
having a size of approximately 1.0 kb.
23. MIG20
A DNA sequence of SEQ ID NO: 89 was deposited with Accession No.
AY871271 into the GenBank database of U.S. National Institutes of Health (NIH)
(Scheduled Release Date: October 1, 2006), and a DNA sequence of the deposited
gene
is similar to that of the Homo sapiens mRNA; cDNA DKFZp686C0390 gene deposited
with Accession No. BX537651 into the database. From this study result, it was
however found that the MIG20 tumor suppressor gene was not expressed at all in
various human tumors including the uterine cancer, while its expression was
significantly increased in various normal tissues.
The DNA sequence of SEQ ID NO: 89 has one open reading frame (ORF)
corresponding to base positions from 8 to 202 of the DNA sequence (base
positions
from 200 to 202 represent a stop codon). Also, the DNA sequence of SEQ ID NO:
89
has another open reading frame corresponding to base positions from 233 to 442
of the
DNA sequence (base positions from 440 to 442 represent a stop codon).
A protein expressed from the gene of the present invention consists of 64
amino
acid residues, and has an amino acid sequence of SEQ ID NO: 90 and a molecular
weight of approximately 7 kDa.
The gene and the protein of the present invention may be separated from human
tissues, or also be synthesized according to the known methods for
synthesizing DNA or
peptide. For example, the gene of the present invention may be screened and
cloned
according to the conventional methods on the basis of the information on the
DNA
sequence set forth in SEQ ID NO: 89. As another example, a 311-bp cDNA
fragment
48
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
(corresponding to base positions from 2,067 to 2,377), which is not expressed
in the
cancer tissue or the cancer cell line but differentially expressed in the
normal tissue, may
be obtained by carrying out a reverse transcription-polymerase chain reaction
(RT-PCR)
on the total RNA extracted from a normal tissue, and a cancer tissue or a
cancer cell line
using a random primer H-AP32 of SEQ ID NO: 91 (5'-AAGCTTCCTGCAA-3') and an
anchored oligo-dT primer of SEQ ID NO: 92 (5'-AAGCTTTTTTTTTTTA-3'), and the
resultant fragment, which is used as the probe, may be plaque-hybridized with
a cDNA
library to obtain a full-length cDNA clone.
It is regarded that the gene of the present invention is overexpressed in the
normal tissues, preferably the uterus, the heart, the skeletal muscle, the
kidney and the
liver to suppress the carcinogenesis. The gene of the present invention is
mainly
overexpressed in these tissues as an mRNA transcript having a size of
approximately 4.4
kb, and mRNA transcripts having sizes of approximately 2.4 kb and 1.5 kb are
also
expressed in addition to the 4.4-kb mRNA transcript.
24. PIG49
The gene of the present invention is a human cancer suppressor gene (GIG49)
having a DNA sequence of SEQ ID NO: 93, which was deposited with Accession No.
AY524047 into the GenBank database of U.S. National Institutes of Health (NIH)
(Scheduled Release Date: December 31, 2005), and a DNA sequence of the
deposited
gene is similar to that of the Homo sapiens transducer of ERBB2, 1(TOB 1) gene
deposited with Accession No. NM005749 into the database. From this study
result, it
was however found that the GIG49 gene was closely related to various human
carcinogenesis. From the study result, it was found that the GIG49 tumor
suppressor
49
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
gene was rarely expressed or not expressed in various human tumors including
the
breast cancer, while its expression was significantly increased in various
normal tissues.
The DNA sequence of SEQ ID NO: 93 has one open reading frame (ORF)
corresponding to base positions from 11 to 1,048 of the DNA sequence (base
positions
from 1,046 to 1,048 represent a stop codon). However, because of degeneracy of
codons, or considering preference of codons for living organisms to express
the gene,
the gene of the present invention may be variously modified in coding region
without
changing an amino acid sequence of the protein expressed from the coding
region, and
also be variously modified or changed in a region except the coding region
within a
range that does not affect the gene expression. Such a modified gene is also
included
in the scope of the present invention. Accordingly, the present invention also
includes
a polynucleotide having substantially the same DNA sequences as the above-
mentioned
gene; and fragments of the gene. The term "substantially the same
polynucleotide"
means a DNA sequence having a sequence homology of at least 80 %, preferably
at
least 90 %, and the most preferably at least 95 %.
A protein expressed from the gene of the present invention consists of 345
amino acid residues, and has an amino acid sequence of SEQ ID NO: 94 and a
molecular weight of approximately 38 kDa. Also, one or more amino acids may be
substituted, added or deleted even in the amino acid sequence of the protein
within a
range that does not affect functions of the protein, and only some of the
protein may be
used depending on their usage. Such a modified amino acid sequence is also
included
in the scope of the present invention. Accordingly, the present invention also
includes
a polypeptide having substantially the same amino acid sequence as the
protein; and
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
fragments thereof. The term "substantially the same polypeptide" means a
polypeptide
having a sequence homology of at least 80 %, preferably at least 90 %, and the
most
preferably at least 95 %.
The gene and the protein of the present invention may be separated from human
tissues, or also be synthesized according to the known methods for
synthesizing DNA or
peptide. For example, the gene of the present invention may be screened and
cloned
according to the conventional methods on the basis of the information on the
DNA
sequence set forth in SEQ ID NO: 93. As another example, a 272-bp cDNA
fragment,
which is not expressed or rarely expressed in the cancer tissue or the cancer
cell line but
differentially expressed in the normal tissue, may be obtained by carrying out
a reverse
transcription-polymerase chain reaction (RT-PCR) on the total RNA extracted
from a
normal tissue, and a cancer tissue or a cancer cell line using a random primer
H-AP 10 of
SEQ ID NO: 95 (5'-AAGCTTCCACGTA-3') and an anchored oligo-dT primer of SEQ
ID NO: 96 (5'-AAGCTTTTTTTTTTTA-3'), and the resultant fragment, which is used
as
the probe, may be plaque-hybridized with a cDNA library to obtain a full-
length cDNA
clone.
The gene prepared thus may be inserted into a vector for expression in the
microorganisms or animal cells, already known in the art, to obtain an
expression vector,
and then DNA of the gene may be replicated in a large quantity or its protein
may be
produced in a commercial quantity by introducing the expression vector into
suitable
host cells, for example Escherichia coli, a MCF-7 cell line, etc. Upon
constructing the
expression vector, expression regulatory sequences such as a promoter and a
terminator,
autonomously replicating sequences, secretion signals, etc. may be suitably
selected and
51
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
combined depending on a kind of the host cell that produces the gene or the
protein.
It is regarded that the gene of the present invention is overexpressed in the
normal tissues, preferably the breast, the muscles, the heart, the kidney, the
liver and the
placenta to suppress the carcinogenesis. The gene of the present invention is
mainly
overexpressed in these tissues as an mRNA transcript having a size of
approximately 2.4
kb. Also, an mRNA transcript having a size of approximately 1.5 kb is
expressed in
addition to the 2.4-kb mRNA transcript.
25. PIG51
The gene of the present invention is a human cancer suppressor gene (PIG51)
having a DNA sequence of SEQ ID NO: 97, which was deposited with Accession No.
AY542308 into the GenBank database of U.S. National Institutes of Health (NIH)
(Scheduled Release Date: December 31, 2005), and a DNA sequence of the
deposited
gene is similar to that of the Homo sapiens TBC1 domain family, member 7 gene
deposited with Accession No. BC007054 into the database. A specific function
of the
gene remains to be specified. From this study result, it was however found
that the
PIG51 gene was closely related to various human carcinogenesis. From the study
result, it was found that the PIG51 tumor suppressor gene was rarely expressed
or not
expressed in various human tumors including the breast cancer, while its
expression was
significantly increased in various normal tissues.
The DNA sequence of SEQ ID NO: 97 has one open reading frame (ORF)
corresponding to base positions from 59 to 802 of the DNA sequence (base
positions
from 800 to 802 represent a stop codon).
A protein expressed from the gene of the present invention consists of 247
52
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
amino acid residues, and has an amino acid sequence of SEQ ID NO: 98 and a
molecular weight of approximately 28 kDa.
The gene and the protein of the present invention may be separated from human
tissues, or also be synthesized according to the known methods for
synthesizing DNA or
peptide. For example, the gene of the present invention may be screened and
cloned
according to the conventional methods on the basis of the information on the
DNA
sequence set forth in SEQ ID NO: 97. As another example, a 211-bp cDNA
fragment,
which is not expressed or rarely expressed in the cancer tissue or the cancer
cell line but
differentially expressed in the normal tissue, may be obtained by carrying out
a reverse
transcription-polymerase chain reaction (RT-PCR) on the total RNA extracted
from a
normal tissue, and a cancer tissue or a cancer cell line using a random primer
H-AP22 of
SEQ ID NO: 99 (5'-AAGCTTTTGATCC-3') and an anchored oligo-dT primer of SEQ
ID NO: 100 (5'-AAGCTTTTTTTTTTTG-3'), and the resultant fragment, which is used
as the probe, may be plaque-hybridized with a cDNA library to obtain a full-
length
cDNA clone.
It is regarded that the gene of the present invention is overexpressed in the
normal tissues, preferably the breast, the heart, the muscles, the thymus, the
spleen, the
kidney, the liver, the placenta and the peripheral blood leukocyte to suppress
the
carcinogenesis. The gene of the present invention is mainly overexpressed in
these
tissues as an mRNA transcript having a size of approximately 1.0 kb.
26. MIG12
The gene of the present invention is a human cancer suppressor gene (MIG12)
having a DNA sequence of SEQ ID NO: 101, which was deposited with Accession
No.
53
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
AY453400 into the GenBank database of U.S. National Institutes of Health (NIH)
(Scheduled Release Date: March 31, 2005), and a DNA sequence of the deposited
gene
is similar to that of the Homo sapiens thymosin, beta 10 gene deposited with
Accession
No. BC016731 into the database. From this study result, it was however found
that the
MIG12 tumor suppressor gene was rarely expressed in various human tumors
including
the lung cancer, while its expression was significantly increased in various
normal
tissues.
The DNA sequence of SEQ ID NO: 101 has one open reading frame (ORF)
corresponding to base positions from 29 to 163 of the DNA sequence (base
positions
from 161 to 163 represent a stop codon).
A protein expressed from the gene of the present invention consists of 44
amino
acid residues, and has an amino acid sequence of SEQ ID NO: 102 and a
molecular
weight of approximately 5 kDa.
The gene and the protein of the present invention may be separated from human
tissues, or also be synthesized according to the known methods for
synthesizing DNA or
peptide. For example, the gene of the present invention may be screened and
cloned
according to the conventional methods on the basis of the information on the
DNA
sequence set forth in SEQ ID NO: 101. As another example, a 161-bp cDNA
fragment,
which is very rarely expressed in the cancer tissue or the cancer cell line
but
differentially expressed in the normal tissue, may be obtained by carrying out
a reverse
transcription-polymerase chain reaction (RT-PCR) on the total RNA extracted
from a
normal tissue, and a cancer tissue or a cancer cell line using a random primer
H-AP 12 of
SEQ ID NO: 103 (5'-AAGCTTGAGTGCT-3') and an anchored oligo-dT primer of SEQ
54
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
ID NO: 104 (5'-AAGCTTTTTTTTTTTC-3'), and the resultant fragment, which is used
as the probe, may be plaque-hybridized with a cDNA library to obtain a full-
length
cDNA clone.
It is regarded that the gene of the present invention is overexpressed in the
normal tissues, preferably the lungs, the brain, the heart, the muscles, the
large intestine,
the thymus, the spleen, the kidney, the liver, the small intestine, the
placenta and the
peripheral blood leukocyte to suppress the carcinogenesis. The gene of the
present
invention is mainly overexpressed in these tissues as an mRNA transcript
having a size
of approximately 0.5 kb, and mRNA transcripts having sizes of approximately
1.0 kb
and 0.8 kb are expressed in addition to the 0.5-kb mRNA transcript.
27. PIG37
The gene of the present invention is a human cancer suppressor gene (PIG37)
having a DNA sequence of SEQ ID NO: 105, which was deposited with Accession
No.
AY513281 into the GenBank database of U.S. National Institutes of Health (NIH)
(Scheduled Release Date: December 31, 2005), and a DNA sequence of the
deposited
gene is similar to that of the Homo sapiens inositol 1,4,5-trisphosphate 3-
kinase B
(ITPKB) gene deposited with Accession No. NM_002221 into the database. From
this
study result, it was however found that the PIG37 tumor suppressor gene was
rarely
expressed in various human tumors including the liver cancer, while its
expression was
significantly increased in the normal liver tissue.
The DNA sequence of SEQ ID NO: 105 has one open reading frame (ORF)
corresponding to base positions from 4 to 1,422 of the DNA sequence (base
positions
from 1,420 to 1,422represent a stop codon).
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
A protein expressed from the gene of the present invention consists of 472
amino acid residues, and has an amino acid sequence of SEQ ID NO: 106 and a
molecular weight of approximately 53 kDa.
The gene and the protein of the present invention may be separated from human
tissues, or also be synthesized according to the known methods for
synthesizing DNA or
peptide. For example, the gene of the present invention may be screened and
cloned
according to the conventional methods on the basis of the information on the
DNA
sequence set forth in SEQ ID NO: 105. As another example, a 263-bp cDNA
fragment,
which is not expressed or rarely expressed in the cancer tissue or the cancer
cell line but
differentially expressed in the normal tissue, may be obtained by carrying out
a reverse
transcription-polymerase chain reaction (RT-PCR) on the total RNA extracted
from a
normal tissue, and a cancer tissue or a cancer cell line using a random primer
H-AP10 of
SEQ ID NO: 107 (5'-AAGCTTCCACGTA-3') and an anchored oligo-dT primer of SEQ
ID NO: 108 (5'-AAGCTTTTTTTTTTTG-3'), and the resultant fragment, which is used
as the probe, may be plaque-hybridized with a cDNA library to obtain a full-
length
cDNA clone. The present inventors inserted the full-length PIG33 cDNA into the
expression vector pBAD/Thio-Topo (Invitrogen, U.A.), and then transformed E.
coli
DH5 a with the resultant expression vector to obtain a transformant, which was
designated E. coli DH5 a/PIG33/pBAD/Thio-Topo.
It is regarded that the gene of the present invention is overexpressed in the
normal tissues, preferably the liver, the brain, the heart, the muscles, the
large intestine,
the thymus, the spleen, the kidney, the small intestine, the placenta and the
lungs to
suppress the carcinogenesis. Also, it is regarded that the gene of the present
invention
56
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
was suppressed in the leukemia, the uterine cancer, the colon cancer, the lung
cancer
and the skin cancer to induce the carcinogenesis. The gene of the present
invention is
mainly overexpressed in these tissues as an mRNA transcript having a size of
approximately 7.0 kb.
28. GIG44
The gene of the present invention is a human cancer suppressor gene 44 (GIG44)
having a DNA sequence of SEQ ID NO: 109, which was deposited with Accession
No.
AY971350 into the GenBank database of U.S. National Institutes of Health (NIH)
(Scheduled Release Date: May 31, 2006), and a DNA sequence of the deposited
gene is
similar to that of the Homo sapiens putative insulin-like growth factor II
associated
protein gene deposited with Accession No. BC042127 into the database. From
this
study result, it was however found that the GIG44 gene was closely related to
various
human carcinogenesis. From the study result, it was found that the GIG44 tumor
suppressor gene was rarely expressed or not expressed in various human tumors
including the breast cancer, while its expression was significantly increased
in various
normal tissues.
The DNA sequence of SEQ ID NO: 109 has one open reading frame (ORF)
corresponding to base positions from 59 to 400 of the DNA sequence (base
positions
from 398 to 400 represent a stop codon).
A protein expressed from the gene of the present invention consists of 113
amino acid residues, and has an amino acid sequence of SEQ ID NO: 110 and a
molecular weight of approximately 12 kDa.
The gene and the protein of the present invention may be separated from human
57
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
tissues, or also be synthesized according to the known methods for
synthesizing DNA or
peptide. For example, the gene of the present invention may be screened and
cloned
according to the conventional methods on the basis of the information on the
DNA
sequence set forth in SEQ ID NO: 109. As another example, a 221-bp cDNA
fragment,
which is not expressed or rarely expressed in the cancer tissue or the cancer
cell line but
differentially expressed in the normal tissue, may be obtained by carrying out
a reverse
transcription-polymerase chain reaction (RT-PCR) on the total RNA extracted
from a
normal tissue, and a cancer tissue or a cancer cell line using a random primer
H-AP 12 of
SEQ ID NO: 111 (5'-AAGCTTGAGTGCT-3') and an anchored oligo-dT primer of SEQ
ID NO: 112 (5'-AAGCTTTTTTTTTTTG-3'), and the resultant fragment, which is used
as the probe, may be plaque-hybridized with a cDNA library to obtain a full-
length
cDNA clone.
It is regarded that the gene of the present invention is overexpressed in the
normal tissues, preferably the breast, the heart, the kidney, the liver, the
placenta and the
lungs to suppress the carcinogenesis. The gene of the present invention is
mainly
overexpressed in these tissues as an mRNA transcript having a size of
approximately 1.0
kb.
29. GIG31
The gene of the present invention is a human cancer suppressor gene 31 (GIG3
1)
having a DNA sequence of SEQ ID NO: 113, which was deposited with Accession
No.
AY971351 into the GenBank database of U.S. National Institutes of Health (NIH)
(Scheduled Release Date: May 31, 2006), and a DNA sequence of the deposited
gene is
similar to that of the Homo sapiens regulator of G-protein signalling 2 gene
deposited
58
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
with Accession No. NM_002923 into the database. From this study result, it was
however found that the GIG31 gene was closely related to various human
carcinogenesis. From the study result, it was found that the GIG31 tumor
suppressor
gene was rarely expressed or not expressed in various human tumors including
the
breast cancer, while its expression was significantly increased in various
normal tissues.
The DNA sequence of SEQ ID NO: 113 has one open reading frame (ORF)
corresponding to base positions from 14 to 649 of the DNA sequence (base
positions
from 647 to 649 represent a stop codon).
A protein expressed from the gene of the present invention consists of 211
amino acid residues, and has an amino acid sequence of SEQ ID NO: 114 and a
molecular weight of approximately 24 kDa.
The gene and the protein of the present invention may be separated from human
tissues, or also be synthesized according to the known methods for
synthesizing DNA or
peptide. For example, the gene of the present invention may be screened and
cloned
according to the conventional methods on the basis of the information on the
DNA
sequence set forth in SEQ ID NO: 113. As another example, a 223-bp cDNA
fragment,
which is not expressed or rarely expressed in the cancer tissue or the cancer
cell line but
differentially expressed in the normal tissue, may be obtained by carrying out
a reverse
transcription-polymerase chain reaction (RT-PCR) on the total RNA extracted
from a
normal tissue, and a cancer tissue or a cancer cell line using a random primer
H-AP4 of
SEQ ID NO: 115 (5'-AAGCTTCTCAACG-3') and an anchored oligo-dT primer of SEQ
ID NO: 116 (5'-AAGCTTTTTTTTTTTA-3'), and the resultant fragment, which is used
as the probe, may be plaque-hybridized with a cDNA library to obtain a full-
length
59
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
cDNA clone.
It is regarded that the gene of the present invention is overexpressed in the
normal tissues, preferably the breast, the heart, the large intestine, the
spleen, the small
intestine, the placenta, the lungs and the peripheral blood leukocyte to
suppress the
carcinogenesis. The gene of the present invention is mainly overexpressed in
these
tissues as an mRNA transcript having a size of approximately 1.4 kb.
Meanwhile, because of degeneracy of codons, or considering preference of
codons for living organisms to express the genes, the genes of the present
invention may
be variously modified in coding regions without changing an amino acid
sequence of the
protein expressed from the coding region, and also be variously modified or
changed in
a region except the coding region within a range that does not affect the gene
expression.
Such a modified gene is also included in the scope of the present invention.
Accordingly, the present invention also includes polynucleotides having
substantially
the same DNA sequences as the above-mentioned genes; and fragments of the
genes.
The term "substantially the same polynucleotide" means a DNA sequence having a
sequence homology of at least 80 %, preferably at least 90 %, and the most
preferably at
least 95 %.
Also, one or more amino acids may be substituted, added or deleted even in the
amino acid sequences of the proteins of the present invention within a range
that does
not affect functions of the proteins, and only some of the proteins may be
used
depending on their usage. Such a modified amino acid sequence is also included
in the
scope of the present invention. Accordingly, the present invention also
includes
polypeptides having substantially the same amino acid sequences as the
proteins; and
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
fragments thereof. The term "substantially the same polypeptide" means a
polypeptide
having sequence homology of at least 80 %, preferably at least 90 %, and the
most
preferably at least 95 %.
In some embodiments, the genes of the present invention prepared thus may be
also inserted into a vector for expression in the microorganisms or animal
cells, already
known in the art, to obtain expression vectors, and then DNA of the genes may
be
replicated in a large quantity or its protein may be produced in a commercial
quantity by
introducing the expression vectors into suitable host cells, for example
Escherichia coli,
a MCF-7 cell line, etc. Upon constructing the expression vectors, expression
regulatory sequences such as a promoter and a terminator, autonomously
replicating
sequences, secretion signals, etc. may be suitably selected and combined
depending on
kinds of the host cells that produce the genes or the proteins.
Especially, the genes of the present invention are differentially expressed
only in
the normal tissues. For example, their gene expressions are slightly detected
or not
detected in the cancer tissues and the cancer cells such as the breast cancer
tissue, the
breast cancer cell line MCF-7, the leukemia cell, the leukemia cell line K562,
the liver
cancer tissue, the liver cancer cell line HepG2, the cervical cancer tissue,
the cervical
cancer cell line, the lung cancer tissue, the metastatic lung cancer tissue
and the lung
cancer cell lines (A549 and NCI-H358), but differentially increased only in
the normal
uterine tissues.
The cancer cell lines introduced with the genes of the present invention
showed
a high mortality, and therefore the genes of the present invention may be
effectively
used for treatment and prevention of the cancer.
61
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
Hereinafter, the present invention will be described in detail referring to
preferred examples. Therefore, the description proposed herein is just a
preferable
example for the purpose of illustrations only, not intended to limit the scope
of the
invention.
Reference Example: Separation of Total RNA
The total RNA samples were separated from fresh tissues or cultured cells
using
the RNeasy total RNA kit (Qiagen Inc., Germany), and the contaminated DNA was
then
removed from the RNA samples using the message clean kit (GenHunter Corp., MA,
U.S.).
Example 1: Separation of Total RNA and Differential Display of mRNA
A differential expression pattern of the gene was investigated in a normal
breast
tissue, a primary breast cancer tissue and a breast cancer cell line, as
follows.
A normal breast tissue sample was obtained from a breast cancer patient during
mastectomy, and a primary breast cancer tissue sample was obtained during
radical
mastectomy from a breast cancer patient who has not been subject to the
radiation
therapy and/or anticancer chemotherapy before the surgical treatment. MCF-7
(American Type Culture Collection; ATCC Number HTB-22) was used as the human
breast cancer cell line. The total RNAs were separated from these tissues and
cells in
the same manner as described in the reference example.
In order to conduct the mRNA differential display of the GIG 15, a bone marrow
tissue was also obtained from a normal person, and a primary leukemic bone
marrow
tissue was obtained from a leukemia patient who has not been previously
subject to the
62
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
anticancer chemotherapy and/or radiation therapy during the bone marrow
biopsy.
K-562 (American Type Cell Collection; ATCC Number CCL-243) was used as the
human chronic myelogenous leukemia cell line in the differential display
method. The
total RNAs were separated from these tissues and cells in the same manner as
described
in the reference example.
Meanwhile, a differential expression pattern of the gene was investigated in a
normal liver tissue, a primary liver cancer tissue and a liver cancer cell
line in the case
of the liver cancer-related genes, as follows.
A normal liver tissue sample and a liver cancer tissue sample were obtained
from a liver cancer patient during the tissue biopsy, and the liver cancer
cell line HepG2
(American Type Culture Collection; ATCC Number HB-8065) was used as the human
liver cancer cell line. The total RNAs were separated from these tissues and
cells in
the same manner as described in the reference example.
Also, a differential expression pattern of the gene of interest was measured
in a
normal exocervical tissue, a primary cervical cancer tissue and a cervical
cancer cell line,
as follows. A normal exocervical tissue sample was obtained from a patient
suffering
from a uterine myoma during hysterectomy, and a primary cervical tumor tissue
sample
and a metastatic iliac lymph node tumor tissue sample were obtained during
radical
hysterectomy from a patient who has not been subject to the radiation therapy
and/or
anticancer chemotherapy before surgical treatment. CUMC-6 (Kim, J. W. et al.,
Gynecol. Oncol. 62: 230-240, 1996) was used as the human cervical cancer cell
line.
The total RNA samples were separated from these tissues and cells in the same
manner
as described in the reference example. The total RNAs were separated from
these
63
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
tissues and cells in the same manner as described in the reference example.
Also, a differential expression pattern of the gene of interest was measured
in a
normal lung tissue, a primary lung cancer tissue, a metastatic lung cancer
tissue and a
lung cancer cell line, as follows. A normal lung tissue sample, a lung cancer
tissue
sample and a metastatic lung cancer tissue sample were obtained from a lung
cancer
patient during surgical operation. The lung cancer cell lines A549 (American
Type
Culture Collection; ATCC Number CCL-185) and NCI-H358 (American Type Culture
Collection; ATCC Number CRL-5807) were used as the human lung cancer cell
line.
The total RNAs were separated from these tissues and cells in the same manner
as
described in the reference example.
A RT-PCR reaction was carried out using each of the total RNA samples
separated from the tissues and the cells according to the modified method as
described
in the disclosure (Liang, P. and Pardee, A. B., Science, 257, 967-971 (1992);
and Liang,
P. et al., Cancer Res., 52, 6966-6968 (1993)), as follows.
1-1. GIG8
0.2 ,tg of the total RNA was reverse-transcribed with an anchored oligo-dT
primer of SEQ ID NO: 4 using a kit (a RNAimage kit, GenHunter), and then a PCR
reaction was carried out in the presence of 0.5 mM [ a-35S]-labeled dATP
(1,200
Ci/mmol) using the same anchored oligo-dT primer and a random 5'-13-mer primer
H-AP33 (RNAimage primer set 5, GenHunter Corporation, U.S.) of SEQ ID NO: 3.
The PCR reaction was conducted under the following conditions: the total 40
amplification cycles consisting of a denaturation step at 95 C for 40
seconds, an
annealing step at 40 C for 2 minutes and an extension step at 72 C for 40
seconds,
64
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
and followed by one extension step at 72 C for 5 minutes. The amplified
fragments
were electrophoresized in a 6 % polyacrylamide gel for DNA base sequence, and
then
autoradiographed.
FIG. 1 shows a PCR result using a random 5'-13-mer primer H-AP33 of SEQ ID
NO: 3 and an anchored oligo-dT primer of SEQ ID NO: 4. In FIG. 1, Lanes 1, 2
and 3
represent a normal breast tissue; Lanes 4, 5 and 6 represent a breast cancer
tissue; and
Lane 7 represents a breast cancer cell line MCF-7. As shown in FIG. 1, it was
confirmed that a 163-bp cDNA fragment (Base positions from 317 to 479 of the
full-length GIG8 gene sequence) was very rarely expressed in the breast cancer
tissue
and the breast cancer cell line, but differentially expressed at an increased
level only in
the normal lung tissue. This cDNA fragment was designated FC33.
A 163-bp band, FC33 fragment, was removed from the dried gell, boiled for 15
minutes to elute the cDNA, and a PCR reaction was then carried out under the
same said
condition using the same primer set as described above to re-amplify the FC33
cDNA,
except that the [ a-35S]-labeled dATP and the 20 It M dNTP were not used
herein.
The re-amplified cDNA fragment FC33 was cloned into an expression vector pGEM-
T
Easy using the TA cloning system (Promega), and then its DNA sequence was
determined using the Sequenase Version 2.0 DNA Sequencing System (United
States
Biochemical Co.).
1-2. GIG 10
0.2 /yg of the total RNA was reverse-transcribed with an anchored oligo-dT
primer of SEQ ID NO: 8 using a kit (a RNAimage kit, GenHunter), and then a PCR
reaction was carried out in the presence of 0.5 mM [ a-35S]-labeled dATP
(1,200
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
Ci/mmol) using the same anchored oligo-dT primer and a random 5'-13-mer primer
H-AP10 (RNAimage primer set 2, GenHunter Corporation, U.S.) of SEQ ID NO: 7.
The PCR reaction was conducted under the following conditions: the total 40
amplification cycles consisting of a denaturation step at 95 C for 40 seconds,
an
annealing step at 40 C for 2 minutes and an extension step at 72 C for 40
seconds,
and followed by one extension step at 72 C for 5 minutes. The amplified
fragments
were electrophoresized in a 6 % polyacrylamide gel for DNA base sequence, and
then
autoradiographed.
FIG. 2 shows a PCR result using a random 5'-13-mer primer H-AP10 of SEQ ID
NO: 7 and an anchored oligo-dT primer of SEQ ID NO: 8. In FIG. 2, Lanes 1, 2
and 3
represent a normal breast tissue; Lanes 4, 5 and 6 represent a breast cancer
tissue; and
Lane 7 represents a breast cancer cell line MCF-7. As shown in FIG. 2, it was
confirmed that a 321-bp cDNA fragment (Base positions from 1,716 to 2,036 of
the
full-length GIG10 gene sequence) was very rarely expressed in the breast
cancer tissue
and the breast cancer cell line, but differentially expressed at an increased
level only in
the normal breast tissue. This cDNA fragment was designated FC42.
A 321-bp band, FC42 fragment, was removed from the dried gell, boiled for 15
minutes to elute the cDNA, and a PCR reaction was then carried out under the
same said
condition using the same primer set as described above to re-amplify the FC42
cDNA,
except that the [ a 35S]-labeled dATP and the 20 u M dNTP were not used
herein.
The re-amplified cDNA fragment FC42 was cloned into an expression vector pGEM-
T
Easy using the TA cloning system (Promega), and then its DNA sequence was
determined using the Sequenase Version 2.0 DNA Sequencing System (United
States
66
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
Biochemical Co.).
1-3. GIG13
0.2 ttg of the total RNA was reverse-transcribed with an anchored oligo-dT
primer of SEQ ID NO: 12 using a kit (a RNAimage kit, GenHunter), and then a
PCR
reaction was carried out in the presence of 0.5 mM [ a-35S]-labeled dATP
(1,200
Ci/mmol) using the same anchored oligo-dT primer and a random 5'-13-mer primer
H-AP5 (RNAimage primer set 1, GenHunter Corporation, U.S.) of SEQ ID NO: 11.
The PCR reaction was conducted under the following conditions: the total 40
amplification cycles consisting of a denaturation step at 95 C for 40
seconds, an
annealing step at 40 C for 2 minutes and an extension step at 72 C for 40
seconds,
and followed by one extension step at 72 C for 5 minutes. The amplified
fragments
were electrophoresized in a 6 % polyacrylamide gel for DNA base sequence, and
then
autoradiographed.
FIG. 3 shows a PCR result using a random 5'-13-mer primer H-AP5 of SEQ ID
NO: 11 and an anchored oligo-dT primer of SEQ ID NO: 12. In FIG. 3, Lanes 1, 2
and
3 represent a normal breast tissue; Lanes 4, 5 and 6 represent a breast cancer
tissue; and
Lane 7 represents a breast cancer cell line MCF-7. As shown in FIG. 3, it was
confirmed that a 347-bp cDNA fragment (Base positions from 3,253 to 3,599 of
the
full-length GIG13 gene sequence) was very rarely expressed in the breast
cancer tissue
and the breast cancer cell line, but differentially expressed at an increased
level only in
the normal breast tissue. This cDNA fragment was designated FC59.
A 347-bp band, FC59 fragment, was removed from the dried gell, boiled for 15
minutes to elute the cDNA, and a PCR reaction was then carried out under the
same said
67
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
condition using the same primer set as described above to re-amplify the FC59
cDNA,
except that the [ a-35S]-labeled dATP and the 20 11 M dNTP were not used
herein.
The re-amplified cDNA fragment FC59 was cloned into an expression vector pGEM-
T
Easy using the TA cloning system (Promega), and then its DNA sequence was
determined using the Sequenase Version 2.0 DNA Sequencing System (United
States
Biochemical Co.).
1-4. GIG15
In order to conduct the mRNA differential display, a bone marrow tissue was
also obtained from a normal person, and a primary leukemic bone marrow tissue
was
obtained from a leukemia patient who has not been previously subject to the
anticancer
chemotherapy and/or radiation therapy during the bone marrow biopsy. K-562
(American Type Cell Collection; ATCC Number CCL-243) was used as the human
chronic myelogenous leukemia cell line in the differential display method. The
total
RNAs were separated from these tissues and cells in the same manner as
described in
the reference example.
A RT-PCR reaction was carried out using each of the total RNA samples
separated from the tissues and the cells according to the modified method as
described
in the disclosure (Liang, P. and Pardee, A. B., Science, 257, 967-971 (1992);
and Liang,
P. et al., Cancer Res., 52, 6966-6968 (1993)), as follows. 0.2 tig of the
total RNA
was reverse-transcribed with an anchored oligo-dT primer of SEQ ID NO: 16
using a kit
(a RNAimage kit, GenHunter), and then a PCR reaction was carried out in the
presence
of 0.5 mM [ a-35S]-labeled dATP (1,200 Ci/mmol) using the same anchored oligo-
dT
primer and a random 5'-13-mer primer H-AP2 (RNAimage primer set 5, GenHunter
68
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
Corporation, U.S.) of SEQ ID NO: 15. The PCR reaction was conducted under the
following conditions: the total 40 amplification cycles consisting of a
denaturation step
at 95 C for 40 seconds, an annealing step at 40 C for 2 minutes and an
extension
step at 72 C for 40 seconds, and followed by one extension step at 72 C for 5
minutes. The amplified fragments were electrophoresized in a 6 %
polyacrylamide gel
for DNA base sequence, and then autoradiographed.
FIG. 4 shows a PCR result using a random 5'-13-mer primer H-AP2 of SEQ ID
NO: 15 and an anchored oligo-dT primer of SEQ ID NO: 16. As shown in FIG. 4,
it
was confirmed that the gene is expressed at a different level in the normal
bone marrow
tissue and the leukemia cell and K-562 cell using the differential display
(DD) method.
As seen in FIG. 4, a 133-bp cDNA fragment, GV2 (Base positions from 212 to 344
of
the full-length GIG15 gene sequence), was very rarely expressed in the
leukemia tissue
and the K-562 cell, but highly expressed only in the normal bone marrow
tissue. This
cDNA fragment was designated GV2. A 133-bp band, GV2 fragment, was removed
from the dried gell, boiled for 15 minutes to elute the cDNA, and a PCR
reaction was
then carried out under the same said condition using the same primer set as
described
above to re-amplify the GV2 cDNA, except that the [ a-35S]-labeled dATP and
the 20
l.t M dNTP were not used herein. The re-amplified cDNA fragment GV2 was cloned
into an expression vector pGEM-T Easy using the TA cloning system (Promega),
and
then its DNA sequence was determined using the Sequenase Version 2.0 DNA
Sequencing System (United States Biochemical Co.).
1-5. GIG16
A differential expression pattern of the gene was investigated in a normal
liver
69
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
tissue, a primary liver cancer tissue and a liver cancer cell line, as
follows.
A normal liver tissue sample and a liver cancer tissue sample were obtained
from a liver cancer patient during the tissue biopsy, and the liver cancer
cell line HepG2
(American Type Culture Collection; ATCC Number HB-8065) was used as the human
liver cancer cell line. The total RNAs were separated from these tissues and
cells in
the same manner as described in the reference example.
A RT-PCR reaction was carried out using each of the total RNA samples
separated from the tissues and the cells according to the modified method as
described
in the disclosure (Liang, P. and Pardee, A. B., Science, 257, 967-971 (1992);
and Liang,
P. et al., Cancer Res., 52, 6966-6968 (1993)), as follows. 0.2 /.tg of the
total RNA
was reverse-transcribed with an anchored oligo-dT primer of SEQ ID NO: 20
using a kit
(a RNAimage kit, GenHunter), and then a PCR reaction was carried out in the
presence
of 0.5 mM [ a 35S]-labeled dATP (1,200 Ci/mmol) using the same anchored oligo-
dT
primer and a random 5'-13-mer primer H-AP8 (RNAimage primer set 1, GenHunter
Corporation, U.S.) of SEQ ID NO: 19. The PCR reaction was conducted under the
following conditions: the total 40 amplification cycles consisting of a
denaturation step
at 95 C for 40 seconds, an annealing step at 40 'C for 2 minutes and an
extension
step at 72 C for 40 seconds, and followed by one extension step at 72 C for 5
minutes. The amplified fragments were electrophoresized in a 6 %
polyacrylamide gel
for DNA base sequence, and then autoradiographed.
FIG. 5 shows a PCR result using a random 5'-13-mer primer H-AP8 of SEQ ID
NO: 19 and an anchored oligo-dT primer of SEQ ID NO: 20. In FIG. 5, Lanes 1, 2
and
3 represent a normal liver tissue; Lanes 4, 5 and 6 represent a liver cancer
tissue; and
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
Lane 7 represents a liver cancer cell line HepG2. - As shown in FIG. 5, it was
F
confirmed that a 213-bp cDNA fragment (Base positions from 867 to 1,079 of the
full-length GIG16 gene sequence) was not expressed in the liver cancer tissue
and the
liver cancer cell line, but differentially expressed only in the normal liver
tissue. This
cDNA fragment was designated HP8.
A 213-bp band, HP8 fragment, was removed from the dried gell, boiled for 15
minutes to elute the cDNA, and a PCR reaction was then carried out under the
same said
condition using the same primer set as described above to re-amplify the HP8
cDNA,
except that the [ a-35S]-labeled dATP and the 20 11 M dNTP were not used
herein.
The re-amplified cDNA fragment HP8 was cloned into an expression vector pGEM-T
Easy using the TA cloning system (Promega), and then its DNA sequence was
determined using the Sequenase Version 2.0 DNA Sequencing System (United
States
Biochemical Co.).
1-6. GIG24
A differential expression pattern of the gene was investigated in a normal
liver
tissue, a primary liver cancer tissue and a liver cancer cell line, as
follows.
A normal liver tissue sample and a liver cancer tissue sample were obtained
from a liver cancer patient during the tissue biopsy, and the liver cancer
cell line HepG2
(American Type Culture Collection; ATCC Number HB-8065) was used as the human
liver cancer cell line. The total RNAs were separated from these tissues and
cells in
the same manner as described in the reference example.
A RT-PCR reaction was carried out using each of the total RNA samples
separated from the tissues and the cells according to the modified method as
described
71
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
in the disclosure (Liang, P. and Pardee, A. B., Science, 257, 967-971 (1992);
and Liang,
P. et al., Cancer Res., 52, 6966-6968 (1993)), as follows. 0.2 ag of the total
RNA
was reverse-transcribed with an anchored oligo-dT primer of SEQ ID NO: 24
using a kit
(a RNAimage kit, GenHunter), and then a PCR reaction was carried out in the
presence
of 0.5 mM [ a-35S]-labeled dATP (1,200 Ci/mmol) using the same anchored oligo-
dT
primer and a random 5'-13-mer primer H-AP7 (RNAimage primer set 1, GenHunter
Corporation, U.S.) of SEQ ID NO: 23. The PCR reaction was conducted under the
following conditions: the total 40 amplification cycles consisting of a
denaturation step
at 95 C for 40 seconds, an annealing step at 40 'C for 2 minutes and an
extension
step at 72 C for 40 seconds, and followed by one extension step at 72 C for 5
minutes. The amplified fragments were electrophoresized in a 6 %
polyacrylamide gel
for DNA base sequence, and then autoradiographed.
FIG. 6 shows a PCR result using a random 5'-13-mer primer H-AP7 of SEQ ID
NO: 23 and an anchored oligo-dT primer of SEQ ID NO: 24. In FIG. 6, Lanes 1, 2
and
3 represent a normal liver tissue; Lanes 4, 5 and 6 represent a liver cancer
tissue; and
Lane 7 represents a liver cancer cell line HepG2. As shown in FIG. 6, it was
confirmed that a 221-bp cDNA fragment (Base positions from 1,057 to 1,277 of
the
full-length GIG42 gene sequence) was not expressed in the liver cancer tissue
and the
liver cancer cell line, but differentially expressed only in the normal liver
tissue. This
cDNA fragment was designated HP71.
A 221-bp band, HP71 fragment, was removed from the dried gell, boiled for 15
minutes to elute the cDNA, and a PCR reaction was then carried out under the
same said
condition using the same primer set as described above to re-amplify the HP71
cDNA,
72
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
except that the [ a-35S]-labeled dATP and the 20 u M dNTP were not used
herein.
The re-amplified cDNA fragment HP71 was cloned into an expression vector pGEM-
T
Easy using the TA cloning system (Promega), and then its DNA sequence was
determined using the Sequenase Version 2.0 DNA Sequencing System (United
States
Biochemical Co.).
1-7. GIG26
0.2 ug of the total RNA was reverse-transcribed with an anchored oligo-dT
primer of SEQ ID NO: 28 using a kit (a RNAimage kit, GenHunter), and then a
PCR
reaction was carried out in the presence of 0.5 mM [ a-35S]-labeled dATP
(1,200
Ci/mmol) using the same anchored oligo-dT primer and a random 5'-13-mer primer
H-AP11 (RNAimage primer set 1, GenHunter Corporation, U.S.) of SEQ ID NO: 27.
The PCR reaction was conducted under the following conditions: the total 40
amplification cycles consisting of a denaturation step at 95 C for 40 seconds,
an
annealing step at 40 'C for 2 minutes and an extension step at 72 C for 40
seconds,
and followed by one extension step at 72 C for 5 minutes. The amplified
fragments
were electrophoresized in a 6 % polyacrylamide gel for DNA base sequence, and
then
autoradiographed.
FIG. 7 shows a PCR result using a random 5'-13-mer primer H-AP11 of SEQ ID
NO: 27 and an anchored oligo-dT primer of SEQ ID NO: 28. In FIG. 7, Lanes 1, 2
and
3 represent a normal liver tissue; Lanes 4, 5 and 6 represent a liver cancer
tissue; and
Lane 7 represents a liver cancer cell line HepG2. As shown in FIG. 7, it was
confirmed that a 204-bp cDNA fragment (Base positions from 1,036 to 1,239 of
the
full-length GIG26 gene sequence) was not expressed in the liver cancer tissue
and the
73
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
liver cancer cell line, but differentially expressed only in the normal liver
tissue. This
cDNA fragment was designated HP 115.
A 204-bp band, HP 115 fragment, was removed from the dried gell, boiled for 15
minutes to elute the cDNA, and a PCR reaction was then carried out under the
same said
condition using the same primer set as described above to re-amplify the HP
115 cDNA,
except that the [ a-35S]-labeled dATP and the 20 lt M dNTP were not used
herein.
The re-amplified cDNA fragment HP 115 was cloned into an expression vector
pGEM-T
Easy using the TA cloning system (Promega), and then its DNA sequence was
determined using the Sequenase Version 2.0 DNA Sequencing System (United
States
Biochemical Co.).
1-8. GIG29
0.2 gg of the total RNA was reverse-transcribed with an anchored oligo-dT
primer of SEQ ID NO: 32 using a kit (a RNAimage kit, GenHunter), and then a
PCR
reaction was carried out in the presence of 0.5 mM [ a-35S]-labeled dATP
(1,200
Ci/mmol) using the same anchored oligo-dT primer and a random 5'-13-mer primer
H-AP3 (RNAimage primer set 1, GenHunter Corporation, U.S.) of SEQ ID NO: 31.
The PCR reaction was conducted under the following conditions: the total 40
amplification cycles consisting of a denaturation step at 95 C for 40 seconds,
an
annealing step at 40 C for 2 minutes and an extension step at 72 C for 40
seconds,
and followed by one extension step at 72 C for 5 minutes. The amplified
fragments
were electrophoresized in a 6 % polyacrylamide gel for DNA base sequence, and
then
autoradiographed.
FIG. 8 shows a PCR result using a random 5'-13-mer primer H-AP3 of SEQ ID
74
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
NO: 31 and an anchored oligo-dT primer of SEQ ID NO: 32. In FIG. 8, Lanes 1, 2
and
3 represent a normal liver tissue; Lanes 4, 5 and 6 represent a liver cancer
tissue; and
Lane 7 represents a liver cancer cell line HepG2. As shown in FIG. 8, it was
confirmed that a 277-bp cDNA fragment (Base positions from 823 to 1,099 of the
full-length GIG29 gene sequence) was not expressed in the liver cancer tissue
and the
liver cancer cell line, but differentially expressed only in the normal liver
tissue. This
cDNA fragment was designated HP3.
A 277-bp band, HP3 fragment, was removed from the dried gell, boiled for 15
minutes to elute the cDNA, and a PCR reaction was then carried out under the
same said
condition using the same primer set as described above to re-amplify the HP3
cDNA,
except that the [ a 35S]-labeled dATP and the 20 u M dNTP were not used
herein.
The re-amplified cDNA fragment HP3 was cloned into an expression vector pGEM-T
Easy using the TA cloning system (Promega), and then its DNA sequence was
determined using the Sequenase Version 2.0 DNA Sequencing System (United
States
Biochemical Co.).
1-9. GIG30
0.2 ,ug of the total RNA was reverse-transcribed with an anchored oligo-dT
primer of SEQ ID NO: 36 using a kit (a RNAimage kit, GenHunter), and then a
PCR
reaction was carried out in the presence of 0.5 mM [ a-35S]-labeled dATP
(1,200
Ci/mmol) using the same anchored oligo-dT primer and a random 5'-13-mer primer
H-AP4 (RNAimage primer set 1, GenHunter Corporation, U.S.) of SEQ ID NO: 35.
The PCR reaction was conducted under the following conditions: the total 40
amplification cycles consisting of a denaturation step at 95 C for 40 seconds,
an
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
annealing step at 40 C for 2 minutes and an extension step at 72 'C for 40
seconds,
and followed by one extension step at 72 'C for 5 minutes. The amplified
fragments
were electrophoresized in a 6 % polyacrylamide gel for DNA base sequence, and
then
autoradiographed.
FIG. 9 shows a PCR result using a random 5'-13-mer primer H-AP4 of SEQ ID
NO: 35 and an anchored oligo-dT primer of SEQ ID NO: 36. In FIG. 9, Lanes 1, 2
and
3 represent a normal breast tissiie; Lanes 4, 5 and 6 represent a breast
cancer tissue; and
Lane 7 represents a breast cancer cell line MCF-7. As shown in FIG. 9, it was
confirmed that a 278-bp cDNA fragment (Base positions from 1,462 to 1,739 of
the
full-length GIG30 gene sequence) was very rarely expressed in the breast
cancer tissue
and the breast cancer cell line, but differentially expressed at an increased
level only in
the normal breast tissue. This cDNA fragment was designated FC48.
A 278-bp band, FC48 fragment, was removed from the dried gell, boiled for 15
minutes to elute the cDNA, and a PCR reaction was then carried out under the
same said
condition using the same primer set as described above to re-amplify the FC48
cDNA,
except that the [ a 35S]-labeled dATP and the 20 u M dNTP were not used
herein.
The re-amplified cDNA fragment FC48 was cloned into an expression vector pGEM-
T
Easy using the TA cloning system (Promega), and then its DNA sequence was
determined using the Sequenase Version 2.0 DNA Sequencing System (United
States
Biochemical Co.).
1-10. GIG32
0.2 ug of the total RNA was reverse-transcribed with an anchored oligo-dT
primer of SEQ ID NO: 40 using a kit (a RNAimage kit, GenHunter), and then a
PCR
76
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
reaction was carried out in the presence of 0.5 mM [ a-35S]-labeled dATP
(1,200
Ci/mmol) using the same anchored oligo-dT primer and a random 5'-13-mer primer
H-AP8 (RNAimage primer set 1, GenHunter Corporation, U.S.) of SEQ ID NO: 39.
The PCR reaction was conducted under the following conditions: the total 40
amplification cycles consisting of a denaturation step at 95 'C for 40
seconds, an
annealing step at 40 C for 2 minutes and an extension step at 72 C for 40
seconds,
and followed by one extension step at 72 C for 5 minutes. The amplified
fragments
were electrophoresized in a 6 % polyacrylamide gel for DNA base sequence, and
then
autoradiographed.
FIG. 10 shows a PCR result using a random 5'-13-mer primer H-AP8 of SEQ ID
NO: 39 and an anchored oligo-dT primer of SEQ ID NO: 40. In FIG. 10, Lanes 1,
2
and 3 represent a normal breast tissue; Lanes 4, 5 and 6 represent a breast
cancer tissue;
and Lane 7 represents a breast cancer cell line MCF-7. As shown in FIG. 10, it
was
confirmed that a 172-bp cDNA fragment tissue (Base positions from 428 to 599
of the
full-length GIG32 gene sequence) was very rarely expressed in the breast
cancer tissue
and the breast cancer cell line, but differentially expressed at an increased
level only in
the normal breast. This cDNA fragment was designated FC82.
A 172-bp band, FC82 fragment, was removed from the dried gell, boiled for 15
minutes to elute the cDNA, and a PCR reaction was then carried out under the
same said
condition using the same primer set as described above to re-amplify the FC82
cDNA,
except that the [ a 35S]-labeled dATP and the 20 u M dNTP were not used
herein.
The re-amplified cDNA fragment FC82 was cloned into an expression vector pGEM-
T
Easy using the TA cloning system (Promega), and then its DNA sequence was
77
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
determined using the Sequenase Version 2.0 DNA Sequencing System (United
States
Biochemical Co.).
1-11. GIG33
0.2 gg of the total RNA was reverse-transcribed with an anchored oligo-dT
primer of SEQ ID NO: 44 using a kit (a RNAimage kit, GenHunter), and then a
PCR
reaction was carried out in the presence of 0.5 mM [ a-35S]-labeled dATP
(1,200
Ci/mmol) using the same anchored oligo-dT primer and a random 5'-13-mer primer
H-AP33 (RNAimage primer set 5, GenHunter Corporation, U.S.) of SEQ ID NO: 43.
The PCR reaction was conducted under the following conditions: the total 40
amplification cycles consisting of a denaturation step at 95 C for 40 seconds,
an
annealing step at 40 C for 2 minutes and an extension step at 72 C for 40
seconds,
and followed by one extension step at 72 C for 5 minutes. The amplified
fragments
were electrophoresized in a 6 % polyacrylamide gel for DNA base sequence, and
then
autoradiographed.
FIG. l l shows a PCR result using a random 5'-13-mer primer H-AP33 of SEQ
ID NO: 43 and an anchored oligo-dT primer of SEQ ID NO: 44. In FIG. 11, Lanes
1, 2
and 3 represent a normal breast tissue; Lanes 4, 5 and 6 represent a breast
cancer tissue;
and Lane 7 represents a breast cancer cell line MCF-7. As shown in FIG. 11, it
was
confirmed that a 182-bp cDNA fragment (Base positions from 216 to 397 of the
full-length GIG33 gene sequence) was very rarely expressed in the breast
cancer tissue
and the breast cancer cell line, but differentially expressed at an increased
level only in
the normal breast tissue. This cDNA fragment was designated FC86.
A 182-bp band, FC86 fragment, was removed from the dried gell, boiled for 15
78
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
minutes to elute the cDNA, and a PCR reaction was then carried out under the
same said
condition using the same primer set as described above to re-amplify the FC86
cDNA,
except that the [ a 35S]-labeled dATP and the 20 u M dNTP were not used
herein.
The re-amplified cDNA fragment FC86 was cloned into an expression vector pGEM-
T
Easy using the TA cloning system (Promega), and then its DNA sequence was
determined using the Sequenase Version 2.0 DNA Sequencing System (United
States
Biochemical Co.).
1-12. GIG34
0.2 gg of the total RNA was reverse-transcribed with an anchored oligo-dT
primer of SEQ ID NO: 48 using a kit (a RNAimage kit, GenHunter), and then a
PCR
reaction was carried out in the presence of 0.5 mM [ a-35S]-labeled dATP
(1,200
Ci/mmol) using the same anchored oligo-dT primer and a random 5'-13-mer primer
H-AP35 (RNAimage primer set 5, GenHunter Corporation, U.S.) of SEQ ID NO: 47.
The PCR reaction was conducted under the following conditions: the total 40
amplification cycles consisting of a denaturation step at 95 C for 40
seconds, an
annealing step at 40 C for 2 minutes and an extension step at 72 C for 40
seconds,
and followed by one extension step at 72 C for 5 minutes. The amplified
fragments
were electrophoresized in a 6 % polyacrylamide gel for DNA base sequence, and
then
autoradiographed.
FIG. 12 shows a PCR result using a random 5'-13-mer primer H-AP35 of SEQ
ID NO: 47 and an anchored oligo-dT primer of SEQ ID NO: 48. In FIG. 12, Lanes
1, 2
and 3 represent a normal breast tissue; Lanes 4, 5 and 6 represent a breast
cancer tissue;
and Lane 7 represents a breast cancer cell line MCF-7. As shown in FIG. 12, it
was
79
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
confirmed that a 205-bp cDNA fragment (Base positions from 343 to 547 of the
full-length GIG34 gene sequence) was very rarely expressed in the breast
cancer tissue
and the breast cancer cell line, but differentially expressed at an increased
level only in
the normal breast tissue. This cDNA fragment was designated FC35.
A 205-bp band, FC42 fragment, was removed from the dried gell, boiled for 15
minutes to elute the cDNA, and a PCR reaction was then carried out under the
same said
condition using the same primer set as described above to re-amplify the FC35
cDNA,
except that the [ a-35S]-labeled dATP and the 20 It M dNTP were not used
herein.
The re-amplified cDNA fragment FC35 was cloned into an expression vector pGEM-
T
Easy using the TA cloning system (Promega), and then its DNA sequence was
determined using the Sequenase Version 2.0 DNA Sequencing System (United
States
Biochemical Co.).
1-13. GIG35
0.2 ug of the total RNA was reverse-transcribed with an anchored oligo-dT
primer of SEQ ID NO: 52 using a kit (a RNAimage kit, GenHunter), and then a
PCR
reaction was carried out in the presence of 0.5 mM [ a-35S]-labeled dATP
(1,200
Ci/mmol) using the same anchored oligo-dT primer and a random 5'-13-mer primer
H-AP3 (RNAimage primer set 1, GenHunter Corporation, U.S.) of SEQ ID NO: 51.
The PCR reaction was conducted under the following conditions: the total 40
amplification cycles consisting of a denaturation step at 95 C for 40
seconds, an
annealing step at 40 C for 2 minutes and an extension step at 72 C for 40
seconds,
and followed by one extension step at 72 C for 5 minutes. The amplified
fragments
were electrophoresized in a 6 % polyacrylamide gel for DNA base sequence, and
then
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
autoradiographed.
FIG. 13 shows a PCR result using a random 5'-13-mer primer H-AP3 of SEQ ID
NO: 51 and an anchored oligo-dT primer of SEQ ID NO: 52. In FIG. 13, Lanes 1,
2
and 3 represent a normal breast tissue; Lanes 4, 5 and 6 represent a breast
cancer tissue;
and Lane 7 represents a breast cancer cell line MCF-7. As shown in FIG. 13, it
was
confirmed that a 212-bp cDNA fragment (Base positions from 1,108 to 1,319 of
the
full-length GIG35 gene sequence) was very rarely expressed in the breast
cancer tissue
and the breast cancer cell line, but differentially expressed at an increased
level only in
the normal breast tissue. This cDNA fragment was designated FC38.
A 212-bp band, FC38 fragment, was removed from the dried gell, boiled for 15
minutes to elute the cDNA, and a PCR reaction was then carried out under the
same said
condition using the same primer set as described above to re-amplify the FC38
cDNA,
except that the [ a 35S]-labeled dATP and the 20 ji M dNTP were not used
herein.
The re-amplified cDNA fragment FC38 was cloned into an expression vector pGEM-
T
Easy using the TA cloning system (Promega), and then its DNA sequence was
determined using the Sequenase Version 2.0 DNA Sequencing System (United
States
Biochemical Co.).
1-14. GIG38
0.2 ug of the total RNA was reverse-transcribed with an anchored oligo-dT
primer of SEQ ID NO: 56 using a kit (a RNAimage kit, GenHunter), and then a
PCR
reaction was carried out in the presence of 0.5 mM [ a-35S]-labeled dATP
(1,200
Ci/mmol) using the same anchored oligo-dT primer and a random 5'-13-mer primer
H-AP12 (RNAimage primer set 2, GenHunter Corporation, U.S.) of SEQ ID NO: 55.
81
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
The PCR reaction was conducted under the following conditions: the total 40
amplification cycles consisting of a denaturation step at 95 C for 40 seconds,
an
annealing step at 40 C for 2 minutes and an extension step at 72 C for 40
seconds,
and followed by one extension step at 72 C for 5 minutes. The amplified
fragments
were electrophoresized in a 6 % polyacrylamide gel for DNA base sequence, and
then
autoradiographed.
FIG. 14 shows a PCR result using a random 5'-13-mer primer H-AP12 of SEQ
ID NO: 55 and an anchored oligo-dT primer of SEQ ID NO: 56. In FIG. 14, Lanes
1, 2
and 3 represent a normal breast tissue; Lanes 4, 5 and 6 represent a breast
cancer tissue;
and Lane 7 represents a breast cancer cell line MCF-7. As shown in FIG. 14, it
was
confirmed that a 172-bp cDNA fragment (Base positions from 328 to 499 of the
full-length GIG38 gene sequence) was very rarely expressed in the breast
cancer tissue
and the breast cancer cell line, but differentially expressed at an increased
level only in
the normal breast tissue. This cDNA fragment was designated FC122.
A 172-bp band, FC 122 fragment, was removed from the dried gell, boiled for 15
minutes to elute the cDNA, and a PCR reaction was then carried out under the
same said
condition using the same primer set as described above to re-amplify the FC
122 cDNA,
except that the [ a-35S]-labeled dATP and the 20 I.t M dNTP were not used
herein.
The re-amplified cDNA fragment FC122 was cloned into an expression vector pGEM-
T
Easy using the TA cloning system (Promega), and then its DNA sequence was
determined using the Sequenase Version 2.0 DNA Sequencing System (United
States
Biochemical Co.).
1-15. GIG39
82
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
0.2 gg of the total RNA was reverse-transcribed with an anchored oligo-dT
primer of SEQ ID NO: 60 using a kit (a RNAimage kit, GenHunter), and then a
PCR
reaction was carried out in the presence of 0.5 mM [ a 35S]-labeled dATP
(1,200
Ci/mmol) using the same anchored oligo-dT primer and a random 5'-13-mer primer
H-AP12 (RNAimage primer set 2, GenHunter Corporation, U.S.) of SEQ ID NO: 59.
The PCR reaction was conducted under the following conditions: the total 40
amplification cycles consisting of a denaturation step at 95 C for 40
seconds, an
annealing step at 40 C for 2 minutes and an extension step at 72 C for 40
seconds,
and followed by one extension step at 72 'C for 5 minutes. The amplified
fragments
were electrophoresized in a 6 % polyacrylamide gel for DNA base sequence, and
then
autoradiographed.
FIG. 15 shows a PCR result using a random 5'-13-mer primer H-AP12 of SEQ
ID NO: 59 and an anchored oligo-dT primer of SEQ ID NO: 60. In FIG. 15, Lanes
1, 2
and 3 represent a normal breast tissue; Lanes 4, 5 and 6 represent a breast
cancer tissue;
and Lane 7 represents a breast cancer cell line MCF-7. As shown in FIG. 15, it
was
confirmed that a 327-bp cDNA fragment (Base positions from 2,533 to 2,859 of
the
full-length GIG39 gene sequence) was very rarely expressed in the breast
cancer tissue
and the breast cancer cell line, but differentially expressed at an increased
level only in
the normal breast tissue. This cDNA fragment was designated FC126.
A 327-bp band, FC126 fragment, was removed from the dried gell, boiled for 15
minutes to elute the cDNA, and a PCR reaction was then carried out under the
same said
condition using the same primer set as described above to re-amplify the FC126
cDNA,
except that the [ a-35S]-labeled dATP and the 20 lt M dNTP were not used
herein.
83
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
The re-amplified cDNA fragment FC 126 was cloned into an expression vector
pGEM-T
Easy using the TA cloning system (Promega), and then its DNA sequence was
determined using the Sequenase Version 2.0 DNA Sequencing System (United
States
Biochemical Co.).
1-16. GIG40
0.2 gg of the total RNA was reverse-transcribed with an anchored oligo-dT
primer of SEQ ID NO: 64 using a kit (a RNAimage kit, GenHunter), and then a
PCR
reaction was carried out in the presence of 0.5 mM [ a 35S]-labeled dATP
(1,200
Ci/mmol) using the same anchored oligo-dT primer and a random 5'-13-mer primer
H-AP7 (RNAimage primer set 1, GenHunter Corporation, U.S.) of SEQ ID NO: 63.
The PCR reaction was conducted under the following conditions: the total 40
amplification cycles consisting of a denaturation step at 95 C for 40
seconds, an
annealing step at 40 C for 2 minutes and an extension step at 72 C for 40
seconds,
and followed by one extension step at 72 C for 5 minutes. The amplified
fragments
were electrophoresized in a 6 % polyacrylamide gel for DNA base sequence, and
then
autoradiographed.
FIG. 16 shows a PCR result using a random 5'-13-mer primer H-AP7 of SEQ ID
NO: 63 and an anchored oligo-dT primer of SEQ ID NO: 64. In FIG. 16, Lanes 1,
2
and 3 represent a normal liver tissue; Lanes 4, 5 and 6 represent a liver
cancer tissue;
and Lane 7 represents a liver cancer cell line HepG2. As shown in FIG. 16, it
was
confirmed that a 275-bp cDNA fragment (Base positions from 3,112 to 3,386 of
the
full-length GIG40 gene sequence) was rarely expressed in the liver cancer
tissue and the
liver cancer cell line, but differentially expressed only in the normal liver
tissue. This
84
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
cDNA fragment was designated HP79.
A 275-bp band, HP79 fragment, was removed from the dried gell, boiled for 15
minutes to elute the cDNA, and a PCR reaction was then carried out under the
same said
condition using the same primer set as described above to re-amplify the HP79
cDNA,
except that the [ a-35S]-labeled dATP and the 20 u M dNTP were not used
herein.
The re-amplified cDNA fragment HP79 was cloned into an expression vector pGEM-
T
Easy using the TA cloning system (Promega), and then its DNA sequence was
determined using the Sequenase Version 2.0 DNA Sequencing System (United
States
Biochemical Co.).
1-17. GIG42
0.2 /Lg of the total RNA was reverse-transcribed with an anchored oligo-dT
primer of SEQ ID NO: 68 using a kit (a RNAimage kit, GenHunter), and then a
PCR
reaction was carried out in the presence of 0.5 mM [ a-35S]-labeled dATP
(1,200
Ci/mmol) using the same anchored oligo-dT primer and a random 5'-13-mer primer
H-AP8 (RNAimage primer set 1, GenHunter Corporation, U.S.) of SEQ ID NO: 67.
The PCR reaction was conducted under the following conditions: the total 40
amplification cycles consisting of a denaturation step at 95 C for 40 seconds,
an
annealing step at 40 C for 2 minutes and an extension step at 72 C for 40
seconds,
and followed by one extension step at 72 C for 5 minutes. The amplified
fragments
were electrophoresized in a 6 % polyacrylamide gel for DNA base sequence, and
then
autoradiographed.
FIG. 17 shows a PCR result using a random 5'-13-mer primer H-AP8 of SEQ ID
NO: 67 and an anchored oligo-dT primer of SEQ ID NO: 68. In FIG. 17, Lanes 1,
2
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
and 3 represent a normal liver tissue; Lanes 4, 5 and 6 represent a liver
cancer tissue;
and Lane 7 represents a liver cancer cell line HepG2. As shown in FIG. 17, it
was
confirmed that a 327-bp cDNA fragment (Base positions from 1,473 to 1,799 of
the
full-length GIG42 gene sequence) was not expressed in the liver cancer tissue
and the
liver cancer cell line, but differentially expressed only in the normal liver
tissue. This
cDNA fragment was designated HP85.
A 327-bp band, HP85 fragment, was removed from the dried gell, boiled for 15
minutes to elute the cDNA, and a PCR reaction was then carried out under the
same said
condition using the same primer set as described above to re-amplify the HP85
cDNA,
except that the [ a-35S]-labeled dATP and the 20 lt M dNTP were not used
herein.
The re-amplified cDNA fragment HP85 was cloned into an expression vector pGEM-
T
Easy using the TA cloning system (Promega), and then its DNA sequence was
determined using the Sequenase Version 2.0 DNA Sequencing System (United
States
Biochemical Co.).
1-18. GIG43
0.2 ,ug of the total RNA was reverse-transcribed with an anchored oligo-dT
primer of SEQ ID NO: 72 using a kit (a RNAimage kit, GenHunter), and then a
PCR
reaction was carried out in the presence of 0.5 mM [ a-35S]-labeled dATP
(1,200
Ci/mmol) using the same anchored oligo-dT primer and a random 5'-13-mer primer
H-AP10 (RNAimage primer set 5, GenHunter Corporation, U.S.) of SEQ ID NO: 71.
The PCR reaction was conducted under the following conditions: the total 40
amplification cycles consisting of a denaturation step at 95 C for 40 seconds,
an
annealing step at 40 C for 2 minutes and an extension step at 72 C for 40
seconds,
86
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
and followed by one extension step at 72 C for 5 minutes. The amplified
fragments
were electrophoresized in a 6 % polyacrylamide gel for DNA base sequence, and
then
autoradiographed.
FIG. 18 shows a PCR result using a random 5'-13-mer primer H-AP10 of SEQ
ID NO: 71 and an anchored oligo-dT primer of SEQ ID NO: 72. In FIG. 18, Lanes
1, 2
and 3 represent a normal breast tissue; Lanes 4, 5 and 6 represent a breast
cancer tissue;
and Lane 7 represents a breast cancer cell line MCF-7. As shown in FIG. 18, it
was
confirmed that a 273-bp cDNA fragment (Base positions from 727 to 999 of the
full-length GIG43 gene sequence) was very rarely expressed in the breast
cancer tissue
and the breast cancer cell line, but differentially expressed at an increased
level only in
the normal breast tissue. This cDNA fragment was designated FC 102.
A 273-bp band, FC102 fragment, was removed from the dried gell, boiled for 15
minutes to elute the cDNA, and a PCR reaction was then carried out under the
same said
condition using the same primer set as described above to re-amplify the FC
102 cDNA,
except that the [ a 35S]-labeled dATP and the 20 u M dNTP were not used
herein.
The re-amplified cDNA fragment FC 102 was cloned into an expression vector
pGEM-T
Easy using the TA cloning system (Promega), and then its DNA sequence was
determined using the Sequenase Version 2.0 DNA Sequencing System (United
States
Biochemical Co.).
1-19. GIG46
A differential expression pattern of the gene of interest was measured in a
normal exocervical tissue, a primary cervical cancer tissue and a cervical
cancer cell line,
as follows. A normal exocervical tissue sample was obtained from a patient
suffering
87
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
from a uterine myoma during hysterectomy, and a primary cervical tumor tissue
sample
and a metastatic iliac lymph node tumor tissue sample were obtained during
radical
hysterectomy from a patient who has not been subject to the radiation therapy
and/or
anticancer chemotherapy before surgical treatment. CUMC-6 (Kim, J. W. et al.,
Gynecol. Oncol. 62: 230-240, 1996) was used as the human cervical cancer cell
line.
The total RNA samples were separated from these tissues and cells in the same
manner
as described in the reference example. The total RNAs were separated from
these
tissues and cells in the same manner as described in the reference example.
A RT-PCR reaction was carried out using each of the total RNA samples
separated from the tissues and the cells according to the modified method as
described
in the disclosure (Liang, P. and Pardee, A. B., Science, 257, 967-971 (1992);
and Liang,
P. et al., Cancer Res., 52, 6966-6968 (1993)), as follows. 0.2 ug of the total
RNA
was reverse-transcribed with an anchored oligo-dT primer of SEQ ID NO: 76
using a kit
(a RNAimage kit, GenHunter), and then a PCR reaction was carried out in the
presence
of 0.5 mM [ a-35S]-labeled dATP (1,200 Ci/mmol) using the same anchored oligo-
dT
primer and a random 5'-13-mer primer H-AP16 (RNAimage primer set 5, GenHunter
Corporation, U.S.) of SEQ ID NO: 75. The PCR reaction was conducted under the
following conditions: the total 40 amplification cycles consisting of a
denaturation step
at 95 C for 40 seconds, an annealing step at 40 C for 2 minutes and an
extension
step at 72 C for 40 seconds, and followed by one extension step at 72 C for 5
minutes. The amplified fragments were electrophoresized in a 6 %
polyacrylamide gel
for DNA base sequence, and then autoradiographed.
FIG. 19 shows a PCR result using a random 5'-13-mer primer H-AP16 of SEQ
88
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
ID NO: 75 and an anchored oligo-dT primer of SEQ ID NO: 76. In FIG. 19, Lane 1
represents a normal exocervical tissue; Lane 2 represents a cervical cancer
tissue; Lane
3 represents a metastatic iliac lymph node tissue; and Lane 4 represents a
cervical cancer
cell line CUMC-6. As shown in FIG. 19, it was confirmed that a 255-bp cDNA
fragment was not expressed in the cervical cancer tissue, the metastatic iliac
lymph node
tissue and the cervical cancer cell line CUMC-6, but differentially expressed
only in the
normal exocervical tissue. This cDNA fragment was designated CA161.
A 255-bp band, CA161 fragment, was removed from the dried gell, boiled for 15
minutes to elute the cDNA, and a PCR reaction was then carried out under the
same said
condition using the same primer set as described above to re-amplify the CA161
cDNA,
except that the [ a-35S]-labeled dATP and the 20 tt M dNTP were not used
herein.
The re-amplified cDNA fragment CA161 was cloned into an expression vector
pGEM-T Easy using the TA cloning system (Promega), and then its DNA sequence
was
determined using the Sequenase Version 2.0 DNA Sequencing System (United
States
Biochemical Co.).
1-20. PIG33
0.2 ug of the total RNA was reverse-transcribed with an anchored oligo-dT
primer of SEQ ID NO: 80 using a kit (a RNAimage kit, GenHunter), and then a
PCR
reaction was carried out in the presence of 0.5 mM [ a 35S]-labeled dATP
(1,200
Ci/mmol) using the same anchored oligo-dT primer and a random 5'-13-mer primer
H-AP2 (RNAimage primer set 1, GenHunter Corporation, U.S.) of SEQ ID NO: 79.
The PCR reaction was conducted under the following conditions: the total 40
amplification cycles consisting of a denaturation step at 95 C for 40
seconds, an
89
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
annealing step at 40 'C for 2 minutes and an extension step at 72 C for 40
seconds,
and followed by one extension step at 72 C for 5 minutes. The amplified
fragments
were electrophoresized in a 6 % polyacrylamide gel for DNA base sequence, and
then
autoradiographed.
FIG. 20 shows a PCR result using a random 5'-13-mer primer H-AP2 of SEQ ID
NO: 79 and an anchored oligo-dT primer of SEQ ID NO: 80. In FIG. 20, Lanes 1,
2
and 3 represent a normal liver tissue; Lanes 4, 5 and 6 represent a liver
cancer tissue;
and Lane 7 represents a liver cancer cell line HepG2. As shown in FIG. 20, it
was
confirmed that a 256-bp eDNA fragment (Base positions from 1,623 to 1,878 of
the
full-length PIG33 gene sequence) was not expressed or rarely expressed in the
liver
cancer tissue and the liver cancer cell line, but differentially expressed
only in the
normal liver tissue. This cDNA fragment was designated HP29.
A 256-bp band, HP29 fragment, was removed from the dried gell, boiled for 15
minutes to elute the cDNA, and a PCR reaction was then carried out under the
same said
condition using the same primer set as described above to re-amplify the HP29
cDNA,
except that the [ a-35S]-labeled dATP and the 20 u M dNTP were not used
herein.
The re-amplified cDNA fragment HP29 was cloned into an expression vector pGEM-
T
Easy using the TA cloning system (Promega), and then its DNA sequence was
determined using the Sequenase Version 2.0 DNA Sequencing System (United
States
Biochemical Co.).
1-21. PIG35
0.2 /Lg of the total RNA was reverse-transcribed with an anchored oligo-dT
primer of SEQ ID NO: 84 using a kit (a RNAimage kit, GenHunter), and then a
PCR
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
reaction was carried out in the presence of 0.5 mM [ a-35S]-labeled dATP
(1,200
Ci/mmol) using the same anchored oligo-dT primer and a random 5'-13-mer primer
H-AP9 (RNAimage primer set 1, GenHunter Corporation, U.S.) of SEQ ID NO: 83.
The PCR reaction was conducted under the following conditions: the total 40
amplification cycles consisting of a denaturation step at 95 C for 40 seconds,
an
annealing step at 40 C for 2 minutes and an extension step at 72 C for 40
seconds,
and followed by one extension step at 72 C for 5 minutes. The amplified
fragments
were electrophoresized in a 6 % polyacrylamide gel for DNA base sequence, and
then
autoradiographed.
FIG. 21 shows a PCR result using a random 5'-13-mer primer H-AP9 of SEQ ID
NO: 83 and an anchored oligo-dT primer of SEQ ID NO: 84. In FIG. 21, Lanes 1,
2
and 3 represent a normal liver tissue; Lanes 4, 5 and 6 represent a liver
cancer tissue;
and Lane 7 represents a liver cancer cell line HepG2. As shown in FIG. 21, it
was
confirmed that a 312-bp cDNA fragment (Base positions from 966 to 1,277 of the
full-length PIG35 gene sequence) was not expressed or rarely expressed in the
liver
cancer tissue and the liver cancer cell line, but differentially expressed
only in the
normal liver tissue. This cDNA fragment was designated HP95.
A 312-bp band, HP95 fragment, was removed from the dried gell, boiled for 15
minutes to elute the cDNA, and a PCR reaction was then carried out under the
same said
condition using the same primer set as described above to re-amplify the HP95
cDNA,
except that the [ a-35S]-labeled dATP and the 20 ji M dNTP were not used
herein.
The re-amplified cDNA fragment HP95 was cloned into an expression vector pGEM-
T
Easy using the TA cloning system (Promega), and then its DNA sequence was
91
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
determined using the Sequenase Version 2.0 DNA Sequencing System (United
States
Biochemical Co.).
1-22. PIG36
0.2 /ag of the total RNA was reverse-transcribed with an anchored oligo-dT
primer of SEQ ID NO: 88 using a kit (a RNAimage kit, GenHunter), and then a
PCR
reaction was carried out in the presence of 0.5 mM [ a 35S]-labeled dATP
(1,200
Ci/mmol) using the same anchored oligo-dT primer and a random 5'-13-mer primer
H-AP9 (RNAimage primer set 1, GenHunter Corporation, U.S.) of SEQ ID NO: 87.
The PCR reaction was conducted under the following conditions: the total 40
amplification cycles consisting of a denaturation step at 95 C for 40 seconds,
an
annealing step at 40 C for 2 minutes and an extension step at 72 C for 40
seconds,
and followed by one extension step at 72 C for 5 minutes. The amplified
fragments
were electrophoresized in a 6 % polyacrylamide gel for DNA base sequence, and
then
autoradiographed.
FIG. 22 shows a PCR result using a random 5'-13-mer primer H-AP9 of SEQ ID
NO: 87 and an anchored oligo-dT primer of SEQ ID NO: 88. In FIG. 22, Lanes 1,
2
and 3 represent a normal liver tissue; Lanes 4, 5 and 6 represent a liver
cancer tissue;
and Lane 7 represents a liver cancer cell line HepG2. As shown in FIG. 22, it
was
confirmed that a 162-bp cDNA fragment (Base positions from 238 to 399 of the
full-length PIG36 gene sequence) was not expressed or rarely expressed in the
liver
cancer tissue and the liver cancer cell line, but differentially expressed
only in the
normal liver tissue. This cDNA fragment was designated HP96.
A 162-bp band, HP96 fragment, was removed from the dried gell, boiled for 15
92
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
minutes to elute the cDNA, and a PCR reaction was then carried out under the
same said
condition using the same primer set as described above to re-amplify the HP96
cDNA,
except that the [ a 35S]-labeled dATP and the 20 u M dNTP were not used
herein.
The re-amplified cDNA fragment HP96 was cloned into an expression vector pGEM-
T
Easy using the TA cloning system (Promega), and then its DNA sequence was
determined using the Sequenase Version 2.0 DNA Sequencing System (United
States
Biochemical Co.).
1-23. MIG20
0.2 gg of the total RNA was reverse-transcribed with an anchored oligo-dT
primer of SEQ ID NO: 92 using a kit (a RNAimage kit, GenHunter), and then a
PCR
reaction was carried out in the presence of 0.5 mM [ a 35S]-labeled dATP
(1,200
Ci/mmol) using the same anchored oligo-dT primer and a random 5'-13-mer primer
H-AP32 (RNAimage primer set 5, GenHunter Corporation, U.S.) of SEQ ID NO: 91.
The PCR reaction was conducted under the following conditions: the total 40
amplification cycles consisting of a denaturation step at 95 C for 40 seconds,
an
annealing step at 40 'C for 2 minutes and an extension step at 72 C for 40
seconds,
and followed by one extension step at 72 C for 5 minutes. The amplified
fragments
were electrophoresized in a 6 % polyacrylamide gel for DNA base sequence, and
then
autoradiographed.
FIG. 23 shows a PCR result using a random 5'-13-mer primer H-AP32 of SEQ
ID NO: 91 and an anchored oligo-dT primer of SEQ ID NO: 92. In FIG. 23, Lane 1
represents a normal exocervical tissue; Lane 2 represents a cervical cancer
tissue; Lane
3 represents a metastatic iliac lymph node tissue; and Lane 4 represents a
cervical cancer
93
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
cell line CUMC-6. As shown in FIG. 23, it was confirmed that a 311 -bp cDNA
fragment was not expressed in the cervical cancer tissue, the metastatic iliac
lymph node
tissue and the cervical cancer cell line CUMC-6, but differentially expressed
only in the
normal exocervical tissue. This cDNA fragment was designated CA324.
A 311-bp band, CA324 fragment, was removed from the dried gell, boiled for 15
minutes to elute the cDNA, and a PCR reaction was then carried out under the
same said
condition using the same primer set as described above to re-amplify the CA324
cDNA,
except that the [ a 35S]-labeled dATP and the 20 u M dNTP were not used
herein.
The re-amplified cDNA fragment CA324 was cloned into an expression vector
pGEM-T Easy using the TA cloning system (Promega), and then its DNA sequence
was
determined using the Sequenase Version 2.0 DNA Sequencing System (United
States
Biochemical Co.).
1-24. PIG49
0.2 ug of the total RNA was reverse-transcribed with an anchored oligo-dT
primer of SEQ ID NO: 96 using a kit (a RNAimage kit, GenHunter), and then a
PCR
reaction was carried out in the presence of 0.5 mM [ a-35S]-labeled dATP
(1,200
Ci/mmol) using the same anchored oligo-dT primer and a random 5'-13-mer primer
H-AP10 (RNAimage primer set 2, GenHunter Corporation, U.S.) of SEQ ID NO: 95.
The PCR reaction was conducted under the following conditions: the total 40
amplification cycles consisting of a denaturation step at 95 C for 40 seconds,
an
annealing step at 40 C for 2 minutes and an extension step at 72 C for 40
seconds,
and followed by one extension step at 72 C for 5 minutes. The amplified
fragments
were electrophoresized in a 6 % polyacrylamide gel for DNA base sequence, and
then
94
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
autoradiographed.
FIG. 24 shows a PCR result using a random 5'-13-mer primer H-AP10 of SEQ
ID NO: 95 and an anchored oligo-dT primer of SEQ ID NO: 96. In FIG. 24, Lanes
1, 2
and 3 represent a normal breast tissue; Lanes 4, 5 and 6 represent a breast
cancer tissue;
and Lane 7 represents a breast cancer cell line MCF-7. As shown in FIG. 24, it
was
confirmed that a 272-bp cDNA fragment (Base positions from 767 to 1,038 of the
full-length PIG49 gene sequence) was very rarely expressed in the breast
cancer tissue
and the breast cancer cell line, but differentially expressed at an increased
level only in
the normal breast tissue. This cDNA fragment was designated FC101.
A 272-bp band, FC 101 fragment, was removed from the dried gell, boiled for 15
minutes to elute the cDNA, and a PCR reaction was then carried out under the
same said
condition using the same primer set as described above to re-amplify the FC
101 cDNA,
except that the [ a-35S]-labeled dATP and the 20 u M dNTP were not used
herein.
The re-amplified cDNA fragment FC 101 was cloned into an expression vector
pGEM-T
Easy using the TA cloning system (Promega), and then its DNA sequence was
determined using the Sequenase Version 2.0 DNA Sequencing System (United
States
Biochemical Co.).
1-25. PIG51
0.2 /ag of the total RNA was reverse-transcribed with an anchored oligo-dT
primer of SEQ ID NO: 100 using a kit (a RNAimage kit, GenHunter), and then a
PCR
reaction was carried out in the presence of 0.5 mM [ a 35S]-labeled dATP
(1,200
Ci/mmol) using the same anchored oligo-dT primer and a random 5'-13-mer primer
H-AP22 (RNAimage primer set 1, GenHunter Corporation, U.S.) of SEQ ID NO: 99.
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
The PCR reaction was conducted under the following conditions: the total 40
amplification cycles consisting of a denaturation step at 95 C for 40 seconds,
an
annealing step at 40 C for 2 minutes and an extension step at 72 'C for 40
seconds,
and followed by one extension step at 72 'C for 5 minutes. The amplified
fragments
were electrophoresized in a 6 % polyacrylamide gel for DNA base sequence, and
then
autoradiographed.
FIG. 25 shows a PCR result using a random 5'-13-mer primer H-AP22 of SEQ
ID NO: 99 and an anchored oligo-dT primer of SEQ ID NO: 100. In FIG. 25, Lanes
1,
2 and 3 represent a normal breast tissue; Lanes 4, 5 and 6 represent a breast
cancer
tissue; and Lane 7 represents a breast cancer cell line MCF-7. As shown in
FIG. 25, it
was confirmed that a 211-bp cDNA fragment (Base positions from 519 to 729 of
the
full-length PIG51 gene sequence) was very rarely expressed in the breast
cancer tissue
and the breast cancer cell line, but differentially expressed at an increased
level only in
the normal breast tissue. This cDNA fragment was designated FC22.
A 211 -bp band, FC22 fragment, was removed from the dried gell, boiled for 15
minutes to elute the eDNA, and a PCR reaction was then carried out under the
same said
condition using the same primer set as described above to re-amplify the FC22
cDNA,
except that the [ a 35S]-labeled dATP and the 20 u M dNTP were not used
herein.
The re-amplified cDNA fragment FC22 was cloned into an expression vector pGEM-
T
Easy using the TA cloning system (Promega), and then its DNA sequence was
determined using the Sequenase Version 2.0 DNA Sequencing System (United
States
Biochemical Co.).
1-26. MIG12
96
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
0.2 gg of the total RNA was reverse-transcribed with an anchored oligo-dT
primer of SEQ ID NO: 104 using a kit (a RNAimage kit, GenHunter), and then a
PCR
reaction was carried out in the presence of 0.5 mM [ a 35S]-labeled dATP
(1,200
Ci/mmol) using the same anchored oligo-dT primer and a random 5'-13-mer primer
H-AP12 (RNAimage primer set 1, GenHunter Corporation, U.S.) of SEQ ID NO: 103.
The PCR reaction was conducted under the following conditions: the total 40
amplification cycles consisting of a denaturation step at 95 C for 40
seconds, an
annealing step at 40 C for 2 minutes and an extension step at 72 C for 40
seconds,
and followed by one extension step at 72 C for 5 minutes. The amplified
fragments
were electrophoresized in a 6 % polyacrylamide gel for DNA base sequence, and
then
autoradiographed.
FIG. 26 shows a PCR result using a random 5'-13-mer primer H-AP12 of SEQ
ID NO: 103 and an anchored oligo-dT primer of SEQ ID NO: 104. In FIG. 26, Lane
1
represents a normal lung tissue; Lane 2 represents a lung cancer tissue; Lane
3
represents a metastatic lung cancer tissue; and Lane 4 represents a lung
cancer cell line
A549. As shown in FIG. 26, it was confirmed that a 161-bp cDNA fragment (Base
positions from 35 to 195 of the full-length MIG12 gene sequence) was rarely
expressed
in the lung cancer tissue, the metastatic lung cancer tissue and the lung
cancer cell line,
but differentially expressed only in the normal lung tissue. This cDNA
fragment was
designated L927.
A 161-bp band, L927 fragment, was removed from the dried gell, boiled for 15
minutes to elute the cDNA, and a PCR reaction was then carried out under the
same said
condition using the same primer set as described above to re-amplify the L927
cDNA,
97
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
except that the [ a-35S]-labeled dATP and the 20 u M dNTP were not used
herein.
The re-amplified cDNA fragment L927 was cloned into an expression vector pGEM-
T
Easy using the TA cloning system (Promega), and then its DNA sequence was
determined using the Sequenase Version 2.0 DNA Sequencing System (United
States
Biochemical Co.).
1-27. PIG37
0.2 ug of the total RNA was reverse-transcribed with an anchored oligo-dT
primer of SEQ ID NO: 108 using a kit (a RNAimage kit, GenHunter), and then a
PCR
reaction was carried out in the presence of 0.5 mM [ a 35S]-labeled dATP
(1,200
Ci/mmol) using the same anchored oligo-dT primer and a random 5'-13-mer primer
H-AP10 (RNAimage primer set 1, GenHunter Corporation, U.S.) of SEQ ID NO: 107.
The PCR reaction was conducted under the following conditions: the total 40
amplification cycles consisting of a denaturation step at 95 C for 40
seconds, an
annealing step at 40 C for 2 minutes and an extension step at 72 C for 40
seconds,
and followed by one extension step at 72 C for 5 minutes. The amplified
fragments
were electrophoresized in a 6 % polyacrylamide gel for DNA base sequence, and
then
autoradiographed.
FIG. 27 shows a PCR result using a random 5'-13-mer primer H-AP10 of SEQ
ID NO: 107 and an anchored oligo-dT primer of SEQ ID NO: 108. In FIG. 27,
Lanes 1,
2 and 3 represent a normal liver tissue; Lanes 4, 5 and 6 represent a liver
cancer tissue;
and Lane 7 represents a liver cancer cell line HepG2. As shown in FIG. 27, it
was
confirmed that a 263-bp cDNA fragment (Base positions from 1,217 to 1,479 of
the
full-length PIG37 gene sequence) was not expressed or rarely expressed in the
liver
98
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
cancer tissue and the liver cancer cell line, but differentially expressed
only in the
normal liver tissue. This cDNA fragment was designated HP102.
A 263-bp band, HP102 fragment, was removed from the dried gell, boiled for 15
minutes to elute the cDNA, and a PCR reaction was then carried out under the
same said
condition using the same primer set as described above to re-amplify the HP
102 cDNA,
except that the [ a-35S]-labeled dATP and the 20 u M dNTP were not used
herein.
The re-amplified cDNA fragment HP 102 was cloned into an expression vector
pGEM-T
Easy using the TA cloning system (Promega), and then its DNA sequence was
determined using the Sequenase Version 2.0 DNA Sequencing System (United
States
Biochemical Co.).
1-28. GIG44
0.2 ug of the total RNA was reverse-transcribed with an anchored oligo-dT
primer of SEQ ID NO: 112 using a kit (a RNAimage kit, GenHunter), and then a
PCR
reaction was carried out in the presence of 0.5 mM [ a-35S]-labeled dATP
(1,200
Ci/mmol) using the same anchored oligo-dT primer and a random 5'-13-mer primer
H-AP12 (RNAimage primer set 5, GenHunter Corporation, U.S.) of SEQ ID NO: 111.
The PCR reaction was conducted under the following conditions: the total 40
amplification cycles consisting of a denaturation step at 95 C for 40
seconds, an
annealing step at 40 C for 2 minutes and an extension step at 72 C for 40
seconds,
and followed by one extension step at 72 C for 5 minutes. The amplified
fragments
were electrophoresized in a 6 % polyacrylamide gel for DNA base sequence, and
then
autoradiographed.
FIG. 28 shows a PCR result using a random 5'-13-mer primer H-AP12 of SEQ
99
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
ID NO: 111 and an anchored oligo-dT primer of SEQ ID NO: 112. In FIG. 28,
Lanes 1,
2 and 3 represent a normal breast tissue; Lanes 4, 5 and 6 represent a breast
cancer
tissue; and Lane 7 represents a breast cancer cell line MCF-7. As shown in
FIG. 28, it
was confirmed that a 221-bp cDNA fragment (Base positions from 179 to 399 of
the
full-length GIG44 gene sequence) was very rarely expressed in the breast
cancer tissue
and the breast cancer cell line, but differentially expressed at an increased
level only in
the normal breast tissue. This cDNA fragment was designated FC123.
A 221-bp band, FC 123 fragment, was removed from the dried gell, boiled for 15
minutes to elute the cDNA, and a PCR reaction was then carried out under the
same said
condition using the same primer set as described above to re-amplify the FC
123 cDNA,
except that the [ a-35S]-labeled dATP and the 20 u M dNTP were not used
herein.
The re-amplified cDNA fragment FC123 was cloned into an expression vector pGEM-
T
Easy using the TA cloning system (Promega), and then its DNA sequence was
determined using the Sequenase Version 2.0 DNA Sequencing System (United
States
Biochemical Co.).
1-29. GIG31
0.2 /ig of the total RNA was reverse-transcribed with an anchored oligo-dT
primer of SEQ ID NO: 116 using a kit (a RNAimage kit, GenHunter), and then a
PCR
reaction was carried out in the presence of 0.5 mM [ a-35S]-labeled dATP
(1,200
Ci/mmol) using the same anchored oligo-dT primer and a random 5'-13-mer primer
H-AP4 (RNAimage primer set 1, GenHunter Corporation, U.S.) of SEQ ID NO: 115.
The PCR reaction was conducted under the following conditions: the total 40
amplification cycles consisting of a denaturation step at 95 C for 40 seconds,
an
100
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
annealing step at 40 C for 2 minutes and an extension step at 72 C for 40
seconds,
and followed by one extension step at 72 C for 5 minutes. The amplified
fragments
were electrophoresized in a 6 % polyacrylamide gel for DNA base sequence, and
then
autoradiographed.
FIG. 29 shows a PCR result using a random 5'-13-mer primer H-AP4 of SEQ ID
NO: 115 and an anchored oligo-dT primer of SEQ ID NO: 116. In FIG. 29, Lanes
1, 2
and 3 represent a normal breast tissue; Lanes 4, 5 and 6 represent a breast
cancer tissue;
and Lane 7 represents a breast cancer cell line MCF-7. As shown in FIG. 29, it
was
confirmed that a 223-bp cDNA fragment (Base positions from 445 to 667 of the
full-length GIG31 gene sequence) was very rarely expressed in the breast
cancer tissue
and the breast cancer cell line, but differentially expressed at an increased
level only in
the normal breast tissue. This cDNA fragment was designated FC47.
A 223-bp band, FC47 fragment, was removed from the dried gell, boiled for 15
minutes to elute the cDNA, and a PCR reaction was then carried out under the
same said
condition using the same primer set as described above to re-amplify the FC47
cDNA,
except that the [ a 35S]-labeled dATP and the 20 u M dNTP were not used
herein.
The re-amplified cDNA fragment FC47 was cloned into an expression vector pGEM-
T
Easy using the TA cloning system (Promega), and then its DNA sequence was
determined using the Sequenase Version 2.0 DNA Sequencing System (United
States
Biochemical Co.).
Example 2: cDNA Library Screening
The cDNA fragments FC33; FC42; FC59; GV2; H-AP8; HP71; HP115; HP3;
FC48; FC82; FC86; FC35; FC38; FC122; FC126; HP79; HP85; FC102; CA161; HP29;
101
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
HP95; HP96; CA324; FC101; FC22; L927; HP102; FC123 and FC47 obtained in
Example 1-1 were labeled according to the method of the disclosure (Feinberg,
A.P. and
Vogelstein, B., Anal. Biochem., 132, 6-13 (1983)) to obtain 32P-labeled probes
FC33;
FC42; FC59; GV2; H-AP8; HP71; HP115; HP3; FC48; FC82; FC86; FC35; FC38;
FC122; FC126; HP79; HP85; FC102; CA161; HP29; HP95; HP96; CA324; FC101;
FC22; L927; P102; FC123 and FC47 cDNA, repectively, and the 32P-labeled probes
were plaque-hybridized with bacteriophage Xgt11 human lung embryonic
fibroblast
cDNA library (Miki, T. et al., Gene, 83, 137-146 (1989)) according to the
method as
described in the disclosure (Sambrook, J. et al., Molecular Cloning: A
Laboratory
manual, New York: Cold Spring Harbor Laboratory (1989)) to obtain full-length
cDNA
clones of the human cancer suppressor gene GIGs.
The full-length cDNA clones were sequenced, and therefore a DNA base
sequence result of the GIG8 was identical with SEQ ID NO: 1. The DNA sequence
of
the GIG8 has an open reading frame encoding 134 amino acid residues, and the
amino
acid sequence derived from the open reading frame was identical with SEQ ID
NO: 2.
The derived protein also had a molecular weight of approximately 15 kDa.
The transformed E. coli strain was incubated in LB broth, and then 1 mM
isopropy-l- 13 -D-thiogalactopyranoside (IPTG) was added to the culture broth,
and
reacted at 37 C for 3 hours to express the GIG8 gene. A protein sample was
obtained from the culture broth, and then SDS-PAGE was conducted with the
protein
sample according to the method as described in the disclosure (Sambrook, J. et
al.,
Molecular Cloning: A Laboratory manual, New York: Cold Spring Harbor
Laboratory
(1989)).
102
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
FIG. 30 is a diagram showing an SDS-PAGE analysis of the GIG8 protein. In
FIG. 30, Lane 1 represents a protein sample before the IPTG induction, and
Lane 2
represents a protein sample after the expression of the GIG8 gene is induced
by IPTG.
As shown in FIG. 30, the expressed GIG8 protein has a molecular weight of
approximately 15 kDa, which corresponds to a molecular weight of a protein
derived
from its DNA sequence.
A DNA base sequence result of the GIG10 was identical with SEQ ID NO: 5.
The DNA sequence of the GIG10 has an open reading frame encoding 665 amino
acid
residues, and the amino acid sequence derived from the open reading frame was
identical with SEQ ID NO: 6. The derived protein also had a molecular weight
of
approximately 73 kDa.
The transformed E. coli strain was incubated in LB broth, and then 1 mM
isopropy-l- j3 -D-thiogalactopyranoside (IPTG) was added to the culture broth,
and
reacted at 37 C for 3 hours to express the GIG10 gene. A protein sample was
obtained from the culture broth, and then SDS-PAGE was conducted with the
protein
sample according to the method as described in the disclosure (Sambrook, J. et
al.,
Molecular Cloning: A Laboratory manual, New York: Cold Spring Harbor
Laboratory
(1989)).
FIG. 31 is a diagram showing an SDS-PAGE analysis of the GIG10 protein. In
FIG. 31, Lane 1 represents a protein sample before the IPTG induction, and
Lane 2
represents a protein sample after the expression of the GIG10 gene is induced
by IPTG.
As shown in FIG. 31, the expressed GIG10 protein has a molecular weight of
approximately 73 kDa, which corresponds to a molecular weight of a protein
derived
103
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
from its DNA sequence.
A DNA base sequence result of the GIG13 was identical with SEQ ID NO: 9.
The DNA sequence of the GIG13 has an open reading frame encoding 1,201 amino
acid
residues, and the amino acid sequence derived from the open reading frame was
identical with SEQ ID NO: 10. The derived protein also had a molecular weight
of
approximately 132 kDa.
The transformed E. coli strain was incubated in LB broth, and then 1 mM
isopropy-1-13-D-thiogalactopyranoside (IPTG) was added to the culture broth,
and
reacted at 37 C for 3 hours to express the GIG13 gene. A protein sample was
obtained from the culture broth, and then SDS-PAGE was conducted with the
protein
sample according to the method as described in the disclosure (Sambrook, J. et
al.,
Molecular Cloning: A Laboratory manual, New York: Cold Spring Harbor
Laboratory
(1989)).
FIG. 32 is a diagram showing an SDS-PAGE analysis of the GIG13 protein. In
FIG. 32, Lane 1 represents a protein sample before the IPTG induction, and
Lane 2
represents a protein sample after the expression of the GIG13 gene is induced
by IPTG.
As shown in FIG. 32, the expressed GIG13 protein has a molecular weight of
approximately 132 kDa, which corresponds to a molecular weight of a protein
derived
from its DNA sequence.
A DNA base sequence result of the GIG 15 was identical with SEQ ID NO: 13.
The DNA sequence of the GIG 15 has an open reading frame encoding 106 amino
acid
residues, and the amino acid sequence derived from the open reading frame was
identical with SEQ ID NO: 14. The derived protein also had a molecular weight
of
104
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
approximately 12 kDa.
The transformed E. coli strain was incubated in LB broth, and then 1 mM
isopropy-1-f3 -D-thiogalactopyranoside (IPTG) was added to the culture broth,
and
reacted at 37 C for 3 hours to express the GIG GIG15 gene. A protein sample
was
obtained from the culture broth, and then SDS-PAGE was conducted with the
protein
sample according to the method as described in the disclosure (Sambrook, J. et
al.,
Molecular Cloning: A Laboratory manual, New York: Cold Spring Harbor
Laboratory
(1989)).
FIG. 33 is a diagram showing an SDS-PAGE analysis of the GIG15 protein. In
FIG. 33, Lane 1 represents a protein sample before the IPTG induction, and
Lane 2
represents a protein sample after the expression of the GIG15 gene is induced
by IPTG.
As shown in FIG. 33, the expressed GIG15 protein has a molecular weight of
approximately 12 kDa, which corresponds to a molecular weight of a protein
derived
from its DNA sequence.
A DNA base sequence result of the GIG16 was identical with SEQ ID NO: 17.
The DNA sequence of the GIG16 has an open reading frame encoding 351 amino
acid
residues, and the amino acid sequence derived from the open reading frame was
identical with SEQ ID NO: 18. The derived protein also had a molecular weight
of
approximately 39 kDa. The resultant full-length GIG16 cDNA was inserted into
the
prokaryotic expression vector pBAD/Thio-Topo (Invitrogen, U.A.), and then E.
coli
DH5 a was transformed with the resultant expression vector to obtain a
transformant,
which was designated E. coli DH5 a/GIG16/pBAD/Thio-Topo.
The transformed E. coli strain was incubated in LB broth, and then 1 mM
105
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
isopropy-1- j3 -D-thiogalactopyranoside (IPTG) was added to the culture broth,
and
reacted at 37 C for 3 hours to express the GIG16 gene. A protein sample was
obtained from the culture broth, and then SDS-PAGE was conducted with the
protein
sample according to the method as described in the disclosure (Sambrook, J. et
al.,
Molecular Cloning: A Laboratory manual, New York: Cold Spring Harbor
Laboratory
(1989)).
FIG. 34 is a diagram showing an SDS-PAGE analysis of the GIG16 protein. In
FIG. 34, Lane 1 represents a protein sample before the IPTG induction, and
Lane 2
represents a protein sample after the expression of the GIG 16 gene is induced
by IPTG.
As shown in FIG. 34, the expressed GIG16 protein has a molecular weight of
approximately 39 kDa, which corresponds to a molecular weight of a protein
derived
from its DNA sequence.
A DNA base sequence result of the GIG24 was identical with SEQ ID NO: 21.
The DNA sequence of the GIG24 has an open reading frame encoding 423 amino
acid
residues, and the amino acid sequence derived from the open reading frame was
identical with SEQ ID NO: 22. The derived protein also had a molecular weight
of
approximately 47 kDa. The resultant full-length GIG24 cDNA was inserted into
the
prokaryotic expression vector pBAD/Thio-Topo (Invitrogen, U.A.), and then E.
coli
DH5 a was transformed with the resultant expression vector to obtain a
transformant,
which was designated E. coli DH5 a/GIG24/pBAD/Thio-Topo.
The transformed E. coli strain was incubated in LB broth, and then 1 mM
isopropy-l- j3 -D-thiogalactopyranoside (IPTG) was added to the culture broth,
and
reacted at 37 C for 3 hours to express the GIG24 gene. A protein sample was
106
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
obtained from the culture broth, and then SDS-PAGE was conducted with the
protein
sample according to the method as described in the disclosure (Sambrook, J. et
al.,
Molecular Cloning: A Laboratory manual, New York: Cold Spring Harbor
Laboratory
(1989)).
FIG. 35 is a diagram showing an SDS-PAGE analysis of the GIG24 protein. In
FIG. 35, Lane 1 represents a protein sample before the IPTG induction, and
Lane 2
represents a protein sample after the expression of the GIG24 gene is induced
by IPTG.
As shown in FIG. 35, the expressed GIG24 protein has a molecular weight of
approximately 47 kDa, which corresponds to a molecular weight of a protein
derived
from its DNA sequence.
A DNA base sequence result of the GIG26 was identical with SEQ ID NO: 25.
The DNA sequence of the GIG26 has an open reading frame encoding 442 amino
acid
residues, and the amino acid sequence derived from the open reading frame was
identical with SEQ ID NO: 26. The derived protein also had a molecular weight
of
approximately 50 kDa. The resultant full-length GIG26 cDNA was inserted into
the
prokaryotic expression vector pBAD/Thio-Topo (Invitrogen, U.A.), and then E.
coli
DH5 a was transformed with the resultant expression vector to obtain a
transformant,
which was designated E. coli DH5 a/GIG26/pBAD/Thio-Topo.
The transformed E. coli strain was incubated in LB broth, and then 1 mM
isopropy-l- 13 -D-thiogalactopyranoside (IPTG) was added to the culture broth,
and
reacted at 37 C for 3 hours to express the GIG26 gene. A protein sample was
obtained from the culture broth, and then SDS-PAGE was conducted with the
protein
sample according to the method as described in the disclosure (Sambrook, J. et
al.,
107
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
Molecular Cloning: A Laboratory manual, New York: Cold Spring Harbor
Laboratory
(1989)).
FIG. 36 is a diagram showing an SDS-PAGE analysis of the GIG26 protein. In
FIG. 36, Lane 1 represents a protein sample before the IPTG induction, and
Lane 2
represents a protein sample after the expression of the GIG26 gene is induced
by IPTG.
As shown in FIG. 36, the expressed GIG26 protein has a molecular weight of
approximately 50 kDa, which corresponds to a molecular weight of a protein
derived
from its DNA sequence.
A DNA base sequence result of the GIG29 was identical with SEQ ID NO: 29.
The DNA sequence of the GIG29 has an open reading frame encoding 349 amino
acid
residues, and the amino acid sequence derived from the open reading frame was
identical with SEQ ID NO: 30. The derived protein also had a molecular weight
of
approximately 38 kDa. The resultant full-length GIG29 cDNA was inserted into
the
prokaryotic expression vector pBAD/Thio-Topo (Invitrogen, U.A.), and then E.
coli
DH5 a was transformed with the resultant expression vector to obtain a
transformant,
which was designated E. coli DH5 a/GIG29/pBAD/Thio-Topo.
The transformed E. coli strain was incubated in LB broth, and then 1 mM
isopropy-l- j3 -D-thiogalactopyranoside (IPTG) was added to the culture broth,
and
reacted at 37 C for 3 hours to express the GIG29 gene. A protein sample was
obtained from the culture broth, and then SDS-PAGE was conducted with the
protein
sample according to the method as described in the disclosure (Sambrook, J. et
al.,
Molecular Cloning: A Laboratory manual, New York: Cold Spring Harbor
Laboratory
(1989)).
108
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
FIG. 37 is a diagram showing an SDS-PAGE analysis of the GIG29 protein. In
FIG. 37, Lane 1 represents a protein sample before the IPTG induction, and
Lane 2
represents a protein sample after the expression of the GIG29 gene is induced
by IPTG.
As shown in FIG. 37, the expressed GIG29 protein has a molecular weight of
approximately 38 kDa, which corresponds to a molecular weight of a protein
derived
from its DNA sequence.
A DNA base sequence result of the GIG30 was identical with SEQ ID NO: 33.
The DNA sequence of the GIG30 has an open reading frame encoding 540 amino
acid
residues, and the amino acid sequence derived from the open reading frame was
identical with SEQ ID NO: 34. The derived protein also had a molecular weight
of
approximately 61 kDa.
The transformed E. coli strain was incubated in LB broth, and then 1 mM
isopropy-l- J3 -D-thiogalactopyranoside (IPTG) was added to the culture broth,
and
reacted at 37 C for 3 hours to express the GIG30 gene. A protein sample was
obtained from the culture broth, and then SDS-PAGE was conducted with the
protein
sample according to the method as described in the disclosure (Sambrook, J. et
al.,
Molecular Cloning: A Laboratory manual, New York: Cold Spring Harbor
Laboratory
(1989)).
FIG. 38 is a diagram showing an SDS-PAGE analysis of the GIG30 protein. In
FIG. 38, Lane 1 represents a protein sample before the IPTG induction, and
Lane 2
represents a protein sample after the expression of the GIG30 gene is induced
by IPTG.
As shown in FIG. 38, the expressed GIG30 protein has a molecular weight of
approximately 61 kDa, which corresponds to a molecular weight of a protein
derived
109
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
from its DNA sequence.
A DNA base sequence result of the GIG32 was identical with SEQ ID NO: 37.
The DNA sequence of the GIG32 has an open reading frame encoding 178 amino
acid
residues, and the amino acid sequence derived from the open reading frame was
identical with SEQ ID NO: 38. The derived protein also had a molecular weight
of
approximately 20 kDa.
The transformed E. coli strain was incubated in LB broth, and then 1 mM
isopropy-l- Ji -D-thiogalactopyranoside (IPTG) was added to the culture broth,
and
reacted at 37 C for 3 hours to express the GIG32 gene. A protein sample was
obtained from the culture broth, and then SDS-PAGE was conducted with the
protein
sample according to the method as described in the disclosure (Sambrook, J. et
al.,
Molecular Cloning: A Laboratory manual, New York: Cold Spring Harbor
Laboratory
(1989)).
FIG. 39 is a diagram showing an SDS-PAGE analysis of the GIG32 protein. In
FIG. 39, Lane 1 represents a protein sample before the IPTG induction, and
Lane 2
represents a protein sample after the expression of the GIG32 gene is induced
by IPTG.
As shown in FIG. 39, the expressed GIG32 protein has a molecular weight of
approximately 20 kDa, which corresponds to a molecular weight of a protein
derived
from its DNA sequence.
A DNA base sequence result of the GIG33 was identical with SEQ ID NO: 41.
The DNA sequence of the GIG33 has an open reading frame encoding 110 amino
acid
residues, and the amino acid sequence derived from the open reading frame was
identical with SEQ ID NO: 42. The derived protein also had a molecular weight
of
110
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
approximately 12 kDa.
The transformed E. coli strain was incubated in LB broth, and then 1 mM
isopropy-1-13 -D-thiogalactopyranoside (IPTG) was added to the culture broth,
and
reacted at 37 C for 3 hours to express the GIG34 gene. A protein sample was
obtained from the culture broth, and then SDS-PAGE was conducted with the
protein
sample according to the method as described in the disclosure (Sambrook, J. et
al.,
Molecular Cloning: A Laboratory manual, New York: Cold Spring Harbor
Laboratory
(1989)).
FIG. 40 is a diagram showing an SDS-PAGE analysis of the GIG33 protein. In
FIG. 40, Lane 1 represents a protein sample before the IPTG induction, and
Lane 2
represents a protein sample after the expression of the GIG33 gene is induced
by IPTG.
As shown in FIG. 40, the expressed GIG33 protein has a molecular weight of
approximately 12 kDa, which corresponds to a molecular weight of a protein
derived
from its DNA sequence.
A DNA base sequence result of the GIG34 was identical with SEQ ID NO: 45.
The DNA sequence of the GIG34 has an open reading frame encoding 177 amino
acid
residues, and the amino acid sequence derived from the open reading frame was
identical with SEQ ID NO: 46. The derived protein also had a molecular weight
of
approximately 20 kDa.
The transformed E. coli strain was incubated in LB broth, and then 1 mM
isopropy-l- J3 -D-thiogalactopyranoside (IPTG) was added to the culture broth,
and
reacted at 37 C for 3 hours to express the GIG34 gene. A protein sample was
obtained from the culture broth, and then SDS-PAGE was conducted with the
protein
111
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
sample according to the method as described in the disclosure (Sambrook, J. et
al.,
Molecular Cloning: A Laboratory manual, New York: Cold Spring Harbor
Laboratory
(1989)).
FIG. 41 is a diagram showing an SDS-PAGE analysis of the GIG34 protein. In
FIG. 41, Lane 1 represents a protein sample before the IPTG induction, and
Lane 2
represents a protein sample after the expression of the GIG34 gene is induced
by IPTG.
As shown in FIG. 41, the expressed GIG34 protein has a molecular weight of
approximately 20 kDa, which corresponds to a molecular weight of a protein
derived
from its DNA sequence.
A DNA base sequence result of the GIG35 was identical with SEQ ID NO: 49.
The DNA sequence of the GIG35 has an open reading frame encoding 437 amino
acid
residues, and the amino acid sequence derived from the open reading frame was
identical with SEQ ID NO: 50. The derived protein also had a molecular weight
of
approximately 50 kDa.
The transformed E. coli strain was incubated in LB broth, and then 1 mM
isopropy-l- j3 -D-thiogalactopyranoside (IPTG) was added to the culture broth,
and
reacted at 37 C for 3 hours to express the GIG35 gene. A protein sample was
obtained from the culture broth, and then SDS-PAGE was conducted with the
protein
sample according to the method as described in the disclosure (Sambrook, J. et
al.,
Molecular Cloning: A Laboratory manual, New York: Cold Spring Harbor
Laboratory
(1989)).
FIG. 42 is a diagram showing an SDS-PAGE analysis of the GIG35 protein. In
FIG. 42, Lane 1 represents a protein sample before the IPTG induction, and
Lane 2
112
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
represents a protein sample after the expression of the GIG35 gene is induced
by IPTG.
As shown in FIG. 42, the expressed GIG35 protein has a molecular weight of
approximately 50 kDa, which corresponds to a molecular weight of a protein
derived
from its DNA sequence.
A DNA base sequence result of the GIG38 was identical with SEQ ID NO: 53.
The DNA sequence of the GIG38 has an open reading frame encoding 153 amino
acid
residues, and the amino acid sequence derived from the open reading frame was
identical with SEQ ID NO: 54. The derived protein also had a molecular weight
of
approximately 17 kDa.
The transformed E. coli strain was incubated in LB broth, and then 1 mM
isopropy-l- J3 -D-thiogalactopyranoside (IPTG) was added to the culture broth,
and
reacted at 37 C for 3 hours to express the GIG38 gene. A protein sample was
obtained from the culture broth, and then SDS-PAGE was conducted with the
protein
sample according to the method as described in the disclosure (Sambrook, J. et
al.,
Molecular Cloning: A Laboratory manual, New York: Cold Spring Harbor
Laboratory
(1989)).
FIG. 43 is a diagram showing an SDS-PAGE analysis of the GIG38 protein. In
FIG. 43, Lane 1 represents a protein sample before the IPTG induction, and
Lane 2
represents a protein sample after the expression of the GIG38 gene is induced
by IPTG.
As shown in FIG. 43, the expressed GIG38 protein has a molecular weight of
approximately 17 kDa, which corresponds to a molecular weight of a protein
derived
from its DNA sequence.
A DNA base sequence result of the GIG39 was identical with SEQ ID NO: 57.
113
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
The DNA sequence of the GIG39 has an open reading frame encoding 928 amino
acid
residues, and the amino acid sequence derived from the open reading frame was
identical with SEQ ID NO: 58. The derived protein also had a molecular weight
of
approximately 103 kDa.
The transformed E. coli strain was incubated in LB broth, and then 1 mM
isopropy-l-J3 -D-thiogalactopyranoside (IPTG) was added to the culture broth,
and
reacted at 37 C for 3 hours to express the GIG39 gene. A protein sample was
obtained from the culture broth, and then SDS-PAGE was conducted with the
protein
sample according to the method as described in the disclosure (Sambrook, J. et
al.,
Molecular Cloning: A Laboratory manual, New York: Cold Spring Harbor
Laboratory
(1989)).
FIG. 44 is a diagram showing an SDS-PAGE analysis of the GIG39 protein. In
FIG. 44, Lane 1 represents a protein sample before the IPTG induction, and
Lane 2
represents a protein sample after the expression of the GIG39 gene is induced
by IPTG.
As shown in FIG. 44, the expressed GIG39 protein has a molecular weight of
approximately 103 kDa, which corresponds to a molecular weight of a protein
derived
from its DNA sequence.
A DNA base sequence result of the GIG40 was identical with SEQ ID NO: 61.
The DNA sequence of the GIG40 has an open reading frame encoding 1,210 amino
acid
residues, and the amino acid sequence derived from the open reading frame was
identical with SEQ ID NO: 62. The derived protein also had a molecular weight
of
approximately 134 kDa. The resultant full-length GIG40 cDNA was inserted into
the
prokaryotic expression vector pBAD/Thio-Topo (Invitrogen, U.A.), and then E.
coli
114
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
DH5 a was transformed with the resultant expression vector to obtain a
transformant,
which was designated E. coli DH5 a/GIG40/pBAD/Thio-Topo.
The transformed E. coli strain was incubated in LB broth, and then 1 mM
isopropy-l-i3-D-thiogalactopyranoside (IPTG) was added to the culture broth,
and
reacted at 37 'C for 3 hours to express the GIG40 gene. A protein sample was
obtained from the culture broth, and then SDS-PAGE was conducted with the
protein
sample according to the method as described in the disclosure (Sambrook, J. et
al.,
Molecular Cloning: A Laboratory manual, New York: Cold Spring Harbor
Laboratory
(1989)).
FIG. 45 is a diagram showing an SDS-PAGE analysis of the GIG40 protein. In
FIG. 45, Lane 1 represents a protein sample before the IPTG induction, and
Lane 2
represents a protein sample after the expression of the GIG40 gene is induced
by IPTG.
As shown in FIG. 45, the expressed GIG40 protein has a molecular weight of
approximately 134 kDa, which corresponds to a molecular weight of a protein
derived
from its DNA sequence.
A DNA base sequence result of the GIG42 was identical with SEQ ID NO: 65.
The DNA sequence of the GIG42 has an open reading frame encoding 609 amino
acid
residues, and the amino acid sequence derived from the open reading frame was
identical with SEQ ID NO: 66. The derived protein also had a molecular weight
of
approximately 69 kDa. The resultant full-length GIG42 cDNA was inserted into
the
prokaryotic expression vector pBAD/Thio-Topo (Invitrogen, U.A.), and then E.
coli
DH5 a was transformed with the resultant expression vector to obtain a
transformant,
which was designated E. coli DH5 a/GIG42/pBAD/Thio-Topo.
115
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
The transformed E. coli strain was incubated in LB broth, and then 1 mM
isopropy-l-f3 -D-thiogalactopyranoside (IPTG) was added to the culture broth,
and
reacted at 37 C for 3 hours to express the GIG42 gene. A protein sample was
obtained from the culture broth, and then SDS-PAGE was conducted with the
protein
sample according to the method as described in the disclosure (Sambrook, J. et
al.,
Molecular Cloning: A Laboratory manual, New York: Cold Spring Harbor
Laboratory
(1989)).
FIG. 46 is a diagram showing an SDS-PAGE analysis of the GIG42 protein. In
FIG. 46, Lane 1 represents a protein sample before the IPTG induction, and
Lane 2
represents a protein sample after the expression of the GIG42 gene is induced
by IPTG.
As shown in FIG. 46, the expressed GIG42 protein has a molecular weight of
approximately 69 kDa, which corresponds to a molecular weight of a protein
derived
from its DNA sequence.
A DNA base sequence result of the GIG43 was identical with SEQ ID NO: 69.
The DNA sequence of the GIG43 has an open reading frame encoding 329 amino
acid
residues, and the amino acid sequence derived from the open reading frame was
identical with SEQ ID NO: 70. The derived protein also had a molecular weight
of
approximately 37 kDa.
The transformed E. coli strain was incubated in LB broth, and then 1 mM
isopropy-l-ii-D-thiogalactopyranoside (IPTG) was added to the culture broth,
and
reacted at 37 C for 3 hours to express the GIG43 gene. A protein sample was
obtained from the culture broth, and then SDS-PAGE was conducted with the
protein
sample according to the method as described in the disclosure (Sambrook, J. et
al.,
116
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
Molecular Cloning: A Laboratory manual, New York: Cold Spring Harbor
Laboratory
(1989)).
FIG. 47 is a diagram showing an SDS-PAGE analysis of the GIG43 protein. In
FIG. 47, Lane 1 represents a protein sample before the IPTG induction, and
Lane 2
represents a protein sample after the expression of the GIG43 gene is induced
by IPTG.
As shown in FIG. 47, the expressed GIG43 protein has a molecular weight of
approximately 37 kDa, which corresponds to a molecular weight of a protein
derived
from its DNA sequence.
A DNA base sequence result of the GIG46 was identical with SEQ ID NO: 73.
The DNA sequence of the GIG46 has an open reading frame encoding 377 amino
acid
residues, and the amino acid sequence derived from the open reading frame was
identical with SEQ ID NO: 74. The derived protein also had a molecular weight
of
approximately 42 kDa.
The resultant full-length GIG46 cDNA clone was inserted into a multi-cloning
site of the prokaryotic expression vector pBAD/thio-Topo (Invitrogen, U.S.) to
obtain a
vector pBAD/thio-Topo/GIG46, and Escherichia coli ToplO (Invitrogen, U.S.) was
then
transformed with the resultant pBAD/thio-Topo/GIG46. The expression protein
HT-Thioredoxin is inserted upstream of the multi-cloning site of the vector
pBAD/thio-Topo. The transformed E. coli strain was incubated in LB broth with
shaking, and the resultant culture broth was diluted 1/100, and then incubated
for 3
hours again. 0.5 mM L-arabinose (Sigma, U.S.) was added to the incubated
culture
broth to induce production of proteins. The E. coli cell in the culture broth
was
sonicated before and after the L-arabinose induction, and then 12 % sodium
dodecyl
117
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
sulphate polyacrylamide gel electrophoresis (SDS-PAGE) was conducted with the
sonicated homogenate. FIG. 48 is a diagram showing an expression pattern of
proteins
of the E. coli Top10 strain transformed with the vector pBAD/thio-Topo/GIG46
using
the SDS-PAGE, wherein a band of a fusion protein having a molecular weight of
approximately 57 kDa was clearly observed after the L-arabinose induction. The
57-kDa fusion protein includes the approximately 15-kDa HT-thioredoxin protein
inserted into the vector pBAD/thio-Topo/GIG46 and the approximately 42-kDa
GIG46
protein.
FIG. 48 is a diagram showing an SDS-PAGE analysis of the GIG46 protein. In
FIG. 48, Lane 1 represents a protein sample before the L-arabinose induction,
and Lane
2 represents a protein sample after the expression of the GIG46 gene is
induced by
L-arabinose.
A DNA base sequence result of the PIG33 was identical with SEQ ID NO: 77.
The DNA sequence of the PIG33 has an open reading frame encoding 664 amino
acid
residues, and the amino acid sequence derived from the open reading frame was
identical with SEQ ID NO: 78. The derived protein also had a molecular weight
of
approximately 75 kDa. The resultant full-length PIG33 cDNA was inserted into
the
prokaryotic expression vector pBAD/Thio-Topo (Invitrogen, U.A.), and then E.
coli
DH5 a was transformed with the resultant expression vector to obtain a
transformant,
which was designated E. coli DH5 a/PIG33/pBAD/Thio-Topo.
The transformed E. coli strain was incubated in LB broth, and then 1 mM
isopropy-1- j3 -D-thiogalactopyranoside (IPTG) was added to the culture broth,
and
reacted at 37 C for 3 hours to express the PIG33 gene. A protein sample was
118
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
obtained from the culture broth, and then SDS-PAGE was conducted with the
protein
sample according to the method as described in the disclosure (Sambrook, J. et
al.,
Molecular Cloning: A Laboratory manual, New York: Cold Spring Harbor
Laboratory
(1989)).
FIG. 49 is a diagram showing an SDS-PAGE analysis of the PIG33 protein. In
FIG. 49, Lane 1 represents a protein sample before the IPTG induction, and
Lane 2
represents a protein sample after the expression of the PIG33 gene is induced
by IPTG.
As shown in FIG. 49, the expressed PIG33 protein has a molecular weight of
approximately 75 kDa, which corresponds to a molecular weight of a protein
derived
from its DNA sequence.
A DNA base sequence result of the PIG35 was identical with SEQ ID NO: 81.
The DNA sequence of the PIG35 has an open reading frame encoding 418 amino
acid
residues, and the amino acid sequence derived from the open reading frame was
identical with SEQ ID NO: 82. The derived protein also had a molecular weight
of
approximately 46 kDa. The resultant full-length PIG35 cDNA was inserted into
the
prokaryotic expression vector pBAD/Thio-Topo (Invitrogen, U.A.), and then E.
coli
DH5 a was transformed with the resultant expression vector to obtain a
transformant,
which was designated E. coli DH5 a/PIG35/pBAD/Thio-Topo.
The transformed E. coli strain was incubated in LB broth, and then 1 mM
isopropy-1-13-D-thiogalactopyranoside (IPTG) was added to the culture broth,
and
reacted at 37 C for 3 hours to express the PIG35 gene. A protein sample was
obtained from the culture broth, and then SDS-PAGE was conducted with the
protein
sample according to the method as described in the disclosure (Sambrook, J. et
al.,
119
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
Molecular Cloning: A Laboratory manual, New York: Cold Spring Harbor
Laboratory
(1989)).
FIG. 50 is a diagram showing an SDS-PAGE analysis of the PIG35 protein. In
FIG. 50, Lane 1 represents a protein sample before the IPTG induction, and
Lane 2
represents a protein sample after the expression of the PIG35 gene is induced
by IPTG.
As shown in FIG. 50, the expressed PIG35 protein has a molecular weight of
approximately 46 kDa, which corresponds to a molecular weight of a protein
derived
from its DNA sequence.
A DNA base sequence result of the PIG 36 was identical with SEQ ID NO: 85.
The DNA sequence of the PIG36 has an open reading frame encoding 108 amino
acid
residues, and the amino acid sequence derived from the open reading frame was
identical with SEQ ID NO: 86. The derived protein also had a molecular weight
of
approximately 13 kDa. The resultant full-length PIG36 cDNA was inserted into
the
prokaryotic expression vector pBAD/Thio-Topo (Invitrogen, U.A.), and then E.
coli
DH5 a was transformed with the resultant expression vector to obtain a
transfonnant,
which was designated E. coli DH5 a /PIG36/pBAD/Thio-Topo.
The transformed E. coli strain was incubated in LB broth, and then 1 mM
isopropy-1-13 -D-thiogalactopyranoside (IPTG) was added to the culture broth,
and
reacted at 37 C for 3 hours to express the PIG36 gene. A protein sample was
obtained from the culture broth, and then SDS-PAGE was conducted with the
protein
sample according to the method as described in the disclosure (Sambrook, J. et
al.,
Molecular Cloning: A Laboratory manual, New York: Cold Spring Harbor
Laboratory
(1989)).
120
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
FIG. 51 is a diagram showing an SDS-PAGE analysis of the PIG36 protein. In
FIG. 51, Lane 1 represents a protein sample before the IPTG induction, and
Lane 2
represents a protein sample after the expression of the PIG36 gene is induced
by IPTG.
As shown in FIG. 51, the expressed PIG36 protein has a molecular weight of
approximately 13 kDa, which corresponds to a molecular weight of a protein
derived
from its DNA sequence.
A DNA base sequence result of the MIG20 was identical with SEQ ID NO: 89.
The DNA sequence of the MIG20 has an open reading frame encoding 64 amino acid
residues, and the amino acid sequence derived from the open reading frame was
identical with SEQ ID NO: 90. The derived protein also had a molecular weight
of
approximately 7 kDa.
The resultant full-length MIG20 cDNA clone was inserted into a multi-cloning
site of the prokaryotic expression vector pBAD/thio-Topo (Invitrogen, U.S.) to
obtain a
vector pBAD/thio-Topo/MIG20, and Escherichia coli ToplO (Invitrogen, U.S.) was
then transformed with the resultant pBAD/thio-Topo/MIG20. The expression
protein
HT-Thioredoxin is inserted upstream of the multi-cloning site of the vector
pBAD/thio-Topo. The transformed E. coli strain was incubated in LB broth with
shaking, and the resultant culture broth was diluted 1/100, and then incubated
for 3
hours again. 0.5 mM L-arabinose (Sigma, U.S.) was added to the incubated
culture
broth to induce production of proteins. The E. coli cell in the culture broth
was
sonicated before and after the L-arabinose induction, and then 12 % sodium
dodecyl
sulphate polyacrylamide gel electrophoresis (SDS-PAGE) was conducted with the
sonicated homogenate. FIG. 52 is a diagram showing an expression pattern of
proteins
121
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
of the E. coli Top 10 strain transformed with the vector pBAD/thio-Topo/MIG20
using
the SDS-PAGE, wherein a band of a fusion protein having a molecular weight of
approximately 22 kDa was clearly observed after the L-arabinose induction. The
22-kDa fusion protein includes the approximately 15-kDa HT-thioredoxin protein
inserted into the vector pBAD/thio-Topo/MIG20 and the approximately 7-kDa
MIG20
protein.
FIG. 52 is a diagram showing an SDS-PAGE analysis of the MIG20 protein. In
FIG. 52, Lane 1 represents a protein sample before the L-arabinose induction,
and Lane
2 represents a protein sample after the expression of the MIG20 gene is
induced by
L-arabinose.
A DNA base sequence result of the PIG49 was identical with SEQ ID NO: 93.
The DNA sequence of the PIG49 has an open reading frame encoding 345 amino
acid
residues, and the amino acid sequence derived from the open reading frame was
identical with SEQ ID NO: 94. The derived protein also had a molecular weight
of
approximately 38 kDa.
The transformed E. coli strain was incubated in LB broth, and then 1 mM
isopropy-1- j3 -D-thiogalactopyranoside (IPTG) was added to the culture broth,
and
reacted at 37 'C for 3 hours to express the PIG49 gene. A protein sample was
obtained from the culture broth, and then SDS-PAGE was conducted with the
protein
sample according to the method as described in the disclosure (Sambrook, J. et
al.,
Molecular Cloning: A Laboratory manual, New York: Cold Spring Harbor
Laboratory
(1989)).
FIG. 53 is a diagram showing an SDS-PAGE analysis of the PIG49 protein. In
122
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
FIG. 53, Lane 1 represents a protein sample before the IPTG induction, and
Lane 2
represents a protein sample after the expression of the PIG49 gene is induced
by IPTG.
As shown in FIG. 53, the expressed PIG49 protein has a molecular weight of
approximately 38 kDa, which corresponds to a molecular weight of a protein
derived
from its DNA sequence.
A DNA base sequence result of the PIG51 was identical with SEQ ID NO: 97.
The DNA sequence of the PIG51 has an open reading frame encoding 247 amino
acid
residues, and the amino acid sequence derived from the open reading frame was
identical with SEQ ID NO: 98. The derived protein also had a molecular weight
of
approximately 28 kDa.
The transformed E. coli strain was incubated in LB broth, and then 1 mM
isopropy-l-i3-D-thiogalactopyranoside (IPTG) was added to the culture broth,
and
reacted at 37 C for 3 hours to express the PIG51 gene. A protein sample was
obtained from the culture broth, and then SDS-PAGE was conducted with the
protein
sample according to the method as described in the disclosure (Sambrook, J. et
al.,
Molecular Cloning: A Laboratory manual, New York: Cold Spring Harbor
Laboratory
(1989)).
FIG. 54 is a diagram showing an SDS-PAGE analysis of the PIG51 protein. In
FIG. 54, Lane 1 represents a protein sample before the IPTG induction, and
Lane 2
represents a protein sample after the expression of the PIG51 gene is induced
by IPTG.
As shown in FIG. 54, the expressed PIG51 protein has a molecular weight of
approximately 28 kDa, which corresponds to a molecular weight of a protein
derived
from its DNA sequence.
123
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
A DNA base sequence result of the MIG12 was identical with SEQ ID NO: 101.
The DNA sequence of the MIG 12 has an open reading frame encoding 44 amino
acid
residues, and the amino acid sequence derived from the open reading frame was
identical with SEQ ID NO: 102. The derived protein also had a molecular weight
of
approximately 5 kDa.
The transformed E. coli strain was incubated in LB broth, and then 1 mM
isopropy-l- f3 -D-thiogalactopyranoside (IPTG) was added to the culture broth,
and
reacted at 37 C for 3 hours to express the MIG12 gene. A protein sample was
obtained from the culture broth, and then SDS-PAGE was conducted with the
protein
sample according to the method as described in the disclosure (Sambrook, J. et
al.,
Molecular Cloning: A Laboratory manual, New York: Cold Spring Harbor
Laboratory
(1989)).
FIG. 55 is a diagram showing an SDS-PAGE analysis of the MIG12 protein. In
FIG. 55, Lane 1 represents a protein sample before the IPTG induction, and
Lane 2
represents a protein sample after the expression of the MIG 12 gene is induced
by IPTG.
As shown in FIG. 55, the expressed MIG12 protein has a molecular weight of
approximately 5 kDa, which corresponds to a molecular weight of a protein
derived
from its DNA sequence.
A DNA base sequence result of the PIG37 was identical with SEQ ID NO: 105.
The DNA sequence of the PIG37 has an open reading frame encoding 472 amino
acid
residues, and the amino acid sequence derived from the open reading frame was
identical with SEQ ID NO: 106. The derived protein also had a molecular weight
of
approximately 53 kDa. The resultant full-length PIG37 cDNA was inserted into
the
124
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
prokaryotic expression vector pBAD/Thio-Topo (Invitrogen, U.A.), and then E.
coli
DH5 a was transformed with the resultant expression vector to obtain a
transformant,
which was designated E. coli DH5 a/PIG37/pBAD/Thio-Topo.
The transformed E. coli strain was incubated in LB broth, and then 1 mM
isopropy-l-f3-D-thiogalactopyranoside (IPTG) was added to the culture broth,
and
reacted at 37 C for 3 hours to express the PIG37 gene. A protein sample was
obtained from the culture broth, and then SDS-PAGE was conducted with the
protein
sample according to the method as described in the disclosure (Sambrook, J. et
al.,
Molecular Cloning: A Laboratory manual, New York: Cold Spring Harbor
Laboratory
(1989)).
FIG. 56 is a diagram showing an SDS-PAGE analysis of the PIG37 protein. In
FIG. 56, Lane 1 represents a protein sample before the IPTG induction, and
Lane 2
represents a protein sample after the expression of the PIG37 gene is induced
by IPTG.
As shown in FIG. 56, the expressed PIG37 protein has a molecular weight of
approximately 53 kDa, which corresponds to a molecular weight of a protein
derived
from its DNA sequence.
A DNA base sequence result of the GIG44 was identical with SEQ ID NO: 109.
The DNA sequence of the GIG44 has an open reading frame encoding 113 amino
acid
residues, and the amino acid sequence derived from the open reading frame was
identical with SEQ ID NO: 110. The derived protein also had a molecular weight
of
approximately 12 kDa.
The transformed E. coli strain was incubated in LB broth, and then 1 mM
isopropy-l-i3 -D-thiogalactopyranoside (IPTG) was added to the culture broth,
and
125
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
reacted at 37 C for 3 hours to express the GIG44 gene. A protein sample was
obtained from the culture broth, and then SDS-PAGE was conducted with the
protein
sample according to the method as described in the disclosure (Sambrook, J. et
al.,
Molecular Cloning: A Laboratory manual, New York: Cold Spring Harbor
Laboratory
(1989)).
FIG. 57 is a diagram showing an SDS-PAGE analysis of the GIG44 protein. In
FIG. 57, Lane 1 represents a protein sample before the IPTG induction, and
Lane 2
represents a protein sample after the expression of the GIG44 gene is induced
by IPTG.
As shown in FIG. 57, the expressed GIG44 protein has a molecular weight of
approximately 12 kDa, which corresponds to a molecular weight of a protein
derived
from its DNA sequence.
A DNA base sequence result of the GIG31 was identical with SEQ ID NO: 113.
The DNA sequence of the GIG31 has an open reading frame encoding 211 amino
acid
residues, and the amino acid sequence derived from the open reading frame was
identical with SEQ ID NO: 114. The derived protein also had a molecular weight
of
approximately 24 kDa.
The transformed E. coli strain was incubated in LB broth, and then 1 mM
isopropy-l-i3 -D-thiogalactopyranoside (IPTG) was added to the culture broth,
and
reacted at 37 C for 3 hours to express the GIG31 gene. A protein sample was
obtained from the culture broth, and then SDS-PAGE was conducted with the
protein
sample according to the method as described in the disclosure (Sambrook, J. et
al.,
Molecular Cloning: A Laboratory manual, New York: Cold Spring Harbor
Laboratory
(1989)).
126
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
FIG. 58 is a diagram showing an SDS-PAGE analysis of the GIG31 protein. In
FIG. 58, Lane 1 represents a protein sample before the IPTG induction, and
Lane 2
represents a protein sample after the expression of the GIG31 gene is induced
by IPTG.
As shown in FIG. 58, the expressed GIG31 protein has a molecular weight of
approximately 24 kDa, which corresponds to a molecular weight of a protein
derived
from its DNA sequence.
Example 3: Northern Blotting of GIG Gene
3-1. GIG8, GIG10 GIG13 GIG30 GIG32 GIG33 GIG34 GIG35 GIG38
GIG39, GIG43, PIG49, PIG51, GIG44 and GIG31
In order to assess expression levels of the GIG and PIG genes, the northern
blottings were carried out, as follows.
gg of each of the total RNA samples obtained from the three normal breast
tissues, the three primary breast cancer tissues and the breast cancer cell
line MCF-7 in
15 Example 1 was denatured and electrophoresized in a 1% formaldehyde agarose
gel, and
then the resultant agarose gels were transferred to nylon membranes
(Boehringer-Mannheim, Germany), respectively. The nylon membranes were then
hybridized at 42 C overnight with the 32P-labeled random prime probes prepared
from
the partial sequences FC33; FC42; FC59; FC48; FC82; FC86; FC35; FC38; FC122;
20 FC126; FC 102; FC 101; FC22; FC123 and FC47 of the full-length GIG cDNAs
using
the Rediprime II random prime labelling system (Amersham, United Kingdom). The
northern blotting procedure was repeated twice; one is that the blots were
quantitified
using the densitometer and the other is that the blots were hybridized with
the 13 -actin
127
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
probe to determine the total amount of mRNA.
FIG. 59 shows the northern blotting result that the GIG8 gene is
differentially
expressed in a normal breast tissue, a primary breast cancer tissue and a
breast cancer
cell line, and a bottom of FIG. 59 shows the northern blotting result obtained
by
hybridizing the same blot with j3 -actin probe. As shown in FIG. 59 and the
bottom of
FIG. 59, it was revealed that the expression level of the GIG8 gene was highly
detected
all in the three samples of the normal breast tissue, but its expression was
significantly
lower or not detected in the three samples of the breast cancer tissue than
the normal
tissue, and very slightly detected even in the one sample of the breast cancer
cell line.
The northern blotting was carried out on the normal human multiple tissue
(Clontech) and the human cancer cell line (Clontech). That is to say, the
northern
blotting was carried out by hybridizing blots, to which each of the total RNA
samples
extracted from the normal tissues and the cancer cell lines was transferred,
in the same
manner as described above, wherein the blots were commercially available from
the
company Clontech (U.S), and the normal tissue is, for example, selected from
the group
consisting of brain, heart, skeletal muscle, colon, thymus, spleen, kidney,
liver, small
intestine, placenta, lungs and peripheral blood leukocyte, and the cancer cell
line is, for
example, selected from the group consisting of a promyelocytic leukemia HL-60,
an
HeLa cervical cancer cell, a chronic myelocytic leukemia cell line K-562,
lymphoblastoid leukemia MOLT-4, a Burkitt's lymphoma Raji, an SW480 colon
cancer
cell, an A549 lung cancer cell and a G361 melanoma cell.
FIG. 88 shows a northern blotting result that the GIG8 gene is differentially
expressed in various normal tissues, and a bottom of FIG. 88 shows a northern
blotting
128
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
result obtained by hybridizing the same blot with J3 -actin probe. As shown in
Fig. 88,
a dominant GIG8 mRNA transcript having a size of approximately 1.3 kb was
overexpressed in the normal tissues such as the brain, the heart, the muscle,
the large
intestine, the thymus, the spleen, the kidney, the liver, the small intestine,
the placenta,
the lungs and the peripheral blood. A GIG8 mRNA transcript having a size of
approximately 2.5 kb was also expressed in the normal tissues such as the
liver and the
peripheral blood at the same time. FIG. 117 shows a northern blotting result
that the
GIG8 gene is differentially expressed in various cancer cell lines, and a
bottom of FIG.
117 shows a northern blotting result obtained by hybridizing the same blot
with
13 -actin probe. As shown in FIG. 117, the GIG8 gene was not expressed in the
tissues
such as the promyelocytic leukemia HL-60, the HeLa cervical cancer cell, the
chronic
myelocytic leukemia cell line K-562, the lymphoblastoid leukemia MOLT-4, the
Burkitt's lymphoma Raji, the SW480 colon cancer cell, the A549 lung cancer
cell and
the G361 melanoma cell. From such a result, it might be seen that the GIG8
gene of
the present invention had the tumor suppresser function in the normal tissues
such as the
breast, the brain, the heart, the muscle, the thymus, the spleen, the kidney,
the liver, the
small intestine, the placenta, the lungs, the large intestine and the
peripheral blood.
FIG. 60 shows the northern blotting result that the GIG10 gene is
differentially
expressed in a normal breast tissue, a primary breast cancer tissue and a
breast cancer
cell line, and a bottom of FIG. 60 shows the northern blotting result obtained
by
hybridizing the same blot with j3 -actin probe. As shown in FIG. 60, it was
revealed
that the expression level of the GIG 10 gene was highly detected all in the
three samples
of the normal breast tissue, but its expression was significantly lower or not
detected in
129
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
the three samples of the breast cancer tissue than the nonnal tissue, and very
slightly
detected even in the one sample of the breast cancer cell line.
The northern blotting was carried out on the normal human multiple tissue
(Clontech) and the human cancer cell line (Clontech). That is to say, the
northern
blotting was carried out by hybridizing blots, to which each of the total RNA
samples
extracted from the normal tissues and the cancer cell lines was transferred,
in the same
manner as described above, wherein the blots were commercially available from
the
company Clontech (U.S), and the normal tissue is, for example, selected from
the group
consisting of brain, heart, skeletal muscle, colon, thymus, spleen, kidney,
liver, small
intestine, placenta, lungs and peripheral blood leukocyte, and the cancer cell
line is, for
example, selected from the group consisting of a promyelocytic leukemia HL-60,
an
HeLa cervical cancer cell, a chronic myelocytic leukemia cell line K-562,
lymphoblastoid leukemia MOLT-4, a Burkitt's lymphoma Raji, an SW480 colon
cancer
cell, an A5491ung cancer cell and a G361 melanoma cell.
FIG. 89 shows a northern blotting result that the GIG10 gene is differentially
expressed in various normal tissues, and a bottom of FIG. 89 shows a northern
blotting
result obtained by hybridizing the same blot with J3 -actin probe. As shown in
Fig. 89,
a dominant GIG10 mRNA transcript having a size of approximately 3.5 kb was
overexpressed in the normal tissues such as the brain, the heart, the muscle,
the thymus,
the spleen, the kidney, the liver, the small intestine, the placenta, the
lungs and the
peripheral blood, but not expressed in the large intestine tissue. A GIG10
mRNA
transcript having a size of approximately 2.2 kb was also expressed in the
normal tissues
such as the heart and the placenta at the same time. FIG. 118 shows a northern
blotting
130
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
result that the GIG10 gene is differentially expressed in various cancer cell
lines, and a
bottom of FIG. 118 shows a northern blotting result obtained by hybridizing
the same
blot with j3 -actin probe. As shown in FIG. 118, the GIG8 gene was not
expressed in
the tissues such as the HeLa cervical cancer cell, the chronic myelocytic
leukemia cell
line K-562, the A549 lung cancer cell and the G361 melanoma cell, but its
expression
was detected in the promyelocytic leukemia HL-60, the lymphoblastoid leukemia
MOLT-4, the Burkitt's lymphoma Raji and the SW480 colon cancer cell.
From such a result, it might be seen that the GIG10 gene of the present
invention
had the tumor suppresser function in the normal tissues such as the breast,
the brain, the
heart, the muscle, the thymus, the spleen, the kidney, the liver, the small
intestine, the
placenta, the lungs and the peripheral blood.
FIG. 61 shows the northern blotting result that the GIG13 gene is
differentially
expressed in a normal breast tissue, a primary breast cancer tissue and a
breast cancer
cell line, and a bottom of FIG. 61 shows the northern blotting result obtained
by
hybridizing the same blot with J3 -actin probe. As shown in FIG. 61, it was
revealed
that the expression level of the GIG 13 gene was highly detected all in the
three samples
of the normal breast tissue, but its expression was significantly lower or not
detected in
the three samples of the breast cancer tissue than the normal tissue, and very
slightly
detected even in the one sample of the breast cancer cell line.
The northern blotting was carried out on the normal human multiple tissue
(Clontech) and the human cancer cell line (Clontech). That is to say, the
northern
blotting was carried out by hybridizing blots, to which each of the total RNA
samples
extracted from the normal tissues and the cancer cell lines was transferred,
in the same
131
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
manner as described above, wherein the blots were commercially available from
the
company Clontech (U.S), and the normal tissue is, for example, selected from
the group
consisting of brain, heart, skeletal muscle, colon, thymus, spleen, kidney,
liver, small
intestine, placenta, lungs and peripheral blood leukocyte, and the cancer cell
line is, for
example, selected from the group consisting of a promyelocytic leukemia HL-60,
an
HeLa cervical cancer cell, a chronic myelocytic leukemia cell line K-562,
lymphoblastoid leukemia MOLT-4, a Burkitt's lymphoma Raji, an SW480 colon
cancer
cell, an A549 lung cancer cell and a G361 melanoma cell.
FIG. 90 shows a northern blotting result that the GIG13 gene is differentially
expressed in various normal tissues, and a bottom of FIG. 90 shows a northern
blotting
result obtained by hybridizing the same blot with J3 -actin probe. As shown in
Fig. 90,
a dominant GIG13 mRNA transcript having a size of approximately 1.3 kb was
overexpressed only in the normal liver tissue. FIG. 119 shows a northern
blotting
result that the GIG13 gene is differentially expressed in various cancer cell
lines, and a
bottom of FIG. 119 shows a northern blotting result obtained by hybridizing
the same
blot with 13 -actin probe. As shown in FIG. 119, the GIG13 gene was not
expressed in
the tissues such as the promyelocytic leukemia HL-60, the HeLa cervical cancer
cell, the
chronic myelocytic leukemia cell line K-562, the lymphoblastoid leukemia MOLT-
4,
the Burkitt's lymphoma Raji, the SW480 colon cancer cell, the A549 lung cancer
cell
and the G361 melanoma cell. From such a result, it might be seen that the
GIG13 gene
of the present invention had the tumor suppresser function in the normal
tissues such as
the breast and the liver.
FIG. 67 shows the northern blotting result that the GIG30 gene is
differentially
132
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
expressed in a normal breast tissue, a primary breast cancer tissue and a
breast cancer
cell line, and a bottom of FIG. 67 shows the northern blotting result obtained
by
hybridizing the same blot with J3 -actin probe. As shown in FIG. 67, it was
revealed
that the expression level of the GIG30 gene was highly detected all in the
three samples
of the normal breast tissue, but its expression was significantly lower or not
detected in
the three samples of the breast cancer tissue than the normal tissue, and very
slightly
detected even in the one sample of the breast cancer cell line.
The northern blotting was carried out on the normal human multiple tissue
(Clontech) and the human cancer cell line (Clontech). That is to say, the
northern
blotting was carried out by hybridizing blots, to which each of the total RNA
samples
extracted from the normal tissues and the cancer cell lines was transferred,
in the same
manner as described above, wherein the blots were commercially available from
the
company Clontech (U.S), and the normal tissue is, for example, selected from
the group
consisting of brain, heart, skeletal muscle, colon, thymus, spleen, kidney,
liver, small
intestine, placenta, lungs and peripheral blood leukocyte, and the cancer cell
line is, for
example, selected from the group consisting of a promyelocytic leukemia HL-60,
an
HeLa cervical cancer cell, a chronic myelocytic leukemia cell line K-562,
lymphoblastoid leukemia MOLT-4, a Burkitt's lymphoma Raji, an SW480 colon
cancer
cell, an A549 lung cancer cell and a G361 melanoma cell.
FIG. 96 shows a northern blotting result that the GIG30 gene is differentially
expressed in various normal tissues, and a bottom of FIG. 96 shows a northern
blotting
result obtained by hybridizing the same blot with j3 -actin probe. As shown in
Fig. 96,
a dominant GIG30 mRNA transcript having a size of approximately 1.9 kb was
133
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
overexpressed in the normal tissues such as the heart, the muscle and the
liver. A
GIG30 mRNA transcript having a size of approximately 1.0 kb was also expressed
in
the normal tissues such as the liver and the peripheral blood at the same
time. FIG.
125 shows a northern blotting result that the GIG30 gene is differentially
expressed in
various cancer cell lines, and a bottom of FIG. 125 shows a northern blotting
result
obtained by hybridizing the same blot with 13 -actin probe. As shown in FIG.
125, the
GIG30 gene was not expressed in the tissues such as the promyelocytic leukemia
HL-60,
the HeLa cervical cancer cell, the chronic myelocytic leukemia cell line K-
562, the
lymphoblastoid leukemia MOLT-4, the Burkitt's lymphoma Raji, the SW480 colon
cancer cell, the A549 lung cancer cell and the G361 melanoma cell. From such a
result,
it might be seen that the GIG30 gene of the present invention had the tumor
suppresser
function in the normal tissues such as the breast, the heart, the muscle and
the liver.
FIG. 68 shows the northern blotting result that the GIG32 gene is
differentially
expressed in a normal breast tissue, a primary breast cancer tissue and a
breast cancer
cell line, and a bottom of FIG. 68 shows the northern blotting result obtained
by
hybridizing the same blot with 13 -actin probe. As shown in FIG. 68, it was
revealed
that the expression level of the GIG32 gene was highly detected all in the
three samples
of the normal breast tissue, but its expression was significantly lower or not
detected in
the three samples of the breast cancer tissue than the normal tissue, and very
slightly
detected even in the one sample of the breast cancer cell line.
The northern blotting was carried out on the normal human multiple tissue
(Clontech) and the human cancer cell line (Clontech). That is to say, the
northern
blotting was carried out by hybridizing blots, to which each of the total RNA
samples
134
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
extracted from the normal tissues and the cancer cell lines was transferred,
in the same
manner as described above, wherein the blots were commercially available from
the
company Clontech (U.S), and the normal tissue is, for example, selected from
the group
consisting of brain, heart, skeletal muscle, colon, thymus, spleen, kidney,
liver, small
intestine, placenta, lungs and peripheral blood leukocyte, and the cancer cell
line is, for
example, selected from the group consisting of a promyelocytic leukemia HL-60,
an
HeLa cervical cancer cell, a chronic myelocytic leukemia cell line K-562,
lymphoblastoid leukemia MOLT-4, a Burkitt's lymphoma Raji, an SW480 colon
cancer
cell, an A549 lung cancer cell and a G361 melanoma cell.
FIG. 97 shows a northern blotting result that the GIG32 gene is differentially
expressed in various normal tissues, and a bottom of FIG. 97 shows a northern
blotting
result obtained by hybridizing the same blot with j3 -actin probe. As shown in
Fig. 97,
a dominant GIG32 mRNA transcript having a size of approximately 4.0 kb was
overexpressed in the normal tissues such as the muscle, the large intestine,
the thymus,
the spleen, the kidney, the placenta, the lungs and the peripheral blood. A
GIG32
mRNA transcript having a size of approximately 1.0 kb was also expressed in
the
normal tissues such as the muscle and the large intestine at the same time.
FIG. 126
shows a northern blotting result that the GIG32 gene is differentially
expressed in
various cancer cell lines, and a bottom of FIG. 126 shows a northern blotting
result
obtained by hybridizing the same blot with j3 -actin probe. As shown in FIG.
126, the
GIG32 gene was expressed in the tissues such as the HeLa cervical cancer cell,
the
A549 lung cancer cell and the G361 melanoma cell, but not expressed in the
promyelocytic leukemia HL-60, the chronic myelocytic leukemia cell line K-562,
the
135
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
lymphoblastoid leukemia MOLT-4, the Burkitt's lymphoma Raji and the SW480
colon
cancer cell.
From such a result, it might be seen that the GIG32 gene of the present
invention
had the tumor suppresser function in the normal tissues such as the breast,
the muscle,
the large intestine, the thymus, the spleen, the kidney, the placenta and the
peripheral
blood.
FIG. 70 shows the northern blotting result that the GIG34 gene is
differentially
expressed in a normal breast tissue, a primary breast cancer tissue and a
breast cancer
cell line, and a bottom of FIG. 70 shows the northern blotting result obtained
by
hybridizing the same blot with j3 -actin probe. As shown in FIG. 70, it was
revealed
that the expression level of the GIG34 gene was highly detected all in the
three samples
of the normal breast tissue, but its expression was significantly lower or not
detected in
the three samples of the breast cancer tissue than the normal tissue, and very
slightly
detected even in the one sample of the breast cancer cell line.
The northern blotting was carried out on the normal human multiple tissue
(Clontech) and the human cancer cell line (Clontech). That is to say, the
northern
blotting was carried out by hybridizing blots, to which each of the total RNA
samples
extracted from the normal tissues and the cancer cell lines was transferred,
in the same
manner as described above, wherein the blots were commercially available from
the
company Clontech (U.S), and the normal tissue is, for example, selected from
the group
consisting of brain, heart, skeletal muscle, colon, thymus, spleen, kidney,
liver, small
intestine, placenta, lungs and peripheral blood leukocyte, and the cancer cell
line is, for
example, selected from the group consisting of a promyelocytic leukemia HL-60,
an
136
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
HeLa cervical cancer cell, a chronic myelocytic leukemia cell line K-562,
lymphoblastoid leukemia MOLT-4, a Burkitt's lymphoma Raji, an SW480 colon
cancer
cell, an A549 lung cancer cell and a G361 melanoma cell.
FIG. 99 shows a northern blotting result that the GIG34 gene is differentially
expressed in various normal tissues, and a bottom of FIG. 99 shows a northern
blotting
result obtained by hybridizing the same blot with 13 -actin probe. As shown in
Fig. 99,
the dominant GIG34 mRNA transcript having a size of approximately 0.6 kb was
overexpressed in the normal tissues such as the brain, the heart, the muscle,
the large
intestine, the thymus, the spleen, the kidney, the liver, the small intestine,
the placenta,
the lungs and the peripheral blood. FIG. 128 shows a northern blotting result
that the
GIG34 gene is differentially expressed in various cancer cell lines, and a
bottom of FIG.
128 shows a northern blotting result obtained by hybridizing the same blot
with
13 -actin probe. As shown in FIG. 128, the GIG34 gene was rarely expressed in
the
tissues such as the promyelocytic leukemia HL-60, the HeLa cervical cancer
cell, the
chronic myelocytic leukemia cell line K-562, the lymphoblastoid leukemia MOLT-
4,
the Burkitt's lymphoma Raji, the SW480 colon cancer cell, the A549 lung cancer
cell
and the G361 melanoma cell.
From such a result, it might be seen that the GIG34 gene of the present
invention
had the tumor suppresser function in the normal tissues such as the breast,
the brain, the
heart, the muscle, the large intestine, the thymus, the spleen, the kidney,
the liver, the
small intestine, the placenta, the lungs and the peripheral blood.
FIG. 71 shows the northern blotting result that the GIG35 gene is
differentially
expressed in a normal breast tissue, a primary breast cancer tissue and a
breast cancer
137
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
cell line, and a bottom of FIG. 71 shows the northern blotting result obtained
by
hybridizing the same blot with j3 -actin probe. As shown in FIG. 71, it was
revealed
that the expression level of the GIG35 gene was highly detected all in the
three samples
of the normal breast tissue, but its expression was significantly lower in the
three
samples of the breast cancer tissue than the normal tissue, and very slightly
detected
even in the one sample of the breast cancer cell line.
The northern blotting was carried out on the normal human multiple tissue
(Clontech) and the human cancer cell line (Clontech). That is to say, the
northern
blotting was carried out by hybridizing blots, to which each of the total RNA
samples
extracted from the normal tissues and the cancer cell lines was transferred,
in the same
manner as described above, wherein the blots were commercially available from
the
company Clontech (U.S), and the normal tissue is, for example, selected from
the group
consisting of brain, heart, skeletal muscle, colon, thymus, spleen, kidney,
liver, small
intestine, placenta, lungs and peripheral blood leukocyte, and the cancer cell
line is, for
example, selected from the group consisting of a promyelocytic leukemia HL-60,
an
HeLa cervical cancer cell, a chronic myelocytic leukemia cell line K-562,
lymphoblastoid leukemia MOLT-4, a Burkitt's lymphoma Raji, an SW480 colon
cancer
cell, an A549 lung cancer cell and a G361 melanoma cell.
FIG. 100 shows a northern blotting result that the GIG35 gene is
differentially
expressed in various normal tissues, and a bottom of FIG. 100 shows a northern
blotting
result obtained by hybridizing the same blot with J3 -actin probe. As shown in
Fig.
100, a GIG35 mRNA transcript having a size of approximately 1.3 kb was also
expressed in the normal tissues such the brain, the heart, the muscle, the
large intestine,
138
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
the thymus, the spleen, the kidney, the liver, the small intestine, the
placenta, the lungs
and the peripheral blood. FIG. 129 shows a northern blotting result that the
GIG35
gene is differentially expressed in various cancer cell lines, and a bottom of
FIG. 129
shows a northern blotting result obtained by hybridizing the same blot with J3
-actin
probe. As shown in FIG. 129, the GIG35 gene was rarely expressed in the
tissues such
as the promyelocytic leukemia HL-60, the HeLa cervical cancer cell, the
chronic
myelocytic leukemia cell line K-562, the lymphoblastoid leukemia MOLT-4, the
Burkitt's lymphoma Raji, the SW480 colon cancer cell, the A549 lung cancer
cell and
the G361 melanoma cell. From such a result, it might be seen that the GIG35
gene of
the present invention had the tumor suppresser function in the normal tissues
such as the
breast, the brain, the heart, the muscle, the thymus, the spleen, the kidney,
the liver, the
small intestine, the placenta, the lungs, the large intestine and the
peripheral blood.
FIG. 72 shows the northern blotting result that the GIG38 gene is
differentially
expressed in a normal breast tissue, a primary breast cancer tissue and a
breast cancer
cell line, and a bottom of FIG. 72 shows the northern blotting result obtained
by
hybridizing the same blot with I3 -actin probe. As shown in FIG. 72, it was
revealed
that the expression level of the GIG38 gene was highly detected all in the
three samples
of the normal breast tissue, but its expression was significantly lower or not
detected in
the three samples of the breast cancer tissue than the normal tissue, and very
slightly
detected even in the one sample of the breast cancer cell line.
The northern blotting was carried out on the normal human multiple tissue
(Clontech) and the human cancer cell line (Clontech). That is to say, the
northern
blotting was carried out by hybridizing blots, to which each of the total RNA
samples
139
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
extracted from the normal tissues and the cancer cell lines was transferred,
in the same
manner as described above, wherein the blots were commercially available from
the
company Clontech (U.S), and the normal tissue is, for example, selected from
the group
consisting of brain, heart, skeletal muscle, colon, thymus, spleen, kidney,
liver, small
intestine, placenta, lungs and peripheral blood leukocyte, and the cancer cell
line is, for
example, selected from the group consisting of a promyelocytic leukemia HL-60,
an
HeLa cervical cancer cell, a chronic myelocytic leukemia cell line K-562,
lymphoblastoid leukemia MOLT-4, a Burkitt's lymphoma Raji, an SW480 colon
cancer
cell, an A549 lung cancer cell and a G361 melanoma cell.
FIG. 101 shows a northern blotting result that the GIG38 gene is
differentially
expressed in various normal tissues, and a bottom of FIG. 101 shows a northern
blotting
result obtained by hybridizing the same blot with 13 -actin probe. As shown in
Fig.
101, a dominant GIG38 mRNA transcript having a size of approximately 0.7 kb
was
overexpressed in the normal tissues such as the heart, the muscle, the kidney,
the liver
and the placenta. GIG38 mRNA transcripts having a size of approximately 1.5 kb
and
2.0 kb were also expressed in the normal tissues such as the heart and the
muscle at the
same time. FIG. 130 shows a northern blotting result that the GIG38 gene is
differentially expressed in various cancer cell lines, and a bottom of FIG.
130 shows a
northern blotting result obtained by hybridizing the same blot with 13 -actin
probe. As
shown in FIG. 130, the GIG38 gene was very rarely expressed or hardly
expressed in the
tissues such as the promyelocytic leukemia HL-60, the HeLa cervical cancer
cell, the
chronic myelocytic leukemia cell line K-562, the lymphoblastoid leukemia MOLT-
4,
the Burkitt's lymphoma Raji, the SW480 colon cancer cell, the A549 lung cancer
cell
140
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
and the G361 melanoma cell. The GIG38 mRNA transcripts having a size of
approximately 1.5 kb and 2.0 kb proven to be expressed in the normal tissues
all were
not expressed in the cancer cell lines. From such a result, it might be seen
that the
GIG38 gene of the present invention had the tumor suppresser function in the
normal
tissues such as the breast, the heart, the muscle, the kidney, the liver and
the placenta.
FIG. 73 shows the northern blotting result that the GIG39 gene is
differentially
expressed in a normal breast tissue, a primary breast cancer tissue and a
breast cancer
cell line, and a bottom of FIG. 73 shows the northern blotting result obtained
by
hybridizing the same blot with J3 -actin probe. As shown in FIG. 73, it was
revealed
that the expression level of the GIG39 gene was highly detected all in the
three samples
of the normal breast tissue, but its expression was significantly lower or not
detected in
the three samples of the breast cancer tissue than the normal tissue, and very
slightly
detected even in the one sample of the breast cancer cell line.
The northern blotting was carried out on the normal human multiple tissue
(Clontech) and the human cancer cell line (Clontech). That is to say, the
northern
blotting was carried out by hybridizing blots, to which each of the total RNA
samples
extracted from the normal tissues and the cancer cell lines was transferred,
in the same
manner as described above, wherein the blots were commercially available from
the
company Clontech (U.S), and the normal tissue is, for example, selected from
the group
consisting of brain, heart, skeletal muscle, colon, thymus, spleen, kidney,
liver, small
intestine, placenta, lungs and peripheral blood leukocyte, and the cancer cell
line is, for
example, selected from the group consisting of a promyelocytic leukemia HL-60,
an
HeLa cervical cancer cell, a chronic myelocytic leukemia cell line K-562,
141
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
lymphoblastoid leukemia MOLT-4, a Burkitt's lymphoma Raji, an SW480 colon
cancer
cell, an A549 lung cancer cell and a G361 melanoma cell.
FIG. 102 shows a northern blotting result that the GIG39 gene is
differentially
expressed in various normal tissues, and a bottom of FIG. 102 shows a northern
blotting
result obtained by hybridizing the same blot with 13 -actin probe. As shown in
Fig.
102, a dominant GIG39 mRNA transcript having a size of 2.4 kb was
overexpressed
only in the liver normal tissue. FIG. 131 shows a northern blotting result
that the
GIG39 gene is differentially expressed in various cancer cell lines, and a
bottom of FIG.
131 shows a northern blotting result obtained by hybridizing the same blot
with
13 -actin probe. As shown in FIG. 131, the GIG39 gene was not expressed in the
tissues such as the promyelocytic leukemia HL-60, the HeLa cervical cancer
cell, the
chronic myelocytic leukemia cell line K-562, the lymphoblastoid leukemia MOLT-
4,
the Burkitt's lymphoma Raji, the SW480 colon cancer cell, the A549 lung cancer
cell
and the G361 melanoma cell. From such a result, it might be seen that the
GIG39 gene
of the present invention had the tumor suppresser function in the normal
tissues such as
the breast and the liver.
FIG. 76 shows the northern blotting result that the GIG43 gene is
differentially
expressed in a normal breast tissue, a primary breast cancer tissue and a
breast cancer
cell line, and a bottom of FIG. 76 shows the northern blotting result obtained
by
hybridizing the same blot with j3 -actin probe. As shown in FIG. 76, it was
revealed
that the expression level of the GIG43 gene was highly detected all in the
three samples
of the normal breast tissue, but its expression was significantly lower or not
detected in
the three samples of the breast cancer tissue than the normal tissue, and very
slightly
142
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
detected even in the one sample of the breast cancer cell line.
The northern blotting was carried out on the normal human multiple tissue
(Clontech) and the human cancer cell line (Clontech). That is to say, the
northern
blotting was carried out by hybridizing blots, to which each of the total RNA
samples
extracted from the normal tissues and the cancer cell lines was transferred,
in the same
manner as described above, wherein the blots were commercially available from
the
company Clontech (U.S), and the normal tissue is, for example, selected from
the group
consisting of brain, heart, skeletal muscle, colon, thymus, spleen, kidney,
liver, small
intestine, placenta, lungs and peripheral blood leukocyte, and the cancer cell
line is, for
example, selected from the group consisting of a promyelocytic leukemia HL-60,
an
HeLa cervical cancer cell, a chronic myelocytic leukemia cell line K-562,
lymphoblastoid leukemia MOLT-4, a Burkitt's lymphoma Raji, an SW480 colon
cancer
cell, an A549 lung cancer cell and a G361 melanoma cell.
FIG. 105 shows a northern blotting result that the GIG43 gene is
differentially
expressed in various normal tissues, and a bottom of FIG. 105 shows a northern
blotting
result obtained by hybridizing the same blot with J3 -actin probe. As shown in
Fig.
105, the dominant GIG43 mRNA transcript having a size of approximately 3.5 kb
was
overexpressed in the normal tissues such as the heart, the kidney, the liver,
the placenta
and the lungs. FIG. 134 shows a northern blotting result that the GIG43 gene
is
differentially expressed in various cancer cell lines, and a bottom of FIG.
134 shows a
northern blotting result obtained by hybridizing the same blot with j3 -actin
probe. As
shown in FIG. 134, the GIG8 gene was not expressed in the tissues such as the
promyelocytic leukemia HL-60, the HeLa cervical cancer cell, the chronic
myelocytic
143
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
leukemia cell line K-562, the lymphoblastoid leukemia MOLT-4, the Burkitt's
lymphoma Raji, the SW480 colon cancer cell, the A549 lung cancer cell and the
G361
melanoma cell. From such a result, it might be seen that the GIG8 gene of the
present
invention had the tumor suppresser function in the normal tissues such as the
breast, the
heart, the kidney, the liver, the placenta and the lungs.
FIG. 82 shows the northern blotting result that the PIG49 gene is
differentially
expressed in a normal breast tissue, a primary breast cancer tissue and a
breast cancer
cell line, and a bottom of FIG. 82 shows the northern blotting result obtained
by
hybridizing the same blot with J3 -actin probe. As shown in FIG. 82, it was
revealed
that the expression level of the PIG49 gene was highly detected all in the
three samples
of the normal breast tissue, but its expression was significantly lower or not
detected in
the three samples of the breast cancer tissue than the normal tissue, and very
slightly
detected even in the one sample of the breast cancer cell line.
The northern blotting was carried out on the normal human multiple tissue
(Clontech) and the human cancer cell line (Clontech). That is to say, the
northern
blotting was carried out by hybridizing blots, to which each of the total RNA
samples
extracted from the normal tissues and the cancer cell lines was transferred,
in the same
manner as described above, wherein the blots were commercially available from
the
company Clontech (U.S), and the normal tissue is, for example, selected from
the group
consisting of brain, heart, skeletal muscle, colon, thymus, spleen, kidney,
liver, small
intestine, placenta, lungs and peripheral blood leukocyte, and the cancer cell
line is, for
example, selected from the group consisting of a promyelocytic leukemia HL-60,
an
HeLa cervical cancer cell, a chronic myelocytic leukemia cell line K-562,
144
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
lymphoblastoid leukemia MOLT-4, a Burkitt's lymphoma Raji, an SW480 colon
cancer
cell, an A549 lung cancer cell and a G361 melanoma cell.
FIG. 111 shows a northern blotting result that the PIG49 gene is
differentially
expressed in various normal tissues, and a bottom of FIG. 111 shows a northern
blotting
result obtained by hybridizing the same blot with J3 -actin probe. As shown in
Fig.
111, a dominant PIG49 mRNA transcript having a size of approximately 2.4 kb
was
overexpressed in the normal tissues such as the heart, the muscle, the kidney,
the liver
and the placenta. A PIG49 mRNA transcript having a size of approximately 1.5
kb
was also expressed in the normal muscle tissue at the same time. FIG. 140
shows a
northern blotting result that the PIG49 gene is differentially expressed in
various cancer
cell lines, and a bottom of FIG. 140 shows a northern blotting result obtained
by
hybridizing the same blot with 13 -actin probe. As shown in FIG. 140, the
PIG49 gene
was not expressed or very rarely expressed in the tissues such as the
promyelocytic
leukemia HL-60, the lymphoblastoid leukemia MOLT-4, the HeLa cervical cancer
cell,
the chronic myelocytic leukemia cell line K-562, the lymphoblastoid leukemia
MOLT-4,
the Burkitt's lymphoma Raji, the SW480 colon cancer cell, the A549 lung cancer
cell
and the G361 melanoma cell.
From such a result, it might be seen that the PIG49 gene of the present
invention
had the tumor suppresser function in the normal tissues such as the breast,
the heart, the
muscle, the kidney, the liver and the placenta.
FIG. 83 shows the northern blotting result that the PIG51 gene is
differentially
expressed in a normal breast tissue, a primary breast cancer tissue and a
breast cancer
cell line, and a bottom of FIG. 83 shows the northern blotting result obtained
by
145
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
hybridizing the same blot with 13 -actin probe. As shown in FIG. 83, it was
revealed
that the expression level of the PIG51 gene was highly detected all in the
three samples
of the normal breast tissue, but its expression was significantly lower or not
detected in
the three samples of the breast cancer tissue than the normal tissue, and very
slightly
detected even in the one sample of the breast cancer cell line.
The northern blotting was carried out on the normal human multiple tissue
(Clontech) and the human cancer cell line (Clontech). That is to say, the
northern
blotting was carried out by hybridizing blots, to which each of the total RNA
samples
extracted from the normal tissues and the cancer cell lines was transferred,
in the same
manner as described above, wherein the blots were commercially available from
the
company Clontech (U.S), and the normal tissue is, for example, selected from
the group
consisting of brain, heart, skeletal muscle, colon, thymus, spleen, kidney,
liver, small
intestine, placenta, lungs and peripheral blood leukocyte, and the cancer cell
line is, for
example, selected from the group consisting of a promyelocytic leukemia HL-60,
an
HeLa cervical cancer cell, a chronic myelocytic leukemia cell line K-562,
lymphoblastoid leukemia MOLT-4, a Burkitt's lymphoma Raji, an SW480 colon
cancer
cell, an A549 lung cancer cell and a G361 melanoma cell.
FIG. 112 shows a northern blotting result that the PIG51 gene is
differentially
expressed in various normal tissues, and a bottom of FIG. 112 shows a northern
blotting
result obtained by hybridizing the same blot with Ji -actin probe. As shown in
Fig.
112, a dominant PIG51 mRNA transcript having a size of approximately 1.0 kb
was
overexpressed in the normal tissues such as the heart, the muscls, the thymus,
the spleen,
the kidney, the liver, the placenta and the peripheral blood. FIG. 141 shows a
northern
146
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
blotting result that the PIG51 gene is differentially expressed in various
cancer cell lines,
and a bottom of FIG. 141 shows a northern blotting result obtained by
hybridizing the
same blot with j3 -actin probe. As shown in FIG. 141, the PIG51 gene was not
expressed in the tissues such as the promyelocytic leukemia HL-60, the HeLa
cervical
cancer cell, the chronic myelocytic leukemia cell line K-562, the
lymphoblastoid
leukemia MOLT-4, the Burkitt's lymphoma Raji, the SW480 colon cancer cell, the
A549 lung cancer cell and the G361 melanoma cell. From such a result, it might
be
seen that the PIG51 gene of the present invention had the tumor suppresser
function in
the normal tissues such as the breast, the heart, the muscle, the thymus, the
spleen, the
kidney, the liver, the placenta and the peripheral blood.
FIG. 86 shows the northern blotting result that the GIG44 gene is
differentially
expressed in a normal breast tissue, a primary breast cancer tissue and a
breast cancer
cell line, and a bottom of FIG. 86 shows the northern blotting result obtained
by
hybridizing the same blot with j3 -actin probe. As shown in FIG. 86, it was
revealed
that the expression level of the GIG44 gene was highly detected all in the
three samples
of the normal breast tissue, but its expression was significantly lower or not
detected in
the three samples of the breast cancer tissue than the normal tissue, and very
slightly
detected even in the one sample of the breast cancer cell line.
The northern blotting was carried out on the normal human multiple tissue
(Clontech) and the human cancer cell line (Clontech). That is to say, the
northern
blotting was carried out by hybridizing blots, to which each of the total RNA
samples
extracted from the normal tissues and the cancer cell lines was transferred,
in the same
manner as described above, wherein the blots were commercially available from
the
147
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
company Clontech (U.S), and the normal tissue is, for example, selected from
the group
consisting of brain, heart, skeletal muscle, colon, thymus, spleen, kidney,
liver, small
intestine, placenta, lungs and peripheral blood leukocyte, and the cancer cell
line is, for
example, selected from the group consisting of a promyelocytic leukemia HL-60,
an
HeLa cervical cancer cell, a chronic myelocytic leukemia cell line K-562,
lymphoblastoid leukemia MOLT-4, a Burkitt's lymphoma Raji, an SW480 colon
cancer
cell, an A549 lung cancer cell and a G361 melanoma cell.
FIG. 115 shows a northern blotting result that the GIG44 gene is
differentially
expressed in various normal tissues, and a bottom of FIG. 115 shows a northern
blotting
result obtained by hybridizing the same blot with j3 -actin probe. As shown in
Fig.
115, a dominant GIG44 mRNA transcript having a size of approximately 1.0 kb
was
overexpressed in the normal tissues such as the brain, the heart, the muscle,
the large
intestine, the thymus, the spleen, the kidney, the liver, the small intestine,
the placenta,
the lung and the leukocyte. A GIG44 mRNA transcript having a size of
approximately
0.5 kb was also expressed in the normal tissues at the same time.
FIG. 144 shows a northern blotting result that the GIG44 gene is
differentially
expressed in various cancer cell lines, and a bottom of FIG. 144 shows a
northern
blotting result obtained by hybridizing the same blot with J3 -actin probe. As
shown
in FIG. 144, the GIG44 gene was very rarely expressed in the tissues such as
the
promyelocytic leukemia HL-60, the HeLa cervical cancer cell, the chronic
myelocytic
leukemia cell line K-562, the lymphoblastoid leukemia MOLT-4, the Burkitt's
lymphoma Raji, the SW480 colon cancer cell, the A549 lung cancer cell and the
G361
melanoma cell. From such a result, it might be seen that the GIG44 gene of the
present
148
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
invention had the tumor suppresser function in the normal tissues such as the
brain, the
heart, the muscle, the large intestine, the thymus, the spleen, the kidney,
the liver, the
small intestine, the placenta, the lung and the leukocyte.
FIG. 87 shows the northern blotting result that the GIG31 gene is
differentially
expressed in a normal breast tissue, a primary breast cancer tissue and a
breast cancer
cell line, and a bottom of FIG. 87 shows the northern blotting result obtained
by
hybridizing the same blot with J3 -actin probe. As shown in FIG. 87, it was
revealed
that the expression level of the GIG31 gene was highly detected all in the
three samples
of the normal breast tissue, but its expression was significantly lower or not
detected in
the three samples of the breast cancer tissue than the normal tissue, and very
slightly
detected even in the one sample of the breast cancer cell line.
The northern blotting was carried out on the normal human multiple tissue
(Clontech) and the human cancer cell line (Clontech). That is to say, the
northern
blotting was carried out by hybridizing blots, to which each of the total RNA
samples
extracted from the normal tissues and the cancer cell lines was transferred,
in the same
manner as described above, wherein the blots were commercially available from
the
company Clontech (U.S), and the normal tissue is, for example, selected from
the group
consisting of brain, heart, skeletal muscle, colon, thymus, spleen, kidney,
liver, small
intestine, placenta, lungs and peripheral blood leukocyte, and the cancer cell
line is, for
example, selected from the group consisting of a promyelocytic leukemia HL-60,
an
HeLa cervical cancer cell, a chronic myelocytic leukemia cell line K-562,
lymphoblastoid leukemia MOLT-4, a Burkitt's lymphoma Raji, an SW480 colon
cancer
cell, an A549 lung cancer cell and a G361 melanoma cell.
149
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
FIG. 116 shows a northern blotting result that the GIG31 gene is
differentially
expressed in various normal tissues, and a bottom of FIG. 116 shows a northern
blotting
result obtained by hybridizing the same blot with J3 -actin probe. As shown in
Fig.
116, a dominant GIG31 mRNA transcript having a size of approximately 1.4 kb
was
overexpressed in the normal tissues such as the breast, the heart, the large
intestine, the
spleen, the small intestine, the placenta, the lung and the leukocyte. FIG.
145 shows a
northern blotting result that the GIG31 gene is differentially expressed in
various cancer
cell lines, and a bottom of FIG. 145 shows a northern blotting result obtained
by
hybridizing the same blot with J3 -actin probe. As shown in FIG. 145, the
GIG31 gene
was not expressed in the tissues such as the promyelocytic leukemia HL-60, the
chronic
myelocytic leukemia cell line K-562, the lymphoblastoid leukemia MOLT-4, the
Burkitt's lymphoma Raji, the SW480 colon cancer cell and the G361 melanoma
cell, but
very rarely expressed in the HeLa cervical cancer cell and the A549 lung
cancer cell.
From such a result, it might be seen that the GIG31 gene of the present
invention had
the tumor suppresser function in the normal tissues such as the breast, the
heart, the
large intestine, the spleen, the small intestine, the placenta, the lung and
the leukocyte.
3-2. GIG15
In order to assess an expression level of the GIG15 gene, the northern
blotting
was carried out, as follows. The total RNA samples were extracted from the
normal
bone marrow tissue, the leukemia bone marrow tissue and the K-562 cell, as
described
in Example 1. 20 gg of each of the total RNA samples was denatured and
electrophoresized in a 1% formaldehyde agarose gel, and then the resultant
agarose gels
were transferred to nylon membranes (Boehringer-Mannheim, Germany),
respectively.
150
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
The nylon membranes were then hybridized at 42 C overnight with the 32P-
labeled
random prime probes prepared from the partial sequence GV2 of the full-length
GIG15
cDNA using the Rediprime II random prime labelling system (Amersham, United
Kingdom). The northern blotting procedure was repeated twice; one is that the
blots
were quantitified using the densitometer and the other is that the blots were
hybridized
with the j3 -actin probe to determine the total amount of mRNA.
FIG. 62 shows the northern blotting result that the GIG15 gene is
differentially
expressed in a normal bone marrow tissue, a leukemia bone marrow tissue and a
K-562
cell, and a bottom of FIG. 62 shows the northern blotting result obtained by
hybridizing
the same blot with 13 -actin probe. As shown in FIG. 62, it was revealed that
the
expression level of the GIG15 gene was highly detected all in the samples of
the normal
bone marrow tissue, but its expression was significantly lower in the samples
of the
leukemia bone marrow tissue than the normal tissue, and slightly detected even
in the
one sample of the leukemia cell line.
The northern blotting was carried out on the normal human multiple tissue
(Clontech) and the human cancer cell line (Clontech). That is to say, the
northern
blotting was carried out by hybridizing blots, to which each of the total RNA
samples
extracted from the normal tissues and the cancer cell lines was transferred,
in the same
manner as described above, wherein the blots were commercially available from
the
company Clontech (U.S), and the normal tissue is, for example, selected from
the group
consisting of brain, heart, skeletal muscle, colon, thymus, spleen, kidney,
liver, small
intestine, placenta, lungs and peripheral blood leukocyte, and the cancer cell
line is, for
example, selected from the group consisting of a promyelocytic leukemia HL-60,
an
151
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
HeLa cervical cancer cell, a chronic myelocytic leukemia cell line K-562,
lymphoblastoid leukemia MOLT-4, a Burkitt's lymphoma Raji, an SW480 colon
cancer
cell, an A549 lung cancer cell and a G361 melanoma cell.
FIG. 91 shows a northern blotting result that the GIG15 gene is differentially
expressed in various normal tissues, and a bottom of FIG. 91 shows a northern
blotting
result obtained by hybridizing the same blot with j3 -actin probe. As shown in
Fig. 91,
a dominant GIG15 mRNA transcript having a size of approximately 0.5 kb was
overexpressed in the normal tissues such as the brain, the heart, the muscle,
the large
intestine, the thymus, the spleen, the kidney, the liver, the small intestine,
the placenta,
the lung and the peripheral blood. A GIG15 mRNA transcript having a size of
approximately 1.0 kb was also expressed in the normal tissues such as the
liver and the
kidney at the same time. FIG. 120 shows a northern blotting result that the
GIG15
gene is differentially expressed in various cancer cell lines, and a bottom of
FIG. 120
shows a northern blotting result obtained by hybridizing the same blot with Ji
-actin
probe. As shown in FIG. 120, the GIG1 5 gene was rarely expressed in the
tissues such
as the promyelocytic leukemia HL-60, the HeLa cervical cancer cell, the
chronic
myelocytic leukemia cell line K-562, the lymphoblastoid leukemia MOLT-4, the
Burkitt's lymphoma Raji, the SW480 colon cancer cell, the A549 lung cancer
cell and
the G361 melanoma cell. From such a result, it might be seen that the GIG15
gene of
the present invention had the tumor suppresser function in the normal tissues
such as the
bone marrow, the brain, the heart, the muscle, the thymus, the spleen, the
kidney, the
liver, the small intestine, the placenta, the lung, the large intestine and
the peripheral
blood.
152
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
3-3. GIG16, GIG24, GIG26, GIG29, GIG40, GIG42, PIG33, PIG35, PIG36,
PIG37
In order to assess an expression level of the GIG and PIG genes, the northern
blottings were carried out, as follows.
20 /ug of each of the total RNA samples obtained from the three normal liver
tissues, the three primary liver cancer tissues and the liver cancer cell line
HepG2 in
Example 1 was denatured and electrophoresized in a 1% formaldehyde agarose
gel, and
then the resultant agarose gels were transferred to nylon membranes
(Boehringer-Mannheim, Germany), respectively. The nylon membranes were then
hybridized at 42 C overnight with the 32P-labeled random prime probes prepared
from
the full-length GIG and PIG cDNAs using the Rediprime II random prime
labelling
system (Amersham, United Kingdom). The northern blotting procedure was
repeated
twice; one is that the blots were quantitified using the densitometer and the
other is that
the blots were hybridized with the J3 -actin probe to determine the total
amount of
mRNA.
FIG. 63 shows the northern blotting result that the GIG16 gene is
differentially
expressed in a normal liver tissue, a primary liver cancer tissue and a liver
cancer cell
line, and a bottom of FIG. 63 shows the northern blotting result obtained by
hybridizing
the same blot with 13 -actin probe. As shown in FIG. 63, it was revealed that
the
expression level of the GIG16 gene was highly detected all in the three
samples of the
normal liver tissue, but its expression was significantly lower in the three
samples of the
liver cancer tissue than the normal tissue, and not detected in the one sample
of the liver
cancer cell line.
153
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
The northern blotting was carried out on the normal human multiple tissue
(Clontech) and the human cancer cell line (Clontech). That is to say, the
northern
blotting was carried out by hybridizing blots, to which each of the total RNA
samples
extracted from the normal tissues and the cancer cell lines was transferred,
in the same
manner as described above, wherein the blots were commercially available from
the
company Clontech (U.S), and the normal tissue is, for example, selected from
the group
consisting of brain, heart, skeletal muscle, colon, thymus, spleen, kidney,
liver, small
intestine, placenta, lungs and peripheral blood leukocyte, and the cancer cell
line is, for
example, selected from the group consisting of a promyelocytic leukemia HL-60,
an
HeLa cervical cancer cell, a chronic myelocytic leukemia cell line K-562,
lymphoblastoid leukemia MOLT-4, a Burkitt's lymphoma Raji, an SW480 colon
cancer
cell, an A549 lung cancer cell and a G361 melanoma cell.
FIG. 92 shows a northern blotting result that the GIG16 gene is differentially
expressed in various normal tissues, and a bottom of FIG. 92 shows a northern
blotting
result obtained by hybridizing the same blot with j3 -actin probe. As shown in
Fig. 92,
the dominant GIG16 mRNA transcript having a size of approximately 2.0 kb was
overexpressed in the normal tissues such as the liver and the kidney.
FIG. 121 shows a northern blotting result that the GIG16 gene is
differentially
expressed in various cancer cell lines, and a bottom of FIG. 121 shows a
northern
blotting result obtained by hybridizing the same blot with Ji -actin probe. As
shown
in FIG. 121, the GIG16 gene was not expressed at all in the tissues such as
the
promyelocytic leukemia HL-60, the HeLa cervical cancer cell, the chronic
myelocytic
leukemia cell line K-562, the lymphoblastoid leukemia MOLT-4, the Burkitt's
154
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
lymphoma Raji, the SW480 colon cancer cell, the A549 lung cancer cell and the
G361
melanoma cell. From such a result, it might be seen that the GIG16 gene of the
present
invention had the tumor suppresser function in the normal tissues such as the
liver and
the kidney. Also, it might be seen that the GIG16 gene of the present
invention had the
tumor suppresser function from the fact that its expression was suppressed
even in the
leukemia, the uterine cancer, the malignant lymphoma, the colon cancer, the
lung cancer
and the skin cancer to induce tumorigenesis.
FIG. 64 shows the northern blotting result that the GIG24 gene is
differentially
expressed in a normal liver tissue, a primary liver cancer tissue and a liver
cancer cell
line, and a bottom of FIG. 64 shows the northern blotting result obtained by
hybridizing
the same blot with 13 -actin probe. As shown in FIG. 64, it was revealed that
the
expression level of the GIG24 gene was highly detected all in the three
samples of the
normal liver tissue, but its expression was significantly lower in the three
samples of the
liver cancer tissue than the normal tissue, and not detected in the one sample
of the liver
cancer cell line.
The northern blotting was carried out on the normal human multiple tissue
(Clontech) and the human cancer cell line (Clontech). That is to say, the
northern
blotting was carried out by hybridizing blots, to which each of the total RNA
samples
extracted from the normal tissues and the cancer cell lines was transferred,
in the same
manner as described above, wherein the blots were commercially available from
the
company Clontech (U.S), and the normal tissue is, for example, selected from
the group
consisting of brain, heart, skeletal muscle, colon, thymus, spleen, kidney,
liver, small
intestine, placenta, lungs and peripheral blood leukocyte, and the cancer cell
line is, for
155
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
example, selected from the group consisting of a promyelocytic leukemia HL-60,
an
HeLa cervical cancer cell, a chronic myelocytic leukemia cell line K-562,
lymphoblastoid leukemia MOLT-4, a Burkitt's lymphoma Raji, an SW480 colon
cancer
cell, an A549 lung cancer cell and a G361 melanoma cell.
FIG. 93 shows a northern blotting result that the GIG24 gene is differentially
expressed in various normal tissues, and a bottom of FIG. 93 shows a northern
blotting
result obtained by hybridizing the same blot with J3 -actin probe. As shown in
Fig. 93,
the dominant GIG24 mRNA transcript having a size of approximately 2.4 kb was
overexpressed in the normal tissues such as the liver, the heart and the
muscle.
FIG. 122 shows a northern blotting result that the GIG24 gene is
differentially
expressed in various cancer cell lines, and a bottom of FIG. 122 shows a
northern
blotting result obtained by hybridizing the same blot with J3 -actin probe. As
shown
in FIG. 122, the GIG24 gene was not expressed at all in the tissues such as
the
promyelocytic leukemia HL-60, the HeLa cervical cancer cell, the chronic
myelocytic
leukemia cell line K-562, the lymphoblastoid leukemia MOLT-4, the Burkitt's
lymphoma Raji, the SW480 colon cancer cell, the A549 lung cancer cell and the
G361
melanoma cell. From such a result, it might be seen that the GIG24 gene of the
present
invention had the tumor suppresser function in the normal tissues such as the
liver, the
heart and the muscle. Also, it might be seen that the GIG24 gene of the
present
invention had the tumor suppresser function from the fact that its expression
was
suppressed even in the leukemia, the uterine cancer, the malignant lymphoma,
the colon
cancer, the lung cancer and the skin cancer to induce tumorigenesis.
FIG. 65 shows the northern blotting result that the GIG26 gene is
differentially
156
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
expressed in a normal liver tissue, a primary liver cancer tissue and a liver
cancer cell
line, and a bottom of FIG. 65 shows the northern blotting result obtained by
hybridizing
the same blot with 13 -actin probe. As shown in FIG. 65, it was revealed that
the
expression level of the GIG26 gene was highly detected all in the three
samples of the
normal liver tissue, but its expression was significantly lower in the three
samples of the
liver cancer tissue than the normal tissue, and not detected in the one sample
of the liver
cancer cell line.
The northern blotting was carried out on the normal human multiple tissue
(Clontech) and the human cancer cell line (Clontech). That is to say, the
northern
blotting was carried out by hybridizing blots, to which each of the total RNA
samples
extracted from the normal tissues and the cancer cell lines was transferred,
in the same
manner as described above, wherein the blots were commercially available from
the
company Clontech (U.S), and the normal tissue is, for example, selected from
the group
consisting of brain, heart, skeletal muscle, colon, thymus, spleen, kidney,
liver, small
intestine, placenta, lungs and peripheral blood leukocyte, and the cancer cell
line is, for
example, selected from the group consisting of a promyelocytic leukemia HL-60,
an
HeLa cervical cancer cell, a chronic myelocytic leukemia cell line K-562,
lymphoblastoid leukemia MOLT-4, a Burkitt's lymphoma Raji, an SW480 colon
cancer
cell, an A549 lung cancer cell and a G361 melanoma cell.
FIG. 94 shows a northern blotting result that the GIG26 gene is differentially
expressed in various normal tissues, and a bottom of FIG. 94 shows a northern
blotting
result obtained by hybridizing the same blot with j3 -actin probe. As shown in
Fig. 94,
the dominant GIG26 mRNA transcript having a size of approximately 2.0 kb was
157
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
overexpressed in the normal liver tissue, and GIG26 mRNA transcripts having a
size of
approximately 2.5 kb and 1.5 kb were also expressed in the kidney, the brain
and the
heart at the same time. FIG. 123 shows a northern blotting result that the
GIG26 gene
is differentially expressed in various cancer cell lines, and a bottom of FIG.
123 shows a
northern blotting result obtained by hybridizing the same blot with j3 -actin
probe. As
shown in FIG. 123, the GIG26 gene was not expressed at all in the tissues such
as the
promyelocytic leukemia HL-60, the HeLa cervical cancer cell, the chronic
myelocytic
leukemia cell line K-562, the lymphoblastoid leukemia MOLT-4, the Burkitt's
lymphoma Raji, the SW480 colon cancer cell, the A549 lung cancer cell and the
G361
melanoma cell. From such a result, it might be seen that the GIG26 gene of the
present
invention had the tumor suppresser function in the normal tissues such as the
liver, the
kidney, the brain and the heart. Also, it might be seen that the GIG16 gene of
the
present invention had the tumor suppresser function from the fact that its
expression was
suppressed even in the leukemia, the uterine cancer, the malignant lymphoma,
the colon
cancer, the lung cancer and the skin cancer to induce tumorigenesis.
FIG. 66 shows the northern blotting result that the GIG29 gene is
differentially
expressed in a normal liver tissue, a primary liver cancer tissue and a liver
cancer cell
line, and a bottom of FIG. 66 shows the northern blotting result obtained by
hybridizing
the same blot with J3 -actin probe. As shown in FIG. 66, it was revealed that
the
expression level of the GIG29 gene was highly detected all in the three
samples of the
normal liver tissue, but its expression was significantly lower in the three
samples of the
liver cancer tissue than the normal tissue, and not detected in the one sample
of the liver
cancer cell line.
158
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
The northern blotting was carried out on the normal human multiple tissue
(Clontech) and the human cancer cell line (Clontech). That is to say, the
northern
blotting was carried out by hybridizing blots, to which each of the total RNA
samples
extracted from the normal tissues and the cancer cell lines was transferred,
in the same
manner as described above, wherein the blots were commercially available from
the
company Clontech (U.S), and the normal tissue is, for example, selected from
the group
consisting of brain, heart, skeletal muscle, colon, thymus, spleen, kidney,
liver, small
intestine, placenta, lungs and peripheral blood leukocyte, and the cancer cell
line is, for
example, selected from the group consisting of a promyelocytic leukemia HL-60,
an
HeLa cervical cancer cell, a chronic myelocytic leukemia cell line K-562,
lymphoblastoid leukemia MOLT-4, a Burkitt's lymphoma Raji, an SW480 colon
cancer
cell, an A549 lung cancer cell and a G361 melanoma cell.
FIG. 95 shows a northern blotting result that the GIG29 gene is differentially
expressed in various normal tissues, and a bottom of FIG. 95 shows a northern
blotting
result obtained by hybridizing the same blot with J3 -actin probe. As shown in
Fig. 92,
the dominant GIG29 mRNA transcript having a size of approximately 1.4 kb was
overexpressed in the normal liver tissue.
FIG. 124 shows a northern blotting result that the GIG29 gene is
differentially
expressed in various cancer cell lines, and a bottom of FIG. 124 shows a
northern
blotting result obtained by hybridizing the same blot with Ji -actin probe. As
shown
in FIG. 124, the GIG29 gene was not expressed at all in the tissues such as
the
promyelocytic leukemia HL-60, the HeLa cervical cancer cell, the chronic
myelocytic
leukemia cell line K-562, the lymphoblastoid leukemia MOLT-4, the Burkitt's
159
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
lymphoma Raji, the SW480 colon cancer cell, the A549 lung cancer cell and the
G361
melanoma cell. From such a result, it might be seen that the GIG29 gene of the
present
invention had the tumor suppresser function in the normal liver tissue. Also,
it might
be seen that the GIG29 gene of the present invention had the tumor suppresser
function
from the fact that its expression was suppressed even in the leukemia, the
uterine cancer,
the malignant lymphoma, the colon cancer, the lung cancer and the skin cancer
to induce
tumorigenesis.
FIG. 74 shows the northern blotting result that the GIG40 gene is
differentially
expressed in a normal liver tissue, a primary liver cancer tissue and a liver
cancer cell
line, and a bottom of FIG. 74 shows the northern blotting result obtained by
hybridizing
the same blot with j3 -actin probe. As shown in FIG. 74, it was revealed that
the
expression level of the GIG40 gene was highly detected all in the three
samples of the
normal liver tissue, but its expression was significantly lower in the three
samples of the
liver cancer tissue than the normal tissue, and not detected in the one sample
of the liver
cancer cell line.
The northern blotting was carried out on the normal human multiple tissue
(Clontech) and the human cancer cell line (Clontech). That is to say, the
northern
blotting was carried out by hybridizing blots, to which each of the total RNA
samples
extracted from the normal tissues and the cancer cell lines was transferred,
in the same
manner as described above, wherein the blots were commercially available from
the
company Clontech (U.S), and the normal tissue is, for example, selected from
the group
consisting of brain, heart, skeletal muscle, colon, thymus, spleen, kidney,
liver, small
intestine, placenta, lungs and peripheral blood leukocyte, and the cancer cell
line is, for
160
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
example, selected from the group consisting of a promyelocytic leukemia HL-60,
an
HeLa cervical cancer cell, a chronic myelocytic leukemia cell line K-562,
lymphoblastoid leukemia MOLT-4, a Burkitt's lymphoma Raji, an SW480 colon
cancer
cell, an A549 lung cancer cell and a G361 melanoma cell.
FIG. 103 shows a northern blotting result that the GIG40 gene is
differentially
expressed in various normal tissues, and a bottom of FIG. 103 shows a northern
blotting
result obtained by hybridizing the same blot with j3 -actin probe. As shown in
Fig.
103, the dominant GIG40 mRNA transcript having a size of approximately 1.5 kb
was
overexpressed in the normal tissues such as the liver, the heart and the
muscle. A
GIG40 mRNA transcript having a size of approximately 5.0 kb was expressed at
the
same time.
FIG. 132 shows a northern blotting result that the GIG40 gene is
differentially
expressed in various cancer cell lines, and a bottom of FIG. 132 shows a
northern
blotting result obtained by hybridizing the same blot with j3 -actin probe. As
shown
in FIG. 132, the GIG40 gene was very rarely expressed in the tissues such as
the
promyelocytic leukemia HL-60, the HeLa cervical cancer cell, the chronic
myelocytic
leukemia cell line K-562, the lymphoblastoid leukemia MOLT-4, the Burkitt's
lymphoma Raji, the SW480 colon cancer cell, the A549 lung cancer cell and the
G361
melanoma cell. From such a result, it might be seen that the GIG40 gene of the
present
invention had the tumor suppresser function in the normal tissues such as the
liver, the
heart and the muscle. Also, it might be seen that the GIG40 gene of the
present
invention had the tumor suppresser function from the fact that its expression
was
suppressed even in the leukemia, the uterine cancer, the malignant lymphoma,
the colon
161
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
cancer, the lung cancer and the skin cancer to induce tumorigenesis.
FIG. 75 shows the northern blotting result that the GIG42 gene is
differentially
expressed in a normal liver tissue, a primary liver cancer tissue and a liver
cancer cell
line, and a bottom of FIG. 75 shows the northern blotting result obtained by
hybridizing
the same blot with j3 -actin probe. As shown in FIG. 75, it was revealed that
the
expression level of the GIG42 gene was highly detected all in the three
samples of the
normal liver tissue, but its expression was significantly lower in the three
samples of the
liver cancer tissue than the normal tissue, and not detected in the one sample
of the liver
cancer cell line.
The northern blotting was carried out on the normal human multiple tissue
(Clontech) and the human cancer cell line (Clontech). That is to say, the
northern
blotting was carried out by hybridizing blots, to which each of the total RNA
samples
extracted from the normal tissues and the cancer cell lines was transferred,
in the same
manner as described above, wherein the blots were commercially available from
the
company Clontech (U.S), and the normal tissue is, for example, selected from
the group
consisting of brain, heart, skeletal muscle, colon, thymus, spleen, kidney,
liver, small
intestine, placenta, lungs and peripheral blood leukocyte, and the cancer cell
line is, for
example, selected from the group consisting of a promyelocytic leukemia HL-60,
an
HeLa cervical cancer cell, a chronic myelocytic leukemia cell line K-562,
lymphoblastoid leukemia MOLT-4, a Burkitt's lymphoma Raji, an SW480 colon
cancer
cell, an A549 lung cancer cell and a G361 melanoma cell.
FIG. 104 shows a northern blotting result that the GIG42 gene is
differentially
expressed in various normal tissues, and a bottom of FIG. 104 shows a northern
blotting
162
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
result obtained by hybridizing the same blot with J3 -actin probe. As shown in
Fig.
104, the dominant GIG42 mRNA transcript having a size of approximately 2.5 kb
was
overexpressed in the normal liver tissue.
FIG. 133 shows a northern blotting result that the GIG42 gene is
differentially
expressed in various cancer cell lines, and a bottom of FIG. 133 shows a
northern
blotting result obtained by hybridizing the same blot with J3 -actin probe. As
shown
in FIG. 133, the GIG42 gene was not expressed at all in the tissues such as
the
promyelocytic leukemia HL-60, the HeLa cervical cancer cell, the chronic
myelocytic
leukemia cell line K-562, the lymphoblastoid leukemia MOLT-4, the Burkitt's
lymphoma Raji, the SW480 colon cancer cell, the A549 lung cancer cell and the
G361
melanoma cell. From such a result, it might be seen that the GIG42 gene of the
present
invention had the tumor suppresser function in the normal liver tissue. Also,
it might
be seen that the GIG42 gene of the present invention had the tumor suppresser
function
from the fact that its expression was suppressed even in the leukemia, the
uterine cancer,
the malignant lymphoma, the colon cancer, the lung cancer and the skin cancer
to induce
tumorigenesis.
FIG. 78 shows the northern blotting result that the PIG33 gene is
differentially
expressed in a normal liver tissue, a primary liver cancer tissue and a liver
cancer cell
line, and a bottom of FIG. 78 shows the northern blotting result obtained by
hybridizing
the same blot with P -actin probe. As shown in FIG. 78, it was revealed that
the
expression level of the PIG33 gene was highly detected all in the three
samples of the
normal liver tissue, but its expression was significantly lower in the three
samples of the
liver cancer tissue than the normal tissue, and not detected in the one sample
of the liver
163
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
cancer cell line.
The northern blotting was carried out on the normal human multiple tissue
(Clontech) and the human cancer cell line (Clontech). That is to say, the
northern
blotting was carried out by hybridizing blots, to which each of the total RNA
samples
extracted from the normal tissues and the cancer cell lines was transferred,
in the same
manner as described above, wherein the blots were commercially available from
the
company Clontech (U.S), and the normal tissue is, for example, selected from
the group
consisting of brain, heart, skeletal muscle, colon, thymus, spleen, kidney,
liver, small
intestine, placenta, lungs and peripheral blood leukocyte, and the cancer cell
line is, for
example, selected from the group consisting of a promyelocytic leukemia HL-60,
an
HeLa cervical cancer cell, a chronic myelocytic leukemia cell line K-562,
lymphoblastoid leukemia MOLT-4, a Burkitt's lymphoma Raji, an SW480 colon
cancer
cell, an A5491ung cancer cell and a G361 melanoma cell.
FIG. 107 shows a northern blotting result that the PIG33 gene is
differentially
expressed in various normal tissues, and a bottom of FIG. 107 shows a northern
blotting
result obtained by hybridizing the same blot with j3 -actin probe. As shown in
Fig.
107, the dominant PIG33 mRNA transcript having a size of approximately 3.0 kb
was
overexpressed in the normal tissues such as the brain, the heart, the skeletal
muscle, the
colon, the thymus, the spleen, the kidney, the liver, the small intestine, the
placenta and
the lung.
FIG. 136 shows a northern blotting result that the PIG33 gene is
differentially
expressed in various cancer cell lines, and a bottom of FIG. 136 shows a
northern
blotting result obtained by hybridizing the same blot with J3 -actin probe. As
shown
164
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
in FIG. 136, the PIG33 gene was not expressed at all in the tissues such as
the HeLa
cervical cancer cell, the chronic myelocytic leukemia cell line K-562, the
lymphoblastoid leukemia MOLT-4, the SW480 colon cancer cell, the A549 lung
cancer
cell and the G361 melanoma cell and rarely expressed in the promyelocytic
leukemia
HL-60 and the Burkitt's lymphoma Raji. From such a result, it might be seen
that the
PIG33 gene of the present invention had the tumor suppresser function in the
normal
tissues such as the brain, the heart, the skeletal muscle, the colon, the
thymus, the spleen,
the kidney, the liver, the small intestine, the placenta and the lung. Also,
it might be
seen that the PIG33 gene of the present invention had the tumor suppresser
function
from the fact that its expression was suppressed even in the leukemia, the
uterine cancer,
the colon cancer, the lung cancer and the skin cancer to induce tumorigenesis.
FIG. 79 shows the northern blotting result that the PIG35 gene is
differentially
expressed in a normal liver tissue, a primary liver cancer tissue and a liver
cancer cell
line, and a bottom of FIG. 79 shows the northern blotting result obtained by
hybridizing
the same blot with 13 -actin probe. As shown in FIG. 79, it was revealed that
the
expression level of the PIG35 gene was highly detected all in the three
samples of the
normal liver tissue, but its expression was significantly lower in the three
samples of the
liver cancer tissue than the normal tissue, and not detected in the one sample
of the liver
cancer cell line.
The northern blotting was carried out on the normal human multiple tissue
(Clontech) and the human cancer cell line (Clontech). That is to say, the
northern
blotting was carried out by hybridizing blots, to which each of the total RNA
samples
extracted from the normal tissues and the cancer cell lines was transferred,
in the same
165
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
manner as described above, wherein the blots were commercially available from
the
company Clontech (U.S), and the normal tissue is, for example, selected from
the group
consisting of brain, heart, skeletal muscle, colon, thymus, spleen, kidney,
liver, small
intestine, placenta, lungs and peripheral blood leukocyte, and the cancer cell
line is, for
example, selected from the group consisting of a promyelocytic leukemia HL-60,
an
HeLa cervical cancer cell, a chronic myelocytic leukemia cell line K-562,
lymphoblastoid leukemia MOLT-4, a Burkitt's lymphoma Raji, an SW480 colon
cancer
cell, an A549 lung cancer cell and a G361 melanoma cell.
FIG. 108 shows a northern blotting result that the PIG35 gene is
differentially
expressed in various normal tissues, and a bottom of FIG. 108 shows a northern
blotting
result obtained by hybridizing the same blot with J3 -actin probe. As shown in
Fig.
108, the dominant PIG35 mRNA transcript having a size of approximately 1.7 kb
was
overexpressed in the normal tissues such as the brain, the heart, the skeletal
muscle, the
liver, the small intestine, the placenta and the lungs.
FIG. 137 shows a northern blotting result that the PIG35 gene is
differentially
expressed in various cancer cell lines, and a bottom of FIG. 137 shows a
northern
blotting result obtained by hybridizing the same blot with J3 -actin probe. As
shown
in FIG. 137, the PIG35 gene was not expressed at all in the tissues such as
the
promyelocytic leukemia HL-60, the HeLa cervical cancer cell, the
lymphoblastoid
leukemia MOLT-4, the Burkitt's lymphoma Raji, the SW480 colon cancer cell, the
A549 lung cancer cell and the G361 melanoma cell, but rarely expressed in the
chronic
myelocytic leukemia cell line K-562. From such a result, it might be seen that
the
PIG35 gene of the present invention had the tumor suppresser function in the
normal
166
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
tissues such as the brain, the heart, the skeletal muscle, the liver, the
small intestine, the
placenta and the lungs. Also, it might be seen that the PIG35 gene of the
present
invention had the tumor suppresser function from the fact that its expression
was
suppressed even in the leukemia, the uterine cancer, the malignant lymphoma,
the colon
cancer, the lung cancer and the skin cancer to induce tumorigenesis.
FIG. 80 shows the northern blotting result that the PIG36 gene is
differentially
expressed in a normal liver tissue, a primary liver cancer tissue and a liver
cancer cell
line, and a bottom of FIG. 80 shows the northern blotting result obtained by
hybridizing
the same blot with J3 -actin probe. As shown in FIG. 80, it was revealed that
the
expression level of the PIG36 gene was highly detected all in the three
samples of the
normal liver tissue, but its expression was significantly lower in the three
samples of the
liver cancer tissue than the normal tissue, and not detected in the one sample
of the liver
cancer cell line.
The northern blotting was carried out on the normal human multiple tissue
(Clontech) and the human cancer cell line (Clontech). That is to say, the
northern
blotting was carried out by hybridizing blots, to which each of the total RNA
samples
extracted from the normal tissues and the cancer cell lines was transferred,
in the same
manner as described above, wherein the blots were commercially available from
the
company Clontech (U.S), and the normal tissue is, for example, selected from
the group
consisting of brain, heart, skeletal muscle, colon, thymus, spleen, kidney,
liver, small
intestine, placenta, lungs and peripheral blood leukocyte, and the cancer cell
line is, for
example, selected from the group consisting of a promyelocytic leukemia HL-60,
an
HeLa cervical cancer cell, a chronic myelocytic leukemia cell line K-562,
167
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
lymphoblastoid leukemia MOLT-4, a Burkitt's lymphoma Raji, an SW480 colon
cancer
cell, an A549 lung cancer cell and a G361 melanoma cell.
FIG. 109 shows a northern blotting result that the PIG36 gene is
differentially
expressed in various normal tissues, and a bottom of FIG. 109 shows a northern
blotting
result obtained by hybridizing the same blot with J3 -actin probe. As shown in
Fig.
109, the dominant PIG36 mRNA transcript having a size of approximately 1.0 kb
was
overexpressed in the normal liver tissue.
FIG. 138 shows a northern blotting result that the PIG36 gene is
differentially
expressed in various cancer cell lines, and a bottom of FIG. 138 shows a
northern
blotting result obtained by hybridizing the same blot with 13 -actin probe. As
shown
in FIG. 138, the PIG36 gene was not expressed at all or rarely expressed in
the tissues
such as the promyelocytic leukemia HL-60, the HeLa cervical cancer cell, the
chronic
myelocytic leukemia cell line K-562, the lymphoblastoid leukemia MOLT-4, the
Burkitt's lymphoma Raji, the SW480 colon cancer cell, the A549 lung cancer
cell and
the G361 melanoma cell. From such a result, it might be seen that the PIG36
gene of
the present invention had the tumor suppresser function in the normal tissues
such as the
liver, the heart, the muscle, the kidney and the placenta. Also, it might be
seen that the
PIG36 gene of the present invention had the tumor suppresser function from the
fact that
its expression was suppressed even in the leukemia, the uterine cancer, the
malignant
lymphoma, the colon cancer, the lung cancer and the skin cancer to induce
tumorigenesis.
FIG. 85 shows the northern blotting result that the PIG37 gene is
differentially
expressed in a normal liver tissue, a primary liver cancer tissue and a liver
cancer cell
168
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
line, and a bottom of FIG. 85 shows the northern blotting result obtained by
hybridizing
the same blot with 13 -actin probe. As shown in FIG. 85, it was revealed that
the
expression level of the PIG37 gene was highly detected all in the three
samples of the
normal liver tissue, but its expression was significantly lower in the three
samples of the
liver cancer tissue than the normal tissue, and not detected in the one sample
of the liver
cancer cell line.
The northern blotting was carried out on the normal human multiple tissue
(Clontech) and the human cancer cell line (Clontech). That is to say, the
northern
blotting was carried out by hybridizing blots, to which each of the total RNA
samples
extracted from the normal tissues and the cancer cell lines was transferred,
in the same
manner as described above, wherein the blots were commercially available from
the
company Clontech (U.S), and the normal tissue is, for example, selected from
the group
consisting of brain, heart, skeletal muscle, colon, thymus, spleen, kidney,
liver, small
intestine, placenta, lungs and peripheral blood leukocyte, and the cancer cell
line is, for
example, selected from the group consisting of a promyelocytic leukemia HL-60,
an
HeLa cervical cancer cell, a chronic myelocytic leukemia cell line K-562,
lymphoblastoid leukemia MOLT-4, a Burkitt's lymphoma Raji, an SW480 colon
cancer
cell, an A549 lung cancer cell and a G361 melanoma cell.
FIG. 114 shows a northern blotting result that the PIG37 gene is
differentially
expressed in various normal tissues, and a bottom of FIG. 114 shows a northern
blotting
result obtained by hybridizing the same blot with Ji -actin probe. As shown in
Fig.
114, the dominant PIG37 mRNA transcript having a size of approximately 7.0 kb
was
overexpressed in the normal tissues such as the brain, the heart, the skeletal
muscle, the
169
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
colon, the thymus, the spleen, the kidney, the liver, the small intestine, the
placenta and
the lung. PIG37 mRNA transcripts having a size of approximately 2.0 and 1.0 kb
were
overexpressed in the normal tissues at the same time.
FIG. 143 shows a northern blotting result that the PIG37 gene is
differentially
expressed in various cancer cell lines, and a bottom of FIG. 143 shows a
northern
blotting result obtained by hybridizing the same blot with J3 -actin probe. As
shown
in FIG. 143, the PIG37 gene was hardly expressed in the tissues such as the
promyelocytic leukemia HL-60, the chronic myelocytic leukemia cell line K-562,
the
lymphoblastoid leukemia MOLT-4, the SW480 colon cancer cell and the A549 lung
cancer cell, but rarely expressed in the HeLa cervical cancer cell, the
Burkitt's
lymphoma Raji and the G361 melanoma cell. From such a result, it might be seen
that
the PIG37 gene of the present invention had the tumor suppresser function in
the normal
tissues such as the brain, the heart, the skeletal muscle, the colon, the
thymus, the spleen,
the kidney, the liver, the small intestine, the placenta and the lungs. Also,
it might be
seen that the PIG37 gene of the present invention had the tumor suppresser
function
from the fact that its expression was suppressed even in the leukemia, the
uterine cancer,
the colon cancer, the lung cancer and the skin cancer to induce tumorigenesis.
3-4. GIG46, MIG20
In order to assess an expression level of the GIG and PIG genes, the northern
blottings were carried out, as follows.
20 ug of each of the total RNA samples obtained from the three normal
exocervical tissues, the three primary cervical cancer tissues and the two
cervical cancer
cell lines as described in Example 1 was denatured and electrophoresized in a
1 %
170
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
formaldehyde agarose gel, and then the resultant agarose gels were transferred
to nylon
membranes (Boehringer-Mannheim, Germany), respectively. The nylon membranes
were then hybridized at 42 C overnight with the 32P-labeled random prime
probes
using the full-length GIG46 and MIG20 cDNAs. The northern blotting procedure
was
repeated twice; one is that the blots were quantitified using the densitometer
and the
other is that the blots were hybridized with the J3 -actin probe to determine
the total
amount of mRNA.
FIG. 77 shows the northern blotting result that the GIG46 gene is
differentially
expressed in a normal exocervical tissue, a primary cervical cancer tissue and
a cervical
cancer cell line, and FIG. 77 is a northern blotting result showing expression
of j3 -actin.
In FIG. 77, Lanes 1 to 3 represent the normal exocervical tissue samples,
Lanes 4 to 6
represent the cervical cancer tissue samples, Lane 7 represents the sample of
the cervical
cancer cell line HeLa, and Lane 8 represents the sample of the cervical cancer
cell line
CUMC-6. As shown in FIG. 77, it was revealed that the expression level of the
GIG46
gene was highly detected all in the three samples of the normal exocervical
tissue, but
its expression level was significantly lower in the three samples of the
cervical cancer
tissue than the normal tissue, and not detected in the two samples of the
cervical cancer
cell lines.
FIG. 106 shows a northern blotting result that the GIG46 gene is
differentially
expressed in various normal tissues, and FIG. 106 shows a northern blotting
result
obtained by hybridizing the same blot with j3 -actin probe. As shown in FIG.
106, a
dominant GIG46 mRNA transcript having a size of approximately 1.5 kb was
overexpressed in the normal tissues such as the uterus, the brain, the heart,
the skeletal
171
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
muscle, the large intestine, the thymus, the spleen, the kidney, the liver,
the small
intestine, the placenta, the lungs and the peripheral blood leukocyte, and a
transcript
having a size of approximately 2.0 kb was also expressed in addition to the
1.5
kb-GIG46 mRNA transcript.
FIG. 135 shows a northern blotting result that the GIG46 gene is
differentially
expressed in various cancer cell lines, and a bottom of FIG. 135 shows a
northern
blotting result obtained by hybridizing the same blot with J3 -actin probe. As
shown
in FIG. 135, the GIG46 gene was rarely expressed in the tissues such as the
promyelocytic leukemia HL-60, the HeLa cervical cancer cell, the chronic
myelocytic
leukemia cell line K-562, the lymphoblastoid leukemia MOLT-4, the Burkitt's
lymphoma Raji, the SW480 colon cancer cell, the A549 lung cancer cell and the
G361
melanoma cell. However, the 2.0 kb-mRNA transcrip proven to be expressed in
the
normal tissues was not expressed in the cancer cell lines.
From such a result, it might be seen that the GIG46 gene of the present
invention
had the tumor suppresser function in the normal tissues such as the uterus,
the brain, the
heart, the skeletal muscle, the large intestine, the thymus, the spleen, the
kidney, the
liver, the small intestine, the placenta, the lungs and the peripheral blood
leukocyte.
FIG. 81 shows the northern blotting result that the MIG20 gene is
differentially
expressed in a normal exocervical tissue, a primary cervical cancer tissue and
a cervical
cancer cell line, and FIG. 81 is a northern blotting result showing expression
of J3 -actin.
In FIG. 81, Lanes 1 to 3 represent the normal exocervical tissue samples,
Lanes 4 to 6
represent the cervical cancer tissue samples, Lane 7 represents the sample of
the cervical
cancer cell line HeLa, and Lane 8 represents the sample of the cervical cancer
cell line
172
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
CUMC-6. As shown in FIG. 81, it was revealed that the expression level of the
MIG20 gene was highly detected all in the three samples of the normal
exocervical
tissue, but its expression level was significantly lower in the three samples
of the
cervical cancer tissue than the normal tissue, and not detected in the two
samples of the
cervical cancer cell lines.
FIG. 110 shows a northern blotting result that the MIG20 gene is
differentially
expressed in various normal tissues, and FIG. 110 shows a northern blotting
result
obtained by hybridizing the same blot with j3 -actin probe. As shown in FIG.
110, a
dominant MIG20 mRNA transcript having a size of approximately 4.4 kb was
overexpressed in the normal tissues such as the heart, the skeletal muscle and
the liver,
and transcripts having sizes of approximately 2.4 kb and 1.5 kb were also
expressed in
addition to the 4.4 kb-MIG20 mRNA transcript.
FIG. 139 shows a northern blotting result that the MIG20 gene is
differentially
expressed in various cancer cell lines, and a bottom of FIG. 139 shows a
northern
blotting result obtained by hybridizing the same blot with 13 -actin probe. As
shown
in FIG. 139, the MIG20 gene was not expressed in the tissues such as the
promyelocytic
leukemia HL-60, the HeLa cervical cancer cell, the chronic myelocytic leukemia
cell
line K-562, the lymphoblastoid leukemia MOLT-4, the Burkitt's lymphoma Raji,
the
SW480 colon cancer cell, the A549 lung cancer cell and the G361 melanoma cell.
From such a result, it might be seen that the MIG2 gene of the present
invention had the
tumor suppresser function in the normal tissues such as the cervix, the heart,
the skeletal
muscle and the liver.
3-5. MIG12
173
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
In order to assess an expression level of the MIG12 gene, the northern
blotting
was carried out, as follows.
20 /ug of each of the total RNA samples obtained from the three normal lung
tissues, the two primary lung cancer tissues, the two metastatic lung cancer
tissues and
the lung cancer cell lines (A549 and NCI-H358) as described in Example 1 was
denatured and electrophoresized in a 1% formaldehyde agarose gel, and then the
resultant agarose gel was transferred to a nylon membrane (Boehringer-
Mannheim,
Germany). The nylon membrane was then hybridized at 42 C overnight with the
32P-labeled random prime probe prepared from the full-length MIG12 cDNA using
the
Rediprime II random prime labelling system (Amersham, United Kingdom). The
northern blotting procedure was repeated twice; one is that the blots were
quantitified
using the densitometer and the other is that the blots were hybridized with
the 13 -actin
probe to determine the total amount of mRNA.
FIG. 84 shows the northern blotting result that the MIG12 gene is
differentially
expressed in a normal lung tissue, a primary lung cancer tissue, a metastatic
lung cancer
tissue and a lung cancer cell line, and a bottom of FIG. 84 shows the northern
blotting
result obtained by hybridizing the same blot with J3 -actin probe. As shown in
FIG.
84, it was revealed that the expression level of the MIG9 gene was highly
detected all in
the three samples of the normal lung tissue, but slightly detected in the two
samples of
the lung cancer tissue, the two samples of the metastatic lung cancer tissue
and the two
samples of the lung cancer cell line.
The northern blotting was carried out on the normal human multiple tissue
(Clontech) and the human cancer cell line (Clontech). That is to say, the
northern
174
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
blotting was carried out by hybridizing blots, to which each of the total RNA
samples
extracted from the normal tissues and the cancer cell lines was transferred,
in the same
manner as described above, wherein the blots were commercially available from
the
company Clontech (U.S), and the normal tissue is, for example, selected from
the group
consisting of brain, heart, skeletal muscle, colon, thymus, spleen, kidney,
liver, small
intestine, placenta, lungs and peripheral blood leukocyte, and the cancer cell
line is, for
example, selected from the group consisting of a promyelocytic leukemia HL-60,
an
HeLa cervical cancer cell, a chronic myelocytic leukemia cell line K-562,
lymphoblastoid leukemia MOLT-4, a Burkitt's lymphoma Raji, an SW480 colon
cancer
cell, an A549 lung cancer cell and a G361 melanoma cell.
FIG. 113 shows a northern blotting result that the MIG12 gene is
differentially
expressed in various normal tissues, and FIG. 113 shows a northern blotting
result
obtained by hybridizing the same blot with J3 -actin probe. As shown in FIG.
113, a
dominant MIG12 mRNA transcript having a size of approximately 0.5 kb was
overexpressed in the normal tissues such as the brain, the heart, the skeletal
muscle, the
colon, the thymus, the spleen, the kidney, the liver, the small intestine, the
placenta, the
lung and the peripheral blood leukocyte. Transcripts having sizes of
approximately 1.0
kb and 0.8 kb were also expressed in the normal tissues such as the heart, the
muscles,
the liver and the kidney in addition to the 0.5 kb-MIG 12 mRNA transcript.
FIG. 142 shows a northern blotting result that the MIG12 gene is
differentially
expressed in various cancer cell lines, and a bottom of FIG. 142 shows a
northern
blotting result obtained by hybridizing the same blot with j3 -actin probe. As
shown
in FIG. 142, the dominant 0.5 kb-MIG12 mRNA transcript expressed in the normal
175
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
tissues was rarely expressed in the tissues such as the promyelocytic leukemia
HL-60,
the HeLa cervical cancer cell, the chronic myelocytic leukemia cell line K-
562, the
lymphoblastoid leukemia MOLT-4, the Burkitt's lymphoma Raji, the SW480 colon
cancer cell, the A549 lung cancer cell and the G361 melanoma cell. From such a
result,
it might be seen that the MIG12 gene of the present invention had the tumor
suppresser
function in the normal tissues such as the brain, the heart, the skeletal
muscle, the colon,
the thymus, the spleen, the kidney, the liver, the small intestine, the
placenta, the lung
and the peripheral blood leukocyte.
Example 4: Construction and Transfection of Expression Vector
4-1. GIG8 GIG10 GIG13, GIG30 GIG32, GIG33, GIG34, GIG35, GIG38,
GIG39, GIG43, PIG49, PIG51, GIG44, GIG31
An expression vector containing each coding region of GIG and PIG genes was
constructed, as follows. Firstly, the full-length cDNA clones prepared in
Example 2
were inserted into a eukaryotic expression vector pcDNA3.1 (Invitrogen, U.S.)
to obtain
expression vectors pcDNA3.1/GIG8; pcDNA3.1/GIG10; pcDNA3.1/GIG13;
pcDNA3.1/GIG30; pcDNA3.1/GIG32; pcDNA3.1/GIG33; pcDNA3.1/GIG34;
pcDNA3.1/GIG35; pcDNA3. 1 /GIG3 8; pcDNA3.1/GIG39; pcDNA3.1/GIG43;
pcDNA3.1/PIG49; pcDNA3.1/PIG51; pcDNA3.1/GIG44 and pcDNA3.1/GIG31,
respectively. Each of the expression vectors was transfected into an MCF-7
breast
cancer cell line using lipofectamine (Gibco BRL), and then incubated in a DMEM
medium containing 0.6 mg/0 of G418 (Gibco) to select transfected cells. At
this time,
the MCF-7 cell transfected with the expression vector pcDNA3.1 devoid of the
GIG
176
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
cDNA was used as the control group.
4-2. GIG 15
An expression vector containing a coding region of the GIG15 gene was
constructed, as follows. Firstly, the full-length GIG15 cDNA clones prepared
in
Example 2 was inserted into a eukaryotic expression vector pcDNA3.1
(Invitrogen,
U.S.) to obtain an expression vector pcDNA3.1/GIG15. The expression vector was
transfected into a K562 leukemia cell line using lipofectamine (Gibco BRL),
and then
incubated in a DMEM medium containing 0.6 mg/mt of G418 (Gibco) to select
transfected cells. At this time, the K562 cell transfected with the expression
vector
pcDNA3.1 devoid of the GIG cDNA was used as the control group.
4-3. GIG16, GIG24 GIG26 GIG29 GIG40 GIG42 PIG33 PIG35 PIG36
PIG37
An expression vector containing each coding region of GIG and PIG genes was
constructed, as follows. Firstly, the full-length cDNA clones GIG16, GIG24,
GIG26,
GIG29, GIG40, GIG42, PIG33, PIG35, PIG36 and PIG37 prepared in Example 2 were
inserted into a eukaryotic expression vector pcDNA3.1 (Invitrogen, U.S.) to
obtain
expression vectors pcDNA3.1/GIG16; pcDNA3.1/GIG24; pcDNA3.1/GIG26;
pcDNA3.1/GIG29; pcDNA3.1/GIG40; pcDNA3.1/GIG42; pcDNA3.1/PIG33;
pcDNA3.1/PIG35; pcDNA3.1/PIG36; and pcDNA3.1/PIG37, respectively. Each of
the expression vectors was transfected into an HepG2 liver cancer cell line
using
lipofectamine (Gibco BRL), and then incubated in a DMEM medium containing 0.6
mg/ini of G418 (Gibco) to select transfected cells. At this time, the HepG2
cell
transfected with the expression vector pcDNA3.1 devoid of the GIG cDNA was
used as
177
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
the control group.
4-4. GIG46, MIG20
An expression vector containing each coding region of the GIG and MIG genes
was constructed, as follows. Firstly, the full-length GIG46 cDNA clones
prepared in
Example 2 was inserted into a eukaryotic expression vector pcDNA3.1
(Invitrogen,
U.S.) to obtain expression vectors pcDNA3.1/GIG46; and pcDNA3.1/MIG20,
respectively. Each of the expression vectors was transfected into an HeLa
cervical
cancer cell line(ATCC CCL-2) using lipofectamine (Gibco BRL), and then
incubated in
a DMEM medium containing 0.6 mg/mt of G418 (Gibco) to select transfected
cells.
At this time, the HeLa cell transfected with the expression vector pcDNA3.1
devoid of
the GIG46 or MIG20 cDNA was used as the control group.
4-5. MIG12
An expression vector containing a coding region of the MIG12 gene was
constructed, as follows. Firstly, the full-length MIG12 cDNA clones prepared
in
Example 2 was inserted into a eukaryotic expression vector pcDNA3.1
(Invitrogen,
U.S.) to obtain an expression vector pcDNA3.1/MIG12. The expression vector was
transfected into an A549 lung cancer cell line using lipofectamine (Gibco
BRL), and
then incubated in a DMEM medium containing 0.6 mgW of G418 (Gibco) to select
transfected cells. At this time, the A549 cell transfected with the expression
vector
pcDNA3.1 devoid of the MIG12 cDNA was used as the control group.
Example 5: Growth Curve of Breast Cancer Cell Transfected with GIG Gene
5-1. GIG8, GIG10 GIG13, GIG30, GIG32, GIG33, GIG34, GIG35, GIG38,
178
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
GIG39, GIG43, PIG49, PIG5 1, GIG44, GIG31
In order to examine effects of the GIG and PIG genes on growth of the breast
cancer cell, the wild-type MCF-7 cell; the MCF-7 breast cancer cells
transfected
respectively with the vectors pcDNA3.1/GIG8; pcDNA3.1/GIG10; pcDNA3.1/GIG13;
pcDNA3.1/GIG30; pcDNA3.1/GIG32; pcDNA3.1/GIG33; pcDNA3.1/GIG34;
pcDNA3.1/GIG35; pcDNA3.1/GIG38; pcDNA3.1/GIG39; pcDNA3.1/GIG43;
pcDNA3.1 /PIG49; pcDNA3.1 /PIG51; pcDNA3.1 /GIG44 and pcDNA3.1 /GIG31
prepared in Example 4; and the MCF-7 cell transfected only with the vector
pcDNA3.1
were incubated to a cell density of 1 x 105 cells/0 in a DMEM medium for 9
days,
respectively. The cells in the culture solutions were isolated from the flask
they attach
to by treatment with trypsin (Sigma), respectively, and then the survived
cells were
counted on days 1, 3, 5, 7 and 9 according to a trypan blue dye exclusion
(Freshney, I.R.,
Culture of Animal Cells, 2nd Ed. A.R. Liss, New York (1987)), respectively.
FIG. 146 is a diagram showing growth curves of the wild-type MCF-7 cell; the
MCF-7 breast cancer cell transfected with the vector pcDNA3.1/GIG8 prepared in
Example 4; and the MCF-7 cell transfected only with the expression vector
pcDNA3.1.
As shown in FIG. 146, it was revealed that the MCF-7 breast cancer cell
transfected
with the vector pcDNA3.1/GIG8 exhibited a higher mortality than those of the
MCF-7
cell transfected with the expression vector pcDNA3.1 and the wild-type MCF-7
cell.
After 9 days of incubation, only approximately 30 % of the MCF-7 breast cancer
cell
transfected with the vector pcDNA3.1/GIG8 was survived when compared to the
wild-type MCF-7 cell. From such a result, it might be seen that the GIG8 gene
suppressed the growth of the breast cancer cell.
179
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
FIG. 147 is a diagram showing growth curves of the wild-type MCF-7 cell; the
MCF-7 breast cancer cell transfected with the vector pcDNA3.1 /GIG 10 prepared
in
Example 4; and the MCF-7 cell transfected only with the expression vector
pcDNA3.1.
As shown in FIG. 147, it was revealed that the MCF-7 breast cancer cell
transfected
with the vector pcDNA3. 1 /GIG 10 exhibited a higher mortality than those of
the MCF-7
cell transfected with the expression vector pcDNA3.1 and the wild-type MCF-7
cell.
After 9 days of incubation, only approximately 40 % of the MCF-7 breast cancer
cell
transfected with the vector pcDNA3.1 /GIG 10 was survived when compared to the
wild-type MCF-7 cell. From such a result, it might be seen that the GIG10 gene
suppressed the growth of the breast cancer cell.
FIG. 148 is a diagram showing growth curves of the wild-type MCF-7 cell; the
MCF-7 breast cancer cell transfected with the vector pcDNA3.1/GIG13 prepared
in
Example 4; and the MCF-7 cell transfected only with the expression vector
pcDNA3.1.
As shown in FIG. 148, it was revealed that the MCF-7 breast cancer cell
transfected
with the vector pcDNA3.1/GIG13 exhibited a higher mortality than those of the
MCF-7
cell transfected with the expression vector pcDNA3.1 and the wild-type MCF-7
cell.
After 9 days of incubation, only approximately 30 % of the MCF-7 breast cancer
cell
transfected with the vector peDNA3.1/GIG13 was survived when compared to the
wild-type MCF-7 cell. From such a result, it might be seen that the GIG13 gene
suppressed the growth of the breast cancer cell.
FIG. 154 is a diagram showing growth curves of the wild-type MCF-7 cell; the
MCF-7 breast cancer cell transfected with the vector pcDNA3.1/GIG30 prepared
in
Example 4; and the MCF-7 cell transfected only with the expression vector
pcDNA3.1.
180
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
As shown in FIG. 154, it was revealed that the MCF-7 breast cancer cell
transfected
with the vector pcDNA3. 1 /GIG3 0 exhibited a higher mortality than those of
the MCF-7
cell transfected with the expression vector pcDNA3.1 and the wild-type MCF-7
cell.
After 9 days of incubation, only approximately 30 % of the MCF-7 breast cancer
cell
transfected with the vector pcDNA3.1/GIG30 was survived when compared to the
wild-type MCF-7 cell. From such a result, it might be seen that the GIG30 gene
suppressed the growth of the breast cancer cell.
FIG. 155 is a diagram showing growth curves of the wild-type MCF-7 cell; the
MCF-7 breast cancer cell transfected with the vector pcDNA3.1/GIG32 prepared
in
Example 4; and the MCF-7 cell transfected only with the expression vector
pcDNA3.1.
As shown in FIG. 155, it was revealed that the MCF-7 breast cancer cell
transfected
with the vector pcDNA3.1/GIG32 exhibited a higher mortality than those of the
MCF-7
cell transfected with the expression vector pcDNA3.1 and the wild-type MCF-7
cell.
After 9 days of incubation, only approximately 50 % of the MCF-7 breast cancer
cell
transfected with the vector pcDNA3.1/GIG32 was survived when compared to the
wild-type MCF-7 cell. From such a result, it might be seen that the GIG32 gene
suppressed the growth of the breast cancer cell.
FIG. 156 is a diagram showing growth curves of the wild-type MCF-7 cell; the
MCF-7 breast cancer cell transfected with the vector pcDNA3.1/GIG33 prepared
in
Example 4; and the MCF-7 cell transfected only with the expression vector
pcDNA3.1.
As shown in FIG. 156, it was revealed that the MCF-7 breast cancer cell
transfected
with the vector pcDNA3.1/GIG33 exhibited a higher mortality than those of the
MCF-7
cell transfected with the expression vector pcDNA3.1 and the wild-type MCF-7
cell.
181
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
After 9 days of incubation, only approximately 70 % of the MCF-7 breast cancer
cell
transfected with the vector pcDNA3.1/GIG33 was survived when compared to the
wild-type MCF-7 cell. From such a result, it might be seen that the GIG33 gene
suppressed the growth of the breast cancer cell.
FIG. 157 is a diagram showing growth curves of the wild-type MCF-7 cell; the
MCF-7 breast cancer cell transfected with the vector pcDNA3.1/GIG34 prepared
in
Example 4; and the MCF-7 cell transfected only with the expression vector
pcDNA3.1.
As shown in FIG. 157, it was revealed that the MCF-7 breast cancer cell
transfected
with the vector pcDNA3.1/GIG34 exhibited a higher mortality than those of the
MCF-7
cell transfected with the expression vector pcDNA3.1 and the wild-type MCF-7
cell.
After 9 days of incubation, only approximately 80 % of the MCF-7 breast cancer
cell
transfected with the vector pcDNA3.1/GIG34 was survived when compared to the
wild-type MCF-7 cell. From such a result, it might be seen that the GIG34 gene
suppressed the growth of the breast cancer cell.
FIG. 158 is a diagram showing growth curves of the wild-type MCF-7 cell; the
MCF-7 breast cancer cell transfected with the vector pcDNA3.1/GIG35 prepared
in
Example 4; and the MCF-7 cell transfected only with the expression vector
pcDNA3.1.
As shown in FIG. 158, it was revealed that the MCF-7 breast cancer cell
transfected
with the vector pcDNA3.1/GIG35 exhibited a higher mortality than those of the
MCF-7
cell transfected with the expression vector pcDNA3.1 and the wild-type MCF-7
cell.
After 9 days of incubation, only approximately 70 % of the MCF-7 breast cancer
cell
transfected with the vector pcDNA3.1/GIG35 was survived when compared to the
wild-type MCF-7 cell. From such a result, it might be seen that the GIG35 gene
182
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
suppressed the growth of the breast cancer cell.
FIG. 159 is a diagram showing growth curves of the wild-type MCF-7 cell; the
MCF-7 breast cancer cell transfected with the vector pcDNA3.1/GIG38 prepared
in
Example 4; and the MCF-7 cell transfected only with the expression vector
pcDNA3.1.
As shown in FIG. 159, it was revealed that the MCF-7 breast cancer cell
transfected
with the vector pcDNA3.1/GIG38 exhibited a higher mortality than those of the
MCF-7
cell transfected with the expression vector pcDNA3.1 and the wild-type MCF-7
cell.
After 9 days of incubation, only approximately 60 % of the MCF-7 breast cancer
cell
transfected with the vector pcDNA3.1/GIG38 was survived when compared to the
wild-type MCF-7 cell. From such a result, it might be seen that the GIG38 gene
suppressed the growth of the breast cancer cell.
FIG. 160 is a diagram showing growth curves of the wild-type MCF-7 cell; the
MCF-7 breast cancer cell transfected with the vector pcDNA3.1/GIG39 prepared
in
Example 4; and the MCF-7 cell transfected only with the expression vector
pcDNA3.1.
As shown in FIG. 160, it was revealed that the MCF-7 breast cancer cell
transfected
with the vector pcDNA3.l/GIG39 exhibited a higher mortality than those of the
MCF-7
cell transfected with the expression vector pcDNA3.1 and the wild-type MCF-7
cell.
After 9 days of incubation, only approximately 40 % of the MCF-7 breast cancer
cell
transfected with the vector pcDNA3.1/GIG39 was survived when compared to the
wild-type MCF-7 cell. From such a result, it might be seen that the GIG39 gene
suppressed the growth of the breast cancer cell.
FIG. 163 is a diagram showing growth curves of the wild-type MCF-7 cell; the
MCF-7 breast cancer cell transfected with the vector pcDNA3.1/GIG43 prepared
in
183
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
Example 4; and the MCF-7 cell transfected only with the expression vector
pcDNA3.1.
As shown in FIG. 163, it was revealed that the MCF-7 breast cancer cell
transfected
with the vector pcDNA3. 1 /GIG43 exhibited a higher mortality than those of
the MCF-7
cell transfected with the expression vector pcDNA3.1 and the wild-type MCF-7
cell.
After 9 days of incubation, only approximately 60 % of the MCF-7 breast cancer
cell
transfected with the vector pcDNA3.1 /GIG43 was survived when compared to the
wild-type MCF-7 cell. From such a result, it might be seen that the GIG43 gene
suppressed the growth of the breast cancer cell.
FIG. 169 is a diagram showing growth curves of the wild-type MCF-7 cell; the
MCF-7 breast cancer cell transfected with the vector pcDNA3.1/PIG49 prepared
in
Example 4; and the MCF-7 cell transfected only with the expression vector
pcDNA3.1.
As shown in FIG. 169, it was revealed that the MCF-7 breast cancer cell
transfected
with the vector pcDNA3.1/PIG49 exhibited a higher mortality than those of the
MCF-7
cell transfected with the expression vector pcDNA3.1 and the wild-type MCF-7
cell.
After 9 days of incubation, only approximately 60 % of the MCF-7 breast cancer
cell
transfected with the vector pcDNA3.1 /PIG49 was survived when compared to the
wild-type MCF-7 cell. From such a result, it might be seen that the PIG49 gene
suppressed the growth of the breast cancer cell.
FIG. 170 is a diagram showing growth curves of the wild-type MCF-7 cell; the
MCF-7 breast cancer cell transfected with the vector pcDNA3.1 /PIG51 prepared
in
Example 4; and the MCF-7 cell transfected only with the expression vector
pcDNA3.1.
As shown in FIG. 170, it was revealed that the MCF-7 breast cancer cell
transfected
with the vector pcDNA3.1 /PIG51 exhibited a higher mortality than those of the
MCF-7
184
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
cell transfected with the expression vector pcDNA3.1 and the wild-type MCF-7
cell.
After 9 days of incubation, only approximately 40 % of the MCF-7 breast cancer
cell
transfected with the vector pcDNA3.1 /PIG51 was survived when compared to the
wild-type MCF-7 cell. From such a result, it might be seen that the PIG51 gene
suppressed the growth of the breast cancer cell.
FIG. 173 is a diagram showing growth curves of the wild-type MCF-7 cell; the
MCF-7 breast cancer cell transfected with the vector pcDNA3.1/GIG44 prepared
in
Example 4; and the MCF-7 cell transfected only with the expression vector
pcDNA3.1.
As shown in FIG. 173, it was revealed that the MCF-7 breast cancer cell
transfected
with the vector pcDNA3.1/GIG44 exhibited a higher mortality than those of the
MCF-7
cell transfected with the expression vector pcDNA3.1 and the wild-type MCF-7
cell.
After 9 days of incubation, only approximately 60 % of the MCF-7 breast cancer
cell
transfected with the vector pcDNA3.1/GIG44 was survived when compared to the
wild-type MCF-7 cell. From such a result, it might be seen that the GIG44 gene
suppressed the growth of the breast cancer cell.
FIG. 174 is a diagram showing growth curves of the wild-type MCF-7 cell; the
MCF-7 breast cancer cell transfected with the vector pcDNA3.1/GIG31 prepared
in
Example 4; and the MCF-7 cell transfected only with the expression vector
pcDNA3.1.
As shown in FIG. 174, it was revealed that the MCF-7 breast cancer cell
transfected
with the vector pcDNA3.1/GIG31 exhibited a higher mortality than those of the
MCF-7
cell transfected with the expression vector pcDNA3.1 and the wild-type MCF-7
cell.
After 9 days of incubation, only approximately 70 % of the MCF-7 breast cancer
cell
transfected with the vector pcDNA3.1 /GIG31 was survived when compared to the
185
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
wild-type MCF-7 cell. From such a result, it might be seen that the GIG31 gene
suppressed the growth of the breast cancer cell.
5-2. GIG15
In order to examine effects of the GIG gene on growth of the leukemia cell,
the
wild-type K562ce11; the K562 leukemia cell transfected respectively by the
vector
pcDNA3.1/GIG15 prepared in Example 4; and the K562 cell transfected only with
the
vector pcDNA3.1 were incubated to a cell density of 1 x 105 cells/mi in a DMEM
medium for 9 days, respectively. The cells in the culture solutions were
isolated from
the flask they attach to by treatment with trypsin (Sigma), respectively, and
then the
survived cells were counted on days 1, 3, 5, 7 and 9 according to a trypan
blue dye
exclusion (Freshney, I.R., Culture of Animal Cells, 2nd Ed. A.R. Liss, New
York
(1987)), respectively.
FIG. 149 is a diagram showing growth curves of the wild-type K562 cell; the
K562 leukemia cell transfected with the vector pcDNA3. 1 /GIG 15 prepared in
Example
4; and the K562 cell transfected only with the expression vector pcDNA3.1. As
shown
in FIG. 149, it was revealed that the K562 cell transfected with the vector
pcDNA3.1/GIG15 exhibited a higher mortality than those of the K562 cell
transfected
with the expression vector pcDNA3.1 and the wild-type K562 cell. After 9 days
of
incubation, only approximately 80 % of the K562 cell transfected with the
vector
pcDNA3.1/GIG15 was survived when compared to the wild-type K562 cell. From
such a result, it might be seen that the GIG15 gene suppressed the growth of
the breast
cancer cell.
5-3. GIG16, GIG24 GIG26 GIG29 GIG40 GIG42 PIG33, PIG35, PIG36
186
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
PIG37
In order to examine effects of the GIG and PIG genes on growth of the liver
cancer cell, the wild-type HepG2 cell; the HepG2 liver cancer cells
transfected
respectively by the vectors pcDNA3.1/GIG16; pcDNA3.1/GIG24; pcDNA3.1/GIG26;
pcDNA3.1/GIG29; pcDNA3.1/GIG40; pcDNA3.1/GIG42; pcDNA3.1/PIG33;
pcDNA3.1/PIG35; pcDNA3.1/PIG36; and pcDNA3.1/PIG37 prepared in Example 4;
and the HepG2 cell transfected only with the vector pcDNA3.1 were incubated to
a cell
density of 1 x 105 cells/O in a DMEM medium for 9 days, respectively. The
cells in
the culture solutions were isolated from the flask they attach to by treatment
with trypsin
(Sigma), respectively, and then the survived cells were counted on days 1, 3,
5, 7 and 9
according to a trypan blue dye exclusion (Freshney, I.R., Culture of Animal
Cells, 2nd
Ed. A.R. Liss, New York (1987)), respectively.
FIG. 150 is a diagram showing growth curves of the wild-type HepG2 cell; the
HepG2 liver cancer cell transfected with the vector pcDNA3.1/GIG16 prepared in
Example 4; and the HepG2 cell transfected only with the expression vector
pcDNA3.1.
As shown in FIG. 150, it was revealed that the HepG2 liver cancer cell
transfected with
the vector pcDNA3.1/GIG16 exhibited a higher mortality than those of the HepG2
cell
transfected with the expression vector pcDNA3.1 and the wild-type HepG2 cell.
After
9 days of incubation, only approximately 70 % of the HepG2 liver cancer cell
transfected with the vector pcDNA3.1/GIG16 was survived when compared to the
wild-type HepG2 cell. From such a result, it might be seen that the GIG16 gene
suppressed the growth of the liver cancer cell.
FIG. 151 is a diagram showing growth curves of the wild-type HepG2 cell; the
187
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
HepG2 liver cancer cell transfected with the vector pcDNA3.1 /GIG24 prepared
in
Example 4; and the HepG2 cell transfected only with the expression vector
pcDNA3.1.
As shown in FIG. 151, it was revealed that the HepG2 liver cancer cell
transfected with
the vector pcDNA3.1/GIG24 exhibited a higher mortality than those of the HepG2
cell
transfected with the expression vector pcDNA3.1 and the wild-type HepG2 cell.
After
9 days of incubation, only approximately 60 % of the HepG2 liver cancer cell
transfected with the vector pcDNA3.1 /GIG24 was survived when compared to the
wild-type HepG2 cell. From such a result, it might be seen that the GIG24 gene
suppressed the growth of the liver cancer cell.
FIG. 152 is a diagram showing growth curves of the wild-type HepG2 cell; the
HepG2 liver cancer cell transfected with the vector pcDNA3.1 /GIG26 prepared
in
Example 4; and the HepG2 cell transfected only with the expression vector
pcDNA3.1.
As shown in FIG. 152, it was revealed that the HepG2 liver cancer cell
transfected with
the vector pcDNA3.l/GIG26 exhibited a higher mortality than those of the HepG2
cell
transfected with the expression vector pcDNA3.1 and the wild-type HepG2 cell.
After
9 days of incubation, only approximately 50 % of the HepG2 liver cancer cell
transfected with the vector pcDNA3.1/GIG26 was survived when compared to the
wild-type HepG2 cell. From such a result, it might be seen that the GIG26 gene
suppressed the growth of the liver cancer cell.
FIG. 153 is a diagram showing growth curves of the wild-type HepG2 cell; the
HepG2 liver cancer cell transfected with the vector pcDNA3.1 /GIG29 prepared
in
Example 4; and the HepG2 cell transfected only with the expression vector
pcDNA3.1.
As shown in FIG. 153, it was revealed that the HepG2 liver cancer cell
transfected with
188
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
the vector pcDNA3.1/GIG29 exhibited a higher mortality than those of the HepG2
cell
transfected with the expression vector pcDNA3.1 and the wild-type HepG2 cell.
After
9 days of incubation, only approximately 70 % of the HepG2 liver cancer cell
transfected with the vector pcDNA3.1/GIG29 was survived when compared to the
wild-type HepG2 cell. From such a result, it might be seen that the GIG29 gene
suppressed the growth of the liver cancer cell.
FIG. 161 is a diagram showing growth curves of the wild-type HepG2 cell; the
HepG2 liver cancer cell transfected with the vector pcDNA3.1/GIG40 prepared in
Example 4; and the HepG2 cell transfected only with the expression vector
pcDNA3.1.
As shown in FIG. 161, it was revealed that the HepG2 liver cancer cell
transfected with
the vector pcDNA3.1 /GIG40 exhibited a higher mortality than those of the
HepG2 cell
transfected with the expression vector pcDNA3.1 and the wild-type HepG2 cell.
After
9 days of incubation, only approximately 80 % of the HepG2 liver cancer cell
transfected with the vector pcDNA3.1 /GIG40 was survived when compared to the
wild-type HepG2 cell. From such a result, it might be seen that the GIG40 gene
suppressed the growth of the liver cancer cell.
FIG. 162 is a diagram showing growth curves of the wild-type HepG2 cell; the
HepG2 liver cancer cell transfected with the vector pcDNA3.1/G1G42 prepared in
Example 4; and the HepG2 cell transfected only with the expression vector
pcDNA3.1.
As shown in FIG. 162, it was revealed that the HepG2 liver cancer cell
transfected with
the vector pcDNA3.1/GIG42 exhibited a higher mortality than those of the HepG2
cell
transfected with the expression vector pcDNA3.1 and the wild-type HepG2 cell.
After
9 days of incubation, only approximately 60 % of the HepG2 liver cancer cell
189
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
transfected with the vector pcDNA3.1/GIG42 was survived when compared to the
wild-type HepG2 cell. From such a result, it might be seen that the GIG42 gene
suppressed the growth of the liver cancer cell.
FIG. 165 is a diagram showing growth curves of the wild-type HepG2 cell; the
HepG2 liver cancer cell transfected with the vector pcDNA3.1/PIG33 prepared in
Example 4; and the HepG2 cell transfected only with the expression vector
pcDNA3.1.
As shown in FIG. 165, it was revealed that the HepG2 liver cancer cell
transfected with
the vector pcDNA3.1/PIG33 exhibited a higher mortality than those of the HepG2
cell
transfected with the expression vector pcDNA3.1 and the wild-type HepG2 cell.
After
9 days of incubation, only approximately 60 % of the HepG2 liver cancer cell
transfected with the vector pcDNA3.1/PIG33 was survived when compared to the
wild-type HepG2 cell. From such a result, it might be seen that the PIG33 gene
suppressed the growth of the liver cancer cell.
FIG. 166 is a diagram showing growth curves of the wild-type HepG2 cell; the
HepG2 liver cancer cell transfected with the vector pcDNA3.1/PIG35 prepared in
Example 4; and the HepG2 cell transfected only with the expression vector
pcDNA3.1.
As shown in FIG. 166, it was revealed that the HepG2 liver cancer cell
transfected with
the vector pcDNA3.1/PIG35 exhibited a higher mortality than those of the HepG2
cell
transfected with the expression vector pcDNA3.1 and the wild-type HepG2 cell.
After
9 days of incubation, only approximately 70 % of the HepG2 liver cancer cell
transfected with the vector pcDNA3.1/PIG35 was survived when compared to the
wild-type HepG2 cell. From such a result, it might be seen that the PIG35 gene
suppressed the growth of the liver cancer cell.
190
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
FIG. 167 is a diagram showing growth curves of the wild-type HepG2 cell; the
HepG2 liver cancer cell transfected with the vector pcDNA3.1/PIG36 prepared in
Example 4; and the HepG2 cell transfected only with the expression vector
pcDNA3.1.
As shown in FIG. 167, it was revealed that the HepG2 liver cancer cell
transfected with
the vector pcDNA3.1/PIG36 exhibited a higher mortality than those of the HepG2
cell
transfected with the expression vector pcDNA3.1 and the wild-type HepG2 cell.
After
9 days of incubation, only approximately 60 % of the HepG2 liver cancer cell
transfected with the vector pcDNA3.1/PIG36 was survived when compared to the
wild-type HepG2 cell. From such a result, it might be seen that the PIG36 gene
suppressed the growth of the liver cancer cell.
FIG. 172 is a diagram showing growth curves of the wild-type HepG2 cell; the
HepG2 liver cancer cell transfected with the vector pcDNA3.1 /PIG37 prepared
in
Example 4; and the HepG2 cell transfected only with the expression vector
pcDNA3.1.
As shown in FIG. 172, it was revealed that the HepG2 liver cancer cell
transfected with
the vector pcDNA3.1/PIG37 exhibited a higher mortality than those of the HepG2
cell
transfected with the expression vector pcDNA3.1 and the wild-type HepG2 cell.
After
9 days of incubation, only approximately 70 % of the HepG2 liver cancer cell
transfected with the vector pcDNA3.1/PIG37 was survived when compared to the
wild-type HepG2 cell. From such a result, it might be seen that the PIG37 gene
suppressed the growth of the liver cancer cell.
5-4. GIG46, MIG20
In order to determine effects of the GIG and MIG genes on growth of the
cervical cancer cell, the normal HeLa cell, the HeLa cervical cancer cell
transfected with
191
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
the GIG46 gene prepared in Example 4, and the HeLa cell transfected only with
the
vector pcDNA3.1 (Invitrogen) were incubated to a cell density of 1 x 105
cells/m~ in a
DMEM medium for 9 days, respectively. The cells in the culture solutions were
isolated from the flask they attach to by treatment with trypsin (Sigma), and
then the
survived cells were counted on days 1, 3, 5, 7 and 9 according to a trypan
blue dye
exclusion (Freshney, I.R., Culture of Animal Cells, 2nd Ed. A.R. Liss, New
York
(1987)).
FIG. 164 is a diagram showing growth curves of the normal HeLa cell; the HeLa
cervical cancer cell transfected with the GIG46 gene prepared in Example 4;
and the
HeLa cell transfected only with the expression vector pcDNA3.1. As shown in
FIG.
164, it was revealed that the HeLa cervical cancer cell transfected with the
GIG46 gene
exhibited a higher mortality when compared to those of the HeLa cell
transfected with
the expression vector pcDNA3.1 and the normal HeLa cell. After 9 days of
incubation,
only 80 % of the HeLa cervical cancer cell transfected with the GIG46 gene was
survived when compared to the normal HeLa cell. From such a result, it might
be seen
that the GIG46 gene suppressed growth of the cervical cancer cell.
FIG. 168 is a diagram showing growth curves of the normal HeLa cell; the HeLa
cervical cancer cell transfected with the MIG20 gene prepared in Example 4;
and the
HeLa cell transfected only with the expression vector pcDNA3.1. As shown in
FIG.
168, it was revealed that the HeLa cervical cancer cell transfected with the
MIG20 gene
exhibited a higher mortality when compared to those of the HeLa cell
transfected with
the expression vector pcDNA3.1 and the normal HeLa cell. After 9 days of
incubation,
only approximately 60 % of the HeLa cervical cancer cell transfected with the
MIG20
192
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
gene was survived when compared to the normal HeLa cell. From such a result,
it
might be seen that the MIG20 gene suppressed growth of the cervical cancer
cell.
5-5. MIG12
In order to determine an effect of the MIG12 gene on growth of the lung cancer
cell, the wild-type A549 cell; the A549 lung cancer cell transfected with the
vector
pcDNA3.1/MIG12 prepared in Example 4; and the A549 cell transfected only with
the
vector pcDNA3.1 were incubated at a cell density of 1 x 105 ce11sW in a DMEM
medium for 9 days, respectively. The cells in the culture solutions were
isolated from
the flask they attach to by treatment with trypsin (Sigma), and then the
survived cells
were counted on days 1, 3, 5, 7 and 9 according to a trypan blue dye exclusion
(Freshney,
I.R., Culture of Animal Cells, 2nd Ed. A.R. Liss, New York (1987)).
FIG. 171 is a diagram showing growth curves of the wild-type A549 cell; the
A5491ung cancer cell transfected by the vector pcDNA3.1/MIG12 prepared in
Example
4; and the A549 cell transfected only by the expression vector pcDNA3.1. As
shown
in FIG. 171, it was revealed that the A549 lung cancer cell transfected by the
vector
pcDNA3.1/MIG12 exhibited a higher mortality when compared to those of the A549
cell transfected by the expression vector pcDNA3.1 and the wild-type A549
cell. After
9 days of incubation, only approximately 70 % of the A549 lung cancer cell
transfected
by the vector pcDNA3.1/MIG12 was survived when compared to the wild-type A549
cell. From such a result, it might be seen that the MIG12 gene suppressed
growth of
the lung cancer cell.
INDUSTRIAL APPLICABILITY
193
CA 02602976 2007-09-26
WO 2006/109941 PCT/KR2006/001174
The GIG, PIG or MIG gene of the present invention may be effectively used for
diagnosing, preventing and treating human cancers.
194