Note: Descriptions are shown in the official language in which they were submitted.
CA 02611540 2016-01-12
28931-45
SUGAR CHAIN-CONTAINING ALBUMIN AS A DRUG CARRIER TO THE LIVER
THECHNICAL FIELD OF THE INVENTION
The present invention relates to a novel glycosylated
albumin protein, wherein a sugar chain is selectively added to
a particular amino acid residue, a production method thereof
and use thereof. More particularly, the present invention
relates to a glycosylated albumin protein wherein a sugar
..zo chain is selectively added to a partial amino acid sequence
contained in a mutant albumin, which partial amino acid
sequence is possibly subject to a glycosylation modification
by a host cell, a DNA encoding the mutant albumin, a
production method of the glycosylated albumin protein,
comprising cultivating eukaryotic cell containing the DNA, and
use of the protein as a drug carrier.
BACKGROUND OF THE INVENTION
Human serum albumin (hereinafter sometimes to be referred
to as "HSA") is widely distributed in the body, including
blood and intercellular fluids. Its primary structure consists
of 585 amino acids, and it is a simple protein having a
molecular weight of about 66.5 kDa, which is free of a sugar
structure. This protein is produced in the liver, mainly
maintains normal osmotic pressure in the bloodstream, and is
responsible for maintaining the liquid content of the blood.
Therefore, HSA is used in various clinical situations for the
treatment of a condition associated with loss of liquid from
the blood vessel, such as surgery, shock, burn,
hypoproteinemia causing edema and the like.
In addition, HSA functions as a carrier of various serum
molecules, and is rich in safety, biocompatibility,
biodegradation property, persistence in blood and the like.
Therefore, it is considered a preferable carrier for a drug
delivery system (DDS) of a drUg having a problem in the
kinetic property.
1
CA 02611540 2007-11-09
The DDS based on an irreversible bond between HSA and a
drug includes a method improving persistence in blood of the
drug bonded utilizing the long half-life of HSA, and a method
using a modified form of HSA as a carrier of the active
transport system. In the former, an attempt has been made to
express a protein or a bioactive peptide having a short half-
life as a hybrid by a gene fusion technique. In the latter, a
method of controlling the physicochemical properties of HSA
such as anionization and cationization, and an attempt to
/o realize accurate kinetic control and cell-specific targeting
by introduction of a recognition element (apparatus) of a
receptor, which is present on the cell surface, such as sugar
structure and peptide have been intensively studied (Lee YC et
al., Biochemisty, 15: 3956-3963, 1976, Opanasopit P et al., Am.
/5 J. Physiol. Gastrointest. Liver Physiol. 280: 879-889, 2001,
Takakura Y et al., Int. J. Pharm. 105: 19-29, 1994, Yamasaki Y
et al., J. Pharmacol. Exp. Ther. 301: 467-477, 2002, Nishikawa
M et al., Am. J. Physiol. Gastrointest. Liver Physiol.
268:G849-G856, 1995, Higuchi Y et al., Int. J. Pharm. 287:
20 147-154, 2004).
It is known that a receptor that recognizes sugar residue
and negative charge is present in the liver. Using this
property, albumin bound with succinic acid, galactose, mannose
and the like is used for targeting the liver.
25 However, for chemical modification of HSA, the following
problems have been pointed out.
(1) The liver does not recognize unless very many sugar
residues are bound;
Galactose-modified albumin is not recognized by the liver
30 unless 10 or more galactoses are bound per albumin molecule
(see Nishikawa M et al., Am. J. Physiol. Gastrointest. Liver
Physiol. 268:G849-G856, 1995).
(2) The cell specificity to liver nonparenchymal cell is low;
Mannose- or fucose-modified albumin is known to be
35 introduced into the both cells of liver endothelial cell and
2
CA 02611540 2007-11-09
kupffer's cell (see Higuchi Y et al., Int. J. Pharm. 287: 147-
154, 2004).
(3) A uniform bound form is difficult to prepare, and
appropriate binding conditions need to be found;
and the like. Accordingly, there is a strong demand for the
development of a method for modifying HSA by a non-chemical
technique.
It is an object of the present invention to provide
uniform glycosylated albumin, particularly serum albumin,
/o which specifically transfer to the liver, particularly
kupffer's cell, thereby providing a drug carrier suitable for
DDS to the liver.
The present inventors have conducted intensive studies in
an attempt to solve the aforementioned problems and succeeded
in preparing a glycosylated HSA with high liver
transferability wherein a high-mannose type sugar chain is
added to the Asn residue of a consensus sequence (Asn-X-
Thr/Ser) by introducing the consensus sequence of an N-linked
sugar chain into a DNA encoding HSA by site-directed
mutagenesis, and cultivating Pichia pastoris transformed with
an expression vector containing the obtained DNA encoding a
mutant HSA, which resulted in the completion of the present
invention.
SUMMARY OF THE INVENTION
Accordingly, the present invention provides the following.
[1] A glycosylated albumin protein comprising a mutant albumin
and a sugar chain, wherein the mutant albumin contains one or
more partial amino acid sequences possibly subject to a
glycosylation modification by an eukaryotic cell, and the
sugar chain is selectively added to the partial amino acid
sequence(s).
[2] The protein of the above-mentioned [1], wherein the sugar
chain is a high-mannose type sugar chain.
[3] The protein of the above-mentioned [1] or [2], wherein at
least one of the partial amino acid sequences is Asn-Xaa-Thr
3
CA 02611540 2007-11-09
or Asn-Xaa-Ser (Xaa is any genetically encoded amino acid).
[4] The protein of the above-mentioned [3], wherein all the
partial amino acid sequences are Asn-Xaa-Thr or Asn-Xaa-Ser
(Xaa is any genetically encoded amino acid).
[5] The protein of any of the above-mentioned [1] to [4],
wherein the albumin is human serum albumin.
[6] The protein of the above-mentioned [5], which has an amino
acid sequence the same as or substantially the same as the
amino acid sequence shown in amino acid numbers 1 - 585 in the
/o amino acid sequence shown in SEQ ID NO: 2, wherein the 63rd
amino acid is Asn and/or the 320th amino acid is Thr or Ser
and/or the 494th amino acid is Asn.
[7] The protein of the above-mentioned [6], wherein at least
the 494th amino acid is Asn.
[8] A DNA encoding a mutant albumin containing one or more
partial amino acid sequences possibly subject to a
glycosylation modification by an eukaryotic cell.
[9] An expression vector comprising the DNA of the above-
mentioned [8], which is under regulation of a promoter
functional in a host eukaryotic cell.
[10] A transformant obtained by introducing the expression
vector of the above-mentioned [9] into a host eukaryotic cell.
[11] The transformant of the above-mentioned [10], wherein the
host eukaryotic cell is a yeast.
[12] The transformant of the above-mentioned [11], wherein the
yeast belongs to the genus Pichia.
[13] A method of producing the protein of the above-mentioned
[1], which comprises culturing the transformant of any of the
above-mentioned [10] - [12] in a medium, and recovering
glycosylated albumin from the obtained culture.
[14] A pharmaceutical agent comprising the protein of any of
the above-mentioned [1] - [7].
[15] A drug carrier to the liver, which comprises the protein
of any of the above-mentioned [1] - [7].
[16] The carrier of the above-mentioned [15], wherein the
4
CA 02611540 2016-01-12
28937-45
target cell is a kupffer's cell.
[17] A pharmaceutical composition comprising a pharmaceutical
compound to be delivered to the liver, and the carrier of the
above-mentioned [15] or [16].
The present invention as claimed relates to:
- a glycosylated albumin protein comprising a mutant
albumin and a sugar chain for use as a drug carrier to the
liver, wherein the mutant albumin (i) has an amino acid
sequence having not less than 90% identity to the amino acid
sequence of wild-type mature human serum albumin (HSA) shown in
amino acids 1-585 of SEQ ID NO:4, (ii) contains one or more
mutations selected from: (a) substitution of Asp residue shown
by amino acid number 63 in the amino acid sequence of SEQ ID
No:4 with Asn residue; (b) substitution of Ala residue shown by
amino acid number 320 in the amino acid sequence of SEQ ID No:4
with Thr residue; and (c) substitution of Asp residue shown by
amino acid number 494 in the amino acid sequence of SEQ ID No:4
with Asn residue; to constitute partial amino acid sequence(s)
that is (are) subject to a glycosylation modification by an
eukaryotic cell, and wherein the sugar chain is selectively
added to the partial amino acid sequence(s) by the eukaryotic
cell containing a DNA encoding the mutant albumin; and
- a pharmaceutical composition comprising a
pharmaceutical compound to be delivered to the liver, and the
glycosylated albumin protein as defined herein, wherein the
pharmaceutical compound is an antioxidative substance or nitric
oxide.
5
CA 02611540 2016-01-12
28937-45
Since the glycosylated albumin of the present
invention is specifically introduced into the liver,
particularly liver nonparenchymal cell, more particularly
kupffer's cell, it can be used as a drug carrier for the cell.
For example, when the glycosylated albumin of the present
invention is bound with an antioxidant or nitric oxide and
administered to hepatic ischemia-reperfusion injury, a superior
treatment effect can be expected. Moreover, since the liver
clearly recognizes even one sugar chain, the albumin can be
used without influencing the original structure and function of
albumin. In addition, since the albumin is a gene recombinant
protein, it is free of a risk of contamination with an unknown
virus and the like, which is a problem specific to blood-
derived preparations, and can be used safely for human body and
the like.
BRIEF DESCRIPTION OF THE DRAWINGS
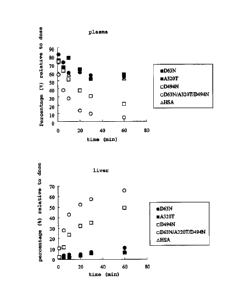
Fig. I is a graph showing the time course changes of
transferability of glycosylated human serum albumin of the
present invention to plasma (upper) and liver (lower) in
Experimental Example 1, wherein the vertical axis shows a
percentage (%) relative to dose and the transverse axis shows
time (min) after administration.
Fig. 2 is a graph showing the transfer of "11n-
labeled, succinic acid-modified (Suc-)bovine serum albumin
(BSA) to plasma (upper), liver (middle) and kidney (lower) when
it was intravenously administered to mouse (see Takakura Y et
al., Int. J. Pharm. 105: 19-29, 1994), wherein n of Suc,-BSA
shows the number of succinic acid bonded to BSA, = shows
0.1 mg/kg, 0 shows 1 mg/kg, V shows 10 mg/kg and V shows
5a
CA 02611540 2016-01-12
28937-45
20 mg/kg each of BSA dose, and the presented data are an
excerpt of a related portion from Takakura Y et al., Int. J.
Pharm. 105: 19-29, 1994 added with the vertical axis.
5b
CA 02611540 2007-11-09
DETAILED DESCRIPTION OF THE INVENTION
Examples of the albumin in the present invention include
serum albumin, egg white albumin and the like, with preference
given to serum albumin. While the origin of albumin is not
particularly limited, for example, human and other warm-
blooded animals (e.g., bovine, monkey, swine, equine, sheep,
goat, canine, feline, rabbit, mouse, rat, hamster, guinea pig,
chicken, quail etc.) can be mentioned. In consideration of use
as a pharmaceutical agent or a carrier of a pharmaceutical
/o compound, preferred is human albumin, more preferred is human
serum albumin (NSA). In the following, the present invention
is sometimes explained in detail by referring to HSA as an
example. Those of ordinary skill in the art can produce and
utilize glycosylated albumin in the same manner based on the
description in the present specification and other known
sequence information of albumin.
The glycosylated albumin of the present invention is a
mutant albumin containing one or more partial amino acid
sequences possibly subject to a glycosylation modification by
an eukaryotic cell, wherein a sugar chain is selectively added
to the partial amino acid sequence. Examples of the "partial
amino acid sequences possibly subject to a glycosylation
modification by an eukaryotic cell" (hereinafter sometimes to
be also referred to as "glycosylation sequence") include, but
are not limited to, Asn-Xaa-Thr or Asn-Xaa-Ser (Xaa is amino
acid genetically coded for and a sugar chain is added to Asn
residue) (hereinafter comprehensively abbreviated as "Asn-Xaa-
Thr/Ser"), which are consensus sequences of an N-linked sugar
chain, Cys-Xaa-Xaa-Gly-Gly-Thr/Ser (Xaa is as defined above,
and a sugar chain is added to Thr/Ser residue), which is a
consensus sequence of 0-linked fucose from among 0-linked
sugar chains, Cys-Xaa-Ser-Xaa-Pro-Cys (Xaa is as defined above,
and a sugar chain is added to Ser residue), which is a
consensus sequence of 0-linked glucose and the like. Preferred
is Asn-Xaa-Thr/Ser, which is a consensus sequence of N-linked
6
CA 02611540 2007-11-09
sugar chain. The number of the glycosylation sequences may be
one or more. While the liver, particularly kupffer's cell,
targeting efficiency is improved as the number of sugar chains
increases, in consideration of the maintenance of the original
physiological function of albumin and the antigenicity problem,
a smaller number of sugar chains to be added is more
advantageous. As mentioned below, the liver targeting function
does not simply depend on the number of sugar chains to be
added, but varies depending on the site of addition. Thus,
/o introduction of a glycosylation sequence into a site highly
contributing to the targeting efficiency achieves superior
targeting efficiency with a small number of sugar chains.
Since natural (wild-type) albumin is a simple protein, it
does not have a partial amino acid sequence that may undergo a
/5 glycosylation modification by an eukaryotic cell. Accordingly,
the glycosylated albumin of the present invention comprises a
mutant amino acid sequence containing the above-mentioned
glycosylation sequence. While the mutant albumin polypeptide
of the present invention may be obtained by any method, for a
20 sugar chain to be selectively added to the glycosylation
sequence in the polypeptide, the mutant albumin polypeptide is
preferably provided by cultivating an eukaryotic cell
containing a DNA encoding the same.
While the eukaryotic cell containing a DNA encoding the
25 mutant albumin can also be obtained, for example, by inducing
a mutation inartificially or artificially (e.g., treatment
with mutagenic agent such as EMS and the like, UV treatment
and the like) in a cell (e.g., hepatocyte and the like)
inherently producing albumin and screening for a cell
30 producing a mutant albumin containing a glycosylation sequence,
it can be more preferably produced by cloning a DNA encoding
albumin, introducing a base sequence encoding a glycosylation
sequence into the DNA by a genetic manipulation, inserting the
obtained mutant DNA into an expression vector containing a
35 promoter functional in a suitable host eukaryotic cell so that
7
CA 02611540 2007-11-09
it will enter the control of the promoter, and transforming
the host eukaryotic cell with the obtained mutant albumin
expression vector.
Examples of the DNA encoding albumin include genomic DNAs
derived from human or other warm-blooded animals, cDNAs
derived from albumin-producing cells (e.g., hepatocyte and the
like), synthetic DNA and the like. The genomic DNA or cDNA
encoding albumin can also be directly amplified by Polymerase
Chain Reaction (hereinafter abbreviated as "PCR method") or
lo Reverse Transcriptase-PCR (hereinafter abbreviated as "RT-PCR
method") using a genomic DNA fraction or a total RNA or mRNA
fraction prepared from the producing cells or tissues (e.g.,
liver and the like) as a template. Alternatively, the genomic
DNA or cDNA encoding albumin can also be cloned by colony or
plaque hybridization method, PCR method and the like from a
genomic DNA library or cDNA library prepared by inserting a
fragment of genomic DNA or total RNA or mRNA prepared from the
above-mentioned cell/tissue into a suitable vector. The vector
to be used for the library may be any of bacteriophage,
plasmid, cosmid, phagemid and the like.
Examples of the DNA encoding albumin include a DNA
containing a base sequence encoding an amino acid sequence the
same as or substantially the same as the amino acid sequence
shown in amino acid numbers 1 - 585 in the amino acid sequence
shown in SEQ ID NO: 4 (wild-type mature HSA) and the like. As
the amino acid sequence substantially the same as the amino
acid sequence shown in amino acid numbers 1 - 585 in the amino
acid sequence shown in SEQ ID NO: 4, an amino acid sequence
having a homology of not less than about 80%, preferably not
less than about 90%, more preferably not less than about 95%,
particularly preferably not less than about 98% with the amino
acid sequence shown in the amino acid sequence shown in amino
acid numbers 1 - 585 in the amino acid sequence shown in SEQ
ID NO: 4 and the like can be mentioned. As used herein, by the
"homology" is meant the proportion (%) of the same amino acid
8
CA 02611540 2007-11-09
and similar amino acid residues relative to the total
overlapping amino acid residues in the optimal alignment when
two amino acid sequences are aligned using a mathematical
algorithm known in the art (preferably, the algorithm is
capable of considering introduction of a gap into one or both
of the sequences for the optimal alignment). The "similar
amino acid" means an amino acid similar in the physicochemical
properties. For example, amino acids classified in the same
group such as aromatic amino acids (Phe, Trp, Tyr), aliphatic
/o amino acids (Ala, Leu, Ile, Val), polar amino acids (Gin, Asn),
basic amino acids (Lys, Arg, His), acidic amino acids (Glu,
Asp), amino acids (Ser, Thr) having a hydroxyl group, amino
acids (Gly, Ala, Ser, Thr, Met) with small side chain and the
like can be mentioned. It is predicted that substitution with
/5 such similar amino acids will not alter protein phenotypes
(namely, preservative amino acid substitution). Specific
examples of preservative amino acid substitution are well
known in the art and are described in various literatures (see
e.g., Bowie et al., Science, 247: 1306-1310 (1990)).
20 The homology of the amino acid sequence in the present
. specification can be calculated using homology calculation
algorithm NCBI BLAST (National Center for Biotechnology
Information Basic Local Alignment Search Tool) under the
following conditions (expectancy=10; allowing gap;
25 matrix=BLOSUM62; filtering=OFF). Other algorithms for
determining the homology of the amino acid sequence include,
for example, the algorithm described in Karlin et al., Proc.
Natl. Acad. Sci. USA, 90: 5873-5877 (1993) [this algorithm is
incorporated in the NBLAST and XBLAST program (version 2.0)
30 (Altschul et al., Nucleic Acids Res., 25: 3389-3402 (1997))],
the algorithm described in Needleman et al., J. Mol. Biol.,
48: 444-453 (1970) [this algorithm is incorporated in the GAP
program in the GCG software package], the algorithm described
in Myers and Miller, CABIOS, 4: 11-17 (1988) [this algorithm
35 is incorporated in the ALIGN program (version 2.0) which is a
9
CA 02611540 2007-11-09
part of the CGC sequence alignment software package], the
algorithm described in Pearson et al., Proc. Natl. Acad. Sci.
USA, 85: 2444-2448 (1988) [this algorithm is incorporated in
the FASTA program in the GCG software package] and the like,
and they can also be used preferably.
More preferably, an amino acid sequence substantially the
same as the amino acid sequence shown in amino acid numbers 1
- 585 in the amino acid sequence shown in SEQ ID NO: 4 has
homology of not less than about 80%, preferably not less than
/0 about 90%, more preferably not less than about 95%,
particularly preferably not less than about 98%, to the amino
acid sequence shown in amino acid numbers 1 - 585 in the amino
acid sequence shown in SEQ ID NO: 4.
A protein containing an amino acid sequence substantially
the same as the amino acid sequence shown in amino acid
numbers 1 - 585 in the amino acid sequence shown in SEQ ID NO:
4 means a protein containing an amino acid sequence
substantially the same as the aforementioned amino acid
sequence shown in amino acid numbers 1 - 585 in the amino acid
sequence shown in SEQ ID NO: 4 and having a substantially
equivalent activity to that of the protein containing the
amino acid sequence shown in amino acid numbers 1 - 585 in the
amino acid sequence shown in SEQ ID NO: 4.
The substantially the equivalent activity includes, for
example, physiological function of albumin (particularly serum
albumin), such as function as a carrier of serum molecules,
function to maintain plasma colloidal osmotic pressure and the
like. The "substantially equivalent" means that the functions
are qualitatively the same. Therefore, the function as a
carrier of the serum molecules and the like is preferably
equivalent, but the quantitative elements such as the level of
the function, molecular weight of the protein and the like may
be different.
In addition, the DNA encoding albumin includes, for
example, DNA encoding a protein containing (1) an amino acid
CA 02611540 2007-11-09
sequence wherein one or more (preferably about 1 - 30, more
preferably about 1 - 10, particularly preferably 1 - several
(2, 3, 4 or 5)) amino acids are deleted from the amino acid
sequence shown in amino acid numbers 1 - 585 of the amino acid
sequence shown in SEQ ID NO: 4, (2) an amino acid sequence
wherein one or more (preferably about 1 - 30, more preferably
about 1 - 10, particularly preferably 1 - several (2, 3, 4 or
5)) amino acids are added to the amino acid sequence shown in
amino acid numbers 1 - 585 in the amino acid sequence shown in
lo SEQ ID NO: 4, (3) an amino acid sequence wherein one or more
(preferably about 1 - 30, more preferably about 1 - 10,
particularly preferably 1 - several (2, 3, 4 or 5)) amino
acids are inserted in the amino acid sequence shown in amino
acid numbers 1 - 585 in the amino acid sequence shown in SEQ
ID NO: 4, (4) an amino acid sequence wherein one or more
(preferably about 1 - 30, more preferably about 1 - 10,
particularly preferably 1 - several (2, 3, 4 or 5)) amino
acids are substituted by other amino acids in the amino acid
sequence shown in amino acid numbers 1 - 585 in the amino acid
sequence shown in SEQ ID NO: 4, or (5) a combination of these,
and the like.
When the amino acid sequence is inserted, deleted or
substituted as mentioned above, the site of the insertion,
deletion or substitution is not particularly limited as long
as the activity of protein is maintained.
More preferably, a DNA encoding albumin (particularly
HSA) includes, for example, a DNA containing the base sequence
shown in base numbers 73 - 1827 in the base sequence shown in
SEQ ID NO: 3, a DNA encoding a protein having a base sequence
hybridizing to the base sequence shown in SEQ ID NO: 3 under
stringent conditions, and having substantially equivalent
activity (e.g., function of serum molecule as a carrier and
the like) to a protein containing the aforementioned amino
acid sequence shown in amino acid numbers 1 - 585 of the amino
acid sequence shown in SEQ ID NO: 4, and the like. As the DNA
11
CA 02611540 2007-11-09
capable of hybridizing to the base sequence shown in SEQ ID
NO: 3 under stringent conditions, for example, a DNA
containing a base sequence having, in an overlapping region, a
homology of not less than about 80%, preferably not less than
about 90%, more preferably not less than about 95%, to the
base sequence shown in base numbers 73 - 1827 in the base
sequence shown in SEQ ID NO: 3 and the like can be used.
The homology of the base sequence in the present
specification can be calculated using a homology calculation
/o algorithm NCBI BLAST (National Center for Biotechnology
Information Basic Local Alignment Search Tool) under the
following conditions (expectancy=10; allowing gap;
filtering=0N; match score=1; mismatch score=-3). Preferable
examples of other algorithms usable for determining homology
of the base sequence include the above-mentioned homology
calculation algorithms for amino acid sequence.
Hybridization can be performed according to a method
known per se or a method according to the method, for example,
the method described in Molecular Cloning, ver. 2 (J. Sambrook
et al., Cold Spring Harbor Lab. Press, 1989) and the like.
When a commercially available library is used, moreover,
hybridization can be performed according to the method
described in the attached instruction manual. Preferably,
hybridization can be performed under high stringent conditions.
The high stringent conditions include, for example, a
hybridization reaction at 45 C in 6xSSC (sodium chloride/sodium
citrate), and washing one or more times at 65 C in 0.2xSSC/0.1%
SDS and the like. Those of ordinary skill in the art can
easily adjust to the desired stringency by appropriately
changing the salt concentration of hybridization solution,
temperature of hybridization reaction, probe concentration,
length of probe, number of mismatches, hybridization reaction
time, salt concentration of washing solution, temperature of
washing and the like.
A DNA encoding albumin (particularly HSA) can be cloned
12
CA 02611540 2007-11-09
by amplifying by PCR method using a synthetic DNA primer
having a part of the base sequence encoding albumin, or
hybridizing DNA incorporated into a suitable expression vector
to with a labeled DNA fragment or synthetic DNA encoding a
part or full region of albumin.
As a method for introducing a base sequence encoding a
glycosylation sequence into a DNA encoding albumin
(particularly HSA) obtained as mentioned above, site-directed
mutagenesis known per se (e.g., Examples below) and the like
lo can be used. A glycosylation sequence-coding sequence may be
introduced into any part of the DNA encoding albumin. In the
case of site-directed mutagenesis using PCR method, wherein,
for example, a base sequence encoding consensus sequence Asn-
Xaa-Thr/Ser of N-linked sugar chain is introduced, the
sequence is preferably introduced into a site encoding the Asn
residue of a DNA encoding albumin or a site encoding the Thr
or Ser residue thereof. To be specific, a base sequence
encoding a consensus sequence of N-linked sugar chain can be
introduced by PCR using a DNA encoding albumin as a template,
and (1) an oligonucleotide complementary to a region
containing a base sequence encoding any Asn-Xaal-Xaa2 site in
the albumin (provided the codon corresponding to Xaa2 is
substituted by a codon encoding Thr or Ser) or (2) an
oligonucleotide complementary to a region containing a base
sequence encoding any Xaal-Xaa2-Thr/Ser site in the albumin
(provided the codon corresponding to Xaal is substituted by a
codon encoding Asn) as one primer. The glycosylation sequence
can be made to be present in the DNA not only by amino acid
substitution as mentioned above, but also by inserting a base
sequence encoding an amino acid (or amino acid sequence) into
a DNA encoding albumin or deleting a base sequence encoding an
amino acid (or amino acid sequence) from the DNA by a similar
method.
In the case of HSA, for example, more preferably, a
consensus sequence of N-linked sugar chain can be introduced
13
CA 02611540 2007-11-09
by substituting Asp residue shown by amino acid number 494 in
the amino acid sequence shown in SEQ ID NO: 4 with Asn residue
(Asn494) (see SEQ ID NO: 2). Glycosylated HSA wherein a sugar
chain is added to Asn494 can be targeted to the liver at an
efficiency equal to more than that of glycosylated albumin
(having a number of sugar chains) obtained by conventionally
known chemical modification, even though the number of sugar
chain in the molecule is only one. In another preferable
embodiment, a consensus sequence of N-linked sugar chain can
/o be introduced by substituting Asp residue shown by amino acid
number 63 in the amino acid sequence shown in SEQ ID NO: 4
with Asn residue (Asn63), or by substituting Ala residue shown
by amino acid number 320 with Thr or Ser residue (Thr/Ser320)
(see SEQ ID NO: 2). In a particularly preferable embodiment,
glycosylated HSA of the present invention can further contain,
in addition to Asn434, one or more glycosylation sequences,
preferably consensus sequence Asn-Xaa-Thr/Ser of N-linked
sugar chain. As a further sugar chain addition site, the
above-mentioned Asn63 and/or Asn318 resulting from the above-
mentioned substitution with Thr/Ser32 can be mentioned.
An expression vector containing a DNA encoding a mutant
albumin containing one or more partial amino acid sequences
possibly subject to a glycosylation modification by an
eukaryotic cell, which has been cloned as mentioned above, can
be produced by ligating the DNA to a downstream of a promoter
in a suitable expression vector using a restriction enzyme and
a DNA ligase.
As the expression vector, bacteriophage such as plasmid
derived from Escherichia coli (e.g., pBR322, pBR325, pUC12,
pUC13), plasmid derived from Bacillus subtilis (e.g., pUB110,
pTP5, pC194), plasmid derived from yeast (e.g., pSH19, pSH15),
X-phage and the like, animal (insect) virus such as retrovirus,
vaccinia virus, baculovirus and the like, pA1-11, pXT1,
pRc/CMV, pRc/RSV, pcDNAI/Neo and the like are used.
The promoter may be any as long as it is an appropriate
14
CA 02611540 2007-11-09
promoter corresponding to the host used for gene expression.
In the present invention, any can be used as a host cell
without any particular limitation as long as it has a
glycosylation modification mechanism to add a sugar chain to
the glycosylation sequence contained in the mutant albumin of
the present invention and, for example, various eukaryotic
cells such as animal cell including mammal, insect cell, plant
cell, yeast cell, fungal cell and the like, or transgenic
animal/plant or insect and the like can be used.
io For example, when the host is a yeast, a PHO5 promoter, a
PGK promoter, a GAP promoter, an ADH promoter and the like are
preferable.
When the host is an animal cell, a promoter derived from
cytomegalovirus (CMV) (e.g., CMV immediate-early promoter), a
promoter derived from human immunodeficiency virus (HIV) (e.g.,
HIV LTR), a promoter derived from Rous sarcoma virus (RSV)
(e.g., RSV LTR), a promoter derived from mouse mammary tumor
virus (MMTV) (e.g., MMTV LTR), a promoter derived from Moloney
murine leukemia virus (MoMLV) (e.g., MMTV LTR), a promoter
derived from simple herpes virus (HSV) (e.g., HSV thymidine
kinase (TI() promoter), a promoter derived from SV40 promoter
(e.g., SV40 early promoter), a promoter derived from Epstein-
Barr virus (EBV), a promoter derived from adeno-associated
virus (AAV) (e.g., AAV p5 promoter), a promoter derived from
adenovirus (AdV) (Ad2 or Ad5 major late promoter) and the like
can be used.
When the host is an insect cell, a polyhedrin promoter, a
P10 promoter and the like are preferable.
As the expression vector, besides those mentioned above,
one containing an enhancer, a splicing signal, a polyA
addition signal, a selection marker, an SV40 replication
origin and the like on demand can be used. As the selection
marker, for example, dihydrofolate reductase (dhfr) gene
[methotrexate (MTX) resistance], ampicillin resistance (Ampr)
gene, neomycin resistance (Neor) gene (G418 resistance) and the
CA 02611540 2007-11-09
like can be mentioned. Particularly, when dhfr-deficient
Chinese hamster (CHO-dhfri cell is used and dhfr gene is used
as a selection marker, the object gene can also be selected in
a thymidine-free medium. Moreover, when the DNA to be inserted
does not contain an initiation codon and a stop codon, a
vector containing an initiation codon (ATG or GTG) and a stop
codon (TAG, TGA, TAA) at the downstream of promoter region and
at the upstream of terminator region, respectively, is
preferably used.
Where necessary, a base sequence encoding a signal
sequence suitable for the host (signal codon) may be added to
the 5' end side of a DNA encoding the mutant albumin. For
example, when the host is a yeast, MFa signal sequence, SUC2
signal sequence and the like can be used. When the host is an
animal cell, insulin signal sequence, a-interferon signal
sequence, antibody molecule signal sequence and the like can
be used. However, since native prepro-sequence of HSA (amino
acid sequence shown by amino acid number -24 to -1 in the
amino acid sequence shown in SEQ ID NO: 4) is known to
function as a secretion signal in most heterologous eukaryotic
cells, a DNA encoding prepro-HSA can also be directly inserted
into an expression vector.
As mentioned above, for example, yeast, insect cell,
insect, animal cell, animal and the like are used as a host.
As the yeast, for example, Saccharomyces cerevisiae AH22,
AH22R-, NA87-11A, DKD-5D, 20B-12, Schizosaccharomyces pombe
NCYC1913, NCYC2036 and the like are used.
As the insect cell, for example, when the virus is AcNPV,
established cell line derived from Spodoptera frugiperda larva
(Spodoptera frugiperda cell; Sf cell), MG1 cell derived from
Trichoplusia ni midgud, High Fiveml cell derived from
Trichoplusia ni egg, cell derived from Mamestra brassicae,
cell derived from Estigmena acrea and the like are used. When
the virus is BmNPV, established cell line derived from
silkworm (Bombyx mori N cell; BmN cell) and the like are used
16
CA 02611540 2007-11-09
as the insect cell. As the Sf cell, for example, Sf9 cell
(ATCC CRL1711), Sf21 cell (both in Vaughn, J.L. et al., In
Vivo, 13, 213-217 (1977)) and the like are used.
As the insect, for example, Bombyx mori larva and the
like are used.
As the animal cell, for example, cell derived from monkey
(e.g., COS-1, COS-7, CV-1, Vero), cell derived from hamster
(e.g., BHK, CHO, CHO-K1, CHO-dhfr), cell derived from mouse
(e.g., NIH3T3, L, L929, CTLL-2, AtT-20), cell derived from rat
/o (e.g., H4IIE, PC-12, 3Y1, NBT-II), cell derived from human
(e.g., HEK293, A549, HeLa, HepG2, HL-60, Jurkat, U937) and the
like are used.
Transformation can be performed according to a known
method depending on the kind of the host.
For example, yeast can be transformed according to the
methods described in Methods in Enzymology, 194, 182-187
(1991), Proc. Natl. Acad. Sci. USA, 75, 1929 (1978) and the
like.
For example, insect cell and insect can be transformed
according to the methods described in Bio/Technology, 6, 47-55
(1988) and the like.
For example, animal cell can be transformed according to
the methods described in Saibo Kogaku, extra issue 8, Shin
Saibo Kogaku Jikken Protocol, 263-267 (1995)(published by
Shujunsha) and Virology, 52, 456 (1973).
The transformant can be cultured according to a known
method depending on the kind of the host.
As the medium, a liquid medium is preferable. The medium
preferably contains a carbon source, a nitrogen source, an
inorganic substance and the like necessary for the growth of
the transformant. Here, as the carbon source, for example,
glucose, dextrin, soluble starch, sucrose and the like can be
used; as the nitrogen source, for example, inorganic or
organic substances such as ammonium salts, nitrate salts, corn
steep liquor, peptone, casein, meat extract, soybean cake,
17
CA 02611540 2007-11-09
potato extract and the like can be used; and as the inorganic
substance, for example, calcium chloride, sodium dihydrogen
phosphate, magnesium chloride and the like can be used. The
medium may contain a yeast extract, vitamins, a growth-
promoting factor and the like. The pH of the medium is
preferably about 5 - 8.
As a medium for cultivating a transformant whose host is
a yeast, for example, Burkholder minimum medium, SD medium
containing 0.5% casamino acid and the like can be mentioned.
The pH of the medium is preferably about 5 - 8. The culture is
generally performed at about 20 C - 35 C for about 24 - 72 hr.
Where necessary, aeration and agitation may also be performed.
As a medium for cultivating a transformant whose host is
an insect cell or a insect, for example, Grace's Insect Medium
is appropriately supplemented with an additive such as
inactivated 10% bovine serum and the like, and the like are
used. The pH of the medium is preferably about 6.2 - 6.4. The
culture is generally performed at about 27 C for about 3 - 5
days. Where necessary, aeration and agitation may also be
performed.
As a medium for cultivating a transformant whose host is
an animal cell, for example, minimum essential medium (MEM),
Dulbecco's Modified Eagle Medium (DMEM), RPMI1640 medium, 199
medium and the like supplemented with about 5 - 20% of fetal
bovine serum are used. The pH of the medium is preferably
about 6 - 8. The culture is generally performed at about 30 C
- 40 C for about 15 - 60 hr. Where necessary, aeration and
agitation may also be performed.
In this manner, Glycosylated albumin can be
intracellularly or extracellularly provided by a transformant.
Since the glycosylated albumin of the present invention
can be preferably used as a carrier molecule capable of
specific transfer to the liver, particularly kupffer's cell,
one wherein a high-mannose type sugar chain having high
affinity for a receptor on the cell surface is added is more
18
CA 02611540 2007-11-09
preferable. Here, the "high-mannose type" means a sugar chain
wherein one or more, preferably two or more, more preferably
three or more, particularly preferably five or more, mannose
molecules are further added to the core sugar chain (including
three mannose molecules). From such aspects, a yeast cell,
permitting addition of only a hyper-mannose type sugar chain
and also permitting addition of a hyper-mannose type sugar
chain having still more mannose molecules than those in animal
cell and the like, is more preferable as a host cell, than
/o animal cell and insect cell capable of a different
glycosylation modification such as those of a complex type and
a mixed type in addition to a high-mannose type. Particularly,
a yeast of the genus Pichia can grow utilizing methanol as a
sole carbon source, and when grown in methanol, an enzyme
necessary for treating methanol and a metabolic intermediate
thereof are disinhibited and expressed. It. is known that the
secretion expression level of a heterologous protein markedly
exceeds that of Saccharomyces yeast when the methanol-
utilizing pathway is used. In fact, production of HSA using
this system is in the phase of practical application (e.g.,
JP-A-6-22784), where HSA of a 10 g order can be produced from
1 L of a medium. In the following, as one of the particularly
preferable embodiments of the present invention, a production
method of the glycosylated albumin of the present invention,
which uses a Pichia yeast as a host cell, is explained.
The vector to be used is not particularly limited as long
as it can be maintained genetically stably by autonomous
replication in a fungus body of yeast of the genus Pichia or
integration into a yeast genome. Examples of the autonomously-
replicable vector include YEp vector, YRp vector, YCp vector
and the like. In addition, examples of the vector to be
integrated into a yeast genome include YIp vector and YRp
vector.
Examples of the promoter functional in the yeast of the
genus Pichia include promoters derived from a yeast, such as
19
CA 02611540 2007-11-09
PHO5 promoter, PGK promoter, GAP promoter, ADH promoter
derived from S. cerevisiae and the like, alcohol oxidase (AOX)
1 promoter, A0X2 promoter, dihydroxyacetone synthase promoter,
P40 promoter, ADH promoter, folic acid dehydrogenase promoter
derived from P. pastoris and the like. In addition, the above-
mentioned promoter derived from a yeast may be a mutant
promoter modified to further improve the gene expression
efficiency, for example, mutant A0X2 (mA0X2) promoter [Ohi et
al., Mol. Gen. Genet., 243, 489-499 (1994); JP-A-4-299984] and
/o the like. Preferably, the promoter is a promoter of an enzyme
gene necessary for treating methanol or a metabolic
intermediate thereof, in order to use a methanol-metabolizing
system in the yeast of the genus Pichia, such as A0X1 promoter,
mA0X2 promoter and the like.
The expression vector containing the DNA encoding mutant
albumin of the present invention preferably further contains
transcription terminator sequence (terminator) functional in a
yeast of the genus Pichia (e.g., A0X1 terminator etc.),
enhancer sequence, selection marker gene usable for selecting
yeast (auxotrophic gene, for example, HIS4, LEU2, ARG4 and
URA3 gene derived from P. pastoris or S. cerevisiae, and the
like, or antibiotic resistance gene, for example, resistance
gene to cycloheximide, G-418, chloramphenicol, bleomycin,
hygromycin etc., and the like) and the like, and when desired,
may contain replicable unit functional in yeast. For
preparation of the vector in a large amount, moreover, the
vector more preferably contains a replicable unit functional
in Escherichia coli and a selection marker gene usable for
selecting Escherichia coli (e.g., resistance gene to
ampicillin and tetracycline etc.)
When the expression vector is of a type incorporated into
a yeast genome, the vector preferably further contains a
sequence homologous to a yeast genome necessary for homologous
recombination. As such homology sequence, the aforementioned
auxotrophic gene sequence can be mentioned. Accordingly, in
CA 02611540 2007-11-09
one preferable embodiment, the expression vector of the
present invention is one wherein an expression cassette of the
above-mentioned mutant albumin is inserted in an auxotrophic
gene (in the present specification, the "expression cassette"
means a unit enabling gene expression, whose minimal unit is a
protein-coding sequence configured under regulation of a
promoter, with preference given to a unit comprising promoter-
protein-coding region-terminator).
The expression vector obtained as mentioned above can be
/0 introduced into the fungus body of the target yeast of the
genus Pichia using, for example, a known transformation
technique such as competent cell method, protoplast method,
calcium phosphate coprecipitation method, polyethylene glycol
method, lithium method, electroporation method, microinjection
method, liposome fusion method, particle gun method and the
like.
While the yeast of the genus Pichia to be used in the
present invention is not particularly limited, for example, P.
pastoris, Pichia acaciae, Pichia angusta, Pichia anomala,
Pichia capsulata, Pichia ciferrii, Pichia etchellsii, Pichia
fabianii, Pichia farinosa, Pichia guilliermondii, Pichia
inositovora, Pichia jadinii, Pichia methanolica, Pichia
norvegensis, Pichia ofunaensis, Pichia pinus and the like can
be used. Preferred is P. pastoris, particularly, auxotrophic
mutant P. pastoris strain (e.g., P. pastoris GTS115 strain
(HIS4) [NNRL Y-15851], P. pastoris GS190 strain (ARG41
[NNRLY-1801], P. pastoris PPF1 (HIS4-= URA4-) [NNRL Y-18017] and
the like).
By cultivating the transformed yeast of the genus Pichia
by a method generally used in the art, glycosylated albumin
can be produced. The medium to be used needs to contain at
least a carbon source and an inorganic or organic nitrogen
source necessary for the growth of the host cell. Examples of
the carbon source include methanol, glycerol, glucose, sucrose,
dextran, soluble starch and the like. In addition, examples of
21
CA 02611540 2007-11-09
the inorganic or organic nitrogen source include ammonium
salts, nitrate salts, amino acid, corn steep liquor, peptone,
casein, meat extract, yeast extract, soybean cake, potato
extract and the like. When desired, moreover, other nutrients,
for example, inorganic salts such as calcium chloride, sodium
dihydrogenphosphate, magnesium chloride and the like, vitamins
such as biotin and the like, antibiotic and the like can be
added.
Examples of the medium to be used include conventional
lo natural medium (e.g., YPD medium, YPM medium, YPG medium etc.)
and synthetic medium. As the pH and culture temperature of the
medium, those suitable for the growth of yeast and production
of albumin are employed. For example, pH of about 5 - about 8
and culture temperature of about 20 C - about 30 C are
preferable. In addition, aeration and agitation are performed
as necessary. The culture is generally performed for about 48
- about 120 hr.
For example, when a promoter whose expression is induced
by methanol, such as A0X1 promoter, mA0X2 promoter and the
like, is used as a promoter functional in the fungus body of a
yeast of the genus Pichia, a method of liquid aeration-
agitation culture using natural medium controlled to pH about
6.0, which contains glycerol as a carbon source for the growth
of fungus body and methanol as albumin expression inducer is
most preferable. When the expression of albumin is not
preferable for the growth of fungus body, a method including
first increasing the amount of fungus body with a carbon
source other than methanol, and inducing the expression of
albumin by addition of methanol is more preferable. In a
culture in a jarfermenter, moreover, a high density culture
method is suitable for the production of albumin. The culture
may be performed by any of batch culture, feeding culture and
continuous culture, with preference given to feeding culture
method. That is, for a certain period, a method including
culturing the host fungus body in a medium (initial medium)
22
CA 02611540 2007-11-09
containing a carbon energy source suitable for the growth
(e.g., glucose etc.) and/or a nutrient source, and confining
albumin in the system until completion of the culture while
additionally supplying a substrate controlling the growth of
the host cell (that is, methanol) to the medium from a certain
point in time according to the situation can be used (see e.g.,
JP-A-3-83595).
Albumin produced in the culture can be isolated and
purified by centrifugation and/or filtration of the culture
/o after completion of the culture to give a culture supernatant
(in the case of secretory expression) or fungus body of yeast
(in the case of expression in fungus body), which is then
treated according to a method known per se. As such method, a
method utilizing the solubility such as salting out, solvent
precipitation and the like; a method mainly utilizing
difference in the molecular weight such as dialysis,
ultrafiltration, gel filtration method, SDS-polyacrylamide gel
electrophoresis and the like; a method utilizing difference in
the electric charge such as ion exchange chromatography and
the like; a method utilizing specific affinity such as
affinity chromatography and the like; a method utilizing
difference in hydrophobicity such as reversed-phase high
performance liquid chromatography and the like; a method
utilizing difference in the isoelectric point such as
isoelectric focusing and the like; and the like can be used.
These methods can be appropriately combined.
Examples of a method for confirming the isolated and
purified glycosylated albumin include known Western blotting
method and the like. In addition, the structure of the
purified glycosylated albumin can be clarified by amino acid
analysis, N-terminal amino acid sequence, primary structure
analysis, sugar chain analysis and the like.
The thus-obtained glycosylated albumin is a uniform
glycoprotein wherein a sugar chain, preferably a high-mannose
type sugar chain, is selectively added to the glycosylation
23
CA 02611540 2007-11-09
sequence of a mutant albumin, and therefore shows high
transferability to the nonparenchymal cells of the liver,
particularly kupffer's cell. Accordingly, the present
invention also provides a drug carrier to the liver, which
contains the above-mentioned glycosylated albumin of the
present invention.
Since the drug carrier of the present invention, which
contains the glycosylated albumin of the present invention
(particularly HSA) as a main component, can be utilized for
/o targeting any pharmaceutical compound that becomes effective
for the prophylaxis and/or treatment on delivery to the liver,
preferably hepatic nonparenchymal cells, particularly
kupffer's cell, to the organ or cell. Examples of such
pharmaceutical compound include antioxidative substances (e.g.,
N-acetylcysteine, ascorbic acid etc.), nitric oxide and the
like. A preparation wherein the pharmaceutical compound is
bound with the glycosylated albumin of the present invention
can be used for the treatment of hepatic ischemia-reperfusion
injury. Moreover, examples of other pharmaceutical compounds
include a hepatic drug such as hepatic fibrosis treatment drug
0K432, and the like. Since albumin itself also has an
antioxidative action, it can be directly used as a
pharmaceutical product having an antioxidative action.
The binding mode of glycosylated albumin and a
pharmaceutical compound is not particularly limited. For
example, covalent bond, hydrogen bond, hydrophobic bond and
the like can be used, with preference given to a covalent bond.
The method for binding albumin with a pharmaceutical compound
is known and, for example, "Drug Delivery System" (1986,
published by CMC) can be referred to.
A pharmaceutical compound-glycosylated albumin conjugate
can be processed into a preparation by a known method
(ultrafiltration, sterilizing by filtration, dispension,
freeze-drying etc.) Specifically, a liquid preparation
containing 5 - 25% of the conjugate and having a pH of about
24
CA 02611540 2007-11-09
6.4 - 7.4 and an osmotic pressure ratio of about 1 can be
mentioned. Where necessary, the preparation can contain
acetyltryptophan or a salt thereof (e.g., sodium salt) and
sodium caprylate as stabilizers. The amount of the stabilizer
to be added is, for example, about 0.01 - 0.2M, preferably
about 0.02 - 0.05M. In addition, the sodium content is, for
example, not more than 3.7 mg/ml. The timing of addition of
the stabilizer is before treatment by ultrafiltration,
sterilizing by filtration, dispension, freeze-drying and the
_to like.
The medical preparation of the present invention obtained
via the above-mentioned steps is considered to have an
extremely slim possibility of contamination with various
microorganisms. As a method for more positively securing the
is aseptic nature of the preparation, inactivation of
contaminating microorganisms can be performed by applying a
heat treatment (pasteurization) after aseptic filling.
By a heat treatment including keeping a preparation
filled in a container per unit dose, irrespective of the kind
20 of the container to be filled in, for example, for not less
than about 30 min in a hot water bath at about 50 C - about
70 C (preferably about 60 C), contaminating microorganisms can
be inactivated sufficiently. The heating time is preferably
about 30 min - about 2 hr.
25 The pharmaceutical preparation can be administered, for
example, as an injection to human, other mammals and the like.
While the dose of the preparation varies depending on the kind
of pharmaceutical compound, administration route, severity of
disease, animal species to be the subject of administration,
30 and drug acceptability, body weight, age and the like of the
administration subject, it is, for example, in the case of a
hepatic ischemia-reperfusion injury therapeutic agent
containing nitric oxide as an active ingredient, generally 0.1
- 30 g/kg/day, preferably 0.5 - 3 g/kg/day, in a nitric oxide
35 amount for an adult, and about 0.1 - 30 mg/kg/day, preferably
CA 02611540 2007-11-09
0.5 - 3 mg/kg/day, in a glycosylated albumin amount for an
adult. This amount is contained in a solution (about 5 - about
ml), and slowly administered by an intravenous injection or
drip intravenous administration.
Albumin (particularly HSA) per se can be used as a
pharmaceutical agent, for example, mainly for the purpose of
rapidly extending plasma during shock, supplementing the
amount of circulating blood, improving hypoproteinemia,
sustaining colloid osmotic pressure and the like. As specific
lo efficacy-effect, it is effective for hypoalbuminemia due to
loss of albumin (burn, nephrosis syndrome etc.) and
suppression of albumin synthesis (hepatic cirrhosis etc.),
hemorrhagic shock and the like. Accordingly, the glycosylated
albumin of the present invention can also be used as a
pharmaceutical agent for improving such disease and condition.
Also in this case, albumin can be processed into an injectable
preparation in the same manner as above.
While the dose of albumin preparation varies depending on
the administration route, severity of disease, animal species
to be the subject of administration, and drug acceptability,
body weight, age and the like of the administration subject,
it is generally 20 - 25 ml of HSA 25% solution (5 - 12.5 g as
HSA) for a single dose to an adult, which is gradually given
by intravenous injection or intravenous drip infusion.
While the present invention is explained in detail in the
following by referring to Examples, the present invention is
not limited by these.
EXAMPLES
Example production of glycosylated albumin
(1) mutation of albumin gene
Using plasmid pPIC9 into which human serum albumin gene
was introduced (hereinafter pPIC9-HSA) as a template, and D63N
sense primer of SEQ ID NO: 5 (5'-
GAGTCAGCTGAAAATTGTAACAAATCACTTCATACCC-3') and D63N antisense
primer of SEQ ID NO: 6 (5'-
26
CA 02611540 2007-11-09
GGGTATGAAGTGATTTGTTACA1TTTTCAGCTGACTC-3') for preparation of
Asn63-linked glycosylated albumin, A320T sense primer of SEQ ID
NO: 7 (5'-GGATGTTTGCAAAAACTATACTGAGGCAAAGG-3') and A320T
antisense primer of SEQ ID NO: 8 (5'-
CCTTTGCCTCAGTATAGTTTTTGCAAACATCC-3') for preparation of Asn318-
linked glycosylated albumin, and D494N sense primer of SEQ ID
NO: 9 (5'-GCTCTGGAAGTCAATGAAACATACGTTCCC-3') and D494N
antisense primer of SEQ ID NO: 10 (5'-
GGGAACGTATGTTTCATTGACTTCCAGAGC-3') for preparation of Asn494-
/0 linked glycosylated albumin as synthetic primers, mutations of
N-linked sugar chain consensus sequences were performed
(QuikChange XL Site-Directed Mutagenesis Kit, Stratagene). As
for mutation reaction conditions, DNA was treated for 30 sec
at 95 C, after which a 12-cycle reaction of denaturation (95 C,
/5 30 sec), annealing (55 C, 1 min) and extension (68 C, 10 min)
was performed. After the reaction, the template plasmid was
digested by Dpn I, and each of obtained pPIC9-HSA(D63N),
pPIC9-HSA(A320T) and pPIC9-HSA(D494N) were transfected into
XL-10-Gold ultracompetent cells to perform transformation. The
20 transformants, which were transfected with the objective
plasmid pPIC9-HSA(D63N), pPIC9-HSA(A320T) or pPIC9-HSA(D494N),
were screened in ampicillin-added medium, and the plasmids
were purified from the obtained transformants (QIAprep Spin
Miniprep Kit, manufactured by QIAGEN). Confirmation of the
25 mutations were performed by ABI Prism 310 Genetic Analyzer
(Applied Biosystems) using D63N sequence primer of SEQ ID NO:
11 (5'-GAAAATTTCGACGCCTTGGTGTTGATTGCC-3') for pPIC9-HSA(D63N),
A320T sequence primer of SEQ ID NO: 12 (5'-
GGCGGACCTTGCCGACTATATCTGTGA-3') for pPIC9-HSA(A320T), and
30 D494N sequence primer of SEQ ID NO: 13 (5'-
GGTCTCAAGAAACCTAGGAAAAGTGGG-3') for pPIC9-HSA(D494N). Moreover,
in order to prepare human serum albumin which was bonded by
sugar chains at all three sites of Asn63, Asn318 and Asn494,
mutation of N-linked sugar chain consensus sequence was
35 performed in the same way using above-prepared pPIC9-HSA(D63N)
27
CA 02611540 2007-11-09
as a template, and A320T sense primer of SEQ ID NO: 7 and
A320T antisense primer of SEQ ID NO: 8 as synthetic primers
(QuikChange XL Site-Directed Mutagenesis Kit, Stratagene).
Using thus prepared pPIC9-HSA(D63N/A320T) as a template, and
D494N sense primer of SEQ ID NO: 9 and D494N antisense primer
of SEQ ID NO: 10 as synthetic primers, mutation was performed
(QuikChange XL Site-Directed Mutagenesis Kit, Stratagene) in
the same way to prepare pPIC9-HSA(D63N/A320T/D494N).
(2) expression of glycosylated human serum albumin
.zo Each of pPIC9-HSA(D63N), pPIC9-HSA(A320T), pPIC9-
HSA(D494N) and pPIC9-HSA(D63N/A320T/D494N) was digested with
restriction enzyme Sal I, purified by phenol extraction and
ethanol precipitation, and subsequently transformed into HIS4
gene locus of Pichia yeast (GS115 strain) by homologous
recombination using an electroporation apparatus (Gene Pulser
II Electroporation System, manufactured by BIO-RAD). The
obtained transformants were cultured in BMMY liquid medium,
and stocked in glycerol after confirmation of expression of
albumin.
(3) purification of glycosylated albumin
The transformed Pichia yeast was cultured in BMGY liquid
medium for 48 hr, and subsequently in BMMY medium for 96 hr as
adding 1% methanol every 12 hr. The yeast was separated by
centrifugation (6,000g x 10 min.), after which the culture
supernatant was dialyzed against 200 mM acetate buffer. Then,
albumin was bonded to Blue Sepharose CL-6B column
(manufactured by Amersham Biosciences), and eluted by
concentration gradient of 0 to 3 M NaCl. Subsequently, this
eluate was dialyzed against 0.65 M ammonium sulfate/100 mM
sodium phosphate buffer (pH 7.0), and passed through HiTrap
Phenyl HP column (manufactured by Amersham Biosciences), and
the nonadsorbed fraction was recovered. After that, defatting
by activated carbon was performed.
Comparative Example 1 production of nonglycosylated (wild-
type) human serum albumin
28
CA 02611540 2007-11-09
Manipulated in the same manner as in Example, except that
mutation of N-linked sugar chain consensus sequence was not
performed, nonglycosylated human serum albumin was expressed
in Pichia yeast, and human serum albumin (HSA) was obtained.
(4) Experimental Example 1
Glycosylated albumin, which was prepared in the same way
as in Example, was labeled with radioactive indium isotope
(mIn) to prepare "In-
glycosylated albumin (D63N, A320T, D494N
and D63N/A320T/D494N).
glycosylated albumin was
administered intravenously through the tail into a mouse
(dose; 1 mg/kg), blood and liver were collected at fixed
intervals after the administration, and albumin concentration
and liver transfer were measured by radiation dose measuring
equipment. As a control, human serum albumin obtained in
/5 Comparative Example 1 was labeled with mIn
("In-human serum
albumin), which was administered into a mouse, and measurement
was carried out in the same way. Proportion of glycosylated
albumin concentration in plasma and liver based on elapsed
time after the administration and dose, namely transferability
to liver (Hepatic accumulation (% of dose)), is shown in Fig.
1.
The result from Fig. 1 illustrates that glycosylated
albumin, particularly D494N and D63N/A320T/D494N, rapidly
vanishes from blood and is actively introduced to liver. Also,
from the remarkable difference of in vivo kinetics among D63N,
A320T and D494N, it is suggested that liver transferability of
glycosylated albumin be largely dependent on the binding site
of a sugar chain, in addition to the sugar density of the
molecular surface so far been proposed.
(5) Experimental Example 2
Charge states of the "In-glycosylated albumin from
Example (D63N, A320T, D494N and D63N/A320T/D494N) and the
human serum albumin from Comparative Example 1 were evaluated
using laser electrophoresis-zeta potential analyzer (LEZA-
500T). As shown in Table 1, significant difference of the
29
CA 02611540 2007-11-09
charge was not found in all of the variants prepared in this
study compared to nonglycosylated albumin (HSA). From this, it
can be said that albumin which has been subjected to
glycosylation modification by eukaryotic cell has little
difference of the charge of protein relative to the one which
has not been subjected to, and sufficiently maintains its
intrinsic properties of the protein.
On the other hand, when the liver transfer (Hepatic
accumulation (% of dose)) at 60 min was read from Fig. 1
io (Table 1), that of glycosylated albumin was 6 - 65 times
higher than that of nonglycosylated albumin (HSA). This proved
that liver transferability was enhanced while maintaining the
properties of albumin protein.
/5 Table 1
various albumin
derived from Comparative HSA -0.311 0.97
genetic Example 1
engineering Example D63N -0.304 10.77
A320T -0.300 5.97
D494N -0.298 49.31
D63N/A320T/D494N -0.302 65.51
chemical Comparative BSA -0.353 1
modification Example 2 Suc28-BSA -0.588 23
(non-patent Suc28-BSA -0.946 63
reference Suc40-BSA -1.277 57
3) Suc46-BSA -1.672 49
Suc54-BSA -1.912 47
Comparative Example 2
Referring to figures presented in non-patent reference 3,
chemically modified albumin was compared (Fig. 2, Table 1).
Table 1 presents values read from Fig. 2. The chemical
20 modification is a result of succinic acid (Suc) modification
(an imide bond with E-amino group of a Lys residue in bovine
serum albumin (BSA)), and "Sucn-BSA" represents a BSA to which
CA 02611540 2015-02-06
28931-45
n succinic acids are bonded.
From experiments using chemically modified albumin (BSA),
it has been shown that negative-charge density on molecular
surface of modified form is important for liver transfer, and
thus it has been recognized until now that the greater the
negative-charge becomes (the more the modification rate
increases), the greater the extent of recognition by the liver
becomes (see non-patent reference 3). However, the result
shows that albumin modified with as much as 20 succinic acid
molecules can acquire liver transferability.
On the other hand, while the charge of the non-modified
BSA was about -0.35, the BSAs modified by succinic acids were
not less than -0.5. Therefore, it is presumable that
chemically modified albumin is heavily affected in its protein
/5 structure and function by the charge change on its molecular
surface.
The glycosylated albumin of the present invention can be
used as a drug carrier for DDS targeting liver nonparenchymal
cells, particularly kupffer's cells. Also, utilizing gene
recombinant proteins and host's glycosylation modification
mechanism, uniform proteins can be produced compared to in
chemical modification methods, and modification operations can
be omitted. Furthermore, there is no risk of contamination of
virus and the like, so it can be safely administered to living
organisms for medical purposes.
While the present invention has been described with
emphasis on preferred embodiments, it is obvious to those
skilled in the art that the preferred embodiments can be
modified. The present invention intends that the present
invention can be embodied by methods other than those described
in detail in the present specification. Accordingly, the present
invention encompasses all modifications encompassed in the
scope of the appended "CLAIMS."
31
CA 02611540 2015-02-06
28931-45
SEQUENCE LISTING IN ELECTRONIC FORM
In accordance with Section 111(1) of the Patent Rules, this
description contains a sequence listing in electronic form in ASCII
text format (file: 28931-45 Seq 17-06-14 v2.txt).
A copy of the sequence listing in electronic form is available from
the Canadian Intellectual Property Office.
The sequences in the sequence listing in electronic form are
reproduced in the following table.
SEQUENCE TABLE
<110> Nipro Corporation
<120> Sugar Chain-Containing Albumin, Production Method Thereof
and Use Thereof
<130> A7649
<140> CA 2611540
<141> 2007-11-09
<160> 13
<170> PatentIn version 3.3
<210> 1
<211> 1827
<212> DNA
<213> Artificial sequence
<220>
<223> DNA encoding mutant ESA having glycosylation site(s)
<220>
<221> CDS
<222> (1)..(1827)
<220>
<221> sig_peptide
<222> (1)..(54)
<220>
<221> mat peptide
<222> (73)..(1827)
<400> 1
atg aag tgg gta acc ttt att tcc ctt ctt ttt ctc ttt agc tcg gct 48
Met Lys Trp Val Thr Phe Ile Ser Leu Leu Phe Leu Phe Ser Ser Ala
-20 -15 -10
32
CA 02611540 2015-02-06
28931-45
=
tat tcc agg ggt gtg ttt cgt cga gat gca cac aag agt gag gtt got 96
Tyr Ser Arg Gly Val Phe Arg Arg Asp Ala His Lys Ser Glu Val Ala
-5 -1 1 5
cat cgg ttt aaa gat ttg gga gaa gaa aat ttc aaa gcc ttg gtg ttg 144
His Arg Phe Lys Asp Leu Gly Glu Glu Asn Phe Lys Ala Leu Val Leu
15 20
att gcc ttt got cag tat ctt cag cag tgt cca ttt gaa gat cat gta 192
Ile Ala Phe Ala Gln Tyr Leu Gln Gln Cys Pro Phe Glu Asp His Val
25 30 35 40
aaa tta gtg aat gaa gta act gaa ttt gca aaa aca tgt gtt got gat 240
Lys Leu Val Asn Glu Val Thr Glu Phe Ala Lys Thr Cys Val Ala Asp
45 50 55
gag tca got gaa aat tgt rac aaa tca ctt cat acc ctt ttt gga gac 288
Glu Ser Ala Glu Asn Cys Xaa Lys Ser Leu His Thr Leu Phe Gly Asp
60 65 70
aaa tta tgc aca gtt gca act ctt cgt gaa acc tat ggt gaa atg gct 336
Lys Leu Cys Thr Val Ala Thr Leu Arg Glu Thr Tyr 61y Glu Met Ala
75 80 85
gac tgc tgt gca aaa caa gaa cot gag aga aat gaa tgc ttc ttg caa 384
Asp Cys Cys Ala Lys Gln Glu Pro Glu Arg Asn Glu Cys Phe Leu Gln
90 95 100
cac aaa gat gac aac cca aac ctc coo cga ttg gtg aga cca gag gtt 432
His Lys Asp Asp Asn Pro Asn Leu Pro Arg Leu Val Arg Pro Glu Val
105 110 115 120
gat gtg atg tgC act got ttt cat gac aat gaa gag aca ttt ttg aaa 480
Asp Val Met Cys Thr Ala Phe His Asp Asn Glu Glu Thr Phe Leu Lys
125 130 135
aaa tac tta tat gaa att gcc aga aga cat cot tac ttt tat gcc cog 528
Lys Tyr Leu Tyr Glu Ile Ala Arg Arg His Pro Tyr Phe Tyr Ala Pro
140 145 150
gaa ctc ctt ttc ttt got aaa agg tat aaa got got ttt aca gaa tgt 576
Glu Leu Leu Phe Phe Ala Lys Arg Tyr Lys Ala Ala Phe Thr Glu Cys
155 160 165
=
tgc caa got got gat aaa got gcc tgc ctg ttg cca aag ctc gat gaa 624
Cys Gln Ala Ala Asp Lys Ala Ala Cys Leu Leu Pro Lys Leu Asp Glu
170 175 180
ctt cgg gat gaa ggg aag got tog tot gcc aaa cag aga ctc aag tgt 672
Lou Arg Asp Glu Gly Lys Ala Ser Ser Ala Lys Gln Arg Leu Lys Cys
185 190 195 200
gcc agt ctc caa aaa ttt gga gaa aga got ttc aaa gca tgg gca gta 720
Ala Ser Leu Gln Lys Phe Gly Glu Arg Ala Phe Lys Ala Trp Ala Val
. 205 210 215
33
CA 02611540 2015-02-06
28931-45
got cgc ctg agc cag aga ttt ccc aaa gct gag ttt gca gaa gtt tcc 768
Ala Arg Leu Ser Gin Arg Phe Pro Lys Ala Glu Phe Ala Glu Val Ser
220 225 230
aag tta gtg aca gat ctt acc aaa gtc cac acg gaa tgc tgc cat gga 816
Lys Leu Val Thr Asp Leu Thr Lys Val His Thr Glu Cys Cys His Gly
235 240 245
gat ctg ctt gaa tgt got gat gac agg gcg gac ctt gcc aag tat atc 864
Asp Leu Leu Glu Cys Ala Asp Asp Arg Ala Asp Leu Ala Lys Tyr Ile
250 255 260
tgt gaa aat caa gat tog atc too agt aaa ctg aag gaa tgc tgt gaa 912
Cys Glu Asn Gin Asp Ser Ile Ser Ser Lys Leu Lys Glu Cys Cys Glu
265 270 275 280
aaa cct ctg ttg gaa aaa too cac tgc att goo gaa gtg gaa aat gat 960
Lys Pro Leu Leu Glu Lys Ser His Cys Ile Ala Glu Val Glu Asn Asp
285 290 295
gag atg cct got gac ttg cct tca tta got got gat ttt gtt gaa agt 1008
Glu Met Pro Ala Asp Leu Pro Ser Leu Ala Ala Asp Phe Val Glu Ser
300 305 310
aag gat gtt tgc aaa aac tat rot gag gca aag gat gtc ttc ctg g9c 1056
Lys Asp Val Cys Lys Asn Tyr Xaa Glu Ala Lys Asp Val Phe Leu Gly
315 320 325
atg ttt ttg tat gaa tat gca aga agg cat cct gat tac tot gtc gtg 1104
Met Phe Leu Tyr Glu Tyr Ala Arg Arg His Pro Asp Tyr Ser Val Val
330 335 340
ctg ctg ctg aga ctt goo aag aca tat gaa acc act cta gag aag tgc 1152
Leu Leu Leu Arg Leu Ala Lys Thr Tyr Glu Thr Thr Leu Glu Lys Cys
345 350 355 360
=
tgt goo got gca gat cct cat gaa tgc tat gcc aaa gtg ttc gat gaa 1200
Cys Ala Ala Ala Asp Pro His Glu Cys Tyr Ala Lys Val Phe Asp Glu
365 370 375
ttt aaa cct ctt gtg gaa gag cct cag aat tta atc aaa caa aat tgt 1248
Phe Lys Pro Leu Val Glu Glu Pro Gin Asn Leu Ile Lys Gin Asn Cys
380 385 390
gag ctt ttt gag cag ctt gga gag tac aaa ttc cag aat gcg cta tta 1296
Glu Leu Phe Glu Gin Leu Gly Glu Tyr Lys Phe Gin Asn Ala Leu Leu
395 . 400 405
gtt cgt tac acc aag aaa gta coo caa gtg tca act cca act ctt gta 1344
Val Arg Tyr Thr Lys Lys Val Pro Gin Val Ser Thr Pro Thr Leu Val
410 415 420
gag gtc tca aga aac cta gga aaa gtg ggc ago aaa tgt tgt aaa cat 1392
Glu Val Ser Arg Asn Leu Gly Lys Val Gly Ser Lys Cys Cys Lys His
425 430 435 440
34
CA 02611540 2015-02-06
28931-45
cot gaa gca aaa aga atg ccc tgt gca gaa gac tat cta tcc gtg gtc 1440
Pro Glu Ala Lys Arg Met Pro Cys Ala Glu Asp Tyr Leu Ser Val Val
445 450 455
ctg aac cag tta tgt gtg ttg cat gag aaa acg cca gta agt gac aga 1488
Leu Asn Gln Leta Cys Val Leu His Glu Lys Thr Pro Val Ser Asp Arg
460 465 470
gtc acc aaa tgc tgc aca gaa tcc ttg gtg aac agg cga cca tgc ttt 1536
Val Thr Lys Cys Cys Thr Glu Ser Leu Val Asn Arg Arg Pro Cys Phe
475 480 485
tca got ctg gaa gtc rat gaa aca tac gtt coo aaa gag ttt aat got 1584
Ser Ala Leu Glu Val Xaa Glu Thr Tyr Val Pro Lys Glu Phe Asn Ala
490 495 500
gaa aca ttc acc ttc cat gca gat ata tgc aca ctt tot gag aag gag 1632
Glu Thr Phe Thr Phe His Ala Asp Ile Cys Thr Leu Ser Glu Lys Glu
505 510 515 520
aga caa atc aag aaa caa act gca ctt gtt gag ctc gtg aaa cac aag 1680
Arg Gln Ile Lys Lys Gln Thr Ala Leu Val Glu Leu Val Lys His Lys
525 530 535
coo aag gca aca aaa gag caa ctg aaa got gtt atg gat gat ttc gca 1728
Pro Lys Ala Thr Lys Glu Gln Leu Lys Ala Val Met Asp Asp Phe Ala
540 545 550
got ttt gta gag aag tgc tgc aag got gac gat aag gag acc tgc ttt 1776
Ala Phe Val Glu Lys Cys Cys Lys Ala Asp Asp Lys Glu Thr Cys Phe
555 560 565
gcc gag gag ggt aaa aaa ctt gtt got gca agt caa got gcc tta ggc 1824
Ala Glu Glu Gly Lys Lys Leu Val Ala Ala Ser Gln Ala Ala Leu Gly
570 575 580
tta 1827
Leu
=
585
<210> 2
<211> 609
<212> PRT
<213> Artificial sequence
<220>
<221> misc_feature
<222> (63)..(63)
<223> The 'Xaa' at location 63 stands for Asp, or Asn.
<220>
<221> misc_feature
<222> (320)..(320)
<223> The 'Xaa' at location 320 stands for Ala, or Thr. =
=
CA 02611540 2015-02-06
28931-45
<220>
<221> misc feature
<222> (494)..(494)
<223> The 'Xaa' at location 494 stands for Asp, or Asn.
<220>
<223> Synthetic Construct
<400> 2
Met Lys Trp Val Thr Phe Ile Ser Leu Leu Phe Leu Phe Ser Ser Ala
-20 -15 -10
Tyr Ser Arg Gly Val Phe Arg Arg Asp Ala His Lys Ser Glu Val Ala
-5 -1 1 5
His Arg Phe Lys Asp Leu Gly Glu Glu Asn Phe Lys Ala Leu Val Leu
15 20
Ile Ala Phe Ala Gin Tyr Leu Gin Gin Cys Pro Phe Glu Asp His Val
25 30 35 40
Lys Leu Val Asn Glu Val Thr Glu Phe Ala Lys Thr Cys Val Ala Asp
45 50 55
Glu Ser Ala Glu Asn Cys Xaa Lys Ser Leu His Thr Leu Phe Gly Asp
60 65 70
Lys Leu Cys Thr Val Ala Thr Leu Arg Glu Thr Tyr Gly Glu Met Ala
75 80 85
Asp Cys Cys Ala Lys Gin Glu Pro Glu Arg Asn Glu Cys Phe Leu Gin
90 95 100
His Lys Asp Asp Asn Pro Asn Leu Pro Arg Leu Val Arg Pro Glu Val
105 110 115 120
Asp Val Met Cys Thr Ala Phe His Asp Asn Glu Glu Thr Phe Leu Lys
125 130 135
Lys Tyr Leu Tyr Glu Ile Ala Arg Arg His Pro Tyr Phe Tyr Ala Pro
140 145 150
Glu Leu Leu Phe Phe Ala Lys Arg Tyr Lys Ala Ala Phe Thr Glu Cys
155 160 165
Cys Gin Ala Ala Asp Lys Ala Ala Cys Lou Leu Pro Lys Leu Asp Glu
170 175 180
Leu Arg Asp Glu Gly Lys Ala Ser Ser Ala Lys Gin Arg Leu Lys Cys
185 190 195 200
Ala Ser Leu Gin Lys Phe Gly Glu Arg Ala Phe Lys Ala Trp Ala Val
205 210 215
=
Ala Arg Leu Ser Gin Arg Phe Pro Lys Ala Glu Phe Ala Glu Val Ser
220 225 230
Lys Leu Val Thr Asp Leu Thr Lys Val His Thr Glu Cys Cys His Gly
235 240 245
Asp Leu Leu Glu Cys Ala Asp Asp Arg Ala Asp Leu Ala Lys Tyr Ile
250 255 260
Cys Glu Asn Gin Asp Ser Ile Ser Ser Lys Leu Lys Glu Cys Cys Glu
265 270 275 280
Lys Pro Leu Leu Glu Lys Ser His Cys Ile Ala Glu Val Glu Asn Asp
285 290 295
Glu Met Pro Ala Asp Leu Pro Ser Leu Ala Ala Asp Phe Val Glu Ser
300 305 310
Lys Asp Val Cys Lys Asn Tyr Xaa Glu Ala Lys Asp Val Phe Lou Gly
315 320 325
Met Phe Lou Tyr Glu Tyr Ala Arg Arg His Pro Asp Tyr Ser Val Val
330 335 340
36
CA 02611540 2015-02-06
28931-45
Leu Leu Leu Arg Leu Ala Lys Thr Tyr Glu Thr Thr Leu Glu Lys Cys
345 350 355 360
Cys Ala Ala Ala Asp Pro His Glu Cys Tyr Ala Lys Val Phe Asp Glu
365 370 375 .
Phe Lys Pro Leu Val Glu Glu Pro Gln Asn Leu Ile Lys Gln Asn Cys
380 385 390
Glu Leu Phe Glu Gln Leu Gly Glu Tyr Lys Phe Gln Asn Ala Leu Leu
395 400 405
Val Arg Tyr Thr Lys Lys Val Pro Gln Val Ser Thr Pro Thr Leu Val
410 415 420
Glu Val Ser Arg Asn Leu Gly Lys Val Gly Ser Lys Cys Cys Lys His
425 430 435 440
Pro Glu Ala Lys Arg Met Pro Cys Ala Glu Asp Tyr Leu Ser Val Val
445 450 455
Leu Asn Gln Leu Cys Val Leu His Glu Lys Thr Pro Val Ser Asp 4g
460 465 470
Val Thr Lys Cys Cys Thr Glu Ser Leu Val Asn Arg Arg Pro Cys Phe
475 480 485
Ser Ala Leu Glu Val Xaa Glu Thr Tyr Val Pro Lys Glu Phe Asn Ala
490 495 500
Glu Thr Phe Thr Phe His Ala Asp Ile Cys Thr Leu Ser Glu Lys Glu
505 510 515 520
Arg Gln Ile Lys Lys Gln Thr Ala Leu Val Glu Leu Val Lys His Lys
525 530 535
Pro Lys Ala Thr Lys Glu Gln Leu Lys Ala Val Met Asp Asp Phe Ala
540 545 550
Ala Phe Val Glu Lys Cys Cys Lys Ala Asp Asp Lys Glu Thr Cys Phe
555 560 565
Ala Glu Glu Gly Lys Lys Leu Val Ala Ala Ser Gln Ala Ala Leu Gly
570 575 580
Leu
585
<210> 3
<211> 1827
<212> DNA
<213> Homo sapiens
=
<220>
<221> CDS
<222> (1)..(1827)
<220>
<221> sig_peptide
<222> (1)..(54)
<220>
<221> mat peptide
<222> (73)..(1827)
<400> 3
atg aag tgg gta acc ttt att tcc ctt ctt ttt ctc ttt agc tcg gct 48
Met Lys Trp Val Thr Phe Ile Ser Leu Leu Phe Leu Phe Ser Ser Ala
-20 -15 -10
37
CA 02611540 2015-02-06
28931-45
tat tcc agg ggt gtg ttt cgt cga gat gca cac aag agt gag gtt gct 96
Tyr Ser Arg Gly Val Phe Arg Arg Asp Ala His Lys Ser Glu Val Ala
-5 -1 1 5
cat cgg ttt aaa gat ttg gga gaa gaa aat ttc aaa gcc ttg gtg ttg 144
His Arg Phe Lys Asp Leu Gly Glu Glu Asn Phe Lys Ala Leu Val Leu
15 20
att gcc ttt gct cag tat ctt cag cag tgt cca ttt gaa gat cat gta 192
Ile Ala Phe Ala Gin Tyr Leu Gin Gin Cys Pro Phe Glu Asp His Val
25 30 35 40
aaa tta gtg aat gaa gta act gaa ttt gca aaa aca tgt gtt gct gat 240
Lys Leu Vsi Asn Glu Val Thr Glu Phe Ala Lys Thr Cys Val Ala Asp
45 50 55
gag tca gct gaa aat tgt gac aaa tca ctt cat acc ctt ttt gga gac 288
Glu Ser Ala Glu Asn Cys Asp Lys Ser Leu His Thr Leu Phe Gly Asp
60 65 70
aaa tta tgc aca gtt gca act ctt cgt gaa acc tat ggt gaa atg gct 336
Lys Leu Cys Thr Val Ala Thr Leu Arg Glu Thr Tyr Gly Glu Met Ala
75 80 85
gac tgc tgt gca aaa caa gaa cct gag aga aat gaa tgc ttc ttg caa 384
Asp Cys Cys Ala Lys Gin Glu Pro Glu Arg Asn Glu Cys Phe Leu Gin
90 95 100
cac aaa gat gac aac cca aac ctc ccc cga ttg gtg aga cca gag gtt 432
His Lys Asp Asp Asn Pro Asn Leu Pro Arg Leu Val Arg Pro Glu Val
105 110 115 120
gat gtg atg tgc act gct ttt cat gac aat gaa gag aca ttt ttg aaa 480
Asp Val Met Cys Thr Ala Phe His Asp Asn Glu Glu Thr Phe Lou Lys
125 130 135
aaa tac tta tat gaa att gcc aga aga cat cct tac ttt tat gcc cog 528
Lys Tyr Leu Tyr Glu Ile Ala Arg Arg His Pro Tyr Phe Tyr Ala Pro
140 145 150
gaa ctc ctt ttc ttt gct aaa agg tat aaa gct gct ttt aca gaa tgt 576
Glu Leu Leu Phe Phe Ala Lys Arg Tyr Lys Ala Ala Phe Thr Glu Cys
155 160 165
tgc caa gct gct gat aaa gct gcc tgc ctg ttg cca aag ctc gat gaa 624
Cys Gin Ala Ala Asp Lys Ala Ala Cys Leu Leu Pro Lys Leu Asp Glu
170 175 180
ctt cgg gat gaa ggg aag gct tog tot gcc aaa cag aga ctc aag tgt 672
Leu Arg Asp Glu Gly Lys Ala Ser Ser Ala Lys Gin Arg Leu Lys Cys
185 190 195 200
gcc agt ctc caa aaa ttt gga gaa aga gct ttc aaa gca tgg gca gta 720
Ala Ser Leu Gin Lys Phe Gly Glu Arg Ala Phe Lys Ala Trp Ala Val
205 210 215
=
38
CA 02611540 2015-02-06
28931-45 =
gct cgc ctg agc cag aga ttt ccc aaa gct gag ttt gca gaa gtt tcc 768
Ala Arg Leu Ser Gin Arg Phe Pro Lys Ala Glu Phe Ala Glu Val Ser
220 225 230
aag tta gtg aca gat ctt acc aaa gtc cac acg gaa tgc tgc cat gga 816
Lys Leu Val Thr Asp Leu Thr Lys Val His Thr Glu Cys Cys His Gly
235 240 245
gat ctg ctt gaa tgt gct gat gac agg gcg gac ctt gcc aag tat atc 864
Asp Leu Leu Glu Cys Ala Asp Asp Arg Ala Asp Leu Ala Lys Tyr Ile
250 255 260
tgt gaa aat caa gat tcg atc tcc agt aaa ctg aag gaa tgc tgt gaa 912
Cys Glu Asn Gin Asp Ser Ile Ser Ser Lys Leu Lys Glu Cys Cys Glu
265 270 275 280
aaa cct ctg ttg gaa aaa tcc cac tgc att gcc gaa gtg gaa aat gat 960
Lys Pro Leu Leu Glu Lys Ser His Cys Ile Ala Glu Val Glu Asn Asp
285 290 295
gag atg cct gct gac ttg cct tca tta gct gct gat ttt gtt gaa agt 1008
Glu Met Pro Ala Asp Leu Pro Ser Leu Ala Ala Asp Phe Val Glu Ser
300 305 310
aag gat gtt tgc aaa aac tat gct gag gca aag gat gtc ttc ctg ggc 1056
Lys Asp Val Cys Lys Asn Tyr Ala Glu Ala Lys Asp Val Phe Leu Gly
315 320 325
atg ttt ttg tat gaa tat gca aga agg cat cct gat tac tct gtc gtg 1104 .
Met Phe Leu Tyr Glu Tyr Ala Arg Arg His Pro Asp Tyr Ser Val Val
330 335 340
ctg ctg ctg aga ctt gcc aag aca tat gaa acc act cta gag aag tgc 1152
Leu Leu Leu Arg Leu Ala Lys Thr Tyr Glu Thr Thr Leu Glu Lys Cys
345 350 355 360
tgt gcc gct gca gat cct cat gaa tgc tat gcc aaa gtg ttc gat gaa 1200
Cys Ala Ala Ala Asp Pro His Glu Cys Tyr Ala Lys Val Phe Asp Glu
365 370 375
=
ttt aaa cct ctt gtg gaa gag cct cag aat tta atc aaa caa aat tgt 1248
Phe Lys Pro Leu Val Glu Glu Pro Gin Asn Leu Ile Lys Gin Asn Cys
380 385 390
gag ctt ttt gag cag ctt gga gag tac aaa ttc cag aat gcg cta tta 1296
Glu Leu Phe Glu Gin Leu Gly Glu Tyr Lys Phe Gin Asn Ala Leu Leu
395 400 405
gtt cgt tac acc aag aaa gta ccc caa gtg tca act cca act ctt gta 1344
Val Arg Tyr Thr Lys Lys Val Pro Gin Val Ser Thr Pro Thr Leu Val
410 415 420 =
gag gtc tca aga aac cta gga aaa gtg ggc agc aaa tgt tgt aaa cat 1392
Glu Val Ser Arg Asn Leu Gly Lys Val Gly Ser Lys Cys Cys Lys His
425 430 435 440
39
CA 02611540 2015-02-06
28931-45
cot gaa gca aaa aga atg ccc tgt gca gaa gac tat cta tcc gtg gtc 1440
Pro Glu Ala Lys Arg Met Pro Cys Ala Glu Asp Tyr Leu Ser Val Val
445 450 455
ctg aac cag tta tgt gtg ttg cat gag aaa acg cca gta agt gac aga 1488
Leu Asn Gln Leu Cys Val Leu His Glu Lys Thr Pro Val Ser Asp Arg
460 465 470
gtc acc aaa tgc tgc aca gaa tcc ttg gtg aac agg cga cca tgc ttt 1536
Val Thr Lys Cys Cys Thr Glu Ser Leu Val Asn Arg Arg Pro Cys Phe
475 480 485
tca got ctg gaa gtc gat gaa aca tac gtt ccc aaa gag ttt aat got 1584
Ser Ala Leu Glu Val Asp Glu Thr Tyr Val Pro Lys Glu Phe Asn Ala
490 495 500
gaa aca ttc acc ttc cat gca gat ata tgc aca ctt tot gag aag gag 1632
Glu Thr Phe Thr Phe His Ala Asp Ile Cys Thr Leu Ser Glu Lys Glu
505 510 515 520
aga caa atc aag aaa caa act gca ctt gtt gag ctc gtg aaa cac aag 1680
Arg Gln Ile Lys Lys Gln Thr Ala Leu Val Glu Leu Val Lys His Lys
. 525 530 535
ccc aag gca aca aaa gag caa ctg aaa get gtt atg gat gat ttc gca 1728
Pro Lys Ala Thr Lys Glu Gln Leu Lys Ala Val Met Asp Asp Phe Ala
540 545 550
got ttt gta gag aag tgc tgc aag got gac gat aag gag acc tgc ttt 1776
Ala Phe Val Glu Lys Cys Cys Lys Ala Asp Asp Lys Glu Thr Cys Phe
555 560 565
gcc gag gag ggt aaa aaa ctt gtt got gca agt caa got gcc tta ggc 1824
Ala Glu Glu Gly Lys Lys Leu Val Ala Ala Ser Gln Ala Ala Leu Gly
570 575 580
tta 1827
Leu
585
<210> 4
<211> 609
<212> PRT
<213> Homo sapiens
<400> 4
Met Lys Trp Val Thr Phe Ile Ser Leu Leu Phe Leu Phe Ser Ser Ala
-20 -15 -10
Tyr Ser Arg Gly Val Phe Arg Arg Asp Ala His Lys Ser Glu Val Ala
-5 -1 1 5
His Arg Phe Lys Asp Leu Gly Glu Glu Asn Phe Lys Ala Leu Val Leu
15 20
Ile Ala Phe Ala Gln Tyr Leu Gln Gln Cys Pro Phe Glu Asp His Val
25 30 35 40
CA 02611540 2015-02-06
28931-45
Lys Leu Val Asn Glu Val Thr Glu Phe Ala Lys Thr Cys Val Ala Asp
45 50 55
Glu Ser Ala Glu Asn Cys Asp Lys Ser Leu His Thr Leu Phe Gly Asp
60. 65 70
Lys Lou Cys Thr Val Ala Thr Leu Arg Glu Thr Tyr Gly Glu Met Ala
75 80 85
Asp Cys Cys Ala Lys Gin Glu Pro Glu Arg Asn Glu Cys Phe Leu Gln
90 95 100
His Lys Asp Asp Asn Pro Asn Leu Pro Arg Leu Val Arg Pro Glu Val
105 110 115 120
Asp Val Met Cys Thr Ala Phe His Asp Asn Glu Glu Thr Phe Leu Lys
125 130 135
Lys Tyr Leu Tyr Glu Ile Ala Arg Arg His Pro Tyr Phe Tyr Ala Pro
140 145 150
Glu Leu Leu Phe Phe Ala Lys Arg Tyr Lys Ala Ala Phe Thr Glu Cys
155 160 165
Cys Gin Ala Ala Asp Lys Ala Ala Cys Leu Leu Pro Lys Leu Asp Glu
170 175 180
Leu Arg Asp Glu Gly Lys Ala Ser Ser Ala Lys Gin Arg Leu Lys Cys
185 190 195 200
Ala Ser Lou Gin Lys Phe Gly Glu Arg Ala Phe Lys Ala Trp Ala Val
205 210 215
Ala Arg Leu Ser Gin Arg Phe Pro Lys Ala Glu Phe Ala Glu Val Ser
220 225 230
Lys Leu Val Thr Asp Leu Thr Lys Val His Thr Glu Cys Cys His Gly
235 . 240 245
Asp Leu Lou Glu Cys Ala Asp Asp Arg Ala Asp Leu Ala Lys Tyr Ile
250 255 260
Cys Glu Asn Gin Asp Ser Ile Ser Ser Lys Leu Lys Glu Cys Cys Glu
265 270 275 280
Lys Pro Leu Leu Glu Lys Ser His Cys Ile Ala Glu Val Glu Asn Asp
285 290 295
Glu Met Pro Ala Asp Leu Pro Ser Lou Ala Ala Asp Phe Val Glu Ser
300 305 310 =
Lys Asp Val Cys Lys Asn Tyr Ala Glu Ala Lys Asp Val Phe Leu Gly
315 320 325
Met Phe Leu Tyr Glu Tyr Ala Arg Arg His Pro Asp Tyr Ser Val Val
330 335 340
Leu Leu Lou Arg Leu Ala Lys Thr Tyr Glu Thr Thr Leu Glu Lys Cys
345 350 355 360
Cys Ala Ala Ala Asp Pro His Glu Cys Tyr Ala Lys Val Phe Asp Glu
365 370 375
Phe Lys Pro Leu Val Glu Glu Pro Gin Asn Leu Ile Lys Gin Asn Cys
380 385 390
Glu Leu Phe Glu Gin Lou Gly Glu Tyr Lys Phe Gin Asn Ala Leu Lou
395 400 405
Val Arg Tyr Thr Lys Lys Val Pro Gin Val Ser Thr Pro Thr Leu Val
410 415 420
Glu Val Ser Arg Asn Leu Gly Lys Val Gly Ser Lys Cys Cys Lys His
425 430 435 440
Pro Glu Ala Lys Arg Met Pro Cys Ala Glu Asp Tyr Leu Ser Val Val
445 450 455
Leu Asn Gin Leu Cys Val Leu His Glu Lys Thr Pro Val Ser Asp Arg
460 465 470
Val Thr Lys Cys Cys Thr Glu Ser Leu Val Asn Arg Arg Pro Cys Phe
475 480 485
41
CA 02611540 2015-02-06
28931-45
Ser Ala Leu Glu Val Asp Glu Thr Tyr Val Pro Lys Glu Phe Asn Ala
490 495 500
Glu Thr Phe Thr Phe His Ala Asp Ile Cys Thr Leu Ser Glu Lys Glu
505 519 515 520
Arg Gln Ile Lys Lys Gln Thr Ala Leu Val Glu Leu Val Lys His Lys
525 530 535
Pro Lys Ala Thr Lys Glu Gln Leu Lys Ala Val Met Asp Asp Phe Ala
540 545 550
Ala Phe Val Glu Lys Cys Cys Lys Ala Asp Asp Lys Glu Thr Cys Phe
555 560 565
Ala Glu Glu Gly Lys Lys Leu Val Ala Ala Ser Gln Ala Ala Leu Gly
570 575 580
Leu
585
=
<210> 5
<211> 37
<212> DNA
<213> Artificial sequence
<220>
<223> Primer
<400> 5
gagtcagctg aaaattgtaa caaatcactt cataccc 37
<210> 6
<211> 37
=
<212> DNA
<213> Artificial sequence
<220>
<223> Primer
<400> 6
gggtatgaag tgatttgtta caattttcag ctgactc 37
<210> 7
<211> 32
<212> DNA
<213> Artificial sequence
<220>
<223> Primer
<400> 7
ggatgtttgc aaaaactata ctgaggcaaa gg 32
<210> 8
<211> 32
<212> DNA
<213> Artificial sequence
42
CA 02611540 2015-02-06
28931-45
<220>
<223> Primer
<400> 8
cctttgcctc agtatagttt ttgcaaacat cc 32
<210> 9
<211> 30
<212> DNA
<213> Artificial sequence
=
<220>
<223> Primer
<400> 9
gctctggaag tcaatgaaac atacgttccc 30
<210> 10
<211> 30
<212> DNA
<213> Artificial sequence
<220>
<223> Primer
<400> 10
gggaacgtat gtttcattga cttccagagc 30
<210> 11
<211> 30
<212> DNA
<213> Artificial sequence
<220>
<223> Primer
<400> 11
gaaaatttcg acgccttggt gttgattgcc 30
<210> 12
<211> 27
<212> DNA
<213> Artificial sequence
<220>
<223> Primer
<400> 12
ggcggacctt gccgactata tctgtga 27
43
CA 02611540 2015-02-06
28931-45
<21o> 13
<211> 27
<212> DNA
<213> Artificial sequence
<220>
<223> Primer
<400> 13
ggtctcaaga,aacctaggaa aagtggg 27
44