Note: Descriptions are shown in the official language in which they were submitted.
CA 02616144 2008-01-25
DEMANDES OU BREVETS VOLUMINEUX
LA PRESENTE PARTIE DE CETTE DENiANDE OU CE BREVETS
COMPREND PLUS D'UN TOME.
CECI EST LE TOME 1 DE 2
NOTE: Pour les tomes additionels, veillea contacter le Bureau Canadien des
Brevets.
JUMBO APPLICATIONS / PATENTS
THIS SECTION OF THE APPLICATION / PATENT CONTAINS MORE
THAN ONE VOLUME.
THIS IS VOLUME 1 OF 2
NOTE: For additional volumes please contact the Canadian Patent Office.
CA 02616144 2008-01-25
1
INSECT OLFACTORY RECEPTORS AND LIGANDS THEREFOR
FIELD OF THE INVENTION
The present invention relates to insect olfactory receptors, as well as
functional
variants and mutants thereof. The invention also relates to polynucleotides
encoding
these olfactory receptors, as well as vectors comprising said polynucleotides.
Furthermore, the present invention relates to methods of identifying odorant
ligands,
methods of identifying compounds that modulate receptor activity, as well as
biosensors comprising said receptors.
BACKGROUND OF THE INVENTION
The detection and location of mating partners and host-plants are critical
insect
behaviors that depend in part on the sense of olfaction. Odorant receptors
(Or) form a
large and diverse family primarily responsible for the molecular recognition
of
olfactory stimuli in the insect's environment. In insects they are expressed
on the
surface of dendrites that extend into a lymph filled interior of sensory hairs
or sensilla.
Buck and Axel (1991) first identified the vertebrate odorant receptors as
members of a
large family of G-protein coupled receptors (GPCR), but because of extreme
sequence
divergence, insect Ors were not discovered until the Drosophila melanogaster
genome was sequenced (Clyne et al., 1999; Gao & Chess, 1999; Vosshall et al.,
1999;
Robertson et al., 2003). Much progress has since been made in our
understanding of
the molecular and neurological mechanisms of insect olfaction, primarily in D.
melanogaster, including the principle that each sensory neuron, characterized
by a
single type of receptor, projects to a single glomerulus (with some notable
exceptions,
for example, see recent reviews on the subject by Rutzler and Zwiebel (2005)
and
Hallem et al., (2006). Using this information it is now possible to interfere
with the
olfactory system by targeting Ors (Larsson et al., 2004), ultimately to
disrupt pest
behaviors for the purpose of developing new insect control technologies. For
example, a mosquito Or that responds to components of human sweat has been
identified (Hallem et al., 2004) and efforts are underway to develop molecular
approaches that alter the olfactory-mediated host seeking behaviors of insect
disease
vectors (http://www.gcgh.org/subcontent.aspx?SecID=389). Despite the progress
being made with dipteran Ors, few odorant receptors have been identified from
the
Lepidoptera, a taxonomic group that includes numerous pest species causing
substantial economic losses to global agriculture and forest production
systems.
The silkworm Bombyx mori is one model for insect olfactory research,
particularly the male moth's legendary sensitivity and selectivity for female
sex
pheromone (Schneider, 1992). Specialization of the male olfactory system
(sexual
CA 02616144 2008-01-25
2
dimorphism) contributes to the male moth's ability to detect minute amounts of
the
female pheromone. While several different types of sensilla are found on male
silkworm antennae (Pophof, 1997), the pheromone sensitive trichoid sensilla
are the
most numerous, with approximately 17,000 on a single male antenna (Heinbockel
and
Kaissling, 1996). Each trichoid sensillum on the male antenna houses two
sensory
neurons tuned to one of the two main pheromone components, bombykol or
bombykal
(Heinbockel and Kaissling, 1996). The large number of pheromone sensitive
neurons
project to enlarged glomeruli (termed the macroglomerular complex) found on
male
(but not female) antennal lobes (reviewed in Hildebrand and Shepherd, 1997; Ai
and
Kanzaki, 2004).
The sexual dimorphism of male moth olfactory systems was used to identify
the first putative pheromone Or genes. The genome sequence of B. mori (Mita et
al.,
2004; Xia et al., 2004), the only lepidopteran genome sequence publicly
available to
date, facilitated the discovery of six Ors that were identified by their amino
acid
similarity with published dipteran Or sequences. Based upon their high levels
of gene
expression in male compared with female antennae they were considered to be
putative pheromone receptors (Sakurai et al., 2004; Krieger et al., 2005;
Nakagawa et
al., 2005). Subsequently it has been demonstrated that BmOrl responds to
bombykol
and bombykal, while BmOr3 selectively responds to bombykal (Sakurai et al.,
2004;
Nakagawa et al., 2005; Grosse-Wilde et al., 2006). Similarly, a total of 18
Heliothis
virescens Ors have been identified from privately owned sequences, including
five
that are expressed at higher levels in male compared with female antennae
(Krieger et
al., 2002; 2004).
The olfactory mechanisms underlying host-plant seeking and selection by
female moths have received less attention, and the female olfactory system is
often
considered to be adapted to detecting general olfactory stimuli. However,
female
silkworm antennae, like males, have large numbers of trichoid sensilla,
approximately
6,000 per antennae (Heinbockel and Kaissling, 1996). One of the two sensory
neurons within the most common female trichoid sensilla responds most strongly
to
benzoic acid (referred to as the benzoic acid cell) while the other responds
to 2,6-
dimethyl-5-hepten-2-ol and linalool (referred to as the terpene cell)
(Priesner, 1979;
Heinbockel and Kaissling, 1996). Insects may use terpenoid odorants to detect
and
discriminate host-plants but the behavioral relevance of benzoic acid
sensitivity in
female silkworm antennae remains unknown (Heinbockel and Kaissling, 1996). In
addition, olfactory neurons from the female trichoid sensilla project to two
lateral
large glomeruli (LLG) that are much larger in the female silkworm antennal
lobe
(Koontz and Schneider, 1987). The antennal lobes of Manduca sexta moths also
have
two large sexually dimorphic glomerui (King et al., 2000). Interestingly, the
lateral
CA 02616144 2008-01-25
3
large female glomerulus (latLFG) of M. sexta responds preferentially to
antennal
stimulation with linalool, particularly the positive enantiomer (Rospars and
Hildebrand, 2000; Reisenman et al., 2004).
There is a need to increase the representation of Ors from the Lepidoptera by
determining additional Ors from the silkworm genome. Furthermore, there is a
need
to identify Or genes that are expressed predominantly in female compared to
male
antennae, a step towards identifying female-specific olfactory pathways that
can
mediate important pest behaviors, such as host-plant seeking and selection for
oviposition.
SUMMARY OF THE INVENTION
The present inventors have identified 41 new olfactory receptors.
Thus, in a first aspect the present invention provides a substantially
purified
and/or recombinant polypeptide comprising amino acids having a sequence as
provided in any one of SEQ ID NOs: 1 to 41, a biologically active fragment of
any
one thereof, or an amino acid sequence which is at least 50% identical to any
one or
more of SEQ ID NOs: I to 41, wherein the polypeptide is an olfactory receptor.
The present inventors have identified olfactory receptors that are highly
expressed in female insects. Accordingly, in a further aspect the present
invention
provides a substantially purified and/or recombinant polypeptide comprising
amino
acids having a sequence as provided in any one of SEQ ID NOs: 12, 23, 38, 39,
40
and 41, a biologically active fragment of any one thereof, or an amino acid
sequence
which is at least 50% identical to any one or more of SEQ ID NOs: 12, 23, 38,
39, 40
and 41, wherein the polypeptide is a female specific olfactory receptor.
Preferably, the polypeptide comprises an amino acid sequence which is at least
90% identical to any one or more of SEQ ID NOs: 12, 23, 38, 39, 40 and 41.
Preferably, the polypeptide can be purified from a Lepidopteran.
In an embodiment, the polypeptide is fused to at least one other polypeptide.
The at least one other polypeptide may be, for example, a polypeptide that
enhances the stability of a polypeptide of the present invention, a
polypeptide that
assists in the purification of the fusion protein, or a label that assists in
the detection of
intracellular signalling by the receptor upon ligand binding.
In a further embodiment, the present invention provides an isolated and/or
exogenous polynucleotide comprising nucleotides having a sequence as provided
in or
complementary to any one of SEQ ID NOs: 42 to 82, a sequence which is at least
50% identical to any one or more of SEQ ID NOs: 42 to 82, a sequence which
hybridizes to any one or more of SEQ ID NOs: 42 to 82, or a sequence which
encodes
a polypeptide of the invention.
CA 02616144 2008-01-25
4
Preferably, the polynucleotide comprises nucleotides having a sequence which
hybridizes to one or more of SEQ ID NOs: 42 to 82 under stringent conditions.
More
preferably, the polynucleotide comprises nucleotides having a sequence which
hybridizes to one or more of SEQ ID NOs: 53, 64, 79, 80, 81 and 82 under
stringent
conditions.
Preferably, the polynucleotide encodes an olfactory receptor.
In an embodiment, the polynucleotide encodes a female specific olfactory
receptor.
Also provided is an oligonucleotide which comprises at least 19 contiguous
nucleotides of a polynucleotide of the invention.
In a further aspect, the present invention provides an isolated and/or
exogenous
polynucleotide which, when present in a cell of an insect, interferes with
chemosensory perception of the insect when compared to a cell of an insect
that lacks
said polynucleotide, wherein the polynucleotide comprises nucleotides having a
sequence as provided in or complementary to any one of SEQ ID NOs: 42 to 82, a
sequence which is at least 50% identical to any one or more of SEQ ID NOs: 42
to 82,
a sequence which hybridizes to any one or more of SEQ ID NOs: 42 to 82, a
sequence
which encodes a polypeptide of the invention, and/or which comprises an
oligonucleotide of the invention.
Examples of such polynucleotide include, but are not limited to, an antisense
polynucleotide, a catalytic polynucleotide and a double stranded RNA.
In an embodiment, the polynucleotide is a catalytic polynucleotide capable of
cleaving a polynucleotide of the invention.
In another embodiment, the polynucleotide is a double stranded RNA
(dsRNA) molecule comprising an oligonucleotide of the invention, wherein the
portion of the molecule that is double stranded is at least 19 basepairs in
length and
comprises said oligonucleotide.
In a further aspect, the present invention provides a vector comprising or
encoding the polynucleotide of the invention.
Preferably, the polynucleotide, or sequence encoding the polynucleotide, is
operably linked to a promoter.
In another aspect, the present invention provides a host cell comprising at
least
one polynucleotide of the invention, and/or at least one vector of the
invention.
In yet another aspect, the present invention provides a process for preparing
a
polypeptide of the invention, the process comprising cultivating a host cell
of the
invention encoding said polypeptide, or a vector of the invention encoding
said
polypeptide, under conditions which allow expression of the polynucleotide
encoding
the polypeptide, and recovering the expressed polypeptide.
CA 02616144 2008-01-25
Also provided is an antibody which specifically binds a polypeptide of the
invention.
In yet another aspect, the present invention provides a composition comprising
a polypeptide of the invention, a polynucleotide of the invention, a vector of
the
5 invention, a host of the invention and/or an antibody of the invention, and
one or more
acceptable carriers.
In another aspect, the present invention provides a kit comprising a
polypeptide of the invention, a polynucleotide of the invention, a vector of
the
invention, a host of the invention, an antibody of the invention, and/or a
composition
of the invention.
In another aspect, the present invention provides a method of identifying a
molecule that binds to a polypeptide of the invention, the method comprising:
i) contacting a polypeptide of the invention with a candidate compound,
ii) determining whether the compound binds the polypeptide.
In a further aspect, the present invention provides a method of identifying a
molecule that binds to a polypeptide of the invention, the method comprising:
a) exposing a polypeptide of the invention to a binding partner which binds
the
polypeptide, and a candidate agent, and
b) assessing the ability of the candidate agent to compete with the binding
partner for binding to the polypeptide.
In one embodiment, the polypeptide comprises amino acids having a sequence
as provided in SEQ ID NO: 12, an amino acid sequence which is at least 50%
identical
to SEQ ID NO:12 or a biologically active fragment thereof, wherein the binding
partner is linalool.
In another embodiment, the polypeptide comprises amino acids having a
sequence as provided in SEQ ID NO:38, an amino acid sequence which is at least
50% identical to SEQ ID NO:38 or a biologically active fragment thereof,
wherein the
binding partner is 2-phenylethanol, benzoic acid, benzaldehyde, ethyl benzoate
and/or
methyl benzoate.
In one embodiment, the polypeptide comprises amino acids having a sequence
as provided in SEQ ID NO:40, an amino acid sequence which is at least 50%
identical
to SEQ ID NO:40 or a biologically active fragment thereof, wherein the binding
partner is benzoic acid.
In an embodiment, the binding partner is an antibody or antigen-binding
fragment thereof.
In a further embodiment, the binding partner is detectably labelled.
In yet another aspect, the present invention provides a method of identifying
a
molecule that binds to a polypeptide of the invention, the method comprising:
CA 02616144 2008-01-25
= 6
i) contacting a protein complex comprising a polypeptide of the invention with
a candidate compound,
ii) determining whether the compound binds the complex.
Preferably, the polypeptide is expressed in a cell.
Preferably, the polypeptide spans the cell membrane.
In an embodiment, the cell is an insect cell. Preferably, the insect cell is
an
olfactory receptor neuron. Preferably, the insect cell is a Lepidopteran cell.
The cell may be in vitro or in vivo.
In a further aspect, the present invention provides a method of identifying a
molecule that modulates the activity of a polypeptide of the invention, the
method
comprising:
i) contacting a cell comprising a polypeptide of the invention with a
candidate
compound,
ii) determining whether the compound modulates a physiologic activity of the
cell.
In a further aspect, the present invention provides a method of identifying a
molecule that modulates the activity of a polypeptide of the invention, the
method
comprising:
i) contacting a first cell comprising a polypeptide of the invention with a
candidate compound,
ii) contacting a second cell lacking the polypeptide with the candidate
compound, and
iii) determining whether the compound modulates a physiologic activity in the
first or second cell,
wherein the first and second cell are the same cell type, and wherein a
compound that
modulates a physiologic activity in the first cell but not the second cell is
a modulator
of the polypeptide.
Preferably, the cell is a cell of an organism.
Preferably, the first cell and second cell are cells of the same cell type
from
two different individuals of an organism of the same species.
Preferably, the organism is a Lepidopteran.
In an embodiment, the physiologic activity is determined by analysing a
behavioural activity of the organism.
In another embodiment, the physiologic activity is G-protein activity.
Preferably, G-protein activity is determined by measuring calcium ion and/or
cyclic
AMP concentration in the cell.
In a further embodiment, the physiologic activity is determined using an
electroolfactogram.
CA 02616144 2008-01-25
7
In yet another aspect, the present invention provides a method of screening
for
a compound that modulates the activity of a polypeptide of the invention, the
method
comprising using the structural coordinates of a crystal of the polypeptide to
computationally evaluate a candidate compound for its ability to bind to the
polypeptide.
Preferably, the compound is an odorant.
In one embodiment, the compound is an antagonist of the physiologic activity.
In another embodiment, the compound is an agonist of the physiologic activity.
Also provided is a compound identified using a method of the invention.
In a further aspect, the present invention provides a method for controlling
an
insect pest, the method comprising exposing the insect pest to an antagonist
of the
invention.
In another aspect, the present invention provides a method for controlling an
insect pest, the method comprising exposing the insect pest to an agonist of
the
invention.
In yet a further aspect, the present invention provides a biosensor comprising
a
polypeptide of the invention.
In another aspect, the present invention provides a transgenic non-human
animal comprising an exogenous polynucleotide, the polynucleotide encoding at
least
one polypeptide of the invention.
In a further aspect, the present invention provides a transgenic non-human
animal comprising an exogenous polynucleotide of the invention, and/or a
polynucleotide encoding therefor.
In yet another aspect, the present invention provides a transgenic plant
comprising an exogenous polynucleotide, the polynucleotide encoding at least
one
polypeptide of the invention.
In a further aspect, the present invention provides a transgenic plant
comprising an exogenous polynucleotide of the invention, and/or a
polynucleotide
encoding therefor.
In yet another aspect, the present invention provides a method for controlling
an insect pest, the method comprising delivering to the insect a
polynucleotide of the
invention, and/or a polynucleotide encoding therefor.
In an embodiment, the polynucleotide is delivered by exposing the insect to a
transgenic plant of the invention, wherein the insect eats the plant.
In a further aspect, the present invention provides a method of controlling
female insect pests, the method comprising exposing the female insect pests to
a
ligand which binds a polypeptide comprising amino acids having a sequence as
provided in any one of SEQ ID NOs: 12, 23, 38, 39, 40 and 41, a biologically
active
CA 02616144 2008-01-25
8
fragment of any one thereof, or an amino acid sequence which is at least 50%
identical to any one or more of SEQ ID NOs: 12, 23, 38, 39, 40 and 41.
In an embodiment, the ligand disrupts mating.
Examples of such ligands include, but are not limited to, linalool, 2-
phenylethanol, benzoic acid, benzaldehyde, ethyl benzoate and methyl benzoate.
In a further embodiment, the insect pest is a Lepidopteran.
As will be apparent, preferred features and characteristics of one aspect of
the
invention are applicable to many other aspects of the invention.
Throughout this specification the word "comprise", or variations such as
"comprises" or "comprising", will be understood to imply the inclusion of a
stated
element, integer or step, or group of elements, integers or steps, but not the
exclusion
of any other element, integer or step, or group of elements, integers or
steps.
The invention is hereinafter described by way of the following non-limiting
Examples and with reference to the accompanying figures.
BRIEF DESCRIPTION OF THE ACCOMPANYING DRAWINGS
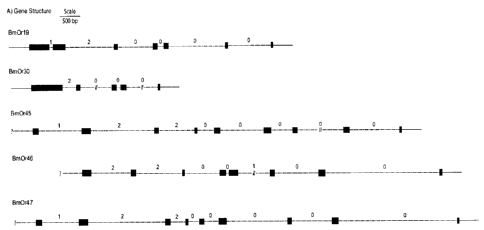
Figure 1 A) Gene structure of five female-biased silkworm Ors, BmOr19, 30 and
45-
47. Each possesses the typical short last exon with a phase 0 intron/exon
boundary.
Solid blocks represent exons, dashed lines denote intron sequence, a break in
the line
indicates that the sequence was not contiguous. The phase of each intron/exon
boundary is indicated numerically above each intron. B) Protein alignment of
five
female-biased silkworm Ors with related Ors from Drosophila melanogaster
(DmOr46a, AAF58834.3) and Anopheles gambiae (AgOrl, AAL35506). The greatest
amino acid similarity occurs at the C-terminus of the protein sequence,
particularly
the typical Ser (sometimes Thr) residue in the seventh codon position of the
last exon.
Figure 2. Neighbor-joining (corrected distance) phylogenetic tree of the
Bombyx mori
and Heliothis virescens Ors identified to date, rooted with lepidopteran
members of
the D. melanogaster Or83b lineage, BmOr2, HvOr2 and Helicoverpa zea Or2
(AAX14773). Krieger et al., (2004) reported 18 Or and 3 Gr H. virescens genes
that
were collectively referred to as chemoreceptors (Crs). Accession numbers for
the
silkworm Ors are listed in Table 2. Percentage bootstrap support (1000
replicates) is
indicated at significant branch points. With the exception of BmOr9 and HvOr6,
all
members of the pheromone receptor subfamily are expresses at higher ratios in
male
compared to female adult moth antennae (Table 3, Figure 3, Sakurai et al.,
2004;
Krieger et al., 2005; Nakagawa et al., 2005).
CA 02616144 2008-01-25
., ~ 9
Figure 3. Silkworm Or gene expression levels in female and male moth antennae
determined by quantitative real-time PCR. Expression levels relative to the
control
gene BmRPS3 were calculated using the equation 2- CT (Livak and Schmittgen,
2001)
and reported on a loglo scale. Abdomen tissue that lacks olfactory sensilla
was
included as a control reference and values were rounded up to a minimum of 10-
4
relative to BmRPS3 for presentation. Ors whose expression was not detected in
adult
moth antennae (BmOr2O, 21, 22, 25 and 42) are not included in the figure.
Figure 4. Silkworm Or gene expression levels in the antennae of individual
moths
determined by quantitative real-time PCR, n = four females and four males.
Expression levels relative to the control gene BmRPS3 were calculated using
the
equation 2" CT (Livak and Schmittgen, 2001) and reported on a loglo scale,
values
were rounded up to a minimum of 10-4 relative to BmRPS3 for presentation.
Statistical significance of differences in expression levels (CT value)
between female
and male antennae was analyzed for each Or gene by nested Analysis of Variance
(three technical replicates nested within each biological replicate).
Asterisks indicate
statistically significant differences, *** indicates p < 0.001 and **
indicates p < 0.01.
Bars on each column represent the lower and upper 95% confidence interval for
the
mean expression value.
Figure 5. Activation of BmOrl9 signaling by linalool.
Figure 6. Activation of BmOr45 signaling by 2-phenylethanol.
Figure 7. Activation of BmOr45 signaling by benzaldehyde.
Figure 8. Activation of BmOr45 signaling by benzoic acid.
Figure 9. Activation of BmOr47 signaling by benzoic acid.
KEY TO THE SEQUENCE LISTING
SEQ ID NO:1 - Polypeptide sequence of BmOr8.
SEQ ID NO:2 - Partial polypeptide sequence of BmOr9.
SEQ ID NO:3 - Polypeptide sequence of BmOr10.
SEQ ID NO:4 - Polypeptide sequence of BmOrl 1.
SEQ ID NO:5 - Polypeptide sequence of BmOrl2.
SEQ ID NO:6 - Partial polypeptide sequence of BmOrl3.
SEQ ID NO:7 - Polypeptide sequence of BmOr14.
CA 02616144 2008-01-25
SEQ ID NO:8 - Polypeptide sequence of BmOr15.
SEQ ID NO:9 - Polypeptide sequence of BmOr16.
SEQ ID NO: 10 - Polypeptide sequence of BmOrl7.
SEQ ID NO:11 - Partial polypeptide sequence of BmOr18.
5 SEQ ID NO:12 - Polypeptide sequence of BmOr19.
SEQ ID NO: 13 - Polypeptide sequence of BmOr20.
SEQ ID NO:14 - Polypeptide sequence of BmOr21.
SEQ ID NO: 15 - Polypeptide sequence of BmOr22.
SEQ ID NO: 16 - Polypeptide sequence of BmOr23.
10 SEQ ID NO: 17 - Polypeptide sequence of BmOr24.
SEQ ID NO: 18 - Polypeptide sequence of BmOr25.
SEQ ID NO: 19 - Polypeptide sequence of BmOr26.
SEQ ID NO:20 - Polypeptide sequence of BmOr27.
SEQ ID NO:21 - Polypeptide sequence of BmOr28.
SEQ ID NO:22 - Polypeptide sequence of BmOr29.
SEQ ID NO:23 - Polypeptide sequence of BmOr30.
SEQ ID NO:24 - Polypeptide sequence of BmOr3 1.
SEQ ID NO:25 - Partial polypeptide sequence of BmOr32.
SEQ ID NO:26 - Polypeptide sequence of BmOr33.
SEQ ID NO:27 - Polypeptide sequence of BmOr34.
SEQ ID NO:28 - Polypeptide sequence of BmOr35.
SEQ ID NO:29 - Polypeptide sequence of BmOr36.
SEQ ID NO:30 - Polypeptide sequence of BmOr37.
SEQ ID NO:31 - Polypeptide sequence of BmOr38.
SEQ ID NO:32 - Polypeptide sequence of BmOr39.
SEQ ID NO:33 - Partial polypeptide sequence of BmOr4O.
SEQ ID NO:34 - Polypeptide sequence of BmOr4l.
SEQ ID NO:35 - Polypeptide sequence of BmOr42.
SEQ ID NO:36 - Partial polypeptide sequence of BmOr43.
SEQ ID NO:37 - Partial polypeptide sequence of BmOr44.
SEQ ID NO:38 - Polypeptide sequence of BmOr45.
SEQ ID NO:39 - Partial polypeptide sequence of BmOr46.
SEQ ID NO:40 - Polypeptide sequence of BmOr47.
SEQ ID NO:41 - Partial polypeptide sequence of BmOr48.
SEQ ID NO:42 - Nucleotide sequence encoding BmOr8.
SEQ ID NO:43 - Nucleotide sequence encoding BmOr9.
SEQ ID NO:44 - Nucleotide sequence encoding BmOr10.
SEQ ID NO:45 - Nucleotide sequence encoding BmOrl 1.
CA 02616144 2008-01-25
11
SEQ ID NO:46 - Nucleotide sequence encoding BmOr12.
SEQ ID NO:47 -Nucleotide sequence encoding BmOr13.
SEQ ID NO:48 - Nucleotide sequence encoding BmOrl4.
SEQ ID NO:49 - Nucleotide sequence encoding BmOr15.
SEQ ID NO:50 - Nucleotide sequence encoding BmOr16.
SEQ ID NO:51 -Nucleotide sequence encoding BmOrl7.
SEQ ID NO:52 - Nucleotide sequence encoding BmOrl8.
SEQ ID NO:53 - Nucleotide sequence encoding BmOr19.
SEQ ID NO:54 - Nucleotide sequence encoding BmOr2O.
SEQ ID NO:55 - Nucleotide sequence encoding BmOr21.
SEQ ID NO:56 - Nucleotide sequence encoding BmOr22.
SEQ ID NO:57 - Nucleotide sequence encoding BmOr23.
SEQ ID NO:58 - Nucleotide sequence encoding BmOr24.
SEQ ID NO:59 - Nucleotide sequence encoding BmOr25.
SEQ ID NO:60 - Nucleotide sequence encoding BmOr26.
SEQ ID NO:61 - Nucleotide sequence encoding BmOr27.
SEQ ID NO:62 - Nucleotide sequence encoding BmOr28.
SEQ ID NO:63 - Nucleotide sequence encoding BmOr29.
SEQ ID NO:64 - Nucleotide sequence encoding BmOr3O.
SEQ ID NO:65 - Nucleotide sequence encoding BmOr3 1.
SEQ ID NO:66 - Nucleotide sequence encoding BmOr32.
SEQ ID NO:67 - Nucleotide sequence encoding BmOr33.
SEQ ID NO:68 - Nucleotide sequence encoding BmOr34.
SEQ ID NO:69 - Nucleotide sequence encoding BmOr35.
SEQ ID NO:70 - Nucleotide sequence encoding BmOr36.
SEQ ID NO:71 - Nucleotide sequence encoding BmOr37.
SEQ ID NO:72 - Nucleotide sequence encoding BmOr38.
SEQ ID NO:73 - Nucleotide sequence encoding BmOr39.
SEQ ID NO:74 - Nucleotide sequence encoding BmOr4O.
SEQ ID NO: 75 - Nucleotide sequence encoding BmOr4 1.
SEQ ID NO:76 - Nucleotide sequence encoding BmOr42.
SEQ ID NO:77 - Nucleotide sequence encoding BmOr43.
SEQ ID NO:78 - Nucleotide sequence encoding BmOr44.
SEQ ID NO:79 - Nucleotide sequence encoding BmOr45.
SEQ ID NO:80 - Nucleotide sequence encoding BmOr46.
SEQ ID NO:81 - Nucleotide sequence encoding BmOr47.
SEQ ID NO:82 - Nucleotide sequence encoding BmOr48.
SEQ ID NO's 83 to 178 - Oligonucleotide primers.
CA 02616144 2008-01-25
12
SEQ ID NO: 179 - Polypeptide sequence of DmOr46a (Genbank: AAF58834.3).
SEQ ID NO: 180 - Polypeptide sequence of AgOrl (Genbank: AAL35506).
DETAILED DESCRIPTION OF THE INVENTION
General Techniques and Definitions
Unless specifically defined otherwise, all technical and scientific terms used
herein shall be taken to have the same meaning as commonly understood by one
of
ordinary skill in the art (e.g., in cell culture, molecular genetics, receptor
biology,
biosensors, immunology, immunohistochemistry, protein chemistry, and
biochemistry).
Unless otherwise indicated, the recombinant protein, cell culture, and
immunological techniques utilized in the present invention are standard
procedures,
well known to those skilled in the art. Such techniques are described and
explained
throughout the literature in sources such as, J. Perbal, A Practical Guide to
Molecular
Cloning, John Wiley and Sons (1984), J. Sambrook et al., Molecular Cloning: A
Laboratory Manual, Cold Spring Harbour Laboratory Press (1989), T.A. Brown
(editor), Essential Molecular Biology: A Practical Approach, Volumes 1 and 2,
IRL
Press (1991), D.M. Glover and B.D. Hames (editors), DNA Cloning: A Practical
Approach, Volumes 1-4, IRL Press (1995 and 1996), and F.M. Ausubel et al.,
(editors), Current Protocols in Molecular Biology, Greene Pub. Associates and
Wiley-
Interscience (1988, including all updates until present), Ed Harlow and David
Lane
(editors) Antibodies: A Laboratory Manual, Cold Spring Harbour Laboratory,
(1988),
and J.E. Coligan et al., (editors) Current Protocols in Immunology, John Wiley
&
Sons (including all updates until present).
As used herein, the term "olfactory receptor", "insect olfactory receptor",
"odorant receptor" or "insect odorant receptor" or variations thereof when
used in
relation to the polypeptides of the invention refers to a polypeptide which,
when
present in a cell of an insect, is involved in chemosensory perception of the
insect.
Preferably, the insect is a Lepidopteran. Preferably, the cell is a neuron,
more
preferably a neuron cell in the antenna of the insect. Furthermore, the term
"olfactory
receptor" or "insect olfactory receptor" when used in relation to the
polypeptides of
the invention refers to a polypeptide which binds an odorant ligand resulting
in a
physiologic response. Furthermore, the term "olfactory receptor", "insect
olfactory
receptor", "odorant receptor" or "insect odorant receptor" or variations
thereof is used
broadly to refer to a polypeptide of the invention in isolation, or as a
complex with at
least one other protein, wherein said other protein is typically also a
receptor molecule
involved in chemosensory perception of an insect. In one embodiment, the at
least
CA 02616144 2008-01-25
13
one other protein is a different olfactory receptor sub-unit when compared to
the
polypeptides of the invention.
As used herein, the term "female specific" means that the polypeptide is
produced at a higher level in adult females than adult males. Preferably, the
female:male expression in antennae is at least 3.0, more preferably 5Ø This
term
also includes mutants of naturally occurring female specific receptors which
have
been mutated. Such mutants are at least 95%, more preferably at least 97%, and
more
preferably at least 99% identical to the naturally produced polypeptide.
"Polynucleotide" refers to a oligonucleotide, nucleic acid molecule or any
fragment thereof. It may be DNA or RNA of genomic or synthetic origin, double-
stranded or single-stranded, and combined with carbohydrate, lipids, protein,
or other
materials to perform a particular activity defined herein.
"Operably linked" as used herein refers to a functional relationship between
two or more nucleic acid (e.g., DNA) segments. Typically, it refers to the
functional
relationship of transcriptional regulatory element to a transcribed sequence.
For
example, a promoter is operably linked to a coding sequence, such as a
polynucleotide
defined herein, if it stimulates or modulates the transcription of the coding
sequence
in an appropriate host cell and/or in a cell-free expression system.
Generally,
promoter transcriptional regulatory elements that are operably linked to a
transcribed
sequence are physically contiguous to the transcribed sequence, i.e., they are
cis-
acting. However, some transcriptional regulatory elements, such as enhancers,
need
not be physically contiguous or located in close proximity to the coding
sequences
whose transcription they enhance.
Compositions of the present invention may include an "acceptable carrier".
Examples of such acceptable carriers include water, saline, Ringer's solution,
dextrose
solution, Hank's solution, and other aqueous physiologically balanced salt
solutions.
Nonaqueous vehicles, such as fixed oils, sesame oil, ethyl oleate, or
triglycerides may
also be used. The exact nature of the "acceptable carrier" will depend on the
use of
the composition. Considering the uses described herein, and the nature of the
component of the invention in the composition, the skilled person can readily
determine suitable a "acceptable carrier(s)" for a particular use.
Polypeptides
By "substantially purified polypeptide" or "purified" we mean a polypeptide
that has generally been separated from the lipids, nucleic acids, other
polypeptides,
and other contaminating molecules with which it is associated in its native
state. It is
preferred that the substantially purified polypeptide is at least 60% free,
more
CA 02616144 2008-01-25
14
preferably at least 75% free, and more preferably at least 90% free from other
components with which it is naturally associated.
The term "recombinant" in the context of a polypeptide refers to the
polypeptide when produced by a cell, or in a cell-free expression system, in
an altered
amount or at an altered rate compared to its native state. In one embodiment
the cell
is a cell that does not naturally produce the polypeptide. However, the cell
may be a
cell which comprises a non-endogenous gene that causes an altered, preferably
increased, amount of the polypeptide to be produced. A recombinant polypeptide
of
the invention includes polypeptides which have not been separated from other
components of the transgenic (recombinant) cell, or cell-free expression
system, in
which it is produced, and polypeptides produced in such cells or cell-free
systems
which are subsequently purified away from at least some other components.
The terms "polypeptide" and "protein" are generally used interchangeably and
refer to a single polypeptide chain which may or may not be modified by
addition of
non-amino acid groups. It would be understood that such polypeptide chains may
associate with other polypeptides or proteins or other molecules such as co-
factors.
The terms "proteins" and "polypeptides" as used herein also include variants,
mutants,
modifications, analogous and/or derivatives of the polypeptides of the
invention as
described herein.
The % identity of a polypeptide is determined by GAP (Needleman and
Wunsch, 1970) analysis (GCG program) with a gap creation penalty=5, and a gap
extension penalty=0.3. The query sequence is at least 25 amino acids in
length, and
the GAP analysis aligns the two sequences over a region of at least 25 amino
acids.
More preferably, the query sequence is at least 50 amino acids in length, and
the GAP
analysis aligns the two sequences over a region of at least 50 amino acids.
More
preferably, the query sequence is at least 100 amino acids in length and the
GAP
analysis aligns the two sequences over a region of at least 100 amino acids.
Even
more preferably, the query sequence is at least 250 amino acids in length and
the GAP
analysis aligns the two sequences over a region of at least 250 amino acids.
Most
preferably, the two sequences are aligned over their entire length.
As used herein a "biologically active" fragment is a portion of a polypeptide
of
the invention which maintains a defined activity of the full-length
polypeptide,
namely be able to act as an olfactory receptor. Biologically active fragments
can be
any size as long as they maintain the defined activity. Preferably,
biologically active
fragments are at least 100, more preferably at least 200, and even more
preferably at
least 350 amino acids in length.
With regard to a defined polypeptide, it will be appreciated that % identity
figures higher than those provided above will encompass preferred embodiments.
CA 02616144 2008-01-25
Thus, where applicable, in light of the minimum % identity figures, it is
preferred that
the polypeptide comprises an amino acid sequence which is at least 55%, more
preferably at least 60%, more preferably at least 65%, more preferably at
least 70%,
more preferably at least 75%, more preferably at least 80%, more preferably at
least
5 85%, more preferably at least 90%, more preferably at least 91%, more
preferably at
least 92%, more preferably at least 93%, more preferably at least 94%, more
preferably at least 95%, more preferably at least 96%, more preferably at
least 97%,
more preferably at least 98%, more preferably at least 99%, more preferably at
least
99.1%, more preferably at least 99.2%, more preferably at least 99.3%, more
10 preferably at least 99.4%, more preferably at least 99.5%, more preferably
at least
99.6%, more preferably at least 99.7%, more preferably at least 99.8%, and
even more
preferably at least 99.9% identical to the relevant nominated SEQ ID NO.
Amino acid sequence mutants of the polypeptides of the present invention can
be prepared by introducing appropriate nucleotide changes into a nucleic acid
of the
15 present invention, or by in vitro synthesis of the desired polypeptide.
Such mutants
include, for example, deletions, insertions or substitutions of residues
within the
amino acid sequence. A combination of deletion, insertion and substitution can
be
made to arrive at the final construct, provided that the final polypeptide
product
possesses the desired characteristics.
Mutant (altered) polypeptides can be prepared using any technique known in
the art. For example, a polynucleotide of the invention can be subjected to in
vitro
mutagenesis. Such in vitro mutagenesis techniques include sub-cloning the
polynucleotide into a suitable vector, transforming the vector into a
"mutator" strain
such as the E. coli XL-1 red (Stratagene) and propagating the transformed
bacteria for
a suitable number of generations. In another example, the polynucleotides of
the
invention, including genes encoding therefor, are subjected to DNA shuffling
techniques as broadly described by Harayama (1998). These DNA shuffling
techniques may include genes related to those of the present invention, such
as
homologous genes encoding olfactory receptors from many different
Lepidopterans.
Products derived from mutated/altered DNA can readily be screened using
techniques
described herein to determine if they have olfactory receptor activity.
In designing amino acid sequence mutants, the location of the mutation site
and the nature of the mutation will depend on characteristic(s) to be
modified. The
sites for mutation can be modified individually or in series, e.g., by (1)
substituting
first with conservative amino acid choices and then with more radical
selections
depending upon the results achieved, (2) deleting the target residue, or (3)
inserting
other residues adjacent to the located site.
CA 02616144 2008-01-25
16
Amino acid sequence deletions generally range from about 1 to 15 residues,
more preferably about 1 to 10 residues and typically about 1 to 5 contiguous
residues.
Substitution mutants have at least one amino acid residue in the polypeptide
molecule removed and a different residue inserted in its place. The sites of
greatest
interest for substitutional mutagenesis include sites identified as important
for
function. Other sites of interest are those in which particular residues
obtained from
various strains or species are identical (see, for example, the protein
alignments
provided herein). These positions may be important for biological activity.
These
sites, especially those falling within a sequence of at least three other
identically
conserved sites, are preferably substituted in a relatively conservative
manner. Such
conservative substitutions are shown in Table 1 under the heading of
"exemplary
substitutions".
Table 1. Exemplary substitutions
Original Exemplary
Residue Substitutions
Ala (A) val; leu; ile; gly
Arg (R) lys
Asn (N) gln; his
Asp (D) glu
Cys (C) ser
Gln (Q) asn; his
Glu (E) asp
Gly (G) pro, ala
His (H) asn; gln
Ile (I) leu; val; ala
Leu (L) ile; val; met; ala; phe
L s (K) arg
Met (M) leu; phe
Phe (F) leu; val; ala
Pro (P) gly
Ser (S) thr
Thr (T) ser
Trp (W) tyr
Tyr (Y) t ; phe
Val (V) ile; leu; met; phe; ala
CA 02616144 2008-01-25
17
In a preferred embodiment a mutant/variant polypeptide has one or two or
three or four or five conservative amino acid changes when compared to a
naturally
occurring polypeptide. Details of conservative amino acid changes are provided
in
Table 1. Sites of particular interest to alter are those which are not
conserved between
two, three or more of the polypeptides described herein. Examples of such
conserved
amino acids are provided in Figure 1B. As the skilled person would be aware,
such
minor changes can reasonably be predicted not to alter the activity of the
polypeptide
when expressed in a recombinant cell or cell free system.
Furthermore, if desired, unnatural amino acids or chemical amino acid
analogues can be introduced as a substitution or addition into the
polypeptides of the
present invention. Such amino acids include, but are not limited to, the D-
isomers of
the common amino acids, 2,4-diaminobutyric acid, a-amino isobutyric acid, 4-
aminobutyric acid, 2-aminobutyric acid, 6-amino hexanoic acid, 2-amino
isobutyric
acid, 3-amino propionic acid, ornithine, norleucine, norvaline,
hydroxyproline,
sarcosine, citrulline, homocitrulline, cysteic acid, t-butylglycine, t-
butylalanine,
phenylglycine, cyclohexylalanine, (3-alanine, fluoro-amino acids, designer
amino
acids such as (3-methyl amino acids, Ca-methyl amino acids, Na-methyl amino
acids,
and amino acid analogues in general.
Also included within the scope of the invention are polypeptides of the
present
invention which are differentially modified during or after synthesis, e.g.,
by
biotinylation, benzylation, glycosylation, acetylation, phosphorylation,
amidation,
derivatization by known protecting/blocking groups, proteolytic cleavage,
linkage to
an antibody molecule or other cellular ligand, etc. These modifications may
serve to
increase the stability and/or bioactivity of the polypeptide of the invention.
Polypeptides of the present invention can be produced in a variety of ways,
including production and recovery of natural polypeptides, production and
recovery of
recombinant polypeptides, and chemical synthesis of the polypeptides. In one
embodiment, an isolated polypeptide of the present invention is produced by
culturing
a cell capable of expressing the polypeptide under conditions effective to
produce the
polypeptide, and recovering the polypeptide. A preferred cell to culture is a
recombinant cell of the present invention. Effective culture conditions
include, but
are not limited to, effective media, bioreactor, temperature, pH and oxygen
conditions
that permit polypeptide production. An effective medium refers to any medium
in
which a cell is cultured to produce a polypeptide of the present invention.
Such
medium typically comprises an aqueous medium having assimilable carbon,
nitrogen
and phosphate sources, and appropriate salts, minerals, metals and other
nutrients,
such as vitamins. Cells of the present invention can be cultured in
conventional
CA 02616144 2008-01-25
18
fermentation bioreactors, shake flasks, test tubes, microtiter dishes, and
petri plates.
Culturing can be carried out at a temperature, pH and oxygen content
appropriate for a
recombinant cell. Such culturing conditions are within the expertise of one of
ordinary skill in the art.
Polynucleotides and Oligonucleotides
By an "isolated polynucleotide", including DNA, RNA, or a combination of
these, single or double stranded, in the sense or antisense orientation or a
combination
of both, dsRNA or otherwise, we mean a polynucleotide which is at least
partially
separated from the polynucleotide sequences with which it is associated or
linked in
its native state. Preferably, the isolated polynucleotide is at least 60%
free, preferably
at least 75% free, and most preferably at least 90% free from other components
with
which they are naturally associated. Furthermore, the term "polynucleotide" is
used
interchangeably herein with the term "nucleic acid".
The term "exogenous" in the context of a polynucleotide refers to the
polynucleotide when present in a cell, or in a cell-free expression system, in
an altered
amount compared to its native state. In one embodiment, the cell is a cell
that does
not naturally comprise the polynucleotide. However, the cell may be a cell
which
comprises a non-endogenous polynucleotide resulting in an altered, preferably
increased, amount of production of the encoded polypeptide. An exogenous
polynucleotide of the invention includes polynucleotides which have not been
separated from other components of the transgenic (recombinant) cell, or cell-
free
expression system, in which it is present, and polynucleotides produced in
such cells
or cell-free systems which are subsequently purified away from at least some
other
components. The exogenous polynucleotide (nucleic acid) can be a contiguous
stretch of nucleotides existing in nature, or comprise two or more contiguous
stretches
of nucleotides from different sources (naturally occurring and/or synthetic)
joined to
form a single polynucleotide. Typically such chimeric polynucleotides comprise
at
least an open reading frame encoding a polypeptide of the invention operably
linked
to a promoter suitable of driving transcription of the open reading frame in a
cell of
interest.
The % identity of a polynucleotide is determined by GAP (Needleman and
Wunsch, 1970) analysis (GCG program) with a gap creation penalty=5, and a gap
extension penalty=0.3. Unless stated otherwise, the query sequence is at least
45
nucleotides in length, and the GAP analysis aligns the two sequences over a
region of
at least 45 nucleotides. Preferably, the query sequence is at least 150
nucleotides in
length, and the GAP analysis aligns the two sequences over a region of at
least 150
nucleotides. More preferably, the query sequence is at least 300 nucleotides
in length
CA 02616144 2008-01-25
19
and the GAP analysis aligns the two sequences over a region of at least 300
nucleotides. Most preferably, the two sequences are aligned over their entire
length.
With regard to the defined polynucleotides, it will be appreciated that %
identity figures higher than those provided above will encompass preferred
embodiments. Thus, where applicable, in light of the minimum % identity
figures, it
is preferred that a polynucleotide of the invention comprises a sequence which
is at
least 55%, more preferably at least 60%, more preferably at least 65%, more
preferably at least 70%, more preferably at least 75%, more preferably at
least 80%,
more preferably at least 85%, more preferably at least 90%, more preferably at
least
91%, more preferably at least 92%, more preferably at least 93%, more
preferably at
least 94%, more preferably at least 95%, more preferably at least 96%, more
preferably at least 97%, more preferably at least 98%, more preferably at
least 99%,
more preferably at least 99.1%, more preferably at least 99.2%, more
preferably at
least 99.3%, more preferably at least 99.4%, more preferably at least 99.5%,
more
preferably at least 99.6%, more preferably at least 99.7%, more preferably at
least
99.8%, and even more preferably at least 99.9% identical to the relevant
nominated
SEQ ID NO.
Polynucleotides of the present invention may possess, when compared to
naturally occurring molecules, one or more mutations which are deletions,
insertions,
or substitutions of nucleotide residues. Mutants can be either naturally
occurring (that
is to say, isolated from a natural source) or synthetic (for example, by
performing site-
directed mutagenesis on the nucleic acid).
The term "stringent hybridization conditions" or "stringent conditions" and
the
like as used herein refers to parameters with which the art is familiar,
including the
variation of the hybridization temperature with length of an polynucleotide or
oligonucleotide. Nucleic acid hybridization parameters may be found in
references
which compile such methods, Sambrook, et al., (supra), and Ausubel, et al.,
(supra).
For example, stringent hybridization conditions, as used herein, can refer to
hybridization at 65 C in hybridization buffer (3.5xSSC, 0.02% Ficoll, 0.02%
polyvinyl pyrrolidone, 0.02% Bovine Serum Albumin, 2.5 mM NaH2PO4 (pH7), 0.5%
SDS, 2 mM EDTA) and washing twice in 0.2xSSC, 0.1% SDS at 65 C, with each
wash step being about 30 min. Alternatively, the nucleic acid and/or
oligonucleotides
(which may also be referred to as "primers" or "probes") hybridize to the
region of the
an insect genome of interest, such as the genome of a Lepidopteran, under
conditions
used in nucleic acid amplification techniques such as PCR.
Oligonucleotides of the present invention can be RNA, DNA, or derivatives of
either. Although the terms polynucleotide and oligonucleotide have overlapping
meaning, oligonucleotide are typically relatively short single stranded
molecules. The
CA 02616144 2008-01-25
= 20
minimum size of such oligonucleotides is the size required for the formation
of a
stable hybrid between an oligonucleotide and a complementary sequence on a
target
nucleic acid molecule. Preferably, the oligonucleotides are at least 15
nucleotides,
more preferably at least 18 nucleotides, more preferably at least 19
nucleotides, more
preferably at least 20 nucleotides, even more preferably at least 25
nucleotides in
length.
Usually, monomers of a polynucleotide or oligonucleotide are linked by
phosphodiester bonds or analogs thereof to form oligonucleotides ranging in
size from
a relatively short monomeric units, e.g., 12-18, to several hundreds of
monomeric
units. Analogs of phosphodiester linkages include: phosphorothioate,
phosphorodithioate, phosphoroselenoate, phosphorodiselenoate,
phosphoroanilothioate, phosphoranilidate, phosphoramidate.
The present invention includes oligonucleotides that can be used as, for
example, probes to identify nucleic acid molecules, or primers to produce
nucleic acid
molecules. Oligonucleotide of the present invention used as a probe are
typically
conjugated with a detectable label such as a radioisotope, an enzyme, biotin,
a
fluorescent molecule or a chemiluminescent molecule.
Probes and/or primers can be used to clone further homologues of the olfactory
receptor cDNAs described herein from other Lepidopteran species. Further
hybridization techniques known in the art can also be used to screen genomic
or
cDNA libraries for such homologues. In addition, techniques such as 5' and 3'
RACE
can be used to determine the entire open reading frame, and the corresponding
full
length protein sequence, of any partial sequences provided herein. In light of
the
present disclosure, the skilled person would find such techniques routine.
Antisense Polynucleotides
The term "antisense polynucleotide" shall be taken to mean a DNA or RNA, or
combination thereof, molecule that is complementary to at least a portion of a
specific
mRNA molecule encoding a polypeptide of the invention and capable of
interfering
with a post-transcriptional event such as mRNA translation. The use of
antisense
methods is well known in the art (see for example, G. Hartmann and S. Endres,
Manual of Antisense Methodology, Kluwer (1999)).
An antisense polynucleotide of the invention hybridises under physiological
conditions to a target polynucleotide (which is fully or partially single
stranded), and
thus are at least capable of forming a double stranded polynucleotide with
mRNA
encoding a protein, such as those proteins provided in SEQ ID NOs: I to 41,
under
normal conditions in a cell.
CA 02616144 2008-01-25
21
Antisense molecules may include sequences that correspond to the structural
genes or for sequences that effect control over the gene expression or
splicing event.
For example, the antisense sequence may correspond to the targeted coding
region of
the genes of the invention, or the 5'-untranslated region (UTR) or the 3'-UTR
or
combination of these. It may be complementary in part to intron sequences,
which
may be spliced out during or after transcription, preferably only to exon
sequences of
the target gene. In view of the generally greater divergence of the UTRs,
targeting
these regions provides greater specificity of gene inhibition.
The length of the antisense sequence should be at least 19 contiguous
nucleotides, preferably at least 50 nucleotides, and more preferably at least
100, 200,
or 500 nucleotides. The full-length sequence complementary to the entire gene
transcript may be used. The length is most preferably 100-2000 nucleotides.
The
degree of identity of the antisense sequence to the targeted transcript should
be at least
90% and more preferably 95-100%. The antisense RNA molecule may of course
comprise unrelated sequences which may function to stabilize the molecule.
Catal tiy c Polynucleotides
The term catalytic polynucleotide/nucleic acid refers to a DNA molecule or
DNA-containing molecule (also known in the art as a "deoxyribozyme") or an RNA
or RNA-containing molecule (also known as a "ribozyme") which specifically
recognizes a distinct substrate and catalyzes the chemical modification of
this
substrate. The nucleic acid bases in the catalytic nucleic acid can be bases
A, C, G, T
(and U for RNA).
Typically, the catalytic nucleic acid contains an antisense sequence for
specific
recognition of a target nucleic acid, and a nucleic acid cleaving enzymatic
activity
(also referred to herein as the "catalytic domain"). The types of ribozymes
that are
particularly useful in this invention are the hammerhead ribozyme (Haseloff
and
Gerlach, 1988, Perriman et al., 1992) and the hairpin ribozyme (Shippy et al.,
1999).
The ribozymes of this invention and DNA encoding the ribozymes can be
chemically synthesized using methods well known in the art. The ribozymes can
also
be prepared from a DNA molecule (that upon transcription, yields an RNA
molecule)
operably linked to an RNA polymerase promoter, e.g., the promoter for T7 RNA
polymerase or SP6 RNA polymerase. Accordingly, also provided by this invention
is
a nucleic acid molecule, i.e., DNA or cDNA, coding for a catalytic
polynucleotide of
the invention. When the vector also contains an RNA polymerase promoter
operably
linked to the DNA molecule, the ribozyme can be produced in vitro upon
incubation
with RNA polymerase and nucleotides. In a separate embodiment, the DNA can be
inserted into an expression cassette or transcription cassette. After
synthesis, the
CA 02616144 2008-01-25
22
RNA molecule can be modified by ligation to a DNA molecule having the ability
to
stabilize the ribozyme and make it resistant to RNase.
As with antisense polynucleotides described herein, catalytic polynucleotides
of the invention should also be capable of hybridizing a target nucleic acid
molecule
(for example an mRNA encoding a polypeptide provided as any one of SEQ ID NOs:
1 to 41) under "physiological conditions", namely those conditions within a
cell
(especially conditions in an insect cell such as a cell of a lepidpoteran).
RNA interference
RNA interference (RNAi) is particularly useful for specifically inhibiting the
production of a particular protein. Although not wishing to be limited by
theory,
Waterhouse et al., (1998) have provided a model for the mechanism by which
dsRNA
can be used to reduce protein production. This technology relies on the
presence of
dsRNA molecules that contain a sequence that is essentially identical to the
mRNA of
the gene of interest or part thereof, in this case an mRNA encoding a
polypeptide
according to the invention. Conveniently, the dsRNA can be produced from a
single
promoter in a recombinant vector or host cell, where the sense and anti-sense
sequences are flanked by an unrelated sequence which enables the sense and
anti-
sense sequences to hybridize to form the dsRNA molecule with the unrelated
sequence forming a loop structure. The design and production of suitable dsRNA
molecules for the present invention is well within the capacity of a person
skilled in
the art, particularly considering Waterhouse et al., (1998), Smith et al.,
(2000), WO
99/32619, WO 99/53050, WO 99/49029, and WO 01/34815.
In one example, a DNA is introduced that directs the synthesis of an at least
partly double stranded RNA product(s) with homology to the target gene to be
inactivated. The DNA therefore comprises both sense and antisense sequences
that,
when transcribed into RNA, can hybridize to form the double-stranded RNA
region.
In a preferred embodiment, the sense and antisense sequences are separated by
a
spacer region that comprises an intron which, when transcribed into RNA, is
spliced
out. This arrangement has been shown to result in a higher efficiency of gene
silencing. The double-stranded region may comprise one or two RNA molecules,
transcribed from either one DNA region or two. The presence of the double
stranded
molecule is thought to trigger a response from an endogenous plant system that
destroys both the double stranded RNA and also the homologous RNA transcript
from
the target plant gene, efficiently reducing or eliminating the activity of the
target gene.
The length of the sense and antisense sequences that hybridise should each be
at least 19 contiguous nucleotides, preferably at least 30 or 50 nucleotides,
and more
preferably at least 100, 200, or 500 or 1000 nucleotides. The full-length
sequence
CA 02616144 2008-01-25
23
corresponding to the entire gene transcript may be used. The lengths are most
preferably 100-2000 nucleotides. The degree of identity of the sense and
antisense
sequences to the targeted transcript should be at least 85%, preferably at
least 90%
and more preferably 95-100%. The RNA molecule may of course comprise unrelated
sequences which may function to stabilize the molecule. The RNA molecule may
be
expressed under the control of a RNA polymerase II or RNA polymerase III
promoter. Examples of the latter include tRNA or snRNA promoters.
Preferred small interfering RNA (`siRNA") molecules comprise a nucleotide
sequence that is identical to about 19-23 contiguous nucleotides of the target
mRNA.
Preferably, the target mRNA sequence commences with the dinucleotide AA,
comprises a GC-content of about 30-70% (preferably, 30-60%, more preferably 40-
60% and more preferably about 45%-55%), and does not have a high percentage
identity to any nucleotide sequence other than the target in the genome of the
insect
(preferably lepidopteran) in which it is to be introduced, e.g., as determined
by
standard BLAST search.
Recombinant Vectors
One embodiment of the present invention includes a recombinant vector,
which comprises at least one isolated polynucleotide molecule of the present
invention, inserted into any vector capable of delivering the polynucleotide
molecule
into a host cell. Such vectors contains heterologous polynucleotide sequences,
that is
polynucleotide sequences that are not naturally found adjacent to
polynucleotide
molecules of the present invention and that preferably are derived from a
species other
than the species from which the polynucleotide molecule(s) are derived. The
vector
can be either RNA or DNA, either prokaryotic or eukaryotic, and typically is a
transposon (such as described in US 5,792,294), a virus or a plasmid.
One type of recombinant vector comprises a polynucleotide molecule of the
present invention operatively linked to an expression vector. The phrase
operatively
linked refers to insertion of a polynucleotide molecule into an expression
vector in a
manner such that the molecule is able to be expressed when transformed into a
host
cell. As used herein, an expression vector includes a DNA or RNA vector that
is
capable of transforming a host cell and of effecting expression of a specified
polynucleotide molecule. Preferably, the expression vector is also capable of
replicating within the host cell. Expression vectors can be either prokaryotic
or
eukaryotic, and are typically viruses or plasmids. Expression vectors of the
present
invention include any vectors that function (i.e., direct gene expression) in
recombinant cells of the present invention, including in bacterial, fungal,
endoparasite, arthropod, animal, and plant cells. Vectors of the invention can
also be
CA 02616144 2008-01-25
24
used to produce the polypeptide in a cell-free expression system, such systems
are
well known in the art.
In particular, expression vectors of the present invention contain regulatory
sequences such as transcription control sequences, translation control
sequences,
origins of replication, and other regulatory sequences that are compatible
with the
recombinant cell and that control the expression of polynucleotide molecules
of the
present invention. In particular, recombinant molecules of the present
invention
include transcription control sequences. Transcription control sequences are
sequences which control the initiation, elongation, and termination of
transcription.
Particularly important transcription control sequences are those which control
transcription initiation, such as promoter, enhancer, operator and repressor
sequences.
Suitable transcription control sequences include any transcription control
sequence
that can function in at least one of the recombinant cells of the present
invention. A
variety of such transcription control sequences are known to those skilled in
the art.
Preferred transcription control sequences include those which function in
bacterial,
yeast, arthropod, nematode, plant or mammalian cells, such as, but not limited
to, tac,
lac, trp, trc, oxy-pro, omp/lpp, rrnB, bacteriophage lambda, bacteriophage T7,
T71ac,
bacteriophage T3, bacteriophage SP6, bacteriophage SPO1, metallothionein,
alpha-
mating factor, Pichia alcohol oxidase, alphavirus subgenomic promoters (such
as
Sindbis virus subgenomic promoters), antibiotic resistance gene, baculovirus,
Heliothis zea insect virus, vaccinia virus, herpesvirus, raccoon poxvirus,
other
poxvirus, adenovirus, cytomegalovirus (such as intermediate early promoters),
simian
virus 40, retrovirus, actin, retroviral long terminal repeat, Rous sarcoma
virus, heat
shock, phosphate and nitrate transcription control sequences as well as other
sequences capable of controlling gene expression in prokaryotic or eukaryotic
cells.
Host Cells
Another embodiment of the present invention includes a recombinant cell
comprising a host cell transformed with one or more recombinant molecules of
the
present invention, or progeny cells thereof. Transformation of a
polynucleotide
molecule into a cell can be accomplished by any method by which a
polynucleotide
molecule can be inserted into the cell. Transformation techniques include, but
are not
limited to, transfection, electroporation, microinjection, lipofection,
adsorption, and
protoplast fusion. A recombinant cell may remain unicellular or may grow into
a
tissue, organ or a multicellular organism. Transformed polynucleotide
molecules of
the present invention can remain extrachromosomal or can integrate into one or
more
sites within a chromosome of the transformed (i.e., recombinant) cell in such
a
manner that their ability to be expressed is retained.
CA 02616144 2008-01-25
Suitable host cells to transform include any cell that can be transformed with
a
polynucleotide of the present invention. Host cells of the present invention
either can
be endogenously (i.e., naturally) capable of producing polypeptides of the
present
invention or can be capable of producing such polypeptides after being
transformed
5 with at least one polynucleotide molecule of the present invention. Host
cells of the
present invention can be any cell capable of producing at least one protein of
the
present invention, and include bacterial, fungal (including yeast), parasite,
nematode,
arthropod, animal and plant cells. Examples of host cells include Salmonella,
Escherichia, Bacillus, Listeria, Saccharomyces, Spodoptera, Mycobacteria,
10 Trichoplusia, BHK (baby hamster kidney) cells, MDCK cells, CRFK cells, CV-1
cells, COS (e.g., COS-7) cells, and Vero cells. Further examples of host cells
are E.
coli, including E. coli K-12 derivatives; Salmonella typhi; Salmonella
typhimurium,
including attenuated strains; Spodoptera frugiperda; Trichoplusia ni; BHK
cells;
MDCK cells; CRFK cells; CV-1 cells; COS cells; Vero cells; and non-tumorigenic
15 mouse myoblast G8 cells (e.g., ATCC CRL 1246). Additional appropriate
mammalian cell hosts include other kidney cell lines, other fibroblast cell
lines (e.g.,
human, murine or chicken embryo fibroblast cell lines), myeloma cell lines,
Chinese
hamster ovary cells, mouse NIH/3T3 cells, LMTK cells and/or HeLa cells.
Particularly preferred host cells are insect cells, especially lepidopteran
cells, as well
20 as nematode cells such as C. elegans cells.
Recombinant DNA technologies can be used to improve expression of a
transformed polynucleotide molecule by manipulating, for example, the number
of
copies of the polynucleotide molecule within a host cell, the efficiency with
which
those polynucleotide molecules are transcribed, the efficiency with which the
resultant
25 transcripts are translated, and the efficiency of post-translational
modifications.
Recombinant techniques useful for increasing the expression of polynucleotide
molecules of the present invention include, but are not limited to,
operatively linking
polynucleotide molecules to high-copy number plasmids, integration of the
polynucleotide molecule into one or more host cell chromosomes, addition of
vector
stability sequences to plasmids, substitutions or modifications of
transcription control
signals (e.g., promoters, operators, enhancers), substitutions or
modifications of
translational control signals (e.g., ribosome binding sites, Shine-Dalgarno
sequences),
modification of polynucleotide molecules of the present invention to
correspond to the
codon usage of the host cell, and the deletion of sequences that destabilize
transcripts.
Transgenic Plants
The term "plant" refers to whole plants, plant organs (e.g. leaves, stems
roots,
etc), seeds, plant cells and the like. Plants contemplated for use in the
practice of the
CA 02616144 2008-01-25
26
present invention include both monocotyledons and dicotyledons. Target plants
include, but are not limited to, the following: cereals (wheat, barley, rye,
oats, rice,
sorghum and related crops); beet (sugar beet and fodder beet); pomes, stone
fruit and
soft fruit (apples, pears, plums, peaches, almonds, cherries, strawberries,
raspberries
and black-berries); leguminous plants (beans, lentils, peas, soybeans); oil
plants (rape,
mustard, poppy, olives, sunflowers, coconut, castor oil plants, cocoa beans,
groundnuts); cucumber plants (marrows, cucumbers, melons); fibre plants
(cotton,
flax, hemp, jute); citrus fruit (oranges, lemons, grapefruit, mandarins);
vegetables
(spinach, lettuce, asparagus, cabbages, carrots, onions, tomatoes, potatoes,
paprika);
lauraceae (avocados, cinnamon, camphor); or plants such as maize, tobacco,
nuts,
coffee, sugar cane, tea, vines, hops, turf, bananas and natural rubber plants,
as well as
ornamentals (flowers, shrubs, broad-leaved trees and evergreens, such as
conifers).
Transgenic plants, as defined in the context of the present invention include
plants (as well as parts and cells of said plants) and their progeny which
have been
genetically modified using recombinant techniques to cause production of at
least one
polypeptide and/or polynucleotide of the present invention in the desired
plant or
plant organ. Transgenic plants can be produced using techniques known in the
art,
such as those generally described in A. Slater et al., Plant Biotechnology -
The
Genetic Manipulation of Plants, Oxford University Press (2003), and P.
Christou and
H. Klee, Handbook of Plant Biotechnology, John Wiley and Sons (2004).
A polynucleotide of the present invention may be expressed constitutively in
the transgenic plants during all stages of development. Depending on the use
of the
plant or plant organs, the polynucleotides may be expressed in a stage-
specific
manner. Furthermore, the polynucleotides may be expressed tissue-specifically.
Regulatory sequences which are known or are found to cause expression of a
polynucleotide of interest in plants may be used in the present invention. The
choice
of the regulatory sequences used depends on the target plant and/or target
organ of
interest. Such regulatory sequences may be obtained from plants or plant
viruses, or
may be chemically synthesized. Such regulatory sequences are well known to
those
skilled in the art.
Constitutive plant promoters are well known. Further to previously mentioned
promoters, some other suitable promoters include but are not limited to the
nopaline
synthase promoter, the octopine synthase promoter, CaMV 35S promoter, the
ribulose-1,5-bisphosphate carboxylase promoter, Adhl-based pEmu, Actl, the SAM
synthase promoter and Ubi promoters and the promoter of the chlorophyll a/b
binding
protein. Alternatively it may be desired to have the transgene(s) expressed in
a
regulated fashion. Regulated expression of the polynucleotides is possible by
placing
the coding sequence under the control of promoters that are tissue-specific,
CA 02616144 2008-01-25
27
developmental-specific, or inducible. Several tissue-specific regulated genes
and/or
promoters have been reported in plants. These include genes encoding the seed
storage proteins (such as napin, cruciferin, (3-conglycinin, glycinin and
phaseolin),
zein or oil body proteins (such as oleosin), or genes involved in fatty acid
biosynthesis
(including acyl carrier protein, stearoyl-ACP desaturase, and fatty acid
desaturases
(fad 2- 1)), and other genes expressed during embryo development (such as
Bce4).
Particularly useful for seed-specific expression is the pea vicilin promoter.
Other
useful promoters for expression in mature leaves are those that are switched
on at the
onset of senescence, such as the SAG promoter from Arabidopsis. A class of
fruit-
specific promoters expressed at or during anthesis through fruit development,
at least
until the beginning of ripening, is discussed in US 4,943,674. Other examples
of
tissue-specific promoters include those that direct expression in leaf cells
following
damage to the leaf (for example, from chewing insects), in tubers (for
example,
patatin gene promoter), and in fiber cells (an example of a developmentally-
regulated
fiber cell protein is E6 fiber.
Several techniques are available for the introduction of an expression
construct
containing a nucleic acid sequence of interest into the target plants. Such
techniques
include but are not limited to transformation of protoplasts using the
calcium/polyethylene glycol method, electroporation and microinjection or
(coated)
particle bombardment. In addition to these so-called direct DNA transformation
methods, transformation systems involving vectors are widely available, such
as viral
and bacterial vectors (e.g. from the genus Agrobacterium). After selection
and/or
screening, the protoplasts, cells or plant parts that have been transformed
can be
regenerated into whole plants, using methods known in the art. The choice of
the
transformation and/or regeneration techniques is not critical for this
invention.
To confirm the presence of the transgenes in transgenic cells and plants, a
polymerase chain reaction (PCR) amplification or Southern blot analysis can be
performed using methods known to those skilled in the art. Expression products
of
the transgenes can be detected in any of a variety of ways, depending upon the
nature
of the product, and include Western blot and enzyme assay. One particularly
useful
way to quantitate expression and to detect replication in different plant
tissues is to
use a reporter gene, such as GUS. Once transgenic plants have been obtained,
they
may be grown to produce plant tissues or parts having the desired phenotype.
The
plant tissue or plant parts, may be harvested, and/or the seed collected. The
seed may
serve as a source for growing additional plants with tissues or parts having
the desired
characteristics.
CA 02616144 2008-01-25
28
Transgenic Non-Human Animals
Techniques for producing transgenic animals are well known in the art. A
useful general textbook on this subject is Houdebine, Transgenic animals -
Generation and Use (Harwood Academic, 1997).
Heterologous DNA can be introduced, for example, into fertilized mammalian
ova. For instance, totipotent or pluripotent stem cells can be transformed by
microinjection, calcium phosphate mediated precipitation, liposome fusion,
retroviral
infection or other means, the transformed cells are then introduced into the
embryo,
and the embryo then develops into a transgenic animal. In a highly preferred
method,
developing embryos are infected with a retrovirus containing the desired DNA,
and
transgenic animals produced from the infected embryo. In a most preferred
method,
however, the appropriate DNAs are coinjected into the pronucleus or cytoplasm
of
embryos, preferably at the single cell stage, and the embryos allowed to
develop into
mature transgenic animals.
Another method used to produce a transgenic animal involves microinjecting a
nucleic acid into pro-nuclear stage eggs by standard methods. Injected eggs
are then
cultured before transfer into the oviducts of pseudopregnant recipients.
Transgenic animals may also be produced by nuclear transfer technology.
Using this method, fibroblasts from donor animals are stably transfected with
a
plasmid incorporating the coding sequences for a binding domain or binding
partner
of interest under the control of regulatory sequences. Stable transfectants
are then
fused to enucleated oocytes, cultured and transferred into female recipients.
Antibodies
The invention also provides monoclonal or polyclonal antibodies to
polypeptides of the invention or fragments thereof. Thus, the present
invention
further provides a process for the production of monoclonal or polyclonal
antibodies
to polypeptides of the invention.
The term "specifically binds" refers to the ability of the antibody to bind to
at
least one polypeptide of the present invention but not other known olfactory
receptors.
In an embodiment, an antibody of the invention is an antagonist of an
olfactory
receptor of the invention.
As used herein, the term "epitope" refers to a region of a polypeptide of the
invention which is bound by the antibody. An epitope can be administered to an
animal to generate antibodies against the epitope, however, antibodies of the
present
invention preferably specifically bind the epitope region in the context of
the entire
polypeptide.
CA 02616144 2008-01-25
29
If polyclonal antibodies are desired, a selected mammal (e.g., mouse, rabbit,
goat, horse, etc.) is immunised with an immunogenic polypeptide of the
invention.
Serum from the immunised animal is collected and treated according to known
procedures. If serum containing polyclonal antibodies contains antibodies to
other
antigens, the polyclonal antibodies can be purified by immunoaffinity
chromatography. Techniques for producing and processing polyclonal antisera
are
known in the art. In order that such antibodies may be made, the invention
also
provides polypeptides of the invention or fragments thereof haptenised to
another
polypeptide for use as immunogens in animals.
Monoclonal antibodies directed against polypeptides of the invention can also
be readily produced by one skilled in the art. The general methodology for
making
monoclonal antibodies by hybridomas is well known. Immortal antibody-producing
cell lines can be created by cell fusion, and also by other techniques such as
direct
transformation of B lymphocytes with oncogenic DNA, or transfection with
Epstein-
Barr virus. Panels of monoclonal antibodies produced can be screened for
various
properties; i.e., for isotype and epitope affinity.
An alternative technique involves screening phage display libraries where, for
example the phage express scFv fragments on the surface of their coat with a
large
variety of complementarity determining regions (CDRs). This technique is well
known in the art.
For the purposes of this invention, the term "antibody", unless specified to
the
contrary, includes fragments of whole antibodies which retain their binding
activity
for a target antigen. Such fragments include Fv, F(ab') and F(ab')2 fragments,
as well
as single chain antibodies (scFv). Furthermore, the antibodies and fragments
thereof
may be humanised antibodies, for example as described in EP-A-239400.
Antibodies of the invention may be bound to a solid support and/or packaged
into kits in a suitable container along with suitable reagents, controls,
instructions and
the like.
In an embodiment, antibodies of the present invention are detectably labeled.
Exemplary detectable labels that allow for direct measurement of antibody
binding
include radiolabels, fluorophores, dyes, magnetic beads, chemiluminescers,
colloidal
particles, and the like. Examples of labels which permit indirect measurement
of
binding include enzymes where the substrate may provide for a coloured or
fluorescent product. Additional exemplary detectable labels include covalently
bound
enzymes capable of providing a detectable product signal after addition of
suitable
substrate. Examples of suitable enzymes for use in conjugates include
horseradish
peroxidase, alkaline phosphatase, malate dehydrogenase and the like. Where not
commercially available, such antibody-enzyme conjugates are readily produced
by
CA 02616144 2008-01-25
techniques known to those skilled in the art. Further exemplary detectable
labels
include biotin, which binds with high affinity to avidin or streptavidin;
fluorochromes
(e.g., phycobiliproteins, phycoerythrin and allophycocyanins; fluorescein and
Texas
red), which can be used with a fluorescence activated cell sorter; haptens;
and the like.
5 Preferably, the detectable label allows for direct measurement in a plate
luminometer,
e.g., biotin. Such labeled antibodies can be used in techniques known in the
art to
detect polypeptides of the invention.
Identification of Compounds that Bind and/or Modulate the Activity Olfactory
10 Receptors
The present invention provides screening methodologies useful in the
identification of compounds which bind to and/or modulate the activity of the
olfactory receptor genes, mRNA and proteins described herein. Such compounds
will
include molecules that agonize or antagonize olfactory receptor function.
15 Screening methodologies to identify compounds that bind and/or modulate the
activity of olfactory receptors are known in the art. Such compounds include
endogenous cellular components which interact with the identified genes and
proteins
in vivo. Thus, cell lysates or tissue homogenates may be screened for proteins
or
other compounds which bind to one of the olfactory receptor genes, mRNA or
20 proteins of the invention.
Alternatively, any of a variety of exogenous compounds, both naturally
occurring and/or synthetic (e. g., libraries of small molecules or peptides),
may be
screened for binding capacity. Binding compounds can include, but are not
limited to,
other cellular proteins. Binding compounds can also include, but are not
limited to,
25 peptides such as, for example, soluble peptides, including, but not limited
to, Ig-tailed
fusion peptides, antibodies such as those described herein, and small organic
or
inorganic molecules. Such compounds can include organic molecules (e. g.,
peptidomimetics) that bind to the receptor and either mimic the activity
triggered by
the natural odorant ligand (namely, agonists); as well as peptides, antibodies
and other
30 organic compounds that mimic the receptor (or a portion thereof) and bind
to and
"neutralize" natural odorant ligand. Such compounds identified in a screen for
binding to the receptor can be assayed for their effects on receptor
signalling.
Particularly useful molecules that bind to and/or modulate olfactory receptor
activity are small molecules, most preferably volatile small molecules, that
function as
odorants. The term "odorant" as employed herein refers to a molecule that has
the
potential to bind to an olfactory receptor. Equivalent terms employed herein
include
"odorant ligand", "odorant molecule" and "odorant compound". The term
"binding"
or "interaction" as used herein with respect to odorant ligands refers to the
interaction
CA 02616144 2008-01-25
31
of ligands with the receptor polypeptide where the ligands may serve as either
agonists and/or antagonists of a given receptor or receptor function. This
effect may
not be direct, but merely by altering the binding of an odorant receptor to
another
ligand. An odorant ligand may thus directly cause a perception of odor (an
agonist),
or may block the perception of odor (an antagonist). An odorant ligand may
include,
but is not limited to, molecules which interact with polypeptides involved in
olfactory
sensation.
Binding of a modulator (ligand) to a receptor of the invention can be examined
in vitro with soluble or solid state reactions, using a full-length receptor
molecule or a
chimeric molecule such as an extracellular domain or transmembrane region, or
combination thereof, of an receptor of the invention covalently linked to a
heterologous signal transduction domain, or a heterologous extracellular
domain
and/or transmembrane region covalently linked to the transmembrane and/or
cytoplasmic domain of an olfactory receptor. Furthermore, ligand-binding
domains of
the protein of interest can be used in vitro in soluble or solid state
reactions to assay
for ligand binding. In numerous embodiments, a chimeric receptor will be made
that
comprises all or part of an olfactory receptor polypeptide, as well an
additional
sequence that facilitates the localization of the olfactory receptor to the
membrane,
such as a rhodopsin, e. g., an N-terminal fragment of a rhodopsin protein.
Ligand binding to an olfactory receptor of the invention, a domain, or
chimeric
protein (also referred to herein as a fusion protein) can be tested in
solution, in a
bilayer membrane, attached to a solid phase, in a lipid monolayer, or in
vesicles.
Binding of a modulator can be tested using, e. g., changes in spectroscopic
characteristics (e. g., fluorescence, absorbance, refractive index)
hydrodynamic (e. g.,
shape), chromatographic, or solubility properties. These assays may involve
displacing a radioactively or fluorescently labeled ligand, and measuring
changes in
intrinsic fluorescence or changes in proteolytic susceptibility, etc.
Methods for screening odorant compounds using olfactory receptors in
neuronal cells are known in the art (WO 98/50081; Duchamp-Viret et al., 1999;
Sato
et al., 1994; Malnic et al., 1999; Zhao et al., 1998). There are also methods
which can
be employed to screen odorant compounds which do not require neuronal cells
and
that are known in the art (US 5,798, 275; Kiefer et al., 1996; Krautwurst et
al., 1998).
The invention provides methods and compositions for expressing the olfactory
receptors of the invention in cells to screen for odorants that can
specifically bind an
olfactory receptor of the invention, and for determining the effect (e. g.,
biochemical
or electrophysiological) of such binding on cell physiology.
Any cell expression system can be used, e. g., insect or mammalian (for
example HEK293, CHO or COS cells) cell expression systems. Cells that normally
CA 02616144 2008-01-25
32
express olfactory receptors can be used, particularly to study the
physiological effect
of an odorant on a cell. Isolation and/or culturing of such cells and their
transformation with the olfactory receptor-expressing sequences of the
invention can
be done with routine methods (Vargas, 1999; Coon et al., 1989).
Several methods of measuring G-protein activity are known to those of skill in
the art and can be used in conjunction with the methods of the present
invention,
including but not limited to measuring calcium ion or cyclic AMP concentration
in the
cells. Such methods are described in Howard et al., (2001), Krautwurst et al.,
(1999),
Chandrashekar et al., (2000), Oda et al., (2000) and Kiely et al., (2007).
To evaluate electrophysiologic effects of ligand binding to cell-expressed
olfactory receptor of the invention, patch-clamping of individual cells can be
done.
Patch-clamp recordings of the olfactory receptor cell membrane can measure
membrane conductances. Some conductances are gated by odorants in the cilia
and
depolarize the cell through cAMP-or IP3-sensitive channels, depending on the
species. Other conductances are activated by membrane depolarization and/or an
increased intracellular Ca2+ concentration (Trotier, 1994).
Changes in calcium ion levels in the cell after exposure of the cell to known
or
potential odorant/ligands can be detected by a variety of means. For example,
cells
can be pre-loaded with reagents sensitive to calcium ion transients.
Techniques for
the measurement of calcium transients are known in the art. For example,
Kashiwayanagi (1996) measured both of inositol 1,4,5-trisphosphate induces
inward
currents and Ca2+ uptake in frog olfactory receptor cells.
In certain specific embodiments, intracellular calcium concentration is
measured in the screening assays of the instant application by using a
Fluorometric
Imaging Plate Reader ("FLIPR") system (Molecular Devices, Inc.), which
provides
the advantages of automated, high-throughput screening, see also Sullivan et
al.,
"Measurement of [Ca2+] i using the fluorometric imaging plate reader (FLIPR)",
p.
125-136, Calcium Signaling Protocols D. G. Lambert, (editor), Humana Press
(1999);
or in US 6,004, 808, which employs Fura-PE3 (Molecular Probes, Inc., Eugene,
OR)
as a stain of calcium ions.
Other physiologic activity mechanisms can also be measured, e. g., plasma
membrane homeostasis parameters (including lipid second messengers), and
cellular
pH changes (see, e. g., Silver, 1998).
Alternatively, in vitro synthesised mRNA coding for a polypeptide of the
invention can be injected into Xenopus oocytes allowing electrophysiological
or
calcium imaging of odorant-driven cell excitation.
A typical principle of the assays used to identify compounds that bind to
olfactory receptors of the invention involves preparing a reaction mixture of
said
CA 02616144 2008-01-25
33
receptor and a test compound under conditions and for a time sufficient to
allow the
two components to interact and bind, thus forming a complex which can be
removed
and/or detected in the reaction mixture. These assays can be conducted in a
variety of
ways. For example, one method to conduct such an assay involves attaching the
receptor or the test substance onto a solid phase and detecting receptor/test
compound
complexes anchored on the solid phase at the end of the reaction. In one
embodiment
of such a method, the receptor can be anchored onto a solid surface, and the
test
compound, which is not anchored, can be labeled, either directly or
indirectly.
In practice, microtiter plates can conveniently be utilized as the solid
phase.
The anchored component can be immobilized by non-covalent or covalent
attachments. Non-covalent attachment can be accomplished by simply coating the
solid surface with a solution of the protein and drying. Alternatively, an
immobilized
antibody, preferably a monoclonal antibody, specific for the protein to be
immobilized
can be used to anchor the protein to the solid surface. The surfaces can be
prepared in
advance and stored.
In order to conduct the assay, the nonimmobilized component is added to the
coated surface containing the anchored component. After the reaction is
complete,
unreacted components are removed (e. g., by washing) under conditions such
that any
complexes formed will remain immobilized on the solid surface. The detection
of
complexes anchored on the solid surface can be accomplished in a number of
ways.
Where the previously nonimmobilized component is pre-labeled, the detection
of label immobilized on the surface indicates that complexes were formed.
Where the
previously nonimmobilized component is not pre-labeled, an indirect label can
be
used to detect complexes anchored on the surface; e. g., using a labeled
antibody
specific for the previously nonimmobilized component (the antibody, in turn,
can be
directly labeled or indirectly labeled with a labeled anti-Ig antibody).
Alternatively, a reaction can be conducted in a liquid phase, the reaction
products separated from unreacted components, and complexes detected; e. g.,
using
an immobilized antibody specific for an olfactory receptor of the invention or
the test
compound to anchor any complexes formed in solution, and a labeled antibody
specific for the other component of the possible complex to detect anchored
complexes.
High throughput screening assays can also be used to identify compounds that
bind and/or modulate an olfactory receptor of the invention. In the high
throughput
assays of the invention, it is possible to screen up to several thousand
different ligands
or modulators in a single day. In particular, each well of a microtiter plate
can be
used to run a separate assay against a selected potential modulator, or, if
concentration
or incubation time effects are to be observed, every 5-10 wells can test a
single
CA 02616144 2008-01-25
34
modulator. Thus, a single standard microtiter plate can assay about 100 (e.
g., 96)
modulators. If 1536 well plates are used, then a single plate can easily assay
from
about 1000 to about 1500 different compounds. It is possible to assay several
plates
per day. More recently, microfluidic approaches to reagent manipulation have
been
developed.
Additionally, methods can be employed which result in the simultaneous
identification of genes which encode proteins interacting with an olfactory
receptor of
the invention. These methods include, for example, probing expression
libraries with
labeled polypeptide of the invention, using this protein in a manner similar
to the well
known technique of antibody probing of ?,gtl l libraries.
One method which detects protein interactions in vivo, the two-hybrid system,
is described in detail for illustration purposes only and not by way of
limitation. One
version of this system has been described (Chien et al., 1991) and is
commercially
available from Clontech (Palo Alto, CA). Briefly, utilizing such a system,
plasmids
are constructed that encode two hybrid proteins: one consists of the DNA-
binding
domain of a transcription activator protein fused to a known protein, in this
case, a
polypeptide of the invention, and the other consists of the activator
protein's activation
domain fused to an unknown protein that is encoded by a cDNA, preferably an
insect
(more preferably a Lepidopteran) antennal or maxillary palp cDNA, which has
been
recombined into this plasmid as part of a cDNA library. The plasmids are
transformed
into a strain of the yeast Saccharomyces cerevisiae that contains a reporter
gene (e. g.,
lacZ) whose regulatory region contains the transcription activator's binding
sites.
Either hybrid protein alone cannot activate transcription of the reporter
gene, the
DNA-binding domain hybrid cannot because it does not provide activation
function,
and the activation domain hybrid cannot because it cannot localize to the
activator's
binding sites. Interaction of the two hybrid proteins reconstitutes the
functional
activator protein and results in expression of the reporter gene, which is
detected by
an assay for the reporter polypeptide.
Protein-Structure Based Desi n~gonists and Antagonists
Computer modeling and searching technologies permit identification of
compounds that can bind olfactory receptors of the invention, including
compounds
that can modulate olfactory receptor activity. The identification of such a
compound
may also allow the active sites or regions of the receptor to be identified.
Such active
sites might typically be odorant ligand binding sites, such as the interaction
domains
of odorant ligands with the receptor.
The three dimensional geometric structure of olfactory receptor or the active
site thereof can be determined. This can be done by known methods, including X-
ray
CA 02616144 2008-01-25
' 35
crystallography, which can determine a complete molecular structure. Solid or
liquid
phase NMR can also be used to determine certain intra-molecular distances
within the
active site and/or in the odorant ligand/receptor complex. Any experimental
method
of structure determination can be used to obtain partial or complete geometric
structures. The geometric structures may be measured with a complexed odorant
ligand, natural or artificial, which may increase the accuracy of the receptor
structure,
or active site structure, that is determined.
Methods of computer based numerical modeling can be used to complete the
structure (e. g., in embodiments wherein an incomplete or insufficiently
accurate
structure is determined) or to improve its accuracy. Any method recognized in
the art
may be used, including, but not limited to, parameterized models specific to
particular
biopolymers such as proteins or nucleic acids, molecular dynamics models based
on
computing molecular motions, statistical mechanics models based on thermal
ensembles, or combined models.
The three-dimensional structure of an olfactory receptor of the invention can
be used to identify antagonists or agonists through the use of computer
modeling
using a docking program such as GRAM, DOCK, or AUTODOCK (Dunbrack et al.,
1997). Computer programs can also be employed to estimate the attraction,
repulsion,
and steric hindrance of a candidate compound to the polypeptide. Generally the
tighter the fit (e.g., the lower the steric hindrance, and/or the greater the
attractive
force) the more potent the potential agonist or antagonist will be since these
properties
are consistent with a tighter binding constant. Furthermore, the more
specificity in the
design of a potential agonist or antagonist the more likely that it will not
interfere with
other proteins.
Initially a potential compound could be obtained, for example, using methods
of the invention such as by screening a random peptide library produced by a
recombinant bacteriophage or a chemical library. A compound selected in this
manner could be then be systematically modified by computer modeling programs
until one or more promising potential compounds are identified.
Such computer modeling allows the selection of a finite number of rational
chemical modifications, as opposed to the countless number of essentially
random
chemical modifications that could be made, and of which any one might lead to
a
useful agonist or antagonist. Each chemical modification requires additional
chemical
steps, which while being reasonable for the synthesis of a finite number of
compounds, quickly becomes overwhelming if all possible modifications needed
to be
synthesized. Thus through the use of the three-dimensional structure and
computer
modeling, a large number of these compounds can be rapidly screened on the
CA 02616144 2008-01-25
36
computer monitor screen, and a few likely candidates can be determined without
the
laborious synthesis of untold numbers of compounds.
For most types of models, standard molecular force fields, representing the
forces between constituent atoms and groups, are necessary, and can be
selected from
force fields known in physical chemistry. Exemplary forcefields that are known
in
the art and can be used in such methods include, but are not limited to, the
Constant
Valence Force Field (CVFF), the AMBER force field and the CHARM force field.
The incomplete or less accurate experimental structures can serve as
constraints on
the complete and more accurate structures computed by these modeling methods.
Alternatively, these methods can be used to identify improved modulating
compounds from an already known modulating compound or odorant ligand. The
composition of the known compound can be modified and the structural effects
of
modification can be determined using the experimental and computer modeling
methods described above applied to the new composition. The altered structure
is then
compared to the active site structure of the compound to determine if an
improved fit
or interaction results. In this manner systematic variations in composition,
such as by
varying side groups, can be quickly evaluated to obtain modified binding
compounds
or odorant ligands of improved specificity or activity.
Further examples of molecular modeling systems are the CHARMm and
QUANTA programs (Polygen Corporation, Waltham, MA). CHARMm performs the
energy minimization and molecular dynamics functions. QUANTA performs the
construction, graphic modelling and analysis of molecular structure. QUANTA
allows
interactive construction, modification, visualization, and analysis of the
behaviour of
molecules with each other.
Biosensors
As used herein, the term "biosensor" means a sensor which converts an
interaction between biomolecules into a signal such as an electric signal, so
as to
measure or detect a target substance. A biosensor of the invention can be any
instrument which detects and/or identifies and/or quantifies odorants or
similar
volatile or non-volatile chemicals. A conventional biosensor is comprised of a
receptor site for recognizing a chemical substance as a detection target and a
transducer site for converting a physical change or chemical change generated
at the
site into an electric signal. Examples of biosensors incorporating receptor
molecules
are well known in the art and include those described in WO 00/70343.
Typically, a
biosensor of the invention will comprise a polypeptide of the invention co-
expressed
with one or more accessory proteins such as a G protein, a sensory
transduction
CA 02616144 2008-01-25
37
mechanism, analogue to digital conversion, digital signal processing, pattern
recognition, decision support and output.
In one embodiment, a fusion polypeptide of the invention comprises a
resonance energy transfer (RET) acceptor and donor. RET is the non-radioactive
transfer of energy from an excited state donor molecule to a ground state
acceptor
molecule. Energy transfer efficiency is dependent on the distance between the
donor
and acceptor, the extent of the spectral overlap and the relative orientation
of the
acceptor and donor dipoles. RET is increasingly being used to monitor inter
and
intra-molecular movements in biological systems. Examples of proteins which
can be
used include, but are not limited to, donors cyan fluorescent protein (CFP)
and Renilla
luciferase (RLuc) and their respective acceptors yellow fluorescent protein
(YFP) and
a variant of green fluorescent protein (GFP2). Fluorescent resonance energy
transfer
(FRET) has previously been used to quantify ligand binding by the a2-
adrenergic
receptor (Vilardaga et al., 2003; Hoffmann et al., 2005), whilst
bioluminescent
resonance energy transfer (BRET) has been used to monitor an intramolecular
conformational change of 0-arrestin following GPCR activation (Charest et al.,
2005).
As the skilled addressee will appreciate, similar receptor fusion proteins to
those
described in Vilardaga et al. (2003), Hoffmann et al. (2005), Charest et al.
(2005) and
US 20060272037 can be used in biosensors of the present invention.
Insect Pests and the Control Thereof
In a preferred embodiment, the insect pest is of the order Lepidoptera.
Examples include, but are not limited to, Achoroia grisella, Acleris
gloverana, Acleris
variana, Adoxophyes orana, Agrotis ipsilon, Alabama argillacea, Alsophila
pometaria, Amyelois transitella, Anagasta kuehniella, Anarsia lineatella,
Anisota
senatoria, Antheraea pernyi, Anticarsia gemmatalis, Archips sp., Argyrotaenia
sp.,
Athetis mindara, Bombyx mori, Bucculatrix thurberiella, Cadra cautella,
Choristoneura sp., Cochylls hospes, Colias eurytheme, Corcyra cephalonica,
Cydia
latiferreanus, Cydia pomonella, Datana integerrima, Dendrolimus sibericus,
Desmia
feneralis, Diaphania hyalinata, Diaphania nitidalis, Diatraea grandiosella,
Diatraea
saccharalis, Ennomos subsignaria, Eoreuma loftini, Esphestia elutella, Erannis
tilaria, Estigmene acrea, Eulia salubricola, Eupocoellia ambiguella,
Eupoecilia
ambiguella, Euproctis chrysorrhoea, Euxoa messoria, Galleria mellonella,
Grapholita molesta, Harrisina americana, Helicoverpa subflexa, Helicoverpa
zea,
Helicoverpa armigera, Heliothis virescens, Hemileuca oliviae, Homoeosoma
electellum, Hyphantia cunea, Keiferia lycopersicella, Lambdina fiscellaria
fiscellaria,
Lambdina fiscellaria lugubrosa, Leucoma salicis, Lobesia botrana, Loxostege
sticticalis, Lymantria dispar, Macalla thyrisalis, Malacosoma sp., Mamestra
CA 02616144 2008-01-25
38
brassicae, Mamestra configurata, Manduca quinquemaculata, Manduca sexta,
Maruca testulalis, Melanchra picta, Operophtera brumata, Orgyia sp., Ostrinia
nubilalis, Paleacrita vernata, Papilio cresphontes, Pectinophora gossypiella,
Phryganidia californica, Phyllonorycter blancardella, Pieris napi, Pieris
rapae,
Plathypena scabra, Platynota flouendana, Platynota stultana, Platyptilia
carduidactyla, Plodia interpunctella, Plutella xylostella, Pontia protodice,
Pseudaletia unipuncta, Pseudoplasia includens, Sabulodes aegrotata, Schizura
concinna, Sitotroga cerealella, Spilonta ocellana, Spodoptera sp.,
Thaurnstopoea
pityocampa, Tinsola bisselliella, Trichoplusia hi, Udea rubigalis, Xylomyges
curiails,
and Yponomeuta padella.
The olfactory receptor genes studied as discussed herein may be used to
identify compounds which interfere with the orientation and mating of a wide
range of
insects, especially Lepidopterans. Thus, the present invention enables the
identification of compositions which disrupt insect mating by selective
inhibition of
specific receptor genes involved in mating attraction.
The identification of receptors for odorants is useful in developing new
insect
repellants and traps for the control of Lepidopterans and other insect pests.
The
olfactory receptor genes studied using the materials, systems and methods of
the
present invention may be used to identify compounds which can be used as
animal
repellants.
The olfactory receptor genes studied using the materials, systems and methods
of the present invention can also be used to identify compounds which attract
specific
insects to a particular location.
Aspects of the present invention are used in various methods which reduce or
eliminate the levels of particular insect pests. Traps may also be utilized
where
trapped insects are killed by toxicant-containing poison baits where the
insect may
consume poisoned bait. The insect attractant compositions so identified may
also be
combined with an insecticide, for example as an insect bait in
microencapsulated
form. Alternatively, or in addition, the insect attractant composition may be
placed
inside an insect trap, or in the vicinity of the entrance to an insect trap.
EXAMPLES
Example 1 - Characterization of novel insect olfactory receptors
Materials and Methods
BmOr bioinformatics
Known insect Ors whose sequences have been entered onto GenBank
(National Center for Biotechnology Information) were used to search for
similar
genes in the silkworm genome sequence. Protein sequences were used to perform
CA 02616144 2008-01-25
39
TBLASTN (Altschul et al., 1997) searches of assembled scaffolds available
through
two internet websites: http://kaikoblast.dna.affrc.go.jp/ (Silkworm Genome
Research
Program, National Institute of Agrobiological Sciences, Japan) and
http://silkworm.genomics.org.cn/ (Beijing Genomics Institute, China). Genomic
scaffold sequences were used to construct Or genes manually in the PAUP text
editor
(Swofford, 2001), using homology with known Or exons and an online program to
predict exon/intron splice sites (SplicePredictor,
http://deepc2.psi.iastate.edu/cgi-
bin/sp.cgi). In some cases a single scaffold did not contain the complete Or
gene;
where possible, 3' RACE was used to resolve the gene sequence (see below).
Divergent silkworm Ors were used in a second round of TBLASTN searches to find
additional genes.
Phylogenetic analysis
Conceptually translated protein sequences from silkworm Or genes identified
in this study along with the 17 H. virescens Ors identified by Krieger et al.,
(2004)
were used to construct a phylogenetic tree. The protein sequences were aligned
using
ClustaIX (Jeanmougin et al., 1998). Amino acid distances were calculated and
multiple amino acid substitutions were corrected for using the maximum
likelihood
model, the BLOSUM62 amino acid exchange matrix, and uniform rates based on the
actual sequences in TREE-PUZZLE v5.0 (Schmidt et al., 2002). Neighbor-joining
followed by a heuristic search was employed to construct phylogenetic trees
using
PAUP* v4.0bl0 software (Swofford, 2001). Bootstrap analysis (n = 1000
replicates)
was performed using uncorrected distances.
3' and 5' RACE
Total RNA was isolated from male and female adult moth antennae (n = 30 to
50 moths) using an RNeasy Mini Kit (Qiagen, Valencia, CA). Total RNA was
quantified by absorption at a wavelength of 260 nm and its quality assessed on
a 1%
agarose gel. For 3' RACE, lst strand cDNA was synthesized from 5 ug of total
RNA
using a SuperScriptTM III First-Strand Synthesis System for RT-PCR kit
(Invitrogen,
Carlsbad, Ca USA) and a custom oligo dT primer, 5'-
GGCCAGTGAATTGTAATACGACTCACTATAGGGAGGCGGT24-3' (SEQ ID
NO:83). First strand cDNA (20 uL final volume) was incubated with 4U of E.
coli
Ribonuclease H (Invitrogen) for 20 min at 37 C. After heat inactivation at 65
C for
15 min, excess dNTPs and primer were removed using Microcon YM100 spin
columns (Millipore Billerica, MA). Two uL of 1S` strand cDNA was used as
template
in a PCR reaction combining a gene-specific forward primer (BmOrs9, 10, 12,
14, 15,
19, 30, 33, 35, 37, 38, 42, 45 and 47) with a reverse primer specific to the
T7 RNA
CA 02616144 2008-01-25
polymerase promoter sequence (5'-
GGCCAGTGAATTGTAATACGACTCACTATAGGGAGGCGG-3' (SEQ ID
NO:84)) on the oligo dT primer used for lst strand cDNA synthesis. The stock
PCR
mix contained: 1X Taq PCR buffer, 2 mM MgC12, 0.5 mM dNTPs, lU of Taq and
5 0.5U of Pfu polymerase per 25 uL (Stratagene, La Jolla, CA), and 0.4 uM
reverse
primer. Two uL of 1 St cDNA and 1 uL of 10 uM gene specific primer (0.4 uM
final
concentration) were added to 22 uL of lx PCR stock mix. PCR reactions were
amplified using a Stratagene RoboCycler set at one cycle of 94 C for 2 min
followed
by 25 cycles of (94 C for 30 sec; 52 C for 30 sec; and 72 C for 2 min). Each
PCR
10 tube was topped up with 25 uL of fresh lx PCR mix and 1 uL of 10 uM gene
specific
primer, amplified for a further 15 cycles (94 C for 30 sec; 52 C for 30 sec;
and 72 C
for 2 min), followed by a final cycle at 72 C for 7 min.
PCR reactions producing a product in the expected size range were gel purified
using a MinElute Gel Extraction Kit (Qiagen, Valencia, CA) and sequenced using
the
15 gene specific primer and the Taq Dye Deoxy Terminator Cycle Sequencing Kit
combined with an automatic DNA sequencer (Applied Biosystems, Foster City,
CA).
5' RACE was performed using the SMARTTM RACE cDNA Amplification Kit
(Clontech, Mountain View, CA) following the manufacturers instructions.
Briefly,
50ng - lug of total antennal cDNA was primed at 70 C for 2 mins with 5' CDS
primer
20 and Smart II oligos (supplied in the kit). First strand cDNA was
synthesised using
PowerScriptTM Reverse Transcriptase (Clontech) at 42 C for 1.5hours. The 1St
starnd
cDNA was used as template in a PCR reaction with gene specific primers
(reverse,
BmOrs45 and 47) and the Clontech universal primer (forward) to produce gene
specific PCR fragment that were sequenced.
Insects
B. mori eggs and artificial diet were purchased from the Carolina Biological
Supply Company (2700 York Road, Burlington, NC). The larvae were reared at 23
C
- 27 C on artificial diet, or white mulberry (Morus alba L.) leaves available
locally
during the summer season. Silkworm pupae were also provided as a gift from
colonies
reared by Dr. M. Goldsmith (University of Rhode Island, Kingston, RI). The
antennae
were dissected from male and female moths one to three days after emergence,
frozen
on dry ice, and stored at -80 C.
Quantitative real-time PCR
Total RNA (isolated from antennae collected from 30-50 female and 30-50
male moths) and lst strand cDNA were prepared as described for 3'RACE, with
two
exceptions: Genomic DNA was digested with DNAseI during on-column total RNA
CA 02616144 2008-01-25
41
purification (Qiagen, Valencia, CA USA) and a second time immediately before
lst
strand cDNA synthesis using the DNA-Free kit (Ambion, WoodwardAustin, TX),
and, 15` strand cDNA was synthesized using the Invitrogen oligo dT(lg) primer.
PCR
primers were designed using ABI Primer Express 2.0 software (Applied
Biosystems)
set to select for an optimal primer annealing temperature of 59 C (58-60 C
range),
amplicon sizes of 50-150 bp, a 3'GC clamp=0 and a minimum and maximum GC
content of 30 and 80%, respectively. In general, primers were designed using
coding
sequence close to the 3' end of the gene, and where possible, primers spanned
an
intron (Table 2). Real-time quantitative PCR was performed using an ABI Prism
7900HT Sequence Detection System (Applied Biosystems) and SYBR Green dye
(SYBR Green PCR Master Mix, Applied Biosystems). The program began with a
single cycle at 50 C for 2 min, followed by a single cycle at 95 C for 10 min
and 40
cycles at (95 C for 15 sec; 60 C for 60 sec). Afterwards, the PCR products
were
heated to 95 C for 15 sec, cooled to 60 C for 15 sec and heated to 95 C for 15
sec to
measure the dissociation curves.
Transcript levels of each Or gene were quantified from total RNA extracted
from male and female adult antennae, and fifth instar larval abdomens, using
the
relative method (Relative Quantitation of Gene Expression, ABI PRISM 7700
Sequence Detection System, User Bulletin #2, Applied Biosystems ). The
efficiency
of each primer set was first validated by constructing a standard curve; a no-
template
control and six lOx serial dilutions of lst strand cDNA were prepared and the
CT value
(the cycle number at which the fluorescence intensity crosses the threshold
line
determined by the ABI Primer Express 2.0 software) of each Or gene calculated
at
each template dose (lx dose = 0.33 uL of lst strand cDNA). The CT value was
plotted
against the log(template dilution) and the slope and r 2 value of each
regression line
calculated. Expression of each Or gene in male and female antennae and larval
abdomens was assessed at a template dose equal to 0.33 uL of lst strand cDNA
per
well. A no-template control was included and all reactions were performed in
triplicate. Dissociation curves were used to assess the purity of the PCR
reactions.
Or gene expression levels were calculated relative to the control gene B. mori
ribosomal protein S3 (BmRPS3, Table 2) using the formula 2- CT (Livak and
Schmittgen, 2001).
CA 02616144 2008-01-25
42
a
...
y, ~. (A ~ 0 z 0 0 0
~
..r
N O~
i-.
Q Q
M "" O M l~ ~v ^
pp ^ Q 17~
00 00
A p
p a Q Q O~ Q O U ~ -
00
Q aQ Q ~o d o~ Q 00 a 0
(J
u o Q Q o~ a a~
uuha
" u~Qdu
~¾ ~ Q~U~
o. d C7 C7 C7
< u ~
Uo~Uu~~U~~Q~
< ¾ d d d ~ U d d ¾ d
8 d~ E~- ~~ d U 0 d H~ Q U d v~ Fd.,
C7~C7~~c~F-FdQuUu ¾u ¾~C7d
C7 U C7 d
U u ¾ ~ ~ Q H H < ¾ o ~ " ¾ u
~ h Q H ~uc~ U~ U U v c-' U
u U u C~7 u U U U U
o U C7 d~~ C7 d ~ . EU-+
CS
o U Q u Q v U
~T,
0
z ~3 ~3 a3 ~3
tn 110 l- 00 m O~ a O p p oo CD 00 O "O ~O !t
Vl - Vl N Vl Vl t(1 tn
p_ rA~, O~ o0
,c oo 00/1 ~ ~/1 ~ ~~=1 ~ ~/~ ~
V
~ d ddd d d d d d d d d w r~a W
m UaaU aaUaaUm U U
z
H N M V~ ~O [~ 00 0~ CD
CA 02616144 2008-01-25
43
` yz z z z ~ y^
o Q
`-" kn
a O~ Q r.=~ N -i o~0 N Q M
W O ^ "" -+ M ~ A -, -- ^ ^
.7: 00
ac~ aQ u aaQ Q~ Q~aQ aQ c~ a~ Q Q
o,~a
U < (3 U
U
U o Q ¾ < < " u ~ < ~ ¾ ¾
d C7 F~- d~ d C7 U d v d c7 v v d
b~ C7 U U u¾ ~ u c7 H~7 ~
dU U V E~. U H U dC7 F" C7 H VO ¾ U C~7 ¾ V U
¾dUQU UUU~U ¾~u c,¾j
~ Q d¾ d U d U d U u~ h du c7
U d U Q U 0 c¾.7 ~ C7 c,7 C~,7 ~~
Uu d c~7 Q v d v~ U c~ Q v v5 ¾ v Q v
H C7 ¾ u 17 U u ~7 u u C7
d F. H U c~
c~ u t7 H c7 Q C7 U U ~- ¾ QQ u~ E-. ~. u d F.,
C7 C7 C7 Q~ d U U U d V Q Q U F- C7 H U U
~: U E-U d U U dc U U CU (~ C7 C7 E- C7 U U U
d v c7 c; H~7 U q E~ y y c.7 H c7 q U c~ C7 P~ c7
a! w a w r~ w ci! w rs:, w rx w a w rx w a: w c~ w c~ w a;
A A A A A A A A A A A
00 OO =--.-,--.-.--.-+ .-,~ .-.-, N N
CA 02616144 2008-01-25
44
~. z ; z ya z z z z
00 M
N ^ M ^
00
00
N
CA a Q Q Q
0~~¾ w Q H~- p.~ o~ a
w ~ ~ C¾7 .w~'. a Q 4' ¾ v
Q~u¾~~¾~d
U U ¾ Q Q
u
U C-~ U U
b ¾ u U U u u
u ¾ ¾ ¾ ¾
¾ C5 ¾ CJ ¾ C. U u d
U~ Uu UU C-~ u 0 UC:
UUUu uUU
t~~~~Udd
E-' C7 d E- F- U U Q U Q
U U C
u U7
u u ~ ~
U u d Q U U U Ud U U H h c7 v U~
E-C7 H ~" H ¾ C~ C7 d~" d d d c7 ¾ d ~~" d F' ¾
C7 C7 d d F= U U C~ C7 C7 U C7 F- C7 U C7 d U U U
U F; U U U , d U U E-; U F~ F" F~ F; C7 F; U~ F~ Q? H
Q Q Q Q Q Q Q Q Q Q Q Q
* *
* *
0G
N N N N N N N M M M M M
CA 02616144 2008-01-25
z z z z z z ~ yz z 0
o n ~ `~ o
00 Q ~ =-~~ ~ ~ ~ Q M w ~ r-ti
o, 00 a ~ N
~Q aaQ kr) aQ
u
o,U a 'Q u 'Q Q o,a
Q Q U U ~ U Cd7 U U U Or
d~~ C7 C7 F- U E-r Ln
U U~ F" F U F- C7 E- E-a tn ¾
U u d d - ¾ ~~ U
U~C7
U~UUF-'~¾¾C7¾ U
~"~~~u¾H~~UU~
U
C7U~uu C~"UUC7F"U U
~E- d¾Q U ¾ U ~.., ~ c7 U~ d ¾ H ¾ C7 C7 C.D
~ Q~Q Q U U~' F~-0 U F¾-U~~ c¾7 U U~ V U U
U ¾ C7 U C7 0 F, ¾ H E" H U U
Q
~U ~ ~~~Q"¾~¾¾~u QQ~
U U u u
U U H¾~ C7 ¾~, ¾ C7 U¾¾ c7 U¾ U U
(-') U- U U U U~ U U~ C7 d U
CD
¾ c~.7 U cd7 ¾Q U d d C5
U ¾ Uu c.7 7
¾¾Q~ C7
c7 c U U d C'? U~; c7 H c~ U
rx w a~ w r~ w rs! w cx w c~ r~. r~ w rs:, w cx w c~ w rx w rs!
Q Q Q Q Q Q Q Q Q Q Q
E-FF-H E-E-E-E-1 H E-H
v~p l- 00
O~ O N M d n
M M M M M
CA 02616144 2008-01-25
46
z
Q Q A Q ~ ^
Cy CY w Q
W_ rWi~ ~ ~ Q
H H C7
C7 Q E-~ U `.'
d d d ~Q
U ¾ Q U CU7 U
~" ~.7 U Q¾ d ~
C
U 0 QU ¾ C~7 o 0 o a~i ~
U Q N N cV ^C
U U .. .~ .~ . ~
QQ U E-~ ~ ~ ~ c~
U C7 c.~7 V CQ7 ~ ~~~~ M
U
U P~
w csw w c~ w r~ a~ a~ .~ ~q .ti
r" 000~~
Q Q ~ f~CGf~ o
C~s
~ ~ d o 0 0~
-izi
~4 r4
d U M Y
~ ~ UC7I
m
kn
CA 02616144 2008-01-25
' 47
Biological replication
The antenna from individual moths (n = 4 female and 4 male moths) were
collected on dry ice and stored separately at -80 C. Total RNA extraction,
genomic
DNA digestion, 1 St cDNA synthesis and gene expression profiling were
performed
separately for each individual using the methods outlined in the above
section.
After normalizing the CT values of each Or to the control gene BmRPS3,
statistical differences between male and female antennae were tested by nested
Analysis of Variance (ANOVA) using the SPSS for Windows Release 11Ø01
statistical package (SPSS inc., Chicago, IL). Technical replicates (n = 3
replicated
wells on each qPCR plate) were nested within each biological replicate (n = 4
males
and n = 4 females).
Results
BmOr bioinformatics
The sequences of seven silkworm Ors have been published. BmOrsl-3 named
by Krieger et al., (2005) and Nakagawa et al., (2005) are synonymous. However,
BmOrs 4-6 named by Krieger et al., (2005) are not synonymous with BmOrs 4-6
named by Nakagawa et al., (2005). BmOrs 4-6 as named by Nakagawa et al.,
(2005)
are used herein since they were published first. The Ors referred to as BmOr4
and
BmOr6 by Krieger et al., (2005) are synonymous with Nakagawa et al., (2005)'s
BmOr5 and BmOr4 respectively. The Or referred to as BmOr5 by Krieger et al.,
(2005) shares 88% amino acid identity with Nakagawa et al., (2005)'s BmOr5
(their
coding regions share 93% nucleotide identity), but since they are located on
different
genomic scaffolds, it is apparent they are different genes. Therefore, Krieger
et al.,
(2005)'s BmOr5, which is a unique gene, is referred to herein as BmOr7 (J.
Krieger,
Personal Communication).
Each new Or identified herein was named consecutively beginning with
BmOr8 (see Table 2 for a summary, including GenBank accession numbers). The
term chemoreceptor (Cr) is used to refer to the superfamily that includes the
odorant
and gustatory receptors (Ors and Grs).
Genomic scaffolds that encode unique Or exons were identified by conducting
TBLASTN searches of the silkworm genome with known insect Or proteins
(http://kaikoblast.dna.affrc.go.jp/, Silkworm Genome Research Program,
National
Institute of Agrobiological Sciences, Japan and
http://silkworm.genomics.org.cn/,
Beijing Genomics Institute, China) (Table 2). Or genes were annotated manually
and
assigned a new number beginning with BmOr8 (Table 2). In several cases the
genomic scaffolds encoded only partial Or gene sequences due to their limited
size
and the presence of numerous intron sequences within the Or genes (BmOrs 45-47
in
CA 02616144 2008-01-25
48
Figure IA for example). The N50 scaffold size (50% of the assembled genome
sequence consists of scaffolds equal to or greater then the N50 value) of the
silkworm
genome is 26.9 kbp (Xia et al., 2004) compared to 990 kbp for the Tribolium
casteneum genome for example (Human Genome Sequencing Center, Baylor College
of Medicine, ftp://ftp.hgsc.bcm.tmc.edu/pub/data/Tcastaneum/Tcas2.0/README-
Tribolium.txt). In some cases 3'and/or 5'RACE was used to amplify and sequence
partial cDNA fragments in order to further annotate the gene structure. The
coding
regions and the translated protein sequences of BmOrs8-48 are presented in
Supplemental Figure S2).
Although a complete assembly is not available, limiting the ability to assess
Or
clustering in the genome sequence, 14 Ors occur as seven tandem pairs on seven
different scaffolds (BmOrs5 & 7, 19 & 20, 21 & 22, 23 & 24, 33 & 34, 38 & 39
and
46 & 48) providing evidence for some Or clustering. Tandem Ors share amino
acid
sequence similarity and group together on the phylogenetic tree (see below)
suggesting they arose from recent gene duplication events. The nucleotide
sequence
of the open reading frame (ORF) of some paired Ors (e.g. BmOrs5 & 7 and 33 &
34)
are almost identical (93 and 97% nucleotide identity, respectively) making it
difficult
to design unique primer sequences that also conform to the parameters required
for
real-time quantitative PCR.
The amino acid sequences of Ors from different insect orders have only low
levels of identity, and tend to form order-specific lineages in phylogenetic
trees (e.g.
Hill et al., 2002; Robertson and Wanner, 2006). However, there are some
typical
traits that can be used to support their designation as Ors. The last three
intron/exon
boundaries of many insect Or genes are spliced in the zero phase (between two
codons rather then within a codon) a trait typical of the insect Ors in
general
(Robertson et al., 2003). The last exon is typically the smallest, and encodes
some of
the few highly conserved amino acid residues, particularly a Ser/Tyr/Ser motif
(Figure
1B). A Ser residue (or in some cases a Thr residue) has been conserved in many
insect Ors at the 7th codon position past the typical phase 0 splice site of
the last
intron/exon boundary. These features appear to have been broadly conserved in
many
(but not all) Or genes found in insect genomes sequenced to date (Robertson et
al.,
2003 and Robertson and Wanner, 2006). Examples include BmOrs19, 30, 45-47,
AgOrl and DmOr46A in Figures IA and IB; these seven genes all have a phase 0
spliced intron/exon boundary located seven codon positions before the
conserved Ser
residue.
CA 02616144 2008-01-25
49
BmOr phylogenetics
Including the seven unique Ors published by Krieger et al., (2005) and
Nakagawa et al., (2005), the total number of silkworm Ors now identified is
48. A
neighbor-joining phylogenetic tree using corrected distances was constructed
using 43
BmOrs (five partial sequences were excluded) and the 18 HvOrs that have been
reported to date (Figure 2). Krieger et al., (2002) reported the sequences of
21
chemoreceptors (Crs); 3 group together phylogenetically with the insect
gustatory
receptors (Grs), and 18 with the insect Ors (Robertson and Wanner, 2006),
which are
referred to herein as HvOrs for consistency.
Two results become evident from this analysis: first, most of the Or lineages
are represented by both species, indicating diversification prior to the
evolution of the
ancestral bombycid and noctuid moths. All lineages with bootstrap support
include
representative Ors from B. mori, and even though fewer Ors have been
identified from
H. virescens (less then half compared to the silkworm), only three Or lineages
lack
HvOrs. Several Ors may be orthologous; examples include BmOrl3 & HvCr8,
BmOrll & HvCr7, BmOrl4 & HvCr20, BmOrl8 & HvCr3, BmOr22 & HvCr19,
BmOr24 & HvCrl2, BmOr25 & HvCr9, BmOr26 & HvCrl7, BmOr41 & HvCr10
(Figure 2).
Second, it is apparent that members of some of the lineages may share
conserved functions (Figure 2). Seven B. mori and six H. virescens Ors group
together in a single lineage that includes the pheromone receptors. All but
two
(BmOr9 and HvCr6) are expressed at higher levels in the male compared to
female
antennae (Table 3; Figure 3; Sakurai et al., 2004; Krieger et al., 2005;
Nakagawa et
al., 2005). Furthermore, the pheromone receptors appear to form two main
lineages,
one that expanded in the bombycids and the other expanded in the noctuids. The
bombykol receptor (BmOrl) may be an ortholog of HvCrl3, a receptor that is
believed to detect the primary component of the H. virescens sex pheromone
(Gohl
and Krieger, 2006). Interestingly, this lineage has expanded in B. mori, while
the
lineage containing bombykal receptor (BmOr3) has expanded in H. virescens.
Similarly, BmOrs 45-48 appear to form a lineage of Ors whose expression is 6-8
times higher in female antennae (Figures 2 and 3, Table 3); whether this
lineage also
includes an orthorlogous Or from H. virescens remains to be determined.
The results of BLAST searches for each of the novel polypeptides performed
when they were identified are provided in Table 4 where the closest known
polypeptide at that time is indicated.
CA 02616144 2008-01-25
Table 3. Ratio of silkworm Or gene expression levels in female compared to
male
adult antennae determined by quantitative real-time PCR. Or gene expression
levels
in female and male antennae were calculated relative to the control gene
BmRPS3
using the equation 2- CT (Livak and Schmittgen, 2001). Or expression values in
5 female antennae were divided by values in the male antennae and are reported
as a
female : male ratio. Ors whose expression was not detected in adult moth
antennae
(BmOr20, 21, 22, 25 and 42) are not included in the table.
BmOr No. female : male expression in anntennae
1 0.0001
2 0.9
3 0.0001
4 0.06
5&7 0.02
6 0.02
8 0.9
9 1.4
10 1.7
11 0.9
12 3.2
13 0.4
14 0.9
15 2.6
16 0.4
17 2.3
18 1.1
19 831.0
21 0.6
24 0.3
26 1.7
27 2.6
28 1.0
29 1.5
30 90.0
31 0.9
32 1.6
33&34 0.2
35 1.7
36 1.7
37 1.8
38 2.0
39 1.0
40 0.7
41 1.4
43 1.1
44 1.2
45 7.0
46 6.0
47 8.6
CA 02616144 2008-01-25
51
0 0 0 R ~ ~ ~ R
0~ 0~ 0 0~0~ 0 0~ 0~0~ 0 0 0 0~0~0~ 0
C 00 N qT .-+ N N N Q) 01 N CO L!1 00 l0 M M N Nqt .-i r-i
G) * + N l!'f It Ill N N N qt .--i 1- M M qt d' Mqt qt N Nqt
U C
L L Q L (n
~../ z N Oo C(- M ~ U
L L E L (6
O O 0 M
~ n. Q ^ O. ~
U) ~ a
u a a ~ a u '
J..i L L O L.n O L L QL
N r1
~~ O 0 U L 0 L (n cn O O Vf O U E
^
O O C C a ~ Q c J a)
U a-+ O N a) - 01 N N
0- In , I Q N O O U n U p NNM pp
L z O U E F ~ M N E L L L ri L L Q rl
O++ N _ po L O O O L O O U) O
aE ~ LL NL L oL aaao aaC'-'~ 0
a) U U C U O O O Q= U O N N N NLn~ a a
UOa) 'w mam UUU~UUO
U
W qt > 01 > c O c U > a) a) L ~ a)
L~ c y, ri (n U a) lI1 w
~ 0 ~1 N(p L(p o O O N
~Qn L - o a 00 a aE i c E o a m vQ~ uQ~ c n c n
~ O O~ L O L L tL0 ~ L N N N C N~> J O C -W 4-1 u tn O a= O ,~ O U U O Ln tn
(A uf cn +~ O
G) O V V U`~ a, O~ O~'- O O O O O~~ O
Ewwwa UQ >o>o¾ EEEoEED oo
O L L > z ~ Z z 0 r+'0 E Z W E W W v Y i r~0 a L~ jj~~ u U U ~ v U Q L ~ a a~
~ ~ E a a U E N N N U a) a) V z .U
> L U a M o m L - L ~ ~ > Q > > ~
i; O~ L~ oE oo 0 0> o a Z L can >
a=i 41aaioo p ~a Qi
a a >>>;o~~z o
' oC z~oz aaa aaE ow,.'
"~ L O Z ~ E a~ L L L a L L ~ ~~
oz u u E E o u o 0 0 0,a ~ oi a
in Q Q u~ v (n a ~ w cn Q Q Q'~- Q Q N O `~
~ o > >- > > ~~z Z z z ~ ~ z
~ u~ ~ z
~o~'~~n ov~ a'QaW ~ EEE~EE
= y,U~, L. ~ a O- t UI Q tn E m m m c C v) ul ~(o E
N a C o U) -w Q4~ C C ( A 4~ c c CM C C >.0 (/)
-o v o c - u ~ u c - v u u c u v ~ c
Uf E m V~ O 4 0 w(n V VI W(n V!n m~ a.~.+ (9 U
x cn = " _ ~ ~ ~ ~ ~ ~ = O v, u,
0 > ~ O .~ N Z M > UQ > ~ O > > > W > > a ~ ~
v~ ~~> m oo ~n m_> v_~ u~ v_~ Vf > v~ v_) ~ oo o >
~ ,= Q ~ ln L L L L L (n
x 0 E L ~+ J-I J-+ J-I a-+ J-+
o ~oo oo Qa O~oo 0 o O~ O Qaoz o~
a - a~ =- c=-
~ o -
N> ~> 2'~S O> 222 O=2 E a v
00 2 = a= _O = _ ~
00
- p=~ =~ ,i.i,i _.i,i,~ C
N d N M ln *"I l11 oo lo to qt N N d' U
-C MIt ~p Uj 00 U O M N Nkp r, M M M N N oo tn^~vj
Q M 0 N~ N V M'~ M M[t M M m M M(~'7 [t ~ O M
00 O L!1 MN N 00 0 00 M I~ oo oo oo M Oo oo = O M
to CO 00 00 V V= pO 00 V d' ~ ap ~ oo > N pp
N O Cp > Ct I, O I~ ~~ I, N I~ N N V= cn M~
r, I r, r, I r, r-,
Q m N
Q V) Q Q Q ~ Q Q Q Q^ Q Q Q4J~ ~
? < Q ~,.~ o X <
~ E r,.n E E E E E~ E E E
E E E a~
~ nnvno,=75 no,nnn~na,nnn~nnn=n~
O
Q 0
41
O.--i N M ~ ln ~p N 00 Q) O+--i N CM qt ln %O N oo
N VW~~ L. L. i i ~ i i i i L N L N N N N N N L
~ 2000 oo oo ooooo oooooooooo
M m p~ E E E E E E E E E E E E E E E E E E E E E
mm mm mm mmmmm mmmmmmmmmm
CA 02616144 2008-01-25
52
~
0~ 0~ ~ 0~ 0~ 0~ 0~ 0 0~ 0~ IR 00-
NIt NN0) 0 t0 N 1DN00 .-+ It t+') tD
NIt N N.--i N .-{ Ln l0 N N It N N .--i
~ ~ '0 L L
'0 U
CO V ~ M CO a)
L Q) Q) L L 0)
a
C ,~ ~
C ~J a O 0 E E a
o M ..
o a E o a) a)
a~ 0 u a) a) II
O O O O L Q u O O L L O O O
N.-I N N 1-1 Z~ p) 1 a' 4
L L L L 0 L Lc~ 0 0 C L 0 L
O O O O Uf E Z O O tn Ln 4) O U O
a a aa ~ 0 E a aE ^ ac a
u u u u o *+ - v u co 0 0 ,~ u L u
L) L) L) CA~ L) a) ce- L~ O i)
12 ci O 0~ -c a c
o O o o u Q~ O O u u Q) O~ O
m(n v) u~ m ~n (7 u(n (n
c c c c~ c ca) a) v c c
~ ~ ~ UNI + M " ~ ~ ^ + 'a L twA N (OJf
O O~
O O O O~ N
E E E E c in 00 E Er. 0 E u E
w w oa) a o ~ w a) o a a c a~ ~ o
~ L L L L O 0 ~~ O L L O~ > t
u u u U 0 0 0 D U O O O uf U i+ u
0 O "- O ~
> > > > Q 0 O > > O Q Q E > >
i+ a=+ i+ i-+ Z 0 6' ~ Z Z O~ a~
~ ~ ~ ~ o ~ ~ Q. a u ~ o
a a 0- aE z 0- aZ E E a) a`~ o.
L L L L fO L L Q L
Q z Q
O O O U) Q O O~ t O~ O
QQ QQ a w ~ aQw a a ~Q E Q
~ ~ ~ ~ F- z LU ~ ~ ~ ~ ~
EE EEaci W~ EEWa a2EaciE
r_a r_o (0 (0 'U^ W f0 ra 0- vUi vUi Q ~p (1I
a) a)
; Z !E a)
+ }
aa aa> ~~ aa~ > > E a> a
(n v Ln ~ m cn m
(A tn VI (A !E f0 O (A tn (0 (n (n :E tn
C C C C y~ C C0-+ 4~ C C y~ C
4J N N 4) O N 4) O O 4) N O N
u u U U E a ~ u u E _ -U U_ U
a)
co a~ a~ a~
ro E
~~ a a Q, ~ ~~ p1 S S U~LJ ~ S ~
> > > ~ O > > O 00 M > > >
_N _~ _N ~ O N W ~ ~ O ~
= ~ L t N ~- ~
O 0 O O-
0 0 Op 00 OO Op O
O O O O 0 --
C_ N N N N C O O N ~r N N~ N
~SS > < = SSa> > SS>S
~__ _ - Q _ _S S __S _
c ~,-~ ,~.~=~ M ,--i,--~'"',~ =~ =~=~,~,~
L6 Lf1 Ln ln M N Ln L!1 M 00 M N Il) 4 In
O M N M fi) 00 N I-~ N Nu) r, Co f'e) N 00 N
4'' M CM M(e)Nr '""I M M M~ qt m Mlqt M
>- CO Cp >Op Op N 0 00 >~ >, 00 0o N I*l CO CO N 00
O "j- O d' It 00 '""i N 01 01 d' "t N 00 00 ~It 00 qt
O O N I , ~ M O O I , I-, M qt N N qt i,
-5 75 - -
OQQ OQQa E E QQmiQ < QQQQ
o o o o .n x
E Es E E E c v ~ c c E E~ E~ E~ E E E E
O U O o O N N O L-O O O 0 4) N L O O N O N N N N
Z A A Z A A A pi A u L c c A A A A pi n0) A A A A
01 O+--i N M,I- Ln t0 f, 00 0) O.--i N M t u1 tD f- 00
L L L L L(L L (L L L L L L L L L L L L
OC OC OC OC OC c O Oc OC 0000000 OC OC Oc OC OC OC OC OC OC O
C C C C C C C E C C C C C C C E C C C E
mmmmmmm m mmmmmmm co mmmm
CA 02616144 2008-01-25
53
BmOr expression patterns
Real-time quantitative PCR was used as an accurate method to survey for sex-
specific Or gene expression in pooled antennal samples collected from 30-50
moths to
blend individual variation. All primer sets were first validated by completing
a
regression analysis of the CT value (the cycle number at which the
fluorescence
intensity crosses the threshold line determined by the ABI Primer Express 2.0
software) versus the log value of 10-fold dilutions of the template. Serial 10-
fold
dilutions will result in log linear slopes equal to 3.3 if the amplification
efficiency is
100% (Relative Quantitation of Gene Expression, ABI PRISM 7700 Sequence
Detection System, User Bulletin #2, Applied Biosystems). The primer sets used
in
this experiment yielded slopes within 10% of 3.3, with r2 values typically
greater then
0.99.
Or gene expression in male and female antennae was normalized to the control
gene BmRPS3 using the equation 2- CT (Livak and Schmittgen, 2001). BmRPS3 was
consistently expressed at similar levels between the different tissues tested,
indicating
that the total RNA quantification was consistent. For example, CT values for
BmRPS3
were 15.2, 16.7 and 16.6 in abdomen, female antennae and male antennae,
respectively. Abdomen tissue from larvae was included as a control reference
since it
lacks any known olfactory sensilla (chemosensilla occur on the adult female
ovipositor on the abdomen). CT values measured in abdominal tissue averaged
31.2
( 1.1, 99% confidence interval) for all Ors tested, while values measured in
antennal
tissue ranged from 18 to 30. Or gene expression values in the abdomen tissue
were
consistently 10-4 times or more lower relative to BmRPS3 (Figure 3, minimum
values
rounded up to 10"4 for presentation). A no-template treatment included in each
experiment controlled for template contamination. PCR products were analyzed
by
agarose gel electrophoresis to exclude contaminating genomic DNA as a source
of
template approximately half of the primer sets spanned an intron) and the
purity of
each PCR product was assessed by analyzing its dissociation curve. Or
expression in
the abdomen was typically 10-4 times that of BmRPS3, while Or expression in
female
and adult male antennae typically ranged from 10-3 to 1 times that of BmRPS3.
BmOr19 and BmOr3O were expressed abundantly in female (but not male)
antennae at large female-biased ratios of 831x and 90x respectively (Table 3,
Figure
3). This pattern of expression is opposite to that of the pheromone receptors
BmOrl
and BmOr3 that are expressed in male but not female antennae (Table 3; Figure
3).
BmOr19 is expressed at very high levels in female antennae, while BmOr3O is
moderately abundant. In the phylogenetic tree (Figure 2) BmOr19 and 30 do not
group together. BmOrs 8, 17, 20-22 and two H. virescens Ors (HvOrsl9 and 21)
form
a lineage with BmOrl9. Expression data is not available for HvCrsl9 and 21,
and the
CA 02616144 2008-01-25
54
other silkworm Ors found within this group were expressed at low levels or
were not
detected, with the exception of BmOrl7 (Figure 3). BmOrs14 and 33 & 34 (a
tandem
pair of Ors that share 97 % amino acid identity) along with two H. virescens
Ors
(HvOrl8 and 20) form a lineage with BmOr3O. BmOrs14 and 33/34 transcripts were
detected in adult moth antennae and no sex-biased expression was observed;
expression data is not available for HvOrs 18 and 20.
Several Or transcripts were moderately more abundant in female compared to
male antennae. In contrast to BmOrs 19 and 30, BmOrs 45-47 were expressed
abundantly in both female and male antennae (46>45>47), but at ratios 6-8
times
higher in female compared to male antennae (Table 3; Figure 3). BmOrs 45-48
all
group together and may represent a lineage of Or genes with female biased
expression
(Figure 2). BmOr12 was also expressed at very high levels in both sexes, and
was
three times more abundant in female antennae. BmOrl2 groups together with two
other silkworm receptors that are expressed at moderately high levels in the
antennae,
BmOrs 13 and BmOrl5 which expressed 2.6 times higher in female antennae (Table
3, Figure 2), and with H. virescens Or8.
The abundance of different Or transcripts relative to BmRPS3 varies over at
least three orders of magnitude (Figure 3). Or gene transcripts that were 1000-
fold
less abundant relative to BmRPS3 are probably associated with a small
proportion of
the antennal sensilla. Conversely, abundant Or transcripts are likely
expressed in
commonly occurring olfactory neurons. For example, transcripts of the two
pheromone receptors BmOrl & 3 were among the most abundant, their levels were
equivalent to that of the control gene BmRPS3. Correspondingly, olfactory
neurons
tuned to the sex pheromone are associated with the most abundant class of
sensilla on
the male antennae (Heinbockel and Kaissling, 1996).
Real-time quantitative PCR accurately detected the sex-biased expression of
five Or genes (BmOrl & 3-6) reported to be expressed at higher levels in male
than
female antennae (Table 3; Sakurai et al., 2004; Krieger et al., 2005). Of
these, BmOrl
and 3 transcripts were highly abundant in male but not female antennae (Table
3),
consistent with their function as pheromone receptors. BmOrs 5 and 6 are the
next
most abundant male-biased Or genes and expression of each is approximately 50x
higher in male antennae. BmOr 4 is expressed at moderate levels in both female
and
male antennae, yielding a moderate male-biased ratio of approximately 17x
(Table 3).
The functions of BmOrs 4-6 have not been characterized. The primer sets used
in this
experiment did not distinguish between BmOrs 5 and 7, or between BmOrs 33 and
34.
The inventors assayed expression levels of the five male biased (BmOrs 1, 3,
4, 5/7, and 6) and five female biased (BmOrsl9, 30, 45, 46 and 47) silkworm Or
genes in antennae collected from individual moths (n = 4 male and 4 female
moths)
CA 02616144 2008-01-25
(Figure 4). In each case, the results from this experiment using biological
replication
supported with statistical significance the male biased expression of BmOrs 1,
3, 4,
5/7, and 6, and the female biased expression of BmOrs19, 30, 45, 46 and 47,
obtained
in the previous experiments that used antennae from pooled individuals
(Figures 2 and
5 4, Table 3). The levels of BmOr2 and BmOr9 were not statistically different
between
male and female antennae, also consistent with previous experiments where a
significant sex-biased ratio was not observed (Table 3).
Discussion
10 The present inventors have identified 41 B. mori Ors bringing the total
number
to 48. The constructed B. mori genes were assigned to the Or family based upon
amino acid similarity with known insects Ors and characteristic gene structure
and
amino acid motifs. Supporting their annotation, the majority of the putative
Or genes
were expressed in the antennae of adult moths. Several of the Or genes may be
15 expressed exclusively in the larval sensory organs, as is the case in D.
melanogaster
(Fishilevich et al., 2005), explaining the failure to detect them in adult
antennae
(BmOrs2O, 22, 23, 25 and 42).
In phylogenetic analyses, insect odorant receptors generally form order-
specific branches reflecting rapid rates of evolution within the gene family
(Robertson
20 and Wanner, 2006). This pattern is also observed between distantly related
groups
within the same order, such as the mosquitoes and flies, one of the earliest
taxonomic
divisions within the Diptera (Hill et al., 2002). This high degree of
divergence in the
amino acid sequences exhibited by the Ors makes it very difficult to use
homology-
based discovery approaches to identify the Ors from insects whose genomes have
not
25 been sequenced.
Regardless, the silkworm Or genes identified herein can be used as probes to
screen cDNA libraries constructed from the antennae of important pest species
found
in other taxonomic families such as the Noctuidae. Homology-based approaches
will
be particularly useful to identify Ors that mediate important pest behaviors
such as
30 host selection, feeding and oviposition.
BmOrl & 3 are not expressed at significant levels in female antennae (Figure
3; Sakurai et al., 2004; Krieger et al., 2005). Absence of a receptor from the
antennae
of one sex (as opposed to expression in both sexes at a biased ratio) suggests
that it
may mediate an olfactory behavior entirely specific to that sex, as is the
case with the
35 silkworm sex pheromones. BmOr6 on the other hand is expressed in both male
and
female antennae, but 50 times more abundantly in male antennae (Table 3),
suggesting that the odor(s) that its detects may mediate behaviors more
prominent in
male moths (such as unidentified female pheromones or conspecific odors). Some
H.
CA 02616144 2008-01-25
56
virescens sensory neurons on the female antennae respond to one of the six
female
produced pheromone components (Hillier et al., 2006), a scenario that could
explain
expression of the receptor in both sexes but at greater levels in male
antennae.
Similarly, several silkworm Or genes are expressed at moderate to large
female-biased ratios. BmOrs45-47 are expressed abundantly in the antennae of
both
sexes, but six to eight times higher in the females. This dimorphism may
reflect an
enhanced sensitivity of female antennae to specific odors used for host plant
discrimination. Interestingly, BmOr19 and BmOr3O are expressed at large female-
biased ratios (approximately 800 and 90 times, respectively). BmOr19 is likely
expressed in a high proportion of olfactory neurons on the female antennae
based
upon the high levels of its transcripts in female antennae.
Comparatively, BmOr3O is less abundant and therefore likely expressed in a
smaller proportion of sensilla on the female antennae. Based on their low
levels of
expression in male antennae, BmOrl9 and BmOr30 may mediate the detection of
female-specific olfactory cues that mediate female-specific behaviors. These
results
are consistent with the existence of two sexually isomorphic glomeruli on the
antennal
lobes of adult B. mori females (Koontz and Schneider, 1987). Similarly, the
antennal
lobes of adult M. sexta females also have two enlarged glomeruli, one of which
responds to linalool (King et al., 2000; Rospars and Hildebrand, 2000;
Reisenman et
al., 2004), a fact that may indicate the conservation of specific olfactory
pathways in
female moths. Koontz and Schneider (1987) suggested that such pathways may
function in the detection of odors related to host plant selection for feeding
and
oviposition or for the detection of male produced sex pheromones. As many as
66%
of the sensilla tested on female H. virescens antennae responded to
conspecific
chemical cues, including pheromones produced by the male that influence mating
behavior (Hillier et al., 2006).
Example 2 - Analysis of receptor-ligand binding
Functional analysis was performed using a calcium imaging assay which has
been described by Kiely et al., (2007). Briefly Spodoptera frugiperda (Sfl9)
cells were
transiently transfected with a pIB vector using Escort IV (Sigma). Transfected
cells
were incubated for 48hours to allow the expression of olfactory receptor
before
calcium imaging of responses to ligands were assessed. Fluo4 was used as a
calcium
indicator and fluorescence images were recorded using a Leitz digital still
camera.
Images were recorded every 10 seconds for 50 seconds after the addition of;
saline (as
a control), the test ligand and lonomycin (to determine maximal fluorescence).
Images were analysed using Metafluor imaging system and AF was calculated for
given concentrations. AF is determined as the ratio of change in fluorescence
from
CA 02616144 2008-01-25
= 57
basal levels after the addition of a ligand to maximum change in fluorescence
from
basal levels after the addition of lonomycin.
EC50 curves were created in Graphprism with all points based on the average
AF of at least 4 individual cells.
Following the same methodology described for the expression and
characterization of EposOr3 (PCT/AU2007/000510), BmOr19, 30, 45 and 47 were
screened for detection of 25 different odours (Table 5) representing a diverse
range of
chemical properties. Each of these molecules are plant volatiles and/or
components of
insect pheromones, and hence uncoupling of signalling will effect insect
behaviour.
Functional assays were carried out at final concentrations of 10"6 and 10"$
for
each odour.
Table 5. Odours tested in calcium aging in assays
a-pinene Citral Limonene
a-terpineol Ethyl-benzoate Linalool
p-cresol Ethyl-butyrate Methyl benzoate
1,4-cineole Ethyl-hexanoate Methyl salicylate
2-phenyl-ethanol Eugenol Myrcene
Benzaldehyde Farnesene Octen-3-ol
Benzoic acid Geraniol Trans-2-hexanal
Butanal Geranyl acetate
Caryophyllene Hexanol
Linalool was the only ligand detected that produced a response from BmOrl9
(Table 6, Figure 5).
Table 6. EC50 values for B. mori Or odours.
Receptor Odour EC50
BmOr19 Linalool (4.69 1.57) x 10"9 M
BmOr45 Benzoic acid (1.44 1.36) x 10-10M
2-phenyl-ethanol (8.89 2.59) x 10-9 M
Benzaldehyde (5.86 1.67) x 10-9 M
BmOr47 Benzoic acid (1.42 1.52) x 10""M
BmOr45 responded most highly to 2-phenylethanol, benzoic acid and
benzaldehyde (Table 6, Figures 6 to 8). Smaller responses were seen for ethyl
benzoate and methyl benzoate although these were inconsistent and EC50 data
was
not obtained.
CA 02616144 2008-01-25
58
BmOr47 responded highly to benzoic acid (Table 6, Figure 9) and only showed
very weak responses to high concentrations of 2-phenyl ethanol and
benzaldehyde.
BmOr3O did not respond to any of the tested ligands.
It will be appreciated by persons skilled in the art that numerous variations
and/or modifications may be made to the invention as shown in the specific
embodiments without departing from the spirit or scope of the invention as
broadly
described. The present embodiments are, therefore, to be considered in all
respects as
illustrative and not restrictive.
All publications discussed and/or referenced herein are incorporated herein in
their entirety.
This application claims priority from US 60/971,133, the entire contents of
which are incorporated herein by reference.
Any discussion of documents, acts, materials, devices, articles or the like
which has been included in the present specification is solely for the purpose
of
providing a context for the present invention. It is not to be taken as an
admission that
any or all of these matters form part of the prior art base or were common
general
knowledge in the field relevant to the present invention as it existed before
the priority
date of each claim of this application.
CA 02616144 2008-01-25
59
REFERENCES
Ai and Kanzaki (2004) J Exp Biol 207:633-644.
Altschul et al (1997) Nucleic Acids Res 25:3389-3402.
Buck and Axel (1991) Cell 65:175-87.
Chandrashekar et al. (2000) Cell 100: 703-711.
Chien et al. (1991) Proc Natl Acad Sci USA 88:9578-9582.
Clyne et al. (1999) Neuron 22:327-338.
Coon et al. (1989) Proc Natl Acad Sci USA 86:1703-1707.
Duchamp-Viret et al. (1999) Science 284:2171-2174.
Dunbrack et al. (1997) Folding and Design 2:R27-42.
Fishilevich et al (2005) Curr Biol 15:2086-2096.
Gao and Chess (1999) Genomics 60:31-39.
Gohl and Krieger (2006) Invert Neurosci 6:13-21.
Grosse-Wilde et al (2006) Chem Senses 31:547-555.
Hallem et al (2006) Annu Rev Entomol 51:113-135.
Hallem et al. (2004) Nature 427:212-213.
Harayama (1998). Trends Biotechnol 16: 76-82.
Haseloff and Gerlach (1988) Nature 334:585-591.
Heinbockel and Kaissling (1996) J Insect Physiol 42:565-578.
Hildebrand and Shepherd (1997) Annu Rev Neurosci. 20:595-63 1.
Hill et al. (2002). Science 298:176-178.
Hillier et al (2006) J Comp Physiol A Neuroethol Sens Neural Behav Physiol
192:199-219.
Howard et al. (2001) Trends Pharmacol Sci. 22:132-140.
Jeanmougin et al (1998) Trends Biochem Sci 23:403-405.
Kashiwayanagi (1996) Biochem. Biophys. Res. Commun. 225:666-671.
Kiefer et al. (1996) Biochemistry 35:16077-16084.
Kiely et al. (2007) J Neurosci Methods 159:189-194.
King et al (2000) J Neurosci 20:2391-2399.
Koontz and Schneider (1987) Cell Tissue Res 249:39-50.
Krautwurst et al. (1998) Ce1195:917-926.
Krieger et al. (2002) Eur J Neurosci 16:619-628.
Krieger et al. (2004) Proc Natl Acad Sci USA 101:11845-11850.
Krieger et al. (2005) Eur J Neurosci. 21:2167-2176.
Larsson et al. (2004) Neuron 43:703-714.
Livak and Schmittgen (2001) Methods 25:402-408.
Malnic et al. (1999) Ce1196:713-723.
CA 02616144 2008-01-25
= 60
Mita et al (2004) DNA Res 11:27-35.
Nakagawa et al. (2005) Science 307:1638-1642.
Needleman and Wunsch (1970) J. Mol. Biol. 48:443-453.
Oda et al. (2000) J Biol Chem. 275:36781-36786.
Perriman et al. (1992) Gene 113: 157-163.
Pophof (1997) Physiological Entomology 22:239-248.
Priesner (1979) Ann Zool Ecol Anim 11:533-546.
Reisenman et al (2004) J Neurosci 24:2602-2611.
Robertson and Wanner (2006) Genome Res, In Press.
Robertson et al. (2003) Proc Nat Acad Sci USA 100: 14537-14542.
Rospars and Hildebrand (2000) Chem Senses 25:119-129.
Rutzler and Zwiebel (2005) J Comp Physiol A Neuroethol Sens Neural Behav
Physiol
191:777-790.
Sakurai et al. (2004) Proc Natl Acad Sci USA 101:16653-16658.
Sato et al. (1994) J. Neurophys. 72:2980-2989.
Schmidt et al (2002) Bioinformatics 18:502-504.
Schneider (1992) Naturvissenschaften 79:241-250.
Shippy et al. (1999) Mol. Biotech. 12: 117-129.
Silver (1998) Methods Cell Biol. 56:237-251.
Smith et al. (2000) Nature 407: 319-320.
Swofford (2001) PAUP*. Phylogenetic Analysis Using Parsimony (*and Other
Methods). Version 4. Sinauer Associates, Sunderland Massachusetts.
Trotier (1994) Semin. Cell Biol. 5:47-54.
Vargas (1999) Chem. Senses 24:211-216.
Vosshall et al (1999) Ce1196:725-736.
Waterhouse et al. (1998). Proc. Natl. Acad. Sci. USA 95: 13959-13964.
Xia et al (2004) Science 306:1937-1940.
Zhao et al. (1998) Science 279:237-242.
CA 02616144 2008-01-25
DEMANDES OU BREVETS VOLUMINEUX
LA PRESENTE PARTIE DE CETTE DEMANDE OU CE BREVETS
COMPREND PLUS D'UN TOME.
CECI EST LE TOME DE _2
NOTE: Pour les tomes additionels, veiilez contacter le Bureau Canadien des
Brevets.
JUMBO APPLICATIONS / PATENTS
THIS SECTION OF THE APPLICATION / PATENT CONTAINS MORE
THAN ONE VOLUME.
THIS IS VOLUME 1 OF 2
NOTE: For additional volumes please contact the Canadian Patent Office.