Note: Descriptions are shown in the official language in which they were submitted.
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
INHIBITION OF VIRAL GENE EXPRESSION USING SMALL
INTERFERING RNA
CROSS-REFERENCE TO RELATED APPLICATIONS
[0001] This application claims the benefit of PCT application
PCT/US2005/032768, filed
September 12, 2005, which claims priority under 35 U.S.C. 119 from U.S.
Provisional
Application No. 60/608,574, filed September 10, 2004, both of which are
incorporated herein by
reference in their entirety.
STATEMENT REGARDING FEDERALLY SPONSORED RESEARCH
OR DEVELOPMENT
[0002] This invention was made in part during work supported by grant no.
5R43AI056611
from the National Institutes of Health. The government may have certain rights
in the invention.
FIELD OF THE INVENTION
[0003] The invention relates to inhibition of viral gene expression, for
example, hepatitis C
IRES-mediated gene expression, with small interfering RNA (shRNA and siRNA).
BACKGROUND OF THE INVENTION
[0004] Treatment and prevention of Hepatitis C virus (HCV) infections remains
a major
challenge for controlling this worldwide health problem; existing therapies
are only partially
effective and no vaccine is currently available. Hepatitis C (HCV) virus
infects more than 170
million people worldwide and is the leading cause of liver transplants.
Existing treatments,
including ribavirin and pegylated interferon alpha, are effective only in
approximately 50 percent
of patients and have substantial side effects. The development of more
effective HCV treatments
is hampered by the lack of a good small animal model, the inability to stably
culture the virus in
tissue culture cells, and the high viral mutation rate [1-3]. The availability
of an HCV replicon
system has allowed the study of HCV replication, host-cell interactions and
evaluation of anti-
viral agents, and more recently, a transgenic chimeric humanized mouse liver
model was
developed that allows full HCV infection [4-7]. Moreover, the use of in vivo
imaging of HCV
1
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
IRES-dependent reporter systems has facilitated efficient evaluation of
delivery and inhibition by
anti-HCV agents in mouse liver over multiple time points using the same
animals [8].
[0005] RNA interference is an evolutionarily conserved pathway that leads to
down-
regulation of gene expression. The discovery that synthetic short interfering
RNAs (siRNAs) of
aboutl9-29 base pairs can effectively inhibit gene expression in mammalian
cells and animals
without activating an immune response has led to a flurry of activity to
develop these inhibitors
as therapeutics [9]. Chemical stabilization of siRNAs results in increased
serum half life [10],
suggesting that intravenous administration may achieve positive therapeutic
outcomes if delivery
issues can be overcome. Furthermore, small hairpin RNAs (shRNA) have also
shown robust
inhibition of target genes in mammalian cells and can be easily expressed from
bacteriophage
(e.g. T7, T3 or SP6) or mammalian (pol III such as U6 or Hi or polII)
promoters, making them
excellent candidates for viral delivery [11].
[0006] Efforts have been made to find effective nucleic acid-based inhibitors
against HCV,
as existing treatments are not fully effective (reviewed in [4, 12]). These
efforts include
traditional antisense oligonucleotides, phosphorodiamidate morpholino
oligomers [8],
ribozymes, and more recently siRNAs. It has been shown that siRNAs can
effectively target
HCV in human tissue culture cells [13-19] and in animal systems [20].
BRIEF SUMMARY OF THE INVENTION
[0007] The invention provides methods, compositions, and kits for inhibition
of IRES-
mediated gene expression in a virus, e.g., hepatitis C virus (HCV).
[0008] For the inhibitory RNA sequences listed in Figs. 4A and 10 and Table
1(e.g., SEQ ID
NOs: 19-26), a complementary sequence is implied, as are sequences unrelated
to the target that
may be appended one or both ends of each strand; for example the 3' ends, as
will be known to
one skilled in the art. The inhibitory (antisense recognition) sequences shown
in Fig. 4A, Fig.
10, and in Table 1 can be incorporated into either shRNA or siRNA. In the case
of shRNA, the
sequence shown is additionally linked to its complementary sequence by a loop
that includes
nucleotide residues usually unrelated to the target. An example of such a loop
is shown in the
shRNA sequences depicted in Fig. 1B and Fig. 1C as well as in Fig. 16A-B. In
the case of both
siRNAs and shRNAs, the strand complementary to the target generally is
completely
complementary, but in some embodiments, the strand complementary to the target
can contain
2
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
mismatches (see, for example, SEQ ID NOs:13, 14, and 15). The sequence can be
varied to
target one or more genetic variants or phenotypes of the virus being targeted
by altering the
targeting sequence to be complementary to the sequence of the genetic variant
or phenotype.
The strand homologous to the target can differ at about 0 to about 5 sites by
having mismatches,
insertions, or deletions of from about 1 to about 5 nucleotides, as is the
case, for example, with
naturally occurring microRNAs. In some embodiments, a sequence can target
multiple viral
strains, e.g., of HCV, although the sequence differs from the target of a
strain at least one
nucleotide (e.g., one, two, or three nucleotides) of a targeting sequence
[0009] In one aspect, the invention provides a composition comprising at least
one small
interfering RNA that is at least partially complementary to, and capable of
interacting with a
polynucleotide sequence of a virus, such that inhibition of viral gene
expression results from the
interaction of the small interfering RNA with the viral target sequence. In
one embodiment, the
composition includes at least one shRNA, for example, comprising, consisting
of, or consisting
essentially of a sequence selected from the group consisting of SEQ ID NO: 12,
SEQ ID NO: 16,
SEQ ID NO:17, and SEQ ID NO:18, or comprising or consisting essentially of a
sequence
selected from the group consisting of SEQ ID NO:27, SEQ ID NO:32, and SEQ ID
NO:33. In
one embodiment, the shRNA comprises, consists of, or consists essentially of
the sequence
depicted in SEQ ID NO: 12. In another embodiment, the composition includes at
least one
siRNA. In one embodiment, the composition includes at least one siRNA or
shRNA, for
example, comprising or consisting essentially of a sequence selected from the
group consisting
of SEQ ID NO: 19, SEQ ID NO:20, SEQ ID NO:21, SEQ ID NO:22, SEQ ID NO:23, SEQ
ID
NO:23, SEQ ID NO:24, SEQ ID NO:25, SEQ ID NO:27, SEQ ID NO:32, and SEQ ID
NO:33.
In some embodiments, the small interfering RNA, e.g., shRNA or siRNA,
interacts with a viral
sequence of about 19 to about 30 nucleotides, or about 19 to about 25
nucleotides, for example,
any of about 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, or 30 nucleotides. In
some embodiments,
the small interfering RNA binds to a hepatitis C virus sequence. In one
embodiment, the small
interfering RNA binds to a sequence within the internal ribosome entry site
(IRES) sequence of a
hepatitis C virus, for example, to the sequence depicted in SEQ ID NO:26
(residues 344-374 of
SEQ ID NO: 11). In one embodiment, the hepatitis C virus is HCV genotype la.
[0010] In some embodiments, a composition of the invention comprises a
pharmaceutically
acceptable excipient, for example, water or saline, and optionally, are
provided in a
3
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
therapeutically effective amount, e.g., for treating HCV infection in a human
or in a non-human
primate such as a chimpanzee or new world monkey. In one embodiment, the
composition is a
pharmaceutical composition comprising, consisting of, or consisting
essentially of at least one
shRNA or siRNA as described herein and a pharmaceutically acceptable
excipient.
[0011] In another aspect, the invention relates to a kit that includes any of
the compositions
described above, and optionally, further includes instructions for use in a
method of inhibiting
gene expression in a virus or treating a viral infection in an individual as
described herein. In
one embodiment, the kit is for use in a method for treating HCV infection in
an individual, such
as a human, and comprises an shRNA comprising, consisting of, or consisting
essentially of a
sequence selected from the group consisting of SEQ ID NO: 12, SEQ ID NO: 16,
SEQ ID NO: 17,
and SEQ ID NO: 18; or comprising or consisting essentially of a sequence
selected from the
group consisting of SEQ ID NO:27, SEQ ID NO:32, and SEQ ID NO:33, or an siRNA
comprising or consisting essentially of a sequence selected from the group
consisting of SEQ ID
NO:19, SEQ ID NO:20, SEQ ID NO:21, SEQ ID NO:22, SEQ ID NO:23, SEQ ID NO:23,
SEQ
ID NO:24, SEQ ID NO:25, SEQ ID NO:27, SEQ ID NO:32, and SEQ ID NO:33, and
optionally
further coinprises instructions for use in a method of inhibiting gene
expression in a hepatitis C
virus, such as HCV genotype la, or instructions for use in a method of
treating a hepatitis C
(such as HCV genotype la) viral infection in an individual, such as a human,
or a non-human
primate such as a chimpanzee.
[0012] In another aspect, the invention provides a method for treatment of a
viral infection in
an individual, such as a mammal, for example, a human or non-human primate.
The method
includes administering to the individual a therapeutically effective amount of
a small interfering
RNA, such as shRNA or siRNA, that is at least partially complementary to and
capable of
binding to a polynucleotide sequence of the virus and a pharmaceutically
acceptable excipient,
such that binding of the small interfering RNA to the viral polynucleotide
sequence inhibits gene
expression in the virus, e.g., decreases the amount of viral expression in the
individual or
decreases the amount of viral expression that would be expected in an
individual that did not
receive the small interfering RNA. In one embodiment, the viral infection
comprises a hepatitis
C virus, such as HCV genotype la. In some embodiments, the virus is selected
from the group
consisting of hepatitis C genotypes la, lb, 2a, and 2b. In some embodiments,
the small
interfering RNA comprises, consists of, or consists essentially of any of the
shRNA or siRNA
4
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
sequences described herein as well as sequences located within five
nucleotides of one of the
siRNA or shRNA sequences described herein. In soine embodiments, the small
interfering RNA
is complementary to a viral sequence of about 19 to about 30 nucleotides, or
about 19 to about
25 nucleotides, for example, any of about 19, 20, 21, 22, 23, 24, 25, 26, 27,
28, 29, or 30
nucleotides. In one embodiment, the virus is a hepatitis C virus, such as HCV
genotype la. In
one embodiment, the small interfering RNA binds to a sequence of about 19 to
about 25
nucleotides within the IRES region of HCV la depicted in SEQ ID NO:26.
Treatment may
include therapy (e.g., amelioration or decrease in at least one symptom of
infection) or cure. In
some embodiments, the shRNA is administered parenterally, for example, by
intravenous
injection or infusion.
[0013] In another aspect, the invention provides a method of inhibiting gene
expression in a
virus, comprising contacting viral RNA or viral mRNA with a small interfering
RNA or
introducing a small interfering RNA into a virus-containing cell, such that
the small interfering
RNA, e.g., shRNA or siRNA, contains a sequence that is at least partially
complementary to a
polynucleotide sequence of the virus and capable of inhibiting viral gene
expression, for
example, by inducing cleavage of viral polynucleotide sequences. In some
embodiments, the
small interfering RNA comprises, consists of, or consists essentially of any
one of the shRNA or
siRNA sequences described herein. In some embodiments, the small interfering
RNA binds to a
viral sequence of about 19 to about 30 nucleotides, or about 19 to about 25
nucleotides, for
example, any of about 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, or 30
nucleotides. In one
embodiment, the virus is a hepatitis C virus, such as HCV la. In one
embodiment, the small
interfering RNA interacts with a sequence of about 19 to about 30 nucleotides
within the IRES
region of HCV genotype la depicted in SEQ ID NO:26 as well as sequences
located within five
nucleotides of one of the siRNA or shRNA sequences described herein. In yet
other
embodiments, at least two small interfering RNAs are introduced into a cell.
[0014] The invention also relates to an RNA sequence that consists of (a) a
first RNA
sequence, such that the first RNA sequence is a sequence illustrated in Fig.
10 or Fig. 16A-B,
e.g., SEQ ID NO:34, SEQ ID NO:35, SEQ ID NO:36, SEQ ID NO:37, SEQ ID NO:38,
SEQ ID
NO:39, SEQ ID NO:40, SEQ ID NO:41, SEQ ID NO:42, SEQ ID NO:43, SEQ ID NO:44,
SEQ
ID NO:45, SEQ ID NO:46, SEQ ID NO:47, SEQ ID NO:48, SEQ ID NO:49, SEQ ID
NO:50,
SEQ ID NO:51; SEQ ID NO:52, SEQ ID NO:53, SEQ ID NO:54, SEQ ID NO:55, SEQ ID
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
NO:56, or a sequence that differs from a foregoing sequence by one, two, or
three nucleotides;
(b) a second RNA sequence that is a complement of the first sequence; (c) a
loop sequence
positioned between the first and second nucleic acid sequence, the loop
sequence consisting of 4-
nucleotides; and (d) optionally, a two nucleotide overhang. In some
embodiments of the
invention, the first RNA sequence is SEQ ID NO:34, SEQ ID NO:35, SEQ ID NO:36,
SEQ ID
NO:37, SEQ ID NO:38, SEQ ID NO:39, SEQ ID NO:40, SEQ ID NO:41, SEQ ID NO:42,
SEQ
ID NO:43, SEQ ID NO:44, SEQ ID NO:45, SEQ ID NO:46, SEQ ID NO:47, SEQ ID
NO:48,
SEQ ID NO:49, SEQ ID NO:50, SEQ ID NO:51; SEQ ID NO:52, SEQ ID NO:53, SEQ ID
NO:54, SEQ ID NO:55, or SEQ ID NO:56. The RNA sequence can, in some cases,
include at
least one modified nucleotide. The loop sequence of an RNA sequence of the
invention can be,
e.g., four nucleotides, five nucleotides, six nucleotides, seven nucleotides,
eight nucleotides, nine
nucleotides, ten nucleotides, or at least ten nucleotides. In some
embodiments, the RNA
sequence is an shRNA and includes an HCV target sequence as described herein
and a
complementary sequence, linked by a loop that includes at least one non-
nucleotide molecule. In
certain embodiments, the loop of the RNA sequence is 3' to a sense strand and
5' to the
complementary antisense strand of the shRNA. In other embodiments, the loop of
the RNA
sequence is 3' to an antisense strand and 5' to the complementary sense strand
of the shRNA. In
some cases, the RNA sequence includes a two nucleotide overhang and the two
nucleotide
overhang is a 3'UU. In some cases, the overhang is one nucleotide, two
nucleotides, three
nucleotides, or more. In some cases, the first RNA sequence is any one of SEQ
ID NOs:57-79,
SEQ ID NO:12, SEQ ID NO: 16, SEQ ID NO: 17, or SEQ ID NO: 18. In some cases,
the RNA
sequence is a sequence illustrated in Fig. 16A-B.
[0015] The invention also relates to a DNA sequence that includes a sequence
encoding an
RNA sequence disclosed herein (e.g., an RNA sequence illustrated in Fig. 10 or
Fig. 16A-B).
The invention also includes an expression vector comprising such a DNA
sequence. Also
included is a retroviral vector that includes such a DNA sequence, e.g., a
retroviral vector that,
upon infection of a cell with the vector, can produce a provirus that can
express an RNA
sequence of the invention, for example, without limitation, an shRNA sequence
illustrated in
Fig. 16A-B.
[0016] In some aspects, the invention relates to a composition that includes
an RNA
sequence as disclosed herein (for example, without limitation, an shRNA
illustrated by Fig. 16A-
6
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
B) and a pharmaceutically acceptable excipient. In some embodiments, the
composition
comprises a vector as disclosed herein and a pharmaceutically acceptable
excipient. In certain
embodiments, a composition of the invention includes at least two RNA
sequences as disclosed
herein.
[0017] In another aspect, the invention includes a method of inhibiting
expression or activity
of a hepatitis C virus. The method includes providing a cell that can express
a hepatitis C virus,
and contacting the cell with an RNA sequence as disclosed herein (non-limiting
examples of
which are illustrated in Fig. 16A-B). The cell can be in a mammal, e.g., a
human or a non-
human primate such as a chimpanzee. In certain embodiments, the cell is
contacted with at least
two different RNA sequences.
[0018] In some aspects, the invention relates to a method that includes
identifying a subject
infected with or suspected of being infected with a hepatitis C virus,
providing to the subject a
therapeutically effective amount of a composition containing one or more
different RNA
sequences disclosed herein. In some embodiments, the method also includes
determining
whether the viral load of the subject is decreased subsequent to providing the
composition to the
subject. In some embodiments, the method also includes determining whether at
least one viral
protein or viral nucleic acid sequence is decreased in the subject subsequent
to providing the
composition to the subject.
[0019] Unless otherwise defined, all technical and scientific terms used
herein have the same
meaning as commonly understood by one of ordinary skill in the art to which
this invention
belongs. Although methods and materials similar or equivalent to those
described herein can be
used in the practice or testing of the present invention, suitable methods and
materials are
described below. All publications, patent applications, patents, and other
references mentioned
herein are incorporated by reference in their entirety. In addition, the
materials, methods, and
examples are illustrative only and not intended to be limiting.
[0020] ' Other features and advantages of the invention will be apparent from
the detailed
description, drawings, and from the claims.
DESCRIPTION OF THE DRAWINGS
[0021] Fig. 1A is a representation of the IRES nucleotide sequence of
hepatitis C genotype
la (see GenBank Accession No. AJ242654). Nucleotides of a target region, 344-
374, are
7
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
underlined. Various regions (indicated in bold) have been successfully
targeted by inhibitors,
including HeptazymeTM ribozyme (siRNA.com; positions 189-207), Chiron 5U5
siRNA [25]
(positions 286-304), ISIS 14803 phosphorothioate antisense oligonucleotide
[34] (positions 330-
349), Mizusawa 331 siRNA [15] (positions 322-340) and a phosphorodiamidate
morpholino
oligomer [8, 35] (positions 344-363). A more complete list of siRNAs that have
been tested to
down-regulate the HCV IRES and other HCV elements can be found in [2, 3].
[0022] Fig. 1B is a representation of RNA sequences of shRNA HCVa-wt (shRNAl)
and
mutated variants thereof resulting from pol III transcription from a U6
promoter of
corresponding DNA templates. Two base pairs (underlined) of HCVa-wt were
altered to create
versions of HCVa-wt containing 1(HCVSNPl or HCVSNP2) or 2 mismatches (HCVa-
mut)
shRNAs as shown.
[0023] Fig. 1C is a representation of the sequences of shRNAs HCVb-wt (sh9),
HCVc-wt
(sh10), and HCVd-wt (shl 1).
[0024] Fig. 1D is a representation of the secondary structure of the HCV IRES
with indicated
target sites for shRNA HCVa-wt, HCVb-wt, HCVc-wt, and HCVd-wt.
[0025] Fig. lE is a schematic representation of the pCDNA3/HCV IRES dual
luciferase
reporter construct used to produce the HCV IRES target as well as the EMCV
IRES control, in
which the IRES from encephalomyocarditis virus replaces the HCV IRES and
therefore lacks
any target for the HCV-directed shRNAs. In each case, firefly luciferase
expression is dependent
on initiation of translation from the IRES sequence, whereas Renilla
luciferase is expressed in a
cap-dependent manner.
[0026] Fig. 1F is a bar graph depicting the results of a screen of shRNAs for
the ability to
inhibit HCV IRES-mediated gene expression in 293FT cells. 293FT cells were
cotransfected
with pCDNA3/HCV IRES dual luciferase reporter construct, pSEAP2 (as a
transfection and
specificity control), and an shRNA (at1 nM) in a well of a 24-well tissue
culture plate. Plasmid
pUC 18 was added to provide a total of 800 ng nucleic acid per well. 48 hours
post-transfection,
cells were lysed and firefly luciferase activity was measured by a
luminometer. All data are the
results of individual, independent experiments performed in triplicate, and
normalized to SEAP.
[0027] Fig. 2A is a bar graph depicting the results of experiments testing
inhibition of HCV-
Il2ES driven gene expression in 293FT cells that were cotransfected with dual
luciferase reporter
and SEAP expressing plasmids and 1 pmole of in vitro transcribed shRNAs. The
target plasmid
8
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
was pCDNA3/HCV IRES dual luciferase reporter (HCV IRES, as shown in Fig. 1E).
Firefly
luciferase activity measured as described in Example 1. Firefly luciferase and
SEAP activities
were normalized to 100.
[0028] Fig. 2B is a bar graph depicting the results of experiments testing HCV
versus EMCB
inhibition in 293FT cells. The data are presented as luciferase activity
divided by SEAP activity
normalized to 100.
[0029] Fig. 2C is a bar graph depicting the results of experiments
demonstrating the effect of
single-base mismatches on potency of shRNAs. Experimental conditions were as
described for
Fig. 2A. SNP1 and SNP2 contained mutated base pairs as shown in Fig. 1B.
[0030] Fig. 2D is a line graph depicting the resulting of experiments testing
dose response of
inhibition of HCV-IRES-driven gene expression by HCVa-wt and mutated (HCVa-
mut) or
control (229) shRNAs. Experimental conditions were as described for Fig. 2A.
The data are
represented as luciferase divided by SEAP normalized to 100. All data are the
results of
individual, independent experiments performed in triplicate.
[0031] Fig. 2E is a line graph depicting the resulting of experiments testing
dose response of
HCVa-wt, HCVa-mut), and 229 shRNAs on gene expression from a dual-luciferase
reporter
lacking shRNA target sites. The procedure was as described for Fig. 2D except
target was
firefly luciferase driven by EMCV IRES instead of HCV IRES.
[0032] Fig. 2F is a reproduction of a Northern blot analysis of co-transfected
293FT cells
treated as follows; 10 g of total RNA isolated from cells transfected with no
inhibitor (lane 1),
229 (lane 2) HCVa-wt (lane 3), or HCVa-mut (lane 4) were separated by
denaturing gel
electrophoresis, transferred to membrane and hybridized sequentially to 32P-
labeled fLuc, SEAP,
or elongation factor 1A (EF1A) cDNA probes. The RNA blot was exposed to a
storage
phosphor screen for visualization and quantitation (BioRad FX Molecular
Imager).
[0033] Fig. 3A is a line graph depicting the results of experiments testing
dose response to
HCVa-wt and HCVa-mut shRNAs using the human hepatocyte cell line, Huh7.
Procedures were
as described for Fig. 2D, except that Huh7 cells were used.
[0034] Fig. 3B is a line graph depicting the results of experiments
demonstrating that HCVa-
wt shRNA does not inhibit a similar target lacking the HCV IRES. Cells were
transfected as in
Fig. 3A except that pCDNA3/EMCV IRES dual luciferase reporter (EMCV IRES) was
added in
place of pCDNA3IHCV IRES dual luciferase reporter (HCV IRES). All data are
presented as
9
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
luciferase activity divided by SEAP. All data were generated from individual,
independent
experiments performed in triplicate.
[0035] Fig. 4A depicts sequences of seven 19 base pair viral recognition
sequences of
synthetic siRNAs and shRNAs contained within the 25 nucleotide target site of
HCV genotype
1A (SEQ IID NO:26) and analysis of their purity on 10% native polyacrylamide
gel stained with
ethidium bromide. siRNAs: sense and antisense strands contained 3'-UU
overhangs; shRNAs:
loop sequences and 3', 5'- end overhangs were identical to those of the 25
base pair shRNAs.
[0036] Fig. 4B is a bar graph depicting the results of experiments in which
RNA inhibitors
(siRNAs and shRNAs) were assayed for inhibition of HCV IRES-mediated gene
expression at an
inhibitor concentration 1 nM in 293 FT cells.
[0037] Fig. 4C is a bar graph depicting the results of experiments in which
RNA inhibitors
were assayed for inhibition of HCV IRES-mediated gene expression at an
inhibitor concentration
of 0.1 nM in 293 FT cells.
[0038] Fig. 5A is a reproduction of IVIS images of mice in which dual
luciferase HCV IRES
reporter plasmid (10 g) and SEAP (added to control for injection efficiency
and nonspecific
inhibition) were co-injected into the tail veins of mice as described in
Example 1 with 100 g of
the indicated HCV shRNA or control 229 shRNA) directly or in the form of 100
g of pol III
expression plasmids expressing shRNA (or pUC18 plasmid as control). At various
time-points
(24, 36, 48, 60, 72, 84 and 100 hours) post-injection, luciferin was
administered
intraperitoneally, and the mice were imaged using the IVIS in vivo imaging
system. Images are
of representative mice from the 84 hour time point.
[0039] Fig. 5B is a graph depicting the quantitated results of experiments
described for Fig.
5A in which there was direct delivery of RNA. Quantitation was performed using
ImageQuantTM software. Each time-point represents the average of 4-5 mice. At
the 96 hour
time point, the mice were bled and the amount of SEAP activity determined by
pNPP assay as
described in Example 1. The quantitated data are presented as luciferase
divided by SEAP
activity, normalized to pUC18 control mice (100%, no error bars shown on pUC18
control for
clarity; error bars are similar to the others shown).
[0040] Fig. 6 is a bar graph depicting the results of experiments in which
shRNA and
phosphorodianli.date morpholino oligomer inhibition of HCV IRES-mediated
reporter gene
expression in mice was compared. Mice were co-injected as described in
experiments for Fig. 5
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
with dual luciferase HCV IRES reporter plasmid and pSEAP with 100 g of the
indicated HCV
shRNA inhibitors or 1 nmole of a morpholino oligonucleotide previously shown
to inhibit HCV
IRES expression construct [8]. The mice were imaged at various times (12
hours, 24 hours, 48
hours, and 144 hours) post-treatment. Data shown are for the 48 hour time
point. The
quantitated data are presented as luciferase and SEAP activities, normalized
to pUC 18 control
(no addition) mice. The results presented are from 3-5 mice per construct.
[0041] Fig 7 is a graph depicting the results of experiments in which BHK-21
cells were
transiently transfected with plasmids expressing an inhibitory shRNA targeting
the nsp-1 gene.
Twenty-four hours after transfection, cells were infected with 10 l of
replication -proficient
GFP-expressing Semliki Forest virus (SFV-GFP-VA7; multiplicity of infection
(MOI) sufficient
for about100% infection) and assayed for virus-mediated GFP expression by flow
cytometry 24
hours after infection. The level of siRNA-mediated suppression was about 35%.
Labels: Nsp 1.
shRNA targeting Nsp-1 gene (nsp-1#2); empty vector, pU6; naYve, uninfected BHK
cells.
[0042] Fig. 8 is a bar graph depicting the results of experiments in which
inhibition of
replication-deficient SFV (SFV-PD713P-GFP) by shRNAs was investigated. BHK-21
cells
were transiently transfected with plasmids expressing inhibitor shRNAs. Forty-
six hours after
transfection, cells were infected with SFV-GFP virus at an MOI of 5 with 8%
PEG in serum-free
media for one hour. Then complete media was added and cells were incubated at
37 C
overnight. Cells were analyzed by flow cytometry at 9, 24, 32, 99, and 125
hours after infection.
For clarity, only three time points are shown (9, 24 and 32 hours). The amount
of inhibition of
each shRNA was normalized to capsid shRNA. Capsid mRNA is not present in this
SFV-GFP
replication-deficient virus and therefore capsid shRNA should have no effect
on GFP expression.
The transfection efficiency for the shRNA expression constructs for this
experiment was about
70%, suggesting that actual viral inhibition is significantly higher than the
levels indicated. The
fifth set of bars (Mixed) refers to a mixture of shRNAs targeting nsp 1-4 and
capsid.
[0043] Fig. 9 is a line graph depicting the results of experiments testing HCV
replicon
inhibition by shRNAs.
[0044] Fig. 10 is a table depicting sequences and results of a screen of
shRNAs for the ability
to inhibit HCV IRES-mediated gene expression in 293FT cells. Cells were
cotransfected (using
LipofectamineTM 2000) with pCDNA3/HCV IRES dual luciferase reporter construct
(40 ng),
pSEAP2 (25 ng, as a transfection and specificity control), and an shRNA (at 1
or 5 nM) in a well
11
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
of a 48-well tissue culture plate. Plasmid pUC18 was added to provide a total
of 400 ng nucleic
acid per well. Forty-eight hours post-transfection, the supernatants were
removed for SEAP
analysis, cells were lysed, and firefly luciferase activity was measured by a
luminometer. All
data are the results of at least two independent experiments performed in
triplicate. SEAP levels
were uniform in all samples. Control experiments to assay specificity of
shRNAs were
performed on mutated pCDNA3/HCV IRES dual luciferase reporter construct as
well, where
C340 (in IRES) was substituted with U.
[0045] Fig. 11 is a diagrammatic representation of 3'-terminal sequence of the
HCV IRES
with segments targeted by shRNAs. Mutation C340->U (used to assay specificity
of shRNAs) is
indicated.
[0046] Fig. 12A is a diagrammatic representation of 5'-termini of HCV IRES and
targeting
positions for six 19-bp shRNAs.
[0047] Fig. 12B is a bar graph depicting the results of a screen of shRNAs for
the ability to
inhibit HCV IRES-mediated gene expression in 293FT cells. Experiments were
conducted as for
Fig. 10; shRNA concentration, 1 nM.
[0048] Fig. 13A is a diagrammatic representation of the sequences of tested
variants of the
depicted 25 base pair shRNA, with the various loop sizes and sequences, as
well as 3'-termini
that were tested.
[0049] Fig. 13B is a bar graph depicting the results of a screen of shRNAs
depicted in
Fig. 13A for the ability to inhibit HCV IRES-mediated gene expression in 293FT
cells.
Experiments were conducted as for those of Fig. 10. shRNA concentration, 1 nM.
(shRNA
sequences are listed in Fig. 16A-B)
[0050] Fig. 14A is a diagrammatic representation of the sequences of tested
variants of the
depicted 19-bp shRNA with the various loop sizes and sequences tested, as well
as 3' termini
that were tested.
[0051] Fig. 14B is a bar graph depicting the results of a screen of shRNAs
depicted in Fig.
14A for the ability to inhibit HCV IRES-mediated gene expression in 293FT
cells. Experiments
were conducted as described for Fig. 10. shRNA concentration, 1 nM. (shRNA
sequences are
listed in Fig. 16A-B).
[0052] Fig. 15 is a bar graph depicting the results of a screen of shRNAs (and
siRNAs) for
the inhibitory activity in the HCV replicon system. Human hepatocytes (AVA5, a
derivative of
12
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
the Huh7 cell line) stably expressing HCV subgenomic replicons, were
transfected with RNA
inhibitors, and the amount of HCV expression was determined. A range of
concentrations was
tested and the concentration of sh/siRNA that resulted in 50% inhibition
(EC50) was determined.
Dark and light bars represent the results of two independent experiments.
[0053] Fig. 16A-B is a table depicting shRNA sequences targeting HCV IRES as
indicated.
ShRNA loops are underlined. Nucleotides indicated by low-case are non-
complementary to the
target.
DETAILED DESCRIPTION OF THE INVENTION
[0054] The invention provides compositions, methods, and kits for inhibiting
viral (e.g.,
hepatitis C) gene expression and/or treating a viral infection in a mammal.
[0055] RNA interference offers a novel therapeutic approach for treating viral
infections.
The present invention provides small interfering RNAs (e.g., shRNAs and
siRNAs) that target a
viral sequence and inhibit (i.e., reduce or eliminate) viral gene expression,
and methods of using
such small interfering RNAs for treatment of a viral infection in a mammal,
such as a human. In
some embodiments, the small interfering RNA constructs of the invention
inhibit gene
expression of a virus by inducing cleavage of viral polynucleotide sequences
within or near the
target sequence that is recognized by the antisense sequence of the small
interfering RNA.
[0056] As used herein, "small interfering RNA" refers to an RNA construct that
contains one
or more short sequences that are at least partially complementary to and can
interact with a
polynucleotide sequence of a virus. Interaction may be in the form of a direct
binding between
complementary (antisense) sequences of the small interfering RNA and
polynucleotide
sequences of the viral target, or in the form of an indirect interaction via
enzymatic machinery
(e.g., a protein complex) that allows the antisense sequence of the small
interfering RNA to
recognize the target sequence. In some cases, recognition of the target
sequence by the small
interfering RNA results in cleavage of viral sequences within or near the
target site that is
recognized by the recognition (antisense) sequence of the small interfering
RNA. The small
interfering RNA can exclusively contain ribonucleotide residues, or the small
interfering RNA
can contain one or more modified residues, particularly at the ends of the
small interfering RNA
or on the sense strand of the small interfering RNA. The term "small
interfering RNA" as used
13
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
herein encompasses shRNA and siRNA, both of which are understood and known to
those in the
art to refer to RNA constructs with particular characteristics and types of
configurations.
[0057] As used herein, "shRNA" refers to an RNA sequence comprising a double-
stranded
region and a loop region at one end forming a hairpin loop. The double-
stranded region is
typically about 19 nucleotides to about 29 nucleotides in length on each side
of the stem, and the
loop region is typically about three to about ten nucleotides in length (and
3'- or 5'-terminal
single-stranded overhanging nucleotides are optional). One example of such an
shRNA, HCVa-
wt shRNA, has a 25 base pair double-stranded region (SEQ ID NO: 12), a ten
nucleotide loop, a
GG extension on the 5' end, and a UU extension on the 3' end. Additional
examples of suitable
shRNAs for use in, e.g., inhibiting HCV expression, are provided throughout
the specification,
e.g., Fig. 16A-B.
[0058] As used herein, "siRNA" refers to an RNA molecule comprising a double-
stranded
region with a 3' overhang of nonhomologous residues at each end. The double-
stranded region
is typically about 18 to about 30 nucleotides in length, and the overhang may
be of any length of
nonhomologous residues, for example, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13,
14, 15, 16 or more
nucleotides. The siRNA can also comprise two or more segments of 19-30 base
pair separated
by unpaired regions. Without committing to any specific theory, the unpaired
regions may
function to prevent activation of innate immunity pathways. One example of
such an siRNA is
HCVa-wt siRNA, which has a 25 base pair double-stranded region (SEQ ID NO:
12), and a UU
extension on each 3' end.
[0059] In one embodiment, a small interfering RNA as described herein
comprises a
sequence complementary to a sequence of the internal ribosome entry site
(IRES) element of
hepatitis C ("HCV"). In one embodiment, the virus is HCV genotype la.
[0060] SiRNA gene inhibition has been shown to robustly inhibit gene
expression in a
number of mammalian systems. Due to its high level of secondary structure, the
HCV Il2ES has
been suggested to be a poor target for siRNAs or shRNAs. Mizusawa reported,
however,
successful targeting of the HCV IRES in 293 and Huh7 tissue culture cells,
reporting 50 and 74
percent knock-down of gene expression, respectively. Similarly, Seo and
coworkers [25]
reported the ability of 100 nM siRNA to inhibit HCV replication (about 85%
knockdown) in 5-2
Huh7 cells. It has now been demonstrated as described herein that small
interfering RNAs
(shRNAs and siRNAs) directed against the 3' end of the HCV IRES, including and
downstream
14
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
of the AUG translation start site, can induce 96 percent knockdown of HCV IRES-
dependent
luciferase expression at 0.3 nM in 293FT cells and 75 percent knockdown at 0.1
nM in Huh7
cells (see Figs. 2D and 3A). Furthermore, direct delivery of shRNA to mouse
liver was shown to
potently inhibit HCV IRES-dependent reporter expression. This is the first
demonstration of
RNAi-mediated gene inhibition in an animal model following direct delivery of
an RNA hairpin
(not expressed in vivo from a plasmid or viral vector). The effectiveness of
shRNA delivered
directly to mouse liver following hydrodynamic injection was surprising in
view of the high
levels of nucleases found in blood. The observation that these shRNAs
effectively knocked
down gene expression in liver indicates that these shRNA inhibitors (1) are
very potent and not
needed at high levels in mouse liver to cause gene inhibition, (2) are
delivered sufficiently
rapidly to the liver, e.g., before they are cleaved by nucleases in quantities
that prevent an
inhibitory effect, or (3) are inherently stable to nuclease degradation (or a
combination of these
characteristics).
[0061] Reports suggest that in vitro-synthesized transcripts from
bacteriophage promoters
potently induce interferon (IFN) alpha and beta due to the presence of an
unnatural 5'
triphosphate [26]. Furthermore, shRNAs expressed from pol III expression
vectors may also
induce IFN [27]. How this interferon induction would affect use of shRNAs in a
clinical setting
for HCV infection is unclear. Current HCV therapy includes treatment with
interferon alpha,
suggesting that if interferon is induced by shRNA, it may have a positive
effect. To date, no
interferon-related side effects appear to have been reported in animals
following administration
of RNAi [3]. Additional concerns have been raised regarding off-target effects
of siRNA as well
as potential cytotoxic effects when siRNAs or shRNAs are delivered by
lentiviral vectors [28].
[0062] The present invention also relates to methods of testing siRNAs and
shRNAs
targeting HCV IRES sequences to identify those sequences having sufficient
activity (e.g., the
highest activity among a selected group of such sequences) to be a candidate
for use as a
treatment. Testing can also include screening for small interfering activities
having undesirable
off-target effects or general cytotoxic effects. Off-target effects include,
without limitation
knockdown of nontargeted genes, inhibition of expression of non-targeted
genes, and
competition with natural microRNA pathways (Birmingham et al., Nat. Methods.
2006 3(3):199-
204; Grimm et al., Nature 2006 441(7092):537-541). Methods of identifying
cytotoxic effects
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
are known in the art (for example, Marques et al., Nat. Biotechnol. 2006
24(5):559-565; Robbins
et al., Nat. Biotechnol. 2006 24(5):566-571).
[0063] The IRES region in the HCV 5'-UTR is highly conserved (92-100%
identical [15, 29-
31]) and has several segments that appear to be invariant, making the IRES a
prime target for
nucleic acid-based inhibitors. The region around the AUG translation
initiation codon is
particularly highly conserved, being invariant at positions +8 to -65 (with
the exception of a
single nucleotide variation at position -2) as observed in over 81 isolates
from various
geographical locations [32]. Despite the conservation of sequence in the IRES
motif, it is
unlikely that targeting a single sequence, even if highly conserved, will be
sufficient to prevent
escape mutants. RNA viruses are known to have high mutation rates due to the
high error rate of
the RNA polymerase and the lack of proofreading activity of that enzyme. On
average, each
time HCV RNA is replicated one error is incorporated into the new strand. This
error rate is
compounded by the prodigious production of viral particles in an active
infection (approximately
a trillion per day in a chronically infected patient) [33]. Therefore, in some
embodiments of the
invention, several conserved sites are targeted or, alternatively, shRNAs as
described herein are
used as a component of a combination treatment, such as with ribaviran and/or
pegylated
interferon. As demonstrated herein, a single mismatch does not completely
block shRNA
activity (see Example 2; Fig. 2D); thus each different shRNA may have some
activity against a
limited number of mutations. Accordingly, the invention includes methods of
inhibiting HCV
expression using an shRNA that may include a mismatch to the target sequence.
The invention
also includes methods of inhibiting HCV expression by administering at least
two different
shRNAs targeting an HCV IRES, such that the shRNAs differ in the targeting
sequences.
[0064] McCaffrey and colleagues reported that a phosphorodiamidate morpholino
oligonucleotide directed against a conserved HCV IRES site at the AUG
translation initiation
site potently inhibits reporter gene expression [8]. The same morpholino
inhibitor was used for
comparison against the shRNA inhibition described herein. It was found that
both the
morpholine and the shRNA targeting the conserved HCV IRES site potently and
robustly
inhibited IRES-dependent gene expression. Four mutations in the morpholino
were required to
block activity, whereas two changes in the shRNA were sufficient, suggesting
greater shRNA
specificity. This potential advantage, coupled with the lack of unnatural
residues in the shRNA
16
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
inhibitor and presumably fewer resultant side effects, are balanced by the
increased stability of
the morpholino oligomer.
[0065] A dual reporter luciferase plasmid was used in which firefly luciferase
(fLuc)
expression was dependent on the HCV II2ES [24]. Expression of the upstream
renilla luciferase
is not HCV IRES-dependent and is translated in a Cap-dependent process. Direct
transfection of
HCV IRES shRNAs, or alternatively shRNAs expressed from poIIII-promoter
vectors,
efficiently blocked HCV IRES-mediated fLuc expression in human 293FT and Huh7
cells.
Control shRNAs containing a double mutation had little or no effect on fLuc
expression, and
shRNAs containing only a single mutation showed partial inhibition. These
shRNAs were also
evaluated in a mouse model where DNA constructs were delivered to cells in the
liver by
hydrodynamic transfection via the tail vein. The dual luciferase expression
plasmid, the
shRNAs, and secreted alkaline phosphatase plasmid were used to transfect cells
in the liver, and
the animals were imaged at time points over 12 to 96 hours. In vivo imaging
revealed that HCV
IRES shRNA directly, or alternatively expressed from a polIlI-plasmid vector,
inhibited HCV
IRES-dependent reporter gene expression; mutant or irrelevant shRNAs had
little or no effect.
These results indicate that shRNAs, delivered as RNA or expressed from viral
or nonviral
vectors, are useful as effective antivirals for the control of HCV and related
viruses.
[0066] Assay of three additional shRNAs targeting different sites on HCV IRES
domain IV
revealed another potent shRNA, HCVd-wt, whose target position is shifted six
nucleotides from
that of HCVa-wt. HCVb-wt and HCVc-wt were much less efficient inhibitors.
[0067] To further investigate local sequence effects on potency, seven in
vitro-transcribed
shRNA constructs comprising a 19 base pair sequence complementary to a
sequence of the HCV
IRES and the corresponding synthetic siRNA comprising the same 19 base pair
sequences,
targeting all possible positions within the 25 base pair site of HCVa-wt (344-
368), were assayed
for inhibitory activity. A 25 base pair synthetic siRNA corresponding to HCVa-
wt shRNA was
also tested. All of the tested constructs exhibited a high level of activity.
In general, 19 base pair
siRNAs were more potent than 19 base pair shRNAs. The most potent, siHCV19-3
was effective
at 1 nM (>90% inhibition), 0.1 nM (about 90% inhibition) and even at a
concentration of 0.01
nM (about 40% inhibition). Thus, 19-25 base pair shRNAs and siRNAs designed to
target the
region 344-374 on the HCV IRES are generally potent inhibitors of HCV
expression, with some
local differences.
17
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
[0068] Small hairpin RNAs of the invention can, optionally, include structures
resulting in
strong noncovalent bonds between the sense and antisense strands of the shRNA.
Examples of
such noncovalent bonds include cross-links mediated by metal ions. Such cross-
links can be
formed between natural or modified nucleotide residues, including, for
example, modified bases,
sugars, and terminal groups, as described in Kazakov and Hecht 2005, Nucleic
Acid-Metal Ion
Interactions. In: King, R. B. (ed.), Eracyclopedia of Inorganic Chemistry. 2nd
ed., Wiley,
Chichester, vol. VI, pp. 3690-3724, e.g., section 5.4.3. Additional non-
limiting examples of
variants of such bonds are found patent application WO 99/09045(US2006074041;
e.g., Fig. 10.
In general the location of cross-linkable nucleotide residues is at the ends
of the complementary
RNA strands that lie in close proximity upon duplex formation. The addition of
certain metal
ions (or metal ion coordination compounds) can result in the cross-linking of
functional groups
that have strong affinity for these metal ions, such as -SH, -SCH3,
phosphorothioates,
imidazolides, o-phenanthrolines, and others. These modified nucleotides are
introduced during
chemical synthesis of the sense and antisense RNA strands. The modified
nucleotides in sense
and antisense strands may either form base pairs or be part of 1-3 nucleotide
overhangs.
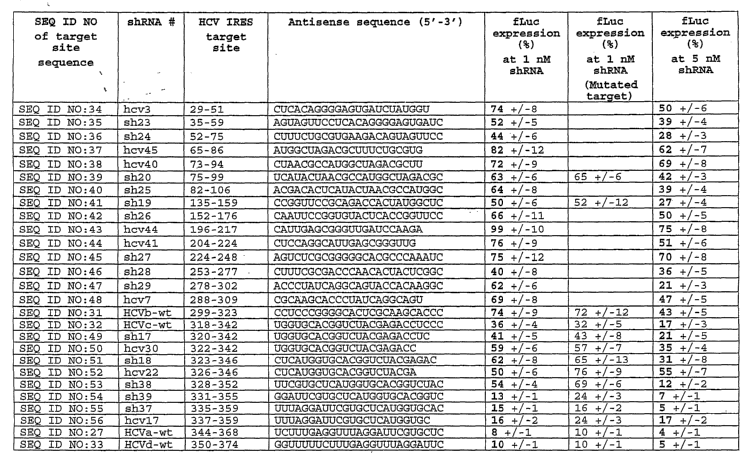
Targeting Sequences
[0069] Examples of targeting sequences are provided throughout the
specification. Non-
limiting examples of targeting sequences are provided in, for example, Table 1
and Fig. 10.
Non-limiting examples of shRNAs and siRNAs incorporating targeting sequences
are found
throughout the specification, e.g., in Fig. 1 and Fig. 16A-B.
Loops
[0070] Effects of size and sequence of loop region of the shRNA were also
investigated. The
loop region of the shRNA stem-loop can be as small as two to three nucleotides
and does not
have a clear upper limit on size; generally, a loop is between four and nine
nucleotides, and is
generally a sequence that does not cause unintended effects, e.g., by being
complementary to a
non-target gene. Highly structured loop sequences such as a GNRA tetraloop can
be used in the
loop region (e.g., as the loop) in an shRNA. The loop can be at either end of
the molecule; that
is, the sense strand can be either 5' or 3' relative to the loop. Also, a
noncomplementary duplex
region (approximately one to six base pairs, for example, four CG base pairs)
can be placed
18
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
between the targeting duplex and the loop, for example to serve as a "CG
clamp" to strengthen
duplex formation. At least 19 base pairs of target-complementary duplex are
needed if a
noncomplementary duplex is used.
[0071] A loop structure can also include reversible linkages such as S-S
bonds, which can be
formed by oxidation of -SH groups introduced into nucleotide residues, e.g.,
as described in
(Earnshaw et al., J. Mol. Biol., 1997, 274: 197-212; Sigurdsson et al. (Thiol-
Containing RNA for
the Study of Structure and Function of Ribozymes. METHODS: A Companion to
Methods in
Enzymology, 1999, 18: 71-77). A non-limiting example of the location for
nucleotide residues
with SH groups is at the ends of the complementary RNA strands that lie in
close proximity upon
duplex formation. Such modified nucleotides are introduced during chemical
synthesis of the
sense and antisense RNA strands of the small interfering RNA. The modified
nucleotides in
sense and antisense strands may either form base pairs or form non-
complementary overhangs of
one to three nucleotides.
[0072] Additional non-limiting examples of loops and their applications, e.g.,
in shRNA and
siRNA targeting HCV, can be found in the Examples.
Termini
[0073] The 3' terminus of an shRNA as described herein can have a non-target-
complementary overhang of two or more nucleotides, for example, UU or dTdT,
however, the
overhangs can be any nucleotide including chemically modified nucleotides
that, for example,
promote enhanced nuclease resistance. In other embodiments, there are one or
zero nucleotides
overhanging on the 3' end.
[0074] The 5' end can have a noncomplementary extension, e.g., two Gs (as
shown in
Fig. 1B), a GAAAAAA sequence, or only one or zero nucleotides extending beyond
the target-
complementary duplex region. In the sequence shown in Fig. 1B, the two 5' G's
are included
primarily for ease of transcription from a T7 promoter.
Additional features
[0075] Additional features that can optionally be included in shRNAs used to
inhibit HCV
expression and that are encompassed by the invention are length variations
between about 19
base pairs and about 30 base pairs for the target complementary duplex region,
small shifts in the
19
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
sequence targeted (generally zero to about two nucleotides, and shifts as
large as about
ten nucleotides in either direction along the target may lie within the
targetable region).
Similarly, mismatches are also tolerated: about one to about two in the
antisense strand and
about one to about nine in the sense strand (the latter destabilizing the
hairpin duplex but not
affecting the strength of binding of the antisense strand to the target; the
number tolerated
depends partly on the length of the target-complementary duplex. As described
herein, an
shRNA having at least seven G-U mismatches within a 29 base pair target-
complementary
duplex region can be used successfully for inhibiting HCV expression, e.g.,
using sequence
targeting the HCV IRES. Note that the two mutations shown in Fig. 1B largely
abrogated
inhibition, but other mutants having mutations in other positions,
particularly if they are closely
spaced and/or near the end, can be better tolerated. Certain variations are
known in the art or
demonstrated in the instant application.
Vectors
[0076] Suitable vectors for producing shRNAs and siRNAs are described herein
and are
known in the art. In non-limiting examples, shRNAs can be expressed using Pol
III promoters
such as U6 or H1, in the context of vectors derived from adeno-associated
virus or lentiviruses.
The human U6 nuclear RNA promoter and human H1 promoter are among the pol III
promoters
for expressing shRNAs.
[0077] One feature that is generally desirable in a vector is relatively
prolonged transgene
expression. Lentiviral vectors are able to transduce nondividing cells and
maintain sustained
long-term expression of transgene. Adeno-associated virus serotype 8 is
considered safe and is
characterized by prolonged transgene expression.
Candidate s1tRNA and siRNA
[0078] In some cases, one or more small interfering RNAs are identified as
having activity
for inhibiting a targeted virus such as HCV. Additional tests can be carried
out to further
characterize the suitability of such RNAs for use, e.g., for inhibiting HCV
expression in an
animal. Animal models can be used for such testing. One non-limiting examples
includes a
mouse model, e.g., as illustrated in Example 3 (infra). Other animal models
suitable for testing
an treatment for HCV are known in the art, for example, using chimpanzees.
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
Metlaods
[0079] The invention relates to methods of inhibiting gene expression in a
virus, comprising
contacting the virus with a small interfering RNA, such as a shRNA or siRNA as
described
herein that comprises a sequence that is at least partially complementary to,
and is capable of
interacting with a polynucleotide sequence of the virus. In some embodiments,
contacting the
virus comprises introducing the small interfering RNA into a cell that
contains the virus, i.e., a
virus infected cell. "Inhibiting gene expression" as used herein refers to a
reduction (i.e.,
decrease in level) or elimination of expression of at least one gene of a
virus. For example,
reduction in expression compared to corresponding cell or animal infected with
the virus. In
some embodiments, inhibition of gene expression is accomplished by cleavage of
the viral target
sequence to which the small interfering RNA binds. Gene expression can be
assayed by
assaying viral RNA or viral protein. In some cases, efficacy of a method (for
example, a
treatment using a composition described herein) is assayed by evaluating an
infected animal for a
decrease in symptoms or a change (e.g., decrease) in the expression or
activity of a protein
associated with viral infection, e.g., a viral protein such as p24, or a host
protein such as an
interferon.
[0080] The invention also relates to methods for treating a viral infection or
for treating a
subject suspected of being infected (including a subject exposed to virus for
prophylactic
treatment) in a mammal, comprising administering to the mammal a composition
comprising a
therapeutically effective amount of a small interfering RNA, such as a shRNA
or siRNA as
described herein that includes a sequence that is at least partially
complementary to, and capable
of interacting with (e.g., hybridizing to under physiological conditions, or
effecting RNAi
activity), a polynucleotide sequence of the virus, e.g., the IRES sequence of
HCV. In some
embodiments, the mammal is human. In one embodiment, the mammal is a human and
the viral
infection is a HCV infection, such as an infection with HCV genotype 1 a, and
the small
interfering RNA comprises a sequence that is at least complementary to a
sequence of the IRES
of the HCV.
[0081] As used herein, a "therapeutically effective amount" is an amount of a
small
interfering RNA that can render a desired therapeutic outcome (e.g., reduction
or elimination of a
viral infection). A therapeutically effective amount may be administered in
one or more doses.
21
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
Non-limiting examples of doses are about 0.1 mg/kg to about 50 mg/kg, e.g.,
about 1 to about 5
mg/kg. Suitable methods of delivery are known in the art and include, for
example, intravenous
administration (e.g., via a peripheral vein of via a catheter). Non-limiting
examples include
delivery via the hepatic artery or the portal vein.
[0082] Generally, in methods for treating a viral infection in a mammal, the
small interfering
RNA is administered with a pharmaceutically acceptable carrier. As used
herein, a
"pharmaceutically acceptable carrier" (also interchangeably termed
"pharmaceutically
acceptable excipient" herein) is a relatively inert substance that facilitates
administration of the
small interfering RNA or RNAs. For example, a carrier can give form or
consistency to the
composition or can act as a diluent. A pharmaceutically acceptable carrier is
biocompatible (i.e.,
not toxic to the host) and suitable for a particular route of administration
for a pharmacologically
effective substance. Suitable pharmaceutically acceptable carriers include but
are not limited to
stabilizing agents, wetting and emulsifying agents, salts for varying
osmolarity, encapsulating
agents, buffers, and skin penetration enhancers. In some embodiments, the
pharmaceutically
acceptable carrier is water or saline. Examples of pharmaceutically acceptable
carriers are
described in Reynington's Pharmaceutical Sciences (Alfonso R. Gennaro, ed.,
18th edition,
1990).
[0083] In methods for treating a viral infection, small interfering RNAs as
described herein
are generally administered parenterally, e.g., subcutaneously, intravenously,
or intramuscularly.
L'onijlositi0)Es
[0084] The invention provides compositions for inhibiting viral gene
expression and/or
treating a viral infection in a mammal comprising at least one small
interfering RNA as
described herein. Compositions of the invention may comprise two or more small
interfering
RNAs as described herein. In accordance with the invention, a small
interfering RNA, e.g.,
shRNA or siRNA, comprises a sequence that is substantially complementary to a
viral
polynucleotide sequence of about 19 to about 30 nucleotides, wherein
interaction of the
substantially complementary sequence of the small interfering RNA with the
polynucleotide
sequence of the virus inhibits viral gene expression, for example, by cleavage
of viral
polynucleotide sequences.
22
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
[0085] In some embodiments, the composition comprises an shRNA that includes a
sequence
selected from the group consisting of SEQ ID NOs: 12, 17, 18, 19, 20, 21, 22,
23, 24, and 25. In
some embodiments, the composition comprises an shRNA that includes one of the
following:
SEQ ID NO:34, SEQ ID NO:35, SEQ ID NO:36, SEQ ID NO:37, SEQ ID NO:38, SEQ ID
NO:39, SEQ ID NO:40, SEQ ID NO:41, SEQ ID NO:42, SEQ ID NO:43, SEQ ID NO:44,
SEQ
ID NO:45, SEQ ID NO:46, SEQ ID NO:47, SEQ ID NO:48, SEQ ID NO:49, SEQ ID
NO:50,
SEQ ID NO:51; SEQ ID NO:52, SEQ ID NO:53, SEQ ID NO:54, SEQ ID NO:55, or SEQ
ID
NO:56 (Table 10). In some embodiments, the composition comprises one or more
shRNAs of
SEQ ID NO:57-1 10. In some embodiments, the composition comprises a siRNA
comprising a
sequence selected from SEQ ID NOs:19, 20, 21, 22, 23, 24, and 25. In other
embodiments, the
composition comprises a siRNA that includes a sequence of SEQ ID NO:34, SEQ ID
NO:35,
SEQ ID NO:36, SEQ ID NO:37, SEQ ID NO:38, SEQ ID NO:39, SEQ ID NO:40, SEQ ID
NO:41, SEQ ID NO:42, SEQ ID NO:43, SEQ ID NO:44, SEQ ID NO:45, SEQ ID NO:46,
SEQ
ID NO:47, SEQ ID NO:48, SEQ ID NO:49, SEQ ID NO:50, SEQ ID NO:51; SEQ ID
NO:52,
SEQ ID NO:53, SEQ ID NO:54, SEQ ID NO:55, or SEQ ID NO:56 (Fig. 10). In some
embodiments, the composition comprises a shRNA or siRNA that binds to, i.e.,
comprises a
sequence substantially complementary to, a sequence of about 19 to about 30
nucleotides within
the IRES element of HCV, for example, HCV genotype la. A composition can
include more
than one different shRNA, e.g., shRNAs targeting different sequences of an
IRES or different
alleles or mutations of a target sequence. An shRNA or siRNA as described
herein can include
more than one of the identified sequences. Certain compositions contain more
than one different
shRNA or siRNA sequences.
[0086] In some embodiments, the invention provides a pharmaceutical
composition
comprising a small interfering RNA as described herein and a pharmaceutically
acceptable
carrier. In some embodiments, the pharmaceutical composition is formulated for
parenteral
administration to a mammal, for example, a human.
[0087] A pharmaceutical composition that includes a short interfering RNA
(e.g., an siRNA
or an shRNA) is formulated to be compatible with its intended route of
administration.
Examples of routes of administration include parenteral, e.g., intravenous,
intradermal,
subcutaneous, inhalation, transdermal (topical), transmucosal, and rectal
administration; or oral.
Solutions or suspensions used for parenteral, intradermal, or subcutaneous
application can
23
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
include the following components: a sterile diluent such as water for
injection, saline solution,
fixed oils, polyethylene glycols, glycerin, propylene glycol or other
synthetic solvents;
antibacterial agents such as benzyl alcohol or methyl parabens; antioxidants
such as ascorbic
acid or sodium bisulfite; chelating agents such as ethylenediaminetetraacetic
acid; buffers such
as acetates, citrates or phosphates and agents for the adjustment of tonicity
such as sodium
chloride or dextrose. pH can be adjusted with acids or bases, such as
hydrochloric acid or
sodium hydroxide. A parenteral preparation can be enclosed in ampoules,
disposable syringes or
multiple dose vials made of glass or plastic.
[0088] Pharmaceutical compositions suitable for injectable use include sterile
aqueous
solutions (where water soluble) or dispersions and sterile powders for the
extemporaneous
preparation of sterile injectable solutions or dispersion. For intravenous
administration, suitable
carriers include physiological saline, bacteriostatic water, Cremophor ELTM
(BASF, Parsippany,
NJ) or phosphate buffered saline (PBS). In all cases, the composition must be
sterile and should
be fluid to the extent that easy syringability exists. It should be stable
under the conditions of
manufacture and storage and must be preserved against the contaminating action
of
microorganisms such as bacteria and fungi. The carrier can be a solvent or
dispersion medium
containing, for example, water, ethanol, polyol (for example, glycerol,
propylene glycol, and
liquid polyetheylene glycol, and the like), and suitable mixtures thereof. The
proper fluidity can
be maintained, for example, by the use of a coating such as lecithin, by the
maintenance of the
selected particle size in the case of dispersion and by the use of
surfactants. Prevention of the
action of microorganisms can be achieved by various antibacterial and
antifungal agents, for
example, parabens, chlorobutanol, phenol, ascorbic acid, thimerosal, and the
like. In some cases,
isotonic agents are included, for example, sugars, or polyalcohols such as
manitol, sorbitol, or
sodium chloride. Prolonged absorption of an injectable composition can be
effected by including
in the composition an agent which delays absorption, for example, aluminum
monostearate or
gelatin.
[0089] Sterile injectable solutions can be prepared by incorporating the
active compound in
the specified amount in an appropriate solvent with one or a combination of
ingredients
enumerated above, as needed, followed by filtered sterilization. Generally,
dispersions are
prepared by incorporating the active compound into a sterile vehicle that
contains a basic
dispersion medium and other ingredients selected from those enumerated above
or others known
24
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
in the art. In the case of sterile powders for the preparation of sterile
injectable solutions, the
preferred methods of preparation are vacuum drying and freeze-drying which
yields a powder of
the active ingredient plus any additional desired ingredient from a previously
sterile-filtered
solution thereof.
[0090] Oral compositions generally include an inert diluent or an edible
carrier. For the
purpose of oral therapeutic administration, the active compound can be
incorporated with
excipients and used in the form of tablets, troches, or capsules, e.g.,
gelatin capsules.
Pharmaceutically compatible binding agents can be included as part of the
composition. The
tablets, pills, capsules, troches and the like can contain any of the
following ingredients, or
compounds of a similar nature: a binder such as microcrystalline cellulose,
gum tragacanth or
gelatin; an excipient such as starch or lactose, a disintegrating agent such
as alginic acid,
Primogel, or corn starch; a lubricant such as magnesium stearate or Sterotes;
a glidant such as
colloidal silicon dioxide; a sweetening agent such as sucrose or saccharin; or
a flavoring agent
such as peppermint, methyl salicylate, or orange flavoring.
[0091] For administration by inhalation, the compounds are delivered in the
form of an
aerosol spray from pressured container or dispenser that contains a suitable
propellant, e.g., a gas
such as carbon dioxide, or a nebulizer.
[0092] Systemic administration can also be by transmucosal or transdermal
means. For
transmucosal or transdermal administration, penetrants appropriate to the
barrier to be permeated
are used in the formulation. Such penetrants are generally known in the art,
and include, for
example, for transmucosal administration, detergents, bile salts, and fusidic
acid derivatives.
Transmucosal administration can be accomplished through the use of nasal
sprays or
suppositories. For transdermal administration, the active compounds are
formulated into
ointments, salves, gels, or creams as generally known in the art.
[0093] The compounds can also be prepared in the form of suppositories (e.g.,
with
conventional suppository bases such as cocoa butter and other glycerides) or
retention enemas
for rectal delivery.
[0094] In one embodiment, the active compounds are prepared with carriers that
will protect
the compound against rapid elimination from the body, such as a controlled
release formulation,
including implants and microencapsulated delivery systems. Biodegradable,
biocompatible
polymers can be used, such as ethylene vinyl acetate, polyanhydrides,
polyglycolic acid,
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
collagen, polyorthoesters, and polylactic acid. Methods for preparation of
such formulations will
be apparent to those skilled in the art. The materials can also be obtained
commercially from
Alza Corporation and Nova Pharmaceuticals, Inc. Liposomal suspensions
(including liposomes
targeted to infected cells with monoclonal antibodies to viral antigens) can
also be used as
pharmaceutically acceptable carriers. These can be prepared according to
methods known to
those skilled in the art, for example, as described in U.S. Patent No.
4,522,811.
[0095] It is advantageous to formulate oral or parenteral compositions in
dosage unit form
for ease of administration and uniformity of dosage. Dosage unit form as used
herein refers to
physically discrete units suited as unitary dosages for the subject to be
treated; each unit
containing a predetermined quantity of active compound calculated to produce
the desired
therapeutic effect in association with the selected pharmaceutical carrier.
[0096] Toxicity and therapeutic efficacy of compounds disclosed herein can be
determined
by pharmaceutical procedures known in the art, for example, in cell cultures
or experimental
animals, e.g., for determining the LD50 (the dose lethal to 50% of the
population) and the ED50
(the dose therapeutically effective in 50% of the population). The dose ratio
between toxic and
therapeutic effects is the therapeutic index and it can be expressed as the
ratio LD50/ED50.
Compounds that exhibit high therapeutic indices are preferred. While compounds
that exhibit
toxic side effects may be used, care should be taken to design a delivery
system that targets such
compounds to the site of affected tissue to minimize potential damage to
uninfected cells and,
thereby, reduce side effects.
[0097] The data obtained from the cell culture assays and animal studies can
be used in
formulating a range of dosage for use in humans. The dosage of such compounds
lies preferably
within a range of circulating concentrations that include the ED50 with little
or no toxicity. The
dosage may vary within this range depending upon the dosage form employed and
the route of
administration utilized. For any compound used in the method of the invention,
the
therapeutically effective dose can be estimated initially from cell culture
assays. A dose may be
formulated in animal models to achieve a circulating plasma concentration
range that includes
the IC50 (i.e., the concentration of the test compound which achieves a half-
maximal inhibition
of symptoms) as determined in cell culture. Such information can be used to
more accurately
determine useful doses in humans. Levels in plasma may be measured, for
example, by high
performance liquid chromatography.
26
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
[0098] The invention also relates to a method of making a medicament for use
in treating a
subject, e.g., for HCV infection. Such medicaments can also be used for
prophylactic treatment
of a subject at risk for or suspected of having an HCV infection.
Kits
[0099] The invention provides kits comprising a small interfering RNA as
described herein.
In some embodiments, the kits also include instructions for use in the methods
for inhibiting viral
gene expression and/or methods for treatment of a viral infection in a mammal
described herein.
Instructions may be provided in printed form or in the form of an electronic
medium such as a
floppy disc, CD, or DVD, or in the form of a website address where such
instructions may be
obtained.
[0100] In some embodiments, the kits include a pharmaceutical composition of
the
invention, for example including at least one unit dose of at least one small
interfering RNA such
as a shRNA or a siRNA, and instructions providing information to a health care
provider
regarding usage for treating or preventing a viral infection. The small
interfering RNA is often
included as a sterile aqueous pharmaceutical composition or dry powder (e.g.,
lyophilized)
composition.
[0101] Suitable packaging is provided. As used herein, "packaging" refers to a
solid matrix
or material customarily used in a system and capable of holding within fixed
limits a
composition of the invention suitable for administration to an individual.
Such materials include
glass and plastic (e.g., polyethylene, polypropylene, and polycarbonate)
bottles, vials, paper,
plastic, and plastic-foil laminated envelopes and the like. If e-beam
sterilization techniques are
employed, the packaging should have sufficiently low density to permit
sterilization of the
contents.
[0102] Kits may also optionally include equipment for administration of a
pharmaceutical
composition of the invention, such as, for example, syringes or equipment for
intravenous
administration, and/or a sterile solution, e.g., a diluent such as water,
saline, or a dextrose
solution, for preparing a dry powder (e.g., lyophilized) composition for
administration.
27
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
Table 1
Listing of Targeting Sequences Disclosed in the Application that can be
Incorporated into
shRNA or siRNA and Examples of such shRNAs and siRNAs
Sequence ID # Antisense sequence (5'-3') Target Examples of
Position on shRNA or siRNA
HCV IRES
SEQ ID NO:27 UCUUUGAGGUUUAGGAUUCGUGCUC 344-368 HCVa-wt shRNA
SEQ ID NO:28 UCUUUGAGGUUUAGGAUUGGUGCUC 344-368 HCVa-SNP1 shRN
SEQ ID NO:29 UCUUUGAGCUUUAGGAUUCGUGCUC 344-368 HCVa-SNP2 shRN
SEQ ID NO:30 UCUUUGAGCUUUAGGAUUGGUGCUC 344-368 HCVa-mut shRNA
SEQ ID NO:31 CCUCCCGGGGCACUCGCAAGCACCC 299-323 HCVb-wt shRNA
SEQ ID NO:32 UGGUGCACGGUCUACGAGACCUCCC 318-342 HCVc-wt shRNA
SEQ ID NO:33 GGUUUUUCUUUGAGGUUUAGGAUUC 350-374 HCVd-wt shRNA
SEQ ID NO:19 AGGUUUAGGAUUCGUGCUC 344-362 siRNA#1, shRNA#
SEQ ID NO:20 GAGGUUUAGGAUUCGUGCU 345-363 siRNA#2, shRNA#
SEQ ID NO:21 UGAGGUUUAGGAUUCGUGC 346-364 siRNA#3, shRNA#
SEQ ID NO:22 UUGAGGUUUAGGAUUCGUG 347-365 siRNA#4, shRNA#
SEQ ID NO:23 UUUGAGGUUUAGGAUUCGU 348-366 siRNA#5, shRNA#
SEQ ID NO:24 CUUUGAGGUUUAGGAUUCG 349-367 siRNA#6, shRNA#
SEQ ID NO:25 UCUUUGAGGUUUAGGAUUC 350-368 siRNA#7, shRNA#
[0103] Fig. 16A-B illustrates examples of shRNAs containing sequence targeting
HCV
IRES, and tested using methods described herein.
EXAMPLES
[0104] The invention is further illustrated by the following examples. The
examples are
provided for illustrative purposes only. They are not to be construed as
limiting the scope or
content of the invention in any way.
28
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
Example 1: Design and Construction of shRNA Expression Cassettes, T7
Transcription Reactions, and Reporter Gene Assays
[0105] Chemically synthesized oligonucleotides were obtained from IDT
(Coralville, IA),
resuspended in RNase- and pyrogen-free water (Biowhittaker), and annealed as
described below.
The following oligonucleotide pairs, for making shRNA, contain a T7 promoter
element (doubly
underlined), sense and antisense HCV IRES sequence and a miR-23 microRNA loop
structure
(reported to facilitate cytoplasmic localization [21, 22]).
[0106] T7-HCVa-wt fw:
5' -taatac gactcactata~ ~ ~agcacgaatcctaaacetca
[0107] aagaCTTCCTGTCAtctttgaggtttaggattcgtgctcTT-3' (SEQ ID NO: 1);
T7-HCVa-wt rev:
5' -AAgagcacgaatcctaaacctcaaagaTGACAGGAA
Gtctttgaggtttaggattcgtgct ccctatagtgagtcgtatta-3'
(SEQ ID NO:2)
[0108] (T7 promoter sequence doubly underlined). T7 transcripts for HCVa-mut
shRNA
were identical with the exception that nucleotide changes (G->C and C->G) were
incorporated
into the synthesized oligonucleotides at the singly underlined residues.
[0109] HCVa-wt shRNA (Fig. 1B) was designed to target the region 344-374 on
the HCV
IRES; HCVb-wt was designed to target the region 299-323 (Fig. 1C); HCVc-wt was
designed to
target the region 318-342 (Fig. 1C); and HCVd-wt was designed to target the
region350-374
(Fig. 1 C).
[0110] ShRNAs #1-7 (targeting positions 344-362, 345-363, 346-364, 347-365,
348-366,
349-367, 350-368 on the HCV IRES; See Fig. 4A, which depicts the 19 base pair
viral
recognition sequences) were in vitro transcribed using the MEGAscript kit
(Ambion) and
contained the same loop sequences and 5', 3'-overhangs as HCVa-wt shRNA.
SiRNAs #1-7
(see Fig. 4A, which depicts the 19 base pair viral recognition sequences) were
chemically
synthesized at Dharmacon (Lafayette, CO) and contained 3'-UU overhangs on both
sense and
antisense strands.
[0111] The oligonucleotide pair used to prepare the control shRNA 229 (which
targets tumor
necrosis factor alpha) is 229-5'-TAATACGACTCACTATAGGGGCG
GTGCCTATGTCTCAGCCTCTTCTCACTTCCTGTCATGAGAAGAGGCTGAGACATAGG
29
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
CACCGCC TT-3' (SEQ ID NO:3)
and 229-3'-AAGGCG GTGCCTATGTC TCAGCC TCT TCTCA TGACAGGAAG TGAGA
AGAGGCTGA GACATAGGCACCCCTATAGTGAGTCGTATTA-5' (SEQ ID NO:4).
Pol III U6 shRNA expression vector construction- design of small hairpin shRNA
expressiott vectoi-s
[0112] Oligonucleotide pairs were incubated together at 95 C for two minutes
in RNA
polymerase buffer (e.g., 120 [tl of each 100 M oligonucleotide in 60 l 5X
annealing buffer
(Promega; 1X = 10 mM Tris-HC1(pH 7.5), 50 mM NaC1) and slowly cooled
(annealed) over 1
hour to less than 40 C. The oligonucleotides were designed to provide 4-base
overhangs for
rapid cloning into Bbsl/BamHl-digested pCRII-U6 plasrnid (Bbs 1 and BamHl
recognition sites
or overhangs are underlined in the oligonucleotide sequences). The pCRII-U6
po1 III expression
plasmid was prepared by subcloning the PCR product obtained from human HT-1080
genomic
DNA using primers and huU6-5' ATCGATCCCCAGTGGAAAGACGCGCAG (SEQ ID NO:5)
and huU6-
3' -GGATCCGAATTCGAAGACCACGGTGTTTCGTCCTTTCCACAA-5'
[0113] (SEQ ID NO:6) [23] into the pCRII vector (Invitrogen) using the TA
cloning kit
(Invitrogen). The cassette consisting of the annealed oligonucleotides
(encoding the HCV Il2ES
shRNA) was ligated into the Bbs1/BantHl-digested pCRII-U6 plasmid. The
expressed shRNA
contains a miR-23 microRNA loop structure to facilitate cytoplasmic
localization [21, 22]. The
final pCRII-U6 constructs were confirmed by sequencing. The primers pairs used
were:
pHCVa-wt 5'-ACG GAGCACGAATCCTAAACCTCAAAGA CTTCCTGTCA
TCTTTGAGGTTTAGGATTCGTGCTC TTTTTTG-3' (SEQ ID NO:7) and 5'-
GATCCAAAAAA GAGCACGAATCCTAAACCTCAAAGA TGACAGGAAG
TCTTTGAGGTTTAGGATTCGTGCTC-3' (SEQ ID NO:8). Oligonucleotides containing the
appropriate sequence changes at the underlined residues (see above) were used
to generate the
pCRII-U6/HCVa-mut (double mutation), HCVsnpl (single change at 5' side) and
HCVsnp2
(single change at 3' end) as depicted in Fig. 1B and described above. The
control pCRII-U6/229
was prepared is similar fashion using the oligonucleotides
5'-ACCGGGCG GTGCCTATGTCTCAGCCTCTTCTCACTTCCTGTCATGAGA
AGAGGCTGAGACATAGGCACCGCCTTTTTT-3' (SEQ ID NO:9) and 3'-
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
GATCAAAAAAGGCGGTGCCTATGTCTCAGCCTCTTCTCATGACAGGAAGTGAGAAG
AGGCTGAGACATAGGCACCGCC-5' (SEQ ID NO:10).
T7 transcription reactions
[0114] Oligonucleotide pairs were incubated at 95 C for two minutes in RNA
polymerase
buffer (e.g., 120 l of each 100 M oligonucleotide in 60 [t15X transcription
buffer (Promega))
and slowly cooled (annealed) over 1 hour to less than 40 C. ShRNA was
transcribed at 42 C for
four hours from 5 M of the resulting annealed double-stranded DNA template
using the
AmpliScribeTM T7 Flash transcription kit (Epicentre Technologies) followed by
purification on a
gel filtration spin column (MicrospinTM G-50, Amersham Biosciences) that had
been thoroughly
washed three times with phosphate buffered saline (PBS) to remove
preservative.
siRNAs
[0115] siRNAs were prepared by annealing chemically synthesized (Dharmacon)
complementary strands of RNA, each containing the appropriate recognition
sequence plus an
(overhanging) UU extension on the 3'end.
Transfections and reporter gene assays
[0116] Human 293FT (Invitrogen) and Huh7 cells (American Type Culture
Collection
(ATCC), Manassas, VA) were maintained in DMEM (Biowhittaker ) with 10% fetal
bovine
serum (HyClone), supplemented with 2 mM L-glutamine and 1mM sodium pyruvate.
The day
prior to transfection, cells were seeded at 1.7 x 105 cells/well in a 24-well
plate, resulting in
about 80% cell confluency at the time of transfection. Cells were transfected
with
LipofectamineTM 2000 (Invitrogen, Carlsbad, CA) following the manufacturer's
instructions.
For the inhibition experiments, 293FT or Huh7 cells were cotransfected (in
triplicate) with 40 ng
pCDNA3/HCV IRES dual luciferase (renilla and firefly) reporter construct, 50
ng pSEAP2-
control plasmid (BD Biosciences Clontech, as transfection controls) and the
indicated amounts
of T7-generated shRNA (typical amount 1 pmole) or pCRII-U6 shRNA expression
construct
(710 ng). Compensatory pUC18 plasmid was added to the transfection mix to give
a final
concentration of 800 ng total nucleic acid per transfection. Forty-eight hours
later, supernatant
was removed, heated at 65 C for 15-30 minutes, and 5-10 l of the supernatant
was added to 150
31
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
l p-nitrophenyl phosphate liquid substrate system (pNPP, Sigma). After a 30-60
minute
incubation at room temperature, samples were read (405 nm) on a Molecular
Devices
Thermomax microplate reader and quantitated using SOFTmax software (Molecular
Devices).
The remaining cells were lysed and luciferase activity measured using the Dual-
Luciferase
Reporter assay system (Promega) and MicroLumat LB 96 P luminometer (Berthold).
Mice
[0117] Six-week old female Balb/c mice were obtained from the animal facility
of Stanford
University. Animals were treated according to the NIH Guidelines for Animal
Care and the
Guidelines of Stanford University.
Mouse lzydrodynamic injections and in vivo imaging
[0118] Hydrodynamic tail vein injections were performed as described by
McCaffrey and
colleagues with minor modifications including omission of RNasin [24]. A total
volume of 1.8
ml of phosphate-buffered saline containing inhibitor (RNA or plasmid), 10 g
of pHCV Dual
Luc plasmid, and 2 g pSEAP2-control plasmid (BD Biosciences Clontech,
contains the SV40
early promoter), was steadily injected into the mouse tail vein over about
five seconds ( N = 4-6
animals per group ). At the indicated times, 100 gl of 30 mg/ml luciferin was
injected
intraperitoneally. Ten minutes following the injection, live anesthetized mice
were analyzed
using the IVIS7 imaging system (Xenogen Corp., Alameda, CA) and the resulting
light emission
data quantitated using LivingImage software (Xenogen). Raw values are reported
as relative
detected light per minute and standard errors of the mean for each group (N=4-
5 animals) are
shown.
Secreted alkaline phosphatase (SEAP) assay
[0119] At day 5, mice were bled through the retro-orbital vein of the eye. The
serum was
separated from blood cells by microcentrifugation, heated at 65 C for 30
minutes to inactivate
endogenous alkaline phosphates, and 5-10 l of the serum was added to 150 l
pNPP liquid
substrate system (see above). After a 30-60 minute incubation at room
temperature, samples
were read (405 nm) and quantitated as described above.
32
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
Example 2: shRNA Inhibition of HCV IRES-Mediated Gene Expression in Human
Tissue Culture Cells
[0120] In this study, short interfering RNAs (shRNAs and siRNAs) designed and
constructed
as in Example 1 to target a conserved region of the hepatitis C IRES were
tested for their ability
to inhibit HCV IRES-mediated reporter expression in human tissue culture
cells.
[0121] Fig. 1A shows the HCV IRES target site (panel A) as well as the HCV
shRNA
resulting from T7 transcription of a template prepared from hybridized
oligonucleotides
containing a T7 promoter sequence and HCV IRES target (Fig. 1B). The
underlined residues are
those that were changed to generate the mutant HCV shRNAs. The shRNAs contain
a mir-23
microRNA loop structure that was previously suggested to facilitate
cytoplasmic localization
[21, 22] and a 25 base pair RNA stem with two nucleotides at the 5' (two
guanines) and 3' (two
uridines) ends that may also hybridize though non Watson-Crick G:U base
pairings. For vector-
delivered shRNAs, overlapping oligonucleotides were subcloned into a poIII
expression vector
(pCRII-U6, see Example 1).
[0122] Three other shRNAs were also designed with the same stem length and
loop sequence
that target nearby positions in Domain IV of the HCV IRES (Fig. 1C). HCVb-wt
shRNA targets
a highly structured region (used as negative control, to compare efficiency),
while HCVc-wt and
HCVd-wt shRNA target regions that are more 'accessible' according to
biochemical footprinting
studies (Fig. 1D; Brown et al., Nucleic Acids Res., 1992, 20:5041-5.). All
RNAs were in vitro
transcribed from dsDNA templates containing a T7 promoter, similar to the HCVa-
wt shRNA.
[0123] To test the effectiveness of the HCV shRNAs to inhibit HCV IRES-
mediated gene
expression, human 293FT or hepatocyte Huh7 cells were co-transfected with
pCDNA3/HCV
IRES dual luciferase expression plasmid, secreted alkaline phosphatase
expression plasmid
(pSEAP2, to control for efficiency of transfection) as well as in vitro
synthesized shRNAs or
alternatively, pol III expression vectors containing the corresponding shRNA
cassettes.
[0124] As seen in Fig. 1F, both HCVa-wt and HCVd-wt shRNAs, which target the
region of
the IRES immediately downstream of the AUG translation start site (positions
344-368 and 350-
374, respectively), strongly inhibit HCV IRES-mediated fLuc expression in
human 293FT cells.
HCVc-wt (targeting 318-342) showed moderate inhibition and HCVb-wt (299-323)
displayed
little if any activity, as expected. Thus, preliminary screening revealed a
potent shRNA, HCVa-
wt, that was chosen for further detailed studies.
33
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
Specificity and potency of inhibition of HCV IRES-naedr'ated gene expression
by shRNAs in
293FT cells
[0125] To further test inhibition of HCV-IRES driven gene expression, 293FT
cells were
cotransfected with dual luciferase reporter and SEAP expressing plasmids as
well as 1 pmole of
in vitro transcribed shRNAs. The target plasmid was pCDNA3/HCV IRES dual
luciferase
reporter (HCV IRES, as shown in Fig. 1E). pUC18 plasmid was added to the
transfection mix to
give a final total nucleic acid concentration of 800 ng per transfection per
well (24-well tissue
culture plates). Forty-eight hours later, supernatant was removed for SEAP
analysis, then cells
were lysed and firefly and renilla (not shown) luciferase activity measured as
described in
Example 1. Firefly luciferase and SEAP activities were normalized to 100.
Results are shown in
Fig. 2A.
[0126] HCVa-wt shRNA targeting the region of the IRES immediately downstream
of the
AUG translation start site strongly inhibited HCV Il2ES-mediated fLuc
expression in both
human 293FT (Fig. 2) and hepatocyte Huh7 (Fig. 3B) cell lines. Little or no
inhibition was
observed using either a mutant shRNA (HCVa-mut) containing two changes in the
pairing of the
RNA hairpin (for mismatch location, see Fig. 1B) or an unrelated TNF (229)
shRNA. The 229
TNF shRNA is highly effective at inhibiting TNF expression (Seyhan et al.,
RNA, 2005, 11:837-
846), suggesting that this shRNA is utilized effectively by the RNAi
apparatus. Single
nucleotide changes in the hairpin region, at either the upstream or downstream
position (SNP1
and SNP2 respectively; see Fig. 2C), had a partial effect.
[0127] Little or no inhibition was observed when the HCV shRNA was targeted to
a similar
dual luciferase construct in which the HCV IRES was replaced by the
encephalomyocarditis
virus (EMCV) IRES (Figs. 2B and 3B). Thus, the data of Fig. 2B illustrate that
HCVa-wt
shRNA does not inhibit a similar target lacking the HCV IRES. In this
experiment, cells were
transfected as for Fig. 2A except that pCDNA3/EMCV dual luciferase reporter
(EMCV IRES)
was used as target in place of pCDNA3/HCV. These data are presented in Fig. 2B
as luciferase
activity divided by SEAP activity normalized to 100.
[0128] To confirm that the shRNAs were acting by degrading their target mRNA,
a Northern
blot analysis was performed (Fig. 2F). Equal amounts of total RNA, isolated
from cells
transfected with no inhibitor or HCVa-wt, HCVmutl/2, or 229 shRNAs, were
separated by gel
34
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
electrophoresis. The separated RNA was transferred to a membrane and
hybridized to
radiolabeled cDNA probes specific for fLuc, SEAP and elongation factor lA
(EF1A). HCVa-wt
shRNA (lane 3) specifically inhibited fLuc mRNA accumulation (63% inhibition
compared to
229 shRNA (lane 2) when corrected for SEAP and EF1A mRNA levels; no inhibition
was
observed for HCVa-inutl/2) (compare lanes 3 and 4) following quantitation by
phosphorimager.
These data demonstrate that the shRNAs were degrading target mRNA.
[0129] Dose response experiments showed that the HCVa-wt shRNA effectively
inhibited
HCV IRES-dependent gene expression at 0.3 nM in 293FT cells (96 percent
inhibition, see Fig.
2D) and 0.1 nM in Huh7 cells (75 percent inhibition, see Fig. 3A).
[0130] To further investigate local sequence effects on potency, seven in
vitro-transcribed 19
bp shRNA and the corresponding synthetic 19 base pair siRNA, targeting all
possible positions
within the 31-base pair site of HCVa (344-374 ; Fig. 4A), were assayed for
inhibitory activity. A
25base pair synthetic siRNA corresponding to HCVa-wt shRNA was also tested.
All of them
exhibited a high level of activity (Fig. 4B). The most potent were siRNA and
shRNA versions of
HCVa as well as siRNA #3, which was effective at 1 nM (>90% inhibition, Fig.
4B) and 0. 1 nM
(about 90% inhibition, Fig. 4C). Thus, 19-25 base pair shRNAs and siRNAs
designed to target
the region 344-374 on the HCV IRES are potent, with some local differences.
Example 3: shRNA Inhibition of HCV IRES-Mediated Gene Expression in a Mouse
Model System
[0131] The ability of the HCV shRNA and HCV shRNA expression plasmid to
inhibit target
gene expression was extended to a mouse model system using hydrodynamic
injection to deliver
the nucleic acids to mouse liver. Fig. 5 shows the results of injecting a
large volume of PBS (1.8
ml) containing pHCV dual Luc, pSEAP2, and shRNAs (10 fold excess over the
target on a rnass
basis of either shRNA or pol III expression vectors expressing the shRNAs)
into the tail veins of
mice (n=4-5 mice). At the time points shown in Fig. 5B, luciferin was injected
intraperitoneally
and the mice were imaged with a high sensitivity, cooled CCD camera. (Fig. 5A
shows
representative mice chosen from each set (4-5 mice per set) at the 84 hour
time point.) At all
time points tested, HCV shRNA robustly inhibited luciferase expression ranging
from 98% (84
hour time point) to 94% (48-hour time point) inhibition compared to mice
injected with pUC18
in place of shRNA inhibitor. Mutant (mut) or control (229) shRNAs had little
or no effect. It
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
should be noted that luciferase activity decreases with time, possibly due to
loss of DNA or
promoter silencing [8] and that the data are normalized within each time point
(see description of
Fig. 5 above).
[0132] Fig. 6 shows a comparison of HCVa-wt shRNA inhibitory activity with a
phosphoramidite morpholino oligomer that was previously shown to effectively
target this same
site [8]. Both the HCVa-wt shRNA and morpholino oligomers effectively blocked
luciferase
expression at all time-points tested. Data are shown for the 48-hour time-
point, where inhibition
was 99.95 and 99.88 percent, respectively for the HCVa-wt shRNA and morpholino
inhibitors.
Example 4: Inhibition of Semliki Forest Virus (SFV ) Using shRNAs
[0133] SFV has been used as a model system for more virulent positive-strand
RNA viruses.
To examine the inhibitory effect of RNAi on SFV growth, shRNAs targeting four
SFV genes
(nsp-1, nsp-2 and nsp-4, and capsid) and one mismatched control for the nsp-4
site were
generated and expressed from a U6 promoter. Their ability to tested their
ability to inhibit the
proliferation of SFV-A7-EGFP, a version of the replication-proficient SFV
strain SFV-A7 that
expresses a eGFP reporter gene [49]. A modest reduction (about 35%) of SFV-GFP
replication
was seen with shRNAs targeting the nsp-1 (Fig. 7) but not nsp-2, nsp-4 or
capsid coding regions,
nor with the mismatched siRNA (not shown).
[0134] A site within the capsid coding region that was previously shown to be
effective on
Sindbis virus [50] was not effective on SFV. The Sindbis-SFV sequence homology
at this site is
only 77%. SFV is a very rapidly growing virus, producing up to 200,000
cytoplasmic RNAs
during its infectious cycle. To see if cells could better protected from a
slower-growing virus,
the effects of these siRNAs on a replication-deficient strain of SFV-GFP were
tested in two
separate experiments. Fig. 8 shows that U6-expressed shRNAs targeting this SFV
strain can
reduce viral expression by >_70% over a time period of up to five days. This
effect was seen with
siRNAs targeting the nonstructural genes nsp-1, nsp-2, and nsp-4 as well as an
siRNA with one
mismatch to nsp-4, but not for the capsid gene (which is lacking in this
crippled virus) or other
controls (Fig. 8). Note that the length of the sequence targeted by the shRNAs
is 29 nucleotides
and the single mismatch used in the nsp-4 mismatch shRNA is apparently not
disruptive for the
RNAi effect. The wide variation in effectiveness of the various shRNAs
underscores the
36
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
importance of a library approach for finding the best siRNAs and shRNAs when
dealing with
rapidly replicating and highly mutagenic viruses such as SFV.
[0135] Dose-response experiments were performed to examine inhibition of an
HCV
replicon system in Huh7 cells by HCVa-wt shRNA and HCVa-mut shRNA as well as a
non-
specific control shRNA (229). The antiviral activity of test compounds was
assayed in the stably
HCV RNA-replicating cell line, AVA5, derived by transfection of the human
hepatoblastoma
cell line, Huh7 (Blight, et al., Science, 2000, 290:1972). RNA-based
inhibitors were co-
transfected with DsRed expression plasmid into cultures that were about 80
percent confluent.
HCV RNA levels were assessed 48 hours after transfection using dot blot
hybridization. Assays
were conducted in triplicate cultures. A total of 4-6 untreated control
cultures, and triplicate
cultures treated with 10, 3, and 1 N/rnl a-interferon (active antiviral with
no cytotoxicity), and
100, 10, and 1 uM ribavirin (no antiviral activity and cytotoxic) served as
positive antiviral and
toxicity controls. The transfection efficiency was estimated by fluorescence
microscopy (DsRed
expression). Both HCV and b-actin RNA levels in triplicate treated cultures
were determined as
a percentage of the mean levels of RNA detected in untreated cultures (6
total). Beta-actin RNA
levels are used both as a measure of toxicity, and to normalize the amount of
cellular RNA in
each sample. A level of 30% or less HCV RNA (relative to control cultures) is
considered to be
a positive antiviral effect, and a level of 50% or less b-actin RNA (relative
to control cultures) is
considered to be a cytotoxic effect. Cytotoxicity is measured using an
established neutral red
dye uptake assay (Korba, B. E. and J. L. Gerin (1992). Use of a standardized
cell culture assay
to determine activities of nucleoside analogs against hepatitis B virus
replication (Antivir. Res.
19:55-70).
[0136] ]Inhibition of an HCV replicon system in Huh7 cells by HCVa-wt shRNA
and
HCVa-mut shRNA as well as an irrelevant control shRNA (229); dose response.
The antiviral
activity of test compounds was assayed in the stably HCV RNA-replicating cell
line, AVA5,
derived by transfection of the human hepatoblastoma cell line, Huh7 (Blight et
al. Science, 2000,
290:1972). RNA-based inhibitors were co-transfected with DsRed expression
plasmid into -80
percent confluent cultures and HCV RNA levels were assessed 48 hours after
transfection using
dot blot hybridization. Assays were conducted in triplicate cultures. A total
of 4-6 untreated
control cultures, and triplicate cultures treated with 10, 3, and 1 IU/ml a-
interferon (active
antiviral with no cytotoxicity), and 100, 10, and 1 uM ribavirin (no antiviral
activity and
37
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
cytotoxic) served as positive antiviral and toxicity controls. The
transfection efficiency was
estimated by fluorescence microscopy (DsRed expression). Both HCV and beta-
actin RNA
levels in triplicate treated cultures were determined as a percentage of the
mean levels of RNA
detected in untreated cultures (6 total). Beta actin RNA levels were used both
as a measure of
toxicity, and to normalize the amount of cellular RNA in each sample. A level
of 30% or less
HCV RNA (relative to control cultures) was considered to be a positive
antiviral effect, and a
level of 50% or less beta-actin RNA (relative to control cultures) was
considered to be a
cytotoxic effect. Cytotoxicity was measured using an established neutral red
dye uptake assay
(Korba et al., Antiviral Res., 1992, 19:55-70). Use of a standardized cell
culture assay to
determine activities of nucleoside analogs against hepatitis B virus
replication (Korba et al., 1992
supra).
Example 5: IdentiBcation of shRNAs that Inhibit HCV IRES-Dependent Gene
Expression
in Tissue Culture Cells
[0137] The ability of in vitro-transcribed small hairpin RNAs (shRNAs) to
inhibit hepatitis C
virus internal ribosome entry site (HCV IRES)-dependent gene expression in
cultured cells was
investigated. As disclosed supra, a 25 base pair shRNA HCVa-wt that targets
the 3' end of the
HCV IRES, near the AUG translation start site (Table 2) was found to be
effective for disrupting
expression of HCV. To assess the ability of co-transfected shRNA constructs to
interfere with
the function of the IRES, a reporter construct (pHCV Dual Luciferase plasmid)
in which firefly
luciferase (fLuc) expression is dependent on the HCV IRES was used (Fig. 1;
Wang et al., Mol.
Ther., 2005, 12:562-568. In these experiments, 293FT cells were cultured and
transfected with a
reporter construct and HCVa-wt or one of the other test sequences as described
in Wang et al.,
2005, supra.
[0138] It was found that at a concentration of 1 nM, HCVa-wt caused 90%
inhibition of
HCV IRES-dependent luciferase expression in 293FT cells (Wang et al., 2005,
supra). In
subsequent experiments, 26 additional shRNAs targeting various regions of the
HCV IlZES were
designed and tested (Fig. 10, Fig. 16A-B); 3 of the 26 were duplicates of
those described above
(HCVb, HCVc, HCVd-wt); 23 were new sequences) to identify additional
inhibitors of HCV.
The goal was to identify shRNAs that can be used either in combination with
HCVa-wt), making
it harder for the virus to develop resistance by mutating the HCVa-wt target
site, or as
38
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
alternatives to HCVa-wt. The shRNAs to be tested were chosen to avoid regions
that vary
among different HCV genotypes. Some test sequences were selected using the
algorithm
available at (e.g., jura.wi.rnit.edu/bioc/siRNAext/, and other test sequences
intentionally targeted
HCV-IRES sequences that, due to their CG content and other characteristics,
would not be
recommended by most algorithms would rule out, such as GC-rich or highly
structured regions.
The shRNAs were generated by in vitro transcription from dsDNA templates using
T7 RNA
polymerase and, to promote transcription efficiency, began with the sequence
5'-pppGGG. This
5' sequence formed an overhang of two to three nucleotides, the exact length
depending on
whether the target site contains one or more guanosine residues at its 5' end
(see Fig. 16A-B). If
the last nucleotide of the RNA sense strand matching a target sequence was
'G,' only two more
Gs had to be added for efficient transcription, and those Gs are single-
stranded on the 5'-end of
the shRNA, not complimentary to the target. If the last nucleotide of the
shRNA sense strand
matching the target, was not a G, then for efficient transcription in the test
systems, three Gs had
to be added that were not complimentary to the target. All shRNAs tested in
this set of
experiments had a duplex stem length of 21-25 base pairs and a 10 nucleotide
loop derived from
microRNA-23, as described for HCVa-wt.
[0139] All of the shRNAs (27 total, including HCVa-wt were assayed for
activity as
described in Wang, 2005. Briefly, human 293FT cells were co-transfected with
pHCV Dual
Luciferase Reporter expression plasmid (Promega, Madison, WI), and a secreted
alkaline
phosphatase expression plasmid (pSEAP2, Clontech, Mountain View, CA) to
control for
efficiency of transfection and possible off-target effects), and shRNA.
Results are shown in Fig.
10. SEAP levels were uniform in all samples, indicating efficient transfection
and the absence of
nonspecific inhibitory or toxic effects, at shRNA concentrations of 1 nM to 5
nM. Most of the
shRNAs displayed only moderate activity (less than 60% inhibition at 1 nM).
Without
committing to any particular theory, this effect is likely because the
targeted areas on IRES are
highly structured. The exceptions were HCVd-wt, sh37, sh39, hcvl7, which
target the IRES
positions near the HCVa-wt site. These shRNAs caused 85-90% inhibition of HCV
IRES
dependent gene expression at 1 nM concentration. The low shRNA concentration
of 1 nM was
chosen to allow easy identification of hyper-functional shRNAs. If the
screening were performed
at 10 nM shRNA, more shRNAs would display high activity; however, significant
nonspecific
inhibition was seen at that concentration in some cases. Thus, the screening
revealed a 44
39
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
nucleotide region (positions 331-374 on the HCV IRES) where five overlapping
shRNAs display
high activity.
Example 6: Effect of Sinmle Base Mismatches on shRNA Activity
[0140] It is desirable that a treatment for HCV be effective against mutated
HCV. To
determine the performance of the RNAs described herein in this regard (e.g.,
shRNAs targeting
HCV IRES), and to address whether off-target effects are problematic, the
sensitivity of selected
shRNA directed against HCV IRES to point mutations in the target sequence was
tested. For
these experiments, a C340->U mutation was introduced in the HCV IRES using the
QuikChange II Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). Of
the 27 shRNAs
that were assayed, nine targeted the mutated region (Fig. 11), therefore their
activity could
theoretically be affected by this mutation. All of these shRNAs were assayed
with the mutated
version of pHCV, along with selected shRNAs targeting other sites as controls.
For all tested
shRNAs, activity was found to be unaffected or slightly decreased compared to
the activity of
original, perfectly matched target (Fig. 10).
[0141] However, in the replicon system, shRNAs were surprisingly found to be
SNP-
sensitive (see below).
Example 7: Fine Mapping of Target Sites
[0142] Six short 19 base pair shRNAs were designed to target a 44 nucleotide
site near the
3'-terminus of the HCV IRES: three targeting nucleotides 331-353 and three
targeting
nucleotides 354-374. These molecules contained 10 nucleotide loops and 5'-GG
and 3'-UU
overhangs. Screening was performed to identify of non-overlapping candidates
that were most
effective among those sequences tested for inhibition of HCV expression. All
six of the shRNAs
tested were able to inhibit activity in the assay system. Three of the six
shRNAs (sh52, sh53,
and sh54) were identified as the most effective (Fig. 12). This does not
preclude the use of those
shRNAs that were less effective in a composition, e.g., for treating HCV, for
example as part of a
composition that includes more than one shRNA and/or siRNA.
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
Example 8: shRNA Design: Effects of Stem Length, Loop Length and Sequence, and
3'-
Terminus
[0143] Additional experiments were performed to test how shRNA design affects
gene
silencing activity. HCVa-wt contained a 25 base pair stem with 5'-GG and 3'-UU
overhangs
(which may form non-canonical base pairs) and a ten nucleotide miR-23 loop. To
test the
importance of these parameters in the effectiveness for inhibition of
expression, each of these
parameters was separately varied (Fig. 13A). The microRNA-23 loop sequence was
initially
selected because it is a naturally occurring sequence (Lagos-Quintana et al.,
Science, 2001,
293:854-258) and was therefore unlikely to be toxic. Two alternative ten
nucleotide loops were
tested, along with loops of six nucleotides, five nucleotides, and four
nucleotides, each in two
versions of a sequence. Neither loop size nor sequence was found to affect the
activity of these
25 base pair shRNAs (Fig. 13B; see Fig. 16A-B for sequences).
[0144] Small hairpin RNAs lacking the 3'-UU terminal sequence (single-stranded
overhang)
had the same efficacy as the parental shRNA containing this feature. Control
shRNA with full-
length (25 nucleotide) sense but short (13 nucleotide) antisense regions had
no activity,
confirming the importance of duplex structure in the targeting sequence.
shRNAs having a 3'-
CC instead of 3'-UU terminus (allowing formation of 2 additional Watson-Crick
base pairs)
were more effective than HCVa-wt for decreasing HCV expression, but also
affected SEAP
levels. This nonspecific inhibition could be a consequence of the longer stem
(27 base pairs),
which can induce genes of the interferon responsive patliway and activate
protein kinase R
(PKR). Surprisingly, moving the loop to the other end of the shRNA resulted in
a dramatic
reduction of activity (15% inhibition at 1 nM instead of 90%). Possible
explanations for this
effect include a shift in the position of Dicer processing (and therefore the
sequence targeted) as
well as a different GC content at the 5'-end.
[0145] Because 19 base pair shRNAs were shown to display potency similar to 25
base pair
shRNA, the effects of loop variations for 19 base pair shRNAs were examined.
The results are
shown in Fig. 14; see Fig. 16A-B for sequences. Loop sizes of 10, 6, 5, and 4
nucleotides were
tested, each in two sequence versions. Sequences containing all loop sizes
demonstrated the
ability to inhibit gene expression. However, in contrast to the results with
25base pair shRNAs,
reduction of loop size, especially below 5 nucleotides, resulted in reduced
activity for the 19 base
41
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
pair shRNAs for both loop sequences tested. Loops of at least 5-6 nucleotides
demonstrated the
most activity.
[0146] Removal of the 3'-UU also resulted in dramatic reduction of activity
for 19 base pair
as well as 20 base pair (but not 25 base pair) shRNAs. Without committing to
any particular
theory, the 3'-UU and 5'-GG may form non-canonical base pairs and the overall
size of shRNA
duplex is impoitant such that the duplex cannot be less than 21 base pairs for
efficient
processing. Thus, for 25 base pair shRNAs, neither the size of the loop nor
the presence of a 3'-
UU matters, whereas these parameters are important for potency of short, e.g.,
19 base pair
shRNAs. Without committing to any particular theory, it may be that Dicer
binds at the termini
prior to processing and does not "sense" the loop in the case of longer
shRNAs, but for 19 base
pair shRNAs the loop is "felt" as Dicer "measures" 19-21 nucleotides from the
ends.
[0147] Accordingly, it was found that 19 base pair shRNAs can be as potent as
25 base pair
shRNAs and 19 base pair siRNA. It was also found that some shRNA molecules
were active at
low concentrations of 0.1-1 nM ("hyper-potent shRNAs." Other groups typically
use 10-25-50-
100 nM siRNA).
[0148] These data demonstrate that sequences that do not include a 3'-UU that
are at least 22
base pairs, e.g., 23 base pairs, 24 base pairs, or 25 base pairs, can be
suitable for inhibition of
HCV expression. Similarly, loop size is not critical for shRNAs that are at
least 22 base pairs in
length.
Example 9: HCV Replicon System
[0149] A number of shRNA and siRNA inhibitors along with negative controls
were used to
transfect human hepatocytes (AVA5, a derivative of the Huh7 cell line) stably
expressing HCV
subgenomic replicons (Blight et al., Science, 2000, 290:5498), and the amount
of HCV
expression was determined. A range of concentrations was tested and the
concentration of RNA
resulting in 50% inhibition (IC50 or EC50) was determined. IC50s from two
independent
experiments are shown side-by-side in Fig. 15. The results generally
correlated with the data
obtained using the fLuc/IRES system in 293 FT cells, with the following
differences: (1) 19 base
pair shRNAs are more potent than 19 base pair siRNAs in the replicon system,
whereas with the
reporter system, 19 base pair siRNAs were more potent than shRNAs; (2) shRNA
HCVa-wt with
point mutations did not demonstrate activity in the replicon system, while it
was effective in
42
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
fLuc/IRES reporter system; (3) in general, 25 base pair siRNA and 25 base pair
shRNA had less
activity than other 19 base pair shRNAs and siRNAs tested. In general, the
IRES and replicon
systems are useful for identification of candidate sequences. Methods of
confirming the efficacy
(e.g., for inhibiting expression of HCV in a subject) of a selected shRNA or
siRNA can be
further tested using methods described herein and methods known in the art.
Other Embodiments
[0150] It is to be understood that while the invention has been described in
conjunction with
the detailed description thereof, the foregoing description is intended to
illustrate and not limit
the scope of the invention, which is defined by the scope of the appended
claims. Other aspects,
advantages, and modifications are within the scope of the following claims.
43
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
REFERENCES
1. Sookoian, S.C., New therapies on the horizon for hepatitis C. Ann Hepatol,
2003. 2: 164-
70.
2. Randall, G. and C.M. Rice, Interfering with hepatitis C virus RNA
replication. Virus Res,
2004. 102: 19-25.
3. Radhakrishnan, S.K., T.J. Layden, and A.L. Gartel, RNA interference as a
new strategy
against viral hepatitis. Virology, 2004. 323: 173-81.
4. Hugle, T. and A. Cerny, Current therapy and new molecular approaches to
antiviral
treatment and prevention of hepatitis C. Rev Med Virol, 2003. 13: 361-71.
5. Blight, K.J., A.A. Kolykhalov, and C.M. Rice, Efficient initiation of
HCVRNA
replication in cell culture. Science, 2000. 290: 1972-4.
6. Pietschmann, T. and R. Bartenschlager, Tissue culture and animal models for
hepatitis C
virus. Clin Liver Dis, 2003. 7: 23-43.
7. Mercer, D.F., D.E. Schiller, J.F. Elliott, D.N. Douglas, C. Hao, A.
Rinfret, W.R. Addison,
K.P. Fischer, T.A. Churchill, J.R. Lakey, D.L. Tyrrell, and N.M. Kneteman,
Hepatitis C
virus replication in mice witlz chimeric human livers. Nat Med, 2001. 7: 927-
33.
8. McCaffrey, A.P., L. Meuse, M. Karimi, C.H. Contag, and M.A. Kay, A potent
and
specific morpholino antisense inhibitor of hepatitis C translation in mice.
Hepatology,
2003. 38: 503-8.
9. Lieberman, J., E. Song, S.K. Lee, and P. Shankar, Interfering with disease:
opportunities
and roadblocks to harnessing RNA interference. Trends Mol Med, 2003. 9: 397-
403.
10. Layzer, J.M., A.P. McCaffrey, A.K. Tanner, Z. Huang, M.A. Kay, and B.A.
Sullenger, In
vivo activity of nuclease-resistant siRNAs. Rna, 2004. 10: 766-7 1.
11. Rossi, J.J. and G.J. Hannon, Unlocking the potential of the human genome
witlz RNA
intef ference. Nature, 2004. 431: 35-43.
12. McHutchison, J.G. and K. Patel, Future therapy of hepatitis C. Hepatology,
2002. 36:
S245-52.
13. Wilson, J.A., S. Jayasena, A. Khvorova, S. Sabatinos, I.G. Rodrigue-
Gervais, S. Arya, F.
Sarangi, M. Harris-Brandts, S. Beaulieu, and C.D. Richardson, RNA interference
blocks
44
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
gene expression and RNA synthesis from hepatitis C replicons propagated in
human liver
cells. Proc Natl Acad Sci U S A, 2003. 100: 2783-8.
14. Randall, G., A. Grakoui, and C.M. Rice, Clearance of replicating hepatitis
C virus
replicon RNAs in cell culture by small itzterferitag RNAs. Proc Natl Acad Sci
U S A,
2003.100: 235-40.
15. Yokota, T., N. Sakamoto, N. Enomoto, Y. Tanabe, M. Miyagishi, S. Maekawa,
L. Yi, M.
Kurosaki, K. Taira, M. Watanabe, and H. Mizusawa, Inhibition of intracellular
hepatitis
C virus replication by synthetic and vector-derived snaall interfering RNAs.
EMBO Rep,
2003. 4: 602-8.
16. Kapadia, S.B., A. Brideau-Andersen, and F.V. Chisari, Interference of
hepatitis C virus
RNA replication by short interfering RNAs. Proc Natl Acad Sci U S A, 2003.
100: 2014-
8.
17. Kronke, J., R. Kittler, F. Buchholz, M.P. Windisch, T. Pietschmann, R.
Bartenschlager,
and M. Frese, Alternative approaches for efficient inhibition of hepatitis C
virus RNA
replication by snaall interfering RNAs. J Virol, 2004. 78: 3436-46.
18. Sen, A., R. Steele, A.K. Ghosh, A. Basu, R. Ray, and R.B. Ray, Inhibition
of hepatitis C
virus protein expression by RNA interference. Virus Res, 2003. 96: 27-35.
19. Zhang, J., O. Yamada, T. Sakamoto, H. Yoshida, T. Iwai, Y. Matsushita, H.
Shimamura,
H. Araki, and K. Shimotohno, Down-regulation of viral replication by
adenoviral-
mediated expression of siRNA against cellular cofactors for hepatitis C virus.
Virology,
2004. 320: 135-43.
20. McCaffrey, A.P., L. Meuse, T.T. Pham, D.S. Conklin, G.J. Hannon, and M.A.
Kay, RNA
interference in adult nzice. Nature, 2002. 418: 38-9.
21. Kawasaki, H. and K. Taira, Short hairpin type of dsRNAs that are
controlled by
tRNA(Val) promoter significantly induce RNAi-ynediated gene silencing in the
cytoplasm
of human cells. Nucleic Acids Res, 2003. 31: 700-7.
22. Lagos-Quintana, M., R. Rauhut, W. Lendeckel, and T. Tuschl, Identification
of novel
genes coding for small expressed RNAs. Science, 2001. 294: 853-8.
23. Qin, X.F., D.S. An, I.S. Chen, and D. Baltimore, Inhibiting HIV-1
infection in huinan T
cells by lentiviral.-nzediated delivefy of small interfering RNA against CCR5.
Proc Natl
Acad Sci U S A, 2003. 100: 183-8.
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
24. McCaffrey, A.P., K. Ohashi, L. Meuse, S. Shen, A.M. Lancaster, P.J.
Lukavslky, P.
Sarnow, and M.A. Kay, Determinants of hepatitis C translational initiation in
vitro, in
cultured cells and rnice. Mol Ther, 2002. 5: 676-84.
25. Seo, M.Y., S. Abrignani, M. Houghton, and J.H. Han, Snaall interfering RNA-
rn.ediated
inhibition of hepatitis C virus replication in the human hepatoma cell line
Huh-7. J Virol,
2003. 77: 810-2.
26. Kim, D.H., M. Longo, Y. Han, P. Lundberg, E. Cantin, and J.J. Rossi,
Interferon
induction by siRNAs and ssRNAs synthesized by phage polynzerase. Nat
Biotechnol,
2004. 22: 321-5.
27. Bridge, A.J., S. Pebernard, A. Ducraux, A.L. Nicoulaz, and R. Iggo,
Induction of an
interferon response by RNAi vectors in mammalian cells. Nat Genet, 2003. 34:
263-4.
28. Fish, R.J. and E.K. Kruithof, Short-term cytotoxic effects and long-term
instability of
RNAi delivered using lentiviral vectors. BMC Mol Biol, 2004. 5: 9.
29. Han, J.H., V. Shyamala, K.H. Richman, M.J. Brauer, B. Irvine, M.S. Urdea,
P_ Tekamp-
Olson, G. Kuo, Q.L. Choo, and M. Houghton, Characterization of the ternainal
regions of
hepatitis C viral RNA: identification of conserved sequences in the 5'
untranslated region
and poly(A) tails at the 3' end. Proc Natl Acad Sci U S A, 1991. 88: 1711-5.
30. Choo, Q.L., K.H. Richman, J.H. Han, K. Berger, C. Lee, C. Dong, C.
Gallegos, D. Coit,
R. Medina-Selby, P.J. Barr, and et al., Genetic organization and diversity of
the hepatitis
C virus. Proc Natl Acad Sci U S A, 1991. 88: 2451-5.
31. Okamoto, H., S. Okada, Y. Sugiyama, K. Kurai, H. Iizuka, A. Machida, Y.
Miyakawa,
and M. Mayumi, Nucleotide sequence of the genomic RNA of hepatitis C virus
isolated
from a hunzan carrier: comparison with reported isolates for conserved and
divergent
regions. J Gen Virol, 1991. 72 ( Pt 11): 2697-704.
32. Bukh, J., R.H. Purcell, and R.H. Miller, Sequence analysis of the 5'
noncoding region of
hepatitis C virus. Proc Natl Acad Sci U S A, 1992. 89: 4942-6.
33. Rice, C.M., Virology: fresh assault on hepatitis C. Nature, 2003. 426: 129-
3 1.
34. Zhang, H., R. Hanecak, V. Brown-Driver, R. Azad, B. Conklin, M.C. Fox, and
K.P.
Anderson, Antisense oligonucleotide inhibition of hepatitis C virus (HCV) gene
expression in livers of naice infected with an HCV-vaccinia virus
recornbinant.
Antimicrob Agents Chemother, 1999. 43: 347-53.
46
CA 02622242 2008-03-11
WO 2007/032794 PCT/US2006/021253
35. Jubin, R., N.E. Vantuno, J.S. Kieft, M.G. Murray, J.A. Doudna, J.Y. Lau,
and B.M.
Baroudy, Hepatitis C virus internal ribosonae entry site (IRES) stem loop IIId
contains a
phylogenetically conserved GGG triplet essential for translation and IRES
folding. J
Virol, 2000. 74: 10430-7.
36. Seyhan AA, Vlassov AV, Ilves H, Egry L, Kaspar RL, Kazakov SA, Johnston
BH.
Complete, gene-specific siRNA libraries: production and expression in
mammalian cells.
RNA. 2005. 11:837-46.
37. Wang Q, Contag CH, lives H, Johnston BH, Kaspar RL. Small hairpin RNAs
efficiently
inhibit hepatitis C IRES-naediated gene expression in human tissue culture
cells and a
mouse aodel. Mol Ther. 2005. 12:562-8
47