Note: Descriptions are shown in the official language in which they were submitted.
CA 02629363 2008-05-12
WO 2007/057402 PCT/EP2006/068464
EMP4 gene
The present invention relates to altering plant development and more
particularly to altering the development of the plant endosperm. It concerns
in
particular nucleic acid molecules which alter the endosperm development.
BACKGROUND OF THE INVENTION
The seed of cereals is one of the most economically important and
scientifically interesting structure in plant biology. It consists of the
embryo and the
io endosperm, the two products of the double fertilization event. The
endosperm
originates from the central cells of the embryo sac, which are fertilized by
one of
the two haploid male gametes, and the embryo originates from the fusion of the
second gamete with the oosphere.
is In maize, the embryo consists of an embryonic axis surrounded by a
single massive cotyledon, the scutellum. The embryonic axis is characterized
by
the presence of a root primordium with the root meristem at the basis, the
scutellar
node, the mesocotyl and the shoot primordium comprising of the apical meristem
and four-five leaf initials. The endosperm functions as an embryonic annex
that
20 sustains the embryo during its development and its germination.
The endosperm, a characteristic formation of Angiosperm seeds, is a
nutritive tissue for the embryo. The maize endosperm originates with series of
free-nuclear divisions, followed by cellularisation and the subsequent
formation of
25 a range of functional cellular domains. This tissue is complex in its
structure and
development, in particular in cereals.
The endosperm is the main storage organ in maize seeds, nourishing the
embryo while the seed develops, and providing nutrients to the seedling on
germination. Thus, the uptake of assimilates by the growing endosperm is a
critical
30 process in seed development.
The central area of the endosperm (first endosperm domain) consists of
large cells with vacuoles, which store the reserves of starch and proteins
(central
starchy endosperm where genes involved in starch and in prolamin storage
CA 02629363 2008-05-12
WO 2007/057402 2 PCT/EP2006/068464
proteins biosynthesis are expressed), whilst the region surrounding the embryo
(ESR that corresponds to the second endosperm domain) is distinguished by
rather small cells, occupied for the major part by cytoplasm. The ESR may have
a
role in embryo nutrition or in establishing a physical barrier between the
embryo
s and the endosperm during seed development.
The Basal Endosperm Transfer Layer (BETL that corresponds to the third
endosperm domain) area is highly specialized to facilitate uptake of solutes
during
grain development. These transfer cells of the basal endosperm have
specialised
internal structures adapted to absorb solutes from the maternal pedicel
tissue, and
io translocate these products to the developing endosperm and embryo. These
transfer cells facilitate nutrient import into the maize kernel.
The fourth endosperm domain consists of the aleurone, which is the outer
layer of the endosperm and accumulates proteins and oil.
is The empty pericarp (emp) phenotype refers to a broad class of defective
kernel (dek) mutants characterized by seeds exhibiting an extreme reduction in
endosperm size, yet possessing a normal pericarp (Sheridan and Neuffer, 1980;
Scanlon et al., 1994; Scanlon et al., 1997).
Scanlon et al. (1994) characterized a group of mutants presenting kernel
20 with little or no endosperm. Such mutants, have aleurone present but no or
little
starchy endosperm.
To date, the molecular basis of only one emp phenotype ('empty
pericarp') has been elucidated: the EMP2 gene, (Fu et al., 2002) which encodes
a
heat-shock response regulator. Absence of this protein in the null mutant
leads to
25 up regulation of hsp genes and is correlated with seed abortion.
The inventors here report a new gene allowing the alteration of the
endosperm and plant development. This gene, EMP4 isolated from Mu tagged
maize lines, encodes a PPR protein that is encoded in the nuclear genome but
30 localised in the mitochondria. PPR proteins are often required for the
maturation of
organellar RNA and thus the EMP4 gene will regulate and will be limiting for
efficient mitochondrial function and energy production at certain
developmental
CA 02629363 2008-05-12
WO 2007/057402 3 PCT/EP2006/068464
stages or in certain cell types. Manipulation of EMP4 levels can thus alter
the
energy status and thus the growth of the cell, tissue, organ or plant either
positively or negatively.
The present invention provides the first example of a maize PPR gene
required for seed development. Moreover, the lesions in the EMP4 gene are
associated with specific developmental defects, which are first recognizable
in the
highly metabolic cells of the endosperm basal transfer layer of emp4 mutants.
The mutation of the EMP4 gene confers a severe reduction in endosperm
io development and a seed lethal phenotype. Endosperm mutants are severely
impaired, with differentiation of the nutrient importing basal endosperm
transfer
tissue being highly irregular. Homozygous mutants affected the general plant
growth.
Such a nucleic acid molecule is particularly useful for enhancing yield via
is overexpression in cells, tissues or organs that are limiting for yield. In
particular,
the maize yield is thought to be limited by the sink strength of the
developing
kernels.
Advantageously, overexpression of the EMP4 gene in kernels or more
specifically in the endosperm will increase sink strength via an increase in
energy
20 production in kernel cells and thus increase seed size.
The overexpression of the EMP4 gene is also useful for increasing plant
growth rate, grain filling, starch and proteins accumulations, speeding seed
development.
25 GENERAL DESCRIPTION
The present invention relates to an isolated nucleic acid molecule encoding a
protein which alters the plant or endosperm development that comprises a
sequence selected from the group consisting of :
30 a) a nucleotide sequence encoding a rp otein consisting of an amino acid
sequence as depicted in SEQ ID No: 2 or an amino acid sequence that is at
least 70% identical to SEQ ID No: 2, and variants thereof;
CA 02629363 2008-05-12
WO 2007/057402 4 PCT/EP2006/068464
b) a nucleotide sequence as depicted in SEQ ID No: 1 or a nucleotide
sequence that is at least 70% identical to SEQ ID No: 1;
c) a sequence hybridizing under stringent conditions with the complementary
strand of a nucleic acid molecule as defined in (a) or (b).
The nucleic acid molecules according to the present invention will be
called EMP4 nucleic acid molecules. The EMP4 nucleic acid molecules alter
plant
development or endosperm development.
The various nucleotide sequences of the invention can be of artificial
origin or not. They may be DNA sequences obtained by screening libraries of
sequences by means of probes produced on the basis of SEQ ID No: 1 or SEQ ID
No: 3. Such libraries can be prepared by conventional techniques of molecular
biology, known to persons skilled in the art.
The nucleotide sequences according to the invention can also be
prepared by chemical synthesis, or by mixed methods including the chemical or
enzymatic modification of sequences obtained by screening banks.
According to an embodiment of the invention, a nucleic acid molecule
which alters the plant or endosperm development consists in SEQ ID No: 1.
According to an other embodiment of the invention, a nucleic acid
molecule that alter the plant or endosperm development consists in a sequence
going from nucleotide 10 (position 10) to nucleotide 1851 (position 1851) of
SEQ
I D No: 1.
According to another embodiment of the invention, a nucleic acid
molecule which alters the plant or endosperm development consists in SEQ ID
No:
4.
As used herein "variants" means that the sequence differs in one or more
positions in comparison with the sequence SEQ ID No: 2 as long as it encodes a
protein altering the plant or endosperm development, and possesses SEQ ID No:
28.
CA 02629363 2008-05-12
WO 2007/057402 5 PCT/EP2006/068464
Such molecules comprise those which are variants of the EMP4 protein
according to the invention and differ, for example, by way of amino acid
and/or
nucleotide deletion(s), insertion(s), substitution(s), addition(s) and/or
recombination(s) or any other modification(s) known in the art either alone or
in
combination from the above described amino acid sequences or their underlying
nucleotide sequence(s). Methods for introducing such modifications in the
nucleic
acid molecules according to the invention are well-known to the person skilled
in
the art. The invention also relates to nucleic acid molecules the sequence of
which
differs from the nucleotide sequence of any of the above-described nucleic
acid
io molecules due to the degeneracy of the genetic code.
The invention further relates to a protein encoded by said nucleic acid
molecules. More specifically, the invention provides a protein that alters the
plant
or endosperm development encoded by a nucleic acid molecule as defined above.
Preferably, a protein according to the invention may comprise, or consist
in an amino acid sequence as depicted in SEQ ID No: 2.
Also preferably, such a protein consists in an amino acid sequence as
depicted in SEQ ID No: 5.
The proteins encoded by the various variants of the above-described
nucleic acid molecules share specific common characteristics, such as
biological
activity, molecular weight, conformation, etc., as well as physical
properties, such
as electrophoretic mobility, chromatographic behavior, sedimentation
coefficients,
pH optimum, temperature optimum, stability, solubility, spectroscopic
properties,
etc.
A nucleic acid molecule "hybridizes" to another nucleic acid molecule,
such as a cDNA, genomic DNA, or RNA, when a single stranded form of the
nucleic acid molecule can anneal to the other nucleic acid molecule under the
3o appropriate conditions of temperature and solution ionic strength (see
Sambrook
et al., 1989).
CA 02629363 2008-05-12
WO 2007/057402 6 PCT/EP2006/068464
Such an hybridizing sequence alters the plant or endosperm development
according to the invention and contains at least 15, 25, 35, 37, 50, 75, 100,
121,
122, 127, 150, 200, 220, 240, 250, 270, 300, 350, 360, 365, 500, 750, 800,
900,
1000, 1100, 1200, 1300, 1400, 1500, 1600, 1700, 1800, 1850 nucleotides.
The invention also encompasses modifications of the DNA sequence as
depicted in SEQ ID No: 1, or of sequence motifs thereof by, e. g., nucleotide
replacements that do not affect the overall structure or the function of the
nucleic
acid molecule altering the plant or endosperm development so that it remains
io capable of altering the plant or endosperm development.
"Homologous nucleic acid sequence", or "homologous DNA sequence",
means any nucleic acid sequence which differs from the sequence of reference
by
a substitution, deletion and/or insertion of one or more nucleotides at
positions
such that these homologous nucleic acid sequences preserve the possibility to
alter plant or endosperm development.
Preferably, when the sequence of reference is SEQ ID No:1, such a
homologous nucleic acid sequence is at least 70 % identical to the sequence
SEQ
ID No: 1, preferably at least 85% identical, more preferably at least 90, 91,
95, 98,
99.9 % identical. Also preferably, the degree of identity is defined by
comparison
with the entire sequence of reference, SEQ ID No: 1.
When the sequence of reference is SEQ ID No:2, such an identical
protein is at least 70 % identical to the sequence SEQ ID No: 2, preferably at
least
79,8% identical, more preferably at least 85, 90, 91, 95, 98, 99.9 %
identical. Also
preferably, the degree of identity is defined by comparison with the entire
sequence of reference, SEQ ID No: 2. The present invention also relates to SEQ
ID No: 5 that is 79.7% identical to SEQ ID No: 2. This percentage of identity
has
been obtained by the method of Needleman and Wunsch.
Homology is generally determined using a sequence analysis software (for
example, the Sequence Analysis Software package of the Genetics Computer
CA 02629363 2008-05-12
WO 2007/057402 7 PCT/EP2006/068464
Group, University of Wisconsin Biotechnology Center, 1710 University Avenue,
Madison, WI 53705). Similar nucleotide sequences are aligned in order to
obtain
the maximum degree of homology (i.e. identity). To this end, it may be
necessary
to artificially introduce gaps in the sequence. Once the optimum alignment has
been achieved, the degree of homology (i.e. identity) is established by
recording
all the positions for which the nucleotides of the two compared sequences are
identical, with respect to the total number of positions.
The preferred method uses the algorithm of Needleman and Wunsch.
In a preferential manner such a homologous nucleic acid sequence
specifically hybridizes to a sequence which is complementary to the sequence
SEQ ID No: 1 under stringent conditions. The parameters defining the
stringency
conditions depend on the temperature at which 50% of the paired strands
is separate (Tm).
For sequences comprising more than 30 bases, Tm is defined by the
equation: Tm = 81.5 + 0.41 (%G+C) + 16.6 Log (concentration in cations) - 0.63
(%formamide) - (600/number of bases) (Sambrook et al., 1989).
For sequences shorter than 30 bases, Tm is defined by the equation: Tm
= 4(G+C) + 2(A+T).
Under appropriate stringency conditions, in which non-specific (aspecific)
sequences do not hybridize, the temperature of hybridization is approximately
between 5 and 30 C, preferably between 5 and 10 C below Tm and hybridization
buffers used are preferably solutions of higher ionic force like a solution
6*SSC for
example.
Particularly, the EMP4 isolated nucleic acid molecule encodes a protein
that alters the plant or endosperm development.
Preferably, the EMP4 nucleic acid molecule altering the plant or
endosperm development consists in SEQ ID No: 1.
CA 02629363 2008-05-12
WO 2007/057402 8 PCT/EP2006/068464
The EMP4 nucleic acid molecules according to the invention can be
isolated from various plant species, notably Angiosperm plants, Monocotyledons
or Dicotyledons and are preferably nucleic acid molecules isolated from a
cereal
or an oily plant. Still preferably, the nucleic acid molecules are isolated
from a
s plant selected from the group consisting of maize, rice, sorghum, wheat,
barley,
rye, brassica napus, pea, sunflower and sugar cane. Still preferably, the
plant is
maize.
It is possible for the person skilled in the art to isolate with the help of
the
io EMP4 nucleotide sequence of the invention, corresponding genes from other
species.
This can be done by conventional techniques known in the art, for
example, by using a sequence as depicted in SEQ ID No: 1 as a hybridization
probe or by designing appropriate PCR primers.
is It is possible to start with coding DNA sequences or Protein sequences
via TBLASTN queries. The approach used to isolate rice EMP4 genes, for
example, is to use the Protein sequence of EMP4, do a TBLASTN with this
sequence against Rice ESTs, then use this EST to find the genomic sequence or
directly use TBLASTN against the rice genome sequence. The same approach is
20 used to isolate other EMP4 genes from other species.
Another object of the present invention is a nucleotide construction,
referred to as an expression cassette comprising an EMP4 nucleic acid molecule
as defined above operatively linked to regulatory elements allowing the
expression
25 in prokaryotic and/or eukaryotic host cells.
"Operatively linked" refers to functional linkage between an EMP4 nucleic
acid molecule (altering the plant or endosperm development) according to the
invention and a promoter sequence (regulatory element having a promoter
activity).
30 The EMP4 nucleic acid molecule can be placed in the sense or antisense
orientation.
CA 02629363 2008-05-12
WO 2007/057402 9 PCT/EP2006/068464
The promoter sequence can be of a heterologous origin.
Any suitable promoter could be used. It could be a constitutive promoter.
It could also be for example a tissue-specific promoter such as a seed-
specific or a
BETL-specific promoter. Numerous tissue-specific promoters are described in
the
s literature and any one of them can be used.
Among constitutive promoters, one can use for example the CsVMV
promoter (Verdaguer et al, 1996), the rice actin promoter (McElroy et aL,
1990),
the CAMV 35 S or the 19S promoter (Kay et aL, 1987).
Preferably, the promoter used allows expression in the endosperm. More
io preferably, the promoter used allows expression in the BETL and still more
preferably, the promoter is BETL-specific.
Advantageously, promoters are for example pMRP1 which is expressed in
the central cell and in the BETL region prior to and after BETL
cellularisation
(Gomez et al (2002)), pMEG1 (Gutierrez-Marcos et al 2004), pBETL1 (Hueros et
is al 1999) and pBETL2 (WO 99/50427) which are BETL-specific promoters.
"BETL-specific promoter" means, as used in the present invention, that
the promoter has a predominant pattern expression in the BETL, and preferably
an exclusive pattern expression in the BETL.
20 The said EMP4 nucleic acid molecule can also be associated with other
regulating elements such as transcription termination sequences (terminators).
By
way of examples of such sequences, it is possible to cite the polyA 35S
terminator
of the cauliflower mosaic virus (CaMV), described in the article of Franck et
al.
(1980) and the NOS terminator corresponding to the region in the non-coding 3'
25 region of the nopaline synthase gene of the Ti-plasmid of the Agrobacterium
tumefaciens nopaline strain (Depicker et al. 1992).
Preferably, the terminator used is the Nos terminator.
According to the invention, the expression cassette, comprising an EMP4
3o nucleic acid molecule as defined above, operatively linked to regulatory
elements
allowing the expression in prokaryotic and/or eukaryotic host cells (such as a
CA 02629363 2008-05-12
WO 2007/057402 10 PCT/EP2006/068464
promoter sequence) may further comprise one or several selection marker genes
for plants, useful for transformation and selection.
In the present invention, the term "selectable marker", "selectable gene",
"selectable marker gene", "selection marker gene", "marker gene" are used
interchangeably.
These selectable markers include, but are not limited to, antibiotic
resistance genes, herbicide resistance genes or visible marker genes. Other
phenotypic markers are known in the art and may be used in this invention.
A number of selective agents and resistance genes are known in the art.
Notably the selectable marker used can be the bar gene conferring
resistance to bialaphos (White et al., 1990), the sulfonamide herbicide Asulam
resistance gene, sul (described in WO 98/49316) encoding a type I
dihydropterate
synthase (DHPS), the nptll gene conferring resistance to a group of
antibiotics
including kanamycin, G418, paromomycin and neomycin (Bevan et al., 1983), the
is hph gene conferring resistance to hygromycin (Gritz et al., 1983), the
EPSPS gene
conferring tolerance to glyphosate (US 5,188,642), the HPPD gene conferring
resistance to isoxazoles (WO 96/38567), the gene encoding for the GUS enzyme,
the green fluorescent protein (GFP), expression of which, confers a
recognisible
physical characteristic to transformed cells, the chloramphenicol transferase
gene,
2o expression of which, detoxifies chloramphenicol.
Advantageously, the selectable marker gene is inserted between a
promoter and a terminator in a second expression cassette. Said second
expression cassette being integrated in the same vector as the expression
25 cassette containing the EMP4 nucleic acid molecule under transcriptional
control
of a promoter according to the invention.
According to this advantageous embodiment, the marker gene is
preferably controlled by a promoter which allows expression in cells, thus
allowing
30 selection of cells or tissue containing the marker at any stage of
development of
the plant. Preferred promoters are the promoter of nopaline synthase gene of
Agrobacterium, the promoter derived from the gene which encodes the 35S
CA 02629363 2008-05-12
WO 2007/057402 11 PCT/EP2006/068464
subunit of cauliflower mosaic virus (CaMV) coat protein, and the rice actin
promoter. However, any other suitable second promoter may be used.
Any terminator may be used. Preferred terminators are the 3'CaMV and
Nos terminator as previously described.
Advantageously, the expression cassette containing the selectable marker
gene is comprised between two Ds elements (transposons) in order for its
removal
at a later stage by interacting with the Ac transposase. This elimination
system is
described in Yoder et al. (1993).
For the transformation step, two vectors could be used, the first one
comprising the expression cassette containing the EMP4 nucleic acid molecule
and the second one comprising the expression cassette containing the
selectable
marker gene. The same host cell being transformed with these two vectors (co-
i5 transformation).
The expression cassettes according to the invention may additionally
contain transit peptide sequences. There are numerous examples in the art of
transit peptides which may be used to deliver a target protein into a plastid
organelle such as the small subunit (SSU) transit peptide of ribulose
biphosphate
carboxylase.
Other elements like introns and enhancers can also be present in the
nucleic sequence of interest in order to improve the expression of the gene of
interest.
Among useful introns, the first intron of maize adh1S can be placed
between the promoter and the coding sequence. This intron when included in a
gene construct increased the expression of the desired protein in maize cells.
One
also can use the 15t intron of the shrunken 1 gene of the maize (Maas et al.,
1991),
the 15t intron of the catalase gene of the bean catalase (CAT-1) (Ohta et al.,
1990),
the 2nd intron of the ST-LS1 gene of potato (Vancanneyt et al. 1990), the DSV
intron of the yellow dwarf virus of tobacco (Morris et al., 1992), the actin-1
intron
(act-1) of rice (McElroy et al., 1990), FAD 2 intron (WO 2006/003186) and
intron 1
of triosephosphate isomerase (TPI) (Snowdon et al., 1996).
CA 02629363 2008-05-12
WO 2007/057402 12 PCT/EP2006/068464
Preferentially, the intron used in the present invention is the Sh1 intron.
The expression cassettes may additionally contain 5' leader sequences.
Such leader sequences can act to enhance translation. Such 5' leader sequences
are known in the art and include, but are not limited to, picornavirus
leaders, for
example, the EMCV leader (Encephalomyocarditis 5' noncoding region) (Elroy-
Stein, Fuerest, and Moss B., 1989) ; potyvirus leaders, for example, the TEV
leader (Tobacco etch Virus) (Allison et al., 1986) ; the human immunoglobulin
heavy-chain binding protein leader (BiP) (Macejack and Sarnow, 1991) ; the
1o untranslated leader from the coat protein mRNA of alfalfa mosaic virus (AMV
RNA
4) (Jobling and Gehrke, 1987) ; the tobacco mosaic virus leader (TMV) (Gallie
et
al., 1989) ; and the maize chlorotic mottle virus leader (MCMV) (Lommel et
al.,
1991). See also, Della-Cioppa et al. (1987). Other methods known to enhance
translation can be utilized, for example introns, and the like.
In preparing the expression cassettes, the various DNA sequences or
fragments may be manipulated, so as to provide DNA sequences or fragments in
the proper orientation and, as appropriate, in the proper reading frame.
Towards
this end, adapters or linkers may be employed to join the DNA fragments and/or
other manipulations may be required to provide convenient restriction sites,
removal of superfluous DNA, removal of restriction sites, or the like. For
this
purpose, in vitro mutagenesis, primer repair, restriction, annealing,
ligation, PCR,
or the like may be employed, where nucleotide insertions, deletions or
substitutions, for example transitions and transversions, may be involved.
These
techniques are well known by those skilled in the art.
Another object of the invention is any nucleotide vector referred to as an
expression vector, such as a plasmid, which can be used for transforming host
cells, characterized in that it contains at least an expression cassette as
defined
3o above. The construction of expression vectors for the transformation is
within the
capability of one skilled in the art following standard techniques.
CA 02629363 2008-05-12
WO 2007/057402 13 PCT/EP2006/068464
The decision as to whether to use a vector, or which vector to use, is
guided by the method of transformation selected, and by the host cell
selected.
Where a naked nucleic acid introduction method is used, then the vector
can be the minimal nucleic acid sequences necessary to confer the desired
phenotype, without the need for additional sequences.
Possible vectors include the Ti plasmid vectors, shuttle vectors designed
merely to maximally yield high numbers of copies, episomal vectors containing
minimal sequences necessary for ultimate replication once transformation has
io occurred, transposon vectors, including the possibility of RNA forms of the
gene
sequences. The selection of vectors and methods to construct them are commonly
known to persons of ordinary skill in the art and are described in general
technical
references (Mullis, KB (1987), Methods in Enzymology).
For other transformation methods requiring a vector, selection of an
is appropriate vector is relatively simple, as the constraints are minimal.
The
apparent minimal traits of the vector are that the desired nucleic acid
sequence be
introduced in a relatively intact state. Thus, any vector which produces a
plant
carrying the introduced DNA sequence should be sufficient. Also, any vector
which
introduces a substantially intact RNA which can ultimately be converted into a
20 stably maintained DNA sequence should be acceptable.
However, any additional attached vector sequences which confer
resistance to degradation of the nucleic acid fragment to be introduced, which
assists in the process of genomic integration or provides a means to easily
select
for those cells or plants which are actually, in fact, transformed are
advantageous
25 and greatly decrease the difficulty of selecting useable transgenic plants.
The vector can exist, for example, in the form of a phage, a plasmid or a
cosmid. The construction of such expression vectors for transformation is well
known in the art and uses standard techniques. Mention may be made of the
methods described by Sambrook et al. (1989).
Another object of the invention is a host cell, containing at least an
expression vector as described above.
CA 02629363 2008-05-12
WO 2007/057402 14 PCT/EP2006/068464
The decision as to whether to use a host cell, or which host cell to use, is
guided by the method of transformation.
The host cell can be any prokaryotic or eukaryotic cell. Any of a large
number of available and well-known host cells may be used in the practice of
this
invention. The selection of a particular host is dependent upon a number of
factors
recognized by the art. These include, for example, compatibility with the
chosen
expression vector, bio-safety and costs. Useful hosts include bacteria such as
E.
coli sp. or Agrobacterium. A plant host cell, may be also used, notably an
Angiosperm plant cell, Monocotyledon as Dicotyledon plant cell, particularly a
io cereal or oily plant cell, and more particularly selected from the group
consisting of
maize, wheat, barley, rice, rape, Brassica napus, sugar cane, sorghum, pea and
sunflower. Still preferably the plant host cell is from maize.
More particularly, the host cell used in carrying out the invention is
Agrobacterium tumefaciens, according to the method described in the article of
An
is et al., 1986, or Agrobacterium rhizogenes, according to the method
described in
the article of Jouanin et al., 1987.
The invention also concerns a transgenic plant, or a part of a transgenic
plant, comprising stably integrated into its genome a nucleic acid molecule
2o according to the present invention operatively linked to regulatory
elements
allowing transcription and/or expression of the nucleic acid molecule in plant
cells.
"Part of a transgenic plant"; according to the present invention, means in
particular
fruit, seed, grain, or pollen.
25 The invention also concerns a transgenic plant, or a part of a transgenic
plant, comprising such a host cell or generated from such a host cell.
Where the plant contains endogenously an EMP4 gene according to the
invention, it will be understood that the transgenic plant according to the
invention
30 comprises an additional "exogenous" EMP4 gene, for instance integrated by
transgenese.
CA 02629363 2008-05-12
WO 2007/057402 15 PCT/EP2006/068464
A whole plant can be regenerated from a single transformed plant cell.
Thus, in a further aspect the present invention provides transgenic plants (or
parts
of them) including nucleic acid molecules in accordance with the invention.
The
regeneration can proceed by known methods.
s The seeds which grow, by fertilization, from this plant also contain this
transgene in their genome.
Advantageously, the transgenic plant obtained can produce grains with a
larger (bigger) endosperm (increased endosperm size) in comparison with a non-
transformed plant. In addition to increases in yield such transgenic plants
may
io have grains with modified starch, oil or protein contents. In particular,
said
modification in starch, oil, or protein contents consists in an increase of
starch, oil,
or protein accumulation.
A plant or part of a plant according to the invention could be a plant or a
part of it from various species, notably an Angiosperm, Monocotyledons as
is Dicotyledons, preferably a cereal or oily plant, and more preferably
selected from
the group consisting of maize, rice, sorghum, wheat, barley, rape, brassica
napus,
sugar cane, and sunflower. Still preferably, the plant is maize.
As used herein, the term "oily plant" denotes a plant that is capable of
producing oil, and preferably that is cultivated for oil production.
20 The hybrid plants obtained by crossing plants according to the invention
also form part of the invention.
The present invention also relates to a protein encoded by a nucleic acid
molecule according to the invention.
25 Preferably, the protein consists of an amino acid sequence as depicted in
SEQ ID No: 2, or being at least 70% identical to SEQ ID No: 2.
Also preferably, such a protein consists in SEQ ID No : 5.
An other object of the invention is a method for obtaining a plant having
30 increased seed size, said method comprising the steps consisting of :
a) transforming at least one plant cell or plant tissue by means of at least a
vector as defined previously;
CA 02629363 2008-05-12
WO 2007/057402 16 PCT/EP2006/068464
b) cultivating the cell(s) or plant tissue thus transformed so as to generate
a
plant containing in its genome at least an expression cassette according to
the invention, whereby said plant has increased seed size.
According to the invention, "increased seed size" means that the
transformed plant have a seed bigger (larger) than a seed from a wild type
plant
(non-transformed plant). Such an increase in seed size is desirable to
increase
seed and or endosperm yield and to improve the germination and vigour of
seedlings.
The invention also relates to a method for increasing plant growth rate, said
method comprising the steps consisting of :
a) transforming at least a plant cell or plant tissue by means of at least a
vector as defined previously;
is b) cultivating the cell(s) or plant tissue thus transformed so as to
generate a
plant containing in its genome at least an expression cassette according to
the invention.
Plant growth rate can be increased with either an improvement in final yield
or earlier flowering and harvest.
The invention also relates to a method for obtaining a plant having
decreased seed moisture content, said method comprising the steps consisting
of
a) transforming at least a plant cell or plant tissue by means of at least a
vector as defined previously;
b) cultivating the cell(s) or plant tissue thus transformed so as to generate
a
plant containing in its genome at least an expression cassette according to
the invention, whereby said plant has decreased seed moisture content.
The over-expression of the EMP4 gene into the endosperm (with the aim of
3o an endosperm promoter) increases the speed of the seed development leading
to
a seed having a lower moisture content than a normal seed (wild type) at
harvest.
This is of interest and useful for reducing drying costs.
CA 02629363 2008-05-12
WO 2007/057402 17 PCT/EP2006/068464
The invention also relates to a method for increasing plant starch and
proteins accumulation, said method comprising the steps consisting of :
a) transforming at least a plant cell or plant tissue by means of at least a
vector as defined previously;
b) cultivating the cell(s) or plant tissue thus transformed so as to generate
a
plant containing in its genome at least an expression cassette according to
the invention, whereby said plant has increased starch and proteins
content.
The invention also relates to a method for increasing grain filling, said
method comprising the steps consisting of :
a) transforming at least a plant cell or plant tissue by means of at least a
vector as defined previously;
is b) cultivating the cell(s) or plant tissue thus transformed so as to
generate a
plant containing in its genome at least an expression cassette according to
the invention.
The invention also relates to a method for speeding seed development,
said method comprising the steps consisting of :
a) transforming at least a plant cell or plant tissue by means of at least a
vector as defined previously;
b) cultivating the cell(s) or plant tissue thus transformed so as to generate
a
plant containing in its genome at least an expression cassette according to
the invention.
The transformation of vegetable cells can be achieved by any one of the
techniques known to one skilled in the art.
It is possible to cite in particular the methods of direct transfer of genes
such as direct micro-injection into plant embryoids (Neuhaus et coll. 1997),
vacuum infiltration (Bechtold at al. 1993) or electroporation (Chupeau et
coll.,
1989) or direct precipitation by means of PEG (Schocher et coll., 1986) or the
CA 02629363 2008-05-12
WO 2007/057402 18 PCT/EP2006/068464
bombardment by gun of particules covered with the plasmidic DNA of interest
(Fromm M et al., 1990).
It is also possible to infect the plant with a bacterial strain, in particular
Agrobacterium. According to one embodiment of the method of the invention, the
s vegetable cells are transformed by a vector according to the invention, the
said
cell host being able to infect the said vegetable cells by allowing the
integration, in
the genome of the latter, of the nucleotide sequences of interest initially
contained
in the above-mentioned vector genome. Advantageously, the above-mentioned
cell host used is Agrobacterium tumefaciens, in particular according to the
method
io described in the article by An et al., (1986), or Agrobacterium rhizogene,
in
particular according to the method described in the article by Guerche et al.
(1987).
For example, the transformation of vegetable cells can be achieved by the
transfer of the T region of the tumour-inducing extra-chromosome circular
plasmid
is of Agrobacterium tumefaciens, using a binary system (Watson et al., 1994).
To do
this, two vectors are constructed. In one of these vectors the T region has
been
eliminated by deletion, with exception of the right and left borders, a marker
gene
being inserted between them to allow selection in the plant cells. The other
partner
of the binary system is an auxiliary plasmid Ti, a modified plasmid which no
longer
2o has any T region but still contains the virulence genes vir necessary to
the
transformation of the vegetable cell.
According to a preferred mode, it is possible to use the method described
by Ishida et al. (1996) for the transformation of Monocotyledons.
According to another protocol, the transformation is achieved according to
25 the method described by Finer et al.(1 992) using the tungsten or gold
particle gun.
Selection :
The engineered plant material may be selected or screened for
transformants (those that have incorporated or integrated the introduced
3o nucleotide construction(s)). Such selection and screening methodologies are
well
known to those skilled in the art. The selection and screening method is
chosen
depending on the marker gene used.
CA 02629363 2008-05-12
WO 2007/057402 19 PCT/EP2006/068464
An isolated transformant may then be regenerated into a plant.
Regeneration :
Normally, regeneration is involved in obtaining a whole plant from the
transformation process. The term "regeneration" as used herein, means growing
a
whole plant cell, a group of plant cells, a plant part or a plant piece (for
example,
from a protoplast, callus, or tissue part).
Methods of regenerating whole plants from plant cells are known in the
art, and the method of obtaining transformed and regenerated plants is not
critical
io to this invention.
In general, transformed plant cells are cultured in an appropriate medium,
which may contain selective agents such as antibiotics, where selectable
markers
are used to facilitate identification, of transformed plant cells. Once callus
forms,
shoot formation can be encouraged by employing appropriate plant hormones in
is accordance with known methods and shoots transferred to rooting medium for
regeneration of plants. The plants may then be used to establish repetitive
generations, either from seeds or using vegetative propagation techniques.
The invention also concerns the use of the transgenic plants obtained
2o according to the invention, or parts of these plants, in particular seeds,
grains, and
fruits for preparing derived products, in particular food products.
The products obtained, whether it be seeds with a higher oil content,
flours of seeds or grains with a higher starch, protein or oil content, also
come
within the scope of the invention.
25 The invention also provides any composition for human or animal food
prepared from the said obtained products.
The present invention also relates to a method for reducing organs
development and particularly for reducing internodes length, said method
30 consisting of obtaining a maize plant having reduced internodes length as
compared to a wild type plant, comprising the step consisting of inhibiting
the
expression of a protein having at least 80% identity with SEQ ID N 2 within a
CA 02629363 2008-05-12
WO 2007/057402 20 PCT/EP2006/068464
maize plant, thus obtaining a plant having reduced internodes length as
compared
to a plant where the expression of SEQ ID N 2 is not inhibited.
A tissue-specific repression of the EMP4 gene is obtained by inhibition of
the EMP4 gene using methods known in the art, as RNAi and antisens technology.
Any other method known by the man skilled in the art is also useful.
The EMP4 gene could be repressed specifically in the internodes or in the
stem by using specific promoters of these organs. In this case for example,
the
EMP4 gene is placed in the antisense orientation and is operatively linked to
a
io promoter specific of the stem or of the internodes.
In addition to the use of the invention to create improved plants via plant
modification via transgenesis the invention can be used to develop molecular
markers to screen for favourable EMP4 alleles in the plant of interest.
Particular
is EMP4 alleles may be linked to desirable agronomic characteristics such as
plant
growth rate, stature and yield.
The present invention will be further understood in view of the annexed
figures and following examples.
Definitions:
emp4 means the emp4 plant mutant, either homozygous or heterozygous for the
mutation (plants containing an EMP4 mutated gene).
emp4/+ and +lemp4 mean plants heterozygous for the emp4 mutation (1 mutated
copy of the EMP4 gene).
emp4/emp4 means plants homozygous for the emp4 mutation (2 mutated copies
of the EMP4 gene).
+/+ means wild type plants.
Sequence Listing:
SEQ ID No: 1: Maize EMP4 cDNA sequence.
SEQ ID No: 2 Maize EMP4 amino acid sequence.
CA 02629363 2008-05-12
WO 2007/057402 21 PCT/EP2006/068464
SEQ ID No: 3 Maize EMP4 genomic sequence.
SEQ ID No: 4 Rice EMP4 cDNA sequence.
SEQ ID No: 5 Rice EMP4 amino acid sequence.
SEQ ID No: 6 Rice EMP4 genomic sequence.
SEQ ID No: 7: Maize EST BQ1 64351.
SEQ ID No: 28 : Consensus sequence between SEQ ID No: 2 and SEQ ID No: 5.
SEQ ID No: 32 : Maize BETL9 promoter sequence (pBETL9).
Table 1 EMP4 oligonucleotide primers and probes used in the following
io examples :
Primer/
Probe SEQ ID No: Primer region Sequence 5'43'
Name
Probel SEQ ID No: 8
Oest2 SEQ ID No: 9 1118 to 1137 TGATGGAGAGGATGCGGG
of maize EMP4 cDNA AG
(forward
oli onucleotide
Oest3 SEQ ID No: 10 1300 to 1320 TGCCTCAATCAGCAAACCC
of maize EMP4 cDNA TG
(reverse
oli onucleotide
Oest5 SEQ ID No: 11 1112 to 1147 TGGAGAGGATGCGGGAGT
of maize EMP4 cDNA GCCGGTGTC
(forward
oli onucleotide
Oest7 SEQ ID No: 12 1172 to 1198 TGGTGATTCGGTTGGCCTG
of maize EMP4 cDNA CAGGCTTG
(forward
oli onucleotide
Oest7re SEQ ID No: 13 1172 to 1198 CAAGCCTGCAGGCCAACC
v of maize EMP4 cDNA GAATCACCA
(reverse
oli onucleotide
CA 02629363 2008-05-12
WO 2007/057402 22 PCT/EP2006/068464
RT5 SEQ ID No: 14 294 to 313 GCACTTCTTCCACTGGTGC
of maize EMP4 cDNA T
(forward
oligonucleotide)
RT5rev SEQ ID No: 15 294 to 316 GGGAGCACCAGTGGAAGA
of maize EMP4 cDNA AGTGC
(reverse
oli onucleotide
ZmH2B SEQ ID No: 16 ATGGCGCCCAAGGCGGAG
5for AAGAAGC
ZmH2B SEQ ID No: 17 CGAGGTGAACTTGGTGACG
5rev GC
SEQ ID No: 18 GAGGGCTGTACATTCTGGG
A
SEQ ID No: 19 TCCTGATCAGTCACGCTGT
C
OmuA SEQ ID No: 20 CTTCGTCCATAATGGCAATT
ATCTC
CP1 SEQ ID No: 21 AGCTGCTCCTTCTTCTCGT
G
TSP1 SEQ ID No: 22 GCACTTCTTCCACTGGTGC
T
EMP4F SEQ ID No: 23 ATGGATCCGACATGTGCAT
or CTCAGTCCGCCACGGG
EMP4R SEQ ID No: 24 CAATGAATTCTATTTCAATT
ev AGCCGG
pMRP1f SEQ ID No: 25 GGGTACCTCGAGATGCATG
or TATTAATTCATTGACACC
pMRP1 r SEQ ID No: 26 GGAAGCTTGCGAGGGGTTA
ev AGTACTACACAAGTTG
Mu3 SEQ ID No: 27
specific
probe
pBETL9 SEQ ID No: 29 CCCTCGAGTTACTCATGAT
forXho GGTCATCTAGG
pBETL9 SEQ ID No: 30 GCTCTAGAGGGTATAACTT
revXba CAACTGTTGACGG
EMP4F SEQ ID No: 31 GCATGTGCATCTCAGTCCG
or2 C
BRIEF DESCRIPTION OF THE DRAWINGS:
CA 02629363 2008-05-12
WO 2007/057402 23 PCT/EP2006/068464
Figure 1 : Maize mutant emp4 seed and plant phenotypes.
(A) Ear segregating for wild-type and emp4 mutant kernels (arrows).
(B) 20 DAP wild-type (left) and mutant (right) kernels.
(C) 30 DAP wild-type (left) and mutant (right) kernels.
(D) Isolated 20 DAP wild-type (left) and mutant (right) embryos.
(E) Comparison of wild type (left) and homozygous emp4 mutant (right) plants
obtained from 30 DAP rescued and cultured embryos.
Scale bars: 5 mm in B and C, 1 mm in D, 5 cm in E.
Figure 2 : Effect of maize emp4 mutation on basal transfer layer and aleurone.
(A-D) Longitudinally section of 14 DAP wild-type (A and C) and mutant (B and
D)
kernels carrying the pBETL1-GUS transgene and showing GUS precipitate in
basal endosperm transfer layer cells. (E-H) Longitudinally section of 20 DAP
wild-
is type (E and G) and emp4 (F and H) kernels carrying the pVP1-GUS transgene
and showing GUS precipitate in embryo and aleurone cells.
C, D, G and H are 10 pm wax sections stained with PAS.
Scale bars: 1 mm in A, B; 200 pm in C, D; 2 mm in E, F; 100 pm in G, H. Arrows
and arrowheads highlight areas of interest.
2o al, aleurone; betl, basal endosperm transfer layer; cse, central starchy
endosperm;
e, embryo; esr, embryo surrounding region; nu, nucelus; p, placento-chalazal
region.
Figure 3 : Effect of maize emp4 mutation on basal endosperm gene expression.
25 mRNA in situ analysis in wild-type (A and C) and mutant (B and D) kernels
at 11
DAP with ZmMegl antisense probe (A and B) and with ZmEsr2 antisense probe
(C and D).
Scale bar: 500 pm.
betl, basal endosperm transfer layer; cse, central starchy endosperm; e,
embryo;
3o esr, embryo surrounding region; p, placento-chalazal region.
Figure 4: DNA gel blot analyses of a Mu3 element linked to emp4 mutation.
CA 02629363 2008-05-12
WO 2007/057402 24 PCT/EP2006/068464
(A) A Mu3 transposon-tagged Pstl fragment (arrows) cosegregating with the
emp4 mutation.
(B) DNA blot hybridized with probel. The blots identified a 2 Kb Pstl band and
an
11.2 kb BamHl band co-segregating with the emp4 mutation (arrows).
Figure 5: Organization of EMP4 gene and protein structure.
(A) Schematic diagram of the EMP4 gene. The location of the Mu3 transposon
insertion of emp4 mutant in the maize coding sequence and position of Mutator
insertions of known emp4 mutants is indicated with arrows. Putative TATA box,
io ATG transcriptional start site and Stop codon are shown. Genomic regions
comprising Probe 1 and EST clone BQ1 64351 are indicated.
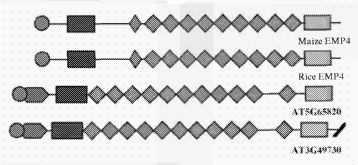
(B) Schematic alignments of maize EMP4, rice EMP4 and Arabidopsis AT3g49730
and AT5g65820 predicted aminoacid products. The PPR motifs and the PLS motif
are indicated in green rhomboids. The putative signal peptide is indicated as
brown circles. The novel N-terminal and C-terminal domains are indicated as
blue
and green boxes, respectively.
(C) Comparison of the nine PPR motifs found in EMP4 with the PPR consensus
sequence. Residues identical to the consensus are shaded in black, and similar
residues are shaded in gray.
(D) Alignment of the novel EMP4 C-terminal domain present in maize EMP4, rice
EMP4 (AC1 35956), and Arabidopsis AT3g49730 and AT5g65820.
(E) Agrobacterium-mediated transient expression in tobacco leaf epidermal
cells of
EMP4-GFP. GFP fluorescence was visualized using a confocal microscope.
Figure 6 : Maize EMP4 gene is expressed in all vegetative and most
reproductive
tissues.
RNA was isolated from leaves, roots and stems (lane 1, 2, 3), anthers (lane
4),
ovaries (lane 5), 6 DAP embryo and endosperm (lane 6 and 7), and 12 DAP
embryo and endosperm (lane 8 and 9). RT-PCR products were detected using
3o EMP4 gene specific primers Oest2 and Oest3 (top gel) (see Tablel) and a
histone2-specific primers (bottom gel).
CA 02629363 2008-05-12
WO 2007/057402 25 PCT/EP2006/068464
Figure 7 : Construct comprising the EMP4 gene under transcriptional control of
the pBETL9 promoter.
Figure 8 : Construct comprising the EMP4 gene under transcriptional control of
the pCsVMV promoter.
EXAMPLES:
The invention will now be described by the way of the following
examples, which should not be construed as in any way limiting the scope of
the
io invention.
EXAMPLE 1 : Phenotypic and genetic characterization of the maize emp4
mutation.
The emp (empty pericarp) phenotype represents a class of defective-
i5 kernel mutants with a severe reduction in endosperm development. Mutant
seeds,
recovered in the selfed progeny of a+/emp plant, appear reduced in size,
devoid
of endosperm material, and flattened by compression of surrounding normal
kernels.
20 Mutant isolation and propagation
The maize emp4 mutation (see figure 5A) has been isolated in the F2 of
a cross between a maize Mutator line and a maize inbred line. This F2 progeny
segregated for normal and defective seeds in a 3:1 ratio thus showing that the
mutant emp4 behaves as a monogenic recessive.
Because the emp4 mutation is a seed lethal mutation, it has been
maintained in heterozygosis and propagated by outcrossing +lemp4 plants to
different maize inbred lines (A188, W64A, H99).
The maize emp4 mutation was mapped onto chromosome 1 L after
crossing heterozygous mutant plants to the whole set of B-A translocations.
Mutants are recognizable as early as 12 DAP, because of a pale,
translucent and collapsed appearance of the caryopsis (Figure 1 A). Comparison
of
CA 02629363 2008-05-12
WO 2007/057402 26 PCT/EP2006/068464
wild-type and mutant sibling kernels at 20 and 30 DAP shows a reduction in
size of
the mutant caryopsis (Figure 1 B and C), whereas the pericarp, being a
maternal
tissue, is not reduced in size (normal size). At 20 DAP, emp4/emp4 embryos
already appear much smaller compared to wild-type siblings (Figure 1 D), at
maturity homozygous emp4 seeds are lethal and do not germinate. However 30
DAP explanted emp4 embryos could be rescued by cultivating on a basal medium,
then transferring seedlings to soil. For embryo rescue experiments, immature
embryos were excised from the caryopsis and cultivated on a synthetic medium
as
described in Consonni et al. (2003). Embryo-rescue seedlings at three leaf
stages
1o were transferred to soil and, after a period of acclimatization, they were
grown in
pots, under standard greenhouse conditions.
Although not significantly altered in their vegetative architecture,
homozygous
mutant plants exhibited delayed growth (Figure 1 E), but were able to reach
reproductive maturity.
Thus the EMP4 gene is required for normal plant development, though
the effect of the emp4 mutation appears to be particularly severe on endosperm
development. EMP4 gene plays an essential, non-redundant role during the
entire
plant life cycle.
EXAMPLE 2 : Endosperm domains morphology are altered in maize emp4 mutant
kernels.
The effect of the mutation on endosperm domain specification and
morphology has been studied by using two approaches, i.e. GUS staining and in
situ hybridization with specific markers.
Example 2.1): Gus marker analysis.
VP1 promoter transcriptional fusion to f3-glucuronidase (GUS) (Costa et
3o al., 2003) and BETL1 promoter transcriptional fusion (Hueros et al., 1999b)
were
introgressed in a maize A188 inbred background for 3 generations.
These maize transgenic lines were crossed to emp4/+ and Fl plants
were genotyped for the presence of f3-glucuronidase (GUS) by PCR using GUS-
CA 02629363 2008-05-12
WO 2007/057402 27 PCT/EP2006/068464
specific oligonucleotides (Gutierrez-Marcos et al., 2004), and backcrossed to
emp4/+ plants.
Kernels were cut longitudinally and GUS was detected histochemically
according to the methods previously described in Costa et al., (2003) and
Gutierrez-Marcos et al., (2004). Wax sections were mounted onto BDH slides and
counterstained with Periodic Acid Schiff's (PAS) Reagent. Polysaccharides
stain
purple-red.
pVP1-GUS plants were introgressed into the emp4 background, thus
io enabling GUS detection of the embryo and aleurone layers. GUS staining was
performed on longitudinally-cut-hand sections of 20 DAP wild-type and mutant
kernels (Figure 2E and 2F). In wild-type seeds, GUS staining was present
throughout the aleurone, extending from the germinal to abgerminal face of the
seed (Figure 2E), whereas GUS staining was detected only in the germinal face
of
is mutant seeds (Figure 2F). Wild-type aleurone usually appeared as a single
layer
of isodiametric cells (Figure 2G) whereas in mutants, it was discontinuous and
contained many irregularly shaped cells (Figure 2H). Interestingly, by using
pVP1-
GUS reporter, the presence of ectopic GUS staining was noted in discrete
portions
of the basal endosperm.
In the maize endosperm, the cells at the chalazal pole are committed to
develop into transfer cells and form a specific region named the BETL (Basal
Endosperm Transfer Layer), presumably involved in nutrient transport from the
maternal phloem to the endosperm. To ascertain whether the emp4 mutation has
an effect on the development of this region, pBETL1-GUS mutant and wild-type
seeds were obtained after introgression of the reporter line into the emp4
background.
Mutant and wild-type maize sibling seeds were analyzed after staining for
GUS (Figure 2A, 2B, 2C and 2D). In wild-type seeds, expression of the
transgene
was confined to the BETL (Figure 2A), where it was evenly distributed in two
or
three adjacent cell layers of the transfer region, decreasing progressively
toward
the center of the endosperm (Figure 2C). By contrast, reporter gene expression
CA 02629363 2008-05-12
WO 2007/057402 28 PCT/EP2006/068464
was irregular in mutant seeds (Figure 2B) and GUS staining was often observed
in
the peripheral-most cells of the basal endosperm (Figure 2D), if at all
present. In
addition, we noticed that some sectors of the emp4 basal endosperm were also
devoid of cell wall ingrowths in emp4 endosperms, when compared to wild-type
s siblings (Figure 2C and D).
These data indicate that the emp4 mutation mainly affects BETL domain
that appears only partially and irregularly developed. Defects in the BETL is
correlated with reduced rates of grain filling (Maitz et al., 2000), which in
some
io cases leads to seed abortion as observed in emp4 mutants.
Sections were counterstained with PAS, which allowed also to
qualitatively compare starch and carbohydrate composition in mutant and
sibling
wild-type kernels. Starch accumulation was notably reduced in the central
starchy
is endosperm (CSE) of emp4 mutants when compared to their wild-type siblings
(Figure 2H and G).
Example 2.2): In situ hybridization with specific markers.
mRNA in situ hybridisation analysis was performed at 11 DAP (Days
2o After Pollination) according to Costa et al., (2003) to determine the
expression of
Zm-Megl and Esr genes, as specific markers for respectively Basal Endosperm
Transfer Layer (BETL) and Embryo Surrounding Region (ESR).
Kernels were trimmed along the medial-lateral axis and immediately fixed
25 in ice-cold FAA, dehydrated in an ethanol series, and embedded in wax.
Sections
were cut at 10-12 pm and affixed onto pre-treated Superfrost Plus slides
(BDH).
Riboprobes were labelled using the DIG RNA labeling mix (Boehringer Mannheim,
catalogue number 1175025) according to manufacturer's instructions, and slides
were hybridised overnight at 50 C. Slides were viewed with a Zeiss AxioPhot
30 microscope under DIC3-5 optics and images were digitally recorded.
CA 02629363 2008-05-12
WO 2007/057402 29 PCT/EP2006/068464
mRNA in situ hybridization analysis were carried on sections of 11 DAP
emp4 and wild-type kernels using an antisense ZmMegl probe (Gutierrez-Marcos
et al., 2004) to detect BETL-specific ZmMegl transcript. ZmMegl transcript was
uniformly distributed throughout the BETL of wild-type kernels (Figure 3A). In
contrast, ZmMegl transcript was irregularly distributed and limited to small
sectors
of emp4 basal endosperms (Figure 3B).
A ZmEsr2 antisense probe has been used to detect all three Esr-specific
transcripts (ZmEsrl, ZmEsr2 and ZmEsr3) (Opsahl-Ferstad et al., 1997). No
io differences were detected in wild-type and sibling emp4 kernels, as in both
instances ZmEsrtranscripts were confined to the ESR (Figure 3C and D).
Together the results from in situs and crosses with GUS lines suggest
that the emp4 mutation affects the correct development of the BETL and
aleurone
is cell layers. It is possible that the metabolic activity of the BETL region
in the
syncitial endosperm phase is important in the differentiation of this region
into
BETL cells. Since the activity of the BETL layer is critical for importation
of
nutrients into the endosperm it is expected that a malformed BETL layer will
lead
to a defective endosperm and impact on aleurone differentiation and starchy
2o endosperm cell number.
EXAMPLE 3 : Co-segregation analysis and molecular characterization of the
maize emp4 mutation.
Since the emp4 mutation was isolated in an active Mutator (Mu)
25 population, cosegregation analysis was performed aiming at the
identification of
the gene. Plants bearing the mutation were outcrossed to W64A, a low Mutator
copy number maize inbred line, to facilitate the molecular analysis and the Fl
plants were selfed to obtain segregating families. DNA extracted from leaves
of
+/emp4 and +/+ single individuals, whose genotype has been ascertained by
30 selfing was compared by Southern analysis.
CA 02629363 2008-05-12
WO 2007/057402 30 PCT/EP2006/068464
DNA Gel Blot Analysis
For cosegregation analysis, maize genomic DNA was extracted from 7-
days old seedlings and leaf tissues, using the urea extraction method known in
the
art. For DNA gel blots 10 ug of digested genomic DNA was separated on 0,8%
agarose gels. DNA fragments were transferred to Hybond N+ membranes
(Amersham Bioscences) with 10X SSC. Membranes were subsequently probed
with a32P-labeled probes prepared from gel-purified restriction fragments
labeled
by random primer extension (Prime-a-gene labeling system; Promega, Madison,
WI).
Hybridization probes were as follows: Mutator3 (Mu3) specific probe
(SEQ ID No: 27) corresponds to the internal Hindlll-Xbal Mu3 cloned fragment.
Probel (SEQ ID No: 8) was obtained from the Mu-tagged genomic clone, and
corresponds to a 401 bp Pstll-Mlu1 restriction fragment of the genomic DNA
flanking Mu3 insertion.
Genomic DNA extracted from leaves of +/emp4 and +/+ individuals
whose genotype had been ascertained by selfing, was compared by Southern blot
analysis. Genomic DNA was digested with Pstl, a methylation-sensitive
endonuclease that cuts once inside the Mu3 element. Hybridization with a Mu3-
specific probe revealed two fragments of approximately 9 kb and 2 kb in length
cosegregating with the mutant phenotype (Figure 4A).
Subsequent hybridization with a different Mu3 probe revealed that the 5'
end of the Mu3 element was included in the 2 kb fragment.
A 11.2 kb polymorphic fragment linked to emp4 was observed also in
BamHldigest. The same polymorphisms were detected in homozygous emp4
plants obtained from embryo rescue.
Cloning of the genomic fragments
A subgenomic library of the polymorphic Mu3 fragment was prepared
with DNA extracted from emp4/+ plants in the pBSSK vector according to
CA 02629363 2008-05-12
WO 2007/057402 31 PCT/EP2006/068464
manufacturer's instructions (Stratagene, La Jolla, CA). By screening this
library
with a Mu3-specific probe, a single hybridizing clone was isolated and the
presence of a Mu3 element and DNA flanking the insertion were determined after
sequencing.
The 2kb Mu3-hybridising Pstl restriction fragment was cloned from a sub
genomic library prepared from size-fractionated Pstl fragments of emp4/+ DNA.
The cloned DNA comprises 1115 bp of genomic sequence flanking the Mu3
insertion and was mapped to the long arm chromosome one, between markers
io UMC140 and UMC106. To verify the identity of the cloned DNA, a 401 bp Pstl-
Mlu1 sequence (probe 1) was used tore-probe the same Pstl digested Southern
gel blot.
In emp4/+ individuals, the same 2 kb Pstl fragment and an additional 11 kb
fragment corresponding to the W64A EMP4 wild-type allele, were identified
(Figure
4B). In wild type, the same 11 kb W64A band was identified, plus a second 10
kb
RFLP (Restriction Fragment Length Polymorphism) band, corresponding to the
Mutator line wild type allele (Figure 4B). Co-segregation analysis was
performed
on a total of 100 plants including individuals from F1 and F2 segregating
families
2o and the data obtained were concordant, indicating that the Mu3 element was
inserted either in the EMP4 gene itself or in a closely linked locus. Southern
blot
analysis on a wide range of inbred lines indicated that EMP4 is a single copy
gene
in maize.
The EMP4 Pst1-Mlu1 probel fragment was genetically mapped by RFLP
(Restriction Fragment Length Polymorphism) using the LHRF population (Lignees
Hautement Recombinantes F2 X F252; Highly Recombinant Inbred Lines F2 X
F252). Mapping data were analysed with the mapmaker/EXP3.0 software (Lander
et al., 1987) with RiSib and Kosambi centiMorgans parameters. This mapping
positioned the maize EMP4 gene on chromosome 1L, between UMC140 and
UMC106. All these data indicate that the Mu3 element was inserted either in
the
EMP4 gene itself or in a tightly linked locus.
CA 02629363 2008-05-12
WO 2007/057402 32 PCT/EP2006/068464
A reverse genetics approach was adopted to verify the identity between
the cloned sequence and the EMP4 gene. To this end, oligonucleotide primers
corresponding to the genomic sequence were employed in combination with Mu
transposon-specific primers for a survey in a'Mutatorgene machine'. This
Mutator
gene machine is a collection of 27,500 plants in which endogeneous Mu elements
have been allowed to transpose at high frequency. Screening for Mu insertions
in
EMP4 was performed on Fl plant material and confirmed for their germinal
status
on F2 plant materials. Mutant screens were performed using a PCR-based method
with an efficient TIR specific primer SEQ ID No: 20 (OmuA) which hybridizes
with
io both the 5' and the 3'TIR and two EMP4 specific primers, SEQ ID No: 21
(CP1)
and SEQ ID No: 22 (TSP1). Four independent germinal Mu insertions were
identified each having a phenotype similar to maize emp4. Complementation
analyses of emp4 heterozygous plants with plants bearing the new insertions
reveals, in all four cases, that the newly identified Mu insertions define
mutations
is that are allelic to emp4. The new mutants were named E2439 (emp4-2), C1220
(emp4-3), C232 (emp4-4) and B2023 (emp4-5) (Figure 5A). These data indicate
that the 1115 bp cloned genomic sequence contains a portion of the EMP4 gene.
EXAMPLE 4 : Maize EMP4 gene is predicted to encode a mitochondria-targeted
20 pentatricopeptide-repeats containing protein.
In order to identify the complete sequence of the maize EMP4 gene the
genomic fragment was used to in an EST database search using BLASTN. This
resulted in the identification of an EST (BQ1 64351) (SEQ ID No: 7) from Zea
mays
immature ears which aligned with 100% homology over a region of overlap of
25 75bp.
Gene specific primers were designed and used in rapid amplification of
cDNA ends (RACE) to generate full-length wild-type EMP4 cDNA. To this aim
poly(A+) RNA was prepared from immature ears. Total RNAs was extracted from
30 maize tissues ground under liquid N2 as previously described and poly(A)+
RNA
was selected by means of the Oligotex kit, (Quiagen, Hilden, Germany) and
following the manufacturer instructions. Reverse transcription and rapid
CA 02629363 2008-05-12
WO 2007/057402 33 PCT/EP2006/068464
amplification of cDNA ends were performed according to the recommended
protocol provided by the SMARTTM RACE cDNA Amplification kit (BD Biosciences
Clontech, Palo Alto, CA). Reaction for the 3'RACE were conducted with the
primer
Oest5 and nested primer Oest7 both designed on the EST BQ1 64351 sequence.
s For the 5'RACE the universal primer provided by the kit was used in
combination
with primer Oest7rev, and with the nested primer RT5rev. The internal cDNA
portion was generated by RT-PCR with the following set of primers: RT5 forward
primer derived from the genomic clone sequence, and Oest3 reverse primer
derived from the EST sequence. The Oest7rev and Oest3 primers were designed
io on the EST BQ164351 sequence whereas RT5 and RT5rev sequence was
deduced from the portion of the genomic clone overlapping with the 5' RACE
product. These amplifications showed that the 5' RACE, internal and 3'RACE
products were derived from the same cDNA.
is In order to obtain the complete genomic sequence of EMP4, the maize
Bacterial Artificial Chromosome (BAC) library ZmF2 (O'Sullivan et al., 2001)
was
screened with the EMP4 Pstl-Mlul 401 bp probe after labelling using a32P-dCTP
(Megaprime DNA labelling system, Amersham). The library filters were pre-
hybridized for 5 hours in hybridization buffer (PerfectHyb Plus, SIGMA, St.
Louis,
20 MO) at 68 C. Hybridization was performed overnight. Filters were washed
twice
for 20 min in 2xSSC, 0.1%SDS and twice for 20 min in 0.2xSSC, 0.1%SDS.
Hybridizing BACs were identified after 24 hour exposure with a Storm 860
Imaging
System (Amersham, Buckinghamshire, UK).
25 3 identical hybridising BACs were isolated and a 5.2 kb region
encompassing EMP4 was subcloned and directly sequenced. The aligned
genomic and cDNA sequences show complete homology and allow the
determination of the gene structure. The EMP4 gene contains a single exon and
a
putative TATA box in position -38 (Figure 5A). The full length maize EMP4 cDNA
30 is 1950 bp (SEQ ID No: 1), excluding the poly-A tail and the maize EMP4
predicted protein contains an open reading frame of 614 amino acids (aa) (SEQ
ID
No: 2).
CA 02629363 2008-05-12
WO 2007/057402 34 PCT/EP2006/068464
The putative ORF was used in a Pfam search to identify any conserved
internal domains. This analysis revealed that EMP4 encodes a PPR protein,
showing high homology to one rice and two Arabidopsis pentatricopeptide repeat
s (PPR) proteins (Figure 5B). A region of the EMP4 protein extending from
residue
209 to residue 525 contains nine putative PPR motifs showing a variable degree
of
conservation (Figure 5C). The PPR motifs are contiguous, except for motifs 7
and
8, which are interrupted by two amino acids. The position of the 9 PPR motifs
was
therefore shifted forward by two amino acids, to allow the alignment with the
PPR
io consensus sequence (Lurin et al., 2004). In addition, a short sequence
preceding
the PPR-tandem repeat motifs was detected, showing significant homology with
the 31 aa long PPR-like short (PLS) motif (Lurin et al., 2004). Two other
domains
were identified in EMP4 sequence, one being located at the N-terminus between
residue 47 and residue 102, and the other at the C-terminus, located between
aa
is 538 and aa 590. The C-terminus domain had only two matches in the Pfam
database to Arabidopsis sequences AT3g49730 and AT5g65820, previously
described by Lurin et al., (2004). To better characterize this domain an
exhaustive
search of the PlantGDB sequences has been performed, which identified strong
conservation of EMP4 with a predicted protein in a rice BAC genomic sequence
20 (AC135956). The alignment of the four C-terminus domains is shown in Figure
5D
and the domain organization of the corresponding proteins is shown in figure
5B.
The domain organization of maize EMP4 and rice AC135956 is most similar
(highly conserved), while the two Arabidopsis sequences contained an
additional
domain at the N-terminus, located between residues 1 and 61 in AT3g49730 and
25 between residues 8 and 69 in AT5g65820 (Figure 5B).
Further, a putative targeting signal peptide has been identified at the N-
terminus of EMP4 and in the three homologous protein sequences using specific
software. A chloroplast subcellular localization signal was predicted for EMP4
sequence and for rice AC1 35956, whereas a mitochondrial localization signal
was
30 predicted for AT3g49730 and AT5g65820 putative gene products. A program
also
predicted a chloroplast localization signal for EMP4 sequence, while no
prediction
was obtained for the other three protein sequences. To experimentally
determine
CA 02629363 2008-05-12
WO 2007/057402 35 PCT/EP2006/068464
the subcellular localization of EMP4, we generated a translational EMP4-GFP
fusion construct, and used this for transient expression studies in tobacco
leaf
epidermal cells. Confocal scanning laser microscopy analysis of leaf samples
showed that the in vivo green fluorescence signal co-localized with
mitochondria
(Figure 5E).
EXAMPLE 5 : Maize EMP4 gene expression analysis.
To compare the level of EMP4 transcript in different plant and seed
tissues, semi-quantitative RT-PCR was performed using total RNA isolated from
io leaves, roots, stems, anthers, ovaries and immature embryos and endosperm.
First strand cDNA were synthesized from 5ug of total RNA using an oligo
d(T) primer following manufacturer's instructions.
For the semi-quantitative RT-PCR, two set of primers were used: the first
comprises the EMP4 specific primers Oest2 and Oest3, the second set was
designed on the sequence of the constitutively expressed histone gene H2B5,
and
used as control (ZmH2B5for and ZmH2B5rev). The amount of cDNA to be added
to each reaction was determined on the basis of the product obtained with the
control reaction. To detect transcript presence in different plant tissues,
2o amplification conditions were as follows: 94 C for 1 min, followed by 15
cycles of
94 C for 30 sec, annealing at 58 C for 20 sec, extension at 72 C for 40 sec
and
final extension at 72 C for 5 min. Products were analyzed on 2% TBE agarose
gel
and confirmed by hybridization. Specificity of the RT-PCR products was
confirmed
by sequence analysis.
Amplification with a set of specific primers for the orp-1 (orange pericarp-
1) gene (Wright et al., 1992) was performed for each experiment to exclude DNA
contamination in RNA samples. These primers (SEQ ID No: 18 and SEQ ID No:
19) are designed across an intron. The obtained products were of the size
3o expected for cDNA amplification.
CA 02629363 2008-05-12
WO 2007/057402 36 PCT/EP2006/068464
By using this procedure, transcripts were detected in all tissues analyzed
except anthers. Higher levels are present in both embryo and endosperm at 6
and
12 DAP (Figure 6). It was also been detected in leaves, roots, stems and
ovaries.
Expression analysis was extended to the later stages of seed
development (15 and 20 DAP). For the transcript level comparison the
amplification conditions using primers Oest2 and Oest3 were as follows: 94'C
for
1 min, followed by 15 cycles (for histone) or 20 cycles (for EMP4) of 94 C for
30
sec, annealing at 58oCfor 20 sec, extension at 72 C for 40 sec and final
extension
at 72 C for 5 min. Products were analyzed on 1.5% TBE agarose gel, stained
with
io Vistra Green (Amersham Bioscences) and densitometrically analyzed using
TyphonTM 9200 scanner and Image Quant Software. A threefold increase in
EMP4 expression was observed at 20 DAP compared to 15 DAP (Figure 6).
This data indicates that the EMP4 gene is transcriptionally active not only
during seed development, where it is detected in both seed compartments, but
in
is most plant tissues. However its transcript level is higher in the
developing kernel
compared to the vegetative tissues tested.
EXAMPLE 6 : EMP4 cellular localization analysis.
20 To generate a translation protein fusion between EMP4 and GFP (Green
Fluorescent Protein), the full length ORF of the maize EMP4 (SEQ ID No: 1) was
amplified by PCR and introduced in the binary vector pGWB5 by GATEWAY in
vitro site-specific recombination (Invitrogen, Carslab, USA). This binary
construct
contains a CaMV 35S promoter that allowed the constitutive expression of the
25 EMP4:GFP, and was introduced into A. tumefaciens strain GV3101 (pMP90)
(Koncz and Schell, 1986) by electroporation.
Agrobacterium tumefaciens mediated transient expression was carried
out in Nicotiana tabacum SR1 (cv Petit Havana). Briefly, a single colony of
the
transformed Agrobacterium was used to inoculate 5 ml of YEB medium,
30 supplemented with 100 g/mL kanamycin, 10 g/ml gentamycin. The bacterial
culture was incubated at 28 C overnight and the bacteria were pelleted by
centrifugation at 2200g for 15 seconds in a microcentrifuge at room
temperature.
CA 02629363 2008-05-12
WO 2007/057402 37 PCT/EP2006/068464
The pellet was washed three times with 1 ml of the infiltration buffer (50 mM
Mes,
pH 5.6, 2 mM Na3PO4, 0.5% glucose [w/v], and 100 pM acetosyringone) (Sigma,
Poole, UK) and then resuspended in 1 ml of the same buffer. The bacterial
suspension was diluted with infiltration buffer to adjust the inoculum
concentration
to 0.1 OD600 value. The inoculum was delivered to the lamina tissues of
tobacco
leaves by gentle pressure infiltration through a small needle puncture created
in
the lower epidermis. The plant was incubated under normal growing conditions
for
48 hours before confocal microscopy analysis. Leaf samples were harvested 48h
after infiltration and protein fusions were visualized using a Zeiss LSM 510
META
io set to measure an emission band of 475 to 525 nm for GFP and an emission
band
of 599 nm for Mitotracker Red for mitochondrial colocalization. The software
LSM
dummy (Zeiss) was used for postacquisition image processing.
EXAMPLE 7 : Expression of the maize EMP4 gene in the BETL region.
is The expression of the EMP4 gene increases during seed development
and seems to be necessary for the correct differentiation and development of
the
BETL. Thus overexpression of EMP4 gene will lead to earlier differentiation of
the
BETL and to a BETL that is larger and more active in the importation of
nutrients
required for seed growth. Therefore EMP4 gene can be overexpressed using
2o BETL-specific promoters, especially those which are active early in BETL
development preferably before BETL cellularisation, in order to increase seed
size
and / or speed of seed development.
Example 7.1) : Maize transformation with ZmMRP1::EMP4 construct.
25 The coding region of the maize EMP4 gene (SEQ ID No: 1) was amplified
from the EMP4 BAC using the primers EMP4For and EMP4Rev. The same
amplification step is also possible by using the primers EMP4For2 and EMP4Rev.
The PCR product was cloned into the vector pGEM-T-EASY (Promega) forming
pBlOS1244 and the EMP4 sequence verified by DNA sequencing. The EMP4
30 coding region was then transferred as an Ncol(blunted), Spel fragment into
Xmnl,
Xbal-cut pENTR1A (Invitrogen) forming the plasmid pEMP4-Entr (also named
pBIOS1245). The MRP1 promoter region (Gomez et al (2002)) is amplified from
CA 02629363 2008-05-12
WO 2007/057402 38 PCT/EP2006/068464
genomic maize DNA using the primers pMRPlfor and pMRPlrev. The PCR
product is cut with Kpnl and Hindlll and cloned into Kpnl, Hindlll-cut
pBIOS664
forming pMRP1-R1 R2 Sac66pA. pBIOS664 is a pBluescript based plasmid that
contains the CsVMV promoter (Verdaguer et al, 1996) fused to the rice actin 5'
s UTR (McElroy et al, 1991) and a terminator sequence derived from the
arabidopsis Sac66 gene (Jenkins et al. (1999)). Between the promoter and
terminator is the GATEWAY R1, R2 cassette (invitrogen). pMRP1-R1 R2 Sac66pA
is then transferred as an Xhol, Pmel fragment into the Xhol, Pmel cut plant
binary
plasmid pBIOS342 forming pMRP1 R1 R2 bin. (pBIOS342 is based on the binary
io vector pSB11(Komari et al (1996)) and contains a gene for selection of
transformed plants). EMP4 is then fused to the MRP1 promoter by performing an
LR clonase reaction between pEMP4-entr and pMRP1 R1 R2 bin. The resulting
binary vector is then transferred to agrobacterium strain LB4404 (pSB1)
according
to Komari et al (1996). Maize cultivar A188 is transformed with these
agrobacterial
is strains essentially as described by Ishida et al (1996).
Analysis of the transformed plants indicates that some plants
overexpress EMP4 and this increased expression is earlier than normal in seed
development. These plants possess seeds that are larger (increased size) than
segregant seeds that lack the transgene which have normal levels of EMP4. The
20 transgenic seeds also mature earlier than wild-type segregants.
Example 7.2): Maize transformation with ZmMeg 1-1:: EMP4 construct.
The coding region of the maize EMP4 gene (SEQ ID No: 1) according to
the invention is usefully expressed in an early stage of the BETL development.
25 EMP4 is fused to the ZmMeg1-1 promoter (Gutierrez-Marcos et al 2004)
by performing an LR clonase reaction between the pEMP4-Entr (also named
pBIOS1245) and pBIOS1027 (pBIOS1027 is a derivative of pBlOS342 containing
the Meg1-1 promoter linked to an R1 R2 GATEWAY cassette). The resulting
binary vector is then transferred to agrobacterium strain LB4404 (pSB1) and
this
30 strain is used to transform maize line A188. Analysis of the transformed
plants
indicates that some plants overexpress EMP4 and this increased expression is
earlier than normal in seed development. These plants possess seeds that are
CA 02629363 2008-05-12
WO 2007/057402 39 PCT/EP2006/068464
larger (increased size) than segregant seeds that lack the transgene which
have
normal levels of EMP4. The transgenic seeds also mature earlier than wild-type
segregants.
Example 7.3): Maize transformation with pBETL 1::EMP4 construct.
The coding region of the maize EMP4 gene (SEQ ID No: 1) according to
the invention is usefully expressed in the BETL area.
EMP4 is fused to the BETL1 promoter (Hueros et al, 1999) by performing
an LR clonase reaction between the plasmids pEMP4-Entr (also named
io pBlOS1245) and pBETL1 R1R2 bin (pBETL1 R1R2 bin is a derivative of
pBlOS342 containing the BETL1 promoter linked to an R1 R2 GATEWAY
cassette). The resulting binary vector is then transferred to agrobacterium
strain
LB4404 (pSB1) and this strain is used to transform maize line A188.
Analysis of the transformed plants indicates that some plants
is overexpress EMP4 and this expression is earlier than normal in seed
development. These plants possess seeds that are larger (increased size) than
segregant seeds that lack the transgene which have normal levels of EMP4. The
transgenic seeds also mature earlier than wild-type segregants.
20 Example 7.4): Maize transformation with p8ETL2::EMP4 construct.
The coding region of the maize EMP4 gene (SEQ ID No: 1) according to
the invention is usefully expressed in the BETL area.
EMP4 is fused to the BETL2 promoter (WO 99/50427) by performing an
LR clonase reaction between the plasmids pEMP4-Entr (also named pBIOS1245)
25 and pBETL2 R1 R2 bin (pBETL2 R1 R2 bin is a derivative of pBlOS342
containing
the BETL2 promoter linked to an R1 R2 GATEWAY cassette). The resulting binary
vector is then transferred to agrobacterium strain LB4404 (pSB1) and this
strain is
used to transform maize line A188.
Analysis of the transformed plants indicates that some plants
30 overexpress EMP4 and this expression is earlier than normal in seed
development. These plants possess seeds that are larger (increased size) than
CA 02629363 2008-05-12
WO 2007/057402 40 PCT/EP2006/068464
segregant seeds that lack the transgene which have normal levels of EMP4. The
transgenic seeds also mature earlier than wild-type segregants.
Example 7.5): Maize transformation with p8ETL9::EMP4 construct.
The coding region of the maize EMP4 gene (SEQ ID No: 1) according to
the invention is usefully expressed in the BETL area. The BETL9 gene has an
expression pattern similar to the BETL1 gene (Hueros et al ,(1995)). However
the
promoter is more highly expressed than that of BETL1. The BETL9 promoter
sequence (pBETL9) is represented by SEQ ID No: 32. The 1911 bp maize BETL9
io promoter was PCRed from genomic DNA of the inbred line F2 using the
primers:pBETL9forXho (SEQ ID No: 29) and pBETL9revXba (SEQ ID No: 30).
These primers introduce an Xhol and an Xbal site 5' and 3' to the BETL9
promoter.
EMP4 was fused to the BETL9 promoter by performing an LR clonase
is reaction between the plasmids pEMP4-Entr (also named pBIOS1245) and
pBlOS960 (pBlOS960 is a derivative of pBlOS342 containing the BETL9 promoter
linked to an R1 R2 GATEWAY cassette and a CsVMV-GFP gene to mark seed
containing the T-DNA). The resulting binary vector (pBIOS 1260 represented by
Figure 7) was then transferred to Agrobacterium strain LB4404 (pSB1) and this
20 strain is used to transform maize line A188.
Analysis of the transformed plants indicates that some plants
overexpress EMP4 and this expression is earlier than normal in seed
development. These plants possess seeds that are larger (increased size) than
segregant seeds that lack the transgene which have normal levels of EMP4. The
25 transgenic GFP expressing seeds also mature earlier than wild-type
segregants.
EXAMPLE 8 - Use of the maize EMP4 gene to modify plant growth rates.
Constitutive overexpression of the maize EMP4 gene can be used to
increase plant growth rate and / or to speed plant development whereas
3o repression of EMP4 expression can reduce plant growth rates and / retard
plant
development. In some enviromental conditions it is advantageous to have high
CA 02629363 2008-05-12
WO 2007/057402 41 PCT/EP2006/068464
growth rates and in others slower growth rates. Manipulation of EMP4 levels
permits plants to be better adapted to the environment and to agronomic
practices.
Example 8.1): Overexpression of the maize EMP4 gene.
The maize EMP4 cDNA (SEQ ID No: 1) is cloned behind the constitutive
promoter pCsVMV (Verdaguer et al, 1996) and cloned into a binary vector for
agrobacterial mediated transformation of maize. This is achieved by an LR
clonase reaction between pEMP4-Entr (also named pBIOS1245) and pBlOS886
(pBlOS886 is a derivative of pBlOS342 containing the CsVMV promoter linked to
io an R1 R2 GATEWAY cassette). The resulting binary vector is pBlOS 1259
represented in Figure 8. Analysis of the transformed plants indicates that
some
plants have increased EMP4 levels. These plants have a faster rate of
development and flower earlier than the control untransformed plants.
is Example 8.2): Repression of the maize EMP4 pene expression.
Methods known in the art such as antisense, partial sense, RNAi can be
used to reduce EMP4 expression. A 500bp maize EMP4 region is selected that
shows no significant homology to other genes. This region is cloned in an
inverted
orientation behind the constitutive CsVMV promoter and cloned into a binary
20 vector for agrobacterial mediated transformation of maize. This is achieved
by an
LR clonase reaction between pEMP4-Entr (also named pBlOS1245) and
pBlOS887 (pBlOS887 is a derivative of pBlOS342 containing the CsVMV
promoter linked to an R2 R1 GATEWAY cassette followed by an intron and a R1
R2 GATEWAY cassette). Analysis of the transformed maize plants indicates that
25 some plants have reduced EMP4 levels. These plants have a slower rate of
development and flower later than the control untransformed plants.
EXAMPLE 9 - Repression of the maize EMP4 gene expression in internodes.
A reduction in plant height is desirable to increase the harvest index and
30 thus yield and also to prevent plant lodging. Ideally this reduction in
height should
be due to reduced growth of stem internodes and not effect growth of other
vegetative organs such as leaves or roots so that photosynthesis and nutrient
and
CA 02629363 2008-05-12
WO 2007/057402 42 PCT/EP2006/068464
water uptake is not compromised. Such promoters from sugarcane (c67 and c51)
are described in WO 01/18211 and a homologue of gene c51 in maize also
exhibits a stem-specific expression pattern. Thus the maize EMP4 antisense
fragment is cloned behind the sugarcane c51 promoter that is preferentially
expressed in stem internodes and cloned into a binary vector for agrobacterial
mediated transformation of maize. Analysis of the maize transformed plants
indicates that some plants have reduced EMP4 levels in internodes. These
plants
are dwarfed with internodes that are reduced in length. Plant seed yield is
improved and lodging reduced.
CA 02629363 2008-05-12
WO 2007/057402 43 PCT/EP2006/068464
REFERENCES
Allison et al. (1986), Virology, 154 :9-20
An et al. (1986), Plant Physiology, 81 :86-91.
s Bechtold et al. (1993), Comptes rendus Academie des Sciences Paris Serie
3,
316:1194-1199.
Bevan et al. (1983), Nature, 304 :184-187.
Chupeau, MC et al. (1989), Biotechnology vol.7: pp. 503-508.
Consonni, G. et al. (2003), Sex Plant Reprod 15, 281-290.
io Costa, L.M. et al. (2003), Development. Oct;130(20):5009-17.
Della-Cioppa et al. (1987), Plant Physiology, 84 :965-968.
Depicker et al. (1992), Mol. Gen. Genet., 235(2-3) :389-396.
Elroy-Stein et al. (1989) Proc Natl Acad Sci U S A. Aug;86(16):6126-30.
Finer J. et al. (1992) Plant Cell Report 11 : 323-328.
is Franck et al. (1980), Cell, 21(1) :285-94.
Fromm M. et al. (1990), Biotechnology, 8:833-839.
Fu et al. (2002) Plant Cell. Dec; 14(12):3119-32.
Gallie, D.R. et al. (1989), Molecular Biology of RNA, pages 237-256.
Gomez et al. (2002), Plant Cell, 14, 599-610.
2o Gritz et al. (1883) Gene. Nov; 25(2-3):179-88.
Guerche et al. (1987), Mol. Gen. Genet., 206 :382
Gutierrez-Marcos JF, et al. (2004), Plant Cell. 16, 1288-301.
Hueros, G. et al. (1999b), Plant Physiol. 121, 1143-1152.
Ishida Y. et al. (1996), Nature Biotechnol. 14, 745-50.
25 Jenkins E. et al. (1999) Plant Cell Environ 22: 159-167.
Jobling, S.A., and Gehrke, L. (1987), Nature, 325 :622-625.
Jouanin et al. (1987), Plant Science, 53 :53-63.
Kay et al. (1987), Science, 236 :1299-1302.
Komari T. et al. (1996), Plant J. 10:165-74.
3o Koncz et al. (1984), EMBO J. May;3(5):1029-37.
Lander E.S. et al. (1987), Genomics 1, 174-181.
Lommel, S.A. et al. (1991), Virology, 81 :382-385.
Lurin et al. (2004), Plant Cell. Aug;16(8):2089-103.
CA 02629363 2008-05-12
WO 2007/057402 44 PCT/EP2006/068464
Maas et al. (1991), Plant Molecular Biology, 16 :199.
Macejack, D.G., and P. Sarnow (1991), Nature, 353 :90-94.
Maitz, M. et al. (2000) Plant J. 23, 29-42.
McElroy et al. (1990), Plant Cell, 2 :163-171.
McElroy et al. (1991) Mol Gen Genet. 231, 150-60.
Morris et al. (1992), Virology, 187 :633
Mullis, KB (1987), Methods in Enzymology 155 :335.
Neuffer, M.G., and Sheridan, F. (1980), Genetics 95, 929-944.
Neuhaus et al. (1987), Theoretical and applied Genet., 75(1):30-36
io Ohta et al. (1990), Plant Cell Physiology, 31 :805.
Opsahl-Ferstad, et al. (1997), Plant J. 12, 235-246.
O'Sullivan, D.M. et al. (2001) TAG 103, 425-432.
Sambrook et al. (1989), Molecular Cloning, A Laboratory Manual, Cold Spring
Harbour Laboratory Press p. 9.54-9.62.
is Scanlon, M.J. et al. (1994) Genetics 136, 281-294.
Scanlon, M.J. et al. (1997) Plant J. 12, 901-909.
Schocher et al. (1986). Biotechnology4:1093-1096.
Sheridan,W.F., and Neuffer M.G. (1980) Genetics 95, 945-960.
Snowdon et al. (1996), Plant Molecular Biology, 31 :689.
20 Vancanneyt et al. (1990) Mol. Gen. Genet. Jan;220(2):245-50.
Verdaguer B. et al. (1996) Plant Mol. Biol. 31, 1129-39.
Watson et al. (1994) Ed. De Boeck Universite, pp 273-292.
White, J. et al. (1990) Nucl. Acid. Res. 18, 1062.
Wright, A.D. et al. (1992) Plant Cell4, 711-719.
25 Yoder et al., (1993) Biotechnology, 12, 263-292.