Note: Descriptions are shown in the official language in which they were submitted.
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
PREPARATION AND USES OF GENE SEQUENCES ENCODING CHIMERICAL
GLYC 0 SYLTRANSFERASES WITH OPTIMIZED GLYC 0 SYLATION ACTIVITY
The present invention relates to the production of gene sequences encoding
chimerical
glycosyltransferases presenting optimized glycosylation activity, and to their
uses in the frame
of the preparation of recombinant proteins of interest by cells transformed
with said
sequences and sequences encoding said recombinant proteins.
Glycosylation of proteins and lipids
Proteins and lipids are major components of cell membranes. Membrane
associated
carbohydrates are exclusively in the form of oligosaccharides covalently
attached to proteins
forming glycoproteins, and to lipids forming glycolipids. Glycoconjugates
(including
glycolipids and glycoproteins) are most often key cell surface molecules which
are considered
to be involved in cell-cell interactions and cell adhesion (Feizi, 1993;
Crocker & Feizi, 1996).
The predominant monosaccharides found in eukaryotic glycoproteins are glucose
(Glc), galactose (Gal), mannose (Man), fucose (Fuc), N-Acetylgalactosamine
(GalNAc), N-
acetylglucosamine (G1cNAc) and sialic acid (Sia) most often as neuraminic acid
(NeuAc)
which may be N-acetylated or N-glycolylated in mammals but only N-acetylated
in humans.
Carbohydrate chains also designated as glycans are linked to the polypeptide
backbone
through either 0-glycosidic or N-glycosidic bonds. The N-glycosidic linkage is
through the
amide group of asparagine. The 0-glycosidic linkage is to the hydroxyl of
serine, threonine or
hydroxylysine. In ser- or thr-type 0-linked glycoproteins, the monosaccharide
directly
attached to the protein is frequently GalNAc while in N-linked glycoproteins,
only GlcNAc
is found.
The N-glycosidic linkage is conserved throughout the eukaryotic kingdom
including
yeast; plants, insects, mammals and humans.. The site of N-linked
glycosylation occurs within
a consensus sequence of amino acids, Asn-X-Ser/Thr (N-X-S(T)), where X is any
amino acid
except proline. When a protein analysis in the public databases is carried
out, it can be shown
that approximately 65% of all the proteins contain at least one such consensus
sequence. N-
linked glycoproteins all contain a common (invariant) pentasaccharide attached
to the
polypeptide. This core consists of three Man and two GlcNAc residues. Antennae
of variable
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
2
sequence are completing the glycan and allowing the classification into three
major N-linked
subclasses:
1. Mannose-rich glycans contain only mannose as terminal sugars.
2. Hybrid glycans contains at least a GlcNAc-Gal antenna.
3. Complex glycans contain from 2 to 5 of those antennae terminated in GlcNAc,
Gal or sialic
acid.
In all eukaryotic cells, N-linked glycoproteins are synthesized co-
translationnally from
polyribosomes bound to the endoplasmic reticulum (ER). Processing of N-linked
glycans
occurs early in the lumen of the ER and continues in the golgi apparatus and
transgolgi
network where glycoproteins achieved their final and functional polymorphism.
Attachment
of 0-linked glycans occurs post-translationally in the golgi apparatus where
glycosylation of
lipids also occurs. Sugars used for both N- and 0-glycosylation are activated
by coupling to
specific nucleotides in the cytoplasm and are imported within the lumen of
organelles through
specific transporters.
Glycosylation is the most sophisticated set of post-translational
modifications which
may occur in proteins .They aimed to: i) control the biological activity of
proteins, ii) signal
proteins for binding lectin-like receptors and/or degradation systems, iii)
address proteins to
the cell surface (secretion), iv) target proteins to cellular compartments, v)
define an
immunological identity (blood groups). As a result, glycosylated proteins
exist as a mixture of
glycoforms whose physico-chemical and biological properties differ from the
product directly
coded by the relevant gene. It is widely admitted that post-translational
modifications of
proteins allow a better adaptation the protein to its biological function
(Helenius & Aebi,
2001).
Many human glycoproteins are of high clinical relevance. For example, on cell
surfaces, they are important for communication between cells, for maintaining
cell structure
and for self-recognition by the immune system.
Glycoproteins are most abundant in soluble forms in biological fluids such as
milk and
blood. In this case, glycans protect the proteins against proteases, increase
their solubility
govern their plasmatic clearence and adress them to target organs.
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
3
Glycosylation Enzymes
Glycosylation reactions are of major biological importance to both prokaryotes
and
eukaryotes and require the coordinated action of a large number of enzymes
designated as
Glycosyltransferases (GTs) (Breton & Imberty, 1999).
Glycosyltransferases (GTs) constitute a large family of enzymes involved in
the
biosynthesis of polysaccharides and glycoconjugates in the prokaryotic and
eukaryotic
kingdoms respectively . Sequences of the prokaryotic enzymes are not
homologous to
mammalian glycosyltransferases while enzymes from human, mammals and
drosophila often
share significant homology. So far, more than a thousand of GTs are known to
mediate a wide
array of biological functions. Developments in the molecular biology of these
enzymes have
revealed an unexpected diversity suggesting that glycosylation process
probably require the
involvment of about 250 genes in a single human living cell (Breton et al.,
2001). About 500
glycosyltransferases including 160 human enzymes have been cloned to date. The
number of
cloned enzymes is increasing in human and may reach 200 shortly (Narimatsu,
2004). GTs
are classified according to the stereochemistry of the reaction substrates and
products as either
retaining or inverting enzymes (Sinnott, 1990). A classification of
glycosyltransferases using
nucleotide diphospho-sugar, nucleotide monophospho-sugars and sugar phosphates
and
related proteins into distinct sequence-based families has been established.
It shows that
human GTs distribute so far into 42 structural families (http://afmb.cnrs-
mrs.fr/CAZY/Homo-
sapiens.html).
Glycosyltransferases catalyze the transfer of sugar residues from a nucleotide-
sugar
(activated donor substrate) to an acceptor (lipid, protein or growing
carbohydrate chain). In
addition to the official classification of enzymes, they can be grouped into
functional families
based on their sequence similarities, which may reflect enzymatic
characteristics such as
donor specificity, acceptor specificity, and linkage specificity between donor
and acceptor.
Based on the sugar they transfer, GTs are named according to the sugar they
transfer such as
N-Acetylglucosaminyltransferase (G1cNAcT), galactosyltransferases (GalT), and,
fucosyltransferases (FucT) or sialyltransferases (SiaT or STs). Owing to the
accumulation of
glycosyltransferase gene data, it is thought that glycosyltransferases have
high specificity for
the type of linkage in either the donor and acceptor substrates as well as for
the nature of the
glycoconjugate acceptor (N-/ 0-linked glycoproteins or lipids).
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
4
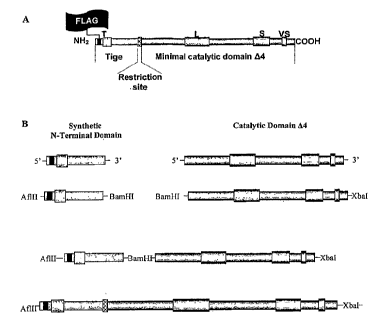
Structure of Glycosyltransferases:
All GTs cloned, so far in vertebrates display the same topology. They are type
II
membrane proteins (N-terminal cytoplasmic domain) composed of four main
domains: a short
cytoplamic tail (CT) at the N-terminal end, a membrane anchor region (TMD) of
10 to 20
amino acids, a stem region (SR) and a large C-terminal catalytic domain (CD)
(Figure 1)
(Paulson et al., 1987). The anchor region acts as a non cleavable signal
peptide and also as a
transmembrane domain (Wickner & Lodisch, 1985), orientating the catalytic
domain of GTs
in the lumenal part of the golgi apparatus. The SR is widely considered as a
flexible region
allowing also the orientation of the CD in the lumenal part of the golgi
apparatus. On a
general way, the CD is reasonably conserved within GTs of various species
whereas the SR
constitutes an hypervariable portion of the transferase. Some of these enzymes
may be
cleaved at their SR by an endogenous protease, or proteases, and secreted out
of the cell
(Paulson & Colley, 1989) to produce milk or blood enzymes. It has been well
documented
that the proteolytic cleavage and secretion of glycosyltransferases are
affected by various
pathological conditions such as malignant transformation and inflammation but
the molecular
mechanisms underlying the cleavage and secretion have not yet been clarified.
GTs and neoglycosylated proteins/lipids have been localized inside the ER and
the
subcompartments of Golgi using both subcellular fractionnation of cellular
membranes and
immunoelectron microscopy (Roth, 1987). Early studies suggest that the
compartmentalization of GTs may reflect the sequence of the oligosaccharide
chain
modification (Kornfeld & Kornfeld, 1985). It was thought that this strict
localization ensure
an optimal biosynthesis of the glycan chains by providing an efficient
vicinity between
enzyme, substrate and sugar nucleotide donor. Further studies showed that many
of
glycosylation enzymes overlap in localization and demonstrated cell-type
specific golgi
subcompartmentation (Colley, 1997). GTs are spread out in the golgi stacks and
this can
differ between cell types for a given protein (Roth, 1991).
Generally, it has been demonstrated that the transmembrane domain (TMD) of
proteins is a determinant key to confer golgi localization essential for at
least GaNAcTs,
GlcNAcTs, GalT, FucT and SiaT to find their acceptors. It was demonstrated
that the flanking
region of TMD and/or the lumenal portion contribute to localize as well
(Munro, 1998; Yang
et al., 1996). Morever, the CT and the SR of GTs specify their in vivo
functional
sublocalisation and stability in the golgi apparatus. Substituting either of
the three domains
(CT, TMD and SR) does not change the catalytic activity of the enzyme but
contribute to alter
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
its distribution in the golgi compartments (Grabenhorst & Conradt, 1999). No
clear specific
targeting sequences have been found over the last decade and only critical
regions of the GTs
have been identified for their compartmentimentalization (Opat et al.,
2001).More recently,
the inventors have found that the soluble catalytic domain of ST6Ga1 followed
a different
5 subcellular route than the full-length enzyme (Ronin, Biochimie 2003).
Among the large number of GTs, the families which are of greatest interest are
those
which are in charge of terminal glycosylation because these sugars play an
important role in
phenomenons of recognition and signalization in humans. Those essentially
include
sialyltransferases(SiaTs), fucosyltransferases (FucTs) and
galactosyltransferases (GalTs).
Galactose and fucose are involved in recognition of blood group (ABH/Lewis)
antigens.
Sialylated oligosaccharides of glycoproteins and glyco lipids are implicated
in many biological
processes such as cell adhesion and receptor recognition in inflamamtion and
cancer
(sialyLewis antigen) as well as neuronal outgrowth (Polysialic antigen)
(Paulson, 1989). The
structural diversity and regulated expression of sialylglycoconjugates appear
to be well
correlated with their functions (Sasaki, 1996).
Sialyltransferases:
The sialic acid family is composed of more than a hundred of derivatives among
which neuraminic acid is the most frequent in mammals and humans.
Sialyltransferases (STs)
catalyze the transfer of a sialic acid residue from its activated form
,cytidyl-monophospho-N-
acetyl-neuraminic acid (CMP-Neu5Ac), to a non-reducing terminal position on a
glycan
acceptor in glycoproteins or glycolipids. The catalyzed reaction is as
follows:
CM P-13-Neu5Ac + HO-acceptor ---> CM P-H + Neu5Ac-41-0-acceptor
Each ST is classified according to the type of linkage established between the
sialic
acid residue and the acceptor substrate specificity (which can be either a
protein or a lipid).
Thus, three groups are distinguished: a2,3-sialyltransferases (a2,3-STs), a2,6-
sialyltransferases (a2,6-STs), and a2,8-sialyltransferases (a2,8-STs). a2,6-
STs transfer a sialic
acid residue at an alpha2,6 position (ST6Ga1) to a galactose residue, or to a
N-Acetyl-D-
galactosamine (ST6Ga1NAc), or to a N-acetyl-D-glucosamine (ST6G1cNAc).
However, the
enzyme involved in the formation of this last type of linkage is still
unknown. The a2,3-STs
transfer a sialic acid to the carbon 3 of galactose (ST3Ga1), and the a2,8-STs
to the carbon 8
of an other acid sialic residue (ST8Sia).
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
6
STs have also been identified in all animals (from birds to humans), in
bacterial cells
and in viruses (Sujino et al., 2000). The family of human STs is composed of
20 different
genes cloned as shown in table 1 (Sasaki, 1996, Ronin 2003). Their substrates
specificities are
as follows.
1) a2,6-STs
= ST6Gal:
ST6Ga1 I transfers a sialic acid residue on the disaccharide Ga1(31-4G1cNAc of
N-
glycans almost (van den Eijnden et al., 1980; Weinstein et al., 1982). It is
expressed in an
ubiquitous manner in human tissues except in testis and brain where it is
expressed a lower
levels (Table 2) (Kitagawa & Paulson, 1994a).
ST6Ga1 II, identified recently (Krzewinski-Recchi et al., 2003; Takashima et
al.,
2002), recognizes the disaccharide Ga1(31-4G1cNAc as acceptor substrate
particularly when it
is on a free oligosaccharide (unknown protein or lipid). The expression
pattern of ST6Ga1 II is
restricted to the brain and shows a low expression level in testis, thyroid,
lymphatic ganglia
and some fetal tissues (Table 2).
= ST6GaNAc:
The second subfamily of a2,6-STs is represented by the group of ST6GaNAc,
which
transfer a sialic acid residue on an N-acetyl galactosamine residue. Six
members have been
identified in human (Table 2).
ST6GaNAc I and ST6Ga1NAc II possess the broadest substrate specificity as they
are
able to transfer a sialic acid on the following 0-glycan structures: Ga1(31-
3Ga1NAc, GaNAca-
O-Ser/Thr, Siaa2-3 Ga1(31-3GalNAca-O-Ser/Thr (Ikehara et al., 1999; Kono et
al., 2000;
Kurosawa et al., 1994; Kurosawa et al., 1996; Harduin-Lepers et al., 2001).
Their expression
pattern is different, ST6Ga1NAc I is expressed in submaxillary and mammary
glands, spleen
and colon whereas ST6Ga1NAc II is expressed in many tissues such as testis and
lactating
mammary glands (Kurosawa et al., 1996; Kurosawa et al., 2000).
ST6GaNAc III, IV, V and VI possess a reduced substrate specificity: they
recognize
only the trisaccharide Siaa2,3Gal(31-3GaNAc. However, it is now established
that
ST6GaNAc III, V and VI catalyze preferentially the formation of the GMlb
glycolipid
(Sjoberg et al., 1996; Lee et al., 1999) whereas ST6Ga1NAc IV catalyzes the
transfer of sialic
acid on 0-glycans (Harduin-Lepers et al., 2000, Ikehara et al., 1999; Lee et
al., 1999,
Okajima et al., 2000). ST6Ga1NAc IV shows restricted substrate specificity
using only the
trisaccharide sequence Siaa2,3Gal(31-3GaNAc and does not discriminate between
a- and (3-
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
7
linked GalNAc. Two different isoforms have been found. Northern-blot analysis
detected a
2.2 kb transcript in various adult tissues and lower levels of expression of
an additional
transcript in brain, heart and skeletal muscle (Harduin-Lepers et al., 2000).
2) a2l 3-STs
They all are ST3Ga1 transferring sialic acid of a galactose residue in protein
or lipid
acceptor.
ST3Ga1 I was cloned from submaxillary glands cDNA library and its ubiquitous
expression (Table 2) was confirmed by Northern-blot (Chang et al., 1995). It
synthesizes
Siaa2,3Galf31-3-Ga1NAc, a structure common to many 0-linked oligosaccharides.
It differs
from other STs in its ability to use glycolipid acceptor substrates in vitro
(Kitagawa &
Paulson, 1994b).
ST3Ga1 II was cloned by Kim et al. (1996) from a liver cDNA library and the
Northern-blot analysis revealed high levels expression in heart, liver and
skeletal muscle,
intermediate levels in thymus, lymph node, appendix and spleen, and lower
levels in lung
peripheral blood lymphocytes (Table 2). ST3Ga1 II can transfer sialic acid
residues on Ga1f31-
3-GalNAc or on the gangliosides (lipid) GM1 or asialo-GM1 as acceptor
substrates
(Giordanengo et al., 1997).
ST3GalIII has been cloned by screening a human placental cDNA library with a
probe
based on sialylmotif, the cDNA encoding ST3Ga1 III was isolated (Kitagawa &
Paulson,
1993). Transcript is abundantly expressed in skeletal muscle and fetal tissue
and lower
expressed in placenta. This enzyme catalyzes the transfer of sialic acid to
galactose-containing
substrates (Table 2).
ST3Ga1 IV is involved in sialylation of 0-linked Ga1f31-3Ga1NAc (Table 2)
(Tetteroo
et al., 1987). There are 5 different mRNAs expressed, and they encode for
identical protein
sequences except at the 5'-ends. These transcripts are produced by a
combination of
alternative splicing. Northern-blot analysis showed that one of them is
specifically expressed
in placenta, testis and ovary, indicating that its expression is independently
regulated from the
others (Kitagawa et al., 1996).
ST3Ga1 V has been isolated from a human cell library of cDNA and called GM3
synthase. A major 2.4 kb transcript is expressed in many tissues particularly
in brain, skeletal
muscle, placenta and testis. It is widely distributed in human brain with
slightly elevated
expression in the cerebral cortex, temporal lobe and putamen. The substrate
specificity of this
enzyme is highly restricted to lacosylceramide as the acceptor (Table 2)
(Ishii et al., 1998).
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
8
ST3Ga1 VI was cloned from a human melanoma cDNA library. This ST exhibits
restricted substrate specificity: it is involved in the synthesis of sialyl-
paragloboside, a
precusor of the sialyl-Lewis X determinant (Okajima et al., 1999). There are 2
forms of
5T3 Gal VI mRNA (called type 1 and 2), differing only in the 5'-untranslated
region. This
enzyme is expressed at similar levels in most tissues (Table 2) (Taniguchi et
al., 2001).
3) a2l 8-STs
ST8Sia I is also called ganglioside GD3 synthase and catalyzes the transfer of
a sialic
acid molecule to the terminal sialic acid of GM3 via an a2,8 linkage (Sasaki
et al., 1994). It
has been shown that this enzyme can also use GM1b, GD1 a and GT1b as acceptor
substrate
(Table 2) (Nakayama et al., 1996; Nara et al., 1996; Watanabe et al., 1996).
ST8Sia II is also called STX. Scheidegger et al. (1995) used sequences of
rodent STX
to clone the human cDNA from a fetal heart library. STX is primarily expressed
in embryonic
tissues and modestly in adult heart, brain and thymus (Angata et al., 1997).
This enzyme
regulates the linkage between neural cell adhesion molecule (NCAM) and
polysialic acid
(PSA) and modulates by this way the adhesive properties of NCAM. STX catalyzes
the
transfer of the first sialic acid via an a2,8 linkage on an other sialic acid
linked in a2,3 or a2,6
(Table 2) (Angata et al., 1997) on a N-Glycan, then it is involved in the
elongation process
making successive a2,8-linkage over the previous residues.
ST8Sia III transfers a sialic acid on Siaa2-3Galf31-4G1cNAc structures inside
N-
glycans and glycolipids such as GM3 (Table 2) (Lee et al., 1998; Yoshida et
al., 1995a;
Yoshida et al., 1995b).
ST8Sia IV, also called PST (Polysialyltransferase), was cloned by Nakayama et
al.
(1995). Northern-blot analysis revealed that PST is expressed in many fetal
tissues and in
adult heart, spleen, thymus and at lower levels in other organs and tissues
(Table 2). This
enzyme also regulates the linkage between neural cell adhesion molecule (NCAM)
and
polysialic acid (PSA). In addition, it is shown that it catalyzes the same
reaction as the STX
(Angata et al., 2000; Nakayam et al., 1995).
ST8Sia V presents a transfer activity towards the gangliosides GM1b, GD1a,
GT1b
and GD3 (Table 2) (Kono et al., 1996).
ST8Sia VI has been cloned quite recently in human and little is known about
it. The
mouse ST8Sia VI possesses a transfer activity of sialic acid on the
NeuAca2,3(6)Ga1f3
structure found at the non reduced end of 0-glycans, N-glycans and free
oligosaccharides
such as the sialyllactose (Table 2) (Takashima et al., 2002).
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
9
4) Structural organization needed for promoting sialyltransferase activity
All members of the STs family possess three conserved region in their CD named
Sialylmotifs: motif L for Large, S for Small and VS for very small (Datta &
Paulson, 1995;
Geremia et al., 1997) based on the comparison of their primary sequences A
survey of the
animal genomes has been published lately (Harduin-Lepers, 2005) to find out
unknown
sequences possessing the sialylmotifs and thus potentially new STs.
The sialylmotif L consisted of 44 or 45 amino acids and contained between 5
invariant
residues among all the human enzymes as shown in Figure 2. This region is in
the center of
the CD. The second sialylmotif S, in the COOH-terminal portion, consisted of
23 amino acids
residues, two of which residues being identical among all the STs (Drickamer,
1993). The
third sialylmotif VS, at the terminal part of the enzyme, consisted of 13
amino acids with two
conserved residues (histidine and glutamate).
It has been demonstrated that the sialylmotif L is involved in the binding of
the donor
substrate CMP-sialic acid (Datta & paulson, 1995). Some amino acids of this
motif can also
participate in the catalytic activity of enzymes (Sasaki, 1996). It has been
proposed that the
sialylmotif S participate to both donor and acceptor binding (Datta et al.,
1998). The precise
role of VS is still unclear but recent studies on STX and PST examined the
functional rule of
the conserved His in this motif (Kitazume-Kawaguchi et al., 2001). The change
of the
Histidine by a Lysine residu affect their catalytic activity showing that the
motif VS is
involved in catalysis. The sialylmotif VS is necessary for optimal catalytic
efficiency and it is
part of the active site, mainly on the acceptor site or at the vicinity of
both donor and acceptor
sugar substrates (Jeanneau et al., 2004). From previous work, it is now clear
that the C-
terminal part of the CD (part of sialylmotif S, motif 3 and sialylmotif VS) is
primarily
dedicated to the recognition of acceptor substrates whereas the N-terminal
part (sialylmotifs L
and part of S) is mostly involved in nucleotide sugar binding (Datta &
Paulson, 1995; Datta et
al., 1998; Laroy et al., 2001; Kitazume-Kawaguchi et al., 2001; Jeanneau et
al., 2004).
However, it cannot been excluded that the Sialylmotif L could also be involved
in acceptor
recognition (Legaigneur et al., 2001).
As mentioned above, the amino acid sequences deduced from the cloned human
sialyltransferase cDNA show the same organization in four domains shared by
many GTs: i) a
short N-terminal cytoplasmic tail (CT; aound 10 amino-acids), ii) a
transmembrane domain
(TMD; around 20 amino acids), iii) a stem region (SR), highly variable in
length and iv) a
catalytic domain (CD; around 300 amino acids.
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
The SR is defined as the peptide region between the TMD and the CD which can
be
removed without altering the enzymatic activity (Paulson 1989, Ronin, 2001,
Vallejo-Ruiz et
al., 2001; Jeanneau et al., 2004). However, the inventors have shown that this
hypervariable
region may be essential to define a fine recognition of the glycan acceptor
5 (tri>bi>tetraantennary) and proposed that it may be involved in a
conformational change to
open the catalytic site (Ronin, 2001). As a result, the catalytic efficiency
is increased by a
factor 35 and the recognition of glycan acceptor is broadened.
The CD is crucial for STs to display enzymatic activity. It contains at least
three
10 highly conserved sequences maintaining identical amino acids positions
found in all
mammalian STs cloned. Those domains are L-, S- and VS-sialylmotifs (Livingston
&
Paulson, 1993; Geremia et al., 1997). An additional domain (motif 3) has also
been recently
isolated between the sialylmotifs S and VS, it contains four highly conserved
residues, with
the following consensus sequence : (H/y)Y(Y/F/W/h)(E/D/q/g). (Capital letters
and lowercase
letters indicate a strong or a low occurence of the amino acid, respectively.)
(Jeanneau et al.,
2004). Many studies descibed in the litterature aim to get insight into
structure/function
relationships in the large STs family and thus particularly to define a
minimal catalytic
domain inside. The minimal CD has been so defined experimentally by site-
directed
mutagenesis or alternatively by sequence alignment and comparison for a few
STs (Vallejo-
Ruiz et al., 2001; Chen & Colley, 2000, Ronin 2003).
Despite the broad definition of the CD and the catalytic activity, delineating
a minimal
CD of STs is still uneasy. ST6Ga1 I is the most studied enzyme among STs and
most of the
research has been realised on it to define both minimal catalytic domain and
catalytic activity.
This is due to the absence of ST6Ga1 in all cells used to produce human
recombinant proteins.
This lack is a technological bottleneck for heterologous systems using non
animal cells (yeast,
insects and plants) since they do not contain any sialic acid. In CHO cells
alternatively, only
a 5T3 activity is expressed.
The delineation between the SR and the CD has never been well defined except
experimentally. Based on bioinformatics, the inventors have designed a
strategy of identifying
the end of the SR for the 3 ST families (Ronin., 2003).
The CD generally coincides with the end of the hypervariable region in the N-
terminal
half of the enzymes (Ronin., 2003). The catalytic domain is therefore assumed
to start around
70-90 residues upstream from the sialylmotif L and around 40-45 residues for
ST6GaNAc III
to VI. The average size of the CD is estimated around 300 ( 20) amino acids,
including the
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
11
sialylmotifs (Jeanneau et al., 2004) (Table 3). The minimal CD has been
defined for a few
STs - either by truncation experiments or sequence comparison ¨ (Legaigneur et
al., 2001;
Vallejo-Ruiz et al., 2001; Chen & Colley, 2000), such as for hST3Ga1 I
(minimal CD
consisting in amino acids 57-340; Vallejo-Ruiz et al., 2001) and hST8Sia IV
(minimal CD
consisting in amino acids 62-359; Angata et al., 2004).
When transfected in CHO cells, the soluble CD of hST6Ga1 I (amino acids 90-
406)
was found to display an enlarged pecificity towards endogenous acceptors as it
follows the
intracellular secretory pathway within the Golgi apparatus. It has thus been
possible to
delineate a minimal CD for hST6Ga1 I containing a critical sequence capable of
displacing the
acceptor recognition from intracellular resident acceptors to cell surface
glycoconjugates
(Donadio et al., 2003). In addition, the soluble secreted CD of hST6Ga1 I
showed increased
transfer efficiency, irrespectively of the branching pattern of the glycan
acceptor (Legaigneur
et al., 2001).
Glycosylation of protein (drugs) produced in heterologous expression
systems
Proteins of therapeutic interest were first extracted from natural sources
such as blood,
placenta, human or animal tissues. However, this approach is limited by the
source, amount
and availability of human tissues and may contain life threatening
contaminants (prions,
oncogenes, viruses...) as well as potential allergens generated by proteins
from animals. With
the rise drug-approved proteins and clinical needs, new approaches have been
developed to
produce proteins using different expression systems.
In most instances, intensive work is currently aiming at humanizing the
glycosylation
pattern of the recombinant proteins to approach the pattern found in the
natural glycoproteins
as closely as possible to improve pharmacokinetics and lower immunogenicity of
the product.
The machinery required for the synthesis, the activation and the introduction
of sialyl
residues is poorly expressed in the various existing recombinant proteins
expression systems.
The recombinant human proteins produced are therefore often under or even not-
sialylated
compared to their native counterparts.
One of the most used systems is the bacteria Escherichia coli (E. coli)
(Swartz, 2001;
Baneyx, 1999), but its main inconvenient is that the human post-traductionnal
modifications,
particularly protein glycosylations are not carried out by this prokaryote
because no such
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
12
glycosyltranferases are expressed in E. coll. This can lead to the reject of
the therapeutic
proteins of interest by the immune system, the reduction of their circulatory
half-life and/or of
their biological activity. The protein produced can eventually be misfolded
and aggregate as
inclusion bodies.
Yeasts and filamentous fungi are also well-known eukaryote expression systems
and
they possess cellular machinery similar to those of human cells. The yeast
produces complex
proteins and is able to carry out several post-traductional modifications such
as simple
glycosylations. However, although the N-glycosylation process performed by
yeast and fungi
is the same as the mammalian process for the initial steps in the endoplasmic
reticulum, no
complex oligosaccharide containing sialic acid, galactose, fucose and N-
acetylgalactosamine
have been found inside the glycoproteins produced by these organisms
(Blanchard, 2004);
both yeast and fungi typically produce mannose-rich glycans by adding up to
100 mannose
residues (in the case of yeast) on the pentasaccharidic core (Tanner & Lehele,
1987;
Herscovics & Orlean, 1993) in the golgi apparatus. Those hypermannosylation
foster an
immune response in human.
Engineering glycosylation in yeast (Hamilton et al., 2003; Roy et al., 2000)
has first
allowed the reduction of the mannoses residues number added. Then, enzymes
which are
necessary to peripheral N-acetylglucosaminylation and galactosylation and have
been added
in these systems (Maras et al., 1999; Bretthauer, 2003; Vervecken et al.,
2004). However, the
addition of the terminal sialic acid is still difficult to achieve, due to the
large number of
enzymes involved in this terminal step.
Proteins expressed in insect cells are properly folded, may undergo post-
traductional
modifications and be secreted. Early N-linked glycosylation carried out by
these cells are
similar to those performed by mammalian cells. However, the glycan structures
obtained in
this case are of the paucimannosidic type i.e truncated-due to the presence of
an undesirable
N-acetylhexosaminidase activity which degrades the neoglycoproteins expressed
during
Baculovirus expression (Blanchard, 2004). In addition, in some insect cells
lines, a1,3/fucose
residues may be found and these residues generally trigger an undesirable
immune response
in human. Thus, the use of this system is restricted to the production
vaccinal antigens.
In insect cells, the GTs catalyzing the transfer of immunogenic sugars have
been
deleted and sialylation could be achieved by adding 3 genes encoding for the N-
acetylglucosamine 2-epimerase, N-acetylneuraminyl lyase and CMP-Neu5Ac
synthase (Jarvis
et al., 1998; Aumiller et al., 2003). A new insect cell line (SfSWT-3)
designed to synthesize
its own CMP-sialic acid has been created The resulting cells express all the 7
mammalian
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
13
genes, can produce CMP-sialic acid and sialylate an heterologous protein when
cultured in a
serum-free growth medium (Aumiller et al., 2003).
As eukaryotic cells, plants exhibit a complex and sophisticated cellular
machinery
which may be used to produce therapeutic proteins. Recombinant proteins
possess a very
good pharmacological quality because plants have many of the enzymes required
for the
maturation of the proteins. However, the glycosylation pathway needs major
adjustments not
to produce allergenic proteins. Indeed, the N-glycosylations in plants
(Lerouge et al., 2000)
are similar to those performed in humans as far as core glycosylation is
concerned. However,
glycans are still lacking sialylated antennae and contain inner (31,2-xylose
and a1,3-fucose.
Both residues are highly immunogenic for human and currently compromise the
approval of
transgenic plants as expression systems for thepareutics.
In plants, the first strategy used aimed at preventing the addition of
allergenic sugars
by preventing proteins to exit the endoplasmic reticulum.As a result, N-linked
glycans can
not mature to complex type. Another strategy was based on the inhibition of
several GTs
inside the golgi apparatus. This inhibition can not be complete and/or can
enter in competition
with the endogenous machinery for maturation.
Mammalian cell expression system, namely CHO cells, is currently the only drug-
approved system to produce recombinant therapeutics. These cells show a major
advantage
because they are able to synthesize complex N-linked glycoproteins of high
molecular weight
and/or multimeric. Mammalian cells naturally express, not only the enzymes
involved in the
synthesis and the transport of the nucleotide-sugars, but also the
glycosyltransferases required
to achieve complex glycosylation of the recombinant proteins with a
satisfactory content in 3-
linked sialic acid. However, there is a lack of some enzymes such as the
a1,3/4-
fucosyltransferases and a2,6-sialytransferases, which realize terminal 0-
linked and N-linked
glycosylation. Moreover, in mammalian cells, the sialylation occurs through N-
glycolyneuraminic acid (NeuGc) which significantly differs also from the N-
Acetylated
derivative (NeuAc) found in human cells
In the case of mammalian cells, work has been performed through the over-
expression
of an a2,3-ST and a (31,4Ga1T (Weikert et a/.,1999); both enzymes are present
in the genome
but their activities are highly variable upon cell culture conditions.. This
led to a wide
variability concerning the presence of the terminal Gal and sialic acid and to
an extensive
microheterogeneity in glycosylated proteins. Work has also been oriented
towards the
optimization of the galactosylation and sialylation (Granbenhorst et al.,
1999) by introducing
an a2,6-ST (Bragonzi et al., 2000) in mammalian cells. A CHO cell line stably
expressing
CA 02653104 2014-05-26
-
14
native rat a2,6-ST has been established. The glycoproteins produced by these
CHO cells
display both a2,6 and a2,3-linked terminal sialic acid residues, similar to
human; the ratio
observed between a2,6 and a2,3-linked terminal sialic acid residues carried
was of 40,4% of
a2,6- and 59,6% a2,3-sialic acid residues, which improved pharmacokinetics in
clearance
studies (Bragonzi etal., 2000). Despite these improvements in humanization of
cells, the ratio
between a2,6 and a2,3-linked terminal sialic acid residues cannot be
controlled and has never
been found even favorable to the 6-activity. It thus appears that first,
sialylation is a critical
step to control glycan structures and secondly, the difficulty resides also in
expressing the
heterologous ST in a specific compartment of the cell (adressing), in
optimizing its activity
provided that the donor substrate for this enzyme is present (carrier).
It is therefore worth noting that terminal sialylation of approved
glycoprotein drugs is
the most difficult step to obtain in all expression systems available so far.
Goal of the invention
The goal of the present invention is to provide a process for generating a
panel of new
gene sequences encoding chimerical membrane enzymes with glycosyltransferase
activity,
and in particular for designing gene sequences encoding innovative chimerical
sialyltransferases to equip cells with a needed sialylating activity and
improve the quality and
yield of recombinant glycosylated proteins.
The invention relates to a process for producing gene sequences encoding
chimerical
membrane glycosyltransferases presenting an optimized glycosylation activity
in cells
transformed with said sequences, when compared with the glycosylation activity
of the
corresponding native glycosyltransferases, i.e. a glycosylation less selective
and/or more
efficient (at least 30¨fold higher than the initial activity of the native
full length
glycosyltransferases) towards the acceptor substrate, said process comprising
the fusion:
- of a first nucleic acid sequence coding for a C-terminal minimal fragment of
the
catalytic domain (CD) of the native full length glycosyltransferase, said C-
terminal minimal
CD fragment displaying a transferase activity (which can be enhanced
at least 30¨fold
higher than the initial activity of to the native CD), said first nucleic
sequence being obtained
by removing nucleotides coding for one or several contiguous aminoacids
extending from the
first aminoacid of the N-terminal end of said CD, and selection of the nucleic
acid encoding
said C-terminal minimal CD fragment which is such that if n represents the
number of
CA 02653104 2014-05-26
contiguous amino acids as defined above which has been deleted, then the
fragment obtained
when deleting at least n+1 contiguous aminoacids as defined above, has no
substantial
transferase activity,
- to a second nucleic acid of variable sequence coding for a transmembrane
peptide
5
chain specifying the anchorage of the glycosyltransferases in intracellular
compartments, and
comprising (or consisting of) in its N-terminal region a cytoplasmic tail (CT)
region located
upstream from a transmembrane domain (TMD), itself located upstream of a stem
region (SR)
or of a fragment of at least 3 contiguous aminoacids of the SR, said SR or
part thereof being
linked to said catalytic domain, optionally via a linker or connection peptide
of at least 2
10
amino acids encoded by a restriction site which does not exist in the
nucleotide sequence
coding for the native CD mentioned above,
provided that at least one of these CT, TMD, SR peptides being different from
the
primary structure of the native corresponding peptides in the native
glycosyltransferase from
which is derived the CD fragment with optimal glycosyltransferase activity as
defined above,
15
this fusion being carried out in such way that the first nucleic acid is
located
downstream from the second nucleic acid and provides a protein product in
which the CD is
in the C-terminal half.
By chimerical membrane glycosyltransferases presenting an optimized
glycosylation
activity in cells transformed with said sequences, when compared with the
glycosylation
activity of the corresponding native glycosyltransferases, one should
understand that said
chimerical membrane glycosyltransferases:
- have a sugar transfer activity less selective towards acceptor substrates
than the
corresponding native glycosyltransferases when tested in vitro with
exogeneous/
commercially available acceptor glycoproteins having known bi-, tri-or
tetraantennary
glycans, i.e. have a in vivo glycosylation activity in cells towards all cell
glycoprotein
acceptors, said glycosylation activity being measurable preferably in
intracellular and cell
surface compartments according to the following general procedure using lectin
SNA binding,
- and/or have a more efficient sugar transfer activity towards their acceptor
substrates
than the corresponding native glycosyltransferases, i.e. show a glycosylation
activity at
least 30¨fold higher than the initial activity of the native full length
glycosyltransferases, said
glycosylation activity being measurable according to the general in vitro
procedure with
exogeneous/ commercially available acceptor glycoproteins having known bi-,
tri-or
tetraantennary glycans.
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
16
The expression "chimerical glycosyltransferases" used above corresponds to a
glycosyltransferase whose full length sequence is not the direct product of a
native gene or a
transcript but has been designed using sequences from other
glycosyltransferases exclusively.
The expression "native glycosyltransferases" used above corresponds to the
sequence of
the full length enzyme as represented by a naturally occurring coding
sequence.
The expression "glycosylation activity" used above corresponds to the
catalytic reaction
of transferring a sugar from a nucleotide donor to an acceptor substrate.
The expression "minimal catalytic domain (CD)" used above corresponds to the C-
terminal peptide domain of a glycosyltransferase sequence which cannot be
deleted further
without loss of transfer activity.
The expression "transmembrane domain (TMD)" used above corresponds to a
peptide
portion composed of a stretch of 17 -24 essentially hydrophobic amino acids
The expression "cytoplasmic tail (CT)" used above corresponds to the N-
terminal
peptide of a glycosyltransferase which may encompass at least more than 3
amino acids
upstream from the TMD.
The expression "stem region (SR)" used above corresponds to a stretch of at
most 246
aminoacids dowstream the TMD/upstream from the CD.
The invention relates more partcicularly to a process as defined above,
characterized in
that the first and second nucleic acids are derived from nucleotide sequences
encoding CD
domains, or CT, TMD, and SR regions, respectively, in glycosyltransferases
from eukaryotic
origin, preferably mammals and humans, said glycosyltransferases being
involved in:
= 0-glycosylation of the proteins in cells, such as N-acetylgalactosaminyl-
, N-
acetylglucosaminyl-,glucosyl-, fucosyl-, galactosyl-, or sialyltransferases,
= N-glycosylation of the proteins in cells such as N-acetylglucosaminyl-,
galactosyl-, fucosyl-, or sialyltransferases,
= Glycosylation of lipids such as the N-acetylgalactosaminyl-, N-
acetylglucosaminyl-, fucosyl-, galactosyl-, or sialyltransferases.
The expression "0-glycosylation" used above corresponds to the biosynthetic
pathway
elaborating monosaccharides or oligosaccharides covalently attached to amino
acids which
are not asparagine residues but can be preferably hydroxy amino acids such as
serine,
threonine or hydroxylysine residues.
The expression "N-glycosylation" used above corresponds to the biosynthetic
pathway
elaborating oligosaccharides attached to asparagine residues within the
consensus tripeptide
Asn-X-Ser/Thr (X being not Proline).
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
17
The expression "galactosyltransferases" used above corresponds to a
glycosyltransferase which transfers a galactose residue from UDP-Gal to an N-
/O-linked
protein or glyco lipid acceptor.
The expression "sialyltransferases" used above corresponds to a
glycosyltransferase
which transfers a sialic acid residue, preferably a derivative of neuraminic
acid from CMP-
NeuAc to a N-/O-linked protein or glyco lipid acceptor.
The invention concerns more particularly a process as defined above,
characterized in
that the first and second nucleic acids are derived from nucleotide sequences
encoding CD
domains, or CT, TMD, and SR regions, respectively in N-
acetylgalactosaminyltransferases,
N-acetylglucosaminyltransferases, glucosyltransferases,
galactosyltransferases,
fucosyltransferases, or sialyltransferases.
The invention concerns more particularly a process as defined above,
characterized in
that the first and second nucleic acids are derived from nucleotide sequences
encoding CD
domains, or CT, TMD, and SR regions, respectively in alpha 6
fucosyltransferases (core
glycosylation), beta 2/4/6 N-acetylglucosaminyltransferases (branching), beta
4
galactosyltransferases, and alpha 3/6/8 sialyltransferases (terminal
glycosylation).
Advantageously, the first and second nucleic acids are derived from nucleotide
sequences encoding CD domains, or CT, TMD, and SR regions, of enzymes
iplicated in the
N-glycosylation pathway.
The invention relates more particularly to a process as defined above,
characterized in
that the first and second nucleic acids are derived from nucleotide sequences
encoding CD
domains, or CT, TMD, and SR regions, respectively, in sialyltransferases.
The invention concerns more particularly a process as defined above,
characterized in
that the first and second nucleic acids are derived from nucleotide sequences
encoding CD
domains, or CT, TMD, and SR regions, respectively, in u2,6-sialyltransferases,
a2,3-
sialyltransferases, or a2,8-sialyltransferases.
Advantageously, nucleotide sequences encoding CT, TMD and/or SR or SR fragment
mentioned above are preferably from synthetic origin.
The expression "u2,6-sialyltransferases" used above corresponds to a
glycosyltransferase able to transfer a sialic acid residue, preferably a
derivative of neuraminic
acid from CMP-NeuAc to the 6-position of a carbohydrate acceptor from the N-/O-
linked
protein or glyco lipid type.
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
18
The
expression "cc2,3 -s ialyltrans feras es" used above corresponds to a
glycosyltransferase able to transfer a sialic acid residue, preferably a
derivative of neuraminic
acid from CMP-NeuAc to the 3-position of an carbohydrate acceptor from the N-
/O-linked
protein or glyco lipid type.
The expression "a2,8-sialyltransferases" used above corresponds to a
glycosyltransferase able to transfer a sialic acid residue, preferably a
derivative of neuraminic
acid from CMP-NeuAc to the 8-position of a sialylated acceptor from the N-/O-
linked protein
or glycolipid type.
The invention relates more particularly to a process as defined above,
characterized in
that the first and second nucleic acids are derived from nucleotide sequences
encoding CD
domains, or CT, TMD, and SR regions, respectively, in:
- o2,6-sialyltransferases chosen among:
* the human I31,4-galactoside o2,6-sialyltransferases I and II (hST6Gal I, and
hST6Gal II) represented by SEQ ID NO : 2, and SEQ ID NO : 4, respectively,
encoded by the nucleotide sequences SEQ ID NO : 1, and SEQ ID NO : 3,
respectively, or
* the human N-acetylgalactosaminide-a2,6-sialyltransferases I to VI
(hST6Ga1NAc I to VI) represented by SEQ ID NO : 6, SEQ ID NO : 8, SEQ ID NO:
10, SEQ ID NO: 12, SEQ ID NO: 14, and SEQ ID NO: 16, respectively, encoded by
the nucleotide sequences SEQ ID NO : 5, SEQ ID NO : 7, SEQ ID NO : 9, SEQ ID
NO: 11, SEQ ID NO : 13, and SEQ ID NO : 15, respectively,
- a2,3-sialyltransferases chosen among the human galactoside -a2,3-
sialyltransferases I
to VI (hST3Gal Ito VI) represented by SEQ ID NO : 18, SEQ ID NO : 20, SEQ ID
NO : 22,
SEQ ID NO : 24, SEQ ID NO : 26, and SEQ ID NO : 28, respectively, encoded by
the
nucleotide sequences SEQ ID NO: 17, SEQ ID NO: 19, SEQ ID NO : 21, SEQ ID NO :
23,
SEQ ID NO : 25, and SEQ ID NO : 27, respectively, or the rat galactoside -
cc2,3-
sialyltransferases I to VI (rST3Gal I to VI), such as the rST3Gal III
represented by SEQ ID
NO : 30 encoded by the nucleotide sequence SEQ ID NO : 29, or any ST from
other animal
origin provided that it shares at least 85% homology with the human enzyme,
- c2,8-sialyltransferases chosen among the human sialic acid-c2,8-
sialyltransferases I
to VI (hST8Sia Ito VI) represented by SEQ ID NO : 32, SEQ ID NO : 34, SEQ ID
NO : 36,
SEQ ID NO : 38, SEQ ID NO : 40, and SEQ ID NO : 42, respectively, encoded by
the
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
19
nucleotide sequences SEQ ID NO : 31, SEQ ID NO : 33, SEQ ID NO : 35, SEQ ID NO
: 37,
SEQ ID NO : 39, and SEQ ID NO : 41, respectively.
The expression "galactoside-u2,6-sialyltransferases" used above corresponds to
a
glycosyltransferase which transfers a sialic acid residue, preferably a
derivative of neuraminic
acid from CMP-NeuAc to the 6-position of a galactosylated acceptor from the N-
/O-linked
protein or lipid type.
The expression "N-acetylgalactosaminide u2,6-sialyltransferases" used above
corresponds to a glycosyltransferase which transfers a sialic acid residue,
preferably a
derivative of neuraminic acid from CMP-NeuAc to the 6-position of a N-
acetylgalactosaminyl residue of a N-/O-linked protein or glycolipid acceptor.
The expression "galactoside u2,3-sialyltransferases" used above corresponds to
a
glycosyltransferase which transfers a sialic acid residue, preferably a
derivative of neuraminic
acid from CMP-NeuAc to the 3-position of a galactosylated N-/O-linked protein
or lipid
acceptor.
The expression "sialic acid c2,8-sialyltransferases" used above corresponds to
a
glycosyltransferase which transfers a sialic acid residue, preferably a
derivative of neuraminic
acid from CMP-NeuAc to a sialylated N-/O-linked protein or glyco lipid
acceptor.
The invention concerns more particularly a process as defined above,
characterized in
that the nucleotide sequence encoding the CD domain comprised in the first
nucleic acid is
chosen among the sequences constituted of, or comprising:
* the nucleotide sequence delimited in its 5' end by the nucleotide located
in position
268 to 330 and in its 3' end by the nucleotide located in position 1218 of SEQ
ID NO: 1, said
nucleotide sequence encoding the polypeptide sequence corresponding to the CD
domain of
hST6Gal I delimited in its N-terminal end by the aminoacid located in position
90 to 110 and
in its C-terminal end by the aminoacid located in position 406 of SEQ ID NO :
2,
* the nucleotide sequence delimited in its 5' end by the nucleotide located
in position
307 to 369 and in its 3' end by the nucleotide located in position 1587 of SEQ
ID NO : 3, said
nucleotide sequence encoding the polypeptide sequence corresponding to the CD
domain of
hST6Gal II delimited in its N-terminal end by the aminoacid located in
position 103 to 123
and in its C-terminal end by the aminoacid located in position 529 of SEQ ID
NO : 4,
* the nucleotide sequence delimited in its 5' end by the nucleotide located
in position
814 to 876 and in its 3' end by the nucleotide located in position 1800 of SEQ
ID NO : 5, said
nucleotide sequence encoding the polypeptide sequence corresponding to the CD
domain of
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
hST6GalNac I delimited in its N-terminal end by the aminoacid located in
position 272 to 292
and in its C-terminal end by the aminoacid located in position 600 of SEQ ID
NO: 6,
* the nucleotide sequence delimited in its 5' end by the nucleotide located
in position
172 to 234 and in its 3' end by the nucleotide located in position 1122 of SEQ
ID NO : 7, said
5 nucleotide sequence encoding the polypeptide sequence corresponding to
the CD domain of
hST6GalNac II delimited in its N-terminal end by the aminoacid located in
position 58 to 78
and in its C-terminal end by the aminoacid located in position 374 of SEQ ID
NO: 8,
* the nucleotide sequence delimited in its 5' end by the nucleotide located
in position
73 to 135 and in its 3' end by the nucleotide located in position 915 of SEQ
ID NO : 9, said
10 nucleotide sequence encoding the polypeptide sequence corresponding to
the CD domain of
hST6GalNac III delimited in its N-terminal end by the aminoacid located in
position 25 to 45
and in its C-terminal end by the aminoacid located in position 305 of SEQ ID
NO : 10,
* the nucleotide sequence delimited in its 5' end by the nucleotide located
in position
61 to 123 and in its 3' end by the nucleotide located in position 906 of SEQ
ID NO : 11, said
15 nucleotide sequence encoding the polypeptide sequence corresponding to
the CD domain of
hST6GalNac IV delimited in its N-terminal end by the aminoacid located in
position 21 to 41
and in its C-terminal end by the aminoacid located in position 302 of SEQ ID
NO : 12,
* the nucleotide sequence delimited in its 5' end by the nucleotide located
in position
121 to 183 and in its 3' end by the nucleotide located in position 1008 of SEQ
ID NO : 13,
20 said nucleotide sequence encoding the polypeptide sequence corresponding
to the CD domain
of hST6GalNac V delimited in its N-terminal end by the amino acid located in
position 41 to
61 and in its C-terminal end by the aminoacid located in position 336 of SEQ
ID NO: 14,
* the nucleotide sequence delimited in its 5' end by the nucleotide located
in position
154 to 216 and in its 3' end by the nucleotide located in position 999 of SEQ
ID NO: 15, said
nucleotide sequence encoding the polypeptide sequence corresponding to the CD
domain of
hST6GalNac VI delimited in its N-terminal end by the aminoacid located in
position 52 to 72
and in its C-terminal end by the aminoacid located in position 333 of SEQ ID
NO : 16,
* the nucleotide sequence delimited in its 5' end by the nucleotide located
in position
145 to 207 and in its 3' end by the nucleotide located in position 1020 of SEQ
ID NO : 17,
said nucleotide sequence encoding the polypeptide sequence corresponding to
the CD domain
of hST3Gal I delimited in its N-terminal end by the aminoacid located in
position 49 to 69
and in its C-terminal end by the aminoacid located in position 340 of SEQ ID
NO : 18,
* the nucleotide sequence delimited in its 5' end by the nucleotide located
in position
175 to 237 and in its 3' end by the nucleotide located in position 1050 of SEQ
ID NO : 19,
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
21
said nucleotide sequence encoding the polypeptide sequence corresponding to
the CD domain
of hST3Ga1 II delimited in its N-terminal end by the aminoacid located in
position 59 to 79
and in its C-terminal end by the aminoacid located in position 350 of SEQ ID
NO : 20,
* the nucleotide sequence delimited in its 5' end by the nucleotide located
in position
199 to 261 and in its 3' end by the nucleotide located in position 1332 of SEQ
ID NO : 21,
said nucleotide sequence encoding the polypeptide sequence corresponding to
the CD domain
of hST3Gal III delimited in its N-terminal end by the aminoacid located in
position 67 to 87
and in its C-terminal end by the aminoacid located in position 444 of SEQ ID
NO : 22,
* the nucleotide sequence delimited in its 5' end by the nucleotide located
in position
79 to 141 and in its 3' end by the nucleotide located in position 987 of SEQ
ID NO : 23, said
nucleotide sequence encoding the polypeptide sequence corresponding to the CD
domain of
hST3Gal IV delimited in its N-terminal end by the aminoacid located in
position 27 to 47 and
in its C-terminal end by the aminoacid located in position 329 of SEQ ID NO :
24,
* the nucleotide sequence delimited in its 5' end by the nucleotide located
in position
136 to 198 and in its 3' end by the nucleotide located in position 1086 of SEQ
ID NO : 25,
said nucleotide sequence encoding the polypeptide sequence corresponding to
the CD domain
of hST3Gal V delimited in its N-terminal end by the aminoacid located in
position 46 to 66
and in its C-terminal end by the aminoacid located in position 362 of SEQ ID
NO : 26,
* the nucleotide sequence delimited in its 5' end by the nucleotide located
in position
73 to 135 and in its 3' end by the nucleotide located in position 993 of SEQ
ID NO : 27, said
nucleotide sequence encoding the polypeptide sequence corresponding to the CD
domain of
hST3Gal VI delimited in its N-terminal end by the aminoacid located in
position 25 to 45 and
in its C-terminal end by the aminoacid located in position 331 of SEQ ID NO :
28,
* the nucleotide sequence delimited in its 5' end by the nucleotide located
in position
103 to 165 and in its 3' end by the nucleotide located in position 1122 of SEQ
ID NO : 29,
said nucleotide sequence encoding the polypeptide sequence corresponding to
the CD domain
of rat ST3Ga1 III delimited in its N-terminal end by the aminoacid located in
position 35 to 55
and in its C-terminal end by the aminoacid located in position 374 of SEQ ID
NO : 30,
* the nucleotide sequence delimited in its 5' end by the nucleotide located
in position
133 to 195 and in its 3' end by the nucleotide located in position 1068 of SEQ
ID NO : 31,
said nucleotide sequence encoding the polypeptide sequence corresponding to
the CD domain
of hST8Sia I delimited in its N-terminal end by the aminoacid located in
position 45 to 65 and
in its C-terminal end by the aminoacid located in position 356 of SEQ ID NO :
32,
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
22
* the nucleotide sequence delimited in its 5' end by the nucleotide located
in position
190 to 252 and in its 3' end by the nucleotide located in position 1125 of SEQ
ID NO : 33,
said nucleotide sequence encoding the polypeptide sequence corresponding to
the CD domain
of hST8Sia II delimited in its N-terminal end by the aminoacid located in
position 64 to 84
and in its C-terminal end by the aminoacid located in position 375 of SEQ ID
NO : 34,
* the nucleotide sequence delimited in its 5' end by the nucleotide located
in position
205 to 267 and in its 3' end by the nucleotide located in position 1140 of SEQ
ID NO : 35,
said nucleotide sequence encoding the polypeptide sequence corresponding to
the CD domain
of hST8Sia III delimited in its N-terminal end by the aminoacid located in
position 69 to 89
and in its C-terminal end by the aminoacid located in position 380 of SEQ ID
NO : 36,
* the nucleotide sequence delimited in its 5' end by the nucleotide located
in position
145 to 207 and in its 3' end by the nucleotide located in position 1077 of SEQ
ID NO : 37,
said nucleotide sequence encoding the polypeptide sequence corresponding to
the CD domain
of hST8Sia IV delimited in its N-terminal end by the aminoacid located in
position 49 to 69
and in its C-terminal end by the aminoacid located in position 359 of SEQ ID
NO : 38,
* the nucleotide sequence delimited in its 5' end by the nucleotide located
in position
211 to 273 and in its 3' end by the nucleotide located in position 1128 of SEQ
ID NO : 39,
said nucleotide sequence encoding the polypeptide sequence corresponding to
the CD domain
of hST8Sia V delimited in its N-terminal end by the aminoacid located in
position 71 to 91
and in its C-terminal end by the aminoacid located in position 376 of SEQ ID
NO : 40,
* the nucleotide sequence delimited in its 5' end by the nucleotide located
in position
277 to 339 and in its 3' end by the nucleotide located in position 1194 of SEQ
ID NO : 41,
said nucleotide sequence encoding the polypeptide sequence corresponding to
the CD domain
of hST8Sia VI delimited in its N-terminal end by the aminoacid located in
position 93 to 113
and in its C-terminal end by the aminoacid located in position 398 of SEQ ID
NO : 42.
The invention relates more particularly to a process as defined above,
characterized in
that the first nucleic acid is the nucleotide sequence SEQ ID NO : 43
corresponding to the
sequence delimited by the nucleotides located in position 268 and 1218 of SEQ
ID NO : 1,
said nucleotide sequence SEQ ID NO : 43 encoding the polypeptide sequence SEQ
ID NO :
44 corresponding to the C-terminal minimal fragment of the CD domain of
hST6Gal I
delimited by the amino acids located in positions 90 to 406 of SEQ ID NO : 2.
The invention concerns more particularly a process as defined above,
characterized in
that:
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
23
- the nucleotide sequence encoding the CT region comprised in the second
nucleic acid
is chosen among:
* the nucleotide sequence SEQ ID NO : 45 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 1 and 27 of SEQ ID
NO : 1, said
nucleotide sequence SEQ ID NO : 45 encoding the polypeptide sequence SEQ ID NO
: 46
corresponding to the CT region of hST6Gal I delimited by the aminoacids
located in positions
1 to 9 of SEQ ID NO :2,
* the nucleotide sequence SEQ ID NO : 47 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 1 and 30 of SEQ ID
NO : 3, said
nucleotide sequence SEQ ID NO : 47 encoding the polypeptide sequence SEQ ID NO
: 48
corresponding to the CT region of hST6Gal II delimited by the aminoacids
located in
positions 1 to 10 of SEQ ID NO : 4,
* the nucleotide sequence SEQ ID NO : 49 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 1 and 42 of SEQ ID
NO : 5, said
nucleotide sequence SEQ ID NO : 49 encoding the polypeptide sequence SEQ ID NO
: 50
corresponding to the CT region of hST6Ga1NAc I delimited by the aminoacids
located in
positions 1 to 14 of SEQ ID NO: 6,
* the nucleotide sequence SEQ ID NO : Si corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 1 and 21 of SEQ ID
NO : 7, said
nucleotide sequence SEQ ID NO : Si encoding the polypeptide sequence SEQ ID NO
: 52
corresponding to the CT region of hST6GalNac II delimited by the aminoacids
located in
positions 1 to 7 of SEQ ID NO: 8,
* the nucleotide sequence SEQ ID NO : 53 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 1 and 24 of SEQ ID
NO : 9, said
nucleotide sequence SEQ ID NO : 53 encoding the polypeptide sequence SEQ ID NO
: 54
corresponding to the CT region of hST6Ga1NAc III delimited by the aminoacids
located in
positions 1 to 8 of SEQ ID NO : 10,
* the nucleotide sequence SEQ ID NO : 55 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 1 and 18 of SEQ ID
NO : 11, said
nucleotide sequence SEQ ID NO : 55 encoding the polypeptide sequence SEQ ID NO
: 56
corresponding to the CT region of hST6Ga1NAc IV delimited by the aminoacids
located in
positions 1 to 6 of SEQ ID NO : 12,
* the nucleotide sequence SEQ ID NO : 57 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 1 and 24 of SEQ ID
NO : 13, said
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
24
nucleotide sequence SEQ ID NO : 57 encoding the polypeptide sequence SEQ ID NO
: 58
corresponding to the CT region of hST6Ga1NAc V delimited by the aminoacids
located in
positions 1 to 8 of SEQ ID NO : 14,
* the nucleotide sequence SEQ ID NO : 59 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 1 and 129 of SEQ ID
NO: 15, said
nucleotide sequence SEQ ID NO : 59 encoding the polypeptide sequence SEQ ID NO
: 60
corresponding to the CT region of hST6Ga1NAc VI delimited by the aminoacids
located in
positions 1 to 43 of SEQ ID NO: 16,
* the nucleotide sequence SEQ ID NO : 61 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 1 and 39 of SEQ ID
NO : 17, said
nucleotide sequence SEQ ID NO : 61 encoding the polypeptide sequence SEQ ID NO
: 62
corresponding to the CT region of hST3Gal I delimited by the amino acids
located in
positions 1 to 13 of SEQ ID NO: 18,
* the nucleotide sequence SEQ ID NO : 63 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 1 and 18 of SEQ ID
NO : 19, said
nucleotide sequence SEQ ID NO : 63 encoding the polypeptide sequence SEQ ID NO
: 64
corresponding to the CT region of hST3Gal II delimited by the amino acids
located in
positions 1 to 6 of SEQ ID NO : 20,
* the nucleotide sequence SEQ ID NO : 65 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 1 and 24 of SEQ ID
NO : 21, said
nucleotide sequence SEQ ID NO : 65 encoding the polypeptide sequence SEQ ID NO
: 66
corresponding to the CT region of hST3Gal III delimited by the amino acids
located in
positions 1 to 8 of SEQ ID NO : 22,
* the nucleotide sequence SEQ ID NO : 67 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 1 and 24 of SEQ ID
NO : 23, said
nucleotide sequence SEQ ID NO : 67 encoding the polypeptide sequence SEQ ID NO
: 68
corresponding to the CT region of hST3Gal IV delimited by the amino acids
located in
positions 1 to 8 of SEQ ID NO : 24,
* the nucleotide sequence SEQ ID NO : 69 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 1 and 15 of SEQ ID
NO : 25, said
nucleotide sequence SEQ ID NO : 69 encoding the polypeptide sequence SEQ ID NO
: 70
corresponding to the CT region of hST3Gal V delimited by the amino acids
located in
positions 1 to 5 of SEQ ID NO : 26,
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
* the nucleotide sequence SEQ ID NO : 71 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 1 and 12 of SEQ ID
NO : 27, said
nucleotide sequence SEQ ID NO : 71 encoding the polypeptide sequence SEQ ID NO
: 72
corresponding to the CT region of hST3Gal VI delimited by the amino acids
located in
5 positions 1 to 4 of SEQ ID NO : 28,
* the nucleotide sequence SEQ ID NO : 73 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 1 and 24 of SEQ ID
NO : 29, said
nucleotide sequence SEQ ID NO : 73 encoding the polypeptide sequence SEQ ID NO
: 74
corresponding to the CT region of ratST3Gal III delimited by the amino acids
located in
10 positions 1 to 8 of SEQ ID NO : 30,
* the nucleotide sequence SEQ ID NO : 75 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 1 and 87 of SEQ ID
NO : 31, said
nucleotide sequence SEQ ID NO : 75 encoding the polypeptide sequence SEQ ID NO
: 76
corresponding to the CT region of hST8Sia I delimited by the amino acids
located in positions
15 1 to 29 of SEQ ID NO: 32,
* the nucleotide sequence SEQ ID NO : 77 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 1 and 18 of SEQ ID
NO : 33, said
nucleotide sequence SEQ ID NO : 77 encoding the polypeptide sequence SEQ ID NO
: 78
corresponding to the CT region of hST8Sia II delimited by the amino acids
located in
20 positions 1 to 6 of SEQ ID NO : 34,
* the nucleotide sequence SEQ ID NO : 79 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 1 and 27 of SEQ ID
NO : 35, said
nucleotide sequence SEQ ID NO : 79 encoding the polypeptide sequence SEQ ID NO
: 80
corresponding to the CT region of hST8Sia III delimited by the amino acids
located in
25 positions 1 to 9 of SEQ ID NO : 36,
* the nucleotide sequence SEQ ID NO : 81 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 1 and 21 of SEQ ID
NO : 37, said
nucleotide sequence SEQ ID NO : 81 encoding the polypeptide sequence SEQ ID NO
: 82
corresponding to the CT region of hST8Sia IV delimited by the amino acids
located in
positions 1 to 7 of SEQ ID NO : 38,
* the nucleotide sequence SEQ ID NO : 83 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 1 and Si of SEQ ID
NO : 39, said
nucleotide sequence SEQ ID NO : 83 encoding the polypeptide sequence SEQ ID NO
: 84
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
26
corresponding to the CT region of hST8Sia V delimited by the amino acids
located in
positions 1 to 17 of SEQ ID NO : 40,
* the nucleotide sequence SEQ ID NO : 85 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 1 and 9 of SEQ ID
NO : 41, said
nucleotide sequence SEQ ID NO : 85 encoding the polypeptide sequence SEQ ID NO
: 86
corresponding to the CT region of hST8Sia VI delimited by the amino acids
located in
positions 1 to 3 of SEQ ID NO : 42,
- the nucleotide sequence encoding the TMD region comprised in the second
nucleic
acid is chosen among:
* the nucleotide sequence SEQ ID NO : 87 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 28 and 78 of SEQ ID
NO : 1, said
nucleotide sequence SEQ ID NO : 87 encoding the polypeptide sequence SEQ ID NO
: 88
corresponding to the TMD region of hST6Gal I delimited by the amino acids
located in
positions 10 to 26 of SEQ ID NO : 2,
* the nucleotide sequence SEQ ID NO : 89 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 31 and 90 of SEQ ID
NO : 3, said
nucleotide sequence SEQ ID NO : 89 encoding the polypeptide sequence SEQ ID NO
: 90
corresponding to the TMD region of hST6Gal II delimited by the amino acids
located in
positions 11 to 30 of SEQ ID NO : 4,
* the nucleotide sequence SEQ ID NO : 91 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 43 and 105 of SEQ
ID NO : 5, said
nucleotide sequence SEQ ID NO : 91 encoding the polypeptide sequence SEQ ID NO
: 92
corresponding to the TMD region of hST6Ga1NAc I delimited by the amino acids
located in
positions 15 to 35 of SEQ ID NO : 6,
* the nucleotide sequence SEQ ID NO : 93 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 22 and 84 of SEQ ID
NO : 7, said
nucleotide sequence SEQ ID NO : 93 encoding the polypeptide sequence SEQ ID NO
: 94
corresponding to the TMD region of hST6Ga1NAc II delimited by the amino acids
located in
positions 8 to 28 of SEQ ID NO: 8,
* the nucleotide sequence SEQ ID NO : 95 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 25 and 84 of SEQ ID
NO : 9, said
nucleotide sequence SEQ ID NO : 95 encoding the polypeptide sequence SEQ ID NO
: 96
corresponding to the TMD region of hST6Ga1NAc III delimited by the amino acids
located in
positions 9 to 28 of SEQ ID NO : 10,
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
27
* the nucleotide sequence SEQ ID NO : 97 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 19 and 81 of SEQ ID
NO : 11, said
nucleotide sequence SEQ ID NO : 97 encoding the polypeptide sequence SEQ ID NO
: 98
corresponding to the TMD region of hST6Ga1NAc IV delimited by the amino acids
located in
positions 7 to 27 of SEQ ID NO: 12,
* the nucleotide sequence SEQ ID NO : 99 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 25 and 87 of SEQ ID
NO : 13, said
nucleotide sequence SEQ ID NO : 99 encoding the polypeptide sequence SEQ ID NO
: 100
corresponding to the TMD region of hST6Ga1NAc V delimited by the amino acids
located in
positions 9 to 29 of SEQ ID NO: 14,
* the nucleotide sequence SEQ ID NO : 101 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 130 and 177 of SEQ
ID NO : 15,
said nucleotide sequence SEQ ID NO : 101 encoding the polypeptide sequence SEQ
ID NO :
102 corresponding to the TMD region of hST6Ga1NAc VI delimited by the
aminoacids
located in positions 44 to 59 of SEQ ID NO: 16,
* the nucleotide sequence SEQ ID NO : 103 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 40 and 102 of SEQ
ID NO : 17,
said nucleotide sequence SEQ ID NO : 103 encoding the polypeptide sequence SEQ
ID NO :
104 corresponding to the TMD region of hST3Gal I delimited by the aminoacids
located in
positions 14 to 34 of SEQ ID NO: 18,
* the nucleotide sequence SEQ ID NO : 105 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 19 and 81 of SEQ ID
NO : 19, said
nucleotide sequence SEQ ID NO: 105 encoding the polypeptide sequence SEQ ID
NO: 106
corresponding to the TMD region of hST3Gal II delimited by the aminoacids
located in
positions 7 to 27 of SEQ ID NO : 20,
* the nucleotide sequence SEQ ID NO : 107 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 25 and 84 of SEQ ID
NO : 21, said
nucleotide sequence SEQ ID NO: 107 encoding the polypeptide sequence SEQ ID
NO: 108
corresponding to the TMD region of hST3Gal III delimited by the amino acids
located in
positions 9 to 28 of SEQ ID NO : 22,
* the nucleotide sequence SEQ ID NO : 109 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 25 and 78 of SEQ ID
NO : 23, said
nucleotide sequence SEQ ID NO: 109 encoding the polypeptide sequence SEQ ID
NO: 110
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
28
corresponding to the TMD region of hST3Ga1 IV delimited by the aminoacids
located in
positions 9 to 26 of SEQ ID NO : 24,
* the nucleotide sequence SEQ ID NO : 111 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 16 and 78 of SEQ ID
NO : 25, said
nucleotide sequence SEQ ID NO: 111 encoding the polypeptide sequence SEQ ID
NO: 112
corresponding to the TMD region of hST3Gal V delimited by the aminoacids
located in
positions 6 to 26 of SEQ ID NO : 26,
* the nucleotide sequence SEQ ID NO : 113 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 13 and 75 of SEQ ID
NO : 27, said
nucleotide sequence SEQ ID NO: 113 encoding the polypeptide sequence SEQ ID
NO: 114
corresponding to the TMD region of hST3Gal VI delimited by the aminoacids
located in
positions 5 to 25 of SEQ ID NO : 28,
* the nucleotide sequence SEQ ID NO : 115 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 25 and 84 of SEQ ID
NO : 29, said
nucleotide sequence SEQ ID NO : 115 encoding the polypeptide sequence SEQ ID
NO : 116
corresponding to the TMD region of rat 5T3 Gal III delimited by the amino
acids located in
positions 9 to 28 of SEQ ID NO: 30,
* the nucleotide sequence SEQ ID NO : 117 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 88 and 144 of SEQ
ID NO : 31,
said nucleotide sequence SEQ ID NO: 117 encoding the polypeptide sequence SEQ
ID NO:
118 corresponding to the TMD region of hST8Sia I delimited by the amino acids
located in
positions 30 to 48 of SEQ ID NO : 32,
* the nucleotide sequence SEQ ID NO : 119 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 19 and 69 of SEQ ID
NO : 33, said
nucleotide sequence SEQ ID NO: 119 encoding the polypeptide sequence SEQ ID
NO: 120
corresponding to the TMD region of hST8Sia II delimited by the amino acids
located in
positions 7 to 23 of SEQ ID NO : 34,
* the nucleotide sequence SEQ ID NO : 121 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 28 and 99 of SEQ ID
NO : 35, said
nucleotide sequence SEQ ID NO: 121 encoding the polypeptide sequence SEQ ID
NO: 122
corresponding to the TMD region of hST8Sia III delimited by the amino acids
located in
positions 10 to 33 of SEQ ID NO : 36,
* the nucleotide sequence SEQ ID NO : 123 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 22 and 60 of SEQ ID
NO : 37, said
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
29
nucleotide sequence SEQ ID NO: 123 encoding the polypeptide sequence SEQ ID
NO: 124
corresponding to the TMD region of hST8Sia IV delimited by the amino acids
located in
positions 8 to 20 of SEQ ID NO : 38,
* the nucleotide sequence SEQ ID NO : 125 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 52 and 114 of SEQ
ID NO : 39,
said nucleotide sequence SEQ ID NO : 125 encoding the polypeptide sequence SEQ
ID NO :
126 corresponding to the TMD region of hST8Sia V delimited by the amino acids
located in
positions 18 to 38 of SEQ ID NO : 40,
* the nucleotide sequence SEQ ID NO : 127 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 10 and 72 of SEQ ID
NO : 41, said
nucleotide sequence SEQ ID NO: 127 encoding the polypeptide sequence SEQ ID
NO: 128
corresponding to the TMD region of hST8Sia VI delimited by the amino acids
located in
positions 4 to 24 of SEQ ID NO : 42,
- the nucleotide sequence encoding the SR region comprised in the second
nucleic acid,
or encoding a fragment of at least 2 amino acids thereof, is chosen among
sequences
constituted of, or comprising:
* the nucleotide sequence delimited in its 5' end by the nucleotide located
in
position 79 and in its 3' end by the nucleotide located in position 267 to 327
of SEQ ID NO :
1, said nucleotide sequence encoding the polypeptide sequence corresponding to
the SR
region of hST6Gal I delimited in its N-terminal end by the amino acid located
in position 27
and in its C-terminal end by the amino acid located in position 89 to 109 of
SEQ ID NO : 2,
* the nucleotide sequence delimited in its 5' end by the nucleotide located
in
position 91 and in its 3' end by the nucleotide located in position 306 to 336
of SEQ ID NO :
3, said nucleotide sequence encoding the polypeptide sequence corresponding to
the SR
region of hST6Gal II delimited in its N-terminal end by the amino acid located
in position 31
and in its C-terminal end by the amino acid located in position 102 to 112 of
SEQ ID NO : 4,
* the nucleotide sequence delimited in its 5' end by the nucleotide located
in
position 106 and in its 3' end by the nucleotide located in position 813 to
873 of SEQ ID NO :
5, said nucleotide sequence encoding the polypeptide sequence corresponding to
the SR
region of hST6Ga1NAc I delimited in its N-terminal end by the aminoacid
located in position
36 and in its C-terminal end by the amino acid located in position 271 to 291
of SEQ ID NO:
6,
* the nucleotide sequence delimited in its 5' end by the nucleotide located
in
position 85 and in its 3' end by the nucleotide located in position 171 to 231
of SEQ ID NO :
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
7, said nucleotide sequence encoding the polypeptide sequence corresponding to
the SR
region of hST6Ga1NAc II delimited in its N-terminal end by the amino acid
located in
position 29 and in its C-terminal end by the amino acid located in position 57
to 77 of SEQ ID
NO : 8,
5 * the nucleotide sequence delimited in its 5' end by the
nucleotide located in
position 85 and in its 3' end by the nucleotide located in position 102 to 132
of SEQ ID NO :
9, said nucleotide sequence encoding the polypeptide sequence corresponding to
the SR
region of hST6Ga1NAc III delimited in its N-terminal end by the amino acid
located in
position 29 and in its C-terminal end by the amino acid located in position 34
to 44 of SEQ ID
10 NO : 10,
* the nucleotide sequence delimited in its 5' end by the nucleotide located
in
position 82 and in its 3' end by the nucleotide located in position 90 to 120
of SEQ ID NO :
11, said nucleotide sequence encoding the polypeptide sequence corresponding
to the SR
region of hST6GaNAc IV delimited in its N-terminal end by the amino acid
located in
15 position 28 and in its C-terminal end by the amino acid located in
position 30 to 40 of SEQ ID
NO: 12,
* the nucleotide sequence delimited in its 5' end by the nucleotide located
in
position 88 and in its 3' end by the nucleotide located in position 120 to 180
of SEQ ID NO :
13, said nucleotide sequence encoding the polypeptide sequence corresponding
to the SR
20 region of hST6Ga1NAc V delimited in its N-terminal end by the amino acid
located in
position 30 and in its C-terminal end by the amino acid located in position 40
to 60 of SEQ ID
NO: 14,
* the nucleotide sequence delimited in its 5' end by the nucleotide located
in
position 178 and in its 3' end by the nucleotide located in position 183 of
SEQ ID NO : 15,
25 said nucleotide sequence encoding the polypeptide sequence corresponding
to the SR region
of hST6Ga1NAc VI delimited in its N-terminal end by the amino acid located in
position 60
and in its C-terminal end by the amino acid located in position 61 of SEQ ID
NO : 16,
* the nucleotide sequence delimited in its 5' end by the nucleotide located
in
position 103 and in its 3' end by the nucleotide located in position 144 to
204 of SEQ ID NO :
30 17, said nucleotide sequence encoding the polypeptide sequence
corresponding to the SR
region of hST3Gal I delimited in its N-terminal end by the amino acid located
in position 35
and in its C-terminal end by the aminoacid located in position 48 to 68 of SEQ
ID NO : 18,
* the nucleotide sequence delimited in its 5' end by the nucleotide located
in
position 82 and in its 3' end by the nucleotide located in position 174 to 234
of SEQ ID NO :
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
31
19, said nucleotide sequence encoding the polypeptide sequence corresponding
to the SR
region of hST3Gal II delimited in its N-terminal end by the aminoacid located
in position 28
and in its C-terminal end by the aminoacid located in position 58 to 78 of SEQ
ID NO : 20,
* the nucleotide sequence delimited in its 5' end by the nucleotide located
in
position 85 and in its 3' end by the nucleotide located in position 198 to 258
of SEQ ID NO:
21, said nucleotide sequence encoding the polypeptide sequence corresponding
to the SR
region of hST3Gal III delimited in its N-terminal end by the amino acid
located in position 29
and in its C-terminal end by the amino acid located in position 66 to 86 of
SEQ ID NO : 22,
* the nucleotide sequence delimited in its 5' end by the nucleotide located
in
position 79 and in its 3' end by the nucleotide located in position 108 to 138
of SEQ ID NO:
23, said nucleotide sequence encoding the polypeptide sequence corresponding
to the SR
region of hST3Gal IV delimited in its N-terminal end by the amino acid located
in position 27
and in its C-terminal end by the amino acid located in position 36 to 46 of
SEQ ID NO : 24,
* the nucleotide sequence delimited in its 5' end by the nucleotide located
in
position 79 and in its 3' end by the nucleotide located in position 135 to 195
of SEQ ID NO:
25, said nucleotide sequence encoding the polypeptide sequence corresponding
to the SR
region of hST3Gal V delimited in its N-terminal end by the amino acid located
in position 27
and in its C-terminal end by the amino acid located in position 45 to 65 of
SEQ ID NO : 26,
* the nucleotide sequence delimited in its 5' end by the nucleotide located
in
position 76 and in its 3' end by the nucleotide located in position 102 to 132
of SEQ ID NO:
27, said nucleotide sequence encoding the polypeptide sequence corresponding
to the SR
region of hST3Gal VI delimited in its N-terminal end by the amino acid located
in position 26
and in its C-terminal end by the amino acid located in position 34 to 44 of
SEQ ID NO : 28,
* the nucleotide sequence delimited in its 5' end by the nucleotide located
in
position 85 and in its 3' end by the nucleotide located in position 105 to 165
of SEQ ID NO:
29, said nucleotide sequence encoding the polypeptide sequence corresponding
to the SR
region of rST3Gal III delimited in its N-terminal end by the amino acid
located in position 29
and in its C-terminal end by the amino acid located in position 35 to 55 of
SEQ ID NO: 30,
* the nucleotide sequence delimited in its 5' end by the nucleotide located
in
position 145 and in its 3' end by the nucleotide located in position 162 to
192 of SEQ ID NO:
31, said nucleotide sequence encoding the polypeptide sequence corresponding
to the SR
region of hST8Sia I delimited in its N-terminal end by the aminoacid located
in position 49
and in its C-terminal end by the aminoacid located in position 54 to 64 of SEQ
ID NO : 32,
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
32
* the nucleotide sequence delimited in its 5' end by the nucleotide located
in
position 70 and in its 3' end by the nucleotide located in position 189 to 249
of SEQ ID NO :
33, said nucleotide sequence encoding the polypeptide sequence corresponding
to the SR
region of hST8Sia II delimited in its N-terminal end by the aminoacid located
in position 24
and in its C-terminal end by the amino acid located in position 63 to 83 of
SEQ ID NO: 34,
* the nucleotide sequence delimited in its 5' end by the nucleotide located
in
position 100 and in its 3' end by the nucleotide located in position 204 to
264 of SEQ ID NO :
35, said nucleotide sequence encoding the polypeptide sequence corresponding
to the SR
region of hST8Sia III delimited in its N-terminal end by the amino acid
located in position 34
and in its C-terminal end by the amino acid located in position 68 to 88 of
SEQ ID NO: 36,
* the nucleotide sequence delimited in its 5' end by the nucleotide located
in
position 61 and in its 3' end by the nucleotide located in position 144 to 204
of SEQ ID NO :
37, said nucleotide sequence encoding the polypeptide sequence corresponding
to the SR
region of hST8Sia IV delimited in its N-terminal end by the amino acid located
in position 21
and in its C-terminal end by the amino acid located in position 48 to 68 of
SEQ ID NO : 38,
* the nucleotide sequence delimited in its 5' end by the nucleotide located
in
position 115 and in its 3' end by the nucleotide located in position 210 to
270 of SEQ ID NO :
39, said nucleotide sequence encoding the polypeptide sequence corresponding
to the SR
region of hST8Sia V delimited in its N-terminal end by the amino acid located
in position 39
and in its C-terminal end by the amino acid located in position 70 to 90 of
SEQ ID NO : 40,
* the nucleotide sequence delimited in its 5' end by the nucleotide located
in
position 73 and in its 3' end by the nucleotide located in position 276 to 336
of SEQ ID NO :
41, said nucleotide sequence encoding the polypeptide sequence corresponding
to the SR
region of hST8Sia VI delimited in its N-terminal end by the amino acid located
in position 25
and in its C-terminal end by the amino acid located in position 92 to 112 of
SEQ ID NO : 42,
* or any fragment of at least 6 nucleotides of the nucleotide sequences
encoding polypeptides sequence corresponding to SR regions mentioned above,
and encoding
at least 2 contiguous amino acids of said SR regions, such as:
** the fragment SEQ ID NO : 129 delimited by the nucleotides located
in positions 106 to 222 of SEQ ID NO : 5, encoding the polypeptide sequence
SEQ ID NO:
130 corresponding to the fragment of the SR region of hST6Ga1NAc I delimited
by the amino
acids located in positions 36 to 74 of SEQ ID NO : 6,
** the fragment SEQ ID NO : 131 delimited by the nucleotides located
in positions 109 to 222 of SEQ ID NO : 5, encoding the polypeptide sequence
SEQ ID NO :
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
33
132 corresponding to the fragment of the SR region of hST6Ga1NAc I delimited
by the amino
acids located in positions 37 to 74 of SEQ ID NO : 6,
** the fragment SEQ ID NO : 133 delimited by the nucleotides located
in positions 106 to 420 of SEQ ID NO : 5, encoding the polypeptide sequence
SEQ ID NO :
134 corresponding to the fragment of the SR region of hST6Ga1NAc I delimited
by the amino
acids located in positions 36 to 140 of SEQ ID NO : 6,
** the fragment SEQ ID NO : 135 delimited by the nucleotides located
in positions 106 to 774 of SEQ ID NO : 5, encoding the polypeptide sequence
SEQ ID NO :
136 corresponding to the fragment of the SR region of hST6Ga1NAc I delimited
by the amino
acids located in positions 36 to 258 of SEQ ID NO : 6,
** the fragment SEQ ID NO : 137 delimited by the nucleotides located
in positions 85 to 138 of SEQ ID NO : 21, encoding the polypeptide sequence
SEQ ID NO :
138 corresponding to the fragment of the SR region of hST3Gal III delimited by
the amino
acids located in positions 29 to 46 of SEQ ID NO : 22,
** the fragment SEQ ID NO : 139 delimited by the nucleotides located
in positions 85 to 138 of SEQ ID NO : 29, encoding the polypeptide sequence
SEQ ID NO :
140 corresponding to the fragment of the SR region of ratST3Gal III delimited
by the amino
acids located in positions 29 to 46 of SEQ ID NO : 30,
* the fragment SEQ ID NO : 141 delimited by the nucleotides located
in positions 70 to 237 of SEQ ID NO : 33, encoding the polypeptide sequence
SEQ ID NO :
142 corresponding to the fragment of the SR region of hST8Sia II delimited by
the amino
acids located in positions 24 to 79 of SEQ ID NO : 34,
* the fragment SEQ ID NO : 143 delimited by the nucleotides located
in positions 61 to 201 of SEQ ID NO : 37, encoding the polypeptide sequence
SEQ ID NO :
144 corresponding to the fragment of the SR region of hST8Sia IV delimited by
the amino
acids located in positions 21 to 67 of SEQ ID NO : 38.
The invention relates more particularly to a process as defined above,
characterized in
that:
- the nucleotide sequence encoding the CT region comprised in the second
nucleic acid
is chosen among:
* the nucleotide sequence SEQ ID NO : 49 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 1 and 42 of SEQ ID
NO : 5, said
nucleotide sequence SEQ ID NO : 49 encoding the polypeptide sequence SEQ ID NO
: 50
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
34
corresponding to the CT region of hST6Ga1NAc I delimited by the amino acids
located in
positions 1 to 14 of SEQ ID NO: 6,
* the nucleotide sequence SEQ ID NO : 65 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 1 and 24 of SEQ ID
NO : 21, said
nucleotide sequence SEQ ID NO : 65 encoding the polypeptide sequence SEQ ID NO
: 66
corresponding to the CT region of hST3Gal III delimited by the amino acids
located in
positions 1 to 8 of SEQ ID NO : 22,
* the nucleotide sequence SEQ ID NO : 73 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 1 and 24 of SEQ ID
NO : 29, said
nucleotide sequence SEQ ID NO : 73 encoding the polypeptide sequence SEQ ID NO
: 74
corresponding to the CT region of ratST3Gal III delimited by the amino acids
located in
positions 1 to 8 of SEQ ID NO : 30,
* the nucleotide sequence SEQ ID NO : 77 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 1 and 18 of SEQ ID
NO : 33, said
nucleotide sequence SEQ ID NO : 77 encoding the polypeptide sequence SEQ ID NO
: 78
corresponding to the CT region of hST8Sia II delimited by the aminoacids
located in
positions 1 to 6 of SEQ ID NO : 34,
* the nucleotide sequence SEQ ID NO : 81 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 1 and 21 of SEQ ID
NO : 37, said
nucleotide sequence SEQ ID NO : 81 encoding the polypeptide sequence SEQ ID NO
: 82
corresponding to the CT region of hST8Sia IV delimited by the amino acids
located in
positions 1 to 7 of SEQ ID NO : 38,
- the nucleotide sequence encoding the TMD region comprised in the second
nucleic
acid is chosen among:
* the nucleotide sequence SEQ ID NO : 91 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 43 and 105 of SEQ
ID NO : 5, said
nucleotide sequence SEQ ID NO : 91 encoding the polypeptide sequence SEQ ID NO
: 92
corresponding to the TMD region of hST6Ga1NAc I delimited by the amino acids
located in
positions 15 to 35 of SEQ ID NO : 6,
* the nucleotide sequence SEQ ID NO : 107 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 25 and 84 of SEQ ID
NO : 21, said
nucleotide sequence SEQ ID NO: 107 encoding the polypeptide sequence SEQ ID
NO: 108
corresponding to the TMD region of hST3Gal III delimited by the amino acids
located in
positions 9 to 28 of SEQ ID NO : 22,
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
* the nucleotide sequence SEQ ID NO : 115 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 25 and 84 of SEQ ID
NO : 29, said
nucleotide sequence SEQ ID NO: 115 encoding the polypeptide sequence SEQ ID
NO: 116
corresponding to the TMD region of rST3Gal III delimited by the amino acids
located in
5 positions 9 to 28 of SEQ ID NO: 30,
* the nucleotide sequence SEQ ID NO : 119 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 19 and 69 of SEQ ID
NO : 33, said
nucleotide sequence SEQ ID NO: 119 encoding the polypeptide sequence SEQ ID
NO: 120
corresponding to the TMD region of hST8Sia II delimited by the amino acids
located in
10 positions 7 to 23 of SEQ ID NO : 34,
* the nucleotide sequence SEQ ID NO : 123 corresponding to the nucleotide
sequence delimited by the nucleotides located in positions 22 and 60 of SEQ ID
NO : 37, said
nucleotide sequence SEQ ID NO: 123 encoding the polypeptide sequence SEQ ID
NO: 124
corresponding to the TMD region of hST8Sia IV delimited by the amino acids
located in
15 positions 8 to 20 of SEQ ID NO :38,
- the nucleotide sequence encoding the SR region or fragment thereof comprised
in the
second nucleic acid, is chosen among:
* the sequence SEQ ID NO : 129 delimited by the nucleotides located in
positions 106 to 222 of SEQ ID NO : 5, encoding the polypeptide sequence SEQ
ID NO : 130
20 corresponding to the fragment of the SR region of hST6Ga1NAc I delimited
by the amino
acids located in positions 36 to 74 of SEQ ID NO : 6,
* the sequence SEQ ID NO : 131 delimited by the nucleotides located in
positions 109 to 222 of SEQ ID NO : 5, encoding the polypeptide sequence SEQ
ID NO: 132
corresponding to the fragment of the SR region of hST6Ga1NAc I delimited by
the amino
25 acids located in positions 37 to 74 of SEQ ID NO : 6,
* the sequence SEQ ID NO : 133 delimited by the nucleotides located in
positions 106 to 420 of SEQ ID NO : 5, encoding the polypeptide sequence SEQ
ID NO : 134
corresponding to the fragment of the SR region of hST6Ga1NAc I delimited by
the amino
acids located in positions 36 to 140 of SEQ ID NO : 6,
30 * the sequence SEQ ID NO : 135 delimited by the nucleotides
located in
positions 106 to 774 of SEQ ID NO : 5, encoding the polypeptide sequence SEQ
ID NO : 136
corresponding to the fragment of the SR region of hST6Ga1NAc I delimited by
the amino
acids located in positions 36 to 258 of SEQ ID NO : 6,
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
36
* the sequence SEQ ID NO : 137 delimited by the nucleotides located in
positions 85 to 138 of SEQ ID NO : 21, encoding the polypeptide sequence SEQ
ID NO : 138
corresponding to the fragment of the SR region of hST3Gal III delimited by the
amino acids
located in positions 29 to 46 of SEQ ID NO : 22,
* the sequence SEQ ID NO : 139 delimited by the nucleotides located in
positions 85 to 138 of SEQ ID NO : 29, encoding the polypeptide sequence SEQ
ID NO : 140
corresponding to the fragment of the SR region of ratST3Gal III delimited by
the amino acids
located in positions 29 to 46 of SEQ ID NO : 30,
* the sequence SEQ ID NO : 141 delimited by the nucleotides located in
positions 70 to 237 of SEQ ID NO : 33, encoding the polypeptide sequence SEQ
ID NO: 142
corresponding to the fragment of the SR region of hST8Sia II delimited by the
amino acids
located in positions 24 to 79 of SEQ ID NO : 34,
* the sequence SEQ ID NO : 143 delimited by the nucleotides located in
positions 61 to 201 of SEQ ID NO : 37, encoding the polypeptide sequence SEQ
ID NO : 144
corresponding to the fragment of the SR region of hST8Sia IV delimited by the
amino acids
located in positions 21 to 67 of SEQ ID NO : 38.
The invention concerns more particularly a process as defined above,
characterized in
that the CT, TMD, SR, or SR fragment peptides comprised in the second nucleic
acid, are
homologous sequences deriving from the same native glycosyltransferase, this
latter being
different from peptides in the native glycosyltransferase from which is
derived the CD
fragment with optimal glycosyltransferase activity as defined above.
The invention relates more particularly to a process as defined above,
characterized in
that the second nucleic acid is chosen among the following sequences:
- the sequence SEQ ID NO : 145 delimited by the nucleotides located in
positions 1 to
222 of SEQ ID NO : 5, containing SEQ ID NO : 49, 91, and 129, and encoding the
polypeptide sequence SEQ ID NO : 146 corresponding to the fragment of
hST6Ga1NAc I
delimited by the amino acids located in positions 1 to 74 of SEQ ID NO : 6,
and containing
the CT, TMD and SR fragment regions of hST6Ga1NAc I corresponding to SEQ ID NO
: 50,
92, and 130, respectively,
- the sequence SEQ ID NO : 147 delimited by the nucleotides located in
positions 1 to
420 of SEQ ID NO : 5, containing SEQ ID NO : 49, 91, and 133, and encoding the
polypeptide sequence SEQ ID NO : 148 corresponding to the fragment of the SR
region of
hST6GalNac I delimited by the aminoacids located in positions 1 to 140 of SEQ
ID NO : 6,
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
37
and containing the CT, TMD and SR fragment regions of hST6Ga1NAc I
corresponding to
SEQ ID NO : 50, 92, and 134, respectively,
- the sequence SEQ ID NO : 149 delimited by the nucleotides located in
positions 1 to
774 of SEQ ID NO : 5, containing SEQ ID NO : 49, 91, and 135, and encoding the
polypeptide sequence SEQ ID NO : 150 corresponding to the fragment of the SR
region of
hST6Ga1NAc I delimited by the amino acids located in positions 1 to 258 of SEQ
ID NO : 6,
and containing the CT, TMD and SR fragment regions of hST6Ga1NAc I
corresponding to
SEQ ID NO : 50, 92, and 136, respectively,
- the sequence SEQ ID NO : 151 delimited by the nucleotides located in
positions 1 to
138 of SEQ ID NO : 21, containing SEQ ID NO : 65, 107, and 137, and encoding
the
polypeptide sequence SEQ ID NO : 152 corresponding to the fragment of the SR
region of
hST3Gal III delimited by the aminoacids located in positions 1 to 46 of SEQ ID
NO : 22, and
containing the CT, TMD and SR fragment regions of hST3Gal III corresponding to
SEQ ID
NO : 66, 108, and 138, respectively,
- the sequence SEQ ID NO : 153 delimited by the nucleotides located in
positions 1 to
138 of SEQ ID NO : 29, containing SEQ ID NO : 73, 115, and 139, and encoding
the
polypeptide sequence SEQ ID NO : 154 corresponding to the fragment of the SR
region of
rST3Gal III delimited by the aminoacids located in positions 1 to 46 of SEQ ID
NO : 30, and
containing the CT, TMD and SR fragment regions of hST3Gal III corresponding to
SEQ ID
NO : 74, 116, and 140, respectively,
- the sequence SEQ ID NO : 155 delimited by the nucleotides located in
positions 1 to
237 of SEQ ID NO : 33, containing SEQ ID NO : 77, 119, and 141, and encoding
the
polypeptide sequence SEQ ID NO : 156 corresponding to the fragment of the SR
region of
hST8Sia II delimited by the amino acids located in positions 1 to 79 of SEQ ID
NO : 34, and
containing the CT, TMD and SR regions of hST8Sia II corresponding to SEQ ID NO
: 78,
120, and 142, respectively,
- the sequence SEQ ID NO : 157 delimited by the nucleotides located in
positions 1 to
201 of SEQ ID NO : 37, containing SEQ ID NO : 81, 123, and 143, and encoding
the
polypeptide sequence SEQ ID NO : 158 corresponding to the fragment of the SR
region of
hST8Sia IV delimited by the aminoacids located in positions 1 to 67 of SEQ ID
NO : 38, and
containing the CT, TMD and SR regions of hST8Sia IV corresponding to SEQ ID NO
: 82,
124, and 144, respectively.
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
38
The invention concerns more particularly a process as defined above,
characterized in
that the CT, TMD, SR, or SR fragment peptides comprised in the second nucleic
acid, are
heterologous sequences deriving from different natural glycosyltransferase
gene or transript.
The invention relates more particularly to a process as defined above,
characterized in
that the second nucleic acid is the sequence SEQ ID NO: 159 corresponding the
fusion of the
nucleotide sequence SEQ ID NO: 177 containing SEQ ID NO : 65 and 107 encoding
the CT
and TMD regions of hST3Gal III corresponding to SEQ ID NO : 66 and 108
respectively,
with the nucleotide sequence SEQ ID NO : 131 encoding the polypeptide sequence
SEQ ID
NO : 132 corresponding to the fragment of the SR region of hST6Ga1NAc I
delimited by the
aminoacids located in positions 37 to 74 of SEQ ID NO : 6, said sequence SEQ
ID NO : 159
encoding the fusion polypeptide SEQ ID NO : 160 between the CT and TMD regions
of
hST3Gal III, on the one hand, and the 37-74 fragment of the SR region of
hST6Ga1NAc I, on
the other hand.
The invention relates more particularly to a process as defined above,
characterized in
that the second nucleic acid is the sequence SEQ ID NO: 179 corresponding the
fusion of the
nucleotide sequence SEQ ID NO : 65 encoding the CT of hST3Gal III
corresponding to SEQ
ID NO : 66, with the nucleotide sequence SEQ ID NO : 119 encoding the TMD
region of
hST8Sia II corresponding to SEQ ID NO : 120, and with the nucleotide sequence
SEQ ID
NO: 129 encoding the polypeptide sequence SEQ ID NO : 130 corresponding to the
fragment
of the SR region of hST6Ga1NAc I delimited by the aminoacids located in
positions 36 to 74
of SEQ ID NO : 6, said sequence SEQ ID NO: 179 encoding the fusion polypeptide
SEQ ID
NO : 180 between the CT region of hST3Gal III, the TMD region of hST8Sia II,
and the 36-
74 fragment of the SR region of hST6Ga1NAc I.
The invention concerns more particularly a process as defined above,
characterized in
that it comprises the fusion of the sequence SEQ ID NO : 43 as the first
nucleic acid, with a
second acid nucleic chosen among:
- the sequence SEQ ID NO: 145, leading to the nucleotide sequence SEQ ID
NO: 161
encoding the fusion protein SEQ ID NO : 162 containing the CT, TMD and SR
fragment
regions of hST6Ga1NAc I corresponding to SEQ ID NO : 50, 92, and 130,
respectively,
linked via a GS linker to the 90-406 C-terminal minimal fragment of the CD
domain of
hST6Gal I,
- the sequence SEQ ID NO: 147, leading to the nucleotide sequence SEQ ID
NO: 163
encoding the fusion protein SEQ ID NO : 164 containing the CT, TMD and SR
fragment
regions of hST6Ga1NAc I corresponding to SEQ ID NO : 50, 92, and 134,
respectively,
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
39
linked via a GS linker to the 90-406 C-terminal minimal fragment of the CD
domain of
hST6Gal I,
- the sequence SEQ ID NO: 149, leading to the nucleotide sequence SEQ ID
NO: 165
encoding the fusion protein SEQ ID NO : 166 containing the CT, TMD and SR
fragment
regions of hST6Ga1NAc I corresponding to SEQ ID NO : 50, 92, and 136,
respectively,
linked via a SR linker to the 90-406 C-terminal minimal fragment of the CD
domain of
hST6Gal I,
- the sequence SEQ ID NO: 151, leading to the nucleotide sequence SEQ ID
NO: 167
encoding the fusion protein SEQ ID NO : 168 containing the CT, TMD and SR
fragment
regions of hST3Gal III corresponding to SEQ ID NO : 66, 108, and 138,
respectively, linked
via a GS linker to the 90-406 C-terminal minimal fragment of the CD domain of
hST6Gal I,
- the sequence SEQ ID NO: 153, leading to the nucleotide sequence SEQ ID
NO: 169
encoding the fusion protein SEQ ID NO : 170 containing the CT, TMD and SR
fragment
regions of ratST3Gal III corresponding to SEQ ID NO : 74, 116, and 140,
respectively, linked
via a SR linker to the 90-406 C-terminal minimal fragment of the CD domain of
hST6Gal I,
- the sequence SEQ ID NO: 155, leading to the nucleotide sequence SEQ ID
NO: 171
encoding the fusion protein SEQ ID NO : 172 containing the CT, TMD and SR
regions of
hST8Sia II corresponding to SEQ ID NO : 78, 120, and 142, respectively, linked
via a KL
linker to the 90-406 C-terminal minimal fragment of the CD domain of hST6Gal
I,
- the sequence SEQ ID NO: 157, leading to the nucleotide sequence SEQ ID NO:
173
encoding the fusion protein SEQ ID NO : 174 containing the CT, TMD and SR
regions of
hST8Sia IV corresponding to SEQ ID NO : 82, 124, and 144, respectively, linked
via a KL
linker to the 90-406 C-terminal minimal fragment of the CD domain of hST6Gal
I,
- the sequence SEQ ID NO: 159, leading to the nucleotide sequence SEQ ID
NO: 175
encoding the fusion protein SEQ ID NO : 176 containing the CT and TMD regions
of
hST3Gal III corresponding to SEQ ID NO : 66 and 108 respectively, and the 37-
74 fragment
of the SR region of hST6GalNac I corresponding to SEQ ID NO: 132, linked via a
GS linker
to the 90-406 C-terminal minimal fragment of the CD domain of hST6Gal I,
- the sequence SEQ ID NO: 179, leading to the nucleotide sequence SEQ ID
NO: 181
encoding the fusion protein SEQ ID NO : 182 containing the CT region of
hST3Gal III, the
TMD region of hST8Sia II, and the 36-74 fragment of the SR region of
hST6Ga1NAc I,
corresponding to SEQ ID NO : 66, 120, and 130 respectively, linked via a GS
linker to the
90-406 C-terminal minimal fragment of the CD domain of hST6Gal I.
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
The invention also relates to gene sequences encoding chimerical
glycosyltransferases
such as obtained according to the process as defined above.
The invention concerns more particularly gene sequences as defined above,
chosen
among:
5 - the sequence SEQ ID NO : 161 encoding the fusion protein SEQ ID NO :
162
containing the CT, TMD and SR fragment regions of hST6Ga1NAc I corresponding
to SEQ
ID NO : 50, 92, and 130, respectively, linked via a GS linker to the 90-406 C-
terminal
minimal fragment of the CD domain of hST6Gal I, said sequence SEQ ID NO : 161
corresponding to the fusion of the sequence SEQ ID NO : 43 and the sequence
SEQ ID NO:
10 145,
- the sequence SEQ ID NO : 163 encoding the fusion protein SEQ ID NO : 164
containing the CT, TMD and SR fragment regions of hST6Ga1NAc I corresponding
to SEQ
ID NO : 50, 92, and 134, respectively, linked via a GS linker to the 90-406 C-
terminal
minimal fragment of the CD domain of hST6Gal I, said sequence SEQ ID NO : 163
15 corresponding to the fusion of the sequence SEQ ID NO : 43 and the
sequence SEQ ID NO:
147,
- the sequence SEQ ID NO : 165 encoding the fusion protein SEQ ID NO : 166
containing the CT, TMD and SR fragment regions of hST6GalNac I corresponding
to SEQ ID
NO : 50, 92, and 136, respectively, linked via a SR linker to the 90-406 C-
terminal minimal
20 fragment of the CD domain of hST6Gal I, said sequence SEQ ID NO : 165
corresponding to
the fusion of the sequence SEQ ID NO : 43 and the sequence SEQ ID NO: 149,
- the sequence SEQ ID NO : 167 encoding the fusion protein SEQ ID NO : 168
containing the CT, TMD and SR fragment regions of hST3Gal III corresponding to
SEQ ID
NO : 66, 108, and 138, respectively, linked via a GS linker to the 90-406 C-
terminal minimal
25 fragment of the CD domain of hST6Gal I, said sequence SEQ ID NO : 167
corresponding to
the fusion of the sequence SEQ ID NO : 43 and the sequence SEQ ID NO: 151,
- the sequence SEQ ID NO : 169 encoding the fusion protein SEQ ID NO : 170
containing the CT, TMD and SR fragment regions of ratST3Gal III corresponding
to SEQ ID
NO : 74, 116, and 140, respectively, linked via a SR linker to the 90-406 C-
terminal minimal
30 fragment of the CD domain of hST6Gal I, said sequence SEQ ID NO : 169
corresponding to
the fusion of the sequence SEQ ID NO : 43 and the sequence SEQ ID NO: 153,
- the sequence SEQ ID NO : 171 encoding the fusion protein SEQ ID NO : 172
containing the CT, TMD and SR regions of hST8Sia II corresponding to SEQ ID NO
: 78,
120, and 142, respectively, linked via a KL linker to the 90-406 C-terminal
minimal fragment
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
41
of the CD domain of hST6Ga1 I, said sequence SEQ ID NO: 171 corresponding to
the fusion
of the sequence SEQ ID NO : 43 and the sequence SEQ ID NO: 155,
- the sequence SEQ ID NO : 173 encoding the fusion protein SEQ ID NO : 174
containing the CT, TMD and SR regions of hST8Sia IV corresponding to SEQ ID NO
: 82,
124, and 144, respectively, linked via a KL linker to the 90-406 C-terminal
minimal fragment
of the CD domain of hST6Gal I, said sequence SEQ ID NO: 173 corresponding to
the fusion
of the sequence SEQ ID NO : 43 and the sequence SEQ ID NO: 157,
- the sequence SEQ ID NO : 175 encoding the fusion protein SEQ ID NO : 176
containing the CT and TMD regions of hST3Gal III corresponding to SEQ ID NO :
66 and
108 respectively, and the 37-74 fragment of the SR region of hST6Ga1NAc I
corresponding to
SEQ ID NO : 132, linked via a GS linker to the 90-406 C-terminal minimal
fragment of the
CD domain of hST6Gal I, said sequence SEQ ID NO: 175 corresponding to the
fusion of the
sequence SEQ ID NO : 43 and the sequence SEQ ID NO: 159,
- the sequence SEQ ID NO : 181 encoding the fusion protein SEQ ID NO : 182
containing the CT region of hST3Gal III, the TMD region of hST8Sia II, and the
36-74
fragment of the SR region of hST6Ga1NAc I, corresponding to SEQ ID NO : 66,
120, and 130
respectively, linked via a GS linker to the 90-406 C-terminal minimal fragment
of the CD
domain of hST6Gal I, said sequence SEQ ID NO : 181 corresponding to the fusion
of the
sequence SEQ ID NO : 43 and sequence SEQ ID NO: 179.
The invention also concerns vectors, such as plasmids , viral or bacterial
constructs,
comprising at least one gene sequence as defined above.
The invention also relates to host eukaryotic cells from yeast, fungi, insect,
plants,
mammalian or human origin, transformed with at least one gene sequence as
defined above,
using at least one vector as mentioned above.
The invention also concerns the use of at least one gene sequence as defined
above, or
of a vector as mentioned above, for the transformation of cells as defined
above, or the use of
transgenic animals obtained from such transformed cells, in the frame of the
production of
recombinant proteins of interest.
The invention also relates to a method for the preparation of recombinant
proteins of
interest comprising the transformation of cells as defined above, with a
vector containing at
least one nucleotide sequence encoding said recombinant proteins of interest.
Preferred recombinant proteins of interest which can be prepared according to
a
method as mentioned above according to the invention are chosen among
hormones,
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
42
enzymes, clotting factors, carbohydrate antigens/serum biomarkers, cytokines,
growth factors,
antibodies or receptors.
Preferred host cells for the preparation of recombinant proteins of interest
as
mentioned above, are chosen among drug-approved cells or organisms, preferably
rodent,
mammalian or human cells.
DESCRIPTION OF THE FIGURES
Figure 1 represents the membrane topology of glycosyltransferases.
Glycosyltransferases are
type II membrane proteins including a cytoplasmic N-terminal tail (CT), a
transmembrane
(TMD) anchor signal followed by a stem region (SR) and a large C-terminal
catalytic domain
(CD).
Figure 2 Amino acid sequence of sialylmotif L, S, and VS in 20 human
sialyltransferases.
Consensus amino acid residues in the all sialyltransferases are shown by bold
letters.
Figure 3 represents schematic distribution of CT, TMD, SR and CD of the rat
ST6Ga1 I
showing key residues which are tyr123 and 7 conserved cys. CT: cytoplasmic
tail (9 amino
acids); TMD: transmembrane domain (17 aa); SR: stem region (70 aa); CD:
catalytic domain
(307 aa); L: sialylmotif L; S: sialylmotif L and VS sialylmotif VS
Figure 4 represents DNA constructs to produce chimera and evaluate the role of
the
cytoplasmic tail, the transmembrane domain in two enzyme isoforms (rST6Gal I
Tyr or Cys
123) respectively (Fenteany & Colley, 2005).
Figure 5 represents sequence alignment of the rat and the human ST6Ga1 I. It
can be noticed
that the N-terminal sequence comprising the CT(1-9), TMD (10-26) is fully
conserved while
the juxtamebrane SR portion(27-89) is more variable although the juxtamembrane
peptide
and especially positively lysine and cysteine residues are wellconserved.
Figure 6 represents the sialic acid pathway in human cells in the context of
overall cellular
glycosylation. The enzymes involved in this sequential process are: (a and b)
UDP-N-
acetylglucosamine / 2-epimerase/N-acetylmannosamine kinase (UDP-G1cNAc 2-
epimerase/ManNAc 6-kinase) - Reactions: (a) epimerase and (b) kinase, (c) N-
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
43
acetylneuraminic acid phosphate synthase; (NANS SAS; N-acetylneuraminic acid
phosphate
synthase) ¨ Reaction and Homo sapiens N-acetylneuraminate pyruvate lyase (NPL;
N-
acetylneuraminate pyruvate lyase) ¨ Reaction, (d) NeuAc 9-phosphatase, (e)
Cytidine 5'-
monophosphate N-acetylneuramininc acid synthetase, (CMP-NeuAc synthetase), (f)
Cytidine
monophosphate-sialic acid transporter (Golgi CMP-NeuAc transporter), and (g)
sialyltransferases.
Of note, the sialic acid derivative shown in this diagram, namely "Neu5Ac," is
widely
considered as the 'human' form of sialic acid. In all other animals, with the
exception of
chickens, there is an additional step in the pathway (shown in this diagram)
where CMP-
Neu5Ac is futher hydroxylated and converted to CMP-Neu5Gc by the enzymatic
action of
CMP-N-Acetylneuraminic Acid Hydroxylase.
Figure 7 represents a schematical overview of the various synthetic chimerical
constructs
generated by the invention.
Figure 7A represents the general construction of a synthetic chimera including
the N-
terminal synthetic domain tagged with the FLAG epitope fused to the minimal
catalytic
domain (CD) through the addition of a restriction site.
Figure 7B shows the ligation between the N-terminal synthetic and the
catalytic
domains using a BamHI restriction site. Note that the constructs are
introduced into the vector
using restriction sites, namely AflII and XbaI distinct from the ligation
site.
Figure 8 shows the topology and characteristic of the pcDNA3.1 (+) vector
(Invitrogen).
Figure 9 represents the constructions of the FLAG-hST6Ga1 I CD and of the
chimeric forms
of this CD fused to several N-Terminal fragments of other sialyltransferases
of variable
length.
Figure 9A corresponds to FLAG-CD.
Figure 9B corresponds to hST8SiaII-79/CD.
Figure 9C corresponds to hST8SiaIV-67/CD.
Figure 9D corresponds to hST6Ga1NAc I-258/CD.
Figure 9E corresponds to r/hST3Ga1 III -46/CD.
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
44
Figure 10 represents the digested minimum catalytic domain of hST6Ga1 I and
its amplified
PCR product to be used in each construction of the synthetic chimera. Samples
were loaded
on a 1,5 % agarose gel with the SmartLadder (SL) nucleic marker.
Figure 10A shows the digested hST6Gal I catalytic domain from CMV-vector,
issued
from the cloning in the laboratory.
Figure 10B shows the PCR product of the minimum catalytic domain at 966 pb.
Figure 10C shows the concentrated PCR product of the minimum catalytic domain
of
hST6Gal I with 5 pL loaded on the gel.
Figure 11 shows an agarose gel (2 %) showing the DNA band of the reconstituted
synthetic
N-terminal region of hST3Gal III. Lane 1 corresponds to a PCR product of 174
pb; lane 2
corresponds to the SmartLadder SF; lane 3 corresponds to 5 pL of the
concentrated PCR
product of 174 pb size.
Figure 12represents the total reconstructed enzyme gene of hST3Gal III/CD.
In Figure 12A, an agarose gel (1.5 %) shows the DNA band of the reconstituted
synthetic hST3Gal III/CD amplified by PCR.
Figure 12B corresponds to 5 pL of the concentrated PCR products of around 1200
pb
in size.
Figure 12C represents the product of the digestion of the recombinant vector
by the
restriction enzymes AflII and XbaI, showing the insertion of the chimera gene
(expected size
1200 pb).
Figure 13 shows an agarose gel (2 %) showing the DNA band of the
reconstituted synthetic
N-terminal region of hST6Ga1NAc 1-74. Lane 1 corresponds to a PCR product of
270 pb; lane
2 corresponds to the SmartLadder SF; lane 3 corresponds to 5 pL of the
concentrated PCR
products of 270 pb size.
Figure 14 represents the complete reconstructed synthetic gene of hST6Ga1NAc I-
74/CD.
Figure 14A shows an agarose gel (1.5 %) with the DNA band of the reconstituted
synthetic hST6Ga1NAc I-74/CD amplified by PCR.
Figure 14B shows 5 pL of the concentrated PCR products of the chimera of
around
1200 pb in size.
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
Figure 14C shows the product of the digestion of the recombinant vector by the
restriction enzymes AflII and XbaI, showing the insertion of the chimera gene
(expected size
1225 pb).
Figure 15 shows an agarose gel (1.5 %) showing the DNA band of the
reconstituted synthetic
5 N-terminal region of hST6Ga1NAc 1-140. Lane 1: SmartLadder SF; lane 2:
PCR product of
468 pb.
Figure 16 shows an agarose gel (1.5 %) showing the DNA band of the
reconstituted synthetic
hybrid gene composed of hST3Gal III-29/37-hST6Ga1NAc 1-74. Lane 1 corresponds
to the
SmartLadder SF; lane 2 corresponds to a PCR product of 249 pb.
10 Figure 17 shows an agarose gel (1.5 %) showing the DNA band of the
reconstituted synthetic
hybrid gene composed of hST3Gal III-29/37-hST6Ga1NAc 1-74 ligated with the CD.
Lane 1
corresponds to the SmartLadder SF; lane 2 corresponds to a PCR product of 1197
pb.
Figure 18 represents the expression of the minimal catalytic domain (CD)
(Panel A)of
hST6Gal I in CHO cells, based on a double labelling with FITC-SNA binding 6-
linked sialic
15 acid and with an anti-FLAG mAb and using confocal microscopy. Transient
expression of the
CD was analysed by anti-FLAG labelling (Panel B) and for SNA-FITC binding
(Panel C).
Panel D shows the superposition of the signal from panels B and C (Donadio et
al., 2003)
Figure 19 shows the expression of two chimeric enzymes, hST8SiaIV-67/CD,
hST8SiaII-
79/CD in CHO cells. Transfected cells were double-labelled with an anti-FLAG
mAb (A and
20 D) and with FITC-SNA (B and E). Overlays are represented in Cand F.
Figure 20 shows the expression of ST3Ga1III/CD and ST3Ga1III -hST6Ga1NAc I-
74/CD in
CHO cells. Transfected cells were double-labelled with an anti-FLAG mAb (A and
D) and
with FITC-SNA (B and E). Overlays are represented in C and F.
Figure 21 shows the expression of hST6Ga1NAc I-74/CD, and 258/CD in CHO cells.
25 Transfected cells were double-labelled with an anti-FLAG mAb (A and D)
and with FITC-
SNA (B and E). Overlays are represented in C and F.
Figure 22 shows an agarose gel (2 %) showing the DNA band of the reconstituted
synthetic N-
terminal region of hST3Ga1 III / hST8Sia II / hST6Ga1NAc I. Lane 1 corresponds
to a PCR
30 product of 240 Pb; lane 2 corresponds to the SmartLadder SF; lane 3
corresponds to 31..t.L of the
240 pb concentrated PCR product.
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
46
Figure 23 represents the product of the digestion of the recombinant vector by
the restriction
enzymes XbaI and BamHI, showing the insertion of the catalytic domain gene
(expected size
960 pb). Sample was loaded on a 1.5 % agarose gel with the SL nucleic marker.
Figure 24 represents the reconstructed enzyme gene of hST3Gal III / hST8Sia II
/
hST6Ga1NAc I / CD.
In figure 24a, an agarose gel (1.5%) shows the product of the digestion of the
recombinant
vector by the restriction enzymes XbaI and AflII, showing the insertion of the
chimera gene
(expected size 1200 pb).
Figure 24b represents the PCR product obtained after the amplification of a
minipreparation
sample, confirming the insertion of the chimera gene.
Figure 25 shows the expression of the chimeric enzyme hST3Gal III / hST8Sia
II /
h5T6Ga1NAc I / CD in CHO cells. Transfected cells were double-labelled with an
anti-FLAG
mAb (A) and with FITC-SNA (B). Overlay is represented in C.
LEGENDS of TABLES 1 to 5
Table 1: Sequences and accession numbers of the 20 sialyltransferases known in
human.
Table 2: Acceptor substrates and expression sites of human sialyltransferases
as
described in Harduin-Lepers et al., 2001 and Jeanneau et al, 2003.
Table 3: Distribution of the 4 peptide domains shared by human STs. A short N-
terminal cytoplasmic tail (CT; around 10 amino-acids), a transmembrane domain
(TMD;
around 20 amino acids), a stem region (SR), highly variable in length and a
catalytic domain
(CD; around 300 amino acids), including the sialylmotifs L, S and VS.
Table 4: Proposed conserved peptides of around 31-85 amino acids identified
within the
hypervariable SR region. 5 motifs were found : motif A common to ST6Ga1 and
ST6Ga1NAc
I, motif B common to ST6Ga1NAc I and ST6Ga1NAc II, motifs C and D common to
all
ST8Sia and motif E present in ST3Ga1 except the ST3Ga1 I and II (Donadio et
al., 2003).
Table 5: Design of the four primer pairs used for PCR amplification.
CA 02653104 2008-11-24
WO 2007/135194 PCT/EP2007/055070
47
HUMAN ACCESSION N GenBank ACCESSION N Swiss
SIALYLTRANSFERASES Prot
ST6Ga1 I A17362 P15907
ST6Ga1 II NM 032528 Q8IUG7
ST6Ga1NAc I NM - 018414 Q9NSC7
ST6Ga1NAc II NM - 006456 Q90J37
ST6Ga1NAc III NM - 152996 Q8NDV1
ST6Ga1NAc IV NM 014403 Q9H4F1
ST6GaINAc V NM 030965 Q9BVH7
ST6Ga1NAc VI AB-035173 Q969X2
ST3Ga1 I L29555 Q11201
ST3Gal II U63090 Q16842
ST3Gal III L23768 Q11203
ST3Ga1 IV L23767 Q11206
ST3Ga1 V AF105026 Q9UNP4
ST3Ga1 VI AF119391 Q9Y274
ST8S1a I L32867 Q92185
ST8Sia II U33551 Q92186
ST8Sia III AF004668 043173
ST8S1a IV L41680 Q92187
ST8Sia V U91641 015466
ST8S1a VI AJ621583 P61647
Table 1
SUBSTITUTE SHEET (RULE 26)
CA 02653104 2008-11-24
WO 2007/135194 PCT/EP2007/055070
48
HUMAN Acceptor Expression
SIALYLTRANSFERASES
ST6Ga1 I Galp1-4G1cNAc Ubiquitous
ST6Ga1 II Galp1-4G1cNAc Brain,
testicules,
thyroid, fetal tissue,
lymphatic ganglia
ST6Ga1NAc I Ga1p1-3Ga1NAc
Submaxillary and
mammary
GalNAca-O-Ser/Thr glands,
spleen, colon
Siaa2-3 Galp1-3GaINAca-
O-Ser/Thr
ST6Ga1NAc II Ga1131- 3GalNAca-0- Lacting
mammary glands,
Ser/Thr testis
Siaa2-3Galp1-3GalNAca-
O-Ser/Thr
ST6Ga1NAc III Siaa2-3Galp1-3Ga1NAc
ST6Ga1NAc IV Siaa2-3GalP1-3GalNAc, Ubiquitous
GMlb
ST6Ga1NAc V Siaa2-3Galp1-3GaINAc,
GMlb
ST6Ga1NAc VI GM1b, GT1b
ST3Ga1 I GalP1-3GalNAc Ubiquitous
ST3Ga1 II Ga1131-3Ga1NAc, GM1, Heart,
liver, skeletal
asialo- muscle,
GM1 thymus,
lymph node,
appendix, salivary
glands, spleen
ST3Gal III Ga1131-3G1cNAc Skeletal
muscle, fetal
Gal31-4G1cNAc tissue
ST3Ga1 IV Gal31-4G1cNAc-0 Placenta, testis, ovary
GalP1-3GalNAc-0
ST3Ga1 V Ga1131-4G1cP-Cer Ubiquitous
ST3Ga1 VI Ga1131-4G1cNAc Heart,
placenta, liver
and most other tissues.
ST8Sia I Siaa2-3Galp1-4G1cp1-0-
Cer, GM3
ST8Sia II SiaGal31-4GalNAc Embryonic
tissues,
brain
ST8Sia III Sia2-3Gal31-4G1cNAc
ST8Sia IV SiaGa1131-4Ga1NAc Brain,
fetal tissues,
adult heart, spleen,
thymus
ST8Sia V GM1b, GD1a, GT1b, GD3
ST8Sia VI NeuAca2,3(6)Gal3
Table 2
SUBSTITUTE SHEET (RULE 26)
0
ESTIMATION OF LENGTH OF DOMAINS
n.)
o
o
--4
Human Total Cytoplasmic Transmembrane Stem
Region Catalytic Domain Sialylmotif L Sialylmotif S
Sialylmotif VS 1--,
Sialyltransferases Tail (Cl') Domain (TMD) (SR; +/-10
AA) (CD; +/-10 AA) (47 AA) (24 AA) (12 AA) t...)
1--,
ST6Ga1 I 406 1-9 (9 AA) 10-26 (17 AA) 27-100
101-406 181-227 320-343 366-378 .6.
(74 AA)
(306 AA)
ST6Ga1 II 529 1-10 (10 AA) 11-30 (20 AA) 31-112
113-529 293-339 433-456 479-491
(82 AA)
(417 AA)
ST6Ga1NAc I 600 1-14 (14 AA) 15-35 (41 AA) 36-281
282-600 362-408 518-541 563-575
(246 AA)
(319 AA)
ST6Ga1NAc II 374 1-7 (7 AA) 8-28 (21 AA) 29-67
68-374 148-194 302-325 347-359
(39 AA)
(307 AA)
OD ST6Ga1NAc III 305 1-8 (8 AA) 9-28 (20 AA) 29-34
35-305 77-123 214-237 273-285
C:
n
CO (6 AA)
(271 AA)
GO ST6Ga1NAc IV 302 1-6(6 AA) 7-27 (21 AA) 28-30
31-302 73-119 210-233 270-282 o
--I
(3 AA) (272 AA) K.)
m
--I ST6Ga1NAc V 336 1-8 (8
AA) 9-29 (21 AA) 30-50 51-336 93-139 230-253
291-303 ul
w
C:H
--I
(21 AA) (286 AA)
.6.
o
[Ti ST6Ga1NAc VI 333 1-43(43 AA) 44-59 (16 AA) 60-61
62-333 105-151 241-264 303-315
OD (2 AA)
(272 AA) K.)
o
=C ST3Ga1 I 340 1-13 (13 AA) 14-34 (21 AA) 35-58
59-340 139-185 266-289 312-324 o
M (24 AA)
(282 AA) op
1
M
H
--I ST3Ga1 II 350 1-6 (6
AA) 7-27(21 AA) 28-68 69-350 149-195 276-299
322-334 H
1
(41 AA)
(282 AA) K.)
JJFl.
C: ST3Ga1 III 444 1-8 (8 AA) 9-28 (20 AA) 29-76
77-444 157-203 299-322 346-358
I¨ (48 AA)
(368 AA)
M ST3Ga1 IV 329 1-8 (8 AA) 9-26(18 AA) 27-36
37-329 117-163 258-281 307-319
ND (10 AA)
(293 AA)
0)
ST3Ga1 V 362 1-5 (5 AA) 6-26 (21 AA) 27-55 (29
AA) 56-362 (307 AA) 136-182 282-305 329-341
ST3Ga1 VI 331 1-4 (4 AA) 5-25 (21 AA) 26-34 (9 AA)
35-331 (297 AA) 115-161 256-279 303-315
ST8Sia I 356 1-29 (29 AA) 30-48(19 AA) 49-54 (6 AA)
55-356 (302 AA) 135-182 272-295 318-330
ST8Sia II 375 1-6 (6 AA) 7-23 (17 AA) 24- 73 (50
AA) 74-375 (302 AA) 157-200 292-315 342-354 .o
n
ST8Sia III 380 1-9 (9 AA) 10-33 (24 AA) 34-78 (45
AA) 79-380 (302 AA) 159-205 298-321 350-362 1-3
ST8Sia IV 359 1-7 (7 AA) 8-20 (13 AA) 21-58 (38
AA) 59-359 (301 AA) 139-185 277-300 327-339 ....-
STBSia V 376 1-17 (17 AA) 18-38 (21 AA) 39-80 (42
AA) 81-376 (296 AA) 93-208 298-321 344-350 00
w
ST8Sia VI 398 1-3 (3 AA) 4-24 (21 AA) 25-102 (78
AA) 103-398 (296 AA) 183-230 320-343 360-378 o
o
--.1
o
u,
Table3
=
--...,
=
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
ENZYMES conserved Motif A
Motif B Motif C Motif D Motif E
region
hST6Gal I 93-165 159-165 X X X X
hST6GaINAc I 259-331 310-315 324-331 X X X __
hST8Sia IV 71-133 X X 71-90 119-133 X
hST3Gal IV 75-106 X X X X 84-91
5
Table 4
SUBSTITUTE SHEET (RULE 26)
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
51
Amplified fragments Primers set PCR product size (Pb) _
hST3Gal III 5' AflII-ST3 186
3'ST3-BamHI
hST6GalNAc 1-74 5' AflII-ST6 270
3 'ST6-BamHI
hST6GaINAc 1-140 5' AfiII-ST6 468
3'ST6-BamHIn 2
hST3Ga1 III-29/37-hST6GaINAc-74 5' AflII-ST3 246
3'ST6-BamHI
Table 5
SUBSTITUTE SHEET (RULE 26)
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
52
Description of the invention
The invention will be further described in the detailed description which
follows.
1 - Introduction
Bases of the invention:
The inventors have focused on the study of hST6Ga1 I because this enzyme
activity is
missing in all host cells used so far for heterologous protein production,
especially in drug-
approved cell systems. The human enzyme has been cloned by the inventors in
Ronin, 2001.
In contrast to the litterature, the inventors were initially able to show that
the full-
legnth enzyme can differentially sialylate acceptor glycoproteins of bi-, tri-
and/or
tetraantennary glycan structure (Ronin 2001) and at that time, the inventors
hypothesized that
within the transferase structure, a steric control located in the
hypervariable SR region should
be exerted on the CD to regulate and naturally constrain enzymatic
specificity. For the
purpose of producing sialylated proteins of biomedical interest, this
regulatory control should
be abolished and as a result, the specificity may be enlarged. Inside the
conserved region (93-
165) of hST6Gal I, an hydrophobic sequence has been noticed : 93- QVWxKDS -
100. The
importance of this sequence has been studied by progessively deleting hST6Gal
I of its N-
terminal part within the conserved region newly identified. The 435, 448, 460
and 482
deleted mutants show an increasing transfer efficiency. Deletions carried out
before and after
the conserved domain (93-165 of ST6Ga1I) showed that the 4100 mutant lose its
catalytic
activity, whereas the 489 (containing the hydrophobic sequence QVWxKDS)
possesses an
optimal catalytic activity. The results clearly indicate that this short
sequence is crucial to
promote activity. This hydrophobic sequence may contribute to local
conformational changes
essential for the active site to promote sialic acid transfer.
A second study has been realized by the inventors aimed at elucidating the
molecular
and cellular basis that govern the acceptor preference of STs (Ronin 2003). As
it was difficult
to delineate the CD from the hypervariable region of the SR, they hypothesized
that the SR of
STs should contain structural features related to an acceptor preference. 53
animal and human
enzymes of known specifity were analyzed by bioinformatics to determine if STs
may share a
peptidic portion to restrict the acceptor recognition to an enzyme subfamily.
A highly
conserved region of around 31-85 amino acids has been identified and inside
this region, 5
motifs were found : motif A common to ST6Ga1 and ST6Ga1NAc I, motif B common
to
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
53
ST6GaNAc I and ST6GaNAc II, motifs C and D common to all ST8Sia and motif E
present
in ST3Ga1 except the ST3Ga1 I and II (Table 4).
Those 5 motifs are typical of a STs subgroup sharing a similar catalytic
activity and
thus involved in the same way to transfer sialic acid. They are located at the
end of the SR
closed to the sialylmotif L and can be considered as part of the CD as key
feature for acceptor
recognition.
Of special interest also, was also the finding that the 489 mutant is
extremely efficient
in CHO cells and that it follows the intracellular pathway from the early
golgi to the trans
golgi/trans golgi network. 489 is expressed along the intracellular routing of
the
glycoptroteins and glycolipids in the entire stacks of the golgi apparatus
(Ronin, 2003). This
work made it possible to delineate the minimum CD for hST6Gal I containing the
crucial
hydrophobic sequence (QVWxKDS) and displacing the acceptor recognition from
intracellular resident acceptors to cell surface glycoconjugates. These
findings gave the
ground of designing novel membrane-anchored chimera which may display a
similar
intracellular trafficking to encounter neosynthesized glycoproteins within the
secretory
pathway as they are packed in engineered host cells when produced as drugs.
The minimum CD of hST6Gal I 489 have been used in the invention as an
"optimized
CD"of enlarged specificity and increased transfer efficiency and will be
further named CD.
The distribution of the sialyltransferases in the golgi apparatus
Golgi localization of GTs is not strictly organized, enzymes are not
distributed and
isolated in a single compartment of the golgi apparatus, most of them overlap
and co-
compartmentalized (Colley, 1997; Berger et al., 1998; Berger, 2002). No clear
retention
signals have been identified yet but enzymes form overlapping gradients across
the stacks
(Opat et al., 2001), and only crucial regions have been identified.
There are two hypothesis concerning retention mechanisms in the golgi : i) the
length
of the TMD drives the golgi retention (bilayer thickness model; Bretscher &
Munro, 1993;
Colley, 1997; Munro, 1998) and ii) the proteins oligomerization leeds to golgi
retention
(Oligomerization/kin recognition hypothesis; Machamer, 1991; Colley, 1997), in
which
authors postulate that enzymes interact in golgi cisterna to form too large
structures to enter in
transport vesicules (Munro, 1998).
The length of the TMD seems also crucial as a retention signal. TMD represents
a key
factor in the retention process, in most of cases it is suficient to promote a
golgi localization
i;e throughout the cis-, medial and trans compartments. Lengthening the TMD of
STs results
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
54
in reduced retention and synthetic TMD (creates by mutagenesis) of 17 leucine
gives golgi
retention whereas one of 23 leucines does not (Munro, 1991, 1995, 1998).
Concerning the
retention signals of the STs, several regions are involved in an independant
manner to retain
enzymes in the golgi apparatus. The TMD with its flanking regions are
sufficient for golgi
retention (Colley, 1997). Other work suggest that no specific sequence in the
TMD are
required for golgi retention, especially in the presence of appropriate spaced
cytoplasmic and
luminal flanking sequences and/or SR (Colley, 1997). The presence of
negatively charged
amino acid sequences on STs, particularly close to the membrane anchor, was
found to
mislocalize the proteins in the golgi apparatus. The presence of FLAG or MYC
epitope, in the
CT, disturbs the golgi localization of ST6Ga1 I, whereas using an additional
spacer sequence,
to space out the negative charges from the TMD, reveals strong advantage for
golgi retention
(Yang et al., 1996). The SR sequences alone appear to function as an other
type of golgi
retention (Colley, 1997).
There is probably more than one retention mechanism, explaining the
colocalization of
the STs inside the trans golgi and the trans golgi network. Moreover the
localization of the
enzymes also depends of the cell type where they are expressed (Colley, 1997).
The TMD of
STs alone may be sufficient for golgi retention in MDCK cells but the same TMD
and its
flanking sequences are clearly required for golgi retention in CHO cells
(Colley, 1997). Of
interest, available data reveal that changing the CT, the TMD and the SR does
not disturb the
catalytic activity of STs, but allow to delocalize them inside the golgi
apparatus (Grabenhorst
et Conradt, 1999).
The most studied enzyme for the golgi retention signal, is the ratST6Gal I
(rST6Gal I),
that has been localized in the trans golgi and the trans golgi network of
hepatocytes (Roth et
al., 1985; Opat et al., 2001) and in post golgi compartments in other cells
(Colley, 1997). Two
natural isoforms of rST6Gal I exist, they differ only by one amino acid at
position 123 in the
CD. This is due to a single A to G nucleotide change, resulting of Tyr to Cys
amino acid
change. No catalytic activity differences were found between the two iso forms
of the enzyme
(Chen et al., 2003). The STcys form is found in the golgi in moderatly
expressing cells and
never at the cell surfaces or cleaved or secreted into the media of COS-1 or
HeLa cells,
whereas the STtyr iso form is found in the golgi, at low levels on the cell
surface and is
cleaved and secreted from COS-1 and HeLa cells (Ma et al., 1997). To explain
the two
different localizations of the proteins, authors have proposed two hypothesis
on the retention
mechanism: bilayer thickness and oligomerization. First, analysis of
lengthening TMD of
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
STcys and tyr suggests that the bilayer thickness is not a predominant process
for the golgi
retention for ST6Ga1 I. Second, analysis of oligomers reveals that 100% and
13% of STcys
and STtyr respectively are insoluble at pH 6.3 (late golgi pH), but at pH 8.0
both isoforms are
soluble. The results suggest that conformational changes and disulfide bounds
formation in
5 the CD represent the basis for the increased ability for STcys to form
oligomers and its stable
loaclization in the golgi apparatus. The nature of the amino acid 123
influences the oligomers
formations (Chen et al., 2000). Moreover, the oligomerization process and the
catalytic
activity depend also on the other seven conserved cystein residues (C24, 139,
181, 332, 359,
361 and 403). The Cys24, inside the TMD is required for dimerization, while
the cystein
10 residues present in the CD are required for trafficking and catalytic
activity. Cys181 and
Cys332 (in sialylmotif L and S, respectively) enzymes are retained in the
endoplasmic
reticulum and are minimally active or inactive respectively. Cys359 and 361
(between
sialylmotifs S and VS) are inactive enzymes without compromising their
localizataion and
trafficking. Cys139 or Cys403 mutated enzymes produce no change of the
catalytic activity
15 and of the golgi localization. The replacement of these two cystein
residues decreases
cleavage and secretion suggesting that they are necessary for signal cleavage
(Qian et al.,
2001) (Figure 3).
Mutants of STtyr enzymes, sharing deletion in the SR have been caracterized:
STtyr44
20 and STtyr45 (deleted of amino acids 32 to 104 and 86 to 104,
respectively) are not active
and/or secreted, STtyr41, 42 and 43 (deleted between amino acids 32 to 86)
represent not
cleavable forms and show an increasing cell surface expression (Kitazume et
al., 1999).
A recent work (Fenteany & Colley, 2005) show that multiple signals are indeed
25 required for rST6Gal I oligomerization and for the golgi localization.
Authors aimed at
reevaluating the role of the CT, the TMD of the two enzyme isoforms (Tyr or
Cys 123). They
realised several DNA constructs to produce chimera as represented in Figure 4.
Lengthening
the transmembrane domain (with more than 17 amino acids) does not enhance the
golgi exit,
it does not change trafficking or golgi localization. Concerning the role of
the CT and the
30 TMD in the STcys isoform, there is a signal composed of 3 Lysine
residues in the CT that are
recognized as export signal out of the ER. Authors show that altering both CT
and TMD
disturb the routing of the enzyme. The only difference between the iso forms
of rST6Gal I is
their capacity to form oligomers according to the pH. For STcys, the oligomers
formation,
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
56
according to the pH, is compromised only when CT and TMD are altered. For
STtyr, it
increases slightly the rate of golgi exit (Fenteany & Colley, 2005).
All these recent findings concerning the rST6Ga1 I can be considered
applicable to the
human ST6Ga1 I since both enzymes share 85% homology and that both CDs are
virtually
identical. Indeed, the two sequences alignment (Figure 5) clearly show that
the seven cysteins
previously described are conserved in the hST6Ga1 I, and the Tyrosine 123 is
also present.
Despite considerable efforts from scientists, no signal in the sequence of STs
or FuTs
could be demonstrated. It is believed that a multifactorial process may slow
down the
enzymes in their way to the surface to target them in appropriate compartments
which are the
Trans Golgi Network for ST6Ga1. Accordingly, the inventors hypothesized that
CT, TMD and
SR may be alternatively exchanged to anchor a defined CD in host cells which
lack the
relevant gene and fail to express the relevant transferase activity. Thus, it
is therefore possible
to generate a wide array of combination as these 3 portions can be selected
out of 55 animal
known genes. Each of them can be potentially fused to 20 known distinct
catalytic domains to
display the ability of transferring sialic acid. This is of definite interest
for the humanisation
of yeast (Pichia pastoris or Shyzosaccharomyces piombae), plant and insects
which do not
express these activities and yet are used to produce recombinant drugs.
Although not tested,
this reasoning may also well apply to FuTs, GalTs, GalNAcTs and GlcNAcs as
they govern
the biosynthesis of glycotopes of high clinical relevance.
Importance of several residues
As previously described, a fourth motif (motif 3) has been identified in the
ST family,
between the S ans the VS motifs. It is composed of four highly conserved amino
acids with
the following consensus sequence: (H/y)Y(Y/F/W/h)(E/D/q/g). Site directed
mutagenesis on
the hST3Gal I showed the functional importance of two amino acids in this
motif: His299 and
Tyr300. Results suggest the importance of aromatic residues, their possible
involvment in
acceptor recognition and their contribution for an optimal catalytic
efficiency. Particularly the
invariant Tyr300 plays a major conformational role. Mutational analysis showed
that mutants
H299A and Y300A display no catalytic activities, whereas mutant Y300F restore
partially the
activity (Jeanneau et al., 2004). This motif is also present in the catalytic
domain of hST6Gal
I. Moreover several cystein residues are of major importance in dimerization
and catalytic
activity as previously evoked (Qian et al., 2001). At least one disulfide
bound has been
evidenced between two conserved cystein residues inside the sialylmotifs L and
S. This link is
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
57
essential to maintain the protein conformation. The same observations revealed
the existence
of this binding in the PST (Angata et al., 2001), moreover, a second disulfide
bound exists
between sialylmotifs and the C-terminal region. Dimerization of the ST6Ga1 I,
via disulfide
bound, has been demonstrated. This enzyme inside the golgi appartus is for
around 20 to 30 %
in dimer form which has a lower activity because its affinity is reduced for
the CMP-NeuAc
(Jeanneau, 2003).
"Autoglycosylations"
GTs are often themselves glycosylated. Few data are available concerning the
state of
STs glycosylation and its importance on the biological function of the
enzymes. It seems that
the N-glycosylation on the ST6Ga1 I is not required for the biological
activity of the enzyme
in vivo. On the other hand, when tested in vitro on mutant, enzymatic activity
is only observed
for the the protein mutated on the first glycosylation site. These results
suggest the existence
of two N-glycans (Asn 146 and Asn 158) that can stabilise and/or prevent the
protein against
degradation. The mutation on the second glycosylation site may leading to
aggregation or
degradation of the protein (Chen & Colley, 2000; Chen et al., 2000). However,
the N-
glycosylation on the ST8Sia I can affect its activity and its subcellular
localization (Martina et
al., 1998). The elimination of the N-glycans reduced its in vivo activity with
less than 10 % of
the initial activity. It has been recently shown that ST8Siall and ST8SiaIV
possess, in
addition to their classical activity, an autopolysialylation activity
(Muhlenhoff et al., 2001).
This autopolysialylation occurs on the N-glycans at the position Asn 74 in the
PST and at the
positions Asn 69 and 219 in the STX. Site directed mutagenesis on these sites
inactive the
enzyme both in vivo and in vitro. As a result, care has been taken to maintain
the Asn 74 and
Asn 69 glycosylation sites when using SR of ST8SiaIV and ST8SiaII respectively
to construct
chimerical genes.
II - Use of the sequences of the invention for introducing new transferase
activity
in expression systems
ha - The role of glycans, their importance in the immune system
Glycans are recognition signals in most living organisms but they are also
structural
key determinants for protein biopotency.
They play a pivotal role in protein folding oligomerization, quality control,
sorting and
transport (Helenius & Aebi, 2001). Glycans represent oligosidic epitopes that
can be
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
58
considered as carbohydrate antigens. The nature of the glycans modifies the
immunoreactivity. Several families of oligosidic antigens exist: ABH blood
groups
determinants, Lewis tissue groups, antigens members of the T family and
antigens specific of
the polysialylated or sulfated cellular adhesion (Legaigneur et al., 1999).
Glycans have
multiple roles: they are also involved in mechanisms of interactions between
cells and
between cells and matrix. Some cell proteins named Lectins specifically
recognize glycanic
structures and act as specific receptors (Gabius et al., 2004). In most cases,
they are
components of cell surface glycoconjugates.
The carbohydrate moieties act as recognition signals in the immune system and
influence the immune recognition in at least two ways: i) the conformation of
the protein is
altered and it modulates their biological function; ii) the oligosaccharides
serve as recognition
determinants. Particularly, sialic acids contribute greatly to both of these
effects because their
highly electronegative and hydrophilic nature may influence the conformation
of sialylated
macromolecules, and this is particularly relevant for glycoproteins. Sialic
acids possess the
ability to act as biological masks to prevent or reduce the accessibility and
play a significant
role on the cell surface in the recognition process of self/non self
discrimination (Pilatte et al.,
1993, Glycobiology 2006 to be added).
Since N-glycans are well represented in serum glycoproteins and many other
tissue
proteins of human body, it is highly desirable that they should not be
antigenic when present
in recombinant glycosylated drugs. However since HuEPO produced in CHO cells
has been
developped as 1st approved drug, it appeared that several terminal sugars may
be antigenic in
humans: polyLacNAc, alphal ,3 Gal and N-Glycolylneuraminic acid. According to
the most
recent regulatory bodies, they should not be present any further in
recombinant therapeutics.
lib - The clearance
Aside from their influence on the physico-chemical properties and the
biological
functions of proteins, glycans also possess a prevailing role on duration of
glycoproteins in
blood. This phenomenon is named metabolic clearance and represents the rate at
which a
natural compound or a drug is removed from the body by the liver or the
kidney. It is defined
as the plasmatic volume purified according to time (mL/min). This mechanism
allows the
organism to eliminate drugs.
Most of the circulating plasmatic proteins belong to the glycoproteins family
sharing
N-linked oligosaccharides and all of them are terminated in sialic acid,
particularly in
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
59
humans. Removal of sialic acid by a neuraminidase drastically decreases the
plasmatic
lifespan of the serum glycoproteins to minutes and promotes their uptake by
the liver (van den
Hamer et al., 1970; Morell et al., 1971). For example, when the pregnancy
hormone (more
recently recombinant HuEPO) is lacking sialic acid, its half-life is reduced
to 2 minutes,
whereas usually, its lifespan is around 48 hours. It is widely known at
present that sialic acid
is required to keep glycoproteins in blood because they prevent the
circulating proteins from
elimination by a sialoglycoprotein receptor (ASGPR or hepatic Lectin) composed
of two
subunits and included in the membranes of hepatocytes (Hudgin et al., 1974;
Kawasaki &
Ashwell, 1976; Bianucci & Chiellini, 2000). This receptor binds galactose or N-
acetylglucosamine residues of the desialylated N-glycans (Meier et al., 2000).
The recognized
glycoproteins are then internalized in endocytic vesicles covered of clathrin
and redirected
into lysosomes where they are degraded (Ashwell & Hardford, 1982).
This step appeared crucial for pharmacokinetic properties of therapeutic
recombinant
glycoproteins as lower organisms are not capable of sialylating proteins and
in CHO cells, the
addition of sialic acid is extremely sensitive to the energetic satus of the
cells in culture
(NeuAc is synthesized by condensation of pyruvate and lactic acid) and is
barely complete.
All the sialylated blockbusters (EPO, IFN, GM-CSF, FSH, Ab...) should be
purified for the
manufacturer to present a high sialic acid content and be controlled for batch-
to-batch
consistency.
Ik - Control of glycosylation & Allergenicity
Sialic acid residues at the terminal position of N-glycans are of major
importance for
therapeutic proteins as the sialylation of the proteins confers important
properties to
glycoproteins. The machinery required for the synthesis, the activation and
the introduction of
sialylated residues is poorly represented in the different recombinant
proteins expression
systems. Then, the recombinant human proteins produced are most often under or
even not-
sialylated compared to their native counterparts. Moreover, if they are
expressed in
mammalian cells, the sialylation may occur through N-glycolylneuraminic acid
(NeuGc)
which significantly differs from the N-Acetylated derivative (NeuAc) as it is
present in
mammalian cells but not in human cells.
Because there is an important issue with sialic acid in the market of
recombinant
drugs, many research teams are working on the modification of the N-
glycosylation process in
the different existing expression systems. Intensive work is aiming at
humanizing the
glycosylation pattern of the recombinant proteins to approach the pattern
found in the natural
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
glycoproteins as closely as possible and improve pharmacokinetics as well as
safety of the
product. Meanwhile, the procedure will tend to reduce the presence of
immunogenic
determinants.In this respect, a very recent study confirmed that 6-linked
sialic acid and 3-
linked sialic acid prevent glycoproteins to bind receptors of the immune
system and generate
5 activation (Glycobiology 2006). Adding 6-linked sialic acid to
recombinant glycosylated
proteins would therefore largely benefit to the safety of the drug and be
major advance in the
field.
Proteins of therapeutic interest were first extracted from natural sources
such as blood,
placenta, human or animal tissues. However, this approach is limited by the
quantity of
10 human tissues available and may bring in important risks of
contamination (viruses, prions,
oncogenes) and/or generate allergic reactions due to traces of animal proteins
or toxins. With
the rise of molecular biology, new approaches have been developped to produce
proteins
using quite different expression systems. A large range of heterologous
expression systems
are available (Andersen & Krummen, 2002) and each of them possess advantages
and
15 disadvantages with a particular attention given to the N-glycosylation
pattern of the
recombinant proteins produced in these systems.
lid- Biosynthesis of N-glycans in known expression systems: needs for
humanization of
the glycosylation pathway
Bacteria
One of the most used system is the bacteria Escherichia coli (E. coli)
(Swartz, 2001;
Baneyx, 1999), but its main inconvenient is that the human post-traductionnal
modifications,
particularly glycosylations, are not realised by this prokaryote because no
such
glycosyltranferases are expressed in E. coli. This can lead to misfolding and
subsequent
reject of the therapeutic proteins of interest by the immune system, reduction
in their lifespan
and biological activity.
No N-glycosylation of the human type has been found so far in this
microorganism.
Yeasts and filamentous fungi
Yeasts and filamentous fungi are also well-established eukaryote expression
systems
and they possess a cellular machinery approaching those of human cells. Yeast
produces
complex proteins and realizes several post-traductional modifications
including
glycosylations. Both yeasts and mushrooms typically produce mannose-rich
glycans by
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
61
adding until 100 mannose residues (concerning yeast) on top of the
pentasaccharidic core
(Tanner & Lehele, 1987; Herscovics & Orlean, 1993). Those hypermannosylation
definitely
foster immune response in human. Moreover, until now, no complex
oligosaccharide
containing sialic acid, galactose, fucose and N-acetylgalactosamine has been
found inside the
glycoproteins produced by these organisms (Blanchard, 2004).
The N-glycosylation process realized by yeasts and mushrooms is similar to the
mammalian process with respect to the initial steps in the ER but the presence
of
polymannans added in the golgi apparatus prevent them to get approval from
regulatory
bodies.
Insect cells
Proteins expressed in insect cells are properly folded, secreted and may
receive post-
traductional modifications. The post-traductional modifications carry out by
these cells are
similar to those realised by mammalian cells, in particular concerning the N-
glycosylation of
the protein. The glycan structures obtained in this case, are however
incomplete and
designated as paucimannose due to the presence of an undesirable N-
acetylglucosaminidase
activity which degrades the neoglycoproteins expressed during Baculovirus
expression
(Blanchard, 2004).
The lack of neuraminic acid in insects is still actively debated (Marchal et
al., 2001,
Lerouge et al 2005). In some insect cells lines, a1,3 linked fucose residues
are found and this
may trigger off an immune respons in human. At present, the use of this system
is therefore
restricted to produce vaccinal antigens.
Transgenic plants
As other eukaryotic cells, plants exhibit a complex and sophisticated cellular
machinery which may be used to produce therapeutic proteins. Recombinant
proteins possess
a very good pharmacological quality because plants express the enzymes
required for the
maturation of the proteins.
However, the glycosylation process needs several adjustments not to produced
allergenic proteins. Indeed, N-glycosylations in plants (Lerouge et al., 2000)
are similar to
those realized in humans as far as core glycosylation is concerned. The
glycosylations are still
lacking sialylated antennae and there is an addition of (31,2-xylose and a1,3-
fucose. Both
residues are highly immunogenic for human and currently considably compromise
the
approval of transgenic plants as expression systems for thepareutics.
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
62
Mammalian cells
Mammalian cells expression system, namely CHO cells, is currently the only
drug-
approved system to produce recombinant therapeutics. Such cells show a major
advantage
because they are able to synthesis complex proteins, achieve complex type N-
glycosylation of
high molecular mass. Mammalian cells naturally use, not only the enzymes
involved in the
synthesis and the transport of the nucleotides-sugars, but also the
glycosyltransferases
required to guarantee a complex glycosylation of heterologous recombinant
proteins with a
high level of a2,3-sialylation. However, there is lack of other enzymes such
as the a1,3/4-
fucosyltransferases and a2,6-sialytransferases, that transfer glycosidic
motifs specific of the
N-glycans of human tissues. In rodents, N-glycolylneuraminic acid is
substantially preferred
in N-glycans , 0-acetylation of N-Acetyl neuraminic acid at position 4, 7, 8
and most often 9
also occurs freqently and each of these derivatives may be potentially
immunogenic in
human. This represents a limitation for the use of mouse cells or lactating
mice/rabbits to
express recombinant proteins (Blanchard, 2004).
He - Glycoengineering
As discussed above, terminal sialylation of approved glycoproteins is the most
difficult step to obtain in all the expression systems available so far. As an
example, no a2,6-
liked sialic acid could be added while it is the prevalent linkage in human
blood. In all
organisms, the enzymatic machinery required is simply missing (Figure 6).
The ultimate goal of research in this field is improving the glycosylation
process, the
yield of production and the quality of the recombinant proteins. The use of
expression
systems genetically modified for glycosylation would allow to produce
glycoproteins with an
exceptional homogeneity in their glycanic structures. Such systems could be
then used to
develop a high level production for proteins of biomedical interest of well-
defined structures.
The glycosylation engineering in yeast (Hamilton et al., 2003; Roy et al.,
2000) has first
prevented the addition of polymannosidic chains. Then, enzymes which are
necessary to
galactosylation and N-acetylglucosaminylation have been added in the systems
(Maras et al.,
1999; Bretthauer, 2003; Vervecken et al., 2004). However, the addition of the
terminal sialic
acid is still difficult to realise, due to the number and location enzymes
involved in this
process but is currently being done in Japan (Figure 6).
In insect cells, the GTs catalyzing the tranfer of immunogenic sugars have
been
removed and the sialylation has been realised by adding 3 genes encoding for
the N-
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
63
acetylglucosamine 2-epimerase, N-acetylneuraminyl lyase and CMP-Neu5Ac
synthase (Jarvis
et al., 1998; Aumiller et al., 2003). Authors have created new insect cell
lines (SfSWT-3)
designed to synthesize their own CMP-sialic acid. The resulting cells express
all the 7
mammalian genes necessary to form an N-glycan, produce CMP-sialic acid and
sialylated a
recombinant protein when cultured in a serum-free growth medium (Aumiller et
al.,
2003).This work has been patented.
In plants, the first strategy used aimed to prevent the addition of allergenic
sugars by
stocking proteins in the ER. But in this case, the glycans can not be of the
complex type. An
other strategy was based on the inhibition of several GTs inside the golgi
apparatus. This
inhibition can be completed and/or can enter in competition with the
endogeneous machinery
for maturation. The addition of the sialyl machinery is also in process.
In the case of mammalian cells, work has been realized through the over-
expression of
an a2,3-ST and a (31,4Gal (Weikert et a/.,1999), these enzymes are both
present in the
genome but their activities vary upon culture conditions. This leads to a wide
variability
concerning the presence of the terminal Gal and sialic acid and also to
extensive
microheterogeneity in glycan structures of secretory proteins.
Researches have been mostly oriented in the optimization of the
galactosylation and
sialylation (Granbenhorst et al., 1999) by introducing an a2,6-ST (Bragonzi et
al., 2000). A
CHO cell line stably expressing full length a2,6-ST has been established long
ago and has
also been disappointing. The ratio observed between a2,6 and a2,3-linked
terminal sialic acid
residues carried was of 40,4% of a2,6- and 59,6% a2,3-sialic acid residues
improving
pharmacokinetics in clearance studies (Bragonzi et al., 2000). Despite
improvment in
humanization of cells, the ratio between a2,6 and a2,3-linked terminal sialic
acid residues
cannot be controlled and is not even favorable to the 6-activity.
In summary, the sialylation is a critical step to control glycan structures in
proteins
produced by genetic engineering. The main difficulty resides not so much in
expressing the
lacking activity but in getting the heterologous production system to work
successfully.
Indeed, 3 main objectives should be met to humanize glycosylation of current
expression
systems: 1) getting the donor substrate and the relevant transporter needed
for this enzyme to
work within the Golgi. 2) getting the enzyme properly compartimentalized to
eventually
compete with endogenoeous sialyltransferase activity. 3) getting the enzyme
able to catalyze
sialic transfer to any relevant acceptor substrate.
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
64
Ilf-Background of the invention:
The strategy of the inventors is based on the consideration that the full
length ST6
enzyme has been so far unable to meet the above objectives, especially the
competition with
the endogenous ST3 activity existing in most mammalian cells, probably because
the gene
product is not inserted in intracellular compartments involved with the
secretory pathway.
The inventors thus developped a procedure which may afford the best
opportunity for a
transferase CD to reach the golgi compartments where the neoglycoproteins are
sorted into
secretory vesicles and/or where they meet the engineered ST before/instead of
the existing
ST3
III ¨ Design of synthetic membrane STs
The present invention consists in a procedure which delivers a panel of
chimeric STs
of known catalytic activity, possessing a membrane anchor to target them to
the Golgi
apparatus of eukaryiotic cells. None of them exist in living cells and are
considered as
"synthetic STs" because their construction does not need any of the DNA
sequence coding for
CT, TMD or SR. The relevant oligonucleotides are within 200pb (60 amino acids)
in length
and could be designed by informatics and obtain commercially. Using PCR
methods, a tagged
synthetic anchor is constructed to code for the N-terminal half of the ST
protein and fused to
the optimized catalytic domain of the hST6GalI.
The invention describes various molecular methods to construct 3 types of
membrane
chimeric STs:
= homologous construct: ST anchor fused to the CD A89 of ST6Ga1. Synthetic
or
eventually copied by PCR. In this case, CT+TMD+SR portions are from the same
ST:
ST3Gal/ST6GalNAc/ST8Sia. The example describes a 200pb sequence
= hybrid construct: CT+TMD are from a ST and the SR from another ST. Here too,
the minimal size is 200pb.
= Heterologous construct: CT, TMD and SR fragments are from different STs.
Here
too, the minimal size is 200pb.
= long construct : a CT+TMD+ SR1 construct of 200pb is prepared and fused
to a
second 5R2 construct of at most 200pb in such a way that 5R2 is dowstream from
SRI. The
dupicated constructed can be further ligated at the 3' end to any other SR
construct as as
repeatedly as needed.
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
The size of 200pb has been based on the observation that the shortest N-
terminal
portion of all human STs is ST6Ga1NAcIII with CT+TMD+SR= 200pb. This enzyme
has
virtually no stem.
At present, the inventors have validated a method to construct synthetic
fragment of
5
DNA and fuse them to a minimal catalytic domain to generate a novel
transferase enzyme of
desired activity. Several representative constructs are available for further
expression. The
synthetic chimeras are active and are being studied in CHO cells using
transient expression
and confocal microscopy.
10 EXAMPLES
Example 1: Producing the minimal catalytic domain (CD) of hST6Ga1 I (SEQ ID
NO:
43 coding for SEQ ID NO : 44)
The human ST6Ga1 I (hST6Gal I) cloned by Legaigneur et al., 2001 was used to
15
amplify the minimal CD cloned into the pFLAG-CMV-vector (Sigma) (Donadio et
al., 2003)
that was used in all the chimera constructs.
The recombinant vector was first digested to verify the presence of the CD of
hST6Gal
I. The running agarose gel reveals the presence of a nucleotidic band at
around 1000 pb,
corresponding to the expected size for the minimal catalytic domain (Figure 10
A-C).
20
The CD corresponding to the amino acids 90 to 406 of hST6Gal I was obtained by
PCR using the following primers:
5 'BamHI-44: 5 '-GAGCCCGGATCCGAGGCCTCCTTC-3 ' and
3 '44-XbaI: 5 ' TAACCCTCTAGATTAGCAGTGAATGGTCCGGAAGC-3 ' .
These primers contain BamH1 (5'BamHI-A.4) and Xbal (3'44-XbaI) restriction
sites
25
and the natural stop codon of the sequence. The BamH1 restriction site was
used to ligate the
CD of hST6Gal 90-406 to the synthetic N-terminal part of other STs to form
chimeric
enzymes. The Xbal restriction site at the 3' end was used to introduce the
chimera inside the
pcDNA3.1 vector (Invitrogen) (Figure 11).
PCR was performed on an 1-Cycler apparatus (Biorad), using 2.5 units of
ProofStart
30
DNA polymerase (Qiagen), 300 i.IM of each dNTP (Sigma Aldrich), 1 i.IM each
primer, 1X of
ProofStart manufacturer buffer (Qiagen), lx of manufacturer Q-Solution
(Qiagen) and 1.5
mM of Mg2+ in a final volume of 50 1 with 200 ng of pFlag-CMV-hST6Gal 1-90-
406. The
reaction was performed as follows: 95 C for 5 min, followed by 40 cycles of 30
s at 94 C, 30
s at 55 C and 1 min at 72 C. The amplified PCR product was analyzed by
electrophoresis on
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
66
a 1,5% agarose gel (amplification of one PCR product at the expected size of
1000 pb; figure
17 B) and purified using the Gel Extraction kit Qiagen according
manufacturers' instructions.
Several PCR amplification products were pooled and concentrated to estimate
the
amount of DNA in an agarose gel (Figure 10 C). Purified PCR products were kept
at -20 C
until used.
Example 2 : Assembling synthetic N-terminal domains
To create a wide array of STs as sorted by bioinformatic analysis, CT, TMD and
SR
from distinct STs were synthesized and assembled.
All the selected N-terminal parts of the chimera contain the 3 typical regions
of the ST
family: the CT, the TMD and the SR. Of note, the SR may be of variable length
and could
originate from synthetic gene duplication. All of these fragments i.e
CT+TMD+SR have been
entirely reconstituted using complementary hybridation and phosphorylation
steps as
described below.
a- Construction of the non-catalytic domain of hST3Gal III (SEO ID NO : 151
encodinz SEO ID NO: 152)
Six sense oligonucleotides and five antisense oligonucleotides corresponding
to part of
the N-terminal region (1 to 138 pb) sequence of hST3Gal III were designed and
synthesized
(Eurogentec). This example shows the method of synthesizing a naturally
occurring sequence
of interest. A FLAG epitope (in Bold and underlined) was added between the
initiating codon
ATG and the second codon (GGA) of the hST3Gal III sequence:
ATG GACTACAAAGACGATGACGACAAG GGA.
One microgram of each internal nucleotide was phosphorylated using 1 unit of
polynucleotide kinase (Eurogentec) in a kinase buffer containing 2 mM of ATP
(Sigma).
Reaction was performed for 60 min at 37 C, and the incubation finally
inactivated for 10 min
at 65 C. All the oligonucleotides were separately denatured for 10 min at 80 C
and then
mixed for matching. Matching was performed overnight with a decreasing
temperature
gradient from 80 C to 20 C. The resulting fragment was then subjected to PCR.
b- Construction of the non-catalytic domain of hST6Ga1NAc 1-74 (SEO ID NO:
145 encodinz SEO ID NO: 146)
One of the shortest stem within the ST family is the stem of the hST6Ga1NAc
III,
which is composed of around 6 amino acids. A possible N-terminal domain of
this ST is of 74
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
67
amino acids (CT, TMD and SR) which correspond to 222 pb. Therefore to
delineate a
construct with the shortest SR, a synthetic N-terminal domain of 222 pb
corresponding to the
74 first amino acids of the hST6Ga1NAc I was reconstituted.
Nine sense oligonucleotides and eight antisense oligonucleotides corresponding
to the
complete N-terminal region (1 to 222 pb) sequence of hST6Ga1NAcI were designed
and
synthesized (Eurogentec). A FLAG sequence (in Bold and underined) was added
between the
initiating codon ATG and the second codon (GGA) of the hST6GaNAc I sequence:
ATG GACTACAAAGACGATGACGACAAG GGA.
The phosphorylation and the matching reactions were performed as previously
described.
c-Synthetic olizonucleotide duplication to assemble the non catalytic domain
hST6Ga1NAc 11-140 (SEO ID NO: 148)
hST6Ga1NAc I was selected as the sialyltransferase exhibiting the longest SR
(246
aas). In this example, the length of the synthetically reconstituted CT+TMD+SR
portion (1- to
35 amino acids residues) was joint to another SR stretch (36-140 amino acids
residues ) to
duplicate the length and be able to add a membrane anchor of 140 amino acids
residues as
reported in Donadio et al., 2003.
d-Construction of a hybrid synthetic domain : hST3Ga1 III 1-28-hST6Ga1NAc 137-
74 (SEO ID NO: 160)
An hybrid N-terminal region containing first the CT and the TMD of the hST3Gal
III
(1 to 28 amino acids residues) and second the begin of the SR of hST6Ga1NAc I
(amino acids
37 to 74) was constructed.
The total length of this synthetic hybrid is of 66 amino acids corresponding
to 198
nucleotides (SEQ ID NO : 159). Nine sense oligonucleotides and eight antisense
oligonucleotides corresponding to the hybride N-terminal region described
above (1 to 225
pb), using the sequences of hST3Gal III/hST6Ga1NAc I, were designed and
synthesized
(Eurogentec). A FLAG epitope (in Bold) was added between the initiating codon
ATG and
the second codon (GGA) of the hST3Gal III sequence:
5'- ATG GACTACAAAGACGATGACGACAAG GGA -3'.
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
68
The phosphorylation and the matching reaction were performed as previously
described.
e-PCR amplification of the synthetic constructs
After the synthetic reconstitution step, the product was amplified by PCR
technique
using specific primers that bring in the desired restriction sites at each end
of the reconstituted
DNA fragment in order to ligate them first with the selected CD and then into
the vector
pcDNA3.1.
The following primers were designated:
5'AflII-ST3: 5'-GAGCCCCTTAAGATGGACTACAAAGACGACGATGACG-3'
and 3' 5T3-BamHI: 5 '-TAAGGGGGATCCGCTAGAGTGACTATACTTACTGGA-3;
5 'AflII-S T6: 5 '-GAGCCCCTTAAGATGGACTACAAAGACGACGATGACG-3 '
and 3' 5T6-BamHI: 5 '-TAAGGGGGATCCGGTTGTGGAGGAACGGGA-3 ';
3' ST6-BamHIn 2: 5 '-TAAGGGGGATCCTCTGGGTGACAGTGTGTTCAC-3 ' .
These primers contain AflII (5'AflII-ST3 and 5'AflII-ST6) and B amHI (3' ST3-
BamHI, 3' ST6-BamHI and 3' ST6-BamHI n 2) restriction sites to ensure the
further
ligations.
PCRs were carried out with the primer pairs set described in Table 5. They
were
performed on a PCR apparatus (I-Cycler, Biorad) with 5 L of the each
synthetic fragments in
a solution containing 2.5 units of ProofStart DNA polymerase (Qiagen), 300 M
of each
dNTP (Sigma Aldrich), 1 M each primer, lx of ProofStart manufacturer buffer
(Qiagen),
1X of manufacturer Q-Solution (Qiagen) and 1.5 mM of Mg2+ in a final volume of
50 L. a
The reaction was performed as follows: 95 C for 5 min, followed by 40 cycles
of 30 s
at 94 C, 30 s at 55 C, 53 C, 53 C and 55 C respectively for hST3Gal III (SEQ
ID NO :
151), hST6GaNAc 1-74 (SEQ ID NO : 145), hST6GaNAc 1-140 (SEQ ID NO : 147) and
hST3Gal III-28/37-hST6Ga1NAc 1-74 (SEQ ID NO: 159), and 1 min at 72 C. The
amplified
PCR product was analyzed by electrophoresis on a 2% agarose gel, according to
the PCR
product size (Table 5):
- hST3 Gal III: only one PCR product was amplified at an estimated size of
170 pb,
which corresponds to the expected size of 174 pb (Figure 11).
- hST6 Gal NAc I: Figure 13 shows one amplified PCR product with an
estimated
size of 270 pb, which corresponds to the expected size of 270 pb .
- hST6Ga1NAc 1-140: a 468 pb nucleotidic sequence was amplified (Figure
15).
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
69
- hST3Ga1 III-28-hST6Ga1NAc 137-74: a 249 pb nucleotide sequence (Figure 17)
was
amplified.
The PCR products were purified using the Gel Extraction kit Qiagen according
manufacturers' instructions.
- The PCR product of hST6Ga1NAc 1-140 was submitted to direct both strand DNA
sequencing by Genome Express (Meylan, France).
- The PCR product of hST3Ga1 III and hST6Ga1NAc 1-74 were ligated to the CD
(SEQ ID NO : 43) before being introduced in the pcDNA3.1 vector (Invitrogen)
to be also
further sequenced by Genome Express (Meylan, France).
- The hST3Ga1 III-28-hST6Ga1NAc 137-74 synthetic fragment was ligated with the
CD (SEQ ID NO : 43) and directly sequenced in both strand prior its
introduction into the
pcDNA3.1 vector (Invitrogen).
f-Ouantification of the PCR products
All the PCR products were pooled and concentrated according to each synthetic
construct. Two volumes ethanol and 0,1 volume of 3 M sodium acetate, pH 5,2
were added to
PCR product samples. The mix was incubated for 30 min at -20 C and centrifuged
20 min at
10000 g at 4 C. The pellets were washed with 100 1_, of 70% ethanol and a
centrifugation
was run for 10 min at 10000 g at 4 C. Then supernatants were removed and the
pellets dried
and resuspended in 50 1_, of purified water. Samples were conserved at -20 C
until use.
Five microliters of the four concentrated synthetic fragments were loaded on a
2% agarose gel
to estimate their quantity (Figures 11, 12, 13 and 14).
Example 3: Assembling non-catalytic N terminal membrane domains with a sialyl-
transferase activity
a-Digestions and purification
All the PCR products (either the synthetic N-terminal constructs or the
catalytic
domain) were digested by the BamHI restriction enzyme.
200 nanograms of the synthetic N-terminal constructs were digested in a
solution
containing 3 units of the BamHI restriction enzyme, 0.1 ps. L-1 of Bovin Serum
Albumin and
1X of the appropriate manufacturer's buffer E (Promega) in a final volume of
20 L.
500 nanograms of the CD were digested with 5 units of BamHI, 0.1 ps.pri of
Bovin
Serum Albumin and 1X of buffer E (Promega) in a final volume of 20 L.
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
Digestions were performed for 90 min at 37 C and the incubation was finally
inactivated for 15 min at 65 C. Digested DNA was purified using the PCR
purification kit
(Qiagen).
5 b-Ligation
Each of the N-terminal fragments were ligated to the CD fragment in a solution
containing 1.5 units of T4 DNA ligase, 1X of the ligation buffer, both
commercialized
(Promega), 62.5 ng, 78.2 ng and 75 ng, respectively for hST3Ga1 III, hST6GaNAc
1-74, and
the hybrid hST3Ga1 III-28-hST6Ga1NAc 137-74 fragment and 100 ng of CD in a
final volume
10 of 20 L. The mix was incubated at 15 C overnight and followed by an
inactivation step for
10 min at 70 C (Figure 17).
c-Amplification of the tagged synthetic insert
To verify the proper ligation, the ligation products were directly submitted
to PCR
15 using the following primers pairs: 5'AflII-ST3/3'44-XbaI, 5'AflII-
ST6/3'44-XbaI and
5'AflII-ST3/3'44-XbaI respectively for the ST3/CD, the ST6Ga1NAc-74/CD and the
hybrid/CD.
Reactions were carried out using 2.5 units of ProofStart DNA polymerase
(Qiagen),
300 M of each dNTP (Sigma Aldrich), 1 M each primer, 1X of ProofStart
manufacturer
20 buffer (Qiagen), lx of manufacturer Q-Solution (Qiagen) and 3 mM of Mg2+
in a final
volume of 50 L. The reaction was performed as follows: 95 C for 5 min,
followed by 40
cycles of 1 min at 94 C, 1 min at 57 C, and 1 min 30 s at 72 C.
The amplified PCR products were analyzed by electrophoresis on a 1,5 % agarose
gel:
- hST3Gal III/CD synthetic insert (SEQ ID NO: 167): a 1200 pb PCR product
was amplified
25 (Figure12), which corresponds to the expected size of the ligated DNA
fragment: ST3Ga1 III
plus CD.
- hST6Ga1NAc I-74/CD insert (SEQ ID NO : 161): a 1200 pb PCR product was
amplified
(Figure 14 ), which corresponds to the expected size of the ligated DNA
fragment :
ST6GaNAc I plus CD that is of 1225 pb exactly.
30 -hST3Ga1111-28/37hST6Ga1NAc-74/CD insert (SEQ ID NO : 175): a 1200 pb
PCR product
was amplified (Figure 17) which corresponds to the expected size of the
ligated DNA
fragment hST3Ga1111-29/hST6Ga1NAc37-74 plus CD.
The amplified PCR products were purified using the Gel Extraction kit Qiagen
according manufacturers' instructions.
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
71
Amplified PCR products of the same synthetic ST were pooled, concentrated and
quantified on a 1,5% agarose gel (Figures 12, 13 and 16), as previously
described above, to
obtained enough quantity of DNA to ligate the chimera into the pcDNA3.1
vector.
d- Cloning the synthetic ST insert into an expression vector
1-Digestion and purification
Synthetic inserts such as the hST3Gal III/CD or hST6Ga1NAc I-74/CD insert were
ligated into the pcDNA3.1 vector into AflII and Xbal resrict ion sites.
The synthetic insert and the pcDNA3.1 vector (Figure 12) (Invitrogen) were
first
digested by the restriction enzyme Xbal:
- for each construction, 500 ng of vector were digested using 2.5 units of
Xbal
(Promega), 0.1 ps.pri of BSA and lx of the appropriate buffer D, supplied by
manufacturer
(Promega), in a final volume of 20 pt, following the manufacturer's
instructions;
- the insert was digested with 2 to 2.5 units of XbaI (Promega), depending
on the
quantity of insert available after PCR (e.g. 200 ng hST3Gal III/CD, hST3Gal
III-
28/hST6Ga1NAc I 37-74/CD and 500 ng of hST6Ga1NAc I-74/CD were respectively
digested
with 2 and 2.5 units of Xb aI), 0.1 ps.pri of BSA and lx of Buffer D
(Promega), in a final
volume of 20 L.
The digestions were performed at 37 C during 60 min and inactivated at 65 C
for 15
min.
The digested products were then submitted to a digestion by the restriction
enzyme
AflII following the manufacturer's instructions (Biolabs): using the
appropriate number
enzyme units, depending on the DNA quantity (2, 2, 2.5 and 2.5 units,
respectively for
hST3Gal III/CD, hST3Gal III-28/hST6Ga1NAc I 37-74/CD, hST6Ga1NAc I-74/CD and
pcDNA3.1), 0.1 ps.pri of BSA and lx of buffer 2 (Biolabs) in a final volume of
50 L. The
digestions were carried out at 37 C for 60 min and inactivated at 65 C for 20
min. The
digestions were purified with the PCR purification kit (Qiagen).
2-Ligation
The ligation conditions were calculated according to the following formula:
iiiiiiiiacgi.gggg444;gggig44gggggf44.4ggg4g4iizfivfiftiiiiiiig
wiiiiiiiiiiiiiiiiiiiiiiiiiiiiiioduipbNettativamoiliti11100111542$11111111111111
1111111111111111111111111111111111111111111111111111111111111111111111111111111
111111111111111yettokiniiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiimii
immilmm111111111111111111111111111111111111111111111111111111111111111111111111
1111111111111111111111111111111111111111111111111111111111111111111111111111111
1111111111111111111111111111111111111111111111111111111111111111111111111111111
1111111111111111111111111111111111111111111111111111111111111111111111111111111
1111111111111111111111111111111111111111111111111111111111111111111112
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
72
where the size of the insert is around 1200 pb for the synthetic construct
(e.g. hST3Ga1
III/CD, hST3Ga1 III-28/hST6Ga1NAc I 37-74/CD or hST6Ga1NAc I-74/CD), the size
of the
vector pcDNA3.1 is of 5428 pb, the quantity of vector used is of 50 ng or 100
ng, and finally
the molar ratio of Insert/Vector is of 3/1.
= Construction of pcDNA3.1/ST3/CD recombinant vector was performed using
33 ng of the digested chimera hST3Ga1 III/CD (SEQ ID NO : 167), 50 ng of
pcDNA3.1
digested vector, 3 units of T4 DNA ligase enzyme (Promega) and lx of the
ligation buffer in
a final volume of 15 L. The mix was incubated overnight at 4 C and the
reaction was
inactivated for 10 min at 70 C.
= The second construction of pcDNA3.1/5T6/CD recombinant vector was
performed using 66,5 ng of the digested chimera hST6Ga1NAc I-74/CD (SEQ ID NO
: 161),
100 ng of pcDNA3.1 digested vector, 3 units of T4 DNA ligase enzyme (Promega)
and 1X of
the ligation buffer in a final volume of 15 L. The mix was incubated
overnight at 15 C and
the reaction was inactivated for 10 min at 70 C.
= The third construction of pCDNA3.1/HYB/CD recombinant vector was
performed using 66.5ng of the digested chimera hST3Gal III-28/hST6Ga1NAc I 37-
74/CD
(SEQ ID NO : 175), 10Ong of pCDNA3.1 vector, 3 units of T4DNA ligase enzyme
(Promega)
and lx of ligation buffer in a fianl volume of 15 L. The mix was incubated
overnight at
15 C and the reaction was inactivated for 10 min at 70 C.
3-Cloning
Competent cells chemical transformation
One Shot TOP10 chemically competent E. coli (Invitrogen) were transformed
with
the recombinant vector following the manufacturer's instructions. Two
microliters of each
ligation reaction were added to 25 pL of chemical competent cells and mixed by
taping
gently. The vials were incubated on ice for 30 min. The chemical
transformations were
performed for exactly 40 s in a 42 C water bath. Vials were removed from the
42 C bath and
place on ice during 2 min. Two hundred and fifty microliters of pre-warmed
S.O.C. medium,
provided by Invitrogen, were added into each vial under sterile conditions.
Then, the mixtures
were shaked at 37 C for exactly 1 h at 225 rpm in a shaking incubator. Each
transformations
were spread on LB agar plates (10 g of bacto-tryptone, 5 g of bacto-yeast
extract, 10 g of
NaC1 and 15 g of agar per liter of solution from GibcoBRL) containing 50 ps/mL
of
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
73
ampicillin under sterile conditions. Plates were inverted, incubated at 37 C
overnight and
maintained at 4 C the next day until the transformants selection.
Amplification
At the end of the day, single colonies were isolated and inoculated into 3 mL
of LB
(10 g of bacto-tryptone, 5 g of bacto-yeast extract, 10 g of NaC1 from Difco)
containing 50
iig/mL of ampicillin. The growth was ensured at 37 C in a shaking incubator
overnight. The
next day, glycerol stocks of cultures were prepared by mixing 0.85 mL of
cultures with 0.15
mL of sterile glycerol and transferred to a cryovial. Glycerol stocks were
stored at -80 C until
sequence verification and further use.
Minipreparation of plasmids
The recombinant vectors were purified by Minipreparation procedure (Maniatis),
1,5
mL of cultures were isolated and centrifuged for 5 min at 10000 g to pellet
the bacteria. Cold
Solution I (5mM glucose, 25 mM Tris-HC1 pH 8,0, 10 mM EDTA pH 8,0) was added
(100
L) to resuspend the pellet, 200 L of Solution 11 (0,2 N NaOH and 1% SDS) and
150 L of
Solution III (3 M potassium and 5 M acetate) were also added. The mixtures
were gently
mixed by vortexing, placed on ice 5 min, and centrifuged at 4 C, 5 min at
15000 g. The
supernatants were taken and 1 volume of isopropanol was added. The
minipreparations were
incubated 5 min at room temperature and centrifuged at 4 C, 5 min at 15000 g.
Then, the
supernaptants were removed. The DNA pellet was dried and resuspended into 50
pL of water.
The quantity of DNA was mesured using a spectrophometer DO apparatus
(Biophotometer,
Eppendorf). At least 30 minipreparations are performed to screen the presence
of the interest
insert for each construction.
One microgram of each of the 30 minipreparations was double digested using
AflII
and Xbal restriction enzyme in order to verify the presence of the insert.
First, the digestion
solution contained 10 units of XbaI (Promega), 0.1 ps. L-1 of BSA and lx of
the appropriate
manufacturer buffer D (Promega) in a final solution of 20 L. Digestions were
performed in a
37 C water bath for 1 h and inactivated at 65 C for 15 min. Samples were
secondly digested
using 10 units of AflII restriction enzyme (Biolabs), 0.1X of BSA and lx of
Buffer 2
(Biolabs) in a final volume of 50 L. Reactions were carried out in a 37 C
water bath for 1 h
and inactivated at 65 C for 20 min. The digested samples were all purified
using the PCR
purification kit (Qiagen). The digestion products were loaded on a 1,5%
agarose gel to detect
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
74
the presence of the chimeric insert. The electrophoresis was run at 100 V
during around 35
min.
In the case of the pcDNA3.1/ST3/CD and pcDNA3.1/ST6/CD, an insert of 1200 pb
was detected (Figure 19 C or 23 C). The size of this insert corresponds
exactly to the
expected size of the reconstituted chimera ST3Ga1 III/CD or ST6Ga1NAc I-74/CD.
The positive clones were fully sequenced to assess the expected inserted DNA
sequence.
Sequencing
The cloning steps were verified by both strands DNA sequencing by Genome
Express
(Meylan, France) using universal primers present inside the vector: T7
Promoter and BGH
Reverse.
The final expected sequences of the three chimeras ST3Ga1 III/CD (SEQ ID NO :
167), hST3Gal III-28/hST6Ga1NAc I 37-74/CD (SEQ ID NO : 175), and ST6Ga1NAc I-
74/CD (SEQ ID NO : 161). The nucleotidic sequence obtained was aligned to the
expected
sequence and the alignment revealed 100 % identity between these three
sequences. The
deduced amino acid sequence was also aligned with the expected sequence of the
chimera
(http://www.infobiogenleservices/analyseq/cgi-bin/alignp_in.p1) and it shows
100 % identity
between two of theses amino acid sequences.
The nucleotide sequence obtained for hST6Ga1NAc 1-140 showed 84% identity with
the theoretical sequence. The missing parts of the sequence are the 36 first
and the 39 last
nucleotides, they both correspond to the primers designed to sequence the DNA
fragment in
both strands. Moreover the amino acids alignment between the expected and the
resulting
sequences showed 76,9% identity. The 12 first and the 24 last amino acids are
not found due
to their correspondences with the specific primers used for the sequencing.
Minipreparation
After the DNA insert sequence verification, the recombinant plasmids were
amplified
in 100 mL cultures to obtain high quantity of each construction
(pcDNA3.1/5T3/CD and
pcDNA3.1/5T6/CD and pcDNA3.1/HYB/CD). A single colony of each plasmid from a
freshly streaked selective plate was picked and inoculated into a starter 3 mL
culture of LB
medium containing the selective antibiotic (ampicillin 50 ps.).1L-1). The
cultures were
incubated overnight at 37 C with vigorous shaking. The started cultures were
diluted into 100
mL of selective LB medium and incubated once again overnight at 37 C with
vigorous
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
shaking. The bacterial cells were harvested and the recombinant vectors were
purified using
the QIAGEN Plasmid Midi Kit (Qiagen) following the manufacturer's
instructions. The
yield was determined with an UV spectrophotometry to quantify the DNA
concentration
(biophotometer, Eppendorf). Each construct was stored at -20 C until used.
5
Example 4 : Functional expression of the synthetic constructs in CHO cells
The expressed inserts are represented in Figure 15.
A-Transient expression of synthetic ST6s in CHO cells
The CHO-Kl cell line was used to express constructions as described by Donadio
et
10 al. (2003). Briefly, cells were grown in Ham medium supplemented with
10% of Fetal Calf
Serum (FCS), fungizone (2.5 ps/mL) and gentamicin (50 ps/mL) at 37 C in a 5%
CO2
incubator. Transfections with FLAG-CMV vector constructs were carried out
using the
LipoFectamine reagent following the recommended procedure of manufacturer,
with 3 ps of
recombinant plasmid DNA. Immunofluorescence experiments were run after 36-48 h
of
15 transfect ion.
Double labelling with FITC-SNA and anti-FLAG mAb was performed as described by
Donadio et al. (2003). Cells were fixed in a 1% para-formaldehyde (PFA
solution, saturated
with 1% BSA, washed with phosphatase-buffered saline (PBS) and incubated with
FITC-
SNA. Cells expressing a 5T6 activity are labeled in green. Samples were washed
twice with
20 PBS and fixed in 1% PFA, further washed with PBS, incubated in 0.05 M
NH4C1 and washed
with PBS. Cells were permeabilized with 0.1% Triton in PBS and further
incubated in PBS
containing 10% goat serum. Anti-FLAG labeling was carried out using the M2
anti-FLAG
mAb (1:600) in PBS containing 5% inactivated goat or horse serum followed by
incubation
with a secondary anti-mouse IgG antibody labeled with Alexa Fluor 568 (1:200)
in the same
25 buffer. Cells expressing the FLAG-tagged sialyltransferase are labeled
in red. After washings,
cells were mounted in Mowiol and kept at -20 C in dark. Confocal microscopy
was
performed with an Olympus or Leica instrument. Confocal images were processed
with a
Metamorph Imaging System. Volumes were originally traced as 24-bit TrueColor
Images and
transferred to Adobe Photoshop as RGB TIFF or JPG files. Under such
conditions,
30 untransfected CHO-Kl cells are not labeled with either of the above
reagents.
B- Enzymatic activity of synthetic ST6s in CHO cells
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
76
B1 Activity of the optimized catalytic domain of hST6Ga1 I
The hST6Ga1 I CD, was cloned into the pFLAG-cytomegalovirus vector (pFLAG-
CMV1) from Sigma, using a preprotrypsinogen signal peptide. This construction
and these
plasmids were previously characterized in vitro by Legaigneur et al. (2001)
and shown to give
an enzymatic protein of the expected size released into the cell medium .This
soluble CD was
found equally active on known exogeneous glycoprotein acceptors of various
degree of
branching.
To generate the minimal CD soluble form of hST6Ga1 I, a PCR fragment encoded
between the
5' (5'-TAATAAAGCTTGAGGCCTCCTTCCAG-3') introducing a HindIII restriction
site and the
3' (5'-CTATTGGATCCTTAGCAGTGAATGGT-3') encoding a BamHI site was
generated. The fragment was then HindIIIIBamHI digested and inserted into the
pFLAg-
CMV1 mammalian expression vector and subcloned into the pBluescript II KS
vector
(Stratagene) for further constructions.
The minimal ST6Ga1 I CD (SEQ ID NO : 44) tagged with the FLAG epitope (Figure
jJ is expressed at a high level in CHO cells (Figure 18 B, D). Its activity
and localization
was characterized using double labeling with anti-FLAG monoclonal Antibody and
FITC-
SNA. Interestingly, the mutant FLAG-CD was highly efficient in sialylating
cell surface
acceptors as shown in Figures 18 C and D.
B2- Functional expression of ST8/CD
CHO-Kl cells were transiently transfected with chimeric forms of hST8SiaIV (1-
67)/CD (SEQ ID NO : 173), hST8Siall (1-79)/CD (SEQ ID NO : 171). Their
activity and
localization were characterized using double labeling with anti-FLAG
monoclonal Antibody
and FITC-SNA, as shown in Figure 19.
Both hST8SiaIV (1-67)/CD (SEQ ID NO : 174) and hST8SiaII (1-79)/CD (SEQ ID
NO : 172) chimeras were found active on endogeneous cell acceptors as the SNA
lectin
reveals an intense cell surface labeling (Figure 19 B, C, E and F), indicating
that the CD is
active independently of the origin of N-Terminal part fused to the catalytic
domain. The a2,6-
sialyltransferase activity is maintained for the both chimeras, indicating
that in the chimerical
enzyme, the information contained in the minimal CD is necessary and
sufficient for acceptor
recognition and transfer efficiency.
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
77
B3-Functional expression of ST3/CD and hybrid ST3-ST6Ga1NAc/CD
CHO-Kl cells were transiently transfected with chimeric constructs of hST3Ga1
III
/CD (SEQ ID NO : 167), and 5T3 1-28-ST6Ga1NAc37-74/CD (SEQ ID NO : 175). Their
activity and localization were characterized using double labeling with anti-
FLAG
monoclonal Antibody and FITC-SNA, as shown in Figure 19.
Again, both chimeras were found active on endogeneous acceptors as SNA binding
reveals an intense cell surface labeling (Figure 19 B, C, E and F). This
indicates that the CD is
active independently of the origin of CT+TMD+SR added. Since a2,6-
sialyltransferase
activity is maintained for both chimeras, it can be also concluded that the
information
contained in the minimal CD is necessary and sufficient for full transfer
efficiency.
Therefore, fusing 5T3/8 sequences upstream from the CD, does not alter the
expression of a 5T6 catalytic activity.Since rat and human 5T3 GalIII and more
generally
rodent and mammalian ST3Ga1 1111 are identical in their N-terminal
(CT+TMD+juxtamembrane SR) portion, it also appears that exchanging any of this
peptide
portion with non-human heterologous has no effect on 5T6 catalytic activity.
Futhermore,
substitution of CT, TMD and SR with heterologous portions selected among
enzymes of 0-
glycosylation like ST GalNAcI does not alter recognition of intracellular
acceptors nor
enzymatic activity.
B4- Functional expression of hST6Ga1NAc 1-74/CD and hST6Ga1NAc 258/CD
A similar approach was used to estimate the expression and activity of
hST6Ga1NAc I
(1-74)/CD (SEQ ID NO : 162) and hST6Ga1NAc I-258/CD (SEQ ID NO : 166) enzymes.
Both chimeric transferases are active: SNA binding revealed a similar intense
cell surface
sialylation and the FLAG labeling shows identical localization within the
Golgi (Figure 21G,
H and I).
It can thus be concluded that the length of the SR region can be widely
variable and
extended up to 258 residues without affecting the functional expression and
intracellular
localization of a chimerical ST6Ga1 catalytic activity. This also further
confirms that the
length of the heterologous N-terminal membrane anchor does not affect the
biological active
conformation of the newly designed sialyltransferase.
Based on the above examples, it can be stated that the process described in
the
invention can abolish the regulatory control of the SR region over the CD
activity. As a result,
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
78
any possible combination of CT+TMD+SR among glycosyltransferases
(sialyltransferases) of
the N- or 0-glycosylation pathway can be selected to equip host cells with a
needed
transferase activity through a relevant CD. Furthermore, the process described
in the
invention is also able to target a needed activity in intracellular
compartments in which
neosynthesized glycoproteins migrate to the cell surface. This finding is of
crucial importance
for properly sorting and secreting sialylated proteins of therapeutic interest
in cells (from
yeast to human) engineered as described in the invention.
Example 5 : Construction of a hybrid synthetic domain: hST3Ga1 III / hST8Sia
II /
hST6GaINAc I, and fusion to the CD of hST6Ga1 I
An hybrid N-terminal region (SEQ ID NO : 180) containing first the CT of
hST3Gal
III (1 to 8 amino acids residues ; SEQ ID NO : 66), then the TMD of hST8Sia II
(7 to 23
amino acids residues; SEQ ID NO : 120) and finally the SR of hST6Ga1NAc I
(amino acids
36 to 74; SEQ ID NO : 130) was constructed. A FLAG epitope (in Bold) was added
between
the initiating codon ATG and the second codon (GGA) of the hST3Gal III
sequence:
5' ¨ ATG GACTACAAAGACGATGACGACAAG GGA ¨3'
The total length of this hybrid is 72 amino acids (SEQ ID NO : 180)
corresponding to
216 nucleotides (SEQ ID NO : 179). Eight sense oligonucleotides and seven anti-
sense
oligonucleotides corresponding to the hybrid N-terminal region described above
(1 to 216
nucleotides), using the sequences of hST3Gal III/hST8Sia II/hST6Ga1NAc I, were
designed
and synthesized (Eurogentec).
The phosphorylation and the matching reaction were performed as previously
described.
PCR amplification of the synthetic construct
After the synthetic reconstitution step, the product was amplified by a high
fidelity
PCR using specific primers that bring in the desired restriction sites at each
end of the
reconstituted DNA fragment in order to ligate it into the pcDNA3.1 vector. The
AflII and
B amHI restriction sites were respectively introduced on the 5' and the 3'
extremities.
The following primers were designated:
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
79
'AflII-S T3 : 5 '-GAGCCCCTTAAGATGGACTACAAAGACGATGACG-3 '
and 3' ST6-BamHIn 3: 5'-TAAGGGGGATCCGGTTGTCCTCCTTGCCCT -3'
The PCR was performed on a PCR apparatus (I-cycler, Biorad) with 5 L of the
5 synthetic fragment in a solution containing lx of ProofStart manufacturer
buffer and lx of
manufacturer Q-Solution, 1.5 mM of Mg2+, 300 M of each dNTP (Sigma Aldrich),
1 M
forward primer AflII-5T3, 1 M reverse primer 5T6-BamHIn 3, 2.5 units of
ProofStart DNA
polymerase (Qiagen) in a final volume of 50 L. The thermocycling profile used
was: 95 C
for 5 minutes, followed by 40 cycles of 30 seconds at 94 C, 30 seconds at 55 C
and 1 minute
at 72 C.
The amplified PCR products were analysed by electrophoresis on a 2% agarose
gel. A
240 pb nucleotidic sequence was amplified which corresponds to the expected
size of the
fragment (Figure 22 lane 1). The PCR products were purified using the Gel
Extraction Kit
(QIA GEN) according to manufacturer's instructions.
Quantification of the PCR product
All PCR products were pooled and concentrated. Two volumes of ethanol and
0.1 volume of 3 M sodium acetate, pH 5.2, were added to the PCR products. The
mix was
incubated for 30 min at -20 C and centrifuged 20 min at 10000g at 4 C. The
pellet was
washed with 100 L of 70 % ethanol, centrifuged again 10 min at 10000g at 4 C.
Then, the
supernatant was removed and the pellet dried and resuspended in 30 L double
distilled
water. DNA sample was conserved at ¨20 C until use.
Three microliters of the concentrated fragment was loaded on a 2% agarose gel
to estimate its
quantity (Figure 22 lane 3).
Ligation of the catalytic domain into an expression vector, pcDNA3.1(+)
1-Digestions and purifications
The nucleotide sequence of the catalytic domain CD of hST6Gal I (SEQ ID NO :
43)
was ligated into the pcDNA3.1 vector into B amHI and XbaI restriction sites.
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
Both pcDNA3.1(+) vector and CD were first digested by the BamHI restriction
enzyme:
500 ng of the CD were digested with 5 units of BamHI (Promega), 0.1 lig.i..1L-
1 of Bovin
Serum Albumin and 1X of buffer E (Promega) in a final volume of 20 4.
5 1 jig of the pcDNA3.1 vector was digested with 10 units of BamHI
(Promega), 0.1 11g41L-1 of
Bovin Serum Albumin and lx of buffer E (Promega) in a final volume of 20 4.
Digestions were performed for 60 min at 37 C and the incubations were finally
inactivated for
15 min at 65 C. Digested products were purified using the PCR purification Kit
(QIAGEN).
10 The digested products were then submitted to a digestion by the
restriction enzyme
XbaI (Promega), following the manufacturer's instructions: using the
appropriate enzyme
units number, depending on DNA quantity (5 and 6, respectively for CD and
pcDNA3.1), 0.1
lig41L-1 of BSA and 1X of buffer D (Promega) in a final volume of 50 4. The
digestions
were carried out at 37 C for 60 min. The digestions were purified using the
PCR purification
15 Kit (QIAGEN).
2-Ligation
The ligation conditions were calculated according to the formula previously
20 mentioned. The size of the insert CD is around 960 pb, the size of the
vector pcDNA3.1 is
5428 pb, the quantity of vector used is 100 ng, and finally the molar ratio of
Insert/Vector is
3/1.
The construction of pcDNA3.1/CD recombinant vector was performed using 53 ng
of
the double digested CD, 100 ng of pcDNA3.1 double digested vector, 6 units of
T4 DNA
25 ligase enzyme (Promega) and lx ligation buffer in a final volume of 20
4. The mix was
incubated overnight at 15 C and the reaction was inactivated for 10 min at 70
C.
3-Cloning
30 Competent cells chemical transformation
One Shot TOP10 Chemically Competent E.coli (Invitrogen, Life Technologies)
were
transformed with the recombinant vector following manufacturer's instructions.
Five
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
81
microliters of the ligation product were added to 50 pL aliquot of TOP 10
cells and mixed by
tapping gently. The mix was incubated on ice for 30 minutes, then heat-shocked
for exactly
30 seconds in a 42 C water bath and placed on ice for 2 min. Two hundred and
fifty
microliters of preheated SOC medium (Invitrogen) were added to the
transformation reaction
and incubated at 37 C for one hour with shaking at 250 rpm. The transformation
reaction was
spread on LB agar plates (composition mentioned previously) containing 50
ps.m1-1 of
ampicillin. Plates were incubated overnight at 37 C.
Amplification
At the end of the next day, single colonies were picked and incubated, with 3
ml of LB
broth (composition mentioned previously) containing 50 ps.m1-1 of ampicillin,
overnight at
37 C with shaking at 250 rpm.
Minipreparation of plamsids
The recombinant vectors were purified from 1.5 mL aliquots of the previous
cultures
using the QIAprep Spin Miniprep Kit provided by QIAGEN and following
manufacturer's
instructions. Twenty-four minipreparations were performed to screen the
presence of the
insert.
Five microliters of each of the 24 minipreparations were double digested using
XbaI
and BamHI restriction enzymes in order to verify the presence of the insert.
First, the digestion solution contained 12 units of XbaI (Promega), 0.1
lig.i..tri of BSA and 1X
of buffer D (Promega) in a final volume of 10 4. Digestions were performed for
2 hours at
37 C. Samples were secondly digested using 20 units of BamHI (Promega) and 1X
of buffer
E (Promega) in a final volume of 20 4. Reactions were carried out overnight at
37 C. The
digestion products were loaded on a 1.5% agarose gel to detect the presence of
the CD. An
insert of around 1000 pb was detected (Figure 23) which corresponds to the
expected size of
the CD (960 pb).
One positive clone has been selected for the next steps of the construction.
Ligation of the N-terminal region into pcDNA3.1/CD recombinant vector
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
82
1-Digestions and purifications
The nucleotide sequence of the N-terminal region (SEQ ID NO : 179) was ligated
into
the pcDNA3.1/CD recombinant vector into BamHI and AflII restrictions sites.
Both N-
terminal fragment and pcDNA3.1/CD recombinant vector were first digested by
the BamHI
restriction enzyme:
300 ng of the N-terminal fragment were digested with 5 units of BamHI
(Promega),
0.1 lig41L-1 of BSA and 1X of buffer E (Promega) in a final volume of 20 4.
2 jig of pcDNA3.1/CD recombinant vector were digested with 20 units of BamHI
(Promega),
0.1 lig41L-1 of BSA and 1X of buffer E (Promega) in a final volume of 20 4.
Digestions were performed for 100 min at 37 C and the incubations were finally
inactivated for 15 min at 65 C. Digested products were purified using the PCR
purification
Kit (QIAGEN).
The digested products were then submitted to a digestion by the restriction
enzyme
AflII (New England BIOLABS), following the manufacturer's instructions: using
the
appropriate enzyme units number, depending on DNA quantity (10 and 20,
respectively for
N-terminal fragment and pcDNA3.1/CD recombinant vector), lx of BSA and lx of
buffer 2
(New England BIOLABS) in a final volume of 50 4. The digestions were carried
out at
37 C for 60 min and the incubations were finally inactivated for 20 min at 65
C. The
digestions were purified using the PCR purification Kit (QIAGEN).
2-Ligation
The ligation conditions were calculated according to the formula previously
mentioned. The size of the insert N-terminal fragment is 240 pb, the size of
the
pcDNA3.1/CD recombinant vector is 6388 pb, the quantity of vector used is 100
ng, and
finally the molar ratio of Insert/Vector is 1/1.
The construction of pcDNA3.1/N-terminal fragment/CD recombinant vector was
performed using 5 ng of the digested N-terminal fragment, 100 ng of
pcDNA3.1/CD digested
recombinant vector, 6 units of T4 DNA ligase enzyme (Promega) and 1X of
ligation buffer in
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
83
a final volume of 20 4. The mix was incubated overnight at 15 C and the
reaction was
inactivated for 10 min at 70 C.
3-Cloning
Competent cells chemical transformation
One Shot TOP10 Chemically Competent E.coli (Invitrogen) were transformed with
the ligation product following manufacturer's instructions and as previously
described.
Amplification
At the end of the next day, single colonies were picked and incubated, with 3
ml of LB
broth (composition mentioned previously) containing 50 ps.m1-1 of ampicillin,
overnight at
37 C with shaking at 250 rpm.
Minipreparation of plasmids
The recombinant vectors were purified from 1.5 mL aliquots of the previous
cultures
using the QIAprep Spin Miniprep Kit provided by QIAGEN following
manufacturer's
instructions. Twenty four minipreparations were performed to screen the
presence of the
insert.
Five microliters of each of the 24 minipreparations were double digested using
Xbal
and AflII restriction enzymes in order to verify the presence of the insert (N-
terminal fragment
+ CD ; SEQ ID NO : 181).
The digestion solution contained 20 units of Xbal (BIOLABS), 20 units of AflII
(BIOLABS), lx of BSA (BIOLABS) and lx of buffer 2 (BIOLABS) in a final volume
of 20
4. Digestions were performed for 2 hours and 30 min at 37 C. The digestion
products were
loaded on a 1.5% agarose gel to detect the presence of both N-terminal region
and CD. An
insert of around 1200 pb was detected (Figure 24a) which corresponds to the
expected size of
the insert.
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
84
A PCR was finally performed on five minipreparation samples to verify the
presence
of the insert DNA (N-terminal fragment followed by the CD: SEQ ID NO: 181).
The PCR
was performed on a PCR apparatus (I-cycler, Biorad) with 1 pL of each of the
minipreparation samples in a solution containing lx of ProofStart manufacturer
buffer and
1X of manufacturer Q-Solution, 3 mM of Mg2+, 300 M of each dNTP (Sigma
Aldrich), 1
M forward primer AflII-5T3, 1 M reverse primer 44-XbaI, 2.5 units of
ProofStart DNA
polymerase (Qiagen) in a final volume of 50 L.
The oligonucleotides included in the reaction were as follows:
5 'AflII-5 T3 : 5 ' -GAGCCCCTTAAGATGGAC TACAAAGAC GAT GAC G-3 '
and 3' A.4-XbaI: 5 ' -TAACCCTCTAGATTAGCAGTGAATGGTCCGGAAGC-3 '
The thermocycling profile used was: 95 C for 5 minutes, followed by 40 cycles
of 1 min at
94 C, 1 min at 57 C and 1 minute 30 s at 72 C. The amplified PCR products were
analysed
by electrophoresis on a 1.5% agarose gel: a 1200 pb nucleotidic sequence was
amplified for
each minipreparation sample which corresponds to the expected size of the
total insert
(Figure 24b).
Sequencing
The cloning steps were verified by both strands DNA sequencing by GENOME
express (Meylan, France) using universal primers present inside the vector: T7
promoter and
BGH reverse.
The nucleotidic sequence obtained was aligned to the expected sequence and the
alignement revealed 100% identity. The deduced amino acid sequence was also
aligned with
the expected sequence SEQ ID NO : 181 of the chimera and it shows 100%
identity.
Midipreparation
After the DNA insert sequence verification, the recombinant plasmid was
amplified in
100 mL culture to obtain high quantity of the construction. Four hundred
microliters of the
corresponding 3 mL LB broth culture mentioned previously were inoculated into
100 mL LB
broth containing 50 ps.m1-1 of ampicillin, and incubated overnight at 37 C
with shaking at
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
250 rpm. The bacterial cells were harvested and the recombinant vectors were
purified using
the QIAGEN Plasmid Midi Kit (Qiagen) following manufacturer's instructions.
The yield was
determined with an UV spectrophotometer to quantify the DNA concentration. The
construct
was stored at ¨20 C.
5
Cell culture : expression of the chimeric enzyme hST3Ga1 III / hST8Sia II /
hST6Ga1NAc I /
CD (SEQ ID NO: 182) in CHO cells
CHO-Kl cells were routinely cultured at 37 C with 5 % of CO2 in DMEM
10 supplemented with 10 % FBS, 2.5 i.tg/mL fungizone and 50 i.tg/mL
gentamicin. The day
before transfection, cells were seeded at a density of 100 000 cells/mL in a 6-
well plate
containing 3 glass coverslips per well. CHO-Kl cells were transiently
transfected with
FuGENE 6 transfection reagent (Roche) using a 3:1 ratio.
15
Following one day of culture, cells were fixed with 1 % paraformaldehyde for
15
minutes at room temperature (RT). Cells were washed three times with PBS,
blocked for
30 minutes at RT with 1% BSA and then incubated with 10 i.tg/mL of SNA-FITC
(vector
laboratories) for one hour at 4 C. Cells were successively washed three times
with PBS, fixed
with 3 % paraformaldehyde for 20 minutes at RT, further washed with PBS and
incubated
20 with 0.05 M NH4C1 for 10 minutes at RT. After washings with PBS,
cells were permeabilized
with 0.25 % Triton X-100 for 5 minutes at RT and blocked with 10 % inactivated
goat serum
and 1% BSA for 30 minutes at RT. Cells were incubated for one hour at RT with
1 i.tg/mL
monoclonal anti-FLAG M2 antibody (Sigma) in PBS containing 5 % inactivated
goat serum
and 0.5% BSA. Cells were washed three times with PBS and incubated for 45
minutes with
25 5.7 g/mL of Alexa Fluor( '594-coupled secondary goat anti-mouse
antibody (Molecular
Probes) in PBS containing 5 % inactivated goat serum and 0.5% BSA. After
washings with
PBS, cells were mounted in Mowiol and analysed using an Olympus microscope
(Figure 25).
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
86
References
Andersen D. C. & Krummen L. (2002) Recombinant protein expression for
therapeutic
applications. Curr. Opin. Biotechnol., 13 (2): 117-23.
Angata K., Nakayama J., Fredette B., Chong K., Ranscht B. & Fukuda M. (1997)
Human
STX polysialyltransferase forms the embryonic form of the neural cell adhesion
molecule.
Tissue-specific expression, neurite outgrowth, and chromosomal localization in
comparison
with another polysialyltransferase, PST. J. Biol. Chem., 272(11):7182-90.
Angata K., Suzuki M., McAuliffe J., Ding Y., Hindsgaul 0. & Fukuda M. (2000)
Differential
biosynthesis of polysialic acid on neural cell adhesion molecule (NCAM) and
oligosaccharide
acceptors by three distinct alpha 2,8-sialyltransferases, ST8Sia IV (PST),
ST8Sia II (STX),
and ST8Sia III. J. Biol. Chem., 275(24):18594-601.
Angata K., Yen T. Y., El-Battari A., Macher B. A. & Fukuda M. (2001) Unique
disulfide
bond structures found in ST8Sia IV polysialyltransferase are required for its
activity. J. Biol.
Chem., 276(18):15369-77.
Ashwell G. & Harford J. (1982) Carbohydrate-specific receptors of the liver.
Annu. Rev.
Biochem., 51:531-54.
Aumiller J. J., Hollister J. R., & Jarvis D. L. (2003) A transgenic insect
cell line engineered to
produce CMP-sialic acid and sialylated glycoproteins. Glycobiology, 13(6):497-
507.
Baneyx F. (1999) Recombinant protein expression in Escherichia coli. Curr.
Opin.
Biotechnol., 10(5):411-21.
Berger E. G. (2002) Ectopic localizations of Golgi glycosyltransferases.
Glycobiology,
12(2):29R-36R.
Berger E.G., Burger P., Borsig L., Malissard M., Felner K.M., Zeng S., Dinter
A. (1998)
Immunodetection of glycosyltransferases: prospects and pitfalls. Adv. Exp.
Med. Biol.,
435:119-32.
Blanchard S. (2004) Ingenierie de glicoside hydrolases pour la glycosylation
des proteines
recombinantes. These de l'universite Joseph Fourier, 272p.
Bragonzi A., Distefano G., Buckberry L.D., Acerbis G., Foglieni C., Lamotte
D., Campi G.,
Marc A., Soria M.R., Jenkins N., Monaco L. (2000) A new Chinese hamster ovary
cell line
expressing alpha2,6-sialyltransferase used as universal host for the
production of human-like
sialylated recombinant glycoproteins. Biochim. Biophys. Acta, 1474(3):273-82.
Breton C. & Imberty A. (1999) Structure/function studies of
glycosyltransferases.
Curr. Opin. Struct. Biol., 9(5):563-71.
Breton C., Mucha J. & Jeanneau C. (2001) Structural and functional features of
glycosyltransferases. Biochimie., 83 (8): 713-8.
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
87
Bretscher M. S. & Munro S. (1993) Cholesterol and the Golgi apparatus.
Science,
261(5126):1280-1.
Bretthauer R.K. (2003) Genetic engineering of Pichia pastoris to humanize N-
glycosylation of
proteins. Trends Biotechnol., 21(11):459-62.
Bianucci A. M. & Chiellini F. (2000) A 3D model for the human hepatic
asialoglycoprotein
receptor (ASGP-R). J. BiomoL Struct. Dyn., 18(3):435-51.
Chang M.L., Eddy R.L., Shows T.B., Lau J.T. (1995) Three genes that encode
human beta-
galactoside alpha 2,3-sialyltransferases. Structural analysis and chromosomal
mapping
studies. Glycobiology, 5 (3):319-25 .
Chen C. & Colley K. J. (2000) Minimal structural and glycosylation
requirements for ST6Ga1
I activity and trafficking. Glycobiology, 10(5):531-83.
Chen C., Ma J., Lazic A., Backovic M., Colley K.J. (2000) Formation of
insoluble oligomers
correlates with ST6Ga1 I stable localization in the golgi. J. Biol. Chem.,
275(18):13819-26.
Chen T.L., Chen C., Bergeron N.Q., Close B.E., Bohrer T.J., Vertel B.M. &
Colley K.J.
(2003) The two rat alpha 2,6-sialyltransferase (ST6Ga1 I) isoforms: evaluation
of catalytic
activity and intra-Golgi localization. Glycobiology, 13(2):109-17.
Colley K. J. (1997) Golgi localization of glycosyltransferases: more questions
than answers.
Glycobiology, 7(1): 1-13 .
Crocker P. R. & Feizi T. (1996) Carbohydrate recognition systems: functional
triads in cell-
cell interactions. Curr. Opin. Struct. Biol., 6(5):679-91.
Datta A. K., Sinha A. & Paulson J. C. (1998) Mutation of the sialyltransferase
S-sialylmotif
alters the kinetics of the donor and acceptor substrates. J. Biol. Chem.,
273(16):9608-14.
Datta A. K. & Paulson J. C. (1995) Sialylmotifs of sialyltransferases. Indian
J. Biochem.
Biophys., 34(1-2):157-65.
Donadio S., Dubois C., Fichant G., Roybon L., Guillemot J.C., Breton C. &
Ronin C. (2003)
Recognition of cell surface acceptors by two human alpha-2,6-
sialyltransferases produced in
CHO cells. Biochimie, 85 (3-4):311-21.
Drickamer K. (1993) A conserved disulphide bond in sialyltransferases.
Glycobiology, 3(1):2-
3.
Feizi T. (1993) Carbohydrate--protein interactions in capillary morphogenesis?
Glycobiology,
3(6):547-8.
Fenteany F. H. & Colley K. J. (2005) Multiple signals are required for
alpha2,6-
sialyltransferase (ST6Ga1 I) oligomerization and Golgi localization. J. Biol.
Chem., (7):5423-
9.
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
88
Gabius H.J., Siebert H.C., Andre S., Jimenez-Barbero J. & Rudiger H. (2004)
Chemical
biology of the sugar code. Chembiochem., 5(6):740-64.
Geremia R.A., Harduin-Lepers A. & Delannoy P. (1997) Identification of two
novel
conserved amino acid residues in eukaryotic sialyltransferases: implications
for their
mechanism of action. Glycobio logy, 7(2):v-vii.
Giordanengo V., Bannwarth S., Laffont C., Van Miegem V., Harduin-Lepers A.,
Delannoy P.
& Lefebvre J.C. (1997) Cloning and expression of cDNA for a human Gal(betal-
3)Ga1NAc
alpha2,3-sialyltransferase from the CEM T-cell line. Eur. J. Biochem.,
247(2):558-66.
Grabenhorst E. & Conradt H. S. (1999) The cytoplasmic, transmembrane, and stem
regions of
glycosyltransferases specify their in vivo functional sublocalization and
stability in the Golgi.
J. Biol. Chem., 274(51):36107-16.
Grabenhorst E., Schlenke P., Pohl S., Nimtz M. & Conradt H.S. (1999) Genetic
engineering
of recombinant glycoproteins and the glycosylation pathway in mammalian host
cells.
Glycoconj. J., 16(2):81-97.
Hamilton S. R., Bobrowicz P., Bobrowicz B., Davidson R. C., Li H., Mitchell
T., Nett J. H.,
Rausch S., Stadheim T. A., Wischnewski H., Wildt S. & Gerngross, T. U. (2003)
Production
of complex human glycoproteins in yeast. Science, 301, 1244-1246.
Harduin-Lepers A., Stokes D.C., Steelant W.F., Samyn-Petit B., Krzewinski-
Recchi M.A.,
Vallejo-Ruiz V., Zanetta J.P., Auge C. & Delannoy P. (2000) Cloning,
expression and gene
organization of a human Neu5Ac alpha 2-3Gal beta 1-3Ga1NAc alpha 2,6-
sialyltransferase:
hST6Ga1NAcIV. Biochem. 1, 352(Pt 1):37-48.
Harduin-Lepers A., Vallejo-Ruiz V., Krzewinski-Recchi M.A., Samyn-Petit B.,
Julien S. &
Delannoy P. (2001) The human sialyltransferase family. Biochimie, 83(8):727-
37.
Helenius A. & Aebi M. (2001) Intracellular functions of N-linked glycans.
Science,
291(5512):2364-9.
Herscovics A. & Orlean P. (1993) Glycoprotein biosynthesis in yeast. FASEB 1,
7(6):540-50.
Hudgin R.L., Pricer W.E. Jr, Ashwell G., Stockert R.J. & Morell A.G. (1974)
The isolation
and properties of a rabbit liver binding protein specific for
asialoglycoproteins. J. Biol. Chem.,
249(17):5536-43.
Ikehara Y., Shimizu N., Kono M., Nishihara S., Nakanishi H., Kitamura T.,
Narimatsu H.,
Tsuji S. & Tatematsu M. (1999) A novel glycosyltransferase with a
polyglutamine repeat; a
new candidate for GD1alpha synthase (ST6Ga1NAc V)(1). FEBS Lett., 463(1-2):92-
6.
Ishii A., Ohta M., Watanabe Y., Matsuda K., Ishiyama K., Sakoe K., Nakamura
M., Inokuchi
J., Sanai Y. & Saito M. (1998) Expression cloning and functional
characterization of human
cDNA for ganglioside GM3 synthase. J. Biol. Chem., 273(48):31652-5.
Jarvis D.L., Kawar Z.S. & Hollister J.R. (1998) Engineering N-glycosylation
pathways in the
baculovirus-insect cell system. Curr. Opin. Biotechnol., 9(5):528-33.
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
89
Jeanneau C. (2003) Bioanalyse et ingenierie de glycosyltransferase. These de
l'universite de
Paris 7, 179p.
Jeanneau C., Chazalet V., Auge C., Soumpasis D.M., Harduin-Lepers A., Delannoy
P.,
Imberty A. & Breton C. (2004) Structure-function analysis of the human
sialyltransferase
ST3Ga1 I: role of n-glycosylation and a novel conserved sialylmotif. J. Biol.
Chem.,
279(14):13461-8.
Kawasaki G. & Ashwell T. (1976) Carbohydrate structure of glycopeptides
isolated from an
hepatic membrane-binding protein specific for asialoglycoproteins. J. Biol.
Chem.,
251(17):5292-9.
Kim Y.J., Kim K.S., Kim S.H., Kim C.H., Ko J.H., Choe I.S., Tsuji S. & Lee
Y.C. (1996)
Molecular cloning and expression of human Gal beta 1,3Ga1NAc alpha 2,3-
sialytransferase
(hST3Gal II). Biochem. Biophys. Res. Commun., 228(2):324-7.
Kitagawa H. & Paulson J. C. (1993) Cloning and expression of human Gal beta
1,3(4)G1cNAc alpha 2,3-sialyltransferase. Biochem. Biophys. Res. Commun.,
194(1):375-82.
Kitagawa H. & Paulson J. C. (1994a) Differential expression of five
sialyltransferase genes in
human tissues. J. Biol. Chem., 269(27):17872-8.
Kitagawa H. & Paulson J. C. (1994b) Cloning of a novel alpha 2,3-
sialyltransferase that
sialylates glycoprotein and glyco lipid carbohydrate groups. J. Biol. Chem.,
269(2):1394-401.
Kitagawa H., Mattei M.G. & Paulson J.C. (1996) Genomic organization and
chromosomal
mapping of the Gal beta 1,3GalNAc/Gal beta 1,4G1cNAc alpha 2,3-
sialyltransferase. J. Biol.
Chem., 271(2):931-8.
Kitazume-Kawaguchi S., Dohmae N., Takio K., Tsuji S. & Colley K.J. (1999) The
relationship between ST6Ga1 I Golgi retention and its cleavage-secretion.
Glycobiology,
9(12):1397-406.
Kitazume-Kawaguchi S., Kabata S. & Arita M. (2001) Differential biosynthesis
of polysialic
or disialic acid Structure by ST8Sia II and ST8Sia IV. J. Biol. Chem.,
276(19):15696-703.
Kono M., Yoshida Y., Kojima N. & Tsuji S. (1996) Molecular cloning and
expression of a
fifth type of alpha2,8-sialyltransferase (ST8Sia V). Its substrate specificity
is similar to that of
SAT-V/III, which synthesize GD1c, GT1a, GQ1b and GT3. J. Biol. Chem.,
271(46):29366-
71.
Kono M., Tsuda T., Ogata S., Takashima S., Liu H., Hamamoto T., Itzkowitz
S.H., Nishimura
S. & Tsuji S. (2000) Redefined substrate specificity of ST6Ga1NAc II: a second
candidate
sialyl-Tn synthase. Biochem. Biophys. Res. Commun., 272(1):94-7.
Kornfeld R. & Kornfeld S. (1985) Assembly of asparagine-linked
oligosaccharides. Annu.
Rev. Biochem., 1985;54:631-64.
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
Krzewinski-Recchi M.A., Julien S., Juliant S., Teintenier-Lelievre M., Samyn-
Petit B.,
Montiel M.D., Mir A.M., Cerutti M., Harduin-Lepers A. & Delannoy P. (2003)
Identification
and functional expression of a second human beta-galactoside alpha2,6-
sialyltransferase,
ST6Ga1 II. Eur. J. Biochem.,270(5):950-61.
5
Kurosawa N., Kojima N., Inoue M., Hamamoto T. & Tsuji S. (1994) Cloning and
expression
of Gal beta 1,3GalNAc-specific GalNAc alpha 2,6-sialyltransferase. J. Biol.
Chem.,
269(29): 19048-53.
10 Kurosawa N., Inoue M., Yoshida Y. & Tsuji S. (1996) Molecular cloning
and genomic
analysis of mouse Galbetal, 3GalNAc-specific GalNAc alpha2,6-
sialyltransferase. J. Biol.
Chem., 1996 Jun 21;271(25):15109-16.
Kurosawa N., Takashima S., Kono M., Ikehara Y., Inoue M., Tachida Y.,
Narimatsu H. &
15 Tsuji S. (2000) Molecular cloning and genomic analysis of mouse GalNAc
alpha2, 6-
sialyltransferase (ST6Ga1NAc I). J. Biochem., 127(5):845-54.
Laroy W., Ameloot P., & Contreras R. (2001) Characterization of
sialyltransferase mutants
using surface plasmon resonance. Glycobiology,11(3):175-82.
Lee Y.C., Kim Y.J., Lee K.Y., Kim K.S., Kim B.U., Kim H.N., Kim C.H. & Do S.I.
(1998)
Cloning and expression of cDNA for a human Sia alpha 2,3Gal beta 1,
4G1cNA:alpha 2,8-
sialyltransferase (hST8Sia III). Arch. Biochem. Biophys., 360(1):41-6.
Lee Y.C., Kaufmann M., Kitazume-Kawaguchi S , Kono M , Takashima S , Kurosawa
N ,
Liu H., Pircher H. & Tsuji S. (1999) Molecular cloning and functional
expression of two
members of mouse NeuAcalpha2,3Galbeta1,3Ga1NAc GalNAcalpha2,6-
sialyltransferase
family, ST6Ga1NAc III and IV. J. Biol. Chem., 274(17):11958-67.
Legaigneur P., Brioude B. & Ronin C. (1999) Antigenes glycanniques
recombinants.
Immunoanal. Biol. Spec., 14 :297-307.
Legaigneur P., Breton C., El Battari A., Guillemot J.C., Auge C., Malissard
M., Berger E.G.
& Ronin C. (2001) Exploring the acceptor substrate recognition of the human
beta-
galactoside alpha 2,6-sialyltransferase. J. Biol. Chem., 276(24):21608-17.
Lerouge P., Bardor M., Pagny S., Gomord V. & Faye L. (2000) N-glycosylation of
recombinant pharmaceutical glycoproteins produced in transgenic plants:
towards an
humanisation of plant N-glycans. Curr. Pharm. Biotechnol., 1(4):347-54.
Livingston B. D. & Paulson J. C. (1993) Polymerase chain reaction cloning of a
developmentally regulated member of the sialyltransferase gene family. J.
Biol. Chem.,
268(16):11504-7.
Ma J., Qian R., Rausa F.M. 3rd & Colley K.J. (1997) Two naturally occurring
alpha2,6-
sialyltransferase forms with a single amino acid change in the catalytic
domain differ in their
catalytic activity and proteolytic processing. J. Biol. Chem., 272(1):672-9.
Machamer C.E. (1991) Golgi retention signals: do membranes hold the key?
Trends Cell
Biol., 1(6):141-4.
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
91
Maras M., Van Die I., Contreras R. & Van den Hondel C. A. M. J. J. (1999)
Filamentous
fungi as production organisms for glycoproteins of bio-medical interest.
Glycoconj. 1, 16:99-
107.
Marchal I., Jarvis D. L., Cacan R. & Verbert A. (2001) Glycoproteins from
insect cells:
sialylated or not? Biol. Chem., 382, 151-159.
Martina J.A., Daniotti J.L. & Maccioni H.J. (1998) Influence of N-
glycosylation and N-
glycan trimming on the activity and intracellular traffic of GD3 synthase. J.
Biol. Chem.,
273(6):3725-31.
Meier M., Bider M.D., Malashkevich V.N., Spiess M. & Burkhard P. (2000)
Crystal structure
of the carbohydrate recognition domain of the H1 subunit of the
asialoglycoprotein receptor.
J. Mol. Biol., 300(4):857-65.
Morell A.G., Gregoriadis G., Scheinberg I.H., Hickman J. & Ashwell G. (1971)
The role of
sialic acid in determining the survival of glycoproteins in the circulation.
J. Biol. Chem.,
246(5): 1461-7.
Muhlenhoff M., Manegold A., Windfuhr M., Gotza B. & Gerardy-Schahn R. (2001)
The
impact of N-glycosylation on the functions of polysialyltransferases. J. Biol.
Chem.,
276(36):34066-73.
Munro S. (1991) Sequences within and adjacent to the transmembrane segment of
alpha-2,6-
sialyltransferase specify Golgi retention. EMBO J., 10(12):3577-88.
Bretscher M.S. & Munro S. (1993) Cholesterol and the Golgi apparatus. Science,
261(5126):1280-1.
Munro S. (1995) An investigation of the role of transmembrane domains in Golgi
protein
retention. EMBO J., 14(19):4695-704.
Munro S. (1998) Localization of proteins to the Golgi apparatus. Trends Cell
Biol.,8(1):11-5.
Nakayama J., Fukuda M.N., Fredette B., Ranscht B. & Fukuda M. (1995)
Expression cloning
of a human polysialyltransferase that forms the polysialylated neural cell
adhesion molecule
present in embryonic brain. Proc. Natl. Acad. Sci. U. S. A., 92(15):7031-5.
Nakayama J., Fukuda M.N., Hirabayashi Y., Kanamori A., Sasaki K., Nishi T. &
Fukuda M.
(1996) Expression cloning of a human GT3 synthase. GD3 AND GT3 are synthesized
by a
single enzyme. J. Biol. Chem., 271(7):3684-91.
Nara K., Watanabe Y., Kawashima I., Tai T., Nagai Y. & Sanai Y. (1996)
Acceptor substrate
specificity of a cloned GD3 synthase that catalyzes the biosynthesis of both
GD3 and
GD1c/GT1a/GQ1b. Eur. J. Biochem., 238(3):647-52.
Narimatsu H. (2004) Construction of a human glycogene library and
comprehensive
functional analysis. Glycoconj. J., 21(1-2): 17-24.
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
92
Okajima T., Fukumoto S., Miyazaki H., Ishida H., Kiso M., Furukawa K., Urano
T. &
Furukawa K. (1999) Molecular cloning of a novel alpha2,3-sialyltransferase
(ST3Ga1 VI) that
sialylates type II lactosamine structures on glycoproteins and glycolipids. J.
Biol. Chem.,
274(17): 11479-86.
Okajima T., Chen H.H., Ito H., Kiso M., Tai T., Furukawa K., Urano T. &
Furukawa K.
(2000) Molecular cloning and expression of mouse GD1alpha/GTlaalpha/GQ1balpha
synthase (ST6GaNAc VI) gene. J. Biol. Chem., 275(10):6717-23.
Opat A.S., van Vliet C. & Gleeson P.A. (2001) Trafficking and localisation of
resident Golgi
glycosylation enzymes. Biochimie, 83(8):763-73.
Paulson J. C. (1989) Glycoproteins: what are the sugar chains for? Trends
Biochem. Sci.,
14(7):272-6.
Paulson J.C., Weinstein J., Ujita E.L., Riggs K.J. & Lai PH. (1987) The
membrane-binding
domain of a rat liver Golgi sialyltransferase. Biochem. Soc. Trans., 15(4):618-
20.
Paulson J.C. & Colley K.J. (1989) Glycosyltransferases. Structure,
localization, and control of
cell type-specific glycosylation. J. Biol. Chem., 264(30):17615-8.
Pilatte Y., Bignon J. & Lambre C.R. (1993) Sialic acids as important molecules
in the
regulation of the immune system: pathophysiological implications of sialidases
in immunity.
Glycobiology, 3(3):201-18.
Qian R., Chen C. & Colley K.J. (2001) Location and mechanism of alpha 2,6-
sialyltransferase
dimer formation. Role of cysteine residues in enzyme dimerization,
localization, activity, and
processing. J. Biol. Chem., 276(31):28641-9.
Reid M.E. & Lomas-Francis C. (2002) Molecular approaches to blood group
identification.
Curr. Opin. Hematol., 9(2):152-9.
Roth J., Taatjes D.J., Lucocq J.M., Weinstein J. & Paulson J.C. (1985)
Demonstration of an
extensive trans-tubular network continuous with the Golgi apparatus stack that
may function
in glycosylation. Cell, 43(1):287-95.
Roth J. (1987) Subcellular organization of glycosylation in mammalian cells.
Biochim.
Biophys. Acta, 906(3):405-36.
Roth J. (1991) Localization of glycosylation sites in the Golgi apparatus
using
immuno labeling and cytochemistry. J. Electron. Microsc. Tech., 17(2): 121-31.
Roy S. K., Chiba Y. & Jigami, Y. (2000) Production of Therapeutic
Glycoproteins through
the Engineering of Glycosylation Pathway in Yeast. Biotechnol. Bioprocess
Eng., 5, 219-226.
Sasaki K. (1996) Molecular cloning and characterization of sialyltransferases.
Trends
Glycosci. Glycotechnol., 8:195-215.
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
93
Sasaki K., Kurata K., Kojima N., Kurosawa N., Ohta S., Hanai N., Tsuji S. &
Nishi T. (1994)
Expression cloning of a GM3-specific alpha-2,8-sialyltransferase (GD3
synthase). J. Biol.
Chem., 269(22): 15950-6.
Scheidegger E.P., Sternberg L.R., Roth J. & Lowe JB. (1995) A human STX cDNA
confers
polysialic acid expression in mammalian cells. J. Biol. Chem., 270(39):22685-
8.
Sinnott M. L. (1990) Catalytic mechanism of enzymic glycosyl transfer. Chem.
Rev., 90,
1171-1202.
Sjoberg E.R, Kitagawa H., Glushka J., van Halbeek H. & Paulson J.C. (1996)
Molecular
cloning of a developmentally regulated N-acetylgalactosamine alpha2,6-
sialyltransferase
specific for sialylated glycoconjugates. J. Biol. Chem., 271(13):7450-9.
Sujino K., Jackson R.J., Chan N.W., Tsuji S. & Palcic M.M. (2000) A novel
viral alpha2,3-
sialyltransferase (v-ST3Ga1 I): transfer of sialic acid to fucosylated
acceptors. Glycobiology,
10(3):313-20.
Swartz J. R. (2001) Advances in Escherichia coli production of therapeutic
proteins. Curr.
Opin. Biotechnol., 12, 195-201.
Takashima S., Ishida H.K., Inazu T., Ando T., Ishida H., Kiso M., Tsuji S. &
Tsujimoto M.
(2002) Molecular cloning and expression of a sixth type of alpha 2,8-
sialyltransferase
(ST8Sia VI) that sialylates 0-glycans. J. Biol. Chem., 277(27):24030-8.
Taniguchi A., Kaneta R., Morishita K. & Matsumoto K. (2001) Gene structure and
transcriptional regulation of human Gal beta1,4(3) GlcNAc alpha2,3-
sialyltransferase VI
(hST3Ga1 VI) gene in prostate cancer cell line. Biochem. Biophys. Res.
Commun., (5):1148-
56.
Tanner W. & Lehele L. (1987) Protein glycosylation in yeast. Biochim. Biophys.
Acta,
906(1):81-99.
Tetteroo P.A., de Heij H.T., Van den Eijnden D.H., Visser F.J., Schoenmarker
E. & Geurts
van Kessel AH. (1987) A GDP-fucose:[Gal beta 1----4]G1cNAc alpha 1----3-
fucosyltransferase activity is correlated with the presence of human
chromosome 11 and the
expression of the Lex, Ley, and sialyl-Lex antigens in human-mouse cell
hybrids. J. Biol.
Chem., 262(33):15984-9.
Vallejo-Ruiz V., Hague R., Mir A.M., Schwientek T., Mandel U., Cacan R.,
Delannoy P. &
Harduin-Lepers A. (2001) Delineation of the minimal catalytic domain of human
Galbetal-
3GalNAc alpha2,3-sialyltransferase (hST3Ga1 I). Biochim. Biophys. Acta,
1549(2):161-73.
van den Eijnden D.H., Joziasse D.H., Dorland L., van Halbeek H., Vliegenthart
J.F. &
Schmid K. (1980) Specificity in the enzymic transfer of sialic acid to the
oligosaccharide
branches of bl- and triantennary glycopeptides of alpha 1-acid glycoprotein.
Biochem.
Biophys. Res. Commun., 92(3):839-45.
CA 02653104 2008-11-24
WO 2007/135194
PCT/EP2007/055070
94
Van Den Hamer C.J., More11 A.G., Scheinberg I.H., Hickman J. & Ashwell G.
(1970)
Physical and chemical studies on ceruloplasmin. IX. The role of galactosyl
residues in the
clearance of ceruloplasmin from the circulation. J. Biol. Chem., 245(17):4397-
402.
Vervecken W., Kaigorodov V., Callewaert N., Geysens S., De Vusser K. &
Contreras R.
(2004) In vivo synthesis of mammalian-like, hybrid-type N-glycans in Pichia
pastoris. Appl.
Environ. Microbiol., 70, 2639-2646.
Watanabe Y., Nara K., Takahashi H., Nagai Y. & Sanai Y. (1996) The molecular
cloning and
expression of alpha 2,8-sialyltransferase (GD3 synthase) in a rat brain. J.
Biochem.,
120(5): 1020-7.
Weikert S., Papac D., Briggs J., Cowfer D., Tom S., Gawlitzek M., Lofgren J.,
Mehta S.,
Chisholm V., Modi N., Eppler S., Carroll K., Chamow S., Peers D,. Berman P. &
Krummen
L. (1999) Engineering Chinese hamster ovary cells to maximize sialic acid
content of
recombinant glycoproteins. Nat. Biotechnol., 17(11): 1116-21.
Weinstein J., de Souza-e-Silva U. & Paulson J.C. (1982) Sialylation of
glycoprotein
oligosaccharides N-linked to asparagine. Enzymatic characterization of a Gal
beta 1 to
3(4)G1cNAc alpha 2 to 3 sialyltransferase and a Gal beta 1 to 4G1cNAc alpha 2
to 6
sialyltransferase from rat liver. J. Biol. Chem., 257(22):13845-53.
Wickner W. T. & Lodisch H. F. (1985) Multiple mechanisms of protein insertion
into and
across membranes. Science, 230(4724):400-7.
Yang W., Pepperkok R., Bender P., Kreis TE. & Storrie B. (1996) Modification
of the
cytoplasmic domain affects the subcellular localization of Golgi glycosyl-
transferases. Eur. J.
Cell Biol., 71(1):53-61.
Yoshida Y., Kojima N. & Tsuji S. (1995a) Molecular cloning and
characterization of a third
type of N-glycan alpha 2,8-sialyltransferase from mouse lung. J. Biochem.,
118(3):658-64.
Yoshida Y., Kojima N., Kurosawa N., Hamamoto T. & Tsuji S. (1995b) Molecular
cloning of
Sia alpha 2,3Gal beta 1,4G1cNAc alpha 2,8-sialyltransferase from mouse brain.
J. Biol.
Chem., 270(24): 14628-33 .