Note: Descriptions are shown in the official language in which they were submitted.
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
1
IL-17C ANTAGONISTS AND METHODS OF USING THE SAME
BACKGROUND OF THE INVENTION
[1] Cytokines are soluble, small proteins that mediate a variety of biological
effects,
including the regulation of the growth and differentiation of many cell types
(see, for example, Arai
et al., Annu. Rev. Biochem. 59:783 (1990); Mosmann, Curr. Opin. Immunol. 3:311
(1991); Paul and
Seder, Cell 76:241 (1994)). Proteins that constitute the cytokine group
include interleukins,
interferons, colony stimulating factors, tumor necrosis factors, and other
regulatory molecules. For
example, human interleukin-17 is a cytokine which stimulates the expression of
interleukin-6,
intracellular adhesion molecule 1, interleukin-8, granulocyte macrophage
colony-stimulating factor,
and prostaglandin E2 expression, and plays a role in the preferential
maturation of CD34+
hematopoietic precursors into neutrophils (Yao et al., J. Immunol. 155:5483
(1995); Fossiez et al., J.
Exp. Med. 183:2593 (1996)). The IL-17 cDNA has been isolated and cloned from
the murine
hybridomas (cytotoxic T lymphocyte antigen 8 (CTLA-8)) and has homology to
open reading frame
13 from the T lymphotropic Herpesvirus saimiri. The human IL-17A gene product
is a protein of
150 amino acids with a molecular weight of 15 kDa, and is secreted as a
disulfide linked homodimer
of 30-35 kDa glycoprotein. Five related cytokines sharing about 20-50%
homology to IL-17A were
subsequently identified. (Rouvier, et al., J. Immunol. 150:5445-5456 (1993);
Yao et al., Gene
168:223 (1996); Fossiez et al., J. Exp. Med. 183:2593 (1996); Moseley et al.
Cytokine and Growth
Factor Reviews 14:155 (2003)).
[2] IL-17C is related to IL-17, having approximately 27% amino acid identity.
See e.g
Li H et al, "Cloning and characterization of IL-17B and IL-17C, two new
members of the IL-17
cytokine family" PNAS 97(2): 773-8 (2000). Although no expression of IL-17C
mRNA is found in
activated T cells, a survey of cytokine induction revealed that IL-17C does
stimulate the release of
tumor necrosis factor a and IL- lb from a THP-1 monocytic cell line, whereas
IL-17A has only a
weak effect in this system. Further, fluorescence activated cell sorter
analysis shows that IL-17C
binds to THP-I cells. IL-17C is not active in an IL-17 assay, nor does it
stimulate IL-6 release from
human fibroblasts or bind to the human IL-17 receptor extracellular domain.
This data shows that
there is a family of IL-17-related cytokines differing in patterns of
expression and proinflammatory
responses that may be transduced through a cognate set of cell surface
receptors. Members of the IL-
17 family have been implicated as factors that contribute to the progression
of various autoimmune
and inflammatory diseases including rheumatoid arthritis and asthma.
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
2
[3] Receptors that bind cytokines are typically composed of one or more
integral
membrane proteins that bind the cytokine with high affinity and transduce this
binding event to the
cell through the cytoplasmic portions of the certain receptor subunits.
Cytokine receptors have been
grouped into several classes on the basis of similarities in their
extracellular ligand binding domains.
Genome-wide homology comparisons led to identification of five ligands and
four receptor paralogs
within the IL-17/IL-17R family, (termed IL-17RA, IL-17RB, IL-17RC, IL17RD and
IL-17RE).
Most of these remain un-paired orphans. Establishment of receptor-ligand pairs
in this family has
been complicated because nearly all IL-17R homologs are represented by
multiple splice variants,
resulting in alternative extracellular domains. The receptor for IL-17A (IL-
17RA) is a single-pass
transmembrane protein of approximately 130 kDa. Emerging data suggests that IL-
17C, like IL-17,
IL-17A and IL-17F, is a pro-inflammatory cytokine causing neutrophilia when
expressed by
intranasal administration and adenoviral infection in mouse lungs.
Specifically, the pro-
inflammatory cytokine IL-17C has a high degree of sequence similarity to IL-
17. While the IL-17A
cytokine is expressed only by T-cells, its receptor is expressed in all
tissues examined to date, a
finding consistent with the pleiotropic activities of IL- 17. The activation
of the receptor by IL-17A
generally results in the induction of other pro-inflammatory cytokines,
through the activation of NF-
.kappa.B. The four additional receptors share partial sequence homology to IL-
17RA. Many of
these IL-17 receptors exist as alternatively spliced isoforms, some of which
may not contain
transmembrane or cytoplasmic domains, and thereby may be acting as soluble
decoy receptors.
(Moseley et al. Cytokine and Growth Factor Reviews 14:155 (2003)). The IL-17
receptors are found
to be widely expressed and exhibit a broad tissue distribution, a finding
consistent with the
pleiotropic activities of IL-17 ligands. Nevertheless, not much is known about
the functions and
interactions of these molecules or of their signal transduction pathways.
[4] IL-17RA receptor activity in humans is dependent on an obligate component;
namely IL-17RC. In humans, the biological activity of IL-17A ligand depends
upon an IL-17RA/IL-
17RC heterodimeric receptor complex. (Toy. Et al., Journal of Immunology,
177:36 (2006)). Other
similar receptor complexes are known in the art. For example, the T-cell
heterodimeric receptor is a
complex of at least seven polypeptide chains, some of which are required for
assembly or transport
of the receptor to the surface, but do not play a direct role in ligand
binding or receptor signaling.
(Carson, et al., Journal of Biol. Chem., 266:7883 (1991)). A further exemplary
complex includes
activity enhancing receptor complexes. For example, soluble IL-6 receptors are
capable of
interacting first with a ligand and then ligand/soluble receptor complex then
binds a membrane
bound IL-6 receptor to activate cell signaling. Although most soluble
receptors are antagonists for
their membrane bound forms, this above receptor model illustrates an agonistic
role for soluble
receptor. (Rose-John, et al., Journal of Leukocyte Biology, 80:227 (2006)).
Thus, there is a need in
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
3
the art for a full understanding of the IL- 17 receptor family members and the
roles that each of these
members plays in signal transduction and, further, in pathogenesis of disease.
[5] The demonstrated in vivo activities of cytokines and their receptors
illustrate the
clinical potential of, and need for, other cytokines, cytokine receptors,
cytokine agonists, and
cytokine antagonists. For example, demonstrated in vivo activities of the pro-
inflammatory cytokine
family illustrates the enormous clinical potential of, and need for
antagonists of pro-inflammatory
molecules.
BRIEF DESCRIPTION OF THE DRAWINGS
[6] Figures IA, IB, IC and ID are graphic representations of the exon
structure of
human IL-17REx1 (SEQ ID NO:2). IL17REx1. --S2- indicates variant S2 (SEQ ID
NO: 113);
-S3- indicates variant S3 (SEQ ID NO:184) and ==S4== indicates variant S4 (SEQ
ID NO:186).
For those amino acids where codon was spliced by exon/intron junction, the
junction was moved to
include the entire codon.
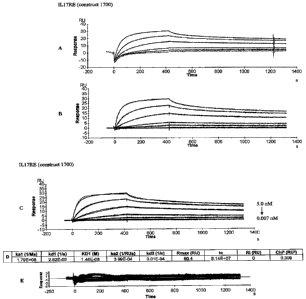
[7] Figure 2: A) Raw curves after subtracting the reference cell sensorgram.
B)
Double-referenced curves with buffer run subtracted and y-intercept set to 0.
C) Curves showing
overlay of bivalent analyte model fitting. Concentrations are 1/3 serial
dilutions from 5nM to
0.007nM. D) Data from the Biacore analysis report. E) Graph of residuals.
[8] Figure 3: A) Raw curves after subtracting the reference cell sensorgram.
B)
Double-referenced curves with buffer run subtracted and y-intercept set to 0.
C) Curves showing
overlay of bivalent analyte model fitting. Concentrations are 1/3 serial
dilutions from 5nM to
0.007nM. D) Data from the Biacore analysis report. E) Graph of residuals.
[9] Figure 4: A) Raw curves after subtracting the reference cell sensorgram.
B)
Double-referenced curves with buffer run subtracted and y-intercept set to 0.
C) Curves showing
overlay of bivalent analyte model fitting. Concentrations are 1/3 serial
dilutions from 5nM to
0.007nM. D) Data from the Biacore analysis report. E) Graph of residuals.
[10] Figure 5: A) Raw curves after subtracting the reference cell sensorgram.
B)
Double-referenced curves with buffer run subtracted and y-intercept set to 0.
C) Curves showing
overlay of bivalent analyte model fitting. Concentrations are 1/3 serial
dilutions from 5nM to
0.007nM. D) Data from the Biacore analysis report. E) Graph of residuals.
[11] Figure 6: A) Raw curves after subtracting the reference cell sensorgram.
B)
Double-referenced curves with buffer run subtracted and y-intercept set to 0.
C) Curves showing
overlay of bivalent analyte model fitting. Concentrations are 1/3 serial
dilutions from 5nM to
0.007nM. D) Data from the Biacore analysis report. E) Graph of residuals.
[12] Figure 7: A) Raw curves after subtracting the reference cell sensorgram.
B)
Double-referenced curves with buffer run subtracted and y-intercept set to 0.
C) Curves showing
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
4
overlay of bivalent analyte model fitting. Concentrations are 1/3 serial
dilutions from 5nM to
0.007nM. D) Data from the Biacore analysis report. E) Graph of residuals.
[13] Figure 8: Representative Calibration Curve of IL17C Binding to the Anti-
IL17C
Immobilized Surface. A) A dilution series of IL17C from 20 - 0.078 nM was
captured onto a high
density anti-IL17C surface, and the resulting sensorgrams were reference (flow
cell #1) subtracted.
The initial rate of association was calculated over a 30 second window
(highlighted).
[14] Figure 9: Dilution Series of IL17RE-FC5 (Construct 1700) with a Constant
Concentration of 5 nM IL17C. A) A dilution series of IL17RE-FC5 (construct
1700) from 20 - 0
nM was premixed with a constant concentration of IL17C (5 nM), then captured
onto a high density
anti-IL17C surface. The resulting sensorgrams were reference (flow cell #1)
subtracted. The initial
rate of association was calculated over a 30 second window. B) Using the
calibration curve
generated in Figure 8B, the initial rate of the association (slope) was used
to back calculate the
concentration of free IL17C in each sample. The concentration of free IL17C
was plotted along the
y-axis while the concentration of IL17RE-FC5 was plotted along the x-axis. The
data was fit with a
1:1 binding model, and a KD was calculated with the Biacore T100
Evaluation software. C) To
optimize the data fit obtained using the Biacore software, the data was
exported into Graphpad
Prism4 software and fit with a four parameter curve. An empirical IC50 value
was calculated from
this data, and this value was used as an approximation of the solution phase
affinity constant
(K. sub. D).
[15] Figure 10: Dilution Series of IL17RE-FC5 (Construct 1702) with a Constant
Concentration of 5 nM IL17C. A) A dilution series of IL17RE-FC5 (construct
1702) from 20 - 0
nM was premixed with a constant concentration of IL17C (5 nM), then captured
onto a high density
anti-IL17C surface. The resulting sensorgrams were reference (flow cell #1)
subtracted. The initial
rate of association was calculated over a 30 second window. B) Using the
calibration curve
generated in Figure 8B, the initial rate of the association (slope) was used
to back calculate the
concentration of free IL17C in each sample. The concentration of free IL17C
was plotted along the
y-axis while the concentration of IL17RE-FC5 was plotted along the x-axis. The
data was fit with a
1:1 binding model, and a KD was calculated with the Biacore T100
Evaluation software. C) To
optimize the data fit obtained using the Biacore software, the data was
exported into Graphpad
Prism4 software and fit with a four parameter curve. An empirical IC50 value
was calculated from
this data, and this value was used as an approximation of the solution phase
affinity constant
(K. sub. D).
DETAILED DESCRIPTION OF THE INVENTION
[16] IL-17C's ability to bind to members of the IL-17R family has been
investigated.
Accordingly, we now report that we have identified IL-17RE as a receptor for
IL-17C. Since
intervention of other IL-17 family members has been proposed as an effective
therapy for several
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
auto-immune diseases, using antagonists of the present invention, which may
block, inhibit, reduce,
antagonize or neutralize the activity of IL-17C or IL-17RE, and which include
soluble IL-17RE
receptors and neutralizing anti-IL-17RE antibodies, may be advantageous. The
present invention
addresses these needs by providing antagonists to pro-inflammatory cytokine IL-
17C. The invention
further provides uses therefor in inflammatory disease, as well as related
compositions and methods.
A) Overview
[17] Immune related and inflammatory diseases are the manifestation or
consequence of
fairly complex, often multiple interconnected biological pathways which in
normal physiology are
critical to respond to insult or injury, initiate repair from insult or
injury, and mount innate and
acquired defense against foreign organisms. Disease or pathology occurs when
these normal
physiological pathways cause additional insult or injury either as directly
related to the intensity of
the response, as a consequence of abnormal regulation or excessive
stimulation, as a reaction to self,
or as a combination of these.
[18] Though the genesis of these diseases often involves multi-step pathways
and often
multiple different biological systems/pathways, intervention at critical
points in one or more of these
pathways can have an ameliorative or therapeutic effect. Therapeutic
intervention can occur by
either antagonism of a detrimental process/pathway or stimulation of a
beneficial process/pathway.
[19] Many immune related diseases are known and have been extensively studied.
Such
diseases include immune-mediated inflammatory diseases (such as rheumatoid
arthritis, immune
mediated renal disease, hepatobiliary diseases, inflammatory bowel disease
(IBD), irritable bowl
syndrome (IBS) psoriasis, and asthma), non-immune-mediated inflammatory
diseases, infectious
diseases, immunodeficiency diseases, neoplasia, etc.
[20] T lymphocytes (T cells) are an important component of a mammalian immune
response. T cells recognize antigens which are associated with a self-molecule
encoded by genes
within the major histocompatibility complex (MHC). The antigen may be
displayed together with
MHC molecules on the surface of antigen presenting cells, virus infected
cells, cancer cells, grafts,
etc. The T cell system eliminates these altered cells which pose a health
threat to the host mammal. T
cells include helper T cells and cytotoxic T cells. Helper T cells proliferate
extensively following
recognition of an antigen-MHC complex on an antigen presenting cell. Helper T
cells also secrete a
variety of cytokines, i.e., lymphokines, which play a central role in the
activation of B cells,
cytotoxic T cells and a variety of other cells which participate in the immune
response.
[21] A central event in both humoral and cell mediated immune responses is the
activation and clonal expansion of helper T cells. Helper T cell activation is
initiated by the
interaction of the T cell receptor (TCR)--CD3 complex with an antigen-MHC on
the surface of an
antigen presenting cell. This interaction mediates a cascade of biochemical
events that induce the
resting helper T cell to enter a cell cycle (the GO to G1 transition) and
results in the expression of a
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
6
high affinity receptor for IL-2 and sometimes IL-4. The activated T cell
progresses through the cycle
proliferating and differentiating into memory cells or effector cells.
[22] In addition to the signals mediated through the TCR, activation of T
cells involves
additional costimulation induced by cytokines released by the antigen
presenting cell or through
interactions with membrane bound molecules on the antigen presenting cell and
the T cell. The
cytokines IL-1 and IL-6 have been shown to provide a costimulatory signal.
Also, the interaction
between the B7 molecule expressed on the surface of an antigen presenting cell
and CD28 and
CTLA-4 molecules expressed on the T cell surface effect T cell activation.
Activated T cells express
an increased number of cellular adhesion molecules, such as ICAM-1, integrins,
VLA-4, LFA-1,
CD56, etc, as is understood by those skilled in the art.
[23] T-cell proliferation in a mixed lymphocyte culture or mixed lymphocyte
reaction
(MLR) is an established indication of the ability of a compound to stimulate
the immune system. In
many immune responses, inflammatory cells infiltrate the site of injury or
infection. The migrating
cells may be neutrophilic, eosinophilic, monocytic or lymphocytic as can be
determined by
histologic examination of the affected tissues. Current Protocols in
Immunology, ed. John E.
Coligan, 1994, John Wiley & Sons, Inc.
[24] Immune related diseases could be treated by suppressing the immune
response.
Using soluble receptors and/or neutralizing antibodies that inhibit molecules
having immune
stimulatory activity would be beneficial in the treatment of immune-mediated
and inflammatory
diseases. Molecules which inhibit the immune response can be utilized
(proteins directly or via the
use of antibody agonists) to inhibit the immune response and thus ameliorate
immune related disease.
[25] The IL-17 cytokine/receptor families appear to represent a unique
signaling system
within the cytokine network that will offer innovative approaches to the
manipulation of immune and
inflammatory responses. As such, antagonists to IL-17C activity, such as IL-
17RE soluble
receptors, soluble receptors comprising one or more IL-17RE domains, soluble
receptors
comprising one or more IL-17RE ligand binding domains, and antibodies
targeting IL-17RE
receptors, are useful in therapeutic treatment of inflammatory diseases,
particularly as
antagonists to IL-17C in the treatment of asthma or psoriasis. Moreover,
antagonists to IL-
17C activity, such as said soluble receptors and antibodies thereto including
anti-human-IL-
17RE monoclonal and neutralizing antibodies, are useful in therapeutic
treatment of other
inflammatory diseases for example as bind, block, inhibit, reduce, antagonize
or neutralize
IL-17C in the treatment of atopic and contact dermatitis, IBD, IBS, colitis,
endotoxemia,
arthritis, rheumatoid arthritis, psoriatic arthritis, adult respiratory
disease (ARD), septic
shock, multiple organ failure, inflammatory lung injury such as asthma,
chronic obstructive
pulmonary disease (COPD), airway hyper-responsiveness, chronic bronchitis,
allergic
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
7
asthma, bacterial pneumonia, psoriasis, eczema, , and inflammatory bowel
disease such as
ulcerative colitis and Crohn's disease, helicobacter pylori infection,
intraabdominal
adhesions and/or abscesses as results of peritoneal inflammation (i.e. from
infection, injury,
etc.), systemic lupus erythematosus (SLE), multiple sclerosis, systemic
sclerosis, nephrotic
syndrome, organ allograft rejection, graft vs. host disease (GVHD), kidney,
lung, heart, etc.
transplant rejection, streptococcal cell wall (SCW)-induced arthritis,
osteoarthritis,
gingivitis/periodontitis, herpetic stromal keratitis, cancers including
prostate, renal, colon,
ovarian, cervical, leukemia, angiogenesis, restenosis and kawasaki disease.
Thus, said
soluble receptors and said antibodies thereto are useful in the manufacture of
a medicament
for treating such immune based disorders.
[26] Cytokine receptors subunits are characterized by a multi-domain structure
comprising a ligand-binding domain and an effector domain that is typically
involved in signal
transduction. Multimeric cytokine receptors include monomers, homodimers
(e.g., PDGF receptor a
a and (3(3 isoforms, erythropoietin receptor, MPL [thrombopoietin receptor],
and G-CSF receptor),
heterodimers whose subunits each have ligand-binding and effector domains
(e.g., PDGF receptor a
0 isoform), and multimers having component subunits with disparate functions
(e.g., IL-2, IL-3, IL-
4, IL-5, IL-6, IL-7, and GM-CSF receptors). Some receptor subunits are common
to a plurality of
receptors. For example, the AIC2B subunit, which cannot bind ligand on its own
but includes an
intracellular signal transduction domain, is a component of IL-3 and GM-CSF
receptors. Many
cytokine receptors can be placed into one of four related families on the
basis of their structures and
functions. Class I hematopoietic receptors, for example, are characterized by
the presence of a
domain containing conserved cysteine residues and the WSXWS motif. Additional
domains,
including protein kinase domains; fibronectin type III domains; and
immunoglobulin domains, which
are characterized by disulfide-bonded loops, are present in certain
hematopoietic receptors. Cytokine
receptor structure has been reviewed by Urdal, Ann. Reports Med. Chem. 26:221-
228, 1991 and
Cosman, C okine 5:95-106, 1993. It is generally believed that under selective
pressure for
organisms to acquire new biological functions, new receptor family members
arose from duplication
of existing receptor genes leading to the existence of multi-gene families.
Family members thus
contain vestiges of the ancestral gene, and these characteristic features can
be exploited in the
isolation and identification of additional family members.
[27] Amongst other inventions, the present invention provides novel uses for a
soluble
receptor, designated "IL-17RE" or "soluble IL-17RE" or "sIL-17RE", all of
which may be used
herein interchangeably, and neutralizing antibodies to IL-17RE cytokine
receptors. The present
invention also provides soluble IL-17RE polypeptide fragments and fusion
proteins, for use in
human inflammatory and autoimmune diseases. The anti- IL-17RE antibodies and
soluble IL-17RE
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
8
receptors of the present invention, including the neutralizing anti-IL-17RE
antibodies of the present
invention, can be used to block, inhibit, reduce, antagonize or neutralize the
activity of IL-17C in the
treatment of inflammation and inflammatory dieases such as psoriasis,
psoriatic arthritis, rheumatoid
arthritis, endotoxemia, inflammatory bowel disease (IBD), IBS, colitis,
asthma, allograft rejection,
immune mediated renal diseases, hepatobiliary diseases, multiple sclerosis,
atherosclerosis,
promotion of tumor growth, or degenerative joint disease and other
inflammatory conditions
disclosed herein.
[28] An illustrative nucleotide sequence that encodes human IL-17REx1 is
provided by
SEQ ID NO:1; the encoded polypeptide is shown in SEQ ID NO:2. Another
illustrative nucleotide
sequence that encodes human IL-17REx2 is provided by SEQ ID NO:4; the encoded
polypeptide is
shown in SEQ ID NO:5. Another illustrative nucleotide sequence that encodes
human IL-17REx3 is
provided by SEQ ID NO:7; the encoded polypeptide is shown in SEQ ID NO:B.
Another illustrative
nucleotide sequence that encodes human IL-17REx4 is provided by SEQ ID NO:10;
the encoded
polypeptide is shown in SEQ ID NO:11. Another illustrative nucleotide sequence
that encodes
human IL-17REx6 is provided by SEQ ID NO:20 the encoded polypeptide is shown
in SEQ ID
NO:21. Yet another illustrative nucleotide sequence that encodes human IL-
17REx13 is provided by
SEQ ID NO:106; the encoded polypeptide is shown in SEQ ID NO:107. Yet another
illustrative
nucleotide sequence that encodes human IL-17REx14 is provided by SEQ ID
NO:108; the encoded
polypeptide is shown in SEQ ID NO:109. Yet another illustrative nucleotide
sequence that encodes
a variant IL-17REs2 is provided by SEQ ID NO:112; the encoded polypeptide is
shown in SEQ ID
NO:113. Yet another illustrative nucleotide sequence that encodes an
engineered soluble human IL-
17RE, designated as "IL-17REs3" is provided by SEQ ID NO:183, the encoded
polypeptide is
shown in SEQ ID NO:184. Yet another illustrative nucleotide sequence that
encodes an engineered
soluble human IL-17RE, designated as "IL-17REs4" is provided by SEQ ID NO:185,
the encoded
polypeptide is shown in SEQ ID NO:186.
[29] Accordingly, the present invention is directed to IL-17RE or IL-17C
antagonists that
block IL-17C from binding and/or signaling through its corresponding receptor
or receptors (such as
an IL-17RE homodimer or IL-17RE-comprising heterodimer as shown in Example
67). Thus, in
preferred embodiments, such antagonists are based on IL-17RE's polypeptide
structure as depicted in
Figure 1. IL-17RE has a large number of splice variants based on the inclusion
or exclusion of
specific exons.
[30] IL-17RE functions as a receptor for IL-17C (SEQ ID NOs:16 & 17). IL-17RE
can
act as a monomer, a homodimer or a heterodimer. Preferably, IL-17RE acts as a
homodimeric
receptor for IL-17C. IL-17RE can also act as a heterodimeric receptor
comprising two subunits, IL-
17RE and another IL-17family member such as IL-17RA, IL-17RB, IL-17RC and IL-
17RD. Thus,
an IL-17RE comprisimg receptor can bind an IL-17-related cytokine, including
IL-17A, IL-17B, IL-
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
9
17C, IL-17D, IL-17E and IL-17F. IL-17RE is disclosed in commonly owned US
Patent Application
No. 10/192,434, and commonly owned WIPO publication WO 03/006,609, both of
which are
incorporated herein in their entirety by reference. Analysis of a human cDNA
clone encoding IL-
17REx1 (SEQ ID NO:1) revealed an open reading frame encoding 667 amino acids
comprising a
putative signal sequence of approximately 23 amino acid residues (amino acid
residues 1-23 of SEQ
ID NO:2 and 1-23 of SEQ ID NO:3), an extracellular ligand-binding domain of
approximately 431
amino acid residues (amino acid residues 24-454 of SEQ ID NO:2 and 24-454 of
SEQ ID NO:3), a
transmembrane domain of approximately 23 amino acid residues (amino acid
residues 455-477 of
SEQ ID NO:2), and an intracellular domain of approximately 190 amino acid
residues (amino acid
residues 478 to 667 of SEQ ID NO:2).
[31] Yet another illustrative nucleotide sequence that encodes a variant human
IL-17RE,
designated as "IL-17REx2" is provided by SEQ ID NO:4, the encoded polypeptide
is shown in SEQ
ID NO:5. Analysis of a human cDNA clone encoding IL-17REx2 revealed an open
reading frame
encoding 589 amino acids (SEQ ID NO:5) comprising a putative signal sequence
of approximately
23 amino acid residues (amino acid residues 1-23 of SEQ ID NO:5 and 1-23 of
SEQ ID NO:6), an
extracellular ligand-binding domain of approximately 353 amino acid residues
(amino acid residues
24-376 of SEQ ID NO:5 and 24-376 of SEQ ID NO:6), a transmembrane domain of
approximately
23 amino acid residues (amino acid residues 377-399 of SEQ ID NO:5), and an
intracellular domain
of approximately 190 amino acid residues (amino acid residues 400 to 589 of
SEQ ID NO:5).
[32] Yet another illustrative nucleotide sequence that encodes a variant human
IL-17RE,
designated as "IL-17REx3" is provided by SEQ ID NO:7, the encoded polypeptide
is shown in SEQ
ID NO:B. Analysis of a human cDNA clone encoding IL-17REx3 revealed an open
reading frame
encoding 609 amino acids (SEQ ID NO:8) comprising a putative signal sequence
of approximately
23 amino acid residues (amino acid residues 1-23 of SEQ ID NO:8), an
extracellular ligand-binding
domain of approximately 373 amino acid residues (amino acid residues 24-396 of
SEQ ID NO:8,
See SEQ ID NO:9), a transmembrane domain of approximately 23 amino acid
residues (amino acid
residues 397-419 of SEQ ID NO:8), and an intracellular domain of approximately
190 amino acid
residues (amino acid residues 420 to 609 of SEQ ID NO:8).
[33] Yet another illustrative nucleotide sequence that encodes a variant human
IL-17RE
which may be a naturally occurring soluble receptor, designated as "IL-17REx4"
is provided by SEQ
ID NO: 10, the encoded polypeptide is shown in SEQ ID NO:11. Analysis of a
human cDNA clone
encoding IL-17REx4 revealed an open reading frame encoding 533 amino acids
(SEQ ID NO:11)
comprising a putative signal sequence of approximately 23 amino acid residues
(amino acid residues
1 to 23 of SEQ ID NO:11), and an extracellular ligand-binding domain of
approximately 510 amino
acid residues (amino acid residues 24-533 of SEQ ID NO:11. See SEQ ID NO: 12).
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
[34] Yet another illustrative nucleotide sequence that encodes a variant human
IL-17RE,
designated as "IL-17REx6" is provided by SEQ ID NO:20, the encoded polypeptide
is shown in
SEQ ID NO:21. Analysis of a human cDNA clone encoding IL-17REx6 revealed an
open reading
frame encoding 627 amino acids (SEQ ID NO:21) comprising a putative signal
sequence of
approximately 23 amino acid residues (amino acid residues 1 to 23 of SEQ ID
NO:21), a cytoplasmic
domain of approximately 192 amino acid residues (amino acid residues 436 to
627 of SEQ ID
NO:21), a transmembrane domain of approximately 21 amino acid residues (amino
acid residues 415
ot 435 of SEQ ID NO:21) and an extracellular ligand-binding domain of
approximately 391 amino
acid residues (amino acid residues 24-414 of SEQ ID NO:21). An IL-17C binding
domain (or ligand
binding domain) comprises approximately 279 amino acid residues (amino acid
residues 136 to 414
of SEQ ID NO:21).
[35] Yet another illustrative nucleotide sequence that encodes a variant human
IL-17RE
which may be a naturally occurring soluble receptor, designated as "IL-17REx7"
is provided by SEQ
ID NO:22, the encoded polypeptide is shown in SEQ ID NO:23.
[36] Yet another illustrative nucleotide sequence that encodes a variant human
IL-17RE,
designated as "IL-17REx13" is provided by SEQ ID NO:106, the encoded
polypeptide is shown in
SEQ ID NO:107. Analysis of a human cDNA clone encoding IL-17REx13 revealed an
open reading
frame encoding 650 amino acids (SEQ ID NO:107) comprising a putative signal
sequence of
approximately 23 amino acid residues (amino acid residues 1 to 23 of SEQ ID
NO:107), a
cytoplasmic domain of approximately 192 amino acid residues (amino acid
residues 459 to 650 of
SEQ ID NO:107), a transmembrane domain of approximately 27 amino acid residues
(amino acid
residues 459 to 458 of SEQ ID NO:107) and an extracellular ligand-binding
domain of
approximately 414 amino acid residues (amino acid residues 24-437 of SEQ ID
NO:107; SEQ ID
NO:122). an IL-17C binding domain (or ligand binding domain) comprises
approximately 279
amino acid residues (amino acid residues 159 to 437 of SEQ ID NO:107).
[37] Yet another illustrative nucleotide sequence that encodes a variant human
IL-17RE
soluble receptor, designated as "IL-17REx14" is provided by SEQ ID NO:108, the
encoded
polypeptide is shown in SEQ ID NO:109. Analysis of a human cDNA clone encoding
IL-17REx14
revealed an open reading frame encoding 414 amino acids (SEQ ID NO:109)
comprising a putative
signal sequence of approximately 23 amino acid residues (amino acid residues 1
to 23 of SEQ ID
NO:109), and an extracellular ligand-binding domain of approximately 391 amino
acid residues
(amino acid residues 24-414 of SEQ ID NO:109). An IL-17C binding domain (or
ligand binding
domain) comprises approximately 279 amino acid residues (amino acid residues
136 to 414 of SEQ
ID NO:109).
[38] Yet another illustrative nucleotide sequence that encodes an engineered
soluble
human IL-17RE, designated as "IL-17REs2" is provided by SEQ ID NO:112, the
encoded
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
11
polypeptide is shown in SEQ ID NO:113. Figure 1 depicts the amino acid
sequence of IL-17REs2 as
compared to IL-17REx1 (SEQ ID NO:2).
[39] Yet another illustrative nucleotide sequence that encodes an engineered
soluble
human IL-17RE, designated as "IL-17REs3" is provided by SEQ ID NO:183, the
encoded
polypeptide is shown in SEQ ID NO: 184. Figure 1 depicts the amino acid
sequence of IL-17REs3 as
compared to IL-17REx1 (SEQ ID NO:2).
[40] Yet another illustrative nucleotide sequence that encodes an engineered
soluble
human IL-17RE, designated as "IL-17REs4" is provided by SEQ ID NO:185, the
encoded
polypeptide is shown in SEQ ID NO: 186. Figure 1 depicts the amino acid
sequence of IL-17REs4 as
compared to IL-17REx1 (SEQ ID NO:2).
[41] Provided herein are IL-17C binding regions. An illustrative example of an
IL-17C
binding region is provided by the polynucleotide of SEQ ID NO:114; the encoded
polypeptide of
SEQ ID NO:115.
[42] Another illustrative example of a binding region is provided by the
polynucleotide
of SEQ ID NO: 116; the encoded polypeptide of SEQ ID NO: 117.
[43] Yet another illustrative example of a binding region is provided by the
polynucleotide shown in SEQ ID NO: 118; the encoded polypeptide shown in SEQ
ID NO: 119.
[44] The present invention also includes variant IL-17RE soluble receptors
that
comprises at least a fragment of IL-17RE and another IL- 17 family member
(i.e. IL-17RA, IL-17RB,
IL-17RC and/or IL-17RD). These variants are described in Example 67 below.
[45] An illustrative nucleotide sequence that encodes a murine IL-17RE is
provided by
SEQ ID NO:13; the encoded polypeptide is shown in SEQ ID NO:14. Analysis of
murine IL-17RE
revealed an extracellular ligand-binding domain of approximately 638 amino
acid residues (amino
acid residues 26-663 of SEQ ID NO:14; SEQ ID NO:15). Murine IL-17RE functions
as a receptor
for murine IL-17C (SEQ ID NOs:18 & 19).
[46] An illustrative nucleotide sequence that encodes a murine IL-17RE variant
is
provided by SEQ ID NO:160; the encoded polypeptide is shown in SEQ ID NO:161.
Analysis of
murine IL-17RE revealed an extracellular ligand-binding domain of
approximately 568 amino acid
residues (amino acid residues 24-591 of SEQ ID NO:161).
[47] Another illustrative nucleotide sequence that encodes a murine IL-17RE is
provided
by SEQ ID NO:110; the encoded polypeptide is shown in SEQ ID NO:111. Analysis
of murine IL-
17RE revealed a cytoplasmic domain of 201 amino acid residues (amino acid
residues 461 to 661 of
SEQ ID NO:111), a transmembrane domain of 22 amino acid residues (amino acid
residues 439 to
460 of SEQ ID NO:111), an extracellular ligand-binding domain of approximately
415 amino acid
residues (amino acid residues 24 to 438 of SEQ ID NO:111). The murine IL-17C
binding domain
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
12
(or ligand binding domain) comprises approximately 275 amino acid residues
(amino acid residues
136 to 410 of SEQ ID NO:111).
[48] Yet another illustrative nucleotide sequence that encodes an engineered
soluble
murine IL-17RE, designated as "mIL-17REs2" is provided by SEQ ID NO:120, the
encoded
polypeptide is shown in SEQ ID NO:121.
[49] Any IL-17RE polypeptide or fragment thereof as described herein which
includes
exon 14 can have the first cysteine (e.g., this cysteine is amino acid residue
429 of SEQ ID NO:2) of
exon 14 substitued with a Serine or any other amino acid which does not form a
disulfide bond.
[50] Any IL-17RE polypeptide or fragment thereof as described herein can have
an
amino acid substitution at one or more of the following Aspargines
corresponding to amino acid
residues 318, 347 and 364 of SEQ ID NO:2 to, for example, Alanine or any other
amino acid which
does not undergo glycosylation.
[51] The IL-17RE gene resides in human chromosome 3p25.3.
[52] Described below are isolated polypeptides comprising an amino acid
sequence that
is at least 70%, at least 80%, or at least 90%, or greater than 95%, or
greater than 99% or more
identical to a reference amino acid sequence of any of SEQ ID NOs:2, 5, 8, 11,
14, 21, 23, 107, 109,
111 or 113 wherein the isolated polypeptide specifically binds with an
antibody that specifically
binds with a polypeptide comprising the amino acid sequence of any of SEQ ID
NOs: 2, 5, 8, 11, 14,
21, 23, 107, 109, 111, 113, 115, 117 or 119. Also described are isolated
polypeptides comprising an
amino acid sequence that is at least 70%, at least 80%, or at least 90%, or
greater than 95%, or
greater than 99% or more identical to a reference amino acid sequence of 24-
589 of SEQ ID NO:5,
wherein the isolated polypeptide specifically binds with an antibody that
specifically binds with a
polypeptide comprising the amino acid sequence of SEQ ID NO:5. Also described
are isolated
polypeptides comprising an amino acid sequence that is at least 70%, at least
80%, or at least 90%, or
greater than 95%, or greater than 99% or more identical to a reference amino
acid sequence of 24-
609 of SEQ ID NO:8, wherein the isolated polypeptide specifically binds with
an antibody that
specifically binds with a polypeptide comprising the amino acid sequence of
SEQ ID NO:B. Also
described herein are isolated polypeptides comprising an amino acid sequence
that is at least 70%, at
least 80%, or at least 90%, or greater than 95%, or greater than 99% or more
identical to a reference
amino acid sequence of 24-533 of SEQ ID NO: 11, wherein the isolated
polypeptide specifically
binds with an antibody that specifically binds with a polypeptide comprising
the amino acid
sequence of SEQ ID NO: 11. Also described herein areisolated polypeptides
comprising an amino
acid sequence that is at least 70%, at least 80%, or at least 90%, or greater
than 95%, or greater than
99% or more identical to any of SEQ ID NOs: 2, 5, 8, 11, 14, 21, 23, 107, 109,
111, 113, 115, 117 or
119, wherein the isolated polypeptide specifically binds with an antibody that
specifically binds with
a polypeptide comprising the amino acid sequence of any of SEQ ID NOs: 2, 5,
8, 11, 14, 21, 23,
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
13
107, 109, 111, 113, 115, 117 or 119. Also described herein areisolated
polypeptides comprising an
amino acid sequence that is at least 70%, at least 80%, or at least 90%, or
greater than 95%, or
greater than 99% or more identical to any of SEQ ID NOs: 2, 5, 8, 11, 14, 21,
23, 107, 109, 111, 113,
115, 117 or 119, wherein the isolated polypeptide specifically binds with an
antibody that
specifically binds with a polypeptide comprising the amino acid sequence of
any of SEQ ID NOs: 2,
5, 8, 11, 14, 21, 23, 107, 109, 111, 113, 115, 117 or 119. Also described
herein areisolated
polypeptides comprising an amino acid sequence that is at least 70%, at least
80%, or at least 90%, or
greater than 95%, or greater than 99% or more identical to a reference amino
acid sequence of 26-
663 of SEQ ID NO:17, wherein the isolated polypeptide specifically binds with
an antibody that
specifically binds with a polypeptide comprising the amino acid sequence of
SEQ ID NO:17.
[53] As is used herein, the terms "at least 70% identical" or "at least 70%
identity"
means that an amino acid sequence shares 70%-100% sequence identity with a
reference
sequence. This range of identity is inclusive of all whole (e.g., 70%, 75%,
79%, 87%, 93%,
98%) or partial numbers (e.g., 72.15, 87.27%, 92.83%, 98.11% - to two
significant figures)
embraced within the recited range numbers, therefore forming a part of this
description..
For example, an amino acid sequence with 200 residues that share 85% identity
with a
reference sequence would have 170 identical residues and 30 non-identical
residues.
Similarly, an amino acid sequence with 235 residues may have 200 residues that
are
identical to a reference sequence, thus the amino acid sequence will be 85.11
% identical to
the reference sequence. Similarly, the terms "at least 80%" and "at least 90%"
are inclusive
of all whole or partial numbers within the recited range. As is used herein,
the terms
"greater than 95% identical" or "greater than 95% identity" means that an
amino acid
sequence shares 95.01%-100% sequence identity with a reference sequence. This
range is
all inclusive as described immediately above. Those ordinarily skilled in the
are will readily
calculate percent identity between an amino acid and a reference sequence.
[54] The present invention also provides isolated polypeptides comprising an
extracellular domain, wherein the extracellular domain comprises an amino acid
sequence with at
least 70% identity to an amino acid sequence selected from the group
consisting of: (a) amino acid
residues 24 to 454 of SEQ ID NO:2, (b) SEQ ID NO:3; (c) amino acid residues 24-
376 of SEQ ID
NO:5; (d) SEQ ID NO:6; (e) amino acid residues 24-396 of SEQ ID NO:8; (f) SEQ
ID NO:9; (g)
amino acid residues 24-533 of SEQ ID NO: 11; (h) SEQ ID NO: 12; (i) amino acid
residues 26-663 of
SEQ ID NO:14; (j) SEQ ID NO:15, (k) one or more of SEQ ID NO:MBD, SEQ ID
NO:2BD or SEQ
ID NO:3BD, wherein the isolated polypeptide specifically binds with an
antibody that specifically
binds with a polypeptide consisting of either the amino acid sequence of any
of SEQ ID NOs: 2, 5, 8,
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
14
11, 14, 21, 23, 107, 109, 111, 113, 115, 117 or 119. Such polypeptides may
further comprise a
transmembrane domain that resides in a carboxyl-terminal position relative to
the extracellular
domain, wherein the transmembrane domain comprises an amino acid sequence
selected from the
group consisting of: (a) amino acid residues 455 to 477 of SEQ ID NO:2; (b)
amino acid residues
377 to 399 of SEQ ID NO:5; or (c) amino acid residues 397 to 419 of SEQ ID
NO:B. These
polypeptides may also comprise an intracellular domain that resides in a
carboxyl-terminal position
relative to the transmembrane domain, and optionally, a signal secretory
sequence that resides in an
amino-terminal position relative to the extracellular domain.
[55] The present invention also includes variant IL-17RE polypeptides, wherein
the
amino acid sequence of the variant polypeptide shares an identity with the
amino acid sequence of
SEQ ID NOs: 2, 5, 8, 11, 14, 21, 23, 107, 109, 111, 113, 115, 117 or 119,
selected from the group
consisting of at least 70% identity, at least 80% identity, at least 90%
identity, at least 95% identity,
or greater than 95% identity, and wherein any difference between the amino
acid sequence of the
variant polypeptide and the amino acid sequence of SEQ ID NOs: 2, 5, 8, 11,
14, 21, 23, 107, 109,
111, 113, 115, 117 or 119is due to one or more conservative amino acid
substitutions.
[56] Moreover, the present invention also provides isolated polypeptides as
disclosed
above that bind IL-17C (e.g., human IL-17C polypeptide sequence as shown in
SEQ ID NO:17).
The human IL-17C polynucleotide sequence is shown in SEQ ID NO:16. The mouse
IL-17C
polynucleotide sequence is shown in SEQ ID NO:18, and corresponding
polyepeptide is shown in
SEQ ID NO:19.
[57] The present invention also provides isolated polypeptides and epitopes
comprising at
least 15 contiguous amino acid residues of an amino acid sequence of SEQ ID
NOs: 2, 5, 8, 11, 14,
21, 23, 107, 109, 111, 113, 115, 117 or 119. Illustrative polypeptides include
polypeptides that either
comprise, or consist of SEQ ID NOs: 2, 5, 8, 11, 14, 21, 23, 107, 109, 111,
113, 115, 117 or 119, an
antigenic epitope thereof, or a functional IL-17C binding fragment thereof.
Moreover, the present
invention also provides isolated polypeptides as disclosed above that bind to,
block, inhibit, reduce,
antagonize or neutralize the activity of IL-17C.
[58] The present invention also includes variant IL-17RE polypeptides, wherein
the
amino acid sequence of the variant polypeptide shares an identity with the
amino acid residues of
SEQ ID NO: SEQ ID NOs: 2, 5, 8, 11, 14, 21, 23, 107, 109, 111, 113, 115, 117
or 119 selected from
the group consisting of at least 70% identity, at least 80% identity, at least
90% identity, at least 95%
identity, or greater than 95% identity, or greater than 99% or more identity,
and wherein any
difference between the amino acid sequence of the variant polypeptide and the
corresponding amino
acid sequence is due to one or more conservative amino acid substitutions.
Such conservative amino
acid substitutions are described herein. Moreover, the present invention also
provides isolated
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
polypeptides as disclosed above that bind to, block, inhibit, reduce,
antagonize or neutralize the
activity of IL-17C.
[59] The present invention further provides antibodies and antibody fragments
that
specifically bind with such polypeptides. Exemplary antibodies include
neutralizing antibodies,
polyclonal antibodies, murine monoclonal antibodies, humanized antibodies
derived from murine
monoclonal antibodies, and human monoclonal antibodies. Illustrative antibody
fragments include
F(ab')z, F(ab)2, Fab', Fab, Fv, scFv, and minimal recognition units.
Neutralizing antibodies
preferably bind IL-17RE such that the interaction of IL-17C with IL-17RE is
blocked, inhibited,
reduced, antagonized or neutralized; anti-IL-17RE neutralizing antibodies such
that the binding of
either IL-17C to IL-17RE is blocked, inhibited, reduced, antagonized or
neutralized are also
encompassed by the present invention. That is, the neutralizing anti-IL-17RE
antibodies of the
present invention can either either bind, block, inhibit, reduce, antagonize
or neutralize IL-17C
singly, or bind, block, inhibit, reduce, antagonize or neutralize IL-17C and
another cytokine, such as
together. The present invention further includes compositions comprising a
carrier and a peptide,
polypeptide, or antibody described herein.
[60] In addition, the present invention provides pharmaceutical compositions
comprising
a pharmaceutically acceptable carrier and at least one of such an expression
vector or recombinant
virus comprising such expression vectors. The present invention further
includes pharmaceutical
compositions, comprising a pharmaceutically acceptable carrier and a
polypeptide or antibody
described herein.
[61] The present invention also contemplates anti-idiotype antibodies, or anti-
idiotype
antibody fragments, that specifically bind an antibody or antibody fragment
that specifically binds a
polypeptide comprising the amino acid sequence of SEQ ID NOs: 2, 5, 8, 11, 14,
21, 23, 107, 109,
111, 113, 115, 117 or 119or a fragment thereof. An exemplary anti-idiotype
antibody binds with an
antibody that specifically binds a polypeptide consisting of any of SEQ ID
NOs: 2, 5, 8, 11, 14, 21,
23, 107, 109, 111, 113, 115, 117 or 119.
[62] The present invention also provides fusion proteins, comprising a IL-17RE
polypeptide and an immunoglobulin moiety. In such fusion proteins, the
immunoglobulin moiety
may be an immunoglobulin heavy chain constant region, such as a human Fc
fragment. The present
invention further includes isolated nucleic acid molecules that encode such
fusion proteins (e.g. SEQ
ID NO:123).
[63] The present invention also provides polyclonal and monoclonal antibodies
that bind
to polypeptides comprising an IL-17RE extracellular domain such as monomeric,
homodimeric,
heterodimeric and multimeric receptors, including soluble receptors. Moreover,
such antibodies can
be used antagonize the binding of IL-17RE ligands, such as IL-17C (SEQ ID
NO:17), to the IL-
17RE receptor.
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
16
[64] These and other aspects of the invention will become evident upon
reference to the
following detailed description. In addition, various references are identified
below and are
incorporated by reference in their entirety.
B) Definitions
[65] In the description that follows, a number of terms are used extensively.
The
following definitions are provided to facilitate understanding of the
invention.
[66] As used herein, "nucleic acid" or "nucleic acid molecule" refers to
polynucleotides,
such as deoxyribonucleic acid (DNA) or ribonucleic acid (RNA),
oligonucleotides, fragments
generated by the polymerase chain reaction (PCR), and fragments generated by
any of ligation,
scission, endonuclease action, and exonuclease action. Nucleic acid molecules
can be composed of
monomers that are naturally-occurring nucleotides (such as DNA and RNA), or
analogs of naturally-
occurring nucleotides (e.g., a-enantiomeric forms of naturally-occurring
nucleotides), or a
combination of both. Modified nucleotides can have alterations in sugar
moieties and/or in
pyrimidine or purine base moieties. Sugar modifications include, for example,
replacement of one or
more hydroxyl groups with halogens, alkyl groups, amines, and azido groups, or
sugars can be
functionalized as ethers or esters. Moreover, the entire sugar moiety can be
replaced with sterically
and electronically similar structures, such as aza-sugars and carbocyclic
sugar analogs. Examples of
modifications in a base moiety include alkylated purines and pyrimidines,
acylated purines or
pyrimidines, or other well-known heterocyclic substitutes. Nucleic acid
monomers can be linked by
phosphodiester bonds or analogs of such linkages. Analogs of phosphodiester
linkages include
phosphorothioate, phosphorodithioate, phosphoroselenoate,
phosphorodiselenoate,
phosphoroanilothioate, phosphoranilidate, phosphoramidate, and the like. The
term "nucleic acid
molecule" also includes so-called "peptide nucleic acids," which comprise
naturally-occurring or
modified nucleic acid bases attached to a polyamide backbone. Nucleic acids
can be either single
stranded or double stranded.
[67] The term "complement of a nucleic acid molecule" refers to a nucleic acid
molecule
having a complementary nucleotide sequence and reverse orientation as compared
to a reference
nucleotide sequence. For example, the sequence 5' ATGCACGGG 3' is
complementary to 5'
CCCGTGCAT 3'.
[68] The term "degenerate nucleotide sequence" denotes a sequence of
nucleotides that
includes one or more degenerate codons as compared to a reference nucleic acid
molecule that
encodes a polypeptide. Degenerate codons contain different triplets of
nucleotides, but encode the
same amino acid residue (i.e., GAU and GAC triplets each encode Asp).
[69] The term "structural gene" refers to a nucleic acid molecule that is
transcribed into
messenger RNA (mRNA), which is then translated into a sequence of amino acids
characteristic of a
specific polypeptide.
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
17
[70] An "isolated nucleic acid molecule" is a nucleic acid molecule that is
not integrated in
the genomic DNA of an organism. For example, a DNA molecule that encodes a
growth factor that has
been separated from the genomic DNA of a cell is an isolated DNA molecule.
Another example of an
isolated nucleic acid molecule is a chemically-synthesized nucleic acid
molecule that is not integrated in
the genome of an organism. A nucleic acid molecule that has been isolated from
a particular species is
smaller than the complete DNA molecule of a chromosome from that species.
[71] A "nucleic acid molecule construct" is a nucleic acid molecule, either
single- or
double-stranded, that has been modified through human intervention to contain
segments of nucleic
acid combined and juxtaposed in an arrangement not existing in nature.
[72] "Linear DNA" denotes non-circular DNA molecules having free 5' and 3'
ends.
Linear DNA can be prepared from closed circular DNA molecules, such as
plasmids, by enzymatic
digestion or physical disruption.
[73] "Complementary DNA (cDNA)" is a single-stranded DNA molecule that is
formed
from an mRNA template by the enzyme reverse transcriptase. Typically, a primer
complementary to
portions of mRNA is employed for the initiation of reverse tra.nscription.
Those skilled in the art also use
the term "cDNA" to refer to a double-stranded DNA molecule consisting of such
a single-stranded DNA
molecule and its complementary DNA strand. The term "cDNA" also refers to a
clone of a cDNA
molecule synthesized from an RNA template.
[74] A "promoter" is a nucleotide sequence that directs the transcription of a
structural gene.
Typically, a promoter is located in the 5' non-coding region of a gene,
proximal to the tra.nscriptional start
site of a structural gene. Sequence elements within promoters that function in
the initiation of
transcription are often characterized by consensus nucleotide sequences. These
promoter elements
include RNA polymerase binding sites, TATA sequences, CAAT sequences,
differentiation-specific
elements (DSEs; McGehee et al., Mol. Endocrinol. 7:551 (1993)), cyclic AMP
response elements
(CREs), serum response elements (SREs; Treisman, Seminars in Cancer Biol. 1:47
(1990)),
glucocorticoid response elements (GREs), and binding sites for other
transcription factors, such as
CRE/ATF (O'Reilly et al., J. Biol. Chem. 267:19938 (1992)), AP2 (Ye et al., J.
Biol. Chem.
269:25728 (1994)), SP1, cAMP response element binding protein (CREB; Loeken,
Gene Expr.
3:253 (1993)) and octamer factors (see, in general, Watson et al., eds.,
Molecular Biology of the
Gene, 4th ed. (The Benjamin/Cummings Publishing Company, Inc. 1987), and
Lemaigre and
Rousseau, Biochem. J. 303:1 (1994)). If a promoter is an inducible promoter,
then the rate of
tra.nscription increases in response to an inducing agent. In contrast, the
rate of tra.nscription is not
regulated by an inducing agent if the promoter is a constitutive promoter.
Repressible promoters are also
known.
[75] A "core promoter" contains essential nucleotide sequences for promoter
function,
including the TATA box and start of transcription. By this definition, a core
promoter may or may
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
18
not have detectable activity in the absence of specific sequences that may
enhance the activity or
confer tissue specific activity.
[76] A "regulatory element" is a nucleotide sequence that modulates the
activity of a core
promoter. For example, a regulatory element may contain a nucleotide sequence
that binds with
cellular factors enabling transcription exclusively or preferentially in
particular cells, tissues, or
organelles. These types of regulatory elements are normally associated with
genes that are expressed
in a "cell-specific," "tissue-specific," or "organelle-specific" manner.
[77] An "enhancef' is a type of regulatory element that can increase the
efficiency of
tra.nscription, regardless of the distance or orientation of the enhancer
relative to the start site of
tra.nscription.
[78] "Heterologous DNA" refers to a DNA molecule, or a population of DNA
molecules,
that does not exist naturally within a given host cell. DNA molecules
heterologous to a particular
host cell may contain DNA derived from the host cell species (i.e., endogenous
DNA) so long as that
host DNA is combined with non-host DNA (i.e., exogenous DNA). For example, a
DNA molecule
containing a non-host DNA segment encoding a polypeptide operably linked to a
host DNA segment
comprising a transcription promoter is considered to be a heterologous DNA
molecule. Conversely,
a heterologous DNA molecule can comprise an endogenous gene operably linked
with an exogenous
promoter. As another illustration, a DNA molecule comprising a gene derived
from a wild-type cell
is considered to be heterologous DNA if that DNA molecule is introduced into a
mutant cell that
lacks the wild-type gene.
[79] A "polypeptide" is a polymer of amino acid residues joined by peptide
bonds,
whether produced naturally or synthetically. Polypeptides of less than about
10 amino acid residues
are commonly referred to as "peptides."
[80] A"protein" is a macromolecule comprising one or more polypeptide chains.
A
protein may also comprise non-peptidic components, such as carbohydrate
groups. Carbohydrates
and other non-peptidic substituents may be added to a protein by the cell in
which the protein is
produced, and will vary with the type of cell. Proteins are defined herein in
terms of their amino acid
backbone structures; substituents such as carbohydrate groups are generally
not specified, but may be
present nonetheless.
[81] A peptide or polypeptide encoded by a non-host DNA molecule is a
"heterologous"
peptide or polypeptide.
[82] A "cloning vectof' is a nucleic acid molecule, such as a plasmid, cosmid,
or
bacteriophage, that has the capability of replicating autonomously in a host
cell. Cloning vectors
typically contain one or a small number of restriction endonuclease
recognition sites that allow insertion
of a nucleic acid molecule in a determinable fashion without loss of an
essential biological function of the
vector, as well as nucleotide sequences encoding a marker gene that is
suitable for use in the
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
19
identification and selection of cells tra.nsformed with the cloning vector.
Marker genes typically include
genes that provide tetracycline resistance or ampicillin resistance.
[83] An "expression vector" is a nucleic acid molecule encoding a gene that is
expressed in a
host cell. Typically, an expression vector comprises a transcription promoter,
a gene, and a tra.nscription
terminator. Gene expression is usually placed under the control of a promoter,
and such a gene is said to
be "operably linked to" the promoter. Similarly, a regulatory element and a
core promoter are operably
linked if the regulatory element modulates the activity of the core promoter.
[84] A "recombinant host" is a cell that contains a heterologous nucleic acid
molecule, such
as a cloning vector or expression vector. In the present context, an example
of a recombinant host is a cell
that produces IL-17RE from an expression vector. In contrast, IL-17RE can be
produced by a cell that
is a "natural source" of IL-17RE, and that lacks an expression vector.
[85] "Integrative transformants" are recombinant host cells, in which
heterologous DNA
has become integrated into the genomic DNA of the cells.
[86] A "fusion protein" is a hybrid protein expressed by a nucleic acid
molecule
comprising nucleotide sequences of at least two genes. For example, a fusion
protein can comprise
at least part of a IL-17RE polypeptide fused with a polypeptide that binds an
affinity matrix. Such a
fusion protein provides a means to isolate large quantities of IL-17RE using
affinity chromatography.
[87] The term "receptor" denotes a cell-associated protein that binds to a
bioactive
molecule termed a "ligand." This interaction mediates the effect of the ligand
on the cell. Receptors
can be membrane bound, cytosolic or nuclear; monomeric (e.g., thyroid
stimulating hormone
receptor, beta-adrenergic receptor) or multimeric (e.g., PDGF receptor, growth
hormone receptor, IL-
3 receptor, GM-CSF receptor, G-CSF receptor, erythropoietin receptor and IL-6
receptor).
Membrane-bound receptors are characterized by a multi-domain structure
comprising an
extracellular ligand-binding domain and an intracellular effector domain that
is typically involved in
signal transduction. In certain membrane-bound receptors, the extracellular
ligand-binding domain
and the intracellular effector domain are located in separate polypeptides
that comprise the complete
functional receptor.
[88] In general, the binding of ligand to receptor results in a conformational
change in the
receptor that causes an interaction between the effector domain and other
molecule(s) in the cell,
which in turn leads to an alteration in the metabolism of the cell. Metabolic
events that are often
linked to receptor-ligand interactions include gene transcription,
phosphorylation,
dephosphorylation, increases in cyclic AMP production, mobilization of
cellular calcium,
mobilization of membrane lipids, cell adhesion, hydrolysis of inositol lipids
and hydrolysis of
phospholipids.
[89] A"soluble receptor" is a receptor polypeptide that is not bound to a cell
membrane.
Soluble receptors are most commonly ligand-binding receptor polypeptides that
lack transmembrane
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
and cytoplasmic domains, and other linkage to the cell membrane such as via
glycophosphoinositol
(gpi). Soluble receptors can comprise additional amino acid residues, such as
affinity tags that
provide for purification of the polypeptide or provide sites for attachment of
the polypeptide to a
substrate, or immunoglobulin constant region sequences. Many cell-surface
receptors have naturally
occurring, soluble counterparts that are produced by proteolysis or translated
from alternatively
spliced mRNAs. Soluble receptors can be monomeric, homodimeric, heterodimeric,
or multimeric,
with multimeric receptors generally not comprising more than 9 subunits,
preferably not comprising
more than 6 subunits, and most preferably not comprising more than 3 subunits.
Receptor
polypeptides are said to be substantially free of transmembrane and
intracellular polypeptide
segments when they lack sufficient portions of these segments to provide
membrane anchoring or
signal transduction, respectively. Soluble receptors of cytokine receptors
generally comprise the
extracellular cytokine binding domain free of a transmsmbrane domain and
intracellular domain. For
example, representative soluble receptors include soluble receptors for IL-17R
as shown in SEQ ID
NOs:3, or 113. It is well within the level of one of skill in the art to
delineate what sequences of a
known cytokine receptor sequence comprise the extracellular cytokine binding
domain free of a
transmsmbrane domain and intracellular domain. Moreover, one of skill in the
art using the genetic
code can readily determine polynucleotides that encode such soluble receptor
polyptides.
[90] The term "secretory signal sequence" denotes a DNA sequence that encodes
a
peptide (a "secretory peptide") that, as a component of a larger polypeptide,
directs the larger
polypeptide through a secretory pathway of a cell in which it is synthesized.
The larger polypeptide
is commonly cleaved to remove the secretory peptide during transit through the
secretory pathway.
[91] An "isolated polypeptide" is a polypeptide that is essentially free from
contaminating cellular components, such as carbohydrate, lipid, or other
proteinaceous impurities
associated with the polypeptide in nature. Typically, a preparation of
isolated polypeptide contains the
polypeptide in a highly purified form, i.e., at least 80% pure, at least 90%
pure, at least 95% pure,
greater than 95% pure, or greater than 99% pure. One way to show that a
particular protein
preparation contains an isolated polypeptide is by the appearance of a single
band following sodium
dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis of the protein
preparation and Coomassie
Brilliant Blue staining of the gel. However, the term "isolated" does not
exclude the presence of the
same polypeptide in alternative physical forms, such as dimers or
alternatively glycosylated or
derivatized forms. As was described above, the term "at least 80% pure" is
inclusive of all whole or
partial numbers from 80% purity to 100% purity. This same applies to "at least
90% pure" and "at
least 95% pure." The term "greater than 95% pure" means 95.01% to 100% purity,
as described
above.
[92] The terms "amino-terminal" and "carboxyl-terminal" are used herein to
denote
positions within polypeptides. Where the context allows, these terms are used
with reference to a
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
21
particular sequence or portion of a polypeptide to denote proximity or
relative position. For
example, a certain sequence positioned carboxyl-terminal to a reference
sequence within a
polypeptide is located proximal to the carboxyl terminus of the reference
sequence, but is not
necessarily at the carboxyl terminus of the complete polypeptide.
[93] The term "expression" refers to the biosynthesis of a gene product. For
example, in the
case of a structural gene, expression involves tra.nscription of the
structural gene into mRNA and the
translation of mRNA into one or more polypeptides.
[94] The term "splice variant" is used herein to denote alternative forms of
RNA
transcribed from a gene. Splice variation arises naturally through use of
alternative splicing sites
within a transcribed RNA molecule, or less commonly between separately
transcribed RNA
molecules, and may result in several mRNAs transcribed from the same gene.
Splice variants may
encode polypeptides having altered amino acid sequence. The term splice
variant is also used herein
to denote a polypeptide encoded by a splice variant of an mRNA transcribed
from a gene.
[95] As used herein, the term "immunomodulator" includes cytokines, stem cell
growth
factors, lymphotoxins, co-stimulatory molecules, hematopoietic factors, an
dthe like, and synthetic
analogs of these molecules.
[96] The term "complement/anti-complement pair" denotes non-identical moieties
that
form a non-covalently associated, stable pair under appropriate conditions.
For instance, biotin and
avidin (or streptavidin) are prototypical members of a complement/anti-
complement pair. Other
exemplary complement/anti-complement pairs include receptor/ligand pairs,
antibody/antigen (or
hapten or epitope) pairs, sense/antisense polynucleotide pairs, and the like.
Where subsequent
dissociation of the complement/anti-complement pair is desirable, the
complement/anti-complement
pair preferably has a binding affinity of less than 109 M-i .
[97] An "anti-idiotype antibody" is an antibody that binds with the variable
region
domain of an immunoglobulin. In the present context, an anti-idiotype antibody
binds with the
variable region of an anti-IL-17RE antibody, and thus, an anti-idiotype
antibody mimics an epitope
of IL-17RE.
[98] An "antibody fragment" is a portion of an antibody such as F(ab')z,
F(ab)2, Fab', Fab,
and the like. Regardless of structure, an antibody fragment binds with the
same antigen that is
recognized by the intact antibody. For example, an anti-IL-17RE monoclonal
antibody fragment binds
with an epitope of IL-17RE.
[99] The term "antibody fragment" also includes a synthetic or a genetically
engineered
polypeptide that binds to a specific antigen, such as polypeptides consisting
of the light chain variable
region, "Fv" fragments consisting of the variable regions of the heavy and
light chains, recombinant
single chain polypeptide molecules in which light and heavy variable regions
are connected by a peptide
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
22
linker ("scFv proteins"), and minimal recognition units consisting of the
amino acid residues that mimic
the hypervariable region.
[100] A "chimeric antibody" is a recombinant protein that contains the
variable domains and
complementary determining regions derived from a rodent antibody, while the
remainder of the antibody
molecule is derived from a human antibody.
[101] "Humanized antibodies" are recombinant proteins in which murine
complementarity
determining regions of a monoclonal antibody have been transferred from heavy
and light variable chains
of the murine immunoglobulin into a human variable domain. Construction of
humanized antibodies for
therapeutic use in humans that are derived from murine antibodies, such as
those that bind to or neutralize
a human protein, is within the skill of one in the art.
[102] As used herein, a "therapeutic agent" is a molecule or atom which is
conjugated to
an antibody moiety to produce a conjugate which is useful for therapy.
Examples of therapeutic
agents include drugs, toxins, immunomodulators, chelators, boron compounds,
photoactive agents or
dyes, and radioisotopes.
[103] A "detectable label" is a molecule or atom which can be conjugated to an
antibody
moiety to produce a molecule useful for diagnosis. Examples of detectable
labels include chelators,
photoactive agents, radioisotopes, fluorescent agents, paramagnetic ions, or
other marker moieties.
[104] The term "affinity tag" is used herein to denote a polypeptide segment
that can be
attached to a second polypeptide to provide for purification or detection of
the second polypeptide or
provide sites for attachment of the second polypeptide to a substrate. In
principal, any peptide or
protein for which an antibody or other specific binding agent is available can
be used as an affinity
tag. Affinity tags include a poly-histidine tract, protein A (Nilsson et al.,
EMBO J. 4:1075 (1985);
Nilsson et al., Methods Enzymol. 198:3 (1991)), glutathione S transferase
(Smith and Johnson, Gene
67:31 (1988)), Glu-Glu affinity tag (Grussenmeyer et al., Proc. Natl. Acad.
Sci. USA 82:7952
(1985)), substance P, FLAG peptide (Hopp et al., Biotechnology 6:1204 (1988)),
streptavidin
binding peptide, or other antigenic epitope or binding domain. See, in
general, Ford et al., Protein
Expression and Purification 2:95 (1991). DNA molecules encoding affinity tags
are available from
commercial suppliers (e.g., Pharmacia Biotech, Piscataway, NJ).
[105] A "naked antibody" is an entire antibody, as opposed to an antibody
fragment,
which is not conjugated with a therapeutic agent. Naked antibodies include
both polyclonal and
monoclonal antibodies, as well as certain recombinant antibodies, such as
chimeric and humanized
antibodies.
[106] As used herein, the term "antibody component" includes both an entire
antibody and
an antibody fragment.
[107] An "immunoconjugate" is a conjugate of an antibody component with a
therapeutic
agent or a detectable label.
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
23
[108] As used herein, the term "antibody fusion protein" refers to a
recombinant molecule
that comprises an antibody component and a IL-17RE polypeptide component.
Examples of an
antibody fusion protein include a protein that comprises a IL-17RE
extracellular domain, and either
an Fc domain or an antigen-binding region (e.g. SEQ ID NO: 123).
[109] A "target polypeptide" or a "target peptide" is an amino acid sequence
that
comprises at least one epitope, and that is expressed on a target cell, such
as a tumor cell, or a cell
that carries an infectious agent antigen. T cells recognize peptide epitopes
presented by a major
histocompatibility complex molecule to a target polypeptide or target peptide
and typically lyse the
target cell or recruit other immune cells to the site of the target cell,
thereby killing the target cell.
[110] An "antigenic peptide" is a peptide which will bind a major
histocompatibility
complex molecule to form an MHC-peptide complex which is recognized by a T
cell, thereby
inducing a cytotoxic lymphocyte response upon presentation to the T cell.
Thus, antigenic peptides
are capable of binding to an appropriate major histocompatibility complex
molecule and inducing a
cytotoxic T cells response, such as cell lysis or specific cytokine release
against the target cell which
binds or expresses the antigen. The antigenic peptide can be bound in the
context of a class I or class
II major histocompatibility complex molecule, on an antigen presenting cell or
on a target cell.
[111] In eukaryotes, RNA polymerase II catalyzes the transcription of a
structural gene to
produce mRNA. A nucleic acid molecule can be designed to contain an RNA
polymerase II
template in which the RNA transcript has a sequence that is complementary to
that of a specific
mRNA. The RNA transcript is termed an "anti-sense RNA" and a nucleic acid
molecule that
encodes the anti-sense RNA is termed an "anti-sense gene." Anti-sense RNA
molecules are capable
of binding to mRNA molecules, resulting in an inhibition of mRNA translation.
[112] An "anti-sense oligonucleotide specific for IL-17RE" or a "IL-17RE anti-
sense
oligonucleotide" or an "antisense RNA modulator" is an oligonucleotide having
a sequence (a)
capable of forming a stable triplex with a portion of the IL-17RE gene, or (b)
capable of forming a
stable duplex with a portion of an mRNA transcript of the IL-17RE gene.
Antisense RNA
oligonucleotides as used herein can refer to both the single stranded
technology and the double
stranded technology, which is more commenly referred to as siRNA.
[113] A "ribozyme" is a nucleic acid molecule that contains a catalytic
center. The term
includes RNA enzymes, self-splicing RNAs, self-cleaving RNAs, and nucleic acid
molecules that
perform these catalytic functions. A nucleic acid molecule that encodes a
ribozyme is termed a
"ribozyme gene."
[114] An "external guide sequence" is a nucleic acid molecule that directs the
endogenous
ribozyme, RNase P, to a particular species of intracellular mRNA, resulting in
the cleavage of the
mRNA by RNase P. A nucleic acid molecule that encodes an external guide
sequence is termed an
"external guide sequence gene."
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
24
[115] The term "variant IL-17RE gene" refers to nucleic acid molecules that
encode a
polypeptide having an amino acid sequence that is a modification of SEQ ID
NOs: 2, 5, 8, 11, 14, 21,
23, 107, 109, 111, 113, 115, 117 or 119. Such variants include naturally-
occurring polymorphisms
of IL-17RE genes, as well as synthetic genes that contain conservative amino
acid substitutions of the
amino acid sequence of SEQ ID NOs: 2, 5, 8, 11, 14, 21, 23, 107, 109, 111,
113, 115, 117 or 119.
Additional variant forms of IL-17RE genes are nucleic acid molecules that
contain substitutions,
insertions or deletions of the nucleotide sequences described herein. A
variant IL-17RE gene can be
identified, for example, by determining whether the gene hybridizes with a
nucleic acid molecule
having the nucleotide sequence of SEQ ID NOs:1, 4, 7, 10, 13, 20, 22, 106,
108, 110 or 112, or any
of their complements, under stringent conditions.
[116] Alternatively, variant IL-17RE genes can be identified by sequence
comparison.
Two amino acid sequences have "100% amino acid sequence identity" if the amino
acid residues of
the two amino acid sequences are the same when aligned for maximal
correspondence. Similarly,
two nucleotide sequences have "100% nucleotide sequence identity" if the
nucleotide residues of the
two nucleotide sequences are the same when aligned for maximal correspondence.
Sequence
comparisons can be performed using standard software programs such as those
included in the
LASERGENE bioinformatics computing suite, which is produced by DNASTAR
(Madison,
Wisconsin). Other methods for comparing two nucleotide or amino acid sequences
by determining
optimal alignment are well-known to those of skill in the art (see, for
example, Peruski and Peruski,
The Internet and the New Biology: Tools for Genomic and Molecular Research
(ASM Press, Inc.
1997), Wu et al. (eds.), "Information Superhighway and Computer Databases of
Nucleic Acids and
Proteins," in Methods in Gene Biotechnology, pages 123-151 (CRC Press, Inc.
1997), and Bishop
(ed.), Guide to Human Genome Computing, 2nd Edition (Academic Press, Inc.
1998)). Particular
methods for determining sequence identity are described below.
[117] Regardless of the particular method used to identify a variant IL-17RE
gene or
variant IL-17RE polypeptide, a variant gene or polypeptide encoded by a
variant gene may be
functionally characterized the ability to bind specifically to an anti-IL-17RE
antibody. A variant IL-
17RE gene or variant IL-17RE polypeptide may also be functionally
characterized the ability to bind
to its ligand, for example, IL-17C, using a biological or biochemical assay
described herein.
[118] The term "allelic variant" is used herein to denote any of two or more
alternative
forms of a gene occupying the same chromosomal locus. Allelic variation arises
naturally through
mutation, and may result in phenotypic polymorphism within populations. Gene
mutations can be
silent (no change in the encoded polypeptide) or may encode polypeptides
having altered amino acid
sequence. The term allelic variant is also used herein to denote a protein
encoded by an allelic
variant of a gene.
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
[119] The term "ortholog" denotes a polypeptide or protein obtained from one
species that
is the functional counterpart of a polypeptide or protein from a different
species. Sequence
differences among orthologs are the result of speciation.
[120] "Paralogs" are distinct but structurally related proteins made by an
organism.
Paralogs are believed to arise through gene duplication. For example, a-
globin, 0-globin, and
myoglobin are paralogs of each other.
[121] The term "substantially similar" when used to describe polypeptide
sequences or
polynucleotide sequences herein means that the two sequences share at least
70% identity over a
corresponding range. More preferably, that percent identity is at least 80%
identity, more preferably
still at least 90% identity, more preferably still at least 95% identity and
most preferably at least 99%
identity. Differences in identity can be due to additions, deletions or
substitutions of residues
in a first sequences compared to a second sequences. Those ordinarily skilled
in the are will
readily calculate percent identity between an amino acid and a reference
sequence.
[122] As is used herein, the terms "at least 70% identical" or "at least 70%
identity"
means that an amino acid sequence shares 70%-100% sequence identity with a
reference
sequence. This range of identity is inclusive of all whole (e.g., 70%, 75%,
79%, 87%, 93%,
98%) or partial numbers (e.g., 72.15, 87.27%, 92.83%, 98.11% - to two
significant figures)
embraced within the recited range numbers, therefore forming a part of this
description..
For example, an amino acid sequence with 200 residues that share 85% identity
with a
reference sequence would have 170 identical residues and 30 non-identical
residues.
Similarly, an amino acid sequence with 235 residues may have 200 residues that
are
identical to a reference sequence, thus the amino acid sequence will be 85.11
% identical to
the reference sequence. Similarly, the terms "at least 80%," "at least 90%,"
"at least 95%"
and "at least 99%" are inclusive of all whole or partial numbers within the
recited range. As
is used herein, the terms "greater than 95% identical" or "greater than 95%
identity" means
that an amino acid sequence shares 95.01%-100% sequence identity with a
reference
sequence. This range is all inclusive as described immediately above.
Differences in
identity can be due to additions, deletions or substitutions of residues in a
first sequences
compared to a second sequences. Those ordinarily skilled in the are will
readily calculate
percent identity between an amino acid and a reference sequence.
[123] The present invention includes functional fragments of IL-17RE genes.
Within the
context of this invention, a "functional fragment" or "fragment" of a IL-17RE
gene refers to a nucleic
acid molecule that encodes a portion of a IL-17RE polypeptide which is a
domain described herein or
at least specifically binds with an anti-IL-17RE antibody.
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
26
[124] Due to the imprecision of standard analytical methods, molecular weights
and
lengths of polymers are understood to be approximate values. When such a value
is expressed as
"about" X or "approximately" X, the stated value of X will be understood to be
accurate to 10%.
C) Production of IL-17RE Polynucleotides or Genes
[125] Nucleic acid molecules encoding a human IL-17RE gene can be obtained by
screening a human cDNA or genomic library using polynucleotide probes based
upon any of SEQ ID
NOs:1, 4, 7, 10, 13, 20, 22, 106, 108, or 112. These screening techniques are
standard and well-
established, and may be accomplished using cloning kits available by
commercial suppliers. See, for
example, Ausubel et al. (eds.), Short Protocols in Molecular Biology, 3rd
Edition, John Wiley & Sons
1995; Wu et al., Methods in Gene Biotechnology, CRC Press, Inc. 1997; Aviv and
Leder, Proc. Nat'l
Acad. Sci. USA 69:1408 (1972); Huynh et al., "Constructing and Screening cDNA
Libraries in Xgt10
and Xgt11," in DNA Cloning: A Practical Approach Vol. I, Glover (ed.), page 49
(IRL Press, 1985);
Wu (1997) at pages 47-52.
[126] Nucleic acid molecules that encode a human IL-17RE gene can also be
obtained
using the polymerase chain reaction (PCR) with oligonucleotide primers having
nucleotide
sequences that are based upon the nucleotide sequences of the identified IL-
17RE gene or cDNA.
General methods for screening libraries with PCR are provided by, for example,
Yu et al., "Use of
the Polymerase Chain Reaction to Screen Phage Libraries," in Methods in
Molecular Biology, Vol.
15: PCR Protocols: Current Methods and Applications, White (ed.), Humana
Press, Inc., 1993.
Moreover, techniques for using PCR to isolate related genes are described by,
for example, Preston,
"Use of Degenerate Oligonucleotide Primers and the Polymerase Chain Reaction
to Clone Gene
Family Members," in Methods in Molecular Biology, Vol. 15: PCR Protocols:
Current Methods and
Applications, White (ed.), Humana Press, Inc. 1993. As an alternative, a IL-
17RE gene can be
obtained by synthesizing nucleic acid molecules using mutually priming long
oligonucleotides and
the nucleotide sequences described herein (see, for example, Ausubel (1995)).
Established
techniques using the polymerase chain reaction provide the ability to
synthesize DNA molecules at
least two kilobases in length (Adang et al., Plant Molec. Biol. 21:1131
(1993), Bambot et al., PCR
Methods and Applications 2:266 (1993), Dillon et al., "Use of the Polymerase
Chain Reaction for the
Rapid Construction of Synthetic Genes," in Methods in Molecular Biology, Vol.
15: PCR Protocols:
Current Methods and Applications, White (ed.), pages 263-268, (Humana Press,
Inc. 1993), and
Holowachuk et al., PCR Methods Appl. 4:299 (1995)). For reviews on
polynucleotide synthesis, see,
for example, Glick and Pasternak, Molecular Biotechnology, Principles and
Applications of
Recombinant DNA (ASM Press 1994), Itakura et al., Annu. Rev. Biochem. 53:323
(1984), and Climie
et al., Proc. Nat'l Acad. Sci. USA 87:633 (1990).
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
27
D) Production ofIL-17RE Gene Variants
[127] The present invention provides a variety of nucleic acid molecules,
including DNA
and RNA molecules, that encode the IL-17RE polypeptides disclosed herein.
Those skilled in the art
will readily recognize that, in view of the degeneracy of the genetic code,
considerable sequence
variation is possible among these polynucleotide molecules. Moreover, the
present invention also
provides isolated soluble monomeric, homodimeric, heterodimeric and multimeric
receptor
polypeptides that comprise at least one IL-17RE receptor subunit that is
substantially homologous to
the receptor polypeptide of any of SEQ ID NOs: 2, 5, 8, 11, 14, 21, 23, 107,
109, 111, 113, 115, 117
or 119. Thus, the present invention contemplates IL-17RE polypeptide-encoding
nucleic acid
molecules comprising degenerate nucleotides of SEQ ID NOs:1, 4, 7, 10, 13, 20,
22, 106, 108, 110,
or 112, and their RNA equivalents.
[128] Those skilled in the art will readily recognize that, in view of the
degeneracy of the
genetic code, considerable sequence variation is possible among these
polynucleotide molecules.
SEQ ID NO:7 is a degenerate nucleotide sequence that encompasses all nucleic
acid molecules that
encode the IL-17RE polypeptide of any of SEQ ID NOs: 2, 5, 8, 11, 14, 21, 23,
107, 109, 111, 113,
115, 117 or 119. Those skilled in the art will recognize that the degenerate
sequence of SEQ ID
NO:7 also provides all RNA sequences encoding any of SEQ ID NOs: 2, 5, 8, 11,
14, 21, 23, 107,
109, 111, 113, 115, 117 or 119, by substituting U for T. Thus, the present
invention contemplates
IL-17RE polypeptide-encoding nucleic acid molecules comprising nucleotide 154
to nucleotide 2229
of SEQ ID NO:1, and their RNA equivalents. Similarly, the IL-17RE degenerate
sequence of SEQ
ID NO:6 also provides all RNA sequences encoding SEQ ID NO:5, by substituting
U for T.
[129] Table 1 sets forth the one-letter codes to denote degenerate nucleotide
positions.
"Resolutions" are the nucleotides denoted by a code letter. "Complement"
indicates the code for the
complementary nucleotide(s). For example, the code Y denotes either C or T,
and its complement R
denotes A or G, A being complementary to T, and G being complementary to C.
Table 1
Nucleotide Resolution Complement Resolution
A A T T
C C G G
G G C C
T T A A
R AIG Y CIT
Y CIT R AIG
M AIC K GIT
K GIT M AIC
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
28
s CIG s CIG
w AIT w AIT
H AICIT D AIGIT
B CIGIT V AICIG
V AICIG B CIGIT
D AIGIT H AICIT
N :]I AAICIGITIJ N AICIGIT
[130] The degenerate codons, encompassing all possible codons for a given
amino acid,
are set forth in Table 2.
Table 2
One Letter Degenerate Codon
Amino Acid Code Codons
Cys C TGCTGT TGY
Ser S AGC AGT TCA TCC TCG TCT WSN
Thr T ACA ACC ACG ACT ACN
Pro P CCA CCC CCG CCT CCN
Ala A GCA GCC GCG GCT GCN
Gly G GGA GGC GGG GGT GGN
Asn N AAC AAT AAY
Asp D GAC GAT GAY
Glu E GAA GAG GAR
Gln Q CAA CAG CAR
His H CAC CAT CAY
Arg R AGA AGG CGA CGC CGG CGT MGN
Lys K AAA AAG AAR
Met M ATG ATG
Ile I ATA ATC ATT ATH
Leu L CTA CTC CTG CTT TTA TTG YTN
Val V GTA GTC GTG GTT GTN
Phe F TTC TTT TTY
Tyr Y TAC TAT TAY
Trp W TGG TGG
Ter . TAA TAG TGA TRR
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
29
AsnIAsp B RAY
G1ulGln Z SAR
Any X NNN
[131] One of ordinary skill in the art will appreciate that some ambiguity is
introduced in
determining a degenerate codon, representative of all possible codons encoding
an amino acid. For
example, the degenerate codon for serine (WSN) can, in some circumstances,
encode arginine
(AGR), and the degenerate codon for arginine (MGN) can, in some circumstances,
encode serine
(AGY). A similar relationship exists between codons encoding phenylalanine and
leucine. Thus,
some polynucleotides encompassed by the degenerate sequence may encode variant
amino acid
sequences, but one of ordinary skill in the art can easily identify such
variant sequences by reference
to the amino acid sequences of SEQ ID NO:3. Variant sequences can be readily
tested for
functionality as described herein.
[132] Different species can exhibit "preferential codon usage." In general,
see, Grantham
et al., Nucl. Acids Res. 8:1893 (1980), Haas et al. Curr. Biol. 6:315 (1996),
Wain-Hobson et al.,
Gene 13:355 (1981), Grosjean and Fiers, Gene 18:199 (1982), Holm, Nuc. Acids
Res. 14:3075
(1986), Ikemura, J. Mol. Biol. 158:573 (1982), Sharp and Matassi, Curr. Opin.
Genet. Dev. 4:851
(1994), Kane, Curr. Opin. Biotechnol. 6:494 (1995), and Makrides, Microbiol.
Rev. 60:512 (1996).
As used herein, the term "preferential codon usage" or "preferential codons"
is a term of art referring
to protein translation codons that are most frequently used in cells of a
certain species, thus favoring
one or a few representatives of the possible codons encoding each amino acid
(See Table 2). For
example, the amino acid threonine (Thr) may be encoded by ACA, ACC, ACG, or
ACT, but in
mammalian cells ACC is the most commonly used codon; in other species, for
example, insect cells,
yeast, viruses or bacteria, different Thr codons may be preferential.
Preferential codons for a
particular species can be introduced into the polynucleotides of the present
invention by a variety of
methods known in the art. Introduction of preferential codon sequences into
recombinant DNA can,
for example, enhance production of the protein by making protein translation
more efficient within a
particular cell type or species. Therefore, the degenerate codon sequences
disclosed herein serve as a
template for optimizing expression of polynucleotides in various cell types
and species commonly
used in the art and disclosed herein. Sequences containing preferential codons
can be tested and
optimized for expression in various species, and tested for functionality as
disclosed herein.
[133] A IL-17RE-encoding cDNA can be isolated by a variety of methods, such as
by
probing with a complete or partial human cDNA or with one or more sets of
degenerate probes based
on the disclosed sequences. A cDNA can also be cloned using the polymerase
chain reaction with
primers designed from the representative human IL-17RE sequences disclosed
herein. In addition, a
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
cDNA library can be used to transform or transfect host cells, and expression
of the cDNA of interest
can be detected with an antibody to IL-17RE polypeptide.
[134] Those skilled in the art will recognize that the sequence disclosed in
SEQ ID NO:1
represents a single allele of human IL-17RE, and that allelic variation and
alternative splicing are
expected to occur. Allelic variants of this sequence can be cloned by probing
cDNA or genomic
libraries from different individuals according to standard procedures. Allelic
variants of the
nucleotide sequences disclosed herein, including those containing silent
mutations and those in
which mutations result in amino acid sequence changes, are within the scope of
the present
invention, as are proteins which are allelic variants of the amino acid
sequences disclosed herein.
cDNA molecules generated from alternatively spliced mRNAs, which retain the
properties of the IL-
17RE polypeptide are included within the scope of the present invention, as
are polypeptides
encoded by such cDNAs and mRNAs. Allelic variants and splice variants of these
sequences can be
cloned by probing cDNA or genomic libraries from different individuals or
tissues according to
standard procedures known in the art.
[135] Using the methods discussed above, one of ordinary skill in the art can
prepare a
variety of polypeptides that comprise a soluble IL-17RE receptor subunit that
is substantially
homologous to either SEQ ID NOs:1, 4, 7, 10, 13, 20, 22, 106, 108, 110 or 112
or that encodes
amino acids of either SEQ ID NOs: 2, 5, 8, 11, 14, 21, 23, 107, 109, 111, 113,
115, 117 or 119, or
allelic variants thereof and retain the ligand-binding properties of the wild-
type IL-17RE receptor.
Such polypeptides may also include additional polypeptide segments as
generally disclosed herein.
[136] Within certain embodiments of the invention, the isolated nucleic acid
molecules can
hybridize under stringent conditions to nucleic acid molecules comprising
nucleotide sequences
disclosed herein. For example, such nucleic acid molecules can hybridize under
stringent conditions
to nucleic acid molecules comprising the nucleotide sequence of any of SEQ ID
NOs: 1, 4, 7, 10, 13,
20, 22, 106, 108, 110 or 112, or to nucleic acid molecules comprising a
nucleotide sequence
complementary to any of SEQ ID NOs: 1, 4, 7, 10, 13, 20, 22, 106, 108, 110 or
112, or fragments
thereof.
[137] In general, stringent conditions are selected to be about 5 C lower than
the thermal
melting point (Tm) for the specific sequence at a defined ionic strength and
pH. The Tm is the
temperature (under defined ionic strength and pH) at which 50% of the target
sequence hybridizes to
a perfectly matched probe. Following hybridization, the nucleic acid molecules
can be washed to
remove non-hybridized nucleic acid molecules under stringent conditions, or
under highly stringent
conditions. See, for example, Sambrook et al., Molecular Cloning: A Laboratory
Manual, Second
Edition (Cold Spring Harbor Press 1989); Ausubel et al., (eds.), Current
Protocols in Molecular
Biology (John Wiley and Sons, Inc. 1987); Berger and Kimmel (eds.), Guide to
Molecular Cloning
Techniques, (Academic Press, Inc. 1987); and Wetmur, Crit. Rev. Biochem. Mol.
Biol. 26:227
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
31
(1990)). Sequence analysis software such as OLIGO 6.0 (LSR; Long Lake, MN) and
Primer
Premier 4.0 (Premier Biosoft International; Palo Alto, CA), as well as sites
on the Internet, are
available tools for analyzing a given sequence and calculating Tm based on
user-defined criteria. It
is well within the abilities of one skilled in the art to adapthybridization
and wash conditions for use
with a particular polynucleotide hybrid.
[138] The present invention also provides isolated IL-17RE polypeptides that
have a
substantially similar sequence identity to the polypeptides of any of SEQ ID
NOs: 2, 5, 8, 11, 14, 21,
23, 107, 109, 111, 113, 115, 117 or 119, or their orthologs. The term
"substantially similar sequence
identity" is used herein to denote polypeptides having at least 70%, at least
80%, at least 90%, at
least 95%, or greater than 95% sequence identity to the sequences shown in any
of SEQ ID NOs: 2,
5, 8, 11, 14, 21, 23, 107, 109, 111, 113, 115, 117 or 119, or their orthologs.
For example, variant and
orthologous IL-17RE receptors can be used to generate an immune response and
raise cross-reactive
antibodies to human IL-17RE. Such antibodies can be humanized, and modified as
described herein,
and used therauputically to treat psoriasis, psoriatic arthritis, IBD, IBS,
colitis, endotoxemia as well
as in other therapeutic applications described herein.
[139] The present invention also contemplates IL-17RE variant nucleic acid
molecules that
can be identified using two criteria: a determination of the similarity
between the encoded
polypeptide with the amino acid sequence of any of SEQ ID NOs: 2, 5, 8, 11,
14, 21, 23, 107, 109,
111, 113, 115, 117 or 119, and a hybridization assay. Such IL-17RE variants
include nucleic acid
molecules (1) that remain hybridized with a nucleic acid molecule having the
nucleotide sequence of
SEQ ID NOs:1, 4, 7, 10, 13, 20, 22, 106, 108, 110 or 112 (or its complement)
under stringent
washing conditions, in which the wash stringency is equivalent to 0.5x - 2x
SSC with 0.1% SDS at
55 - 65 C, and (2) that encode a polypeptide having at least 70%, at least
80%, at least 90%, at least
95%, or greater than 95% or 99%, sequence identity to the amino acid sequence
of any of SEQ ID
NOs: 2, 5, 8, 11, 14, 21, 23, 107, 109, 111, 113, 115, 117 or 119.
Alternatively, IL-17RE variants
can be characterized as nucleic acid molecules that: (1) remain hybridized
with a nucleic acid
molecule having the nucleotide sequence of SEQ ID NOs:1, 4, 7, 10, 13, 20, 22,
106, 108, 110 or
112 (or its complement) under highly stringent washing conditions, in which
the wash stringency is
equivalent to 0.1x - 0.2x SSC with 0.1% SDS at 50 - 65 C, and (2) encode a
polypeptide having at
least 70%, at least 80%, at least 90%, at least 95% or greater than 95%, or
99% or greater, sequence
identity to the amino acid sequence of any of SEQ ID NOs: 2, 5, 8, 11, 14, 21,
23, 107, 109, 111,
113, 115, 117 or 119.
[140] Percent sequence identity is determined by conventional methods. See,
for example,
Altschul et al., Bull. Math. Bio. 48:603 (1986), and Henikoff and Henikoff,
Proc. Natl. Acad. Sci.
USA 89:10915 (1992). Briefly, two amino acid sequences are aligned to optimize
the alignment
scores using a gap opening penalty of 10, a gap extension penalty of 1, and
the "BLOSUM62"
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
32
scoring matrix of Henikoff and Henikoff (ibid.) as shown in Table 3 (amino
acids are indicated by
the standard one-letter codes). The percent identity is then calculated as:
([Total number of identical
matches]/ [length of the longer sequence plus the number of gaps introduced
into the longer sequence
in order to align the two sequences])(100).
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
33
Table 3
A R N D C Q E G H I L K M F P S T W Y V
A 4
R -1 5
N -2 0 6
D -2 -2 1 6
C 0 -3 -3 -3 9
Q -1 1 0 0 -3 5
E -1 0 0 2 -4 2 5
G 0 -2 0 -1 -3 -2 -2 6
H -2 0 1 -1 -3 0 0 -2 8
I -1 -3 -3 -3 -1 -3 -3 -4 -3 4
L -1 -2 -3 -4 -1 -2 -3 -4 -3 2 4
K -1 2 0 -1 -3 1 1 -2 -1 -3 -2 5
M -1 -1 -2 -3 -1 0 -2 -3 -2 1 2 -1 5
F -2 -3 -3 -3 -2 -3 -3 -3 -1 0 0 -3 0 6
P -1 -2 -2 -1 -3 -1 -1 -2 -2 -3 -3 -1 -2 -4 7
S 1 -1 1 0 -1 0 0 0 -1 -2 -2 0 -1 -2 -1 4
T 0 -1 0 -1 -1 -1 -1 -2 -2 -1 -1 -1 -1 -2 -1 1 5
W -3 -3 -4 -4 -2 -2 -3 -2 -2 -3 -2 -3 -1 1 -4 -3 -2 11
Y -2 -2 -2 -3 -2 -1 -2 -3 2 -1 -1 -2 -1 3 -3 -2 -2 2 7
V 0 -3 -3 -3 -1 -2 -2 -3 -3 3 1 -2 1 -1 -2 -2 0 -3 -1 4
[141] Those skilled in the art appreciate that there are many established
algorithms
available to align two amino acid sequences. The "FASTA" similarity search
algorithm of Pearson
and Lipman is a suitable protein alignment method for examining the level of
identity shared by an
amino acid sequence disclosed herein and the amino acid sequence of a putative
IL-17RE variant.
The FASTA algorithm is described by Pearson and Lipman, Proc. Nat'l Acad. Sci.
USA 85:2444
(1988), and by Pearson, Meth. Enzymol. 183:63 (1990). Briefly, FASTA first
characterizes sequence
similarity by identifying regions shared by the query sequence (e.g., any of
SEQ ID NOs: 2, 5, 8, 11,
14, 21, 23, 107, 109, 111, 113, 115, 117 or 119) and a test sequence that have
either the highest
density of identities (if the ktup variable is 1) or pairs of identities (if
ktup=2), without considering
conservative amino acid substitutions, insertions, or deletions. The ten
regions with the highest
density of identities are then rescored by comparing the similarity of all
paired amino acids using an
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
34
amino acid substitution matrix, and the ends of the regions are "trimmed" to
include only those
residues that contribute to the highest score. If there are several regions
with scores greater than the
"cutoff' value (calculated by a predetermined formula based upon the length of
the sequence and the
ktup value), then the trimmed initial regions are examined to determine
whether the regions can be
joined to form an approximate alignment with gaps. Finally, the highest
scoring regions of the two
amino acid sequences are aligned using a modification of the Needleman-Wunsch-
Sellers algorithm
(Needleman and Wunsch, J. Mol. Biol. 48:444 (1970); Sellers, SIAMJ. Appl.
Math. 26:787 (1974)),
which allows for amino acid insertions and deletions. Illustrative parameters
for FASTA analysis
are: ktup=1, gap opening penalty=10, gap extension penalty=l, and substitution
matrix=BLOSUM62. These parameters can be introduced into a FASTA program by
modifying the
scoring matrix file ("SMATRIX"), as explained in Appendix 2 of Pearson, Meth.
Enzymol. 183:63
(1990).
[142] FASTA can also be used to determine the sequence identity of nucleic
acid
molecules using a ratio as disclosed above. For nucleotide sequence
comparisons, the ktup value can
range between one to six, preferably from three to six, most preferably three,
with other parameters
set as described above.
[143] The present invention includes nucleic acid molecules that encode a
polypeptide
having a conservative amino acid change, compared with an amino acid sequence
disclosed herein.
For example, variants can be obtained that contain one or more amino acid
substitutions of any of
SEQ ID NOs: 2, 5, 8, 11, 14, 21, 23, 107, 109, 111, 113, 115, 117 or 119, in
which an alkyl amino
acid is substituted for an alkyl amino acid in a IL-17RE amino acid sequence,
an aromatic amino
acid is substituted for an aromatic amino acid in a IL-17RE amino acid
sequence, a sulfur-containing
amino acid is substituted for a sulfur-containing amino acid in a IL-17RE
amino acid sequence, a
hydroxy-containing amino acid is substituted for a hydroxy-containing amino
acid in a IL-17RE
amino acid sequence, an acidic amino acid is substituted for an acidic amino
acid in a IL-17RE
amino acid sequence, a basic amino acid is substituted for a basic amino acid
in a IL-17RE amino
acid sequence, or a dibasic monocarboxylic amino acid is substituted for a
dibasic monocarboxylic
amino acid in a IL-17RE amino acid sequence. Among the common amino acids, for
example, a
"conservative amino acid substitution" is illustrated by a substitution among
amino acids within each
of the following groups: (1) glycine, alanine, valine, leucine, and
isoleucine, (2) phenylalanine,
tyrosine, and tryptophan, (3) serine and threonine, (4) aspartate and
glutamate, (5) glutamine and
asparagine, and (6) lysine, arginine and histidine. The BLOSUM62 table is an
amino acid
substitution matrix derived from about 2,000 local multiple alignments of
protein sequence
segments, representing highly conserved regions of more than 500 groups of
related proteins
(Henikoff and Henikoff, Proc. Nat'l Acad. Sci. USA 89:10915 (1992)).
Accordingly, the
BLOSUM62 substitution frequencies can be used to define conservative amino
acid substitutions
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
that may be introduced into the amino acid sequences of the present invention.
Although it is
possible to design amino acid substitutions based solely upon chemical
properties (as discussed
above), the language "conservative amino acid substitution" preferably refers
to a substitution
represented by a BLOSUM62 value of greater than -1. For example, an amino acid
substitution is
conservative if the substitution is characterized by a BLOSUM62 value of 0, 1,
2, or 3. According
to this system, preferred conservative amino acid substitutions are
characterized by a BLOSUM62
value of at least 1(e.g., 1, 2 or 3), while more preferred conservative amino
acid substitutions are
characterized by a BLOSUM62 value of at least 2 (e.g., 2 or 3). Particular
variants of IL-17RE are
characterized by having at least 70%, at least 80%, at least 90%, at least 95%
or greater than 95% or
99% or greater sequence identity to the corresponding amino acid sequence
(e.g., any of SEQ ID
NOs: 2, 5, 8, 11, 14, 21, 23, 107, 109, 111, 113, 115, 117 or 119), wherein
the variation in amino
acid sequence is due to one or more conservative amino acid substitutions.
[144] Conservative amino acid changes in a IL-17RE gene can be introduced, for
example,
by substituting nucleotides for the nucleotides recited in SEQ ID NOs:1, 4, 7,
10, 13, 20, 22, 106,
108, 110 or 112. Such "conservative amino acid" variants can be obtained by
oligonucleotide-
directed mutagenesis, linker-scanning mutagenesis, mutagenesis using the
polymerase chain reaction,
and the like (see Ausubel (1995); and McPherson (ed.), Directed Mutagenesis: A
Practical Approach
(IRL Press 1991)). A variant IL-17RE polypeptide can be identified by the
ability to specifically
bind anti-IL-17RE antibodies.
[145] The proteins of the present invention can also comprise non-naturally
occurring
amino acid residues. Non-naturally occurring amino acids include, without
limitation, trans-3-
methylproline, 2,4-methanoproline, cis-4-hydroxyproline, trans-4-
hydroxyproline, N-methylglycine,
allo-threonine, methylthreonine, hydroxyethylcysteine,
hydroxyethylhomocysteine, nitroglutamine,
homoglutamine, pipecolic acid, thiazolidine carboxylic acid, dehydroproline, 3-
and 4-methylproline,
3,3-dimethylproline, tert-leucine, norvaline, 2-azaphenylalanine, 3-
azaphenylalanine, 4-
azaphenylalanine, and 4-fluorophenylalanine. Several methods are known in the
art for
incorporating non-naturally occurring amino acid residues into proteins. For
example, an in vitro
system can be employed wherein nonsense mutations are suppressed using
chemically aminoacylated
suppressor tRNAs. Methods for synthesizing amino acids and aminoacylating tRNA
are known in
the art. Transcription and translation of plasmids containing nonsense
mutations is typically carried
out in a cell-free system comprising an E. coli S30 extract and commercially
available enzymes and
other reagents. Proteins are purified by chromatography. See, for example,
Robertson et al., J. Am.
Chem. Soc. 113:2722 (1991), Ellman et al., Methods Enzymol. 202:301 (1991),
Chung et al., Science
259:806 (1993), and Chung et al., Proc. Nat'l Acad. Sci. USA 90:10145 (1993).
[146] In a second method, translation is carried out in Xenopus oocytes by
microinjection
of mutated mRNA and chemically aminoacylated suppressor tRNAs (Turcatti et
al., J. Biol. Chem.
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
36
271:19991 (1996)). Within a third method, E. coli cells are cultured in the
absence of a natural
amino acid that is to be replaced (e.g., phenylalanine) and in the presence of
the desired non-
naturally occurring amino acid(s) (e.g., 2-azaphenylalanine, 3-
azaphenylalanine, 4-azaphenylalanine,
or 4-fluorophenylalanine). The non-naturally occurring amino acid is
incorporated into the protein
in place of its natural counterpart. See, Koide et al., Biochem. 33:7470
(1994). Naturally occurring
amino acid residues can be converted to non-naturally occurring species by in
vitro chemical
modification. Chemical modification can be combined with site-directed
mutagenesis to further
expand the range of substitutions (Wynn and Richards, Protein Sci. 2:395
(1993)).
[147] A limited number of non-conservative amino acids, amino acids that are
not encoded
by the genetic code, non-naturally occurring amino acids, and unnatural amino
acids may be
substituted for IL-17RE amino acid residues.
[148] Essential amino acids in the polypeptides of the present invention can
be identified
according to procedures known in the art, such as site-directed mutagenesis or
alanine-scanning
mutagenesis (Cunningham and Wells, Science 244:1081 (1989), Bass et al., Proc.
Nat'l Acad. Sci.
USA 88:4498 (1991), Coombs and Corey, "Site-Directed Mutagenesis and Protein
Engineering," in
Proteins: Analysis and Design, Angeletti (ed.), pages 259-311 (Academic Press,
Inc. 1998)). In the
latter technique, single alanine mutations are introduced at every residue in
the molecule, and the
resultant mutant molecules are tested for biological activity to identify
amino acid residues that are
critical to the activity of the molecule. See also, Hilton et al., J. Biol.
Chem. 271:4699 (1996).
[149] Although sequence analysis can be used to further define the IL-17RE
ligand
binding region, amino acids that play a role in IL-17RE binding activity (such
as binding of IL-17RE
to Il-17C, or to an anti-IL-17RE antibody) can also be determined by physical
analysis of structure,
as determined by such techniques as nuclear magnetic resonance,
crystallography, electron
diffraction or photoaffinity labeling, in conjunction with mutation of
putative contact site amino
acids. See, for example, de Vos et al., Science 255:306 (1992), Smith et al.,
J. Mol. Biol. 224:899
(1992), and Wlodaver et al., FEBS Lett. 309:59 (1992).
[150] Multiple amino acid substitutions can be made and tested using known
methods of
mutagenesis and screening, such as those disclosed by Reidhaar-Olson and Sauer
(Science 241:53
(1988)) or Bowie and Sauer (Proc. Nat'l Acad. Sci. USA 86:2152 (1989)).
Briefly, these authors
disclose methods for simultaneously randomizing two or more positions in a
polypeptide, selecting
for functional polypeptide, and then sequencing the mutagenized polypeptides
to determine the
spectrum of allowable substitutions at each position. Other methods that can
be used include phage
display (e.g., Lowman et al., Biochem. 30:10832 (1991), Ladner et al., U.S.
Patent No. 5,223,409,
Huse, international publication No. WO 92/06204, and region-directed
mutagenesis (Derbyshire et
al., Gene 46:145 (1986), and Ner et al., DNA 7:127, (1988)). Moreover, IL-17RE
labeled with biotin
or FITC can be used for expression cloning of IL-17RE ligands.
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
37
[151] Variants of the disclosed IL-17RE nucleotide and polypeptide sequences
can also be
generated through DNA shuffling as disclosed by Stemmer, Nature 370:389
(1994), Stemmer, Proc.
Nat'l Acad. Sci. USA 91:10747 (1994), and international publication No. WO
97/20078. Briefly,
variant DNA molecules are generated by in vitro homologous recombination by
random
fragmentation of a parent DNA followed by reassembly using PCR, resulting in
randomly introduced
point mutations. This technique can be modified by using a family of parent
DNA molecules, such
as allelic variants or DNA molecules from different species, to introduce
additional variability into
the process. Selection or screening for the desired activity, followed by
additional iterations of
mutagenesis and assay provides for rapid "evolution" of sequences by selecting
for desirable
mutations while simultaneously selecting against detrimental changes.
[152] Mutagenesis methods as disclosed herein can be combined with high-
throughput,
automated screening methods to detect activity of cloned, mutagenized
polypeptides in host cells.
Mutagenized DNA molecules that encode biologically active polypeptides, or
polypeptides that bind
with anti-IL-17RE antibodies, can be recovered from the host cells and rapidly
sequenced using
modern equipment. These methods allow the rapid determination of the
importance of individual
amino acid residues in a polypeptide of interest, and can be applied to
polypeptides of unknown
structure.
[153] The present invention also includes "functional fragments" or
"fragments" of IL-
17RE polypeptides and nucleic acid molecules encoding such functional
fragments. Routine
deletion analyses of nucleic acid molecules can be performed to obtain
functional fragments of a
nucleic acid molecule that encodes a IL-17RE polypeptide. As an illustration,
DNA molecules
having the nucleotide sequence of SEQ ID NOs:1, 4, 7, 10, 13, 20, 22, 106,
108, 110 or 112 can be
digested with Ba131 nuclease to obtain a series of nested deletions. The
fragments are then inserted
into expression vectors in proper reading frame, and the expressed
polypeptides are isolated and
tested for the ability to bind anti-IL-17RE antibodies. One alternative to
exonuclease digestion is to
use oligonucleotide-directed mutagenesis to introduce deletions or stop codons
to specify production
of a desired fragment. Alternatively, particular fragments of a IL-17RE gene
can be synthesized
using the polymerase chain reaction.
[154] This general approach is exemplified by studies on the truncation at
either or both
termini of interferons have been summarized by Horisberger and Di Marco,
Pharmac. Ther. 66:507
(1995). Moreover, standard techniques for functional analysis of proteins are
described by, for
example, Treuter et al., Molec. Gen. Genet. 240:113 (1993), Content et al.,
"Expression and
preliminary deletion analysis of the 42 kDa 2-5A synthetase induced by human
interferon," in
Biological Interferon Systems, Proceedings of ISIR-TNO Meeting on Interferon
Systems, Cantell
(ed.), pages 65-72 (Nijhoff 1987), Herschman, "The EGF Receptor," in Control
of Animal Cell
Proliferation, Vol. 1, Boynton et al., (eds.) pages 169-199 (Academic Press
1985), Coumailleau et
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
38
al., J. Biol. Chem. 270:29270 (1995); Fukunaga et al., J. Biol. Chem.
270:25291 (1995); Yamaguchi
et al., Biochem. Pharmacol. 50:1295 (1995), and Meisel et al., Plant Molec.
Biol. 30:1 (1996).
[155] The present invention also contemplates functional fragments of a IL-
17RE gene
that have amino acid changes, compared with an amino acid sequence disclosed
herein. A variant
IL-17RE gene can be identified on the basis of structure by determining the
level of identity with
disclosed nucleotide and amino acid sequences, as discussed above. An
alternative approach to
identifying a variant gene on the basis of structure is to determine whether a
nucleic acid molecule
encoding a potential variant IL-17RE gene can hybridize to a nucleic acid
molecule comprising a
nucleotide sequence, such as SEQ ID NOs:1, 4, 7, 10, 13, 20, 22, 106, 108,
110, or 112.
[156] The present invention also includes using functional fragments of IL-
17RE
polypeptides, antigenic epitopes, epitope-bearing portions of IL-17RE
polypeptides, and nucleic acid
molecules that encode such functional fragments, antigenic epitopes, epitope-
bearing portions of IL-
17RE polypeptides. For example, such IL-17RE fragments include polypeptides
encoded by SEQ
ID NOs:115, 117 or 119. These fragments encode binding domains of IL-17RE and
are used to
generate polypeptides for use in generating antibodies and binding partners
that bind, block, inhibit,
reduce, antagonize or neutralize activity of IL-17C. A"functional" IL-17RE
polypeptide or
fragment thereof as defined herein is characterized by its ability to block,
inhibit, reduce, antagonize
or neutralize IL-17C inflammatory, proliferative or differentiating activity,
by its ability to induce or
inhibit specialized cell functions, or by its ability to bind specifically to
an anti-IL-17RE antibody,
cell, or IL-17C. As previously described herein, IL-17RE is characterized by a
unique cytokine
receptor structure and domains as described herein. Thus, the present
invention further contemplates
using fusion proteins encompassing: (a) polypeptide molecules comprising one
or more of the
domains described above; and (b) functional fragments comprising one or more
of these domains.
The other polypeptide portion of the fusion protein may be contributed by
another cytokine receptor,
such as IL-17RA, IL-17RB, IL-17RC, IL-17RD, IL-17RE, or by a non-native and/or
an unrelated
secretory signal peptide that facilitates secretion of the fusion protein.
[157] The present invention also provides polypeptide fragments or peptides
comprising
an epitope-bearing portion of a IL-17RE polypeptide described herein. Such
fragments or peptides
may comprise an "immunogenic epitope," which is a part of a protein that
elicits an antibody
response when the entire protein is used as an immunogen. Immunogenic epitope-
bearing peptides
can be identified using standard methods (see, for example, Geysen et al.,
Proc. Nat'l Acad. Sci. USA
81:3998 (1983)).
[158] In contrast, polypeptide fragments or peptides may comprise an
"antigenic epitope,"
which is a region of a protein molecule to which an antibody can specifically
bind. Certain epitopes
consist of a linear or contiguous stretch of amino acids, and the antigenicity
of such an epitope is not
disrupted by denaturing agents. It is known in the art that relatively short
synthetic peptides that can
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
39
mimic epitopes of a protein can be used to stimulate the production of
antibodies against the protein
(see, for example, Sutcliffe et al., Science 219:660 (1983)). Accordingly,
antigenic epitope-bearing
peptides, antigenic peptides, epitopes, and polypeptides of the present
invention are useful to raise
antibodies that bind with the polypeptides described herein, as well as to
identify and screen anti-IL-
17RE monoclonal antibodies that are neutralizing, and that may bind, block,
inhibit, reduce,
antagonize or neutralize the activity of IL-17C. Such neutralizing monoclonal
antibodies of the
present invention can bind to an IL-17RE antigenic epitope. Hopp/Woods
hydrophilicity profiles
can be used to determine regions that have the most antigenic potential within
any of SEQ ID NOs:
2, 5, 8, 11, 14, 21, 23, 107, 109, 111, 113, 115, 117 or 119 (Hopp et al.,
Proc. Natl. Acad.
Sci.78:3824-3828, 1981; Hopp, J. Immun. Meth. 88:1-18, 1986 and Triquier et
al., Protein
Engineering 11:153-169, 1998). The profile is based on a sliding six-residue
window. Buried G, S,
and T residues and exposed H, Y, and W residues were ignored. In IL-17RE these
regions can be
determined by one of skill in the art. Moreover, IL-17RE antigenic epitopes
within any of SEQ ID
NOs: 2, 5, 8, 11, 14, 21, 23, 107, 109, 111, 113, 115, 117 or 119 as predicted
by a Jameson-Wolf
plot, e.g., using DNASTAR Protean program (DNASTAR, Inc., Madison, WI) serve
as preferred
antigenic epitpoes, and can be determined by one of skill in the art. The
results of this analysis
indicated that SEQ ID NOs: 115 ("antigenic peptide 1"), 117 ("antigenic
peptide 2"), 119 ("antigenic
peptide 3"), and the following amino acid sequences of SEQ ID NO:6 would
provide suitable
antigenic peptides: amino acids 51 to 59 ("antigenic peptide 4"), amino acids
72 to 83 ("antigenic
peptide 5"), 91 to 97 ("antigenic peptide 6"), amino acids 174 to 180
("antigenic peptide 7"), and
amino acids 242 to 246 ("antigenic peptide 8"). The present invention
contemplates the use of any
one of, or any sub-combinations thereof, of antigenic peptides 1 to 8 to
generate antibodies to IL-
17RE. The present invention also contemplates polypeptides comprising at least
one of antigenic
peptides 1 to 8. For instance, antigenic peptides 1 and 2 may be combined to
generate a polypeptide
useful in generating an antibody antagonist of the the present invention.
[159] In preferred embodiments, antigenic epitopes to which neutralizing
antibodies of the
present invention bind would contain residues of any of SEQ ID NOs:2, 3, 5, 6,
8, 9, 11, 12, 14, 15,
21, 23, 107, 109, 111, 113, 115, 117, or 119 that are important to ligand-
receptor binding, for
example, with IL-17RE and IL-17C. Most preferably, antigenic epitopes to which
neutralizing
antibodies of the present invention bind would contain residues of any of SEQ
ID NOs: 115, 117, or
119.
[160] Antigenic epitope-bearing peptides and polypeptides can contain at least
four to ten
amino acids, at least ten to fifteen amino acids, or about 15 to about 30
amino acids of an amino acid
sequence disclosed herein. Such epitope-bearing peptides and polypeptides can
be produced by
fragmenting a IL-17RE polypeptide, or by chemical peptide synthesis, as
described herein.
Moreover, epitopes can be selected by phage display of random peptide
libraries (see, for example,
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
Lane and Stephen, Curr. Opin. Immunol. 5:268 (1993), and Cortese et al., Curr.
Opin. Biotechnol.
7:616 (1996)). Standard methods for identifying epitopes and producing
antibodies from small
peptides that comprise an epitope are described, for example, by Mole,
"Epitope Mapping," in
Methods in Molecular Biology, Vol. 10, Manson (ed.), pages 105-116 (The Humana
Press, Inc.
1992), Price, "Production and Characterization of Synthetic Peptide-Derived
Antibodies," in
Monoclonal Antibodies: Production, Engineering, and Clinical Application,
Ritter and Ladyman
(eds.), pages 60-84 (Cambridge University Press 1995), and Coligan et al.
(eds.), Current Protocols
in Immunology, pages 9.3.1 - 9.3.5 and pages 9.4.1 - 9.4.11 (John Wiley & Sons
1997).
[161] For any IL-17RE polypeptide, including variants and fusion proteins, one
of
ordinary skill in the art can readily generate a fully degenerate
polynucleotide sequence encoding
that variant using the information set forth in Tables 1 and 2 above.
Moreover, those of skill in the
art can use standard software to devise IL-17RE variants based upon the
nucleotide and amino acid
sequences described herein.
[162] Production ofIL-17RE Polypentides
[163] The polypeptides of the present invention, including full-length
polypeptides; soluble
monomeric, homodimeric, heterodimeric and multimeric receptors; full-length
receptors; receptor
fragments (e.g. ligand-binding fragments and antigenic epitopes), functional
fragments, and fusion
proteins, can be produced in recombinant host cells following conventional
techniques. To express a IL-
17RE gene, a nucleic acid molecule encoding the polypeptide must be operably
linked to regulatory
sequences that control transcriptional expression in an expression vector and
then, introduced into a host
cell. In addition to transcriptional regulatory sequences, such as promoters
and enhancers, expression
vectors can include translational regulatory sequences and a marker gene which
is suitable for selection
of cells that carry the expression vector.
[164] Expression vectors that are suitable for production of a foreign protein
in eukaryotic
cells typically contain (1) prokaryotic DNA elements coding for a bacterial
replication origin and an
antibiotic resistance marker to provide for the growth and selection of the
expression vector in a
bacterial host; (2) eukaryotic DNA elements that control initiation of
transcription, such as a
promoter; and (3) DNA elements that control the processing of transcripts,
such as a transcription
termination/polyadenylation sequence. As discussed above, expression vectors
can also include
nucleotide sequences encoding a secretory sequence that directs the
heterologous polypeptide into
the secretory pathway of a host cell. For example, an IL-17RE expression
vector may comprise a IL-
17RE gene and a secretory sequence derived from any secreted gene.
[165] IL-17RE proteins of the present invention may be expressed in mammalian
cells.
Examples of suitable mammalian host cells include African green monkey kidney
cells (Vero; ATCC
CRL 1587), human embryonic kidney cells (293-HEK; ATCC CRL 1573), baby hamster
kidney
cells (BHK-21, BHK-570; ATCC CRL 8544, ATCC CRL 10314), canine kidney cells
(MDCK;
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
41
ATCC CCL 34), Chinese hamster ovary cells (CHO-K1; ATCC CCL61; CHO DG44
(Chasin et al.,
Som. Cell. Molec. Genet. 12:555, 1986)), rat pituitary cells (GH1; ATCC
CCL82), HeLa S3 cells
(ATCC CCL2.2), rat hepatoma cells (H-4-II-E; ATCC CRL 1548) SV40-transformed
monkey
kidney cells (COS-1; ATCC CRL 1650) and murine embryonic cells (NIH-3T3; ATCC
CRL 1658).
[166] For a mammalian host, the transcriptional and translational regulatory
signals may
be derived from mammalian viral sources, for example, adenovirus, bovine
papilloma virus, simian
virus, or the like, in which the regulatory signals are associated with a
particular gene which has a
high level of expression. Suitable transcriptional and translational
regulatory sequences also can be
obtained from mammalian genes, for example, actin, collagen, myosin, and
metallothionein genes.
[167] Transcriptional regulatory sequences include a promoter region
sufficient to direct
the initiation of RNA synthesis. Suitable eukaryotic promoters include the
promoter of the mouse
metallothionein I gene (Hamer et al., J. Molec. Appl. Genet. 1:273 (1982)),
the TK promoter of
Herpes virus (McKnight, Cell 31:355 (1982)), the SV40 early promoter (Benoist
et al., Nature
290:304 (1981)), the Rous sarcoma virus promoter (Gorman et al., Proc. Nat'l
Acad. Sci. USA
79:6777 (1982)), the cytomegalovirus promoter (Foecking et al., Gene 45:101
(1980)), and the
mouse mammary tumor virus promoter (see, generally, Etcheverry, "Expression of
Engineered
Proteins in Mammalian Cell Culture," in Protein Engineering: Principles and
Practice, Cleland et
al. (eds.), pages 163-181 (John Wiley & Sons, Inc. 1996)).
[168] Alternatively, a prokaryotic promoter, such as the bacteriophage T3 RNA
polymerase promoter, can be used to control IL-17RE gene expression in
mammalian cells if the
prokaryotic promoter is regulated by a eukaryotic promoter (Zhou et al., Mol.
Cell. Biol. 10:4529
(1990), and Kaufman et al., Nucl. Acids Res. 19:4485 (1991)).
[169] In certain embodiments, a DNA sequence encoding a IL-17RE soluble
receptor
polypeptide, or a fragment of IL-17RE polypeptide is operably linked to other
genetic elements
required for its expression, generally including a transcription promoter and
terminator, within an
expression vector. The vector will also commonly contain one or more
selectable markers and one
or more origins of replication, although those skilled in the art will
recognize that within certain
systems selectable markers may be provided on separate vectors, and
replication of the exogenous
DNA may be provided by integration into the host cell genome. Selection of
promoters, terminators,
selectable markers, vectors and other elements is a matter of routine design
within the level of
ordinary skill in the art. Many such elements are described in the literature
and are available through
commercial suppliers. Multiple components of a soluble receptor complex can be
co-transfected on
individual expression vectors or be contained in a single expression vector.
Such techniques of
expressing multiple components of protein complexes are well known in the art.
[170] An expression vector can be introduced into host cells using a variety
of standard
techniques including calcium phosphate transfection, liposome-mediated
tra.nsfection, microprojectile-
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
42
mediated delivery, electroporation, and the like. The transfected cells can be
selected and propagated to
provide recombinant host cells that comprise the expression vector stably
integrated in the host cell
genome. Techniques for introducing vectors into eukaryotic cells and
techniques for selecting such
stable transformants using a dominant selectable marker are described, for
example, by Ausubel (1995)
and by Murray (ed.), Gene Transfer and Expression Protocols (Humana Press
1991).
[171] For example, one suitable selectable marker is a gene that provides
resistance to the
antibiotic neomycin. In this case, selection is carried out in the presence of
a neomycin-type drug,
such as G-418 or the like. Selection systems can also be used to increase the
expression level of the
gene of interest, a process referred to as "amplification." Amplification is
carried out by culturing
transfectants in the presence of a low level of the selective agent and then
increasing the amount of
selective agent to select for cells that produce high levels of the products
of the introduced genes. A
suitable amplifiable selectable marker is dihydrofolate reductase (DHFR),
which confers resistance
to methotrexate. Other drug resistance genes (e.g., hygromycin resistance,
multi-drug resistance,
puromycin acetyltransferase) can also be used. Alternatively, markers that
introduce an altered
phenotype, such as green fluorescent protein, or cell surface proteins such as
CD4, CD8, Class I
MHC, placental alkaline phosphatase may be used to sort transfected cells from
untransfected cells
by such means as FACS sorting or magnetic bead separation technology.
[172] IL-17RE polypeptides can also be produced by cultured mammalian cells
using a
viral delivery system. Exemplary viruses for this purpose include adenovirus,
retroviruses,
herpesvirus, vaccinia virus and adeno-associated virus (AAV). Adenovirus, a
double-stranded DNA
virus, is currently the best studied gene transfer vector for delivery of
heterologous nucleic acid (for
a review, see Becker et al., Meth. Cell Biol. 43:161 (1994), and Douglas and
Curiel, Science &
Medicine 4:44 (1997)). Advantages of the adenovirus system include the
accommodation of
relatively large DNA inserts, the ability to grow to high-titer, the ability
to infect a broad range of
mammalian cell types, and flexibility that allows use with a large number of
available vectors
containing different promoters.
[173] By deleting portions of the adenovirus genome, larger inserts (up to 7
kb) of
heterologous DNA can be accommodated. These inserts can be incorporated into
the viral DNA by
direct ligation or by homologous recombination with a co-transfected plasmid.
An option is to delete
the essential El gene from the viral vector, which results in the inability to
replicate unless the El
gene is provided by the host cell. Adenovirus vector-infected human 293 cells
(ATCC Nos. CRL-
1573, 45504, 45505), for example, can be grown as adherent cells or in
suspension culture at
relatively high cell density to produce significant amounts of protein (see
Gamier et al., Cytotechnol.
15:145 (1994)).
[174] IL-17RE can also be expressed in other higher eukaryotic cells, such as
avian,
fungal, insect, yeast, or plant cells. The baculovirus system provides an
efficient means to introduce
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
43
cloned IL-17RE genes into insect cells. Suitable expression vectors are based
upon the Autographa
californica multiple nuclear polyhedrosis virus (AcMNPV), and contain well-
known promoters such
as Drosophila heat shock protein (hsp) 70 promoter, Autographa californica
nuclear polyhedrosis
virus immediate-early gene promoter (ie-1) and the delayed early 39K promoter,
baculovirus p10
promoter, and the Drosophila metallothionein promoter. A second method of
making recombinant
baculovirus utilizes a transposon-based system described by Luckow (Luckow, et
al., J. Virol.
67:4566 (1993)). This system, which utilizes transfer vectors, is sold in the
BAC-to-BAC kit (Life
Technologies, Rockville, MD). This system utilizes a transfer vector, PFASTBAC
(Life
Technologies) containing a Tn7 transposon to move the DNA encoding the IL-17RE
polypeptide
into a baculovirus genome maintained in E. coli as a large plasmid called a
"bacmid." See, Hill-
Perkins and Possee, J. Gen. Virol. 71:971 (1990), Bonning, et al., J. Gen.
Virol. 75:1551 (1994), and
Chazenbalk, and Rapoport, J. Biol. Chem. 270:1543 (1995). In addition,
transfer vectors can include
an in-frame fusion with DNA encoding an epitope tag at the C- or N-terminus of
the expressed IL-
17RE polypeptide, for example, a Glu-Glu epitope tag (Grussenmeyer et al.,
Proc. Nat'l Acad. Sci.
82:7952 (1985)). Using a technique known in the art, a transfer vector
containing a IL-17RE gene is
transformed into E. coli, and screened for bacmids which contain an
interrupted lacZ gene indicative
of recombinant baculovirus. The bacmid DNA containing the recombinant
baculovirus genome is
then isolated using common techniques.
[175] The illustrative PFASTBAC vector can be modified to a considerable
degree. For
example, the polyhedrin promoter can be removed and substituted with the
baculovirus basic protein
promoter (also known as Pcor, p6.9 or MP promoter) which is expressed earlier
in the baculovirus
infection, and has been shown to be advantageous for expressing secreted
proteins (see, for example,
Hill-Perkins and Possee, J. Gen. Virol. 71:971 (1990), Bonning, et al., J.
Gen. Virol. 75:1551 (1994),
and Chazenbalk and Rapoport, J. Biol. Chem. 270:1543 (1995). In such transfer
vector constructs, a
short or long version of the basic protein promoter can be used. Moreover,
transfer vectors can be
constructed which replace the native IL-17RE secretory signal sequences with
secretory signal
sequences derived from insect proteins. For example, a secretory signal
sequence from Ecdysteroid
Glucosyltransferase (EGT), honey bee Melittin (Invitrogen Corporation;
Carlsbad, CA), or
baculovirus gp67 (PharMingen: San Diego, CA) can be used in constructs to
replace the native IL-
17RE secretory signal sequence.
[176] The recombinant virus or bacmid is used to transfect host cells.
Suitable insect host
cells include cell lines derived from IPLB-Sf-21, a Spodoptera frugiperda
pupal ovarian cell line,
such as Sf9 (ATCC CRL 1711), Sf21AE, and Sf21 (Invitrogen Corporation; San
Diego, CA), as well
as Drosophila Schneider-2 cells, and the HIGH FIVEO cell line (Invitrogen)
derived from
Trichoplusia ni (U.S. Patent No. 5,300,435). Commercially available serum-free
media can be used
to grow and to maintain the cells. Suitable media are Sfg00 IIT"" (Life
Technologies) or ESF 921TM
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
44
(Expression Systems) for the Sf9 cells; and Ex-cellO405T"" (JRH Biosciences,
Lenexa, KS) or
Express FiveOT"" (Life Technologies) for the T. ni cells. When recombinant
virus is used, the cells
are typically grown up from an inoculation density of approximately 2-5 x 105
cells to a density of 1-
2 x 106 cells at which time a recombinant viral stock is added at a
multiplicity of infection (MOI) of
0.1 to 10, more typically near 3.
[177] Established techniques for producing recombinant proteins in baculovirus
systems
are provided by Bailey et al., "Manipulation of Baculovirus Vectors," in
Methods in Molecular
Biology, Volume 7.= Gene Transfer and Expression Protocols, Murray (ed.),
pages 147-168 (The
Humana Press, Inc. 1991), by Patel et al., "The baculovirus expression
system," in DNA Cloning 2:
Expression Systems, 2nd Edition, Glover et al. (eds.), pages 205-244 (Oxford
University Press 1995),
by Ausubel (1995) at pages 16-37 to 16-57, by Richardson (ed.), Baculovirus
Expression Protocols
(The Humana Press, Inc. 1995), and by Lucknow, "Insect Cell Expression
Technology," in Protein
Engineering: Principles and Practice, Cleland et al. (eds.), pages 183-218
(John Wiley & Sons, Inc.
1996).
[178] Fungal cells, including yeast cells, can also be used to express the
genes described
herein. Yeast species of particular interest in this regard include
Saccharomyces cerevisiae, Pichia
pastoris, and Pichia methanolica. Suitable promoters for expression in yeast
include promoters from
GAL] (galactose), PGK (phosphoglycerate kinase), ADH (alcohol dehydrogenase),
AOXI (alcohol
oxidase), HIS4 (histidinol dehydrogenase), and the like. Many yeast cloning
vectors have been
designed and are readily available. These vectors include YIp-based vectors,
such as YIp5, YRp
vectors, such as YRp 17, YEp vectors such as YEp 13 and YCp vectors, such as
YCp 19. Methods for
transforming S. cerevisiae cells with exogenous DNA and producing recombinant
polypeptides
therefrom are disclosed by, for example, Kawasaki, U.S. Patent No. 4,599,311,
Kawasaki et al., U.S.
Patent No. 4,931,373, Brake, U.S. Patent No. 4,870,008, Welch et al., U.S.
Patent No. 5,037,743,
and Murray et al., U.S. Patent No. 4,845,075. Transformed cells are selected
by phenotype
determined by the selectable marker, commonly drug resistance or the ability
to grow in the absence
of a particular nutrient (e.g., leucine). A suitable vector system for use in
Saccharomyces cerevisiae
is the POT] vector system disclosed by Kawasaki et al. (U.S. Patent No.
4,931,373), which allows
transformed cells to be selected by growth in glucose-containing media.
Additional suitable
promoters and terminators for use in yeast include those from glycolytic
enzyme genes (see, e.g.,
Kawasaki, U.S. Patent No. 4,599,311, Kingsman et al., U.S. Patent No.
4,615,974, and Bitter, U.S.
Patent No. 4,977,092) and alcohol dehydrogenase genes. See also U.S. Patents
Nos. 4,990,446,
5,063,154, 5,139,936, and 4,661,454.
[179] Transformation systems for other yeasts, including Hansenula polymorpha,
Schizosaccharomyces pombe, Kluyveromyces lactis, Kluyveromyces fi agilis,
Ustilago maydis, Pichia
pastoris, Pichia methanolica, Pichia guillermondii and Candida maltosa are
known in the art. See,
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
for example, Gleeson et al., J. Gen. Microbiol. 132:3459 (1986), and Cregg,
U.S. Patent No.
4,882,279. Aspergillus cells may be utilized according to the methods of
McKnight et al., U.S.
Patent No. 4,935,349. Methods for transforming Acremonium chrysogenum are
disclosed by Sumino
et al., U.S. Patent No. 5,162,228. Methods for transforming Neurospora are
disclosed by
Lambowitz, U.S. Patent No. 4,486,533.
[180] For example, the use of Pichia methanolica as host for the production of
recombinant proteins is disclosed by Raymond, U.S. Patent No. 5,716,808,
Raymond, U.S. Patent
No. 5,736,383, Raymond et al., Yeast 14:11-23 (1998), and in international
publication Nos. WO
97/17450, WO 97/17451, WO 98/02536, and WO 98/02565. DNA molecules for use in
transforming P. methanolica will commonly be prepared as double-stranded,
circular plasmids,
which are preferably linearized prior to transformation. For polypeptide
production in P.
methanolica, the promoter and terminator in the plasmid can be that of a P.
methanolica gene, such
as a P. methanolica alcohol utilization gene (AUG1 or AUG2). Other useful
promoters include those
of the dihydroxyacetone synthase (DHAS), formate dehydrogenase (FMD), and
catalase (CAT)
genes. To facilitate integration of the DNA into the host chromosome, it is
preferred to have the
entire expression segment of the plasmid flanked at both ends by host DNA
sequences. A suitable
selectable marker for use in Pichia methanolica is a P. methanolica ADE2 gene,
which encodes
phosphoribosyl-5-aminoimidazole carboxylase (AIRC; EC 4.1.1.21), and which
allows ade2 host
cells to grow in the absence of adenine. For large-scale, industrial processes
where it is desirable to
minimize the use of methanol, host cells can be used in which both methanol
utilization genes
(AUG1 and A UG2) are deleted. For production of secreted proteins, host cells
can be deficient in
vacuolar protease genes (PEP4 and PRB1). Electroporation is used to facilitate
the introduction of a
plasmid containing DNA encoding a polypeptide of interest into P. methanolica
cells. P.
methanolica cells can be transformed by electroporation using an exponentially
decaying, pulsed
electric field having a field strength of from 2.5 to 4.5 kV/cm, preferably
about 3.75 kV/cm, and a
time constant (t) of from 1 to 40 milliseconds, most preferably about 20
milliseconds.
[181] Expression vectors can also be introduced into plant protoplasts, intact
plant tissues, or
isolated plant cells. Methods for introducing expression vectors into plant
tissue include the direct
infection or co-cultivation of plant tissue with Agrobacterium tumefaciens,
microprojectile-mediated
delivery, DNA injection, electroporation, and the like. See, for example,
Horsch et al., Science 227:1229
(1985), Klein et al., Biotechnology 10:268 (1992), and Miki et al.,
"Procedures for Introducing Foreign
DNA into Plants," in Methods in Plant Molecular Biology and Biotechnology,
Glick et al. (eds.), pages
67-88 (CRC Press, 1993).
[182] Alternatively, IL-17RE genes can be expressed in prokaryotic host cells.
Suitable
promoters that can be used to express IL-17RE polypeptides in a prokaryotic
host are well-known to
those of skill in the art and include promoters capable of recognizing the T4,
T3, Sp6 and T7
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
46
polymerases, the PR and PL promoters of bacteriophage lambda, the trp, recA,
heat shock, lacUV5,
tac, lpp-lacSpr, phoA, and lacZ promoters of E. coli, promoters of B.
subtilis, the promoters of the
bacteriophages of Bacillus, Streptomyces promoters, the int promoter of
bacteriophage lambda, the
bla promoter of pBR322, and the CAT promoter of the chloramphenicol acetyl
transferase gene.
Prokaryotic promoters have been reviewed by Glick, J. Ind. Microbiol. 1:277
(1987), Watson et al.,
Molecular Biology of the Gene, 4th Ed. (Benjamin Cummins 1987), and by Ausubel
et al. (1995).
[183] Suitable prokaryotic hosts include E. coli and Bacillus subtilus.
Suitable strains of
E. coli include BL21(DE3), BL21(DE3)pLysS, BL21(DE3)pLysE, DH1, DH4I, DH5,
DH5I,
DH5IF, DH5IMCR, DHIOB, DHIOB/p3, DHIIS, C600, HB101, JM101, JM105, JM109,
JM110,
K38, RR1, Y1088, Y1089, CSH18, ER2151, and ER1647 (see, for example, Brown
(ed.), Molecular
Biology Labfax (Academic Press 1991)). Suitable strains of Bacillus subtilus
include BR151,
YB886, MI119, M1120, and B170 (see, for example, Hardy, "Bacillus Cloning
Methods," in DNA
Cloning: A Practical Approach, Glover (ed.) (IRL Press 1985)).
[184] When expressing a IL-17RE polypeptide in bacteria such as E. coli, the
polypeptide
may be retained in the cytoplasm, typically as insoluble granules, or may be
directed to the
periplasmic space by a bacterial secretion sequence. In the former case, the
cells are lysed, and the
granules are recovered and denatured using, for example, guanidine
isothiocyanate or urea. The
denatured polypeptide can then be refolded and dimerized by diluting the
denaturant, such as by
dialysis against a solution of urea and a combination of reduced and oxidized
glutathione, followed
by dialysis against a buffered saline solution. In the latter case, the
polypeptide can be recovered
from the periplasmic space in a soluble and functional form by disrupting the
cells (by, for example,
sonication or osmotic shock) to release the contents of the periplasmic space
and recovering the
protein, thereby obviating the need for denaturation and refolding.
[185] Methods for expressing proteins in prokaryotic hosts are well-known to
those of skill
in the art (see, for example, Williams et al., "Expression of foreign proteins
in E. coli using plasmid
vectors and purification of specific polyclonal antibodies," in DNA Cloning 2:
Expression Systems,
2nd Edition, Glover et al. (eds.), page 15 (Oxford University Press 1995),
Ward et al., "Genetic
Manipulation and Expression of Antibodies," in Monoclonal Antibodies:
Principles and
Applications, page 137 (Wiley-Liss, Inc. 1995), and Georgiou, "Expression of
Proteins in Bacteria,"
in Protein Engineering: Principles and Practice, Cleland et al. (eds.), page
101 (John Wiley & Sons,
Inc. 1996)).
[186] Standard methods for introducing expression vectors into bacterial,
yeast, insect, and
plant cells are provided, for example, by Ausubel (1995).
[187] General methods for expressing and recovering foreign protein produced
by a
mammalian cell system are provided by, for example, Etcheverry, "Expression of
Engineered Proteins in
Mammalian Cell Culture," in Protein Engineering: Principles and Practice,
Cleland et al. (eds.), pages
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
47
163 (Wiley-Liss, Inc. 1996). Standard techniques for recovering protein
produced by a bacterial
system is provided by, for example, Grisshammer et al., "Purification of over-
produced proteins
from E. coli cells," in DNA Cloning 2: Expression Systems, 2nd Edition, Glover
et al. (eds.), pages
59-92 (Oxford University Press 1995). Established methods for isolating
recombinant proteins from
a baculovirus system are described by Richardson (ed.), Baculovirus Expression
Protocols (The
Humana Press, Inc. 1995).
[188] As an alternative, polypeptides of the present invention can be
synthesized by
exclusive solid phase synthesis, partial solid phase methods, fragment
condensation or classical
solution synthesis. These synthesis methods are well-known to those of skill
in the art (see, for
example, Merrifield, J. Am. Chem. Soc. 85:2149 (1963), Stewart et al., "Solid
Phase Peptide
Synthesis" (2nd Edition), (Pierce Chemical Co. 1984), Bayer and Rapp, Chem.
Pept. Prot. 3:3
(1986), Atherton et al., Solid Phase Peptide Synthesis: A Practical Approach
(IRL Press 1989),
Fields and Colowick, "Solid-Phase Peptide Synthesis," Methods in Enzymology
Volume 289
(Academic Press 1997), and Lloyd-Williams et al., Chemical Approaches to the
Synthesis of
Peptides and Proteins (CRC Press, Inc. 1997)). Variations in total chemical
synthesis strategies,
such as "native chemical ligation" and "expressed protein ligation" are also
standard (see, for
example, Dawson et al., Science 266:776 (1994), Hackeng et al., Proc. Nat'l
Acad. Sci. USA 94:7845
(1997), Dawson, Methods Enzymol. 287: 34 (1997), Muir et al, Proc. Nat'l Acad.
Sci. USA 95:6705
(1998), and Severinov and Muir, J. Biol. Chem. 273:16205 (1998)).
[189] Peptides and polypeptides of the present invention comprise at least
six, at least nine,
or at least 15 contiguous amino acid residues of any of SEQ ID NOs:2, 5, 8,
11, 14, 21, 23, 107, 109,
113, 115, 117, or 119. As an illustration, polypeptides can comprise at least
six, at least nine, or at
least 15 contiguous amino acid residues of of any of SEQ ID NOs: 2, 5, 8, 11,
14, 21, 23, 107, 109,
113, 115, 117, or 119. Within certain embodiments of the invention, the
polypeptides comprise 20,
30, 40, 50, 100, or more contiguous residues of these amino acid sequences.
Nucleic acid molecules
encoding such peptides and polypeptides are useful as polymerase chain
reaction primers and probes.
[190] Moreover, IL-17RE polypeptides and fragments thereof can be expressed as
monomers, homodimers, heterodimers, or multimers within higher eukaryotic
cells. Such cells can
be used to produce IL-17RE monomeric, homodimeric, heterodimeric and
multimeric receptor
polypeptides that comprise at least one IL-17RE polypeptide ("IL-17RE-
comprising receptors" or
"IL-17RE-comprising receptor polypeptides"), or can be used as assay cells in
screening systems.
Within one aspect of the present invention, a polypeptide of the present
invention comprising the IL-
17RE extracellular domain is produced by a cultured cell, and the cell is used
to screen for ligands
for the receptor, including the natural ligand, IL-17C, or even agonists and
antagonists of the natural
ligand. To summarize this approach, a cDNA or gene encoding the receptor is
combined with other
genetic elements required for its expression (e.g., a transcription promoter),
and the resulting
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
48
expression vector is inserted into a host cell. Cells that express the DNA and
produce functional
receptor are selected and used within a variety of screening systems. Each
component of the
monomeric, homodimeric, heterodimeric and multimeric receptor complex can be
expressed in the
same cell. Moreover, the components of the monomeric, homodimeric,
heterodimeric and
multimeric receptor complex can also be fused to a transmembrane domain or
other membrane
fusion moiety to allow complex assembly and screening of transfectants as
described above.
[191] To assay the IL-17C antagonist polyepeptides and antibodies of the
present
invention, mammalian cells suitable for use in expressing IL-17RE-comprising
receptors or other
receptors known to bind IL-17C and transducing a receptor-mediated signal
include cells that express
other receptor subunits that may form a functional complex with IL-17RE. It is
also preferred to use
a cell from the same species as the receptor to be expressed. Within a
preferred embodiment, the cell
is dependent upon an exogenously supplied hematopoietic growth factor for its
proliferation.
Preferred cell lines of this type are the human TF-1 cell line (ATCC number
CRL-2003) and the
AML-193 cell line (ATCC number CRL-9589), which are GM-CSF-dependent human
leukemic cell
lines and BaF3 (Palacios and Steinmetz, Cell 41: 727-734, (1985)) which is an
IL-3 dependent
murine pre-B cell line. Other cell lines include BHK, COS-1 and CHO cells.
Suitable host cells can
be engineered to produce the necessary receptor subunits or other cellular
component needed for the
desired cellular response. This approach is advantageous because cell lines
can be engineered to
express receptor subunits from any species, thereby overcoming potential
limitations arising from
species specificity. Species orthologs of the human receptor cDNA can be
cloned and used within
cell lines from the same species, such as a mouse cDNA in the BaF3 cell line.
Cell lines that are
dependent upon one hematopoietic growth factor, such as GM-CSF or IL-3, can
thus be engineered
to become dependent upon another cytokine that acts through the IL-17RE
receptor, such as IL-17C.
[192] Cells expressing functional receptor are used within screening assays. A
variety of
suitable assays are known in the art. These assays are based on the detection
of a biological response
in a target cell. One such assay is a cell proliferation assay. Cells are
cultured in the presence or
absence of a test compound, and cell proliferation is detected by, for
example, measuring
incorporation of tritiated thymidine or by colorimetric assay based on the
metabolic breakdown of 3-
(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) (Mosman, J.
Immunol. Meth.
65: 55-63, (1983)). An alternative assay format uses cells that are further
engineered to express a
reporter gene. The reporter gene is linked to a promoter element that is
responsive to the receptor-
linked pathway, and the assay detects activation of transcription of the
reporter gene. A preferred
promoter element in this regard is a serum response element, or SRE. See,
e.g., Shaw et al., Cell
56:563-572, (1989). A preferred such reporter gene is a luciferase gene (de
Wet et al., Mol. Cell.
Biol. 7:725, (1987)). Expression of the luciferase gene is detected by
luminescence using methods
known in the art (e.g., Baumgartner et al., J. Biol. Chem. 269:29094-29101,
(1994); Schenborn and
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
49
Goiffin, Promega Notes 41:11, 1993). Luciferase activity assay kits are
commercially available
from, for example, Promega Corp., Madison, WI. Target cell lines of this type
can be used to screen
libraries of chemicals, cell-conditioned culture media, fungal broths, soil
samples, water samples,
and the like. For example, a bank of cell-conditioned media samples can be
assayed on a target cell
to identify cells that produce ligand. Positive cells are then used to produce
a cDNA library in a
mammalian expression vector, which is divided into pools, transfected into
host cells, and expressed.
Media samples from the transfected cells are then assayed, with subsequent
division of pools, re-
transfection, subculturing, and re-assay of positive cells to isolate a cloned
cDNA encoding the
ligand.
[193] An additional screening approach provided by the present invention
includes the use
of hybrid receptor polypeptides. These hybrid polypeptides fall into two
general classes. Within the
first class, the intracellular domain of IL-17RE, is joined to the ligand-
binding domain of a second
receptor. A second class of hybrid receptor polypeptides comprise the
extracellular (ligand-binding)
domain of IL-17RE (e.g. SEQ ID NO:3, amino acid residues 24-376 of SEQ ID
NO:5, amino acid
residues 24-396 of SEQ ID NO:8, SEQ ID NO:12, amino acid residues 24-414 of
SEQ ID NO:21,
SEQ ID NO:22, SEQ ID NO:122, amino acid residues 24-414 of SEQ ID NO:109, SEQ
ID NO:113,
SEQ ID NO:115, SEQ ID NO:117, or SEQ ID NO:119) with an intracellular domain
of a second
receptor, preferably a hematopoietic cytokine receptor, and a transmembrane
domain. Hybrid IL-
17RE monomers, homodimers, heterodimers and multimers of the present invention
receptors of this
second class are expressed in cells known to be capable of responding to
signals transduced by the
second receptor. Together, these two classes of hybrid receptors enable the
identification of a
responsive cell type for the development of an assay for detecting IL-17C.
Moreover, such cells can
be used in the presence of IL-17C to assay the soluble receptor antagonists of
the present invention
in a competition-type assay. In such assay, a decrease in the proliferation or
signal transduction
activity of IL-17C in the presence of a soluble receptor of the present
invention demonstrates
antagonistic activity. Moreover IL-17RE-soluble receptor binding assays, and
cell-based assays, can
also be used to assess whether a soluble receptor binds, blocks, inhibits,
reduces, antagonizes or
neutralizes IL-17C activity.
F) Production of IL-17RE Fusion Proteins and Conjugates
[194] One general class of IL-17RE analogs are variants having an amino acid
sequence
that is a mutation of the amino acid sequence disclosed herein. Another
general class of IL-17RE
analogs is provided by anti-idiotype antibodies, and fragments thereof, as
described below.
Moreover, recombinant antibodies comprising anti-idiotype variable domains can
be used as analogs
(see, for example, Monfardini et al., Proc. Assoc. Am. Physicians 108:420
(1996)). Since the
variable domains of anti-idiotype IL-17RE antibodies mimic IL-17RE, these
domains can provide
IL-17RE binding activity. Methods of producing anti-idiotypic catalytic
antibodies are known to
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
those of skill in the art (see, for example, Joron et al., Ann. N YAcad. Sci.
672:216 (1992), Friboulet
et al., Appl. Biochem. Biotechnol. 47:229 (1994), and Avalle et al., Ann. N Y
Acad. Sci. 864:118
(1998)).
[195] Another approach to identifying IL-17RE analogs is provided by the use
of
combinatorial libraries. Methods for constructing and screening phage display
and other
combinatorial libraries are provided, for example, by Kay et al., Phage
Display of Peptides and
Proteins (Academic Press 1996), Verdine, U.S. Patent No. 5,783,384, Kay, et.
al., U.S. Patent No.
5,747,334, and Kauffinan et al., U.S. Patent No. 5,723,323.
[196] IL-17RE polypeptides have both in vivo and in vitro uses. As an
illustration, a
soluble form of IL-17RE can be added to cell culture medium to inhibit the
effects of the IL-17RE
ligand (i.e. IL-17C) produced by the cultured cells.
[197] Fusion proteins of IL-17RE can be used to express IL-17RE in a
recombinant host,
and to isolate the produced IL-17RE. As described below, particular IL-17RE
fusion proteins also
have uses in diagnosis and therapy. One type of fusion protein comprises a
peptide that guides a IL-
17RE polypeptide from a recombinant host cell. To direct a IL-17RE polypeptide
into the secretory
pathway of a eukaryotic host cell, a secretory signal sequence (also known as
a signal peptide, a
leader sequence, prepro sequence or pre sequence) is provided in the IL-17RE
expression vector.
While the secretory signal sequence may be derived from IL-17RE, a suitable
signal sequence may
also be derived from another secreted protein or synthesized de novo. The
secretory signal sequence
is operably linked to a IL-17RE-encoding sequence such that the two sequences
are joined in the
correct reading frame and positioned to direct the newly synthesized
polypeptide into the secretory
pathway of the host cell. Secretory signal sequences are commonly positioned
5' to the nucleotide
sequence encoding the polypeptide of interest, although certain secretory
signal sequences may be
positioned elsewhere in the nucleotide sequence of interest (see, e.g., Welch
et al., U.S. Patent No.
5,037,743; Holland et al., U.S. Patent No. 5,143,830).
[198] Although the secretory signal sequence of IL-17RE or another protein
produced by
mammalian cells (e.g., tissue-type plasminogen activator signal sequence, as
described, for example,
in U.S. Patent No. 5,641,655) is useful for expression of IL-17RE in
recombinant mammalian hosts,
a yeast signal sequence is preferred for expression in yeast cells. Examples
of suitable yeast signal
sequences are those derived from yeast mating phermone a-factor (encoded by
the MFal gene),
invertase (encoded by the SUC2 gene), or acid phosphatase (encoded by the PHO5
gene). See, for
example, Romanos et al., "Expression of Cloned Genes in Yeast," in DNA Cloning
2: A Practical
Approach, 2"d Edition, Glover and Hames (eds.), pages 123-167 (Oxford
University Press 1995).
[199] IL-17RE soluble receptor polypeptides can be prepared by expressing a
truncated
DNA encoding the extracellular domain, for example, a polypeptide which
contains SEQ ID NO:6,
or the corresponding region of a non-human receptor. It is preferred that the
extracellular domain
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
51
polypeptides be prepared in a form substantially free of transmembrane and
intracellular polypeptide
segments. To direct the export of the receptor domain from the host cell, the
receptor DNA is linked
to a second DNA segment encoding a secretory peptide, such as a t-PA secretory
peptide. To
facilitate purification of the secreted receptor domain, a C-terminal
extension, such as a poly-
histidine tag, substance P, F1agTM peptide (Hopp et al., Biotechnology 6:1204-
1210, (1988); available
from Eastman Kodak Co., New Haven, CT) or another polypeptide or protein for
which an antibody
or other specific binding agent is available, can be fused to the receptor
polypeptide. Moreover, IL-
17RE antigenic epitopes from the extracellular cytokine binding domains are
also prepared as
described above.
[200] In an alternative approach, a receptor extracellular domain of IL-17RE
or other
cytokine receptor component can be expressed as a fusion with immunoglobulin
heavy chain
constant regions, typically an Fc fragment, which contains two constant region
domains and a hinge
region but lacks the variable region (See, Sledziewski, AZ et al., US Patent
No. 6,018,026 and
5,750,375). Immunoglobulin heavy chain constant regions are known to those
skilled in the art, and
include Fc5, Fc10, IgGl, IgG2, IgG3, IgG4, IgA, IgD, IgM and IgE. The soluble
IL-17RE
polypeptides of the present invention include such fusions. One such fusion is
shown in SEQ ID
NOs:100 and 102; and 123 and 124. Such fusions are typically secreted as
multimeric molecules
wherein the Fc portions are disulfide bonded to each other and two receptor
polypeptides are arrayed
in closed proximity to each other. Fusions of this type can be used to
affinity purify the cognate
ligand from solution, as an in vitro assay tool, to block, inhibit or reduce
signals in vitro by
specifically titrating out ligand, and as antagonists in vivo by administering
them parenterally to bind
circulating ligand and clear it from the circulation. To purify ligand, a IL-
17RE-Ig chimera is added
to a sample containing the ligand (e.g., cell-conditioned culture media or
tissue extracts) under
conditions that facilitate receptor-ligand binding (typically near-
physiological temperature, pH, and
ionic strength). The chimera-ligand complex is then separated by the mixture
using protein A, which
is immobilized on a solid support (e.g., insoluble resin beads). The ligand is
then eluted using
conventional chemical techniques, such as with a salt or pH gradient. In the
alternative, the chimera
itself can be bound to a solid support, with binding and elution carried out
as above. The chimeras
may be used in vivo to regulate inflammatory responses including acute phase
responses such as
serum amyloid A (SAA), C-reactive protein (CRP), and the like. Chimeras with
high binding
affinity are administered parenterally (e.g., by intramuscular, subcutaneous
or intravenous injection).
Circulating molecules bind ligand and are cleared from circulation by normal
physiological
processes. For use in assays, the chimeras are bound to a support via the Fc
region and used in an
ELISA format.
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
52
[201] To assist in isolating anti-IL-17RE and binding partners of the present
invention, an
assay system that uses a ligand-binding receptor (or an antibody, one member
of a complement/ anti-
complement pair) or a binding fragment thereof, and a commercially available
biosensor instrument
(BlAcore, Pharmacia Biosensor, Piscataway, NJ) may be advantageously employed.
Such receptor,
antibody, member of a complement/anti-complement pair or fragment is
immobilized onto the
surface of a receptor chip. Use of this instrument is disclosed by Karlsson,
J. Immunol. Methods
145:229-40, 1991 and Cunningham and Wells, J. Mol. Biol. 234:554-63, 1993. A
receptor,
antibody, member or fragment is covalently attached, using amine or sulfhydryl
chemistry, to
dextran fibers that are attached to gold film within the flow cell. A test
sample is passed through the
cell. If a ligand, epitope, or opposite member of the complement/anti-
complement pair is present in
the sample, it will bind to the immobilized receptor, antibody or member,
respectively, causing a
change in the refractive index of the medium, which is detected as a change in
surface plasmon
resonance of the gold film. This system allows the determination of on- and
off-rates, from which
binding affinity can be calculated, and assessment of stoichiometry of
binding. Alternatively,
ligand/receptor binding can be analyzed using SELDI(TM) technology (Ciphergen,
Inc., Palo Alto,
CA). Moreover, BIACorE technology, described above, can be used to be used in
competition
experiments to determine if different monoclonal antibodies bind the same or
different epitopes on
the IL-17RE polypeptide, and as such, be used to aid in epitope mapping of
neutralizing antibodies
of the present invention that bind, block, inhibit, reduce, antagonize or
neutralize IL-17C.
[202] Ligand-binding receptor polypeptides can also be used within other assay
systems
known in the art. Such systems include Scatchard analysis for determination of
binding affinity (see
Scatchard, Ann. NY Acad. Sci. 51: 660-72, 1949) and calorimetric assays
(Cunningham et al.,
Science 253:545-48, 1991; Cunningham et al., Science 245:821-25, 1991).
[203] The present invention further provides a variety of other polypeptide
fusions and
related multimeric proteins comprising one or more polypeptide fusions. For
example, a soluble IL-
17RE receptor can be prepared as a fusion to a dimerizing protein as disclosed
in U.S. Patents Nos.
5,155,027 and 5,567,584. Preferred dimerizing proteins in this regard include
immunoglobulin
constant region domains, e.g., IgGyl, and the human ic light chain.
Immunoglobulin-soluble IL-
17RE fusions can be expressed in genetically engineered cells to produce a
variety of multimeric IL-
17RE receptor analogs. Auxiliary domains can be fused to soluble IL-17RE
receptor to target them
to specific cells, tissues, or macromolecules (e.g., collagen, or cells
expressing the IL-17RE ligand,
IL-17C). A IL-17RE polypeptide can be fused to two or more moieties, such as
an affinity tag for
purification and a targeting domain. Polypeptide fusions can also comprise one
or more cleavage
sites, particularly between domains. See, Tuan et al., Connective Tissue
Research 34:1-9, 1996.
[204] In bacterial cells, it is often desirable to express a heterologous
protein as a fusion
protein to decrease toxicity, increase stability, and to enhance recovery of
the expressed protein. For
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
53
example, IL-17RE can be expressed as a fusion protein comprising a glutathione
S-transferase
polypeptide. Glutathione S-transferease fusion proteins are typically soluble,
and easily purifiable
from E. coli lysates on immobilized glutathione columns. In similar
approaches, a IL-17RE fusion
protein comprising a maltose binding protein polypeptide can be isolated with
an amylose resin
column, while a fusion protein comprising the C-terminal end of a truncated
Protein A gene can be
purified using IgG-Sepharose. Established techniques for expressing a
heterologous polypeptide as a
fusion protein in a bacterial cell are described, for example, by Williams et
al., "Expression of
Foreign Proteins in E. coli Using Plasmid Vectors and Purification of Specific
Polyclonal
Antibodies," in DNA Cloning 2: A Practical Approach, 2"d Edition, Glover and
Hames (Eds.), pages
15-58 (Oxford University Press 1995). In addition, commercially available
expression systems are
available. For example, the PINPOINT Xa protein purification system (Promega
Corporation;
Madison, WI) provides a method for isolating a fusion protein comprising a
polypeptide that
becomes biotinylated during expression with a resin that comprises avidin.
[205] Peptide tags that are useful for isolating heterologous polypeptides
expressed by
either prokaryotic or eukaryotic cells include polyHistidine tags (which have
an affinity for nickel-
chelating resin), c-myc tags, calmodulin binding protein (isolated with
calmodulin affinity
chromatography), substance P, the RYIRS tag (which binds with anti-RYIRS
antibodies), the Glu-
Glu tag, and the FLAG tag (which binds with anti-FLAG antibodies). See, for
example, Luo et al.,
Arch. Biochem. Biophys. 329:215 (1996), Morganti et al., Biotechnol. Appl.
Biochem. 23:67 (1996),
and Zheng et al., Gene 186:55 (1997). Nucleic acid molecules encoding such
peptide tags are
available, for example, from Sigma-Aldrich Corporation (St. Louis, MO).
[206] Another form of fusion protein comprises a IL-17RE polypeptide and an
immunoglobulin heavy chain constant region, typically an Fc fragment, which
contains two or three
constant region domains and a hinge region but lacks the variable region. As
an illustration, Chang et
al., U.S. Patent No. 5,723,125, describe a fusion protein comprising a human
interferon and a human
immunoglobulin Fc fragment. The C-terminal of the interferon is linked to the
N-terminal of the Fc
fragment by a peptide linker moiety. An example of a peptide linker is a
peptide comprising
primarily a T cell inert sequence, which is immunologically inert. An
exemplary peptide linker has
the amino acid sequence: GGSGG SGGGG SGGGG S (SEQ ID NO:25). In this fusion
protein, an
illustrative Fc moiety is a human y4 chain, which is stable in solution and
has little or no complement
activating activity. Accordingly, the present invention contemplates a IL-17RE
fusion protein that
comprises a IL-17RE moiety and a human Fc fragment, wherein the C-terminus of
the IL-17RE
moiety is attached to the N-terminus of the Fc fragment via a peptide linker,
such as a peptide
comprising the amino acid sequence of SEQ ID NOs:2, 5, 8, 11, 14, 21, 23, 107,
109, 113, 115, 117,
119, or 122. The IL-17RE moiety can be a IL-17RE molecule or a fragment
thereof. For example, a
fusion protein can comprise the amino acid of SEQ ID NO:3 and an Fc fragment
(e.g., a human Fc
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
54
fragment) (SEQ ID NO:100), SEQ ID NO:6 and an Fc fragment (SEQ ID NO:102), SEQ
ID NO:122
and an Fc fragment (e.g., a human Fc fragment), SEQ ID NO:109 and an Fc
fragment (e.g., a human
Fc fragment), SEQ ID NO: 113 and an Fc fragment (e.g., a human Fc fragment)
(SEQ ID NO: 124),
SEQ ID NO: 115 and an Fc fragment (e.g., a human Fc fragment), SEQ ID NO:117
and an Fc
fragment (e.g., a human Fc fragment), and SEQ ID NO: 119 and an Fc fragment
(e.g., a human Fc
fragment).
[207] In a preferred embodiment of the invention, an amino acid linker may be
included
between the soluble IL-17RE and the Fc domains. Additionally, an alternative
secretion leader may
be used in place of the native IL-17RE leader.
[208] One skilled in the art would also recognize that the IL-17RE
polypeptides disclosed
herein may be fused to a number of different Fc domains (e.g. Fc4, Fc5, Fc10
or any other variation
thereof).
[209] In another variation, a IL-17RE fusion protein comprises an IgG
sequence, a IL-
17RE moiety covalently joined to the aminoterminal end of the IgG sequence,
and a signal peptide
that is covalently joined to the aminoterminal of the IL-17RE moiety, wherein
the IgG sequence
consists of the following elements in the following order: a hinge region, a
CH2 domain, and a CH3
domain. Accordingly, the IgG sequence lacks a CHi domain. The IL-17RE moiety
displays a IL-
17RE activity, as described herein, such as the ability to bind with a IL-17RE
ligand. This general
approach to producing fusion proteins that comprise both antibody and
nonantibody portions has
been described by LaRochelle et al., EP 742830 (WO 95/21258).
[210] Fusion proteins comprising a IL-17RE moiety and an Fc moiety can be
used, for
example, as an in vitro assay tool. For example, the presence of a IL-17RE
ligand in a biological
sample can be detected using a IL-17RE-immunoglobulin fusion protein, in which
the IL-17RE
moiety is used to bind the ligand, and a macromolecule, such as Protein A or
anti-Fc antibody, is
used to bind the fusion protein to a solid support. Such systems can be used
to identify agonists and
antagonists that interfere with the binding of a IL-17RE ligands, e.g., IL-
17C, to its receptor.
[211] Other examples of antibody fusion proteins include polypeptides that
comprise an
antigen-binding domain and a IL-17RE fragment that contains a IL-17RE
extracellular domain.
Such molecules can be used to target particular tissues for the benefit of IL-
17RE binding activity.
[212] The present invention further provides a variety of other polypeptide
fusions. For
example, part or all of a domain(s) conferring a biological function can be
swapped between IL-
17RE of the present invention with the functionally equivalent domain(s) from
another member of
the cytokine receptor family. Polypeptide fusions can be expressed in
recombinant host cells to
produce a variety of IL-17RE fusion analogs. A IL-17RE polypeptide can be
fused to two or more
moieties or domains, such as an affinity tag for purification and a targeting
domain. Polypeptide
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
fusions can also comprise one or more cleavage sites, particularly between
domains. See, for
example, Tuan et al., Connective Tissue Research 34:1 (1996).
[213] Fusion proteins can be prepared by methods known to those skilled in the
art by
preparing each component of the fusion protein and chemically conjugating
them. Alternatively, a
polynucleotide encoding both components of the fusion protein in the proper
reading frame can be
generated using known techniques and expressed by the methods described
herein. General methods
for enzymatic and chemical cleavage of fusion proteins are described, for
example, by Ausubel
(1995) atpages 16-19 to 16-25.
[214] IL-17RE binding domains can be further characterized by physical
analysis of
structure, as determined by such techniques as nuclear magnetic resonance,
crystallography, electron
diffraction or photoaffinity labeling, in conjunction with mutation of
putative contact site amino
acids of IL-17RE ligand agonists. See, for example, de Vos et al., Science
255:306 (1992), Smith et
al., J. Mol. Biol. 224:899 (1992), and Wlodaver et al., FEBS Lett. 309:59
(1992).
[215] The present invention also contemplates chemically modified IL-17RE
compositions, in which a IL-17RE polypeptide is linked with a polymer.
Illustrative IL-17RE
polypeptides are soluble polypeptides that lack a functional transmembrane
domain, such as a
polypeptide comprising any of SEQ ID NOs: 2, 5, 8, 11, 14, 21, 23, 107, 109,
113, 115, 117, 119, or
122. Typically, the polymer is water soluble so that the IL-17RE conjugate
does not precipitate in an
aqueous environment, such as a physiological environment. An example of a
suitable polymer is one
that has been modified to have a single reactive group, such as an active
ester for acylation, or an
aldehyde for alkylation. In this way, the degree of polymerization can be
controlled. An example of
a reactive aldehyde is polyethylene glycol propionaldehyde, or mono-(CI-C10)
alkoxy, or aryloxy
derivatives thereof (see, for example, Harris, et al., U.S. Patent No.
5,252,714). The polymer may be
branched or unbranched. Moreover, a mixture of polymers can be used to produce
IL-17RE
conjugates.
[216] IL-17RE conjugates used for therapy can comprise pharmaceutically
acceptable
water-soluble polymer moieties. Suitable water-soluble polymers include
polyethylene glycol
(PEG), monomethoxy-PEG, mono-(CI-C10)alkoxy-PEG, aryloxy-PEG, poly-(N-vinyl
pyrrolidone)PEG, tresyl monomethoxy PEG, PEG propionaldehyde, bis-succinimidyl
carbonate
PEG, propylene glycol homopolymers, a polypropylene oxide/ethylene oxide co-
polymer,
polyoxyethylated polyols (e.g., glycerol), polyvinyl alcohol, dextran,
cellulose, or other
carbohydrate-based polymers. Suitable PEG may have a molecular weight from
about 600 to about
60,000, including, for example, 5,000, 12,000, 20,000 and 25,000. A IL-17RE
conjugate can also
comprise a mixture of such water-soluble polymers.
[217] One example of a IL-17RE conjugate comprises a IL-17RE moiety and a
polyalkyl
oxide moiety attached to the N-terminus of the IL-17RE moiety. PEG is one
suitable polyalkyl
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
56
oxide. As an illustration, IL-17RE can be modified with PEG, a process known
as "PEGylation."
PEGylation of IL-17RE can be carried out by any of the PEGylation reactions
known in the art (see,
for example, EP 0 154 316, Delgado et al., Critical Reviews in Therapeutic
Drug Carrier Systems
9:249 (1992), Duncan and Spreafico, Clin. Pharmacokinet. 27:290 (1994), and
Francis et al., Int J
Hematol 68:1 (1998)). For example, PEGylation can be performed by an acylation
reaction or by an
alkylation reaction with a reactive polyethylene glycol molecule. In an
alternative approach, IL-
17RE conjugates are formed by condensing activated PEG, in which a terminal
hydroxy or amino
group of PEG has been replaced by an activated linker (see, for example,
Karasiewicz et al., U.S.
Patent No. 5,382,657).
[218] PEGylation by acylation typically requires reacting an active ester
derivative of PEG
with a IL-17RE polypeptide. An example of an activated PEG ester is PEG
esterified to N-
hydroxysuccinimide. As used herein, the term "acylation" includes the
following types of linkages
between IL-17RE and a water soluble polymer: amide, carbamate, urethane, and
the like. Methods
for preparing PEGylated IL-17RE by acylation will typically comprise the steps
of (a) reacting a IL-
17RE polypeptide with PEG (such as a reactive ester of an aldehyde derivative
of PEG) under
conditions whereby one or more PEG groups attach to IL-17RE, and (b) obtaining
the reaction
product(s). Generally, the optimal reaction conditions for acylation reactions
will be determined
based upon known parameters and desired results. For example, the larger the
ratio of PEG:IL-
17RE, the greater the percentage of polyPEGylated IL-17RE product.
[219] The product of PEGylation by acylation is typically a polyPEGylated IL-
17RE
product, wherein the lysine c-amino groups are PEGylated via an acyl linking
group. An example of
a connecting linkage is an amide. Typically, the resulting IL-17RE will be at
least 95% mono-, di-,
or tri-pegylated, although some species with higher degrees of PEGylation may
be formed depending
upon the reaction conditions. PEGylated species can be separated from
unconjugated IL-17RE
polypeptides using standard purification methods, such as dialysis,
ultrafiltration, ion exchange
chromatography, affinity chromatography, and the like.
[220] PEGylation by alkylation generally involves reacting a terminal aldehyde
derivative
of PEG with IL-17RE in the presence of a reducing agent. PEG groups can be
attached to the
polypeptide via a -CH2-NH group.
[221] Moreover, anti-IL-17RE antibodies or antibody fragments of the present
invention
can be PEGylated using methods in the art and described herein.
[222] Derivatization via reductive alkylation to produce a monoPEGylated
product takes
advantage of the differential reactivity of different types of primary amino
groups available for
derivatization. Typically, the reaction is performed at a pH that allows one
to take advantage of the
pKa differences between the c-amino groups of the lysine residues and the a-
amino group of the N-
terminal residue of the protein. By such selective derivatization, attachment
of a water-soluble
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
57
polymer that contains a reactive group such as an aldehyde, to a protein is
controlled. The
conjugation with the polymer occurs predominantly at the N-terminus of the
protein without
significant modification of other reactive groups such as the lysine side
chain amino groups. The
present invention provides a substantially homogenous preparation of IL-17RE
monopolymer
conjugates.
[223] Reductive alkylation to produce a substantially homogenous population of
monopolymer IL-17RE conjugate molecule can comprise the steps of: (a) reacting
a IL-17RE
polypeptide with a reactive PEG under reductive alkylation conditions at a pH
suitable to permit
selective modification of the a-amino group at the amino terminus of the IL-
17RE, and (b) obtaining
the reaction product(s). The reducing agent used for reductive alkylation
should be stable in aqueous
solution and able to reduce only the Schiff base formed in the initial process
of reductive alkylation.
Illustrative reducing agents include sodium borohydride, sodium
cyanoborohydride, dimethylamine
borane, trimethylamine borane, and pyridine borane.
[224] For a substantially homogenous population of monopolymer IL-17RE
conjugates,
the reductive alkylation reaction conditions are those that permit the
selective attachment of the
water-soluble polymer moiety to the N-terminus of IL-17RE. Such reaction
conditions generally
provide for pKa differences between the lysine amino groups and the a-amino
group at the N-
terminus. The pH also affects the ratio of polymer to protein to be used. In
general, if the pH is
lower, a larger excess of polymer to protein will be desired because the less
reactive the N-terminal
a-group, the more polymer is needed to achieve optimal conditions. If the pH
is higher, the
polymer:IL-17RE need not be as large because more reactive groups are
available. Typically, the pH
will fall within the range of 3 to 9, or 3 to 6. This method can be employed
for making IL-17RE-
comprising homodimeric, heterodimeric or multimeric soluble receptor
conjugates.
[225] Another factor to consider is the molecular weight of the water-soluble
polymer.
Generally, the higher the molecular weight of the polymer, the fewer number of
polymer molecules
which may be attached to the protein. For PEGylation reactions, the typical
molecular weight is
about 2 kDa to about 100 kDa, about 5 kDa to about 50 kDa, or about 12 kDa to
about 25 kDa. The
molar ratio of water-soluble polymer to IL-17RE will generally be in the range
of 1:1 to 100:1.
Typically, the molar ratio of water-soluble polymer to IL-17RE will be 1:1 to
20:1 for
polyPEGylation, and 1:1 to 5:1 for monoPEGylation.
[226] General methods for producing conjugates comprising a polypeptide and
water-
soluble polymer moieties are known in the art. See, for example, Karasiewicz
et al., U.S. Patent No.
5,382,657, Greenwald et al., U.S. Patent No. 5,738, 846, Nieforth et al.,
Clin. Pharmacol. Ther.
59:636 (1996), Monkarsh et al., Anal. Biochem. 247:434 (1997)). This method
can be employed for
making IL-17RE-comprising homodimeric, heterodimeric or multimeric soluble
receptor conjugates.
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
58
[227] The present invention contemplates compositions comprising a peptide or
polypeptide, such as a soluble receptor or antibody described herein. Such
compositions can further
comprise a carrier. The carrier can be a conventional organic or inorganic
carrier. Examples of
carriers include water, buffer solution, alcohol, propylene glycol, macrogol,
sesame oil, corn oil, and
the like.
G) Isolation of IL-17RE Polypeptides
[228] The polypeptides of the present invention can be purified to at least
80% purity, to at
least 90% purity, to at least 95% purity, or greater than 95%, or greater than
99% purity with respect
to contaminating macromolecules, particularly other proteins and nucleic
acids, and free of infectious
and pyrogenic agents. The polypeptides of the present invention may also be
purified to a
pharmaceutically pure state, which is greater than 99.9% pure. In certain
preparations, purified
polypeptide is substantially free of other polypeptides, particularly other
polypeptides of animal
origin.
[229] Fractionation and/or conventional purification methods can be used to
obtain
preparations of IL-17RE purified from natural sources (e.g., human tissue
sources), synthetic IL-
17RE polypeptides, and recombinant IL-17RE polypeptides and fusion IL-17RE
polypeptides
purified from recombinant host cells. In general, ammonium sulfate
precipitation and acid or
chaotrope extraction may be used for fractionation of samples. Exemplary
purification steps may
include hydroxyapatite, size exclusion, FPLC and reverse-phase high
performance liquid
chromatography. Suitable chromatographic media include derivatized dextrans,
agarose, cellulose,
polyacrylamide, specialty silicas, and the like. PEI, DEAE, QAE and Q
derivatives are suitable.
Exemplary chromatographic media include those media derivatized with phenyl,
butyl, or octyl
groups, such as Phenyl-Sepharose FF (Pharmacia), Toyopearl butyl 650 (Toso
Haas,
Montgomeryville, PA), Octyl-Sepharose (Pharmacia) and the like; or polyacrylic
resins, such as
Amberchrom CG 71 (Toso Haas) and the like. Suitable solid supports include
glass beads, silica-
based resins, cellulosic resins, agarose beads, cross-linked agarose beads,
polystyrene beads, cross-
linked polyacrylamide resins and the like that are insoluble under the
conditions in which they are to
be used. These supports may be modified with reactive groups that allow
attachment of proteins by
amino groups, carboxyl groups, sulfhydryl groups, hydroxyl groups and/or
carbohydrate moieties.
[230] Examples of coupling chemistries include cyanogen bromide activation, N-
hydroxysuccinimide activation, epoxide activation, sulfhydryl activation,
hydrazide activation, and
carboxyl and amino derivatives for carbodiimide coupling chemistries. These
and other solid media
are well known and widely used in the art, and are available from commercial
suppliers. Selection of
a particular method for polypeptide isolation and purification is a matter of
routine design and is
determined in part by the properties of the chosen support. See, for example,
Affinity
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
59
Chromatogi aphy: Principles & Methods (Pharmacia LKB Biotechnology 1988), and
Doonan,
Protein Purification Protocols (The Humana Press 1996).
[231] Additional variations in IL-17RE isolation and purification can be
devised by those
of skill in the art. For example, anti-IL-17RE antibodies, obtained as
described below, can be used to
isolate large quantities of protein by immunoaffinity purification.
[232] The polypeptides of the present invention can also be isolated by
exploitation of
particular properties. For example, immobilized metal ion adsorption (IMAC)
chromatography can
be used to purify histidine-rich proteins, including those comprising
polyhistidine tags. Briefly, a gel
is first charged with divalent metal ions to form a chelate (Sulkowski, Trends
in Biochem. 3:1
(1985)). Histidine-rich proteins will be adsorbed to this matrix with
differing affinities, depending
upon the metal ion used, and will be eluted by competitive elution, lowering
the pH, or use of strong
chelating agents. Other methods of purification include purification of
glycosylated proteins by
lectin affinity chromatography and ion exchange chromatography (M. Deutscher,
(ed.), Meth.
Enzymol. 182:529 (1990)). Within additional embodiments of the invention, a
fusion of the
polypeptide of interest and an affinity tag (e.g., maltose-binding protein, an
immunoglobulin domain)
may be constructed to facilitate purification. Moreover, the ligand-binding
properties of IL-17RE
extracellular domain can be exploited for purification, for example, of IL-
17RE-comprising soluble
receptors; for example, by using affinity chromatography wherein IL-17C ligand
is bound to a
column and the IL-17RE-comprising receptor is bound and subsequently eluted
using standard
chromatography methods.
[233] IL-17RE polypeptides or fragments thereof may also be prepared through
chemical
synthesis, as described above. IL-17RE polypeptides may be monomers or
multimers; glycosylated
or non-glycosylated; PEGylated or non-PEGylated; and may or may not include an
initial methionine
amino acid residue.
H) Production of Antibodies to IL-17RE Proteins
[234] Antibodies to IL-17RE can be obtained, for example, using the product of
a IL-17RE
expression vector or IL-17RE isolated from a natural source as an antigen.
Particularly useful anti-
IL-17RE antibodies "bind specifically" with IL-17RE. Antibodies are considered
to be specifically
binding if the antibodies exhibit at least one of the following two
properties: (1) antibodies bind to
IL-17RE with a threshold level of binding activity, and (2) antibodies do not
significantly cross-react
with polypeptides related to IL-17RE.
[235] With regard to the first characteristic, antibodies specifically bind if
they bind to a
IL-17RE polypeptide, peptide or epitope with a binding affinity (Ka) of 106 M-
1 or greater, preferably
10' M-1 or greater, more preferably 108 M-1 or greater, and most preferably
109 M-1 or greater. The
binding affinity of an antibody can be readily determined by one of ordinary
skill in the art, for
example, by Scatchard analysis (Scatchard, Ann. NYAcad. Sci. 51:660 (1949)).
With regard to the
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
second characteristic, antibodies do not significantly cross-react with
related polypeptide molecules,
for example, if they detect IL-17RE, but not presently known polypeptides
using a standard Western
blot analysis. Examples of known related polypeptides include known cytokine
receptors.
[236] Anti-IL-17RE antibodies can be produced using antigenic IL-17RE epitope-
bearing
peptides and polypeptides. Antigenic epitope-bearing peptides and polypeptides
of the present
invention contain a sequence of at least nine, or between 15 to about 30 amino
acids contained within
any of SEQ ID NOs: 2, 5, 8, 11, 14, 21, 23, 107, 109, 113, 115, 117, 119, or
122, or another amino
acid sequence disclosed herein. However, peptides or polypeptides comprising a
larger portion of an
amino acid sequence of the invention, containing from 30 to 50 amino acids, or
any length up to and
including the entire amino acid sequence of a polypeptide of the invention,
also are useful for
inducing antibodies that bind with IL-17RE. It is desirable that the amino
acid sequence of the
epitope-bearing peptide is selected to provide substantial solubility in
aqueous solvents (i.e., the
sequence includes relatively hydrophilic residues, while hydrophobic residues
are typically avoided).
Moreover, amino acid sequences containing proline residues may be also be
desirable for antibody
production.
[237] As an illustration, potential antigenic sites in IL-17RE were identified
using the
Jameson-Wolf method, Jameson and Wolf, CABIOS 4:181, (1988), as implemented by
the
PROTEAN program (version 3.14) of LASERGENE (DNASTAR; Madison, WI). Default
parameters were used in this analysis.
[238] The Jameson-Wolf method predicts potential antigenic determinants by
combining
six major subroutines for protein structural prediction. Briefly, the Hopp-
Woods method, Hopp et
al., Proc. Nat'l Acad. Sci. USA 78:3824 (1981), was first used to identify
amino acid sequences
representing areas of greatest local hydrophilicity (parameter: seven residues
averaged). In the
second step, Emini's method, Emini et al., J. Virology 55:836 (1985), was used
to calculate surface
probabilities (parameter: surface decision threshold (0.6) = 1). Third, the
Karplus-Schultz method,
Karplus and Schultz, Naturwissenschaften 72:212 (1985), was used to predict
backbone chain
flexibility (parameter: flexibility threshold (0.2) = 1). In the fourth and
fifth steps of the analysis,
secondary structure predictions were applied to the data using the methods of
Chou-Fasman, Chou,
"Prediction of Protein Structural Classes from Amino Acid Composition," in
Prediction of Protein
Structure and the Principles of Protein Conformation, Fasman (ed.), pages 549-
586 (Plenum Press
1990), and Garnier-Robson, Gamier et al., J. Mol. Biol. 120:97 (1978) (Chou-
Fasman parameters:
conformation table = 64 proteins; a region threshold = 103; 0 region threshold
= 105; Gamier-
Robson parameters: a and 0 decision constants = 0). In the sixth subroutine,
flexibility parameters
and hydropathy/solvent accessibility factors were combined to determine a
surface contour value,
designated as the "antigenic index." Finally, a peak broadening function was
applied to the antigenic
index, which broadens major surface peaks by adding 20, 40, 60, or 80% of the
respective peak value
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
61
to account for additional free energy derived from the mobility of surface
regions relative to interior
regions. This calculation was not applied, however, to any major peak that
resides in a helical region,
since helical regions tend to be less flexible. Hopp/Woods hydrophilicity
profiles can be used to
determine regions that have the most antigenic potential within SEQ ID NO:6
(Hopp et al., Proc.
Natl. Acad. Sci.78:3824-3828, 1981; Hopp, J. Immun. Meth. 88:1-18, 1986 and
Triquier et al.,
Protein EnOneering 11:153-169, 1998). The profile is based on a sliding six-
residue window.
Buried G, S, and T residues and exposed H, Y, and W residues were ignored.
Moreover, IL-17RE
antigenic epitopes within SEQ ID NO:6 as predicted by a Jameson-Wolf plot,
e.g., using DNASTAR
Protean program (DNASTAR, Inc., Madison, WI) serve as preferred antigenic
epitopes, and can be
determined by one of skill in the art. Such antigenic epitopes include SEQ ID
NOs: 115 ("antigenic
peptide 1"), 117 ("antigenic peptide 2"), 119 ("antigenic peptide 3"), and the
following amino acid
sequences of SEQ ID NO:6 would provide suitable antigenic peptides: amino
acids 51 to 59
("antigenic peptide 4"), amino acids 72 to 83 ("antigenic peptide 5"), 91 to
97 ("antigenic peptide
6"), amino acids 174 to 180 ("antigenic peptide 7"), and amino acids 242 to
246 ("antigenic peptide
8"). The present invention contemplates the use of any one of antigenic
peptides X to Y to generate
antibodies to IL-17RE or as a tool to screen or identify neutralizing
monoclonal antibodies of the
present invention. The present invention also contemplates polypeptides
comprising at least one of
antigenic peptides 1 to 5. The present invention contemplates the use of any
antigenic peptides or
epitopes described herein to generate antibodies to IL-17RE, as well as to
identify and screen anti-
IL-17RE monoclonal antibodies that are neutralizing, and that may bind, block,
inhibit, reduce,
antagonize or neutralize the activity of IL-17C.
[239] Moreover, suitable antigens also include the IL-17RE polypeptides
comprising a IL-
17RE cytokine binding, or extracellular domain disclosed above in combination
with another
cytokine extracellular domain, such as a class I or II cytokine receptor
domain, such as those that
may form soluble IL-17RE heterodimeric or multimeric polypeptides, and the
like.
[240] Polyclonal antibodies to recombinant IL-17RE protein or to IL-17RE
isolated from
natural sources can be prepared using methods well-known to those of skill in
the art. See, for
example, Green et al., "Production of Polyclonal Antisera," in Immunochemical
Protocols (Manson,
ed.), pages 1-5 (Humana Press 1992), and Williams et al., "Expression of
foreign proteins in E. coli
using plasmid vectors and purification of specific polyclonal antibodies," in
DNA Cloning 2:
Expression Systems, 2nd Edition, Glover et al. (eds.), page 15 (Oxford
University Press 1995). The
immunogenicity of a IL-17RE polypeptide can be increased through the use of an
adjuvant, such as
alum (aluminum hydroxide) or Freund's complete or incomplete adjuvant.
Polypeptides useful for
immunization also include fusion polypeptides, such as fusions of IL-17RE or a
portion thereof with
an immunoglobulin polypeptide or with maltose binding protein. The polypeptide
immunogen may
be a full-length molecule or a portion thereof. If the polypeptide portion is
"hapten-like," such
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
62
portion may be advantageously joined or linked to a macromolecular carrier
(such as keyhole limpet
hemocyanin (KLH), bovine serum albumin (BSA) or tetanus toxoid) for
immunization.
[241] Although polyclonal antibodies are typically raised in animals such as
horses, cows,
dogs, chicken, rats, mice, rabbits, guinea pigs, goats, or sheep, an anti-IL-
17RE antibody of the
present invention may also be derived from a subhuman primate antibody.
General techniques for
raising diagnostically and therapeutically useful antibodies in baboons may be
found, for example, in
Goldenberg et al., international patent publication No. WO 91/11465, and in
Losman et al., Int. J.
Cancer 46:310 (1990).
[242] Alternatively, monoclonal anti-IL-17RE antibodies can be generated.
Rodent mono-
clonal antibodies to specific antigens may be obtained by methods known to
those skilled in the art
(see, for example, Kohler et al., Nature 256:495 (1975), Coligan et al.
(eds.), Current Protocols in
Immunology, Vol. 1, pages 2.5.1-2.6.7 (John Wiley & Sons 1991) ["Coligan"],
Picksley et al.,
"Production of monoclonal antibodies against proteins expressed in E. coli,"
in DNA Cloning 2:
Expression Systems, 2nd Edition, Glover et al. (eds.), page 93 (Oxford
University Press 1995)).
[243] Briefly, monoclonal antibodies can be obtained by injecting mice with a
composition
comprising a IL-17RE gene product, verifying the presence of antibody
production by removing a
serum sample, removing the spleen to obtain B-lymphocytes, fusing the B-
lymphocytes with
myeloma cells to produce hybridomas, cloning the hybridomas, selecting
positive clones which
produce antibodies to the antigen, culturing the clones that produce
antibodies to the antigen, and
isolating the antibodies from the hybridoma cultures.
[244] In addition, an anti-IL-17RE antibody of the present invention may be
derived from a
human monoclonal antibody. Human monoclonal antibodies are obtained from
transgenic mice that have
been engineered to produce specific human antibodies in response to antigenic
challenge. In this
technique, elements of the human heavy and light chain locus are introduced
into strains of mice derived
from embryonic stem cell lines that contain targeted disruptions of the
endogenous heavy chain and light
chain loci. The transgenic mice can synthesize human antibodies specific for
human antigens, and the
mice can be used to produce human antibody-secreting hybridomas. Methods for
obtaining human
antibodies from transgenic mice are described, for example, by Green et al.,
Nature Genet. 7:13 (1994),
Lonberg et al., Nature 368:856 (1994), and Taylor et al., Int. Immun. 6:579
(1994).
[245] Monoclonal antibodies can be isolated and purified from hybridoma
cultures by a
variety of well-established techniques. Such isolation techniques include
affinity chromatography
with Protein-A Sepharose, size-exclusion chromatography, and ion-exchange
chromatography (see,
for example, Coligan at pages 2.7.1-2.7.12 and pages 2.9.1-2.9.3; Baines et
al., "Purification of
Immunoglobulin G(IgG)," in Methods in Molecular Biology, Vol. 10, pages 79-104
(The Humana
Press, Inc. 1992)).
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
63
[246] For particular uses, it may be desirable to prepare fragments of anti-IL-
17RE
antibodies. Such antibody fragments can be obtained, for example, by
proteolytic hydrolysis of the
antibody. Antibody fragments can be obtained by pepsin or papain digestion of
whole antibodies by
conventional methods. As an illustration, antibody fragments can be produced
by enzymatic
cleavage of antibodies with pepsin to provide a 5S fragment denoted F(ab')z.
This fragment can be
further cleaved using a thiol reducing agent to produce 3.5S Fab' monovalent
fragments. Optionally,
the cleavage reaction can be performed using a blocking group for the
sulfhydryl groups that result
from cleavage of disulfide linkages. As an alternative, an enzymatic cleavage
using pepsin produces
two monovalent Fab fragments and an Fc fragment directly. These methods are
described, for
example, by Goldenberg, U.S. patent No. 4,331,647, Nisonoff et al., Arch
Biochem. Biophys. 89:230
(1960), Porter, Biochem. J. 73:119 (1959), Edelman et al., in Methods in
Enzymology Vol. 1, page
422 (Academic Press 1967), and by Coligan at pages 2.8.1-2.8.10 and 2.10.-
2.10.4.
[247] Other methods of cleaving antibodies, such as separation of heavy chains
to form
monovalent light-heavy chain fragments, further cleavage of fragments, or
other enzymatic, chemical
or genetic techniques may also be used, so long as the fragments bind to the
antigen that is
recognized by the intact antibody.
[248] For example, Fv fragments comprise an association of VH and VL chains.
This
association can be noncovalent, as described by Inbar et al., Proc. Nat'l
Acad. Sci. USA 69:2659
(1972). Alternatively, the variable chains can be linked by an intermolecular
disulfide bond or cross-
linked by chemicals such as glutaraldehyde (see, for example, Sandhu, Crit.
Rev. Biotech. 12:437
(1992)).
[249] The Fv fragments may comprise VH and VL chains which are connected by a
peptide
linker. These single-chain antigen binding proteins (scFv) are prepared by
constructing a structural
gene comprising DNA sequences encoding the VH and VL domains which are
connected by an
oligonucleotide. The structural gene is inserted into an expression vector
which is subsequently
introduced into a host cell, such as E. coli. The recombinant host cells
synthesize a single
polypeptide chain with a linker peptide bridging the two V domains. Methods
for producing scFvs
are described, for example, by Whitlow et al., Methods: A Companion to Methods
in Enzymology
2:97 (1991) (also see, Bird et al., Science 242:423 (1988), Ladner et al.,
U.S. Patent No. 4,946,778,
Pack et al., Bio/Technology 11:1271 (1993), and Sandhu, supra).
[250] As an illustration, a scFV can be obtained by exposing lymphocytes to IL-
17RE
polypeptide in vitro, and selecting antibody display libraries in phage or
similar vectors (for instance,
through use of immobilized or labeled IL-17RE protein or peptide). Genes
encoding polypeptides
having potential IL-17RE polypeptide binding domains can be obtained by
screening random peptide
libraries displayed on phage (phage display) or on bacteria, such as E. coli.
Nucleotide sequences
encoding the polypeptides can be obtained in a number of ways, such as through
random
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
64
mutagenesis and random polynucleotide synthesis. These random peptide display
libraries can be
used to screen for peptides which interact with a known target which can be a
protein or polypeptide,
such as a ligand or receptor, a biological or synthetic macromolecule, or
organic or inorganic
substances. Techniques for creating and screening such random peptide display
libraries are known
in the art (Ladner et al., U.S. Patent No. 5,223,409, Ladner et al., U.S.
Patent No. 4,946,778, Ladner
et al., U.S. Patent No. 5,403,484, Ladner et al., U.S. Patent No. 5,571,698,
and Kay et al., Phage
Display of Peptides and Proteins (Academic Press, Inc. 1996)) and random
peptide display libraries
and kits for screening such libraries are available commercially, for instance
from CLONTECH
Laboratories, Inc. (Palo Alto, CA), Invitrogen Inc. (Carlsbad, CA), New
England Biolabs, Inc.
(Beverly, MA), and Pharmacia LKB Biotechnology Inc. (Piscataway, NJ). Random
peptide display
libraries can be screened using the IL-17RE sequences disclosed herein to
identify proteins which
bind to IL-17RE.
[251] Another form of an antibody fragment is a peptide coding for a single
complementarity-determining region (CDR). CDR peptides ("minimal recognition
units") can be
obtained by constructing genes encoding the CDR of an antibody of interest.
Such genes are
prepared, for example, by using the polymerase chain reaction to synthesize
the variable region from
RNA of antibody-producing cells (see, for example, Larrick et al., Methods: A
Companion to
Methods in Enzymology 2:106 (1991), Courtenay-Luck, "Genetic Manipulation of
Monoclonal
Antibodies," in Monoclonal Antibodies: Production, Engineering and Clinical
Application, Ritter et
al. (eds.), page 166 (Cambridge University Press 1995), and Ward et al.,
"Genetic Manipulation and
Expression of Antibodies," in Monoclonal Antibodies: Principles and
Applications, Birch et al.,
(eds.), page 137 (Wiley-Liss, Inc. 1995)).
[252] Alternatively, an anti-IL-17RE antibody may be derived from a
"humanized"
monoclonal antibody. Humanized monoclonal antibodies are produced by
transferring mouse
complementary determining regions from heavy and light variable chains of the
mouse
immunoglobulin into a human variable domain. Typical residues of human
antibodies are then
substituted in the framework regions of the murine counterparts. The use of
antibody components
derived from humanized monoclonal antibodies obviates potential problems
associated with the
immunogenicity of murine constant regions. General techniques for cloning
murine immunoglobulin
variable domains are described, for example, by Orlandi et al., Proc. Nat'l
Acad. Sci. USA 86:3833
(1989). Techniques for producing humanized monoclonal antibodies are
described, for example, by
Jones et al., Nature 321:522 (1986), Carter et al., Proc. Nat'l Acad. Sci. USA
89:4285 (1992),
Sandhu, Crit. Rev. Biotech. 12:437 (1992), Singer et al., J. Immun. 150:2844
(1993), Sudhir (ed.),
Antibody Engineering Protocols (Humana Press, Inc. 1995), Kelley, "Engineering
Therapeutic
Antibodies," in Protein Engineering: Principles and Practice, Cleland et al.
(eds.), pages 399-434
(John Wiley & Sons, Inc. 1996), and by Queen et al., U.S. Patent No. 5,693,762
(1997).
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
[253] Moreover, anti-IL-17RE antibodies or antibody fragments of the present
invention
can be PEGylated using methods in the art and described herein.
[254] Polyclonal anti-idiotype antibodies can be prepared by immunizing
animals with
anti-IL-17RE antibodies or antibody fragments, using standard techniques. See,
for example, Green
et al., "Production of Polyclonal Antisera," in Methods In Molecular Biology:
Immunochemical
Protocols, Manson (ed.), pages 1-12 (Humana Press 1992). Also, see Coligan at
pages 2.4.1-2.4.7.
Alternatively, monoclonal anti-idiotype antibodies can be prepared using anti-
IL-17RE antibodies or
antibody fragments as immunogens with the techniques, described above. As
another alternative,
humanized anti-idiotype antibodies or subhuman primate anti-idiotype
antibodies can be prepared
using the above-described techniques. Methods for producing anti-idiotype
antibodies are described,
for example, by Irie, U.S. Patent No. 5,208,146, Greene, et. al., U.S. Patent
No. 5,637,677, and
Varthakavi and Minocha, J. Gen. Virol. 77:1875 (1996).
[255] An anti-IL-17RE antibody can be conjugated with a detectable label to
form an anti-IL-
17RE immunoconjugate. Suitable detectable labels include, for example, a
radioisotope, a fluorescent
label, a chemiluminescent label, an enzyme label, a bioluminescent label or
colloidal gold. Methods of
making and detecting such detectably-labeled immunoconjugates are well-known
to those of ordinary
skill in the art, and are described in more detail below.
[256] The detectable label can be a radioisotope that is detected by
autoradiography. Isotopes
that are particularly useful for the purpose of the present invention are 3H
1251, 1311, 35S and 14C.
[257] Anti-IL-17RE immunoconjugates can also be labeled with a fluorescent
compound.
The presence of a fluorescently-labeled antibody is determined by exposing the
immunoconjugate to
light of the proper wavelength and detecting the resultant fluorescence.
Fluorescent labeling compounds
include fluorescein isothiocyanate, rhodamine, phycoerytherin, phycocyanin,
allophycocyanin, o-phthal-
dehyde and fluorescamine.
[258] Alternatively, anti-IL-17RE immunoconjugates can be detectably labeled
by coupling
an antibody component to a chemiluminescent compound. The presence of the
chemiluminescent-tagged
immunoconjugate is determined by detecting the presence of luminescence that
arises during the course
of a chemical reaction. Examples of chemiluminescent labeling compounds
include luminol, isoluminol,
an aromatic acridinium ester, an imidazole, an acridinium salt and an oxalate
ester.
[259] Similarly, a bioluminescent compound can be used to label anti-IL-17RE
immunoconjugates of the present invention. Bioluminescence is a type of
chemiluminescence found in
biological systems in which a catalytic protein increases the efficiency of
the chemiluminescent reaction.
The presence of a bioluminescent protein is determined by detecting the
presence of luminescence.
Bioluminescent compounds that are useful for labeling include luciferin,
luciferase and aequorin.
[260] Alternatively, anti-IL-17RE immunoconjugates can be detectably labeled
by linking an
anti-IL-17RE antibody component to an enzyme. When the anti-IL-17RE-enzyme
conjugate is
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
66
incubated in the presence of the appropriate substrate, the enzyme moiety
reacts with the substrate to
produce a chemical moiety which can be detected, for example, by
spectrophotometric, fluorometric or
visual means. Examples of enzymes that can be used to detectably label
polyspecific immunoconjugates
include 0-galactosidase, glucose oxidase, peroxidase and alkaline phosphatase.
[261] Those of skill in the art will know of other suitable labels which can
be employed in
accordance with the present invention. The binding of marker moieties to anti-
IL-17RE antibodies can
be accomplished using standard techniques known to the art. Typical
methodology in this regard is
described by Kennedy et al., Clin. Chim. Acta 70:1 (1976), Schurs et al.,
Clin. Chim. Acta 81:1 (1977),
Shih et al., Int'l J. Cancer 46:1101 (1990), Stein et al., Cancer Res. 50:1330
(1990), and Coligan, supra.
[262] Moreover, the convenience and versatility of immunochemical detection
can be
enhanced by using anti-IL-17RE antibodies that have been conjugated with
avidin, streptavidin, and
biotin (see, for example, Wilchek et al. (eds.), "Avidin-Biotin Technology,"
Methods In Enzymology,
Vol. 184 (Academic Press 1990), and Bayer et al., "Immunochemical Applications
of Avidin-Biotin
Technology," in Methods In Molecular Biology, Vol. 10, Manson (ed.), pages 149-
162 (The Humana
Press, Inc. 1992).
[263] Methods for performing immunoassays are well-established. See, for
example, Cook
and Self, "Monoclonal Antibodies in Diagnostic Immunoassays," in Monoclonal
Antibodies: Production,
Engineering, and Clinical Application, Ritter and Ladyman (eds.), pages 180-
208, (Cambridge
University Press, 1995), Perry, "The Role of Monoclonal Antibodies in the
Advancement of
Immunoassay Technology," in Monoclonal Antibodies: Principles and
Applications, Birch and Lennox
(eds.), pages 107-120 (Wiley-Liss, Inc. 1995), and Diamandis, Immunoassay
(Academic Press, Inc.
1996).
[264] The present invention also contemplates kits for performing an
immunological
diagnostic assay for IL-17RE gene expression. Such kits comprise at least one
container comprising
an anti-IL-17RE antibody, or antibody fragment. A kit may also comprise a
second container
comprising one or more reagents capable of indicating the presence of IL-17RE
antibody or antibody
fragments. Examples of such indicator reagents include detectable labels such
as a radioactive label, a
fluorescent label, a chemiluminescent label, an enzyme label, a bioluminescent
label, colloidal gold, and
the like. A kit may also comprise a means for conveying to the user that IL-
17RE antibodies or
antibody fragments are used to detect IL-17RE protein. For example, written
instructions may state
that the enclosed antibody or antibody fragment can be used to detect IL-17RE.
The written material
can be applied directly to a container, or the written material can be
provided in the form of a
packaging insert.
I) Use of Anti-IL-17RE Antibodies to Antagonize IL-17RE Binding to IL-17C
[265] Alternative techniques for generating or selecting antibodies useful
herein include in
vitro exposure of lymphocytes to soluble IL-17RE receptor polypeptides or
fragments thereof, such
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
67
as antigenic epitopes, and selection of antibody display libraries in phage or
similar vectors (for
instance, through use of immobilized or labeled soluble IL-17RE receptor
polypeptides or fragments
thereof, such as antigenic epitopes). Genes encoding polypeptides having
potential binding domains
such as soluble IL-17RE receptor polypeptides or fragments thereof, such as
antigenic epitopes can
be obtained by screening random peptide libraries displayed on phage (phage
display) or on bacteria,
such as E. coli. Nucleotide sequences encoding the polypeptides can be
obtained in a number of
ways, such as through random mutagenesis and random polynucleotide synthesis.
These random
peptide display libraries can be used to screen for peptides that interact
with a known target that can
be a protein or polypeptide, such as a ligand or receptor, a biological or
synthetic macromolecule, or
organic or inorganic substances. Techniques for creating and screening such
random peptide
display libraries are known in the art (Ladner et al., US Patent NO.
5,223,409; Ladner et al., US
Patent NO. 4,946,778; Ladner et al., US Patent NO. 5,403,484 and Ladner et
al., US Patent NO.
5,571,698) and random peptide display libraries and kits for screening such
libraries are available
commercially, for instance from Clontech (Palo Alto, CA), Invitrogen Inc. (San
Diego, CA), New
England Biolabs, Inc. (Beverly, MA) and Pharmacia LKB Biotechnology Inc.
(Piscataway, NJ).
Random peptide display libraries can be screened using the soluble IL-17RE
receptor polypeptides
or fragments thereof, such as antigenic epitope polypeptide sequences
disclosed herein to identify
proteins which bind to IL-17RE-comprising receptor polypeptides. These
"binding polypeptides,"
which interact with soluble IL-17RE-comprising receptor polypeptides, can be
used for tagging cells;
for isolating homolog polypeptides by affinity purification; they can be
directly or indirectly
conjugated to drugs, toxins, radionuclides and the like. These binding
polypeptides can also be used
in analytical methods such as for screening expression libraries and
neutralizing activity, e.g., for
binding, blocking, inhibiting, reducing, antagonizing or neutralizing
interaction between IL-17C and
IL-17RE, or viral binding to a receptor. The binding polypeptides can also be
used for diagnostic
assays for determining circulating levels of soluble IL-17RE-comprising
receptor polypeptides; for
detecting or quantitating soluble or non-soluble IL-17RE-comprising receptors
as marker of
underlying pathology or disease. These binding polypeptides can also act as
"antagonists" to block
or inhibit soluble or membrane-bound IL-17RE monomeric receptor or IL-17RE
homodimeric,
heterodimeric or multimeric polypeptide binding (e.g. to ligand) and signal
transduction in vitro and
in vivo. Again, these binding polypeptides serve as anti-IL-17RE monomeric
receptor or anti-IL-
17RE homodimeric, heterodimeric or multimeric polypeptides and are useful for
inhibiting IL-17C
activity, as well as receptor activity or protein-binding. Antibodies raised
to the natural receptor
complexes of the present invention, and IL-17RE-epitope-binding antibodies,
and anti-IL-17RE
neutralizing monoclonal antibodies may be preferred embodiments, as they may
act more
specifically against the IL-17RE and can inhibit IL-17C. Moreover, the
antagonistic and binding
activity of the antibodies of the present invention can be assayed in an IL-
17C proliferation, signal
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
68
trap, luciferase or binding assays in the presence of IL-17C, and IL-17RE-
comprising soluble
receptors, and other biological or biochemical assays described herein.
[266] Antibodies to soluble IL-17RE receptor polypeptides (e.g., antibodies to
SEQ ID
NO: 2, 5, 8, 11, 14, 21, 23, 107, 109, 113, 115, 117, 119, or 122) or
fragments thereof, such as
antigenic epitopes may be used for inhibiting the inflammatory effects of IL-
17C in vivo, for
theraputic use against inflammation and inflammatory dieases such as
psoriasis, psoriatic arthritis,
rheumatoid arthritis, endotoxemia, inflammatory bowel disease (IBD), IBS,
colitis, asthma, allograft
rejection, immune mediated renal diseases, hepatobiliary diseases, multiple
sclerosis, atherosclerosis,
promotion of tumor growth, or degenerative joint disease and other
inflammatory conditions
disclosed herein; tagging cells that express IL-17RE receptors; for isolating
soluble IL-17RE-
comprising receptor polypeptides by affinity purification; for diagnostic
assays for determining
circulating levels of soluble IL-17RE-comprising receptor polypeptides; for
detecting or quantitating
soluble IL-17RE-comprising receptors as marker of underlying pathology or
disease; in analytical
methods employing FACS; for screening expression libraries; for generating
anti-idiotypic
antibodies that can act as IL-17C agonists; and as neutralizing antibodies or
as antagonists to bind,
block, inhibit, reduce, or antagonize IL-17RE receptor function, or to bind,
block, inhibit, reduce,
antagonize or neutralize IL-17C activity in vitro and in vivo. Suitable direct
tags or labels include
radionuclides, enzymes, substrates, cofactors, biotin, inhibitors, fluorescent
markers,
chemiluminescent markers, magnetic particles and the like; indirect tags or
labels may feature use of
biotin-avidin or other complement/anti-complement pairs as intermediates.
Antibodies herein may
also be directly or indirectly conjugated to drugs, toxins, radionuclides and
the like, and these
conjugates used for in vivo diagnostic or therapeutic applications. Moreover,
antibodies to soluble
IL-17RE-comprising receptor polypeptides, or fragments thereof may be used in
vitro to detect
denatured or non-denatured IL-17RE-comprising receptor polypeptides or
fragments thereof in
assays, for example, Western Blots or other assays known in the art.
[267] Antibodies to soluble IL-17RE receptor or soluble IL-17RE homodimeric,
heterodimeric or multimeric receptor polypeptides are useful for tagging cells
that express the
corresponding receptors and assaying their expression levels, for affinity
purification, within
diagnostic assays for determining circulating levels of receptor polypeptides,
analytical methods
employing fluorescence-activated cell sorting. Moreover, divalent antibodies,
and anti-idiotypic
antibodies may be used as agonists to mimic the effect of the IL-17RE ligand,
IL-17C.
[268] Antibodies herein can also be directly or indirectly conjugated to
drugs, toxins,
radionuclides and the like, and these conjugates used for in vivo diagnostic
or therapeutic
applications. For instance, antibodies or binding polypeptides which recognize
soluble IL-17RE
receptor or soluble IL-17RE homodimeric, heterodimeric or multimeric receptor
polypeptides can be
used to identify or treat tissues or organs that express a corresponding anti-
complementary molecule
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
69
(i.e., a IL-17RE-comprising soluble or membrane-bound receptor). More
specifically, antibodies to
soluble IL-17RE-comprising receptor polypeptides, or bioactive fragments or
portions thereof, can
be coupled to detectable or cytotoxic molecules and delivered to a mammal
having cells, tissues or
organs that express the IL-17RE-comprising receptor such as IL-17RE-expressing
cancers.
[269] Suitable detectable molecules may be directly or indirectly attached to
polypeptides
that bind IL-17RE-comprising receptor polypeptides, such as "binding
polypeptides," (including
binding peptides disclosed above), antibodies, or bioactive fragments or
portions thereof. Suitable
detectable molecules include radionuclides, enzymes, substrates, cofactors,
inhibitors, fluorescent
markers, chemiluminescent markers, magnetic particles and the like. Suitable
cytotoxic molecules
may be directly or indirectly attached to the polypeptide or antibody, and
include bacterial or plant
toxins (for instance, diphtheria toxin, Pseudomonas exotoxin, ricin, abrin and
the like), as well as
therapeutic radionuclides, such as iodine-131, rhenium- 188 or yttrium-90
(either directly attached to
the polypeptide or antibody, or indirectly attached through means of a
chelating moiety, for
instance). Binding polypeptides or antibodies may also be conjugated to
cytotoxic drugs, such as
adriamycin. For indirect attachment of a detectable or cytotoxic molecule, the
detectable or
cytotoxic molecule can be conjugated with a member of a complementary/
anticomplementary pair,
where the other member is bound to the binding polypeptide or antibody
portion. For these
purposes, biotin/streptavidin is an exemplary complementary/ anticomplementary
pair.
[270] In another embodiment, binding polypeptide-toxin fusion proteins or
antibody-toxin
fusion proteins can be used for targeted cell or tissue inhibition or ablation
(for instance, to treat
cancer cells or tissues). Alternatively, if the binding polypeptide has
multiple functional domains
(i.e., an activation domain or a ligand binding domain, plus a targeting
domain), a fusion protein
including only the targeting domain may be suitable for directing a detectable
molecule, a cytotoxic
molecule or a complementary molecule to a cell or tissue type of interest. In
instances where the
fusion protein including only a single domain includes a complementary
molecule, the anti-
complementary molecule can be conjugated to a detectable or cytotoxic
molecule. Such domain-
complementary molecule fusion proteins thus represent a generic targeting
vehicle for celUtissue-
specific delivery of generic anti-complementary-detectable/ cytotoxic molecule
conjugates.
[271] In another embodiment, IL-17RE binding polypeptide-cytokine or antibody-
cytokine fusion proteins can be used for enhancing in vivo killing of target
tissues (for example,
spleen, pancreatic, blood, lymphoid, colon, and bone marrow cancers), if the
binding polypeptide-
cytokine or anti- IL-17RE receptor antibody targets the hyperproliferative
cell (See, generally,
Hornick et al., Blood 89:4437-47, 1997). The described fusion proteins enable
targeting of a
cytokine to a desired site of action, thereby providing an elevated local
concentration of cytokine.
Suitable anti-IL-17RE monomer, homodimer, heterodimer or multimer antibodies
target an
undesirable cell or tissue (i.e., a tumor or a leukemia), and the fused
cytokine mediates improved
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
target cell lysis by effector cells. Suitable cytokines for this purpose
include interleukin 2 and
granulocyte-macrophage colony-stimulating factor (GM-CSF), for instance.
[272] Alternatively, IL-17RE receptor binding polypeptides or antibody fusion
proteins
described herein can be used for enhancing in vivo killing of target tissues
by directly stimulating a
IL-17RE receptor-modulated apoptotic pathway, resulting in cell death of
hyperproliferative cells
expressing IL-17RE-comprising receptors.
J) Therapeutic Uses of Polypeptides Having IL-17RE Activity or Antibodies to
IL-17RE
[273] Amino acid sequences having soluble IL-17RE activity can be used to
modulate the
immune system by binding IL-17RE ligands IL-17C, and thus, preventing the
binding of IL-17RE
ligand with endogenous IL-17RE receptor. IL-17RE antagonists, such as soluble
IL-17RE or anti-
IL-17RE antibodies, can also be used to modulate the immune system by
inhibiting the binding of
IL-17RE ligand with the endogenous IL-17RE receptor. Accordingly, the present
invention includes
the use of proteins, polypeptides, and peptides having IL-17RE activity (such
as soluble IL-17RE
polypeptides, IL-17RE polypeptide fragments, IL-17RE analogs (e.g., anti-IL-
17RE anti-idiotype
antibodies), and IL-17RE fusion proteins) to a subject which lacks an adequate
amount of this
polypeptide, or which produces an excess of IL-17RE ligand. IL-17RE
antagonists (e.g., anti-IL-
17RE antibodies) can be also used to treat a subject which produces an excess
of either IL-17RE
ligand or IL-17RE. Suitable subjects include mammals, such as humans. For
example, such IL-
17RE polypeptides and anti-IL-17RE antibodies are useful in binding, blocking,
inhibiting, reducing,
antagonizing or neutralizing IL-17C, in the treatment of inflammation and
inflammatory dieases such
as psoriasis, psoriatic arthritis, rheumatoid arthritis, endotoxemia,
inflammatory bowel disease (IBD),
IBS, colitis, asthma, allograft rejection, immune mediated renal diseases,
hepatobiliary diseases,
multiple sclerosis, atherosclerosis, promotion of tumor growth, or
degenerative joint disease and
other inflammatory conditions disclosed herein.
[274] Within preferred embodiments, the soluble receptor form of IL-17RE, (SEQ
ID
NOs:3, 6, 9, 12, 15, 21, 23, 109, 113, 115, 117, 119, or 122) is a monomer,
homodimer, heterodimer,
or multimer that binds to, blocks, inhibits, reduces, antagonizes or
neutralizes IL-17C in vivo.
Antibodies and binding polypeptides to such IL-17RE monomer, homodimer,
heterodimer, or
multimers also serve as antagonists of IL-17RE activity, and as IL-17C as
described herein.
[275] Thus, particular embodiments of the present invention are directed
toward use of
soluble IL-17RE and anti-IL-17RE antibodies as antagonists in inflammatory and
immune diseases
or conditions such as psoriasis, psoriatic arthritis, atopic dermatitis,
inflammatory skin conditions,
rheumatoid arthritis, inflammatory bowel disease (IBD), IBS, Crohn's Disease,
diverticulosis,
asthma, pancreatitis, type I diabetes (IDDM), pancreatic cancer, pancreatitis,
Graves Disease, colon
and intestinal cancer, autoimmune disease, sepsis, organ or bone marrow
transplant; inflammation
due to endotoxemia, trauma, sugery or infection; amyloidosis; splenomegaly;
graft versus host
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
71
disease; and where inhibition of inflammation, immune suppression, reduction
of proliferation of
hematopoietic, immune, inflammatory or lymphoid cells, macrophages, T-cells
(including Thl and
Th2 cells), suppression of immune response to a pathogen or antigen, or other
instances where
inhibition of IL-17C or another IL- 17 family member or cytokine is desired.
[276] Moreover, antibodies or binding polypeptides such as soluble receptors
that bind IL-
17RE polypeptides described herein, and IL-17RE polypeptides themselves are
useful to:
[277] Block, inhibit, reduce, antagonize or neutralize signaling via either IL-
17C or the IL-
17C receptor (e.g. IL-17RE) in the treatment of acute inflammation,
inflammation as a result of
trauma, tissue injury, surgery, sepsis or infection, and chronic inflammatory
diseases such as asthma,
inflammatory bowel disease (IBD), IBS, chronic colitis, splenomegaly,
rheumatoid arthritis,
recurrent acute inflammatory episodes (e.g., tuberculosis), and treatment of
amyloidosis, and
atherosclerosis, Castleman's Disease, asthma, and other diseases associated
with the induction of
acute-phase response.
[278] Block, inhibit, reduce, antagonize or neutralize signaling via either IL-
17C or the IL-
17C receptor (e.g. IL-17RE) in the treatment of autoimmune diseases such as
IDDM, multiple
sclerosis (MS), systemic Lupus erythematosus (SLE), myasthenia gravis,
rheumatoid arthritis, IBS
and IBD to prevent or inhibit signaling in immune cells (e.g. lymphocytes,
monocytes, leukocytes).
Alternatively antibodies, such as monoclonal antibodies (MAb) to IL-17RE-
comprising receptors,
can also be used as an antagonist to deplete unwanted immune cells to treat
autoimmune disease.
Asthma, allergy and other atopic disease may be treated with a MAb of the
present invention against,
for example, the IL-17RE binding domain (as described in any of SEQ ID NOs:
115, 117 or 119) to
inhibit the immune response or to deplete offending cells. Blocking,
inhibiting, reducing, or
antagonizing signaling via lL-17RE, using the soluble receptors, polypeptides
and antibodies of the
present invention, may also benefit diseases of the pancreas, kidney,
pituitary and neuronal cells.
IDDM, NIDDM, pancreatitis, and pancreatic carcinoma may benefit.
[279] IL-17RE may serve as a target for MAb therapy of cancer where an
antagonizing
MAb inhibits cancer growth and targets immune-mediated killing. (Holliger P,
and Hoogenboom, H:
Nature Biotech. 16: 1015-1016, 1998). MAbs to soluble IL-17RE may also be
useful to treat
nephropathies such as glomerulosclerosis, membranous neuropathy, amyloidosis
(which also affects
the kidney among other tissues), renal arteriosclerosis, glomerulonephritis of
various origins,
fibroproliferative diseases of the kidney, as well as kidney dysfunction
associated with SLE, IDDM,
type II diabetes (NIDDM), renal tumors and other diseases.
[280] 3) Agonize, enhance, increase or initiate signaling via the IL-17C
receptor (e.g. IL-
17RE) in the treatment of autoimmune diseases such as IDDM, MS, SLE,
myasthenia gravis,
rheumatoid arthritis, IBS and IBD. Anti-IL-17RE neutralizing and monoclonal
antibodies may
signal lymphocytes or other immune cells to differentiate, alter
proliferation, or change production of
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
72
cytokines or cell surface proteins that ameliorate autoimmunity. Specifically,
modulation of a T-
helper cell response to an alternate pattern of cytokine secretion may deviate
an autoimmune
response to ameliorate disease (Smith JA et al., J. Immunol. 160:4841-4849,
1998). Similarly,
agonistic anti-soluble IL-17RE monomers, homodimers, heterodimers and multimer
monoclonal
antibodies may be used to signal, deplete and deviate immune cells involved in
asthma, allergy and
atopoic disease. Signaling via lL-17RE may also benefit diseases of the
pancreas, kidney, pituitary
and neuronal cells. IDDM, NIDDM, pancreatitis, and pancreatic carcinoma may
benefit. IL-17RE
may serve as a target for MAb therapy of pancreatic cancer where a signaling
MAb inhibits cancer
growth and targets immune-mediated killing (Tutt, AL et al., J Immunol. 161:
3175-3185, 1998).
Similarly renal cell carcinoma may be treated with monoclonal antibodies to IL-
17RE-comprising
soluble receptors of the present invention.
[281] Soluble IL-17RE polypeptides described herein can be used to bind,
block, inhibit,
reduce, antagonize or neutralize IL-17C activity, in the treatment of
autoimmune disease, atopic
disease, NIDDM, pancreatitis and kidney dysfunction as described above. A
soluble form of IL-
17RE, such as IL-17REs2 (SEQ ID NO:113) may be used to promote an antibody
response mediated
by Th cells and/or to promote the production of IL-4 or other cytokines by
lymphocytes or other
immune cells.
[282] The soluble IL-17RE-comprising receptors of the present invention are
useful as
antagonists of IL-17C. Such antagonistic effects can be achieved by direct
neutralization or binding
of IL-17C. In addition to antagonistic uses, the soluble receptors of the
present invention can bind
IL-17C and act as carrier proteins for IL-17C cytokine, in order to transport
the ligand to different
tissues, organs, and cells within the body. As such, the soluble receptors of
the present invention can
be fused or coupled to molecules, polypeptides or chemical moieties that
direct the soluble-receptor-
ligand complex to a specific site, such as a tissue, specific immune cell, or
tumor. For example, in
acute infection or some cancers, benefit may result from induction of
inflammation and local acute
phase response proteins by the action of IL-17C. Thus, the soluble receptors
of the present invention
can be used to specifically direct the action of IL-17C. See, Cosman, D. C
okine 5: 95-106, 1993;
and Fernandez-Botran, R. Exp. Opin. Invest. Drugs 9:497-513, 2000.
[283] Moreover, the soluble receptors of the present invention can be used to
stabilize IL-
17C, to increase the bioavailability, therapeutic longevity, and/or efficacy
of IL-17C by stabilizing it
from degradation or clearance, or by targeting the ligand to a site of action
within the body. For
example the naturally occurring IL-6/soluble IL-6R complex stabilizes IL-6 and
can signal through
the gp130 receptor. See, Cosman, D. supra., and Fernandez-Botran, R. supra..
Moreover, IL-17RE
may be combined with a cognate ligand such as IL-17C to comprise a
ligand/soluble receptor
complex. Such complexes may be used to stimulate responses from cells
presenting a companion
receptor subunit. The cell specificity of IL-17RE/ligand complexes may differ
from that seen for the
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
73
ligand administered alone. Furthermore the complexes may have distinct
pharmacokinetic properties
such as affecting half-life, dose/response and organ or tissue specificity. IL-
17RE/IL-17C complexes
thus may have agonist activity to enhance an immune response or stimulate
mesangial cells or to
stimulate hepatic cells. Alternatively only tissues expressing a signaling
subunit the heterodimerizes
with the complex may be affected analogous to the response to IL6/IL6R
complexes (Hirota H. et al.,
Proc. Nat'l. Acad. Sci. 92:4862-4866, 1995; Hirano, T. in Thomason, A. (Ed.)
"The Cytokine
Handbook", 3d Ed., p. 208-209). Soluble receptor/cytokine complexes for IL-12
and CNTF display
similar activities.
[284] Moreover, inflammation is a protective response by an organism to fend
off an
invading agent. Inflammation is a cascading event that involves many cellular
and humoral
mediators. On one hand, suppression of inflammatory responses can leave a host
immunocompromised; however, if left unchecked, inflammation can lead to
serious complications
including chronic inflammatory diseases (e.g., psoriasis, arthritis,
rheumatoid arthritis, multiple
sclerosis, inflammatory bowel disease and the like), septic shock and multiple
organ failure.
Importantly, these diverse disease states share common inflammatory mediators.
The collective
diseases that are characterized by inflammation have a large impact on human
morbidity and
mortality. Therefore it is clear that anti-inflammatory proteins, such as IL-
17RE, and anti-IL-17RE
antibodies, could have crucial therapeutic potential for a vast number of
human and animal diseases,
from asthma and allergy to autoimmunity and septic shock.
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
74
Arthritis
[285] Arthritis, including osteoarthritis, rheumatoid arthritis, arthritic
joints as a result of
injury, and the like, are common inflammatory conditions which would benefit
from the therapeutic
use of anti-inflammatory proteins, such as IL-17RE soluble polypeptides and
MAbs of the present
invention. For example, rheumatoid arthritis (RA) is a systemic disease that
affects the entire body
and is one of the most common forms of arthritis. It is characterized by the
inflammation of the
membrane lining the joint, which causes pain, stiffness, warmth, redness and
swelling. Inflammatory
cells release enzymes that may digest bone and cartilage. As a result of
rheumatoid arthritis, the
inflamed joint lining, the synovium, can invade and damage bone and cartilage
leading to joint
deterioration and severe pain amongst other physiologic effects. The involved
joint can lose its
shape and alignment, resulting in pain and loss of movement.
[286] Rheumatoid arthritis (RA) is an immune-mediated disease particularly
characterized
by inflammation and subsequent tissue damage leading to severe disability and
increased mortality.
A variety of cytokines are produced locally in the rheumatoid joints. Numerous
studies have
demonstrated that IL-1 and TNF-alpha, two prototypic pro-inflammatory
cytokines, play an
important role in the mechanisms involved in synovial inflammation and in
progressive joint
destruction. Indeed, the administration of TNF-alpha and IL-1 inhibitors in
patients with RA has led
to a dramatic improvement of clinical and biological signs of inflammation and
a reduction of
radiological signs of bone erosion and cartilage destruction. However, despite
these encouraging
results, a significant percentage of patients do not respond to these agents,
suggesting that other
mediators are also involved in the pathophysiology of arthritis (Gabay,
Expert. Opin. Biol. Ther.
2 2:135-149, 2002). One of those mediators could be IL-17C, and as such a
molecule that binds or
inhibits IL-17C activity, such as soluble IL-17RE, IL-17RE polypeptides, or
anti-IL-17RE antibodies
or binding partners, could serve as a valuable therapeutic to reduce
inflammation in rheumatoid
arthritis, and other arthritic diseases.
[287] There are several animal models for rheumatoid arthritis known in the
art. For
example, in the collagen-induced arthritis (CIA) model, mice develop chronic
inflammatory arthritis
that closely resembles human rheumatoid arthritis. Since CIA shares similar
immunological and
pathological features with RA, this makes it an ideal model for screening
potential human anti-
inflammatory compounds. The CIA model is a well-known model in mice that
depends on both an
immune response, and an inflammatory response, in order to occur. The immune
response comprises
the interaction of B-cells and CD4+ T-cells in response to collagen, which is
given as antigen, and
leads to the production of anti-collagen antibodies. The inflammatory phase is
the result of tissue
responses from mediators of inflammation, as a consequence of some of these
antibodies cross-
reacting to the mouse's native collagen and activating the complement cascade.
An advantage in
using the CIA model is that the basic mechanisms of pathogenesis are known.
The relevant T-cell
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
and B-cell epitopes on type II collagen have been identified, and various
immunological (e.g.,
delayed-type hypersensitivity and anti-collagen antibody) and inflammatory
(e.g., cytokines,
chemokines, and matrix-degrading enzymes) parameters relating to immune-
mediated arthritis have
been determined, and can thus be used to assess test compound efficacy in the
CIA model (Wooley,
Curr. Opin. Rheum. 3:407-20, 1999; Williams et al., Immunol. 89:9784-788,
1992; Myers et al., Life
Sci. 61:1861-78, 1997; and Wang et al., Immunol. 92:8955-959, 1995).
[288] The administration of soluble IL-17RE comprising polypeptides (IL-17RE),
such as
IL-17RE-Fc4 or other IL-17RE soluble and fusion proteins to these CIA model
mice is used to
evaluate the use of soluble IL-17RE as an antagonist to IL-17C used to
ameliorate symptoms and
alter the course of disease. Moreover, results showing inhibition of IL-17C by
a soluble IL-17RE
polypeptide or anti-IL-17RE antibody of the present invention would provide
proof of concept that
other IL-17C antagonists, such as soluble IL-17RE or neutralizing antibodies
thereto, can also be
used to ameliorate symptoms and alter the course of disease. Furthermore, the
systemic or local
administration of soluble IL-17RE comprising polypeptides, such as IL-17RE-Fc4
or other IL-17C
soluble receptors (e.g., IL-17RE; SEQ ID NO:3, 6, 9, 12, 15, 21, 23 109, 113,
115, 117, 119, or 122)
and anti-IL-17RE antibodies, and fusion proteins can potentially suppress the
inflammatory response
in RA. By way of example and without limitation, the injection of 10 - 100 ug
soluble IL-17RE-Fc
per mouse (one to seven times a week for up to but not limited to 4 weeks via
s.c., i.p., or i.m route
of administration) can significantly reduce the disease score (paw score,
incident of inflammation, or
disease). Depending on the initiation of IL-17RE-Fc administration (e.g. prior
to or at the time of
collagen immunization, or at any time point following the second collagen
immunization, including
those time points at which the disease has already progressed), IL-17RE can be
efficacious in
preventing rheumatoid arthritis, as well as preventing its progression. Other
potential therapeutics
include IL-17RE polypeptides, anti-IL-17RE antibodies, or anti IL-17C
antibodies or binding
partners, and the like.
2. Endotoxemia
[289] Endotoxemia is a severe condition commonly resulting from infectious
agents such
as bacteria and other infectious disease agents, sepsis, toxic shock syndrome,
or in
immunocompromised patients subjected to opportunistic infections, and the
like. Therapeutically
useful of anti-inflammatory proteins, such as IL-17RE polypeptides and
antibodies of the present
invention, could aid in preventing and treating endotoxemia in humans and
animals. IL-17RE
polypeptides, or anti-IL-17RE antibodies or binding partners, could serve as a
valuable therapeutic to
reduce inflammation and pathological effects in endotoxemia.
[290] Lipopolysaccharide (LPS) induced endotoxemia engages many of the
proinflammatory mediators that produce pathological effects in the infectious
diseases and LPS
induced endotoxemia in rodents is a widely used and acceptable model for
studying the
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
76
pharmacological effects of potential pro-inflammatory or immunomodulating
agents. LPS, produced
in gram-negative bacteria, is a major causative agent in the pathogenesis of
septic shock (Glausner et
al., Lancet 338:732, 1991). A shock-like state can indeed be induced
experimentally by a single
injection of LPS into animals. Molecules produced by cells responding to LPS
can target pathogens
directly or indirectly. Although these biological responses protect the host
against invading
pathogens, they may also cause harm. Thus, massive stimulation of innate
immunity, occurring as a
result of severe Gram-negative bacterial infection, leads to excess production
of cytokines and other
molecules, and the development of a fatal syndrome, septic shock syndrome,
which is characterized
by fever, hypotension, disseminated intravascular coagulation, and multiple
organ failure (Dumitru et
al. Cell 103:1071-1083, 2000).
[291] These toxic effects of LPS are mostly related to macrophage activation
leading to the
release of multiple inflammatory mediators. Among these mediators, TNF appears
to play a crucial
role, as indicated by the prevention of LPS toxicity by the administration of
neutralizing anti-TNF
antibodies (Beutler et al., Science 229:869, 1985). It is well established
that lug injection of E. coli
LPS into a C57B1/6 mouse will result in significant increases in circulating
IL-6, TNF-alpha, IL-1,
and acute phase proteins (for example, SAA) approximately 2 hours post
injection. The toxicity of
LPS appears to be mediated by these cytokines as passive immunization against
these mediators can
result in decreased mortality (Beutler et al., Science 229:869, 1985). The
potential
immunointervention strategies for the prevention and/or treatment of septic
shock include anti-TNF
mAb, IL-1 receptor antagonist, LIF, IL-10, and G-CSF.
[292] The administration of soluble IL-17RE comprising polypeptides, such as
IL-17RE-
Fc5, IL-17RE-Fc10 or other IL-17RE soluble and fusion proteins to these LPS-
induced model may
be used to to evaluate the use of IL-17RE to ameliorate symptoms and alter the
course of LPS-
induced disease. Moreover, results showing inhibition of IL-17C by IL-17RE
provide proof of
concept that other IL-17C antagonists, such as soluble IL-17RE or antibodies
thereto, can also be
used to ameliorate symptoms in the LPS-induced model and alter the course of
disease. The model
will show induction of IL-17C by LPS injection and the potential treatment of
disease by IL-17RE
polypeptides. Since LPS induces the production of pro-inflammatory factors
possibly contributing to
the pathology of endotoxemia, the neutralization of IL-17C activity or other
pro- inflammatory
factors by an antagonist IL-17RE polyepeptide can be used to reduce the
symptoms of endotoxemia,
such as seen in endotoxic shock. Other potential therapeutics include IL-17RE
polypeptides, anti-
IL-17RE antibodies, or binding partners, and the like.
3. Inflammatory Bowel Disease IBD
[293] In the United States approximately 500,000 people suffer from
Inflammatory Bowel
Disease (IBD) which can affect either colon and rectum (Ulcerative colitis) or
both, small and large
intestine (Crohn's Disease). The pathogenesis of these diseases is unclear,
but they involve chronic
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
77
inflammation of the affected tissues. IL-17RE polypeptides, anti-IL-17RE
antibodies, or binding
partners, could serve as a valuable therapeutic to reduce inflammation and
pathological effects in
IBD and related diseases.
[294] Ulcerative colitis (UC) is an inflammatory disease of the large
intestine, commonly
called the colon, characterized by inflammation and ulceration of the mucosa
or innermost lining of
the colon. This inflammation causes the colon to empty frequently, resulting
in diarrhea. Symptoms
include loosening of the stool and associated abdominal cramping, fever and
weight loss. Although
the exact cause of UC is unknown, recent research suggests that the body's
natural defenses are
operating against proteins in the body which the body thinks are foreign (an
"autoimmune reaction").
Perhaps because they resemble bacterial proteins in the gut, these proteins
may either instigate or
stimulate the inflammatory process that begins to destroy the lining of the
colon. As the lining of the
colon is destroyed, ulcers form releasing mucus, pus and blood. The disease
usually begins in the
rectal area and may eventually extend through the entire large bowel. Repeated
episodes of
inflammation lead to thickening of the wall of the intestine and rectum with
scar tissue. Death of
colon tissue or sepsis may occur with severe disease. The symptoms of
ulcerative colitis vary in
severity and their onset may be gradual or sudden. Attacks may be provoked by
many factors,
including respiratory infections or stress.
[295] Although there is currently no cure for UC available, treatments are
focused on
suppressing the abnormal inflammatory process in the colon lining. Treatments
including
corticosteroids immunosuppressives (eg. azathioprine, mercaptopurine, and
methotrexate) and
aminosalicytates are available to treat the disease. However, the long-term
use of
immunosuppressives such as corticosteroids and azathioprine can result in
serious side effects
including thinning of bones, cataracts, infection, and liver and bone marrow
effects. In the patients in
whom current therapies are not successful, surgery is an option. The surgery
involves the removal of
the entire colon and the rectum.
[296] There are several animal models that can partially mimic chronic
ulcerative colitis.
Some of the most widely used models are the oxazolone and the 2,4,6-
trinitrobenesulfonic
acid/ethanol (TNBS) induced colitis models, which induce chronic inflammation
and ulceration in
the colon. When oxazolone or TNBS is introduced into the colon of susceptible
mice via intra-rectal
instillation, it induces T-cell mediated immune response in the colonic
mucosa, in this case leading to
a massive mucosal inflammation characterized by the dense infiltration of T-
cells and macrophages
throughout the entire wall of the large bowel. Moreover, this histopathologic
picture is accompanies
by the clinical picture of progressive weight loss (wasting), bloody diarrhea,
rectal prolapse, and
large bowel wall thickening (Neurath et al. Intern. Rev. Immunol. 19:51-62,
2000).
[297] Another colitis model uses dextran sulfate sodium (DSS), which induces
an acute
colitis manifested by bloody diarrhea, weight loss, shortening of the colon
and mucosal ulceration
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
78
with neutrophil infiltration. DSS-induced colitis is characterized
histologically by infiltration of
inflammatory cells into the lamina propria, with lymphoid hyperplasia, focal
crypt damage, and
epithelial ulceration. These changes are thought to develop due to a toxic
effect of DSS on the
epithelium and by phagocytosis of lamina propria cells and production of TNF-
alpha and IFN-
gamma. Despite its common use, several issues regarding the mechanisms of DSS
about the
relevance to the human disease remain unresolved. DSS is regarded as a T cell-
independent model
because it is observed in T cell-deficient animals such as SCID mice.
[298] The administration of soluble IL-17RE or other IL-17RE soluble and
fusion proteins
to these TNBS or DSS models can be used to evaluate the use of soluble IL-17RE
to ameliorate
symptoms and alter the course of gastrointestinal disease. Moreover, the
results showing inhibition
of IL-17C by IL-17RE provide proof of concept that other IL-17C antagonists,
such as soluble IL-
17RE or antibodies thereto, can also be used to ameliorate symptoms in the
colitis/IBD models and
alter the course of disease.
4. Psoriasis
[299] Psoriasis is a chronic skin condition that affects more than seven
million Americans.
Psoriasis occurs when new skin cells grow abnormally, resulting in inflamed,
swollen, and scaly
patches of skin where the old skin has not shed quickly enough. Plaque
psoriasis, the most common
form, is characterized by inflamed patches of skin ("lesions") topped with
silvery white scales.
Psoriasis may be limited to a few plaques or involve moderate to extensive
areas of skin, appearing
most commonly on the scalp, knees, elbows and trunk. Although it is highly
visible, psoriasis is not a
contagious disease. The pathogenesis of the diseases involves chronic
inflammation of the affected
tissues. IL-17RE polypeptides, anti-IL-17RE antibodies, or binding partners,
could serve as a
valuable therapeutic to reduce inflammation and pathological effects in
psoriasis, other inflammatory
skin diseases, skin and mucosal allergies, and related diseases.
[300] Psoriasis is a T-cell mediated inflammatory disorder of the skin that
can cause
considerable discomfort. It is a disease for which there is no cure and
affects people of all ages.
Psoriasis affects approximately two percent of the populations of European and
North America.
Although individuals with mild psoriasis can often control their disease with
topical agents, more
than one million patients worldwide require ultraviolet or systemic
immunosuppressive therapy.
Unfortunately, the inconvenience and risks of ultraviolet radiation and the
toxicities of many
therapies limit their long-term use. Moreover, patients usually have
recurrence of psoriasis, and in
some cases rebound, shortly after stopping immunosuppressive therapy.
[301] IL-17RE soluble receptor polypeptides and antibodies thereto may also be
used
within diagnostic systems for the detection of circulating levels of IL-17C
ligand, and in the
detection of IL-17C associated with acute phase inflammatory response. Within
a related
embodiment, antibodies or other agents that specifically bind to IL-17RE
soluble receptors of the
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
79
present invention can be used to detect circulating receptor polypeptides;
conversely, IL-17RE
soluble receptors themselves can be used to detect circulating or locally-
acting IL-17C polypeptides.
Elevated or depressed levels of ligand or receptor polypeptides may be
indicative of pathological
conditions, including inflammation or cancer. Moreover, detection of acute
phase proteins or
molecules such as IL-17C can be indicative of a chronic inflammatory condition
in certain disease
states (e.g., asthma, psoriasis, rheumatoid arthritis, colitis, IBD).
Detection of such conditions serves
to aid in disease diagnosis as well as help a physician in choosing proper
therapy.
[302] In addition to other disease models described herein, the activity of
soluble IL-17RE
and/or anti-IL-17RE antibodies on inflammatory tissue derived from human
psoriatic lesions can be
measured in vivo using a severe combined immune deficient (SCID) mouse model.
Several mouse
models have been developed in which human cells are implanted into
immunodeficient mice
(collectively referred to as xenograft models); see, for example, Cattan AR,
Douglas E, Leuk. Res.
18:513-22, 1994 and Flavell, DJ, Hematological Oncology 14:67-82, 1996. As an
in vivo xenograft
model for psoriasis, human psoriatic skin tissue is implanted into the SCID
mouse model, and
challenged with an appropriate antagonist. Moreover, other psoriasis animal
models in ther art may
be used to evaluate IL-17C antagonists, such as human psoriatic skin grafts
implanted into AGR129
mouse model, and challenged with an appropriate antagonist (e.g., see, Boyman,
O. et al., J. Exp.
Med. Online publication #20031482, 2004, incorporated hereing by reference).
Soluble IL-17RE or
anti-IL-17RE antibodies that bind, block, inhibit, reduce, antagonize or
neutralize the activity of IL-
17C are preferred antagonists, however, anti-IL-17C, soluble IL-17RE, as well
as other IL-17C
antagonists can be used in this model. Similarly, tissues or cells derived
from human colitis, IBD,
arthritis, or other inflammatory lestions can be used in the SCID model to
assess the anti-
inflammatory properties of the IL-17C antagonists described herein.
[303] Therapies designed to abolish, retard, or reduce inflammation using
soluble IL-
17RE, anti-IL-17RE antibodies or its derivatives, agonists, conjugates or
variants can be tested by
administration of anti-IL-17RE antibodies or soluble IL-17RE compounds to SCID
mice bearing
human inflammatory tissue (e.g., psoriatic lesions and the like), or other
models described herein.
Efficacy of treatment is measured and statistically evaluated as increased
anti-inflammatory effect
within the treated population over time using methods well known in the art.
Some exemplary
methods include, but are not limited to measuring for example, in a psoriasis
model, epidermal
thickness, the number of inflammatory cells in the upper dermis, and the
grades of parakeratosis.
Such methods are known in the art and described herein. For example, see
Zeigler, M. et al. Lab
Invest 81:1253, 2001; Zollner, T. M. et al. J. Clin. Invest. 109:671, 2002;
Yamanaka, N. et al.
Microbio.l Immunol. 45:507, 2001; Raychaudhuri, S. P. et al. Br. J. Dermatol.
144:931, 2001;
Boehncke, W. H et al. Arch. Dermatol. Res. 291:104, 1999; Boehncke, W. H et
al.. J. Invest.
Dermatol. 116:596, 2001; Nickoloff, B. J. et al. Am. J. Pathol. 146:580, 1995;
Boehncke, W. H et al.
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
J. Cutan. Pathol. 24:1, 1997; Sugai, J., M. et al. J. Dermatol. Sci. 17:85,
1998; and Villadsen L.S. et
al. J. Clin. Invest. 112:1571, 2003. Inflammation may also be monitored over
time using well-
known methods such as flow cytometry (or PCR) to quantitate the number of
inflammatory or
lesional cells present in a sample, score (weight loss, diarrhea, rectal
bleeding, colon length) for IBD,
paw disease score and inflammation score for CIA RA model. For example,
therapeutic strategies
appropriate for testing in such a model include direct treatment using soluble
IL-17RE, anti-IL-17RE
antibodies, other IL-17C antagonists or related conjugates or antagonists
based on the disrupting
interaction of soluble IL-17RE with its ligand IL-17C, or for cell-based
therapies utilizing soluble
IL-17RE or anti-IL-17RE antibodies or its derivatives, agonists, conjugates or
variants.
[304] Moreover, psoriasis is a chronic inflammatory skin disease that is
associated with
hyperplastic epidermal keratinocytes and infiltrating mononuclear cells,
including CD4+ memory T
cells, neutrophils and macrophages (Christophers, Int. Arch. Allergy Immunol.,
110:199, 1996). It is
currently believed that environmental antigens play a significant role in
initiating and contributing to
the pathology of the disease. However, it is the loss of tolerance to self-
antigens that is thought to
mediate the pathology of psoriasis. Dendritic cells and CD4+ T cells are
thought to play an important
role in antigen presentation and recognition that mediate the immune response
leading to the
pathology. We have recently developed a model of psoriasis based on the
CD4+CD45RB transfer
model (Davenport et al., Internat. Immunopharmacol., 2:653-672). Soluble IL-
17RE or anti-IL-
17RE antibodies of the present invention are administered to the mice.
Inhibition of disease scores
(skin lesions, inflammatory cytokines) indicates the effectiveness of IL-17C
antagonists in psoriasis,
e.g., anti-IL-17RE antibodies or IL-17RE soluble receptors.
5. Atopic Dermatitis.
[305] AD is a common chronic inflammatory disease that is characterized by
hyperactivated cytokines of the helper T cell subset 2 (Th2). Although the
exact etiology of AD is
unknown, multiple factors have been implicated, including hyperactive Th2
immune responses,
autoimmunity, infection, allergens, and genetic predisposition. Key features
of the disease include
xerosis (dryness of the skin), pruritus (itchiness of the skin),
conjunctivitis, inflammatory skin
lesions, Staphylococcus aureus infection, elevated blood eosinophilia,
elevation of serum IgE and
IgGl, and chronic dermatitis with T cell, mast cell, macrophage and eosinophil
infiltration.
Colonization or infection with S. aureus has been recognized to exacerbate AD
and perpetuate
chronicity of this skin disease.
[306] AD is often found in patients with asthma and allergic rhinitis, and is
frequently the
initial manifestation of allergic disease. About 20% of the population in
Western countries suffer
from these allergic diseases, and the incidence of AD in developed countries
is rising for unknown
reasons. AD typically begins in childhood and can often persist through
adolescence into adulthood.
Current treatments for AD include topical corticosteroids, oral cyclosporin A,
non-corticosteroid
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
81
immunosuppressants such as tacrolimus (FK506 in ointment form), and interferon-
gamma. Despite
the variety of treatments for AD, many patients' symptoms do not improve, or
they have adverse
reactions to medications, requiring the search for other, more effective
therapeutic agents. The
soluble IL-17RE polypeptides and anti-IL-17RE antibodies of the present
invention, including the
neutralizing anti-human IL-17RE antibodies of the present invention, can be
used to neutralize IL-
17C in the treatment of specific human diseases such as atoptic dermatitis,
inflammatory skin
conditions, and other inflammatory conditions disclosed herein.
6. Irritable Bowel Syndrome ("IBS")
[307] Irritable bowel syndrome represents a disease characterized by abdominal
pain or
discomfort and an erratic bowel habit. IBS patients can be characterized into
three main groups
based on bowel habits: those with predominantly loose or frequent stools,
those with predominantly
hard or infrequent stools, and those with variable or normal stools (Talley et
al., 2002). Altered
intestinal motility, abnormalities in epithelial function, abnormal transit of
stool and gas, and stress,
may contribute to symptoms, while visceral hypersensitivity is a key feature
in most patients.
Genetic factors affecting pain-signaling and disturbances in central
processing of afferent signals are
postulated to predispose individuals to IBS following specific environmental
exposures. Studies have
also demonstrated that inflammatory responses in the colon may contribute to
increased sensitivity of
smooth muscle and enteric nerves and therefore perturb sensory-motor functions
in the intestine
(Collins et al., 2001). There is clinical overlap between IBS and IBD, with
IBS-like symptoms
frequently reported in patients before the diagnosis of IBD, and a higher than
expected IBS
symptoms in patients in remission from established IBD. Thus, these conditions
may coexist with a
higher than expected frequency, or may exist on a continuum, with IBS and IBD
at different ends of
the same spectrum. However, it should be noted that in most IBS patients,
colonic biopsy specimens
appear normal. Nevertheless, IBS significantly affects a very large number of
individuals (U.S.
prevalence in 2000, approximately 16 million individuals), resulting in a
total cost burden of 1.7
billion dollars (year 2000). Thus, among the most prevalent and costly
gastrointestinal diseases and
disorders, IBS is second only to gastroesophageal reflux disease (GERD). Yet
unlike GERD,
treatment for IBS remains unsatisfactory (Talley et al., 2002; Farhadi et al.,
21001; Collins et al.,
2001), demonstrating that IBS clearly represents an unmet medical need.
[308] Converging disease models have been proposed that postulate an enhanced
responsiveness of neural, immune or neuroimmune circuits in the central
nervous system (CNS) or in
the gut to central (psychosocial) or peripheral (tissue irritation,
inflammation, infection)
perturbations of normal homeostasis (Talley et al., 2002). This enhanced
responsiveness results in
dysregulation of gut motility, epithelial function (immune, permeability), and
visceral
hypersensitivity, which in turn results in IBS symptoms.
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
82
[309] There may be a role for a number of different molecules in the
pathogenesis of IBS
including a role for molecules that stimulate neurons and those that are
involved in initiation of
inflammatory process. A number of molecules are known to be linked to possible
activity on
neurons due to their direct expression by neurons or expression of their
receptors on neurons,
including IL-17D, IL-17B and IL-3 1. Moreover, a number of IL- 17 family
members and related
molecules have been associated with inflammation in the gut, including IL-17A,
IL-17C, IL-17F, IL-
23 and IL-31.
[310] Efficacy of inhibitors of these molecules could be tested in vivo in
animal models of
disease. Several animal models have been proposed that mimic key features of
IBS and involve
centrally targeted stimuli (stress) or peripherally targeted stimuli
(infection, inflammation). Two
examples of in vivo animal models that can be used to determine the
effectiveness of inhibitors in the
treatment of IBS are (i) models focusing on primary CNS-directed pathogeneisis
of IBS (stress
models), and (ii) models focusing on gut-directed inducers of stress (i.e. gut
inflammation, infection
or physical stress). It should be noted however, that events within the CNS or
in the gastrointestinal
(GI) tract do not occur in isolation and that symptoms of IBS most likely
result from a complex
interaction between signals from the CNS on the GI and vice versa.
K) Pharmaceutical Use of IL-17RE
[311] For pharmaceutical use, the soluble IL-17RE or anti-IL-17RE antibodies
of the
present invention are formulated for parenteral, particularly intravenous or
subcutaneous, delivery
according to conventional methods. Intravenous administration will be by bolus
injection, controlled
release, e.g, using mini-pumps or other appropriate technology, or by infusion
over a typical period
of one to several hours. In general, pharmaceutical formulations will include
a hematopoietic protein
in combination with a pharmaceutically acceptable vehicle, such as saline,
buffered saline, 5%
dextrose in water or the like. Formulations may further include one or more
excipients,
preservatives, solubilizers, buffering agents, albumin to provent protein loss
on vial surfaces, etc.
When utilizing such a combination therapy, the cytokines may be combined in a
single formulation
or may be administered in separate formulations. Methods of formulation are
well known in the art
and are disclosed, for example, in Remington's Pharmaceutical Sciences,
Gennaro, ed., Mack
Publishing Co., Easton PA, 1990, which is incorporated herein by reference.
Therapeutic doses will
generally be in the range of 0.1 to 100 mg/kg of patient weight per day,
preferably 0.5-20 mg/kg per
day, with the exact dose determined by the clinician according to accepted
standards, taking into
account the nature and severity of the condition to be treated, patient
traits, etc. Determination of
dose is within the level of ordinary skill in the art. The proteins will
commonly be administered over
a period of up to 28 days following chemotherapy or bone-marrow transplant or
until a platelet count
of >20,000/mm3, preferably >50,000/mm3, is achieved. More commonly, the
proteins will be
administered over one week or less, often over a period of one to three days.
In general, a
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
83
therapeutically effective amount of soluble IL-17RE or anti-IL-17RE antibodies
of the present
invention is an amount sufficient to produce a clinically significant increase
in the proliferation
and/or differentiation of lymphoid or myeloid progenitor cells, which will be
manifested as an
increase in circulating levels of mature cells (e.g. platelets or
neutrophils). Treatment of platelet
disorders will thus be continued until a platelet count of at least
20,000/mm3, preferably
50,000/mm3, is reached. The soluble IL-17RE or anti-IL-17RE antibodies of the
present invention
can also be administered in combination with other cytokines such as IL-3, -6
and -11; stem cell
factor; erythropoietin; G-CSF and GM-CSF. Within regimens of combination
therapy, daily doses
of other cytokines will in general be: EPO, 150 U/kg; GM-CSF, 5-15 lg/kg; IL-
3, 1-5 lg/kg; and G-
CSF, 1-25 lg/kg. Combination therapy with EPO, for example, is indicated in
anemic patients with
low EPO levels.
[312] Generally, the dosage of administered soluble IL-17RE (or IL-17RE analog
or
fusion protein) or anti-IL-17RE antibodies will vary depending upon such
factors as the patient's age,
weight, height, sex, general medical condition and previous medical history.
Typically, it is
desirable to provide the recipient with a dosage of soluble IL-17RE or anti-IL-
17RE antibodies
which is in the range of from about 1 pg/kg to 10 mg/kg (amount of agent/body
weight of patient),
although a lower or higher dosage also may be administered as circumstances
dictate.
[313] Administration of soluble IL-17RE or anti-IL-17RE antibodies to a
subject can be
intravenous, intraarterial, intraperitoneal, intramuscular, subcutaneous,
intrapleural, intrathecal, by
perfusion through a regional catheter, or by direct intralesional injection.
When administering
therapeutic proteins by injection, the administration may be by continuous
infusion or by single or
multiple boluses.
[314] Additional routes of administration include oral, mucosal-membrane,
pulmonary,
and transcutaneous. Oral delivery is suitable for polyester microspheres, zein
microspheres,
proteinoid microspheres, polycyanoacrylate microspheres, and lipid-based
systems (see, for example,
DiBase and Morrel, "Oral Delivery of Microencapsulated Proteins," in Protein
Delivery: Physical
Systems, Sanders and Hendren (eds.), pages 255-288 (Plenum Press 1997)). The
feasibility of an
intranasal delivery is exemplified by such a mode of insulin administration
(see, for example,
Hinchcliffe and Illum, Adv. Drug Deliv. Rev. 35:199 (1999)). Dry or liquid
particles comprising
soluble IL-17RE or anti-IL-17RE antibodies can be prepared and inhaled with
the aid of dry-powder
dispersers, liquid aerosol generators, or nebulizers (e.g., Pettit and
Gombotz, TIBTECH 16:343
(1998); Patton et al., Adv. Drug Deliv. Rev. 35:235 (1999)). This approach is
illustrated by the
AERX diabetes management system, which is a hand-held electronic inhaler that
delivers aerosolized
insulin into the lungs. Studies have shown that proteins as large as 48,000
kDa have been delivered
across skin at therapeutic concentrations with the aid of low-frequency
ultrasound, which illustrates
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
84
the feasibility of trascutaneous administration (Mitragotri et al., Science
269:850 (1995)).
Transdermal delivery using electroporation provides another means to
administer a molecule having
IL-17RE binding activity (Potts et al., Pharm. Biotechnol. 10:213 (1997)).
[315] A pharmaceutical composition comprising a soluble IL-17RE or anti-IL-
17RE
antibody can be formulated according to known methods to prepare
pharmaceutically useful
compositions, whereby the therapeutic proteins are combined in a mixture with
a pharmaceutically
acceptable carrier. A composition is said to be a "pharmaceutically acceptable
carrier" if its
administration can be tolerated by a recipient patient. Sterile phosphate-
buffered saline is one
example of a pharmaceutically acceptable carrier. Other suitable carriers are
well-known to those in
the art. See, for example, Gennaro (ed.), Remington's Pharmaceutical Sciences,
19th Edition (Mack
Publishing Company 1995).
[316] For purposes of therapy, soluble IL-17RE or anti-IL-17RE antibody
molecules and a
pharmaceutically acceptable carrier are administered to a patient in a
therapeutically effective
amount. A combination of a therapeutic molecule of the present invention and a
pharmaceutically
acceptable carrier is said to be administered in a "therapeutically effective
amount" if the amount
administered is physiologically significant. An agent is physiologically
significant if its presence
results in a detectable change in the physiology of a recipient patient. For
example, an agent used to
treat inflammation is physiologically significant if its presence alleviates
the inflammatory response.
[317] A pharmaceutical composition comprising IL-17RE (or IL-17RE analog or
fusion
protein) or neutralizing anti-IL-17RE antibody can be furnished in liquid
form, in an aerosol, or in
solid form. Liquid forms, are illustrated by injectable solutions and oral
suspensions. Exemplary
solid forms include capsules, tablets, and controlled-release forms. The
latter form is illustrated by
miniosmotic pumps and implants (Bremer et al., Pharm. Biotechnol. 10:239
(1997); Ranade,
"Implants in Drug Delivery," in Drug Delivery Systems, Ranade and Hollinger
(eds.), pages 95-123
(CRC Press 1995); Bremer et al., "Protein Delivery with Infusion Pumps," in
Protein Delivery:
Physical Systems, Sanders and Hendren (eds.), pages 239-254 (Plenum Press
1997); Yewey et al.,
"Delivery of Proteins from a Controlled Release Injectable Implant," in
Protein Delivery: Physical
Systems, Sanders and Hendren (eds.), pages 93-117 (Plenum Press 1997)).
[318] Polypeptides and antibodies can be encapsulated within liposomes using
standard
techniques of protein microencapsulation (see, for example, Anderson et al.,
Infect. Immun. 31:1099
(1981), Anderson et al., Cancer Res. 50:1853 (1990), and Cohen et al.,
Biochim. Biophys. Acta
1063:95 (1991), Alving et al. "Preparation and Use of Liposomes in
Immunological Studies," in
Liposome Technology, 2nd Edition, Vol. III, Gregoriadis (ed.), page 317 (CRC
Press 1993), Wassef
et al., Meth. Enzymol. 149:124 (1987)). As noted above, therapeutically useful
liposomes may
contain a variety of components. For example, liposomes may comprise lipid
derivatives of
poly(ethylene glycol) (Allen et al., Biochim. Biophys. Acta 1150:9 (1993)).
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
[319] The present invention also contemplates chemically modified polypeptides
having
binding IL-17RE activity such as IL-17RE monomeric, homodimeric, heterodimeric
or multimeric
soluble receptors, and IL-17RE antagonists, for example anti-IL-17RE
antibodies or binding
polypeptides, or neutralizing anti-IL-17RE antibodies, which a polypeptide is
linked with a polymer,
as discussed above.
[320] As an illustration, pharmaceutical compositions may be supplied as a kit
comprising
a container that comprises a polypeptide with a IL-17RE extracellular domain,
e.g., IL-17RE
monomeric, homodimeric, heterodimeric or multimeric soluble receptors, or a IL-
17RE antagonist
(e.g., an antibody or antibody fragment that binds a IL-17RE polypeptide, or
neutralizing anti-IL-
17RE antibody). Therapeutic polypeptides can be provided in the form of an
injectable solution for
single or multiple doses, or as a sterile powder that will be reconstituted
before injection.
Alternatively, such a kit can include a dry-powder disperser, liquid aerosol
generator, or nebulizer
for administration of a therapeutic polypeptide. Such a kit may further
comprise written information
on indications and usage of the pharmaceutical composition. Moreover, such
information may
include a statement that the IL-17RE composition is contraindicated in
patients with known
hypersensitivity to IL-17RE.
[321] A pharmaceutical composition comprising anti-IL-17RE antibodies or
binding
partners (or anti-IL-17RE antibody fragments, antibody fusions, humanized
antibodies and the like),
or IL-17RE soluble receptor, can be furnished in liquid form, in an aerosol,
or in solid form. Liquid
forms, are illustrated by injectable solutions, aerosols, droplets,
topological solutions and oral
suspensions. Exemplary solid forms include capsules, tablets, and controlled-
release forms. The
latter form is illustrated by miniosmotic pumps and implants (Bremer et al.,
Pharm. Biotechnol.
10:239 (1997); Ranade, "Implants in Drug Delivery," in Drug Delivery Systems,
Ranade and
Hollinger (eds.), pages 95-123 (CRC Press 1995); Bremer et al., "Protein
Delivery with Infusion
Pumps," in Protein Delivery: Physical Systems, Sanders and Hendren (eds.),
pages 239-254 (Plenum
Press 1997); Yewey et al., "Delivery of Proteins from a Controlled Release
Injectable Implant," in
Protein Delivery: Physical Systems, Sanders and Hendren (eds.), pages 93-117
(Plenum Press
1997)). Other solid forms include creams, pastes, other topological
applications, and the like.
[322] Liposomes provide one means to deliver therapeutic polypeptides to a
subject
intravenously, intraperitoneally, intrathecally, intramuscularly,
subcutaneously, or via oral
administration, inhalation, or intranasal administration. Liposomes are
microscopic vesicles that
consist of one or more lipid bilayers surrounding aqueous compartments (see,
generally, Bakker-
Woudenberg et al., Eur. J. Clin. Microbiol. Infect. Dis. 12 (Suppl. 1):S61
(1993), Kim, Drugs 46:618
(1993), and Ranade, "Site-Specific Drug Delivery Using Liposomes as Carriers,"
in Drug Delivery
Systems, Ranade and Hollinger (eds.), pages 3-24 (CRC Press 1995)). Liposomes
are similar in
composition to cellular membranes and as a result, liposomes can be
administered safely and are
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
86
biodegradable. Depending on the method of preparation, liposomes may be
unilamellar or
multilamellar, and liposomes can vary in size with diameters ranging from 0.02
m to greater than
m. A variety of agents can be encapsulated in liposomes: hydrophobic agents
partition in the
bilayers and hydrophilic agents partition within the inner aqueous space(s)
(see, for example, Machy
et al., Liposomes In Cell Biology And Pharmacology (John Libbey 1987), and
Ostro et al., American
J. Hosp. Pharm. 46:1576 (1989)). Moreover, it is possible to control the
therapeutic availability of
the encapsulated agent by varying liposome size, the number of bilayers, lipid
composition, as well
as the charge and surface characteristics of the liposomes.
[323] Liposomes can adsorb to virtually any type of cell and then slowly
release the
encapsulated agent. Alternatively, an absorbed liposome may be endocytosed by
cells that are
phagocytic. Endocytosis is followed by intralysosomal degradation of liposomal
lipids and release
of the encapsulated agents (Scherphof et al., Ann. N.Y. Acad. Sci. 446:368
(1985)). After
intravenous administration, small liposomes (0.1 to 1.0 m) are typically
taken up by cells of the
reticuloendothelial system, located principally in the liver and spleen,
whereas liposomes larger than
3.0 m are deposited in the lung. This preferential uptake of smaller
liposomes by the cells of the
reticuloendothelial system has been used to deliver chemotherapeutic agents to
macrophages and to
tumors of the liver.
[324] The reticuloendothelial system can be circumvented by several methods
including
saturation with large doses of liposome particles, or selective macrophage
inactivation by
pharmacological means (Claassen et al., Biochim. Biophys. Acta 802:428
(1984)). In addition,
incorporation of glycolipid- or polyethelene glycol-derivatized phospholipids
into liposome
membranes has been shown to result in a significantly reduced uptake by the
reticuloendothelial
system (Allen et al., Biochim. Biophys. Acta 1068:133 (1991); Allen et al.,
Biochim. Biophys. Acta
1150:9 (1993)).
[325] Liposomes can also be prepared to target particular cells or organs by
varying
phospholipid composition or by inserting receptors or ligands into the
liposomes. For example,
liposomes, prepared with a high content of a nonionic surfactant, have been
used to target the liver
(Hayakawa et al., Japanese Patent 04-244,018; Kato et al., Biol. Pharm. Bull.
16:960 (1993)). These
formulations were prepared by mixing soybean phospatidylcholine, a-tocopherol,
and ethoxylated
hydrogenated castor oil (HCO-60) in methanol, concentrating the mixture under
vacuum, and then
reconstituting the mixture with water. A liposomal formulation of
dipalmitoylphosphatidylcholine
(DPPC) with a soybean-derived sterylglucoside mixture (SG) and cholesterol
(Ch) has also been
shown to target the liver (Shimizu et al., Biol. Pharm. Bull. 20:881 (1997)).
[326] Alternatively, various targeting ligands can be bound to the surface of
the liposome,
such as antibodies, antibody fragments, carbohydrates, vitamins, and transport
proteins. For
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
87
example, liposomes can be modified with branched type galactosyllipid
derivatives to target
asialoglycoprotein (galactose) receptors, which are exclusively expressed on
the surface of liver cells
(Kato and Sugiyama, Crit. Rev. Ther. Drug Carrier Syst. 14:287 (1997);
Murahashi et al., Biol.
Pharm. Bull. 20:259 (1997)). Similarly, Wu et al., Hepatology 27:772 (1998),
have shown that
labeling liposomes with asialofetuin led to a shortened liposome plasma half-
life and greatly
enhanced uptake of asialofetuin-labeled liposome by hepatocytes. On the other
hand, hepatic
accumulation of liposomes comprising branched type galactosyllipid derivatives
can be inhibited by
preinjection of asialofetuin (Murahashi et al., Biol. Pharm. Bull. 20:259
(1997)). Polyaconitylated
human serum albumin liposomes provide another approach for targeting liposomes
to liver cells
(Kamps et al., Proc. Nat'l Acad. Sci. USA 94:11681 (1997)). Moreover, Geho, et
al. U.S. Patent No.
4,603,044, describe a hepatocyte-directed liposome vesicle delivery system,
which has specificity for
hepatobiliary receptors associated with the specialized metabolic cells of the
liver.
[327] In a more general approach to tissue targeting, target cells are
prelabeled with
biotinylated antibodies specific for a ligand expressed by the target cell
(Harasym et al., Adv. Drug
Deliv. Rev. 32:99 (1998)). After plasma elimination of free antibody,
streptavidin-conjugated
liposomes are administered. In another approach, targeting antibodies are
directly attached to
liposomes (Harasym et al., Adv. Drug Deliv. Rev. 32:99 (1998)).
[328] Anti-IL-17RE neutralizing antibodies and binding partners with IL-17C
binding
activity, or IL-17RE soluble receptor, can be encapsulated within liposomes
using standard
techniques of protein microencapsulation (see, for example, Anderson et al.,
Infect. Immun. 31:1099
(1981), Anderson et al., Cancer Res. 50:1853 (1990), and Cohen et al.,
Biochim. Biophys. Acta
1063:95 (1991), Alving et al. "Preparation and Use of Liposomes in
Immunological Studies," in
Liposome Technology, 2nd Edition, Vol. III, Gregoriadis (ed.), page 317 (CRC
Press 1993), Wassef
et al., Meth. Enzymol. 149:124 (1987)). As noted above, therapeutically useful
liposomes may
contain a variety of components. For example, liposomes may comprise lipid
derivatives of
poly(ethylene glycol) (Allen et al., Biochim. Biophvs. Acta 1150:9 (1993)).
[329] Degradable polymer microspheres have been designed to maintain high
systemic
levels of therapeutic proteins. Microspheres are prepared from degradable
polymers such as
poly(lactide-co-glycolide) (PLG), polyanhydrides, poly (ortho esters),
nonbiodegradable ethylvinyl
acetate polymers, in which proteins are entrapped in the polymer (Gombotz and
Pettit, Bioconjugate
Chem. 6:332 (1995); Ranade, "Role of Polymers in Drug Delivery," in Drug
Delivery Systems,
Ranade and Hollinger (eds.), pages 51-93 (CRC Press 1995); Roskos and
Maskiewicz, "Degradable
Controlled Release Systems Useful for Protein Delivery," in Protein Delivery:
Physical Systems,
Sanders and Hendren (eds.), pages 45-92 (Plenum Press 1997); Bartus et al.,
Science 281:1161
(1998); Putney and Burke, Nature Biotechnology 16:153 (1998); Putney, Curr.
Opin. Chem. Biol.
2:548 (1998)). Polyethylene glycol (PEG)-coated nanospheres can also provide
carriers for
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
88
intravenous administration of therapeutic proteins (see, for example, Gref et
al., Pharm. Biotechnol.
10:167 (1997)).
[330] Other dosage forms can be devised by those skilled in the art, as shown,
for
example, by Ansel and Popovich, Pharmaceutical Dosage Forms and Drug Delivery
Systems, 5th
Edition (Lea & Febiger 1990), Gennaro (ed.), Remington's Pharmaceutical
Sciences, 19th Edition
(Mack Publishing Company 1995), and by Ranade and Hollinger, Drug Delivery
Systems (CRC
Press 1996).
[331] The present invention also contemplates antisense RNA modulators
targeting IL-
17RE, more preferably targeting the extracellylar binding domain of IL-17RE,
and most preferably
targeting a ligand binding domain of IL-17RE. Preparation and delivery of
antisense RNA
modulators is known in the art. Antisense modulators of IL-17RE or its ligands
or precursor
molecules can be prepared and delivered as described by Crooke, Antisense Drug
Technology:
Principles, Strategies, and Applications, 2"d Edition (CRC 2007). In brief,
short oligonucleotide
molecules are constructed having sequence complementarity to an mRNA encoding
IL-17RE or one
of its ligands or precursors thereof. The molecules are typically from about 8
consecutive
nucleobases in length to about 50 consecutive nucleobases in length, though 20
nucleobases is more
typical. Preferably, but not necessarily the antisense molecules are modified
to include one or more
phosphorothioate linkages to retard degredation of the antisense compound, one
or more modified
sugar residues or a combination thereof. Sugar residue modifications include,
but are not limited to,
2'-methoxyethoxy (2'-O--CH2CH2OCH3, also known as 2'-O-(2-
methoxyethyl) or 2'-
MOE) (Martin et al., Helv. Chim. Acta, 1995, 78, 486-504). See, e.g., United
States Publication No.:
20030220282. Similar to antisense RNA modulators are siRNA modulators, which
are double
stranded; one strand being antisense to the target nucleic acid and the other
strand sense to the target
nucleic acid. Preparation and delivery of siRNA modulators is also well know
in the art. See e.g.,
United States Publication No.: 20050176667. As is used herein, both the
classical antisense RNA
modulator and the siRNA modulator are referred to as antisense RNA modulators.
[332] The present invention also contemplates chemically modified anti-IL-17RE
antibody
or binding partner, for example anti-IL-17RE antibodies or IL-17RE soluble
receptor, linked with a
polymer, as discussed above.
[333] The present invention contemplates compositions of anti-IL-17C
antibodies, and
methods and therapeutic uses comprising an antibody, peptide or polypeptide
described herein. Such
compositions can further comprise a carrier. The carrier can be a conventional
organic or inorganic
carrier. Examples of carriers include water, buffer solution, alcohol,
propylene glycol, macrogol,
sesame oil, corn oil, and the like.
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
89
L) Production of Transgenic Mice
[334] Transgenic mice can be engineered to over-express either the IL-17C or
the IL-17RE
gene in all tissues or under the control of a tissue-specific or tissue-
preferred regulatory element.
These over-producers can be used to characterize the phenotype that results
from over-expression,
and the transgenic animals can serve as models for human disease caused by
excess IL-17C or IL-
17RE. Transgenic mice that over-express any of these also provide model
bioreactors for production
of IL-17RE, such as soluble IL-17RE, in the milk or blood of larger animals.
Methods for producing
transgenic mice are well-known to those of skill in the art (see, for example,
Jacob, "Expression and
Knockout of Interferons in Transgenic Mice," in Overexpression and Knockout of
Cytokines in
Transgenic Mice, Jacob (ed.), pages 111-124 (Academic Press, Ltd. 1994),
Monastersky and Robl
(eds.), Strategies in Transgenic Animal Science (ASM Press 1995), and Abbud
and Nilson,
"Recombinant Protein Expression in Transgenic Mice," in Gene Expression
Systems: Using Nature
for the Art of Expression, Fernandez and Hoeffler (eds.), pages 367-397
(Academic Press, Inc.
1999)).
[335] For example, a method for producing a transgenic mouse that expresses a
IL-17RE
gene can begin with adult, fertile males (studs) (B6C3f1, 2-8 months of age
(Taconic Farms,
Germantown, NY)), vasectomized males (duds) (B6D2f1, 2-8 months, (Taconic
Farms)),
prepubescent fertile females (donors) (B6C3f1, 4-5 weeks, (Taconic Farms)) and
adult fertile females
(recipients) (B6D2f1, 2-4 months, (Taconic Farms)). The donors are acclimated
for one week and
then injected with approximately 8 IU/mouse of Pregnant Mare's Serum
gonadotrophin (Sigma
Chemical Company; St. Louis, MO) I.P., and 46-47 hours later, 8 IU/mouse of
human Chorionic
Gonadotropin (hCG (Sigma)) I.P. to induce superovulation. Donors are mated
with studs subsequent
to hormone injections. Ovulation generally occurs within 13 hours of hCG
injection. Copulation is
confirmed by the presence of a vaginal plug the morning following mating.
[336] Fertilized eggs are collected under a surgical scope. The oviducts are
collected and
eggs are released into urinanalysis slides containing hyaluronidase (Sigma).
Eggs are washed once in
hyaluronidase, and twice in Whitten's W640 medium (described, for example, by
Menino and
O'Claray, Biol. Reprod. 77:159 (1986), and Dienhart and Downs, Zygote 4:129
(1996)) that has been
incubated with 5% COz, 5% Oz, and 90% N 2 at 37 C. The eggs are then stored in
a 37 C/5% CO 2
incubator until microinjection.
[337] Ten to twenty micrograms of plasmid DNA containing a IL-17RE encoding
sequence is linearized, gel-purified, and resuspended in 10 mM Tris-HC1 (pH
7.4), 0.25 mM EDTA
(pH 8.0), at a final concentration of 5-10 nanograms per microliter for
microinjection. For example,
the IL-17RE encoding sequences can encode a polypeptide comprising any of SEQ
ID NOs:3, 6, 9,
12, 15, 21, 23, 109, 113, 115, 117, 119, or 122.
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
[338] Plasmid DNA is microinjected into harvested eggs contained in a drop of
W640
medium overlaid by warm, COZ-equilibrated mineral oil. The DNA is drawn into
an injection needle
(pulled from a 0.75mm ID, Imm OD borosilicate glass capillary), and injected
into individual eggs.
Each egg is penetrated with the injection needle, into one or both of the
haploid pronuclei.
[339] Picoliters of DNA are injected into the pronuclei, and the injection
needle withdrawn
without coming into contact with the nucleoli. The procedure is repeated until
all the eggs are
injected. Successfully microinjected eggs are transferred into an organ tissue-
culture dish with pre-
gassed W640 medium for storage overnight in a 37 C/5% CO 2 incubator.
[340] The following day, two-cell embryos are transferred into pseudopregnant
recipients.
The recipients are identified by the presence of copulation plugs, after
copulating with vasectomized
duds. Recipients are anesthetized and shaved on the dorsal left side and
transferred to a surgical
microscope. A small incision is made in the skin and through the muscle wall
in the middle of the
abdominal area outlined by the ribcage, the saddle, and the hind leg, midway
between knee and
spleen. The reproductive organs are exteriorized onto a small surgical drape.
The fat pad is stretched
out over the surgical drape, and a baby serrefine (Roboz, Rockville, MD) is
attached to the fat pad
and left hanging over the back of the mouse, preventing the organs from
sliding back in.
[341] With a fine transfer pipette containing mineral oil followed by
alternating W640 and
air bubbles, 12-17 healthy two-cell embryos from the previous day's injection
are transferred into the
recipient. The swollen ampulla is located and holding the oviduct between the
ampulla and the bursa,
a nick in the oviduct is made with a 28 g needle close to the bursa, making
sure not to tear the
ampulla or the bursa.
[342] The pipette is transferred into the nick in the oviduct, and the embryos
are blown in,
allowing the first air bubble to escape the pipette. The fat pad is gently
pushed into the peritoneum,
and the reproductive organs allowed to slide in. The peritoneal wall is closed
with one suture and the
skin closed with a wound clip. The mice recuperate on a 37 C slide warmer for
a minimum of four
hours.
[343] The recipients are returned to cages in pairs, and allowed 19-21 days
gestation.
After birth, 19-21 days postpartum is allowed before weaning. The weanlings
are sexed and placed
into separate sex cages, and a 0.5 cm biopsy (used for genotyping) is snipped
off the tail with clean
scissors.
[344] Genomic DNA is prepared from the tail snips using, for example, a QIAGEN
DNEASY kit following the manufacturer's instructions. Genomic DNA is analyzed
by PCR using
primers designed to amplify a IL-17RE gene or a selectable marker gene that
was introduced in the
same plasmid. After animals are confirmed to be transgenic, they are back-
crossed into an inbred
strain by placing a transgenic female with a wild-type male, or a transgenic
male with one or two
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
91
wild-type female(s). As pups are born and weaned, the sexes are separated, and
their tails snipped
for genotyping.
[345] To check for expression of a transgene in a live animal, a partial
hepatectomy is
performed. A surgical prep is made of the upper abdomen directly below the
zyphoid process.
Using sterile technique, a small 1.5-2 cm incision is made below the sternum
and the left lateral lobe
of the liver exteriorized. Using 4-0 silk, a tie is made around the lower lobe
securing it outside the
body cavity. An atraumatic clamp is used to hold the tie while a second loop
of absorbable Dexon
(American Cyanamid; Wayne, N.J.) is placed proximal to the first tie. A distal
cut is made from the
Dexon tie and approximately 100 mg of the excised liver tissue is placed in a
sterile petri dish. The
excised liver section is transferred to a 14 ml polypropylene round bottom
tube and snap frozen in
liquid nitrogen and then stored on dry ice. The surgical site is closed with
suture and wound clips,
and the animal's cage placed on a 37 C heating pad for 24 hours post
operatively. The animal is
checked daily post operatively and the wound clips removed 7-10 days after
surgery. The expression
level of IL-17RE mRNA is examined for each transgenic mouse using an RNA
solution
hybridization assay or polymerase chain reaction.
[346] In addition to producing transgenic mice that over-express IL-17C or IL-
17RE, it is
useful to engineer transgenic mice with either abnormally low or no expression
of any of these
genes. Such transgenic mice provide useful models for diseases associated with
a lack of IL-17C or
IL-17RE. As discussed above, IL-17RE gene expression can be inhibited using
anti-sense genes,
ribozyme genes, or external guide sequence genes. To produce transgenic mice
that under-express
the IL-17RE gene, such inhibitory sequences are targeted to IL-17RE mRNA.
Methods for
producing transgenic mice that have abnormally low expression of a particular
gene are known to
those in the art (see, for example, Wu et al., "Gene Underexpression in
Cultured Cells and Animals
by Antisense DNA and RNA Strategies," in Methods in Gene Biotechnology, pages
205-224 (CRC
Press 1997)).
[347] An alternative approach to producing transgenic mice that have little or
no IL-17RE
gene expression is to generate mice having at least one normal IL-17RE allele
replaced by a
nonfunctional IL-17RE gene. One method of designing a nonfunctional IL-17RE
gene is to insert
another gene, such as a selectable marker gene, within a nucleic acid molecule
that encodes IL-17RE.
Standard methods for producing these so-called "knockout mice" are known to
those skilled in the
art (see, for example, Jacob, "Expression and Knockout of Interferons in
Transgenic Mice," in
Overexpression and Knockout of Cytokines in Transgenic Mice, Jacob (ed.),
pages 111-124
(Academic Press, Ltd. 1994), and Wu et al., "New Strategies for Gene
Knockout," in Methods in
Gene Biotechnology, pages 339-365 (CRC Press 1997)).
[348] Mouse myeloma cell lines 347.72.1.2 and 347.24.3.4 were deposited with
the
American Type Culture Collection (10801 University Boulevard, Manassas, VA
20110-2209) on
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
92
March 13, 2007. Clone designation number 347.72.1.2 was assigned Patent
Deposit Designation No.
PTA-8233. Clone designation number 347.24.3.4 was assigned Patent Deposit
Designation No.
PTA-8234. These deposits will be maintained under the terms of the Budapest
Treaty on the
International Recognition of the Deposit of Microorganisms for the Purposes of
Patent Procedure.
The deposit was made merely as a convenience for those of skill in the art and
is not an admission
that a deposit is required under 35 U.S.C. 112.
[349] The invention provides an isolated antibody or antibody fragmet that
binds a
polypeptide, wherein the polypeptide comprises the extracellular domain of an
IL-17RE molecule, or
fragment thereof (e.g., extracellular domain of IL-17REx2 or IL-17REx3),
wherein the polypeptide
is capable of binding the antibody produced by the hybridoma selected from a)
the hybridoma of
clone designation number 347.72.1.2 (ATCC Patent Deposit Designation PTA-
8233); or b) the
hybridoma of clone designation number 347.24.3.4 (ATCC Patent Deposit
Designation PTA-8234).
[350] The invention provides isolated antisera containing an antibody or
antibody
fragment that binds a polypeptide, wherein the polypeptide comprises the
extracellular domain of an
IL-17RE molecule, or fragment thereof (e.g., extracellular domain of IL-17REx2
or IL-17REx3), and
wherein the polypeptide is capable of binding the antibody produced by the
hybridoma selected
from: a) the hybridoma of clone designation number 347.72.1.2 (ATCC Patent
Deposit Designation
PTA-8233); or b) the hybridoma of clone designation number 347.24.3.4 (ATCC
Patent Deposit
Designation PTA-8234).
[351] The invention provides an isolated antibody produced by a hybridoma
selected from
ATCC Patent Deposit Designation PTA-8233 or ATCC Patent Deposit Designation
PTA-8234,
wherein the antibody reduces the pro-inflammatory activity of IL-17C by
neutralizing IL-17C from
binding and signally via endogenous IL-17RE.
[352] The invention provides a hybridoma of ATCC Patent Deposit Designation
PTA-
8233 and the antibody produced by the hybridoma. The invention provides a
hybridoma of ATCC
Patent Deposit Designation PTA-8234 and the antibody produced by the
hybridoma.
[353] Hybridomas expressing monoclonal antibodies to IL-17RE were produced
using
methods similar to those described above were deposited with the American Type
Tissue Culture
Collection (ATCC; Manassas VA) patent depository as original deposits under
the Budapest Treaty
and were given the following ATCC Accession Nos: clone 347.72.1.2 (ATCC Patent
Deposit
Designation PTA-8233, deposited on March 13, 2007); and clone 347.24.3.4 (ATCC
Patent Deposit
Designation PTA-8234, deposited on March 13, 2007).
[354] In some embodiments, the antibody-producing cells are the hybridomas
expressing
monoclonal antibodies to IL-17RE that were deposited with the American Type
Tissue Culture
Collection (ATCC; Manassas VA) patent depository as original deposits under
the Budapest Treaty
and were given the following ATCC Accession Nos: clone 347.72.1.2 (ATCC Patent
Deposit
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
93
Designation PTA-8233, deposited on March 13, 2007); and clone 347.24.3.4 (ATCC
Patent Deposit
Designation PTA-8234, deposited on March 13, 2007).
[355] Hybridomas expressing monoclonal antibodies to IL-17RE were deposited
with the
American Type Tissue Culture Collection (ATCC; Manassas VA) patent depository
as original
deposits under the Budapest Treaty and were given the following ATCC Accession
Nos: clone
347.72.1.2 (ATCC Patent Deposit Designation PTA-8233, deposited on March 13,
2007); and clone
347.24.3.4 (ATCC Patent Deposit Designation PTA-8234, deposited on March 13,
2007).
[356] The invention is further illustrated by the following non-limiting
examples.
EXAMPLES
EXAMPLE 1
Human IL-17RE Tissue Distribution in Tissue Panels Using PCR
[357] The Human Rapid-Scan cDNA panel represents 24 adult tissues and is
arrayed at 4
different concentrations called 1X, lOX, 100X, and 1000X (Origen, Rockville,
MD.). The "1000x
and 100x" levels were screened for IL-17RE transcription using PCR. The sense
primer was
zc39334, (5' AGGCCCTGCCACCCACCTTC 3') (SEQ ID NO:26) located in a cDNA area
corresponding to the 5' untranslated region. The antisense primer was zc39333,
(5'-
CGAGGCACCCCAAGGATTTCAG-3') (SEQ ID NO:27) located in a cDNA area corresponding
to
the 3' untranslated region. PCR was applied using pfu turbo polymerase and the
manufacturer's
recommendations (Stratagene, La Jolla, CA) except for using rediload dye,
(Research Genetics, Inc.,
Huntsville, AL) a wax hot start, (Molecular Bioproducts Inc. San Diego, CA)
and 10% (final
concentration) DMSO. The amplification was carried out as follows: 1 cycle at
94 C for 4 minutes,
40 cycles of 94 C for 30 seconds, 51 C for 30 seconds and 72 C for 3 minutes,
followed by 1 cycle
at 72 C for 7 minutes. About 10 1 of the PCR reaction product was subjected
to standard agarose
gel electrophoresis using a 1% agarose gel. Following electrophoresis, the
gels were Southern
blotted and the membranes hybridized by standard methods using a 32P isotope-
labeled
oligonucleotide, zc40458 (5'-TCTCTGACTCTGCTGGGATTGG-3') (SEQ ID NO:28) which
maps
to the cDNA area in the translated region, just downstream of the start codon.
X ray film
autoradiography revealed IL-17RE-specific amplicons only in colon, lung,
stomach, placenta, and
bone marrow.
EXAMPLE 2
Cloning of Human IL-17REx1
[358] Human IL-17REx1 (SEQ ID NO:l) was cloned by PCR using lOng of a human
hacat cell line (skin-derived) amplified plasmid cDNA library template and
primers 5'
CGAGGCACCCCAAGGATTTCAG 3'(SEQ ID NO: 179) and 5' AGGCCCTGCCACCCACCTTC
3' (SEQ ID NO: 180) and pfu ultra polymerase according to the manufacturer's
recommendations.
These primers map to the 5' and 3' utr regions of human IL-17RE cDNA. The
resulting products
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
94
were subjected to a preparative low melt agarose TAE gel electrophoresis and
the approximately
1.3-2.5 KB region size-selectively purified and then liquefied using the
gelase method. (Epicenter)
This template was then diluted 1:50 in sterile water and luL amplified using
pfu ultra polymerase by
nested PCR, using 5' CGTACGGGCCGGCCACCATGGGGAGCTCCAGACTGGCA 3' (SEQ ID
NO:181) containing a Fsel restriction site and 5'
TGACGAGGCGCGCCTCAACCTAGGTCTGC
AAGT 3'(SEQ ID NO: 182) containing an Ascl restriction site. These primers
amplify just the
translated region of human IL-17RE. The resulting products were desalted and
the primers
eliminated utilizing a chromaspin 100 column (Clontech) and then digested with
Fsel and Ascl
restriction enzymes, size-selected on a low melt agarose gel for approximately
1.3-2.5 KB fragments.
Fragments were ligated into a pZMPl l expression vector's Fsel/Ascl
restriction sites. Clone's DNA
inserts were subjected to sequencing analysis, revealing clone d2, which was
designated IL-17REx1
(SEQ ID NO:1)
EXAMPLE 3
Cloning of Human IL-17REx2, IL-17REx3 and IL-17REx4
[359] Human IL-17REx2 (SEQ ID NO:4), IL-17REx3 (SEQ ID NO:7), and IL-17REx4
(SEQ ID NO:10) were cloned by PCR using lul of a human adult skin cDNA
(clontech) template
and the following primers:
5'CGAGGCACCCCAAGGATTTCAG3' (SEQ ID NO:162 ) and
5'AGGCCCTGCCACCCACCTTC3' (SEQ ID NO: 163)
and pfu ultra polymerase according to the manufacturer's recommendations.
These primers map to the
5' and 3' utr regions of human IL-17RE cDNA. The resulting products were
subjected to a preparative
low melt agarose TAE gel electrophoresis and the approximately 1.3-2.5 KB
region size-selectively
purified and then liquefied using the gelase method. (Epicenter) This template
was then diluted 1:50 in
sterile water and luL amplified using pfu ultra polymerase by nested PCR,
using 5'CGTACGGGCCG
GCCACCATGGGGAGCTCCAGACTGGCA3' (SEQ ID NO:164) containing a Fsel restriction
site
and 5' TGACGAGGCGCGCCTCAACCTAGGTCTGCAAGT 3' (SEQ ID NO:165) containing an
Ascl restriction site. These primers amplify just the translated region of
human IL-17RE. The resulting
products were desalted and the primers eliminated utilizing a chromaspin 100
column (Clontech) and
then digested with Fsel and Ascl restriction enzymes, size-selected on a low
melt agarose gel for
approximately 1.3-2.5 KB fragments. Fragments were ligated into a pZMPll
expression vector's
Fsel/Ascl restriction sites. Clone's DNA inserts were subjected to sequencing
analysis, revealing
clones Fl, F5, and F6, which were designated IL-17REx2 (SEQ ID NO:4), IL-
17REx3 (SEQ ID
NO:7), and IL-17REx4 (SEQ ID NO: 10) respectively.
EXAMPLE 4
Cloning of Human IL-17REx6 and x13
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
[360] Briefly, cDNA obtained from human colon from a patient with active
Crohn's
disease was used as a template. One micro liter of the above template was
amplified by PCR, using
primers 39333 5' CGAGGCACCCCAAGGATTTCAG 3'(SEQ ID NO:53) and 39334, 5'
AGGCCCTGCCACCCACCTTC 3' (SEQ ID NO:54) and pfu ultra polymerase according to
the
manufacturer's recommendations. These primers map to the 5' and 3' utr regions
of human IL-17RE
cDNA. The resulting products were subjected to a preparative low melt agarose
TAE gel
electrophoresis and the -1.3-2.5 KB region size-selectively purified and then
liquefied using the
gelase method. (Epicenter) Two micro liters of the purified fragments were
amplified using pfu ultra
polymerase by nested PCR, using ZC 39429, 5'CGTACGGGCCGGCCACCATGGGGAGCTCCAG
ACTGGCA3' (SEQ ID NO:65) containing a Fsel restriction site and zc 39433, 5'
TGACGAGGCGCGCCTCAACCTAGGTCTGCAAGT 3' (SEQ ID NO:66) containing an Ascl
restriction site. These primers amplify just the translated region of human IL-
17RE. The resulting
products were then digested with Fsel and Ascl restriction enzymes, size-
selected on a low melt
agarose gel for -1.3-2.5 KB fragments and cloned in and expression vector,
pZMP11. IL-17RE
positive clones were identified using colony lifts of the resulting colonies
and hybridized to a
radiolabeled oligomer, zc 39948, 5'TTTCGCCACCTGCCCCACTGGAACACCCGCTGTCC3'
(SEQ ID NO:67) One hundred human IL-17RE positive colonies were sent for DNA
sequence
determination, revealing a variety of different IL-17RE cDNAs including human
IL-17REx6 (SEQ
ID NOs:20 and 21) and human IL-17REx13 (SEQ ID NOs:106 and 107).
EXAMPLE 5
Cloning of Murine IL-17RE
[361] A putative full-length mouse cDNA sequence for IL-17RE was identified
through
computational and bioinformatical methods, using homology to the sequence of
human IL-17RE
(SEQ ID NO:6). This sequence was used in a Blast query to identify potential
full-length mouse
clones to purchase through vendors of IMAGE consortium clones. In this manner,
clones
corresponding to IMAGE ID numbers 5319489, 4457159, 6311568, and 4482367 were
purchased
(American Type Culture Collection, Manassas, VA) and sequenced in their
entirety. Analysis of
these sequences led to the identification of two isoforms of this gene
designated murine IL-17REx5
(SEQ ID NOs: 68 and 69) and murine IL-17REx6 (SEQ ID NOs:13 and 14).
EXAMPLE 6
Cloning of Murine IL-17REx15
[362] To clone murine IL-17REx15 (SEQ ID NOs:110 and 111), total RNA was
extracted
from the colons of mice with artificially induced colitis (described below in
Example 42) This RNA
was reverse transcribed into first strand cDNA using standard methods.
Approximately 50ng cDNA
was amplified by PCR using primers 51388 5' CCTGCCCCTGCCTGCGGAGTT 3' (SEQ ID
NO:70) and 51387, 5' GTTGCTACACAGGCTGAGGCTACA 3' (SEQ ID NO:71) and pfu ultra
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
96
polymerase according to the manufacturer's recommendations. The resulting
products were
subjected to a preparative low melt agarose TAE gel electrophoresis and the -
1.3-2.5 KB region
size-selectively purified and then liquefied using the gelase method.
(Epicenter) Approximately .5uL
of the purified fragments were amplified using pfu ultra polymerase by nested
PCR, using the same
primers described above and Advantage2 2 polymerase, (Clontech) to add 5' T
overhangs, which
enabled sub-cloning them in pCR4TOPO. (Invitrogen) Amplicons were size
selected as above again
prior to sub-cloning. Positives were identified using colony lifts and
hybridized to a radiolabeled
oligomer 51602. 5' CTACCAAGGCTCAACCAATAGTCCCTGTGGTTTC 3'. (SEQ ID NO:72)
One hundred mouse IL-17RE positive colonies were sent for DNA sequence
determination,
revealing a variety of different IL-17RE cDNAs including murine IL-17REx15
(SEQ ID NOs: I10
and I11).
EXAMPLE 7
Cloning of Human IL-17C
[363] A fragment of a putative IL-17C cDNA was identified through
computational means
and the PCR primers zc18634 (5'atgaggaccgctatccacagaagc 3') (SEQ ID NO:29) and
zc18635
(5'ggacgtggatgaactcggtgtgg 3') (SEQ ID NO:30) were synthesized and used to
survey by PCR a
number of potential cloning sources for IL-17C. PCR conditions were are
follows: Takara ExTaq
polymerase and buffer (Takara, Otsu, Shiga, Japan) were used in 50u1 PCR
reactions with 5u1
marathon cDNA templates made from RNAs from salivary gland, spinal cord, MCF-7
cell line,
CaCo2 cell line, T47D cell line, Molt-4 cell line, and prostate, using a
Marathon cDNA
Amplification Kit (Clontech, Palo Alto, CA) according to the manufacturer's
instructions. Also,
each reaction contained 2.5u1 lOX PCR buffer, 2.5u1 Redi-Load, (Invitrogen,
Carlsbad, CA), 2u1
2.5mM GeneAmp dNTPs (Applied Biosystems, Foster City, CA) 0.5u1 ExTaq, 0.5u1
of 20pm/ul
zc18634 and zc18635, and water to 50 ul. Cycling conditions were: 94 C 1', 30
cycles of 94 C 20",
68 C 1', followed by one cycle of 72 C 7'.
[364] PCR products were subjected to agarose gel electrophoresis and the -
200bp
fragment was excised from the gel and purified using a Qiaquick Gel extraction
spin column
(Qiagen, Valencia, CA) according to the manufacturer's directions. This
fragment was then
sequenced to verify it as IL-17C. Standard 5' and 3' nested RACE reactions
were then performed on
DNA from an amplified in-house fetal lung library to generate overlapping PCR
fragments, the
sequence of which enabled the elucidation of the complete open reading frame
plus some 5' and 3'
untranslated sequence of IL-17C.
[365] Finally, zc21607 (5'gcacacctggcggcaccatgac3') (SEQ ID NO:31) and zc21597
(5'ctgtcctccagacacggggaatg3') (SEQ ID NO:32) were used to generate by PCR a
cDNA containing
the complete open reading frame plus some 3' untranslated region of IL-17C
from DNA of an
amplified in-house fetal lung library. PCR conditions were are follows:
Advantage 2 PCR reagents
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
97
(Clontech, Palo Alto, CA) were used in a 50u1 PCR reaction with 5u1 template,
5u1 l OX PCR buffer,
5u1 Redi-Load, (Invitrogen, Carlsbad, CA), 4u1 2.5mM GeneAmp dNTPs (Applied
Biosystems,
Foster City, CA), lul Advantage 2 polymerase mix, 5u1 GC-melt (Clontech, Palo
Alto, CA), 2.5u1
DMSO, lul of 20pm/ul zc21607 and zc21597, and water to 50u1. Cycling
conditions were: 94 C 1',
25 cycles of 94 C 20", 68 C 1'30", followed by one cycle of 72 C 5'. The PCR
product was
subjected to agarose gel electrophoresis and the -770bp fragment was excised
from the gel and
purified using a Qiaquick Gel extraction spin column (Qiagen, Valencia, CA)
according to the
manufacturer's directions.
[366] The fragment was subcloned into a TA cloning vector, PCR2.1 (Invitrogen,
Carlsbad, CA), according to the manufacturer's instructions, sequenced, and
compared to the
sequences of the overlapping RACE products and existing human public genome
sequence to
identify potential PCR errors. A correct clone was archived and used for
additional research
applications.
EXAMPLE 8
Identification and Cloning of Murine IL-17C
[367] Based on the NCBI Mus musculus mRNA accession # XM 146558 and in-house
computational gene prediction models, the cDNA for mouse IL17C was generated
by PCR of the
predicted exons from mouse genomic DNA (Clonetech Cat. # 6650-1, lot #
0050310). Exon 2 PCR
product was generated using primers 49910: 5'TCACTGTGATGAGTCTCCTGCTTCTAG3'
(SEQ
ID N0:73) and 44991: 5'GTGTCGATGCGATATCTCCATGGTGAGA3' (SEQ ID NO:74). Exon
3 PCR product was generated using primers 49912: 5'GAGATATCGCATCGACACAGATGAGA
ACC3' (SEQ ID NO:75) and 49913: 5'TCACTGTGTAGACCTGGGAAGA3' (SEQ ID NO:76).
Exon 1 and the entire cDNA was then amplified in a cross-over PCR reaction
using the PCR
products for exons 2 and 3 along with primers 49959:
5'GCCACCATGGCCACCGTCACCGTCAC
TGTGATGAGTCTCCTGCTT3' (SEQ ID NO:77). The resulting PCR product that encoded
murine
IL-17C (SEQ ID NO:19) was cloned into PCR II Blunt TOPO vector for sequence
verification.
EXAMPLE 9
Expression of IL-17C Using Adenovirus Constructs
Generation of Untagged Recombinant Adenovirus
[368] The protein coding region of human IL-17C (SEQ ID NO:16) was amplified
by
PCR using primers that added Fsel and Ascl restriction sties at the 5' and 3'
termini respectively.
PCR primers ZC21925 (5'cacacaggccggccaccatgacgctcctccccggcctcc3') (SEQ ID
NO:37) and
ZC21922 (5'cacacaggcgcgccttcacactgaacggggcagcacgc3') (SEQ ID NO:38) were used
with a
pCR2.1 ta plasmid containing the full-length murine IL-17C cDNA in a PCR
reaction as follows:
one cycle at 95 C for 5 minutes, followed by 18 cycles at 95 C for 0.5 minute,
58 C for 0.5 minute,
and 72 C for 0.5 minute, followed by 72 C for 7 minutes, followed by a 4 C
soak. The PCR
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
98
reaction product was loaded onto a 1.2% (low melt) SEAPLAQUE GTG (FMC
BioProducts;
Rockland, ME) gel in TAE buffer. The IL-17C PCR product was excised from the
gel, melted at
65 C, phenol extracted twice and then ethanol precipitated. The PCR product
was then digested with
Fsel-Ascl, phenoUchloroform extracted, ethanol precipitated, and rehydrated
(Tris/EDTA, pH 8).
[369] The IL-17C fragment was then ligated into the Fsel-Ascl sites of a
modified
pAdTrack CMV (He et al., Proc. Nat'l Acad. Sci. USA 95:2509 (1998)). This
construct also contains
the green fluorescent protein (GFP) marker gene. The CMV promoter driving GFP
expression was
replaced with the SV40 promoter and the SV40 polyadenylation signal was
replaced with the human
growth hormone polyadenylation signal. In addition, the native polylinker was
replaced with Fsel,
EcoRV, and Ascl sites. This modified form of pAdTrack CMV was named pZyTrack.
Ligation was
performed using the FAST-LINK DNA ligation and screening kit (EPICENTRE
TECHNOLOGIES;
Madison, WI). Clones containing the IL-17C cDNA were identified by standard
mini prep
procedures. In order to linearize the plasmid, approximately 5 g of the
pZyTrack IL-17C plasmid
were digested with Pmel. Approximately 1 g of the linearized plasmid was
cotransformed with
200ng of supercoiled pAdEasy (He et al., Proc. Nat'l Acad. Sci. USA 95:2509
(1998)) into BJ5183
cells. The co-transformation was performed with a BIO-RAD GENE PULSER (BIO-RAD
laboratories, Inc.; Hercules, CA) at 2.5kV, 200 ohms and 25mFa. The entire co-
transformation was
plated on four LB plates containing 25 g/ml kanamycin. The smallest colonies
were picked and
expanded in LB/kanamycin and recombinant adenovirus DNA identified by standard
DNA miniprep
procedures. Digestion of the recombinant adenovirus DNA with Fsel-Ascl
confirmed the presence
of IL-17C. The recombinant adenovirus miniprep DNA was transformed into DH10B
competent
cells and DNA prepared using a QIAGEN maxi prep kit as per kit instructions.
Transfection of 293A Cells with Recombinant DNA
[370] Approximately 5 g of recombinant adenoviral DNA were digested with PacI
enzyme for three hours at 37 C in a reaction volume of 100 1 containing 20-30U
of PacI. The
digested DNA was extracted twice with an equal volume of phenol/chloroform and
precipitated with
ethanol. The DNA pellet was resuspended in 5 1 distilled water. A T25 flask of
QBI-293A cells
(Quantum Biotechnologies, Inc.; Montreal, Quebec, Canada), inoculated the day
before and grown to
60-70% confluence, were transfected with the PacI digested DNA. The Pacl-
digested DNA was
diluted up to a total volume of 50 1 with sterile HBS (150 mM NaC1, 20 mM
HEPES). In a separate
tube, 25 1 DOTAP (1 mg/ml; Roche Molecular Biochemicals; Indianapolis, IN)
were diluted to a
total volume of I00 1 with HBS. The DNA was added to the DOTAP, mixed gently
by pipeting up
and down, and left at room temperature for 15 minutes. The medium was removed
from the 293A
cells and washed with 5 ml serum-free MEMalpha (LIFE TECHNOLOGIES, Inc;
Rockville, MD)
containing ImM sodium pyruvate (LIFE TECHNOLOGIES, Inc), 0.1 mM MEM non-
essential
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
99
amino acids (LIFE TECHNOLOGIES, Inc) and 25mM HEPES buffer (LIFE TECHNOLOGIES,
Inc). Five milliliters of serum-free MEM were added to the 293A cells and held
at 37 C. The
DNA/lipid mixture was added drop-wise to the T25 flask of 293A cells, mixed
gently and incubated
at 37 C for 4 hours. After four hours, the medium containing the DNA/lipid
mixture was aspirated
off and replaced with 5 ml complete MEM containing 5% fetal bovine serum. The
transfected cells
were monitored for green fluorescent protein (GFP) expression and formation of
foci.
[371] Seven days after transfection of 293A cells with the recombinant
adenoviral DNA,
the cells expressed the GFP protein and started to form foci. These foci are
viral "plaques" and the
crude viral lysate was collected by using a cell scraper to collect all of the
293A cells. The lysate
was transferred to a 50 ml conical tube. To release most of the virus
particles from the cells, three
freeze/thaw cycles were done in a dry ice/ethanol bath and a 37 C water bath.
Amplification of Recombinant Adenovirus (rAdV)
[372] The crude lysate was amplified ("primary amplification") to obtain a
working stock
of IL-17C rAdV lysate. Two hundred milliliters of crude rAdV lysate were added
to each of ten 10
cm plates of nearly confluent (80-90%) 293A cells, which had been set up 20
hours previously. The
plates were monitored for 48 to 72 hours for cytopathic effect under the white
light microscope and
expression of GFP under the fluorescent microscope. When all of the 293A cells
showed cytopathic
effect, this primary amplification stock lysate was collected and freeze/thaw
cycles performed as
described above.
[373] Secondary amplification of IL-17C rAdV was obtained as follows. Twenty
15 cm
tissue culture dishes of 293A cells were prepared so that the cells were 80-
90% confluent. All but 20
milliliters of 5% MEM media was removed, and each dish was inoculated with 300-
500 ml primary
amplified rAdv lysate. After 48 hours, the 293A cells were lysed from virus
production and this
lysate was collected into 250 ml polypropylene centrifuge bottles and the rAdV
purified.
AdV/cDNA Purification
[374] NP-40 detergent was added to a final concentration of 0.5% to the
bottles of crude
lysate to lyse all cells. Bottles were placed on a rotating platform for 10
minutes, agitating as fast as
possible without displacing the bottles. The debris was pelleted by
centrifugation at 20,000xg for 15
minutes. The supernatant was transferred to 250 ml polycarbonate centrifuge
bottles, and 0.5 volume
of 20% PEG8000/2.5 M NaC1 solution was added. The bottles were shaken
overnight on ice. The
bottles were centrifuged at 20,000xg for 15 minutes and supernatant discarded
into a bleach solution.
The precipitated virus/PEG appeared as a white precipitate located in two
vertical lines along the
wall of the bottle on either side of the spin mark. Using a sterile cell
scraper, the precipitate from
two bottles was resuspended in 2.5 ml PBS. The virus solution was placed in 2
ml microcentrifuge
tubes and centrifuged at 14,000xg in the microfuge for 10 minutes to remove
any additional cell
debris. The supernatant from the 2 ml microcentrifuge tubes was transferred
into a 15 ml
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
100
polypropylene snapcap tube and adjusted to a density of 1.34 g/ml with cesium
chloride (CsC1). The
volume of the virus solution was estimated and 0.55 g/ml of was CsC1 added.
The CsC1 was
dissolved and 1 ml of this solution weighed 1.34 g. The solution was
transferred polycarbonate
thick-walled centrifuge tubes 3.2 ml and spun at 80,000 rpm (348,000xg) for 3-
4 hours at 25 C in a
Beckman Optima TLX micro-ultracentrifuge with the TLA-100.4 rotor. The virus
formed a white
band. Using wide-bore pipette tips, the virus band was collected.
[375] The virus from the gradient has a large amount of CsC1 which must be
removed
before it can be used with cells. Pharmacia PD-10 columns prepacked with
SEPHADEX G-25M
(Amersham Pharmacia Biotech, Inc; Piscataway, NJ) were used to desalt the
virus preparation. The
column was equilibrated with 20 ml of PBS. The virus was loaded and allowed to
run into the
column. Five milliliters of PBS were added to the column and fractions of 8-10
drops collected. The
optical densities of 1:50 dilutions of each fraction were determined at 260 nm
on a
spectrophotometer. A clear absorbance peak was present between fractions 7-12.
These fractions
were pooled and the optical density (OD) of a 1:10 dilution determined. The
following formula was
used to convert OD into virus concentration: (OD at 260 nm)(10)(1.1 x 1012) =
virions/ml. The
OD of a 1:10 dilution of the IL-17C rAdV was 0.27 giving a virus concentration
of 2.8 X 1012
virions/ml.
[376] To store the virus, glycerol was added to the purified virus to a final
concentration of
15%, mixed gently but effectively, and stored in aliquots at -80 C.
Tissue Culture Infectious Dose at 50% CPE (TCID 50) Viral Titration Assay
[377] A protocol developed by Quantum Biotechnologies, Inc. (Montreal, Quebec,
Canada) was followed to measure recombinant virus infectivity. Briefly, two 96-
well tissue culture
plates were seeded with 1x104 293A cells per well in MEM containing 2% fetal
bovine serum for
each recombinant virus assayed. After 24 hours, 10-fold dilutions of each
virus from 1x10-2 to
1 x 10-14 were made in MEM containing 2% fetal bovine serum. One hundred
microliters of each
dilution were placed in each of 20 wells. After five days at 37 C, wells were
read either positive or
negative for cytopathic effect, and a value for "plaque forming units/ml"
(PFU) is calculated.
[378] The TCID50 formulation was produced as per Quantum Biotechnologies,
Inc.,
above. The titer is determined from a plate where virus used is diluted from
10 2 to 10 14, and read
five days after the infection. At each dilution a ratio (R) of positive wells
for cytopathic effect per
the total number of wells is determined.
[379] To calculate the titer of the undiluted virus sample, factor "F" was
first calculated, as
l+d(S-0.5), where "S" is the sum of the ratios (R), and "d" is loglO of the
dilution series (e.g., "d" is
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
101
equal to one for a ten-fold dilution series). The titer of the undiluted
sample is calculated as: 10(1+F)
= TCID50/ml. To convert TCID50/ml to pfu/ml, 0.7 is subtracted from the
exponent in the
calculation for titer (T).
[380] Using this method, the IL-17C adenovirus had a titer of 1.3x1010 pfu/ml.
EXAMPLE 10
Construction of Mammalian Expression Vectors that Express Human IL-17RE
[381] An expression vector was prepared for the expression of the soluble,
extracellular
domain of the human IL-17RE polypeptide, IL-17RECHIS, wherein the construct is
designed to
express a IL-17RE polypeptide comprised of the predicted initiating methionine
and truncated
adjacent to the predicted transmembrane domain, and with a C-terminal HIS tag:
5'GGCTCAGGATCTGGTGGCGGCCATCACCACCATCATCACTAAATCTAGA3' (SEQ ID
NO: 78).
[382] A 1160 bp PCR generated IL-17RE DNA fragment was created using ZC50282:
5'GAAGAACGTCTCTCATGGGGAGCTCCAGACTGGCAGC3' (SEQ ID NO:79) and ZC50283:
5'GAAGAACGTCTCTAGCCGTGTCTGTAAGAGACATCCGGAC3' (SEQ ID NO:80) as PCR
primers to add Esp3I restriction sites and Tgo reagents (Roche, Applied
Sciences, Indianapolis, IN).
A plasmid containing the IL-17RE cDNA (Clonetrack ID#100989) was used as a
template. PCR
amplification of the IL-17RE fragment was performed as follows: One cycle of
94C for 2 minutes;
then fifteen cycles at 94 C for 30 seconds, 65 C for 30 seconds, 72 C for 1
minute, followed by one
cycle of 72 C for 5 minutes and then a 4 C hold. The reaction was purified
using QlAquick PCR
purification kit (Qiagen, Santa Clarita, Ca.) and digested with Esp3I
(Fermentas, Hanover, MD)
following manufacturer's protocol. The reaction was purified using QlAquick
PCR purification kit
(Qiagen, Santa Clarita, Ca.) according the manufacturer's instructions.
[383] The excised DNA was subcloned into plasmid pExpress47 which had been cut
with
Eco31I (Fermentas, Hanover, MD). The pExpress47 vector uses the native IL-17RE
signal peptide
and attaches the HIS tag: 5'GGCTCAGGATCTGGTGGCGGCCATCACCACCATCATCACTAAA
TCTAGA3' (SEQ ID NO:125) to the C-terminus of the extracellular portion of the
IL-17RE
polypeptide-encoding polynucleotide sequence. Plasmid pExpress47, is a entry
vector containing
pDONR221 backbone, Kozak, Eco31I sites for ORF cloning, for seamless ligation
to 3' His tag and
Cassette A (Invitrogen) between cloning sites. The plasmid also has a pUC
origin of replication, a
mammalian selectable marker expression unit.
[384] About 10 1 of the restriction digested IL-17RE insert and about 75ng of
the digested
vector were ligated using the Fast link ligation kit (EPICENTRE technologies
(Madison, WI). Two
microliter of the ligation reaction was transformed into One shot MAX
efficiency DH10B-T1
competent cells (Invitrogen, Carlsbad, California) according to manufacturer's
direction and plated
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
102
onto LB plates containing 25 g/ml Kanamycin, and incubated overnight.
Colonies were submitted
for sequencing in 5 ml liquid cultures of individual colonies. The insert
sequence of clones was
verified by sequence analysis.
[385] An LR reaction was set up using LR reaction kit (Invitrogen, Carlsbad,
California),
about 300ng of pExpress 4 expression vector and about 100-300ng of IL-
17RE/pexpress47 entry
clone. Plasmid pExpress4, is a expression vector made by cloning Gateway
conversion cassette A
into the Nru I site of pEXPRESS-01; a standard vector; modular design;
Promoter (Kpn I/ Mfe); poly
A (Xba I/ Hind III); Zeo selection marker (Hind III/ Bgl II); E. coli Ori (Bgl
II/ Kpn I); Gene Amp
cassette (Sfi I/ Sap I). The reaction contained 4 15X LR reaction buffer, 1 1
of Topoisomerase, 4 1
of LR Clonase enzyme mix and TE buffer for a final volume of 20. Incubated for
1 hour at 25 C,
then 2 1 proteinase K added and incubated at 37 C for 10 minutes. One
microliter of the LR reaction
was transformed into One shot MAX efficiency DH10B-T1 competent cells
(Invitrogen, Carlsbad,
California) according to manufacturer's direction and plated onto LB plates
containing 50 g/ml
Kanamycin, and incubated overnight. Colonies were screened by PCR and
simultaneously
inoculating 100 1 of LB broth.
[386] PCR was set up using the following: Advantage 2 reagents (BD Biosciences
Clontech, Palo Alto, CA) and ZC5020: 5'CACTGGAGTGGCAACTTCCAG3' (SEQ ID NO:126)
and ZC14063: 5'CACCAGACATAATAGCTGACAGACT3' (SEQ ID NO:127) as PCR primers.
PCR amplification of the IL-17RE was performed as follows: One cycle of 94C
for 2 minutes; then
35 cycles at 94 C for 30 seconds, 62 C for 30 seconds, 72 C for 2 minute,
followed by one cycle of
72 C for 5 minutes and then a 4 C hold. A band of the predicted size 1468bp
was visualized by 4%
agarose gel electrophoresis. 5m1 liquid culture was inoculated with the 100 1
LB clone mix and left
ON at 37 C with shaking.
[387] A mini prep was done using a QlAprep spin Miniprep kit (Qiagen, Santa
Clarita,
Ca.) according the manufacturer's instructions.
EXAMPLE 11
Construction of Mammalian Expression Vectors that Express Human IL-17C
[388] An expression vector was prepared for the expression of human IL-17C
polypeptide,
IL-17CCHIS, wherein the construct is designed to express a IL-17C polypeptide
comprised of the
predicted initiating methionine to the last amino acid minus the stop codon
and with a C-terminal
HIS tag: 5' GGCTCAGGATCTGGTGGCGGCCATCACCACCATCATCACT AAATCTAGA3'
(SEQ ID NO: 128).
[389] A 594 bp PCR generated IL-17C DNA fragment was created using ZC80204"
5'GAAGAACGTCTCTCATGACGCTCCTCCCCGGCCTCC3' (SEQ IDNO:129) and ZC80300:
5'GAAGAACGTCTCTAGCCCACTGAACGGGGCAGCACGCAGGTG3' (SEQ ID NO:130) as
PCR primers to add Esp3I restriction sites and Tgo reagents (Roche, Applied
Sciences, Indianapolis,
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
103
IN) with or without DMSO ( Sigma, ST. Louis, MO). A plasmid containing the IL-
17C cDNA
(Clonetrack ID#100527) was used as a template. PCR amplification of the IL-17C
fragment was
performed as follows: PCR amplification of the IL-17C fragment was performed
as follows: One
cycle of 94C for 2 minutes; then three cycles at 94 C for 15 seconds, 45 C for
30 seconds, 72 C for
2.5 minutes, then nine cycles at 94 C for 15 seconds, 63 C for 30 seconds, 72
C for 2.5 minutes;
followed by one cycle of 72 C for 5 minutes and then a 4 C hold.. The reaction
was purified using
QlAquick PCR purification kit (Qiagen, Santa Clarita, Ca.) and digested with
Esp3I (Fermentas,
Hanover, MD) following manufacturer's protocol. The reaction was purified
using QlAquick PCR
purification kit (Qiagen, Santa Clarita, Ca.) according the manufacturer's
instructions.
[390] The excised DNA was subcloned into plasmid pExpress47 which had been cut
with
Eco31I (Fermentas, Hanover, MD). The pExpress47 vector uses the native IL-17C
signal peptide
and attaches the HIS tag: 5'GGCTCAGGATCTGGTGGCGGCCATCACCACCATCA
TCACTAAATCTAGA3' (SEQ ID NO:131) to the IL-17C polypeptide-encoding
polynucleotide
sequence. Plasmid pExpress47, is a entry vector containing pDONR221 backbone,
Kozak, Eco31I
sites for ORF cloning, for seamless ligation to 3' His tag and Cassette A
(Invitrogen) between
cloning sites. The plasmid also has a pUC origin of replication, a mammalian
selectable marker
expression unit.
[391] About 10 1 of the restriction digested IL-17C insert and about 75ng of
the digested
vector were ligated using the Fast link ligation kit (EPICENTRE technologies
(Madison, WI). Two
microliter of the ligation reaction was transformed into One shot MAX
efficiency DH10B-T1
competent cells (Invitrogen, Carlsbad, California) according to manufacturer's
direction and plated
onto LB plates containing 25 g/ml Kanamycin, and incubated overnight.
Colonies were submitted
for sequencing in 5 ml liquid cultures of individual colonies. The insert
sequence of clones was
verified by sequence analysis.
[392] An LR reaction was set up using LR reaction kit (Invitrogen, Carlsbad,
California),
about 300ng of pExpress 4 expression vector and about 100-300ng of IL-
17C/pexpress47 entry
clone. Plasmid pExpress4, is a expression vector made by cloning Gateway
conversion cassette A
into the Nru I site of pEXPRESS-01; a standard vector; modular design;
Promoter (Kpn I/ Mfe); poly
A (Xba I/ Hind III); Zeo selection marker (Hind III/ Bgl II); E. coli Ori (Bgl
II/ Kpn I); Gene Amp
cassette (Sfi I/ Sap I). The reaction contained 4 15X LR reaction buffer, 1 1
of Topoisomerase, 4 1
of LR Clonase enzyme mix and TE buffer for a final volume of 20. Incubated for
1 hour at 25 C,
then 2 1 proteinase K added and incubated at 37 C for 10 minutes. One
microliter of the LR reaction
was transformed into One shot MAX efficiency DH10B-T1 competent cells
(Invitrogen, Carlsbad,
California) according to manufacturer's direction and plated onto LB plates
containing 50 g/ml
Kanamycin, and incubated overnight. Colonies were screened by PCR and
simultaneously
inoculating 100 1 of LB broth.
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
104
[393] PCR was set up using the following: Advantage 2 reagents (BD Biosciences
Clontech, Palo Alto, CA) and ZC5020: 5'CACTGGAGTGGCAACTTCCAG3' (SEQ ID NO:132)
and ZC14063: 5'CACCAGACATAATAGCTGACAGACT3' (SEQ ID NO:133) as PCR primers.
PCR amplification of the IL-17C was performed as follows: One cycle of 94C for
2 minutes; then 35
cycles at 94 C for 30 seconds, 62 C for 30 seconds, 72 C for 2 minute,
followed by one cycle of
72 C for 5 minutes and then a 4 C hold. A band of the predicted size 942bp was
visualized by
agarose gel electrophoresis. 5m1 liquid culture was inoculated with the 100 1
LB clone mix and left
ON at 37 C with shaking. Glycerol stock archieved at -80 C. Plate was struck
with glycerol stock
and left ON at 37 C. A 5m1 liquid culture was inoculated with clone and left
ON at 37 C with
shaking. 5m1 ON culture used to inoculate 500m1 of liquid culture, left ON at
37 C with shaking.
[394] A mega prep was done using a QlAfilter plasmid mega kit ( Qiagen, Santa
Clarita,
Ca.) according to an optimized protocols based on manufacturer's instructions.
EXAMPLE 12
Construction of Mammalian Expression Vector for Murine IL-17C
[395] An expression vector was prepared for the expression of mouse IL-17C
polypeptide,
IL-17CCHIS, wherein the construct is designed to express a IL-17C polypeptide
comprised of the
predicted initiating methionine to the last amino acid minus the stop codon,
and with a C-terminal
HIS tag, 5'GGCTCAGGATCTGGTGGCGGCCATCACCACCATCATCACTA AATCTAGA3'
(SEQ ID NO:134).
[396] A 620 bp PCR generated IL-17C DNA fragment was created using ZC50745:
5'GAAGCCGAAGACTTCATGGCCACCGTCACCGTCACT3' (SEQ ID NO:135) and ZC50743:
5'GAAGCCGAAGACTTAGCCCTGTGTAGACCTGGGAAGAA3' (SEQ ID NO:136) as PCR
primers to add Bbsl restriction sites and Tgo reagents (Roche, Applied
Sciences, Indianapolis, IN)
plus 10% DMSO ( Sigma, ST. Louis, MO). A plasmid containing the IL-17C cDNA
(Clonetrack
ID#101619) was used as a template. PCR amplification of the IL-17C fragment
was performed as
follows: One cycle of 94C for 2 minutes; then three cycles at 94 C for 15
seconds, 45 C for 30
seconds, 72 C for 2.5 minutes, then nine cycles at 94 C for 15 seconds, 63 C
for 30 seconds, 72 C
for 2.5 minutes; followed by one cycle of 72 C for 5 minutes and then a 4 C
hold. The reaction was
purified using QlAquick PCR purification kit (Qiagen, Santa Clarita, Ca.) and
digested with Bbsl
(Fermentas, Hanover, MD) following manufacturer's protocol. The reaction was
gel extracted using
QlAquick gel extraction kit (Qiagen, Santa Clarita, Ca.) according the
manufacturer's instructions.
[397] The excised DNA was subcloned into plasmid pExpress47 which had been cut
with
Eco31I (Fermentas, Hanover, MD). The pExpress47 vector uses the native IL-17C
signal peptide
and attaches the HIS tag: 5'GGCTCAGGATCTGGTGGCGGCCATCACCACCATCA
TCACTAAATCTAGA3' (SEQ ID NO:137) to the C-terminus of the IL-17C polypeptide-
encoding
polynucleotide sequence. Plasmid pExpress47, is a entry vector containing
pDONR221 backbone,
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
105
Kozak, Eco31I sites for ORF cloning, for seamless ligation to 3' His tag and
Cassette A (Invitrogen)
between cloning sites. The plasmid also has a pUC origin of replication, a
mammalian selectable
marker expression unit.
[398] About 10 1 of the restriction digested IL-17C insert and about 75ng of
the digested
vector were ligated using the Fast link ligation kit (EPICENTRE technologies
(Madison, WI). Two
microliter of the ligation reaction was transformed into One shot MAX
efficiency DH10B-T1
competent cells (Invitrogen, Carlsbad, California) according to manufacturer's
direction and plated
onto LB plates containing 25 g/ml Kanamycin, and incubated overnight.
Colonies were submitted
for sequencing in 5 ml liquid cultures of individual colonies. The insert
sequence of clones was
verified by sequence analysis.
[399] An LR reaction was set up using LR reaction kit (Invitrogen, Carlsbad,
California),
about 300ng of pExpress 4 expression vector and about 100-300ng of IL-
17C/pexpress47 entry
clone. Plasmid pExpress4, is a expression vector made by cloning Gateway
conversion cassette A
into the Nru I site of pEXPRESS-01; a standard vector; modular design;
Promoter (Kpn I/ Mfe); poly
A (Xba I/ Hind III); Zeo selection marker (Hind III/ Bgl II); E. coli Ori (Bgl
II/ Kpn I); Gene Amp
cassette (Sfi I/ Sap I). The reaction contained 4 15X LR reaction buffer, 1 1
of Topoisomerase, 4 1
of LR Clonase enzyme mix and TE buffer for a final volume of 20. Incubated for
1 hour at 25 C,
then 2 1 proteinase K added and incubated at 37 C for 10 minutes. One
microliter of the LR reaction
was transformed into One shot MAX efficiency DH10B-T1 competent cells
(Invitrogen, Carlsbad,
California) according to manufacturer's direction and plated onto LB plates
containing 50 g/ml
Kanamycin, and incubated overnight. Colonies were screened by PCR and
simultaneously
inoculating 100 1 of LB broth.
[400] PCR was set up using the following: Advantage 2 reagents (BD Biosciences
Clontech, Palo Alto, CA) and ZC5020: 5'CACTGGAGTGGCAACTTCCAG3' (SEQ ID NO:138)
and ZC14063: 5'CACCAGACATAATAGCTGACAGACT3' (SEQ ID NO:139) as PCR primers.
PCR amplification of the IL-17C was performed as follows: One cycle of 94C for
2 minutes; then 35
cycles at 94 C for 30 seconds, 62 C for 30 seconds, 72 C for 2 minute,
followed by one cycle of
72 C for 5 minutes and then a 4 C hold. A band of the predicted size 934bp was
visualized by
agarose gel electrophoresis. 5m1 liquid culture was inoculated with the 100 1
LB clone mix and left
ON at 37 C with shaking.
[401] A mini prep was done using a QlAprep spin Miniprep kit ( Qiagen, Santa
Clarita,
Ca.) according the manufacturer's instructions.
EXAMPLE 13
Transfection and Expression of Soluble Human IL-17C
[402] On day 1, .5L of shake flask cultured 293f cells (Invitrogen, Carlsbad,
CA Cat#
R790-07), passage 5-post thaw at 2.4e6 c/m1, were seeded into 4.5 L of
Freestyle 293 Expression
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
106
Medium (Invitrogen, Carlsbad, CA Cat# 12338-026) in a Wave Biotech reactor
(Wave Biotech, Cat#
cell bag 20L/O). 25 mls of a Penicillin-Streptomicin (Invitrogen, Carlsbad, CA
Cat# 1507-063)
mixture was also added at this time. The cells were cultured at 37 C with
ambient airflow @.2 LPM
supplemented with 6% C02. The reactor was rocked 25 times per minute with an
angle setting of
9.5. These settings were utilized for the entire length of the culture.
[403] On day 4, 4.7 L of fresh Freestyle 293 media w/ 5mls/L of Penicillin-
Streptomicin
mixture was then added to the reactor, for a final volume of 9.7L. The cells
were then transfected as
follows: .8mg/ml mega prep. plasmid DNA (MPET construct #889, IL-17CcH6), as
described in the
above Example, was obtained. Two 120 ml aliquots of Optimem media (Invitrogen,
Carlsbad, CA
Cat# 31985-070) were prewarmed to 37 C. Into one Optimem aliquot, 10 mls of
the DNA prep was
added and mixed. Into the other Optimem aliquot 10.5 mls of Lipofectimine 2000
(Invitrogen,
Carlsbad, CA Cat# 11668-019) was added and mixed. The two aliquot mixtures
were added
together, mixed and incubated for 30 minutes at room temp., with occasional
mixing. The
Lipofectimine 2000/DNA mixture was then added to the reactor.
[404] After 96 hrs post transfection, the culture was harvested, the cells
spun out of the
media for 10 minutes @ 4000 G's in a Beckman Coulter Avanti J-HC centrifuge.
The conditioned
media was then passed consecutively through a 1.2 and .2um Millipore Opticap
filter set (Millipore
Bedford MA. Cat#s KW1904HB3, KWSSL4HB3). The filtered media was then purified
by known
methods.
EXAMPLE 14
Transfection and Expression of Soluble Murine IL-17C
[405] On day 1, 1.25L of shake flask cultured 293f cells (Invitrogen,
Carlsbad, CA Cat#
R790-07) passage 22-post thaw at 2e6 c/ml, were seeded into 8.15 L of
Freestyle 293 Expression
Medium (Invitrogen, Carlsbad, CA Cat# 12338-026) in a Wave Biotech reactor
(Wave Biotech, Cat#
cell bag 20L/O). The cells were cultured at 37 C with ambient airflow @.2 LPM
supplemented
with 6% C02. The reactor was rocked 25 times per minute with an angle setting
of 9.5. These
settings were utilized for the entire length of the culture. On day 4, 700 mls
of the culture was
extracted and discarded. 1.4L of fresh Freestyle 293 media was then added for
a final volume of
10L. On day 5, 2.6 L of media was extracted and discarded. 1.4L of fresh
Freestyle 293 media was
added for a final volume of 8.8L @ 2e6 c/ml and the cells were transfected as
follows: mega prep
plasmid DNA (MPET construct #1280, IL 17CmcH6) @ 1.88mg/ml was obtained as
described
herein. Two 150 ml aliquots of DMEM media (Invitrogen, Carlsbad, CA Cat#
119092) were
prewarmed to 37 C. Into one DMEM aliquot, 9.4 mls of the DNA prep was added
and mixed. Into
the other DMEM aliquot 17.6 mls of a 1 mg/ml solution of PEI
(Polyethyleneimine, Linear 25kDa.
Cat# 23966. Polysciences, Inc. Warrington PA.) mixture was added and mixed.
The two mixtures
were incubated separately at room temperature for 5 minutes, then added
together, mixed and
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
107
incubated for 20 minutes at room temperature with occasional mixing. The
PEI/DNA mixture was
then added to the reactor. Fifty mls of a Penicillin-Streptomicin mixture was
also added at this time
(Invitrogen, Carlsbad, CA Cat# 1507-063).
[406] After 96 hrs post transfection, the culture was harvested, the cells
spun out of the
media for 10 minutes @ 4000 G's in a Beckman Coulter Avanti J-HC centrifuge.
The conditioned
media was then passed consecutively through a 1.2 and .2um Millipore Opticap
filter set (Millipore
Bedford MA. Cat#s KW1904HB3, KWSSL4HB3). The filtered media was then purified
by known
methods.
EXAMPLE 15
IL-17RE Luminex Assay
[407] Oligonucleotides specific to unique intron/exon junctions for IL-17RE
splice
variants can be designed for use in a Luminex microsphere-based assay to
measure levels of splice
variant specific mRNAs. However, it is not possible to design a specific oligo
to IL-17REx1, as it
contains no unique intron/exon junction that the other splice variants lack.
For example, IL-17REx2
(SEQ ID NO:4), zc49789 (5'gcctcccacacgaggaagctgctgc 3') (SEQ ID NO:39) is
synthesized with a
5' amine Uni-Link group and its complementary antisense oligonucleotide
zc49890
(5'gcagcagcttcctcgtgtgggaggc3') (SEQ ID NO:40) was synthesized with a 5'
biotin group for
monitoring coupling efficiency later in the protocol. IL-17REx3 (SEQ ID NO:7)
has three unique
intron/exon junctions relative to the other IL-17RE splice variants, therefore
it is necessary to design
three sense oligonucleotides, zc49790 (5'tggactcacaaaggacccgagttct3') (SEQ ID
NO:41), zc49891
(5'gcctctgttattccagtctggtggg3') (SEQ ID NO:42), and zc49892
(5'ccccgttgaagaccgtgtgggaggc3')
(SEQ ID NO:43), each with a 5' amine Uni-Link group and their complementary
antisense 5' biotin
labeled control oligonucleotides, zc49791 (5'cccaccagactggaataacagaggc3') (SEQ
ID NO:44),
zc49792 (5'gcctcccacacggtcttcaacgggg3') (SEQ ID NO:45), and zc49724
(5'agaactcgggtcctttgtgagtcca3') (SEQ ID NO:46). IL-17REx4 (SEQ ID NO:10)
specific sense
oligonucleotide zc49793 (5'tgctgtgtcctgctccatgcttcac3') (SEQ ID NO:47) is
synthesized with a 5'
amine Uni-Link group and it's 5' biotin labeled antisense complement, zc49729,
(5'gtgaagcatggagcaggacacagca3') (SEQ ID NO:48) is also synthesized. To assess
the efficiency of
the RNA amplification step in amplifying long mRNAs, oligos are designed to
the first and last
exons of IL-17RE, which are common to all known splice variants. For the first
exon of IL-17RE,
zc49794 (5'tctgactctgctgggattggctttc3') (SEQ ID NO:49) is synthesized with a
5' amine Uni-Link
group and it's complementary antisense oligonucleotide zc49893
(5'gaaagccaatcccagcagagtcaga3')
(SEQ ID NO:50) is synthesized with a 5' biotin group. For the last exon of IL-
17RE, zc49795
(5'tgctgctgctgtggagcggcgccga3') (SEQ ID NO:51) is synthesized with a 5' amine
Uni-Link group
and it's complement zc49894 (5'tcggcgccgctccacagcagcagca3') (SEQ ID NO:52).
The ratio of the
measurements of the first and last exons can be used to qualitatively assess
the impact of measuring
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
108
the levels of a sequence target that is not near the 3' end of an mRNA, such
as the unique intron/exon
junction specific to IL-17REx2.
[408] Each sense oligonucleotide is coupled to specific xMAPTM Multi Analysis
Carboxylated Microspheres (Luminex Corporation, Austin, TX) as follows: stock
microspheres are
resuspended by vortex and sonication for approximately 20 seconds, 200 1
(2.5x106 microspheres)
are transferred to a microfuge tube and pelleted by microcentrifugation at
>8000 x g for 1-2 minutes.
Supernatents are removed and the microsphere pellets are resuspended in 50u1
of 0.1M MES (2(N-
Morpholino) ethanesulfonic acid, Sigma, St. Louis, MO), ph4.5, by vortex and
sonication. A fresh
solution of lOmg/ml EDC carbodimide HCL (1-Ethyl-3- (3-dimethylaminopropyl)
carbodimide HC1,
Pierce, Rockford, IL) is prepared in dHzO and 2.5u1 of this solution is added
to the microspheres,
vortexed and incubated at room temperature 30 minutes in the dark. A second
fresh solution of
10mg/ml EDC is prepared, 2.5u1 is added to the microspheres, and incubation in
the dark for 30
minutes is repeated. A third iteration of the EDC addition and incubation is
optional. lml of 0.02%
Tween20 (Polyoxyethylenesorbitan monolaurate, Sigma, St. Louis, MO) is added
to the coupled
microspheres and mixed by vortexing and pelleted by microcentrifugation. The
supernatent is
removed and the microsphere pellets are resuspended in lml of 0.1% SDS (Lauryl
Sulfate, Sigma,
St. Louis, MO) by vortexing and pelleted by centrifugation. The supernatent is
removed and pellets
are resuspended in 100 1 of TE, ph 8.0 by vortexing and sonication for about
20 seconds.
Microspheres are enumerated by using a hemacytometer and stored at 4 C in the
dark until use.
[409] Coupling and hybridization efficiency of the microspheres is evaluated
by mixing
the coupled microspheres with the biotin labeled complementary oligonucleotide
as follows: The
coupled microspheres are resuspended by vortex and sonication for about 20
seconds, and a working
mixture is prepared by diluting coupled microsphere stocks to 150
microspheres/ul in 1.5X TMAC
hybridization buffer (4.5M TMAC (Sigma, St. Louis, MO) 0.15% Sarkosyl, 0.75mM
Tris-HC1, pH8
(Sigma, St. Louis, MO), 6mM EDTA, pH 8.0 (Gibco, Grand Island, NY). To each
sample or
background well in a MicroAmp optical 96 well reaction plate (Applied
Biosystems, Foster City,
CA) 33.3u1 of coupled microspheres is added, and to each background well is
added 16.67u1 TE, pH
8Ø The appropriate biotinylated complementary oligonucleotide over a range
from 5 to 200
femtomoles, adjusted to a final volume of 16.7u1, is added to each sample
well, the plate is sealed
and reactions are mixed with a plate shaker at 400rpm. Plates are incubated at
94 C for 3 minutes,
then 55 C for 15 minutes. A vacuum manifold (Millipore Corporation, Billerica,
MA) is used to
remove unbound oligonucleotides and the plate is washed 3 times with 100uUwell
wash buffer (1mM
PBS, 0.01% Tween 20), removing the buffer each time by vacuum filtration.
Fresh reporter mix is
prepared by diluting streptavidin-R-phycoerythrin conjugate (Molecular Probes,
Eugene, OR) to
4ug/ml in wash buffer, 75u1 is added to each well, the assay plate is covered
with foil and mixed on a
plate shaker at 1100 rpm for 30 seconds, then incubated at room temperature
for 15 minutes at 400
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
109
rpm. The plate is then washed 3X to remove unbound streptavidin-PE, and
samples are resuspended
in a final volume of 75u1 wash buffer. 50u1 are then analyzed on a Bio-Plex
Array Reader (BioRad
Laboratories, Inc, Hercules, CA).
[410] Approximately 2 X 106 U937 cells are plated and stimulated with 20ng/ml
PMA and
20ng/ml PMA + 0.5 ug/ml ionomycin for 6,11 and 24 hours. - 2 x 106 THP1 cells
are stimulated
with PMA at 100ng/ml for 12, 24 and 48 hours. Cells are harvested and total
RNA is purified using a
Qiagen (Valencia, CA) RNeasy kit according to the manufacturer's instructions
with the optional
DNAse step incorporated into the protocol. The RNA is DNAsed using DNA-free
reagents (Ambion,
Inc, Austin, TX) according to the manufacturer's instructions. The quality of
the RNA is assessed by
running an aliquot on an Agilent Bioanalyzer. If the RNA is significantly
degraded, it is not used for
subsequent assays for IL-17RE mRNAs. Presence of contaminating genomic DNA is
assessed by a
PCR assay on an aliquot of the RNA with zc37263 (5'gaattacaccctctggagagtgg 3')
and zc37264 (5'
gaatttcggacaatccagtactc 3'), primers that amplify a single site in genomic DNA
within an intron at
the cathepsin Z gene locus. The PCR conditions for the contaminating genomic
DNA assay are as
follows: 2.5u1 l OX buffer and 0.5u1 Advantage 2 cDNA polymerase mix (BD
Biosciences Clontech,
Palo Alto, CA), 2u12.5mM dNTP mix (Applied Biosystems, Foster City, CA), 2.5u1
lOX Rediload
(Invitrogen, Carlsbad, CA), and 0.5u1 20uM zc37263 and zc37264, in a final
volume of 25 ul.
Cycling parameters are 94 C 20", 40 cycles of 94 C 20" 62 C 20" 72 C 1' and
one cycle of 72 C 7'.
lOul of each reaction is subjected to agarose gel electrophoresis and gels are
examined for presence
of a PCR product from contaminating genomic DNA. Only RNAs that appear to be
free of
contaminating genomic DNA are used in subsequent assays for IL-17RE splice
variant mRNAs.
[411] 5 g of each RNA to be assayed for IL-17RE splice variants using coupled
Luminex
microspheres is first amplified using an Ambion MessageAmpTM aRNA Kit (Ambion
Incorporated,
Austin, TX) according to the manufacturer's instructions, but modifying the In-
Vitro transcription
step synthesizing the antisense RNA such that labeled dNTPs (biotin-l6-UTP and
biotin-ll-CTP,
Perkin-Elmer Life Sciences, Boston, MA) are used instead of the dNTPs provided
with the kit.
Levels of IL-17RE splice variant mRNAs are determined in each amplified RNA
sample as follows:
appropriate housekeeping gene control oligonucleotide coupled microsphere and
IL-17RE splice
variant specific oligonucleotide coupled microspheres are used to prepare a
working microsphere
mixture by diluting the coupled microsphere stocks to 5000 per 33.3u1 in 1.5X
TMAC hybridization
buffer; the total volume being 33.3u1 multiplied by the number of sample and
background wells to be
tested. Mix this working microsphere solution by vortex and sonication for
about 20 seconds. To
each background well, add 16.7u1 TE, pH 8.0, and to each sample well add 5ug
of the amplified
biotinylated RNA, which is first heated to 94 C for 35 minutes and iced, in a
volume of 16.7u1 TE,
pH 8Ø To each sample and background well is added 33.3u1 of the working
microsphere mixture,
and wells are mixed by pipetting up and down, and shaking briefly on a plate
shaker. The plate is
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
110
sealed and incubated at 94 C for 10 minutes to denature the amplified
biotinylated RNA, then
incubated at 60 C in a shaking incubator for 5 hours with gentle rocking. The
reactions are
transferred to a microtiter plate, a vacuum manifold is used to separate the
unbound nucleotides and
wash the plate, and reporter mix is incubated with the samples as described
above. Plates are then
washed and counted in a Bio-Plex Array Reader as described above.
[412] Results may demonstrate that in comparison to THP1, IL-17RE transcripts
in U937
cells are expressed at a much higher level, regardless of the presence or
absence of PMA.
Additionally, significant expression of each splice variant IL-17REx2, x3 and
x4 is observed. By
inference the variant IL-17REx1 and/or possible splice variants that are as
yet undescribed are also
highly expressed in U937 relative to THP1, because of the high levels of
expression of the last exon.
EXAMPLE 16
Northern and Dot Blot Analysis of IL-17RE
[413] Northern and dot blot analyses were performed using Human Multiple
Tissue Blots
I, II, and III and the Human RNA Master Blot (CLONTECH Laboratories, Inc.,
Palo Alto, CA). A
1.4kb DNA fragment was generated by digesting DNA of a IL-17REx1 (SEQ ID NO:
1) cDNA with
EcoRl and Not, followed by gel electrophoresis and purification of the
fragment using Qiaquick Gel
Extraction reagents and protocol. (Qiagen, Valencia, CA). The DNA fragment
encompassed the
sequence encoding amino acids #257-690 of SEQ ID NO:2 and is predicted to
hybridze to all known
splice variants of IL-17RE. The fragment was radioactively labeled using the
Redi-Prime II kit
(Stratagene, La Jolla, CA) according to the manufacturer's protocol. The probe
was purified using a
MicroSpin S-200 HR spin column (Amersham, Arlington Heights, IL) according to
the
manufacturer's instructions. Salmon sperm DNA (Stratagene, La Jolla, CA) and
Cot-1 DNA
(Invitrogen, Carlsbad, CA) were boiled 5', snap-chilled on ice, added to
ExpressHyb (CLONTECH)
at 100 g/ml and 6 g/ml, respectively, and used as prehybridization and
hybridization solutions for
the blots. Prehybridization took place for 3 hours at 55 C. The radioactively
labeled DNA fragment
was boiled 5', snap-chilled on ice and added to the blots at 1 X 106 cpm/ml
hybridization solution.
Hybridization took place overnight at 55 C. Following hybridization, the blots
were washed as
follows: twice in 2XSSC, 0.1% SDS at room temperature, one time in 2X SSC,
0.1% SDS at 65 C,
followed by one 20' wash in O.1X SSC, 0.1% SDS at 65 C. The blots were exposed
to film
overnight The results are illustrated in the figures below, and demonstrate IL-
17RE mRNA is widely
expressed, being most strongly expressed in stomach, pancreas and expressed to
a lesser extent in
prostate, thyroid, trachea, salivary gland, liver, kidney, small intestine,
lung, fetal lung, fetal thymus,
placenta, mammary gland, heart, cerebellum, caudate nucleus, and colon. In
contrast, there is little or
no expression in whole brain, skeletal muscle, spleen, thymus, testis, ovary,
peripheral blood
leukocytes, spinal cord, lymph node, adrenal gland, uterus, bladder, fetal
whole brain, fetal heart,
fetal liver, fetal spleen, and bone marrow.
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
111
EXAMPLE 17
Northern, Dot Blot and Disease Array Analysis of IL-17C
[414] Northern, dot blot, and disease array analyses were performed using
Human
Multiple Tissue Blots I and III, Human Fetal Multiple Tissue Blot II, Human
RNA Master Blot,
Cancer Profiling Array II, Blood Disease Profiling Array, Autoimmune Disease
Profiling Array, and
the Cancer Cell Line Profiling Array. (CLONTECH Laboratories, Inc., Palo Alto,
CA). A-770bp
DNA fragment was generated by digesting IL-17C cDNA with EcoRl, followed by
gel
electrophoresis and purification of the fragment using Qiaquick Gel Extraction
reagents and
protocol. (Qiagen, Valencia, CA). The DNA fragment encompassed the sequence
encoding the
complete open reading frame of IL-17C. The fragment was radioactively labeled
using the Redi-
Prime II kit (Stratagene, La Jolla, CA) according to the manufacturer's
protocol. The probe was
purified using a MicroSpin S-200 HR spin column (Amersham, Arlington Heights,
IL) according to
the manufacturer's instructions. Salmon sperm DNA (Stratagene, La Jolla, CA)
and Cot-1 DNA
(Invitrogen, Carlsbad, CA) were boiled 5', snap-chilled on ice, added to
ExpressHyb (CLONTECH)
at 100 ug/ml and 6 ug/ml, respectively, and used as prehybridization and
hybridization solutions for
the blots. Prehybridization took place overnight at 55 C. The radioactively
labeled DNA fragment
was boiled 5', snap-chilled on ice and added to the blots at 1 X 106 cpm/ml
hybridization solution.
Hybridization took place overnight at 55 C. Following hybridization, the blots
were washed as
follows: twice in 2XSSC, 0.1% SDS at room temperature, one time in 2X SSC,
0.1% SDS at 65 C,
followed by one 20' wash in 0.1X SSC, 0.1% SDS at 65 C. The blots were exposed
to film with
intensifying screens for six days.
[415] The results generally demonstrate that IL-17C mRNA is not widely or
highly
expressed. A transcript of -1.4kb is visible in fetal lung, but no IL-17C
transcript is present in fetal
brain, fetal liver, or fetal kidney. In adult tissues a transcript of -4.8kb
is visible in heart and two
transcripts of -5kb and 3kb are visible in skeletal muscle. In contrast, no IL-
17C transcript is
observable in brain, placenta, lung, liver, kidney, pancreas, stomach,
thyroid, spinal cord lymph
node, trachea, adrenal gland or bone marrow. In the cancer profiling array, IL-
17C is relatively
absent in normal and tumor cDNAs from multiple patients with cancer of the
breast, ovary, colon,
stomach, lung, kidney, bladder, vulva, prostate, trachea, uterus, cervix,
rectum, thyroid gland, testis,
skin and pancreas cancer. However, slightly higher IL-17C hybridization is
observable in the normal
liver and small intestine from several patients with cancers of those same
tissues. In the
Autoimmune and Blood Disease profiling arrays, IL-17C mRNA can be seen to be
slightly increased
in the CD 19 (primarily B-cell) fraction of the blood across the board in
normal and diseased patients,
relative to the levels of IL-17C mRNA in CD14 (primarily monocye), CD3
(primarily T cell),
Mononuclear cells and Polymorphonuclear cells Interestingly, the IL-17C mRNA
levels appear to be
further elevated in the CD19 blood fraction in patients with Multiple
Sclerosis, Von Willebrand's
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
112
Disease, Lupus Anticoagulans, Takayasu's Arthritis, Idiopathic
Thrombocytopenic Purpura,
Hodgkin's disease, and Chronic Myelogenous Leukemia, relative to normal
patient CD19 blood
fraction levels of IL-17C. In the cancer cell line profiling array, IL-17C
again is not highly or widely
expressed, but it is visible in a scattered few cell lines under certain
conditions. MDA-MB-435S
stimulated with Cytochalasine D and U-87 MG stimulated with Demecolcine,
Miomycine,
Actinomycin D and Cyclohexamide all appear to express low levels of IL-17C
mRNA while the
other 24 cell lines combined with stimulation conditions express little to no
IL-17C mRNA.
EXAMPLE 18
Transient Expression of IL-17RE
[416] Human IL-17REx1 (SEQ ID NO:1) and x2 (SEQ ID NO:4) cDNAs were placed in
a
dicistronic expression vector, pzmpl1. The cDNAs were inserted downstream of
the cmv promoter,
followed by an IRES site and a cDNA for the cell surface marker, human CD8.
CD8 expression
correlates with transcription of the inserted cDNA and can be used to facs
sort for CD8 cells and ask
if that population correlates with binding events, vs the non-CD8 population.
[417] 293FB suspension cells were seeded into 125 ml tissue culture erlenmeyer
fermenter
flasks at a density of 106 cells/ml in lOml fresh Freestyle 293 expression
medium (Invitrogen). 10 g
of IL-17REx1-pzmpll, IL-17REx2-pzmpll and empty pzmpll vector were transfected
into these
cells using lipofectamine 2000 (Invitrogen) 24-78 hours after transfection,
cells were used in the
binding experiments, as provided herein.
EXAMPLE 19
Creation of a Stable nih3t3 Assay Clone Expressing ap/nfkb Transcription
Factor
[418] Murine nih3t3 cells were stably transfected with the kz142 apl/nfkb
reporter
construct containing a neomycin-selectible marker. The Neo resistant
transfection pool was plated at
clonal density. Clones were isolated using cloning rings and screened by
luciferase assay using the
human IL-17C ligand as an inducer. Clones with the highest mean fluorescence
intensity (MFI) (via
apl/NfkB luciferase) and the lowest background were selected. A stable
transfectant cell line was
selected and called nih3t3/kz142.8.
EXAMPLE 20
Murine nih3t3 Cells Express IL-17RE
[419] Two-step PCR analysis of nih3t3 RNA demonstrated that these cells are
positive for
IL-17RE transcription, consistent with their signaling response to IL-17C
being mediated through
this receptor. First strand cDNA was prepared from total RNA isolated from
nih3t3 cells using
standard methods. PCR was applied using hot star polymerase and the
manufacturer's
recommendations, (Qiagen, Valencia, CA) except for utilizing 10% DMSO final
concentration. The
primers utilized included sense primer, zc40413 (5' tgcgcccggatcctacagaagc 3')
(SEQ ID NO:55)
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
113
and antisense primer, zc 40412 (5'gcacctcgggcagcaaatcaaag 3') (SEQ ID NO:56)
Agarose gel
electrophoresis revealed a single, robust amplicon of the expected size.
EXAMPLE 21
Creation of Cell Lines with Recombinant Over-Expression of
IL-17RE Splice Variants
[420] Stable recombinant over expression of human IL-17RE facilitates
identification of
its ligand by increasing sensitization of target cells to activation and
binding by its ligand. This
phenomenon has been observed for homologs of IL-17RE. Ligand activation
occurred with far
lower concentrations than that seen in the same cells, lacking recombinant
receptor over expression.
This activation phenomenon was observed in a murine nih3t3/kz142.8 cell line,
which was shown to
express these receptors endogenously. Ligand binding studies were done in
recombinant IL-17RE
over expressing baby hamster kidney cells (BHK570).
Stable over expression of human and mouse IL-17RE in the murine assay cell
line,nih3t3/kz142.8
[421] Murine nih3t3/kz142.8 (Example 17) were shown to produce endogenous IL-
17RE
mRNA by PCR (Example 18). These cells were transfected with cDNAs of human IL-
17REx1 (SEQ
ID NO:1), IL-17REx2 (SEQ ID NO:4) IL-17REx3 (SEQ ID NO:7), IL-17REx6 (SEQ ID
NO:20),
IL-17REX13 (SEQ ID NO:106) and mouse IL-17REx6 (SEQ ID NO:13) in pZMP11, a
dicistronic
expression vector with a CMV promoter driving transcription inserted cDNA
transcription, followed
by an IRES, followed by a cDNA for human CD8. CD8 expressing cells can be
selected for and
correlated with expression of the inserted cDNAs. Pzmp11 has a methotrexate
resistance gene.
(dihydrofolate reductase) Transfections were performed using a commercially
available kit and the
manufacturer's recommendations. (Mirus, Madison,Wl. Cat. #MIR218) Cells were
placed in 1 M
mtx amended growth medium to select for the expression constructs containing
the human and
mouse IL-17RE transgenes. After selection, transfection pools were generated,
and called
nih3t3/kz142.8/hcytor21x1, nih3t3/kz142.8/hcytor21x2,
nih3t3/kz142.8/hcytor21x3,
nih3t3/kz142.8/hcytor21x6, nih3t3/kz142.8/hcytor21x13 and
nih3t3/kz142.8/mcytor21x6.
Stable over expression of human and mouse IL-17RE in the baby hamster kidney
cell line, (BHK570)
[422] Baby Hamster Kidney cells (BHK570) were chosen for recombinant over-
expression
of IL-17RE for binding studies. These cells were transfected with cDNAs of
human IL-17REx1
(SEQ ID NO:1), IL-17REx2 (SEQ ID NO:4) IL-17REx3 (SEQ ID NO:7), IL-17REx6 (SEQ
ID
NO:20), IL-17REX13 (SEQ ID NO:106) and mouse IL-17REx6 (SEQ ID NO:13) in
pZMPll, a
dicistronic expression vector with a CMV promoter driving transcription
inserted cDNA
transcription, followed by an IRES, followed by a cDNA for human CD8. CD8
expressing cells can
be selected for and correlated with expression of the inserted cDNAs. Pzmpll
has a methotrexate
resistance gene. (dihydrofolate reductase) Transfections were performed using
a commercially
available kit and the manufacturer's recommendations. (Mirus, Madison,Wl. Cat.
#MIR218) Cells
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
114
were placed in 1 M mtx amended growth medium to select for the expression
constructs containing
the human and mouse IL-17RE transgenes. After selection, transfection pools
were generated, and
called BHK/hcytor21x1, BHK/hcytor21x2, BHK/hcytor21x3, BHK/hcytor21x6,
BHK/hcytor21x13,
and BHK/mcytor21x6.
EXAMPLE 22
Distribution of IL-17RE mRNA in Cell Line Panels Using PCR
[423] Total RNA was purified from resting and stimulated cell lines grown in-
house and
purified using a Qiagen (Valencia, CA) RNeasy kit according to the
manufacturer's instructions, or
an acid-phenol purification protocol (Chomczynski and Sacchi, Analytical
Biochemistry, 162:156-9,
1987). The quality of the RNA was assessed by running an aliquot on an Agilent
Bioanalyzer. If the
RNA was significantly degraded, it was not used for subsequent creation of
first strand cDNA.
Presence of contaminating genomic DNA was assessed by a PCR assay on an
aliquot of the RNA
with zc41011 (5'ctctccatccttatctttcatcaac3') (SEQ ID NO: 57) and zc41012
(5'ctctctgctggctaaacaaaacac3') (SEQ ID NO:58), primers that amplify a single
site of intergenic
genomic DNA. The PCR conditions for the contaminating genomic DNA assay were
as follows:
2.5u1 lOX buffer and 0.5u1 Advantage 2 cDNA polymerase mix (BD Biosciences
Clontech, Palo
Alto, CA), 2u1 2.5mM dNTP mix (Applied Biosystems, Foster City, CA), 2.5u1 lOX
Rediload
(Invitrogen, Carlsbad, CA), and 0.5u1 20uM zc41011 and zc41012, in a final
volume of 25 ul.
Cycling parameters were 94 C 20", 40 cycles of 94 C 20" 60 C 1'20" and one
cycle of 72 C 7'.
lOul of each reaction was subjected to agarose gel electrophoresis and gels
were examined for
presence of a PCR product from contaminating genomic DNA. If contaminating
genomic DNA was
observed, the total RNA was DNAsed using DNA-free reagents (Ambion, Inc,
Austin, TX)
according to the manufacturer's instructions, then retested as described
above. Only RNAs which
appeared to be free of contaminating genomic DNA were used for subsequent
creation of first strand
cDNA.
[424] 20 ug total RNA from 82 human cell lines were each brought to 98u1 with
H20, then
split into two 49u1 aliquots, each containing l0ug total RNA, and placed in
two 96-well PCR plates.
To each aliquot was added reagents for first strand cDNA synthesis (Invitrogen
First Strand cDNA
Synthesis System, Carlsbad, CA): 20u1 25mM MgC1z, lOul lOX RT buffer, lOul
0.1M DTT, 2u1
oligo dT, 2u1 RNAseOut. Then, to one aliquot from each cell line 2u1
Superscript II Reverse
Transcriptase was added, and to the corresponding cell line aliquot 2u1 H20
was added to make a
minus Reverse Transcriptase negative control. All samples were incubated as
follows: 25 C 10',
42 C 50', 70 C 15 `. Samples were arranged in deep well plates and diluted to
1.7m1 with H20. A
Multipette (Saigan) robot was used to aliquot 16.5u1 into each well of a 96-
well PCR plate multiple
times, generating numerous one-use PCR panels of the cell lines, which were
then sealed and stored
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
115
at -20 C. Each well in these panels represents first strand cDNA from
approximately 100ng total
RNA. The 82 cell lines were spread across two panels, array #118A and #118B.
[425] Quality of first strand cDNA on the panels was assessed by a multiplex
PCR assay
on one set of the panels using primers to two widely expressed, but only
moderately abundant genes,
CLTC (clathrin) and TFRC (transferrin receptor C). 0.5u1 each of Clathrin
primers zc42901
(5'ctcatattgctcaactgtgtgaaaag 3') (SEQ ID NO: 59), zc42902
(5'tagaagccacctgaacacaaatctg3') (SEQ
ID NO:60), and TFRC primers zc42599 (5'atcttgcgttgtatgttgaaaatcaatt3') (SEQ ID
NO:61), zc42600
(5'ttctccaccaggtaaacaagtctac3') (SEQ ID NO:62), were mixed with 2.5u1 lOX
buffer and 0.5u1
Advantage 2 cDNA polymerase mix (BD Biosciences Clontech, Palo Alto, CA), 2u1
2.5mM dNTP
mix (Applied Biosystems,, Foster City, CA), 2.5u1 lOX Rediload (Invitrogen,
Carlsbad, CA), and
added to each well of a panel of array#118A and array #118B. Cycling
parameters were as follows:
94 C 20", 35 cycles of 94 C 20", 67 C 80", and one cycle of 72 C 7'. lOul of
each reaction was
subjected to agarose gel electrophoresis and gels were scored for the presence
of a robust PCR
product for each gene specific to the +RT wells for each cell line.
[426] Expression of mRNA in the human first strand cDNA panels for IL-17RE was
assayed by PCR with sense oligo zc40450 (5'tcctgcctctcctcctcatagtca3') (SEQ ID
NO:63) and
antisense oligo zc40454 (5'ccaggatcaagagccccaggtgtc3') (SEQ ID NO:64) under
these PCR
conditions per sample: 2.5u1 lOX buffer and 0.5u1 advantage 2 cDNA polymerase
mix (BD
Biosciences Clontech, Palo Alto, CA), 2u1 2.5mM dNTP mix (Applied Biosystems,
), 2.5u1 10X
Rediload (Invitrogen, Carlsbad, CA), and 0.5u120uM each sense and antisense
primer. Primers were
predicted to pick up all known splice variants of IL-17RE, but they did not
necessarily distinguish
between each variant. Cycling conditions were 94 C 20", 35 cycles of 94 C 20",
69 C 2'30", and
one cycle of 72 C 7'. lOul of each reaction was subjected to agarose gel
electrophoresis and gels
were scored for positive or negative expression of IL-17RE. Results showed
widespread expression
of IL-17REmRNA in cell lines by this assay. IL-17RE was consistently and
usually strongly positive
in U-937(unstimulated and stimulated with PMA or PMA/Ionomycin), B-lymphomas
(DOHH-2
Ramos, Granta-519 and RL), and several cell lines from the digestive system
(CaCO2, CaCO2
differentiated, HCT-15, and HCT-116). Overall, samples that were positive for
IL-17RE were: L363,
A375, CTB-1 +PMA/Ionomycin, TF1, ARH77, G-361, MacLLC + PMA/Ionomycin, DOHH-2,
REH, HaCat, Ramos, Granta-519, RL, Hs294T, HL60 + butyric acid, AsPC-1, A-172.
Hep G2, U937
+ PMA/Ionomycin, TrBMEC, HepG2 + IL6, U937 + PMA, ME180, ARPE, A-549, U937,
CaCO2,
MRC-5, PC-3, CaCO2 differentiated, DLD-1, SKLU-1, Int407, HCT116, and HCT15.
EXAMPLE 23
Distribution of Murine IL-17RE mRNA in Murine Cell Line
Panels Using PCR
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
116
[427] Total RNA was purified from 60 resting and stimulated cell lines grown
in-house
and purified using a Qiagen (Valencia, CA) RNeasy kit according to the
manufacturer's instructions,
an acid-phenol purification protocol (Chomczynski and Sacchi, Analytical
Biochemistry, 162:156-9,
1987), or a Trizol reagent protocol (Invitrogen, Carlsbad, CA). 5ug of total
RNA from each cell line
was arranged in a deep well 96-well plate, 125u1 3M NaOAc and 100u1 Pellet
Paint (Novagen,
Madison, WI)) were added to each well, then the final volume was adjusted to
1.25m1 with H20. A
Multipette (Saigan) robot was used to aliquot 25u1 of the RNA mixture followed
by 75u1 EtOH into
each well of a 96-well PCR plate multiple times, generating numerous one-use
RT PCR panels of the
cell lines, each well with 100ng total RNA in EtOH. Panels were then sealed
and stored at -20 C.
The arrangement and content of the samples on this array are detailed below in
Table 1. RT PCR
screening was performed by first centrifuging a panel in a Qiagen (Valencia,
CA) 96-well centrifuge
for 10' at 6000 RPM. Supernatant was removed by inverting the plate onto
absorbent paper. RNA
pellets were washed with 100u1 70% EtOH, followed by a 5' centrifugation at
6000 RPM.
Supernatant was again removed and plates allowed to air-dry until the
remaining EtOH was
evaporated.
[428] Expression of IL-17REm mRNA in the mouse cell line RNA panels was
assayed by
RT PCR with sense oligo SEQ ID NO:81 and antisense oligo SEQ ID NO:82 using
Superscript One-
Step RT PCR reagents (Invitrogen, Carlsbad, CA). RNA pellets were resuspended
in a total volume
of 25u1/well reaction mix that contained 2.5u1 lOX Rediload (Invitrogen,
Carlsbad, CA), 12.5u1 2X
Reaction Mix, 0.5u1 of 20pmoUul sense oligo, 0.5u1 of 20pmoUul antisense
oligo, 0.5u1 RT/Platinum
Taq and 8.5u1 sterile water. Cycling conditions were:l cycle at 52 C for 30
minutes, 1 cycle at 94 C
for 2 minutes, 35 cycles at 94 C for 30 seconds, 55 C for 30 seconds and 72 C
for 1 minute,
followed by a final cycle at 72 C for 7 minutes. 10u1 of each reaction was
subjected to agarose gel
electrophoresis and gels were scored for positive or negative expression of IL-
17REm. The primers
were predicted to pick up all known splice variants and not produce a product
on contaminating
genomic DNA. Results indicated presence of IL-17RE mRNA in 14 cell lines, most
representing
lines of pancreatic origin: pik10, pik15, pik18, pik 34, pid14, pid20 5FU-17
and 5FU-19. IL-17RE
mRNA was also present in C2C 12, a skeletal muscle myoblast cell line, RAW
264.7, a monocyte cell
line, SAG-5/22-6, a salivary gland cell line, and AML, a liver cell line. In
constrast, IL-17REm RNA
was not expressed in T or B lymphocyte cell lines, embryonic cell lines,
adipocyte cell lines,
osteoblast and osteoclast cell lines, and hypothalamus cell lines. There were
also 10 pancreatic cell
lines and 4 salivary gland cell lines that did not express IL-17RE.
EXAMPLE 24
Construction of Mammalian Soluble IL-17REx1 Expression Constructs that Express
IL-
17REx1CEE, IL-17REx1CHIS, and IL-17REx1CFLAG Tagged Proteins
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
117
[429] An expression construct containing the extracellular domain of human IL-
17REx1
with a C-terminal tag, either Glu-Glu (CEE), six His (CHIS), or FLAG (CFLAG),
is constructed via
PCR and homologous recombination using a DNA fragment encoding IL-17REx1 (SEQ
ID NO: 83)
and the expression vector pZMP20.
[430] The PCR fragment encoding IL-17REx1CEE contains a 5' overlap with the
pZMP20 vector sequence in the optimized tissue plasminogen activator pre-pro
secretion leader
sequence coding region, the IL-17REx1 extracellular domain coding region (SEQ
ID NO: 84), the
Glu-Glu tag (Glu Glu Tyr Met Pro Met Glu) coding sequence, and a 3' overlap
with the pZMP20
vector in the poliovirus internal ribosome entry site region. The PCR
amplification reaction uses the
following 5' oligonucleotide (GTTTCGCTCAGCCAGGAAATCCATGCCGAGTTGAGACGCTTC
CGTAGAGCTGGGATTGGCTTTCGCCAC) (SEQ ID NO:85), the following 3' oligonucleotide
(CAACCCCAGAGCTGTTTTAAGGCGCGCCTCTAGATTATTCCATGGGCATGTATTCTTCGT
AAGAGACATCTGGACACA) (SEQ ID NO:86), and a previously generated DNA clone of IL-
17REx1 as the template (SEQ ID NO:83).
[431] The PCR amplification reaction condition is as follows: 1 cycle, 94 C,
5 minutes;
35 cycles, 94 C, 1 minute, followed by 55 C, 2 minutes, followed by 72 C, 3
minutes; 1 cycle, 72
C, 10 minutes. The PCR reaction mixture is run on a 1% agarose gel and the DNA
fragment
corresponding to the expected size is extracted from the gel using a
QlAquickT"" Gel Extraction Kit
(Qiagen, Cat. No. 28704).
[432] Plasmid pZMP20 is a mammalian expression vector containing an expression
cassette having the chimeric CMV enhancer/MPSV promoter, a BglII site for
linearization prior to
yeast recombination, an otPA signal peptide sequence, an internal ribosome
entry element from
poliovirus, the extracellular domain of CD8 truncated at the C-terminal end of
the transmembrane
domain; an E. coli origin of replication; a mammalian selectable marker
expression unit comprising
an SV40 promoter, enhancer and origin of replication, a DHFR gene, and the
SV40 terminator; and
URA3 and CEN-ARS sequences required for selection and replication in S.
cerevisiae.
[433] The plasmid pZMP20 is digested with BglII prior to recombination in
yeast with the
gel extracted IL-17REx1CEE PCR fragment. One hundred 1 of competent yeast (S.
cerevisiae) cells
are combined with 10 l of the IL-17REx1CEE insert DNA and 100 ng of BglII
digested pZMP20
vector, and the mix is transferred to a 0.2 cm electroporation cuvette. The
yeast/DNA mixture is
electropulsed using power supply (BioRad Laboratories, Hercules, CA) settings
of 0.75 kV (5
kV/cm), co ohms, and 25 F. Six hundred 1 of 1.2 M sorbitol is added to the
cuvette, and the yeast is
plated in 100 l and 300 l aliquots onto two URA-D plates and incubated at 30
C. After about 72
hours, the Ura+ yeast transformants from a single plate are resuspended in 1
ml H20 and spun briefly
to pellet the yeast cells. The cell pellet is resuspended in 0.5 ml of lysis
buffer (2% Triton X-100,
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
118
1% SDS, 100 mM NaC1, 10 mM Tris, pH 8.0, 1 mM EDTA). The five hundred l of
the lysis
mixture is added to an Eppendorf tube containing 250 l acid-washed glass
beads and 300 l phenol-
chloroform, is vortexed for 3 minutes, and spun for 5 minutes in an Eppendorf
centrifuge at
maximum speed. Three hundred 1 of the aqueous phase is transferred to a fresh
tube, and the DNA
is precipitated with 600 l ethanol, followed by centrifugation for 30 minutes
at maximum speed.
The tube is decanted and the pellet is washed with 1 mL of 70% ethanol. The
tube is decanted and
the DNA pellet is resuspended in 30 l 10 mM Tris, pH 8.0, 1 mM EDTA.
[434] Transformation of electrocompetent E. coli host cells (DH12S) is done
using 5 l of
the yeast DNA preparation and 50 l of E. coli cells. The cells are
electropulsed at 2.0 kV, 25 F,
and 400 ohms. Following electroporation, 1 ml SOC (2% BactoTM Tryptone (Difco,
Detroit, MI),
0.5% yeast extract (Difco), 10 mM NaC1, 2.5 mM KC1, 10 mM MgC1z, 10 mM MgS04,
20 mM
glucose) is added and then the cells are plated in 50 l and 200 l aliquots
on two LB AMP plates
(LB broth (Lennox), 1.8% BactoTM Agar (Difco), 100 mg/L Ampicillin).
[435] The inserts of three DNA clones for the construct are subjected to
sequence analysis
and one clone containing the correct sequence is selected. Large-scale plasmid
DNA is isolated using
a commercially available kit (QIAGEN Plasmid Mega Kit, Qiagen, Valencia, CA)
according to
manufacturer's instructions.
[436] The same process is used to prepare the IL-17REx1 with a C-terminal his
tag,
composed of Gly Ser Gly Gly His His His His His His (SEQ ID NO:87) (IL-
17REx1CHIS) or the C-
terminal FLAG tag, composed of Gly Ser Asp Tyr Lys Asp Asp Asp Asp Lys (SEQ ID
NO:88) (IL-
17REx1CFLAG). To prepare these constructs, the following 3' oligonucleotide
(CAACCCCAGAGCTGTTTTAAGGCGCGCCTCTAGATTAGTGATGGTGATGGTGATGTCCA
CCAGATCCGTAAGAGACATCTGGACACA) (SEQ ID NO:89) is used to generate IL-
17REx1CHIS or the 3' oligonucleotide (CAACCCCAGAGCTGTTTTAAGGCGCGCCTCTAGAT
TACTATCATCATCATCCTTATAATCGGATCCGTAAGAGACATCTGGACACA) (SEQ ID NO:
90) is used to generate IL-17REx1CFLAG.
EXAMPLE 25
Transfection and Expression of Soluble IL-17REx1 Receptor Expression
Constructs that
Express the IL-17REx1CEE, IL-17REx1CHIS, and IL-17REx1CFLAG C-Terminal Tagged
Proteins
[437] Three sets of 200 g of each of the soluble IL-17REx1 tagged expression
constructs,
as described in Example 22, are separately digested with 200 units of Pvul at
37 C for three hours,
precipitated with isopropyl alcohol, and centrifuged in a 1.5 mL microfuge
tube. The supernatant is
decanted off the pellet, and the pellet is washed with 1 mL of 70% ethanol and
allowed to incubate
for 5 minutes at room temperature. The tube is spun in a microfuge for 10
minutes at 14,000 RPM
and the supernatant is decanted off the pellet. The pellet is then resuspended
in 750 l of CHO cell
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
119
tissue culture medium in a sterile environment, allowed to incubate at 60 C
for 30 minutes, and is
allowed to cool to room temperature. Approximately 5 x 106 CHO cells are
pelleted in each of three
tubes and are resuspended using the DNA-medium solution. The DNA/cell mixtures
are placed in a
0.4 cm gap cuvette and electroporated using the following parameters; 950 F,
high capacitance, at
300 V. The contents of the cuvettes are then removed, pooled, and diluted to
25 mLs with CHO cell
tissue culture medium and placed in a 125 mL shake flask. The flask is placed
in an incubator on a
shaker at 37 C, 6% COz with shaking at 120 RPM.
[438] The CHO cells are subjected to nutrient selection followed by step
amplification to
200 nM methotrexate (MTX), and then to 1 M MTX. Tagged protein expression is
confirmed by
Western blot, and the CHO cell pool is scaled-up for harvests for protein
purification.
EXAMPLE 26
Construction of Mammalian Soluble IL-17REx2 Expression Constructs that Express
IL-
17REx2CEE, IL-17REx2CHIS, and IL-17REx2CFLAG Tagged Proteins
[439] An expression construct containing the extracellular domain of human IL-
17REx2
with a C-terminal tag, either Glu-Glu (CEE), six His (CHIS), or FLAG (CFLAG)
(Example 22), is
constructed via PCR and homologous recombination using a DNA fragment encoding
IL-17REx2
(SEQ ID NO:91) and the expression vector pZMP20.
[440] The PCR fragment encoding IL-17REx2CEE contains a 5' overlap with the
pZMP20 vector sequence in the optimized tissue plasminogen activator pre-pro
secretion leader
sequence coding region, the IL-17REx2 extracellular domain coding region (SEQ
ID NO: 92), the
Glu-Glu tag (Glu Glu Tyr Met Pro Met Glu) coding sequence, and a 3' overlap
with the pZMP20
vector in the poliovirus internal ribosome entry site region. The PCR
amplification reaction uses the
5' oligonucleotide (GTTTCGCTCAGCCAGGAAATCCATGCCGAGTTGAGACGCTTCCGTAGA
GCTGGGATTGGCTTTCGCCAC) (SEQ ID NO:93), the 3' oligonucleotide (CAACCCCAGAGCT
GTTTTAAGGCGCGCCTCTAGATTATTCCATGGGCATGTATTCTTCGTAAGAGACATCTGG
ACACA) (SEQ ID NO:94), and a previously generated DNA clone of IL-17REx2 as
the template
(SEQ ID NO: 91).
[441] The PCR amplification reaction condition is as follows: 1 cycle, 94 C,
5 minutes;
35 cycles, 94 C, 1 minute, followed by 55 C, 2 minutes, followed by 72 C, 3
minutes; 1 cycle, 72
C, 10 minutes. The PCR reaction mixture is run on a 1% agarose gel and the DNA
fragment
corresponding to the expected size is extracted from the gel using a
QlAquickT"" Gel Extraction Kit
(Qiagen, Cat. No. 28704).
[442] Plasmid pZMP20 is a mammalian expression vector containing an expression
cassette having the chimeric CMV enhancer/MPSV promoter, a BglII site for
linearization prior to
yeast recombination, an otPA signal peptide sequence, an internal ribosome
entry element from
poliovirus, the extracellular domain of CD8 truncated at the C-terminal end of
the transmembrane
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
120
domain; an E. coli origin of replication; a mammalian selectable marker
expression unit comprising
an SV40 promoter, enhancer and origin of replication, a DHFR gene, and the
SV40 terminator; and
URA3 and CEN-ARS sequences required for selection and replication in S.
cerevisiae.
[443] The plasmid pZMP20 is digested with BglII prior to recombination in
yeast with the
gel extracted IL-17REx2CEE PCR fragment. One hundred 1 of competent yeast (S.
cerevisiae) cells
are combined with 10 l of the IL-17REx2CEE insert DNA and 100 ng of BglII
digested pZMP20
vector, and the mix is transferred to a 0.2 cm electroporation cuvette. The
yeast/DNA mixture is
electropulsed using power supply (BioRad Laboratories, Hercules, CA) settings
of 0.75 kV (5
kV/cm), co ohms, and 25 F. Six hundred 1 of 1.2 M sorbitol is added to the
cuvette, and the yeast is
plated in 100 l and 300 l aliquots onto two URA-D plates and incubated at 30
C. After about 72
hours, the Ura+ yeast transformants from a single plate are resuspended in 1
ml H20 and spun briefly
to pellet the yeast cells. The cell pellet is resuspended in 0.5 ml of lysis
buffer (2% Triton X-100,
1% SDS, 100 mM NaC1, 10 mM Tris, pH 8.0, 1 mM EDTA). The five hundred 1 of
the lysis
mixture is added to an Eppendorf tube containing 250 1 acid-washed glass
beads and 300 l phenol-
chloroform, is vortexed for 3 minutes, and spun for 5 minutes in an Eppendorf
centrifuge at
maximum speed. Three hundred 1 of the aqueous phase is transferred to a fresh
tube, and the DNA
is precipitated with 600 l ethanol, followed by centrifugation for 30 minutes
at maximum speed.
The tube is decanted and the pellet is washed with 1 mL of 70% ethanol. The
tube is decanted and
the DNA pellet is resuspended in 30 l 10 mM Tris, pH 8.0, 1 mM EDTA.
[444] Transformation of electrocompetent E. coli host cells (DH12S) is done
using 5 l of
the yeast DNA preparation and 50 l of E. coli cells. The cells are
electropulsed at 2.0 kV, 25 F,
and 400 ohms. Following electroporation, 1 ml SOC (2% BactoTM Tryptone (Difco,
Detroit, MI),
0.5% yeast extract (Difco), 10 mM NaC1, 2.5 mM KC1, 10 mM MgC1z, 10 mM MgS04,
20 mM
glucose) is added and then the cells are plated in 50 l and 200 l aliquots
on two LB AMP plates
(LB broth (Lennox), 1.8% BactoTM Agar (Difco), 100 mg/L Ampicillin).
[445] The inserts of three DNA clones for the construct are subjected to
sequence analysis
and one clone containing the correct sequence is selected. Large-scale plasmid
DNA is isolated using
a commercially available kit (QIAGEN Plasmid Mega Kit, Qiagen, Valencia, CA)
according to
manufacturer's instructions.
[446] The same process is used to prepare the IL-17REx2 with a C-terminal his
tag,
composed of Gly Ser Gly Gly His His His His His His (SEQ ID NO:95) (IL-
17REx2CHIS) or the C-
terminal FLAG tag, composed of Gly Ser Asp Tyr Lys Asp Asp Asp Asp Lys (SEQ ID
NO:96) (IL-
17REx2CFLAG). To prepare these constructs, the 3' oligonucleotide
(CAACCCCAGAGCTGTTTT
AAGGCGCGCCTCTAGATTAGTGATGGTGATGGTGATGTCCACCAGATCCGTAAGAGACA
TCTGGACACA) (SEQ ID NO:97) is used to generate IL-17REx2CHIS or the 3'
oligonucleotide
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
121
(CAACCCCAGAGCTGTTTTAAGGCGCGCCTCTAGATTACTTATCATCATCATCCTTATAAT
CGGATCCGTAAGAGACATCTGGACACA) (SEQ ID NO:98) is used to generate IL-
17REx2CFLAG. IL-17C with a 6HIS tag is similarly prepared ("IL-17C CH6").
EXAMPLE 27
Transfection and Expression of Soluble IL-17REx2 Receptor Expression
Constructs that
Express the IL-17REx2CEE, IL-17REx2CHIS, and IL-17REx2CFLAG C-Terminal Tagged
Proteins
[447] Three sets of 200 g of each of the soluble IL-17REx2 tagged expression
constructs,
described in Example 24, are separately digested with 200 units of Pvul at 37
C for three hours,
precipitated with isopropyl alcohol, and centrifuged in a 1.5 mL microfuge
tube. The supernatant is
decanted off the pellet, and the pellet is washed with 1 mL of 70% ethanol and
allowed to incubate
for 5 minutes at room temperature. The tube is spun in a microfuge for 10
minutes at 14,000 RPM
and the supernatant is decanted off the pellet. The pellet is then resuspended
in 750 l of CHO cell
tissue culture medium in a sterile environment, allowed to incubate at 60 C
for 30 minutes, and is
allowed to cool to room temperature. Approximately 5 x 106 CHO cells are
pelleted in each of three
tubes and are resuspended using the DNA-medium solution. The DNA/cell mixtures
are placed in a
0.4 cm gap cuvette and electroporated using the following parameters; 950 F,
high capacitance, at
300 V. The contents of the cuvettes are then removed, pooled, and diluted to
25 mLs with CHO cell
tissue culture medium and placed in a 125 mL shake flask. The flask is placed
in an incubator on a
shaker at 37 C, 6% COz with shaking at 120 RPM.
[448] The CHO cells are subjected to nutrient selection followed by step
amplification to
200 nM methotrexate (MTX), and then to 1 M MTX. Tagged protein expression is
confirmed by
Western blot, and the CHO cell pool is scaled-up for harvests for protein
purification.
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
122
EXAMPLE 28
Transfection and Expression of IL-17C Expression Constructs that
Express the IL-17C-CEE, IL-17C-CHIS, and IL-17C-CFLAG
C-Terminal Tagged Proteins
[449] Expression constructs of IL-17C fusion or tagged constructs were used to
transfect
baby hamster kidney cells (BHK) by the lipofectamine method. Specifically,
1x106 BHK cells were
seeded on to a 100 mm dish in Dulbeccos Modified Eagle Media (DMEM) containing
10% fetal
bovine serum, 10 mM Hepes, pH 7.2 and incubated overnight at 37 C. The
attached cells were
rinsed with 10 ml of Serum Free Media(SFM): DMEM/F12(Ham) media(1:1) which
also contained
mM Hepes, 1 ug/ml insulin, 4 ng/ml selenium dioxide, 25 uM ferric citrate. A
16 ug aliquot of
an expression construct containing the cDNA for IL-17C -CEE was complexed with
35 ul of
lipofectamine (Invitrogen, Inc.) in 1.2 ml of SFM for 20 minutes and then
following dilution with
SFM, applied to the plated BHK cells. Following a 5 hr incubation at 37 C, 6.5
mls of DMEM
containing 10% fetal bovine serum was added. The cells were cultured overnight
at 37 C in a
humidified tissue culture incubator. Approximately 24 hrs after transfection,
the cell media was
replaced with fresh DMEM containing 10% fetal bovine serum and also containing
1 uM
methotrexate (MTX). After 7 days in 1 uM MTX, the MTX concentration was
increased to 10 uM
and the cells were allowed to grow for an additional 7-10 days. The cells were
maintained in culture
to recover the MTX resistant clones and the media was evaluated for the
expression of IL-17C-CEE
by polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting using an
anti-EE peptide
antibody (EE peptide = Glu-Glu tag (Glu Glu Tyr Met Pro Met Glu). The IL-17C-
CEE producing
cells were then scaled-up for production of recombinant protein. It is well
known in the art that for
this process, expression contructs containing alternative fusion proteins such
as Fc sequences or
other tag sequences (His, Flag, etc.) may be substituted for the EE peptide
sequenced described here.
EXAMPLE 29
Purification of IL-17C CEE from BHK Cells
[450] Conditioned media from BHK cells expressing IL-17C-CEE (Example 26) was
0.2
.micro.m sterile filtered and then loaded on to an anti-EE peptide (EE peptide
= Glu-Glu tag (Glu
Glu Tyr Met Pro Met Glu) antibody affinity column by loading at 4 C. Prior to
loading the pH the
conditioned media and the anti-EE antibody-column were adjusted to pH 7.4.
[451] Following the loading of media on to the column, the column was washed
with 10
column volumes of 20mM Tris, 500mM NaC1, pH7.4. Bound protein was then eluted
with 3 column
volumes of phosphate buffered saline containing 0.5 mg/ml of EE peptide (Glu
Glu Tyr Met Pro Met
Glu). Fractions were collected and were analyzed via SDS-PAGE Coomassie
staining. Fractions
containing IL-17C-CEE were pooled and concentrated approximately 10-fold using
a l OkD
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
123
molecular weight cutoff Ultrafree- 15 membrane concentrator (Millipore,
Bedford, MA) according to
the manufacturer's instructions.
[452] The concentrated sample was then subjected to size exclusion
chromatography on a
Sephacryl-S100 column (16/60) (Pharmacia, Piscataway, NJ) equilibrated in 10
mM sodium
phosphate, 150 mM NaC1, pH 7.2. The eluted protein was collected in 3 ml
fractions which and
were analyzed via SDS-PAGE Coomassie staining. The fractions containing pure
IL-17C-CEE were
pooled and following 0.22 um sterile filtration, the protein was aliquoted and
stored at -80 C until
use. N-terminal sequencing of the pure protein confirmed its identity as IL-
17C. The analysis of the
recombinant protein shows an N-terminal sequence of the mature protein,
lacking the signal
sequence, begins at Histidine- 19 and has a molecular weight of 20663 which
includes the C-terminal
EE-tag.
EXAMPLE 30
Purification of His-tagged IL-17C from 293F Transient Cells
[453] The following procedure was used to purify both human and murine forms
of IL17C
having polyhistidine fused at their carboxy-termini. The purification was
performed at 4 C. About
L of conditioned media from 293F cells transfected with His-tagged IL-17C was
concentrated to
1.6 L using Pellicon 2 5k filters (Millipore, Bedford, MA). Imidazole and NaC1
were added to the
1.6 L media to a final concentration of 15 mM and 0.5 M respectively. A Talon
(BD Biosciences)
column with a 5 mL bed-volume was packed and equilibrated with 20 mM NaPi, 15
mM Imidazole,
0.5 M NaC1, pH 7.5. The media was loaded onto the column at a flow-rate of 1.7
mL/min then
washed with 10 CV of the equilibration buffer. His-tagged IL17C was eluted
from the column with
mM NaPi, 0.5 M NaC1, 0.5 M Imidazole, pH 7.4 at a flow-rate of 1 mL/min. 2 mL
fractions were
collected and analyzed for the presence of His-tagged IL17C by Coomassie-
stained SDS-PAGE.
[454] Talon column elution pool was concentrated from 12 mL to 1 mL using an
Amicon
Ultra 5k centrifugal filter (Millipore, Bedford, MA). A Superdex 75 column
with a bed-volume of
121 mL was equilibrated with 50 mM NaPi, 109 mM NaC1, pH 7.3, and the 1 mL
sample was
injected into the column at a flow-rate of 0.5 mL/min. 2 mL fractions were
collected and analyzed
for the presence of His-tagged IL17C by Coomassie-stained SDS-PAGE. Fractions
containing pure
His-tagged IL17C were pooled and concentrated to 2 mL, sterile-filtered
through a 2^m Acrodisc
filter (Pall Corporation), and stored at -80 C. Concentration of the final
sample was determined by
BCA (Pierce, Rockford, IL).
EXAMPLE 31
IL-17RE FclO Fusion Protein Expression Constructs
[455] Expression plasmids containing either IL-17REx1-C(Fc10) (SEQ ID NO:99;
SEQ
ID NO:100) or IL-17REx2-C(Fc10) (SEQ ID NO:101; SEQ ID NO:102) were
constructed via
homologous recombination using DNA fragments encoding the gene of interest and
the expression
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
124
vector pZMP40. Fragments of polynucleotide sequence of IL-17REx1 (SEQ ID NO:1)
and IL-
17REx2 (SEQ ID NO:4) were generated by PCR amplification using primer zc48706
(SEQ ID
NO:103), zc48707 (SEQ ID NO:104) and zc48708 (SEQ ID NO:105).
[456] The fragments for both IL-17REx1 and IL-17REx2 both contained the
extracellular
domain of their respective coding regions, which was made using previously
generated clones of
either IL-17REx1 or IL-17REx2 as templates. The fragments both included a 5'
overlap with a
partial pZMP40 vector sequence, either the IL-17REx1 or IL-17REx2 segment, a
linker sequence, a
Caspase-3 cleavage site, and a linker region encoding the first 5 amino acids
of Fc 10 followed by a
3' overlap containing a partial pZMP40 vector sequence. PCR conditions: 1
cycle, 94 C, 5 minutes;
35 cycles, 94 C, 1 minute, followed by 55 C, 2 minutes, followed by 72 C, 3
minutes; 1 cycle,
72 C, 10 minutes.
[457] The PCR reaction mixtures were run on a 1% agarose gel and a band
corresponding
to the sizes of the inserts were gel-extracted using a QlAquickT"" Gel
Extraction Kit (Qiagen, Cat. No.
28704).
[458] The plasmid pZMP40, which was cut with Bg1II, was used in a
recombination
reaction using either one or the other of the PCR insert fragments. Plasmid
pZMP40 is a mammalian
expression vector containing an expression cassette having the MPSV promoter,
multiple restriction
sites for insertion of coding sequences, and an Fc9 coding region; an E. coli
origin of replication; a
mammalian selectable marker expression unit comprising an SV40 promoter,
enhancer and origin of
replication, a DHFR gene, and the SV40 terminator; and URA3 and CEN-ARS
sequences required
for selection and replication in S. cerevisiae. It was constructed from pZP9
(deposited at the
American Type Culture Collection, 10801 University Boulevard, Manassas, VA
20110-2209, under
Accession No. 98668) with the yeast genetic elements taken from pRS316
(deposited at the
American Type Culture Collection, 10801 University Boulevard, Manassas, VA
20110-2209, under
Accession No. 77145), an internal ribosome entry site (IRES) element from
poliovirus, and the
extracellular domain of CD8 truncated at the C-terminal end of the
transmembrane domain.
[459] One hundred microliters of competent yeast (S. cerevisiae) cells were
independently
combined with 10 l of the insert DNA and 100ng of cut pZMP40 vector, and the
mix was
transferred to a 0.2-cm electroporation cuvette. The yeast/DNA mixture was
electropulsed using
power supply (BioRad Laboratories, Hercules, CA) settings of 0.75 kV (5
kV/cm), co ohms, and 25
F. Six hundred 1 of 1.2 M sorbitol was added to the cuvette, and the yeast
was plated in a 100- 1
and 300 1 aliquot onto two URA-D plates and incubated at 30 C. After about 72
hours, the Ura+
yeast transformants from a single plate were resuspended in 1 ml H20 and spun
briefly to pellet the
yeast cells. The cell pellet was resuspended in 0.5 ml of lysis buffer (2%
Triton X-100, 1% SDS,
100 mM NaC1, 10 mM Tris, pH 8.0, 1 mM EDTA). The five hundred microliters of
the lysis
mixture was added to an Eppendorf tube containing 250 l acid-washed glass
beads and 300 l
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
125
phenol-chloroform, was vortexed for 3 minutes, and spun for 5 minutes in an
Eppendorf centrifuge at
maximum speed. Three hundred microliters of the aqueous phase was transferred
to a fresh tube, and
the DNA was precipitated with 600 l ethanol (EtOH) and 30 1 3M sodium
acetate, followed by
centrifugation for 30 minutes at maximum speed. The tube was decanted and the
pellet was washed
with 1 mL of 70% ethanol. The tube was decanted and the DNA pellet was
resuspended in 30 l TE.
[460] Transformation of electrocompetent E. coli host cells (DH12S) was done
using 5 l
of the yeast DNA prep and 50 1 of cells. The cells were electropulsed at 2.0
kV, 25 F, and 400
ohms. Following electroporation, 1 ml SOC (2% BactoTM Tryptone (Difco,
Detroit, MI), 0.5% yeast
extract (Difco), 10 mM NaC1, 2.5 mM KC1, 10 mM MgC1z, 10 mM MgS04, 20 mM
glucose) was
added and then the cells were plated in a 50 l and 200 l aliquot on two LB
AMP plates (LB broth
(Lennox), 1.8% BactoTM Agar (Difco), 100 mg/L Ampicillin).
[461] The inserts of three clones for the construct was subjected to sequence
analysis and
one clone for each construct, containing the correct sequence, was selected.
Larger scale plasmid
DNA was isolated using a commercially available kit (QIAGEN Plasmid Mega Kit,
Qiagen,
Valencia, CA) according to manufacturer's instructions.
[462] Three sets of 200 g of the IL-17REx1-C(Fc10) construct (SEQ ID NO:99)
were
each digested with 200 units of Pvu I at 37 C for three hours and then were
precipitated with IPA
and spun down in a 1.5 mL microfuge tube. The supernatant was decanted off the
pellet, and the
pellet was washed with 1 mL of 70% ethanol and allowed to incubate for 5
minutes at room
temperature. The tube was spun in a microfuge for 10 minutes at 14,000 RPM and
the supernatant
was decanted off the pellet. The pellet was then resuspended in 750 l of PF-
CHO media in a sterile
environment, and allowed to incubate at 60 C for 30 minutes. 5E6 APFDXBII
cells were spun
down in each of three tubes and were resuspended using the DNA-media solution.
The DNA/cell
mixtures were placed in a 0.4 cm gap cuvette and electroporated using the
following parameters:
950 F, high capacitance, and 300 V. The contents of the cuvettes were then
removed, pooled, and
diluted to 25 mLs with PF-CHO media and placed in a 125 mL shake flask. The
flask was placed in
an incubator on a shaker at 37 C, 6% C02, and shaking at 120 RPM. Protein
expression was
confirmed via western blot.
[463] The cell line was subjected to nutrient selection followed by step
amplification to
lOOnM methotrexate (MTX), then to 500nM MTX. Step amplification was followed
by a CD8 cell
sort. The CD8 cell sort was accomplished by taking a stable 500 nM MTX
amplified pool and
staining approximately 5E6 cells with a monoclonal FITC anti-CD8 antibody (BD
PharMingen, cat#
30324X) using manufacturers recommended concentration. The stained cells were
processed and
sorted on a FACS Aria (BD) flow cytometer. The top 5% of cells were collected
and outgrown.
EXAMPLE 32
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
126
Phosphoprotein Assay for Detection of Receptor-Ligand Interactions
[464] Specific receptor-ligand binding results in activation of intracellular
signaling
pathways that can be detected in several different ways. Within minutes of
specific receptor-ligand
binding, changes occur in the phosphorylation state of kinases and
transcription factors within the
signaling pathways that result in activation or inactivation of downstream
cellular responses
including proliferation, apoptosis, cell adhesion, inflammatory responses,
etc. Activation of these
signaling pathways can be detected through use of antibodies that specifically
recognize the
phosphorylated forms of the kinases or transcription factors. The changes in
phosphoprotein levels
can be detected and quantitated by Western blotting, by standard ELISA
methods, or in multiplexed
immunoassays using commercial kits based on Luminex detection technology, such
as the BioRad
Bio-Plex Suspension Array System.
[465] The BioRad Bio-Plex assay system is a bead based assay system similar to
a capture
sandwich immunoassay. Antibody directed against the desired target protein,
(total transcription
factor or kinase) is covalently coupled to internally dyed fluorescent beads.
Coupled beads are
allowed to react with lysate containing the target protein. After a series of
washes to remove unbound
protein, a biotinylated detection antibody specific for a different epitope,
directed against the
phosphorylated form of the target protein (phosphorylated transcription factor
or kinase) is added.
This results in formation of a sandwich around the target protein.
Streptavidin-phycoerythrin is
added to bind the biotinylated detection antibody. Antibodies coupled to beads
with different
fluorescent dyes can be run separately or in combination so that multiple
target proteins can be
measured simultaneously on the BioRad Bio-Plex Suspension Array System in
combination with the
BioRad Bio-Plex ManagerTM 3.0 software. Up to 100 different target proteins
can be assayed
simultaneously in this fashion. An example of a multiplexed assay format is
the simultaneous
measurement of phosphorylated forms of ERK1/2, JNK, p38 MapKinase, Akt, ATF-2,
STAT-3, and
Ix(3a.
[466] The binding and activation of IL-17RE by IL-17C or other specific
ligands can be
detected by using cell lines endogenously expressing the receptor (as
determined by RT-PCR).
Alternatively, cells overexpressing a transfected IL-17RE receptor can be used
(NIH3T3/KZ142.8
cells overexpressing a transfected IL-17RE receptor, as in Example 17).
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
127
Treatment of cells
[467] Cell lines expressing endogenous or transfected IL-17RE are plated at
5000
cells/well in 96 well tissue culture plates and grown overnight in complete
growth medium. Cells are
cultured for an additiona124 hours in serum free growth medium and then
treated for 7 and 15
minutes with IL-17C at varying concentrations up to 300 ng/mL. Additionally,
cells can be
incubated in the presence of known cytokines or growth factors in combination
with the IL-17RE
ligand(s) (IL-17C) to look at the ability of the IL-17RE ligand to enhance or
inhibit the signal
transduction of known factors.
Lysate Preparation
[468] Following incubation, cells are washed with 100 uL/well ice-cold wash
buffer, put
on ice, and 50 uL/well lysis buffer is added (BioPlex Cell Lysis Kit, Catalog#
171-304012). Lysates
are pipetted up and down five times while on ice, and then agitated on a
microplate platform shaker
at 300 rpm at 4 C for 20 minutes. Plates are centrifuged at 4 C for 20 minutes
at 4500 rpm.
Supernatants are collected and transferred to a new microtiter plate for
storage at -20 C until time of
phosphoprotein assay. The protein concentration in the lysate is determined
using BioRad's DC
protein assay or any standard method of determining total protein
concentration. Samples are
adjusted to 200-900 ug/mL total protein by addition of lysis buffer as needed.
Bio-Plex (Luminex) Phosphoprotein assay
[469] Capture beads (50uL/well) (beads coupled to primary antibody for
transcription
factor of interest) are added to 50 uL of lysate in a microtiter plate. The
aluminum foil covered plate
is incubated overnight at room temperature, with shaking at 300 rpm. The plate
is transferred plate
to microtiter vacuum apparatus and washed three times with assay buffer. After
addition of 25
uL/well detection antibody, the aluminum foil covered plate is incubated at
room temperature for 30
min, at 300 rpm. The plate is filtered and washed three times with assay
buffer. Streptavidin-PE (50
uL/well) is added and the aluminum foil covered plate is incubated at room
temperature for 15
minutes, with shaking at 300 rpm. The plate is filtered and washed two times
with bead resuspension
buffer. After the final wash, beads are resuspended in 125 uL/well of bead
suspension buffer, shaken
for 30 seconds, and read on Bio-Plex Suspension Array System according to
manufacturers
instructions. Data is analyzed using Bio-Plex Manager software. Changes in the
level of any of the
phosphorylated transcription factors present in the lysate are indicative of a
specific receptor-ligand
interaction.
Western analysis of phosphoprotein
[470] Lysate prepared as described above can also be analyzed using standard
Western
blotting protocols and probed using phosphorylation state specific antibodies.
A receptor-ligand
interaction between IL-17RE and IL-17C (or other ligands) can be demonstrated
by change in the
intensity of the band of phosphorylated transcription factor present on the
gel.
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
128
EXAMPLE 33
Binding of Human IL-17C to Human IL-17REx1 and IL-17REx2,
and Human IL-17R
[471] 293fb cells that had been transfected with expression vectors encoding
human IL-
17REx1 and IL-17REx2 were assessed for their ability to bind biotinylated
human IL-17C. Control
transfections included 1) no DNA transfection, 2) vector only (pzmpl l), and
3) human IL17R. 106
cells were removed from transfected suspension cultures at day 1, day 2, day
3, and day 5. Cells were
pelleted and resuspended in 100u1 of staining media (SM), which is HBSS plus 1
mg/ml bovine
serum albumin (BSA), 10 mM Hepes, and 0.1% sodium azide (w/v). Biotinylated
human IL-17C
was incubated with the cells on ice for 45 minutes at a concentration of 1
ug/ml. An APC conjugated
anti-human CD8 antibody (BD Pharmingen; cat.#555369) was also added at 1:25
dilution. After 30
minutes, excess cytokine and antibody was washed away with SM and the cells
were incubated with
a 1:100 dilution of streptavidin conjugated to phycoerythrin (SA-PE; BD
Pharmingen; cat# 554061)
for 30 minutes on ice. Excess SA-PE was washed away and cells were analyzed by
flow cytometry.
[472] The amount of cytokine binding was detected from the change in the mean
fluorescence intensity of the cytokine staining relative to negative controls -
1) no DNA transfection
and 2) vector only. From this analysis, we fmd that human IL-17C binds both
the human IL-17REx1
and IL-17REx2, although binding to IL-17REx2 is significantly greater. Binding
was also seen to
human IL-17R.
[473] Baby hamster kidney cells were then transfected with expression vectors
as
described above, except that the cells were then subjected to methotrexate
drug selection to
selectively grow out only cells that had been transfected. Stable cell lines
were established and these
were assayed for CD8 expression and for binding of biotinylated IL-17C as
above. Consistent with
results obtained in analysis of transient transfections, only those BHK cell
lines that expressed IL-
17REx2 and xl forms bound to IL-17C, with x2 binding IL-17C better than the xl
form.
EXAMPLE 34
IL-17C Binding to IL-17RE Variants
[474] BHK cells stably transfected with human and mouse IL-17RE splice
variants were
plated and grown to confluency in T-75 flasks. Cells were lifted off using a
non-protease reagent
such as Versene (Invitrogen 15040-066), pelleted, and resuspended in a
staining reagent (HBSS +
1%BSA + 0.1% NaAzide + 10mM HEPES) at 2 x 10e7 cells/ml and aliquoted to a 96-
well Costar
plate. IL-17C that has been labeled with biotin was independently added to
cells at a concentration of
lug/ml. The cell/ligand mixture can be incubated for lhr at 4 degrees. The
wells were washed lx in
staining reagent, and incubated in a secondary reagent containing staining
reagent plus Streptavidin-
PE (BD Pharmingen 554061) at a 1:100 ratio. The wells were incubated at 4 C in
the dark for 1 hr,
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
129
followed by a 2x wash in staining media. The cells were then resuspended in a
1:1 mixture of
staining media and Cytofix (BD Bioscience 554655) and incubated 10 minutes at
RT. The cells were
analysed by Flow Cytometry and by gating on the PE positive events for cells
that bound IL-17C.
[475] The results were as follows: IL-17RE splice variants that bound human IL-
17C are
IL-17REx1, IL-17REx2, IL-17REx6, IL-17REx13, and murine IL-17REx6. IL-17RE
splice variants
that bound murine IL-17C were as follows; IL-17REx1, IL-17REx2, IL-17REx4, IL-
17RE-S2, IL-
17REx6, IL-17REx13, and murine IL-17REx6. Murine IL-17C also bound murine IL17-
RA.
Furthermore, IL-17REx1, IL-17REx2, and IL-17REx3 did not bind any of the
following: IL-17A,
IL-17B, IL-17D, and IL-17F (all biotinylated human forms).
[476] 293F cells transiently transfected with IL-17RE splice variants using
Lipofectamine2000 (Invitrogen 11668-027) were stained as described above. The
IL-17RE splice
variants were engineered with the extra-cellular domain C-terminally linked to
a Flag Tag and GPI
linkage domain, as described in Examples 22 and 24. The Flag Tag was detected
with an anti-Flag-
FITC antibody at 1:100 (Sigma F-4049) following staining guidelines described
above.
[477] The results were as follows: IL-17RE splice variants that bound human IL-
17C are
IL-17REx1, IL-17REx2, IL-17RE-S2, IL-17REx4, IL-17REx6, IL-17REx13, and murine
IL-
17REx6. To a lesser extent, IL-17REx3 also bound human IL-17C.
EXAMPLE 35
Distribution of mRNA in cell line panels using PCR
[478] Human cell lines were grown in-house, some of which were treated with
various
agents as follows: PMA (phorbol-12-myristate-13-acetate) at lOng/ml plus
lonomycin at 0.5ug/ml
for 4 hours (these cell lines are labeled as "activated"), TNF alpha lOng/ml
for 48 hours, LPS
(Lipopolysaccharide) at 100ng/ml for 24 hours, SEB (Staphlyococcus enterotoxin
B) at lug/ml for
24 hours, and CTX (cholera toxin) at 50nM for 24 hours. RNA was purified using
a Qiagen
(Valencia, CA) RNeasy kit according to the manufacturer's instructions, or an
acid-phenol
purification protocol (Chomczynski and Sacchi, Analytical Biochemistry,
162:156-9, 1987). The
quality of the RNA was assessed by running an aliquot on an Agilent
Bioanalyzer. If the RNA was
significantly degraded, it was not used for subsequent creation of first
strand cDNA. Presence of
contaminating genomic DNA was assessed by a PCR assay on an aliquot of the RNA
with zc4 1011:
5'CTCTCCATCCTTATCTTTCATCAAC3'(SEQ ID NO: 140) and zc41012: 5'CTCTCTGCTGGCT
AAACAAAACAC3' (SEQ ID NO:141), primers that amplify a single site of
intergenic genomic
DNA. The PCR conditions for the contaminating genomic DNA assay were as
follows: 2.5u1 lOX
buffer and 0.5u1 Advantage 2 cDNA polymerase mix (BD Biosciences Clontech,
Palo Alto, CA), 2u1
2.5mM dNTP mix (Applied Biosystems, Foster City, CA), 2.5u1 10X Rediload
(Invitrogen,
Carlsbad, CA), and 0.5u1 20uM zc41011 and zc41012, in a final volume of 25 ul.
Cycling
parameters were 94 C 20", 40 cycles of 94 C 20" 60 C 1'20" and one cycle of 72
C 7'. 1 0u1 of each
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
130
reaction was subjected to agarose gel electrophoresis and gels were examined
for presence of a PCR
product from contaminating genomic DNA. If contaminating genomic DNA was
observed, the total
RNA was DNAsed using DNA-free reagents (Ambion, Inc, Austin, TX) according to
the
manufacturer's instructions, then retested as described above. Only RNAs which
appeared to be free
of contaminating genomic DNA were used for subsequent creation of first strand
cDNA.
[479] 20 ug total RNA from 90 cell lines were each brought to 98u1 with H20,
then split
into two 49u1 aliquots, each containing l0ug total RNA, and placed in two 96-
well PCR plates. To
each aliquot was added reagents for first strand cDNA synthesis (Invitrogen
First Strand cDNA
Synthesis System, Carlsbad, CA): 20u1 25mM MgC1z, lOul lOX RT buffer, lOul
0.1M DTT, 2u1
oligo dT, 2u1 RNAseOut. Then, to one aliquot from each cell line 2u1
Superscript II Reverse
Transcriptase was added, and to the corresponding cell line aliquot 2u1 H20
was added to make a
minus Reverse Transcriptase negative control. All samples were incubated as
follows: 25 C 10',
42 C 50', 70 C 15". Samples were arranged in deep well plates and diluted to
1.7m1 with H20. A
Multipette (Saigan) robot was used to aliquot 16.5u1 into each well of a 96-
well PCR plate multiple
times, generating numerous one-use PCR panels of the cell lines, which were
then sealed and stored
at -20 C. Each well in these panels represents first strand cDNA from
approximately 100ng total
RNA. The 180 samples are spread across two 96 well panels, array #119.01 and
#119.02. Quality of
first strand cDNA on the panels was assessed by a multiplex PCR assay on one
set of the panels
using primers to two widely expressed, but only moderately abundant genes,
CLTC (clathrin) and
TFRC (transferrin receptor C). 0.5u1 each of Clathrin primers zc42901: 5'CTCA
TATTGCTCAACTGTGTGAAAAG3' (SEQ ID NO: 142), zc42902: 5'TAGAAGCCACCTGAAC
ACAAATCTG3' (SEQ ID NO: 143), and TFRC primers zc42599: 5'ATCTTGCGTTGTATGTTGA
AAATCAATT3' (SEQ ID NO: 144), zc42600: 5'TTCTCCACCAGGTAAACAAGTCTAC3' (SEQ
ID NO:145), were mixed with 2.5u1 l OX buffer and 0.5u1 Advantage 2 cDNA
polymerase mix (BD
Biosciences Clontech, Palo Alto, CA), 2u1 2.5mM dNTP mix (Applied Biosystems,,
Foster City,
CA), 2.5u1 lOX Rediload (Invitrogen, Carlsbad, CA), and added to each well of
a panel of
array#119.01 and array #119.02. Cycling parameters were as follows: 94 C 20",
35 cycles of 94 C
20", 67 C 80", and one cycle of 72 C 7'. lOul of each reaction was subjected
to agarose gel
electrophoresis and gels were scored for the presence of a robust PCR product
for each gene specific
to the +RT wells for each cell line.
[480] Expression of mRNA in the first strand cDNA panels for IL-17C was
assayed by
PCR with sense oligo zc26004: 5'cactgctactcggctgaggaactgc3' (SEQ ID NO: 146)
and antisense
oligo zc20996: `5ttctgtggatagcggtcctcatc3' (SEQ ID NO:147) under these PCR
conditions per
sample: 2.5u1 lOX buffer and 0.5u1 advantage 2 cDNA polymerase mix (BD
Biosciences Clontech,
Palo Alto, CA), 2u1 2.5mM dNTP mix (Applied Biosystems, ), 2.5u1 lOX Rediload
(Invitrogen,
Carlsbad, CA), and 0.5u1 20uM each sense and antisense primer. Cycling
conditions were 94 C 2',
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
131
35 cycles of 94 C 30", 68 C 30", 72 C 1', and one cycle of 72 C 7'. lOul of
each reaction was
subjected to agarose gel electrophoresis and gels were scored for positive or
negative expression of
IL-17C.
[481] Results showed some cell lines had differential expression of IL-17C
depending on
whether they were treated with an agent. Cell lines which were negative in the
resting state and
positive for IL-17C in the activated or treated state were: the bone marrow
AML cell line KG-1, the
NHBE (normal human bronchial epithelial primary cells) cell line treated with
TNF alpha, LPS, or
SEB, and the U-937 monocyte cell line. Conversely, the Tanoue ALL B-cell line
and the Hodgkin's
lymphoma cell line KM-H2 appeared positive in the resting state while the
activated cell line RNA
was negative for IL-17C.
[482] Cell lines that were positive for IL-17C in both the stimulated and
resting states
were: DU-4475, U698, MN60, AML-193, DB, NK-92, Molt-4, UT-7, WeRI-Rb.1, CCRF-
HSB2,
and NCI-H929. Finally, the cell lines tested only in the resting state which
were positive for IL-17C
mRNA were: NCI-H716, NCI-H295R, MDA-MB-468, JAR, NIH: OVCAR-3, Sup-B15, NCI-
H69,
HEL-299, IMR-90, NIC-H292, BEAS2B, U2OS, HFLS-OA, MG-63, 5637, HK-2, Daudi,
and Hut
78.
[483] The overall results show that IL-17C is constitutively expressed in many
cell lines,
including several immune system-related cell lines, but there are a few cell
lines that begin
expressing IL-17C mRNA in response to activation by various agents. Of
particular interest is the
response of the bronchial primary epithelial cell line NHBE in producing IL-
17C mRNA when
treated with TNF alpha, LPS and SEB, which are all considered pro-inflammatory
compounds. This
suggests that IL-17C plays a role in the setting of inflammation.
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
132
EXAMPLE 36
Murine IL-17RE mRNA is Regulated in Select Tissues in Murine Models of Disease
Compared
to Non-Disease Tissues
Experimental Protocol
[484] Tissues were obtained from the following murine models of disease:
Colitis, Asthma,
Experimental Allergic Encephalomyelitis (EAE), Psoriasis and Collagen Induced
Arthritis (CIA).
Animal models were run following standard procedures and included appropriate
non-diseased
controls. Colitis was induced by dextran sulfate sodium (DSS) in the drinking
water and the tissues
isolated from the model included distal colon, proximal colon and mesenteric
lymph nodes. Asthma
was induced by sensitization and intranasal challenge to the antigen
ovalbumin. The tissues isolated
included lung, spleen and lymph node. EAE was induced by immunizing with MOG35-
55 peptide in
RIBI adjuvant. Tissues isolated included brain, cervical, lymph node, and
spinal cord. Psoriasis was
induced by adoptive transfer of naive T cells into minor histocompatibility
mismatched or syngeneic
immunocompromised mice. Tissues isolated included lesional skin and adjacent
skin. CIA was
induced by collagen injections and tissues isolated included foot and
popliteal lymph node. RNA was
isolated from all tissues using standard procedures. In brief, tissues were
collected and immediately
frozen in liquid N2 and then transferred to -80 C until processing.
[485] For processing, tissues were placed in Qiazol reagent (Qiagen, Valencia,
CA) and
RNA was isolated using the Qiagen RNAeasy kit according to manufacturer's
recommendations.
Expression of murine IL-17RE mRNA was measured with multiplex real-time
quantitative RT-PCR
method (TaqMan) and the ABI PRISM 7900 sequence detection system (PE Applied
Biosystems). IL-
17RE mRNA levels were normalized to the expression of the murine hypoxanthine
guanine
physphoribosyl transferase mRNA and determined by the comparative threshold
cycle method (User
Bulletin 2; PE Applied Biosystems). The primers and probe for murine IL-17RE
included forward
primer 5' CCACTCACACCCTGCGAAA (SEQ ID NO:148), reverse primer 5'
GCAAGTCCACATTCTCCAGGAT (SEQ ID NO:149), and probe ACCATCCTTCTGACTCCTGTG
CTGTGG (SEQ ID NO:150).
Results
[486] Murine IL-17RE mRNA expression was detected in all tissues tested.
Highest levels
of expression were observed in the colon, skin, lung, and foot tissues. Lower
levels of expression
were found in brain, spinal cord, lymph node, and spleen tissues. IL-17RE mRNA
was increased in
the spinal cord tissue from animals in the EAE model compared to non-diseased
controls. IL-17RE
mRNA was increased approximately 3.75 fold in animals with mild disease score
and approximately
2.8 fold in animals with severe disease scores. Murine IL-17RE mRNA was
decreased in tissues
from an acute model of DSS colitis compared to tissues from non-diseased
controls. IL-17RE
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
133
mRNA was decreased approximately 2.2 fold in the distal colon and
approximately 2.8 fold in the
proximal colon compared to non-diseased controls.
[487] Accordingly, one skilled in the art would recognize that since IL-17RE
expression is
increased in such diseases, a IL-17RE antagonist, such as the soluble
receptors and MAbs of the
present invention, would be useful in the treatment of these diseases.
EXAMPLE 37
IL-17RE is Regulated in Inflamed Large Intestine Sections of Patients with
Ulcerative Colitis
and Crohn's Disease
[488] Human IL-17RE mRNA is regulated in inflamed large intestine sections of
patients
with ulcerative colitis and Crohn's disease compared to large intestine
sections from normal control
patients.
Experimental Protocol
[489] Tissues were obtained from inflamed and un-inflamed large intestine
sections of
patients with Crohn's disease, ulcerative colitis or normal control patients.
RNA was isolated using
standard procedures. Expression of human IL-17RE mRNA was measured with
multiplex real-time
quantitative RT-PCR method (TaqMan) and the ABI PRISM 7900 sequence detection
system (PE
Applied Biosystems). IL-17RE mRNA levels were normalized to the expression of
the human
hypoxanthine guanine physphoribosyl transferase mRNA and determined by the
comparative threshold
cycle method (User Bulletin 2; PE Applied Biosystems). The primers and probe
for human IL-17RE
included forward primer 5' TCAGCGTGCGTCTTTGTCA (SEQ ID NO:151), reverse primer
5'
GGCCCCCAGACACAATTTT (SEQ ID NO: 152), and probe CATAGGGACTGCTCAGCTCTTCA
CACTCCA (SEQ ID NO:153).
Results
[490] Human IL-17RE mRNA expression was detected in all large intestine
samples
tested. IL-17RE mRNA was decreased 2.1 fold in the large intestine of patients
with ulcerative
colitis compared to the large intestines from normal patients. IL-17RE mRNA
was decreased in
large intestine samples from patients with Crohn's disease. IL-17RE mRNA was
decreased 1.5 fold
compared to normal patients with no disease.
[491] The decrease in IL-17RE expression may be explained by loss of IL-17RE-
expressing cells from the mucosal epithelium. For example, a rat colitis model
(reference Scand J
Gastroenterol. 2000 Oct;35(10):1053-9.) involving administration of dextran
sulfate sodium (DSS)
supports this hypothesis in demonstrating decreased epithelial cell survival
60 minutes after
administration of DSS and shedding of the epithelium 2 days after
administration.
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
134
EXAMPLE 38
Murine IL-17C mRNA is Regulated in Select Tissues in Murine Models of Disease
Compared to
Non-Disease Tissues
[492] Murine IL-17C mRNA is regulated in select tissues in murine models of
disease
compared to non-diseased controls.
Experimental Protocol
[493] Tissues were obtained from the following murine models of disease:
Colitis,
Asthma, Experimental Allergic Encephalomyelitis (EAE), Psoriasis and Collagen
Induced Arthritis
(CIA). Animal models were run following standard procedures and included
appropriate non-
diseased controls. Colitis was induced by dextran sulfate sodium (DSS) in the
drinking water and
the tissues isolated from the model included distal colon, proximal colon and
mesenteric lymph
nodes. Asthma was induced by sensitization and intranasal challenge to the
antigen ovalbumin. The
tissues isolated included lung, spleen and lymph node. EAE was induced by
immunizing with
MOG35-55 peptide in RIBI adjuvant. Tissues isolated included brain, cervical,
lymph node, and
spinal cord. Psoriasis was induced by adoptive transfer of naive T cells into
minor histocompatibility
mismatched or syngeneic immunocompromised mice. Tissues isolated included
lesional skin and
adjacent skin. CIA was induced by collagen injections and tissues isolated
included foot and
popliteal lymph node. RNA was isolated from all tissues using standard
procedures. In brief, tissues
were collected and immediately frozen in liquid N2 and then transferred to -80
C until processing.
For processing, tissues were placed in Qiazol reagent (Qiagen, Valencia, CA)
and RNA was isolated
using the Qiagen Rneasy kit according to manufacturer's recommendations.
Expression of murine
IL-17C mRNA was measured with multiplex real-time quantitative RT-PCR method
(TaqMan) and
the ABI PRISM 7900 sequence detection system (PE Applied Biosystems). IL-17C
mRNA levels
were normalized to the expression of the murine hypoxanthine guanine
physphoribosyl transferase
mRNA and determined by the comparative threshold cycle method (User Bulletin
2; PE Applied
Biosystems). The primers and probe for murine IL-17C included forward primer
5'
TGGAGATATCGCATCGACACA (SEQ ID NO:154), reverse primer 5' GCATCCACGACACAA
GCATT (SEQ ID NO:155), and probe CCGCTACCCACAGAAGCTGGCG (SEQ ID NO:156).
[494] Results: Murine IL-17C mRNA expression was detected in all tissues
tested.
Highest levels of expression were observed in the lymph node, colon, skin,
lung, foot and spleen
tissues. Lower levels of expression were found in brain and spinal cord
tissues. IL-17C mRNA was
increased in whole foot tissue from mice in the CIA model of arthritis
compared to foot tissue from
non-diseased controls. IL-17C mRNA was increased approximately 6.6 fold in
animals scored with
mild disease, approximately 9.1 fold in animals scored with mid level disease
and approximately 5
fold in animals with severe disease. IL-17C mRNA was increased in the spinal
cord tissue from
animals in the EAE model compared to non-diseased controls. IL-17C mRNA was
increased
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
135
approximately 2.05 fold in animals with mild disease score and approximately
2.9 fold in animals
with severe disease scores. Murine IL-17C mRNA was increased in tissues from a
acute model of
DSS colitis compared to tissues from non-diseased controls. IL-17C mRNA was
increased
approximately 2.8 fold in the distal colon and approximately 1.9 fold in the
proximal colon
compared to non-diseased controls.
EXAMPLE 39
IL-17C is Regulated in Inflamed Large Intestine Sections of Patients with
Ulcerative Colitis and
Crohn's Disease
[495] Human IL-17C mRNA is regulated in inflamed large intestine sections of
patients
with Crohn's disease.
Experimental Protocol
[496] Tissues were obtained from inflamed and un-inflamed large intestine
sections of
patients with Crohn's disease, Ulcerative Colitis or normal control patients.
RNA was isolated using
standard procedures. Expression of human IL-17C mRNA was measured with
multiplex real-time
quantitative RT-PCR method (TaqMan) and the ABI PRISM 7900 sequence detection
system (PE
Applied Biosystems). IL-17C mRNA levels were normalized to the expression of
the human
hypoxanthine guanine physphoribosyl transferase mRNA and determined by the
comparative
threshold cycle method (User Bulletin 2; PE Applied Biosystems). The primers
and probe for human
IL-17C included forward primer: 5' atg agg acc gct atc cac aga 3' (SEQ ID
NO:157), reverse primer:
5' ccc gtc cgt gca tcg a3' (SEQ ID NO: 158), and probe: tgg cct tcg ccg agt
gcc tg (SEQ ID NO: 159).
Results
[497] Human IL-17C mRNA expression was detected in all large intestine samples
tested.
IL-17C mRNA was increased in large intestine samples from patients with
Crohn's disease. IL-17C
mRNA was increased approximately 7.7 fold compared to normal patients with no
disease. IL-17C
mRNA was increased in the large intestine of some but not all patients with
Ulcerative colitis
compared to the large intestines from normal patients.
EXAMPLE 40
IL-17C Functional Response on IL-17RE Transfectants
[498] NIH-3T3/KZ142 cells were stably transfected with human IL-17REx1, human
IL-
17REx2, and human IL-17REx6 receptor splice variants. As described herein,
each cell line was
treated for 7 and 15 minutes with a dose response of human IL-17C (SEQ ID
NO:17), mouse IL-17C
(SEQ ID NO:19), and appropriate controls. The human IL-17REx1 transfectants
were analyzed with
only human IL-17C. Human and mouse IL-17C induced a dose dependent response in
phosphorylated IxB-a in the lines overexpressing human IL-17REx1 (n=3), human
IL-17REx2
(n=3), and human IL-17REx6 (n=2) splice variants and gave no response in
untransfected NIH-
3T3/KZ142 cells (n=3). At the 7 minute time point human IL-17C gave a maximum
response of
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
136
4.68 fold at 300 ng/mL while mouse IL-17C gave a maximum response of 5.22 fold
at 300 ng/mL on
the NIH-3T3/humanIL-17REx2 line. Similarly human IL-17C gave a maximum
response of 3.04
fold while mouse IL-17C gave a maximum response of 2.92 fold on the NIH-
3T3/humanIL-17REx6
line. At the 15 minute time point human IL-17C (A903G) gave a maximum response
of 2.54 fold at
100 ng/mL on the NIH-3T3/humanIL-17REx1 line.
EXAMPLE 41
Luciferase Reporter Assay To Determine IL-17C Activity on NIH3T3/KZ142.8 Cells
and
NIH3T3/KZ142.8 Tranfected with human IL-17REx1 and IL-17REx2 Splice Variants
[499] Day 1: NIH3T3/KZ142.8 (NIH3T3 cells stably transfected with a inducible
NFkB/AP1 luciferase reporter), and these same cells additionally stably
transfected with IL-17RE
receptor splice variants human IL-17REx1, IL-17REx2, or IL-17REX6 were plated
at 5000
cells/well in solid white tissue culture 96 well plates (Cat. #3917. Costar)
in DMEM high glucose,
5%FBS, 1mM Na Pyruvate, 1xG418, and luM MTX. (MTX is omitted in the
NIH3T3/KZ142.8
parental cell line growth medium). Plates were cultured overnight at 37 C, 5%
C02.
[500] Day 2: Growth media was replaced with DMEM high glucose, 1mM Na
Pyruvate,
0.1% BSA, and 25mM Hepes (Assay medium) and plates were incubated overnight at
37 C, 5%C02
overnight.
[501] Day 3: Human IL-17C, mouse IL-17C, and appropriate control proteins were
serially diluted in assay medium. The human IL-17REx1 transfectants were
analyzed with only
human IL-17C. Spent medium was removed from cells, and each concentration of
test ligand or
control protein was added to triplicate wells for final assay concentrations
of 0, 0.1, 1, 10 and 100
ng/ml. Following incubation for 4 hr at 37 C, 5%C02, assay medium was removed
and 25uUwell of
lx lysis buffer (Promega cat #E1531) was added. Plates were incubated for 10
minutes at room
temperature then read on a Berthold microplate luminometer using 3 seconds of
integration and 40u1
of luciferase substrate (Promega cat #E4550).
[502] Both human and mouse IL-17C induced luciferase reporter gene expression
by 2-
fold or greater in cells over expressing the IL-17REX2 and IL-17REX6 splice
variants. No induction
was observed in the parental NIH3T3/KZ142.8 cells. Thus, one skilled in the
art would recognize
that the binding and cellular signaling produced by IL-17C, that occurs only
in cells where IL-17RE
receptor splice variants are over-expressed, is evidence of a specific
receptor-ligand interaction
between IL- 17C andIL-17RE.
EXAMPLE 42
Efficacy of Soluble IL-17RE in Disease Models
[503] Based on the expression patterns for IL-17C and IL-17RE, one skilled in
the art
would recognize that modulation of the interaction between these two molecules
would have
biological activity in the following disease models. Such modulation could be
facilitated using an Fc
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
137
fusion protein with an IL-17RE polypeptide disclosed herein (e.g. any of SEQ
ID NOs: 100, 102 or
124).
Soluble IL-17RE Efficacy in a Murine Model of Asthma
[504] A murine model of asthma is induced by sensitization and challenge with
the DerPl
antigen or with ovalbumin. Mice can be sensitized by intra-peritoneal
injection with antigen in alum
and then challenged by intra-nasal administration of antigen.
[505] To demonstrate efficacy of soluble IL-17RE mice can be treated at
challenge with
recombinant IL-17RE. Lung inflammation can be assessed at various time points
post challenge by
quantitation of inflammatory cells in lavage fluid, by measurement of airway
hyper responsiveness
and by pathological analysis. In vivo efficacy of IL-17RE will be demonstrated
by a reduction in the
migration of inflammatory cells into the lung and by alterations in lung
pathology and airway hyper
responsiveness.
Soluble IL-17RE Efficacy in a Murine Model of Collagen Induced Arthritis
[506] The model can be used to investigate mechanisms of disease and potential
therapeutics for rheumatoid arthritis. Mice can be immunized with chick type
II collagen in
Complete Freunds Adjuvant on day -21 and with chick type II collagen in
Incomplete Freunds
Adjuvant on day 0 in the base of the tail. Disease progression can be scored
daily after the second
immunization and is assessed by collecting qualitative clinical scores (scale
0-3) and caliper
measurements of paw thickness. Clinical scores can be assessed as follows:
normal toes and paw;
0.5 - a single toe is inflamed; 1- Two or more toes are inflamed or the top of
the foot is inflamed; 2 -
The Top of the foot and the arch (till the ankle) are inflamed (excluding the
ankle); 3 - The whole
foot including the ankle is inflamed.
[507] To demonstrate efficacy of soluble IL-17RE mice can be treated with
recombinant
IL-17RE by intraperitoneal, intramuscular, subcutaneous, or intravenous
injection prior to
immunization or during the progression of disease. In vivo efficacy of IL-17RE
can be demonstrated
by a reduction in the progression of disease as judged by a decrease in
clinical symptoms, a reduction
in paw swelling, a reduction in inflammatory infiltrates as measured by
histopathology, and/or
reductions in bone/cartilage degradation in the leg as measured by
histopathology.
Soluble IL-17RE Efficacy in a Murine Model of EAE
[508] EAE is used to investigate mechanisms of disease and potential
therapeutics for
multiple sclerosis in animal models. It can be induced in C57BL/6 mice using
rMOG protein or
MOG35-55 peptide, or SJL mice with proteolipid protein peptide(s). To induce
EAE mice can be
immunized subcutaneously on day 0 with a rMOG/complete Freund's adjuvant
(CFA), MOG35-55
peptide/RIBI, or PLP/CFA emulsion, followed by treatment on day 0 and/or day 2
with an intra-
venous injection of pertussis toxin. Disease progression can be monitored by
clinical score and by
weight loss starting after pertussis toxin injection. Clinical scores are
based on the animals tail tone,
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
138
posture and gait as follows:O - healthy,l - tail weakness (tip of tail does
not curl), 2 - tail paralysis
(unable to hold tail upright), 3 - tail paralysis and mild waddle, 4 - tail
paralysis and severe waddle, 5
- tail paralysis and paralysis of one limb, 6 - tail paralysis and paralysis
of ANY 2 limbs ,7 -
tetrapareisis (a1141imbs paralyzed), 8 - moribund or dead.
[509] To demonstrate efficacy of soluble IL-17RE mice can be treated with
recombinant
IL-17RE prior to immunization or during the progression of disease. In vivo
efficacy of IL-17RE
can be demonstrated by a reduction in the progression of disease as judged by
a decrease in clinical
symptoms, by an amelioration of weight loss and by a reduction in inflammatory
infiltrates in the
brain as measured by histopathology.
Soluble IL-17RE Efficacy in a Murine Model of Experimental Colitis
[510] Colitis models can be induced in the mouse and used to evaluate the
mechanisms of
efficacy of therapeutics in human disease.
[511] Mice can be treated with a solution of dextran sulfate sodium (DSS)
administered ad
libitum in drinking water. DSS can be administered in such a way as to induce
either acute or
chronic disease. Disease progression can be monitored by loss of weight and by
disease activity
index (DAI) scores, composed of percent body weight loss, stool consistency
(where 0 = normal, 2 =
soft stool, 4 = diarrhea) and hemocult (where 0 = normal, 2 = no visible blood
on anus or in feces,
but blue color on Hemocult slide, 4 = visible blood on anus or in feces). In
the chronic form of this
model progression and regression of disease can be measured using these
criteria. In vivo efficacy of
IL-17RE can be demonstrated by a reduction in the progression of disease using
the above criteria
and by a reduction in inflammatory infiltrates in the gut as measured by
pathology.
[512] A hapten induced model of colitis can be used to study Th2 mediated
colitis. In this
model mice are sensitized by topical application of oxazalone or TNBS on day 0
and challenged by
intrarectal administration of oxazalone or TNBS on day 6. Disease progression
can be monitored by
loss of weight and by disease activity index (DAI) scores, composed of percent
body weight loss,
stool consistency (where 0 = normal, 2 = soft stool, 4 = diarrhea) and
hemocult (where 0 = normal, 2
= no visible blood on anus or in feces, but blue color on Hemocult slide, 4 =
visible blood on anus or
in feces). In vivo efficacy of IL-17RE can be demonstrated by a reduction in
the progression of
disease using the above criteria and by a reduction in inflammatory
infiltrates in the gut as measured
by histopathology.
EXAMPLE 43
Construction of IL-17RE Variant Extracellular Domains in a Vector that
Allowing expression
of a carboxy-terminal epitope tag and GPI mediated, plasma membrane anchorage
[513] Expression of IL-17RE extracellular (ECD) domains fused to a carboxy-
terminal
FLAG epitope tag and anchored to cell plasma membranes via a GPI linker allows
ligand binding
studies to be normalized to protein expression levels. The commercial
mammalian expression
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
139
vector pVAC2 (Invivogen, SanDiego, CA) allows for the fusion of ECD's to the
32 amino acid
carboxy-terminal domain of human placental alkaline phosphatase (PLAP). During
processing of the
pro-peptide as it transits the Golgi, a transaminase cleaves this PLAP domain
and simultaneously
adds a GPI tail thus providing a hydrophobic anchor for the ECD in the cell
membrane. Each of the
following IL-17RE ECD splice variants was cloned into the commercial mammalian
expression
vector pVAC2 utilizing the vector's BamH1 and EcoRl sites such that the PLAP
fragment was kept
in frame. The FLAG epitope sequence is commonly used and there are monoclonal
antibodies
commercially available. The epitope sequence was coded for in the each
antisense oligonucleotide
utilized in the PCR reactions that generated the ECD's. The fragments for
human IL-17REx1, human
IL-17REx2, human IL-17REx3, human IL-17REx6, human IL-17REx13, human IL-
17REs3, human
IL-17REs4 and murine IL-17REx6 were generated by PCR using previously
generated clones as
templates. The regions of difference between these clones lay internal to the
oligos thus all PCR
reactions utilized the same oligonucleotide pair as shown in SEQ ID NO: 166
and SEQ ID NO: 167.
The human IL-17RE-S2 clone was generated using human IL-17REx2 as template and
a different
sense oligonucleotide as shown by SEQ ID NO:168 but the same antisense primer.
A murine version
of IL-17REx6 was generated using a previously cloned template and the primers
as shown in SEQ
ID NO:169 and SEQ ID NO:170. Due to the presence of an internal EcoRl site,
PCR products were
digested with the restriction enzyme Esp3I that left cohesive ends matching
EcoRl and BamHl. The
digested and purified products were successfully ligated into pVAC2 and
sequenced yielding:
pVAC2-human IL-17REx1, (SEQ ID NO:171), pVAC2-IL-17REx2, (SEQ ID NO:172),
pVAC2-
hIL-17REx3, (SEQ ID NO:173), pVAC2-hIL-17REx6, (SEQ ID NO:174), pVAC2-hIL-
17REx13,
(SEQ ID NO:175), pVAC2-mIL-17REx6, (SEQ ID NO:176), pVAC2-hIL-17REs2, (SEQ ID
NO: 177).
EXAMPLE 44
IL-17RE FclO Fusion Protein Expression Constructs
[514] An expression plasmid containing IL-17REx2-C(Fc10) with a native leader
was
constructed from a previously described, optimized TPA leader version (Example
29; SEQ ID
NO:101 and SEQ ID NO:102) This was accomplished by exchanging an approximately
530 bp
EcoRI fragment from the TPA leader version, for an approximately 480 bp EcoRl
fragment from a
full length human IL-17REx2 pzmpll dicistronic expression construction
described in Example 16.
The two expression constructions in question share a vector-derived EcoRl site
just upstream of the
insert, on one hand, and a IL-17RE insert-derived EcoRI site, on the other
hand. Several clones
resulting from this genetic engineering event were sequenced and a clone with
a correctly oriented
EcoRl fragment was selected for expression. This native leader version of IL-
17REx2-C(Fc10) is
called mpet 1330 (SEQ ID NO:178).
EXAMPLE 45
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
140
Expression of GPI anchored, IL-17RE Variant Extracellular Domains in Mammalian
Cells
[515] The assessment of the ligand binding characteristics of cytokine
receptors can be
facilitated through the expression of their extracellular domains tethered to
the surface of cells via a
GPI linker. The following constructs, previously described in Example 41, were
transiently
expressed in 293f cells (Invitrogen, Carlsbad, CA) for 48-96 hours, harvested
by centrifugation and
utilized for ligand binding analysis by FACS: pVAC2-hIL-17REx1, pVAC2-hIL-
17REx2, pVAC2-
hIL-17REx3, pVAC2-hIL-17REx6, pVAC2-hIL-17REx13, pVAC2-hIL-17REs2 and pVAC2-
mIL-
17REx6.
[516] On day 1, 25m1 of shake flask cultured, low passage 293f cells were
seeded into
100m1 of Freestye Expression Medium (Invitrogen, Carlsbad, CA) in a 500m1
Erlenmeyer,
polycarbonate TC flask (Corning, Corning NY) at a density of approximately
0.7e6 cells/ml. The
cells were cultured at 37 C with ambient airflow @.2 LPM supplemented with 6%
C02, affixed to
an orbital shaker rotating at 90rpm. These settings were utilized for the
entire length of the culture.
On day 2, the cells were counted using a haemocytometer, centrifuged at 800g,
resuspended in fresh
Freestyle media to 1.0e6 cells/ml and divided into 20 125 ml Erlenmeyer,
polycarbonate TC flask
(Corning, Corning NY) at 10m1/flask and transfected as follows. l0ug of
plasmid DNA prepared
using either a miniprep or maxiprep Qiagen kit (Valencia,CA) following the
manufacturer's
suggested procedures was diluted into 200 microliters of Optimem media
(Invitrogen, Carlsbad, CA).
Simultaneously, 12.5 microliters of Lipofectamine2000 transfection reagent
(Invitrogen, Carlsbad,
CA) was mixed with 200 microliters of Optimem. After both mixtures had
incubated for 5 minutes
at room temperature they were mixed by pipetting and incubated at room
temperature an additional
30 minutes. Each DNA-lipid mixture was then added to a 125m1 flask of cells.
Thus transfected
cells were incubated for 48-96, harvested and washed into PBS+azide/BSA by
centrifugation and
utilized for FACS based binding studies. Receptor expression levels were
assessed by measurement
of a FLAG epitope specific antibody and biotinylated IL17C binding compared to
the nonspecific
binding seen in cells transfected with an unmodified pVAC2 "empty" vector.
EXAMPLE 46
Human IL-17RE Polyclonal Antibodies
[517] Anti-IL-17RE polyclonal antibodies are prepared by immunizing 2 female
New
Zealand white rabbits with either: the purified mature recombinant human IL-
17RE polypeptide
produced from 293 cells (ZytoRl-293), purified recombinant human IL-17REs2, or
subdomains
thereof, including SEQ ID NOs:113, 115, 117 or 119 containing a C-terminal tag
fusion to facilitate
purification (e.g. His, FLAG, EE, Fc).
[518] Alternatively, a IL-17RE-MBP fusion protein, produced in E.coli, which
utilizes the
extracellular domain sequence of IL-17RE fused to the Maltose-binding protein
(MBP), or synthetic
peptides containing a portion of the peptide sequence found in the
extracellular domain of human IL-
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
141
17RE with an additional Cys added to the N-teminus or C-terminus of the
peptides to facilitate
conjugation. The peptides and fusion proteins are conjugated by methods known
in the art (e.g.
Maleimide Activated Supercarrier System, No 77656, or Pharmalink Kit No 77158,
Pierce
Biotechnology, Rockland IL) to a carrier protein such as BSA and KLH to
increase the antigenicity
of the peptide or fusion-protein. The rabbits were each given an initial
intraperitoneal (ip) injection
of 200 g of purified protein in Complete Freund's Adjuvant followed by
booster IP injections of
100 g peptide in Incomplete Freund's Adjuvant every three weeks. Seven to ten
days after the
administration of the second booster injection (3 total injections), the
animals were bled and the
serum was collected. The animals were then boosted and bled every three weeks.
[519] The human IL-17RE-specific polyclonal antibodies are affinity purified
from the
immune rabbit serum using a CNBr-SEPHAROSE 4B protein column (Pharmacia LKB)
that was
prepared using 10 mg of the specific antigen purified recombinant protein
human IL-17RE-293 or
peptide per gram of CNBr-SEPHAROSE, followed by 20X dialysis in PBS overnight.
Human IL-
17RE-specific antibodies are characterized by ELISA using 500ng/ml of the
purified recombinant
protein human IL-17RE-293 as antibody target. The lower limit of detection
(LLD) of the rabbit anti-
human IL-17RE affinity purified antibody is usually 10-500 pg/ml on its
specific purified
recombinant antigen human IL-17RE-293. Alternatively, the serum can be
processed to isolate the
IgG fraction by Protein A-affinity chromatography or other methods known in
the art.
[520] The human IL-17RE-specific polyclonal antibodies are characterized for
their ability
to bind the IL-17RE-Fc protein in an ELISA format or to specifically bind IL-
17RE transfected
NIH3T3, 293 or BHK cells or to block the induction of luciferase in IL-17C
treated NIH3T3 cells
which contain an NFkB-sensitive luciferase reporter construct and have also
been transfected with
IL-17RE. The ability of IL-17RE directed polyclonal antibodies to inhibit the
binding of purified
recombinant human IL-17C to IL-17RE-Fc protein or IL-17RE transfected NIH3T3,
293 or BHK
cells or to inhibit the bioactivity of IL-17C in the NIH3T3/IL-17RE/NFkB-
luciferase bioassay would
be evidence of the ability of the IL-17RE specific antibody to antagonize the
bioactivity of human
IL-17C.
EXAMPLE 47
Generation of Murine Anti-Human IL-17RE Monoclonal Antibodies
A. Immunization for generation of anti-IL-17RE Antibodies
1. Soluble IL-17RE-Fc
[521] Six to twelve week old intact or IL-17RE knockout mice are immunized by
intraperitoneal injection with 50-100 ug of soluble human IL-17RE-mFc protein
mixed 1:1 (v:v)
with Ribi adjuvant (Sigma) on a biweekly schedule. Seven to ten days following
the third
immunization, blood samples are taken via retroorbital bleed, the serum
harvested and evaluated for
its ability to inhibit the binding of IL-17C to IL-17RE in neutralization
assays and to stain IL-17RE
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
142
transfected versus transfected P815 or NIH3T3 cells in a FACS staining assay
or on a FMAT system.
Mice are continued to be immunized and blood samples taken and evaluated as
described above until
neutralization titers reached a plateau. At that time, mice with the highest
neutralization titers are
injected intravenously with 25-50 ug of soluble IL-17RE-Fc protein in PBS.
Three days later, the
spleen and lymph nodes from these mice are harvested and used for hybridoma
generation, for
example using mouse myeloma (P3-X63-Ag8.653.3.12.11) cells or other
appropriate cell lines in the
art, using standard methods known in the art (e.g. see Kearney, J.F. et al.,
Jlmmunol. 123:1548-50,
1979; and Lane, R.D. Jlmmunol Methods 81:223-8. 1985.
2. Soluble IL-17RE, IL-17RE-CEE, IL-17RE-His, IL-17RE-FLAG
[522] Six to twelve week old intact or IL-17RE knockout mice are immunized by
intraperitoneal injection with 50-100 ug of soluble human soluble IL-17RE, IL-
17RE-CEE, IL-
17RE-His, IL-17RE-FLAG protein mixed 1:1 (v:v) with Ribi adjuvant (Sigma) on a
biweekly
schedule. Seven to ten days following the third immunization, blood samples
were taken via
retroorbital bleed, the serum harvested and evaluated for its ability to
inhibit the binding of IL-17C to
IL-17RE-Fc, human soluble IL-17RE, IL-17RE-CEE, IL-17RE-His, or IL-17RE-FLAG
in
neutralization assays and to stain IL-17RE transfected versus transfected P815
or NIH3T3 cells in a
FACS staining assay or on a FMAT system. Mice are continued to be immunized
and blood samples
taken an evaluated as described above until neutralization titers reached a
plateau. At that time, mice
with the highest neutralization titers were injected intravenously with 25-50
ug of soluble IL-17RE-
Fc protein in PBS. Three days later, the spleen and lymph nodes from these
mice are harvested and
used for hybridoma generation, for example using mouse myeloma (P3-X63-
Ag8.653.3.12.11) cells
or other appropriate cell lines in the art, using standard methods known in
the art (e.g. see Kearney,
J.F. et al., Jlmmunol. 123:1548-50, 1979; and Lane, R.D. Jlmmunol Methods
81:223-8. 1985.
3. Soluble IL-17REdomains
[523] Six to twelve week old intact or IL-17RE knockout mice are immunized by
intraperitoneal injection with 50-100 ug of soluble purified recombinant human
IL-17RE domain
HUIL-17REs2 (SEQ ID NO: 113), or the subdomains thereof (e.g. SEQ ID NOs: 115,
117 or 119)
containing a C-terminal tag fusion to facilitate purification (e.g. His, FLAG,
EE, Fc) conjugated by
methods known in the art (e.g. Pharmalink Immunogen Kit No 77158, Pierce
Biotechnology,
Rockland IL) to a carrier protein such as BSA and KLH to increase the
antigenicity. The pure
protein is mixed 1:1 (v:v) with Ribi adjuvant (Sigma) on a biweekly schedule.
Seven to ten days
following the third immunization, blood samples were taken via retroorbital
bleed, the serum
harvested and evaluated for its ability to inhibit the binding of IL-17C to IL-
17RE-Fc, human soluble
IL-17RE, IL-17RE-CEE, IL-17RE-His, or IL-17RE-FLAG in neutralization assays
and to stain IL-
17RE transfected versus transfected P815 or 293 cells in a FACS staining assay
or on a FMAT
system. Mice are continued to be immunized and blood samples taken an
evaluated as described
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
143
above until neutralization titers reached a plateau. At that time, mice with
the highest neutralization
titers were injected intravenously with 25-50 ug of soluble IL-17RE protein
antigen in PBS. Three
days later, the spleen and lymph nodes from these mice are harvested and used
for hybridoma
generation, for example using mouse myeloma (P3-X63-Ag8.653.3.12.11) cells or
other appropriate
cell lines in the art, using standard methods known in the art (e.g. see
Kearney, J.F. et al., J
Immunol. 123:1548-50, 1979; and Lane, R.D. Jlmmunol Methods 81:223-8. 1985.
4. P815 transfectants that express IL-17RE
[524] Six to ten week old female DBA/2 mice are immunized by intraperitoneal
injection
1-5 x 106 irradiated, transfected cells every 2-3 weeks. In this approach, no
animals develop and die
of ascites tumor. Instead, animals are monitored for a neutralizing immune
response to IL-17RE in
their serum as outlined above, starting with a bleed after the second
immunization. Once
neutralization titers have reached a maximal level, the mice with highest
titers are given a pre-fusion,
intraperitoneal injection of 5 x 106 irradiated cells and four days later, the
spleen and lymph nodes
from these mice are harvested and used for hybridoma generation, for example
using mouse
myeloma (P3-X63-Ag8.653.3.12.11) cells or other appropriate cell lines in the
art, using standard
methods known in the art (e.g. see Kearney, J.F. et al., J Immunol. 123:1548-
50, 1979; and Lane,
R.D. Jlmmunol Methods 81:223-8. 1985.
B. Screening the Hybridoma Fusions for Antibodies that bind IL-17RE and
inhibit the Binding
of IL-17C to IL-17RE
[525] Four different primary screens are performed on the hybridoma
supernatants at 8-10
days post-fusion. For the first assay, antibodies in supernatants were tested
for their ability to bind to
plate bound soluble IL-17RE-Fc, human soluble IL-17RE, IL-17RE-CEE, IL-17RE-
His, or IL-
17RE-FLAG protein by ELISA using HRP-conjugated goat anti mouse kappa and anti-
lambda light
chain second step reagents to identify bound mouse antibodies. To demonstrate
specificity for the IL-
17RE portion of the IL-17RE fusion proteins, positive supernatants in the
initial assay are evaluated
on an irrelevant protein fused to the same murine Fc region (mG2a), EE
sequence, His sequence, or
FLAG sequence. Antibody in those supernatants that bound to IL-17RE-fusion
protein and not he
irrelevant muFc or other proteins containing fusion protein sequence were
deemed to be specific for
IL-17RE. For the second assay, antibodies in all hybridoma supernatants were
evaluated by ELISA
for their ability to inhibit the binding of biotinylated human IL-17C to plate
bound IL-17RE-Fc or
other IL-17RE-fusion proteins.
[526] All supernatants containing antibodies that bound specifically to IL-
17RE, whether
they inhibited the binding of IL-17C to IL-17RE or not in the ELISA assay, are
subsequently tested
for their ability to inhibit the binding of IL-17C to IL-17RE transfected
NIH3T3, 293 or BHK cells
or normal human epithelial cells. All supernatants that are neutralization
positive in the IL-17C
neutralization assays are subsequently evaluated for their ability to stain IL-
17RE transfected
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
144
NIH3T3, 293 or BHK cells by FACS analysis. This analysis is designed to
confirm that inhibition of
IL-17C binding to IL-17RE, was indeed due to the antibody that specifically
binds the IL-17RE
receptor. Additionally, since the FACS analysis in performed with an anti-IgG
second step reagent,
specific FACS positive results indicate that the neutralizing antibody is
likely to be of the IgG class.
By these means, a master well is identified that binds IL-17RE in a plate
bound ELISA, inhibits the
binding of IL-17C to IL-17RE in the ELISA based inhibition assay, blocks the
interaction of IL-17C
with IL-17RE transfected NIH3T3, 293 or BHK cells, respectively, and is
positive for the staining of
IL-17RE transfected NIH3T3, 293 or BHK cells with an anti-mouse IgG second
step reagent.
[527] The third assay consists of NIH/3T3 cells containing an NFkB sensitive
luciferase
reporter construct and which have also been transfected with IL-17RE and can
therefore respond to
IL-17C treatment. These cells respond to IL-17C treatment by increasing the
expression of
luciferase which can then be assayed by standard methods known in the art. The
specific monoclonal
antibody to IL-17RE is assayed by its ability to, for example, inhibit IL17C-
stimulated luciferase
production by these cells.
[528] The fourth assay consists of primary human epithelial cells or cell
lines of human
origin such as U937, HCT15, DLD-1 or Caco2 cells which express IL-17RE and
respond to IL-17C
treatment. The specific monoclonal antibody is assayed by its ability to, for
example, inhibit IL17C
stimulated chemokine or cytokine production by these cells. Chemokine or
cytokine production is
assayed in response to IL-17C using commercially available ELISA assay kits
(e.g. R&D Systems,
Minneapolis, MN). Alternatively, the phospho-IkB levels in the IL-17C
responsive cells can be
monitored using phosphorylation specific antibodies available for this purpose
(BioRad, Richmond,
CA). The inhibition of IL-17C mediated phospho-IkB production would be a
measure of IL-17RE
antagonist activity by the monoclonal antibody.
[529] Cloning Anti-IL-17RE Specific Antibody Producing Hybridomas
[530] Hybridoma cell lines producing a specific anti-IL-17RE mAb that
neutralizes the
binding of IL-17C to appropriately transfected BaF3 or BHK cells are cloned by
a standard low-
density dilution (less than 1 cell per well) approach. Approximately 5-7 days
after plating, the clones
are screened by ELISA on, for example, plate bound human IL-17RE-Fc followed
by a retest of
positive wells by ELIDA on irrelevant Fc containing fusion protein as
described above. Selected
clones, whose supernatants bind to IL-17RE-Fc and not the irrelevant Fc
containing fusion protein,
are further confirmed for specific antibody activity by repeated both
neutralization assays as well as
the FACS analysis. All selected IL-17RE antibody positive clones are cloned a
minimum of two
times to help insure clonality and to assess stability of antibody production.
Further rounds of
cloning are performed and screened as described until, preferably, at least
95% of the resulting
clones are positive for neutralizing anti-IL-17RE antibody production.
D. Biochemical Characterization of the Molecule Recoznized by Anti-IL-17RE
mAbs
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
145
[531] Biochemical confirmation that the target molecule, IL-17RE, recognized
by the
putative anti-IL-17RE mAbs is indeed IL-17RE is performed by standard
immunoprecipitation
followed by SDS-PAGE analysis or western blotting procedures, both employing
soluble membrane
preparations from IL-17RE transfected versus untransfected Baf3 or BHK cells.
Moreover, soluble
membrane preparations of non-transfected cell lines that express IL-17RE are
used to show that
mAbs recognize the native receptor chain as well as the transfected one.
Alternatively, the mAbs are
tested for their ability to specifically immunoprecipitate or western blot the
souble IL-17RE-Fc
protein.
EXAMPLE 48
Neutalization of Human IL-17RE by Sera from Mice Injected with P815 Cells
Transfected with
Human IL-17RE
[532] Using a cell based neutralization assay, serum from mice injected with
human IL-
17RE transfected P815 cells, as described herein, is added as a serial
dilution at 1%, 0.5%, 0.25%,
0.13%, 0.06%, 0.03%, 0.02% and 0% The assay plates are incubated at 37 C, 5%
COz for 4 days at
which time Alamar Blue (Accumed, Chicago, IL) is added at 20 .micro.l/well.
Plates are again
incubated at 37 C, 5% COz for 4-16 hours. Differences in Alamar Blue
conversion shows that serum
from the animals can neutralize the signaling of Il-17C through human IL-17RE.
[533] Results from this assay can provide additional evidence that effectively
blocking IL-
17RE binding, blocking, inhibiting, reducing, antagonizing or neutralizing IL-
17C activity, for
example via a neutralizing monoclonal antibody to IL-17RE of the present
invention, could be
advantageous in reducing the effects of IL-17C in vivo and may reduce IL-17C
associated
inflammation, such as that seen in, psoriasis, IBD, colitis, chronic
obstructive pulmonary disease,
cystic fibrosis, arthritis, asthma, psoriatic arthritis, atopic dermatitis or
other inflammatory diseases.
Peptide Synthesis
[534] Peptide IL-17RE-1.1 [CIEASYLQEDTVRRKK-amide] andpeptide IL-17RE-2.1[
ISHKGLRSKRTQPSDPETWESC] were synthesized with Fmoc chemistry on a mode1433A
Peptide
Synthesizer (Applied Biosystems). Fmoc-Amide or Fmoc-Cys (Trt)- Wang resin
(AnaSpec) (0.25
mmol) was used as the initial support resins, respectively. A mixture of 2-(1H-
Benzotriazol-1-yl)-
1,1,3,3-Tetramethyluronium hexafluorophosphate (HBTU), 1-Hydroxybenzotriazole
(HOBt), N,N-
Diisopropylethyamine, N-Methylpyrrolidone, Dichloromethane (Applied Biosystems
and Piperidine
(Aldrich Chemical Co.) were used as synthesis reagents. The peptide was
cleaved from the solid
support wit 95% trifluoroacetic acid(TFA). Purification of the peptide was
performed by RP-HBLC
using a Vydac C18, 10-15 micron, 50 x 250 mm preparative column with
water/acetonitrile/ TFA
gradients. Eluted fractions from the column were collected and analyzed for
purity by analytical RP-
HPLC. Pooled fractions were lyophilized to dryness and resuspended in 10%
acetonitrile, 1% acetic
acid, then re-lyophilized to dryness in a Falcon tube of known weight.
Analytical HPLC and mass
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
146
spectrometry (MS) were performed before the final dry-down. Overall synthesis
yields were 30-33
%.
EXAMPLE 49
Construction of an IL-17RE pvac2-neo Construct with Geneticin Drug Resistance
[535] To circumvent the need to use co-transfections of the IL-17RE-pvac2
constructions
described in example 40 which lack a mammalian cell selectable marker and to
possibly increase the
stability of the transfection pools. Pvac2-neo was constructed by deleting the
pvac2 prokaryotic
zeocin transcription region contained within a 437 bp Pstl/Sfil fragment and
replacing it with a
neomycin acetyl transferase cDNA coupled to both its native prokaryotic
promoter, in addition to an
SV40 promoter for expression in mammalian cells. This geneticin resistance
conferring DNA
segment was obtained using a vector called pHZ1 (SEQ ID NO:194) as a template
and PCR using
SEQ ID NO:195 and SEQ ID NO:196 and pfu Ultra and other than the addition of
10% DMSO,
using the manufacturer's recommendations. Eight PCR reactions were performed.
Amplicons were
pooled and purified using chromaspin 100 (Clontech) columns and then digested
using standard
methods with Xhol and HinDIII, and gel purified. These two fragments were
ligated together using
linkers and standard methods, electroporated into E.coli. Colonies were used
to make plasmid
preparations and a correct clone identified using DNA sequencing. This new
vector is called pvac2-
neo and is shown in SEQ ID NO: 187.
EXAMPLE 50
Construction of IL-17RE Extracellular Domains in a Pvac2neo Vector
[536] Example 43 describes the expression of IL-17RE extracellular domains
(ECD) in a
commercial expression vector called pVAC2. To facilitate the transfections
needed to obtain stable
pools of IL-17RE ECD and possibly stabilize their long term expression for the
duration of the
radioligand binding studies to be performed, the same constructions described
in Example 43 were
modified to confer resistance to a mammalian cell selectable drug called
geneticin. Example 49
provides some details of this drug resistance gene and how pvac2 was converted
to a new plasmid
called pvac2neo. New IL-17RE-pvac2neo constructions to be made by conversion
of the pvac2
constructs described previously, with their open reading frames match that of
the pvac2 versions.
Pvac2neo (SEQ ID NO: 187) has unique Kpnl and Nhel sites and these sites do
not cleave within any
IL-17RE variant, thus these sites were chosen for the conversion of the IL-
17RE-pvac2 constructs
into IL-17RE-pvac2neo constructs. Briefly, pvac2neo was digested with Kpnl and
Nhel restriction
enzymes and the approximately 3KB fragment containing the prokaryotic origin
of replication,
neomycin acetyl transferase gene and promoter, and SV40 promoter gel purified.
Next the all of the
IL-17RE-pvac2 constructs listed above were Kpnl/Nhel digested and each had an -
3KB fragment
gel purified. In each case the pvac2neo Kpnl/Nhel fragment was ligated to
these IL-17RE-pvac2
Kpnl/Nhel fragments and electroporated into E.coli using standard methods and
plated on 25ug/ml
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
147
kanamycin-amended Luria agar. Several kanamycin resistant clones representing
each of the IL-
17RE variant-pvac2neo constructs listed above were sequenced, and clones
identified matching that
of their pvac2, kanamycin sensitive counterparts. Additionally, IL-17REs3 (SEQ
ID NO:203) and
IL-17REs4 (SEQ ID N2 10) was also constructed.
EXAMPLE 51
Construction of a Mammalian Soluble IL-17REs3 Expression Construct
[537] Mutagenesis, protein engineering and binding studies (as described
herein)
demonstrated that a truncated IL-17RE extra-cellular domain, designated IL-
17REs3 binds robustly
to biotinylated human IL-17C using flow cytometry. In order to evaluate
whether IL-17REs3 can be
secreted, an expression construct was made containing the extra-cellular
domain of human IL-
17REs3 with a carboxy-terminal Fc type tag placed into the mammalian
expression vector pZMP40.
This construction was called IL-17REs3-Fc10 (SEQ ID NO:188) and it was
constructed as follows.
IL-17REs3-Fc10 only differs from IL-17REs2-Fc10 in one small region, roughly
near its N-
terminus. Thus IL-17REs2-Fc10 was used as an intermediate to make IL-17REs3-
Fc10 by digestion
with Fsel and BstEII restriction enzymes to remove this variable region. The
remaining region to be
retained in the large Fsel/BstEII fragment of IL- 17REs2-Fc 10 (SEQ ID NO: 189
and 190) containing
the vector backbone and ECD region was gel purified using low melt agarose gel
electrophoresis and
then liquefied using gelase enzyme. (Epicentre) Next the second fragment
needed, the IL-17REs3-
specific Fsel/BstEII fragment, was obtained by digesting the previously
described IL-17REs3-
pvac2neo plasmid with Fsel/BstEII and purifying this 221 bp fragment using the
same method as
described for the first fragment just above. These two fragments were ligated
together and
electroporated into E.coli using standard techniques. Transformants were
screened by colony PCR
using sense primer zc39200 (SEQ ID NO:191) which corresponds to the optimized
TPA leader and
antisense primer zc40455 (SEQ ID NO:192) which corresponds to IL-17RE using
standard
conditions. PCR products were separated on a 2% agarose TBE gel and the
matching transformants
with the expected band were analized by DNA sequencing and a clone identified
with the expected
sequence. The DNA sequencing validated IL-17REs3-FC10 clone was used to
inoculate a 500m1
Luria broth culture amended to 100ug/ml ampicillin and a plasmid megaprep
(Qiagen) procedure
was employed to purify a high quality plasmid preparation for use in
expression analysis.
[538] Plasmid pZMP40 is a mammalian expression vector containing an expression
cassette having the chimeric CMV enhancer/MPSV promoter, a BglII site for
linearization prior to
yeast recombination, an otPA signal peptide sequence, an internal ribosome
entry element from
poliovirus, the extracellular domain of CD8 truncated at the C-terminal end of
the transmembrane
domain; an E. coli origin of replication; a mammalian selectable marker
expression unit comprising
an SV40 promoter, enhancer and origin of replication, a DHFR gene, and the
SV40 terminator; and
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
148
URA3 and CEN-ARS sequences required for selection and replication in S.
cerevisiae, and is the
scaffolding for the MPET 1122 construct.
[539] The inserts of three DNA clones for the construct are subjected to
sequence analysis
and one clone containing the correct sequence is selected. Large-scale plasmid
DNA is isolated using
a commercially available kit (QIAGEN Plasmid Mega Kit, Qiagen, Valencia, CA)
according to
manufacturer's instructions. The soluble protein was produced using common
mammalian protein
production cells CHOduxbll and 293fb using standard transfection procedures as
previously
described in previous Examples.
EXAMPLE 52
Construction of a Mammalian Soluble IL-17REs4 Expression Construct
[540] Mutagenesis, protein engineering and binding studies demonstrated that a
truncated
IL-17RE extra-cellular domain, designated IL-17REs4 binds robustly to
biotinylated human IL-17C
using flow cytometry. In order to evaluate whether IL-17REs4 can be secreted,
an expression
construct was made containing the extra-cellular domain of human IL-17REs4
with a carboxy-
terminal Fc type tag placed into the mammalian expression vector pZMP40 were
constructed. This
construction was called IL-17REs4-FC10 (SEQ ID NO:193) and it was constructed
as follows. IL-
17REs4-Fc10 only differs from IL-17REs2-Fc10 in one small region, roughly near
its N-terminus.
Thus IL-17REs2-Fc10 was used as an intermediate to make IL-17REs4-Fc10 by
digestion with Fsel
and BstEII restriction enzymes to remove this variable region. The remaining
region to be retained is
the large Fsel/BstEII fragment of IL-17REs2-Fc10 containing the vector
backbone and ECD region
was gel purified using low melt agarose gel electrophoresis and then liquefied
using gelase enzyme.
(Epicentre) Next the second fragment needed, the IL-17REs4-specific
Fsel/BstEII fragment, was
obtained by digesting the previously described IL-17REs4-pvac2 plasmid with
Fsel/BstEII and
purifying this 244 bp fragment as described for the first fragment just above.
These two fragments
were ligated together and electroporated into E.coli using standard
techniques. Transformants were
screened by colony PCR using sense primer zc39200 (SEQ ID NO:191) which
corresponds to the
optimized TPA leader and antisense primer zc40455 (SEQ ID NO:192) which
corresponds to IL-
17RE using standard conditions. PCR products were separated on a 2% agarose
TBE gel and the
matching transformants with the expected band were analized by DNA sequencing
and a clone
identified with the expected sequence. The DNA sequencing validated IL-17REs4-
Fc10 clone was
used to inoculate a 500m1 Luria broth culture amended to 100ug/ml ampicillin
and a plasmid
megaprep (Qiagen) procedure was employed to purify a high quality plasmid
preparation for use in
expression analysis.
[541] Plasmid pZMP40 is a mammalian expression vector containing an expression
cassette having the chimeric CMV enhancer/MPSV promoter, a BglII site for
linearization prior to
yeast recombination, an otPA signal peptide sequence, an internal ribosome
entry element from
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
149
poliovirus, the extracellular domain of CD8 truncated at the C-terminal end of
the transmembrane
domain; an E. coli origin of replication; a mammalian selectable marker
expression unit comprising
an SV40 promoter, enhancer and origin of replication, a DHFR gene, and the
SV40 terminator; and
URA3 and CEN-ARS sequences required for selection and replication in S.
cerevisiae, and is the
scaffolding for the MPET 1122 construct.
[542] The inserts of three DNA clones for the construct are subjected to
sequence analysis
and one clone containing the correct sequence is selected. Large-scale plasmid
DNA is isolated using
a commercially available kit (QIAGEN Plasmid Mega Kit, Qiagen, Valencia, CA)
according to
manufacturer's instructions. The soluble protein was produced using common
mammalian protein
production cells CHOduxbll and 293fb using standard transfection procedures as
previously
described in herein.
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
150
EXAMPLE 53
Multiple Independently Generated Lots of Soluble IL-17REs2-Fc10 Block IL-17C
Mediated
Activation of Ix(3 in IL-17RE Transfected NIH3T3 Cells
[543] On day one NIH-3T3/KZ142 cells stably transfected with the human IL-
17REx2
receptor were plated out at 1x104 cells/well in growth media (DMEM, 5% fetal
bovine serum, 1%
Sodium Pyruvate, 1 M MTX) in 96-well, flat-bottom tissue culture plates. On
day two cells were
switched to assay media (DMEM, 0.1% BSA, 10mM HEPES). On day three a sub-
maximal
concentration (EC90, effective concentration at 90 percent) of human IL-17C
(huIL-17C) was
combined with a dose range of the human IL-17REs2 soluble receptor (Fc-10
fusion; SEQ ID
NOs: 189 and 190) and incubated together at 37 C for 30 minutes in assay media
prior to addition to
cells. Following pre-incubation, treatments were added to the plates
containing the cells and
incubated together at 37 C for 15 minutes.
[544] Following incubation, cells were washed with ice-cold wash buffer and
put on ice to
stop the reaction according to manufacturer's instructions (BIO-PLEX Cell
Lysis Kit, BIO-RAD
Laboratories, Hercules, CA). 50 L/well lysis buffer was added to each well;
lysates were pipetted
up and down five times while on ice, then agitated on a microplate platform
shaker for 20 minutes at
300 rpm and 4 C. Plates were centrifuged at 4500 rpm at 4 C for 20 minutes.
Supernatants were
collected and transferred to a new micro titer plate for storage at -20 C.
[545] Capture beads (BIO-PLEX Phospho-Ix(3-a Assay, BIO-RAD Laboratories) were
combined with 50 L of 1:1 diluted lysates and added to a 96-well filter plate
according to
manufacture's instructions (BIO-PLEX Phosphoprotein Detection Kit, BIO-RAD
Laboratories). The
aluminum foil-covered plate was incubated overnight at room temperature, with
shaking at 300 rpm.
The plate was transferred to a microtiter vacuum apparatus and washed three
times with wash buffer.
After addition of 25 L/well detection antibody, the foil-covered plate was
incubated at room
temperature for 30 minutes with shaking at 300 rpm. The plate was filtered and
washed three times
with wash buffer. Streptavidin-PE (50 L/well) was added, and the foil-covered
plate was incubated
at room temperature for 15 minutes with shaking at 300 rpm. The plate was
filtered and washed two
times with bead resuspension buffer. After the final wash, beads were
resuspended in 125 L/well of
bead suspension buffer, shaken for 30 seconds, and read on an array reader
(BIO-PLEX, BIO-RAD
Laboratories) according to the manufacture's instructions. Data was analyzed
using analytical
software (BIO-PLEX MANAGER 3.0, BIO-RAD Laboratories). Decreases in the level
of the
phosphorylated Ix(3-a transcription factor present in the lysates were
indicative of neutralization of
the IL-17REx2 receptor-ligand interaction.
[546] For huIL-17C the EC90 concentration was determined to be 5nM. Run in
combination with a dose-response of the human IL-17REs2 soluble receptor, the
IC50 (inhibitory
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
151
concentration at 50%) was determined multiple times. The results for IC50 (nM)
for the multiple
independently generated lots of IL-17REs2-Fc10 were: 5.1, 3.7, 3.1 and 2.7.
EXAMPLE 54
Construction of a IL-17RE-S3-Pvac2neo Expression Construct and Its Use for the
Establishment of Recombinant BHK 570 Cells for Use in Binding Assays
[547] Mutagenesis, protein engineering and binding studies have suggested that
a IL-
17REx2 extracellular domain without amino acids M1-R76, R375 and H376,
designated IL-17REs3
has robust binding affinity. This construction was called IL-17RE-s3-pvac2neo
(SEQ ID NO:203)
and it was constructed as follows.
A) Construction of IL- 17RE-s3 -pvac2neo
[548] IL-17REs3-pvac2neo differs from IL-17REs2-pvac2 (described above) in
several
ways, including having an optimized tpa leader (amino acid residues 1-35 of
SEQ ID NO:212) in
place of the s2's native leader, a somewhat larger ECD, and in having a
geneticin selectable marker
in its vector (pvac2neo) backbone. The construction of IL-17REs3-pvac2neo
involved making some
intermediates. The otpa IL-17REs2-pvac2neo intermediate was constructed using
a 3 part ligation.
The first part was a Pvac2neoAF intermediate, which was constructed prior to
this 3 part ligation as
follows. The multiple cloning site of pvac2neo was modified to contain an Fsel
site. This was
accomplished by digestion of pvac2neo with BamHI and EcoRl, and purifying the
large fragment
minus the short fragment between. A linker was synthesized with BamHI/EcoRI
cohesive ends.
This linker created several restriction sites, including Fsel. These linkers
included sense oligo SEQ
ID NO:197 and antisense oligo SEQ ID NO:198. The linkers were annealed and
then ligated to the
pvac2neo BamHI/EcoRI fragment, and electroporated into E.coli, creating an
intermediate called
pvac2neo-AF. Pvac2neo AF was prepared for the 3 part ligation by digesting it
with Fsel/Nhel
restriction enzymes and purifying the large fragment containing the majority
of the plasmid using the
gelase method. (Epicentre) An approximately 805 bp Fsel/BsrGI fragment
containing the opta leader
and part of IL-17REs2 was purified from IL-17REs2-FC10 and an approximately
300 bp
BsrGl/Nhel fragment purified from hIL-17REs2-pvac2. These 3 fragments were
ligated together
creating the intermediate otpa-IL-17REs2-pvac2neo.
[549] IL-17REs3-pvac2neo was constructed using the above otpa IL-17REs2-
pvac2neo as
an intermediate. It was digested with Fsel and BsrG1 and the large fragment
containing the vector
backbone gel purified. Next hIL-17REx2-pvac2neo was digested with BamHI and
BsrGI and the
742 bp BamHI/BsrGI fragment gel purified. Then, utilizing IL-17REs2-pvac2neo
as a template,
sense primer SEQ ID NO:199 and antisense primer SEQ ID NO:200 were used to
create an
approximately 136 bp amplicon using pfu ultra polymerase and the
manufacturer's
recommendations. PCR amplicons were pooled and purified through chromaspin 100
columns
(Amicon) and digested with Fsel and BamHI restriction enzymes and then gel
purified. The 3
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
152
fragments were ligated together and electroporated into E.coli using standard
methods, creating otpa-
IL-17REs3-pvac2neo. Colonies were screened by PCR using hot star polymerase
(Qiagen) primers
SEQ ID NO:201 and SEQ ID NO:202. Colonies whose amplicons were of the expected
1216 bp
were sent for DNA isolation and DNA sequencing, which subsequently identified
a correct clone.
Large-scale plasmid DNA was isolated using a commercially available kit
(QIAGEN Plasmid Mega
Kit, Qiagen, Valencia, CA) and purified according to manufacturer's
instructions. BHK570 cells
were transfected with construct otpa-IL-17RE-s3-pvac2neo (SEQ ID NO:203) using
a(Mirus)
NIH3T3 plasmid transfection kit according to the manufacturer's
recommendations. A stable pool
was generated using selection in 500ug/ml geneticin amended growth medium for
use in binding
studies.
EXAMPLE 55
Construction of a IL-17REs4-Pvac2 Expression Construct and Its Use for the
Establishment of
Recombinant BHK 570 Cells for Use in Binding Assays
[550] Mutagenesis, protein engineering and binding studies have suggested that
a IL-
17REx2 extracellular domain without amino acids M1-G96, designated IL-17REs4
has robust
binding affinity. This construction was called IL-17REs4-pvac2 and it was
constructed as follows.
A) Construction of IL- 17REs4-pvac2.
[551] IL-17REs4-pvac2 differs from IL-17REs2-pvac2 in several ways, including
having
an optimized tpa leader in place of the s2's native IL-17RE leader and having
a somewhat larger
ECD. The construction of IL-17REs4-pvac2 involved making some intermediates.
The otpa IL-
17REs2-pvac2 intermediate was constructed using a 3 part ligation. The first
part was a Pvac2-AF
intermediate, which was constructed prior to this 3 part ligation as follows.
The multiple cloning site
of pvac2 was modified to contain an Fsel site. This was accomplished by
digestion of pvac2 with
BamHI and EcoRl, and purifying the large fragment minus the short fragment
between. A linker
was synthesized with BamHI/EcoRI cohesive ends. This linker created several
restriction sites,
including Fsel. These linkers included sense oligo SEQ ID NO:204 and antisense
oligo SEQ ID
NO:205. The linkers were annealed and then ligated to the pvac2 BamHI/EcoRI
fragment, and
electroporated into E.coli, creating an intermediate called pvac2AF. Pvac2AF
was prepared for the 3
part ligation by digesting it with Fsel/Nhel restriction enzymes and purifying
the large fragment
containing the majority of the plasmid using the gelase method. (Epicentre) An
approximately 800
bp Fsel/BsrGI fragment containing the opta leader and part of IL-17REs2 was
purified from IL-
17REs2-FC10 and a 304 bp BsrGl/Nhel fragment purified from hIL-17REs2-pvac2.
These 3
fragments were ligated together creating the intermediate otpa-IL-17REs2-
pvac2.
[552] IL-17REs4-pvac2 was constructed using the above otpa IL-17REs2-pvac2 as
an
intermediate. It was digested with Fsel and Nhel and the large fragment
containing the vector
backbone gel purified. Synthetic oligonucleotides SEQ ID NOs:206 and 207were
annealed, in
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
153
addition, synthetic oligonucleotides SEQ ID NO:208 (phophorylated) and SEQ ID
NO:209 were
annealed. The DNA fragment and the two pairs of annealed oligos described
above were ligated
together and electroporated into E.coli using standard methods. Several
colonies were used to
generate small scale plasmid preparations. Plasmids were digested with BamHI
and clones
producing the expected 919 bp fragment were sent for DNA sequencing. DNA
sequence
determination identified a clone with the correct sequence. This positive was
called otpa-IL-
17REs4-pvac2 (SEQ ID NO:210). Large-scale plasmid DNA is isolated using a
commercially
available kit (QIAGEN Plasmid Mega Kit, Qiagen, Valencia, CA) according to
manufacturer's
instructions. A co-transfection was necessary to generate stable, recombinant
otpa-IL-17REs4-pvac2
BHK570 cells because the pvac2 vector lacks a mammalian drug selection. Thus,
a 10- fold excess
of otpa-IL-17REs4-pvac2 plasmid was co-transfected along with a pHZ1 plasmid
which confers
resistance to geneticin in mammalian cells. The transfection was accomplished
using a(Mirus)
NIH3T3 plasmid transfection kit according to the manufacturer's
recommendations. A stable pool
was generated using selection in 500ug/ml geneticin amended growth medium for
use in binding
studies.
EXAMPLE 56
IL-17C Binding to IL-17REs3 and IL-17REs4
[553] IL-17REs3 and IL-17REs4 were stable expressed on the surface of BHK
cells using
the pVAC2 expression system. This system uses a GPI anchor to tether the
molecule to the surface
of the cell, facilitating binding studies. Stably transfected cells were
harvested from flasks using a
non protease reagent such as Versene (Invitrogen 15040-066) and resuspended in
staining media
(HBSS +1%BSA +0.1%NaAzide +10mM HEPES). Cells were incubated for 30-60 minutes
at 4 C
with biotinylated mouse or human IL-17C at a concentration of lug/ml. Cells
were pelleted and
washed in staining media and the presence of bound IL-17C was detected by
staining for 30-60
minutes at 4 C with streptavidin-PE. After washing in staining media cells
were analysed by flow
cytometry. Binding of IL-17C to transfected receptors on the cell surface was
measured by mean
PE fluorescence. Using this system the following interactions were detected:
mouse IL-17C binds
specifically to IL-17REs3 and IL-17REs4; and human IL-17C binds specifically
to IL-17REs3 and
IL-17REs4.
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
154
EXAMPLE 57
Soluble IL-17RE Exhibits Efficacy in Ex Vivo Organ Cultures or Organotypic
Cultures
[554] Tissues can be collected from normal and diseased organ systems and
cultured ex
vivo in the presence or absence of various mitogens or growth factors.
Examples of such
organotypic culture systems include synovial explant cultures from normal
individuals and from
patients with joint disease, and differentiating cultures of normal or
psoriaitic skin. Biological
activity in these systems can be assessed by measuring a number of parameters,
including
proliferation, cytokine, chemokine or growth factor release, changes in gene
expression or alterations
in differentiation state. These ex vivo culture systems allow the activity of
exogenous therapeutic
molecules to be measured in a system that mimics some of the features of
normal or diseased tissue.
Any of the soluble IL-17RE polypeptides disclosed herein (i.e. IL-17REs2, IL-
17REs3 or IL-17REs4
as non-limiting examples) can be added to these ex vivo systems to show
efficacious alterations in
the production of inflammatory cytokines, chemokines or growth factors and
beneficial alterations in
proliferation, gene expression or differentiation state.
EXAMPLE 58
Soluble IL-17RE Blocks Disease Progression in the Xenograft Mouse Psoriasis
Transplantation
Model
[555] Psoriasis is a common human disorder which is best studied in the mouse
using a
xenograft transplantation model. In this model full thickness punch biopsies
of lesional skin are
collected from patients with psoriasis and control skin harvested from normal
volunteers. Using
established skin grafting technologies skin biopsies are transplanted onto the
dorsal surface of SCID
mice. In a variation of the model engraftment can be accompanied by the
adoptive transfer of
peripheral blood mononuclear cells from normal or psoriatic individuals. Mice
are subsequently
maintained in a pathogen free environment and graft beds inspected for take
(ie persistence) of the
grafted material. When psoriatic skin is transplanted in this manner it will
maintain its characteristic
disease appearance, allowing administered therapeutics to be evaluated for
efficacy in disease
modulation. Two to three weeks post transplantation mice with healthy
persistent grafts are divided
into experimental treatment groups. Animals subsequently receive either a
soluble IL-17RE
polypeptide disclosed herein (i.e. IL-17REs2, IL-17REs3 or IL-17REs4 as non-
limiting examples)
delivered using a series of dosing schedules or routes of delivery, or are
treated with vehicle.
Animals are typically treated and monitored for 5-7 weeks. Clinical
observations and assessments
are made during this period of treatment. At the end of the study disease
severity in the engrafted
psoriatic tissue is assessed by histopathological analysis, including
measurements of epidermal
thickening, of the disregulation of the epidermal architecture and of
keratinocyte proliferation using
the Ki67 marker. Efficacy of the soluble IL-17RE polypeptide in this system
may be distinguished
by the presence of a normal well differentiated epidermis of appropriate
thickness and with
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
155
proliferating keratinocytes restricted to the epidermal basal layer when
compared to animals treated
with vehicle alone.
EXAMPLE 59
Soluble IL-17RE Blocks Disease Progression in the Mouse IBD Model
[556] Experimental colitis can be induced in Scid or Rag-deficient mice by the
transfer of
syngeneic CD4+ T cells which have been enriched for the expression of a number
of cell surface
markers. CD4+ T cells which are able to induce disease in this model include
the CD45RBhi
population, CD62L positive population and CD25 negative populations. Adoptive
transfer of these
cells into syngeneic Scid or Rag-deficient mice results in the development of
experimental colitis.
Disease progression in this model can be measured using a number of
parameters, including weight
loss, histology and immunohistochemistry on gut tissues at post-mortem, colon
length and weight,
the presence of blood in the stools and the presence of inflammatory cytokines
in stool samples and
in the circulation. Animals undergoing experimental colitis in this adoptive
transfer model can be
treated with any of the IL-17RE polypeptides of the present invention using
either therapeutic or
prophylactic treatment regimens and a variety of dosing schedule. The
amelioration of disease
progression or development in these soluble-receptor treated mice can be
assessed using the
aforementioned markers of disease incidence.
EXAMPLE 60
Effect of IL-17C on Human Small Airway Epithelial Cells (SAEC)
[557] Human small airway epithelial cells (SAEC) were treated with human IL-
17C and
48hr supernatants were collected. These supernatants were assayed and showed a
dose-dependent
induction of G-CSF as shown in Table 4 below:
Table 4
Fold Induction
in 48hr supernatants
G-CSF
SAEC treated with:
huIL-17C 100 ng/ml 2.75
25 ng/ml 2.54
6.25 ng/ml 1.36
1.56 ng/ml 1.46
[558] SAEC were also treated with 0.067 - 150 nM doses of the soluble receptor
IL-
17REs2 in combination with 25 ng/ml human IL-17C (ligand and soluble receptor
were incubated
together for 30 minutes at 37 C before adding to cells), and 48hr supernatants
collected. These
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
156
supernatants showed decreased G-CSF, demonstrating that the soluble receptor
IL-17REs2 was able
to neutralize the activity of human IL-17C induction of this cytokine, with
IC50 as shown in Table 5
below:
Table 5
Soluble IL17REs2Fc10 receptor
neutralizes activity of huIL-17C IC50 of IL17REs2Fc10
to induce G-CSF: (nM)
huIL-17C 25 ng/ml:
lot A1712F 3.2
lot A1721F 3.7
lot A1730F 4.5
EXAMPLE 61
Efficacy of the Soluble IL-17RE Polypeptides in Human IBD Samples via
Epithelial Barrier
Function
[559] Maintenance of epithelial barrier integrity is a critical factor in the
preservation of a
healthy gastrointestinal tract. Experimental evidence suggests that leakiness
of the epithelial barrier
in the gut may contribute to the development of IBD. Immune cells located in
the intestinal lamina
propria generally interact with intestinal epithelial cells via cell to cell
contact or production of
soluble factors to maintain immune surveillance and contribute to epithelial
barrier integrity.
However, prolonged or dysregulated immune-mediated inflammation may contribute
to defects in
epithelial barrier cell integrity and function. The following study is
designed to measure the direct
effect(s) of T cell-derived IL-17C on epithelial barrier integrity.
[560] In this example, intestinal epithelial cell lines, like Caco-2 cells,
are differentiated on
semipermeable membranes and co-cultured on the basolateral side with either T
cells or monocytes
derived from biopsies from IBD patients or normal individuals. Epithlelial
monolayer integrity is
monitored over time using assessment of transepithelial electrical resistance
or resistance of the
monolayer to dye diffusion. Decreases in transepithial resistance of
monolayers in co-cultures would
suggest a disruption in the monolayer induced by the activity of the T cells
or monocytes in the co-
culture. Inhibitors of IL-17C such as the soluble polypeptides of the present
invention (i.e. IL-
17REs2, IL-17REs3 or IL-17REs4 as non-limiting examples) could be used to
determine the relative
contribution of IL-17C to the disruption of the epithelial monolayer and test
whether inhibitors of IL-
17C would be effective in maintaining epithelial barrier integrity. Prevention
of epithelial monolayer
disruption induced by activated T cells by such molecules would suggest that
the soluble IL-17RE
polypeptides of the present invention may be effective for the therapeutic
treatment of IBD in
humans. Co-culture systems could also be generated using monolayers formed by
primary epithelium
from IBD patients to determine whether these cells are more sensitive to IL-
17C compared to
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
157
epithelial cells derived from healthy individuals. If so, these data would
suggest that inhibiting IL-
17C would be a suitable strategy for the therapeutic treatment of IBD.
EXAMPLE 62
Effects of IL-17C on Lamina PropPria T cells and Monocytes/Macrophages from
Normal and
Human IBD Samples
[561] Dysregulated or sustained immune-mediated inflammation may contribute to
the
symptoms and pathology associated with IBD by way of tissue damage or
permanent skewing to
inappropriate or prolonged immune responses. This model can determine the
potential down-stream
consequences of exposure of disease-associated T cells and monocytes to IL-17C
which may be
present in the immediate environmental cytokine mileu of the intestinal
tissue.
[562] Therapeutics that would be efficacious in human IBD in vivo would work
in the
above ex vivo models by inhibiting and/or neutralizing the production and/or
presence of
inflammatory mediators (including but not limited to IL-lb, IL-4, IL-5, IL-6,
IL-8, IL-12, IL-13, IL-
15, IL-17 A and F, IL-18, IL-23, TNF-a, IFN-g, MIP family members, MCP-1, G-
and GM-CSF,
etc.).
[563] In this model, T cells and monocytes/macrophages are isolated from
biopsy samples
by carefully mincing biopsies with scissors in HBSS, treating with collagense
and Dispase II and
incubating for 1 hr at 37oC in a shaker. The cell suspension is filtered
through nylon mesh to
remove debris and cell clumps and washed multiple times in HBSS. T cells and
macrophage/monocytes can be isolated using direct cell sorting or bead-
depletion/enrichment
protocols. Isolated cells are incubated in the presence of IL-17C. This
induces the production of
inflammatory mediators by T cells and monocytes/macrophages or results in
skewing subsequent T
cell responses to highly pro-inflammatory responses. Comparisons between the
types of
inflammatory mediators produced by cells from IBD patients and those from
cells of normal
individuals can be made and might suggest that T cells and
monocyte/macrophages from IBD
patients produce a more pro-inflammatory profile in the presence of IL-17C.
The addition of a
soluble polypeptide of the present invention (i.e. IL-17REs2, IL-17REs3 or IL-
17REs4 as non-
limiting examples) to neutralize the production of downstream inflammatory
mediators induced by
IL-17C suggests that such soluble IL-17RE polypeptides may be efficacious in
the therapeutic
treatment of patients with IBD.
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
158
EXAMPLE 63
Efficacy of the Soluble IL-17RE Polypeptides in
Irritable Bowl Syndrome ("IBS"): CNS-Directed Pathogenesis
[564] A model focusing on primary CNS-directed pathogenesis of IBS which
employs
stress stimuli to induce symptoms characteristic of IBS. The neonatal
psychosocial stress model
mimics some clinical features associated with IBS patients including visceral
hyperalgesia, diarrhea
and stress-sensitivity. Daily separation of the litter from their mothers for
180 minutes each day
during postnatal days 4-18 will result in an alteration of maternal behaviour
and significantly reduce
times of the licking/grooming behaviour. The stress on the neonates results in
permanent changes in
the CNS resulting in altered stress-induced visceral and somatic pain
sensitivity. Colonic motor
function in response to stress is enhanced in these animals and preliminary
data shows evidence of
increased intestinal permeability (Mayer et al., 2002). Treatment with a
soluble polypeptide of the
present invention (i.e. IL-17REs2, IL-17REs3 or IL-17REs4 as non-limiting
examples) and
subsequent analysis of colonic motor function, epithelial permeability and
response to stress stimuli
could determine efficacy in this animal model of IBS. Decreases in the
incidence of symptoms
following treatment with these inhibitors would suggest potential efficacy in
the treatment of IBS.
EXAMPLE 64
Efficacy of the Soluble IL-17RE in
Irritable Bowl Syndrome ("IBS"): Primary Gut-Directed Inducers of Stress
[565] This is a model focusing on primary gut-directed inducers of stress (ie.
gut
inflammation, infection or physical stress). Animal studies have indicated
that low-grade
inflammation or immune activation may be a basis for altered motility, and/or
afferent and epithelial
function of the gut (Mayer et al,. 2002). In this model, daily colon
irritation is produced in neonatal
animals (days 8-21) in the form of daily intracolonic injection of mustard
oil. Mustard oil is a neural
stimulant and has been shown to induce visceral hyperalgesia following
intracolonic administration.
This model mimics key features of the IBS including visceral hypersensitivity
and alteration in
bowel habits. Animals also present with diarrhea or constipation, a key
feature of IBS patients
(Mayer et al., 2002; Kimball et al., 2005). A soluble polypeptide of the
present invention (i.e. IL-
17REs2, IL-17REs3 or IL-17REs4 as non-limiting examples) could be delivered to
determine
changes in the development of symptoms associated with this model. Decreases
in the incidence or
magnitude of visceral hypersensitivity and altered gut motility following
therapeutic treatment with
our inhibitors would suggest a potential for these molecules to be efficacious
in the treatment of IBS.
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
159
EXAMPLE 65
Soluble IL-17RE Modulates In Vivo Cytokine Production in a Murine Model of
Asthma
[566] Female BALB/c mice aresensitized i.p. with OVA in alum (l0ug/50% alum)
on days
0 and 7. On days 13 and 14 mice are treated intranasally with 150 ug of
soluble IL-17RE (such as
IL-17REs2, IL-17REs3 or IL-17REs4) or with vehicle. On day 14 and 15 mice aree
challenged
intranasally with OVA (20 ug/50u1 PBS). 24h and 48 h after the last intranasal
challenge mice are
euthanised. Bronchoalveolar lavage fluid is collected and cytokine levels
measured by Luminex
analysis. OVA sensitization and challenge in this model results in the
accumulation of a number of
immune related cytokines in bronchoalveolar lavage fluid. Treatment of mice
with any of the IL-
17RE polypeptides of the present invention result in a statistically
significant decrease in the levels
of several of these cytokines when compared with vehicle treated controls. The
following cytokines
are statistically significantly decreased at 24 or 48hrs post OVA challenge:
24hrs: RANTES, MIP1a;
and 48hrs: ILlbeta, IL4, IL5, IL13, IFNg, RANTES, MIP1a, IP10, GCSF. Thus,
therapeutic
manipulation of these cytokines in vivo is anticipated to have a beneficial
therapeutic effect.
EXAMPLE 66
IL-17C Induces Activation of the NFxB Pathway in NIH3T3 Cells Transfected with
Murine IL-
17REx6
[567] NIH3T3 cells were transfected with a IL-17REx6 expression construct, in
which the
CMV promoter was used to drive expression of IL-17REx6, with a zeocin
resistance gene included
as a selectable marker. Cells were transfected using the Mirus 3T3
transfection kit according to
manufacturer's instruction. Zeocin selection resulted in the establishment of
a population of zeocin
resistant cells. These cells were then stimulated with either mouse or human
IL-17C. IL-17C
mediated activation through IL-17REx6 was measured by the phosphorylation of
IkappaB in IL-
17RE transfected cells when compared with parental non-transfected cells. Non-
transfected NIH3T3
cells showed no dose dependent phosphorylation of IkappaB in response to
treatment with either
mouse or human IL-17C. In contrast NIH3T3 transfected with IL-17REx6 showed a
dose dependent
increase in IkappaB phosphorylation when stimulated with either mouse or human
IL-17C. These
data indicate that IL-17C is able to activate cells through the mouse IL-
17REx6 splice variant and
that both mouse and human IL-17C are able to interact productively with the
mouse IL-17REx6
receptor.
EXAMPLE 67
IL-17RE Variants and Constructs
[568] A series of constructs was created to comprise one or more fragments of
the IL-
17RE polypeptide. These constructs further comprise additional polypeptide
fragments, such as,
without limitation, leader fragments and immunoglobulin heavy chain constant
region fragments.
The IL-17RE fragments of the current constructs share at least a substantially
similar sequence
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
160
identity to SEQ ID NO: 2. Constructs are described below by identifying the
fragment portions of
said construct beginning at the n-terminus (amino acid residue no. 1) and
reading towards the c-
terminus. For the IL-17RE fragments of the constructs, the numbering is based
on the IL-17REx1
splice variant polypeptide (SEQ ID NO: 2) unless otherwise noted. For
convenience, the Fc
sequences that can be used interchangeably throughout have been included in
TABLE 6, below.
TABLE 6: Fc Polynucleotide Sequences and Polypeptide Sequences:
Fragment aa SEQ Polypeptide Sequence
Name ID NO:
nt SEQ Polynucleotide Sequence
ID NO:
Fc5 216
EPKSSDKTHTCPPCPAPEAEGAPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHED
Fragment PEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSN
KALPSSIEKTISKAKGQPREPQVYTLPPSRDELTKNQVSLTCLVKGFYPSDIAVE
WESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEALHNH
YTQKSLSLSPGK
215
GAGCCCAAATCTTCAGACAAAACTCACACATGCCCACCGTGCCCAGCACCTGAAG
CCGAGGGGGCACCGTCAGTCTTCCTCTTCCCCCCAAAACCCAAGGACACCCTCAT
GATCTCCCGGACCCCTGAGGTCACATGCGTGGTGGTGGACGTGAGCCACGAAGAC
CCTGAGGTCAAGTTCAACTGGTACGTGGACGGCGTGGAGGTGCATAATGCCAAGA
CAAAGCCGCGGGAGGAGCAGTACAACAGCACGTACCGTGTGGTCAGCGTCCTCAC
CGTCCTGCACCAGGACTGGCTGAATGGCAAGGAGTACAAGTGCAAGGTCTCCAAC
AAAGCCCTCCCATCCTCCATCGAGAAAACCATCTCCAAAGCCAAAGGGCAGCCCC
GAGAACCACAGGTGTACACCCTGCCCCCATCCCGGGATGAGCTGACCAAGAACCA
GGTCAGCCTGACCTGCCTGGTCAAAGGCTTCTATCCCAGCGACATCGCCGTGGAG
TGGGAGAGCAATGGGCAGCCGGAGAACAACTACAAGACCACGCCTCCCGTGCTGG
ACTCCGACGGCTCCTTCTTCCTCTACAGCAAGCTCACCGTGGACAAGAGCAGGTG
GCAGCAGGGGAACGTCTTCTCATGCTCCGTGATGCATGAGGCTCTGCACAACCAC
TACACGCAGAAGAGCCTCTCCCTGTCTCCGGGTAAA
Fc10 218
EPKSSDKTHTCPPCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHED
Fragment PEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSN
KALPAPIEKTISKAKGQPREPQVYTLPPSRDELTKNQVSLTCLVKGFYPSDIAVE
WESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEALHNH
YTQKSLSLSPGK
217
GAGCCCAAATCTTCAGACAAAACTCACACATGCCCACCGTGCCCAGCACCTGAAC
TCCTGGGGGGACCGTCAGTCTTCCTCTTCCCCCCAAAACCCAAGGACACCCTCAT
GATCTCCCGGACCCCTGAGGTCACATGCGTGGTGGTGGACGTGAGCCACGAAGAC
CCTGAGGTCAAGTTCAACTGGTACGTGGACGGCGTGGAGGTGCATAATGCCAAGA
CAAAGCCGCGGGAGGAGCAGTACAACAGCACGTACCGTGTGGTCAGCGTCCTCAC
CGTCCTGCACCAGGACTGGCTGAATGGCAAGGAGTACAAGTGCAAGGTCTCCAAC
AAAGCCCTCCCAGCCCCCATCGAGAAAACCATCTCCAAAGCCAAAGGGCAGCCCC
GAGAACCACAGGTGTACACCCTGCCCCCATCCCGGGATGAGCTGACCAAGAACCA
GGTCAGCCTGACCTGCCTGGTCAAAGGCTTCTATCCCAGCGACATCGCCGTGGAG
TGGGAGAGCAATGGGCAGCCGGAGAACAACTACAAGACCACGCCTCCCGTGCTGG
ACTCCGACGGCTCCTTCTTCCTCTACAGCAAGCTCACCGTGGACAAGAGCAGGTG
GCAGCAGGGGAACGTCTTCTCATGCTCCGTGATGCATGAGGCTCTGCACAACCAC
TACACGCAGAAGAGCCTCTCCCTGTCTCCGGGTAAA
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
161
[569] Construct 1543 (SEQ ID NOs: 219 and 220) comprises a leader fragment
from
amino acid position 1 to amino acid position 35 of said construct, an IL-17RE
fragment from amino
acid position 36 to amino acid position 332 of said construct and an fc5
fragment form amino acid
position 333 to amino acid position 564 of said construct. Said leader
fragment is an otPA pre-pro
signal secretion sequence. Said IL-17RE fragment is the amino acid sequence
from positions 155-
451 of the IL-17REx1 polypeptide. Construct 1542 (SEQ ID NOs: 259 and 260)
comprises an IL-
17RE fragment from amino acid position 1 to amino acid position 533 and an Fc5
fragment from
amino acid position 534 to amino acid position 765. Said IL-17RE fragment is
the amino acid
sequence from position 1 to 533 of the IL-17REx4 polypeptide. Constructs
disclosed herein may
further comprise amino acid insertions, deletions and substitutions as
compared to the sequences of
the referenced splice variants (e.g., IL-17REx1, IL-17REx4 or other references
polyprptide), and
these alternate sequence constructs are within the spirit of the current
invention. Applicants
hereinbelow provide a non-limiting description of a plurality of constructs
comprising IL-17RE
fragments.
[570] In one embodiment, the construct further comprises an addition of
a(G1y4Ser)3
linker insertion. In this embodiment, the construct (termed construct 1572
(SEQ ID NOs: 221 and
222)) comprises an otPA leader fragment from amino acid position 1 to amino
acid position 35, an
IL-17RE fragment from amino acid position 36 to amino acid position 332; a
linker from amino acid
position 333 to amino acid position 347 and an Fc5 fragment from amino acid
position 348 to amino
acid position 579. Said IL-17RE fragment of the 1572 construct is the amino
acid sequence from
position 155-451 of the IL-17REx1 polypeptide. Further constructs comprising
linker additions
include construct 1470 (SEQ ID NOs: 251 and 252), construct 1532 (SEQ ID NOs:
253 and 254)
and construct 1533 (SEQ ID NOs: 255 and 256). Construct 1470 comprises an otPA
leader fragment
from amino acid position 1 to amino acid position 35, an IL-17RE fragment from
amino acid
position 36 to amino acid position 312, said IL-17RE fragment corresponding to
the amino acid
sequence at position 176-452 of IL-17REx1, a linker fragment from position 313
to 330 and an Fc5
fragment from amino acid position 331 to amino acid position 562. Construct
1532 comprises an
otPA leader fragment from amino acid position 1 to amino acid position 35, an
IL-17RE fragment
from amino acid position 36 to amino acid position 333, said IL-17RE fragment
corresponding to the
amino acid sequence at amino acid position 155 to 452 of IL-17REx1, a linker
fragment from
position 334 to 351 and an Fc5 fragment from amino acid position 352 to amino
acid position 583.
Construct 1533 comprises an otPA leader fragment from amino acid position 1 to
amino acid
position 35, an IL-17RE fragment from amino acid position 36 to amino acid
position 337, said IL-
17RE fragment corresponding to the amino acid sequence at amino acid position
151 to 452 of IL-
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
162
17REx1, a linker fragment from position 338 to 355 and an Fc5 fragment from
amino acid position
356 to amino acid position 587.
[571] In a further alternative embodiment of the construct, the IL-17RE
fragment
comprises a domain extension which adds Tyr Arg His residues at said
fragment's c-terminus. Thus,
this alternative construct (termed construct 1574 (SEQ ID NOs: 223 and 224))
comprises an otPA
leader fragment from amino acid position 1 to amino acid position 35, an IL-
17RE fragment from
amino acid position 36 to amino acid position 335 (corresponding to the amino
acid sequence from
position 155-454 of the IL-17REx1 polypeptide) and an Fc5 fragment from amino
acid position 336
to amino acid position 567. Construct 1653 (SEQ ID NOs: 277 and 278) comprise
an IL-17RE
fragment, wherein said IL-17RE fragment is a sequence from exon 7 through exon
12 of IL-17REx1
and wherein said IL-17RE fragment further comprises the addition of a
glutamine residue at its n-
terminus. Said Q residue is added to the n-terminus of the IL-17RE fragment to
facilitate
cleavage/processing of a leader sequence, such as an optimized tissue
plasminogen activator or an
otPA leader sequence. This construct further comprises an Fc5 fragment.
Construct 1660 (SEQ ID
NOs: 285 and 286) comprise an IL-17RE fragment, wherein said IL-17RE fragment
is a sequence
from exon 5 through exon 10 of IL_17REx1 and wherein said IL-17RE fragment
further comprises
the addition of an alanine residue and a glycine residue at its n-terminus.
Said A and G residues are
added to the n-terminus to facilitate cleavage/processing of a leader
sequence. It is further notable
that the A residue corresponds to the residue that is two positions upstream
from exon 5 in the IL-
17REx1 polypeptide sequence, but at one position upstream the native IL-17REx1
Glutamine residue
has been substituted with the glycine residue.
[572] Constructs 1528 (SEQ ID NOs: 257 and 258) and 1543 (SEQ ID NOs: 219 and
220)
comprise IL-17RE fragments that have deletion bearing amino acid sequences
corresponding to S2
and S3 polypeptide regions of IL-17REx1, respectively (See Figure 1).
Construct 1528 comprises an
otPA leader fragment at amino acid position 1 to amino acid position 35, an IL-
17RE fragment at
amino acid position 36 to amino acid position 309, said IL-17RE fragment
corresponding to the
amino acid sequence from position 178 to 451 of the IL-17REx1 polypeptide, and
an Fc5 fragment at
amino acid position 310 to amino acid position 541. The S2 polypeptide
illustrated in figure 1
corresponds to the amino acid sequence at position 176 to 454 of IL-17REx1,
thus said IL-17RE
fragment of construct 1528 bears a 2 amino acid residue deletion at the n-
terminus of the fragment
and a 3 amino acid residue deletion at the c-terminus of the fragment when
aligned with the S2
polypeptide. Construct 1543 is discussed above. The IL-17RE fragment of
construct 1543
corresponds to the amino acid sequence from position 155 to 451 of the IL-
17REx1 polypeptide.
The S3 polypeptide illustrated in figure 1 corresponds to the amino acid
sequence at position 155 to
454 of IL-17REx1, thus said IL-17RE fragment of construct 1543 bears a 3 amino
acid residue
deletion at the c-terminus of said fragment when aligned with the S3
polypeptide.
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
163
[573] In a further embodiment, constructs comprise one or more substitutions
in the IL-
17RE fragment wherein an Asn residue is substituted with an Ala residue. Said
substitutions are N-
linked glycosylation substitution mutations corresponding to the N-linked
motifs of said IL-17RE
fragment. Construct 1585 (SEQ ID NOs: 225 and 226)) comprises an otPA fragment
from amino
acid position 1 to amino acid position 35, an IL-17RE fragment from amino acid
position 36 to
amino acid position 332, said IL-17RE fragment further comprising N199A, and
an Fc5 fragment
from amino acid position 333 to amino acid position 564. Said IL-17RE fragment
of construct 1585
is the amino acid sequence from position 155 to 451 of IL-17REx1 but
comprising N318A, with
reference to said same polypeptide. A still further construct comprises an N-
linked glycosylation
substitution mutation at the second N-linked motif of its IL-17RE fragment.
This construct (termed
construct 1586 (SEQ ID NOs: 227 and 228)) comprises an otPA fragment from
amino acid position
1 to amino acid position 35, an IL-17RE fragment from amino acid position 36
to amino acid
position 332, and further comprising N228A, and an Fc5 fragment from amino
acid position 333 to
amino acid position 564. Said IL-17 fragment corresponds to the amino acid
sequence from position
155 to 451 of IL-17REx1, with N347A. An additional embodiment comprises an N-
linked
glycosylation substitution mutation at a third N-linked motif (counting the
direction from N-terminus
to C-terminus) of its IL-17RE fragment. This construct is termed 1587 (SEQ ID
NOs: 229 and 230)
and comprises an otPA leader fragment from amino acid position 1 to amino acid
position 35, an IL-
17RE fragment from amino acid position 36 to amino acid position 332 and an
Fc5 fragment from
amino acid position 333 to amino acid position 564. Said IL-17RE fragment
further comprises
N245A. Said IL-17RE fragment corresponds to the amino acid sequence from
position 155 to 451 of
IL-17REx1, with N364A. Construct 1588 (SEQ ID NOs: 231 and 232) comprises N-
linked
glycosylation substitution mutations at all three of the above described N-
linked motifs. Thus,
construct 1588 comprises an otPA leader fragment from amino acid position 1 to
amino acid position
35, an IL-17RE fragment from amino acid position 36 to amino acid position 332
and an Fc5
fragment from amino acid position 333 to amino acid position 564. Said IL-17RE
fragment further
comprises N199A, N228A and N245A. Said IL-17RE fragment corresponds to the
amino acid
sequence from position 155 to 451 of IL-17REx1, with N318A, N347A and N364A.
Constructs
comprises these and other amino acid substitutions are well within the scope
of the instantly
disclosed discovery.
[574] Applicants have further made constructs comprising IL-17RE fragments,
said IL-
17RE fragments further comprising substitutions and/or additions wherein the
substituted or added
amino acid sequences come from other cytokine receptors; preferably other IL-
17 receptor family
members. One exemplary construct (termed construct 1497 (SEQ ID NO: 233 and
234)) comprises
an IL-17 receptor fragment from amino acid position 1 to amino acid position
476 and an Fc5
fragment from amino acid position 477 to amino acid position 708. Amino acid
position 1 to amino
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
164
acid position 199 of said IL- 17 receptor fragment corresponds to exon 1 to
exon 6 of an IL-17RA
polypeptide, while amino acid position 200 to amino acid position 476
corresponds to exon 5 to exon
16 of IL-17REx1. Alternatively, construct 1529 (SEQ ID NOs: 235 and 236)
comprises an IL-17
receptor fragment at amino acid position 1 to amino acid position 450 and an
Fc5 fragment at amino
acid position 451 to amino acid position 682. Amino acid position 1 to amino
acid position 176 of
said IL- 17 receptor fragment corresponds to exon 1 to exon 6 of IL-17RB while
amino acid position
177 to amino acid position 450 corresponds to exon 5 to exon 16 of IL-17REx1.
A similar construct
was designed with IL-17RD exons substituted and added to IL-17RE exons to
generate an IL- 17
receptor fragment. This construct was termed 1530 (SEQ ID NOs: 237 and 238)
and comprises an
IL- 17 receptor fragment from amino acid position 1 to amino acid position 472
and an Fc5 fragment
from amino acid position 473 to amino acid position 704. Amino acid position 1
to amino acid
position 198 of said IL-17 receptor fragment corresponds to exon 1 to exon 6
of IL-17RD while
amino acid position 199 to amino acid position 472 corresponds to exon 5 to
exon 15 of IL-17REx1.
Construct 1570 (SEQ ID NOs: 261 and 262) comprises an IL-17RA fragment,
wherein said IL-
17RA fragment sequence is exon 1 through exon 6 of IL-17RA, and an IL-17RE
fragment, wherein
said IL-17RE fragment sequence is from the start of S3 polypeptide region of
IL-17REx1 through
exon 15 of said IL-17REx1. Construct 1578 (SEQ ID NOs: 263 and 264) comprises
a first IL-17RA
fragment, wherein said first IL-17RA fragment sequence is exon 1 through exon
6 of IL-17RA, an
IL-17RE fragment, wherein said IL-17RE fragment's amino acid sequence is from
the start site of
the S3 polypeptide region of IL-17REx1 through exon 11 of said IL-17REx1, and
a second IL-17RA
fragment, wherein said second IL-17RA fragment sequence is exon7 through exon
10 of IL-17RA.
Construct 1579 (SEQ ID NOs: 265 and 266) comprises exon 1 through exon 6 of IL-
17RA, exon 6
through exon 11 of IL-17REx1 and exon 7 through exon 10 of IL-17RA. Construct
1583 (SEQ ID
NOs: 267 and 268) comprises exon 1 through exon 6 of IL-17RA, from the S3
polyprptide site
through exon 8 of IL-17REx1, exon 7 through exon 9 of IL-17RA and exon 12
through exon 15 of
IL-17REx1. Construct 1666 (SEQ ID NOs: 269 and 270) comprises from the native
leader sequence
of IL-17RA through exon 5 of said IL-17RA and exon 6 through exon 15 of IL-
17REx1. Construct
1667 (SEQ ID NOs: 271 and 272) comprises exon 1 through exon 10 of IL-17RA, an
IL-17RE
fragment, wherein said fragment is the amino acid sequence from amino acid
position 176 to amino
acid position 451 of IL-17REx1, and an fc5 fragment. Construct 1664 (SEQ ID
NOs: 273 and 274)
comprises exon 1 through exon 5 of IL-17RB, an IL-17RE fragment, wherein said
fragment is the
amino acid sequence from amino acid position 176 to amino acid position 451 of
IL-17REx1, and an
fc5 fragment. Construct 1668 (SEQ ID NOs: 275 and 276) comprises exon 1
through exon 5 of IL-
17RD, an IL-17RE fragment, wherein said fragment is the amino acid sequence
from amino acid
position 176 to amino acid position 451 of IL-17REx1, and an fc5 fragment.
Construct 1703 (SEQ
ID NOs: 287 and 288) comprises exon 1 through exon 5 of IL-17RA, and an IL-
17RE fragment,
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
165
wherein said fragment is the amino acid sequence from the start sight of the
S3 polyprptide of IL-
17REx1 through exon 11 of said IL-17REx1. Said construct can be conjugated
with any Fc
molecule, including, but not limited to, Fc5 and Fc10. IL-17RA sequences are
known in the art.
(See, e.g., Yao, Z et al, Cytokine, 9(11):794 (1997)). IL-17RB sequences are
known in the art. (See
e.g., Shi, Y et al, J. Biol. Chem. 275(25):19167 (2000)). IL-17RD sequences
are known in the art.
(See e.g., GenBank Accession No.: NM017563.1 and GI:24308146). Additional
substitutions
and/or additions of cytoking receptors to the IL-17RE fragment of a construct
are contemplated by
the instantly disclosed discovery.
[575] Constructs comprising an IL-17RE fragment and a fusion tag addition can
be made.
Fusion tags are useful for facilitating, amongst other things, polypeptide
purification, polypeptide
identification, and polypeptide post-translational modification to effect
structure or function (e.g.,
folding, export, phosphorylation, bond formation) using validated technology
(See e.g., Stevens,
Structure, 8, R177-R185 (2000)). Fusion tags can be either C-terminal, N-
terminal or internal, as is
known in the art. In one example, construct 1589 (SEQ ID NOs: 239 and 240) was
designed
comprising an IL-17RE fragment and an N-terminal His-tag. Said IL-17RE
fragment is from amino
acid residue 1 to amino acid residue 454 of said construct 1589, and has an
amino acid sequence that
corresponds to exon 1 to exon 15 of IL-17REx1. Said His-tag is a 6X HIS from
amino acid position
455 to amino acid position 460. Other fusion tags and methods of making fusion
polypeptides are
well known to those ordinarily skilled in the art.
[576] Additional constructs include, but are not limited to, the following.
Construct 1661
(SEQ ID NOs: 279 and 280) comprises an IL-17RE amino acid sequence
corresponding to position
90 to position 267 of IL-17REx1 and an Fc5 fragment. Construct 1652 (SEQ ID
NOs: 281 and 282)
comprises an IL-17RE amino acid sequence corresponding to position 176 to 357
of IL-17REx1 and
an Fc5 fragment. Construct 1654 (SEQ ID NOs: 283 and 284) comprises an IL-17RE
amino acid
corresponding to position 246 to 432 of IL-17REx1 and an Fc5 fragment.
[577] As is illustrated in Figure 1, the extracellular binding domain of IL-
17RE comprises
the amino acids of exon 1 through exon 15 and the first five amino acid
residues of exon 16. Figure
1 uses IL-17REx1 splice variant for illustration purposes only and does not
limit the application of
applicants' discovery to this IL-17RE polypeptide. Constructs were designed to
comprise an IL-
17RE extracellular domain ("ecd") fragment. Construct 1640 (SEQ ID NOs: 289
and 290)
comprises an IL-17RE ecd fragment, wherein said ecd fragment is the amino acid
sequence from
exon 1 through exon 15 of IL-17REx1. Construct 1641 (SEQ ID NOs: 291 and 292)
comprises an
IL-17RE ecd fragment, wherein said ecd fragment is the amino acid sequence
from exon 2 through
exon 15 of IL-17REx1. Construct 1642 (SEQ ID NOs: 293 and 294) comprises an IL-
17RE ecd
fragment, wherein said ecd fragment is the amino acid sequence from exon 3
through exon 15 of IL-
17REx1. Construct 1643 (SEQ ID NOs: 295 and 296) comprises an IL-17RE ecd
fragment, wherein
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
166
said ecd fragment is the amino acid sequence from exon 4 through exon 15 of IL-
17REx1. Construct
1644 (SEQ ID NOs: 297 and 298) comprises an IL-17RE ecd fragment, wherein said
ecd fragment is
the amino acid sequence from exon 5 through exon 15 of IL-17REx1, and further
comprises the
addition of AG residues to the n-terminus end of said ecd fragment. A
description of this AG
addition is found above in the discussion of Construct 1660. Construct 1645
(SEQ ID NOs: 299 and
300) comprise an IL-17RE ecd fragment, wherein said ecd fragment is the amino
acid sequence from
exon 6 through exon 15 of IL-17REx1. Construct 1646 (SEQ ID NOs: 301 and 302)
comprises an
IL-17RE ecd fragment, wherein said ecd fragment is the amino acid sequence
from exon 7 through
exon 15 of IL-17REx1, and further comprises a Q residue added to the n-
terminus of said ecd
fragment. The description of this Q residue addition is found above in the
discussion of construct
1653. Construct 1647 (SEQ ID NOs: 303 and 304) comprise an IL-17RE ecd
fragment, wherein said
ecd fragment is the amino acid sequence from exon 8 through exon 15 of IL-
17REx1. Construct
1648 (SEQ ID NOs: 305 and 306) comprise an IL-17RE ecd fragment, wherein said
ecd fragment is
the amino acid sequence from exon 9 through exon 15 of IL-17REx1. Construct
1669 (SEQ ID
NOs: 307 and 308) comprise an IL-17RE ecd fragment, wherein said ecd fragment
is the amino acid
sequence from exon 1 through exon 14 of IL-17REx1. Construct 305 (SEQ ID NOs:
247 and 248)
comprise an IL-17RE ecd fragment, wherein said ecd fragment is the amino acid
sequence from
amino acid position 176 to amino acid position 409 of IL-17REx1.
[578] Applicants have further identified three ligand binding domains within
the IL-17RE
ecd. With reference to IL-17REx1, the three ligand binding domains are as
follows: (1) from the S3
start site through exon 8; (2) from exon 9 through exon 11; and (3) from exon
12, through exon 15
and including the valine and the serine residues at the very n-terminal
portion of exon 16. Applicants
have further designed constructs to comprise one or more of these ligand
binding domains.
Construct 1700 (SEQ ID NOs: 241 and 242) comprises an IL-17RE fragment,
wherein the amino
acid sequence of said IL-17RE fragment corresponds to the amino acid sequence
that is from the S3
start site through exon 8 of IL-17REx1, and a serine residue and a valine
residue have been added to
the n-terminus of said IL-17RE fragment, in that order, to facilitate leader
cleavage efficiency. Said
construct further comprises an otPA leader fragment, an Fc5 fragment or both.
One example of a
construct comprising said IL-17RE fragment, an otPA fragment and an Fc5
fragment is SEQ ID NO:
312 found in TABLE 7. Construct 1699 (SEQ ID NOs: 309 and 310) comprises an IL-
17RE
fragment, wherein the amino acid sequence of said IL-17RE fragment corresponds
to the amino acid
sequence that is from exon 9 through exon 11 of IL-f17REx1. Said construct
further comprises an
otPA leader fragment, an Fc5 fragment or both. One example of a construct
comprising said IL-
17RE fragment, an otPA fragment and an Fc5 fragment is SEQ ID NO: 313 found in
TABLE 7.
Construct 1701 (SEQ ID NOs: 243 and 244) comprises an IL-17RE fragment,
wherein the amino
acid sequence of said IL-17RE fragment corresponds to the amino acid sequence
that is from the S3
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
167
start site through exon 11 of IL-17REx1, and a serine residue and a valine
residue have been added
to the n-terminus of said IL-17RE fragment, in that order, to facilitate
leader cleavage efficiency.
Said construct further comprises an otPA leader fragment, an Fc5 fragment or
both. One example of
a construct comprising said IL-17RE fragment, an otPA fragment and an Fc5
fragment is SEQ ID
NO: 314 found in TABLE 7. Construct 1702 (SEQ ID NOs: 245 and 246) comprises
an IL-17RE
fragment, wherein the amino acid sequence of said IL-17RE fragment corresponds
to the amino acid
sequence that is from the S3 start site through exon 15 of IL-17REx1, with the
exception that the
Cysteine residue corresponding to position 439 of IL-17REx1 was converted to a
Serine residue in
construct 1702 to provide for an even number of total cysteine residues in
said construct;
additionally, a serine residue and a valine residue have been added to the n-
terminus of said IL-17RE
fragment, in that order, to facilitate leader cleavage efficiency. Said
construct further comprises an
otPA leader fragment, an Fc5 fragment or both. One example of a construct
comprising said IL-
17RE fragment, and otPA fragment and an Fc5 fragment is SEQ ID NO: 315 found
in TABLE 7.
Table 7
SEQ ID
NO: Sequence
312
MDAMKRGLCCVLLLCGAVFVSLSQEIHAELRRFRRSVTQPSDPETWESLP
RLDSQRHGGPEFSFDLLPEARAIRVTISSGPEVSVRLCHQWALECEELSS
PYDVQKIVSGGHTVELPYEFLLPCLCIEASYLQEDTVRRKKCPFQSWPEA
EPKSSDKTHTCPPCPAPEAEGAPSVFLFPPKPKDTLMISRTPEVTCVWD
VSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLN
GKEYKCKVSNKALPSSIEKTISKAKGQPREPQVYTLPPSRDELTKNQVSL
TCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKS
RWQQGNVFSCSVMHEALHNHYTQKSLSLSPGK
313
MDAMKRGLCCVLLLCGAVFVSLSQEIHAELRRFRRYGSDFWKSVHFTDYS
QHTQMVMALTLRCPLKLEAALCQRHDWHTLCKDLPNATARESDGWYVLEK
VDLHPQLCFKFSFGNSSHVECPHQTEPKSSDKTHTCPPCPAPEAEGAPSV
FLFPPKPKDTLMISRTPEVTCVWDVSHEDPEVKFNWYVDGVEVHNAKTK
PREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKALPSSIEKTISKAK
GQPREPQVYTLPPSRDELTKNQVSLTCLVKGFYPSDIAVEWESNGQPENN
YKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEALHNHYTQKS
LSLSPGK
314
MDAMKRGLCCVLLLCGAVFVSLSQEIHAELRRFRRYSVTQPSDPETWESL
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
168
PRLDSQRHGGPEFSFDLLPEARAIRVTISSGPEVSVRLCHQWALECEELS
SPYDVQKIVSGGHTVELPYEFLLPCLCIEASYLQEDTVRRKKCPFQSWPE
AGSDFWKSVHFTDYSQHTQMVMALTLRCPLKLEAALCQRHDWHTLCKDLP
NATARESDGWYVLEKVDLHPQLCFKFSFGNSSHVECPHQTEPKSSDKTHT
CPPCPAPEAEGAPSVFLFPPKPKDTLMISRTPEVTCVWDVSHEDPEVKF
NWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSN
KALPSSIEKTISKAKGQPREPQVYTLPPSRDELTKNQVSLTCLVKGFYPS
DIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSC
SVMHEALHNHYTQKSLSLSPGK
315
MDAMKRGLCCVLLLCGAVFVSLSQEIHAELRRFRRYSVTQPSDPETWESL
PRLDSQRHGGPEFSFDLLPEARAIRVTISSGPEVSVRLCHQWALECEELS
SPYDVQKIVSGGHTVELPYEFLLPCLCIEASYLQEDTVRRKKCPFQSWPE
AGSDFWKSVHFTDYSQHTQMVMALTLRCPLKLEAALCQRHDWHTLCKDLP
NATARESDGWYVLEKVDLHPQLCFKFSFGNSSHVECPHQTGSLTSWNVSM
DTQAQQLILHFSSRMHATFSAAWSLPGLGQDTLVPPVYTVSQARGSSPVS
LDLIIPFLRPGSCVLVWRSDVQFAWKHLLCPDVSEPKSSDKTHTCPPCPA
PEAEGAPSVFLFPPKPKDTLMISRTPEVTCVWDVSHEDPEVKFNWYVDG
VEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKALPSS
IEKTISKAKGQPREPQVYTLPPSRDELTKNQVSLTCLVKGFYPSDIAVEW
ESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEA
LHNHYTQKSLSLSPGK
EXAMPLE 68
Effects of Soluble Il-17RE on Mice with LPS Induced Endotoxemia
[579] BALBc mice received two intranasal deliveries each of 250mg IL17REs2Fc10
(which is an IL-17REx2 s2 polypeptide splice variant fused at its c-terminus
to an Fc10 fragment) at
T-24hrs and T--2hrs. At TOhrs animal were challenged with 60mg LPS, delivered
intranasally. At
6hrs and 24hrs mice were euthanised and the contents of the lungs
(bronchoalveolar lavage fluid,
BALF), harvested. Control animals were processed in an identical manner, but
received vehicle in
place of IL17REs2Fc10. Each experimental group contained 5 animals.
[580] Cytokine and chemokine levels were measured in the BALF of individual
animals
by Luminex analysis. Data was analysed by Unpaired T-test. The following
cytokines showed
statistically significant decreases in the LPS-induced BALF of animals treated
with IL17REs2Fc10
when compared with LPS-induced BALF from vehicle treated controls:IL-la, GM-
CSF, IL-5, MCP-
1, MIP-1a, KC, G-CSF. Since LPS induced endotoxemia engages many of the
proinflammatory
mediators that produce pathological effects in the infectious diseases and LPS
induced endotoxemia
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
169
in rodents is a widely used and acceptable model for studying the
pharmacological effects of
potential pro-inflammatory or immunomodulating agents, the present results
indicate that the soluble
IL-17RE polypeptides of the present invention are potent immunomodulating
agents and thus useful
in treating and/or reducing the symptoms of multiple inflammatory diseases.
EXAMPLE 69
Soluble Constructs Comprising At Least One Binding Domain.
[565] Applicants have characterized the IL-17RE polypeptide, including its
Extracellular
Ligand Binding Domain, Transmembrane Domain and Intracellular Signaling
Domain. Applicants
have identified that the Extracellular Ligand Binding Domain comprises a
signaling sequence, a first
ligand binding domain, a second ligand binding domain and a third ligand
binding domain.
Applicants have used this discovery to design a series of constructs
comprising an amino acid residue
sequence with substantial similarity to one or more of said binding domains.
Moreover, Applicants
have designed constructs that further comprise one or more of immunoglobulin
heavy chain constant
regions, leader sequences, linker regions, or protease enhancer regions.
Applicants have further
designed (i) polynucleotide sequences encoding said polypeptide constructs;
(ii) expression vectors
comprising said polynucleotide sequences of (i); and (iii) cell lines that
have been transformed with
said expression vectors of (ii) and which are capable of expressing said
polypeptide constructs.
Applicants have expressed said polypeptide constructs from said transformed
cell lines, recovered the
purified polypeptide constructs and used said purified polypeptide constructs
to perform ligand
binding assays (e.g., BIACOR, Biacor AB, a GE Healthcare, Piscataway, NJ).
Thus, said polypeptide
constructs are useful for binding ligand, said ligand including, but not
limited to IL-17C. Said
polypeptide constructs may also be useful for preparing medicaments to treat
autoimmune disorders in
patients needing such said treatment. For example, pharmaceutical compositions
comprising said
polypeptide constructs may be useful to treat patients having multiple
sclerosis, inflammatory bowel
disease or rheumatoid arthritis. Said polypeptide constructs may also be
useful as in vitro diagnostic
systems. Kits comprising said polypeptide constructs, either alone or as
pharmaceutical compositions,
may also be prepared and will include additional kitting components. Those
ordinarily skilled in the
art will readily generate polypeptide constructs as described herein and
within the spirit of this
disclosure. Said constructs, as well as the encoding polynucleotides, vectors,
cell lines, methods for
providing medicaments, pharmaceutical compositions, kits and methods for
treating autoimmune
disorders or providing diagnosis therewith are within the spirit of this
disclosure.
[566] Illustration of ligand binding domains with reference to IL-17REx1. IL-
17REx1 is a
667 amino acid polypeptide and is illustrated along with splicing variants S2,
S3 and S4, in FIGS l a-d
Herein, IL-17REx1 (SEQ ID NOs 1 and 2) is used as the reference sequence when
discussing the
current discoveries, however, this reference is for convenience and not
limitation. The IL-17REx1
reference sequence comprises an extracellular ligand binding domain that
begins at amino acid
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
170
residue 24 and continues to amino acid residue 454. The extracellular ligand
binding domain further
comprises a first ligand binding domain, a second ligand binding domain and a
third ligand binding
domain. These ligand binding domains are described herein above, and for
convience, are called out
in the following TABLE 8
Table 8
Ligand Position Relative SEQUENCE SEQ
Binding to SEQ ID NO: 2 ID
Domain NO:
First From the S3 start TQPSDPETWESLPRLDSQRHGGPEFSFDLLPEARAIRV 249
site through exon TISSGPEVSVRLCHQWALECEELSSPYDVQKIVSGGHT
8 VELPYEFLLPCLCIEASYLQEDTVRRKKCPFQSWPEA
Second From exon 9 YGSDFWKSVHFTDYSQHTQMVMALTLRCPLKLEAALCQRH 310
through exon 11 DWHTLCKDLPNATARESDGWYVLEKVDLHPQLCFKFSFGN
SSHVECPHQT
Third From exon 12 GSLTSWNVSMDTQAQQLILHFSSRMHATFSAAWSLPGLGQD 250
through exon 15 TLVPPVYTVSQARGSSPVSLDLIIPFLRPGSCVLVWRSDVQ
and includes the V FAWKHLLCPDVS
residue and S
residue at the n-
terminus of exon
16
[567] Construct 1700 is designed to preferably comprise a leader sequence, a
ligand binding
domain and an Fc fragment. The ligand binding domain of construct 1700
preferably comprises an
amino acid sequence that is substantially similar to the first ligand binding
domain of SEQ ID NO:2,
more preferably at leat 90% identical to the first ligand binding domain of
SEQ ID NO: 2 and most
preferably is SEQ ID NO: 242. SEQ ID NO: 241 is a polynucleotide that encodes
SEQ ID NO: 242.
Applicants cloned SEQ ID NO: 241 into a suitable expression vector, as
described above, said
expression vector further comprising a polynucleotide encoding a leader
sequence and a
polynucleotide encoding an Fc fragment. Preferably, said leader sequence is an
otPA leader
sequence. Preferably, said Fc fragment is Fc5. A CHO cell line was transformed
with said expression
vector comprising said leader sequence, said ligand binding domain and said fc
fragment. Purified
polypeptide construct 1700 was expressed from this cell line and was harvested
from the cell media.
An HPLC assay confirmed that the harvested 1700 construct was present in the
cell media at about 6
mg/L. Purified polypeptide construct 1700 was then analyzed via a BIACOR
binding assay, as
described below, and it was determined that construct 1700 is capable of
binding IL-17C ligand.
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
171
[568] Construct 1699 is designed to preferably comprise a leader sequence, a
ligand binding
domain and an Fc fragment. The ligand binding domain of construct 1699
preferably comprises an
amino acid sequence that is substantially similar to the second ligand binding
domain of SEQ ID NO:
2, more preferably at least 90% identical to said second ligand binding domain
of SEQ ID NO: 2, and
most preferably is SEQ ID NO: 310. SEQ ID NO: 309 is a polynucleotide sequence
that encodes
SEQ ID NO: 310. Applicants cloned SEQ ID NO: 309 into a suitable expression
vector, said vector
further comprising a polynucleotide encoding a leader sequence and a
polynucleotide encoding an Fc
fragment. Preferably, said leader sequence is is an otPA leader sequence.
Preferably, said Fc
fragment is Fc5. A CHO cell line was transformed with said expression vector
comprising said leader
sequence, said ligand binding domain and said fc fragment. Purified
polypeptide construct 1699 was
expressed from this cell line and was harvested from the cell media. An HPLC
assay confirmed that
the harvested 1699 construct was present in the cell and the expressed
construct was then analyzed via
a BIACOR binding assay, as described below. Construct 1699 is capable of
binding IL-17C ligand.
This construct was similarly transformed into a CHO cell line, expressed and
purified from the cell
media supernatant. BIACOR binding assays indicated that this construct binds
the IL-17C ligand.
[569] Construct 1701 is designed to preferably comprise a leader sequence, a
first and a
second ligand binding domain and an Fc fragment. The first ligand binding
domain of construct 1701
preferably comprises an amino acid sequence that is substantially similar to
the first ligand binding
domain of SEQ ID NO:2, more preferably at leat 90% identical to the first
ligand binding domain of
SEQ ID NO: 2 and most preferably is SEQ ID NO: 249. The second ligand binding
domain of
construct 1701 preferably comprises an amino acid sequence that is
substantially similar to the second
ligand binding domain of SEQ ID NO:2, more preferably at leat 90% identical to
the second ligand
binding domain of SEQ ID NO: 2 and most preferably is SEQ ID NO: 310. In a
preferred
embodiment, said first and second ligand binding domains of construct 1701
comprises an amino acid
sequence that is substantially similar to SEQ ID NO: 244, more preferably, at
least 90% identical to
SEQ ID NO: 244 and most preferably is SEQ ID NO: 244. SEQ ID NO: 243 is a
polynucleotide that
encodes SEQ ID NO: 244. Applicants cloned SEQ ID NO: 243 into a suitable
expression vector, said
vector further comprising a polynucleotide encoding a leader sequence and a
polynucleotide encoding
an Fc fragment. Preferably, said leader sequence is is an otPA leader
sequence. Preferably, said Fc
fragment is Fc5. A CHO cell line was transformed with said expression vector
comprising said leader
sequence, said ligand binding domain and said fc fragment. Purified
polypeptide construct 1701 was
expressed from this cell line and was harvested from the cell media. An HPLC
assay confirmed that
the harvested 1701 construct was present in the cell and the expressed
construct was then analyzed via
a BIACOR binding assay, as described below. Construct 1701 is capable of
binding IL-17C ligand.
A hybrid version of this construct was designed to include a leader sequence
from IL-17RA, and
lacking the Ser residue and Val residue at the amino terminus of the ligand
binding domain. A
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
172
preferred embodiment of this hybrid polypeptide construct is was named
Construct 1703 (SEQ ID
NO: 288).
[570] Construct 1702 is designed to preferably comprise a leader sequence, a
first, a second
and a third ligand binding domain and an Fc fragment. The first ligand binding
domain of construct
1702 preferably comprises an amino acid sequence that is substantially similar
to the first ligand
binding domain of SEQ ID NO:2, more preferably at leat 90% identical to the
first ligand binding
domain of SEQ ID NO: 2 and most preferably is SEQ ID NO: 249. The second
ligand binding
domain of construct 1702 preferably comprises an amino acid sequence that is
substantially similar to
the second ligand binding domain of SEQ ID NO:2, more preferably at leat 90%
identical to the
second ligand binding domain of SEQ ID NO: 2 and most preferably is SEQ ID NO:
310. The third
ligand binding domain of construct 1702 preferably comprises an amino acid
sequence that is
substantially similar to SEQ ID NO: 250, more preferably is at least 90%
identical to SEQ ID NO:
250 and most preferably is SEQ ID NO: 250. In a preferred embodiment, said
first, second and third
ligand binding domains of construct 1702 comprises an amino acid sequence that
is substantially
similar to SEQ ID NO: 246, more preferably, at least 90% identical to SEQ ID
NO: 246 and most
preferably is SEQ ID NO: 246. In construct 1702, a Cysteine residue in
corresponding to Cys429 of
SEQ ID NO: 2 was substituted with a Serine. This substitution evens out the
number of Cysteine
residues in construct 1702 thus providing for more consistent Cysteine bond
paring in the soluble
polypeptide receptor. SEQ ID NO: 245 is a polynucleotide that encodes SEQ ID
NO: 244.
Applicants cloned SEQ ID NO: 245 into a suitable expression vector, said
vector further comprising a
polynucleotide encoding a leader sequence and a polynucleotide encoding an Fc
fragment.
Preferably, said leader sequence is is an otPA leader sequence. Preferably,
said Fc fragment is Fc5.
A CHO cell line was transformed with said expression vector comprising said
leader sequence, said
ligand binding domain and said fc fragment. Purified polypeptide construct
1702 was expressed from
this cell line and was harvested from the cell media. An HPLC assay confirmed
that the harvested
1702 construct was present in the cell and the expressed construct was then
analyzed via a BIACOR
binding assay, as described below. Construct 1702 is capable of binding IL-17C
ligand.
[571] Western blots were performed to identify the ratio of construct
expressed into the
supernatants to construct remaining within a transformed cell line. As
described above, constructs
1699, 1700, 1701 and 1702 were each cloned into an independent CHO cell line.
Briefly, 200
micrograms of purified plasmids encoding constructs 1699, 1700, 1701 and 1702
were each digested
using with 240 units of BSTB1 enzyme in 300 microliter vol. in NEB buffer #4
to linearize. The
linearized plasmids were extracted by phenol:chloroform, Sodium Acetate/EtOH
precipitation. Said
extracted plasmids were resuspended in 400 microliter of a protein free CHO
media (EX-Ce11325 PF
CHO Media, SAFC Biosciences, Sigma, St. Louis, MO, Catalog No.: 14340C) and
incubated for 10
min at 37 deg C. to solubilize.
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
173
[572] For each electroporation: 1e7 5XS.A. DXB11 cells (passage 48) in 0.8 mls
EX-Cell
325 media containing 200 micrograms of linearized plasmid were electroporated
at .3kV @ 950
.micro.F in a 4mM gap cuvette and the contents of electroporation chamber were
added to 25 mls of
EX-Cell 325 media in a 125m1 shakeflask w/ 500 micro.l of Pen/Strep solution
addition and a cell
concentration of 4e5 cells/ml. At day 6 post electroporation cells were spun
out of media at 1200 rpm
for 5min in a tabletop centrifuge. The cell pellet was resuspended in 25m1s
fresh EX-Ce11325 media
w/ 200 nM Methotrexate and incubated for 24 hours in an incubator with
settings at 37 deg. C, with
6% CO2. Cells were repeatedly pelleted and resuspended to from 3e5 to 5e5
cells per ml every
three to four days using 25m1s fresh EX-Cell 325 media w/ from about 200 nM
Methotrexate to about
500nM Methotrexate. After Passage 5 post transfection, the cell culture was
centrifuged to pellet the
cells. An aliquot of the supernatant and an equivalent amount of cells were
prepared for western
analysis (lml and lml equivalent, respectively).
[573] Samples were run denatured, non-reduced at 10 microliter/sample/lane on
a NuPage
4-12% Bis-Tris gels w/ MES buffer (Invitrogen, Carlsbad, CA). Cell aliquot
samples were prepared
by resuspending each frozen lml equivalent cell pellet in 250 microliter of
Lysis buffer (150 mM
NaC1, 50mM Tris pH 8, 1% NP-40 w/ 1 protease inhibitor tablet/ lOml of buffer
(Roche Complete
Mini-EDTA free)). The resuspended cell pellets were sonicated for 5 sec. at
setting of "4." followed
by the addition of 750 microliter of additional buffer for a volume of 1
ml/sample. The resuspension
volume was microcentrifuged at 13K rpm for 5 min to pellet any cellular debris
and the supernatant
was removed and saved as "sol. fraction." Each of the media aliquots and the
and sol. fraction
aliquots were combined with4 microliter of 4x loading buffer to 10 microliter
of sample and then
incubated for 15 min at 75 deg C. 10 microliters of the sample/loading buffer
mixture was loaded on
the gel and run 35 min at 200 volts. The gel was then transferred to a 0.45
micrometer nitrocellulose
membrane and incubated at 25V for 65 minutes. The transferred sample was then
blocked with
WesternA/10% milk solution followed by a 1:2000 dilution of Jackson Labs Gt
Anti Hu IgG-HRP in
WesternA/2.5% milk solution O/N @ 4 deg C.. The blot was washed three times in
WesternA
solution followed by incubation with Pierce SuperSignal West Pico
Chemoluminescent Substrate.
Construct was identified in both the supernatant samples and the cell pellet
samples for all of the
constructs. Thus, the constructs are produced and expressed and can be
purified from an aliquot of
cell media and/or from a cell lysate. Purified construct is then available for
use in a diagnostic assay,
for the manufacture of a medicament, as a pharmaceutical composition or in
methods of treatment
using said pharmaceutical composition of said purified construct.
Additionally, antibodies to said
constructs can be produced and said antibodies are useful for the manufacture
of a medicament, as a
pharmaceutical composition, in methods of treating disorders using a
pharmaceutical composition or
in a diagnostic assay.
EXAMPLE 70
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
174
Comparison of Relative Affinities of Human IL-17C Binding to Multiple
IL-17RE Constructs Using Surface Plasmon Resonance
[574] This Example illustrates that the identified ligand binding domains (SEQ
ID NOs:
249, 310 and 250) each form a complex with IL17C ligand. Through surface
plasmon resonance
(SPR), the differences in kinetic constants of five distinct IL17RE constructs
to IL17C were
measured. These values were compared to identify the three ligand binding
domains and further
identify receptor:ligand interactions for the ligand binding domains
collectively and alone.
[575] SPR allows for the real-time detection of protein:protein interactions .
A protein of
interest is covalently bound to flow cells on a sensor chip used in the
machine; this bound protein is
referred to as the "ligand." A second protein, termed the "analyte," is then
flowed over the cells to
determine if the two proteins exhibit binding properties. Because signal
detected in the machine is
directly proportional to mass, an increase in response signifies that the
analyte is binding to the ligand.
[576] Two proteins are serially bound to the chip for this assay, which
reduces the number
of chips required for the experiment. An.alpha.-human Fc antibody is first
immobilized on the chip.
The five IL17RE constructs being analyzed are Fc5 fusions, so the receptors
bind to the immobilized
antibody and are used as ligands. Due to the non-covalent nature of this
antibody:Fc interaction, it is
possible to remove the IL17RE proteins from the chip by regenerating with an
acidic solution. This
allows the user to perform assays with multiple IL17RE constructs while using
one chip.
[577] IL17C CH6 (SEQ ID NO: 17 fused at its c-terminus to a 6His tag, also
called
Construct 889) is the analyte utilized to determine the binding properties of
the IL17RE polyprotein
constructs. Multiple concentrations of the analyte are used in the
performances to yield a broad range
of curves.
[578] All IL17 polyproteins usedin the study were produced as described
herein. CHO
cells were used for expression of the five IL17RE Fc5 constructs, and IL17C
CH6 is the product of
293F cell expression. Protein lots were purified using Protein A columns,
correct construct molecular
weights were confirmed via size-exclusion chromatography equipped with
multiangle light scattering
and quasi-elastic light scattering detectors (SEC-MALS) analyses, and
quantification was
accomplished with amino acid analyses.
[579] IL17 ligand proteins are dimmers, and for IL17C ligand the
homodimerization occurs
naturally upon expression. The IL17RE Fc5 constructs are dimeric due to the
Fc5 fusion partner.
This dimeric nature causes both the analyte and the ligand to be bivalent.
Accordingly, fits using the
Biacore software are done with the bivalent analyte model.
[580] Equipment and Reagents used are as follows: SPC analysis was performed
using a
Biacore T100 Analyzer and Biacore Control and Evaluation software. The sensor
chip was a CM4
(Biacore BR-1005-34, Lot 1162158). HBS-EP Buffer is essentially; 10mM HEPES,
150mM NaC1,
3mM EDTA, 0.005% surfactant P20, pH 7.4 (Biacore, BR-1003-68), with added
lmg/mL BSA
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
175
(Proliant Biologicals, BB42900101) An amine coupling kit (Biacore, BR-1000-50)
was also used, as
was 10mM sodium acetate, pH 5.0 (Biacore, BR-1003-51), Goat anti-human IgG FCg
specific
antibody, 2.4 mg/mL (Jackson ImmunoResearch, #109-005-008, Lot 70187), and
Hydrochloric acid
(J.T.Baker, 9535-01, Lot V08057). The IL-17C ligand used as well as the IL-
17RE constructs used
are indicated in Table 9, and are further described herein.
Table 9: Protein Reagents
SEQ ID [AAA] MW
Protein Construct NO: (mg/mL)
kDa
IL17C CH6 dimer 889 17 0.64 42.04
IL17RE Fc5 dimer 1543 220 1.83 119.34
IL17RE Fc5 dimer 1700 242 1.81 78.16
IL17RE Fc5 dimer 1702 246 2.42 119.68
IL17RE Fc5 dimer 1699 310 0.94 76.26
IL17RE Fc5 dimer 1701 (a) 244 1.85 99.10
IL17RE Fc5 dimer 1701(b) 244 1.51 99.10
[581] All Biacore assays were performed at 25.deg.C and samples were stored at
4.deg.C.
Immobilization: Goat anti-human IgG antibody was diluted in 10 mM sodium
acetate pH 5.0 to a
working concentration of 20.micro.g/mL. This antibody was immobilized onto a
CM4 sensor chip
using a mixture of 0.4M N-ethyl-N'-(3-diethylamino-propyl) carbodiimide (EDC)
and 0.1M N-
hydroxysuccinimide (NHS) found in the amine coupling kit from Biacore. A level
of 5000 response
units (RU) was targeted for all four flow cells. After immobilization of the
antibody, the remaining
active sites on the flow cells were blocked with 1M ethanolamine
hydrochloride. Non-specifically
bound protein was removed by washing with 50mM NaOH. The final immobilization
levels on the
four flow cells were 3787 RU, 3923 RU, 3619 RU, and 3666 RU, respectively.
[582] Capture: Six different IL17RE protein lots were used as ligand
molecules. In each of
two analysis cycles, three IL17RE constructs were bound to flow cells 2-4,
leaving flow cell 1 as a
reference cell with only goat anti-human IgG antibody bound to check for non-
specific binding to
IL17C ligand. After analysis of the first three IL17RE constructs was
completed, the three remaining
IL17RE constructs were captured on flow cells 2-4. Samples were diluted to
various concentrations
(Table 10) in the same HBS-EP-BSA running buffer as was used for the analysis
performance.
[583] A target RU of approximately 100 was sought in loading the capture
ligand proteins,
which gives theoretical analyte binding Rmax. values between 33 and 49
RU. The formula for
determining Rmax. is Ligand Load (RU) * (analyte molecular weight/ligand
molecular weight) *
stoichiometry. Rmax. values between 50 and 150 are suggested by Biacore,
but lower values are
recommended for more accurate results should it be known that the ligand and
the analyte bind with at
least moderate affinity. Adequate binding between IL17C and three of the
IL17RE constructs was
shown to be the case in previous Biacore studies. Stoichiometry was assumed to
be one, because two
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
176
binding sites of the IL17RE Fc5 ligand can bind to the two sites on the
dimeric IL17C analyte,
leading to a nominal 1:1 interaction.
Table 10: Capture Ligand Loads
Concentration Load Time Avg. Ligand Flow Theoretical
Construct /mL (seconds) Load RU Cell Rmax (RU)
1543 2.5 48 94 2 33
1700 10 20 77 3 41
1702 10 60 94 4 33
1699 5 20 88 2 49
1701 (a) 5 30 90 3 38
1701 (b) 5 25 95 4 40
[584] Injection of Analyte: IL17C CH6 was diluted to 5nM using an HBS-EP-BSA
buffer.
Further 1/3 serial dilutions were made to obtain seven total samples of
decreasing molarity, thus
ranging from 5nM to 0.007nM. These samples were injected in order of
increasing molarity over all
four flow cells at 50.micro.L/min for 420 seconds to allow for comparison of
three IL17RE constructs
(flow cells 2-4) to nonspecific binding that had occurred on the reference
cell (flow cell 1).
Dissociation was allowed to occur for 900 seconds. A second, replicate set of
injections of increasing
molarity followed, including a OnM buffer sample, using the same IL17C samples
in order to check
reproducibility of results. Five injections of buffer were flowed over the
cells prior to IL17C to allow
for subtraction of background noise.
[585] Regeneration: The IL17RE capture surfaces and the reference surface were
regenerated after each analyte injection cycle with two 15 micro.L injections
of 20 mM HC1 at 50
.micro.L/min. This removed the captured IL17RE Fc5 but left the .alpha.-human
Fc antibody intact
on the chip.
[586] Data Analysis: Analyses were performed using the Biacore T100 Evaluation
Software. Individual flow cell graphs were assessed to ensure that the
regeneration step provided
consistent baseline stability of the binding surface throughout the sequence
of injections. The level of
non-specific binding of the IL17C analyte to the reference surface (flow cell
1) was checked and
confirmed to be minimal. Binding curves were processed by subtraction of the
control surface
sensorgram and by subtraction of the sensorgram of the last of the five buffer
injections to eliminate
instrument background noise. Because IL17C has a dimeric structure, the
resulting binding curves
were globally fit to the bivalent kinetic binding model. RI values were set to
zero for all fits. (Figures
2-7).
[587] The bivalent analyte model gives two values for ka and kd. The
first,
kal and kdl, are values given for the monovalent affinity of the
interaction. The second set
of values, ka2 and kd2, pertain to the avidity of the interaction.
Ka2 is in units of
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
177
1/RUs, so it is not incorporated into the final KD value. The numbers
reported for KD are
products of kdl/ kal and are hence termed "KDl."
[588] Constructs 1543 and 1700 initially yielded fits that did not match with
the concave
shape of the dissociation curve. This suggests that the model was finding a
local minimum for
Rmax that was not ideal for all of the curves. To adjust this, the
resulting values for kal,
kdl, ka2 and kd2 from the fit for construct 1702 were input as
the starting values for the
model to work from to perform the fit. This resulted in a better overall fit.
Construct 1702 was not
modified to perform the bivalent analyte fit. Construct 1699 had low responses
for all curves. The
lines for the duplicates of 0.02nM and of 0.06nM were below the OnM buffer
line, so they were
excluded from the final fit. Also, the second of the 5nM duplicates was not a
clean line, it was also
removed. This left four concentrations that had duplicate curves. Reported
values for this construct
are not included in the summary table below (TABLE 11). The response was too
minimal for kinetic
values to be accurately determined. Both 1701 constructs exclude the results
for the 0.06nM curves.
These curves were extreme outliers and were not used for any part of the
analyses. Both 0.02nM
curves were omitted for the final fits because they were below the OnM buffer
line.
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
178
Table 11 Kinetic Constants
construct Protein
51 28 370
1543 IL17RE Fc5 dimer 6E-9 4E+6 2E-2 66.5 0.552 0 0 0
23 14 200
1700 IL17RE Fc5 dimer 1E-8 2E+6 3E-2 60.4 0.309 0 0 0
12
1702 IL17RE Fc5 dimer 6E-9 3E+6 2E-2 69.1 0.644 0 60 790
1699 IL17RE Fc5 dimer Binding levels too low for accurate assessment.
18
1701 IL17RE Fc5 dimer 2E-8 6E+5 1E-2 33.8 0.14 0 52 350
23
1701 IL17RE Fc5 dimer 2E-8 7E+5 1E-2 42.4 0.181 0 61 440
[589] Results: Figures 2-7 show the final curves obtained from the runs. T-
values greater
than 10 are considered to indicate that the variables measured are
significant. Graphs of residual
values found in Figures 2-7 demonstrate that there is not much disparity
between the actual curves
obtained from the assays and the curves applied by the fit.
[590] For accurate measurements of kd, Biacore suggests allowing time for
50% of the
complexes to dissociate. Interactions with a kd of 1x10-2 have a
half-life of 69.3 seconds;
interactions with a kd of 1x10-3 reach half dissociation after 11.55
minutes. The
dissociation time of 900 seconds for these assays is appropriate for the
kd values obtained.
Results for ka2 and kd2, whose significance is described in the data
analysis section, can be
found in Figures 2-7.
[591] Four of the five IL17RE constructs produced soluble receptor with
comparable
binding affinities for human IL17C with equilibrium dissociation constants
(kD) in the range of
1-10nM. All four of these moderate binders produce protein starting at a furin
site (or an optimized
furin site) identified in exon 5. Constructs 1543 and 1702 contain two
fibronectin-like ligand binding
domains as well as a third ligand binding domain defined by exons 12, 14, and
15. Construct 1701
contains the two fibronectin-like ligand binding domains but no third ligand
binding domain. The
fourth construct, 1700, consists of only the first fibronectin-like ligand
binding domain. Construct
1699 is comprised solely of the second fibronectin-like ligand binding domain.
The small amount of
1699 that was isolated demonstrated minimal levels of binding to IL17C. These
results demonstrate
that the first fibronectin-like ligaand binding domain is the minimal
functional ligand binding domain
for soluble IL17RE binding to IL17C.
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
179
EXAMPLE 71
Measurement of the Solution Affinity Dissociation Constant (KD) of IL17RE-FC5
from
Constructs 1700 and 1702 for IL17C by Surface Plasmon Resonance
[592] In this second SPR study, the solution affinity dissociation constant
(KD) of
IL17RE-FC5 was determined for IL17C. Two versions of IL17RE-FC5 (construct
1700 and
construct 1702) were analyzed. The IL17C used in this study was the human form
of the protein
(SEQ ID NO.: 17). This data complements the above results of a kinetic binding
study.
[593] The dissociation constant (KD) for IL17RE-FC5 binding to IL17C was
determined via solution-phase affinity experiments. For these studies, capture
onto the immobilized
surface was performed under mass transport limited conditions. Utilizing a low
flow rate and a high
density of immobilized ligand on the capture surface (anti-IL17C and IL17RE-
FC5 were directly
coupled to separate flow cells of the chip to compare different capture
strategies), the rate of binding
of IL17C to the immobilized surface was proportional to the concentration of
IL17C in solution.
There were two parts to this experimental design: First, a calibration curve
was generated by allowing
known concentrations of IL17C to flow over the immobilized capture surface.
The initial rate of
binding to the surface was plotted versus IL17C concentration. This curve was
used to back calculate
the concentration free IL17C in the next step. Second, a dilution series of
IL17RE-FC5 constructs
was combined with a constant concentration of IL17C. Flowing this mixture over
the immobilized
surface allowed the free IL17C (uncomplexed with IL17RE-FC5) to bind to the
surface. In this
experiment the binding of IL17C to the capture surface competes with the its
binding to IL17RE-FC5,
and so this experiment is designed to avoid IL17C simultaneously binding both
the capture surface
and IL17RE-FC5 in solution. The initial rate of this binding was determined,
and the concentration of
free IL17C in the mixture was calculated from the calibration curve. The
concentration of free IL17C
was plotted versus the concentration of IL17RE-FC5, and the solution phase
KD was calculated
via the Biacore T100 Evaluation software. The data was imported into Graphpad
Prism4 software and
fit with a four parameter curve. An empirical IC50 value was calculated from
this data, and this value
was used as an approximation of the solution phase affinity constant
(KD). The results of this
study are reported as a range of KD values obtained with Biacore T100
Evaluation software and
calculated from the four parameter fit of the data.
[594] The materials used for this assay includes the following: Biacore T100
Analyzer;
Biacore T100 Control and Evaluation software (version 1.1.1); Graphpad Prism4
software (San
Diego, CA); Sensor chip CM4 (Biacore, # BR-1005-34); HBS-EP Buffer; 10 mM
HEPES, 150 mM
NaC1, 3 mM EDTA, 0.005% surfactant P20, pH 7.4 (Biacore, BR-1006-69); Amine
coupling kit
(Biacore, BR-1000-50); 10 mM sodium acetate, pH 5.0 (Biacore, BR-1003-51);
recombinant human
IL17RE-FC5 (Construct 1700 at 1.81 mg/mL in 50mM sodium phosphate, 109 mM
NaC1, pH 7.3 and
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
180
Construct 1702 at 2.42 mg/mL in 50mM sodium phosphate, 109 mM NaC1, pH 7.3);
and recombinant
human IL17C (Construct 889 at 0.64 mg/mL in 35 mM sodium phosphate, 120 mM
NaC1, pH 7.2).
[595] All studies were run at 25 C, and the samples were stored at 8 C in the
autosampler.
Immobilization: Mouse anti-human-IL17C monoclonal antibody was generated and
immobilized
onto flow cell #3 of a CM4 sensor chip using the amine coupling kit. The anti-
IL-17C antibody must
be an antibody that binds only the IL-17C ligand, and not one that binds IL-
17C when complexed
with an IL-17RE soluble receptor. The antibody was diluted in 10 mM sodium
acetate pH 5.0 to a
concentration of 100 .micro.g/mL. After immobilization of the antibody, the
active sites on the flow
cell were blocked with ethanolamine. The final immobilization level was 4628
RU. Recombinant
human IL17RE-FC5 (construct 1543) was diluted to 100 .micro.g/mL in 10 mM
sodium acetate pH
5.0 and immobilized to flow cell #4 of a CM4 sensor chip using the amine
coupling kit. After
immobilization of the protein, the active sites on the flow cell were blocked
with ethanolamine. The
final immobilization level was 5646 RU. Flow cell 1 was used as the reference
cell for these studies.
It was activated and blocked with ethanolamine.
[596] Calibration Curve of Human IL17C: IL17C was injected over flow cells 1-4
in series
to allow for comparative analysis of binding of the IL17C to the anti-IL17C
monoclonal antibody
capture surface (flow ce113) relative to binding to the blank control surface
(flow cell 1). In addition,
binding of the IL17C to the 1543 construct capture surface (flow ce114) was
also determined relative
to binding to the control surface (flow cell 1). Serial 1:2 dilutions of IL17C
from 20 nM to 0.078 nM
were made in HBS-EP buffer containing 1 mg/mL BSA. Single injections of each
concentration were
performed. The IL17C was injected at 10 .micro.L/min for 3 minutes
(association time). By
performing these studies with a low flow rate and a high density of
immobilized ligand on the capture
surface, the binding of IL17C to the surface occurred under mass transport
limited conditions. During
the initial portion of the association phase, the rate of binding to the
immobilized surface was
proportional to the concentration of IL17C in solution. A representative
figure of IL17C binding to
the anti-IL17C monoclonal antibody (flow cell 3-1) is shown in Figure 8A.
Immediately after the
start of sample injection, the slope of this association was calculated for a
period of 30 seconds. The
results were plotted as function of IL17C concentration to generate a
calibration curve (Figure 8B).
[597] Dilution Curve of IL17RE-FC5 with a Constant Concentration of Human
IL17C:
IL17RE-FC5 from constructs 1700 and 1702 were diluted from 20 - 0 nM and mixed
with a constant
nM concentration of IL17C. This mixture was injected over flow cells 1-4 in
series to allow for
comparative analysis of binding of the free IL17C (uncomplexed with IL17RE-
FC5) to the capture
surfaces (anti-IL17C on flow cell 3 and IL17RE-FC5 on flow cell 4) relative to
the blank control
surface (flow cell 1). Data was obtained for both the anti-IL17C and the
IL17RE-FC5 capture
surfaces; however, only the anti-IL17C results are detailed in this example.
Upon analysis, identical
solution affinity dissociation constants were obtained with both capture
surfaces. The resulting
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
181
reference-subtracted sensorgrams of free-IL17C binding to the anti-IL17C
capture surface (flow cell
3-1) are shown in Figures 9A (construct 1700) and 10A (construct 1702). The
initial rate of
association onto the capture surface was calculated over a 30 second window.
[598] Using the calibration curve generated in Figure 8B, the initial rate of
the association
(slope) was used to back calculate the concentration of free IL17C in each
sample, and this back-
calculated concentration was plotted versus the concentration of IL17RE-FC5.
The data was fit with
a 1:1 binding model using the Biacore T100 Evaluation software, and a solution
affinity KD was
calculated (Figures 9B and lOB for constructs 1700 and 1702, respectively).
Since the fit to the data
was poor using the Biacore software, the data was exported into Graphpad
Prism4 software and fit
with a four parameter curve. An empirical IC50 value was calculated, and this
value was used as an
approximation of the solution affinity KD (Figures 9C and 10C for
constructs 1700 and 1702,
respectively). These results have been summarized in Table 12.
Table 12: Dissociation Constants
KD (Calculated with IC50 (Calculated with Four
ILl7RE-FC5 a 1:1 Binding Model, Parameter Fit to Data,
Construct Biacore T100 Evaluation Graphpad Prism4 Software)
Software)
1700 2.9 nM 6.3 nM
1702 1.2 nM 4.4 nM
[599] Regeneration: The capture surface was regenerated after each injection
cycle of
analyte via two 30 micro.L injections of 20 mM HC1 at 50 .micro.L/min. This
removed the IL17C
from the chip surface. Data Analysis: Data analysis was performed with the
Biacore T100
Evaluation software. Baseline stability was assessed to ensure that the
regeneration step provided a
consistent binding surface throughout the sequence of injections. The level of
non-specific binding of
the IL17C to the control surface was checked and confirmed to be minimal.
Binding curves were
processed by subtraction of the control surface (flow cell 1).
[600] For the solution affinity binding experiments with IL17RE-FC5 and IL17C,
two
different capture surfaces were used. An anti- IL17C monoclonal antibody was
directly coupled to
flow cell 3, while IL17RE-FC5 was coupled to flow cell 4. Both of these
capture surfaces detected
only free IL17C (uncomplexed with IL17RE-FC5). Binding to the surface was
performed at 10
.micro.L/min in order to maximize mass transport effects. A calibration curve
of IL17C was
generated by injecting various concentrations of IL17C over the capture
surfaces and calculating the
initial rate of binding. Next, samples containing a constant 5 nM IL17C and
variable concentrations
of IL17RE-FC5 were injected over the surface, and the concentration of free
IL17C in the sample was
calculated from the calibration curve. The Biacore software plots the
concentration of IL17C versus
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
182
the concentration of IL17RE-FC5, and calculates a KD. The fit that the Biacore
software applies to
the data is based on a 1:1 binding model. However, the IL17RE-FC5-IL17C
interaction is bivalent,
and the resulting fit is not perfect. Examination of the fit (Figures 2B and
3B) demonstrates that the
Biacore software underestimates the KD of this interaction (the curve is
shifted to the left of the data
points). In an attempt to better fit a curve to the data, the values were
exported into GraphPad Prism
software and an four-parameter fit of the data was performed (Figures 9C and
10C). While no
assumptions are made about the binding model, the curve better fits the data
points, and an empirical
IC50 was obtained as an estimate of the KD. The two KD values
(obtained from the two
analysis methods) are used to bracket the expected KD of the interaction.
For construct 1700 the
dissociation constant is reported between 2.9-6.3 nM, while for construct 1702
it is reported as
between 1.2-4.4 nM. These values are consistent with the KD reported
above.
[601] Solution affinity dissociation constants (KD) were calculated for
IL17RE-FC5
binding to IL17C. IL17RE-FC5 constructs 1700 and 1702 were analyzed and found
to have
comparable KD values. The KD of construct 1700 was found to be
between 2.9 - 6.3 nM,
while the KD of construct 1702 was calculated to be between 1.2 - 4.4 nM.
These values are
consistent with the dissociation constants obtained via kinetic analysis and a
bivalent analyte model
(above).
EXAMPLE 72
Production of Monoclonal Antibodies to IL-17RE from Hybridoma Cell Lines
[602] Mouse anti-human IL17RE monoclonal antibodies (mAb) were generated using
mouse myeloma cell line designated P3-X63-Ag8.653/ATCC/STR.
1. Procedure
1.1 Immunizations
[603] Ten 6-8 week old BALB/c mice (Charles River Laboratories, Wilmington,
MA) were
immunized with human IL17RE. The mice were initially immunized by subcutaneous
injection with
65 g of purified, recombinant human IL17RE (produced in BHK cells) fused at
the C-terminal with a
FLAG tag (A1709F, extracellular domain (ECD) of IL-17REx2 (SEQ ID NO.: 311)
fused at the C-
terminus with a FLAG tag) conjugated with BSA in combination with Emulsigen -
P adjuvant as per
manufacturer's instructions. Following the initial immunization each of the
mice received an
additional 65 g of human IL17REx2 ECD in Emulsigen -P adjuvant via the
subcutaneous route for
a total of 4 boosts. Seven days after the third and fourth immunizations the
mice were bled via the
retro orbital plexus and the serum was assessed for the immune response to
IL17RE.
1.2 Development of assay formats and selection of fusion animals
1.2.1 Capture Assay
[604] The ability of polyclonal anti-IL17RE antibodies in the immune sera to
bind to
IL17RE Fc (A1712F, ECD of IL-17REx2, del M1-R375 and del G97-G580) and Fc10
was assessed
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
183
using a "capture" style ELISA assay. In this assay, wells of 96 well
polystyrene ELISA plates were
first coated with 100 L/well of goat anti-mouse IgG, Fc specific antibody
(Jackson ImmunoResearch
Laboratories, West Grove, PA) at a concentration of 1 g/mL in Coating Buffer
(0.1M Na2CO3, pH
9.6).
[605] Plates were incubated overnight at 4 C after which unbound antibody was
aspirated
and the plates washed twice with 300 L/well of Wash Buffer (PBS-Tween defined
as 0. 137M NaC1,
0.0022M KC1, 0.0067M Na2HPO4, 0.0020M KH2PO4, 0.05% v/w polysorbate 20, pH
7.2). Wells
were blocked with 200 L/well of Blocking Buffer (PBS-Tween plus 1% w/v bovine
serum albumin
(BSA)) for 60 minutes at room temperature (RT), aspirated and the plates
washed twice with 300
L/well of PBS-Tween. Serial 10-fold dilutions (in 1% BSA in PBS-Tween) of the
sera were
prepared beginning with an initial dilution of 1:1000 and ranged to
1:1,000,000. Duplicate samples of
each dilution were then transferred to the assay plate, 100 L/well, in order
to bind mouse IgG in the
sera to the assay plate through the Fc portion of the molecule. Normal mouse
sera served as a
negative control. Following a one-hour incubation at Room Temperature (RT),
the wells were
aspirated and the plates washed twice as described above. Biotinylated IL17RE
Fc (A1712F, which is
amino acid residues 176 to 452 of IL-17REx1 fused with an Fc fragment and
further biotinylated, 6:1
molar ratio of biotin: protein) at a concentration of 500 ng/mL was then added
to the wells, 100
L/well. Following a one-hour incubation at RT, unbound biotinylated IL17RE was
aspirated from
the wells and the plates washed twice. Horseradish peroxidase labeled
streptavidin (Pierce, Rockford,
IL) at a concentration of 500 ng/mL was then added to each well, 100 L /well,
and the plates
incubated at RT for 1 hour. After removal of unbound HRP-SA, the plates were
washed twice, 100
L /well of tetramethyl benzidine (TMB) (BioFX Laboratories, Owings Mills, MD)
was added to
each well and the plates incubated for 3 minutes at RT. Color development was
stopped by the
addition of 100 L /well of 450nm TMB Stop Reagent (BioFX Laboratories, Owings
Mills, MD) and
the absorbance values of the wells were read on a Molecular Devices Spectra
MAX 340 instrument at
450nm.
1.2.1.1 Direct Assay
[606] The ability of IL17RE Fc to bind polyclonal anti-mouse IL17RE antibodies
in the
immune sera was assessed using a"direct" style ELISA assay. In this assay,
wells of 96 well
polystyrene ELISA plates were first coated with 100 L /well of IL17RE Fc
(A1712F, ECD of IL-
17REx2, del M1-R375 and del G97-G580) and Fc10 at a concentration of 1 g/mL
in Coating Buffer
(0.1M Na2CO3, pH 9.6). Plates were incubated overnight at 4 C after which
unbound receptor was
aspirated and the plates washed twice with 300 L /well of Wash Buffer (PBS-
Tween defined as
0.137M NaC1, 0.0022M KC1, 0.0067M Na2HPO4, 0.0020M KH2PO4, 0.05% v/w
polysorbate 20, pH
7.2). Wells were blocked with 200 L /well of Blocking Buffer (PBS-Tween plus
1% w/v bovine
serum albumin (BSA)) for 1 hour, after which the plates were washed twice with
Wash Buffer. Serial
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
184
10-fold dilutions (in 1% BSA in PBS-Tween) of the sera were prepared beginning
with an initial
dilution of 1:1000 and ranged to 1:1,000,000. Duplicate samples of each
dilution were then
transferred to the assay plate, 100 L /well, in order to bind polyclonal anti-
mouse IL17RE in the
sera. Following 1 hour incubation at RT, the wells were aspirated and the
plates washed twice as
described above. Horseradish peroxidase labeled Goat anti Mouse IgG, Fc
specific (Jackson
ImmunoResearch Laboratories, West Grove, PA) at a dilution of 1:5000 was then
added to each well,
100 L /well, and the plates incubated at RT for 1 hour. After removal of
unbound HRP conjugated
antibody, the plates were washed twice, 100 L /well of tetra methyl benzidine
(TMB) (BioFX
Laboratories, Owings Mills, MD) added to each well and the plates incubated
for 3 minutes at RT.
Color development was stopped by the addition of 100 L /well of 450nm TMB
Stop Reagent
(BioFX Laboratories, Owings Mills, MD) and the absorbance values of the wells
were read on a
Molecular Devices Spectra MAX 340 instrument at 450nm.
1.3 Fusion 347
[607] Four days after a final boost, spleen and lymph nodes of titer bearing
mice were
harvested, prepared into a single cell suspension and fused with myeloma cell
line at a 2:1 lymphoid
cell: myeloma cell ratio with PEG 1500 using standard methods known in the
art. The fusion mixture
was distributed into 96 well flat-bottomed plates. Fusion plates were fed
three times with a 70%
replacement of media and assayed ten days after plating of the fusion
utilizing Capture and Direct
assay formats.
1.4 Selection of Master Wells
[608] Fusion plates were screened for IL17RE binding activity with specific
antibodies as
described above in section 1.2.1 and 1.2.2 except that sera was replaced with
undiluted hybridoma
supernatant. Seventy-eight positive wells were identified from both assay
formats. Master wells were
expanded into culture in 24 well plates, supernatant from subconfluent
cultures harvested and cells
cryopreserved. Supernatants were confirmed in both capture and direct assay
formats and ten master
wells were selected for expansion. Further evaluation of the established
antibodies as potential
FACS, IHC and neutralization was then accomplished.
1.5 Cloning
[609] Four master wells (347.24, 347.32, 347.59 and 347.72) were considered
for cloning.
Cells were seeded in 96 well microtiter cell culture plates using a standard
low-density dilution (1 cell
per well) approach and monoclonality was assessed by microscopic examination
of wells for a single
foci of growth prior to assay. Six days post-plating, all wells on the plates
were screened by Direct
ELISA.
1.5.1 Selection of First Round Clones
[610] Supernatant from 6 wells that was both positive for specific mAb and
originated from
wells with only a single colony of hybridoma growth was collected from each
cloning set and
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
185
rescreened at 10 fold dilutions utilizing the Direct ELISA format in order to
identify a clone with
superior binding properties to IL17RE. Two appropriate clones were selected
and banked. One clone
was forwarded into second cloning process.
1.5.2 Selection of Second Round Clones
[611] Supernatant from approximately 6 wells that was both positive for
specific mAb and
originated from wells with only a single colony of hybridoma growth was
collected from each cloning
set and rescreened as in 3.5.1. The screen yielded final hybridoma lines:
347.32.5.5; 347.59.2.5;
347.24.3.4; 347.72.1.2. The mouse IgG isotype of the mAb produced by each of
these hybridomas
was determined using Mouse Monoclonal Antibody IsoStrip test (Roche Applied
Science).
347.32.5.5 IgG2b kappa; 347.59.2.5 IgGl kappa; 347.24.3.4 IgG2b kappa;
347.72.1.2 IgG2a kappa.
1.6 Scale up for Monoclonal Antibody Production
[612] Upon selection of a final clone for antibody production, hybridoma cell
line was
removed from liquid nitrogen storage, quickly thawed and washed once in growth
media to remove
all DMSO (dimethyl sulfoxide). The cells were resuspended in growth media and
if necessary,
weaned from any extraneous additives prior to scale up. Once weaned, the cells
were expanded to the
appropriate volume to achieve the requested target. The cell suspension was
harvested, cells removed
and the supernatant filtered through a 0.2 m filtration system. The
supernatant pool was submitted
for purification, labeled and dispensed into aliquots.
2. Results
[613] Monoclonal antibodies specific for IL17RE were generated, and their
utility to
perform as FACS, Assay, Western Blot reagents as well as neutralizing
antibodies has been identified.
Table 13: Specificity of anti IL17RE mAbs
Clone ID Lot # ATCC Neutralizer FACS Assay Western
Deposit Blot
Designation
347.24.3.4 E10135 PTA-8234 + + + +
347.32.5.5 E10133 NA + +
347.59.2.5 E10134 NA + +
347.72.1.2 E10136 PTA-8233 + + +
Example 73
Monoclonal antibodies raised against human IL17RE are able to block IL17C
mediated
activation of Ix(3 in IL17RE transfected NIH3T3
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
186
[614] On day one NIH-3T3/KZ142 cells stably transfected with the human IL-
17REx2
receptor were plated out at 1x104 cells/well in growth media (DMEM, 5% fetal
bovine serum, 1%
Sodium Pyruvate, 1 M MTX) in 96-well, flat-bottom tissue culture plates. On
day two cells were
switched to assay media (DMEM, 0.1% BSA, 10mM HEPES). On day three cells were
preincubated
for 30 minutes with monoclonal antibodies to IL17RE at a range of
concentrations. Cells were then
stimulated with a sub-maximal concentration (EC90, effective concentration at
90 percent) of human
IL-17C and incubated at 37 C for 15 minutes.
[615] Following incubation, cells were washed with ice-cold wash buffer and
put on ice to
stop the reaction according to manufacturer's instructions (BIO-PLEX Cell
Lysis Kit, BIO-RAD
Laboratories, Hercules, CA). Fifty L/well lysis buffer was added to each
well; lysates were pipetted
up and down five times while on ice, then agitated on a microplate platform
shaker for 20 minutes at
300 rpm and 4 C. Plates were centrifuged at 4500 rpm at 4 C for 20 minutes.
Supernatants were
collected and transferred to a new micro titer plate for storage at -20 C.
[616] Capture beads (BIO-PLEX Phospho-Ix(3-a Assay, BIO-RAD Laboratories) were
combined with 50 L of 1:1 diluted lysates and added to a 96-well filter plate
according to
manufacture's instructions (BIO-PLEX Phosphoprotein Detection Kit, BIO-RAD
Laboratories). The
aluminum foil-covered plate was incubated overnight at room temperature, with
shaking at 300 rpm.
The plate was transferred to a microtiter vacuum apparatus and washed three
times with wash buffer.
After addition of 25 L/well detection antibody, the foil-covered plate was
incubated at room
temperature for 30 minutes with shaking at 300 rpm. The plate was filtered and
washed three times
with wash buffer. Streptavidin-PE (50 L/well) was added, and the foil-covered
plate was incubated
at room temperature for 15 minutes with shaking at 300 rpm. The plate was
filtered and washed two
times with bead resuspension buffer. After the final wash, beads were
resuspended in 125 L/well of
bead suspension buffer, shaken for 30 seconds, and read on an array reader
(BIO-PLEX, BIO-RAD
Laboratories) according to the manufacture's instructions. Data was analyzed
using analytical
software (BIO-PLEX MANAGER 3.0, BIO-RAD Laboratories). Decreases in the level
of the
phosphorylated Iic(3-a transcription factor present in the lysates were
indicative of neutralization of the
IL-17REx2 receptor-ligand interaction.
[617] For huIL-17C the EC90 concentration was determined to be lOOng/ml.
Antibody
E10135, raised against human IL17RE, was able to block this huIL17C mediated
activation of
huIL17REx2 transfected cells, with an IC50 (inhibitory concentration at 50%)
of 99ng/ml.
Example 74
Neutralization of IL17C binding to IL17RE transfected cells using mAbs to
huIL17RE
Assay Background: FACS binding assay to show neutralization of IL17C bindinz
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
187
[618] Introduction: BHK/pVAC2neo transfected cells expressing splice variant
human
IL17REx2 or mouse IL17REx6 bind IL17C. The ability of an antibody to
neutralize binding of
biotinylated IL17C is measured by FACS analysis in this assay.
[619] Method: Cells and mAb are pre-incubated for 30-40 min. Biotinylated
IL17C was
added and incubated for another 30-60 min, followed by a Streptavidin-PE
conjugate. Fluorescence
was measured using an LSRII flow cytometer. Neutralization of binding of IL17C
was shown by a
decrease in PE Mean fluorescence.
Summary of Results
[620] Monoclonal antibody to huIL17RE (E10135) demonstrated neutralization of
binding
of biotinylated human IL17C (50ng/ml) to cells expressing the human IL17REx2
splice variant at
doses of 1-5 g/ml, and also showed neutralization of binding of biotinylated
mouse IL17C
(50ng/ml) to cells with the mouse IL17REx6 splice variant at these same doses.
[621] Monoclonal antibody to human IL17RE (E10134) neutralized binding of
biotinylated
human IL17C (50ng/ml) to cells with the human IL17REx2 splice variant at doses
of 1-5 g/ml. No
neutralization of binding of mouse IL17C to the mouse IL17REx6 splice variant
was detected with
this monoclonal antibody.
Example 75
Neutralization of IL17C induction of G-CSF in SAEC cells by mAb to huIL17RE
Assay Backaound: Bioassay for Neutralization of huIL-17C Induction of Cytokine
in Human Small
AirwE Epithelial Cells (SAEC)
[622] Introduction: Treatment of SAEC with huIL-17C induces the production of
cytokine
huG-CSF. The ability of an antibody or soluble receptor to neutralize huIL-17C
in inducing these
cytokines is measured in this bioassay.
[623] Method: SAEC (cells and growth media purchased from Cambrex) were plated
at
8,000 cells/well in 96-well flat bottom tissue culture multi-well plates, and
placed in a 37 C, 5% COz
incubator. The following day, cells were pre-treated with a dose range of
antibody to huIL17RE for
30 minutes at 37 C. Following pretreatment cells were stimulated with 25ng/ml
huIL-17C. Duplicate
or triplicate wells were set up for each dose. After 48-72 hr, supernatants
were collected, and stored
at (-80) C if not used directly. Before taking supernatants, wells were
scanned by inverted
microscope to eliminate wells containing cells of questionable viability.
Supernatants were assayed
for cytokines huG-CSF in a singleplex bead-based assay system (Bio-Rad
Laboratories), and IC50
determined.
Summary of Results
[624] Monoclonal antibody to huIL17RE (E10135) demonstrated neutralization of
huIL17C
(25ng/ml) induction of G-CSF in SAEC at a concentration of 1 g/ml.
Example 76
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
188
Neutralization of IL17C induction of G-CSF in SAEC cells
by soluble receptor huIL17RE.S3-Fc5
Assay Backaound: Bioassay for Neutralization of huIL-17C Induction of Cytokine
in Human Small
AirwE Epithelial Cells (SAEC)
[625] Introduction: Treatment of SAEC with huIL-17C induces the production of
cytokine
huG-CSF. The ability of an antibody or soluble receptor to neutralize huIL-17C
in inducing these
cytokines is measured in this bioassay.
[626] Method: SAEC (cells and growth media purchased from Cambrex) were plated
at
8,000 cells/well in 96-well flat bottom tissue culture multi-well plates, and
placed in a 37 C, 5% COz
incubator. The following day, cells were pre-treated with a dose range of the
soluble receptor for 30
minutes at 37 C. After pretreatment cells were stimulated with 25ng/ml huIL-
17C. Duplicate or
triplicate wells were set up for each dose. After 48-72 hr, supernatants were
collected, and stored at (-
80) C if not used directly. Before taking supernatants, wells were scanned by
inverted microscope to
eliminate wells containing cells of questionable viability. Supernatants were
assayed for cytokines
huG-CSF in a singleplex bead-based assay system (Bio-Rad Laboratories).
Summary of Results
[627] Soluble receptor huIL17RE.S2-Fc10, which is the S2 splice variant of IL-
17REx1
fused with Fc10, (lots A1712F, A1721F, A1730F, A1757F, and A1758F) was shown
to neutralize
huIL17C (25ng/ml) induction of G-CSF in SAEC, with IC50 doses of 1.7-4.5nM.
Soluble
IL17RE.S3Fc5, which is the S3 splice variant of IL-17REx1 fused with Fc5, (lot
A1859F) also
neutralized this affinity, with an activity that appears comparable to that of
the huIL17RE.S2-Fc10
molecule: At the lowest dose of IL17RE.S3Fc5 tested (0.067nm), 23%
neutralization was still
observed.
Example 77
Monoclonal antibodies raised against mouse IL17C are able to block IL17C
mediated activation
of IK(3 in IL17RE transfected NIH3T3
[628] On day one NIH-3T3/KZ142 cells stably transfected with the human IL-
17REx2
receptor were plated out at 1x104 cells/well in growth media (DMEM, 5% fetal
bovine serum, 1%
Sodium Pyruvate, 1 M MTX) in 96-well, flat-bottom tissue culture plates. On
day two cells were
switched to assay media (DMEM, 0.1% BSA, 10mM HEPES). On day three a sub-
maximal
concentration (EC90, effective concentration at 90 percent) of human IL-17C
(huIL-17C) was
combined with a dose range of the anti-murine IL17C mAb and incubated together
at 37 C for 30
minutes in assay media prior to addition to cells. Following pre-incubation,
treatments were added to
the plates containing the cells and incubated together at 37 C for 15 minutes.
[629] Following incubation, cells were washed with ice-cold wash buffer and
put on ice to
stop the reaction according to manufacturer's instructions (BIO-PLEX Cell
Lysis Kit, BIO-RAD
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
189
Laboratories, Hercules, CA). Fifty L/well lysis buffer was added to each
well; lysates were pipetted
up and down five times while on ice, then agitated on a microplate platform
shaker for 20 minutes at
300 rpm and 4 C. Plates were centrifuged at 4500 rpm at 4 C for 20 minutes.
Supernatants were
collected and transferred to a new micro titer plate for storage at -20 C.
[630] Capture beads (BIO-PLEX Phospho-Ix(3-a Assay, BIO-RAD Laboratories) were
combined with 50 L of 1:1 diluted lysates and added to a 96-well filter plate
according to
manufacture's instructions (BIO-PLEX Phosphoprotein Detection Kit, BIO-RAD
Laboratories). The
aluminum foil-covered plate was incubated overnight at room temperature, with
shaking at 300 rpm.
The plate was transferred to a microtiter vacuum apparatus and washed three
times with wash buffer.
After addition of 25 L/well detection antibody, the foil-covered plate was
incubated at room
temperature for 30 minutes with shaking at 300 rpm. The plate was filtered and
washed three times
with wash buffer. Streptavidin-PE (50 L/well) was added, and the foil-covered
plate was incubated
at room temperature for 15 minutes with shaking at 300 rpm. The plate was
filtered and washed two
times with bead resuspension buffer. After the final wash, beads were
resuspended in 125 L/well of
bead suspension buffer, shaken for 30 seconds, and read on an array reader
(BIO-PLEX, BIO-RAD
Laboratories) according to the manufacture's instructions. Data was analyzed
using analytical
software (BIO-PLEX MANAGER 3.0, BIO-RAD Laboratories). Decreases in the level
of the
phosphorylated Iic(3-a transcription factor present in the lysates were
indicative of neutralization of the
IL-17REx2 receptor-ligand interaction.
[631] For huIL-17C the EC90 concentration was determined to be lOOng/ml.
Antibody
E10132, raised against mouse IL17C, was able to block this huIL17C mediated
activation of
huIL17REx2 transfected cells, with an IC50 (inhibitory concentration at 50%)
of 800 ng/ml.
Example 78
Neutralization of IL17C induction of G-CSF in SAEC cells by mAb to muIL17C
Assay Backaound: Bioassay for Neutralization of huIL-17C Induction of Cytokine
in Human Small
AirwE Epithelial Cells (SAEC)
[632] Introduction: Treatment of SAEC with huIL-17C induces the production of
cytokine
huG-CSF. The ability of an antibody or soluble receptor to neutralize huIL-17C
in inducing these
cytokines is measured in this bioassay.
[633] Method: SAEC (cells and growth media purchased from Cambrex) were plated
at
8,000 cells/well in 96-well flat bottom tissue culture multi-well plates, and
placed in a 37 C, 5% COz
incubator. The following day, cells were treated with a dose range of the
antibody in combination
with 25ng/ml huIL-17C (where ligand and anti-muIL17C mAb were incubated
together for 30
minutes at 37 C before adding to the cells). Duplicate or triplicate wells
were set up for each dose.
After 48-72 hr, supernatants were collected, and stored at (-80) ~C if not
used directly. Before taking
supernatants, wells were scanned by inverted microscope to eliminate wells
containing cells of
CA 02666842 2009-04-17
WO 2008/049070 PCT/US2007/081812
190
questionable viability. Supernatants were assayed for cytokines huG-CSF in a
singleplex bead-based
assay system (Bio-Rad Laboratories).
Summary of Results
[634] Monoclonal antibody to muIL17C(E10132) demonstrated neutralization of
huIL17C
(25ng/ml) induction of G-CSF in SAEC, at a concentration of 5 g/ml.
[635] From the foregoing, it will be appreciated that, although specific
embodiments of the
invention have been described herein for purposes of illustration, various
modifications may be made
without deviating from the spirit and scope of the invention. Accordingly, the
invention is not limited
except as by the appended claims.