Note: Descriptions are shown in the official language in which they were submitted.
CA 02680909 2009-09-11
WO 2008/112749 PCT/US2008/056636
METHODS OF DETERIVIINING ACUTE MYELOID LEUKEMIA RESPONSE
TO TREATMENT WITH FARNESYLTRANSFERASE
Background of the Invention
Currently there is no method available to predict response to
famesyltransferase
inhibitors. Tipifarnib was the first=farnesyltransferase inhibitor (FTI) to be
tested in the clinic.
Rowinsky et al. (2006). It has demonstrated significant activity in
hematological disorders
including AML, MM, MDS and CML, with complete response rates in AML and MDS of
up to
approximately 15%. Mesa et al. (2006); Karp et al. (2001); Lancet et al.
(2007); Fenaux et al.
(2007); and Harousseau et al. (2007). FTIs function by competitively
inhibiting the addition of a
farnesyl moiety to a number of important signaling molecules including Ras.
Rowinsky et al.
(2006); and Cox et al. (2002).
Some molecules, such as Ras, that are implicated in cancers must be
farnesylated by the
farnesyl transferase enzyme in order to interact with the inner leaflet of the
plasma membrane of
the cell and become involved in various signaling pathways. Ras is not the
only protein
implicated in cancer that has a CAAX box that is prenylated. Farnesyl
transferase inhibitors
(FTIs) are therapeutic agents that inhibit the covalent attachment of the
carbon farnesyl moieties
to the C-terminal CAAX motif of various proteins. They have utility in the
treatment of cancers
and proliferative disorders such as leukemia. Acute myelogenous leukemia (AML)
is among the
diseases that can most beneficially be addressed with FTIs.
As is true in the case of many treatment regimens, some patients respond to
treatment
with FTIs and others do not. Prescribing the treatment to a patient who is
unlikely to respond to
it is not desirable. Thus, it would be useful to know how a patient could be
expected to respond
to such treatment before a drug is administered so that non-responders would
not be
unnecessarily treated and so that those with the best chance of benefiting
from the drug are
properly treated and monitored. Further, of those who respond to treatment,
there may be
varying degrees of response. Treatment with therapeutics other than FTIs or
treatment with
therapeutics in addition to FTIs may be beneficial for those patients who
would not respond to
FTIs or in whom response to FTIs alone is less than desired.
Historically, the mutation status of the ras gene was considered to be a
candidate
biomarker for patient response to FTIs. This rationale was based on pre-
clinical evidence that
FTIs could block Ras-transformed cells, and that specific point mutations
within ras genes cause
1
CA 02680909 2009-09-11
WO 2008/112749 PCT/US2008/056636
constitutive activation of the Ras pathway in many cancers. End et al. (2001)
Reuter et al.
(2000); and Bos et al. (1989). Since it is generally accepted that tumors are
heavily reliant on the
activation of one or two pathways ("oncogene addiction" hypothesis), it
follows that patients
whose tumors are promoted by a particular pathway should respond to drugs that
inhibit that
pathway. Weinstein et al. (2006). However, pathways can be activated by
multiple events and it
has been found that Ras can be up-regulated in the absence of activating Ras
mutations. Ehmann
et al. (2006). Furthermore, no correlation between ras mutations and response
to FTIs has been
demonstrated in clinical studies. Karp et al. (2001); and 20070048782. Indeed,
while several
early clinical studies focused on cancers that exhibited high frequencies of
ras mutations the
response rate was disappointingly low in those trials. Mesa (2006); Rao et al.
(2004); and Van
Cutwem et al. (2004).
Summary of the Invention
We analyzed bone marrow from 67 patients from a phase 2 study of
famesyltransferase
inhibition with tipifarnib (R115777, ZARNESTRA8), in older adults with
previously untreated,
poor-risk acute myeloid leukemia (AML) for N-Ras mutations, global gene
expression, and/or
quantitative PCR (qPCR) of specific genes. Microarray profiling identified a
two-gene
expression ratio (RASGRPI:APTX) which provided the greatest accuracy for
predicting response
to tipifarnib. We demonstrated that this classifier could predict response to
tipifarnib in an
independent set of 54 samples from relapsed or refractory AML, with a NPV and
PPV of 92%
and 28%, respectively (odds ratio of 4.4). Therefore, in both newly diagnosed
and relapsed or
refractory AML, this classifier improves the overall response rate by
approximately 50% while
maintaining a high NPV, and significantly improves patient overall survival.
The two-gene
classifier was also validated by qPCR in thirty AML samples from the same
clinical study
demonstrating a negative predictive value (NPV) and positive predictive value
(PPV) of 81 %
and 50%, respectively (odds ratio of 4.3). These data indicate that a simple
two-gene expression
assay may have utility in diagnosing a population of AML patients who are more
likely to
respond to tipifarnib.
Microarray technology has been utilized to identify gene expression profiles
that are
predictive of response or resistance to a number of different therapeutic
modalities in a variety of
cancers, including chemotherapies or endocrine therapies in breast cancer,
diffuse large b-cell
lymphoma and leukemia. Ma et al. (2004); Chang et al. (2003) Jansen et al.
(2005); Potti et al.
2
CA 02680909 2009-09-11
WO 2008/112749 PCT/US2008/056636
(2006); Shipp et al. (2002); Rosenwald et al. (2002); Lossos et al. (2004);
Yeoh et al. (2002) and
Holleman et al. (2004). We have previously used gene expression profiling to
identify molecular
predictors of response to tipifarnib in relapsed or refractory AML.
20070048782. Here we have
extended this work to newly diagnosed AML which has led to the identification
of a two-gene
expression ratio (RASGRPI:APTX) that is predictive of clinical outcome. We
further show that
this classifier can be assayed by qPCR and that it also has predictive utility
in relapsed or
refractory AML.
Brief Description of the Drawings
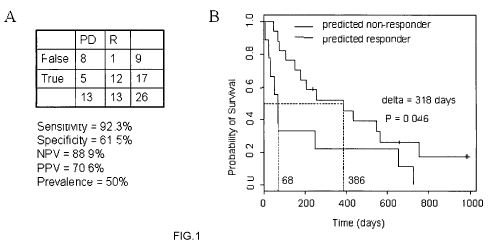
Figure 1 depicts the performance of the RASGRPI gene as a predictor of
response to
tipifarnib in AML. The accuracy rates (A) and Kaplan-Meier survival curves (B)
using the
RASGRPI gene classifier in newly diagnosed AML.
Figure 2 depicts the performance of the RASGRPI:APTX gene pair as a predictor
of
response to tipifarnib in AML. The overall survival of newly diagnosed AML
patients (A) and
relapsed/refractory AML patients (C) stratified with the 2-gene classifier are
plotted using
Kaplan-Meier analysis. The accuracy rates of the two-gene classifier in newly
diagnosed AML
(B) and relapsed/refractory AML (D) are shown.
. Figure 3 depicts the performance of RASGRPI:APTX gene classifier using qPCR.
(A)
The normalized RASGRPI:APTX Ct values for 20 responders and 10 patients with
progressive
disease. The 20 independent samples and 10 training samples that were run on
microarray are
shown separately. Horizontal bars indicate group means. (B) The accuracy rates
of the
RASGRPI gene classifier in newly diagnosed AML for all 30 patients are shown
using a cutoff
of 0 was used to stratify patients. (C) The associated overall survival of the
stratified patients are
plotted using Kaplan-Meier analysis.
Figure 4 depicts the performance of the RASGRP 1 gene as a predictor of
response to
tipifarnib in relapsed and refractory AML. The accuracy rates (A) and Kaplan
Meier survival
curves (B) using the RASGRPI gene classifier in relapsed/refractory AML.
Figure 5 depicts the overall survival of non-FTI treated AML patients
stratified with the
RASGRPI:APTX gene expression ratio. Three cDNA probes for both RASGRPI and
APTX
were present in the available data set. We first calculated the mean value for
each gene and then
calculated the RASGRPI:APTX ratio of these values. Patients whose ratio was
above I were
3
CA 02680909 2009-09-11
WO 2008/112749 PCT/US2008/056636
classified as progressors and those with a ratio below 1 were classified as
responders.
Kaplan-Meier analysis was then performed.
Figure 6 depicts the correlation of Affymetrix and qPCR data. Nine RNA samples
that
were analyzed on both the Affymetrix GeneChip and by qPCR were compared by
linear
regression analysis.
Detailed Description of the Invention
The therapeutic agents referred to in this specification are FTIs. They take
on a multitude
of forms but share the essential inhibitory function of interfering with or
lessening the
farnesylation of proteins implicated in cancer and proliferative diseases.
Preferably, the FTIs are
those indicated for the treatment of leukemias such as AML. A patient who
responds to an FTI
is one in whom a reduction of more than 50% of blast cells is seen in bone
marrow following
treatment with the FTI.
Numerous FTIs are within the scope of the invention and include those
described in
5,976,851; 5,972,984; 5,972,966; 5,968,965; 5,968,952; 6,187,786; 6,169,096;
6,037,350;
6,177,432; 5,965,578; 5,965,539; 5,958,939; 5,939,557; 5,936,097; 5,891,889;
5,889,053;
5,880,140; 5,872,135; 5,869,682; 5,861,529; 5,859,015; 5,856,439; 5,856,326;
5,852,010;
5,843,941; 5,807,852; 5,780,492; 5,773,455; 5,767,274; 5,756,528; 5,750,567;
5,721,236;
5,700,806; 5,661,161; 5,602,098; 5,585,359; 5,578,629; 5,534,537; 5,532,359;
5,523,430;
5,504,212; 5,491,164; 5,420,245; and 5,238,922. Non-peptidal, so-called "small
molecule"
therapeutics are preferred. More preferred FTIs are quinolines or quinoline
derivatives such as:
7-(3-chlorophenyl)-9-[(4-chlorophenyl)-IH-imidazol-l-ylmethyl]-2,3-dihydr- o-
1H,5H-
benzo[ij]quinolizin-5-one,
7-(3-chlorophenyl)-9-[(4-chlorophenyl)-1 H-imidazol-l-ylmethyl]-1,2-dihydr-o-
4H-
pyrrolo[3,2,1-ij]quinoline-4-one,
8-[amino(4-chlorophenyl)(1-methyl-1 H-imidazol-5-yl),methyl]-6-(3-chloroph-
enyl)-1,2-
dihydro-4H-pyrrolo[3,2,1-ij]quinolin-4-one, and
8-[amino(4-chlorophenyl)(1-methyl-1 H-imidazol-5-yl)methyl]-6-(3-chlorophe-
nyl)-2,3-
dihydro-IH,5H-benzo[ij]quinolizin-5-one. The most preferred FTI is (B)-6-
[amino(4-
chlorophenyl)(1-methyl-lH-imidazol-5-yl)methyl]-4-(3-ch- lorophenyl)-I-methyl-
2(1.H)-
quinolinone).
4
CA 02680909 2009-09-11
WO 2008/112749 PCT/US2008/056636
In the aspect of the invention comprising treating leukemia with FTIs and
other
therapeutic agents, the therapeutic agents referred to in this specification
are those that have an
effect on the biological pathway explicated through the gene expression
analysis of leukemic
cells subjected to treatment with quinilone-based FTIs.
The mere presence of nucleic acid sequences having the potential to express
proteins or
peptides ("genes") within the genome is not determinative of whether a protein
or peptide is
expressed in a given cell. Whether or not a given gene capable of expressing
proteins or peptides
does so and to what extent such expression occurs, if at all, is determined by
a variety of
complex factors. Irrespective of difficulties in understanding and assessing
these factors,
assaying gene expression can provide useful information about the cellular
response to a given
stimulus such as the introduction of a drug or other therapeutic agent.
Relative indications of the
degree to which genes are active or inactive can be found in gene expression
profiles. The gene
expression profiles of this invention are used to identify and treat patients
who will likely benefit
from a given therapy or exclude patients from a.given therapy where the
patient likely would
experience little or no beneficial response to the drug or therapy.
Preferred methods for establishing gene expression profiles (including those
used to
arrive at the explication of the relevant biological pathways) include
determining the. amount of
RNA that is produced by a gene that can code for a protein or peptide. This is
accomplished by
reverse transcription PCR (RT-PCR), competitive RT-PCR, real time RT-PCR,
differential
display RT-PCR, Northern Blot analysis and other related tests. While it is
possible to conduct
these techniques using individual PCR reactions, it is best to amplify copy
DNA (cDNA) or copy
RNA (cRNA) produced from mRNA and analyze it via microarray. A number of
different array
configurations and methods for their production are known to those of skill in
the art and are
described in U. S. Patents such as: U. S. Pat. Nos. 5,445,934; 5,532,128;
5,556,752; 5,242,974;
5,384,261; 5,405,783; 5,412,087; 5,424,186; 5,429,807; 5,436,327; 5,472,672;
5,527,681;
5,529,756; 5,545,531; 5,554,501; 5,561,071; 5,571,639; 5,593,839; 5,599,695;
5,624,711;
5,658,734; and 5,700,637.
Microarray technology allows for the measurement of the steady-state mRNA
level of
thousands of genes simultaneously thereby presenting a powerful tool for
identifying the effect
of FTIs on cell biology and the likely effect of treatment based on analysis
of such effects. Two
microarray technologies are currently in wide use. The first are cDNA arrays
and the second are
CA 02680909 2009-09-11
WO 2008/112749 PCT/US2008/056636
oligonucleotide arrays. Although differences exist in the construction of
these chips, essentially
all downstream data analysis and output are the same. The product of these
analyses are
typically measurements of the intensity of the signal received from a labeled
probe used to detect
a cDNA sequence from the sample that hybridizes to a nucleic acid sequence at
a known location
on the microarray. Typically, the intensity of the signal is proportional to
the quantity of cDNA,
and thus mRNA, expressed in the sample cells. A large number of such
techniques are available
and useful. Preferred methods for determining gene expression can be found in
6,271,002;
6,218,122; 6,218,114; and 6,004,755.
Analysis of the expression levels is conducted by comparing such intensities.
This is best
done by generating a ratio matrix of the expression intensities of genes in a
test sample versus
those in a control sample. For instance, the gene expression intensities from
a tissue that has
been treated with a drug can be compared with the expression intensities
generated from the
same tissue that has not been treated with the drug. A ratio of these
expression intensities
indicates the fold-change in gene expression between the test and control
samples.
Gene expression profiles can also be displayed in a number of ways. The most
common
method is to arrange a ratio matrix into a graphical dendogram where columns
indicate test
samples and rows indicate genes. The data is arranged so genes that have
similar expression
profiles are proximal to each other. The expression ratio for each gene is
visualized as a color.
For example, a ratio less than one (indicating down-regulation) may appear in
the blue portion of
the spectrum while a ratio greater than one (indicating up-regulation) may
appear as a color in
the red portion of the spectrum. Commercially available computer software
programs are
available to display such data including "OMNIVIZ PRO" software from Batelle
and "TREE
VIEW" software from Stanford
The genes that are differentially expressed are either up regulated or down
regulated in
diseased cells following treatment with an FTI. Up regulation and down
regulation are relative
terms meaning that a detectable difference (beyond the contribution of noise
in the system used
to measure it) is found in the amount of expression of the genes relative to
some baseline. In this
case, the baseline is the measured gene expression of the untreated diseased
cell. The genes of
interest in the treated diseased cells are then either up regulated or down
regulated relative to the
baseline level using the same measurement method. Preferably, levels of up and
down
regulation are distinguished based on fold changes of the intensity
measurements of hybridized
6
CA 02680909 2009-09-11
WO 2008/112749 PCT/US2008/056636
microarray probes. A 1.5 fold difference is preferred for making such
distinctions. That is,
before a gene is said to be differentially expressed in treated versus
untreated diseased cells, the
treated cell is found to yield at least 1.5 times more, or 1.5 times less
intensity than the untreated
cells. A 1.7 fold difference is more preferred and a 2 or more fold difference
in gene expression
measurement is most preferred.
A portfolio of genes is a set of genes grouped so that information obtained
about them
provides the basis for making a clinically relevant judgment such as a
diagnosis, prognosis, or
treatment choice. In this case, the judgments supported by the portfolios
involve the treatment of
leukemias with FTI's. Portfolios of gene expression profiles can be comprised
of combinations
of genes.
One method of the invention involves comparing gene expression profiles for
various
genes to determine whether a person is likely to respond to the use of a
therapeutic agent.
Having established the gene expression profiles that distinguish responder
from non-responder,
the gene expression profiles of each are fixed in a medium such as a computer
readable medium
as described below. A patient sample is obtained that contains diseased cells
(such as
hematopoietic blast cells in the case of AML) is then obtained. Sample RNA is
then obtained
and amplified from the diseased patient cell and a gene expression profile is
obtained, preferably
via micro-array, for genes in the appropriate portfolios. The expression
profiles of the samples
are then compared to those previously determined as responder and non-
responder. If the sample
expression patterns are consistent with an FTI responder expression pattern
then treatment with
an FTI could be indicated (in the absence of countervailing medical
considerations). If the
sample expression patterns are consistent with an FTI non-responder expression
pattern then
treatment with an FTI would not be indicated. Preferably, consistency of
expression pattems is
determined based on intensity measurements of micro-array reading as described
above.
In similar fashion, gene expression profile analysis can be conducted to
monitor
treatment response. In one aspect of this method, gene expression analysis as
described above is
conducted on a patient treated with an FTI at various periods throughout the
course of treatment.
If the gene expression patterns are consistent with a responder then the
patient's therapy is
continued. If it is not, then the patient's therapy is altered as with
additional therapeutics such as
tyrosine kinase inhibitor, changes to the dosage, or elimination of FTI
treatment. Such analysis
7
CA 02680909 2009-09-11
WO 2008/112749 PCT/US2008/056636
permits intervention and therapy adjustment prior to detectable clinical
indicia or in the face of
otherwise ambiguous clinical indicia.
It is possible to attain ambiguous results in which some gene expression
profiles are
recorded that are in some respects indicative of a responder and in other
respects indicative of a
non-responder. For example, the profiles may show that three genes are up-
regulated consistent
with a responder but that another gene is not up-regulated as would ordinarily
be the case for a
responder. In such a case, statistical algorithms can be applied to determine
the probability that
the patient will respond or not respond to the drug. Statistical algorithms
suitable for this
purpose are well known and are available.
Articles of this invention are representations of the gene expression profiles
useful for
treating, diagnosing, prognosticating, staging, and otherwise assessing
diseases that are reduced
to a medium that can be automatically read such as computer readable media
(magnetic, optical,
and the like). The articles can also include instructions for assessing the
gene expression profiles
in such media. For example, the articles may comprise a CD ROM having computer
instructions
for comparing gene expression profiles of the portfolios of genes described
above. The articles
may also have gene expression profiles digitally recorded therein so that they
may be compared
with gene expression data from patient samples. Alternatively, the profiles
can be recorded in
different representational format. A graphical recordation is one such format.
FIG. I shows an
example of the graphical display of such a recordation. Clustering algorithms
such as those
incorporated in "OMNIVIZ" and "TREE VIEW" computer programs mentioned above
can best
assist in the visualization of such data.
Additional articles according to the invention are nucleic acid arrays (e.g.
cDNA or
oligonucleotide arrays), as described above, configured to discem the gene
expression profiles of
the invention.
Using clustering analysis (including the algorithms mentioned above) one can
compare
the expression levels of patient samples to establish regulatory relationships
among genes with a
certain statistical confidence. A dynamic map was constructed based upon such
expression data.
Such a genetic network map is useful for drug discovery. For example, once
basic genes of
interest were identified, a list of potential up-stream regulatory genes was
found using such a
genetic network map. The genes so identified or their expression products were
then analyzed
8
CA 02680909 2009-09-11
WO 2008/112749 PCT/US2008/056636
for their use as drug targets. In some embodiments, the regulatory function of
the particular
genes identified was used to identify therapeutics for use in treating
leukemia.
The regulation of transcription, RNA processing and RNA editing are all
accomplished
by proteins which are coded by their own genes. In addition, DNA sequences can
exert
long-range control over the expression of other genes by positional effects.
Therefore, the
expression of genes is often regulated by the expression of other genes. Those
regulatory genes
are called upstream genes, relative to the regulated or down-stream genes. In
a simple regulatory
pathway: A++>B-->C++>D where: A, B, C, D are genes ++ up-regulates -- down-
regulates
Gene A is an up-stream gene of gene B and B is an up-stream gene of C. One of
skill in the art
would appreciate that the network is frequently looped and inter-connected. In
some instances,
the expression of a gene is regulated by its own product as either a positive
or negative feedback.
Cluster analysis methods were used to group genes whose expression level is
correlated.
Methods for cluster analysis are described in detail in Harfigan (1975)
Clustering Algorithms,
NY, John Wile and Sons, Inc, and Everritt, (1980) Cluster Analysis 2nd. Ed.
London Heineman
Educational books, Ltd. Path analysis was used to decompose relations among
variables and for
testing causal models for the genetic networks. Multiple primary targets of a
drug in leukemic
cells were identified as were drugs/drug classes useful in treating such
cells. According to the
current invention, drugs are any compounds of any degree of complexity that
perturb a biological
system.
The biological effect of a drug may be a consequence of drug-mediated changes
in the
rate of transcription or degradation of one or more species of RNA, the rate
or extent of
translation or post-translational processing of one or more polypeptides, the
rate or extent of the
degradation of one or more proteins, the inhibition or stimulation of the
action or activity of one
or more proteins, and so forth. In addition to the FTI's that are preferred,
the preferred drugs of
this invention are those that modulate the MAPK/ERK signaling pathways, TGF-
(3, WNT or
apoptotic pathways. These include, without limitation, tyrosine kinase
inhibitors, MEK kinase
inhibitors, P13K kinase inhibitors, MAP kinase inhibitors, apoptosis
modulators and
combinations thereof. Exemplary drugs that are most preferred among these are
the
"GLEEVEC" tyrosine kinase inhibitor of Novartis, U-0126 MAP kinase inhibitor,
PD-098059
MAP kinase inhibitor, SB-203580 MAP kinase inhibitor, and antisense, ribozyme,
and
DNAzyme Bcl-XL anti-apoptotics. Examples of other useful drugs include,
without limitation,
9
CA 02680909 2009-09-11
WO 2008/112749 PCT/US2008/056636
the calanolides of 6,306,897; the substituted bicyclics of 6,284,764; the
indolines of 6,133,305;
and the antisense oligonucleotides of 6,271,210.
As noted, the drugs of the instant invention can be therapeutics directed to
gene therapy
or antisense therapy. Oligonucleotides with sequences complementary to an mRNA
sequence
can be introduced into cells to block the translation of the mRNA, thus
blocking the function of
the gene encoding the mRNA. The use of oligonucleotides to block gene
expression is
described, for example, in, Strachan and Read, Human Molecular Genetics, 1996.
These antisense molecules may be DNA, stable derivatives of DNA such as
phosphorothioates or methylphosphonates, RNA, stable derivatives of RNA such
as 2'-0-
alkylRNA, or other antisense oligonucleotide mimetics. Antisense molecules may
be introduced
into cells by microinjection, liposome encapsulation or by expression from
vectors harboring the
antisense sequence.
In the case of gene therapy, the gene of interest can be ligated into viral
vectors that
mediate transfer of the therapeutic DNA by infection of recipient host cells.
Suitable viral
vectors include retrovirus, adenovirus, adeno-associated virus, herpes virus,
vaccinia virus, polio
virus and the like. Alternatively, therapeutic DNA can be transferred into
cells for gene therapy
by non-viral techniques including receptor-mediated targeted DNA transfer
using ligand-DNA
conjugates or adenovirus-ligand-DNA conjugates, lipofection membrane fusion or
direct
microinjection. These procedures and variations thereof are suitable for ex
vivo as well as in
vivo gene therapy. Protocols for molecular methodology of gene therapy
suitable for use with
the gene is described in Gene Therapy Protocols, edited by Paul D. Robbins,
Human press,
Totowa NJ, 1996.
Pharmaceutically useful compositions comprising the drugs of this invention
may be
formulated according to known methods such as by the admixture of a
pharmaceutically
acceptable carrier. Examples of such carriers and methods of formulation may
be found in
Remington's Pharmaceutical Sciences. To form a pharmaceutically acceptable
composition
suitable for effective administration, such compositions will contain an
effective amount of the
drug. The effective amount of the drug may vary according to a variety of
factors such as the
individual's condition, weight, sex and age. Other factors include the mode of
administration.
The pharmaceutical compositions may be provided to the individual by a variety
of routes such
as subcutaneous, topical, oral and intramuscular.
CA 02680909 2009-09-11
WO 2008/112749 PCT/US2008/056636
The drugs of this invention include chemical derivatives of the base molecules
of the
drug. That is, they may contain additional chemical moieties that are not
normally a part of the
base molecule. Such moieties may improve the solubility, half-life,
absorption, etc. of the base
molecule. Alternatively the moieties may attenuate undesirable side effects of
the base molecule
or decrease the toxicity of the base molecule. Examples of such moieties are
described in a
variety of texts, such as Remington's Pharmaceutical Sciences.
Compounds identified according to the methods disclosed herein may be used
alone at
appropriate dosages defined by routine testing in order to obtain optimal
inhibition or activity
while minimizing any potential toxicity. In addition, co-administration or
sequential
administration of other agents may be desirable.
The drugs of this invention can be administered in a wide variety of
therapeutic dosage
forms in conventional vehicles for administration. For example, the drugs can
be administered in
such oral dosage forms as tablets, capsules (each including timed release and
sustained release
formulations), pills, powders, granules, elixirs, tinctures, solutions,
suspensions, syrups and
emulsions, or by injection. Likewise, they may also be administered in
intravenous (both bolus
and infusion), intraperitoneal, subcutaneous, topical with or without
occlusion, or intramuscular
form, all using forms well known to those of ordinary skill in the
pharmaceutical arts. An
effective but non-toxic amount of the compound desired can be employed as a
modulating agent.
The daily dosage of the products may be varied over a wide range from 0.01 to
1,000 mg
per patient, per day. For oral administration, the compositions are preferably
provided in the
form of scored or unscored tablets containing 0.01, 0.05, 0.1, 0.5, 1.0, 2.5,
5.0, 10.0, 15.0, 25.0,
and 50.0 milligrams of the active ingredient for the symptomatic adjustment of
the dosage to the
patient to be treated. An effective amount of the drug is ordinarily supplied
at a dosage level of
from about 0.0001 mg/kg to about 100 mg/kg of body weight per day. The range
is more
particularly from about 0.001 mg/kg to 10 mg/kg of body weight per day. The
dosages are
adjusted when combined to achieve desired effects. On the other hand, dosages
of these various
agents may be independently optimized and combined to achieve a synergistic
result wherein the
pathology is reduced more than it would be if either agent were used alone.
Advantageously, compounds or modulators used in the present invention may be
administered in a single daily dose, or the total daily dosage may be
administered in divided
doses of two, three or four times daily. Furthermore, compounds or modulators
for the present
11
CA 02680909 2009-09-11
WO 2008/112749 PCT/US2008/056636
invention can be administered in intranasal form via topical use of suitable
intranasal vehicles, or
via transdermal routes, using those forms of transdermal skin patches well
known to those of
ordinary skill in that art. To be administered in the form of a transdermal
delivery system, the
dosage administration will, of course, be continuous rather than intermittent
throughout the
dosage regimen.
For combination treatment with more than one active agent, where the active
agents are
in separate dosage formulations, the active agents can be administered
concurrently, or they each
can be administered at separately staggered times.
The dosage regimen utilizing the compounds or modulators in the present
invention is
selected in accordance with a variety of factors including type, species, age,
weight, sex and
medical condition of the patient; the severity of the condition to be treated;
the route of
administration; the renal and hepatic function of the patient; and the
particular drug employed. A
physician or veterinarian of ordinary skill can readily determine and
prescribe the effective
amount of the drug required to prevent, counter or arrest the progress of the
condition. Optimal
precision in achieving concentrations of drug within the range that yields
efficacy without
toxicity requires a regimen based on the kinetics of the drug's availability
to target sites. This
involves a consideration of the distribution, equilibrium, and elimination of
a drug.
The drugs of this invention can form the active ingredient, and are typically
administered
in admixture with suitable pharmaceutical diluents, excipients or carriers
(collectively referred to
herein as "carrier" materials) suitably selected with respect to the intended
form of
administration, that is, oral tablets, capsules, elixirs, syrups and the like,
and consistent with
conventional pharmaceutical practices.
For instance, for oral administration in the form of a tablet or capsule, the
active drug
component can be combined with an oral, non-toxic pharmaceutically acceptable
inert carrier
such as ethanol, glycerol, water and the like. Moreover, when desired or
necessary, suitable
binders, lubricants, disintegrating agents and coloring agents can also be
incorporated into the
mixture. Suitable binders include, without limitation, starch, gelatin,
natural sugars such as
glucose or beta-lactose, corn sweeteners, natural and synthetic gums such as
acacia, tragacanth or
sodium alginate, carboxymethylcellulose, polyethylene glycol, waxes and the
like. Lubricants
used in these dosage forms include, without limitation, sodium oleate, sodium
stearate,
magnesium stearate, sodium benzoate, sodium acetate, sodium chloride and the
like.
12
CA 02680909 2009-09-11
WO 2008/112749 PCT/US2008/056636
Disintegrators include, without limitation, starch, methyl cellulose, agar,
bentonite, xanthan gum
and the like.
For liquid forms the active drug component can be combined in suitably
flavored
suspending or dispersing agents such as the synthetic and natural gums, for
example, tragacanth,
acacia, methyl-cellulose and the like. Other dispersing agents that may be
employed include
glycerin and the like. For parenteral administration, sterile suspensions and
solutions are desired.
Isotonic preparations, which generally contain suitable preservatives, are
employed when
intravenous administration is desired.
The drugs in the present invention can also be administered in the form of
liposome
delivery systems, such as small unilamellar vesicles, large unilamellar
vesicles and multilamellar
vesicles. Liposomes can be formed from a variety of phospholipids, such as
cholesterol,
stearylamine or phosphatidylcholines.
Drugs in the present invention may also be delivered by the use of monoclonal
antibodies
as individual carriers to which the compound molecules are coupled. The drugs
in the present
invention may also be coupled with soluble polymers as targetable drug
carriers. Such polymers
can include polyvinyl-pyrrolidone, pyran copolymer, polyhydroxypropylmethacryl-
amidephenol,
polyhydroxy-ethylaspartamidephenol, or polyethyl-eneoxidepolylysine
substituted with
palmitoyl residues. Furthermore, the drugs in the present invention may be
coupled to a class of
biodegradable polymers useful in achieving controlled release of a drug, for
example, polylactic
acid, polyepsilon caprolactone, polyhydroxy butyric acid, polyorthoesters,
polyacetals,
polydihydro-pyrans, polycyanoacrylates and cross-linked or amphipathic block
copolymers of
hydrogels.
For oral administration, the drugs may be administered in capsule, tablet, or
bolus form
or alternatively they can be mixed with feed. The capsules, tablets, and
boluses are comprised of
the active ingredient in combination with an appropriate carrier vehicle such
as starch, talc,
magnesium stearate, or di-calcium phosphate. These unit dosage forms are
prepared by
intimately mixing the active ingredient with suitable finely-powdered inert
ingredients including
diluents, fillers, disintegrating agents, and/or binders such that a uniform
mixture is obtained. An
inert ingredient is one that will not react with the drugs and which is non-
toxic to the animal
being treated. Suitable inert ingredients include starch, lactose, talc,
magnesium stearate,
vegetable gums and oils, and the like. These formulations may contain a widely
variable amount
13
CA 02680909 2009-09-11
WO 2008/112749 PCT/US2008/056636
of the active and inactive ingredients depending on numerous factors such as
the size and type of
the animal species to be treated and the type and severity of the infection.
The active ingredient
may also be administered by simply mixing the compound with the feedstuff or
by applying the
compound to the surface of the foodstuff.
The compounds or modulators may alternatively be administered parenterally via
injection of a formulation consisting of the active ingredient dissolved in an
inert liquid carrier.
Injection may be either intramuscular, intraluminal, intratracheal, or
subcutaneous. The
injectable formulation consists of the active ingredient mixed with an
appropriate inert liquid
carrier. Acceptable liquid carriers include the vegetable oils such as peanut
oil, cotton seed oil,
sesame oil and the like as well as organic solvents such as solketal, glycerol
formal and the like.
As an alternative, aqueous parenteral formulations may also be used. The
vegetable oils are the
preferred liquid carriers. The formulations are prepared by dissolving or
suspending the active
ingredient in the liquid carrier such that the final formulation contains from
0.005 to 10% by
weight of the active ingredient.
All references cited herein are hereby incorporated herein by reference. The
invention is
further illustrated by the following non-limiting examples.
EXAMPLE I
Materials and Methods
Clinical Evaluation
The current study utilized 67 bone marrow samples collected from an open
label,
multicenter, non-comparative phase 2 study investigating the efficacy and
safety of
farnesyltransferase inhibition with tipifarnib (R115777, ZARNESTRA ) in 158
older adults with
previously untreated, poor-risk AML. The clinical results have been published
elsewhere.
Lancet et al. (2006).
Sample Collection and Processing
Bone marrow samples were collected from consenting patients before treatment
with
tipifarnib and mononuclear cells were processed on site. Bone marrow aspirates
were diluted
with PBS and centrifuged with ficoll-diatrizoate (1.077g/ml). Enriched
leukemic blood cells
were washed twice with PBS, resuspended in FBS with 10% DMSO and immediately
frozen at -
70 C to -80 C. Total RNA was extracted from cell samples using the Trizol Kit
(Qiagen, Santa
Clarita, CA). RNA quality was determined by assessing the presence of
ribosomal bands on an
14
CA 02680909 2009-09-11
WO 2008/112749 PCT/US2008/056636
Agilent Bioanalyzer. Good quality samples were further processed for
microarray analysis. DNA
was isolated from the same sample of Trizol-processed bone marrow as per the
manufacturers
instructions (Qiagen, Santa Clarita, CA). Samples were assayed for global gene
expression, N-
Ras mutations, and/or qPCR of specific genes (Fig 1).
N-Ras mutational status
Analysis of activating mutations in N-ras was determined by PCR and RFLP
analysis as
previously described. End et al. (2001). Exons 1 and 2 of the N-ras gene were
simultaneously
amplified in a single multiplex reaction and an aliquot was used for a second
round of PCR.
Resistance to cleavage at natural or primer induced restriction enzyme sites
in second-round
amplicons indicated the presence of a mutation that had abolished the site at
the loci being
analyzed. Restriction enzymes for the analysis of specific loci were Bsl I (N-
ras codons 12 and
13), Msc I (N-ras codon 61, positions 1 and 2), and Bfa I (N-ras codon 61,
position 3). Reactions
were digested overnight and PCR products were analyzed on an Agilent
Bioanalyzer.
Microarray analysis
Synthesis of cDNA and cRNA were performed according to Affymetrix (Santa
Clara,
CA) protocols. Since the yield of many samples was low two rounds of linear
amplification was
performed as previously described. 20070048782. For hybridization, I 1 g of
cRNA were
fragmented randomly by incubation at 94 C for 35 min in 40 mM Tris-acetate, pH
8.1, 100 mM
potassium acetate, and 30 mM magnesium acetate. Fragmented cRNA was hybridized
to U133A
arrays at 45 C for 16 h in a rotisserie oven set at 60 rpm. Following
hybridization, arrays were
washed (with 6x SSPE and 0.5x SSPE containing Triton X-100 (0.005%)), and
stained with
streptavidin-phycoerythrin (SAPE; Molecular Probes, Eugene, OR).
Quantification of bound
labeled probe was conducted using the Agilent G2500A GeneArray scanner
(Agilent
Technologies, Palo Alto, CA).
The total fluorescence intensity for each array was scaled to the uniform
value of 600.
Chip performance was quantified by calculating a signal to noise ratio (raw
average
signal/noise). Chips were removed from further analysis if their signal-to-
noise ratio was less
than 20 or if the present calls on the chip was less than 30%. Genes were only
included in further
analysis if they were called "present" in at least 10% of the chips.
Approximately 12,000
Affymetrix probe sets remained following this cut-off. The quality of the gene
expression data
were further controlled by identifying outliers based on principal components
analysis and by
CA 02680909 2009-09-11
WO 2008/112749 PCT/US2008/056636
analyzing the normal distributions of the gene intensities (Partek Pro V5.1).
The microarray data
have been deposited in NCBIs Gene Expression Omnibus (GEO,
http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series
accession number
GSE',X_}iXX.
Response definitions
Response to tipifarnib is reported in the clinical paper and was defined as
patients who
had a complete response (CR), a partial response (PR), or hematological
improvement (HI).
Lancet et al. (2006). Briefly, HI was defined as any bone marrow blast count
less than 5% or a
reduction in bone marrow blasts by at least half. Progressive disease (PD) was
defined as either
>50% increase in bone marrow or circulating blast % from baseline, or new
appearance of
circulating blasts (on at least 2 consecutive occasions). Stable disease (SD)
was defined as any
response not meeting CR, PR, HI, or PD criteria.
Statistical analysis
Receiver Operator Characteristic (ROC) analysis was utilized to test the
overall
predictive value of individual genes and/or multigene classifiers. The
following gene filtering
criteria were used to identify genes differentially expressed between
responders and patients with
progressive disease: Specificity for identifying "responder" with 100%
sensitivity >= 40%, T-
test p value (log2 transformed data with unequal variance) < 0.05, fold change
> 2. The genes
that passed these criteria were ranked by AUC (Area under the ROC curve).
To build a classifier the response score was used to calculate each patient's
chance to
response to tipifarnib therapy. The score was defined as the linear
combination of weighted
expression signals with the t statistic as the weight. The threshold was
determined from the ROC
curve of the training set to ensure 100% sensitivity and the highest
specificity. To determine how
many genes needed to be included in the predictor, leave-one-out cross
validation (LOOCV) was
carried out. The response scores for the `left-out' samples based on different
numbers of genes
were recorded. The performances of the predictors with different numbers of
genes were
assessed based on misclassification error rate, sensitivity, specificity, p
values measuring the
separation of Kaplan-Meier curves of the two predicted groups. And the best
predictor was
selected accordingly.
The Top Scoring Pair (TSP) algorithm was first introduced by Geman et al.
(2004). In
essence, the algorithm ranks all the gene pairs (genes i and j) based on the
absolute difference
16
CA 02680909 2009-09-11
WO 2008/112749 PCT/US2008/056636
(Dij) in the frequency of event where gene i has higher expression value than
gene j in samples
among class C 1 to C2. In the cases of there are multiple top scoring pairs
(all sharing the same
Dij), we select the top pair by a secondary rank score that measures the
magnitude to which
inversions of gene expression levels occur from one class to the other within
a pair of genes. The
top pair with highest frequency of absolute Dij > 2 fold in all samples will
be selected as
candidate pair. The candidate pair was then assessed in an independent testing
data set.
Leave-one-out cross validation (LOOCV) was carried out in the training data
set to
evaluate how the algorithm perform. The performances of the predictors were
assessed based on
maximum misclassification error rate. All the statistical analyses were done
using R (R
Development Core Team, 2006).
Real-Time Quantitative RT-PCR
For each sample, 1 g of total RNA (as assessed by OD260) was reversed
transcribed
using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems,
Foster City, CA)
according to the manufacturers instructions. Samples were then incubated at 25
C for 10
minutes and then 37 C for 30 minutes for optimum RNA conversion. QPCR was
performed
using the ABI Prism 7900HT sequence detection system (Applied Biosystems,
Foster City, CA)
with all samples run in triplicate. Each reaction contained 5 l TaqMan
Universal PCR Master
Mix containing UNG (Applied Biosystems, Foster City, CA), 4.5 l of cDNA
template and 0.5
l of 20 x Assay on Demand Gene Expression Assay Mix or 9 pmol of both forward
and reverse
primer and 2.5 pmol of probe (Applied Biosystems, Foster City, CA), in a total
reaction volume
of 10 l. All primer, probe sets were chosen due to the small amplicon size
(less than 100
nucleotides) and FAM fluorogenic probes were used. Primers and probes used
were APTX
(product number 4331182 Applied Biosystems) and RASGRPI (product number
4351372
Applied Biosystems). The RASGRPI:APTX expression ratio was calculated by
normalizing the
raw Ct values by subtracting the mean Ct from the sample set, dividing by the
standard
deviation, and then calculating the difference of the normalized Ct values of
each gene (APTX-
RASGRPl ). Ma et al. (2004).
Results
This study examined gene expression profiles of leukemic bone marrow samples
from
patients enrolled in a Phase 2 clinical trial of the farnesyltransferase
inhibitor tipifamib in elderly
patients with previously untreated poor-risk acute myelogenous leukemia.
Lancet et al. (2006).
17
CA 02680909 2009-09-11
WO 2008/112749 PCT/US2008/056636
Bone marrow from 67 patients was collected before treatment with tipifarnib
and leukemic
myeloid cells were enriched by Ficoll-density centrifugation (Table 1). Good
quality total RNA
from 13 responders (9 CR, 4 HI), 8 stable disease and 13 progressive disease
patients was
amplified, labeled, and hybridized to the Affymetrix U133A GeneChip. A total
of 30 samples
were evaluated by qPCR for validation of specific genes and 32 samples were
evaluated for N-
Ras mutational status.
Table 1. Comparison of profiled patients.
Parameter All treated patients PGx profiled patients
Total patients, n 158 67
microarray assay, n 34
qPCR assay, n 30
N-Ras assay, n 32
N-Ras mutation, n (%) 11(34)
median age, y (range) 74 (34-85) 73 (63-85)
sex, n male (%) 95(60) 41(61)
Prior MDS, yes (%) 119 (75) 48 (72)
CR, no. (%) 22(14) 14(21)
PR, no. (%) 3 (2) 1(2)
HI, no. ( /a) 12 (8) 7 (10)
SD, no. (%) 50(32) 15 (22)
PD, no. (%) 58 (37) 30 (44)
NE, no. (%) 13 (8) 0(0)
CR = complete response; PR = partial response; HI = hematological improvement,
SD = stable disease, PD =
progressive disease, NE = not evaluable; PGx = pharmacogenomics
Ras mutational status and patient outcome
DNA from the bone marrow of 32 AML patients was screened for N-Ras activating
mutations (codons 12, 13, 61). Thirty-four percent (11/32) of patients
exhibited N-Ras mutations
with one patient having mutations at multiple codons (Table 2). There was no
statistically
significant correlation between N-Ras mutational status and response to
tipifarnib or overall
survival.
Table 2.
SUBJID RESPONSE N-Ras Mutation OS Alive Microarray qPCR SEX AGE Prior MDS
100101 HI ND 378 NO ND YES MALE 68 NO
100104 PD ND 728 NO YES YES FEMALE 63 NO
100109 PD ND 68 NO YES YES FEMALE 81 NO
100110 CR ND 983 YES YES YES FEMALE 74 NO
100112 PD ND 169 NO ND YES FEMALE 69 YES
100113 CR ND 211 NO ND YES MALE 82 YES
100116 PD ND 14 NO ND YES FEMALE 72 YES
100121 SD ND 252 NO YES ND MALE 72 YES
18
CA 02680909 2009-09-11
WO 2008/112749 PCT/US2008/056636
100204 SD N-12 493 NO ND ND FEMALE 69 YES
100205 PD WT 754 NO YES ND MALE 74 YES
100208 PD WT 29 NO YES ND MALE 76 YES
100209 PD N61(1,2) 209 NO YES ND MALE 73 YES
100210 PD N-12, N-13 654 NO YES ND MALE 68 YES
100212 SD N-12 1200 YES ND ND MALE 70 YES
100213 CR WT 257 NO YES ND FEMALE 81 YES
100214 CR N-13 395 NO ND ND FEMALE 73 YES
100215 SD WT 54 NO ND ND MALE 82 NO
100216 SD N-13 116 NO ND ND MALE 77 YES
100302 PD N-12 48 NO YES ND FEMALE 73 NO
100307 HI WT 179 NO YES ND MALE 68 YES
100310 SD WT 242 NO ND ND FEMALE 76 YES
100316 SD WT 273 NO ND ND FEMALE 66 NO
100317 PD WT 39 NO ND ND MALE 76 NO
100319 SD WT 233 NO YES ND MALE 71 NO
100320 HI WT 374 NO ND ND FEMALE 78 NO
100322 CR WT 237 YES YES ND MALE 73 YES
100324 HI WT 248 NO YES ND MALE 85 YES
100330 HI N-12 153 NO YES ND FEMALE 67 NO
100333 SD N-12 364 NO YES ND MALE 65 YES
100336 CR N-12 67 NO YES ND MALE 80 YES
100337 PD WT 38 NO ND ND MALE 72 YES
100338 PD N-12 8 NO YES ND MALE 78 NO
100339 PD WT 25 NO YES ND MALE 75 NO
100340 SD WT 32 NO ND ND FEMALE 83 NO
100341 CR WT 433 NO YES ND MALE 67 YES
100604 SD WT 64 NO YES ND MALE 63 YES
100605 PD WT 74 NO ND ND MALE 67 YES
101008 CR WT 548 NO YES ND MALE 82 NO
101021 CR ND 991 YES YES YES FEMALE 69 YES
101025 CR ND 735 YES ND YES MALE 70 YES
101029 PD ND 64 NO ND YES MALE 70 YES
101038 SD ND 151 NO YES ND FEMALE 75 YES
101039 PD ND 50 NO ND YES FEMALE 85 YES
101043 SD ND 200 NO YES ND FEMALE 79 YES
101046 PD ND 53 NO YES YES FEMALE 66 YES
101049 CR WT 564 NO YES ND MALE 65 YES
101057 CR WT 386 NO YES ND MALE 85 YES
101067 PD ND 88 NO ND YES FEMALE 76 YES
101069 PD ND 94 NO ND YES MALE 81 YES
101075 HI ND 659 YES YES YES MALE 71 YES
101077 SD ND 574 YES YES ND FEMALE 75 YES
101078 PD ND 190 NO ND YES FEMALE 77 NO
101079 PD ND 429 NO ND YES FEMALE 70 YES
101083 PD ND 71 NO ND YES MALE 73 YES
101091 CR ND 671 YES ND YES MALE 71 YES
101092 PD ND 136 NO ND YES FEMALE 69 YES
101094 HI ND 579 YES ND YES MALE 65 YES
101095 PD ND 108 NO YES YES MALE 82 YES
19
CA 02680909 2009-09-11
WO 2008/112749 PCT/US2008/056636
101096 CR ND 390 YES ND YES MALE 69 YES
101101 PD ND 91 NO ND YES MALE 69 YES
101102 PD ND 76 NO YES YES MALE 69 YES
101103 PD ND 29 NO ND YES FEMALE 80 NO
101108 PR ND 123 NO NO YES MALE 70 YES
101109 SD ND 656 YES YES ND MALE 68 YES
101114 PD ND 69 NO YES YES MALE 72 YES
101121 PD ND 43 NO ND YES MALE 78 NO
101122 PD ND 44 NO ND YES FEMALE 80 NO
ND = not determined; WT = wildtype; CR = complete response; PR = partial
response; HI = hematological
improvement, SD = stable disease, PD = progressive disease, OS = Overall
survival.
Identification of predictive genes from the newly diagnosed AML cohort
We next aimed to identify genes predictive of response to tipifarnib in the
newly
diagnosed AML population. To this end we performed discovery experiments in
the 13
responders (9 CR and 4 HI) and 13 patients with progressive disease. Patients
with stable disease
were not utilized in this analysis since these patients cannot be clearly
defined as either
responders or non-responders. Using the same approach as was utilized for
identifying markers
for relapsed and refractory AML (20070048782) we identified 45 probesets
(corresponding to 38
unique genes) that were predictive of response (Table 3). The selection
criteria aimed at
identifying genes that would predict responders with a high sensitivity
(approaching 100%) with
a specificity cut-off of 40% and a mean gene expression difference of at least
two-fold. The
genes were ranked based on the area under the curve (AUC) defined from a
receiver operator
characteristic (ROC) analysis of the training set. This value represents the
overall predictive
value of the gene with an AUC of 1.0 indicating perfect classification. Each
gene was first tested
on the training set using a LOOCV method. The top gene, the RAS guanyl-
releasing protein 1
(RASGRPI ), showed an AUC of 0.95.
CA 02680909 2009-09-11
WO 2008/112749 PCT/US2008/056636
N N-- ~- O O Ol r- IO \p V'1 V1 /1 W1 Vl Vl V1 Vl 7 M M M M M N N
U Q, p, p~ p~ L1 O~ p~ Q~ 00 OO GG 00 Op CO CO 00 00 CO 00 00 00 00 GO 00 00
00 00 00 GO CO
¾ O O O O O O O O O O O O O O O O O O O O O O O O O O O O O O
~ Vl V1 ~ Vl N 7 ---oo ~ V'l l, M N c+ - O -~ ~/1 ~ 00 ~/1 ~O O 00 00 M
O M-- oo ~n co --M~n ~ O~t O o0 00 --~ O~ Q~ O p~ a. l~ ln 7 O~n --O
. . . . . . . . . . . . . . . . . . . . . . . . . .
U 7 N N M N N t~l N N M N N M N N N N M N M fV N N N N N N N N N
CL
O oo v'1 ~D 7 M o0 1~ v^ V' V t~ ~n v'~ N oo O V' t~ O O~ .-= 7 00 ~o vl .-=
~p ~n o0
V O l/1 11p o0 r l- c0 vl n r [l vl
~D C' N 7 7 7 ~ 7 M M M M M M M M M M M N M M M M M M M M
y V O, I- l~ N O, N v1 ~O N V -11- Cl 7 7 N ~p 10 10 N 'Ir N -,T ~O O, ~p 7\p -
IT V'
I-c r r I~q lxl~ oo \4~ r r .p n 1~ ~ ll:~ I*: V: lp r 10 n I*: Ip c r V: r v
a ei o 0 0 0 0 0 0 0 0 0 0 0 0 0 0 o c o O 0 0 0 0 0 0 0 0 0 0 0
\p V1 Vl V N V 7 Ct V V' M 'IT M M M1~3- M M M M M N m Hl M M M M M M
~ O O O O O O O O O O O O O O O O O O O O O O O O O O O O O O
W Ccl Ccl R] Cil Li7 W W W W W[i7 W W Gi] W W W W W Li] W W W W W W W W W
~ 7~ 7 --M 1~ p~ Vl fV ~V O O [~ Vl M 7 Q~ O~ ~/1 (V ~/1 M GO M (V M M 7 ~ --M
7 ~p 7 N N r [-: 'p lp I- O~ O~ N V= =-.= Co O~ lD lp =-= '"D Vl 7 7 M.-= - c0
N
; 7 DO ^ M - N M N t+l - 1- ~ - - 1- ^ ^ ^ ^-~ ^ ^' M ~ M --~ \p V1 --Q
^ G ~ N
e ca
_ `v o`~n L - s y
n a s
8 R X`o a E
CL E r- Q v~ w E
C -
L O T axi p - t p E >`
v a E
0 '~ O V ~~ t T V cia
ej
8
+.+ O n. ~ O oA X u N
,~ n O y -p bA C a) N F N OD 7)
O c U c a y c c c- c _ c L o a y
` y" O O F' N - ` E Y ` V E
C ~'3 U a c a cc o ~ ~~
a s_ N H = N Z tz
c q n n ~ c o r > Z C_
o"
L R U d U ~=C 1 V y p N t3 S y > T Y Y'p O R a ^ e7 cU3
Q U ip U OA C C n d C! ~ G y U a ry c"' .
o v V U p c~s Ov p Z ~ ~ y~ L 0 O Y C ~ X ~ O'O O
? - N ~ O O V Q N
T o p.
0 0 o o~ E o ~~' c
V F' on o =~ c n c L s c L T s c c :: N
c ~ o v p~ ? n_ o v- V a~ c o~ ~ a 8 o x o
Q~ c Mp 0 c o'u ~' c i c"i s Lo 8 8 a ecn .? i~~~ E
L cL a.5 U s n~ 8 n E en F nS n.E .~ V 2 r A on . _ ? ~= n =,
y o
a~ ~ - r Q N
V)~ v o n cL ~_ 0 L~ 2 v a ~ p Q r~ X vi
F p = r¾n ~ O n. m 1~ X v W Q<~ Q C7 x x~ r~ a 2--Qr m cG ¾ p c
d G V cG U a U F~ T ~ F~ a ~ S CN7 aF~ Q a v v V~ S Q V v F-t U S a F
~
14- M a a v,~ ia c`s ca v~~ `a ~n~ `n ca rn~ ce ~n~ ca ie x~ x~ ia v~~ ia x~ a
cs x
O y cn I I I I I I I I I I I c~ I I I I I 1 1 I I I I I I I 1 I a 1
~ O oo t- p\ - QN oo M O V oo ~ N -[~ 00 O -- - - 00 00 .-= 0o N O+ I~ V ~ O
O, N CO M 7 H1 -\p M M 00 f- 00 N
X 7 01 O, f~ V1 V1 00 - O o0 N M M Ql O, 0
~~n O 'V V v^ ~/1 O O - oo M oo O~ M - fV p~ ~D V' ~p O~ V Q~ et O M M ac N vl
Q R o ~ r 'U O V' M GO M 00 V' M ~ O O l~ In N-- I- N M O, Vl [~ I- \p 00 t-
pl
~ O
L O
> - O NN - O - O O O - - 00 - - - O NO NN N N N O O N - N O O N- O NN N O O O
M N
r C N N N N N N N N N M N N N N N N
CA 02680909 2009-09-11
WO 2008/112749 PCT/US2008/056636
0
~
~
~
v
on
~
U
~
U_
~
U
~
=--o o O\ O, 00 00 r ~O ~O V' M M N Y
cc oo co co r r r r-: r r r r r- r r
O O O O O O o o O O O C o O O
V V1 -= V'1 N 'O V Q, Vl 00 v1 M -- --p, Q)
~ ~ O- O --O O -- N>
N N M N 7 N N N N N N N M N N
r~ r r N~O rn v~ v~ -~n N O O O
. . . . . . . . . . . . . . . M M M N N N N N N M N N N N N ~
V' V' V N ~0 ~O N ~O 7 ~O ~O N 7 N'ct
O o O o O o O O O O O O O o O ¾
M M M N N N N M N M N N N N N=V
O O O O O O O O O O O O O o O
W fil W LU Cil W W W W ti7 Cz~ !i] Lcl W W L
O~ 00 r O, O O ~ M O~ O N ~ N O O ~
~D 7^ O N-- O O~ O~n =--M ~O U
7 M V -~ ~ -+ ~ ~D =-- ~D N ^' R M N
~
U
Q
ca
oD p N
O Q N
~ O
8
y 0 7
D
E - U
C y C
T ~ N p
Eo
oD 0
a~ v to O U
_ = O C r U
y v ~
_ vi y U C w
V ~ U U L U U' U
O R O O > R V 2 ~
R.~ R R ~'u U M L a =
'O ~ 'D vOi O a x C ~
L~ af D U 2 d U c~i cd
E8 ~ T G G ~ cR~i cLQ
C c~ e~ o~ o =_ ti ~I
U
oD OD a oq O ~ C E.~
C C C L .. U O ~` O C O
O 7 T 7 R ` G a ~ ~ 'J
x O N pp O O 7 OA
E E 6) O a eD O cU U U c=C
.~ ._. E ~n G.C U cCJ V cv c6 ~ ~ V
M =~
C] N_ F F U
G t N~ C~ ~^ Ca7 a ~z
4 Z_
d v F v C~ J~~ U~ CJ < 4~ cC =-
O "
R cJ R V
U
xl x~ xl xl xl x~ c s ~`a I ~ . , I x~ a s a d
O I I I I I I I I I NI I I I I 'c7
Lo r v x-- n a o0 0 0o r- o o n II c
r ^ M N V ~O O+ O O N v-~ O O~ oo U O
Q V' M 00 01 0 'V 01 ~O G1 0 ~O o0 7 ~ a
7 o Q+ r ~/1 ~/1 V 7 O\ ~/1 V'1 ~/1 1~ N M
~ N N N N N N N N N N N N N N N V) L
CA 02680909 2009-09-11
WO 2008/112749 PCT/US2008/056636
We then examined whether increasing the number of genes in the classifier
improved its predictive value. Using the LOOCV approach and then plotting
sensitivity,
specificity, and overall error rate of each classifier, it was found that the
top gene alone
provided the best predictive value (data not shown). Adding genes to the
classifier in a
linear fashion did not improve its predictive value. Using a cutoff that
biases for high
sensitivity, the LOOCV demonstrated that the expression of the RASGRPI gene
allowed
for a NPV 88.9%, and a PPV of 70.6%, with an overall predictive accuracy of
76.9% (Fig
1 A). In addition, Kaplan Meier analysis showed a significant difference in
median overall
survival of the responders (386 days) and those with progressive disease (68
days) (Fig
I B). Over expression of this single gene therefore predicted response to
tipifarnib in
newly diagnosed AML with a high negative predictive value.
Identification of a Top Scoring Pair classifier
The predictive value of RASGRPI was not improved if additional genes were
added to the classifier using a linear approach. We thus utilized an
alternative gene
selection algorithm to select genes that would improve the predictive value of
RASGRPl
alone. To this end we utilized the Top Scoring Pair (TSP) algorithm to
identify the best
pair of genes that would provide the greatest predictive accuracy. Geman et
al. (2004).
This approach was utilized to exploit the greatest difference in expression
between two
genes and may be useful when aiming to develop a qPCR based diagnostic assay.
The
TSP from the training set was RASGRPI and aprataxin (APTX). RASGRPI and APTX
were over- and under-expressed in responders, respectively. A robust LOOCV
showed
that this top scoring pair (TSP) provided 85.7% NPV and 91.7% PPV in the
training set
of samples with an overall error rate of only 8% (Fig 2A). The difference in
overall
survival between predicted responders and non-responders was 357 days (Fig
2B). These
data demonstrate that the model-building algorithm has a low associated
prediction error
rate.
Validation of the RASGRPI:APTX classifier in an independent set of relapsed or
refractory AML.
We next performed external validation of the TSP classifier in an independent
microarray dataset comprising of 54 relapsed/refractory AML patient samples.
20070048782. Importantly, a diagnostic assay that aims to predict response to
a cancer
23
CA 02680909 2009-09-11
WO 2008/112749 PCT/US2008/056636
therapy should have a high sensitivity (and negative predictive value) since
it is important
to capture as many potential responders as possible. Therefore, to define an
appropriate
cutoff for testing the TSP classifier we considered the need to obtain a high
sensitivity of
predicting responders while maintaining an acceptable level of specificity. In
the training
set, the level of specificity that could be achieved ranged from approximately
30% to
100% when the sensitivity was set at 100% to 80%, respectively. To ensure the
classifier
would predict as many responders as possible we tested a conservative cutoff
that
provided a specificity of approximately 60% in the training set. When this
cutoff was
applied to the independent testing set of relapsed/refractory AML, the
RASGRPI:APTX
gene classifier stratified responders with 92% NPV and 27.6% PPV (compared to
18.5%
prevalence) (Fig 3C). The associated odds ratio for being a responder was
4.38. While
this was similar to the predictive accuracy of RASGRPI alone, the application
of the TSP
classifier demonstrated a better NPV and an improved difference in overall
survival of 98
days between predicted responders and progressors (Fig 3D), compared to only
56 days
for RASGRPI (Fig 4).
QPCR validation of the RASGRPI:APTX expression ratio
A two-gene expression ratio allows the use of a more clinically relevant qPCR
detection system. Thirty samples (20 PD, 6 CR, 3 HI and I PR) provided enough
total
RNA for qPCR. Therefore, the RASGRPI:APTX gene expression ratio was evaluated
as a
predictor of response to tipifarnib using TaqMan qPCR in these 30 samples (10
responders, 20 progressive disease) from the newly diagnosed AML clinical
study. Nine
of these samples had been assayed on the microarray platform, however 21 had
not been
utilized in the discovery set due to poor quality RNA. Therefore, two thirds
of this test set
was comprised of completely independent samples. Evaluation of the 9 samples
indicated
there was good correlation (r = 0.74) of the RASGRPI:APTX expression ratio
between
the two platforms (Fig 6). Using a cut-point of 0, the two-gene classifier
correctly
predicted the treatment outcome in 20 of the 30 patients with PPV and NPV of
50% and
81%, respectively (Fig 3). The median overall survival of the predicted
resistant patients
was 82 days while those classified as responders had a median value of 295
days (Fig
3C).
24
CA 02680909 2009-09-11
WO 2008/112749 PCT/US2008/056636
The RASGRPI:APTX classifier does not have prognostic utility independent of
FTI
treatment.
We tested the two-gene expression ratio in an independent microarray dataset
of
116 AML patients treated with chemotherapeutic regimes. Bullinger et al.
(2004). When
the RASGRPI:APTX classifier was applied to this set of patients, utilizing a
similar cut-
off as for the tipifarnib-treated population, no significant separation in
overall survival
was seen (Fig 5). Nor were significant survival differences observed when a
range of
other cut-offs was utilized (Table 4). This indicated that the RASGRPI:APTX
classifier
specifically stratifies patients who have been treated with tipifarnib and is
not relevant to
non-FTIs. On the other hand when the prognostic signature defined by Bullinger
et al.
was applied to our set of relapsed and refractory AML patients there was a
clear
stratification in terms of overall survival.
Table 4.
cutoff p value Responders median Progressors median OS No. Responders No.
Progressors
OS
0.5 0.956 336 414 13 103
0.6 0.342 672 374 24 92
0.7 0.266 511 335 34 82
0.8 0.269 511 326 47 69
0.9 0.101 540 316 . 57 59
I 0.215 483' 326 64 52
2 0.795 374 570 94 22
3 0.209 346 909 104 12
OS = overall survival
Kaplan-Meier analysis is shown in Supplementary Fig. 2 for highlighted (bold)
cut-off.
DISCUSSION
Stratification of patient populations to predict therapeutic response is
becoming
increasingly valuable in the clinical management of cancer patients. For
example,
companion diagnostics are required for the stratification of patients being
treated with
targeted therapies such as trastuzumab (Herceptin, Genentech) in metastatic
breast
cancer, and cetuximab (Erbitux, Merck) in colorectal cancer. Seidman et al.
(2001); and
Moroni et al. (2005). Predictive biomarkers are also being utilized for
imatinib (Gleevec,
Novartis) in gastrointestinal stromal tumors, and for erlotinib (Tarceva, OSI
Pharmaceuticals) and gefitinib (Iressa, Astra-Zeneca) in lung cancer. Burger
et al.
(2005); Tsao et al. (2005); and Lynch et al. (2004). Currently there is no
method
available to predict response to an FTI in any indication. To identify genes
that are
CA 02680909 2009-09-11
WO 2008/112749 PCT/US2008/056636
associated with greater sensitivity to the FTI, tipifarnib, we performed gene
expression
profiling of leukemic bone marrow samples from a phase 2 study in elderly
patients with
previously untreated poor-risk AML. Lancet et al. (2006). Importantly, an
assay that
aims to predict response to an oncology therapy should have a high NPV since
it is
important to capture as many potential responders as possible. Therefore,
using criteria
to identify markers that predict response with high sensitivity, we identified
45 genes in
the newly diagnosed AML bone marrow samples that were differentially expressed
between responders and non-responders.
While we found no significant correlation with N-Ras mutations or baseline
phosphorylation status of ERK or AKT and response to tipifarnib (Lancet et al.
(2006)),
we did identify genes predictive of response to tipifarnib that are involved
in Ras
activation including, PTPN6 (a protein tyrosine phosphatase that is
farnesylated and was
down-regulated in responders), CD3D, TRATI, LTB, TNFRSFI7, TNFSF13, and
RASGRPI. Chen et al. (2005); Stone (2006); and Delgado (2000). It is well
known that
activation of the Ras pathway can be caused by other events outside of
constitutive
activation of the Ras protein itself. Illmer et al. (2005); and Solit et al.
(2006). Indeed,
N-Ras and K-Ras have been identified in their activated state in AML in the
absence of
activating mutations. Ehmann et al. (2006). It is therefore plausible that Ras
deregulation is an important target of tipifarnib in AML regardless of Ras
mutational
status. Watters et al. (2006). In support of this, Feldkamp et al. (2001)
demonstrated that
isotype-specific Ras.GTP levels correlates with response to the FTI SCH66336
regardless
of Ras activating mutations.
RASGRPI was the most robust single predictive gene expression marker with an
overall predictive accuracy of 77% in the cross-validated training set.
RASGRPI is a
guanine nucleotide exchange factor (GEF) that specifically activates Ras.
Stone (2006).
Expression of RASGRPI has been found in brain, T-cells, cells of monocytic
lineage,
and primitive hematopoietic precursors. Kawasaki et al. (1998) Ebinu et al.
(1998); and
Tognon et al. (1998). Interestingly, another RASGRP (RASGRP4) was previously
identified as a potential oncogene in AML (Reuther et al. (2002)), however,
our data is
the first to examine and demonstrate expression of RASGRPI in AML cells in
addition to
implicating it's importance in response to FTIs.
26
CA 02680909 2009-09-11
WO 2008/112749 PCT/US2008/056636
We found that the combination of RASGRPl and APTX provided the most robust
predictive accuracy (approximately 89%) for a multi-gene classifier. APTX is
involved in
DNA excision repair and was found to be down-regulated in responders. Ahel et
al.
(2006). This two-gene classifier showed predictive utility in the discovery
set of newly
diagnosed AML when a cross validation was performed, with a NPV and PPV of
identifying responders of 86% and 92%, respectively. However, cross-validation
only
provides a model of performance and thus testing of an independent data set
was
performed to provide bona fide accuracy of response prediction. To this end we
also
examined microarray data from an independent set of relapsed/refractory AML
and
performed qPCR on independent samples from the newly diagnosed AML clinical
study.
A simple qPCR-based diagnostic assay has wider utility in the clinic than gene
expression microarrays due to the ability to assay poor quality clinical
samples that may
not be profiled by current microarray technologies. In a set of 30 samples (20
of which
had not been profiled by microarray) from the newly diagnosed AML population,
we
demonstrated that the RASGRPI:APTX expression ratio can be reliably detected
with
qPCR regardless of sample quality. The classifier demonstrated a NPV and PPV
of 81%
and 50%, respectively and provided a clear overall survival advantage for
those patients
predicted to be responders. Clearly, it will be important to profile larger
datasets in
future studies to further validate the use of a two-gene qPCR assay.
In the absence of a larger independent set of newly diagnosed AML samples we
also utilized 54 relapsed or refractory AML samples from our previous
investigation as
an independent testing set (20070048782). Surprisingly, even though the
samples were
from a biologically distinct population of AML patients the two-gene
classifier showed
good stratification of responders and non-responders with a NPV of 92% and a
PPV of
28%. Since the prevalence of responders in that dataset was 18% this
represents an
improvement of overall response of approximately 50%. Furthermore, the
stratified
predicted responders had a median overall survival that was approximately 3-
fold longer
than patients predicted to be resistant to tipifarnib. Importantly we found no
association
with the two-gene classifier and patient prognosis in an independent set of
AML patients
who were treated with chemotherapeutics. This indicated that the current
classifier
27
CA 02680909 2009-09-11
WO 2008/112749 PCT/US2008/056636
specifically predicts response to tipifarnib treatment. Further work needs to
be done to
clarify whether the RASGRPI:APTX expression ratio'has utility for other
classes of FTIs.
How might increased RASGRPI expression lead to sensitivity to FTIs?
RASGRPI has been shown to activate H-RAS and N-Ras, but not K-Ras, exclusively
on
the golgi apparatus. Bivona et al. (2003); and Perez de Castro et al. (2004).
Further, K-
Ras and N-Ras can be alternatively geranylgeranylated following
farnesyltransferase
inhibition. Whyte et al. (1997). H-Ras on the other hand is only farnesylated
and this
may explain the observation that tumors transformed with H-Ras are more
sensitive than
those transformed with N-, or K-Ras. End et al. (2001); and Lubet et al.
(2006). Thus it
is possible that aberrant expression of RASGRPI in AML leads to activation of
the N-
Ras and H-Ras pathways but it is the blockage of H-Ras that is causing the
anti-
tumorigenic effect. Therefore, while H-Ras activating mutations have not been
identified
in AML, the specific activation of H-Ras pathways by other means (such as Ras-
specific
GEFs) may still be a target of FTIs in certain tumors.
We previously identified AKAP13 as being predictive of resistance to
tipifarnib
in relapsed/refractory AML. 200700448782. Interestingly, AKAP13 is also a GEF,
but
activates the Rho pathway. Sterpetti et al. (1999). However, whilst showing
utility in
relapsed or refractory AML, expression of AKAP13 did not demonstrate
predictive utility
in newly diagnosed AML. This may be because the population of leukemic cells
that
over-express AKAP13 is absent in newly diagnosed disease and only proliferates
in late
stage AML. The other question that arises is why over-expression of the
RASGRPI GEF
increases sensitivity while over-expression of the AKAP13 GEF increases
resistance to
tipifamib? Rho GEFs have been found to drive cellular transformation in a Ras-
independent fashion. Reuther et al. (2001); and Sahai et al. (2002). Thus, one
hypothesis
is that AKAP13 activates a downstream compensatory pathway in RhoA while
RAGRPI
activates Ras, a clear target of FTIs. More biochemical analyses will need to
be done to
investigate this model. Nevertheless, the identification of two GEFs playing
opposing
roles in responsiveness to an FTI does highlight the importance of this class
of small
GTPase activators in FTI-mediated therapy. It also highlights the need for
multiple
markers in predicting response to targeted therapies across a wide range of
diseases and
disease subtypes. As GEFs are increasingly becoming attractive drug targets it
may also
28
CA 02680909 2009-09-11
WO 2008/112749 PCT/US2008/056636
be of interest to investigate combination therapies of FTIs and inhibitors of
specific
GEFs.
In summary, we have identified and validated a two-gene expression ratio that
can
be assayed using simple qPCR. The classifier has predictive utility in both
newly
diagnosed and relapsed or refractory AML, and improves the overall response
rate by
approximately 50% while maintaining a high NPV. In addition, stratification
with this
classifier significantly improves patient overall survival. Our data compare
favorably to
the use of FDA-approved companion diagnostics for targeted cancer therapies
such as
Herceptin. For instance, it has been demonstrated that stratification of
metastatic breast
cancer patients with over-expression of Her2/Neu improves the overall response
to
Herceptin and paclitaxel combination therapy from approximately 59% to 69% or
75%
when using the HercepTest or PathVysion tests, respectively. Seidman et al.
(2001). Our
data therefore indicates that a simple two-gene expression assay may have
utility in
diagnosing a population of AML patients who are more likely to respond to
tipifarnib.
29
CA 02680909 2009-09-11
WO 2008/112749 PCT/US2008/056636
References
Ahel et al. (2006) The neurodegenerative disease protein aprataxin resolves
abortive
DNA ligation intermediates Nature 443:713-716
Bivona et al. (2003) Phospholipase Cgamma activates Ras on the Golgi apparatus
by
means of RasGRPI Nature 424:694-698
Bos (1989) ras oncogenes in human cancer: a review Cancer Res 49:4682-4689
Bullinger et al. (2004) Use of gene-expression profiling to identify
prognostic subclasses
in adult acute myeloid leukemia N Engl J Med 350:1605-1616
Burger et al. (2005) Activating mutations in c-KIT and PDGFRalpha are
exclusively
found in gastrointestinal stromal tumors and not in other tumors
overexpressing these
imatinib mesylate target genes Cancer Biol Ther 4:1270-1274
Chang et al. (2003) Gene expression profiling for the prediction of
therapeutic response
to docetaxel in patients with breast cancer Lancet 362:362-369
Chen et al. (2005) FLT3/ITD mutation signaling includes suppression of SHP-1 J
Biol
Chem 280:5361-5369
Cox et al (2002) Farnesyltransferase inhibitors: promises and realities Curr
Opin
Pharmacol 2:388-393
Ebinu et al. (1998) RasGRP, a Ras guanyl nucleotide- releasing protein with
calcium-
and diacylglycerol-binding motifs Science 280:1082-1086
Ehmann et al. (2006) Detection of N-RAS and K-RAS in their active GTP-bound
form in
acute myeloid leukemia without activating RAS mutations Leuk Lymphoma 47: l
387-
1391
End et al. (2001) Characterization of the antitumor effects of the selective
farnesyl
protein transferase inhibitor Rl 15777 in vivo and in vitro Cancer Res 61:131-
137
Feldkamp et al. (2001) Isotype-specific RasGTP-levels predict the efficacy of
farnesyl
transferase inhibitors against human astrocytomas regardless of Ras mutational
status
Cancer Res 61:4425-4431
Geman et al. (2004) Classifying gene expression profiles from pairwise mRNA
comparisons Stat Appl Genet Mol Biol 3:30
Holleman et al. (2004) Gene-expression patterns in drug-resistant acute
lymphoblastic
leukemia cells and response to treatment N Engl J Med 351:533-542
Illmer et al. (2005) Activation of the RAS pathway is predictive for a
chemosensitive
phenotype of acute myelogenous leukemia blasts Clin Cancer Res 11:3217-322
Jansen et al. (2005) Molecular classification of tamoxifen-resistant breast
carcinomas by
gene expression profiling J Clin Oncol 23:732-740
Karp et al. (2001) Clinical and biologic activity of the farnesyltransferase
inhibitor
R115777 in adults with refractory and relapsed acute leukemias: a phase 1
clinical-
laboratory correlative trial Blood 97:3361-3369
Kawasaki et al. (1998) A Rap guanine nucleotide exchange factor enriched
highly in the
basal ganglia Proc Natl Acad Sci 95:13278-13283
Lancet et al. (2006) A phase lI study of the farnesyltransferase inhibitor
tipifarnib in
poor-risk and elderly patients with previously untreated acute myelogenous
leukemia
Blood 2:2
Lossos et al. (2004) Prediction of survival in diffuse large-B-cell lymphoma
based on the
expression of six genes N Engl J Med 350:1828-1837
CA 02680909 2009-09-11
WO 2008/112749 PCT/US2008/056636
Lubet et al. (2006) Effects of the farnesyl transferase inhibitor R115777
(Zarnestra) on
mammary carcinogenesis: prevention, therapy, and role of HaRas mutations Mol
Cancer Ther 5:1073-1078
Lynch et al. (2004) Activating mutations in the epidermal growth factor
receptor
underlying responsiveness of non-small-cell lung cancer to gefitinib N Engi J
Med
350:2129-2139
Ma et al. (2004) A two-gene expression ratio predicts clinical outcome in
breast cancer
patients treated with tamoxifen Cancer Cell 5:607-616
Mesa et al (2006) Tipifarnib: farnesyl transferase inhibition at a crossroads
Expert Rev
Anticancer Ther 6:313-319
Moroni et al. (2005) Gene copy number for epidermal growth factor receptor
(EGFR) and
clinical response to antiEGFR treatment in colorectal cancer: a cohort study
Lancet
Oncol 6:279-286
Perez de Castro et al. (2004) A Ras activation in Jurkat T cells following low-
grade
stimulation of the T-cell receptor is specific to N-Ras and occurs only on the
Golgi
apparatus Mol Cell Biol 24:3485-3496
Potti et al. (2006) Genomic signatures to guide the use of chemotherapeutics
Nat Med
12:1294-1300
Rao et al. (2004) Phase Ill Double-Blind Placebo-Controlled Study of Farnesyl
Transferase Inhibitor R115777 in Patients With Refractory Advanced Colorectal
Cancer J Clin Oncol 22:3950-3957
Reuter et al. (2000) Targeting the Ras signaling pathway: a rational,
mechanism-based
treatment for hematologic malignancies? Blood 96:1655-1669
Reuther et al. (2001) Leukemia-associated Rho guanine nucleotide exchange
factor, a
Dbl family protein found mutated in leukemia, causes transformation by
activation of
RhoA J Biol Chem 276:27145-27151
Reuther et al. (2002) RasGRP4 is a novel Ras activator isolated from acute
myeloid
leukemia J Biol Chem 277:30508-30514
Rosenwald et al. (2002) The use of molecular profiling to predict survival
after
chemotherapy for diffuse large-B-cell lymphoma N Engl J Med 346:1937-1947
Rowinsky et al (1999) Ras protein farnesyltransferase: A strategic target for
anticancer
therapeutic development J Clin Oncol 17:3631-3652
Sahai et al. (2002) RHO-GTPases and cancer Nat Rev Cancer 2:133-142
Seidman et al. (2001) Weekly trastuzumab and paclitaxel therapy for metastatic
breast
cancer with analysis of efficacy by HER2 immunophenotype and gene
amplification J
Clin Oncol 19:2587-2595
Ship et al. (2002) Diffuse large B-cell lymphoma outcome prediction by gene-
expression
profiling and supervised machine leaming Nat Med 8:68-74
Solit et al. (2006) BRAF mutation predicts sensitivity to MEK inhibition
Nature 439:358-
362
Sterpetti et al. (1999) Activation of the Lbc Rho exchange factor proto-
oncogene by
truncation of an extended C terminus that regulates transformation and
targeting Mol
Cell Biol 19:1334-1345
Stone (2006) Regulation of Ras in lymphocytes: get a GRP Biochem Soc Trans
34:858-
861
Tognon et al. (1998) Regulation of RasGRP via a phorbol ester-responsive C 1
domain
31
CA 02680909 2009-09-11
WO 2008/112749 PCT/US2008/056636
Mo1 Cell Biol 18:6995-7008
Tsao et al. (2005) Erlotinib in lung cancer - molecular and clinical
predictors of outcome
N Engl J Med 353:133-144
Van Cutsem et al. (2004) Phase III trial of gemcitabine plus tipifarnib
compared with
gemcitabine plus placebo in advanced pancreatic cancer J Clin Oncol 22:1430-
1438
Waters et al. (2006) Developing gene expression signatures of pathway
deregulation in
tumors Mol Cancer Ther 5:2444-2449
Weinstein et al. (2006) Mechanisms of disease: Oncogene addiction--a rationale
for
molecular targeting in cancer therapy Nat Clin Pract Oncol 3:448-457
White et al. (1997) K- and N-Ras are geranylgeranylated in cells treated with
farnesyl
protein transferase inhibitors J Biol Chem 272:14459-14464
Yeoh et al. (2002) Classification, subtype discovery, and prediction of
outcome in
pediatric acute lymphoblastic leukemia by gene expression profiling Cancer
Cell
1:133-143
32
CA 02680909 2009-09-11
WO 2008/112749 PCT/US2008/056636
Sequences
Probe Set ID
205590 at
ggattcaaaaggtgtcacagtccacttaattagtcaaattagcaatggctaaacagtatcaagtactgcagaatttatc
actgaaatg
gataagaggaaatagtttagtcacaggtttttacagtccagcaagggccaaagaggtatagtatacaagttaatagtat
ttgtgttga
gcaacatggggctagtgggatcacagaaatctggaaaaaaaaaaaaaaaggctttggcttatcaagcctagtgtaaatt
tctgcat
ctcacacgactttagtttggccaggtatttatctgccaaaacaaggacaaatcttgttgtattaacagcagggtcactt
ctcattttcttt
gctgacttacctttttactgaccgttgtgaatttctgtctcaaa
217028_at
attgatgtgtgtctaggcaggacctgtggccaagttcttagttgctgtatgtctcgtggtaggactgtagaaaagggaa
ctgaacat
tccagagcgtgtagtgaatcacgtaaagctagaaatgatccccagctgtttatgcatagataatctctccattcccgtg
gaacgtttt
tcctgttcttaagacgtgattttgctgtagaagatggcacttataaccaaagcccaaagtggtatagaaatgctggttt
ttcagttttca
ggagtgggttgatttcagcacc
206687_s_at
gggcctggactgtgacattgacatccagaagaccatccagatggtgcgggcgcagcgctcgggcatggtgcagacggag
gc
gcagtacaagttcatctacgtggccatcgcccagttcattgaaaccactaagaagaagctggaggtcctgcagtc
gcagaaggg
ccaggagtcggagtacgggaacatcacctatcccccagccatgaagaatgcccatgccaaggcctcccgcacctcgtcc
aaa
cacaaggaggatgtgtatgagaacctgcacactaagaacaagagggaggagaaagtgaagaagcagcggtcagcagaca
a
ggagaagagcaagggttccctcaagaggaagtgagcggtgctgtcctcaggtggccatgcctcagccctgaccctgtgg
aag
catttcgcgatggacagactcacaacctgaacctaggagtgcccc
2I0439_at
gcttctgaagcagccaatgtcgatgcaacaacatttgtaactttaggtaaactgggattatgttgtagtttaacatttt
gtaactgtgtg
cttatagtttacaagtgagacccgatatgtcattatgcatacttatattatcttaagcatgtgtaatgctggatgtgta
cagtacagtact
taacttgtaatttgaatctagtatggtgttctgttttcagctgacttggacaacctgactggctttgcacaggtgttcc
ctgagttgtttg
caggtttctgtgtgtggggtggggtatggggaggagaaccttcatggtggcccacctggcctggttgtccaagctgtgc
ctcgac
acatcctcatcccaagcatgggacacctcaagatgaataataattcacaaaatttctgtgaaatcaaatccagttttaa
gaggagcc
acttatcaaagagat
206641_at
atttctttggcagttttcgtgctaatgtttttgctaaggaagataagctctgaaccattaaaggacgagtttaaaaaca
caggatcagg
tctcctgggcatggctaacattgacctggaaaagagcaggactggtgatgaaattattcttccgagaggcctcgagtac
acggtg
gaagaatgcacctgtgaagactgcatcaagagcaaaccgaaggtcgactctgaccattgctttccactcccagctatgg
aggaa
ggcgcaaccattcttgtcaccacgaaaacgaatgactattgcaagagcctgccagctgctttgagtgctacggagatag
agaaat
caatttctgctaggtaattaaccatttcgactcgagcagtgccactttaaaaatcttttgtcagaatagatgatgtgtc
agatctctttag
gatgactgtatttttcagttgccgatacagctttttgtcctctaactg
213539_at
gggaacactgctctcagacattacaagactggacctgggaaaacgcatcctggacccacgaggaatatataggtgtaat
ggga
cagatatatacaaggacaaagaatctaccgtgcaagttcattatcgaatgtgccagagctgtgtggagctggatccagc
caccgt
ggctggcatcattgtcactgatgtcattgccactctgctccttgctttgggagtcttctgctttgctggacatgagact
ggaaggctgt
ctggggctgccgacacacaagctctgttgaggaatgaccaggtctatcagcccctccgagatcgagatgatgctcagta
cagcc
accttggaggaaactgggctcggaacaagtgaacctgagactggtggcttctagaagcagccattaccaactgtacct
208018_s_at
gctgatggagatcgtcacctacggccggatcccttacccagggatgtcaaaccctgaagtgatccgagctctggagcgt
ggata
ccggatgcctcgcccagagaactgcccagaggagctctacaacatcatgatgcgctgctggaaaaaccgtccggaggag
cgg
ccgaccttcgaatacatccagagtgtgctggatgacttctacacggccacagagagccagtaccaacagcagccatgat
aggg
aggaccagggcagggcagggggtgcccaggtggtggctcgaaggtggctccagcaccatccgccagggcccacaccccc
t
tcctactcccagacacccaccctcgcttcagccacagtttcctcatctgtccagtgggtaggttggactggaaaatctc
tttttgact
cttgcaatccacaatctgacattctcaggaagcccccaagttgatatttctatt
203063_at
33
CA 02680909 2009-09-11
WO 2008/112749 PCT/US2008/056636
ctcagaatcccagaggcagtcccagcctcagaacccaggataggaaatgggtgtgtttagtggggaaagggacggggtg
cag
acggcagggccagtatggggccccctccctctcctctcctctcctatggtgagcccagcgtgggcaccgggccgtctca
gccg
tgttcccagggctgggaggacagctctggcccttcttaggcctagcctcgtcccaagctaaatgtaagccagttgggct
gtgttaa
aggaagcagtgtttttggtttgattctgcctctgtagctcaaggggggcagcccccagagtcctgtgcattctgccaag
gctccat
agctttgccaaatgcacggagctctgccattccggtgcagtgcaggccttgcgaagggtttatctgcgttcgtctcggt
gggcttct
cctgcatgggagttgtgttcctgtgcaagggggagctttgctcaggacaggatgactgtcttccctattcttagggaca
agtccca
agatgccagaaaggcagtctcccaagga
208130_s_at
tatggccaggcctttgactgtggatgagattgtgggccaggccttcatcttcctcatcgctggctatgaaatcatcacc
aacacact
ttcttttgccacctacctactggccaccaaccctgactgccaagagaagcttctgagagaggtagacgtttttaaggag
aaacaca
tggcccctgagttctgcagcctcgaggaaggcctgccctatctggacatggtgattgcagagacgctgaggatgtaccc
gccag
ctttcagattcacacgggaggcagctcaggactgcgaggtgctggggcagcgcatccccgcaggcgctgtgctagagat
ggc
cgtgggtgccctgcaccatgaccctgagcactggccaagcccggagaccttcaaccctgaaaggtaccgctgcagctag
aatc
caaatctgccctaggtccaaaaaatggtgtctatatcaagatcgtatcccgctgacacagaaggctgccgg
216834at
atgaaactgattacaacaggctgtaagaatcaaagtcaactgacatctatgctacatattattatatagtttgtactga
gctattgaagt
cccattaacttaaagtatatgttttcaaattgccattgctactattgcttgtcggtgtattttattttattgtttttga
ctttggaagagatgaa
ctgtgtatttaacttaagctattgctcttaaaaccagggatcagaatatatttgtaagttaaatcattggtgctaataa
taaatgtggattt
tgtattaaaatatatagaagcaatttctgtttacatgtccttgctacttttaaaaacttgcatttattcctcagatttt
213388_at
cctctttctcaatctataacctttgtaggcatgcatttataccagcatgttttataaattatgagtttctatctgtgtc
catgaagtcttacta
gttttcacttaaacttttgtgggttgttaagaagaattaaagtgattcataacttcacgcttgaacctgggaggtggag
gttgcagtga
gccgagatcatgccattgcactccagcctgggcaacaagagtgaaactctgtcttaaataaataaataaagtggttcat
aacatca
gatgaagaaggaggtgagtgatatgttaaatgatcagaaacttggcattacattatttccaggaccatttccctaccaa
agctgtgt
atttttcatttcttcatggcactgtgctgttaatttctgtta
38487_at
acagttgtggttagccgtatcattgtgtgggacatcatggccttcaatggcatcatccatgctctggccagccccctcc
tggcaccc
ccacagcccnagncagtgntggcgcctgaagccccacctgtggcggcaggcgnnnnnnnnnnnnnnnnnnnnnnnnc
actgcttggcttggtggccggagctctctacctccgtgcccgaggcaagcccangggctttggcttctctgccttccag
gcggaa
nnnnnnnnnnnnnnnnnnnnnnnnnnnnngcaagaagggaccaaccccaccctggtctctgtccccaaccctgtctttg
gcagcgacaccttttgtgaacccttcgatgactcactgctggaggaggacttccctgacacccagaggatcctcacagt
caagt
210982_s_at
gaaggagacggtctggcggcttgaagaatttggacgatttgccagctttgaggctcaaggtgcattggccaacatagct
gtgga
caaagccaacttggaaatcatgacaaagcgctccaactatactccgatcaccaatgacaagttcaccccaccagtggtc
aatgtc
acgtggcttcgaaatggaaaacctgtcaccacaggagtgtcagagacagtcttcctgcccagggaagaccaccttttcc
gcaag
ttccactatctccccttcctgccctcaactgaggacgtttacgactgcagggtggagcactggggcttggatgagcctc
ttctcaa
gcactgggagtttgatgctccaagccctctcccagagactacagagaacgtggtgtgtgccctgggcctgactgtgggt
ctggt
gggcatcattattgggaccatc
210321at
gccaagtggaccacagctgtgcggcctctcaggctacctagcagcaaggcccaggtgaagccagggcagctgtgcagtg
tg
gctggctggggttatgtctcaatgagcactttagcaaccacactgcaggaagtgttgctgacagtgcagaaggactgcc
agtgtg
aacgtctcttccatggcaattacagcagagccactgagatttgtgtgggggatccaaagaagacacagaccggtttcaa
ggggg
actccggggggcccctcgtgtgtaaggacgtagcccaaggtattctctcctatggaaataaaaaagggacacctccagg
agtct
acatcaaggtctcacacttcctgccctggataaagagaacaatgaagcgcctctaacagcaggcatgagactaaccttc
ctctgg
gcctgaccatctctgggacagaggcaagaatccccaagggtg
217147_s_at
tctcctttctcaccaatgggcaatagcccataattgaaataaatttctgattgaaaggtataggaaacattaaaatgca
ttactaagag
aagtaatataattttcttacaaagtatttttcccaaagatagctttactatttcaaaaattgtcaaattaatgcatgct
ccttacaacaaac
34
CA 02680909 2009-09-11
WO 2008/112749 PCT/US2008/056636
aaatatcaaaaagagtttaggaattctactagccagagatagtcacttggagaaactttctatatatccttctaaatat
ttttctgggca
tgctcatgtatgtacatcagttgtttctttttattttgaaccaaaaatgtggtttcttttgtacacattacttaaactt
tctttccagtcaacaa
tatattgtggatttattttcactgttatatttaactatatataaatacgcatatattgtaattttaatgtctgcttagc
accccactgataacc
aaatcacag
206298_at
aagctgcggaactctgaacgggcgcgggaggatgcggagaggaggaaccagctgttgcagagggaaatggaggagtttt
ttt
cgaccctaggaagcttgactgttggggcaaaaggtgccagggccccaaagtaaaaggaatggcagagctcacttctgta
ccac
gtctgctggtctccagccttgtatggagttagaagcgtctgtatctctggagcagccaggcgctctggagccagctgga
gagag
agagatcctgatacctctgtggggactgtggggacttttgggaccccacacactccaggtgggatcagatgctgctcca
accatg
cagttcctggtgagggtcagaaggggacggtaccaagagcagcgcttagcccttacccaggaaatatccttcatggcca
caga
aatggagggcgcccaggatccaggcagccaccgggaacagtcagctttcttta
202990 at
agagcagatttccactgcaggcaccgaagcctcggggacaggcaatatgaagttcatgctaaatggggccctaactatc
ggga
ccatggatggggccaatgtggaaatggcagaagaagctggggaagagaacctgttcatctttggcatgaggatagatga
tgtg
gctgctttggacaagaaagggtacgaggcaaaagaatactatgaggcacttccagagctgaagctggtcattgatcaaa
ttgac
aatggctttttttctcccaagcagcctgacctcttcaaagatatcatcaacatgctattttatcatgacaggtttaaag
tctttgcagact
acgaagcctatgtcaagtgtcaagataaagtgagtcagctgtacatgaatccaaaggcctggaacacaatggtactcaa
aaacat
agctgcctcggggaaattctccagtgaccgaacaattaaagaatatgcccaaaacatctggaacgtggaaccttcagat
ctaaa
221671_x_at
caacaccgtgacaattggcctccgggggccactttcggcggagggaccaaggtggagatcaaacgaactgtggctgcac
cat
ctgtcttcatcttcccgccatctgatgagcagttgaaatctggaactgcctctgttgtgtgcctgctgaataacttcta
tcccagagag
gccaaagtacagtggaaggtggataacgccctccaatcgggtaactcccaggagagtgtcacagagcaggacagcaagg
ac
agcacctacagcctcagcagcaccctgacgctgagcaaagcagactacgagaaacacaaagtctacgcctgcgaagtca
ccc
atcagggcctgagctcgcccgtcacaaagagcttcaacaggggagagtgttagagggagaagtgcccccacctgctcct
cagt
tccagcctgaccccctcccatcctttggcctctgaccctttttccacaggggacctacccctattgcggtcctccagct
catctttca
cctcacccccctcctcctccttggctttaattatgc
221651x at
gttatcctgtcacttttggccaggggaccaagctggagatcaaacgaactgtggctgcaccatctgtcttcatcttccc
gccatctg
atgagcagttgaaatctggaactgcctctgttgtgtgcctgctgaataacttctatcccagagaggccaaagtacagtg
gaaggtg
gataacgccctccaatcgggtaactcccaggagagtgtcacagagcaggacagcaaggacagcacctacagcctcagca
gc
accctgacgctgagcaaagcagactacgagaaacacaaagtctacgcctgcgaagtcacccatcagggcctgagctcgc
ccg
tcacaaagagcttcaacaggggagagtgttagagggagaagtgcccccacctgctcctcagttccagcctgaccccctc
ccatc
ctttggcctctgaccctttttccacaggggacctacccctattgcggtcctccagctcatctttcacctcacccccctc
ctcctccttg
gctttaattatgc
207651_at
ttgccttgtaattcgacagctctacagaaacaaagataatgaaaattacccaaatgtgaaaaaggctctcatcaacata
cttttagtg
accacgggctacatcatatgctttgttccttaccacattgtccgaatcccgtataccctcagccagacagaagtcataa
ctgattgct
caaccaggatttcactcttcaaagccaaagaggctacactgctcctggctgtgtcgaacctgtgctttgatcctatcct
gtactatca
cctctcaaaagcattccgctcaaaggtcactgagacttttgcctcacctaaagagaccaaggctcagaaagaaaaatta
agatgt
gaaaataatgcataaaagacaggattttttgtgctaccaattctggccttactgga
202988sat
gtgaacagcttggccttttttgggtgtcttgacaggccaagaagaacaaatgactcagaaccggattaacatgaaagtt
atccagg
cgcagagttgaagaagcataagcaagcaagacaaaaacagagagaccgcaaggaggaagatctgtggtactgtcataaa
aaa
cagtggagctctgtattagaaaagcccctcagaactgggaaggccaggtaactctagttacacagaaactggtactaaa
gtctat
caaactgattacacagactgtaagaattcaaagtcaactgacatctatgctacatatattatatagtttgtacttgact
atgagccatta
acttaaagcatatgtttcaaatagccattgctactattccttgtccggtgtaattttattttattgtttttactttgga
agagatgaactgtgt
atttaacttaagctattgctcttaaaaccaggg
213418_at
CA 02680909 2009-09-11
WO 2008/112749 PCT/US2008/056636
ggttcatgaagccgagcagtacaaggctgaggatgaggcccagagggacagagtggctgccaaaaactcgctggaggcc
ca
tgtcttccatgtgaaaggttctttgcaagaggaaagccttagggacaagattcccgaagaggacaggcgcaaaatgcaa
gacaa
gtgtcgggaagtccttgcctggctggagcacaaccagctggcagagaaggaggagtatgagcatcagaagagggagctg
ga
gcaaatctgtcgccccatcttctccaggctctatggggggcctggtgtccctgggggcagcagttgtngcnctcaagcc
cncca
gggggaccccagcaccggccccatcattgaggaggttgattgaatggcccttcgtgataagtcagctgtgactgtcagg
gctat
gctatgggccttctagactgtcttctatgatcctgcccttcagagatga
209901_x_at
ccagcatctgctgagctatgagccaaaccagggatttacagggaggaaaagctttcggactgctgaaggcccagcagga
aga
gaggctggatgagatcaacaagcaattcctagacgatcccaaatatagcagtgatgaggatctgccctccaaactggaa
ggctt
caaagagaaatacatggagtttgaccttaatggaaatggcgatattgatatcatgtccctgaaacgaatgctggagaaa
cttggag
tccccaagactcacctagagctaaagaaattaattggagaggtgtccagtggctccggggagacgttcagctaccctga
ctttct
caggatgatgctgggcaagagatctgccatcctaaaaatgatcctgatgtatgaggaaaaagcgagagaaaaggaaaag
ccaa
caggccccccagccaagaaagctatctct
205488 at
cagccacacgcgaaggtgaccttaaacttttacagctgacggaaaaagcaaaaattaacaaatatgtgactatccttca
tctacct
aaaaagggggatgatgtgaaaccaggaaccatgtgccaagttgcagggtgggggaggactcacaatagtgcatcttggt
ccga
tactctgagagaagtcaatatcaccatcatagacagaaaagtctgcaatgatcgaaatcactataattttaaccctgtg
attggaatg
aatatggtttgtgctggaagcctccgaggtggaagagactcgtgcaatggagattctggaagccctttgttgtgcgagg
gtgttttc
cgaggggtcacttcctttggccttgaaaataaatgcggagaccctcgtgggcctggtgtctatattcttctctcaaaga
aacacctc
aactgga
217022_s-at
tcaagtgggaagagcgctgttcaaggaccacctgagcgtgacctctgtggctgctacagcgtgttccagtgtcctgccg
ggctgt
gccgagccatggaaccatggggagaccttcacttgcactgctgcccaccccgagttgaagaccccactaaccgccaaca
tcac
aaaatccggaaacacattccggcccgaggtccacctgctgccgccgccgtcggaggagctggccctgaacgagctggtg
ac
gctgacgtgcctggcacgtggcttcagcccaaggatgtgctggttcgctggctgcaggggtcacaggagctgccccgcg
aga
agtacctgacttgggcatcccggcaggagcccagccagggcaccaccaccttcgctgtgaccagcatactgcgcgtggc
agc
cgaggactggaagaagggggacaccttctcctgcatggtgggccacgaggccctgccgctggccttcacacagaagacc
atc
gaccgcttggcgggtaaacccacccatgtcaatgtgtctgttgtcatggcgga
207339_s_at
gcaggggctaggctgggagacgacgaaggaacaggcgtttctgacgagcgggacgcagttctcggacgccgaggggctg
g
cgctcccgcaggacggcctctattacctctactgtctcgtcggctaccggggccgggcgccccctggcggcggggaccc
cca
gggccgctcggtcacgctgcgcagctctctgtaccgggcggggggcgcctacgggccgggcactcccgagctgctgctc
ga
gggcgccgagacggtgactccagtgctggacccggccaggagacaagggtacgggcctctctggtacacgagcgtgggg
tt
cggcggcctggtgcagctccggaggggcgagagggtgtacgtcaacatcagtcaccccgatatggtggacttcgcgaga
gg
gaagaccttctttggggccgtgatggtggggtgagggaatatgagtgcgtggtgcgagtgcgtgaatattgggggcccg
gac
206337 at
gtgggagtggcctgaagagtcctctgaatgaaccttctggcctcccacagactcaaatgctcagaccagctcttccgaa
aacca
ggccttatctccaagaccagagatagtggggagacttcttggcttggtgaggaaaagcggacatcagctggtcaaacaa
actct
ctgaacccctccctccatcgttttcttcactgtcctccaagccagcgggaatggcagctgccacgccgccctaaaagca
cactca
tcccctcacttgccgcgtcgccctcccaggctctcaacaggggagagtgtggtgtttcctgcaggccaggccagctgcc
tccgc
gtgatcaaagccacactctgggctccagagtggggatgacatgcactcagctcttggctccactgggatgggaggagag
gaca
agggaaatgtcaggggcggggagggtgacagtggccgcccaaggccacgagcttgttctttgttctttgtcacagggac
tgaa
aacctctcctcatgttctgctttcgattcgttaagagagcaacattttacccacaca
208894 at
cgatcaccaatgtacctccagaggtaactgtgctcacgaacagccctgtggaactgagagagcccaacgtcctcatctg
tttcat
agacaagttcacccca
39729 at
36
CA 02680909 2009-09-11
WO 2008/112749 PCT/US2008/056636
acacaattaggctggctaacggatagtgagcttgtgcccctgcctaggtngcctgtgctgggtgtccancctgtgcccc
cancct
gggtgcccnnnnnnnnnnnnnnnnnnnggccagacctgcccctccaaactccacagtatgggaccctggagggntannn
nnnnnnnnnnnnatgcctccacctagaagntgaatagtgacgccctcccccaagcccacccagccgcacacaggcctag
a
ggtaaccaataaagt
209500xat
catggagctccgaattcttgcgtgtgtgtagatgaggggcgggggacgggcgccaggcattgttcagacctggtcgggg
ccca
ctggaagcatccagaacagcaccaccatctagcggccgctcgagggaagcacccgccggttggccgaagtccacgaagc
cg
ccctctgctagggaaaacccctggttctccatgccacacctctctccaggtgccctctgcctcttcaccccacaagaag
ccttatc
ctacgtccttctctccatctatcggaccccagtttccatcactatctccagagatgtagctattatgcgcccgtctaca
gggggtgcc
cgacgatgacggtgccttcgcagtcaaattactcttcgggtcccaaggtttggctttcacgcgctccattgccccggcg
tggcag
gccattccaagcccttccgggctggaactggtgtcggaggagcctcgggtgtatcgtacgccctggtgttggtgttgcc
tcactc
ctctgagctcttctttctgatcaagcc
214677_x_at
tcataagtgacttctacccgggagccgtgacagtggcctggaaggcagatagcagccccgtcaaggcgggagtggagac
cac
cacaccctccaaacaaagcaacaacaagtacgcggccagcagctatctgagcctgacgcctgagcagtggaagtcccac
aga
agctacagctgccaggtcacgcatgaagggagcaccgtggagaagacagtggcccctacagaatgttcataggttctca
accc
tcac
210314xat
catggagctccgaattcttgcgtgtgtgtagatgaggggcgggggacgggcgccaggcattgttcagacctggtcgggg
ccca
ctggaagcatccagaacagcaccaccatctagcggccgctcgagggaagcacccgccggttggccgaagtccacgaagc
cg
ccctctgctagggaaaacccctggttctccatgccacacctctctccaggtgccctctgcctcttcaccccacaagaag
ccttatc
ctacgtccttctctccatctatcggaccccagtttccatcactatctccagagatgtagctattatgcgcccgtctaca
gggggtgcc
cgacgatgacggtgccttcgcagtcaatttactcttcgggtcccaaggtttggctttcacgcgctccattgccccggcg
tggcagg
ccattccaaggccttccgggctggaactggtgtcggaggagcctcgggtgtatcgtacgccctggtgttggtgttgcct
cactcct
ctgagctcttctttctgatcaagcc
209138_x_at
tctctgggctccaggctgaggacgaggctgattattactgctgctcatatgcaggtagttacactgtggttttcggcgg
agggacc
aaactgaccgtcctaggtcagcccaaggctgccccctcggtcactctgttcccgccctcctctgaggagcttcaagcca
acaag
gccacactggtgtgtctcataagtgacttctacccgggagccgtgacagtggcctggaaggcagatagcagccccgtca
aggc
gggagtggagaccaccacaccctccaaacaaagcaacaacaagtacgcggccagcagctatctgagcctgacgcctgag
ca
gtggaagtcccacagaagctacagctgccaggtcacgcatgaagggagcaccgtggagaagacagtggcccctacagaa
tg
tt
207831xat
tgaggactggctgatgcccattctggaccagatggtgatggagcagaacacagagggtgtaaagtggacgccttctaag
atgat
cgcccggctgggcaaggagatcaacaacccagagtccgtgtattactgggcccagaagaaccacatccctgtgtttagt
cccg
cacttacagacggctcgctgggcgacatgatcttcttccattcctacaagaacccgggcctggtcctggacatcgttga
gggtgc
ccgaccagacgaggctgtctcctggggcaagatccgggtggatgcacagcccgtcaaggtctatgctgacgcctccctg
gtctt
ccccctgcttgtggctgaaacctttgcccagaagatggatgccttcatgcatgagaagaacgaggactgagcggctgcg
gtccc
aggaaggtcttaccccctcttctatttattaatttgcagacccagcccctcccctactttttggtcagctacgtctcta
gaa
215121 x at
aagccaacaaggccacactggtgtgtctcataagtgacttctacccgggagccgtgacagtggcctggaaggcagatag
cagc
cccgtcaaggcgggagtggagaccaccacaccctccaaacaaagcaacaacaagtacgcggccagcagctanctgagcc
tg
acgcctgagcagtggaagtcccacanaagctacagctgccaggtcacgcatgaagggagcaccgtggagaagacagtgg
cc
cctacagaatgttcataggttctn nanccctcancccccn
nccacgggagactagagctgcaggatcccaggggaggggtct
ctcctcccaccccaaggcatcaagcccttctccctgcactcnataaacccncaataaatatnctcattngntcaancag
aaannn
nn
nnnnannnnattttttttctcacataaattgctagcctccccggggttctcagtgtggggtacagggaattctgcaccc
agtgt
gaaaatcacccaagggagaggctcacagcctccctgagtcatctcaccagaggg
215946_x_at
37
CA 02680909 2009-09-11
WO 2008/112749 PCT/US2008/056636
gcaacatgcaggttcctgctccagcgnggctcctggactggcnccngntgctgnccnngggggtttcaatccaagcata
attc
agtgaagcatgtgtttggcagngggacccagctcacngttttaggtcagcccaagncnaccccntcggtcantctgttc
ctgcc
gtcctntgaggagcnccaagccaacaaggccacactggtgtgtctcatgaatnacttnnnncngggaatcttganggtg
acct
ggaaggcagatggtacccncatcacccagngcgtggagangaccacgccnctccaaacagagcaacancaagtacatgg
c
cagcagctacctgagcctgacgcccgagcagtggaggtcccgcagaagctacagctgccaggtcatgcatgaagggagc
ac
tgcagagaagacggtggcccctgcagaatgttcataggttcccagcccccaccccacccacaggggcctggagctgcag
gat
cccaggggaggcgtc
204069_at
aagccttacagttatcctgcaagggacaggaaggtctgatttgcaggatttttagagcattaaaataactatcaggcag
aagaatct
ttcttctcgcctaggatttcagccatgcgcgcgctctctctctttctctctcttttcctctctctccctctttctagcc
tggggcttgaattt
gcatgtctaattcatttactcaccatatttgaattggcctgaacagatgtaaatcgggaaggatgggaaaaactgcagt
catcaaca
atgattaatcagctgttgcaggcagtgtcttaaggagactggtaggaggaggcatggaaaccaaaaggccgtgtgttta
gaagc
ctaattgtcacatcaagcatcattgtccccatgcaacaaccaccaccttatacatcacttcctgttttaagcagctcta
aaacatagac
tgaagatttatttttaatatgttgactttatttctgagcaaagcatcggtcatgtgtgtattttttcatagtcccacct
tggagcatttatg
204698_at
caagttcatccggcctgagggagagatcaccgattacagaacccgggtcagcggggtcacccctcagcacatggtgggg
gcc
acaccatttgccgtggccaggctagagatcctgcagctcctgaaaggcaagctggtggtgggtcatgacctgaagcacg
acttc
caggcactgaaagaggacatgagcggctacacaatctacgacacgtccactgacaggctgttgtggcgtgaggccaagc
tgg
accactgcaggcgtgtctccctgcgggtgctgagtgagcgcctcctgcacaagagcatccagaacagcctgcttggaca
cagc
tcggtggaagatgcgagggcaacgatggagctctatcaaatctcccagagaatccgagcccgccgagggctgccccgcc
tgg
ctgtgtcagactgaagccccatccagcccgttccgcagggactagaggctttcggctttttgggaca
209906 at
gaaagcaaggcagtccattcagggaattctggaggcagccttcagtgaggagctcacacgttccacccactgtccctca
aacaa
tgtcatttcagaaagaaatagtacaactgtgtgaaaatgtggagcagccaacaagcaggggctcttaggcaatcacata
gtgaaa
gtttataagaggatgaagtgatatggtgagc
agcggacttcaaaaactgtcaaagaatcaatccagcggttctcaaacggtacac
agactattgacatcagcatcacctagaaacttgttagaaatgcaaattctcaagccgcatcccagacttgctgaatcgg
aatctctg
ggggttgggacccagcaagggcacttaacaaacccccgtttctgattaatgctaaatgtaagaatcattgtaaacatta
gttctattt
ctatcccaaactaagc
205608_s_at
agagcagcctgatcttacacggtgctgatttcagcactaaagatgctgataatgacaactgtatgtgcaaatgtgccct
catgttaa
caggaggatggtggtttgatgcttgtggcccctccaatctaaatggaatgttctatactgcgggacaaaaccatggaaa
actgaat
gggataaagtggcactacttcaaagggccc
agttactccttacgttccacaactatgatgattcgacctttagatttttgaaagcgca
atgtcagaagcgattatgaaagcaacaaagaaatccggagaagctgccaggtgagaaactgtttgaaaacttcagaagc
aaac
aatattgtctcccttccagcaataagtggtagttatgtgaagtcaccaaggttcttgaccgtgaatctggagccgtttg
agttcacaa
gagtctctacttggggtgacagtgctcacgtggctcgactatagaaaactccactgactgtcgggctttaaaaagggaa
gaaact
gctgagcttgctgtgcttcaaactactact
205927_s_at
tccacacacggccaggcctgtttatctacactgctgcccactcctctctccagctccacatgctgtacctggatcattc
tgaagcaa
attccgagcattacatcattttgtccataaatatttctaacatccttaaatatacaatcggaattcaagcatctcccat
tgtcccacaaat
gtttggctgtttttgtagttggattgtttgtattaggattcaagcaaggcccatatattgcatttatttgaaatgtctg
taagtctctttccat
ctacagagtttagcacatttgaacgttgctggttgaaatcccgaggtgtcatttgacatggttctctgaacttatcttt
cctataaaatg
gtagttagatctggaggtctgattttgtggcaaaaatacttcctaggtggtgctgggtacttcttgttgcatcctgtca
ggaggcaga
taatgctggtgcctctctattggtaatgttaagactgctgggtgggtttggagttcttggc
21505I x at
tgctgaaaaccctccagtcagcgcttatcccttctgctctctcccctcacccagagaaatacatggagtttgaccttaa
tggaaatg
gcgatattgatatcatgtccctgaaacgaatgctggagaaacttggagtccccaagactcacctagagctaaagaaatt
aattgga
gaggtgtccagtggctccggggagacgttcagctaccctgactttctcaggatgatgctgggcaagagatctgccatcc
taaaaa
38
CA 02680909 2009-09-11
WO 2008/112749 PCT/US2008/056636
tgatcctgatgtatgaggaaaaagcgagagaaaaggaaaagccaacaggccccccagccaagaaagctatctctgagtt
gcc
ctgatttgaagggaaaagggatgatgggattgaaggggcttctaatgacccagatatgg
205609_at
gtttaccatcaagtcttttttatatttatgtgtctgtattctacccctttttgccttacaagtgatatttgcaggtatt
ataccatttttctattctt
ggtggcttcttcatagcaggtaagcctctccttctaaaaacttctcaactgttttcatttaagggaaagaaaatgagta
ttttgtcctttt
gtgttcctacagacactttcttaaaccagtttttggataaagaatactatttccaaactcatattacaaaaacaaaata
aaataataaaa
aaagaaagcatgatatttactgttttgttgtctgggtttgagaaatgaaatattgtttccaattatttataataaatca
gtataaaatgtttt
atgattgttatgtgtattatgtaatacgtacatgtttatggcaatttaacatgtgtattcttttcatttaattgtttca
gaataggataattagg
tattcgaattttgtctttaaaattcatgtggtttctatgcaaagttcttcatatcatcaca
202890 at
aatgatggaatgttgactgtgtttggcacacaggacacggaccttcatggaagtccttgctctgcgtggcatctgtcag
cttttcac
ctttcattcttattcttcacttttgctgctgagcctagctgtacaaacttgcactttcatttgctaatataaattcagt
tttattttaccatttta
gagactactaatgattaaatgtagaaggagagggtgcacatgtttttatgtggagtgtttaaaagataaatttatacca
ctgtaatgtg
cagcttttattaaaagagaaattggttgaactgctaggttgaatgagagacttcatctattggactattttttttaatc
caggcatatggt
ctttagtaatggcttgtaatttgtgaaaacattaatttgggggttttccctgttttcagttgtccatgtacacatagtc
attatattagaaaa
gaaagctgttcaacaaacttgtttaatttgtttaaatcaacatagcatgaaacaccaaat
203485_at
aagcagtcgaccgcacttatggtaatcagttttgtataacttaaaataattaaataaatgaataaatccaaaacaaaca
tgcagtactt
ttgttgtatgggattggtgggctgatttacatgtatggttactaaaaagtaccagcatgttaactttattacaatttgt
attactttctctgt
agttcctaatggattcaattacggactctggatatttgcactt
39