Note: Descriptions are shown in the official language in which they were submitted.
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
COMPOSITIONS, METHODS AND KITS FOR THE DIAGNOSIS OF
CARRIERS OF MUTATIONS IN THE BRCA1 AND BRCA2 GENES AND
EARLY DIAGNOSIS OF CANCEROUS DISORDERS ASSOCIATED
WITH MUTATIONS IN BRCAI AND BRCA2 GENES
Field of the Invention
The invention relates to early diagnosis of cancerous disorders. More
particularly, the invention relates to compositions methods and kits based on
measuring differential expression of at least one marker gene, for the
diagnosis
of carriers of mutations in the BRCA1 and BRCA2 genes and thereby, the
diagnosis of cancerous disorders associated therewith, specifically, of
ovarian
and breast cancer.
Background of the Invention
All publications mentioned throughout this application are fully incorporated
herein by reference, including all references cited therein.
Diagnostic markers are important for early diagnosis of many diseases, as well
as predicting response to treatment, monitoring treatment and determining
prognosis of such diseases.
Mutations in the breast and ovarian cancer susceptibility genes BRCA1 and
BRCA2 are found in a high proportion of multiple-case families with breast and
ovarian cancer [Antoniou, A.C. et al. Genetic Epidemiology 25:190-202 (2003)].
Carriers of mutations in BRCA1 or BRCA2 genes have up to 80% lifetime risk
of developing breast and ovarian cancers and elevated risk of developing other
types of cancer, such as prostate and pancreas. Mutations in the BRCA1 gene
account for 50% of familial breast cancer cases. Mutations in BRCA2 account
for 30% of familial breast cancer cases and are also linked to male breast
cancer.
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
2
About 80% of all alterations in BRCAI and BRCA2 tumors are frameshift or
nonsense mutations, and yield a truncated protein product [Breast cancer
Information Core-BIC at http://www. nhgri.nih. gov/Intramural_ research
/Lab_transfer/Bic]. The types of mutation differ in distribution depending on
ethnicity and geographic location. There is increasing evidence that
hereditary
cancer syndromes resulting from germline mutations in cancer susceptibility
genes lead to organ-specific cancers with distinct histological phenotypes.
The
hereditary breast tumors that result from germline BRCA1 and BRCA2
mutations exemplify this phenomenon. In recent years, it has been
demonstrated that BRCA1 and BRCA2 breast carcinomas differs from sporadic
breast cancer of age-matched controls and from non-BRCA1 /2 familial breast
carcinomas in their morphological, immunophenotypic and molecular
characteristics [Phillips K.A. Journal of Clinical Oncology 18:107s-112s
(2000)].
The structurally distinct proteins encoded by BRCA1 and BRCA2 regulate
numerous cellular functions, including DNA repair, chromosomal segregation,
gene transcription, cell-cycle arrest and apoptosis. BRCA1 and BRCA2 are
considered to be ,gatekeepers,,: genes which, when mutated or abnormally
expressed, cause disruption of normal cell biology, interrupt cell division or
death control, and promote the outgrowth of cancer cells. Recent reports have
provided insight into the role of BRCA1 and BRCA2 in the cellular response to
DNA damage [Tutt A. et al. The EMBO Journal 20:4704-4716 (2001)]. Several
groups have demonstrated that BRCA1- or BRCA2-deficient rodent cells or
human tumors are specifically deficient in DNA repair via homologous
recombination, whereas, when measured, non-homologous recombination
remains intact after double-strand DNA breaks. Moreover, the correlation
between BRCA1 or BRCA2 mutation and alterations in p53, HER 2 and Myc
gene expression as well as alterations in cell-cycle regulation have been
shown
in breast carcinoma patients [Venkitaraman AR. Journal of Cell Science.
114:3591-8 (2005)]. Together, these data imply that accumulation of somatic
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
3
genetic changes during tumor progression may follow a unique pathway in
individuals genetically predisposed to cancer.
As mentioned above, BRCA1 and BRCA2 proteins maintain genomic stability
through an involvement in DNA repair processes. Mutations in BRCA1 and
BRCA2 seem to predispose cells to an increased risk of mutagenesis and
transformation after exposure to radiation. It was shown recently that normal
human fibroblasts and lymphoblastoid cells with heterozygous BRCAl and
BRCA2 mutations seem to have increased radiosensitivity [Buchholz T.A. et al.
International Journal of Cancer 97:557-561 (2002)]. Previous study of the
present inventors on short-term lymphocyte cultures, provided additional
evidence that heterozygous mutation carriers have a different response to DNA
damage compared with non-carriers [Kote-Jarai Z. et al. British Journal of
Cancer 94:308-310 (2006)]. The characterization of BRCA1/2 RNA expression
profile of human fibroblasts from healthy mutation carriers has been described
using spotted cDNA microarray [Kote-Jarai Z. et al. Clinical Cancer Research
12:3896-901 (2006)]. This study shows a significant difference in gene
expression profiling in heterozygous BRCA1 and BRCA2 mutation carriers as
compared to non-carriers following induced DNA damage caused by exposure
to irradiation.
The present invention discloses marker genes differentially expressed in
lymphocytes from BRCA1 and BRCA2 carriers versus non-carriers following
irradiation stress. These marker genes are used by the compositions, kits and
methods of the invention as a tool for detecting carriers and thereby for
early
detection of proliferative disorders and particularly, of breast and ovarian
carcinomas.
It is therefore one object of the invention to provide a simple diagnostic
composition comprising at least one detecting molecule specific for
quantitative
determination of the expression profile of a collection of marker genes.
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
4
It is another object of the invention to provide a simple, inexpensive, and
clear
test to distinguish between BRCA1 or BRCA2 genes mutation carriers and
non-carriers.
As indicated above, carriers of mutations in BRCAI or BRCA2 genes exhibit
increased predisposition to cancerous disorders Therefore, another object of
the
invention is to provide diagnostic method for early detection of cancerous
disorders associated with mutations in these genes, particularly of breast and
ovarian cancer. This method is based on quantitative determination of the
expression of at least one marker gene described by the invention.
A further object of the invention is to provide diagnostic kit for detection
of
carriers of BRCA1 and BRCA2 gene mutations and thereby the diagnosis of
cancerous disorders associated with mutations in BRCA1 or BRCA2 genes.
These and other objects of the invention will become apparent as the
description proceeds.
Summary of the Invention
In a first aspect, the invention relates to a composition comprising at least
one
detecting molecule specific for determination of the expression of at least
one
marker gene. More specifically, the detecting molecules used by the
composition of the invention may be either isolated detecting nucleic acid
molecule or isolated detecting amino acid molecule. Such detecting molecules
are specific for a marker gene selected from the group consisting of:
RAB3GAP1, RAB3 GTPase activating protein subunit 1 (catalytic); NFAT5,
nuclear factor of activated T-cells 5, tonicity-responsive; MRPS6,
mitochondrial
ribosomal protein S6; AUH, AU RNA binding protein/enoyl-Coenzyme A
hydratase; MID11P1, MIDl interacting protein 1(gastrulation specific G12
homolog (zebrafish)); RGS16, regulator of G-protein signaling 16; MARCH7,
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
membrane-associated ring finger (C3HC4) 7; NR3C1, nuclear receptor
subfamily 3, group C, member 1(glucocorticoid receptor); ELFl, E74-like factor
1(ets domain transcription factor); RPS6KB1, ribosomal protein S6 kinase,
70kDa, polypeptide 1; STAT5A, signal transducer and activator of
transcription 5A; YTHDF3, YTH domain family, member 3; DNAJC12, DnaJ
(Hsp40) homolog, subfamily C, member 12; IFI44L, interferon-induced protein
44-like; SARS, seryl-tRNA synthetase; SMURF2, SMAD specific E3 ubiquitin
protein ligase 2; SFRS18, splicing factor, arginine/serine-rich 18; NR4A2,
nuclear receptor subfamily 4, group A, member 2; CDKN1B, cyclin-dependent
kinase inhibitor 1B (p27, Kip1); and EIF3D, eukaryotic translation initiation
factor 3, subunit D, as set forth in Table 4.
It should be appreciated that the composition of the invention is specifically
used for determining the level of expression of at least one of the marker
gene
indicated by the invention in a biological test sample of a mammalian subject.
According to one preferred embodiment, the composition of the invention is
specifically applicable for the detection of at least one mutation in at least
one
of BRCA1 and BRCA2 genes in a biological sample of a mammalian subject.
This particular composition comprises at least one isolated oligonucleotide
which specifically hybridizes to a nucleic acid sequence of RNA products of at
least one marker gene. These marker genes were shown by the invention as
exhibiting a differential expression in samples obtained from BRCA1 or
BRCA2 carriers under irradiation stress. Differential expression of at least
one
of the marker genes reflects the existence of at least one mutation in any one
of
BRCA1 /2 and therefore is indicative of an increased genetic predisposition of
said subject to a cancerous disorder associated with mutations in any one of
BRCA1 or BRCA2.
In another aspect, the invention relates to a method for the detection of at
least
one mutation in at least one of BRCA1 and BRCA2 genes in a biological sample
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
6
of a mammalian subject. The method of the invention comprises the steps of:
(a) determining the level of expression of at least one of the marker genes
identified by the invention as set for the in Table 4, in the test sample and
in a
suitable control sample; (b) optionally, determining the level of expression
of at
least one control gene in the test sample and in a suitable control sample.
(c) comparing the level of expression as obtained by step (a) of each of the
marker genes in the test sample with the level of expression in the control
sample; and optionally (d) comparing the level of expression as obtained by
step (c) of each of the control genes in said test sample with the level of
expression in the control sample.
It should be appreciated that the detection of differential expression of the
marker genes in the tested sample as compared to a control sample, indicates
that the tested subject is a carrier of at least one gene mutation in at least
one
of BRCAI and BRCA2.
Another aspect of the invention relates to a kit comprising:
(a) means for obtaining a sample of a mammalian subject;
(b) at least one detecting molecule or a collection of at least two detecting
molecules specific for determination of the expression of at least one marker
gene or a collection of at least two marker genes. According to a particular
embodiment, these marker genes are the genes described by the invention as
set forth in Table 4; (c) optionally, at least one detecting molecule or a
collection of at least two detecting molecules specific for determination of
the
expression of at least one control reference gene or a collection of at least
two
control reference genes. (d) at least one control sample that may be at least
one
of a negative control sample and a positive control sample;
(e) instructions for carrying out the detection and quantification of
expression
of the marker genes and optionally, of the control reference gene in the
tested
and control samples;
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
7
(f) instructions for evaluating the differential expression of the marker gene
in
the tested sample and optionally, of a control reference gene, in the test
sample
as compared to a control sample.
According to a particular embodiment, the kit of the invention maybe
specifically applicable for detecting of at least one of BRCAI aiid BRCA 2
mutations in a mammalian subject.
These and other aspects of the invention will become apparent by the liand of
the following figures ancl examples.
Brief Description of the Figures
Figure lA-1 C. Heat map of gene expression profile of lymphocytes from
BRCA1 mutation carriers and control non-carriers (A) or BRCA2 carriers and
control non-carriers (B). Data analysis by Expression Console Software
(Affymetrix) represented in Figure (C) Only the genes expressed in
significantly distinct manner (with p-value <0.05) were selected for analysis.
Abbreviations: cont. (control).
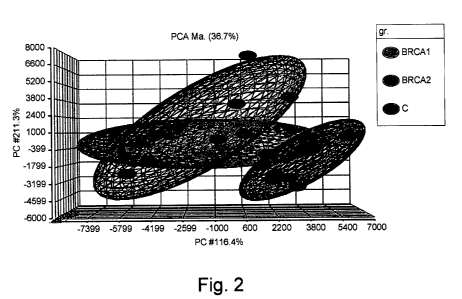
Figure 2. Principal components analysis (PCA) of gene profile in BRCA1 and
BRCA2 niutation carriers and control. Abbreviations: gr. (group), C (control),
Ma (mapping).
Figure 3A-3C. ANOVA analysis of BRCA1 (yellow), BRCA2 (blue) and control
(red) gene expression.
Fig. 3A. Clustering of the whole gene set. Note the homogenous clustering of
BRCA2 as compared to the somewhat more heterogeneous clustering of
BRCA1. Fig. 3B. An enlargement of a sample cluster Fig. 3C. Cluster of 17.
genes that were significantly under-expressed in BRCA1 in comparison to
BRCA2 and control.
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
8
Figure 4A-4B. Graphic presentation of functional groups of all genes having
differentionl expression in samples of BRCA1 mutation carriers. F'ig. 4A
demonstrate genes which are up regulated as compared to a non-carrier control
and Fig. 4B demonstrate genes which are down regulated in BRCAl mutation
sample. Abbreviations: bin. (binding), sig. (signal), trans. (transducer), ac.
(activity), tm. (transineinbrane), Rec.(receptor), Ag. (antigen), reg.
(regulator),
Ha. (heavy), met. (metal), pr. (protein), Unf. (unfolded), Enz (enzymatic).
Figure 5A-5B. Graphic presentation of functional groups of all genes having
differentionl expression in samples of BRCA2 mutation carriers. Fig. 5A
demonstrate genes which are up regulated as compared to a non-carrier control
and Fig. 5B demonstrate genes which are down regulated in BRCA2 mutation
sample. Abbreviations: bin. (binding), ac. (activity), cat. (catalytic), nuc.
(nucleotide), pr. (protein), Enz (enzymatic), kin. (kinase), sin. (single),
str.
(strand), lip. (lipid), cons. (constituent), stru. (structured).
Figure 6. Gene Ontology analysis of the genes differentially expressed, with
most similar gene expression consistent into each,group.
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
9
Detailed Description of the Invention
The present invention discloses characterization of the gene expression
profile
in freshly cultured lymphocytes obtained from healthy women as compared to
carriers of mutations in either BRCA1 or BRCA2. This comparison revealed
significant differences in gene expression profile between BRCA1 and BRCA2
mutation groups and the control group. Thus, according to a first aspect, the
invention relates to a composition comprising at least one detecting molecule
or
a collection of at least two detecting molecules specific for determination of
the
expression of at least one marker gene or a collection of at least two marker
genes. More specifically, these marker genes may be selected from the group
consisting of: RAB3GAP1, RAB3 GTPase activating protein subunit 1
(catalytic); NFAT5, nuclear factor of activated T-cells 5, tonicity-
responsive;
MRPS6, mitochondrial ribosomal protein S6; AUH, AU RNA binding
protein/enoyl-Coenzyme A hydratase; MID lIP 1, MID 1 interacting protein 1
(gastrulation specific G12 homolog (zebrafish)); RGS16, regulator of G-protein
signaling 16; MARCH7, membrane-associated ring finger (C3HC4) 7; NR3C1,
nuclear receptor subfamily 3, group C, member 1(glucocorticoid receptor);
ELF1, E74-like factor 1 (ets domain transcription factor); RPS6KB1, ribosomal
protein S6 kinase, 70kDa, polypeptide 1; STAT5A, signal transducer and
activator of transcription 5A; YTHDF3, YTH domain family, member 3;
DNAJC12, DnaJ (Hsp40) homolog, subfamily C, member 12; IFI44L,
interferon-induced protein 44-like; SARS, seryl-tRNA synthetase; SMURF2,
SMAD specific E3 ubiquitin protein ligase 2; SFRS18, splicing factor,
arginine/serine-rich 18; NR4A2, nuclear receptor subfamily 4, group A,
member 2; CDKN1B, cyclin-dependent kinase inhibitor 1B (p27, Kipl); and
EIF3D, eukaryotic translation initiation factor 3, subunit D, and are as set
forth in Table 4, or any collection or combination thereof. It should be noted
that the composition of the invention may be specifically applicable for
determining the level of expression (also referred to herein as "profiling" or
"expression pattern") of at least one of said marker genes in a biological
test
sample of a mammalian subject.
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
According to one embodiment, the detecting molecules are specific for
quantitative or qualitative determination of expression of said marker genes.
Preferably, the detecting molecules used by the invention may be specifically
suitable for quantitative determination of expression of any of the marker
genes used by the composition of the invention, as set forth in Table 4.
According to one embodiment, the detecting molecule used by the composition
of the invention may be an isolated nucleic acid molecule or an isolated amino
acid molecule. It should be appreciated that the composition of the invention
may comprises both, nucleic acid based detecting molecules and amino acid
based detecting molecules. Thus, the invention further contemplates the use of
a combination of proteins or polypeptides in combination with polynucleotides
so as to measure one or more products of one or more of the marker genes of
the invention, in any combination thereof.
As used herein, "nucleic acid(s)" is interchangeable with the term
"polynucleotide(s)" and it generally refers to any polyribonucleotide or poly-
deoxyribonucleotide, which may be unmodified RNA or DNA or modified RNA
or DNA or any combination thereof. "Nucleic acids" include, without
limitation,
single- and double-stranded nucleic acids. As used herein, the term "nucleic
acid(s)" also includes DNAs or RNAs as described above that contain one or
more modified bases. Thus, DNAs or RNAs with backbones modified for
stability or for other reasons are "nucleic acids". The term "nucleic acids"
as it
is used herein embraces such chemically, enzymatically or metabolically
modified forms of nucleic acids, as well as the chemical forms of DNA and RNA
characteristic of viruses and cells, including for example, simple and complex
cells. A "nucleic acid" or "nucleic acid sequence" may also include regions of
single- or double- stranded RNA or DNA or any combinations.
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
11
As used herein, the term "oligonucleotide" is defined as a molecule comprised
of
two or more deoxyribonucleotides and/or ribonucleotides, and preferably more
than three. Its exact size will depend upon many factors which in turn, depend
upon the ultimate function and use of the oligonucleotide. The
oligonucleotides
may be from about 8 to about 1,000 nucleotides long. Although oliognucleotides
of 5 to 100 nucleotides are useful in the invention, preferred
oligonucleotides
range from about 5 to about 15 bases in length, from about 5 to about 20 bases
in length, from about 5 to about 25 bases in length, from about 5 to about 30
bases in length, from about 5 to about 40 bases in length or from about 5 to
about 50 bases in length. More specifically, the detecting oligonucleotides
molecule used by the composition of the invention may comprise any one of 5,
6, 7, 8, 9, 10, 11, 12, 13, 1, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26,
27, 28,
29, 30, 35, 40, 45, 50 bases in length.
As indicated above, the detecting molecules of the invention may be amino acid
based molecules that may be referred to as protein/s or polypeptide/s. As used
herein, the terms "protein" and "polypeptide" are used interchangeably to
refer
to a chain of amino acids linked together by peptide bonds. In a specific
embodiment, a protein is composed of less than\200, less than 175, less than
150, less than 125, less than 100, less than 50, less than 45, less than 40,
less
than 35, less than 30, less than 25, less than 20, less than 15, less than 10,
or
less than 5 amino acids linked together by peptide bonds.
In another embodiment, a protein is composed of at least 200, at least 250, at
least 300, at least 350, at least 400, at least 450, at least 500 or more
amino
acids linked together by peptide bonds.
According to one specifically preferred embodiment, the isolated detecting
nucleic acid molecule comprised within the composition of the invention may be
an isolated oligonucleotide which specifically or/and selectively hybridizes
to a
nucleic acid sequence of the RNA products of at least one marker gene selected
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
12
from the group consisting of: RAB3GAP1, RAB3 GTPase activating protein
subunit 1(catalytic); NFAT5, nuclear factor of activated T-cells 5, tonicity-
responsive; MRPS6, mitochondrial ribosomal protein S6; AUH, AU RNA
binding protein/enoyl-Coenzyine A hydratase; MIDIIPI, MID1 interacting
protein 1(gastrulation. specific G12 homolog (zebrafish)); RGS16, regulator of
G-protein signaling 16; MARCH7, meinbrane-associated ring finger (C3HC4) 7;
NR3C1, nuclear receptor subfamily 3, group C, member 1(glucocorticoid
receptor); ELF1, E74-like factor 1(ets domain transcription factor); RPS6KB1,
ribosomal protein S6 kinase, 70kDa, polypeptide 1; STAT5A, signal transducer
and activator of transcription 5A; YTHDF3, YTH domain family, member 3;
DNAJC12, DnaJ (Hsp40) homolog, subfamily C, member 12; IFI44L,
interferon-induced protein 44-like; SARS, seryl-tRNA synthetase; SMURF2,
SMAD specific E3 ubiquitin protein ligase 2; SFRS18, splicing factor,
arginine/serine-rich 18; NR4A2, nuclear receptor subfamily 4, group A,
member 2; CDKNIB, cyclin-dependent kinase inhibitor 1B (p27, Kipl); and
EIF3D, eukaryotic translation initiation factor 3, subunit D, as set forth in
Table 4.
As used herein, the term "hybridize" refers to process that two complementary
nucleic acid strands anneal to each other under appropriately stringent
conditions. Hybridizations are typically and preferably conducted with probe-
length nucleic acid molecules, preferably 5-200 nucleotides in length, 5-100,
5-
50, 5-40, 5-30 or 5-20.
As used herein "selective or specific hybridization" in the context of this
invention refers to a hybridization which occurs as between a polynucleotide
encompassed by the invention and an RNA product of the marker gene of the
invention, wherein the hybridization is such that the polynucleotide binds to
the RNA products of the marker gene of the invention preferentially to any
RNA products of other gene products in the tested sample. In a preferred
embodiment a polynucleotide which "selectively hybridizes" is one which
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
13
hybridizes with a selectivity of greater than 60%, greater than 70%, greater
than 80%, greater than 90% and most preferably on 100% (i.e. cross
hybridization with other RNA species preferably occurs at less than 40%, less
than 30%, less than 20%, less than 10%). As would be understood to a person
skilled in the art, a detecting polynucleotide which "selectively hybridizes"
to
the RNA product of a marker gene of the invention can be determined taking
into account the length and composition.
As used herein, "specifically hybridizes", "specific hybridization" refers to
hybridization which occurs when two nucleic acid sequences are substantially
complementary (at least about 60% complementary over a stretch of at least 5
to 25 nucleotides, preferably at least about 70%, 75%, 80% or 85%
complementary, more preferably at least about 90% complementary, and most
preferably, about 95% complementary).
The measuring of the expression of the RNA product of any one of the marker
genes and combination of marker genes of the invention, can be done by using
those polynucleotides as detecting molecules, which are specific and/or
selective for the RNA products of the marker genes of the invention to
quantitate the expression of the RNA product. In a specific embodiment of the
invention, the polynucleotides which are specific and/or selective for the RNA
products may be probes or primers. It should be further appreciated that the
composition of the invention may comprise as an oligonucleotide-based
detection molecule, both, primers and probes.
The term, "primer", as used herein refers to an oligonucleotide, whether
occurring naturally as in a purified restriction digest or produced
synthetically,
which is capable of acting as a point of initiation of synthesis when placed
under conditions in which synthesis of a primer extension product, which is
complementary to a nucleic acid strand, is induced, i.e., in the presence of
nucleotides and an inducing agent such as a DNA polymerase and at a suitable
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
14
temperature and pH. The primer may be either single-stranded or double-
stranded and must be sufficiently long to prime the synthesis of the desired
extension product in the presence of the inducing agent. The exact length of
the primer will depend upon many factors, including temperature, source of
primer and the method used. For example, for diagnostic applications,
depending on the complexity of the target sequence, the oligonucleotide primer
typically contains 10-30 or more nucleotides, although it may contain fewer
nucleotides. More specifically, the primer used by the composition of the
invention may comprise 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23,
24,
25, 26, 27, 28, 29 or 30 nucleotides. The factors involved in determining the
appropriate length of primer are readily known to one of ordinary skill in the
art.
As used herein, the term "probe" means oligonucleotides and analogs thereof
and refers to a range of chemical species that recognize polynucleotide target
sequences through hydrogen bonding interactions with the nucleotide bases of
the target sequences. The probe or the target sequences may be single- or
double-stranded RNA or single- or double- stranded DNA or a combination of
DNA and RNA bases. A probe is at least 5 or preferably, 8 nucleotides in
length
and less than the length of a complete gene. A probe may be 5, 6, 7, 8, 9, 10,
11,
12., 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30,
40, 50, 75,
100, 150, 200, 250, 400, 500 and up to 2000 nucleotides in length as long as
it
is less than the full length of the target gene. Probes can include
oligonucleotides modified so as to have a tag which is detectable by
fluorescence, chemiluminescence and the like. The probe can also be modified
so as to have both a detectable tag and a quencher molecule, for example
TaqMan and Molecular Beacon probes, that will be described in detail
below.
The oligonucleotides and analogs thereof may be RNA or DNA, or analogs of
RNA or DNA, commonly referred to as antisense oligomers or antisense
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
oligonucleotides. Such RNA or DNA analogs comprise but are not limited to 2-
'O-alkyl sugar modifications, methylphosphonate, phosphorothiate,
phosphorodithioate, formacetal, 3-thioformacetal, sulfone, sulfamate, and
nitroxide backbone modifications, and analogs wherein the base moieties have
been modified. In addition, analogs of oligomers may be polymers in which the
sugar moiety has been modified or replaced by another suitable moiety,
resulting in polymers which include, but are not limited to, morpholino
analogs
and peptide nucleic acid (PNA) analogs.
Probes may also be mixtures of any of the oligonucleotide analog types
together
or in combination with native DNA or RNA. At the same time, the
oligonucleotides and analogs thereof may be used alone or in combination with
one or more additional oliognucleotides or analogs thereof.
According to another preferred embodiment, when the detecting molecule is an
oligonucleotide, the expression level of any of the marker genes may be
determined using at least one nucleic acid amplification assay, such as a Real-
Time PCR, micro arrays, PCR, in situ Hybridization or Comparative Genomic
Hybridization.
The term "amplify" with respect to nucleic acid sequences refers to methods
that increase the representation of a population of nucleic acid sequences in
a
sample. Nucleic acid amplification methods, such as PCR, isothermal methods,
rolling circle methods, etc., are well known to the skilled artisan. More
specifically, as used herein, the term "amplified", when applied to a nucleic
acid sequence, refers to a process whereby one or more copies of a particular
nucleic acid sequence is generated from a template nucleic acid, preferably by
the method of polymerase chain reaction. "Polymerase chain reaction" or "PCR"
refers to an in vitro method for amplifying a specific nucleic acid template
sequence. The PCR reaction involves a repetitive series of temperature cycles
and is typically performed in a volume of 50-100 l. The reaction mix
comprises
dNTPs (each of the four deoxynucleotides dATP, dCTP, dGTP, and dTTP),
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
16
primers, buffers, DNA polymerase, and nucleic acid template. The PCR
reaction comprises providing a set of polynucleotide primers wherein a first
primer contains a sequence complementary to a region in one strand of the
nucleic acid template sequence and primes the synthesis of a complementary
DNA strand, and a second primer contains a sequence complementary to a
region in a second strand of the target nucleic acid sequence and primes the
synthesis of a complementary DNA strand, and amplifying the nucleic acid
template sequence employing a nucleic acid polymerase as a template-
dependent polymerizing agent under conditions which are permissive for PCR
cycling steps of (i) annealing of primers required for amplification to a
target
nucleic acid sequence contained within the template sequence, (ii) extending
the primers wherein the nucleic acid polymerase synthesizes a primer
extension product. "A set of polynucleotide primers", "a set of PCR primers"
or
`pair of primers" can comprise two, three, four or more primers.
Real time nucleic acid amplification and detection methods are efficient for
sequence identification and quantification of a target since no pre-
hybridization amplification is required. Amplification and hybridization are
combined in a single step and can be performed in a fully automated, large-
scale, closed-tube format.
Methods that use hybridization-triggered fluorescent probes for real time PCR
are based either on a quench-release fluorescence of a probe digested by DNA
Polymerase (e.g., methods using TaqMan , MGB- TaqMan ) or on a
hybridization-triggered fluorescence of intact probes (e.g., molecular
beacons,
and linear probes). In general, the probes are designed to hybridize to an
internal region of a PCR product during annealing stage (also referred to as
amplicon). For those methods utilizing TaqMan and MGB- TaqMan the 5'-
exonuclease activity of the approaching DNA Polymerase cleaves a probe
between fluorophore and quencher thus releasing fluorescence.
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
17
Thus, a "real time PCR" assay providing dynamic fluorescence detection of
amplified marker gene products produced in a PCR amplification reaction.
During PCR, the amplified products created using suitable primers hybridize
to probe nucleic acids (TaqMan probe, for example), which may be labeled
according to some embodiments with both a reporter dye and a quencher dye.
When these two dyes are in close proximity, i.e. both are present in an intact
probe oligonucleotide, the fluorescence of the reporter dye is suppressed.
However, a polymerase, such as AmpliTaq GoldTM., having 5'-3' nuclease
activity can be provided in the PCR reaction. This enzyme cleaves the
fluorogenic probe if it is bound specifically to the target nucleic acid
sequences
between the priming sites. The reporter dye and quencher dye are separated
upon cleavage, permitting fluorescent detection of the reporter dye. Upon
excitation by a laser provided, e.g., by a sequencing apparatus, the
fluorescent
signal produced by the reporter dye is detected and/or quantified. The
increase
in fluorescence is a direct consequence of amplification of target nucleic
acids
during PCR.
The method and hybridization assays using self-quenching fluorescence probes
with and/or without internal controls for detection of nucleic acid
application
products are known in the art, for example, U.S. Pat. Nos. 6,258,569;
6,030,787; 5,952,202; 5,876,930; 5,866,336; 5,736,333; 5,723,591; 5,691,146;
and 5,538,848.
More particularly, QRT-PCR (Quantitative RT-PCR), which is quantitative in
nature, can also be performed to provide a quantitative measure of gene
expression levels. In QRT-PCR reverse transcription and PCR can be
performed in two steps, or reverse transcription combined with PCR can be
performed. One of these techniques, for which there are commercially available
kits such as TaqMan (Perkin Elmer, Foster City, CA) , is performed with a
transcript-specific antisense probe. This probe is specific for the PCR
product
(e.g. a nucleic acid fragment derived from a gene) and is prepared with a
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
18
quencher and fluorescent reporter probe with complex to the 5' end of the
oligonucleotide. Different fluorescent markers are attached to different
reporters, allowing for measurement of at least two products in one reaction.
When Taq DNA polymerase is activated, it cleaves off the fluorescent reporters
of the probe bound to the template by virtue of its 5 -to-3' exonuclease
activity.
In the absence of the quenchers, the reporters now fluoresce. The color change
in the reporters is proportional to the amount of each specific product and is
measured by a fluorometer; therefore, the amount of each color is measured
and the PCR product is quantified. The PCR reactions can be performed in any
solid support, for example, slides, microplates, 96 well plates, 384 well
plates
and the like so that samples derived from many individuals are processed and
measured simultaneously. The TaqMan system has the additional advantage
of not requiring gel electrophoresis and allows for quantification when used
with a standard curve.
A second technique useful for detecting PCR products quantitatively without is
to use an intercolating dye such as the commercially available QuantiTect
SYBR Green PCR (Qiagen, Valencia California)\ RT-PCR is performed using
SYBR green as a fluorescent label which is incorporated into the PCR product
during the PCR stage and produces a flourescense proportional to the amount
of PCR product.
Both TaqMan and QuantiTect SYBR systems can be used subsequent to
reverse transcription of RNA. Reverse transcription can either be performed in
the same reaction mixture as the PCR step (one-step protocol) or reverse
transcription can be performed first prior to amplification utilizing PCR (two-
step protocol).
Additionally, other systems to quantitatively measure mRNA expression
products are known including Molecular Beacons which uses a probe having
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
19
a fluorescent molecule and a quencher molecule, the probe capable of forming a
hairpin structure such that when in the hairpin form, the fluorescence
molecule is quenched, and when hybridized the flourescense increases giving a
quantitative measurement of gene expression.
In one embodiment, these polynucleotide-based detection molecules of the
invention may be in the form of nucleic acid probes which can be spotted onto
an array to measure RNA from the sample of a subject to be diagnosed.
As defined herein, a"nucleic acid array" refers a plurality of nucleic acids
(or
"nucleic acid members"), optionally attached to a support where each of the
nucleic acid members is attached to a support in a unique pre- selected and
defined region. These nucleic acid sequences are used herein as detecting
nucleic acid molecules. In one embodiment, the nucleic acid member attached
to the surface of the support is DNA. In a preferred embodiment, the nucleic
acid member attached to the surface of the support is either cDNA or
oligonucleotides. In another preferred embodiment, the nucleic acid member
attached to the surface of the support is cDNA synthesized by polymerase
chain reaction (PCR). In another preferred embodiment, a "nucleic acid array"
refers to a plurality of unique nucleic acid detecting molecules attached to
~. ~
nitrocellulose or other membranes used in Southern and/or Northern blotting
techniques.
For oligonucleotide-based arrays, the selection of oligonucleotides
corresponding to the gene of interest which are useful as probes is well
understood in the art.
More particularly it is important to choose regions which will permit
hybridization to the target nucleic acids. Factors such as the Tm of the
oligonucleotide, the percent GC content, the degree of secondary structure and
the length of nucleic acid are important factors.
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
According to this embodiment, the detecting molecule may be in the form of
probe corresponding and thereby hybridizing to any region or part of the
marker gene. For example, these probes may be a set of corresponding 5' ends
or a set of corresponding 3' ends or a set of corresponding internal coding
regions. Of course, mixtures of a 5' end of one. gene may be used as a target
or a
probe in combination with a 3' end of another gene to achieve the same result
of measuring the levels of expression of the marker gene.
As used herein, the "5' end" refers to the end of an mRNA up to the first 1000
nucleotides or one third of the mRNA (where the full length of the mRNA does
not include the poly A tail), starting at the first nucleotide of the mRNA.
The
"5' region" of a gene refers to a polynucleotide (double-stranded or single-
stranded) located within or at the 5' end of a gene, and includes, but is not
limited to, the 5' untranslated region, if that is present, and the 5' protein
coding region of a gene. The 5' region is not shorter than 8 nucleotides in
length and not longer than 1000 nucleotides in length. Other possible lengths
of the 5' region include but are not limited to 10, 20, 25, 50, 100, 200, 400,
and
500 nucleotides.
As used herein, the "3' end" refers to the end of an mRNA up to the last 1000
nucleotides or one third of the mRNA, where the 3' terminal nucleotide is that
terminal nucleotide of the coding or untranslated region that adjoins the poly-
A tail, if one is present. That is, the 3' end of an mRNA does not include the
poly-A tail, if one is present. The "3' region" of a gene refers to a
polynucleotide
(double-stranded or single-stranded) located within or at the 3' end of a
gene,
and includes, but is not limited to, the 3' untranslated region, if that is
present,
and the 3' protein coding region of a gene. The 3' region is not shorter than
8
nucleotides in length and not longer than 1000 nucleotides in length. Other
possible lengths of the 3' region include but are not limited to 10, 20, 25,
50,
100, 200, 400, and 500 nucleotides. As used herein, the "internal coding
region"
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
21
of a gene refers to a polynucleotide (double- stranded or single-stranded)
located between the 5' region and the 3' region of a gene as defined herein.
The "internal coding region" is not shorter than 8 nucleotides in length and
not
longer than 1000 nucleotides in length. Other possible lengths of the
"internal
coding region" include but are not limited to 10, 20, 25, 50, 100, 200, 400,
and
500 nucleotides. The 5', 3' and internal regions are non-overlapping and may,
but need not be contiguous, and may, but need not, add up to the full length
of
the corresponding gene.
As indicated above, assay based on micro array may involve attaching or
spotting of the probes in a solid support. As used herein, the terms
"attaching"
and "spotting" refer to a process of depositing a nucleic acid onto a
substrate to
form a nucleic acid array such that the nucleic acid is stably bound to the
substrate via covalent bonds, hydrogen bonds or ionic interactions.
As used herein, "stably associated" or "stably bound" refers to a nucleic acid
that is stably bound to a solid substrate to form an array via covalent bonds,
hydrogen bonds or ionic interactions such that the nucleic acid retains its
unique pre-selected position relative to all other nucleic acids that are
stably
associated with an array, or to all other pre-selected regions on the solid
substrate under conditions in which an array is typically analyzed (i.e.,
during
one or more steps of hybridization, washes, and/or scanning, etc.).
As used herein, "substrate" or "support" or "solid support" when referring to
an
array refers to a material having a rigid or semi-rigid surface. The support
may
be biological, non-biological, organic, inorganic, or a combination of any of
these, existing as particles, strands, precipitates, gels, sheets, tubing,
spheres,
beads, containers, capillaries, pads, slices, films, plates, slides, chips,
etc.
Often, the substrate is a silicon or glass surface, (poly)tetrafluoroethylene,
(poly) vinylidendifluoride, polystyrene, polycarbonate, a charged membrane,
such as nylon or nitrocellulose, or combinations thereof. Preferably, at least
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
22
one surface of the substrate will be substantially flat. The support may
optionally contain reactive groups, including, but not limited to, carboxyl,
amino, hydroxyl, thiol, and the like. In one embodiment, the support is
optically transparent.
It should be noted that other nucleic acid based assays may be used for
quantitative measurement of the marker genes expression level. For example,
Nuclease protection assays (including both ribonuclease protection assays and
Si nuclease assays) can be used to detect and quantitate the RNA products of
the marker genes of the invention. In nuclease protection assays, an antisense
probe (labeled with, e.g., radiolabeled or nonisotopic) hybridizes in solution
to
an RNA sample. Following hybridization, single-stranded, unhybridized probe
and RNA are degraded by nucleases. An acrylamide gel is used to separate the
remaining protected fragments.
It should be further noted that a standard Northern blot assay can also be
used
to ascertain an RNA transcript size and the relative amounts of RNA products
of the marker gene of the invention, in accordance with conventional Northern
hybridization techniques known to those persons of ordinary skill in the art.
The invention further contemplates the use of amino acid based molecules such
as proteins or polypeptides as detecting molecules disclosed herein and would
be known by a person skilled in the art to measure the protein products of the
marker genes of the invention. Techniques known to persons skilled in the art
(for example, techniques such as Western Blotting, Immunoprecipitation,
ELISAs, protein microarray analysis and the like can then be used to measure
the level of protein products corresponding to the marker genes of the
invention. As would be understood to a person skilled in the art, the measure
of
the level of expression of the protein products of the marker genes of the
invention requires a protein which specifically and/or selectively binds to
one
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
23
or more of the protein products corresponding to each marker genes of the
invention.
Thus, according to a particular embodiment, the invention provides an
alternative composition comprising as the detection molecule, an isolated
amino acid molecule. Accordingly, such detection molecule may be an isolated
polypeptide which binds selectively and specifically to a protein product of
at
least one marker gene selected from the group consisting of RAB3GAP1, RAB3
GTPase activating protein subunit 1(catalytic); NFAT5, nuclear factor of
activated T-cells 5, tonicity-responsive; MRPS6, mitochondrial ribosomal
protein S6; AUH, AU RNA binding protein/enoyl-Coenzyme A hydratase;
MID1IP1, MID1 interacting protein 1 (gastrulation specific G12 homolog
(zebrafish)); RGS16, regulator of G-protein signaling 16; MARCH7, membrane-
associated ring finger (C3HC4) 7; NR3C1, nuclear receptor subfamily 3, group
C, member 1 (glucocorticoid receptor); ELF1, E74-like factor 1(ets domain
transcription factor); RPS6KB1, ribosomal protein S6 kinase, 70kDa,
polypeptide 1; STAT5A, signal transducer and activator of transcription 5A;
YTHDF3, YTH domain family, member 3; DNAJC12, DnaJ (Hsp40) homolog,
subfamily C, member 12; IFI44L, interferon-induced protein 44-like; SARS,
seryl-tRNA synthetase; SMURF2, SMAD specific E3 ubiquitin protein ligase 2;
SFRS18, splicing factor, arginine/serine-rich 18; NR4A2, nuclear receptor
subfamily 4, group A, member 2; CDKNIB, cyclin-dependent kinase inhibitor
1B (p27, Kipl); and EIF3D, eukaryotic translation initiation factor 3, subunit
D, as set forth in Table 4.
"selectively binds" in the context of proteins encompassed by the invention
refers to the specific interaction of a any two of a peptide, a protein, a
polypeptide an antibody, wherein the interaction preferentially occurs as
between any two of a peptide, protein, polypeptide and antibody preferentially
as compared with any other peptide, protein, polypeptide and antibody. For
example, when the two molecules are protein molecules, a structure on the
first
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
24
molecule recognizes and binds to a structure on the second molecule, rather
than to other proteins. "Selective binding", as the term is used herein, means
that a molecule binds its specific binding partner with at least 2-fold
greater
affinity, and preferably at least 10-fold, 20-fold, 50-fold, 100-fold or
higher
affinity than it binds a non- specific molecule.
According to a specifically preferred embodiment, such detecting molecule may
be an isolated and purified antibody specific for the protein product of any
of
the marker genes used by the invention.
The term "antibody" also encompasses antigen-binding fragments of an
antibody. The term "antigen-binding fragment" of an antibody (or simply
"antibody portion," or "fragment"), as used herein, refers to one or more
fragments of a full-length antibody that retain the ability to specifically
bind to
a polypeptide encoded by one of the marker genes of the invention, or the
control reference genes. Examples of binding fragments encompassed within
the term "antigen- binding fragment" of an antibody include (i) a Fab
fragment,
a monovalent fragment consisting of the VL, VH, CL and CHI domains; (ii) a
F(ab')2 fragment, a bivalent fragment comprising two Fab fragments linked by
a disulfide bridge at the hinge region; (iii) a Fd fragment consisting of the
VH
and CH1 domains; (iv) a Fv fragment consisting of the VL and VH domains of a
single arm of an antibody, (v) a dAb fragment, which consists of a VH domain;
and (vi) an isolated complementarity determining region (CDR). Furthermore,
although the two domains of the Fv fragment, VL and VH, are coded for by
separate genes, they can be joined, using recombinant methods, by a synthetic
linker that enables them to be made as a single protein chain in which the VL
and VH regions pair to form monovalent molecules (known as single chain Fv
(scFv). Such single chain antibodies are also intended to be encompassed
within the term "antigen-binding fragment" of an antibody. These antibody
fragments are obtained using conventional techniques known to those with
skill in the art, and the fragments are screened for utility in the same
manner
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
as are intact antibodies. The antibody is preferably monospecific, e.g., a
monoclonal antibody, or antigen-binding fragment thereof. The term
"monospecific antibody" refers to an antibody that displays a single binding
specificity and affinity for a particular target, e.g., epitope. This term
includes
a "monoclonal antibody" or "monoclonal antibody composition", which as used
herein refer to a preparation of antibodies or fragments thereof of single
molecular composition.
It should be recognized that the antibody can be a human antibody, a chimeric
antibody, a recombinant antibody, a humanized antibody, a monoclonal
antibody, or a polyclonal antibody. The antibody can be an intact immuno
globulin, e.g., an IgA, IgG, IgE, IgD, 1gM or subtypes thereof. The antibody
can
be conjugated to a functional moiety (e.g., a compound which has a biological
or
chemical function. The antibody of the invention interacts with a polypeptide,
encoded by one of the marker genes of the invention, with high affinity and
specificity.
Were the detection molecule is an antibody, the expression of any of the
marker genes may be determined according to a specific embodiment, using an
immunoassay such as for example, an ELISA, a RIA, a slot blot, a dot blot,
immunohistochemical assay, FACS, a radio-imaging assay or a Western blot. It
should be noted that any combination of these assays may be also applicable.
Immuno assays for a protein of interest typically comprise incubating a
biological sample of a detectably labeled antibody capable of identifying a
protein of interest, and detecting the bound antibody by any of a number of
techniques well-known in the art.
As discussed in more detail, below, the term "labeled" can refer to direct
labeling of the antibody via, e.g., coupling (i.e., physically linking) a
detectable
substance to the antibody, and can also refer to indirect labeling of the
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
26
antibody by reactivity with another reagent that is directly labeled. Examples
of indirect labeling include detection of a primary antibody using a
fluorescenfly labeled secondary antibody.
It should be appreciated that all the detecting molecules (either nucleic acid
based or amino acid based) used by any of the compositions of the invention
are
isolated and/or purified molecules. As used herein, "isolated" or "purified"
when
used in reference to a nucleic acid means that a naturally occurring sequence
has been removed from its normal cellular (e.g., chromosomal) environment or
is synthesized in a non-natural environment (e.g., artificially synthesized).
Thus, an "isolated" or "purified" sequence may be in a cell-free solution or
placed in a different cellular environment. The term "purified" does not imply
that the sequence is the only nucleotide present, but that it is essentially
free
(about 90-95% pure) of non- nucleotide material naturally associated with it,
and thus is distinguished from isolated chromosomes. As used herein, the
terms "isolated" and "purified" in the context of a proteinaceous agent (e.g.,
a
peptide, polypeptide, protein or antibody) refer to a proteinaceous agent
which
is substantially free of cellular material and in some embodiments,
substantially free of heterologous proteinaceous agents (i.e. contaminating
proteins) from the cell or tissue source from which it is derived, or
substantially free of chemical precursors or other chemicals when chemically
synthesized. The language "substantially free of cellular material" includes.
preparations of a proteinaceous agent in which the proteinaceous agent is
separated from cellular components of the cells from which it is isolated or
recombinantly produced. Thus, a proteinaceous agent that is substantially free
of cellular material includes preparations of a proteinaceous agent having
less
than about 30%, 20%, 10%, or 5% (by dry weight) of heterologous proteinaceous
agent (e.g. protein, polypeptide, peptide, or antibody; also referred to as a
"contaminating protein"). When the proteinaceous agent is recombinantly
produced, it is also preferably substantially free of culture medium, i.e.
culture
medium represents less than about 20%, 10%, or 5% of the volume of the
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
27
protein preparation. When the proteinaceous agent is produced by chemical
synthesis, it is preferably substantially free of chemical precursors or other
chemicals, i.e., it is separated from chemical precursors or other chemicals
which are involved in the synthesis of the proteinaceous agent. Accordingly,
such preparations of a proteinaceous agent have less than about 30%, 20%,
10%, 5% (by dry weight) of chemical precursors or compounds other than the
proteinaceous agent of interest. Preferably, proteinaceous agents disclosed
herein are isolated.
As used herein the term "product of the marker gene" or "products of the
marker genes of the invention" refers to the RNA and/or the protein expressed
by the marker gene of the invention. In the case of RNA it refers to the RNA
transcripts transcribed from the marker gene of the invention. In the case of
protein it refers to proteins translated from the genes corresponding to the
marker gene of the invention. The "RNA product of a marker gene of the
invention" includes mRNA transcripts, and/or specific spliced variants of
mRNA whose measure of expression can be used as a marker gene in
accordance with the teachings disclosed herein. The "protein product of a
marker gene of the invention" includes proteins translated from the RNA
products of the marker genes of the invention. -
As shown by the following examples, samples obtained from carriers of
mutations in at least one of BRCA1 and BRCA2 genes exhibit differential
expression of at least one of said marker genes as compared to control samples
obtained from non-carrier subjects. Therefore, the composition of the
invention
may be used for detecting carriers of BRCA1 and BRCA2 gene mutations.
Thus, the invention further provides a diagnostic composition for the
detection
of at least one mutation in at least one of BRCA1 and BRCA 2 genes in a
biological sample of a mammalian subject. This particular diagnostic
composition comprises at least one isolated oligonucleotide or a collection of
at
least two isolated detecting oligonucleotides which specifically hybridizes to
a
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
28
nucleic acid sequence of RNA products of at least one marker gene or a
collection of at least two marker genes. More specifically, such marker genes
may be selected from the group consisting of RAB3GAP1, RAB3 GTPase
activating protein subunit 1(catalytic); NFAT5, nuclear factor of activated T-
cells 5, tonicity-responsive; MRPS6, mitochondrial ribosomal protein S6; AUH,
AU RNA binding protein/enoyl-Coenzyme A hydratase; MIDIIP1, MID1
interacting protein 1 (gastrulation specific G12 homolog (zebrafish)); RGS16,
regulator of G-protein signaling 16; MARCH7, membrane-associated ring
finger (C3HC4) 7; NR3C1, nuclear receptor subfamily 3, group C, member 1
(glucocorticoid receptor); ELF1, E74-like factor 1(ets domain transcription
factor); RPS6KB1, ribosomal protein S6 kinase, 70kDa, polypeptide 1;
STAT5A, signal transducer and activator of transcription 5A; YTHDF3, YTH
domain family, member 3; DNAJC12, DnaJ (Hsp40) homolog, subfamily C,
member 12; IFI44L, interferon-induced protein 44-like; SARS, seryl-tRNA
synthetase; SMURF2, SMAD specific E3 ubiquitin protein ligase 2; SFRS18,
splicing factor, arginine/serine-rich 18; NR4A2, nuclear receptor subfamily 4,
group A, member 2; CDKNIB, cyclin-dependent kinase inhibitor 1B (p27,
Kipl); and EIF3D, eukaryotic translation initiation factor 3, subunit D, as
set
forth in Table 4.
It should be noted that these marker genes were shown by the invention as
exhibiting a differential expression in lymphocytes from samples obtained from
BRCA1 or BRCA2 carriers under irradiation stress. Differential expression of
at least one of the marker genes of the invention as compared to a control
sample reflects the existence of at least one mutation in any one of BRCA1 and
BRCA2 and may therefore be indicative of an increased genetic predisposition
of said subject to a cancerous disorder, disease or condition associated with
mutations in any one of BRCA1 or BRCA2.
According to one specific embodiment, the invention provides a diagnostic
composition for the detection of at least one mutation of BRCA1 gene in a
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
29
biological sample of a subject. This particular diagnostic composition
comprises
at least one isolated oligonucleotide or a collection of at least two isolated
oligonucleotides which specifically hybridizes to a nucleic acid sequence of
RNA
products of at least one marker gene or a collection of at least two marker
genes selected from the group consisting of AUH, AU RNA binding
protein/enoyl-Coenzyme A hydratase; RGS16, regulator of G-protein signaling
16; MARCH7, menibrane-associated ring finger (C3HC4) 7; DNAJC12, DnaJ
(Hsp40) homolog, subfamily C, member 12; IFI44L, interferon-induced protein
44-like; SARS, seryl-tRNA synthetase; and SMURF2, SMAD specific E3
ubiquitin protein ligase 2.
It should be further appreciated that in case of detection of BRCAI mutation,
the marker gene may be selected from even a larger group of genes
demonstrated by the invention as having most consistent gene expression
patterns among all the samples. These genes are represented by genes 1 to 16
of the list disclosed by Table 2. In yet another embodiment, marker genes for
BRCAI gene mutations may be selected form genes exhibiting differential
expression of about 1.5 folds. Such genes may be selected from any of the
genes
set forth in Table 5.
In yet another alternative specific embodiment, the invention provides a
composition for the detection of at least one mutation of BRCA2 gene in a
biological sample of said subject. This particular composition comprises at
least
one isolated oligonucleotide or a collection of at least two isolated
oligonucleotides which specifically hybridizes to a nucleic acid sequence of
RNA
products of at least one marker gene or a collection of at least two marker
genes selected from the group consisting of RAB3GAP1, RAB3 GTPase
activating protein subunit 1 (catalytic); NFAT5, nuclear factor of activated T-
cells 5, tonicity-responsive; MRPS6, mitochondrial ribosomal protein S6;
MIDIIPI, MID1 interacting protein 1 (gastrulation specific G12 homolog
(zebrafish)); MARCH7, membrane-associated ring finger (C3HC4) 7; NR3C1,
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
nuclear receptor subfamily 3, group C, member 1(glucocorticoid receptor);
ELF1, E74-like factor 1(ets domain transcription factor); RPS6KB1, ribosomal
protein S6 kinase, 70kDa, polypeptide 1; STAT5A, signal transducer and
activator of transcription 5A; YTHDF3, YTH domain family, member 3;
SFRS18, splicing factor, arginine/serine-rich 18; NR4A2, nuclear receptor
subfamily 4, group A, member 2; CDKN1B, cyclin-dependent kinase inhibitor
1B (p27, Kipl); and EIF3D, eukaryotic translation initiation factor 3, subunit
D.
It should be further appreciated that in case of detection of BRCA2 mutations,
the marker gene may be selected from even a larger group of genes
demonstrated by the invention as having most consistent gene expression
patterns among all the samples. These genes are represented by genes 17 to 37
of the list disclosed by Table 2. In yet another embodiment, marker genes for
BRCA2 gene mutations may be selected form genes exhibiting differential
expression of about 2 folds. Such genes may be selected from any of the genes
set forth in Table 6.
According to one embodiment, the diagnostic compositions of the invention are
specifically used for detection of at lease one mutation in any one of BRCA1
and BRCA2 genes, comprises a nucleic acid based detection molecule. In such
case, the expression of the marker genes may be determined using a nucleic
acid amplification assay selected from the group consisting of a Real-Time
PCR, microarrays, PCR, in situ Hybridization and Comparative Genomic
Hybridization.
According to a specifically preferred embodiment, the composition of the
invention may comprise detecting molecules specifically adopted for Real Time
PCR assay as described herein before.
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
31
It should be further appreciated that these specific diagnostic compositions
of
the invention may comprise an amino-acid based detecting molecule, for
example, an isolated antibody. In such case, the expression of the marker
genes
may be determined by immuno assays, as described above.
According to a specifically preferred embodiment, the diagnostic composition
of
the invention may be used for detecting at least one mutation in any one of
BRCA1 and BRCA2 genes. Existence of mutations in any of these genes may be
indicative of an increased genetic predisposition of a subject to a cancerous
disorder associated with mutations in any one of BRCA1 and/or BRCA2.
According to one embodiment, this cancerous disorder may be breast, ovarian,
pancreas or prostate carcinoma. More specifically, such carcinoma may be any
one of breast carcinoma and ovarian carcinoma.
Thus, according to a preferred embodiment, the composition of the invention
may be applicable for detection, and preferably for early detection of breast
cancer. Breast cancer is a cancer of the glandular breast tissue. Worldwide,
breast cancer is the fifth most common cause of cancer death (after lung
cancer, stomach cancer, liver cancer, and colon cancer). In 2005, breast
cancer
~
caused 502,000 deaths (7% of cancer deaths; almost 1% of all deaths)
worldwide. Among women worldwide, breast cancer is the most common
cancer. It should be indicated that pathological and clinical categories of
breast
cancer are encompassed by the invention and include ductal carcinoma (65-
90%), Lobular carcinoma 10%, Inflammatory breast cancer, Medullary
carcinoma of the breast, Colloid carcinoma, Papillary carcinoma and
Metaplastic carcinoma.
Early breast cancer can in some cases present as breast pain (mastodynia) or a
painful lump. Since the advent of breast mammography, breast cancer is most
frequently discovered as an asymptomatic nodule on a mammogram, before
any symptoms are present. A lump under the arm or above the collarbone that
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
32
does not go away may be present. When breast cancer associates with skin
inflammation, this is known as inflammatory breast cancer. In inflammatory
breast cancer, the breast tumor itself is causing an inflammatory reaction of
the skin, and this can cause pain, swelling, warmth, and redness throughout
the breast. Changes in the appearance or shape of the breast can raise
suspicions of breast cancer.
Another reported symptom complex of breast cancer is Paget's disease of the
breast. This syndrome presents as eczematoid skin changes at the nipple, and
is a late manifestation of an underlying breast cancer.
Most breast symptoms do not turn out to represent underlying breast cancer.
Benign breast diseases such as fibrocystic mastopathy, mastitis, functional
mastodynia, and fibroadenoma of the breast are more common causes of breast
symptoms. The appearance of a new breast symptom should be taken seriously
by both patients and their doctors, because of the possibility of an
underlying
breast cancer at almost any age.
Occasionally, breast cancer presents as metastatic disease, that is, cancer
that
,. ~
has spread beyond the original organ. Metastatic breast cancer will cause
symptoms that depend on the location of metastasis.
Moreover, it should be noted that each marker gene of the present invention,
is
described herein as a marker for detection of carriers of BRCAI or BRCA2 gene
mutations, and therefore may be regarded as a potential marker for breast
cancer. The marker genes of the invention might optionally be used alone or in
combination with one or more other breast cancer marker genes described
herein, and/or in combination with known markers for breast cancer, including
but not limited to Calcitonin, CA15-3 (Mucinl), CA27-29, TPA, a combination of
CA 15-3 and CEA, CA 27.29 (monoclonal antibody directed against MUCl),
Estrogen 2 (beta), HER-2 (c-erbB2), and/or in any combination thereof.
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
33
In yet another embodiment, the compositions of the invention may be
applicable for the diagnosis of ovarian carcinoma. Ovarian cancer is the most
common cause of cancer death from gynecologic tumors in the United States.
Early disease causes minimal, nonspecific, or no symptoms. Therefore, most
patients are diagnosed in an advanced stage. Overall, prognosis for these
patients remains poor. Standard treatment involves aggressive debulking
surgery followed by chemotherapy.
Ovarian carcinoma can spread by local extension, lymphatic invasion,
intraperitoneal implantation, hematogenous dissemination, and
transdiaphragmatic passage. Intraperitoneal dissemination is the most
common and recognized characteristic of ovarian cancer. Malignant cells can
implant anywhere in the peritoneal cavity but are more likely to implant in
sites of stasis along the peritoneal fluid circulation.
It should be noted that in some embodiments, the marker genes of the
invention or any polypeptides and/or polynucleotides derived therefrom may be
used in the diagnosis of ovarian cancer, alone or in combination with one or
~. ~
more polypeptides and/or polynucleotides of this invention, and/or in
combination with known markers for ovarian cancer, including but not limited
to CEA, CA125 (Mucin 16), CA72-4TAG, CA-50, CA 54-61, CA-195 and CA 19-9
in combination with CA-125, and/or in combination with the known protein(s)
associated with the indicated polypeptide or polynucleotide, as described
herein.
According to another embodiment, the dagnostic composition of the invention
may be used for detection of prostate carcinoma. Prostate cancer is an
important growing health problem, presenting a challenge to urologists,
radiologists, and oncologists. Prostate cancer is the most common
nondermatologic cancer, yet despite this frequent occurrence, the clinical
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
34
course is often unpredictable. Most prostate cancers are slow growing and do
not manifest themselves during the man's lifetime. Approximately 95% of
prostate cancers are adenocarcinomas that develop in the acini of the
prostatic
ducts. Other rare histopathologic types of prostate cancer occur in
approximately 5% of patients, these include small cell carcinoma, mucinous
carcinoma, endometrioid carcinoma (prostatic ductal carcinoma), transitional
cell carcinoma, squamous cell carcinoma, basal cell carcinoma, adenoid cystic
carcinoma (basaloid), signet-ring cell carcinoma, and neuroendocrine
carcinoma.
Still further, the composition of the invention may be useful for the
diagnosis of
pancreatic carcinoma.
Pancreatic cancer is the fourth leading cause of death from cancer in the
United States. The disease is slightly more common in men than in women,
and risk increases with age.
The cause is unknown, but it is more common in smokers and in obese
individuals. There is controversy as to whether type 2 diabetes is a risk
factor
for pancreatic cancer. A small number of cases are known to be related to
syndromes that are passed down through families. Pancreatic cancers can arise
from both the exocrine and endocrine portions of the pancreas. Of pancreatic
tumors, 95% develop from the exocrine portion of the pancreas, including the
ductal epithelium, acinar cells, connective tissue, and lymphatic tissue.
According to optional but preferred embodiments of the present invention, any
marker gene according to the present invention may optionally be used alone
or in combination. Such a combination may optionally comprise a plurality of
marker genes described herein, optionally including any subcombination of
marker genes, and/or a combination featuring at least one other marker genes,
for example a known marker gene. Furthermore, such a combination may
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
optionally and preferably be used as described above with regard to
determining a ratio between a quantitative or semi-quantitative measurement
of any marker gene described herein to any other marker gene described
herein, and/or any other known marker gene, and/or any other marker. As
used herein, "a plurality of "a collection of' "a combination of' or "a set of
refers to more than two, for example, 3 or more, 4 or more, 5 or more, 6 or
more, 7 or more, 8 or more, 9 or more and 10 or more. The present invention
thus encompasses any combination of the genes described by Table 4. For
example, a combination of 11 or more, 12 or more, 13 or more, 14 or more, 15
or
more, 16 or more, 17 or more, 18 or more, 19 or more and 20 or more genes.
According to one optional embodiment, the compositions described by the
invention or any components thereof, specifically, the detecting molecules may
be attached to a solid support. The solid support may include polymers, such
as
polystyrene, agarose, Sepharose, cellulose, glass, glass beads and
magnetizable
particles of cellulose or other polymers. The solid-support can be in the form
of
large or small beads, chips or particles, tubes, plates, or other forms.
A particular and non-limiting example of a diagnostic composition for
detecting
carriers of BRCAI and BRCA2 gene mutations, may comprises at least one or a
collection of at least two detecting molecules specific for at least one of
the
marker genes as set for the in Table 4. It should be noted that preferred
detecting molecules may be probes and primers derived from these genes. More
specifically, such primers and probes are suitable for Real-Time RT-PCR
reaction, specifically, the TaqMan reaction as described by the examples.
According to a particularly specific embodiment, such primers and probes may
be derived from any of the amplicons as presented by Table 4.
In yet another optional embodiment, any of the compositions of the invention
optionally further comprise at least one detecting molecule or a collection of
at
least two detecting molecules specific for determination of the expression of
at
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
36
least one control reference gene. Such reference control genes may be for
example, RPS9, HSPCB, Eukaryotic 18S-rRNA and R-actin.
According to another aspect, the invention relates to a method for the
detection
of at least one mutation in at least one of BRCA1 and BRCA 2 genes in a
biological test sample of a mammalian subject. The method of the invention
comprises the steps of: (a) determining the level of at least one marker gene
in
the test biological sample and in a suitable control sample. In a particular
embodiment, these marker genes may be selected from the group consisting of:
RAB3GAP1, RAB3 GTPase activating protein subunit 1(catalytic); NFAT5,
nuclear factor of activated T-cells 5, tonicity-responsive; MRPS6,
mitochondrial
ribosomal protein S6; AUH, AU RNA binding protein/enoyl-Coenzyme A
hydratase; MIDIIP1, MID1 interacting protein 1(gastrulation specific G12
homolog (zebrafish)); RGS16, regulator of G-protein signaling 16; MARCH7,
membrane-associated ring finger (C3HC4) 7; NR3C1, nuclear receptor
subfamily 3, group C, member 1(glucocorticoid receptor); ELF1, E74-like factor
1 (ets domain transcription factor); RPS6KB1, ribosomal protein S6 kinase,
70kDa, polypeptide 1; STAT5A, signal transducer and activator of
transcription 5A; YTHDF3, YTH domain family, member 3; DNAJC12, DnaJ
(Hsp40) homolog, subfamily C, member 12; IFI44L, interferon-induced protein
44-like; SARS, seryl-tRNA synthetase; SMURF2, SMAD specific E3 ubiquitin
protein ligase 2; SFRS18, splicing factor, arginine/serine-rich 18; NR4A2,
nuclear receptor subfamily 4, group A, member 2; CDKNIB, cyclin-dependent
kinase inhibitor 1B (p27, Kipl); and EIF3D, eukaryotic translation initiation
factor 3, subunit D, as set forth in Table 4; (b) optionally, determining the
level
of expression of at least one control gene in the test sample and in a
suitable
control sample. According to a specific embodiment, the control gene may be at
least one of RPS9, HSPCB, Eukaryotic 18S-rRNA and (3-actin; (c) comparing
the level of expression as obtained by step (a) of each of the marker genes in
the test sample with the level of expression in the control sample; and
optionally (d) comparing the level of expression as obtained by step (b) of
each
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
37
of the control genes in the test sample with the level of expression in the
control sample.
It should be appreciated that the detection of a difference in the level of
expression of at least one of the marker genes in the test sample as compared
to a control sample according to step (c), may indicate that the test subject
is a
carrier of at least one mutation in at least one of BRCAI and BRCA2 genes.
Moreover, it should be noted that were control genes are also examined,
detection of no difference in the level of expression of the control genes in
the
test sample as compared to the control sample according to step (d), and a
differential expression of the marker genes, even reinforce the indication
that
the test sample is of a carrier of BRCA1 /2 gene mutation.
As used herein in this specific embodiment, the term "marker gene" refers to a
gene that is differentially regulated as between a carrier of mutations in any
one of BRCA1 or BRCA2 genes and a non-carrier individual.
"Differentially expressed" can also include a measurement of the RNA or
protein encoded by the marker gene of the invention in a sample or plurality
of
samples as compared with the amount or level of RNA or protein expression in
a second sample or population or plurality of samples, specifically, a control
sample of non-carrier subject. Differential expression can be determined as
described herein and as would be understood by a person skilled in the art.
The
term "differentially expressed" or "changes or difference in the level of
expression" refers to an increase or decrease in the ineasurable expression
level
of a given marker gene as measured by the amount of RNA and/or the amount
of protein in a sample as compared with the measurable expression level of a
given marker gene in a second sample, specifically, a control sample. The term
"differentially expressed" or "changes or differences in the level of
expression"
can also refer to an increase or decrease in the measurable expression level
of a
given marker gene in a population of samples as compared with the
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
38
measurable expression level of a marker gene in a second population of
samples, for example, a control sample obtained from a non-carrier subject. As
used herein, "differentially expressed" can be measured using the ratio of the
level of expression of a given marker gene(s) as compared with the mean
expression level of the given marker gene(s) of a control sample wherein the
ratio is not equal to 1Ø Differentially expressed can also be measured using
p-
value. When using p-value, a marker gene is identified as being differentially
expressed as between a first and second population when the p-value is less
than 0.1. More preferably the p-value is less than 0. 05. Even more preferably
the p-value is less than 0.01. More preferably, the p-value is less than
0.005.
Most preferably, the p-value is less than 0.001. When determining
differentially expression on the basis of the ratio, an RNA or protein is
differentially expressed if the ratio of the level of expression in a first
sample
as compared with a second sample is greater than or less than 1Ø For
example, a ratio of greater than 1.2, 1.5, 1.7, 2, 3, 4, 5, 6, 7, 8, 9, 10, 20
or a
ratio less than 1, for example 0.8, 0.6, 0.4, 0.2, 0.1. 0.05. In another
specific
embodiment of the invention, a nucleic acid transcript is differentially
expressed if the ratio of the mean of the level of expression of a first
population
.as compared with the mean level of expression of the second population is
greater than or less than 1Ø For example, a ratio of greater than 1.2, 1.5,
1.7,
2, 3, 4, 5, 6, 7, 8, 9, 10, 20, or a ratio less than 1, for example 0.9, 0.8,
0.6, 0.4,
0.3, 0.2, 0.1, 0.05 or 0.01. In another embodiment of the invention, a nucleic
acid transcript is differentially expressed if the ratio of its level of
expression in
a first sample as compared with the mean of the second population is greater
than or less than 1.0 and includes for example, a ratio of greater than 1.2,
1.5,
1.7, 2, 3, 4, 5, 6, 7, 8, 9, 10, 20, or a ratio less than 1, for example 0.9,
0.8, 0.6,
0.4, 0.3, 0.2, 0.1, 0.05 or 0.01.
More specifically, "Differentially increased expression" or "up regulation"
refers
to genes which demonstrate at least 10% or more, for example, 20%, 30%, 40%,
or 50%, 60%, 70%, 80%, 90% or more, or 1.1 fold, 1.2 fold, 1.4 fold, 1.6 fold,
1.8
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
39
fold, or more increase in gene expression (as measured by RNA expression or
protein expression), relative to a control sample.
"Differentially decreased expression" or "down regulation" refers to genes
which demonstrate at least 10% or more; for example, 20%, 30%, 40%, or 50%,
60%, 70%, 80%, 90% or a less than 1.0 fold, 0.8 fold, 0.6 fold, 0. 4 fold, 0.2
fold,
0.1 fold or less decrease in gene expression (as measured by RNA expression or
protein expression), relative to a control.
It should be further noted that in case the expression level of more than one
marker gene is examined by the diagnostic method of the invention, it may
reflect and result in "gene expression pattern" or "gene expression profile"
of
the diagnosed individual. As used herein, a "gene expression pattern" or "gene
expression profile" indicates the combined pattern of the results of the
analysis
of the level of expression of at least one, preferably, at least two or more
marker genes of the invention including 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13,
14, 15,
16, 17, 18, 19, 20, 21 or more or all of the markers of the invention. A gene
expression pattern or gene expression profile can result from the measurement
of expression of the RNA or protein products of the marker genes of the
invention and can be done using any known technique. For example,
techniques to measure expression of the RNA products of the marker genes of
the invention includes, PCR based methods (including RT- PCR) and non PCR
based method as well as micro-array analysis. To measure protein products of
the marker genes of the invention, techniques include western blotting and
ELISA analysis.
More particularly, according to this embodiment, determination of the
expression of the marker genes as well as of the control genes may be
performed by the following steps:
The first step (i) involves providing an array comprising:
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
(A) at least one detecting nucleic acid or amino acid molecule specific for
determination of the expression of at least one of said marker genes. The
detecting molecule may be a set of primers, a probe or both or alternatively
or
additionally, an antibody. It should be noted that each of said detecting
molecules is located in a defined position in said array. Optionally, said
array
may further comprise (B) at least one detecting nucleic acid or amino acid
molecule specific for determination of the expression of at least one of the
control genes. Each of the detecting molecules is located in a defined
position in
the array;
The second step (ii), involves contacting aliquots of the test sample and
particularly, nucleic acids '(RNA samples) or protein product prepared from
the
irradiated lymphocytes, and aliquots of the control samples with the detecting
molecules (primers, probes or both or antibodies) comprised in said array of
(i)
under conditions allowing for detection of the expression of the marker genes
and the control genes in both the test and the control samples. The third step
(iii), involves determining the level of the expression of the marker genes
and
optionally, the control genes in the test and control samples by suitable
means.
Preferably, by Real Time-PCR or micro-arrays, as indicated in detail herein
before.
The term "array" as used by the methods and kits of the invention refers to an
"addressed" spatial arrangement of the detecting molecules specific for the
marker genes of (A) and, optionally, the detecting molecules specific for the
control genes of (B). Each "address" of the array is a predetermined specific
spatial region containing a detecting molecule. For example, an array may be
a plurality of vessels (test tubes), plates (or even different predetermined
locations in one plate or one slide, micro-wells in a micro-plate each
containing
a different detecting molecule. An array may also be any solid support holding
in distinct regions (dots, lines, columns) different detecting molecules. The
array preferably includes built-in appropriate controls, for example, regions
without the sample, regions without any detecting molecules, regions without
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
41
either, namely with solvent and reagents alone. Solid support used for the
array of the invention will be described in more detail herein after, in
connection with the kits provided by the invention.
Reference to "determining" as used by the methods of the present invention,
includes estimating, quantifying, calculating or otherwise deriving a level of
expression of the marker or control genes by measuring an end point indication
that may be for example, the appearance of a detectable product.
It should be appreciated that the detection step may be performed using the
tested sample as obtained from the tested subject, or alternatively, may be
performed using any constituent or material derived or prepared therefrom. As
a non-limiting example, it should be noted that the method of the invention
further encompasses the use of nucleic acid molecules and or proteins prepared
from the tested sample.
According to one preferred embodiment the detecting molecule used for the
diagnostic method of the invention may be an isolated nucleic acid molecule or
an isolated amino acid molecule, or any combination thereof.
According to one alternative and preferred embodiment, the method of the
invention uses as a detecting molecule an isolated nucleic acid molecule. More
specifically, such nucleic acid molecule may be an isolated oligonucleotide
which specifically hybridizes to a nucleic acid sequence of the RNA products
of
at least one marker gene selected from the group consisting of: RAB3GAP1,
RAB3 GTPase activating protein subunit 1(catalytic); NFAT5, nuclear factor
of activated T-cells 5, tonicity-responsive; MRPS6, mitochondrial ribosomal
protein S6; AUH, AU RNA binding protein/enoyl-Coenzyme A hydratase;
MIDIIPI, MIDl interacting protein 1(gastrulation specific G12 homolog
(zebrafish)); RGS16, regulator of G-protein signaling 16; MARCH7, membrane-
associated ring finger (C3HC4) 7; NR3C1, nuclear receptor subfamily 3, group
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
42
C, member 1(glucocorticoid receptor); ELF1, E74-like factor 1 (ets domain
transcription factor); RPS6KB1, ribosomal protein S6 kinase, 70kDa,
polypeptide 1; STAT5A, signal transducer and activator of transcription 5A;
YTHDF3, YTH domain family, member 3; DNAJC12, DnaJ (Hsp40) homolog,
subfamily C, member 12; IFI44L, interferon-induced protein 44-like; SARS,
seryl-tRNA synthetase; SMURF2, SMAD specific E3 ubiquitin protein ligase 2;
SFRS18, splicing factor, arginine/serine-rich 18; NR4A2, nuclear receptor
subfamily 4, group A, member 2; CDKNIB, cyclin-dependent kinase inhibitor
1B (p27, Kipl); and EIF3D, eukaryotic translation initiation factor 3, subunit
D, as set forth in Table 4.
It should be appreciated that further genes may serve as marker genes by the
method of the invention. For example, any of the genes demonstrating a
significant differential expression listed in Table 2.
Accordingly, the detecting molecule specific for the control reference genes
may
be therefore an isolated nucleic acid molecule, and preferably, an isolated
oligonucleotide which specifically hybridizes to a nucleic acid sequence of
the
RNA products of at least one control reference gene. Examples for possible
control genes may be RPS9, HSPCB, Eukaryotic 18S-rRNA and (3-actin.
According to a specifically preferred embodiment, the oligonucleotide used as
a
detecting molecule by the method of the invention may be for example, a pair
of
primers, a nucleotide probe or any combination thereof.
In a specific embodiment, the primers and probes used by the method of the
invention may be selected from the amplicons defined by Table 4. Nevertheless,
it should be appreciated that any region of such marker genes may be used as
an amplicon and therefore as a possible region for targeting primers and
probes.
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
43
Accordingly, the expression of the marker gene and of the control reference
gene may be determined according to a preferred embodiment, using a nucleic
acid amplification assay such as Real Time PCR, micro arrays, PCR, in situ
Hybridization and Comparative Genomic Hybridization, as described in detail
herein before.
According to an alternative embodiment, the method of the invention uses an
isolated amino acid molecule as the detecting molecule. Such detecting
molecule may be therefore an isolated polypeptide which binds selectively to
the protein product of at least one marker gene selected from the group
consisting ofRAB3GAP1, RAB3 GTPase activating protein subunit 1
(catalytic); NFAT5, nuclear factor of activated T-cells 5, tonicity-
responsive;
MRPS6, mitochondrial ribosomal protein S6; AUH, AU RNA binding
protein/enoyl-Coenzyme A hydratase; MIDIIP1, MID1 interacting protein 1
(gastrulation specific G12 homolog (zebrafish)); RGS16, regulator of G-protein
signaling 16; MARCH7, inembrane-associated ring finger (C3HC4) 7; NR3C1,
nuclear receptor subfamily 3, group C, member 1(glucocorticoid receptor);
ELF1, E74-like factor 1 (ets domain transcription factor); RPS6KB1, ribosomal
protein S6 kinase, 70kDa, polypeptide 1; STAT5A, signal transducer and
activator of transcription 5A; YTHDF3, YTH domain family, member 3;
DNAJC12, DnaJ (Hsp40) homolog, subfamily C, member 12; IFI44L,
interferon-induced protein 44-like; SARS, seryl-tRNA synthetase; SMURF2,
SMAD specific E3 ubiquitin protein ligase 2; SFRS18, splicing factor,
arginine/serine-rich 18; NR4A2, nuclear receptor subfamily 4, group A,
member 2; CDKN1B, cyclin-dependent kinase inhibitor 1B (p27, Kipl); and
EIF3D, eukaryotic translation initiation factor 3, subunit D, as set forth in
Table 4.
Accordingly, the detecting molecule for the control reference genes may be an
isolated polypeptide which binds selectively to a protein product of at least
one
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
44
control reference gene. For example, RPS9, HSPCB, Eukaryotic 18S-rRNA and
(3-actin.
According to a specifically preferred embodiment, the detecting molecule used
by the method of the invention, may be an isolated antibody. In such case, the
expression may be determined using an immunoassay selected from the group
consisting of an ELISA, a RIA, a slot blot, a dot blot, immunohistochemical
assay, FACS, a radio-imaging assay or a Western blot, as described herein
before.
According to a particular embodiment, the invention provides a specific method
for the detection of at least one mutation of BRCA1 gene in a biological
sample
of a tested subject. According to this particular embodiment, the marker gene
or a collection of at least two marker genes may be selected from the group
consisting of: AUH, AU RNA binding protein/enoyl-Coenzyme A hydratase;
RGS16, regulator of G-protein signaling 16; MARCH7, membrane-associated
ring finger (C3HC4) 7; DNAJC12, DnaJ (Hsp40) homolog, subfamily C,
member 12; IFI44L, interferon-induced protein 44-like; SARS, seryl-tRNA
synthetase; and SMURF2, SMAD specific E3 ubiquitin protein ligase 2.
It should be further appreciated that in case of detection of BRCA1 mutation,
the marker gene may be selected from even a larger group of genes
demonstrated by the invention as having most consistent gene expression
patterns among all the samples. These genes are represented by genes 1 to 16
of the list disclosed by Table 2. In yet another embodiment, marker genes for
BRCA1 gene mutations may be selected form genes exhibiting differential
expression of about 1.5 folds. Such genes may be selected from any of the
genes
set forth in Table 5.
According to another particular embodiment, the invention provides a specific
method for the detection of at least one mutation of BRCA2 gene in a
biological
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
sample of a tested subject. According to this particular embodiment, the
marker gene or a collection of at least two marker genes may be selected from
the group consisting of: RAB3GAP1, RAB3 GTPase activating protein subunit
1 (catalytic); NFAT5, nuclear factor of activated T-cells 5, tonicity-
responsive;
MRPS6, mitochondrial ribosomal protein S6; MID1IP1, MID1 interacting
protein 1 (gastrulation specific G12 homolog (zebrafish)); MARCH7,
membrane-associated ring finger (C3HC4) 7; NR3C1, nuclear receptor
subfamily 3, group C, member 1(glucocorticoid receptor); ELF1, E74-like factor
1 (ets domain transcription factor); RPS6KB1, ribosomal protein S6 kinase,
70kDa, polypeptide 1; STAT5A, signal transducer and activator of
transcription 5A; YTHDF3, YTH domain family, member 3; SFRS18, splicing
factor, arginine/serine-rich 18; NR4A2, nuclear receptor subfamily 4, group A,
member 2; CDKNIB, cyclin-dependent kinase inhibitor 1B (p27, Kipl); and
EIF3D, eukaryotic translation initiation factor 3, subunit D.
It should be further appreciated that in case of detection of BRCA2 mutations,
the marker gene may be selected from even a larger group of genes
demonstrated by the invention as having most consistent gene expression
patterns among all the samples. These genes are represented by genes 17 to 37
of the list disclosed by Table 2. In yet another embodiment, marker genes for
BRCA2 gene mutations may be selected form genes exhibiting differential
expression of about 2 folds. Such genes may be selected from any of the genes
set forth in Table 6.
The present invention relates, in some enibodiments, to diagnostic assays,
which in some embodiments, utilizes a biological sample taken from a subject
(patient, in some embodiments or carrier), which for example may comprise
any biological sample, such as body fluid or secretion including but not
limited
to seminal plasma, blood, serum, urine, prostatic fluid, seminal fluid, semen,
the external secretions of the skin, respiratory, intestinal, and
genitourinary
tracts, tears, cerebrospinal fluid, sputtun, saliva, milk, peritoneal fluid,
pleural
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
46
fluid, cyst fluid, secretions of the breast ductal system (and/or lavage
thereof),
broncho alveolar lavage, lavage of the reproductive.system, lavage of any
other
part of the body or system in the body, samples of any organ including but not
limited to lung, colon, ovarian and/or breast tissue, feaces or a tissue
sample,
any cells derived therefrom, or any combination thereof. In some embodiments,
the term encompasses samples of in vitro or ex vivo cell culture or cell
culture
constituents. The sample can optionally be diluted with a suitable eluant
before contacting the sample with the detecting molecule/s of the invention
and/or performing any other diagnostic assay.
As used herein, "patient", "subject", "carrier" or "individual" refers to a
mammal, preferably human, that is diagnosed by the method of the invention.
According to a particular and specific optional embodiment, were the sample
used comprises cells obtained from the tested subject, the method of the
invention may comprise an additional step. The additional step includes
induction of DNA damage by treating the cells with an agent inducing such
damage. This may be performed by exposing the cells to irradiation as
demonstrated by the following examples. It should be noted that such
additional step may be referabl as a
p y performed preliminary step prior to
determination of the expression levels or profile of the marker genes or the
control reference genes.
Thus, according to a specific and particular embodiment, the invention
provides a method for the detection of at least one mutation in at least one
of
BRCA1 and BRCA2 genes in a biological test sample of a mammalian subject.
According to this embodiment, the diagnostic method comprises the steps of:
(a) providing a nucleic acid sample prepared from lymphocytes of a tested
mammalian subject and a nucleic acid sample obtained from lymphocytes of a
suitable control. It should be noted that in order to induce DNA damage, the
lymphocytes were irradiated prior to nucleic acid preparation;
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
47
(b) determining the level of expression of at least one of the marker genes
identified by the invention, in said test sample and in a suitable control
sample.
Optionally, (c) determining the level of expression of at least one control
gene
in said test sample and in a suitable control sample, wherein said at least
one
control gene may be any one of RPS9, HSPCB, Eukaryotic 18S-rRNA and ~3-
actin;
(d) comparing the level of expression as obtained by step (b) of each of the
marker genes in the test sample with the level of expression in the control
sample; and optionally, (e) comparing the level of expression as obtained by
the
optional step (c) of each of the control genes in said test sample with the
level
of expression in the control sample.
It should be noted that detecting a difference in the level of expression, or
as
also indicated by the invention a "differential expression" of at least one of
the
marker genes in the test sample as compared to the control sample according
to step (c), is indicative of that the tested subject is a carrier of at least
one
mutation in at least one of BRCAI and BRCA2 genes.
It should be further appreciated, that when the control reference gene are
also
examined, no difference in the level of expression of the control genes is
expected when the tested sample is compared to a control sample according to
step (d). Therefore, a differential expression in the marker genes and no
difference in the expression of the control genes, indicates that the tested
subject is a carrier of at least one gene mutation in at least one of BRCA1
and
BRCA2.
More particularly, according to this embodiment, determination of the
expression of the marker genes and optionally, of the control genes may be
performed by the following steps:
The first step (i), involves providing an array comprising:
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
48
(A) at least one detecting nucleic acid molecule specific for determination of
the
expression of at least one of said marker genes. The detecting nucleic acid
inolecule may be a set of primers, a probe or both. It should be noted that
each
of said detecting molecules is located in a defined position in said array;
and
optionally,
(B) at least one detecting nucleic acid molecule specific for determination of
the
expression of at least one of the control genes. Each of the detecting nucleic
acid molecules is located in a defined position in the array.
The second step (ii), involves contacting aliquots of the test sample and
particularly, nucleic acids (RNA samples) product prepared from the irradiated
lymphocytes, and aliquots of the control sample with the detecting nucleic
acid
molecules (primers, probes or both) comprised in said array of (i) under
conditions allowing for detection of the expression of the marker genes and
the
control genes in both the test and the control samples; and
The third step (iii), involves determining the level of the expression of the
marker genes and optionally, the control genes in the test and control samples
by suitable means. Preferably, by Real Time-PCR or micro-arrays, as indicated
in detail herein before.
It should be further noted that the detection of a mutation in any one of
BRCA1 or BRCA2 genes by the method of the invention may be an indicative of
an increased genetic predisposition of the diagnosed subject to a cancerous
disorder associated with at least one mutation in at least one of BRCA1 and
BRCA2 genes.
Such cancerous disorders may be for example, breast, ovarian, pancreas and
prostate carcinoma.
It should thus be appreciated that the method of the invention may provide
early detection of such cancerous disorders. Therefore, the invention may be
applicable and therefore provides a diagnostic method for the diagnosis,
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
49
preferably, early detection of breast, ovarian, pancreas and prostate
carcinoma,
and particularly of breast carcinoma and ovarian carcinoma.
It should be thus noted that this invention may provides diagnostic methods
optionally applicable in the selection of a particular therapy, or
optimization of
a given therapy for a disease, disorder or condition.
Another aspect of the invention relates to a kit comprising:
(a) means for obtaining a sample of a mammalian subject;
(b) at least one detecting molecule or a collection of at least two detecting
molecules specific for determination of the expression of at least one marker
gene or a collection of at least two marker genes selected from the group
consisting of: RAB3GAP1, RAB3 GTPase activating protein subunit 1
(catalytic); NFAT5, nuclear factor of activated T-cells 5, tonicity-
responsive;
MRPS6, mitochondrial ribosomal protein S6; AUH, AU RNA binding
protein/enoyl-Coenzyme A hydratase; MIDIIPI, MID1 interacting protein 1
(gastrulation specific G12 homolog (zebrafish)); RGS16, regulator of G-protein
signaling 16; MARCH7, membrane-associated ring finger (C3HC4) 7; NR3C1,
nuclear receptor subfamily 3, group C, member 1(glucocorticoid receptor);
ELF1, E74-like factor 1(ets domain transcription factor); RPS6KB1, ribosomal
protein S6 kinase, 70kDa, polypeptide 1; STAT5A, signal transducer and
activator of transcription 5A; YTHDF3, YTH domain family, member 3;
DNAJC12, DnaJ (Hsp40) homolog, subfamily C, member 12; IFI44L,
interferon-induced protein 44-like; SARS, seryl-tRNA synthetase; SMURF2,
SMAD specific E3 ubiquitin protein ligase 2; SFRS18, splicing factor,
arginine/serine-rich 18; NR4A2, nuclear receptor subfamily 4, group A,
member 2; CDKNIB, cyclin-dependent kinase inhibitor 1B (p27, Kipl); and
EIF3D, eukaryotic translation initiation factor 3, subunit D, as set forth in
Table 4.
(c) optionally, at least one detecting molecule or a collection of at least
two
detecting molecules specific for determination of the expression of at least
one
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
control reference gene or a collection of at least two control reference
genes.
According to a specific embodiment, these control reference genes may be
selected from the group consisting of: RPS9, HSPCB, Eukaryotic 18S-rRNA
and R-actin;
(d) at least one control sample that may be at least one of a negative control
sample and a positive control sample;
(e) instructions for carrying out the detection and quantification of
expression
of the marker genes and of the control reference gene in the tested sample;
(f) instructions for evaluating the differential expression of the marker gene
in
the tested sample and optionally of a control reference gene in the sample as
compared to the expression of the marker gene and optionally control reference
gene in the control sample.
It should be noted that the detecting molecule of the marker genes (b) or the
control genes (c), maybe provided by the kit of the invention attached or
connected to a solid support or to an array, as described herein before.
As used herein, the term "control" or "control sample" includes positive or
negative controls. In the context of this invention the term "positive
control"
refers to one or more samples isolated from an individual or group of
individuals who are classified as carrier of mutations in any one of BRCA1 or
BRCA2 genes. The term "negative control" refers to one or more samples
isolated from an individual or group of individuals who are classified as non-
carrier of mutations in any one of BRCA1 or BRCA2 genes.
According to an alternative or additional embodiment, instead of control
samples, the kit of the invention may comprise a standard curve/s illustrating
the expression of the marker genes and optionally of the control genes in
control samples.
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
51
According to one embodiment, the detecting molecule comprised within the kit
of the invention may be an isolated nucleic acid molecule or an isolated amino
acicl molecule, or any combination thereof.
According to one specific and preferred embodiment, the detecting molecule
com prised within the kit of the invention may be an isolated nucleic acid
molecule. Such molecule may be preferably, an isolated oligonucleotide which
specifically hybridizes to a nucleic acid sequence of the RNA products of at
least one marker gene selected from the group consisting of: RAB3GAP1, RAB3
GTPase activating protein subunit 1(catalytic); NFAT5, nuclear factor of
activated T-cells 5, tonicity-responsive; MRPS6, mitochondrial ribosomal
protein S6; AUH, AU RNA binding protein/enoyl-Coenzyme A hydratase;
MIDIIPI, MID1 interacting protein 1(gastrulation specific G12 homolog
(zebrafish)); RGS16, regulator of G-protein signaling 16; MARCH7, membrane-
associated ring finger (C3HC4) 7; NR3C1, nuclear receptor subfamily 3, group
C, iilember 1 (glucocorticoid receptor); ELF1, E74-like factor 1(ets domain
transcription factor); RPS6KB1, ribosomal protein S6 kinase, 70kDa,
polypeptide 1; STAT5A, signal transducer and activator of transcription 5A;
YTHDF3, YTH domain family, member 3; DNAJC12, DnaJ (Hsp40) homolog,
subfamily C, member 12; IFI44L, interferon-induced protein 44-like; SARS,
seryl-tRNA synthetase; SMURF2, SMAD specific E3 ubiquitin protein ligase 2;
SFRS18, splicing factor, arginine/serine-rich 18; NR4A2, nuclear receptor
subfamily 4, group A, member 2; CDKNIB, cyclin-dependent kinase inhibitor
1B (p27, Kipl); and EIF3D, eukaryotic translation initiation factor 3, subunit
D, as set forth in Table 4. It should be noted that the marker genes may be
also selected from any of the genes listed in Table 2.
Accordingly, the kit of the invention may therefore comprises as the detecting
molecule for the control reference genes, an oligonucleotide which
specifically
hybridizes to a nucleic acid sequence of the RNA products of at least one
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
52
control reference gene selected from the group consisting of: RPS9, HSPCB,
Eukaryotic 18S-rRNA and (3-actin.
According to a preferred embodiment, such oligonucleotide may be a pair of
printers or nucleotide probe or any combination, mixture or collection
thereof.
According to such specific and particular embodiment, the primers and probes
used by the kit of the invention may be derived from regions of the genes that
are also defined as amplicons (selected regions for amplification). Examples
for
amplicons used are demonstrated by Table 4, which also discloses partial
sequences of the amplicons (SEQ ID NO. 25 to 48) used in the following
Examples. It should be appreciated that primers and probes may be derived
from any other amplicon in the listed marker genes described by the invention.
According to another preferred optional embodiment, the kit of the invention
may further comprise at least one reagent for performing a nucleic acid
amplification based assay. Such nucleic acid amplification assay may be any
one of PCR, Real Time PCR, micro arrays, in situ Hybridization and
Comparative Genomic Hybridization.
~. ~
According to an alternative embodiment, the detecting molecule comprised
within the kit of the invention may be an isolated amino acid molecule, for
example, an isolated polypeptide which binds selectively to the protein
product
of at least one marker gene selected from the group consisting of: RAB3GAP1,
RAB3 GTPase activating protein subunit 1(catalytic); NFAT5, nuclear factor
of activated T-cells 5, tonicity-responsive; MRPS6, mitochondrial ribosomal
protein S6; AUH, AU RNA binding protein/enoyl-Coenzyme A hydratase;
MIDIIP1, MIDl interacting protein 1 (gastrulation specific G12 homolog
(zebrafish)); RGS16, regulator of G-protein signaling 16; MARCH7, membrane-
associated ring finger (C3HC4) 7; NR3C1, nuclear receptor subfamily 3, group
C, member 1(glucocorticoid receptor); ELFl, E74-like factor 1(ets domain
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
53
transcription factor); RPS6KB1, ribosoinal protein S6 kinase, 70kDa,
polypeptide 1; STAT5A, signal transducer and activator of transcription 5A;
YTHDF3, YTH domain family, member 3; DNAJC12, DnaJ (Hsp40) homolog,
subfamily C, member 12; IFI44L, interferon-induced protein 44-like; SARS,
seryl-tRNA synthetase; SMURF2, SMAD specific E3 ubiquitin protein ligase 2;
SFRS18, splicing factor, arginine/serine-rich 18; NR4A2, nuclear receptor
subfamily 4, group A, member 2; CDKNIB, cyclin-dependent kinase inhibitor
IB (p27, Kipl); and EIF3D, eukaryotic translation initiation factor 3, subunit
D, as set forth in Table 4.
Accordingly, the detecting molecule specific for the control reference genes
may
be an isolated polypeptide which binds selectively to the protein product of
at
least one control reference gene selected from the group consisting of RPS9,
HSPCB, Eukaryotic 18S-rRNA and P-actin.
According to a specifically preferred embodiment such detecting molecule may
be an isolated antibody.
It should be noted that the kit of the invention may optionally further
~.
comprises at least one reagent for performing an immuno assay, such as
ELISA, a RIA, a slot blot, a dot blot, immunohistochemical assay, FACS, a
radio-imaging assay, Western blot or any combination thereof.
According to a preferred embodiment, the kits provided by the invention may
further comprise suitable means and reagents for preparing or isolating at
least one of nucleic acids and amino acids from the examined sample.
As shown by the following examples, the marker genes of the invention
demonstrate a clear differential expression in carries of BRCA1/2 gene
mutations. Thus, the invention further provides a particular kit for detecting
of
at least one mutation in at lest of BRCAI and BRCA2 genes in a mammalian
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
54
test subject. This particular kit of the invention comprises: (a) means for
obtaining a sample of said subject; (b) at least one detecting molecule or a
collection of at least two detecting molecules specific for determination of
the
expression of at least one marker gene or a collection of at least two marker
genes. According to a particular embodiment, these marker genes may be
selected from the group consisting of: RAB3GAP1, RAB3 GTPase activating
protein subunit 1(catalytic); NFAT5, nuclear factor of activated T-cells 5,
tonicity-responsive; MRPS6, mitochondrial ribosomal protein S6; AUH, AU
RNA binding protein/enoyl-Coenzyme A hydratase; MID1IP1, MIDl
interacting protein 1(gastrulation specific G12 homolog (zebrafish)); RGS16,
regulator of G-protein signaling 16; MARCH7, membrane-associated ring
finger (C3HC4) 7; NR3C1, nuclear receptor subfamily 3, group C, member 1
(glucocorticoid receptor); ELF1, E74-like factor 1(ets domain transcription
factor); RPS6KB1, ribosomal protein S6 kinase, 70kDa, polypeptide 1;
STAT5A, signal transducer and activator of transcription 5A; YTHDF3, YTH
domain family, member 3; DNAJC12, DnaJ (Hsp40) homolog, subfamily C,
member 12; IFI44L, interferon-induced protein 44-like; SARS, seryl-tRNA
synthetase; SMURF2, SMAD specific E3 ubiquitin protein ligase 2; SFRS18,
splicing factor, arginine/serine-rich 18; NR4A2, nuclear receptor subfamily 4,
group A, member 2; CDKNIB, cyclin-dependent kinase inhibitor 1B (p27,
Kip1); and EIF3D, eukaryotic translation initiation factor 3, subunit D, as
set
forth in Table 4.
(c) optionally, at least one detecting molecule or a collection of at least
two
detecting molecules specific for determination of the expression of at least
one
control reference gene or a collection of at least two control reference
genes.
According to a specific embodiment, these control reference genes may be
selected from the group consisting of: RPS9, HSPCB, Eukaryotic 18S-rRNA
and (3-actin; (d) at least one control sample that may be at least one of a
negative control sample and a positive control sample, Alternatively or
additionally, the kit of the invention may comprise a standard curve/s
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
illustrating the expression of the marker genes and optionally of the control
genes in a control sample.
(e) instructions for carrying out the detection and quantification of
expression
of the marker genes and of the control reference gene in the tested sample;
(f) instructions for evaluating the differential expression of the marker gene
in
the tested sample and optionally of a control reference gene in the sample as
compared to the expression of the marker gene and optionally control reference
gene in the control sample.
According to one embodiment, the negative control may be obtained from a
non-carrier subject and a positive control may be obtained from a subject
which
is a carrier of at least one mutation in at least one of BRCA1 and BRCA2
genes.
According to one embodiment, the detecting molecule comprised within the kit
of the invention may be an isolated nucleic acid molecule or an isolated amino
acid molecule, or any combination thereof.
According to one specific and preferred embodiment, the detecting molecule
comprised within the kit of the invention may be an isolated nucleic acid
molecule. Such molecule may be preferably, an isolated oligonucleotide which
specifically hybridizes to a nucleic acid sequence of the RNA products of at
least one of the marker gene of the invention, as set forth in Table 4.
Accordingly, the kit of the invention may therefore comprises as the detecting
molecule for the control reference genes, an oligonucleotide which
specifically
hybridizes to a nucleic acid sequence of the RNA products of at least one
control reference gene selected from the group consisting of: RPS9, HSPCB,
Eukaryotic 18S-rRNA and (3-actin.
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
56
According to a preferred embodiment, such oligonucleotide may be a pair of
primers or nucleotide probe or any combination, mixture or collection thereof.
According to such specific and particular embodiment, the primers and probes
used by the kit of the invention may be derived from regions of the genes that
are also defined as amplicons (selected regions for amplification). Examples
for
amplicons used are demonstrated by Table 4, which also discloses partial
sequences of the amplicons (SEQ ID NO. 25 to 48) used in the following
Examples. It should be appreciated that primers and probes may be derived
from any other amplicon in the listed marker genes described by the invention.
In another embodiment, the present invention relates in part to kits
comprising sufficient materials for performing one or more of the diagnostic
methods described by the invention. In preferred embodiments, a kit includes
one or more materials selected from the following group in an amount
sufficient to perform at least one assay.
Thus, according to another optional embodiment, the kit of the invention may
further comprise at least one reagent for performing a nucleic acid
amplification based assay. Such nucleic acid amplification assay may be any
one of Real Time PCR, micro arrays, PCR, in situ Hybridization and
Comparative Genomic Hybridization.
Control nucleic acid members may be present on the array including nucleic
acid members comprising oligonucleotides or nucleic acids corresponding to
genomic DNA, housekeeping genes, vector sequences, plant nucleic acid
sequence, negative and positive control genes, and the like. Control nucleic
acid
members are calibrating or control genes whose function is not to tell whether
a particular "key" gene of interest is expressed, but rather to provide other
useful information, such as background or basal level of expression.
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
57
Preferred control samples may be selected from HSPCB, RPS9, RPL32 and (3-
actin. Optionally, other control nucleic acids may be spotted on the array and
used as target expression control nucleic acids.
According to an alternative embodiment, the detecting molecule comprised
within the kit of the invention may be an isolated amino acid molecule, for
example, an isolated polypeptide, which binds selectively to the protein
product
of at least one of the marker genes of the invention, as set forth in Table 4.
Accordingly, the detecting molecule specific for the control reference genes
may
be an isolated polypeptide which binds selectively to the protein product of
at
least one control reference gene selected from the group consisting of RPS9,
HSPCB, Eukaryotic 18S-rRNA and R-actin.
According to a specifically preferred embodiment such detecting molecule may
be an isolated antibody.
It should be noted that the kit of the invention may therefore further
comprises
at least one reagent for performing an immuno.assay, such as ELISA, a RIA, a
slot blot, a dot blot, immunohistochemical assay, FACS, a radio-imaging assay,
Western blot or any combination thereof.
According to a preferred embodiment, the kits provided by the invention may
further comprise suitable means and reagents for preparing or isolating at
least one of nucleic acids and amino acids from said sample.
The invention further provides specific kit for the detection of at least one
mutation of BRCAI gene in a biological sample of a subject, according to a
preferred embodiment, such kit may comprises detection molecule specific for a
marker gene or a collection of at least two marker genes. These specific genes
exhibiting a differential expression in BRCA1 carriers may be selected from
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
58
the group consisting of: AUH, AU RNA binding protein/enoyl-Coenzyme A
hydratase; RGS16, regulator of G-protein signaling 16; MARCH7, membrane-
associated ring finger (C3HC4) 7; DNAJC12, DnaJ (Hsp40) homolog, subfamily
C, member 12; IFI44L, interferon-induced protein 44-like; SARS, seryl-tRNA
synthetase; and SMURF2, SMAD specific E3 ubiquitin protein ligase 2.
It should be further appreciated that in case of detection of BRCAI mutation,
the marker gene may be selected from genes demonstrated by the invention as
exhibiting differential expression of about 1.5 folds. Such genes may be
selected from any of the genes set forth in Table 5.
Still further, the invention also provides a specific kit for the detection of
at
least one mutation of BRCA2 gene in a biological sample of a subject.
According to a preferred embodiment, such kit may comprises detection
molecule specific for a marker gene or a collection of at least two marker
genes.
These specific genes exhibiting a differential expression in BRCA2 carriers
may be selected from the group consisting of: RAB3GAP1, RAB3 GTPase
activating protein subunit 1 (catalytic); NFAT5, nuclear factor of activated T-
cells 5, tonicity-responsive; MRPS6, mitochondrial ribosomal protein S6;
MID1IP1, MIDl interacting protein 1(gastrulation specific G12 homolog
(zebrafish)); MARCH7, membrane-associated ring finger (C3HC4) 7; NR3C1,
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor);
ELF1, E74-like factor 1 (ets domain transcription factor); RPS6KB1, ribosomal
protein S6 kinase, 70kDa, polypeptide 1; STAT5A, signal transducer and
activator of transcription 5A; YTHDF3, YTH domain family, member 3;
SFRS18, splicing factor, arginine/serine-rich 18; NR4A2, nuclear receptor
subfamily 4, group A, member 2; CDKN1B, cyclin-dependent kinase inhibitor
1B (p27, Kipi); and EIF3D, eukaryotic translation initiation factor 3, subunit
D.
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
59
It should be further appreciated that in case of detection of BRCA2 mutations,
the marker gene may be selected from genes demonstrated by the invention as
exhibiting differential expression of about 2 folds. Such genes may be
selected
from any of the genes set forth in Table 6.
It should be noted that detection of a mutation in any one of BRCA1 or BRCA2
genes may be an indicative of an increased genetic predisposition of the
carrier
subject to a cancerous disorder associated with mutations in at least one of
BRCAI and BRCA2.
Such cancerous disorder may be any disorder of the group consisting of:
breast,
ovary, pancreas and prostate carcinomas. Therefore, the kits of the invention
may be applicable for the detection and preferably, the early detection of
such
cancerous disorders, particularly of breast carcinoma and ovarian carcinoma.
More specifically, for nucleic acid micoarray kits, the kits may generally
comprise probes attached to a support surface. The probes may be labeled with
a detectable label. In a specific embodiment, the probes are specific for an
exon(s), an intron(s), an exon junction(s), or an exon-intron junction(s)), of
RNA
products of 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18,19
and 20 or
more or any combination of the marker genes of the invention. The microarray
kits may comprise instructions for performing the assay and methods for
interpreting and analyzing the data resulting from the performance of the
assay. The kits may also comprise hybridization reagents and/or reagents
necessary for detecting a signal produced when a probe hybridizes to a target
nucleic acid sequence. Generally, the materials and reagents for the
microarray kits are in one or more containers. Each component of the kit is
generally in its own a suitable container.
For Real-Time RT-PCR kits, the kits generally comprise pre-selected primers
specific for particular RNA products (e.g., an exon(s), an intron(s), an exon
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
junction(s), and an exon- intron junction(s)) of 1, 2, 3, 4, 5, 6, 7, 8, 9,
10, 11, 12,
13, 14, 15, 16, 17, 18,19, 20, 21 or all or any combination of the marker
genes of
the invention. The RT-PCR kits may also comprise enzymes suitable for
reverse transcribing and/or amplifying nucleic acids (e.g., polymerases such
as
Taq), and deoxynucleotides and buffers needed for the reaction mixture for
reverse transcription and amplification. The RT- PCR kits may also comprise
probes specific for RNA products of 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13,
14, 15,
16, 17, 18, 19, 20 or more or all, or any combination of the marker genes of
the
invention. The probes may or may not be labeled with a detectable label (e.g.,
a
fluorescent label). Each component of the RT-PCR kit is generally in its own
suitable container. Thus, these kits generally comprise distinct containers
suitable for each individual reagent, enzyme, primer and probe. Further, the
RT- PCR kits may comprise instructions for performing the assay and methods
for interpreting and analyzing the data resulting from the performance of the
assay.
For antibody based kits, the kit can comprise, for example: (1) a first
antibody
(which may or may not be attached to a support) which binds to protein of
interest (e.g., a protein product of 1, 2, 3, 4, 5, 6, 7, all or any
combination of
the marker genes of the invention); and, optionally, (2) a second, different
antibody which binds to either the protein, or the first antibody and is
conjugated to a detectable label (e. g., a fluorescent label, radioactive
isotope or
enzyme).
The antibody-based kits may also comprise beads for conducting an immuno-
precipitation assay.
Each component of the antibody-based kits is generally in its own suitable
container. Thus, these kits generally comprise distinct containers suitable
for
each antibody. Further, the antibody-based kits may comprise instructions for
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
61
performing the assay and methods for interpreting and analyzing the data
resulting from the performance of the assay.
It should be thus appreciated that any of the kits of the invention may
optionally further comprises solid support, such as plates, beads, tube or
containers. These may be specifically adopted for performing different
detection steps or any nucleic acid amplification based assay or immuno assay,
as described for example by the method of the invention. It should be further
noted that any substance or ingredient comprised within any of the kits of the
invention may be attached, embedded, connected or linked to any solid support.
It should be noted that any of the detecting molecules used by the
compositions, methods and kits of the invention may be labeled by a detectable
label. The term "detectable label" as used herein refers to a composition or
moiety that is detectable by spectroscopic, photochemical, biochemical,
immunochemical, electromagnetic, radiochemical, or chemical means such as
fluorescence, chemifluoresence, or chemiluminescence, or any other
appropriate means. Preferred detectable labels are fluorescent dye molecules,
or fluorochromes, such fluorescein, phycoerythrin, CY3, CY5, allophycocyanine,
Texas Red, peridenin chlorophyll, cyanine, FAM, JOE, TAMRA, tandem
conjugates such as phycoerythrin-CY5, and the like. These examples are not
meant to be limiting.
It is to be understood that any polynucleotide or polypeptide or any
combination thereof described by the invention may be useful as a marker for a
disease, disorder or condition, and such use is to be considered a part of
this
invention.
It should be appreciated that all method and kits described herein, preferably
comprises any of the compositions of the invention.
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
62
It should be recognized that the nucleic acid sequences and/or amino acid
sequences used by the kits of the present invention relate, in some
embodiments, to their isolated form, as isolated polynucleotides (including
for
all transcripts), oligonucleotides (including for all segments, amplicons and
primers), peptides (including for all tails, bridges, insertions or heads,
optionally including other antibody epitopes as described herein) and/or
polypeptides (including for all proteins). It should be noted that the terms
"oligonucleotide" and "polynucleotide", or "peptide" and "polypeptide", may
optionally be used interchangeably.
According to a specifically preferred embodiment, the marker genes used by
any of the compositions, methods and kits of the invention may be selected
from the genes as set forth in Table 4.
According to another embodiment, the marker gene may be RAB3 GTPase
activating protein subunit 1 (catalytic).
Members of the RAB3 protein family are implicated in regulated exocytosis of
neurotransmitters and hormones. RAB3GAP, which is involved in regulation of
RAB3 activity, is a heterodimeric complex consisting of a 130-kD catalytic
subunit and a 150-kD noncatalytic subunit. RAB3GAP specifically converts
active RAB3-GTP to the inactive form RAB3-GDP. NCBI accession number:
NM_012233.1, as also denoted by SEQ ID NO. 1. It should be noted that the
assay ID of this marker gene (by Applied Biosysteins) is Hs00326824_m1_
According to another embodiment, the marker gene may be Nuclear factor of
activated T-cells 5, tonicity-responsive.
The NFAT5 gene is widely transcribed and encodes a protein of 1,455 aa. In
contrast to the conventional NFAT proteins, NFAT1-4, which shows high and
moderate sequence identity in their DNA-binding and N-terminal regulatory
domains, respectively, NFAT5 exhibits a clear relation to NFAT proteins only
in its Rel-like DNA-binding domain. The DNA-binding specificity of NFAT5 is
similar to that of NFATl, but the NFAT5 DNA-binding domain differs from the
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
63
DNA-binding domains of NFAT1-4 in that it does not cooperate with Fos/Jun
at NFAT:AP-1 composite sites. A striking feature of NFAT5 is its constitutive
nuclear localization that is not modified on cellular activation. Taken
together,
our data indicate that NFAT5 is a target of signaling pathways distinct from
those that regulates NFAT1-4, and that it is likely to modulate cellular
processes in a wide variety of cells. NCBI accession number: NM_006599, as
also denoted by SEQ ID NO. 2. It should be noted that the assay ID of this
marker gene (by Applied Biosystems) is Hs00232437_ml.
According to another embodiment, the marker gene may be Mitochondrial
ribosomal protein S6.
The nuclear encoded MRPS6, a building block of the human mitoribosome of
the oxidative phosphorylation system (OXPHOS).
The expression of a gene encoding the mitochondria ribosomal protein S6
(MRPS6) had the highest combined mean fold change and topped the list of
regulated genes.
Multiregional gene expression profiling identifies MRPS6 _ as a possible
candidate gene for Parkinson's disease. NCBI accession number:
NM_032476.2, as also, denoted by SEQ ID NO. 3. It should be noted,that the
assay ID of this marker gene (by Applied Biosystems) is Hs00606808 _ml.
According to another embodiment, the marker gene may be AU RNA binding
protein/enoyl-Coenzyme A hydratase. AUH gene encodes an RNA-binding
protein with intrinsic enzymatic activity. It was suggested, that Its
hydratase
and AU-binding functions are located on different domains within a single
polypeptide.
It was shown that 3-methylglutaconyl-CoA hydratase, a key enzyme of leucine
degradation, is encoded by the AUH gene. NCBI accession number:
NM001698, as also denoted by SEQ ID NO. 4. It should be noted that the
assay ID of this marker gene (by Applied Biosysteins) is H s00156044_ml.
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
64
According to another embodiment, the marker gene may be MID 1 interacting
protein 1 (gastrulation specific G12-like (zebrafish)).
MID1 is a gene which encodes a TRIM/RBCC protein that is anchored to the
microtubules. The association of Midlwith the cytoskeleton is regulated by
dynamic phosphorylation, through the interaction with the alpha4 subunit of
phosphatase 2A (PP2A). Midl acts as an E3 ubiquitin ligase, regulating PP2A
degradation on microtubules.
NCBI accession number: NM021242, as also denoted by SEQ ID NO. 5. It
should be noted that the assay ID of this marker gene (by Applied Biosystems)
is Hs00221999 ml.
According to another embodiment, the marker gene may be Regulator of G-
protein signaling 16. Members of the 'regulator of G protein signaling' (RGS)
gene family encode proteins that stimulate the GTPase activities of G protein
alpha-subunits. RGS16 is widely expressed as an approximately 2.4-kb mRNA
and that its expression is induced by mitogenic signals. Over-expression of
RGS16 inhibits G protein-coupled mitogenic signal transduction and activation
of the mitogen-activated protein kinase (MAPK) signaling cascade. NCBI
accession number: NM_002928.3, as also denoted by SEQ ID NO. 6. It should
be noted that the assay ID of this marker gene (by Applied Biosystems) is
Hs00161399_ml.
According to another embodiment, the marker gene may be Membrane-
associated ring finger (C3HC4) 7. The MARCH-family of proteins regulates
endocytosis of cell surface receptors (e.g., transferrin receptor,
histocompatibility antigens and Fas; type I as well as type II transmembrane
domains) via ubiquitination. A RING finger consists of a double ring structure
containing 8 metal binding cysteine and histidine residues that coordinate two
zinc ions. RING fingers of E3 ligases can be formed by different
configurations
of histidine and cysteine residues. The most frequently found `classical'
C3HC4
RING domains are involved in many different cellular events. Examples are c-
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
CBL, which functions in ubiquitin-dependent lysosomal trafficking and
BRCA1, which affects cell cycle progression through its ligase activity via a
mechanism that is still elusive. RING fingers with a C3H2C3 configuration are
found in membrane associated E3 ligases catalyzing ubiquitination of
degradation substrates occurring in the secretory pathway, especially the ER,
and endolysosomal compartments. NCBI accession number: NM022826.2, as
also denoted by SEQ ID NO. 7. It should be noted that the assay ID of this
marker gene (by Applied Biosystems) is. Hs00224521_in l.
According to another embodiment, the marker gene may be Nuclear receptor
subfamily 3(glucocorticoid receptor) (NR3C1). Of the 2 isoforms of the
glucocorticoid receptor generated by alternative splicing, GR-alpha is a
ligand-
activated transcription factor that, in the hormone-bound state, modulates the
expression of glucocorticoid-responsive genes by binding to a specific
glucocorticoid response element (GRE) DNA sequence. In contrast, GR-beta
does not bind glucocorticoids and is transcriptionally inactive. It was
demonstrated that GR-beta is able to inhibit the effects of hormone-activated
GR-alpha on a glucocorticoid-responsive reporter gene in a concentration-
dependent manner. The inhibitory effect appeared to be due to competition for
GRE target sites. Since RT-PCR analysis showed expression of GR-beta mRNA
in multiple human tissues, GR-beta may be a physiologically and
pathophysiologically relevant endogenous inhibitor of glucocorticoid action
and
may participate in defining the sensitivity of tissues to glucocorticoids.
NCBI
accession number: X03348, as also denoted by SEQ ID NO. 8. It should be
noted that the assay ID of this marker gene (by Applied Biosystems) is
Hs00353740_ml
According to another embodiment, the marker gene may be E74-like factor 1
(ets domain transcription factor). E74-like factor (1 ELFl) is a lymphoid-
specific ETS transcription factor that regulates inducible gene expression
during T cell activation. Wang et al. (1993) demonstrated that ELFl contains a
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
66
sequence motif that is highly related to the RB (retinoblastoma) binding sites
of several viral oncoproteins and binds to the pocket region of RB both in
vitro
and in vivo. Other results demonstrated that RB interacts specifically with
this
lineage-restricted ETS transcription factor. The interaction may be important
for the coordination of lineage-specific effector function. NCBI accession
number: NM172373, as also denoted by SEQ ID NO. 9. It should be noted that
the assay ID of this marker gene (by Applied Biosystems) is Hs00152844_ml.
According to another embodiment, the marker gene may be Similar to
ribosomal protein S6 kinase, polypeptide 1(RPS6KB1).
RPS6KB1 mediates the rapid phosphorylation of ribosomal protein S6 on
multiple serine residues in response to insulin or several classes of
mitogens.
Acquisition of S6 protein kinase catalytic function is restricted to the most
extensively phosphorylated polypeptides. In mammals, mammalian target of
rapamycin cooperates with P13K-dependent effectors in a biochemical
signaling pathway to regulate the size of proliferating cells. NCBI accession
number: NM003161, as also denoted by SEQ ID NO. 10. It should be noted
that the assay ID of this marker gene (by Applied Biosystems) is
Hs00177357_ml.
According to another embodiment, the marker gene may be Signal transducer
and activator of transcription 5A. STATs, such as STAT5, are proteins that
serve the dual function of signal transducers and activators of transcription
in
cells exposed to signaling polypeptides. More than 30 different polypeptides
cause STAT activation in various mammalian cells. STAT5 was identified as
the protein most notably induced in response to T-cell activation with IL2.
They hypothesized that STAT5 may govern the effects of IL2 during the
immune response. NCBI accession number: NM_003152, as also denoted by
SEQ ID NO. 11. It should be noted that the assay ID of this marker gene (by
Applied Biosystems) is Hs00559643_ml.
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
67
According to another embodiment, the marker gene may be YTH domain
family, member 3 [Mehrle, A, et al. Nucleic Acids Res. 1; 34 (Database
issue):D415-8. Related Articles, Links (2006)]. NCBI accession number:
NM152758.4, as also denoted by SEQ ID NO. 12. It should be noted that the
assay ID of this marker gene (by Applied Biosysteins) is Hs00405590_ml.
More particularly, according to one embodiment, such marker gene may be
DnaJ (Hsp40) hoinolog, subfamily C, member 12. DnaJ/HSP40 proteins, which
are molecular chaperones of HSP70 proteins, contain all or a combination of 4
domains: an N-terminal J domain; a glycine/phenylalanine (G/F)-rich domain;
a central repeat region (CRR), and a weakly conserved C-terminal domain. The
J domain, which is believed to mediate interaction with HSP70 proteins,
contains a highly conserved histidine-proline-aspartate (HPD) tripeptide. J
domain-only proteins are members of a subclass of the HSP40/DnaJ family
that possess the J domain as well as a highly conserved C terminus, but lack
the G/F-rich and CRR domain. NCBI accession number NM_021800, as also
denoted by SEQ ID NO. 13. It should be noted that the assay ID of this marker
gene (by Applied Biosystems) is Hs00222318_ml.
According to another embodiment, the marker gene may be Interferon-induced
protein 44-like (IFI44L). The biological function of this gene is unknown.
[Suzuki, Y. et al., Gene. 24:200(1-2):149-56 (1997)]. NCBI accession number:
NM006820, as also denoted by SEQ ID NO. 14. It should be noted that the
assay ID of this marker gene (by Applied Biosysteins) is Hs00199115_ml.
According to another embodiment, the marker gene may be Seryl-tRNA
synthetase. The human seryl-tRNA synthetase has been expressed in E. coli,
purified (95% pure as determined by SDS/PAGE). The human seryl-tRNA
synthetase sequence (514 amino acid residues) shows significant sequence
identity with seryl-tRNA synthetases from E. coli (25%), Saccharomyces
cerevisiae (40%), Arabidopsis thaliana (41%) and Caenorhabditis elegans
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
68
(60%). The functional studies show that the enzyme aininoacylates calf liver
tRNA and prokaryotic E. coli tRNA. NCBI accession number: NM_006513, as
also denoted by SEQ ID NO. 15. It should be noted that the assay ID of this
marker gene (by Applied Biosystems) is Hs00197856_ml.
According to another embodiment, the marker gene may be SMAD specific E3
ubiquitin protein ligase 2. Ubiquitin-mediated proteolysis regulates the
activity of diverse receptor systems. SMAD specific E3 ubiquitin protein
ligase
2 (SMURF2) associated constitutively with SMAD7. Western blot analysis
showed that SMURF2 selectively regulated the expression of SMAD2 and, to
some extent, SMAD1, but not SMAD3, through a ubiquitination- and
proteasome-dependent degradation process catalyzed by the HECT ligase.
It was found that telomere attrition in human fibroblasts induced SMURF2
upregulation, and this upregulation was sufficient to produce the senescence
phenotype. Infection of early passage fibroblasts with retrovirus carrying
SMURF2 led to morphologic and biochemical alterations characteristic of
senescence, including altered gene expression and reversal of cellular
immortalization by TERT. It was further showed that SMURF2 activated
senescence through the RB (180200) and p53 pathways. NCBI accession
number: NM022739.3, as also denoted by SEQ ID NO. 16. It should be noted
that the assay ID of this marker gene (by Applied Biosystems) is
Hs00224203_ml.
In yet another embodiment, the inarker gene may be splicing factor,
arginine/serine-rich 18 (SFRS18). This gene has an undefined function. NCBI
accession number: NM_032870.2, as also denoted by SEQ ID NO. 17. It should
be noted that the assay ID of this marker gene (by Applied Biosystems) is
Hs00369090_ml.
According to another embodiment, the marker gene may be Nuclear receptor
subfamily 4, group A, member 2. Nuclear receptor subfamily 4, group A,
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
69
member 2, is a gene encoding a member of the steroid/thyroid hormone family
of receptors. The receptor, called NOT (nuclear receptor of T cells) by them,
has
all of the structural features of steroid/thyroid hormone receptors but is
rapidly
and only very transiently expressed after cell activation. NURRl and PITX3
cooperatively promoted terminal maturation of murine and human embryonic
stem cell. NCBI accession number: NM_006186, as also denoted by SEQ ID
NO. 18. It should be noted that the assay ID of this marker gene (by Applied
Biosystems) is Hs00428691_ml.
According to another embodiment, the marker gene may be Cyclin-dependent
kinase inhibitor 1B -CDKNIB (p27, Kipl).
Cyclin-dependent kinase (CDK) activation requires association with cyclins
(e.g., CCNE1) and phosphorylation by CAK (CCNH), and leads to cell
proliferation. Inhibition of cellular proliferation occurs upon association of
CDK inhibitor (e.g., CDKNIB) with a cyclin-CDK complex. It was showed that
expression of CCNEl-CDK2 at physiologic levels of ATP results in
phosphorylation of CDKNIB at thrl87, leading to elimination of CDKNIB
from the cell and progression of the cell cycle from Gl to S phase. At low ATP
levels, the inhibitory functions of CDKNIB are enhanced, thereby arresting
cell proliferation.
NCBI accession number: BC001971, as also denoted by SEQ ID NO. 19. It
should be noted that the assay ID of this marker gene (by Applied Biosystems)
isHs00153277 m 1.
According to another embodiment, the marker reference gene may be
Eukaryotic translation initiation factor 3, subunit 7 zeta, 66/67kDa.
Eukaryotic initiation factor-3 (eIF3), the largest of the eIFs, is a
multiprotein
complex of approximately 600 kD that binds to the 40S ribosome and helps
maintain the 40S and 60S ribosomal subunits in a dissociated state. It is also
thought to play a role in the formation of the 40S initiation complex by
interacting with the ternary complex of eIF2/GTP/methionyl-tRNA, and by
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
promoting mRNA binding. NCBI accession number: NM_003753, as also
denoted by SEQ ID NO. 20. It should be noted that the assay ID of this marker
gene (by Applied Biosystems) is Hs00388727_ml.
According to one embodiment, the control reference gene may be Eukaryotic
18S rRNA. NCBI accession number: X03205.1, as also denoted by SEQ ID NO.
21. It should be noted that the assay ID of this marker gene (by Applied
Biosystems) is Hs99999901sl_
According to another embodiment, the control reference gene may be ribosomal
protein S9. NCBI accession number: NM_001013.3, as also denoted by SEQ ID
NO. 22. It should be noted that the assay ID of this marker gene (by Applied
Biosystems) is Hs02339426_gl.
According to another embodiment, the control reference gene may be Actin,
beta.
Interaction of phospholipase D with actin microfilaments regulates cell
proliferation, vesicle trafficking, and secretion. Localization of beta-actin
mRNA to sites of active actin polymerization modulates cell migration during
embryogenesis, differentiation, and possibly carcinogenesis. In
immunoprecipitation studies of embryonic fibroblasts from wild type and
knockout mice deficient in the arginylation enzyme Atel (607103), Karakozova
et al. (2006) found that approximately 40% of intracellular beta-actin is
arginylated in vivo. Karakozova et al. (2006) found that arginylation of beta-
actin regulates cell motility. Mammalian cytoplasmic actins are the products
of
2 different genes and differ by many amino acids from muscle actin. NCBI
accession number: NM_001101, as also denoted by SEQ ID NO. 23. It should
be noted that the assay ID of this marker gene (by Applied Biosystems) is
Hs99999903_ml.
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
71
According to another embodiment, the control reference gene may be heat
shock protein 90kDa alpha (cytosolic), class B member 1.
NCBI accession number: NM_007355.2, as also denoted by SEQ ID NO. 24. It
should be noted that the assay ID of this marker gene (by Applied Biosystems)
is Hs00607336_gH.
According to another embodiment, the marker gene may be Sorting nexin 2.
The sorting nexins constitute a large conserved family of hydrophilic
molecules
that interact with a variety of receptor types. These molecules contain an
approximately 100-amino acid region termed the phox homology (PX) domain.
NCBI accession number: AF043453.
According to another embodiment, the marker gene may be Hypothetical
protein MGC4504. MGC4504 is a homolog protein of ChaC, cation transport
regulator homolog 1(E. coli) CHAC 1. CHAC 1 molecular function is regulation
of cellular ion concentrations which is necessary to sustain a multitude of
physiological processes including pH balance and ion homeostasis. NCBI
accession number: NM024111, PubMed ID: 12460671.
According to another embodiment, the marker gene may be Granulysin.
Cytolytic T lymphocytes (CTLs) are required for protective immunity against
intracellular pathogens. CTLs that kill infected cells through the granule-
exocytosis pathway may release 1 or more effector molecules with the capacity
to kill the intracellular microbial pathogen directly showed that granulysin
is a
critical effector molecule of the antimicrobial activity of CTLs. Granulysin
is a
protein present in cytotoxic granules of CTLs and natural killer (NK) cells.
Amino acid sequence comparison indicated that granulysin is a member of the
saposin-like protein (SAPLIP) family. Granulysin is located in the cytotoxic
granules of T cells, which are released upon antigen stimulation. NCBI
accession number: NM 006433.
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
72
According to another embodiment, the marker gene may be Serine
hydroxymethyltransferase 2 (mitochondrial). The enzyme serine
hydroxymethyltransferase (SHMT is a pyridoxal phosphate-dependent enzyme
that catalyzes the reversible interconversion of serine and H4PteGlu to
glycine
and 5,10-CH2-H4PteGlu with generating of one-carbon units. SHMT is present
in both the mitochondria (mSHMT) and the cytoplasm (cSHMT) in mammalian
cells. The human SHMT cDNAs encoding the two isozymes have been isolated
and the genes localized to chromosomes 12q13 and 17p11.2, respectively.
Currently, the metabolic role of the individual SHMT isozymes is not clearly
understood. The central role of SHMT isozymes in producing one-carbon-
substituted folate cofactors has suggested that the regulation of these
enzymes
may influence cell growth and proliferation and that they may be targets for
the development of antineoplastic agents. NCBI accession number:
NM_005412.
According to another embodiment, the marker gene may be Annexin A2.
Annexin II, a major cellular substrate of the tyrosine kinase encoded by the
SRC oncogene belongs to the annexin family of Ca(2+)-dependent phospholipid-
and membrane-binding proteins. By screening a cDNA expression library
generated from highly purified human osteoclast-like multinuclear cells (MNC)
formed in long-term bone marrow cultures, a candidate clone that stimulated
MNC formation was identified. Sequence analysis showed that this cDNA
encoded annexin II. Further studies yielded results suggesting that ANX2 is an
autocrine factor that enhances osteoclast formation and bone resumption, a
previously unknown function for this molecule. NCBI accession number:
BC001388.
According to another embodiment, the marker gene may be BTB and CNC
homology, basic leucine zipper transcription factor 2. Members of the small
Maf family are basic region leucine zipper (bZIP) proteins that can function
as
transcriptional activators or repressors. Oyake et al. (1996) identified mouse
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
73
cDNAs encoding Bach1 (602751) and Bach2. Both Bach proteins contain a BTB
(broad complex-tramtrack-bric-a-brac) or POZ (poxvirus and zinc finger)
protein-interaction domain and a CNC (Cap'n'collar) -type bZIP domain.
Bachl and Bach2 functioned as transcription repressors in transfection assays
using fibroblast cells, but they functioned as a transcriptional activator and
repressor, respectively, in cultured erythroid cells. Gel shift analysis by
Sasaki
et al. (2000) showed that when overexpressed, BACH2 binds to MAF
recognition elements (MARE). Over expression also resulted in a loss of
clonogenic activity. BACH2/CA-1 microsatellite analysis indicated that loss of
heterozygosity occurred in 5 of 25 non-Hodgkin lymphoma patients. NCBI
accession number: NM 021813.
According to another embodiment, the marker gene may be E2F transcription
factor 2. The ability of Myc to induce S phase and apoptosis requires distinct
E2F activities. Hence, the induction of specific E2F activities is an
essential
component in the MYC pathways that control cell proliferation and cell fate
decisions. The retinoblastoma tumor suppressor (Rb) pathway is believed to
have a critical role in the control of cellular proliferation by regulating
E2F
activities. E2F1, E2F2, and E2F3 belong to a subclass of E2F factors thoughtto
act as transcriptional activators important for progression through the G1/S
transition. NCBI accession number: NM 004091.
According to another embodiment, the marker gene may be Major
histocompatibility complex, class II, DQ beta 1. The genes for the heteromeric
major histocompatibility complex class II proteins, the alpha and beta
subunits, are clustered in the 6p2l.3 region. It was suggested that the
structure of the DQ molecule, in particular residue 57 of the beta-chain,
specifies the autoimmune response against insulin-producing islet cells that
leads to insulin-dependent diabetes mellitus. The extremely high
polymorphism of HLA class II transznembrane heterodimers is due to a few
hypervariable segments present in the most external domain of their alpha and
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
74
beta chains. Some changes in amino acid sequence are critical in disease
susceptibility associations as well as the ability to present processed
antigens
to T cells. In addition to insulin-dependent diabetes mellitus, an increased
frequency of specific alleles at the DQB1 locus has been claimed for
narcolepsy,
pemphigus vulgaris, and ocular cicatricial pemphigoid. It was found that HLA-
DQB1 genotypes encoding aspartate-57 are associated with 3-beta-
hydroxysteroid dehydrogenase autoiminunity in Premature ovarian failure.
NCBI accession number: NM 002123.
According to another embodiment, the marker gene may be Tensin 3.
Tensin 3 is a cytoplasmic phosphoprotein that localized to integrin-mediated
focal adhesions. It binds to actin filaments and contains a phosphotyrosine-
binding (PTB) domain, which interacts with the cytoplasmic tails of integrin.
In addition, tensin has a Src Homology 2 (SH2) domain capable of interacting
with tyrosine-phosphorylated proteins. Furthermore, several factors induce
tyrosine phosphorylationof tensin. Thus, tensin functions as a platform for
dis/assembly of signaling complexes at focal adhesions by recruiting tyrosine-
phosphorylated signaling molecules through the SH2 domain, and also by
providing interaction sites for other SH2-containing proteins. An elevated
expression of tensin 3 was demonstrated during tumor angiogenesis, so it
serves as tumor endothelial marker (TEM). NCBI accession number:
NM_022748.
According to another embodiment, the marker gene may be Lysosomal-
associated membrane protein 2. The lysosomal membrane plays a vital role in
the function of lysosoines by sequestering numerous acid hydrolases that are
responsible for the degradation of foreign materials and for specialized
autolytic functions. LAMP2 is glycoprotein that constitutes a significant
fraction of the total lysosomal membrane glycoproteins. It consists of
polypeptides of about 40 kD, with 16 to 20 N-linked saccharides attached to
the
core polypeptides. LAMP2 is thought to protect the lysosomal membrane from
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
proteolytic enzymes within lysosomes and to act as a receptor for proteins to
be
imported into lysosomes. NCBI accession number: NM_013995.
According to another embodiment, the marker gene may be Retinoblastoma-
like 2 (p130). Retinoblastoma-like 2 is transcription factor, which shown to
related to DNA-dependent cell cycle regulation and to negative regulation of
progression through cell cycle. RBL2 is essential for telomere length control
in
human fibroblasts, with loss of either protein leading to longer telomeres. It
was proposed that RBL2 forms a complex with RAD50 through RINT1 to block
telomerase-independent telomere lengthening. NCBI accession number:
NM 005611.
According to another embodiment, the marker gene may be Interleukin 15
receptor, alpha. Interleukin-2 (IL2) and interleukin-15 (IL15) are cytokines
with overlapping but distinct biologic effects. Their receptors share 2
subunits,
the IL2R beta and gamma chains, which are essential for signal transduction.
The IL2 receptor requires an additional IL2-specific alpha subunit (IL2RA) for
high-affinity IL2 binding. Confocal microscopy demonstrated that full-length
IL15RA was associated primarily with the nuclear membrane, with part of the
receptor having an intranuclear localization. It was shown that the
IL15/IL15RA complex has enhanced effects on T-cell survival compared with
IL15 alone. NCBI accession number: NM 002189.
According to another embodiment, the marker gene may be Cyclin H.
The cdk-activating kinase (CAK) is a multi-subunit protein which
phosphorylates and thus activates certain cyclin-dependent protein kinases in
the regulation of cell cycle progression. Presence of the CAK complex as a
distinct component of TFIIH, suggesting a link between TFIIH (by the
phosphorylation of CDC2 or CDK2) and the processes of transcription, DNA
repair, and cell cycle progression.
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
76
Phosphorylation of mammalian cyclin H by CDK 8 represses both the ability of
TFIIH to activate transcription and its C-terminal kinase activity. In
addition,
mimicking CDK8 phosphorylation of cyclin H in vivo has a dominant-negative
effect on cell growth. NCBI accession number: NM_001239.
According to another embodiment, the marker gene may be: Stromal antigen 2.
A multi-subunit complex, termed cohesin, is likely to be a central player in
sister chromatid cohesion. STAD is mammalian analog of Smc1p, Smc3p, and
Scclp. Smclp and Smc3p belong to a large family of chromosomal ATPases
(the structural maintenance of chromosomes [SMC] 1 family), members of
which are involved in many aspects of higher order chromosome architecture
and dynamics.
NCBI accession number: BC001765.
According to another embodiment, the marker gene may be Ring finger protein
11.
The RING finger is a C3HC4-type zinc finger motif, and members of RING
finger proteins are mostly nuclear proteins and the motif is involved in both
protein-DNA and protein-protein interactions. Some members of the RING
finger family have been implicated in carcinogenesis and cell transformation.
For example, a RING finger protein, BRCA1, is a tumor suppressor in an early
onset breast cancer. The approximate corresponding cytogenetic location of the
ring finger protein llgene is on chromosome lp3l-p32 region. This region is
frequently involved in deletions and chromosomal translocations observed in T-
cell acute lymphoblastic leukemia (T-ALL). NCBI accession number:
AB024703.
According to another embodiment, the marker gene may be Cyclin T2.
Cyclin T2 is a part of positive transcription elongation factor b (P-TEFb)
which
is thought to facilitate the transition from abortive to productive elongation
by
phosphorylating the C-terminal domain (CTD) of the largest subunit of RNA
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
77
polymerase II. cDNAs encoding human cyclins T1 and T2 was identified.
Immunoprecipitation studies demonstrated that CDK9 is complexed with the
cyclins Tl and T2 in HeLa cell nuclear extracts. Approximately 80% of CDK9 is
complexed with cyclin T1, 10% with cyclin T2a and 10% with T2b. Each
complex is an active P-TEFb molecule that can phosphorylate the CTD of RNA
polymerase II and cause the transition from abortive elongation into
productive elongation. When expressed in mammalian cells, all 3 CDK9/cyclin
T combinations strongly activated a CMV promoter. Northern blot analysis
revealed that cyclin T2 was expressed as multiple mRNAs in all human tissues
tested. NCBI accession number: NM 001241.
It should be further appreciated that in case of detection of BRCA1 mutation,
the marker gene may be selected from genes demonstrated by the invention as
exhibiting differential expression of about 1.5 folds. Therefore, according to
one
embodiment, such genes may be selected from any of the genes set forth in
Table 5 (disclosed at the end of the Examples).
In another embodiment, it should recognized that in case of detection of
BRCA2 mutations, the marker gene may be selected from genes demonstrated
by the invention as exhibiting differential expression of about 2 folds.
Therefore, according to another embodiment, such genes may be selected from
any of the genes set forth in Table 6 (disclosed at the end of the Examples).
All technical and scientific terms used herein should be understood to have
the
meaning commonly understood by a person skilled in the art to which this
invention belongs, as well as any other specified description.
The following references provide one of skill with a general definition of
many
of the terms used in this invention: Singleton et al., Dictionary of
Microbiology
and Molecular Biology (2nd ed. 1994); The Cambridge Dictionary of Science
and Technology (Walker ed., 1988); The Glossary of Genetics, 5th Ed., R.
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
78
Rieger et al. (eds.), Springer Verlag (1991); and Hale & Marham, The
Harper Collins Dictionary of Biology (1991). All of these are hereby
incorporated by reference as if fully set forth herein.
Disclosed and described, it is to be understood that this invention is not
limited
to the particular examples, methods steps, and compositions disclosed herein
as such methods steps and compositions may vary somewhat. It is also to be
understood that the terminology used herein is used for the purpose of
describing particular embodiments only and not intended to be limiting since
the scope of the present invention will be limited only by the appended claims
and equivalents thereof.
It must be noted that, as used in this specification and the appended claims,
the singular forms "a", "an" and "the" include plural referents unless the
content clearly dictates otherwise.
Throughout this specification and the Examples and claims which follow,
unless the context requires otherwise, the word "comprise", and variations
such as "comprises" and "comprising", will be understood to imply the
inclusion
of a stated integer or step or group of integers or steps but not the
exclusion of
any other integer or step or group of integers or steps.
The following examples are representative of techniques employed by the
inventors in carrying out aspects of the present invention. It should be
appreciated that while these techniques are exemplary of preferred
embodiments for the practice of the invention, those of skill in the art, in
light
of the present disclosure, will recognize that numerous modifications can be
made without departing from the spirit and intended scope of the invention.
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
79
Examples
Experimental procedures
Samples information
Fresh blood samples were obtained from proven unaffected carriers of BRCAI
mutations, 8 unaffected carriers of BRCA2 mutations, and healthy age-
matched control women with no individual or family history of cancer.
Individuals heterozygous for BRCA1 and BRCA2 germline mutations were
identified from the BRCAI and BRCA2 predictive testing program in the
Institute of Cancer Research Royal Marsden Foundation NHS Trust, Cancer
Genetics Carrier Clinic, London, UK and from the Cancer Genetic Clinic of
Hadassah University Medical Center, Jerusalem, Israel. Fresh blood samples
were collected from unaffected BRCAI12 heterozygous gene mutation carriers
and healthy age-matched control women with no individual or family history of
cancer. The mutations in the BRCA1 /2 carriers are listed in Table 1. Written
informed consent was obtained from all participating individuals prior to
inclusion into the study, and the study protocol was approved by the Royal
Marsden Locoregional Ethics Committee.
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
Table 1. Characteristics of mutations in BRCAI and BRCA2 genes in carriers
used for the present invention
Gene Mutation
BRCA1 5382 inc C
BRCAl Del AG i 85
BRCAl 185 del AG
BRCA1 3875 del 4
BRCAl Ins C5382
BRCA1 A>T 1182
BRCAl 44184 del
BRCA1 185 del AG
BRCAl 3450 del CAAG
BRCA2 5950 del CT
BRCA2 del TT65o3
BRCA2 C>T96 10
BRCA2 del CA995
BRCA2 6503 del TT
BRCA2 4075del GT
BRCA2 del CA995
BRCA2 6174del T
Samples preparation and RNA extraction
Lymphocytes were collected from blood samples using LymphoPrep kit
(Sigma), short-term cultured for 6 days and irradiated with 8 Gray (Gy) at a
high dose rate (0.86 Gy/min) using Ortovoltage X-ray machine. One hour later
RNA was extracted using Qiagene EZ RNA kit according to manufacturer's
instruction for further analysis. The integrity of all RNA samples was
verified
by 2% agarose gel electrophoresis before use in microarray experiments.
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
81
Microarray assay
Gene expression profile of the lymphocytes from BRCA1 /2 mutation carriers
and non- carriers was performed using Affyinetrix GeneChip Human Genome
U133A 2.0 oligonucleotide arrays. Total RNA from each sample was used to
prepare biotinylated target RNA. Briefly, 5 g was used to generate first-
strand cDNA by using a T7-linked oligo(dT) primer. After second-strand
synthesis, in vitro transcription was performed with biotinylated UTP and CTP
(Affymetrix), resulting in approximately 300-fold amplification of mRNA. The
target eDNA generated from each sample was processed as per the
manufacturer's recommendation using an Affymetrix GeneChip instrument
system. For this reason, spike controls were added to 15 gg of fragmented
cRNA before overnight hybridization. Arrays were then washed and stained
with streptavidin- phycoerythrin, before being scanned on an Affymetrix
GeneChip scanner. A complete description of these procedures is available at:
http://www.affvmetrix.com/supi)ort/technical/manual/exl)ression manual.affx.
After scanning, array images were assessed by eye to confirm scanner
alignment and the absence of significant bubbles or scratches on the chip
surface. The 3'/5' ratios for GAPDH and beta-actin were confirmed to be within
acceptable limits and BioB spike controls were found to be present on all
chips,
with BioC, BioD, and CreX also present in increasing intensity. When scaled to
a target intensity of 150 (using Affymetrix MAS 5.0 array analysis software),
scaling factors for. all arrays were within acceptable limits as were
background,
Q values and mean intensities
Data analysis
Data analysis was done using GeneSpring GX software (Agilent technologies).
Background adjustment, quantile normalization and summarization were done
using RMA methodology. The relative expression data for each probe set then
generated by normalizes each gene to the median of its own expression
intensities across the entire experiment set (per gene normalization). Control
probes and genes whose expression does not change across the experiment
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
82
were removed out from the list before statistical analysis was performed.
Differentially expressed genes were analyzed by One-way Welch ANOVA, with
p-value cutoff of 0.05 after Benjamini and Hochberg False Discovery Rate
multiple testing correction. Average linkage hierarchical clustering of the
different experimental samples was obtained for selected genes using Pearson
correlation as similarity measure. Bootstrapping analysis was carried out for
the assessment of the robustness of the cluster dendogram topology. Cluster
members were categorized according to their biological functions using The
Database for Annotation, Visualization and Integrated Discovery (DAVID)
tools [Dennis G. Jr. et al. Genome Biol. 4(5): 3 (2003)]. Pathway express tool
[Khatri P. et al. Nucleic Acids Res. 33(Web Server issue):W762-5 (2005)] was
used to characterize the responsive genes on molecular interactions networks
in regulatory pathways.
TaqMan quantitative gene expression measitrement
To validate the results obtained by the Affymetrix U133 A chips, the inventors
have performed TaqMan verification for expression of 42 selected genes in all
experimental samples, using an Applied Biosysteins7900 HT Micro Fluidic
Card System.
The measurements were performed using an ABI PRISMI 7900HT Sequence
Detection Systems described in the products User Guide
(http://www.appliedbiosystems.com, CA, USA).
TaqMan Arrays' 384-wells are pre-loaded with TaqMan Gene Expression
Assays. Each TaqMan Array evaluates from one to eight cDNA samples
generated in a reverse transcription step using random primers on 7900HT
Systems. The TaqMan Array functions as an array of reaction vessels for the
PCR step. Relative levels of gene expression are determined from the
fluorescence data generated during PCR sing the ABI PRISM 7900HT
Sequence Detection System or Applied Biosystems 7900HT Fast Real-Time
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
83
PCR System Relative Quantitation software. The TaqMan array technology
allows multiple targets to be analyzed per sample with very few pipetting
steps, streamlining reaction set-up time, and eliminating the need for liquid
handling robotics. TaqMan arrays provide a standardized format for gene
expression studies that permits direct comparison of results across different
researchers and laboratories.
cDNA samples for the PCR reaction were prepared by performing reverse
transcription of the RNA samples using the High Capacity cDNA Reverse
Transcription Kit (Applied Biosystems). For this reaction 2 g of total RNA in
a single 20 L reaction used to obtain up to 10 g of single stranded cDNA
from
a single reaction. 100 ng of cDNA samples loaded to plastic tube together with
RNase-free water up to total volume of 50 gl and add 50g1 of TaqMan
Universal PCR Master Mix (2X). 100 L of the desired sample-specific PCR
reaction mix loaded into the fill reservoir on the Place the TaqMan Array
plate. The samples were loaded in triplicates. The TaqMan Array plates with
loaded samples centrifuged at 1200 rpm for 1-2 minutes to ensure complete
distribution of the sample specific PCR reaction mix. Following the
centrifugation the plate were sealed to isolate the wells of a TaqMan Array
after it is loaded with cDNA samples and master mix. The sealer uses a
precision stylus assembly (carriage) to seal the main fluid distribution
channels of the array. After sealing procedure, the TaqMan Arrays were
running on the 7900HT instrument.
The extracted delta Ct values (which represent the expression normalized to
the ribosomal 18S expression) were grouped according to the resistance pattern
of the cell lines. Then, the Student's t-test was performed to compare the
expression values in the resistant cell lines to the sensitive cell lines.
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
84
Example 1
Expression profile of BRCA1 and BRCA2 mutations carriers
In order to identify marker genes potentially applicable for early diagnosis
and
prognosis of breast cancer patients, and specifically of BRCA1/2 mutation
carriers, the inventors analyzed expression profiles of control samples
compares to samples of BRCAI12 mutation carriers under irradiation stress.
Therefore, fresh blood samples were obtained from seventeen proven
unaffected carriers of BRCA1 mutations, ten unaffected carriers of BRCA2
mutations and twelve healthy age-matched control women with no individual
or family history of cancer (were tested negatively for mutations in the
BRCA1 /2 genes). Lymphocytes prepared from the samples were irradiated,
and subsequently RNA extracted from these samples was analyzed using
Affimetrix oligonucleotide arrays assay, as described in experimental
procedures.
To examine potential relationships between the expression profiles of control
and BRCA112 mutation carrier samples, microarray analyses were performed.
The Affymetrix GeneChip Human Genome U133A 2.0 Array was probed using
cDNA obtained from lymphocytes from nine proven unaffected carriers of
BRCA1, eight BRCA2 carriers and from ten non-carrier healthy women. For
each sample an individual chip was used. Hybridization experiments were
carried on RNA extracted from lymphocytes before irradiation and 1 hour
following exposure to 8 Gy of ionizing irradiation.
No significant differences in gene expression profiles were detected in a
preliminary study using RNA from non-irradiated lymphocytes from the three
groups (data not shown). This result is consistent with findings in previous
studies [Kote-Jarai Z. et al. Clin. Cancer Res. 12(13):3896-901 (2006); Kote-
Jarai Z. et al. Clin. Cancer Res. 10(3):958-63 (2004)]. Following irradiation,
differences in gene expression profiles between the three groups were
observed.
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
Data was processed used the MAS 5.0 and RMA algorithm to provide a
baseline expression level and detection for each probe set. For each probe
set,
the ratio between expression level of the BRCA1 /2 mutation carriers and
control samples was calculated. The results of the expression analysis are
demonstrated in Figure 1.
The rule-based clustering method used on the probe sets, demonstrated
significantly different expression pattern between either BRCA1 or BRCA2
group as compared to control group (p-value <_ 0.05). For each probe set, the
ratio between expression level of the mutation carriers and control samples
was calculated. Each probe set was graded as increased, decreased or
unchanged. As shown in Figure 1, clustering of up-regulated genes in BRCA2
mutation carrier group when compared to control group can be clearly observed
(Figure 1) and a sharp distinction in gene expression pattern in BRCA2
mutation carriers is demonstrated. Moreover, expression patterns within a
BRCA2 mutation group were highly conserved among all samples (Figure 1B),
whereas gene expression profile of BRCA1 mutation carrier samples is much
less homogenous (Figure lA).
The results of the principal components analysis (PCA) of these genes as
demonstrated by Figure 2, strengthen aforementioned findings. PCA clearly
indicate that samples from BRCA2 mutation carriers are well assembled
together and almost completely separated from either control or BRCA1
groups. By contrast, BRCA1 group represents more variable expression
patterns.
Founder mutations 185delAG, and 5382insC in the BRCA1 gene and 6174delT
in the BRCA2 gene are common among Jewish Ashkenazi population (-0.5%)
[Simard J. et al. Natural Genetics 8:392-398 (1994); Takahashi H. et al.
Cancer Research 55:2998-3002 (1995); Tonin P. et al. American Journal of
Human Genetics 57:189-189 (1995)]. In the general Ashkenazi population, the
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
86
carrier frequencies of these mutations were estimated to be -0.9% for 185delAG
[Struewing J.P. et al. Natural Genetics 11:198-200 (1995)], 0.9%-1.5 % for
6174de1T, and 0.13% for 5382insC [Benjamin B. et al. Natural Genetics 14:185-
187 (1996); Oddoux C. et al. Natural Genetics 14:188-190 (1996)]. In more-
detailed analysis shown in Figure 3, there was a clear-cut distinction between
up-regulated genes in the BRCA2 mutation carrier group and the control
group. Moreover, expression patterns within the BRCA2 mutation group were
highly conserved among all samples. In contrast, the gene expression profile
in
BRCA1 mutation carrier samples was less homogeneous, but still showed a
clearly distinct gene expression pattern from that of the control group. This
is
exemplified in Figure 3C where a set of genes which were down regulated in
BRCA1 is displayed in comparison to both BRCA2 and control cells. In total,
137 probe sets in BRCA1 and 1345 probe sets in BRCA2 mutation carriers
were differentially expressed (p <_ 0.05) when compared to the control
samples.
Using a 5% false discovery rate [Reiner A. D. Yekutieli and Y. Benjamini.
Bioinformatics 19(3):368-75 (2003)], the number of BRCA2 differentiated genes
was reduced to 596. This method was not applicable for the BRCA1 group due
to the lower homogeneity of the samples.
Example 2
Selection of specific genes demonstrating differential expression in
BRCA1/2 carriers as compared to healthy controls
The inventors have further analyzed the results differential expression of
different genes in BRCA1/2 carriers. Therefore, an additional filtration of
the
probe stets list was applied. The selection criterion was a signal that is
differentially expressed by at least two-fold between the tested groups (BRCA1
or BRCA2 carriers vs. controls). As result of this selection, a set of 86
genes in
BRCA1 carriers and 97 genes in BRCA2 carriers was established. These genes
were analyzed for Gene Ontology (GO) annotations. The results for BRCA2
mutations revealed that genes related to the gene expression regulation
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
87
pathways, DNA repair processes, cell cycle regulation and cancer possess the
highest score. As shown by Figure 4(BRCAl) and Figure 5 (BRCA2), the next
largest group of genes is related to the hematological system functioning and
defense system.
Genes expressing differentially at most samples of each of the groups were
further selected (genes performing higher alterations only in a part of the
patients in each group, were not chosen). From these genes only those with the
most consistent pattern of expression in all samples within the same group
were chosen. This selection resulted in a list of 38 genes shown in Table 2.
Interestingly, the function of most of the genes which differed between the
BRCA1 carrier mutation and the control group is related to transcription
regulation processes and DNA binding, as illustrated by Figure 6.
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
88
Table 2 List of the genes demonstrating most consistent gene expression
patterns among all the samples. These genes were demonstrated to be
differentially expressed among the tested groups using the Kruskal-Wallis
test.
Gene Gene Title
Symbol
1 DNAJC12 DnaJ (Hsp40) homolog, subfamily C,
member 12
2 SNX2 Sorting nexin 2
3 MGC4504 Hypothetical protein MGC4504
4 IFI44L Interferon-induced protein 44-like
GNLY Granulysin
6 SHMT2 Serine hydroxymethyltransferase 2
(mitochondrial)
7 PROSC Proline synthetase co-transcribed homolog
(bacterial)
8 FBXL8 Seryl-tRNA synthetase
9 AUH AU RNA binding protein/enoyl-Coenzyme
A hydratase
ANXA2 Annexin A2
11 BACH2 BTB and CNC homology 1, basic leucine
zipper transcription factor 2
12 SMURF2 SMAD specific E3 ubiquitin protein ligase
2
13 E2F2 E2F transcription factor 2
14 HLA-DQB1 Major histocompatibility complex, class II,
DQ beta 1
RGS16 Regulator of G-protein signalling 16
16 TNS3 Tensin 3
17 LAMP2 lysosomal-associated membrane protein 2
18 RBL2 Retinoblastoma-like 2 (p130)
19 C3HC4) 7 Membrane-associated ring finger (C3HC4)
7
C\)
NR3C1 Nuclear receptor subfamily 3, group C,
member 1
21 ELF1 E74-like factor 1 (ets domain transcription
factor)
22 RPS6KB1 RPS6KB1
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
89
23 TMEM30A Transmembrane protein 30A
24 STAT5A Signal transducer and activator of
transcription 5A
25 IL15RA Interleukin 15 receptor, alpha
26 CCNH Cyclin H
27 YTHDF3 YTH domain family, member 3
28 STAG2 Stromal antigen 2
29 RAB3GAP1 RAB3 GTPase activating protein subunit 1
(catalytic) .
30 RNF11 Ring finger protein 11
31 MRPS6 Mitochondrial ribosomal protein S6
32 NFAT5 Nuclear factor of activated T-cells 5,
tonicit -responsive
33 PKCs Protein Kinase C epsilon
34 NR4A2 Nuclear receptor subfamily 4, group A,
member 2
35 CCNT2 Cyclin T2
36 CDKNIB Cyclin-dependent kinase inhibitor 1B (p27,
Ki 1)
37 MID 1IP 1 MID 1 interacting protein 1(gastrulation
specific G12-like (zebrafish))
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
Example 3
Real-time'RT- PCR validation analysis of the selected transcripts
The inventors next performed a real time RT-PCR analysis of those thirty-
eight transcripts which were identified as being differentially expressed
between the three groups (presented by Table 2). In this analysis a larger
number of samples were tested: seventeen samples in the BRCA1 group, ten
from the BRCA2 group and twelve samples of non-carriers of mutations. Five
known housekeeping genes which were similarly expressed in the three groups
were served as internal controls. In total, forty three genes were tested by
TaqMan gene cards RT-PCR. As shown by Table 3, twenty genes out of the
forty tree examined, demonstrate a significantly differential expression in
BRCA1 or BRCA2 mutation carriers and control samples, with the p<0.05
threshold. These genes were therefore defined as the "marker genes". The
control housekeeping genes showed no significant difference between the
BRCA1 /2 and non-carrier control samples. Table 4 presents a summary of the
amplicon chosen for each of the marker genes, as well as partial sequences
thereof.
One of the differentially expressed genes checked had the lowest P value and
highest fold change. This gene was defined as the RAB-23 gene, a small
GTPase which acts as a negative regulator of the hedgehog signaling pathway
[Marcos I. et al. Int. J. Mol. Med. 12(6):983-7 (2003)].
RAB23 was proposed as modulating the activities of the Gli transcription
factors, perhaps by directly affecting their nucleocytoplasmic trafficking.
Interestingly, RAB23 was shown to be overexpressed and/or activated in
hepatocellular carcinoma [Liu Y.J. et al. World J. Gastroenterol. 13(7):1010-7
(2007)]. It is worthwhile mentioning that some of the genes found by the
present invention as being differentially expressed in BRCA1/2 mutation
carriers are know as involve in ubiquitination. Among them are the axotrophin
(MARCH7), a stem cell gene which can regulate immune tolerance and the
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
91
SMURF2, which participates in TGK signaling, causes degradation of the
RUNX transcription factor [Kaneki H. et al. J. Biol. Chem. 281(7):4326-33
(2006)] and induces cell senescence. Another differentially expressed gene,
ELF1, was found to be downregulated in mammary cancer in mice [Hu Y. et al.
Cancer Res. 64(21):7748-55 (2004)]. Other genes in this list regulate cell
cycle
or are DNA binding proteins.
Table 3. Twenty one genes that were significantly upregulated in BRCA1 and
BRCA2 mutation carriers as compared to the control group. The P value
between BRCA2 and control was calculated by Wilcoxon Rank-Sums test (P
values between BRCA1 and control are not shown.
Gene BRCA1/control BRCA21control P VALUE
RAB-23GAP1 2.267857 2.70063 0.0009
NFAT5 1.820479 2.303268 0.0036
MRPS6 2.089909 2.321377 0.0041
AUH 2.16 2.2 0.0046
MIDIIPI 1.945218 2.187696 0.0075
YTHDF3 1.509434 1.957825 0.0145
MARCH7 1.460844 1.927102 0.0147
ELF 1 1.75 2 0.015
STAT5A 1.48776 1.933504 0.016
C6orfl 11 1.55 1.9 0.025
NR3C1 1.356322 1.79716 0.0257
NR4A2 1.494553 1.869281 0.0275
1F144L 1.77931 1.842596 0.0297
RPL32 1.54199 1.860155 0.0333
SARS 1.759104 1.795918 0.0348
RP6K-IBI 1.494767 1.825334 0.0377
CDK-1N1B 1.68 1.81 0.041
RGS 16 1.364034 1.736471 0.0463
DNAJC12 1.68 1.71 0.0491
EIF3S7 1.57 1.93 0.0491
SMURF2 1.487574 1.782943 0.0491
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
92
Table 4. list of marker genes differentially expressed in carriers of BRCA1 /2
gee mutations
Gene
name RefSeq Partial sequence of
replicon
~ E u _ ^
a a~ E=
W
RAB3
GTPase
activating GAATGCCCAGAGGGC
RA133GAP I protein NM_012233.1 85 23-24 2738 TGCAGCTATG
subunit 1 SEQ ID NO. 1
(catalytic) SEQ ID NO.25
nuclear
factor of
NFAT5 activated NM_006599 2352 GACACTGGCGGTGGA
70 5-6 CTGCGTAGGG
2 T-cells 5, SEQ ID NO.2
tonicity-
responsive SEQ ID NO.26
mitochond GCAGCACAACAGAGG
MRPS6 rial NM_032476.2 CGGGTATTTC
3 ribosomal SEQ ID N0.3 100 2-3 359
protein S6 SEQ ID NO.27
AU RNA
binding
protein/en 878 TTTTTACCTCAGGGAC
AUH oyl- NM_001698 61 CTGTTGCAA
4 Coenzynie SEQ ID NO.4 ~ 8
A SEQ ID NO. 28
hydratase
MIDI
MID 1 IP 1 interacting
protein I
(gastrulati AGAGGAGGCCAGGGC
on specific NM_021242 70 1-2 596 TCGACCCACA
G 12 SEQ ID NO.5
lioniolog SEQ ID NO.29
(zebi-afish)
6 RGS16 regulator NM 002928.3 108 1-2 203
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
93
of G- SEQ ID NO.6 TGCCTGGAGAGAGCC
protein AAAGAGTTCA
signaling
16 SEQ ID NO.30
membrane AAAAGAGAGCCTCCT
MARCH7 -associated NM022826.2 TTTAGAGGAC
7 ring finger SEQ ID NO.7 102 5-6 1738
(C3HC4) 7 SEQ ID NO.31
nuclear
receptor
subfamily
NR3CI 3, group AATGAACCTGGAAGC
8 j, member SEQ ID NO.8 7' 4-5 1602 TCGAAAAACA
(glucocorti SEQ ID NO.32
coid
receptor)
E74-like NM_172373.2
factor I
SEQ ID NO.9
9 ELFI (ets GGATGAACGACAGCT
doniain 76 1-2 301 TGGTGATCCA
transcripti
on factor) SEQ ID NO. 33
ribosomal
protein S6
RPS6KB1 kinase, NM_003161.2 97 AAGACACTGCCTGCTT
70kDa, SEQ ID NO.10 6-7 690 TTACTTGGC
polypeptid
e'l SEQ ID NO.34
signal
transducer
and 17-18 ACTCCTGTGCTGGCTA
11 STAT5A activator NM_003152.2 85 2706 AAGCTGTTG
of SEQ ID NO. i l
transcripti SEQ ID NO.35
on 5A
YTH GGAAGCCATGCGTAG
YTHDF3 domain NM152758.4 118 GGAGAGAAAT
12 family, SEQ ID N0.12 4-5 2044
member 3 SEQ ID NO. 36
13 DNAJCI2 DnaJ NM_021800 82 3-4 467 CAGTGAAGACGTCAA
(Hs 40) SEQ ID NO.13 TGCACTGGGT
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
94
homolog,
subfamily SEQ ID NO.37
C, member
12
interferon- CATAACCGAGCGGTA
14 IFI44L induced NM_006820 124 4-5 900 TAGGATATAT
protein 44- SEQ ID NO.14
like SEQ ID NO.38
GCGACGATGTAGATT seryl- 15 SARS tRNA NM_006513 101 17 216 TCGGGCAGAC
synthetase SEQ ID NO.15
SEQ ID NO.39
SMAD GGAGCGCCCAACACG
specific E3 ACCGGCATCC
16 SMURF2 ubiquitin NM_022739.3 100
protein SEQ ID NO.16 7 8 960
SEQ ID NO.40
ligase 2
splicing CAGGATCCAAGCCAG
factor, 3-4 ATTGATTGGG
SFRS18 arginine/se NM_032870.2
17 rine-rich SEQ ID NO.17 56 316
18 SEQ ID NO.41
nuclear
receptor TGGACTATTCCAGGTT
18 NR4A2 subfaniily NM_006186 69 CCAGGCGAA
4, group SEQ ID NO.18 5-6 1491
A, SEQ ID NO.42
niember 2
cyclin-
dependent
CDKNIB TGCAACCGACGATTCT
kinase BC001971 TCTACTCAA
19 inhibitor SEQ ID N0.19 71 1-2 857
I B (p27
SEQ ID NO.43
Kip 1)
eukaryotic
translation GAGTGGGATTCCAGG
20 EIF3D initiation NMSEQ ID NO.20 .20 132 12-13 1367 CACTGTAATG
factor 3,
subunit D SEQ ID NO.44
21 18S Eukaryotic X03205.1 TGGAGGGCAAGTCTG
18S rRNA SEQ ID NO.21 187 609 GTGCCAGCAG
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
SEQ ID NO.45
GCGCCATATCAGGGT
22 RPS9 ribosonial NM001013.3 CCGCAAGCAG
protein S9 SEQ ID NO.22 156 4-5 467
SEQ ID NO.46
CCTTTGCCGATCCGCC
23 ACTB actin, beta NM_001101.2 171 1-1 49 GCCCGTCCA
SEQ ID NO.23 SEQ ID NO.47
heatshock
protein GCATGATCAAGCTAG
90kDa GTCTAGGTAT
24 HSPCB alpha NM 007355.2 155 11-12 2142
(cytosolic) SEQ ID NO.24 SEQ ID NO.48
, class B
member I
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
96
Example 4
GO analysis of differentially expressed genes between BRCA1/2
mutation carriers versus non-carriers
Gene Ontology analysis was performed on a list of genes which had different
expression patterns for either the BRCA1 or the BRCA2 groups as compared to
the control group. Analysis in the BRCA2 mutation group, revealed that most
of the genes are related to gene expression regulation pathways involved in
DNA repair processes (i.e. DNAJ, RAD51), cell cycle regulation (i.e. cyclin H,
Kip1), cancer associated (i.e. RPS6KB1, RBL2) and apoptosis. Furthermore, a
number of these genes were shown to function together (for example SMURF2
and RNF11). Mutations in BRCAI have been shown to impair the homologous
repair of double stranded breaks in the DNA, and the BRCA1 protein has been
shown to function in cell cycle regulation. Therefore, these results might be
relevant to the function of BRCA1 and BRCA2. The next largest group of genes
is related to the hematological system functioning and the immune system (i.e.
HLA-DQB1, Granulysin), as can be expected when tested in lymphocytes. It
have been previously shown that BRCA1 regulates targets of the innate
immune system dependent on IFN signaling [Buckley N.E. et al. Mol. Cancer
Res. 5(3):261-70 (2007)].
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
97
Table 5. Genes differentially expressed (with p<0.05 and >1.5 fold) in BRCA1
gene mutation carriers.
Affimetrix ID Representative Gene Title
Public ID
204972_at NM_016817 2'-5'-oligoadenylate synthetase 2, 69/71kDa
202672_s_at NM_001674 activating transcription factor 3
222108 at AC004010 adhesion molecule with Ipg-like domain 2
201000_at NM_001605 alanyl-tRNA synthetase
213503 x at BE908217 annexin A2
201590 x at NM 004039 annexin A2
201525_at NM_001647 apolipoprotein D
203747_at NM_004925 aquaporin 3
205047_s_at NM_001673 asparagine synthetase
211852 s at AF106861 attractin
211725_s_at BC005884 BH3 interacting domain death agonist ; BH3
interacting domain death
agonist
211190 x at AF054817 CD84 antigen (leukocyte antigen)
218085 at NM_015961 chromatin modifying protein 5
220235 s at NM 018372 chromosome 1 open reading frame 103
206707 x at NM 015864 chromosome 6 open reading frame 32
218325 s at NM 022105 death associated transcription factor 1
Systematic Genbank Description
222154 s at AK002064 DNA polymerase-transactivated protein 6
200880 at AL534104 DnaJ (Hsp40) homolog, subfamily A, member 1
200881 s at AL534104 DnaJ (Hsp40) homolog, subfamily A, member 1
209015 s at BC002446 DnaJ (Hsp40) homolog, subfamily B, member 6
219551 at NM 018456 ELL associated factor 2
37145_at M85276 Granulysin
206976_s_at NM_006644 heat shock 105kDa/110kDa protein 1
208744 x at D86956 heat shock 105kDa/110kDa protein 1
200799 at NM 005345 heat shock 70kDa protein lA
200800 s at NM 005345 heat shock 70kDa protein 1A ; heat shock
70kDa protein 1B
202581 at NM 005346 heat shock 70kDa protein 1B
211968_s_at A1962933 heat shock 90kDa protein 1, alpha
215933_s_at Z21533 hematopoietically expressed homeobox
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
98
220387_s_at NM_007071 HERV-H LTR-associating 3
211597_s_at AB059408 homeodomain-only protein ; homeodomain-only
protein
203914_x_at NM_000860 hydroxyprostaglandin dehydrogenase 15-(NAD)
205404_at NM_005525 hydroxysteroid (11-beta) dehydrogenase 1
213674_x_at A1858004 immunoglobulin heavy constant delta
214973_x_at AJ275469 immunoglobulin heavy constant delta
211798_x_at AB001733 immunoglobulin lambda joining 3
211881_x_at AB014341 immunoglobulin lambda joining 3
205786_s_at NM_000632 integrin, alpha M (complement component
receptor 3, alpha; also known as CD11b (p170),
macrophage antigen alpha polypeptide);
integrin, alpha M (complement component
receptor 3, alpha; also known as CD11b (p170),
macrophage antigen alpha polypeptide)
219209 at NM 022168 interferon induced with helicase C domain 1
208436_s_at NM_004030 interferon regulatory factor 7
202220 at NM 014949 KIAA0907
212714_at AL050205 La ribonucleoprotein domain family, member 4
221274_s_at NM_030805 lectin, mannose-binding 2-like ; lectin,
mannose-binding 2-like
205569_at NM_014398 lysosomal-associated membrane protein 3
209199_s_at L08895 MADS box transcription enhancer factor 2,
polypeptide C
(m oc te enhancer factor 2C)
209200_at AL536517 MADS box transcription enhancer factor 2,
polypeptide C
(m oc te enhancer factor 2C)
213537_at AI128225 major histocompatibility complex, class II, DP
alpha 1
209823_x_at M17955 major histocompatibility complex, class II, DQ
beta 1
208306_x_at NM_021983 Major histocompatibility complex, class II, DR
beta 3
201475_x_at NM_004990 methionine-tRNA synthetase
213733_at BF740152 myosin IF
210218_s_at U36501 nuclear antigen SplOO
219165_at NM_021630 PDZ and LIM domain 2 (mystique)
204286_s_at NM_021127 phorbol-12-myristate-13-acetate-induced
protein 1
210617_at U87284 phosphate regulating endopeptidase homolog,
X-linked (hypophosphatemia, vitamin D
resistant rickets)
201397_at NM_006623 phosphoglycerate dehydrogenase
202446 s_at A1825926 phospholipid scramblase 1
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
99
202430_s_at NM_021105 phospholipid scramblase 1
220892_s_at NM_021154 phosphoserine aminotransferase 1
205267_at NM_006235 POU domain, class 2, associating factor 1
201703_s_at NM_002714 protein phosphatase 1, regulatory subunit 10
219412_at NM022337 RAB38, member RAS oncogene family
212125_at NM_002883 Ran GTPase activating protein 1
214369_s_at A1688812 RAS guanyl releasing protein 2 (calcium and
DAG-re ulated)
206220_s_at NM_007368 RAS p21 protein activator 3 -
209325_s_at U94829 regulator of G-protein signalling 16
213566_at NM_005615 ribonuclease, RNase A family, k6 ; ribonuclease,
RNase A family, k6
213502_x_at AA398569 similar to bK246H3.1 (immunoglobulin lambda-
like polypeptide 1,
re-B-cell s ecific)
213820_s_at T54159 START domain containing 5
209999_x_at AB005043 suppressor of cytokine signaling 1
209307 at AB014540 SWAP-70 protein
216180_s_at AK026758 synaptojanin 2
222010_at BF224073 t-complex 1
220558_x_at NM_005705 tetraspanin 32
210176_at AL050262 toll-like receptor 1
200629_at NM_004184 tryptophanyl-tRNA synthetase
213361 at AW129593 tudor domain containing 7
201535_at NM_007106 ubiquitin-like 3
206133 at NM 017523 XIAP associated factor-1
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
100
Table 6. Genes differentially expressed (with p<0.05 and >2 fold) in BRCA2
gene mutation carriers.
Affimetrix Representative Gene Title
ID Public ID
201963_at NM_021122 acyl-CoA synthetase long-chain family member 1
208002_s_at NM_007274 acyl-CoA thioesterase 7
215728_s_at AL031848 acyl-CoA thioesterase 7
200734_s_at BG341906 ADP-ribosylation factor 3
211622_s_at M33384 ADP-ribosylation factor 3 ; ADP-ribosylation factor 3
221589_s_at AW612403 Aldehyde dehydrogenase 6 family, member Al
208859_s_at AI650257 alpha thalassemia/mental retardation syndrome X-
linked (RAD54 homolog, S. cerevisiae)
203566_s_a.t NM_000645 amylo-1, 6-glucosidase, 4- alpha- glucanotransferase
(glycogen debranching enzyme, glycogen storage
disease type III)
200602_at NM_000484 amyloid beta (A4) precursor protein (peptidase nexin-
II, Alzheimer disease)
213106_at A1769688 ATPase, aminophospholipid transporter (APLT), Class
I, type 8A, member 1
207521_s_at AF068220 ATPase, Ca++ transporting, ubiquitous
201242_s_at BC000006 ATPase, Na+/K+ transporting, beta 1 polypeptide
203140_at NM001706 B-cell CLL/lymphoina 6 (zinc finger protein 51) ; B-cell
CLL/lymphoma 6 (zinc finger protein 51)
221478_at AL132665 BCL2/adenovirus E1B 19kDa interacting protein 3-
like; BCL2/adenovirus E1B 19kDa interacting protein
3-like
218090_s_at NM_018117 bromodomain and WD repeat domain containing 2
214450_at NM_001335 cathepsin W (lymphopain) ; cathepsin W (lymphopain)
218871 x at NM 018590 chondroitin sulfate Ga1NAcT-2
205583 s at NM 024810 Chromosome X open reading frame 45
205518_s_at NM_003570 cytidine monophosphate-N-acetylneuraminic acid
hydroxylase (CMP-N-acetylneuraminate
monooxygenase)
221628_s_at AF326966 cytokine-like nuclear factor n-pac
213998_s_at AW188131 DEAD (Asp-Glu-Ala-Asp) box polypeptide 17
212107 s at BE561014 DEAH (Asp-Glu-Ala-His) box polypeptide 9
Systematic Genbank Description
204646_at NM000110 dihydropyrimidine dehydrogenase
219237_s_at NM_024920 DnaJ (Hsp40) homolog, subfamily B, member 14
201693_s_at NM_001964 early growth response 1
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
101
206115at NM_004430 early growth response 3
209004_s_at AF142481 F-box and leucine-rich repeat protein 5
201540 at NM 001449 four and a half LIM domains 1
206492_at NM_002012 fragile histidine triad gene
215001_s_at AL161952 glutamate-ammonia ligase (glutamine synthetase)
208798xat AF204231 golgi autoantigen, golgin subfamily a, 8A
212525_s_at AA760862 H2A histone family, member X
202979_s_at NM021212 HCF-binding transcription factor Zhangfei
213359_at W74620 Heterogeneous nuclear ribonucleoprotein D (AU-rich
element RNA binding protein 1, 37kDa)
214753_at AW084068 Hypothetical gene CGO12
221899_at A1809961 Hypothetical gene CGO12
213375_s_at N80918 Hypothetical gene CGO18
218051_s_at NM_022908 Hypothetical protein FLJ12442
213212_x_at AI632181 Hypothetical protein LOC161527
213931_at A1819238 inhibitor of DNA binding 2, dominant negative helix-
loop-helix protein ; inhibitor of DNA binding 2B,
dominant negative helix-loop-helix protein
203607_at NM014937 inositol polyphosphate-5-phosphatase F
203628_at H05812 insulin-like growth factor 1 receptor
38892 at D87077 KIAA0240
203049_s_at NM_014639 KIAA0372
207719_x_at NM_014812 KIAA0470
212633_at AL132776 KIAA0776
218219_s_at NM018697 LanC lantibiotic synthetase component C-like 2
(bacterial)
205668_at NM_002349 lymphocyte antigen 75
213975_s_at AV711904 lysozyme (renal amyloidosis) ; leukocyte
immunoglobulin-like receptor, subfamily B (with TM
and ITIM dornains), member 1
220615_s_at NM_018099 male sterility domain containing 1
201755_at NM006739 MCM5 minichromosome maintenance deficient 5, cell
division cycle 46 (S. cerevisiae)
213158_at BG251521 MRNA; cDNA DKFZp586B211 (from clone
DKFZp586B211)
201467_s_at AI039874 NAD(P)H dehydrogenase, quinone 1
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
102
205005_s_at AW293531 N-myristoyltransferase 2
205006_s_at NM_004808 N-myristoyltransferase 2
201577_at , NM000269 non-metastatic cells 1, protein (NM23A) expressed in
216321_s_at X03348 nuclear receptor subfamily 3, group C, member 1
(glucocorticoid receptor)
207564_x_at NM_003605 0-linked N-acetylglucosamine (G1cNAc) transferase
(UDP-N-acetylglucosamine:polypeptide-N-
acetylglucosaminyl transferase)
201246_s_at NM_017670 OTU domain, ubiquitin aldehyde binding 1
201490_s_at NM005729 peptidylprolyl isomerase F (cyclophilin F)
209422_at AY027523 PHD finger protein 20
218640_s_at BF439250 pleckstrin homology domain containing, family F (with
FYVE domain) meinber 2
207002_s_at NM_002656 pleiomorphic adenoma gene-like 1
222273_at A1419423 poly(A) polymerase gamma
212016_s_at AA679988 Polypyrimidine tract binding protein 1
211791_s_at AF044253 potassium voltage-gated channel, shaker-related
subfamily, beta member 2
201300_s_at NM_000311 prion protein (p27-30) (Creutzfeld-Jakob disease,
Gerstmann-Strausler-Scheinker syndrome, fatal
familial insomnia)
208988_at BE675843 PR01880 protein
218668_s_at NM_021183 RAP2C, member of RAS oncogene family
221524_s_at AL138717 Ras-related GTP binding D
209285_s_at N38985 retinoblastoma-associated protein 140
205407_at NM021111 reversion-inducing-cysteine-rich protein with kazal
motifs
201167_x_at NM_004309 Rho GDP dissociation inhibitor (GDI) alpha
213350_at BF680255 Ribosomal protein S11
209889_at AF274863 SEC31-like 2 (S. cerevisiae)
201996_s_at AL524033 spen homolog, transcriptional regulator (Drosophila)
203455_s_at NM_002970 spermidine/spermine Ni-acetyltransferase
210592_s_at M55580 spermidine/spermine N1-acetyltransferase
210172_at D26121 splicing factor 1
215113_s_at AK000923 SUM01/sentrin/SMT3 specific peptidase 3
213510_x_at AW194543 TL132 protein
CA 02692803 2010-01-07
WO 2009/007958 PCT/IL2008/000934
103
212983_at NM_005343 v-Ha-ras Harvey rat sarcoma viral oncogene homolog
209348_s_at BF508646 v-maf musculoaponeurotic fibrosarcoma oncogene
homolog (avian)
220118_at NM_014383 zinc finger and BTB domain containing 32
203739_at NM_006526 zinc finger protein 217
212774 at AJ223321 zinc finger protein 238
221645_s_at M27877 zinc finger protein 83 (HPFl)
214670_at AA653300 zinc finger with KRAB and SCAN domains 1
In summary, identification of marker genes differentially expressed in
carriers
of BRCA1/2 gene mutations as compared to non-carrier controls, by the present
invention, demonstrate the feasibility of using such marker genes or any
combination thereof in the diagnosis of carriers.
It will be evident to those skilled in the art that the invention is not
limited to
the details of the foregoing illustrative examples and that the present
invention may be embodied in other specific forms without departing from the
essential attributes thereof, and it is therefore desired that the present
embodiments and examples be considered in all respects as illustrative and not
restrictive, reference being made to the appended claims, rather than to the
foregoing description, and all changes which come within the meaning and
range of equivalency of the claims are therefore intended to be embraced
therein.