Note: Descriptions are shown in the official language in which they were submitted.
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
METHODS FOR ENHANCING MUSCLE PERFORMANCE AND TONE
CROSS-REFERENCE TO RELATED APPLICATIONS
[0001] This application claims the benefit of US Provisional
Application No. 61/080,841, filed July 15, 2008, herein
incorporated by reference. This application is a continuation-in-
part of and claims priority to U.S. Application No. 11/966,851, and
International Application No. PCT/US2007/089124, both filed
December 28, 2008, the disclosures of which is incorporated herein
by reference.
ACKNOWLEDGEMENT OF GOVERNMENT SUPPORT
[0002] This work was supported by National Institutes of Health
Grant No. 1 F32 AR053803-01 (NRSA Fellowship). Therefore, the
Government of the United States has certain rights in this
invention.
FIELD OF THE INVENTION
[0003] This disclosure concerns the use of agonists of AMP-
activated protein kinase (AMPK) for improving exercise and
modifying energy metabolism in a subject. The disclosure also
provides methods of treating muscle wasting diseases and disorders
and promoting muscle tone in sedentary subjects. The disclosure
also relates to a combination of AMPK and peroxisome proliferator-
activated receptor (PPAR)b agonists for improving exercise
performance in a subject, and methods for identifying compounds
that modulate gene expression profiles associated with muscle tone,
endurance or performance.
BACKGROUND
[0004] Skeletal muscle is an adaptive tissue composed of
multiple myofibers that differ in their metabolic and contractile
properties including oxidative slow-twitch (type I), mixed
oxidative/glycolytic fast- twitch (type Ha) and glycolytic fast-
twitch (type Hb) myofibers (Fluck et ai., Rev. Physiol. Biochem.
Pharmacol., 146:159-216, 2003; Pette and Staron, Microsc. Res.
Tech., 50:500-509, 2000). Type I muscle fibers preferentially
express enzymes that oxidize fatty acids, contain slow isoforms of
contractile proteins and are more resistant to fatigue than are
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
glycolytic muscle fibers (Fluck et al., Rev. Physiol. Biochem.
Pharmacol., 146:159-216, 2003; Pette and Staron, Microsc. Res.
Tech., 50:500-509, 2000). Type II fibers preferentially metabolize
glucose and express the fast isoforms of contractile proteins
(Fluck et al., Rev. Physiol. Biochem. Pharmacol, 146:159-216, 2003;
Pette and Staron, Microsc. Res. Tech., 50:500-509, 2000).
[0005] Endurance exercise training triggers a complex
remodeling program in skeletal muscle that progressively enhances
performance in athletes such as marathon runners, mountain climbers
and cyclists. This involves changes in metabolic programs and
structural proteins within the myofibers that alter the energy
substrate utilization and contractile properties that act to reduce
muscle fatigue (Fluck et al., Rev. Physiol. Biochem. Pharmacol,
146:159-216, 2003; Pette and Staron, Microsc. Res. Tech., 50:500-
509, 2000). Training based adaptations in the muscle are linked to
increases in the expression of genes involved in the slow-twitch
contractile apparatus, mitochondrial respiration and fatty acid
oxidation (Holloszy and Coyle, J. Appl. Physiol. , 56:831-838,
1984; Booth and Thomason, Physiol. Rev. , 71:541-585, 1991; Schmitt
et al., Physiol. Genomics, 15:148-157, 2003; Yoshioka et al., FASEB
J., 17:1812-1819, 2003; Mahoney et al., FASEB J., 19:1498-1500,
2005; Mahoney and Tarnopolsky, Phys. Med. Rehabil Clin. N. Am.,
16:859-873, 2005; Siu et al., J. Appl Physiol, 97:277-285, 2004;
Gamier et al., FASEB J., 19:43-52, 2005; Short et al., J. Appl
Physiol, 99:95-102, 2005; Timmons et al., FASEB J., 19:750-760,
2005). Such exercise training-related adaptations can improve
performance and protect against obesity and related metabolic
disorders (Wang et al., P. Biol, 2:e294, 2004; Koves et al., J.
Biol. Chem., 280:33588-33598, 2005). Moreover, skeletal muscles
rich in oxidative slow-twitch fibers are resistant to muscle
wasting (Minnaard et al., Muscle Nerve. 31: 339-48, 2005).
SUMMARY
[0006] The disclosure also provide a method of treating a
muscle wasting disease, muscle atrophy or aging comprising
administering to a subject a composition comprising an AMPK
agonist, wherein the muscle tone, mass or endurance is promoted.
2
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
In one embodiment, the method comprises treating a subject that may
have an immobilized limb or which may be immobilized due to other
medical treatments to promote or maintain muscle tone in the
subject.
[0007] The disclosure demonstrates that unexpected finding that
orally active AMPK agonists are sufficient as a single agent to
improve exercise endurance by nearly 45% in non-exercised subjects.
[0008] The disclosure provides a method for enhancing an
exercise effect in a subject, comprising administering to a subject
an AMP kinase (AMPK) agonist wherein an exercise effect is
enhanced. The AMPK agonist can be any AMPK agonist, derivatives,
salts or esters thereof. In one embodiment, the AMPK agonist is
AICAR. The method can further comprise administering to the
subject an effective amount of a PPAR5 agonist (e.g., GW1516),
thereby further enhancing the exercise effect in the subject. The
subject can be a racing animal including a human, equine, or
canine.
[0009] The disclosure further comprises a method for
identifying the use of performance-enhancing substances in an
exercise-trained subject comprising determining in a biological
sample taken from an exercise-trained subject the presence of an
AMPK agonist and/or expression of one or more molecules listed in
Tables 2, 4 or 6.
[0010] The disclosure also provides a composition comprising an
AMPK agonist and a PPAR6 agonist in a pharmaceutically acceptable
carrier. The composition may be an energy supplement, beverage,
food product or pharmaceutical. The AMPK agonist or PPAR agonist
can be a salt, ester, prodrug, precursor or derivative thereof.
[0011] This disclosure illustrates that, despite expectations
to the contrary, pharmacological activation of AMPK or endogenous
PPAR6 in adults, promote remodeling of skeletal muscle to an
oxidative phenotype or increase running endurance in such subjects.
In addition, agonist-induced activation of endogenous PPAR6 in
combination with exercise led to a unique "gene expression
signature" in skeletal muscle, which was distinct from the gene
expression profile obtained by either exercise or drug intake
3
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
alone, and revealed direct interactions between PPARf and exercise-
induced kinases (such as AMPK al and/or AMPK 2).
[0012] These and other discoveries described herein serve as
the basis for disclosed methods. For example, it can now be
appreciated that PPAR6 agonists (e.g., GW1516) used in combination
with exercise can enhance exercise-induced effects, such as to
improve exercise endurance (e.g., running endurance) even more than
may be achieved by exercise alone. In another example, the
expression of one or more genes and/or proteins that are uniquely
regulated by the combination of exercise and PPAR5 agonist
administration can be used to identify subjects using drugs to
enhance exercise performance. In still other examples, the newly
identified protein complexes, including PPAR5 and exercise-induced
kinases (such as AMPK al and/or AMPK cc2), can be used to identify
agents that have potential to affect PPAR6-regulated gene networks
and the corresponding downstream biochemical and/or physiological
effects.
[0013] The foregoing and other features will become more
apparent from the following detailed description of several
embodiments, which proceeds with reference to the accompanying
figures.
BRIEF DESCRIPTION OF THE FIGURES
[0014] FIG. 1A is a series of bar graphs showing the effects of
orally administered PPAR5 agonist (GW1516) on mRNA expression
levels of three biomarkers of fatty acid oxidation, uncoupling
protein 3 (UCP3), carnitine palmitoyltransferase I (mCPT I), and
pyruvate dehydrogenase kinase, isoenzyme 4 (PDK4), in quadriceps
muscle isolated from sedentary vehicle-treated (V), sedentary
GW1516-treated (GW), sedentary VP16-PPAR5 transgenic (TG), and
sedentary wild-type littermates of VP16-PPAR5 transgenic mice (WT).
Data are presented as mean SEM of N=4-9 mice each analyzed in
triplicate. * Represents a statistically significant difference
between V and GW1516 groups (p<0.05, unpaired student's t-test), or
TG and WT groups (p<0.05, unpaired student's t-test).
[0015] FIG. 1B-D are a series of bar graphs showing the
regulation of oxidative genes UCP3, mCPT I, and PDK4 by GW1516 (GW)
4
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
in wild-type (WT) and PPAR6 null (KO) primary muscle cells. *
represents statistical significance between V and indicated groups
(p<0.05, One Way ANOVA; post hoc: Dunnett's Multiple Comparison
Test).
[0016] FIG. 1E is a series of bar graphs showing running
endurance of vehicle-treated sedentary (V; open bars) and GW1516-
treated sedentary (GW; black bars) mice before (Week 0) and after
(Week 5) treatment. Running endurance is quantified by the amount
of time for which (left panel) or the distance (right panel)
animals in each group ran on the treadmill. Data is represented as
mean SD values from N=6 mice.
[0017] FIGS. 2A-C show the effects of administration of a PPAR5
agonist, GW1516, on the gastrocnemius muscle of sedentary (V or GW)
or trained (Tr or Tr+GW) mice. FIG. 2A shows digital images of
representative meta-chromatically stained frozen cross-sections of
gastrocnemius muscle from vehicle-treated, sedentary (V), GW1516-
treated, sedentary (GW), vehicle-treated, exercised (Tr) and
GW1516-treated, exercised (Tr+GW) mice. Type I (slow oxidative)
fibers are darkly stained. FIG. 2B is a bar graph showing the
percentage of type I fibers (as a percentage of the total fibers)
in V, GW, Tr, and Tr+GW gastrocnemius (N=3). FIG. 2C is a bar graph
showing the fold change in mitochondrial DNA to nuclear DNA ratio
in V (left bar), GW (left center bar), Tr (right center bar), and
Tr+GW (right bar) groups of mice (N=9). Data in (B) and (C) are
presented as mean SEM. In each bar graph, * represents a
statistical difference between V and the group(s) indicated by
asterisk (p<0.05, One-Way ANOVA; post hoc: Dunnett's Multiple
Comparison Test).
[0018] FIGS. 3A-D are a series of bar graphs showing gene
expression in quadriceps muscle isolated from V, GW, Tr and Tr+GW
groups. FIG. 3A shows the relative gene expression levels of
biomarkers for fatty acid oxidation (UCP3, mCPT I, PDK4; from left
to right). (B) shows the relative gene expression levels of
biomarkers for fatty acid storage (SCD1, FAS, SREBPIc). (C) shows
the relative gene expression levels of biomarkers for fatty acid
uptake (FAT/CD36, LPL). Data is presented as mean SEM of N=9
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
mice, each analyzed in triplicate. * represents statistically
significant difference between V and the group(s) indicated by
asterisk (p<0.05, One Way ANOVA; post hoc: Dunnett's Multiple
Comparison Test). (D) shows images of Western blots illustrating
protein expression levels of oxidative biomarkers (myoglobin, UCP3,
CYCS, SCD1) and loading control (tubulin) in protein lysates
prepared from quadriceps (N=3).
[0019] FIG. 4 shows a graph of muscle triglyceride levels in
gastrocnemius muscle of V, GW, Tr and Tr+GW mice. Data is presented
as mean SEM of N=9 mice, each analyzed in triplicate. *
represents statistical significance between V and group(s)
indicated by asterisk (*p<0.05, One Way ANOVA; post hoc:Dunnett's
Multiple Comparison Test).
[0020] FIGS. 5A-C are bar graphs showing the effects of GW1516
treatment on running endurance in exercise-trained mice. Bar graphs
of the (A) time and (B) distance that vehicle- (V; open bars) and
GW1516-treated (GW; black bars) mice ran on a treadmill before
(Week 0) and after (Week 5) exercise training. Data is represented
as mean SD of N=6 mice. *** represents statistically significant
difference between V and GW groups (p<0.001; One Way ANOVA; post
hoc:Tukey's Multiple Comparison Test). (C) is a bar graph showing
epididymal white adipose to body weight ratio in V, GW, Tr and
Tr+GW mice. Data is presented as mean SEM of N=9 mice, each
analyzed in triplicate. * represents statistical significance
between V and group(s) indicated by asterisk (*p<0.05, One Way
ANOVA; post hoc:Dunnett's Multiple Comparison Test).
[0021] FIG. 5D shows digital images of H&E-stained cross-
sections of epididymal white adipose from V, GW, Tr and Tr+GW mice.
Similar results were obtained from N=3 mice. * represents
statistical significance between V and group(s) indicated by
asterisk (*p<0.05, One Way ANOVA; post hoc:Dunnett's Multiple
Comparison Test).
[0022] FIG. 6 shows a Venn diagram comparing GW, Tr and Tr+GW
target genes identified in microarray analysis of quadriceps. Also
shown are a classification of target genes in Tr+GW mice and
relative expression of 48 unique TR+GW target genes in GW, TR,
6
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
TR+GW, and VP16-PPARdelta muscles. Data is an average of N=3
samples in each group. The selection criteria used a p<0.05 on
Bonferroni's multiple comparison test and a fold change greater
than 1.5.
[0023] FIG. 7A is a series of Western blot images showing AMPK
activation by exercise. The levels of phospho-AMPK (phospho-AMPK)
and total- AMPK in quadriceps muscle of sedentary (Sed/C57B1) and
exercise-trained (Tr/C57B1) mice (N=5-7) are shown.
[0024] FIG. 7B is a series of Western blot images showing AMPK
activation by VP16-PPARd over-expression. The levels of phospho-
AMPK (phospho-AMPK) and total- AMPK in quadriceps muscle of
sedentary wild-type or transgenic mice (Sed/WT or Sed/TG) are
shown.
[0025] FIGS. 8A-C show the synergistic regulation of muscle
gene expression by PPAR6 and AMPK. (A) Venn diagram comparing GW,
Al, and AI+GW target genes identified in microarray analysis of
quadriceps. Data is an average of N=3 samples in each group. The
selection criteria used a p<0.05 on Bonferroni's multiple
comparison test and fold change greater than 1.5. (B) Comparison of
Tr+GW and AI+GW dependent gene signatures identified in quadriceps.
Data is an average of N=3 samples in each group. The selection
criteria used is similar to one used in FIG. 8A. (C) Classification
of 52 targets that were common to Tr+GW and AI+GW gene signatures.
[0026] FIGS. 9A-H show the expression of (A) UCP3, (B) mCPT I,
(C) PDK4, (D) SCD1, (E) ATP citrate lyase, (F) HSL, (G) mFABP, and
(H) LPL transcripts in quadriceps of mice treated with vehicle (V),
GW 1516 (GW), AICAR (AT) and the combination of the two drugs (GW+
Al) for 6 days. Data is presented as mean SEM of N=6 mice in each
group, analyzed in triplicate. * Indicates statistically
significant difference between V and indicated groups (p<0.05, One
Way ANOVA; post hoc: Dunnett's Multiple Comparison Test).
[0027] FIGS. 1OA-L demonstrate the AMPK-PPAR5 interaction. (A-
D) show the expression of metabolic genes in wild type and PPAR6
null (KO) primary muscle cells treated with V, GW, Al and GW+AI
(bars from left to right) for 24 hours. In (E-F, J), AD293 cells
were transfected with PPAR6+RXRa+Tk-PPRE along with control vector,
7
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
AMPK al, cc2 and/or PGC in as indicated above. (E) Induction of
basal PPAR6 transcriptional activity by AMPK al or cc2. (F) Dose-
dependent induction of PPAR6 transcriptional activity is enhanced
by AMPKal (closed circle) or AMPK cc2 (closed square) compared to
control (open triangle). In (G-I, K), AD293 cells were transfected
and processed as indicated. (G-H) Representative blot showing co-
immunoprecipitation of transfected (G) or endogenous (H) AMPK with
Flag-PPAR5. (I) Metabolic p32 labeling of PPAR6 in AD293 cells
transfected as described. (J) Synergistic regulation of basal (V)
and ligand (GW) dependent PPAR5 transcriptional activity by AMPK
cc2 subunit and PGC in. (K) Co- immunoprecipitation of PPAR6 but
not AMPK a2 subunit with Flag-PGCla. (L) Model depicting exercise-
PPAR6 interaction in re -programming muscle genome.
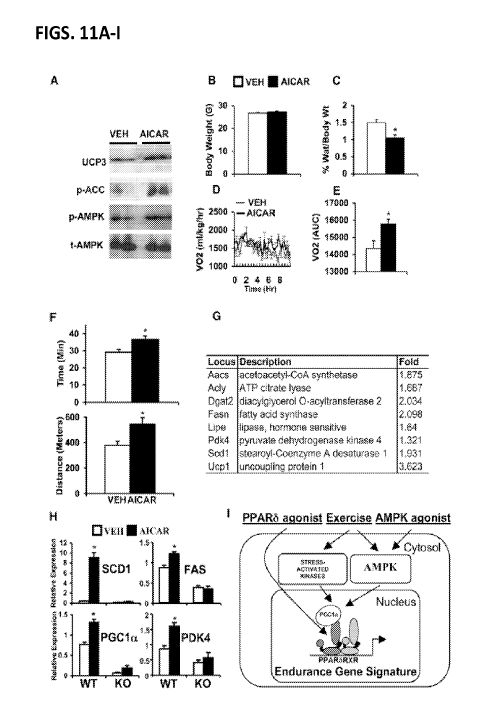
[0028] Figure 11A-I shows that AICAR increases running
endurance. (A-F) C57B1/6J mice were treated with vehicle (open bars
or thin lines) or AICAR (500 mg/kg/day, 4 weeks) (closed bars or
thick lines). (A) Representative immunoblots showing levels of
UCP3, phospho-acetyl CoA carboxylase (ACC), phospho-AMPK, and
total-AMPK in quadriceps. (B) Average body weight. (C) Percent
epididymal fat mass to body weight ratio. (D) Oxygen consumption
rates (mg/kg/hr) measured over 12 hr period. (E) Data in (D)
represented as AUC. (F) Running endurance measured as a function of
time (upper panel) and distance (lower panel). (G) Representative
oxidative genes induced by AICAR treatment (250 mg/kg/day, 6 days).
(H) Expression of oxidative biomarkers (Scdl, Fasn, Ppargcla, Pdk4)
in wild-type and PPARd null primary myoblast treated with vehicle
(open bars) or AICAR (closed bars) for 72 hr. (I) Model depicting
the interaction between exercise and AMPK-PPARd in reprogramming
muscle genome. Data in (B) and (C) (n = 10), (D) and (E) (n = 4),
(F) (n =15-20), and (H) (n = 9) are presented as mean SEM, and *
indicates statistical significance (p <0.05, unpaired student's t
test).
SEQUENCE INFORMATION
[0029] Nucleic acid and amino acid sequences may be referred to
herein by GenBank accession number. It is understood that the
8
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
sequences given such GenBank accession numbers are incorporated by
reference as they existed and were known as of December 29, 2007.
DETAILED DESCRIPTION
[0030] Unless specifically noted otherwise herein, the
definitions of the terms used are standard definitions used in the
art of pharmaceutical sciences. As used in the specification and
the appended claims, the singular forms "a," "an" and "the" include
plural referents unless the context clearly dictates otherwise.
Thus, for example, reference to "a pharmaceutical carrier" includes
mixtures of two or more such carriers, and the like.
[0031] Also, the use of "or" means "and/or" unless stated
otherwise. Similarly, "comprise," "comprises," "comprising"
"include," "includes," and "including" are interchangeable and not
intended to be limiting.
[0032] It is to be further understood that where descriptions
of various embodiments use the term "comprising," those skilled in
the art would understand that in some specific instances, an
embodiment can be alternatively described using language
"consisting essentially of" or "consisting of."
[0033] Unless defined otherwise, all technical and scientific
terms used herein have the same meaning as commonly understood to
one of ordinary skill in the art to which this disclosure belongs.
Although any methods and reagents similar or equivalent to those
described herein can be used in the practice of the disclosed
methods and compositions, the exemplary methods and materials are
now described.
[0034] All publications mentioned herein are incorporated
herein by reference in full for the purpose of describing and
disclosing the methodologies, which are described in the
publications, which might be used in connection with the
description herein. The publications discussed above and
throughout the text are provided solely for their disclosure prior
to the filing date of the disclosure. Nothing herein is to be
construed as an admission that the inventors are not entitled to
antedate such disclosure by virtue of prior disclosure.
9
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
[0035] Given the numerous benefits of exercise on general
health, identification of orally active agents that mimic or
potentiate the genetic effects of endurance exercise is a long-
standing, albeit elusive, medical goal. High doses of certain
natural extracts such as resveratrol can improve endurance (Lagouge
et al., 2006). The aerobic effects of resveratrol are thought to
depend on activation of SIRT1-PGCla coactivator complex in skeletal
muscle. However, the downstream transcriptional factor(s) targeted
by SIRT1/PGCla in mediating these effects are not known. More
importantly, both SIRT1/PGC1a and resveratrol activate multiple
targets, and thus whether there is a specific signaling pathway
that can be selectively activated by a synthetic drug to improve
endurance is not known.
[0036] Exercise training activates a number of transcriptional
regulators and serine-threonine kinases in skeletal muscles that
contribute to metabolic reprogramming (Bassel-Duby and Olson,
2006). Overexpression of a constitutively active PPAR5 (VP16-PPAR5)
in skeletal muscles of transgenic mice preprograms an increase in
oxidative muscle fibers, enhancing running endurance by nearly 100%
in untrained adult mice (Wang et al., 2004). One of the best
understood serine-threonine kinases is AMP-activated protein kinase
(AMPK), a master regulator of cellular and organism metabolism.
[0037] AMP kinase agonists such as AICAR have been studied for
insulin regulation, diabetes and obesity. However, AMP kinases
have not previously been demonstrated to promote muscle tone or to
improve endurance or exercise. The disclosure provides that AMPK
agonists provide a beneficial effect for muscle wasting diseases
and disorders, benefits to sedentary subject or immobilized limbs
and in combination with PPARd agonist an unexpected synergistic
effect.
[0038] The disclosure demonstrates that PPAR6 agonist such as,
for example, GW1516 (shown to be bioactive in humans) enables mice
to run 60%-75% longer and further than the nontreated controls
only; however, such an effect is only seen when administration of a
PPAR5 agonist is combined with exercise training. This "super-
endurance phenotype" is linked to a transcriptional boost provided
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
by exercise-activated AMPK resulting in a novel endurance gene
signature (see, e.g., Figure 10L)
[0039] The disclosure also demonstrated that this super-
endurance phenotype can be obtained, in the absence of exercise
training, by orally active AMPK agonist and that such an AMPK
agonist is sufficient as a single agent to improve running
endurance by nearly 45% in nonexercised subjects.
[0040] AMP-activated protein kinase (AMPK) and AMPK kinase
(AMPKK) comprise a protein kinase cascade. The AMPK cascade
regulates fuel production and utilization intracellularly. For
example, low cellular fuel (e.g., an increase in AMP concentration)
increase AMPK activity. Once activated, AMPK functions either to
conserve ATP or to promote alternative methods of ATP generation.
[0041] 5'AMP-activated protein kinase or AMPK consists of three
proteins (subunits) that together make a functional enzyme that
plays a role in cellular energy homeostasis. It is expressed in a
number of tissues, including the liver, brain, and skeletal muscle.
Activation of AMPK has been shown to activate hepatic fatty acid
oxidation and ketogenesis, inhibit cholesterol synthesis,
lipogenesis, and triglyceride synthesis, inhibit adipocyte
lipolysis and lipogenesis, stimulate skeletal muscle fatty acid
oxidation and muscle glucose uptake, and modulate insulin secretion
by pancreatic beta-cells.
[0042] Triggering the activation of AMPK can be carried out
with increasing concentrations of AMP. The y subunit of AMPK
undergoes a conformational change so as to expose the active site
(Thr-172) on the a subunit. The conformational change of the y
subunit of AMPK can be accomplished under increased concentrations
of AMP. Increased concentrations of AMP will give rise to the
conformational change on the y subunit of AMPK as two AMP bind the
two Bateman domains located on that subunit. This role of AMP is
demonstrated in experiments that show AMPK activation via an AMP
analogue 5-amino-4-imidazolecarboxamide ribotide (ZMP) which is
derived from 5-amino-4-imidazolecarboxamide riboside (AICAR).
[0043] As muscles contract, ATP is hydrolyzed, forming ADP. ADP
then helps to replenish cellular ATP by donating a phosphate group
11
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
to another ADP, forming an ATP and an AMP. As more AMP is produced
during muscle contraction, the AMP:ATP ratio dramatically
increases, leading to the allosteric activation of AMPK.
[0044] Various AMPK agonist are known in the art. Methods and
compositions comprising such AMPK agonist are provided herein. The
use of such AMPK agonist improves muscle tone and muscle mass, as
well as improve endurance compared to subjects not receiving such
AMPK agonists. Various AMPK agonists are described herein and are
known in the art. In one embodiment, the AMPK agonist comprises an
AICAR compound. Other compounds useful in the method of the
disclosure include analogs of AICAR (such as those disclosed in U.
S. Patent No. 5,777,100, hereby incorporated by reference herein)
and prodrugs or precursors of AICAR (such as those disclosed in U.
S. Patent No. 5,082,829, hereby incorporated by reference herein),
which increase the bicavailability of AICAR, all of which are well-
known to those of ordinary skill in the art. Other activators of
AMPK include those described in U.S. Patent Publication No.
20060287356 to Iyengar et al. (the disclosure of which is
incorporated herein by reference). Conventionally known AMPK-
activating compounds include, in addition to the aforementioned
leptin, adiponectin, and metformin, AICAR (5-aminoimidazole-4-
carboxamide). Other AMPK agonists include, but are not limited to,
DRL-16536 (Dr. Reddy's/Perlecan Pharma), BG800 compounds
(Betagenon), furan-2-carboxylic acid derivative (Hanall, KR; see
also Int'l. Application Publ. WO/2008/016278, incorporated herein
by reference), A-769662 (Abbott) (structure I; see also, Cool et
al., Cell Metabol. 3:403-416, 2006); AMPK agonist under development
by Metabasis as set forth in Int'l. Publication No. WO/2006/033709;
MT-39 series of compounds (Mercury Therapeutics); and AMPK agonist
under development by TransTech Pharma.
12
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
HO
O'H
C N,
H
(I)
[0045] AICAR, for example, is taken into the cell and converted
to ZMP, an AMP analog that has been shown to activate AMPK. ZMP
acts as an intracellular AMP mimic, and, when accumulated to high
enough levels, is able to stimulate AMPK activity (Corton, J. M.
et.al. Eur. J. Biochem. 229: 558 (1995)). However, ZMP also acts as
an AMP mimic in the regulation of other enzymes, and is therefore
not a specific AMPK activator (Musi, N. and Goodyear, L. J. Current
Drug Targets--Immune, Endocrine and Metabolic Disorders 2:119
(2002)).
[0046] The disclosure provides methods for stimulating an
"exercise conditioned state" in a subject. The method includes
administering to a subject an AMPK agonist in an amount sufficient
to simulate an energy deficient state in a subject. By "energy
deficient state" refers to a state in which the y subunit of AMPK
undergoes a conformation change, there is increased catabolism of
fat stores in a subject or there is conservation of ATP energy
stores or a metabolic state found in an exercising individual. The
exercise conditioned state can be accomplished in the absence of
exercise using the AMPK agonist of the disclosure (an "exercise-
free conditioning"). However, it will be recognized, that although
administration of an AMPK agonist can promote an exercise
conditioned state, it may be desirable or appropriate for a subject
to perform exercise conditioning even with the administration of an
AMPK agonist.
13
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
[0047] Stimulating and exercise conditioned states not only has
benefits to athletic training, but also provides benefits to
subject who, do to injury, disease or disorder, are unable to
exercise a limb/muscle or where the subject is sedentary or
immobilized. By stimulating an exercise conditioned state the
subject can maintain muscle tone and/or mass in the limb or muscle
and promote health or recovery.
[0048] Exercise is known to have many effects on subjects that
perform it. Exercise effects at the molecular, biochemical, and/or
cellular levels (e.g., modified regulation of genes and/or gene
networks and corresponding proteins involved in energy substrate
utilization and contractile properties of muscle) form the basis of
physiological effects that are observed at the tissue, organ,
and/or whole body levels (e.g., increased cardiorespiratory
endurance, muscular strength, muscular endurance, and/or
flexibility, and/or improvements in body appearance).
[0049] In general terms, exercise is the performance of some
physical activity. A single episode (also referred to as a bout) of
physical activity is performed for a particular duration and at a
particular intensity. If more than one bout of exercise is
performed, separate bouts of exercise may have the same or
different durations and/or the same or different intensities.
[0050] In some embodiments, a single bout of exercise may last
for up to 30 minutes, up to 45 minutes, up to 60 minutes, up to 90
minutes, up to 2 hours, up to 2.5 hours, up to 3 hours, or even
longer. Typically, in the absence of a prior exercise history,
repeated episodes of physical activity are needed to achieve an
exercise-induced effect (such as, increased aerobic capacity or
increase running endurance) for a PPAR5 agonist to be effective.
However, the administration of an AMPK agonist eliminates that need
for exercise to see an effective exercise-induced or promoting
effect.
[0051] Thus, in some disclosed methods, no exercise is needed
when an AMPK agonist is taken alone or prior to an PPAR5 agonist.
However, in some instances bouts of physical activity may be
repeated within a single day; for instance, up to 2 bouts of
14
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
exercise per day, up to 3 bouts of exercise per day, up to 4 bouts
of exercise per day, up to 5 bouts of exercise per day, or even
more bouts per day. Some professional athletes or racing mammals
may exercise in repeated bouts for a total of 8 hours or more a
day. In other method embodiments, bouts (or repeated bouts) of
exercise are performed on a daily basis, 6 times per week, 5 times
per week, 4 times per week or 3 times per week. In at least some of
the disclosed methods, exercise may continue for at least 2 weeks,
for at least 4 weeks, for at least 6 weeks, for at least 3 months,
for at least 6 months, for at least 1 year, for at least 3 years,
or indefinitely (for the lifetime of the subject).
[0052] Exercise generally is performed at an intensity that is
more than the usual (e.g., average, median, normal standard, or
normoactive) activity for a subject, and/or at or less than the
maximum activity achievable by a subject performing a particular
exercise. Any known indicator of physical performance can be used
to determine whether a subject is performing more than a usual
amount of activity, including, for instance, measuring heart rate,
repetition rate (e.g., revolutions per second, minutes per mile,
lifts per minute, and many others), and/or force output. In some
methods, a bout of exercise is performed at sub-maximal intensity;
for instance, at about 10% maximal intensity, 25% maximal
intensity, 50% maximal intensity, or 75% maximal intensity. In
other methods, a bout of exercise is performed at 40%-50% maximal
heart rate, 50%-60% maximal heart rate, 60%-70% maximal heart rate,
or 750-80o maximal heart rate, where maximum heart rate for a human
subject is calculated as: 220 bps - (age of the subject).
[0053] Exercise is generally grouped into three types: (i)
flexibility exercise (such as, stretching), which is believed to,
at least, improve the range of motion of muscles and joints; (ii)
aerobic exercise; and (iii) anaerobic exercise (such as, weight
training, functional training or sprinting) which is believed to,
at least, increase muscle strength and mass.
[0054] Aerobic exercise refers to a physical activity in which
oxidative or aerobic metabolism (as compared to glycolytic or
anaerobic metabolism) substantially predominates in exercised
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
skeletal muscles. In particular method embodiments, a subject
performs one or more aerobic exercises. Exemplary aerobic exercises
include, without limitation, aerobics, calisthenics, cycling,
dancing, exercise machines (rowing machine, cycling machine (e.g.,
inclined or upright), climbing machine, elliptical trainers, and/or
skiing machines), basketball, football, baseball, soccer, footbag,
housework, jogging, martial arts, massage, pilates, rowing,
running, skipping, swimming, walking, yoga, boxing, gymnastics,
badminton, cricket, track and field, golf, ice hockey, lacrosse,
rugby, tennis, or combinations thereof.
[0055] The disclosed methods contemplate enhancing any known or
observable effect of exercise (such as an aerobic exercise, like
walking or running). In particular methods, running endurance
(e.g., running distance and/or running time) is enhanced. In
another embodiment, the methods and compositions are useful for
treating a subject having muscle immobility, muscle wasting disease
or disorder or a sedentary subject. In one such embodiment, muscle
tone or mass is improved or maintained in a subject having muscle
immobility, muscle wasting disease or disorder or a sedentary
activity compared to a subject that does not receive a composition
of the disclosure (e.g., an AMPK agonist, or a combination of an
AMPK agonist or PPAR agonist). In one embodiment, the compositions
and methods of the disclosure can reduce muscle loss or the rate of
muscle loss in a subject having a muscle wasting disease by 50,
10%, 20%, 30%, 40%, 50%, 60%, 70%, 80%, 90% or more compared to a
subject not receiving a composition of the disclosure.
[0056] Muscle weakness, tone, and atrophy result from a number
of diseases and disorders including denervation or prolonged muscle
disuse. When deprived of regular exercise, muscle fibers lose both
bulk and length, producing a visible loss of muscle size and
contour and apparent emaciation or deformity in the affected area.
Even slight atrophy usually causes some loss of motion or power.
Atrophy usually results from neuromuscular disease or injury.
However, muscle tone, atrophy and weakness may also stem from
certain metabolic, cardiovascular or endocrine disorders and
prolonged immobility. Some muscle atrophy also occurs with aging.
16
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
The compositions of the disclosure are useful for treating such
muscle weakness, tone and atrophy comprising administering an AMPK
agonist alone or in combination with a PPARd agonist.
[0057] Muscle wasting disease includes muscle weakness and
atrophy that typically begin in a limb (e.g., hand, arm, or leg).
Eventually, weakness and atrophy spread to the trunk, neck, tongue,
larynx, pharynx, and legs; progressive respiratory muscle weakness
leads to respiratory insufficiency. Other findings include muscle
flaccidity, fasciculations, hyperactive deep tendon reflexes,
slight leg muscle spasticity, dysphagia, impaired speech, excessive
drooling, and depression.
[0058] Fibrous scar tissue formation, pain, and loss of serum
proteins from severe burns can limit muscle movement, resulting in
atrophy. Muscle atrophy is a late sign of irreversible ischemia,
along with contractures, paralysis, and loss of pulses. Herniated
disk can cause muscle weakness, disuse, and ultimately, atrophy.
Hypercortisolism may cause limb weakness and eventually atrophy.
Hypothyroidism can cause weakness and atrophy of proximal limb
muscles. Injuries that result in immobilization of a limb can lead
to muscle atropy and muscle wasting including, but not limited to,
meniscal tears or broken bones or other cartilage injuries
resulting from prolonged knee or limb immobility. Multiple
sclerosis is a degenerative disease that cause arm and leg atrophy
as a result of chronic progressive weakness; spasticity and
contractures may also develop. Osteoarthritis eventually causes
atrophy proximal to involved joints as a result of progressive
weakness and disuse. Parkinson's disease causes muscle rigidity,
weakness, and disuse producing muscle atrophy. Peripheral nerve
trauma or injury to or prolonged pressure on a peripheral nerve
leads to muscle weakness and atrophy. Peripheral neuropathy can
lead to muscle weakness that progresses slowly to flaccid paralysis
and eventually atrophy. Distal extremity muscles are generally
affected first. Associated findings include loss of vibration
sense; paresthesia, hyperesthesia, or anesthesia in the hands and
feet; mild to sharp, burning pain; anhidrosis; glossy red skin; and
diminished or absent deep tendon reflexes. Damaged spinal nerve
17
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
roots can cause muscle atrophy as well as weakness. Rheumatoid
arthritis causes muscle atrophy in the late stages of this
disorder, as joint pain and stiffness decrease range of motion and
discourage muscle use. Spinal cord injury or trauma can produce
severe muscle weakness and flaccid, then spastic, paralysis,
eventually leading to atrophy. Stroke may produce contralateral or
bilateral weakness and eventually atrophy of the arms, legs, face,
and tongue. Associated signs and symptoms depend on the site and
extent of vascular damage and may include dysarthria, aphasia,
ataxia, apraxia, agnosia, and ipsilateral paresthesia or sensory
loss. Prolonged steroid therapy can interfere with muscle
metabolism and can lead to atrophy, most prominently in the limbs.
As mentioned above, prolonged immobilization from bed rest, casts,
splints, or traction may cause muscle weakness and atrophy. Any of
these diseases or disorder can be treated with a composition or
combination of compositions of the disclosure. Other muscle
wasting disease or disorders are recognized in the art.
[0059] The disclosure provides compositions and methods useful
for treating such disease and disorders by promoting or maintaining
muscle mass or tone. For example, administering an AMPK agonist
such as AICAR alone or in combination with a PPARd agonist can
promote muscle tone or mass. In healthy subjects this can
contribute to an enhanced exercise effect. In subjects with a
potential for losing muscle mass or having a muscle wasting disease
or disorder the compositions and methods of the disclosure can slow
or eliminate the rate of such muscle atrophy.
[0060] Enhancing an exercise effect (such as running endurance)
means that such effect is improved in a subject more than would
have occurred by exercise alone. In some method embodiments, an
enhanced exercise effect is determined by discontinuing
administration of an AMPK agonist or a PPAR6 agonist in the subject
and observing (e.g., qualitatively or quantitatively) a reduction
in the exercise effect of interest (e.g., aerobic endurance, such
as running endurance). In some instances, an exercise effect of
interest such as a AMPK-agonist induced effect or the PPAR5-
enhanced portion of which is lost upon discontinuance of an AMPK
18
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
agonist or PPAR6 agonist administration, will be reduced by at
least about 50, by at least about 100, by at least about 20%, by at
least about 30%, or by at least about 50% as compared to the
magnitude of the effect with exercise alone.
[0061] The disclosed methods can be performed in any subject
capable of performing physical activity {e.g., aerobic exercise).
In some method embodiments, a subject is a living multi-cellular
vertebrate organism {e.g., human and/or non-human animals). In
other exemplary methods, a subject is a mammal (including humans
and/or non-human mammals such as veterinary or laboratory mammals)
or, in more particular examples, a racing mammal (such as a horse,
a dog, or a human). In still other methods, a subject is an adult,
an exercise-trained subject, or a healthy subject. Some
representative adult, human subjects are 16 years old or older, 18
years old or older, or 21 years old or older. In some embodiment,
the subject does not perform any routine exercise. In some
embodiment, some representative exercised- trained subjects have
performed physical activity (such described in detail above) for at
least 4 weeks, for at least 6 weeks, for at least 3 months, or for
at least 6 months. In some examples the subject is healthy, for
example, is a subject in which no known disease or disorder has
been diagnosed or would be apparent after reasonable inquiry to an
ordinarily skilled physician in the field to which the disease or
disorder pertains.
[0062] As described more fully below, the AMPK agonist may be
administered orally, parenterally, intramuscularly, intravascularly
or by any appropriate route. A subject can be any mammalian
subject (e.g., equine, canine, or human). An AMPK agonist is
particularly useful in combination with agents that promote muscle
fiber development and growth. Examples of such agents include
agonists of the PPAR family of proteins.
[0063] PPARs are members of the nuclear receptor superfamily of
ligand-inducible transcription factors. They form heterodimers with
retinoid X receptors (RXRs) and bind to consensus DNA sites
composed of direct repeats of hexameric DNA sequences separated by
1 bp. In the absence of ligand, PPAR-RXR heterodimers recruit
19
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
corepressors and associated histone deacetylases and chromatin-
modifying enzymes, silencing transcription by so-called active
repression (Ordentlich et al., Curr. Top. Microbiol. Immunol,
254:101-116, 2001; Jepsen and Rosenfeld, J. Cell Sci, 115:689-698,
2002; Privalsky, Ann. Rev. Physiol, 66:315-360, 2004). Ligand
binding induces a conformational change in PPAR-RXR complexes,
releasing repressors in exchange for coactivators. Ligand-activated
complexes recruit the basal transcriptional machinery, resulting in
enhanced gene expression. PPARs bind to lower-affinity ligands
generated from dietary fat or intracellular metabolism. In keeping
with their roles as lipid sensors, ligand-activated PPARs turn on
feed-forward metabolic cascades to regulate lipid homeostasis via
the transcription of genes involved in lipid metabolism, storage,
and transport.
[0064] Three PPAR isotypes exist in mammals: a (also known as
NR1C1), y (also known as NR1C3), and b (also known as R or NR1C2).
PPAR5 is expressed in most cell types with relative abundance
(Smith, Biochem. Soc. Trans., 30(6): 1086- 1090, 2002), which led
to early speculation that it may serve a "general housekeeping
role" (Kliewer et al., Proc. Natl. Acad. Sci U. S. A, 91:7355-7359,
1994). More recently, PPARb transgenic mouse models and discoveries
aided by the development of high-affinity PPAR5 agonists have
revealed PPAR6 as a key transcriptional regulator with effects in
diverse tissues including fat, skeletal muscle, and the heart (for
review see, e.g., Barish et al., J. Clin. Invest., 116(3):590-597,
2006).
[0065] Targeted expression of a constitutively active PPAR5
receptor (VP16-PPARb) transgene in rodent skeletal muscle promoted
remodeling of skeletal muscle to an oxidative phenotype and
increased running endurance in unexercised adult mice (PLoC Biol,
2:e294, 2004). The observed PPARb-mediated reprogramming of muscle
fibers involved the increased expression of genes related to fatty
acid oxidation, mitochondrial respiration, oxidative metabolism,
and slow-twitch contractile apparatus (Wang et al., PLoC. Biol.,
2:e294, 2004). These VP16-PPAR5 transgenic mice, who had a
phenotype similar to endurance-trained athletes, but who had had no
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
exercise training, suggest that pharmacological activation of
endogenous PPAR5 in an adult, sedentary subject might provide an
exercise effect without the actual exercise. Given the numerous
benefits of exercise on general health, identification of orally
active agents that mimic the effects of exercise is a long
standing, albeit elusive medical goal.
[0066] Disclosed herein are methods for enhancing an exercise
effect or promote/maintain muscle tone or mass in a subject
comprising administering an AMPK agonist sufficient to produce an
exercise effect, enhance mitochondrial expression or activity. In
one embodiment, the subject is a an exercising subject. In another
embodiment, the subject is a sedentary subject. In another
embodiment, the subject is immobilized or has an immobilized limb.
The disclosure further includes administering to the subject an
effective amount of a PPAR5 agonist (e.g., GW1516). The exercise
effect that is enhanced can be, for example, improved running
endurance (such as, improved running distance or improved running
time or a combination thereof, increased fatty acid oxidation in at
least one skeletal muscle of the subject, and/or body fat {e.g.,
white adipose tissue) reduction). In some method embodiments, a
subject is a mammal (such as a racing mammal, like a horse, a dog,
or a human), and/or an adult, and/or an exercise-trained subject.
In other exemplary methods, a PPAR6 agonist is administered on the
same day(s) on which the AMPK agonist is administered or on the
same day upon which physical exertion will be performed. In
another embodiment, the combination of an AMPK agonist and PPAR
agonist are administered on the same day as a physical exertion
will be performed. In some methods, administration of the AMPK
agonist is by oral administration, intravenous injection,
intramuscular injection, and/or subcutaneous injection. In other
method embodiments, the effective amount of the AMPK agonist is
from about 1 mg per day to about 20 mg per day in a single dose or
in divided doses. In some methods, administration of the PPAR6
agonist is by oral administration, intravenous injection,
intramuscular injection, and/or subcutaneous injection. In other
method embodiments, the effective amount of the PPAR6 agonist is
21
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
from about 1 mg per day to about 20 mg per day in a single dose or
in divided doses.
[0067] The disclosed methods envision the use of any PPAR5
agonist. Preferably such agonist would be non-toxic in the subject
to which it is administered. Exemplary PPARS agonists include
GW1516, L- 165041 (as described by, e.g., Leibowitz et al., FEBS
Lett., 473(3):333-336, 2000), any one or more compounds described
in PCT Publication Nos. WO/2006/018174, WO/2005/113506,
WO/2005/105754, WO/2006/041197, WO/2006/032023, WO/01/00603,
WO/02/092590, WO/97/28115, WO/97/28149, WO/97/27857, WO/97/28137,
WO/97/27847, and/or WO/98/27974, and/or a published U.S. national
phase application or issued U.S. patent corresponding to any of the
foregoing (each of which is expressly incorporated herein by
reference). Moreover, other PPAR5 agonists can be identified using
the methods described, for example, in PCT Publication No.
WO/1998/049555 or any corresponding published U.S. national phase
application or issued U.S. patent (each of which is expressly
incorporated herein by reference).
[0068] In a specific example, the PPAR5 agonist is GW1516 (also
referred to in the art as GW501516). GW1516 is (2-methyl-4(((4-
methyl-2-(4- trifluoromethylphenyl)-1,3-thiazol-5-
yl)methyl)sulfanyl)phenoxy)acetic acid as has been shown to be is
bioactive in humans (Sprecher et al., Arterioscler. Thromb. Vase.
Biol. 27(2): 359-65, 2007). In specific examples, GW1516 is
administered orally, for example 1 mg - 20 mg/day, such as 2.5 mg
or 10 mg per day.
[0069] Disclosed herein are methods for enhancing or
stimulating one or more exercise effects by combining, at least, an
AMPK agonist with physical activity and/or administration of one or
more PPAR6 agonists.
[0070] The disclosed methods envision the use of any method of
administration, dosage, and/cr formulation of an AMPK agonist alone
or in combination with a PPARS agonist that has the desired outcome
of enhancing an exercise effect in a subject receiving the
formulation, including, without limitation, methods of
22
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
administration, dosages, and formulations well known to those of
ordinary skill in the pharmaceutical arts.
[0071] AMPK agonist of the disclosure may be administered in
the form of a drug to a human or an animal. Alternatively, the AMPK
agonist may be incorporated into a variety of foods and beverages
or pet foods so as to be consumed by humans or animals. The AMPK
agonist may be applied to a common food or beverage; or may be
applied to a functional food or beverage, a food for a subject
suffering a disease, or a food for specified health use, the food
(or beverage) bearing a label thereon indicating that it has a
physiological function; for example, energy supplement, exercise
enhancer or the like.
[0072] The AMPK agonist alone or in combination with a PPAR6
agonist may be formulated into a drug product; for example, a
peroral solid product such as a tablet or a granule, or a peroral
liquid product such as a solution or a syrup.
[0073] Modes of administering an AMPK agonist (or a formulation
including a PPAR6 agonist) in a disclosed method include, but are
not limited to, intrathecal, intradermal, intramuscular,
intraperitoneal (ip), intravenous (iv), subcutaneous, intranasal,
epidural, intradural, intracranial, intraventricular, and oral
routes. In a specific example, the AMPK agonist or AMPK agonist and
PPAR6 agonist is administered orally. Other convenient routes for
administration of an AMPK agonist (or a formulation including a
PPAR5 agonist) include for example, infusion or bolus injection,
topical, absorption through epithelial or mucocutaneous linings
(for example, oral mucosa, rectal and intestinal mucosa, and the
like) ophthalmic, nasal, and transdermal. Administration can be
systemic or local. Pulmonary administration also can be employed
(for example, by an inhaler or nebulizer), for instance using a
formulation containing an aerosolizing agent.
[0074] In specific method embodiments, it may be desirable to
administer an AMPK agonist or an AMPK agonist and PPAR6 agonist
locally. This may be achieved by, for example, local or regional
infusion or perfusion, topical application (for example, wound
dressing), injection, catheter, suppository, or implant (for
23
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
example, implants formed from porous, non-porous, or gelatinous
materials, including membranes, such as sialastic membranes or
fibers), and the like.
[0075] In other embodiments, a pump (such as a transplanted
minipump) may be used to deliver an AMPK agonist or a combination
of an AMPK agonist and a PPAR6 agonist (or a formulation including
a PPAR5 agonist) (see, e.g., Langer Science 249, 1527, 1990; Sefton
Crit. Rev. Biomed. Eng. 14, 201, 1987; Buchwald et al., Surgery 88,
507, 1980; Saudek et al., N. Engl. J. Med. 321, 574, 1989). In
another embodiment, an AMPK agonist (or a formulation including a
PPARO agonist) is delivered in a vesicle, in particular liposomes
(see, e.g., Langer, Science 249, 1527, 1990; Treat et al., in
Liposomes in the Therapy of Infectious Disease and Cancer, Lopez-
Berestein and Fidler (eds.), Liss, N. Y., pp. 353-365, 1989).
[0076] In yet another method embodiment, an AMPK agonist alone
or in combination with a PPAR6 agonist can be delivered in a
controlled-release formulation. Controlled-release systems, such as
those discussed in the review by Langer (Science 249, 1527 1990),
are known. Similarly, polymeric materials useful in controlled-
released formulations are known (see, e.g., Ranger et al.,
Macromol. ScL Rev. Macromol. Chem. 23, 61, 1983; Levy et al.,
Science 228, 190, 1985; During et al., Ann. Neurol. 25, 351, 1989;
Howard et al., J. Neurosurg. 71, 105, 1989). For example, an
agonists may be coupled to a class of biodegradable polymers useful
in achieving controlled release of a compound, including polylactic
acid, polyglycolic acid, copolymers of polylactic and polyglycolic
acid, polyepsilon caprolactone, polyhydroxy butyric acid,
polyorthoesters, polyacetals, polydihydropyrans, polycyanoacrylates
and cross- linked or amphipathic block copolymers of hydrogels.
[0077] The disclosed methods contemplate the use of any dosage
form of an AMPK agonist alone or in combination with a PPAR6
agonist (or formulation containing the same) that delivers the
agonist(s) and achieves a desired result. Dosage forms are commonly
known and are taught in a variety of textbooks, including for
example, Allen et al., Ansel's Pharmaceutical Dosage Forms and Drug
Delivery Systems, Eighth Edition, Philadelphia, PA:Lippincott
24
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
Williams & Wilkins, 2005, 738 pages. Dosage forms for use in a
disclosed method include, without limitation, solid dosage forms
and solid modified-release drug delivery systems (e.g., powders and
granules, capsules, and/or tablets); semi-solid dosage forms and
transdermal systems (e.g., ointments, creams, and/or gels);
transdermal drug delivery systems; pharmaceutical inserts (e.g.,
suppositories and/or inserts); liquid dosage forms (e.g., solutions
and disperse systems); and/or sterile dosage forms and delivery
systems (e.g., parenterals, and/or biologies). Particular exemplary
dosage forms include aerosol (including metered dose, powder,
solution, and/or without propellants); beads; capsule (including
conventional, controlled delivery, controlled release, enteric
coated, and/or sustained release); caplet; concentrate; cream;
crystals; disc (including sustained release); drops; elixir;
emulsion; foam; gel (including jelly and/or controlled release);
globules; granules; gum; implant; inhalation; injection; insert
(including extended release); liposomal; liquid (including
controlled release); lotion; lozenge; metered dose (e.g., pump);
mist; mouthwash; nebulization solution; ocular system; oil;
ointment; ovules; powder (including packet, effervescent, powder
for suspension, powder for suspension sustained release, and/or
powder for solution); pellet; paste; solution (including long
acting and/or reconstituted); strip; suppository (including
sustained release); suspension (including lente, ultre lente,
reconstituted); syrup (including sustained release); tablet
(including chewable, sublingual, sustained release, controlled
release, delayed action, delayed release, enteric coated,
effervescent, film coated, rapid dissolving, slow release);
transdermal system; tincture; and/or wafer. Typically, a dosage
form is a formulation of an effective amount (such as a
therapeutically effective amount) of at least one active
pharmaceutical ingredient (such as an AMPK agonist or PPAR5
agonist) with pharmaceutically acceptable excipients and/or other
components (such as one or more other active ingredients). An aim
of a drug formulation is to provide proper administration of an
active ingredient (such as an AMPK agonist alone or in combination
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
with a PPAR6 agonist) to a subject. A formulation should suit the
mode of administration. The term "pharmaceutically acceptable"
means approved by a regulatory agency of the federal or a state
government or listed in the U.S. Pharmacopoeia or other generally
recognized pharmacopoeia for use in animals, and, more
particularly, in humans. Excipients for use in exemplary
formulations include, for instance, one or more of the following:
binders, fillers, disintegrants, lubricants, coatings, sweeteners,
flavors, colorings, preservatives, diluents, adjuvants, and/or
vehicles. In some instances, excipients collectively may constitute
about 5%-95% of the total weight (and/or volume) of a particular
dosage form.
[0078] Pharmaceutical excipients can be, for instance, sterile
liquids, such as water and/or oils, including those of petroleum,
animal, vegetable, or synthetic origin, such as peanut oil, soybean
oil, mineral oil, sesame oil, and the like. Water is an exemplary
carrier when a formulation is administered intravenously. Saline
solutions, blood plasma medium, aqueous dextrose, and glycerol
solutions can also be employed as liquid carriers, particularly for
injectable solutions. Oral formulations can include, without
limitation, pharmaceutical grades of mannitol, lactose, starch,
magnesium stearate, sodium saccharine, cellulose, magnesium
carbonate, and the like. A more complete explanation of parenteral
pharmaceutical excipients can be found in Remington, The Science
and Practice of Pharmacy, 19th Edition, Philadelphia, PA:Lippincott
Williams & Wilkins, 1995, Chapter 95. Excipients may also include,
for example, pharmaceutically acceptable salts to adjust the
osmotic pressure, lipid carriers such as cyclodextrins, proteins
such as serum albumin, hydrophilic agents such as methyl cellulose,
detergents, buffers, preservatives and the like. Other examples of
pharmaceutical excipients include starch, glucose, lactose,
sucrose, gelatin, malt, rice, flour, chalk, silica gel, sodium
stearate, glycerol monostearate, talc, sodium chloride, dried skim
milk, glycerol, propylene, glycol, water, ethanol, and the like. A
formulation, if desired, can also contain minor amounts of wetting
or emulsifying agents, or pH buffering agents.
26
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
[0079] A dosage regimen utilizing a PPAR6 agonist is selected
in accordance with a variety of factors including type, species,
age, weight, sex and physical condition of the subject; the route
of administration; and/or the particular PPAR5 agonist formulation
employed. An ordinarily skilled physician or veterinarian can
readily determine an effective amount of a PPAR6 agonist (or
formulation thereof) useful for enhancing an exercise effect in a
subject.
[0080] In some embodiments involving oral administration, oral
dosages of an AMPK agonist alone or in combination with a PPAR5
agonist will generally range between about 0.001 mg per kg of body
weight per day (mg/kg/day) to about 100 mg/kg/day, and such as
about 0.01-10 mg/kg/day (unless specified otherwise, amounts of
active ingredients are on the basis of a neutral molecule, which
may be a free acid or free base). For example, an 80 kg subject
would receive between about 0.08 mg/day and 8 g/day, such as
between about 0.8 mg/day and 800 mg/day. A suitably prepared
medicament for once a day administration would thus contain between
0.08 mg and 8 g, such as between 0.8 mg and 800 mg. In some
instance, formulation including an AMPK agonist alone or in
combination with a PPAR5 agonist may be administered in divided
doses of two, three, or four times daily. For administration twice
a day, a suitably prepared medicament as described above would
contain between 0.04 mg and 4 g, such as between 0.4 mg and 400 mg.
Dosages outside of the aforementioned ranges may be necessary in
some cases. Examples of daily dosages that may be given in the
range of 0.08 mg to 8 g per day include 0.1 mg, 0.5 mg, 1 mg, 2.5
mg, 5 mg, 10 mg, 25 mg, 50 mg, 100 mg, 200 mg, 300 mg, 400 mg, 500
mg, 600 mg, 800 mg, 1 g, 2 g, 4 g and 8 g. These amounts can be
divided into smaller doses if administered more than once per day
(e.g., one-half the amount in each administration if the drug is
taken twice daily).
[0081] For some method embodiments involving administration by
injection (e.g., intravenously or subcutaneous injection), a
subject would receive an injected amount that would deliver the
active ingredient in approximately the quantities described above.
27
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
The quantities may be adjusted to account for differences in
delivery efficiency that result from injected drug forms bypassing
the digestive system. Such quantities may be administered in a
number of suitable ways, e.g. large volumes of low concentrations
of active ingredient during one extended period of time or several
times a day, low volumes of high concentrations of active
ingredient during a short period of time, e.g. once a day.
Typically, a conventional intravenous formulation may be prepared
which contains a concentration of active ingredient of between
about 0.01-1.0 mg/ml, such as for example 0.1 mg/ml, 0.3 mg/ml, or
0.6 mg/ml, and administered in amounts per day equivalent to the
amounts per day stated above. For example, an 80 kg subject,
receiving 8 ml twice a day of an intravenous formulation having a
concentration of active ingredient of 0.5 mg/ml, receives 8 mg of
active ingredient per day.
[0082] In other method embodiments, an AMPK agonist (or a
formulation thereof) can be administered at about the same dose
throughout a treatment period, in an escalating dose regimen, or in
a loading-dose regime (for example, in which the loading dose is
about two to five times a maintenance dose). In some embodiments,
the dose is varied during the course of an AMPK agonist formulation
usage based on the condition of the subject receiving the
composition, the apparent response to the composition, and/or other
factors as judged by one of ordinary skill in the art. In some
embodiments long-term administration of an AMPK agonist or
combination therapy (or formulation thereof) is contemplated, for
instance in order to effect sustained enhancement of an exercise
effect (such as aerobic endurance, e.g., running endurance).
[0083] Also disclosed herein are methods for identifying the
use of performance-enhancing substances in an exercise-trained
subject, which include determining in a biological sample taken
from an exercise-trained subject (e.g., a skeletal muscle biopsy)
the presence of ZMP or other non-naturally occurring AMP analog
and/or the expression of the molecules listed in Table 2 or listed
in Table 4, or a subset thereof, such as expression of at least 1,
28
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
at least 5, at least 10, at least 20, at least 40 of the molecules
listed in Table 2 or in Table 4.
[0084] In some methods for identifying the use of performance-
enhancing substances in an exercise-trained subject the presence of
a ZMP or AMP analog will be measured alone or in combination with
whether (i) expression is upregulated in one or more of (such as at
least 5, at least 10, at least 20, at least 35, or all of) adipose
differentiation related protein; stearoyl-Coenzyme A desaturase 2;
acetyl-Coenzyme A acetyltransferase 2; ATP citrate lyase;
adiponectin, C1Q and collagen domain containing; diacylglycerol 0-
acyltransferase 2; lipase, hormone sensitive; monoglyceride lipase;
resistin; CD36 antigen; fatty acid binding protein 4, adipocyte;
lipoprotein lipase; microsomal glutathione S-transferase 1; GPI-
anchored membrane protein 1; dual specificity phosphatase 7;
homeodomain interacting protein kinase 3; insulin-like growth
factor binding protein 5; protein phosphatase 2 (formerly 2A),
regulatory subunit A (PR 65), beta isoform; protein tyrosine
phosphatase-like (proline instead of catalytic arginine); member b;
CCAAT/enhancer binding protein (C/EBP), alpha; nuclear receptor
subfamily 1, group D, member 2(Reverb-b); transferring; archain 1;
solute carrier family 1 (neutral amino acid transporter), member 5;
RIKEN cDNA 1810073N04 gene; haptoglobin; retinol binding protein 4,
plasma; phosphoenolpyruvate carboxykinase 1, cytosolic; cell death-
inducing DFFA-like effector c; interferon, alpha-inducible protein
27; carbonic anhydrase 3; cysteine dioxygenase 1, cytosolic; DNA
segment, Chr 4, Wayne State University 53, expressed; dynein
cytoplasmic 1 intermediate chain 2; Kruppel-like factor 3 (basic);
thyroid hormone responsive SPOT14 homolog (Rattus); cytochrcme
P450, family 2, subfamily e, polypeptide 1; complement factor D
(adipsin); and/or transketolase; and/or (ii) expression is
downregulated in one or more of gamma-glutamyl carboxylase; 3-
oxoacid CoA transferase 1; solute carrier family 38, member 4;
annexin A7; CD55 antigen; RIKEN cDNA 1190002H23 gene; fusion,
derived from t(12;16) malignant liposarcoma (human); lysosomal
membrane glycoprotein 2; and/or neighbor of Punc El 1, such as 1,
2, 3, 4, 5, 6, 7, 8 or 9 of these molecules.
29
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
[0085] Exemplary methods for identifying the use of
performance-enhancing substances in an exercise-trained subject
involve determining protein expression and/or determining
expression of a gene encoding the protein. Such methods are routine
in the art. In some examples, the level of protein or nucleic acid
expression is quantified. Methods of identifying an agent having
potential to enhance exercise performance in a subject also are
disclosed herein. Such methods can include (i) providing a first
component comprising a PPAR5 receptor or an AMPK-binding fragment
thereof; (ii) providing a second component comprising an AMP-
activated protein kinase (AMPK), AMPKal, AMPKa2, or a PPAR6-binding
fragment of any thereof; (iii) contacting the first component and
the second component with at least one test agent under conditions
that would permit the first component and the second component to
specifically bind to each other in the absence of the at least one
test agent; and (iv) determining whether the at least one test
agent affects the specific binding of the first component and the
second component to each other. An effect on specific binding of
the first component and the second component to each other
identifies the at least one test agent as an agent having potential
to enhance exercise performance in a subject.
[0086] The use of performance-enhancing substances (PES),
particularly by children and professional athletes, has been in the
news because of potential adverse health consequences and the
arguable effects that such practices have on moral development of
the individual and on fair athletic competition for all (Committee
on Sports Medicine and Fitness, Reginald L. Washington, MD,
Chairperson, Pediatrics, 115(4): 1103- 1106, 2005). One of the
discoveries provided herein is that certain genes (and/or the
proteins encoded thereby) are uniquely regulated by a combination
of exercise and a pharmaceutical agent (a PPAR6 agonist) that
results in enhanced physical performance (see Table 2). In some
cases, the particular genes (and/or proteins encoded thereby) were
up- or down-regulated by the combined treatment but were not
affected by either intervention alone. In other cases, the
particular genes (and/or proteins encoded thereby) were not
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
affected by the combined treatment but were up- or down-regulated
by one or both intervention when practiced alone. The unique
regulation of these genes (and/or the encoded proteins) makes them
useful markers (either alone or in any combination) for identifying
exercising subjects who are taking (or receiving) PES.
[0087] A PES is any substance taken in nonpharmacologic doses
specifically for the purpose of improving sports performance (e.g.,
by increasing strength, power, speed, or endurance (ergogenic) or
by altering body weight or body composition). Exemplary PES include
the following: (i) pharmacologic agents (prescription or
nonprescription) taken in doses that exceed the recommended
therapeutic dose or taken when the therapeutic indication(s) are
not present (e.g., using decongestants for stimulant effect, using
bronchodilators when exercise-induced bronchospasm is not present,
increasing baseline methylphenidate hydrochloride dose for athletic
competition); (ii) agents used for weight control, including
stimulants, diet pills, diuretics, and laxatives, when the user is
in a sport that has weight classifications or that rewards
leanness; (iii) agents used for weight gain, including over-the-
counter products advertised as promoting increased muscle mass;
(iv) physiologic agents or other strategies used to enhance oxygen-
carrying capacity, including erythropoietin and red blood cell
transfusions (blood doping); (v) any substance that is used for
reasons other than to treat a documented disease state or
deficiency; (vi) any substance that is known to mask adverse
effects or detectability of another performance-enhancing
substance, and/or (vii) nutritional supplements taken at super
physiologic doses or at levels greater than required to replace
deficits created by a disease state, training, and/or participation
in sports. In one example the PES is as AMPK agonist (e.g., one
that provides an MP analog) or GW1516.
[0088] For example, to quantify the levels of an AMPK agonist
such as AICAR in a sample of body fluid such as serum, as sensitive
LC-MS/MS assay can be used. The assay utilizes a structurally
related analog to the AMPK activator as an internal standard. For
example, the adenosine analog tubercidin (4-amino-7-beta-D-
31
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
ribufuranosyl-7H-pyrrclo[2,3-d]pyrimindine; 7-beta-D-riburuanosyl-
7H-pyrrolo[2, 3-d]pyrimindin-4-amine; 7-deazaadenosine) is
structurally related to AICAR and can be used as the internal
standard when assaying for AICAR. Samples containing a known
concentration of the internal standard can be directly analyzed by
LC-MS/MS. The substance of interest and the internal standard can
be resolved on a LC-MS/MS (tandem mass spec) system, for example,
suing a hydrophobic column with a solvent or acidified solvent
gradient, and detected as positive ions using selective reaction
monitoring. Other analyzers and detection methods known in the art
can be used. A standard curve over one two , three or four orders
of magnitude constructed using agonist-spiked serum can be
constructed to facilitate quantification of AICAR levels.
[0089] The biomarkers of substance-induced performance
enhancement identified herein and useful in a disclosed method
include one or more (or any combination of) of an AMP analog or the
genes (and/or proteins encoded thereby) listed in Table 2, and in
some examples listed in Table 4. In particular method embodiments,
at least 2, at least 3, at least 5, at least 7, at least 10, at
least 15, at least 20, at least 30, or at least 40 of the genes
(and/or proteins encoded thereby) listed in Table 2 (or Table 4)
are detected in a disclosed method. In one example at least one
gene (and/or protein encoded thereby) from each class listed in
Table 2 (e.g., cytokines, fat metabolism) is analyzed.
[0090] In one embodiment, upregulated expression is detected
for one or more of the following genes (or proteins encoded
thereby): adipose differentiation related protein; stearoyl-
Coenzyme A desaturase 2; acetyl-Coenzyme A acetyltransferase 2; ATP
citrate lyase; adiponectin, C1Q and collagen domain containing;
diacylglycerol 0-acyltransferase 2; lipase, hormone sensitive;
monoglyceride lipase; resistin; CD36 antigen; fatty acid binding
protein 4, adipocyte; lipoprotein lipase; microsomal glutathione S-
transferase 1; GPI- anchored membrane protein 1; dual specificity
phosphatase 7; homeodomain interacting protein kinase 3; insulin-
like growth factor binding protein 5; protein phosphatase 2
(formerly 2A), regulatory subunit A (PR 65), beta isoform; protein
32
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
tyrosine phosphatase-like (proline instead of catalytic arginine);
member b; CCAAT/enhancer binding protein (C/EBP), alpha; nuclear
receptor subfamily 1, group D, member 2(Reverb-b); transferring;
archain 1; solute carrier family 1 (neutral amino acid
transporter), member 5; RIKEN cDNA 1810073N04 gene; haptoglcbin;
retinol binding protein 4, plasma; phosphoenolpyruvate
carboxykinase 1, cytosolic; cell death-inducing DFFA-like effector
c; interferon, alpha-inducible protein 27; carbonic anhydrase 3;
cysteine dioxygenase 1, cytosolic; DNA segment, Chr 4, Wayne State
University 53, expressed; dynein cytoplasmic 1 intermediate chain
2; Kruppel-like factor 3 (basic); thyroid hormone responsive SPOT14
homolog (Rattus); cytcchrome P450, family 2, subfamily e,
polypeptide 1; complement factor D (adipsin); and/or transketolase.
In particular method embodiments, upregulation of at least 2, at
least 3, at least 5, at least 7, at least 10, at least 15, at least
20, at least 30, or at least 38 of the foregoing genes (and/or
proteins encoded thereby) are detected in a disclosed method. In
other method embodiments, downregulated expression is detected in
one or more of the following genes (and/or proteins encoded
thereby): gamma- glutamyl carboxylase; 3-oxoacid CoA transferase 1;
solute carrier family 38, member 4; annexin A7; CD55 antigen; RIKEN
cDNA 1190002H23 gene; fusion, derived from t(12;16) malignant
liposarcoma (human); lysosomal membrane glycoprotein 2; and/or
neighbor of Punc El 1. In particular method embodiments,
downregulation of at least 2, at least 3, at least 5, or at least 7
of the foregoing genes (and/or proteins encoded thereby) are
detected in a disclosed method.
[0091] In still other method embodiments, a combination of
upregulated genes (and/or proteins encoded thereby) and
downregulated genes (and/or proteins encoded thereby) as described
above is detected in a sample from a subject (such as, an exercised
or exercise-trained subject).
[0092] Yet other method embodiments involve the detection in a
sample of a combination of an AMP analog (e.g., ZMP) and one or
more of the above-described upregulated genes (and/or proteins
encoded thereby) and/or the above-described downregulated genes
33
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
(and/or proteins encoded thereby), and/or the above-described
exercise-regulated genes that are not affected by exercise combined
with PPAR5 administration.
[0093] Disclosed methods may be used for detecting PES use in
any subject capable of taking or receiving one or more such PES. In
some method embodiments, a subject is a living multi-cellular
vertebrate organism (e.g., human and/or non-human animals). In
other exemplary methods, a subject is a mammal (including humans
and/or non-human mammals) or, in more particular examples, a racing
mammal (such as a horse, a dog, or a human). In still other
methods, a subject is an exercise-trained subject. Some
representative exercised-trained subjects have performed physical
activity (such described in detail above) for at least 4 weeks, for
at least 6 weeks, for at least 3 months, or for at least 6 months.
Other exercise-trained subjects may be student athletes and/or
professional athletes (including, in some examples, non-human
professional athletes, such as race horses and/or racing dogs).
[0094] Any sample from a subject (e.g., a biological sample) in
which can be detected an AMP analog and/or one or more genes and/or
proteins uniquely regulated by exercise in combination with PPAR5
agonist intake (as described in detail throughout this
specification) is contemplated for use in a disclosed method.
Exemplary samples for use in a disclosed method include blood,
saliva, urine, muscle biopsy (e.g., skeletal muscle biopsy), cheek
swab, fecal sample, sweat, and/or sperm.
[0095] Methods of detecting the expression of genes and/or
proteins in a sample (e.g., biological sample) are very well known
(see, e.g., U.S. Patent Nos. 6,911,307; 6,893,824; 5,972,692;
5,972,602; 5,776,672; 7,031,847; 6,816,790; 6,811,977; 6,806,049;
6,203,988; and/or 6,090,556). In particular embodiments,
expression of one or more genes identified herein can be detected
by any method of nucleic acid amplification (such as, polymerase
chain reaction (PCR) or any adaptation thereof, ligase chain
reaction, transcription-based amplification systems, cycling probe
reaction, QR replicase amplification, strand displacement
amplification, and/or rolling circle amplification), solid-surface
34
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
hybridization assays (such as Northern blot, dot blot, gene chips,
and/or reversible target capture), solution hybridization assays
(such as MAP technology (which uses a liquid suspension array of
100 sets of 5.5 micron probe-conjugated beads, each internally dyed
with different ratios of two spectrally distinct fluorophores to
assign it a unique spectral address)), and/or in situ
hybridization. Various of the foregoing nucleic acid detection
methods are described in detail in the review by Wolcott (Clin.
Microbiol. Rev., 5(4):370-386, 1992). Other detailed and long-
established protocols for practicing some such nucleic acid
detection methods are found in Sambrook et al., Molecular Cloning:
A Laboratory Manual, 2nd edition, Cold Spring Harbor Laboratory
Press, 1989; Sambrook et al., Molecular Cloning: A Laboratory
Manual, 3rd edition, Cold Spring Harbor Press, 2001; Ausubel et
al., Current Protocols in Molecular Biology, Greene Publishing
Associates, 1992 (and Supplements to 2000); and/or Ausubel et al.,
Short Protocols in Molecular Biology: A Compendium of Methods from
Current Protocols in Molecular Biology, 4th edition, Wiley & Sons,
1999.
[0096] In other embodiments, expression of one or more proteins
encoded by corresponding genes identified herein can be detected by
Western blot, immunohistochemistry, immunoprecipitation, antibody
microarrays, ELISA, and/or by functional assay (e.g., kinase assay,
ATPase assay, substrate (or ligand) binding assay, protein-protein
binding assay, or other assay suitable for measuring a particular
protein function).
[0097] If the pattern of expression identified in the test
subject is similar to that shown in Table 2 (e.g., the genes shown
as upregulated and downregulated in Table 2 are observed in the
subject to be upregulated and downregulated, respectively), this
indicates that the subject is taking a PES, such as a PPAR6 agonist
(e.g., GW1516). In contrast, if the pattern of expression
identified in the test subject is different to that shown in Table
2 (e.g., the genes shown as upregulated and downregulated in Table
2 are observed in the subject to be not differentially expressed or
show a different pattern of regulation), this indicates that the
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
subject is not taking a PES, such as a PPAR6 agonist (e.g., GIN
1516).
[0098] This disclosure identifies a previously unknown protein-
protein interaction between PPAR5 and particular exercise-induced
kinases (e.g., AMPK, such as the AMPKal and/or AMPKa2 subunit(s) of
AMPK). The interaction between PPAR6 and AMPK may have important
functional outcomes, such as enhancing exercise performance (e.g.,
aerobic exercise performance, such as running endurance) in a
subject.
[0099] The foregoing discoveries enable methods for identify
agents, e.g., having potential to enhance exercise performance
(e.g., aerobic exercise performance, such as running endurance) in
a subject. In some such methods, agents that affect (e.g., enhance,
weaken, or substantially disrupt) the protein-protein interaction
are identified. In other such methods, agents that affect (e.g.,
increase, decrease, or substantially eliminate) AMPK-dependent
phosphorylation of a PPAR5 complex are identified.
[00100] An "agent" is any substance or any combination of
substances that is useful for achieving an end or result; for
example, a substance or combination of substances useful for
modulating a protein activity associated with AMPK activation
cascade (e.g., AMPK-dependent phosphorylation of a PPAR5 complex),
or useful for modifying or affecting a protein-protein interaction
(e.g., PPAR5- AMPK interaction) or ATP metabolism. Any agent that
has potential (whether or not ultimately realized) to modulate any
aspect of the PPAR6- AMPK interaction disclosed herein is
contemplated for use in the screening methods of this disclosure.
[00101] Exemplary agents include, but are not limited to,
peptides such as, for example, soluble peptides, including but not
limited to members of random peptide libraries (see, e.g., Lam et
al., Nature, 354:82-84, 1991; Houghten et al., Nature, 354:84-86,
1991), and combinatorial chemistry-derived molecular library made
of D- and/or L-configuration amino acids, phosphopeptides
(including, but not limited to, members of random or partially
degenerate, directed phosphopeptide libraries; see, e.g., Songyang
et al., Cell, 72:767-778, 1993), antibodies (including, but not
36
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
limited to, polyclonal, monoclonal, humanized, anti-idiotypic,
chimeric or single chain antibodies, and Fab, F(ab')2 and Fab
expression library fragments, and epitope-binding fragments
thereof), small organic or inorganic molecules (such as, so-called
natural products or members of chemical combinatorial libraries),
molecular complexes (such as protein complexes), or nucleic acids.
[00102] Libraries (such as combinatorial chemical libraries)
useful in the disclosed methods include, but are not limited to,
peptide libraries (see, e.g., U.S. Pat. No. 5,010,175; Furka, Int.
J. Pept. Prot. Res., 37:487-493, 1991; Houghton et al., Nature,
354:84-88, 1991; PCT Publication No. WO 91/19735), encoded peptides
(e.g., PCT Publication WO 93/20242), random bio-oligomers (e.g.,
PCT Publication No. WO 92/00091), benzodiazepines (e.g., U.S. Pat.
No. 5,288,514), diversomers such as hydantoins, benzodiazepines and
dipeptides (Hobbs et al., Proc. Natl. Acad. Sci. USA, 90:6909-6913,
1993), vinylogous polypeptides (Hagihara et al., J. Am. Chem. Soc,
114:6568, 1992), nonpeptidal peptidomimetics with glucose
scaffolding (Hirschmann et al., J. Am. Chem. Soc, 114:9217-9218,
1992), analogous organic syntheses of small compound libraries
(Chen et al., J. Am. Chem. Soc, 116:2661, 1994), oligocarbamates
(Cho et al., Science, 261: 1303, 1003), and/or peptidyl
phosphonates (Campbell et al., J. Org. Chem., 59:658, 1994),
nucleic acid libraries (see Sambrook et al. Molecular Cloning, A
Laboratory Manual, Cold Springs Harbor Press, N. Y., 1989; Ausubel
et al., Current Protocols in Molecular Biology, Green Publishing
Associates and Wiley Interscience, N.Y., 1989), peptide nucleic
acid libraries (see, e.g., U.S. Pat. No. 5,539,083), antibody
libraries (see, e.g., Vaughn et al., Nat. Biotechnol, 14:309-314,
1996; PCT App. No. PCT/US96/10287), carbohydrate libraries (see,
e.g., Liang et al., Science, 274:1520-1522, 1996; U.S. Pat. No.
5,593,853), small organic molecule libraries (see, e.g.,
benzodiazepines, Baum, C&EN, Jan 18, page 33, 1993; isoprenoids,
U.S. Pat. No. 5,569,588; thiazolidionones and methathiazones, U.S.
Pat. No. 5,549,974; pyrrolidines, U.S. Pat. Nos. 5,525,735 and
5,519,134; morpholino compounds, U.S. Pat. No. 5,506,337;
benzodiazepines, 5,288,514) and the like.
37
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
[00103] Libraries useful for the disclosed screening methods can
be produce in a variety of manners including, but not limited to,
spatially arrayed multipin peptide synthesis (Geysen, et al., Proc
Natl. Acad. Sci., 81(13):3998-4002, 1984), "tea bag" peptide
synthesis (Houghten, Proc Natl. Acad. Sci., 82(15):5131-5135,
1985), phage display (Scott and Smith, Science, 249:386-390, 1990),
spot or disc synthesis (Dittrich et al., Bioorg. Med. Chem. Lett.,
8(17):2351-2356, 1998), or split and mix solid phase synthesis on
beads (Furka et al., Int. J. Pept. Protein Res., 37(6):487-493,
1991; Lam et al., Chem. Rev., 97 (2):411-448, 1997). Libraries may
include a varying number of compositions (members), such as up to
about 100 members, such as up to about 1000 members, such as up to
about 5000 members, such as up to about 10,000 members, such as up
to about 100,000 members, such as up to about 500,000 members, or
even more than 500,000 members.
[00104] In one embodiment, high throughput screening methods
involve providing a combinatorial chemical or peptide library
containing a large number of potential therapeutic compounds (e.g.,
affectors of AMPK-PPARb protein-protein interactions). Such
combinatorial libraries are then screened in one or more assays as
described herein to identify those library members (particularly
chemical species or subclasses) that display a desired
characteristic activity (such as increasing or decreasing an AMPK-
PPARb protein-protein interaction). The compounds thus identified
can serve as conventional "lead compounds" or can themselves be
used as potential or actual therapeutics. In some instances, pools
of candidate agents may be identify and further screened to
determine which individual or subpools of agents in the collective
have a desired activity. PPARb forms a protein-protein interaction
with AMPK or one or more of its subunits (such as AMPKal and/or
AMPKcc2). Agents that affect (e.g., increase or decrease) an AMPK-
PPARb interaction or AMP-dependent phosphorylation of a PPARb
complex may have the effect of enhancing exercise performance
(e.g., aerobic exercise performance, such as running endurance) in
a subject and, therefore, are desirable to identify.
38
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
[00105] In screening methods described here, tissue samples,
isolated cells, isolated polypeptides, and/or test agents can be
presented in a manner suitable for high-throughput screening; for
example, one or a plurality of isolated tissue samples, isolated
cells, or isolated polypeptides can be inserted into wells of a
microtitre plate, and one or a plurality of test agents can be
added to the wells of the microtitre plate. Alternatively, one or a
plurality of test agents can be presented in a high-throughput
format, such as in wells of microtitre plate (either in solution or
adhered to the surface of the plate), and contacted with one or a
plurality of isolated tissue samples, isolated cells, and/or
isolated polypeptides under conditions that, at least, sustain the
tissue sample or isolated cells or a desired polypeptide function
and/or structure. Test agents can be added to tissue samples,
isolated cells, or isolated polypeptides at any concentration that
is not lethal to tissues or cells, or does not have an adverse
effect on polypeptide structure and/or function. It is expected
that different test agents will have different effective
concentrations. Thus, in some methods, it is advantageous to test a
range of test agent concentrations.
[00106] Disclosed methods envision, as appropriate, the use of
PPAR5 or AMPK (such as AMPKal or AMPKcc2) or functional fragments
of any thereof as contained, independently, in a subject, one or a
plurality of cells or cellular extracts, one or a plurality of
tissue or tissue extracts, or as an isolated polypeptide. PPAR6
ligand optionally is included (or is omitted) in disclosed methods.
[00107] A "direct association" between two or more polypeptides
(such as, PPAR6 and AMPK (such as AMPKal or AMPKa2) is
characterized by physical contact between at least a portion of the
interacting polypeptides that is of sufficient affinity and
specificity that, for example, immunoprecipitation of one of the
polypeptides also will specifically precipitate the other
polypeptide; provided that the immunoprecipitating antibody does
not also affect the site(s) involved in the interaction. A direct
association between polypeptides also may be referred to as a
"protein-protein interaction." The binding of one polypeptide to
39
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
another in a protein-protein interaction {e.g., PPARb to AMPK (or
AMPKal and/or AMPKa2) and vice versa) is considered "specific
binding". Agents that affect an AMPK-PPAR6 interaction can be
identified by a variety of assays, including solid-phase or
solution-based assays. In an exemplary solid-phase assay, PPARb or
an AMPK-binding fragment thereof and AMPK or a subunit thereof
(such as AMPKal and/or AMPKa2) or a PPARb-binding fragment thereof
are mixed under conditions in which PPAR6 and AMPK (or its
subunit(s) or functional fragments) normally interact {e.g., co-
immunoprecipitate). One of the binding partners is labeled with a
marker such as biotin, fluorescein, EGFP, or enzymes to allow easy
detection of the labeled component. The unlabeled binding partner
is adsorbed to a support, such as a microtiter well or beads. Then,
the labeled binding partner is added to the environment where the
unlabeled binding partner is immobilized under conditions suitable
for interaction between the two binding partners. One or more test
compounds, such as compounds in one or more of the above-described
libraries, are separately added to individual microenvironments
containing the interacting binding partners. Agents capable of
affecting the interaction between the binding partners are
identified, for instance, as those that increase or decrease (e.g.,
increase) retention or binding of the signal (i.e., labeled binding
partner) in the reaction microenvironment, for example, in a
microtiter well or on a bead for example. As discussed previously,
combinations of agents can be evaluated in an initial screen to
identify pools of agents to be tested individually, and this
process is easily automated with currently available technology.
[00108] In other embodiments, solution phase selection can be
used to screen large complex libraries for agents that specifically
affect protein-protein interactions (see, e.g., Boger et al.,
Bioorg. Med. Chem. Lett., 8(17):2339-2344, 1998); Berg et al.,
Proc. Natl. Acad. Sci., 99(6):3830-3835, 2002). In one such
example, each of two proteins that are capable of physical
interaction (for example, PPARb (or AMPK-binding fragments thereof)
and AMPK or AMPKal or AMPKa2 (or PPARb-binding fragments of any
thereof) are labeled with fluorescent dye molecule tags with
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
different emission spectra and overlapping adsorption spectra. When
these protein components are separate, the emission spectrum for
each component is distinct and can be measured. When the protein
components interact, fluorescence resonance energy transfer (FRET)
occurs resulting in the transfer of energy from a donor dye
molecule to an acceptor dye molecule without emission of a photon.
The acceptor dye molecule alone emits photons (light) of a
characteristic wavelength. Therefore, FRET allows one to determine
the kinetics of two interacting molecules based on the emission
spectra of the sample. Using this system, two labeled protein
components are added under conditions where their interaction
resulting in FRET emission spectra. Then, one or more test
compounds, such as compounds in one or more of the above-described
libraries, are added to the environment of the two labeled protein
component mixture and emission spectra are measured. An increase in
the FRET emission, with a concurrent decrease in the emission
spectra of the separated components indicates that an agent (or
pool of candidate agents) has affected (e.g., enhanced) the
interaction between the protein components.
[00109] Interactions between PPAR5 (or AMPK-binding fragments
thereof) and AMPK or AMPKal or AMPKa2 (or PPARb-binding fragments
of any thereof) also can be determined (e.g., quantified) by co-
immunoprecipitation of the relevant component polypeptides (e.g.,
from cellular extracts), by GST-pull down assay (e.g., using
purified GST-tagged bacterial proteins), and/or by yeast two-hybrid
assay, each of which methods is standard in the art. Conducting any
one or more such assays in the presence and, optionally, absence of
a test compound can be used to identify agents that improve or
enhance (or, in other embodiments, decrease or inhibit) the
interaction between PPAR6 (or AMPK-binding fragments thereof) and
AMPK or AMPKal or AMPKa2 (or PPAR6-binding fragments of any
thereof) in the presence of a test compound as compared to in the
absence of the test compound or as compared to some other standard
or control.
[00110] In certain embodiments, one or more AMPK (such as AMPKal
and/or AMPKcc2)-binding fragments of PPAR6 and/or one or more
41
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
PPAR6-binding fragments of AMPK (such as AMPKal and/or AMPKa2) are
used. Polypeptide fragments having the desired binding activities
can be identified by making a series of defined PPAR6 fragments
and/or AMPK (such as AMPKal or AMPKa2) fragments using methods
standard in the art. For example, cDNA encoding the protein(s) of
interest (e.g., PPAR6 or AMPK) can be serially truncated from the
3' or 5' end (provided that a start codon is engineered into 5'
truncations) using conveniently located restriction enzyme sites
(or other methods) and leaving intact (or otherwise correcting) the
proper reading frame. Conveniently, a nucleic acid sequence
encoding an epitope tag (such as a FLAG tag) is placed in frame
with (and substantially adjacent to) the truncated protein-encoding
sequence to produce a nucleic acid sequence encoding an epitope-
tagged protein fragment. The epitope-tagged protein fragment can be
expressed in any convenient expression system (such as a bacterial
expression system), isolated or not, and mixed with a sample
containing a protein or other protein fragment to which the
epitope-tagged protein fragment may bind. An antibody specific for
the tag (or other region of the protein fragment) can be used to
immunoprecipitate the fragment of interest together with any
protein(s) or protein fragment(s) that bind to it. Protein(s) or
protein fragment(s) that bind to the epitope-tagged protein
fragment of interest can be particular identified, e.g., by Western
blot.
[00111] In particular methods, the formation of a PPARf-AMPK
(such as AMPKal and/or AMPKcc2) complex (including complexes
including one or both of PPAR5-binding AMPK fragments and/or AMPK-
binding PPAR6 fragments) or the affinity of PPAR6 (or AMPK-binding
fragments thereof) and AMPK (or PPARb-binding fragments thereof)
for each other is increased when the amount of such complex or the
binding affinity is at least 5%, at least 100, at least 20%, at
least 30%, at least 50%, at least 100% or at least 250% higher than
a control measurement (e.g., in the same test system prior to
addition of a test agent, or in a comparable test system in the
absence of a test agent). In other particular methods, the
formation of a PPAR6-AMPK (such as AMPKal and/or AMPKcc2) complex
42
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
(including complexes including one or both of PPAR6-binding AMPK
fragments and/or AMPK-binding PPAR5 fragments) or the affinity of
PPAR6 (or AMPK-binding fragments thereof) and AMPK (or PPAR6-
binding fragments thereof) for each other is decreased when the
amount of such complex or the binding affinity is at least 5%, at
least 10%, at least 20%, at least 30%, at least 50%, at least 100%
or at least 250% lower than a control measurement (e.g., in the
same test system prior to addition of a test agent, or in a
comparable test system in the absence of a test agent).
[00112] Disclosed are methods of screening test agents for those
that affect (e.g., increase or decrease) AMPK (e.g., AMPKal and/or
AMPKa2)-dependent phosphorylation of the PPAR5 complex. Agents that
affect AMPK-dependent phosphorylation of the PPAR6 complex can be
identified by a variety of assays, such adaptations of solid-phase-
or solution-based assays described above, where the end point to be
detected is phosphorylation of one or more components of the PPAR6
complex.
[00113] Methods for detecting protein phosphorylation are
conventional (see, e.g., Gloffke, The Scientist, 16(19):52, 2002;
Screaton et al., Cell, 119:61-74, 2004) and detection kits are
available from a variety of commercial sources (see, e.g., Upstate
(Charlottesville, VA, USA), Bio-Rad (Hercules, CA, USA), Marligen
Biosciences, Inc. (Ijamsville, MD, USA), Calbiochem (San Diego, CA,
USA). Briefly, phosphcrylated protein (e.g., phosphorylation of one
or more components of the PPAR5 complex) can be detected using
stains specific for phosphorylated proteins in gels. Alternatively,
antibodies specific phosphorylated proteins can be made or
commercially obtained. Antibodies specific for phosphorylated
proteins can be, among other things, tethered to the beads
(including beads having a particular color signature) or used in
ELISA or Western blot assays.
[00114] In one example, a PPAR6 complex (or a fragment thereof
containing an AMPK phcsphorylation site) and AMPK or one or more of
it subunits (such as AMPKal and/or AMPKcc2) or functional fragments
thereof that are capable of phosphorylation are mixed under
conditions whereby a PPAR6 complex is phosphorylated by AMPK. A
43
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
PPAR6 complex is adsorbed to a support, such as a microtiter well
or beads. Then, AMPK (or its one or more subunits (such as AMPKal
and/or AMPKcc2) or phosphorylation-capable fragments thereof) is
added to the environment where the complex is immobilized. A
phosphate donor typically is also included in the environment. The
phosphate to be donated, optionally, can be labeled. One or more
test compounds, such as compounds in one or more of the above-
described libraries, are separately added to the individual
microenvironments. Agents capable of affecting AMPK-dependent
phosphorylation are identified, for instance, as those that enhance
(or inhibit) phosphorylation of immobilized PPAR6 complex. In
embodiments involving a labeled phosphate donor, phosphorylation of
immobilized PPAR5 complex can be determined by retention or binding
of a labeled phosphate in the reaction microenvironment, for
example, in a microtiter well or on a bead for example. In other
embodiments, such reactions can take place in solution (i.e., with
no immobilized components), PPAR5 complex can be isolated from the
solution (e.g., by immunoprecipitation with PPARS- specific or
phosphate-specific antibodies), and its level of phosphorylation in
the presence (and, optionally, absence) of one of more test agents
determined as previously discussed. In yet another embodiment, the
phosphorylation of an AMPK is measured, wherein an agent that
modulates AMPK activity is thus identified as an AMPK agonist.
[00115] In particular methods, the phosphorylation of a PPAR5
complex is increased when such posttranslational modification is
detectably measured or when such posttranslational modification is
at least 20%, at least 30%, at least 50%, at least 100% or at least
250% higher than control measurements (e.g., in the same test
system prior to addition of a test agent, or in a comparable test
system in the absence of a test agent, or in a comparable test
system in the absence of AMPK).
[00116] In particular methods, the phosphorylation of PPAR5
complex is decreased when such posttranslational modification is
detestably reduced or when such posttranslational modification is
at least 20%, at least 30%, at least 50%, at least 100% or at least
250% lower than control measurements (e.g., in the same test system
44
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
prior to addition of a test agent, or in a comparable test system
in the absence of a test agent, or in a comparable test system in
the absence of AMPK).
[00117] A PPAR5 polypeptide useful in a disclosed screening
method is any known PPAR5 receptor. Also useful in the disclosed
screening methods are homologs, functional fragments, or functional
variants of a PPAR5 that retains at least AMPK-binding activity as
described herein for a prototypical PPAR6 polypeptide (see Example
6).
The amino acid sequences of prototypical PPAR5 polypeptides (and
PPARO-encoding nucleic acid sequences) are well known. Exemplary
PPAR5 amino acid sequences and PPAR5-encoding nucleic acid
sequences are described, for instance, in U.S. Patent No.
5,861,274, and U.S. Pat. Appl. Pub. No. 20060154335 (each of which
is expressly incorporated herein by reference), and in GenBank
Accession Nos. NP 035275 (GI:33859590)(Mus musculus amino acid
sequence); NM 011145.3 (GI:89001112)(Mus musculus nucleic acid
sequence); NP 006229 (GI:5453940)(Homo sapiens amino acid
sequence); NM 006238.3 (GI:89886454)(Homo sapiens nucleic acid
sequence); NP 037273 (GI:69S13S4)(Rattus norvegicus amino acid
sequence); NM 013141.1 (GL6981383) (Rattus norvegicus nucleic acid
sequence); NP 990059 (gi45382025) (Gallus gallus amino acid
sequence) or NM 204728.1 (GI:45382024)(Gallus gallus nucleic acid
sequence). In some method embodiments, a PPAR5 homolog or
functional variant shares at least 60% amino acid sequence identity
with a prototypical PPAR6 polypeptide; for example, at least 75%,
at least 80%, at least 85%, at least 90%, at least 95%, or at least
98% amino acid sequence identity with an amino acid sequence as set
forth in U.S. Patent No. 5,861,274, U.S. Pat. Appl. Pub. No.
20060154335, or GenBank Accession No. NP 035275 (GI:33859590)(Mus
musculus amino acid sequence); NP 006229 (GI:5453940)(Homo sapiens
amino acid sequence); NP 037273 (G'I.69$13$4)(Rattus norvegicus
amino acid sequence); or NP 990059 (gi45382025) {Gallus gallus
amino acid sequence). In other method embodiments, a PPAR5 homolog
or functional variant has one or more conservative amino acid
substitutions as compared to with a prototypical PPAR6 polypeptide;
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
for example, no more than 3, 5, 10, 15, 20, 25, 30, 40, or 50
conservative amino acid changes compared to an amino acid sequence
as set forth in U.S. Patent No. 5,861,274, U.S. Pat. Appl. Pub. No.
20060154335, or GenBank Accession No. NP 035275
(GI:33859590)(M***Unknown: ***s musculus amino acid sequence);
NP 006229 (GI:5453940)(Homo sapiens amino acid sequence); NP 037273
(GI.69$13$4)(Rattus ncrvegicus amino acid sequence); or NP 990059
(gi45382025) (Gallus gallus amino acid sequence).
[00118] Some method embodiments involve a PPAR5 functional
fragment (such as an AMPK-binding fragment), which can be any
portion of a full-length known PPAR6 polypeptide, including, e.g.,
about 20, about 30, about 40, about 50, about 75, about 100, about
150 or about 200 contiguous amino acid residues of same; provided
that the fragment retains a PPAR5 function of interest (e.g., AMPK
binding). PPAR6 encompasses known functional motifs (such as
ligand-binding domain, a DNA-binding domain, and a transactivation
domain).
[00119] Mammalian AMP-activated kinase (AMPK) is a
heterotrimeric protein composed of 1 alpha subunit, 1 beta subunit,
and 1 gamma subunit. There are, at least, two known isoforms of the
alpha subunit (al and a2). AMPKal and AMPKa2 have 90% amino acid
sequence identity within their catalytic cores but only 61% in
their C-terminal tails (see Online Mendelian Inheritance in Man
(OMIM) Database Accession No. 602739; publicly available at the
following website: ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=602739).
[00120] An AMPK (such as AMPKal and/or AMPKa2) polypeptide
useful in a disclosed screening method is any known AMPK protein or
subunit thereof (such as AMPKal and/or AMPKa2). Also useful in the
disclosed screening methods are homologs, functional fragments, or
functional variants of an AMPK protein or subunit thereof (such as
AMPKal and/or AMPKa2) that retains at least PPAR6-binding activity
as described herein (see Example 6). The amino acid sequences of
prototypical AMPK subunits (such as AMPKal and/or AMPKa2) (and
nucleic acids sequences encoding prototypical AMPK subunits (such
as AMPKal and/or AMPKa2)) are well known. Exemplary AMPKal amino
acid sequences and the corresponding nucleic acid sequences are
46
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
described, for instance, in GenBank Accession Nos. NM 206907.3
(GI:94557298)(Homo sapiens transcript variant 2 REFSEQ including
amino acid and nucleic acid sequences); NM 006251.5
(GI:94557300)(Homo sapiens transcript variant 1 REFSEQ including
amino acid and nucleic acid sequences); NM 001013367.3
(GI:94681060)(Mus musculus REFSEQ including amino acid and nucleic
acid sequences); NMJ)01039603.1 (GI:88853844)(Gallus gallus REFSEQ
including amino acid and nucleic acid sequences); and NM 019142.1
(GI: 11862979XRaJfWS norvegicus REFSEQ including amino acid and
nucleic acid sequences). Exemplary AMPKa2 amino acid sequences and
the corresponding nucleic acid sequences are described, for
instance, in GenBank Accession Nos. NM 006252.2 (GI:46877067)(Homo
sapiens REFSEQ including amino acid and nucleic acid sequences);
NM 178143.1 (GI:54792085)(Mus musculus REFSEQ including amino acid
and nucleic acid sequences); NM 001039605.1 (GI:88853850)(Gallus
gallus REFSEQ including amino acid and nucleic acid sequences); and
NM 214266.1 (GI:47523597)(Mus musculus REFSEQ including amino acid
and nucleic acid sequences).
[00121] In some method embodiments, a homolog or functional
variant of an AMPK subunit shares at least 60% amino acid sequence
identity with a prototypical AMPKal and/or AMPKa2 polypeptide; for
example, at least 75%, at least 80%, at least 85%, at least 90%, at
least 95%, or at least 98% amino acid sequence identity with an
amino acid sequence as set forth in the GenBank Accession Nos.
NM 206907.3; NM 006251.5; NMJ)01013367.3; NM 001039603.1;
NM 019142.1; NM 006252.2; NM 178143.1; NM 001039605.1; or
NM 214266.1. In other method embodiments, a homolog or functional
variant of an AMPK subunit has one or more conservative amino acid
substitutions as compared to a prototypical AMPKal and/or AMPKa2
polypeptide; for example, no more than 3, 5, 10, 15, 20, 25, 30,
40, or 50 conservative amino acid changes compared to an amino acid
sequence as set forth in as set forth in GenBank Accession Nos.
NM 206907.3; NM 006251.5; NM 001013367.3; NM 001039603.1;
NM 019142.1; NM 006252.2; NM 178143.1; NM 001039605.1; or
NM 214266.1. Exemplary conservative amino acid substitutions have
been previously described herein.
47
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
[00122] Some method embodiments involve a functional fragment of
AMPK or a subunit thereof (such as AMPKal and/or AMPKa2), including
a PPAR5-binding fragment or a fragment with PPAR6 phosphorylation
activity. Functional fragments of AMPK or a subunit thereof (such
as AMPKal and/or AMPKa2) can be any portion of a full-length or
intact AMPK polypeptide complex or subunit thereof (such as AMPKal
and/or AMPKa2), including, e.g., about 20, about 30, about 40,
about 50, about 75, about 100, about 150 or about 200 contiguous
amino acid residues of same; provided that the fragment retains at
least one AMPK (or AMPKal and/or AMPKa2) function of interest
(e.g., PPAR6 binding and/or PPAR6 phosphorylation activity).
Protein-protein interactions between PPAR6 and AMPK are believed to
involve, at least, an AMPKa subunit (such as AMPKal and/or AMPKa2).
Moreover, because PPAR6 specifically binds both AMPKal and AMPKa2
(see Example 6), such interaction likely is mediated by the
portions of these AMPKa isoforms that share the most sequence
homology (as discussed above). Accordingly, in some method
embodiments, an AMPK PPAR6-binding fragment includes a functional
fragment encompassing (or consisting of) the catalytic core domain
of an alpha subunit of AMPK (such as AMPKal and/or AMPKa2).
[00123] An "isolated" biological component (such as a
polynucleotide, polypeptide, or cell) has been purified away from
other biological components in a mixed sample (such as a cell or
tissue extract). For example, an "isolated" polypeptide or
polynucleotide is a polypeptide or polynucleotide that has been
separated from the other components of a cell in which the
polypeptide or polynucleotide was present (such as an expression
host cell for a recombinant polypeptide or polynucleotide).
[00124] The term "purified" refers to the removal of one or more
extraneous components from a sample. For example, where recombinant
polypeptides are expressed in host cells, the polypeptides are
purified by, for example, the removal of host cell proteins thereby
increasing the percent of recombinant polypeptides in the sample.
Similarly, where a recombinant polynucleotide is present in host
cells, the polynucleotide is purified by, for example, the removal
48
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
of host cell polynucleotides thereby increasing the percent of
recombinant polynuclectide in the sample.
[00125] Isolated polypeptides or nucleic acid molecules,
typically, comprise at least 50%, at least 60%, at least 70%, at
least 80%, at least 90%, at least 95% or even over 99% (w/w or w/v)
of a sample.
[00126] Polypeptides and nucleic acid molecules are isolated by
methods commonly known in the art and as described herein. Purity
of polypeptides or nucleic acid molecules may be determined by a
number of well-known methods, such as polyacrylamide gel
electrophoresis for polypeptides, or agarose gel electrophoresis
for nucleic acid molecules.
[00127] The similarity between two nucleic acid sequences or
between two amino acid sequences is expressed in terms of the level
of sequence identity shared between the sequences. Sequence
identity is typically expressed in terms of percentage identity;
the higher the percentage, the more similar the two sequences.
[00128] Methods for aligning sequences for comparison are well
known in the art. Various programs and alignment algorithms are
described in: Smith and Waterman, Adv. Appl. Math. 2:482, 1981;
Needleman and Wunsch, J. Mol. Biol. 48:443, 1970; Pearson and
Lipman, Proc. Natl. Acad. ScL USA 85:2444, 1988; Higgins and Sharp,
Gene 73:237-244, 1988; Higgins and Sharp, CABIOS 5:151-153, 1989;
Corpet et al., Nucleic Acids Research 16:10881-10890, 1988; Huang,
et al., Computer Applications in the Biosciences 8:155-165, 1992;
Pearson et al., Methods in Molecular Biology 24:307-331, 1994;
Tatiana et al., (1999), FEMS Microbiol. Lett., 174:247-250, 1999.
Altschul et al. present a detailed consideration of sequence
alignment methods and homology calculations (J. Mol. Biol. 215:403-
410, 1990). The National Center for Biotechnology Information
(NCBI) Basic Local Alignment Search Tool (BLASTTM, Altschul et al.,
J. Mol. Biol. 215:403-410, 1990) is available from several sources,
including the National Center for Biotechnology Information (NCBI,
Bethesda, MD) and on the Internet, for use in connection with the
sequence-analysis programs blastp, blastn, blastx, tblastn and
tblastx. A description of how to determine sequence identity using
49
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
this program is available on the internet under the help section
for BLAST'[00129] For comparisons of amino acid sequences of greater than
about 30 amino acids, the "Blast 2 sequences" function of the
BLAST' (Blastp) program is employed using the default BLOSUM62
matrix set to default parameters (cost to open a gap [default = 5];
cost to extend a gap [default = 2]; penalty for a mismatch [default
= -3]; reward for a match [default = 1]; expectation value (E)
[default = 10.0]; word size [default = 3]; number of one-line
descriptions (V) [default = 100]; number of alignments to show (B)
[default = 100]). When aligning short peptides (fewer than around
30 amino acids), the alignment should be performed using the Blast
2 sequences function, employing the PAM30 matrix set to default
parameters (open gap 9, extension gap 1 penalties). Proteins with
even greater similarity to the reference sequences will show
increasing percentage identities when assessed by this method.
[00130] For comparisons of nucleic acid sequences, the "Blast 2
sequences" function of the BLASTTT (Blastn) program is employed
using the default BLOSUM62 matrix set to default parameters (cost
to open a gap [default = 11]; cost to extend a gap [default = 1];
expectation value (E) [default = 10.0]; word size [default = 11];
number of one-line descriptions (V) [default = 100]; number of
alignments to show (B) [default = 100]). Nucleic acid sequences
with even greater similarity to the reference sequences will show
increasing percentage identities when assessed by this method.
[00131] Specific binding refers to the particular interaction
between one binding partner (such as a binding agent) and another
binding partner (such as a target). Such interaction is mediated by
one or, typically, more noncovalent bonds between the binding
partners (or, often, between a specific region or portion of each
binding partner). In contrast to non-specific binding sites,
specific binding sites are saturable. Accordingly, one exemplary
way to characterize specific binding is by a specific binding
curve. A specific binding curve shows, for example, the amount of
one binding partner (the first binding partner) bound to a fixed
amount of the other binding partner as a function of the first
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
binding partner concentration. As the first binding partner
concentration increases under these conditions, the amount of the
first binding partner bound will saturate. In another contrast to
non-specific binding sites, specific binding partners involved in a
direct association with each other (e.g., a protein-protein
interaction) can be competitively removed (or displaced) from such
association (e.g., protein complex) by excess amounts of either
specific binding partner. Such competition assays (or displacement
assays) are very well known in the art.
[00132] The disclosure also provides methods for identifying
agents useful for effecting muscle tone or mass. The disclosure
provides endurance gene signatures (see, e.g., Table 2 and Table 4)
comprising genes that are modulated in the presence of AICAR or
GW1516, or a combination of AICAR and GW1516. Such gene signature
are useful for identifying agents that provides an increase in
muscle tone or mass and thereby can modulate physical endurance. A
GW-AI endurance gene signature refers to a set of genes described
in Table 4 or a subset thereof. A GW-TR endurance gene signature
refers to a set of genes described in Table 2 or a subset thereof.
[00133] As depicted in Figures 6 and 8, an overlap of gene
expression profile for each of the agonists is provided. An agent
to be tested can be administered to a subject and the gene
expression profile measure in a muscle sample (e.g., a biopsy) or
other biological sample. Where the gene expression profile
comprises a set of the endurance gene signatures (e.g., the
overlapping 52 genes associated with GW1516 and AICAR
administration, see Table 4) or a subset thereof such an agent can
be identified as an agent or drug useful for treating or modifying
muscle activity.
EXAMPLES
[00134] The following examples are provided to illustrate
certain particular features and/or embodiments. These examples
should not be construed to limit the invention to the particular
features or embodiments described.
Example 1
51
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
[00135] Administration of PPAR6 agonist does not enhance
physical performance in non-exercised subjects. Wang et a.
previously demonstrated that skeletal muscle-specific expression of
a constitutively active form of PPAR5 receptor resulted in
transgenic mice with skeletal muscles that had an increased number
of slow, oxidative (type I) muscle fibers and markedly increased
running endurance (Wang et al., PLoC Biol., 2:e294, 2004). This
Example demonstrates that administration of a PPAR6 agonist
(GW1516) to non-transgenic mice also results in the expression in
skeletal muscle of some biomarkers of oxidative metabolism.
However, in unexpected contrast to the results obtained by genetic
activation of the PPAR5 pathway, PPAR5 activation by
pharmacological treatment did not modify fiber-type composition of
skeletal muscle, nor improve running endurance in non-transgenic,
sedentary (also referred to as "non-exercised" or "untrained")
mice. Male C57B/6J mice (8 wks old) were obtained from Jackson
Laboratory and housed in the Salk Institute animal care facility.
The animals were acclimated to their surroundings for one week
prior to experimentation, and had access at all times to standard
mouse chow and water ad libitum.
[00136] Mice were acclimated to moderate treadmill running (10
m/min for 15 min) every other day for 1 week. After acclimation,
basal running endurance was determined by placing each mouse on a
treadmill, gradually increasing the speed from 0 to 15 m/min, and
maintaining 15 m/min until the mouse was exhausted. The time and
distance run until exhaustion were recorded as the basal endurance
values (Week 0).
[00137] Mice then were treated once per day for 4 weeks with
vehicle or the PPAR5 agonist, GW 1516 (5 mg/kg). Treatments were
administered orally. During the treatment period, mice were housed
in standard laboratory cages and received only the amount of
physical activity that could be had by normal movements about such
cage.
[00138] Animals were euthanized by carbon dioxide asphyxiation
72 hours after the final treatment. Gastrocnemius and quadriceps
muscles were isolated, frozen and stored at -80 C for future
52
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
analysis. Total RNA was prepared from quadriceps muscle using
TRIzoLl reagent (Invitrogen, Carlsbad, CA, USA) in conformance with
manufacturer's instructions. Real time quantitative PCR (QPCR) was
used to determine expression levels of uncoupling protein 3 (UCP3),
muscle carnitine palmitoyl transferase I (mCPT I) and pyruvate
dehydrogenase kinase 4 (PDK4) using primers known to those of
ordinary skill in the art.
[00139] As shown in FIG. lA, four weeks of GW1516 treatment
induced the expression of UCP3, mCPT I, and PDK4, in quadriceps
muscle of treated mice (compare V to GW). These changes in gene
expression were detected as early as 4 days after treatment and
with drug concentrations ranging from 2-5 mg/kg/day.
[00140] Moreover, in the gene expression studies, maximal
effects of PPAR5 activation were detected in pre-dominantly fast-
twitch (quadricep and gastrocnemius) but not slow- twitch (soleus)
muscles.
[00141] Using primary muscle cells cultured from wild type and
PPAR6 null mice (Chawla et al., Proc. Natl. Acad. ScL U SA. 100(3):
1268-73, 2003; Man et al., J. Invest. Dermatol. 2007; Rando and
Blau, J. Cell. Biol. 125(6): 1275-87, 1994), it was confirmed that
the induction of oxidative genes by GW1516 is mediated via
activation of PPAR5 in skeletal muscles (FIGS. 1B-D). Moreover,
this is similar to the expression changes found in the same gene
set in muscles from mice expressing the constitutively active VP1(3-
PPAR5 transgene (Wang et al., PLoC Biol, 2:e294, 2004) (FIG. IA,
see TG). Collectively, these results indicate that pharmacological
activation of PPAR5 can initiate an oxidative response in adult
skeletal muscle.
[00142] Expression of biomarkers characteristic of an oxidative
phenotype in skeletal muscle, typically, has been correlated with
increased oxidative performance (e.g., increased running endurance)
of such skeletal muscle. This correlation was observed, for
instance, in the VP1(3-PPARb transgenic mouse (Wang et al., PLoC
Biol., 2:e294, 2004). For this and other reasons, it was expected
that GW1516 treatment similarly would increase running performance.
Accordingly, to determine the functional effects of ligand, age and
53
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
weight matched cohorts of treated and control mice were subjected
to an endurance treadmill performance test before (week 0) and
after (week 5) treatment. Following four weeks of treatment and
housing in standard laboratory cages without additional exercise,
the running endurance of GW1516-treated and control mice again was
determined as described above. Remarkably, and despite expectations
for improvement, GW1516-treated mice did not significantly differ
from controls in either the time spent or distance run on the
treadmill prior to exhaustion (FIG. lE). Furthermore, long-term
drug treatment of up to 5 months also did not change running
endurance.
[00143] These results indicate that although in non-trained
adult muscle pharmacological activation of PPAR6 induces some
transcriptional changes, it fails to alter either fiber type
composition or endurance. In summary, pharmacologic activation of
the PPAR6 genetic program in adult C57B1/6J mice is insufficient to
promote a measurable enhancement of treadmill endurance.
Example 2
[00144] Administration of PPARS agonist remodels skeletal muscle
in exercised-trained subjects. Fiber type proportions in skeletal
muscle are believed to be determined by heredity and environmental
factors, such as physical activity level (Simoneau and Bouchard,
FASEB J., 9(11): 1091-1095, 1995; Larsson and Ansved, Muscle Nerve,
8(8):714-722, 1985). Endurance exercise training is known to
remodel the skeletal muscle by increasing type I slow-twitch
fibers, oxidative enzymes, and mitochondrial density, which
progressively alter performance (Holloszy et al., J. Appl. Physiol.
56:831-8, 1984; Booth et al., Physiol Rev. 71:541-85, 1991; Schmitt
et al., . Physiol. Genomics. 15:148-57, 2003; Yoshioka et al.,
FASEB J. 17:1812-9, 2003; Mahoney et al., Phys. Med. Rehabil Clin.
N. Am. 16:859-73, 2005; Mahoney et al., FASEB J. 19:1498-500, 2005;
Siu et al., J. Appl Physiol. 97:277-85, 2004; Gamier et al., FASEB
J. 19:43-52, 2005; Short et al., J Appl Physiol. 99:95-102, 2005;
Timmons et al., FASEB J. 19: 750-60, 2005). This example
demonstrates that PPAR5 agonist treatment influences skeletal
muscle on a molecular level.
54
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
[00145] To determine whether co-administration of GW1516 in the
context of endurance exercise can enhance changes in fiber type
composition and mitochondrial biogenesis, the effect of GW1516
treatment on muscle fiber-type composition was determined by meta-
chromatic staining of cryo-sections of gastrocnemius as described
by Wang et al (Phys Biol, 2:e294, 2004). Meta-chromatic staining
was used, following a routine myofibrillar ATPase reaction, to
demonstrate quantitative differences in phosphate deposition among
different skeletal muscle fiber types and, thereby, differentiate
skeletal muscle fiber types (Doriguzzi et al., Histochem.,
79(3):289-294, 1983; Ogilvie and Feeback, Stain Technol, 65(5):231-
241, 1990). In this assay, muscle fibers with high ATPase activity
{e.g., type I (slow oxidative) muscle fibers) are darkly stained.
[00146] As shown in FIG. 2A, there was no significant difference
in the proportion of type I (slow, oxidative) muscle fibers in the
gastrocnemius muscles of vehicle- and GW1516-treated sedentary
mice. In contrast, hind limb muscles of VP16-PPAR5 transgenic mice
exhibited an increased number of type I muscle fibers when assayed
by monochromatic staining. In trained mice, GW1516 increased the
proportion of type I fibers (by -38%) compared to the vehicle-
treated sedentary mice (FIGS. 2A and 2B). Therefore, administration
of a PPAR6 agonist (e.g., GW1516) alone to sedentary subjects does
not significantly affect the number of type I (slow-twitch,
oxidative) muscle fibers in hind limb muscles, but can increase the
number of type I muscle fibers in hind limb muscles of trained
subjects.
[00147] In addition to its effects on the fiber type, exercise
training increased skeletal muscle mitochondrial biogenesis, which
can be measured as a function of mitochondrial DNA expression
levels using quantitative real time PCR (QPCR). Mitochondrial DNA
expression levels were determined in muscles of V, GW, Tr, and
GW+Tr subjects using quantitative real time PCR. As shown in FIG.
2C, similar to type I fiber changes, mitochondrial DNA expression
was not changed by drug alone but was increased by approximately
50% with the combination of exercise and GW1516 treatment (FIG.
2C). Such an increase is known to contribute to enhanced endurance
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
capacity (e.g., Holloszy, Med. Sci. Sports 7:155-64, 1975). Slow-
twitch and fast- twitch muscle fiber types also can be
distinguished by myosin isoform expression (Gauthier and Lowey, J.
Cell Biol. 81:10-25, 1979; Fitzsimons and Hoh, Biochem. J. 193:229-
33, 1981). Myosin isoform expression in skeletal muscle adapts to
various conditions, such as changes in muscle mechanics, muscle
innervation, or exercise paradigm (for review, see, e.g., Baldwin
and Haddad, J. Appl. Physiol., 90(1):345-57, 2001; Baldwin and
Haddad, Am. J. Phys. Med. Rehabil., 81(11 Suppl):S40-51, 2002;
Parry, Exerc. Sport Sci Rev., 29(4): 175-179, 2001). The effect of
GW1516 administration on myosin heavy chain (MHC) expression (MHC
I, MHC Ha, MHC lib) was determined by methods known to those of
ordinary skill in the art. GW1516 treatment in sedentary mice
increased the expression of MHC I (a marker of slow-twitch,
oxidative muscle fibers) and decreased the expression of MHC lib (a
marker of fast-twitch, glycolytic muscle fibers) as compared to
vehicle-treated, control mice. In comparison, GW1516 treatment did
not alter the expression of MHC Ha (a marker of fast-twitch
oxidative/glycolytic muscle fibers) in sedentary mice. Therefore,
at least at the transcriptional level, the PPAR5 agonist was
capable of inducing some proteins characteristic of a slow-twitch
muscle fiber phenotype.
[00148] Expression of constitutively active PPARb in the
skeletal muscles of VP1(3-PPAR6 transgenic mice resulted in a "long-
distance running phenotype" with "profound and coordinated
increases in oxidative enzymes, mitochondrial biogenesis and
production of specialized type I fiber contractile proteins-the
three hallmarks of muscle fiber type switching" (Wang at al., PLoC
Biol., 2:e294, 2004). In contrast, pharmacological activation of
PPARb in normal subjects only partially recapitulated VPl(3-PPAR5
transgenesis by regulating some metabolic genes. Markedly,
administration of a PPARb agonist to sedentary subjects did not
lead to a change in fiber type specification (as measured by
monochromatic staining) or enhance exercise endurance. Transgenic
over- expression of activated PPARb at birth pre-programs the
nascent myofibers to trans- differentiate into slow-twitch fibers,
56
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
thus imparting a high basal endurance capacity to adult transgenic
mice. In contrast, since fiber type specification is completed
prior to exposure of adults to PPAR6 agonist, the potential
plasticity of muscle to drug treatment alone is virtually non-
existent.
[00149] This example illustrates that the genetic or
pharmacologic activation of the PPAR5 regulatory program in
skeletal muscles of adult, sedentary subjects does not have the
same outcome. The ability to genetically manipulate skeletal muscle
specification by activation of the PPAR5 receptor in a transgenic
mouse from early development in the absence of exercise is not
necessarily predictive of the result of pharmacologically
activating the PPAR6 program in the sedentary, normal adult. The
cellular "template" for PPAR5 effects on skeletal muscle is very
different in a normal subject as compared to a genetically
engineered transgenic subject. For example, in a normal adult,
muscle fiber specification of individual muscle groups is already
determined and the connections between muscle fibers and spinal
motor neurons are established prior to pharmacological activation
of the PPAR5-regulated program. In the transgenic mouse, the
constitutively active PPAR6 transgene is active all the while
muscle fiber specification is being determined and connections
between muscle fibers and motor neurons are being made. In
addition, the effects of activation of endogenous PPARS receptor by
a single daily dose of a PPAR5 agonist, which is expect to have a
transient peak exposure followed by clearance, likely are much
different from the effects of the constitutive activation of a
VP1(3-PPAR6 transgene.
Example 3
[00150] The combination of PPAR6 agonist treatment and exercise
training significantly affected fatty acid metabolism and markers
of fatty acid oxidation. In addition to affecting the contractile
apparatus of skeletal muscle, exercise training also increases
skeletal muscle mitochondrial density (e.g., Freyssenet et al.,
Arch. Physiol. Biochem., 104(2): 129-141, 1996). This Example
illustrates that PPAR6 agonist treatment (e.g., GW1516) in
57
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
exercise-trained subjects affected fatty acid metabolism in
exercised muscle. The effects of GW1516 treatment and exercise,
alone or in combination, on components of the oxidative metabolism
of fatty acids were determined by measuring gene expression levels
of selective biomarkers for fatty acid R-oxidation (FAO). Male
C57B/6J mice (8-10 wks old) were randomly divided into four groups
(nine per group): (i) vehicle-treated and sedentary (V), (ii)
GW1516-treated and sedentary (GW), (iii) vehicle-treated and
exercise trained (Tr) and (iv) GW1516-treated and exercise trained
(GW+Tr). Mice in all groups were acclimated to moderate treadmill
running and basal running endurance was determined as described in
Example 1. Thereafter, mice in the exercise-trained groups received
four weeks (5 days/week) of exercise training on a treadmill
inclined at 5 degrees. Intensity and time of training were
gradually increased. At the end of four weeks, all exercise-trained
mice were running for 50 min/day at 18 m/min. Vehicle or GW1516 was
administered to the respective exercise-treated or sedentary groups
as described in Example 1. Unless otherwise noted, V, GW, Tr and
GW+Tr subjects described in this and the examples below were
similarly treated. At the end of the drug treatment and/or training
protocol (Week 5) 6 mice per group were subjected to the running
test. These interventions do not affect body weight and food intake
in mice. RNA was prepared real time quantitative PCR performed as
described in Example 1.
[00151] Confirming the results obtained in Example 1, UCP3, mCPT
I, and PDK4 were upregulated by GW1516 but showed no further
induction with exercise (see FIGS. 1A and 3A). Unexpectedly, a
second set of genes were identified that showed no response to
exercise or GW1516 alone but were robustly induced by the
combination. This intriguing response profile includes a series of
genes involved in the regulation of fatty acid storage (such as
steroyl-CoA-desaturase (SCD1), fatty acyl coenzyme A synthase (FAS)
and serum response element binding protein Ic (SREBPIc)) and fatty
acid uptake [such as the fatty acid transporter (FAT/CD36) and
lipoprotein lipase (LPL)] adding a new set of target genes to
exercise and drug treated mice (FIGS. 3B, 3C and 6A-C).
58
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
[00152] In addition to gene expression, protein expression was
determined for selective oxidative biomarkers including myoglobin,
UCP3, cytochrome c (CYCS) and SCD1, using Western blotting. Protein
homogenates were prepared from quadriceps muscle, separated by SDS
polyacrylamide gel electrophoresis, transferred to blotting
membrane and probed with antibodies specific for myoglobin (Dako),
UCP3 (Affinity Bioreagents), cytochrome c (Santacruz) SCD1
(Santacruz), and, as a loading control, tubulin (Sigma). A robust
up regulation of myoglobin, UCP3, cytochrome c, and SCD1 protein
expression was observed with combined exercise and GW1516 treatment
in comparison to treatment with the PPAR6 agonist or exercise alone
(FIG. 3D).
[00153] Altered triglycerides can be used to access changes in
muscle oxidative capacity. Muscle triglyceride (mTG) content was
measured as previously described (Wang et al., PLoC Biol, 2:e294,
2004) using a kit from Thermo Electron Corporation. As shown in
FIG. 4, mTG content was comparable between vehicle and GW1516-
treated sedentary mice and was substantially increased in muscle of
mice receiving only exercise training. In contrast, dramatic
increase in triglycerides in exercised muscle was completely
reversed in GW1516-treated exercise trained mice, indicating
increased fat utilization (FIG. 4). Gene and/or protein expression
that is induced by a combination of exercise and drug treatment
(e.g., PPAR5 agonist administration) but not by either input alone
is believed to be a new discovery. This type of response can be
used to further characterize the intersection of pharmacologic and
physiologic genetic networks. For example, one or more genes and/or
proteins uniquely regulated by one or more drugs {e.g., PPAR6
agonists) and exercise can be used as markers, for instance, of
illicitly boosting performance in professional and/or amateur
athletes.
Example 4
[00154] Administration of PPAR6 agonist enhances the physical
performance of exercise-trained subjects. As described in Example
1, although GW1516 treatment induces wide- spread genomic changes
associated with oxidative metabolism, nonetheless alone it failed
59
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
to increase running endurance. This finding was unexpected because
it was known that constitutive activation of the PPARb gene network
(in the VP1(3-PPAR6 transgenic mouse) lead to a distance-running
phenotype (familiarly, a "marathon mouse"). On the other hand, as
surprisingly shown in Example 3, PPARb agonist (e.g., GW1516)
treatment in conjunction with exercise produced an enriched
remodeling program that included a series of transcriptional and
post-translational adaptations in the skeletal muscle. This
indicates that exercise training serves as a trigger to unmask a
set of PPAR5 target genes. This Example provides methods used to
demonstrate that administration of a PPARb agonist (e.g., GW1516)
surprisingly improves physical performance in exercised (trained)
subjects.
[00155] Male C57B/6J mice (8-10 wks old) were randomly divided
into four groups (nine per group): (i) vehicle-treated and
sedentary (V), (ii) GW1516-treated and sedentary (GW), (iii)
vehicle-treated and exercise trained (Tr) and (iv) GW1516-treated
and exercise trained (GW+Tr), acclimated to moderate treadmill
running as described in Example 1, and exercise-trained as
described in Example 3. At the end of the drug treatment and/or
training protocol (Week 5) 6 mice per group were subjected to the
running test.
[00156] At the end of the drug treatment and/or training
protocol (Week 5), running endurance of six mice per group was
determined in the same manner as was basal running endurance. No
follow-up endurance tests were performed on three mice in each
group to confirm that changes observed in the skeletal muscle were
not due to the acute run, but were related to the exercise
training.
[00157] As shown in FIGS. 5A and 5B, the same dose and duration
of GW1516 treatment that failed to alter running endurance in
sedentary mice, when paired with 4 weeks of exercise training,
increases running time by 68% and running distance by 70% over
vehicle-treated trained mice (FIGS. 5A and 5B, compare Week 5).
Comparison of running time and distance before (week 0) and after
(week 5) exercise and drug treatment revealed a 100% increment in
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
endurance capacity for individual mice, underscoring the robustness
of the combination paradigm (FIGS. 5 A and 5B). In contrast, the
same exercise protocol without concurrent GW1516 treatment did not
significantly increase running endurance in C57B1/6J mice.
Hematoxylin and eosin (H&E) staining of white adipose tissue
paraffin sections was performed as previously described (Wang et
al., PLoS Biol., 2:e294, 2004; Wang et al., Cell, 113:159-70,
2003). As shown in FIG. 5C, GW1516 treatment in combination with
exercise produced a significant (32%) reduction in the epididymal
fat to body weight ratio, which was further evident in the
decreased cross-sectional area of the adipocytes in the same group
(FIG. 5D). Therefore, the combined effects of GW1516 and exercise
are not restricted to muscle.
[00158] Using the methods described in Example 2, it was also
demonstrated that the combination of GW1516 treatment and exercise
training significantly increased the number of type I muscle fibers
in exercised muscle. However, combining GW1516 treatment with
exercise did not induce additional changes in MHC I and MHC lib
expression. Therefore, although orally administered PPAR5 agonist
(GW1516) alone is capable of inducing the expression of at least
some of the contractile proteins in the PPAR6-regulated gene
network (see Example 5) the transcriptional effect observed was not
sufficient to induce a post-transcriptional change in fiber-type
composition as was observed by meta-chromatic staining in GW1516-
treated, exercised mice.
[00159] This Example illustrates that PPAR6 agonist (e.g.,
GW1516) treatment unexpectedly augments the performance of aerobic
exercise (e.g., running distance and endurance) in an exercised
subject. Endurance exercise is known to channel extra-muscular fat
to muscle triglyceride stores by inducing adipose tissue lipolysis
to meet increased oxidative demands (Despres et al., Metabolism,
33:235-9, 1984; Mauriege et al., Am. J. Physiol, 273:E497-506,
1997; Mader et al., Int. J. Sports Med., 22:344-9, 2001; Schmitt et
al., Physiol. Genomics, 15:148-57, 2003; Schrauwen-Hinderling et
al., J. Clin. Endocrinol. Metab., 88:1610-6, 2003). In addition,
the induction of FAO components and selective up-regulation of
61
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
fatty acid storage and up-take components in GW15i6-treated,
exercised mice described in Example 3 indicate enhanced
mobilization of fat as fuel in skeletal muscle.
[00160] Therefore, combined exercise and GW1516 treatment
dramatically increases muscle oxidative capacity in subjects, for
example by increasing local fatty acid synthesis and/or mobilizing
fatty acid stores from adipose tissue.
[00161] This is the first demonstration of how an orally active
PPAR5 agonist and exercise can co-operatively re -program the
muscle genome and raise endurance limits.
Example 5
[00162] The combination of PPAR5 agonist treatment and exercise
training produced a unique gene expression signature. A
comprehensive study of the skeletal muscle transcriptional program
in V, GW, Tr and Tr+GW mice was conducted using microarray
analysis. AffymetrixTM high-density oligonucleotide array mouse
genome 430A 2.0 chips were used.
[00163] Preparation of in vitro transcription products,
oligonucleotide array hybridization, and scanning were performed in
conformance with AffymetrixTM-provided protocols. To minimize
discrepancies due to variables, the raw expression data were scaled
by using AffymetrixTM MICROARRAY SUITETM 5.0 software, and pairwise
comparisons were performed. The trimmed mean signal of all probe
sets was adjusted to a user-specified target signal value (200) for
each array for global scaling. No specific exclusion criteria were
applied. Additional analyses were performed using the freeware
program BULLFROG 7 (available on the internet Barlow-
LockhartBrainMapNIMHGrant.org) and the Java-based statistical tool
VAMPIRE (Hsiao et al., Bioinformatics, 20:3108-3127, 2004).
[00164] Genome-wide analysis of the quadriceps muscle revealed
that GW1516 treatment, exercise, and the combination regulated 96,
113 and 130 genes, respectively (FIG. 6). Approximately 50% of the
target genes regulated by GW1516 or exercise alone were the same,
demonstrating that PPAR6 activation of the gene network partially
mimics exercise effects on the same network.
62
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
[00165] The 130 genes regulated by the combination of GW1516
treatment and exercise training and a classification of each such
gene are shown in Table 1. The 130 regulated genes included 30 fat
metabolism genes, 5 oxygen carriers, 5 mitochondrial genes, 3
carbohydrate metabolism genes, 15 signal transduction genes, 16
transcription genes, 10 transport genes, 3 steroid biogenesis
genes, 5 heat shock genes, 2 angiogenesis genes, 5 proliferation
and apoptosis genes, 2 cytokines, and 29 others. The majority of
the genes in the exercise-trained/GW1516-treated (GW+Tr) gene
signature shown in Table 1 were induced (109/130). The 109
upregulated genes are shown in non-bold font in Table 1 (final
column >1). Down-regulated genes are shown in bold italics in Table
1 (final column <1).
63
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
TABLE 1
Genes regulated by GW 1516 treatment and exercise training
FEATURE LOCUS DESCRIPTION GW +Tr
ANGIOGENESIS
1417130s at Angptl4 angiopoietin-like 4 5.495
1418762_at Cd55 CD55 aat gex 0.56
CARBOHYDRATE METABOLISM
1449088at Fbp2 fructose bisphosphatase 2 2.808
1423439_at Pekl phosphoenolpyruvate carboxykinase 1, cytosolic 3.518
1434499aat Ldhb lactate dehydrogenase B 2.541
PROLIFERATION & APOPTOSIS
1425621at Trim35 tripartite motif-containing 35 1.856
1418003 at 1190002H23Rik RLIENtDNA 1190002H23 gene 0.543
1448272at Btg2 B-cell translocation gene 2, anti-proliferative 1.601
1452260_at Cidec cell death-inducing DFFA-like effector c 4.771
1417956 at Cidea cell death-inducing DNA fragmentation factor, alpha 49.625
subunit-like effector A
CYTOKINES
1426278_at Ifi27 interferon, alpha-inducible protein 27 1.714
1421239at I16st interleukin 6 signal transducer 1.972
FAT METABOLISM
1448318_at Adt adipose differentiation related protein 2.009
1424729at BC054059 cDNA sequence BC054059 5.08
1424937at 2310076L09Rik RIKEN cDNA 2310076L09 gene 1.868
1450010 at Hsdl7bl2 hydroxysteroid (17-beta) dehydrogenase 12 2.376
1415965at Scdl stearoyl-CoenzymeAdesaturase1 6.494
1415822at Scd2 stearoyl-CoenzymeA desaturase 2 1.849
1423828at Fasn fatly acid synthase 6.323
1455061a at Acaa2 acetyl-Coenzyme A acyltransferase 2 (mitochondrial 3- 1.926
oxoacyl-Coenzyme A thiolase)
1448987_at Acadl acetyl-Coenzyme A dehydrogenase, long-chain 2.549
1422651at Adipoq adiponectin, C1Q and collagen domain containing 3.082
1422820_at Lipe lipase, hormone sensitive 3.032
1449964aat Mlycd malonyl-CoA decarboxylase 1.781
1426785s at Mgll monoglyceride lipase 1.907
1420658at Ucp3 uncoupling protein 3 (mitochondrial, proton carrier) 2.943
1425326at Acly ATP citrate lyase 2.606
1460409_at Cptla carnitine palmitoyltransferase 1a, liver 2.753
1422677at Dgat2 diacylglycerol O-acyltransferase 2 2.784
1456702 x at Gge gamma-glutmnyl carbmylase 0.575
1425834aat Gpam glycerol-3-phosphate acyltransferase, mitochondrial 2.207
1417273at Pdk4 pyruvate dehydrogenase kinase, isoenzyme 4 2.27
1449182 at Retn resistin 4.114
1435630s at Acat2 acetyl-Coenzyme A acetyltransferase 2 1.625
1425829aat Abcbla ATP-binding cassette, sub-family B (MDR/TAP), member 10.322
1A
1423166at Cd36 CD36 antigen 1.584
1422811 at Slc27al solute carrier family 27 (fatty acid transporter), member 1
3.58
1416023at Fabp3 fatty acid binding protein 3, muscle and heart 1.833
1424155at Fabp4 fatty acid binding protein 4, adipocyte 2.189
1431056aat Lpl lipoprotein lipase 1.659
1422432at Dbi diazepam binding inhibitor 1.936
1422811_at Slc27al solute carrier family 27 (fatty acid transporter), 1 3.58
HEAT SHOCK RESPONSE
1448881at Hp haptoglobin 1.679
1427126 at Hspalb heat shock protein lB 8.845
1438902_a_at Hsp90aal heat shock protein 90 kDa alpha (cytosolic), class A
1.513
member 1
64
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
TABLE 1-continued
Genes regulated by GW 1516 treatment and exercise training
FEATURE LOCUS DESCRIPTION GW + Tr
1431274aat Hspa9a heat shock protein 9A 1.61
1416755at Dnajbl DnaJ (Hsp40) homolog, subfamily B, member 1 3.59
MISCELLANEOUS
1460256at Cara carbonic anhydrase 3 2.339
1415841at Dyncli2 dynein cytoplasmic 1 intermediate chain 2 1.705
1432344aat Aplp2 amyloid beta (A4) precursor-like protein 2 1.937
1416429a at Cat catalase 1.82
1418306at Crybbl crystallin, beta B1 2.457
1448842at Cdol cysteine dioxygenase 1, cytosolic 3.266
1434503 s at Lamp2 lysosomal membrane glycoprotea 2 0.608
1416473_a at Nope neigbborofPuncEll 0.452
1453527aat Neurl neuralized-like homolog (Drosophila) 1.941
1451603 at Rtbdn retbindin 2.32
1453724aat Serpinfl serine (or cysteine) peptidase inhibitor, Glade F, member
1 7.765
1448680 at Serpinala seriae (or cysteiae) proteinase inbitor, dadeA, member
0.396
1a
1427285sat Surf4 surfeit gene 4 2.091
1424737at Thrsp thyroid hormone responsive SPOT14 homolog (Rattus) 2.685
1431609a at Acp5 acid phosphatase 5, tartrate resistant 3.91
1448538aat D4Wsu53e DNA segment, Chr 4, Wayne State University 53, 1.586
expressed
1452406x at erytluord differentiation regulator 1 0.619
fusion, derived from t(12;16) malignant lipacarcwna
1451286 s at Fus (human) 0.605
1425552at Hiplr huntingtin interacting protein 1 related 1.75
1428091 at 1ab17 kelcb-like 7 (Drosopln7a)
1429360at KID Kruppel-like factor 3 (basic) 1.901
1449413 at Mpvl7l Mpv17 transgene, kidney disease mutant-like 1.988
1451667at C530043G21Rik RIKEN cDNA C530043G21 gene 1.5
1425865a at Lig3 ligase III, DNA, ATP-dependent 2.693
1415994 at Cyp2el cytochrome P450, family 2, subfamily e, polypeptide 1 2.941
1417867 at Cfd complement factor D (adipsin) 2.828
1451015 at Tkt transketolase 2.256
1432344aat Aplp2 amyloid beta (A4) precursor-like protein 2 1.937
1419487at Mybph Myosin binding protein H 1.578
MITOCHONDRIAL PROTEINS
1436750 a at Oxctl 3-axoadd CoAiransferase 1 0.574
1415897aat Mgstl microsomal glutathione S-transferase 1 1.916
1434970 a at Mnp115 mitoclandrialrnbosmmalproteunLIS 0.61
1423109sat Slc25a20 solute carrier family 25 (mitochondrial 1.865
carnitine/acylcarnitine translocase), member 20
1416014 at Abcel ATP4mdbngcasseue, sub-amiy E (OABP), member I 0.556
OXYGEN CARRIERS
1448348at Gpiapl GPI-anchored membrane protein 1 1.83
1451203at Mb myoglobin 1.578
1428111 at S1c38a4 slate carrier family 38, member 4 0.579
1428361xat Hba-al hemoglobin alpha, adult chain 1 1.632
1417184 sat Hbb-b2lHbb-y hemoglobin, beta adult minor chain hemoglobin Y,,beta-
1.626
like embryonic chain
SIGNAL TRANSDUCTION
1416137 at Anxa7 ammexiiA7 0.544
1455918at Adrb3 adrenergic receptor, beta 3 3.83
1417163 at Dusp10 dual specificityphosplatae 10 0.579
1452097_a_at Dusp7 dual specificity phosphatase 7 1.661
1419191at Hipk3 homeodomain interacting protein kinase 3 1.694
1448152at Igf2 insulin-like growth factor 2 1.635
1422313a at Igfbp5 insulin-like growth factor binding protein 5 1.772
1428265at Ppp2rlb protein phosphatase 2 (formerly 2A), regulatory subunit A
2.509
(PR 65), beta isoform
1438562 a at Ptpn2 Protein tyrosunepbasphatoce, non-rceeptr type 2 0.432
1449342at Ptplb protein tyrosine phosphatase-like (proline instead of 2.38
catalytic arginine), member b
1422119 at Rab5b RAB5B, member RAS oncogene family 1.603
1437016 x at Rap2c RAM, memberofRASoneogenefamMy 0.601
1425444aat Tgfbr2 transforming growth factor, beta receptor II 2.13
1431164 at Rragd Ras-related GTP binding D 2.101
1420816at Ywhag 3-monooxygenase/tryptophan 5-monooxygenase activation 1.87
protein, gamma polypeptide
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
TABLE 1-continued.
Genes regulated by GW1516 treatment and exercise training
FEATURE LOCUS DESCRIPTION GW + Tr
STEROID BIOGENESIS
1418601_at Aldhla7 aldehyde dehydrogenase family 1, subfamilyA7 3.862
1426225at Rbp4 retinol binding protein 4, plasma 2.065
1455913 x at TYr trarchyre i 0.026
TRANSCRIPTION
1417794at Zfp261 zinc finger protein 261 1.847
1424731at Nlel notchless homolog 1 (Drosophila) 1.831
1454791_a_at Rbbp4 retinoblastoma binding protein 4 2.865
1460281at Ash 15 ankyrin repeat and SOCS box-containing protein 15 1.78
1449363 at Atf3 activating transcription factor 3 1.802
1418982at Cebpa CCAAT/enhancer binding protein (C!EBP), alpha 2.168
1417065_at Egrl early growth response 1 2.577
1434831 a at Faxn3a fort lead box 03a 0.634
1415899at Junb Jun-B oncogene 1.792
1421554_at Lmxla LIM homeobox transcription factor 1 alpha 4.106
1416959_at Nrld2 nuclear receptor subfamily 1, group D, member 2(Reverb-b)
1.794
1450749 aat Nr4a2 nuclear receptor subfamily 4, group A, member 2 (NURR1)
1.776
1460215at Rpol-4 RNA polymerase 1-4 2.498
1417719 at Sap30 sm3 associated polypeptide @551
1420892_at Wnt7b wingless-related MMTV integration site 713 4.449
1423100_at Fos FBI osteosarcoma oncogene 3.9
TRANSPORT PROTEINS
1427222 Cat Svp2 seminal vesicle protein 2 t 014
1456124_x_at SvsS seminal vestide secretion 5 Q 095
1425546 a at Trf transferrin 1.907
1423743 at Arent archain 1 1.617
1451771 at Tpcnl two pore channel 1 2.842
1416629_at Slc1a5 solute carrier family 1 (neutral amino acid transporter),
1.939
member 5
1420295 x at Clcn5 chloride channel 5 2.333
1417839 at CldnS claudin 5 1.545
1425260_at Albl albumin 1 x245
1434617 xat 1810073N04Rik RIKEN cDNA 1810073N04 gene 2.326
Data is average of N=3 samples in each group (p<0.05).
[00166] Surprisingly, the combination of GW1516 treatment and
exercise established a unique gene expression pattern ("a GW+TR
profile") that was neither an amalgamation nor a complete overlap
of the two interventions (FIG. 6). This unique signature included
48 target genes (Table 2) not regulated by GW1516 and exercise
alone and excluded 74 genes regulated by GW1516 or exercise alone
(some of which are shown in Table 3). This signature for the
combination of GW1516 treatment and exercise (Table 2) was highly
enriched in genes encoding regulatory enzymes for energy
homeostasis, angiogenesis, oxygen transport, signal transduction,
66
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
transcription and substrate transport, which are processes that are
involved in endurance adaptation. Particularly, a predominance of
genes involved in oxidative metabolism, is selectively up-regulated
by combined exercise and drug treatment (see unbolded genes in
Tables 1 and 2). In addition, several stress-related genes
activated by either intervention, including heat shock proteins,
metallothioneins and other stress biomarkers (Table 3) are not
changed by the combination possibly reflecting a potential
lessening of exercise-based damage.
TABLE 2
Gene targets unique to combined GW 1516 treatment and exercise training.
DESCRIPTION LOCUS GW + Tr
ANGIOGENESIS
CD55 antigen Cd55 0.56
CARBOHYDRATE METABOLISM
phosphoenolpyruvate carboxykinase 1, cytosolic Pckl 3.518
CYTOKINES
interferon, alpha-inducible protein 27 Ifi27 1.714
67
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
TABLE 2-continued
Gene targets unique to combined GW 1516 treatment and exercise training.
DESCRIPTION LOCUS GW + Tr
FAT METABOLISM
adipose differentiation related protein Adrp 2.009
stearoyl-Coenzyme A desaturase 2 Scd2 1.849
acetyl-Coenzyme A acetyltransferase 2 Acat2 1.625
ATP citrate lyase Acly 2.606
adiponectin, C1Q and collagen domain containing Adipoq 3.082
diacylglycerol O-acyltransferase 2 Dgat2 2.784
gumma-glutmnyl oarboiylose Ggcx 0.575
lipase, hormone sensitive Lipe 3.032
monoglyceride lipase Mgll 1.907
resistin Retn 4.114
CD36 antigen Cd36 1.584
fatty acid binding protein 4, adipocyte Fabp4 2.189
lipoprotein lipase Lpl 1.659
HEAT SHOCK RESPONSE
haptoglobin Hp 1.679
MITOCHONDRLAL PROTEINS
3-aroadd CoA eranaferase 1 Oxctl 0.574
microsomal glutathione S-transferase 1 Mgstl 1.916
OTHERS
carbonic anhydrase 3 Cara 2.339
cysteine dioxygenase 1, cytosolic Cdol 3.266
DNA segment, Chr 4, Wayne State University 53, expressed D4Wsu53e 1.586
dynein cytoplasmic 1 intermediate chain 2 Dyncli2 1.705
Fus 0.605
Kruppel-like factor 3 (basic) KIf3 1.901
lysosomal membrane glycoprotein 2 Lamp2 0.608
neighbor ofFuncEll Nope 0.452
thyroid hormone responsive SPOT14 homolog (Rattus) Thrsp 2.685
cytochrome P450, family 2, subfamily e, polypeptide 1 Cyp2el 2.941
complement factor D (adipsin) Cfd 2.828
transketolase Tkt 2.256
OXYGEN CARRIERS
GPI-anchored membrane protein 1 Gpiapl 1.83
solute carrier family 38, member 4 S1c38a4 0.579
PROLIFERATION & APOPTOSIS
BUOY cDNA1190002H23geee 1190002H23Rik 0.543
cell death-inducing DFFA-like effector c Cidec 4.771
SIGNAL TRANSDUCTION
amrezbz A7 Anxa7 0.544
dual specificity phosphatase 7 Dusp7 1.661
homeodomain interacting protein kinase 3 Hipk3 1.694
insulin-like growth factor binding protein 5 Igfbps 1.772
protein phosphatase 2 (formerly 2A), regulatory subunit A (PR 65), beta
Ppp2rlb 2.509
isoform
protein tyrosine phosphatase-like (proline instead of catalytic arginine),
Ptplb 2.38
member b
STEROID BIOGENESIS
retired binding protein 4, plasma Rbp4 2.065
TRANSCRIPTION
CCAATienhancer binding protein (C/EBP), alpha Cebpa 2.168
nuclear receptor subfamily 1, group D, member 2(Reverb-b) Nrld2 1.794
TRANSPORT
transferrin Trf 1.907
archain 1 Arent 1.617
solute carrier family 1 (neutral amino acid transporter), member 5 SlclaS
1.939
RIKEN cDNA 1810073N04 gene 1810073NO4Rik 2.326
68
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
TABLE 3
Gene targets regulated by GW 1516 treatment or exercise training alone.
FEATURE LOCUS DESCRIPTION GW Tr GW + Tr
Hspbl heat shock protein 1 1.815 1.965
1451284_at Hspb7 heat shock protein family, 7 (cardiovascular) 3.414 1.753 -
1422943 aat Dnajal DnaJ (Hsp40) homolog, subfamily A, 1 1.545
1421290_at Hsp110 heat shock protein 110 1.587
1416288 at Serpinhl serine (or cysteine) peptidase inhibitor, IT, 1 2.198
1423566_a_at Dnaja4 DnaJ (Hsp40) homolog, subfamily A, 4 1.756 1.545
1417872at Mtl metallothionein 1 2.364
1424596_s_at Mt2 metallothionein 2 2.151
1416157 at Cryab crystallin, alpha B 1.561 1.52
1423139 at Crygf crystallin, gamma F 1.801 3.56
1448830 at Smad3 MAD homolog 3 (Drosophila) 1.841 1.886
1450637_a_at Ankrdl ankyrin repeat domain 1 (cardiac muscle) 4.235
1416029at Tnfrsfl2a TNF receptor superfamily, 12a 1.759 1.782
1426464at Jun Jun oncogene 1.521
Data is average of N=3 samples in each group (p<0.05)
[00167] Thirty-two percent of the GW+Tr-regulated genes encode
enzymes of metabolic pathways such as fatty acid
biosynthesis/storage (e.g., FAS, SCD 1 & 2), uptake [e.g.,
FAT/CD36, fatty acid binding proteins (FABP) and LPL] and oxidation
[e.g., adiponectin, hormone sensitive lipase (HSL), PDK4, UCP3];
and carbohydrate metabolism [e.g., fructose bisphosphate 2 (FBP2),
phosphoenolpyruvate carboxykinase 1 (PEPCK1), lactate dehydrogenase
B], which along with oxygen transporters and mitochondrial proteins
form the largest class of genes directly linked to muscle
performance (Ikeda et al., Biochem. Biophys. Res. Commun. 296:395-
400, 2002; Achten and Jeukendrup, Nutrition. 20:716-27, 2004;
Hittel et al., J. Appl. Physiol. 98: 168-79, 2005; Civitarese et
al., Cell Metab. 4:75-87, 2006; Nadeau et al., FASEB J. 17:1812-9,
2006; Kiens, Physiol. Rev. 86:205-43, 2006; Yamauchi et al., Nat.
Med. 8:1288-95, 2006). Unexpectedly, established PPARa target genes
fatty acyl-CoA oxidase and medium chain acyl-CoA dehydrogenase
(MCAD) were not represented in the signature. All but four of these
metabolic genes were induced, which indicated a general increase in
oxidative capacity of skeletal muscle in exercise-trained subjects
that received GW1516 treatment.
[00168] Other genes regulated in quadriceps muscle by the
combination of exercise and GW1516 treatment encoded proteins
69
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
involved in pathways such as angiogenesis (e.g., angiopoietin-like
4 protein/also a known regulator of lipid metabolism), (e.g.,
adrenergic receptor R3, insulin-like growth factor, insulin-like
growth factor binding protein 5), transcription (e.g., C/EBP a,
Reverb R, NURR1) and substrate transport (e.g., transferrin,
chloride channel 5) (Nagase et al., J. Clin. Invest. 97:2898-904,
1996; Singleton and Feldman, Neurobiol. Dis. 8:541-54, 2001; Adams,
J. Appl. Physiol. 93:1159-67, 2002; Centrella et al., Gene. 342:
13-24, 2004; Lundby et al., Eur. J. Appl Physiol. 96: 363-9, 2005;
Mahoney et al., FASEB J. 19: 1498-500, 2005; Mahoney et al., Phys.
Med. Rehabil Clin. N. Am. 16: 859-73, 2005; Ramakrishnan et al., J.
Biol. Chem. 280:8651-9, 2005). Without wishing to be bound to a
particular theory, such other genes are likely involved, at least
in part, in muscle remodeling and increased endurance observed in
GW1516-treated, exercise-trained subjects.
[00169] Interestingly, comparative expression analysis of the 48
gene subset of the endurance signature (Table 2), but not of either
intervention alone, revealed a striking similarity to 'untrained'
VP16-PPAR5 transgenic mice. This observation confirms the primary
dependence of the 48 genes on PPAR5 and indicates that exercise-
generated signals may function to synergize PPAR5 transcriptional
activity to levels comparable to transgenic over-expression.
Therefore, exercise cues along with PPAR5 agonist may function to
hyper-activate receptor transcriptional activity to re-program of
adult muscle.
[00170] Genes and/or proteins uniquely affected (e.g., up-
regulated or down-regulated or not substantially regulated) by
exercise in the presence of one or more pharmaceutical agents
(e.g., PPAR5 agonists) can be used as markers, for instance, of
"drug doping" in exercise-trained subjects (e.g., athletes). It is
expected that the unique set of 48 genes regulated by GW+Tr, but
not GW1516 treatment or exercise training alone, can be used to
identify exercised subjects who have received a variety
performance-enhancing drugs.
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
Example 6
[00171] PPARb directly interacts with exercise-activated
kinases, p44/42 MAPK and AMPK. Exercise training is known to
activate kinases, such as p44/42 MAPK and AMPK, which regulate gene
expression in skeletal muscle (Chen et al., Diabetes, 52:2205-12,
2003; Goodyear et al., Am. J. Physiol, 271:E403-8, 1996). AMPK
affects skeletal muscle gene expression and oxidative metabolism
(Chen et al., Diabetes. 52: 2205-12, 2003, Reznick et al., J.
Physiol. 574: 33-9, 2006). The interaction between exercise-
regulated kinases and PPAR5 signaling is described in this Example.
The levels of phospho-p44/42 MAPK and phospho-AMPK a subunit and
total AMPK were determined in protein homogenates of quadriceps
muscle by Western blot. Antibodies specific for phospho-p44/42
MAPK, phospho- and total-AMPK al antibodies were obtained from Cell
Signaling. The phospho- specific AMPK al antibody recognizes the
key activating threonine in the activation loop.
[00172] Active forms of both kinases (phospho-p44/42 MAPK and
phospho-AMPK a subunit) were expressed at higher levels in the
quadriceps muscles of exercised mice relative to the sedentary
controls (FIG. 7A). Previous reports claim that PPARb is not
required for activation of AMPK by GW1516 in cultured cells (Kramer
et al., Diabetes. 54(4): 1157-63, 2005 and Kramer et al., J. Biol
.Chem. 282(27): 19313-2, 2007). In contrast, it was observed that
GW1516 failed to activate p44/42 or AMPK in either sedentary or
trained muscles, which indicated that PPARb-regulated effects are
downstream to the exercise-induced signals that activate these
kinases. Furthermore, AMPK appears to be constitutively active in
muscles of VP16-PPAR6 transgenic mice in absence of exercise or
drug (FIG. 7B). These results indicate that synergy is AMPK and
PPARb co-dependent.
[00173] If synergy is AMPK and PPARb co-dependent, selective co-
activation of AMPK and PPARb would induce gene expression changes
that mimic those triggered by combined exercise and PPAR6 as well
as VP16-PPAR6 over-expression. To demonstrate this, transcriptional
changes induced in skeletal muscle by combined exercise and GW1516
treatment (as described in Example 5) were compared to that of
71
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
combined AMPK activator (the cell permeable AMP analog AICAR; 250
mg/kg/day, i.p.) and GW1516 (5 mg/kg/day, oral gavage) treatment
for 6 days. Genome analysis was performed using the methods
described in Example 5.
[00174] Simultaneous GW1516 and AICAR treatment for 6 days
created a unique gene expression signature in the quadriceps of
untrained C57B1/6J mice (FIG. 8A, which includes target genes
associated with translation, protein processing, amino acid
metabolism, fat metabolism, oxygen carriers, carbohydrate
metabolism, signal transduction, transcription, transport, steroid
biogenesis, heat shock response, angiogenesis, proliferation and
apoptosis, cytokines, contractile proteins, stress, and others)
that shares 40% of the genes with that of combined GW1516 treatment
and exercise (FIG. 8B). Classification of the 52 genes common to
the two signatures (combined PPAR5 activation and exercise or PPAR5
and AMPK co-activation) (listed in Table 4) revealed that the
majority of the targets were linked to oxidative metabolism.
72
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
TABLE 4
Targets common to exercise-PPARS and AMPK-PPARS gene signatures.
DESCRIPTION LOCUS Tr + GW AI + GW
ANGIOGENESIS
angiopoietin-like 4 Angptl4 5.495 2.917
APOPTOSIS
cell death-inducing DFFA-like effector c Cidec 4.771 1.838
cell death-inducing DNA fragmentation factor, alpha Cidea 49.625 1.842
subunit-like effector A
CARBOHYDRATE METABOLISM
lactate dehydrogenase B Ldhb 2.541 1.917
fructose bisphosphatase 2 Fbp2 2.808 2.478
FAT METABOLISM
stearoyl-Coenzyme A desaturase 1 Scdl 6.494 1.78
fatty acid binding protein 3, muscle and heart Fabp3 1.833 1.5
pyruvate dehydrogenase kinase, ioenzyme 4 Pdk4 2.27 2.486
uncoupling protein 3 (mitochondrial, proton carrier) Ucp3 2.943 2.792
adiponectin, C1Q and collagen domain containin Adipoq 3.082 1.56
diacylglycerol O-acyltransferase 2 Dgat2 2.784 2.14
solute carrier family 27 (fatty acid transporter), member 1 S1c27a1 3.58 2.195
lipase, hormone sensitive Lipe 3.032 1.746
solute carrier family 25 (mitochondrial Slc25a20 1.704 1.697
carnitine/acylcarnitine translocase), member 20
CD36 antigen Cd36 1.584 1.513
phosphoenolpyruvate carboxykinase 1, cytosolic Pckl 3.518 1.781
fatty acid synthase Fasn 6.323 2.24
fatty acid binding protein 4, adipocyte Fabp4 2.189 1.81
monoglyceride lipase Mgll 1.907 1.51
acetyl-Coenzyme A acetyltransferase 2 Acat2 1.625 1.563
acetyl-Coenzyme A dehydrogenase, long-chain Acadl 2.549 1.992
resistin Retn 4.114 1.756
malonyl-CoA decarboxylase Mlycd 1.781 1.962
transketolase Tkt 2.256 1.983
ATP citrate lyase Acly 2.458 1.91
HEAT SHOCK
heat shock protein 90 kDa alpha (cytosolic), class A member 1 Hsp90aal 1.455
0.616
DnaJ (Hsp40) homolog, subfamily B, member 1 Dnajbl 3.59 0.604
CYTOKINES
interferon, alpha-inducible protein 27 Ifi27 1.714 1.537
OTHER
sarcolipin Sln 0.363 4.576
thyroid hormone responsive SPOT14 homolog (Rattus) Thrsp 2.685 1.766
RIKEN cDNA 2310076L09 gene 2310076L09Rik 1.868 2.117
myosin, heavy polypeptide 2, skeletal muscle, adult Myh2 2.194 1.797
surfeit gene 4 Surf4 2.091 0.654
73
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
TABLE 4-continued
Targets common to exercise-PPARS and AMPK-PPAR6 gene signatures.
DESCRIPTION LOCUS Tr + GW AI + GW
acid phosphatase 5, tartrate resistant AcpS 3.91 1.477
serine (or cysteine) proteinase inhibitor, Glade A, member la Serpinala 0.396
3.891
cysteine dioxygenase 1, cytosolic Cdol 3.266 1.678
erythroid differentiation regulator 1 0.619 1.805
RIKEN cDNA 1810073N04 gene 1810073N04R1k 2.326 1.628
superoxide dismutase 3, extracellular Soda 1.606 1.617
complement factor D (adipsin) Cfd 2.828 1.5
cytochrome P450, family 2, subfamily e, polypeptide 1 Cyp2el 2.941 1.743
catalase cat 1.728 1.902
early growth response 1 Egrl 2.577 0.65
OXYGEN CARRIER
hemoglobin, beta adult minor chain hemoglobin Y, beta- Hbb-b2 lHbb-y 1.626
1.503
like embryonic chain
STEROID BIOGENESIS
retinol binding protein 4, plasma Rbp4 2.065 2.225
SIGNAL TRANSDUCTION
adreneyrgic receptor, beta 3 Adrb3 3.83 1.56
protein tyrosine phosphatase-like (proline instead of catalytic Ptplb 2.38
1.569
arginine), member b
dual specificity phosphatase 7 Dusp7 1.661 1.672
TRANSCRIPTION
nuclear receptor subfamily 4, group A, member 2 Nr4a2 1.776 0.437
TRANSPORT
solute carrier family 1 (neutral amino acid transporter), SlclaS 1.939 1.511
member 5
two pore channel 1 Tpcnl 2.842 1.487
seminal vesicle secretion 5 SvsS 0.095 2.243
Data is average of N=3 samples in each group (p<0.05).
[00175] Quantitative expression analysis of selective oxidative
genes (eight of those listed in Table 4) was determined in
quadriceps of mice treated with vehicle (V), GW 1516 (GW,
5mg/kg/day), AICAR (AI, 250 mg/kg/day) and the combination of the
two drugs (GW+AI) for 6 days using the methods described in Example
1. As shown in FIGS. 9A-H, several of these biomarkers including
PDK4, SCD1, ATP citrate lyase, HSL, mFABP and LPL were induced in a
synergistic fashion by GW1516 and AICAR in the quadriceps (FIGS.
9C-9H). Intriguingly, synergism was undetectable in UCP3 and mCPT I
(FIGS. 9A and B). These genes were induced in quadriceps of
74
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
untrained VP16-PPAR5 mice, where AMPK is constitutively active
(Table 5).
TABLE 5
Selective oxidative genes induced in muscle by combined
PPARO and AMPK activation as well as
VP16-PPAR6 over-expression
Description Locus CJ + Al VP-PPAR6
ATP citrate lyase Acly 1.648 3.095
carnitine palmitoyltransferase 1b, Cptlb 1.371 1.678
muscle
fatty acid binding protein 3, muscle Fabp3 1.447 5.904
and heart
fatty acid synthase Fasn 2.24 2.749
lipoprotein lipase Lpl 1.113 1.72
lipase, hormone sensitive Lipe 1.746 2.203
pyruvate dehydrogenase kinase, Pdk4 2.486 5.06
Coenzyme 4
stearoyl-Coenzyme A desaturase 1 Scdl 1.78 7.353
uncoupling protein 3 Ucp3 2.792 4.107
[00176] Collectively, these results demonstrate that while
interaction between AMPK and PPAR5 may substantially contribute to
re-programming of the skeletal muscle transcriptome during
exercise, additional changes may involve cross-talk between other
components of the exercise signaling network and PPAR5. In summary,
PPAR5 and exercise synergistically regulate running endurance.
Although not bound by theory, kinase activation may influence PPAR5
signaling during exercise in establishing an "endurance gene
expression signature" that effectively enhances performance.
Example 7
[00177] AMPK increases transcription activation by PPAR5. The
genetic synergism described in Example 6 indicates that AMPK
directly regulates the transcriptional activity of PPAR6 in
skeletal muscles. To demonstrate this, an analysis of the effects
of GW1516 and AICAR on gene expression in primary muscle cells
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
isolated from wild type and PPAR6 null mice was performed. Primary
muscle cells were isolated from wild type and PPAR5 null mice as
previously described (Rando and Blau, J. Cell. Biol. 125(6): 1275-
87, 1994). Skeletal muscle C2C12 cells were cultured in DMEM
containing 20% serum and penicillin/streptomycin cocktail. For
differentiation, cells at 80% confluence were switched to a
differentiation medium (DMEM + 2% serum) for 4 days to obtain
differentiated myotubules. Cells were treated with vehicle, GW1516,
AICAR, or GW1516 + AICAR (GW: 0.1 pM; AICAR: 500 pM) for 24 hours.
RNA expression of UCP3, PDK4, LPL, and HSL was determined using
real time quantitative PCR as described in Example 1. As shown in
FIGS. 1OA-D, synergism is dependent on PPAR5 and lost in the null
cells. Similar synergistic regulation of gene expression by GW1516
and AICAR was also observed in differentiated C2C12 cells. These
results show that AMPK activation may enhance ligand-dependent
transcriptional effects of PPAR6 in muscles. To more directly
address this, reporter gene expression assays were utilized.
[00178] AD 293 cells were cultured in DMEM containing 10% serum
and an antibiotic cocktail. Cells were transfected with one or more
of CMX-Flag, CMX-Flag PPAR5, CMX-Tk-PPRE, or CMX-(3GAL, or an hAMPK
(al and a2 subunits, Origene) expression vector using
Lipofectamin&M 2000 in accordance with the manufacturer's
instructions. Anti-Flag antibody-conjugated beads were incubated
overnight at 40C with lysates from transfected cells. Flag-tagged
protein or protein complexes were immunoprecipitated by separating
the beads from non-bound materials. The beads were washed in ice-
cold lysis buffer followed by extraction in Laemmli buffer. For co-
immunoprecipitation experiments SDS was excluded from the lysis
buffer. Western blotting was performed with antibodies specific for
the Flag tag or AMPK a subunit(s).
[00179] Co-transfection of either catalytic AMPK al or cc2
subunits, but not control vector, with PPAR5 increased the basal
(FIG. 10E) and GW1516-dependent transcriptional activity (FIG. 10F)
of PPAR5 in inducing a PPRE-driven reporter gene in AD293 cells.
AMPK over-expression or GW1516 treatment did not change reporter
activity in transfections excluding the PPAR6 expression vector
76
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
negating the possibility of an effect via RXR. Additional results
indicate that AMPK may modulate PPARb transcriptional activity by
directly interacting with the receptor. In AD293 cells co-
transfected with Flag-PPARb and with either catalytic AMPK al or a2
subunits, both of the subunits co-immunoprecipitated as a complex
with Flag- PPARb (FIG. 10G). Furthermore, Flag-PPARb also cc-
immunoprecipitated endogenous AMPKa subunits from AD293 cells
confirming a direct physical interaction between the nuclear
receptor and the kinase (FIG. 10H). Despite physical interaction,
AMPK failed to increase PPAR5 phosphorylation.
[00180] While potential AMPK phosphorylation sites were found in
PPARb, none of these sites were phosphorylated by AMPK in in vitro
kinase assays. This was further confirmed by measuring the p32
labeling of PPARb in AD 293 cells in the presence or absence of
AMPK. AD 293 cells were transfected with PPARb and hAMPk (al or a2
subunit) expression vectors as described above. Forty-eight hours
after transfection, the cells were washed three times with
phosphate-free DMEM and incubated with 32P-orthophosphate in
phosphate-free DMEM for 20 hours (100 pCi/5 ml). Cells were washed
three times with ice-cold phosphate-free DMEM and lysed in ice-cold
lysis buffer.
[00181] As shown in FIG. 101, overall PPARb phosphorylation is
not increased by AMPK in vivo. However, co-transfection of AMPKcc2
and co-activator PGCla (a known phosphorylation target of AMPK) co-
operatively interact to further induce both the basal and ligand-
dependent transcriptional activity of PPARb (FIG. 10J). Strikingly,
no significant physical interaction between Flag-PGCla and AMPK
(FIG. 10K) was detected, both of which independently interacted
with PPARb. Collectively, these observations indicate that AMPK may
be present in a transcriptional complex with PPARb where it can
potentiate receptor activity via direct protein-protein interaction
and/or by phosphorylating co-activators such as PGCla.
[00182] These results indicate that AMPK directly interacts with
PPARb and dramatically increases basal and ligand-dependent
transcription via the receptor. Despite physical interaction, AMPK
does not phosphorylate PPARb. AMPK and its substrate PGCla
77
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
synergistically increased PPAR5 transcription, indicating indirect
regulation of receptor by AMPK via co-regulator modification.
[00183] The conclusion that exercise-activated AMPK interacts
with PPARS in regulating gene expression in vivo is strengthened by
the observation that treatment of animals with AICAR (AMPK
activator) and GW1516 creates a gene signature in skeletal muscle
that replicates up to 40% of the genetic effects of combined
exercise and GW1516 treatment (see Table 4). Moreover, several
candidate genes from this signature are synergistically induced by
GW1516 and AICAR in wild type but not in PPAR5 null primary muscle
cells, demonstrating that the interactive effects of the two drugs
are mediated through PPAR5. While 45% of the commonly regulated
genes are linked to oxidative metabolism, additional common targets
relevant to muscle performance include angiogenic, signal
transduction and glucose sparing genes (Table 4). It is possible
that the portion of the PPAR6-exercise signature that is
independent of PPAR5-AMPK interaction (FIG. 8B) may depend on
cross-talk between the receptor and other exercise signal
transducers such as MAPK, calcineurin/NFAT and SIRT 1. These
possibilities are summarized in FIG. 10L, where AMPK and additional
components of the signaling network are proposed to interact with
liganded PPAR5 to generate a muscle endurance gene signature and
enhanced endurance adaptation.
[00184] The data show that synthetic PPAR5 activation alone
induces a set of genomic changes that fail to alter the preset
muscle architecture and endurance levels in adult mice. However,
the combination of PPAR6 activation with exercise brings about
novel transcriptional changes, potentially via interaction with
kinases such as AMPK (as depicted in FIG. 10L), re-setting the
muscle transcriptome to a phenotype that dramatically enhances
muscle performance.
Example 8
[00185] The data demonstrate that pharmacologic activation of
PPAR5 in adult mice can increase running endurance in conjunction
with exercise signals. The central role for AMPK in this process is
especially underscored by the observations that it is both robustly
78
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
stimulated by exercise as well as constitutively active in muscles
of VP16-PPARd transgenic mice that exhibit endurance without
exercise. Further, AMPK can integrate multiple transcriptional
programs by interacting not only with PPAR5 but also other
transcriptional regulators of metabolism (e.g., PGClo:, PPARa) (Hong
et al., 2003; Leff, 2003; Bronner et al., 2004; Jaager et al.,
2007). This raises the interesting question as to whether chemical
activation of AMPK is sufficient to increase running endurance
without exercise.
[00186] To test this idea C57B/6J mice were treated with AICAR
(500 mg/kg/day) for 4 weeks. AICAR increased phosphorylation of
AMPK a subunit and acetyl CoA carboxylase (ACC) and increased
expression of UCP3 in quadriceps, confirming effective activation
of AMPK signaling (Figure 11A). Interestingly, 4 weeks of drug
treatment decreased the ratio of epididymal fat mass to body weight
and increased oxygen consumption without changing body weight
(Figures 11B-11E), supporting the speculation that AICAR may
positively regulate endurance. Indeed, in a treadmill endurance
test, ACIAR-treated mice ran longer (230) and further (440) than
did vehicle-treated mice, revealing that increase in endurance can
be achieved without exercise (Figure 11F). Furthermore, global gene
expression analysis of quadriceps revealed that AICAR treatment
alone upregulated a set of 32 genes linked to oxidative metabolism
(Figure 11G and Table 6). Notably, 30 of these 32 genes were also
upregulated in VP16- PPAR5 transgenic mice, suggesting that
stimulation of oxidative genes by AMPK may depend on PPAR6 (Table
6).
Table 6: Oxidative genes induced by AMPK agonist AICAR as well as
by transgenic over-expression of VPl6-PPARbin quadriceps of
untrained mice. Data is average of N=3 mice in each group (p<0.5).
79
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
Locus Description AICAR VP-PPARd
A acs _ac:~o cet,l Co. sti hen -e ` 75 2 771
Achy ATP citrate lease 1s 8^ . ' `
M pac 122
.2 3de ~'Iat l ina:se t ;S 1173
"Poe E - 1,403 959
(A36 CD 3S antigen w,6 3.66
Ce 3 s ;'Ã ox l{) Sti lii[:31:: 3
eryr1.S 51 1.85`.?
e12 41 atll .i duciit~~ DN 1 Llayilientatiol
Cidea f'.Ictor, alpha Rislunit-1 ke e `fec:tos A 5.021 56
D"2ai:` 11.~t ylyceroi C #- 34~ 1 i'ans '.fS 2 034 147
Elo=rl6 ELOVL family member 5 2.134 4.43
Fa.bp4 fatty ^acit Ã,intlilla protein idipocyte 1 779# 1.:3
a.sn fifty ei w,itlla e 9$ 'L 49
Lop 1, _)ti -t 047 70
? ` 3
Lie lipase, 1,64
I. l li oprateiii 1ipzl3e 8 i 185
X12.11 1&ion LTI~'ce i.de lipase 503 2.. ' 15
Mgst,1 ]I icro maJ gll tathioli4 ti'illy~ei"i3 1 ?02 l.9 05
\riol uucle aI rec tt?I' is eractin protein 1 I 7, 4 1.8`7
Pcl zl3t}~ hoeriC);'t4'T'Fii ate Li ll)i~;4 r' T 6' 169
PCx Pvate caiboxyÃase 93 -
Pdk4 `rp ovate fl.hydi'ogenase X11` se., iioei3z role 4 .21 5,'t)5
pi:SreFsom 1 oli e#atry.';ac5v9ted1ei:.eptor
2,334
Retn reS'lstill 1.z 51 L i'541
1 REF' rco 1- oei z`:3T A desaturase 1 11, P. i 1 353
-hd ^uu.4cinZtc fileÃ1y' rogefnase comilplex. su~_fliiit: D ?4} 1.'4 `
SEm 11I uitles~itle, .: 1 13 1.494
SIc?S3 L 1.348
Stilt li4i sfF.li i 3nsfe õ=e family is i 2:121 1,996
Tspo t&':#E'is1o '..z?3' rot in 1.4441 _21 33
Uc ].1 unconp1i:1 , protein I i 7 Lulcoup1in. protein 3 11_25,S 4.107
[00187] To test this possibility, wild-type and PPAR5 null
primary muscle cells were used. Treatment of wild-type primary
cells with AICAR (for 72 hr) increased expression of key oxidative
biomarker genes (Scdl, fasn [FAS], Ppargc la, Pdk4) (Figure 11H).
In contrast, AICAR failed to increase the expression of the above
genes in PPAR5 null cells, demonstrating the requirement of the
receptor for transcriptional effects of AMPK on oxidative genes.
[00188] The data show that the AMP-mimetic AICAR can increase
endurance in sedentary mice by genetically reprogramming muscle
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
metabolism in a PPAR6-dependent manner. The data also demonstrate
that PPAR6 agonist in combination with exercise synergistically
induces fatigue resistant type I fiber specification and
mitochondrial biogenesis, ultimately enhancing physical
performance. These changes correlate with an unexpected but
interesting establishment of a muscle endurance gene signature that
is unique to the drug-exercise paradigm. Such a signature is an
outcome of molecular crosstalk and perhaps a physical association
between exercise-activated AMPK and PPAR5. These findings identify
a novel pharmacologic strategy to reprogram muscle endurance by
targeting AMPK- PPAR6 signaling axis with orally active ligands.
[00189] The AMPK activator AICAR increased oxygen consumption
and endurance in untrained adult mice in part by stimulating PPAR6-
dependent oxidative genes. Despite a demonstrated role for PPAR5 in
endurance, 4 week treatment with a potent and selective agonist
failed to alter either fiber type composition or endurance,
revealing that direct and pharmacologic activation of PPAR5 is
insufficient to enhance running performance. In contrast,
transgenic overexpression of activated PPAR5 at birth preprograms
the nascent myofibers to transdifferentiate into slow-twitch
fibers, thus imparting a high basal endurance capacity to adult
transgenic mice. Apparently, once fiber type specification is
complete in adults, the potential plasticity of muscle to synthetic
activation of a single transcriptional pathway is constrained.
[00190] Along these lines, the unexpected yet successful
reprogramming of endurance in untrained adults with synthetic AMP-
mimetic might be linked to the ability of AMPK to simultaneously
target multiple transcriptional programs governed by its substrates
such as PGCla, PPARa and PPAR5, triggering a genetic effect akin to
exercise (Hong et al., 2003; Leff, 2003; Bronner et al., 2004;
Jager et al., 2007).
[00191] Interestingly, the recalcitrance of adult skeletal
muscle endurance to manipulation by PPAR6 agonist alone is relieved
by combining drug treatment with exercise. Indeed, this strategy
generates an endurance gene signature that is unique from either
paradigm alone, reflecting a crosstalk between exercise and PPAR6
81
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
signaling. Although exercise activates a cascade of signaling
events, AMPK is likely central to this genetic adaptation for
several reasons. First, AMPK is a metabolic sensor that detects low
ATP levels (such as occur during exercise) and in turn increases
oxidative metabolism (Mu et al., 2001; Reznick and Shulman, 2006).
Second, long-term effects of AMPK are in part mediated via
regulation of gene expression (Reznick and Shulman, 2006). Third,
exercise induces activation and nuclear import of AMPK, where it
can potentially interact with transcription factors.
[00192] And finally, transgenic mice defective for AMPK
activation exhibit reduced voluntary exercise (Mu et al., 2001;
Thomson et al., 2007), making it an attractive exercise cue that
modulates receptor signaling.
[00193] The notion that exercise-activated AMPK interacts with
PPAR5 in regulating gene expression is supported by the
demonstration that AMPK associates with PPAR6 and dramatically
increases basal and ligand-dependent transcription via the
receptor. Despite physical interaction, AMPK does not induce PPAR6
phosphorylation in metabolic labeling studies.
[00194] Interestingly, AMPK and its previously reported
substrate PGCla synergistically increased PPAR6 transcription,
suggesting indirect regulation of receptor function by AMPK via
coregulator modification. Nevertheless, it could be a possible
regulation of PPAR5 by AMPK via direct protein-protein interaction.
[00195] Indeed, regulation of other transcription factors by
AMPK via similar mechanisms has been previously demonstrated (Hong
et al., 2003; Leff, 2003; Bronner et al., 2004). A physiological
validation of AMPK- PPAR6 interaction comes from the observation
that GW1516 and AICAR (AMPK activator) synergistically induce
several endurance-related genes in wild-type but not in PPARf null
primary muscle cells. More importantly, treatment of animals with
AICAR and GW1516 creates a gene signature in skeletal muscle that
replicates up to 40% of the genetic effects of combined exercise
and GW1516 treatment. Notably, the shared genes between the two
profiles are linked to oxidative metabolism, angiogenesis, and
82
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
glucose sparing, pathways that are directly relevant to muscle
performance.
[00196] Although not all genes regulated by either exercise or
exercise- PPAR5 interaction are AMPK dependent, two key findings
assign a critical role for the kinase in promoting endurance
compared to other known exercise signals (Bassel-Duby and Olson,
2006; Goodyear et al., 1996; Lagouge et al., 2006). First, AMPK is
constitutively active in VP16- PPAR6 transgenic muscles that
exhibit endurance without exercise. Second, AMPK activation by
AICAR was sufficient to increase running endurance without
additional exercise signals. Strikingly, the majority of the
oxidative genes (30 out of 32) upregulated by AICAR are active in
super-endurance VP16-PPARd mice and perhaps are the core set of
genes required to improve muscle performance.
[00197] Interestingly, AICAR failed to induce oxidative gene
expression in PPAR6 null muscle cells, indicting the requirement of
PPAR5, at least for regulation of oxidative metabolism by AMPK.
Collectively, these findings demonstrate a molecular partnership
between AMPK and PPAR6 in reprogramming skeletal muscle
transcriptome and endurance (Figure 11I) that can be readily
exploited by orally active AMPK drugs to replace exercise.
[00198] In humans, endurance exercise leads to physiological
adaptations in the cardiopulmonary, endocrine, and neuromuscular
systems (Jones and Carter, 2000; Lucia et al., 2001). Although the
investigation focused on skeletal muscle, extramuscular effects of
PPARd, AMPK, and exercise may also contribute to increased
endurance. Although potentiation of extramuscular adaptations by
PPARd and AMPK agonists remains to be studied, drug treatment can
reduce epididymal fat mass, possibly conferring additional systemic
benefits. It is noteworthy that PPAR5 is important for normal
cardiac contractility, as well as for the endocrine function of
adipose tissue (Wang et al., 2003; Cheng et al., 2004). Similarly,
the activation of AMPK by metformin is thought to mediate its
ability to lower blood glucose levels (Shaw et al., 2005). In
addition to increasing performance in athletes, exercise has
beneficial effects in a wide range of pathophysiological
83
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
conditions, such as respiratory disorders, cardiovascular
abnormalities, type 2 diabetes, and cancer risk. The disclosure
demonstrates that synthetic PPAR6 activation and exercise-and, more
importantly, AMPK activation alone-provide a robust transcriptional
cue that reprograms the skeletal muscle genome and dramatically
enhances endurance. The disclosure provides a strategy for
reorganizing the preset genetic imprint of muscle (as well as other
tissues) with exercise mimetic drugs has therapeutic potential in
treating certain muscle diseases such as wasting and frailty as
well as obesity where exercise is known to be beneficial.
Example 9
[00199] Enhancing exercise effect in a subject. This example
describes methods that can be used to increase or enhance an
exercise in a healthy mammalian subject. Although specific
conditions are described, one skilled in the art will appreciate
that minor changes can be made to such conditions.
[00200] Healthy adult human subjects perform aerobic exercise
(e.g., running) for at least 30 minutes (e.g., 30-90 minutes) for
at least 3-4 days per week (e.g., 3-7 days per week) for at least 2
weeks (e.g., at least 4-12 weeks). The exercise is performed at
400-50% maximal heart rate, 500-60% maximal heart rate, 601-70%
maximal heart rate, or 75%-80% maximal heart rate, where maximum
heart rate for a human subject is calculated as: 220 bps - (age of
the subject).
[00201] During or after performing aerobic exercise as described
above, the subjects are orally administered GW1516 [(2-methyl-
4(((4-methyl-2-(4- trifluoromethylphenyl)-1,3-thiazol-5-
yl)methyl)sulfanyl)phenoxy)acetic acid] at a dose of 1 to 20 mg per
day, such as 2.5 or 10 mg per day. Subjects can continue to perform
aerobic exercise while receiving GW1516. The subject can receive
GW1516 for a period of at least 2 weeks, such as at least 4 weeks.
[00202] The exercise effect achieved in the treated subjects
(e.g., running endurance) can be compared to such an effect in
untreated subjects. Exercise effect can be measured using methods
known in the art, such as measuring aerobic or running endurance
(for example measuring distance run until exhaustion or amount of
84
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
time to run a particular distance). In some instances, the exercise
effect of interest is increased in treated subjects by at least 5 ,
such as at least 10% as compared to untreated subjects.
Example 10
[00203] Identifying performance enhancing substances in an
exercise-trained subject. This example describes methods that can
be used to identify performance- enhancing substances in an
exercised-trained subject. A biological sample obtained from a
healthy adult human is analyzed to determine if the subject is
taking a PES (e.g., GW1516) by analyzing expression of one or more
of the molecules (nucleic acids or proteins) listed in Table 2 or
Table 4. Suitable biological samples include samples containing
genomic DNA or RNA (including mRNA) or proteins obtained from cells
of a subject, such as those present in peripheral blood, urine,
saliva, tissue biopsy, or buccal swab. For example, a biological
sample of the subject can be assayed for a change in expression
(such as an increase or decrease) of any combination of at least
four molecules (nucleic acids or proteins) listed in Table 2 or 4,
such as any combination of at least 10, at least 20, at least 30,
or at least 40 of those listed in Table 2 or 4, for example all of
those listed in Table 2 or 4.
[00204] Methods of isolating nucleic acid molecules from a
biological sample are routine, for example using PCR to amplify the
molecules from the sample, or by using a commercially available kit
to isolate mRNA or cDNA. However, nucleic acids need not be
isolated prior to analysis. Nucleic acids can be contacted with an
oligonucleotide probe that will hybridize under stringent
conditions with one or more nucleic acid molecule listed in Table 2
or 4. The nucleic acids which hybridize with the probe are then
detected and quantified. The sequence of the oligonucleotide probe
can bind specifically to a nucleic acid molecule represented by the
sequences listed in Table 2 or 4.
[00205] Increased or decreased expression of the molecules
listed in Table 2 or 4 can be detected by measuring the cellular
levels of mRNA. mRNA can be measured using techniques well known in
the art, including for instance Northern analysis, RT-PCR and mRNA
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
in situ hybridization. Details of mRNA analysis procedures can be
found, for instance, in provided examples and in Sambrook et al.
(ed.), Molecular Cloning: A Laboratory Manual, 2nd ed., vol. 1-3,
Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 1989.
[00206] Oligonucleotides specific to sequences listed in Table 2
or 4 can be chemically synthesized using commercially available
machines. These oligonucleotides can then be labeled, for example
with radioactive isotopes (such as 32P) or with non-radioactive
labels such as biotin (Ward and Langer et al., Proc. Natl. Acad.
Sci. USA 78:6633-57, 1981) or a fluorophore, and hybridized to
individual DNA samples immobilized on membranes or other solid
supports by dot- blot or transfer from gels after electrophoresis.
These specific sequences are visualized, for example by methods
such as autoradiography or fluorometric (Landegren et al., Science
242:229 '-37 ', 1989) or colorimetric reactions (Gebeyehu et al.,
Nucleic Acids Res. 15:4513-34, 1987).
[00207] Analyzing Proteins in the biological sample can also be
analyzed. In some examples, proteins are isolated using routine
methods prior to analysis.
[00208] In one example, surface-enhanced laser desorption-
ionization time-of-flight (SELDI-TOF) mass spectrometry is used to
detect changes in differential protein expression, for example by
using the ProteinChipTM (Ciphergen Biosystems, Palo Alto, CA). Such
methods are well known in the art (for example see U.S. Pat. No.
5,719,060; U.S. Pat. No. 6,897,072; and U.S. Pat. No. 6,881,586).
SELDI is a solid phase method for desorption in which the analyte
is presented to the energy stream on a surface that enhances
analyte capture or desorption. Therefore, in a particular example,
the chromatographic surface includes antibodies that recognize
proteins listed in Table 2 or 4. Antigens present in the sample can
recognize the antibodies on the chromatographic surface. The
unbound proteins and mass spectrometric interfering compounds are
washed away and the proteins that are retained on the chromato
graphic surface are analyzed and detected by SELDI-TOF. The MS
profile from the sample can be then compared using differential
protein expression mapping, whereby relative expression levels of
86
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
proteins at specific molecular weights are compared by a variety of
statistical techniques and bioinformatic software systems.
[00209] In another examples, the availability of antibodies
specific to the molecules listed in Table 2 or 4 facilitates the
detection and quantification of proteins by one of a number of
immunoassay methods that are well known in the art, such as those
presented in Harlow and Lane (Antibodies, A Laboratory Manual,
CSHL, New York, 1988). Methods of constructing such antibodies are
known in the art. Any standard immunoassay format (such as ELISA,
Western blot, or RIA assay) can be used to measure protein levels.
Immunohistochemical techniques can also be utilized for protein
detection and quantification. For example, a tissue sample can be
obtained from a subject, and a section stained for the presence of
the desired protein using the appropriate specific binding agents
and any standard detection system (such as one that includes a
secondary antibody conjugated to horseradish peroxidase). General
guidance regarding such techniques can be found in Bancroft and
Stevens (Theory and Practice of Histological Techniques, Churchill
Livingstone, 1982) and Ausubel et al. (Current Protocols in
Molecular Biology, John Wiley & Sons, New York, 1998).
[00210] For the purposes of detecting or even quantifying
protein or nucleic acid expression, expression in the test sample
can be compared to levels found in cells from a subject who has not
taken a PES. Alternatively, the pattern of expression identified in
the test subject can be compared to that shown in Table 2 or 4.
[00211] For example, if the test sample shows a pattern of
expression similar to that in Table 2 or 4 (e.g., the genes shown
as upregulated and downregulated in Table 2 or 4 are observed in
the subject to be upregulated and downregulated, respectively),
this indicates that the subject is taking a PES, such as a PPAR5
agonist (e.g., GW1516). In contrast, If the pattern of expression
identified in the test subject is different to that shown in Table
2 or 4 (e.g., the genes shown as upregulated and downregulated in
Table 2 or 4 are observed in the subject to be not differentially
expressed or show a different pattern of regulation), this
87
CA 02710764 2010-06-23
WO 2009/086526 PCT/US2008/088466
indicates that the subject is not taking a PES, such as a PPAR6
agonist (e.g., GW1516).
[00212] A significant increase in the non-bolded proteins listed
in Table 2 in the cells of a test subject compared to the amount of
the same protein found in normal human cells is usually at least 2-
fold, at least 3-fold, at least 4-fold or greater difference.
Substantial overexpression of the non-bolded proteins listed in
Table 2 in the subject's sample can be indicative of the subject
taking a PES. Similarly, a significant decrease in the bolded
proteins listed in Table 2 in the cells of a test subject compared
to the amount of the same protein found in normal human cells is
usually at least 2-fold, at least 3-fold, at least 4-fold or
greater difference.
[00213] Substantial underexpression of the bolded proteins
listed in Table 2 in the subject's sample can be indicative of the
subject taking a PES.
[00214] While this disclosure has been described with an
emphasis upon particular embodiments, it will be obvious to those
of ordinary skill in the art that variations of the particular
embodiments may be used and it is intended that the disclosure may
be practiced otherwise than as specifically described herein.
Accordingly, this disclosure includes all modifications encompassed
within the spirit and scope of the disclosure as defined by the
following claims:
88