Note: Descriptions are shown in the official language in which they were submitted.
CA 02728736 2010-12-20
WO 2009/155505 PCT/US2009/047937
MICROVESICLE-BASED COMPOSITIONS AND METHODS
[0001] This application claims priority to our US provisional application with
the serial
number 61/074,218, filed June 20, 2008, which is incorporated by reference
herein.
Field of The Invention
[0002] The field of the invention is detection and/or analysis of RNA in
microvesicles and
their use in diagnosis, prognosis, and/or research of diseases and other
conditions.
Background of the Invention
[0003] Microvesicles were historically regarded as cellular debris with no
apparent function.
However, a growing body of experimental data has suggested that microvesicles
have
numerous biological activities. For example, platelet-derived microvesicles
were shown to
stimulate selected cells via surface proteins on the microvesicles (e.g.,
Thromb. Haemost.
(1999), 82:794, or J. Biol. Chem. (1999), 274:7545). In other examples,
specific effects of
bioactive lipids in platelet microvesicles on certain target cells were
reported (e.g., J. Biol.
Chem. (2001), 276: 19672; or Cardiovasc. Res. (2001), 49(5):88). In still
further examples,
platelet microvesicles increased adhesion of mobilized CD34+ endothelial cells
by transfer of
certain microvesicle surface components to the mobilized cells (e.g., Blood
(2001), 89:3143).
[0004] More recently, microvesicles have also been shown to comprise RNA that
at least in
part appeared to reflect the RNA content of the cell from which they
originate. Microvesicles
have also been shown to have significant biological effect on other cells,
probably due to the
RNA present in the microvesicles, and various examples and aspects for such
microvesicles
are described in our commonly owned International application (W02005/121369),
which
expressly forms part of this application.
[0005] In further known reports, microvesicles were described as including non-
coding
miRNA (microRNA) that could potentially interfere or regulate gene expression
in cells with
which such microvesicles merge (PLoS November 2008, Vol. 3(l 1), e3694). Other
reports
discuss in vitro cell-to-cell signaling via exosomal RNA (Cell Adh Migr 1:3,
156-158; 2007;
Cancer Immunol Immunother 2006 Jul;55(7):808-18; Blood. 2007 Oct 1;110(7):2440-
8.). It
was also shown that while some exosomal RNA was functional and translatable in
a recipient
cell, many of the RNA molecules present in the exosomes were not present in
the cytoplasm
of cells from which the exosomes were though to have originated (Nat Cell
Biol. 2007
1
CA 02728736 2010-12-20
WO 2009/155505 PCT/US2009/047937
Jun;9(6):654-9). U.S. Pat. No. 6,916,634 teaches that while RNA is generally
instable in
serum and readily hydrolyzed by RNAses, other RNA is resistant to RNAse
attack,
presumably due to its varying association with circulating particles.
Remarkably, the '634
patent elaborates on the chemically and structurally highly diverse nature of
the RNA
associated particles (presumably due to their diverse origin and manner of
generation) and
thus concludes that serum RNA is best isolated in an indiscriminate manner.
Thus, even
though membrane associated or vesiculated RNA has more recently been reported,
there is a
large body of contradictory data and hypotheses with respect to the nature,
quality,
availability, and origin/manner of generation of microvesicles.
[0006] Consequently, the enormous diagnostic potential of RNA-containing
microvesicles
has not been fully recognized in the art, and there is still a need for
microvesicle-based
diagnostic compositions and methods in which RNA from microvesicles is
enriched and
differentiated to so obtain a result that is indicative of the condition of
tissue or organ from
which the microvesicle originated.
Summary of the Invention
[0007] According to the present inventive subject matter, microvesicles are
employed in
various diagnostic, prognostic, and/or analytic compositions and methods in
which specific
RNA content of microvesicles and optionally at least one more additional
information
bearing component of the microvesicle are used to obtain cell-, tissue-, organ-
, and/or
disease-specific information.
[0008] In one especially preferred aspect of the inventive subject matter,
method of analyzing
a biological sample (e.g., plasma or serum) of a mammal in which microvesicles
are obtained
from a living donor mammal (preferably human) that include a plurality of
distinct RNA
molecules. The microvesicles are then enriched and differentiated to produce a
primary result
based on one or more distinct RNA molecules, a secondary result based on at
least two
distinct RNA molecules, and/or a ternary result based on sub-segregated
proxysomes and one
or more distinct RNA molecules. Of course, it should be appreciated that the
RNA analysis
in the differentiation may include analysis of a single RNA, of at least two
distinct RNAs,
and in some aspects even an entire RNA profile (typically obtained by analysis
of an array of
multiple distinct DNA or RNA). The so obtained results are then correlated
with a diagnosis
(e.g., cancer or a clinical stage of a cancer, including pre-cancerous stages)
or
prognosis/diagnosis of a condition of the mammal.
2
CA 02728736 2010-12-20
WO 2009/155505 PCT/US2009/047937
[0009] Most typically, the distinct RNA molecule(s) in the primary result
is/are RNA that is
overexpressed, underexpressed, and/or mutated, wherein the change in
expression and/or the
mutation is characteristic for the condition. Similarly, one of the two
distinct RNA molecules
in the secondary result is an RNA that are overexpressed, underexpressed,
and/or mutated,
wherein the change in expression and/or the mutation is characteristic for the
condition, while
the other of the distinct RNA molecules in the secondary result is an RNA that
is uniquely
expressed in a specific tissue or organ of the mammal. The sub-segregated
proxysome
population is preferably obtained by isolating the proxysome population based
on a surface
molecule specific for origin of the proxysome population, and the distinct RNA
molecule in
the ternary result is RNA that is overexpressed, underexpressed, and/or
mutated, wherein the
change in expression and/or the mutation is characteristic for the condition.
[0010] With respect to the RNA that is overexpressed, underexpressed, and/or
mutated, it is
typically preferred that the RNA encodes MMP11, BCAR1, ERBB2, MK167, PLAU,
and/or
TP53 where the condition is breast cancer; or encodes FGFR1, KRAS2, TGFBR2,
MAP2K4,
and/or CDKN2A where the condition is pancreatic cancer; or encodes KLK3,
ERBB2, FGF8,
PSCA, and/or CAVlwhere the condition is prostate cancer; or encodes BAX,
SLC2A1,
PTGS2, MUC1, and/or RUNX3 where the condition is gastric cancer; or encodes
BCL10,
PAP, SPARC, CD44, and/or TP53 where the condition is liver cancer; or encodes
a stem cell
marker, and especially an adult stem cell marker (e.g., CD33, CD44, CXCR4,
CXCR4+, lin-,
CD45-, Oct-4, Nanog, SCAT, 7-AAD), where the condition is a cancer.
[0011] With respect to the RNA that is uniquely expressed in the specific
tissue or organ it is
preferred that the RNA encodes DCD, SCGB2A2, and/or ANKRD30A where the
specific
tissue or organ is breast tissue; or encodes UCN3, IPF1, and/or REG1B where
the specific
tissue or organ is a pancreas; or encodes UPK3A, SEMG 1, and/or PRAC where the
specific
tissue or organ is prostate tissue; or encodes GAST, GKN1, and/or TFF2 where
the specific
tissue or organ is gastric tissue; or encodes GYS2, F9, and/or HRG where the
specific tissue
or organ is a liver.
[0012] It is further generally preferred that the microvesicles are enriched
via aggregation by
interlinking the microvesicles with an interlinking composition (e.g., annexin
V, fibrin, or an
antibody or antibody fragment against a tetraspanin, ICAM-1 1, or CD86).
Consequently,
microvesicles especially contemplated herein have a membrane composition such
that
phosphatidylserine is on the outside of the membrane. It is further generally
preferred that
3
CA 02728736 2010-12-20
WO 2009/155505 PCT/US2009/047937
the primary or secondary result are obtained using a quantitative rtPCR for
the distinct RNA
molecule or via microarray technology.
[0013] Thus, viewed from a different perspective, a method of staging a
mammalian
neoplasm (which also includes staging of precancerous lesions or growth) will
include a step
of obtaining a whole blood fraction (e.g., serum or plasma) that includes a
plurality of
microvesicles comprising a plurality of distinct RNA molecules. In another
step, the
microvesicles are enriched (e.g., via centrifugation and/or aggregation) and
differentiated
(e.g., using quantitative rtPCR) to produce a primary result based on at least
one distinct
RNA molecule, a secondary result based on at least two distinct RNA molecules,
and/or a
ternary result based on a sub-segregated proxysome population and at least one
distinct RNA
molecule. The results are then correlated with a stage of the neoplasm in the
mammal,
typically via comparison with a reference result.
[0014] Among other neoplasms, especially contemplated neoplasms include acute
lymphoblastic leukemia, bladder cancer, breast carcinoma, cervical cancer,
colorectal cancer,
lung cancer, ovarian cancer, and pancreatic adenocarcinoma. Therefore, one of
the at least
two distinct RNA molecules in the secondary result or one of the distinct RNA
molecules in
the primary or ternary result will be an RNA that encodes ERBB2.
[0015] Various objects, features, aspects and advantages of the present
invention will become
more apparent from the following detailed description of preferred embodiments
of the
invention.
Brief Description of The Drawing
[0016] Figure 1 is a graph depicting the results for in vitro expression of
Her2-RNA in
selected cell lines.
[0017] Figure 2A is a photograph of an agarose gel with Her2-RNA amplification
products
of RNA from microvesicles that were isolated from culture supernatants of the
selected cell
lines.
[0018] Figure 2B is a graph depicting the quantitative difference of the
amplification
products of Figure 2A.
4
CA 02728736 2010-12-20
WO 2009/155505 PCT/US2009/047937
[0019] Figure 3A is a photograph of an agarose gel with Her2-RNA amplification
products
of RNA from microvesicles that were isolated from murine serum of mice that
developed
tumors derived from one of the selected cell lines.
[0020] Figure 2B is a graph depicting the quantitative difference of the
amplification
products of Figure 3A.
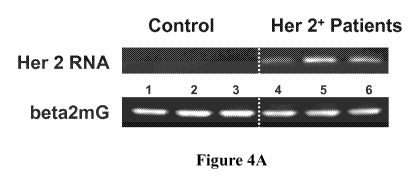
[0021] Figure 4A is a photograph of an agarose gel with Her2-RNA amplification
products
of RNA from microvesicles that were isolated from human sera of patients with
confirmed
breast cancer diagnosis.
[0022] Figure 4B is a graph depicting the quantitative difference of the
amplification
products of Figure 3A.
Detailed Description
[0023] The inventor has discovered that mammalian, and especially human
microvesicles can
be employed as a proxy diagnostic tool in the analysis and/or diagnosis of a
cell, tissue,
organ, or even multi-organ system where RNA and/or multiple non-RNA components
(e.g.,
soluble and/or membrane proteins, lipids, etc.) from the microvesicles are
correlated with
status and/or health of the cell, tissue, organ, or even multi-organ system
from which the
microvesicle originated. Based on the inventor's findings that the RNA content
of the
microvesicles is entirely or almost entirely representative of the cellular
RNA of the cell from
which the microvesicle originated, numerous uses are now envisioned. As
microvesicles are
shed by all cells in relatively high numbers, and as microvesicles are also
present in equally
high number in blood, the inventor concluded that, inter alia, gene
expression,
soluble/membrane proteins, and membrane composition of various cells, tissues,
and organs
can be easily determined by analyzing the corresponding microvesicles, where
those
microvesicles are obtained from a biological fluid, and especially blood.
[0024] The term "microvesicle" as used herein refers to a membranaceus
particle having a
diameter (or largest dimension where the particle is not spheroid) of between
about 10 nm to
about 5000 nm, more typically between 30 nm and 1000 nm, and most typically
between
about 50 nm and 750 nm, wherein at least part of the membrane of the
microvesicle is
directly obtained from a cell. Therefore, especially contemplated
microvesicles include those
that are shed from a donor cell, and will typically also include exosomes.
Therefore, and
CA 02728736 2010-12-20
WO 2009/155505 PCT/US2009/047937
depending on the manner of generation (e.g., membrane inversion, exocytosis,
shedding, or
budding), the microvesicles contemplated herein may exhibit different
surface/lipid
characteristics. Viewed from a different perspective, the microvesicles
suitable for use in the
present inventive subject matter may originate from cells membrane inversion,
exocytosis,
shedding, blebbing, or budding. Most typically, microvesicles suitable for use
herein will be
identifiable by having phosphatidylserine, a tetraspanin, ICAM-1, and/or CD86
on the outer
surface. In contrast, a liposome made from isolated lipids will not include
lipids or other
components obtained from a cell-membrane, and is not considered a microvesicle
under the
definition used herein. Therefore, most typically, contemplated microvesicles
will have a
lipid bilayer structure and are not multi-lamellar.
[0025] Moreover, it is preferred that the microvesicles are generated from a
differentiated
cell, and most preferably from a terminally differentiated cell (i.e., a cell
that has reached the
end of its differentiation pathway). Therefore, microvesicles generated by
blast cells,
progenitor cells, and stem cells are excluded in at least some of the
embodiments herein.
However, and especially where the microvesicles are generated from a cancer
cell, and where
that cancer cell is derived from an adult stem cell (as opposed to an
embryonic stem cell) less
differentiated cells are also deemed suitable sources for the microvesicles.
It is especially
preferred that the microvesicles are derived from cells that are diseased
(e.g., neoplastic cell,
infected cell, cell in an infected organ) or subject to an abnormal condition
(e.g., metabolic
abnormality, cell exposed to a drug, and more typically a drug that
preferentially affects the
cell relative to other cells, or dietary toxin, or damaged by free radicals).
In still further
contemplated aspects, the microvesicles are generated from a healthy, but
ageing cell, i.e., a
cell that has reached at least 50%, and more typically at least 70% of its
ordinary number of
cell divisions (Hayflick limit). It should be noted that synaptosomes
(vesicular entities in the
synaptic gap, but not present in the general circulation) and microvesicles
that are formed
from platelets are expressly excluded.
[0026] Thus, the RNA content of the microvesicles is thought to be
characteristic of a
particular condition (e.g., disease, age, stress, response to a chemical
compound, senescence,
inflammation, infection [e.g., viral, microbial, parasitic], immune status,
regeneration,
rejection, etc.). Non-RNA information bearing components in microvesicles
contemplated
herein include various proteins (e.g., receptor, ligand, glycoprotein, etc.)
that are associated
with the microvesicle, which may be at least partially embedded in the
membrane (or even be
6
CA 02728736 2010-12-20
WO 2009/155505 PCT/US2009/047937
entirely enclosed), specific lipids or glycolipids that are associated with
the microvesicle,
and/or a further nucleic acid (DNA and/or RNA) associated with the
microvesicle. Thus, and
especially where the non-RNA information bearing component is specific to a
particular
organ, tissue, and/or cell, analysis of the condition of the particular organ,
tissue, and/or cell
can be obtained in a highly simplified and even multiplexed manner.
Consequently, and
among other advantages, it should be especially appreciated that a serum or
blood based test
can be performed that provides (even multiple) organ specific results without
the need for an
otherwise required organ-specific biopsy. Still further, it should be noted
that the methods
contemplated herein will allow virtually unlimited and repeated real time
access to an
expression profile of the same tissue or organ.
[0027] Consequently, methods according to the inventive subject mater will
typically include
the following sequence (in which one or more steps may be combined):
Isolation -Differentiation -Analysis.
[0028] More specifically, and in one particularly preferred example, the
inventor
contemplates a method of analyzing a biological sample of a mammal in which a
plurality of
microvesicles that include distinct RNA molecules is first obtained from a
living donor
mammal. The microvesicles are then enriched and differentiated to produce a
primary result
based on one or more distinct RNA molecules (which are most typically a RNA
that are
uniquely expressed and/or mutated as a function of the condition of the cell
from which it
derived), a secondary result based on at least two distinct RNA molecules
(with one of the
molecules being RNA that is uniquely expressed in a specific tissue or organ,
and with the
other molecule being RNA that is uniquely expressed and/or mutated as a
function of the
condition of the cell from which it derived) and/or a ternary result based on
a sub-segregated
proxysome population (e.g., based on a specific surface marker of the cell
from which the
microvesicle was derived) and one or more distinct RNA molecules (most
typically a RNA
that is uniquely expressed and/or mutated as a function of the condition of
the cell from
which it derived). The primary, secondary, and/or ternary results are then
correlated with a
diagnosis or prognosis of a condition of the mammal.
[0029] Most typically, the microvesicles are obtained from whole blood, serum,
plasma, or
any other biological fluid, including urine, milk, tears, spinal fluid,
amniotic fluid, etc., which
are preferably obtained from a living mammal, and most preferably within the
time limit
7
CA 02728736 2010-12-20
WO 2009/155505 PCT/US2009/047937
acceptable for processing biological fluids for later clinical analysis.
Alternatively, in less
preferred aspects, microvesicles may also be obtained from stored materials
(e.g., biological
fluids, tissues, organs, etc.), wherein the time between obtaining the
biological fluid, tissue,
or organ, and enrichment of the microvesicles from the sample may be at least
12 hours, at
least 24 hours, at least 2-5 days, or even at least one or more weeks. Such
storage may even
include storage at reduced temperature (e.g., 4 C) or even storage in frozen
form. Similarly,
microvesicles may also be obtained from an in vitro source, and most typically
from cell or
tissue culture, or even organ culture. In yet further contemplated aspects, it
should be noted
that deceased donors are also deemed suitable as a source for the
microvesicles. Most
commonly, however, microvesicles will be obtained from blood, which may be
immediately
or within a few hours (less than 12 hours) processed to form serum or plasma,
which is then
either stored, shipped, and/or further processed o enrich the microvesicles.
[0030] With respect to isolation or enrichment of microvesicles it is
contemplated that all
known manners of isolation of microvesicles are deemed suitable for use
herein. As used
herein, the terms "isolation" or "isolating" in conjunction with microvesicles
are
interchangeably used with the terms "enrichment" or "enriching", and refer to
one or more
process steps that result in an increase of the fraction of microvesicles in a
sample as
compared to the fraction of microvesicles in the obtained biological sample.
Thus,
microvesicles may be purified to homogeneity, purified to at least 90% (with
respect to non-
microvesicle particulate matter), less preferably at least 80%, even less
preferably at least
50%, and least preferably at least 20% (or even less). For example, physical
properties of
microvesicles/proxysomes may be employed to separate them from a medium or
other source
material, and especially preferred physical methods include separation on the
basis of
electrical charge (e.g., electrophoretic separation), size (e.g., filtration,
molecular sieving,
etc), density (e.g., regular or gradient centrifugation), Svedberg constant
(e.g., sedimentation
with or without external force, etc). Alternatively, or additionally,
isolation may be based on
one or more biological properties, and especially suitable isolation methods
may employ
surface markers (e.g., for precipitation, reversible binding to solid phase,
FACS separation,
specific ligand binding, non-specific ligand binding such as annexin V, etc.).
In yet further
contemplated methods, the microvesicles may also be fused using chemical
and/or physical
methods, including PEG-induced fusion and/or ultrasonic fusion.
8
CA 02728736 2010-12-20
WO 2009/155505 PCT/US2009/047937
[0031] Viewed from a different perspective, enriching can be done in a general
and non-
selective manner (typically including serial centrifugation), and may be
performed by
aggregation where the microvesicles are interlinked with an interlinking
composition (e.g.,
annexin V, fibrin, or an antibody or fragment thereof against at least one of
a tetraspanin,
ICAM-1, and CD86). Alternatively, enriching can be done in a more specific and
selective
manner (e.g., using tissue or cell specific surface markers). For example,
specific surface
markers may be used in immunoprecipitation, FACS sorting, bead-bound ligands
for
magnetic separation etc. Such isolation or enrichment will advantageously
allow obtaining
cell-, tissue-, organ-, and/or disease specific information without the need
for direct access to
the cell, tissue, or organ under investigation. The specific RNA that is
enclosed in a general
population of microvesicles is therefore generally termed "vesiculated RNA",
while RNA of
a specific type of microvesicle (e.g., microvesicle generated by hepatocyte)
is termed "proxy
RNA", and microvesicles that are specific to a single type of origin are
referred to as
"proxysomes". Such proxysomes will consequently have an RNA content and
membrane
composition (especially in terms of surface markers, but also to at least some
degree in terms
of lipid composition) that is consistent with the RNA content of a cell from
which the
proxysome is produced.
[0032] Therefore, and especially where the microvesicles/proxysomes are
isolated from a
biological source (e.g., whole blood or serum) or mixed cell or tissue
culture, it should be
noted that the isolation may produce a heterogeneous population of
microvesicles/proxysomes with respect to the cell-/tissue-, and/or organ-type
from which the
microvesicles/proxysomes were produced. On the other hand, and especially
where a specific
surface marker was used in the isolation of the microvesicles/proxysomes, the
isolated
population may already be homogenous with respect to the cell-/tissue-, and/or
organ-type
from which the microvesicles/proxysomes were produced. In still further
contemplated
aspects, the mechanism of release of the vesicles from the cell may be used to
further
differentiate, even among proxysomes. However, it should be noted that all
vesicular
architectures (e.g., inside-out, etc.) are deemed suitable for use herein.
[0033] As a population of microvesicles obtained from a biological fluid will
typically
represent a plurality of cells, tissues, and organs, differentiation of the
heterogeneous
population is often desirable to obtain a cell, tissue, or organ specific
result. Differentiation
9
CA 02728736 2010-12-20
WO 2009/155505 PCT/US2009/047937
[0034] First, where a condition (e.g., cancer, infection, senescence,
inflammation, etc.) of a
cell, tissue, or organ is associated with a unique and distinguishable
expression profile (e.g.,
over- or underexpression) of a gene and/or with a specific mutation, a
meaningful result may
be based on the expression profile and/or specific sequence of one or more
distinct RNA
molecules without further normalization of the signal. For example, where the
condition is
breast cancer, several genes are often misregulated, and presence of high
quantities of
ERBB2 will be indicative of breast cancer. Similarly, certain types of
leukemia (CML) are
associated with a specific fusion mutant (Bcr/Abl) that is specifically
associated with the
leukemia. Thus, a primary result can be obtained based on the expression
profile and/or
specific sequence of one or more distinct RNA molecules without physical sub-
segregation of
the microvesicles.
[0035] Second, a condition (e.g., cancer, infection, senescence, inflammation,
etc.) of a cell,
tissue, or organ may be associated with a unique and distinguishable cell,
tissue, or organ
type and may therefore be specifically characterized by normalizing an
expression profile
and/or specific sequence of one or more distinct RNA molecules against the
expression
profile and/or specific sequence of one or more further distinct RNA molecules
that are
uniquely present or expressed in the specific cell, tissue, or organ. Thus, a
secondary result
can be obtained based on the expression profile and/or specific sequence of at
least two
distinct RNA molecules without physical sub-segregation of the microvesicles.
[0036] Third, a condition (e.g., cancer, infection, senescence, inflammation,
etc.) of a cell,
tissue, or organ may be associated with a unique and distinguishable cell,
tissue, or organ
type and may therefore be specifically characterized by physical sub-
segregation (infra) of
the microvesicles prior to analysis of the expression profile and/or specific
sequence of one or
more distinct RNA molecules in the sub-segregated microvesicles.
[0037] Therefore, in some aspects of the inventive subject matter,
differentiation is
preferably done using one or more additional information bearing components in
and/or on
the microvesicle to correlate the result with the additional information to so
obtain
normalized cell-, tissue-, organ-, and/or disease-specific information. Viewed
from a
different perspective, differentiation may therefore include a sub-segregation
of a subset of
CA 02728736 2010-12-20
WO 2009/155505 PCT/US2009/047937
proxysomes from the microvesicles, and/or a determination of a reference
marker that is
specific to a subset of proxysomes of the isolated population. It should be
especially
appreciated that the differentiation may also include detection or use of a
component (or
portion thereof) on a microvesicle or proxysome that is otherwise not
accessible on the cell.
Such is especially true where the microvesicle or proxysome has an inside-out
architecture
that exposes on the outside of the microvesicle or proxysome the component
that has an
orientation normally oriented towards the interior of the cell from which the
vesicle is
produced.
[0038] For example, where differentiation is based on molecular analysis of
gene
expression of a gene that is significantly associated with a particular
disease, differentiation
may be based on the unique sequence, expression profile, mutation, or other
character of a
particular RNA and thus will typically not require physical subsegregation
and/or
normalization against other RNA and/or protein markers. Thus, in at least some
instances,
presence, abundance, and/or sequence of one or more RNA molecules in the
microvesicle
population can be directly attributed to a specific disease or condition of a
cell, tissue, or
organ. For example, where a patient is suspected of breast cancer the
presence, specific
sequence, and/or expression rate of a relevant oncogene (e.g., BRCA, Her2/neu,
etc.) in a
microvesicle will be useful as a proxy marker for the cancer as such
particular RNA is
normally not present in measurable quantities in microvesicles.
[0039] Alternatively, and especially where an RNA under investigation is
present in diseased
(or otherwise compromised) as well as healthy cells, tissues, or organ,
differentiation may be
based on normalization of the RNA under investigation against one or more
other
constituent parts of the microvesicles/proxysomes. Therefore, it should be
appreciated that
differentiation can be performed on a non-segregated pool of microvesicles,
and even on the
source material where the microvesicles are present. In such case, it is
generally preferred
that the additional information bearing component is specific to a particular
subset of
proxysomes. For example, it is of interest to analyze microvesicles from
mammary gland
tissue, a gene may be identified that is uniquely (greater or equal 90%) or
predominantly
(greater or equal 75%) expressed in the mammary gland tissue. As the proxysome
RNA
content is an at least partial, and in many cases a complete representation of
the RNA content
of the mammary gland cell from which the proxysome is derived, the mammary
gland
proxysome is expected also to comprise the uniquely or predominantly expressed
gene. For
11
CA 02728736 2010-12-20
WO 2009/155505 PCT/US2009/047937
example, it is known that dermcidin is >95% selectively expressed in mammary
gland tissue
and therefore also expected to be uniquely present in mammary gland
proxysomes. There are
numerous genes known in the art that are uniquely or predominantly expressed
in specific
cells and organs, and suitable genes with such expression can be found, inter
alia, at the
TIGER database (ht(p://bioinfo.wiliner.jha.edu/tiger/; incorporated by
reference herein).
[0040] Alternatively, or additionally, numerous known non-nucleic acid
components can be
used to differentiate and may include disease specific markers (e.g., tumor
specific markers
such as CEA), organ specific markers (e.g., PSA), and/or cell-specific markers
(e.g., CD4 for
T-helper cells), etc. These additional information bearing components may then
be used for
physical subsegregation in a manner as described above. However, it is also
noted that so
long as these additional information bearing components are specific to the
desired
subpopulation of proxysomes, no physical subsegregation may be required, and
the RNA
information (infra) may be normalized or otherwise parameterized using the
additional
information bearing component. Thus, it should be recognized that molecular
analysis for
differentiation can be done with high specificity to a particular cell type,
tissue type, and/or
organ type.
[0041] Physical subsegregation of a subset of proxysomes may be performed
using
antibodies or other binding agents that binds selectively to a component that
is specific to
particular cell, tissue, or organ. Such components are typically cell-, tissue-
, or organ-
specific marker, receptors, structural components, glycoproteins, CDs, etc.,
all of which may
be further derivatized. Suitable organ, tissue, and cell specific surface
markers are well
known in the art and can be found in numerous publications (e.g., for natural
receptors see
e.g., Cell Surface Receptors: A Short Course on Theory and Methods by Lee E.
Limbird
Springer; 2nd ed. edition (December 31, 1995), ISBN-10: 0792338391; for
synthetic peptides
to selected cells, see e.g., Curr Opin Chem Biol. 2000 Feb;4(l):16-21). Thus,
it should be
appreciated that subsegregation can be performed specific to a cell type,
tissue type, and/or
organ type.
[0042] Most typically, physical subsegregation will involve use of a solid
phase to which a
compound is bound that (preferably releasably) binds to one or more of the
component that is
specific to particular cell, tissue, or organ. For example, where the solid
phase is a multi-well
plate, proxysomes may be separated into separate wells. Where the solid phase
is a magnetic
or color coded bead, the proxysomes may be isolated using a magnet or light
activated sorter.
12
CA 02728736 2010-12-20
WO 2009/155505 PCT/US2009/047937
Similarly, the solid phase may be a dipstick or membrane and separation will
(preferably
releasably) bind the proxysomes on the solid phase. In still further
contemplated aspects,
subsegregation may also be performed on an array in which antibodies are
coupled to the
surface of the array in predetermined positions to so specifically bind
distinct proxysomes in
distinct and predetermined positions on the array. Detection of the bound
proxysomes may
then be performed in a non-specific manner (e.g., using a labeled form of
annexin V) or a
specific manner (e.g., using a labeled antibody against another surface marker
of the
proxysome).
[0043] Of course, it should be recognized that subsegregation may be done
after isolation of
a population of microvesicles, but may also be directly performed on the
source material of
the microvesicles/proxysomes. In such case, the steps of isolation and
subsegregation are
combined. Most typically, and depending on the surface marker used, the
subsegregation will
produce a subpopulation of proxysomes that is substantially homogeneous (i.e.,
at least 70%,
more typically at least 80%, most typically at least 90%) with respect to the
tissue from
which they originate (e.g., microvesicle from liver, prostate, mammary gland,
etc.). It should
further be especially appreciated that the subsegregation may also be
performed (e.g., without
RNA analysis) to determine numerical ratios and/or absolute numbers of
microvesicles/proxysomes.
[0044] The specific RNA of the proxysome of interest can most typically be
analyzed using
all known manners of RNA analysis, and particularly preferred manners include
rtPCR,
quantitative PCR (rt or otherwise), primer extension analysis (with or without
prior
amplification), and all solid-phase based methods (e.g., using arrays,
magnetic or color coded
beads, etc.). While not limiting to the inventive subject matter, it is
generally preferred that
the specific RNA is (1) native to the cell, tissue or organ, i.e., not
introduced (directly or
indirectly) by a pathogen, and/or (2) specific or indicative to a
predisposition, condition,
disease, response to a stimulus, and/or pathogen. For example, suitable
specific RNAs
include genes that are specifically and exclusively expressed or overexpressed
in cells,
tissues, or organs affected with a neoplastic disease, a metabolic disease,
inflammation,
senescence, hypoxia, an infection, and/or inheritable disease.
[0045] For example, especially suitable specific RNAs are those known to have
an
association with a particular disease and especially with cancer. There are
numerous such
RNAs and genes known in the art, and all of those are deemed suitable for use
herein. Among
13
CA 02728736 2010-12-20
WO 2009/155505 PCT/US2009/047937
other sequences, RNA specific for breast cancer includes RNA that encodes MMP
11,
BCAR1, ERBB2, MK167, PLAU, and/or TP53, RNA specific for pancreatic cancer
includes
RNA that encodes FGFR1, KRAS2, TGFBR2, MAP2K4, and/or CDKN2A, RNA specific for
prostate cancer includes RNA that encodes KLK3, ERBB2, FGF8, PSCA, and/or
CAV1, or
RNA specific for gastric cancer includes RNA that encodes BAX, SLC2A1, PTGS2,
MUC1,
and/or RUNX3, RNA specific for liver cancer includes RNA that encodes BCL10,
PAP,
SPARC, CD44, and/or TP53, and RNA found for various cancers includes RNA that
encodes
adult stem cell markers (e.g., CD33, CD44, CXCR4, CXCR4+, lin-, CD45-, Oct-4,
Nanog,
SCAT, and/or 7-AAD. Further numerous cancer related genes/RNA can be found in
various
publicly or commercially available database (e.g.,
fittp:,,'/www.cancei-itid,,x.orgIgEewebi,'clirik3O.lit,in, incorporated by
reference herein). In still
further especially preferred aspects, it is contemplated that more than one
specific RNA is
measured to so improve a diagnostic finding. For example, where paired or
grouped markers
are analyzed, ERBB2 and VEGFR may be measured in a population of proxysomes
from
mammary gland tissue.
[0046] Of course, it should be noted that the RNA of interest need not be
limited to an RNA
that is associated with a particular disease, but that suitable information
may be generated
from analysis of the expression pattern on of or more distinct RNAs. For
example, it is
contemplated that total RNA from a non-segregated microvesicle population is
analyzed on a
chip of other multiplexed platform to so arrive at a genome wide expression
profile. Such
expression profile may then be used as a basis for diagnosis of a condition or
disease where
an expression profile of a healthy individual is known. Consequently, it
should be appreciated
that for the first time, one or more systemic expression profiles may be
obtained from the
same individual (or group of individuals) in a manner that does not require
multiple biopsies.
Similarly, and especially where the RNA analysis is normalized against a
specific tissue or
performed on a sub-segregated population of microvesicles, tissue specific
expression
profiles may be obtained from the same individual (or group of individuals) in
a manner that
does not require biopsies.
[0047] While not expressly excluded, it is generally preferred that the RNA is
an RNA that is
coding and can give rise to a protein via ribosomal translation. Therefore,
most typically
RNA will include a 3'-polyA tail and may further include a 5'-cap structure
(typically 7-
methylated guanine nucleotide bound to mRNA via 5'-5' triphosphate group, but
less
14
CA 02728736 2010-12-20
WO 2009/155505 PCT/US2009/047937
common structures also deemed suitable and include methylation of the 2'
hydroxy-groups of
the first 3 ribose sugars of the 5' end of the mRNA). Furthermore, it should
also be noted that
while mRNA is deemed especially useful in conjunction with the teachings
presented herein,
other forms of RNA are also expressly contemplated herein. For example,
suitable RNA for
analysis include single stranded and double stranded RNA, heterogenous RNA
(hnRNA),
small interfering RNA (siRNA), transfer RNA (tRNA), ribosomal RNA (rRNA),
mitochondrial RNA (mtRNA), capped RNA and uncapped RNA, polyadenylated and non-
polyadenylated RNA, etc.
[0048] Analysis of the specific RNA and the additional information bearing
component is
based on the premise that each proxysome will have at least one RNA specific
for a
disease/condition and at least one additional representative component of the
cell from which
that proxysome was produced (e.g., RNA of a tissue-specific expressed gene, a
cytoplasmic
component, a membrane composition, membrane associated component (typically
comprising a protein). Together, the specific RNA and the additional component
(which may
be another RNA that is cell- or tissue specific, a protein, a lipid, or any
combination thereof)
are then employed to analyze a condition (e.g., disease or predisposition,
age, reaction to
physical or chemical stimulus [e.g., drug, food, radiation, etc.]) in a highly
cell-, tissue-,
and/or organ-specific manner without the need to directly sample the cell-,
tissue-, and/or
organ. Analysis may be typically be specific with respect to a set of markers.
However,
analysis may also be analyzed as a function of time to see fluctuations in
total microvesicles
and/or proxysomes or proxysome populations. Analysis may be focused on time
course,
quantity, and/or association with further markers. Thus, analytic results will
typically include
variants, quantities, and/or or time course expression of the specific RNA,
which will most
typically be correlated with one or more results obtained from the
differentiation (which may
be a quantitative measure of mRNA, a peptide, a membrane component, etc.).
[0049] With respect to analysis of the additional information bearing
component it should be
appreciated that those components can be analyzed (or used) in all known
manners. For
example, where the additional information bearing component comprises a
peptide,
especially preferred manners include use of antibodies (or fragments thereof),
ligands
(synthetic or natural), etc., all of which may be labeled or otherwise
modified for qualitative
or quantitative analysis. Where the additional information bearing component
is a
polysaccharide, lectins may be used, and where the additional information
bearing
CA 02728736 2010-12-20
WO 2009/155505 PCT/US2009/047937
component is a lipid, specific ligands (e.g., annexin V) may be used for
quantification.
Moreover, and especially where the additional information bearing component is
a compound
other than a nucleic acid, it is contemplated that the additional information
bearing
component may be used to sub-segregate a population of microvesicle into a
population that
is enriched in one specific type of microvesicle (enrichment preferably >80%).
[0050] Depending on the particular RNA and manner of differentiation, it
should be
appreciated that the manner of correlation of the results to a specific
diagnosis or condition
may vary to at least some degree. For example, where the analysis of RNA
relies on
quantification and/or sequence of a disease-specific gene, correlation may
advantageously be
done by comparison of the result(s) with result(s) obtained from a
microvesicle population
obtained from a reference patient (which may be healthy, or representative to
a specific
disease stage or state). Similar correlation can be performed for tests in
which the RNA is
normalized against another RNA or non-RNA compound from a microvesicle, or is
normalized mcg of total RNA/mL of serum, total RNA per mg of MVs proteins
isolated from
1 mL of serum, or other preferably fixed parameter. In like manner, where the
RNA is
obtained from a population of proxysomes, correlation is typically performed
against a
known standard that is indicative for a particular disease or condition. Of
course, it should be
appreciated that the comparison of the results may be performed one a single
RNA basis, on
the basis of two or more RNA results, or in at least some cases, on a genome
or organ-
specific basis. In such case, the correlation will include comparison of
multiple RNA-
specific results with multiple reference results, and all possible cross-
relations thereof.
[0051] Thus, it should be appreciated that the inventor especially
contemplates a method of
confirming and/or staging a mammalian neoplasm in which in one step a whole
blood
fraction is obtained that includes a plurality of microvesicles comprising a
plurality of distinct
RNA molecules. In another step the microvesicles are enriched and
differentiated to so
produce a primary result based on one or more distinct RNA molecules, a
secondary result
based on at least two distinct RNA molecules, and/or a ternary result based on
a sub-
segregated proxysome population and at least one of the plurality of distinct
RNA molecules.
In yet another step, the primary, secondary, and/or ternary results are
correlated with a stage
of the neoplasm in the mammal.
[0052] Generally all neoplasms are deemed suitable for use herein (supra),
however, it is
particularly preferred that the neoplasm is characterized by overexpression
(relative to
16
CA 02728736 2010-12-20
WO 2009/155505 PCT/US2009/047937
healthy cell) of ERRB2. Consequently, suitable neoplasms include acute
lymphoblastic
leukemia, bladder cancer, brain cancer, breast carcinoma, cervical cancer,
colorectal cancer,
lung cancer, ovarian cancer, and pancreatic adenocarcinoma. RNA coding for
ERRB2
overexpression can be quantified in numerous manners, however, it is generally
preferred that
the step of differentiating comprises quantitative rtPCR and/or reverse
transcription and solid
phase (e.g., microarray) hybridization. As already noted above, it is
generally preferred that
the step of correlating comprises comparison of the primary, secondary, and/or
ternary results
with one or more reference results that are characteristic of a healthy cell
and/or a diseased
cell, tissue or organ in a specific diseased, compromised, or aged state.
[0053] Viewed from a different perspective, contemplated uses of diagnostic
tests and
compositions include diagnosis, predisposition, and prognosis of the metabolic
state of a cell,
tissue, and/or organ, analysis of the response of a cell, tissue, and/or organ
to a physical and
more typically chemical stimulus, and/or diagnosis or determination of
predisposition for a
disease or disorder in patient harboring a cell, tissue, and/or organ from
which the
proxysomes were analyzed. In especially preferred uses, the methods according
to the
inventive subject matter are employed in the diagnosis of cancer, a pre-
cancerous condition
or predisposition, and/or a clinical stage of a cancer. Additionally, it
should be noted that
contemplated methods are also deemed suitable for prenatal diagnosis as fetal
microvesicles
have been identified in (Kidney International, 2007; 72(9):1095-102) amniotic
fluid,
placenta, and possibly also blood of the pregnant mother.
[0054] In still further contemplated uses, it should be appreciated that
microvesicle/proxysome compositions and methods according to the inventive
subject matter
can be used in bioinformatic analysis in which 'normal' proxysome quantity and
composition
is acquired and then compared to proxysome quantity and composition from cells
treated
with one or more agents or conditions. Wile contemplated compositions and
methods
typically reply on already known specific RNAs and additional markers and
information
bearing components, it should be appreciated that new specific RNAs and new
information
bearing component can be relatively easy obtained. For example, a cell culture
of diseased
cells or cells from a disease model can be propagated in vitro. The
supernatant is then
collected and microvesicles are isolated from the supernatant. In an optional
step, protein is
extracted from the microvesicles and antibodies generated. The antibody
collection is then
subtracted against normal tissue to obtain disease specific antibodies.
Alternatively
17
CA 02728736 2010-12-20
WO 2009/155505 PCT/US2009/047937
corresponding experiments can be performed in silico based on expression
profiling, or
subtractive nucleic acid libraries of diseased and corresponding healthy cells
can be
generated.
[0055] In still further contemplated aspects, the microvesicle diagnostic
methods presented
herein can be advantageously employed in drug discovery where multiple cell
cultures are
separately exposed to multiple drugs. Genomics and proteomics analyses can be
performed
as known in the art. However, in preferred aspects, proxysomes can be
collected from the
individual cultures and analyzed as described above. The so obtained results
can then be
followed in vivo to ascertain that in vitro findings correlate with prior in
vitro results.
Moreover, as the microvesicles can be collected multiple times from a single
animal and as a
single collection is sufficient to analyze distinct reactions in multiple
organs without
sacrificing speed or accuracy (or the animal), drug discovery using proxysomes
dramatically
increases throughput in drug discovery. While it is generally preferred that
analysis is
performed using specific RNA(a) and additional information bearing components,
it is also
contemplated that the microvesicle/proxysome components can be analyzed in
relative and
absolute proportions to obtain a measure for a particular clinical parameter.
It should still
further be noted that while contemplated methods and compositions are
particularly useful for
clinical diagnostic purposes and R&D, the methods and compositions according
to the
inventive subject matter will may also be advantageously employed in
personalized medicine
and personalized nutrition in which the effect of administration of
nutraceuticals (and
medications) on microvesicles and proxysomes can be monitored in a highly
simplified
manner.
[0056] It should be appreciated that contemplated tests allow for
standardization, which is
one of the more significant hurdles in clinical diagnosis. In one aspect of
the inventive subject
matter, the RNA signal is normalized against a second signal, typically
derived from the
additional information bearing components (e.g., total signal from proxysome
membrane
[e.g., via labeled annexin V]). Alternatively, a range of normal can be
established from a
clinically healthy population, again using a signal from an additional
information bearing
component. For example, one could employ a known microvesicle ELISA (which
measures
amount of PS, phosphatidylserine), which is highly correlated with
microvesicles. There is
already normal range established in healthy individuals (about l ONm/ml),
which may serve
as a normalization signal. In such assay, RNA Her2 could be calculated per
total
18
CA 02728736 2010-12-20
WO 2009/155505 PCT/US2009/047937
microvesicles, measured PS, or HER2 RNA signal per (phosphatidylserine -
Platelet
microvesicles phosphatidylserine). If using realtime PCR, one would compare
the number of
cycles per phosphatidylserine. In case off an arras, the array signal is
correlated to per
phosphatidylserine.
Examples
[0057] The following is provided to illustrate exemplary methods, conditions,
and
embodiments in connection with contemplated methods and compositions, but
should not be
deemed limiting the inventive subject matter.
Detection Of Her-2 Expression In Breast Cancer Cell Lines
[0058] Real time PCR: Three cancer cell lines were cultured in normal cell
culture conditions
(37 C, 5% C02) according to respective ASTM standard protocols. Total RNA was
isolated
from the cells using the commercially available RNeasy isolation kit (QUIAGEN)
according
to manufacturer's protocol. After isolation, the concentration of total RNA
was determined by
UV spectroscopy using well established methods. The concentration of RNA was
adjusted
for all groups to 50 ng/mcl. So prepared RNA was reverse transcribed to cDNA
using a
commercially available reverse transcription kit (Applied Biosystems)
following the
manufacturer's protocol. This cDNA was then used for real time PCR studies.
The real time
PCR reaction contained Sybr green master mix, forward primer, reverse primer,
water, and
respective cDNA samples, and the following HER-2 primers were used (5'- to 3'-
end):
[0059] Forward: ATTTCTGCCGGAGAGCTTTGAT (SEQ ID NO:1)
[0060] Reverse: CCGGCCATGCTGAGATGTATAG (SEQ ID NO:2)
[0061] Real time PCR reaction was performed with using Real Time PCR System
ABI 7500
(Applied Biosystems), using following settings: 50 C 2 min, 95 C 5 min, 95 C
15 sec, 60 C
60 sec; Last two cycles were repeated 40 times. Relative quantitation of Her-2
mRNA
expression was calculated with the comparative Ct method. The relative
quantitation value of
target, normalized to an endogenous control B2MG gene and relative to a
calibrator, is
expressed as 2- ct (fold difference), where ACt=Ct of target gene, -Ct of
endogenous control
gene (B2MG), and AACt = ACt of samples for target gene- ACt of calibrator for
the target
gene.
19
CA 02728736 2010-12-20
WO 2009/155505 PCT/US2009/047937
[0062] To avoid the possibility of amplifying contaminating DNA (i) all the
primers for real
time PCR were designed with an intron sequence inside cDNA to be amplified,
(ii) all
reactions were performed with appropriate negative controls (template-free
controls), (iii) a
uniform amplification of the products was rechecked by analyzing the melting
curves of the
amplified products (dissociation graphs), and (iv) gel electrophoresis was
performed to
confirm the correct size of the amplification and the absence of unspecific
bands. The results
for this experiment are shown in Figure 1. As can be readily taken from the
Figure, and as
assessed via rtPCR, SKOV3 exhibited significant expression of Her-2 whereas
Her-2
expression in T47D cells was moderate and nearly undetectable in MCF cells.
This finding
correlates well with published literature data on Her-2 expression in these
cell lines.
[0063] Regular PCR: Using RNA isolated as described above, a regular PCR was
performed.
Briefly, RNA was reverse transcribed to cDNA with using Applied Biosystem
reverse
transcription kit following the manufacturer's protocol, and the so prepared
cDNA was used
for PCR studies. Regular PCR reaction used the following Her-2 primers (5'- to
3'- end):
[0064] Forward: GTGACAGCAGAGGATGGAACAC (SEQ ID NO:3)
[0065] Reverse: CGCCATTGTGCAGAATTCG (SEQ ID NO:4)
[0066] To avoid the possibility of amplifying contaminating DNA (i) all the
primers for PCR
were designed with an intron sequence inside cDNA to be amplified, and the
(ii) reactions
were performed with appropriate negative controls (template-free controls).
The PCR
products were visualized using a 1.5% agarose gel. Once more, Her-2 was
detectable in the
mRNA level in all three cell lines, but SKOV3 cell lines showed highest
expression, and
T47D cells and MCF cells showed expression on very low level. Negative
controls were
performed using RNA without reverse transcription, and PCR products were not
detected.
Detection Of Her-2 In Microvesicles from Supernatants of the Cell Lines
[0067] Isolation of Microvesicles: The tumor derived microvesicles were
isolated from
culture conditioned media. Briefly, the supernatant was spun at about 850 g
for 10 minutes at
4 C. The supernatant was collected and the pellets were discarded. The
supernatants were
again spun at 24,000 g for 2 hr at 4 C and the supernatant was discarded. PBS
supplemented
with HEPES (5mM) was added to pellet that now contained microvesicles and the
resuspended microvesicles were transferred to Eppendorf tubes (1.6 ml) in
which they were
CA 02728736 2010-12-20
WO 2009/155505 PCT/US2009/047937
spun at maximum speed for 60 min at 4 C. The supernatant was discarded and
PBS
supplemented with HEPES was added to the pellet, spun again at maximum speed
for 60 min
at 4 C. The so obtained pellet was resuspended pellet in a small volume of
PBS
supplemented with HEPES (usually 100-200 mcl).
[0068] PCR Analysis: Using the methods described for cells above, total RNA
was isolated
from the microvesicles, and real time PCR and regular PCR were performed. When
using
real time PCR, Her-2 was detected only in microvesicles from the SKOV cell
culture but not
from T47D and MCF cell culture. However, when using regular PCR, Her-2 was
detectable
on the mRNA level from all three cell cultures by agarose gel electrophoresis
as can be seen
in Figure 2A. Not surprisingly, a quantitative analysis of the agarose gel
electrophoresis
showed that microvesicles from the SKOV cell culture provided the strongest
her-2 signal,
which was followed by the T47D cell culture, which was in turn followed by the
MCF cell
culture. A quantitative graph illustrating these results is shown in Figure
2B. It should
therefore be appreciated that cellular expression levels of Her-2 are
paralleled by
microvesicular Her-2 RNA quantities detected in the culture supernatants of
the respective
cell lines.
Detection Of Her-2 In Microvesicles From Murine Serum Of Mice Harboring
SKOV Derived Tumors
[0069] Mice with SKOV-derived Tumors: Athymic nude mide were injected with
3x106
SKOV3 cells using standard protocol to establish solid tumors in these mice.
After three
weeks, presence of palpable/measurable tumors was confirmed, and serum was
collected
from the animals as well as from control mice without tumors (no SKOV
injection).
[0070] The microvesicles were isolated from the serum following the general
protocol as
outlined above. The total RNA obtained from the microvesicles was evaluated
for the
presence of Her-2 RNA using the PCR protocols as provided above. PCR products
were
quantified and checked via agarose gel electrophoresis and exemplary results
are shown in
Figure 3A (to ensure analysis of equal amounts, control PCR was performed
using beta-2-
microglobulin as standard). A graphic representation of the Her-2
quantification of
corresponding rtPCR is shown in Figure 3B. Based on these results, it should
be appreciated
that SKOV-tumor derived microvesicles that were generated in vivo were not
only positive
21
CA 02728736 2010-12-20
WO 2009/155505 PCT/US2009/047937
for Her-2 RNA, but that such RNA can also be detected from a relatively small
blood sample
with a high degree of sensitivity and specificity.
Detection Of Her-2 In Microvesicles From Human Serum Of Patients Diagnosed
With Her-2 Positive Breast Cancer
[0071] To validate the concept of using microvesicles as proxy diagnostic
markers for
diseased or otherwise distressed cells, blood samples were analyzed from
patients diagnosed
with Stage I and II breast cancer that was characterized by
immunohistochemical assay as
Her-2 positive at level 3+.
[0072] Microvesicles were isolated from actual patient and control sera
following the
general protocol as outlined above, and total RNA obtained from the
microvesicles was
evaluated for the presence of Her-2 RNA using the PCR protocols as provided
above. The
PCR products were quantified and checked via agarose gel electrophoresis and
the results for
these experiments are shown in Figure 4A (to ensure analysis of equal amounts
control PCR
was again performed using beta-2-microglobulin as standard). Remarkably, none
of the
control sera from healthy volunteers showed any detectable Her-2 RNA as tested
by rtPCR
and normal PCR, but all three of the patients with confirmed breast cancer
exhibited a
significant and strong signal. Moreover, while patients in lane 4 and 6 were
staged at Stage I,
the patent in lane 5 was diagnosed at Stage II, which correlated with the
strongest signal. The
corresponding graphic representation of the Her-2 quantification is shown in
Figure 4B.
Based on these results, it should be appreciated that microvesicles can act as
sensitive proxy
diagnostic tools for the cells from which they originate. Still further, it is
noted that such
analysis can be performed not only the without radiation burden from
radiographic analyses,
but also in a manner that entirely avoids any biopsies
[0073] It should be apparent to those skilled in the art that many more
modifications besides
those already described are possible without departing from the inventive
concepts herein.
The inventive subject matter, therefore, is not to be restricted except in the
spirit of the
appended claims. Moreover, in interpreting both the specification and the
claims, all terms
should be interpreted in the broadest possible manner consistent with the
context. In
particular, the terms "comprises" and "comprising" should be interpreted as
referring to
elements, components, or steps in a non-exclusive manner, indicating that the
referenced
elements, components, or steps may be present, or utilized, or combined with
other elements,
22
CA 02728736 2010-12-20
WO 2009/155505 PCT/US2009/047937
components, or steps that are not expressly referenced. Where the
specification claims refers
to at least one of something selected from the group consisting of A, B, C
.... and N, the text
should be interpreted as requiring only one element from the group, not A plus
N, or B plus
N, etc. Furthermore, where a definition or use of a term in a reference, which
is incorporated
by reference herein is inconsistent or contrary to the definition of that term
provided herein,
the definition of that term provided herein applies and the definition of that
term in the
reference does not apply.
23