Note: Descriptions are shown in the official language in which they were submitted.
CA 02733021 2011-02-25
DEMANDES OU BREVETS VOLUMINEUX
LA PRESENTE PARTIE DE CETTE DEMANDE OU CE BREVETS
COMPREND PLUS D'UN TOME.
CECI EST LE TOME'-1- DE _2
NOTE: Pour les tomes additionels, veillez contacter le Bureau Canadien des
Brevets.
JUMBO APPLICATIONS / PATENTS
THIS SECTION OF THE APPLICATION / PATENT CONTAINS MORE
THAN ONE VOLUME-
THIS IS VOLUME 1 OF 2
NOTE: For additional volumes please contact the Canadian Patent Office.
CA 02733021 2011-02-25
CLEAVAGE KIT, AND GENE THERAPY BY USING THE SAME AND
NUCLEIC ACID CLEAVAGE DETECTION APPARATUS
BACKGROUND OF THE INVENTION
Field of Invention
[0001] The present invention relates a kit and a cleavage detection apparatus,
and,
in particular, a kit and a cleavage detection apparatus with high sequence
specificity.
The present invention also relates a gene therapy by using the nucleic acid
cleavage
kit.
Related Art
[0002] Nucleic acid cleavage is a reaction of breakage in the nucleic acid
sequence
imposed by external force. It can be applied to clinical healthcare,
biotechnology or
other related fields. Previously, various types of conventional nucleic acid
cleavage
tools have been used. Although they may be slightly different in composition,
restriction enzymes with high specificity have been widely used in recognition
and
cleavage on nucleic acids.
[0003] However, as the expansion of the application scope of nucleic acid
cleavage,
the sequence-specificity provided by restriction enzymes has been insufficient
to meet
the demand. For example, several important researches and medical fields, such
as
gene transformation, genetic mapping and gene therapy, are fairly expecting
the
development of a highly specific nucleic acid cleavage tool.
[0004] In particular, the lengths of the nucleic acid sequences in higher
organisms
like human beings are approximately 105-107 base pairs. To create a site-
specific
cleavage on these nucleic acids, it has to use a cleavage tool possessing the
ability of
t
CA 02733021 2011-02-25
recognizing at least 8 to 15 base pairs. However, conventional restriction
enzymes
cannot recognize more than 8 base pairs and are incompetent for nucleic acid
cleavage.
Similarly, for implementing a genetic analysis on a genome (generally about
105-107
base pairs in length) or nucleic acids with high sequence similarity (for
example
heterochromatin), the basic demand of the sequence-specific recognition
ability
provided by a nucleic acid cleavage tool should be more than 8 base pairs
correspondingly. The conventional nucleic acid cleavage tools used restriction
enzymes as the functional components are certainly insufficient for certain
application.
[0005] Besides, in order to improve the ability of nucleic acid sequence
recognition,
conventional methods have indicated oligonucleotides, which can bind to other
nucleic acid segments containing corresponding sequences, can provide highly
sequence-specific recognition for particular nucleic acid sequences. However,
certain feature of the oligonucleotides was only applied as a nucleic acid
probe for
detecting whether particular sequences exist in an organism but not applied in
cooperating with nucleic acid cleavage technique.
[0006] Therefore, it is an important subject of the invention to provide a
nucleic
acid cleavage tool with high sequence-specificity.
SUMMARY OF THE INVENTION
[0007] In view of foregoing, the present invention is to provide a nucleic
acid
cleavage tool with high sequence-specificity, a method for gene therapy by
using the
same and a nucleic acid cleavage detection apparatus.
[0008] To achieve the above, a nucleic acid cleavage kit in accordance with
the
present invention includes a carrier, an oligonucleotide and a nucleic acid
cleavage
2
CA 02733021 2011-02-25
agent. The nucleic acid cleavage kit functions on a target nucleic acid. The
first
end of the oligonucleotide is bound with the carrier to recognize at least
partial
sequence of the target nucleic acid. The nucleic acid cleavage agent is bound
to the
second end of the oligonucleotide to cleave the target nucleic acid.
[0009] In one embodiment of the present invention, the nucleic acid cleavage
kit is
applied to a gene therapy.
[0010] In one embodiment of the present invention, the target nucleic acid is
an in
vitro nucleic acid.
[0011] In one embodiment of the present invention, the carrier is a
nanoparticle.
[0012] In one embodiment of the present invention, the carrier includes a base
and
a bonding layer. The bonding layer is at least disposed on a partial surface
of the
base, and the first end of the oligonucleotide is bound to the bonding layer
of the
carrier.
[0013] In one embodiment of the present invention, the base is a flat
substrate, a
microplate, a spherical particle, a columnar container, a box-shaped
container, a
plate-shaped container or a cylindrical container.
[0014] In one embodiment of the present invention, the target nucleic acid is
a
single-strand nucleic acid and the oligonucleotide recognizes the target
nucleic acid
by forming a double helix with the partial sequence of the target nucleic
acid.
[0015] In one embodiment of the present invention, the target nucleic acid is
a
double-strand nucleic acid and the oligonucleotide recognizes the target
nucleic acid
by forming a triple helix with the partial sequence of the target nucleic
acid.
[0016] In one embodiment of the present invention, the oligonucleotide is a 10-
mer
to 30-mer oligonucleotide.
3
CA 02733021 2011-02-25
[0017] In one embodiment of the present invention, the oligonucleotide
substantially consists of polypurines or modified polypurines and the partial
sequence
of the target nucleic acid substantially consists of polypyrimidines.
[0018] In one embodiment of the present invention, the oligonucleotide
substantially consists of polypyrimidines or modified polypyrimidines and the
partial
sequence of the target nucleic acid substantially consists of polypurines.
[0019] In one embodiment of the present invention, the nucleic acid cleavage
agent
is a photoactivated nucleic acid cleavage agent.
[0020] To achieve the above, a nucleic acid cleavage detection apparatus
includes
the aforementioned a nucleic acid cleavage kit and a nucleic acid detection
kit. The
nucleic acid cleavage detection apparatus functions on a target nucleic acid.
The
nucleic acid detection kit detects at least one nucleic acid fragment derived
from the
nucleic acid after the cleavage of the nucleic acid cleavage kit.
[0021] In one embodiment of the present invention, the nucleic acid detection
kit is
an electrophoresis device or a nucleic acid amplification device.
[0022] To achieve the above, a method for gene therapy in accordance with the
present invention includes administrating to a subject an effective amount of
a nucleic
acid cleavage composition to cleave a target nucleic acid. The nucleic acid
cleavage
composition includes a nanoparticle, an oligonucleotide and a nucleic acid
cleavage
agent. The first end of the oligonucleotide is bound with the nanoparticle to
recognize at least partial sequence of the target nucleic acid, and the
nucleic acid
cleavage agent is bound to the second end of the oligonucleotide to cleave the
target
nucleic acid.
[0023] In one embodiment of the present invention, the target nucleic acid is
a
4
CA 02733021 2011-02-25
single-strand nucleic acid, and the oligonucleotide recognizes the target
nucleic acid
by forming a double helix with the partial sequence of the target nucleic
acid.
[0024] In one embodiment of the present invention, the target nucleic acid is
a
double-strand nucleic acid, and the oligonucleotide recognizes the target
nucleic acid
by forming a triple helix with the partial sequence of the target nucleic
acid.
[0025] In one embodiment of the present invention, the oligonucleotide is a 10-
mer
to 30-mer oligonucleotide.
[0026] In one embodiment of the present invention, the oligonucleotide
substantially consists of polypurines or modified polypurines, and the partial
sequence
of the target nucleic acid substantially consists of polypyrimidines.
[0027] In one embodiment of the present invention, the oligonucleotide
substantially consists of polypyrimidines or modified polypyrimidines and the
partial
sequence of the target nucleic acid substantially consists of polypurines.
[0028] In one embodiment of the present invention, the nucleic acid cleavage
agent
is a photoactivated nucleic acid cleavage agent.
[0029] In summary, the nucleic acid cleavage kit, the method for gene therapy
by
using the same and the nucleic acid cleavage detection apparatus in accordance
with
the present invention specifically recognize the partial sequence of the
target nucleic
acid by the oligonucleotide and thereby cleave the target nucleic acid by the
nucleic
acid cleavage agent. Because the oligonucleotide can recognize longer
sequence, the
nucleic acid cleavage kit can perform its cleavage function with high sequence
specificity for nucleic acid in vivo and in vitro. Comparing to prior art, the
present
invention provides a novel nucleic acid cleavage kit and a method for gene
therapy by
using the same to overcome the issue that the application scope of nucleic
acid
CA 02733021 2011-02-25
cleavage is limited by the insufficient recognition ability of restriction
enzymes. It is
beneficial for achieving the goal of clinical application and reducing side
effects. In
addition, the present invention also provides a novel nucleic acid cleavage
tool for in
vitro usage, which can recognize longer sequence and be easily modified and
manufactured such that it expands the application scope of nucleic acid
cleavage and
reduces production cost.
BRIEF DESCRIPTION OF THE DRAWINGS
[0030] The invention will become more fully understood from the detailed
description and accompanying drawings, which are given for illustration only,
and
thus are not limitative of the present invention, and wherein:
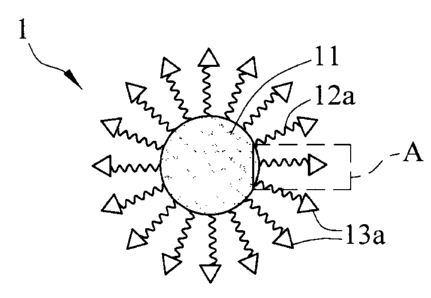
[0031] FIG 1 is a schematic figure of the nucleic acid cleavage kit in
accordance
with the first embodiment of the present invention;
[0032] FIG 2 is a partial enlarged figure of the region A shown in FIG. 1;
[0033] FIG 3 is a flow chart of the cleavage process of the nucleic acid
cleavage
kit with high sequence specificity in accordance with the first embodiment of
the
present invention;
[0034] FIG 4 is a cross-sectional figure of the nucleic acid cleavage kit in
accordance with the second embodiment of the present invention;
[0035] FIG. 5 is a partial enlarged figure of the region A shown in FIG 4;
[0036] FIG. 6a flow chart of the cleavage process of the nucleic acid cleavage
kit
with high sequence specificity in accordance with the second embodiment of the
present iinvention;
6
CA 02733021 2011-02-25
[00371 FIG 7a is a top view of the nucleic acid cleavage kit in accordance
with the
third embodiment of the present invention;
[00381 FIG. 7b is a cross-sectional figure along the section line E shown in
FIG 7a;
[0039] FIG. 8a is a schematic figure of the nucleic acid cleavage kit filled
in a
column in accordance with the fourth embodiment of the present invention;
[00401 FIG 8b is a partial enlarged figure of the region F shown in FIG 8a;
[0041] FIG. 9 is a block diagram of the nucleic acid cleavage detection
apparatus in
accordance with the embodiment of the present invention;
[0042] FIG 10 is a synthesis flow chart of the nucleic acid cleavage kit in
accordance with the first embodiment of the present invention;
[00431 FIG l la is a microscopic image of the 2-B2 cells after the treatment
of the
nucleic acid cleavage kit in accordance with the first embodiment of the
present
invention;
[0044] FIG 11 b is an electrophoregram of the genomic DNA PCR products in
accordance with the first embodiment of the present invention;
[00451 FIG Ile is a bar diagram illustrating the signal strength of the
products
shown in FIG 11 b;
[00461 FIG 12 is a synthesis flow chart of the nucleic acid cleavage kit in
accordance with the third embodiment of the present invention;
[0047] FIG 13a is a schematic diagram showing the target nucleic acid and the
useful sites thereof in accordance with the second experiment of the present
invention;
[0048] FIG 13b is an electrophoregram of the target nucleic acids respectively
cleaved by the restriction enzymes and nucleic acid cleavage kit in accordance
with
7
CA 02733021 2011-02-25
the fourth experiment of the present invention;
[0049] FIG 14 is a diagram illustrating the auto-sequencing result adjacent to
the
triple helix on the target nucleic acid in accordance with the fourth
experiment of the
present invention; and
[0050] FIG. 15 is a sequencing list indicating the relative information of the
primer
used in the present invention.
DETAILED DESCRIPTION OF THE INVENTION
[0051] The terms "recognition" or "recognize" as used herein is a
characteristic
that one nucleic acid can distinguish a target nucleic acid from others by
forming of
base-pairings (bps) to combine the nucleic acid and the target nucleic acid
into one.
Because the formation of base-pairings depends on whether two nucleic acids
can
form chemical bonds between the nitrogenous bases of their nucleotides at the
corresponding positions, a candidate nucleic acid without a complementary
sequence
cannot complete the aforementioned base-paring process. The term "cleavage" or
"cleave" as used herein is to generate at least one gap or nick on a
continuous
nucleotide sequence. As for function, the cleavage reaction can result in the
loss
function of an original gene containing the continuous nucleotide sequence.
[0052] The nucleic acid cleavage kit in accordance with the present invention
can
provide a cleavage reaction with high sequence specificity for particular
nucleic acids.
The application field of the nucleic acid cleavage kit is for example but not
limited to
gene therapy or sequence recombination.
[0053] In one embodiment of the present invention, the nucleic acid cleavage
kit is
applied to gene therapy. Meanwhile, the target nucleic acid is an in vivo
target
8
CA 02733021 2011-02-25
nucleic acid. The target nucleic acid can be for example but not limited to a
whole
genome or a part thereof, an artificial or synthetic nucleic aid, the
aforementioned
nucleic acid with modification or a natural or synthetic nucleoid. In
addition, the
target nucleic acid for the nucleic acid cleavage kit also can be an in vitro
target
nucleic acid including a whole genome or a part thereof purified from an
organism, a
natural nucleic acid purified from a cell, a synthetic nucleic acid, the
aforementioned
nucleic acid with modification, or a natural or synthetic nucleoid.
[0054] The target nucleic acid further can be a nucleic acid unit with a
biological
function such as a gene, a gene fragment or a promoter, a enhancer or a
poly(A) tail of
a gene. In one embodiment of the present invention, the nucleic acid can be a
nucleic acid unit of the double-stranded deoxyoligonucleotide chain with a
biological
function. The aforementioned gene with a biological function can express a
polypeptide or a protein directly or indirectly involved in a pathogenic
mechanism of
a genetic disease or gene mutation. The genetic disease can be for example but
not
limited to leukemia, diabetes, Huntington's disease, and the gene mutation is
considered to induce various cancers.
[0055] However, to be noted, if the target nucleic acid is the aforementioned
nucleic acid unit with a biological function, the length of the target nucleic
acid may
include certain sequence of the nucleic acid unit with biological function and
the
adjacent sequences. It is to provide enough binding length for the
oligonucleotide so
as to prevent miscleavage occurring.
[0056] FIRST EMBODIMENT
[0057] The following and accompanying figures describe the nucleic acid
cleavage
kit in accordance with the first embodiment of the present invention.
9
CA 02733021 2011-02-25
[0058] FIG 1 is a cross-sectional figure of the nucleic acid cleavage kit in
accordance with the present invention, and FIG 2 is the partial enlarged
figure of FIG
1. As shown in FIG 1 and FIG. 2, the nucleic acid cleavage kit 1 in accordance
with
the first embodiment of the present invention includes a carrier 11, an
oligonucleotide
12a and a nucleic acid cleavage agent 13a. The nucleic acid cleavage kit 1
functions
on a target nucleic acid (not shown in the figure). The first end of the
oligonucleotide 12a is bound with the carrier 11 to recognize at least partial
sequence
of the target nucleic acid. The nucleic acid cleavage 13a agent is bound to
the
second end of the oligonucleotide 12a to cleave the target nucleic acid.
[0059] In the present invention, the carrier I1 is a nanoparticle. In more
detailed,
the carriers 11 can be nanoparticles with uniform size, shape and/or chemical
composition or nanoparticles with different size, shape and/or chemical
composition.
To be noted, when the carrier 11 is a nanoparticle, the nucleic acid cleavage
kit 1 can
be considered as a sort of nucleic acid cleavage compositions, in particular
for
medical use.
[0060] The aforementioned nanoparticle can be for example but not limited to
an
aggregate particle, an isotropic nanoparticle such as a solid spheral
nanoparticle or a
hollow spheral nanoparticle, an anti-isotropic nanoparticle such as an
anisotropic
conical, rectangular or rhombic nanoparticle, a dendrimer or a composite
nanoparticle
such as a core-shell nanoparticle. In the present embodiment, the carrier 11
is a solid
spheral nanoparticle.
[0061] In addition, when the carrier 11 is a nanoparticle, the mean particle
diameter
of the carrier 11 is about 1 to 100 nanometers. In the present embodiment, the
mean
particle diameter of the carrier 11 is about 10 to 30 nanometers.
[0062] Similarly, when the carrier 11 is a nanoparticle, the material of the
carrier 11
CA 02733021 2011-02-25
can include metal and/or magnetic material. The metal can be for example Au,
Ag,
Pd, Pt, Ni, Al, In, Ti, Cu, Fe, Co, Zn, Sn, Cr and other metal easily to form
into
spherical shape. The magnetic material is substantially material with
paramagnatism
and thereby can be attracted or repulsed by an external magnetic force, for
example
ferrous oxide or nickel oxide. It benefits the operation and recycling of the
nucleic
acid cleavage kit 1. In the present embodiment, the carrier 11 is a gold
nanoparticle,
which has the features of good biocompatibility and in vitro stability.
[0063] The material of the aforementioned nanoparticle can also include
semiconductor or inorganic material. The semiconductor can be cadmium
selenide,
cadmium sulfide, or zinc sulfide-coated cadmium selenide or cadmium sulfide.
In
addition, the inorganic material can be silicon or silicon dioxide. The
organic
material included in the aforementioned nanoparticle can be poly(lactide-co-
glycolide)
(PLGA).
[0064] The oligonucleotide 12a can be for example an oligonucleotide derived
from purification, polymerase chain reaction (PCR) or chemical synthesis. The
sequence of the oligonucleotide 12a is about 10 to 30 mers in length, and
preferably is
about 11 to 15 mers in length. As for its molecular composition, the
oligonucleotide
12a can be a RNA oligonucleotide, a DNA oligonucleotide or the aforementioned
oligonucleotide with modifications.
[0065] The first end of the oligonucleotide 12a is connected to the carrier
11. As
shown in FIG. 1 and FIG. 2, in the present invention, the oligonucleotide 12a
and the
carrier 11 are connected with a covalent bond. As shown in FIG. 2, in more
detailed,
the 5' end of the oligonucleotide 12a includes a thiol group connected to the
carrier 11
with a covalent bond. However, other connection type between the
oligonucleotide
12a and the carrier 11 can be used as well, for example but not limited
hydrogen bond,
I1
CA 02733021 2011-02-25
van der Waals' forces or static electric interaction.
[0066] In the present embodiment, the oligonucleotide 12a substantially
consists of
polypurines to form a triple helix structure with at least partial sequence of
the target
nucleic acid. The triple helix structure is called as a triplex-forming
oligonucleotide
(TFO). Correspondingly, the at least partial sequence of the target nucleic
acid
substantially consists of polypyrimidines. In contrast, in another embodiment
of the
present invention, the oligonucleotide 12a substantially consists of
polypyrimidines
and the at least partial sequence of the target nucleic acid substantially
consists of
polypurines.
[0067] The term "purine" as used herein substantially indicates an adenine or
a
guanine. However, it further includes a modified adenine or guanine or a
synthesized analogue of adenine or guanine. In addition, the term "pyrimidine"
as
used herein substantially indicates a thymine, a cytosine or a uracil.
However, it
further includes a modified thymine, cytosine or uracil or a synthesized
analogue of
athymine, a cytosine or a uracil.
[0068] As for the modification, the major modification sites of the purine or
pyrimidine can be classified to three portions: the nucleobase, the sugar and
the
phosphate backbone. The modified product or synthesized analogue can be for
example but not limited to 7-deaza-2'-deoxyxanthosine, 2'-deoxy-6-
thioguanosine,
5-fluoro-deoxyuracil, 2' -deoxynebularine, 5-methylcytosine,
5-propargylamino-2'-deoxyuridine, nucleotides containing 2'-methoxylated
riboses,
5-propynyldeoxyuridine, nucleotides with riboses replaced by 2'-
aminoethylribose
and analog thereof, 02', 04'-methylene-linked nucleic acid, locked nucleic
acid
(LNA) monomer, 02', 04'-ethylene linked nucleic acid (ENA) monomer and peptide
nucleic acid (PNA) (Please reference Chan et al., Triplex DNA: fundamentals,
12
CA 02733021 2011-02-25
advances, and potential applications for gene therapy, J. Mol. Med., 75, 267-
282,
1997.)
[0069] In addition, the term "triple helix" as used herein is a sort of helix
structure
formed by the interaction between the oligonucleotide 12a and a double-
stranded
target nucleic acid including the corresponding sequences, which can form
chemical
bonds between the nucleobases. In more detailed, the interaction occurs at the
major
grooves or minor grooves of the double-stranded target nucleic acid.
[0070] To be noted, the nucleic acid cleavage kit 1 in accordance with the
present
invention can include the carrier I1 connected by more than one sort of the
oligonucleotides 12a instead of one such that it can simultaneously react with
more
than one sort of the target nucleic acid.
[0071] The nucleic acid cleavage agent 13a is used to cleave the target
nucleic acid,
and the selection of the nucleic acid cleavage agent can depend on the
molecular
composition of the target nucleic acid. Preferably, the nucleic acid is for
example
but not limited to a synthesized deoxyribonucleic acid cleavage agent.
[0072] In the present embodiment, the nucleic acid cleavage agent 13a is a
photoactivated nucleic acid cleavage agent such as arylhydrazone (shown in FIG
2).
Other nucleic acid cleavage agents can be for example azidoproflavine,
azidophenacyl
or azido or ellipticine derivatives activated by a light source with a
wavelength longer
than 300 nanometers.
[0073] The second end of the oligonucleotide 12a is connected to the nucleic
acid
cleavage agent. As shown in FIG 1 and FIG 2, in the present embodiment, the
oligonucleotide 12a and the nucleic acid cleavage agent 13a are connected by a
covalent bond. In more detailed, as shown in FIG 2, the 3' end of the
13
CA 02733021 2011-02-25
oligonucleotide 12a includes an amino group connected to the nucleic acid
cleavage
agent 13a by a covalent bond.
[0074] In another aspect of the present embodiment, the first end of the
oligonucleotide 12a can further be connected to the carrier 11 via a spacer.
The
spacer such as poly(ethylene glycol) (PEG) can adjust the water solubility of
the
nucleic acid cleavage kit 1.
[0075] The following and accompanying FIG. 3 are taken for an example to
describe the reaction mechanism of the nucleic acid cleavage kit 1 in
accordance with
the first embodiment.
[0076] The nucleic acid cleavage kit 1 recognizes the at least partial
sequence of
the target nucleic acid by the oligonucleotide 12a, and then cleaves the
target nucleic
acid by the nucleic acid cleavage agent 13a. As shown in FIG. 3, when the
nucleic
acid cleavage kit 1 functions on the target nucleic acid B, the
oligonucleotide 12a
recognizes the at least partial sequence of the target nucleic acid B. In
addition, the
structure of the target nucleic acid B is a double helix. Therefore, a triple
helix
structure (shown in region C) is formed by the formation of chemical bonds
between
the oligonucleotide 12a and the at least partial sequence of the target
nucleic acid B.
[0077] If the target nucleic acid B is a single-stranded nucleic acid, the
structure
formed by the oligonucleotide 12a and the target nucleic acid B turns into a
double
helix. After the oligonucleotide 12a recognizes the target nucleic acid, the
nucleic
acid cleavage agent 13a functions and generates a breakage at the desired
cleavage
site D on the target nucleic acid B with a continuous sequence. In more
detailed, the
desired cleavage site D can be on base pair within or adjacent to the region C
of the
target nucleic acid B. In the present embodiment, the desired cleavage site D
is on
the base pair within the region C of the target nucleic acid B such that the
nucleic acid
14
CA 02733021 2011-02-25
cleavage kit 1 cleaves the target nucleic acid B into two nucleic acid
fragments B I
and B2.
[0078] SECOND EMBODIMENT
[0079] The following and accompanying figures describe the nucleic acid
cleavage
kit in accordance with the second embodiment of the present invention.
[0080] FIG 4 is a cross-sectional figure of a nucleic acid cleavage kit in
accordance
with the second embodiment of the present invention, and FIG 5 is a partial
enlarged
figure of the region A' of FIG. 4. As shown in FIG 4 and FIG 5, the nucleic
acid
cleavage kit 4 includes a carrier 41, an oligonucleotide 12b and a nucleic
acid
cleavage agent 13b. The nucleic acid cleavage kit 4 functions on a target
nucleic
acid. The first end of the oligonucleotide 12b is bound with the carrier 41 to
recognize at least partial sequence of the target nucleic acid. The nucleic
acid
cleavage agent 13b is bound to the second end of the oligonucleotide 12b to
cleave the
target nucleic acid.
[0081] As shown in FIG 4 and FIG 5, in the present embodiment, the carrier 41
includes a base 411 and a bonding layer 412. The bonding layer 412 is at least
disposed on a partial surface of the base 411. In more detailed, the bonding
layer
412 is at least disposed on the partial surface of the base 411 by for example
but not
limited to coating.
[0082] The base 411 can be a flat substrate, a microplate, a spherical
particle, a
columnar container, a box-shaped container, a plate-shaped container, a
cylindrical
container or other two-dimensional/three-dimensional configurations. In the
present
embodiment, the base 411 is a flat substrate (shown in FIG. 5).
[0083] In addition, the material of the bonding layer 412 can include metal or
CA 02733021 2011-02-25
magnetic material. The metal can be for example Au, Ag, Pd, Pt, Ni, Al, In,
Ti, Cu,
Fe, Co, Zn, Sri, Cr and other metal easily formed into flat layer-shaped. In
the
present embodiment, the bonding layer 412 preferably includes Au. The magnetic
material substantially refers to material with paramagnatism such as ferrous
oxide or
nickel oxide. The bonding layer 412 can also include semiconductor or
inorganic
material. In the present embodiment, the semiconductor can be for example but
not
limited to cadmium selenide, cadmium sulfide, or zinc sulfide-coated cadmium
selenide or cadmium sulfide. In addition, the inorganic material can be
silicon or
silicon dioxide.
[0084] The technical characteristics of the oligonucleotide 12b and the
nucleic acid
cleavage agent 13b are similar to those of the oligonucleotide 12a and the
nucleic acid
cleavage agent 13a in the first embodiment such that the detailed description
thereof
will be omitted.
[0085] The following and accompanying FIG 6 are taken for an example to
describe the reaction mechanism of the nucleic acid cleavage kit 4 in
accordance with
the second embodiment of the present invention.
[0086] In the present embodiment, the nucleic acid cleavage kit 4 recognizes
the at
least partial sequence of the target nucleic acid by the oligonucleotide 12b,
and then
cleaves the target nucleic acid by the nucleic acid cleavage agent 13b. As
shown in
FIG 6, when the nucleic acid cleavage kit 4 is close to the at least partial
sequence of
the target nucleic acid B', the oligonucleotide 12b is capable of recognizing
at least
partial sequence of the target nucleic acid. Because the target nucleic acid
B' is a
double helix, a triple helix (shown in region C') is formed by the formation
of
chemical bonds between the oligonucleotide 12b and the at least partial
sequence of
the target nucleic acid B'.
16
CA 02733021 2011-02-25
[0087] If the target nucleic acid B' is a single-stranded nucleic acid, the
structure
formed by the oligonucleotide 12b and the target nucleic acid B' is a double
helix
structure. After the oligonucleotide 12b recognizes the target nucleic acid,
the
nucleic acid cleavage agent 13b functions and generates a breakage at the
desired
cleavage site D' on the target nucleic acid B' with a continuous sequence. In
more
detailed, the desired cleavage site D' can be on the base pair within or
adjacent to the
region C' of the target nucleic acid B'. In the present embodiment, the
desired
cleavage site D' is on the base pair within the region C' of the target
nucleic acid B'
such that the nucleic acid cleavage kit 4 cleaves the target nucleic acid B
into two
nucleic acid fragments B1 and B2.
[0088] Furthermore, in the present embodiment, because the nucleic acid
cleavage
kit 4 can include a certain amount of the oligonucleotides and the nucleic
acid
cleavage agents, the nucleic acid cleavage kit 4 can be used for the sequence
detection
of large amounts of nucleic acids
[0089] THIRD EMBODIMENT
[0090] The following and accompanying related figures are taken for an example
to
describe the third embodiment of the present invention.
[0091] FIG 7a is a side view of the nucleic acid cleavage kit 7 in accordance
with
the third embodiment of the present invention, and FIG. 7b is a cross-
sectional figure
along the section line E shown in FIG 7A. As shown in FIG 7a and FIG. 7b, the
nucleic acid cleavage kit 7 includes a carrier 71, an oligonucleotide 12c and
a nucleic
acid cleavage agent 13c. The nucleic acid cleavage kit 7 functions on a target
nucleic acid. The first end of the oligonucleotide 12c is bound with the
carrier 71 to
recognize at least partial sequence of the target nucleic acid. The nucleic
acid
cleavage agent 13c is bound to the second end of the oligonucleotide 12c to
cleave the
17
CA 02733021 2011-02-25
target nucleic acid.
[0092] As shown in FIG 7b, in the present embodiment, the carrier 71 includes
a
base 711 and a bonding layer 712. The base 711 is a microplate such as a 12-
well
plate, and the bonding layer 712 is disposed on the bottom surface of each
well 77 of
the base 711 by for example but not limited to coating.
[0093] In the present embodiment, the technical characteristics of the nucleic
acid,
the bonding layer 712, the oligonucleotide 12c and the nucleic acid cleavage
agent
13c are similar to those of the nucleic acid, the bonding layer 412, the
oligonucleotide
12a and the nucleic acid cleavage agent 13a in the first embodiment such that
the
detailed description thereof will be omitted.
[0094] In addition, the cleavage process of the nucleic acid cleavage kit 7 in
the
present embodiment to the target nucleic acid is similar to the cleavage
process of the
nucleic acid cleavage kit 4 shown in FIG 6 such that the detailed description
thereof
will be omitted. To be noted, because the nucleic acid cleavage kit 7 includes
12
separated wells 77 (shown in FIG. 7a), each of the wells 77 can independently
can be
bound with the same or different oligonucleotides 12c and the nucleic acid
cleavage
agents 13c after the bonding layer 712 is disposed on the bottom surface of
each of
the wells 77 of the base 711.
[0095] FOURTH EMBODIMENT
[00961 The following and accompanying FIG 8 are taken for an example to
describe the fourth embodiment of the present embodiment.
[0097] FIG 8a is a schematic figure of a nucleic acid cleavage kit 8 in
accordance
with the fourth embodiment of the present invention, and FIG 8b is a partial
enlarged
figure of the region F shown in FIG. 8a. To be noted, the nucleic acid
cleavage kit 8
18
CA 02733021 2011-02-25
is filled in a column 84. As shown in FIG 8a and FIG 8b, the nucleic acid
cleavage
kit 8 includes a carrier 81, an oligonucleotide 12d and a nucleic acid
cleavage agent
13d. The nucleic acid cleavage kit 8 functions on a target nucleic acid. The
first
end of the oligonucleotide 12d is bound with the carrier 81 to recognize at
least partial
sequence of the target nucleic acid. The nucleic acid cleavage agent 13d is
bound to
the second end of the oligonucleotide 12d to cleave the target nucleic acid.
[0098] As shown in FIG 8a, a chromatographic column 84 is filled with the
nucleic
acid cleavage kit 8 in accordance with the present embodiment. In addition, as
shown in FIG. 8b, the carrier 81 includes a base 811 and a bonding layer 812.
The
base 811 is a spheroid particle, and the bonding layer 812 is disposed on the
surface of
the base 811 by for example but not limited to coating.
[0100] In the present embodiment, the technical characteristics of the target
nucleic
acid, the bonding layer 812, the oligonucleotide 12d and the nucleic acid
cleavage
agent 13d are similar to those of the aforementioned target nucleic acid,
bonding layer
412, oligonucleotide 12b and nucleic acid cleavage agent 13b such that the
detailed
description thereof will be omitted.
[0101] Similarly, the cleavage process of the nucleic acid cleavage kit 8 in
accordance with the present embodiment to the. target nucleic acid is similar
to the
cleavage process of the nucleic acid cleavage kit 4 shown in FIG. 6 such that
the
detailed description thereof will be omitted. As shown in FIG. 8a, because the
chromatographic column 84 can be filled with the nucleic acid cleavage kits 8
with
the same or different oligonucleotides 12d in any proportion, it can provide
large
amounts of nucleic acid the sequence detection for at least one sort of target
nucleic
acids specifically.
[0102] The nucleic acid cleavage kit I in accordance with the present
invention also
19
CA 02733021 2011-02-25
can be further processed into a pharmaceutical composition. In the first
embodiment,
the nucleic acid cleavage kit I can further include at least one acceptable
pharmaceutical carrier, for example but not limited to microcrystalline
cellulose,
mannitol, glucose, dried skim milk, starch, polyvinylprrolidone or a
combination
thereof.
[0103] In addition, the nucleic acid cleavage kit 1 in the pharmaceutical
composition
form can be administered to a subject in need for example but limited to
orally,
parentally, by inhalation spray , topically, nasally or via an implanted
reservoir,
microinjection or a gene gun. In more detailed, the term "parenteral" as used
herein
includes subcutaneous, intracutaneous, intravenous, intramuscular,
intraarticular,
intraarterial, intrasynovial or infrasternal injection or infusion techniques.
[0104] In summary, because the nucleic acid cleavage kit in accordance with
the
present invention can recognize the at least partial sequence of the target
nucleic acid
by the oligonucleotide and then cleave the target nucleic acid by the nucleic
acid
cleavage agent, it can provide cleavage reaction to the target nucleic acid
with high
sequence specificity by recognizing more than 10 nucleotide units in
operation. In
gene therapy, it can prevent the generation of undesired cleavage to affect
the quality
of healthcare or induce side effects. Furthermore, a large amount of the
oligonucleotides and the nucleic acid cleavage agent can be connected to the
carrier to
provide sufficient cleavage reactions in a use, and thereby the nucleic acid
cleavage
kit in accordance with the present invention is suitable for treating a large
amount of
nucleic acids. In addition, since the carrier can be a nanoparticle, it helps
users to
deliver the nucleic acid cleavage kit to a target site or recycle the nucleic
acid
cleavage kit from human body efficiently, and can further combine with image
technique to track the nucleic acid cleavage kit.
CA 02733021 2011-02-25
[0105] The nucleic acid cleavage kit of the present invention can be applied
to
cleave nucleic acids in vitro for preventing the generation of undesired
fragments
derived from the nucleic acids from affecting consequential analysis as well.
The
use of nucleic acid cleavage agents produced from general chemical synthesis
can
reduce the production cost for cleaving target nucleic acids and is more
convenient for
cleavage and detection. Moreover, the nucleic acid cleavage kit is flexible
for
application resulting from various sorts and modification sites of the
chemical
synthesized nucleic acid cleavage agents.
[0106] The oligonucleotide of the nucleic acid cleavage kit in accordance with
the
present invention can further be customized depending on target nucleic acids.
In
addition, the manufacturing process of the nucleic acid cleavage kit is simple
such
that it contributes to the expansion of nucleic acid cleavage market.
[0107] The following and accompanying related figures are taken for an example
to
describe a nucleic acid cleavage detection apparatus in accordance one
embodiment of
the present invention.
[0108] As shown in FIG. 9, the nucleic acid cleavage detection device 9 in
accordance the present invention functions on a target nucleic acid, and it
includes a
nucleic acid cleavage kit 91 and a nucleic acid detection kit 92. The nucleic
acid
cleavage kit 91 includes a carrier, a oligonucleotide, and a nucleic acid
cleavage agent.
The first end of the oligonucleotide is bound with the carrier to recognize at
least
partial sequence of the target nucleic acid, and the nucleic acid cleavage
agent is
bound with a second end of the oligonucleotide to cleave the target nucleic
acid.
Because the nucleic acid cleavage kit 91 and the elements thereof are similar
to the
aforementioned nucleic acid cleavage kit 1 and the elements thereof, the
detail
description will be omitted.
21
CA 02733021 2011-02-25
[0109] The nucleic acid detection kit 92 detects at least one nucleic acid
fragment
derived from the nucleic acid after the cleavage of the nucleic acid cleavage
kit 91.
The detection assay provided by the nucleic acid detection kit 92 can be set
up by
applying a physical and/or chemical factor to the nucleic acid fragment to
observe the
result of the nucleic acid fragment response to the physical and/or chemical
factor in
accordance with the length of the nucleic acid fragment. To be noted, the
nucleic
acid detection kit 92 is for example but not limited to an electrophoresis
device or a
nucleic acid amplification device.
[0110] Accordingly, the nucleic acid cleavage detection device in accordance
with
the present invention can provide cleavage reaction with high sequence
specificity and
further detect the at least one nucleic acid fragment after the cleavage of
the nucleic
acid cleavage kit. It can collects detection data directly and thereby
validate
real-time results.
[0111] The following takes the first embodiment of the present invention for
an
example to describe a method for gene therapy by administering the nucleic
acid
cleavage kit.
[0112] The method for gene therapy in accordance with the first embodiment
includes administering to a subject in need an effective amount of a nucleic
acid
cleavage composition to cleave a target nucleic acid. The nucleic acid
cleavage
composition includes a nanoparticle, an oligonucleotide and a nucleic acid
cleavage
agent. The first end of the oligonucleotide is bound with the nanoparticle to
recognize at least partial sequence of the target nucleic acid and the nucleic
acid
cleavage agent is bound to the second end of the oligonucleotide to cleave the
target
nucleic acid. Because the aforementioned nucleic acid cleavage composition and
the
elements thereof are similar to the nucleic acid cleavage kit and the elements
thereof
22
CA 02733021 2011-02-25
disclosed in the present invention, the detailed description will be omitted.
[0113] The term "subject in need" used herein includes a subject, who has a
disease,
or a symptom of either diseases, or a predisposition toward the disease, needs
to be
treated with gene therapy with the purpose to cure, heal, alleviate, relieve,
alter,
remedy, ameliorate, improve, or affect the disease, the symptoms of the
disease, or the
predisposition toward the disease.
[0114] In addition, in present embodiment, the method can be considered as an
upstream step to provide cleavage reaction with high sequence specificity in
gene
therapy, and then combines with corresponding sequential steps to complete the
entire
gene therapy process for different diseases specifically. The sequential steps
can be
for example but not limited to inserting a normal gene, replacing a
defect/mutated
gene with a normal gene or suppressing/knocking out an overexpressed gene.
Generally, the last step of the gene therapy process is manipulating a DNA
repair
mechanism to repair the breakage on the target nucleic acid and then restore
the target
nucleic acid as the original structure thereof.
[0115] Therefore, the method for gene therapy in accordance with the present
invention can provide cleavage reaction with high sequence specificity for the
target
nucleic acid such that the method is able to perform gene therapy at the
specific site
on the target nucleic acid precisely to repair defect or mutated nucleic acid
sequences.
Since the method is contributive to prevent the non-specificity of gene
therapy
occurring on the nucleic acids in organisms, it improves the quality of
healthcare and
reduces side effects during the period of treatment.
[0116] The following and accompanying figures take a number of experiments for
examples to describe the manufacturing method and the cleavage mechanism of
the
nucleic acid cleavage kits in accordance with the embodiments of the present
23
CA 02733021 2011-02-25
invention.
[0117] Experiment 1: the manufacture of the nucleic acid cleavage kit in
accordance
with the first embodiment
[0118] The following takes a synthesis experiment for an example to describe
the
manufacturing method of the nucleic acid cleavage kit in accordance with the
present
invention.
[0119] As shown in FIG. 10, the carrier 11 (a gold nanoparticle) was provided
by
using sodium citrate reduction (please reference to Frens, Cx Controlled
nucleation for
the regulation of the particle size in monodisperse gold suspensions., Nat.
Phys. Sci.,
241, 20-22, 1973, and Grabar, et al., Preparation and characterization of Au
colloid
monolayers., Anal. Chem., 67, 735-743, 1995).
[0120] In the meantime, each of the oligonucleotides 12 was prepared by
modifying
the 5' end (the first ends) with a thiol group. The carrier 11 was incubated
with the
oligonucleotides 12 in the molar ratio of 1:100, and the mixture was gently
shaken (<
1 Hz) at 4 C with 10mM phosphate buffers for 24 hrs. on an orbital shaker and
then
processed through a gradient of salt concentration (0.3M NaCl/10mM phosphate
buffer) to connect the oligonucleotides 12 to the carrier 11 by forming
covalent bonds
on the thiol groups.
[0121] The nucleic acid cleavage agents 13 were provided to connect to the 3'
ends
(the second ends) of the oligonucleotides 12. The 3' ends of the
oligonucleotides 12
were modified with amino groups in advance, and the nucleic acid cleavage
agents 13
were N-((3-maleimidopropyloxy)-succinimide ester hydrazones (BMPSs) compounds.
The carrier 11 connected with the oligonucleotides 12 was mixed with the
nucleic
acid cleavage agents 13 at room temperature for 2 hrs. The carrier 11 was
incubated
24
CA 02733021 2011-02-25
with the nucleic acid cleavage agents 13 in the molar ratio of 1:100, and
thereby
formed the nucleic acid cleavage kit 1. The nucleic acid cleavage kit 1 was
resuspended with phosphate buffer and then stored at 4 C in the dark for
further use.
[01221 Experiment 2: the in vivo cleavage reaction with high sequence
specificity
generated by the nucleic acid cleavage kit in accordance with the first
embodiment
101231 Human cervical carcinoma (HeLa) cell line is taken for an example and
was
established as an enhanced green fluorescent protein (EGFP) HeLa cell model
(also
called as 2-B2 cells in the following) for observing the in vivo cleavage
reaction with
high sequence specificity generated by the nucleic acid cleavage kit.
[01241 HeLa cell line was cultured with DMEM medium contained 10% fetal bovine
serum (FBS) and 1% antibiotics (PSF) at 37 C under a 5% humidified CO2
incubator.
For EGFP expression, the plasmids were constructed and then transferred into
HeLa
cells. The two single-stranded sequences recognized by the oligonucleotide
anneal
with each other by decreasing the temperature to form a double-stranded insert
with 5'
EcoRI and 3' BamHI sequence over-hangings. The double-stranded insert was then
ligated with pEGFP-N1 vector (CLONTECH Laboratories, Inc.) between EcoRI and
BamHI restriction site to form the plasmid, which can be recognized by the
oligonucleotide and express EGFP. To be noted, the oligonucleotide was able to
recognize upstream of EGFP coding sequence, and two sequences were in the same
reading frame. Then, the plasmid was validated by DNA sequencing.
[01251 The vector containing the oligonucleotide recognition sequence and EGFP
expression sequence was transiently transfected into the aforementioned HeLa
cells
incubated in a constant temperature incubator by Lipofectamine 2000
(Invitrogen)
method following manufacturer's protocol. Upon the stage 60% confluent on a
CA 02733021 2011-02-25
culture plate, HeLa cells were harvested for transfection. Then a single
colony was
picked from successfully transfected HeLa cells and screened by 600ug/ml G418
(Sigma, A1720) for one month to collect desired 2-B2 cells.
[0126] The 2-B2 cells were seeded into 6-well microplate with 1x105 density
then
incubated for 24 hours. A nucleic acid cleavage kit was added into each well
to the
concentration of 0.5uM. Waiting for 5 hours to make sure the 2-B2 cells uptake
the
nucleic acid cleavage kit. The 2-B2 cells are exposed in the 460nm blue LED
for 15
mins to activate the nucleic acid cleavage reaction. After the light exposure,
the
2-B2 cells were cultured for 17 hours in an appropriate manner to degrade the
EGFP
protein originally existing in the 2-B2 cells before processing with the
nucleic acid
cleavage kit. Then, the result was observed by two techniques: microscopy and
genomic DNA PCR.
[0127] FIG, 11 shows the level of EGFP expression of 2-B2 cells treated with
the
nucleic acid cleavage kit under microscopy. The 2-B2 cells were respectively
photographed in bright field and with a 509 nm emission filter, which can
absorb all
sorts of light except green fluorescence. As shown in FIGG. IIa, the green
fluorescence signal of the 2-B2 cells treated with the nucleic acid cleavage
kit is
weaker than one without the treatment. It is due to the cleavage reaction
performed
by the nucleic acid cleavage kit to reduce the expression level of EGFP.
[0128] Meanwhile, the genomic DNA PCR technique was used to examine the in
vivo cleavage reaction with high sequence specificity generated by the nucleic
acid
cleavage kit. Total genomic DNAs of the 2-B2 cells with or without the nucleic
acid
cleavage kit treatment were isolated by Genomic DNA mini kit (Geneaid, GB
100).
General standard reagents were used for all genomic DNA PCR reactions. Two
sets
of primers were used herein. The first set was primers for sequencing part of
EGFP
26
CA 02733021 2011-02-25
expression sequence for (the part of the sequence expressing EGFP)
(5'-CCTACGGCGTGCAGTGCTTCAGC-3' (SEQ ID NO:1 in FIG. 15) and
5'-CGGCGAGCTGCACGCTGCCGTCCTC-3' (SEQ ID NO:2 in FIG. 15), also
called as E primers in the following). The second set was primers for
sequencing the
oligonucleotide recognition sequence (5'-TACCGGACTCAGATCTCGAGCTCA-3'
(SEQ ID NO:3 in FIG 15), also called as T primers in the following).
[0129] The PCR cycle program initiated at 95 C for 5', followed by 30 cycles
of
95 C for 45", 60 C for 30", 72 C for 45", and finally at 72 C for 5'.
Amplified PCR
products were electrophoresed on 2% agarose gels and visualized by using UV
fluorescence.
[0130] As shown in FIG llb, comparing to the amount of the T-primer induced
products from 2-B2 cells processed without the nucleic acid cleavage kit
treatment
(marked with "2-B2" in FIG. I Ib) and the products processed with the light
exposure
but no nucleic acid cleavage kit treatment (marked with "2-B2(+)" in FIG lib),
the
products with the nucleic acid cleavage kit treatment were at extremely low
expression and thereby it indicated that the 2-B2 cells processed with the
nucleic acid
cleavage kit cannot synthesize the products containing the T primer sequence
after
genomic DNA PCR. The genomic DNA PCR products containing the E primers
were used for the control groups. FIG. 11c is a bar diagram showing the
electrophoresis result obtained in FIG. 11 b in accordance with the signal
strength of
the products and the data of the 2-B2 cells without any processing was used
for
standardization.
[0131] Experiment 3: the manufacture of the nucleic acid cleavage kit in
accordance
with the second embodiment
[0132] As shown in FIG. 12, the carrier 41 was generated by coating the
bonding
27
CA 02733021 2011-02-25
layer 412 on the base 411. The base 411 is a flat substrate. The bonding layer
412
was synthesized via reduction of gold salt by sodium citrate, and then
disposed on the
base 411 to form the carrier 41 by coating (please reference to Frens, G.
Controlled
nucleation for the regulation of the particle size in monodisperse gold
suspensions,
Nat. Phys. Sci., 241, 20-22, 1973, and Grabar, et al., Preparation and
characterization
of Au colloid monolayers., Anal. Chem., 67, 735-743, 1995).
[0133] Then, the oligonucleotides 12b were connected to the bonding layer 412,
and
the connection method can refer to the connection method between the
oligonucleotides 12a and the carrier 11 described in the first experiment such
that the
detailed description thereof can be omitted. The method of connecting the
nucleic
acid cleavage reagent 13b to the oligonucleotide 12b can also be refer to the
first
experiment such that the detailed description thereof can be omitted.
[0134] Experiment 4: the cleavage with high sequence specificity of the
nucleic acid
cleavage kit in accordance with the fourth embodiment
[0135] In the present experiment, the target nucleic acid was the pGEM-T easy
plasmid (Promega, Madison, WI, USA) containing the oligonucleotide recognition
sequence 100 ng (nanogram), and the control group was pGEM-T easy plasmid
without the oligonucleotide recognition sequence 100 ng. Both of the target
nucleic
acids and the control group have been created a Nae I site, and then were
cleaved by
restriction enzyme cleavage technique to form linear plasmid nucleic acids
with its
original length respectively. In more detailed, the Nae I site were located at
2,710 in
both of the target nucleic acids and the control group.
[0136] Then, each of the target nucleic acids and the control group separately
mixed
with the nucleic acid cleavage kit in Tris-base buffer at 37 C for 72 hrs.
The molar
ratio of the target nucleic acids/control group to the oligonucleotide is 1:1.
Because
28
CA 02733021 2011-02-25
the nucleic acid cleavage agents in the present experiment were hydrazone
compounds, the reactions were sequentially exposed to 302 nm ultraviolet light
for 30
mins for activation of the cleavage. In more detailed, the source of the 302
rim
ultraviolet light is an F8T5 UV -B lamp (16-watt, peak wavelength at 302 nm).
[0137] The products derived from the target nucleic acids and the control
group were
purified after cleavage respectively, and then the purified products were
separated in a
2.0% agarose gel and visualized by using an imaging system (BioSpectrum AC
[formerly ACI AutoChemi]; UVP, Inc., Upland, CA, USA). Furthermore, the
2689-bps DNA fragment bands derived from the target nucleic acids after
cleavage
were then isolated and sequenced (ABI Prism 3730 DNA Sequencer; AME Bioscience
A/S, Toroed, Norway).
[0138] As shown in FIG. 13a, the target nucleic acids and the control group
have
been respectively cleaved by Nae I restriction enzyme to form linear plasmid
nucleic
acids previously, and then interacted with the nucleic acid cleavage kit. The
oligonucleotides included in the nucleic acid cleavage kit can recognize the
at least
partial sequence of the target nucleic acid with high specificity such that
each of the
oligonucleotides bound to the at least partial sequence of the target nucleic
acid to
form a triple helix structure (60-87). After photoactivation induced by the
ultraviolet
light, the nucleic acid cleavage agents were able to cleave downstream of the
3' end of
the triple helix and thereby cleaved each of the complete linear plasmid
nucleic acids
into two fragments. As lane 8 shown in FIG 13, the larger one is about 2,689-
bps in
length, and the smaller one is about 356-bps in length. In contrast, the
control group
was observed with the length of original 3,045 bps (as indicated in lane 4)
because it
is without the oligonucleotide recognition sequence.
[0139] Primers were designed based on the 311 bps downstream from the
29
CA 02733021 2011-02-25
oligonucleotide recognition sequence of the plasmids so that primers can be
used for
auto-sequencing of the target nucleic acids treated with the target nucleic
acid
cleavage kit (ABI Prism 3730 DNA sequencer; AME Bioscience AJS, Toroed,
Norway). The primer sequence is 5'-AGCGAGTCAGTGAGCGAGGA-3' (SEQ ID
NO:4 in FIG. 15) in this experiment. As shown in FIG 14, comparing the
auto-sequencing result with the original sequence of the target nucleic acid
before the
treatment of the nucleic acid cleavage kit, each of the post-cleavage
sequences was
truncated 12 bps downstream from the 3' end of the triple helix (374) at which
the
auto-sequencing process stopped. It indicated that certain position was one of
the
ends of the target nucleic acid after the cleavage of generated by the nucleic
acid
cleavage kit. It further confirmed that the nucleic acid cleavage kit of the
present
invention can perform a cleavage reaction with high sequence specificity.
[01401 Although the invention has been described with reference to specific
embodiments, this description is not meant to be construed in a limiting
sense.
Various modifications of the disclosed embodiments, as well as alternative
embodiments, will be apparent to persons skilled in the art. It is, therefore,
contemplated that the appended claims will cover all modifications that fall
within the
true scope of the invention.
CA 02733021 2011-02-25
DEMANDES OU BREVETS VOLUMINEUX
LA PRESENTE PARTIE DE CETTE DEMANDE OU CE BREVETS
COMPREND PLUS D'UN TOME.
CECI EST LE TOME I DE 2
NOTE: Pour les tomes additionels, veillez contacter le Bureau Canadien des
Brevets.
JUMBO APPLICATIONS / PATENTS
THIS SECTION OF THE APPLICATION / PATENT CONTAINS MORE
THAN ONE VOLUME.
THIS IS VOLUME 1 OF 2
NOTE: For additional volumes please contact the Canadian Patent Office.