Note: Descriptions are shown in the official language in which they were submitted.
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
TLR 7 Ligand and Uses Thereof
FIELD OF THE INVENTION
The present invention relates to the field of immunotherapy. The present
invention provides
a TLR7 ligand and its use in therapeutic applications. Specifically, the
present application
provides an RNA oligonucleotide comprising a G:U wobble base pair and its use
in treating
disease such as viral infections, immune disorders and cancer.
--10- -BACKGROUND-OF THE INVENTION- -
The family of Toll like receptors (TLRs) is involved in the detection of most
classes of
pathogens. TLRs on the cell surface recognize conserved molecules of bacteria
fungi and
parasites that are "foreign" to the host organism [1]. A structurally closely
related subset of
TLRs has been shown to contribute to the detection of viruses [2-5]. Viruses
derive all their
components from the infected host cell and therefore do not contain "foreign"
molecules.
Detection of viruses instead hinges on the strategic localization of TLRs that
recognize viral
nucleic acids in endosomes and lysosomes. These cellular compartments are not
usually
accessed by host nucleic acids, but are traversed by most viruses during their
infectious
cycles. The endosomal TLRs 3, 7/8 and 9 have been shown to recognize double
stranded
(ds), single stranded (ss) or short double stranded RNA and DNA, respectively
[2,6-8]. In
humans, the function to recognize viral ssRNA is shared by TLR7 and TLR8 [7],
and the
expression pattern of both receptors appears mutually exclusive, and is
largely confined to
immune cell subsets. Among human PBMC, TLR7 is expressed by plasmacytoid
dendritic
cells (PDCs) and by B cells, while monocytes and myeloid dendritic cells
express TLR8
[9,10]. In contrast, mice deficient in TLR7 were found entirely unable to
respond to ssRNA
ligands, indicating that mouse TLR8 is either inactive, or has a non-immune
function
[11,12]. The first defined TLR7/8 ligands were imidazoquinoline derivates and
C8 and/or N7
modified analogs of guanosine, small molecules known to induce an antiviral
response
[11,13-15]. Most of these compounds (e.g. resiquimod R-848, 3M-003) activate
both, TLR7
and TLR8. Others, including imiquimod R-837, 3M-001 and loxoribine, are TLR7
selective,
and mainly stimulate PDC to produce IFN-a, or preferentially activate TLR8 (3M-
002), and
induce monocytes to secrete TNF- a and IL-12. The differential cytokine
profiles are of
relevance for the therapeutic use of these compounds. The first drug of this
class is 5%
imiquimod cream, which is approved for the topical treatment of genital warts
caused by
human papillomavirus (HPV), as well as of basal cell carcinoma and actinic
keratosis [16].
Recognition of cognate RNA ligands by TLR7/8 has been studied mostly using
short
1
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
synthetic oligoribonucleotides (ORNs) [17-22]. To achieve stimulation, ORN
must be
formulated in a complex with lipid transfection reagents or polycations to
enable delivery to
the endosomal compartment [6,7,21,23]. It was noted that ORN lacking uridines
were not
immunostimulatory, and most ORNs rich in guanosines and uridines were
stimulatory for
both, TLR7 and TLR8 [7]. Stimulatory sequence motifs were reported [21],
however a
systematic evaluation of sequence motifs has not been performed. Based on the
observation that homopolymeric uridine (pU) and ORNs consisting of 21 uridine
repeats
show TLR7 ligand activity, it was proposed that bona fide RNA recognition
motifs may not
exist [18]. However, a recent publication confirmed that some ORN sequences
lacking
guanosines selectively activate TLR8 [20]. Other studies found that compared
to the single
- - stranded-ORNs,-complementary- strands in the siRNA duplex show reduced
TLR8 activity
[24]. The biological impact of a differential recognition of RNA by TLR7 and
TLR8 is
unknown, and selective activation of human TLR7 by RNA has so far not been
reported.
Natural ssRNAs including mRNA and viral RNA do not present as a single strand
under
physiological conditions but form secondary structures in which the majority
of bases are
paired in double helical stems to minimize free energy [25]. Besides the
standard Watson-
Crick base pairing RNA secondary structure additionally contains wobble (i.e.
non-
canonical) base pairs formed by guanosines and uridines [26]. G=U
(guanosine=uridine)
base pairs can functionally substitute for Watson-Crick base pairs, as they
are nearly
isomorphic, and of comparable thermodynamic stability [27]. RNA stem
structures
containing G=U wobble base pairs are present in virtually all functional RNA
classes and
are therefore a hallmark of ssRNA secondary structure. Moreover, in a wide
range of
biological processes, unique structural, chemical and conformational
characteristics mark
sites containing G=U base pairs for recognition by proteins [27,28].
Here, we examined whether RNA secondary structural elements, in particular
those that
form G=U wobble base pairs, influence ssRNA recognition. We found that short
RNA stems
were highly immunostimulatory, and selectively activated TLR7 when they
contained at
least one G=U wobble base pair. The G=U base pair therefore constitutes the so
far
unappreciated minimal structural motif sufficient to confer TLR7 agonist
activity in single
stranded RNA. In addition, we identified the first phosphodiester RNA ligand,
which
selectively stimulates TLR7 on its own, obviating the need for formulation
with transfection
reagents such as polycations.
Therefore, there is a need in the art to better understand the mechanism by
which TLR7
distinguishes between self and non-self RNA. Specifically, there is a need in
the art to
provide molecules that are specifically recognized by TLR7 and/or activate
TLR7. The
2
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
provision of such molecules allows for the provision of immunostimulatory
nucleic acid
molecules, which are useful for the production of type I IFN in vitro and in
vivo and for
treating various diseases, which can be alleviated or even eradicated by type
I IFN, such as
viral infections, immune disorders and cancers.
SUMMARY OF THE INVENTION
The present invention relates to a method for preparing a RNA polynucleotide
or
oligonucleotide (poly/oligonucleotide), which is capable of inducing an immune
response,
preferably an anti-viral response, more preferably a type I IFN response,
comprising the
-steps-of:
(a) identifying a nucleotide sequence which allows for the formation of at
least one
substantially, preferably fully double-stranded section, wherein the at least
one
double-stranded section comprises at least one G:U base pair, and wherein the
at
least one double-stranded section has a stability that is comparable to that
of a
double-stranded section composed of at least 4, preferably 6 to 11, G:C base
pairs
and at least one, preferably one G:U, base pair or is comparable to a double-
stranded
section composed of at least 8, preferably 10 to 21, A:U base pairs and at
least one,
preferably one G:U, base pair,
(b) producing an RNA poly/oligonucleotide having the nucleotide sequence
identified in
(a), and
(c) optionally testing the ability of the RNA produced in step (b) to induce a
type I IFN
response.
The present invention further relates to a single-stranded or double-stranded
RNA
poly/oligonucleotide comprising at least one substantially, preferably fully
double-stranded
section, wherein the at least one double-stranded section comprises at least
one G:U base
pair, and wherein the at least one double-stranded section has a stability
that is comparable
to that of a double-stranded section composed of at least 4, preferably 6 to
11, G:C base
pairs and at least one, preferably one G:U, base pair or a fully double-
stranded section
composed of at least 8, preferably 10 to 21, A:U base pairs and at least one,
preferably one
G:U, base pair, wherein the poly/oligonucleotide is capable of inducing an
immune
response, preferably an anti-viral response, more preferably a type I IFN
response
The present invention further relates to single-stranded RNA
poly/oligonucleotide
comprising at least one substantially, preferably fully double-stranded
section, wherein the
3
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
at least one double-stranded section has a structure defined by the following
general
formula I:
XnG/UVmNoWmU/GYn (Formula I),
wherein if Xn is followed by G then Wm is followed U, or if Xn is followed by
U then Wm is
followed by G (in order to form a wobble base pair), wherein 2 s n <_ 12, 2:5
m s 12 and 2:5
o s 12; X defines any base that forms Watson- Crick base pairs with
corresponding bases in
Y; V defines any base that forms Watson- Crick base pairs with corresponding
bases in W
in an RNA stem structure; N is any base in a loop; and wherein the total
length of the RNA
defined by Formula I is preferably 15 to 45 bases.
- In a further embodiment, the RNA poly/oligonucleotides of the invention is a
single-stranded
RNA poly/oligonucleotide comprising at least one substantially, preferably
fully double-
stranded section wherein the one strand or a portion of a strand that forms
the at least one
double-stranded section are composed of two RNA-strands having a structure
defined by
the following general formulas II and III,
5'Xõ G/UVm 3'(Formula II) 5'oWmU/GYM 3' (Formula I11),
wherein G/U and U/G are selected that a wobble base pair forms, 2 <_ n s 12, 2
s m <_ 12
and 2:5 o :S 12; X defines any base that forms a Watson- Crick base pair with
corresponding
bases in Y; V defines any base that forms a Watson- Crick base pair with
corresponding
bases in W in an RNA stem structure; N is any base in a loop; and wherein the
total length
of the RNA strands defined by Formula II and III is preferably 5 to 45 bases.
Preferably, X
represents Gs or Cs that form Watson-Crick base pairs with corresponding G and
C bases
in Y, and V represents Gs or Cs that form Watson-Crick base pairs with
corresponding
bases in W.
In one embodiment of the invention, the one strand or a portion of a strand
that forms the at
least one substantially, preferably fully double-stranded section of the
herein defined RNA
poly/oligonucleotide contains n base pairs and preferably consists of p G:U
and q G:C
basepairs, wherein p is an integer less than or equal to n, wherein q is an
integer less than
or equal to n-1, and wherein p+q=n and n is an integer of at least 5,
preferably 6-15.
In a further embodiment of the invention, the one strand or a portion of a
strand that forms
the at least one substantially, preferably fully double-stranded section of
the herein defined
RNA poly/oligonucleotide contains n base pairs and preferably consists of p
G:U and q A:U,
wherein p is an integer less than or equal to n, wherein q is an integer less
than or equal to
n-1, and wherein p+q=n and n is an integer of at least 9, preferably 12-26.
4
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
In a further embodiment, the poly/oligonucleotide of the invention is fully or
partially double-
stranded, whereby one strand in the double-strand contains, preferably
consists of, n G's,
and the other strand in the double-strand contains, preferably consists of, p
U's and q C's,
wherein n, p and q are integers greater than zero, wherein p+q=m, wherein m is
an integer
up to 25, preferably equal to or greater than 5 and less than or equal to 9,
and wherein n is
equal to or greater than m.
In a further embodiment, the poly/oligonucleotide of the invention is single-
stranded and
contains, preferably consists of, a 5' portion and a 3' portion, one of the
portions further
contains, preferably consists of, n G's, wherein the other portion contains,
preferably
consists of, p U's and q C's, wherein n, p and q are integers greater than
zero, wherein
p+q=m, wherein m is an integer up to 25, preferably equal to or greater than 5
and less
than or equal to 9, and wherein n is equal to or greater than m.
In a further embodiment, the poly/oligonucleotide of the invention is fully or
partially double-
stranded, wherein one strand in the double-strand may further contain,
preferably consist
of, n U's, wherein the other strand in the double-strand contains, preferably
consists of, p
G's and q A's, wherein n, p and q are integers greater than zero, wherein
p+q=m, wherein
m is an integer up to 25, preferably equal to or greater than 11 and less than
or equal to 21,
and wherein n is equal to or greater than m.
In a further embodiment, the poly/oligonucleotide of the invention is single-
stranded and
may further contain, preferably consist of, a 5' portion and a 3' portion,
wherein one of the
portions contains, preferably consists of, n U's, and the other portion
contains, preferably
consists of, p G's and q A's. n, p and q are integers greater than zero,
wherein p+q=m,
wherein m is an integer up to 25, preferably equal to or greater than 11 and
less than or
equal to 21, and wherein n is equal to or greater than m.
In one embodiment of the invention the herein defined RNA
poly/oligonucleotide, wherein
one strand or a portion of a strand that makes up the at least one
substantially, preferably
fully double-stranded section contains n base pairs and preferably consists of
p G:U and q
G:C basepairs and r A:U basepairs, wherein p is an integer less than or equal
to n, wherein
q is an integer less than or equal to n-1, and r is an integer less or equal
to n-1, and
wherein p+q+r=n and n is an integer of at least 5, preferably 6-26.
In a further embodiment, the RNA poly/oligonucleotide of the invention is a
single-stranded
poly/oligonucleotide which has at least one stem-and-loop structure, and the
at least one
5
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
double-stranded section contained therein is the stem of the at least one
single-stranded
poly/oligonucleotide.
The invention further relates to the RNA poly/oligonucleotide as defined
above, wherein n is
greater than 100, preferably greater than 1000, more preferably greater than
2000, most
preferably between 3000 and 5000.
In a preferred embodiment, the invention relates to RNA poly/oligonucleotide
as defined
above that show selective TLR7 and/or TLR8 activity. In a further embodiment
the RNA
poly/oligonucleotide prepared by the method defined herein or the RNA
poly/oligonucleotide
described herein does not contain AU base pairs and no unpaired and shows
selective
TLR7 activity.In a more preferred embodiment of the invention the RNA
poly/oligonucleotide
as defined above that show selective TLR7 activity without the need of
complexation to
other reagents, especially an RNA poly/oligonucleotide as defined above,
wherein the
poly/oligonucleotide is substantially, preferably fully or partially double-
stranded, wherein
one strand in the double-strand contains, preferably consists of, n G's,
wherein n is an
integer between 20 and 100, and wherein the other strand in the double-strand
contains,
preferably consists of, p U's and q C's, wherein n, p and q are integers
greater than zero,
wherein p+q=m, wherein m is an integer up to 20, preferably equal to or
greater than 5 and
less than or equal to 9, and wherein n is equal to or greater than m.
In an alternative embodiment, the oligonucleotides show selective TLR7
activity without the
need of complexation to an RNA poly/oligonucleotide as defined above, wherein
n is
greater than 100, preferably greater than 1000, more preferably greater than
2000, most
preferably between 3000 and 5000.
In a preferred embodiment of the invention, the RNA poly/oligonucleotide as
defined above,
comprises one G:U base pair in each double-stranded section, and the G:U base
pair is in
the center of each double-stranded section.
In a further embodiment of the invention a RNA poly/oligonucleotide is
provided showing
selective TLR8 activity, wherein the G nucleoside in the G:U wobble base pair
is adjacent
on each side to C nucleosides.
In a further embodiment of the invention a RNA poly/oligonucleotide is
provided comprising
at least one U that is not involved in RNA base pairing, wherein the
poly/oligonucleotide is
6
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
capable of inducing an immune response, preferably a type I IFN response,
wherein the
immune response is enhanced by the addition of exogenous G nucleoside.
Moreover, the invention provides a pharmaceutical composition comprising at
least one
poly/oligonucleotide as defined above.
In one embodiment of the invention, the pharmaceutical composition further
comprises at
least one agent selected from an immunostimulatory agent, an anti-viral agent,
an anti-
bacterial agent, an anti-tumor agent, IFN-a, and IFN-R.
The invention further provides an in vitro method for inducing type I IFN
production in a cell,
comprising the steps of: contacting a cell with at least one
poly/oligonucleotide as defined
above, optionally mixed with a complexation agent, wherein the cell expresses
TLR7 and is
capable of producing an immune response, preferably an anti-viral response
upon TLR7
activation.
BRIEF DESCRIPTION OF THE FIGURES
Figure 1: Induction of IFN-a by polyU strands is enhanced by polyG counter-
strands.
Human PBMCs (A,C) or human PDCs (B,D) were stimulated with ssRNA or dsRNA
complexed with Poly-L-Arginine. dsRNA was generated by hybridizing two
required ssRNA
oligonucleotides (pG, pU, pA, G21 or U21) in equal quantities (w/w). 20 h
after stimulation,
cell culture supernatants were assed for IFN-a by ELISA. Data shown are
representative of
3 independent experiments presented as mean standard error of mean (s.e.m.)
(.* P s
0.05, and ** P:5 0.01: t-test).
Figure 2: RNA stem structures containing a single G:U base pair are strong
inducers of
type I IFN. (A-C, E) Human PBMC were stimulated with either 1 pg/ml, 0,3 pg/ml
or 0,1
pg/ml or (A-C) or 200 ng, 60 ng or 20 ng RNA (E) complexed with Poly-L-
Arginine. The
cells were stimulated with either ssRNA or dsRNA. dsRNAs were prepared by
hybridizing
various short ssRNA (timer to 21mer, ORNs) to pU (A,B) or pG (C) in equal
quantities
(w/w). (D) 8mer ssRNA (CCCCUCCC) was titrated to a constant amount of pG
(ratio 1/1,
1/0.2, 1/0.05, 1/0.02, 1/0.01) as indicated in the figure. Human PBMC were
stimulated with
2 pg/ml, 0,6 pg/ml or 0,2 pg/ml dsRNA and as control with pG or R805 alone. 20
h after
stimulation, cell culture supernatants were assed for IFN-a by ELISA. Data
shown are
representative for 4 independent experiments.
7
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
Figure 3: Recognition of G:U base pairs is mediated by TLR7. (A) human PBMCs
were
pre-incubated for 30 min with different concentrations of chloroquine as
indicated in the
figure. pG+R805 and pU+1602 were comlexed with Poly-L-Arginine to stimulate
the cells.
As control, PBMC were transfected with pl:C or stimulated with CpG-DNA 2216.
(B) Induction of IFN-a in murine PDC derived from bone marrow cultures in
vitro by pG:pU
and pG+R805. Addition of R1602 strongly enhanced IFN-a levels. (C) Wild-type
mice were
injected i.v. with 25 pg pG, pG+R805, in complex with Dotap. Serum samples
were taken
3, 5 and 8 h after stimulation and serum IFN-a was analyzed by ELISA. The
lines show
cytokine kinetics in individual mice(D) IFN-a-inducing activity of RNAs CpG
oligonucleotide
2336, pU, oligoU (U21) and G:U base pair-containing RNAs (pG+ORN805 and
pU+ORN1602) in FLT3-L PDCs from wild-type (WT) and TLR7" mice that were
stimulated
in vitro with ssRNA or dsRNA complexed to Dotap. 20 h after stimulation, cell
culture
supernatants were assed for IFN-a by ELISA. CpG-DNA 2336 was used as positive
control
in a concentration of 3 pg/ml
Figure 4: G:U base pairs selectively activate TLR7. Human PDC (A, C, E) or
human
monocytes (B, D, F) were stimulated with ssRNA or dsRNA complexed with Poly-L-
Arginine. Supernatants were harvested 20 h after stimulation and IFN-a was
analyzed by
ELISA. Figure (G) shows the preferred secondary structure of ORNs containing a
G=U base
pair.
Figure 5: RNAs containing G and U that are unable to form a G=U base pair are
inert:
Human PDCs were stimulated with RNA complexed to Poly-l-Arginine. After 20 h,
IFN-a
was measured in the supernatants by ELISA. Data shown are representative of 3
independent experiments. (B) Secondary structure of oligonucleotides used in
(A). It is
apparent that even with the overall base composition conserved,
oligonucleotides are inert
when a G=U base pair cannot form. (C) RNA modifications that affect the
integrity of the
G=U base pair interfere with stimulatory activity. Human PDC were stimulated
with RNA
pG+R805 containing one single uridine (R805), phosphothioated-uridine (R805
PTO-U), 2'-
0-methyluridine (R805 2'-O-Me) or pseudouridine (R805 y) in complex with Poly-
L-
Arginine. After 20 h of culture, IFN-a was measured in the supernatants by
ELISA. Data
shown are representative of 3 independent experiments. Modifications of the
Uridine in the
G=U base pair abrogate activity. Shown are modifications to Pseudouridine and
to 2-0-
Methyl-Uridine. In contrast, phosphorthioate modifications that affect the
phosphodiester
linkage to neighboring bases is tolerated.
8
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
Figure 6: G:U base pair forming complexes comprising polyG and R805 are able
to
activate human PDCs without transfection agents. (A) Human PDCs were
stimulated with
pG+R805 either complexed with Poly-L-Arginine (black bar) or without
complexation (white
bar) in the indicated amount. 20 h after stimulation, cell culture
supernatants were assed
for IFN-a by ELISA. Data shown are representative of 5 independent
experiments. (B)
shows the uptake of fluorescent R805 alone, together with polyA, or together
with pG in
human PDCs incubated with fluorescent R805 in the culture supernatant. (C)
Human PDCs
were isolated from Buffy Coats and stimulated with G=U base pair containing
RNA
produced by R805 hybridized to pG in varying lengths (pG, G42, G21, G8). RNA
was given
into the supernatant of PDCs in concentrations of 25 pg/ml, 12.5 pg/ml, 6.25
pg/ml or 3.125
pg/ml. After 20 h, IFN-d was measured in the supernatants. This shows that
oligo G strands
(in particular G42) paired with R805 can be active without added transfection
reagent, but
the activity is much reduced compared to pG.
Figure 7: Addition of guanosine nucleosides strongly enhances the type I IFN
response to
poly uridine RNA. Human PDC were stimulated with pU (A, C) or with U21s (B)
complexed
to Poly-L-Arginine. (A, B) At the same time guanosine-nucleosides (G) in
different
concentrations (0.005 mM, 0.05 mM or 0.25 mM) were added to stimulated or
untreated
PDCs. (C) Different nucleosides (G, cytidine (GQ' , uridine (lQ or adenosine
(J) were added
in a concentration of 0.25 mM to stimulated or untreated PDCs. After 20 h,
cell culture
supernatants were assessed for IFN-a by ELISA. Data shown are representative
of 3
independent experiments.
Figure 8: The length of the RNA strand influences the stimulatory activity of
untransfected,
but not of transfected G=U base pair containing RNA. Human PDC were isolated
from Buffy
Coats and stimulated with G=U base pair containing RNA produced by R805
hybridized to
pG in varying lengths (pG, G42, G21, G8). (A) RNA was given into the
supernatant of PDC in
concentrations of 25 pg/ml, 12.5 pg/ml, 6.25 pg/ml or 3.125 pg/ml. (B) RNA was
complexed
to Poly-I-Arginin and complexes were added into the supernatants in different
concentrations (0.3 pg/ml or 1 pg/ml). (A,B) After 20 h, IFN-a was measured in
the
supernatants. Data shown are representative of 2 independent experiments.
Figure 9: Preferential recognition of G=U base pairs is evident over a wide
range of doses
and in differing RNA contexts. PBMC were isolated from Buffy coats and
stimulated with
varying concentrations of RNA complexed to Poly-I-Arginin. After 20 h IFN-a
was measured
in the supernatants. (A) pU compared to pU+pG and pU+R1 602 (B) U21 compared
to
U21+G21 (C) 9.2s RNA compared to R2115 and R2117 hairpins.
9
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
Figure 10: GU base pairs containing RNA specificly induces IFN-a and IP-10 and
not
proinflammatory cyokines. PBMC were isolated from Buffy coats and stimulated
with 1
pg/ml RNA containing GU base pairs. As positive control, the TLR7/8 isRNA 9.2s
and as
negantive control RNA R2127 was used. RNAs were complexed to Poly-l-Arginine
and
added to the supernatants. After 20 h, cytokines ((A,C) IFN-a, (B,F) IL-12p70,
(D) IP-10
and (F) IL-6 were measured by ELISA.
Figure 11: The presence of pG does not per se impair TLR8 mediated recognition
of RNA.
PBMC were isolated from Buffy coats and stimulated with either isRNA 9.2s
alone (1 pg/ml
- or 05 N9/ml) or isRNA9.2s in presence of O G (0,5 pg/m1-9.2s RNA and 0.5
N9/ml PG).
isRNA 9.2s and pG were mixed before compexation to Poly-l-Arginine. After 20 h
IL-12 p70
was measured in the supernatants.
Figure 12: Enhanced stimulation by RNAs containing G=U base pairs is not
mediated by
preferential uptake of RNA containing multiple guanosines. In contrast to
other RNA
oligonucleotides, untransfeced G21 is readily taken up by PDC and monocytes.
However,
upon complexation with Poly-l-Arginine (pArg) all forms of RNA are taken up
equally well.
Moreover, while constituting the strongest immune stimulus (Figure S5) uptake
of
fluoresencent U21 was rather reduced when hybridized to G21. Cy3 fluorescence
labeled
RNAs were either given untransfected or in complex with to PBMC, and incubated
for 1 h.
Subsequently PBMC were washed and stained for cytometric analysis. Cells that
took up
fluorescent RNA are shown as % positive PDC (BDCA-2+) and Monocytes (CD14+).
Data
shown is representative of three independent experiments.
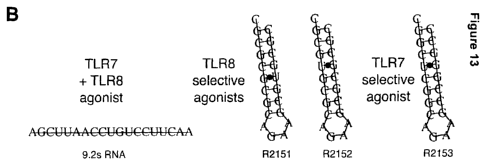
Figure 13: Evidence that some RNA hairpins containing G=U base pairs act as
selective
TLR8 agonists. (A) PBMC were isolated from Buffy coats and stimulated with the
RNAs
shown in (B) in compex with Poly-I-Arginine. After 20 h IFN-a, IL-12 p70 and
IL-6 were
measured in the supernatants by cytokine Elisa. R2151 and R2152 are examples
for
selective TLR8 agonists.
Figure 14: In cells that express both functional TLR7 and TLR8 RNAs containing
G=U
wobble base pairs retain TLR7 selective activity. PMA activated THP-1 cells
express TLR8,
and upon stimulation with IFN-g (100 U/ml) also upregulate functional TLR7.
(A) THP-1
cells were stimulated with the small molecule agonists R848 (TLR7/8 agonist)
C1087 (TLR7
agonist), or RNAs R2153, R2116 (TLR7 agonists) R2127 (inert), and 9.2s (TLR7/8
agonist).
After 20 h TNF-a was measured in the culture supernatant by ELISA. (B) Knock-
down of
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
TLR7 using 3 different lentiviral shRNA constructs in PMA/IFN-g treated THP-1
cells
represses TLR7 but not TLR8 mRNA. Lentiviral vectors without (empty) or with
nonsense
shRNA (scrambled) were used as negative controls. (C) THP-1 cells treated as
in described
in (B) were stimulated with small molecule agonists (R848, C1087 and
C1075[TLR8
agonist]), or the indicated RNAs in compex with Poly-l-Arginine. After 20 h
TNF-a was
measured in the culture supernatant by ELISA. Data shown are representative of
2 (A) or 3
(B, C) independent experiments.
DETAILED DESCRIPTION OF THE INVENTION
Early studies showed that TLR7 was activated by genomic RNA from ssRNA viruses
[4, 6,
17]. Further studies showed that short synthetic ssRNAs were able to stimulate
TRL7 in a
similar manner as single-stranded viral RNA [2, 6, 7, 21, 24].
Even though the stimulation of TLR7 by short ssRNAs has been described as
being
sequence-dependent (Hornung et al, 2004), a large number of ssRNAs with very
different
sequences have been reported to stimulate TLR7. Furthermore, even though
uridine-rich
sequences have been reported to be a molecular motif recognized by TLR7
(Diebold SS et
al, 2006), it has also been reported that ORN lacking uridines were not
immunostimulatory,
and most ORN rich in guanosines and uridines were stimulatory for both, TLR7
and TLR8.
While long RNA molecules activate both, TLR7 and TLR8, particular short
oligoribonucleotide (ORN) that lack guanosines fail to activate TLR7. The
structural basis
for this distinction is unknown. Moreover, the biological impact of a
differential recognition of
RNA by TLR7 and TLR8 is unknown, and selective activation of human TLR7 by RNA
has
so far not been reported.
Surprisingly, the present inventors found that it was not the GU or U content
of a RNA
which determines its TLR7-activating and/or IFN-a-inducing activity, rather,
it was the
presence of a G:U wobble base pair, in particular, a G:U base pair in the
context of a
double-stranded structure. The inventors found that a single G=U wobble base
pair within
otherwise non-stimulatory RNA is sufficient to provide full TLR7 agonist
activity. G=U base
pairs form naturally in the secondary structure assumed by single-stranded
RNA.
Elimination of the G=U wobble base pair abolished TLR7 activity even when the
overall
base composition was maintained. The inventor showed that RNAs that form a G=U
base
pair are able to induce high levels of type I IFN secretion by plasmacytoid
dendritic cells
(PDC), but do not activate human monocytes. Moreover, phosphodiester ORN that
form a
G=U base pair when hybridized with poly guanosine (pG) show high and selective
TLR7
11
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
activity without the need of complexation to other reagents. The
identification of the minimal
structural motif for TLR7 allows the design of RNA-based TLR7 selective
agonists that act
independently of additional components comparable to CpG oligonucleotides for
TLR9.
Furthermore, the present inventors found that an optimal IFN-a-inducing
activity was
observed when a G:U base pair was placed in the center of a double-stranded
structure
which has stability comparable to that of a double-stranded structure formed
by 4-8 G:C
base pairs or 10-20 A:U base pairs in addition to the G:U base pair.
Furthermore, most of
the ssRNAs disclosed in the prior art, which activated TLR7 also activated
TLR8 (review
Heil et al., 2004).
In summary, the present inventors found for the first time RNA poly- and
oligonucleotides,
which activated TLR7 specifically without activating TLR8. These TLR7-specific
ligands are
characterized by a G:U base pair in the center of a double-stranded structure
which is
formed by 4-8 G:C base pairs in addition to the G:U base pair. The present
inventors found
that short RNA stems were highly immunostimulatory, and selectively activated
TLR7 when
they contained at least one G=U wobble base pair. The G=U base pair therefore
constitutes
the so far unappreciated minimal structural motif sufficient to confer TLR7
agonist activity in
single stranded RNA.
Moreover, because TLR7 is located in the endosomal compartments of certain
immune
cells, immunostimulatory RNAs have to be delivered to the endosomes for them
to be able
to stimulate TLR7. Complexation or transfection reagents, such as cationic
polypeptides,
are required for the cellular uptake and delivery of RNAs into the endosomes.
Furthermore,
since RNA with phosphodiester backbone is subject to hydrolysis as well as
ribonuclease
degradation, complexation or transfection agents are required for stabilizing
RNA prior to its
arrival in the target cell or subcellular compartment.
The present inventors found for the first time that certain TLR7-specifc RNA
ligands having
phosphodiester backbone can be taken up by the cells, enter the endosomal
compartments
and activate TLR7 without being complexed with a complexation or transfection
reagent.
Some of these ligands are composed of a long poly-guanosine (polyG or pG)
strand (3000-
5000 nucleotides) and short ssRNA 6-8 nucleotides in length containing one U
in the center
of a stretch of 5-7 C's.
Thus, the inventors identified the first phosphodiester RNA ligand, which
selectively
stimulates TLR7 on its own, and therefore obviating the need for formulation
with
transfection reagents such as polycations.
12
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
The findings of the present inventors made it possible for the first time to
develop TLR7-
specific and highly active RNA agents which can be used for stimulating type I
IFN
production in vitro and in vivo and for treating or preventing diseases which
can be treated
or prevented by type I IFN, such as viral infections, immune disorders and
cancer.
Furthermore, the discovery of immunostimulatory RNAs, which do not require a
complexation or transfection agent for delivery in vitro and in vivo offers
great new research
and clinical possibilities.
Definition
As used herein, "a" and "an" refer to not only a single individual, but also a
group or species
of entities unless otherwise noted.
All terms used herein bear the meanings that are established in the art unless
otherwise
noted. Techniques disclosed herein can be performed by a person skilled in the
art
following the present description and/or established protocols, such as those
disclosed in
Molecular Cloning: A Laboratory Manual (Sambrook et al., 1989, Cold Spring
Harbour
Laboratory, New York), Current Protocols in Molecular Biology (Ausubel et al.,
2007, John
Wiley & Sons, New York), and Current Protocols in Immunology (Coligan et al.,
2007, John
Wiley & Sons, New York).
As used herein, "oligonucleotide" refers to a nucleic acid molecule having 2-
100 nucleotides
and "polynucleotide" refers to a nucleic acid molecule having more than 100
nucleotides.
All sequences are in the 5'->3' direction unless otherwise noted. n
consecutive nucleotides
X is written as Xn when n is an integer greater than zero. For example,
XXXYXXXXXZXX
is written as X3YX5ZX2. When n is greater than 100, Xn can also be written as
polyX or pX.
Method for Preparing a Polynucleotide or an Oligonucleotide
The present invention provides a method for preparing an RNA polynucleotide or
oligonucleotide (hereinafter "poly/oligonucleotide") which is capable of
inducing an immune
response, preferably an anti-viral response, more preferably a type I IFN
response, more
specifically, an IFN-a response, comprising the steps of:
(a) identifying a nucleotide sequence which allows for the formation of at
least one
double-stranded section, wherein the at least one full double-stranded section
13
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
comprises at least one G:U base pair, and wherein the at least one double-
stranded
section has a stability that is comparable to that of a double-stranded
section
composed of at least 4, preferably 4 to 8, G:C base pairs and at least one,
preferably one G:U base pair or is comparable to a double-stranded section
composed of at least 8, preferably 10 to 20, A:U base pairs and at least one,
preferably one G:U, base pair;
(b) producing an RNA poly/oligonucleotide having the nucleotide sequence
identified in
(a); and
(c) optionally testing the ability of the RNA produced in step (b) to induce a
type I IFN
response.
The poly/oligonucleotide may be produced by any suitable means known in the
art, such as
chemical synthesis and in vitro transcription. In the case of a double-
stranded
poly/oligonucleotide, the two strands may be produced by the same or different
methods.
The ability of a poly/oligonucleotide to induce a type I IFN response can be
tested by any
suitable methods known in the art, such as those disclosed in the Examples.
Polynucleotides and Oligonucleotides
The present invention provides an RNA poly/oligonucleotide, which is capable
of inducing
an immune response, preferably an antiviral response, more preferably a type I
IFN
response, most preferably an IFN-a response, obtained by the method described
above.
In particular, the present invention provides a poly/oligonucleotide which
comprises at least
one substantially, preferably fully double-stranded section, wherein the at
least one double-
stranded section comprises at least one G:U base pair, and wherein at least
one double-
stranded section has a stability that is comparable to that of a double-
stranded section
composed of at least 4, preferably 6 to 11, G:C base pairs and at least one,
preferably one
G:U, base pair or a double-stranded section composed of at least 10,
preferably 10 to 21,
A:U base pairs and at least one, preferably one G:U, base pair.
By "fully double-stranded", it is meant that the double-stranded section does
not contain
mismatches that compromise a minimal stability of the double-strand of said
section. A
section is fully double-stranded when the sequences of the two stretches of
nucleic acid
forming the section are 100% complementary to each other. Two nucleotides are
said to
be complementary to each other if they can form a base pair, either a Waston-
Crick base
pair (A-U, G-C) or a wobble base pair (U-G, U-A, I-A, I-U, I-C).
14
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
By "substiantially fully double-stranded", it is meant that the double-
stranded section as
defined above may contain mismatches or single base insertions up to 10% of
the total
number of bases, provided the that the substiantially fully double-stranded
section
maintains a minimal stability of the double-strand of said section.
The stability of a substantially, preferably fully double-stranded section can
be determined
by a skilled person using methods known in the art. In a preferred embodiment,
the stability
is determined by the software DNamelt
(http://www.bioinfo.rpi.edu/applications/hybrid/)
based on Markham N R & Zuker M (2005) and Markham NR & Zuker M (2008).
By "comparable" it is meant that the stability of a substantially, preferably
fully double-
stranded section is 75%-125%, preferably 80-120%, more preferably 90-110%,
most
preferably 95-105% of that of a double-stranded section composed of at least
4, preferably
6 to 11, G:C base pairs and at least one, preferably one G:U, base pair or a
section
composed of at least 8, preferably 10 to 21, A:U base pairs and at least one,
preferably one
G:U, base pair. Preferably, the stability of a substantially, preferably fully
double-stranded
section is within a range with the lower limit being 75%, preferably 80%, more
preferably
90%, most preferably 95% of the stability of a double-stranded section
composed of 4 G:C
base pairs and one G:U base pair or a section composed of 10 A:U base pairs
and one G:U
base pair and the upper limit being 125%, preferably 120%, more preferably
110%, most
preferably 105% of the stability of a double-stranded section composed of 8
G:C base pairs
and one G:U base pair or a section composed of 20 A:U base pairs and one G:U
base pair.
In one embodiment, the one strand or a portion of a strand that forms the at
least one fully
double-stranded section contains, preferably consists of, n G's, and the other
strand or a
portion of a strand that forms the same at least one fully double-stranded
section contains,
preferably consists of, p U's and q C's, wherein n is an integer equal to or
greater than 5
and less than or equal to 9, wherein p is an integer less than or equal to n,
wherein q is an
integer, and wherein p+q=n.
In one embodiment, the one strand or a portion of a strand that forms the at
least one fully
double-stranded section contains, preferably consists of, n U's, and the other
strand or a
portion of a strand that forms the same at least one fully double-stranded
section contains,
preferably consists of, p G's and q A's, wherein n is an integer equal to or
greater than 11
and less than or equal to 21, wherein p is an integer less than or equal to n,
wherein q is an
integer, and wherein p+q=n.
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
In one embodiment, each fully double-stranded section comprises only one G:U
base pair
in the center of the fully double-stranded section. When the fully double-
stranded section is
composed of an odd number (2n+1) of base pairs, the center of the section is
base pair
number n+1. When the fully double-stranded section is composed of an even
number, 2n,
of base pairs, the center of the section can be either base pair number n or
n+1.
In one embodiment, the poly/oligonucleotide is a double-stranded
poly/oligonucleotide.
In a preferred embodiment, the double-stranded poly/oligonucleotide is fully
double-
stranded. By "fully double-stranded" it is meant within the context of this
preferred
embodiment that the two strands forming the poly/oligonucleotide have the same
length
and have sequences which are 100% complementary to each other.
In one embodiment, one strand in the double strand contains, preferably
consists of, n G's,
and the other strand in the double strand contains, preferably consists of, p
U's and q C's,
wherein n is an integer equal to or greater than 5 and less than or equal to
9, wherein p is
an integer less than or equal to n, wherein q is an integer, and wherein
p+q=n. In a
preferred embodiment, p equals 1 and U is in the center of the strand
containing or
consisting of U and C's.
In a preferred embodiment, one strand in the double strand contains,
preferably consists of,
n U's, and the other strand in the double strand contains, preferably consists
of, p G's and q
A's, wherein n is an integer equal to or greater than 11 and less than or
equal to 21,
wherein p is an integer less than or equal to n, wherein q is an integer, and
wherein p+q=n.
In a preferred embodiment, p equals 1 and G is in the center of the strand
containing or
consisting of G and A's.
In one embodiment of the invention the herein defined RNA
poly/oligonucleotide, wherein
one strand or a portion of a strand that makes up the at least one
substantially, preferably
fully double-stranded section contains n base pairs and preferably consists of
p G:U and q
G:C basepairs and r A:U basepairs, wherein p is an integer less than or equal
to n, wherein
q is an integer less than or equal to n-1, and r is an integer less or equal
to n-1, and
wherein p+q+r=n and n is an integer of at least 5, preferably 6-26.
The position of the G-U base pairs in the double stranded
poly/oligonucleotides as defined
herein is not limited and may be at any position well-off the center in the
double stranded
section, preferably the G-U base pairs are located in the center in the double
stranded
section.
16
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
The double-stranded section preferably contains uridines (one or multiple)
exclusively in the
form of G=U base pairs.
Specific Examples of a fully double-stranded poly/oligonucleotide include:
G5 + C2UC2 (CCUCC),
G6 + C3UC2 (CCCUCC),
G6 + C2UC3 (CCUCCC),
G7 + C3UC3 (CCCUCCC),
G8 + C4UC3 (CCCCUCCC),
G8 + C3UC4 (CCCUCCCC),
G9 + C4UC4 (CCCCUCCCC),
U11 + A5GA5,
U12 + A6GA5,
U12 + A5GA6,
U13 + A6GA6,
U14 +A7GA6,
U14 + A6GA7i
U15 +A7GA7i
U16 + A8GA7,
U16 + A7GA8,
U17 + A8GA8,
U18 + A9GA8,
U18 +A8GA9,
U19 +A9GA9,
U20 + A10GA9 ,
U20 + A9GA10 ,
U21 + A10GA10,
U21 + A5GA1OGA4 .
In another preferred embodiment, the double-stranded poly/oligonucleotide is
partially
double-stranded. By "partially double-stranded" it is meant that the two
strands forming the
poly/oligonucleotide have different lengths, sequences which are not 100%
complementary
to each other, or both. In other words, the fully double-stranded section is
connected with a
single-stranded structure at one or both ends.
In one embodiment, one strand in the double strand contains, preferably
consists of, n G's,
and the other strand in the double strand contains, preferably consists of, p
U's and q C's,
17
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
wherein n, p and q are integers greater than zero, wherein p+q=m, wherein m is
an integer
up to 25, preferably equal to or greater than 5 and less than or equal to 9,
and wherein n is
greater than m.
In one embodiment, n is less than or equal to 100. In another embodiment, n is
greater
than 100, preferably greater than 1000. In a poly/oligonucleotide preparation
containing
more than one poly/oligonucleotide molecule, n may be different in different
poly/oligonucleotide molecules. In other words, a poly/oligonucleotide
preparation of the
present invention may contain a mixture of molecules, which have different
length in the
one strand which contains or consists of n G's. In a preferred embodiment, the
G-
-containing--strand-is- a poly=guanosine (polyG or pG) containing- about-3000-
5000-G-8---
obtained by conventional enzymatic processes, such as polynucleotide
phosphorylase
(PNPase) enzymatic reaction. In a preferred embodiment, p equals 1 and U is in
the center
of the strand containing or consisting of U's and C's.
In one embodiment, one strand in the double strand contains, preferably
consists of, n U's,
and the other strand in the double strand contains, preferably consists of, p
G's and q A's,
wherein n, p and q are integers greater than zero, wherein p+q=m, wherein m is
an integer
up to 25, preferably equal to or greater than 11 and less than or equal to 21,
and wherein n
is greater than m.
In one embodiment, n is less than or equal to 100. In another embodiment, n is
greater
than 100, preferably greater than 1000. In a poly/oligonucleotide preparation
containing
more than one poly/oligonucleotide, n may be different in different
poly/oligonucleotide
molecules. In other words, a poly/oligonucleotide preparation of the present
invention may
contain a mixture of molecules, which have different length in the one strand
which contains
or consists of n U's. In a preferred embodiment, the U-containing strand is a
poly-uridine
(polyU or pU) comprising about 3000-5000 U's obtained by conventional
enzymatic
processes, such as polynucleotide phosphorylase (PNPase). In a preferred
embodiment, p
equals 1 and G is in the center of the strand containing or consisting of G
and A's.
Examples of a partially double-stranded poly/oligonulceotide include:
Gn + C2UC2 (CCUCC), wherein n is an integer greater than 5;
Gn + C3UC2 (CCCUCC), wherein n is an integer greater than 6;
Gn + C2UC3 (CCUCCC), wherein n is an integer greater than 6;
Gn + C3UC3 (CCCUCCC), wherein n is an integer greater than 7;
18
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
Gn + C4UC3 (CCCCUCCC), wherein n is an integer greater than 8;
Gn + C3UC4 (CCCUCCCC), wherein n is an integer greater than 8;
Gn + C4UC4 (CCCCUCCCC), wherein n is an integer greater than 9;
Gn + C2UC3UC2 (CCUCCCUCC) , wherein n is an integer greater than 9
Un + A4G4, wherein n is an integer greater than 9;
Un + A4G5, wherein n is an integer greater than 10;
Un + A5GA4, wherein n is an integer greater than 10;
Un + A5GA5 , wherein n is an integer greater than 11;
Un + A6GA5, wherein n is an integer greater than 12;
Un + A5GA6, wherein n is an integer greater than 12;
-Un-+ A6GA6 , wherein n is-an-integer greater than 13;
Un + A7GA6, wherein n is an integer greater than 14;
Un + A6GA7, wherein n is an integer greater than 14;
Un + A7GA7 , wherein n is an integer greater than 15;
Un + A8GA7, wherein n is an integer greater than 16;
Un + A,GA8 , wherein n is an integer greater than 16;
Un + A8GA8 , wherein n is an integer greater than 17;
Un + A9GA8 , wherein n is an integer greater than 18;
Un + A8GA9, wherein n is an integer greater than 18;
Un + A9GA9 , wherein n is an integer greater than 19;
Un + A,0GA9 , wherein n is an integer greater than 20;
Un + A9GA10, wherein n is an integer greater than 20;
Un + A,0GA10 , wherein n is an integer greater than 21;
Un + A5GA,OGA4 , wherein n is an integer greater than 21;
Preferably, n is greater than 1000, more preferably greater than 3000, most
preferably
about 3000-5000 in the examples above. Most preferably, Gn is polyG containing
about
3000-5000 G's and obtained from conventional enzymatic processes, such as
polynucleotide phosphorylase (PNPase) enzymatic reaction.
In one embodiment, the poly/oligonucleotide is a single-stranded
poly/oligonucleotide.
In a preferred embodiment, the single-stranded poly/oligonucleotide is
completely self-
complementary. By "completely self-complementary" it is meant that the 5' half
of the
molecule is 100% complementary to the 3' half of the molecule. As a result,
the molecule
forms a single fully double-stranded section.
19
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
In one embodiment, one of the 5' and 3' halves contains, preferably consists
of, n G's, and
the other half contains, preferably consists of, p U's and q C's, wherein n is
an integer equal
to or greater than 5 and less than or equal to 9, wherein p is an integer less
than or equal to
n, wherein q is an integer, and wherein p+q=n. In a preferred embodiment, p
equals 1 and
U is in the center of the half containing or consisting of U and C's.
In one embodiment, one of the 5' and 3' halves contains, preferably consists
of, n U's, and
the other half contains, preferably consists of, p G's and q A's, wherein n is
an integer equal
to or greater than 11 and less than or equal to 21, wherein p is an integer
less than or equal
to n, wherein q is an integer, and wherein p+q=n. In a preferred embodiment, p
equals 1
and G is in the center of the half containing or consisting of G and A's.
Examples of a completely self-complementary single-stranded
poly/oligonucleotide include:
C2UC2G5
C3UC2G6,
C2UC3G6,
C3UC3G7,
C4UC3G8,
C3UC4G8,
C4UC4G9,
G5C2UC2,
G6C3UC2,
G6C2UC3,
G7C3UC3,
GSC4UC3,
G8C3UC4,
G9C4UC4,
A5GA5U11 ,
A6GA5U12,
A5GA6U12,
A6GA6U13,
A7GA6U14,
A6GA7U14,
A7GA7U15,
A8GA7U16,
A7GA8U16,
A8GA8U17,
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
A9GA8U18,
A8GA9U18,
A9GA9U19 ,
A1oGA9U20,
A9GA10U20 ,
A1oGA10U21
A5GA10GA4U21 ,
U11A5GA5 ,
U 12A6GA5 ,
U 12A5GA6 ,
U13A6GA6,
U14A7GA6,
U14A6GA7,
U 15A7GA7 ,
U16A8GA7,
U 16A7GA8 ,
U 17A8GA8 ,
U18A9GA8,
U 18A8GA9 ,
U, 9A9GA9 ,
U2oA1oGA9 ,
U2oA9GA1o ,
U2,A,oGA,o
U21A5GA10GA4.
In another preferred embodiment, the single-stranded poly/oligonucleotide has
at least one
stem-and-loop structure, and the at least one fully double-stranded section is
the stem of
the at least one single-stranded poly/oligonucleotide.
The formation of a stem-and-loop structure can be readily predicted by a
person skilled in
the art on the basis of the nucleotide sequence of the poly/oligonucleotide
and
experimentally verified by methods known in the art. For example, a ssRNA
oligonculeotide
can be digested with a single-strand-specific RNase and analysed on a
denaturing gel.
The exact size and the sequence of the loop are not critical; it is only
critical that the loop
does not adversely affect the formation and the stability of the stem.
21
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
The present invention relates to poly/oligonucleotides having a structure
defined by the
following general formula I:
XnG/UVmNoWmU/GYn (Formula I),
wherein if Xn is followed by G then Wm is followed U, or if Xn is followed by
U then
Wm is followed by G (in order to form a wobble base pair) , wherein
2sn<_12,2<_m<_12and 2<_o512;
X defines any base that forms a Watson-Crick base pair with corresponding
bases in Y;
V defines any base that forms a Watson-Crick base pair with corresponding
bases in W in
an RNA stem structure, and
N is any base in a loop.
Also encompassed are repeats of the base paired cassettes. The total length of
the RNA
containing the pattern of Formula I is preferably 15 to 45 bases.
Preferred poly/oligonucleotides of the invention have a structure defined by
the following
general formula II and III, defining two separate RNA strands containing the
cassettes:
5'XnG/UVm 3'(Formula II) 5'oWmU/GYn 3' (Formula III),
wherein G/U and U/G are selected that a wobble base pair forms,
with 2:5 n:5 12, 2:5 m:5 12 and 2:5 o:5 12;
X defines any base that forms a Watson-Crick base pair with corresponding
bases in Y;
V defines any base that forms a Watson-Crick base pair with corresponding
bases in W in
an RNA stem structure, and
N is any base in a loop,
The total lenght of these separate RNA strands of Formula II and III is
preferably 5 bases to
45 bases.
Most preferred poly/oligonucleotides of the invention having at least one stem-
and-loop
structure have a structure defined by the following general formula 11 and
III, defining two
separate RNA strands containing the cassettes:
5'XnG/UVm 3'(Formula II) 5'oWmU/GYn 3' (Formula III),
22
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
wherein G/U and U/G are selected that a wobble base pair forms,
with 2sns 12,2sms 12and25os 12;
with X denoting Gs or Cs that form Watson-Crick bases pairs with corresponding
G and C
bases in Y, and V represents Gs or Cs that form Watson-Crick base pairs with
corresponding bases in W.
Repeats of the base paired cassettes (i.e. multiple G=U base pairs in double
stranded
structures) will have activity.
A preferred total length of these separate RNA strands is 5 bases to 45 bases.
In one embodiment of the RNA poly/oligonucleotide of the invention, the one
strand or a
portion of a strand that forms the at least one substantially, preferably
fully double-stranded
section contains n base pairs and preferably consists of p G:U and q G:C
basepairs,
wherein p is an integer less than or equal to n, wherein q is an integer less
than or equal to
n-1, and wherein p+q=n and n is an integer of at least 5, preferably 6-15.
In a further embodiment, RNA poly/oligonucleotide of the invention, the one
strand or a
portion of a strand that forms the at least one substantially, preferably
fully double-stranded
section contains n base pairs and preferably consists of p G:U and q A:U,
wherein p is an
integer less than or equal to n, wherein q is an integer less than or equal to
n-1, and
wherein p+q=n and n is an integer of at least 9, preferably 12-26.
The design of such RNA poly/oligonucleotide as defined above allows the
generation of
RNAs having TLR7 and TLR8 activity.
In one embodiment, one of the two portions of the single-stranded
poly/oligonucleotide
which make up the at least one fully double-stranded stem structure contains,
preferably
consists of, n G's, and the other portion which forms the same at least one
double-stranded
stem structure contains, preferably consists of, p U's and q C's, wherein n is
an integer
equal to or greater than 5 and less than or equal to 9, wherein p is an
integer less than or
equal to n, wherein q is an integer, and wherein p+q=n. In a preferred
embodiment, p
equals 1 and U is in the center of the portion containing or consisting of U
and C's.
In a specific embodiment, the single-stranded poly/oligonucleotide contains,
preferably
consists of, two portions, a 5' portion and a 3' portion, wherein one of the
two portions
contains, preferably consists of, n G's, wherein the other portion contains,
preferably
23
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
consists of, p U's and q C's, wherein n, p and q are integers greater than
zero, wherein
p+q=m, wherein m is an integer up to 25, preferably equal to or greater than 5
and less
than or equal to 9, and wherein n is greater than m. In one embodiment, n is
less than or
equal to 100. In another embodiment, n is greater than 100, preferably greater
than 1000,
more preferably greater than 3000, most preferably on the order of 3000-5000.
In certain
embodiments, n may be different in different molecules in a
poly/oligonucleotide preparation
of the present invention. In a preferred embodiment, p equals 1 and U is in
the center of
the portion containing or consisting of U and C's.
In one embodiment, one of the two portions of the single-stranded
poly/oligonucleotide
which-make-up--the at least-one fully -double-stranded- stem structure-
contains;- preferably
consists of, n U's, and the other portion which forms the same at least one
fully double-
stranded stem structure contains, preferably consists of, p G's and q A's,
wherein n is an
integer equal to or greater than 11 and less than or equal to 21, wherein p is
an integer less
than or equal to n, wherein q is an integer, and wherein p+q=n. In a preferred
embodiment,
p equals 1 and G is in the center of the portion containing or consisting of G
and A's.
In a specific embodiment, the single-stranded poly/oligonucleotide contains,
preferably
consists of, two portions, a 5' portion and a 3' portion, wherein one of the
two portions
contains, preferably consists of, n U's, and the other portion contains,
preferably consists
of, p G's and q A's, wherein n, p and q are integers greater than zero,
wherein p+q=m,
wherein m is an integer up to 25, preferably equal to or greater than 11 and
less than or
equal to 21, and wherein n is greater than m. In one embodiment, n is less
than or equal to
100. In another embodiment, n is greater than 100, preferably greater than
1000, more
preferably greater than 3000, most preferably on the order of 3000-5000. In
certain
embodiments, n may be different in different molecules in a
poly/oligonucleotide preparation
of the present invention. In a preferred embodiment, p equals 1 and G is in
the center of
the portion containing or consisting of G and A's.
Examples of a single-stranded poly/oligonucleotide having at least one stem-
and-loop
structure include:
C2UC2Gn, wherein n is an integer equal to or greater than 4;
preferably greater than 8 to 9.
C3UC2Gn, wherein n is an integer greater than 6;
C2UC3Gn, wherein n is an integer greater than 6;
C3UC3Gn, wherein n is an integer greater than 7;
24
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
C4UC3Gn, wherein n is an integer greater than 8;
C3UC4Gn, wherein n is an integer greater than 8;
C4UC4Gn, wherein n is an integer greater than 9;
GnC2UC2, wherein n is an integer greater than 5;
GnC3UC2, wherein n is an integer greater than 6;
GnC2UC3i wherein n is an integer greater than 6;
GnC3UC3, wherein n is an integer greater than 7;
GnC4UC3, wherein n is an integer greater than 8;
GnC3UC4, wherein n is an integer greater than 8;
GnC4UC4, wherein n is an integer greater than 9;
e " s n, wherein n is an-An eger greater an 14;-
A6GA5Un, wherein n is an integer greater than 12;
A5GA6Un, wherein n is an integer greater than 12;
A6GA6Un, wherein n is an integer greater than 13;
A7GA6Un, wherein n is an integer greater than 14;
A6GA7Un, wherein n is an integer greater than 14;
A7GA7Un, wherein n is an integer greater than 15;
A8GA7Un, wherein n is an integer greater than 16;
A7GA8Un, wherein n is an integer greater than 16;
A8GA8Un, wherein n is an integer greater than 17;
A9GA8Un, wherein n is an integer greater than 18;
A8GA9Un, wherein n is an integer greater than 18;
A9GA9Un, wherein n is an integer greater than 19;
A,0GA9Un, wherein n is an integer greater than 20;
A9GA10Un, wherein n is an integer greater than 20;
A10GAt0Un, wherein n is an integer greater than 21;
A5GA10GA4Un, wherein n is an integer greater than 21;
UnA5GA5, wherein n is an integer greater than 11;
UnA6GA5, wherein n is an integer greater than 12;
UnA5GA6, wherein n is an integer greater than 12;
UnA6GA6, wherein n is an integer greater than 13;
UnA,GA6, wherein n is an integer greater than 14;
UnA6GA7, wherein n is an integer greater than 14;
UnA7GA7, wherein n is an integer greater than 15;
UnA8GA7, wherein n is an integer greater than 16;
UnA7GA8, wherein n is an integer greater than 16;
UnA8GA8, wherein n is an integer greater than 17;
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
UnA9GA8, wherein n is an integer greater than 18;
UnA8GA9, wherein n is an integer greater than 18;
UnA9GA9, wherein n is an integer greater than 19;
UnA1OGA9, wherein n is an integer greater than 20;
UnA9GA10, wherein n is an integer greater than 20;
UnA10GA10, wherein n is an integer greater than 21;
UnA5GA10GA4i wherein n is an integer greater than 21.
Preferably, n is greater than 1000, more preferably 3000, even more preferably
on the order
of 3000-5000 in the examples above.
The present invention also provides RNA poly/oligonucleotides that have the
properties of
activating TLR7 via the herein described G=U containing cassettes, but do not
require
additional facilitators of cellular uptake (e.g. lipids, poly cations,
transfection reagents).
These RNA poly/oligonucleotides form at least one G=U cassette when hybridized
or
connected via a loop to a highly G rich strand. G rich strands are either
obtained by
standard synthesis (preferred length 20 - 100 bp), or by enzymatic synthesis
with an
optimal length between 1000 and 5000 bases. Such strands of due to the
enzymatic
process poorly defined length are commonly synthesized by polynucleotide
phosphorylase
(PNPase). It is likely that the presence of few non-G bases (e.g. >: 1/6 of
the bases could be
I, A, U, C) will not interfere with the properties of the G rich strand. It is
also expected that
modifications known to stabilize RNA (e.g. desoxinucleotides, phosphothioates,
etc.) will
not restrict activity, and may thus be desirable.
The present invention also provides Um + Gn which is capable of inducing an
immune
response, in particular anti-viral response, more particular a type I IFN
response, more
specifically, an IFN-a response, wherein both, m and n, are integers between
1000-5000,
and wherein m is greater than or equal to n. These nucleic acids are generated
enzymatically by Polynucleotide Phosphorylase (PNPase).
In preferred embodiments, the poly/oligonucleotide of the invention is
specifically
recognized by and/or specifically activates TLR7 without being recognized by
or activating
TLR8.
Activitity conferred by the G:U base pair can be readily determined by testing
the RNA
poly/oligonucleotide of the invention (such as those consisting exclusively of
G=U and G:C
base pairs), whether the immune stimulatory activity is lost when either one
of the following
26
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
mutations is applied: a) The U of all G=U base pairs and the G of flanking G:C
base pairs
swap positions (in this test the overall base composition is maintained) or b)
The G of all
G=U base pairs is mutated to A.
Binding of a poly/oligonucleotide to TLR7 and/or TLR8 can be readily
determined by
methods known in the art. For example, a poly/oligonucleotide conjugated to a
label (e.g., a
radioactive molecule, a fluorescent molecule, an enyzme) can be delivered to
cells
expressing TLR7 and/or TLR8, and the binding of the labelled
poly/oligonucleotide to TLR7
and/or TLR8 may be determined using techniques such as gel electrophoresis,
microscopy,
X-ray chrystallography. Activation of TLR7 and/or TLR8 by a
poly/oligonucleotide can also
be readily determined by methods known in the art. For example, cells
expressing TLR7
and/or TLR8 can be stimulated with a poly/oligonucleotide, and the production
of type I IFN
(in particular, IFN-(x) and IL-12 can be used as readouts for the activation
of TLR7 and
TLR8, respectively. PDCs obtained from wild-type, TLR7"" and TLR8-- mice are
ideal for
such tests. Activation of TLR7 and/or TLR8 upon stimulation by a
poly/oligonucleotide can
also readily determined by detecting Overexpression of human and/or mouse TLR7
or
TLR8 in non-immune reporter cells (e.g. HEK293T cells). Gene silencing may be
used to
reduce TLR7/8 expression. Gene silencing activity may be combined with the
present
structural requirements. Within the 16 - 21 bp siRNA duplex that usually
consists
exclusively of Watson-Crick base pairs G:U base pairs can be introduced
without an
expected penalty to the silencing activity, as long as the alterations to
introduce G=U base
pairs are limited to the sense strand.
The G=U base pair containing cassettes as defined herein may also be inserted
into RNAs
that exert an additional function, for example siRNA, ribozymes, aptamers.
TLR7-specific ligands include poly/oligonucleotides in which one strand or a
portion of a
strand that forms the at least one fully double-stranded section contains,
preferably consists
of, n G's, and the other strand or a portion of a strand that forms the same
at least one fully
double-stranded section contains, preferably consists of, p U's and q C's,
wherein n is an
integer equal to or greater than 5 and less than or equal to 9, wherein p is
an integer less
than or equal to n, wherein q is an integer, and wherein p+q=n.
The poly/oligonucleotide may contain any naturally-occurring, synthetic,
modified
nucleotides, or a mixture thereof, as long as the synthetic and/or modified
nucleotides do
not compromise (i.e., reduce) the type I IFN-inducing activity of the
poly/oligonucleotide.
The poly/oligonucleotide may contain any naturally-occurring, synthetic,
modified
27
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
internucleoside linkages, or a mixture thereof, as long as the linkages do not
compromise
the type I IFN-inducing activity of the poly/oligonucleotide.
In a further embodiment of the invention a RNA poly/oligonucleotide is
provided showing
selective TLR8 activity, wherein the U nucleoside in the G:U wobble base pair
is adjacent
on each side to G bases. The RNA poly/oligonucleotide showing selective TLR8
activity
may be single stranded or partially double stranded. Preferably the RNA
poly/oligonucleotide having G nucleoside in the G-U wobble base pair adjacent
to C
nucleoside at each side of the G:U wobble base pair, forms a hairpin structure
as defined
herein, wherein n is between 5 and 100, preferably 20 and 100, and. Said RNA
poly/oligonucleotide shows highly selective TLR8 activity. Said RNA
poly/oligonucleotide
showing selective TLR8 activity, preferably has at least 2 adjacent base pairs
on each side
of the G:U wobble base pair. The term "selective TLR8 activity" as used herein
means the
RNA poly/oligonucleotide as defined above shows TLR8 activity but no TLR7
activity.
TLR7-specific ligands also include poly/oligonucleotides capable of inducing
an immune
response that comprise at least one uridine, preferably polyuridine, that
is/are not involved
in RNA base pairing, wherein the immune response, preferably a type I IFN
response, is
enhanced by the addition of exogenous G nucleoside, whereby base pairing with
guanosine
nucleoside enhances the activity of uridine to activate.
Various methods for producing poly/oligonucleotides are known in the art,
including, but are
not limited to, chemical synthesis and in vitro transcription.
The poly/oligonucleotide may be modified covalently or non-covalently to
improve its
chemical stability, resistance to nuclease degradation, ability to across
cellular and/or
subcellular membranes, target (organ, tissue, cell type, subcellular
compartment)-
specificity, pharmacokinetic properties, biodistribution, or any combinations
thereof. For
example, phosphorothioate linkage(s) and/or pyrophosphate linkage(s) may be
introduced
to enhance the chemical stability and/or the nuclease resistance of an RNA
poly/oligonucleotide. In another example, the poly/oligonucleotide may be
covalently linked
to one or more lipophilic group(s) or molecule(s), such as a lipid or a lipid-
based molecule,
preferably cholesterol or a derivative thereof. Preferably, the modification
does not comprise
the type I IFN-inducing activity of the poly/oligonucleotide. Alternatively, a
reduction in the
type I IFN-inducing activity of the poly/oligonucleotide caused by the
modification is off-set
by an increase in the stability and/or delivery and/or other properties as
described above.
28
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
The poly/oligonucleotide of the present invention may have one or more of the
features
described above in any combinations.
The present invention also provides a poly/oligonucleotide preparation
comprising at least
one poly/oligonucleotide of the present invention. The poly/oligonucleotide
preparation of
the present invention may comprise two or more different
poly/oligonucleotides. Preferably,
the two or more different poly/oligonucleotides comprised in the same
preparation share the
same fully double-stranded section and have similar structure. The variations
in the
chemical compositions of the poly/oligonucleotides in a preparation may result
from the
method of preparation.
Pharmaceutical Composition
The present invention provides a pharmaceutical composition comprising at
least one of the
RNA polynucleotide or oligonucleotide (poly/oligonucleotide) of the invention
described
above and a pharmaceutically acceptable carrier.
By "at least one" it is meant that one or more poly/oligonucleotide
preparation(s) of the
same or different poly/oligonucleotide(s) can be used together.
In one embodiment, the pharmaceutical composition further comprises an agent,
which
facilitates the delivery of the poly/oligonucleotide into a cell, in
particular, into the endosomal
compartments of the cell.
In one embodiment, the delivery agent is a complexation agent, which forms a
complex with
the poly/oligonucleotide and facilitates the delivery of the
poly/oligonucleotide into cells.
Complexation agents are also referred to as "transfection agents" in the art.
Any
complexation agent, which is compatible with the intended use of the
pharmaceutical
composition can be employed. Examples for transfection agents are
Protamine(s).
Examples of complexation agents include polymers and biodegradable
microspheres. The
polymer is preferably a cationic polymer, more preferably a cationic lipid.
Examples of a
polymer include polyethylenimine (PEI) such as in vivo-jetPEITM (PolyPlus) and
collagen
derivatives. Examples of biodegradable microspheres include liposomes,
virosomes,
stable-nucleic-acid-lipid particles (SNALPs), ISCOMATRIX (CSL Limited), and
poly (D,L-
lactide-co-glycolide) copolymer (PLGA) microspheres.
29
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
In another embodiment, the delivery agent is a virus, preferably a replication-
deficient virus.
The poly/oligonucleotide to be delivered is contained in the viral capsule and
the virus may
be selected based on its target specificity. Examples of useful viruses
include
polymyxoviruses which target upper respiratory tract epithelia and other
cells, hepatitis B
virus which targets liver cells, influenza virus which targets epithelial
cells and other cells,
adenoviruses which targets a number of different cell types, papilloma viruses
which targets
epithelial and squamous cells, herpes virus which targets neurons,
retroviruses such as HIV
which targets CD4+ T cells, dendritic cells and other cells, modified Vaccinia
Ankara which
targets a variety of cells, and oncolytic viruses which target tumor cells.
Examples of
oncolytic viruses include naturally occurring wild-type Newcastle disease
virus, attenuated
strains of reovirus, vesicular stomatitis virus (VSV), and genetically
engineered mutants of-
herpes simplex virus type 1 (HSV-1), adenovirus, poxvirus and measles virus.
In addition to being delivered by a delivery agent, the poly/oligonucleotide
and/or the
pharmaceutical composition can be delivered into cells via physical means such
as
electroporation, shock wave administration, ultrasound triggered transfection,
and gene gun
delivery with gold particles.
The pharmaceutical composition may further comprise another agent such as an
agent that
stabilizes the oligonucleotide. Examples of a stabilizing agent include a
protein that
complexes with the oligonucleotide to form an iRNP, chelators such as EDTA,
salts, and
RNase inhibitors.
In certain embodiments, the pharmaceutical composition further comprises one
or more
pharmaceutically active therapeutic agent(s). Examples of a pharmaceutically
active agent
include immunostimulatory agents, anti-viral agents, antibiotics, anti-fungal
agents, anti-
parasitic agents, anti-tumor agents, cytokines, chemokines, growth factors,
anti-angiogenic
factors, chemotherapeutic agents, antibodies and gene silencing agents.
Preferably, the
pharmaceutically active agent is selected from the group consisting of an
immunostimulatory agent, an anti-viral agent and an anti-tumor agent. The more
than one
pharmaceutically active agents may be of the same or different category.
The pharmaceutical composition may be formulated in any way that is compatible
with its
therapeutic application, including intended route of administration, delivery
format and
desired dosage. Optimal pharmaceutical compositions may be formulated by a
skilled
person according to common general knowledge in the art, such as that
described in
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
Remington's Pharmaceutical Sciences (18th Ed., Gennaro AR ed., Mack Publishing
Company, 1990).
The pharmaceutical composition may be formulated for instant release,
controlled release,
timed-release, sustained release, extended release, or continuous release.
The pharmaceutical composition may be administered by any route known in the
art,
including, but not limited to, topical, enteral and parenteral routes,
provided that it is
compatible with the intended application. Topic administration includes, but
is not limited
to, epicutaneous, inhalational, intranasal, vaginal administration, enema, eye
drops, and ear
drops. Enteral administration includes, but is not limited to, oral, rectal
administration and
administration through feeding tubes. Parenteral administration includes, but
is not limited
to, intravenous, intraarterial, intramuscular, intracardiac, subcutaneous,
intraosseous,
intradermal, intrathecal, intraperitoneal, transdermal, transmucosal, and
inhalational
administration. In case of tumours or cancer, a preferred route of
administration is an intra
or peritumoral injection.
In a preferred embodiment, the pharmaceutical composition is for local (e.g.,
mucosa, skin)
applications, such as in the form of a spray (i.e., aerosol) preparation.
The pharmaceutical composition may be used for prophylactic and/or therapeutic
purposes.
For example, a spray (i.e., aerosol) preparation may be used to strengthen the
anti-viral
capability of the nasal and the pulmonary mucosa.
The present invention also relates to prodrugs of the herein defined RNA
poly/oligonucleotides having immune-stimmulatory activity. An Example for a
suitable
prodrug is a linear RNA which is designed to form dsRNA sections containing
G=U as a
central secondary structure. Further prodrugs of the RNA poly/oligonucleotide
as defined
herein, include RNA poly/oligonucleotides which are chemically modified in
either the G or
the U in a fashion that in an organism the RNA poly/oligonucleotide is
reverted to the active
G and/or U, for example derivates of G and/or U that form an G=U equivalent.
The optimal dosage, frequency, timing and route of administration can be
readily
determined by a person skilled in the art on the basis of factors such as the
disease or
condition to be treated, the severity of the disease or condition, the age,
gender and
physical status of the patient, and the presence or absence of previous
treatment.
31
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
In Vitro Applications
The present application provides the in vitro use of the RNA
poly/oligonucleotide of the
invention described above. In particular, the present application provides the
use of at least
one poly/oligonucleotide for inducing an anti-viral response, in particular, a
type I IFN
response, more specifically, an IFN-a response, in vitro.
The present invention provides an in vitro method for stimulating an anti-
viral response, in
particular, a type I IFN response, more specifically, an IFN-a response in a
cell, comprising
the steps of:
---- - -- --- --- -- -
--(a) mixing at least one poIY /oligonucleotide of the i- nvention described
above
with a complexation agent; and
(b) contacting a cell with the mixture of (a), wherein the cell expresses TLR7
and is capable of producing an anti-viral response upon TLR7 activation.
The present invention provides an alternative in vitro method for stimulating
an anti-viral
response, in particular, a type I IFN response, more specifically, an IFN-a
response in a
cell, comprising the steps of: contacting a cell with at least one
poly/oligonucleotide of the
invention described above, which is optionally mixed with a complexation
agent, wherein
the cell expresses TLR7 and is capable of producing an anti-viral response
upon TLR7
activation.
The cells may express TLR7 endogenously and/or exogenously from an exogenous
nucleic
acid (RNA or DNA). The exogenous DNA may be a plasmid DNA, a viral vector, or
a
portion thereof. The exogenous DNA may be ingerated into the genome of the
cell or may
exist extra-chromosomally. The cells include, but are not limited to, primary
immune cells,
primary non-immune cells, and cell lines. Immune cells include, but are not
limited to,
peripheral blood mononuclear cells (PBMC), plasmacytoid dendritric cells
(PDC), myeloid
dendritic cells (MDC), macrophages, monocytes, B cells, natural killer cells,
granulocytes,
CD4+ T cells, CDB+ T cells, and NKT cells. Among these cells, plasmacytoid
dendritric
cells (PDC), which are a subpopulation of peripheral blood mononuclear cells
(PBMC),
express TLR7 endogenously. Non-immune cells include, but are not limited to,
fibroblasts,
endothelial cells, epithelial cells, and tumor cells. Cell lines may be
derived from immune
cells or non-immune cells, which do or do not express TLR7 endogenously.
Whether a cell expresses TLR7 can be readily determined by a skilled person
using
standard techniques such as western blotting, RT-PCR, and northern blotting.
32
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
Whether a cell is capable of producing an anti-viral response upon TLR7
activation can be
readily determined by a skilled person using a known TLR7 ligand and standard
assays for
cytokine detection such as ELISA, quantitative or semi-quantitative RT-PCR.
In Vivo Applications
The present application provides the in vivo use of the RNA
poly/oligonucleotide of the
invention described above.
-1n particular, the-present application provides at-least -one RNA-
poly/oligonucleotide- for
inducing an anti-viral response, in particular, a type I IFN response, more
specifically, an
IFN-a response, in a vertebrate animal, in particular, a mammal. The present
application
further provides at least one RNA poly/oligonucleotide for preventing and/or
treating a
disease and/or disorder in a vertebrate animal, in particular, a mammal, in
medical and/or
veterinary practice. The present application further provides RNA
poly/oligonucleotides for
enhancing humoral immunity. The invention also provides RNA
poly/oligonucleotides
enhancing antigen-specific T-cell response. The invention also provides at
least one RNA
poly/oligonucleotide for use as a vaccine adjuvant. The RNA
poly/oligonucleotides are
further useful as prophylactic or therapeutic vaccines.
Furthermore, the present application provides the use of at least one RNA
poly/oligonucleotide for the preparation of a pharmaceutical composition for
inducing an
anti-viral response, in particular, a type I IFN response, more specifically,
an IFN-a
response, in a vertebrate animal, in particular, a mammal. The present
application further
provides the use of at least one RNA poly/oligonucleotide for the preparation
of a
pharmaceutical composition for preventing and/or treating a disease and/or
disorder in a
vertebrate animal, in particular, a mammal, in medical and/or veterinary
practice.
The disease or disorder is one that can be prevented or treated by type I IFN.
The
diseases and/or disorders include, but are not limited to, infections,
tumors/cancers, and
immune disorders.
Infections include, but are not limited to, viral infections, bacterial
infections, anthrax,
parasitic infections, fungal infections and prion infection.
33
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
Viral infections include, but are not limited to, infections by hepatitis C,
hepatitis B, influenza
virus, herpes simplex virus (HSV), human immunodeficiency virus (HIV),
respiratory
syncytial virus (RSV), vesicular stomatitis virus(VSV), cytomegalovirus (CMV),
poliovirus,
encephalomyocarditis virus (EMCV), human papillomavirus (HPV) and smallpox
virus. In
one embodiment, the infection is an upper respiratory tract infection caused
by viruses
and/or bacteria, in particular, flu, more specifically, bird flu.
Bacterial infections include, but are not limited to, infections by
streptococci, staphylococci,
E. coli, and pseudomonas. In one embodiment, the bacterial infection is an
intracellular
bacterial infection, which is an infection by an intracellular bacterium such
as mycobacteria
- (tuberculosis), --chlamydia,-mycoplasma,-listeria,- and a facultative
intracelluar -bacterium
such as Staphylococcus aureus.
Parasitic infections include, but are not limited to, worm infections, in
particular, intestinal
worm infection.
In a preferred embodiment, the infection is a viral infection or an
intracellular bacterial
infection. In a more preferred embodiment, the infection is a viral infection
by hepatitis C,
hepatitis B, influenza virus, RSV, HPV, HSV1, HSV2, and CMV.
Tumors include both, benign and malignant tumors (i.e., cancer).
Cancers include, but are not limited to biliary tract cancer, brain cancer,
breast cancer,
cervical cancer, choriocarcinoma, colon cancer, endometrial cancer, esophageal
cancer,
gastric cancer, intraepithelial neoplasm, leukemia, lymphoma, liver cancer,
lung cancer,
melanoma, myelomas, neuroblastoma, oral cancer, ovarian cancer, pancreatic
cancer,
prostate cancer, rectal cancer, sarcoma, skin cancer, testicular cancer,
thyroid cancer and
renal cancer.
In certain embodiments, the cancer is selected from hairy cell leukemia,
chronic
myelogenous leukemia, cutaneous T-cell leukemia, chronic myeloid leukemia, non-
Hodgkin's lymphoma, multiple myeloma, follicular lymphoma, malignant melanoma,
squamous cell carcinoma, renal cell carcinoma, prostate carcinoma, bladder
cell
carcinoma, breast carcinoma, ovarian carcinoma, non-small cell lung cancer,
small cell lung
cancer, hepatocellular carcinoma, basaliom, colon carcinoma, cervical
dysplasia, and
Kaposi's sarcoma (AIDS-related and non-AIDS related).
34
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
Immune disorders include, but are not limited to, allergies, autoimmune
disorders, and
immunodeficiencies. Allergies inlclude, but are not limited to, respiratory
allergies, contact
allergies and food allergies.
Autoimmune diseases include, but are not limited to, multiple sclerosis,
diabetes mellitus,
arthritis (including rheumatoid arthritis, juvenile rheumatoid arthritis,
osteoarthritis, psoriatic
arthritis), encephalomyelitis, myasthenia gravis, systemic lupus
erythematosis, autoimmune
thyroiditis, dermatisis (including atopic dermatitis and eczematous
dermatitis), psoriasis,
Siogren's Syndrome, Crohn's disease, aphthous ulcer, iritis, conjunctivitis,
keratoconjunctivitis, ulcerative colitis, asthma, allergic asthma, cutaneous
lupus
erythematosus, scleroderma, vaginitis, proctitis, drug eruptions, leprosy
reversal reactions,
erythema nodosum leprosum, autoimmune uveitis, allergic encephalomyelitis,
acute
necrotizing hemorrhagic encephalopathy, idiopathic bilateral progressive
sensorineural
hearing loss, aplastic anemia, pure red cell anemia, idiopathic
thrombocytopenia,
polychondritis, Wegener's granulomatosis, chronic active hepatitis, Stevens-
Johnson
syndrome, idiopathic sprue, lichen planus, Graves' disease, sarcoidosis,
primary biliary
cirrhosis, uveitis posterior, and interstitial lung fibrosis.
Immunodeficiencies include, but are not limited to, spontaneous
immunodeficiency,
acquired immunodeficiency (including AIDS), drug-induced immunodeficiency or
immunosuppression (such as that induced by immunosuppressants used in
transplantation
and chemotherapeutic agents used for treating cancer), and immunosuppression
caused by
chronic hemodialysis, trauma or surgical procedures.
In a preferred embodiment, the immune disorder is multiple sclerosis.
In certain embodiments, the poly/oligonucleotide is used in combination with a
delivery
agent. In one embodiment, the delivery agent is a complexation agent, wherein
the
poly/oligonucleotide and the complexation agent form a complex. In another
embodiment,
the delivery agent is a replication-deficient virus, wherein the
poly/oligonucleotide is
encapsulated in the viral capsule.
In other embodiments, the poly/oligonucleotide is used without a delivery
agent.
In certain embodiments, the poly/oligonucleotide, with or without a delivery
agent, is used in
combination with one or more pharmaceutically active agents such as
immunostimulatory
agents, anti-viral agents, antibiotics, anti-fungal agents, anti-parasitic
agents, anti-tumor
agents, cytokines, chemokines, growth factors, anti-angiogenic factors,
chemotherapeutic
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
agents, antibodies and gene silencing agents. Preferably, the pharmaceutically
active
agent is selected from the group consisting of an immunostimulatory agent, an
anti-viral
agent and an anti-tumor agent. The more than one pharmaceutically active
agents may be
of the same or different category.
In one embodiment, the poly/oligonucleotide, with or without a delivery agent,
is used in
combination with an anti-viral vaccine, an anti-bacterial vaccine, and/or an
anti-tumor
vaccine, wherein the vaccine can be prophylactic and/or therapeutic. The
poly/oligonucleotide can serve as a vaccine adjuvant.
In one embodiment, the pharmaceutical composition is for use in combination
with one or
more prophylactic and/or therapeutic treatments of diseases and/or disorders
such as
infection, tumor, and immune disorders. The treatments may be pharmacological
and/or
physical (e.g., surgery, radiation).
In a further embodiment, the poly/oligonucleotide comprising at least one
uridine that is not
involved in base pairing, is used in combination with exogenous G nucleoside.
The term
"used in combination" as used herein means simultaneously or consecutively by
suitable
route of administration as defined herein above.
Vertebrate animals include, but are not limited to, fish, amphibians, birds,
and mammals.
Mammals include, but are not limited to, rats, mice, cats, dogs, horses,
sheep, cattle, cows,
pigs, rabbits, non-human primates, and humans. In a preferred embodiment, the
mammal
is human.
The present invention is illustrated by the following Examples. The Examples
are for
illustration purpose only and shall not be construed to limit the scope of the
invention.
36
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
EXAMPLES
Materials and Methods
Reagents
The RNA oligonucleotides or oligoribonucleotides (ORNs, sequences detailed in
Table 1)
were obtained from Eurogentec or Biomers. The CpG-containg DNA
oligonucleotides or
oligodeoxyribonucleotides (ODNs) CpG-ODN 2216 and CpG-ODN 2336 were purchased
from Invitrogen. Polyriboinosinic:polyribocytidilic acid (pl:C) and
Homopolymers poly G/U/A
__was obtained from-Sigma-Aldrich.-ORNs were resuspended-in sterile endotoxin-
free DEPC__
treated water (Invitrogen), and CpG-ODNs were resuspended in 0.9 % NaCl
aqueous
solution (Braun). To prevent contamination, all oligonucleotides were stored
and handled
under aseptic conditions. The sequences of the oligonucleotides are listed in
Table I and II.
Nucleic acids were suspended in sterile endotoxin free DEPC-water (Invitrogen,
Karlsruhe).
Unless indicated otherwise, dsRNA oligonucleotides (ds ORNs) were generated by
mixing
two ssRNA oligonucleotides (ss ORNs) in a ratio of 1:1 (w/w) at a
concentration of 1 pg/pl.
Unless indicated otherwise, nucleic acids were hybridized at a ratio of 1:1
(w/w).
The term "ORN" and "R" are used interchangeably in the following to signify an
RNA
oligonucleotide. For example, R805 and ORN805 refer to the same RNA
oligonucleotide.
Table I.
ORN Sequence (5'->3') SEQ Figure
ID NO:
A8 AAAAAAAA 1 2
C8 CCCCCCCC 2 2
G8 GGGGGGGG 3
G21 GGGGGGGGGGGGGGGGGGGGG 4 1,2,4,6
U21 5 3,4
U21S U* U* U* U* U* U* U* U* U* U* U* 6 1
U* U* U* U* U* U* U* U* U* U*
A21 7 2
C21 CCCCCCCCCCCCCCCCCCCCC 8 2
CCCUCC 9 2
CCCUCCC 10 2
R805 ccccuccc 11 2-6
37
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
cccccucccc 12 2
CCCCCCUCCCCC 13 2
CCCCCCCCUCCCCCCC 14 2
CCCCCCCCCCUCCCCCCCCCC 15 2
AAAAGAAA 16 2
AAAAAAGAAAAA 17 2
AAAAAAAGAAAAAA 18 2
R1602 AAAAAAAAGAAAAAAA 19 2-4
AA-kAAAAAAAGA-kAAAAAAAA 20 2
AAAAAG GAAAA 21 2
9.2s AGCUUAACCUGUCCUUCAA 22 3-5
R17 GGCAUUCUUAUUCUUACGG 23 3-4
R1917 GGCAUUCUUAUUCUUACGG 24 4
R2115 CCCUCCCCGGGGGGGGGGGGG 25 4-5
R2116 GGGGGGGGGGGGGCCCUCCCC 26 4
R2117 CCCUCCGGGGGGGGGGGGGGG 27 4
R3101 CCCUCCCCGGGGGGGGGGGGGGGGGGGGGGG 28 4
R2127 CCCCCCCCGGGGGGGGGUGGG 29 5
R2151 CGCGCGCGCAGAAGCGUGCGC 30 13
R2152 CGCGUGCGCAGAAGCGCGCGC 31 13
R2153 CGGCUCGGCAGAAGCCGGGCC 32 13
R2161 CGCCUGGGCAGAAGCCCGGGC 33 14
R805PTO-U CCCC*U*CCC 34 5
R805 2'-O-Me-U CCCCmUCCC 35 5
R805 ij CCCC14JCCC 36 5
CpG-ODN
CpG 2216 G*G*GGGACGATCGTCG*G*G*G*G*G 37 3,4
CpG 2336 G*G*G*G*ACGACGTCGTGGG*G*G*G*G 38 3
* phosphothioate bond
Isolation of human PBMCs, PDCs and human monocytes
Human PBMCs were prepared from blood of healthy male and female donors. Fresh
Buffy-
Coats were obtained from the Institute of Experimental Hematology and
Transfusion
Medicine of the University Bonn (Bonn, Germany). PBMCs were purified by
biocoll density
gradient centrifugation (Biochrom AG). PDC were isolated by using the MACS
CD304
(BDCA-4/Neuropilin-1)-MicroBead Kit (Miltenyi, Bergisch Gladbach). Monocytes
were
38
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
prepared by depleting PDC and using the Monocyte Isolation Kit II (Miltenyi).
The purity of
PDCs and monocytes was determined by antibody (obtained from BD Bioscience,
Heidelberg and Miltenyi) staining followed by flow cytometric (FACS) analysis
and was
typically >90%.
Generation of FLT3-L PDCs from mouse bone marrow
Cells were isolated from bone marrow of wild-type or TLRT'- mice. Cells were
cultured in
RPMI 1640 supplemented with 10% FCS, 0.1 mM MEM non-essential amino acids, 1mM
MEM sodium pyruvate (all from Gibco), 2mM L-glutamine (Cambrex BioWhittaker),
100
- - - U/ml--Penicillin, 100-pg/ml Streptomycin (PAA-Laboratories -GrnbH) -and
20 -rYg/r"hl--FMS
related tyrosine kinase 3 ligand (rFLT3-L) R&D systems. After 7 days, non-
adherent cells
were collected and the PDCs were isolated by MACS-separation using B220-
MicroBeads
(Miltenyi).
Cell culture
Human cells were cultured in RPMI 1640 (Gibco) supplemented with 2% human
serum
(Cambrex, BioWhittaker), 2 mM L-glutamine (Cambrex, BioWhittaker), 100 U/ml
Penicillin
and 100 pg/ml Streptomycin (PAA Laboratories GmbH) in 96-well flat-bottem
plates. The
density of human PBMCs and human monocytes was 4x105 cells/200 pl per well,
and the
density of human PDC was 4x104 cells/200 pl per well. Mouse PDCs were cultured
in 96-
well flat-bottom plates in RPMI 1640 supplemented with 10% FCS, 0.1 mM MEM non-
essential amino acids, 1 mM MEM sodium pyruvate (all from Gibco), 2 mM L-
glutamine
(Cambrex BioWhittaker), 100 U/ml Penicillin and 100 pg/ml Streptomycin (PAA
Laboratories GmbH). The density of mouse pDC was 4x104 cells/200 pl per well.
In vitro cell stimulation
Stimulations were performed in triplicate in 96-well flat-bottom plates (Nunc,
Langenselbold)
with a 200 pl total volume, and incubated for 20 h at 37 C, 5 % CO2. Unless
where
indicated otherwise, homopolymers, ORN or CpG-ODN were complexed to the
polycation
Poly-l-arginine (P4663 by Sigma-Aldrich) and used at a final concentration of
1 pg/ml. Poly
I:C was transfected with TranslT-LT1 (Mirus, Madison). Mouse PDC were
transfected with
ORN or CpG-ODN using the Dotap liposomal transfection reagent (Roth,
Karlsruhe).
Human cells, were cultured in RPMI 1640 (Gibco, Karlsruhe) supplemented with
2% human
serum (Cambrex BioWhittaker, New Jersey), 2 mM L-glutamine (Cambrex
BioWhittaker),
39
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
100 U/ml Penicillin and 100 pg/ml Streptomycin (PAA Laboratories GmbH,
Pasching). Cells
were plated in 96 well plates at a density of 4x105 cells/200 pl for human
PBMC and
monocytes, and 4x104 cells/200 pl for PDC. Mouse PDC were cultured in RPMI
1640
supplemented with 10 % FCS, 0,1 mM MEM non-essential amino acids, 1 mM MEM
sodium pyruvate (all from Gibco), 2 mM L-glutamine (Cambrex BioWhittaker), 100
U/ml
Penicillin , 100 pg/ml Streptomycin (PAA Laboratories GmbH) at a density of
4x104
cells/200 pl. Supernatants were removed and assayed for cytokine production.
In vivo stimulation
Wild-type mice were injected i.v. with 25 pg ORN formulated with 125 pg Dotap.
Mice were
-bled-3 h, 5 h and-8- h post-injection via the tail vein. Serum was
purifiedand-the 1FN-alevel--
was measured.
Detection of cytokine
Supernatants from human or mouse cells were harvested 20 h after stimulation.
If not used
immediately, supernatants were frozen at -20 C until used. IFN-a and/or IL-
12p70 were
analysed in the supernatant by ELISA. The human IFN-a ELISA was purchased from
Bender MedSystems (Vienna), and the human IL-12p70 ELISA from BD Biosciences.
Murine IFN-a in the supernatants of stimulated mouse cells or in mice sera was
measured
by the mouse IFN-a ELISA Kit (PBL Biomedical Laboratories).
Fluorescence microscopy
Uptake of fluorescence labelled RNA by PDC was analysed using a Observer D1
fluorescence microscope (Zeiss, Gottingen). Cells were incubated for 2.5 h
with Cy3-
labelled ORN R805 6 pg/ml either alone or together with 6 pg/ml pA or pG. PDC
were then
stained with anti-BDCA-1 (CD303, Miltenyi) followed by the secondary antibody
goat anti
mouse Alexa 488 (Invitrogen). Nuclei were visualized using Hoechst 33342
(Hoechst,
Frankfurt).
Statistics
Results are expressed as mean standard error of mean (s.e.m.). Group
comparisons
were carried out using a two-tailed Student's t-test with an expected unequal
variance.
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
Example 1. Poly uridine (polyU, pU)-stimulated IFN-a induction is enhanced by
the
formation of double strands via G:U base pairing.
Human peripheral blood mononuclear cells (PBMCs) and purified PDCs were
stimulated
with pU, pG, either in the form of an ssRNA or dsRNAThe levels of IFN-a
induced by RNA
with uridine repeats of varying lengths were compared to those obtained by
other
polyribonucleotides, either in the form of single stranded (ss) or as double
stranded (ds)
RNA. Among the ssRNAs, polyU was found to stimulate IFN-a production from both
human
PBMCs (Fig. 1 A) and human PDCs (Fig 1 B), whereas polyguanosine (pG), poly
adenosine
(pA), poly cytidine (pC) were inactive (data not shown). The formation of
dsRNA between
pU and pA greatly reduced or almost completely abolished the IFN-a-inducing
activity of
single-stranded pU. In stark contrast, the formation of dsRNA between pU and
pG
significantly and consistently enhanced the IFN-a-inducing activity of single-
stranded pU,
although the relative number of U within pU:pG was reduced by 50 % compared to
pU
alone (Figure 1 B). Similar results were obtained when oligo uridine (oligoU)
21 nucleotides
in length (U21) and oligo guanosine (oligoG) (G21) were tested (Fig. 1C & D).
These findings are very surprising since it has long been established that
TLR7 recognizes
single-stranded RNA.
Example 2. The presence of a single G:U base pair in RNA stem structure is
sufficient to
induce high levels of type I IFN.
To decipher the basis for the increase in IFN-a-inducing activity observed
with pU:pG,
21 mer ORNs were designed by the inventors comprising A's and one or two G(s).
As
observed before, the pairing with oligo A 21 nucleotides in length (A21) led
to a reduction in
the IFN-a-inducing activity of pU and the pairing with oligo G 21 nucleotides
in length (G21)
led to an increase in the IFN-a-inducing activity of pU, further supporting a
contribution of
G=U base pairs. Surprisingly, when a 21 mer ORN containing 20 A's and 1 G or
19 A's and
2 G's were paired with pU, the IFN-a-inducing activity of the dsRNA was
significantly higher
than that of single-stranded pU, and was comparable or even higher than the
partially
double-stranded G21:pU (Figure 2A). Of note, none of the 21 mers tested, on
its own, was
able to induce IFN-a. This observation shows that G=U base pairs represent the
molecular
structure that is recognized by TLR7.
Subsequently, we designed RNA oligonucleotides of different lengths consisting
of A's
around a central G, and tested whether the number of neighbouring base pairs
influenced
the IFN-a-inducing activity of the G:U base pair. We found that, when paired
with polyU, a
range of ORNs, having 7-8 A's flanking the central G, showed IFN-a-inducing
activity
41
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
(Figure 2B). The G=U containing dsRNA pU+R1 602 was selected for subsequent
experiments (Figure 2A).
While polyU on its own was able to induce type I IFN in PBMCs, polyG was
inactive, but
may become stimulatory when hybridized to ORN that introduce G=U base pairs.
(Figure
1A). It was tested whether G:U base pairing would render the otherwise
inactive polyG
active in inducing IFN-a. ORNs consisting of two stretches of C's around a
central U were
designed by the inventorsSurprisingly, the highest IFN-a-inducing activity was
observed
when pG was paired with ORNs containing 2-5 C's flanking the central G; any
further
increase in the number of flanking C's resulted in a drastic decrease and
eventual
abolishment of the IFN-a-inducing activity of the partially double-stranded
RNA (Figure 2C).
From these data, the inventors hypothesized that the IFN-a-inducing activity
of a partially
double-stranded RNA is determined by the stability of the double-stranded RNA
structures
surrounding the G:U base pair, with G:C base pair providing three hydrogen
bonds, and
U:A base pair providing two hydrogen bonds. Both, too low and too high,
stability comprise
or eliminate the IFN-a-inducing activity of the partially double-stranded RNA
oligonucleotide. To confirm our hypothesis, the software DNamelt
(http://www.bioinfo.rpi.edu/applications/hybrid/) assigned similar stability
to the double-
stranded portion of the two most active partially double-stranded RNAs tested,
pU+ORN1602 (AAAAAAAAGAAAAAAA) and pG+ORN805 (CCCCUCCC) (data not
shown).
Furthermore, the IFN-a-inducing activity of partially double stranded RNAs
formed by pG
and ORN805 (CCCCUCCC) mixed at different weight ratios was tested. When pG and
ORN805 were mixed at a ratio of 1:1 (w/w), it was expected that the resulting
RNA would
be mostly double-stranded. A step-wise reduction in the amount of ORN805 would
be
expected to result in a step-wise increase in abundance of single-stranded
structures. We
found that maximal IFN-a-inducing activity was maintained up to a pG:ORN805
ratio of
20:1 (w/w). IFN-a-inducing activity was maintained down to a ratio of 100:1
which
corresponds to one G=U base pair in 80 to 100 bases of RNA. This indicates
that few short
RNA stems (8mer) containing very few G=U wobble base pairs within long ssRNA
are
sufficient for detection by TLR7 in PDC.
Taken together, these results suggest that ssRNAs comprising short double-
stranded
structures containing G:U base pairs, rather than dsRNAs, are capable of
inducing the
production of type I IFN from human PBMCs and PDCs.
42
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
Example 3. Mechanism of G:U base pair-mediated stimulation.
Type I IFN production in response to ssRNA oligonucleotides in both, human and
mouse,
PDCs has been shown to depend on TLR7 [7]. Toll-like receptors able to
recognize nucleic
acid ligands are located in the endosomal compartment and their activation can
be blocked
by chloroquine, which inhibits the acidification of the endosomes. In
contrast, the
recognition of nucleic acids by cytosolic helicases such as RIG-I (retinoic
inducible gene I)
and MDA-5 (melanoma differentiation antigen 5) are not sensitive to
chloroquine (Gitlin L et
al, 2006).
To investigate whether the recognition of RNA containing G:U base pairs was
TLR-
dependent, human PBMCs that were pre-incubated with an increasing amount of
chloroquine were stimulated. As expected, TLR9-dependent recognition of CpG
ODN was
chloroquine-sensitive, whereas the MDA-5-mediated recognition of poly(I:C)
(pl:C) was not.
It was found that IFN-a production induced by both polyU:R1602 and polyG:R805
were
sensitive to chloroquine (Figure 3A), indicating that their recognition of
RNAs containing
G=U base pairs, i.e. pU+R1 602 or pG+R805 was mediated by an endosomal TLR.
Subsequently, mouse gene knock-out models were prepared to confirm the
requirement of
TLR7; consistent with the results in human immune cells (Figure 1 and 2).
Next, pG:pU and
pG+R805 were used to induce IFN-a in murine PDC derived from bone marrow
cultures in
vitro (Figure 3B). Addition of R1602 strongly enhanced IFN-a levels. Injection
of G=U
containing RNAs in complex with Dotap in mice in vivo induced a systemic type
I IFN
response (Figure 3C). Finally, using FLT3-L PDCs from wild-type (WT) and
TLR7"" mice
and stimulated them in vitro with ssRNA or dsRNA complexed to Dotap, it was
found that
IFN- a -inducing activity of RNAs that contained G=U base pairs required TLR7
while this
was not the case for the CpG oligonucleotide 2336 (positive control), which is
recognized
by TLR9. Whereas pU, oligoU (U21) and G:U base pair-containing RNAs (pG+ORN805
and
pU+ORN1602) induced IFN-a production in WT PDCs, they failed to do so in
TLR7"" PDCs
(Figure 3D). .
These results indicate that IFN-a production induced by G:U base pair-
containing RNA is
mediated by TLR7. Furthermore, these results confirm the notion that a single
G:U base
pair is sufficient to activate TLR7 (Fig. 3C, U21+ORN1602).
Example 4. Selective activation of human PDC by RNA containing G:U base pairs.
Most of the published RNA ligands activate both TLR7 and TLR8 As a result,
stimulation of
PBMCs with these RNA ligands leads to TLR7-dependent production of IFN-a in
PDCs as
43
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
well as TLR8-dependent production of IL-12 mostly in monocytes (review
Bekeredjan-Ding
et al, 2005 and 2006). Therefore, the ability of different RNA molecules to
induce IFN-a in
PDC and IL-12 in monocytes (Figure 4A and B) was compared. The cytokine
production in
human PDCs and monocytes in response to a published RNA ligand (9.2s) and RNA
containing G:U base pairs was measured. It was found that, while pU, pU+R1
602, U21 and
R17 (identical to R1917) induced both, IFN-a in PDC and IL-12 in monocytes,
CpG ODN,
pG+R805 selectively activated PDC but not monocytes, indicating that pG+R805
exclusively activates TLR7 but not TLR8. TLR7 selectivity was seen for all
duplex forming
RNA molecules that contain at least one single G=U base pair and in which the
corresponding single strands did not activate PDC or monocytes (Figure 4).
Selective
activation of PDC was also achieved with single strand ORN (R3101, R2115,
R2116,
R2117) that form a stem loop structure containing at least one single G=U
wobble base pair
(Figure 4C, D, E, F, G). In this setup, a G=U containing stem structure of 6
bp was sufficient
to elicit a potent IFN-a response without inducing IL-12p70 in monocytes
(Figure 4E, F), a
further indication that the G=U base pair is selectively recognized by human
TLR7 but not
by TLR8 (Figure 4F).
ORNs that contained a single uridine, form a duplex or a stem loop structure
but do not
form a G=U wobble base pair did not induce IFN-a, confirming the strict G=U
wobble base
pair specificity of selective TLR7 ligands (Figure 5 A, B). Moreover, chemical
modification of
the uridine within R805 to pseudouridine or 2'-O-Me-Uridine largely abrogated
activity of the
complex with pG (Figure 5 C).
These data suggest that TLR7 and 8 recognize different molecular motifs and it
is possible
to design TLR7-specific RNA ligands.
Example 5. RNA containing G:U base pairs stimulates human PDC in the absence
of
transfection reagents.
Stimulation of TLR7 by RNA oligonucleotides has been shown to depend on the
use of
complexation or transfection reagents, which facilitate the cellular uptake
and delivery of
RNA ligands to TLR7 which is confined to the endosomal compartment. In
addition,
complexation with poly-cations protects RNA from degradation (review Diebold
SS et al,
2004, Heil et al, 2004 and Heil et al 2003). Naked RNA is inherently unstable
because its
phosphodiester backbone is susceptible to both alkaline hydrolysis and
degradation by
ubiquitous RNases. Because most cellular RNases preferentially degrade ssRNA,
it was
tested whether stem structures containing a G=U base pair were able to
stimulate PDC also
44
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
in the absence of complexation reagents. Surprisingly, "naked" polyG+R805 not
complexed
with any complexation or transfection agent was not only active in inducing
high levels of
type I IFN, it was as active as polyG+R805 complexed with poly L-arginine
(Fig. 6A) using
the same quantities. Using R805 hybridized to short synthetic pG ORN (42mer,
21 mer,
8mer) it was observed that that ability to induce IFN-a correlated with
overall RNA length
(Figure 6C). In order to test whether the pG polymer indeed facilitated the
uptake of
hybridized ORN R805, human PDC was incubated with fluorescent R805 in the
culture
supernatant. It was found that PDC did not take up any R805 by itself, or
together with poly
A (Figure 613). In contrast, when R805 was added together with pG, it was
readily taken up
by all PDC in the sample. To our knowledge, polyG+R805 is the first example of
a
phosphodiester--RNA; which is able-to stimulate TLR7 without complex formation
with a
transfection reagent.
Fluorescence microscopy indicates that, indeed, polyG+R805 entered the
endosomal
compartments of monocytes without the aid of any complexation agent (Fig. 6B).
Example 6. Addition of guanosine nucleosides strongly enhances the type I IFN
response
to poly uridine RNA.
Human PDC were isolated from PBMC by MACS enrichment using BDCA-4 Microbeads.
40.000 PDC per well of a 96 well plate were stimulated with 200 ng pU (A, C)
or with U21 s
(B) complexed to Poly-L-Arginine. (A, B) At the same time guanosine-
nucleosides (G) in
different concentrations (0.005 mM, 0.05 mM or 0.25 mM) were added to
stimulated or
untreated PDCs. (C) Different nucleosides (G, cytidine (C), uridine (U) or
adenosine (A))
were added in a concentration of 0.25 mM to stimulated or untreated PDCs. All
Nucleosides
used were from Sigma-Aldrich (A 9251 Adenosine; C 9505 Cytidine; G 6752
Guanosine; U
3750 Uridine) After 20 h, cell culture supernatants were assessed for IFN-a by
ELISA. Data
shown are representative of 3 independent experiments. Given that high
concentrations of
a combination of G and U nucleosides but not the single nucleosides were
reported to
stimulate cytokine release in PBMC (Heil et al. 2004), and the guanosine
analogue
loxoribine is a known TLR7 specific stimulus (Heil et al. 2003), we next
examined whether
the IFN-a inducing activity of poly uridines (pU, U21s) is influenced by the
addition of
monomeric nucleosides. The addition of the nucleoside guanosine dose
dependently
increased the ability of pU and U21s to induce IFN-a in PDC (Fig. 7A and B).
No increased
activity of pU or U21s was seen for the other three nucleosides cytidine,
guanosine and
adenosine (Fig. 7C).
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
Together these results demonstrate that base pairing with guanosine enhances
the activity
of uridine to activate TLR7 in PDC; furthermore they show that polyuridine
RNAs lack TLR7
activity if uridines are bound to adenosine and not guanosine, suggesting that
not the mere
uridine content of RNA per se, but rather binding of uridine to guanosine
determines the
TLR7 ligand activity of double stranded RNA.
Example 7. Evidence that some RNA hairpins containing G=U base pairs act as
selective
TLR8 agonists.
(A) PBMC were isolated from Buffy coats and stimulated with the RNAs shown in
(B) in
complex with Poly-l-Arginine: After 20 -h -IFN-a, 1L-12 p70- and--IL-6- were -
measured in- the
supernatants by cytokine Elisa. R2151 CGCGCGCGCAGAAGCGUGCGC and R2152
CGCGUGCGCAGAAGCGCGCGC are examples for selective TLR8 agonists.These data
show, that also cells expressing TLR8, as for example human monocytes, can
selectively
be activated by RNA oligos that contain a single uridine in a G=U wobble base
pair. In this
case, RNA sequences that showed selective TLR8 activation the U involved in
the G=U
base pair was preferentially flanked by G on either side.
Example 8. In cells that express both functional TLR7 and TLR8 RNAs containing
G=U
wobble base pairs retain TLR7 selective activity.
THP-1 cells were maturated with 300ng/ml PMA for 4 h in a 10cm dish, then
harvested,
washed with PBS twice and plated at 100.000 cells/well of a 96well Plate. PMA
activated
THP-1 cells express TLR8, and upon stimulation with IFN-g (100 U/ml) for 8 h
also
upregulate functional TLR7 (adapted from Gantier et al. TLR7 is involved in
sequence-
specific sensing of single-stranded RNAs in human macrophages. J Immunol
(2008) vol.
180 (4) pp. 2117-24). (A) THP-1 cells were stimulated with the small molecule
agonists
R848 (TLR7/8 agonist) C1087 (TLR7 agonist), or RNAs R2153, R2116 (TLR7
agonists)
R2127 (inert), and 9.2s (TLR7/8 agonist). After 20 h TNF-a was measured in the
culture
supernatant by ELISA. (B) Knock-down of TLR7 using 3 different lentiviral
shRNA
constructs in PMA/IFN-g treated THP-1 cells represses TLR7 but not TLR8 mRNA.
THP-1
cells were infected with shRNA lentiviruses targeting TLR7 and with control
viruses. Stable
lines were obtained and maintained by antibiotic selection with Puromycin
(2pg/ml) for 10
days. Lentiviral vectors without (empty) or with nonsense shRNA (scrambled)
were used as
negative controls. THP-1 cells were treated as described in A and harvested
after 8h
treatment with IFN-g. RNA was extracted and TLR7/8 expression normalized to
GAPDH
levels was analyzed by SYBR Green real-time RT-PCR. (C) THP-1 cells treated as
in
46
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
described in (B) were stimulated with small molecule agonists (R848, C1087 and
C1075[TLR8 agonist]), or the indicated RNAs in complex with Poly-I-Arginine
(R2161
CGCCUGGGCAGAAGCCCGGGC). After 20 h TNF-a was measured in the culture
supernatant by ELISA. Data shown are representative of 2 (A) or 3 (B, C)
independent
experiments. These data show that the strictly TLR7 selective activation via
RNA oligos
containing an immune stimulatory G=U base pairs is maintained even when
functional TLR8
is expressed within the same cell.
47
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
REFERENCES
1. Uematsu S, Akira S: Toll-Like receptors (TLRs) and their ligands. Handb
ExpPharmacol 2008:1-20.
2. Alexopoulou L, Holt AC, Medzhitov R, Flavell RA: Recognition of double-
stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature
2001, 413:732-738.
3. Krug A, Luker GD, Barchet W, Leib DA, Akira S, Colonna M: Herpes simplex
virus type 1 activates murine natural interferon-producing cells through
toll-like receptor 9. Blood 2004, 103:1433-1437.
4. Lund J, Sato A, Akira S, Medzhitov R, Iwasaki A: Toll-like receptor 9-
mediated recognition of Herpes simplex virus-2 by plasmacytoid dendritic
cells. J Exp Med 2003, 198:513-520.
5. Lund JM, Alexopoulou L, Sato A, Karow M, Adams NC, Gale NW, Iwasaki A,
Flavell RA: Recognition of single-stranded RNA viruses by Toll-like
receptor 7. Proc Nat/ Acad Sci U S A 2004, 101:5598-5603.
6. Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C: Innate antiviral
responses by means of TLR7-mediated recognition of single-stranded
RNA. Science 2004, 303:1529-1531.
7. Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, Lipford
G,
Wagner H, Bauer S: Species-specific recognition of single-stranded RNA
via toll-like receptor 7 and 8. Science 2004, 303:1526-1529.
8. Hemmi H, Takeuchi 0, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M,
Hoshino K, Wagner H, Takeda K, et al.: A Toll-like receptor recognizes
bacterial DNA. Nature 2000, 408:740-745.
9. Hornung V, Rothenfusser S, Britsch S, Krug A, Jahrsdorfer B, Giese T,
Endres
S, Hartmann G: Quantitative expression of toll-like receptor 1-10 mRNA in
48
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
cellular subsets of human peripheral blood mononuclear cells and
sensitivity to CpG oligodeoxynucleotides. J Immunol 2002, 168:4531-4537.
10. Kadowaki N, Ho S, Antonenko S, Malefyt RW, Kastelein RA, Bazan F, Liu YJ:
Subsets of human dendritic cell precursors express different toll-like
receptors and respond to different microbial antigens. J Exp Med 2001,
194:863-869.
11. Jurk M, Heil F, Vollmer J, Schetter C, Krieg AM, Wagner H, Lipford G,
Bauer S:
Human TLR7 or TLR8 independently confer responsiveness to the antiviral
compound R-848. Nat Immunol 2002, 3:499.
12. Ma Y, Li J, Chiu I, Wang Y, Sloane JA, Lu J, Kosaras B, Sidman RL, Volpe
JJ,
Vartanian T: Toll-like receptor 8 functions as a negative regulator of neurite
outgrowth and inducer of neuronal apoptosis. J Cell Biol 2006, 175:209-215.
13. Gorden KB, Gorski KS, Gibson SJ, Kedl RM, Kieper WC, Qiu X, Tomai MA,
Alkan SS, Vasilakos JP: Synthetic TLR agonists reveal functional
differences between human TLR7 and TLR8. J Immunol2005, 174:1259-
1268.
14. Heil F, Ahmad-Nejad P, Hemmi H, Hochrein H, Ampenberger F, Gellert T,
Dietrich H, Lipford G, Takeda K, Akira S, et al.: The Toll-like receptor 7
(TLR7)-
specific stimulus loxoribine uncovers a strong relationship within the
TLR7, 8 and 9 subfamily. Eur J Immunol 2003, 33:2987-2997.
15. Lee J, Chuang TH, Redecke V, She L, Pitha PM, Carson DA, Raz E, Cottam
HB: Molecular basis for the immunostimulatory activity of guanine
nucleoside analogs: activation of Toll-like receptor 7. Proc Natl Acad Sci U
S A 2003, 100:6646-6651.
16. Wagstaff AJ, Perry CM: Topical imiquimod: a review of its use in the
management of anogenital warts, actinic keratoses, basal cell carcinoma
and other skin lesions. Drugs 2007, 67:2187-2210.
49
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
17. Barchet W, Krug A, Cella M, Newby C, Fischer JA, Dzionek A, Pekosz A,
Colonna M: Dendritic cells respond to influenza virus through TLR7- and
PKR-independent pathways. Eur J Immunol 2005, 35:236-242.
18. Diebold SS, Massacrier C, Akira S, Paturel C, Morel Y, Reis e Sousa C:
Nucleic
acid agonists for Toll-like receptor 7 are defined by the presence of uridine
ribonucleotides. Eur J Immunol 2006, 36:3256-3267.
19. Eberle F, Giessler K, Deck C, Heeg K, Peter M, Richert C, Dalpke AH:
Modifications in Small Interfering RNA That Separate Immunostimulation
from RNA Interference. J Immunol2008, 180:3229-3237.
20. Forsbach A, Nemorin JG, Montino C, Muller C, Samulowitz U, Vicari AP, Jurk
M,
Mutwiri GK, Krieg AM, Lipford GB, et al.: Identification of RNA Sequence
Motifs Stimulating Sequence-Specific TLR8-Dependent Immune
Responses. J Immunol 2008, 180:3729-3738.
21. Hornung V, Guenthner-Biller M, Bourquin C, Ablasser A, Schlee M, Uematsu
S,
Noronha A, Manoharan M, Akira S, de Fougerolles A, et al.: Sequence-specific
potent induction of IFN-alpha by short interfering RNA in plasmacytoid
dendritic cells through TLR7. Nat Med 2005, 11:263-270.
22. Judge AD, Sood V, Shaw JR, Fang D, McClintock K, MacLachlan I: Sequence-
dependent stimulation of the mammalian innate immune response by
synthetic siRNA. Nat Biotechnol 2005, 23:457-462.
23. Scheel B, Teufel R, Probst J, Carralot JP, Geginat J, Radsak M, Jarrossay
D,
Wagner H, Jung G, Rammensee HG, et al.: Toll-like receptor-dependent
activation of several human blood cell types by protamine-condensed
mRNA. Eur J Immunol 2005, 35:1557-1566.
24. Sioud M: Single-stranded small interfering RNA are more
immunostimulatory than their double-stranded counterparts: a central role
for 2'-hydroxyl uridines in immune responses. Eur J Immunol 2006, 36:1222-
1230.
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
25. Mathews DH, Turner DH: Prediction of RNA secondary structure by free
energy minimization. Curr Opin Struct Biol 2006, 16:270-278.
26. Masquida B, Westhof E: On the wobble GoU and related pairs. RNA 2000,
6:9-15.
27. Varani G, McClain WH: The G x U wobble base pair. A fundamental building
block of RNA structure crucial to RNA function in diverse biological
systems. EMBO Rep 2000, 1:18-23.
28. Xu D, Landon T, Greenbaum NL, Fenley MO: The electrostatic characteristics
of G.U wobble base pairs. Nucleic Acids Res 2007, 35:3836-3847.
29. Diebold SS, Montoya M, Unger H, Alexopoulou L, Roy P, Haswell LE, Al-
Shamkhani A, Flavell R, Borrow P, Reis e Sousa C: Viral infection switches
non-plasmacytoid dendritic cells into high interferon producers. Nature
2003, 424:324-328.
30. Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, Flavell RA, Diamond
MS,
Colonna M: Essential role of mda-5 in type I IFN responses to
polyriboinosinic:polyribocytidylic acid and encephalomyocarditis
picornavirus. Proc Natl Acad Sci U S A 2006, 103:8459-8464.
31. Bekeredjian-Ding I, Roth SI, Gilles S, Giese T, Ablasser A, Hornung V,
Endres
S, Hartmann G: T cell-independent, TLR-induced IL-12p70 production in
primary human monocytes. J Immunol 2006, 176:7438-7446.
32. Bekeredjian-Ding IB, Wagner M, Hornung V, Giese T, Schnurr M, Endres S,
Hartmann G: Plasmacytoid dendritic cells control TLR7 sensitivity of naive
B cells via type I IFN. J Immunol2005, 174:4043-4050.
33. Barrat FJ, Meeker T, Gregorio J, Chan JH, Uematsu S, Akira S, Chang B,
Duramad 0, Coffman RL: Nucleic acids of mammalian origin can act as
endogenous ligands for Toll-like receptors and may promote systemic
lupus erythematosus. J Exp Med 2005, 202:1131-1139.
51
CA 02744346 2011-05-19
WO 2010/105819 PCT/EP2010/001686
34. Krieg AM, Vollmer J: Toll-like receptors 7, 8, and 9: linking innate
immunity
to autoimmunity. Immunol Rev 2007, 220:251-269.
35. Lau CM, Broughton C, Tabor AS, Akira S, Flavell RA, Mamula MJ, Christensen
SR, Shlomchik MJ, Viglianti GA, Rifkin IR, et al.: RNA-associated
autoantigens activate B cells by combined B cell antigen receptor/Toll-like
receptor 7 engagement. J Exp Med 2005, 202:1171-1177.
36. Hattermann K, Picard S, Borgeat M, Leclerc P, Pouliot M, Borgeat P: The
Toll-
like receptor 7/8-ligand resiquimod (R-848) primes human neutrophils for
leukotriene B4, prostaglandin E2 and platelet-activating factor
biosynthesis. FASEB J 2007, 21:1575-1585.
37. Chuang TH, Ulevitch RJ: Cloning and characterization of a sub-family of
human toll-like receptors: hTLR7, hTLR8 and hTLR9. Eur Cytokine Netw
2000, 11:372-378.
38. Palecanda A, Kobzik L: Receptors for unopsonized particles: the role of
alveolar macrophage scavenger receptors. Curr Mol Med 2001, 1:589-595.
52