Note: Descriptions are shown in the official language in which they were submitted.
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
1
LACCASES AND METHODS OF USE THEREOF AT LOW TEMPERATURE
PRIORITY
[01] The present application claims priority to U.S. Provisional Patent
Application Serial Nos.
61/140,724, filed on December 24, 2008, 61/154,882, filed on February 24,
2009, and
61/237,532, filed on August 27, 2009, each of which is incorporated by
reference in its entirety.
TECHNICAL FIELD
[02] The present systems, compositions, and methods relate to laccase enzymes
and nucleic
acid sequences encoding such laccase enzymes. The laccase enzymes may be
employed in
conjunction with mediators in improved methods for modifying the color of
denim fabrics.
BACKGROUND
[03] Laccases are copper-containing phenol oxidizing enzymes that are known to
be good
oxidizing agents in the presence of oxygen. Laccases are found in microbes,
fungi, and higher
organisms. Laccase enzymes are used for many applications, including pulp and
paper
bleaching, treatment of pulp waste water, de-inking, industrial color removal,
bleaching in
laundry detergents, oral care teeth whiteners, and as catalysts or
facilitators for polymerization
and oxidation reactions.
[04] Laccases can be utilized for a wide variety of applications in a number
of industries,
including the detergent industry, the paper and pulp industry, the textile
industry and the food
industry. In one application, phenol oxidizing enzymes are used as an aid in
the removal of
stains, such as food stains, from clothes during detergent washing. Most
laccases exhibit pH
optima in the acidic pH range while being inactive in neutral or alkaline pHs.
[05] Laccases are known to be produced by a wide variety of fungi, including
species of the
genii Aspergillus, Neurospora, Podospora, Botrytis, Pleurotus, Fornes,
Phlebia, Trametes,
Polyporus, Stachybotrys, Rhizoctonia, Bipolaris, Curvularia, Amerosporium,
Lentinus,
Myceliophtora, Coprinus, Thielavia, Cerrena, Streptomyces, and Melanocarpus.
However, there
remains a need for laccases having different performance profiles in various
applications.
[06] For many applications, the oxidizing efficiency of a laccase can be
improved through the
use of a mediator, also known as an enhancing agent. Systems that include a
laccase and a
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
2
mediator are known in the art as laccase-mediator systems (LMS). The same
compounds can
also be used to activate or initiate the action of laccase.
[07] There are several known mediators for use in a laccase-mediator system.
These include
HBT (1-hydroxybenzotriazole), ABTS [2,2'- azinobis(3- ethylbenzothiazoline-6-
sulfinic acid)],
NHA (N-hydroxyacetanilide), NEIAA (N-acetyl-N-phenylhydroxylamine), HBTO (3-
hydroxy
1,2,3-benzotriazin-4(3H)-one), and VIO (violuric acid). In addition, there are
several compounds
containing NH-OH or N-O groups that have been found to be useful as mediators.
[08] Functional groups and substituents have large effects on mediator
efficiency. Even
within the same class of compounds, a substituent can change the laccase
specificity towards a
substrate, thereby increasing or decreasing mediator efficacy greatly. In
addition, a mediator
may be effective for one particular application but unsuitable for another
application. Another
drawback for current mediators is their tendency to polymerize during use.
Thus, there is a need
to discover efficient mediators for specific applications. One such
application is the bleaching of
textiles, wherein it is also important that the mediators are not unduly
expensive or hazardous.
Other applications of the laccase-mediator system are given below.
[09] Methods of use for laccases at low temperatures would provide a benefit
in terms of
energy savings, for example, in textile processing methods where energy input
for heating of
processing baths could be reduced. Development of methods in which laccase
enzymes are used
at low temperatures for applications such as enzymatic bleaching would be
desirable.
SUMMARY
[10] Described are enzymatic oxidation systems, compositions, and methods,
involving
laccases. In one aspect, a textile processing method is provided, comprising
contacting a textile
with a laccase enzyme and, optionally, a mediator at a temperature less than
40 C, for a length
of time and under conditions sufficient to cause a color modification of the
textile. In some
embodiments, the color modification is selected from lightening of color,
change of color,
change in color cast, reduction of redeposition/backstaining, and bleaching.
In some
embodiments, the temperature is from about 20 C to less than 40 C. In some
embodiments, the
temperature is from about 20 to about 35 C. In some embodiments, the
temperature is from
about 20 C to about 30 C. In some embodiments, the temperature is from about
20 C to about
23 C. In some embodiments, the temperature is 20 C, 21 C9 22 C9 23 C9 24 C9 25
C9 26 C9
27 C9 28 C9 29 C9 30 C9 31 C9 32 C9 33 C9 34 C, or 35 C. In some embodiments,
the
temperature is the ambient temperature of tap water.
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
3
[11] In some embodiments, the textile is indigo-dyed denim. In some
embodiments, the
textile is sulfur-dyed denim. In some embodiments, the denim is desized and/or
stonewashed
prior to or simultaneously with contacting the textile with the laccase enzyme
and the mediator.
In some embodiments, the stonewashing and contacting the textile with the
laccase enzyme and
the mediator occur in the same bath.
[12] In some embodiments, the method further comprises contacting the textile
with a
cellulase enzyme, simultaneously or sequentially with contacting the textile
with the laccase
enzyme and the mediator. In some embodiments, contacting the textile with the
cellulase
enzyme and contacting the textile with the laccase enzyme and the mediator are
performed
sequentially, and wherein contacting the textile with the cellulase enzyme is
performed prior to
contacting the textile with the laccase enzyme and the mediator. In some
embodiments,
contacting the textile with the cellulase enzyme and contacting the textile
with the laccase
enzyme and the mediator are performed sequentially in the same bath without
draining the bath
between contacting the textile with a cellulase enzyme and contacting the
textile with the laccase
enzyme and the mediator.
[13] In some embodiments, contacting the textile with the cellulase enzyme and
contacting
the textile with the laccase enzyme and the mediator are performed a
temperature less than
40 C. In some embodiments, the temperature is from about 20 C to less than 40
C. In some
embodiments, the temperature is from about 20 to about 35 C. In some
embodiments, the
temperature is from about 20 C to about 30 C. In some embodiments, the
temperature is from
about 20 C to about 23 C. In some embodiments, the temperature is 20 C, 21 C9
22 C9 23 C9
24 C9 25 C9 26 C9 27 C9 28 C9 29 C9 30 C9 31 C9 32 C9 33 C9 34 C, or 35 C. In
some
embodiments, the temperature is the ambient temperature of tap water.
[14] In some embodiments, the cellulase enzyme acts synergistically with the
laccase enzyme
to produce a textile with a greater degree of lightening of color of the
textile, change in color,
change in color cast, reduction of redoposition/backstaining, and/or
bleaching. In some
embodiments, the cellulase enzyme acts additively with the laccase enzyme to
produce a textile
with a greater degree of lightening of color of the textile, change in color,
change in color cast,
reduction of redoposition/backstaining, and/or bleaching in comparison to an
identical method in
which cellulase is not included.
[15] In some embodiments, the laccase is a microbial laccase. In some
embodiments, laccase
is from a Cerrena species. In some embodiments, the laccase is from Cerrena
unicolor. In
some embodiments, the laccase is laccase D from C. unicolor.
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
4
[16] In some embodiments, the laccase has an amino acid sequence that is at
least 70%
identical to an amino acid sequence selected from the group consisting of SEQ
ID NO: 2, SEQ
ID NO: 4, SEQ ID NO: 6, SEQ ID NO: 8, SEQ ID NO: 10, SEQ ID NO: 12, SEQ ID NO:
14,
SEQ ID NO: 16, SEQ ID NO: 18, SEQ ID NO: 19, and SEQ ID NO: 20. In some
embodiments,
the laccase has an amino acid sequence that is at least 80% identical to an
amino acid sequence
selected from the group consisting of SEQ ID NO: 2, SEQ ID NO: 4, SEQ ID NO:
6, SEQ ID
NO: 8, SEQ ID NO: 10, SEQ ID NO: 12, SEQ ID NO: 14, SEQ ID NO: 16, SEQ ID NO:
18,
SEQ ID NO: 19, and SEQ ID NO: 20. In some embodiments, the laccase has an
amino acid
sequence that is at least 90%, or even at least 95%, identical to an amino
acid sequence selected
from the group consisting of SEQ ID NO: 2, SEQ ID NO: 4, SEQ ID NO: 6, SEQ ID
NO: 8,
SEQ ID NO: 10, SEQ ID NO: 12, SEQ ID NO: 14, SEQ ID NO: 16, SEQ ID NO: 18, SEQ
ID
NO: 19, and SEQ ID NO: 20.
[17] In some embodiments, the laccase has an amino acid sequence that is at
least 70%
identical to SEQ ID NO: 19 or SEQ ID NO: 20. In some embodiments, the laccase
has an
amino acid sequence that is at least 80% identical to SEQ ID NO: 19 or SEQ ID
NO: 20. In
some embodiments, the laccase has an amino acid sequence that is at least 90%
identical to SEQ
ID NO: 19 or SEQ ID NO: 20. In some embodiments, the laccase has an amino acid
sequence
that is at least 95% identical to SEQ ID NO: 19 or SEQ ID NO: 20.
[18] In some embodiments, the laccase enzyme and the mediator are provided
together in a
ready-to-use composition. In some embodiments, the laccase enzyme and the
mediator are
provided in a solid form. In some embodiments, the laccase enzyme and the
mediator are
provided as granules. In particular embodiments, the mediator is
syringonitrile.
[19] In another aspect, laccases, nucleic acid sequences encoding such
laccases, and vectors
and host cells for expressing the laccases are provided. The laccases can be
used at low
temperatures in methods in which a reduction of energy input would be
desirable, such as textile
processing. In some embodiments, the laccase enzyme comprises, consists of, or
consists
essentially of the amino acid sequence depicted in any of SEQ ID NOs: 2, 4, 6,
8, 10, 12, 14, 16,
18, 19, or 20, or an amino acid sequence having at least about 60%, 65%, 70%,
80%, 90%, 91%,
92%,93%,94%, 95%, 96%, 97%,98%,99%, or even 99.5%, identical to any of SEQ ID
NOs:
2, 4, 6, 8, 12, 14, 16, 18, 19, or 20. In particular embodiments, the laccase
has an amino acid
sequence that is at least 70%, 80%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%,
98%, 99%, or
even 99.5%, identical to SEQ ID NO: 19 or SEQ ID NO: 20. In still more
particular
embodiments, the laccase has the amino acid sequence SEQ ID NO: 19 or SEQ ID
NO: 20.
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
Preferably, such polypeptides have laccase enzymatic activity, which can be
determined, e.g.,
using the assays described, herein.
[20] In another aspect, a composition comprising a laccase enzyme comprising,
consisting of,
or consisting essentially of any of the aforementioned amino acid sequences is
provided. In
5 some embodiments, the composition further comprises a buffering system to
maintain the pH of
the composition at about 5.5 to about 6.5 in solution. In some embodiments,
the composition
further comprises a mediator. The mediator may be selected from, e.g.,
acetosyringone,
syringaldehyde, syringamide, methyl syringamide, 2-hydroxyehyl syringamide,
methyl
syringate, dimethylsyringamide, shrine acid, and 4-hydroxy-3,5-
dimethoxybenzonitrile
(syringonitrile). In one embodiment, the mediator is 4-hydroxy-3,5-
dimethoxybenzonitrile. In
some embodiments, the composition is in a solid form. In some embodiments, the
laccase
enzyme and the mediator are provided together in a ready-to-use composition.
In some
embodiments, the laccase enzyme and the mediator are provided in a solid form.
In some
embodiments, the laccase enzyme and the mediator are provided as granules. In
particular
embodiments, the mediator is syringonitrile.
[21] In some embodiments, the laccase enzyme is used at a pH of about 5 to
about 7, a
temperature of about 20 C to about 30 C, a liquor ratio of about 5:1 to about
10:1, and for a time
period of about 15 to about 60 minutes.
[22] These and other aspects and embodiments of the present system,
compositions, and
methods will be apparent from the description and accompanying figures.
BRIEF DESCRIPTION OF THE FIGURES
[23] Figure 1 shows the effects of modifying the color of stonewashed denim
with laccase
enzymes at different temperatures, as described in Example 1.
[24] Figure 2 shows the effects of laccase and mediator ratios on modifying
the color of
stonewashed denim, as described in Example 2.
[25] Figure 3 shows the effect of temperature on modifying the color of
stonewashed denim
with a "ready to use" laccase composition, as described in Example 3.
[26] Figure 4 shows the effect of temperature on color-modifying performance
of laccase
enzymes on stonewashed denim, as described in Example 3.
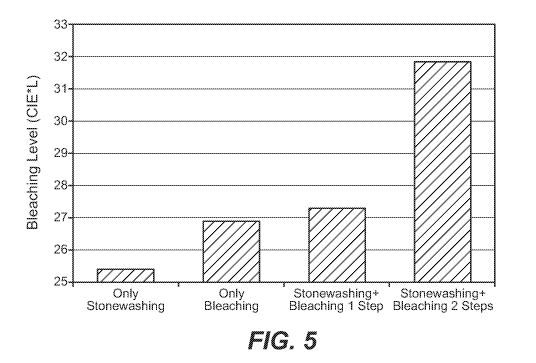
[27] Figure 5 shows the effect of cellulase treatment in combination with
laccase-mediated
color modification, as described in Examples 4-6.
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
6
DETAILED DESCRIPTION
[28] Described are enzymatic oxidation systems, compositions, and methods,
involving
laccases. The systems, compositions, and methods are useful, for example, for
low-temperature
processing of textiles to affect color modification. Such processing uses less
energy than
conventional textile processing technologies, and involves more
environmentally-friendly
chemical reagents. Various aspects and embodiments of the systems,
compositions, and
methods are to be described.
Definitions
[29] Unless defined otherwise herein, all technical and scientific terms used
herein have the
same meaning as commonly understood by one of ordinary skill in the art.
Singleton et al.,
DICTIONARY OF MICROBIOLOGY AND MOLECULAR BIOLOGY, 2D ED., John Wiley
and Sons, New York (1994), and Hale and Marham, THE HARPER COLLINS DICTIONARY
OF BIOLOGY, Harper Perennial, N.Y. (1991) provide a general dictionary of many
of the terms
used herein. The following terms are defined for additional clarity.
[30] As used herein, the term "enzyme" refers to a protein that catalyzes a
chemical reaction.
The catalytic function of an enzyme constitutes its "enzymatic activity" or
"activity." An
enzyme is typically classified according to the type of reaction it catalyzes,
e.g., oxidation of
phenols, hydrolysis of peptide bonds, incorporation of nucleotides, etc.
[31] As used herein, the term "substrate" refers to a substance (e.g., a
chemical compound) on
which an enzyme performs its catalytic activity to generate a product.
[32] As used herein, a "laccase" is a multi-copper containing oxidase (EC
1.10.3.2) that
catalyzes the oxidation of phenols, polyphenols, and anilines by single-
electron abstraction, with
the concomitant reduction of oxygen to water in a four-electron transfer
process.
[33] As used herein, "variant" proteins encompass related and derivative
proteins that differ
from a parent/reference protein by a small number of amino acid substitutions,
insertions, and/or
deletions. In some embodiments, the number of different amino acid residues is
any of about 1,
2, 3, 4, 5, 10, 20, 25, 30, 35, 40, 45, or 50. In some embodiments, variants
differ by about 1 to
about 10 amino acids residues. In some embodiments, variant proteins have at
least about 35%,
40%, 45%, 50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 91%, 92%, 93%, 94%,
95%,
96%, 97%, 98%, 99%, or even 99.5% amino acid sequence identity to a
parent/reference protein.
[34] As used herein, the term "analogous sequence" refers to a polypeptide
sequence within a
protein that provides a similar function, tertiary structure, and/or conserved
residues with respect
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
7
to a sequence within a parent/reference protein. For example, in structural
regions that contain
an alpha helix or a beta sheet structure, replacement amino acid residues in
an analogous
sequence maintain the same structural feature. In some embodiments, analogous
sequences
result in a variant protein that exhibits a similar or improved function with
respect to the parent
protein from which the variant is derived.
[35] As used herein, a "homologous protein" or "homolog" refers to a protein
(e.g., a laccase
enzyme) that has a similar function (e.g., enzymatic activity) and/or
structure as a reference
protein (e.g., a laccase enzyme from a different source). Homologs may be from
evolutionarily
related or unrelated species. In some embodiments, a homolog has a quaternary,
tertiary and/or
primary structure similar to that of a reference protein, thereby potentially
allowing for
replacement of a segment or fragment in the reference protein with an
analogous segment or
fragment from the homolog, with reduced disruptiveness of structure and/or
function of the
reference protein in comparison with replacement of the segment or fragment
with a sequence
from a non-homologous protein.
[36] As used herein, "wild-type," "native," and "naturally-occurring" proteins
are those found
in nature. The terms "wild-type sequence" refers to an amino acid or nucleic
acid sequence that
is found in nature or naturally occurring. In some embodiments, a wild-type
sequence is the
starting point of a protein engineering project, for example, production of
variant proteins.
[37] As used herein, a "signal sequence" refers to a sequence of amino acids
bound to the N-
terminal portion of a protein, and which facilitates the secretion of the
mature form of the
protein from the cell. The mature form of the extracellular protein lacks the
signal sequence
which is cleaved off during the secretion process.
[38] As used herein, the term "culturing" refers to growing a population of
microbial cells
under suitable conditions in a liquid, semi-solid, or solid medium for
expressing a polypeptide of
interest. In some embodiments, culturing is conducted in a vessel or reactor,
as known in the art.
[39] As used herein, the term "derivative" refers to a protein that is derived
from a
parent/reference protein by addition of one or more amino acids to either or
both the N- and C-
terminal end(s), substitution of one or more amino acid residues at one or a
number of different
sites in the amino acid sequence, deletion of one or more amino acid residues
at either or both
ends of the protein or at one or more sites in the amino acid sequence, and/or
insertion of one or
more amino acids at one or more sites in the amino acid sequence. The
preparation of a protein
derivative is often achieved by modifying a DNA sequence which encodes for the
native protein,
transformation of that DNA sequence into a suitable host, and expression of
the modified DNA
sequence to form the derivative protein.
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
8
[40] As used herein, the term "expression" refers to the process by which a
polypeptide is
produced based on the nucleic acid sequence of a gene. The process includes
both transcription
and translation.
[41] As used herein, the term "expression vector" refers to a DNA construct
containing a
DNA coding sequence (e.g., gene sequence) that is operably linked to one or
more suitable
control sequence(s) capable of effecting expression of the coding sequence in
a host. Such
control sequences include a promoter to effect transcription, an optional
operator sequence to
control such transcription, a sequence encoding suitable mRNA ribosome binding
sites, and
sequences which control termination of transcription and translation. The
vector may be a
plasmid, a phage particle, or simply a potential genomic insert. Once
transformed into a suitable
host, the vector may replicate and function independently of the host genome,
or may, in some
instances, integrate into the genome itself.
[42] As used herein, the term "host cell" refers to a cell or cell line into
which a recombinant
expression vector for production of a polypeptide may be transfected,
transformed, or otherwise
introduced for expression of a polypeptide. Host cells include progeny of a
single host cell, and
the progeny may not necessarily be identical (in morphology or in total
genomic DNA
complement) to the parent cell due to natural, accidental, or deliberate
mutation. A host cell
may be bacterial or fungal. A host cell includes a cell transfected or
transformed in vivo with an
expression vector.
[43] As used herein, the term "introduced," in the context of inserting a
nucleic acid sequence
into a cell includes "transfection," "transformation," and "transduction," and
refers to the
incorporation of a nucleic acid sequence into a eukaryotic or prokaryotic
cell, wherein the
nucleic acid sequence is incorporated into the genome of the cell (e.g.,
chromosome, plasmid,
plastid, or mitochondrial DNA), converted into an autonomous replicon, or
transiently
expressed.
[44] As used herein, "cleaning compositions" and "cleaning formulations" refer
to
compositions that may be used for the removal of undesired compounds from
items to be
cleaned, such as fabrics, dishes, contact lenses, hair (shampoos), skin (soaps
and creams), teeth
(mouthwashes, toothpastes), and other solid and surfaces. The terms encompass
any
materials/compounds selected for the particular type of cleaning composition
desired and the
form of the product (e.g., liquid, gel, granule, or spray composition), as
long as the composition
is compatible with the enzyme(s) used in the composition. The specific
selection of cleaning
composition materials are readily made by considering the surface, item or
fabric to be cleaned,
and the desired form of the composition for the cleaning conditions during
use.
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
9
[45] The terms further refer to any composition that is suitable for cleaning,
bleaching,
disinfecting, and/or sterilizing a object and/or surface. It is intended that
the terms include, but
are not limited to detergent compositions (e.g., liquid and/or solid laundry
detergents and fine
fabric detergents; hard surface cleaning formulations, such as for glass,
wood, ceramic and metal
counter tops and windows; carpet cleaners; oven cleaners; fabric fresheners;
fabric softeners;
and textile and laundry pre-spotters, as well as dish detergents).
[46] Indeed, the terms "cleaning compositions" and "cleaning formulations"
include (unless
otherwise indicated) granular or powder-form all-purpose or heavy-duty washing
agents,
especially cleaning detergents; liquid, gel or paste-form all-purpose washing
agents, especially
the so-called heavy-duty liquid (HDL) types; liquid fine-fabric detergents;
hand dishwashing
agents or light duty dishwashing agents, especially those of the high-foaming
type; machine
dishwashing agents, including the various tablet, granular, liquid and rinse-
aid types for
household and institutional use; liquid cleaning and disinfecting agents,
including antibacterial
hand-wash types, cleaning bars, mouthwashes, denture cleaners, car or carpet
shampoos,
bathroom cleaners; hair shampoos and hair-rinses; shower gels and foam baths
and metal
cleaners; as well as cleaning auxiliaries such as bleach additives and "stain-
stick" or pre-treat
types.
[47] As used herein, the terms "detergent composition" and "detergent
formulation" are used
in reference to mixtures that are intended for use in a wash medium for the
cleaning of soiled
objects. In some embodiments, the term is used in reference to laundering
fabrics and/or
garments (e.g., "laundry detergents"). In alternative embodiments, the term
refers to other
detergents, such as those used to clean dishes, cutlery, etc. (e.g.,
"dishwashing detergents"). In
addition to enzyme(s), "detergent compositions" and "detergent formulations"
encompasses
detergents that contain surfactants, builders, bleaching agents, bleach
activators, bluing agents
and fluorescent dyes, caking inhibitors, masking agents, enzyme activators,
antioxidants, and
solubilizers.
[48] As used herein, the phrase "detergent stability" refers to the stability
of an enzyme, and
optionally an associated substrate or mediator, in a detergent composition. In
some
embodiments, the stability is assessed during the use of the detergent, while
in other
embodiments, the term refers to the stability of a detergent composition
during storage.
[49] As used herein the term "hard surface cleaning composition," refers to
detergent
compositions for cleaning hard surfaces such as floors, walls, tiles,
stainless steel vessels (e.g.,
fermentation tanks), bath and kitchen fixtures, and the like. Such
compositions may be provided
in any form, including but not limited to solids, liquids, emulsions, and the
like.
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
[50] As used herein, the term "dishwashing composition" refers to all forms of
compositions
for cleaning dishes, including but not limited to granular and liquid forms.
[51] As used herein, the term "disinfecting" refers to the removal or killing
of microbes,
including fungi, bacteria, spores, and the like.
5 [52] As used herein, the term "fabric cleaning composition" refers a form of
detergent
composition for cleaning fabrics, including but not limited to, granular,
liquid and bar forms.
[53] As used herein, the terms "polynucleotide," "nucleic acid," and
"oligonucleotide," are
used interchangeably to refers to a polymeric form of nucleotides of any
length and any three-
dimensional structure, whether single- or multi-stranded (e.g., single-
stranded, double-stranded,
10 triple-helical, etc.), which contain deoxyribonucleotides, ribonucleotides,
and/or analogs or
modified forms of deoxyribonucleotides or ribonucleotides, including modified
nucleotides or
bases or their analogs. Because the genetic code is degenerate, more than one
codon may be
used to encode a particular amino acid. Any type of modified nucleotide or
nucleotide analog
may be used, so long as the polynucleotide retains the desired functionality
under conditions of
use, including modifications that increase nuclease resistance (e.g., deoxy,
2'-O-Me,
phosphorothioates, etc.). Labels may also be incorporated for purposes of
detection or capture,
for example, radioactive or nonradioactive labels or anchors, e.g., biotin.
The term
polynucleotide also includes peptide nucleic acids (PNA). Polynucleotides may
be naturally
occurring or non-naturally occurring. A sequence of nucleotides may be
interrupted by non-
nucleotide components. One or more phosphodiester linkages may be replaced by
alternative
linking groups. For example, phosphate may be replaced by P(O)S ("thioate"),
P(S)S
("dithioate"), (O)NR2 ("amidate"), P(O)R, P(O)OR', CO or CH2 ("formacetal"),
in which each R
or R' is independently H or substituted or unsubstituted alkyl (1-20 C)
optionally containing an
ether (-0-) linkage, aryl, alkenyl, cycloalkyl, cycloalkenyl or araldyl. Not
all linkages in a
polynucleotide need be identical. Polynucleotides may be linear or circular or
comprise a
combination of linear and circular portions.
[54] As used herein, the terms "polypeptide, "protein," and "peptide," refer
to a composition
comprised of amino acids (i.e., amino acid residues). The conventional one-
letter or three-letter
codes for amino acid residues are used. A polypeptide may be linear or
branched, may comprise
modified amino acids, and may be interrupted by non-amino acids. The terms
also encompass
an amino acid polymer that has been modified naturally or by intervention; for
example,
disulfide bond formation, glycosylation, lipidation, acetylation,
phosphorylation, or any other
manipulation or modification, such as conjugation with a labeling component.
Also included
within the definition are, for example, polypeptides containing one or more
analogs of an amino
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
11
acid (including, for example, unnatural amino acids, etc.), as well as other
modifications known
in the art.
[55] As used herein, the term "primer" refers to an oligonucleotide, whether
occurring
naturally, e.g., as in a purified restriction fragment, or produced
synthetically, which is capable
of acting as a point of initiation of nucleic acid synthesis when incubated
with a complementary
nucleic acid in the presence of nucleotides and polymerase at a suitable
temperature and pH.
The primer is preferably single stranded but may alternatively be double
stranded. If double
stranded, the primer is first treated to separate its strands before being
used to prepare extension
products. Preferably, the primer is an oligodeoxyribonucleotide. The exact
lengths of the
primers will depend on many factors, including temperature, source of primer
and the use of the
method.
[56] As used herein, the terms "recovered," "isolated," "purified," and
"separated" refer to a
material (e.g., a protein, nucleic acid, or cell) that is removed from at
least one component with
which it is naturally-associated, or associated as the result of heterologous
expression.
[57] As used herein, the term "textile(s)" refers to fibers, yams, fabrics,
garments, and non-
woven materials. The term encompasses textiles made from natural and synthetic
(e.g.,
manufactured) materials, as well as natural and synthetic blends. The term
"textile" refers to
both unprocessed and processed fibers, yarns, woven or knit fabrics, non-
wovens, and garments.
In some embodiments, a textile contains cellulose.
[58] As used herein, the phrase "textile(s) in need of processing" refers to a
textile that needs
to be desized, scoured, bleached, and/or biopolished to produce a desired
effect.
[59] As used herein, the phrase "textile(s) in need of color modification"
refers to a textile
that needs to be altered with respect to it color. These textiles may or may
not have been already
subjected to other treatments. Similarly, these textiles may or may not need
subsequent
treatments.
[60] As used herein, the term "fabric" refers to a manufactured assembly of
fibers and/or
yarns that has substantial surface area in relation to its thickness and
sufficient cohesion to give
the assembly useful mechanical strength.
[61] As used herein, the term "color modification" refers to a change in the
chroma,
saturation, intensity, luminance, and/or tint of a color associated with a
fiber, yarn, fabric,
garment, or non-woven material, collectively referred to as textile materials.
Without being
limited to a theory, it is proposed that color modification results from the
modification of
chromaphores associated with a textile material, thereby changing its visual
appearance. The
chromophores may be naturally-associated with the material used to manufacture
a textile (e.g.,
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
12
the white color of cotton) or associated with special finishes, such as dying
or printing. Color
modification encompasses chemical modification to a chromophore as well as
chemical
modification to the material to which a chromophore is attached. Examples of
color
modification include fading, bleaching, and altering tint. A particular color
modification to
indigo-dyed denim is fading to a "vintage look," which has a less intense
blue/violet tint and
more subdued grey appearance than the freshly-dyed denim.
[62] As used herein, the term "bleaching" refers to the process of treating a
textile material
such as a fiber, yarn, fabric, garment or non-woven material to produce a
lighter color. This
term includes the production of a brighter and/or whiter textile, e.g., in the
context of a textile
processing application, as well as lightening of the color of a stain, e.g.,
in the context of a
cleaning application.
[63] As used herein, the terms "size" and "sizing" refer to compounds used in
the textile
industry to improve weaving performance by increasing the abrasion resistance
and strength of a
yarn. Size is usually made of starch or starch-like compounds.
[64] As used herein, the terms "desize" and "desizing" refer to the process of
eliminating/removing size (generally starch) from a textile, usually prior to
applying special
finishes, dyes or bleaches.
[65] As used herein, the term "desizing enzyme(s)" refers to an enzyme used to
remove size.
Exemplary enzymes are amylases, cellulases, and mannanases.
[66] As used herein, the term "% identity" refers to the level of nucleic acid
sequence identity
between a nucleic acid sequence that encodes a laccase as described herein and
another nucleic
acid sequence, or the level of amino acid sequence identity between a laccase
enzyme as
described herein and another amino aid sequence. Alignments may be performed
using a
conventional sequence alignment program. Exemplary levels of nucleic acid and
amino acid
sequence identity include, but are not limited to, at least 60%, at least 65%,
at least 70%, at least
75%, at least 80%, at least 85%, at least 90%, at least 91%, at least 92%, at
least 93%, at least
94%, at least 95%, at least 96%, at least 97%, at least 98%, or even at least
99%, or more,
sequence identity to a given sequence, e.g., the coding sequence for a laccase
or the amino acid
sequence of a laccase, as described herein.
[67] Exemplary computer programs that can be used to determine identity
between two
sequences include, but are not limited to, the suite of BLAST programs, e.g.,
BLASTN,
BLASTX, and TBLASTX, BLASTP and TBLASTN, publicly available on the Internet at
www.ncbi.nlm.nih.gov/BLAST. See also, Altschul, et al., 1990 and Altschul, et
al., 1997.
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
13
[68] Sequence searches are typically carried out using the BLASTN program when
evaluating
a given nucleic acid sequence relative to nucleic acid sequences in the
GenBank DNA
Sequences and other public databases. The BLASTX program is preferred for
searching nucleic
acid sequences that have been translated in all reading frames against amino
acid sequences in
the GenBank Protein Sequences and other public databases. Both BLASTN and
BLASTX are
run using default parameters of an open gap penalty of 11.0, and an extended
gap penalty of 1.0,
and utilize the BLOSUM-62 matrix. (See, e.g., Altschul, et al., 1997.)
[69] An alignment of selected sequences in order to determine "% identity"
between two or
more sequences, may be performed using, for example, the CLUSTAL-W program in
Mac Vector version 6.5, operated with default parameters, including an open
gap penalty of 10.0,
an extended gap penalty of 0.1, and a BLOSUM 30 similarity matrix.
[70] As used herein, the terms "chemical mediator" and "mediator" are used
interchangeably
to refer to a chemical compound that functions as a redox mediator to shuttle
electrons between
an enzyme exhibiting oxidase activity (e.g., a laccase) and a secondary
substrate or electron
donor. Such chemical mediators are also known in the art as "enhancers" and
"accelerators."
[71] As used herein, the terms "draining" or "dropping" with respect to a bath
in which textile
materials are present refers to fully or partially releasing/emptying the
solvent and reagents
present in a bath. Draining a bath is typically performed between process
steps such that the
solvent and reagents present in one processing step do not interfere with a
subsequent processing
step. Draining may be accompanied by one or more rinse steps to further remove
such the
solvent and reagents.
[72] As used herein, the terms "secondary substrate" and "electron donor" are
used
interchangeably to refer to a dye, pigment (e.g., indigo), chromophore (e.g.,
polyphenolic,
anthocyanin, or carotenoid), or other secondary substrate to and from which
electrons can be
shuttled by an enzyme exhibiting oxidase activity.
[73] The following abbreviations/acronyms have the following meanings unless
otherwise
specified:
EC enzyme commission
EDTA ethylenediaminetetraacetic acid
kDa kiloDalton
MW molecular weight
w/v weight/volume
w/w weight/weight
v/v volume/volume
wt% weight percent
C degrees Centigrade
H2O water
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
14
dH2O or DI deionized water
dIH2O deionized water, Milli-Q filtration
g or gm gram
g microgram
mg milligram
kg kilogram
L and l microliter
mL and ml milliliter
mm millimeter
m micrometer
M molar
mm millimolar
M micromolar
U unit
sec and " second
min and' minute
hr hour
eq. equivalent
N normal
RTU ready-to-use
U Unit
owg on weight of goods
CIE International Commission on Illumination
[74] Numeric ranges are inclusive of the numbers defining the range. The
singular articles
"a," "an," "the," and the like, include the plural referents unless otherwise
clear from context.
Unless otherwise specified, polypeptides are written in the standard N-
terminal to C-terminal
direction and polynucleotides are written in the standard 5' to 3' direction.
It is to be understood
that the particular methodologies, protocols, and reagents described, are not
intended to be
limiting, as equivalent methods and materials can be used in the practice or
testing of the present
compositions and methods. Although the description is divided into sections to
assist the reader,
section heading should not be construed as limiting and the description in one
section may apply
to another. All publications cited herein are expressly incorporated by
reference.
Laccase and Laccase Related Enzymes
[75] The enzymatic oxidation systems, compositions, and methods include one or
more
laccases or laccase-related enzymes, herein collectively referred to as
"laccases" or "laccase
enzymes." Such laccases include any laccase enzyme encompassed by EC 1.10.3.2,
according
to the Nomenclature Committee of the International Union of Biochemistry and
Molecular
Biology (IUBMB). Laccase enzymes from microbial and plant origin are known in
the art. A
microbial laccase enzyme may be derived from bacteria or fungi (including
filamentous fungi
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
and yeasts). Suitable examples include a laccase derived or derivable from a
strain of
Aspergillus, Neurospora (e.g., N. crassa), Podospora, Botrytis, Collybia,
Cerrena (e.g., C.
unicolor), Stachybotrys, Panus (e.g., P. rudis), Thielavia, Fomes, Lentinus,
Pleurotus, Trametes
(e.g., T. villosa, and T. versicolor), Rhizoctonia (e.g., R. solani), Coprinus
(e.g., C. plicatilis and
5 C. cinereus), Psatyrella, Myceliophthora (e.g., M. thermonhila),
Schytalidium, Phlebia (e.g., P.
radita (WO 92/01046)), or Coriolus (e.g., C. hirsutus (JP 2238885)),
Spongipellis, Polyporus,
Ceriporiopsis subvermispora, Ganoderma tsunodae, and Trichoderma.
[76] A laccase may be produced by culturing a host cell transformed with a
recombinant
DNA vector that includes nucleotide sequences encoding the laccase. The DNA
vector may
10 further include nucleotide sequences permitting the expression of the
laccase in a culture
medium, and optionally allowing the recovery of the laccase from the culture.
[77] An expression vector containing a polynucleotide sequence encoding a
laccase enzyme
may be transformed into a suitable host cell. The host cell may be a fungal
cell, such as a
filamentous fungal cell, examples of which include but are not limited to
species of Trichoderma
15 [e.g., T reesei (previously classified as T. longibrachiatum and currently
also known as
Hypocrea jecorina], T. viride, T. koningii, and T. harzianum), Aspergillus
(e.g., A. niger, A.
nidulans, A. oryzae, and A. awamori), Penicillium, Humicola (e.g., H. insolens
and H. grisea),
Fusarium (e.g., F. graminum and F. venenatum), Neurospora, Hypocrea, and
Mucor. A host
cell for expression of a laccase enzyme may also be from a species of Cerrena
(e.g., C.
unicolor). Fungal cells may be transformed by a process involving protoplast
formation and
transformation of the protoplasts followed by regeneration of the cell wall
using techniques
known in the art.
[78] Alternatively, the host organism may from a species of bacterium, such as
Bacillus [e.g.,
B. subtilis, B. licheniformis, B. lentus, B. (now Geobacillus)
stearothermophilus, and B. brevis],
Pseudomonas, Streptomyces (e.g., S. coelicolor, S. lividans), or E. coli. The
transformation of
bacterial cells may be performed according to conventional methods, e.g., as
described in
Maniatis, T. et al., "Molecular Cloning: A Laboratory Manual," Cold Spring
Harbor, 1982. The
screening of appropriate DNA sequences and construction of vectors may also be
carried out by
standard procedures (cf. supra).
[79] The medium used to culture the transformed host cells may be any
conventional medium
suitable for growing the host cells. In some embodiments, the expressed enzyme
is secreted into
the culture medium and may be recovered therefrom by well-known procedures.
For example,
laccases may be recovered from a culture medium as described in U.S. Patent
Publication No.
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
16
2008/0196173. In some embodiments, the enzyme is expressed intracellularly and
is recovered
following disruption of the cell membrane.
[80] In particular embodiments, the expression host may be Trichoderma reesei
with the
laccase coding region under the control of a CBH1 promoter and terminator
(see, e.g., U.S.
Patent No. 5,861,271). The expression vector may be, e.g., pTrex3g, as
disclosed in U.S. Patent
No. 7,413,887. In some embodiments, laccases are expressed as described in
U.S. Patent
Publication Nos. 2008/0196173 or 2009/0221030.
[81] The following laccase genes and laccases are described in U.S.
Publication No.
2008/0196173:
A. Cerrena laccase Al gene from CBS 115.075 strain
Polynucleotide sequence (SEQ ID NO: 1):
ATGAGCTCAA AGCTACTTGC TCTTATCACT GTCGCTCTCG TCTTGCCACT 50
AGGCACCGAC GCCGGCATCG GTCCTGTTAC CGACTTGCGC ATCACCAACC 100
AGGATATCGC TCCAGATGGC TTCACCCGAC CAGCGGTACT AGCTGGGGGC 150
ACATTCCCTG GAGCACTTAT TACCGGTCAG AAGGTATGGG AGATCAACTT 200
GGTTGAATAG AGAAATAAAA GTGACAACAA ATCCTTATAG GGAGACAGCT 250
TCCAAATCAA TGTCATCGAC GAGCTTACCG ATGCCAGCAT GTTGACCCAG 300
ACATCCATTG TGAGTATAAT TTAGGTCCGC TCTTCTGGCT ATCCTTTCTA 350
ACTCTTACCG TCTAGCATTG GCACGGCTTC TTTCAGAAGG GATCTGCGTG 400
GGCCGATGGT CCTGCCTTCG TTACTCAATG CCCTATCGTC ACCGGAAATT 450
CCTTCCTGTA CGACTTTGAT GTTCCCGACC AACCTGGTAC TTTCTGGTAC 500
CATAGTCACT TGTCTACTCA ATATTGCGAT GGTCTTCGTG GCCCGTTCGT 550
TGTATACGAT CCAAAGGATC CTAATAAACG GTTGTACGAC ATTGACAATG 600
GTATGTGCAT CATCATAGAG ATATAATTCA TGCAGCTACT GACCGTGACT 650
GATGCTGCCA GATCATACGG TTATTACCCT GGCAGACTGG TACCACGTTC 700
TCGCAAGAAC TGTTGTCGGA GTCGCGTAAG TACAGTCTCA CTTATAGTGG 750
TCTTCTTACT CATTTTGACA TAGGACACCC GACGCAACCT TGATCAACGG 800
TTTGGGCCGT TCTCCAGACG GGCCAGCAGA TGCTGAGTTG GCTGTCATCA 850
ACGTTAAACG CGGCAAACGG TATGTTATTG AACTCCCGAT TTCTCCATAC 900
ACAGTGAAAT GACTGTCTGG TCTAGTTATC GATTTCGTCT GGTCTCCATC 950
TCATGTGACC CTAATTACAT CTTTTCTATC GACAACCATT CTATGACTGT 1000
CATCGAAGTC GATGGTGTCA ACACCCAATC CCTGACCGTC GATTCTATTC 1050
AAATCTTCGC AGGCCAACGA TACTCGTTCG TCGTAAGTCT CTTTGCACGA 1100
TTACTGCTTC TTTGTCCATT CTCTGACCTG TTTAAACAGC TCCATGCCAA 1150
CCGTCCTGAA AACAACTATT GGATCAGGGC CAAACCTAAT ATCGGTACGG 1200
ATACTACCAC AGACAACGGC ATGAACTCTG CCATTCTGCG ATACAACGGC 1250
GCACCTGTTG CGGAACCGCA AACTGTTCAA TCTCCCAGTC TCACCCCTTT 1300
GCTCGAACAG AACCTTCGCC CTCTCGTGTA CACTCCTGTG GTATGTTTCA 1350
AAGCGTTGTA ATTTGATTGT GGTCATTCTA ACGTTACTGC GTTTGCATAG 1400
CCTGGAAACC CTACGCCTGG CGGCGCCGAT ATTGTCCATA CTCTTGACTT 1450
GAGTTTTGTG CGGAGTCAAC ATTCGTAAAG ATAAGAGTGT TTCTAATTTC 1500
TTCAATAATA GGATGCTGGT CGCTTCAGTA TCAACGGTGC CTCGTTCCTT 1550
GATCCTACCG TCCCCGTTCT CCTGCAAATT CTCAGCGGCA CGCAGAATGC 1600
ACAAGATCTA CTCCCTCCTG GAAGTGTGAT TCCTCTCGAA TTAGGCAAGG 1650
TCGTCGAATT AGTCATACCT GCAGGTGTCG TCGGTGGACC TCATCCGTTC 1700
CATCTCCATG GGGTACGTAA CCCGAACTTA TAACAGTCTT GGACTTACCC 1750
GCTGACAAGT GCATAGCATA ACTTCTGGGT CGTGCGAAGT GCCGGAACCG 1800
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
17
ACCAGTACAA CTTTAACGAT GCCATTCTCC GAGACGTCGT CAGTATAGGA 1850
GGAACCGGGG ATCAAGTCAC CATTCGTTTC GTGGTATGTT TCATTCTTGT 1900
GGATGTATGT GCTCTAGGAT ACTAACCGGC TTGCGCGTAT AGACCGATAA 1950
CCCCGGACCG TGGTTCCTCC ATTGCCATAT CGACTGGCAC TTGGAAGCGG 2000
GTCTCGCTAT CGTATTTGCA GAGGGAATTG AAAATACTGC TGCGTCTAAT 2050
TTAACCCCCC GTACGCGGTT TCCCTCACAT CCTGGAGCTA AGCAGCTTAC 2100
TAACATACAT TTGCAGAGGC TTGGGATGAG CTTTGCCCGA AGTATAACGC 2150
GCTCAGCGCA CAAAAGAAGG TTGCATCTAA GAAAGGCACT GCCATCTAAT 2200
TTTTGTAACA AACAAGGAGG GTCTCTTGTA CTTTTATTGG GATTTCTTTC 2250
TTGGGGTTTA TTGTTAAACT TGACTCTACT ATGTTTGGAA GACGAAAGGG 2300
GCTCGCGCAT TTATATACTA TCTCTCTTGG CATCACCTGC AGCTCAATCC 2350
TTCAACCACC TAA 2363
Translated protein sequence (SEQ ID NO: 2):
MSSKLLALIT VALVLPLGTD AGIGPVTDLR ITNQDIAPDG FTRPAVLAGG 50
TFPGALITGQ KGDSFQINVI DELTDASMLT QTSIHWHGFF QKGSAWADGP 100
AFVTQCPIVT GNSFLYDFDV PDQPGTFWYH SHLSTQYCDG LRGPFVVYDP 150
KDPNKRLYDI DNDHTVITLA DWYHVLARTV VGVATPDATL INGLGRSPDG 200
PADAELAVIN VKRGKRYRFR LVSISCDPNY IFSIDNHSMT VIEVDGVNTQ 250
SLTVDSIQIF AGQRYSFVLH ANRPENNYWI RAKPNIGTDT TTDSGMNSAI 300
LRYNGAPVAE PQTVQSPSLT PLLEQNLRPL VYTPVPGNPT PGGADIVHTL 350
DLSFDAGRFS INGASFLDPT VPVLLQILSG TQNAQDLLPP GSVIPLELGK 400
VVELVIPAGV VGGPHPFHLH GHNFWVVRSA GTDQYNFNDA ILRDVVSIGG 450
TGDQVTIRFV TDNPGPWFLH CHIDWHLEAG LAIVFAEGIE NTAASNLTPQ 500
AWDELCPKYN ALSAQKKLNP STT 523
B. Cerrena laccase A2 gene from CBS 154.29 strain
Polynucleotide sequence (SEQ ID NO: 3):
ATGAGCTCAA AGCTACTTGC TCTTATTACT GTCGCTCTCG TCTTGCCACT 50
AGGCACTGAC GCCGGCATCG GTCCTGTTAC CGACTTGCGC ATCACCAACC 100
AGGATATCGC TCCAGATGGC TTCACCCGAC CAGCTGTACT GGCTGGGGGC 150
ACATTCCCCG GAGCACTGAT TACCGGTCAG AAGGTATGGG AGATCGATTT 200
CGTTGAATAG AGAAATACAA CTGAAAACAA ATTCTTATAG GGAGACAGCT 250
TCCAAATCAA TGTCATCGAC GAGCTTACCG ATGCCAGCAT GTTGACCCAG 300
ACATCCATTG TGAGTATAAT ATGGGTCCGC TCTTCTAGCT ATCCTTTCTA 350
ACTCTTACCC TCTAGCATTG GCACGGCTTC TTTCAGAAGG GATCTGCGTG 400
GGCCGATGGT CCTGCCTTCG TTACTCAATG TCCTATCGTC ACCGGAAATT 450
CCTTCCTGTA CGACTTTGAT GTCCCCGACC AACCTGGTAC TTTCTGGTAC 500
CATAGTCACT TGTCTACTCA ATATTGCGAT GGTCTTCGGG GCCCGTTCGT 550
TGTATACGAT CCAAAGGATC CTAATAAACG GTTGTACGAC ATTGACAATG 600
GTATGTGCAT CATCATAAAA ATATAATTCA TGCAGCTACT GACCGCGACT 650
GATGCTGCCA GATCATACGG TTATTACCCT GGCAGACTGG TACCACGTTC 700
TCGCACGAAC TGTTGTCGGA GTCGCGTAAG TACAGTCTGA CTTATAGTGG 750
TCTTCTTACT CATTTTGACA TAGGACACCC GACGCAACCT TGATCAACGG 800
TTTGGGCCGT TCTCCAGACG GGCCAGCAGA TGCTGAGTTG GCTGTCATCA 850
ACGTTAAACG CGGCAAACGG TATGTCATTG AACTCCCGAT TTCTCCATTC 900
ACATTGAAAT GACTGTCTGG TCTAGTTATC GATTCCGTCT GGTCTCCATC 950
TCATGTGACC CTAATTACAT CTTTTCTATC GACAACCATT CTATGACTGT 1000
CATCGAAGTC GATGGTGTCA ACACCCAATC CCTGACCGTC GATTCTATCC 1050
AAATCTTCGC AGGCCAACGC TACTCGTTCG TCGTAAGTCT CTTTGAATGG 1100
TTGGTGCTTT TTCTGTCCAT TCTCTAACCT GTTTATACAG CTCCATGCCA 1150
ACCGTCCTGA AAACAACTAT TGGATCAGGG CCAAACCTAA TATCGGTACG 1200
GATACTACCA CAGACAACGG CATGAACTCT GCCATTCTGC GATACAACGG 1250
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
18
CGCACCTGTT GCGGAACCGC AAACTGTTCA ATCTCCCAGT CTCACCCCTT 1300
TGCTCGAACA GAACCTTCGC CCTCTCGTGT ACACTCCTGT GGTATGTTTC 1350
AAAGCGTTGT AATTTGATTG TGGTCATTCT AACGTTACTG CCTTTGCACA 1400
GCCTGGAAAT CCTACGCCTG GCGGGGCCGA TATTGTCCAT ACTCTTGACT 1450
TGAGTTTTGT GCGGAGTCAA CATTCGTAAA GATAAGAGTG TTTCTAATTT 1500
CTTCAATAAT AGGATGCTGG TCGCTTCAGT ATCAACGGTG CCTCGTTCCT 1550
TGATCCTACC GTCCCTGTTC TCCTGCAAAT TCTCAGCGGC ACGCAGAATG 1600
CACAAGATCT ACTCCCTCCT GGAAGTGTGA TTCCTCTCGA ATTAGGCAAG 1650
GTCGTCGAAT TAGTCATACC TGCAGGTGTT GTCGGTGGAC CTCATCCGTT 1700
CCATCTCCAT GGGGTACGTA ACCCGAACTT ATAACAGTCT TGGACTTACC 1750
CGCTGACAAG TGTATAGCAT AACTTCTGGG TCGTGCGAAG TGCCGGAACC 1800
GACCAGTACA ACTTTAACGA TGCCATTCTC CGAGACGTCG TCAGTATAGG 1850
AGGAACCGAG GATCAAGTCA CCATTCGATT CGTGGTATAT ACTTCATTCT 1900
TGTGGATGTA TGTGCTCTAG GATACTAACT GGCTTGCGCG TATAGACCGA 1950
TAACCCCGGA CCGTGGTTCC TCCATTGCCA TATCGACTGG CACTTGGAAG 2000
CGGGTCTCGC TATCGTATTT GCAGAGGGAA TTGAAAATAC TGCTGCGTCT 2050
AATCCAACCC CCCGTATGCG GTTTCCCACA CATTCTGAAT CTAAGCAGCT 2100
TACTAATATA CATTTGCAGA GGCTTGGGAT GAGCTTTGCC CGAAGTATAA 2150
CGCGCTCAAC GCACAAAAGA AGGTTGCATC TAAGAAAGGC ACTGCCATCT 2200
AATCCTTGTA ACAAACAAGG AGGGTCTCTT GTACTTTTAT TGGGATTTAT 2250
TTCTTGGGGT TTATTGTTCA ACTTGATTCT ACTATGTTTG GAAGTAGCGA 2300
TTACGAAAGG GGCTTGCGCA TTTATATACC ATCTTTCTTG GCACCACCTG 2350
CAGCTCAATC CTTCAACCAC CTAA 2374
Translated protein sequence (SEQ ID NO: 4):
MSSKLLALIT VALVLPLGTD AGIGPVTDLR ITNQDIAPDG FTRPAVLAGG 50
TFPGALITGQ KGDSFQINVI DELTDASMLT QTSIHWHGFF QKGSAWADGP 100
AFVTQCPIVT GNSFLYDFDV PDQPGTFWYH SHLSTQYCDG LRGPFVVYDP 150
KDPNKRLYDI DNDHTVITLA DWYHVLARTV VGVATPDATL INGLGRSPDG 200
PADAELAVIN VKRGKRYRFR LVSISCDPNY IFSIDNHSMT VIEVDGVNTQ 250
SLTVDSIQIF AGQRYSFVLH ANRPENNYWI RAKPNIGTDT TTDNGMNSAI 300
LRYNGAPVAE PQTVQSPSLT PLLEQNLRPL VYTPVPGNPT PGGADIVHTL 350
DLSFDAGRFS INGASFLDPT VPVLLQILSG TQNAQDLLPP GSVIPLELGK 400
VVELVIPAGV VGGPHPFHLH GHNFWVVRSA GTDQYNFNDA ILRDVVSIGG 450
TEDQVTIRFV TDNPGPWFLH CHIDWHLEAG LAIVFAEGIE NTAASNPTPQ 500
AWDELCPKYN ALNAQKKLNP STT 523
C. Cerrena laccase B 1 gene from CBS 115.075 strain
Polynucleotide sequence (SEQ ID NO: 5):
ATGTCTCTTC TTCGTAGCTT GACCTCCCTC ATCGTACTAG TCATTGGTGC 50
ATTTGCTGCA ATCGGTCCAG TCACTGACCT ACATATAGTG AACCAGAATC 100
TCGACCCAGA TGGTTTCAAC CGCCCCACTG TACTCGCAGG TGGTACTTTC 150
CCCGGTCCTC TGATTCGTGG TAACAAGGTA CGCTTCATAA CCGCCCTCCG 200
TAGACGTAGG CTTCGGCTGA CATGACCATC ATCTGTAGGG AGATAACTTT 250
AAAATTAATG TGATTGACGA CTTGACAGAG CACAGTATGC TCAAGGCTAC 300
GTCCATCGTA AGTCCCTGAT TAACGTTTCA CCTGGTCATA TCGCTCAACG 350
TCTCGAAGCA CTGGCATGGG TTCTTCCAGA AGGGAACCAA CTGGGCCGAT 400
GGCCCCGCCT TTGTCACCCA ATGTCCTATC ACATCAGGAA ACGCCTTCCT 450
GTATGATTTC AACGTTCCGG ACCAAGCTGG TACTTTCTGG TACCACAGCC 500
ATCTCTCTAC ACAGTATTGT GACGGTCTTC GTGGTGCCTT TGTCGTCTAT 550
GATCCTAATG ATCCCAACAA GCAACTCTAT GATGTTGATA ACGGCAAGTT 600
CCTTGCATAT TTCATTTCTA TCATATCCTC ACCTGTATTG GCACAGAAAG 650
CACCGTGATT ACCTTGGCTG ATTGGTATCA TGCCCTTGCT CAGACTGTCA 700
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
19
CTGGTGTCGC GTGAGTGACA AATGGCCCTC AATTGTTCAC ATATTTTCCT 750
GATTATCATA TGATAGAGTA TCTGATGCAA CGTTGATCAA CGGATTGGGA 800
CGTTCGGCCA CCGGCCCCGC AAATGCCCCT CTGGCGGTCA TCAGTGTCGA 850
GCGGAATAAG AGGTCAGTTC CATAATTATG ATTATTTCCC GCGTTACTTC 900
CTAACAATTA TTTTTGTATC CCTCCACAGA TATCGTTTCC GATTGGTTTC 950
TATTTCTTGC GACCCTAACT TTATTTTCTC AATTGACCAC CACCCAATGA 1000
CCGTAATTGA GATGGACGGT GTTAATACCC AATCTATGAC CGTAGATTCG 1050
ATCCAAATAT TCGCAGGTCA ACGATATTCA TTTGTCGTAG GTTATTATAA 1100
ACTGCCCACC GATCATCTCT CACGTAACTG TTATAGATGC AAGCCAACCA 1150
ACCAGTTGGA AATTATTGGA TCCGCGCTAA ACCTAATGTT GGGAACACAA 1200
CTTTCCTTGG AGGCCTGAAC TCCGCTATAT TACGATATGT GGGAGCCCCT 1250
GACCAAGAAC CGACCACTGA CCAAACACCC AACTCTACAC CGCTCGTTGA 1300
GGCGAACCTA CGACCCCTCG TCTATACTCC TGTGGTATGT TGTTCTCGTT 1350
ACATATACCA AACCTAATAT GAAGACTGAA CGGATCTACT AGCCGGGACA 1400
GCCATTCCCT GGCGGTGCTG ATATCGTCAA GAACTTAGCT TTGGGTTTCG 1450
TACGTGTATT TCACTTCCCT TTTGGCAGTA ACTGAGGTGG AATGTATATA 1500
GAATGCCGGG CGTTTCACAA TCAATGGAGC GTCCCTCACA CCTCCTACAG 1550
TCCCTGTACT ACTCCAGATC CTCAGTGGTA CTCACAATGC ACAGGATCTT 1600
CTCCCAGCAG GAAGCGTGAT CGAACTTGAA CAGAATAAAG TTGTCGAAAT 1650
CGTTTTGCCC GCTGCGGGCG CCGTTGGCGG TCCTCATCCT TTTCACTTAC 1700
ATGGTGTAAG TATCAGACGT CCTCATGCCC ATATTGCTCC GAACCTTACA 1750
CACCTGATTT CAGCACAATT TCTGGGTGGT TCGTAGCGCC GGTCAAACCA 1800
CATACAATTT CAATGATGCT CCTATCCGTG ATGTTGTCAG TATTGGCGGT 1850
GCAAACGATC AAGTCACGAT CCGATTTGTG GTATGTATCT CGTGCCTTGC 1900
ATTCATTCCA CGAGTAATGA TCCTTACACT TCGGGTTCTC AGACCGATAA 1950
CCCTGGCCCA TGGTTCCTTC ACTGTCACAT TGACTGGCAT TTGGAGGCTG 2000
GGTTCGCTGT AGTCTTTGCG GAGGGAATCA ATGGTACTGC AGCTGCTAAT 2050
CCAGTCCCAG GTAAGACTCT CGCTGCTTTG CGTAATATCT ATGAATTTAA 2100
ATCATATCAA TTTGCAGCGG CTTGGAATCA ATTGTGCCCA TTGTATGATG 2150
CCTTGAGCCC AGGTGATACA TGA 2173
Translated protein sequence (SEQ ID NO: 6):
MSLLRSLTSL IVLVIGAFAA IGPVTDLHIV NQNLDPDGFN RPTVLAGGTF 50
PGPLIRGNKG DNFKINVIDD LTEHSMLKAT SIHWHGFFQK GTNWADGPAF 100
VTQCPITSGN AFLYDFNVPD QAGTFWYHSH LSTQYCDGLR GAFVVYDPND 150
PNKQLYDVDN GNTVITLADW YHALAQTVTG VAVSDATLIN GLGRSATGPA 200
NAPLAVISVE RNKRYRFRLV SISCDPNFIF SIDHHPMTVI EMDGVNTQSM 250
TVDSIQIFAG QRYSFVMQAN QPVGNYWIRA KPNVGNTTFL GGLNSAILRY 300
VGAPDQEPTT DQTPNSTPLV EANLRPLVYT PVPGQPFPGG ADIVKNLALG 350
FNAGRFTING ASLTPPTVPV LLQILSGTHN AQDLLPAGSV IELEQNKVVE 400
IVLPAAGAVG GPHPFHLHGH NFWVVRSAGQ TTYNFNDAPI RDVVSIGGAN 450
DQVTIRFVTD NPGPWFLHCH IDWHLEAGFA VVFAEGINGT AAANPVPAAW 500
NQLCPLYDAL SPGDT 515
D. Cerrena laccase B2 gene from CBS 154.29 strain
Polynucleotide sequence (SEQ ID NO: 7):
CACCGCGATG TCTCTTCTTC GTAGCTTGAC CTCCCTCATC GTACTAGCCA 50
CTGGTGCATT TGCTGCAATC GGTCCAGTCA CCGACCTACA TATAGTGAAC 100
CAGAATCTCG CCCCAGATGG TTTAAACCGC CCCACTGTAC TCGCAGGTGG 150
TACTTTCCCC GGTCCTCTGA TTCGTGGTAA CAAGGTACGC TTCATAACCG 200
CCCTCCGTAG ACGTAGGCTT CGGCTGACAT GACCATCATC TGTAGGGAGA 250
TAACTTTAAA ATTAATGTGA TTGACGACTT GACAGAACAC AGTATGCTCA 300
AGGCTACGTC CATTGTAAGT CCCTGATTAA CGTTTCACCT GGTCATATCG 350
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
CTCAACGTCT CGAAGCACTG GCATGGGTTC TTCCAGAAGG GAACCAACTG 400
GGCCGATGGC CCCGCCTTTG TCACCCAATG TCCTATCACA TCAGGAAACG 450
CCTTCTTGTA TGATTTCAAC GTTCCGGACC AAGCTGGTAC TTTCTGGTAC 500
CACAGCCATC TCTCYACACA GTATTGTGAC GGTCTTCGTG GTGCCTTTGT 550
5 CGTCTATGAT CCTAATGATC CCAACAAGCA ACTCTATGAT GTTGATAACG 600
GCAAGTCCCT TGCATATTTC AGTTCTATCA TATCCTCACC TGTATTGGCA 650
CAGAAAGCAC CGTGATTACC TTGGCTGATT GGTATCATGC CCTTGCTCAG 700
ACTGTCACTG GTGTCGCGTG AGTGACAAAT GGCCCTTAAT TGTTCACATA 750
TTTTCCTGAT TATCATATGA TAGAGTATCT GATGCAACGT TGATCAACGG 800
10 ATTGGGACGT TCGGCCACCG GCCCCGCAAA TGCCCCTCTG GCGGTCATCA 850
GTGTCGAGCG GAATAAGAGG TCAGTTCCAT AATTATGATT ATTTCCCGCG 900
TTACTTCCTA ACGATTATTT TTGTATCCCT CCACAGATAT CGTTTCCGAT 950
TGGTTTCTAT TTCTTGCGAC CCTAACTTTA TTTTCTCAAT TGACCACCAC 1000
CCAATGACCG TAATTGAGAT GGACGGTGTT AATACCCAAT CTATGACCGT 1050
15 AGATTCGATC CAAATATTCG CAGGTCAACG ATATTCATTT GTCGTAGGTT 1100
ATTATAAACT GCCCACCGAT CATCTCTCAC GTAACTGTTA TAGATGCAAG 1150
CCAACCAACC AGTTGGAAAT TATTGGATCC GYGCTAAACC TAATGTTGGG 1200
AACACAACTT TCCTTGGAGG CCTGAACTCC GCTATATTAC GATATGTGGG 1250
AGCCCCTGAC CAAGAACCGA CCACTGACCA AACACCCAAC TCTACACCGC 1300
20 TCGTCGAGGC GAACCTACGT CCCCTCGTCT ATACTCCTGT GGTATGTTGT 1350
TCTCGTTACA TATACCAAAC CTAATATGAG GACTGAACGG ATCTACTAGC 1400
CGGGACAGCC ATTCCCTGGC GGTGCTGATA TCGTCAAGAA CTTAGCTTTG 1450
GGTTTCGTAC GTGTATTTCA CTTCCCTTTT GGCAGTAACT GAGGTGGAAT 1500
GTATATAGAA TGCCGGGCGT TTCACAATCA ATGGAACATC CTTCACACCT 1550
CCTACAGTCC CTGTACTACT CCAGATCCTC AGTGGTACTC ACAATGCACA 1600
GGATCTTCTT CCAGCAGGAA GCGTGATCGA ACTTGAACAG AATAAAGTTG 1650
TCGAAATCGT TCTGCCCGCT GCGGGCGCCG TTGGCGGTCC TCATCCTTTC 1700
CACTTACATG GTGTAAGTAT CAGACGTCCT CATGCCTATA TTGCTCCGAA 1750
CCTTACACAC CTGATTTCAG CACAATTTCT GGGTGGTTCG TAGCGCCGGT 1800
CAAACCACAT ACAATTTCAA TGATGCTCCT ATCCGTGATG TTGTCAGTAT 1850
TGGCGGTGCA AACGATCAAG TCACGATCCG ATTTGTGGTA TGTATCTCGT 1900
GCCTTGCATT CATTCCACGA GTAATGATCC TTACACTTCG GGTTCTCAGA 1950
CCGATAACCC TGGCCCATGG TTCCTTCACT GTCACATTGA CTGGCATTTG 2000
GAGGCTGGGT TCGCTGTAGT CTTTGCGGAG GGAATCAATG GCACTGCAGC 2050
TGCTAATCCA GTCCCAGGTA AGACTCTCGC TGCTTTGCGT AATATCTATG 2100
AATTTAAAGC ATATCAATTT GCAGCGGCTT GGAATCAATT GTGCCCGTTG 2150
TATGATGCCT TGAGCCCAGG TGATACATGA TTACTCGTAG CTGTGCTTTC 2200
TTATACATAT TCTATGGGTA TATCGGAGTA GCTGTACTAT AGTATGTACT 2250
ATACTAGGTG GGATATGYTG ATGTTGATTT ATATAATTTT GTTTGAAGAG 2300
TGACTTTATC GACTTGGGAT TTAGCCGAGT ACATACTGAT CTCTCACTAC 2350
AGGCTTGTTT TGTCTTTGGG CGCTTACTCA ACAGTTGACT GTTTTTGCTA 2400
TTACGCATTG AACCGCATTC CGGTCYGACT CGTGTCCTCT ACTGTGACTT 2450
GTATTGGCAT TCTAGCACAT ATGTCTCTTA CCTATAGGAA CAATATGTCT 2500
CAACACTGTT CCAAAACCTG CGTAAACCAA ATATCGTCCA TCAGATCAGA 2550
TCATTAACAG TGCCGCACTA ACCTAATACA CTGGCARGGA CTGTGGAAAT 2600
CCCTATAAAT GACCTCTAGA CCGTGAGGTC ATTGCAAGGT CGCTCTCCTT 2650
GTCAAGATGA CCC 2663
Translated protein sequence (SEQ ID NO: 8):
MSLLRSLTSL IVLATGAFAA IGPVTDLHIV NQNLAPDGLN RPTVLAGGTF 50
PGPLIRGNKG DNFKINVIDD LTEHSMLKAT SIHWHGFFQK GTNWADGPAF 100
VTQCPITSGN AFLYDFNVPD QAGTFWYHSH LSTQYCDGLR GAFVVYDPND 150
PNKQLYDVDN GNTVITLADW YHALAQTVTG VAVSDATLIN GLGRSATGPA 200
NAPLAVISVE RNKRYRFRLV SISCDPNFIF SIDHHPMTVI EMDGVNTQSM 250
TVDSIQIFAG QRYSFVMQAN QPVGNYWIRA KPNVGNTTFL GGLNSAILRY 300
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
21
VGAPDQEPTT DQTPNSTPLV EANLRPLVYT PVPGQPFPGG ADIVKNLALG 350
FNAGRFTING TSFTPPTVPV LLQILSGTHN AQDLLPAGSV IELEQNKVVE 400
IVLPAAGAVG GPHPFHLHGH NFWVVRSAGQ TTYNFNDAPI RDVVSIGGAN 450
DQVTIRFVTD NPGPWFLHCH IDWHLEAGFA VVFAEGINGT AAANPVPAAW 500
NQLCPLYDAL SPGDT 515
E. Cerrena laccase B3 gene (partial) from ATCC20013 strain
Polynucleotide sequence (SEQ ID NO: 9):
GTGGGGGCGG ATCCCTAACT GTTTCGAATC GGCACCGAAG TATGCAGGTG 50
TGACGGAGAT GAGGCGTTTT TTCATCTTCC ACTGCAGTAT AAAATGTCTC 100
AGGTAACGTC CAGCTTTTTG TACCAGAGCT ACCTCCAAAT ACCTTTACTC 150
GCAAAGGTTT CGCGATGTCT CTTCTTCGTA GCTTGACCTC CCTCATCGTA 200
CTAGCCACTG GTGCATTTGC TGCAATCGGT CCAGTCACTG ACCTACATAT 250
AGTGAACCAG AATCTCGCCC CAGATGGTTT CAACCGCCCC ACTGTACTCG 300
CAGGTGGTAC TTTCCCCGGT CCTCTGATTC GTGGTAACAA GGTACGCTTC 350
ATAACCGCCC TCCGTAGACG TAGGCTTCGG CTGACATGAC CATCATCTGT 400
AGGGAGATAA CTTTAAAATT AATGTGATTG ACGACTTGAC AGAACACAGT 450
ATGCTCAAGG CCACGTCCAT TGTAAGTCCC TGATTAACGT TTCACCTGGT 500
CATATCGCTC AACGTCTCGA AGCACTGGCA TGGGTTCTTC CAGAAGGGAA 550
CCAACTGGGC CGATGGCCCC GCCTTTGTCA CCCAATGTCC TATCACATCA 600
GGAAACTCCT TCCTGTATGA TTTCAACGTT CCGGACCAAG CTGGTACTTT 650
CTGGTACCAC AGCCATCTCT CTACACAGTA TTGTGACGGT CTTCGTGGTG 700
CCTTTGTCGT CTATGATCCT AATGATCCCA ACAAGCAACT CTATGATGTT 750
GATAACGGCA AGTCCCTTGC ATATTTCATT TCTATCATAT CCTCACCTGT 800
ATTGGCACAG AAAGCACCGT GATTACCTTG GCTGATTGGT ATCATGCCCT 850
TGCTCAGACT GTCACTGGTG TCGCGTGAGT GACAAATGGC CCTCAATTGT 900
TCACATATTT TCCTGATTAT CATATGATAG AGTATCTGAT GCAACGTTGA 950
TCAACGGATT GGGACGTTCG GCCACCGGCC CCGCAAATGC CCCTCTGGCG 1000
GTCATCAGTG TCGAGCGGAA TAAGAGGTCA GTTCCATAAT TATGATTATT 1050
TCCCGCGTTA CTTCCTAACA ATTATTCTTG TATCCCTCCA CAGATATCGC 1100
TTCCGATTGG TGTCTATTTC TTGCGACCCT AACTTTATTT TCTCAATTGA 1150
TCACCACCCA ATGACCGTAA TTGAGATGGA CGGTGTTAAT ACCCAATCTA 1200
TGACCGTAGA TTCGATCCAA ATATTCGCAG GTCAACGATA TTCATTTGTC 1250
GTAGGTTATT ATAAACTGCC CACCGATCAT CTCTCACGTA ACTGTTATAG 1300
ATGCAAGCCA ACCAACCRGT TGGAAATTAT TGGATCC 1337
Translated protein sequence (SEQ ID NO: 10):
MSLLRSLTSL IVLATGAFAA IGPVTDLHIV NQNLAPDGFN RPTVLAGGTF 50
PGPLIRGNKG DNFKINVIDD LTEHSMLKAT SIHWHGFFQK GTNWADGPAF 100
VTQCPITSGN SFLYDFNVPD QAGTFWYHSH LSTQYCDGLR GAFVVYDPND 150
PNKQLYDVDN GKTVITLADW YHALAQTVTG VAVSDATLIN GLGRSATGPA 200
NAPLAVISVE RNKRYRFRLV SISCDPNFIF SIDHHPMTVI EMDGVNTQSM 250
TVDSIQIFAG QRYSFVMQAN QPVGNYWI 278
F. Cerrena laccase C gene (partial) from CBS 154.29 strain
Polynucleotide sequence (SEQ ID NO: 11):
TGCAATCGGA CCGGTBGCTG ACCTTCACAT TACGGACGAT ACCATTGCCC 50
CCGATGGTTT CTCTCGTCCT GCTGTTCTCG CTGGCGGGGG TTTCCCTGGC 100
CCTCTCATCA CCGGAAACAA GGTAATGCCT AATGGTTGCG TCTTTGTTGG 150
TGCTCTCATT CATCCACGAC ATTTTGTACC AGGGCGACGC CTTTAAACTC 200
AATGTCATCG ATGAACTAAC GGACGCATCC ATGCTGAAGY CGACTTCCAT 250
CGTAAGTCTC GCTGTATTGC TCCTTGAGCC ATTTCATTGA CTATAACTAC 300
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
22
AACCAGCACT GGCATGGATT CTTCCAAAAG GGTACTAATT GGGCAGATGG 350
TCCCGCTTTT GTGAACCAAT GCCCCATCAC CACGGGAAAC TCCTTCTTGT 400
ACGACTTCCA GGTTCCTGAT CAAGCTGGTA AGCATGAGAT TACACTAGGA 450
AAGTTTAATT TAATAACTAT TCAATCAGGA ACCTACTGGT ATCATAGTCA 500
TTTGTCTACG CAATACTGTG ATGGTCTCAG AGGTGCATTC GTTGTCTACG 550
ACCCTTCAGA TCCTCACAAG GATCTCTACG ACGTCGACGA CGGTGAGCTT 600
TGCTTTTTTC ATTGGTATCC ATTATCGCTC ACGTGTCATT ACTGCGCCAC 650
AGAAAGTACC GTCATCACTT TGGCTGATTG GTATCATACT TTGGCTCGTC 700
AGATTGTTGG CGTTGCGTGA GTAGTCTTGT ACCGACTGAA ACATATTCCA 750
GTTGCTGACT TCCCCACAGC ATTTCTGATA CTACCTTGAT AAACGGTTTG 800
GGCCGCAATA CCAATGGTCC GGCTGATGCT GCTCTTGCTG TGATCAATGT 850
TGACGCTGGC AAACGGTGTG TCCAGATTAC TATACTCCCC ATGACGTCTC 900
AATGCTGATG TGTACTACTT CCAGGTACCG TTTCCGTCTT GTTTCCATAT 950
CCTGTGACCC CAATTGGGTA TTCTCGATTG ACAACCATGA CTTTACGGTC 1000
ATTGAAGTCG ATGGTGTTAA CAGTCAACCT CTCAACGTCG ATTCTGTTCA 1050
GATCTTCGCC GGACAACGTT ACTCGTTCGT 1080
Translated protein sequence (SEQ ID NO: 12):
AIGPVADLHI TDDTIAPDGF SRPAVLAGGG FPGPLITGNK GDAFKLNVID 50
ELTDASMLKX TSIHWHGFFQ KGTNWADGPA FVNQCPITTG NSFLYDFQVP 100
DQAGTYWYHS HLSTQYCDGL RGAFVVYDPS DPHKDLYDVD DESTVITLAD 150
WYHTLARQIV GVAISDTTLI NGLGRNTNGP ADAALAVINV DAGKRYRFRL 200
VSISCDPNWV FSIDNHDFTV IEVDGVNSQP LNVDSVQIFA GQRYSF 246
G. Cerrena laccase D1 gene from CBS154.29 strain
Polynucleotide sequence (SEQ ID NO: 13):
GATTCTAATA GACCAGGCAT ACCAAGAGAT CTACAGGTTG ACAGACCATT 50
CTTCTAGGCG GCATTTATGC TGTAGCGTCA GAAATTATCT CTCCATTTGT 100
ATCCCACAGG TCCTGTAATA ACACGGAGAC AGTCCAAACT GGGATGCCTT 150
TTTTCTCAAC TATGGGCGCA CATAGTCTGG ACGATGGTAT ATAAGACGAT 200
GGTATGAGAC CCATGAAGTC AGAACACTTT TGCTCTCTGA CATTTCATGG 250
TTCACACTCT CGAGATGGGA TTGAACTCGG CTATTACATC GCTTGCTATC 300
TTAGCTCTGT CAGTCGGAAG CTATGCTGCA ATTGGGCCCG TGGCCGACAT 350
ACACATTGTC AACAAAGACC TTGCTCCAGA TGGCGTACAA CGTCCAACCG 400
TGCTTGCCGG AGGCACTTTT CCTGGGACGT TGATCACCGG TCAGAAAGTA 450
AGGGATATTA GTTTGCGTCA AAGAGCCAAC CAAAACTAAC CGTCCCGTAC 500
TATAGGGTGA CAACTTCCAG CTCAATGTCA TCGATGATCT TACCGACGAT 550
CGGATGTTGA CGCCAACTTC CATTGTGAGC CTATTATTGT ATGATTTATC 600
CGAATAGTTT CGCAGTCTGA TCATTGGATC TCTATCGCTA GCATTGGCAC 650
GGTTTCTTCC AGAAGGGAAC CGCTTGGGCC GACGGTCCCG CCTTCGTAAC 700
TCAGTGCCCT ATAATAGCAG ATAACTCTTT TCTGTATGAC TTCGACGTCC 750
CAGACCAAGC TGGTACTTTC TGGTATCATA GTCATCTATC CACTCAGTAC 800
TGTGACGGTT TACGTGGTGC CTTCGTTGTG TACGATCCTA ACGATCCTCA 850
CAAAGACCTA TACGATGTTG ATGACGGTGG GTTCCAAATA TTTGTTCTGC 900
AGACATTGTA TTGACGGTGT TCATTATAAT TTCAGAGAGC ACCGTGATTA 950
CCCTTGCGGA TTGGTACCAT GTTCTCGCCC AGACCGTTGT CGGCGCTGCG 1000
TGAGTAACAC ATACACGCGC TCCGGCACAC TGATACTAAT TTTTTTTTAT 1050
TGTAGCACTC CTGATTCTAC CTTGATCAAC GGGTTAGGCC GTTCACAGAC 1100
CGGACCCGCT GATGCTGAGC TGGCTGTTAT CAGCGTTGAA CATAACAAAC 1150
GGTATGTCAT CTCTACCCAG TATCTTCTCT CCTGCTCTAA TTCGCTGTTT 1200
CACCATAGAT ACCGTTTCCG TTTGGTTTCG ATTTCGTGCG ACCCCAACTT 1250
TACCTTCTCC GTTGATGGTC ATAATATGAC TGTCATCGAA GTCGATGGTG 1300
TCAACACACG ACCCCTGACC GTTGACTCTA TTCAAATCTT CGCCGGACAG 1350
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
23
AGGTATTCCT TTGTCGTAAG TTAATCGATA TATTCTCCTT ATTACCCCTG 1400
TGTAATTGAT GTCAATAGCT CAATGCTAAC CAACCCGAAG ACAATTACTG 1450
GATCCGTGCT ATGCCAAACA TCGGTAGAAA TACAACAACA CTGGACGGAA 1500
AGAATGCCGC TATCCTTCGA TACAAGAATG CTTCTGTAGA AGAGCCCAAG 1550
ACCGTTGGGG GCCCCGCTCA ATCCCCGTTG AATGAAGCGG ACCTGCGTCC 1600
ACTCGTACCT GCTCCTGTGG TATGTCTTGT CGCGCTGTTC CATCGCTATT 1650
TCATATTAAC GTTTTGTTTT TGTCAAGCCT GGAAACGCTG TTCCAGGTGG 1700
CGCAGACATC AATCACAGGC TTAACTTAAC TTTCGTACGT ACACCTGGTT 1750
GAAACATTAT ATTTCCAGTC TAACCTCTCT TGTAGAGTAA CGGCCTCTTC 1800
AGCATCAACA ACGCCTCCTT CACTAATCCT TCGGTCCCCG CCTTATTACA 1850
AATTCTGAGC GGTGCTCAGA ACGCTCAAGA TTTACTTCCA ACGGGTAGTT 1900
ACATTGGCCT TGAACTAGGC AAGGTTGTGG AGCTCGTTAT ACCTCCTCTG 1950
GCAGTTGGAG GACCGCACCC TTTCCATCTT CATGGCGTAA GCATACCACA 2000
CTCCCGCAGC CAGAATGACG CAAACTAATC ATGATATGCA GCACAATTTC 2050
TGGGTCGTCC GTAGTGCAGG TAGCGATGAG TATAACTTTG ACGATGCTAT 2100
CCTCAGGGAC GTCGTRAGCA TTGGAGCGGG GACTGATGAA GTCACAATCC 2150
GTTTCGTGGT ATGTCTCACC CCTCGCATTT TGAGACGCAA GAGCTGATAT 2200
ATTTTAACAT AGACCGACAA TCCGGGCCCG TGGTTCCTCC ATTGCCATAT 2250
TGATTGGCAT TTGGAGGCAG GCCTTGCCAT CGTCTTCGCT GAGGGCATCA 2300
ATCAGACCGC TGCAGCCAAC CCAACACCCC GTACGTGACA CTGAGGGTTT 2350
CTTTATAGTG CTGGATTACT GAATCGAGAT TTCTCCACAG AAGCATGGGA 2400
TGAGCTTTGC CCCAAATATA ACGGGTTGAG TGCGAGCCAG AAGGTCAAGC 2450
CTAAGAAAGG AACTGCTATT TAAACGTGGT CCTAGACTAC GGGCATATAA 2500
GTATTCGGGT AGCGCGTGTG AGCAATGTTC CGATACACGT AGATTCATCA 2550
CCGGACACGC TGGGACAATT TGTGTATAAT GGCTAGTAAC GTATCTGAGT 2600
TCTGGTGTGT AGTTCAAAGA GACAGCCCTT CCTGAGACAG CCCTTCCTGA 2650
GACAGCCCTT CCTGAGACGT GACCTCCGTA GTCTGCACAC GATACTYCTA 2700
AATACGTATG GCAAGATGAC AAAGAGGAGG ATGTGAGTTA CTACGAACAG 2750
AAATAGTGCC CGGCCTCGGA GAGATGTTCT TGAATATGGG ACTGGGACCA 2800
ACATCCGGA 2809
Translated protein sequence (SEQ ID NO: 14):
MGLNSAITSL AILALSVGSY AAIGPVADIH IVNKDLAPDG VQRPTVLAGG 50
TFPGTLITGQ KGDNFQLNVI DDLTDDRMLT PTSIHWHGFF QKGTAWADGP 100
AFVTQCPIIA DNSFLYDFDV PDQAGTFWYH SHLSTQYCDG LRGAFVVYDP 150
NDPHKDLYDV DDGGTVITLA DWYHVLAQTV VGAATPDSTL INGLGRSQTG 200
PADAELAVIS VEHNKRYRFR LVSISCDPNF TFSVDGHNMT VIEVDGVNTR 250
PLTVDSIQIF AGQRYSFVLN ANQPEDNYWI RAMPNIGRNT TTLDGKNAAI 300
LRYKNASVEE PKTVGGPAQS PLNEADLRPL VPAPVPGNAV PGGADINHRL 350
NLTFSNGLFS INNASFTNPS VPALLQILSG AQNAQDLLPT GSYIGLELGK 400
VVELVIPPLA VGGPHPFHLH GHNFWVVRSA GSDEYNFDDA ILRDVVSIGA 450
GTDEVTIRFV TDNPGPWFLH CHIDWHLEAG LAIVFAEGIN QTAAANPTPQ 500
AWDELCPKYN GLSASQKVKP KKGTAI 526
H. Cerrena laccase D2 gene from CBS115.075 strain
Polynucleotide sequence (SEQ ID NO: 15):
GATCTGGACG ATGGTATATA AGACGATGGT ATGAGACCCA TGAAGTCTGA 50
ACACTTTTGC TCTCTGACAT TTCATGGTTC ATACTCTCGA GATGGGATTG 100
AACTCGGCTA TTACATCGCT TGCTATCTTA GCTCTGTCAG TCGGAAGCTA 150
TGCTGCAATT GGGCCCGTGG CCGACATACA CATTGTCAAC AAAGACCTTG 200
CTCCAGATGG TGTACAACGT CCAACCGTGC TCGCCGGAGG CACTTTTCCT 250
GGGACGTTGA TCACCGGTCA GAAAGTAAGG AATATTAGTT TGCGTCAAAG 300
AGCCAACCAA AATTAACCGT CCCGTCCCAT AGGGTGACAA CTTCCAGCTC 350
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
24
AATGTCATTG ATGATCTTAC CGACGATCGG ATGTTGACAC CAACTTCCAT 400
TGTGAGCCTA TTATTGTATG ATTTATCCGT ATAGTTTCTC AGTCTGATCA 450
TTGGCTCTCT ATCGCTAGCA TTGGCACGGT TTCTTCCAGA AGGGAACCGC 500
TTGGGCCGAC GGTCCCGCCT TCGTAACTCA GTGCCCTATA ATAGCAGATA 550
ACTCTTTTCT GTATGACTTC GACGTCCCCG ACCAAGCTGG TACTTTCTGG 600
TATCATAGTC ATCTATCCAC TCAGTACTGT GACGGTTTAC GTGGTGCCTT 650
CGTTGTGTAC GATCCTAACG ATCCTCACAA AGACCTATAC GATGTTGATG 700
ACGGTGGGTT CCAAATACTT GACCAAGAAA CATTATATTG ATAGTATCCA 750
CTCTGATTTT CAGAGAGCAC CGTGATTACC CTTGCGGATT GGTACCATGT 800
TCTCGCCCAG ACCGTTGTCG GCGCTGCGTG AGTAACACAT ACACGCGCTC 850
CGGCACACTG ATACTAATTT TTTATTGTAG CACTCCTGAT TCTACCTTGA 900
TCAACGGGTT AGGCCGTTCA CAGACCGGAC CCGCTGATGC TGAGCTGGCT 950
GTTATCAGCG TTGAACATAA CAAACGGTAT GTCATCTCTA CCCATTATCT 1000
TCTCTCCTGC TTTAATTCGC TGTTTCACCA TAGATACCGA TTCCGTTTGG 1050
TTTCGATTTC GTGCGACCCC AACTTTACCT TCTCCGTTGA TGGTCATAAT 1100
ATGACTGTCA TCGAAGTCGA CGGTGTCAAC ACACGACCCC TGACCGTTGA 1150
CTCTATTCAA ATCTTCGCCG GACAGAGGTA TTCCTTTGTC GTAAGTTAAT 1200
CGATATATTC TCCCTATTAC CCCTGTGTAA TTGATGTCAA CAGCTCAATG 1250
CTAACCAACC CGACGACAAT TACTGGATCC GTGCTATGCC AAACATCGGT 1300
AGAAATACAA CAACACTGGA CGGAAAGAAT GCCGCTATCC TTCGATACAA 1350
GAATGCTTCT GTAGAAGAGC CCAAGACCGT TGGGGGCCCC GCTCAATCCC 1400
CGTTGAATGA AGCGGACCTG CGTCCACTCG TACCTGCTCC TGTGGTATGT 1450
CTTGTCGTGC TGTTCCATCG CTATTTCATA TTAACGTTTT GTTTTTGTCA 1500
AGCCTGGAAA CGCTGTTCCA GGTGGCGCAG ACATCAATCA CAGGCTTAAC 1550
TTAACTTTCG TACGTACACC TGGTTGAAAC ATTATATTTC CAGTCTAACC 1600
TCTTGTAGAG TAACGGCCTT TTCAGCATCA ACAACGCCTC CTTCACTAAT 1650
CCTTCGGTCC CCGCCTTATT ACAAATTCTG AGCGGTGCTC AGAACGCTCA 1700
AGATTTACTT CCAACGGGTA GTTACATTGG CCTTGAACTA GGCAAGGTTG 1750
TGGAGCTCGT TATACCTCCT CTGGCAGTTG GAGGACCGCA CCCTTTCCAT 1800
CTTCATGGCG TAAGCATACC ACACTCCCGC AGCCAGAATG ACGCAAACTA 1850
ATCATGATAT GCAGCACAAT TTCTGGGTCG TCCGTAGTGC AGGTAGCGAT 1900
GAGTATAACT TTGACGATGC TATCCTCAGG GACGTCGTGA GCATTGGAGC 1950
GGGGACTGAT GAAGTCACAA TCCGTTTCGT GGTATGTCTC ACCCCTCGCA 2000
TTTTGAGACG CAAGAGCTGA TATATTTTAA CATAGACCGA CAATCCGGGC 2050
CCGTGGTTCC TCCATTGCCA TATTGATTGG CATTTGGAGG CAGGCCTTGC 2100
CATCGTCTTC GCTGAGGGCA TCAATCAGAC CGCTGCAGCC AACCCAACAC 2150
CCCGTACGTG ACACTGAGGG TTTCTTTATA GTGCTGGATT ACTGAATCGA 2200
GATTTCTCCA CAGAAGCATG GGATGAGCTT TGCCCCAAAT ATAACGGGTT 2250
GAGTGCGAGC CAGAAGGTCA AGCCTAAGAA AGGAACTGCT ATTTAAACG 2299
Translated protein sequence (SEQ ID NO: 16):
MGLNSAITSL AILALSVGSY AAIGPVADIH IVNKDLAPDG VQRPTVLAGG 50
TFPGTLITGQ KGDNFQLNVI DDLTDDRMLT PTSIHWHGFF QKGTAWADGP 100
AFVTQCPIIA DNSFLYDFDV PDQAGTFWYH SHLSTQYCDG LRGAFVVYDP 150
NDPHKDLYDV DDGGTVITLA DWYHVLAQTV VGAATPDSTL INGLGRSQTG 200
PADAELAVIS VEHNKRYRFR LVSISCDPNF TFSVDGHNMT VIEVDGVNTR 250
PLTVDSIQIF AGQRYSFVLN ANQPDDNYWI RAMPNIGRNT TTLDGKNAAI 300
LRYKNASVEE PKTVGGPAQS PLNEADLRPL VPAPVPGNAV PGGADINHRL 350
NLTFSNGLFS INNASFTNPS VPALLQILSG AQNAQDLLPT GSYIGLELGK 400
VVELVIPPLA VGGPHPFHLH GHNFWVVRSA GSDEYNFDDA ILRDVVSIGA 450
GTDEVTIRFV TDNPGPWFLH CHIDWHLEAG LAIVFAEGIN QTAAANPTPQ 500
AWDELCPKYN GLSASQKVKP KKGTAI 526
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
1. Cerrena laccase E gene (partial) from CBS154.29 strain
Polynucleotide sequence (SEQ ID NO: 17):
TGCAATCGGA CCGGTGGCCG ACCTCAAGAT CGTAAACCGA GACATTGCAC 50
5 CTGACGGTTT TATTCGTCCC GCCGTTCTCG CTGGAGGGTC GTTCCCTGGT 100
CCTCTCATTA CAGGGCAGAA AGTACGTTAC GCTATCTCGG TGCTTTGGCT 150
TAATTAAACT ATTTGACTTT GTGTTCTCTT AGGGGAACGA GTTCAAAATC 200
AATGTAGTCA ATCAACTGAC CGATGGTTCT ATGTTAAAAT CCACCTCAAT 250
CGTAAGCAGA ATGAGCCCTT TGCATCTCGT TTTATTGTTA ATGCGCCCAC 300
10 TATAGCATTG GCATGGATTC TTCCAGAAGG GAACAAACTG GGCAGACGGT 350
CCTGCGTTCG TGAACCAATG TCCAATCGCC ACGAACAATT CGTTCTTGTA 400
TCAGTTTACC TCACAGGAAC AGCCAGGTGA GTATGAGATG GAGTTCATCC 450
GAGCATGAAC TGATTTATTT GGAACCTAGG CACATTTTGG TACCATAGTC 500
ATCTTTCCAC ACAATACTGC GATGGTTTGC GAGGGCCACT CGTGGTGTAT 550
15 GACCCACAAG ACCCGCATGC TGTTCTCTAC GACGTCGACG ATGGTTCGTA 600
CTTCGCATAT CCACGCTCGC TTTCATACAA TGTAAACTTT GTTCCTCCAG 650
AAAGTACAAT CATCACGCTC GCGGATTGGT ATCATACCTT GGCTCGGCAA 700
GTGAAAGGCC CAGCGTAAGG CACTTTAGTG TTTCCTCATA GTCCAAGAAA 750
TTCTAACACG CCTTCTTCAT CAGGGTTCCT GGTACGACCT TGATCAACGG 800
20 GTTGGGGCGT CACAACAATG GTCCTCTAGA TGCTGAACTA GCGGTGATCA 850
GTGTTCAAGC CGGCAAACGG CAAGTTCAAT TCACACTTTT CACTCTGTAC 900
CTTCTTCCTG ACATTCTTTT CTTGTAGTTA CCGCTTCCGC CTGATTTCAA 950
TTTCATGCGA TCCCAACTAC GTATTCTCCA TTGATGGCCA TGATATGACT 1000
GTCATCGAAG TGGATAGTGT TAACAGTCAA CCTCTCAAGG TAGATTCTAT 1050
25 CCAAATATTT GCAGGTCAGA GATATTCGTT CGTGGTGAGT CAGATCAGGG 1100
CATATCCTTT TGTCGATACG TCATTGACCA TATAATGCTA CAAGCTGAAT 1150
GCCAACCAAC CAG 1163
Translated protein sequence (SEQ ID NO: 18):
AIGPVADLKI VNRDIAPDGF IRPAVLAGGS FPGPLITGQK GNEFKINVVN 50
QLTDGSMLKS TSIHWHGFFQ KGTNWADGPA FVNQCPIATN NSFLYQFTSQ 100
EQPGTFWYHS HLSTQYCDGL RGPLVVYDPQ DPHAVLYDVD DESTIITLAD 150
WYHTLARQVK GPAVPGTTLI NGLGRHNNGP LDAELAVISV QAGKRQVQFT 200
LFTLYRFRLI SISCDPNYVF SIDGHDMTVI EVDSVNSQPL KVDSIQIFAG 250
QRYSFVLNAN QP 262
A Laccase D enzyme having the following amino acid sequence (SEQ ID NO: 19;
signal
sequence in italics) may be used in the methods described herein:
MGLNSAITSL AILALSVGSY AAIGPVADLH IVNKDLAPDG VQRPTVLAGG 50
TFPGTLITGQ KGDNFQLNVI DDLTDDRMLT PTSIHWHGFF QKGTAWADGP 100
AFVTQCPIIA DNSFLYDFDV PDQAGTFWYH SHLSTQYCDG LRGAFVVYDP 150
NDPHKDLYDV DDGGTVITLA DWYHVLAQTV VGAATPDSTL INGLGRSQTG 200
PADAELAVIS VEHNKRYRFR LVSISCDPNF TFSVDGHNMT VIEVDGVNTR 250
PLTVDSIQIF AGQRYSFVLN ANQPEDNYWI RAMPNIGRNT TTLDGKNAAI 300
LRYKNASVEE PKTVGGPAQS PLNEADLRPL VPAPVPGNAV PGGADINHRL 350
NLTFSNGLFS INNASFTNPS VPALLQILSG AQNAQDLLPT GSYIGLELGK 400
VVELVIPPLA VGGPHPFHLH GHNFWVVRSA GSDEYNFDDA ILRDVVSIGA 450
GTDEVTIRFV TDNPGPWFLH CHIDWHLEAG LAIVFAEGIN QTAAANPTPQ 500
AWDELCPKYN GLSASQKVKP KKGTAI 526
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
26
The mature processed form of this polypeptide is as follows (SEQ ID NO: 20):
AIGPVADLEIVNKDLAPDGVQRPTVLAGGTFPGTLITGQKGDNFQLNVIDDLTDDRMLTPTS
IEWEGFFQKGTAWADGPAFVTQCPIIADNSFLYDFDVPDQAGTFWYESELSTQYCDGLRGAF
VVYDPNDPEKDLYDVDDGGTVITLADWYEVLAQTVVGAATPDSTLINGLGRSQTGPADAELA
VISVEENKRYRFRLVSISCDPNFTFSVDGENMTVIEVDGVNTRPLTVDSIQIFAGQRYSFVL
NANQPEDNYWIRAMPNIGRNTTTLDGKNAAILRYKNASVEEPKTVGGPAQSPLNEADLRPLV
PAPVPGNAVPGGADINHRLNLTFSNGLFSINNASFTNPSVPALLQILSGAQNAQDLLPTGSY
IGLELGKVVELVIPPLAVGGPEPFELEGENFWVVRSAGSDEYNFDDAILRDVVSIGAGTDEV
TIRFVTDNPGPWFLECHIDWELEAGLAIVFAEGINQTAAANPTPQAWDELCPKYNGLSASQK
VKPKKGTAI
[82] In some embodiments, laccase enzymes suitable for use in the present
compositions and
methods are mature polypeptides that lack a signal sequence that may be used
to direct secretion
of a full-length polypeptide from a cell. A suitable mature polypeptide may
have at least 60%,
at least 65%, at least 70%, at least 75%, at least 80%, at least 85%, at least
90%, at least 91%, at
least 92%, at least 93%, at least 94%, at least 95%, at least 96%, at least
97%, at least 98%, or
even at least 99%, or more, amino acid sequence identity to an amino acid
sequence selected
from the group consisting of SEQ ID NO: 2, SEQ ID NO: 4, SEQ ID NO: 6, SEQ ID
NO: 8,
SEQ ID NO: 10, SEQ ID NO: 12, SEQ ID NO: 14, SEQ ID NO: 16, SEQ ID NO: 18, SEQ
ID
NO: 19, and SEQ ID NO: 20. Preferably, such polypeptides have enzymatic
laccase activity, as
determined using the assays and procedures described, herein.
[83] In some embodiments, laccase enzymes suitable for use in the present
compositions and
methods are truncated with respect to a full-length or mature parent/reference
sequence. Such
truncated polypeptides may be generated by the proteolytic degradation of a
full-length or
mature polypeptide sequence or by engineering a polynucleotide to encode a
truncated
polypeptide. Exemplary polypeptides are truncated at the amino and/or carboxyl-
terminus with
respect to an amino acid sequence selected from the group consisting of SEQ ID
NO: 2, SEQ ID
NO: 4, SEQ ID NO: 6, SEQ ID NO: 8, SEQ ID NO: 10, SEQ ID NO: 12, SEQ ID NO:
14, SEQ
ID NO: 16, SEQ ID NO: 18, SEQ ID NO: 19, and SEQ ID NO: 20. The truncation may
be of a
small number, e.g., 1, 2, 3, 4, 5, 6, 7, 8, 9, or 10 amino acid residues, or
of entire structural or
functional domains. A suitable truncated polypeptide may have at least 60%, at
least 65%, at
least 70%, at least 75%, at least 80%, at least 85%, at least 90%, at least
91%, at least 92%, at
least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least
98%, or even at least
99%, or more, amino acid sequence identity to the corresponding portion of one
or more of the
above-references amino acid sequences. Preferably, such polypeptides have
enzymatic laccase
activity, as determined using the assays and procedures described, herein.
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
27
Mediators
[84] In some embodiments, the enzymatic oxidation systems, compositions, and
methods
further include one or more chemical mediator agents that enhance the activity
of the laccase
enzyme. A mediator (also called an enhancer or accelerator) is a chemical that
acts as a redox
mediator to effectively shuttle electrons between the enzyme exhibiting
oxidase activity and a
dye, pigment (e.g., indigo), chromophore (e.g., polyphenolic, anthocyanin, or
carotenoid, for
example, in a colored stain), or other secondary substrate or electron donor.
[85] In some embodiments the chemical mediator is a phenolic compound, for
example,
methyl syringate, or a related compound, as described in, e.g., PCT
Application Nos. WO
95/01426 and WO 96/12845. The mediator may also be an N-hydroxy compound, an N-
oxime
compound, or an N-oxide compound, for example, N-hydroxybenzotriazole,
violuric acid, or N-
hydroxyacetanilide. The mediator may also be a phenoxazine/phenothiazine
compound, for
example, phenothiazine-10-propionate. The mediator may further be 2,2'-
azinobis-(3-
ethylbenzothiazoline-6-sulfonic acid) (ABTS). Other chemical mediators are
well known in the
art, for example, the compounds disclosed in PCT Application No. WO 95/01426,
which are
known to enhance the activity of a laccase. The mediator may also be
acetosyringone, methyl
syringate, ethyl syringate, propyl syringate, butyl syringate, hexyl
syringate, or octyl syringate.
[86] In some embodiments, the mediator is 4-cyano-2,6-dimethoxyphenol, 4-
carboxamido-
2,6-dimethoxyphenol or an N-substituted derivative thereof such as, for
example, 4-(N-methyl
carboxamido)-2,6-dimethoxyphenol, 4-[N-(2-hydroxyethyl) carboxamidol-2,6-
dimethoxyphenol, or 4-(N,N-dimethyl carboxamido)-2,6-dimethoxyphenol.
[87] In some embodiments, the mediator is described by the following formula:
B-O
HO
A
_P7
C-0
in which A is a group such as -R, -D, -CH=CH-D, -CH=CH-CH=CH-D, -CH=N-D, -N=N-
D, or -N=CH-D, D is selected from the group consisting of -CO-E, -S02-E, -CN, -
NXY, and
-N+XYZ, E is -H, -OH, -R, -OR, or -NXY, and X,Y, and Z are independently
selected from -
H, -OH, -OR, and -R; where R is a C1 - C16 alkyl, preferably a Cl -C8 alkyl,
which alkyl may be
saturated or unsaturated, branched or unbranched and optionally substituted
with a carboxy,
sulfo or amino group; and B and C are independently selected from Cm Hem+1 ; 1
< m < 5.
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
28
[88] In some embodiments, A in the above mentioned formula is -CN or -CO-E,
wherein E
may be -H, -OH, -R, -OR, or -NXY, where X and Y are independently selected
from -H, -OH,
-OR, and -R, where R is a C1 -C16 alkyl, preferably a C1 -C8 alkyl, which
alkyl may be saturated
or unsaturated, branched or unbranched and optionally substituted with a
carboxy, sulfo or
amino group; and B and C are independently selected from Cm Hem+1; 1 < m < 5.
In some
embodiments, the mediator is 4-hydroxy-3,5-dimethoxybenzonitrile (also
referred to as
"syringonitrile" or "SN").
[89] Note that in the above mentioned formula, A may be placed meta to the
hydroxy group,
instead of being placed in the para position as shown.
[90] For applications such as textile processing, the mediator may be present
in a
concentration of about 0.005 to about 1,000 pmole per g denim, about 0.05 to
about 500 pmole
per g denim, about 0.1 to about 100 pmole per g denim, about 1 to about 50
mole per g denim,
or about 2 to about 20 mole per g denim.
[91] The mediators may be prepared by methods known to the skilled artisan,
such as those
disclosed in PCT Application Nos. WO 97/11217 and WO 96/12845 and U.S. Patent
No.
5,752,980. Other suitable mediators are described in, e.g., U.S. Patent
Publication No.
2008/0189871.
Methods of Use
[92] The present systems and compositions can be use in applications where
enzymatic
laccase activity is useful or desirable. Among these applications/methods is
color modification
of a substrate, which may be associated with a textile. In some embodiments,
such methods
include incubation of a laccase enzyme with a suitable substrate at a low
temperature, for
example, about 40 C or less. In some embodiments, the temperature is between
about 20 C and
about 40 C. In some embodiments, the temperature is between about 20 to about
35 C. In
some embodiments, the temperature is about 20 C, 25 C, 30 C, or 35 C. In some
embodiments,
the temperature is the ambient temperature of tap water, for example, about 20
C to about 23 C.
The temperature may be maintained within a narrow range or allowed to
fluctuate without
significantly affecting the performance of the system and compositions.
[93] The methods contemplate the use of one or more of the laccases described
herein. In
some embodiments, the laccase is from a Cerrena species, such as C. unicolor.
In some
embodiments, the laccase comprises, consists of, or consists essentially of
the amino acid
sequence of any of the C. unicolor laccase enzymes described herein, or an
amino acid sequence
having any of at least about 35%, 40%, 45%, 50%, 55%, 60%, 65%, 70%, 75%, 80%,
85%,
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
29
90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or even 99.5% identity to
any of the
C. unicolor laccase enzymes described herein, and having laccase enzymatic
activity.
[94] In some embodiments, the systems and methods are used in a textile
processing method,
for example a method for modifying the color of a textile product, including,
e.g., fibers, yarns,
cloth, or complete garments. Generally, the methods involve contacting the
textile with a
laccase and a mediator for a length of time, and under conditions, sufficient
to result in at least
one (i.e., one or more) measurable effects selected from, e.g., a change in
color, a change in
color cast, lightening, bleaching, fading, and/or a reduction of
redeposition/backstaining. In
some embodiments, the methods are used to impart a "vintage look" to dyed
denim products. In
the case of indigo-dyed denim, the vintage look has a less intense blue/violet
tint and more
subdued grey appearance than the freshly-dyed denim. In the case of sulfur-
dyed denim, the
vintage look is faded without the brown tint that can result from hypochlorite
treatment.
Accordingly, while an aspect of the color modification obtained using laccases
can be
characterized as a "bleaching" affect, this term does not fully describe the
color modifications
possible using laccases.
[95] Textiles provided for color modification may be a cellulosic textiles or
blends of
cellulosic and synthetic fibers. In some embodiments, the textile is denim
dyed with indigo
and/or a sulfur-based dye. In a particular embodiment, the textile is dyed
with indigo, and the
laccase enzyme and mediator are used to oxidize the indigo to isatin. The
denim may optionally
be desized and/or stonewashed prior to color modification with the laccase
enzyme.
[96] Generally, given the same amount of abrasion in a textile processing
method, denim
strength is reduced to a greater degree at a higher temperature, compared to a
lower temperature.
Because the present methods can be performed at lower temperatures compared to
conventional
methods, they have the advantage of reducing the damage to textiles during
processing
compared to conventional methods. Moreover, laccase enzymes generally do not
react with
cellulosic textile fibers to reduce their strength during processing.
Accordingly, in some
embodiments, the present methods do not affect the physical strength of the
denim, or reduce the
loss of physical strength compared to conventional methods. Where the denim is
stretch denim
comprising, e.g., elastane or spandex, and the present methods do not affect
the stretch
performance of the fabric, or reduce the loss of stretch performance compared
to conventional
methods.
[97] In some embodiments, the laccase is used in a textile processing method
in combination
with at least one other enzyme. Where such processing is simultaneous,
enzymatic treatment
may be performed at a low temperature as described herein. Where the
processing is sequential,
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
the laccase may be used at a low temperature as described herein, and the
other enzyme(s) may
optionally also be used at a low temperature. In some embodiments, the laccase
is used in
combination with a cellulase enzyme, either simultaneously or sequentially. In
one
embodiment, the textile is contacted with the laccase and cellulase
simultaneously. In another
5 embodiment, the textile is contacted with the laccase and cellulase
sequentially. In one
embodiment, the textile is contacted with the cellulase first to effect
"stonewashing," and then
with the laccase to affect color modification. In another embodiment, the
textile is contacted
with the laccase first, and then with the cellulase. Where cellulase and
laccase treatments are
sequential, the two processing steps can be performed in the same bath, and
without draining the
10 bath between treatments. Such methods are referred to as "single-bath"
methods.
[98] Suitable cellulases may be derived from microorganisms which are known to
be capable
of producing cellulolytic enzymes, such as, e.g., species of Humicola,
Thermomyces, Bacillus,
Trichoderma, Fusarium, Myceliophthora, Phanerochaete, Irpex, Scytalidium,
Schizophyllum,
Penicillium, Aspergillus or Geotricum. Known species capable for producing
celluloytic
15 enzymes include Humicola insolens, Fusarium oxysporum or Trichoderma
reesei. Non-limiting
examples of suitable cellulases are disclosed in U.S. Patent No. 4,435,307;
European patent
application No. 0 495 257; PCT Patent Application No. WO 91/17244; and
European Patent
Application No. EP-A2-271 004, all of which are incorporated herein by
reference.
[99] In some embodiments, enzymatic "stonewashing" using a cellulase,
bleaching using an
20 aryl esterase, and color modification using a laccase, can be combined to
provide a
comprehensive enzymatic textile processing system. Such a system allows a
textile processor to
produce textiles with a wide variety of finishes without the need to use
conventional textile
processing chemical.
[100] Laccases can also be used in other aspects of textile manufacturing,
generally including
25 aspects of treatment, processing, finishing, polishing, production of
fibers, or the like. In
addition to modifying the color of dyed denim, laccases can be used in de-
coloring dyed waste
(including indigo-dyed waste), in fabric dyeing, in textile bleaching work-up,
in fiber
modification; in achieving enhanced fiber or fabric properties, and the like.
[101] In further embodiments, the present systems and compositions may also be
used in a
30 method for modifying the color of wool. For example, European Patent No. EP
0 504 005
discloses that laccases can be used for dyeing wool. Laccases can also be used
in the leather
industry. For example, laccases can be used in the processing of animal hides
including but not
limited to de-hairing, liming, bating and/or tanning of hides.
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
31
[102] The present systems and compositions may also be used in a method for
modifying the
color of pulp or paper products. Such methods involve contacting the pulp or
paper product in
need of color modification with a laccase as described, herein, for a length
of time and under
conditions sufficient for color modification to occur. In particular
embodiments, the color
modification is bleaching.
[103] The present systems and compositions may also be used in a method for
hair color
modification. Laccases have reportedly been found to be useful for hair dyeing
(see, e.g., WO
95/33836 and WO 95/33837). Such methods involve contacting the hair having a
color to be
modified with the laccase for a length of time and under conditions suitable
for changing the
color of the hair.
[104] The present systems and compositions may also be used in the field of
waste-water
treatment. For example, laccases can be used in decolorization of colored
compounds; in
detoxification of phenolic components; for anti-microbial activity (e.g., in
water recycling); in
bio-remediation; etc.
[105] The present systems and compositions may also be used in the
depolymerization of high-
molecular-weight aggregates, deinking waste paper, the polymerization of
aromatic compounds,
radical-mediated polymerization and cross-linking reactions (e.g., paints,
coatings, biomaterials),
the activation of dyes, and coupling organic compounds.
[106] The present systems and compositions may also be used in a cleaning
composition or
component thereof, or in a detergent for use in a cleaning method. For
example, laccases can be
used in the cleaning, treatment or care of laundry items such as clothing or
fabric; in the cleaning
of household hard surfaces; in dish care, including machine dishwashing
applications; and in
soap bars and liquids and/or synthetic surfactant bars and liquids. The
enzymes presented herein
can be useful, for example, in stain removal/de-colorization, and/or in the
removal of odors,
and/or in sanitization, etc. Laccase mediators can be used as sanitization and
antimicrobial
agents (e.g., wood protection, detergents), independently of or in conjunction
with laccase
enzymes.
[107] Laccases can be used in other aspects of field of personal care. For
example, laccases
can be used in the preparation of personal products for humans such as
fragrances, and products
for skin care, hair care, oral hygiene, personal washing and deodorant and/or
antiperspirants, for
humans. Laccases can be useful, for example, in hair dyeing and/or bleaching,
nails dyeing
and/or bleaching; skin dyeing and/or bleaching; surface modification (e.g., as
coupling reagent);
as an anti-microbial agent; in odor removal; teeth whitening; etc. Laccases
can be used in the
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
32
field of contact lens cleaning. For example, laccases can be used in the
cleaning, storage,
disinfecting, and/or preservation of contact lenses.
[108] Laccases can be used in the field of bio-materials. For example,
laccases can be used as
bio-catalysts for various organic reactions; and/or in connection with
biopolymers; in connection
with packaging; in connection with adhesives; in surface modification
(activation and coupling
agent); in production of primary alcohols; in connection with biosensors
and/or organic
syntheses; etc. Laccases are capable of oxidizing a wide variety of colored
compounds having
different chemical structures, using oxygen as the electron acceptor.
[109] The present systems and compositions may also be used for the removal of
lignin from
lignocellulose-containing material (e.g., the delignification of pulp), the
bleaching of
lignocellulose-containing material (i.e. the enzymatic de-inking of recycled
paper) and/or the
treatment of waste water arising from the manufacture of paper or cellulose.
Such processes
may use a laccase enzyme in combination with adding or metering-in non-
aromatic redox agents
plus phenolic and/or non-phenolic aromatic redox compounds, the phenolic and
non-phenolic
units of the lignin either being oxidized directly by the action of these
phenolic and/or non-
phenolic aromatic compounds, or the lignin being oxidized by other phenolic
and/or non-
phenolic compounds produced by the oxidizing action of these compounds.
[110] Laccases can be used in other aspects relating to pulp and paper. For
example, laccases
can be used in the manufacture of paper pulps and fluff pulps from raw
materials such as wood,
bamboo, and cereal rice straw; the manufacture of paper and boards for
printing and writing,
packaging, sanitary and other technical uses; recycling of cellulose fiber for
the purpose of
making paper and boards; and the treatment of waste products generated by and
treated at pulp
or paper mills and other facilities specifically dedicated to the manufacture
of paper, pulp, or
fluff. Laccases can be useful, for example, in wood processing; in pulp
bleaching; in wood fiber
modification; in bio-glue (lignin activation) for MDF manufacturing; for
enhanced paper
properties; in ink removal; in paper dyeing; in adhesives (e.g. lignin based
glue for particle- or
fiber boards); etc.
[111] Laccases can be used in the field of feed. For example, the laccases can
be used as a
feed additive alone or as part of a feed additive with the aim to increase the
nutritional value of
feed for any kind of animals such as chicken, cows, pigs, fish and pets;
and/or as a processing
aid to process plant materials and food industry by products with the aim to
produce
materials/products suitable as feed raw materials.
[112] Laccases can be used in the field of starch processing. For example,
laccases can be used
in the processing of a substrate including starch and/or grain to glucose
(dextrose) syrup,
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
33
fructose syrup or any other syrup, alcohol (potable or fuel) or sugar. Such
starch processing may
include processing steps such as liquefaction, saccharification,
isomerization, and de-branching
of a substrate.
[113] Laccases can be used in the field of food. For example, laccases can be
used in the
preparation, processing, or as an active ingredient in foods such as yellow
fat, tea based
beverages, culinary products, bakery, and frozen foods for human consumption.
Laccases can be
used, for example, as a bread improver, in food preservation, as an oxygen
scavenger, etc.
Laccases can be used for reducing or eliminating the microbial load of various
foods (e.g.,
meats) or feed.
[114] Any of the methods or uses for laccases described herein may be
performed at a low
temperature, e.g., at a temperature lower than about 40 C, e.g., less than
about 40 C, less than
about 37 C, less than about 35 C, less than about 32 C, less than about 30 C,
less than about
27 C, less than about 25 C, and less than about 22 C. Exemplary temperature
ranges are from
about 20 C to less than about 40 C. Exemplary temperatures are 20 C, 21 C9 22
C9 23 C9
24 C9 25 C9 26 C9 27 C9 28 C9 29 C9 30 C9 31 C9 32 C9 33 C9 34 C, or 35 C. In
some
embodiments, the temperature is at room temperature or the ambient temperature
of tap water,
for example, about 20 C to about 23 C.
[115] Any of the methods or uses for laccases described herein may be
performed using any of
the laccase enzymes described herein, e.g., laccases from Cerrena unicolor. In
some
embodiments, laccases are used at a concentration of about 0.005 to about 5000
mg/liter, about
0.05 to about 500 mg/liter, about 0.1 to about 100 mg/liter, or about 0.5 to
about 10 mg/liter. In
some denim processing embodiments, a laccase is used at a concentration of
about 0.005 to
about 5000 mg/kg of denim, about 0.05 to about 500 mg/kg of denim, about 0.1
to about 100
mg/kg of denim, or about 0.5 to about 10 mg/kg of denim. In some embodiments,
a laccase is
used at a pH of about 5 to about 7, about 5.5 to about 6.5, about 5 to about
6, or about 6.
Exemplary pH values are about 5.0, 5.1, 5.2, 5.3, 5.4, 5.5, 5.6, 5.7, 5.8,
5.9, 6.0, 6.1, 6.2, 6.3,
6.4, 6.5, 6.6, 6.7, 6.8, 6.9, and 7Ø
Ready to Use Compositions and Kits
[116] As described above, the present compositions include one or more
laccases, and
optionally one or more mediators. In some embodiments, the compositions
comprise a
polypeptide comprising, consisting of, or consisting essentially of an amino
acid sequence
selected from SEQ ID NO: 2, SEQ ID NO: 4, SEQ ID NO: 6, SEQ ID NO: 8, SEQ ID
NO: 10,
SEQ ID NO: 12, SEQ ID NO: 14, SEQ ID NO: 16, SEQ ID NO: 18, SEQ ID NO: 19, SEQ
ID
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
34
NO: 20, or a variant or fragment, thereof. In particular embodiments the
compositions comprise
a polypeptide comprising, consisting of, or consisting essentially of an amino
acid sequence
selected from SEQ ID NO: 19 and 20, or a variant or fragment, thereof.
Preferably, such
polypeptides have enzymatic laccase activity, which can be determined using
the assays and
procedures described, herein
[117] Such composition can also be provided in the form of a "ready to use"
(RTU)
composition comprising, consisting of, or consisting essentially of a laccase
enzyme and a
mediator. In some embodiments, the mediator is selected from acetosyringone,
syringaldehyde,
syringamide, methyl syringamide, 2-hydroxyethyl syringamide, methyl syringate,
syringonitrile,
dimethylsyringamide, and syringic acid. In one embodiment, the mediator is
syringonitrile (4-
hydroxy-3,5-dimethoxybenzonitrile). The RTU composition may further contain
one or more
compounds to provide a pH buffer when the composition is in solution. For
example, in some
embodiments, the composition contains monosodium phosphate and adipic acid as
a buffering
system. The RTU composition may be in a solid, granular form for ease of
storage and
transportation. The composition is then diluted with water to provide an
aqueous solution for
use, e.g., as described. RTU compositions may also include any number of
additional reagents,
such as dispersants, surfactant, blockers, polymers, preservatives, and the
like.
[118] The following examples are provided to illustrate the systems,
compositions, and
methods, and should in no way be construed as limiting. Other aspects and
embodiments will be
apparent to the skilled person in view of the description.
EXAMPLES
[119] The following enzyme nomenclature is used in the Examples:
Trade name Description
PRIMAGREENR EcoWhite 1 Mycobacterium smegmatis perhydrolase, S54V variant
of SEQ ID NO: 1
PRIMAGREENR EcoFade LT Cerrena unicolor laccase and syringonitrile in a dry
formulation
OPTISIZE 160 amylase Amylase from Bacillus amyloliquefaciens
INDIAGE Neutra L Endoglucanase from Streptomyces sp. 1 1AG8
INDIAGE 2XL Cellulase from Trichoderma reesei
INDIAGE SUPER GX Cellulase from Trichoderma reesei
NOVOPRIME 268 Laccase from Aspergillus oryzae
NOVOPRIME F258 Methyl syringate
DENILITE II S Laccase from Aspergillus oryzae and methyl syringate
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
Example 1 - Effect of temperature on laccase-mediated color modification of
stonewashed
denim
Enzyme
5 [120] Granular Laccase D enzyme from Cerrena unicolor (38,000 U/g) was used
in this
experiment. One laccase unit is defined as the amount of laccase activity that
oxidizes 1 nmol of
ABTS substrate per second under conditions of an assay based on the ability of
laccase enzyme
to oxidize ABTS (2,2'-azinobis(3-ethylbenzthiazoline-6-sulfonate)) into its
corresponding stable
cation radical, ABTS . Accumulation of the radical causes the ABTS to turn a
dark green color
10 and an increase in absorbance at 420 nm. The color formation is
proportional to laccase activity
and is monitored against a laccase standard.
Mediator
[121] 4-hydroxy-3,5-dimethoxybenzonitrile (syringonitrile, SN) was purchased
from Punjab
Chemicals & Crop Protection Limited (Mumbai, India).
15 Procedure
[122] 12 denim legs weighing approximately 3 kg (total) were desized in a
Unimac UF 50
washing machine under the following conditions:
= Desizing for 15 minutes at 10:1liquor ratio 50 C with 0.5 g/1(15 g) of
OPTISIZE 160
amylase (Genencor) and 0.5 g/1(15 g) of a non-ionic surfactant [e.g., Rucogen
BFA
20 (Rudolf Chemie) or Ultravon RW (Huntsman)].
= 2 cold rinses for 5 minutes at 30:1 liquor ratio.
[123] Following desizing, the denim was stonewashed in a Unimac UF 50 washing
machine
under the following conditions:
= Cold rinse for 5 minutes at 10:1 liquor ratio.
25 = Stonewashing for 60 minutes at 10:1 liquor ratio 55 C with 1 kg of pumice
stone, pH 4.5
(1 g/1 tri-sodium citrate dihydrate and 1 g/1 citric acid monohydrate) and 1.2
g/1
INDIAGE 2XL cellulase (Genencor).
= 2 cold rinses for 5 minutes at 30:1 liquor ratio.
[124] After stonewashing, laccase treatment was performed in a Unimac UF 50
washing
30 machine according to the following process:
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
36
= 30 minutes at 10:1 liquor ratio, with either (i) C. unicolor laccase D and
syringonitrile at
pH 6 (0.7 g/1 monosodium phosphate and 0.17 g/1 adipic acid) and temperatures
of 40 C,
30 C, or 23 C or (ii) NOVOPRIME Base 268 and NOVOPRIME F258 at pH 4.8
(0.29 g/1 monosodium phosphate and 0.56 g/1 of adipic acid) and temperatures
of 40 or
30 C.
= 2 cold rinses for 5 minutes at 30:1 liquor ratio.
Evaluation of denim legs
[125] The amount of color modification, reported as "bleaching," of denim legs
was evaluated
after laccase treatment with a Minolta Chromameter CR 310 in the CIE Lab color
space with a
D 65 light source. The CIE color space, also known as the CIELUV color space,
was adopted
by the International Commission on Illumination (CIE) in 1976, and involves
the values L*, u*,
and v* calculated as follows:
t* = 116 ( ` 16 wl Een 0 008Ã 56
yo- ypi
v* i3L*'(v --Vo)
wh era
Y: TE EsIEniuhis iral lit! ~' i IrEstEE1'Euhis, va uo ~i' i C= can a'so
he use_ )
u', s/ : Ghrornaticity ooordinates from the CIE 1976
UGS diaglain
Ye, ikv, Vo: Tristirnulus valEie Y (or Yi c and
chromatic.:ity coordinates u', Vof the perfect
E-etlo-Ainq difluser.
[126] For each denim leg, 8 measurements were taken and the results from the
12 legs (96
measurements total) were averaged. The results are shown in Tables 1 and 2 and
in Figure 1.
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
37
Table 1. Results using C. unicolor laccase and syringonitrile
C. unicolor laccase Syringonitrile Bleaching Standard
concentration, g/l concentration, Temp., C level,
deviation
(U/ml) g/l (mM) CIE* Lab
0.54 0.07 40 38.3/-1.2/-12.0 0.5/0.1/0.1
(20.5) (0.39)
0.3 0.07 40 37.6/-0.5/-12.3 0.6/0.1/0.1
(11.4) (0.39)
0.15 0.07 40 36.4/-0.2/-12.8 0.5/0.1/0.1
(5.7) (0.39)
0.54 0.07
(20.5) (0.39) 30 36.2/-0.2/-12.8 0.5/0.1/0.1
0.3 0.07
(11.4) (0.39) 30 36.1/-0.2/-13.0 0.5/0.1/0.1
0.15 0.07 30 35.3/0.0/-13.3 0.5/0.1/0.1
(5.7) (0.39)
0.15 0.07 23 (no steam) 34.0/0.3/-13.5 0.6/0.1/0.1
(5.7) (0.39)
Table 2. Results using A. oryzae laccase from and methyl syringate
NOVOPRIME NOVOPRIME Bleaching Standard
Base 268 conc., g/1 F258 conc., g/l Temp., C level, deviation
(mM) CIE* Lab
0.47 0.07 40 36.2/-0.5/-11.2 0.6/0.1/0.2
(0.33)
0.27 0.07 40 36.5/-0.4/-11.4 0.6/0.1/0.2
(0.33)
0.15 0.07 40 35.7/-1.0/-11.9 0.5/0.1/0.2
(0.33)
0.15 0.07 30 33.9/0.1/-12.6 0.5/0.1/0.2
(0.33)
[127] The results show the effectiveness of C. unicolor laccase and
syringonitrile in affecting a
color change of stonewashed denim.
Example 2 - Effect of the laccase:mediator ratio on color modification of
stonewashed
denim
Procedure
[128] 12 denim legs weighing approximately 3 kg (total) were desized and
stonewashed as
described in Example 1. After stonewashing, laccase treatment was performed in
a Unimac UF
50 washing machine according to the following process:
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
38
= C. unicolor laccase D and syringonitrile, 30 minutes at 10:1 liquor ratio,
pH 6 (0.7 g/l
monosodium phosphate and 0.17 g/l adipic acid) at 40 C.
= 2 cold rinses for 5 minutes at 30:1 liquor ratio.
Evaluation of denim legs
[129] Color modification of denim legs was evaluated as described in Example
1. The results
are shown in Table 3 and Figure 2.
Table 3. Results using C. unicolor laccase and syringonitrile in different
ratios
C. unicolor laccase Mediator Bleaching Standard
concentration, g/l concentration, Temp., C level,
(U/ml) g/l (mM) CIE* Lab deviation
0.15 0.07 40 36.4/-0.2/-12.8 0.5/0.1/0.1
(5.7) (0.39)
0.15 0.08 40 37.0/-0.4/-12.7 0.5/0.1/0.1
(5.7) (0.44)
0.15 0.1
(5.7) (0.55) 40 37.1/-0.4/-12.7 0.6/0.1/0.1
[130] The results show that the ratio of laccase enzyme to mediator can be
manipulated to alter
color modification.
Example 3 - Effect of temperature on color modification performance of
composition
containing laccase and mediator on stonewashed denim
[131] For the purpose of investigating laccase-mediated color modification
performance at low
temperature, a "ready-to-use" (RTU) composition was prepared as shown in Table
4. The
monosodium phosphate and adipic acid provide a buffering function at about pH
6 in an
application of use as described below.
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
39
Table 4. Ready-to-use formulation
Component % w/w
Monosodium phosphate (anhydrous) 70
Adipic acid 7
C. unicolor laccase D granules (38,000 U/g) 15
Syringonitrile 8
Procedure
[132] 12 denim legs weighing approximately 3 kg (total) were desized and
stonewashed as
described in Example 1. After stonewashing, laccase treatment was performed in
a Unimac UF
50 washing machine according to the following process:
= 30 minutes at 10:1 liquor ratio at 30 C or without incoming steam (i.e.,
temperature of
21-22 C) with the RTU laccase composition described above or DENILITE II S
(Novozymes) at concentrations and temperatures as described in the Tables 5
and 6,
below.]
= 2 cold rinses for 5 minutes at 30:1 liquor ratio.
Evaluation of denim legs
[133] Color modification of denim legs was evaluated as described in Example
1. The results
are shown in Tables 5 and 6 and in Figures 3 and 4.
Table 5. Results using C. unicolor RTU composition
RTU laccase, Temp., 'C Bleaching level, Standard
% owg* CIE* Lab deviation
1 30 35.1/-0.7/-13.5 0.6/0.1/0.2
3 30 38.5/-1.3/-12.6 0.7/0.1/0.2
1 21-22 33.3/-0.4/-13.6 0.6/0.1/0.1
3 21-22 37.2/-1.013.3 0.7/0.1/0.1
* "owg" = on weight of goods
Table 6. Results using an A. oryzae laccase RTU composition
DENILITE II S, Temp., 'C Bleaching level, Standard
% owg CIE* Lab deviation
3 30 36.1/-1.3/-10.9 0.6/0.1/0.2
3 21-22 33.8/-0.8/-12.1 0.5/0.1/0.2
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
[134] The results show that a C. unicolor laccase RTU composition provides
superior color
modification at low temperature compared to conventional commercial laccase
compositions.
Example 4 - One-step stonewashing and color modification at 30 C
5 [135] 12 denim legs weighing approximately 3 kg (total) were desized in a
Unimac UF 50
washing machine as described in Example 1.
[136] Following desizing, the denim was stonewashed and bleached in a Unimac
UF 50
washing machine under the following conditions:
= 30 minutes, 30 C at 10:1 liquor ratio, pH 6, (i) 0.4 % owg INDIAGE Super GX
10 cellulase (Genencor) + 3% owg RTU laccase composition described in Example
3 (i.e.,
"stonewashing + bleaching 1-step") or (ii) INDIAGE Super GX cellulase, alone
(i.e.,
"stonewashing only").
= 2 cold rinses for 5 minutes at 30:1 liquor ratio. No pumice stones were
used. The results
are shown in Table 7 and Figure 5.
Table 7. Results of one-step stonewashing and color modification
Temp. Bleaching level, Standard
( C) CIE*Lab deviation
Stonewashing only 30 25.4/1.3/-12 0.3/0.1/0.3
Stonewashing+ bleaching, 1 step 30 27.3/0.6/-12.2 0.5/0.2/0.2
[137] The results show that color modification can be achieved using laccase
and cellulase
simultaneously.
Example 5 - Two-step stonewashing and color modification at 30 C
[138] 12 denim legs weighing approximately 3 kg (total) were desized in a
Unimac UF 50
washing machine as described in Example 1.
[139] Following desizing, the denim was stonewashed in a Unimac UF 50 washing
machine
under the following conditions:
= 30 minutes, 30 C at 10:1 liquor ratio, pH 5.5, 0.4% owg INDIAGE Super GX
cellulase
(Genencor)
[140] Following stonewashing, the denim was bleached in a Unimac UF 50 washing
machine
under the following conditions:
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
41
= 30 minutes, 30 C at 10:1 liquor ratio, pH 6, 3% owg RTU laccase composition
described
in Example 3.
= 2 cold rinses for 5 minutes at 30:1 liquor ratio. No pumice stones were
used.
[141] The results are shown in Table 8 and Figure 5. The two-step stonewashing
and color
modification results were compared to the results for stonewashing alone as
described in
Example 4.
Table 8. Results of two-step stonewashing and color modification
Temp. Bleaching level,
( C) CIE* L a b Standard deviation
Stonewashing only 30 25.4/1.3/-12 0.3/0.1/0.3
Stonewashing + bleaching, 2 steps 30 31.9/-0.3/-12.9 0.6/0.2/0.1
[142] The results show that color modification by laccase treatment can be
achieved following
stonewashing.
Example 6 - Laccase-mediated color modification of denim at 300 without
stonewashing
[143] 12 denim legs weighing approximately 3 kg (total) were desized in a
Unimac UF 50
washing machine as described in Example 1.
[144] Following desizing, the denim was bleached in a Unimac UF 50 washing
machine under
the following conditions:
= 30 minutes, 30 C at 10:1 liquor ratio, pH 6, 3% owg RTU laccase composition
described
in example 3.
= 2 cold rinses for 5 minutes at 30:1 liquor ratio. No pumice stones were
used.
[145] The results are shown in Table 9 and Figure 5. The color modification
results were
compared to the results for stonewashing alone as described in Example 4.
Table 9. Results of color modification without stonewashing
Temp. ( C) Bleaching level, Standard deviation
CIE* Lab
Stonewashing only 30 25.4/1.3/-12 0.3/0.1/0.3
Bleaching, no stonewashing 30 26.9/0.7/12.1 0.5/0.1/0.2
[146] The results show that the amount of color modification produced by
laccase treatment
without stonewashing is higher than with stonewashing alone.
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
42
Example 7 - Stonewashing and color modification with cellulase and laccase in
a single-
bath bath process without pumice stones
[147] This Example shows that effective stonewashing and color modification
can be obtained
using laccase and cellulase in a single-bath process.
Enzyme
[148] PRIMAGREEN EcoFade LT 100 laccase (Batch No. 780913616, 6,292 GLacU/g).
Procedure
[149] Starting material was desized denim weighing approximately 3 kg (ballast
+ 2 legs for
evaluation).
[150] The denim was stonewashed in a Renzacci LX 22 washing machine under the
following
conditions:
= 40 minutes, 50 C at 10:1 liquor ratio, pH 6.5 0.4 % owg of INDIAGE Neutra L
cellulase (Batch No. 40105358001 activity 5197 NPCNU/g) (Genencor).
= After stonewashing 1 leg was taken out and dried for evaluation.
= Following stonewashing, and without draining (i.e., dropping) the bath, the
second
denim leg was subjected to color modification under the following conditions:
= 40 minutes, 40 C at 10:1 liquor ratio and 1 % owg of RTU PRIMAGREEN EcoFade
LT
100
= 2 cold rinses for 3 minutes
= The denim was dried in an industrial dryer
Evaluation of denim legs
[151] Color modification and stonewashing on denim legs were evaluated after
laccase
treatment and after cellulase treatment with a Minolta Chromameter CR 310 in
the CIE Lab
color space with a D 65 light source. Six measurements were taken for each
leg, and the results
were averaged.
[152] The results are summarized in Table 10. The amount of color modification
obtained
with sequential (i.e., two-step) addition of cellulase and laccase in a single
bath was greater than
that obtained by adding cellulase and laccase at the same time as in Example
4.
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
43
Table 10
Bleaching level, Standard deviation
CIE*Lab
Stonewashing 27.8/1.1/-13.2 0.3/0.1/0.1
Stonewashing+ bleaching, single bath 34.7/0.0/-12.2 0.5/0.1/0.1
[153] The results show that the amount of color modification obtained with
sequential (i.e.,
two-step) addition of cellulase and laccase in a single bath is greater than
that obtained by
adding cellulase and laccase at the same time as in Example 4.
Example 8 - Color modification with laccase and pumice stones
[154] This Example shows that effective stonewashing and color modification
can be obtained
using pumice stones and a laccase-mediator system in a single-bath process.
Enzyme
[155] PRIMAGREEN EcoFade LT 100 laccase (Batch No. 7809136160, 6,292
GLacU/g).
Procedure
[156] 12 denim legs weighing approximately 3 kg (total) were desized in a
Unimac UF 50
washing machine as described in Example 1.
[157] Following desizing, the denim was stonewashed in a Unimac UF 50 washing
machine
under the following conditions:
= 30 minutes, 30 C at 10:1 liquor ratio, 3 kg of pumice stone, with 3 %
PRIMAGREEN
EcoFade LT100 (Genencor). The blank/control was performed only with stones in
water.
= 2 cold rinses for 5 minutes at 30:1 liquor ratio.
Evaluation of denim legs
[158] Color modification on denim legs were evaluated after laccase treatment
and after the
stonewashing treatment with a Minolta Chromameter CR 310 in the CIE Lab color
space with a
D 65 light source, as before. The average of eight measurements taken on the
outside of each
leg were reported as the Bleaching level. The average of four measurements
taken on the inside
of each leg were reported as the Backstaining level.
[159] The results are summarized in Tables 11 and 12.
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
44
Table 11
Bleaching level, Standard deviation
CIE*Lab
Stonewashing 25.5/1.1/-11.4 0.3/0.1/0.2
Color modification 28.7/0.3/-12.0 0.6/0.1/0.1
Table 12
Backstaining level, Standard deviation
CIE*Lab
Stonewashing 50.6/-1.2/-5.5 0.4/0.1/0.3
Stonewashing+ color modification, 52.2/-1.2/-4.0 0.4/0.1/0.3
single bath
[160] The results show that laccase treatment provides color modification even
if pumice
stones are present, and further shows reduction/removal of backstaining.
Example 9 - Stonewashing and color modification of sulphur dyed garments.
[161] The test garments were made of 100% cotton Twill fabric dyed with
sulphur khaki
brown dye. 21 garments weighing approximately 7 kg (total) were stonewashed in
a 25 kg belly
washer (36 rpm) under the following conditions:
= 45 minutes, 55 C at 18:1 liquor ratio, pH 4.5 at 1 g/l of INDIAGE 2XL
= 1 cold rinse for 3 minutes at 12:1 liquor ratio. No pumice stones were used.
= After washing the garments were dried for evaluation
= 3 garments (approximately 1 kg, total) stonewashed as described above were
treated with
PRIMAGREEN EcoFade LT 100 under the following conditions:
= 15, 30 or 45 minutes, 40 C at 50:1 liquor ratio and 1, 2 or 3 g/1 of
PRIMAGREEN
EcoFade LT 100. The blank/control was performed with the garment washed for
15, 30
or 45 min with only water.
= 1 cold rinse for 3 minutes.
= The denim was dried in an industrial dryer.
Evaluation of denim legs
[162] Color modification and stonewashing of sulphur dyed garments were
evaluated after
laccase treatment and after the stonewashing treatment with a Minolta
Chromameter CR 310 in
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
the CIE Lab color space with a D 65 light source, as above. For each garment
10 measurements
were taken and the results were averaged.
[163] The results are summarized in Tables 13
5 Table 13
Treatment CIE*Lab
Before treatment 40.2/2.2/20.8
Whole complex cellulase 43.7/2.1/19.2
Blank, 15 min 44.9/1.3/18.3
Blank, 30 min 46.1/1.3/19.1
Blank, 45 min 46.6/1.3/18.5
PRIMAGREENR Ecofade (1 g/1) 15 min 45.1/2.9/16.5
PRIMAGREENR Ecofade (1 g/1) 30 min 45.5/3.3/16.8
PRIMAGREENR Ecofade (1 g/1) 45 min 44.7/3.2/16.4
PRIMAGREENR Ecofade (2 g/1) 15 min 44.7/3.3/15.9
PRIMAGREENR Ecofade (2 g/1) 30 min 45.3/3.5/15.9
PRIMAGREENR Ecofade (2 g/1) 45 min 45.0/3.5/15.6
PRIMAGREENR Ecofade (3 g/1) 15 min 44.4/3.4/15.5
PRIMAGREENR Ecofade (3 g/1) 30 min 44.6/3.6/15.7
PRIMAGREENR Ecofade (3 g/1) 45 min 45.0/3.6/15.4
[164] The results show that the a and the b values of the color space
significantly change
compared to the untreated fabric, as well as to the blank. The modification to
the cast of the
garments is visible by eye.
Example 10 - Color modification of sulphur dyed garments without stonewashing
[165] 3 garments made of 100% cotton Twill fabric dyed with sulphur khaki
brown dye and
weighing approximately 1 kg (total) were treated in a 5 kg belly washer (36
rpm) under the
following conditions:
= 15, 30 or 45 minutes, 40 C at 40:1 liquor ratio and 1, 2 or 3 g/1 of
PRIMAGREEN
EcoFade LT 100. The blank/control was performed with the garment washed for
15, 30
or 45 min with just water.
= 1 cold rinses for 3 minutes
= The denim was dried in an industrial dryer
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
46
Evaluation of denim legs
[166] Color modification and stonewashing on sulphur dyed garment were
evaluated after
laccase treatment and after the stonewashing treatment with a Minolta
Chromameter CR 310 in
the CIE Lab color space with a D 65 light source. For each garment 10
measurements were
taken and the results were averaged.
[167] The results are summarized in Table 14
Table 14
Treatment CIE*Lab
Before treatment 40.2/2.2/20.8
Blank 15 min 41.1/2.0/19.7
Blank 30 min 41.9/2.1/20.4
Blank 45 min 42.4/2.1/20.29
PRIMAGREENR Ecofade (1 g/1) 15 min 41.1/3.7/17.6
PRIMAGREENR Ecofade (1 g/1) 30 min 41.5/4.2/18.6
PRIMAGREENR Ecofade (1 g/1) 45 min 41.6/4.0/18.1
PRIMAGREENR Ecofade (2 g/1) 15 min 40.4/3.9/17.0
PRIMAGREENR Ecofade (2 g/1) 30 min 41.2/4.2/17.3
PRIMAGREEN Ecofade (2 g/1) 45 min 41.6/4.3/17.3
PRIMAGREEN Ecofade (3 g/1) 15 min 40.5/4.0/16.6
PRIMAGREEN Ecofade (3 g/1) 30 min 41.0/4.2/17.1
PRIMAGREEN Ecofade (3 g/1) 45 min 40.6/4.3/17.0
[168] The results show that the a and the b values of the color space
significantly change
compared to the untreated fabric as well as to the blank. The modification to
the cast of the
garments is visible by eye.
Example 11 - Stonewashing and bleaching performance with cellulase and laccase
in a
single-bath process in the presence of surfactant and pumice stone
Enzyme
[169] PRIMAGREEN EcoFade LT 100 laccase (Batch No. 780913616, 6,292 GLacU/g).
Procedure
[170] 12 denim garments weighing 10 kg (total) and dyed with pure indigo were
desized in a
Tupesa front loading machine (36 rpm) under the following conditions:
CA 02747813 2011-06-20
WO 2010/075402 PCT/US2009/069229
47
= 10 minutes, 40 C at 10:1 liquor ratio, pH 7, and 0.5 g/1 of lubricant, 0.2
g/1 of dispersant
(non ionic surfactant), and 0.2 g/1 of polyester blocker (non ionic
hydrophilic co-
polymer).
[171] Following desizing, the denim was de stonewashed under the following
conditions:
= 30 minutes, 47 C at 5:1 liquor ratio, pH 6 with 7 kg of pumice stones 4 %
owg of
INDIAGE Super GX cellulase (Genencor). 1 garment was taken out for evaluation
= Following stonewashing, and without draining (dropping) the bath, the denim
was
bleached under the following conditions:
= 30 minutes, 47 C at 5:1 liquor ratio and 2% owg of RTU PRIMAGREEN EcoFade
LT
100.
= 2 cold rinses for 2 minutes at 1:50 liquor ratio
= The denim was dried in an industrial dryer
Evaluation of denim legs
[172] Color modification and stonewashing on denim were evaluated after
laccase treatment
and after cellulase treatment with a Minolta Chromameter CR 310 in the CIE Lab
color space
with a D 65 light source. For each leg 8 measurements were taken and the
results were
averaged.
[173] The results are summarized in Table 15.
Bleaching level Standard
CIE*Lab deviation
Stonewashing 27.6/0.6/-12.0 0.5/0.1/0.1
Stonewashing+ bleaching single bath in 32.3/-0.1/-12.8 0.7/0.1/0.1
presence of surfactant and pumice stone
[174] The results show that color modification by laccase treatment occurs in
the presence of
pumice stones and in the presence of a surfactant.
[175] The aspects, embodiments, and examples described herein are for
illustrative purposes
only. Various modifications will be apparent to the skilled person, and are
included within the
spirit and purview of this application, and the scope of the appended claims.
All publications
and patent documents cited herein are hereby incorporated by reference in
their entirety.