Note: Descriptions are shown in the official language in which they were submitted.
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
1
Infectious Clones of Torque Teno Virus
Field of the Invention
The present invention is directed to novel nucleotide and amino acid
sequences of Torque teno virus ("TTV"), including novel genotypes thereof, all
of
which are useful in the preparation of vaccines for treating and preventing
diseases in swine and other animals. Vaccines provided according to the
practice of the invention are effective against multiple swine TTV genotypes
and
isolates. Diagnostic and therapeutic polyclonal and monoclonal antibodies are
also a feature of the present invention, as are infectious clones useful in
the
propagation of the virus and in the preparation of vaccines. Of particular
importance, there are disclosed vaccines that comprise, as antigen, the
expressed protein of single TTV open reading frames, most particularly from
ORF1 or ORF2, and also fragments of the full length ORF1 and ORF2-encoded
proteins.
Background of the Invention
Torque Teno Virus ("TTV"), also referred to as transfusion-transmitted
virus, is generally assigned to the Circoviridae family. It is generally
recognized
that TTV was first isolated from human transfusion patients (see for example,
Nishizawa et al., Biochem. Biophys. Res. Comm. vol. 241, 1997, pp.92-97).
Subsequently, TTV or TTV-like viruses have been identified from other
mammals, including swine, and numerous strains or isolates have been
published (see for example, McKeown et al. Vet. Microbiol. vol. 104, 2004, pp
113-117).
Subsequent work as shown that TTV and TTV-like viruses are very
common; however the pathogenesis of TTV, and the contributions it may make to
other disease states (for example, those caused by other viruses and bacteria)
remains unclear. For example, TTV infections appear to be common in humans,
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-2-
including even in healthy individuals, and such infections are often
asymptomatic,
and may remain for years. In addition, the general inability to propagate the
virus
in cell culture, and a lack of any clear mechanistic disease models, have made
any overall characterization of TTV biology difficult. Notwithstanding that
TTV
viremia is elevated in human patients afflicted with other viral
diseases,(such as
hepatitis or HIV/AIDS), there is also considerable medical literature
suggesting
that TTVs are, in fact, avirulent, and await any clear actual association with
known disease states. See, for example, Biagini et al., Vet. Microbiol. vol.
98,
2004, pp. 95-101.
In regard of swine, the situation is similar. There is considerable work
suggesting that TTV infection is associated with, and contributes to, numerous
diseases such as porcine circovirus disease (and its various clinical
manefestations, such as postweaning multisystemic wasting syndrome and
respiratory disease complicated by lung lesions), and PRRSV- associated
disease (porcine respiratory and reproductive syndrome virus). See for example
published international patent applications WO 2008/150275 and WO
2008/127279. Krakowka et al. also report on an often fatal disease in pigs
referred to as PDNS (porcine dermatitis and neuropathy syndrome) which is
described as a manifestation of disseminated intravascular coagulation, and
for
which combined infection by serotype 1 TTV and PRRSV virus was possibly
implicated (Am. J. Vet Res, vol 69(12), 2008, pp. 1615-1622. PDNS disease was
also correlated with porcine circovirus disease (notably PCV-2) and also with
bacterial infections. Accordingly, while considerable work has been
accomplished, there remains little work that definitively correlates porcine
TTV
infection with specific pathologies. Nonetheless, it has become reasonably
clear
that TTV infection can potentiate numerous disease states. Accordingly, there
is
a need for various classes of TTV reagents, such as high affinity antibodies,
and
for example, peptide fragments of TTV or whole virions that are highly
immunizing, both to further our understanding of overall TTV biology and to
vaccinate, directly or indirectly, against numerous disease states to which
TTV
may contribute.
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-3-
Thus, although the possibility exists that TTV is the principle causative
factor of diseases in swine, it seems more likely that numerous swine diseases
either require the presence of more than one virus, or that the primary effect
of
certain "primary" pathogens is potentiated by TTV infection. As stated, the
possibility exists that numerous diseases of swine can be treated or lessened
by
administering anti-TTV agents to affected or potentially affected animals.
Notwithstanding the well established interest in TTV, effective vaccines have
not
emerged.
TTV is a small, non-enveloped virus comprised of negative polarity, single-
strand circularized DNA. The genome includes three major open reading frames,
ORF1, ORF2 and ORF3, which overlap, and ORF1 encodes the capsid protein.
(Biagini et al., supra). For a detailed discussion thereof, please see the
following
references, which are incorporated by referenece: Kakkola et al., Virology,
vol.
382 (2008), pp. 182-189; Mushahwar et al., Proc. Natl. Acad. Sci, USA, vol 96,
(1999) pp. 3177-3182; and T. Kekarainen and J. Segales, "Torque teno virus
infection in the pig and its potential role as a model of human infection",
The
Veterinary Journal, accepted December 13, 2007 for 2008.
Despite the relatively simple genome, it has been generally very difficult to
propagate the virus in cell culture or by other in vitro methods. The present
invention is directed to recombinant constructs whereby TTV can be propagated
in vitro, including via infectious clones. More particularly, the invention is
directed
to the discovery that effective vaccines can in fact be made from TTV, most
particularly when the TTV antigen is the expression product of a single ORF,
or a
fragment thereof. In a preferred embodiment, the invention provides for ORFI
protein vaccines.
Summary of the Invention
The present invention provides a method of treating or preventing a
disease or disorder in an animal caused by infection with torque teno virus
(TTV),
including disease states that are directly caused by TTV, and disease states
contributed to or potentiated by TTV. In a preferred example, the animal
treated
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-4-
is a swine. Disease states in swine that may be potentiated by TTV, and which
may also be treated or prevented according to the practice of the invention,
include those caused by or associated with porcine circovirus (PCV), and
porcine
reproductive and respiratory syndrome virus (PRRS).
The present invention also includes the option to administer a combination
vaccine, that is, a bivalent or multivalent combination of antigens, which may
include live, modified live, or inactivated antigens against the non-TTV
pathogen,
with appropriate choice of adjuvant.
Based in part upon the unique TTV amino acid sequences as disclosed
herein, the present invention also provides a diagnostic kit for
differentiating
between porcine animals vaccinated with the above described TTV vaccines and
porcine animals infected with field strains of TTV.
Representative embodiments of the invention include an isolated
polynucleotide sequence that comprises a polynucleotide selected from the
group consisting of:
(a,) the DNA of genotype 2 sequence TTV13 (SEQ ID NO: 1); the DNA
genotype 2 sequence TTV10 (SEQ ID NO: 2); or a fragment thereof than
encodes the TTV capsid protein or a fragment of said protein;
(a2) the DNA of a genotype 1 sequence selected from the group
consisting of ttvgl-7 (SEQ ID NO: 4), ttvGT1-17 (SEQ ID NO: 5), ttvGT1-21
(SEQ ID NO: 6), ttvgtl-27 (SEQ ID NO: 3), ttvgtl-178 (SEQ ID NO: 7) or a
fragment thereof than encodes the TTV capsid protein or a fragment of said
protein;
(b) the complement of any sequence in (a);
(c) a polynucleotide that hybridizes with a sequence of (a) or (b) under
stringent conditions defined as hybriding to filter bound DNA in 0.5M NaHPO4,
7% SDS, 1 mM EDTA at 65 C, and washing in 0.1 xSSC/0.1 % SDS at 68 C;
(d) a polynucleotide that is at least 70% identical to the polynucleotide of
(a) or (b);
(e) a polynucleotide that is at least 80% identical to the polynucleotide of
(a) or (b);
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-5-
(f) a polynucleotide that is at least 90% identical to the polynucleotide of
(a) or (b); and
(g) a polynucleotide that is at least 95% identical to the polynucleotide of
(a) or (b).
The invention further provides RNA polynucleotide molecules that are the
complement of any such DNA polynucleotide sequence, and vectors and
plasmids for the expression of any such RNA or DNA polynucleotides, and for
TTV virus that is expressed from such nucleotide sequences, wherein said virus
is live, or fully or partially attenuated.
The invention also provides a DNA vaccine that comprises a
polynucleotide sequence as aforementioned, and corresponding nucleotide
sequences that function as infectious clones.
The invention provides a polypeptide encoded by any of the open reading
frames of the genotype 2 TTV13 (SEQ ID NO:1) or genotype 2 TTV10 (SEQ ID
NO: 2) polynucleotides, or a polypeptide that is at least 90% identical
thereto, or
to a fragment thereof, including the option that additional otherwise
identical
amino acids are replaced by conservative substitutions.
The invention also provides a polypeptide encoded by any of the open
reading frames of the (all sertotype 1) ttvgl-7 (SEQ ID NO:10), ttvGT1-17 (SEQ
ID NO:11), ttvGT1-21 (SEQ ID NO:12), ttvgtl-27 (SEQ ID NO:13), and ttvgtl-
178 (SEQ ID NO:9) ORF1 polynucleotides, or a polypeptide that is at least 90%
identical thereto, or to a fragment thereof, including the option that
additional
otherwise identical amino acids are replaced by conservative substitutions.
Despite continued failures as reported in the art, to provide effective
vaccines against TTV (or to limit the ability of TTV to potentiate other
diseases),
the present invention provides for such effective vaccines, which preferably
comprise a polypeptide resultant from expression of a single TTV open reading
frame, or a mixture thereof. In a preferred embodiment, the polypeptide is
expressed from ORF1, and preferred mixtures include a combination of the
polypeptides of ORF1 and ORF2, and ORF1 and ORF3.
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-6-
In a further preferred embodiment, and taking advantage of the substantial
polypeptide sequence information disclosed herein, there are further provided
polypeptide vaccines wherein the antigen is defined by (a) the first 100 N-
terminal amino acids of the capsid protein of TTV1 3 (SEQ NO:1) or TTV1 0 (SEQ
ID NO:2); or (b) an amino acid sequence that is at least 90 percent identical
thereto; or (c) an arginine rich region thereof.
Brief Description of the Drawings
Figure 1 (panels A and B) shows detection of ORF1 protein by immunological
methods.
Figure 2 evidences successful expression of codon-optimized TTVg1 ORF1
protein in E. coli, with a 6X His tag for affinity purification
Figure 3 provides a vector map for the Chromos construct pcTV-TTV1-7 ORF1
(plus yeast invertase) expression plasmid from which is expressed (following
integration into an artificial chromosome in CHO cells) vaccinating ORF1
protein.
Figure 4 provides a phylogenetic tree for various TTV strains including a
compilation of percent identities.
Figure 5 (panels A, B and C) provides identification of in-common arginine
rich
regions of ORF1 proteins as expressed from various TTV isolates.
Figure 6 provides a vector map for TTVg1-178 as assembled.
Figure 7 demonstrates that Chromos-expressed g1TTV ORF1 significantly
reduced lung lesions compared to the challenge controls and reduces the
magitude and duration of g1TTV viremia, again compared to the challenge
controls.
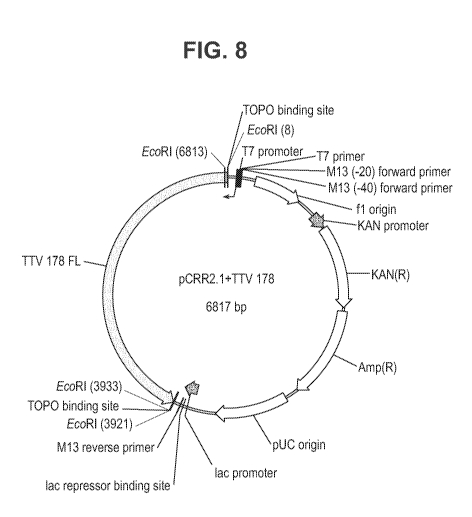
Figure 8 provides a vector map for the pCR2.1+TTVg1-178 construct that
contains a ttvgl-178 strain full length infectious clone.
Brief Description of the Sequence Listings
SEQ ID NO:1 provides the genotype gt2 TTV 10 DNA sequence.
SEQ ID NO:2 provides the genotype 2 gt2 TTV 13 DNA sequence.
SEQ ID NO:3 provides the genotype 1 ttvgtl-27 DNA sequence.
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-7-
SEQ ID NO:4 provides the genotype 1 ttvgtl-7 DNA sequence.
SEQ ID NO:5 provides the genrotype 1 ttvgtl-17 DNA sequence.
SEQ ID NO:6 provides the genotype 1 ttvgtl-21 DNA sequence.
SEQ ID NO:7 provides the genotype 1 ttvgl-178 DNA sequence
SEQ ID NO:8 provides the amino acid sequence of TTV strain AY823991 ORF1.
SEQ ID NO:9 provides the amino acid sequence of TTV strain ttvgtl-178 ORF1
(TTV genotype 1).
SEQ ID NO:10 provides the amino acid sequence of TTV strain ttvgtl-7 ORF1.
SEQ ID NO:11 provides the amino acid sequence of TTV strain ttvgtl-17 ORF1.
SEQ ID NO:12 provides the amino acid sequence of TTV strain ttvgtl-21 ORF1.
SEQ ID NO:13 provides the amino acid sequence of TTV strain ttvgtl-27 ORF1.
SEQ ID NO:14 provides the amino acid sequence of TTV strain gt2 TTV10 ORF1
(genotype 2).
SEQ ID NO:15 provides the amino acid sequence of TTV strain gt2 TTV13 ORF1
SEQ ID NO:16 provides the DNA sequence of known strain AY823991
(genotype 2).
SEQ ID NO:17 provides the DNA sequence of known strain AY823990
(genotype 1).
SEQ ID NO:18 provides the 76057-3 TTV capsid encoding sequence, codon
optimized for E. coli. as cloned into the pUC57 GenScript vector.
SEQ ID NO:19 provides the 76057-4 TTV capsid encoding sequence, codon
optimized for E. coli. as cloned into the Invitrogen pET101/D-TOPO expression
plasmid.
SEQ ID NO:20 provides the 76057-5 TTV capsid encoding sequence, codon
optimized for Saccharomyces cerevisiae as cloned into the pUC57 GenScript
vector.
SEQ ID NO:21 provides the DNA sequence for a construct that encodes ttvgtl-7
ORF1 with a yeast invertase expression tag (YI).
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-8-
SEQ ID NO:22 provides a ttvgtl peptide sequence (numbering based on the
corresponding AY823990 sequence) from the ORF1 capsid protein
corresponding to residues 167-185, which is used with the C-terminal AA in
amidated form.
SEQ ID NO:23 provides a ttvgtl peptide sequence (numbering based on the
corresponding AY823990 sequence) from the ORF1 capsid protein
corresponding to residues 459-479.
SEQ ID NO:24 provides a ttvgtl peptide sequence (numbering based on the
corresponding AY823990 sequence) from the ORF1 capsid protein
corresponding to residues 612-637.
SEQ ID NO:25 provides the amino acid sequence of TTV strain AY823990
ORF1.
SEQ ID NOS:26-29 define primer sequences.
In connection with the descriptors for the sequences, those familiar with
the art will recognize that numerous slightly different abbreviations are
commonly
used interchangeably for specific serotypes, for example, g1TTV, TTVg1,
genotype 1 TTV, serotype 1 TTV, gt1TTV, and the like. A similar situation
exists
for genotype 2.
Detailed Description of the Invention
The following definitions and introductory matters are applicable in the
specification.
The terms "porcine" and "swine" are used interchangeably herein and
refer to any animal that is a member of the family Suidae such as, for
example, a
pig. "Mammals" include any warm-blooded vertebrates of the Mammalia class,
including humans.
An "infectious DNA molecule", for purposes of the present invention, is a
DNA molecule that encodes the necessary elements for viral replication,
transcription, and translation into a functional virion in a suitable host
cell.
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-9-
Likewise, an "isolated polynucleotide molecule" refers to a composition of
matter comprising a polynucleotide molecule of the present invention purified
to
any detectable degree from its naturally occurring state, if any.
For purposes of the present invention, the nucleotide sequence of a
second polynucleotide molecule (either RNA or DNA) is "homologous" to the
nucleotide sequence of a first polynucleotide molecule , or has "identity" to
said
first polynucleotide molecule, where the nucleotide sequence of the second
polynucleotide molecule encodes the same polyaminoacid as the nucleotide
sequence of the first polynucleotide molecule as based on the degeneracy of
the
genetic code, or when it encodes a polyaminoacid that is sufficiently similar
to the
polyaminoacid encoded by the nucleotide sequence of the first polynucleotide
molecule so as to be useful in practicing the present invention. Homologous
polynucleotide sequences also refers to sense and anti-sense strands, and in
all
cases to the complement of any such strands. For purposes of the present
invention, a polynucleotide molecule is useful in practicing the present
invention,
and is therefore homologous or has identity, where it can be used as a
diagnostic
probe to detect the presence of TTV virus or viral polynucleotide in a fluid
or
tissue sample of an infected pig, e.g. by standard hybridization or
amplification
techniques. Generally, the nucleotide sequence of a second polynucleotide
molecule is homologous to the nucleotide sequence of a first polynucleotide
molecule if it has at least about 70% nucleotide sequence identity to the
nucleotide sequence of the first polynucleotide molecule as based on the
BLASTN algorithm (National Center for Biotechnology Information, otherwise
known as NCBI, (Bethesda, Maryland, USA) of the United States National
Institute of Health). In a specific example for calculations according to the
practice of the present invention, reference is made to BLASTP 2.2.6 [Tatusova
TA and TL Madden, "BLAST 2 sequences- a new tool for comparing protein and
nucleotide sequences." (1999) FEMS Microbiol Lett. 174:247-250.]. Briefly, two
amino acid sequences are aligned to optimize the alignment scores using a gap
opening penalty of 10, a gap extension penalty of 0.1, and the "blosum62"
scoring matrix of Henikoff and Henikoff (Proc. Nat. Acad. Sci. USA
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-10-
89:10915-10919. 1992). The percent identity is then calculated as: Total
number
of identical matches x 100/ divided by the length of the longer
sequence+number
of gaps introduced into the longer sequence to align the two sequences.
Preferably, a homologous nucleotide sequence has at least about 75%
nucleotide sequence identity, even more preferably at least about 80%, 85%,
90% and 95% nucleotide sequence identity. Since the genetic code is
degenerate, a homologous nucleotide sequence can include any number of
"silent" base changes, i.e. nucleotide substitutions that nonetheless encode
the
same amino acid.
A homologous nucleotide sequence can further contain non-silent
mutations, i.e. base substitutions, deletions, or additions resulting in amino
acid
differences in the encoded polyaminoacid, so long as the sequence remains at
least about 70% identical to the polyaminoacid encoded by the first nucleotide
sequence or otherwise is useful for practicing the present invention. In this
regard, certain conservative amino acid substitutions may be made which are
generally recognized not to inactivate overall protein function: such as in
regard
of positively charged amino acids (and vice versa), lysine, arginine and
histidine;
in regard of negatively charged amino acids (and vice versa), aspartic acid
and
glutamic acid; and in regard of certain groups of neutrally charged amino
acids
(and in all cases, also vice versa), (1) alanine and serine, (2) asparagine,
glutamine, and histidine, (3) cysteine and serine, (4) glycine and proline,
(5)
isoleucine, leucine and valine, (6) methionine, leucine and isoleucine, (7)
phenylalanine, methionine, leucine, and tyrosine, (8) serine and threonine,
(9)
tryptophan and tyrosine, (10) and for example tyrosine, tyrptophan and
phenylalanine.
Homologous nucleotide sequences can be determined by comparison of
nucleotide sequences, for example by using BLASTN, above. Alternatively,
homologous nucleotide sequences can be determined by hybridization under
selected conditions. For example, the nucleotide sequence of a second
polynucleotide molecule is homologous to SEQ ID NO:1 (or any other particular
polynucleotide sequence) if it hybridizes to the complement of SEQ ID NO:1
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-11-
under moderately stringent conditions, e.g., hybridization to filter-bound DNA
in
0.5 M NaHPO4, 7% sodium dodecyl sulfate (SDS), 1 mM EDTA at 65 C, and
washing in 0.2xSSC/0.1 % SDS at 42 C (see Ausubel et al editors, Protocols in
Molecular Biology, Wiley and Sons, 1994, pp. 6Ø3 to 6.4.10), or conditions
which will otherwise result in hybridization of sequences that encode a TTV
virus
as defined below. Modifications in hybridization conditions can be empirically
determined or precisely calculated based on the length and percentage of
guanosine/cytosine (GC) base pairing of the probe. The hybridization
conditions
can be calculated as described in Sambrook, et al., (Eds.), Molecular Cloning:
A
Laboratory Manual, Cold Spring Harbor Laboratory Press: Cold Spring Harbor,
New York (1989), pp. 9.47 to 9.51.
In another embodiment, a second nucleotide sequence is homologous to
SEQ ID NO:1 (or any other sequence of the invention) if it hybridizes to the
complement of SEQ ID NO:1 under highly stringent conditions, e.g.
hybridization
to filter-bound DNA in 0.5 M NaHPO4, 7% SDS, 1 mM EDTA at 65 C, and
washing in 0.1 xSSC/0.1 % SDS at 68 C, as is known in the art.
It is furthermore to be understood that the isolated polynucleotide
molecules and the isolated RNA molecules of the present invention include both
synthetic molecules and molecules obtained through recombinant techniques,
such as by in vitro cloning and transcription.
Polypetides and Polynucleotides of the Invention
Representative embodiments of the invention include an isolated
polynucleotide sequence that comprises a polynucleotide selected from the
group consisting of:
(a,) the DNA of genotype 2 sequence TTV13 (SEQ ID NO: 1); the DNA
genotype 2 sequence TTV10 (SEQ ID NO: 2); or a fragment thereof than
encodes the TTV capsid protein or a fragment of said protein;
(a2) the DNA of a genotype 1 sequence selected from the group
consisting of ttvgl-7 (SEQ ID NO: 4), ttvGT1-17 (SEQ ID NO: 5), ttvGT1-21
(SEQ ID NO: 6), ttvgtl-27 (SEQ ID NO: 3), ttvgtl-178 (SEQ ID NO: 7) or a
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-12-
fragment thereof than encodes the TTV capsid protein or a fragment of said
protein;
(b) the complement of any sequence in (a);
(c) a polynucleotide that hybridizes with a sequence of (a) or (b) under
stringent conditions defined as hybriding to filter bound DNA in 0.5M NaHPO4,
7% SDS, 1 mM EDTA at 65 C, and washing in 0.1 xSSC/0.1 % SDS at 68 C;
(d) a polynucleotide that is at least 70% identical to the polynucleotide of
(a) or (b);
(e) a polynucleotide that is at least 80% identical to the polynucleotide of
(a) or (b);
(f) a polynucleotide that is at least 90% identical to the polynucleotide of
(a) or (b); and
(g) a polynucleotide that is at least 95% identical to the polynucleotide of
(a) or (b).
The invention also provides a polypeptide encoded by any of the open
reading frames of the genotype 2 TTV13 (SEQ ID NO:1) or genotype 2 TTV10
(SEQ ID NO:2) polynucleotides, or a polypeptide that is at least 90% identical
thereto, or to a fragment thereof, including the option that additional
otherwise
identical amino acids are replaced by conservative substitutions.
The invention also provides a polypeptide encoded by any of the open
reading frames of the (all sertotype 1) ttvgl-7 (SEQ ID NO:10), ttvGT1-17 (SEQ
ID NO:11), ttvGT1-21 (SEQ ID NO:12), ttvgtl-27 (SEQ ID NO:13), and ttvgtl-
178 (SEQ ID NO:9) ORF1 polynucleotides, or a polypeptide that is at least 90%
identical thereto, or to a fragment thereof, including the option that
additional
otherwise identical amino acids are replaced by conservative substitutions.
In a preferred embodiment, the polypeptide is expressed from ORF1, and
preferred mixtures include a combination of the polypeptides of ORF1 and ORF2,
and ORF1 and ORF3.
In a further preferred embodiment, there are further provided TTV
polypeptide-based vaccines wherein the antigen is defined by:
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-13-
(a) the first 300 N-terminal amino acids of the ORF1 capsid protein of TTV13
(SEQ NO: 1) or TTV10 (SEQ ID NO:2); or (b) an amino acid sequence that is at
least 90 percent identical thereto;
(b) the first 200 N-terminal amino acids of the ORF1 capsid protein of TTV13
(SEQ NO: 1) or TTV10 (SEQ ID NO:2); or (b) an amino acid sequence that is at
least 90 percent identical thereto;
(c) the first 100 N-terminal amino acids of the ORF1 capsid protein of TTV1 3
(SEQ NO: 1) or TTV10 (SEQ ID NO:2); or (b) an amino acid sequence that is at
least 90 percent identical thereto;
(d) the first 300 N-terminal amino acids of the ORF1 capsid protein of any of
(all
sertotype 1) ttvgl-7 (SEQ ID NO:10), ttvGT1-17 (SEQ ID NO:11), ttvGT1-21
(SEQ ID NO:12), ttvgtl-27 (SEQ ID NO:13), and ttvgtl-178 (SEQ ID NO:9) or a
polypeptide that is at least 90% identical thereto;
(e) the first 200 N-terminal amino acids of the ORF1 capsid protein of any of
(all
sertotype 1) ttvgl-7 (SEQ ID NO:10), ttvGT1-17 (SEQ ID NO:11), ttvGT1-21
(SEQ ID NO:12), ttvgtl-27 (SEQ ID NO:13), and ttvgtl-178 (SEQ ID NO:9) or a
polypeptide that is at least 90% identical thereto; and
(f) the first 100 N-terminal amino acids of the ORF1 capsid protein of any of
(all
sertotype 1) ttvg1-7 (SEQ ID NO:10), ttvGT1-17 (SEQ ID NO:11), ttvGT1-21
(SEQ ID NO:12), ttvgtl-27 (SEQ ID NO:13), and ttvgtl-178 (SEQ ID NO:9) or a
polypeptide that is at least 90% identical thereto.
Further Genetic Manipulations
The DNA and amino acid sequence information provided by the present
invention also makes possible the systematic analysis of the structure and
function of the viral genes and their encoded gene products. Knowledge of a
polynucleotide encoding a viral gene product of the invention also makes
available anti-sense polynucleotides which recognize and hybridize to
polynucleotides encoding a polypeptide of the invention, or a fragment
thereof.
Full length and fragment anti-sense polynucleotides are useful in this
respect.
The worker of ordinary skill will appreciate that fragment anti-sense
molecules of
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-14-
the invention include (i) those which specifically recognize and hybridize to
a
specific RNA (as determined by sequence comparison of DNA encoding a viral
polypeptide of the invention as well as (ii) those which recognize and
hybridize to
RNA encoding variants of the encoded proteins. Antisense polynucleotides that
hybridize to RNA/DNA encoding other TTV peptides are also identifiable through
sequence comparison to identify characteristic, or signature sequences for the
family of molecules. Such techniques (see Example 8) are further of use in the
study of antigenic domains in TTV polypeptides, and may also be used to
distinguish between infection of a host animal with remotely related non-TTV
members of the Circoviridae.
Example 4 provides guidance as to effective codon optimization for
enhanced expression in yeast and E. coli for the constructs of the invention.
Vaccine formulations
Vaccines of the present invention can be formulated following accepted
convention to include acceptable carriers for animals, including humans (if
applicable), such as standard buffers, stabilizers, diluents, preservatives,
and/or
solubilizers, and can also be formulated to facilitate sustained release.
Diluents
include water, saline, dextrose, ethanol, glycerol, and the like. Additives
for
isotonicity include sodium chloride, dextrose, mannitol, sorbitol, and
lactose,
among others. Stabilizers include albumin, among others. Other suitable
vaccine vehicles and additives, including those that are particularly useful
in
formulating modified live vaccines, are known or will be apparent to those
skilled in
the art. See, e.g., Remington's Pharmaceutical Science, 18th ed., 1990, Mack
Publishing, which is incorporated herein by reference.
Vaccines of the present invention may further comprise one or more
additional immunomodulatory components such as, e.g., an adjuvant or cytokine,
among others. Non-limiting examples of adjuvants that can be used in the
vaccine of the present invention include the RIBI adjuvant system (Ribi Inc.,
Hamilton, MT), alum, mineral gels such as aluminum hydroxide gel, oil-in-water
emulsions, water-in-oil emulsions such as, e.g., Freund's complete and
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-15-
incomplete adjuvants, Block copolymer (CytRx, Atlanta GA), QS-21 (Cambridge
Biotech Inc., Cambridge MA), SAF-M (Chiron, Emeryville CA), AMPHIGEN
adjuvant, saponin, Quil A or other saponin fraction, monophosphoryl lipid A,
ionic
polysaccharides, and Avridine lipid-amine adjuvant. Non-limiting examples of
oil-
in-water emulsions useful in the vaccine of the invention include modified
SEAM62 and SEAM 1/2 formulations. Modified SEAM62 is an oil-in-water
emulsion containing 5% (v/v) squalene (Sigma), 1% (v/v) SPAN 85 detergent
(ICI
Surfactants), 0.7% (v/v) TWEEN 80 detergent (ICI Surfactants), 2.5% (v/v)
ethanol, 200 pg/ml Quil A, 100 pg/ml cholesterol, and 0.5% (v/v) lecithin.
Modified
SEAM 1/2 is an oil-in-water emulsion comprising 5% (v/v) squalene, 1 % (v/v)
SPAN 85 detergent, 0.7% (v/v) Tween 80 detergent, 2.5% (v/v) ethanol, 100
pg/ml Quil A, and 50 pg/ml cholesterol. Other immunomodulatory agents that can
be included in the vaccine include, e.g., one or more interleukins,
interferons, or
other known cytokines.
Additional adjuvant systems permit for the combination of both T-helper
and B-cell epitopes, resulting in one or more types of covalent T-B epitope
linked
structures, with may be additionally lipidated, such as those described in
WO 2006/084319, W02004/014957, and W02004/014956.
In a preferred embodiment of the present invention, ORFI TTV protein, or
other TTV proteins or fragments thereof, is formulated with 5% AMPHIGEN .
Vaccines of the present invention can optionally be formulated for
sustained release of the virus, infectious DNA molecule, plasmid, or viral
vector
of the present invention. Examples of such sustained release formulations
include virus, infectious DNA molecule, plasmid, or viral vector in
combination
with composites of biocompatible polymers, such as, e.g., poly(lactic acid),
poly(lactic-co-glycolic acid), methylcellulose, hyaluronic acid, collagen and
the
like. The structure, selection and use of degradable polymers in drug delivery
vehicles have been reviewed in several publications, including A. Domb et al.,
1992, Polymers for Advanced Technologies 3: 279-292, which is incorporated
herein by reference. Additional guidance in selecting and using polymers in
pharmaceutical formulations can be found in texts known in the art, for
example
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-16-
M. Chasin and R. Langer (eds), 1990, "Biodegradable Polymers as Drug Delivery
Systems" in: Drugs and the Pharmaceutical Sciences, Vol. 45, M. Dekker, NY,
which is also incorporated herein by reference. Alternatively, or
additionally, the
virus, plasmid, or viral vector can be microencapsulated to improve
administration and efficacy. Methods for microencapsulating antigens are well-
known in the art, and include techniques described, e.g., in U.S. Patent
3,137,631; U.S. Patent 3,959,457; U.S. Patent 4,205,060; U.S. Patent
4,606,940;
U.S. Patent 4,744,933; U.S. Patent 5,132,117; and International Patent
Publication WO 95/28227, all of which are incorporated herein by reference.
Liposomes can also be used to provide for the sustained release of virus,
plasmid, viral protein, or viral vector. Details concerning how to make and
use
liposomal formulations can be found in, among other places, U.S. Patent
4,016,100; U.S. Patent 4,452,747; U.S. Patent 4,921,706; U.S. Patent
4,927,637;
U.S. Patent 4,944,948; U.S. Patent 5,008,050; and U.S. Patent 5,009,956, all
of
which are incorporated herein by reference.
An effective amount of any of the above-described vaccines can be
determined by conventional means, starting with a low dose of virus, viral
protein
plasmid or viral vector, and then increasing the dosage while monitoring the
effects. An effective amount may be obtained after a single administration of
a
vaccine or after multiple administrations of a vaccine. Known factors can be
taken
into consideration when determining an optimal dose per animal. These include
the species, size, age and general condition of the animal, the presence of
other
drugs in the animal, and the like. The actual dosage is preferably chosen
after
consideration of the results from other animal studies (see, for example,
Examples
2 and 3 below).
One method of detecting whether an adequate immune response has been
achieved is to determine seroconversion and antibody titer in the animal after
vaccination. The timing of vaccination and the number of boosters, if any,
will
preferably be determined by a doctor or veterinarian based on analysis of all
relevant factors, some of which are described above.
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-17-
The effective dose amount of virus, protein, infectious DNA molecule,
plasmid, or viral vector, of the present invention can be determined using
known
techniques, taking into account factors that can be determined by one of
ordinary
skill in the art such as the weight of the animal to be vaccinated. The dose
amount
of virus of the present invention in a vaccine of the present invention
preferably
ranges from about 101 to about 109 pfu (plaque forming units), more preferably
from about 102 to about 108 pfu, and most preferably from about 103 to about
107
pfu. The dose amount of a plasmid of the present invention in a vaccine of the
present invention preferably ranges from about 0.1 g to about 100mg, more
preferably from about 1 g to about 10mg, even more preferably from about 10 g
to about 1 mg. The dose amount of an infectious DNA molecule of the present
invention in a vaccine of the present invention preferably ranges from about
0.1 g
to about 100mg, more preferably from about 1 g to about 10mg, even more
preferably from about 10 g to about 1 mg. The dose amount of a viral vector of
the
present invention in a vaccine of the present invention preferably ranges from
about 101 pfu to about 109 pfu, more preferably from about 102 pfu to about
108
pfu, and even more preferably from about 103 to about 107 pfu. A suitable
dosage
size ranges from about 0.5 ml to about 10 ml, and more preferably from about 1
ml
to about 5 ml.
Suitable doses for viral protein or peptide vaccines according to the practice
of the present invention range generally from 1 to 50 micrograms per dose, or
higher amounts as may be determined by standard methods, with the amount of
adjuvant to be determined by recognized methods in regard of each such
substance. In a preferred example of the invention relating to vaccination of
swine,
an optimum age target for the animals is between about 1 and 21 days, which at
pre-weening, may also correspond with other scheduled vaccinations such as
against Mycoplasma hyopneumoniae. Additionally, a preferred schedule of
vaccination for breeding sows would include similar doses, with an annual
revaccination schedule.
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-18-
Antibodies
Also contemplated by the present invention are anti-TTV antibodies (e.g.,
monoclonal and polyclonal antibodies, single chain antibodies, chimeric
antibodies, humanized, human, porcine, and CDR-grafted antibodies, including
compounds which include CDR sequences which specifically recognize a TTV
polypeptide of the invention. The term "specific for" indicates that the
variable
regions of the antibodies of the invention recognize and bind a TTV
polypeptide
exclusively (i.e., are able to distinguish a single TTV polypeptide from
related
polypeptides despite sequence identity, homology, or similarity found in the
family of polypeptides), and which are permitted (optionally) to interact with
other
proteins (for example, S. aureus protein A or other antibodies in ELISA
techniques) through interactions with sequences outside the variable region of
the antibodies, and in particular, in the constant region of the Ab molecule.
Screening assays to determine binding specificity of an antibody of the
invention
are well known and routinely practiced in the art. For a comprehensive
discussion of such assays, see Harlow et al. (Eds), Antibodies A Laboratory
Manual; Cold Spring Harbor Laboratory; Cold Spring Harbor, NY (1988), Chapter
6. Antibodies that recognize and bind fragments of the TTV polypeptides of the
invention are also contemplated, provided that the antibodies are first and
foremost specific for, as defined above, a TTV polypeptide of the invention
from
which the fragment was derived.
For the purposes of clarity, "antibody" refers to an immunoglobulin
molecule that can bind to a specific antigen as the result of an immune
response
to that antigen. Immunoglobulins are serum proteins composed of "light" and
"heavy" polypeptide chains having "constant" and "variable" regions and are
divided into classes (e.g., IgA, IgD, IgE, IgG, and IgM) based on the
composition
of the constant regions. Antibodies can exist in a variety of forms including,
for
example, as, Fv, Fab', F(ab')2, as well as in single chains, and include
synthetic
polypeptides that contain all or part of one or more antibody single chain
polypeptide sequences.
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-19-
Diagnostic Kits
The present invention also provides diagnostic kits. The kit can be
valuable for differentiating between porcine animals naturally infected with a
field
strain of a TTV virus and porcine animals vaccinated with any of the TTV
vaccines described herein. The kits can also be of value because animals
potentially infected with field strains of TTV virus can be detected prior to
the
existence of clinical symptoms and removed from the herd, or kept in isolation
away from naive or vaccinated animals. The kits include reagents for analyzing
a sample from a porcine animal for the presence of antibodies to a particular
component of a specified TTV virus. Diagnostic kits of the present invention
can
include as a component a peptide or peptides from ORF1, 2, or 3 which is
present in a field strain but not in a vaccine of interest, or vice versa, and
selection of such suitable peptide domains is made possible by the extensive
amino acid sequencing as provided for in Examples 1 and 2 of the
Specification.
As is known in the art, kits of the present invention can alternatively
include as a
component a peptide which is provided via a fusion protein. The term "fusion
peptide" or "fusion protein" for purposes of the present invention means a
single
polypeptide chain consisting of at least a portion of a TTV virus protein,
preferably of ORF1, and a heterologous peptide or protein.
Examples
Example 1 Cloning of swine TTV complete genome
A. TTV genotype 2.
DNA was purified from porcine serum using a DNA blood mini kit (Qiagen)
per manufacturer's protocol. DNA was eluted from the columns in 50 L Tris-
EDTA buffer. DNA was then amplified via random primed rolling circle
amplification. Briefly, 5 uL of purified DNA and 100 ng random hexamers
(Invitrogen) were then added to 71 pl water and heated at 95C for 3 min and
cooled on ice. One mM dNTP's, 100 ng random hexamers (Invitrogen), 1X
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-20-
phi29 polymerase buffer and 1 pL of phi29 polymerase were then added and the
reaction was incubated overnight at 30C.
One-fifth total volume was digested with EcoRl and electrophoresed on
0.8% E-gel (Invitrogen) to detect presence of 2.7 kB fragment. EcoRl digested
material was purified using a Qiagen PCR purification kit following
manufacturer's protocol, and ligated into an EcoRl digested/shrimp alkaline
phosphatase-treated pGem3zf(+) vector (Promega). Ligated DNA was used to
transform chemically competent E. coli DH5a. Transformed E. coli was selected
on LB/amp agar plates.
Plasmid DNA was isolated from transformed colonies and digested with
EcoRl to confirm presence of an approximately 2.7 kB insert. Four clones (4,
7,
10 and 13) were selected and submitted to ACGT, Inc. for sequencing.
Alignment of sequence data indicated that clones 10 and 13 demonstrated
homology to TTV published sequence and aligned more closely to TTV genotype
2 than genotype 1. These clones were subsequently named TTV1 0 and TTV1 3.
Analyses of sequencing data for PAH TTV genotype 2.
Nucleotide Alignment of TTV13 (SEQ ID NO:2) and TTV10 (SEQ ID NO:1) to
published TTV genotype 2 AY823991 DNA sequence (SEQ ID NO:16).
AY823991 (1) TCATGACAGGGTTCACCGGAAGGGCTGCAAAA-TTACAGCTAAAACCACA
TTV13 (1) TAATGACAGGGTTCACCGGAAGGGCTGCAAAA-TTACAGCTAAAACCACA
TTV10 (1) TAATGACAGGGTTC-CAGGAAGTGCTGCAAAAATTACAGCTAAAACCACA
AY823991 (50) AGT-CTAACACAATAAACCACAAAGTATTACAGGAAACTGCAATAAATTT
TTV13 (50) AAT-CTAACACAATAAACCACAAAATATTACAGGAAACTGCAATAAATTT
TTV10 (50) ACTACTTACACAT--AACCACAAAATATTTCAGGAAACTGCAATAATTTT
AY823991 (99) AGAAATAAGTTACACATAACCACCA------------AACCACAGGAAAC
TTV13 (99) AGAAATAAATTACACATAACCACCA------------AACCACAGGAAAC
TTV10 (98) CAACACACATTGCACAAAACCACAAGATATCAACATAAACCACAGGAAAC
AY823991 (137) TGTGCAAAAAAGAGGAAATAAATTTCATTGGCTGGGCCTGAAGTCCTCAT
TTV13 (137) TCTGCAAAAAAGAGGAAATAAATTTCATTGGCTGGTCCATAAGTCCTCAT
TTV10 (148) TCTGCAAAAAAGAGGAAGTAAATGCTATTGGCTAAATCTGAAGTCTTCAT
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-21-
AY823991 (187) TAGAATAATAAAAGAACCAATCAGAAGAACTTCCTCTTTTAGAGTATATA
TTV13 (187) TAGAATACAAAAAGAACCAATCAGAAACACTTCCTCTTTTAGAGTATATA
TTV10 (198) TAGCATACACAACCAACCAATCAGAAACACTTCCTCATTTGAAGTATATA
AY823991 (237) AGTAAGTGCGCAGACGAATGGCTGAGTTTATGCCGCTGGTGGTAGACACG
TTV13 (237) AGTAAGTGCGCAGACGAATGGCTGAGTTTATGCCGCTGGTGGTAGACACG
TTV10 (248) AGTAAATGCGCAGACGAATGGCTGAGTTTATGCCGCTGGTGGTAGACACG
AY823991 (287) AACAGAGCTGAGTGTCTAACCGCCTGGGCGGGTGCCGGAGCTCCTGAGAG
TTV13 (287) AACAGAGCTGAGTGTCTAACCGCCTGGGCGGGTGCCGGAGCTCCTGAGAG
TTV10 (298) AACAGAGCTGAGTGTCTAACCGCCTGGGCGGGTGCCGGAGCTCCAGAGAG
AY823991 (337) CGGAGTCAAGGGGCCTATCGGGCAGGCGGTAATCCAGCGGAACCGGGCCC
TTV13 (337) CGGAGTCAAGGGGCCTATCGGGCAGGCGGTAATCCAGCGGAACCGGGCCC
TTV10 (348) CGGAGTCAAGGGGCCTATCGGGCGGGCGGTAATCCAGCGGAACCGGGCCC
AY823991 (387) CCC-TCGATGGAAGAAAGATGGCTGACGGTAGCGTACTGCGCACACGGAT
TTV13 (387) CCCCTCCATGGAAGAAAGATGGCTGACGGTAGCGTACTGCGCCCACGGAT
TTV10 (398) CCC-TCCATGGAGGAGAGATGGCTGACGGTAGCGTACGCCGCCCACGGAT
AY823991 (436) TATTCTGCAGCTGTAAAGACCCGAAAAAACATCTTGAAAAATGCCTTACA
TTV13 (437) TATTCTGCGACTGTAAAGACCCGAAAAAACATCTTGAAAAATGCCTTACA
TTV10 (447) TATTCTGCGCCTGCAGTAAGCCCAAAGACCACCTTGAAAAATGCCTTTCC
AY823991 (486) GACGCTATCGCAGACGCCGAAGAAGACCGACACGGAGATGGAGGCACCGG
TTV13 (487) GACGCTATCGCAGACGCCGAAGGAGACCGACAAGAAGATGGAGGCACCGG
TTV10 (497) ACCGCTATCGCCGACGCCGAAGGAGACCCACCAGGAGATGGAGGAGAAGG
AY823991 (536) AGGTGGAGACGCTACTTTCGATATCGGTATCGACGCGCTCCTCGCCGCCG
TTV13 (537) AGGTGGAGACGCTACTTTCGATATCGGTATCGACGCGCTCCTCGCCGCCG
TTV10 (547) AGGTTCCAGCGCTACTTTCGATATCGGTATAGACGCGCTCCTCGCCGCCG
AY823991 (586) CCG --- CACAAAGGTAAGGAGACGGAGG --- AAAAAAGCTCCGGTCATAC
TTV13 (587) CCG --- CACAAAGGTAAGGAGACGGAGG --- AGGAAAGCTCCGGTCATAC
TTV10 (597) CCGACGCTACAAGGTAAGGAGACGGAGGGTTAAAAAGGCTCCGGTCATTC
AY823991 (630) AATGGTTCCCTCCTAGCCGGAGAACCTGCCTCATAGAGGGATTTTGGCCG
TTV13 (631) AATGGAACCCTCCTAGCCGGAGGACCTGCCTCATAGAGGGGTTCTGGCCG
TTV10 (647) AATGGTTCCCCCCAACAGTCAGAAACTGTTTTATCAAGGGAATCTGGCCG
AY823991 (680) TTGAGCTACGGACACTGGTTCCGTACCTGTCTCCCCTTTAGGCGGTTAAA
TTV13 (681) TTGAGCTACGGACACTGGTTCCGTACCTGTCTCCCCTTTAGAAGAAAAAA
TTV10 (697) TTGAGCTACGGACACTGGCTCCGTACCTGTCTCCCTATGAGAAAAGAAAA
AY823991 (730) TGGACTAGTATTCCCGGGTGGAGGTTGTGACTGGAGCCAGTGGAGTTTAC
TTV13 (731) TGGACTAATATTTACGGGAGGAGGTTGTGACTGGACTCAGTGGAGCTTAC
TTV10 (747) CGGACTCATATTCCTAGGAGGTGGCATAGACTGGACTGTCTGGAGTTTAC
AY823991 (780) AAAACCTTTACAATGAAAAACTTAACTGGAGAAATATATGGACAGCTAGT
TTV13 (781) AAAACCTTTATCATGAAAAACTAAACTGGAGAAATATATGGACAGCTAGT
TTV10 (797) AGAATCTATACCATGAAAAACTAAACTGGAGGAATGTGTGGACTTCTTCA
AY823991 (830) AATGTTGGAATGGAATTCGCTAGATTTTTAAAAGGAAAGTTTTACTTTTT
TTV13 (831) AACGTGGGAATGGAATTCGCTAGATTTTTAAAAGGAAAATTCTACTTTTT
TTV10 (847) AATGATGGCATGGAGTTCGCTAGATTCAGATATGCAAAGTTTAAATTTTT
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-22-
AY823991 (880) CAGACATCCATGGAGAAATTATATAATAACTTGGGATCAAGATATACCAT
TTV13 (881) TAGACATCCTTGGAGAAACTATATAGTGACTTGGGATCAGGACATTCCTT
TTV10 (897) TAGACACACAACCAGATCCTACGTAGTAACATGGGACCAAGACATACCAT
AY823991 (930) GCAGGCCACTACCTTATCAAAACCTGCATCCACTCCTAATGCTACTAAAA
TTV13 (931) GTAAACCTTTACCATATCAGAACTTACACCCATTATTAATGCTATTAAAA
TTV10 (947) GTAAACCTTTACCATACACAAATTTACATCCATTTGTAATGCTTCTAAAA
AY823991 (980) AAACAGCACAAAATTGTACTTTCACAGCAAAACTGTAACCCAAACAGAAA
TTV13 (981) AAACAACACAAATTAGTACTCTCACAACAAAACTGTAACCCTAACAGAAA
TTV10 (997) AAACATCATAAAGTAGTTCTAAGCAAACAAGACTGTAATCCTAGAAAAAT
AY823991 (1030) ACAAAAACCTGTCACATTAAAATTCAAACCTCCGCCAAAACTAACATCAC
TTV13 (1031) ACAAAAACCTGTAACTTTAAAATTCAGACCGCCACCAAAACTAACTTCAC
TTV10 (1047) GGACAAACCAGTCACCTTAAAAATAAAGCCACCACCAAAACTCACATCAC
AY823991 (1080) AATGGAGACTAAGTAGAGAATTAGCAAAGATGCCACTAATAAGACTTGGA
TTV13 (1081) AATGGAGACTAAGTAGAGAATTAGCAAAAATGCCACTCATTAGACTAGGA
TTV10 (1097) AGTGGAGACTAAGCAGAGAATTATCAAAAATACCGCTCTTAAGACTAGGA
AY823991 (1130) GTAAGCTTTATAGACCTAACAGAACCATGGGTAGAAGGGTGGGGAAATGC
TTV13 (1131) GTTAGTTTTATAGACTTAACAGAACCGTGGCTAGAAGGTTGGGGAAATGC
TTV10 (1147) GTTTCTTTAATAGACTTCAGAGAACCATGGGTTGAAGGTTTTGGAAATGC
AY823991 (1180) ATTTTATTCCGTGCTAGGATATGAAGCAGTAAAAGACCAAGGACACTGGT
TTV13 (1181) ATTTTACTCAGTACTAGGATATGAAGCCATAAAAGAACAAGGACACTGGT
TTV10 (1197) ATTCTTTAGTACTTTAGGATATGAAGCAGATAAAAGCAATTTAAAAACAA
AY823991 (1230) CAAACTGGACACAAATAAAATACTATTGGATCTATGACACGGGAGTAGGA
TTV13 (1231) CAAATTGGTCACAAATTAAATATTACTGGATATATGATACAGGAGTAGGA
TTV10 (1247) GCGCTTGGTGCCAATGTAAATACTTCTGGATATATGATACCGGAGTAAAT
AY823991 (1280) AATGCAGTATATGTTATACTATTAAAAAAAGACGTTACTGATAATCCAGG
TTV13 (1281) AATGCTGTATATGTAGTTATGCTAAAACAAGATGTAGACGACAACCCAGG
TTV10 (1297) AATCATGTATATGTAGTCATGTTAAACAAAGACGCAGGAGATAATGCAGG
AY823991 (1330) AAACATGGCAACAACCTTTAAAGCATCAGGAGGACAGCATCCAGATGCAA
TTV13 (1331) AAAAATGGCATCAACATTTAAAACAACTCAGGGACAACATCCCAATGCTA
TTV10 (1347) AGACCTAATAACAA------------------------ATCAAAACTCAA
AY823991 (1380) TAGATCACATTGAATTGATAAACCAAGGATGGCCTTACTGGTTATACTTT
TTV13 (1381) TAGATCACATAGAATTAATAAATGAAGGATGGCCGTACTGGTTATACTTT
TTV10 (1373) TAGCACACATAGAACAGATAGGAGAAGGTTATCCATACTGGTTATATTTT
AY823991 (1430) TATGGTAAAAGTGAACAAGACATTAAAAAAGAGGCACAC --- AGCGCAGA
TTV13 (1431) TTTGGTAAAAGTGAACAAGACATAAAAAAGGAAGCACAT --- AGCGCTGA
TTV10 (1423) TTTGGAAGATCTGAAAGAGACTTAAAAGCACTAGCAACTTCAAACACAAA
AY823991 (1477) AATATCAAGAGAATATACTAGAGACCCAAAATCTAAAAAACTAAAAATAG
TTV13 (1478) AATAGCAAGAGAATATGCTACAAATCCAAAATCAAAAAAACTAAAAATAG
TTV10 (1473) CATAAGAAACGAATTCAATACTAATCCTAACAGCAAAAAATTAAAAATAG
AY823991 (1527) GAATAGTAGGATGGGCATCTTCAAACTACACAACAACAGGCAGTGATCAA
TTV13 (1528) GAATAGTAGGATGGGCATCCTCTAACTTCACAACACCAGGCAGTTCACAA
TTV10 (1523) CTGTAATAGGATGGGCTAGCAGTAACAACACAGCACAAGATAGTACACAA
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-23-
AY823991 (1577) AACAGTGGTGGATCAACATCAGCTATACAAGGTGGATATGTAG-----CA
TTV13 (1578) AACTCAGGGGGAAATATAGCAGCAATACAAGGAGGATACGTAG-----CA
TTV10 (1573) ---------GGAGCGAATACTCCAATAGAAGGAACATATTTAATATCACA
AY823991 (1622) TATGC-AGG-GTCCGGGGTCA--------TAGGAGCAGGGTCAATAGGAA
TTV13 (1623) TGGGC-AGGAGGACAAGGAAAACTAAATCTAGGAGCAGGATCAATAGGAA
TTV10 (1614) TGTGCTACAAACATCAGGACATACAG --- CAGGAGCAGCACAAATAAATA
AY823991 (1662) ATTTATATCAACAAGGATGGCCATCTAATCAAAACTGGCCTAATACAAAC
TTV13 (1672) ATTTGTACCAACAAGGATGGCCATCAAATCAAAACTGGCCAAATACAAAC
TTV10 (1661) ACCTATTCGCCTCTGGATGGCCTAACTCTCAAAACTATCCACCTTTAAAT
AY823991 (1712) AGAGACAAAACAAACTTTGACTGGGGAATACGAGGACTATGTATACTCAG
TTV13 (1722) AGAGACGAAACTAACTTTGATTGGGGACTCAGATCACTTTGTATACTAAG
TTV10 (1711) CTAGACAAAAACAACTTTGACTGGGGAAAAAGAGCGCTATGTATACTAAG
AY823991 (1762) AGATAACATGCACTTAGGAAGCCAAGAATTAGATGATGAATGCACAATGC
TTV13 (1772) AGATAACATGCAATTAGGAAATCAAGAATTAGATGATGAATGTACCATGC
TTV10 (1761) AAACAACATGAAAATTGGAAACCAAAATTTAGATGATGAGACCACTATGT
AY823991 (1812) TCACATTGTTCGGACCCTTTGTAGAAAAAGCAAATCCAATATTTGCAACA
TTV13 (1822) TCTCACTCTTTGGACCTTTTGTAGAAAAAGCAAATCCAATATTTGCAACA
TTV10 (1811) TTGCCCTCTTCGGACCCTTGGTAGAAAAAGCAAA-CTGGGAAGGCCTAGA
AY823991 (1862) ACAGACCCTAAATTCTTTAAACCTGAACTCAAAGACTATAATATAATCAT
TTV13 (1872) ACAGACCCTAAATACTTTAAACCAGAACTAAAAGACTATAATTTAATCAT
TTV10 (1860) AAAAATACCAGAA--CTAAAACCAGAACTCAAAGACTATAATATCTTAAT
AY823991 (1912) GAAATATGCCTTTAAATTTCAGTGGGGAGGACATGGCACAGAAAGATTTA
TTV13 (1922) GAAATATGCCTTTAAATTCCAGTGGGGAGGACATGGCACAGAAAGATTTA
TTV10 (1908) GAGATATAACTTTCGCTTTCAGTGGGGCGGACACGGAACAGAGACCTTCA
AY823991 (1962) AAACCAACATCGGAGACCCCAGCACCATACCCTGCCCCTTCGAACCCGGG
TTV13 (1972) AAACAACCATCGGAGACCCCAGCACCATACCCTGCCCCTTCGAACCCGGG
TTV10 (1958) AAACAAGTATTGGAGACCCCAGCCAAATACCCTGTCCCTACGGACCAGGT
AY823991 (2012) GACCGCTTCCA-CAGCGGGATACAAGACCCCTCCAAGGTACAAAACACCG
TTV13 (2022) GACCGCTTCCA-CAGCGGGATACAAGACCCCTCCAAGGTACAAAACACCG
TTV10 (2008) GAAGCCCCCCAACACCTTGTCAGGA-ACCCCTCCAAGGTACACGAGGGGG
AY823991 (2061) TCCTCAACCCCTGGGACTATGACTGTGATGGGATTGTTAGAAAAGATACT
TTV13 (2071) TCCTCAACCCCTGGGACTATGACTGTGATGGGATTGTTAGAAAAGATACT
TTV10 (2057) TCCTCAATGCGTGGGATTATGACTATGATGGAATTGTTAGAAAAGACACT
AY823991 (2111) CTCAAAAGACTTCTCGAACTCCCCACAGAGACAGAGGAGGAGGAGAAGGC
TTV13 (2121) CTCAAAAGACTTCTCGAACTCCCCACAGAGACAGAGGAGGAGGAGAAGGC
TTV10 (2107) CTCAAAAGACTGCTTGCCATCCCCACAGACTC --- GGAGGAGGAGAAAGC
AY823991 (2161) GTACCCACTCCTTGGACAAAAAACAGAGAAAGAGCCATTATCAGACTCCG
TTV13 (2171) GTACCCACTCCTTGGACAAAAAACAGAGAAAGAGCCATTATCAGACTCCG
TTV10 (2154) GTACCCGCTCGCTGGACCCAAAACAGAGAAATTGCCCTCCTCAGACGAAG
AY823991 (2211) ACGAAGAGAGCGTTATCTCAAGCACGAGCAGTGGATCCTCTCAAGAA---
TTV13 (2221) ACGAAGAGAGCGTTATCTCAAGCACGAGCAGTGGATCCGATCAAGAA---
TTV10 (2204) AAGGAGAGAGCGATATCAGTTCTTCGAGCGACTCATCGACGCAAGAAAGC
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-24-
AY823991 (2258) GAAGAAACGCAGAGAC --- GAAGACACCACAAGCCAAGCAAGCGACGACT
TTV13 (2268) GAAGAGACGCAGAGAC --- GAAAGCACCACAAGCCAAGCAAGCGACGACT
TTV10 (2254) GAAGAAGAGAAGAGATACAGAAGACGACACAAGCCCTCAAAGCGAAGACT
AY823991 (2305) CCTCAAGCACCTCCAGCGGGTGGTAAAGAGGATGAAAACACTGTGATAGA
TTV13 (2315) CCTCAAGCACCTCCAGCGGGTGGTAAAGAGGATGAAAACACTGTGATAGA
TTV10 (2304) CCTCCAGCATGTCCAGCGACTGGTGAAGAGATTCAGGACCCT --- ATAGA
AY823991 (2355) TAAATATAGAAACCTAGCAGACCCCTCACTCAATGTCACAGGACACATGG
TTV13 (2365) TAAATACAGAAACCTAGCAGACCCCTCACTCAATGTCACAGGACACATGG
TTTV10 (2351) CAAATACAGAAACTTAGCAGACCCCTCATTAAATGTCACAGGACATTTTG
AY823991 (2405) AAAAATTCATGCAGTTACATATTCAAAACGTACAAGAAATAAGAGCTAAA
TTV13 (2415) AAAAATTCATGCAACTACATATCCAAAACATACAAGAAATAAGAGCTAAA
TTV10 (2401) AACACTTCTGCCGCTTACACTATAAAAACATAGCAGAAATCAGAGCTAGA
AY823991 (2455) AATGCTAAAAAATCCCTCAATAAACTTTACTTTTCTGATTAATAGCGGCC
TTV13 (2465) AATGCTAAAAAATCCCTCAATAAACTTTACTTTTCTGATTAATAGCGGCC
TTV10 (2451) AATGCCAAAAAAAACCTCAATAAACTATACTTTTCAGACTAAAAGAAG--
AY823991 (2505) TCCTGTGTCCAACCTATTTTTCCTAAACCCCTTCAAAATGGCGGGCGGGA
TTV13 (2515) TCCTGTGTCCAATCTATTTTTTTAAACACCCTTCAAAATGGCGGGAGGGA
TTV10 (2499) TTT--------ATTTCTTTATTTAAAACACC-------------------
AY823991 (2555) CACAAAATGGCGGAGGGACTAAGGGGGGGGCAAGCCCCCCTNNNNNNNNN
TTV13 (2565) CACAAAATGGCGGAGGGACTAAGGG-----------------TGNNNNNN
TTV10 (2522) -----------------ACTA----------------------GAGGGCG
AY823991 (2605) NNNNNNNNNNNNNNNNNNGGGGGGCGACCCCCCCGCACCCCCCCCTGCGG
TTV13 (2598) NNNNNNNNNTAGGCTCTTCG---------CCCCCGCACCCCCCC-TGCGG
TTV10 (2533) TAGCGGGGGGGGGACC-------------CCCCTGCACCCCCCCATGCGG
AY823991 (2655) GGGCTCCGCCCCCTGCACCCCCGGGAGGGGGGGAAACCCCCCCTCAACCC
TTV13 (2638) GGGCTCCGCCCCCTGCACCCCCGGGAGGGGGGGAAACCCCCCCTCAACCC
TTV10 (2570) GGGCTCCGCCCCCTGCACCCCCGGGAGGGGGGGAAACCCCCCCTCAACCC
AY823991 (2705) CCCGCGGGGGG-CAAGCCCCCCTGCACCCCCC-
TTV13 (2688) CCCGCGGGGGG-CAAGCCCCCCTGCACCCCCC-
TTV10 (2620) CCCGCGGGGGGGCAAGCCCCCCTGCACCCCCCC
AB076001 AY823990 AY823991 TTV13 TTV10
AB076001 72 49 49 48
AY823990 48 48 48
AY823991 92 76
TTV13 76
TTV10
Nucleotide Identity
AY823991 TTV13 TTV10
AY823991 92 76
TTV13 76
TTV 13 shows 92% identity when compared with previously published
AY823991 sequence. However, TTV10 only show 76% similarity between either
AY823991 or TTV13 and may be considered a separate genotype.
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-25-
Amino Acid Alignment of PAH TTV genotype ORF1 for TTV10 (SEQ ID NO:14)
and TTV13 (SEQ ID NO:15) with AY823991 ORF1 (SEQ ID NO:8).
AY823991 Orfl (1) MPYRRYRRRRRRPTRRWRHRRWRRYFRYRYRRAPRRRR-TKVRRR-RKKA
TTV100rf1 (1) MPFHRYRRRRRRPTRRWRRRRFQRYFRYRYRRAPRRRRRYKVRRRRVKKA
TTV130RF1 (1) MPYRRYRRRRRRPTRRWRHRRWRRYFRYRYRRAPRRRR-TKVRRR-RRKA
AY823991 Orfl (49) PVIQWFPPSRRTCLIEGFWPLSYGHWFRTCLPFRRLNGLVFPGGGCDWSQ
TTV100rf1 (51) PVIQWFPPTVRNCFIKGIWPLSYGHWLRTCLPMRKENGLIFLGGGIDWTV
TTV130RF1 (49) PVIQWNPPSRRTCLIEGFWPLSYGHWFRTCLPFRRKNGLIFTGGGCDWTQ
AY823991 Orfl (99) WSLQNLYNEKLNWRNIWTASNVGMEFARFLKGKFYFFRHPWRNYIITWDQ
TTV100rf1 (101) WSLQNLYHEKLNWRNVWTSSNDGMEFARFRYAKFKFFRHTTRSYVVTWDQ
TTV130RF1 (99) WSLQNLYHEKLNWRNIWTASNVGMEFARFLKGKFYFFRHPWRNYIVTWDQ
AY823991 Orfl (149) DIPCRPLPYQNLHPLLMLLKKQHKIVLSQQNCNPNRKQKPVTLKFKPPPK
TTV100rf1 (151) DIPCKPLPYTNLHPFVMLLKKHHKVVLSKQDCNPRKMDKPVTLKIKPPPK
TTV130RF1 (149) DIPCKPLPYQNLHPLLMLLKKQHKLVLSQQNCNPNRKQKPVTLKFRPPPK
AY823991 Orfl (199) LTSQWRLSRELAKMPLIRLGVSFIDLTEPWVEGWGNAFYSVLGYEAVKDQ
TTV100rf1 (201) LTSQWRLSRELSKIPLLRLGVSLIDFREPWVEGFGNAFFSTLGYEADKSN
TTV130RF1 (199) LTSQWRLSRELAKMPLIRLGVSFIDLTEPWLEGWGNAFYSVLGYEAIKEQ
AY823991 Orfl (249) GHWSNWTQIKYYWIYDTGVGNAVYVILLKKDVTDNPGNMATTFKASGGQH
TTV100rf1 (251) LKTSAWCQCKYFWIYDTGVNNHVYVVMLNKDAGDNAGDLITNQNS-----
TTV130RF1 (249) GHWSNWSQIKYYWIYDTGVGNAVYVVMLKQDVDDNPGKMASTFKTTQGQH
AY823991 Orfl (299) PDAIDHIELINQGWPYWLYFYGKSEQDIKKEAHS-AEISREYTRDPKSKK
TTV100rf1 (296) ---IAHIEQIGEGYPYWLYFFGRSERDLKALATSNTNIRNEFNTNPNSKK
TTV130RF1 (299) PNAIDHIELINEGWPYWLYFFGKSEQDIKKEAHS-AEIAREYATNPKSKK
AY823991 Orfl (348) LKIGIVGWASSNYTTTGSDQNSGG-STSAIQGGYVAYAGSG---VIGAGS
TTV100rf1 (343) LKIAVIGWASSNNTAQDSTQGANTPIEGTYLISHVLQTSGH---TAGAAQ
TTV130RF1 (348) LKIGIVGWASSNFTTPGSSQNSGG-NIAAIQGGYVAWAGGQGKLNLGAGS
AY823991 Orfl (394) IGNLYQQGWPSNQNWPNTNRDKTNFDWGIRGLCILRDNMHLGSQELDDEC
TTV100rf1 (390) INNLFASGWPNSQNYPPLNLDKNNFDWGKRALCILRNNMKIGNQNLDDET
TTV130RF1 (397) IGNLYQQGWPSNQNWPNTNRDETNFDWGLRSLCILRDNMQLGNQELDDEC
AY823991 Orfl (444) TMLTLFGPFVEKANPIFATTDPKFFKPELKDYNIIMKYAFKFQWGGHGTE
TTV100rf1 (440) TMFALFGPLVEKAN-WEGLEKIPELKPELKDYNILMRYNFRFQWGGHGTE
TTV130RF1 (447) TMLSLFGPFVEKANPIFATTDPKYFKPELKDYNLIMKYAFKFQWGGHGTE
AY823991 Orfl (494) RFKTNIGDPSTIPCPFEPGDRFHSGIQDPSKVQNTVLNPWDYDCDGIVRK
TTV100rf1 (489) TFKTSIGDPSQIPCPYGPGEAPQHLVRNPSKVHEGVLNAWDYDYDGIVRK
TTV130RF1 (497) RFKTTIGDPSTIPCPFEPGDRFHSGIQDPSKVQNTVLNPWDYDCDGIVRK
AY823991 Orfl (544) DTLKRLLELPTETEEEEKAYPLLGQKTEKEPLSDSDEESVISSTSSGSSQ
TTV100rf1 (539) DTLKRLLAIPTDSEEE-KAYPLAGPKTEKLPSSDEEGESDISSSSDSSTQ
TTV130RF1 (547) DTLKRLLELPTETEEEEKAYPLLGQKTEKEPLSDSDEESVISSTSSGSDQ
AY823991 Orfl (594) EEETQR--RRHHKPSKRRLLKHLQRVVKRMKTL--
TTV100rf1 (588) ESEEEKRYRRRHKPSKRRLLQHVQRLVKRFRTL--
TTV130RF1 (597) EEETQR--RKHHKPSKRRLLKHLQRVVKRMKTL--
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-26-
Amino Acid Alignment of TTV10 TTV13 ORF with Published Sequence
AY823991 Orf1 TTV10Orf1 TTV1 3ORF1
AY823991 OrfI 65 92
TTV10Orf1 66
TTVI3ORF1
On the amino acid level, TTV10 ORF demonstrates only 65% homology to
the published sequence and may represent a unique phenotype of TTV
Cloning of TTV -genotype 2 ORF1 for baculovirus expression
Based on sequence data derived above, primers were designed to clone
the ORF from TTV10 and TTV13 for expression in baculovirus using the
Invitrogen Gateway system. Primer sequences were:
For TTV13 ORF: Ttvl3Rev1211: 5' cgt act cga gtc aca gtg ttt tca tcc (SEQ ID
NO:26); TTV13For1211: 5' cta ggt acc atg cct tac aga cgc tat (SEQ ID NO:27)
For TTV10 ORF: ttlOforl207: 5' cta ggt acc atg cct ttc cac cgc tat (SEQ ID
NO:28) and ttvrev1207: cgt act cga get ata ggg tcc tga at (SEQ ID NO:29)
Since the EcoRl cloning into pGem resulted in interrupting the reading
frame of the ORF1, the TTV insert in pGem was isolated by EcoRl digestion, gel-
purified and re-circularized using standard ligation conditions. Following an
overnight ligation at 4 C, ligase was inactivated at 65 C, and the reaction
was
purified using QuiQuick purification kit (Qiagen) following the manufacturer's
protocol.
TTVORF13 was the PCR amplified using re-circularized TTV13 genomic
DNA with Expand Hi-Fidelity enzyme (Roche) using the above described
TTV13 forward and reverse primers (0.15 M each), 0.2 mM dNTP's in 1X Hi
Fidelity enzyme buffer. PCR conditions were:1 cycle at 4 min, 95 C; 35 cycles
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-27-
with 94 C, 15 sec denaturation, 55 C, 30 sec anneal, and 68 C 1.5 min
extension; and 1 cycle of 72 C, 7 min extension.
Similarly, TTVORF10 was PCR amplified using re-circularized TTV10
genomic DNA with Expand Hi-Fidelity enzyme (Roche) using the above
described TTV10 forward and reverse primers (0.15 M each) 0.2 mM dNTP's in
1X Hi Fidelity enzyme buffer. PCR conditions were:1 cycle at 4 min, 95 C; 35
cycles with 94 C, 15 sec denaturation, 56 C, 30 sec anneal, and 68 C 1.5 min
extension; and 1 cycle of 72 C, 7 min extension.
PCR products were purified using QiaQuick PCR purification kit (Qiagen)
following the manufacturer's protocol. Both PCR TTV10Orf1 anfd TTV13Orf1
products and the Gateway entry plasmid, pENTR3C, were digested with Kpnl.
Digested DNA was purified using QlAquick PCR amplification kit and
subsequently digested with Xhol. Following QlAquick purifications, the TTV10
ORF1 or the TTV13ORF1 DNAs were ligated into pENTR3C using standard
ligation procedures. Following a 2 hour ligation at room temperature, ligated
DNA was used to transform chemically competent E. coli DHSa. Transformed
colonies were selected using Kanamycin. Plasmid was purified from transformed
E.coli and ORF1 DNA insertion was verified by restriction fragment analysis.
pENTR3C plasmids containing TTV10 ORF1 or TTV13 ORF1 were then
inserted into Invitrogen destination vectors pDEST10 or pDEST 20 encoding a
His6X or a GST protein N-terminal to the TTV Orfl reading frame. Recombinant
pDEST vectors containing the open reading frame of TTV Orfl were used to
transform DH1OBac E. coli. Recombinant bacmid DNA was isolated and used
for transfection of SF9 cells following standard protocol. Recombinant
baculovirus containing the native Orfl were isolated by plaque purification.
Confirmation of recombinant baculovirus was performed using PCR.
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-28-
Native TTVOrf1 construction for baculovirus expression.
Standard PCR was used to incorporate a BamH1 restriction site upstream
from the initiation codon in TTV10 Orfl or an Xbal restriction site upstream
from
the initiation codon in TTV Orfl3. These constructs were cloned into pFastBac
transfer vector and used to transform E. coli DH1 OBac. The resultant
recombinant bacmids were subsequently used to transfect SF9 cells.
Recombinant baculovirus containing the native Orfl were isolated by plaque
purification. Confirmation of recombinant baculovirus was performed using PCR.
Cloning of TTV genotype 2 ORF1 for E. coli expression.
Full-length TTVOrf10 was also cloned into a PGex-6p-1 vector for
expression of a GST-fusion protein in a bacterial system. The TTV ORFs contain
an arginine rich amino terminus. To determine if protein production could be
increased in a bacterial expression system, the arginine rich segment was
removed from TTVOrf13 at a convenient restriction site (EcoRl) located at
nucleotide 368 of the Orfl open reading frame and was in frame with the GST
coding region of pGex-6p-1. This clone resulted in the removal of 100 amino-
terminal amino acids containing a highly enriched arginine segment.
B. TTV genotype 1.
Total cellular DNA from porcine bone marrow was amplified by rolling
circle amplification following procedures described above, except that single-
stranded binding protein was added to improve the efficiency of the
amplification
reaction. Amplification products were digested with EcoRl, purified using a
QlAquick PCR purification kit (Qiagen), and ligated into pGem3zf(+) vector
which
had been previously treated with shrimp alkaline phosphatase. Recombinant
vector containing putative TTV genomic DNA was selected based on restriction
digests with EcoRl and/or BamH1. Plasmids containing approximately 2.7 kB
inserts were purified and submitted to ACGT, Inc. for sequencing of the ORF1
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-29-
sequences to confirm genotype. The complete genome, i.e. the region
containing the high G/C rich region, was not sequenced to entirety.
Analyses of sequencing data for PAH TTV genotype 1
Nucleotide alignment of PAH TTV7 (SEQ ID NO:4), TTV17 (SEQ ID NO:5),
TTV21 (SEQ ID NO:6), and TTV27 (SEQ ID NO:3) with published sequence,
AY823990 (SEQ ID NO:17).
AY823990 (1) TACACTTTGGGGTTCAGGAGGCTCAATTTGGCTCGCTTCGCTCGCACCAC
ttvgl-7 (1) TACACTTCCGGGTTCAGGAGGCTCAATTTGGCTCGCTTCGCTCGCACCAC
ttvGT1-17 (1) TACACTTCCGGGTTCAGGAGGCTCAATTTGGCTCGCTTCGCTCGCACCAC
ttvGT1-21 (1) TACACTTCCGGGTTCAGGAGGCTCAATTTGGCTCGCTTCGCTCGCACCAC
ttvgtl-27 (1) TACACTTCCGGGTTCAGAGGGCTCAATTTGGCTCGCTTCGCTCGCACCAC
AY823990 (51) GTTTGCTGCCAGGCGGACCTGATTGAAGACTGAAAACCGTTAAATTCAAA
ttvgl-7 (51) GTTTGCTGCCAGGCGGACCTGTTTGAAGACTGAAAACCGTTAAATTCAAA
ttvGT1-17 (51) GTTTGCTGCCAAGCGGACCTGATTGAAGACTGAAAACCGTTACATTCAAA
ttvGT1-21 (51) GTTTGCTGCCAGGCGGACCTGATTGAAGACTGAAAACCGTTAAATTCAAA
ttvgtl-27 (51) GTTTGCTGCCAGGCGGACCTGATTGAAGACTGAAAACCGTTAAGTTCAAA
AY823990 (101) ATTGAAAAGGGCGGGCAAA-ATGGCGGACAGGGGGCGGAGTTTATGCAAA
ttvgl-7 (101) TTTGAAATTGGCGGT-AAACATGGCGGAAGGGGGGCGGAGTATATGCAAA
ttvGT1-17 (101) TTTGAAAATGGCGCCCAAACATGGCGGATGTGGG-CGGAGTATATGCAAA
ttvGT1-21 (101) TTTGAAATTGGCGGT-AAATATGGCGGAAGGGGGGCGGAGTATATGCAAA
ttvgtl-27 (101) TTTGAAAATGGCGCCCAAACATGGCGGAG-GGGGGCGGAGTTTATGCAAA
AY823990 (150) TTAATTTATGCAAAGTAGGAGGAGCTCGATTTTAATTTATGCAAAGTAGG
ttvgl-7 ... (150) TTAATTTATGCAAAGTAGGAGGAGCTCGATTTTAATTTATGCAAAGTAGG
ttvGT1-17... (150) TTAATTTATGCAAAGTAGGAGGAGCTCGATTTTAATTTATGCAAAGTAGG
ttvGT1-21... (150) TTAATTTATGCAAAGTAGGAGGAGCTCGATTTTAATTTATGCAAAGTAGG
ttvgtl-27... (150) TTAATTTATGCAAAGTAGGAGGAGCTCCATTTTAATTTATGCAAAGTAGG
AY823990 (200) AGGAGTCAAATCTGATTGGTCGGGAGCTCAAGTCCTCATTTGCATAGGGT
ttvgl-7 ... (200) AGGAGTCAAATCTGATTGGTCGGGAGCTCAAGTCCTCATTTGCATAGGGT
ttvGT1-17... (200) AGGAGTCACTTCTGATTGGTCGGGAACTCAAGCCCTCATTTGCATAGGGT
ttvGT1-21... (200) AGGAGTCAAATCTGATTGGTCGGGAGCTCAAGTCCTCATTTGCATAGGGT
ttvgtl-27... (200) AGGAGTCACTTCTGATTGGTCGGGAGCTCAAGTCCTCATTTGCATAGGGT
AY823990 (250) GTAACCAATCAGAATTAAGGCGTTCCCACGAAAGCGAATATAAGTAGGTG
ttvgl-7 ... (250) GTAACCAATCAGAATTAAGGCGTGCCCACTAAAGTGAATATAAGTAAGTG
ttvGT1-17... (250) GTAACCAATCAGAATTAAGGCGTTCCCCGTGAAGTGAATATAAGTAAGTA
ttvGT1-21... (250) GTAACCAATCAGAATTAAGGCGTGCCCACTAAAGTGAATATAAGTGAGTG
ttvgtl-27... (250) GTAACCAATCAAACTTAAGGCGTTCCCACTAAAGTGAATATAAGTAAGTG
AY823990 (300) AGGTTCCGAATGGCTGAGTTTATGCCGCCAGCGGTAGACAGAACTGTCTA
ttvgl-7 ... (300) CAGTTCCGAATGGCTGAGTTTATGCCGCCAGCGGTAGACAGAACTGTCTA
ttvGT1-17... (300) AAGTTCCGAATGGCTGAGTTTATGCCGCCAGCGGTAGACAGAACTGTCTA
ttvGT1-21... (300) CAGTTCCGAATGGCTGAGTTTATGCCGCCAGCGGTAGACAGAACTGTCTA
ttvgtl-27... (300) CGGTTCCGAATGGCTGAGTTTATGCCGCCAGCGGTAGACAGAACTGTCTA
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-30-
AY823990 (350) GCGACTGGGCGGGTGCCGGAGGATCCCTGATCCGGAGTCAAGGGGCCTAT
ttvgl-7 ... (350) GCGACTGGGCGGGTGCCGGAGGATCCCAGATCCGGAGTCAAGGGGCCTAT
ttvGT1-17... (350) GCGACTGGGCGGGTGCCGAAGGATCCCAGATCCGGAGTCAAGGGGCCTAT
ttvGT1-21... (350) GCGACTGGGCGGGTGCCGGAGGATCCCAGATCCGGAGTCAAGGGGCCTAT
ttvgtl-27... (350) GCGACTGGGCGGGTGCCGGAGGATCCCTGATCCGGAGTCAAGGGGCCTAT
AY823990 (400) CGGGCAGGAGCAGCTAGGCGGAGGGCCTATGCCGGAACACTGGGAGGAAG
ttvgl-7 ... (400) CGGGCAGGAGCAGCTGAGCGGAGGGCCTATGCCGGAACACTGGGAGGAGG
ttvGT1-17... (400) CGGGCAGGAGCAGCTGAGCGGAGGGCCTATGCCGGAACACTGGGAGGAGG
ttvGT1-21... (400) CGGGCAGGAGCAGCTGAGCGGAGGGCCTATGCCGGAACACTGGGAAGAGG
ttvgtl-27... (400) CGGGCAGGAGCAGCTGAGCGGAGGGCCTATGCCGGAACACTGGGAAGAAG
AY823990 (450) CCTGGTTGGAAGCTACCAAGGGCTGGCACGATCTCGACTGCCGCTGCGGT
ttvgl-7 ... (450) CCTGGTTGGAAGCTACCAAGGGCTGGCACGACCTTGACTGCCGCTGCGGT
ttvGT1-17... (450) CCTGGTTGGAAGCTACCAAGGGCTGGCACGACCTCGACTGCCGCTGCGGT
ttvGT1-21... (450) CCTGGTTGGAAGCTACCAAGGGCTGGCACGACCTTGACTGCCGCTGCGGT
ttvgtl-27... (450) CCTGGTTGGAAGCTACCAAGGGCTGGCACGACTTAGACTGCCGCTGCGGT
AY823990 (500) AACTGGCAGGACCACCTATGGCTCCTACTCGCCGATGGAGACGCCGCTTT
ttvgl-7 ... (500) AATTGGCAAGACCACCTATGGCTTTTGCTCGCCGATGGAGACGCCGCTTT
ttvGT1-17... (500) AACTGGCAAGACCACCTATGGCTCCTGCTCGCCGATGGAGACGCGGCTTT
ttvGT1-21... (500) AATTGGCAAGACCACCTATGGCTTTTGCTCGCCGATGGAGACGCCGCTTT
ttvgtl-27... (500) AACTGGCAGGACCACCTATGGCTCCTACTCGGCGATGGAGACGCCGCTTT
AY823990 (550) GGCCGCCGCCGTAGACGCTATAGAAAGAGACGCTATGGCTGGAGACGACG
ttvgl-7 ... (550) GGCCGCCGCCGTAGACGCTATAGAAAGAGACGCTATGGATGGAGGAGACG
ttvGT1-17... (550) GGCCGCCGCCGTAGACGCTATAGAAAGAGACGCTGGGGCTGGAGAAGGCG
ttvGT1-21... (550) GGCCGCCGCCGTAGACGCTATAGAAAGAGACGCTATGGATGGAGGAGACG
ttvgtl-27... (550) GGCCGCCGCCGTAGACGCTATAGAAAGAGACGCTATGGCTGGAGAAGACG
AY823990 (600) CTACTACCGCTACAGGCCGCGTGACTATCGGCGACGATGGCTGGTAAGGA
ttvgl-7 ... (600) CTACTACCGCTACAGACCGCGTTACTATCGGAGACGATGGCTGGTAAGGA
ttvGT1-17... (600) CTACTGGAGATACCGACCGCGTTACCGTCGGCGCAGATGGCTGGTAAGGA
ttvGT1-21... (600) CTACTACCGCTACAGACCGCGTTACTATCGGAGACGATGGCTGGTAAGGA
ttvgtl-27... (600) CTACTACCGCTACAGACCGCGTTACTATCGGAGACGATGGCTGGTAAGGA
AY823990 (650) GAAGGCGGCGTTCCGTCTACCGTAGAGGTGGACGTAGAGCGCGCCCCTAC
ttvgl-7 ... (650) GAAGGCGGCGTTCCGTCTACCGACGAGGTGGACGTAGAGCGCGCCCCTAC
ttvGT1-17... (650) GAAGGCGGCGTTCCGTCTACCGAAGAGGTGGACGTAGAGCGCGCCCCTAC
ttvGT1-21... (650) GAAGGCGGCGTTCCGTCTACCGACGAGGTGGACGTAGAGCGCGCCCCTAC
ttvgtl-27... (650) GAAGGCGGCGTTCCGTCTACCGTAGAGGTGGACGTAGAGCGCGCCCCTAC
AY823990 (700) CGA----CTG--TTTAATCCAAAAGTAATGCGGAGAGTAGTAATTAGGGG
ttvgl-7 ... (700) CGCATTTCTGCCTTTAATCCGAAAGTAATGCGTAGAGTAGTGATTAGAGG
ttvGT1-17... (700) CGTATTTCTGCTTTTAATCCAAAAATAATGCGGAGAGTAGTAATAAGGGG
ttvGT1-21... (700) CGCATTTCTGCCTTTAATCCGAAAGTAATGCGTAGAGTAGTGATTAGAGG
ttvgtl-27... (700) CGGGTATCTGCCTTTAACCCCAAAGTAATGCGGAGAGTAGTAATAAGGGG
AY823990 (744) GTGGTGGCCTATTTTACAATGCTTAAAAGGACAGGAGGCACTAAGATATA
ttvgl-7 ... (750) GTGGTGGCCAATACTGCAGTGCCTAAAAGGTCAGGAATCACTAAGATACA
ttvGT1-17... (750) ATGGTGGCCAATCCTACAATGTCTAAGAGGACAGGAATCACTAAGATATA
ttvGT1-21... (750) GTGGTGGCCAATACTGCAGTGCCTAAAAGGTCAGGAATCACTAAGATACA
ttvgtl-27... (750) GTGGTGGCCAATACTACAGTGCTTAAAAGGACAGGAATCGCTGAGATATA
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-31-
AY823990 (794) GACCTCTACAGTGGGACACAGAGAGACAGTGGAGAGTGAGATCAGACTTC
ttvgl-7 ... (800) GACCACTTCAGTGGGACGTAGAGAAAAGCTGGAGAATAAACACAACTCTT
ttvGT1-17... (800) GACCGTTACAGTGGGACGTAGAAAAAAGCTGGAGAATAAAGACAGACTTA
ttvGT1-21... (800) GACCACTTCAGTGGGACGTAGAGAAAAGCTGGAGAATAAACACAACTCTT
ttvgtl-27... (800) GACCACTACAGTGGGACACAGAAAGACAGTGGAGAGTGAGACAAGACTTC
AY823990 (844) GAAGACCAGTACGGATACCTCGTACAATACGGGGGAGGTTGGGGAAGTGG
ttvgl-7 ... (850) GAGGACAACTATGGATACTTAGTACAGTATGGAGGTGGTTGGGGTAGCGG
ttvGT1-17... (850) GAAGACAACTACGGCTACTTAGTACAGTACGGAGGAGGTTGGGGGAGCGG
ttvGT1-21... (850) GAGGACAACTATGGATACTTAGTACAGTATGGAGGTGGTTGGGGTAGCGG
ttvgtl-27... (850) GAGGATCAATACGGATACCTGGTGCAATACGGTGGAGGTTGGGGAAGTGG
AY823990 (894) TGATGTGACACTTGAAGGTCTCTACCAAGAGCACTTATTGTGGAGAAACT
ttvgl-7 ... (900) AGAGGTAACACTGGAGGGGCTGTATCAGGAGCACCTACTATGGAGAAACT
ttvGT1-17... (900) AGAGGTGACTCTAGAAGGACTGTACCAGGAACACCTACTATGGAGAAATT
ttvGT1-21... (900) AGAGGTAACACTGGAGGGGCTGTATCAGGAGCACCTACTATGGAGAAACT
ttvgtl-27... (900) TGATGTGACACTAGAGGGACTATACCAGGAACACTTACTATGGAGAAATT
AY823990 (944) CTTGGTCTAAAGGAAACGATGGAATGGACCTAGTAAGATACTTTGGATGT
ttvgl-7 ... (950) CTTGGTCAAAAGGAAACGATGGGATGGACTTAGTGAGATACTTCGGCTGC
ttvGT1-17... (950) CATGGTCAAAAGGAAATGATGGAATGGATCTAGTAAGATACTTCGGCTGC
ttvGT1-21... (950) CTTGGTCAAAAGGAAACGATGGGATGGACTTAGTGAGATACTTCGGCTGC
ttvgtl-27... (950) CCTGGTCAAAAGGAAATGATGGCATGGACTTAGTGAGATACTTTGGCTGT
AY823990 (994) GTAGTATACCTATATCCACTAAAGGACCAGGACTATTGGTTCTGGTGGGA
ttvgl-7 ... (1000) ATAGTATATCTATATCCGTTAAAAGATCAAGACTACTGGTTTTGGTGGGA
ttvGT1-17... (1000) ATAGTATACCTGTACCCACTGAAAGATCAGGACTACTGGTTTTGGTGGGA
ttvGT1-21... (1000) ATAGTATATCTATATCCGTTAAAAGATCAGGACTACTGGTTTTGGTGGGA
ttvgtl-27... (1000) GTGGTATACCTCTACCCACTTAAAGATCAGGACTATTGGTTCTGGTGGGA
AY823990 (1044) CACGGACTTCAAAGAATTATATGCAGAAAACATAAAGGAATACAGCCAAC
ttvgl-7 ... (1050) CACAGATTTTAAAGAATTATATGCAGAGAGTATCAAAGAATACTCACAGC
ttvGT1-17... (1050) CACAGACTTTAAGGAACTCTATGCAGAAAGTATTAAGGAGTACTCACAAC
ttvGT1-21... (1050) CACAGATTTTAAGGAATTATATGCAGAGAGTATCAAAGAATACTCACAGC
ttvgtl-27... (1050) CACTGACTTTAAAGAGCTATACGCAGAAAACATAAAAGAATACAGCCAAC
AY823990 (1094) CATCAGTAATGATGATGGCAAAAAGAACAAGAATAGTAATAGCCAGAGAA
ttvgl-7 ... (1100) CATCTGTAATGATGATGGCAAAAAGAACAAAAATAGTGATCGCAAGAAGT
ttvGT1-17... (1100) CATCAGTAATGATGATGGCAAAAAAAACAAAAATTGTAATAGCGAGAAGT
ttvGT1-21... (1100) CATCTGTAATGATGATGGCAAAAAGAACAAAAATAGTGATCGCAAGAAGT
ttvgtl-27... (1100) CATCAGTAATGATGATGGCAAAAAGAACTAGAATAGTAATAGCGAGAGAC
AY823990 (1144) AGGGCACCACATAGAAGAAAAGTAAGAAAAATATTTATTCCGCCACCTTC
ttvgl-7 ... (1150) AGAGCCCCACATAGAAGGAAGGTACGCAGAATTTTCATACCGCCTCCAAG
ttvGT1-17... (1150) AGGGCACCACACAGACGAAAAGTAAGAAAAATATTCATACCGCCACCAAG
ttvGT1-21... (1150) AGAGCCCCACATAGAAGGAAGGTACGCAGAATTTTCATACCGCCTCCAAG
ttvgtl-27... (1150) AGAGCTCCACATAGAAGAAAAGTGAGAAAAATATTCATCCCACCACCATC
AY823990 (1194) GAGAGACACAACACAGTGGCAGTTTCAGACAGATTTCTGCAATAGAAAGT
ttvgl-7 ... (1200) TAGAGACACGACACAGTGGCAATTTCAAACTGACTTTTGCAATAGACCAC
ttvGT1-17... (1200) TAGAGACACTACACAATGGCAATTTCAAACAGAGTTCTGCAACAAACCAC
ttvGT1-21... (1200) TAGAGACACGACACAGTGGCAATTTCAAACTGACTTTTGCAATAGACCAC
ttvgtl-27... (1200) AAGAGACACTACGCAGTGGCAGTTTCAGACAGACTTCTGTAATAGGAAGC
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-32-
AY823990 (1244) TATTTACGTGGGCAGCTGGTCTAATAGACATGCAAAAACCGTTCGATGCT
ttvgl-7 ... (1250) TATTCACATGGGCTGCAGGACTCATAGACCTCCAAAAACCATTTGACGCA
ttvGT1-17... (1250) TATTCACTTGGGCTGCAGGACTAATAGACCTCCAAAAGCCATTTGACGCA
ttvGT1-21... (1250) TATTCACATGGGCTGCAGGACTCATAGACCTCCAAAAACCATTTGACGCA
ttvgtl-27... (1250) TATTTACCTGGGCGGCAGGACTAATAGACATGCAAAAACCCTTTGATGCC
AY823990 (1294) AATGGAGCCTTTAGAAATGCTTGGTGGCTGGAACAGAGAAATGATCAGGG
ttvgl-7 ... (1300) AACGGTGCGTTCAGAAATGCCTGGTGGTTAGAACAGAGAAACGAGGCAGG
ttvGT1-17... (1300) AACGGAGCTTTTAGAAATGCGTGGTGGTTAGAACAGAGAAATGAGGCAGG
ttvGT1-21... (1300) AACGGTGCGTTCAGAAATGCCTGGTGGTTAGAACAGAGAAACGAGGCAGG
ttvgtl-27... (1300) AACGGAGCTTTTAGAAATGCGTGGTGGCTGGAGCAGAGAACGGAACAGGG
AY823990 (1344) AGAAATGAAATACATAGAACTGTGGGGAAGAGTACCCCCACAAGGAGATT
ttvgl-7 ... (1350) AGAAATGAAATACATAGAGCTATGGGGTAGAGTACCACCCCAGGGGGACA
ttvGT1-17... (1350) AGAGATGAAATACATAGAATTATGGGGGAGAGTCCCACCGCAAGGAGACA
ttvGT1-21... (1350) AGAAATGAAATACATAGAGCTATGGGGTAGAGTACCACCCCAGGGGGACA
ttvgtl-27... (1350) TGAAATGAAGTACATAGAACTGTGGGGAAGAGTGCCCCCACAAGGAGACT
AY823990 (1394) CAGAGCTGCCCAAAAAAAAAGAATTCTCCACAGGAACAG---ATAACCCA
ttvgl-7 ... (1400) CGGAATTACCCGTTCAAACAGAATTCCAAAAACCCTCGGGATATAACCCA
ttvGT1-17... (1400) CAGAATTGCCGGCCCAAAAAGAATTCCAGAAACCAGACGGGTATAACCCA
ttvGT1-21... (1400) CGGAATTACCCCTTCAAACAGAATTCCAAAAACCCTCGGGATATAACCCA
ttvgtl-27... (1400) CAGAACTACCCAAGAAAAGTGAATTCACAACAGCTACAG --- ACAATAAA
AY823990 (1441) AACTACAATGTTCAGGACAATGAGGAGAAAAACATATACCCCATTATAAT
ttvgl-7 ... (1450) AAATACTACGTAAACCCGGGGGAGGAAAAACCAATCTACCCAGTAATAAT
ttvGT1-17... (1450) AAATACTATGTGCAGGCAGGAGAGGAAAAACCTATATATCCAATAATAAT
ttvGT1-21... (1450) AAATACTACGTAAACCCGGGGGAGGAAAAACCAATCTACCCAGTAATAAT
ttvgtl-27... (1447) AACTACAATGTGAATGACGGTGAGGAAAAACCTATATACCCCATAATTAT
AY823990 (1491) ATACGTAGACCAAAAAGATCAAAAACCAAGAAAAAAGTACTGCGTATGTT
ttvgl-7 ... (1500) ATACGTAGACATGAAAGACCAAAAACCAAGAAAAAAGTACTGCGTCTGCT
ttvGT1-17... (1500) TTACGTAGACAAAAAAGATCAGAAAGCAAGAAAGAAATACTGTGTCTGTT
ttvGT1-21... (1500) ATACGTAGACATGAAAGACCAAAAACCAAGAAAAAAGTACTGCGTCTGCT
ttvgtl-27... (1497) ATACGTAGACCAAAAAGACCAAAAACCAAGGAAAAAGTACTGTGTATGTT
AY823990 (1541) ATAATAAGACCCTCAACAGATGGAGACTAGGACAGGCAAGTACTCTAAAG
ttvgl-7 ... (1550) ACAACAAGACGCTTAACAGGTGGCGCAGCGCTCAAGCAAGCACATTAAAA
ttvGT1-17... (1550) ACAATAAGACACTAAACAGATGGAGAGCAGCACAAGCAAGTACCCTAAAA
ttvGT1-21... (1550) ACAACAAGACGCTTAACAGGTGGCGCAGCGCTCAGGCAAGCACATTAAAA
ttvgtl-27... (1547) ACAACAAAACTCTGAACAGGTGGAGATTAGGACAAGCGAGTACTCTAAAA
AY823990 (1591) ATAGGAAACCTGAAAGGACTAGTACTAAGACAGCTGATGAATCAAGAAAT
ttvgl-7 ... (1600) ATTGGTGACTTGCAGGGGCTAGTATTGAGACAGCTAATGAACCAAGAAAT
ttvGT1-17... (1600) ATAGGAGACCTGCAAGGACTAGTACTAAGACAATTAATGAACCAGGAAAT
ttvGT1-21... (1600) ATTGGTGACTTGCAGGGGCTAGTATTGAGACAGCTAATGAACCAAGAAAT
ttvgtl-27... (1597) ATAGGAAACCTGAAAGGACTAGTGCTAAGACAGTTGATGAACCAAGAGAT
AY823990 (1641) GACGTATATATGGAAAGAAGGAGAATACAGTGCCCCCTTTGTACAAAGGT
ttvgl-7 ... (1650) GACATACACATGGAAAGAAGGAGAATTTACCAATGTATTCCTGCAGAGGT
ttvGT1-17... (1650) GACATATATTTGGAAAGAGGGAGAGTTCACAAACGTATTCCTGCAAAGGT
ttvGT1-21... (1650) GACATACACATGGAAAGAAGGAGAATTTACAAATGTATTCCTGCAAAGGT
ttvgtl-27... (1647) GACTTACATATGGAAGGAAGGAGAGTACAGCTCACCATTTGTACAAAGGT
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-33-
AY823990 (1691) GGAAAGGCAGCAGATTCGCTGTGATAGACGCAAGAAAGGCAGACCAAGAA
ttvgl-7 ... (1700) GGAGAGGTTTCAGATTAGCAGTAATAGACGCAAGAAAGGCAGACACAGAA
ttvGT1-17... (1700) GGAAAGGCTTCAGACTAGCAGTCATAGACGCCAGAAAGGGAGACACAGAA
ttvGT1-21... (1700) GGAGAGGTTTCAGATTAGCAGTAATAGACGCTAGAAAGGCAGACACAGAA
ttvgtl-27... (1697) GGAAAGGAAGCAGATTTGTTGTGATAGACGCAAGAAAGGCTGACCAGGAA
AY823990 (1741) AACCCGAAAGTATCAACATGGCCAATTGAGGGAACGTGGAACACACAGGA
ttvgl-7 ... (1750) AACCCGACAGTCCAAACTTGGAAGGTGGACGGACAGTGGAACACACAAGG
ttvGT1-17... (1750) AATCCAACAGTACAAACATGGAAAGTAGACGGAAACTGGAACACTAGTGG
ttvGT1-21... (1750) AACCCGACAGTCCAAACTTGGAAGGTGGACGGACAGTGGAACACACAAGG
ttvgtl-27... (1747) AATCCCAAAGTATCTACATGGCCAATAGAGGGAGTGTGGAACACACAGGG
AY823990 (1791) CACAGTACTGAAGGATGTATTCGGTATTAACTTGCAAAATCAACAATTTA
ttvgl-7 ... (1800) GACAGTGCTTAAAGAGGTTTTCAATATAAACCTGAATAATGAACAGATGA
ttvGT1-17... (1800) AACAGTACTACAAGAAGTGTTCGGCATAAACCTCACCCAACAACAAATGA
ttvGT1-21... (1800) GACAGTTCTTAAAGAGGTTTTCAATATAAACCTGAATAATGAACAGATGA
ttvgtl-27... (1797) TACAGTACTTAAGGATGTATTCCAGATTGACTTAAACAGTACTAATTTCA
AY823990 (1841) GGGCGGCGGACTTTGGTAAACTCACACTACCAAAATCACCGCATGACTTA
ttvgl-7 ... (1850) GACAGGCAGACTTTGGAAAACTAAACTTACCAAAATCCCCGCACGACATT
ttvGT1-17... (1850) GGGCATCGGACTTTGCTAAGCTAACACTACCAAAATCGCCACATGACATT
ttvGT1-21... (1850) GACAGGCAGACTTTGGAAAACTAAACTTACCAAAATCCCCGCACGACATT
ttvgtl-27... (1847) GAGCGGCAGACTTTGGAAAACTAACACTACCAAAATCACCGCACGACTTA
AY823990 (1891) GACTTCGGTCACCACAGCAGATTTGGGCCATTTTGTGTGAAAAATGAACC
ttvgl-7 ... (1900) GACTTTGGACACCACAGTAGATTTGGACCTTTCTGTGTAAAAAACGAACC
ttvGT1-17... (1900) GACTTTGGACACCACAGTAGATTTGGGCCATTTTGTGTCAAAAACGAACC
ttvGT1-21... (1900) GACTTTGGACACCACAGTAGATTTGGACCTTTCTGTGTAAAAAACGAACC
ttvgtl-27... (1897) GACTTCGGACATCACAGTAGATTCGGACCATTCTGTGTGAAAAATGAACC
AY823990 (1941) ACTGGAGTTTCAGGTATACCCTCCAGAACCAACTAACTTGTGGTTTCAGT
ttvgl-7 ... (1950) ACTGGAGTTTCAACTAACAGCCCCAGAGCCAACTAACCTGTGGTTTCAGT
ttvGT1-17... (1950) GCTGGAGTTTCAACTAACCGCTCCAGAACCTATTAATCTTTGGTTTCAGT
ttvGT1-21... (1950) ACTGGAGTTTCAACTAACAGCCCCAGAGCCAACTAACCTGTGGTTTCAGT
ttvgtl-27... (1947) ACTGGAATTTCAGGTATACCCGCCAGAACCCACTAACCTGTGGTTTCAGT
AY823990 (1991) ACAGATTTTTCTTTCAGTTTGGAGGTGAATACCAACCCCCCACAGGAATC
ttvgl-7 ... (2000) ACAAATTTCTGTTTCAGTTTGGAGGTGAATACCAACCACCAACAGGCATC
ttvGT1-17... (2000) ACAAATTTCTCTTTCAGTTTGGAGGTGAATACCAACCACCAACAGGCATC
ttvGT1-21... (2000) ACAAATTTCTGTTTCAGTTTGGAGGTGAATACCAACCACCAACAGGCATC
ttvgtl-27... (1997) ACAGATTTTTCTTTCAGTTTGGAGGTGAATACCAACCCCCCACAGGAATC
AY823990 (2041) CGGGATCCATGCGTTGATACACCAGCCTATCCTGTGCCGCAGTCAGGAAG
ttvgl-7 ... (2050) CGCGATCCCTGCGCTGATAACCCAGCCTATCCTGTGCCGCAGTCAGGAAG
ttvGT1-17... (2050) CGCGATCCCTGCGCTGATAACCAACCCTATCCTGTGCCGCAGTCAGGAAG
ttvGT1-21... (2050) CGCGATCCCTGCGCTGATAACCCAGCCTATCCTGTGCCGCAGTCAGGAAG
ttvgtl-27... (2047) CGCGATCCATGCGTTGATACACCAGCCTATCCTGTGCCGCAGTCAGGAAG
AY823990 (2091) TATTACACACCCCAAATTCGCCGGAAAAGGAGGAATGCTCACGGAAACAG
ttvgl-7 ... (2100) TATTACACACCCCAAATTCGCCGGAAAAGGCGGCATGCTCACGGAAACAG
ttvGT1-17... (2100) TATTACACACCCAAAATTCGCCGGGAAAGGAGGAATGCTCACGGAAACAG
ttvGT1-21... (2100) TATTACACACCCCAAATTCGCCGGAAAAGGCGGCATGCTCACGGAAACAG
ttvgtl-27... (2097) TATTACACACCCCAAATTCGCCGGAAAAGGCGGAATGCTCACGGAAACAG
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-34-
AY823990 (2141) ACCGTTGGGGTATCACTGCTGCCTCTTCCAGAGCCCTCAGTGCAGATACA
ttvgl-7 ... (2150) ACCGTTGGGGTATCACTGCTGCCTCTTCCCGAACCCTCAGTGCAGATACA
ttvGT1-17... (2150) ACCGTTGGGGTATCACTGCTGCCTCTTCCAGAGCCCTCAGTGCAGATACA
ttvGT1-21... (2150) ACCGTTGGGGTATCACTGCTGCCTCTTCCCGAGCCCTCAGTGCAGATACA
ttvgtl-27... (2147) ACCGTTGGGGTATCACTCCTGCCTCTACCAGAGCCCTCTGTGCAGATACA
AY823990 (2191) CCCACAGAGGCAGCGCAAAGTGCACTTCTCCGAGGGGACTCGGAAGCGAA
ttvgl-7 ... (2200) CCCACGGAAGCAACGCAAAGTGCACTTCTCCGAGGGGACTCGGAAAAGAA
ttvGT1-17... (2200) CCCACGGAGGCAGCGCAAAGTGCACTTCTCCGAGGGGACTCGGAAAAGAA
ttvGT1-21... (2200) CCCACGGAAGCAACGCAAAGTGCACTTCTCCGAGGGGACTCGGAAAAGAA
ttvgtl-27... (2197) CCCACAGAAGCAACGCAGAGTGCACTTCTCCGAGGGGACTCGGAAAAGAA
AY823990 (2241) AGGAGAGGAAACCGAGGAAACCGCGTCATCGTCCAGTATCACGAGTGCCG
ttvgl-7 ... (2250) AGGAGAGGAAACCGAGGAAACCTCGTCATCGTCCAGTATCACGAGTGCCG
ttvGT1-17... (2250) AGGAGAGGAAACCGAGGAAACCACGTCATCGTCCAGTATCACGAGTGCCG
ttvGT1-21... (2250) AGGAGAGGAAACCGAGGAAACCTCGTCATCGTCCAGTATCACGAGTGCCG
ttvgtl-27... (2247) AGGAGAGGAAACCGAGGAAACCACGTCATCGTCCAGTATCACGAGTGCCG
AY823990 (2291) AAAGCTCTACTGAGGGAGATGGATCGTCTGATGATGAAGAGACAATCAGA
ttvgl-7 ... (2300) AAAGCTCTACTGAAGGAGATGGATCGTCTGATGATGAAGAGACAATCAGA
ttvGT1-17... (2300) AAAGCTCTACTGAAGGAGATGGATCGTCTGATGATGAAGAGACAATCCGA
ttvGT1-21... (2300) AAAGCTCTACTGAAGGAGATGGATCGTCTGATGATGAAGAGACAATCAGA
ttvgtl-27... (2297) AAAGCTCTACTGAGGGAGATGGATCGTCTGATGATGAAGAGACAGTCAGA
AY823990 (2341) CGCAGAAGGAGGACCTGGAAGCGACTCAGACGAATGGTCAGAGAGCAGCT
ttvgl-7 ... (2350) CGCCGAAGGAGGACCTGGAAGCGACTCAGACGGATGGTCCGAGAGCAGCT
ttvGT1-17... (2350) CGCAGAAGGAGGACCTGGAAGCGACTCCGACGAATGGTCAGAGAGCAGCT
ttvGT1-21... (2350) CGCCGAAGGAGGACCTGGAAGCGACTCAGACGGATGGTCCGAGAGCAGCT
ttvgtl-27... (2347) CGCCGAAGGAGGACCTGGAAGCGACTCAGACGAATGGTCCGAGAGCAGCT
AY823990 (2391) TGACCGACGAATGGACCACAAGCGACAGCGACTTCATTGACACCCCCATA
ttvgl-7 ... (2400) TGACCGACGAATGGACCACAAGCGACAGCGACTTCATTGACACCCCCATT
ttvGT1-17... (2400) TGACCGACGAATGGACCACAAGCGACAGCGACTTCATTGACACCCCCATA
ttvGT1-21... (2400) TGACCGACGAATGGACCACAAGCGACAGCGACTTCATTGACACCCCCATT
ttvgtl-27... (2397) TGACCGACGAATGGACCACAAGCGACAGCGACTTCATTGACACCCCCATT
AY823990 (2441) AGAGAAAGATGCCTCAATAAAAAACAAAAGAAACGCTAAACAGTGTCCGA
ttvgl-7 ... (2450) AAACAGAGATGCCTCAATAAAAAACAAAAGAAACGCTAAGCAGTGTCC-C
ttvGT1-17... (2450) AGAGAACGATGCCTGAATAAAAAACAAAAAAAACGCTACACAGTGTCCGC
ttvGT1-21... (2450) AGACAGAGATGCCTCAATAAAAAGCAAAAGAAACGCTAAACAGTGTCC-C
ttvgtl-27... (2447) AGAGACAGATGCCTCAATAAAAAGCAAAAGAAACGCTAAACTGCCTCCGC
AY823990 (2491) TTACTAATGGGGGGGGGTCCGGGGGGGGCTTGCCCCCCCGCAAGCTGGGT
ttvgl-7 ... (2499) TATTATTTTGGGGGG--TCCGGGGGGGGCTTGCCCCCCCGTAAGCTGGGT
ttvGT1-17... (2500) TTATTTGTAGGGGGGG-TCCGGGGGGGGCTTGCCCCCCCGTAAGCTGGGT
ttvGT1-21... (2499) TATTACTTTGGGGGGG-TCCGGGGGGGGCTTGCCCCCCCGTAAGCTGTGT
ttvgtl-27... (2497) TTATTTTTTGGGGGG--TCCGGGGGGGGCTTGCCCCCCCGAAAGCTGGGT
AY823990 (2541) TACCGCACTAACTCCCTGCCAAGTGAAACTCGGGGACGAGTGAGTGCGGG
ttvgl-7 ... (2547) TACCGCACTAACTCCCTGCCAAGTGAAACTCGGGGACGAGTGAGTGCGGG
ttvGT1-17... (2549) TGCCGCACTAACTCCCTGCCAAGTGAAACTCGGGGACGAGTGAGTGCGGG
ttvGT1-21... (2548) TACCGCACTAACTCCCTGCCAAGTGAAACTCGGGGACGAGTGAGTGCGGG
ttvgtl-27... (2545) TACCGCACTAACTCCCTGCCAAGTGAAACTCGGGGACGAGTGAGTGCGGG
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-35-
AY823990 (2591) ACATCCCGTGTAATGGCTACATAACTACCCGGCTTTGCTTCGACAGTGGC
ttvgl-7 ... (2597) ACATCCCGTGTAATGGCTACATAACTACCCGGCTTTGCTTCGACAGTGGC
ttvGT1-17... (2599) ACATCCCGTGTAATGGCTACATAACTACCCGGCTTTGCTTCGACAGTGGC
ttvGT1-21... (2598) ACATCCCGTGTAATGGCTACATAACTACCCGGCTTTGCTTCCACAGTGGC
ttvgtl-27... (2595) ACATCCCGTGTAATGGCTACATAACTACCCGGCTTTGCTTCGACAGTGGC
AY823990 (2641) CGTGGCTCGACCCTCACACAACACTGCAGGTAGGGGGCGCAATTGGGATC
ttvgl-7 ... (2647) CGTGGCTCGACCCTCACACAACACTGCAGGTAGGGGGCGCAATTGTGATC
ttvGT1-17... (2649) CGTGGCTCGACCCTCACACAACAATGCAGGTAGGGGGCGCAATTGGGATC
ttvGT1-21... (2648) CGTGGCTCGACCCTCACACAACACTGCAGGTAGGGGGCGCAATTGGGATC
ttvgtl-27... (2645) CGTGGCTCGACCCTCACACAACACTGCAGATAGGGGGCGCAATTGGGATC
AY823990 (2691) GTTAGAAAACTATGGCC--GAGCATGGGGG CCAACC
ttvgl-7 ... (2697) GTTAGAAAACTATGGCCCGGAGCATGG-CCCCCCAAAC------CCCCCC
ttvGT1-17... (2699) GTTAGAAAACTATGGCCCG-AGCATGGGCCCCCCAAAA------CCCCCC
ttvGT1-21... (2698) GTTAGAAAACTATGGCCCCAAGCATGG-CCCA--AAAC------CCCCCC
ttvgtl-27... (2695) GTTAGAAAACTATGGCC--GAGCATGGGCCCCCACAAA-----CCCCCCC
AY823990 (2739) CCCCCGGTGGGGGGGCCAAGGCCCCCCCTACACCCCCCCATGGGGGGCTG
ttvgl-7 ... (2740) TTGCCCGGGGCTGTGCCCCGGACCCCC-----------------------
ttvGT1-17... (2742) TTGCCCGGGGCTGTGCCCCGGACCCCC-----------------------
ttvGT1-21... (2739) TT-CCCGGGGCTGTGCCCCGGACCCCC-----------------------
ttvgtl-27... (2738) CTGCCCGGGGCTGTGCCCCGGACCCCCC----------------------
AY823990 (2789) CCGCCCCCCAAACCCCCCGCGTCGGATGGGGGGGGCTGCGCCCCCCCCAA
ttvgl-7 ... (2767) --------------------------------------------------
ttvGT1-17... (2769) --------------------------------------------------
ttvGT1-21... (2765) --------------------------------------------------
ttvgtl-27... (2766) --------------------------------------------------
AY823990 (2839) ACCCCCCTTGCCCGGGGCTGTGCCCCGGACCCCC
ttvgl-7 ... (2767) ----------------------------------
ttvGT1-17... (2769) ----------------------------------
ttvGT1-21... (2765) ----------------------------------
ttvgtl-27... (2766) ----------------------------------
Nucleotide Identity among PAH TTV's and Published Sequence
AY823990 ttv 1-7 ttvGT1 -17 ttvGT1 -21 ttvgtl -27
AY823990 85 87 85 91
ttv 1-7 89 99 86
ttvGT1-17 89 86
ttvGT1-21 86
ttvgtl -27
TTVgt1-27 demonstrates the greatest homology with published sequence,
AY823990, demonstrating 91 % identity. TTVgt1-7,17, and 21 demonstrate 85-
87% identity. TTVgt1-7 and TTVgt1-21 share 99% nucleotide identity
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-36-
Orfl Amino Acid Alignment
The following provides a comparison of the published AY823990
sequence (SEQ ID NO:25) to the corresponding amino acid sequences for TTV7
(SEQ ID NO:10), TTV17 (SEQ ID NO:11), TTV21 (SEQ ID NO:12), and TTV27
(SEQ ID NO:13)
AY823990 (1) MAPTRRWRRRFGRRRRRYRKRRYGWRRRYYRYRPRDYRRRWLVRRRRRSV
Ttvgl-70rfl (1) MAFARRWRRRFGRRRRRYRKRRYGWRRRYYRYRPRYYRRRWLVRRRRRSV
Ttgl-170rfl (1) MAPARRWRRGFGRRRRRYRKRRWGWRRRYWRYRPRYRRRRWVVRRRRRSV
Ttgl-270rfl (1) MAPTRRWRRRFGRRRRRYRKRRYGWRRRYYRYRPRYYRRRWLVRRRRRSV
ttgl-21Orf1 (1) MAFARRWRRRFGRRRRRYRKRRYGWRRRYYRYRPRYYRRRWLVRRRRRSV
AY823990 (51) YRRGGRRARPYRL--FNPKVMRRVVIRGWWPILQCLKGQEALRYRPLQWD
Ttvgl-70rfl (51) YRRGGRRARPYRISAFNPKVMRRVVIRGWWPILQCLKGQESLRYRPLQWD
Ttgl-170rfl (51) YRRGGRRARPYRISAFNPKIMRRVVIRGWWPILQCLRGQESLRYRPLQWD
Ttgl-270rfl (51) YRRGGRRARPYRVSAFNPKVMRRVVIRGWWPILQCLKGQESLRYRPLQWD
ttgl-21Orf1 (51) YRRGGRRARPYRISAFNPKVMRRVVIRGWWPILQCLKGQESLRYRPLQWD
AY823990 (99) TERQWRVRSDFEDQYGYLVQYGGGWGSGDVTLEGLYQEHLLWRNSWSKGN
Ttvgl-70rfl (101) VEKSWRINTTLEDNYGYLVQYGGGWGSGEVTLEGLYQEHLLWRNSWSKGN
Ttgl-170rfl (101) VEKSWRIKTDLEDNYGYLVQYGGGWGSGEVTLEGLYQEHLLWRNSWSKGN
Ttgl-270rfl (101) TERQWRVRQDFEDQYGYLVQYGGGWGSGDVTLEGLYQEHLLWRNSWSKGN
ttgl-21Orf1 (101) VEKSWRINTTLEDNYGYLVQYGGGWGSGEVTLEGLYQEHLLWRNSWSKGN
AY823990 (149) DGMDLVRYFGCVVYLYPLKDQDYWFWWDTDFKELYAENIKEYSQPSVMMM
Ttvgl-70rfl (151) DGMDLVRYFGCIVYLYPLKDQDYWFWWDTDFKELYAESIKEYSQPSVMMM
Ttgl-170rfl (151) DGMDLVRYFGCIVYLYPLKDQDYWFWWDTDFKELYAESIKEYSQPSVMMM
Ttgl-270rf1 (151) DGMDLVRYFGCVVYLYPLKDQDYWFWWDTDFKELYAENIKEYSQPSVMMM
ttgl-210rfl (151) DGMDLVRYFGCIVYLYPLKDQDYWFWWDTDFKELYAESIKEYSQPSVMMM
AY823990 (199) AKRTRIVIARERAPHRRKVRKIFIPPPSRDTTQWQFQTDFCNRKLFTWAA
Ttvgl-70rfl (201) AKRTKIVIARSRAPHRRKVRRIFIPPPSRDTTQWQFQTDFCNRPLFTWAA
Ttgl-170rfl (201) AKKTKIVIARSRAPHRRKVRKIFIPPPSRDTTQWQFQTEFCNKPLFTWAA
Ttgl-270rf1 (201) AKRTRIVIARDRAPHRRKVRKIFIPPPSRDTTQWQFQTDFCNRKLFTWAA
ttgl-210rfl (201) AKRTKIVIARSRAPHRRKVRRIFIPPPSRDTTQWQFQTDFCNRPLFTWAA
AY823990 (249) GLIDMQKPFDANGAFRNAWWLEQRNDQGEMKYIELWGRVPPQGDSELPKK
Ttvgl-70rfl (251) GLIDLQKPFDANGAFRNAWWLEQRNEAGEMKYIELWGRVPPQGDTELPVQ
Ttgl-170rfl (251) GLIDLQKPFDANGAFRNAWWLEQRNEAGEMKYIELWGRVPPQGDTELPAQ
tgl-270rf1 (251) GLIDMQKPFDANGAFRNAWWLEQRTEQGEMKYIELWGRVPPQGDSELPKK
ttgl-21Orf1 (251) GLIDLQKPFDANGAFRNAWWLEQRNEAGEMKYIELWGRVPPQGDTELPLQ
AY823990 (299) KEFSTGT-DNPNYNVQDNEEKNIYPIIIYVDQKDQKPRKKYCVCYNKTLN
Ttvgl-70rfl (301) TEFQKPSGYNPKYYVNPGEEKPIYPVIIYVDMKDQKPRKKYCVCYNKTLN
Ttgl-170rfl (301) KEFQKPDGYNPKYYVQAGEEKPIYPIIIYVDKKDQKARKKYCVCYNKTLN
Ttgl-270rf1 (301) SEFTTAT-DNKNYNVNDGEEKPIYPIIIYVDQKDQKPRKKYCVCYNKTLN
ttgl-21Orf1 (301) TEFQKPSGYNPKYYVNPGEEKPIYPVIIYVDMKDQKPRKKYCVCYNKTLN
AY823990 (348) RWRLGQASTLKIGNLKGLVLRQLMNQEMTYIWKEGEYSAPFVQRWKGSRF
Ttvgl-70rfl (351) RWRSAQASTLKIGDLQGLVLRQLMNQEMTYTWKEGEFTNVFLQRWRGFRL
Ttgl-170rfl (351) RWRAAQASTLKIGDLQGLVLRQLMNQEMTYIWKEGEFTNVFLQRWKGFRL
Ttgl-270rf1 (350) RWRLGQASTLKIGNLKGLVLRQLMNQEMTYIWKEGEYSSPFVQRWKGSRF
ttgl-21Orf1 (351) RWRSAQASTLKIGDLQGLVLRQLMNQEMTYTWKEGEFTNVFLQRWRGFRL
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-37-
AY823990 (398) AVIDARKADQENPKVSTWPIEGTWNTQDTVLKDVFGINLQNQQFRAADFG
Ttvgl-70rfl (401) AVIDARKADTENPTVQTWKVDGQWNTQGTVLKEVFNINLNNEQMRQADFG
Ttgl-170rfl (401) AVIDARKGDTENPTVQTWKVDGNWNTSGTVLQEVFGINLTQQQMRASDFA
Ttgl-270rf1 (400) VVIDARKADQENPKVSTWPIEGVWNTQGTVLKDVFQIDLNSTNFRAADFG
ttgl-21Orf1 (401) AVIDARKADTENPTVQTWKVDGQWNTQGTVLKEVFNINLNNEQMRQADFG
AY823990 (448) KLTLPKSPHDLDFGHHSRFGPFCVKNEPLEFQVYPPEPTNLWFQYRFFFQ
Ttvgl-70rfl (451) KLNLPKSPHDIDFGHHSRFGPFCVKNEPLEFQLTAPEPTNLWFQYKFLFQ
Ttgl-170rfl (451) KLTLPKSPHDIDFGHHSRFGPFCVKNEPLEFQLTAPEPINLWFQYKFLFQ
Ttgl-270rf1 (450) KLTLPKSPHDLDFGHHSRFGPFCVKNEPLEFQVYPPEPTNLWFQYRFFFQ
ttgl-210rfl (451) KLNLPKSPHDIDFGHHSRFGPFCVKNEPLEFQLTAPEPTNLWFQYKFLFQ
AY823990 (498) FGGEYQPPTGIRDPCVDTPAYPVPQSGSITHPKFAGKGGMLTETDRWGIT
Ttvgl-70rfl (501) FGGEYQPPTGIRDPCADNPAYPVPQSGSITHPKFAGKGGMLTETDRWGIT
Ttgl-170rfl (501) FGGEYQPPTGIRDPCADNQPYPVPQSGSITHPKFAGKGGMLTETDRWGIT
Ttgl-270rf1 (500) FGGEYQPPTGIRDPCVDTPAYPVPQSGSITHPKFAGKGGMLTETDRWGIT
ttgl-210rfl (501) FGGEYQPPTGIRDPCADNPAYPVPQSGSITHPKFAGKGGMLTETDRWGIT
AY823990 (548) AASSRALSADTPTEAAQSALLRGDSEAKGEETEETASSSSITSAESSTEG
Ttvgl-70rfl (551) AASSRTLSADTPTEATQSALLRGDSEKKGEETEETSSSSSITSAESSTEG
Ttgl-170rfl (551) AASSRALSADTPTEAAQSALLRGDSEKKGEETEETTSSSSITSAESSTEG
Ttgl-270rf1 (550) PASTRALCADTPTEATQSALLRGDSEKKGEETEETTSSSSITSAESSTEG
ttgl-210rfl (551) AASSRALSADTPTEATQSALLRGDSEKKGEETEETSSSSSITSAESSTEG
AY823990 (598) DGSSDDEETIRRRRRTWKRLRRMVREQLDRRMDHKRQRLH-
Ttvgl-70rfl (601) DGSSDDEETIRRRRRTWKRLRRMVREQLDRRMDHKRQRLH-
Ttgl-170rfl (601) DGSSDDEETIRRRRRTWKRLRRMVREQLDRRMDHKRQRLH-
Ttgl-270rf1 (600) DGSSDDEETVRRRRRTWKRLRRMVREQLDRRMDHKRQRLH-
ttgl-210rfl (601) DGSSDDEETIRRRRRTWKRLRRMVREQLDRRMDHKRQRLH-
AY823990ORF1 ttv 1-7ORF1 ttv tl-16ORF1 ttv tl-270RF1 ttv 1-21ORF1
A 823990ORF1 87 86 95 87
ttv 1-7ORF1 93 87 100
ttv tl-17ORF1 85 93
ttvgtl-270RF1 87
ttvgl-21ORF1
Hydrophobicity plots of the proteins demonstrate 5 areas of hydrophilicity,
which may indicate surface-exposed regions that are potentially antigenic. Two
of these regions are at the amino terminus and at the carboxy terminus, and
are
both arginine-rich and highly conserved. A highly conserved hydrophilic region
between amino acids 190 and 232 was observed and may potentially serve as
antigenic site. The remaining hydrophilic regions between amino acids 295 and
316, and between amino acids 415 and 470 are also be antigenic.
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-38-
Additionally, it has been determined that the putative start codons for
ORF1 and coding region are as follows: ttvgtl-27 nt 517-2435; ttvgl-7 nt 517-
2435; ttvgtl-17 nt 517-2436; ttvgtl-21 nt 517-2439; ttv10 nt 487-2346; and
ttv13 nt 477-2363. The putative start codons for ORF 2 and coding region are
as
follows: ttvgtl -27 nt 428-646; ttvg 1-7 nt 428-643; ttvgtl -17 nt 428-643;
ttvgtl -
21 nt 428-646; ttv10 nt 404-610; and ttv13 nt 394-597.
TTV ORF1 protein expression utilizing recombinant baculovirus
A series of experiments was then undertaken to express the genotype 2
TTV ORF1 protein utilizing insect cells and recombinant baculovirus.
Optimization of protein expression was conducted with three cell lines (SF9,
SF21 and Hi Five), multiple media configurations (ExCell 420, SF900 III SFM,
Express Five SFM), various cell densities (5e5, 1e6, 2e6 and 4e6 cells/ml),
and
various multiplicities of infection (0.005, 0.1, 0.5, 2.0), and the resultant
cultures
were monitored daily over a seven day post infection period.
The processes were monitored for cell density and viability, and infection
was monitored through monitoring of cell size and virus titration. Protein
expression was monitored through SDS-PAGE, Coomassie gel analysis and
Western blotting. To ensure proper control, negative and positive controls
were
maintained throughout all experiments. Although all experiments were able to
confirm expression of the target protein, optimal conditions were found when
utilizing SF9 cells maintained in ExCell 420 media (Sigma, SAFC) with a cell
density of 2x106 cells/ml and an MOI of 0.1, with the process terminated
following
a three day infection. The majority of the recombinant expressed protein can
be
located within the cell pellet although some resides in the resultant
supernatant.
Confirmation of protein expression with Western Blotting (GST-tag)
As the Invitrogen destination vectors (pDEST10) contained a GST protein
N-terminal to the TTV Orfl reading frame, a resultant GST-ORF1 fusion protein
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-39-
of approximately 95kD was generated, which was detected using a commercially
available rabbit anti-GST (CALBIOCHEM) antibody. Of the 95kD fusion protein,
approximately 68kD is considered to be ORF1 and 25kD to be the GST protein.
No commercial antibody was available for standardized detection of TTV ORF1
protein, which necessitated the use of the anti-GST antibody.
Production of rabbit anti-TTV ORF1 antibody
Due to the initial lack of availability of known TTV reagents, efforts were
undertaken to produce anti-TTV ORF1 antibodies. Following the optimized
expression protocol for preparing the TTV ORF1 recombinant protein, the
resultant material was further purified utilizing the commercially available
Baculogold GST purification kit. Purified TTV10 and TTV13 ORF1 protein was
then utilized to hyperimmunize rabbits for the subsequent production of
antibodies against the ORF1 recombinant protein.
In regard of protein detection, Figure 1A sample lanes were as follows
(from right to left)
Samples:
1 SeeBlue Plus 2
2 ORF1 TTV13 d.3 1 e6 cell/ml (GST purified pellet)
3 ORF1 TTV13 d.3 1 e6 cell/ml (GST unbound)
4 ORF1 TTV13 d.3 2e6 cell/ml (GST purified pellet)
5 ORF1 TTV13 d.3 2e6 cell/ml (GST unbound)
6 ORF1 TTV13 d.3 4e6 cell/ml (GST purified pellet)
7 ORF1 TTV13 d.3 4e6 cell/ml (GST unbound)
8 ORF1 TTV13 d.3 4e6 cell/ml (GST purified supe)
9 ORF1 TTV13 d.3 4e6 cell/ml (GST unbound)
10 ORF1 TTV13 d.3 4e6 cell/ml untreated supe.)
11 ORF1 TTV13 d.3 4e6 cell/ml untreated cell pellet
12 SF9 Negative Control d.3 pellet 1e6 cell/ml
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-40-
Lanes 2, 4 and 6 demonstrate the purified 95kD TTV13 ORF1 fusion
protein which was later utilized for the rabbit immunization, see Figure 1A.
Detection of Native TTV ORF1 utilizing the Rabbit anti-ORF1 protein
Additional expression experiments were conducted with the native TTV
ORF1 recombinant baculovirus. This recombinant baculovirus was constructed
without a 6xHis or GST fusion tag and hence requires a specific anti-TTV ORF1
antibody. Consequently, post expression Western blot analysis was conducted
utilizing the rabbit anti-TTV ORF1 antibody to confirm expression of the
native
protein, and to confirm the reagent reactivity. Western blot analysis
demonstrated a faint reaction at approximately 69kD, which is approximately
the
predicted size of TTV ORF1 as well as reaction to an additional band at
approximately 49kD (see Figure 1 B). The 49kD protein band is unknown. The
faint banding at 69kD is assumed to be a function of either low protein
expression in the native TTV ORF1 construct or poor antibody yield from the
rabbit immunization. It should be noted that no purification of the antigen or
antibody was conducted in this particular analysis. Lane 5 (see the arrows in
Figure 1 B) demonstrates a unique reaction to a -69kD and 49kD protein in the
native TTV ORF1 expression utilizing anti-TTV ORF1 rabbit polyclonal antibody.
Accordingly, there was demonstrated binding of antibody to capsid protein
as antigen, herein the antigen provided only TTV sequence and was not tagged.
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-41-
Figure 6B sample lanes were as follows (from right to left)
Samples:
1 SeeBlue Plus 2
2 g1TTV standard 1:50 diluted
3 g2TTV13 ORF1 Native 2DPI cell/supe 0.005 MOI
4 g2TTV13 ORF1 Native 3DPI cell/supe 0.005 MOI
g2TTV13 ORF1 Native 4DPI cell/supe 0.005 MOI
6 g2TTV13 ORF1 Native 2DPI cell/supe 0.1 MOI
7 g2TTV13 ORF1 Native 3DPI cell/supe 0.1 MOI
8 g2TTV13 ORF1 Native 4DPI cell/supe 0.1 MOI
9 g2TTV13 ORF1 Native 2DPI cell/supe 2.0 MOI
g2TTV13 ORF1 Native 3DPI cell/supe 2.0 MOI
11 g2TTV13 ORF1 Native 4DPI cell/supe 2.0 MOI
12 SF9 Neg. Control 4DPP cell/supe
5 Example 2 Backpassaging
A liver was collected aseptically from a caesarean-derived, colostrum
deprived (CDCD) pig. The liver tissue was tested for g1 and g2 TTV in unique
qPCR assays and confirmed to be positive for only g1TTV. A 10% (wt/vol) liver
homogenate was then prepared in media containing antibiotics and antimycotics.
10 Finally, the homogenate was clarified by centrifugation, designated as
g1TTVp0
and frozen at -70C. The resulting g1TTV homogenate was tested to be free of
extraneous viruses, bacteria and mycoplasma via routine testing. Following
satisfactory testing, two milliliters of freshly thawed g1TTVp0 was IP
inoculated
into each of six 11-day old gnotobiotic piglets. At approximately 12 days post-
inoculation the pigs were euthanized and the bone marrow, spleen and livers
were aseptically collected. Each of the resulting livers were confirmed by
qPCR
to be rich in g1TTV and negative for g2TTV. Liver homogenates were then
prepared from each of the resulting livers as aforementioned, labeled and
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-42-
aliquoted as g1TTVp1 and placed at -70C. A further second passage (g1TTVp2)
was created from g1TTVp1.
Example 3 Evaluation of the Efficacy of Three Torque Teno Virus (TTV)
Vaccines in Young Pigs
The present study was conducted to evaluate the efficacy of three TTV
vaccine candidates administered at -7 days of age, and again at weaning (-21
days of age) followed by a challenge at -5 weeks of age.
This study provided a preliminary immunogenicity evaluation in pigs
injected intramuscularly with formulations for TTV. As previously mentioned,
TTV is a small, non-enveloped virus with a single-stranded circular DNA genome
of negative polarity. The genome includes an untranslated region and at least
three major overlapping open reading frames. Porcine TTV is ubiquitous and
PCR-detection of the virus in serum samples collected from various
geographical
regions shows prevalence in pigs ranging from 33 to 100%. McKeown et al., Vet.
Microbiol. (2004) 104:113-117. Krakowka et al., AJVR (2008) 69: 1623-1629,
reported that gl-TTV inoculated pigs had no clinical signs but developed
interstitial pneumonia, transient thymic atrophy, membranous
glomerulonephropathy and modest lymphocytic to histiocytic infiltrates in the
liver
after inoculation. The present study provided a comparison of three different
formulations of TTV vaccines, and evaluated if any of these prototype
formulations can be numerically or statistically differentiated when compared
to
challenge control groups.
Materials and Methods
Animals: Six clinically healthy, crossbred pregnant, PRRSV and M hyo
seronegative females without a history of disease caused by PRRSV or M hyo
(or vaccination against the same organisms) were sourced from Lincoln Trail /
Puregenic Pork, Alton, IL, and transported to the Pfizer Animal Health
Research
Farm in Richland, MI at approximately 3 weeks pre-partum. If necessary, sows
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-43-
were induced to farrow within a 2 or 3 day period using injectable
prostaglandin
(Lutalyse ). Normal piglets from these sows were allotted to study according
to
the allotment design. Pigs were randomized to treatment by litter and each
litter
had at least one piglet assigned to each treatment.
Housing: During the vaccination phase, pigs were housed with their mother with
no cross-fostering, in BL-2 isolation facilities. Pigs remained housed by
litter until
the time of 2nd vaccination. Post-second vaccination pigs were moved to a
further facility and housed in two rooms (one room contains NTX (non-
vaccinated
and non-challenge controls) animals, the second room vaccinates), and each
room contains 4 or 8 pigs per pen.
Feed: Following farrowing, sows were fed a lactating sow diet as appropriate.
Piglets accessed creep feed and milk replacer prior to weaning. Once weaned,
piglets were feed an age-appropriate diet offering free choice. Water was
available to all animals ad libitum.
Allotment/Randomization: Pigs were randomized to treatment by litter. Each
litter had at least one piglet assigned to each treatment.
Study Design
TX Inoculum Dose/Route Vacc. Days Challenge/Route # of
Pigs
NTX* NA NA NA NA -10
Chromos g1TTV
T01 ORF1 recombinant IM -10
protein
Baculovirus g2TTV -7 days of
T02 ORF1 recombinant IM age and at g1TTV passl/IP -10
protein weaning
Inactivated challenge (Day 21)
T03 virus g1TTVp1. IM -10
T04 Mock IM -10
* Minimum of 1 NTX pig from each litter
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-44-
Masking: Vaccine was masked using a numeric code prior to vaccination. The
investigator, vaccine administrator and study personnel were masked to
treatment and did not have access to the masking code unless treatment
information was required for the welfare of an animal.
Investigational Veterinary Products
Table 1. IVP Formulation
T01 True Name: Chromos g1TTV ORF1 recombinant protein
Serial Number: # 117473-185C
Dosage/Formulation: 2 mL; formulated to contain equal volumes of g1TTV
ORF1 recombinant protein and sterile 5% Amphigen
diluent.
T02 True Name: Baculovirus g2TTV ORF1 recombinant protein killed
subunit vaccine
Serial Number: # 117473-185B
Dosage/Formulation: 2 mL; formulated to contain equal volumes of g2TTV
ORF1 recombinant protein and sterile 5% Amphigen
diluent.
T03 True Name: Torque Teno Vaccine, g1TTVp1 Killed Virus
Serial Number: # 117473-185D
Dosage/Formulation: 2 mL; formulated to contain equal volumes of g1TTVp1
KV antigen and sterile 5% Amphigen diluent.
T04 True Name: Mock (Placebo)
Serial Number: # 117473-185A
Dosage/Formulation: 2 mL; formulated to contain equal volumes of
Phosphate buffered saline and sterile 5% Amphigen
diluent.
Challenge Material Preparation: g1TTV pass 1 was derived from liver
homogenate tested positive (7.6x10e8 to 1.6x10e9 DNA copies/2mL) for g1TTV
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-45-
and negative for g2TTV by qPCR. An appropriate number of bottles were
removed from the freezer and thawed shortly before challenge. An aliquot was
then removed from one of the bottles, and held for retitration at a later
time.
Challenge stock was transported on ice to the research facility and maintained
on
ice during the challenge procedure. A challenge dose equals 2.0mL of stock
solution (2.0 mL intraperitoneal). The dose was delivered to each pig is
therefore
expected to be 7.6x10e8 to 1.6x10e9 DNA copies/2mL. Following challenge, an
aliquot of challenge stock was kept for titration to confirm challenge dose.
General Health Observations: Animals were observed daily by a qualified
individual and general health observations were recorded.
Body Weights: All pigs were weighed Day 0, the day of challenge (Day 28) and
at necropsy. All weights were recorded.
Vaccination: At approximately 7 days of age (Day 0), -10 randomly allotted
pigs
per treatment group (Groups T01 thru T04) were vaccinated as described in
Table 1. Pigs were injected in the right neck with a single dose syringe (2.0
mL
intramuscular (IM) dose) of IVP, or a 2 mL IM dose of control according to
allotment. A second dose of the same IVP or control was administered in the
left
neck at the time of weaning (-21 days of age).
Blood Sampling: Prior to Day 0, Day 14 (prior to vaccination) and Day 28 prior
to
challenge (as well as Day 31, 34, 37, and 40), a blood sample was collected,
using 5 mL or 9 mL serum separator tubes (dependant on body weight), from all
pigs for g1TTV status (qPCR-Pfizer-VMRD Laboratory Sciences). Serum
samples were aliquoted by site personnel to at least three separate tubes and
were stored at -80 C.
Table 2: g1TTV qPCR analysis to be performed on sera by time point
Study Day DO D14 D28 D31 D34 D37 D40
gPCR g1TTV X X X X X X X
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-46-
Challenge: At -5 weeks of age, piglets were inoculated with a 2.0 mL (IP or
IN)
dose of a TTV isolate according to allotment. Challenge material was shipped
to
the facility identified by a treatment code for masking purposes.
Rectal Temperatures post challenge were recorded once per day on Day 28 prior
to challenge as well as Day 31, 34, 37, and 40.
Necropsies: On Day 40 all animals were euthanized and necropsied. Upon
necropsy, lung lesions were scored using the following methods: 1) a numeric
score (0, 1, 2, 3) and 2) the percentage of consolidation for each lobe (left
cranial, left middle, left caudal, right cranial, right middle, right caudal,
and
accessory) was scored and recorded as percent of lobe observed with lesions.
Liver, kidney, thymus and lymph nodes were also scored. A blood sample was
also taken prior to euthanasia. Tissues were collected as indicated in the
following table:
Sample Type Collection Method Test Location of
Lab
Inguinal,
mesenteric Formalin fixed sample
and bronchial
lymph nodes Borgess
Formalin fixed tissue Hospital
Thymus Formalin fixed sample sections will be examined
for histologic lesions. University:
Formalin fixed and sterile sample
Spleen tissue samples (kidney, Sterile Tissue samples processing
spleen, liver) for Histology
Formalin fixed and sterile will be processed for DNA
tissue samples isolation and quantitative
Liver
(kidney, PCR analysis of gl-TTV Pfizer Animal
spleen, liver) and g2-TTV. Health
Formalin fixed and sterile (qPCR)
Kidney tissue samples (kidney,
spleen, liver)
Formalin Formalin fixed sample
Inflated Lung
In regard of assessment of safety and/or efficacy, no confounding
secondary disease conditions were detected. Animals were vaccinated and
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-47-
challenged according to protocol. In regard of outcome criteria, reduction in
any
or all of the following were used: decreased gross or microscopic lesions;
decreased viremia by qPCR; and decreased incidence of fever, weight loss or
death, two-sided tests.
Method of Analysis
Upon necropsy, lung lesions were scored using the following methods:
1) a numeric score (0=no lesions, 1=mild lesions, 2=moderate lesions, 3=severe
lesions) and 2) the percentage of consolidation for each lobe (left cranial,
left
middle, left caudal, right cranial, right middle, right caudal, and accessory)
was
scored and recorded as percent of lobe observed with lesions.
The percentage of total lung with lesions was transformed and analyzed
with a general linear mixed model with fixed effects, treatment, and random
effect
litter. Linear combinations of the parameter estimates were used in a priori
contrasts after testing for a significant (P<_0.10) treatment effect. The 10%
level
of significance (P<_0.10) was used to assess statistical differences.
qPCR data will be transformed prior to analysis with an appropriate log
transformation. The transformed titers will be analyzed using a general linear
repeated measures mixed model analysis. Pairwise treatment comparisons will
be made at each time point if the treatment or treatment by time point
interaction
effect is significant (P<_0.10). Treatment least squares means, 90% confidence
intervals, the minimum and maximum will be calculated and back-transformed for
each time point. Descriptive statistics, means, standard deviations, and
ranges,
will be calculated for each treatment and day of study, pre-challenge.
Study Results and Discussion
Lung Lesions
Although the overall percent lung lesions observed was low throughout all
treatment groups, significant differences were found. T01 (Chromos expressed
g1TTV ORF1) yielded significantly lower lung lesions when compared to both the
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-48-
T02 (Baculovirus expressed g2TTV ORF1) and T04 (Challenge controls). Since
the challenge virus was comprised of infectious g1TTV, it may not be
surprising
that the genotype 2 ORF1 from Baculovirus did not provide very substantially
lower lung lesions as compared to the challenge controls. It is however
interesting to note that while not substantial, it did offer numerically lower
lung
lesion scores compared to the challenge controls, thereby indicating that some
level of cross protection is possible between different TTV genotypes upon
optimization of dose and adjuvant selection. It was surprising that the
inactivated
challenge virus (T03, g1TTVp1 Killed Virus) did not offer cross-protection
against
the live g1TTV challenge virus as evidenced by the lack of any statistical
difference between T03 and T04. This surprising lack of cross protection
further
enhances the veterinary importance of novel vaccines of the invention, such as
g1TTVORF1 (T01 Chromos).
Back Lower Upper
transform Standard 90% 90%
Is mean error confidence confidence
Number % lung % lung limit limit Range
of with with of of % lung
Treatment animals lesions lesions mean mean with lesions
T01 11 0.9 0.74 0.0 3.2 0 to 7.65
T02 11 1.5 1.07 0.1 4.3 0 to 12.3
T03 11 2.0 1.23 0.3 5.1 0.1 to 8.6
T04 11 2.0 1.25 0.3 5.2 0.18to7.1
Contrast 2-tailed 1)-value (1) significance of 2-tailed 1)-value
T01 vs T02 0.2167 N.S.
T01 vs T02 0.0472
T01 vs. T04 0.0389 *
T02 vs T03 0.5394 N.S.
T02 vs T04 0.4955 N.S.
T02 vs. T04 0.9454 N.S.
(1) P-Values > 0.10 are designated as "N.S." (Not Significant) and P-Values <
or = 0.10
are designated as "*" (Signficant).
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-49-
g1TTV gPCR
Analysis of the TTV qPCR viremia data (Figure 7) reveals that T01
(Chromos g1TTV ORF1) has numerically lower TTV qPCR values as compared
to T04 (Challenge controls). There exists a decrease in viremia magnitude and
duration, which along with a reduction in lung lesions are indicators of
efficacy.
In addition, T02 (Baculovirus g2TTV ORF1) demonstrates a numerical reduction
in viremia magnitude and duration compared to T04 (Challenge controls) but for
a shorter period of time. This combined with the numerically lower lung
lesions
indicates that some genotypic cross protection (g2TTV ORF1 vaccine vs g1TTV
challenge virus) was observed. One can suggest that with an optimized dose
and adjuvant that broad genotypic cross protection can be realized. It is also
interesting to note that (T03) g1TTVp1 KV offered no reduction in TTV qPCR
viremia when compared to the challenge controls. This observation in
conjunction with the lung lesion data further illustrate the novel finding
that the
recombinantly expressed g1TTV ORF1 (T01) provides efficacy as a vaccine.
Example 4 Codon optimization and recombinant expression g1TTV ORF1 as a
full length protein with a 6His tag, and detection thereof by an antibody.
The TTVg1 nucleotide sequence was submitted to GenScript (Piscataway,
New Jersey, USA) for codon optimization and gene synthesis for both E. coli
and
Saccharomyces cerevisiae. In both cases, the codon optimized gene was cloned
into the GenScript pUC57 vector, as product. The GenScript OptimumGeneTM
codon optimization analysis involves analysis of numerous parameters including
codon usage bias,GC content, CpG dinucleotide content, mRNA secondary
structure, identification of possible cryptic splicing sites, presence of
premature
polyA sites, internal chi sites and ribosomal binding sites, negative CpG
islands,
RNA instability motifs (ARE), inhibition sites (INS), repeat sequences of
various
kinds (including direct, reverse and dyad), and also restriction sites that
may
interfere with cloning. Translational performance may be additionally improved
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-50-
via translational initiation Kozak sequences, Shine-Dalgarno sequences, and to
increase efficiency of translational termination via stop codons.
SEQ ID NO: NOS 18-20 provide TTV capsid gene that were codon
optimized for both Escherichia coli (NOS: 18-19) and Saccharomyces cerevisiae
(NO: 20). The sequences for E. coli are very similar, however, to clone the
gene
into the commercial pET101/D-TOPO expression vector (Invitrogen) to create
76057-4 (SEQ ID NO:19), additional CA nucleotides had to be added at the N-
terminus. The pET101/D-TOPO expression vector also has a C-terminal V5 tag
and 6X-His for purification, although the sequences for 76057-3 (SEQ ID NO:18)
and 76057-4 are otherwise identical. The expressed codon-optimized TTVg1
protein is approximately 68 kD in size, relative to the 63kD protein, due to
the
addition of a 10 amino acid protective peptide at the amino terminus, and 32
amino acids corresponding to the V5 epitope and a 6X His tag at the carboxy
terminus (Figure 2).
The sequence for 76057-5 (SEQ ID NO: 20) has been codon optimized for
S. cerevisiae, and it thus differs slightly from the E. coli sequences. In
addition,
this sequence lacks a 10 amino acid protective peptide at the N-terminus
(which
was added to the E. coli sequence), and it also has flanking restriction
endonuclease sites, Notl at the N-terminus and Aatll at the C-terminus, for
subcloning of the gene into yeast vectors.
Aditionally, it should be noted that the protective peptide of ten amino
acids was added to N-terminus of the TTVg1 sequence for expression in E. coli.
since this has been shown to increase protein stablility when fused to the
amino
terminus. Restriction sites have been engineered such that the peptide can be
removed for evaluation of the full length protein. Expression of the codon
optimized TTVg1 was evaluated in the pET101/D-TOPO vector with and without
the protective peptide N-terminal fusion. The TTVg1 sequence codon optimized
for S. cerevisiae was also subcloned into a pESC-Trp vector with the potential
for
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-51-
producing suface-expressed protein in yeast that can be used to elicit an
antibody response in vivo.
Example 5. TTV Peptide Coniuaation and Antibody Production (Polyclonal and
Monoclonal)
Rabbit polyclonal antibodies were raised against Baculovirus expressed
g2 TTV GST-ORF1 protein prepared in Example 2. Two rabbits were
hyperimmunized, but only one rabbit responded. The rabbit antiserum cross-
reacts to various preparations of g1 TTV whole virus that was propagated in
pigs
and also reacts against the immunizing antigen, Baculovirus expressed g2TTV
ORF1. The rabbit antibody did not, however, respond to the E.coli expressed
g2TTV ORF1 that had the 100 A.A. N-terminal arginine-rich region removed from
the amino terminus as described in Example 2. This may suggest that a major
antigenic epitope may be in the 100 amino acid region that was missing in the
truncated g2 TTV ORF1, and that there is homology between g1 and g2 TTV in
this region.
Monoclonal antibodies can be generated against full-length g1 TTV ORF1,
or other g1 TTV antigens. Other potential immunizing antigens include g1 TTV
whole virus, g2 TTV GST-ORF1 (Baculo), g1 TTV GST-truncated ORF1 (E. coli),
and g2 TTV GST-truncated ORF1 (E. coli). A peptide library can be generated to
identify linear epitopes that are antigenic. For example, 18mer peptides, with
a
1 OAA overlap, can be utilized to cover the TTV genome. The peptides can then
be utilized in Western blots or ELISA's to determine their overall reactivity
to the
g1TTV ORF1 or g2TTV ORF1 monoclonal and/or polyclonal antibodies so that
immunogenic domains can further be identified.
Rabbit polyclonal antibodies may also be raised against three g1 TTV
ORF1 peptides cross-linked to KLH, and subsequently screened using peptide-
ovalbumin conjugates. The peptide-KLH conjugates can also be used to
produce monoclonal antibodies. In this respect, in one embodiment, multiple g1
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-52-
TTV ORF1 peptides copies may be conjugated together, including from different
strains.
In particular examples, once peptides were generated (CPC Scientific),
they were then conjugated to KLH or ovalbumin (by the Proteos Co). The KLH-
conjugated peptides were used for immunization of rabbits, while the Ovalbumin
conjugated peptides are used for screening the serum (i.e., to detect
antibodies
to the peptides and not the carrier protein).
Example 6 Peptide Sequences for Polyclonal Antibody Generation
The following peptide sequences were chosen from TTVg1 (numbering
based on AY823990) for polyclonal antibody generation, and represent SEQ ID
NOS; 22-24 respectively.
1. [L167C]TTV(167-185)-NH2: CKDQDYWFWWDTDFKELYA-NH2 (19 aa, pl 4)
2. TTV(459-479): DFGHHSRFGPFCVKNEPLEFQ (21 aa, pl 6.9)
3. [Cys612]-TTV( 612-637):
CTWKRLRRMVREQLDRRMDHKRQRLH (26 aa, pl 13)
Each of the three peptides has a single cysteine residue present in the
sequence to enable selective peptide coupling to a carrier protein. In
[L167C]TTV(167-185)-NH2 and [Cys612]-TTV( 612-637), an extra cysteine
residue was added at the N-terminus, while in TTV(459-479) there is a native
cys
present at position 470. Additionally, [L167C]TTV(167-185)-NH2 has an amidated
C-terminus to yield a less acidic peptide. The peptides were selected based on
sequence identity for different TTV isolates. Additionally, the C-terminal
fragment
[Cys612]-TTV( 612-637) appears to be surface exposed. The peptides were
custom made by solid phase peptide synthesis at CPC Scientific and obtained
with >95% purity.
A further and highly preferred peptide is constructed by using the peptide
sequence corresponding to residues 601-620 of SEQ ID NO:9 (20AA, pl 13)
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-53-
except that a cysteine residue is used at the N-terminus in replacement for
Asn
601. This peptide is also likely surface exposed in the native protein.
Example 7 TTV g1 ORF1 protein expression using the Chromos system
The Chromos ACE system is a protein expression platform that consists of
three main components. The first component is a neutral, functional mammalian
artificial chromosome called the Platform ACE, which resides in the genetic
material of a modified Chinese Hamster Ovary (CHO) cell line. The second
component is the ACE targeting vector, which is a plasmid used for loading
target genes onto the Platform ACE. The third element is a site-specific,
unidirectional integrase, which catalyzes the direct and specific loading of
the
target gene onto the Platform ACE. Additional information concerning the ACE
System can be found of the website of Chromos Molecular Systems, Inc. of
Canada, or by contacting the company directly at 604-415-7100 where the
technology is available for license.
The Chromos ACE system has a number of significant advantages over
traditional protein production platforms. The first of these is speed. The
Chromos ACE system allows for the rapid, efficient and reproducible insertion
of
selected genes. The second advantage is expression. High level constituitive
protein expression is achieved over time. A third advantage is stability. The
Chromos ACE system allows selective and controlled protein expression.
Briefly,
restriction sites were added to both ends of the TTV7 ORF1 g1DNA using PCR.
Additionally, the sequence for yeast invertase was added to the 5' end of a
separate PCR preparation. The amplified sequences were then treated with
restriction enzymes and sub-cloned into the plasmid pCTV927. The DNA
sequence was verified by ACGT Inc. CHk2 (Chinese Hamster Ovary) cells were
then transfected with the plasmids using Lipofectamine 2000 (Invitrogen), and
selective pressure was added using hygromycin B. Ten single-cell clones were
analyzed for TTV protein production using SDS PAGE and Western Blotting.
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-54-
More specifically, the ACE Targeting Vector pCTV-TTV7ORF1+YI was
generated as follows (see Figure 3). The gene TTV7ORF1 was obtained as a
PCR product. A primer was designed to contain the yeast invertase secretion
signal and the restriction site EcoRV at the 5' end of the gene. A second
primer
was designed to contain the restriction site Kpnl at the 3' end of the gene.
These
sequences were added to the gene TTV7ORF1 using the polymerase chain
reaction. The modified gene was then subcloned into the ACE Targeting Vector
ATVCHS4Hyg, which contained a hygromycin resistance marker suitable for
downstream antibiotic selection. The new plasmid was named pCTV-
TTV7ORF 1 +YI .
The plasmids pCTV-TTV7ORF1+YI and pSIO343, which coded for
TTV7ORF1/yeast invertase and the unidirectional lambda integrase,
respectively,
were transfected into the Chk2 cell line, which contained the Platform ACE.
The
transfected cells were named Chk2-TTV7ORF1+YI. These cells were seeded in
96-well plates and monitored for the formation of single-cell clones. Media
containing Hygromycin was added to each 96-well plate to select for cell
clones
that contained the ACE targeting vector. Once single-cell clones were
identified,
twelve of them were expanded into 24-well plates, and then to 6-well plates.
Finally, the clones were expanded into suspension cell culture. Culture Chk2-
TTV7ORF1+Yl #75 was used to generate cell-free supernatant for subsequent
experimental vaccine preparation.
Figure 7 demonstrates that Chromos-expressed g1TTV ORF1 significantly
reduced lung lesions compared to the challenge controls, and reduced the
numerical magitude and duration of g1TTV viremia, again compared to the
challenge controls. Vaccination was at Day 0 and 14, with challenge at Day 28.
The geometric mean of detected g1TTV copies was reported exponentially, i.e.
1.00 E+00 is 1, 4.25E+00 is 4.25, and 4.42E+01 is 44.2.
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-55-
Example 8 Nuclear Localization Signals
Figures 4 and 5 provides a 7-way amino acid alignment of ORF1 (capsid
proteins) from 5 TTV gt1 viruses of the present invention and two TTV gt2 (or
gt2-like) viruses of the invention. There are, of course, many gaps and
mismatches because the gt1 capsids are only about 22.3 to 23.2% identical to
the gt2 capsids. The five gt1 capsids are 85.6 to 99.7% identical, however,
among themselves. The two gt2 capsids (TTV10 and TTV13) are similarly
66.8% identical.
Two known types of NLS signals (Pat7 and Pat4, see US Patent
7,544,362, for example) were identified by inspection. In Figure 5, the The
NLS
signals are underlined. Note the all seven capsids contain multiple NLS of
both
pat7 and pat4 type. Some are conserved between genotypes, some within a
genotype, and some are not conserved. Most are near the N-terminus, where
they tend to form overlapping poly-NLS regions. Numerous of these arginine -
rich motifs are substantially immunogenic in mammals, and peptides containing
them are useful in the generation of anti-TTV antibodies.
Example 9 Clone Fragments for infectious clone construction
The following provides a basis for the construction from overlapping
clones of TTV genotype 1 strain ttvgtl-178 (see SEQ ID NO:7) for which the
amino acid sequence is shown as SEQ ID NO:9.
In summary, two TTV fragments (1900bp and 2200bp), which together
span the entire TTV circular genome, were separately cloned into separate pCR
2.1 TA (Invitrogen) cloning vectors. The clone fragments were as follows:
Clone
1: 680s to 2608a=-1900bp, and Clone 2: 1340s to 764a= -2200bp.
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-56-
In order to accomplish this, PCR primers were designed using the
consensus sequence that was generated from strains of the present invention
(ttvgtl-27, -7, -17 and -21), and also from published sequences (AY823990(g1)
and AB076001-(Sd-TTV31)). Primer pairs that correspond to the sequence at
680s and 2608a or 1340s and 764a were used to amplify PCR products from
DNA that was extracted from liver homogenate samples of pigs infected with TTV
challenge strain. These PCR fragments were cloned into Invitrogen's pCR2.1-
TOPO TA vector using directions that were supplied with the kit. Clones were
subsequently used to generate DNA sequences across the entire 2880 base
genome and the sequence was found to be 86% homologous to published
sequences GQ120664.1 and AY823990.1.
The fully correct sequences will now be combined for construction of a full
length infectious clone.
Example 10 Infectious clone for g1TTV
Cloning of g1TTV dsDNA fragments. g1TTV is a single-stranded DNA (ssDNA)
virus. Fragments of g1TTV are converted to double-stranded DNA (dsDNA)
using polymerase chain reaction (PCR). The dsDNA fragments of g1TTV are
then cloned into pUC-based plasmid cloning vectors and transformed into E.
coli.
The fragments of g1TTV are less than 1 full-length dsDNA equivalent of the
g1TTV genome.
Amplification of g1TTV dsDNA concatemers. Concatemers of full-length g1TTV
dsDNA genome equivalents are generated using X29 polymerase amplification
kits (e.g., illustra TempliPhi). Full-length g1TTV dsDNA fragments are
generated
by digestion of the concatemers at appropriate restriction endonucleases (RE)
sites. These full-length g1TTV dsDNA fragments can be cloned into plasmid
vectors. Alternatively, the concatemers or the uncloned fragments (resulting
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-57-
from RE digestion) can be used without immediate cloning in subsequent
molecular biology constructions (see below).
Tandem duplications of the g1TTV genome. Plasmid constructs encoding
tandem duplications of the g1TTV genome are next generated. The tandem
duplications in the constructs are approximately greater than 1.2 copies of
full-
length dsDNA equivalents of the the g1TTV genome. The tandem duplications in
plasmids are generated using (1) subcloning employing appropriate RE sites,
(2)
PCR assembly of tandem duplications, or (3) other molecular biology methods.
The templates for the generation of the tandem duplications are the g1TTV
dsDNA fragments and/or the full-length g1TTV dsDNA clones (yielded by X29
polymerase amplification).
In vivo recombination and generation of g1TTV virus. The tandem duplication
plasmid constructs are not identical to the g1TTV virus. The tandem
duplication
constructs are dsDNA while the virus is ssDNA, the constructs encode >1.2 full-
length dsDNA equivalents of the g1TTV genome while the virus has only one full-
length equivalent, the construct contains interrupting plasmid sequences while
the virus has only viral sequences. To generate the bona fide g1TTV virus, the
tandem duplication plasmid constructs are introduced into pigs (by
inoculation,
injection, electroporation, or other methods of introduction) or introduced
into
tissue culture cells (by transfection, electroporation, or other methods of
introduction) where the plasmid construct recombines at homologous sequences
to regenerate a unit-length dsDNA equivalent of the g1TTV genome. The dsDNA
equivalent of the g1TTV genome is a presumed replicative intermediate of the
g1TTV viral life cycle. The presence of this presumed dsDNA replicative
intermediate will lead to the production of the bona fide ssDNA g1TTV.
Enabling in vivo generation of g1TTV virus by co-transfection of g1TTV ORF-
expressing constructs. It is expected that a circular dsDNA g1TTV genome
would be capable of yielding virus production. In the unexpected event that
the
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-58-
dsDNA form of g1TTV is not replication-competent, the immediate expression of
a g1TTV ORF may be required for the initiation of g1TTV replication from the
dsDNA replicative intermediate. Plasmid constructs directing in vivo
transcription
of g1TTV ORFs can be made, such as the fusion of transcriptional promoters
(e.g., CMV) to g1TTV ORFs. Alternatively, plasmid constructs directing the in
vitro generation of g1TTV ORF transcripts can be made, such as the fusion of
transcriptional promoters (e.g., T7) to g1TTV ORFs followed by use of in vitro
transcription kits. Either g1TTV ORF-expressing plasmids or g1TTV ORF-
expressing RNA transcripts can be co-injected into pigs or co-transfected into
cells along with the tandem duplication plasmid constructs to yield g1TTV
virus.
Detection of g1TTV virus production. To date, whole g1TTV virus cannot be
propagated in tissue culture cells. The generation of g1TTV virus is detected
by
immune reagents (e.g., a-g1TTV antibody) or by molecular methods (e.g.,
qPCR).
Example 11 Provision of TTV1-178 Clone in pCR2.1 Vector
Total DNA was isolated (DNEasy Blood and Tissue Kit, Qiagen, Valencia,
CA ) from a frozen liver homogenate sample (200 microliters) derived from a
prior TTV challenge study. The DNA was then PCR-amplified using forward and
reverse primers selected to overlap at the unique EcoRl site of the swine TTV
1-
178 genome. The forward primer: for TTVg1-178 was selected as positions 1399
to1428 (ACGG... CCAA) from SEQ ID NO:7, and the reverse primer:was
selected to correspond to base positions 1443 (5')-to 1416 (3') ATAT... TTGT
(opposite strand) from SEQ ID NO:7. PCR conditions were as follows: 1 cycle of
denaturation at 94 C for 1 minute; 35 cycles of 94 C, 30 seconds; 55 C, 30
sec;
72 C, 3 minutes; followed by a final 10 minute extension at 72 C. The
resultant
-2.8kb fragment was cloned into pCR2.1 vector using a TOPO TA cloning kit.
(Invitrogen, Carlsbad, CA). Upon sequence verification, this plasmid (named
CA 02775277 2012-03-23
WO 2011/046634 PCT/US2010/031373
-59-
pCR2.1+TTV 178) was found to contain the entire TTV 1-178 genome
sequence.
pCR2.1+TTV 178 vector was then linearized with EcoRl in order to
release the full length TTV genome, which was then transfected into human
embryonic kidney (293), baby hamster kidney (BHK-21), swine testicular (ST)
and porcine kidney (PK) cells lines using Lipofectin (Invitrogen, Carlsbad,
CA).
The transfection was allowed to proceed for a total of 5 days at which time
the cells were fixed, and then used for IFA staining to determine if the TTV
DNA
provided expression of ORF 1 protein. IFA staining was accomplished with
rabbit polyclonal sera that was raised against a peptide corresponding to a C-
terminal region of the capsid protein (residues 601-620 in SEQ ID NO:9),
except
that the N terminal residue thereof (Asn 601) was replaced with a cysteine
residue. The results indicate that the TTV-transfected DNA successfully
expressed the ORF-1 protein in at least 293, BHK-21, ST and PK cells lines.