Note: Descriptions are shown in the official language in which they were submitted.
CA 02775278 2012-04-23
ARRESTIN BIOSEN R
TECHNICAL FIELD
[0001] Described herein is a novel biosensor and method suitable for
monitoring activation
of receptors and signaling molecules. More specifically, the use of a modified
arrestin as a
biosensor to monitor the activation state of receptors is described, such as G
protein-coupled
receptors (GPCR). Advantageously, the biosensor and method allow for a highly
sensitive
and quantitative assay that can be used in large-scale screening analyses.
BACKGROUND
[0002] The largest class of cell surface receptors in mammalian genomes is the
superfamily
of G protein-coupled receptors (GPCRs). GPCRs are proteins that span the
membrane of a
cell and relay the information provided by numerous Iigands, e.g. hormones and
neurotransmitters, into intracellular signalling pathways. GPCRs are thus the
targets of many
clinically important drugs, with approximately half of all current
prescription drugs acting
through GPCRs (Drews J (1996) Genomic sciences and the medicine of tomorrow.
Nat
Biotechnol 14: 1516-1518). Examples of GPCRs are many and include beta-2
adrenergic
receptor (a2-AR), Frizzled 4 (Fz4), V2-vasopressin receptor (V2R), V1a
vasopressin receptor
(VIaR), 6-opioid receptor (6-OR), platelet-activating factor receptor (PAFR),
CC chemokine
receptor type 5 (CCR5), and angiotensin receptor type 1a (AT1aR).
[0003] GRCRs relay the information encoded by the ligand (e.g. hormones and
neurotransmitters) through the activation of G proteins and intracellular
effector molecules. G
proteins are heterotrimeric proteins, consisting of an alpha, a beta, and a
gamma subunit.
The three G-subunits are non-covalently bound together and the G protein as a
whole binds
to the inside surface of the cell membrane and associates with the GPCR.
Starting in such
conformation, the G-alpha subunit is complexed to GDP (guanosine diphosphate).
When a
ligand binds to a domain of the GPCR accessible from the outside of the cell
membrane, a
conformational change in the GPCR occurs, which in turn prompts the exchange
of the GDP
for a molecule of guanosine triphosphoate (GTP) on the G-alpha subunit, and
activates the
G-protein. The G-protein's a subunit, together with the bound GTP, can then
dissociate from
the R and y subunits to further affect intracellular signaling proteins or
target functional
proteins directly, depending on the a subunit type (e.g. Gas, Gal/o, Gaq/1 1,
Gal 2/13).
CA 02775278 2012-04-23
2
[0004] In order to turn off this response by GPCRs to stimulus, or adapt to a
persistent
stimulus, the activated GPCRs are inactivated. This inactivation may be
achieved, in part, by
the binding of a soluble protein, R-arrestin (Q-arr), which uncouples the
receptor from the
downstream G protein after the receptor is phosphorylated by a G protein-
coupled receptor
kinase (GRK). More specifically, through their binding to agonist-occupied,
GRK-
phosphorylated receptors, 13-arrs prevent further coupling to G proteins and
promote GPCR
endocytosis, thus leading to decreased signalling efficacy.
[0005] Despite our growing understanding of the diversity in GPCR signaling
mechanisms,
drug efficacy is often defined only in terms of the regulation of the
classical G protein
signaling. Within this framework, agonists are defined as drugs that stabilize
an active
receptor conformation that induces G protein activation, whereas inverse
agonists favor an
inactive receptor state that reduces spontaneous G protein signaling. The
question arises as
to whether this paradigm may be transferred to drug effects generated through
the formation
of metastable complexes involving scaffolding proteins such as 3-arr. Because
all studies
describing R-arr-mediated MAPK signalling have concentrated on agonist drugs,
little is
known of how ligands that are commonly classified as inverse agonists may
regulate the
scaffold assembly that is crucial for such signalling.
[0006] In one study (Azzi et al, 2003), this question was addressed by
assessing whether
{32-adrenergic receptor ((32AR) and V2 vasopressin receptor (V2R) ligands with
proven
inverse efficacy on adenylyl cyclase (AC) activity could also regulate MAPK
activation via
receptor-mediated scaffold formation. It was found that, despite being inverse
agonists in the
AC pathway, the R2AR (IC1118551 and propranolol) and V2R (SR121463A) induced
the
recruitment of [3-arr leading to the activation of the ERK cascade. Such
observations indicate
that the same drug acting on a unique receptor can have opposite efficacies
depending on
the signaling pathway considered.
[0007] The above study relied on the use of a bimolecular bioluminescence
resonance
energy transfer (BRET) assay. It was used to assess f -arrestin recruitment to
(32AR or V2R.
Fusion proteins consisting of GFP10 variant (GFP) covalently attached to the
carboxyl tail of
the receptor of interest ([32AR-GFP; V2R-GFP) were co-expressed with [3-
arrestin 2 fused at
its carboxyl terminus to Rluc (R-arrestin-Rluc). After incubation of the
transfected cells with
different ligands, coelenterazine 400a (Perkin-Elmer, Wellesley, MA, USA) was
added and
CA 02775278 2012-04-23
3
readings were collected using a modified top-count apparatus (BRETCount,
Packard) that
allows the sequential integration of the signals detected at 370-450 nm and
500-530 nm. The
BRET signal was determined by calculating the ratio of the light emitted by
the Receptor-
GFP (500-530 nm) over the light emitted by the 0-arrestin2-Rluc (370-450 nm).
The values
were corrected by subtracting the background signal detected when the 0-
arrestin2-Rluc
construct was expressed alone.
[0008] While the results elicited from the above study were instructive, a
necessary feature
involved the construction of fusion proteins that included the receptors of
interest. Ideally, a
method could be devised in which receptor activation might be observed without
first having
to modify the receptors that are to be studied. Other features of such a
method that would
make it highly desirable for research and development endeavors include the
following: (1) a
high level of sensitivity; (2) an ability to provide quantitative results; (3)
adaptability for use in
large scale screening analyses; (4) an assay that requires the expression of a
single
recombinant construct; and (5) a biosensor based on an intramolecular RET
signal.
[0009] Resonance energy transfer (abbreviated RET, and also referred to as
Forster
resonance energy transfer), is a mechanism describing energy transfer between
two
chromophores, having overlapping emission/absoprtion spectra. When the two
chromophores (the "donor" and the "acceptor"), are within 10-100 A of one
another and their
transition dipoles are appropriately oriented, the donor chromophore is able
to transfer its
excited-state energy to the acceptor chromophore through nonradiative dipole-
dipole
coupling. When both chromophores are fluorescent, the term typically used is
"fluorescence
resonance energy transfer" (abbreviated FRET). In bioluminescence resonance
energy
transfer (BRET), the donor chromophore of the RET pair, rather than being a
fluorophore, is
a bioluminescent molecule, typically luciferase. In the presence of a
substrate,
bioluminescence from the donor excites the acceptor fluorophore through the
same Forster
resonance energy transfer mechanism described above (Xu, Y. et al., PNAS,
96:151-156
(1999)).
[0010] There is a need for a simpler method to measure receptor activity in
living cells. The
present invention seeks to meet this and related needs.
CA 02775278 2012-04-23
4
SUMMARY
[0011] A resonance energy transfer (RET) biosensor comprising an arrestin
tagged with a
first and a second chromophore, wherein said first chromophore is a
fluorophore and said
second chromophore is a fluorophore or a bioluminophore.
[0012] Further, there is described herein a method of identifying candidate
molecules that
bind to a receptor comprising screening candidate molecules for activation of
the biosensor.
Uses of the biosensor for assaying receptor activity are described, as well as
kits for
evaluating receptor binding.
[0013] Other objects, advantages and features of the present invention will
become more
apparent upon reading of the following non-restrictive description of
preferred embodiments
thereof, given by way of example only with reference to the accompanying
figures.
BRIEF DESCRIPTION OF THE FIGURES
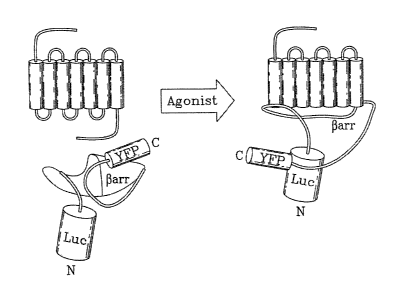
[0014] FIGURE 1: Double-brilliance A-arr. Schematic diagram illustrating how
agonist-
promoted conformational rearrangement of R-arr can be measured as changes in
BRET
using double-brilliance [3-arr. Luc and YFP are represented by cylinders
proportional to their
sizes, but their real orientation is unknown.
[0015] FIGURE 2: Functionality of double-brilliance R-arr. HEK293 (A-C) or COS
(D) cells
were transiently transfected with the indicated plasmids. (A) Cells incubated
or not In the
presence of saturating concentrations of specific agonists (02-AR, 10 mM
isoproterenol
(ISO); V2R, 1 mM arginine vasopressin (AVP)). Localization of Luc-P-arr-YFP
and Myc-
tagged receptors was analysed by confocal fluorescence microscopy. (B) Agonist-
induced
recruitment of R-arr measured using intermolecular BRET2. tl/2=half-time of
maximal R-arr
recruitment. (C) Dose-dependent recruitment of R-arr to the receptors measured
in
intermolecular BRET2 following 2 min stimulation with the agonist.
EC50=concentration of
agonist producing half-maximal 13-arr recruitment. (D) Cells treated or not
for 15 min with the
specific agonists at 37 C and cell-surface receptor levels measured by enzyme-
linked
immunosorbent assay (ELISA). Receptor endocytosis is defined as the loss of
cell-surface
immunoreactivity and is expressed as a percentage of total immunoreactivity
measured
under basal conditions. Expression levels of (3-arr were controlled using
western blot (data
CA 02775278 2012-04-23
not shown). Data are the mean t s.e.m. of at least three independent
experiments. *P<0.05
between treatment and each individual control condition. Mock, non-transfected
cells.
[0016] FIGURE 3: AVP-induced conformational change of R-arr monitored by
intramolecular
BRET1. HEK293 cells were transfected with the indicated plasmids and BRET was
measured at 25 C in the presence of coelenterazine h. (A) Specificity of
agonist-induced G-
arr intramolecular BRET1. (B) Real-time BRET measurements of the agonist-
induced R-arr
conformational change. tl/2=half-time of maximal conformational change of R-
arr. (C) Dose-
dependent agonist-promoted increase of [3-arr intramolecular BRET1. Cells were
stimulated
with increasing concentrations of AVP for 4 min. EC50=concentration of AVP
producing half-
maximal conformational change of R-arr. Data are the ' mean t s.e.m. of at
least three
independent experiments. *P<0.01 between treated and control condition.
[0017] FIGURE 4: Agonist-promoted conformational change of a phosphate
insensitive
Rarrestin mutant. HEK293 cells were transfected with V2R and either Luc-Barr-
YFP or Luc-
(3arr(R169E)-YFP. Cells were stimulated or not for 10 min with 1 pM AVP prior
the addition of
5 pM coelenterazine h (Molecular Probe) and performing the intramolecular
BRET1
measurements using a Multilabel Reader Mithras LB 940 (Berthold Technologies).
The
BRET signal was determined by calculating the ratio of the light emitted by
YFP over the light
emitted by Luc following the addition of coelenterazine h. The values were
corrected by
subtracting the background BRET signals detected when Luc-Parr was expressed
alone.
Inset, AVP-induced BRET increase. Data represent the mean t SEM of three
independent
experiments. * indicates p<0.02 between treatment and each individual control
condition.
[0018] FIGURE 5: Double-brilliance Q-arr monitors the activation of many
GPCRs. HEK293
cells were transfected with Luc-f3-ar -YFP and either pcDNA3.1 or plasmids
encoding the
indicated receptors. (A) Agonist-Induced translocation of Luc-(3-arr-YFP
measured following
treatment with 1 mM of the specific agonists (f32-AR, ISO; V1aR, AVP; 5-OR,
SNC80; PAFR,
PAF; CCR5, hRANTES; AT1aR, angiotensin II). (B) Agonist-induced conformational
change
of Luc-R-arr-YFP measured following 10 min stimulation with the specific
agonists
mentioned in (A). BRET1 was measured using a Multilabel Reader Mithras LB 940
(Berthold
Technologies). The BRET signal was determined by calculating the ratio of the
light emitted
by YFP over the light emitted by Luc following the addition of coelenterazine
h. Data are the
CA 02775278 2012-04-23
6
mean s.e.m. of three independent experiments. *P<0.05 between treatment and
each
individual control condition.
[0019] FIGURE 6: Agonist-promoted conformational change of constitutively
activated
parrestin mutants. HEK293 cells were transfected with V2R and either Luc-parr-
YFP or Luc-
(3arr (3A)-YFP or Luc-Parr (IV)-YFP. Cells were stimulated or not for 10 min
with 1 pM AVP
prior to the addition of 5 pM coelenterazine h and performing the BRET
measurements as
described in the previous figure. Inset, AVP-induced BRET increase. The BRET
signal was
determined by calculating the ratio of the light emitted by YFP over the light
emitted by Luc
following the addition of coelenterazine h. The values were corrected by
subtracting the
background BRET signals detected when Luc-Parr was expressed alone. Data
represent the
mean SEM of two independent experiments. * Indicates p<0.05 between
treatment and
each individual control condition.
[0020] FIGURE 7: Conformational change of parrestin induced by ligands of
different
efficacies. HEK293 cells transiently co-expressing the V2R and Luc-Parr-YFP
were subjected
to real-time BRET measurements in the presence or absence of two different V2R
ligands.
The basal BRET signals were subtracted from each condition to express the data
as ligand-
induced BRET increase. The figure shows the detection of conformational
changes of Luc-
parr-YFP in time, reflected by the increase in BRET signal, as Induced by the
V2R agonist
AVP or the inverse agonist SR121463. No BRET increase was observed when cells
were
incubated in the presence of the carrier alone (non-stimulated). The fact that
the observed
increase in BRET signal induced by SR121463 is significantly lower than that
induced by
AVP treatment can be correlated with the smaller SR121463-mediated recruitment
of
parrestin to the V2R when compared to AVP, as reported previously (Azzi et at,
2003).
[0021] FIGURE 8: parrestin-dependant endocytosis beyond GPCRs. (A) Endocytosis
of the
receptor Frizzled 4 (Fz4) stimulated by Wnt5a is orchestrated by parrestin 2,
in a manner that
is dependent upon the phosphorylation of the adaptor protein Dishevelled 2
(Dvl2) by protein
kinase C (PKC). (B) Endocytosis of the RII and Rill receptor subtypes of TGF-
01 is
orchestrated by parrestin 2, and facilitated by the phosphorylation of Rill by
Ril. (C)
Endocytosis of the IGF1 receptor is orchestrated by parrestin (Modified from
Lefkowitz &
Whalen, 2004.).
CA 02775278 2012-04-23
7
[00221 FIGURE 9: Characterization of BRET2-RArrestin double-brilliance
sensors. (A)
Structure and activation: BRET1 and BRET2-[3Arrestin double brilliance (db)
sensors are
unimolecular with BRET tags in N-and C-terminus of a central RArrestin core.
The linkers
separating the BRET1 and BRET2 tags from RArrestin differ in both length and
composition.
For the BRET1 sensor the structure is: BRET donor (Rluc)-Linkerl- [3Arrestin-
Linker2-BRETI
acceptor (YFP) and for the BRET2 sensors: Structure: BRET2 acceptor (sCFP3A,
mAmetrine or GFP10)-Linker3- RArrestin- Linker4- BRET2 donor (Rlucll). For
BRETI, the
Rluc substrate is coelenterazine H, whereas for BRET2, the Rluc substrate is
deep-blue
coelentrazine. All versions of the (iArrestinl and 2 db are conformational
sensors. However,
following GPCR activation by an agonist (illustrated as a triangle), changes
in RArrestin
conformation lead to a decreased BRET signal for the BRET2 sensors while it
leads to an
increased BRET signal with the BRET1 sensor configuration (see Figures 1-7).
(B) Kinetics
and dose-responses measured in BRET2 with the BRET2-(3ARR1 and 2 db sensors,
in
response to V2R activation by its agonist AVP: at 100nM for the kinetics or at
increasing
concentrations of AVP for dose-response experiments. (C) PARR db sensor to
characterize
ligands of different efficacies. Hek293 cells transiently expressing both
AT1aR and GFP10-
{3arrl-Rlucil db sensor, were stimulated with a full (Angll) or partial
agonists and responses
were evaluated as a BRET2 signal modulation. a) Dose-dependent ligand-promoted
decrease of (3arrestin intramolecular BRET2 signal after a 25 min stimulation.
Data are the
mean +/- S.E.M. of 3 independent experiments. b) Agonist-promoted BRET
changes. Cells
were treated for 25 min with 1 pM Angil or 10pM of the partial agonists. Data
represent mean
+/- S.E.M. of 4 independent experiments. One-way ANOVA followed by Tukey's
multiple
comparison post-hoc test (Angll as reference) was used to assess statistical
significance. *,
p < 0.05, """*, p < 0.001. Angll= Angiotensin 2 octapeptide, SVdF: Angll
analog with
Sar,,Val5,D-Phe8 substitutions at the indicated amino acid positions in the
octapeptide, SII:
Angll analog with Sar,,lle4ille8 substitutions at the indicated amino acid
positions in the
octapeptide, SBpA: Angll analog with Sar,,Bpa8 substitutions at the indicated
amino acid
positions in the octapeptide, SIVI: Angli analog with Sar,,lle8 substitutions
at the indicated
amino acid positions in the octapeptide, DVG: Angli analog with Asp,,Val5,Gly8
substitutions
at the indicated amino acid positions in the octapeptide.
[0023] FIGURE 10: Z'-factor evaluation for both BRETI- and BRET2-[3Arrestin
sensors.
HEK293 cells transiently expressing both V2R and the double-brilliance sensor,
were
CA 02775278 2012-04-23
8
exposed to either 100nM AVP or a control vehicle, for 25-35 min. BRET ratios
are
represented for each individual well of a 96-well plate. Z'-factors were
calculated as
described in (Ji-Hu Zhang et al. 1999 J Biomol Screen, 4; 67). A Z'-factor
between 0.4 and 1
is considered a robust assay.
DETAILED DESCRIPTION
[0024] A resonance energy transfer (RET) biosensor is described herein which
comprises an
arrestin tagged with a first and a second chromophore. The first chromophore
is a
fluorophore and said second chromophore is a fluorophore or a bioluminophore.
For
example, the arrestin may be 0-arrestin-1 (arrestin-2), 0-arrestin-2 (arrestin-
3), arrestin-1 or
arrestin-4.
[0025] The second chromophore may be a bioluminophore and the RET is
bioluminescence
resonance energy transfer (BRET), in certain embodiments. An exemplary
bioluminophore
may be Renilla luciferase or a mutant form of Renilla luciferase. A specific
bioluminophore
could be Riucil (A55T/C124A/M185V).
[0026] The fluorophore may be a fluorescent protein, for example green
fluorescent protein
or a variant thereof. Specifically, the fluorophore could be YFP, mAmetrine,
cyan fluorescent
protein (CFP), or GFP10.
[0027] Specific exemplary biosensors may comprise Luc-R-arr--YFP, YFP-Barr-
Luc, Luc-0-
arr(3A)-YFP, Luc-R-arr(IV)-YFP, Luc-J3arr(R169E)-YFP, Ametrine-hpar1-Rlucil,
Ametrine-
hRarr2-RluciI, CFP-hlarrl-Rlucil, CFP-h[3arr2-Rlucil, GFPI0-h[3arrl-Riucli, or
GFP10-
h(3arr2-Rlucil.
[0028] In certain embodiments, the fluorophore may be positioned at the N-
terminus of the
arrestin and the bioluminophore at the C-terminus of the arrestin. Further, in
such an
embodiment, the fluorophore can be green fluorescent protein or a variant
thereof.
Specifically, the fluorophore might be YFP, mAmetrine, cyan fluorescent
protein (CFP), or
GFP10. The bioluminophore, in certain embodiments, can be Renilla luciferase
or a mutant
form thereof, for example Rlucl I (A55T/C124A/M 185V).
CA 02775278 2012-04-23
9
[0029] The biosensor may have a fluorophore at the C-terminus of the arrestin
and the
bioluminophore at the N-terminus of the arrestin. In this case, the
fluorophore may be green
fluorescent protein or a variant thereof, such as YFP, mAmetrine, cyan
fluorescent protein
(CFP), or GFP10. The bioluminophore of such an embodiment may be Renilla
luciferase or
a mutant form thereof.
[0030] In certain embodiments of the biosensor, one of the fluorophore or
bloluminophore
may be linked to a position between the C and N-termini. For example, the
bioluminophore
may be linked to the C-terminus of the arrestin while the fluorophore may be
linked internally
to the arrestin. In such exemplary embodiments, the fluorophore may be YFP,
mAmetrine,
cyan fluorescent protein (CFP), or GFP10, and the bioluminophore could be
Renilla
luciferase or a mutant form thereof.
[0031] In certain embodiments of the biosensor, both said first and said
second
chromophores may be fluorophores and the RET may be fluorescence resonance
energy
transfer (FRET). For example, the fluorophore donor can be CFP or a variant
thereof, while
the the fluorophore acceptor could be a YFP or a variant thereof. YFP may be a
non-
circularly permuted sYFP2 or a circularly permuted sYFP2.
[0032] A method of identifying candidate molecules is described herein, which
that bind to a
receptor. The method may comprise (a) screening candidate molecules for
activation of a
biosensor in which the second chromophore Is a bioluminophore and the RET is
bioluminescence resonance energy transfer (BRET), and in which the fluorophore
is at the
C-terminus of the arrestin and the bioluminophore Is at the N-terminus of the
arrestin, so as
to determine a candidate population; and (b) screening this candidate
population for
activation using a further biosensor to identify candidate molecules that bind
to receptors. In
this case, the further biosensor used to screen the candidate population
determined in (a) is
one in which the second chromophore is a bioluminophore and the RET is
bioluminescence
resonance energy transfer (BRET), and in which the fluorophore is at the N-
terminus of the
arrestin and the bioluminophore is at the C-terminus of the arrestin.
[0033] In the above method, the receptor may be a Frizzled protein receptor or
a G protein-
coupled receptor (GPCR). An exemplary receptor may be Frizzled 4(Fz4), R2AR,
VI
vasopressin receptor (V1aR), V2 vasopressin repressor (V2R), delta opioid
receptor,
CA 02775278 2012-04-23
platelet-activating factor receptor, CC chemokine receptor type 5, or
angiotensin receptor
type 1 a.
[0034] In the method described above, the activation of the biosensor in (a)
may be
observed by an increase in BRET signal. The activation of the biosensor in (b)
may result in
a decrease in BRET signal. The candidate molecule that binds the receptor may
be an
agonist, inverse agonist, partial agonist, antagonist or an allosteric
regulator.
[0035] The above-described biosensor may used for assaying receptor activity.
Further, the
biosensors as described in the above method, in parts (a) and (b), may be used
together for
identifying agonists, inverse agonists, partial agonists, antagonists or
allosteric regulators.
[0036] A kit is described herein for evaluating receptor binding. The kit may
comprise the
biosensor described in part (a) of the method described above, together with
the biosensor
described in part (b) of the method above. Such a kit may be used for
evaluating receptor
binding to identify candidate molecules that bind to the receptor, and thus
may include
instructions for use in the above.
[0037] In the method described above, the way in which molecules can be
screened may
comprise identifying an agonist or inverse agonist for the receptor by
incubating (i) cells co-
expressing the receptor and the biosensor with a potential agonist or inverse
agonist, or (ii)
an isolated receptor and the biosensor with a potential agonist or inverse
agonist. Following
this, a suitable substrate may be added to detect bioluminescence resonance
energy transfer
(BRET) in said biosensor. The BRET signal can then be detected and compared
with a
BRET signal obtained under similar conditions in the absence of the potential
agonist or
inverse agonist. The potential agonist or inverse agonist may thus be
identified as an
agonist or inverse agonist if a change in BRET signal level is observed.
[0038] In this method, the BRET signal may be evaluated by detecting light
emissions at
about 440-510 nm and at about 510-570 nm. As a further option, the BRET signal
may be
evaluated by detecting light emissions at about 320-490 nm and at about 490-
550 nm. The
receptor may be a Frizzled protein receptor or a G protein-coupled receptor.
Further,
receptor may be chosen from the exemplary group of: Frizzled 4 (Fz4), 02AR, V1
vasopressin receptor (V1 aR), V2 vasopressin receptor (V2R), delta-opioid
receptor (SOR),
CA 02775278 2012-04-23
11
platelet-activating factor receptor (PAFR), CC chemokine receptor type 5
(CCR5), and
angiotensin receptor type Ia (AT1aR).
DEFINITIONS:
[0039] Unless otherwise defined, the terms used in the present description
have the
meanings that would be understood by a person of skill in the art.
[0040] Ligand: A molecule which may be but is not restricted to a hormone,
neurotransmitor, chemical compound, drug, or diagnostic agent that binds to a
receptor and
has an agonistic, inverse agonistic, antagonistic or allosteric effect on the
receptor. Ligands
may be further classified as follows (for a more detailed summary, see Wilson,
Keith et al.
(Eds.), Principles and Techniques of Biochemistry and Molecular Biology, 7th
Edition (2010),
Chapter 17, incorporated by reference herein):
a) Agonist: a ligand that has the same or similar effect as a hormone,
neurotransmitter
or signaling molecule or a group of hormones, neurotransmitters or signaling
molecules activating a receptor, by binding to the same natural receptor. A
partial
agonist is a type of agonist that with lower intrinsic activity than a full
agonist and that
produces a lower maximum effect. Examples of agonists include:
i. Angiotensin II: The active form of angiotensin. An octapeptide found in
blood, it
is synthesised from angiotensin I and quickly destroyed. Angiotensin II causes
profound vasoconstriction with resulting increase in blood pressure. It is an
agonist of the angiotensin receptor.
ii. AVP: arginine vasopressin, vasopressin containing arginine, as that from
most
mammals, including man. This hormone controls water reabsorbtion by the kidney
and is also known as the antidiuretic hormone.
iii. ISO: isoproterenol, a synthetic beta-adrenergic receptor agonist which
controls
peripheral vasoconstriction, bronchodilation and increased cardiac rate,
contractility and output.
CA 02775278 2012-04-23
12
iv. SNC80: 4-[( R)-a-((2S,5R)-4-allyl-2,5-dimethyi-1-piperazinyl)-3-
methoxybenzyl]-
N,N-diethylbenzamide. An agonist of the delta-opioid receptor that possesses
anti-nociceptive action.
v. PAF: platelet-activating factor; a hormone that regulates platelet
aggregation. It is
an agonist of the PAF receptor.
vi. hRANTES: human RANTES (regulated upon activation, normal T cell expressed
and secreted) is a chemoattractant for monocytes and T cells. It is an agonist
of
the chemokine receptors: CCR1, CCR3, CCR5 and GPR75.
vii. Wnt5a: Ligand for members of the frizzled family of seven transmembrane
receptors.
viii.IGF1: insulin-like growth factor 1 (also known as somatomedin C), a
hormone
homologous to proinsulin.
ix. TGF-01: Transforming Growth Factor-beta1, a multifunctional peptide that
controls proliferation, differentiation, and other functions in many cell
types. Many
cells synthesize TGF-beta 1 and essentially all of them have specific
receptors for
this peptide. TGF-beta 1 regulates the actions of many other peptidic growth
factors and determines a positive or negative direction of their effects.
b) Inverse agonist: a ligand that produces an effect opposite to that of an
agonist by
occupying the same receptor. Examples include:
i. SR121463: SR121463 is a selective, orally active, non-peptide antagonist of
vasopressin (AVP) V2 receptors, with powerful aquaretic, properties in various
animal species and humans. SR121463 also behaves as an inverse agonist in
cells expressing constitutively active human V2 receptor.
c) Antagonist: a ligand that counteracts the effect of another ligand (agonist
or inverse
agonist) acting on a receptor by binding to the same receptor, thus blocking
or
dampening the ability of the agonist to bind (also called competitive
antagonist).
Neutral antagonists have affinity but no efficacy for their cognate receptors.
CA 02775278 2012-04-23
13
d) Allosteric regulator: a ligand that modulates receptor activity through
binding at a
site that is different from that bound by orthosteric ligands (i.e. endogenous
ligands).
Allosteric regulators may have an antagonistic or agonistic effect.
[0041] Chromophore: A small molecule, or a part of a larger molecule, that is
responsible
for the spectral band of the molecule.
[0042] Blosensor: A type of biomolecular probe that measures the presence or
concentration of biological molecules, biological structures, activity state
etc., by translating a
biochemical interaction at the probe surface into a quantifiable physical
signal such as light
or electric pulse.
[0043] Receptor: A popular and generally accepted hypothesis that appears to
explain
many pharmacodynamic phenomena holds that specialized protein molecules on the
surfaces of cells provide a "fit" for an intrinsic molecule (such as a hormone
or
neurotransmitter) or a drug such that when that molecule occupies (binds to)
that area, it
leads to a biochemical or physiologic response. This Idea is often compared to
the operation
of a lock (receptor) by a key (Iigand). Examples of GPCR receptors include:
a) 12-AR: beta-2 adrenergic receptor
b) Frizzled 4 (Fz4): a seven transmembrane receptor that selectively
recognizes
hormones of the Wnt family.
C) V2R: Vasopressin V2 receptor
d) VIaR: Vasopressin V1a receptor
e) 5-OR: b-opioid receptor
f) PAFR: platelet-activating factor receptor
g) CCR5: CC chemokine receptor type 5
h) AT1aR: angiotensin receptor type 1a
CA 02775278 2012-04-23
14
(0044] Signalling molecule: a membrane or soluble protein involved in the
transaction of
signals in cells initiated by hormones, neurotransmitters or synthetic
ligands.
[0045] Identity as known in the art, is a relationship between two or more
polypeptide
sequences, as determined by comparing the sequences. In the art, "identity"
also means the
degree of sequence relatedness between polypeptides as determined by the match
between
strings of such sequences. "Identity" and "similarity" can be readily
calculated by known
methods, including, but not limited to, those described in (Computational
Molecular Biology,
Lesk, A. M., Ed., Oxford University Press, New York, 1988; Biocomputing:
Informatics and
Genome Projects, Smith, D. W., Ed., Academic Press, New York, 1993; Computer
Analysis
of Sequence Data, Part I, Griffin, A. M., and Griffin, H. G., Eds., Humana
Press, New Jersey,
1994; Sequence Analysis in Molecular Biology, von Heinje, G., Academic Press,
1987; and
Sequence Analysis Primer, Gribskov, M. and Devereux, J., Eds., M Stockton
Press, New
York, 1991; and Carillo, H., and Lipman, D., SIAM J Applied Math., 48: 1073
(1988).
[0046] By way of example, a polypeptide sequence may be identical to the
reference
sequence, that is be 100% identical, or it may Include up to a certain integer
number of
amino acid residue alterations as compared to the reference sequence such that
the %
identity is less than 100%. Such alterations are selected from: at least one
conservative or
non-conservative amino acid residue substitution, deletion, or insertion, and
wherein said
alterations may occur at the amino- or carboxy-terminal positions of the
reference
polypeptide sequence or anywhere between those terminal positions,
interspersed either
individually among the amino acid residues in the reference sequence or in one
or more
contiguous groups within the reference sequence. The number of amino acid
residue
alterations for a given % identity is determined by multiplying the total
number of amino acids
in the reference polypeptide by the numerical percent of the respective
percent identity
(divided by 100) and then subtracting that product from said total number of
amino acids in
the reference polypeptide.
[0047] Conservative amino acid variants can also comprise non-naturally
occurring amino
acid residues. Non-naturally occurring amino acids include, without
limitation, trans-3-
methylproline, 2,4-methanoproline, cis-4-hydroxyproline, trans-4-
hydroxyproline, N-methyl-
glycine, allothreonine, methylthreonine, hydroxy-ethylcysteine,
hydroxyethylhomocysteine,
nitro-glutamine, homoglutamine, pipecolic acid, thiazolidine carboxylic acid,
dehydroproline,
CA 02775278 2012-04-23
3- and 4-methyiproline, 3,3-dimethylproline, tert-leucine, norvaiine, 2-
azaphenyl-alanine, 3-
azaphenylalanine, 4-azaphenylalanine, and 4-fluorophenylalanine. Several
methods are
known in the art for incorporating non-naturally occurring amino acid residues
into proteins.
For example, an in vitro system can be employed wherein nonsense mutations are
suppressed using chemically aminoacylated suppressor tRNAs. Methods for
synthesizing
amino acids and aminoacylating tRNA are known in the art. Transcription and
translation of
plasmids containing nonsense mutations is carried out in a cell-free system
comprising an E.
coli S30 extract and commercially available enzymes and other reagents.
Proteins are
purified by chromatography. (Robertson, etal., J. Am. Chem. Soc, 113:
2722,1991; Ellman, et
a!., Methods Enzymoi, 202: 301, 1991; Chung, etal., Science, 259: 806-9, 1993;
and Chung,
et a!, Proc. Nati. Acad. Sci. USA, 90: 10145-9, 1993). In a second method,
translation is
carried out in Xenopus oocytes by microinjection of mutated mRNA and
chemically
aminoacylated suppressor tRNAs (Turcatti, at a!, J. Biol. Chem., 271: 19991-8,
1996). Within
a third method, E. coli cells are cultured in the absence of a natural amino
acid that is to be
replaced (e.g., phenylalanine) and in the presence of the desired non-
naturally occurring
amino acid(s) (e.g., 2-azaphenylalanine, 3-azaphenylalanine, 4-
azaphenylalanine, or 4-
fluorophenylalanine). The non-naturally occurring amino acid is incorporated
into the protein
in place of its natural counterpart. (Koide, et al, Biochem., 33: 7470-6,
1994). Naturally
occurring amino acid residues can be converted to non-naturally occurring
species by in vitro
chemical modification. Chemical modification can be combined with site-
directed
mutagenesis to further expand the range of substitutions (Wynn, et a!. Protein
Sci., 2: 395-
403, 1993).
[0048] Variant: refers to a polypeptide or polynucleotide that differs from a
reference
polypeptide or polynucleotide, but retains essential properties. A typical
variant of a
polypeptide differs in amino acid sequence from another, reference
polypeptide. Generally,
differences are limited so that the sequences of the reference polypeptide and
the variant are
closely similar overall and, in many regions, identical. A variant and
reference polypeptide
may differ in amino acid sequence by one or more modifications (e.g.,
substitutions,
additions, and/or deletions). A variant of a polypeptide includes
conservatively modified
variants. A substituted or inserted amino acid residue may or may not be one
encoded by the
genetic code. A variant of a polypeptide may be naturally occurring, such as
an allelic
variant, or it may be a variant that is not known to occur naturally.
CA 02775278 2012-04-23
16
[0049] Modifications and changes can be made in the structure of the
polypeptides of this
disclosure and still obtain a molecule having similar characteristics as the
polypeptide
(e.g., a conservative amino acid substitution). For example, certain amino
acids can
be substituted for other amino acids in a sequence without appreciable loss of
activity. Because it is the interactive capacity and nature of a polypeptide
that defines
that polypeptide's biological functional activity, certain amino acid residue
substitutions can be made in a polypeptide sequence and nevertheless obtain a
polypeptide with like properties.
[0050] In one aspect, such variants have at least 60%, at least 70%, at least
80%, at least
85%, at least 90%, at least 91, at least 92%, at least 93%, at least 94%, at
least 95%, at
least 96%, at least 97%, at least 98%, or at least 99% sequence identity with
the reference
polypeptide or polynucleotide.
ARRESTIN:
[0051] A receptor could either be constitutively active or inactive and,
ligands such agonists,
inverse agonists and allosteric modulators are known to modulate this
activity. The
interaction of arrestins, including a-arrestin (Garr), with receptors, such as
but not limited to
GPCRs, is a reflection of the receptor activity. The beta-arrestins belong to
the family of
arrestins. It is generally accepted that there are 4 arrestins in mammals:
arrestins 1-4.
Arrestin-1 and arrestin-4 are visual arrestins whereas arrestin-2 and arrestin-
3 are widely
distributed in all tissues and correspond to beta-arrestin-1 and beta-arrestin-
2 respectively.
The interaction of R-arrestins with receptors or other proteins, such as a
beta2-adaptin, has
an impact on the conformation of the (3-arrestins. This change in conformation
is linked to its
property of interacting with effectors of signaling pathways and receptor
endocytosis. This
characteristic of arrestin is conserved throughout evolution of eukaryotic
organisms and, is
the basis for the unimolecular RET-based conformational sensors: a-arrestinl
(also known
as Arrestin-2 or R-arrestin) and R-arrestin 2 (also known as Arrestin-3)
double brilliance
sensors to monitor receptor activity. A conformational change in 13-arrestin
is monitored
through a modulation of the RET signal.
[0052] The arrestins exemplified herein are human f3-arrestins (hparr) and
mutants and
variants thereof; however, other arrestins are contemplated, including
proteins having at
CA 02775278 2012-04-23
17
least 60%, at least 70%, at least 80%, at least 85%, at least 90%, at least
92%, at least 94%,
at least 96%, at least 98%, or at least 99% sequence identity with h(3arr1,
h3arr2, or with
another arrestin, wherein such proteins interact with GPCRs.
RESONANCE ENERGY TRANSFER ASSAYS:
[0053] Resonance energy transfer (abbreviated RET, and also referred to as
Forster
resonance energy transfer) is a mechanism describing energy transfer between
two
chromophores, having overlapping emission/absoprtion spectra. When the two
chromophores (the "donor" and the "acceptor"), are within 10-100 A (Angstroms)
of one
another and their transition dipoles are appropriately oriented, the donor
chromophore is able
to transfer its excited-state energy to the acceptor chromophore through non-
radiative dipole-
dipole coupling.
[0054] Bioluminescence Resonance Energy Transfer (BRET) Assay is a proximity
assay
based on the non-radiative transfer of energy between a donor bioluminophore
(bioluminescent enzyme (ex: luciferase)) and an acceptor fluorophore (ex: GFP
or YFP).
[0055] As used herein, BRET1 uses coelenterazine h as the luciferase substrate
(i.e.
bioluminescent initiator molecule) and YFP and its variants as the energy
acceptor. BRET2
uses coelenterazine 400a (Perkin-Elmer, Wellesley, MA, USA and, Biotium Inc,
Hayward,
CA, USA) as the luciferase substrate and CFP, GFP2, GFP10, Tsapphire or
mAmetrine as
the energy acceptor. BRET1 and BRET2 represent different variants of BRET that
are based
on the use of different, luminescent enzymes, luciferase substrates and
different fluorescent
proteins. The difference between the BRET1 and BRET2 biosensors as used herein
also
incorporates differences in both the linkers used to join the chromophores to
the beta arrestin
molecules and the orientation of the chromophores relative to each other (i.e.
to which
terminal of beta-arrestin are they linked.
[0056] Each version of BRET typically uses a different coelenterazine to be
able to excite the
acceptor at different wavelengths. Typically, the acceptor for BRET1 is a YFP
and for
BRET3 is an OFP (Abhijit De, Pritha Ray , Andreas Markus Loening and Sanjiv
Sam
Gambhir, BRET3: a red-shifted bioluminescence resonance energy transfer (BRET)-
based
integrated platform for imaging protein-protein interactions from single live
cells and living
animals The FASEB Journal. 23(8): 2702-2709, incorporated by reference herein)
For
CA 02775278 2012-04-23
18
BRET2 the acceptor is typically any fluorophore that can be excited close to
400nM such as
BFP, Cyan, GFP or mKeima (RFP).
[0057] (i) bioluminophore: The bioluminophore in the BRET assay is a protein,
that
catalyzes the reaction of a substrate (i.e. a bioluminescent initiator
molecule) producing
bioluminescence.
[0058] Luciferase is an example of a protein that catalyzes the oxidation of
its substrate (ex:
coelenterazine) producing light, and can be used as a bioluminophore. As used
herein,
luciferases refer to an enzyme that catalyzes a bioluminescent reaction (a
reaction that
produces bioluminescence). In representative embodiments, the subject
luciferase
polypeptides are polypeptide sequences of the Renilla reniformis wild-type and
mutant
luciferases, which are known and reported in Lorenz et al., Proc. Natl. Acad.
Sci. USA (1991)
88:4438-4442, Loening et al., Protein Eng Des Sel. (2006) 19(9):391-400, and
also reported
in U.S. Pat. No. 6,451,549 as SEQ ID NOS: 1 and 2, and in U.S. Pat. No.
7,842,469, the
disclosure of which is herein incorporated by reference.
[0059] In representative embodiments, the subject luciferase polypeptides may
also be
mutants (also referred to as variants herein) of wild-type luciferases found
in Renilla species
(e.g., Renilla koellikeri; Renilla muelleri and Renilla reniformis, where in
representative
embodiments, the mutant luciferase is a mutant of the Renilla reniformis wild-
type luciferase).
The term "mutant" is employed broadly to refer to a protein that differs in
some way from a
reference wild-type protein, where the subject protein retains at least one
biological property
of the reference wild-type (e.g., naturally occurring) protein. The term
"biological property" of
the subject proteins includes, but is not limited to, spectral properties,
such as emission
maximum, quantum yield, and brightness (e.g., as compared to the wild-type
protein or
another reference protein such as firefly luciferase from P. pyralis), and the
like; in vivo
and/or in vitro stability (e.g., half-life); and the like. In particular, the
mutants (or variants)
retain luciferase activity (e.g., catalyze the conversion of a coelenterazine
substrate into a
luminescent product in the presence of molecular oxygen). Mutants of the
disclosure include
single amino acid changes (point mutations), deletions of one or more amino
acids (point-
deletions), N-terminal truncations, C-terminal truncations, insertions, and
the like.
CA 02775278 2012-04-23
19
[0060] For purposes of the disclosure, a naturally occurring luciferase is a
reference wild
type luciferase for a given mutant if the amino acid sequences of the wild-
type and the
mutant have high identity over at least the length of the mutant (e.g., at
least about 90%, at
least about 95%, at least about 97%, at least about 98%, at least about 99% or
higher) but
will not have complete sequence identity in representative embodiments.
[0061] In representative embodiments, the mutants encoded by the subject
polynucleotides
exhibit increased light output as compared to their corresponding reference
wild-type protein.
Specifically, the subject mutants have at least enhanced light output with a
given
coelenterazine substrate as compared to their corresponding reference wild
type. For
purposes of the present disclosure, increased light output is determined by
evaluating at
least one of the kinetics and quantum yield of a given mutant using a
convenient assay
known to those of skill in the art. In representative embodiments in which the
subject
polynucleotides encode a mutant of Renilla luciferase that exhibits enhanced
light output, the
encoded mutant may include a substitution at at least one of the following
positions: C124;
K136; M185, and S287. In one aspect the Renilla luciferase mutant has the
following
substitutions: C124A and M185V. In another aspect the Renilla luciferase
mutant has the
following substitutions: A55T, C1 24A and M1 85V, and is referred to herein as
Rlucll. These
mutations and variations thereof are known (see Loening et al., Protein Eng
Des Sel. (2006)
19(9):391-400 and US 7,842,469, both of which are incorporated herein), and
are
contemplated for use herein. Examples of Renilla luciferase proteins
contemplated herein
include proteins that have an amino acid-sequence selected from:
Rluc WT
MTSKVYDPEQRKRM ITGPQW WARCKQMNVLDS FI NYYDSEKHAENAVI FLHGNAASSYLW
RHWPHIEPVARCIIPDLIGMGKSGKSGNGSYRLLDHYKYLTAWFELLNLPKKI IFVGHDWGA
CLAFHYSYEHQDKIKAIVHAESWDVIESWDEWPDIEEDIALIKSEEGEKMVLENNFFVETML
PSKIMRKLEPEEFAAYLEPFKEKGEVRRPTLSWPREIPLVKGGKPDWQIVRNYNAYLRASD
DLPKMFIESDPGFFSNAIVEGAKKFPNTEFVKVKGLHFSQEDAPDEMGKYIKSFVERVLKNE
Q
Rlucll (A55T/C124A/M185V)
MTSKVYDPEQRKRMITGPQWWARCKQMNVLDSFINYYDSEKHAENAVIFLHGNATSSYLW
RHWPHIEPVARCIIPDLIGMGKSGKSGNGSYRLLDHYKYLTAWFELLNLPKKIIFVGHDWGA
CA 02775278 2012-04-23
ALAFHYSYEHQDKIKAIVHAESWDVIESWDEWPDIEEDIALIKSEEGEKMVLENNFFVETVLP
SKIMRKLEPEEFAAYLEPFKEKGEVRRPTLSWPREIPLVKGGKPDWQIVRNYNAYLRASDD
LPKMFIESDPGFFSNAIVEGAKKFPNTEFVKVKGLHFSQEDAPDEMGKYIKSFVERVLKNEQ*
RIuc8 (A55T/C124A/S130A1K136R/A143M/M 185V/M253L/S287L)
MASKVYDPEQRKRM ITGPQWWARCKQMNVLDSFINYYDSEKHAENAVI FLHGNATSSYLW
RHWPHIEPVARCIIPDLIGMGKSGKSGNGSYRLLDHYKYLTAWFELLNI,PKKIIFVGHDWGA
ALAFHYAYEHQDRIKAIVHMESWDVIESWDEWPDIEEDIALIKSEEGEKMVLENNFFVETVL
PSKIMRKLEPEEFAAYLEPFKEKGEVRRPTLSWPREIPLVKGGKPDWQIVRNYNAYLRASD
DLPKLFIESDPGFFSNAIVEGAKKFPNTEFVKVKGLHFLQEDAPDEMGKYIKSFVERVLKNEQ
portions thereof, mutants thereof, variants thereof, or conservative variants
thereof. Other
luciferase variants known in the art include those disclosed in US
2009/0136998,
incorporated by reference herein.
[0062] In representative embodiments, the mutant luciferase polynucleotides
encoded by the
nucleic acids are mutants of luciferase polynucleotides that employ a
coelenterazine as a
substrate, where the term coelenterazine refers collectively to native
coelenterazine, as well
as analogues thereof, where representative coelenterazine analogues of
interest include, but
are not limited to: benzyl-coelenterazine; coelenterazine-cp; coelenterazine-
n; bis-deoxy-
coelenterazine (also known as coelenterazine 400a and DeepBlue-
coelenterazine); and the
like.
[0063] In addition to the above-described specific subject polynucleotide
compositions, also
of interest are homologues of the above-sequences. With respect to homologues
of the
subject polynucleotide, the source of homologous genes may be any species of
plant or
animal, or the sequence may be wholly or partially synthetic. In certain
embodiments,
sequence similarity between homologues is at least about 20%, at least about
25%, and may
be 30%, 35%, 40%, 50%, 60%, 70% or higher, including 75%, 80%, 85%, 90% and
95% or
higher. Sequence similarity is calculated based on a reference sequence, which
may be a
subset of a larger sequence, such as a conserved motif, coding region,
flanking region, and
the like. A reference sequence will usually be at least about 18 nt long, more
usually at least
about 30 nt long, and may extend to the complete sequence that is being
compared.
Algorithms for sequence analysis are known in the art, such as BLAST,
described in Altschul
CA 02775278 2012-04-23
21
et al. (1990), J. Mol. Biol. 215:403-10 (using default settings, e.g.
parameters w=4 and
T=17). The sequences provided herein are used for recognizing related and
homologous
nucleic acids in database searches.
[0064] (ii) fluorophore: The fluorophore in the BRET assay is a fluorescent
protein.
[0065] Green fluorescent protein ("GFP") is a 238 amino acid residues
polypeptide with
amino acid residues 65 to 67 involved in the formation of the chromophore,
which does not
require additional substrates or cofactors to fluoresce (see, e.g, Prasher at
al, 1992, Gene
111:229-233; Yang at a/, 1996, Nature Biotechnol. 14:1252-1256; and Cody et
al, 1993,
Biochemistry 32:1212-1218). Thus, in one embodiment, such a fluorophore is a
green
fluorescent protein (GFP) (referring to native Aequorea green fluorescent
protein), and
variants thereof.
[0066] A broad range of fluorescent protein genetic variants have been
developed over the
past several years that feature fluorescence emission spectral profiles
spanning almost the
entire visible light spectrum. Such variants of the GFP gene have been found
useful to
enhance expression and to modify excitation and fluorescence. Extensive
mutagenesis
efforts in the original jellyfish protein have resulted in fluorescent probes
that range in color
from blue to yellow. For example, substitution of a serine at position 65 to
either alanine,
glycine, isoleucine, or threonine results in mutant GFPs with a shift in
excitation maxima and
greater fluorescence than wild type protein when excited at 488 nm (see, e.g,
Heim et al,
1995, Nature 373:663-664; U.S. Pat. No. 5,625,048; Delagrave et al, 1995,
Biotechnology
13:151-154; Cormacketal, 1996, Gene 173:33-38; and Cramer at al, 1996, Nature
Biotechnol. 14:315-319). Longer wavelength fluorescent proteins, emitting in
the orange and
red spectral regions, have been developed from the marine anemone Discosoma
striata and
reef corals belonging to the class Anthozoa. Still other species produce
similar proteins
having cyan, green, yellow, orange, red, and far-red fluorescence emission.
Thus, in another
embodiment, GFPs are isolated from organisms other than the jellyfish, such
as, but not
limited to, the sea pansy, Renilla reriformis, or are variants thereof.
[0067] Thus, a fluorophore, as used herein, includes wild type green
fluorescent protein and
its variants, as well as fluorescent proteins and variants from other species.
Such
CA 02775278 2012-04-23
22
fluorophores are many, and are known to those of skill in the art. They
include, but are not
limited to:
= Green Fluorescent Proteins include GFP (wt), EGFP, Emerald, Superfolder GFP,
Azami Green, mWasabi, TagGFP, TurboGFP, AcGFP, ZsGreen, T-Sapphire.
= Blue Fluorescent Proteins include Blue Fluorescent Protein (BFP), EBFP,
EBFP2,
Azurite, GFP2, GFP10, and mTagBFP;
= Cyan Fluorescent Proteins include Cyan Fluorescent Protein (CFP), ECFP,
mECFP,
Cerulean, CyPet, AmCyanl, Midori-Ishi Cyan, TagCFP, mCFPmm, and mTFP1
(Teal);
= Yellow Fluorescent Proteins include Yellow Fluorescent Protein (CFP), EYFP,
Topaz,
Venus, mCitrine, YPet, TagYFP, PhiYFP, ZsYellowl, and mBanana;
= Orange Fluorescent Proteins include Orange Fluorescent Protein (OFP),
Kusabira
Orange, Kusabira Orange2, mOrange, mOrange2, dTomato, dTomato-Tandem,
TagRFP, TagRFP-T, DsRed, DsRed2, DsRed-Express (T1), DsRed-Monomer, and
mTangerine; and
= Red Fluorescent Proteins include Red Fluorescent Protein (RFP), mRuby,
mApple,
mStrawberry, AsRed2, mRFP1, JRed, mCherry, HcRedl, mRaspberry, dKeima-
Tandem, HcRed-Tandem, mPlum, tdTomato, and AQ143.
[0068] Both green and yellow fluorescent proteins have been genetically
engineered to
create circular permutations of the original sequences that enable fusions to
amino acids far
removed from the normal amino and carboxy termini (abbreviated cpGFP and
cpYFP).
[0069] The choice of a suitable fluorophore for use in a BRET assay will be
known to one of
skill in the art. In one embodiment, fluorophores include green fluorescent
protein - wild type
(GFP-wt), yellow fluorescent protein (YFP), Venus, Topaz, ZsYellow1, mOrange2,
mKeima,
blue fluorescent protein (BFP), cyan fluorescent protein (CFP), Tsapphire,
mAmetrine, green
fluorescent protein-2 (GFP2) and green fluorescent protein-10 (GFP10), or
variants thereof.
Fluorescent proteins having an excitation peak close to 400 nm may be
particularly suitable.
CA 02775278 2012-04-23
23
More particular examples of fluorophores include mAmetrine, cyan fluorescent
protein (CFP),
and GFP10.
[0070] Fluorescence Resonance Energy Transfer (FRET) Assay. Similar to BRET,
FRET
involves the transfer of energy from an excited donor fluorophore to an
adjacent acceptor
fluorophore. For example, CFP and YFP, two color variants of GFP, can be used
as donor
and acceptor, respectively.
[0071] (i) fluorophore: The fluorophores in the FRET assay are fluorescent
proteins, having
the same properties as the fluorophore as defined above for the BRET assay.
[0072] Two fluorophores are employed in FRET, one as donor and one as
acceptor. The
term "donor fluorophore-acceptor pair," as used herein, means a donor
fluorophore and an
acceptor that has an absorbance spectrum overlapping the emission spectrum of
the donor
fluorophore. Where the first member of the pair is a donor fluorophore, the
second member
of the pair will be an acceptor. Where the first member of the pair is an
acceptor, the second
member of the pair will be a donor fluorophore.
[0073] Any of a number of fluorophore combinations can be selected for use in
the FRET
embodiment described herein (see for example, Pesce et al., eds, Fluorescence
Spectroscopy, Marcel Dekker, New York, 1971; White et al., Fluorescence
Analysis: A
practical Approach, Marcel Dekker, New York, 1970; Handbook of Fluorescent
Probes and
Research Chemicals, 6th Ed, Molecular Probes, Inc., Eugene, Oreg., 1996; which
are
incorporated herein by reference). In general, a preferred donor fluorophore
is selected that
has a substantial spectrum of the acceptor fluorophore. Furthermore, it may
also be
desirable in certain applications that the donor have an excitation maximum
near a laser
frequency such as Helium-Cadmium 442 nM, Argon 488 nM, Nd:YAG 532 nm, He-Ne
633
nm, etc. In such applications the use of intense laser light can serve as an
effective means to
excite the donor fluorophore. In certain preferred embodiments, the acceptor
fluorophore has
a substantial overlap of its excitation spectrum with the emission spectrum of
the donor
fluorophore. In some cases, the wavelength maximum of the emission spectrum of
the
acceptor moiety is preferably at least 10 nm greater than the wavelength
maximum of the
excitation spectrum of the donor moiety. Additional examples of useful FRET
labels include,
e.g., those described in U.S. Pat. Nos. 5,654,419, 5,688,648, 5,853,992,
5,863,727,
CA 02775278 2012-04-23
24
5,945,526, 6,008,373, 6,150,107, 6,177,249, 6,335,440, 6,348,596, 6,479,303,
6,545,164,
6,849,745, 6,696,255, and 6,908,769 and Published U.S. Patent Application Nos.
2002/0168641, 2003/0143594, and 2004/0076979, the disclosures of which are
incorporated
herein by reference.
[0074] As indicated above, the donor and acceptor fluorophores should be
capable of
forming a FRET pair. Many suitable fluorophore pairs are familiar to those of
skilled in the art.
In one embodiment, the FRET pair comprises one of CFP-YFP, GFP-mRFP1, YFP-
mRFP1,
GFP-RFP, sCFP3A-sYFP2, as well as sCFP3A in combination with circular
permutations of
sYFP2 (such as cp145, cp173, and cp229).
[0075] Circular permutations can be made by PCR. Such permutations have been
described
(see US 6,699,687 and Takeharu Nagai, Shuichi Yamada, Takashi Tominaga,
Michinori
Ichikawa and Atsushi Miyawaki (2004) PNAS 101(29): 10554-10559, incorporated
by
reference herein). They create variants of a FRET sensor with different
orientations of the
donor vs acceptor's chromophore.
LINKERS
[0076] The chromophores are each attached to the beta-arrestin molecule
through
independent linkers. Linkers may be employed to provide the desired
conformation of the
BRET/FRET label chromophores within the labeled compound, e.g., including the
separation
between chromophores in a BRET/FRET pair. The linkers may be bound to the C-
terminal,
the N-terminal, or at an intermediate position.
[0077] In one embodiment, the linkers are peptide linkers, typically ranging
from 2 to 30
amino acids in length. The composition and length of each of the linkers may
be chosen
depending on various properties desired such as flexibility and aqueous
solubility. For
instance, the peptide linker may comprise relatively small amino acid
residues, including, but
not limited to, glycine; small amino acid residues may reduce the steric bulk
and increase the
flexibility of the peptide linker. The peptide linker may also comprise polar
amino acids,
including, but not limited to, serine. Polar amino acid residues may increase
the aqueous
solubility of the peptide linker. Furthermore, programs such as Globplot 2.3
(http://globplot.embl.de/cgiDict.py), may be used to help determine the degree
of disorder
and globularity, thus also their degree of flexibility.
CA 02775278 2012-04-23
[0078] By way of example, contemplated linkers include
GDLRRALENSHASAGYQACGTGS and CLEDPRVPVAT. Short and relatively flexible linkers
include GSAGT and KLPAT. Longer 15-residues long linkers, such as
GSAGTGSAGTGSAGT (=3xGSAGT linker) and KLPATKLPATKLPAT (=3xKLPAT linker) are
also contemplated; these latter 2 linkers are predicted (Globplot 2.3:
http://globplot.embl.de/cgiDict.py) to be disordered and non-globular
sequences, and thus
flexible. Alpha-helix structured rigid linker, REAAAREAAAREAAAR (16-residues
long), is
also contemplated.
[0079] The linkers may be attached to the beta-arrestin at the N-terminus, the
C-terminus, or
between the two termini of the beta-arrestin. When attaching in between the
two termini, the
linker may be attached, for instance to the first or second loop of the beta-
arrestin.
METHODS
[0080] Expression vectors. Plasmids encoding Flag-AT1 aR, CCR5 (Pleskoff et
al, 1997)
and Myc-PAFR (Marrache et a!, 2002) were provided by S. Meloche, N. Heveker
and S.
Chemtob, respectively (Universit6 de Montreal, Quebec, Canada) and WT R-arr2
was a
generous gift from S. Marullo (Institut Cochin, Paris). Myc-V2R and HA-V1aR
(Terrillon eta!,
2003), Myc-02-AR (Hebert et al, 1996), Myc-b-OR (Petaja-Repo et a!, 2002), V2R-
GFP
(Charest & Bouvier, 2003), 02-AR-GFP (Mercier et al, 2002).
[0081] BRET1 biosensors: 0-arr2-YFP (Angers et al, 2000) and Luc- 0-arr2
(Perroy et a!,
2003) have been described previously. Luc- 13-arr-YFP was generated by
subcloning the
coding sequence of enhanced YFP in-frame at the C terminus of (3-arr2 in
pcDNA3.1-Luc-
R-arr2, yielding Luc- R-arr-YFP with flexible spacers of 23 as between Luc and
R-arr, and 10
as between R-arr and YFP. Mutation of arginine 169 into glutamate in Luc-R-arr
(R169E)-
YFP was generated by PCR site-directed mutagenesis using Luc-(3-arr-YFP. It
should be
noted that while the construct described here is specific for Luc- 3-arr-YFP,
a construct
leading to the production of a YFP-R-arr-Luc biosensor is feasible. Moreover,
the resulting
biosensor, YFP-R-arr-Luc, would be expected to function in the same manner as
Luc-(3-arr-
YFP. Similarly, DNA constructs may be devised for the specific expression of
Luc-3-arr-
GFP, GFP-[i-arr-Luc biosensors, and variants thereof.
CA 02775278 2012-04-23
26
[0082] BRET2 biosensors: Acceptor-beta-arrl/2-Rlucll, with Acceptor being
either
mAmetrine, sCFP3A or GFP10, were derived from previously published GFP10-EPAC-
RlucIl
fusion protein (Leduc at al. JPET 2009) by excising the EPAC coding sequence
with Acc651-
Hindill restriction enzymes and replacing it with a PCR-amplified coding
sequence of human
beta-arrestinl or beta-arrestin2. Sequence integrity was confirmed by DNA
sequencing.
[0083] Cell culture. Human embryonic kidney 293 (HEK293) cells and simian
kidney
fibroblast (COS) cells were maintained as described previously (Charest &
Bouvier, 2003).
Cells were transfected with the indicated plasmids using the calcium phosphate
precipitation
method (Sambrook et al, 1989) or the FuGENE 6 transfection reagent (Roche
Applied
Science, Laval, Canada) according to the manufacturer's protocol. The
experiments were
performed 48 h after transfection.
[0084] Fluorescence microscopy. To detect Myc-02-AR and Myc-V2R, cells were
incubated with anti-Myc 9E10 monoclonal antibody (ascite fluid from our core
facility) for 1 h
at 4 C and then treated with the appropriate agonist (Sigma, Oakville, Canada)
for 2 or 30
min at 37 C. Cells were then fixed and permeabilized before adding Texas-red-
conjugated
secondary antibody (Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA). The
samples
were analysed by confocal laser-scanning microscopy using a Leica TCS SP1.
Measurements were as follows: YFP (green), Aex= 488 nm, Aem= 540/25nm; Texas
red
(red), Aex= 568 nm, Aem= 610/30nm.
[0085] BRET assays. Assessment of R-arr recruitment in BRET was performed as
described previously (Charest & Bouvier, 2003). Briefly, cells were
distributed in 96-well
microplates (Corning, Corning, USA) and incubated with or without agonist for
the indicated
time at 25 C. The appropriate Luc substrate was added to a final concentration
of 5 mM,
either simultaneously with the agonist (time course) or following agonist
treatment (single
measurement or dose dependency), and readings were collected using a
Multilabel Reader
Mithras LB 940 (Berthold Technologies, Bad Wildbad, Germany). To detect BRET1
between
Luc and YFP, coelenterazine h (Molecular Probes, Burlington, Canada) was used
as
substrate and light emission was detected at approximately 460-500nm (Luc) and
approximately 510-550nm (YFP), whereas for BRET2 detection (Luc and GFP),
coelenterazine 400a (Perkin-Elmer, Wellesley, MA, USA or Biotium Inc, Hayward,
CA, USA)
and filters at approximately 330-470nm (Luc) and approximately 495-535nm
(GFP2/GFP10)
CA 02775278 2012-04-23
27
were used. (Broadly speaking, ranges for the detection of light emission for
BRET1 are
approximately 440-510nm (Luc) and 510-570nm (YFP), while those for BRET2 are
approximately 320-490nm (Luc) and 490-550nm (GFP)). Those for BRET3 are ????.
The
BRET signal was determined by calculating the ratio of the light emitted by
the fluorescent
acceptor and the light emitted by Luc. The values were corrected by
subtracting the
background BRET signals detected when Luc-R-arr was expressed alone.
Expression levels
of the different receptors transfected were verified by enzyme-linked
immunosorbent assay
(ELISA) (Charest & Bouvier, 2003).
[0086] Receptor endocytosis assay. Receptor endocytosis was measured by ELISA
as
described previously (Charest & Bouvier, 2003).
100871 Z'-factor determination. HEK293T cells were cultured in DMEM
supplemented with
10% fetal bovine serum, 100 units/ml penicillin and streptomycin (Wisent Inc).
3.0x106 cells
were seeded in 10cm dishes. Transient transfection was performed using
polyethylenelmine
(PEI; Polysciences) at a DNA:PEI ratio. 24h post-transfection, cells were
detached, seeded
in pretreated poly-L-ornithine hydrobromide (Sigma-Aldrich) 96-well white
plates at 50,000
cells per well, and re-incubated at 37 C for an additional 24h before being
processed. Cells
were washed once with Tyrode's buffer directly in the 96-well plates and
incubated in buffer
with or without 100nM of AVP for 25 to 35 min. Coelenterazine 400A was added
to a final
concentration of 5pM in Tyrode's buffer 5min before reading. Readings were
collected as a
sequential integration of the signals detected in the 480 i 20 and 530 20nm
window for the
Rlucll Renilla luciferase and GFP10 light emissions, respectively. The BRET
signal was
determined by calculating the ratio of the light intensity emitted by the
GFP10 over the light
intensity emitted by the RLucll.
RESULTS
[0088] Double-brilliance Q-arr sensor (BRET1): Inspired by previous reports of
intramolecular fluorescence resonance energy transfer (FRET)-based biosensors
(Zhang et
al, 2002) showing that resonance energy transfer (RET) is sensitive to changes
in the
relative positions of the donor and acceptor molecules, the feasibility of
monitoring whether
conformational changes of R-arr using an intramolecular BRET approach was
assessed. A
double-brilliance R-arr was engineered in which Luc was fused to the N
terminus of R-arr2
CA 02775278 2012-04-23
28
and YFP to its C terminus, yielding Luc- P-arr-YFP (Fig 1). To test the
functionality of Luc-
P-arr-YFP, the ability of this molecule to be recruited to agonist-stimulated
class A (receptors
interacting transiently with Parr) 02-adrenergic receptor (P2-AR) and class B
(receptors
interacting stably with Parr) V2 vasopressin receptor (V2R) by fluorescence
microscopy was
determined. As shown in Fig 2A, agonist stimulation led to rapid translocation
of Luc- -arr-
YFP to the plasma membrane, colocalizing with Myc-tagged 02-AR and V2R (Myc-
P2-AR;
Myo-V2R). The patterns of Luc- P-arr-YFP interaction were consistent with
those observed
for class A (transient P-arr interaction) and B (stable P-arr association)
receptors in similar
experiments using a P-arr-green fluorescent protein (GFP) conjugate (Oakley at
a/, 2000).
Indeed, whereas Luc-f3-arr-YFP was recruited to both P2-AR and V2R after 2 min
of
stimulation, it returned to the cytoplasm after 30 min in Myc-P2-AR-expressing
cells but
remained colocalized with Myc-V2R in endocytic vesicles.
[0089] To quantitatively assess the recruitment of Luc-P-arr-YFP to agonist-
activated
GPCRs, an intermolecular BRET2 assay that takes advantage of the different
spectral
properties of Luc substrates that allow energy transfer to different
fluorescent acceptors
(Milligan, 2004) was used. Luc-P-arr-YFP was transiently coexpressed with the
receptors,
and the agonist-induced BRET2 between Luc- P-arr-YFP and either 02-AR-GFP or
V2R-
GFP was measured in the presence of DeepBlueCTM coelenterazine, allowing
transfer of
energy to GFP. As shown in Fig 2, agonist stimulation promoted a time-
dependent (Fig 2B)
and dose-dependent (Fig 2C) increase in BRET2, reflecting the recruitment of
Luc-13-arr-
YFP to the receptors. Similar kinetics and EC50 were obtained for the
recruitment of both
Luce-arr-YFP and Luc-f3-arr, indicating that double-brilliance P-arr is as
efficiently recruited
to the receptors as the singly conjugated construct. It should be noted that,
although the
maximum agonist-promoted BRET increase observed with the class A 02-AR is less
than
that observed with the class B V2R, the stability of the signals was similar,
indicating that the
signal observed with 02-AR reflects a steady state corresponding to constant
association and
dissociation of P-arr from the activated receptors.
[0090] To assess the biological activity of Luc- P-arr-YFP, its capacity to
promote receptor
endocytosis in COS cells, which express low endogenous levels of P-arr, was
tested. As
shown in Fig 2D, agonist-promoted P2-AR and V2R endocytosis was considerably
increased
when overexpressing Luc-P-arr-YFP. Even though this increase in receptor
endocytosis was
CA 02775278 2012-04-23
29
not as pronounced as that obtained by the overexpression of wild-type (WT) i3-
arr, it
suggests that Luc-R-arr-YFP retains significant biological activity.
[0091] Agonist-induced conformational changes of R-arr: To assess whether Luc-
R-arr-
YFP could be used to monitor the conformational rearrangement of parr upon
receptor
activation, the construct was expressed with and without V2R, and BRET was
measured in
the presence of coelenterazine h, allowing transfer of energy to YFP. As shown
In Fig 3A, an
important basal BRET signal could be measured in cells transfected with Luc-(3-
arr-YFP,
reflecting the proximity of the energy donor and acceptor in the construct.
Arginine
vasopressin (AVP) stimulation of cells coexpressing V2R led to a significant
increase in
BRET, suggesting movement of Luc and YFP relative to each other. To rule out
the
possibility that this increased signal results from intermolecular BRET
between individual
Luc-R-arr-YFP molecules brought together through oligomerization (Hirsch et
al, 1999) or
clustering at the plasma membrane, the occurrence of BRET in cells transiently
expressing
Luc-p-arr and (3-arr-YFP was determined. In transfection conditions leading to
equivalent
fluorescence and luminescence levels as those obtained in Luc-P-arr-YFP-
expressing cells,
coexpression of Luc-f3-arr and R-arr-YFP led to the detection of only a
marginal basal BRET
that could not be modulated by V2R stimulation (Fig 3A). This observation
demonstrates that
the AVP-induced increase in BRET signal observed in cells transfected with Luc-
(3-arr-YFP
results from a change in intramolecular BRET. As variations in RET can reflect
changes in
both the distance and orientation between the energy donor and acceptor
molecules
(Andrews & Demidov, 1999), the observed agonist-promoted increase in the Luc -
f3-arr-YFP
intramolecular BRET could indicate that the N terminus and C terminus are
either brought
closer or are in a more permissive BRET orientation following activation.
[0092] To further characterize the agonist-induced change in the conformation
of (3-arr, the
kinetics and dose dependency of AVP-mediated BRET increase were assessed. Real-
time
BRET measurements show a time-dependent AVP-induced conformational change of R-
arr,
with half-time of maximal BRET increase (t1/2) of 5.1 1.5 min (Fig 3B). The
kinetics are
significantly slower (P < 0.02) than that of the AVP-induced recruitment of 13-
arr (tl/2=0.8
0.2 min; Fig 2B, right panel), suggesting that the conformational change
observed in Luc-13-
arr-YFP occurs after its initial recruitment to the activated V2R. The
difference in kinetics
cannot result from inter-experimental variations because similar results were
obtained when
the two events were measured in the same cell population expressing V2R-GFP
and Luc-[3-
CA 02775278 2012-04-23
arr-YFP (data not shown). Despite the difference in kinetics, the efficacy of
AVP to induce a
conformational change in Luc-P-arr-YFP (Fig 3C) was similar to that observed
for R-arr
recruitment (Fig 2C, right panel), indicating that these two events are
directly linked and
reflect the binding affinity of V2R for AVP (KD -1 x 10-9 M).
[0093] The observed kinetic lag between P-arr recruitment and its
conformational change
could be consistent with the proposal that inactive P-arr is first recruited
to the activated
GPCR where its interaction with the GRK-phosphorylated residues subsequently
induces the
release of its C-tail (Gurevich & Gurevich, 2003). Alternatively, such a lag
could indicate that
the intramolecular BRET changes observed with Luc-[i-arr-YFP result from the
subsequent
recruitment of P-arr-interacting proteins (e.g. clathrin and AP2 or signalling
proteins such as
c-Src, Raft, ERK1/2, ASK1 and JNK3) to the receptor-bound P-arr (Lefkowitz &
Whalen,
2004). Interestingly, a P-arr (R169E) mutant shown to bind to GPCRs in a
phosphorylation-
independent manner, probably as a result of a constitutively open conformation
(Kovoor et al,
1999) resulted, when inserted between Luc and YFP (Luc- P-arr(R169E)-YFP), in
basal and
AVP-stimulated BRET signals similar to those observed with WT Luc-P-arr-YFP
(Fig 4). This
indicates that the engagement of Parr by the activated receptor can be
detected by the
double brilliance Parr independently of the phosphorylation state of the
receptor.
[0094] A general biosensor to monitor GPCR activity: To assess whether Luc-P-
arr-YFP
could be used as a general GPCR activity sensor, a determination of whether
its agonist-
induced conformational change could be promoted by other receptors was made,
particularly
those of class A, which are believed to interact only transiently with P-arr.
Recruitment of
Luc-p-arr-YFP and agonist promoted intramolecular BRET were assessed in cells
coexpressing different receptors of class A (p2-AR, V1 vasopressin receptor
(V1 aR), d-opioid
receptor (6-OR)) and class B (platelet-activating factor receptor (PAFR), CC
chemokine
receptor type 5 (CCR5), angiotensin receptor type 1a (AT1aR)). As shown in Fig
5A, agonist
stimulation efficiently induced the recruitment of Luc-p-arr YFP to the plasma
membrane,
with the expected interaction patterns for all class A (transient) and class B
(stable)
receptors. In all cases, activation of Luc-P-arr-YFP mediated by class A and B
receptors
was accompanied by a significant increase in BRET (Fig 5B). Interestingly,
although the
kinetics and stability of the BRET increase were found to be similar for
receptors of class A
and B (data not shown), a tendency of class A receptors to induce smaller BRET
increases
was observed. As previously noted when comparing the BRET-detected recruitment
of P-arr
CA 02775278 2012-04-23
31
to class A P2-AR and class B V2R (Fig 2B), this probably indicates that the
BRET assays
provide a steady-state signal reflecting continuous rounds of association-
dissociation cycles.
In any case, these results suggest that Luc-P-arr-YFP can be used as a general
biosensor
to monitor GPCR activity and that the interaction can be monitored for
extended periods of
time making it compatible with its use in high through put screening assays
that request long
lived signals. When compared with the intermolecular BRET-based P-arr
recruitment assays
(Angers et al, 2000; Bertrand at al, 2002), double-brilliance P-arr avoids the
difficulty of
expressing the appropriate ratio of energy donor and acceptor constructs and
allows the
study of unmodified GPCRs.
[0095] The interaction of P-arr with the GRK-phosphorylated GPCRs is thought
to induce the
release of P-arr's C-tail and the opening of its structure (Gurevich and
Gurevich 2003),
subsequently leading to the recruitment of Parrestin-interacting proteins
(Lefkowitz and
Whalen 2004). To assess if the conformational change of P-arr detected with
the double
brilliance P-arr could also be detected using Parr mutants believed to be
constitutively in the
open state, assessment was made of the agonist-promoted BRET signal of two
other P-arr
mutants (P-arr(3A): 1387A, V388A, F389A; P-arr(IV): 1387A, V388A),inserted
between Luc
and YFP (Luc-Parr(3A)-YFP and Luc-Parr(IV)-YFP). These mutant P-arrs are
believed to be
constitutively active due to the disruption of the polar core keeping
Parrestin In a closed and
inactive conformation (Gurevich 1998). As shown in Fig 6, while the basal BRET
signal
observed with each Luc-Parr-YFP constitutively active mutant (Luc-Parr(3A)-YFP
and Luc-
Parr(IV)-YFP) was found to be similar to that of wild-type Luc-Parr-YFP, the
agonist-induced
BRET increase was significantly reduced by the mutations (Fig 6, inset).
[0096] In addition to agonists, the activity of ligands with inverse agonist
efficacy towards
specific signalling pathways can be detected by the double brilliance P-arr.
As shown in Fig
7, the V2R inverse agonist SR121463 that inhibits cyclic AMP production can
promote an
increase in the BRET signal in cells co-expressing wild type V2R and Luc-(3-
arr-YFP.
[0097] Double brilliance P-arr may also prove to be an effective tool in the
study of the
increasingly diverse roles played by P-arr, such as its involvement with
receptors other than
GPCRs and diverse signaling molecules in different systems (Fig. 8). A list of
some of the
proteins that have been shown to interact with Parr and which activity could
be monitored by
double brilliance P-arr is presented in Table 1. The spectrum of receptors
capable of utilizing
CA 02775278 2012-04-23
32
P-arr for endocytosis via clathrin binding sites has significantly increased
(Lefkowitz and
Whalen 2004a). For exemple, P-arr appears to be required for engulfing
Frizzled-4, an
atypical seven-transmembrane domain receptor, through interaction with the
adaptor protein
Dishevelled-2 phosphorylated by PKC (Chen et al. 2003a); for the endocytosis
of receptors
with serine/threonine kinase activity such as the transforming growth factor 0
receptor (TGF-
PR), in a manner dependent on the phosphorylation of Rill by RII (Chen et al.
2003b); as well
as for the endocytosis of the IGF1 receptor, in a manner that is independent
from its
phosphorylation (Dalle et al. 2001). This indicates that the Parr double
brillance could be a
general biosensor of the activity of many distinct receptors and signalling
molecules.
Table 1: List of proteins capable of interacting with P-arrestin
Binding Protein -arrestin isoform jyjLe of protein
Clathrin -arr 1, 2 trafficking
AP2 -arr 1, 2 trafficking
NSF -arr I trafficking
ARF6 -arr 2 1 Small G/GEFs
ARNO -arr 2 Small G/GEFs
Rai-GDS -arr 1, 2 Small G/GEFs
RhoA -arr 1 Small G/GEFs
MAPK cascade components: Signaling
ASK1 P-arr 1, 2
c-Raf-1 P-arr 1, 2
JNK3 P-arr 2, 1
ERK2 -arr 1, 2
Nonreceptor tyrosine kinases: signaling
c-Src P-arr 1, 2
Yes P-arr I
Hck P-arr 1
Fgr -arr 1
Others: signaling
Mdm2 P-arr 1, 2
IKBa P-arr 1, 2
PDE4D family P-arr 1, 2
Dishevelled P-arr 1, 2
PP2A -arr I
(Lefkowitz & Shenoy, 2005)
[0098] Double-brilliance 3-arr sensor (BRET2): In addition to the constructs
described
above, the following BRET2-based beta-arrestin 1 and 2 sensors: Acceptor
(mAmetrine;
CA 02775278 2012-04-23
33
sCFP3A; GFP10)-GSAGT-RArrestinl/2-KLPAT-Rlucll were also made (Fig 9a), using
similar
techniques, namely:
Ametrine-h(3arl r-Rlucll,
Ametrine-hparr2-Rlucl I,
UP-hparrl-Rlucll,
CFP-h3arr2-RluclI,
GFP10-h3arr1-Rlucll, and
GFP10-hF3arr2-Rlucll.
[0099] An enhanced Rluc variant (RIuc Ii) was used with the BRET2 versions as
it provides a
sustained and a stronger signal with coelenterazine 400a (200-400 times) than
with the WT
Rluc. The orientation of the BRET tags and nature of the linkers in these
constructs differ
from the BRET1 version. In contrast to the BRET1 sensors, these structure
leads to a
decrease BRET signal in response to an agonist-promoted GPCR activation. As
shown in
figure 9B and 2B, this inversion of the BRET signal between the BRET1 and
BRET2 versions
of the beta-arrestin db sensors still lead to similar kinetics and dose-
responses to an AVP
stimulation of V2R. Both beta-arrestin1 and 2 sensors are functional and give
similar
responses for the same receptor activation (Fig 9B).
[00100] The inversion of the signal between BRET1 and BRET2 provides a user
with
complementary tools that could be useful for high throughput. For example,
having sensors
with different responses (such as BRET1 and BRET2) could be useful to identify
compounds
that were mislabeled as active (ie. false positives) due to their colour and
not because of
their activity or binding.
[00101] In certain embodiments when a change in the orientation of the BRET
tags
(fluorophores and chromophores), leads to a difference in BRET response,
advantages may
be realized. Orientation of the tags and possibly the linkers as well, can be
adjusted and
result in a change in the nature of the BRET signal (increase or decrease).
Such modulation
of the BRET response through the orientation of the tags could provide the
user with a
complementary tool that can be used in screening assays, especially In high
throughput
screening assays, for example.
CA 02775278 2012-04-23
34
[00102] Both orientations of the BRET tags lead to sensors with similar Z'
factors
(reflecting the robustness of the assay) that could be adapted to a high
throughput screening
assay (HTS). The observed responses need not be attributed to the nature of
the BRET
tags. The nature of the tags may not necessarily cause the different BRET
responses
observed with the BRET1 vs BRET2 sensors.
[00103] The orientation of the donor vs acceptor may be a possible cause of
the
inversion of causing this inversion of the sensor's response. A BRET2
construct made with
the same orientation of the BRET1 construct shows a consistent increase in
BRET upon
stimulation.
[00104] The selection of a linker may impact the amplitude of the sensor's
response.
[00105] Having sensors with responses going in different directions could be
useful in
the validation process of an HTS screen as they could be used to identify
compounds
mislabelled as active because of their color and not because of their
activity.
[00106] The Z'-factor Is a reflection of the robustness of an assay and should
vary
depending on the experimental conditions and receptor used. With cells
transiently
expressing both V2R and sensors a Z'-factor between 0.43 and 0.63 (fig 10) was
obtained
for both BRET1 and BRET2 versions of the beta-arrestin db sensors, providing a
robust
assay for monitoring GPCR activation with both full and partial agonists (fig
9C). Using cell
lines stably expressing both receptor and sensor, an even better Z'-factor is
expected and
thus be sufficient to develop a high throughput screening (HTS) assay. Even
though the
response of the BRET1 vs BRET2 sensors is going in different directions,
similar Z' factors
are obtained.
[00107]
[00108] Since the fluorescent energy transfer is based on stimulatory
principles such
as BRET, a biosensor as described herein based on FRET instead of BRET would
also be
expected to function and is included herein.
[00109] In summary, the above is believed to be the first real-time monitoring
of
agonist-promoted conformational changes of 13-arr in living cells using a
double-brilliance S-
CA 02775278 2012-04-23
arr intramolecular BRET-based biosensor. The conformational rearrangement of
the [i-arr
molecule and its interaction with other proteins reflects its transition from
an inactive state to
a biologically active state that follows its initial recruitment to activated
GPCRs and involves
the relative movement of the C-and N-terminus leading to a change in the BRET
signal Beta-
arrestin db sensors offer a robust assay for GPCR activation and
characteristics
(unimolecular structure, ratiometric signal and recruited to most GPCRs) that
could be
amenable to large-scale screening campaigns. In conclusion, double brilliance
R-arr
represents the first intramolecular BRET-based biosensor that allows the
monitoring of
protein conformational changes. This should lead the way to the development of
similar tools
to study other proteins believed to undergo significant conformational
rearrangement linked
to their function.
[00110] Although the present invention has been described by way of specific
embodiments and examples thereof, with a particular focus on G protein-coupled
receptors,
it should be noted that it will be apparent to persons skilled in the art that
modifications may
be applied to the present particular embodiments described.
LIST OF REFERENCES:
1. Andrews DL, Demidov AA (1999) Resonance Energy Transfer. Chichester, UK:
Wiley
2. Angers S, Salahpour A, Joly E, Hilairet S, Chelsky D, Dennis M, Bouvier M
(2000)
Detection of R2-adrenergic receptor dimerization in living cells using
bioluminescence
resonance energy transfer (BRET). Proc Natl Acad Sci USA 97: 3684-3689
3. Azzi M, Charest PG, Angers S, Rousseau G, Kohout T, Bouvier M, Pineyro G
(2003)
Arrestin-mediated activation of MAPK by inverse agonists reveals distinct
active
conformations for G protein-coupled receptors. PNAS 100: 11406-11411
4. Bertrand L, Parent S, Caron M, Legault M, Joly E, Angers S, Bouvier M,
Brown M, Houle
B, Menard L (2002) The BRET2/arrestin assay in stable recombinant cells: a
platform to
screen for compounds that interact with G protein-coupled receptors (GPCRs). J
Receptor Signal Transduction Res 22: 533-541
CA 02775278 2012-04-23
36
5. Charest PG, Bouvier M (2003) Palmitoylation of the V2 vasopressin receptor
carboxyl tail
enhances Q-arrestin recruitment leading to efficient receptor endocytosis and
ERK1/2
activation. J Biol Chem 278: 41541-41551
6. Chen W, Kirkbride KC, How T, Nelson CD, Mo J, Frederick JP, Wang XF,
Lefkowitz RJ,
Blobe GC (2003) Beta-arrestin 2 mediates endocytosis to type III TFG-beta
receptor and
down-regulation of its signalling. Science 301 (5638): 1394-1397
7. Dalle S, Ricketts W, Imamura T, Voilenweider P, Olefsky JM (2001) Insulin
and insulin-
like growth factor I receptors utilize different G protein signalling
components. J Biol
Chem 276 (19): 15688-15695
8. Gurevich W, Benovic JL (1993) Visual arrestin interaction with rhodopsin.
Sequential
multisite binding ensures strict selectivity toward lightactivated
phosphorylated rhodopsin.
J Biol Chem 268: 11628-11638
9. Gurevich W, Gurevich EV (2003) The new face of active receptor bound
arrestin attracts
new partners. Structure (Camb) 11: 1037-1042
10. Han M, Gurevich W, Vishnivetskiy SA, Sigler PB, Schubert C (2001) Crystal
structure of
R-arrestin at 1.9 A : possible mechanism of receptor binding and membrane
translocation. Structure (Camb) 9: 869-880
11. Hebert TE, Moffett S, Morello JP, Loisel TP, Bichet DG, Barret C, Bouvier
M (1996) A
peptide derived from a R2-adrenergic receptor transmembrane domain inhibits
both
receptor dimerization and activation. J Biol Chem 271: 16384-16392
12. Hirsch JA, Schubert C, Gurevich W, Sigler PB (1999) The 2.8A crystal
structure of
visual arrestin: a model for arrestin's regulation. Cell 97: 257-269
13. Kovoor A, Celver J, Abdryashitov RI, Chavkin C, Gurevich W (1999) Targeted
construction of phosphorylation-independent R-arrestin mutants with
constitutive activity
in cells. J Biol Chem 274: 6831-6834
CA 02775278 2012-04-23
37
14. Leduc M, Breton B, Gales C, Le Gouill C, Bouvier M, Chemtob S, Heveker N
(2009)
Functional selectivity of natural and synthetic prostaglandin EP4 receptor
ligands J
Pharmacol Exp Ther. 331(1):297-307
15. Lefkowitz RJ, Whalen EJ (2004) (3-anrestins: traffic cops of cell
signalling. Curr Opin Cell
Biol 16: 162-168
16. Lefkowitz RJ, Shenoy SK (2005) Transduction of Receptor Signals by (3-
arrestins.
Science 308: 512-517
17. Loening AM, Fenn TD, Wu AM, Gambhir SS. (2006) Consensus guided
mutagenesis of
Renilla luciferase yields enhanced stability and light output. Protein Eng Des
Sel.
19(9):391-400.
18. Lin FT, Miller WE, Luttrell LM, Lefkowitz RJ (1999) Feedback regulation of
R-arrestinl
function by extracellular signal-regulated kinases. J Biol Chem 274: 15971-
15974
19. Lin FT, Chen W, Shenoy S, Cong M, Exum ST, Lefkowitz RJ (2002)
Phosphorylation of
0-arrestin2 regulates its function in internalization of b(2)-adrenergic
receptors.
Biochemistry 41: 10692-10699
20. Luttrell LM, Lefkowitz RJ (2002) The role of R-arrestins in the
termination and
transduction of G-protein-coupled receptor signals. J Cell Sci 115: 455-465
21. Marrache AM et a/ (2002) Proinflammatory gene induction by
plateletactivating factor
mediated via its cognate nuclear receptor. J Immunol 169: 6474-6481
22. Mercier JF, Salahpour A, Angers S, Breit A, Bouvier M (2002) Quantitative
assessment of
b1- and 32-adrenergic receptor homo- and heterodimerization by bioluminescence
resonance energy transfer. J Biol Chem 277: 44925-44931
23. Milligan G (2004) Applications of bioluminescence- and fluorescence
resonance energy
transfer to drug discovery at G protein-coupled receptors. Eur J Pharm Sci 21:
397-405
24. Oakley RH, Laporte SA, Holt JA, Caron MG, Barak LS (2000) Differential
affinities of
visual arrestin, R-arrestin1, and R-arrestin2 for G proteincoupled receptors
delineate two
major classes of receptors. J Biol Chem 275: 17201-17210
CA 02775278 2012-04-23
38
25. Oakley RH, Laporte SA, Holt JA, Barak LS, Caron MG (2001) Molecular
determinants
underlying the formation of stable intracellular G proteincoupled receptor-p-
arrestin
complexes after receptor endocytosis. J Biol Chem 276: 19452-19460
26. Perroy J, Adam L, Qanbar R, Chenier S, Bouvier M (2003)
Phosphorylationindependent
desensitization of GABA(B) receptor by GRK4. EMBO J 22: 3816-3824
27. Petaja-Repo UE, Hogue M, Bhalla S, Laperriere A, Morello JP, Bouvier M
(2002) Ligands
act as pharmacological chaperones and increase the efficiency of d-opioid
receptor
maturation. EMBO J 21: 1628-1637
28. Pleskoff 0, Treboute C, Brelot A, Heveker N, Seman M, Alizon M (1997)
Identification of
a chemokine receptor encoded by human cytomegalovirus as a cofactor for HIV-1
entry.
Science 276: 1874-1878
29. Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory
Manual. New
York: Cold Spring Harbor Laboratory Press
30. Terrillon S, Durroux T, Mouillac B, Breit A, Ayoub MA, Taulan M, Jockers
R, Barberis C,
Bouvier M (2003) Oxytocin and vasopressin V1 a and V2 receptors form
constitutive
homo- and heterodimers during biosynthesis. Mol Endocrinol 17: 677-691
31. Vishnivetskiy SA, Hirsch JA, Velez MG, Gurevich YV, Gurevich W (2002)
Transition of
arrestin into the active receptor-binding state requires an extended
interdomain hinge. J
Biol Chem 277: 43961-43967
32. Xiao K, Shenoy SK, Nobles K, Lefkowitz RJ (2004) Activation dependent
conformational
changes in R-arrestin 2. J Biol Chem 279: 55744-55753.
33. Zhang J, Campbell RE, Ting AY, Tsien RY (2002) Creating new fluorescent
probes for
cell biology. Nat Rev Mol Cell Biol 3: 906-918Zhang JH, Chung TD, Oldenburg
KR.
(1999) A Simple Statistical Parameter for Use in Evaluation and Validation of
High
Throughput Screening Assays. J Biomol Screen. 4(2):67-73.