Note: Descriptions are shown in the official language in which they were submitted.
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
IMPROVED COMPLEMENT RECEPTOR 2 (CR2) TARGETING GROUPS
CROSS-REFERENCE TO RELATED APPLICATIONS
[0001] This application claims priority benefit to U.S. Provisional Patent
Application No.
61/345,035, filed May 14, 2010, the disclosure of which is hereby incorporated
herein by
reference in its entirety.
TECHNICAL FIELD
[0002] This application pertains to improved compositions for targeted
delivery of
therapeutics, including complement inhibitors, to sites of inflammation.
STATEMENT REGARDING FEDERALLY SPONSORED RESEARCH OR
DEVELOPMENT
[0003] This invention was made in part during work supported by Grant No. RO1-
CA53617
from the National Institutes of Health. The government has certain rights in
the invention.
BACKGROUND
[0004] Human complement receptor 2 (CR2/CD21) is a 145 kiloDalton ("kDa")
transmembrane protein comprised of 15 or 16 short consensus repeat (SCR)
extracellular
domains, a 28 amino acid single pass transmembrane domain and a short 34 amino
acid
intracellular domain (1-5). Each of the extracellular SCRs comprises
approximately 60-70
amino acid residues and is connected by linker regions of three to eight amino
acid residues.
All SCRs contain a number of conserved amino acid residues including four
cysteine
residues, which form a pattern of disulfide bridges connecting Cysl-Cys3 and
Cys2-Cys4.
CR2 is primarily present on B cells, where it is found in complex with other
membrane
proteins that promote normal humoral and cellular immune responses (6-9).
Using the most
distally located (i.e., amino-terminal) SCR domains, SCR1-2, CR2 binds four
classes of
ligands - complement component 3 (C3) proteolytic fragments iC3b, C3dg and C3d
(10, 11);
1
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
the Epstein-Barr virus (EBV) glycoprotein gp350/220 (gp350) (12-14); the low
affinity IgE
receptor CD23 (15, 16); and the cytokine interferon alpha (IFNa) (17-19).
[0005] The primary role of CR2 is to function as a B cell co-receptor for
antigen-mediated B
cell activation through enhanced signal transduction (20, 21). This function
is carried out
through co-ligation via C3d and surface IgM, when C3d is covalently attached
to an antigen
(22-28). CR2 is also the obligate cellular receptor for EBV through its
envelope surface
glycoprotein gp350 (12, 20, 29-3 1). Actual cellular EBV infection is achieved
after the
ligation of CR2 to gp350 tethers the virus close enough to the cell surface
(14, 32, 33),
allowing viral gp42 to bind human leukocyte antigen class II molecules (34,
35) and
subsequently triggering host cell fusion via three additional viral
glycoproteins gB, gH and
gL (36-38). IFNa has been shown to be a ligand of CR2, though the physiologic
importance
of this interaction remains unclear (17-19). It has been suggested, however,
that IFNa and
CR2 may be involved in the development of the autoimmune disease systemic
lupus
erythematosus (39-41).
[0006] Mutagenesis studies along with structural studies of the CR2-gp350
interaction have
suggested residues on CR2 that are required for the interaction (20, 42, 43).
ELISA and flow
cytometry was used to test candidate CR2 mutants for the binding of gp350 and
CR2 (20, 42,
43). In recent studies specific residues on CR2 which were found to have a
deleterious effect
on gp350-binding when mutated included R13, S15, R28, R36, K41, K57, K67, R83
and R89
(42, 43). In separate work residues P8-S 15 within the first conserved inter-
cysteine region of
SCR1 and the linker region between SCR1 and SCR2 were also highlighted as
being essential
for gp350-binding to occur (20). These data, in conjunction with separate
mutagenesis
analyses targeting the gp350 molecule were used to drive an in silico model of
the CR2-
gp350 interaction utilizing the soft docking program HADDOCK (43-45). This
analysis
suggested that the primary interaction on CR2 was between SCR1 and the linker
region
joining SCR1 to SCR2, and for gp350, the linker region between domain 1 and
domain 2
(43).
[0007] CR2 has been suggested as a receptor for IFNa by the finding that IFNa
mimics both
gp350 and C3d binding, and the observation that all three ligands bind a
similar region on
CR2 (18, 19). The mimicry was shown to be functional as well (18). After both
the C3d and
IFNa structures were solved, the putative CR2 binding sequence was found to
have similar
2
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
structural motifs. IFNa has been described as being able to bind to multiple
forms of CR2
from full length to SCR1-2, although to varying degrees (17). Though CR2 has
been shown
to be a receptor for IFNa, the IFNa binding site within CR2 SCR1-2 is unknown.
[0008] Further analysis of CR2 interactions with known ligands to identify
specific amino
acid residues involved in binding to these ligands would enable the design of
modified CR2
molecules with defined binding specificity for each known CR2 ligand (e.g., C3
proteolytic
fragments iC3b, C3dg and C3d; EBV glycoprotein gp350; CD23; and IFNa..
BRIEF SUMMARY
[0009] Provided herein are compositions and methods directed to soluble
proteins which can
selectively deliver modulators of complement activity. Targeted delivery of
these modulators
is accomplished by selectively mutating particular amino acids in a targeting
protein portion
of the composition corresponding to at least the first two N-terminal SCR
domains of CR2.
Depending on the particular combination of mutations introduced into the
targeting portion, a
complement activity modulator can be selectively delivered to particular
ligands of CR2 at
sites where complement system activation or suppression is desired.
[0010] Accordingly, in one aspect, provided herein are soluble compositions
comprising a
construct, wherein the construct comprises: (a) a complement receptor 2 (CR2)
portion
comprising a CR2 protein of SEQ ID NO:1 or a biologically active fragment
thereof, wherein
the CR2 portion contains at least the first two N-terminal SCR domains of the
CR2 protein;
and (b) a complement modulator portion; wherein the CR2 portion contains at
least one
amino acid substitution at an amino acid residue selected from the group
consisting of: N11,
R36, K41, Y64 and K67. In certain embodiments, the construct is a fusion
protein.
[0011] In certain embodiments, the complement modulator portion comprises a
complement
inhibitor or biologically-active fragment thereof. In certain embodiments, the
complement
inhibitor or biologically active fragment thereof is selected from the group
consisting of
human membrane complement protein (MCP)(SEQ ID NO: 10), human decay
accelerating
factor (DAF) (SEQ ID NO: 11), mouse DAF (SEQ ID NO: 12), mouse complement
receptor 1-
related gene/protein y (Crry) (SEQ ID NO:4), human CD59 (SEQ ID NO:3), mouse
CD59
isoform A (SEQ ID NO:6), mouse CD59 isoform B (SEQ ID NO:7), human complement
3
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
receptor 1 (CR1) (SEQ ID NO:9), human factor H (SEQ ID NO:5), and mouse factor
H (SEQ
ID NO:8).
[0012] In certain embodiments, the complement inhibitor comprises human
membrane
complement protein (MCP) (SEQ ID NO: 10) or a biologically-active fragment
thereof. In
certain embodiments, the biologically active fragment of human MCP (SEQ ID NO:
10) is
selected from the group consisting of SCR 1-4 (amino acids 35-285 of SEQ ID
NO: 10),
SCR 1-4 plus the serine/threonine-rich domain (amino acids 35-326 of SEQ ID
NO: 10), and
the extracellular domain of MCP (amino acids 35-343 of SEQ ID NO: 10). In
certain
embodiments, the complement inhibitor comprises human DAF (SEQ ID NO: 11) or a
biologically-active fragment thereof. In certain embodiments, the biologically
active
fragment of human DAF (SEQ ID NO: 11) is selected from the group consisting of
SCR 1-4
(amino acids 25-285 of SEQ ID NO: 11) and SCR 1-4 plus the O-glycosylated
serine/threonine-rich domain (amino acids 25-353 of SEQ ID NO: 11). In certain
embodiments, the complement inhibitor comprises mouse DAF (SEQ ID NO: 12) or a
biologically-active fragment thereof. In certain embodiments, the biologically
active
fragment of mouse DAF (SEQ ID NO: 12) is selected from the group consisting of
SCR 1-4
(amino acids 35-286 of SEQ ID NO: 12) and SCR 1-4 plus the O-glycosylated
serine/threonine-rich domain (amino acids 35-362 of SEQ ID NO: 12). In certain
embodiments, the complement inhibitor comprises Crry (SEQ ID NO:4) or a
biologically-
active fragment thereof. In certain embodiments, the biologically-active
fragment of Crry
(SEQ ID NO:4) is selected from the group consisting of SCR 1-5 (amino acids 41-
400 of SEQ
ID NO:4) and the extracellular domain mouse Crry protein (amino acids 41-405
of SEQ ID
NO:4). In certain embodiments, the complement inhibitor comprises human CD59
(SEQ ID
NO:3) or a biologically-active fragment thereof. In certain embodiments, the
biologically-
active fragment of human CD59 (SEQ ID NO:3) comprises the extracellular domain
of
human CD59lacking its GPI anchor (amino acids 26-101 of SEQ ID NO:3). In
certain
embodiments, the complement inhibitor comprises mouse CD59 isoform A (SEQ ID
NO:6)
or a biologically-active fragment thereof. In certain embodiments, the
biologically-active
fragment of mouse CD59 isoform A (SEQ ID NO:6) comprises the extracellular
domain of
mouse CD59, isoform A lacking its GPI anchor (amino acids 24-95 of SEQ ID
NO:6). In
certain embodiments, the complement inhibitor comprises mouse CD59 isoform B
(SEQ ID
NO:7) or a biologically-active fragment thereof. In certain embodiments, the
biologically-
active fragment of mouse CD59 isoform B (SEQ ID NO:7) comprises the
extracellular
4
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
domain of mouse CD59, isoform B lacking its GPI anchor (amino acids 24-103 of
SEQ ID
NO:7). In certain embodiments, the complement inhibitor comprises human CR1
(SEQ ID
NO:9) or a biologically-active fragment thereof. In certain embodiments, the
biologically-
active fragment of human CR1 (SEQ ID NO:9) is selected from the group
consisting of
SCR1-3 (amino acids of 42-234 of SEQ ID NO:9), SCR1-4 (amino acids 42-295 of
SEQ ID
NO:9), SCR1-10 (amino acids 42-684 of SEQ ID NO:9), SCR8-10 (amino acids of
491-684
of SEQ ID NO:9), SCR 8-11 (amino acids 491-745 of SEQ ID NO:9), SCR15-17
(amino
acids of 941-1134 of SEQ ID NO:9), SCR15-18 (amino acids 941-1195 of SEQ ID
NO:9),
and SCR22-28 (amino acids 1394-1842 of SEQ ID NO:9). In certain embodiments,
the
complement inhibitor comprises human factor H (SEQ ID NO:5) or a biologically-
active
fragment thereof. In certain embodiments, the biologically-active fragment of
human factor H
(SEQ ID NO:5) is selected from the group consisting of SCR 1-4 (amino acids 21-
262 of SEQ
ID NO:5), SCR1-5 (amino acids 21-320 of SEQ ID NO:5), SCR1-8 (amino acids 21-
507 of
SEQ ID NO:5), and SCR1-18 (amino acids 21-1104 of SEQ ID NO:5). In certain
embodiments, the complement inhibitor comprises mouse factor H (SEQ ID NO:8)
or a
biologically-active fragment thereof. In certain embodiments, the biologically-
active
fragment of mouse factor H (SEQ ID NO:8) is selected from the group consisting
of SCR 1-4
(amino acids 19-264 of SEQ ID NO:8), SCR 1-5 (amino acids 19-322 of SEQ ID
NO:8),
SCR1-8 (amino acids 19-507 of SEQ ID NO:8), and SCR1-18 (amino acids 19-1109
of SEQ
ID NO:8).
[0013] In certain embodiments, the complement modulator portion comprises a
complement
activator or biologically-active fragment thereof. In certain embodiments, the
complement
activator or biologically-active fragment thereof is selected from the group
consisting of
human IgGi, human IgGi Fc domain, human IgM, human IgM Fc domain, mouse IgG3,
mouse IgG3 Fc domain, mouse IgM, mouse IgM Fc domain, and cobra venom factor
(CVF).
[0014] In certain embodiments, the construct exhibits decreased binding
affinity for EBV-
gp350 or IFNa compared to a construct in which the CR2 or biologically active
fragment
thereof does not contain any amino acid substitution. In certain embodiments,
the construct
exhibits decreased binding affinity for EBV-gp350 compared to a construct in
which the CR2
or biologically active fragment thereof does not contain any amino acid
substitution. In
certain embodiments, the CR2 or biologically active fragment thereof contains
at least one
amino acid substitution of an amino acid residue selected from the group
consisting of: N11,
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
R36, K41, Y64 and K67. In certain embodiments, the construct exhibits
decreased binding
affinity for IFNa compared to a construct in which the CR2 or biologically
active fragment
thereof does not contain any amino acid substitution. In certain embodiments,
the CR2 or
fragment thereof contains at least one amino acid substitution to an amino
acid residue
selected from the group consisting of: S42 and K50.
[0015] In another aspect, provided herein are methods of reducing the binding
affinity of a
construct comprising: (a) a complement receptor 2 (CR2) portion comprising a
CR2 protein
of SEQ ID NO:1 or a biologically active fragment thereof, wherein the CR2
portion contains
at least the first two N-terminal SCR domains of the CR2 portion; and (b) a
complement
modulator portion, for EBV-gp350, comprising mutating at least one amino acid
residue
selected from the group consisting of N11, R36, K41, Y64 and K67.
[0016] In another aspect, provided herein are methods of reducing the binding
affinity of a
contruct comprising: (a) a complement receptor 2 (CR2) portion comprising a
CR2 protein of
SEQ ID NO:1 or a biologically active fragment thereof, wherein the CR2 portion
contains at
least the first two N-terminal SCR domains of the CR2 portion; and (b) a
complement
modulator portion, for IFNa, comprising mutating at least one amino acid
residue selected
from the group consisting of S42 and K50.
BRIEF DESCRIPTION OF THE DRAWINGS
[0017] Figure 1. NMR titration analysis reveals that SCR1 and SCR2 of CR2 are
both
involved in ligating gp350. Two superimposed 1H-15N Transverse Relaxation
Optimized
Spectroscopy-Heteronuclear Single Quantum Coherence (TROSY-HSQC) spectra of
15N-
labled CR2 SCR1-2 (0.6 mM in 1/3X PBS) collected during titration with
increasing amounts
of gp350. Black, no gp350 and grey, saturating amounts of gp350. Inset,
detailed view of
chemical shift change. The numbering scheme used here for CR2 is based on the
amino acid
sequence for the mature protein.
[0018] Figure 2. NMR titration analysis reveals that SCR1 and SCR2 of CR2 are
both
involved in ligating IFNa. Five superimposed 1H-15N TROSY-HSQC spectra of 15N-
labled
6
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
CR2 SCR1-2 (0.6 mM in 1/3X PBS) collected during titration with increasing
amounts of
IFNa.
[0019] Figure 3. NMR derived CR2-ligand binding residue comparison. Histogram
illustrates chemical shift changes induced in the backbone amides of CR2 SCR1-
2 upon
binding C3d, IFNa or gp350.
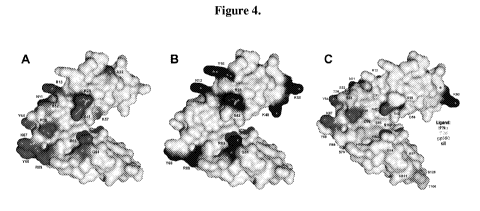
[0020] Figures 4A, 4B and 4C. Surface representation of CR2 SCR1-2 x-ray
crystal
structure in its ligand-bound state (C3d not shown) with NMR-determined ligand
binding residues. Figure 4A. NMR-determined gp350 binding residues. Gray
residues
represent residues unaffected by gp350 titration. The black residues on SCR 1,
the linker
region and SCR2 represent residues involved in gp350 binding to CR2 SCR 1-2.
Figure 4B.
NMR determined IFNa binding residues. Gray residues represent residues
unaffected by
IFNa titration. The black residues on SCR1, the linker region and SCR2
represent residues
involved in IFNa binding to CR2 SCR 1-2. Figure 4C. NMR determined ligand
unique and
shared binding residues. The black residues represent residues that are
uniquely involved in
CR2 binding to IFNa and gp350. The dark grey residues represent residues that
are uniquely
involved in CR2 binding to C3d. The light grey residues represent residues
that are involved
in all three CR2 ligand binding events.
[0021] Figure 5. HADDOCK CR2-gp350 docking model with NMR derived CR2-gp350
ligand binding residues highlighted. Model from Young, et al (43). Black
ribbons represent
gp350 and light grey represents glycosyl-groups that decorate the surface of
gp350. Dark
grey ribbons represent CR2 SCR 1-2. Inset, magnified view of theoretical side-
chain
interactions between NMR derived binding residues and gp350 mapped on the
docking
model of Young, et al.
[0022] Table 1. CR2 binding constants from NMR titrations and ITC. Shown are
weak
and upper limit to tight binding constants for CR2-ligand interactions
determined using NMR
titrations monitoring chemical shift changes. Also shown are CR2-ligand
binding constants
determined using ITC. UL, upper limit. ITC, isothermal titration calorimetry.
[0023] Table 2. Comparison of CR2 ligand binding residues. Shown are residues
involved in each CR2-ligand binding interaction. Residues with an asterisk are
unique to the
respective binding interaction.
7
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
BRIEF DESCRIPTION OF THE SEQUENCES
[0024] SEQ ID NO:1 is the complete amino acid sequence of human complement
receptor 2
(CR2).
[0025] SEQ ID NO:2 is the complete amino acid sequence of short consensus
repeat (SCR)
domains 1 and 2 of human CR2.
[0026] SEQ ID NO:3 is the complete amino acid sequence of human CD59 protein.
[0027] SEQ ID NO:4 is the complete amino acid sequence of mouse complement
receptor 1-
related gene/protein y (Crry).
[0028] SEQ ID NO:5 is the complete amino acid sequence of human factor H.
[0029] SEQ ID NO:6 is the complete amino acid sequence of mouse CD59A protein.
[0030] SEQ ID NO:7 is the complete amino acid sequence of mouse CD59B protein.
[0031] SEQ ID NO:8 is the complete amino acid sequence of mouse factor H.
[0032] SEQ ID NO:9 is the complete amino acid sequence of human complement
receptor 1
(CR1).
[0033] SEQ ID NO:10 is the complete amino acid sequence of human membrane
cofactor
protein (MCP).
[0034] SEQ ID NO: 11 is the complete amino acid sequence of human decay
accelerating
factor (DAF/CD55).
[0035] SEQ ID NO: 12 is the complete amino acid sequence of mouse decay
accelerating
factor (DAF/CD55).
[0036] SEQ ID NO: 13 is the complete amino acid sequence of cobra venom factor
(CVF)
from the monocled cobra (Naja kaouthia).
[0037] SEQ ID NO: 14 is the complete amino acid sequence of the human IgGi
heavy chain,
C domain.
8
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
[0038] SEQ ID NO: 15 is the complete amino acid sequence of the human IgG1
light chain, C
domain.
[0039] SEQ ID NO: 16 is the complete amino acid sequence of the Fc domain of
human IgGi.
[0040] SEQ ID NO: 17 is the complete amino acid sequence of human IgM heavy
chain, C
domain.
[0041] SEQ ID NO: 18 is the complete amino acid sequence of human IgM light
chain, C
domain.
[0042] SEQ ID NO: 19 is the complete amino acid sequence of the Fc domain of
human IgM.
[0043] SEQ ID NO:20 is the complete amino acid sequence of mouse IgG3 heavy
chain, C
domain.
[0044] SEQ ID NO:21 is the complete amino acid sequence of mouse IgG3 light
chain, C
domain.
[0045] SEQ ID NO:22 is the complete amino acid sequence of mouse IgG3 Fc
domain.
[0046] SEQ ID NO:23 is the complete amino acid sequence of mouse IgM heavy
chain, C
domain.
[0047] SEQ ID NO:24 is the complete amino acid sequence of mouse IgM light
chain, C
domain.
[0048] SEQ ID NO:25 is the complete amino acid sequence of mouse IgM Fc
domain.
[0049] SEQ ID NO:26 is a linking sequence between the first two N-terminal
SCRs of
human CR2.
[0050] SEQ ID NO:27 is a linking sequences between the first two N-terminal
SCRs of
human CR2.
[0051] SEQ ID NO:28 is a linking sequence between the fourth and the fifth N-
terminal short
consensus repeat domains of human CR2.
9
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
DETAILED DESCRIPTION
[0052] Complement is an important component of immunity, but inappropriate and
excessive
activation of the complement system is involved in numerous pathological and
inflammatory
conditions. Complement activation products that mediate tissue injury are
generated at
various points in the complement pathway. Complement activation on cell
surfaces results in
the cleavage of serum complement component 3 (C3) and the covalent attachment
of C3
fragments that serve as opsonins for immune effector cells to the cell
surfaces. The resulting
C3 fragments include C3a, a soluble peptide that is a potent anaphylatoxin,
and C3b, a
component of the alternative complement pathway C3 convertase. Later in the
pathway,
serum complement component 5 (C5) is cleaved to release soluble CSa, another
potent
anaphylatoxin and chemoattractant with a wide range of bioactive properties.
Cleavage of C5
also initiates formation of the membrane attack complex (MAC), a cytolytic
protein complex
that assembles in cell membranes, ultimately resulting in lysis of opsonized
cells.
[0053] Complement component 3 (C3) is a zymogen. Intact C3 circulates at high
concentrations (1-2 mg/ml). M. Janzi et al., Mol. Cell. Proteomics (2005)
4(12):1942-1947.
During complement activation, whole C3 is cleaved to form C3b, a component of
the
alternative complement pathway C3 convertase, which becomes covalently bound
to target
surfaces. Endogenous complement regulatory proteins inactivate tissue-bound
C3b to form
iC3b and eventually the 35 kilodalton ("kD") C3d fragment. The C3d fragment
remains fixed
to tissues and serves as a durable marker of complement-mediated inflammation.
I. Leivo et
al., J. Cell. Biol. (1986) 103:1091-1100.
[0054] Targeted delivery of complement inhibitors to sites of complement
activation and
disease can improve their efficacy. Since complement plays an important role
in host defense
and the shaping of immunity, as well as in immune homeostatic mechanisms such
as immune
complex catabolism and apoptotic cell clearance, targeted delivery of
complement inhibitors
reduces potentially serious side effects resulting from systemic complement
inhibition,
particularly long-term complement inhibition.
General Techniques
[0055] The practice of the present invention will employ, unless otherwise
indicated,
conventional techniques of molecular biology, microbiology, cell biology,
biochemistry, and
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
nucleic acid chemistry which are well known to those skilled in the art. Such
techniques are
explained fully in the literature, such as, Molecular Cloning: A Laboratory
Manual, second
edition (Sambrook et al., 1989) and Molecular Cloning: A Laboratory Manual,
third edition
(Sambrook and Russell, 2001), (jointly referred to herein as "Sambrook");
Current Protocols
in Molecular Biology (F.M. Ausubel et al., eds., 1987, including supplements
through 2001);
PCR: The Polymerase Chain Reaction (Mullis et al., eds., 1994); Current
Protocols in
Nucleic Acid Chemistry John Wiley & Sons, Inc., New York, 2000), Handbook of
Experimental Immunology, 4th edition (D. M. Weir & C. C. Blackwell, eds.,
Blackwell
Science Inc., 1987); and Gene Transfer Vectors for Mammalian Cells (J. M.
Miller & M. P.
Calos, eds., 1987).
Complement Receptor 2
[0056] Human complement receptor 2, also referred to as CD21 (CR2/CD21)(SEQ ID
NO:1
and SEQ ID NO:2), is a -145 kD transmembrane protein of the C3 binding protein
family
comprising 15 or 16 short consensus repeat (SCR) domains, structural units
characteristic of
such proteins. CR2 is expressed on mature B cells and follicular dendritic
cells, and plays an
important role in humoral immunity. J. Hannan et al., Biochem. Soc. Trans.
(2002) 30:983-
989; K.A. Young et al., J. Biol. Chem. (2007) 282(50):36614-36625. CR2 protein
does not
bind intact C3 protein, but binds its breakdown products, including the C3b,
iC3b, and C3d
cleavage fragments, via a binding site located within the first two amino-
terminal short
consensus repeats ("SCRs 1-2") of the CR2 protein. Consequently, the SCR 1-2
domain of
CR2 discriminates between cleaved (i.e., activated) forms of C3 and intact
circulating C3. As
a targeting group, SCRs 1-2 of CR2 are therefore able to discriminate between
circulating C3
and the C3 fragments generated during complement activation. While the
affinity of CR2 for
C3d is only 620-658 nM Q. Hannan et al., Biochem. Soc. Trans. (2002) 30:983-
989; J.M.
Guthridge et al., Biochem. (2001) 40:5931-5941), the avidity of CR2 for
clustered C3d makes
it an effective method of targeting molecules to sites of complement
activation.
[0057] Cleavage of C3 results initially in the generation and deposition of
C3b on the
activating cell surface. The C3b fragment is involved in the generation of
enzymatic
complexes that amplify the complement cascade. On a cell surface, C3b is
rapidly converted
to inactive iC3b, particularly when deposited on a host surface containing
regulators of
complement activation (i.e., most host tissue). Even in the absence of
membrane-bound
11
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
complement regulators, substantial levels of iC3b are formed because of the
action of serum
factor H and serum factor I. iC3b is subsequently digested to the membrane-
bound fragments
C3dg and then C3d by factor I and other proteases and cofactors, but this
process is relatively
slow. Thus, the C3 ligands for CR2 are relatively long lived once they are
generated and are
present in high concentrations at sites of complement activation.
Definitions
[0058] General reference to "the composition" or "compositions" includes and
is applicable
to compositions of the invention.
[0059] As used herein, the singular form of the articles "a," "an," and "the"
includes plural
references unless indicated otherwise. For example, the phrase "a biologically
active CR2
fragment" includes one or more biologically active CR2 fragments.
[0060] Reference to "about" a value or parameter herein includes (and
describes)
embodiments that are directed to that value or parameter per se. For example,
description
referring to "about X" includes description of "X."
[0061] It is understood that aspects and embodiments of the invention
described herein
include consisting and/or consisting essentially of aspects and embodiments.
[0062] As used herein, the term "individual" refers to a vertebrate,
preferably a mammal,
more preferably a human. Mammals include, but are not limited to, research
animals,
domestic animals, farm animals, sport animals, pets, primates, mice and rats.
In certain
embodiments, the individual is human. In certain embodiments, the individual
is an
individual other than a human. In certain embodiments, the individual is an
animal model for
the study of a disease in which the alternative complement pathway is
implicated.
[0063] It is intended that every maximum numerical limitation given throughout
this
specification includes every lower numerical limitation, as if such lower
numerical
limitations were expressly written herein. Every minimum numerical limitation
given
throughout this specification will include every higher numerical limitation,
as if such higher
numerical limitations were expressly written herein. Every numerical range
given throughout
this specification will include every narrower numerical range that falls
within such broader
numerical range, as if such narrower numerical ranges were all expressly
written herein.
12
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
Amino Acid Substitutions
[0064] Twenty amino acids are commonly found in proteins. Those amino acids
can be
grouped into nine classes or groups based on the chemical properties of their
side chains.
Substitution of one amino acid residue for another within the same class or
group is referred
to herein as a "conservative" substitution. Conservative amino acid
substitutions can
frequently be made in a protein without significantly altering the
conformation or function of
the protein. Substitution of one amino acid residue for another from a
different class or group
is referred to herein as a "non-conservative" substitution. In contrast, non-
conservative amino
acid substitutions tend to disrupt conformation and function of a protein.
Table 3: Example of amino acid classification
Small/Aliphatic residues: ly, Ala, Val, Leu, Ile
Cyclic Imino Acid: Pro
Hydroxyl-containing Residues: Ser, Thr
Acidic Residues: Asp, Glu
Amide Residues: sn, Gln
Basic Residues: Lys, Arg
Imidazole Residue: His
Aromatic Residues: he, Tyr, Trp
Sulfur-containing Residues: Met, Cys
[0065] In certain embodiments, the conservative amino acid substitution
comprises
substituting any of glycine (G), alanine (A), isoleucine (I), valine (V), and
leucine (L) for any
other of these aliphatic amino acids; serine (S) for threonine (T) and vice
versa; aspartic acid
(D) for glutamic acid (E) and vice versa; glutamine (Q) for asparagine (N) and
vice versa;
lysine (K) for arginine (R) and vice versa; phenylalanine (F), tyrosine (Y)
and tryptophan
(W) for any other of these aromatic amino acids; and methionine (M) for
cysteine (C) and
13
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
vice versa. Other substitutions can also be considered conservative, depending
on the
environment of the particular amino acid and its role in the three-dimensional
structure of the
protein. For example, glycine (G) and alanine (A) can frequently be
interchangeable, as can
alanine (A) and valine (V). Methionine (M), which is relatively hydrophobic,
can frequently
be interchanged with leucine and isoleucine, and sometimes with valine. Lysine
(K) and
arginine (R) are frequently interchangeable in locations in which the
significant feature of the
amino acid residue is its charge and the differing pKs of these two amino acid
residues are
not significant. Still other changes can be considered "conservative" in
particular
environments (see, e.g., BIOCHEMISTRY at pp. 13-15, 2"d ed. Lubert Stryer ed.
(Stanford
University); Henikoff et al., Proc. Nat'l Acad. Sci. USA (1992) 89:10915-
10919; Lei et al., J.
Biol. Chem. (1995) 270(20):11882-11886).
[0066] In certain embodiments, the non-conservative amino acid substitution
comprises
substituting any of glycine (G), alanine (A), isoleucine (I), valine (V), and
leucine (L) for any
of serine (S), threonine (T), aspartic acid (D), glutamic acid (E), glutamine
(Q), asparagine
(N), lysine (K), arginine (R), phenylalanine (F), tyrosine (Y), tryptophan
(W), methionine
(M), cysteine (C), histidine (H), and proline (P). In certain embodiments, the
non-
conservative amino acid substitution comprises substituting any of serine (S)
and threonine
(T) for any of glycine (G), alanine (A), isoleucine (I), valine (V), leucine
(L), aspartic acid
(D), glutamic acid (E), glutamine (Q), asparagine (N), lysine (K), arginine
(R), phenylalanine
(F), tyrosine (Y), tryptophan (W), methionine (M), cysteine (C), histidine (H)
and proline (P).
In certain embodiments, the non-conservative amino acid substitution comprises
substituting
any of aspartic acid (D)and glutamic acid (E) for any of glycine (G), alanine
(A), isoleucine
(I), valine (V), leucine (L), serine (S), threonine (T), glutamine (Q),
asparagine (N), lysine
(K), arginine (R), phenylalanine (F), tyrosine (Y), tryptophan (W), methionine
(M), cysteine
(C), histidine (H), and proline (P). In certain embodiments, the non-
conservative amino acid
substitution comprises substituting any of glutamine (Q) and asparagine (N)
for any of
glycine (G), alanine (A), isoleucine (I), valine (V), leucine (L), serine (S),
threonine (T),
aspartic acid (D), glutamic acid (E), lysine (K), arginine (R), phenylalanine
(F), tyrosine (Y),
tryptophan (W), methionine (M), cysteine (C), histidine (H), and proline (P).
In certain
embodiments, the non-conservative amino acid substitution comprises
substituting any of
lysine (K) and arginine (R) for any of glycine (G), alanine (A), isoleucine
(I), valine (V),
leucine (L), serine (S), threonine (T), aspartic acid (D), glutamic acid (E),
glutamine (Q),
asparagine (N), phenylalanine (F), tyrosine (Y), tryptophan (W), methionine
(M), cysteine
14
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
(C), histidine (H), and proline (P). In certain embodiments, the non-
conservative amino acid
substitution comprises substituting any of phenylalanine (F), tyrosine (Y),
and tryptophan
(W) for any of glycine (G), alanine (A), isoleucine (I), valine (V), leucine
(L), serine (S),
threonine (T), aspartic acid (D), glutamic acid (E), glutamine (Q), asparagine
(N), lysine (K),
arginine (R), methionine (M), cysteine (C), histidine (H), and proline (P). In
certain
embodiments, the non-conservative amino acid substitution comprises
substituting any of
methionine (M) and cysteine (C) for any of glycine (G), alanine (A),
isoleucine (I), valine
(V), leucine (L), serine (S), threonine (T), aspartic acid (D), glutamic acid
(E), glutamine (Q),
asparagine (N), lysine (K), arginine (R), phenylalanine (F), tyrosine (Y),
tryptophan (W),
histidine (H), and proline (P). In certain embodiments, the non-conservative
amino acid
substitution comprises substituting histidine (H) for any of glycine (G),
alanine (A),
isoleucine (I), valine (V), leucine (L), serine (S), threonine (T), aspartic
acid (D), glutamic
acid (E), glutamine (Q), asparagine (N), lysine (K), arginine (R),
phenylalanine (F), tyrosine
(Y), tryptophan (W), methionine (M), cysteine (C), and proline (P). In certain
embodiments,
the non-conservative amino acid substitution comprises substituting proline
(P) for any of
glycine (G), alanine (A), isoleucine (I), valine (V), leucine (L), serine (S),
threonine (T),
aspartic acid (D), glutamic acid (E), glutamine (Q), asparagine (N), lysine
(K), arginine (R),
phenylalanine (F), tyrosine (Y), tryptophan (W), methionine (M), cysteine (C),
and histidine
(H).
Modulators of Complement Activity
[0067] As used herein, the term "complement modulator" refers to a compound,
composition,
or protein that modulates (e.g., inhibits or activates) complement activity or
a biologically
active fragment thereof. A complement modulator can be a complement inhibitor
or a
complement activator.
[0068] As used herein, the term "complement inhibitor" refers to any compound,
composition, or protein that reduces or eliminates complement activity or a
biologically
active fragment thereof. The reduction in complement activity may be
incremental (e.g., a
10%, 20%, 30%, 40%, 50%, 60%, 70%, 80%, or 90% reduction in activity) or
complete. A
complement inhibitor may be a soluble or membrane-bound protein such as, for
example,
membrane cofactor protein (MCP), decay accelerating factor (DAF/CD55), CD59,
mouse
complement receptor 1-related gene/protein y (Crry), human complement receptor
1 (CR1)
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
and factor H, or an antibody specific for a component of a complement pathway
such as, for
example, eculizumab (an anti-C5 antibody marketed under the trade name Soliris
),
pexelizumab (a single-chain antibody (scFv) comprising the antigen-binding
fragment of
eculizumab), an anti-factor B antibody (such as the monoclonal antibody 1379
produced by
ATCC Deposit No. PTA-6230), an anti-properdin antibody, an anti-factor D
antibody, and
the like. Alternatively, a complement inhibitor may be a small molecule or a
linear or cyclic
peptide such as, for example, compstatin, N-acetylaspartylglutamic acid
(NAAGA), and the
like.
[0069] As used herein the term "complement activator" refers to any compound,
composition, or protein that increases or activates complement activity or a
biologically
active fragment thereof. The increase in complement activity may be
incremental (e.g., a
10%, 20%, 30%, 40%, 50%, 60%, 70%, 80%, or 90% increase in activity). A
complement
activator may be a soluble or membrane-bound protein such as, for example,
human Ig
isotype Gi (IgGi), human Ig isotype M (IgM), mouse Ig isotype G3 (IgG3), and
mouse IgM
Fc, as well as cobra venom factor (CVF) and biologically-active fragments
thereof, such as
the Fc domain of Ig proteins, such as human IgGi Fc domain, human IgM Fc
domain, mouse
IgG3 Fc domain, and mouse IgM Fc domain. Complement activators may also
include, for
example, hybrid CVF molecules comprising a CVF portion and a complement
component 3
(C3) portion, such as those described in Fritzinger et al., "Functional
characterization of
human C3/cobra venom factor hybrid proteins for therapeutic complement
depletion,"
Develop. Comp. Immunol. 33(1):105-116 (2009). Those hybrids comprise proteins
in which
the 113 or 315 C-terminal residues of C3 were replaced with corresponding CVF
sequences.
Complement Inhibitor Proteins
[0070] Provided herein are soluble compositions comprising a construct,
wherein the
construct comprises: (a) a complement receptor 2 (CR2) portion comprising a
CR2 protein of
SEQ ID NO:1 or a biologically active fragment thereof, wherein the CR2 portion
contains at
least the first two N-terminal SCR domains of the CR2 protein; and (b) a
complement
inhibitor portion; wherein the CR2 portion contains at least one amino acid
substitution. In
certain embodiments, the construct is a fusion protein. A number of endogenous
soluble and
membrane-bound proteins that inhibit complement have been identified. These
complement
inhibitor proteins include, but are not limited to, membrane cofactor protein
(MCP), decay
16
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
accelerating factor (DAF/CD55), CD59, mouse complement receptor 1-related
gene/protein y
(Crry), human complement receptor 1 (CR1) and factor H. In certain
embodiments, the
complement modulator portion of the construct comprises a complement inhibitor
or
biologically active fragment thereof. In certain embodiments, the complement
inhibitor is
selected from the group consisting of human MCP, human DAF, mouse DAF, human
CD59,
mouse CD59 isoform A, mouse CD59 isoform B, mouse Crry protein, human CR1,
human
factor H, or mouse factor H, or a biologically active fragment thereof.
Membrane Cofactor Protein (MCP)
[0071] As used herein, the term "membrane cofactor protein," "MCP," or "CD46"
refers to a
widely distributed C3b/C4b-binding cell surface glycoprotein which inhibits
complement
activation on host cells and serves as a cofactor for the factor I-mediated
cleavage of C3b and
C4b, including homologs thereof. T.J. Oglesby et al., J. Exp. Med. (1992)
175:1547-1551.
MCP belongs to a family known as the regulators of complement activation
("RCA"). Family
members share certain structural features, comprising varying numbers of short
consensus
repeat (SCR) domains, which are typically between 60 and 70 amino acids in
length.
Beginning at its amino-terminus, MCP comprises four SCRs, a
serine/threonine/proline-
enriched region, an area of undefined function, a transmembrane hydrophobic
domain, a
cytoplasmic anchor and a cytoplasmic tail. It is understood that species and
strain variations
exist for the disclosed peptides, polypeptides, and proteins, and that human
MCP or
biologically active fragments thereof encompasses all species and strain
variations.
[0072] SEQ ID NO: 10 represents the full-length human MCP amino acid sequence
(see, e.g.,
UniProtKB/Swiss-Prot. Accession No. P15529). Amino acids 1-34 correspond to
the signal
peptide, amino acids 35-343 correspond to the extracellular domain, amino
acids 344-366
correspond to the transmembrane domain, and amino acids 367-392 correspond to
the
cytoplasmic domain. In the extracellular domain, amino acids 35-96 correspond
to SCR 1,
amino acids 97-159 correspond to SCR 2, amino acids 160-225 correspond to SCR
3, amino
acids 226-285 correspond to SCR 4, and amino acids 302-326 correspond to the
serine/threonine-rich domain. It is understood that species and strain
variations exist for the
disclosed peptides, polypeptides, and proteins, and that MCP or biologically
active fragments
thereof encompasses all species and strain variations. As used herein, the
term "biologically
active" fragment of MCP refers to any soluble fragment lacking both the
cytoplasmic domain
17
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
and the transmembrane domain, including fragments comprising, consisting
essentially of or
consisting of 1, 2, 3, or 4 SCR domains, with or without the serine/threonine-
rich domain,
having some or all the complement inhibitory activity of the full-length human
MCP protein.
In certain embodiments, the complement inhibitor portion comprises full-length
human MCP
(amino acids 35-392 of SEQ ID NO: 10), the extracellular domain of human MCP
(amino
acids 35-343 of SEQ ID NO: 10), or SCRs 1-4 of human MCP (amino acids 35-285
of SEQ
ID NO:10).
[0073] In one aspect, there is provided a soluble composition comprising a
construct, wherein
the construct comprises: (a) a complement receptor 2 (CR2) portion comprising
a CR2
protein of SEQ ID NO:1 or a biologically active fragment thereof, wherein the
CR2 portion
contains at least the first two N-terminal SCR domains of the CR2 protein; and
(b) MCP;
wherein the CR2 portion contains at least one amino acid substitution. In some
embodiments,
the CR2 portion selectively binds one or more C3 proteolytic fragments
selected from the
group consisting of C3d, iC3dg, C3dg, and a cell-bound fragment of C3b that
binds to the
two N-terminal SCR domains of CR2. In some embodiments, the CR2 portion
selectively
binds one or more C3 proteolytic fragments selected from the group consisting
of C3d,
iC3dg, C3dg, and a cell-bound fragment of C3b that binds to the two N-terminal
SCR
domains of CR2 and also selectively binds EBV gp350. In some embodiments, the
CR2
portion selectively binds one or more C3 proteolytic fragments selected from
the group
consisting of C3d, iC3dg, C3dg, and a cell-bound fragment of C3b that binds to
the two N-
terminal SCR domains of CR2 and also selectively binds IFNa. In some
embodiments, the
CR2 portion selectively binds IFNa. In some embodiments, the CR2 portion
selectively
binds IFNa and also selectively binds EBV gp350. In some embodiments, the CR2
portion
selectively binds EBV gp350. As used herein, "selectively binds" means that a
construct
exhibits enhanced binding to one ligand and/or decreased binding to a
different ligand. For
example, "selectively binds" can mean: 1) that a construct which has been
altered has a
binding affinity for a first ligand that is similar to the binding affinity of
the unaltered
construct for the first ligand whereas the construct that has been altered has
a lower affinity
for a second ligand than does the unaltered construct, and therefore the
altered construct
"selectively binds" the first ligand as compared to the binding of the
unaltered construct to the
two ligands; 2) that a construct which has been altered has increased binding
to a first ligand
as compared to the binding of the unaltered construct to the first ligand
while retaining
binding to a second ligand that is similar to the binding of the unaltered
construct to the
18
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
second ligand, and therefore the construct "selectively binds" the first
ligand as compared to
the binding of the unaltered construct to the two ligands; or 3) that a
construct which has been
altered has increased binding to a first ligand as compared to the binding of
the unaltered
construct to the first ligand and also has a lower binding affinity for a
second ligand as
compared to the binding of the unaltered construct to the second ligand, and
therefore the
altered construct "selectively binds" the first ligand and not the second
ligand as compared to
the binding of the unaltered construct to the two ligands. In certain
embodiments, the
construct is a fusion protein. In certain embodiments, the complement
inhibitor portion of the
construct comprises full-length human MCP (SEQ ID NO: 10). In certain
embodiments, the
complement inhibitor portion of the construct comprises a biologically active
fragment of
human MCP (SEQ ID NO: 10). In certain embodiments, the biologically active
fragment of
human MCP is selected from the group consisting of SCR 1-4 (amino acids 35-285
of SEQ ID
NO: 10), SCR 1-4 plus the serine/threonine-rich domain (amino acids 35-326 of
SEQ ID
NO: 10), and the extracellular domain of MCP (amino acids 35-343 of SEQ ID NO:
10).
Decay Accelerating Factor (DAF)
[0074] Decay accelerating factor, also referred to as CD55 (DAF/CD55) (SEQ ID
NO: 11 and
SEQ ID NO: 12), is a -70 kiloDalton (kDa) membrane-bound glycoprotein which
inhibits
complement activation on host cells. Like several other complement regulatory
proteins,
DAF comprises several approximately 60 amino acid repeating motifs termed
short
consensus repeats (SCR).
[0075] As used herein, the term "decay accelerating factor," "DAF," or "CD55"
refers to a
seventy kilodalton ("kD") membrane glycoprotein comprising four short
consensus repeat
(SCR) domains followed by a heavily O-glycosylated serine/threonine-rich
domain at the C-
terminus that elevates the molecule from the membrane surface, followed by a
glycosylphosphatidylinositol ("GPI") anchor. DAF protects the cell surface
from complement
activation by dissociating membrane-bound C3 convertases that are required to
cleave
complement protein C3 and to amplify the complement cascade. DAF prevents
assembly or
accelerates decay of both the C3- and C5-convertases of the alternative and
classical
complement pathways.
[0076] SEQ ID NO: 11 represents the full-length human DAF amino acid sequence
(see, e.g.,
UniProtKB/Swiss-Prot. Accession No. P08173); SEQ ID NO: 12 represents the full-
length
19
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
mouse DAF amino acid sequence (see, e.g., UniProtKB/Swiss-Prot. Accession No.
Q61475).
In the human DAF sequence, amino acids 1-34 correspond to the signal peptide,
amino acids
35-353 appear in the mature protein, and amino acids 354-381 are removed from
the
polypeptide after translation. Within the mature protein, amino acids 35-96
correspond to
SCR 1, amino acids 96-160 correspond to SCR 2, amino acids 161-222 correspond
to SCR 3,
amino acids 223-285 correspond to SCR 4, and amino acids 287-353 correspond to
the 0-
glycosylated serine/threonine-rich domain. The GPI anchor is attached to human
DAF at a
serine at position 353. In the mouse DAF sequence, amino acids 1-34 correspond
to the
signal peptide, amino acids 35-362 appear in the mature protein, and amino
acids 363-390 are
removed from the polypeptide after translation. Within the mature protein,
amino acids 35-96
correspond to SCR 1, amino acids 97-160 correspond to SCR 2, amino acids 161-
222
correspond to SCR 3, amino acids 223-286 correspond to SCR 4, and amino acids
288-362
correspond to the O-glycosylated serine/threonine-rich domain. The GPI anchor
is attached to
mouse DAF at a serine at position 362. It is understood that species and
strain variations exist
for the disclosed peptides, polypeptides, and proteins, and that DAF or
biologically active
fragments thereof encompasses all species and strain variations. As used
herein, the term
"biologically active" fragment of DAF refers to any fragment of DAF lacking a
GPI anchor
and/or the amino acid to which it is attached (i.e., Ser-353), including any
fragments of the
full-length DAF protein comprising, consisting essentially of or consisting of
1, 2, 3, or 4
SCR domains, with or without the O-glycosylated serine/threonine-rich domain,
having some
or all the complement inhibitory activity of the full-length DAF protein.
[0077] In one aspect, there is provided a soluble composition comprising a
construct, wherein
the construct comprises: (a) a complement receptor 2 (CR2) portion comprising
a CR2
protein of SEQ ID NO:1 or a biologically active fragment thereof, wherein the
CR2 portion
contains at least the first two N-terminal SCR domains of the CR2 protein; and
(b) DAF;
wherein the CR2 portion contains at least one amino acid substitution. In some
embodiments,
the CR2 portion selectively binds one or more C3 proteolytic fragments
selected from the
group consisting of C3d, iC3dg, C3dg, and a cell-bound fragment of C3b that
binds to the
two N-terminal SCR domains of CR2. In some embodiments, the CR2 portion
selectively
binds one or more C3 proteolytic fragments selected from the group consisting
of C3d,
iC3dg, C3dg, and a cell-bound fragment of C3b that binds to the two N-terminal
SCR
domains of CR2 and also selectively binds EBV gp350. In some embodiments, the
CR2
portion selectively binds one or more C3 proteolytic fragments selected from
the group
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
consisting of C3d, iC3dg, C3dg, and a cell-bound fragment of C3b that binds to
the two N-
terminal SCR domains of CR2 and also selectively binds IFNa. In some
embodiments, the
CR2 portion selectively binds IFNa. In some embodiments, the CR2 portion
selectively
binds IFNa and also selectively binds EBV gp350. In some embodiments, the CR2
portion
selectively binds EBV gp350. As used herein, "selectively binds" means that a
construct
exhibits enhanced binding to one ligand and/or decreased binding to a
different ligand. For
example, "selectively binds" can mean: 1) that a construct which has been
altered has a
binding affinity for a first ligand that is similar to the binding affinity of
the unaltered
construct for the first ligand whereas the construct that has been altered has
a lower affinity
for a second ligand than does the unaltered construct, and therefore the
altered construct
"selectively binds" the first ligand as compared to the binding of the
unaltered construct to the
two ligands; 2) that a construct which has been altered has increased binding
to a first ligand
as compared to the binding of the unaltered construct to the first ligand
while retaining
binding to a second ligand that is similar to the binding of the unaltered
construct to the
second ligand, and therefore the construct "selectively binds" the first
ligand as compared to
the binding of the unaltered construct to the two ligands; or 3) that a
construct which has been
altered has increased binding to a first ligand as compared to the binding of
the unaltered
construct to the first ligand and also has a lower binding affinity for a
second ligand as
compared to the binding of the unaltered construct to the second ligand, and
therefore the
altered construct "selectively binds" the first ligand and not the second
ligand as compared to
the binding of the unaltered construct to the two ligands. In certain
embodiments, the
construct is a fusion protein. In certain embodiments, the complement
inhibitor portion of the
construct comprises full-length human DAF. In certain embodiments, the
complement
inhibitor portion of the construct comprises a biologically active fragment of
human DAF
(SEQ ID NO: 11). In certain embodiments, the biologically active fragment of
human DAF is
selected from the group consisting of SCR 1-4 (amino acids 25-285 of SEQ ID
NO: 11) and
SCR 1-4 plus the O-glycosylated serine/threonine-rich domain (amino acids 25-
353 of SEQ
ID NO: 11). In certain embodiments, the complement inhibitor portion of the
construct
comprises full-length mouse DAF (SEQ ID NO: 12). In certain embodiments, the
complement
inhibitor portion of the construct comprises a biologically active fragment of
mouse DAF. In
certain embodiments, the biologically active fragment of mouse DAF is selected
from the
group consisting of SCR 1-4 (amino acids 35-286 of SEQ ID NO: 12) and SCR 1-4
plus the 0-
glycosylated serine/threonine-rich domain (amino acids 35-362 of SEQ ID NO:
12).
21
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
CD59
[0078] As used herein, the term "CD59" refers to a membrane-bound 128 amino
acid
glycoprotein that potently inhibits the membrane attack complex (MAC) of
complement.
CD59 acts by binding to the C8 and/or C9 components of the MAC during
assembly,
ultimately preventing incorporation of the multiple copies of C9 required for
complete
formation of the osmolytic pore at the heart of the MAC. CD59 is both N- and 0-
glycosylated. The N-glycosylation comprises primarily of bi- or tri-antennary
structures with
and without lactosamine and outer arm fucose residues, with variable
sialylation present at
some sites. Like DAF, CD59 is anchored in the cell membrane by a
glycosylphosphatidylinositol ("GPI") anchor, which is attached to an
asparagine at amino
acid 102. Soluble forms of CD59 (sCD59) have been produced, but they generally
have low
functional activity in vitro, particularly in the presence of serum,
suggesting that unmodified
sCD59 has little or no therapeutic efficacy. See, e.g., S. Meri et al.,
"Structural composition
and functional characterization of soluble CD59: heterogeneity of the
oligosaccharide and
glycophosphoinositol (GPI) anchor revealed by laser-desorption mass
spectrometric
analysis," Biochem. J. 316:923-935 (1996).
[0079] SEQ ID NO:3 represents the full-length human CD59 amino acid sequence
(see, e.g.,
UniProtKB/Swiss-Prot. Accession No. P13987); SEQ ID NO:6 represents the full-
length
mouse CD59 sequence, isoform A (see, e.g., UniProtKB/Swiss-Prot. Accession No.
055186); SEQ ID NO:7 represents the full-length mouse CD59 sequence, isoform B
(see,
e.g., UniProtKB/Swiss-Prot. Accession No. P58019). In the human CD59 sequence,
amino
acids 1-25 of SEQ ID NO:3 correspond to the leader peptide, amino acids 26-102
of SEQ ID
NO:3 correspond to the mature protein, and amino acids 103-128 of SEQ ID NO:3
are
removed after translation. The GPI anchor is attached to CD59 at an asparagine
at position
102 of SEQ ID NO:3. In isoform A of the mouse CD59 sequence, amino acids 1-23
of SEQ
ID NO:6 correspond to the leader peptide, amino acids 24-96 of SEQ ID NO:6
correspond to
the mature protein, and amino acids 97-123 of SEQ ID NO:6 are removed after
translation.
The GPI anchor is attached to CD59 at a serine at position 96 of SEQ ID NO:6.
In isoform B
of the mouse CD59 sequence, amino acids 1-23 of SEQ ID NO:7 correspond to the
leader
peptide, amino acids 24-104 of SEQ ID NO:7 correspond to the mature protein,
and amino
acids 105-129 of SEQ ID NO:7 are removed after translation. The GPI anchor is
attached to
CD59 at an asparagine at position 104 of SEQ ID NO:7. It is understood that
species and
22
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
strain variations exist for the disclosed peptides, polypeptides, and
proteins, and that CD59 or
biologically active fragments thereof encompasses all species and strain
variations.
[0080] As used herein, the term "biologically active" fragment of human CD59
refers to any
fragment of human CD59 lacking a GPI anchor and/or the amino acid to which it
is attached
(i.e., Asn-102), including any fragments of the full-length human CD59 protein
having some
or all the complement inhibitory activity of the full-length CD59 protein; and
the term
"biologically active" fragment of mouse CD59 refers to any fragment of mouse
CD59
isoform A or isoform B lacking a GPI anchor and/or the amino acid to which it
is attached
(i.e., Ser-96 of isoform A, or Asp-104 of isoform B) , including any fragments
of either full-
length mouse CD59 protein isoform having some or all the complement inhibitory
activity of
the full-length CD59 protein.
[0081] In one aspect, there is provided a soluble composition comprising a
construct, wherein
the construct comprises: (a) a complement receptor 2 (CR2) portion comprising
a CR2
protein of SEQ ID NO:1 or a biologically active fragment thereof, wherein the
CR2 portion
contains at least the first two N-terminal SCR domains of the CR2 protein; and
(b) CD59;
wherein the CR2 portion contains at least one amino acid substitution. In
certain
embodiments, the construct is a fusion protein. In some embodiments, the CR2
portion
selectively binds one or more C3 proteolytic fragments selected from the group
consisting of
C3d, iC3dg, C3dg, and a cell-bound fragment of C3b that binds to the two N-
terminal SCR
domains of CR2. In some embodiments, the CR2 portion selectively binds one or
more C3
proteolytic fragments selected from the group consisting of C3d, iC3dg, C3dg,
and a cell-
bound fragment of C3b that binds to the two N-terminal SCR domains of CR2 and
also
selectively binds EBV gp350. In some embodiments, the CR2 portion selectively
binds one
or more C3 proteolytic fragments selected from the group consisting of C3d,
iC3dg, C3dg,
and a cell-bound fragment of C3b that binds to the two N-terminal SCR domains
of CR2 and
also selectively binds IFNa. In some embodiments, the CR2 portion selectively
binds IFNa.
In some embodiments, the CR2 portion selectively binds IFNa and also
selectively binds
EBV gp350. In some embodiments, the CR2 portion selectively binds EBV gp350.
As used
herein, "selectively binds" means that a construct exhibits enhanced binding
to one ligand
and/or decreased binding to a different ligand. For example, "selectively
binds" can mean: 1)
that a construct which has been altered has a binding affinity for a first
ligand that is similar
to the binding affinity of the unaltered construct for the first ligand
whereas the construct that
23
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
has been altered has a lower affinity for a second ligand than does the
unaltered construct,
and therefore the altered construct "selectively binds" the first ligand as
compared to the
binding of the unaltered construct to the two ligands; 2) that a construct
which has been
altered has increased binding to a first ligand as compared to the binding of
the unaltered
construct to the first ligand while retaining binding to a second ligand that
is similar to the
binding of the unaltered construct to the second ligand, and therefore the
construct
"selectively binds" the first ligand as compared to the binding of the
unaltered construct to the
two ligands; or 3) that a construct which has been altered has increased
binding to a first
ligand as compared to the binding of the unaltered construct to the first
ligand and also has a
lower binding affinity for a second ligand as compared to the binding of the
unaltered
construct to the second ligand, and therefore the altered construct
"selectively binds" the first
ligand and not the second ligand as compared to the binding of the unaltered
construct to the
two ligands. In certain embodiments, the complement inhibitor portion of the
construct
comprises full-length human CD59 (SEQ ID NO:3). In certain embodiments, the
complement
inhibitor portion of the construct comprises a biologically active fragment of
human CD59
(SEQ ID NO:3). In certain embodiments, the biologically active fragment of
human CD59
comprises the extracellular domain of human CD59 lacking its GPI anchor (amino
acids 26-
101 of SEQ ID NO:3). In certain embodiments, the complement inhibitor portion
of the
construct comprises full-length mouse CD59, isoform A (SEQ ID NO:6). In
certain
embodiments, the complement inhibitor portion of the construct comprises a
biologically
active fragment of mouse CD59, isoform A (SEQ ID NO:6). In certain
embodiments, the
biologically active fragment of mouse CD59, isoform A comprises the
extracellular domain
of mouse CD59, isoform A lacking its GPI anchor (amino acids 24-95 of SEQ ID
NO:6). In
certain embodiments, the complement inhibitor portion of the construct
comprises full-length
mouse CD59, isoform B (SEQ ID NO:7). In certain embodiments, the complement
inhibitor
portion of the construct comprises a biologically active fragment of mouse
CD59, isoform B
(SEQ ID NO:7). In certain embodiments, the biologically active fragment of
mouse CD59,
isoform B comprises the extracellular domain of mouse CD59, isoform B lacking
its GPI
anchor (amino acids 24-103 of SEQ ID NO:7).
Mouse Complement Receptor 1-related Gene/protein Y (Crry)
[0082] As used herein, the term "mouse complement receptor 1-related
gene/protein y" or
"Crry" refers to a membrane-bound mouse glycoprotein that regulates complement
24
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
activation, including homologs thereof. Crry regulates complement activation
by serving as a
cofactor for complement factor I, a serine protease which cleaves C3b and C4b
deposited on
host tissue. Crry also acts as a decay-accelerating factor, preventing the
formation of C4b2a
and C3bBb, the amplification convertases of the complement cascade.
[0083] SEQ ID NO:4 represents the full-length mouse Crry protein amino acid
sequence.
Amino acids 1-40 correspond to the leader peptide, amino acids 41-483 of SEQ
ID NO:4
correspond to the mature protein, comprising amino acids 41-405 of SEQ ID
NO:4,
corresponding to the extracellular domain, amino acids 406-426 of SEQ ID NO:4,
corresponding to the transmembrane domain, and amino acids 427-483 of SEQ ID
NO:4,
corresponding to the cytoplasmic domain. In the extracellular domain, amino
acids 83-143 of
SEQ ID NO:4 correspond to SCR 1, amino acids 144-205 of SEQ ID NO:4 correspond
to
SCR2, amino acids 206-276 of SEQ ID NO:4 correspond to SCR3, amino acids 277-
338 of
SEQ ID NO:4 correspond to SCR4, and amino acids 339-400 of SEQ ID NO:4
correspond to
SCRS. It is understood that species and strain variations exist for the
disclosed peptides,
polypeptides, and proteins, and that mouse Crry protein or biologically active
fragments
thereof encompasses all species and strain variations. As used herein, the
term "biologically
active" fragment of mouse Crry protein refers to refers to any soluble
fragment of mouse
Crry lacking the transmembrane domain and the cytoplasmic domain, including
fragments
comprising, consisting essentially of or consisting of 1, 2, 3, 4, or 5 SCR
domains, including
any fragments of the full-length mouse Crry protein having some or all the
complement
inhibitory activity of the full-length Crry protein.
[0084] In one aspect, there is provided a soluble composition comprising a
construct, wherein
the construct comprises: (a) a complement receptor 2 (CR2) portion comprising
a CR2
protein of SEQ ID NO:1 or a biologically active fragment thereof, wherein the
CR2 portion
contains at least the first two N-terminal SCR domains of the CR2 protein; and
(b)Crry;
wherein the CR2 portion contains at least one amino acid substitution. In
certain
embodiments, the construct is a fusion protein. In some embodiments, the CR2
portion
selectively binds one or more C3 proteolytic fragments selected from the group
consisting of
C3d, iC3dg, C3dg, and a cell-bound fragment of C3b that binds to the two N-
terminal SCR
domains of CR2. In some embodiments, the CR2 portion selectively binds one or
more C3
proteolytic fragments selected from the group consisting of C3d, iC3dg, C3dg,
and a cell-
bound fragment of C3b that binds to the two N-terminal SCR domains of CR2 and
also
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
selectively binds EBV gp350. In some embodiments, the CR2 portion selectively
binds one
or more C3 proteolytic fragments selected from the group consisting of C3d,
iC3dg, C3dg,
and a cell-bound fragment of C3b that binds to the two N-terminal SCR domains
of CR2 and
also selectively binds IFNa. In some embodiments, the CR2 portion selectively
binds IFNa.
In some embodiments, the CR2 portion selectively binds IFNa and also
selectively binds
EBV gp350. In some embodiments, the CR2 portion selectively binds EBV gp350.
As used
herein, "selectively binds" means that a construct exhibits enhanced binding
to one ligand
and/or decreased binding to a different ligand. For example, "selectively
binds" can mean: 1)
that a construct which has been altered has a binding affinity for a first
ligand that is similar
to the binding affinity of the unaltered construct for the first ligand
whereas the construct that
has been altered has a lower affinity for a second ligand than does the
unaltered construct,
and therefore the altered construct "selectively binds" the first ligand as
compared to the
binding of the unaltered construct to the two ligands; 2) that a construct
which has been
altered has increased binding to a first ligand as compared to the binding of
the unaltered
construct to the first ligand while retaining binding to a second ligand that
is similar to the
binding of the unaltered construct to the second ligand, and therefore the
construct
"selectively binds" the first ligand as compared to the binding of the
unaltered construct to the
two ligands; or 3) that a construct which has been altered has increased
binding to a first
ligand as compared to the binding of the unaltered construct to the first
ligand and also has a
lower binding affinity for a second ligand as compared to the binding of the
unaltered
construct to the second ligand, and therefore the altered construct
"selectively binds" the first
ligand and not the second ligand as compared to the binding of the unaltered
construct to the
two ligands. In certain embodiments, the complement inhibitor portion of the
construct
comprises full-length mouse Crry protein (SEQ ID NO:4). In certain
embodiments, the
complement inhibitor portion of the construct comprises a biologically active
fragment of
mouse Crry protein (SEQ ID NO:4). In certain embodiments, the biologically
active fragment
of mouse Crry protein is selected from the group consisting of SCR1-5 (amino
acids 41-400
of SEQ ID NO:4) and the extracellular domain of mouse Crry protein (amino
acids 41-405 of
SEQ ID NO:4).
Complement Receptor 1 (CR1)
[0085] As used herein, the term "complement receptor 1," "CR1," or "CD35"
refers to a
human gene encoding a protein of 2039 amino acids, with a predicted molecular
weight of
26
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
220 kilodaltons ("kD"), including homologs thereof. The gene is expressed
principally on
erythrocytes, monocytes, neutrophils, and B cells, but is also present on some
T lymphocytes,
mast cells, and glomerular podocytes. CR1 protein is typically expressed at
between 100 and
1000 copies per cell. CR1 is the main system for processing and clearance of
complement-
opsonized immune complexes. CR1 negatively regulates the complement cascade,
mediates
immune adherence and phagocytosis, and inhibits both the classic and
alternative
complement pathways. The full-length CR1 protein comprises a 42 amino acid
signal
peptide, an extracellular domain of 1930 amino acids, a 25 amino acid
transmembrane
domain, and a 43 amino acid C-terminal cytoplasmic domain. The extracellular
domain of
CR1 has 25 potential N-glycosylation signal sequences, and comprises 30 short
consensus
("SCR") domains, also known as complement control protein (CCP) repeats, or
sushi
domains, each 60 to 70 amino acids long. The sequence homology between SCRs
ranges
between 60-99 percent. The 30 SCR domains are further grouped into four longer
regions
termed long homologous repeats ("LHRs"), each encoding approximately 45 kD
segments of
the CR1 protein, designated LHR-A, -B, -C, and -D. The first three comprise
seven SCR
domains each, while LHR-D comprises 9 SCR domains. The active sites on the
extracellular
domain of CR1 protein include a C4b-binding site with lower affinity for C3b
in SCRs 1-4
comprising amino acids 42-295, a C3b-binding site with lower affinity for C4b
in SCRs 8-11
comprising amino acids 490-745, a C3b-binding site with lower affinity for C4b
in SCRs 15-
18 comprising amino acids 940-1196, and a Clq-binding site in SCRs 22-28
comprising
amino acids 1394-1842.
[0086] SEQ ID NO:9 represents the full-length human CR1 amino acid sequence
(see, e.g.,
UniProtKB/Swiss-Prot. Accession No. P17927). Amino acids 1-41 correspond to
the signal
peptide, amino acids 42-2039 correspond to the mature protein, comprising
amino acids 42-
1971, corresponding to the extracellular domain, amino acids 1972-1996,
corresponding to
the transmembrane domain, and amino acids 1997-2039, corresponding to the
cytoplasmic
domain. In the extracellular domain, amino acids 42-101 correspond to SCR 1,
102-163
correspond to SCR2, amino acids 164-234 correspond to SCR3, amino acids 236-
295
correspond to SCR4, amino acids 295-355 correspond to SCRS, amino acids 356-
418
correspond to SCR6, amino acids 419-489 correspond to SCR7, amino acids 491-
551
correspond to SCR8, amino acids 552-613 correspond to SCR9, amino acids 614-
684
correspond to SCR10, amino acids 686-745 correspond to SCR11, amino acids 745-
805
correspond to SCR12, amino acids 806-868 correspond to SCR13, amino acids 869-
939
27
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
correspond to SCR14, amino acids 941-1001 correspond to SCR15, amino acids
1002-1063
correspond to SCR16, amino acids 1064-1134 correspond to SCR17, amino acids
1136-1195
correspond to SCR18, amino acids 1195-1255 correspond to SCR 19, amino acids
1256-1318
correspond to SCR 20, amino acids 1319-1389 correspond to SCR 21, amino acids
1394-
1454 correspond to SCR 22, amino acids 1455-1516 correspond to SCR 23, amino
acids
1517-1587 correspond to SCR 24, amino acids 1589-1648 correspond to SCR 25,
amino
acids 1648-1708 correspond to SCR 26, amino acids 1709-1771 correspond to SCR
27,
amino acids 1772-1842 correspond to SCR 28, amino acids 1846-1906 correspond
to SCR
29, amino acids 1907-1967 correspond to SCR 30. It is understood that species
and strain
variations exist for the disclosed peptides, polypeptides, and proteins, and
that CR1 protein or
biologically active fragments thereof encompasses all species and strain
variations. As used
herein, the term "biologically active" fragment of CR1 protein refers to
refers to any soluble
fragment of CR1 lacking the transmembrane domain and the cytoplasmic domain,
including
fragments comprising, consisting essentially of or consisting of 1, 2, 3, 4,
5, 6, 7, 8, 9, 10, 11,
12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, or 30
SCR domains,
including any fragments of the full-length CR1 protein having some or all the
complement
inhibitory activity of the full-length CR1 protein.
[0087] In one aspect, there is provided a soluble composition comprising a
construct, wherein
the construct comprises: (a) a complement receptor 2 (CR2) portion comprising
a CR2
protein of SEQ ID NO:1 or a biologically active fragment thereof, wherein the
CR2 portion
contains at least the first two N-terminal SCR domains of the CR2 protein; and
(b) CR1;
wherein the CR2 portion contains at least one amino acid substitution. In some
embodiments,
the CR2 portion selectively binds one or more C3 proteolytic fragments
selected from the
group consisting of C3d, iC3dg, C3dg, and a cell-bound fragment of C3b that
binds to the
two N-terminal SCR domains of CR2. In some embodiments, the CR2 portion
selectively
binds one or more C3 proteolytic fragments selected from the group consisting
of C3d,
iC3dg, C3dg, and a cell-bound fragment of C3b that binds to the two N-terminal
SCR
domains of CR2 and also selectively binds EBV gp350. In some embodiments, the
CR2
portion selectively binds one or more C3 proteolytic fragments selected from
the group
consisting of C3d, iC3dg, C3dg, and a cell-bound fragment of C3b that binds to
the two N-
terminal SCR domains of CR2 and also selectively binds IFNa. In some
embodiments, the
CR2 portion selectively binds IFNa. In some embodiments, the CR2 portion
selectively
binds IFNa and also selectively binds EBV gp350. In some embodiments, the CR2
portion
28
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
selectively binds EBV gp350. As used herein, "selectively binds" means that a
construct
exhibits enhanced binding to one ligand and/or decreased binding to a
different ligand. For
example, "selectively binds" can mean: 1) that a construct which has been
altered has a
binding affinity for a first ligand that is similar to the binding affinity of
the unaltered
construct for the first ligand whereas the construct that has been altered has
a lower affinity
for a second ligand than does the unaltered construct, and therefore the
altered construct
"selectively binds" the first ligand as compared to the binding of the
unaltered construct to the
two ligands; 2) that a construct which has been altered has increased binding
to a first ligand
as compared to the binding of the unaltered construct to the first ligand
while retaining
binding to a second ligand that is similar to the binding of the unaltered
construct to the
second ligand, and therefore the construct "selectively binds" the first
ligand as compared to
the binding of the unaltered construct to the two ligands; or 3) that a
construct which has been
altered has increased binding to a first ligand as compared to the binding of
the unaltered
construct to the first ligand and also has a lower binding affinity for a
second ligand as
compared to the binding of the unaltered construct to the second ligand, and
therefore the
altered construct "selectively binds" the first ligand and not the second
ligand as compared to
the binding of the unaltered construct to the two ligands. In certain
embodiments, the
construct is a fusion protein. In certain embodiments, the complement
inhibitor portion of the
construct comprises full-length human CR1 protein (SEQ ID NO:9). In certain
embodiments,
the complement inhibitor portion of the construct comprises a biologically
active fragment of
human CR1 protein (SEQ ID NO:9). In certain embodiments, the biologically
active
fragment of human CR1 protein is selected from the group consisting of SCR 1-3
(amino
acids of 42-234 of SEQ ID NO:9), SCR1-4 (amino acids 42-295 of SEQ ID NO:9),
SCR1-10
(amino acids 42-684 of SEQ ID NO:9), SCR8-10 (amino acids of 491-684 of SEQ ID
NO:9),
SCR 8-11 (amino acids 491-745 of SEQ ID NO:9), SCR15-17 (amino acids of 941-
1134 of
SEQ ID NO:9), SCR15-18 (amino acids 941-1195 of SEQ ID NO:9), and SCR22-28
(amino
acids 1394-1842 of SEQ ID NO:9).
Factor H (FH)
[0088] As used herein, the term "complement factor H," "factor H," or "FH"
refers to
complement factor H, a single polypeptide chain plasma glycoprotein, including
homologs
thereof. The protein is composed of 20 conserved short consensus repeat (SCR)
domains of
approximately 60 amino acids, arranged in a continuous fashion like a string
of beads,
29
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
separated by short linker sequences of 2-6 amino acids each. Factor H binds to
C3b,
accelerates the decay of the alternative pathway C3-convertase (C3bBb), and
acts as a
cofactor for the proteolytic inactivation of C3b. In the presence of factor H,
proteolysis by
factor I results in the cleavage and inactivation of C3b. Factor H has at
least three distinct
binding domains for C3b, which are located within SCRs 1-4, SCRs 5-8, and SCRs
19-20.
Each site of factor H binds to a distinct region within the C3b protein: the N-
terminal sites
bind to native C3b; the second site, located in the middle region of factor H,
binds to the C3c
fragment and the site located within SCR19 and 20 binds to the C3d region. In
addition,
factor H also contains binding sites for heparin, which are located within SCR
7, SCRs 5-12,
and SCR 20 of factor H and overlap with those of the C3b binding sites.
Structural and
functional analyses have shown that the domains for the complement inhibitory
activity of
factor H are located within the first four N-terminal SCR domains.
[0089] SEQ ID NO:5 represents the full-length human factor H amino acid
sequence (see,
e.g., UniProtKB/Swiss-Prot. Accession No. P08603); SEQ ID NO:8 represents the
full-length
mouse factor H amino acid sequence (see, e.g., UniProtKB/Swiss-Prot. Accession
No.
P06909). In the human factor H sequence, amino acids 1-18 of SEQ ID NO:5
correspond to
the signal peptide, and amino acids 19-1231 of SEQ ID NO:5 correspond to the
mature
protein. Within that protein, amino acids 21-80 of SEQ ID NO:5 correspond to
SCR 1, amino
acids 85-141 of SEQ ID NO:5 correspond to SCR 2, amino acids 146-205 of SEQ ID
NO:5
correspond to SCR 3, amino acids 210-262 of SEQ ID NO:5 correspond to SCR 4,
and amino
acids 267-320 of SEQ ID NO:5 correspond to SCR 5. In the mouse factor H
sequence, amino
acids 1-18 of SEQ ID NO:8 correspond to the signal peptide, and amino acids 19-
1234 of
SEQ ID NO:8 correspond to the mature protein. Within that protein, amino acids
19-82 of
SEQ ID NO:8 correspond to SCR 1, amino acids 83-143 of SEQ ID NO:8 correspond
to SCR
2, amino acids 144-207 of SEQ ID NO:8 correspond to SCR 3, amino acids 208-264
of SEQ
ID NO:8 correspond to SCR 4, and amino acids 265-322 of SEQ ID NO:8 correspond
to SCR
5. It is understood that species and strain variations exist for the disclosed
peptides,
polypeptides, and proteins, and that factor H or biologically active fragments
thereof
encompasses all species and strain variations.
[0090] As used herein, the term "biologically active" fragment of factor H
refers to any
portion of a factor H protein having some or all the complement inhibitory
activity of the full-
length factor H protein, and includes, but is not limited to, factor H
fragments comprising
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
SCRs 1-4, SCRs 1-5, SCRs 1-8, SCRs 1-18, SCRs 19-20, or any homolog of a
naturally-
occurring factor H or fragment thereof, as described in detail below. In
certain embodiments,
the biologically active fragment of factor H has one or more of the following
properties: (1)
binding to C-reactive protein (CRP), (2) binding to C3b, (3) binding to
heparin, (4) binding to
sialic acid, (5) binding to endothelial cell surfaces, (6) binding to cellular
integrin receptor,
(7) binding to pathogens, (8) C3b co-factor activity, (9) C3b decay-
acceleration activity, and
(10) inhibiting the alternative complement pathway.
[0091] In one aspect, there is provided a soluble composition comprising a
construct, wherein
the construct comprises: (a) a complement receptor 2 (CR2) portion comprising
a CR2
protein of SEQ ID NO:1 or a biologically active fragment thereof, wherein the
CR2 portion
contains at least the first two N-terminal SCR domains of the CR2 protein; and
(b) FH;
wherein the CR2 portion contains at least one amino acid substitution. In some
embodiments,
the CR2 portion selectively binds one or more C3 proteolytic fragments
selected from the
group consisting of C3d, iC3dg, C3dg, and a cell-bound fragment of C3b that
binds to the
two N-terminal SCR domains of CR2. In some embodiments, the CR2 portion
selectively
binds one or more C3 proteolytic fragments selected from the group consisting
of C3d,
iC3dg, C3dg, and a cell-bound fragment of C3b that binds to the two N-terminal
SCR
domains of CR2 and also selectively binds EBV gp350. In some embodiments, the
CR2
portion selectively binds one or more C3 proteolytic fragments selected from
the group
consisting of C3d, iC3dg, C3dg, and a cell-bound fragment of C3b that binds to
the two N-
terminal SCR domains of CR2 and also selectively binds IFNa. In some
embodiments, the
CR2 portion selectively binds IFNa. In some embodiments, the CR2 portion
selectively
binds IFNa and also selectively binds EBV gp350. In some embodiments, the CR2
portion
selectively binds EBV gp350. As used herein, "selectively binds" means that a
construct
exhibits enhanced binding to one ligand and/or decreased binding to a
different ligand. For
example, "selectively binds" can mean: 1) that a construct which has been
altered has a
binding affinity for a first ligand that is similar to the binding affinity of
the unaltered
construct for the first ligand whereas the construct that has been altered has
a lower affinity
for a second ligand than does the unaltered construct, and therefore the
altered construct
"selectively binds" the first ligand as compared to the binding of the
unaltered construct to the
two ligands; 2) that a construct which has been altered has increased binding
to a first ligand
as compared to the binding of the unaltered construct to the first ligand
while retaining
binding to a second ligand that is similar to the binding of the unaltered
construct to the
31
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
second ligand, and therefore the construct "selectively binds" the first
ligand as compared to
the binding of the unaltered construct to the two ligands; or 3) that a
construct which has been
altered has increased binding to a first ligand as compared to the binding of
the unaltered
construct to the first ligand and also has a lower binding affinity for a
second ligand as
compared to the binding of the unaltered construct to the second ligand, and
therefore the
altered construct "selectively binds" the first ligand and not the second
ligand as compared to
the binding of the unaltered construct to the two ligands. In certain
embodiments, the
construct is a fusion protein. In certain embodiments, the complement
inhibitor portion of the
construct comprises full-length human (SEQ ID NO:5) or mouse (SEQ ID NO:8)
factor H. In
certain embodiments, the complement inhibitor portion of the construct
comprises a
biologically active fragment of human (SEQ ID NO:5) or mouse (SEQ ID NO:8)
factor H. In
certain embodiments, the biologically active fragment of human factor H (SEQ
ID NO:5) is
selected from the group consisting of SCRs 1-4 (amino acids 21-262 of SEQ ID
NO:5), SCRs
1-5 of factor H (amino acids 21-320 of SEQ ID NO:5), SCRs 1-8 of factor H
(amino acids
21-507 of SEQ ID NO:5), and SCRs 1-18 of factor H (amino acids 21-1104 of SEQ
ID
NO:5). In certain embodiments, the biologically active fragment of mouse
factor H (SEQ ID
NO:8) is selected from the group consisting of SCRs 1-4 (amino acids 19-264 of
SEQ ID
NO:8), SCRs 1-5 of factor H (amino acids 19-322 of SEQ ID NO:8), SCRs 1-8 of
factor H
(amino acids 19-507 of SEQ ID NO:8), and SCRs 1-18 of factor H (amino acids 19-
1109 of
SEQ ID NO:8). In certain embodiments, the biologically active fragment of
human (SEQ ID
NO:5) or mouse (SEQ ID NO:8) factor H comprises (and in certain embodiments
consists of
or consists essentially of) at least the first four N-terminal SCR domains of
factor H,
including for example, at least any of the first 5, 6, 7, 8, 9, 10, 11, 12,
13, 14, 15, 16, 17, 18,
or more N-terminal SCR domains of factor H.
Complement Activator Proteins
[0092] Provided herein are soluble compositions comprising a construct,
wherein the
construct comprises: (a) a complement receptor 2 (CR2) portion comprising a
CR2 protein of
SEQ ID NO:1 or a biologically active fragment thereof, wherein the CR2 portion
contains at
least the first two N-terminal SCR domains of the CR2 protein; and (b) a
complement
activator portion; wherein the CR2 portion contains at least one amino acid
substitution. In
certain embodiments, the construct is a fusion protein. A number of endogenous
soluble
proteins that activate complement have also been identified. These complement
activators
32
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
include, but are not limited to, various immunoglobulin (Ig) proteins,
including human Ig
isotype Gi (IgGi), human Ig isotype M (IgM), mouse Ig isotype G3 (IgG3), and
mouse IgM
Fc, as well as cobra venom factor (CVF) and biologically-active fragments
thereof. The
complement activating activity of Ig proteins has been localized to the Fc
domain. Therefore
biologically-active fragments of complement-activating human and mouse Ig
proteins include
the Fc domain, such as human IgG1 Fc domain, human IgM Fc domain, mouse IgG3
Fc
domain, and mouse IgM Fc domain.
Immuno2lobulin Proteins
[0093] As used herein, the term "antibody" or "immunoglobulin" refers to
glycoproteins of
the immunoglobulin (Ig) superfamily of proteins. An antibody or immunoglobulin
(Ig)
molecule is tetrameric, comprising two identical light chain polypeptides and
two identical
heavy chain polypeptides. The two heavy chains are linked together by
disulfide bonds, and
each heavy chain is linked to a light chain by a disulfide bond. Each full-
length Ig molecule
contains at least two binding sites for a specific target or antigen.
[0094] The immune system produces several different classes of Ig molecules
(isotypes),
including IgA, IgD, IgE, IgG, and IgM, each distinguished by the particular
class of heavy
chain polypeptide present: alpha (a) found in IgA, delta (8) found in IgD,
epsilon (E) found
in IgE, gamma (y) found in IgG, and mu ( ) found in IgM. There are at least
five different y
heavy chain polypeptides (isotypes) found in IgG. In contrast, there are only
light chain
polypeptide isotypes, referred to as kappa (x) and lambda (X) chains. The
distinctive
characteristics of antibody isotypes are defined by sequences of the constant
domains of the
heavy chain.
[0095] An IgG molecule comprises two light chains (either x or X form) and two
heavy
chains (y form) bound together by disulfide bonds. The K and X forms of IgG
light chain both
contain a domain of relatively variable amino acid sequences, called the
variable region
(variously referred to as a "VL-," "VK ," or "Vx-region") and a domain of
relatively conserved
amino acid sequences, called the constant region (CL-region). Similarly, each
IgG heavy
chain contains a variable region (VH-region) and one or more conserved
regions: a complete
IgG heavy chain contains three constant domains ("CHI-," "CH2-," and "CH3-
regions") and a
hinge region. Within each VL- or VH-region, hypervariable regions, also known
as
33
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
complementarity-determining regions ("CDR"), are interspersed between
relatively
conserved framework regions ("FR"). Generally, the variable region of a light
or heavy chain
polypeptide contains four FR and three CDR arranged in the following order
along the
polypeptide: NH2-FR I -CDR I -FR2-CDR2-FR3-CDR3-FR4-COOH. Together the CDR and
FR determine the three-dimensional structure of the IgG binding site and thus,
the specific
target protein or antigen to which that IgG molecule binds. Each IgG molecule
is dimeric,
able to bind two antigen molecules. Cleavage of a dimeric IgG with the
protease papain
produces two identical antigen-binding fragments ("Fab"') and an "Fc" fragment
or Fc
domain, so named because is readily crystallized.
[0096] In some embodiments, the composition comprises a construct, wherein the
construct
comprises: (a) a complement receptor 2 (CR2) portion comprising a CR2 protein
of SEQ ID
NO:1 or a biologically active fragment thereof, wherein the CR2 portion
contains at least the
first two N-terminal SCR domains of the CR2 protein; and (b) a complement
activator
portion selected from the group consisting of: human Ig isotype Gi (IgGi),
human Ig isotype
M (IgM), mouse Ig isotype G3 (IgG3), mouse IgM Fc, human IgGi Fc domain, human
IgM Fc
domain, mouse IgG3 Fc domain, and mouse IgM Fc domain, and wherein the CR2
portion
contains at least one amino acid substitution. In some embodiments, the CR2
portion
selectively binds one or more C3 proteolytic fragments selected from the group
consisting of
C3d, iC3dg, C3dg, and a cell-bound fragment of C3b that binds to the two N-
terminal SCR
domains of CR2. In some embodiments, the CR2 portion selectively binds one or
more C3
proteolytic fragments selected from the group consisting of C3d, iC3dg, C3dg,
and a cell-
bound fragment of C3b that binds to the two N-terminal SCR domains of CR2 and
also
selectively binds EBV gp350. In some embodiments, the CR2 portion selectively
binds one
or more C3 proteolytic fragments selected from the group consisting of C3d,
iC3dg, C3dg,
and a cell-bound fragment of C3b that binds to the two N-terminal SCR domains
of CR2 and
also selectively binds IFNa. In some embodiments, the CR2 portion selectively
binds IFNa.
In some embodiments, the CR2 portion selectively binds IFNa and also
selectively binds
EBV gp350. In some embodiments, the CR2 portion selectively binds EBV gp350.
As used
herein, "selectively binds" means that a construct exhibits enhanced binding
to one ligand
and/or decreased binding to a different ligand. For example, "selectively
binds" can mean: 1)
that a construct which has been altered has a binding affinity for a first
ligand that is similar
to the binding affinity of the unaltered construct for the first ligand
whereas the construct that
has been altered has a lower affinity for a second ligand than does the
unaltered construct,
34
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
and therefore the altered construct "selectively binds" the first ligand as
compared to the
binding of the unaltered construct to the two ligands; 2) that a construct
which has been
altered has increased binding to a first ligand as compared to the binding of
the unaltered
construct to the first ligand while retaining binding to a second ligand that
is similar to the
binding of the unaltered construct to the second ligand, and therefore the
construct
"selectively binds" the first ligand as compared to the binding of the
unaltered construct to the
two ligands; or 3) that a construct which has been altered has increased
binding to a first
ligand as compared to the binding of the unaltered construct to the first
ligand and also has a
lower binding affinity for a second ligand as compared to the binding of the
unaltered
construct to the second ligand, and therefore the altered construct
"selectively binds" the first
ligand and not the second ligand as compared to the binding of the unaltered
construct to the
two ligands. In certain embodiments, the construct is a fusion protein. In
certain
embodiments, the complement activator portion of the construct comprises an Ig
protein or
biologically-active fragment thereof. In certain embodiments, the Ig protein
or biologically-
active fragment thereof comprises human IgGi, human IgGi Fc domain, human IgM,
human
IgM Fc domain, mouse IgG3, mouse IgG3 Fc domain, mouse IgM, and mouse IgM Fc
domain.
Cobra Venom Factor (CVF)
[0097] As used herein, the terms "cobra venom factor," "CVF," and "C3b
(Cobra)" refer to
the non-toxic, complement-activating component of cobra venom. Like naturally
occurring
human C3b, CVF (SEQ ID NO: 13) forms a complex, or convertase, with complement
components Factor B and Factor D. This CVFBbD convertase is capable of
activating C3 in a
wide variety of species via the alternative complement pathway. CVFBbD
convertase is
Factor H-resistant and is therefore not blocked through the activity of Factor
I or CR1 and
can convert nearly 100% of the C3 to C3 and C5 fragments. Levels of iC3b, C3a,
SC5b-9,
C5a and the Factor B cleavage product Bb are all extremely high in CVF treated
sera. The
cloning and sequencing of CVF from the monocled cobra (Naja kaouthia) was
reported in
Fritzinger, et al., "Molecular cloning and derived primary structure of cobra
venom factor,"
Proc. Nat'l Acad. Sci. USA 91(26):12775-779 (1994); the sequence was deposited
in the
GenBank database under Accession Number U09969. Both the Fritzinger et al.
reference and
the sequence deposited in GenBank under Accession Number U09969 are hereby
incorporated herein by reference. The terms "cobra venom factor," "CVF," and
"C3b
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
(Cobra)" also refer to hybrid CVF molecules comprising a CVF portion and a
complement
component 3 (C3) portion, such as those described in Fritzinger et al.,
"Functional
characterization of human C3/cobra venom factor hybrid proteins for
therapeutic complement
depletion," Develop. Comp. Immunol. 33(1):105-116 (2009), which is
incorporated herein by
reference. Those hybrids comprise proteins in which the 113 or 315 C-terminal
residues of
C3 were replaced with corresponding CVF sequences. Both hybrids formed stable
convertases that exhibited C3-cleaving activity, although at different rates.
Neither convertase
cleaved C5. Both convertases showed partial resistance to inactivation by
factors H and I,
allowing them to deplete complement in human serum.
[0098] In one aspect, the composition comprises a construct, wherein the
construct
comprises: (a) a complement receptor 2 (CR2) portion comprising a CR2 protein
of SEQ ID
NO:1 or a biologically active fragment thereof, wherein the CR2 portion
contains at least the
first two N-terminal SCR domains of the CR2 protein; and (b) CVF. In some
embodiments,
the CR2 portion selectively binds one or more C3 proteolytic fragments
selected from the
group consisting of C3d, iC3dg, C3dg, and a cell-bound fragment of C3b that
binds to the
two N-terminal SCR domains of CR2. In some embodiments, the CR2 portion
selectively
binds one or more C3 proteolytic fragments selected from the group consisting
of C3d,
iC3dg, C3dg, and a cell-bound fragment of C3b that binds to the two N-terminal
SCR
domains of CR2 and also selectively binds EBV gp350. In some embodiments, the
CR2
portion selectively binds one or more C3 proteolytic fragments selected from
the group
consisting of C3d, iC3dg, C3dg, and a cell-bound fragment of C3b that binds to
the two N-
terminal SCR domains of CR2 and also selectively binds IFNa. In some
embodiments, the
CR2 portion selectively binds IFNa. In some embodiments, the CR2 portion
selectively
binds IFNa and also selectively binds EBV gp350. In some embodiments, the CR2
portion
selectively binds EBV gp350. As used herein, "selectively binds" means that a
construct
exhibits enhanced binding to one ligand and/or decreased binding to a
different ligand. For
example, "selectively binds" can mean: 1) that a construct which has been
altered has a
binding affinity for a first ligand that is similar to the binding affinity of
the unaltered
construct for the first ligand whereas the construct that has been altered has
a lower affinity
for a second ligand than does the unaltered construct, and therefore the
altered construct
"selectively binds" the first ligand as compared to the binding of the
unaltered construct to the
two ligands; 2) that a construct which has been altered has increased binding
to a first ligand
as compared to the binding of the unaltered construct to the first ligand
while retaining
36
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
binding to a second ligand that is similar to the binding of the unaltered
construct to the
second ligand, and therefore the construct "selectively binds" the first
ligand as compared to
the binding of the unaltered construct to the two ligands; or 3) that a
construct which has been
altered has increased binding to a first ligand as compared to the binding of
the unaltered
construct to the first ligand and also has a lower binding affinity for a
second ligand as
compared to the binding of the unaltered construct to the second ligand, and
therefore the
altered construct "selectively binds" the first ligand and not the second
ligand as compared to
the binding of the unaltered construct to the two ligands. In certain
embodiments, the
construct is a fusion protein.
Compositions of the Invention
Amino acid residues in CR2 involved in selective binding to particular ligands
[0099] Structural analyses of CR2 binding to EBV gp350, IFNa, and C3d
described herein
(see Example 1) identified a number of CR2 amino acid residues that are
important for
binding of each of those ligands. While some of those amino acid residues are
important for
binding of all three ligands, others are important for binding one specific
ligand (e.g., EBV
gp350, IFNa, or C3d).
[0100] For example, residues determined to be important for the CR2-EBV gp350
binding
interaction are N11, R13, A22, R28, S32, R36, K41, K57, Y64, K67, Y68, R83,
G84 and R89
(see Table 2). Residues determined to be important for the CR2-EBV gp350
binding
interaction but not the CR2-C3d or the CR2-IFNa binding interaction are N11,
R36, K41,
Y64, and K67 (see Table 2). Residues determined to be important for the CR2-
IFNa binding
interaction are R13, Y16, R28, S42, K48, K50, Y68, R83, G84 and R89 (see Table
2).
Residues determined to be important for the CR2-IFNa binding interaction but
not the CR2-
C3d or CR2-EBV gp350 binding interaction are S42 and K50 (see Table 2).
Residues
determined to be important for the CR2-C3d binding interaction are 19, R13,
Y16, A22, R28,
Y29, C31, S32, G33, T34, K48, D56, K57, Y68, S70, R83, G84, R89, H90, D92,
S93, A97,
T100, N101, 5109, and 5128 (see Table 2). Residues determined to be important
for the
CR2-C3d binding interaction but not the CR2-gp350 or the CR2-IFNa binding
interaction are
19, Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97, T100, N101, 5109, and
5128 (see
Table 2). The amino acid residues determined to be important for the CR2-C3d
binding
interaction are likely important for the CR2 binding interaction with other
cell surface-bound
37
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
C3 fragments such as C3dg, and iC3b. Residues determined to be important for
the CR2-
gp350, the CR2-IFNa, and the CR2-C3d binding interactions are R13, Y16, A22,
R28, S32,
K48, K57, Y68, R83, G84, and R89. Because these amino acid residues are
important for
binding interactions between CR2 and more than one of its ligands, mutating
amino acid
residues at one or more of those positions may also improve the targeting
capability of the
CR2 portion for C3d and/or other CR2 ligands.
[0101] The improved targeting groups described herein, comprising at least one
or more
mutations in the various amino acid residues important for the binding
interactions between
CR2 and EBV-gp350, CR2 and IFNa, and/or CR2 and C3d can be used to deliver a
complement modulator (e.g., a complement inhibitor or complement activator) to
any
physiological site (e.g., a site of inflammation) suitable for the use of the
CR2 targeting
groups described in US Patent Publication No. 2008/0267980 Al, US Patent
Publication No.
US 2008/0221011 Al, and US Patent Publication No. 2005/0260198 Al, all of
which are
incorporated herein by reference.
[0102] Mutation of CR2 amino acid residues determined to be important for a
binding
interaction between CR2 and a specific ligand will likely decrease the binding
affinity of CR2
for that specific ligand while leaving the binding affinity of CR2 for its
other ligands
relatively unaffected. For example, mutation of at least one of amino acid
residues N11, R36,
K41, Y64, and K67 in CR2 (SEQ ID NO: 1) will likely reduce the binding
affinity of CR2 for
EBV gp350 while leaving its binding affinity for C3d and IFNa unchanged.
Similarly,
mutation of at least one of amino acid residues S42 and K50 will likely reduce
the binding
affinity of CR2 for IFNa while leaving its binding affinity for C3d and gp350
unchanged.
Mutation of at least one of amino acid residues 19, Y29, C31, G33, T34, D56,
S70, H90, D92,
S93, A97, T100, N101, S 109, and S128 will likely reduce the binding affinity
of CR2 for
C3d while leaving its binding affinity for IFNa and gp350 unchanged.
[0103] Provided herein are soluble compositions comprising a construct,
wherein the
construct comprises: (a) a complement receptor 2 (CR2) portion comprising a
CR2 protein of
SEQ ID NO:1 or a biologically active fragment thereof, wherein the CR2 portion
contains at
least the first two N-terminal SCR domains of the CR2 protein; and (b) a
complement
modulator portion; wherein the CR2 portion contains at least one amino acid
substitution. In
certain embodiments, the construct is a fusion protein. In some embodiments
the construct
selectively binds to one or more C3 proteolytic fragments but does not bind to
or has reduced
binding affinity for IFNa or EBV gp350. In some embodiments, the construct
selectively
38
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
binds to one or more C3 proteolytic fragments and IFNa but does not bind to or
has reduced
binding affinity for EBV gp350. In some embodiments the construct selectively
binds to one
or more C3 proteolytic fragments and EBV gp350 but does not bind to or has
reduced
binding affinity for IFNa. In some embodiments, the construct selectively
binds to IFNa but
does not bind to or has reduced binding affinity for one or more C3
proteolytic fragments and
EBV gp350. In some embodiments, the construct selectively binds to IFNa and
EBV gp350
but does not bind to or has reduced binding affinity for one or more C3
proteolytic fragments.
In some embodiments, the construct selectively binds to EBV gp350 but does not
bind to or
has reduced binding affinity for IFNa and one or more C3 proteolytic
fragments.
[0104] In certain embodiments, the CR2 portion contains at least one amino
acid substitution
at an amino acid residue selected from the group consisting of: N11, R36, K41,
S42, K50,
Y64 and K67. In certain embodiments, the CR2 portion contains at least two
amino acid
substitutions at an amino acid residue selected from the group consisting of:
N11, R36, K41,
S42, K50, Y64 and K67. In certain embodiments, the CR2 portion contains at
least three
amino acid substitutions at an amino acid residue selected from the group
consisting of: N11,
R36, K41, S42, K50, Y64 and K67. In certain embodiments, the CR2 portion
contains at least
four amino acid substitutions at an amino acid residue selected from the group
consisting of:
N11, R36, K41, S42, K50, Y64 and K67. In certain embodiments, the CR2 portion
contains
at least five amino acid substitutions at an amino acid residue selected from
the group
consisting of: N11, R36, K41, S42, K50, Y64 and K67. In certain embodiments,
the CR2
portion contains at least six amino acid substitutions at an amino acid
residue selected from
the group consisting of: N11, R36, K41, S42, K50, Y64 and K67. In certain
embodiments,
the CR2 portion contains at least seven amino acid substitutions at an amino
acid residue
selected from the group consisting of: N11, R36, K41, S42, K50, Y64 and K67.
In any of the
above embodiments, the at least one, two, three, four, five, six, or seven
amino acid
substitutions may be conservative substitutions. In any of the above
embodiments, the at least
one, two, three, four, five, six, or seven amino acid substitutions may be non-
conservative
substitutions.
[0105] In certain embodiments, the CR2 portion contains at least one amino
acid substitution
at an amino acid residue selected from the group consisting of: N11, R36, K41,
Y64 and
K67, and the CR2 portion has decreased binding affinity for EBV gp350. In
certain
embodiments, the CR2 portion contains at least two amino acid substitutions at
an amino acid
residue selected from the group consisting of: N11, R36, K41, Y64 and K67, and
the CR2
39
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
portion has decreased binding affinity for EBV gp350. In certain embodiments,
the CR2
portion contains at least three amino acid substitutions at an amino acid
residue selected from
the group consisting of: N11, R36, K41, Y64 and K67, and the CR2 portion has
decreased
binding affinity for EBV gp350. In certain embodiments, the CR2 portion
contains at least
four amino acid substitutions at an amino acid residue selected from the group
consisting of:
N11, R36, K41, Y64 and K67, and the CR2 portion has decreased binding affinity
for EBV
gp350. In certain embodiments, the CR2 portion contains at least five amino
acid
substitutions at an amino acid residue selected from the group consisting of:
N11, R36, K41,
Y64 and K67, and the CR2 portion has decreased binding affinity for EBV gp350.
In any of
the above embodiments, the at least one, two, three, four, or five amino acid
substitutions
may be conservative substitutions. In any of the above embodiments, the at
least one, two,
three, four, or five amino acid substitutions may be non-conservative
substitutions.
[0106] In certain embodiments, the CR2 portion contains at least one amino
acid substitution
at an amino acid residue selected from the group consisting of: S42 and K50,
and the CR2
portion has decreased binding affinity for IFNa. In certain embodiments, the
CR2 portion
contains at least two amino acid substitutions at an amino acid residue
selected from the
group consisting of: S42 and K50, and the CR2 portion has decreased binding
affinity for
IFNa. In any of the above embodiments, the at least one or two amino acid
substitutions may
be conservative substitutions. In any of the above embodiments, the at least
one or two amino
acid substitutions may be non-conservative substitutions.
[0107] In certain embodiments, the CR2 portion comprises a polypeptide that
contains some
or all of the ligand binding sites of the CR2 protein, and includes, but is
not limited to, full-
length CR2 proteins (such as human CR2 as shown in SEQ ID NO: 1), soluble CR2
proteins
(such as a CR2 fragment comprising the extracellular domain of CR2) and other
biologically
active fragments of CR2, such as a CR2 fragment comprising SCR1-2 (SEQ ID
NO:2), or
any homolog of a naturally occurring CR2 or fragment thereof, as described in
detail below.
In certain embodiments, the CR2 portion has one or more of the following
properties of CR2:
(1) the ability to selectively bind to the C3 proteolytic fragment C3d , (2)
the ability to
selectively bind to the C3 proteolytic fragment iC3b, (3) the ability to
selectively bind to the
C3 proteolytic fragment C3dg, (4) the ability to selectively bind to one or
more cell-bound
fragments of the C3 proteolytic fragment C3b that bind to the two N-terminal
SCR domains
of CR2, (5) the ability to selectively bind to EBV gp350, and (6) the ability
to selectively
bind to IFNa.
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
[0108] In certain embodiments, the CR2 portion comprises the first two N-
terminal SCR
domains of CR2 (amino acids 23 through 146 of SEQ ID NO:2). In certain
embodiments, the
CR2 portion comprises the first three N-terminal SCR domains of CR2 (amino
acids 23
through 212 of SEQ ID NO: 1). In certain embodiments, the CR2 portion
comprises the first
four N-terminal SCR domains of CR2 (amino acids 23 through 271 of SEQ ID NO:
1). In
certain embodiments, the CR2 portion comprises (and in some embodiments
consists of or
consists essentially of) at least the first two N-terminal SCR domains of CR2,
including for
example at least any of the first 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, or
15 SCR domains of
CR2.
[0109] In certain embodiments, the CR2 portion of the constructs described
herein comprises
(and in some embodiments consists of or consists essentially of) at least 1,
2, 3, 4, 5, 6, 7, or
more amino acid substitutions. In certain embodiments, the amino acid
substitutions may be
conservative substitutions. In certain embodiments, the amino acid
substitutions may be non-
conservative substitutions. In certain embodiments, the amino acid
substitutions may be a
mixture of conservative and non-conservative substitutions.
Compositions for Targeted Delivery of Complement Modulators to Areas of
Complement System Activation
[0110] In some aspects, there is provided a soluble composition capable of
targeted delivery
of a complement modulator to sites of complement system activation comprising
a construct,
wherein the construct comprises: (a) a complement receptor 2 (CR2) portion
comprising a
CR2 protein of SEQ ID NO:1 or a biologically active fragment thereof, wherein
the CR2
portion contains at least the first two N-terminal SCR domains of the CR2
protein; and (b) a
complement modulator portion; wherein the CR2 portion contains at least one
amino acid
substitution that decreases binding affinity of the CR2 portion for EBV gp350.
In certain
embodiments, the CR2 portion contains at least one amino acid substitution at
an amino acid
residue selected from the group consisting of: N11, R36, K41, Y64 and K67 and
does not
bind to, or has decreased binding affinity for, EBV gp350. In certain
embodiments, the CR2
portion contains at least two amino acid substitutions at amino acid residues
selected from the
group consisting of: N11, R36, K41, Y64 and K67 and does not bind to, or has
decreased
binding affinity for, EBV gp350. In certain embodiments, the CR2 portion
contains at least
three amino acid substitutions at amino acid residues selected from the group
consisting of:
N11, R36, K41, Y64 and K67 and does not bind to, or has decreased binding
affinity for,
EBV gp350. In certain embodiments, the CR2 portion contains at least four
amino acid
41
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
substitutions at amino acid residues selected from the group consisting of:
N11, R36, K41,
Y64 and K67 and does not bind to, or has decreased binding affinity for, EBV
gp350. In
certain embodiments, the CR2 portion contains five amino acid substitutions at
amino acid
residues N11, R36, K41, Y64 and K67 and does not bind to, or has decreased
binding affinity
for, EBV gp350. In any of the above embodiments, the at least one, two, three,
four, or five
amino acid substitutions may be conservative substitutions. In any of the
above embodiments,
the at least one, two, three, four, or five amino acid substitutions may be
non-conservative
substitutions. In certain embodiments, the CR2 portion contains one or more
substitutions in
amino acids from the group consisting of: NI IA, R36A, K41A, Y64A, and K67A.
In certain
embodiments, the construct is a fusion protein. In some embodiments, the CR2
portion
selectively binds to one or more proteins from the group consisting of: C3d,
iC3b, C3dg, one
or more cell-bound fragments of C3b that bind to the two N-terminal SCR
domains of CR2,
CD23, and IFNa. In some embodiments, the construct does not bind to EBV gp350.
In other
embodiments, the construct has decreased binding affinity to EBV gp350. In
some
embodiments, the at least one amino acid substitution decreases the binding
affinity of the
CR2 portion for EBV gp350 by any of about 20%, 25%, 30%, 35%, 40%, 45%, 50%,
55%,
60%, 65%, 70%, 75%, 80%, 85%, 90%, 95%, or 100%, inclusive, as well as any
numerical
value in between these percentages. In some embodiments, the complement
modulator
portion is a complement inhibitor. In some of these embodiments, the
complement inhibitor
is selected from the group consisting of: MCP, DAF, CD59, Crry, CR1, and FH.
In other
embodiments, the complement modulator portion is a complement activator. In
some of
these embodiments, the complement activator is selected from the group
consisting of:
human Ig isotype Gi (IgGi), human Ig isotype M (IgM), mouse Ig isotype G3
(IgG3), mouse
IgM Fc, human IgGi Fc domain, human IgM Fc domain, mouse IgG3 Fc domain, mouse
IgM
Fc domain, and CVF.
[0111] In some aspects, there is provided a soluble composition capable of
targeted delivery
of a complement modulator to sites of complement system activation comprising
a construct,
wherein the construct comprises: (a) a complement receptor 2 (CR2) portion
comprising a
CR2 protein of SEQ ID NO:1 or a biologically active fragment thereof, wherein
the CR2
portion contains at least the first two N-terminal SCR domains of the CR2
protein; and (b) a
complement modulator portion; wherein the CR2 portion contains at least one
amino acid
substitution that decreases binding affinity of the CR2 portion for IFNa. In
certain
embodiments, the CR2 portion contains at least one amino acid substitution at
an amino acid
42
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
residue selected from the group consisting of S42 and K50 and does not bind
to, or has
decreased binding affinity for, IFNa. In certain embodiments, the CR2 portion
contains at
least two amino acid substitutions at amino acid residues S42 and K50 and does
not bind to,
or has decreased binding affinity for, IFNa. In any of the above embodiments,
the at least
one or two amino acid substitutions may be conservative substitutions. In any
of the above
embodiments, the at least one or two amino acid substitutions may be non-
conservative
substitutions. In certain embodiments, the CR2 portion contains one or more
substitutions in
amino acids from the group consisting of: S42A and K50A. In certain
embodiments, the
construct is a fusion protein. In some embodiments, the CR2 portion
selectively binds to one
or more proteins from the group consisting of: C3d, iC3b, C3dg, one or more
cell-bound
fragments of C3b that bind to the two N-terminal SCR domains of CR2, CD23, and
EBV
gp350. In some embodiments, the construct does not bind to IFNa. In other
embodiments,
the construct has decreased binding affinity to IFNa. In some embodiments, the
at least one
amino acid substitution decreases the binding affinity of the CR2 portion for
IFNa by any of
about 20%, 25%, 30%, 35%, 40%, 45%, 50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%,
90%,
95%, or 100%, inclusive, as well as any numerical value in between these
percentages. In
some embodiments, the complement modulator portion is a complement inhibitor.
In some
of these embodiments, the complement inhibitor is selected from the group
consisting of:
MCP, DAF, CD59, Crry, CR1, and FH. In other embodiments, the complement
modulator
portion is a complement activator. In some of these embodiments, the
complement activator
is selected from the group consisting of: human Ig isotype Gi (IgGi), human Ig
isotype M
(IgM), mouse Ig isotype G3 (IgG3), mouse IgM Fc, human IgG1 Fc domain, human
IgM Fc
domain, mouse IgG3 Fc domain, mouse IgM Fc domain, and CVF.
[0112] In some aspects, there is provided a soluble composition capable of
targeted delivery
of a complement modulator to sites of complement system activation comprising
a construct,
wherein the construct comprises: (a) a complement receptor 2 (CR2) portion
comprising a
CR2 protein of SEQ ID NO:1 or a biologically active fragment thereof, wherein
the CR2
portion contains at least the first two N-terminal SCR domains of the CR2
protein; and (b) a
complement modulator portion; wherein the CR2 portion contains at least two
amino acid
substitutions that decrease binding affinity of the CR2 portion for EBV gp350
and IFNa. In
certain embodiments, the CR2 portion contains at least one amino acid
substitution at an
amino acid residue selected from the group consisting of: S42 and K50, further
contains at
least one amino acid substitution at an amino acid residue selected from the
group consisting
43
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
of: N11, R36, K41, Y64 and K67, and does not bind to, or has decreased binding
affinity for,
EBV gp350 and IFNa. In certain embodiments, the CR2 portion contains at least
one amino
acid substitution at an amino acid residue selected from the group consisting
of: S42 and
K50, further contains at least two amino acid substitutions at amino acid
residues selected
from the group consisting of: N11, R36, K41, Y64 and K67, and does not bind
to, or has
decreased binding affinity for, EBV gp350 and IFNa. In certain embodiments,
the CR2
portion contains at least one amino acid substitution at an amino acid residue
selected from
the group consisting of: S42 and K50, further contains at least three amino
acid substitutions
at amino acid residues selected from the group consisting of: N11, R36, K41,
Y64 and K67,
and does not bind to, or has decreased binding affinity for, EBV gp350 and
IFNa. In certain
embodiments, the CR2 portion contains at least one amino acid substitution at
an amino acid
residue selected from the group consisting of: S42 and K50, further contains
at least four
amino acid substitutions at amino acid residues selected from the group
consisting of: N11,
R36, K41, Y64 and K67, and does not bind to, or has decreased binding affinity
for, EBV
gp350 and IFNa. In certain embodiments, the CR2 portion contains at least one
amino acid
substitution at an amino acid residue selected from the group consisting of:
S42 and K50,
further contains five amino acid substitutions at amino acid residues N11,
R36, K41, Y64 and
K67, and does not bind to, or has decreased binding affinity for, EBV gp350
and IFNa. In
any of the above embodiments, the at least two amino acid substitutions may be
conservative
substitutions. In any of the above embodiments, the at least two amino acid
substitutions may
be non-conservative substitutions. In certain embodiments, the CR2 portion
contains one or
more substitutions in an amino acid from the group consisting of: NI IA, R36A,
K41A,
Y64A, and K67A and a mutation from the group consisting of S42A and K50A. In
certain
embodiments, the construct is a fusion protein. In some embodiments, the CR2
portion
selectively binds to one or more proteins from the group consisting of: C3d,
iC3b, C3dg, one
or more cell-bound fragments of C3b that bind to the two N-terminal SCR
domains of CR2,
and CD23. In some embodiments, the construct does not bind to IFNa and EBV
gp350. In
other embodiments, the construct has decreased binding affinity to IFNa and
EBV gp350. In
some embodiments, the at least two amino acid substitutions decrease the
binding affinity of
the CR2 portion for IFNa and EBV gp350 by any of about 20%, 25%, 30%, 35%,
40%, 45%,
50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 95%, or 100%, inclusive, as well
as any
numerical value in between these percentages. In some embodiments, the
complement
modulator portion is a complement inhibitor. In some of these embodiments, the
44
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
complement inhibitor is selected from the group consisting of: MCP, DAF, CD59,
Crry, CR1,
and FH. In other embodiments, the complement modulator portion is a complement
activator. In some of these embodiments, the complement activator is selected
from the
group consisting of: human Ig isotype Gi (IgGi), human Ig isotype M (IgM),
mouse Ig
isotype G3 (IgG3), mouse IgM Fc, human IgG1 Fc domain, human IgM Fc domain,
mouse
IgG3 Fc domain, mouse IgM Fc domain, and CVF.
[0113] In some aspects, there is provided a soluble composition capable of
targeted delivery
of a complement modulator to sites of complement system activation comprising
a construct,
wherein the construct comprises: (a) a complement receptor 2 (CR2) portion
comprising a
CR2 protein of SEQ ID NO:1 or a biologically active fragment thereof, wherein
the CR2
portion contains at least the first two N-terminal SCR domains of the CR2
protein; and (b) a
complement modulator portion; wherein the CR2 portion contains at least three
amino acid
substitutions that decrease binding affinity of the CR2 portion for EBV gp350
and IFNa. In
certain embodiments, the CR2 portion contains amino acid substitutions at
amino acid
residues S42 and K50, further contains at least one amino acid substitution at
an amino acid
residue selected from the group consisting of: N11, R36, K41, Y64 and K67, and
does not
bind to, or has decreased binding affinity for, EBV gp350 and IFNa. In certain
embodiments, the CR2 portion contains amino acid substitutions at amino acid
residues S42
and K50, further contains at least two amino acid substitutions at amino acid
residues
selected from the group consisting of: N11, R36, K41, Y64 and K67, and does
not bind to, or
has decreased binding affinity for, EBV gp350 and IFNa. In certain
embodiments, the CR2
portion contains amino acid substitutions at amino acid residues S42 and K50,
further
contains at least three amino acid substitutions at amino acid residues
selected from the group
consisting of: N11, R36, K41, Y64 and K67, and does not bind to, or has
decreased binding
affinity for, EBV gp350 and IFNa. In certain embodiments, the CR2 portion
contains amino
acid substitutions at amino acid residues S42 and K50, further contains at
least four amino
acid substitutions at amino acid residues selected from the group consisting
of: N11, R36,
K41, Y64 and K67, and does not bind to, or has decreased binding affinity for,
EBV gp350
and IFNa. In certain embodiments, the CR2 portion contains amino acid
substitutions at
amino acid residues S42 and K50, further contains five amino acid
substitutions at amino
acid residues N11, R36, K41, Y64 and K67, and does not bind to, or has
decreased binding
affinity for, EBV gp350 and IFNa. In any of the above embodiments, the at
least three amino
acid substitutions may be conservative substitutions. In any of the above
embodiments, the at
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
least three amino acid substitutions may be non-conservative substitutions. In
certain
embodiments, the CR2 portion contains one or more substitutions in an amino
acid from the
group consisting of: NI IA, R36A, K41A, Y64A, and K67A and mutations S42A and
K50A.
In certain embodiments, the construct is a fusion protein. In some
embodiments, the CR2
portion selectively binds to one or more proteins from the group consisting
of: C3d, iC3b,
C3dg, one or more cell-bound fragments of C3b that bind to the two N-terminal
SCR
domains of CR2, and CD23. In some embodiments, the construct does not bind to
IFNa and
EBV gp350. In other embodiments, the construct has decreased binding affinity
to IFNa and
EBV gp350. In some embodiments, the at least three amino acid substitutions
decrease the
binding affinity of the CR2 portion for IFNa and EBV gp350 by any of about
20%, 25%,
30%, 35%, 40%, 45%, 50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 95%, or 100%,
inclusive, as well as any numerical value in between these percentages. In
some
embodiments, the complement modulator portion is a complement inhibitor. In
some of
these embodiments, the complement inhibitor is selected from the group
consisting of: MCP,
DAF, CD59, Crry, CR1, and FH. In other embodiments, the complement modulator
portion
is a complement activator. In some of these embodiments, the complement
activator is
selected from the group consisting of: human Ig isotype Gi (IgGi), human Ig
isotype M
(IgM), mouse Ig isotype G3 (IgG3), mouse IgM Fc, human IgG1 Fc domain, human
IgM Fc
domain, mouse IgG3 Fc domain, mouse IgM Fc domain, and CVF.
Compositions for Targeted Delivery of Complement Modulators to Sites of
Epstein
Barr Virus infection
[0114] In some aspects, there is provided a soluble composition capable of
targeted delivery
of a complement modulator to sites of Epstein Barr Virus infection comprising
a construct,
wherein the construct comprises: (a) a complement receptor 2 (CR2) portion
comprising a
CR2 protein of SEQ ID NO:1 or a biologically active fragment thereof, wherein
the CR2
portion contains at least the first two N-terminal SCR domains of the CR2
protein; and (b) a
complement modulator portion; wherein the CR2 portion contains at least one
amino acid
substitution that decreases binding affinity of the CR2 portion for one or
more C3 proteolytic
fragments. In certain embodiments, the CR2 portion contains at least one amino
acid
substitution at an amino acid residue selected from the group consisting of:
19, Y29, C31,
G33, T34, D56, S70, H90, D92, S93, A97, T100, N101, 5109, and 5128 and does
not bind to,
or has decreased binding affinity for, one or more C3 proteolytic fragments.
In certain
embodiments, the CR2 portion contains at least two amino acid substitutions at
amino acid
46
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
residues selected from the group consisting of: 19, Y29, C31, G33, T34, D56,
S70, H90, D92,
S93, A97, T100, N101, 5109, and S128 and does not bind to, or has decreased
binding
affinity for, one or more C3 proteolytic fragments. In certain embodiments,
the CR2 portion
contains at least three amino acid substitutions at amino acid residues
selected from the group
consisting of: 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97, T100,
N101, S109,
and S 128 and does not bind to, or has decreased binding affinity for, one or
more C3
proteolytic fragments. In certain embodiments, the CR2 portion contains at
least four amino
acid substitutions at amino acid residues selected from the group consisting
of: 19, Y29, C3 1,
G33, T34, D56, S70, H90, D92, S93, A97, T100, N101, 5109, and 5128 and does
not bind to,
or has decreased binding affinity for, one or more C3 proteolytic fragments.
In certain
embodiments, the CR2 portion contains at least five amino acid substitutions
at amino acid
residues selected from the group consisting of: 19, Y29, C31, G33, T34, D56,
S70, H90, D92,
S93, A97, T100, N101, 5109, and 5128 and does not bind to, or has decreased
binding
affinity for, one or more C3 proteolytic fragments. In certain embodiments,
the CR2 portion
contains at least six amino acid substitutions at amino acid residues selected
from the group
consisting of: 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97, T100,
N101, S109,
and S 128 and does not bind to, or has decreased binding affinity for, one or
more C3
proteolytic fragments. In certain embodiments, the CR2 portion contains at
least seven amino
acid substitutions at amino acid residues selected from the group consisting
of: 19, Y29, C3 1,
G33, T34, D56, S70, H90, D92, S93, A97, T100, N101, 5109, and 5128 and does
not bind to,
or has decreased binding affinity for, one or more C3 proteolytic fragments.
In certain
embodiments, the CR2 portion contains at least eight amino acid substitutions
at amino acid
residues selected from the group consisting of: 19, Y29, C31, G33, T34, D56,
S70, H90, D92,
S93, A97, T100, N101, 5109, and 5128 and does not bind to, or has decreased
binding
affinity for, one or more C3 proteolytic fragments. In certain embodiments,
the CR2 portion
contains at least nine amino acid substitutions at amino acid residues
selected from the group
consisting of: 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97, T100,
N101, S109,
and S 128 and does not bind to, or has decreased binding affinity for, one or
more C3
proteolytic fragments. In certain embodiments, the CR2 portion contains at
least ten amino
acid substitutions at amino acid residues selected from the group consisting
of: 19, Y29, C3 1,
G33, T34, D56, S70, H90, D92, S93, A97, T100, N101, 5109, and 5128 and does
not bind to,
or has decreased binding affinity for, one or more C3 proteolytic fragments.
In certain
embodiments, the CR2 portion contains at least eleven amino acid substitutions
at amino acid
47
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
residues selected from the group consisting of: 19, Y29, C31, G33, T34, D56,
S70, H90, D92,
S93, A97, T100, N101, 5109, and S128 and does not bind to, or has decreased
binding
affinity for, one or more C3 proteolytic fragments. In certain embodiments,
the CR2 portion
contains at least twelve amino acid substitutions at amino acid residues
selected from the
group consisting of: 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97,
T100, N101,
S 109, and S 128 and does not bind to, or has decreased binding affinity for,
one or more C3
proteolytic fragments. In certain embodiments, the CR2 portion contains at
least thirteen
amino acid substitutions at amino acid residues selected from the group
consisting of: 19,
Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97, T100, N101, 5109, and 5128
and does
not bind to, or has decreased binding affinity for, one or more C3 proteolytic
fragments. In
certain embodiments, the CR2 portion contains at least fourteen amino acid
substitutions at
amino acid residues selected from the group consisting of: 19, Y29, C31, G33,
T34, D56,
S70, H90, D92, S93, A97, T100, N101, S 109, and S 128 and does not bind to, or
has
decreased binding affinity for, one or more C3 proteolytic fragments. In
certain
embodiments, the CR2 portion contains fifteen amino acid substitutions at
amino acid
residues 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97, T100, N101,
S109, and
S 128 and does not bind to, or has decreased binding affinity for, one or more
C3 proteolytic
fragments. In some embodiments, the one or more C3 proteolytic fragments are
selected
from the group consisting of C3d, iC3b, C3dg, and one or more cell-bound
fragments of C3b
that bind to the two N-terminal SCR domains of CR2. In any of the above
embodiments, the
at least one, two, three, four, five, six, seven, eight, nine, ten, eleven,
twelve, thirteen,
fourteen, or fifteen amino acid substitutions may be conservative
substitutions. In any of the
above embodiments, the at least one, two, three, four, five, six, seven,
eight, nine, ten, eleven,
twelve, thirteen, fourteen, or fifteen amino acid substitutions may be non-
conservative
substitutions. In certain embodiments, the construct is a fusion protein. In
some
embodiments, the CR2 portion selectively binds to one or more proteins from
the group
consisting of: CD23, EBV gp350, and IFNa. In some embodiments, the construct
does not
bind to one or more C3 proteolytic fragments. In other embodiments, the
construct has
decreased binding affinity to one or more C3 proteolytic fragments. In some
embodiments,
the at least one amino acid substitution decreases the binding affinity of the
CR2 portion for
one or more C3 proteolytic fragments by any of about 20%, 25%, 30%, 35%, 40%,
45%,
50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 95%, or 100%, inclusive, as well
as any
numerical value in between these percentages. In some embodiments, the
complement
48
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
modulator portion is a complement inhibitor. In some of these embodiments, the
complement inhibitor is selected from the group consisting of: MCP, DAF, CD59,
Crry, CR1,
and FH. In other embodiments, the complement modulator portion is a complement
activator. In some of these embodiments, the complement activator is selected
from the
group consisting of: human Ig isotype Gi (IgGi), human Ig isotype M (IgM),
mouse Ig
isotype G3 (IgG3), mouse IgM Fc, human IgG1 Fc domain, human IgM Fc domain,
mouse
IgG3 Fc domain, mouse IgM Fc domain, and CVF.
[0115] In some aspects, there is provided a soluble composition capable of
targeted delivery
of a complement modulator to sites of Epstein Barr Virus infection comprising
a construct,
wherein the construct comprises: (a) a complement receptor 2 (CR2) portion
comprising a
CR2 protein of SEQ ID NO:1 or a biologically active fragment thereof, wherein
the CR2
portion contains at least the first two N-terminal SCR domains of the CR2
protein; and (b) a
complement modulator portion; wherein the CR2 portion contains at least two
amino acid
substitutions that decrease binding affinity of the CR2 portion for one or
more C3 proteolytic
fragments and IFNa. In certain embodiments, the CR2 portion contains at least
one amino
acid substitution at an amino acid residue selected from the group consisting
of: S42 and
K50, further contains at least one amino acid substitution at an amino acid
residue selected
from the group consisting of: 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93,
A97, T100,
N101, S 109, and S 128, and does not bind to, or has decreased binding
affinity for, one or
more C3 proteolytic fragments and IFNa. In certain embodiments, the CR2
portion contains
at least one amino acid substitution at an amino acid residue selected from
the group
consisting of: S42 and K50, further contains at least two amino acid
substitutions at amino
acid residues selected from the group consisting of: 19, Y29, C31, G33, T34,
D56, S70, H90,
D92, S93, A97, T100, N101, S 109, and S 128, and does not bind to, or has
decreased binding
affinity for, one or more C3 proteolytic fragments and IFNa In certain
embodiments, the
CR2 portion contains at least one amino acid substitution at an amino acid
residue selected
from the group consisting of: S42 and K50, further contains at least three
amino acid
substitutions at amino acid residue selected from the group consisting of: 19,
Y29, C31, G33,
T34, D56, S70, H90, D92, S93, A97, T100, N101, S 109, and S 128, and does not
bind to, or
has decreased binding affinity for, one or more C3 proteolytic fragments and
IFNa. In
certain embodiments, the CR2 portion contains at least one amino acid
substitution at an
amino acid residue selected from the group consisting of: S42 and K50, further
contains at
least four amino acid substitutions at amino acid residues selected from the
group consisting
49
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
of: 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97, T100, N101, S109,
and S128,
and does not bind to, or has decreased binding affinity for, one or more C3
proteolytic
fragments and IFNa. In certain embodiments, the CR2 portion contains at least
one amino
acid substitution at an amino acid residue selected from the group consisting
of: S42 and
K50, further contains at least five amino acid substitutions at amino acid
residues selected
from the group consisting of: 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93,
A97, T100,
N101, S 109, and S 128, and does not bind to, or has decreased binding
affinity for, one or
more C3 proteolytic fragments and IFNa. In certain embodiments, the CR2
portion contains
at least one amino acid substitution at an amino acid residue selected from
the group
consisting of: S42 and K50, further contains at least six amino acid
substitutions at amino
acid residues selected from the group consisting of: 19, Y29, C31, G33, T34,
D56, S70, H90,
D92, S93, A97, T100, N101, S 109, and S 128, and does not bind to, or has
decreased binding
affinity for, one or more C3 proteolytic fragments and IFNa. In certain
embodiments, the
CR2 portion contains at least one amino acid substitution at an amino acid
residue selected
from the group consisting of: S42 and K50, further contains at least seven
amino acid
substitutions at amino acid residues selected from the group consisting of:
19, Y29, C3 1,
G33, T34, D56, S70, H90, D92, S93, A97, T100, N101, 5109, and 5128, and does
not bind
to, or has decreased binding affinity for, one or more C3 proteolytic
fragments and IFNa. In
certain embodiments, the CR2 portion contains at least one amino acid
substitution at an
amino acid residue selected from the group consisting of: S42 and K50, further
contains at
least eight amino acid substitutions at amino acid residues selected from the
group consisting
of: 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97, T100, N101, S109,
and S128,
and does not bind to, or has decreased binding affinity for, one or more C3
proteolytic
fragments and IFNa. In certain embodiments, the CR2 portion contains at least
one amino
acid substitution at an amino acid residue selected from the group consisting
of: S42 and
K50, further contains at least nine amino acid substitutions at amino acid
residues selected
from the group consisting of: 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93,
A97, T100,
N101, S 109, and S 128, and does not bind to, or has decreased binding
affinity for, one or
more C3 proteolytic fragments and IFNa. In certain embodiments, the CR2
portion contains
at least one amino acid substitution at an amino acid residue selected from
the group
consisting of: S42 and K50, further contains at least ten amino acid
substitutions at amino
acid residues selected from the group consisting of: 19, Y29, C31, G33, T34,
D56, S70, H90,
D92, S93, A97, T100, N101, S 109, and S 128, and does not bind to, or has
decreased binding
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
affinity for, one or more C3 proteolytic fragments and IFNa. In certain
embodiments, the
CR2 portion contains at least one amino acid substitution at an amino acid
residue selected
from the group consisting of: S42 and K50, further contains at least eleven
amino acid
substitutions at amino acid residues selected from the group consisting of:
19, Y29, C31, G33,
T34, D56, S70, H90, D92, S93, A97, T100, N101, 5109, and 5128, and does not
bind to, or
has decreased binding affinity for, one or more C3 proteolytic fragments and
IFNa. In
certain embodiments, the CR2 portion contains at least one amino acid
substitution at an
amino acid residue selected from the group consisting of: S42 and K50, further
contains at
least twelve amino acid substitutions at amino acid residues selected from the
group
consisting of: 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97, T100,
N101, S109,
and S 128, and does not bind to, or has decreased binding affinity for, one or
more C3
proteolytic fragments and IFNa. In certain embodiments, the CR2 portion
contains at least
one amino acid substitution at an amino acid residue selected from the group
consisting of:
S42 and K50, further contains at least thirteen amino acid substitutions at
amino acid residues
selected from the group consisting of: 19, Y29, C31, G33, T34, D56, S70, H90,
D92, S93,
A97, T100, N101, 5109, and 5128, and does not bind to, or has decreased
binding affinity
for, one or more C3 proteolytic fragments and IFNa. In certain embodiments,
the CR2
portion contains at least one amino acid substitution at an amino acid residue
selected from
the group consisting of: S42 and K50, further contains at least fourteen amino
acid
substitutions at amino acid residues selected from the group consisting of:
19, Y29, C3 1,
G33, T34, D56, S70, H90, D92, S93, A97, T100, N101, 5109, and 5128, and does
not bind
to, or has decreased binding affinity for, one or more C3 proteolytic
fragments and IFNa. In
certain embodiments, the CR2 portion contains at least one amino acid
substitution at an
amino acid residue selected from the group consisting of: S42 and K50, further
contains
fifteen amino acid substitutions at amino acid residues 19, Y29, C31, G33,
T34, D56, S70,
H90, D92, S93, A97, T100, N101, 5109, and 5128, and does not bind to, or has
decreased
binding affinity for, one or more C3 proteolytic fragments and IFNa. In some
embodiments,
the one or more C3 proteolytic fragments are selected from the group
consisting of C3d,
iC3b, C3dg, and one or more cell-bound fragments of C3b that bind to the two N-
terminal
SCR domains of CR2. In any of the above embodiments, the at least two amino
acid
substitutions may be conservative substitutions. In any of the above
embodiments, the at least
two amino acid substitutions may be non-conservative substitutions. In certain
embodiments,
the CR2 portion contains a substitution of an amino acid from the group
consisting of S42A
51
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
and K50A. In certain embodiments, the construct is a fusion protein. In some
embodiments,
the CR2 portion selectively binds to one or more proteins from the group
consisting of: CD23
and EBV gp350. In some embodiments, the construct does not bind to IFNa and
one or more
C3 proteolytic fragments. In other embodiments, the construct has decreased
binding affinity
to IFNa and one or more C3 proteolytic fragments. In some embodiments, the at
least two
amino acid substitutions decrease the binding affinity of the CR2 portion for
IFNa and one or
more C3 proteolytic fragments by any of about 20%, 25%, 30%, 35%, 40%, 45%,
50%, 55%,
60%, 65%, 70%, 75%, 80%, 85%, 90%, 95%, or 100%, inclusive, as well as any
numerical
value in between these percentages. In some embodiments, the complement
modulator
portion is a complement inhibitor. In some of these embodiments, the
complement inhibitor
is selected from the group consisting of: MCP, DAF, CD59, Crry, CR1, and FH.
In other
embodiments, the complement modulator portion is a complement activator. In
some of
these embodiments, the complement activator is selected from the group
consisting of:
human Ig isotype Gi (IgGi), human Ig isotype M (IgM), mouse Ig isotype G3
(IgG3), mouse
IgM Fc, human IgGi Fc domain, human IgM Fc domain, mouse IgG3 Fc domain, mouse
IgM
Fc domain, and CVF.
[0116] In some aspects, there is provided a soluble composition capable of
targeted delivery
of a complement modulator to sites of complement system activation comprising
a construct,
wherein the construct comprises: (a) a complement receptor 2 (CR2) portion
comprising a
CR2 protein of SEQ ID NO:1 or a biologically active fragment thereof, wherein
the CR2
portion contains at least the first two N-terminal SCR domains of the CR2
protein; and (b) a
complement modulator portion; wherein the CR2 portion contains at least three
amino acid
substitutions that decrease binding affinity of the CR2 portion for one or
more C3 proteolytic
fragments and IFNa. In certain embodiments, the CR2 portion contains amino
acid
substitutions at amino acid residues S42 and K50, further contains at least
one amino acid
substitution at an amino acid residue selected from the group consisting of:
19, Y29, C3 1,
G33, T34, D56, S70, H90, D92, S93, A97, T100, N101, S 109, and S 128, and does
not bind
to, or has decreased binding affinity for, one or more C3 proteolytic
fragments and IFNa. In
certain embodiments, the CR2 portion contains amino acid substitutions at
amino acid
residues S42 and K50, further contains at least two amino acid substitutions
at amino acid
residues selected from the group consisting of: 19, Y29, C31, G33, T34, D56,
S70, H90,
D92, S93, A97, T100, N101, S 109, and S 128, and does not bind to, or has
decreased binding
affinity for, one or more C3 proteolytic fragments and IFNa. In certain
embodiments, the
52
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
CR2 portion contains amino acid substitutions at amino acid residues S42 and
K50, further
contains at least three amino acid substitutions at amino acid residues
selected from the group
consisting of: 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97, T100,
N101, S109,
and S 128, and does not bind to, or has decreased binding affinity for, one or
more C3
proteolytic fragments and IFNa. In certain embodiments, the CR2 portion
contains amino
acid substitutions at amino acid residues S42 and K50, further contains at
least four amino
acid substitutions at amino acid residues selected from the group consisting
of: 19, Y29, C3 1,
G33, T34, D56, S70, H90, D92, S93, A97, T100, N101, 5109, and 5128, and does
not bind
to, or has decreased binding affinity for, one or more C3 proteolytic
fragments and IFNa. In
certain embodiments, the CR2 portion contains amino acid substitutions at
amino acid
residues S42 and K50, further contains at least five amino acid substitutions
at amino acid
residues selected from the group consisting of: 19, Y29, C31, G33, T34, D56,
S70, H90,
D92, S93, A97, T100, N101, S 109, and S 128, and does not bind to, or has
decreased binding
affinity for, one or more C3 proteolytic fragments and IFNa. In certain
embodiments, the
CR2 portion contains amino acid substitutions at amino acid residues S42 and
K50, further
contains at least six amino acid substitutions at amino acid residues selected
from the group
consisting of: 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97, T100,
N101, S109,
and S 128, and does not bind to, or has decreased binding affinity for, one or
more C3
proteolytic fragments and IFNa. In certain embodiments, the CR2 portion
contains amino
acid substitutions at amino acid residues S42 and K50, further contains at
least seven amino
acid substitutions at amino acid residues selected from the group consisting
of: 19, Y29, C3 1,
G33, T34, D56, S70, H90, D92, S93, A97, T100, N101, 5109, and 5128, and does
not bind
to, or has decreased binding affinity for, one or more C3 proteolytic
fragments and IFNa. In
certain embodiments, the CR2 portion contains amino acid substitutions at
amino acid
residues S42 and K50, further contains at least eight amino acid substitutions
at amino acid
residues selected from the group consisting of: 19, Y29, C31, G33, T34, D56,
S70, H90,
D92, S93, A97, T100, N101, S 109, and S 128, and does not bind to, or has
decreased binding
affinity for, one or more C3 proteolytic fragments and IFNa. In certain
embodiments, the
CR2 portion contains amino acid substitutions at amino acid residues S42 and
K50, further
contains at least nine amino acid substitutions at amino acid residues
selected from the group
consisting of: 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97, T100,
N101, S109,
and S 128, and does not bind to, or has decreased binding affinity for, one or
more C3
proteolytic fragments and IFNa. In certain embodiments, the CR2 portion
contains amino
53
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
acid substitutions at amino acid residues S42 and K50, further contains at
least ten amino acid
substitutions at amino acid residues selected from the group consisting of:
19, Y29, C3 1,
G33, T34, D56, S70, H90, D92, S93, A97, T100, N101, 5109, and 5128, and does
not bind
to, or has decreased binding affinity for, one or more C3 proteolytic
fragments and IFNa. In
certain embodiments, the CR2 portion contains amino acid substitutions at
amino acid
residues S42 and K50, further contains at least eleven amino acid
substitutions at amino acid
residues selected from the group consisting of: 19, Y29, C31, G33, T34, D56,
S70, H90,
D92, S93, A97, T100, N101, S 109, and S 128, and does not bind to, or has
decreased binding
affinity for, one or more C3 proteolytic fragments and IFNa. In certain
embodiments, the
CR2 portion contains amino acid substitutions at amino acid residues S42 and
K50, further
contains at least twelve amino acid substitutions at amino acid residues
selected from the
group consisting of: 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97,
T100, N101,
S 109, and S 128, and does not bind to, or has decreased binding affinity for,
one or more C3
proteolytic fragments and IFNa. In certain embodiments, the CR2 portion
contains amino
acid substitutions at amino acid residues S42 and K50, further contains at
least thirteen amino
acid substitutions at amino acid residues selected from the group consisting
of: 19, Y29, C3 1,
G33, T34, D56, S70, H90, D92, S93, A97, T100, N101, 5109, and 5128, and does
not bind
to, or has decreased binding affinity for, one or more C3 proteolytic
fragments and IFNa. In
certain embodiments, the CR2 portion contains amino acid substitutions at
amino acid
residues S42 and K50, further contains at least fourteen amino acid
substitutions at amino
acid residues selected from the group consisting of: 19, Y29, C31, G33, T34,
D56, S70, H90,
D92, S93, A97, T100, N101, S 109, and S 128, and does not bind to, or has
decreased binding
affinity for, one or more C3 proteolytic fragments and IFNa. In certain
embodiments, the
CR2 portion contains amino acid substitutions at amino acid residues S42 and
K50, further
contains fifteen amino acid substitutions at amino acid residues 19, Y29, C31,
G33, T34, D56,
S70, H90, D92, S93, A97, T100, N101, 5109, and 5128, and does not bind to, or
has
decreased binding affinity for, one or more C3 proteolytic fragments and IFNa.
In some
embodiments, the one or more C3 proteolytic fragments are selected from the
group
consisting of C3d, iC3b, C3dg, and one or more cell-bound fragments of C3b
that bind to the
two N-terminal SCR domains of CR2. In any of the above embodiments, the at
least three
amino acid substitutions may be conservative substitutions. In any of the
above embodiments,
the at least three amino acid substitutions may be non-conservative
substitutions. In certain
embodiments, the CR2 portion contains substitutions of amino acids S42A and
K50A. In
54
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
certain embodiments, the construct is a fusion protein. In some embodiments,
the CR2
portion selectively binds to one or more proteins from the group consisting
of: CD23 and
EBV gp350. In some embodiments, the construct does not bind to IFNa and one or
more C3
proteolytic fragments. In other embodiments, the construct has decreased
binding affinity to
IFNa and one or more C3 proteolytic fragments. In some embodiments, the at
least three
amino acid substitutions decreases the binding affinity of the CR2 portion for
IFNa and one
or more C3 proteolytic fragments by any of about 20%, 25%, 30%, 35%, 40%, 45%,
50%,
55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 95%, or 100%, inclusive, as well as
any
numerical value in between these percentages. In some embodiments, the
complement
modulator portion is a complement inhibitor. In some of these embodiments, the
complement inhibitor is selected from the group consisting of: MCP, DAF, CD59,
Crry, CR1,
and FH. In other embodiments, the complement modulator portion is a complement
activator. In some of these embodiments, the complement activator is selected
from the
group consisting of: human Ig isotype Gi (IgGi), human Ig isotype M (IgM),
mouse Ig
isotype G3 (IgG3), mouse IgM Fc, human IgGi Fc domain, human IgM Fc domain,
mouse
IgG3 Fc domain, mouse IgM Fc domain, and CVF.
Compositions for Targeted Delivery of Complement Modulators to Sites of
Interferon
alpha Production
[0117] In some aspects, there is provided a soluble composition capable of
targeted delivery
of a complement modulator to sites of IFNa production comprising a construct,
wherein the
construct comprises: (a) a complement receptor 2 (CR2) portion comprising a
CR2 protein of
SEQ ID NO:1 or a biologically active fragment thereof, wherein the CR2 portion
contains at
least the first two N-terminal SCR domains of the CR2 protein; and (b) a
complement
modulator portion; wherein the CR2 portion contains at least two amino acid
substitutions
that decrease binding affinity of the CR2 portion for one or more C3
proteolytic fragments
and EBV gp350. In certain embodiments, the CR2 portion contains at least one
amino acid
substitution at an amino acid residue selected from the group consisting of:
N11, R36, K41,
Y64 and K67, further contains at least one amino acid substitution at an amino
acid residue
selected from the group consisting of: 19, Y29, C31, G33, T34, D56, S70, H90,
D92, S93,
A97, T100, N101, S 109, and S 128, and does not bind to, or has decreased
binding affinity
for, one or more C3 proteolytic fragments and EBV gp350. In certain
embodiments, the CR2
portion contains at least one amino acid substitution at an amino acid residue
selected from
the group consisting of: N11, R36, K41, Y64 and K67, further contains at least
two amino
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
acid substitutions at amino acid residues selected from the group consisting
of: 19, Y29, C3 1,
G33, T34, D56, S70, H90, D92, S93, A97, T100, N101, S 109, and S 128, and does
not bind
to, or has decreased binding affinity for, one or more C3 proteolytic
fragments and EBV
gp350. In certain embodiments, the CR2 portion contains at least one amino
acid substitution
at amino acid residues selected from the group consisting of: N11, R36, K41,
Y64 and K67,
further contains at least three amino acid substitutions at amino acid
residues selected from
the group consisting of: 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97,
T100,
N101, S 109, and S 128, and does not bind to, or has decreased binding
affinity for, one or
more C3 proteolytic fragments and EBV gp350. In certain embodiments, the CR2
portion
contains at least one amino acid substitution at an amino acid residue
selected from the group
consisting of: N11, R36, K41, Y64 and K67, further contains at least four
amino acid
substitutions at amino acid residues selected from the group consisting of:
19, Y29, C31, G33,
T34, D56, S70, H90, D92, S93, A97, T100, N101, 5109, and 5128, and does not
bind to, or
has decreased binding affinity for, one or more C3 proteolytic fragments and
EBV gp350. In
certain embodiments, the CR2 portion contains at least one amino acid
substitution at amino
acid residues selected from the group consisting of: N11, R36, K41, Y64 and
K67, further
contains at least five amino acid substitutions at amino acid residues
selected from the group
consisting of: 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97, T100,
N101, S109,
and S 128, and does not bind to, or has decreased binding affinity for, one or
more C3
proteolytic fragments and EBV gp350. In certain embodiments, the CR2 portion
contains at
least one amino acid substitution at amino acid residues selected from the
group consisting
of: N11, R36, K41, Y64 and K67, further contains at least six amino acid
substitutions at
amino acid residues selected from the group consisting of: 19, Y29, C31, G33,
T34, D56,
S70, H90, D92, S93, A97, T100, N101, 5109, and 5128, and does not bind to, or
has
decreased binding affinity for, one or more C3 proteolytic fragments and EBV
gp350. In
certain embodiments, the CR2 portion contains at least one amino acid
substitution at an
amino acid residue selected from the group consisting of: N11, R36, K41, Y64
and K67,
further contains at least seven amino acid substitutions at amino acid
residues selected from
the group consisting of: 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97,
T100,
N101, S 109, and S 128, and does not bind to, or has decreased binding
affinity for, one or
more C3 proteolytic fragments and EBV gp350. In certain embodiments, the CR2
portion
contains at least one amino acid substitution at an amino acid residue
selected from the group
consisting of: N11, R36, K41, Y64 and K67, further contains at least eight
amino acid
56
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
substitutions at amino acid residues selected from the group consisting of:
19, Y29, C31, G33,
T34, D56, S70, H90, D92, S93, A97, T100, N101, S 109, and S 128, and does not
bind to, or
has decreased binding affinity for, one or more C3 proteolytic fragments and
EBV gp350. In
certain embodiments, the CR2 portion contains at least one amino acid
substitution at an
amino acid residue selected from the group consisting of: N11, R36, K41, Y64
and K67,
further contains at least nine amino acid substitutions at amino acid residues
selected from the
group consisting of: 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97,
T100, N101,
S 109, and S 128, and does not bind to, or has decreased binding affinity for,
one or more C3
proteolytic fragments and EBV gp350. In certain embodiments, the CR2 portion
contains at
least one amino acid substitution at an amino acid residue selected from the
group consisting
of: N11, R36, K41, Y64 and K67, further contains at least ten amino acid
substitutions at
amino acid residues selected from the group consisting of: 19, Y29, C31, G33,
T34, D56,
S70, H90, D92, S93, A97, T100, N101, 5109, and 5128, and does not bind to, or
has
decreased binding affinity for, one or more C3 proteolytic fragments and EBV
gp350. In
certain embodiments, the CR2 portion contains at least one amino acid
substitution at amino
acid residues selected from the group consisting of: N11, R36, K41, Y64 and
K67, further
contains at least eleven amino acid substitutions at an amino acid residue
selected from the
group consisting of: 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97,
T100, N101,
S 109, and S 128, and does not bind to, or has decreased binding affinity for,
one or more C3
proteolytic fragments and EBV gp350. In certain embodiments, the CR2 portion
contains at
least one amino acid substitution at an amino acid residue selected from the
group consisting
of: N11, R36, K41, Y64 and K67, further contains at least twelve amino acid
substitutions at
amino acid residues selected from the group consisting of: 19, Y29, C31, G33,
T34, D56,
S70, H90, D92, S93, A97, T100, N101, 5109, and 5128, and does not bind to, or
has
decreased binding affinity for, one or more C3 proteolytic fragments and EBV
gp350. In
certain embodiments, the CR2 portion contains at least one amino acid
substitution at an
amino acid residue selected from the group consisting of: N11, R36, K41, Y64
and K67,
further contains at least thirteen amino acid substitutions at amino acid
residues selected from
the group consisting of: 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97,
T100,
N101, S 109, and S 128, and does not bind to, or has decreased binding
affinity for, one or
more C3 proteolytic fragments and EBV gp350. In certain embodiments, the CR2
portion
contains at least one amino acid substitution at an amino acid residue
selected from the group
consisting of: N11, R36, K41, Y64 and K67, further contains at least fourteen
amino acid
57
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
substitutions at amino acid residues selected from the group consisting of:
19, Y29, C31, G33,
T34, D56, S70, H90, D92, S93, A97, T100, N101, S 109, and S 128, and does not
bind to, or
has decreased binding affinity for, one or more C3 proteolytic fragments and
EBV gp350. In
certain embodiments, the CR2 portion contains at least one amino acid
substitution at an
amino acid residue selected from the group consisting of: N11, R36, K41, Y64
and K67,
further contains fifteen amino acid substitutions at amino acid residues 19,
Y29, C31, G33,
T34, D56, S70, H90, D92, S93, A97, T100, N101, 5109, and 5128, and does not
bind to, or
has decreased binding affinity for, one or more C3 proteolytic fragments and
EBV gp350. In
some embodiments, the one or more C3 proteolytic fragments are selected from
the group
consisting of C3d, iC3b, C3dg, and one or more cell-bound fragments of C3b
that bind to the
two N-terminal SCR domains of CR2. In any of the above embodiments, the at
least two
amino acid substitutions may be conservative substitutions. In certain
embodiments, the CR2
portion contains one or more substitutions of amino acids from the group
consisting of:
NI IA, R36A, K41A, Y64A, and K67A. In any of the above embodiments, the at
least two
amino acid substitutions may be non-conservative substitutions. In certain
embodiments, the
construct is a fusion protein. In some embodiments, the CR2 portion
selectively binds to one
or more proteins from the group consisting of: CD23 and IFNa. In some
embodiments, the
construct does not bind to EBV gp350 and one or more C3 proteolytic fragments.
In other
embodiments, the construct has decreased binding affinity to EBV gp350 and one
or more C3
proteolytic fragments. In some embodiments, the at least two amino acid
substitutions
decreases the binding affinity of the CR2 portion for EBV gp350 and one or
more C3
proteolytic fragments by any of about 20%, 25%, 30%, 35%, 40%, 45%, 50%, 55%,
60%,
65%, 70%, 75%, 80%, 85%, 90%, 95%, or 100%, inclusive, as well as any
numerical value
in between these percentages. In some embodiments, the complement modulator
portion is a
complement inhibitor. In some of these embodiments, the complement inhibitor
is selected
from the group consisting of: MCP, DAF, CD59, Crry, CR1, and FH. In other
embodiments,
the complement modulator portion is a complement activator. In some of these
embodiments, the complement activator is selected from the group consisting
of: human Ig
isotype Gi (IgGi), human Ig isotype M (IgM), mouse Ig isotype G3 (IgG3), mouse
IgM Fc,
human IgGi Fc domain, human IgM Fc domain, mouse IgG3 Fc domain, mouse IgM Fc
domain, and CVF.
[0118] In some aspects, there is provided a soluble composition capable of
targeted delivery
of a complement modulator to sites of IFNa production comprising a construct,
wherein the
58
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
construct comprises: (a) a complement receptor 2 (CR2) portion comprising a
CR2 protein of
SEQ ID NO:1 or a biologically active fragment thereof, wherein the CR2 portion
contains at
least the first two N-terminal SCR domains of the CR2 protein; and (b) a
complement
modulator portion; wherein the CR2 portion contains at least three amino acid
substitutions
that decrease binding affinity of the CR2 portion for one or more C3
proteolytic fragments
and EBV gp350. In certain embodiments, the CR2 portion contains at least two
amino acid
substitutions at amino acid residues selected from the group consisting of:
N11, R36, K41,
Y64 and K67, further contains at least one amino acid substitution at an amino
acid residue
selected from the group consisting of: 19, Y29, C31, G33, T34, D56, S70, H90,
D92, S93,
A97, T100, N101, S 109, and S 128, and does not bind to, or has decreased
binding affinity
for, one or more C3 proteolytic fragments and EBV gp350. In certain
embodiments, the CR2
portion contains at least two amino acid substitutions at an amino acid
residue selected from
the group consisting of: N11, R36, K41, Y64 and K67, further contains at least
two amino
acid substitutions at an amino acid residue selected from the group consisting
of: 19, Y29,
C31, G33, T34, D56, S70, H90, D92, S93, A97, T100, N101, S109, and S128, and
does not
bind to, or has decreased binding affinity for, one or more C3 proteolytic
fragments and EBV
gp350. In certain embodiments, the CR2 portion contains at least two amino
acid
substitutions at amino acid residues selected from the group consisting of:
N11, R36, K41,
Y64 and K67, further contains at least three amino acid substitutions at amino
acid residues
selected from the group consisting of: 19, Y29, C31, G33, T34, D56, S70, H90,
D92, S93,
A97, T100, N101, S 109, and S 128, and does not bind to, or has decreased
binding affinity
for, one or more C3 proteolytic fragments and EBV gp350. In certain
embodiments, the CR2
portion contains at least two amino acid substitutions at amino acid residues
selected from the
group consisting of: N11, R36, K41, Y64 and K67, further contains at least
four amino acid
substitutions at amino acid residues selected from the group consisting of:
19, Y29, C31, G33,
T34, D56, S70, H90, D92, S93, A97, T100, N101, S 109, and S 128, and does not
bind to, or
has decreased binding affinity for, one or more C3 proteolytic fragments and
EBV gp350. In
certain embodiments, the CR2 portion contains at least two amino acid
substitutions at amino
acid residues selected from the group consisting of: N11, R36, K41, Y64 and
K67, further
contains at least five amino acid substitutions at amino acid residues
selected from the group
consisting of: 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97, T100,
N101, S109,
and S 128, and does not bind to, or has decreased binding affinity for, one or
more C3
proteolytic fragments and EBV gp350. In certain embodiments, the CR2 portion
contains at
59
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
least two amino acid substitutions at amino acid residues selected from the
group consisting
of: N11, R36, K41, Y64 and K67, further contains at least six amino acid
substitutions at
amino acid residues selected from the group consisting of: 19, Y29, C31, G33,
T34, D56,
S70, H90, D92, S93, A97, T100, N101, 5109, and 5128, and does not bind to, or
has
decreased binding affinity for, one or more C3 proteolytic fragments and EBV
gp350. In
certain embodiments, the CR2 portion contains at least two amino acid
substitutions at amino
acid residues selected from the group consisting of: N11, R36, K41, Y64 and
K67, further
contains at least seven amino acid substitutions at amino acid residues
selected from the
group consisting of: 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97,
T100, N101,
S 109, and S 128, and does not bind to, or has decreased binding affinity for,
one or more C3
proteolytic fragments and EBV gp350. In certain embodiments, the CR2 portion
contains at
least two amino acid substitutions at amino acid residues selected from the
group consisting
of: N11, R36, K41, Y64 and K67, further contains at least eight amino acid
substitutions at
amino acid residues selected from the group consisting of: 19, Y29, C31, G33,
T34, D56,
S70, H90, D92, S93, A97, T100, N101, 5109, and 5128, and does not bind to, or
has
decreased binding affinity for, one or more C3 proteolytic fragments and EBV
gp350. In
certain embodiments, the CR2 portion contains at least two amino acid
substitutions at amino
acid residues selected from the group consisting of: N11, R36, K41, Y64 and
K67, further
contains at least nine amino acid substitutions at amino acid residues
selected from the group
consisting of: 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97, T100,
N101, 5109,
and S 128, and does not bind to, or has decreased binding affinity for, one or
more C3
proteolytic fragments and EBV gp350. In certain embodiments, the CR2 portion
contains at
least two amino acid substitutions at amino acid residues selected from the
group consisting
of: N11, R36, K41, Y64 and K67, further contains at least ten amino acid
substitutions at
amino acid residues selected from the group consisting of: 19, Y29, C31, G33,
T34, D56,
S70, H90, D92, S93, A97, T100, N101, 5109, and 5128, and does not bind to, or
has
decreased binding affinity for, one or more C3 proteolytic fragments and EBV
gp350. In
certain embodiments, the CR2 portion contains at least two amino acid
substitutions at amino
acid residues selected from the group consisting of: N11, R36, K41, Y64 and
K67, further
contains at least eleven amino acid substitutions at amino acid residues
selected from the
group consisting of: 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97,
T100, N101,
S 109, and S 128, and does not bind to, or has decreased binding affinity for,
one or more C3
proteolytic fragments and EBV gp350. In certain embodiments, the CR2 portion
contains at
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
least two amino acid substitutions at amino acid residues selected from the
group consisting
of: N11, R36, K41, Y64 and K67, further contains at least twelve amino acid
substitutions at
amino acid residues selected from the group consisting of: 19, Y29, C31, G33,
T34, D56,
S70, H90, D92, S93, A97, T100, N101, 5109, and 5128, and does not bind to, or
has
decreased binding affinity for, one or more C3 proteolytic fragments and EBV
gp350. In
certain embodiments, the CR2 portion contains at least two amino acid
substitutions at amino
acid residues selected from the group consisting of: N11, R36, K41, Y64 and
K67, further
contains at least thirteen amino acid substitutions at amino acid residues
selected from the
group consisting of: 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97,
T100, N101,
S 109, and S 128, and does not bind to, or has decreased binding affinity for,
one or more C3
proteolytic fragments and EBV gp350. In certain embodiments, the CR2 portion
contains at
least two amino acid substitutions at amino acid residues selected from the
group consisting
of: N11, R36, K41, Y64 and K67, further contains at least fourteen amino acid
substitutions
at amino acid residues selected from the group consisting of: 19, Y29, C31,
G33, T34, D56,
S70, H90, D92, S93, A97, T100, N101, 5109, and 5128, and does not bind to, or
has
decreased binding affinity for, one or more C3 proteolytic fragments and EBV
gp350. In
certain embodiments, the CR2 portion contains at least two amino acid
substitutions at amino
acid residue selected from the group consisting of: N11, R36, K41, Y64 and
K67, further
contains fifteen amino acid substitutions at amino acid residues 19, Y29, C31,
G33, T34, D56,
S70, H90, D92, S93, A97, T100, N101, 5109, and 5128, and does not bind to, or
has
decreased binding affinity for, one or more C3 proteolytic fragments and EBV
gp350. In
some embodiments, the one or more C3 proteolytic fragments are selected from
the group
consisting of C3d, iC3b, C3dg, and one or more cell-bound fragments of C3b
that bind to the
two N-terminal SCR domains of CR2. In any of the above embodiments, the at
least three
amino acid substitutions may be conservative substitutions. In any of the
above embodiments,
the at least three amino acid substitutions may be non-conservative
substitutions. In certain
embodiments, the CR2 portion contains one or more substitutions of amino acids
from the
group consisting of: N11A, R36A, K41A, Y64A, and K67A. In certain embodiments,
the
construct is a fusion protein. In some embodiments, the CR2 portion
selectively binds to one
or more proteins from the group consisting of: CD23 and IFNa. In some
embodiments, the
construct does not bind to EBV gp350 and one or more C3 proteolytic fragments.
In other
embodiments, the construct has decreased binding affinity to EBV gp350 and one
or more C3
proteolytic fragments. In some embodiments, the at least three amino acid
substitutions
61
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
decreases the binding affinity of the CR2 portion for EBV gp350 and one or
more C3
proteolytic fragments by any of about 20%, 25%, 30%, 35%, 40%, 45%, 50%, 55%,
60%,
65%, 70%, 75%, 80%, 85%, 90%, 95%, or 100%, inclusive, as well as any
numerical value
in between these percentages. In some embodiments, the complement modulator
portion is a
complement inhibitor. In some of these embodiments, the complement inhibitor
is selected
from the group consisting of: MCP, DAF, CD59, Crry, CR1, and FH. In other
embodiments,
the complement modulator portion is a complement activator. In some of these
embodiments, the complement activator is selected from the group consisting
of: human Ig
isotype Gi (IgGi), human Ig isotype M (IgM), mouse Ig isotype G3 (IgG3), mouse
IgM Fc,
human IgG1 Fc domain, human IgM Fc domain, mouse IgG3 Fc domain, mouse IgM Fc
domain, and CVF.
[0119] In some aspects, there is provided a soluble composition capable of
targeted delivery
of a complement modulator to sites of IFNa production comprising a construct,
wherein the
construct comprises: (a) a complement receptor 2 (CR2) portion comprising a
CR2 protein of
SEQ ID NO:1 or a biologically active fragment thereof, wherein the CR2 portion
contains at
least the first two N-terminal SCR domains of the CR2 protein; and (b) a
complement
modulator portion; wherein the CR2 portion contains at least four amino acid
substitutions
that decrease binding affinity of the CR2 portion for one or more C3
proteolytic fragments
and EBV gp350. In certain embodiments, the CR2 portion contains at least three
amino acid
substitutions at amino acid residues selected from the group consisting of:
N11, R36, K41,
Y64 and K67, further contains at least one amino acid substitution at an amino
acid residue
selected from the group consisting of: 19, Y29, C31, G33, T34, D56, S70, H90,
D92, S93,
A97, T100, N101, S 109, and S 128, and does not bind to, or has decreased
binding affinity
for, one or more C3 proteolytic fragments and EBV gp350. In certain
embodiments, the CR2
portion contains at least three amino acid substitutions at amino acid
residues selected from
the group consisting of: N11, R36, K41, Y64 and K67, further contains at least
two amino
acid substitutions at amino acid residues selected from the group consisting
of: 19, Y29, C3 1,
G33, T34, D56, S70, H90, D92, S93, A97, T100, N101, S 109, and S 128, and does
not bind
to, or has decreased binding affinity for, one or more C3 proteolytic
fragments and EBV
gp350. In certain embodiments, the CR2 portion contains at least three amino
acid
substitutions at amino acid residues selected from the group consisting of:
N11, R36, K41,
Y64 and K67, further contains at least three amino acid substitutions at amino
acid residues
selected from the group consisting of: 19, Y29, C31, G33, T34, D56, S70, H90,
D92, S93,
62
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
A97, T100, N101, 5109, and 5128, and does not bind to, or has decreased
binding affinity
for, one or more C3 proteolytic fragments and EBV gp350. In certain
embodiments, the CR2
portion contains at least three amino acid substitutions at amino acid
residues selected from
the group consisting of: N11, R36, K41, Y64 and K67, further contains at least
four amino
acid substitutions at amino acid residues selected from the group consisting
of: 19, Y29, C3 1,
G33, T34, D56, S70, H90, D92, S93, A97, T100, N101, 5109, and 5128, and does
not bind
to, or has decreased binding affinity for, one or more C3 proteolytic
fragments and EBV
gp350. In certain embodiments, the CR2 portion contains at least three amino
acid
substitutions at amino acid residues selected from the group consisting of:
N11, R36, K41,
Y64 and K67, further contains at least five amino acid substitutions at amino
acid residues
selected from the group consisting of: 19, Y29, C31, G33, T34, D56, S70, H90,
D92, S93,
A97, T100, N101, 5109, and 5128, and does not bind to, or has decreased
binding affinity
for, one or more C3 proteolytic fragments and EBV gp350. In certain
embodiments, the CR2
portion contains at least three amino acid substitutions at amino acid
residues selected from
the group consisting of: N11, R36, K41, Y64 and K67, further contains at least
six amino
acid substitutions at amino acid residues selected from the group consisting
of: 19, Y29, C3 1,
G33, T34, D56, S70, H90, D92, S93, A97, T100, N101, 5109, and 5128, and does
not bind
to, or has decreased binding affinity for, one or more C3 proteolytic
fragments and EBV
gp350. In certain embodiments, the CR2 portion contains at least three amino
acid
substitutions at amino acid residues selected from the group consisting of:
N11, R36, K41,
Y64 and K67, further contains at least seven amino acid substitutions at amino
acid residues
selected from the group consisting of: 19, Y29, C31, G33, T34, D56, S70, H90,
D92, S93,
A97, T100, N101, 5109, and 5128, and does not bind to, or has decreased
binding affinity
for, one or more C3 proteolytic fragments and EBV gp350. In certain
embodiments, the CR2
portion contains at least three amino acid substitutions at amino acid
residues selected from
the group consisting of: N11, R36, K41, Y64 and K67, further contains at least
eight amino
acid substitutions at amino acid residues selected from the group consisting
of: 19, Y29, C3 1,
G33, T34, D56, S70, H90, D92, S93, A97, T100, N101, 5109, and 5128, and does
not bind
to, or has decreased binding affinity for, one or more C3 proteolytic
fragments and EBV
gp350. In certain embodiments, the CR2 portion contains at least three amino
acid
substitutions at amino acid residues selected from the group consisting of:
N11, R36, K41,
Y64 and K67, further contains at least nine amino acid substitutions at amino
acid residues
selected from the group consisting of: 19, Y29, C31, G33, T34, D56, S70, H90,
D92, S93,
63
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
A97, T100, N101, 5109, and 5128, and does not bind to, or has decreased
binding affinity
for, one or more C3 proteolytic fragments and EBV gp350. In certain
embodiments, the CR2
portion contains at least three amino acid substitutions at amino acid
residues selected from
the group consisting of: N11, R36, K41, Y64 and K67, further contains at least
ten amino
acid substitutions at amino acid residues selected from the group consisting
of: 19, Y29, C3 1,
G33, T34, D56, S70, H90, D92, S93, A97, T100, N101, 5109, and 5128, and does
not bind
to, or has decreased binding affinity for, one or more C3 proteolytic
fragments and EBV
gp350. In certain embodiments, the CR2 portion contains at least three amino
acid
substitutions at amino acid residues selected from the group consisting of:
N11, R36, K41,
Y64 and K67, further contains at least eleven amino acid substitutions at
amino acid residues
selected from the group consisting of: 19, Y29, C31, G33, T34, D56, S70, H90,
D92, S93,
A97, T100, N101, 5109, and 5128, and does not bind to, or has decreased
binding affinity
for, one or more C3 proteolytic fragments and EBV gp350. In certain
embodiments, the CR2
portion contains at least three amino acid substitutions at amino acid
residues selected from
the group consisting of: N11, R36, K41, Y64 and K67, further contains at least
twelve amino
acid substitutions at amino acid residues selected from the group consisting
of: 19, Y29, C3 1,
G33, T34, D56, S70, H90, D92, S93, A97, T100, N101, 5109, and 5128, and does
not bind
to, or has decreased binding affinity for, one or more C3 proteolytic
fragments and EBV
gp350. In certain embodiments, the CR2 portion contains at least three amino
acid
substitutions at amino acid residues selected from the group consisting of:
N11, R36, K41,
Y64 and K67, further contains at least thirteen amino acid substitutions at
amino acid
residues selected from the group consisting of: 19, Y29, C31, G33, T34, D56,
S70, H90, D92,
S93, A97, T100, N101, S 109, and S 128, and does not bind to, or has decreased
binding
affinity for, one or more C3 proteolytic fragments and EBV gp350. In certain
embodiments,
the CR2 portion contains at least three amino acid substitutions at amino acid
residues
selected from the group consisting of: N11, R36, K41, Y64 and K67, further
contains at least
fourteen amino acid substitutions at amino acid residues selected from the
group consisting
of: 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97, T100, N101, S109,
and S128,
and does not bind to, or has decreased binding affinity for, one or more C3
proteolytic
fragments and EBV gp350. In certain embodiments, the CR2 portion contains at
least three
amino acid substitutions at amino acid residues selected from the group
consisting of: N11,
R36, K41, Y64 and K67, further contains fifteen amino acid substitutions at
amino acid
residues 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97, T100, N101,
S109, and
64
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
S 128, and does not bind to, or has decreased binding affinity for, one or
more C3 proteolytic
fragments and EBV gp350. In some embodiments, the one or more C3 proteolytic
fragments
are selected from the group consisting of C3d, iC3b, C3dg, and one or more
cell-bound
fragments of C3b that bind to the two N-terminal SCR domains of CR2. In any of
the above
embodiments, the at least four amino acid substitutions may be conservative
substitutions. In
any of the above embodiments, the at least four amino acid substitutions may
be non-
conservative substitutions. In certain embodiments, the CR2 portion contains
one or more
substitutions in amino acids from the group consisting of: NI IA, R36A, K4 IA,
Y64A, and
K67A In certain embodiments, the construct is a fusion protein. In some
embodiments, the
CR2 portion selectively binds to one or more proteins from the group
consisting of: CD23
and IFNa. In some embodiments, the construct does not bind to EBV gp350 and
one or more
C3 proteolytic fragments. In other embodiments, the construct has decreased
binding affinity
to EBV gp350 and one or more C3 proteolytic fragments. In some embodiments,
the at least
four amino acid substitutions decreases the binding affinity of the CR2
portion for EBV
gp350 and one or more C3 proteolytic fragments by any of about 20%, 25%, 30%,
35%,
40%, 45%, 50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 95%, or 100%,
inclusive, as
well as any numerical value in between these percentages. In some embodiments,
the
complement modulator portion is a complement inhibitor. In other embodiments,
some of
these complement inhibitor is selected from the group consisting of: MCP, DAF,
CD59, Crry,
CR1, and FH. In other embodiments, the complement modulator portion is a
complement
activator. In some of these embodiments, the complement activator is selected
from the
group consisting of: human Ig isotype Gi (IgGi), human Ig isotype M (IgM),
mouse Ig
isotype G3 (IgG3), mouse IgM Fc, human IgGi Fc domain, human IgM Fc domain,
mouse
IgG3 Fc domain, mouse IgM Fc domain, and CVF.
[0120] In some aspects, there is provided a soluble composition capable of
targeted delivery
of a complement modulator to sites of IFNa production comprising a construct,
wherein the
construct comprises: (a) a complement receptor 2 (CR2) portion comprising a
CR2 protein of
SEQ ID NO:1 or a biologically active fragment thereof, wherein the CR2 portion
contains at
least the first two N-terminal SCR domains of the CR2 protein; and (b) a
complement
modulator portion; wherein the CR2 portion contains at least five amino acid
substitutions
that decrease binding affinity of the CR2 portion for one or more C3
proteolytic fragments
and EBV gp350. In certain embodiments, the CR2 portion contains at least four
amino acid
substitutions at amino acid residues selected from the group consisting of:
N11, R36, K41,
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
Y64 and K67, further contains at least one amino acid substitution at an amino
acid residue
selected from the group consisting of: 19, Y29, C31, G33, T34, D56, S70, H90,
D92, S93,
A97, T100, N101, 5109, and 5128, and does not bind to, or has decreased
binding affinity
for, one or more C3 proteolytic fragments and EBV gp350. In certain
embodiments, the CR2
portion contains at least four amino acid substitutions at amino acid residues
selected from
the group consisting of: N11, R36, K41, Y64 and K67, further contains at least
two amino
acid substitutions at amino acid residues selected from the group consisting
of: 19, Y29, C3 1,
G33, T34, D56, S70, H90, D92, S93, A97, T100, N101, 5109, and 5128, and does
not bind
to, or has decreased binding affinity for, one or more C3 proteolytic
fragments and EBV
gp350. In certain embodiments, the CR2 portion contains at least four amino
acid
substitutions at amino acid residues selected from the group consisting of:
N11, R36, K41,
Y64 and K67, further contains at least three amino acid substitutions at amino
acid residues
selected from the group consisting of: 19, Y29, C31, G33, T34, D56, S70, H90,
D92, S93,
A97, T100, N101, 5109, and 5128, and does not bind to, or has decreased
binding affinity
for, one or more C3 proteolytic fragments and EBV gp350. In certain
embodiments, the CR2
portion contains at least four amino acid substitutions at amino acid residues
selected from
the group consisting of: N11, R36, K41, Y64 and K67, further contains at least
four amino
acid substitutions at amino acid residues selected from the group consisting
of: 19, Y29, C3 1,
G33, T34, D56, S70, H90, D92, S93, A97, T100, N101, 5109, and 5128, and does
not bind
to, or has decreased binding affinity for, one or more C3 proteolytic
fragments and EBV
gp350. In certain embodiments, the CR2 portion contains at least four amino
acid
substitutions at amino acid residues selected from the group consisting of:
N11, R36, K41,
Y64 and K67, further contains at least five amino acid substitutions at amino
acid residues
selected from the group consisting of: 19, Y29, C31, G33, T34, D56, S70, H90,
D92, S93,
A97, T100, N101, 5109, and 5128, and does not bind to, or has decreased
binding affinity
for, one or more C3 proteolytic fragments and EBV gp350. In certain
embodiments, the CR2
portion contains at least four amino acid substitutions at amino acid residues
selected from
the group consisting of: N11, R36, K41, Y64 and K67, further contains at least
six amino
acid substitutions at amino acid residues selected from the group consisting
of: 19, Y29, C3 1,
G33, T34, D56, S70, H90, D92, S93, A97, T100, N101, 5109, and 5128, and does
not bind
to, or has decreased binding affinity for, one or more C3 proteolytic
fragments and EBV
gp350. In certain embodiments, the CR2 portion contains at least four amino
acid
substitutions at amino acid residues selected from the group consisting of:
N11, R36, K41,
66
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
Y64 and K67, further contains at least seven amino acid substitutions at amino
acid residues
selected from the group consisting of: 19, Y29, C31, G33, T34, D56, S70, H90,
D92, S93,
A97, T100, N101, 5109, and 5128, and does not bind to, or has decreased
binding affinity
for, one or more C3 proteolytic fragments and EBV gp350. In certain
embodiments, the CR2
portion contains at least four amino acid substitutions at amino acid residues
selected from
the group consisting of: N11, R36, K41, Y64 and K67, further contains at least
eight amino
acid substitutions at amino acid residues selected from the group consisting
of: 19, Y29, C3 1,
G33, T34, D56, S70, H90, D92, S93, A97, T100, N101, 5109, and 5128, and does
not bind
to, or has decreased binding affinity for, one or more C3 proteolytic
fragments and EBV
gp350. In certain embodiments, the CR2 portion contains at least four amino
acid
substitutions at amino acid residues selected from the group consisting of:
N11, R36, K41,
Y64 and K67, further contains at least nine amino acid substitutions at amino
acid residues
selected from the group consisting of: 19, Y29, C31, G33, T34, D56, S70, H90,
D92, S93,
A97, T100, N101, 5109, and 5128, and does not bind to, or has decreased
binding affinity
for, one or more C3 proteolytic fragments and EBV gp350. In certain
embodiments, the CR2
portion contains at least four amino acid substitutions at amino acid residues
selected from
the group consisting of: N11, R36, K41, Y64 and K67, further contains at least
ten amino
acid substitutions at amino acid residues selected from the group consisting
of: 19, Y29, C3 1,
G33, T34, D56, S70, H90, D92, S93, A97, T100, N101, 5109, and 5128, and does
not bind
to, or has decreased binding affinity for, one or more C3 proteolytic
fragments and EBV
gp350. In certain embodiments, the CR2 portion contains at least four amino
acid
substitutions at amino acid residues selected from the group consisting of:
N11, R36, K41,
Y64 and K67, further contains at least eleven amino acid substitutions at
amino acid residues
selected from the group consisting of: 19, Y29, C31, G33, T34, D56, S70, H90,
D92, S93,
A97, T100, N101, 5109, and 5128, and does not bind to, or has decreased
binding affinity
for, one or more C3 proteolytic fragments and EBV gp350. In certain
embodiments, the CR2
portion contains at least four amino acid substitutions at amino acid residues
selected from
the group consisting of: N11, R36, K41, Y64 and K67, further contains at least
twelve amino
acid substitutions at amino acid residues selected from the group consisting
of: 19, Y29, C3 1,
G33, T34, D56, S70, H90, D92, S93, A97, T100, N101, 5109, and 5128, and does
not bind
to, or has decreased binding affinity for, one or more C3 proteolytic
fragments and EBV
gp350. In certain embodiments, the CR2 portion contains at least four amino
acid
substitutions at amino acid residues selected from the group consisting of:
N11, R36, K41,
67
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
Y64 and K67, further contains at least thirteen amino acid substitutions at
amino acid
residues selected from the group consisting of: 19, Y29, C31, G33, T34, D56,
S70, H90, D92,
S93, A97, T100, N101, S 109, and S 128, and does not bind to, or has decreased
binding
affinity for, one or more C3 proteolytic fragments and EBV gp350. In certain
embodiments,
the CR2 portion contains at least four amino acid substitutions at amino acid
residues selected
from the group consisting of: N11, R36, K41, Y64 and K67, further contains at
least fourteen
amino acid substitutions at amino acid residues selected from the group
consisting of: 19,
Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97, T100, N101, 5109, and 5128,
and does
not bind to, or has decreased binding affinity for, one or more C3 proteolytic
fragments and
EBV gp350. In certain embodiments, the CR2 portion contains at least four
amino acid
substitutions at amino acid residues selected from the group consisting of:
N11, R36, K41,
Y64 and K67, further contains fifteen amino acid substitutions at amino acid
residues 19,
Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97, T100, N101, 5109, and 5128,
and does
not bind to, or has decreased binding affinity for, one or more C3 proteolytic
fragments and
EBV gp350. In some embodiments, the one or more C3 proteolytic fragments are
selected
from the group consisting of C3d, iC3b, C3dg, and one or more cell-bound
fragments of C3b
that bind to the two N-terminal SCR domains of CR2. In any of the above
embodiments, the
at least five amino acid substitutions may be conservative substitutions. In
any of the above
embodiments, the at least five amino acid substitutions may be non-
conservative
substitutions. In certain embodiments, the CR2 portion contains one or more
substitutions of
amino acids from the group consisting of: N11A, R36A, K41A, Y64A, and K67A In
certain
embodiments, the construct is a fusion protein. In some embodiments, the CR2
portion
selectively binds to one or more proteins from the group consisting of: CD23
and IFNa. In
some embodiments, the construct does not bind to EBV gp350 and one or more C3
proteolytic fragments. In other embodiments, the construct has decreased
binding affinity to
EBV gp350 and one or more C3 proteolytic fragments. In some embodiments, the
at least
five amino acid substitutions decreases the binding affinity of the CR2
portion for EBV
gp350 and one or more C3 proteolytic fragments by any of about 20%, 25%, 30%,
35%,
40%, 45%, 50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 95%, or 100%,
inclusive, as
well as any numerical value in between these percentages. In some embodiments,
the
complement modulator portion is a complement inhibitor. In some of these
embodiments,
the complement inhibitor is selected from the group consisting of: MCP, DAF,
CD59, Crry,
CR1, and FH. In other embodiments, the complement modulator portion is a
complement
68
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
activator. In some of these embodiments, the complement activator is selected
from the
group consisting of: human Ig isotype Gi (IgGi), human Ig isotype M (IgM),
mouse Ig
isotype G3 (IgG3), mouse IgM Fc, human IgGi Fc domain, human IgM Fc domain,
mouse
IgG3 Fc domain, mouse IgM Fc domain, and CVF.
[0121] In some aspects, there is provided a soluble composition capable of
targeted delivery
of a complement modulator to sites of IFNa production comprising a construct,
wherein the
construct comprises: (a) a complement receptor 2 (CR2) portion comprising a
CR2 protein of
SEQ ID NO:1 or a biologically active fragment thereof, wherein the CR2 portion
contains at
least the first two N-terminal SCR domains of the CR2 protein; and (b) a
complement
modulator portion; wherein the CR2 portion contains at least six amino acid
substitutions that
decrease binding affinity of the CR2 portion for one or more C3 proteolytic
fragments and
EBV gp350. In certain embodiments, the CR2 portion contains five amino acid
substitutions
at amino acid residues N11, R36, K41, Y64 and K67, further contains at least
one amino acid
substitution at an amino acid residue selected from the group consisting of:
19, Y29, C3 1,
G33, T34, D56, S70, H90, D92, S93, A97, T100, N101, S 109, and S 128, and does
not bind
to, or has decreased binding affinity for, one or more C3 proteolytic
fragments and EBV
gp350. In certain embodiments, the CR2 portion contains five amino acid
substitutions at
amino acid residues N11, R36, K41, Y64 and K67, further contains at least two
amino acid
substitutions at amino acid residues selected from the group consisting of:
19, Y29, C31, G33,
T34, D56, S70, H90, D92, S93, A97, T100, N101, S 109, and S 128, and does not
bind to, or
has decreased binding affinity for, one or more C3 proteolytic fragments and
EBV gp350. In
certain embodiments, the CR2 portion contains five amino acid substitutions at
amino acid
residues N11, R36, K41, Y64 and K67, further contains at least three amino
acid substitutions
at amino acid residues selected from the group consisting of: 19, Y29, C31,
G33, T34, D56,
S70, H90, D92, S93, A97, T100, N101, S 109, and S 128, and does not bind to,
or has
decreased binding affinity for, one or more C3 proteolytic fragments and EBV
gp350. In
certain embodiments, the CR2 portion contains five amino acid substitutions at
amino acid
residues N11, R36, K41, Y64 and K67, further contains at least four amino acid
substitutions
at amino acid residues selected from the group consisting of: 19, Y29, C31,
G33, T34, D56,
S70, H90, D92, S93, A97, T100, N101, S 109, and S 128, and does not bind to,
or has
decreased binding affinity for, one or more C3 proteolytic fragments and EBV
gp350. In
certain embodiments, the CR2 portion contains five amino acid substitutions at
amino acid
residues N11, R36, K41, Y64 and K67, further contains at least five amino acid
substitutions
69
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
at amino acid residues selected from the group consisting of: 19, Y29, C31,
G33, T34, D56,
S70, H90, D92, S93, A97, T100, N101, 5109, and 5128, and does not bind to, or
has
decreased binding affinity for, one or more C3 proteolytic fragments and EBV
gp350. In
certain embodiments, the CR2 portion contains five amino acid substitutions at
amino acid
residues N11, R36, K41, Y64 and K67, further contains at least six amino acid
substitutions
at amino acid residues selected from the group consisting of: 19, Y29, C31,
G33, T34, D56,
S70, H90, D92, S93, A97, T100, N101, 5109, and 5128, and does not bind to, or
has
decreased binding affinity for, one or more C3 proteolytic fragments and EBV
gp350. In
certain embodiments, the CR2 portion contains five amino acid substitutions at
amino acid
residues N11, R36, K41, Y64 and K67, further contains at least seven amino
acid
substitutions at amino acid residues selected from the group consisting of:
19, Y29, C31, G33,
T34, D56, S70, H90, D92, S93, A97, T100, N101, 5109, and 5128, and does not
bind to, or
has decreased binding affinity for, one or more C3 proteolytic fragments and
EBV gp350. In
certain embodiments, the CR2 portion contains five amino acid substitutions at
amino acid
residues N11, R36, K41, Y64 and K67, further contains at least eight amino
acid substitutions
at amino acid residues selected from the group consisting of: 19, Y29, C31,
G33, T34, D56,
S70, H90, D92, S93, A97, T100, N101, 5109, and 5128, and does not bind to, or
has
decreased binding affinity for, one or more C3 proteolytic fragments and EBV
gp350. In
certain embodiments, the CR2 portion contains five amino acid substitutions at
amino acid
residues N11, R36, K41, Y64 and K67, further contains at least nine amino acid
substitutions
at amino acid residues selected from the group consisting of: 19, Y29, C31,
G33, T34, D56,
S70, H90, D92, S93, A97, T100, N101, 5109, and 5128, and does not bind to, or
has
decreased binding affinity for, one or more C3 proteolytic fragments and EBV
gp350. In
certain embodiments, the CR2 portion contains five amino acid substitutions at
amino acid
residues N11, R36, K41, Y64 and K67, further contains at least ten amino acid
substitutions
at amino acid residues selected from the group consisting of: 19, Y29, C31,
G33, T34, D56,
S70, H90, D92, S93, A97, T100, N101, 5109, and 5128, and does not bind to, or
has
decreased binding affinity for, one or more C3 proteolytic fragments and EBV
gp350. In
certain embodiments, the CR2 portion contains five amino acid substitutions at
amino acid
residues N11, R36, K41,Y64 and K67, further contains at least eleven amino
acid
substitutions at amino acid residues selected from the group consisting of:
19, Y29, C31, G33,
T34, D56, S70, H90, D92, S93, A97, T100, N101, 5109, and 5128, and does not
bind to, or
has decreased binding affinity for, one or more C3 proteolytic fragments and
EBV gp350. In
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
certain embodiments, the CR2 portion contains five amino acid substitutions at
amino acid
residues N11, R36, K41, Y64 and K67, further contains at least twelve amino
acid
substitutions at amino acid residues selected from the group consisting of:
19, Y29, C31, G33,
T34, D56, S70, H90, D92, S93, A97, T100, N101, S109, and S128, and does not
bind to, or
has decreased binding affinity for, one or more C3 proteolytic fragments and
EBV gp350. In
certain embodiments, the CR2 portion contains five amino acid substitutions at
amino acid
residues N11, R36, K41, Y64 and K67, further contains at least thirteen amino
acid
substitutions at amino acid residues selected from the group consisting of:
19, Y29, C31, G33,
T34, D56, S70, H90, D92, S93, A97, T100, N101, S 109, and S 128, and does not
bind to, or
has decreased binding affinity for, one or more C3 proteolytic fragments and
EBV gp350. In
certain embodiments, the CR2 portion contains five amino acid substitutions at
amino acid
residues N11, R36, K41, Y64 and K67, further contains at least fourteen amino
acid
substitutions at amino acid residues selected from the group consisting of:
19, Y29, C31, G33,
T34, D56, S70, H90, D92, S93, A97, T100, N101, S 109, and S 128, and does not
bind to, or
has decreased binding affinity for, one or more C3 proteolytic fragments and
EBV gp350. In
certain embodiments, the CR2 portion contains five amino acid substitutions at
amino acid
residues N11, R36, K41, Y64 and K67, further contains fifteen amino acid
substitutions at
amino acid residues 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97,
T100, N101,
S 109, and S 128, and does not bind to, or has decreased binding affinity for,
one or more C3
proteolytic fragments and EBV gp350. In some embodiments, the one or more C3
proteolytic fragments are selected from the group consisting of C3d, iC3b,
C3dg, and one or
more cell-bound fragments of C3b that bind to the two N-terminal SCR domains
of CR2. In
any of the above embodiments, the at least six amino acid substitutions may be
conservative
substitutions. In any of the above embodiments, the at least six amino acid
substitutions may
be non-conservative substitutions. In certain embodiments, the CR2 portion
contains one or
more substitutions of amino acids from the group consisting of: NI IA, R36A,
K41A, Y64A,
and K67A In certain embodiments, the construct is a fusion protein. In some
embodiments,
the CR2 portion selectively binds to one or more proteins from the group
consisting of: CD23
and IFNa. In some embodiments, the construct does not bind to EBV gp350 and
one or more
C3 proteolytic fragments. In other embodiments, the construct has decreased
binding affinity
to EBV gp350 and one or more C3 proteolytic fragments. In some embodiments,
the at least
six amino acid substitutions decreases the binding affinity of the CR2 portion
for EBV gp350
and one or more C3 proteolytic fragments by any of about 20%, 25%, 30%, 35%,
40%, 45%,
71
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 95%, or 100%, inclusive, as well
as any
numerical value in between these percentages. In some embodiments, the
complement
modulator portion is a complement inhibitor. In some of these embodiments, the
complement inhibitor is selected from the group consisting of: MCP, DAF, CD59,
Crry, CR1,
and FH. In other embodiments, the complement modulator portion is a complement
activator. In some of these embodiments, the complement activator is selected
from the
group consisting of: human Ig isotype Gi (IgGi), human Ig isotype M (IgM),
mouse Ig
isotype G3 (IgG3), mouse IgM Fc, human IgGi Fc domain, human IgM Fc domain,
mouse
IgG3 Fc domain, mouse IgM Fc domain, and CVF.
Other CR2 Substitutions for Targeted Delivery of Complement Modulators
[0122] In any embodiments of any of the compositions described herein, the CR2
portion can
further contain at least one, two, three, four, five, six, seven, eight, nine,
ten, or eleven
additional amino acid substitutions at other positions in the CR2 portion. In
certain
embodiments, the CR2 portion further contains at least one amino acid
substitution selected
from the group consisting of: R13, Y16, A22, R28, S32, K48, K57, Y68, R83,
G84, and
R89. In certain embodiments, the CR2 portion further contains at least two
amino acid
substitutions selected from the group consisting of: R13, Y16, A22, R28, S32,
K48, K57,
Y68, R83, G84, and R89. In certain embodiments, the CR2 portion further
contains at least
three amino acid substitutions selected from the group consisting of: R13,
Y16, A22, R28,
S32, K48, K57, Y68, R83, G84, and R89. In certain embodiments, the CR2 portion
further
contains at least four amino acid substitutions selected from the group
consisting of: R13,
Y16, A22, R28, S32, K48, K57, Y68, R83, G84, and R89. In certain embodiments,
the CR2
portion further contains at least five amino acid substitutions selected from
the group
consisting of: R13, Y16, A22, R28, S32, K48, K57, Y68, R83, G84, and R89. In
certain
embodiments, the CR2 portion further contains at least six amino acid
substitutions selected
from the group consisting of: R13, Y16, A22, R28, S32, K48, K57, Y68, R83,
G84, and
R89. In certain embodiments, the CR2 portion further contains at least seven
amino acid
substitutions selected from the group consisting of: R13, Y16, A22, R28, S32,
K48, K57,
Y68, R83, G84, and R89. In certain embodiments, the CR2 portion further
contains at least
eight amino acid substitutions selected from the group consisting of: R13,
Y16, A22, R28,
S32, K48, K57, Y68, R83, G84, and R89. In certain embodiments, the CR2 portion
further
contains at least nine amino acid substitutions selected from the group
consisting of: R13,
Y16, A22, R28, S32, K48, K57, Y68, R83, G84, and R89. In certain embodiments,
the CR2
72
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
portion further contains at least ten amino acid substitutions selected from
the group
consisting of: R13, Y16, A22, R28, S32, K48, K57, Y68, R83, G84, and R89. In
certain
embodiments, the CR2 portion further contains at least eleven amino acid
substitutions
selected from the group consisting of: R13, Y16, A22, R28, S32, K48, K57, Y68,
R83, G84,
and R89. In any of the above embodiments, the at least one, two, three, four,
five, six, seven,
eight, nine, ten or eleven amino acid substitutions may be conservative
substitutions. In any
of the above embodiments, the at least one, two, three, four, five, six,
seven, eight, nine, ten
or eleven amino acid substitutions may be non-conservative substitutions.
Linker Proteins
[0123] In any of the embodiments described herein, the construct comprising a
CR2 portion
or a biologically active fragment thereof and a complement inhibitor portion
comprising
human CD59, mouse CD59 isoform A, mouse CD59 isoform B, mouse Crry protein,
human
factor H, mouse factor H, human CR1, human MCP, human DAF or mouse DAF or a
biologically active fragment thereof also includes an amino acid linker
sequence linking the
CR2 portion and the complement inhibitor portion (e.g., human CD59, mouse CD59
isoform
A, mouse CD59 isoform B, mouse Crry protein, human factor H, mouse factor H,
human
CR1, human MCP, human DAF or mouse DAF or a biologically active fragment
thereof).
[0124] In any of the embodiments described herein, the construct comprising a
CR2 portion
or a biologically active fragment thereof and a complement activator portion
comprising
human IgGI, human IgM, mouse IgG3, mouse IgM, or CVF or a biologically-active
fragment
thereof also includes an amino acid linker sequence linking the CR2 portion
and the
complement activator portion (e.g., human IgGI, human IgM, mouse IgG3, mouse
IgM, or
CVF or a biologically active fragment thereof).
[0125] Examples of linker sequences are known in the art, and include, for
example,
(Gly4Ser), (Gly4Ser)2, (Gly4Ser)3, (G1y3Ser)4, (SerGly4), (SerGly4)2,
(SerGly4)3, and
(SerGly4)4. Linking sequences can also comprise "natural" linking sequences
found between
different domains of complement factors. For example, VSVFPLE (SEQ ID NO:26)
or
EYFNKYSS (SEQ ID NO:27), the linking sequence between the first two N-terminal
short
consensus repeat domains of human CR2, can be used. In some embodiments, the
linking
sequence between the fourth and the fifth N-terminal short consensus repeat
domains of
human CR2 (EEIF) (SEQ ID NO:28) is used. In some embodiments, the linker
sequence
comprises at least about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15,
16, 17, 18, 19, 20, 21,
73
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
22, 23, 24, 25, 30, 35, 40, 45, 50, 60, 70, 80, 90, or 100 amino acids,
inclusive, as well as any
numerical value in between these numbers.
Pharmaceutical Compositions
[0126] In another aspect, provided herein are pharmaceutical compositions
comprising any of
the constructs and/or fusion proteins described herein. Pharmaceutical
compositions
comprising any of the constructs and/or fusion proteins described herein are
generally
formulated as sterile, substantially isotonic pharmaceutical solutions in full
compliance with
all Good Manufacturing Practice (GMP) regulations of the U.S. Food and Drug
Administration. In certain embodiments, the composition is free of pathogen.
For injection,
the pharmaceutical compositions can be in the form of liquid solutions, for
example in
physiologically compatible buffers such as Hank's Balanced Salt Solution,
Phosphate-
Buffered Saline or Ringer's solution. In addition, the pharmaceutical
compositions provided
herein can be in solid form and redissolved or resuspended immediately prior
to use.
Lyophilized compositions are also contemplated.
[0127] For oral administration, the pharmaceutical compositions can take the
form of, for
example, tablets or capsules prepared by conventional means with
pharmaceutically
acceptable excipients such as binding agents (e.g., pregelatinized maize
starch,
polyvinylpyrrolidone or hydroxypropyl methylcellulose); fillers (e.g.,
lactose,
microcrystalline cellulose or calcium hydrogen phosphate); lubricants (e.g.,
magnesium
stearate, talc or silica); disintegrants (e.g., potato starch or sodium starch
glycolate); or
wetting agents (e.g., sodium lauryl sulfate). Liquid preparations for oral
administration can
take the form of, for example, solutions, syrups or suspensions, or they can
be presented as a
dry product for constitution with water or other suitable vehicle before use.
Such liquid
preparations can be prepared by conventional means with pharmaceutically
acceptable
additives such as suspending agents (e.g., sorbitol syrup, cellulose
derivatives or
hydrogenated edible fats); emulsifying agents (e.g., lecithin or acacia); non-
aqueous vehicles
(e.g., fractionated oil, oily esters, ethyl alcohol or fractionated vegetable
oils); and
preservatives (e.g., methyl or propyl-p-hydroxybenzoates or sorbic acid). The
preparations
can also contain buffer salts, flavoring, coloring and sweetening agents as
appropriate.
[0128] In certain embodiments, the compositions are formulated in accordance
with routine
procedures as a pharmaceutical composition adapted for injection. In certain
embodiments,
the pharmaceutical compositions provided herein are formulated for
intravenous,
intraperitoneal, or intraocular injection. Typically, compositions for
injection are solutions in
74
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
sterile isotonic aqueous buffer. Where necessary, the composition may also
include a
solubilizing agent and a local anesthetic such as lignocaine to ease pain at
the site of the
injection. Generally, the ingredients are supplied either separately or mixed
together in unit
dosage form, for example, as a dry lyophilized powder or water-free
concentrate in a
hermetically sealed container such as an ampoule or sachette indicating the
quantity of active
agent. Where the composition is to be administered by infusion, it can be
dispensed with an
infusion bottle containing sterile pharmaceutical grade water or saline. Where
the
composition is administered by injection, an ampoule of sterile water for
injection or saline
can be provided so that the ingredients may be mixed prior to administration.
[0129] The pharmaceutical compositions may further comprise additional
ingredients, for
example preservatives, buffers, tonicity agents, antioxidants and stabilizers,
nonionic wetting
or clarifying agents, viscosity-increasing agents, and the like.
[0130] Suitable preservatives for use in a solution include polyquaternium- 1,
benzalkonium
chloride, thimerosal, chlorobutanol, methyl paraben, propyl paraben,
phenylethyl alcohol,
disodium-EDTA, sorbic acid, benzethonium chloride, and the like. Typically
(but not
necessarily) such preservatives are employed at a level of from 0.00 1% to
1.0% by weight.
[0131] Suitable buffers include boric acid, sodium and potassium bicarbonate,
sodium and
potassium borates, sodium and potassium carbonate, sodium acetate, sodium
biphosphate and
the like, in amounts sufficient to maintain the pH at between about pH 6 and
pH 8, and
preferably, between about pH 7 and pH 7.5.
[0132] Suitable tonicity agents include dextran 40, dextran 70, dextrose,
glycerin, potassium
chloride, propylene glycol, sodium chloride, and the like, such that the
sodium chloride
equivalent of the injectable solution is in the range 0.9 plus or minus 0.2%.
[0133] Suitable antioxidants and stabilizers include sodium bisulfite, sodium
metabisulfite,
sodium thiosulfite, thiourea and the like. Suitable wetting and clarifying
agents include
polysorbate 80, polysorbate 20, poloxamer 282 and tyloxapol. Suitable
viscosity-increasing
agents include dextran 40, dextran 70, gelatin, glycerin,
hydroxyethylcellulose,
hydroxmethylpropylcellulose, lanolin, methylcellulose, petrolatum,
polyethylene glycol,
polyvinyl alcohol, polyvinylpyrrolidone, carboxymethylcellulose and the like.
[0134] The pharmaceutical compositions may be suitable for a variety of modes
of
administration described herein, including for example systemic or localized
administration.
The pharmaceutical compositions can be in the form of injectable solutions or
in a form
suitable for oral administration. The pharmaceutical compositions described
herein can be
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
packaged in single unit dosages or in multidosage forms. In certain
embodiments, the
pharmaceutical compositions are suitable for administration to an individual,
a vertebrate, a
mammal, or a human by any route of administration described herein, including
oral
administration or intravenous injection.
Methods of the Invention
Methods of Making a Targeted Construct for Complement System Modulation
[0135] Provided herein are methods for making a construct that selectively
binds to one or
more ligands of CR2, wherein the method comprises mutating one or more amino
acids in a
complement receptor 2 (CR2) portion of the construct, and wherein the
construct comprises
(a) a complement receptor 2 (CR2) portion comprising a CR2 protein of SEQ ID
NO:1 or a
biologically active fragment thereof, wherein the CR2 portion contains at
least the first two
N-terminal SCR domains of the CR2 protein; and (b) a complement modulator
portion. In
certain embodiments, the construct is a fusion protein. In some embodiments
the construct
selectively binds to one or more C3 proteolytic fragments but does not bind to
or has reduced
binding affinity for IFNa or EBV gp350. In some embodiments, the construct
selectively
binds to one or more C3 proteolytic fragments and IFNa but does not bind to or
has reduced
binding affinity for EBV gp350. In some embodiments the construct selectively
binds to one
or more C3 proteolytic fragments and EBV gp350 but does not bind to or has
reduced
binding affinity for IFNa. In some embodiments, the construct selectively
binds to IFNa but
does not bind to or has reduced binding affinity for one or more C3
proteolytic fragments and
EBV gp350. In some embodiments, the construct selectively binds to IFNa and
EBV gp350
but does not bind to or has reduced binding affinity for one or more C3
proteolytic fragments.
In some embodiments, the construct selectively binds to EBV gp350 but does not
bind to or
has reduced binding affinity for IFNa and one or more C3 proteolytic
fragments.
[0136] In one aspect, there is provided a method for making a construct that
selectively binds
to one or more C3 proteolytic fragments but does not selectively bind to EBV
gp350 or IFNa,
wherein the method comprises (a) mutating one or more amino acids in a
complement
receptor 2 (CR2) portion of the construct from the group consisting of: N11,
R36, K41, Y64
and K67; and (b) mutating one or more amino acids in a complement receptor 2
(CR2)
portion of the construct from the group consisting of: S42 and K50, wherein
the construct
comprises (i) a complement receptor 2 (CR2) portion comprising a CR2 protein
of SEQ ID
NO:1 or a biologically active fragment thereof, wherein the CR2 portion
contains at least the
76
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
first two N-terminal SCR domains of the CR2 protein; and (ii) a complement
modulator
portion. In some embodiments, the one or more mutations in the complement
receptor 2
(CR2) portion of the construct are mutations to the amino acid alanine. In
some
embodiments, the method further comprises mutating one or more amino acids in
the
complement receptor 2 (CR2) portion of the construct selected from the group
consisting of:
R13, Y16, A22, R28, S32, K48, K57, Y68, R83, G84, and R89. In some
embodiments, the
method comprises mutating any of 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13,
14, 15, 16, 17, or 18
amino acids in the complement receptor 2 (CR2) portion of the construct.
[0137] In one aspect, there is provided a method for making a construct that
selectively binds
to one or more C3 proteolytic fragments and EBV gp350, but does not
selectively bind to
IFNa, wherein the method comprises mutating one or more amino acids in a
complement
receptor 2 (CR2) portion of the construct from the group consisting of: S42
and K50, wherein
the construct comprises (a) a complement receptor 2 (CR2) portion comprising a
CR2 protein
of SEQ ID NO:1 or a biologically active fragment thereof, wherein the CR2
portion contains
at least the first two N-terminal SCR domains of the CR2 protein; and (b) a
complement
modulator portion. In some embodiments, the one or more mutations in the
complement
receptor 2 (CR2) portion of the construct are mutations to the amino acid
alanine. In some
embodiments, the method further comprises mutating one or more amino acids in
the
complement receptor 2 (CR2) portion of the construct selected from the group
consisting of:
R13, Y16, A22, R28, S32, K48, K57, Y68, R83, G84, and R89. In some
embodiments, the
method comprises mutating any of 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, or 13
amino acids in the
complement receptor 2 (CR2) portion of the construct.
[0138] In one aspect, there is provided a method for making a construct that
selectively
binds to one or more C3 proteolytic fragments and IFNa, but does not
selectively bind to
EBV gp350, wherein the method comprises mutating one or more amino acids in a
complement receptor 2 (CR2) portion of the construct from the group consisting
of: N11,
R36, K41, Y64 and K67, wherein the construct comprises (a) a complement
receptor 2 (CR2)
portion comprising a CR2 protein of SEQ ID NO:1 or a biologically active
fragment thereof,
wherein the CR2 portion contains at least the first two N-terminal SCR domains
of the CR2
protein; and (b) a complement modulator portion. In some embodiments, the one
or more
mutations in the complement receptor 2 (CR2) portion of the construct are
mutations to the
amino acid alanine. In some embodiments, the method further comprises mutating
one or
more amino acids in the complement receptor 2 (CR2) portion of the construct
selected from
77
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
the group consisting of: R13, Y16, A22, R28, S32, K48, K57, Y68, R83, G84, and
R89. In
some embodiments, the method comprises mutating any of 1, 2, 3, 4, 5, 6, 7, 8,
9, 10, 11, 12,
13, 14, 15, or 16 amino acids in the complement receptor 2 (CR2) portion of
the construct.
[0139] In one aspect, there is provided a method for making a construct that
selectively binds
to IFNa but does not selectively bind to one or more C3 proteolytic fragments
and EBV
gp350, wherein the method comprises (a) mutating one or more amino acids in a
complement
receptor 2 (CR2) portion of the construct from the group consisting of: N11,
R36, K41, Y64
and K67; and (b) mutating one or more amino acids in a complement receptor 2
(CR2)
portion of the construct from the group consisting of: 19, Y29, C31, G33, T34,
D56, S70,
H90, D92, S93, A97, T100, N101, S109, and S128, wherein the construct
comprises (i) a
complement receptor 2 (CR2) portion comprising a CR2 protein of SEQ ID NO:1 or
a
biologically active fragment thereof, wherein the CR2 portion contains at
least the first two
N-terminal SCR domains of the CR2 protein; and (ii) a complement modulator
portion. In
some embodiments, the one or more mutations in the complement receptor 2 (CR2)
portion
of the construct are mutations to the amino acid alanine. In some embodiments,
the method
further comprises mutating one or more amino acids in the complement receptor
2 (CR2)
portion of the construct selected from the group consisting of: R13, Y16, A22,
R28, S32,
K48, K57, Y68, R83, G84, and R89. In some embodiments, the method comprises
mutating
any of 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20,
21, 22, 23, 24, 25, 26,
27, 28, 29, 30, or 31 amino acids in the complement receptor 2 (CR2) portion
of the
construct.
[0140] In one aspect, there is provided a method for making a construct that
selectively binds
to EBV gp350 and IFNa, but does not selectively bind to one or more C3
proteolytic
fragments, wherein the method comprises mutating one or more amino acids in a
complement receptor 2 (CR2) portion of the construct from the group consisting
of: 19, Y29,
C31, G33, T34, D56, S70, H90, D92, S93, A97, T100, N101, S109, and S128,
wherein the
construct comprises (a) a complement receptor 2 (CR2) portion comprising a CR2
protein of
SEQ ID NO:1 or a biologically active fragment thereof, wherein the CR2 portion
contains at
least the first two N-terminal SCR domains of the CR2 protein; and (b) a
complement
modulator portion. In some embodiments, the one or more mutations in the
complement
receptor 2 (CR2) portion of the construct are mutations to the amino acid
alanine. In some
embodiments, the method further comprises mutating one or more amino acids in
the
complement receptor 2 (CR2) portion of the construct selected from the group
consisting of:
78
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
R13, Y16, A22, R28, S32, K48, K57, Y68, R83, G84, and R89. In some
embodiments, the
method comprises mutating any of 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13,
14, 15, 16, 17, 18,
19, 20, 21, 22, 23, 24, 25, or 26 amino acids in the complement receptor 2
(CR2) portion of
the construct.
[0141] In one aspect, there is provided a method for making a construct that
selectively binds
to EBV gp350 but does not selectively bind to one or more C3 proteolytic
fragments and
IFNa, wherein the method comprises (a) mutating one or more amino acids in a
complement
receptor 2 (CR2) portion of the construct from the group consisting of: S42
and K50; and (b)
mutating one or more amino acids in a complement receptor 2 (CR2) portion of
the construct
from the group consisting of: 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93,
A97, T100,
N101, S 109, and S 128, wherein the construct comprises (i) a complement
receptor 2 (CR2)
portion comprising a CR2 protein of SEQ ID NO:1 or a biologically active
fragment thereof,
wherein the CR2 portion contains at least the first two N-terminal SCR domains
of the CR2
protein; and (ii) a complement modulator portion. In some embodiments, the one
or more
mutations in the complement receptor 2 (CR2) portion of the construct are
mutations to the
amino acid alanine. In some embodiments, the method further comprises mutating
one or
more amino acids in the complement receptor 2 (CR2) portion of the construct
selected from
the group consisting of: R13, Y16, A22, R28, S32, K48, K57, Y68, R83, G84, and
R89. In
some embodiments, the method comprises mutating any of 1, 2, 3, 4, 5, 6, 7, 8,
9, 10, 11, 12,
13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, or 28 amino acids
in the complement
receptor 2 (CR2) portion of the construct.
Methods for Reducing the Binding Affinity for or Altering the Binding Kinetics
of
the CR2 Portion of the Construct for One or More Ligands
[0142] In another aspect, provided herein are methods for reducing the binding
affinity of the
CR2 portion of any of the constructs disclosed herein for one or more CR2
ligands, the
method comprising introducing one or more mutations in the amino acid sequence
of the CR2
portion of the construct wherein the one or more mutations reduces binding
affinity of the
CR2 portion of the construct for one or more CR2 ligands.
[0143] In certain aspects, there is provided a method for reducing the binding
affinity of the
CR2 portion of any of the constructs disclosed herein for EBV gp350 and IFNa,
the method
comprising mutating at least one amino acid residue in the CR2 portion
selected from the
group consisting of: N11, R36, K41, S42, K50, Y64 and K67. In certain
embodiments, the
method comprises mutating at least two amino acid residues in the CR2 portion
selected from
79
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
the group consisting of: N11, R36, K41, S42, K50, Y64 and K67. In certain
embodiments,
the method comprises mutating at least three amino acid residues in the CR2
portion selected
from the group consisting of: N11, R36, K41, S42, K50, Y64 and K67. In certain
embodiments, the method comprises mutating at least four amino acid residues
in the CR2
portion selected from the group consisting of: N11, R36, K41, S42, K50, Y64
and K67. In
certain embodiments, the method comprises mutating at least five amino acid
residues in the
CR2 portion selected from the group consisting of: N11, R36, K41, S42, K50,
Y64 and K67.
In certain embodiments, the method comprises mutating at least six amino acid
residues in
the CR2 portion selected from the group consisting of: N11, R36, K41, S42,
K50, Y64 and
K67. In certain embodiments, the method comprises mutating seven amino acid
residues in
the CR2 portion selected from the group consisting of: N11, R36, K41, S42,
K50, Y64 and
K67. In certain embodiments, the at least one, two, three, four, five, six or
seven mutations
are conservative amino acid substitutions. In certain embodiments, the at
least one, two,
three, four, five, six or seven mutations are non-conservative amino acid
substitutions. In
some embodiments, the binding affinity of the construct for EBV gp350 and/or
IFNa is
reduced by any of about 20%, 25%, 30%, 35%, 40%, 45%, 50%, 55%, 60%, 65%, 70%,
75%, 80%, 85%, 90%, 95%, or 100%, inclusive, as well as any numerical value in
between
these percentages.
[0144] In certain aspects, there is provided a method for reducing the binding
affinity of the
CR2 portion of any of the constructs disclosed herein for EBV gp350, the
method comprising
mutating at least one amino acid residue selected from the group consisting of
N11, R36,
K41, Y64 and K67. In certain embodiments, the method comprises mutating at
least two
amino acid residues selected from the group consisting of N11, R36, K41, Y64
and K67. In
certain embodiments, the method comprises mutating at least three amino acid
residues
selected from the group consisting of N11, R36, K41, Y64 and K67. In certain
embodiments,
the method comprises mutating at least four amino acid residues selected from
the group
consisting of N11, R36, K41, Y64 and K67. In certain embodiments, the method
comprises
mutating at least five amino acid residues selected from the group consisting
of N11, R36,
K41, Y64 and K67. In certain embodiments, the at least one, two, three, four,
or five
mutations may be conservative amino acid substitutions. In certain
embodiments, the at least
one, two, three, four, or five mutations may be non-conservative amino acid
substitutions. In
some embodiments, the binding affinity of the construct for EBV gp350 is
reduced by any of
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
about 20%, 25%, 30%, 35%, 40%, 45%, 50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%,
90%,
95%, or 100%, inclusive, as well as any numerical value in between these
percentages.
[0145] In certain aspects, there is provided a method for reducing the binding
affinity of the
CR2 portion of any of the constructs disclosed herein for IFNc , the method
comprising
mutating at least one amino acid residue selected from the group consisting of
S42 and K50.
In certain embodiments, the method comprises mutating two amino acid residues
selected
from the group consisting of S42 and K50. In certain embodiments, the at least
one or two
mutations may be conservative amino acid substitutions. In certain
embodiments, the at least
one or two mutations may be non-conservative amino acid substitutions. In some
embodiments, the binding affinity of the construct for IFNa is reduced by any
of about 20%,
25%, 30%, 35%, 40%, 45%, 50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 95%, or
100%, inclusive, as well as any numerical value in between these percentages.
[0146] In another aspect, the method for reducing the binding affinity of the
CR2 portion of
any of the constructs disclosed herein for one or more CR2 ligands further
comprises
mutating at least one, two, three, four, five, six, seven, eight, nine, ten,
or eleven amino acids
residues at other positions in the CR2 portion. In certain embodiments, the
CR2 portion
optionally further contains at least one amino acid substitution selected from
the group
consisting of: R13, Y16, A22, R28, S32, K48, K57, Y68, R83, G84, and R89. In
certain
embodiments, the CR2 portion optionally further contains at least two amino
acid
substitutions selected from the group consisting of: R13, Y16, A22, R28, S32,
K48, K57,
Y68, R83, G84, and R89. In certain embodiments, the CR2 portion optionally
further
contains at least three amino acid substitutions selected from the group
consisting of: R13,
Y16, A22, R28, S32, K48, K57, Y68, R83, G84, and R89. In certain embodiments,
the CR2
portion optionally further contains at least four amino acid substitutions
selected from the
group consisting of: R13, Y16, A22, R28, S32, K48, K57, Y68, R83, G84, and
R89. In
certain embodiments, the CR2 portion optionally further contains at least five
amino acid
substitutions selected from the group consisting of: R13, Y16, A22, R28, S32,
K48, K57,
Y68, R83, G84, and R89. In certain embodiments, the CR2 portion optionally
further
contains at least six amino acid substitutions selected from the group
consisting of: R13,
Y16, A22, R28, S32, K48, K57, Y68, R83, G84, and R89. In certain embodiments,
the CR2
portion optionally further contains at least seven amino acid substitutions
selected from the
group consisting of: R13, Y16, A22, R28, S32, K48, K57, Y68, R83, G84, and
R89. In
certain embodiments, the CR2 portion optionally further contains at least
eight amino acid
81
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
substitutions selected from the group consisting of: R13, Y16, A22, R28, S32,
K48, K57,
Y68, R83, G84, and R89. In certain embodiments, the CR2 portion optionally
further
contains at least nine amino acid substitutions selected from the group
consisting of: R13,
Y16, A22, R28, S32, K48, K57, Y68, R83, G84, and R89. In certain embodiments,
the CR2
portion optionally further contains at least ten amino acid substitutions
selected from the
group consisting of: R13, Y16, A22, R28, S32, K48, K57, Y68, R83, G84, and
R89. In
certain embodiments, the CR2 portion optionally further contains eleven amino
acid
substitutions selected from the group consisting of: R13, Y16, A22, R28, S32,
K48, K57,
Y68, R83, G84, and R89. In any of the above embodiments, the at least one,
two, three, four,
five, six, seven, eight, nine, ten or eleven amino acid substitutions may be
conservative
substitutions. In any of the above embodiments, the at least one, two, three,
four, five, six,
seven, eight, nine, ten or eleven amino acid substitutions may be non-
conservative
substitutions.
[0147] In another aspect, provided herein are methods of altering the binding
kinetics of the
CR2 portion of the construct for C3, C3(H20) or cell surface-bound C3
proteolytic
fragments, such as C3d, C3dg, and iC3b, relative to other CR2 ligands (e.g.,
EBV gp350 and
IFNa). In certain embodiments, the altering improves the binding kinetics of
the CR2 portion
of the construct for cell surface-bound proteolytic fragments of C3, such as
C3d, C3dg, and
iC3b, and the method comprises mutating at least one amino acid residue in the
CR2 portion
of the construct selected from the group consisting of N11, R36, K41, S42,
K50, Y64 and
K67. In certain embodiments, the altering improves the binding kinetics of the
CR2 portion
of the construct for cell surface-bound proteolytic fragments of C3, such as
C3d, C3dg, and
iC3b, and the method comprises mutating at least two amino acid residues in
the CR2 portion
of the construct selected from the group consisting of N11, R36, K41, S42,
K50, Y64 and
K67. In certain embodiments, the altering improves the binding kinetics of the
CR2 portion
of the construct for cell surface-bound proteolytic fragments of C3, such as
C3d, C3dg, and
iC3b, and the method comprises mutating at least three amino acid residues in
the CR2
portion of the construct selected from the group consisting of N11, R36, K41,
S42, K50, Y64
and K67. In certain embodiments, the altering improves the binding kinetics of
the CR2
portion of the construct for cell surface-bound proteolytic fragments of C3,
such as C3d,
C3dg, and iC3b, and the method comprises mutating at least four amino acid
residues in the
CR2 portion of the construct selected from the group consisting of N11, R36,
K41, S42, K50,
Y64 and K67. In certain embodiments, the altering improves the binding
kinetics of the CR2
82
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
portion of the construct for cell surface-bound proteolytic fragments of C3,
such as C3d,
C3dg, and iC3b, and the method comprises mutating at least five amino acid
residues in the
CR2 portion of the construct selected from the group consisting of N11, R36,
K41, S42, K50,
Y64 and K67. In certain embodiments, the altering improves the binding
kinetics of the CR2
portion of the construct for cell surface-bound proteolytic fragments of C3,
such as C3d,
C3dg, and iC3b, and the method comprises mutating at least six amino acid
residues in the
CR2 portion of the construct selected from the group consisting of N11, R36,
K41, S42, K50,
Y64 and K67. In certain embodiments, the altering improves the binding
kinetics of the CR2
portion of the construct for cell surface-bound proteolytic fragments of C3,
such as C3d,
C3dg, and iC3b, and the method comprises mutating at least seven amino acid
residues in the
CR2 portion of the construct selected from the group consisting of N11, R36,
K41, S42, K50,
Y64 and K67. In certain embodiments, the cell surface-bound fragment of C3 is
selected
from the group consisting of C3d, C3dg, and iC3b. In certain embodiments, the
cell surface-
bound fragment of C3 is C3d. In certain embodiments, the cell surface-bound
fragment of C3
is C3dg. In certain embodiments, the cell surface-bound fragment of C3 is
iC3b. In certain
embodiments, the at least one, two, three, four, five, six, or seven mutations
may be
conservative amino acid substitutions. In certain embodiments, the at least
one, two, three,
four, five, six, or seven mutations may be non-conservative amino acid
substitutions. In
some embodiments, the altering improves the binding kinetics of the CR2
portion of the
construct for cell surface-bound proteolytic fragments of C3, such as C3d,
C3dg, and iC3b by
any of at least 20%, 25%, 30%, 35%, 40%, 45%, 50%, 55%, 60%, 65%, 70%, 75%,
80%,
85%, 90%, 95%, or 100%.
[0148] In certain aspects, the altering worsens the binding kinetics of the
CR2 portion of
the construct for C3, C3(H20), and/or cell surface-bound C3 proteolytic
fragments, such as,
but not limited to, C3d, C3dg, and iC3b, and the method comprises mutating at
least one
amino acid residue selected from the group consisting of 19, Y29, C31, G33,
T34, D56, S70,
H90, D92, S93, A97, T100, N101, 5109, and 5128. In certain aspects, the
altering worsens
the binding kinetics of the CR2 portion of the construct for C3, C3(H20),
and/or cell surface-
bound C3 proteolytic fragments, such as, but not limited to, C3d, C3dg, and
iC3b, and the
method comprises mutating at least two amino acid residues selected from the
group
consisting of 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97, T100,
N101, 5109,
and S 128. In certain aspects, the altering worsens the binding kinetics of
the CR2 portion of
the construct for C3, C3(H20), and/or cell surface-bound C3 proteolytic
fragments, such as,
83
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
but not limited to, C3d, C3dg, and iC3b, and the method comprises mutating at
least three
amino acid residues selected from the group consisting of 19, Y29, C31, G33,
T34, D56, S70,
H90, D92, S93, A97, T100, N101, 5109, and 5128. In certain aspects, the
altering worsens
the binding kinetics of the CR2 portion of the construct for C3, C3(H20),
and/or cell surface-
bound C3 proteolytic fragments, such as, but not limited to, C3d, C3dg, and
iC3b, and the
method comprises mutating at least four amino acid residues selected from the
group
consisting of 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97, T100,
N101, 5109,
and S 128. In certain aspects, the altering worsens the binding kinetics of
the CR2 portion of
the construct for C3, C3(H20), and/or cell surface-bound C3 proteolytic
fragments, such as,
but not limited to, C3d, C3dg, and iC3b, and the method comprises mutating at
least five
amino acid residues selected from the group consisting of 19, Y29, C31, G33,
T34, D56, S70,
H90, D92, S93, A97, T100, N101, 5109, and 5128. In certain aspects, the
altering worsens
the binding kinetics of the CR2 portion of the construct for C3, C3(H20),
and/or cell surface-
bound C3 proteolytic fragments, such as, but not limited to, C3d, C3dg, and
iC3b, and the
method comprises mutating at least six amino acid residues selected from the
group
consisting of 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97, T100,
N101, 5109,
and S 128. In certain aspects, the altering worsens the binding kinetics of
the CR2 portion of
the construct for C3, C3(H20), and/or cell surface-bound C3 proteolytic
fragments, such as,
but not limited to, C3d, C3dg, and iC3b, and the method comprises mutating at
least seven
amino acid residues selected from the group consisting of 19, Y29, C31, G33,
T34, D56, S70,
H90, D92, S93, A97, T100, N101, 5109, and 5128. In certain aspects, the
altering worsens
the binding kinetics of the CR2 portion of the construct for C3, C3(H20),
and/or cell surface-
bound C3 proteolytic fragments, such as, but not limited to, C3d, C3dg, and
iC3b, and the
method comprises mutating at least eight amino acid residues selected from the
group
consisting of 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97, T100,
N101, 5109,
and S 128. In certain aspects, the altering worsens the binding kinetics of
the CR2 portion of
the construct for C3, C3(H20), and/or cell surface-bound C3 proteolytic
fragments, such as,
but not limited to, C3d, C3dg, and iC3b, and the method comprises mutating at
least nine
amino acid residues selected from the group consisting of 19, Y29, C31, G33,
T34, D56, S70,
H90, D92, S93, A97, T100, N101, 5109, and S128. In certain aspects, the
altering worsens
the binding kinetics of the CR2 portion of the construct for C3, C3(H20),
and/or cell surface-
bound C3 proteolytic fragments, such as, but not limited to, C3d, C3dg, and
iC3b, and the
method comprises mutating at least ten amino acid residues selected from the
group
84
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
consisting of 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97, T100,
N101, 5109,
and S 128. In certain aspects, the altering worsens the binding kinetics of
the CR2 portion of
the construct for C3, C3(H20), and/or cell surface-bound C3 proteolytic
fragments, such as,
but not limited to, C3d, C3dg, and iC3b, and the method comprises mutating at
least eleven
amino acid residues selected from the group consisting of 19, Y29, C31, G33,
T34, D56, S70,
H90, D92, S93, A97, T100, N101, 5109, and S128. In certain aspects, the
altering worsens
the binding kinetics of the CR2 portion of the construct for C3, C3(H20),
and/or cell surface-
bound C3 proteolytic fragments, such as, but not limited to, C3d, C3dg, and
iC3b, and the
method comprises mutating at least twelve amino acid residues selected from
the group
consisting of 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97, T100,
N101, 5109,
and S 128. In certain aspects, the altering worsens the binding kinetics of
the CR2 portion of
the construct for C3, C3(H20), and/or cell surface-bound C3 proteolytic
fragments, such as,
but not limited to, C3d, C3dg, and iC3b, and the method comprises mutating at
least thirteen
amino acid residues selected from the group consisting of 19, Y29, C31, G33,
T34, D56, S70,
H90, D92, S93, A97, T100, N101, 5109, and S128. In certain aspects, the
altering worsens
the binding kinetics of the CR2 portion of the construct for C3, C3(H20),
and/or cell surface-
bound C3 proteolytic fragments, such as, but not limited to, C3d, C3dg, and
iC3b, and the
method comprises mutating at least fourteen amino acid residues selected from
the group
consisting of 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97, T100,
N101, 5109,
and S 128. In certain aspects, the altering worsens the binding kinetics of
the CR2 portion of
the construct for C3, C3(H20), and/or cell surface-bound C3 proteolytic
fragments, such as,
but not limited to, C3d, C3dg, and iC3b, and the method comprises mutating
fifteen amino
acid residues selected from the group consisting of 19, Y29, C31, G33, T34,
D56, S70, H90,
D92, S93, A97, T100, N101, 5109, and 5128. In certain embodiments, the at
least one, two,
three, four, five, six, seven, eight, nine, ten, eleven, twelve, thirteen,
fourteen, or fifteen
mutations may be conservative amino acid substitutions. In certain
embodiments, the at least
one, two, three, four, five, six, seven, eight, nine, ten, eleven, twelve,
thirteen, fourteen, or
fifteen mutations may be non-conservative amino acid substitutions. In some
embodiments,
the altering worsens the binding kinetics of the CR2 portion of the construct
for cell surface-
bound proteolytic fragments of C3, such as C3d, C3dg, and iC3b by any of about
20%, 25%,
30%, 35%, 40%, 45%, 50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 95%, or 100%,
inclusive, as well as any numerical value in between these percentages.
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
Methods of Treating Complement-Associated Diseases or Conditions
[0149] Provided herein are methods of treating a complement-associated disease
or condition
in an individual comprising administering to the individual any of the
compositions described
herein. As used herein, an "individual" can be a vertebrate, a mammal, or a
human.
Specifically, as used herein, a "mammal" can be a nonhuman primate, mouse,
rat, pig, dog,
cat, monkey, cow, or horse. It is understood that administration of the
composition to the
individual can have the effect of, but is not limited to, reducing the
symptoms of the
condition, a reduction in the severity of the condition, or the complete
ablation of the
condition.
Methods of Treatment by Inhibiting Complement Activity
[0150] In some aspects, there is provided a method of treating a complement-
associated
disease or condition in an individual comprising administering to the
individual any of the
compositions described herein, wherein the complement modulator portion of the
composition comprises a complement inhibitor or biologically-active fragment
thereof, and
wherein the administration of the composition inhibits complement activity. In
some
embodiments, the composition selectively binds to one or more C3 proteolytic
fragments but
does not bind to, or has reduced binding affinity for, IFNa and EBV gp350. In
some
embodiments, the composition selectively binds to one or more C3 proteolytic
fragments and
EBV gp350 but does not bind to, or has reduced binding affinity for, IFNa. In
some
embodiments, the composition selectively binds to one or more C3 proteolytic
fragments and
IFNa but does not bind to, or has reduced binding affinity for, EBV gp350. In
some
embodiments, the composition selectively binds to IFNa but does not bind to,
or has reduced
binding affinity for, EBV gp350 and one or more C3 proteolytic fragments. In
some
embodiments, the composition selectively binds to IFNa and EBV gp350 but does
not bind
to, or has reduced binding affinity for, one or more C3 proteolytic fragments.
In some
embodiments, the composition selectively binds to EBV gp350 but does not bind
to, or has
reduced binding affinity for, one or more C3 proteolytic fragments and IFNa.
In some
embodiments, the one or more C3 proteolytic fragments are selected from the
group
consisting of C3d, iC3b, C3dg, and one or more cell-bound fragments of C3b
that bind to the
two N-terminal SCR domains of CR2.
[0151] In some aspects, there is provided a method of treating a complement-
associated
disease or condition in an individual comprising administering to the
individual any of the
86
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
compositions described herein, wherein the complement modulator portion of the
composition comprises a complement inhibitor or biologically-active fragment
thereof,
wherein the administration of the composition inhibits complement activity;
and wherein the
complement associated disease or condition is an inflammatory condition. In
some
embodiments, the complement associated disease or condition can include an
inflammatory
condition such as, but not limited to, asthma, systemic lupus erythematosus,
rheumatoid
arthritis, reactive arthritis, spondylarthritis, systemic vasculitis, insulin
dependent diabetes
mellitus, multiple sclerosis, experimental allergic encephalomyelitis,
Sjogren's syndrome,
graft versus host disease, inflammatory bowel disease including Crohn's
disease, ulcerative
colitis, ischemia reperfusion injury, myocardial Infarction, Alzheimer's
disease, transplant
rejection (allogeneic and xenogeneic), thermal trauma, any immune complex-
induced
inflammation, glomerulonephritis, myasthenia gravis, cerebral lupus, Guillain-
Barre
syndrome, vasculitis, systemic sclerosis, anaphylaxis, catheter reactions,
atheroma, infertility,
thyroiditis, adult respitory distress syndrome (ARDS), post-bypass syndrome,
hemodialysis,
juvenile rheumatoid, Behcets syndrome, hemolytic anemia, pemphigus, bullous
pemphigoid,
stroke, atherosclerosis, and scleroderma. In some embodiments, the complement
inhibitor is
selected from the group consisting of: MCP, DAF, CD59, Crry, CR1, and FH. In
some
embodiments, the composition selectively binds to one or more C3 proteolytic
fragments and
EBV gp350 but does not bind to, or has reduced binding affinity for, IFNa. In
some
embodiments, the composition selectively binds to one or more C3 proteolytic
fragments and
IFNa but does not bind to, or has reduced binding affinity for, EBV gp350. In
some
embodiments, the composition selectively binds to IFNa but does not bind to,
or has reduced
binding affinity for, EBV gp350 and one or more C3 proteolytic fragments. In
some
embodiments, the composition selectively binds to IFNa and EBV gp350 but does
not bind
to, or has reduced binding affinity for, one or more C3 proteolytic fragments.
In some
embodiments, the composition selectively binds to EBV gp350 but does not bind
to, or has
reduced binding affinity for, one or more C3 proteolytic fragments and IFNa.
In some
embodiments, the one or more C3 proteolytic fragments are selected from the
group
consisting of C3d, iC3b, C3dg, and one or more cell-bound fragments of C3b
that bind to the
two N-terminal SCR domains of CR2.
[0152] In some aspects, there is provided a method of treating a complement-
associated
disease or condition in an individual comprising administering to the
individual any of the
compositions described herein, wherein the complement modulator portion of the
87
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
composition comprises a complement inhibitor or biologically-active fragment
thereof,
wherein the administration of the composition inhibits complement activity,
and wherein the
complement associated disease or condition is a viral infection. In some
embodiments, the
viral infection can include, but is not limited to, Influenza virus A,
Influenza virus B,
Respiratory syncytial virus, Dengue virus, Yellow fever virus, Ebola virus,
Marburg virus,
Lassa fever virus, Eastern Equine Encephalitis virus, Japanese Encephalitis
virus, St. Louis
Encephalitis virus, Murray Valley fever virus, West Nile virus, Rift Valley
fever virus,
Rotavirus A, Rotavirus B, Rotavirus C, Sindbis virus, Hantavirus. In some
aspects, the
complement associated disease or condition is a result of the response of an
individual to a
viral vector. In certain embodiments, the viral vector includes, but is not
limited to,
adenovirus, vaccinia virus, adeno associated virus, modified vaccinia ancara
virus,
cytomegalovirus, or any other viral vector known in the art. In some
embodiments, the
complement inhibitor is selected from the group consisting of: MCP, DAF, CD59,
Crry, CR1,
and FH. In some embodiments, the composition selectively binds to one or more
C3
proteolytic fragments and EBV gp350 but does not bind to, or has reduced
binding affinity
for, IFNa. In some embodiments, the composition selectively binds to one or
more C3
proteolytic fragments and IFNa but does not bind to, or has reduced binding
affinity for, EBV
gp350. In some embodiments, the composition selectively binds to IFNa but does
not bind to,
or has reduced binding affinity for, EBV gp350 and one or more C3 proteolytic
fragments. In
some embodiments, the composition selectively binds to IFNa and EBV gp350 but
does not
bind to, or has reduced binding affinity for, one or more C3 proteolytic
fragments. In some
embodiments, the composition selectively binds to EBV gp350 but does not bind
to, or has
reduced binding affinity for, one or more C3 proteolytic fragments and IFNa.
In some
embodiments, the one or more C3 proteolytic fragments are selected from the
group
consisting of C3d, iC3b, C3dg, and one or more cell-bound fragments of C3b
that bind to the
two N-terminal SCR domains of CR2. In some embodiments, the CR2 portion of the
composition has one or more mutations selected from the group consisting of:
19, Y29, C3 1,
G33, T34, D56, S70, H90, D92, S93, A97, T100, N101, 5109, and 5128 and does
not bind to,
or has reduced binding efficiency for, one or more C3 proteolytic fragments.
In some
embodiments, the CR2 portion of the composition has one or more mutations
selected from
the group consisting of: N11, R36, K41, Y64 and K67 and does not bind to, or
has reduced
binding efficiency for, EBV gp350. In some embodiments, the CR2 portion of the
composition has one or more mutations selected from the group consisting of:
S42A and
88
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
K50A and does not bind to, or has reduced binding efficiency for, IFNa. In
some
embodiments, the CR2 portion of the composition has one or more mutations
selected from
the group consisting of: 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97,
T100,
N101, S109, S128, Nil, R36, K41, Y64 and K67 and does not bind to, or has
reduced
binding efficiency for, one or more C3 proteolytic fragments and EBV gp350. In
some
embodiments, the CR2 portion of the composition has one or more mutations
selected from
the group consisting of: 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97,
T100,
N101, S 109, S 128, S42A and K50A and does not bind to, or has reduced binding
efficiency
for, one or more C3 proteolytic fragments and IFNa. In some embodiments, the
CR2 portion
of the composition has one or more mutations selected from the group
consisting of: N11,
R36, K41, Y64 K67, S42A and K50A and does not bind to, or has reduced binding
efficiency
for, EBV gp350 and IFNa.
[0153] In some aspects, there is provided a method of treating a complement-
associated
disease or condition in an individual comprising administering to the
individual any of the
compositions described herein, wherein the complement modulator portion of the
composition comprises a complement inhibitor or biologically-active fragment
thereof,
wherein the administration of the composition inhibits complement activity,
and wherein the
complement associated disease or condition is a fungal infection. It is
understood in the art
that Candida express a CR3-like protein that has similar binding properties as
CR2. The
Candida CR3-like protein appears to be involved in pathogenesis. Therefore, an
embodiment
of the invention is directed to a method of treating an individual with a
fungal infection,
wherein the treatment blocks fungal-"CR3" function as well as inhibits
complement,
comprising administering to a subject any of the compositions described
herein, wherein the
complement modulator portion of the composition comprises a complement
inhibitor or
biologically-active fragment thereof. In some embodiments, the complement
inhibitor is
selected from the group consisting of: MCP, DAF, CD59, Crry, CR1, and FH. In
some
embodiments, the composition selectively binds to one or more C3 proteolytic
fragments and
EBV gp350 but does not bind to, or has reduced binding affinity for, IFNa. In
some
embodiments, the composition selectively binds to one or more C3 proteolytic
fragments and
IFNa but does not bind to, or has reduced binding affinity for, EBV gp350. In
some
embodiments, the composition selectively binds to IFNa but does not bind to,
or has reduced
binding affinity for, EBV gp350 and one or more C3 proteolytic fragments. In
some
embodiments, the composition selectively binds to IFNa and EBV gp350 but does
not bind
89
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
to, or has reduced binding affinity for, one or more C3 proteolytic fragments.
In some
embodiments, the composition selectively binds to EBV gp350 but does not bind
to, or has
reduced binding affinity for, one or more C3 proteolytic fragments and IFNa.
In some
embodiments, the one or more C3 proteolytic fragments are selected from the
group
consisting of C3d, iC3b, C3dg, and one or more cell-bound fragments of C3b
that bind to the
two N-terminal SCR domains of CR2. In some embodiments, the CR2 portion of the
composition has one or more mutations selected from the group consisting of:
19, Y29, C3 1,
G33, T34, D56, S70, H90, D92, S93, A97, T100, N101, 5109, and 5128 and does
not bind to,
or has reduced binding efficiency for, one or more C3 proteolytic fragments.
In some
embodiments, the CR2 portion of the composition has one or more mutations
selected from
the group consisting of: N11, R36, K41, Y64 and K67 and does not bind to, or
has reduced
binding efficiency for, EBV gp350. In some embodiments, the CR2 portion of the
composition has one or more mutations selected from the group consisting of:
S42A and
K50A and does not bind to, or has reduced binding efficiency for, IFNa. In
some
embodiments, the CR2 portion of the composition has one or more mutations
selected from
the group consisting of: 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97,
T100,
N101, S109, S128, Nil, R36, K41, Y64 and K67 and does not bind to, or has
reduced
binding efficiency for, one or more C3 proteolytic fragments and EBV gp350. In
some
embodiments, the CR2 portion of the composition has one or more mutations
selected from
the group consisting of: 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97,
T100,
N101, S 109, S128, S42A and K50A and does not bind to, or has reduced binding
efficiency
for, one or more C3 proteolytic fragments and IFNa. In some embodiments, the
CR2 portion
of the composition has one or more mutations selected from the group
consisting of: N11,
R36, K41, Y64 K67, S42A and K50A and does not bind to, or has reduced binding
efficiency
for, EBV gp350 and IFNa.
[0154] Apoptosis occurring during normal development is noninflammatory and is
involved
in induction of immunological tolerance. Although apoptotic cell death can be
inflammatory
depending on how it is activated and in what cell types (for example,
therapeutic agents that
ligate Fas are able to induce inflammation), necrotic cell death results in a
sustained and
powerful inflammatory response mediated by released cell contents and by
proinflammatory
cytokines released by stimulated phagocytes. Apoptotic cells and vesicles are
normally
cleared by phagocytes, thus preventing the pro-inflammatory consequences of
cell lysis. In
this context, it has been shown that apoptotic cells and apoptotic bodies
directly fix
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
complement, and that complement can sustain an anti-inflammatory response due
to
opsonization and enhanced phagocytosis of apoptotic cells.
[0155] Inflammation is involved in nonspecific recruitment of immune cells
that can
influence innate and adaptive immune responses. Modulating complement
activation during
apoptosis-based tumor therapy to inhibit phagocytic uptake of apoptotic
cells/bodies
enhances the Inflammatory/innate immune response within the tumor environment.
In
addition, apoptotic cells can be a source of immunogenic self antigens and
uncleared
apoptotic bodies can result in autoimmunization. In addition to creating an
enhanced
immuno-stimulatory environment, modulating complement at a site in which tumor
cells
have been induced to undergo apoptosis further augments or triggers specific
immunity
against a tumor to which the host is normally tolerant.
[0156] Accordingly, in some aspects, there is provided a method of enhancing
the outcome
of an apoptosis-based therapy (e.g., gene therapy with adenovirus expressing
Fas ligand) in
an individual comprising administering to the individual any of the
compositions described
herein, wherein the complement modulator portion of the composition comprises
a
complement inhibitor or biologically-active fragment thereof and wherein the
administration
of the composition inhibits complement activity. In some embodiments, the
complement
inhibitor is selected from the group consisting of: MCP, DAF, CD59, Crry, CR1,
and FH. In
some embodiments, the composition selectively binds to one or more C3
proteolytic
fragments and EBV gp350 but does not bind to, or has reduced binding affinity
for, IFNa. In
some embodiments, the composition selectively binds to one or more C3
proteolytic
fragments and IFNa but does not bind to, or has reduced binding affinity for,
EBV gp350. In
some embodiments, the composition selectively binds to IFNa but does not bind
to, or has
reduced binding affinity for, EBV gp350 and one or more C3 proteolytic
fragments. In some
embodiments, the composition selectively binds to IFNa and EBV gp350 but does
not bind
to, or has reduced binding affinity for, one or more C3 proteolytic fragments.
In some
embodiments, the composition selectively binds to EBV gp350 but does not bind
to, or has
reduced binding affinity for, one or more C3 proteolytic fragments and IFNa.
In some
embodiments, the one or more C3 proteolytic fragments are selected from the
group
consisting of C3d, iC3b, C3dg, and one or more cell-bound fragments of C3b
that bind to the
two N-terminal SCR domains of CR2. In some embodiments, the CR2 portion of the
composition has one or more mutations selected from the group consisting of:
19, Y29, C3 1,
G33, T34, D56, S70, H90, D92, S93, A97, T100, N101, 5109, and 5128 and does
not bind to,
91
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
or has reduced binding efficiency for, one or more C3 proteolytic fragments.
In some
embodiments, the CR2 portion of the composition has one or more mutations
selected from
the group consisting of: N11, R36, K41, Y64 and K67 and does not bind to, or
has reduced
binding efficiency for, EBV gp350. In some embodiments, the CR2 portion of the
composition has one or more mutations selected from the group consisting of:
S42A and
K50A and does not bind to, or has reduced binding efficiency for, IFNa. In
some
embodiments, the CR2 portion of the composition has one or more mutations
selected from
the group consisting of: 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97,
T100,
N101, S109, S128, N11, R36, K41, Y64 and K67 and does not bind to, or has
reduced
binding efficiency for, one or more C3 proteolytic fragments and EBV gp350. In
some
embodiments, the CR2 portion of the composition has one or more mutations
selected from
the group consisting of: 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97,
T100,
N101, S109, S128, S42A and K50A and does not bind to, or has reduced binding
efficiency
for, one or more C3 proteolytic fragments and IFNa. In some embodiments, the
CR2 portion
of the composition has one or more mutations selected from the group
consisting of: N11,
R36, K41, Y64 K67, S42A and K50A and does not bind to, or has reduced binding
efficiency
for, EBV gp350 and IFNa.
Methods of Treatment by Enhancing Complement Activity
[0157] In some aspects, there is provided a method of treating a complement-
associated
disease or condition in an individual comprising administering to the
individual any of the
compositions described herein, wherein the complement modulator portion of the
composition comprises a complement activator or biologically-active fragment
thereof, and
wherein the administration of the composition enhances complement activity. In
some
embodiments, enhancing complement activity can have the effect of, but is not
limited to,
reducing the symptoms of the condition, a reduction in the severity of the
condition, or the
complete ablation of the condition. In some embodiments, the composition
selectively binds
to one or more C3 proteolytic fragments and EBV gp350 but does not bind to, or
has reduced
binding affinity for, IFNa. In some embodiments, the composition selectively
binds to one or
more C3 proteolytic fragments and IFNa but does not bind to, or has reduced
binding affinity
for, EBV gp350. In some embodiments, the composition selectively binds to IFNa
but does
not bind to, or has reduced binding affinity for, EBV gp350 and one or more C3
proteolytic
fragments. In some embodiments, the composition selectively binds to IFNa and
EBV gp350
but does not bind to, or has reduced binding affinity for, one or more C3
proteolytic
92
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
fragments. In some embodiments, the composition selectively binds to EBV gp350
but does
not bind to, or has reduced binding affinity for, one or more C3 proteolytic
fragments and
IFNa. In some embodiments, the one or more C3 proteolytic fragments are
selected from the
group consisting of C3d, iC3b, C3dg, and one or more cell-bound fragments of
C3b that bind
to the two N-terminal SCR domains of CR2.
[0158] In some aspects, there is provided a method of treating a complement-
associated
disease or condition in an individual comprising administering to the
individual any of the
compositions described herein, wherein the complement modulator portion of the
composition comprises a complement activator or biologically-active fragment
thereof,
wherein the administration of the composition enhances complement activity,
and wherein
the complement-associated disease or condition is cancer. A representative but
non-limiting
list of cancers that the disclosed complement enhancing compositions can be
used to treat
includes: lymphoma, B cell lymphoma, T cell lymphoma, mycosis fungoides,
multiple
myeloma, Hodgkin's Disease, myeloid leukemia, bladder cancer, brain cancer,
nervous
system cancer, head and neck cancer, squamous cell carcinoma of head and neck,
kidney
cancer, lung cancers such as small cell lung cancer and non-small cell lung
cancer, urothelial
carcinomas, adenocarcinomas, sarcomas, gliomas, high grade gliomas, blastomas,
neuroblastomas, plasmacytomas, histiocytomas, adenomas, hypoxic tumors,
myelomas,
AIDS-related lymphomas or sarcomas, metastatic cancers,
neuroblastoma/glioblastoma,
ovarian cancer, pancreatic cancer, prostate cancer, skin cancer, liver cancer,
melanoma,
squamous cell carcinomas of the mouth, throat, larynx, and lung, colon cancer,
cervical
cancer, cervical carcinoma, breast cancer, and epithelial cancer, renal
cancer, genitourinary
cancer, pulmonary cancer, esophageal carcinoma, head and neck carcinoma, large
bowel
cancer, hematopoietic cancers, testicular cancer, colon and rectal cancers,
stomach cancer,
prostatic cancer, Waldenstroms disease or pancreatic cancer. In other
embodiments, the
complement-associated disease or condition is a precancerous condition such
as, but not
limited to, cervical and anal dysplasias, other dysplasias, severe dysplasias,
hyperplasias,
atypical hyperplasias, and neoplasias. In certain embodiments, the complement
activator is
selected from the group consisting of: human Ig isotype Gi (IgGi), human Ig
isotype M
(IgM), mouse Ig isotype G3 (IgG3), mouse IgM Fc, human IgGi Fc domain, human
IgM Fc
domain, mouse IgG3 Fc domain, mouse IgM Fc domain, and CVF. In some
embodiments,
the composition selectively binds to one or more C3 proteolytic fragments and
EBV gp350
but does not bind to, or has reduced binding affinity for, IFNa. In some
embodiments, the
93
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
composition selectively binds to one or more C3 proteolytic fragments and IFNa
but does not
bind to, or has reduced binding affinity for, EBV gp350. In some embodiments,
the
composition selectively binds to IFNa but does not bind to, or has reduced
binding affinity
for, EBV gp350 and one or more C3 proteolytic fragments. In some embodiments,
the
composition selectively binds to IFNa and EBV gp350 but does not bind to, or
has reduced
binding affinity for, one or more C3 proteolytic fragments. In some
embodiments, the
composition selectively binds to EBV gp350 but does not bind to, or has
reduced binding
affinity for, one or more C3 proteolytic fragments and IFNa. In some
embodiments, the one
or more C3 proteolytic fragments are selected from the group consisting of
C3d, iC3b, C3dg,
and one or more cell-bound fragments of C3b that bind to the two N-terminal
SCR domains
of CR2. In some embodiments, the CR2 portion of the composition has one or
more
mutations selected from the group consisting of: 19, Y29, C31, G33, T34, D56,
S70, H90,
D92, S93, A97, T100, N101, S 109, and S 128 and does not bind to, or has
reduced binding
efficiency for, one or more C3 proteolytic fragments. In some embodiments, the
CR2 portion
of the composition has one or more mutations selected from the group
consisting of: N11,
R36, K41, Y64 and K67 and does not bind to, or has reduced binding efficiency
for, EBV
gp350. In some embodiments, the CR2 portion of the composition has one or more
mutations
selected from the group consisting of: S42A and K50A and does not bind to, or
has reduced
binding efficiency for, IFNa. In some embodiments, the CR2 portion of the
composition has
one or more mutations selected from the group consisting of: 19, Y29, C31,
G33, T34, D56,
S70, H90, D92, S93, A97, T100, N101, 5109, 5128, N11, R36, K41, Y64 and K67
and does
not bind to, or has reduced binding efficiency for, one or more C3 proteolytic
fragments and
EBV gp350. In some embodiments, the CR2 portion of the composition has one or
more
mutations selected from the group consisting of: 19, Y29, C31, G33, T34, D56,
S70, H90,
D92, S93, A97, T100, N101, 5109, 5128, S42A and K50A and does not bind to, or
has
reduced binding efficiency for, one or more C3 proteolytic fragments and IFNa.
In some
embodiments, the CR2 portion of the composition has one or more mutations
selected from
the group consisting of: N11, R36, K41, Y64 K67, S42A and K50A and does not
bind to, or
has reduced binding efficiency for, EBV gp350 and IFNa.
[0159] In some aspects, there is provided a method of treating a complement-
associated
disease or condition in an individual comprising administering to the
individual any of the
compositions described herein, wherein the complement modulator portion of the
composition comprises a complement activator or biologically-active fragment
thereof,
94
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
wherein the administration of the composition enhances complement activity,
and wherein
the complement-associated disease or condition is a viral infection. A
representative but non-
limiting list of viral infections that the disclosed complement enhancing
compositions can be
used to treat includes: Herpes simplex virus type-1, Herpes simplex virus type-
2,
Cytomegalovirus, Epstein-Barr virus, Varicella-zoster virus, Human herpesvirus
6, Human
herpesvirus 7, Human herpesvirus 8, Variola virus, Vesicular stomatitis virus,
Hepatitis A
virus, Hepatitis B virus, Hepatitis C virus, Hepatitis D virus, Hepatitis E
virus, Rhinovirus,
Coronavirus, Influenza virus A, Influenza virus B, Measles virus,
Polyomavirus, Human
Papilomavirus, Respiratory syncytial virus, Adenovirus, Coxsackie virus,
Dengue virus,
Mumps virus, Poliovirus, Rabies virus, Rous sarcoma virus, Yellow fever virus,
Ebola virus,
Marburg virus, Lassa fever virus, Eastern Equine Encephalitis virus, Japanese
Encephalitis
virus, St. Louis Encephalitis virus, Murray Valley fever virus, West Nile
virus, Rift Valley
fever virus, Rotavirus A, Rotavirus B, Rotavirus C, Sindbis virus, Human T-
cell Leukemia
virus type-1, Hantavirus, Rubella virus, Simian Immunodeficiency virus, Human
Immunodeficiency virus type-1, and Human Immunodeficiency virus type-2. In
certain
embodiments, the complement activator is selected from the group consisting
of: human Ig
isotype Gi (IgGi), human Ig isotype M (IgM), mouse Ig isotype G3 (IgG3), mouse
IgM Fc,
human IgG1 Fc domain, human IgM Fc domain, mouse IgG3 Fc domain, mouse IgM Fc
domain, and CVF. In some embodiments, the composition selectively binds to one
or more
C3 proteolytic fragments and EBV gp350 but does not bind to, or has reduced
binding
affinity for, IFNa. In some embodiments, the composition selectively binds to
one or more
C3 proteolytic fragments and IFNa but does not bind to, or has reduced binding
affinity for,
EBV gp350. In some embodiments, the composition selectively binds to IFNa but
does not
bind to, or has reduced binding affinity for, EBV gp350 and one or more C3
proteolytic
fragments. In some embodiments, the composition selectively binds to IFNa and
EBV gp350
but does not bind to, or has reduced binding affinity for, one or more C3
proteolytic
fragments. In some embodiments, the composition selectively binds to EBV gp350
but does
not bind to, or has reduced binding affinity for, one or more C3 proteolytic
fragments and
IFNa. In some embodiments, the one or more C3 proteolytic fragments are
selected from the
group consisting of C3d, iC3b, C3dg, and one or more cell-bound fragments of
C3b that bind
to the two N-terminal SCR domains of CR2. In some embodiments, the CR2 portion
of the
composition has one or more mutations selected from the group consisting of:
19, Y29, C31,
G33, T34, D56, S70, H90, D92, S93, A97, T100, N101, 5109, and 5128 and does
not bind to,
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
or has reduced binding efficiency for, one or more C3 proteolytic fragments.
In some
embodiments, the CR2 portion of the composition has one or more mutations
selected from
the group consisting of: N11, R36, K41, Y64 and K67 and does not bind to, or
has reduced
binding efficiency for, EBV gp350. In some embodiments, the CR2 portion of the
composition has one or more mutations selected from the group consisting of:
S42A and
K50A and does not bind to, or has reduced binding efficiency for, IFNa. In
some
embodiments, the CR2 portion of the composition has one or more mutations
selected from
the group consisting of: 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97,
T100,
N101, S109, S128, N11, R36, K41, Y64 and K67 and does not bind to, or has
reduced
binding efficiency for, one or more C3 proteolytic fragments and EBV gp350. In
some
embodiments, the CR2 portion of the composition has one or more mutations
selected from
the group consisting of: 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97,
T100,
N101, S109, S128, S42A and K50A and does not bind to, or has reduced binding
efficiency
for, one or more C3 proteolytic fragments and IFNa. In some embodiments, the
CR2 portion
of the composition has one or more mutations selected from the group
consisting of: N11,
R36, K41, Y64 K67, S42A and K50A and does not bind to, or has reduced binding
efficiency
for, EBV gp350 and IFNa.
[0160] In some aspects, there is provided a method of treating a complement-
associated
disease or condition in an individual comprising administering to the
individual any of the
compositions described herein, wherein the complement modulator portion of the
composition comprises a complement activator or biologically-active fragment
thereof,
wherein the administration of the composition enhances complement activity,
and wherein
the complement-associated disease or condition is a bacterial infection. A
representative but
non-limiting list of bacterial infections that the disclosed complement
enhancing
compositions can be used to treat includes bacterial infection by: M.
tuberculosis, M. bovis,
M. bovis strain BCG, BCG substrains, M. avium, M. intracellulare, M.
africanum, M.
kansasii, M. marinum, M. ulcerans, M. avium subspecies paratuberculosis,
Nocardia
asteroides, other Nocardia species, Legionella pneumophila, other Legionella
species,
Salmonella typhi, other Salmonella species, Shigella species, Yersinia pestis,
Pasteurella
haemolytica, Pasteurella multocida, other Pasteurella species, Actinobacillus
pleuropneumoniae, Listeria monocytogenes, Listeria ivanovii, Brucella abortus,
other
Brucella species, Cowdria ruminantium, Chlamydia pneumoniae, Chlamydia
trachomatis,
Chlamydia psittaci, Coxiella burnetti, other Rickettsial species, Ehrlichia
species,
96
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
Staphylococcus aureus, Staphylococcus epidermidis, Streptococcus pyogenes,
Streptococcus
agalactiae, Bacillus anthracis, Escherichia coli, Vibrio cholerae,
Campylobacter species,
Neisseria meningitidis, Neisseria gonorrhea, Pseudomonas aeruginosa, other
Pseudomonas
species, Haemophilus Influenzae, Haemophilus ducreyi, other Hemophilus
species,
Clostridium tetani, other Clostridium species, Yersinia enterolitica, and
other Yersinia
species. In certain embodiments, the complement activator is selected from the
group
consisting of: human Ig isotype Gi (IgGi), human Ig isotype M (IgM), mouse Ig
isotype G3
(IgG3), mouse IgM Fc, human IgGi Fc domain, human IgM Fc domain, mouse IgG3 Fc
domain, mouse IgM Fc domain, and CVF. In some embodiments, the composition
selectively binds to one or more C3 proteolytic fragments and EBV gp350 but
does not bind
to, or has reduced binding affinity for, IFNa. In some embodiments, the
composition
selectively binds to one or more C3 proteolytic fragments and IFNa but does
not bind to, or
has reduced binding affinity for, EBV gp350. In some embodiments, the
composition
selectively binds to IFNa but does not bind to, or has reduced binding
affinity for, EBV
gp350 and one or more C3 proteolytic fragments. In some embodiments, the
composition
selectively binds to IFNa and EBV gp350 but does not bind to, or has reduced
binding
affinity for, one or more C3 proteolytic fragments. In some embodiments, the
composition
selectively binds to EBV gp350 but does not bind to, or has reduced binding
affinity for, one
or more C3 proteolytic fragments and IFNa. In some embodiments, the one or
more C3
proteolytic fragments are selected from the group consisting of C3d, iC3b,
C3dg, and one or
more cell-bound fragments of C3b that bind to the two N-terminal SCR domains
of CR2. In
some embodiments, the CR2 portion of the composition has one or more mutations
selected
from the group consisting of: 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93,
A97, T100,
N101, S 109, and S 128 and does not bind to, or has reduced binding efficiency
for, one or
more C3 proteolytic fragments. In some embodiments, the CR2 portion of the
composition
has one or more mutations selected from the group consisting of: N11, R36,
K41, Y64 and
K67 and does not bind to, or has reduced binding efficiency for, EBV gp350. In
some
embodiments, the CR2 portion of the composition has one or more mutations
selected from
the group consisting of: S42A and K50A and does not bind to, or has reduced
binding
efficiency for, IFNa. In some embodiments, the CR2 portion of the composition
has one or
more mutations selected from the group consisting of: 19, Y29, C31, G33, T34,
D56, S70,
H90, D92, S93, A97, T100, N101, 5109, 5128, N11, R36, K41, Y64 and K67 and
does not
bind to, or has reduced binding efficiency for, one or more C3 proteolytic
fragments and
97
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
EBV gp350. In some embodiments, the CR2 portion of the composition has one or
more
mutations selected from the group consisting of: 19, Y29, C31, G33, T34, D56,
S70, H90,
D92, S93, A97, T100, N101, 5109, 5128, S42A and K50A and does not bind to, or
has
reduced binding efficiency for, one or more C3 proteolytic fragments and IFNa.
In some
embodiments, the CR2 portion of the composition has one or more mutations
selected from
the group consisting of: N11, R36, K41, Y64 K67, S42A and K50A and does not
bind to, or
has reduced binding efficiency for, EBV gp350 and IFNa.
[0161] In some aspects, there is provided a method of treating a complement-
associated
disease or condition in an individual comprising administering to the
individual any of the
compositions described herein, wherein the complement modulator portion of the
composition comprises a complement activator or biologically-active fragment
thereof,
wherein the administration of the composition enhances complement activity,
and wherein
the complement-associated disease or condition is a parasitic infection. A
representative but
non-limiting list of parasitic infections that the disclosed complement
enhancing
compositions can be used to treat includes: Toxoplasma gondii,
Plasmodiumfalciparum,
Plasmodium vivax, Plasmodium malariae, other Plasmodium species., Trypanosoma
brucei,
Trypanosoma cruzi, Leishmania major, other Leishmania species, Schistosoma
mansoni,
other Schistosoma species, and Entamoeba histolytica. In certain embodiments,
the
complement activator is selected from the group consisting of: human Ig
isotype Gi (IgGi),
human Ig isotype M (IgM), mouse Ig isotype G3 (IgG3), mouse IgM Fc, human IgGi
Fc
domain, human IgM Fc domain, mouse IgG3 Fc domain, mouse IgM Fc domain, and
CVF.
In some embodiments, the composition selectively binds to one or more C3
proteolytic
fragments and EBV gp350 but does not bind to, or has reduced binding affinity
for, IFNa. In
some embodiments, the composition selectively binds to one or more C3
proteolytic
fragments and IFNa but does not bind to, or has reduced binding affinity for,
EBV gp350. In
some embodiments, the composition selectively binds to IFNa but does not bind
to, or has
reduced binding affinity for, EBV gp350 and one or more C3 proteolytic
fragments. In some
embodiments, the composition selectively binds to IFNa and EBV gp350 but does
not bind
to, or has reduced binding affinity for, one or more C3 proteolytic fragments.
In some
embodiments, the composition selectively binds to EBV gp350 but does not bind
to, or has
reduced binding affinity for, one or more C3 proteolytic fragments and IFNa.
In some
embodiments, the one or more C3 proteolytic fragments are selected from the
group
consisting of C3d, iC3b, C3dg, and one or more cell-bound fragments of C3b
that bind to the
98
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
two N-terminal SCR domains of CR2. In some embodiments, the CR2 portion of the
composition has one or more mutations selected from the group consisting of:
19, Y29, C3 1,
G33, T34, D56, S70, H90, D92, S93, A97, T100, N101, 5109, and 5128 and does
not bind to,
or has reduced binding efficiency for, one or more C3 proteolytic fragments.
In some
embodiments, the CR2 portion of the composition has one or more mutations
selected from
the group consisting of: N11, R36, K41, Y64 and K67 and does not bind to, or
has reduced
binding efficiency for, EBV gp350. In some embodiments, the CR2 portion of the
composition has one or more mutations selected from the group consisting of:
S42A and
K50A and does not bind to, or has reduced binding efficiency for, IFNa. In
some
embodiments, the CR2 portion of the composition has one or more mutations
selected from
the group consisting of: 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97,
T100,
N101, S109, S128, Nil, R36, K41, Y64 and K67 and does not bind to, or has
reduced
binding efficiency for, one or more C3 proteolytic fragments and EBV gp350. In
some
embodiments, the CR2 portion of the composition has one or more mutations
selected from
the group consisting of: 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93, A97,
T100,
N101, S 109, S128, S42A and K50A and does not bind to, or has reduced binding
efficiency
for, one or more C3 proteolytic fragments and IFNa. In some embodiments, the
CR2 portion
of the composition has one or more mutations selected from the group
consisting of: N11,
R36, K41, Y64 K67, S42A and K50A and does not bind to, or has reduced binding
efficiency
for, EBV gp350 and IFNa.
[0162] In some aspects, there is provided a method of treating a complement-
associated
disease or condition in an individual comprising administering to the
individual any of the
compositions described herein, wherein the complement modulator portion of the
composition comprises a complement activator or biologically-active fragment
thereof,
wherein the administration of the composition enhances complement activity,
and wherein
the complement-associated disease or condition is a fungal infection. A
representative but
non-limiting list of fungal infections that the disclosed complement enhancing
compositions
can be used to treat includes: Candida albicans, Cryptococcus neoformans,
Histoplama
capsulatum, Aspergillus fumigatus, Coccidiodes immitis, Paracoccidiodes
brasiliensis,
Blastomyces dermitidis, Pneuomocystis carnii, Penicillium marneffi, and
Alternaria
alternate. In certain embodiments, the complement activator is selected from
the group
consisting of: human Ig isotype Gi (IgGi), human Ig isotype M (IgM), mouse Ig
isotype G3
(IgG3), mouse IgM Fc, human IgGi Fc domain, human IgM Fc domain, mouse IgG3 Fc
99
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
domain, mouse IgM Fc domain, and CVF. In some embodiments, the composition
selectively binds to one or more C3 proteolytic fragments and EBV gp350 but
does not bind
to, or has reduced binding affinity for, IFNa. In some embodiments, the
composition
selectively binds to one or more C3 proteolytic fragments and IFNa but does
not bind to, or
has reduced binding affinity for, EBV gp350. In some embodiments, the
composition
selectively binds to IFNa but does not bind to, or has reduced binding
affinity for, EBV
gp350 and one or more C3 proteolytic fragments. In some embodiments, the
composition
selectively binds to IFNa and EBV gp350 but does not bind to, or has reduced
binding
affinity for, one or more C3 proteolytic fragments. In some embodiments, the
composition
selectively binds to EBV gp350 but does not bind to, or has reduced binding
affinity for, one
or more C3 proteolytic fragments and IFNa. In some embodiments, the one or
more C3
proteolytic fragments are selected from the group consisting of C3d, iC3b,
C3dg, and one or
more cell-bound fragments of C3b that bind to the two N-terminal SCR domains
of CR2. In
some embodiments, the CR2 portion of the composition has one or more mutations
selected
from the group consisting of: 19, Y29, C31, G33, T34, D56, S70, H90, D92, S93,
A97, T100,
N101, S 109, and S 128 and does not bind to, or has reduced binding efficiency
for, one or
more C3 proteolytic fragments. In some embodiments, the CR2 portion of the
composition
has one or more mutations selected from the group consisting of: N11, R36,
K41, Y64 and
K67 and does not bind to, or has reduced binding efficiency for, EBV gp350. In
some
embodiments, the CR2 portion of the composition has one or more mutations
selected from
the group consisting of: S42A and K50A and does not bind to, or has reduced
binding
efficiency for, IFNa. In some embodiments, the CR2 portion of the composition
has one or
more mutations selected from the group consisting of: 19, Y29, C31, G33, T34,
D56, S70,
H90, D92, S93, A97, T100, N101, 5109, 5128, N11, R36, K41, Y64 and K67 and
does not
bind to, or has reduced binding efficiency for, one or more C3 proteolytic
fragments and
EBV gp350. In some embodiments, the CR2 portion of the composition has one or
more
mutations selected from the group consisting of: 19, Y29, C31, G33, T34, D56,
S70, H90,
D92, S93, A97, T100, N101, 5109, 5128, S42A and K50A and does not bind to, or
has
reduced binding efficiency for, one or more C3 proteolytic fragments and IFNa.
In some
embodiments, the CR2 portion of the composition has one or more mutations
selected from
the group consisting of: N11, R36, K41, Y64 K67, S42A and K50A and does not
bind to, or
has reduced binding efficiency for, EBV gp350 and IFNa.
100
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
EXAMPLES
[0163] The examples, which are intended to be purely exemplary of the
invention and should
therefore not be considered to limit the invention in any way, also describe
and detail aspects
and embodiments of the invention discussed above. The foregoing examples and
detailed
description are offered by way of illustration and not by way of limitation.
All publications,
patent applications, and patents cited in this specification are herein
incorporated by reference
as if each individual publication, patent application, or patent were
specifically and
individually indicated to be incorporated by reference. In particular, all
publications cited
herein are expressly incorporated herein by reference for the purpose of
describing and
disclosing compositions and methodologies which might be used in connection
with the
invention. Although the foregoing invention has been described in some detail
by way of
illustration and example for purposes of clarity of understanding, it will be
readily apparent to
those of ordinary skill in the art in light of the teachings of this invention
that certain changes
and modifications may be made thereto without departing from the spirit or
scope of the
appended claims.
EXAMPLE 1: IDENTIFICATION OF AMINO ACID RESIDUES IMPORTANT FOR
CR2 BINDING INTERACTIONS WITH EBV gp350 AND IFNa.
Experimental Methods
Expression and purification of recombinant proteins
[0164] Human CR2 SCR1-2 for NMR and isothermal titration calorimetry ("ITC")
studies
was expressed in Pichia pastoris using a BioF1oTM 110 Fermenter (New Brunswick
Scientific, Edison, NJ) as previously described (46). Briefly, a single colony
was grown up in
ml Pichia basal salt medium containing 1% glycerol (BMG, per liter: 85%
phosphoric acid
26.7 ml, calcium sulfate 0.93 g, potassium sulfate 18.2 g, magnesium sulfate-
heptahydrate
14.9 g, potassium hydroxide 4.13 g, glycerol 10.0 g, distilled, deionized
water to 1 liter)
overnight at 30 C and 250 rpm, expanded to 50 ml BMG (24 hrs) and finally
expanded to
300 ml BMG (24 hrs). The inoculation culture was centrifuged at 2500xg 25 C
and
resuspended in 30 ml BMG. The 30 ml inoculation culture was used to inoculate
1 L of
minimal Pichia basal salt medium containing 40 g of glycerol. Dissolved 02
concentration
was maintained at 40%, the temperature at 30 C and the pH at 5.0 using 2 M
KOH. Initial
feeds were batch glycerol feeds; transition to methanol was eased by a
methanol injection
101
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
before an exponential methanol feed profile was initiated. Methanol induction
lasted for two
days, after which the culture was centrifuged to remove cellular debris. The
supernatant was
exchanged into 10 mM formate pH 4.0 before being passed over an SP-Sepharose
column (2
x 5 mL SP HiTrapTM columns, GE Biosciences, Pittsburgh, PA) followed by a CR2
affinity
column, generated in-house by binding GST-C3d to a GSTrapTM column (GE
Biosciences,
Pittsburgh, PA). CR2 was eluted along an increasing linear NaCl gradient, 0-
1.0 M in 1/3X
Phosphate Buffered Saline (PBS, 1.6 mM MgC12, 0.9 mM KC1, 0.5 mM KH2PO4, 45.6
mM
NaCl, 2.7 mM Na2HPO4 pH 7.4). Finally, CR2 SCR 1-2 was purified by size
exclusion
chromatography. Purity and identity of CR2 was monitored via SDS-PAGE, Western
blot
analysis and mass spectrometry. Both 15N and 15N-13C isotopically labeled
proteins were
prepared using this strategy. For 15N isotopically labeled CR2 15N-Ammonium
sulfate was
used. For 15N-13C isotopically labeled CR2 15N -Ammonium sulfate, 13C -
glycerol and 13C -
methanol were used. Isotopically enriched chemicals were purchased from Isotec
Inc.,
Miamisburg, OR
[0165] Human CR2 SCR 1-2 for ITC studies was generated using the pMAL-P2XTM
expression system (New England Biolabs, Ipswich, MA) in E. coli as previously
described
(42, 43). Ampicillin-resistant colonies were used to start overnight cultures
that were
expanded to 1 L and grown at 37 C until an A600 of 0.3 was obtained. Cultures
were induced
with 0.3 mM isopropyl- (3-D-thiogalactoside (IPTG) at 30 C overnight before
harvesting by
centrifugation. Harvested pellets were resuspended in amylose column buffer
(20 mM Tris-
HC1, pH 7.4, 0.2 M NaCl, 1 mM EDTA) and lysed by sonication. Lysate was
clarified by
centrifugation and applied to an amylose resin column (New England
Biosciences, Ipswich,
MA). Bound MBP-CR2 SCR 1-2 was eluted from the column using amylose column
elution
buffer (amylose column buffer plus 10 mM maltose). Finally, the MBP-CR2 SCR 1-
2 was
purified by size exclusion chromatography. Purity and identity of MBP-CR2 was
monitored
via SDS-PAGE and Western blot analysis.
[0166] Human C3d for ITC studies was generated using the pGEXTM expression
system (GE
Healthcare, Piscataway, NJ) in E. coli as previously described (47). Briefly,
ampicillin-
resistant colonies were used to start overnight cultures that were expanded to
1 L and grown
at 37 C until an A600 of 0.3 was achieved. Cultures were induced with 0.3 mM
IPTG at 30 C
overnight before harvesting by centrifugation. Harvested pellets were
resuspended in GST
column buffer (50 mM Tris-HC1, pH 8.0, 250 mM NaCl, 1 mM EDTA) and lysed by
sonication. Lysate was clarified by centrifugation and applied to a GSTrapTM
column (GE
102
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
Biosciences, Pittsburgh, PA). C3d was cleaved from the column by digesting
with 50 U of
thrombin overnight at 4 C and subsequently purified by size exclusion
chromatography.
Purity of C3d was monitored via SDS-PAGE.
[0167] Purification of a truncated construct of EBV gp350 comprising residues
1-470 of the
wild-type protein for NMR titrations and ITC studies was performed as
previously described
(46). gp350 was produced by infecting Sf9 insect cells with the gp350-packaged
baculovirus
particles (pVI-Bac Transfer vector, C-terminal polyhistidine tag) at a
multiplicity of infection
(MOI) of 3. The baculoviral supernatant was concentrated, buffered with 10 mM
Tris-HC1
with 10 mM imidazole pH 7.4 and applied to a 5 mL HiTrapTM column (GE
Biosciences,
Pittsburgh, PA) and subsequently eluted with a linear imidazole gradient.
Purity and identity
of gp350 were monitored via SDS-PAGE and Western blot analysis.
[0168] Human IFNa for NMR titrations and ITC studies was generated using the
pMALTM
expression system (New England Biolabs, Ipswich, MA) in E. coli as previously
described
(48). Ampicillin-resistant colonies were used to start overnight cultures that
were expanded
to 1 L and grown at 37 C until an A600 of 0.3 was obtained. Cultures were
induced with 0.3
mM IPTG at 25 C overnight before harvesting by centrifugation. Harvested
pellets were
resuspended in amylose column buffer (20 mM Tris-HC1, pH 7.4, 0.2 M NaCl, 1 mM
EDTA)
and lysed by sonication. Lysate was clarified by centrifugation and applied to
an amylose
resin column (New England Biosciences, Ipswich, MA). Bound MBP-IFNa was eluted
from
the column using amylose column elution buffer (amylose column buffer plus
10mM
maltose). After elution the MBP tag was cleaved overnight at 4 C with Factor
Xa (New
England Biosciences, Ipswich, MA). Finally, IFNa was purified by size
exclusion
chromatography. Purity and identity of IFNa was monitored via SDS-PAGE and
Western
blot analysis.
NMR Analysis
[0169] NMR experiments were carried out on Varian 600, 800 and 900 MHz magnets
housed in the Rocky Mountain Regional NMR facility at the University of
Colorado Denver
School of Medicine (UCDSOM) campus (600 and 900 MHz) and in the W. M. Keck
High
Field NMR Facility at the University of Colorado Boulder campus (800 MHz). The
uniformly 15N-13C labeled SCR1-2 domains of CR2 in 1/3X PBS were used to
sequentially
assign the 15N-TROSY-HSQC (49) by using HNCACB (50), CBCA(CO)NH (51) and 15N
edited NOESY-HSQC (52) three-dimensional spectra. The NMR data was processed
with
103
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
nmrPipe (53) and analyzed with ccpNMR (54). Chemical shift changes were
monitored
using ccpNMR by overlaying TROSY-HSQC spectra from free CR2 SCR1-2 and CR2
SCR1-2 with increasing concentrations of either EBV gp350 or IFNa.
Isothermal Titration Calorimetry ("ITC ") Analysis
[0170] ITC experiments were carried out on a Microcal VP-ITC (GE Healthcare,
Piscataway,
NJ) housed in the Biophysics Core facility on the UCDSOM campus. CR2 SCR1-2 in
U3X
PBS was used in titration experiments carried out at 20 C. Each titration
experiment
consisted of a 5 l injection followed by 26 injections of 10 l of graded
concentrations of
C3d, gp350 or IFNa. Data was analyzed using the software provided by the
manufacturer
(Origin, version 7.0 MicroCal) using either single site or two site binding
models (55).
Chemical Shift Analysis
[0171] Using previously described resonance assignments (48), full-length
ligands EBV
gp350 and IFNa were titrated into uniformly 15N-labeled CR2 SCR 1-2 samples
and the 1H-
15N chemical shifts were monitored (Figures 1 - 3). Titration with EBV gp350
yielded a
single mode of binding characterized by the disappearance and reappearance of
specific
resonances, indicative of a tight binding interaction. The residues on CR2 SCR
1-2 exhibiting
chemical shift changes with EBV gp350 were N11, R13, A22, R28, S32, R36, K41,
K57,
Y64, K67, Y68, R83, G84 and R89. Those residues encompass the SCR1, SCR2 and
the
inter-SCR linker region between SCR 1-2 of CR2 (Figures 3 and 4A). Chemical
shift change
magnitudes are shown in Figure 3. These results suggest that the inter-SCR
linker between
SCR 1-2 and a ridge on SCR1 play the most important role in ligating gp350 to
CR2 (Figure
3). Since this interaction is under slow exchange on the NMR time scale, only
an upper limit
Kd can be calculated. The Kd was calculated using the minimal observed
chemical shift
difference between free and bound resonances (about 60 Hz); assuming a
diffusion-limited on
rate of _108 M-1 s-1, an upper limit to the binding constant was calculated as
-60 M (Table I).
[0172] Full length IFNa was also titrated into a uniformly 15N-labeled CR2
SCR1-2 samples
and the 1H-15N chemical shifts were monitored (Figure 2). Titration with the
cytokine IFNa
yielded a single mode of binding similar to that of gp350 ligation and thus a
tight interaction.
The residues on CR2 SCR1-2 exhibiting chemical shift changes are R13, Y16,
R28, S42,
K48, K50, Y68, R83, G84 and R89. These residues encompass the SCR1, SCR2 and
the
inter SCR linker region of CR2 (Figures 3 and 4B). Chemical shift change
magnitudes are
shown in figure 3. These results suggest that IFNa binding surface is similar
to that of the
104
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
C3d binding surface (Figure 3). Similar to the gp350 chemical shift changes,
the chemical
shift changes for the IFNa suggest a tighter than visible via the NMR time
scale; the upper
limit Kd was calculated as before to be -70 M (Table I).
[0173] For comparison, unique and shared residues on CR2 required for ligation
by C3d,
gp350 and IFNa are shown in Figure 4C. Change in magnitude of chemical shift
for each
ligation state is shown in Figure 3.
Results
Chemical Shift Analysis
[0174] Using previously described resonance assignments (48), full-length
ligands EBV
gp350 and IFNa were titrated into uniformly 15N-labeled CR2 SCR 1-2 samples
and the 1H-
15N chemical shifts were monitored (Figures 1 - 3). Titration with EBV gp350
yielded a
single mode of binding characterized by the disappearance and reappearance of
specific
resonances, indicative of a tight interaction. The residues on CR2 SCR 1-2
exhibiting
chemical shift changes are N11, R13, A22, R28, S32, R36, K41, K57, Y64, K67,
Y68, R83,
G84 and R89. These residues encompass the SCR I, SCR2 and the inter SCR linker
region of
CR2 (Figures 3 and 4A). Chemical shift change magnitudes are shown in Figure
3. These
results suggest that the inter SCR linker and a ridge on SCR1 play the most
important role in
ligating gp350 to CR2 (Figure 3). Since this interaction is under slow
exchange on the NMR
time scale, only an upper limit Kd can be calculated. The Kd was calculated
using the
minimal observed chemical shift difference between free and bound resonances
(about 60
Hz); assuming a diffusion-limited on rate of -10 X 8 M-ls-1, an upper limit to
the binding
constant was calculated as -60 M (Table I).
[0175] Full length IFNa was also titrated into a uniformly 15N-labeled CR2
SCR1-2 samples
and the 1H-15N chemical shifts were monitored (Figure 2). Titration with the
cytokine IFNa
yielded a single mode of binding similar to that of gp350 ligation and thus a
tight interaction.
The residues on CR2 SCR1-2 exhibiting chemical shift changes are R13, Y16,
R28, S42,
K48, K50, Y68, R83, G84 and R89. These residues encompass the SCR1, SCR2 and
the
inter-SCR linker region between SCR 1-2 of CR2 (Figures 3 and 4B). Chemical
shift change
magnitudes are shown in Figure 3. These results suggest that IFNa binding
surface is similar
to that of the C3d binding surface (Figure 3). Similar to the gp350 chemical
shift changes, the
chemical shift changes for the IFNa suggest a tighter binding interaction than
visible via the
NMR time scale; the upper limit Kd was calculated as before to be -70 M
(Table I). For
105
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
comparison, unique and shared residues on CR2 required for ligation by C3d,
gp350 and
IFNa are shown in Figure 4C. Change in magnitude of chemical shift for each
ligation state
is shown in Figure 3.
Thermodynamics of CR2-ligand interactions
[0176] ITC was used to determine binding affinities of CR2-ligand
interactions. Consistent
with the NMR chemical shift analyses, the interaction between CR2 and C3d was
determined
to be a two site binding based on the goodness of fit of a two site binding
model rather than a
single site binding model. The two affinities are 0.13 0.05 M and 160 20
M. The
interaction between CR2 and gp350 was fit using a single site binding model
which yielded
an affinity of 0.014 0.009 M. The interaction between CR2 and IFNa was fit
using a
single site binding model yielding an affinity of 0.035 0.008 M. The
results of all
thermodynamic parameters from NMR and ITC derived affinities are listed in
Table 1.
Discussion
[0177] The studies described herein used two approaches to study CR2-ligand
interactions
with EBV gp350 and IFNa in the fluid phase: (1) NMR spectroscopy experiments
in which
full-length gp350 or IFNa was titrated into 15N labeled CR2 SCR1-2 and
chemical shifts
were monitored; and (2) ITC to further characterize and determine binding
constants for each
CR2-ligand interaction.
[0178] Previous analyses showed that both SCR1 and 2 were needed for the
binding of
gp350 (1, 12, 20, 56, 57). Furthermore, it was also reported that specific
areas of SCR1-2
were important in binding gp350 (20, 58). These areas were between the first
and second, the
second and third cysteine residues of SCR1, and the second half of SCR2; amino
acids
included R89 to R96 and T100 to S128 in SCR2 (58). Interactions with the
linker was also
inferred by the finding that the introduction of a glycosylation site into the
linker eliminated
gp350, but not C3d binding (20, 31, 57). More recently it has been shown that
there are
specific interacting amino acids on the surface of CR2 SCR1-2. Mutagenesis
studies
suggested that residues R13, S15, R28, R36, K41, K56, K67, R83 and R89 are the
most
important residues in the CR2-gp350 interaction (42, 43). In addition, using
HADDOCK, a
model of interaction was determined where the linker region between domains
one and two
of gp350 interacts with the linker between SCR1 and SCR2 of CR2 (43).
[0179] However, although there have been suggestions of important regions and
more
recently amino acids that are important in the interaction between CR2 and
gp350, there has
106
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
been no physical evidence of these interactions occurring. The data described
herein now
illustrates amino acid residues important for the CR2-gp350 interaction
(Figure 4A).
Residues determined to be important to the CR2-gp350 interaction are N11, R13,
A22, R28,
S32, R36, K41, K57, Y64, K67, Y68, R83, G84 and R89. Since there are multiple
interactions within the linker region it is possible to imagine a
rearrangement of SCR
domains about the linker region upon binding gp350 and thus allowing for all
contact points
to be met. If that is the case, some of these residues identified herein as
important for
interaction might be involved in structural rearrangement upon binding and not
intimate
amino acid contact sites. Some resonances disappear due to the large size of
the ligated
complex, approximately 110 kDa, and the resultant increased tumbling time;
therefore,
alternative labeling techniques are necessary to observe such resonances.
[0180] This data appears to confirm that the linker region is important in the
CR2-gp350
interaction. The linker interaction has been shown to be important in
mutagenesis-derived
data as well as in the soft dock model from HADDOCK (43). The linker region
between
SCR1 and SCR2 is eight amino acids, one of the longest in SCR-containing
proteins, and
thus is likely to be flexible enough to mediate multiple ligand interactions.
Unlike the CR2-
C3d interaction, our data suggests that two residues in the linker region, K67
and Y68, are
important in the CR2-gp350 interaction. Thus with both a charged residue, K67,
and a
hydroxyl-containing residue, Y68, it is likely that the interaction with the
linker is stronger in
the CR2-gp350 interaction than with the CR2-C3d interaction. This information
provides a
start to defining how CR2 can mediate multiple specific ligand interactions.
[0181] As with the CR2-C3d interaction (59, 60), the CR2-gp350 interaction is
likely driven
largely by electrostatic interactions as is evident by the large number of
charged residues
among those important in the CR2-gp350 interaction. The majority of these
charged residues
are found on SCR1, suggesting that this domain plays a more significant role
in the CR2-
gp350 interaction. Interestingly R83 was also determined to be important in
the CR2-gp350,
although many other residues around R83 were not shown to exhibit changes in
chemical
shift during the CR2-C3d interaction. This data, along with the weak
interaction found in the
CR2-C3d interaction could signify that the R83 interaction is more important
in the initial
electrostatic attraction of gp350 to CR2 than to significant amino-acid
contacts. The charged
residues that were identified in this study agree with the HADDOCK model (42,
43). Again,
as with the CR2-C3d NMR binding map we have found that there are more residues
than just
charged residues involved in the CR2-gp350 interaction. Specifically, three
hydroxyl-
107
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
containing amino acids (S32, Y64 and Y68) are important in the CR2-gp350
interaction.
These side chain interactions are likely hydrogen bond interactions. This new
data suggests
that the CR2-gp350 and CR2-C3d binding sites are likely similar which explains
why the
ligands cross compete, yet there are substantial differences which begin to
explain how
selective binding occurs.
[0182] The HADDOCK model fits well with the NMR determined CR2-gp350 binding
residues (Figure 5). All but two residues, K57 and A22, are found within the
hypothetical
binding face derived from the HADDOCK model. The A22 chemical shift could
likely be
due to a slight conformation change in SCR1 upon CR2 binding gp350. Whereas
the K57
interaction described by NMR could be used to drive a different and
potentially lower energy
docking model, as it was not utilized as an active residue in the simulated
docking approach
of Young, et al (43).
[0183] The CR2-IFNa interaction has been characterized in several ways. The
first started
with investigating sequence similarities between proposed CR2 binding sites on
C3d and
gp350 (19). To further confirm the potential binding interaction, antibodies
raised against
peptide sequences of the proposed CR2 binding site on IFNa were found to
inhibit the CR2-
C3d interaction in cell binding assays. It was also found that IFNa binding to
CR2 inhibits
CR2-C3d complex formation in cell-binding assays. In addition, it was found
that IFNa
inhibited the capping of CR2 by gp350, thus acting as an anti-viral inhibitor
of early phase
infection from EBV (18). More recently a biophysical study has been completed
on the
thermodynamic properties of CR2-ligand interactions, thus indicating the
physical binding of
CR2 and IFNa (17). The data presented herein further defines a binding site or
binding
surface for the CR2-IFNa interaction. Using NMR titration studies, the
following amino
acids were identified as being involved in the CR2-IFNa interaction-R13, Y16,
R28, S42,
K48, K50, Y68, R83, G84 and R89.
[0184] As with other CR2-ligand interactions, the CR2-IFNa interaction is
largely driven by
electrostatic interactions. The CR2-IFNa interaction is likely the closest
related to the C3d
interaction, since the proposed CR2 binding motifs of C3d and IFNa are the
closest. In
addition, the same linker region residue, Y68, appears to undergo significant
perturbations
upon addition of either C3d and IFNa, as well as the same overall layout of
residues involved
in both interactions (Figure 4C).
[0185] Thermodynamic studies of CR2-ligand interactions have yielded slightly
differing
results (Table 2). As reported previously, the CR2-C3d interaction has been
described as
108
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
either being a two site or a single site binding interaction (17, 48). The ITC
data presented
herein best fit a two-site model with a weaker Kd of 160 M and a tighter
interaction of 0.13
M. This Kd is fairly close to the previously determined Kd from a surface
plasmon
resonance (SPR)-based biophysical study (17). Using ITC, we are now able for
the first time
to measure in the fluid phase the two separate affinities for the two unique
binding events.
The CR2-C3d interaction is unique in that all other characterized CR2-ligand
interactions fit
a simple one to one binding isotherm. In contrast, the current ITC study of
the CR2-gp350
interaction best fit a single binding isotherm with a Kd of 0.014 M, an
affinity only slightly
tighter than the previously reported Kd of 0.077 M determined by SPR. The
difference in
affinities here could be due to the differing experimental conditions of the
respective studies.
Finally, the ITC data for the CR2-IFNa interaction best fit a single binding
isotherm with a
Kd of 0.036 M, an affinity in excellent agreement with the previously
reported Kd of 0.042
M determined by SPR. Again, the difference is likely due to the difference in
buffers used
as well as differences in each assay, with the ITC experiments using CR2 and
IFNa purely in
solution, while the SPR studies used CR2 fixed to a solid support. The rank
order of binding
strength makes sense in that both IFNa and gp350 binding both inhibit C3d
binding to CR2,
which has been previously reported (17, 18).
References
1. Fingeroth, J. D., Clabby, M. L., and Strominger, J. D. (1988) J. Virol. 62,
1442-1447.
2. Fujisaku, A., Harley, J. B., Frank, M. B., Gruner, B. A., Frazier, B., and
Holers, V. M.
(1989) J. Biol. Chem. 264, 2118-2125.
3. Moore, M. D., Cooper, N. R., Tack, B. F., and Nemerow, G. R. (1987) Proc.
Nat'l Acad.
Sci. USA 84, 9194-9198.
4. Weis, J. J., Fearon, D. T., Klickstein, L. B., Wong, W. W., Richards, S.
A., de Bruyn Kops,
A., Smith, J. A., and Weis, J. H. (1986) Proc. Nat'lAcad. Sci. USA 83, 5639-
5643.
5. Weis, J. J., Toothaker, L. E., Smith, J. A., Weis, J. H., and Fearon, D. T.
(1988) J. Exp.
Med. 167, 1047-1066.
6. Ahearn, J. M., and Fearon, D. T. (1989) Adv. Immunol. 46, 183-219.
7. Cooper, N. R., Moore, M. D., and Nemerow, G. R. (1988) Ann. Rev. Immunol.
6, 85-113.
109
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
8. Holers, V. M. (1995) Mosby, 363-391.
9. Tolnay, M., and Tsokos, G. C. (1998) Clin. Immunol. Immunopathol. 88, 123-
132.
10. Lida, K., Nadler, L., and Nussenzweig, V. (1983) J. Exp. Med. 158, 1021-
1033.
11. Weis, J. J., Tedder, T. F., and Fearon, D. T. (1984) Proc. Nat'l Acad.
Sci. USA 81, 881-
885.
12. Fingeroth, J. D., Weis, J. J., Tedder, T. F., Strominger, J. L., Biro, P.
A., and Fearon, D.
T. (1984) Proc. Nat'lAcad. Sci. USA 81, 4510-4514.
13. Nemerow, G. R., Houghten, R. A., Moore, M. D., and Cooper, N. R. (1989)
Cell 56, 369-
377.
14. Nemerow, G. R., Wolfert, R., McNaughton, M. E., and Cooper, N. R. (1985)
J. Virol. 55,
347-351.
15. Aubry, J. P., Pochon, S., Gauchat, J. F., Nueda-Marin, A., Holers, V. M.,
Graber, P.,
Siegfried, C., and Bonnefoy, J. Y. (1994) J. Immunol. 152, 5806-5813.
16. Aubry, J. P., Pochon, S., Graber, P., Jansen, K. U., and Bonnefoy, J. Y.
(1992) Nature
358, 505-507.
17. Asokan, R., Hua, J., Young, K. A., Gould, H. J., Hannan, J. P., Kraus, D.
M., Szakonyi,
G., Grundy, G. J., Chen, X. S., Crow, M. K., and Holers, V. M. (2006) J.
Immunol. 177, 383-
394.
18. Delcayre, A. X., Lotz, M., and Lernhardt, W. (1993) J. Virol. 67, 2918-
2921.
19. Delcayre, A. X., Salas, F., Mathur, S., Kovats, K., Lotz, M., and
Lernhardt, W. (1991)
EMBO J. 10, 919-926.
20. Martin, D. R., Yuryev, A., Kalli, K. R., Fearon, D. T., and Ahearn, J. M.
(1991) J. Exp.
Med. 174, 1299-1311.
21. Tuveson, D. A., Ahearn, J. M., Matsumoto, A. K., and Fearon, D. T. (1991)
J. Exp. Med.
173, 1083-1089.
110
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
22. Bohnsack, J. F., and Cooper, N. R. (1988) J. Immunol. 141, 2569-2576.
23. Carter, R. H., and Fearon, D. T. (1989) J. Immunol. 143, 1755-17608.
24. Carter, R. H., and Fearon, D. T. (1992) Science 256, 105-107.
25. Carter, R. H., Spycher, M. 0., Ng, Y. C., Hoffman, R., and Fearon, D. T.
(1988) J.
Immunol. 141, 457-463.
26. Dempsey, P. W., Allison, M. E., Akkaraju, S., Goodnow, C. C., and Fearon,
D. T. (1996)
Science 271, 348-350.
27. Luxembourg, A. T., and Cooper, N. R. (1994) J. Immunol. 153, 4448-4457.
28. Lyubchenko, T., dal Porto, J., Cambier, J. C., and Holers, V. M. (2005) J.
Immunol. 174,
3264-3272.
29. Ahearn, J. M., Hayward, S. D., Hickey, J. C., and Fearon, D. T. (1988)
Proc. Nat'l Acad.
Sci. USA 85, 9307-9311.
30. Tanner, J., Weis, J., Fearon, D., Whang, Y., and Kieff, E. (1987) Cell 50,
203-213.
31. Lowell, C. A., Klickstein, L. B., Carter, R. H., Mitchell, J. A., Fearon,
D. T., and Ahearn,
J. M. (1989) J. Exp. Med. 170, 1931-1946.
32. Moore, M. D., DiScipio, R. G., Cooper, N. R., and Nemerow, G. R. (1989) J.
Biol. Chem.
264, 20576-20582.
33. Nemerow, G. R., Mold, C., Schwend, V. K., Tollefson, V., and Cooper, N. R.
(1987) J.
Virol 61, 1416-1420.
34. Mullen, M. M., Haan, K. M., Longnecker, R., and Jardetzky, T. S. (2002)
Mol. Cell 9,
375-385.
35. Spriggs, M. K., Armitage, R. J., Comeau, M. R., Strockbine, L., Farrah,
T., Macduff, B.,
Ulrich, D., Alderson, M. R., Mullberg, J., and Cohen, J. I. (1996) J. Virol.
70, 5557-5563.
36. Haan, K. M., Lee, S. K., and Longnecker, R. (2001) Virology 290, 106-114.
111
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
37. Haddad, R. S., and Hutt-Fletcher, L. M. (1989) J. Virol. 63, 4998-5005.
38. Molesworth, S. J., Lake, C. M., Borza, C. M., Turk, S. M., and Hutt-
Fletcher, L. M.
(2000) J. Virol. 74, 6324-6332.
39. Baechler, E. C., Batliwalla, F. M., Karypis, G., Gaffney, P. M., Ortmann,
W. A., Espe, K.
J., Shark, K. B., Grande, W. J., Hughes, K. M., Kapur, V., Gregersen, P. K.,
and Behrens, T.
W. (2003) Proc. Nat'l Acad. Sci. USA 100, 2610-2615.
40. Santiago-Raber, M. L., Baccala, R., Haraldsson, K. M., Choubey, D.,
Stewart, T. A.,
Kono, D. H., and Theofilopoulos, A. N. (2003) J. Exp. Med. 197, 777-788.
41. Takahashi, K., Kozono, Y., Waldschmidt, T. J., Berthiaume, D., Quigg, R.
J., Baron, A.,
and Holers, V. M. (1997) J. Immunol. 159, 1557-1569.
42. Young, K. A., Chen, X. S., Holers, V. M., and Hannan, J. P. (2007) J.
Biol. Chem. 282,
36614-36625.
43. Young, K. A., Herbert, A. P., Barlow, P. N., Holers, V. M., and Hannan, J.
P. (2008) J.
Virol.
44. Dominguez, C., Boelens, R., and Bonvin, A. M. (2003) J. Am. Chem. Soc.
125, 1731-
1737.
45. Szakonyi, G., Klein, M. G., Hannan, J. P., Young, K. A., Ma, R. Z.,
Asokan, R., Holers,
V. M., and Chen, X. S. (2006) Nature Struct. Mol. Biol. 13, 996-1001.
46. Guthridge, J. M., Rakstang, J. K., Young, K. A., Hinshelwood, J., Aslam,
M., Robertson,
A., Gipson, M. G., Sarrias, M. R., Moore, W. T., Meagher, M., Karp, D.,
Lambris, J. D.,
Perkins, S. J., and Holers, V. M. (2001) Biochemistry 40, 5931-59419.
47. Hannan, J. P., Young, K. A., Guthridge, J. M., Asokan, R., Szakonyi, G.,
Chen, X. S., and
Holers, V. M. (2005) J. Mol. Biol. 346, 845-858.
48. Kovacs, J. M., Hannan, J. P., Eisenmesser, E. Z., and Holers, V. M. (2009)
J. Biol. Chem.
284, 9513-9520.
112
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
49. Pervushin, K., Riek, R., Wider, G., and Wuthrich, K. (1997) Proc. Nat'l
Acad. Sci. USA
94, 12366-12371.
50. Wittkekind, M. a. L. M. (1993) J. Magn. Reson. 101, 201-205.
51. Grzesiek, S. a. A. B. (1992) J. Magn. Reson. 96, 432-440.
52. Zuiderweg, E. R., and Fesik, S. W. (1989) Biochemistry 28, 2387-2391.
53. Delaglio, F., Grzesiek, S., Vuister, G. W., Zhu, G., Pfeifer, J., and Bax,
A. (1995) J.
Biomol. NMR 6, 277-293.
54. Vranken, W. F., Boucher, W., Stevens, T. J., Fogh, R. H., Pajon, A.,
Llinas, M., Ulrich,
E. L., Markley, J. L., lonides, J., and Lane, E. D. (2005) Proteins 59, 687-
696.
55. Wiseman, T., Williston, S., Brandts, J. F., and Lin, L. N. (1989) Anal.
Biochem. 179, 131-
137.
56. Carel, J. C., Myones, B. L., Frazier, B., and Holers, V. M. (1990) J.
Biol. Chem. 265,
12293-12299.
57. Prota, A. E., Sage, D. R., Stehle, T., and Fingeroth, J. D. (2002) Proc.
Nat'l Acad. Sci.
USA 99, 10641-10646.
58. Molina, H., Brenner, C., Jacobi, S., Gorka, J., Carel, J. C., Kinoshita,
T., and Holers, V.
M. (1991) J. Biol. Chem. 266, 12173-12179.
59. Morikis, D., and Lambris, J. D. (2004) J. Immunol. 172, 7537-7547.
60. Zhang, L., Mallik, B., and Morikis, D. (2007) J. Mol. Biol. 369, 567-583.
EXAMPLE 2: BINDING ASSAYS.
Experimental Methods
Expression and purification of recombinant proteins.
[0186] Native human and mutagenized CR2 SCR 1-2 for binding studies is
expressed in
Pichia pastoris using a BioF1oTM 110 Fermenter (New Brunswick Scientific,
Edison, NJ) as
113
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
previously described (46) and as set forth in Example 1 above. Briefly, a
single colony is
grown up in 5 ml Pichia basal salt medium containing 1% glycerol (BMG, per
liter: 85%
phosphoric acid 26.7 ml, calcium sulfate 0.93 g, potassium sulfate 18.2 g,
magnesium sulfate-
7H20 14.9 g, potassium hydroxide 4.13 g, glycerol 10.0 g, distilled, deionized
water to 1
liter) overnight at 30 C and 250 rpm, expanded to 50 ml BMG (24 hrs) and
finally expanded
to 300 ml BMG (24 hrs). The inoculation culture is centrifuged at 2500xg 25 C
and
resuspended in 30 ml BMG. The 30 ml inoculation culture is used to inoculate 1
L of
minimal Pichia basal medium containing 40 g of glycerol. Dissolved 02
concentration is
maintained at 40%, the temperature at 30 C and the pH at 5.0 using 2 M KOH.
Initial feeds
are batch glycerol feeds; transition to methanol is eased by a methanol
injection before an
exponential methanol feed profile is initiated. Methanol induction lasts for
two days, after
which the culture is centrifuged to remove cellular debris. The supernatant is
exchanged into
mM Formate pH 4.0 before being passed over an SP-Sepharose column (2 x 5 mL SP
HiTrapTM columns, GE Biosciences, Pittsburgh, PA) followed by a CR2 affinity
column,
generated in-house by binding GST-C3d to a GSTrapTM column (GE Biosciences,
Pittsburgh,
PA). CR2 is eluted along an increasing linear NaCl gradient, 0-1.0 M in 1/3X
Phosphate
Buffered Saline (PBS, 1.6 mM MgC12, 0.9 mM KC1, 0.5 mM KH2PO4, 45.6 mM NaCl,
2.7
mM Na2HPO4 pH 7.4). Finally, native human and mutant CR2 SCR 1-2 is purified
by size
exclusion chromatography. Purity and identity of CR2 is monitored via SDS-
PAGE, Western
blot analysis and mass spectrometry.
[0187] Human C3d for binding studies is generated using the pGEXTM expression
system
(GE Healthcare, Piscataway, NJ) in E. coli as previously described (47) and as
set forth in
Example 1 above. Briefly, ampicillin-resistant colonies are used to start
overnight cultures
that are expanded to 1 L and grown at 37 C until an A600 of 0.3 is achieved.
Cultures are
induced with 0.3 mM IPTG at 30 C overnight before harvesting by
centrifugation. Harvested
pellets are resuspended in GST column buffer (50 mM Tris-HC1, pH 8.0, 250 mM
NaCl, 1
mM EDTA) and lysed by sonication. Lysate is clarified by centrifugation and
applied to a
GSTrapTM column (GE Biosciences, Pittsburgh, PA). C3d is cleaved from the
column by
digesting with 50 U of thrombin overnight at 4 C and subsequently purified by
size exclusion
chromatography. Purity of C3d is monitored via SDS-PAGE.
[0188] EBV gp350 is isolated from infected cells in culture by immunoaffinity
chromatography using commercially available anti-gp350 antibodies and standard
procedures. Human IFNa is purchased from commercial suppliers.
114
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
Binding assays.
[0189] Binding of native human CR2 SCR1-2 to EBV gp350, IFNa, and C3d is
compared to
that of human CR2 SCR1-2 variants containing one or more mutations in amino
acids
determined to be essential for binding interactions between CR2 and either EBV
gp350 or
IFNa by enzyme-linked immunosorbent assay (ELISA). Initial binding assays are
performed
with alanine-containing CR2 variants because it eliminates the side chain
beyond the f3-
carbon of the amino acid without altering the main chain conformation (as
glycine or proline
sometimes does) or imposing extreme electrostatic or steric effects. The
following CR2
SCR1-2 variants are tested for binding to EBV gp350 and to C3d in separate
experiments:
SCR1-2 NI IA, SCR1-2 NI IA +R36A, SCR1-2 NI IA + R36A + K41A, SCR1-2 NI IA +
R36A + K41A +Y64A, and SCR1-2 NI IA + R36A + K41A + Y64A + K67A. The
following CR2 SCR1-2 variants are tested for binding to IFNa and C3d in
separate
experiments: S42A and S42A + K50A. The effect of the amino acid substitutions
on CR2
binding kinetics and other properties (e.g., binding specificity) is assessed
by comparing the
binding of CR2 SCR1-2 variants to either EBV gp350 or IFNa and C3d to binding
of native
CR2 SCR1-2 to the same ligands. Alternative conservative or non-conservative
amino acid
substitutions are tested in subsequent experiments.
[0190] Microwell ELISA plates (Corning Life Sciences, New York, N.Y., USA) are
coated
overnight at 4 C with an appropriate amount of purified EBV gp350, IFNa, or
C3d obtained
as described above in 0.1 M NaHCO3, pH 8.6. The coated plates are then washed
with 1X
PBS three times for one minute each, and then incubated with 200 l of
blocking solution (5
mg/ml bovine serum albumin (BSA) in PBST) at 37 C for 1 hour. Each well is
washed with
PBST (80 mM Na2HPO4, 20 mM NaH2PO4, 100 mM NaCl, 0.05%-0.1% (v/v) Tween-20)
six
times for one minute each. Next, the plates are incubated with increasing
concentrations of
native human CR2 SCR1-2 and various CR2 SCR1-2 variants in PBS for 1 hour and
30
minutes at 37 C.
[0191] After the binding reaction is complete, the plates are again washed
with PBST six
times for one minute each. Next, the plates are incubated with anti-CR2
primary antibody,
washed six times for one minute each with blocking solution, and then
incubated with the
secondary antibody, horseradish peroxidase-conjugated mouse anti-human IgG
specific for
Fcy (Jackson ImmunoResearch Laboratories, Inc.) diluted 1:10,000 in PBST+5
mg/ml BSA,
at room temperature for 1 hour. The plates are again washed six times with
PBST+5 mg/ml
115
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
BSA at room temperature, and developed by the addition of 100 l of 3,3',5,5'-
tetramethylbenzidine (TMB) solution, prepared in 50 mM citrate phosphate.
After the
addition of the TMB solution, the plates are incubated for thirty minutes at
room temperature,
and then absorbance is measured at 450 nm with an MRX Microplate Reader (Dynex
Technologies) according to the manufacturer's instructions. Binding curves are
constructed
from the raw data, and binding affinities are estimated.
[0192] Alternatively, binding affinities are determined more precisely by
surface plasmon
resonance analysis using, for example, a BIACoreTM 4000 SPR system (Biacore
Life
Sciences, GE Healthcare, Piscataway, NJ). Like the ELISA, microwell plates are
coated with
either EBV gp350, IFNa, or C3d, and then incubated with increasing
concentrations of native
human CR2 SCR1-2 or the CR2 SCR1-2 variants. Unlike ELISAs, however, SPR
measures
binding affinities directly, and does not require use of a detectable label.
[0193] The SCR1-2 NI IA, SCR1-2 NI IA + R36A, SCR1-2 NI IA + R36A + K41A, SCR1-
2 NI IA + R36A + K41A + Y64A, and SCR1-2 NI IA + R36A + K41A + Y64A + K67A
variants are tested for binding affinity for EBV gp350 and are observed to
have progressively
less binding affinity for EBV gp350 while maintaining essentially the same
binding affinity
for C3d as native CR2 SCR 1-2. The SCR 1-2 S42A and SCR 1-2 S42A + K50A
variants are
tested for binding affinity for IFNa and are observed to have progressively
less binding
affinity for IFNa while maintaining essentially the same binding affinity for
C3d as native
CR2 SCR1-2.
SEQUENCES
SEQ ID NO:1 [complete amino acid sequence of human complement receptor 2
(CR2)]:
MGAAGLLGVFLALVAPGVLGISCGSPPPILNGRISYYSTPIAVGTVIRYSCSGTFRLIGE
KSLLCITKDKVDGTWDKPAPKCEYFNKYSSCPEPIVPGGYKIRGSTPYRHGDSVTFAC
KTNFSMNGNKS V WCQANNMWGPTRLPTCV S VFPLECPALPMIHNGHHTSENVGSIA
PGLSVTYSCESGYLLVGEKIINCLSSGKWSAVPPTCEEARCKSLGRFPNGKVKEPPILR
VGVTANFFCDEGYRLQGPPSSRC VIAGQGV AWTKMPVCEEIFCPSPPPILNGRHIGNS
LANV SYGSIVTYTCDPDPEEGVNFILIGESTLRCTVDS QKTGTW SGPAPRCELSTSAV
QCPHPQILRGRMV SGQKDRYTYNDTVIFACMFGFTLKGS KQIRCNAQGTWEPSAPV
CEKECQAPPNILNGQKEDRHM VRFDPGTS IKYSCNPGYVLVGEES IQCTSEGV WTPP
VPQCKVAACEATGRQLLTKPQHQFVRPD VNS SCGEGYKLSGS VYQECQGTIPWFME
IRLC KEITCPPPP V IY NGAHTG S S LED FPYGTT V TYTCNPGPERG V EFS LIGE S TIRC TS N
116
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
DQERGTWSGPAPLCKLSLLAVQCSHVHIANGYKISGKEAPYFYNDTVTFKCYSGFTL
KGS S QIRCKRDNTWDPEIPVCEKGCQPPPGLHHGRHTGGNTVFFV S GMTVDYTCDP
GYLLVGNKS IHCMPSGNWSPSAPRCEETCQHVRQSLQELPAGSRVELV NTS CQDGY
QLTGHAYQMCQDAENGIWFKKIPLCKV IHCHPPPVIVNGKHTGMMAENFLYGNEV S
YECDQGFYLLGEKNCSAEVILKAWILERAFPQCLRS LCPNPEV KHGYKLNKTHSAYS
HNDIVY VDCNPGFIMNGSRVIRCHTDNTW VPGVPTCIKKAFIGCPPPPKTPNGNHTGG
NIARFSPGMSILYSCDQGYLVVGEPLLLCTHEGTWSQPAPHCKEVNCSSPADMDGIQ
KGLEPRKMYQYGAV VTLECEDGYMLEGSPQS QCQSDHQWNPPLA VCRSRSLAPVL
CGIAAGLILLTFLIV ITLYV IS KHRERNYYTDTS QKEAFHLEAREVYS VDPYNPAS
SEQ ID NO:2 [amino acid sequence of short consensus repeat (SCR) domains 1 and
2 of
human CR2] :
ISCGSPPPILNGRISYYSTPIAVGTVIRYSCSGTFRLIGEKS LLCITKD KVDGTWDKPAP
KCEYFNKYS SCPEPIV PGGYKIRGSTPYRHGDS VTFACKTNFSMNGNKS V WCQANN
MWGPTRLPTCVS
SEQ ID NO:3 [amino acid sequence of human CD59 protein]:
MGIQGGSVLFGLLLVLAVFCHSGHSLQCYNCPNPTADCKTAVNCSSDFDACLITKAG
LQVYNKCWKFEHCNFND VTTRLRENELTYYCCKKDLCNFNEQLENGGTSLSEKTV L
LLVTPFLAAAWSLHP
SEQ ID NO:4 [amino acid sequence of mouse complement receptor 1-related
gene/protein y
(Crry)] :
MEV S S RS S EPLDP V W LLV AFGRGG V KLEV LLLFLLPFTLGELRGGLGKHGHTV HREP
AVNRLCADS KRWSGLPV SAQRPFPMGHCPAPS QLPSAKPINLTDESMFPIGTYLLYEC
LPGYIKRQFSITCKQDSTWTSAEDKCIRKQCKTPSDPENGLVHVHTGIQFGSRINYTC
NQGYRLIGSSSAVCVITDQSVDWDTEAPICEWIPCEIPPGIPNGDFFSSTREDFHYGMV
VTYRCNTDARGKALFNLVGEPS LYCTSNDGEIGV WSGPPPQCIELNKCTPPPYVENA
VMLSENRSLFSLRDIVEFRCHPGFIMKGASSVHCQSLNKWEPELPSCFKGVICRLPQE
MSGFQKGLGMKKEYYYGENVTLECEDGYTLEGSSQSQCQSDGSWNPLLAKCVSRSI
SGLIV GIFIGIIVFILV IIVFIWMILKYKKRNTTDEKYKEVGIHLNYKEDSCVRLQSLLTS
QENSSTTSPARNSLTQEVS
SEQ ID NO:5 [amino acid sequence of human factor H]:
MRLLAKIICLMLWAIC VAEDCNELPPRRNTEILTGS W SDQTYPEGTQAIYKCRPGYRS
117
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
LGN V IM V CRKGEW V ALNPLRKCQKRPCGHPGDTPFGTFTLTGGN V FEYG V KA V YT
CNEGYQLLGEINYRECDTDGWTNDIPICEVVKCLPVTAPENGKIVSSAMEPDREYHF
GQAVRFVCNSGYKIEGDEEMHCSDDGFWSKEKPKCVEISCKSPDVINGSPISQKIIYK
ENERFQYKCNMGYEYSERGDAVCTESGWRPLPSCEEKSCDNPYIPNGDYSPLRIKHR
TGDEITYQCRNGFYPATRGNTAKCTSTGWIPAPRCTLKPCDYPDIKHGGLYHENMRR
PYFPVAVGKYYSYYCDEHFETPSGSYWDHIHCTQDGWSPAVPCLRKCYFPYLENGY
NQNYGRKFVQGKSIDVACHPGYALPKAQTTVTCMENGWSPTPRCIRVKTCSKSSIDI
ENGFISES QYTYALKEKAKYQCKLGYVTADGETSGSITCGKDGW SAQPTCIKSCDIPV
FMNARTKNDFTWFKLNDTLDYECHDGYES NTGSTTGS IVCGYNGWSDLPICYEREC
ELPKIDVHLVPDRKKDQYKVGEVLKFSCKPGFTIVGPNSVQCYHFGLSPDLPICKEQV
QSCGPPPELLNGNVKEKTKEEYGHSEVVEYYCNPRFLMKGPNKIQCVDGEWTTLPV
CIVEESTCGDIPELEHGWAQLSSPPYYYGDS VEFNCSESFTMIGHRSITCIHGVWTQLP
QCVAIDKLKKCKSSNLIILEEHLKNKKEFDHNSNIRYRCRGKEGWIHTVCINGRWDPE
VNCSMAQIQLCPPPPQIPNSHNMTTTLNYRDGEKVSVLCQENYLIQEGEEITCKDGR
W QS IPLC V EKIPCS QPPQIEHGTINS S RS S QESYAHGTKLS YTCEGGFRIS EENETTCYM
GKWSSPPQCEGLPCKSPPEISHGVVAHMSDSYQYGEEVTYKCFEGFGIDGPAIAKCL
GEKW SHPPSCIKTDCLS LPS FENAIPMGEKKD VYKAGEQVTYTCATYYKMDGASNV
TCINSRWTGRPTCRDTSC VNPPTV QNAYIV SRQMS KYPSGERVRYQCRSPYEMFGDE
EVMCLNGNWTEPPQCKDSTGKCGPPPPIDNGDITSFPLS VYAPAS S VEYQCQNLYQL
EGNKRITCRNGQWSEPPKCLHPCV ISREIMENYNIALRWTAKQKLYSRTGES VEFVC
KRGYRLSSRSHTLRTTCWDGKLEYPTCAKR
SEQ ID NO:6 [amino acid sequence of mouse CD59A protein]:
MRAQRGLILLLLLLAVFCSTAVSLTCYHCFQPVVSSCNMNSTCSPDQDSCLYAVAG
MQVYQRCWKQSDCHGEIIMDQLEETKLKFRCCQFNLCNKSDGSLGKTPLLGTSVLV
AILNLCFLSHL
SEQ ID NO:7 [amino acid sequence of mouse CD59B protein]:
M RA QRG LILLLLLLA V FC S TA V S LKCYNC F QF V S S C KINTTC S PNLD S C LYA V
AGR Q
VYQQCW KLSDCNSNYIMSRLD V AGIQS KCCQWGLCNKNLDGLEEPNNAETS SLRKT
ALLGTSVLVAILKFCF
SEQ ID NO:8 [amino acid sequence of mouse factor H]:
MRLSARIIWLILWTVCAAEDCKGPPPRENSEILSGSWSEQLYPEGTQATYKCRPGYRT
118
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
LGTIV KVCKNGKW VASNPSRICRKKPCGHPGDTPFGSFRLAVGS QFEFGAKV VYTCD
DGYQLLGEIDYRECGADGWINDIPLCEVVKCLPVTELENGRIVSGAAETDQEYYFGQ
V VRFECNSGFKIEGHKEIHCSENGLW SNEKPRC VEILCTPPRVENGDGINV KPVYKEN
ERYHYKCKHGYVPKERGDAVCTGSGWS S QPFCEEKRCSPPYILNGIYTPHRIIHRSDD
EIRYECNYGFYPVTGSTV S KCTPTGWIPVPRCTLKPCEFPQFKYGRLYYEESLRPNFP
VSIGNKYSYKCDNGFSPPSGYSWDYLRCTAQGWEPEVPCVRKCVFHYVENGDSAY
WEKVYV QGQSLKV QCYNGYS LQNGQDTMTCTENGW SPPPKCIRIKTCSASDIHIDN
GFLS ES S S IYA LNRET S YRC KQGY V TNTGEIS G S ITC LQNG W S P QP S C IKS CD MP
V FEN
SITKNTRTWFKLNDKLDYECLVGFENEYKHTKGSITCTYYGWSDTPSCYERECSVPT
LDRKLV V SPRKEKYRVGDLLEFSCHSGHRVGPDS V QCYHFGW SPGFPTCKGQVASC
APPLEILNGEINGAKKVEYSHGEVVKYDCKPRFLLKGPNKIQCVDGNWTTLPVCIEEE
RTCGDIPELEHGSAKCS VPPYHHGDS VEFICEENFTMIGHGS V SCISGKWTQLPKCVA
TDQLEKCRV LKSTGIEAIKPKLTEFTHNSTMDYKCRDKQEYERS ICINGKWDPEPNCT
SKTSCPPPPQIPNTQVIETTVKYLDGEKLSVLCQDNYLTQDSEEMVCKDGRWQSLPR
CIEKIPCS QPPTIEHGS INLPRS SEERRDSIES S SHEHGTTFSYVCDDGFRIPEENRITCYM
GKWSTPPRCVGLPCGPPPS IPLGTV S LELESYQHGEEVTYHCSTGFGIDGPAFIICEGG
KWSDPPKCIKTDCD VLPTV KNAIIRGKS KKSYRTGEQVTFRCQSPYQMNGSDTVTCV
NSRWIGQPVCKDNSCVDPPHVPNATIVTRTKNKYLHGDRVRYECNKPLELFGQVEV
MCENGIWTEKPKCRDSTGKCGPPPPIDNGDITSLSLPVYEPLS S VEYQCQKYYLLKGK
KTITCTNGKW SEPPTCLHAC VIPENIMESHNIILKWRHTEKIYSHSGEDIEFGCKYGYY
KARDSPPFRTKCINGTINYPTCV
SEQ ID NO:9 [amino acid sequence of human complement receptor 1 (CR1)]:
MGASSPRSPEPVGPPAPGLPFCCGGSLLAV V VLLALPVAWGQCNAPEWLPFARPTNL
TDEFEFPIGTYLNYECRPGYSGRPFSIICLKNSVWTGAKDRCRRKSCRNPPDPVNGMV
HVIKGIQFGS QIKYSCTKGYRLIGS S SATCIISGDTVIWDNETPICDRIPCGLPPTITNGD
FISTNRENFHYGS V VTYRCNPGSGGRKVFELVGEPSIYCTSNDDQVGIWSGPAPQCIIP
NKCTPPNVENGILVSDNRSLFSLNEVVEFRCQPGFVMKGPRRVKCQALNKWEPELPS
CSRVCQPPPD VLHAERTQRDKDNFSPGQEV FYSCEPGYDLRGAAS MRCTPQGDWSP
AAPTCEVKSCDDFMGQLLNGRVLFPVNLQLGAKVDFVCDEGFQLKGSSASYCVLAG
MESLWNS S VPVCEQIFCPSPPVIPNGRHTGKPLEVFPFGKAVNYTCDPHPDRGTSFDLI
GESTIRCTSDPQGNGV WS SPAPRCGILGHCQAPDHFLFAKLKTQTNASDFPIGTSLKY
ECRPEYYGRPFSITCLDNLV W S SPKD VCKRKSCKTPPDPVNGM VHVITDIQ VGSRINY
SCTTGHRLIGHSSAECILSGNAAHWSTKPPICQRIPCGLPPTIANGDFISTNRENFHYGS
119
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
V VTYRCNPGSGGRKVFELVGEPSIYCTSNDD QVGIWSGPAPQCIIPNKCTPPNVENGI
LVSDNRSLFSLNEVVEFRCQPGFVMKGPRRVKCQALNKWEPELPSCSRVCQPPPDVL
HAERTQRDKDNFSPGQEVFYSCEPGYDLRGAASMRCTPQGDW SPAAPTCEV KSCDD
FMGQLLNGRVLFPVNLQLGAKVDFVCDEGFQLKGSSASYCVLAGMESLWNSSVPV
CEQIFCPSPPV IPNGRHTGKPLEVFPFGKAV NYTCDPHPDRGTSFDLIGESTIRCTSDPQ
GNGV WS SPAPRCGILGHCQAPDHFLFAKLKTQTNASDFPIGTSLKYECRPEYYGRPFS
ITCLDNLVWSSPKDVCKRKSCKTPPDPVNGMVHVITDIQVGSRINYSCTTGHRLIGHS
SAECILSGNTAHW STKPPICQRIPCGLPPTIANGDFISTNRENFHYGS V VTYRCNLGSR
GRKVFELVGEPSIYCTSNDDQVGIWSGPAPQCIIPNKCTPPNVENGILVSDNRSLFSLN
EV VEFRCQPGFVMKGPRRV KCQALNKWEPELPSCSRVCQPPPEILHGEHTPSHQDNF
SPGQEVFYSCEPGYDLRGAASLHCTPQGD WSPEAPRCAV KSCDDFLGQLPHGRV LFP
LNLQLGAKVSFVCDEGFRLKGSS VSHCVLVGMRSLWNNS VPVCEHIFCPNPPAILNG
RHTGTPSGDIPYGKEISYTCDPHPDRGMTFNLIGESTIRCTSDPHGNGV W S SPAPRCEL
S VRAGHCKTPEQFPFASPTIPINDFEFPVGTSLNYECRPGYFGKMFSISCLENLVWSS V
EDNCRRKSCGPPPEPFNGMVHINTDTQFGSTVNYSCNEGFRLIGSPSTTCLVSGNNVT
WDKKAPICEIISCEPPPTISNGDFYSNNRTSFHNGTVVTYQCHTGPDGEQLFELVGERS
IYCTS KDD Q V G V W S S PPPRCIS TNKCTAPEV ENAIRV PGNRS FFS LTEIIRFRC QPGFV
M VGSHTV QCQTNGRWGPKLPHCSRVCQPPPEILHGEHTLSHQDNFSPGQEV FYSCEP
SYDLRGAASLHCTPQGDW SPEAPRCTV KSCDDFLGQLPHGRVLLPLNLQLGAKV SF
VCDEGFRLKGRSASHCVLAGMKALWNSSVPVCEQIFCPNPPAILNGRHTGTPFGDIP
YGKEISYACDTHPDRGMTFNLIGESSIRCTSDPQGNGVWSSPAPRCELSVPAACPHPP
KIQNGHYIGGHV SLYLPGMTISYTCDPGYLLVGKGFIFCTDQGIWS QLDHYCKEVNC
SFPLFMNGIS KELEMKKVYHYGDYVTLKCEDGYTLEGSPW S QCQADDRWDPPLAK
CTSRAHDALIVGTLSGTIFFILLIIFLSWIILKHRKGNNAHENPKEVAIHLHSQGGSSVH
PRTLQTNEENSRVLP
SEQ ID NO: 10 [amino acid sequence of human membrane cofactor protein (MCP)]:
MEPPGRRECPFPS WRFPGLLLAAMV LLLYSFSDACEEPPTFEAMELIGKPKPYYEIGE
RVDYKCKKGYFYIPPLATHTICDRNHTWLPVSDDACYRETCPYIRDPLNGQAVPANG
TYEFGYQMHFICNEGYYLIGEEILYCELKGSVAIWSGKPPICEKVLCTPPPKIKNGKHT
FSEVEVFEYLDAVTYSCDPAPGPDPFSLIGESTIYCGDNS VWSRAAPECKV VKCRFPV
VENGKQISGFGKKFYYKATVMFECDKGFYLDGSDTIVCDSNSTWDPPVPKCLKVLPP
SSTKPPALSHS VSTSSTTKSPASSASGPRPTYKPPVSNYPGYPKPEEGILDSLDVW VIA
VIVIAIV VGVAVICV VPYRYLQRRKKKGTYLTDETHREVKFTSL
120
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
SEQ ID NO: 11 [amino acid sequence of human decay accelerating factor
(DAF/CD55)]:
MTVARPSVPAALPLLGELPRLLLLVLLCLPAVWGDCGLPPDVPNAQPALEGRTSFPE
DTVITYKCEESFVKIPGEKDSVICLKGSQWSDIEEFCNRSCEVPTRLNSASLKQPYITQ
NYFPVGTV VEYECRPGYRREPSLSPKLTCLQNLKW STAVEFCKKKSCPNPGEIRNGQI
DVPGGILFGATISFSCNTGYKLFGSTSSFCLISGSSVQWSDPLPECREIYCPAPPQIDNGI
IQGERDHYGYRQS VTYACNKGFTMIGEHSIYCTVNNDEGEWSGPPPECRGKSLTS KV
PPTV QKPTTVNVPTTEV SPTS QKTTTKTTTPNAQATRSTPV SRTTKHFHETTPNKGSG
TTSGTTRLLSGHTCFTLTGLLGTLVTMGLLT
SEQ ID NO:12 [amino acid sequence of mouse decay accelerating factor
(DAF/CD55)]:
MIRGRAPRTRPS PPPPLLPLLS LS LLLLS PT V RGDCGPPPDIPNARPILGRHS KFAEQS K
VAYSCNNGFKQVPDKSNIVVCLENGQWSSHETFCEKSCVAPERLSFASLKKEYLNM
NFFPVGTIVEYECRPGFRKQPPLPGKATCLEDLVWSPVAQFCKKKSCPNPKDLDNGH
INIPTGILFGSEIFNSCNPGYRLVGVSSTFCSVTGNTVDWDDEFPVCTEIHCPEPPKINN
GIMRGESDSYTYSQVVTYSCDKGFILVGNASIYCTVSKSDVGQWSSPPPRCIEKSKVP
TKKPTINVPSTGTPSTPQKPTTES VPNPGDQPTPQKPSTV KV SATQHVPVTKTTVRHPI
RTSTDKGEPNTGGDRYIYGHTCLITLTVLHV MLS LIGYLT
SEQ ID NO: 13 [amino acid sequence of CVF from Naja kaouthia]:
MERMALYLV AALLIGFPGS SHGALYTLITPA VLRTDTEEQILVEAHGDSTPKQLDIFV
HDFPRKQKTLFQTRVDMNPAGGMLVTPTIEIPAKEVSTDSRQNQYV V VQVTGPQVR
LEKV VLLSYQS SFLFIQTDKGIYTPGSPVLYRVFSMDHNTS KMNKTVIVEFQTPEGILV
SSNSVDLNFFWPYNLPDLVSLGTWRIVAKYEHSPENYTAYFDVRKYVLPSFEVRLQP
SEKFFYIDGNENFHV SITARYLYGEEV EGVAFV LFGV KIDDAKKS IPDSLTRIPIIDGDG
KATLKRDTFRSRFPNLNELV GHTLYAS VTVMTESGSDMV VTEQSGIHIVAS PYQIHFT
KTPKYFKPGMPYELTVYVTNPDGSPAAHVPV V SEAFHSMGTTLSDGTAKLILNIPLN
AQSLPITVRTNHGDLPRERQATKSMTAIAYQTQGGSGNYLHVAITSTEIKPGDNLPVN
FNVKGNANSLKQIKYFTYLILNKGKIFKVGRQPRRDGQNLVTMNLHITPDLIPSFRFV
AYYQVGNNEIV ADS V W VD V KDTCMGTLV V KGDNLIQMPGAAMKIKLEGDPGARV
GLVAVDKAVYVLNDKYKIS QAKIWDTIEKSDFGCTAGSGQNNLGV FEDAGLALTTS
TNLNTKQRSAAKCPQPANRRRRSSVLLLDSNASKAAEFQDQDLRKCCEDVMHENP
MGYTCEKRAKYIQEGDACKAAFLECCRYIKGVRDENQRESELFLARDDNEDGFIADS
DIISRSDFPKSWLWLTKDLTEEPNSQGISSKTMSFYLRDSITTWVVLAVSFTPTKGICV
AEPYEIRVMKVFFIDLQMPYS V VKNEQVEIRAILHNYVNEDIYVRVELLYNPAFCSAS
121
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
TKGQRYRQQFPIKALSSRAVPFVIVPLEQGLHDVEIKAS VQEALWSDGVRKKLKV VP
EGV QKS IVTIV KLDPRAKGVGGTQLEVIKARKLDDRVPDTEIETKIIIQGDPVAQIIENS
IDGS KLNHLIITPSGCGEQNMIRMAAPV IATYYLDTTEQWETLGINRRTEA VNQIVTG
YAQQMVYKKADHSYAAFTNRASSSWLTAYVVKVFAMAAKMVAGISHEIICGGVR
WLILNRQQPDGAFKENAPVLSGTMQGGIQGAEEEVYLTAFILV ALLES KTICNDYVN
SLDSSIKKATNYLLKKYEKLQRPYTTALTAYALAAADQLNDDRVLMAASTGRDHW
EEYNAHTHNIEGTSYALLALLKMKKFDQTGPIVRWLTDQNFYGETYGQTQATVMAF
QALAEYEIQMPTHKDLNLDITIELPDREVPIRYRINYENALLARTVETKLNQDITVTAS
GDGKATMTILTFYNAQLQEKANVCNKFHLNV S VENIHLNAMGAKGALMLKICTRY
LGEVDSTMTIIDISMLTGFLPDAEDLTRLSKGVDRYISRYEVDNNMAQKVAVIIYLNK
VSHSEDECLHFKILKHFEVGFIQPGSVKVYSYYNLDEKCTKFYHPDKGTGLLNKICIG
NVCRCAGETCS SLNHQERID VPLQIEKACETNVDYVYKTKLLRIEEQDGNDIY VMD V
LEV IKQGTDENPRAKTHQYIS QRKCQEALNLKVNDDYLIWGSRSDLLPTKD KISYIIT
KNTWIERWPHEDECQEEEFQKLCDDFAQFSYTLTEFGCPT
SEQ ID NO: 14 [amino acid sequence of the human IgGi heavy chain, C domain]:
ASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVL
QS SGLYSLS S V VTVPS S SLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTCPPCPAP
ELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKT
KPREEQYNSTYRV VS VLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREP
QVYTLPPSRDELTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGS
FFLYSKLTVDKSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGK
SEQ ID NO: 15 [amino acid sequence of the human IgG1 light chain, C domain]:
TVAAPS VFIFPPSDEQLKSGTAS V VCLLNNFYPREAKVQWKVDNALQSGNS QES VTE
QDS KDSTYS LS STLTLS KADYEKHKV YACEVTHQGLS SPVTKSFNRGEC
SEQ ID NO:16 [amino acid sequence of the Fc domain of human IgGi]:
EPKSCDKTHTCPPCPAPELLGGPS VFLFPPKPKDTLMISRTPEVTCV V VDVSHEDPEV
KFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKA
LPAPIEKTISKAKGQPREPQVYTLPPSRDELTKNQVSLTCLVKGFYPSDIAVEWESNG
QPENNYKTTPPVLDSDGPFFLYSKLTVDKSRWQQGNVFSCSVMHEALHNHYTQKSL
SLSPGK
122
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
SEQ ID NO: 17 [amino acid sequence of human IgM heavy chain, C domain]:
GSASAPTLFPLVSCENSPSDTSSVAVGCLAQDFLPDSITLSWKYKNNSDISSTRGFPSV
LRGGKYAATSQVLLPSKDVMQGTDEHV VCKVQHPNGNKEKNVPLPVIAELPPKVS V
FVPPRDGFFGNPRKS KLICQATGFSPRQIQ V S WLREGKQVGSGVTTDQV QAEAKESG
PTTYKVTSTLTIKESDWLGQSMFTCRVDHRGLTFQQNASSMCVPDQDTAIRVFAIPPS
FASIFLTKSTKLTCLVTDLTTYDSVTISWTRQNGEAVKTHTNISESHPNATFSAVGEAS
ICEDDWNSGERFTCTVTHTDLPSPLKQTISRPKGVALHRPDVYLLPPAREQLNLRESA
TITCLVTGFSPADVFVQWMQRGQPLSPEKYVTSAPMPEPQAPGRYFAHSILTVSEEE
WNTGETYTCVAHEALPNRVTERTVDKSTGKPTLYNVSLVMSDTAGTCY
SEQ ID NO: 18 [amino acid sequence of human IgM light chain, C domain]:
GSASAPTLFPLVSCENSPSDTSSVAVGCLAQDFLPDSITLSWKYKNNSDISSTRGFPSV
LRGGKYAATSQVLLPSKDVMQGTDEHVVCKVQHPNGNKEKNVPLPV
SEQ ID NO:19 [amino acid sequence of the Fc domain of human IgM]:
GSASAPTLFPLVSCENSPSDTSSVAVGCLAQDFLPDSITFSWKYKNNSDISSTRGFPSV
LRGGKYAATSQVLLPSKDVMQGTDEHV VCKVQHPNGNKEKNVPLPVIAELPPKVS V
FVPPRDGFFGNPRS KS KLICQATGFSPRQIQ V S WLREGKQVGSGVTTDQV QAEAKES
GPTTYKVTSTLTIKESDWLSQSMFTCRVDHRGLTFQQNASSMCVPDQDTAIRVFAIPP
SFASIFLTKSTKLTCLVTDLTTYDSVTISWTRQNGEAVKTHTNISESHPNATFSAVGEA
SICEDDWNSGERFTCTVTHTDLPSPLKQTIS RPKGVALHRPD VYLLPPAREQLNLRES
ATITCLVTGFSPAD VFV QWMQRGQPLSPEKYVTSAPMPEPQAPGRYFAHSILTV SEEE
WNTGETYTCVVAHEALPNRVTERTVDKSTGKPTLYNVSLVMSDTAGTCY
SEQ ID NO:20 [amino acid sequence of mouse IgG3 heavy chain]:
TTTAPSVYPLVPGCSDTSGSSVTLGCLVKGYFPEPVTVKWNYGALSSGVRTVSSVLQ
SGFYSLSSLVTVPSSTWPSQTVICNVAHPAS KTELIKRIEPRIPKPSTPPGSSCPPGNILG
GPSVFIFPPKPKDALMISLTPKVTCVVVDVSEDDPDVHVSWFVDNKEVHTAWTQPRE
AQYNSTFRVVSALPIQHQDWMRGKEFKCKVNNKALPAPIERTISKPKGRAQTPQVYT
IPPPREQMSKKKVSLTCLVTNFFSEAISVEWERNGELEQDYKNTPPILDSDGTYFLYS
KLTVDTDS WLQGEIFTCS V VHEALHNHHTQKNLSRSP
SEQ ID NO:21 [amino acid sequence of mouse IgG3 light chain]:
IVLTQSPAIMSASPGEKVTMTCRASSSVRSSYLHWYQQKPGSSPKLWIYSTSNLASGV
PVRFSGSGSGTSYSLTISSVEAEDAATYYCQQYDSSPSITFGAGTKLELK
123
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
SEQ ID NO:22 [amino acid sequence of mouse IgG3 Fc domain]:
EPRIPKPSTPPGS SCPPGNILGGPS VFIFPPKPKDALMIS LTPKVTC V V VD VSEDDPD VH
VSWFVDNKEVHTAWTQPREAQYNSTFRVVSALPIQHQDWMRGKEFKCKVNNKALP
APIERTISKPKGRAQTPQVYTIPPPREQMSKKKVSLTCLVTNFFSEAISVEWERNGELE
QDYKNTPPILDSDGTYFLYSKLTVDTDSWLQGEIFTCS V VHEALHNHHTQKNLSRSP
GK
SEQ ID NO:23 [amino acid sequence of mouse IgM heavy chain, C domain]:
S QSFPNVFPLV SCESPLSDKNLV AMGCLARDFLPSTISFTWNYQNNTEV IQGIRTFPTL
RTGGKYLATS QVLLSPKS ILEGSDEYLVCKIHYGGKNRDLH VPIPA VAEMNPNVNVF
VPPRDGFSGPAPRKS KLICEATNFTPKPITV S WLKDGKLVESGFTTDPVTIENKGSTPQ
TYKV ISTLTISEIDWLNLNVYTCRVDHRGLTFLKNV S STCAASPSTDILTFTIPPSFADIF
LSKSANLTCLVSNLATYETLNISWASQSGEPLETKIKIMESHPNGTFSAKGVASVCVE
DWNNRKEFVCTVTHRDLPSPQKKFISKPNEVHKHPPAVYLLPPAREQLNLRESATVT
CLVKGFSPADISVQWLQRGQLLPQEKYVTSAPMPEPGAPGFYFTHSILTVTEEEWNS
GETYTCVVGHEALPHLVTERTVDKSTGKPTLYNVSLIMSDTGGTCY
SEQ ID NO:24 [amino acid sequence of mouse IgM light chain, C domain]:
S QSFPNVFPLV SCESPLSDKNLV AMGCLARDFLPSTISFTWNYQNNTEV IQGIRTFPTL
RTGGKYLATS QVLLSPKS ILEGSDEYLVCKIHYGGKNRDLH VPIP
SEQ ID NO:25 [amino acid sequence of mouse IgM Fc domain]:
ASPSTDILTFTIPPSFADIFLS KSANLTCLV SNLATYETLNISWAS QSGEPLETKIKIMES
HPNGTFSAKGVASVCVEDWNNRKEFVCTVTHRDLPSPQKKFISKPNEVHKHPPAVY
LLPPAREQLNLRESATVTCLV KGFSPADIS V QWLQRGQLLPQEKYV TSAPMPEPGAP
GFYFTHSILTVTEEEWNSGETYTCVVGHEALPHLVTERTVDKSTGKPTLYNVSLIMS
DTGGTCY
SEQ ID NO:26 [linking sequence between the first two N-terminal short
consensus repeat
domains of human CR2]:
VSVFPLE
SEQ ID NO:27 [linking sequence between the first two N-terminal short
consensus repeat
domains of human CR2]:
EYFNKYSS
124
CA 02799192 2012-11-09
WO 2011/143637 PCT/US2011/036552
SEQ ID NO:28 [linking sequence between the fourth and the fifth N-terminal
short consensus
repeat domains of human CR2]:
EEIF
125