Note: Descriptions are shown in the official language in which they were submitted.
CA 02828878 2013-11-08
PREDICTING GASTROENTEROPANCREATIC NEUROENDOCRINE
NEOPLASMS (GEP-NENs)
Cross-Reference to Related Applications
[0001] This application claims benefit of United States application No.
61/448,137,
filed on March 1, 2011.
Reference to Sequence Listing
[0002] This description contains a sequence listing in electronic form in
ASCII text
format. A copy of the sequence listing in electronic fount is available from
the Canadian
Intellectual Property Office.
Technical Field
[0003] The invention described herein relates to gastroenteropancreatic
neuroendocrine neoplasm (GEP-NEN) biomarkers and agents, systems, and kits for
detecting
the same, and associated GEP-NEN diagnostic and prognostic methods, such as
detection,
prediction, staging, profiling, classification, and monitoring treatment
efficacy and other
outcomes.
Background Art
[0004] Gastroenteropancreatic neuroendocrine neoplasm (GEP-NEN, also called
Gastroenteropancreatic (GEP) neuroendocrine tumor and neuoroendocrine tumor
(NET)) is
the second most prevalent malignant tumor of the gastrointestinal (GI) tract
in the U.S., more
prevalent than gastric, esophageal, pancreatic, and hepatobiliary neoplasms,
with an incidence
of about 2.5-5 cases per 100,000. Incidence and prevalence have increased
between 100 and
600 percent in the U.S. over the last thirty years, with no increase in
survival.
1
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
[0005] Heterogeneity and complexity of GEP-NENs has made diagnosis, treatment,
and
classification difficult. These neoplasms lack several mutations commonly
associated with other
cancers; microsatellite instability is largely absent. See Tannapfel A,
Vomschloss S, Karhoff D,
et al., "BRAF gene mutations are rare events in gastroenteropancreatic
neuroendocrine tumors,"
Am J Clin Pathol 2005;123(2):256-60; Kidd M, Eick G, Shapiro MD, et al.
Microsatellite
instability and gene mutations in transforming growth factor-beta type II
receptor are absent in
small bowel carcinoid tumors," Cancer 2005;103(2):229-36. Individual
histopathologic
subtypes associate with distinct clinical behavior, yet there is no
definitive, generally accepted
pathologic classification or prediction scheme, hindering treatment
development.
[0006] Existing diagnostic and prognostic approaches include imaging (e.g., CT
and MRI),
histology, and detection of some gene products. Available methods are limited,
for example, by
low sensitivity and/or specificity, and inability to detect early-stage
disease. GEP-NENs often
go undiagnosed until they are metastatic and often untreatable.
[0007] There is a need for specific and sensitive methods and agents for
detection of GEP-
NENs, including early-stage GEP-NENs, for example, for use in diagnosis,
prognosis,
prediction, staging, classification, treatment, monitoring, and risk
assessment, and for
investigating and understanding molecular factors of pathogenesis, malignancy,
and
aggressiveness of this disease. For example, such methods and agents are
needed that can be
performed simply, rapidly, and at relatively low cost. Provided herein are
methods,
compositions, and combinations that meet these needs.
Summary
[0008] In one aspect, the present invention relates to gastroenteropancreatic
neuroendocrine
neoplasm (GEP-NEN) biomarkers, the detection of which may be used in
diagnostic, prognostic
and predictive methods. Among the provided objects are GEP-NEN biomarkers,
panels of the
biomarkers, agents for binding and detecting the biomarkers, kits and systems
containing such
agents, and methods and compositions for detecting the biomarkers, for
example, in biological
samples, as well as prognostic, predictive, diagnostic, and therapeutic uses
thereof.
[0009] Provided are agents, sets of agents, and systems containing the agents
for GEP-NEN
prognosis, detection and diagnosis. Typically, the systems include a plurality
of agents (e.g., set
of agents), where the plurality specifically binds to and/or detects a
plurality of GEP-NEN
biomarkers in a panel of GEP-NEN biomarkers. Typically, the agents are
isolated polypeptides
2
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
or polynucleotides which specifically bind to one or more GEP-NEN biomarkers.
For example,
provided are sets of isolated polynucleotides and polypeptides that bind to a
panel of GEP-NEN
biomarkers, and methods and uses of the same.
[0010] Also provided are prognostic, diagnostic and predictive methods and
uses of the
agents, compositions, systems, and kits for GEP-NEN and associated conditions,
syndromes and
symptoms. For example, provided are methods and uses for detection, diagnosis,
classification,
prediction, therapeutic monitoring, prognosis, or other evaluation of GEP-NEN
or an outcome,
stage or level of aggressiveness or risk thereof, or associated condition. In
some embodiments,
the methods are performed by determining the presence, absence, expression
levels, or
expression profile of a GEP-NEN biomarker, more typically a plurality of GEP-
NEN
biomarkers, such as a panel of biomarkers, and/or comparing such information
with normal or
reference expression levels or profiles or standards. Thus, in some
embodiments, the methods
are carried out by obtaining a biological test sample and detecting the
presence, absence,
expression levels, or expression profile of a GEP-NEN biomarker as described
herein, more
typically of a panel of at least two of the provided GEP-NEN biomarkers. For
example, the
methods can be performed with any of the systems of agents, e.g.,
polynucleotides or
polypeptides, provided herein. For example, the methods generally are carried
out using one or
more of the provided systems.
[0011] Provided are methods, agents and compositions for detection of and
distinguishing
between a number of different GEP-NEN types, stages, and sites (for example,
pancreatic vs.
small intestine GEP-NEN). In one aspect, differentiating between sites can
provide prognostic
information or help identify the GEP-NEN. Thus, in some embodiments, the
methods
distinguish between small intestine NENs (SI-NENs) and pancreatic NENs (PI-
NENs).
Exemplary GEP-NEN types and stages include metastatic and primary GEP-NEN, GEP-
NENs
that are or are not responsive to various treatment approaches, and various
GEP-NENs sub-
types, including well-differentiated NET (WDNET), primary well differentiated
nuroendocrine
carcinoma (WDNEC), primary poorly differentiated neuroendocrine tumor (PDNET),
primary
poorly differentiated NEC (PDNEC), metastatic WDNET (WDNET MET), metastatic
WDNEC
(WDNEC MET) metastatic PDNEC (PDNEC MET) and metastatic PDNET (PDNET MET).
[0012] In one aspect, the provided methods and compositions may be used to
specifically
and sensitively detect GEP-NENs, such as early-stage, primary, or asymptomatic
GEP-NENs; in
some aspects, the methods and compositions may be used to predict disease
progression,
3
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
treatment response, and metastasis. Methods and compositions provided herein
are useful for
diagnosis, prognosis, prediction (i.e., prediction of metastases in early-
stage and primary GEP-
NENs), staging, classification, treatment, monitoring, assessing risk, and
investigating molecular
factors associated with GEP-NEN disease.
[0013] Provided are such methods capable of being carried out quickly, simply,
and at
relatively low cost, as compared to other diagnostic and prognostic methods.
[0014] Provided are methods and compositions that are useful for defining gene
expression-
based classification of GEP-NENs, and thus are useful for allowing the
prediction of malignancy
and metastasis, such as in early stage disease or using histologically
negative samples, providing
accurate staging, facilitating rational therapy, and in developing large
validated clinical datasets
for GEP-NEN-specific therapeutics.
[0015] The GEP-NEN biomarkers include biomarkers, the expression of which is
different
in or is associated with the presence or absence of GEP-NEN, or is different
in or is associated
with a particular classification, stage, aggressiveness, severity, degree,
metastasis, symptom,
risk, treatment responsiveness or efficacy, or associated syndrome. The panel
of GEP-NEN
biomarkers typically includes at least 2 GEP-NEN biomarkers, typically at
least 3 biomarkers.
In some embodiments, the panel of biomarkers includes at least 1, 2, 3, 4, 5,
6, 7, 8, 9, 10, 11,
12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30,
31, 32, 33, 34, 35, 36, 37,
38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56,
57, 58, 59. 60, 61, 62, 63,
64, 65, 66, 67, 68, 69, 70, 71, 72. 73, 74, 75, 80, 85, 90, 95, or 100 or more
biomarkers, or
includes at or about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16,
17, 18, 19, 20, 21, 22, 23,
24, 25, 26, 27, 28, 29, 30, 31, 32. 33, 34, 35, 36, 37, 38, 39, 40, 41, 42,
43, 44, 45. 46, 47, 48, 49,
50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68,
69, 70, 71, 72, 73, 74, 75,
80, 85, 90, 95, or 100 GEP-NEN biomarkers, or more.
[0016] For example, in some aspects, the panel of biomarkers includes at least
3, at least 11,
at least 21, or at least 29 biomarkers, at least 51 biomarkers, or at least 75
more biomarkers. In a
particular example, the panel contains at least 51 biomarkers or about 51
biomarkers or 51
biomarkers. Because the systems contain a plurality of agents (generally
polypeptides or
polynucleotides) that specifically bind to or hybridize to the biomarkers in
the panel, the number
of biomarkers generally relates to the number of agents in a particular
system. For example,
among the provided systems is a system that contains 51 agents, which
specifically hybridize to
or bind to a panel of 51 GEP-NEN biomarkers, respectively.
4
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
[0017] In some aspects, the panel of biomarkers includes at least 1, 2, 3, 4,
5, 6, 7, 8, 9, 10,
11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21,22, 23, 24, 25, 26, 27, 28, 29, 30,
31, 32. 33, 34, 35, 36,
37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, or 51, and/or all of
the following groups of
gene products, including polynucleotides (e.g., transcripts) and polypeptides:
[0018] AKAP8L, ATP6Vl H, BNIP3L, C21 orf7, COMMD9, ENPP4, FAM13A, FLJ10357,
GLT8D1, HDAC9, HSF2, LE01, MORF4L2, NOL3, NUDT3, OAZ2, PANK2, PHF21A,
PKD1, PLD3, PQBP1, RNF41. RSF1, RTN2, SMARCD3, SPATA7, SST1, SST3, SST4, SSTS,
TECPR2, TRMT112, VPS13C, WDFY3, ZFHX3, ZXDC, ZZZ3. APLP2. CD59, ARAF1,
BRAF1, KRAS, and RAF1 gene products:
[0019] AKAP8L, ATP6V1H, BNIP3L, C21orf7, COMMD9, ENPP4, FAM13A, FLJ10357,
GLT8D1, HDAC9, HSF2, LE01, MORF4L2, NOL3, NUDT3, OAZ2, PANK2, PHF21A,
PKD1, PLD3, PQBP1, RNF41, RSF1, RTN2, SMARCD3, SPATA7, SST1, SST3, SST4, SSTS,
TECPR2, TRMT112, VPS13C, WDFY3, ZFHX3, ZXDC, and ZZZ3 gene products: and
[0020] APLP2, ARAF1, BRAF, CD59, CTGF, FZD7, Ki67, KRAS. NAP1L1, PNMA2,
RAF1, TPH1, VMAT1, and VMAT2 gene products.
[0021] In some examples, the panel of biomarkers includes AKAP8L, ATP6V1H,
BNIP3L,
C21orf7, COMMD9, ENPP4, FAM13A, FLJ10357, GLT8D1, HDAC9, HSF2, LE01,
MORF4L2, NOL3, NUDT3, OAZ2, PANK2, PHF21A, PKD1, PLD3, PQBP1, RNF41, RSF1,
RTN2, SMARCD3, SPATA7, SST1, SST3, SST4, SSTS, TECPR2, TRMT112, VPS13C,
WDFY3, ZFHX3, ZXDC. and ZZZ3 gene products.
[0022] In some examples, the panel of biomarkers includes APLP2, ARAF1, BRAF.
CD59,
CTGF, FZD7, Ki67, KRAS, NAP1L1, PNMA2, RAF1, TPH1, VMAT1, and VMAT2 gene
products.
[0023] In some examples, the panel of biomarkers includes AKAP8L, APLP2,
ARAF1,
ATP6V1H, BNIP3L, BRAF, C21orf7, CD59, COMMD9, CTGF, ENPP4, FAM13A, F1110357,
FZD7, GLT8D1, HDAC9, HSF2, Ki67, KRAS, LE01, MORF4L2, NAP1L1, NOL3, NUDT3,
OAZ2, PANK2, PHF21A, PKD1, PLD3, PNMA2, PQBP1, RAF1, RNF41, RSF1, RTN2,
SMARCD3, SPATA7, SST1, SST3, SST4, SSTS, TECPR2, TPH1, TRMT112, VMAT1,
VMAT2, VPS13C, WDFY3, ZFHX3, ZXDC, and ZZZ3 gene products.
[0024] In some examples, the panel of biomarkers includes an APLP2 gene
product, a CD59
gene product, an ARAF1 gene product, a BRAF1 gene product, a KRAS gene
product, or a
RAF1 gene product.
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
[0025] In some examples, the panel of GEP-NEN biomarkers includes APLP2,
ARAF1,
BRAF, CD59, CTGF, FZD7, Ki67, KRAS, NAPIL1, PNMA2, RAF1, TPH1, and VMAT2 gene
products; or the panel of GEP-NEN biomarkers includes APLP2, ARAF1, BRAF1,
CD59,
KRAS, RAFl , CXCL14, GRIA2, HOXC6, NKX2-3, OR51E1, PNMA2, PTPRN2, SCG5,
SPOCK1, X2BTB48, CgA, CTGF, FZD7, Ki-67, Kissl, MAGE-D2, MTA1, NAP] Ll , NRP2,
Tphl, VMAT1, VMAT2 and Survivin gene products.
[0026] In some examples, it further includes further includes a gene product
selected from
the group consisting of MAGE-D2, MTA1, Survivin, Kissl, HOXC6, NRP2, X2BTB48,
CXCL14., GRIA2, NKX2-3, OR51E1, CTGF, PTPRN2, SPOCK1, and SCG5 gene products.
[0027] In other examples, the panel of GEP-NEN biomarkers includes at least 2,
3, 4, 5, 6, 7,
8,9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27,
28, 29 of, or includes
each of, the biomarkers in one, more, or all of the following groups of gene
products, including
polynucleotides (e.g., transcripts) and polypeptides:
[0028] (a) APLP2, ARAF1, BRAF1. CD59, KRAS, RAF1, CXCL14, GRIA2, HOXC6,
NKX2-3, OR51E1, PNMA2, PTPRN2, SCG5, SPOCK1, X2BTB48, CgA, CTGF, FZD7, Ki-67,
Kissl, MAGE-D2, MTA1, NAPIL1, NRP2, Tphl, VMAT1, VMAT2, and Survivin gene
products; (b) MAGE-D2, MTA1, NAP1L1, Ki67, Survivin, FZD7, Kissl, NRP2,
X2BTB48,
CXCL14, GRIA2, NKX2-3, OR51E1, PNMA2, SPOCK1, HOXC6. CTGF, PTPRN2, SCG5,
and Tph gene products; (c) ARAF1, BRAF, CTGF, FZD7, Ki67, KRAS, NAP1L1, PNMA2,
RAF1, TPH1, and VMAT2 gene products; (d) CXCL14, GRIA2, HOXC6, Ki-67, Kiss],
MAGE-D2, MTA1, NAP1L1, NKX2-3, OR51E1, PTPRN2, SCG5. SPOCK1, and X2BTB48
gene products; (e) CXCL14, GRIA2, HOXC6, NKX2-3, OR51E1, PNMA2, PTPRN2, SCG5,
SPOCK1, and X2BTB48 gene products; (f) APLP2, ARAF1, BRAF, CD59, CTGF, FZD7,
Ki67, KRAS, NAP1L1, PNMA2, RAF1, TPH1, and VMAT2 gene products; (g) APLP2,
ARAF1, BRAF1, CD59, KRAS, and RAF1 gene products; and/or (h) ARAF1, BRAF1,
CD59,
KRAS, RAF1, CXCL14, GRIA2, HOXC6, NKX2-3, OR51E1, PNMA2, PTPRN2, SCG5,
SPOCK1, X2BTB48, CgA, CTGF, FZD7, Ki-67, Kissl, MAGE-D2, MTA1, NAP1L1, NRP2,
Tphl, VMAT1, VMAT2 and Survivin gene products.
[0029] In some examples, the biomarkers include at least 2, 3, 4, 5, 6, 7, 8,
9, 10, 11, 12, 13,
14, 15, 16, 17, 18, 19, 20, 21, 22. 23, 24, 25, 26, 27, 28, 29, 30, 31, 32,
33, 34, 35. 36, 37, 38, 39,
40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, or 51 of the following gene
products (the term gene
product including, for example, polynucleotides (e.g., transcripts) and
polypeptides): AKAP8L.
6
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
APLP2, ARAF1, ATP6V1H, BNIP3L, BRAF, C21orf7, CD59, COMMD9, CTGF, ENPP4,
FAM13A, F1110357, FZD7, GLT8D1, HDAC9, HSF2, Ki67, KRAS, LE01, MORF4L2,
NAP1L1, NOL3, NUDT3, OAZ2, PANK2, PHF21A, PKD1, PLD3, PNMA2, PQBP1, RAF1,
RNF41, RSF1, RTN2, SMARCD3, SPATA7, SST1, SST3, SST4, SSTS, TECPR2, TPH1,
TRMT11 2, VMAT1, VMAT2, VPS13C, WDFY3, ZFHX3, ZXDC, and ZZZ3 gene products.
[0030] In some aspects, the biomarkers include AKAP8L, APLP2, ARAF1, ATP6V1H,
BNIP3L, BRAF, C21orf7, CD59, COMMD9, CTGF, ENPP4, FAM13A, FLJ10357, FZD7,
GLT8D1, HDAC9, HSF2, Ki67, KRAS, LE01, MORF4L2, NAP1L1, NOL3, NUDT3, OAZ2,
PANK2, PHF21A, PKD1, PLD3, PNMA2, PQBP1, RAF1, RNF41, RSF1, RTN2, SMARCD3,
SPATA7, SST1, SST3, SST4, SSTS, TECPR2, TPH1, TRMT112, VMAT1, VMAT2, VPS13C,
WDFY3, ZFHX3, ZXDC, and ZZZ3 gene products.
[0031] In some examples, the biomarkers include at least 1, 2, 3, 4, 5, 6, 7,
8, 9, 10, 11, 12,
13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31,
32, 33, 34. 35, 36, 37, or
38 of the following gene products (the term gene product including, for
example,
polynucleotides (e.g., transcripts) and polypeptides): AKAP8L, ATP6V1H,
BNIP3L, C21orf7.
COMMD9, ENPP4, FAM13A, FU10357, GLT8D1, HDAC9, HSF2, LE01, MORF4L2, NOL3,
NUDT3, OAZ2, PANK2, PHF21A, PKD1, PLD3, PQBP1, RNF41, RSF1, RTN2, SMARCD3,
SPATA7, SST1, SST3, SST4, SSTS, TECPR2, TRMT112, VPS13C, WDFY3, ZFHX3, ZXDC,
and ZZZ3 gene products.
[0032] In some aspects, the biomarkers include AKAP8L, ATP6V1H, BNIP3L,
C21orf7,
COMMD9, ENPP4, FAM13A, FU10357, GLT8D1, HDAC9, HSF2, LE01, MORF4L2, NOL3,
NUDT3, OAZ2, PANK2, PHF21A, PKD1, PLD3, PQBP1, RNF41, RSF1, RTN2, SMARCD3,
SPATA7, SST1, SST3, SST4, SSTS, TECPR2, TRMT112, VPS13C, WDFY3, ZFHX3, ZXDC,
and ZZZ3.
[0033] In some examples, the biomarkers include at least two GEP-NEN
biomarkers
selected from among APLP2, ARAF1, BRAF1, CD59, KRAS, RAF1, CXCL14, GRIA2,
HOXC6, NKX2-3, OR51E1, PNMA2, PTPRN2, SCG5, SPOCK1, X2BTB48, CgA, CTGF,
FZD, Ki-67, Kissl, MAGE-D2, MTA1, NAP1L1, NRP2, Tphl, VMAT1, VMAT2, and
Survivin
gene products.
[0034] In one embodiment, the plurality of GEP-NEN biomarkers includes an
APLP2 gene
product or a CD59 gene product. In one embodiment, the GEP-NEN biomarkers
include an
APLP2 gene product. In one embodiment, they include a CD59 gene product.
7
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
[0035] In one embodiment, the GEP-NEN biomarkers include an APLP2, CD59,
ARAF1,
BRAF1, KRAS or RAF1 gene product.
[0036] In some embodiments, the panel of GEP-NEN biomarkers includes an APLP2,
ARAF1, BRAF, CD59, KRAS, or RAF1 gene product or a GTGF, FZD7, Ki67, NAP1L1,
PNMA2, TPH1, or VMAT2 gene product. In some embodiments, the panel of GEP-NEN
biomarkers includes a PNMA2 gene product. In some embodiments, the panel of
GEP-NEN
biomarkers includes a VMAT2 gene product. In some embodiments, the panel of
GEP-NEN
biomarkers includes a CgA, CXCL14, GRIA2, HOXC6, Kissl, MAGE-D2, MTA1, NKX2-3,
NRP2, OR51E1, PTPRN2, SCG5, SPOCK1, survivin, VMAT1, or X2BTB48 gene product.
In
other embodiments, the panel includes a PNMA2 biomarker. In some embodiments,
the panel
includes a VMAT2 biomarker.
[0037] In some embodiments, the panel includes APLP2, ARAF1, BRAF, CD59, CTGF,
FZD7, Ki67, KRAS, NAP1L1, PNMA2, RAF1, TPH1, and VMAT2 gene products; or
includes
MAGE-D2, MTA1, NAP1L1, Ki67, Survivin, FZD7, Kissl, NRP2, X2BTB48, CXCL14,
GRIA2, NKX2-3, OR51E1, PNMA2, SPOCK1, HOXC6, CTGF, PTPRN2, SCG5, CgA, and
Tph gene products. In one aspect, the biomarkers includes at least one of or
includes each of the
following biomarkers: APLP2, ARAFI, BRAF1, CD59, KRAS, RAF1, CXCL14, GRIA2,
HOXC6, NKX2-3, 0R51E1, PNMA2, PTPRN2, SCG5, SPOCK1, X2BTB48, CTGF, FZD7,
Ki-67, Kissl, MAGE-D2, MTA1, NAP1L1, NRP2, Tphl, VMAT1, VMAT2õS'urvivin and
X2BTB48 gene products. In one such embodiment, the biomarkers further include
a CgA gene
product.
[0038] In one embodiment, the GEP-NEN biomarkers include one or more gene
products
having a nucleotide sequence with at least at or about or at or about 90, 91,
92, 93, 94, 95, 96,
97, 98, 99 % identity to or 100 % identity to (i.e., having a nucleotide
sequence of) SEQ ID NO:
1, or to SEQ ID NO: 1, from nucleotide residues 158-2449; SEQ ID NO: 2, or to
SEQ ID NO: 2
from nucleotide residues 195-2015 SEQ ID NO: 3 or to SEQ ID NO: 1 from
nucleotide residues
62-2362; SEQ ID NO: 4 or to SEQ ID NO: 4, from nucleotide residues 278-664;
SEQ ID NO: 5,
or to SEQ ID NO: 5 from nucleotide residues 1 to 1374; SEQ ID NO: 6, or to SEQ
ID NO: 6
from nucleotide residues 207-1256; SEQ ID NO: 7, or to SEQ ID NO: 7 from
nucleotide
residues 466-801; SEQ ID NO: 8, or to SEQ ID NO: 8 from nucleotide residues 62-
1786; SEQ
ID NO: 9, or to SEQ ID NO: 9 from nucleotide residues 460-3111; SEQ ID NO: 10,
or to SEQ
ID NO: 10 from nucleotide residues 113-820; SEQ ID NO: 11, or to SEQ ID NO: 11
from
8
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
nucleotide residues 196-9966; SEQ ID NO: 12, or to SEQ ID NO: 12 from
nucleotide residues
155-571; SEQ ID NO: 13, or to SEQ ID NO: 13 from nucleotide residues 182-751;
SEQ ID NO:
14, or to SEQ ID NO: 14 from nucleotide residues 100-1920; SEQ ID NO: 15, or
to SEQ ID
NO: 15 from nucleotide residues 188-2335; S EQ ID NO: 16, or to SEQ ID NO: 16
from
nucleotide residues 413-1588; SEQ ID NO: 17, or to SEQ ID NO: 17 from
nucleotide residues
200-1294; SEQ ID NO: 18, or to SEQ ID NO: 18 from nucleotide residues 792-
3587; SEQ ID
NO: 19, or to SEQ ID NO: 19 from nucleotide residues 145-1101; SEQ ID NO: 20,
or to SEQ
ID NO: 20 from nucleotide residues 771-1865; SEQ ID NO: 21, or to SEQ ID NO:
21 from
nucleotide residues 122-3169; SEQ ID NO: 22, or to SEQ ID NO: 22 from
nucleotide residues
416-2362; or to SEQ ID NO: 22 SEQ ID NO: 23, or to SEQ ID NO: 23 from
nucleotide residues
118-756; SEQ ID NO: 24, or to SEQ ID NO: 24 from nucleotide residues 152-1471;
SEQ ID
NO: 25, or to SEQ ID NO: 25 nucleotide residues 2811-2921, 3174-3283, 5158-
5275, 11955-
12044, or to SEQ ID NO: 34; SEQ ID NO: 26, or to SEQ ID NO: 26 from nucleotide
residues
27-1361; SEQ ID NO: 27, or to SEQ ID NO: 27 from nucleotide residues 472-2049;
SEQ ID
NO: 28, or to SEQ ID NO: 28 from nucleotide residues 32-1576; SEQ ID NO: 29,
or to SEQ ID
NO: 29 from nucleotide residues 467-1801; SEQ ID NO: 105, or to SEQ ID NO: 105
from
nucleotide residues 122-1456; SEQ ID NO: 201, or to SEQ ID NO: 201 from
nucleotide
residues 100-2040; SEQ ID NO: 204, or to SEQ ID NO: 240, from nucleotide
residues 293-
1744; SEQ ID NO: 205, or to SEQ ID NO: 205, from nucleotide residues 125-784;
SEQ ID NO:
206, or to SEQ ID NO: 206 from nucleotide residues 278-1006; SEQ ID NO: 207,
or to SEQ ID
NO: 207 from nucleotide residues 38-508; SEQ ID NO: 208, or to SEQ ID NO: 208
from
nucleotide residues 260-1621; SEQ ID NO: 209, or to SEQ ID NO: 209 from
nucleotide
residues 281-1126; SEQ ID NO: 210, or to SEQ ID NO: 210 from nucleotide
residues 30-4589;
SEQ ID NO: 211, or to SEQ ID NO: 211 from nucleotide residues 852-1967; SEQ ID
NO: 212,
or to SEQ ID NO: 212 from nucleotide residues 362-2128; SEQ ID NO: 213, or to
SEQ ID NO:
213 from nucleotide residues 188-1798; SEQ ID NO: 215, or to SEQ ID NO: 215
from
nucleotide residues 17-2017; SEQ ID NO: 217, or to SEQ ID NO: 217 from
nucleotide residues
505-1371; SEQ ID NO: 218, or to SEQ ID NO: 218 from nucleotide residues 194-
853; SEQ ID
NO: 219, or to SEQ ID NO: 219 from nucleotide residues 319-837; SEQ ID NO:
220, or to SEQ
ID NO: 220 from nucleotide residues 216-311 and 313-786; SEQ ID NO: 221, or to
SEQ ID
NO: 221 from nucleotide residues 312-1151; SEQ ID NO: 222, or to SEQ ID NO:
222 from
nucleotide residues 625-2667; SEQ ID NO: 223, or to SEQ ID NO: 223 from
nucleotide
9
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
residues 210-13117, or to the sequence referenced at GenBank gi Number
205360961 or to that
sequence from nucleotide residues 210-13118; SEQ ID NO: 224, or to SEQ ID NO:
224 from
nucleotide residues 399-1871; SEQ ID NO: 225, or to SEQ ID NO: 225 from
nucleotide
residues 122-919; SEQ ID NO: 227, or to SEQ ID NO: 227 from nucleotide
residues 320-1273;
SEQ ID NO: 228, or to SEQ ID NO: 228 from nucleotide residues 121-4446; SEQ ID
NO: 229,
or to SEQ ID NO: 229 from nucleotide residues 229-1866: SEQ ID NO: 232, or to
SEQ ID NO:
232 from nucleotide residues 102-1553: SEQ ID NO: 233, or to SEQ ID NO: 233
from
nucleotide residues 176-1879; SEQ ID NO: 234, or to SEQ ID NO: 234 from
nucleotide
residues 618-1793; SEQ ID NO: 235, or to SEQ ID NO: 235 from nucleotide
residues 526-1782;
SEQ ID NO: 236, or to SEQ ID NO: 236 from nucleotide residues 65-1231; SEQ ID
NO: 237,
or to SEQ ID NO: 237 from nucleotide residues 89-1183; SEQ ID NO: 238, or to
SEQ ID NO:
238 from nucleotide residues 227-4030; SEQ ID NO: 239, or to SEQ ID NO: 239
from
nucleotide residues 104-1969; SEQ ID NO: 240, or to SEQ ID NO: 240 from
nucleotide
residues 94-612, SEQ ID NO: 243, or to SEQ ID NO: 243 from nucleotide residues
409-10988,
SEQ ID NO: 244, or to SEQ ID NO: 244 from nucleotide residues 130-8499, SEQ ID
NO: 245,
or to SEQ ID NO: 245 from nucleotide residues 55-2187, and/or SEQ ID NO: 246,
or to SEQ ID
NO: 246 from nucleotide residues 477-3188.
[0039] Among the provided methods, agents, and systems are those that are able
to classify
or detect a GEP-NEN in a human blood sample. In some embodiments, the provided
systems
and methods can identify or classify a GEP-NEN in a human blood sample; in
some
embodiments, it can differentiate between a subject with GEP-NEN and a subject
with another
type of gastrointestinal (GI) cancer (or other cancer) or can determine the
site of a GEP-NEN,
e.g., by differentiating between a subject with small intestinal NEN and a
subject with a
pancreatic NEN. In some examples, the systems can provide such information
with a
specificity, sensitivity, and/or accuracy of at least 75 %, 80 %, 85 %, 90 %,
91 %, 92 %, 93 %,
94 %, 95 %, 96 %, 97 %, 98 %, 99 % or 100 %, e.g., at least 80 %.
[0040] In some embodiments, the system can predict treatment responsiveness
to, or
determine whether a patient has become clinically stable following, or is
responsive or non-
responsive to, a GEP-NEN treatment, such as a surgical intervention or drug
therapy (for
example, somatostatin analog therapy). In some cases, the methods and systems
do so with a
specificity, sensitivity, and/or accuracy of at least 75 %, 80 %, 85 %, 90 %,
91 %, 92 %, 93 %,
94 %, 95 %, 96 %, 97 %, 98 %, 99 % or 100 %, e.g., with at least 90 %
accuracy. In some cases,
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
it can differentiate between treated and untreated GEP-NEN with a specificity,
sensitivity, and/or
accuracy of at least 75 %, 80 %, 85 %, 90 %, 91 %, 92 %, 93 %, 94%, 95 %, 96
%, 97 %, 98 cio,
99 % or 100 %, e.g., with a sensitivity and specificity of at least 85 %.
[0041] In some cases, it can determine diagnostic or prognostic information
regarding a
subject previously diagnosed with GEP-NEN, for example, whether the subject
has stable
disease, progressive disease, or is in complete remission (for example, would
be clinically
categorized as having stable disease, progressive disease, or being in
complete remission).
[0042] In some embodiments, the agents for detecting the biomarkers (e.g., the
sets of
polynucleotide or polypeptide agents), and uses thereof, are capable of
distinguishing between
the presence and absence of GEP-NEN in a biological sample, between GEP-NEN
and other
intestinal and mucosal samples, such as enterochromaffin (EC) and small
intestinal (SI) mucosal
samples and GEP-NEN samples, between metastatic or aggressive and primary GEP-
NEN
samples, and/or between specific classes or subtypes of GEP-NENs.
[0043] In some embodiments, the methods distinguish between GEP-NEN and other
cancers, such as adenocarcinomas, including gastrointestinal adenocarcinoma or
one of the
breast, prostate, or pancreas, or a gastric or hepatic cancer, such as
esophageal, pancreatic,
gallbladder, colon, or rectal cancer. In other embodiments, the methods and
systems
differentiate between GEP-NENs of different sites, such as between GEP-NENs of
the small
intestine and those of the pancreas.
[0044] In one embodiment, the set of agents distinguishes between
enterochromaffin (EC)
and small intestinal (SI) mucosa. In one aspect, the panel of GEP-NEN
biomarkers comprises
CTGF, CXCL14, FZD7, Kissl, FZD, Kissl, NKX2-3, PNMA2, PTPRN2, SCG5. SPOCK1,
and
X2BTB48 gene products. In another embodiment, the system or set of agents
distinguishes
between Adenocarcinoma and GEP-NEN, such as an adenocarcinoma and a GEP-NEN
sample.
In one aspect, the panel of GEP-NEN biomarkers comprises at least sixteen GEP-
NEN
biomarkers, including a CgA gene product. In another embodiment, the system or
set of agents
distinguishes between primary and metastatic GEP-NEN disease. In one aspect of
this
embodiment, the panel of GEP-NEN biomarkers includes at least eighteen GEP-NEN
biomarkers.
[0045] In some embodiments, the system or set of agents or use of the same
distinguishes
between one or more various sub-types of GEP-NEN, and/or contains agents that
bind to or
detect a set of biomarkers the expression profile of which or the summed
expression (e.g.
11
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
vectorally summed expression) of which differs significantly between the
various sub-types. In
one aspect of this embodiment, the system distinguishes between primary PDNEC
and primary
WDNET; in one example, the panel of biomarkers includes CXCL14 and MAGE-D2
gene
products. In another embodiment, the system distinguishes between primary
PDNEC and
primary WDNEC; in one example, the panel of GEP-NEN biomarkers includes three
biomarkers, including a PTPRN2 gene product. In another embodiment, the system
distinguishes between primary PDNEC and primary PDNET; in one example, the
panel of
biomarkers includes MTA1 and PNMA2 gene products. In another embodiment, the
system
distinguishes between primary PDNET and primary WDNET; in one example, the
panel of NE
biomarkers includes at least four biomarkers. In another embodiment, the
system distinguishes
between primary WDNEC and primary WDNET; in one example, the set contains at
least 21
biomarkers.
[0046] In another embodiment, the system distinguishes between metastatic sub-
types of
GEP-NEN, such as between metastatic WDNEC and metastatic WDNET, for example,
where
the panel contains at least three biomarkers, including a CXCL14 gene product;
between
metastatic PDNEC and metastatic WDNEC, for example, where the set of
biomarkers includes
at least four biomarkers, including a NAP1L1 gene product; between metastatic
PDNEC and
metastatic WDNET, for example, where the panel of GEP-NEN biomarkers includes
at least six
biomarkers, for example, including a NRP2 gene product.
[0047] In one aspect, the system is able to classify or detect a GEP-NEN in a
human blood
sample or human saliva sample. In one aspect, the human sample is whole blood
or nucleic acid
or protein prepared from whole blood, without first sorting or enriching for
any particular
population of cells. In one aspect, the system includes agents that bind to
biomarkers in a panel
of at least 29 GEP-NEN biomarkers.
[0048] In some aspects, the methods and systems provide such diagnostic,
differentiation,
detection, predictive, or prognostic information or determination as described
above with a
greater sensitivity, specificity, or accuracy compared with another diagnostic
method, such as
available detection or diagnosis method, such as the detection of circulating
CgA levels.
[0049] In some embodiments, in addition to the agents that bind the GEP-NEN
biomarkers,
the provided systems contain one or more agents that bind to gene products for
use in
normalization or as controls, for example, housekeeping gene products,
including any one or
more of: ACTB, TOX4, TPT1 and TXNIP gene products;
12
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
[0050] housekeeping gene products, including any one or more of: 18S, GAPDH,
ALG9,
SLC25A3, VAPA, TXNIP, ADD3, DAZAP2, ACTG1, ACTB, ACTG4B, ARF1, HUWEl,
MORF4L1 RHOA, SERPI, SKP1, TPT1, TOX4, TFCP2, and ZNF410, gene products;
[0051] housekeeping genes including any one or more of 18S, GAPDH, ALG9,
SLC25A3,
VAPA, TXNIP. ADD3, DAZAP2, ACTG1, ACTB, ACTG4B, ARF1, HUWEl , MORF4L1
RHOA, SERP 1 , S KP1, TPT1, and TOX4 gene products; or
[0052] housekeeping genes including any one or more of: ALG9, TFCP2, ZNF410,
18S, and
GAPDH gene products.
[0053] In some embodiments, the system distinguishes between enterochromaffin
(EC) and
small intestinal (SI) mucosa and the panel of GEP-NEN biomarkers further
includes CTGF,
CXCL14, F1D7, Kissl, FZD, Kissl, NKX2-3, PNMA2, PTPRN2, SCG5, SPOCK1, and
X2BTB48 gene products. In another embodiment, the panel of GEP-NEN biomarkers
includes
MAGE-D2, MTA1, NAP1L1, Ki67, Survivin, FZD7, Kissl, NRP2, X2BTB48, CXCL14,
GRIA2, NKX2-3, OR51E1, PNMA2, SPOCKI, HOXC6, CgA, CTGF, PTPRN2, SCG5, and
Tphl gene products. In another embodiment, the system distinguishes between
Adenocarcinoma and GEP-NEN and includes a set of polynucleotides or
polypeptides that
specifically hybridize to a panel of sixteen or more GEP-NEN biomarkers,
including a CgA gene
product.
[0054] In some embodiments, the methods and systems determine the presence,
absence,
expression levels, or expression profile indicates the presence, absence,
classification, prognosis,
risk, responsiveness to treatment, aggressiveness, severity, or metastasis of
the GEP-NEN. For
example, in one aspect, the presence, absence, expression levels, or
expression profile detected
in the test sample indicates the efficacy of a GEP-NEN treatment. In one
aspect, the detected
presence, absence, expression levels, or expression profile distinguishes
between primary
PDNEC and primary WDNET and the panel of biomarkers includes CXCL14 and MAGE-
D2
gene products; in other aspects, it distinguishes between primary PDNEC and
primary WDNEC
and the panel of biomarkers includes three biomarkers, including a PTPRN2 gene
product; in
another aspect, it distinguishes between primary PDNEC and primary PDNET, and
the panel of
biomarkers includes MTA1 and PNMA2 gene products; in another aspect, it
distinguishes
between primary PDNET and primary WDNET or in primary PDNET and primary WDNEC,
and the panel of biomarkers includes at least four biomarkers; in another
aspect, it distinguishes
between primary WDNEC and primary WDNET, the panel of biomarkers includes
twenty-one
13
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
biomarkers; in another aspect, it distinguishes between metastatic WDNEC and
metastatic
WDNET and the panel of biomarkers includes at least three biomarkers,
including a CXCL14
gene product; in another aspect, it distinguishes between metastatic PDNEC and
metastatic
WDNEC and the panel comprises at least four biomarkers, including a NAP] Ll
gene product; in
another aspect, it distinguishes between metastatic PDNEC and metastatic WDNET
and the
panel comprises at least six biomarkers, including a NRP2 gene product.
[0055] The biological test sample used with the methods can be any biological
sample, such
as tissue, biological fluid, or other sample, including blood samples, such as
plasma, serum,
whole blood, huffy coat, or other blood sample, tissue, saliva, serum, urine,
or semen sample. In
some aspects, the sample is obtained from blood. Often, the test sample is
taken from a GEP-
NEN patient.
[0056] In some embodiments, the methods further include comparing the
expression levels
or expression profile or presence or absence of the biomarkers detected in the
test sample to a
normal or reference level of expression or a normal or reference expression
profile, or a standard
value of expression level, amount, or expression profile, or the presence (or
more typically the
absence) of detection in a reference or normal sample.
[0057] In some such embodiments, the methods include a step of obtaining a
normal or
reference sample and detecting the presence, absence, expression levels, or
expression profile of
the panel of GEP-NEN biomarkers in the normal sample, typically carried out
prior to the
comparison step. In one aspect, this further step determines a normal or
reference level of
expression Or a normal or reference expression profile, which can be compared
to the expression
level or profile detected in the test biological sample.
[0058] In some cases, statistical analysis is performed to determine whether
there is a
difference, such as a significant difference, between the expression levels
detected in the test
biological sample and the normal or reference sample, or other standard or
reference expression
level. For example, a difference may be considered significant where there is
a p value of less
than 0.05 or where there is a 2 standard deviation. Other methods for
determining significance
are known in the art.
[0059] The normal or reference sample may be from a healthy patient or a
patient who has
GEP-NEN. Where the test sample is from a patient with GEP-NEN, the normal or
reference
sample or level may be from the same or a different patient. For example, the
normal or
reference sample may be from the GEP-NEN patient from a tissue, fluid or cell
not expected to
14
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
contain GEP-NEN or GEP-NEN cells. On another aspect, the normal or control
sample is from
the GEP-NEN patient before or after therapeutic intervention, such as after
surgery or chemical
intervention. In another aspect, the reference or normal sample is from a
tissue or fluid that
corresponds to the GEP-NEN or metastasis of the test sample, from a healthy
individual, such as
normal EC or SI sample, or normal liver, lung, bone, blood, saliva, or other
bodily fluid, tissue,
or biological sample. In another embodiment, the test sample is from a
metastatis, plasma, or
whole blood or other fluid of a GEP-NEN patient and the reference sample is
from primary
tumor or sorted tumor cells.
[0060] In one aspect, the test biological sample is from a GEP-NEN patient
prior to
treatment and the normal or reference sample is from the GEP-NEN patient after
treatment. In
another aspect, the normal or reference sample is from a non-metastatic tissue
of the GEP-NEN
patient.
[0061] In other aspects, the test sample is from blood and the test biological
sample is from
the GEP-NEN patient after treatment and the reference sample is from the same
GEP-NEN
patient as the test biological sample, prior to treatment; the reference
sample is from a tissue or
fluid not containing GEP-NEN cells; the reference sample is from a healthy
individual; the
reference sample is from a cancer other than GEP-NEN; the reference sample is
from an EC cell
or SI tissue; the test biological sample is from a metastatic GEP-NEN and the
reference sample
is from a non-metastatic GEP-NEN; or the reference sample is from a GEP-NEN of
a different
classification compared to the GEP-NEN patient from which the test biological
sample is
obtained.
[0062] The agents can be any agents for detection of biomarkers, and typically
are isolated
polynucleotides or isolated polypeptides or proteins, such as antibodies, for
example, those that
specifically hybridize to or bind to a panel of GEP-NEN biomarkers including
at least 21 GEP-
NEN biomarkers.
[0063] In some embodiments, the methods are performed by contacting the test
sample with
one of the provided agents, more typically with a plurality of the provided
agents, for example,
one of the provided systems, such as a set of polynucleotides that
specifically bind to the panel
of GEP-NEN biomarkers. In some embodiments, the set of polynucleotides
includes DNA,
RNA, cDNA, PNA, genomic DNA, or synthetic oli2onucleotides. In some
embodiments, the
methods include the step of isolating RNA from the test sample prior to
detection, such as by
RT-PCR, e.g., QPCR. Thus, in some embodiments, detection of the GEP-NEN
biomarkers,
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
such as expression levels thereof, includes detecting the presence, absence,
or amount of RNA.
In one example, the RNA is detected by PCR or by hybridization.
[0064] In one aspect, the polynucleotides include sense and antisense primers,
such as a pair
of primers that is specific to each of the GEP-NEN biomarkers in the panel of
biomarkers. In
one aspect of this embodiment, the detection of the GEP-NEN biomarkers is
carried out by PCR,
typically quantitative or real-time PCR. For example, in one aspect, detection
is carried out by
producing cDNA from the test sample by reverse transcription; then amplifying
the cDNA using
the pairs of sense and antisense primers that specifically hybridize to the
panel of GEP-NEN
biomarkers, and detecting products of the amplification. In some embodiments,
the GEP-NEN
biomarkers include naRNA, cDNA, or protein.
[0065] In some embodiments, the methods are capable of detecting low-volume
GEP-NENs,
early-stage GEP-NENs, micrometastes, circulating GEP-NEN cells, and/or other
instances of
GEP-NEN that are difficult to detect by available methods, such as imaging or
detection of
available biomarkers such as GEP-NEN. For example, in some embodiments, the
sample is a
blood sample, such as a whole blood sample, and the method detects at least at
or about three
GEP-NEN cells per milliliter (mL) of whole blood.
[0066] In some aspects the methods further comprise statistical analysis and
analysis using
predictive models such as mathematical algorithms. In one example, the methods
include
computing a mean expression level for the panel of GEP-NEN biomarkers in the
test biological
sample. In one aspect of this embodiment, the computing is carried out by
vectorally summing
the detected expression levels for each of the plurality of GEP-NEN
biomarkers. In some
aspects, the mean expression level is compared to a reference mean expression
level, such as one
obtained by performing the methods on a reference or normal sample. Often the
comparison
reveals a significant difference in the mean expression levels in the test
sample compared to the
mean reference expression levels. In some aspects, the detected expression or
expression
profiles are sufficiently different, such as significantly different or
sufficiently up or down-
regulated, where there is a p value of less than at or about 0.05, or a
difference of + standard
deviation, or an S value of - 0.4, with S <-0.4 or S > 0.4, or other known
method, such as those
described herein. In some aspects, the expression, such as mean expression,
mean summed
expression or expression profile detected and/or determined in the test
biological sample
correlates with that of another GEP-NEN sample, such as where the test sample
is a whole blood
or other biological fluid sample, and the amount correlates with that of a GEP-
NEN tissue or
16
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
purified cell population. For example, with an R2 of at least about 0.4, 0.5,
0.6, 0.7, 0.8, 0.9, or
1.
[0067] In one embodiment, the method identifies the presence or absence,
classification, or
stage of GEP-NEN with between 80 % to 100 %, such as at or about or at least
at or about 80,
81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, or
100% predictive
value, sensitivity, or specificity. In some embodiments, the methods include a
step of
compressing the detected expression levels of the biomarkers from the test
biological sample.
Typically, the compression is carried out to determine the expression profile
of the panel of
biomarkers.
[0068] In some embodiments, the test biological sample is a whole blood or
saliva sample
from a GEP-NEN patient and expression levels or expression profile detected or
determined for
the test biological sample correlate(s) with the expression levels or
expression profile for the
same GEP-NEN biomarkers for a GEP-NEN tissue sample or purified GEP-NEN cell
sample
obtained from the same patient, with an R2 of at least about 0.4.
[0069] In some embodiments, the methods include steps for analyzing the data
using a
predictive algorithm, model, and/or topographical analysis. In some examples,
the predictive
algorithm is support vector machines (SVM), linear discriminant analysis
(LDA). K-nearest
neighbor (KNN) or naïve Bayes (NB). In some examples, the predictive algorithm
is support
vector machines (SVM), linear discriminant analysis (LDA), or K-nearest
neighbor (KNN). In
other examples, the algorithm is decision tree, SVM, RDA, or Perceptron, or
other model known
in the art or described herein. In one aspect, the model or algorithm
determines the presence,
absence, metastatic or non-metastatic nature of a GEP-NEN, or distinguishes
between two or
more classes of GEP-NEN with a misclassification rate of between 0.05 to 0.
[0070] Also among the provided are methods are methods for detecting
neuroendocrine
tumor cells in blood, by obtaining a blood sample; and contacting the blood
sample with one or
more agents, which specifically binds to a panel of GEP-NEN biomarkers, which
includes at
least two GEP-NEN biomarkers, wherein the method detects at least at or about
one, two, three,
four, or five cells per mL of blood. GEP-NEN cells per mL blood.
[0071] Also among the provided embodiments are methods for enriching or
isolating GEP-
NEN cells from fluids and mixtures of cells, such as plasma, blood, buffy
coat, cell culture,
biological fluid, or other cell preparation. In one aspect of this embodiment
the method is
carried out by contacting the mixture of cells with an agent that specifically
binds to a GEP-NEN
17
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
biomarker and purifying cells which bind to the agent. In one aspect of this
embodiment, the
biomarker is CD164. In one aspect, the biomarker is a polypeptide biomarker.
In one such
aspect, the agent is an antibody that specifically binds to the biomarker,
such as a CD164
antibody. In some embodiments, the purification is by FACS or column
purification or any
other known method for purifying cells based on affinity. In one aspect, the
contacting further
includes contacting the cells with another GEP-NEN-specific agent. In some
aspects, the
method enriches or isolates at least at or about one, two, three, four, or
five cells per mL of
blood.
[0072] Also provided are methods and uses of the provided biomarkers, agents,
systems and
detection methods for use in GEP-NEN treatment and treatment monitoring. For
example,
provided are methods using the diagnostic, predictive, and detection methods
described above in
conjunction with GEP-NEN treatment, such as to assay a sample obtained from a
subject
undergoing treatment or who was previously undergoing treatment for GEP-NEN.
In one
embodiment, such methods are carried out by obtaining a sample from such a
patient and
detecting or determining the presence or absence of expression, expression
levels, or expression
profile of a GEP-NEN biomarker, typically a panel of GEP-NEN biomarkers, in
the sample. In
one aspect, the method includes first providing a treatment to the patient. In
such methods, the
biomarker or panels generally is or are detected using an agent or system as
provided herein,
such as those described above. In some aspects, the method further includes,
prior to providing
the treatment, determining a pre-treatment amount, presence, absence,
expression levels, or
expression profile in a sample from the patient of the biomarker or panel of
biomarkers. Thus,
in some examples, the pre-treatment amount, presence, absence, expression
levels, or expression
profile is or are compared to the amount, presence, absence, expression
levels, or expression
profile determined or detected in the patient after treatment.
[0073] In some cases, this analysis determines that there is a difference in
expression levels
between the pre-treatment expression levels and the post-treatment expression
levels, which can
indicate the efficacy of the treatment. In some cases, the method further
includes determining
expression amount, presence, absence, levels, or profiles of the biomarkers in
the patient or a
sample from the patient at a later time. Such methods can further include
comparing the
information from the later time to that originally detected or determined.
This information, for
example, a difference between the expression amounts, presence, absence,
levels, or profiles
levels can indicate information about whether the individual has been
responsive to treatment,
18
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
for example, can indicate recurrence, lack of treatment responsiveness, stable
disease, or
progressive disease.
[0074] In some embodiments, such methods provide the advantage of providing
more
sensitive, specific, or accurate information compared to available diagnostic
methods, such as
detection of CgA levels in the serum or other sample. Thus, in one example,
the methods
provide the indicated diagnostic, prognostic, or predictive information in a
case where the CgA
expression levels are not significantly different in the samples assayed, for
example, between the
pre-treatment and post-treatment samples and/or the sample taken at a later
timepoint, or
between the test sample and the normal sample.
[0075] In some cases, the treatment is discontinued or modified based on the
determination
from the methods. The methods may be performed in an iterative fashion, with
treatment
reevaluated or modified according to the expression levels or profiles or
comparisons. Thus, in
some embodiments, the methods further include discontinuing the treatment or
modifying the
treatment provided to the patient, for example, based on the information
determined by the
diagnostic approach. In some cases, the comparison and/or expression amount,
presence,
absence, levels, or profile indicates the presence of a GEP-NEN
micrometastasis in the patient.
In one example, one or more of the samples taken from the patient is or was
determined to be
free of GEP-NEN, GEP-NEN metastases, or GEP-NEN recurrence by another
diagnostic
method, such as by histology or detection of CgA alone.
[0076] In other embodiments, the treatment methods are carried out by
obtaining a first
sample from a GEP-NEN patient and detecting expression levels of a panel of
GEP-NEN
biomarkers in the first sample; providing a treatment to the patient;
obtaining a second sample
from the GEP-NEN patient and detecting expression levels of the panel of GEP-
NEN
biomarkers in the second sample; and comparing the expression levels detected
in the first
sample to those detected in the second sample. In one aspect, the method
further includes
determining whether there is a difference in expression levels between the
first and second
samples, for example, determining that there is such a difference. In one
example the difference
indicates the efficacy of the treatment. In a further embodiment, the method
further includes
obtaining a third sample from the patient, detecting the expression levels in
the third sample and
comparing them to the expression levels in the first or second sample. In some
cases, the
comparison indicates the presence of a metastasis, such as micrometastasis. In
some
embodiments, one or more of the samples is taken from a patient determined to
be free of GEP-
1 9
CA2828878
NEN, GEP-NEN metastases, or GEP-NEN recurrence by another assay, such as
detection of
CgA alone, imaging, or histology, yet the methods detect the presence of GEP-
NEN, GEP-
NEN metastases, or GEP-NEN recurrence in the same sample.
[0077] In one example, the method further includes determining that there is a
difference
between expression levels detected in the second and third samples, where the
difference
indicates a recurrence or lack of treatment responsiveness. In some aspects,
the levels of CgA
expression in the second sample are not significantly different compared to
those in the first
sample or the third sample.
[077A] The invention disclosed and claimed herein pertains to a system for
gastroenteropancreatic neuroendocrine neoplasm (GEP-NEN) analysis comprising:
(a) a set of
isolated polynucleotides or isolated polypeptides that specifically hybridize
or bind to
gastroenteropancreatic neuroendocrine neoplasm (GEP-NEN) biomarkers in a
biological
sample from a subject, wherein the GEP-NEN biomarkers comprise AKAP8L, COMMD9,
Ki67, MORF4L2, OAZ2, SST1, SST3, TECPR2, ZFHX3, ZXDC, CD59, BRAF, and RAF1
gene products; (b) written instructions for detecting the expression levels or
expression profile
of the GEP-NEN biomarkers in the sample and comparing the expression levels or
expression
profile of the GEP-NEN biomarkers detected in the biological sample to a
reference level or a
reference expression profile of the GEP-NEN biomarkers; and c) written
instructions for
generating a report, wherein the report determines treatment responsiveness
to, or determines
whether the subject has become clinically stable following surgical
intervention or somatostatin
analog therapy for GEP-NEN, with at least 90% accuracy.
1077B1 The invention disclosed and claimed herein also pertains to a method
for evaluating
response to therapy of a gastroenteropancreatic neuroendocrine neoplasm (GEP-
NEN) or GEP-
NEN cell, comprising: (a) contacting a biological sample from a subject with a
set of
polynucleotides that specifically hybridize or bind to GEP-NEN biomarkers
AKAP8L,
COMMD9, Ki67, MORF4L2, OAZ2, SST1, SST3, TECPR2, ZFHX3, ZXDC, CD59, BRAF,
and RAF1 wherein the biological sample is a tissue, a blood or a plasma
sample; (b) detecting
the hybridization or binding of the GEP-NEN biomarkers to the set of
polynucleotides, thereby
detecting the expression levels or expression profile of the GEP-NEN
biomarkers in the
biological sample from the subject; (c) comparing the expression levels or
expression profile of
CA 2828878 2020-01-29
=
the detected GEP-NEN biomarkers to a reference level or a reference expression
profile of the
GEP-NEN biomarkers in a reference sample from the same subject prior to
treatment; and
(d) based on differences between the expression levels or the expression
profile of the detected
GEP-NEN biomarkers and the reference level or the reference profile of the GEP-
NEN
biomarkers, determining treatment responsiveness to, or determining whether
the subject has
become clinically stable following, surgical intervention or somatostatin
analog therapy for
GEP-NEN, with at least 90% accuracy.
20a
CA 2828878 2019-07-12
CA 2828878
Brief Description of the Drawings
[0078] FIG. 1. Gene expression distribution across Normal, Localized and
Malignant
tissues. Expression of individual genes (listed above individual graphs)
across samples was
compared to average expression in the normal enterochomaffin (EC) cell and
assigned to
Upregulated, Downregulated, or Baseline class. Each graph shows results for
normal,
malignant, and localized tissues, from left to right. Ellipsoids correspond to
a +2 Standard
Deviations (SD) threshold. All p-values:p <0.05.
[0079] FIG. 2. Principal Component (PC) Analysis of primary Small Intestinal
Neuroendocrine Tumors, metastases and normal EC cells. Ln-normalized real-time
PCR
expression levels of indicated biomarkers, reduced to 3 PCs, representing
75.6% variance in
primary tumor subtypes and normal EC cell preparations (2A) and 712% variance
in primary
tumor subtypes and corresponding metastases (2C). For primary tumors and
normal EC cells,
three groups of genes with similar expression patterns were observed (2B),
with two groups
identified in corresponding metastases (2D).
[0080] FIG. 3: Similarity Matrix using Pearson Correlation of the marker gene
expressions in primary Small Intestinal Neuroendocrine Tumors and normal EC
cells.
Ln-normalized real-time PCR expression levels of indicated genes plotted onto
X- and Y-axis.
[0081] FIG. 4: Density Map of distributions between normal EC cells and Small
Intestinal Neuroendocrine Tumors. Expression levels of indicated transcripts
as identified by
FS plotted on X- and Y-axes, with normal and neoplastic samples scattered
according to
respective gene pair expressions, distribution densities based on average
Euclidean distance
(difference in expression) between samples were colorized green (normal) and
red (neoplastic).
Blue areas indicate a region of transition between normal and neoplastic
groups.
20b
CA 2828878 2018-06-11
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
[0082] FIG. 5: Decision Tree classifying primary Small Intestinal
Neuroendocrine
Tumors. Expression levels of NAP ILI and Ki-67 were identified as principle
discriminators in
the Decision Tree classifier using Feature Selection. The model was
constructed by correlating
values of WAPITI and Ki-67 to primary tumor subtypes. Percentages in
parenthesis indicate the
occurrence frequencies of primary small intestinal neuroendocrine tumor
subtypes.
[0083] FIG. 6: Density Map of distributions between primary Small Intestinal
Neuroendocrine Tumors and their metastases. Expression levels of Kiss],
NAP1L1, MAGE-
D2, and CgA transcripts as identified by the FS algorithm plotted on X- and Y-
axis. Primary
Small Intestinal Neuroendocrine tumor subtypes (WDNETs, WDNECs, PDNECs) and
respective metastases (METs) scattered according to their respective gene pair
expressions (6A-
C). Distribution densities based on the average Euclidean distance (difference
in expression)
between samples were colorized blue (primary tumors) and red (metastases).
Green areas
indicate a region of transition between primary tumor subtypes and respective
metastases.
[0084] FIG. 7: Evaluation of the classifier performance in the test and
training sets.
Graph shows percentage of correctly-validated samples in training and test
sets, showing Normal
EC cells cross-validated with the 77% accuracy and predicted in an independent
test set with
76% accuracy (p = 0.84). Localized NETs were cross-validated with 78% accuracy
and
predicted with 63% accuracy in the test set (p = 0.25). Malignant NETs were
cross-validated
with 83% accuracy and predicted with 83% accuracy in an independent set (p =
0.80)
[0085] FIG. 8: Principal component analysis (PCA) and expression of marker
genes in
NETs, adenocarcinomas and normal tissues. 8A. Transcript expressions of the 21
marker
gene panel reduced to 3 principal components that capture most of the variance
(83%) within the
dataset. Each centroid (average expression) corresponds to the transcript
expression profile of
the sample as given by its principal component vector. In this representation,
proximity of
separation between centroids is indicative of the degree of similarity. Thus,
the marker gene
panel can successfully distinguish adenocarcinomas (breast, colon, pancreas),
normal SI mucosa,
normal EC cells, and primary and metastatic NET subtypes. Of note, normal EC
cells have a
substantially different genetic profile to normal SI mucosa and neoplastic
tissue. 8B. An analysis
of the proportion of samples that express each of the marker genes,
demonstrated that
significantly more NET samples (>95%) were positive for 16 of the marker genes
compared to
adenocarcinomas (AC). Genes highly expressed in both tumor types included
CTGF, FZD7,
NRP2, PNMA2 and survivin. NML=normal SI mucosa, NML_EC=normal EC cell,
21
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
MET=metastasis, WDNET=well differentiated NET; WDNEC=well differentiated
neuroendocrine carcinoma; PDNET=poorly differentiated NET; PDNEC=poorly
differentiated
NEC. *p<0.002 Si NETs versus adenocarcinomas (Fisher's exact test).
[0086] FIG. 9: Heatmap of correlation coefficients and relationship network of
highly
correlated gene pairs. 9A. Pearson's correlation coefficients (R2) for each
gene across all tissue
types were calculated and represented as a heatmap with the lowest value (-
0.03) represented in
black, medium (0.4) in dark grey, and highest (1) in light grey. 9B. A network
of co-expression
was constructed such that transcript pairs with R2>0.40 were connected by an
edge. Actual R2
values are superimposed on every edge.
[0087] FIG. 10: Volcano plots of gene ranks and significance (p) values for a
t-test. 10A.
A two-sample t-test was computed to identify differentially expressed genes in
1) EC cells,
normal SI mucosa, and primary and metastatic tissues; 2) primary NET subtypes;
3) metastatic
NET subtypes. In normal EC cells compared to normal SI mucosa, transcript
expression of the
classic neuroendocrine marker Tphl, was significantly higher (p<0.001, S=0.7).
10B. Compared
to normal SI mucosa, neoplastic tissue expressed higher transcript levels of
CgA and GRIA-2,
however CgA expression was not significantly altered (p=0.07, S=0.39) between
neoplastic
tissue and normal EC cells. 10C. There were no differentially expressed genes
between all
metastases as a group and all the different primary NET subtypes when analyzed
as a group.
10D. There were no differentially expressed transcripts between PDNET-PDNEC
and WDNET-
PDNEC, and in WDNEC-PDNEC, MAGE-D2 was the only significant marker (p=0.009,
S=1.03). CgA, Kiss], NRP2, and Tphl were differentially expressed between all
metastasis
subtypes.
[0088] FIG. 11: Transcript expression in whole blood. Housekeeping genes (ALG-
9,
TFCP2, ZNF410) identified in plasma after Trizol mRNA isolation (11A) and the
QIAamp RNA
Blood Mini Kit approach (11B), showing identification of housekeeping genes in
significantly
more samples (8/15 versus 2/15, p=0.05) after isolation with the QIAamp RNA
Blood Mini Kit
approach. Transcript expression levels of the same 3 housekeeping genes and 11
NET biomarker
genes were evaluated by PCR in mRNA prepared from whole blood from 3 healthy
donors
(normal samples), showing highly correlated detected gene expression levels
across samples
(11C).
[0089] FIG. 12. Average combined transcript expression of each gene
(12A).Transcripts
exhibited a low variability: 0.04-0.45 (median 0.12). Principal component
analysis of average
22
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
marker gene expression for all samples (12B). The mathematical algorithms
(SVM, LDA,
KNN and NB) identified that the correct calls were made for times 0, 30mins, 1
hr, 2 hr and 4 hr.
Inconsistent call rates occurred between 8 ¨ 48 hrs indicating the optimal
time for storage in a
refrigerator prior to freezing is 0-4 hrs.
[0090] FIG. 13: Identification of the most appropriate housekeeping gene and
determination of the effect of feeding on ALG-9 transcripts in whole blood.
Transcript
expression of 5 housekeeping genes was evaluated in 5 healthy controls. ALG-9
was identified
to have the least variation (13A). ALG-9 expression was measured as a function
of time after
feeding, showing no significant alteration after feeding (up to 4 hrs) (13B).
[0091] FIG. 14: Topological analysis of candidate house-keeping genes
mapped to the blood
interactome (7,000 genes, 50,000 interactions): Degree (14A), Betweeness (14B)
and Clustering
(14C). Genes with the lowest values in each category included TXNIP, ACTB,
TOX4 and
TPTI. Analysis of blood- and tissue-associated house-keeping genes identified
potential
candidate genes for normalization protocols.
[0092] FIG. 15: Raw CT values plotted as a function of either tissue-derived
or blood-
derived candidate house-keeping genes. Genes with the least variation included
ALG9, ARF1,
ATG4B, RHDA, and SKPl. Mean and SD are included. A value of 40 was assigned to
samples
with no amplification. Samples with no gene expression are given a value of 40
(e.g., 4 samples
amplified using MORF4L1). Analysis of candidate house-keeping genes identified
a relatively
small number (n=6) that exhibited low variability and were candidates for
development of
normalization protocols.
[0093] FIG. 16: M-values for each of the candidate house-keeping genes
calculated using
the geNorm program. ALG9 was the most stable of the tissue-derived genes. Nine
of the 10
blood-derived genes (except SERPI) were considered robust. Robust markers
(dotted boxes).
[0094] FIG. 17: PCR efficiency curves plotted for each of the candidate house-
keeping
genes. Efficient amplification occurs between 0.9-1Ø Values lower than 0.9
indicate sub-
optimal primer binding and inefficient amplification. Values above 1.0
identify over-
amplification, presumably through less than specific primer binding. Genes
with appropriate
efficiency included 18S, ALG9, and TPT I. Mean SD. n=3. A small number of
candidates
(n=3) exhibited efficacy as house-keeping genes.
[0095] FIG. 18: Variance in amplification kinetics for the house-keeping and
target genes.
Values ¨0.1 demonstrate similar PCR efficiencies and indicate the house-keeper
can be used in
23
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
comparative CT methods. ALG9 was the only house-keeping gene to exhibit an
acceptable
efficiency for normalization protocols.
[0096] FIG. 19: Variance in target gene expression in normal samples using
either a geNorm
protocol (using 18S, ALG9 and GAPDH as house-keeping genes) or AACT with ALG9.
The
latter exhibited a significantly lower co-efficient of variation for each of
the target genes and
¨60% of genes exhibited a normal distribution. *p<0.004 (Mann-Whitney test).
The optimal
method for normalization was AACT
[0097] FIG. 20: Identification of tissue-associated genes from U133A and
HUGE arrays.
PCA of GEP-NENs compared to other neoplasia (breast, colon, prostate and
liver) identified the
transcriptome was most similar to Crohn's Disease (20A). Subtraction of
transcript expression
associated with other neoplasia identified a specific GEP-NEN gene signature
(modeled as an
interactome ¨ 20B). Back-analysis to the tissue arrays identified 21 novel
markers which
differentiated control from GEP-NENs both by hierarchical cluster analysis
(20C) and principal
component analysis (PCA) (20D). SI-NENs exhibit a different transcript
spectrum to other
cancers. A NEN-specific gene signature is identifiable, which can
differentiate these tumors
from control samples.
[0098] FIG. 21: Gene expression profiles in the Blood (21A,D), "In-house"
(21B, E), and
Public datasets (21C, F). Analysis of transcript expression identified that
samples from both
GEP-NEN tissue and blood could be differentiated from controls. This indicates
that each of
these compartments contain a definable GEP-NEN molecular fingerprint that can
be measured
and used to distinguish tumors from controls.
[0099] FIG. 22: Correlation profiles of transcript changes in blood and tissue
samples. Both
tissue databases were highly correlated (R=0.59, 22A) but lower correlations
were noted
between blood transcriptomes and either the "In-house" dataset (R=-0.11, 22B)
or the Public
dataset (R=-0.05, 22C). The common genes identified in both the tissue and
blood samples
provided a group of candidate marker transcripts which we then examined in
blood.
[00100] FIG. 23: A: Correlated and anti-correlated biological processes in GEP-
NEN
transcriptomes from peripheral blood and tumor tissue samples. B & C: Eighty-
five genes
associated with tumor function (intracellular signaling and transcription and
regulation of cell
death), were up-regulated in both tissue and blood samples. This group was
considered to
represent evaluable candidate circulating biomarkers.
24
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
[00101] FIG. 24: Expression of the 22 genes with low paralog numbers (0-3) are
-3 times
more central in the blood interactome compared to all other genes (-6,000
genes). This group of
specific genes, with few relatives, is present in blood and can be considered
as potential markers
of neuroendocrine neoplasia.
[00102] FIG. 25: FACS of AO/APC-CD164 dual stained whole blood from a patient
with
metastatic NETs. Flow cytometric analysis following AO (acridine orange) /APC-
CD164 dual-
staining of whole blood from a patient with metastatic NET showed a distinct
population of cells
consistent in size with NET cells (P1, arrow: 25A) exhibiting the
characteristic AO/APC
positivity of NETs (25B). This population of cells was collected (25C);
immunostaining with
anti-TPH confirmed that the cells were NET cells (25C-inset).
[00103] FIG. 26: Relationship between whole blood PCR marker levels and FACS-
collected circulating NETs and tissue. Whole blood expression levels of
biomarker transcripts
were highly correlated (p<0.0001) with FACS-sorted samples (representing
circulating tumor
cells (26A)) and with tissue (26B), confirming that whole blood is an
appropriate compartment
for measuring NET transcripts.
[00104] FIG. 27: ROCs and sensitivity and specificities for predicting NETs.
ROCs and
AUCs for selected genes (top 3 panels) and summed transcripts (V1) (bottom
left) were
calculated in Yale samples (NETs and controls) as described in Example 5D. Use
of predicted
cut-offs were tested in NETs from Berlin and Uppsala and sensitivities and
specificities are
provided (bottom right).
[00105] FIG. 28: Reproducibility studies of target genes in blood. The
reproducibility of
the marker gene ALG9 and the target gene, FZD7, demonstrated high correlation:
R2: 0.92-0.97,
p<0.0001 (28A-B). Intra- and inter-assay reproducibilities were high for
normalized FDZ7
(28C-D, CV = 2.28-3.95%); no differences were noted between normalized FZD7 in
controls
and tumor samples (28D), demonstrating the significant reproducibility of
blood measurements.
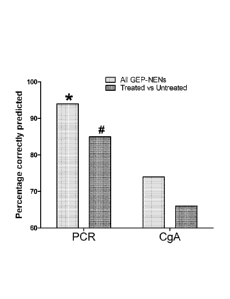
[00106] FIG. 29: Performance of the marker gene panel in differentiating
Normal tissue from
GEP-NENs (Treated and Untreated). All four mathematical algorithms exhibited
similar
performance metrics of -88%.
[00107] FIG. 30: Correct call rates for each of the mathematical algorithms
(SVM, LDA,
KNN and Bayes) in each of the four independent sets. The 51 marker panel had
significantly
more correct calls (20% better) than either the 25 or 13 panel subsets.
Increasing the number of
the marker genes increased the sensitivity of detecting GEP-NENs in blood.
Mean SEM.
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
*p<0.008 vs. 13 and 25 panels (Yates value 6.8-14.7;# Fishers 2-tailed exact
probability test
<0.005).
[00108] FIG. 31: Alterations in blood PCR marker levels and CgA during
surgical
resection. Tumor excision significantly reduced expression levels of a panel
of biomarkers
("PCR") as described in Example 5H, when measured 2 weeks post-operation
(31A). CgA levels
were variable (31B). horizontal = mean. n=9 patients.
[00109] FIG. 32: Alterations in blood PCR marker levels and CgA during
Octreotide
LAR therapy. Octreotide LAR significantly reduced blood expression levels of
the panel of
biomarkers ("PCR") as described in Example 5H; expression remained suppressed
over the time
course (32A). CgA levels were variable before being reduced by 6 months (32B).
*p<0.02
versus BEFORE. #p=0.06 vs. 1 MONTH. Horizontal line = mean. MON = month. n=8
patients.
[00110] FIG. 33: Alterations in blood PCR marker levels and CgA after
CryoAblation.
Expression levels of CgA and a 13-biomarker panel (PCR+) in patient SK before
and at
various times following cryoablation, as described in Example 5H, with changes
in biomarker
expression correlating with the appearance of micrometastases.
[00111] FIG. 34: Alterations in blood PCR marker levels and CgA during
surgical
resection and Octreotide LAR therapy. Expression levels of CgA and an NET
biomarker
panel levels in patient BG, as described in Example 5H, measured out to 2
weeks post-operation
and following Octreotide LAR.
[00112] FIG. 35: Overall percentage correct calls for patients in complete
remission (Group
I: complete responders [CR], n=12), considered clinically as exhibiting stable
disease (SD)
following surgery (n=42, Group II ¨ SD-Sx) or after treatment with long-acting
somatostatin
analogs (LAR: n=78. Group III ¨ SD-LAR). *This includes pasireotide: n=1 and
everolimus:
n=4). The PCR test exhibited between 90-100% correct call rates for treated
samples.
[00113] FIG. 36: Mathematical analyses including SVM, LDA, KNN and Bayes
demonstrated that the 13 marker panel could differentiate between stable and
progressive disease
with sensitivities of ¨73%.
[00114] FIG. 37: Comparison of CgA DAKO levels across control and GEP-NEN
(both
untreated and treated) blood samples (n=130). Differences were noted between
untreated and
treated samples using the Student's t-test (37A) or non-parametric analyses
(37B). Red crosses
represent outliers (37A) and the y-scale is transformed logarithmically for
visualization purposes
26
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
(37A, B). CgA levels can consistently distinguish between normal and untreated
groups but
exhibit significant overlap with treated samples.
[00115] FIG. 38: Utility of CgA ELISA for correctly detecting CEP-NENs and
differentiating treated from untreated samples. Using 19 U/L as a cut-off (as
per DAKO criteria),
the overall percentage correct calls for GEP-NENs and controls was 70%, and
the performance
metrics were better in untreated patients compared to treated patients
(sensitivity 63% vs. 45%).
CgA levels best identify untreated patients and samples from individuals with
no disease
(controls).
[00116] FIG. 39: Comparison of correct call rates for circulating CgA DAKO
levels and the
individual algorithms using the PCR-based approach across control and GEP-NEN
(both
untreated and treated) blood samples (n=130). Call rates were significantly
higher for the PCR-
based test (-90-95% for each of the algorithms) compared to ¨50% of CgA (FIG.
39A).
Inclusion of CgA values in the algorithm did not increase the correct call
rates, and was
associated with a decrease in correct calls for the KNN algorithm (FIG. 39B).
[00117] FIG. 40: PCR score for a blood sample obtained from a normal control
(black dotted
line: PCR Score = 15, called "normal") and from Case 1 presenting with
mesenteric metastases
(red dotted line: PCR Score 68: called "tumor untreated". The population
distributions are in
solid lines. This provides an illustration of the relationship between the
algorithm calls and a
Score (Transcript index).
[00118] FIG. 41: PCA of SI-NENs (n=46) and PNENs (n=18) identified that the 51
marker
panel could differentiate between pancreatic NENs and small bowel NENs. (41A).
A variety of
mathematical algorithms including SVM, LDA, KNN and Bayes demonstrated these
two tumor
groups could be differentiated with an overall sensitivity of ¨92% (FIG. 41B).
The signature for
SI-NENs is different to PNENs.
[00119] FIG. 42: PCA of GEP-NENs (n=64) and GI cancers (n=42) identified that
the 51
marker panel could differentiate between the two neoplasia types (FIG. 42A).
Mathematical
analyses including SVM, LDA, KNN and Bayes demonstrated these two tumor types
could be
differentiated with sensitivities of ¨83% (FIG. 42B). The signature for GEP-
NENs is different
to GI cancers.
[00120] FIG. 43: Comparison of the PCR-based approach and CgA DAKO levels
across
control and CEP-NEN (both untreated and treated) blood samples (n=130). Call
rates were
significantly higher for the PCR-based test for identifying either a CEP-NEN
or for
27
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
differentiating between treated and untreated samples. *p<0.0005 vs. CgA,
#p<0.02 vs. CgA.
The PCR blood test is significantly more accurate than measurement of CgA
levels to detect
tumors and differentiate treated from non-treated patients.
Detailed description
A. Definitions
[00121] Unless otherwise defined, all terms of art, notations and other
scientific terminology
used herein are intended to have the meanings commonly understood by those of
skill in the art
to which this invention pertains. In some cases, terms with commonly
understood meanings are
defined herein for clarity and/or for ready reference, and the inclusion of
such definitions herein
should not necessarily be construed to represent a substantial difference over
what is generally
understood in the art. The techniques and procedures described or referenced
herein are
generally well understood and commonly employed using conventional methodology
by those
skilled in the art, such as, for example, the widely utilized molecular
cloning methodologies
described in Sambrook et al., Molecular Cloning: A Laboratory Manual 2nd.
edition (1989)
Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. As appropriate,
procedures
involving the use of commercially available kits and reagents are generally
carried out in
accordance with manufacturer defined protocols and/or parameters unless
otherwise noted.
[00122] As used herein, the term "GEP-NEN biomarker" and "NET biomarker" refer
synonymously to a biological molecule, such as a gene product, the expression
or presence of
which (e.g., the expression level or expression profile) on its own or as
compared to one or more
other biomarkers (e.g., relative expression) differs (i.e., is increased or
decreased) depending on
the presence, absence, type, class, severity, metastasis, location, stage,
prognosis, associated
symptom, outcome, risk, likelihood of treatment responsiveness, or prognosis
of GEP-NEN
disease, or is associated positively or negatively with such factors or the
prediction thereof.
[00123] As used herein, the term "polynucleotide" or nucleic acid molecule
means a
polymeric form of nucleotides of at least 10 bases or base pairs in length,
either ribonucleotides
or deoxynucleotides or a modified form of either type of nucleotide, and is
meant to include
single and double stranded forms of DNA. As used herein, a nucleic acid
molecule or nucleic
acid sequence of the invention that serves as a probe in a microarray analysis
preferably
28
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
comprises a chain of nucleotides, more preferably DNA and/or RNA. In other
embodiments a
nucleic acid molecule or nucleic acid sequence of the invention comprises
other kinds of nucleic
acid structures such as for instance a DNA/RNA helix, peptide nucleic acid
(PNA), locked
nucleic acid (LNA) and/or a ribozyme. Hence, as used herein the term "nucleic
acid molecule"
also encompasses a chain comprising non-natural nucleotides, modified
nucleotides and/or non-
nucleotide building blocks which exhibit the same function as natural
nucleotides.
[00124] As used herein, the term "polypeptide" means a polymer of at least 10
amino acids.
Throughout the specification, standard three letter or single letter
designations for amino acids
are used.
[00125] As used herein, the terms "hybridize," "hybridizing." "hybridizes,"
and the like, used
in the context of polynucleotides, are meant to refer to conventional
hybridization conditions,
preferably such as hybridization in 50% formamide/6XSSC/0.1% SDS/100 pg/m1
ssDNA, in
which temperatures for hybridization are above 37 degrees C and temperatures
for washing in
0.1XSSC/0.1% SDS are above 55 degrees C, and most preferably to stringent
hybridization
conditions.
[00126] In the context of amino acid sequence comparisons, the term "identity"
is used to
express the percentage of amino acid residues at the same relative position
which are the same.
Also in this context, the term "homology" is used to express the percentage of
amino acid
residues at the same relative positions which are either identical or are
similar, using the
conserved amino acid criteria of BLAST analysis, as is generally understood in
the art. Further
details regarding amino acid substitutions, which are considered conservative
under such
criteria, are provided below.
[00127] Additional definitions are provided throughout the subsections which
follow.
B. GEP-NEN disease and biomarkers
[00128] Diagnosis and prognosis of GEP-NEN has been difficult, in part due to
the prosaic
symptoms and syndromes of the disease, such as carcinoid syndrome, diarrhea,
flushing,
sweating, bronchioconstruction, GI bleeding, cardiac disease, intermittent
abdominal pain, which
often remain silent for years. Available diagnostic methods include anatomical
localization,
such as by imaging, e.g., X-ray, gastrointestinal endoscopy, abdominal
computed tomography
(CT), combined stereotactic radiosurgery (SRS) / CT, and MRI, and detection of
some gene
products. Known methods are limited, for example by low specificity and/or
sensitivity and/or
29
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
in the ability to detect early-stage disease. Detection of single biomarkers
has not been entirely
satisfactory, for example, to identify malignancy in human blood samples and
predict complex
outcomes like fibrosis and metastasis. See Michiels S. Koscielny S, Hill C, -
Interpretation of
microarray data in cancer," Br J Cancer 2007;96(8):1155-8. Limitations in
available methods
have contributed to difficulties in pathological classification, staging, and
prediction, treatment
developing and monitoring therapeutic effects. Among the embodiments provided
herein are
methods and compositions that address these limitations.
[00129] In one aspect, the provided invention relates to the detection and
identification of
GEP-NEN biomarkers and panels of such biomarkers, for example, in biological
samples.
Provided are methods and compositions (e.g., agents, such as polynucleotides),
for detecting,
determining expression levels of, and recognizing or binding to the
biomarkers, in biological
samples, typically blood samples, and for detecting and analyzing expression
profiles
(signatures) of panels of biomarkers. Also provided are compositions and
combinations
containing the agents, including sets (panels) of agents, systems, and kits,
for use in the provided
methods.
[00130] Also provided are methods and compositions for the detection,
enrichment, isolation,
and purification of GEP-NEN cells, e.g., circulating GEP-NEN cells (CNCs), for
example, from
a blood sample, culture, cell mixture, fluid, or other biological sample,
based on the expression
of one or more of the GEP-NEN biomarkers.
[00131] Also provided are models and biomathematical algorithms, e.g.,
supervised learning
algorithms, and methods using the same, for prediction, classification, and
evaluation of GEP-
NEN and associated outcomes, for example, predicting degree of risk,
responsiveness to
treatment, metastasis or aggressiveness, and for determining GEP-NEN sub-type.
[00132] Detection of the biomarkers using the provided embodiments is useful
for improving
GEP-NEN diagnostics and prognostics, and to inform treatment protocols. In
some aspects,
detection of the biomarkers and/or expression levels by the provided
embodiments confirms or
indicates the presence, absence, stage, class, location, sub-type,
aggressiveness, malignancy,
metastasis, prognosis, or other outcome of GEP-NEN, or a GEP-NEN cell, such as
a circulating
GEP-NEN cell (CNC). The provided methods and compositions may be used for
tumor
localization, and for predicting or detecting metastases, micrometastases, and
small lesions,
and/or for determining degree of risk, likelihood of recurrence, treatment
responsiveness or
remission, and informing appropriate courses of treatment. For example,
detecting the
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
biomarkers, e.g., in circulation may be used to detect early-stage and primary
GEP-NENs (e.g.,
to identify GEP-NEN disease or metastases in a patient previously deemed -
negative" by
another approach, such as anatomic localization).
[00133] The provided methods and compositions may be used for designing,
implementing,
and monitoring treatment strategies, including patient-specific treatment
strategies. In one
example, detected expression levels of the GEP-NEN biomarkers serve as
surrogate markers for
treatment efficacy, e.g., to monitor the effects of surgical therapy (e.g.,
removal of tumors),
targeted medical therapy (e.g., inhibition of tumor secretion/ proliferation),
and other therapeutic
approaches, by detecting remission or recurrence of tumors, even in the form
of small
micrometastases. The methods also may be used in evaluating clinical symptoms
and outcomes,
and for histological grading and molecular characterization of GEP-NENs.
C. GEP-NEN biomarkers
[00134] The provided biomarkers including GEP-NEN biomarkers, and panels
(sets) of the
same. Among the provided GEP-NEN biomarkers are gene products, such as DNA,
RNA, e.g.,
transcripts, and protein, which are differentially expressed in GEP-NEN
disease, and/or in
different stages or sub-types of GEP-NEN, or in different GEP-NEN tumors, such
as gene
products differentially expressed in metastatic versus primary tumors, tumors
with different
degrees of aggressiveness, high versus low-risk tumors, responsive versus non-
responsive
tumors, tumors exhibiting different pathological classifications and/or
likelihood of response to
particular courses of treatment, as well as those associated with features of
GEP-NEN disease,
stage, or type, or with neuroendocrine cells or related cell-types.
[00135] For example, the biomarkers include gene products whose expression is
associated
with or implicated in tumorogenicity, metastasis, or hormone production, or a
phenotype of
primary or metastatic GEP-NEN, such as adhesion, migration, proliferation,
apoptosis,
metastasis, and hormone secretion, and those associated with neoplasia or
malignancy in
general. The biomarkers also include gene products expressed in related normal
tissues, such as
neuroendocrine cells, the small intestine (SI) mucosa, and enterochromaffin
(EC) cells.
[00136] Among the biomarkers are GEP-NEN cell secretion products, including
hormones
and amines, e.g., gastrin, ghrelin, pancreatic polypeptide, substance P,
histamine, and serotonin,
and growth factors such as tumor growth factor-beta (TGF-I3) and connective
tissue growth
31
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
factor (CTGF), which are detectable in the circulation. Secretion products can
vary with tumor
sub-type and origin.
[00137] In one example, the biomarkers are gene products associated with
regulatory
genotypes (i.e., adhesion, migration, proliferation, apoptosis, metastasis,
and/or hormone
secretion) that underlay various GEP-NEN subtypes, stages, degrees of
aggressiveness, or
treatment responsiveness.
[00138] Also among the GEP-NEN biomarkers are gene products differentially
expressed in
primary GEP-NENs and hepatic metastases as compared to normal small bowel
mucosa and
pure preparations of EC cells See Modlin etal., "Genetic differentiation of
appendiceal tumor
malignancy: a guide for the perplexed," Ann Surg 2006;244(1):52-60; Kidd M, et
al., "The role
of genetic markers, NAP 1L1, MAGE-D2 and MTAL in defining small intestinal
carcinoid
neoplasia." Annals of Surgical Oncology 2006;13:253-62; Kidd M etal., "Q RT-
PCR detection
of Chromogranin A: A new standard in the identification of neuroendocrine
tumor disease,"
Annals of Surgery 2006;243:273-80.
[00139] The GEP-NEN biomarkers include: AKAP8L (A kinase (PRKA) anchor protein
8-
like), ATP6V1H (ATPase, H+ transporting, lysosomal 50/57kDa, VI subunit H),
BNIP3L
(BCL2/adenovirus ElB 19kDa interacting protein 3-like), C2lorf7 (chromosome 21
open
reading frame 7), COMMD9 (COMM domain containing 9), ENPP4 (ectonucleotide
pyrophosphatase/phosphodiesterase 4). FAM13A (family with sequence similarity
13, member
A). FLJ10357 (Rho guanine nucleotide exchange factor 40), GLT8D1
(glycosyltransferase 8
domain containing 1). HDAC9 (histone deacetylase 9), HSF2 (heat shock
transcription factor 2),
LE01 (Pafl/RNA polymerase II complex component, homolog (S. cerevisiae)),
MORF4L2
(MORF4L2 mortality factor 4 like 2), NOL3 (nucleolar protein 3 (apoptosis
repressor with
CARD domain)), NUDT3 (nudix (nucleoside diphosphate linked moiety X)-type
motif 3),
OAZ2 (ornithine decarboxylase antizyme 2), PANK2 (pantothenate kinase 2),
PHF21A (PHD
finger protein 21A), PKD1 (polycystic kidney disease 1 (autosomal dominant)),
PLD3
(phospholipase D family, member 3), PQBP1 (polyglutamine binding protein 1),
RNF41
(polyglutamine binding protein 1), RSF1 (remodeling and spacing factor 1),
RTN2 (reticulon 2),
SMARCD3 (SWI/SNF related, matrix associated, actin dependent regulator of
chromatin,
subfamily d, member 3p), SPATA7 (spermatogenesis associated 7). SST1
(somatostatin receptor
1), SST3 (somatostatin receptor 3), SST4 (somatostatin receptor 4), SSTS
(somatostatin receptor
5), TECPR2 (tectonin beta-propeller repeat containing 2), TRMT112 (tRNA
methyltransferase
32
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
11-2 homolog (S. cerevisiae)), VPS13C (vacuolar protein sorting 13 homolog C
(S. cerevisiae)),
WDFY3 (WD repeat and FYVE domain containing 3), ZFHX3 (zinc finger homeobox
3),
ZXDC (ZXD family zinc finger C), ZZZ3 (zinc finger, ZZ-type containing 3),
Amyloid beta
(A4) precursor-like protein 2 (APLP2); v-raf murine sarcoma 3611 viral
oncogene homolog
(ARAF1); v-raf murine sarcoma viral oncogene homolog BI (BRAF1); CD59;
Chromogranin A
(CgA, also called parathyroid secretory protein 1, CHGA); connective tissue
growth factor
(CTGF); chemokine (C-X-C motif) ligand 14 (CXCL14); frizzled homolog 7 (FZD7);
glutamate
receptor, ionotropic, AMPA 2 (GRIA2); homeobox C6 (HOXC6); Ki-67; KiSS-1
metastasis-
suppressor (Kiss 1); v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog
(KRAS); melanoma
antigen family D, 2 (MAGE-D2); metastasis associated 1 (MTA1); nucleosome
assembly
protein 1-like 1 (NAP1L1); NK2 transcription factor related, locus 3 (e.g.,
Homo Sapiens NK2
transcription factor related. locus 3 (Drosophila)) (NKX2-3); neuropilin 2
(NRP2); olfactory
receptor, family 51, subfamily E. member 1 (0R51E1); paraneoplastic antigen
MA2 (PNMA2);
protein tyrosine phosphatase, receptor type, N polypeptide 2 (PTPRN2); v-raf-1
murine
leukemia viral oncogene homolog 1 (RAF1); secretogranin V (7B2 protein)
(SCG5);
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1
(SPOCK1); apoptosis
inhibitor survivin gene (BIRC5; API4; EPR-I) (Survivin); tryptophan
hydroxylase 1 (TPH1),
solute carrier family 18 (vesicular monoamine), member 1 (VMAT1); solute
carrier family 18
(vesicular monoamine), member 2 (VMAT2); and X2BTB48 (serpin peptidase
inhibitor, clade A
(alpha-1 antiproteinase, antitrypsin), member 10), including gene products
typically human gene
products, including transcripts, mRNA, cDNA, coding sequences, proteins and
polypeptides, as
well as polynucleotides (nucleic acids) encoding the proteins and
polypeptides, including
naturally occurring variants, e.g., allelic variants, splice variants,
transcript variants, and single
nucleotide polymorphism (SNP) variants. For example, the biomarkers include
polynucleotides,
proteins, and polypeptides having the sequences disclosed herein, and
naturally occurring
variants thereof.
[00140] The GEP-NEN biomarkers further include CD164. In another aspect, the
biomarkers
include NALP, e.g., products of the caspase-3 activating apoptosis gene and
apoptotic marker,
NALP.
[00141] APLP2 biomarkers include human APLP2 gene products, including natural
variants,
e.g., allelic variants, and homologs and analogs thereof. In one example, the
APLP2 biomarker
is a polynucleotide having the nucleotide sequence set forth in SEQ ID NO: 1
(referenced at
33
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
GenBank gi number 214010177) or containing a protein-coding portion thereof,
(e.g., the open
reading frame at nucleotides 158-2449 of SEQ ID NO: 1), a natural variant
thereof, or a protein
encoded by such a polynucleotide.
[00142] The ARAF1 biomarkers include human ARAFl gene products, including
natural
variants, e.g., allelic variants, and homologs and analogs thereof. In one
example, the ARAFl
biomarker is a polynucleotide having the nucleotide sequence set forth in SEQ
ID NO: 2
(referenced at GenBank gi number 283484007), or containing a protein-coding
portion thereof,
(e.g., the open reading frame at nucleotides 195-2015 of SEQ ID NO: 2), a
natural variant
thereof, or a protein encoded by such a polynucleotide.
[00143] The BRAF1 biomarkers include BRAF1 gene products, including natural
variants,
e.g., allelic variants, and homologs and analogs thereof. In one example, the
BRAF1 biomarker
is a polynucleotide having the nucleotide sequence set forth in SEQ ID NO: 3
(referenced at
GenBank gi number 187608632), or containing a protein-coding portion thereof,
(e.g., the open
reading frame at nucleotides 62-2362 of SEQ ID NO: 3), a natural variant
thereof, or a protein
encoded by such a polynucleotide.
[00144] The CD59 biomarkers include human CD59 gene products, including
natural
variants, e.g., allelic variants, and homologs and analogs thereof. In one
example, the CD59
biomarker is a polynucleotide having the nucleotide sequence set forth in SEQ
ID NO: 4
(referenced at GenBank gi number 187829037), or containing a protein-coding
portion thereof,
(e.g., the open reading frame at nucleotides 278-664 of SEQ ID NO: 4), a
natural variant thereof,
or a protein encoded by such a polynucleotide.
[00145] The CgA biomarkers include human CGA or CHGA gene products, including
natural
variants, e.g., allelic variants, and homologs and analogs thereof. In one
example, the CgA
biomarker is a polynucleotide having the nucleotide sequence set forth in SEQ
ID NO: 5
(referenced at GenBank gi number 33990769), or containing a protein-coding
portion thereof,
(e.g., the open reading frame at nucleotides 1 to 1374 of SEQ ID NO: 5), a
natural variant
thereof, or a protein encoded by such a polynucleotide. Human CgA encodes a
water soluble
acidic glycoprotein stored in the secretory granules of neuroendocrine cells
and detectable in
plasma.
[00146] The CTGF biomarkers include human CTGF gene products, including
natural
variants, e.g., allelic variants, and homologs and analogs thereof. In one
example, the CTGF
biomarker is a polynucleotide having the nucleotide sequence set forth in SEQ
ID NO: 6
34
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
(referenced at GenBank gi number 98986335), or containing a protein-coding
portion thereof,
(e.g., the open reading frame at nucleotides 207-1256 of SEQ ID NO: 6), a
natural variant
thereof, or a protein encoded by such a polynucleotide.
[00147] The CXCL14 biomarkers include human CXCL1 4 gene products, including
natural
variants, e.g., allelic variants, and homologs and analogs thereof. In one
example, the CXCL14
biomarker is a polynucleotide having the nucleotide sequence set forth in SEQ
ID NO: 7
(referenced at GenBank gi number 208022628), or containing a protein-coding
portion thereof,
(e.g., the open reading frame at nucleotides 466-801of SEQ ID NO: 7), a
natural variant thereof,
or a protein encoded by such a polynucleotide.
[00148] The FZD7 biomarkers include human FZD7 gene products, e.g., Homo
sapiens
frizzled homolog 7 (Drosophila) (FDZ7), including natural variants, e.g.,
allelic variants, and
homologs and analogs thereof. In one example, the FDZ7 biomarker is a
polynucleotide having
the nucleotide sequence set forth in SEQ ID NO: 8 (referenced at GenBank gi
number 4503832),
or containing a protein-coding portion thereof, (e.g., the open reading frame
at nucleotides 62-
1786of SEQ ID NO: 8 a natural variant thereof, or a protein encoded by such a
polynucleotide.
[00149] The GRIA2 biomarkers include human GRIA2 gene products, including
natural
variants, e.g., allelic variants, and homologs and analogs thereof. In one
example, the GRIA2
biomarker is a polynucleotide having the nucleotide sequence set forth in SEQ
ID NO: 9
(referenced at GenBank gi number 134304849), or containing a protein-coding
portion thereof,
(e.g., the open reading frame at nucleotides 460-3111 of SEQ ID NO: 9), a
natural variant
thereof, or a protein encoded by such a polynucleotide.
[00150] The homeobox C6 (HOXC6) biomarkers include human HOXC6 gene products,
including natural variants, e.g., allelic variants, and homologs and analogs
thereof. In one
example, the HOXC6 biomarker is a polynucleotide having the nucleotide
sequence set forth in
SEQ ID NO: 10 (referenced at GenBank gi number 93141222) or containing a
protein-coding
portion thereof, (e.g., the open reading frame at nucleotides 113-820 of SEQ
ID NO: 10), a
natural variant thereof, or a protein encoded by such a polynucleotide.
[00151] The Ki67 biomarkers include human Ki67 gene products, including
natural variants,
e.g., allelic variants, and homologs and analogs thereof. In one example, the
Ki67 biomarker is a
polynucleotide having the nucleotide sequence set forth in SEQ ID NO: 11
(referenced at
GenBank gi number 225543213)or containing the coding region thereof (e.g.,
nucleotides 196-
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
9966) of SEQ ID NO: 11), a natural variant thereof, or a protein encoded by
such a
polynucleotide.
[00152] The Kissl biomarkers include human KISS] gene products, including
natural
variants, e.g., allelic variants, and homologs and analogs thereof. In one
example, the KISS1
biomarker is a polynucleotide having the nucleotide sequence set forth in SEQ
ID NO: 12
(referenced at GenBank gi number 116829963), or containing a protein-coding
portion thereof,
(e.g., the open reading frame at nucleotides 155-571 of SEQ ID NO: 12), a
natural variant
thereof, or a protein encoded by such a polynucleotide.
[00153] The KRAS biomarkers include human KRAS gene products, including
natural
variants, e.g., allelic variants, and homologs and analogs thereof. In one
example, the KRAS
biomarker is a polynucleotide having the nucleotide sequence set forth in SEQ
ID NO: 13
(referenced at GenBank gi number 34485724) or containing the coding region
thereof (e.g.,
nucleotides 182-751 of SEQ ID NO: 13), a natural variant thereof, or a protein
encoded by such
a polynucleotide.
[00154] The MAGE-D2 biomarkers include human MAGE-D2 gene products, including
natural variants, e.g., allelic variants, and homologs and analogs thereof. In
one example, the
MAGE-D2 biomarker is a polynucleotide having the nucleotide sequence set forth
in SEQ ID
NO: 14 (referenced at GenBank gi number 29171703) or containing a protein-
coding portion
thereof, (e.g., the open reading frame at nucleotides 100-1920 of SEQ ID NO:
14), a natural
variant thereof, or a protein encoded by such a polynucleotide. MAGE-D2
encodes an adhesion-
associated protein.
[00155] The MTA1 biomarkers include human MTA1 gene products, including
natural
variants, e.g., allelic variants, and homologs and analogs thereof. In one
example, the MTA1
biomarker is a polynucleotide having the nucleotide sequence set forth in SEQ
ID NO: 15
(referenced at GenBank gi number 115527079) or containing the coding region
thereof (e.g.,
nucleotides 188-2335 of SEQ ID NO: 15), a natural variant thereof, or a
protein encoded by such
a polynucleotide. MTA, an estrogen-antagonistic breast cancer malignancy gene,
has been used
to identify progressive (metastatic) disease in other tumors including breast,
hepatocellular,
esophageal, gastric, and colorectal carcinomas.
[00156] The NAP1L1 biomarkers include human NAP1L1 gene products, including
natural
variants, e.g., allelic variants, and homologs and analogs thereof. In one
example, the NAPIL1
biomarker is a polynucleotide having the nucleotide sequence set forth in SEQ
ID NO: 16
36
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
(referenced at GenBank gi number 219842231) or containing a protein-coding
portion thereof,
(e.g., the open reading frame at nucleotides 413-1588 of SEQ ID NO: 16), a
natural variant
thereof, or a protein encoded by such a polynucleotide. NAP 1L1 is a mitosis-
regulatory gene
encoding a nuclear protein involved in chromatin assembly and DNA replication.
[00157] The NKX2-3 biomarkers include human NKX2-3 gene products, including
natural
variants, e.g., allelic variants, and homologs and analogs thereof. In one
example, the NKX2-3
biomarker is a polynucleotide having the nucleotide sequence set forth in SEQ
ID NO: 17
(referenced at GenBank gi number 148746210) or containing a protein-coding
portion thereof,
(e.g., the open reading frame at nucleotides 200-1294 of SEQ ID NO: 17), a
natural variant
thereof, or a protein encoded by such a polynucleotide.
[00158] The NRP2 biomarkers include human NRP2 gene products, including
natural
variants, e.g., allelic variants, and homologs and analogs thereof. In one
example, the NRP2
biomarker is a polynucleotide having the nucleotide sequence set forth in SEQ
ID NO: 18
(referenced at GenBank gi number 41872561) or containing a protein-coding
portion thereof,
(e.g., the open reading frame at nucleotides 792-3587 of SEQ ID NO: 18), a
natural variant
thereof, or a protein encoded by such a polynucleotide.
[00159] The OR51E1 biomarkers include human ORS 1E1 gene products, including
natural
variants, e.g., allelic variants, and homologs and analogs thereof. In one
example, the OR51E1
biomarker is a polynucleotide having the nucleotide sequence set forth in SEQ
ID NO: 19
(referenced at GenBank gi number 205277377) or containing a protein-coding
portion thereof,
(e.g., the open reading frame at nucleotides 145-1101 of SEQ ID NO: 19), a
natural variant
thereof, or a protein encoded by such a polynucleotide.
[00160] The PNMA2 biomarkers include human PNMA2 gene products, including
natural
variants, e.g., allelic variants, and homologs and analogs thereof. In one
example, the PNMA2
biomarker is a polynucleotide having the nucleotide sequence set forth in SEQ
ID NO: 20
(referenced at GenBank gi number 156766040) or containing a protein-coding
portion thereof,
(e.g., the open reading frame at nucleotides 771-1865 of SEQ ID NO: 20), a
natural variant
thereof, or a protein encoded by such a polynucleotide.
[00161] The PTPRN2 biomarkers include human PTPRN2 gene products, including
natural
variants, e.g., allelic variants, and homologs and analogs thereof. In one
example, the PTPRN2
biomarker is a polynucleotide having the nucleotide sequence set forth in SEQ
ID NO: 21
(referenced at GenBank gi number 194097439) or containing a protein-coding
portion thereof,
37
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
(e.g., the open reading frame at nucleotides 122-3169 of SEQ ID NO: 21), a
natural variant
thereof, or a protein encoded by such a polynucleotide.
[00162] The RAF1 biomarkers include human RAE] gene products, including
natural
variants, e.g., allelic variants, and homologs and analogs thereof. In one
example, the RAF]
biomarker is a polynucleotide having the nucleotide sequence set forth in SEQ
ID NO: 22
(referenced at GenBank gi number 189458830) or containing a protein-coding
portion thereof,
(e.g., the open reading frame at nucleotides 416-2362 of SEQ ID NO: 22), a
natural variant
thereof, or a protein encoded by such a polynucleotide.
[00163] The SCG5 biomarkers include human SCG5 gene products, including
natural
variants, e.g., allelic variants, and homologs and analogs thereof. In one
example, the SCG5
biomarker is a polynucleotide having the nucleotide sequence set forth in SEQ
ID NO: 23
(referenced at GenBank gi number 221139784) or containing a protein-coding
portion thereof,
(e.g., the open reading frame at nucleotides 118-756 of SEQ ID NO: 23), a
natural variant
thereof, or a protein encoded by such a polynucleotide.
[00164] The SPOCK1 biomarkers include human SPOCK1 gene products, including
natural
variants, e.g., allelic variants, and homologs and analogs thereof. In one
example, the SPOCK1
biomarker is a polynucleotide having the nucleotide sequence set forth in SEQ
ID NO: 24
(referenced at GenBank gi number 82659117) or containing a protein-coding
portion thereof,
(e.g., the open reading frame at nucleotides 152-1471 of SEQ ID NO: 24), a
natural variant
thereof, or a protein encoded by such a polynucleotide.
[00165] The Survivin biomarkers include human Survivin gene products,
including natural
variants, e.g., allelic variants, and homologs and analogs thereof. In one
example, the Survivin
biomarker is a polynucleotide having the nucleotide sequence set forth in SEQ
ID NO: 25
(referenced at GenBank gi number 59859877) or containing a protein-coding
portion thereof,
(e.g., the open reading frame at nucleotides 122-550 of SEQ ID NO: 25) or a
polynucleotide
having the protein-coding sequence (SEQ ID NO: 34) of nucleotides 2811-2921,
3174-3283,
5158-5275, 11955-12044 of GenBank gi number 2315862), a natural variant
thereof, or a protein
encoded by such a polynucleotide.
[00166] The TPH1 biomarkers include human TPH1 gene products, including
natural
variants, e.g., allelic variants, and homologs and analogs thereof. In one
example, the TPH1
biomarker is a polynucleotide having the nucleotide sequence set forth in SEQ
ID NO: 26
(referenced at GenBank gi number 226342925) or containing a protein-coding
portion thereof
38
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
(e.g., the open reading frame at nucleotides 27-1361 of SEQ ID NO: 26), a
natural variant
thereof, or a protein encoded by such a polynucleotide. TPH1 encodes an enzyme
produced by
enterochromaffin (EC) cells of the GI tract, important for the production of
serotonin.
[00167] The VMATI biomarkers include human VMATI gene products, including
natural
variants, e.g., allelic variants, and homologs and analogs thereof. In one
example, the VMATI
biomarker is a polynucleotide having the nucleotide sequence set forth in SEQ
ID NO: 27
(referenced at GenBank gi number 215272388) or containing the coding region
thereof (e.g.,
nucleotides 472-2049 of SEQ ID NO: 27), a natural variant thereof, or a
protein encoded by
such a polynucleotide.
[00168] The VMAT2 biomarkers include human VMAT2 gene products, including
natural
variants, e.g., allelic variants, and homologs and analogs thereof. In one
example, the VMAT2
biomarker is a polynucleotide having the nucleotide sequence set forth in SEQ
ID NO: 28
(referenced at GenBank gi number 141803164) or containing the coding region
thereof (e.g.,
nucleotides 32-1576 of SEQ ID NO: 28), a natural variant thereof, or a protein
encoded by such
a polynucleotide.
[00169] The X2BTB48 biomarkers include human serpin peptidase inhibitor. clade
A (alpha-
1 antiproteinase, antitrypsin), member 10) gene products, including natural
variants, e.g., allelic
variants, and homologs and analogs thereof. In one example, the X2BTB48
biomarker is a
polynucleotide having the nucleotide sequence set forth in SEQ ID NO: 29
(referenced at
GenBank gi number 154759289) or containing the coding region thereof (e.g.,
nucleotides 467-
1801 of SEQ ID NO: 29), a natural variant thereof, such as the nucleotide
sequence referenced at
GenBank gi number 154759290 (SEQ ID NO: 105) Or a coding sequence thereof,
e.g., the
coding sequence thereof at nucleotides 122-1456, or a protein encoded by such
polynucleotides,
such as the protein having the amino acid sequence referenced at GenBank gi
number 7705879.
[00170] The AKAP8L (A kinase (PRKA) anchor protein 8-like) biomarkers include
human
AKAP8L gene products, including natural variants, e.g., allelic variants, and
homologs and
analogs thereof. In one example, the AKAP8L biomarker is a polynucleotide
having the
nucleotide sequence set forth in SEQ ID NO: 201 (referenced at GenBank gi
number 49472840),
or containing a protein-coding portion thereof, e.g., the coding sequence
thereof of nucleotides
100-2040 of SEQ ID NO: 201, a natural variant thereof, or a protein encoded by
such a
polynucleotide.
39
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
[00171] The ATP6V1H (ATPase, H+ transporting, lysosomal 50/57kDa, VI subunit
H)
biomarkers include human ATP6V1H gene products, including natural variants,
e.g., allelic
variants, and homologs and analogs thereof. In one example, the ATP6V1H
biomarker is a
polynucleotide having the nucleotide sequence set forth in SEQ ID NO: 204
(referenced at
GenBank gi number 47717103), or containing a protein-coding portion thereof,
e.g., the coding
sequence thereof of nucleotides 293-1744, a natural variant thereof, or a
protein encoded by such
a polynucleotide.
[00172] The BNIP3L (BCL2/adenovirus ElB 19kDa interacting protein 3-like)
biomarkers
include human BNIP3L gene products, including natural variants, e.g., allelic
variants, and
homologs and analogs thereof. In one example, the BNIP3L biomarker is a
polynucleotide
having the nucleotide sequence set forth in SEQ ID NO: 205 (referenced at
GenBank gi number
47078259), or containing a protein-coding portion thereof, e.g., the coding
sequence thereof at
nucleotides 125-784 of SEQ ID NO: 205, a natural variant thereof, or a protein
encoded by such
a polynucleotide.
[00173] The C21orf7 (chromosome 21 open reading frame 7) biomarkers include
human
C21or17 gene products, including natural variants, e.g., allelic variants, and
homologs and
analogs thereof. In one example, the C2lorf7 biomarker is a polynucleotide
having the
nucleotide sequence set forth in SEQ ID NO: 206 (referenced at GenBank gi
number 31542267),
or containing a protein-coding portion thereof, e.g., the coding sequence
thereof at nucleotides
278-1006 of SEQ ID NO: 206, a natural variant thereof, or a protein encoded by
such a
polynucleotide.
[00174] The COMMD9 (COMM domain containing 9) biomarkers include human ATP6V1H
gene products, including natural variants, e.g., allelic variants, and
homologs and analogs
thereof. In one example, the COMMD9 biomarker is a polynucleotide having the
nucleotide
sequence set forth in SEQ ID NO: 207 (referenced at GenBank gi number
156416006), or
containing a protein-coding portion thereof, e.g., the coding sequence thereof
at nucleotides 38-
508 of SEQ ID NO: 207, a natural variant thereof, or a protein encoded by such
a
polynucleotide.
[00175] The ENPP4 (ectonucleotide pyrophosphatase/phosphodiesterase 4)
biomarkers
include human ENPP4 gene products, including natural variants, e.g., allelic
variants, and
homologs and analogs thereof. In one example, the ENPP4 biomarker is a
polynucleotide
having the nucleotide sequence set forth in SEQ ID NO: 208 (referenced at
GenBank gi number
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
194688140), or containing a protein-coding portion thereof, e.g., the coding
sequence thereof of
nucleotides 260-1621 of SEQ ID NO: 208, a natural variant thereof, or a
protein encoded by
such a polynucleotide.
[00176] The FAMI 3A (family with sequence similarity 13, member A) biomarkers
include
human FAM13A gene products, including natural variants, e.g., allelic
variants, and homologs
and analogs thereof. In one example, the FAM13A biornarker is a polynucleotide
having the
nucleotide sequence set forth in SEQ ID NO: 209 (referenced at GenBank gi
number
283806631), or containing a protein-coding portion thereof, e.g., the coding
sequence thereof at
nucleotides 281-1126 of SEQ ID NO: 209, a natural variant thereof, or a
protein encoded by
such a polynucleotide.
[00177] The FLJ10357 (Rho guanine nucleotide exchange factor 40) biomarkers
include
human ARHGEF40 gene products, including natural variants, e.g., allelic
variants, and
homologs and analogs thereof. In one example, the F1110357 biomarker is a
polynucleotide
having the nucleotide sequence set forth in SEQ ID NO: 210 (referenced at
GenBank gi number
50843836), or containing a protein-coding portion thereof, e.g., the coding
sequence thereof of
nucleotides 30-4589 of SEQ ID NO: 210, a natural variant thereof, or a protein
encoded by such
a polynucleotide.
[00178] The GLT8D1 (glycosyltransferase 8 domain containing 1) biomarkers
include human
GLT8D lgene products, including natural variants, e.g., allelic variants, and
homologs and
analogs thereof. In one example, the GLT8D1 biomarker is a polynucleotide
having the
nucleotide sequence set forth in SEQ ID NO: 211 (referenced at GenBank gi
number 58331224),
or containing a protein-coding portion thereof, e.g., the coding sequence
thereof of nucleotides
852-1967 of SEQ ID NO: 211, a natural variant thereof, or a protein encoded by
such a
polynucleotide.
[00179] The HDAC9 (histone deacetylase 9) biomarkers include human HDAC9 gene
products, including natural variants, e.g., allelic variants, and homologs and
analogs thereof. In
one example, the HDAC9 biomarker is a polynucleotide having the nucleotide
sequence set forth
in SEQ ID NO: 212 (referenced at GenBank gi number 323423043), or containing a
protein-
coding portion thereof, e.g., the coding sequence thereof at nucleotides 362-
2128 of SEQ ID
NO: 212, a natural variant thereof, or a protein encoded by such a
polynucleotide.
[00180] The HSF2 (heat shock transcription factor 2) biomarkers include human
HSF2 gene
products, including natural variants, e.g., allelic variants, and homologs and
analogs thereof. In
41
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
one example, the HSF2 biomarker is a polynucleotide having the nucleotide
sequence set forth
in SEQ ID NO: 213 (referenced at GenBank gi number 207113145), or containing a
protein-
coding portion thereof, e.g., the coding sequence thereof of nucleotides 188-
1798 of SEQ ID
NO: 213, a natural variant thereof, or a protein encoded by such a
polynucleotide.
[00181] The LE01 (Pafl /RNA polymerase IT complex component, homolog (S.
cerevisiae))
biomarkers include human LE01 gene products, including natural variants, e.g.,
allelic variants,
and homologs and analogs thereof. In one example, the LE01 biomarker is a
polynucleotide
having the nucleotide sequence set forth in SEQ ID NO: 215 (referenced at
GenBank gi number
37059738), or containing a protein-coding portion thereof, e.g., the coding
sequence thereof at
nucleotides 17-2017 of SEQ ID NO: 215, a natural variant thereof, or a protein
encoded by such
a polynucleotide.
[00182] The MORF4L2 (MORF4L2 mortality factor 4 like 2) biomarkers include
human
MORF4L2 gene products, including natural variants, e.g.. allelic variants, and
homologs and
analogs thereof. In one example, the MORF4L2 biomarker is a polynucleotide
having the
nucleotide sequence set forth in SEQ ID NO: 217 (referenced at GenBank gi
number
215490020, or containing a protein-coding portion thereof, e.g., the coding
sequence thereof at
nucleotides 505-1371, a natural variant thereof, or a protein encoded by such
a polynucleotide.
[00183] The NOL3 (nucleolar protein 3 (apoptosis repressor with CARD domain))
biomarkers include human NOL3 gene products, including natural variants, e.g.,
allelic variants,
and homologs and analogs thereof. In one example, the NOL3 biomarker is a
polynucleotide
having the nucleotide sequence set forth in SEQ ID NO: 218 (referenced at
GenBank gi number
297632351), or containing a protein-coding portion thereof, e.g., the coding
sequence thereof at
nucleotides 194-853, a natural variant thereof, or a protein encoded by such a
polynucleotide.
[00184] The NUDT3 (nudix (nucleoside diphosphate linked moiety X)-type motif
3)
biomarkers include human NUDT3 gene products, including natural variants,
e.g., allelic
variants, and homologs and analogs thereof. In one example, the NUDT3
biomarker is a
polynucleotide having the nucleotide sequence set forth in SEQ ID NO: 219
(referenced at
GenBank gi number 322302838), or containing a protein-coding portion thereof,
e.g., the coding
sequence thereof at nucleotides 319-837 of SEQ ID NO: 219, a natural variant
thereof, or a
protein encoded by such a polynucleotide.
[00185] The OAZ2 (omithine decarboxylase antizyme 2) biomarkers include human
OAZ2
gene products, including natural variants, e.g., allelic variants, and
homologs and analogs
42
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
thereof. In one example, the OAZ2 biomarker is a polynucleotide having the
nucleotide
sequence set forth in SEQ ID NO: 220 (referenced at GenBank gi number
161377456), or
containing a protein-coding portion thereof, e.g., the coding sequence thereof
at nucleotides 216-
311 and 313-786, or a natural variant thereof, or a protein encoded by such a
polynucleotide.
[00186] The PANK2 (pantothenate kinase 2) biomarkers include human PANK2 gene
products, including natural variants, e.g., allelic variants, and homologs and
analogs thereof. In
one example, the PANK2 biomarker is a polynucleotide having the nucleotide
sequence set forth
in SEQ ID NO: 221 (referenced at GenBank gi number 85838514), or containing a
protein-
coding portion thereof, e.g., the coding sequence thereof at nucleotides 312-
1151, a natural
variant thereof, or a protein encoded by such a polynucleotide.
[00187] The PHF21A (PHD finger protein 21A) biomarkers include human PHF21A
gene
products, including natural variants, e.g., allelic variants, and homologs and
analogs thereof. In
one example, the PHF21A biomarker is a polynucleotide having the nucleotide
sequence set
forth in SEQ ID NO: 222 (referenced at GenBank gi number 156546893), or
containing a
protein-coding portion thereof, e.g., the coding sequence thereof at
nucleotides 625-2667, a
natural variant thereof, or a protein encoded by such a polynucleotide.
[00188] The PKD1 (polycystic kidney disease 1 (autosomal dominant)) biomarkers
include
human PKD1 gene products, including natural variants, e.g., allelic variants,
and homologs and
analogs thereof. In one example, the PKD1 biomarker is a polynucleotide having
the nucleotide
sequence set forth in SEQ ID NO: 223, or the sequence referenced at GenBank gi
Number
205360961, or containing a protein-coding portion thereof of such a sequence,
e.g., the coding
sequence thereof at nucleotides 210-13118 or nucleotides 210-13117 of GenBank
gi Number
205360961 or SEQ ID NO: 223, a natural variant thereof, or a protein encoded
by such a
polynucleotide.
[00189] The PLD3 (phospholipase D family, member 3) biomarkers include human
PLD3
gene products, including natural variants, e.g., allelic variants, and
homologs and analogs
thereof. In one example, the PLD3 biomarker is a polynucleotide having the
nucleotide
sequence set forth in SEQ ID NO: 224 (referenced at GenBank gi number
166197669), or
containing a protein-coding portion thereof, e.g., the coding sequence thereof
at nucleotides 399-
1871 of SEQ ID NO: 224, a natural variant thereof, or a protein encoded by
such a
polynucleotide.
43
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
[00190] The PQBP1 (polyglutamine binding protein 1) biomarkers include human
PQBP1
gene products, including natural variants, e.g., allelic variants, and
homologs and analogs
thereof. In one example, the PQBP1 biomarker is a polynucleotide having the
nucleotide
sequence set forth in SEQ ID NO: 225 (referenced at GenBank gi number
74027246), or
containing a protein-coding portion thereof, e.g., the coding sequence thereof
at nucleotides 122-
919, a natural variant thereof, or a protein encoded by such a polynucleotide.
[00191] The RNF41 (polyglutamine binding protein 1) biomarkers include human
RNF41
gene products, including natural variants, e.g., allelic variants, and
homologs and analogs
thereof. In one example, the RNF41 biomarker is a polynucleotide having the
nucleotide
sequence set forth in SEQ ID NO: 227 (referenced at GenBank gi number
338827617), or
containing a protein-coding portion thereof, e.g., the coding sequence thereof
at nucleotides 320-
1273 of SEQ ID NO: 227, a natural variant thereof, or a protein encoded by
such a
polynucleotide.
[00192] The RSF1 (remodeling and spacing factor 1) biomarkers include human
RSF1 gene
products, including natural variants, e.g., allelic variants, and homologs and
analogs thereof. In
one example, the RSF1 biomarker is a polynucleotide having the nucleotide
sequence set forth in
SEQ ID NO: 228 (referenced at GenBank gi number 38788332), or containing a
protein-coding
portion thereof, e.g., the coding sequence thereof at nucleotides 121-4446 of
SEQ ID NO: 228, a
natural variant thereof, or a protein encoded by such a polynucleotide.
[00193] The RTN2 (reticul on 2) biomarkers include human RTN2 gene products,
including
natural variants, e.g., allelic variants, and homologs and analogs thereof. In
one example, the
RTN2 biomarker is a polynucleotide having the nucleotide sequence set forth in
SEQ ID NO:
229 (referenced at GenBank gi number 46255010), or containing a protein-coding
portion
thereof, e.g., the coding sequence thereof at nucleotides 229-1866 of SEQ ID
NO: 229, a natural
variant thereof, or a protein encoded by such a polynucleotide.
[00194] The SMARCD3 (SWI/SNF related, matrix associated, actin dependent
regulator of
chromatin, subfamily d, member 3p) biomarkers include human SMARCD3 gene
products,
including natural variants, e.g., allelic variants, and homologs and analogs
thereof. In one
example, the SMARCD3 biomarker is a polynucleotide having the nucleotide
sequence set forth
in SEQ ID NO: 232 (referenced at GenBank gi number 51477701), or containing a
protein-
coding portion thereof, e.g., the coding sequence thereof at nucleotides 102-
1553, a natural
variant thereof, or a protein encoded by such a polynucleotide.
44
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
[00195] The SPATA7 (spermatogenesis associated 7) biomarkers include human
SPATA7
gene products, including natural variants, e.g., allelic variants, and
homologs and analogs
thereof. In one example, the SPATA7 biomarker is a polynucleotide having the
nucleotide
sequence set forth in SEQ ID NO: 233 (referenced at GenBank gi number
295789142), or
containing a protein-coding portion thereof, e.g., the coding sequence thereof
at nucleotides 176-
1879 of SEQ ID NO: 233, a natural variant thereof, or a protein encoded by
such a
polynucleotide.
[00196] The SST1 (somatostatin receptor 1) biomarkers include human SST1 gene
products,
including natural variants, e.g., allelic variants, and homologs and analogs
thereof. In one
example, the SST1 biomarker is a polynucleotide having the nucleotide sequence
set forth in
SEQ ID NO: 234 (referenced at GenBank gi number 33946330), or containing a
protein-coding
portion thereof, e.g., the coding sequence thereof at nucleotides 618-1793 of
SEQ ID NO: 234, a
natural variant thereof, or a protein encoded by such a polynucleotide.
[00197] The SST3 (somatostatin receptor 3) biomarkers include human SST3 gene
products,
including natural variants, e.g., allelic variants, and homologs and analogs
thereof. In one
example, the SST3 biomarker is a polynucleotide having the nucleotide sequence
set forth in
SEQ ID NO: 235 (referenced at GenBank gi number 44890055), or containing a
protein-coding
portion thereof, e.g., the coding sequence thereof at nucleotides 526-1782 of
SEQ ID NO: 235, a
natural variant thereof, or a protein encoded by such a polynucleotide.
[00198] The SST4 (somatostatin receptor 4) biomarkers include human SST3 gene
products,
including natural variants, e.g., allelic variants, and homologs and analogs
thereof. In one
example, the SST3 biomarker is a polynucleotide having the nucleotide sequence
set forth in
SEQ ID NO: 236 (referenced at GenBank gi number 149944553), or containing a
protein-coding
portion thereof, e.g., the coding sequence thereof at nucleotides 65-1231, a
natural variant
thereof, or a protein encoded by such a polynucleotide.
[00199] The SSTS (somatostatin receptor 5) biomarkers include human SST3 gene
products,
including natural variants, e.g., allelic variants, and homologs and analogs
thereof. In one
example, the SST3 biomarker is a polynucleotide having the nucleotide sequence
set forth in
SEQ ID NO: 237 (referenced at GenBank gi number 289547751), or containing a
protein-coding
portion thereof, e.g., the coding sequence thereof at nucleotides 89-1183, a
natural variant
thereof, or a protein encoded by such a polynucleotide.
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
[00200] The TECPR2 (tectonin beta-propeller repeat containing 2) biomarkers
include human
TECPR2 gene products, including natural variants, e.g., allelic variants, and
homologs and
analogs thereof. In one example, the TECPR2 biomarker is a polynucleotide
having the
nucleotide sequence set forth in SEQ ID NO: 238 (referenced at GenBank gi
number
289547516), or containing a protein-coding portion thereof, e.g., the coding
sequence thereof at
nucleotides 227-4030 of SEQ ID NO: 238, a natural variant thereof, or a
protein encoded by
such a polynucleotide.
[00201] The TRMT112 (tRNA methyltransferase 11-2 homolog (S. cerevisiae))
biomarkers
include human TRMT112 gene products, including natural variants, e.g., allelic
variants, and
homologs and analogs thereof. In one example, the TRMT112 biomarker is a
polynucleotide
having the nucleotide sequence set forth in SEQ ID NO: 241 (referenced at
GenBank gi number
7705476), or containing a protein-coding portion thereof, e.g., the coding
sequence thereof at
nucleotides 36-413 of SEQ ID NO: 241, a natural variant thereof, or a protein
encoded by such a
polynucleotide.
[00202] The VPS13C (vacuolar protein sorting 13 homolog C (S. cerevisiae))
biomarkers
include human VPS13C gene products, including natural variants, e.g., allelic
variants, and
homologs and analogs thereof. In one example, the VPS13C biomarker is a
polynucleotide
having the nucleotide sequence set forth in SEQ ID NO: 242 (referenced at
GenBank gi number
308081495), or containing a protein-coding portion thereof, e.g., the coding
sequence thereof at
nucleotides 92-10978, a natural variant thereof, or a protein encoded by such
a polynucleotide.
[00203] The WDFY3 (WD repeat and FYVE domain containing 3) biomarkers include
human WDFY3 gene products, including natural variants, e.g., allelic variants,
and homologs
and analogs thereof. In one example, the WDFY3 biomarker is a polynucleotide
having the
nucleotide sequence set forth in SEQ ID NO: 243, or the sequence referenced at
GenBank gi
number 195972885, or containing a protein-coding portion thereof, e.g., the
coding sequence at
nucleotides 409-10988 of SEQ ID NO: 243 or GenBank gi number 195972885, a
natural variant
thereof, or a protein encoded by such a polynucleotide.
[00204] The ZFHX3 (zinc finger homeobox 3) biomarkers include human ZFHX3 gene
products, including natural variants, e.g., allelic variants, and homologs and
analogs thereof. In
one example, the ZFHX3 biomarker is a polynucleotide having the nucleotide
sequence set forth
in SEQ ID NO: 244 (referenced at GenBank gi number 258613986), or containing a
protein-
46
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
coding portion thereof, e.g., the coding sequence thereof at nucleotides 130-
8499 of SEQ ID
NO: 244, a natural variant thereof, or a protein encoded by such a
polynucleotide.
[00205] The ZXDC (ZXD family zinc finger C) biomarkers include human ZXDC gene
products, including natural variants, e.g., allelic variants, and homologs and
analogs thereof. In
one example, the ZXDC biomarker is a polynucleotide having the nucleotide
sequence set forth
in SEQ ID NO: 245 (referenced at GenBank gi number 217035098), or containing a
protein-
coding portion thereof, e.g., the coding sequence thereof at nucleotides 55-
2187 of SEQ ID NO:
245, a natural variant thereof, or a protein encoded by such a polynucleotide.
[00206] The ZZZ3 (zinc finger, ZZ-type containing 3) biomarkers include human
ZZZ3 gene
products, including natural variants, e.g., allelic variants, and homologs and
analogs thereof. In
one example, the ZZZ3 biomarker is a polynucleotide having the nucleotide
sequence set forth
in SEQ ID NO: 246 (referenced at GenBank gi number 141803158), or containing a
protein-
coding portion thereof, e.g., the coding sequence thereof at nucleotides 477-
3188 of SEQ ID
NO: 246, a natural variant thereof, or a protein encoded by such a
polynucleotide.
[00207] In some aspects, the provided methods and compositions detect a GEP-
NEN
biomarker; in some examples, the provided methods and compositions detect
panels of GEP-
NEN biomarkers, including two or more GEP-NEN biomarkers, such as at least 2,
3, 4, 5, 6, 7,
8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26,27,
28, 29, 30, 31, 32, 33,
34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52,
53, 54, 55. 56, 57, 58, 59,
60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 80, 85, 90,
95, or 100 or more
biomarkers.
[00208] For example, provided are methods and compositions that detect at
least 1, 2, 3, 4, 5,
6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21,22, 23, 24, 25, 26,
27, 28, 29, 30, 31, 32,
33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, or 51.
and/or all of the
following sets of biomarkers:
[00209] AKAP8L, ATP6V1H, BNIP3L, C21orf7, COMMD9, ENPP4, FAM13A, FLJ10357,
GLT8D1, HDAC9, HSF2, LE01, MORF4L2, NOL3, NUDT3, OAZ2, PANK2, PHF21A,
PKD1, PLD3, PQBP1, RNF41, RSF1, RTN2, SMARCD3, SPATA7, SST1, SST3, SST4, SSTS,
TECPR2, TRMT112, VPS13C, WDFY3, ZFHX3, ZXDC, ZZZ3, APLP2, CD59. ARAF1,
BRAF1, KRAS, and RAF1 gene products;
[00210] AKAP8L, ATP6V1H, BNIP3L, C21orf7, COMMD9, ENPP4, FAMI3A, FLJI0357,
GLT8D1, HDAC9, HSF2, LEOI, MORF4L2, NOL3, NUDT3, OAZ2, PANK2, PHF2IA,
47
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
PKD1, PLD3, PQBP1, RNF41, RSF1, RTN2, SMARCD3, SPATA7, SST1, SST3, SST4, SSTS,
TECPR2, TRMT112, VPS13C, WDFY3, ZFHX3, ZXDC, and ZZZ3 gene products; and
[00211] APLP2, ARAF1, BRAF, CD59, CTGF, FZD7, Ki67, KRAS, NAP1LI, PNMA2,
RAF1, TPH1, VMAT1, and VMAT2 gene products.
[00212] Also provided are methods and compositions that detect at least 2, 3,
4, 5, 6, 7, 8, 9,
10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, or
29 of the following
sets of biomarkers:
[00213] APLP2, ARAF1, BRAF1, CD59, CgA, CTGF, CXCL14, FZD7, GRIA2, HOXC6,
Ki-67; Kissl, KRAS, MAGE-D2, MTA1, NAP1L1, NKX2-3, NRP2, OR51E1, PNMA2,
PTPRN2, RAF1, SCG5, SPOCK1, Survivin, TPH1, VMAT1, VMAT2); and X2BTB48;
[00214] APLP2, ARAF1, BRAF1, CD59, KRAS, RAF1, CXCL14, GRIA2, HOXC6, NKX2-
3, OR51E1, PNMA2, PTPRN2, SCG5, SPOCK1, and X2BTB48;
[00215] CXCL14, GRIA2, HOXC6, NKX2-3, OR51E1, PNMA2, PTPRN2, SCG5, SPOCK1,
and X2BTB48; or CgA (chromogranin A), CTGF, FZD7 (frizzled homolog 7), Ki-67
(a marker
of proliferation), Kissl (Kissl metastasis suppressor), MAGE-D2 (melanoma
antigen family
D2), MTA1 (metastasis-associated 1), NAPIL1, NRP2 (neuropilin 2), Tphl, VMAT1,
VMAT2.
and Survivin.
[00216] In some aspects, the panels further include CD164.
[00217] In some aspects, they further include NALP or other known biomarkers.
[00218] In some embodiments, the panel of polynucleotides further includes one
or more
polynucleotide able to specifically hybridize to "housekeeping," or reference
genes, for example,
genes for which differences in expression is known or not expected to
correlate with differences
in the variables analyzed, for example, with the presence or absence of GEP-
NEN or other
neoplastic disease, differentiation of various GEP-NEN sub-types, metastasis,
mucosal or other
tissue types, prognostic indications, and/or other phenotype, prediction, or
outcome. In some
aspects, expression levels of such housekeeping genes are detected and used as
an overall
expression level standards, such as to normalize expression data obtained for
GEP-NEN
biomarkers across various samples.
[00219] Housekeeping genes are well known in the art. Typically, the
housekeeping genes
include one or more genes characterized as particularly appropriate for
analyzing GEP-NEN
samples, such as ALG9, TFCP2 and ZNF410. See Kidd M, et al., "GeneChip, geNorm
and
Gastrointestinal tumors: novel reference genes for real-time PCR." Physiol
Genomics
48
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
2007;30:363-70. Other housekeeping genes and polynucleotides are well known in
the art and
include glyceraldehyde-3-phosphate dehydrogenase (GAPDH), hypoxanthine
phosphoribosyltransferase (HPRT) and 18S RNA.
[00220] The ALG9 housekeeping genes include human ALG9 (asparagine-linked
glycosylation 9, alpha-1,2-mannosyltransferase homolog) gene products,
including natural
variants, e.g., allelic variants, and homologs and analogs thereof. In one
example, the ALG9
housekeeper is a polynucleotide having the nucleotide sequence set forth in
SEQ ID NO: 35 and
referenced at GenBank gi no.: 118026920or containing the coding region thereof
of nucleotides
100-1956 of SEQ ID NO: 35, a natural variant thereof, or a protein encoded by
such a
polynucleotide.
[00221] The TFCP2 housekeeping genes include human TFCP2 (transcription factor
CP2)
gene products, including natural variants, e.g., allelic variants, and
homologs and analogs
thereof. In one example, the TFCP2 housekeeper is a polynucleotide having the
nucleotide
sequence set forth in SEQ ID NO: 36 and referenced at GenBank gi no.
291219872, or
containing the coding region thereof at nucleotides 722-2230 of SEQ ID NO: 36,
a natural
variant thereof, or a protein encoded by such a polynucleotide.
[00222] The ZNF410 housekeeping genes include human ZNF410 (zinc finger
protein 410)
gene products, including natural variants, e.g., allelic variants, and
homologs and analogs
thereof. In one example, the ZNF410 housekeeper is a polynucleotide having the
nucleotide
sequence set forth in SEQ ID NO: 37 and referenced at GenBank gi no. 10863994,
or containing
the coding region thereof at nucleotides 183-1619 of SEQ ID NO: 37, a natural
variant thereof,
or a protein encoded by such a polynucleotide.
[00223] The GAPDH housekeeping genes include human GAPDH (glyceraldehyde-3-
phosphate dehydrogenase) gene products, including natural variants, e.g.,
allelic variants, and
homologs and analogs thereof. In one example, the GAPDH housekeeper is a
polynucleotide
having the nucleotide sequence set forth in SEQ ID NO: 38 and referenced at
GenBank gi
number 83641890 or containing the coding region thereof at nucleotides 103-
1110 of SEQ ID
NO: 38, a natural variant thereof, or a protein encoded by such a
polynucleotide.
[00224] The 18S housekeeping genes include human 18S (Eukaryotic 18S rRNA),
including
natural variants, e.g., allelic variants, and homologs and analogs thereof. In
one example, the
18S housekeeper is a polynucleotide having the nucleotide sequence set forth
in SEQ ID NO: 39
and referenced at GenBank gi number 36162 or a natural variant thereof.
49
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
[00225] The HPRT housekeeping genes include human HPRT gene products,
including
natural variants, e.g., allelic variants, and homologs and analogs thereof. In
one example, the
HPRT housekeeper is a polynucleotide having the nucleotide sequence set forth
in SEQ ID NO:
40, and referenced at GenBank gi no. 164518913, or containing the coding
region thereof at
nucleotides 168-824 of SEQ ID NO: 40, a natural variant thereof, or a protein
encoded by such a
polynucleotide.
[00226] The SLC25A3 housekeeping genes include human SLC25A3 (solute carrier
family
25 (mitochondrial carrier; phosphate carrier). member 3) gene products,
including natural
variants, e.g., allelic variants, and homologs and analogs thereof. In one
example, the SLC25A3
housekeeper is a polynucleotide having the nucleotide sequence set forth in
SEQ ID NO: 247
and referenced at GenBank gi no.: 223718119, or containing a coding region
thereof, e.g., the
coding sequence thereof at nucleotides 121-1209 of SEQ ID NO: 247, a natural
variant thereof,
or a protein encoded by such a polynucleotide.
[00227] The VAPA housekeeping genes include human VAPA ((vesicle-associated
membrane protein)-associated protein A) gene products, including natural
variants, e.g., allelic
variants, and homologs and analogs thereof. In one example. the VAPA
housekeeper is a
polynucleotide having the nucleotide sequence set forth in SEQ ID NO: 248 and
referenced at
GenBank gi no.: 94721249, or containing a coding region thereof, e.g., the
coding sequence
thereof at nucleotides 300-1184 of SEQ ID NO: 248, a natural variant thereof,
or a protein
encoded by such a polynucleotide.
[00228] The TXNIP housekeeping genes include human TXNIP (thioredoxin
interacting
protein) gene products, including natural variants, e.g., allelic variants,
and homologs and
analogs thereof. In one example, the TXNIP housekeeper is a polynucleotide
having the
nucleotide sequence set forth in SEQ ID NO: 249 and referenced at GenBank gi
no.: 171184420,
or containing a coding region thereof, e.g., the coding sequence thereof at
nucleotides 342-1517
of SEQ ID NO: 249, a natural variant thereof, or a protein encoded by such a
polynucleotide.
[00229] The ADD3 housekeeping genes include human ADD3 (adducin 3 (gamma))
gene
products, including natural variants, e.g., allelic variants, and homologs and
analogs thereof. In
one example, the ADD3 housekeeper is a polynucleotide having the nucleotide
sequence set
forth in SEQ ID NO: 250 and referenced at GenBank gi no.: 62912451, or
containing a coding
region thereof, e.g., the coding sequence thereof at nucleotides 377-2497 of
SEQ ID NO: 250, a
natural variant thereof, or a protein encoded by such a polynucleotide.
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
[00230] The DAZAP2 housekeeping genes include human DAZAP2 (DAZ-associated
protein
2) gene products, including natural variants, e.g., allelic variants, and
homologs and analogs
thereof. In one example, the DAZAP2 housekeeper is a polynucleotide having the
nucleotide
sequence set forth in SEQ ID NO: 251 and referenced at GenBank gi no.:
211904132 or
containing a coding region thereof, e.g., the coding sequence thereof at
nucleotides 1 85-691 , a
natural valiant thereof, or a protein encoded by such a polynucleotide.
[00231] The ACTG1 housekeeping genes include human ACTG1 (actin, gamma 1) gene
products, including natural variants, e.g., allelic variants, and homologs and
analogs thereof. In
one example, the ACTG1 housekeeper is a polynucleotide having the nucleotide
sequence set
forth in SEQ ID NO: 252 and referenced at GenBank gi no.: 316659408, or
containing a coding
region thereof, e.g., the e.g., the coding sequence thereof at nucleotides 259-
1386 of SEQ ID
NO: 252, a natural variant thereof, or a protein encoded by such a
polynucleotide.
[00232] The ACTB housekeeping genes include human ACTB (actin, beta) gene
products,
including natural variants, e.g., allelic variants, and homologs and analogs
thereof. In one
example, the ACTB housekeeper is a polynucleotide having the nucleotide
sequence set forth in
SEQ ID NO: 200 and referenced at GenBank gi no.: 168480144, or containing a
coding region
thereof, e.g., the coding sequence thereof at nucleotides 85-1212 of SEQ ID
NO: 200, a natural
variant thereof, or a protein encoded by such a polynucleotide.
[00233] The ATG4B housekeeping genes include human ACG4B (autophagy related 4
homolog B (S. cerevisiae)) gene products, including natural variants, e.g.,
allelic variants, and
homologs and analogs thereof. In one example, the ACTG4B housekeeper is a
polynucleotide
having the nucleotide sequence set forth in SEQ ID NO: 203, or containing a
coding region
thereof, e.g., the coding sequence thereof at nucleotides 104-1285 of SEQ ID
NO: 203, or the
sequence referenced at GenBank gi no.: 47132610, a natural variant thereof, or
a protein
encoded by such a polynucleotide.
[00234] The ARF1 housekeeping genes include human ARF1 (ADP-ribosylation
factor 1)
gene products, including natural variants, e.g., allelic variants, and
homologs and analogs
thereof. In one example, the ARF1 housekeeper is a polynucleotide having the
nucleotide
sequence set forth in SEQ ID NO: 202 and referenced at GenBank gi no.:
66879659 or
containing a coding region thereof, e.g., the coding sequence thereof at
nucleotide residues 229-
774, a natural variant thereof, or a protein encoded by such a polynucleotide.
51
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
[00235] The HUWE1 housekeeping genes include human HUWE1 (HECT, UBA and WWE
domain containing 1) gene products, including natural variants, e.g., allelic
variants, and
homologs and analogs thereof. In one example, the HUWE1 housekeeper is a
polynucleotide
having the nucleotide sequence set forth in SEQ ID NO: 214 and referenced at
GenBank gi no.:
195963314, or containing the coding region thereof, e.g., the coding sequence
thereof at
nucleotides 403-13527, a natural variant thereof, or a protein encoded by such
a polynucleotide.
[00236] The MORF4L1 housekeeping genes include human M0RF4L1 (mortality factor
4
like 1) gene products, including natural variants, e.g., allelic variants, and
homologs and analogs
thereof. In one example, the MORF4L1 housekeeper is a polynucleotide having
the nucleotide
sequence set forth in SEQ ID NO: 216 and referenced at GenBank gi no.:
45643136, or
containing a coding region thereof, e.g., the coding sequence at nucleotides
189-1160, a natural
variant thereof, or a protein encoded by such a polynucleotide.
[00237] The RHOA housekeeping genes include human RHOA (ras homolog gene
family,
member A) gene products, including natural variants, e.g., allelic variants,
and homologs and
analogs thereof. In one example, the RHOA housekeeper is a polynucleotide
having the
nucleotide sequence set forth in SEQ ID NO: 226 and referenced at GenBank gi
no.: 50593005,
or containing a coding region thereof, e.g., the coding sequence thereof at
nucleotides 277-858, a
natural variant thereof, or a protein encoded by such a polynucleotide.
[00238] The SERPI housekeeping genes include human SERPI (stress-associated
endoplasmic reticulum protein 1) gene products, including natural variants,
e.g., allelic variants,
and homologs and analogs thereof. In one example, the SERPI housekeeper is a
polynucleotide
having the nucleotide sequence set forth in SEQ ID NO: 230 and referenced at
GenBank gi no.:
109809760, or containing a coding region thereof, e.g., the coding sequence
thereof at
nucleotides 507-707, a natural variant thereof, or a protein encoded by such a
polynucleotide.
[00239] The SKP1 housekeeping genes include human SKP1 (S-phase kinase-
associated
protein 1) gene products, including natural variants, e.g., allelic variants,
and homologs and
analogs thereof. In one example, the SKP1 housekeeper is a polynucleotide
having the
nucleotide sequence set forth in SEQ ID NO: 231 and referenced at GenBank gi
no.: 160420325,
or containing a coding region thereof, e.g., the coding sequence thereof at
nucleotides 180-662, a
natural variant thereof, or a protein encoded by such a polynucleotide.
[00240] The TOX4 housekeeping genes include human TOX4 (TOX high mobility
group box
family member 4) gene products, including natural variants, e.g., allelic
variants, and homologs
52
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
and analogs thereof. In one example, the TOX4 housekeeper is a polynucleotide
having the
nucleotide sequence set forth in SEQ ID NO: 239 and referenced at GenBank gi
no.: 99077116,
or containing a coding region thereof, e.g., the coding sequence thereof at
nucleotides 104-1969,
a natural variant thereof, or a protein encoded by such a polynucleotide.
[00241] The TPT1 housekeeping genes include human TPT1 (tumor protein,
translati onall y-
controlled 1) gene products, including natural variants, e.g., allelic
variants, and homologs and
analogs thereof. In one example, the TPT1 housekeeper is a polynucleotide
having the
nucleotide sequence set forth in SEQ ID NO: 240 and referenced at GenBank gi
no.: 141801911,
or containing a coding region thereof, e.g., the coding sequence thereof at
nucleotides 94-612, a
natural variant thereof, or a protein encoded by such a polynucleotide.
D. Methods and agents for detecting the GEP-NEN biomarkers. tumors, and
cells
[00242] Also provided are methods, compositions, and systems, for the
detection of the GEP-
NEN biomarkers and for identifying, isolating, and enriching tumors and cells
that express the
GEP-NEN biomarkers. For example, provided are agents, sets of agents, and
systems for
detecting the GEP-NEN biomarkers and methods for use of the same, including
for diagnostic
and prognostic uses.
1. Agents and systems for detecting the biomarkers
[00243] In one embodiment, the agents are proteins, polynucleotides or other
molecules
which specifically bind to or specifically hybridize to the GEP-NEN
biomarkers. The agents
include polynucleotides, such as probes and primers, e.g. sense and antisense
PCR primers,
having identity or complementarity to the polynucleotide biomarkers, such as
mRNA, and
proteins, such as antibodies, which specifically bind to such biomarkers. Sets
and kits
containing the agents, such as agents specifically hybridizing to or binding
the panel of
biomarkers, also are provided.
[00244] Thus, the systems, e.g., microarrays, sets of polynucleotides, and
kits, provided
herein include those with nucleic acid molecules, typically DNA
oligonucleotides, such as
primers and probes, the length of which typically varies between 15 bases and
several kilo bases,
such as between 20 bases and 1 kilobase, between 40 and 100 bases, and between
50 and 80
nucleotides or between 20 and 80 nucleotides. In one aspect, most (i.e. at
least 60% of) nucleic
acid molecules of a nucleotide microarray, kit, or other system, are capable
of hybridizing to
GEP-NEN biomarkers.
53
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
[00245] In one example, systems containing polynucleotides that specifically
hybridize to the
biomarkers, e.g., nucleic acid microarrays, are provided to detect and measure
changes in
expression levels and determine expression profiles of the biomarkers
according to the provided
methods. Among such systems, e.g., microarrays. are those comprising
polynucleotides able to
hybridize to at least as at least 2, 3, 4, 5, 6, 7, 8, 9, 10,11, 12, 13, 14,
15, 16, 17, 18, 9, 20, 21,
22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40,
41, 42, 43, 44, 45, 46, 47,
48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66,
67, 68, 69, 70, 71, 72, 73,
74, 75, 80, 85, 90, 95, or 100 or more biomarkers, such as to at least 1, 2,
3, 4, 5, 6, 7, 8, 9, 10,
11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21,22, 23, 24, 25, 26, 27, 28, 29, 30,
31, 32, 33, 34, 35, 36,
37, 38, 39, 40, 41, 42, 43, 44, 45. 46, 47, 48, 49, 50, or 51, and/or all of
the following sets of
biomarkers:
[00246] AKAP8L, ATP6V1H, BNIP3L, C21orf7. COMMD9, ENPP4, FAM13A, FLJ10357,
GLT8D1, HDAC9, HSF2, LE01, MORF4L2, NOL3, NUDT3, OAZ2, PANK2. PHF21A,
PKD1, PLD3, PQBP1. RNF41, RSFI, RTN2, SMARCD3, SPATA7, SST1, SST3, SST4, SSTS,
TECPR2, TRMT112, VPS13C, WDFY3, ZFHX3, ZXDC, ZZZ3. APLP2. CD59. ARAF1,
BRAF1, KRAS, and RAF1 gene products;
[00247] AKAP8L, ATP6V1H, BN1P3L, C21orf7, COMMD9, ENPP4, FAM13A, FLJ10357,
GLT8D1, HDAC9, HSF2, LE01, MORF4L2, NOL3, NUDT3, OAZ2, PANK2, PHF21A,
PKD1, PLD3, PQBP1, RNF41, RSF1, RTN2, SMARCD3, SPATA7, SST1, SST3, SST4, SSTS,
TECPR2, TRMT11 2, VPS13C, WDFY3, ZFHX3, ZXDC, and ZZZ3 gene products; and
[00248] APLP2, ARAF1, BRAF, CD59, CTGF, FZD7, Ki67, KRAS, NAP1L1, PNMA2,
RAF1, TPH1, VMAT1, and VMAT2 gene products; or
[00249] at least 2, 3, 4, 5, 6, 7, 8. 9. 10, 11, 12, 13, 14, 15, 16, 17. 18,
19, 20, 21, 22. 23, 24,
25, 26, 27, 28, or 29 of APLP2, ARAF1, BRAF1, CD59, CgA, CTGF, CXCL14, FZD7,
GRIA2,
HOXC6, Ki-67; Kissl, KRAS, MAGE-D2, MTA1, NAP1L1, NKX2-3, NRP2, OR51E1,
PNMA2, PTPRN2, RAF1, SCG5, SPOCK1, Survivin, TPH1, VMAT1. VMAT2); and
X2BTB48; or of the biomarkers APLP2. ARAF1, BRAF1. CD59, KRAS, RAF1, CXCL14,
GRIA2, HOXC6, NKX2-3, OR51E1, PNMA2, PTPRN2, SCG5, SPOCK1, and X2BTB48; or of
the biomarkers CXCL14, GRIA2, HOXC6, NKX2-3, OR51E1, PNMA2. PTPRN2, SCG5,
SPOCK1, and X2BTB48.
[00250] In some aspects, at least 60%, or at least 70%, at least 80%, or more,
of the nucleic
acid molecules of the system, e.g., micromay, are able to hybridize to
biomarkers in the panel of
54
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
biomarkers. In one example, probes immobilized on such nucleotide microarrays
comprise at
least 2, and typically at least 3, 4, 5, 6, 7. 8, 9, 10, 11, 12. 13, 14, 15,
16, 17, 18, 19, 20, 21, 22,
23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41,
42, 43, 44, 45, 46, 47, 48,
49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67,
68, 69, 70, 71, 72, 73, 74,
75, 80, 85, 90, 95, or 100 or more biomarkers, such as to at least 1, 2, 3, 4,
5. 6. 7, 8, 9, 10, 11,
12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30,
31, 32, 33, 34, 35, 36, 37,
38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, or 51, or more nucleic
acid molecules able to
hybridize to at least 2, 3, 4. 5, 6, 7, 8,9, 10, 11, 12, 13, 14, 15, 16, 17,
18, 19, 20, 21, 22, 23. 24,
25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43,
44, 45, 46, 47, 48, 49, 50,
51, 52, 53, 54, 55, 56, 57, 58, 59. 60, 61, 62, 63, 64, 65, 66, 67, 68, 69,
70, 71, 72. 73, 74, 75, 80,
85, 90, 95, or 100 or more biomarkers, such as to at least 1, 2, 3, 4, 5, 6,
7, 8, 9, 10, 11, 12, 13,
14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32,
33, 34, 35, 36, 37, 38, 39,
40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, or 51, or more of the biomarkers,
where each of the
nucleic acid molecules is capable of specifically hybridizing to a different
one of the biomarkers,
such that at least that many different biomarkers can be bound.
[00251] In one example, the remaining nucleic acid molecules, such as 40% or
at most 40%
of the nucleic acid molecules on the microarray or in the set of
polynucleotides are able to
hybridize to a set of reference genes or a set of normalization genes (such as
housekeeping
genes), for example, for normalization in order to reduce systemic bias.
Systemic bias results in
variation by inter-array differences in overall performance, which can be due
to for example
inconsistencies in array fabrication, staining and scanning, and variation
between labeled RNA
samples, which can be due for example to variations in purity. Systemic bias
can be introduced
during the handling of the sample in a microarray experiment. To reduce
systemic bias, the
determined RNA levels are preferably corrected for background non-specific
hybridization and
normalized.
[00252] The use of such reference probes is advantageous but not mandatory. In
one
embodiment a set of polynucleotides or system. e.g., microarray. is provided
wherein at least
90% of the nucleic acid sequences are able to hybridize to the GEP-NEN
biomarkers; further
embodiments include such systems and sets in which at least 95% or even 100%
of the
polynucleotides hybridize to the biomarkers.
[00253] Disclosed in the Examples are exemplary suitable polynucleotides, such
as PCR
primers. Other nucleic acid probes and primers, able to hybridize to different
regions of the
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
biomarkers are of course also suitable for use in connection with the provided
systems, kits and
methods.
2. Detection of the biomarkers
[00254] Also provided are methods for detecting and quantifying the
biomarkers, including
detecting the presence, absence, amount or relative amount. such as expression
levels or
expression profile of the biomarkers. Typically, the methods are nucleic acid
based methods, for
example, measuring the presence, amount or expression levels of biomarker mRNA
expression.
Such methods typically are carried out by contacting polynucleotide agents to
biological
samples, such as test samples and normal and reference samples, for example,
to quantify
expression levels of nucleic acid biomarkers (e.g., mRNA) in the samples.
[00255] Detection and analysis of biomarkers according to the provided
embodiments can be
performed with any suitable method known in the art. For example. where the
biomarkers are
RNA biomarkers, RNA detection and quantification methods are used.
[00256] Exemplary methods for quantifying or detecting nucleic acid expression
levels, e.g.,
mRNA expression, are well known, and include northern blotting and in situ
hybridization
(Parker and Barnes, Methods in Molecular Biology 106:247-283, 1999); RNAse
protection
assays (Hod, Biotechniques 13:852-854, 1992); and quantitative or semi-
quantitative reverse
transcription polymerase chain reaction (RT-PCR) (Weis et al., Trends in
Genetics 8:263-264,
1992), representative methods for sequencing-based gene expression analysis
include Serial
Analysis of Gene Expression (SAGE), and gene expression analysis by massively
parallel
signature sequencing (MPSS).
[00257] Therefore, in one embodiment, expression of the biomarker or biomarker
panel
includes RNA expression; the methods include determining levels of RNA of the
biomarkers,
such as RNA obtained from and/or present in a sample of a patient, and
performing analysis,
diagnosis, or predictive determinations based upon the RNA expression levels
determined for
the biomarkers or panel of biomarkers.
[00258] RNA samples can be processed in numerous ways. as is known to those in
the art.
Several methods are well known for isolation of RNA from samples, including
guanidinium
thiocyanate-phenol-chloroform extraction, which may be carried out using the
TRIZOL
reagent, a proprietary formulation (see Chomczynski P, Sacchi N (2006). "The
single-step
method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform
extraction:
twenty-something years on". Nat Protoc 1 (2): 581-5). In this method, TRIZOL
is used to
56
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
extract RNA and DNA; chloroform and centrifugation are used to separate RNA
from other
nucleic acids, followed by a series of washes with ethanol for cleanup of the
RNA sample.
[00259] The RNA samples can be freshly prepared from cells or tissues at the
moment of
harvesting; alternatively, they can be prepared from samples that stored at -
70 C until processed
for sample preparation. Alternatively, tissues or cell samples can be stored
under and/or
subjected to other conditions known in the art to preserve the quality of the
RNA, including
fixation for example with formalin or similar agent; and incubation with RNase
inhibitors such
as RNAsin@ (Pharmingen) or RNasecure0 (Ambion): aqueous solutions such as
RNAlater@
(Assuragen), Hepes-Glutamic acid buffer mediated Organic solvent Protection
Effect (HOPE),
and RCL2 (Alphelys); and non-aqueous solutions such as Universal Molecular
Fixative (Sakura
Finetek USA Inc.). A chaotropic nucleic acid isolation lysis buffer (Boom
method, Boom et al,
J Clin Microbiol. 1990; 28:495-503) may also be used for RNA isolation.
[00260] In one embodiment, RNA is isolated from buffy coat by incubating
samples with
TRIZOL , followed by RNA clean-up. RNA is dissolved in diethyl pyrocarbonate
water and
measured spectrophotometrically, and an aliquot analyzed on a Bioanalyzer
(Agilent
Technologies, Palo Alto, CA) to assess the quality of the RNA (Kidd M, et al.
"The role of
genetic markers--NAP1L1, MAGE-D2, and MTA1--in defining small-intestinal
carcinoid
neoplasia," Ann Stag Oncol 2006;13(2):253-62). In another embodiment, RNA is
isolated from
plasma using the QIAamp RNA Blood Mini Kit; in some cases, this method allows
better
detection by real-time PCR of significantly more housekeeping genes from
plasma compared to
the TRIZOL@ approach. In another embodiment, RNA is isolated directly from
whole blood,
for example, using the QIAamp RNA Blood Mini Kit in a similar manner.
[00261] Methods for isolating RNA from fixed, paraffin-embedded tissues as the
RNA source
are well-known and generally include mRNA isolation, purification, primer
extension and
amplification (for example: T. E. Godfrey et al,. J. Molec. Diagnostics 2: 84-
91 [2000]; K.
Specht et al., Am. J. Pathol. 158: 419-29 [2001]). In one example, RNA is
extracted from a
sample such as a blood sample using the QIAamp RNA Blood Mini Kit RNA.
Typically, RNA
is extracted from tissue, followed by removal of protein and DNA and analysis
of RNA
concentration. An RNA repair and/or amplification step may be included, such
as a step for
reverse transcription of RNA for RT-PCR.
[00262] Expression levels or amounts of the RNA biomarkers may be determined
or
quantified by any method known in the art, for example, by quantifying RNA
expression relative
57
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
to housekeeping gene or with relation to RNA levels of other genes measured at
the same time.
Methods to determine RNA levels of genes are known to a skilled person and
include, but are
not limited to, Northern blotting, (quantitative) PCR, and microarray
analysis.
[00263] Northern blotting may be performed for quantification of RNA of a
specific
biomarker gene or gene product, by hybridizing a labeled probe that
specifically interacts with
the RNA, following separation of RNA by gel electrophoresis. Probes are for
example labeled
with radioactive isotopes or chemiluminescent substrates. Quantification of
the labeled probe
that has interacted with said nucleic acid expression product serves as a
measure for determining
the level of expression. The determined level of expression can be normalized
for differences in
the total amounts of nucleic acid expression products between two separate
samples with for
instance an internal or external calibrator by comparing the level of
expression of a gene that is
known not to differ in expression level between samples or by adding a known
quantity of RNA
before determining the expression levels.
[00264] For RT-PCR, biomarker RNA is reverse transcribed into cDNA. Reverse
transcriptase polymerase chain reaction (RT-PCR) is, for example, performed
using specific
primers that hybridize to an RNA sequence of interest and a reverse
transcriptase enzyme.
Furthermore, RT-PCR can be performed with random primers, such as for instance
random
hexamers or decamers which hybridize randomly along the RNA, or oligo d(T)
which hybridizes
to the poly(A) tail of mRNA, and reverse transcriptase enzyme.
[00265] In some embodiments, RNA expression levels of the biomarkers in a
sample, such as
one from a patient suffering from or suspected of suffering from GEP-NEN or
associated
symptom or syndrome, are determined using quantitative methods such as by real-
time rt-PCR
(qPCR) or microarray analysis. In some embodiments, quantitative Polymerase
Chain Reaction
(QPCR) is used to quantify the level of expression of nucleic acids. In one
aspect, detection and
determining expression levels of the biomarkers is carried out using RT-PCR,
GeneChip
analysis, quantitative real-time PCR (Q RT-PCR), or carcinoid tissue
microarray (TMA)
immunostaining/quantitation, for example, to compare biomarker RNA, e.g.,
mRNA, or other
expression product, levels in different sample populations, characterize
patterns of gene
expression, to discriminate between closely related mRNAs, and to analyze RNA
structure.
[00266] In one example, QPCR is performed using real-time PCR (RTPCR), where
the
amount of product is monitored during the amplification reaction, or by end-
point
measurements, in which the amount of a final product is determined. As is
known to a skilled
58
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
person, rtPCR is for instance performed by the use of a nucleic acid
intercalator, such as for
example ethidium bromide or SYBR Green I dye, which interacts which all
generated double
stranded products resulting in an increase in fluorescence during
amplification, or for instance by
the use of labeled probes that react specifically with the generated double
stranded product of the
gene of interest. Alternative detection methods that can be used are provided
by amongst other
things dendrimer signal amplification, hybridization signal amplification, and
molecular
beacons.
[00267] In one embodiment, reverse transcription on total RNA is carried out
using the High
Capacity cDNA Archive Kit (Applied Biosystems (ABI), Foster City, CA)
following the
manufacturer's suggested protocol (briefly, using 2 micrograms of total RNA in
50 microliters
water, mixing with 50 uL of 2XRT mix containing Reverse Transcription Buffer,
deoxynucleotide triphosphate solution, random primers, and Multiscribe Reverse
Transcriptase).
RT reaction conditions are well known. In one example, the RT reaction is
performed using the
following thermal cycler conditions: 10 mins, 25 C; 120 min., 37 C (see Kidd
M. etal., -The
role of genetic markers--NAP1L1 , MAGE-D2, and MTA1--in defining small-
intestinal carcinoid
neoplasia." Ann Surg Oncol 2006;13(2):253-62).
[00268] For measurement of individual transcript levels, in one embodiment,
Assays-on-
DemandTM products are used with the ABI 7900 Sequence Detection System
according to the
manufacturer's suggestions (see Kidd M, Eick G. Shapiro MD. et al.
Microsatellite instability
and gene mutations in transforming growth factor-beta type II receptor are
absent in small bowel
carcinoid tumors. Cancer 2005;103(2):229-36). In one example, cycling is
performed under
standard conditions, using the TaqMan Universal PCR Master Mix Protocol, by
mixing cDNA
in 7.2 uL water, 0.8 uL 20=Assays-on-Demand primer and probe mix and 8 uL of
2X TaqMan
Universal Master mix, in a 384-well optical reaction plate, under the
following conditions: 50 C,
2 min.; 95 C; 10 min.; 50 cycles at 95 C for 15 min., 60 for 1 min (see Kidd
M, et al., "The
role of genetic markers--NAP1L1, MAGE-D2, and MTA1--in defining small-
intestinal carcinoid
neoplasia," Ann Surg Oncol 2006;13(2):253-62).
[00269] Typically, results from real-time PCR are normalized, using internal
standards and/or
by comparison to expression levels for housekeeping genes. For example, in one
embodiment,
Raw ACT (delta CT = change in cycle time as a function of amplification) data
from QPCR as
described above is normalized using well-known methods, such as geNorm (see
Vandesompele
59
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
J, De Preter K, Pattyn F, et al. Accurate normalization of real-time
quantitative RT-PCR data by
geometric averaging of multiple internal control genes. Genome Biol
2002;3(7):RESEARCH0034). Normalization by house-keeping gene expression levels
is also
well-known. See Kidd M, et al., "GeneChip, geNorm, and gastrointestinal
tumors: novel
reference genes for real-time PCR," Physiol Genomics 2007;30(3):363-70.
[00270] Microarray analysis involves the use of selected nucleic acid
molecules that are
immobilized on a surface. These nucleic acid molecules, termed probes, are
able to hybridize to
nucleic acid expression products. In a preferred embodiment the probes are
exposed to labeled
sample nucleic acid, hybridized, washed and the (relative) amount of nucleic
acid expression
products in the sample that are complementary to a probe is determined.
Microarray analysis
allows simultaneous determination of nucleic acid expression levels of a large
number of genes.
In a method according to the invention it is preferred that at least 5 genes
according to the
invention are measured simultaneously.
[00271] Background correction can be performed for instance according to the
"offset"
method that avoids negative intensity values after background subtraction.
Furthermore,
normalization can be performed in order to make the two channels on each
single array
comparable for instance using global loess normalization, and scale
normalization which ensures
that the log-ratios are scaled to have the same median-absolute-deviation
(MAD) across arrays.
[00272] Protein levels may, for example, be measured using antibody-based
binding assays.
Enzyme labeled, radioactively labeled or fluorescently labeled antibodies may
be used for
detection of protein. Exemplary assays include enzyme-linked immunosorbent
assays (ELISA),
radio-immuno assays (RIA), Western Blot assays and immunohistochemical
staining assays.
Alternatively, in order to determine the expression level of multiple proteins
simultaneously
protein arrays such as antibody-arrays are used.
[00273] Typically, the biomarkers and housekeeping markers are detected in a
biological
sample, such as a tissue or fluid sample, such as a blood, such as whole
blood, plasma, serum,
stool, urine, saliva, tears, serum or semen sample, or a sample prepared from
such a tissue or
fluid, such as a cell preparation, including of cells from blood, saliva, or
tissue, such as intestinal
mucosa, tumor tissue, and tissues containing and/or suspected of containing
GEP-NEN
metastases or shed tumor cells, such as liver, bone, and blood. In one
embodiment, a specific
cell preparation is obtained by fluorescence-activated cell sorting (FACS) of
cell suspensions or
fluid from tissue or fluid, such as mucosa, e.g., intestinal mucosa, blood or
buffy coat samples.
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
[00274] In some embodiments, the sample is taken from a GEP-NEN patient, a
patient
suspected of having GEP-NEN, a patient having and/or suspected of having
cancer generally, a
patient exhibiting one or more GEP-NEN symptoms or syndromes or determined to
be at-risk
for GEP-NEN, or a GEP-NEN patient undergoing treatment or having completed
treatment,
including patients whose disease is and/or is thought to be in remission.
[00275] In other embodiments, the sample is taken from a human without GEP-NEN
disease,
such as a healthy individual or an individual with a different type of cancer,
such as an
adenocarcinoma, for example, a gastrointestinal adenocarcinoma or one of the
breast, prostate,
or pancreas, or a gastric or hepatic cancer, such as esophageal, pancreatic,
gallbladder, colon, or
rectal cancer.
[00276] In some examples, the methods and systems distinguish between GEP-NEN
and
other cancers, such as adenocarcinomas, including gastrointestinal
adenocarcinoma or one of the
breast, prostate, or pancreas, or a gastric or hepatic cancer, such as
esophageal, pancreatic,
gallbladder, colon, or rectal cancer. In other embodiments, the methods and
systems
differentiate between GEP-NENs of different sites, such as between GEP-NENs of
the small
intestine and those of the pancreas. Such embodiments are useful, for example,
to determine the
primary location of a tumor where it is unknown and to determine prognosis
(particularly
because GEP-NEN tumors can exhibit significantly different prognosis depending
upon site of
origin). In some embodiments, the methods and systems differentiate between
GEP-NENs of
different sites, e.g., pancreatic and small intestinal tumors, with at least
80, 85, 90, 91, 92, or
greater accuracy. In other embodiments, the methods can diagnose or detect
adenocarcinomas
with neuroendocrine components.
[00277] In some embodiments, the sample is taken from the GEP-NEN tumor or
metastasis.
In other embodiments, the sample is taken from the GEP-NEN patient, but from a
tissue or fluid
not expected to contain GEP-NEN or GEP-NEN cells: such samples may be used as
reference or
normal samples. Alternatively, the normal or reference sample may be a tissue
or fluid or other
biological sample from a patient without GEP-NEN disease, such as a
corresponding tissue,
fluid or other sample, such as a normal blood sample, a normal small
intestinal (SI) mucosa
sample, a normal enterochromaffin (EC) cell preparation.
[00278] In some embodiments, the sample is a whole blood sample. As
neuroendocrine
tumors metastasize, they typically shed cells into the blood. Accordingly,
detection of the panels
of GEP-NEN biomarkers provided herein in plasma and blood samples may be used
for
61
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
identification of GEP-NENs at an early time point and for predicting the
presence of tumor
metastases, e.g., even if anatomic localization studies are negative.
Accordingly, the provided
agents and methods are useful for early diagnosis.
[00279] Thus, in some embodiments, the methods can identify a GEP-NEN
molecular
signature or expression profile in I mt. or about 1 mL of whole blood. In some
aspects, the
molecular signature or expression profile is stable for up to four hours (for
example, when
samples are refrigerated 4-8 C following phlebotomy) prior to freezing. In
one aspect, the
approach able to diagnose, prognose or predict a given GEP-NEN-associated
outcome using a
sample obtained from tumor tissue is also able to make the same diagnosis,
prognosis, or
prediction using a blood sample.
[00280] A number of existing detection and diagnostic methodologies require 7
to 10 days to
produce a possible positive result, and can be costly. Thus, in one aspect,
the provided methods
and compositions are useful in improving simplicity and reducing costs
associated with GEP-
NEN diagnosis, and make early-stage diagnosis feasible.
[00281] Thus in one example, the biomarkers are detected in circulation, for
example by
detection in a blood sample, such as a serum, plasma, cells, e.g., peripheral
blood mononuclear
cells (PBMCs), obtained from buffy coat, or whole blood sample.
[00282] Tumor-specific transcripts have been detected in whole blood in some
cancers. See
Sieuwerts AM, et at., "Molecular characterization of circulating tumor cells
in large quantities of
contaminating leukocytes by a multiplex real-time PCR," Breast Cancer Res
Treat
2009;118(3):455-68 and Mimori K. et al., "A large-scale study of MT1-MMP as a
marker for
isolated tumor cells in peripheral blood and bone marrow in gastric cancer
cases," Ann Surg
Oncol 2008;15(10):2934-42.
[00283] The CellSearchTM CTC Test (Veridex LLC) (described by Kahan L.,
"Medical
devices; immunology and microbiology devices; classification of the
immunomagnetic
circulating cancer cell selection and enumeration system. Final rule," Fed Re
gist 2004;69:26036-
8) uses magnetic beads coated with EpCAM- specific antibodies that detects
epithelial cells (CK-
8/18/19) and leukocytes (CD45), as described by Sieuwerts AM, Kraan J, Bolt-de
Vries J, et al.,
"Molecular characterization of circulating tumor cells in large quantities of
contaminating
leukocytes by a multiplex real-time PCR," Breast Cancer Res Treat
2009;118(3):455-68. This
method has been used to detect circulating tumor cells (CTCs), and monitoring
disease
progression and therapy efficacy in metastatic prostate (Danila DC, Heller G,
Gignac GA, et al.
62
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
Circulating tumor cell number and prognosis in progressive castration-
resistant prostate cancer.
Clin Cancer Res 2007;13(23):7053-8), colorectal (Cohen SJ, Alpaugh RK, Gross
S, et al.
Isolation and characterization of circulating tumor cells in patients with
metastatic colorectal
cancer. Clin Colorectal Cancer 2006;6(2):125-32,.and breast (Cristofanilli M,
Budd GT, Ellis
MJ, et al., Circulating tumor cells, disease progression, and survival in
metastatic breast cancer.
N Erigl .1 Med 2004;351(8):781-91).
[00284] This and other existing approaches have not been entirely satisfactory
for detection of
GEP-NEN cells, which can exhibit variable expression and/or not express
cytokeratin (See Van
Eeden S, et al, Classification of low-grade neuroendocrine tumors of midgut
and unknown
origin," Hum Pathol 2002;33(11):1126-32; Cal YC, et al., "Cytokeratin 7 and 20
and thyroid
transcription factor 1 can help distinguish pulmonary from gastrointestinal
carcinoid and
pancreatic endocrine tumors," Hum Pathol 2001;32(10):1087-93, and studies
described herein,
detecting EpCAM transcript expression in two of twenty-nine GEP-NEN samples).
[00285] Factors to consider in the available detection methods for circulating
tumor cells are
relatively low numbers of the cells in peripheral blood, typically about 1 per
106 peripheral blood
mononuclear cells (PBMCs) (see Ross AA, et al. "Detection and viability of
tumor cells in
peripheral blood stem cell collections from breast cancer patients using
immunocytochemical
and clonogenic assay techniques," Blood 1993;82(9):2605-10), and the potential
for leukocyte
contamination. See Sieuwerts AM, et al. -Molecular characterization of
circulating tumor cells
in large quantities of contaminating leukocytes by a multiplex real-time PCR."
Breast Cancer
Res Treat 2009;118(3):455-68; Mimori K, et al) and technical complexity of
available
approaches. These factors can render available methods not entirely
satisfactory for use in the
clinical laboratory.
[00286] In some embodiments, Neuroendocrine cells are FACS-sorted to
heterogeneity, using
known methods, following acridine orange (AO) staining and uptake, as
described Kidd M. et
al.. "Isolation, Purification and Functional Characterization of the Mastomys
EC cell," Am J
Physiol 2006;291:G778-91; Modlin IM, et a/.,`The functional characterization
of normal and
neoplastic human enterochromaffin cells," .1 Clin Endocrinol Metab
2006;91(6):2340-8.
[00287] In some embodiments, the provided detection methods are used to
detect, isolate, or
enrich for the GEP-NEN cells and/or biomarkers in two to three mL of blood or
less. The
methods are performed using standard laboratory apparatuses and thus are
easily performed in
63
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
the clinical laboratory setting. In one example, a readout is obtained within
12 hours, at an
average cost of approximately 20-30 per sample.
E. Diagnostic, Prognostic, and Predictive Uses
[00288] Also provided are diagnostic, prognostic, and predictive uses for the
agents and
detection methods provided herein, such as for the diagnosis, prognosis, and
prediction of GEP-
NEN, associated outcomes, and treatment responsiveness. For example, available
GEP-NEN
classification methods are limited, in part due to incorrect classifications
and that individual
lesions or tumors can evolve into different GEP-NEN sub-types or patterns,
and/or contain more
than one GEP-NEN sub-type. Known classification frameworks are limited, for
example, in the
ability to predict response to treatment or discriminate accurately between
tumors with similar
histopathologic features that may vary substantially in clinical course and
treatment response,
and to predict treatment responsiveness.
[00289] For example, the World Health Organization (WHO) classification
criteria, adopted
in 2000, distinguish between well differentiated NETs (WDNETs) (benign
behavior or uncertain
malignant potential), well differentiated neuroendocrine carcinomas (low-grade
malignancy)
(WDNECs), poorly differentiated neuroendocrine tumors (PDNETs) (medium grade
malignancy), and poorly differentiated (usually small cell) NECs (PDNECs)
(high-grade
malignancy), based on size, proliferative rate, localization, differentiation,
and hormone
production. Metastatic sub-types follow the same nomenclature and
classification strategy
(MET-WDNET; MET-WDNEC, MET-PDNET, MET-PDNEC). Proposed alternatives to
classification can be subjective. There is a need for molecular or gene-based
classification
schemes. The provided methods and systems, including GEP-NEN- specific
predictive gene-
based models, address these issues, and may be used in identifying and
analyzing molecular
parameters that are predictive of biologic behavior and prediction based on
such parameters.
[00290] Among the provided diagnostic, prognostic, and predictive methods are
those which
employ statistical analysis and biomathematical algorithms and predictive
models to analyze the
detected information about expression of GEP-NEN biomarkers and other markers
such as
housekeeping genes. In some embodiments, expression levels, detected binding
or other
information is normalized and assessed against reference value(s), such as
expression levels in
normal samples or standards. Provided embodiments include methods and systems
for
classification and prediction of GEP-NENs using the detected and measured
infon-nation about
64
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
the expression of the GEP-NEN biomarkers, for example, in classification,
staging, prognosis,
treatment design, evaluation of treatment options, and prediction of GEP-NEN
disease
outcomes, e.g., predicting development of metastases.
Detection and diagnosis of GEP-NEN
[00291] In some embodiments, the methods are used to establish GEP-NEN
diagnosis, such
as diagnosis or detection of early-stage disease or metastasis, define or
predict the extent of
disease, identify early spread or metastasis, predict outcome or prognosis,
predict progression,
classify disease, monitor treatment responsiveness, detect or monitor for
recurrence, and to
facilitate early therapeutic intervention. For example, among the provided
methods and
algorithms are those for use in classification, staging, prognosis, treatment
design, evaluation of
treatment options, and prediction of GEP-NEN disease outcomes, e.g.,
predicting development
of metastases.
[00292] In one embodiment, the methods, algorithms and models are useful for
diagnostic
surveillance, such as routine surveillance. In some embodiments, the methods,
algorithms and
models provide for early diagnosis; in one aspect, the methods are capable of
detection of low-
volume tumors, and detection of circulating tumor cells, including at early
stages of disease,
such as detection of as few as at or about 3 circulating GEP-NEN cells per
milliliter of blood. In
some embodiments, early detection allows early therapeutic intervention, at a
time when
therapies are more effective, which can improve survival rates and disease
outcomes.
[00293] For example, in one embodiment, the methods useful for early detection
of the
recurrence and/or metastasis of GEP-NEN, such as after treatment for example
following
surgical or chemical intervention. In some aspect, the methods are performed
weekly or
monthly following therapeutic intervention, for example, on human blood
samples. In some
aspects, the methods are capable of detecting micrometastases that are too
small to be detected
by conventional means, such as by imaging methods. For example, in one aspect
the methods
are capable of detecting metastases less than one centimeter (cm), such as at
or about 1, 0.9, 0.8,
0.7, 0.6, 0.5, 0.4. 0.3, 0.2, or 0.1 cm metastases. such as in the liver.
[00294] For example, among the provided methods and systems are those that
determine the
presence or absence (or both) of a GEP-NEN in a subject or sample with a
correct call rate of
between 56 and 92 %, such as at least or at least about a 65. 70, 75, 80, 81,
82. 83, 84, 85, 86,
87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, or 100 % correct call
rate. In some cases, the
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
methods are useful for diagnosis with a specificity or sensitivity of at least
or at least about 70,
75, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97,
98, 99, or 100 %.
[00295] In other aspects, the methods are capable of detecting the recurrence,
metastasis, or
spread of GEP-NEN following treatment or during initial di saease progression
at an earlier stage
as compared with other diagnostic methods, such as imaging and detection of
available
biomarkers. In some aspects, the detected expression levels and/or expression
signature of the
biomarkers correlate significantly with the progression of disease, disease
severity or
aggressiveness, lack of responsiveness of treatment, reduction in treatment
efficacy, GEP-NEN-
associated events, risk, prognosis, type or class of GEP-NEN or disease stage.
[00296] For example, in some embodiments, the methods are capable of
predicting or
monitoring the effects of therapeutic intervention. In one aspect, the methods
provide are
capable of detecting an improvement in disease as a result of treatment sooner
or more
effectively than available methods for detection and diagnosis, such as
detection of tumors and
metastasis by imaging and detection of available biomarkers, such as CgA.
Development and monitoring of treatment and therapeutic uses
[00297] Among the provided embodiments are methods that use the provided
biomarkers and
detection thereof in treatment development, strategy, and monitoring,
including evaluation of
response to treatment and patient-specific or individualized treatment
strategies that take into
consideration the likely natural history of the tumor and general health of
the patient.
[00298] GEP-NEN management strategies include surgery¨for cure (rarely
achieved) or
cytoreduction¨radiological intervention¨for example, by chemoembolisation or
radiofrequency ablation¨chemotherapy, cryo ablation, and treatment with
somatostatin and
somatostatin analogues (such as Sandostatin LARO (Octreotide acetate
injection)) to control
symptoms caused by released peptides and neuroamines, CPET-CT, and met
resection.
Biological agents, including interferon, and hormone therapy, and somatostatin-
tagged
radionucleotides are under investigation.
[00299] In one example, Cryoablation liberates GEP-NEN tissue for entry into
the blood,
which in turn induces symptoms. as described by Mazzaglia PJ, et al.,
"Laparoscopic
radiofrequency ablation of neuroendocrine liver metastases: a 10-year
experience evaluating
predictors of survival," Surgery 2007;142(1):10-9.
[00300] Chemotherapeutic agents, e.g., systemic cytotoxic chemotherapeutic
agents, include
etoposide, cisplatin, 5-fluorouracil, streptozotocin, doxorubicin; vascular
endothelial growth
66
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
factor inhibitors, receptor tyrosine kinase inhibitors (e.g., sunitinib,
sorafenib, and vatalanib),
and mammalian target of rapamycin (mTOR) inhibitors (e.g., temsirolimus and
everolimus), and
combinations thereof, for example to treat disseminated and/or poorly
differentiated disease.
Other treatment approaches are well known.
[00301] In some embodiments, the detection and diagnostic methods are used in
conjunction
with treatment, for example, by performing the methods weekly or monthly
before and/or after
treatment. In some aspects, the expression levels and profiles correlate with
the progression of
disease, ineffectiveness or effectiveness of treatment, and/or the recurrence
or lack thereof of
disease. In some aspects, the expression information indicates that a
different treatment strategy
is preferable. Thus, provided herein are therapeutic methods, in which the GEP-
NEN biomarker
detection methods are performed prior to treatment, and then used to monitor
therapeutic effects.
[00302] At various points in time after initiating or resuming treatment,
significant changes in
expression levels or expression profiles of the biomarkers (e.g., as compared
to expression or
expression profiles before treatment, or at some other point after treatment,
and/or in a normal or
reference sample) indicates that a therapeutic strategy is or is not
successful, that disease is
recurring, or that a different therapeutic approach should be used. In some
embodiments, the
therapeutic strategy is changed following performing of the detection methods,
such as by
adding a different therapeutic intervention, either in addition to or in place
of the current
approach, by increasing or decreasing the aggressiveness or frequency of the
current approach,
or stopping or reinstituting the treatment regimen.
[00303] In another aspect, the detected expression levels or expression
profile of the
biomarkers identifies the GEP-NEN disease for the first time or provides the
first definitive
diagnosis or classification of GEP-NEN disease. For example, in some aspects
the method
distinguishes between one or more of GEP-NEN classifications, such as WDNEC,
WDNET,
PDNEC, PDNET, and metastatic forms thereof, and/or distinguishes between GEP-
NEN and
other cancers, including other intestinal cancers. In some aspects of this
embodiment, a
treatment approach is designed based upon the expression levels or expression
profiles, and/or
the determined classification. The methods include iterative approaches,
whereby the biomarker
detection methods are followed by initiation or shift in therapeutic
intervention, followed by
continued periodic monitoring, reevaluation, and change, cessation, or
addition of a new
therapeutic approach, optionally with continued monitoring.
67
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
[00304] In some aspects, the methods and systems determine whether or not the
assayed
subject is responsive to treatment, such as a subject who is clinically
categorized as in complete
remission or exhibiting stable disease. In some aspects, the methods and
systems determine
whether or not the subject is untreated (or treatment-naive, i.e., has not
received treatment) or is
non-responsive (i.e., clinically categorized as "progressive." For example,
methods are provided
for distinguishing treatment-responsive and non-responsive patients, and for
distinguishing
patients with stable disease or those in complete remission, and those with
progressive disease.
In various aspects, the methods and systems make such calls with at least at
or about 65, 70, 75,
80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98,
99, or 100 % correct call
rate (i.e., accuracy), specificity, or sensitivity.
[00305] In some aspects, the sensitivity or correct call rate for the
diagnostic or predictive or
prognostic outcome is greater than, e.g., significantly greater than, that
obtained using a known
diagnosis or prognostic method, such as detection and measurement of
circulating CgA or other
single protein.
Statistical analysis, mathematical algorithms and predictive models
[00306] Typically, the diagnostic, prognostic, and predictive methods include
statistical
analysis and mathematical modeling. Thus, provided are supervised learning
algorithms useful
for the construction of predictive models, based on the GEP-NEN biomarkers
identified herein,
and methods and uses thereof for the prediction and classification of GEP-
NENs.
[00307] Any of a number of well-known methods for evaluating differences in
gene
expression may be used. Such methods range from simple comparisons of mean
expression
levels in each population e.g., using ANOVA (which is limited as the relevance
of changes are
complex to quantify) to mathematical analyses that are based on topographic,
pattern-recognition
based protocols e.g., support vector machines (SVM) (Noble WS. What is a
support vector
machine? Nat Biotechnol. 2006;24(12): 1565-7). Machine-learning based
techniques are
typically desirable for developing sophisticated, automatic, and/or objective
algorithms for
analyzing high-dimensional and multimodal biomedical data.
[00308] In some examples, SVM a variant of the supervised learning algorithm
is used in
connection with the provided methods and systems. SVMs have been used to
predict the
grading of astrocytomas with a >90% accuracy, and prostatic carcinomas with an
accuracy of
74-80% (Glotsos D, Tohka J, Ravazoula P. Cavouras D, Nikiforidis G. Automated
diagnosis of
brain tumours astrocytomas using probabilistic neural network clustering and
support vector
68
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
machines. Int J Neural Syst 2005;15(1-2): 1-11; Glotsos D, Tohka J, Ravazoula
P, Cavouras D,
Nikiforidis G. Automated diagnosis of brain tumours astrocytomas using
probabilistic neural
network clustering and support vector machines. Int J Neural Syst 2005;15(1-
2): 1-11).
[00309] Other algorithms for use with the provided methods and systems include
linear
discriminant analysis (LDA), naïve Bayes (NB), and K-nearest neighbor (KNN)
protocols. Such
approaches are useful for identifying individual or multi-variable alterations
in neoplastic
conditions (Drozdov I, Tsoka S, Ouzounis CA, Shah AM. Genome-wide expression
patterns in
physiological cardiac hypertrophy. BMC Genomics. 2010;11: 55; Freeman TC,
Coldovsky L.
Brosch M, et al. Construction, visualisation, and clustering of transcription
networks from
microarray expression data. PLoS Comput Biol 2007;3(10): 2032-42; Zampetaki A,
Kiechl S.
Drozdov I, et al. Plasma microRNA profiling reveals loss of endothelial miR-
126 and other
microRNAs in type 2 diabetes. Circ Res. 2010;107(6): 810-7. Epub 2010 Jul 22;
Dhawan M,
Selvaraja S, Duan ZH. Application of committee kNN classifiers for gene
expression profile
classification. Int J Bioinform Res Appl. 2010;6(4): 344-52; Kawarazaki S,
Taniguchi K,
Shirahata M, et al. Conversion of a molecular classifier obtained by gene
expression profiling
into a classifier based on real-time PCR: a prognosis predictor for gliomas.
BMC Med
Genomics. 2010;3: 52; Vandebriel RJ, Van Loveren H, Meredith C. Altered
cytokine (receptor)
mRNA expression as a tool in immunotoxicology. Toxicology. 1998;130(1): 43-67;
Urgard E,
Vooder T, Vosa U, et al. Metagenes associated with survival in non-small cell
lung cancer.
Cancer Inform. 2011;10: 175-83. Epub 2011 Jun 2; Pimentel M, Amichai M, Chua
K, Braham
L. Validating a New Genomic Test for Irritable Bowel Syndrome Gastroenterology
2011;140
(Suppl 1): S-798; Lawlor G, Rosenberg L, Ahmed A, et al. Increased Peripheral
Blood GATA-3
Expression in Asymptomatic Patients With Active Ulcerative Colitis at
Colonoscopy.
Gastroenterology 2011;140 (Suppl 1)).
[00310] In some embodiments, the provided methods and systems analyze
expression of the
GEP-NEN biomarkers as a group, with outputs dependent on an expression
signature, such as
expression signatures or profiles that are distinct between normal or
reference samples and
samples obtained from a subject with a "GEP-NEN." In such embodiments, pattern
recognition
protocols generally are used. Such approaches are useful, for example, to
identify malignant
signatures and signaling pathways in GEP-NEN tumor tissue (such as those
described in
Drozdov I, Kidd M, Nadler B, et al. Predicting neuroendocrine tumor
(carcinoid) neoplasia using
gene expression profiling and supervised machine learning. Cancer.
2009;115(8): 1638-50) and
69
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
determining whether individual plasma samples were obtained from normal
control or GEP-
NENs (for example, as described in Modlin IM, Gustafsson BI, Drozdov I, Nadler
B, Pfragner
R, Kidd M. Principal component analysis, hierarchical clustering, and decision
tree assessment
of plasma mRNA and hormone levels as an early detection strategy for small
intestinal
neuroendocrine (carcinoid) tumors. Ann Surg Oncol 2009;16(2): 487-98)..
[00311] Methods using the predictive algorithms and models use statistical
analysis and data
compression methods, such as those well known in the art. For example,
expression data may be
transformed, e.g., ln-transformed, and imported into a statistical analysis
program, such as
Partek Genomic Suite ("Partek," Partek Genomics SuiteTM, ed. Revision 6.3
St. Louis:
Partek Inc, 2008) or similar program, for example. Data are compressed and
analyzed for
comparison.
[00312] Statistical analyses include determining mean (M), e.g., geometric
mean, of gene
expression levels for individual sample types, standard deviations (SD) among
types of samples,
Geometric Fold Change (FC) between different sample types or conditions,
calculated as the
ratio of geometric means for two groups of samples or values, comparison of
expression levels
by 2-tailed Fisher's test, or two-sample t-test, e.g., to identify biomarker
genes differentially
expressed between various samples and tissue types. Analysis of Variance
(ANOVA) is used to
evaluate differences in biomarker expression levels between expression in
different samples
and/or values. In one example, a two-class unpaired algorithm is implemented,
such as by
expression levels from a test and normal sample or reference value defining
the two groups.
[00313] Whether differences in expression levels, amounts, or values are
deemed significant
may be determined by well-known statistical approaches, and typically is done
by designating a
threshold for a particular statistical parameter, such as a threshold p-value
(e.g., p < 0.05),
threshold S-value (e.g., 0.4, with S < -0.4 or S > 0.4), or other value, at
which differences are
deemed significant, for example, where expression of a biomarker is considered
significantly
down- or up-regulated, respectively, among two different samples, for example,
representing
two different GEP-NEN sub-types, tumors, stages, localizations,
aggressiveness, or other aspect
of GEP-NEN or normal or reference sample.
[00314] In one aspect, the algorithms, predictive models, and methods are
based on
biomarkers expressed from genes associated with regulatory genotypes (i.e.,
adhesion,
migration, proliferation, apoptosis, metastasis, and hormone secretion)
underlying various GEP-
NEN subtypes.
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
[00315] In one aspect, the methods apply the mathematical formulations,
algorithms or
models identify specific cutoff points, for example, expression levels or
amounts, which
distinguish between normal and GEP-NEN samples, between GEP-NEN and other
cancers, and
between various sub-types, stages, and other aspects of disease or disease
outcome. In another
aspect, the methods are used for prediction, classification, prognosis, and
treatment monitoring
and design. In one aspect, the predictive embodiments are useful for
identifying molecular
parameters predictive of biologic behavior, and prediction of various GEP-NEN-
associated
outcomes using the parameters. In one aspect of these embodiments, machine
learning
approaches are used, e.g., to develop sophisticated, automatic and objective
algorithms for the
analysis of high-dimensional and multimodal biomedical data.
Compression of data and determining expression profiles
[00316] For the comparison of expression levels or other values, and to
identify expression
profiles (expression signatures) or regulatory signatures based on GEP-NEN
biomarker
expression, data are compressed. Compression typically is by Principal
Component Analysis
(PCA) or similar technique for describing and visualizing the structure of
high-dimensional data.
PCA allows the visualization and comparison of GEP-NEN biomarker expression
and
determining and comparing expression profiles (expression signatures,
expression patterns)
among different samples, such as between normal or reference and test samples
and among
different tumor types.
[00317] In some embodiments, expression level data are acquired, e.g., by real-
time PCR, and
reduced or compressed, for example, to principal components.
[00318] PCA is used to reduce dimensionality of the data (e.g., measured
expression values)
into uncorrelated principal components (PCs) that explain or represent a
majority of the variance
in the data, such as about 50. 60, 70, 75, 80, 85, 90, 95 or 99 % of the
variance.
[00319] In one example, the PCA is 3-component PCA, in which three PCs are
used that
collectively represent most of the variance, for example, about 75%. 80%, 85%,
90%, or more
variance in the data (Jolliffe IT, "Principle Component Anlysis," Springer,
1986).
[00320] PCA mapping, e.g., 3-component PCA mapping is used to map data to a
three
dimensional space for visualization, such as by assigning first (1st), second
(2nd) and third (3rd)
PCs to the x-, y-, and z-axes, respectively.
[00321] PCA may be used to determine expression profiles for the biomarkers in
various
samples. For example, reduced expression data for individual sample types
(e.g., each tumor
71
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
type, sub-type or grade, or normal sample type) are localized in a PCA
coordinate system and
localized data used to determine individual transcript expression profiles or
signatures.
[00322] In one aspect, the expression profile is determined for each sample by
plotting or
defining a centroid (center of mass; average expression), corresponding to or
representing the
sample's individual transcript expression profile (regulatory signature), as
given by the principal
component vector, as determined by PCA for the panel of biomarkers.
[00323] Generally, two centroids or points of localization separated by a
relatively large
distance in this coordinate system represent two relatively distinct
transcript expression profiles.
Likewise, relatively close centroids represent relatively similar profiles. In
this representation,
the distance between centroids is inversely equivalent to the similarity
measure (greater distance
= less similarity) for the different samples, such that large distances or
separation between
centroids indicates samples having distinct transcript expression signatures.
Proximity of
centroids indicates similarity between samples. For example, the relative
distance between
centroids for different GEP-NEN tumor samples represents the relative
similarity of their
regulatory signatures or transcript expression profiles.
Correlation, linear relationships and regulatory clusters
[00324] In one aspect, the statistical and comparative analysis includes
determining the
inverse correlation between expression levels or values for two biomarkers. In
one example, this
correlation and the cosine of the angle between individual expression vectors
(greater angle =
less similarity), is used to identify related gene expression clusters
(Gabriel KR, "The biplot
graphic display of matrices with application to principal component analysis,"
Biometrika
1971;58(3):453).
[00325] In some embodiments, there is a linear correlation between expression
levels of two
or more biomarkers, and/or the presence or absence of GEP-NEN, sub-type,
stage, or other
outcome. In one aspect, there is an expression-dependant correlation between
the provided
GEP-NEN biomarkers and characteristics of the biological samples, such as
between biomarkers
(and expression levels thereof) and various GEP-NEN sub-types (primary or
metastatic), normal
versus GEP-NEN samples, and/or primary versus metastatic or aggressive
disease.
[00326] Pearson's Correlation (PC) coefficients (R2) may be used to assess
linear
relationships (correlations) between pairs of values, such as between
expression levels of a
biomarker for different biological samples (e.g., tumor sub-types) and between
pairs of
biomarkers. This analysis may be used to linearly separate distribution in
expression patterns,
72
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
by calculating PC coefficients for individual pairs of the biomarkers (plotted
on x- and y- axes of
individual Similarity Matrices). Thresholds may be set for varying degrees of
linear correlation,
such as a threshold for highly linear correlation of (R2> 0.50, or 0.40).
Linear classifiers can be
applied to the datasets. In one example, the correlation coefficient is 1Ø
[00327] In one embodiment, regulatory clusters are determined by constructing
networks of
correlations using statistical analyses, for example, to identify regulatory
clusters composed of
subsets of the panel of biomarkers. In one example, PC correlation
coefficients are determined
and used to construct such networks of correlations. In one example, the
networks are identified
by drawing edges between transcript pairs having R2 above the pre-defined
threshold. Degree of
correlation can provide information on reproducibility and robustness.
Predictive Models and Supervised Learning Algorithms
[00328] Also provided herein are objective algorithms, predictive models, and
topographic
analytical methods, and methods using the same, to analyze high-dimensional
and multimodal
biomedical data, such as the data obtained using the provided methods for
detecting expression
of the GEP-NEN biomarker panels. As discussed above, the objective algorithms,
models, and
analytical methods include mathematical analyses based on topographic, pattern-
recognition
based protocols e.g., support vector machines (SVM) (Noble WS. What is a
support vector
machine? Nat Biotechnol. 2006;24(12): 1565-7), linear discriminant analysis
(LDA), naïve
Bayes (NB), and K-nearest neighbor (KNN) protocols, as well as other
supervised learning
algorithms and models, such as Decision Tree, Perceptron, and regularized
discriminant analysis
(RDA), and similar models and algorithms well-known in the art (Gallant SI,
"Perceptron-based
learning algorithms," Perceptron -based learning algorithms 1990;1(2):179-91).
[00329] In some embodiments, biomarker expression data is analyzed in
biological samples,
using feed-forward neural networks; best transcripts-predictors are selected.
[00330] In some embodiments, Feature Selection (FS) is applied to remove the
most
redundant features from a dataset, such as a GEP-NEN biomarker expression
dataset. FS
enhances the generalization capability, accelerates the learning process, and
improves model
interpretability. In one aspect, FS is employed using a "greedy forward"
selection approach,
selecting the most relevant subset of features for the robust learning models.
(Peng H, Long F,
Ding C, "Feature selection based on mutual information: criteria of max-
dependency, max-
relevance, and mm-redundancy," IEEE Transactions on Pattern Analysis and
Machine
Intelligence, 2005;27(8):1226-38).
73
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
[00331] In some embodiments, Support Vector Machines (SVM) algorithms are used
for
classification of data by increasing the margin between the n data sets
(Cristianini N, Shawe-
Taylor J. An Introduction to Support Vector Machines and other kernel-based
learning methods.
Cambridge: Cambridge University Press, 2000).
[00332] In some embodiments, the predictive models include Decision Tree,
which maps
observations about an item to a conclusion about its target value (Zhang H,
Singer B. Recursive
Partitioning in the Health Sciences," (Statistics for Biology and Health):
Springer, 1999.). The
leaves of the tree represent classifications and branches represent
conjunctions of features that
devolve into the individual classifications. It has been used effectively (70-
90%) to predict
prognosis of metastatic breast cancer (Yu L et al "TGF-beta receptor-activated
p38 MAP kinase
mediates Smad-independent TGF-beta responses.," Embo J 2002;21(14):3749-59),
as well as
colon cancer (Zhang H et al "Recursive partitioning for tumor classification
with gene
expression microarray data.," Proc Nall Acad Sci USA 2001;98(12):6730-5.), to
predict the
grading of astrocytomas (Glotsos D et al "Automated diagnosis of brain tumours
astrocytomas
using probabilistic neural network clustering and support vector machines.,"
Int J Neural Syst
2005;15(1-2):1-11.) with a >90% accuracy, and prostatic carcinomas with an
accuracy of 74-
80% (Mattfeldt T et al. -Classification of prostatic carcinoma with artificial
neural networks
using comparative genomic hybridization and quantitative stereological data.,"
Pathol Res Pract
2003;199(12):773-84.). The efficiency of this technique has been measured by
10-fold cross-
validation (Pirooznia M et al "A comparative study of different machine
learning methods on
microarray gene expression data.," BMC Genomics 2008;9 Suppl 1:S13.).
[00333] The predictive models and algorithms further include Perceptron, a
linear classifier
that forms a feed forward neural network and maps an input variable to a
binary classifier
(Gallant SI. "Perceptron-based learning algorithms," Perceptron-based learning
algorithms
1990;1(2):179-91). It has been used to predict malignancy of breast cancer
(Markey MK et al.
"Perceptron error surface analysis: a case study in breast cancer diagnosis.,"
Comput Biol Med
2002;32(2):99-109). In this model, the learning rate is a constant that
regulates the speed of
learning. A lower learning rate improves the classification model, while
increasing the time to
process the variable (Markey MK et al. "Perceptron error surface analysis: a
case study in breast
cancer diagnosis.," Comput Biol Med 2002;32(2):99-109). In one example, a
learning rate of
0.05 is used. In one aspect, a Perceptron algorithm is used to distinguish
between localized or
74
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
primary tumors and corresponding metastatic tumors. In one aspect, three data
scans are used to
generate decision boundaries that explicitly separate data into classes.
[00334] The predictive models and algorithms further include Regularized
Discriminant
Analysis (RDA), which can be used as a flexible alternative to other data
mining techniques,
including Linear and Quadratic Discriminant Analysis (LDA, QDA) (Lifien RH,
Farid H,
Donald BR. "Probabilistic disease classification of expression-dependent
proteomic data from
mass spectrometry of human serum.," J Comput Biol 2003;10(6):925-46.;
Cappellen D, Luong-
Nguyen NH, Bongiovanni S, et al. "Transcriptional program of mouse osteoclast
differentiation
governed by the macrophage colony-stimulating factor and the ligand for the
receptor activator
of NFkappa B.," J Biol Chem 2002;277(24):21971-82.). RDA's regularization
parameters, 7 and
X, are used to design an intermediate classifier between LDA and QDA. QDA is
performed
when y=0 and X=0 while LDA is performed when y=0 and X=1 (Picon A, Gold LI,
Wang J,
Cohen A, Friedman E. A subset of metastatic human colon cancers expresses
elevated levels of
transforming growth factor betal. Cancer Epidemiol Biomarkers Prey
1998;7(6):497-504).
[00335] To reduce over-fitting, RDA parameters are selected to minimize cross-
validation
error while not being equal 0.0001, thus forcing RDA to produce a classifier
between LDA,
QDA, and L2 (Pima I, Aladjem M., -Regularized discriminant analysis for face
recognition,"
Pattern Recognition 2003;37(9):1945-48). Finally, regularization itself has
been used widely to
overcome over-fitting in machine learning (Evgeniou T, Pontil M, Poggio T.
"Regularization
Networks and Support Vector Machines.," Advances in Computational Math
2000;13(1):1-50.;
Ji S, Ye J. Kernel "Uncorrelated and Regularized Discriminant Analysis: A
Theoretical and
Computational Study.," IEEE Transactions on Knowledge and Data Engineering
2000;20(10):1311-21.).
[00336] In one example, regularization parameters are defined as y = 0.002 and
X. = 0. In one
example, for each class pair, S-values are assigned to all transcripts which
are then arranged by a
decreasing S-value. RDA is performed, e.g., 21 times, such that the Nth
iteration consists of top
N scoring transcripts. Error estimation can be carried out by a 10-fold cross-
validation of the
RDA classifier. This can be done by partitioning the tissue data set into
complementary subsets,
performing the analysis on one subset (called the training set), and
validating the analysis on the
other subset (called the validation set or testing set).
Calculating misclassification error
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
[00337] In one example, misclassification error is averaged to reduce
variability in the overall
predictive assessment, which can provide a more accurate approach to error
estimation
compared to other approaches, including bootstrapping and leave-one-out cross-
validation
(Kohavi R. "A study of cross-validation and bootstrap for accuracy estimation
and model
selection.," Proceedings of the Fourteenth International Joint Conference on
Artificial
Intelligence, 1995 ;2(12): 1137-43.).
[00338] In one example, selection for tissue classification is performed, for
example, by
computing the rank score (S) for each gene and for each class pair as:
1Pc2
[00339] =
Jci
aC I C2
[00340] where and pc, represent means of first and second class
respectively and a( .J and
ifyõ are inter-class standard deviations. A large S value is indicative of a
substantial differential
expression ("Fold Change") and a low standard deviation ("transcript
stability") within each
class. Genes may be sorted by a decreasing S-value and used as inputs for the
regularized
discriminant analysis algorithm (RDA).
[00341] The algorithms and models may be evaluated, validated and cross-
validated, for
example, to validate the predictive and classification abilities of the
models, and to evaluate
specificity and sensitivity. In one example, radial basis function is used as
a kernel, and a 10-
fold cross-validation used to measure the sensitivity of classification
(Cristianini N, Shawe-
Taylor J. "An Introduction to Support Vector Machines and other kernel-based
learning
methods.," Cambridge: Cambridge University Press, 2000.). Various
classification models and
algorithms may be compared by the provided methods, for example, using
training and cross-
validation, as provided herein, to compare performance of the predictive
models for predicting
particular outcomes.
[00342] Embodiments of the provided methods, systems, and predictive models
are
reproducible, with high dynamic range, can detect small changes in data, and
are performed
using simple methods, at low cost, e.g., for implementation in a clinical
laboratory.
F. KITS
[00343] For use in the diagnostic, prognostic, predictive, and therapeutic
applications
described or suggested above, kits and other articles of manufacture are
provided. In some
76
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
embodiments, the kits include a carrier, package, or packaging,
compartmentalized to receive
one or more containers such as vials, tubes, plates, and wells, in which each
of the containers
includes one of the separate elements for use in the methods provided herein,
and in some
aspects further include a label or insert with instructions for use, such as
the uses described
herein. In one example, the individual containers include individual agents
for detection of the
GEP-NEN biomarkers as provided herein; in some examples, individual containers
include
agents for detection of housekeeping genes and/or normalization.
[00344] For example, the container(s) can comprise an agent, such as a probe
or primer,
which is or can be detectably labeled. Where the method utilizes nucleic acid
hybridization for
detection, the kit can also have containers containing nucleotide(s) for
amplification of the target
nucleic acid sequence. Kits can comprise a container comprising a reporter,
such as a biotin-
binding protein, such as avidin or streptavidin, bound to a reporter molecule,
such as an
enzymatic, fluorescent, or radioisotope label; such a reporter can be used
with, e.g., a nucleic
acid or antibody.
[00345] The kits will typically comprise the container(s) described above and
one or more
other containers associated therewith that comprise materials desirable from a
commercial and
user standpoint, including buffers, diluents, filters, needles, syringes;
carrier, package, container,
vial and/or tube labels listing contents and/or instructions for use, and
package inserts with
instructions for use.
[00346] A label can be present on or with the container to indicate that the
composition is
used for a specific therapeutic or non-therapeutic application, such as a
prognostic, prophylactic,
diagnostic or laboratory application, and can also indicate directions for
either in vivo or in vitro
use, such as those described herein. Directions and or other information can
also be included on
an insert(s) or label(s) which is included with or on the kit. The label can
be on or associated
with the container. A label a can be on a container when letters, numbers or
other characters
forming the label are molded or etched into the container itself; a label can
be associated with a
container when it is present within a receptacle or carrier that also holds
the container, e.g., as a
package insert. The label can indicate that the composition is used for
diagnosing, treating,
prophylaxing or prognosing a condition, such as GEP-NEN.
[00347] In another embodiment, an article(s) of manufacture containing
compositions, such as
amino acid sequence(s), small molecule(s), nucleic acid sequence(s), and/or
antibody(s), e.g.,
materials useful for the diagnosis, prognosis, or therapy of GEP-NEN is
provided. The article of
77
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
manufacture typically comprises at least one container and at least one label.
Suitable containers
include, for example, bottles, vials, syringes, and test tubes. The containers
can be formed from
a variety of materials such as glass, metal or plastic. The container can hold
amino acid
sequence(s), small molecule(s), nucleic acid sequence(s), cell population(s)
and/or antibody(s).
In one embodiment, the container holds a polynucleotide for use in examining
the mRNA
expression profile of a cell, together with reagents used for this purpose. In
another
embodiment a container comprises an antibody, binding fragment thereof or
specific binding
protein for use in evaluating protein expression of GEP-NEN biomarkers in
cells and tissues. Or
for relevant laboratory, prognostic, diagnostic, prophylactic and therapeutic
purposes;
indications and/or directions for such uses can be included on or with such
container, as can
reagents and other compositions or tools used for these purposes.
[00348] The article of manufacture can further comprise a second container
comprising a
pharmaceutically-acceptable buffer, such as phosphate-buffered saline,
Ringer's solution and/or
dextrose solution. It can further include other materials desirable from a
commercial and user
standpoint, including other buffers, diluents, filters, stirrers. needles,
syringes, and/or package
inserts with indications and/or instructions for use.
EXAMPLES
[00349] Various aspects of the invention are further described and illustrated
by way of the
several examples which follow, none of which are intended to limit the scope
of the invention.
EXAMPLE 1: DETECTION AND DETERMINING EXPRESSION LEVELS OF
BIOMARKERS IN GEP-NEN AND NORMAL SAMPLES
Sample preparation, RNA extraction, real-time PCR
[00350] Normal and neoplastic samples were obtained for detection and
determination of
GEP-NEN biomarker expression levels by real-time PCR. Normal samples included
twenty-
seven (27) normal small intestinal (SI) mucosa samples (NML), and thirteen
(13) normal human
enterochromaffin (EC) cell preparations (NML_EC; obtained through fluorescence-
activated cell
sorting (FACS) of normal mucosa; >98% pure EC cells (Modlin IM ei al., "The
functional
characterization of normal and neoplastic human enterochromaffin cells," J
Clin Endocrinol
Metab 2006;91(6):2340-8).
78
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
[00351] Neoplastic samples included fifty-three (53) primary SI GEP-NENs and
twenty-one
(21) corresponding liver metastases collected from a frozen biobank (all
tissues microdissected).
The GEP-NEN samples were obtained from patients enrolled according to
protocols approved
by the Institutional Review Board of Yale University. Each was classified as
functional, with
greater than 80% pure neoplastic cells and as positive for TPHl , confirming
it was EC cell-
derived (Modlin IM et al., "The functional characterization of normal and
neoplastic human
enterochromaffin cells," J Clin Endocrinol Meiab 2006;91(6):2340-8). Patient
samples also
were collected from adenocarcinomas of the breast (n=53), colon (n=21), and
pancreas (n=16).
[00352] Primary GEP-NENs were classified pathologically according to the 2000
World
Health Organization (WHO) standard, as well differentiated NETs ((WDNETs)
(n=26) (benign
behavior or uncertain malignant potential)); well differentiated
neuroendocrine carcinomas
((WDNECs) (n=20) (low-grade malignancy)); poorly differentiated neuroendocrine
tumors
((PDNETs) (n=5) (medium grade malignancy)); and poorly-differentiated
(typically small-cell)
neuroendocrine carcinomas ((PDNECs) (n=2) (high grade malignancy)). Metastatic
GEP-NEN
tissue samples (metastases; MET) (collected from liver resections from
corresponding tumor
types), were classified using a similar standard, as: WDNET MET (n=6), WDNEC
MET (n=12),
and PDNEC MET (n=3). Metastatic PDNETs (PDNET METs) are classified using the
same
method.
[00353] For real-time PCR, RNA was extracted from various normal and
neoplastic samples
(27 samples of normal SI mucosa, 13 preparations of normal human EC cells, 53
primary SI
GEP-NENs, 21 corresponding liver metastases, and 53 adenocarcinoma samples)
using TRIzol0
reagent (ready-to-use, monophasic solution of phenol and guanidine
isothiocyanate;
InvitrogenTM, Carlsbad, California).
[00354] Transcript expression levels were measured by real-time PCR using
Assays-on-
DemandTM gene expression products and the ABI 7900 Sequence Detection System
(both from
Applied Biosystems) according to the manufacturer's suggestions (Kidd M et al,
"Microsatellite
instability and gene mutations in transforming growth factor-beta type II
receptor are absent in
small bowel carcinoid tumors," Cancer 2005;103(2):229-36). Cycling was
performed under
standard conditions using the TaqMan0 Universal PCR Master Mix Protocol
(Applied
Biosystems).
[00355] GEP-NEN biornarkers were detected and expression levels measured by
real-time
PCR, using sets of polynucleotide primer pairs, where each set contained
primer pairs designed
79
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
to specifically bind to and amplify a panel of GEP-NEN biomarkers. The GEP-NEN
biomarker
panel included products (transcripts) of genes implicated in typical primary
and metastatic GEP-
NEN phenotypes, for example, genes involved in adhesion, migration,
proliferation, apoptosis,
metastasis, and hormone secretion. and neuroendocrine marker genes.
Housekeeping gene
(ALG9, TFCP2 and ZNF410) expression levels also were measured. Raw ACT values
for
biomarker expression were normalized using the geNorm algorithm (Vandesompele
J et al.,
"Accurate normalization of real-time quantitative RT-PCR data by geometric
averaging of
multiple internal control genes," Genome Biol 2002;3(7):RESEARCH0034) and the
housekeeping expression levels.
[00356] Normalized data were natural log (1n)-transformed for compression and
imported into
Partek Genomic Suite (Partek, "Partek Genomics SuiteTM," ed. Revision 6.3
St. Louis:
Partek Inc, 2008). Mean gene expression levels (M) of the various biomarker
transcripts and
Standard Deviations (SD) were calculated. All statistical computations were
carried out using R
2.9 language for statistical computing (R Development Core Team. R, "A
language and
environment for statistical computing," Vienna, Austria: R Foundation for
Statistical
Computing, 2008).
Detection and determination of transcript expression levels of a 9-biomarker
panel
[00357] Expression levels were determined by real-time PCR as described above
using a set
of primer pairs specific for a panel of nine GEP-NEN biomarkers (MAGE-D2,
MTAI, NAPILI,
Ki-67,S'urvivin, FZD7, Kiss], NRP2, and CgA (see Kidd M etal., "The role of
genetic markers--
NAP1L , MAGE-D2, and MTA1--in defining small-intestinal carcinoid neoplasia,"
Ann Surg
Oncol 2006;13(2):253-62; Kidd M et al., "Q RT-PCR detection of chromogranin A:
a new
standard in the identification of neuroendocrine tumor disease," Ann Surg
2006;243(2):273-80)
transcripts. The sequences of the primer pairs are listed in Table 1A, below,
with other
information about primer pairs listed in Table 1B. Expression of the nine
biomarkers
(transcripts) was measured in samples from primary SI GEP-NEN (AKA SI NET) (n
= 53),
corresponding liver metastases (ti = 21), and normal EC cell preparations (n =
13). Expression
levels in the tumor samples were compared for each biomarker to corresponding
average
expression levels in the normal enterochomaffin (EC) cell preparations. Based
on this
CA 02828878 2013-08-30
WO 2012/119013
PCT/US2012/027351
comparison, expression levels in the tumor samples were classified as
Upregulated,
Downregulated, or Baseline.
Table 1: Sets of primers for GEP-NEN biomarkers and housekeeping genes
Table 1A: Primer sequences
GEP-NEN
Biomarker Forward SEQ SEQ
Reverse Primer
or Primer ID ID
sequence
Housekee sequence NO: NO:
ping Gene
5'- 5'-
AKAP8L gaagcatctgaaga 106 atgagggaggacttcttg 107
ccatgg-3' ga-3'
5'- 5'-
APLP2 cggtgccgaagag 41 ctctctcggcattgaaaat 42
aaagtga-3' c-3'
5'- 5'-
ARAF1 ctcatcgacgtggc 43 gtggatgatgttcttggcat 44
ccggca-3' -3'
5'- 5'-
ATP6V1H caggtccgctataa 108 ggcagcggtctggggct 109
tgctct-3' gct-3'
5'- 5'-
BNIP3L cagagtagttccag 110 aaacatgatctgcccatctt 111
aggcag-3' c-3'
5'- 5'-
BRAF1 cctcttcggctgcg 45 gtgtcaacttaatcatttgt- 46
gaccct-3 3'
5'- 5'-
C210RF7 attactgtgcccgtg 112 gaaagaccaaaggaatg 113
gaaat-3' gag-3'
5'- 5'-
CD59 ggctgctgctcgtc 47 ttgggttaggacagttgta 48
ctggct-3' g-3'
5'- 5'-
CgA aacggatcctttcca 49 ctgagagttcatcttcaaa 50
ttctg-3' a-3'
COMMD
ctcaaaaacctgct 114 gtggcagagagatctgatt 115
9
gacaaa-3' t-3'
CTGF tgcgaagctgacct 51 ttttgggagtacggatgca 52
ggaaga-3' c-3'
5'- 5'-
CXCL14 aagcgcttcatcaa 53 tgaggtttttcaccctattc- 54
gtggta-3' 3'
81
CA 02828878 2013-08-30
WO 2012/119013
PCT/US2012/027351
GEP-NEN
Biomarker Forward SEQ SEQ
Reverse Primer
or Primer ID ID
Housekee sequence NO: sequence
NO:
ping Gene
5'- 5'-
ENPP4 gctggacaattgtg 116 aaatggatgcatactagg 117
ctaaatg-3' ca-3'
FAM131
A tcagcgagggcga 118 tccgcgatggcaaactgc 119
acaagag-3' tc-3'
5'- 5'-
FLJ10357 acacaaactggag 120 tgttccagaggctgctgca 121
aatggtc-3' g-3'
5'- 5'-
FZD7 gatgcgggacccc 55 cagcagcgccagcacca 56
ggcgcgg-3' ggg-3'
5'- 5'-
GLT8D1 aacttcctcagcttg 122 atttgggacaaagtctata 123
agcag-3' g-3'
5'- 5'-
GRIA2 aagtttgcatacctc 57 tttcagcagcagaatccag 58
tatga-3' ca-3'
5'- 5'-
HDAC9 gcagaagcaatac 124 tgcttcagttgttcaataga 125
cagcagc-3' -3'
1
5' 5'
HOXC6 ccagatttacccctg 59 cgagtagatctggcggcc 60
gatgc-3' gc-3'
5'- 5'-
HSF2 aggaagacaattta 126 gatgtaatctgtgggattc 127
gcatag-3' a-3'
5'- 5'-
Ki-67 gcacgtcgtgtctc 61 gacacacgccttcttttcaa 62
aagatc-3' -3'
5'- 5'-
Kiss1 gtggcctctgtggg 63 ctccccgggggccagga 64
gaattc-3' ggc-3'
H
5'-
5'-tcatcttttctttatgtttt-
KRAS tcaggacttagcaa 65 66
3'
gaagtt -3'
5'- 5'-
LE01 ggaaggcgagga 128 gagctttatcttcttctgat- 129
gtccatca-3' 3'
MAGE-
D2 gaatcaggatactc 67 actctgatcactgctgcca 68
ggccca-3' t-3'
82
CA 02828878 2013-08-30
WO 2012/119013
PCT/US2012/027351
GEP-NEN
Biomarker Forward SEQ SEQ
Reverse Primer
or Primer ID ID
Housekee sequence NO: sequence
NO:
ping Gene
5'-
MORF4L
2 gcaaagaattctgc 130 aattttacaagaaaaagac 131
atctct-3' t-3.
5'- 5'-
MTA1 ggcggtacgcaag 69 ggacacgcttttcacggg 70
ccgctgg-3' gtc-3'
5'- 5'-
NAPILI ggtctaccttctgctt 71 tcttgtaagaactaaaattg
72
ccct-3' -3'
5'- 5'-
NKX2,-3 aaggaacatgaag 73 ccgccttgcagtctccgg 74
aggagcc-3' ccg-3'
5'- 5'-
NOL3 ccgtgttggcctcc 132 ggcgtttccgctcgcggtc 133
aggtcc-3' g-3'
5'- 5'-
NRP2 agccctctacttttc 75 agccagcatctttggaatt 76
aagaca-3' ca-3'
5'- 5'-
NUDT3 t2gcagcagttcgt 134 aactaatcttcccaatgtcc 135
gaagtc-3' -3'
5' 5'
OAZ2 ctcccaccctgagc 136 gtgcctgcagcactggag 137
agagcc-3' -3'
5'- 5'-
OR51E1 ctggaggaagact 77 caccatcatgaagaagct 78
ggacaaag-3' gaa-3'
5'- 5'-
PANK2 agtacgagatattta 138 ctggccaggtcctctttact 139
tggag-3' -3'
5'- 5'-
PHF21A ggaagaagcaattc 140 ttgtcgttcttgttttaaat- 141
catggc-3' 3'
5'- 5'-
PKD1 agcctgaccgtgtg 142 gccttgcaggacacacac 143
gaaggc-3' tc-3'
5'-
-
5'
PLD3 accctcaccaacaa 144 145
t2acac-3'
cggacgttcacgccct-3'
5'- 5'-
PNMA2 gggtccaagccgc 79 ccttccactctgggaccca 80
cctgctg-3' g-3'
83
CA 02828878 2013-08-30
WO 2012/119013
PCT/US2012/027351
GEP-N EN
Biomarker Forward SEQ SEQ
Reverse Primer
or Primer ID ID
Housekee sequence NO: sequence
NO:
ping Gene
5'- 5' -
PQB 1 caagagaggcatc 146 cgtcatagtcctcggcaat 147
ctcaaac-3' g-3'
5'- 5'-
PTPRN2 gcagcgcctgcgc 81 tcacatactgagtatagtc 82
gtggcgt-3' a-3'
5'- 5'-
RAF 1 gacatccacaccta 83 ctg attcgctgtg acttcg a 84
atgtcca-3' a-3'
5'- 5' -
RNF41 gaacagggaacct 148 gttacatcataccccatgtc 149
gcccccc-3' -3'
5' -
-
5'
RSF1 aaaaatgtggcctt 150 151
cactatcgcaagagtc-3'
ccaaac-3'
5'- 5' -
RTN2 gtgattggtctattc 152 aactgattggttcaccaac 153
acc at-3 ' ccc-3'
SMARCD
gctgcaggactccc 154 ggctgtgaggcgctggg 155
3
atgaca-3' gaa-3'
1
5' 5'
SP ATA7 tgcaagaggactaa 156 aaataggcacggtggacc 157
gcatgg-3' at-3'
5'- 5'-
SCG5 ctcctttacgagaa 85 acattatccagtctctgtcc 86
gatgaa-3 -3'
5'- 5'-
SPOCK1 cctgtgtgtcagcc 87 gtgtttctgggccacgttc 88
gcaagc-3' c-3'
5'- 5' -
SSTI acggcatggagga 158 atgaaagagatcaggatg 159
gccaggg-3' gc-3'
H
5'- 5' -
SST3 gcagggctggccg 160 gaccagcgagttacccag 161
tcagtgg-3' ca-3'
5'- 5' -
SST4 cgggggcgagga 162 cccgcgcgtccccgggc 163
agggctgg-3' ccc-3'
5'- 5' -
SSTS cctctggggagcg 164 caagcgctttcgggtgtct 165
acttttc-3' t-3'
84
CA 02828878 2013-08-30
WO 2012/119013
PCT/US2012/027351
GEP-NEN
Biomarker Forward SEQ SEQ
Reverse Primer
or Primer ID ID
Housekee sequence NO: sequence
NO:
ping Gene
5'- 5'-
S urvivin ctggactttcctcca 89 ccgcagtttcctcaaattct
90
ggagtt-3' -3'
5'- 5' -
TECPR2 aggcgagcagtgg 166 ttatgcagacgggttctaa 167
aagtgtg-3' a-3'
5'- 5'-
Tphl gaagagcaagtctc 91 aacaaaaatctcaaattct 92
attttttc-3' g-3'
5'- 5' -
TRMT1 12 cttacccacaatctg 168 ggcagatacggacctcg 169
ctgagct-3' gtg-3'
5'- 5'-
VMAT1 ctaacagctgccaa 93 ctgcagcctttatggaaga 94
tacctc a-3' gg-3'
5'- 5'-
VMAT2 ctgaaggacccgta 95 gcgatgcccatgtttgcaa 96
catcct-3' ag-3'
5'- 5' -
VPS13C aagtttaagggcca 170 cagggaacattgcacctg 171
ggttgt-3' gt-3'
1
5' 5'
WDFY3 aagtcctagaaatg 172 cttctgaatcactgctgtcc 173
caggaa-3' -3'
5'- 5'-
X2BTB48 gtgaactctcagct 97 tgcccctttcatcaacttca 98
actgga-3' -3'
5'- 5' -
ZFHX3 gaggagcttgctaa 174 gaatctgtcagctccttctc 175
ggacca-3' -3'
5'- 5' -
ZXDC gcgcccttacaag 176 gaaaacagggcactca 177
tgtg act-3' ctgt-3' H
5'- 5' -
ZZZ3 gaaagtggatttgt 178 tggatgggttctatgcca 179
gcaaca-3' ca-3'
5'- 5'-
ALG9 tttgtgagctgtattt 99 caacccaaacttcttgcac
100
gtg a-3' a-3'
5'- 5'-
TFCP2 aatctgtggccctg 101 gattcctgacaaacataaa 102
cagatgg-3' tg-3'
CA 02828878 2013-08-30
WO 2012/119013
PCT/US2012/027351
GEP-NEN
Biomarker Forward SEQ SEQ
Reverse Primer
or Primer ID ID
Housekee sequence NO: sequence
NO:
ping Gene
5'- 5 '-
ZNF410 cgttcctttgctg ag 103 ccactctgagagaaggtc
104
tattc-3' ttcc-3'
5'- 5'-
18S tacctggttgatcct 30 cgeccgtcggcatgtatta 31
gccag-3' g-3'
5'- 5'-
GAPDH atttggtcgtattgg 32 gaatcatattggaacatgt 33
gcgcc-3' a-3'
5'- 5' -
ACTB accgccgagaccg 180 gcccggggggcatcgtc 181
cgtccgc-3' gee-3'
5'- 5' -
ARF1 ggagaccccgcct 182 tgaccatgcagaattgatc 183
agcatag-3' g-3'
5'- 5' -
ATG4B gagctccttggcgg 184 ctgcaggaaacgcagtg 185
tccaca-3' gcg-3'
5'- 5' -
HUWE1 cacgttttggataca 186 ttggtccgctgctgtgtg a 187
ctcat-3' a-3'
1
5' 5'
MORF4L
1 gtgctgtgaggtct 188 tcaggcactgccagctcta 189
gcgggc-3' c-3'
5'- 5' -
RHOA gcacacaaggcgg 190 ctctgccttcttcaggtttc- 191
gagctag-3' 3'
5'- 5' -
SERP 1 ctggttattggctct 192 catgcccatcctgatact- 193
cttca-3' 3'
5'- 5' -
SKP1 ctaggatgtcttcca 194 gcaatatatttaaaactaag 195
gcctc-3 ' -3' -1
5'- 5' -
TOX4 gaactcagtatagt 196 gtgccaccccctaggctc 197
gccaac-3' aa-3'
5'- 5' -
TPT1 atggtcagtaggac 198 atggttcatgacaatatcg 199
agaagg-3' a-3'
86
(:::)
Table 1B: other information
t-e
=
_______________________________________________________________________________
________________________________________ -,
NO
GEP-NEN Biomarker
Amplicon produced
--..
1..,
NCBI Primer Pair SEQ ID NO: using
forward and sl-Sk
or Housekeeping Gene
UniGene =
________________ Chromosom RefSeq reverse
primers ,..,
ID
c...e
e location
Exon
Symbol Name Fwd Rev Length
Position
Boundary
Eukaryotic 18S X03205.
18S 30 31 187
1-1 1-187
rRNA 1
Chr.7:
Hs.52064 NM 001
ACTB Actin, beta 5566779- 180 181
170 1-1 1-170
0 101
5570232
n
asparagine-
linked
o
Chr. 11 -
N.)
glycosylation 11s.50385
NM 024 99 co
N.)
ALG9 111652919 - 100 68
4-5 541-600
9, alpha-1,2- 0 740.2
op
111742305
op
mannosyltransf
--4
co
erase homolog
iv
A kinase
o
Chr.19: NM 014
(PRKA) Hs.39980
u4
AKAP8L 15490859¨ 371 106 107 75
12-13 1596-1670 1
anchor protein 0
o
15529833
co
8-like
wi
amyloid beta
o
Chr. 11 -
(A4) Hs.37024 NM 001
APLP2 129939716- 41 42 102
14-15 2029-2132
precursor-like 7 142276.1
130014706
protein 2
v-raf murine
Chr. X -
sarcoma 3611 Hs.44664 NM 001 A 1
ARAF1 47420578- --' 44 74
10-11 1410-1475
viral oncogene 1 654.3
homolog
47431320
't
en
ADP-
3
ribosylation Chr.1:
Hs.28622 NM 001
ci)
ARF1 factor 1, 228270361¨ 182 183
122 5-5 1231-1352
1 024226
transcript 228286913
1..,
r..)
variant 3
-o's
i,..)
-4
Co.e
87
¨
ATG4
autophagy
0
related 4
Chr.2: NJ
Hs.28361 NM 013
=
ATG4B homolog B (S. 242577027¨ 184 185
110 7-8 586-695 ..,
0 325
LV
cerevisiae),
242613272 --,
1..,
transcript
sl-Sk
variant 1
=
,..k
c...)
ATPase, H+
ATP6V1
transporting, Chr.8: Hs.49173 NM 015
lysosomal 54628115¨ 941 108 109
102 13-14 1631-1732
H 7
50/571(Da, V1, 54755850
Subunit II
baculoviral
Chr. 17 -
Survivin TAP repeat- Hs.51452 AB1544
76210277- 89 90 78
3-4 473-551
(BIRC5) containing 5 7 16.1
76221716
n
(Survivin)
BCF2/adenovi
Chr.8:
o
rus E1B 19kDa Hs.13122 NM 004
iv
BNIP3L 26240523¨ 110 111 69
2-3 374-342 co
interacting 6 331
N.)
26270644
co
protein 3-like
co
--4
v-raf murine
co
Chr. 7 -
sarcoma viral Hs.55006 NM 004
n.)
BRAF 140433812- _ 45 46 77
1-2 165-233 o
oncogene 1 333.4
i-A
140624564
u4
homolog B1
1
o
chromosome
co
Chr.21: NM 020
C210RF 21 open IIs.22280
wi
30452873¨ 152 112 113 76
611-686 o
7 reading frame 2
30548204
7
CD59
molecule, Chr. 11 -
Hs.27857 NM 203
CD59 complement 33724556- õ 48
70 3-4 193-264
3 331.2 ''
regulatory 33758025
protein
*L:1
chromogranin
en
Chr. 14 -
3
A (parathyroid Hs.15079 NM 001
CgA 93389445- do 50 115
4-5 451-557
secretory 3 275.3
93401638
i..)
protein 1)
1..,
COMM COMM Chr.11: Hs.27983 NM 001
t.)
114 115 85
2-3 191-275 -o's
D9 domain 36293842¨ 6 101653
-4
Co.e
88
¨
containing 9 36310999
connective
Chr. 6 - 0
Hs.41003 NM 001
ci ts.)
LIGE tissue growth 132269316- 52
60 4-5 929-990 =
7 901.2 'r
...,
factor 132272518
LV
......
chemokine (C
Chr. 5 1..,
Hs.48344 NM 004
sl-Sk
CXCL14 X-C motif) 134906369- 53 54 73
3-4 742-816 =
4 887.4
,..,
ligand 14 134914969
c...e
ectonucleotide
Chr.6:
Hs.64349 NM 014
pyrophosphata
ENPP4 46097701 ¨ 116 117 82
3-4 1221-1303
se/phosphodies 7 936
46114436
terase 4
family with
sequence
Chr.3:
FAM131 similarity 131, Hs.59130 NM 001
184053717 ¨ 118 119 64 4-5
498-561
A member A, 7 171093
n
184064063
transcript
variant 2
oro
co
Rho guanine
Ni
co
nucleotide
Chr.14:
co
FLJ1035 exchange NM 21538527¨
--.]
Hs.35125 120 121 102 16-17 3557-
3658 co
7 factor (GEF) 071
21558036
Ni
40
0
i-A
(ARHGEF40)
u.)
1
frizzled Oh. 2 -
o
Hs.17385 NM 003
co
FZD7 homolog 7 202899310- c, 56 70
1-1 1-70
9 507.1 l'i'
wi
(Drosophila)
202903160 o
glyceraldehyde Chr. 12 -
Hs.54457 NM_002
GAPDH -3-phosphate 6643657 - 37 33
122 3-4 132-254
7 046.3
dehydrogenase 6647536
glycosyltransfe
rase 8 domain Chr.3:
Hs.29730 NM 001
GLT8D1 containing 1, 52728504¨ 177 123
87 4-5 924-1010
'TJ
transcript 52740048 4 010983
en
variant 3
glutamate
Chr. 4-
-
ci)
receptor. NM 001
, Ne
GRIA2 158141736 ¨ IIs.32763 7 58
71 3-4 898-970
ionotropic. 083619.1
l' ' 1..,
158287227
r..)
AMPA 2
-o-
r.)
-..1
Co.e
89
¨
histone
Chr.7:
deacetylase 9,
Hs.19605 NM001 0
HDAC9 18535369- _ 124 125 69
11-12 1777-1845
transcript 4 204144
ts.)
= 19036993
..)
variant 6
LV
......
Chr. 12 -
1..,
Hs.54904 NM 153
HOXc6 homeobox C6 54410642- co 60
87 2-3 863-951 sl-Sk
0 693.3 ¨
=
54424607
,..,
c...)
heat shuck
transcription Chr.6:
IIs.15819 NM 004
HSF2 factor 2, 1 227 20 6 9 6 - 126 127 82
10-11 1 3 24 - 1 4 05
506
transcript 122754264
variant 1
IIECT, UBA
Chr.X:
and \AWE Hs.13690 NM 031
HUWEl 53559063- - 186 187 68
67-68 10405-10472
domain 5 407
53713673
n
containing 1
antigen
Chr. 10 -
o
identified by
H Ni
s.68982 NM 001
co
Ki-67 129894923 - 62 78
6-7 556-635 Ni
monoclonal 3 145966.1 61
op
129924655
op
antibody Ki-67 .
s4 . .
co
KiSS-1 Chr. 1 -
NM 002
Ni
KISS1 metastasis- 204159469- Hs.95008 63 64 71
2-3 777-299 o
256.3
F-A
suppressor 204165619
u.)
1
v-Ki-ras2
o
co
Kirsten rat Chr. 12 -
1
Hs.50503 NM 004
KRAS sarcoma viral
25358180- c,,)
65 66 130
4-5 571-692 o
3 985.3
oncogene 25403854
homolog
Leo 1,
Pafl/RNA
polymerase 11 Chr.15:
Hs.56766 NM 138
LF01 complex 52230222-
2 792 128 129 122
10-11 1753-1874
component 52263958
'TJ
en homolog (S.
cerevisiae)
melanoma Chr. X -
ci)
MAGE- Hs.52266 NM_014
antigen family 54834171 - 67 68 90
3-4 591-682
D7 5 599.4
1..,
D, 2 54842445
-4
Co.e
90
¨
mortality
Chr.15:
MORF4 factor 4 like 1, Hs.37450 NM 006
0
79165172¨ 188 189 62
1-1 35-96
Li transcript 3 791
ts.)
79190075
=
variant 1
..,
LV
mortality
---,
1..,
Chr.X:
MORF4 factor 4 like 2, Hs.32638 NM 001
sl-Sk
102930426¨ 130 131 153
5-5 1294-1447 =
L2 transcript 7 142418
,..k
102943086
c...e
variant 1
Chr. 14 -
metastasis IIs.52562 NM 004
MTA1 105886186- 69 70 86
16-17 1771-1838
associated 1 9 689.3
105937062 nucleosome Chr. 12 -
Hs.52459 NM 139
NAP1L1 assembly 76438672- 7, 72 139
16-16 1625-1764
9 207.2 ' '
protein 1-like 1 76478738
NK2
transcription
Chr. 10 - n
Hs.24327 NM 145
NKX2-3 factor related, 101292690- 7,1
74 95 1-2 512-608 Ni
2 285.2 ' l'
o
locus 3 101296281
co
(Drosophila)
Ni
co
nucleolar
co
s.]
protein 3
co
(apoptosis
iv
Chr.16:
o
repressor with Hs.51366
NM 001 F-A
NOL3 67204405¨ 133 118
1-2 131-248 u.)
1
CARD 7 185057 132
67209643
o
domain),
co
(,,)1
transcript
o
variant 3
Chr. 2 -
Hs.47120 NM_018
NRP2 neuropilin 2 206547224- 75
76 81 1-2 824-906
0 534.3
206662857
nudix
(nucleoside
Chr.6:
diphosphate 34255997 ¨ _ Hs.18888 NM 006
NUDT3 134 135 62
2-3 500-561 *L:1
linked moiety 2 703
en
34360441
3
X)-type motif
3
ci)
(..)
ornithine Chr.15:
Hs.71381 NM 002
1..,
OAZ2 decarboxylase 64979773¨ 136
137 96 1-2 189-284 s.)
6 537
antizyme 2 64995462
-o-
r.)
--.1
Co.e
91 ¨
olfactory
receptor. Chr. 11 -
0
Hs.47003 NM_152
OR51E1 family 51, 4665156 - 77 78 97
1-2 55-153 Is.)
8 430.3
=
subfamily E, 4676718
,...
IN=J
member 1
--,
1--,
Chr.20:
sl-Sk
pantothenate Hs.51685 NM 024
=
PANK7 3869486¨ 138 139 126
4-5 785-910 ,..,
kinase 2 9 960
c...e
3904502
PHD finger
Chr.11:
protein 21A, Hs.50245 NM 001
PHF7 IA 45950870¨ 140 141 127
16-17 2241-2367
transcript 8 101802
46142985
variant 1
polycystic
kidney disease
Chr.16:
M-0 00
PRD1 2138711¨ Hs.75813 142 143
110 16-17 7224-7333 n
dominant), 296
2185899
transcript
o
variant 2
NiCO
Ni
phospholipase
op
1) Mmily, Chr.19:
op
IIs.25700
NM 001 s.]
PLD3 member 3, 40854332¨ 144 145 104
6-7 780-883 co
8 031696
transcript 40884390
Ni
o
variant 1
F-A
u.)
polyglutamine
1
Chr.X:
o
POI31 binding protein 487551,5 Hs.53438 NM 001
146 147 68
2-3 157-224 co
1, transcript 4 032381
wi
48760422
o
variant 2
Chr. 8 -
paraneoplastic Hs.59183 NM 007
PNMA2 76362196 - 70 80 60
3-3 283-343
antigen MA2 8 257.5 f '
26371483
protein
tyrosine
Chr. 7 -
pTpRN2 phosphatase,
157331750 ¨ Hs.49078 NM 157331750¨
81 82 75
2-3 307-383 'TJ
receptor type, 9 842.2
en
158380482
N polypeptide
2
ci)
is.)
v-raf-1 murine Chr. 3 -
Hs.15913
NM 002 1..,
RAFI leukemia viral 12625100- 83 84
90 7-8 1186-1277 ts.)
0 880.3
-o-
oncogene 12705700
ts.)
---)
Co.e
'JI
92
¨
homolog 1
0
_______________________________________________________________________________
________________________________________ ts.)
=
ras homolog Chr.3:
..,
IIs.24707 NM 001
LV
RHOA gene family, 49396578¨ 190 191
62 4-5 651-712
member A 49449526
sl-Sk
ring finger
=
Chr.12:
,..,
c...e
protein 41, IIs.52450 NM 001
RNF41 56598285¨ 148 149 72
2-3 265-336
transcript 2 242826
56615735
variant 4
remodeling Chr.11:
Hs.42022 NM 016
RSF1 and spacing 77377274¨ 150 151
60 7-8 2804-2863
9 578
factor 1 77531880 1
_______________________________________________________________________________
________
reticulon 2, Chr.19:
NM 005
RTN2 transcript 45988550¨ Hs.47517 152 153 87 9-10 1681-
1766
619
variant 1 46000313
o
secretogranin Chr. 15 -
o
Hs.15654 NM 001
SCG5 V (7B2 32933870- 25 86 84
5-6 616-701 N.)
0 144757.1 ¨
protein) 32989298
mc
oo
co
stress-
--ti
associated Chr.3:
co
Hs.51832 NM 014
SERP endoplasmic 150259780¨ 192 193
79 2-3 626-704 no
6 445
0
reticulum 150264428
r-A
6.)
protein 1
i
o
S-phase
co
tx)1
kinase-
Chr.5:
o
associated Hs.17162 NM 006
SKP1 133492082¨ 194 195 140
5-5 1821-1960
protein 1, 6 930
133512724
transcript
variant 1
SW1/SNE
related, matrix
associated,
*L:1
actin Chr.7:
en
SMARC Hs.64706 NM 001
3
dependent 150936059¨ 154 155 109
8-9 986-1094
D3 7 003801
regulator of 150974231
cio
t..)
chromatin,
1..,
subfamily d,
r..)
member 3,
t,..)
---.1
Co.e
93
¨
transcript
variant 3
0
_______________________________________________________________________________
______________________________________ ls.)
=
spermatogenes
..,
Chr.14:
IV
SPATA7 is associated 7. gss519gs Hs.52551 NM 001
156 157 81
1-2 160-241 ---,
..,
transcript 8 040428 .1..g
88904804
variant 2
=
c...e
sparc/osteonect
in, cwcv and
Chr. 5 -
kazal-like lls.59613 NM 004 87
S POCK] 136310987- 88 63
4-5 465-529
domains 6 598.3
136835018
proteoglyean
(testican) 1
Chr.14:
somatostatin Hs./4816 NM 001
SST1 38677204- 158 159 85
3-3 724-808
receptor 1 0 049
38682268
n
Chr.22:
somatostatin Hs./2599 NM 001
o
SST3 37602245- 160 161 84
1-'7 637-720 No
receptor 3 5 051
37608353
mc
oo
Chr.20:
co
somatostatin Hs.67384 NM 001
.4
SST4 23016057 - 162 163 104
1-1 91-194 co
receptor 4 6 052
/3017314
No
o
somatostatin
r-A
Chr.16:
u4
receptor 5, Hs.44984 NM 001
1
SST5 1122756 - 164 165 157
1-1 1501-1657 o
transcript 0 053 co
1131454
variant 1
wi
o
tectonin beta-
propeller
Chr.14:
repeat Hs. NM 001
TECPR2 102829300- 166 167 61
12-13 3130-3191
containing 2, 102968818 7 172631
transcript
variant 2
Chr. 12 -
transcription NM 005
't
TFCP2 51488620- Hs.48849 65,733 101 102 91 11-12
1560-1652 n
factor CP2
51566664
TOX high
c/o
Chr.14:
t..)
mobility group lls.55591 NM _014
TOX4 21945335- 196 197 145
5-5 441-585 ..,
box family 0 828
t..)
/1967319
member 4
t..)
-...1
CAI
'JI
94
-
Chr. 11 -
tryptophan Hs.59I99 NM 004 0 1
TPH1 18042538- 92 145
1-2 73-219
hydroxylase 1 9 179.2 'r
0
18062309
NJ
=
tumor protein,
Chr.13: ..,
Hs.37459 NM-003 198
LV
TPTI translationally- 45911304¨ 199
131 3-3 196-321 --...
..,
6 295
controlled 1 45915297
sl-Sk
=
tRNA
,..k
methyltransfer
Chr.11: t...e
TRMT11 Hs.33357 NM 016
ase 11-2 64084163¨ 168 169 91
1-2 45-135
2 9 404
homolog (S. 64085033
cerevisiae)
solute carrier
family 18 Chr. 8 -
Hs.15832 NM 003 93 VMAT1 (vesicular 20002366 - 2
053.3 94 102 1-2 93-196
monoamine), 20040717
n
member 1
solute carrier
o
family 18 Chr. 10 -
Ni
co
Hs.59699 NM 003
Ni
VMAT2 (vesicular 119000716- a,
2 054.3 "l' 96 60
9-10 896-957
co
monoamine),
119037095 co
--4
co
member 2
vacuolar
Nio
i-A
protein sorting
Chr.15:
u.)
13 homolog C Hs.51166 NM 001
1
VPS13C 62144588¨ 170 171 65
69-70 9685-9749 o
(S. cerevisiae), 8
018088 co
' 62352647
wi
transcript
o
variant 2B
WD repeat and Chr.4:
Hs.48011 NM 014
WDFY3 FYVE domain 85590690¨ _ 172 173
81 64-65 10190-10270
6 991
containing 3 85887544
serpin
peptidase
'TJ
en
inhibitor, clade
Chr. 14 - 3
X2BTB4 Hs.11862 NM 001
A (alpha-1 94749650- 07 98 80
4-5 1305-1224
8 0 100607.1
'' ci)
antiproteinase,
94759596 i..)
antitrypsin),
1..,
t,..)
member 10
-o-
t,..)
--..1
Co.e
95
¨
zinc finger
Chr.16:
homeobox 3, Hs.59829 NM _001
ZFHX3 72816784¨ 174 175 68
5-6 886-953
1'4
7 164766
transcript
73092534
variant B
Chr. 14 -
Hs.17086 NM_ 103 104 102
7-8 1134-1237021 zinc finger
74353586 - 71\1F410
protein 410 9 188.1
74398803
zinc finger C, Chr.3:
Hs.44004 NM 001
ZXDC transcript
126156444-
176 177 61
1-2 936-1001
9 040653
variant 2 126194762
2909-2971
62
13-14
Chr.1:
Hs.48050 NM 015
178 179
zinc finger,
78030190 ¨
534
ZZZ3 ZZ-type
6
containing 3 78148343
Ni
co
Ni
oo
Ni
IJ
oo
co
co
96
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
[00358] The results are presented in FIG. 1, with each of the figures nine
panels showing
average expression levels for an individual biomarker in normal EC (left),
malignant/metastatic
(center) and localized (right) samples. Ellipsoids correspond to a 2 Standard
Deviation (SD)
threshold. All p-values: p < 0.05. The results demonstrate significantly
higher expression levels
of MAGE-D2. MTA1, NAP1L1, Ki-67. FZD7, CgA and NRP2, and reduced levels of
survivin and
Kiss] in SI GEP-NEN (AKA SI NETs), confirming differential expression of these
GEP-NEN
biomarkers in GEP-NEN samples compared to normal cells, and between different
GEP-NEN
tumor grades.
Detection and expression level determination for transcripts of 21-biomarker
panel
[00359] Quantitative real-time PCR (QPCR) was carried out as described above,
using a set
of primer pairs to measure expression levels of transcripts from a 21-gene GEP-
NEN biomarker
panel (including MAGE-D2, MTA1, NAP1L1, Ki67, Survivin, FZD7, Kissl, NRP2,
X2BTB48.
CXCL14, GRIA2, NKX2-3, OR51E1, PNMA2, SPOCK1, HOXC6. CTGF, PTPRN2. SCG5,
and Tphl). The primer sequences and information are listed in Tables 1A and
1B, above.
Expression of the 21 biomarkers was measured in 167 human tissue samples,
including normal
EC cell (n=13) normal SI mucosa (n=27), and primary (n=53) and metastatic
(n=21 liver METs)
GEP-NEN subtype and 53 adenocarcinoma (colon, breast, and pancreatic) samples.
This study
demonstrated that each of the 21 biomarkers is significantly differentially
expressed in GEP-
NEN tumor samples.
[00360] For each of the 21 biomarkers, the proportion of GEP-NEN samples
versus
adenocarcinoma samples in which transcript levels were detected was calculated
and compared
using a 2-tailed Fisher's test (GraphPad Prizm 4; FIG. 8B: *p<0.002 SI GEP-
NENs versus
adenocarcinomas (Fisher's exact test)). As shown in FIG. 8B, a significantly
higher proportion
(>95%) of the GEP-NEN samples in this study expressed (i.e., were positive
for) 16 of the 21
GEP-NEN biomarker genes (76%), as compared to adenocarcinomas (p<0.002). Genes
highly
expressed in both tumor types included CTGF, FZD7, NRP2. PNMA2 and survivirt.
[00361] In contrast to different GEP-NEN sub-types, the various normal EC cell
samples
exhibited homogeneous transcript expression, with low transcript variation
(57%) between
samples. Different neoplastic SI GEP-NEN (a.k.a. SI NET) subtypes showed
heterogeneity at
97
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
the transcript level, indicating that different GEP-NEN sub-types could be
differentiated by
detecting and determining expression levels of transcripts in the 21-biomarker
panel.
EXAMPLE 2: Principal Component Analysis (PCA)
[00362] After natural log (ln)-transformation, and importation into Partek0
Genomic Suite,
Principal Component Analysis (PCA) was performed to describe the structure of
the high-
dimensional expression data. PCA allowed visualization and comparison of
transcript
expression patterns among various samples (e.g., normal, neoplastic, GEP-NEN
vs. other tumor,
GEP-NEN subtypes, primary vs. metastatic/malignant). PCA reduced
dimensionality of the
expression data¨obtained with each of the nine-biomarker and twenty-one
biomarker panels¨
to three uncorrelated principal components (PCs), which explained most
variations (Jolliffe IT,
"Principle Component Anlysis," Springer, 1986.). PCA mapping was visualized in
a 3-
dimentional space, with the first (14). second (2nd) and third (3rd) PCs
assigned to the x-, y-, and
z-axes, respectively.
[00363] For the nine and the twenty-one gene panels, average expression data
for various
samples were superimposed in this PCA coordinate system. The centroid (center
of mass
(average expression)) of each sample represented its individual transcript
expression profile
(regulatory signature) as given by the principal component vector. In this
representation, the
distance between centroids inversely equivalent to the similarity measure
(greater distance = less
similarity). Thus, large distances or separation between centroids indicated
samples with distinct
transcript expressions signatures; proximity of centroids indicated similarity
between samples.
For example, distance between centroids for different tumor type samples
represented the
relative similarity of their regulatory signatures (transcript expression
levels).
9-biomarker panel
[00364] PCA was carried out, as described above, for the real-time PCR
expression data for
the nine-gene biomarker panel (MAGE-D2, MTA1 , NAP1L1 , Ki-67, Survivin, FZD7,
Kiss] .
NRP2. and CgA). Three PCs (PC#1, PC#2, PC#3) reflected most of the expression
variance
between primary SI GEP-NENs, normal EC cell preparations, and respective
metastases.
Reduced data were mapped to a three dimensional space (FIG. 2). As shown in
FIG. 2, for
primary SI GEP-NENs and normal EC cell preparations, PC#1, PC#2, and PC#3
represented
98
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
31.7%, 26.5%, and 17.4% of the variance, respectively; overall, the three PCs
represented 75.6%
of the variance.
[00365] The three PCs represented 75.6% of variance for primary tumor subtypes
and normal
EC cell preparations (FIG. 2A), and 73.2% of variance for primary GEP-NEN
tumor subtypes
and corresponding metastases (FIG. 2C). For metastases, PC#1 , PC#2, and PC#3
represented
40.4%, 19.9%, and 12.9% of the variance, respectively; overall 73.2% of the
variance in the data
was represented by all 3 PCs (FIG. 2C).
[00366] The inverse correlation between biomarker expression levels and the
cosine of the
angle between individual expression vectors (greater angle = less similarity)
was used to identify
related gene expression clusters. The clusters are shown in FIG. 2B for
primary SI GEP-NENs
((1) CgA, NRP2, NAP1L1, FZD7; (2) MAGE-D2, MTA1, Kiss]; and (3) Ki-67,
Survivin)) and in
FIG. 2D for corresponding metastases ((1) NAP1L1, FZD7, CgA, Survivin, Ki-67,
Kiss]; (2)
MTA1, MAGE-D2, NRP2) (Gabriel KR, "The biplot graphic display of matrices with
application
to principal component analysis," Biometrika 1971;58(3):453).
21-biomarker panel PCA
[00367] PCA also was carried out as described above for the 21-biomarker panel
(MAGE-D2,
MTA1. NAPIL1, Ki67, Survivin, FZD7, Kissl, NRP2, X2BTB48, CXCL14, GRIA2, NKX2-
3,
OR51E1, PNMA2, SPOCK1, HOXC6, CTGF, PTPRN2, SCG5, and Tph1). Three principal
components captured most of the variance (83%) within the dataset. Reduced
data were mapped
to a three dimensional space.
[00368] FIG. 8A shows a comparison of expression profiles for GEP-NENs
(including
various primary and metastatic sub-types). adenocarcinomas (sub-types), and
normal tissues (EC
and SI). As shown, centroids for the three adenocarcinoma types were separate
from those for
both normal SI mucosa and neoplastic GEP-NEN tissue subtypes. This observation
confirms
significant difference in expression levels in this other (epithelial) tumor
type, shown using the
Fisher's exact test, described above (FIG. 8B). The various neoplastic (SI GEP-
NEN) subtypes
displayed heterogeneous expression profiles, showing they could be
distinguished using this
panel of biomarkers.
[00369] All normal EC preparations displayed homogeneous transcript expression
(with low
variation (57%) within samples). Further, the normal sample expression
profiles were
99
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
substantially different compared to those of other tissue types, including
normal SI mucosa. The
genetic profile for normal EC cells was substantially different compared to
normal SI mucosa
and neoplastic tissue.
[00370] The results demonstrate differences in expression profiles for the GEP-
NEN
biomarkers and distinct regulatory expression signatures for primary SI GEP-
NEN tumor
subtypes, normal EC cell preparations, and SI GEP-NEN metastases. This study
confirms that
measuring expression of the 21 biomarkers can successfully distinguish between
GEP-NEN sub-
types, adenocarcinoma types, normal SI mucosa, normal EC cells, and between
primary and
metastatic GEP-NEN subtypes.
EXAMPLE 3: Tumor profiling and analysis.
[00371] Statistical analyses and tumor profiling were performed on transformed
expression
data obtained with the nine- and twenty-one biomarker panels described above.
9-biomarker panel
[00372] Mean (M) transcript expression levels and standard deviations (SD)
were calculated
for the nine-biomarker panel, for primary tumor subtypes and normal EC cell
preparations.
Mean normal expression of CgA (MNormal = -9.2. SD = 4.2), Ki-67 (M
Normal ¨ -Normal = -4.5. SDNormai =
1.1), Kiss] (M
Normal - -Normal = 4.0,- SDNormal = 3.2), NAP1L1 (M
Normal - -Normal = - 8 .3 , SDNormal = 1.1), NRP2
(MNormal = -9.37 SD = 3.8), and Survivin (MNormai= -6.0, SDNormal = 1.0) was
significantly
different compared to mean expression in primary tumors, both overall (All
Tumors) and among
individual subtypes. Seep values and Fold Change (FC), listed in Table 2,
below. Transcript
expression level measurements were reevaluated in a subset of samples (n=35).
The data were
highly correlated (R2=0.93, p=0.001), demonstrating this approach was both
highly reproducible
and robust.
[00373] Analysis of Variance (ANOVA) was carried out to evaluate differences
in
biomarker expression levels between tumor and normal samples, and between
normal samples
and individual tumor subtype samples. Specifically, ANOVA compared expression
between
normal EC cell preparations and primary tumor tissues, and between normal EC
cell
preparations and individual primary tumor types (Table 2). A two-class
unpaired algorithm was
implemented, with tumor sample data (total or individual sub-type) and normal
sample
expression data defining the two groups. As there were no missing values in
the datasets,
100
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
imputation was unnecessary. For each biomarker transcript, geometric Fold
Change (FC) was
calculated as the ratio of geometric means for the Tumor Group and the Normal
Group.
[00374] Biomarker genes calculated to have differences in expression between
normal and
tumor groups, with p<0.05, were considered significantly changed. Transcripts
with p<0.05 and
absolute FC>2.0 were considered differentially expressed between groups. CgA,
FZD7, Ki-67,
NAP1L1, NRP2, and Survivin were significantly altered in WDNETs compared to
non-nal EC
cell preparations. Transcript levels of CgA, Ki-67, MAGE-D2, and NRP2 were
significantly
changed in WDNECs. PDNETs displayed alternatively expressed levels of CgA, Ki-
67,
NAP1L1, NRP2, and Survivin. Finally, PDNECs were different only in expressions
of NAP1L1
and NRP2.
Table 2:
ANOVA comparing biomarker expression levels in SI-GEP-NENs, and individual SI-
GEP-
NEN sub-types, to expression levels in normal EC cell samples
All Tumors vs. Normal WDNET vs. Normal WDNEC vs. Normal PDNET vs. Normal PDNEC
vs. Normal
Gene P IV P FC P FC P FL' P Fe
CgA 1.3x10-4 17.7 1.05x10-4 28.3 0.03 8.3 0.01 13.5 NS 20.5
FZD7 0.05 3.6 0.02 5.9 NS -1.1 NS 5.5 NS
6.9
Ki-67 1.1x10-3 -3.5 0.01 -3.0 0.02 -3.5 2.7x10-3 -
5.5 NS -3.7
Kiss! 0.02 -3.9 0.05 -3.7 NS -4.5 NS -4.4 NS
-1.8
MAGE-D2 NS 1.2 NS -1.6 6.4x10-4 5.3 NS 1.6 NS -
1.8
MTA1 NS -1.2 NS -1.5 NS 1.1 NS 1.1 NS -1.6
NAP1L1 4.7x10-5 13.7 4.1x1(16 24.8 NS 2.9
7.4x1(14 17.3 0.01 26.9
NRP2 2.2x10-8 39.5 1.6x10-6 31.5 2.3x10 33.7
1.9x10-6 82.08 5.0x10-3 47.1
Survivin 0.01 -3.5 0.04 -3.1 NS -3.1 0.02 -5.1 NS
-5.07
WDNET = Well Differentiated Neuroendocrine Tumors, WDNEC = Well Differentiated
Neuroendocrine
Carcinomas, PDNET = Poorly Differentiated Neuroendocrine Tumors, PDNEC =
Poorly Differentiated
Neuroendocrine Carcinomas; NS = pX).05, FC = Fold Change
[00375] Pearson's Correlation (PC) coefficients (R2) were calculated for the
nine-biomarker
panel to assess linear relationships between pairs of biomarkers and between
tumor sub-type
differentiation and expression of the biomarkers. The distribution of
biomarker expression
among primary GEP-NEN subtypes and normal EC samples was linearly separated by
calculating PC coefficients for individual pairs of the biomarkers (plotted on
x- and y- axes of
individual Similarity Matrices shown in FIG. 3). The study determined highly
linear (R2> 0.50)
correlation of expression for four pairs of biomarkers (MTAI :MAGE-D2,
MTAI:Kissl,
101
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
FZD7:NAP1L1. and Survivin:Ki-67 (highly correlated (R2 > 0.50)). Additionally,
distribution of
expression profiles for WDNETs, WDNEC, and PDNETs was linearly correlated to
pair-wise
expressions of Kissl:Survivin, TZD7:1VAPIL1, Survivin:MIA1, and M1A1 :MAGE-D2,
indicating a linear classifier could be applied to the dataset. The data
further suggest an
expression-dependant correlation between the biomarkers and primary tumor
subtypes.
21-biornarker panel
[00376] Pearson's Correlation (PC) coefficients were used to identify linear
relationships
between expression levels of biomarkers in the 21-gene panel. PC coefficients
were calculated
for each pair of the 21 biomarkers, across all tissue types (FIG. 9A). FIG. 9
shows the results in
a heatmap, with the pairs with the lowest (-0.03), medium (0.4), and highest
(1) correlations
indicated in black, dark grey, and light grey, respectively. The 21-biomarker
panel contained 27
highly correlated (R2>0.40) transcript pairs, with the highest correlation
coefficient (R2=1.00)
between MTA1, NRP2, and Kiss].
[00377] From these data, a network of correlations was constructed by drawing
an edge
between any transcript pair having an R2 above a pre-defined threshold
(R2>0.40) (FIG. 9B,
with actual R2 values superimposed on each edge). As shown in FIG. 9B, five
distinct
regulatory clusters were identified within the network, each having a unique
set of biomarkers:
(1) MAGE-D2, NRP2, Kiss], MTA1, and CgA (most tightly-connected cluster (every
R2-
value>0.79)); (2) GRIA2, 0R51E1õS'POCK1, and SCGS: (3), CXCL14, NKX2-3, HOXC6,
CTGF, PTPRN2; (4) NAP ILL FZD7 , and PNMA2; and (5) Survivin and Tphl . In
FIG. 9B, the
R2 values are superimposed on individual edges. The lowest R2- value is 0.40
within each
cluster; the highest value is 1Ø The results demonstrate expression levels
of the panel of
biomarkers are biologically relevant to GEP-NEN.
[00378] A two-sample t-test computation was used to identify biomarker genes
that are
differentially expressed between: 1) EC cells, normal SI mucosa, and primary
and metastatic
tissues; 2) primary GEP-NEN subtypes; and 3) metastatic GEP-NEN subtypes (FIG.
10).
[00379] Calculated S-values for each subset ranged from -1.4 to 1.1. Based on
the number of
genes (n=21) and the sample size (n=114). the threshold for statistical
significance for the S-
value was set at 0.4 (Nadler B, "Discussion of "On consistency and sparsity
for principal
component analysis in high dimensions," Journal of the American Statistical
Association
2009;104:694-97). Transcripts with S <-0.4 or S > 0.4, and p<0.05, were
considered
102
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
significantly down- or up-regulated, respectively. Results are presented in
FIG. 10, with
volcano plots of gene ranks and significance (p) values for the t-test.
[00380] FIG. 10A shows the comparison between normal SI mucosa, normal EC
cells and SI
GEP-NENs. Compared to normal mucosa, transcript expression of the classic
neuroendocrine
marker Tphl . was significantly higher (p<0.001. S=0.7; FIG. 10A) in SI GEP-
NEN samples.
Compared to normal SI mucosa, neoplastic tissue expressed higher transcript
levels of CgA and
GRIA2 (FIG. 10B); expression of CgA was not significantly altered (p=0.07,
S=0.39) between
neoplastic tissue and normal EC cells.
[00381] FIG. 10B shows the comparison between all GEP-NEN (tumor) samples and
all
normal samples, all metastatic GEP-NEN samples and all normal samples, and all
metastatic
GEP-NEN samples and all primary GEP-NEN samples. None of the biomarker
transcripts were
differentially expressed in the collective metastatic GEP-NEN samples, when
analyzed as an
entire group, compared to the collective primary GEP-NEN samples, analyzed as
a group.
[00382] FIG. 10C shows the comparison between primary GEP-NEN subtypes and all
metastases as a group. No biomarker transcripts were differentially expressed
in PDNET
samples as compared to PDNEC samples (PDNET-PDNEC). or WDNET samples as
compared
to PDNEC (WDNET-PDNEC) samples. Between WDNEC and PDNEC (WDNEC-PDNEC),
MAGE-D2 was the only significant differentiating marker (p=0.009. S=1.03: FIG.
10C).
[00383] FIG. 1011 shows comparison between primary tumors and metastatic
subtypes. CgA,
Kissl, NRP2. and Tphl were differentially expressed between all metastasis
subtypes (FIG.
10D).
EXAMPLE 4: Predictive Models for classifying GEP-NENs
[00384] Expression levels of GEP-NEN biomarkers obtained in the studies in
Examples 1-4
were further analyzed with supervised learning algorithms and models,
including Support Vector
Machines (SVM), Decision Tree, Perceptron, and regularized discriminant
analysis RDA
(Gallant SI, "Perceptron-based learning algorithms," Perceptron-based learning
algorithms
1990;1(2):179-91)).
EXAMPLE 4A: Prediction and modeling with detected expression of the nine-
biomarker panel
[00385] Expression data obtained in the nine-biomarker study were analyzed
using the
Feature Selection (FS) classification model. The model was employed using a
"greedy forward"
103
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
selection approach, selecting the most relevant subset of features for the
robust learning models,
as described by Peng H, Long F, Ding C, "Feature selection based on mutual
information:
criteria of max-dependency, max-relevance, and min-redundancy," IEEE
Transactions on
Pattern Analysis and Machine Intelligence, 2005;27(8):1226-38.
[00386] FS determined that for this study, expression levels of NAP1 F7D7,
Kiss 1 and
MAGE-D2 were the best variables (of the nine biomarkers) for SVM
classification. Thus, SVM
was carried out by comparing expression levels for these biomarkers in normal
EC cell
preparations (n=13) and primary SI-GEP-NENs (n=36). For SVM, radial basis
function was
used as a kernel and a 10-fold cross-validation was used to measure the
sensitivity of
classification. See Cristofanilli M et al. "Circulating tumor cells, disease
progression, and
survival in metastatic breast cancer." N Engl J Med 2004. The results are
shown in Table 3,
below, and in FIG. 4. As shown, SMV predicted SI GEP-NENs in this study with
100%
sensitivity. and 92% class specificity; normal EC cell preparations were
accurately predicated
with 77% sensitivity, with a class specificity of 100%.
Table 3: Class predictions produced by the Support Vector Machines
classification model
using transcript expression levels of NAP1L1, 171)7, Kissl, and MAGE-D2
True Normal True Tumor Class Precision
Predicted Normal 10 0 100%
Predicted Tumor 3 36 92%
Class Recall 77% 100%
[00387] Density maps in FIG. 4 display distributions between SI GEP-NENs and
normal EC
cells, colorized to the density of the samples produced differential zones
that depended on the
individual gene expressions. Expression levels of NRP2, MAGE-D2, Kiss], and
EZD7
transcripts as identified by the Feature Selection algorithm were plotted on
the X- and Y-axis.
Normal and neoplastic sample data were scattered according to their respective
gene pair
expressions. Distribution densities based on average Euclidean distance
(difference in
expression) between samples were colorized green (normal) and red
(neoplastic). Blue areas
indicate a region of transition between normal and neoplastic groups. The
distinct separation
between normal EC cells and primary small intestinal tumors indicates the
utility of the selected
transcripts as malignancy markers.
104
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
[00388] Feature Selection identified NAP1L1 and Ki-67 expression levels as
principle
discriminators in the Decision Tree classifier. Based on this result, Decision
Trees classification
model was constructed on expression data for individual primary SI GEP-NEN
subtypes by
correlating NAM,' and Ki-67 expression level values to the corresponding
expression levels for
primary tumor subtypes, as determined above. The results are displayed in FIG.
5, with the
leaves of the tree representing classifications and branches representing
conjunctions of features
that devolve into the individual classifications. A 10-fold cross-validation
was used to measure
the efficiency of this technique, as described by Pirooznia M, el al., "A
comparative study of
different machine learning methods on microarray gene expression data," BMC
Genomies
2008;9 Suppl 1:S13. Percentages shown in parenthesis in FIG. 5 indicate the
occurrence
frequencies of primary SI GEP-NEN subtypes. As shown in Table 4. below,
Decision Trees
classification predicted WDNETs in this study with 78% sensitivity and 82%;
predicted
WDNECs in this study with 78% sensitivity and 64%; and predicted PDNETs in
this study with
71% sensitivity and 63% specificity. With the nine biomarker panel, PDNECs
were
misclassified in this study as either WDNETs or PDNETs. (FIG. 5; Table 4).
Table 4: Class predictions produced by the Decision Trees classification model
using
transcript expression of Ki-67 and NAND.
True True True True Class
WDNET WDNEC PDNET PDNEC Precision
Predicted
14 1 1 1 82%
WDNET
Predicted
WDNEC 3 7 1 0 64%
Predicted
PDNET 1 1 5 1 63%
Predicted
PDNEC 0 0 0 0 0%
Class Recall 78% 78% 78% 0%
[00389] ANOVA was performed to identify transcripts differentially expressed
in primary SI
GEP-NEN subtypes and corresponding metastases (Table 5). Significant gain of
Kiss]
(p<0.005) was associated with metastasis in all tumor subtypes.
Table 5: ANOVA results across Small Intestinal Neuroendocrine Tumor subtypes
and
corresponding metastases.
WDNET vs. WDNET WDNEC vs. WDNEC PDNEC vs. PDNEC
105
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
MET MET MET
Gene p FC FC p FC
Kissl 5.7x10-7 52.8 1.2x10-7 81.2 0.004 41.6
MAGE-
5.2x10 3 5.6 NS -1.04 0.03 10.4
D2
CgA 0.02 9.08 0.01 12.4 0.08 21.1
Ki-67 NS 2.7 0.02 3.7 NS 1.5
MTA1 0.02 2.8 NS 1.1 NS 4.4
Survivin NS 4.02 0.05 4.4 NS 6.1
EZD7 NS 1.7 1.8x10-3 27.2 NS 1.2
NAP1L1 NS 1.1 0.01 12.05 NS -1.9
NRP2 NS 1.2 NS -1.6 NS -1.4
MET = Metastasis; FC = Fold Change; "p" =p value; NS =p 0.05
[00390] Detected expression levels of MAGE-D2, NAP1L1, and Kiss] (as
identified by FS)
were analyzed in primary and corresponding metastatic WDNETs, using SVM to
construct a
classifier. To evaluate expression of biomarkers as compared to metastatic
potential of primary
tumors, samples were plotted in correlation with the selected gene expression
levels and
distribution densities were colorized to outline the separation of primary and
metastatic samples
(FIG. 6A).
[00391] WDNETs and metastatic WDNET results scattered according to their
respective gene
pair expressions, with distribution densities based on the average Euclidean
distance (difference
in expression) between samples colorized blue (primary tumors) and red
(metastases), green
areas indicating a region of transition between primary and metastatic
tumors). As shown,
WDNETs and WDNET METs were predicted with 100% sensitivity and specificity.
WDNET
could be predicted to metastasize if transcript levels of 1) NAP1L1 > -2.71
and Kiss] > -2.50; 2)
NAP1L1 > -3.82 and MA GE-D2 > -4.42; 3) MAGE-D2 > -3.21 and Kissl > -2.12.
[00392] A perceptron classifier (Markey MK et al., "Perceptron error surface
analysis: a case
study in breast cancer diagnosis," Comput Biol Med 2002;32(2):99-109) of 0.05
was used to
distinguish between localized tumors and the corresponding metastases. This
methodology has
been shown to effectively predict malignancy of breast cancer (Markey MK et
at., -Perceptron
error surface analysis: a case study in breast cancer diagnosis" (2omput Biol
Med 2002;32(2):99-
109). A Perceptron classifier (using three data scans to generate the decision
boundaries that
explicitly separate data into classes, with a learning rate of 0.05) was used
to predict metastases
of WDNECs and PDNECs.
[00393] The FS algorithm predicted that NAP1L1 and Kiss] were highly expressed
specifically in WDNEC METs and that CgA was highly expressed specifically in
PDNEC
METs. Metastatic potential of primary tumors was visualized by plotting
expressions of
106
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
featured genes and colorizing the distribution densities of primary tumors and
their metastases.
Data are presented in FIG. 6B and FIG. 6C, showing data from primary tumor
subtypes and
respective metastases scattered according to their respective gene pair
expressions, with
distribution densities based on the average Euclidean distance (difference in
expression) between
samples colorized blue (primary tumors) and red (metastases), and green areas
indicating regions
of transition between primary tumor subtypes and respective metastases. WDNECs
were
predicted to metastasize with values of NAP1L1 > -5.28 and Kiss] > -2.83,
while PDNECs could
be predicted to metastasize when CgA> -3.5. These results show distinct
separation of primary
SI GEP-NEN subtypes and the respective metastases, demonstrating the utility
of the provided
biomarkers as metastasis markers.
EXAMPLE 4B: Evaluating classification and predictive capabilities of the nine-
bioniarker panel
[00394] To evaluate classification and predictive capabilities using the nine-
biomarker panel,
real-time PCR was performed on samples obtained from an independent set of SI
GEP-NEN
tissues (n=37), including normal EC cell preparations (n=17), localized SI GEP-
NENs (n=8),
and malignant SI GEP-NENs (n=12), to measure the marker gene transcript
expression. All
WDNETs were considered as "localized" while other tumor subtypes were
considered
"malignant". Assessment of linearly correlated transcript pairs identified a
pattern similar to the
training set whereas MTA1:MAGE-D2, MTA1:Kissl, FZD7:NAP1L1, and Survivin:Ki-67
transcript pairs were highly correlated (R2>0.50). The trained SVM model was
applied to
differentiate normal EC cell preparations from neoplasia with 76% accuracy.
[00395] The results (shown in FIG. 7) indicated that in this study (using
subsets of the nine-
biomarker panel), normal EC cells were cross-validated with only 77% accuracy
and predicted
in an independent test set with 76% accuracy (p = 0.84). Localized GEP-NENs
were cross-
validated with only 78% accuracy and predicted with 63% accuracy in the test
set (p = 0.25).
Malignant GEP-NENs were cross-validated with only 83% accuracy and predicted
with 83%
accuracy in an independent set (p = 0.80). The Decision tree model could
predict localized and
malignant GEP-NENs with only 63% and 83% accuracy respectively (FIG. 7). The F-
test
statistic was computed to confirm that the classification results of the
training and the
independent sets were not significantly different. The p-values for normal,
localized, and
malignant subgroups were 0.84, 0.25, and 0.80 respectively.
107
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
EXAMPLE 4C: Prediction and modeling using expression levels from the 21-
biomarker panel
[00396] A regularized discriminant analysis (RDA) algorithm was designed and
applied to
expression data for the twenty-one biomarker panel (MAGE-D2, MTA1, NAP1L1,
Ki67,
Survivin, FZD7, Kissl, NRP2, X2BTB48, CXCL14, GRIA2, NKX2-3, OR51E1, PNMA2,
SPOCK1, HOXC6, CTGF. PTPRN2, SCG5, and Tphl), described above. Gene selection
for
tissue classification was performed by computing the rank score (S) for each
gene and for each
class pair as:
S =1,-/c2 mucil
aci +ac2
[00397] where /dm and itc, represent means of first and second class
respectively and o-ci and
0-C2 are inter-class standard deviations. A large S value was indicative of a
substantial
differential expression ("Fold Change") and a low standard deviation
("transcript stability")
within each class. Genes were sorted by a decreasing S-value and used as
inputs for the RDA.
[00398] RDA's regularization parameters, y and X, were used to design an
intermediate
classifier between LDA (performed when y=0 and X=1) and QDA (performed when
y=0 and
X=0) (Picon A, Gold LI, Wang 1, Cohen A, Friedman E. A subset of metastatic
human colon
cancers expresses elevated levels of transforming growth factor betal . Cancer
Epidemiol
Biomarkers Prey 1998;7(6):497-504). To reduce over-fitting, RDA parameters
were selected to
minimize cross-validation error while not being equal 0.0001, thus forcing RDA
to produce a
classifier between LDA, QDA, and L2 (Pima I, Aladjem M. Regularized
discriminant analysis
for face recognition. Pattern Recognition 2003;37(9):1945-48).
[00399] Regularization parameters were defined as y = 0.002 and k = 0. For
each class pair,
S-values were assigned to expression data for individual transcripts, which
were then arranged
by a decreasing S-value. RDA was performed 21 times, such that the Nth
iteration consisted of
top N scoring transcripts. Error estimation was done by a 10-fold cross-
validation of the RDA
classifier, by partitioning the tissue data set into complementary subsets,
performing the analysis
on one subset (called the training set), and validating the analysis on the
other subset (called the
validation set or testing set). This operation was performed for all
permutations of test-train sets
and misclassification error was averaged to reduce variability in the overall
predictive
assessment.
108
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
EXAMPLE 4D: Sensitive, accurate mathematical classification of unknown tissues
and GEP-
NENs, differentiation of GEP-NEN sub-Apes and staging of GEP-NENs using
expression data
from a twentv-one-biomarker panel
[00400] This RDA algorithm was applied to expression data obtained as
described above for
the panel of 21-biomarkers (MAGE-D2, MTA1, NAP1L1, Ki67, Survivin, FZD7, Kiss
1, NRP2,
X2BTB48, CXCL14, GRIA2, NKX2-3, OR51E1, PNMA2, SPOCK1, HOXC6, CTGF,
PTPRN2, SCG5, and Tphl). The algorithm was used to distinguish samples of
unknown tissue
types (ECs (normal enterochromaffin cells); "Normal" (normal small intestinal
mucosa);
"Tumor" (aggregation of primary and metastatic GEP-NEN s and carcinomas
(NET/NEC)); and
primary WDNET; WDNEC; PDNET; PDNEC), for mathematical classification of GEP-
NENs,
as follows.
[00401] For each sample, it first was determined whether the tissue was normal
or neoplastic.
Tissues deemed neoplastic then were assessed to determine whether they were
primary or
metastatic. GEP-NEN subtypes (primary or metastatic) then were characterized.
The RDA
algorithm was applied in every step using the same set of learning parameters
(y=0.002 and
k=0). Performance of the classifier was measured by calculating
misclassification rate (overall
proportion of false-positives between any two classes).
[00402] Results are shown in Tables 6A-C (listing misclassification rate
versus numbers of
gene (biomarker) transcripts detected, beginning with the highest ranked
transcript for each
distinction).
Table 6A: Misclassification Rates versus number of transcripts detected
(normal vs. GEP-NEN; primary vs. Metastasis)
Misclassification Rates
Number of Normal SI Mucosa vs.
Transcripts EC Cells vs. Tumor Tumor Primary vs. Metastasis
1 0.08 0.21 0.28
2 0.06 0.15 0.27
3 0.06 0.16 0.22
4 0.05 0.17 0.23
0.02 0.17 0.18
6 0.01 0.12 0.19
109
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
7 0.01 0.07 0.18
8 0 0.09 0.14
9 0 0.06 .14.
0 0.07 0.11
11 0.01 0.05 0.12
12 0.01 0.04 0.07
13 0 0.03 0.08
14 0.01 0.03 0.05
0 0.02 0.03
16 0 0.01 0.02
17 0 0.01 0.02
18 0 0.01 0
19 0 0.01 0
0 0 0.02
21 0 0 0.02
Table 6B: Misclassification rates versus number of transcripts detected
(primary GEP-
NENs)
Number of
Misclassification Rates
Transcripts
PDNEC PDNEC PDNEC PDNET PDNET
vs. vs. vs. vs. vs.
1 WDNET VVDNEC PDNET WDNET VVDNEC
2 0.07 0.09 0.14 0.16 0.2
3 0.04 0 0 0.29 0.2
4 0 0 0 0.16 0.08
5 0 0 0 0.04 0.05
6 0 0 0 0 0
7 0 0 0 0 0
8 0 0 0 0 0
9 0 0 0 0 0
10 0 0 0 0 0
11 0 0 0 0 0
12 0 0 0 0 0
13 0 0 0 0 0
14 0 0 0 0 0
15 0 0 0 0 0
16 0 0 0 0 0
17 0 0 0 0 0
18 0 0 0 0 0
19 0 0 0 0 0
110
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
Number of
Misclassification Rates
Transcripts
20 0 0 0 0 0
21 0 0 0 0 0
Table 6C: Misclassification rates versus number of transcripts detected
(metastatic GEP-
NENs)
Misclassification Rates
WDNEC MET PDNEC MET PDNEC MET
Number of vs. vs. vs.
Transcripts WDNET MET WDNEC MET WDNET MET
1 0.17 0.2 0.22
2 0.28 0.27 0.22
3 0.06 0 0.11
4 0.06 0 0
0.06 0 0
6 0 0 0
7 0 0 0
8 0 0 0
9 0 0 0
0 0 0
11 0 0 0
12 0 0 0
13 0 0 0
14 0 o 0
0 0 0
16 0 0 0
17 0 0 0
18 0 0 0
19 0 0 0
0 0 0
21 0 0 0
[00403] As shown in Tables 6A-C, the methods and RDA algorithm was able to
detect the
presence, stage, and classification (sub-type), with zero misclassification
rates across pair-wise
iterations of normal EC cells, normal small intestinal mucosa, and GEP-NEN
subtypes.
[00404] As shown in Table 6A, the RDA algorithm distinguished normal EC cells
from
neoplastic tissue. Detection and analysis of expression levels of just the
single highest ranked
biomarker transcript (PNMA2) was able to make this distinction with a
misclassification rate of
0.08; detection and analysis of the respective single highest ranked biomarker
(CO) was able to
1 1 1
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
make the distinction between normal SI mucosa from neoplastic tissue with a
misclassification
rate of 0.21 (Table 6A).
[00405] Applying the method to data for pluralities of biomarkers (detecting
expression of
levels of biomarker panels and applying the RDA algorithm to the data) was
able to detect and
distinguish GEP-NENs from normal samples with zero misclassification.
Distinguishing EC
cells from tumor tissue with a misclassification rate of zero was achieved
using a panel of eight
(8) biomarker transcripts. Distinguishing normal SI mucosa from tumor tissue
with a
misclassification rate of zero was achieved using a panel of twenty (20)
biomarker transcripts
(Table 7). In this study, misclassification rates were higher with fewer
transcripts. These
results demonstrate the biomarker specificity for different tissue groups and
confirm the ability
of the present methods to detect GEP-NEN disease and distinguish GEP-NEN
tissue from
different normal tissue types, with high specificity.
[00406] Likewise, applying the RDA algorithm to expression levels of panels of
biomarkers
could determine with 100% accuracy whether an unknown tissue sample was
primary or
metastatic. For this determination, expression levels were detected for
eighteen (18) biomarker
transcripts and the data included in the RDA model (Table 7), with higher
misclassification rates
using fewer transcripts. Detecting expression of and applying the algorithm to
only the highest
ranked transcript (MAGE-D2) distinguished primary and metastatic samples with
a 0.28
misclassification rate. (Table 6A).
[00407] Primary GEP-NEN subtypes also could be differentiated with 100 %
accuracy using
the RDA algorithm. Misclassification rates when only the single highest ranked
transcripts were
detected ranged from 0.07 (PTPRN2, for distinguishing between PDNEC and WDNEC)
to 0.37
(NRP2, for distinguishing between WDNEC and WDNET). Applying the RDA algorithm
to
expression levels of all 21 biomarker transcripts, the methods distinguished
between WDNETs
and WDNECs with a zero misclassification rate (Table 6B), with higher
misclassification rates
using fewer biomarkers.
[00408] As shown in Table 6C the RDA algorithm also was used to distinguish
with 100%
accuracy between metastatic GEP-NEN subtypes. Misclassification rates with
only the single
highest ranked transcripts were 0.22 (CXCL14, for distinguishing between WDNET
MET and
WDNEC MET), 0.2 (NAP1L1, for distinguishing between PDNEC MET and WDNEC MET),
and 0.17 (NRP2, for distinguishing between PDNEC MET and WDNET MET),
respectively
(Table 6C).
112
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
[00409]
Table 7. Misclassification rates with detection of various numbers of
transcripts; achieving
minimal misclassification; SVM, decision trees (DT), and multi-layer
perceptron (MLP)
classifiers.
Number of transcripts to
Lowest misclassification rate achieve lowest
Sample/Class Distinguished in this example
misclassification rate in this
example
SVM DT MLP SVM DT MLP
EC vs. Normal SI Mucosa 0.02 0.05 0 14 21 8
EC vs. Tumor 0.01 0.03 0 18 3 5
Normal SI Mucosa vs. Tumor 0.14 0.14 0.01 16 21 13
Primary vs. Metastasis 0.19 0.19 0.14 3 2 7
PDNEC vs. WDNET 0.07 0.07 0 21 21 4
PDNEC vs. WDNEC 0.09 0.09 0 21 21 2
PDNEC vs. PDNET 0.14 0.28 0.14 21 21 3
PDNET vs. WDNET 0.16 0.16 0.03 21 1 11
PDNET vs. WDNEC 0 0.20 0 19 21 10
WDNEC vs. WDNET 0.02 0.26 0.02 21 21 16
WDNEC MET vs. WDNET
0.11 0.33 0.11 3 21 12
MET
PDNEC MET vs. WDNEC
0.20 0.20 0 21 21 12
MET
PDNEC MET vs. WDNET
0.22 0.33 0 21 21 14
MET
"Normal" = normal small intestinal mucosa;
"Tumor"= aggregation of primary and metastatic NETs and carcinomas (NET/NEC)
[00410] Table 8 summarizes the numbers of in NET biomarkers able to
distinguish between
indicated samples using the RDA algorithm in this example. In this example,
all 21 biomarkers
distinguished WDNEC from WDNET with zero misclassification (higher
misclassification with
fewer transcripts). By contrast, as few as two biomarkers could differentiate
between PDNEC
and WDNET (MAGE-D2, CXCL14), and between PDNEC and PDNET (PTPRN2, MTA1) with
zero misclassification. In this example, 11 biomarkers distinguished normal
enterochromaffin
(EC) cells from normal SI mucosa with zero misclassification (PNMA2, CXCL14,
PTPRN2,
Tphl, FZD7, CTGF, X2BTB48, NKX2-3,SCG5, Kiss 1 õSPOCKI, with a higher
misclassification
rate using fewer biomarkers). Fewer transcripts were able to distinguish
normal EC cells from
neoplastic tissue (n=8, PNMA2, Tphl, PTPRN2, SCG5, SPOCK1, X2BTB48, GRIA2,
OR51E1).
Expression of twenty of the biomarkers (with the exception of CXCL14) could
differentiate
113
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
normal SI mucosa from neoplastic tissue with zero misclassification (higher
misclassification
rates with fewer transcripts).
Table 8: Numbers of biomarker transcripts used for pairwise distinctions with
zero
classification rate by RDA
Number of Transcripts that
Distinction achieved a Zero Misclassification
Rate
EC vs. Normal 11
EC vs. Tumor 8
Normal vs. Tumor 20
Primary vs. Metastasis 18
PDNEC vs. WDNET 3
PDNEC vs. WDNEC 2
PDNEC vs. PDNET 2
PDNET vs. WDNET 4
PDNET vs. WDNEC 4
WDNEC vs. WDNET 21
3
WDNEC MET vs. WDNET MET
4
PDNEC MET vs. WDNEC MET
6
PDNEC MET vs. WDNET MET
[00411] Finally, SVM, decision trees (DT), and MLP classifiers were applied,
as described
above, using data for transcripts of the twenty-one biomarker panel, in a
similar fashion as RDA.
The performance of RDA was compared to performance of SVM, decision trees, and
multi-layer
perceptron (MLP), for classification of GEP-NEN subtypes by detecting
expression of the
twenty-one biomarker panel. All classifiers were subject to the training and
cross-validation
protocol outlined in Example 4A. Misclassification rates were calculated
(Table 7). SVM was
able to achieve a zero misclassification to distinguish PDNET from WDNEC.
Decision trees
distinguished with misclassification rates ranging from 0.03 (between EC and
Tumor sample) to
0.33 (between WDNEC MET and WDNET MET, and between PDNEC MET and WDNET
MET). Somewhat comparable to RDA, the MLP classifier produced zero
misclassification rates
with 7/13 iterations, with a high overall accuracy. The RDA approach was most
reliable in this
example with the 21 marker gene panel, achieving zero misclassification rates
in all iterations.
114
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
EXAMPLE 5: Detection of circulating GEP-NEN cells (CNC) and identification of
biomarker
transcripts (mRNA) from plasma
[00412] Circulating GEP-NEN cells (CNCs) were detected in human blood using
the
provided methods and biomarkers. For this process, human blood samples
(plasma, buffy coat,
and whole blood) were obtained and subjected to staining, cell sorting, and
real-time PCR (to
detect GEP-NEN biomarkers and housekeeping genes).
EXAMPLE 5A: Sample preparation and RNA isolation from plasma, bulb, coat, and
whole
blood
[00413] In the following studies for detection of biomarkers in human plasma
and buffy coat,
human blood samples were obtained from a blood databank, with samples from
healthy controls
(n=85) or patients (n=195) who had been treated for GEP-NEN disease, at Yale
New Haven
Hospital, Uppsala or Berlin. See Kidd M, et al., -CTGF, intestinal stellate
cells and carcinoid
fibrogenesis," World J Gastroenterol 2007;13(39):5208-16. Five mL of blood
were collected in
tubes containing ethylenediaminetetraacetic acid (EDTA). Plasma was separated
from huffy
coat following 2 spin cycles (5 min at 2,000rpm) and then stored at -80 C
prior to nucleic acid
isolation and/or hormone (CgA) analysis.
RNA isolation from various blood samples
[00414] For isolation of RNA from buffy coat, samples were incubated with
TRIZOL ,
followed by RNA clean-up. RNA was dissolved in diethyl pyrocarbonate water and
measured
spectrophotometrically, and an aliquot analyzed on a Bioanalyzer (Agilent
Technologies, Palo
Alto, CA) to assess the quality of the RNA (Kidd M, et al. "The role of
genetic markers--
NAP ILI, MAGE-D2, and MTA1--in defining small-intestinal carcinoid neoplasia,"
Ann Surg
Oncol 2006;13(2):253-62).
[00415] For isolation of RNA from GEP-NEN patient and control plasma, the
Q1Aamp RNA
Blood Mini Kit was used (FIG. 11A), which in this study allowed real-time PCR
detection of
housekeeping genes in significantly more samples compared to the TRIZOL
approach (FIG.
11B) (8/15 versus 2/15, p-=0.05). For isolation of RNA directly from whole
blood, the QIAamp
RNA Blood Mini Kit was used, following the manufacturer's guidelines.
115
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
Stability and reproducibility of samples
[00416] The blood test is based on identifying the GEP-NEN molecular signature
in lml of
whole blood, collected in an EDTA-tube. It was determined that the gene
signature is stable for
up to four hours (refrigeration at 4-8 C, following phlebotomy) prior to
freezing (FIG. 13). It is
unaffected by fasting/feeding. Analysis of inter-assay reproducibility (same
samples processed
on separate days) ranged from 98.8-99.6% while intra-assay reproducibility was
99.1-99.6%.
[00417] These studies identify that the gene signature is highly reproducible
(-99%), is stable
for up to four hours in refrigeration (prior to freezing) and is unaffected by
fasting/feeding.
Real-time PCR
[00418] Total RNA obtained from plasma, buffy coat, and whole blood as
described above
was subjected to reverse transcription with the High Capacity cDNA Archive Kit
(Applied
Biosystems (AB1), Foster City, CA) following the manufacturer's suggested
protocol. Briefly, 2
micrograms of total RNA in 50 microliters of water was mixed with 50 uL of
2XRT mix
containing Reverse Transcription Buffer, deoxynucleotide triphosphate
solution, random
primers, and Multiscribe Reverse Transcriptase. The RT reaction was performed
in a thermal
cycler for 10 mins at 25 C followed by 120 mins at 37 C, as described by Kidd
M, etal., "The
role of genetic markers--NAP1L1, MAGE-D2, and MTA1--in defining small-
intestinal carcinoid
neoplasia," Ann Surg Oncol 2006;13(2):253-62. Transcript levels of the marker
genes were
measured using Assays-on-DemandTM products and the ABI 7900 Sequence Detection
System
according to the manufacturer's suggestions (see Kidd M. Eick ('i, Shapiro MD,
et al.
Microsatellite instability and gene mutations in transforming growth factor-
beta type II receptor
are absent in small bowel carcinoid tumors. Cancer 2005;103(2):229-36).
[00419] Cycling was performed under standard conditions, using the TaqMan
Universal
PCR Master Mix Protocol. Briefly. complementary DNA in 7.2 uL of water was
mixed with 0.8
uL of 20 Assays-on-Demand primer and probe mix and 8 uL of 2X TaqMan Universal
Master
mix in a 384-well optical reaction plate. The following PCR conditions were
used: 50 C for 2
mins and then 95 C for 10 mins, followed by 50 cycles at 95 C for 15 mins and
60 for 1 min,
as described by Kidd M, etal., "The role of genetic markers--NAP1L1, MAGE-D2,
and MTA1-
-in defining small-intestinal carcinoid neoplasia," Ann Surg Oncol
2006;13(2):253-62. Raw ACT
(delta CT = change in cycle time as a function of amplification) normalized
using geNorm (see
116
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
Vandesompele J, De Preter K, Pattyn F, et al. Accurate normalization of real-
time quantitative
RT-PCR data by geometric averaging of multiple internal control genes. Genome
Biol
2002;3(7):RESEARCH0034), and expression of the house-keeping genes ALG9, TFCP2
and
ZNE410. See Kidd M, et aL, "GeneChip, geNorm, and gastrointestinal tumors:
novel reference
genes for real-time PCR," Physiol Genomics 2007;30(3):363-70. Normalized data
were natural
log (1n)-transformed for compression. ALG-9 was used as the housekeeping gene,
its expression
detected and used to normalize GEP-NEN biomarker expression data.
[00420] For statistical analysis, all computations were carried out using R
2.9 language for
statistical computing. See R Development Core Team. R: A language and
environment for
statistical computing Vienna, Austria: R Foundation for Statistical Computing,
2008. GraphPad
(Prizm 4) and SPSS16.0 were used for all statistical analyses, by receiver-
operator characteristic
(ROC) curves, Fisher's exact test and/or ANOVA, using 2-tailed tests, with
p<0.05, considered
significant.
EXAMPLE 5B: Detection of housekeeping genes and detection in whole blood
[00421] Transcript expression levels of three (3) housekeeping genes (ALG9,
TFCP2 and
ZNF410) were determined in mRNA isolated using the TRT7OL approach, described
above,
from huffy coat from five healthy donors. All three genes were detected with
ACT levels
between 30 and 35. Sequences and information for exemplary primer pairs are
listed in Tables
1A and 1B.
[00422] Transcript expression levels of the same 3 housekeeping genes and 11
GEP-NEN
biomarker genes were evaluated in mRNA prepared from whole blood from 3
healthy donors
(normal samples). For this process, mRNA was isolated, cDNA synthesized, and
PCR
performed by different people on different days, on separate plates, using
independently-
prepared reagents, made on different days. Detected gene expression levels
across samples were
highly correlated (FIG. 11C; R>0.99, p<0.0001).
[00423] Expression of 5 housekeeping genes (18S, ALG9, GAPDH, TFCP2 and
ZNF410) was
detected by real-time PCR on mRNA isolated using TR1ZOL , from whole blood
samples,
from 5 healthy donors. Primer pairs are listed in Tables 1A and 1B. In this
study, ALG9
expression was the least variable between samples (coefficient of variation =
1.6%) (FIG. 13A).
ALG9 transcript levels were determined by real-time PCR on RNA isolated from
whole blood
117
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
from five healthy control patients, before and at thirty-minute intervals
after feeding. The results
showed that expression levels of ALG9 were not significantly altered up to 4
hours
postprandially (as determined by nANOVA: p>0.05) (FIG. 13B). These results
demonstrate
that detection of gene products according to the provided methods produces
consistent results
and is useful for comparison of data from patient samples acquired and
prepared on different
days by distinct investigators.
Delineation of A House-Keeping Gene
[00424] To identify the most useful house-keeping genes for normalization, a
panel (n=19) of
candidate markers was examined that comprised those identified from GEP-NEN
tissue (n=9),
and those through screening of the GEP-NEN blood transcriptomes (n=10). In
order to select
"house-keeper" markers, a number of criteria were used including: topological
importance when
mapped to the blood interactome (7,000 genes, 50,000 interactions)_ENREF_3,
stability (M-
value) following real-time PCR, and efficiency of transcription in the blood.
In addition, the
presence of efficiencies between the target genes and the house-keeping gene
were examined.
Such a correlation supports a relative quantitation-based algorithm for
calculation. The 19 genes
included in the analysis were tissue-derived: 18S, GAPDH, ALG9, 5LC25A3, VAPA,
TXNIP,
ADD3, DAZAP2, ACTG I, and blood microarray-derived: ACTB, ACTG4B, ARF1, HUWE1,
MORF4L1 RHOA, SERPI, SKP1, TPT1, and TOX4. Targets that were considered
appropriate
house-keepers exhibited >3 characteristics.
Topological importance in blood microarray
[00425] Three topological features were examined: -Degree" = number of
connections in
each gene; "Betweenness" = importance of a gene in signal transduction, and
"Clustering" =
clustering coefficient or the extent to which a gene's neighbors are
interconnected. A high
"degree" indicates many connections per gene, a high "betweenness" indicates a
more critical
role in the information flow within the interactome while a "high" clustering
coefficient means
that more of a gene's neighbors are connected to each other. The most
appropriate gene would
have low values for Degree, Betweeness and Clustering. Genes that fulfill all
these
characteristics include ACTB, TOX4, TPT1 and TXNIP (FIG. 14A-C). The order of
genes is:
118
CA 02828878 2013-08-30
WO 2012/119013
PCT/US2012/027351
[00426] TXNIP=ACTB=TOX4=TPT1>ALG9=ARF1>GAPDH>DAZAP2>VAPA=ATG4B=
HUWEI=MORF4L1=RHOA=SERP 1 >ADD3.
Variability (Coefficient of variation and M-value)
[00427] Two approaches were used to assess variation in house-keeping gene
expression,
firstly variability and secondly robustness (the -M" value) measured by
geNorm. Raw CT values
were examined for variation (FIG. 15) and whether expression passed the D'
Agostino and
Pearson normality test (Table 9).
Table 9. Candidate House-keeping genes and normality of expression
118S ACTG1 ADD3 ALG9 DAZAP GAPDH SI,C25A3 TXNIP VAPA
2
CV 17.9% 11.03% 13.21% 6.93% 10.01% 10.36% 18.43% 15.09% 14.09%
DP N
test
AC TB ARF1 ATGB FILINVE MORF4 RHDA SERP 1 SKP1 TOX4
TPT I
4 1 Li
V 9.27% 5.1% 6.9% 8.39% 9.74% 7.14% 9.33%
4.36% 4.34% 7.65
DP N
test
CV = coefficient of variation, DP = D'Agostino and Pearson omnibus normality
test. N = not normally distributed,
Y = passed the normality test.
[00428] Variability analysis identified that A1G9, ARF1, ATG4B, RHDA and SKP I
were
the least variable genes. Genes selected by geNorm as showing the least
variation between
samples (and hence the greatest stability or robust expression) are indicated
in FIG. 16. The
"M"-value is a measure of gene stability and defined as the average pair-wise
variation of a
particular gene with all other potential reference genes. The most stable
genes included: ALG9,
ACTB, ARF1, ATG4B, HUWE4, MORF4L1, RHDA, SKP1, TPT1 and TOX4.
PCR efficiency
[00429] PCR efficiency was examined to evaluate which candidate house-keeping
genes
fulfilled adequate amplification criteria. This was undertaken in two
independent samples using
a standard curve (dilution: 2000 - 0.01ng/u1). The PCR efficiency was
calculated using the Fink
equation:
1 1 9
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
Efficiency = 10^ (-1/slope) ¨ 1
[00430] Analysis identified that 18S and ALG9 were the most efficiently
transcribed tissue-
derived genes while TPT1 was the most efficiently transcribed blood-derived
candidate house-
keeping gene (FIG. 17).
Efficacy of amplification compared to target genes
[00431] Finally, the amplification kinetics of the target and reference genes
were examined
for similarities. This is a necessary pre-requisite for any appropriate PCR
amplification protocol
otherwise a correction factor is required in quantitation algorithms to deal
with over-estimated
expression calculations. It is also important for any comparative CT method
e.g., MCT
particularly as estimations from raw data are more accurate than from standard
curves.
[00432] In general, a house-keeping gene is considered appropriate if the
difference in CT for
the target-reference gene across a series of dilutions is <0.1. One house-
keeping gene identified
that shared similar PCR efficacies with target genes was ALG9 (FIG. 18).
[00433] None of the blood-microarray derived candidate house-keeping genes
exhibited the
appropriate features necessary to act as a house-keeping gene. ALG9, the
tissue-derived
candidate house-keeping gene, in contrast, exhibited low variability (M-value
and DP test),
appropriate topological features, was efficiently transcribed and shared
similar amplification
features with the target genes of interest. This gene was therefore selected
as an appropriate
house-keeping gene to normalize circulating tumor transcripts.
Target Normalization
[00434] There are a two major methods for normalizing target gene expression:
absolute and
relative quantitation. The former requires a standard curve (and therefore
uses up plate space), is
more labor-intensive and is less accurate than protocols based on raw CT
values. This study
focused on relative quantitation approaches. A number of algorithms have been
developed for
relative quantification including the Gentle model, the Pfaffl model, models
based on
amplification plots, Q-Gene and geNorm. The majority of methods include
mechanisms to
estimate for differences in PCR efficiencies, use multiple house-keepers e.g.,
geNorm, or can
only be commercially acquired (e.g., qBase)Lus from Biogazelle). One method
that is easy to use
and does not require estimation factors is the AACT protocol. This is a
mathematical model that
120
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
calculates changes in gene expression as a relative fold difference between an
experimental and
calibrator sample. It is dependent on similar amplification efficiencies for
the house-keeper and
target genes (a feature identified for ALG9), requires the amplification of
small PCR products
(<150bprs ¨ a feature of Applied Biosystems Taqman), and a PCR method that has
been
optimized (e.g., starting concentration of target has been established). The
AACT approach was
selected for normalization of the 51 candidate genes in peripheral blood. The
utility of this
approach was demonstrated when this method (MCT normalization with ALG9) was
compared
to geNorm (normalization with 18S, ALG9 and GAPDH) (FIG. 19).
[00435] The variation in target gene expression was significantly lower in
control samples
using a AACT protocol (p<0.004 vs. geNorm) while the majority of targets
exhibited a normal
distribution (62% versus 0%, D'Agostino and Pearson omnibus normality test)
following
normalization with ALG9. A AACT protocol (with ALG9) has been shown to
successfully
normalize target expression in GEP-NEN tumor tissue. A AACT approach using
ALG9 as a
house-keeping gene was identified to be the most appropriate normalization
protocol for the 51
candidate GEP-NEN marker genes. Accordingly, this approach was selected to
profile transcript
expression in blood samples.
Identification of. Candidate Tumor Marker Genes
[00436] To identify potential marker genes, both tissue- and blood-based
tissue microarrays
were from GEP-NEN samples as resources to detect candidate marker genes. Gene
selection was
optimized by applying and developing a number of biomathematical algorithms.
[00437] Initially, GEP-NEN (obtained from the small intestine) transcriptomes
were analyzed
and compared this to normal small intestinal mucosa (U133A chips, n=8 tumors
and n=4
controls). Using dCHIP (lower bound fold change >1.2-fold, unpaired t-test,
and hierarchical
clustering based on Pearson correlation) 1,451 up-regulated genes in tumor
samples were
identified. Thirty-two candidate markers were chosen based on level of up-
regulation (>3-fold,
e.g., NAP1L1), known biological processes (proliferation e.g., Ki67; survival
e.g., survivin), and
clinical significance (e.g., somatostatin receptor expression, CgA). In a
separate study, PCR-
based expression in tumor tissue of nine of these candidate markers were
confirmed as predictive
of GEP-NEN malignancy. In the current study, the 32 candidate genes were
examined further
and 17 were included in the final gene panel.
121
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
[00438] As a second strategy, two micromay data sets of tumor tissue (HUGE and
U133A, a
total of n=30 tumors and n=10 controls) were utilized, and compared GEP-NENs
(obtained from
small intestinal sites) to other tumors (breast, colon, prostate and liver
cancers) from publicly
available databases. Small bowel material from Crohn's disease, which is known
to perturb local
neuroendocrine cell activity and is associated with SI-NEN risk, was also
assessed to further
delineate the overall GEP-NEN gene landscape and help identify candidate
markers. In order to
assess the relationships of the genes involved, a graphic theoretical analysis
of gene co-
expression networks was constructed. This approach determined that the "GEP-
NEN" gene
network (generated by integrating the two platforms. U133A and HUGE) consisted
of 6,244
genes and 46,948 links. The gene network was highly modular (i.e. genes tended
to organize into
interconnected communities) and therefore contained genes that were
functionally related (as
they occurred within the same community). An unbiased community detection
algorithm
identified 20 communities (collections of related genes) with >20 genes each.
Enrichment of
each gene community for biological processes identified terms including
'Oxidation reduction'
(Cluster 1/2). 'Immune response' (Cluster 5), and 'Cell cycle' (Cluster 18).
Of importance was
identification that the GEP-NEN gene network was topologically distinct from
other common
cancers (but shared similarity) to Crohn's disease (FIG. 20A). The latter may
reflect the known
proliferation of neuroendocrine cells in Crohn's disease.
[00439] The topological distinction reflected unique connectivity patterns
around each gene in
the interactome providing information that a panel of genes or gene-
interactions may be specific
to the tumor (GEP-NEN). Such a tumor-specific signature was generated by
eliminating gene-
gene interactions found in breast, colon, prostate, and liver cancer gene
networks from the GEP-
NEN gene network. The resulting GEP-NEN- specific signature yielded 124 genes
and 150
interactions (FIG. 20B).
[00440] Mapping these 124 GEP-NEN- specific genes back to the U133A tissue-
based
microarray identified that 41 genes were differentially expressed, of which 21
were up-regulated
(FIG. 20C) and could differentiate between GEP-NENs and controls (FIG. 20D),
These 21 up-
regulated genes were examined further, and 12 were included in the final gene
panel.
[00441] As a third strategy, circulating GEP-NEN transcriptomes were examined
to identify
additional candidate markers. For these studies, peripheral blood
transcriptomes (n=7 controls,
n=7 GEP-NENs) were compared to the "In-house" tissue array (n=3 controls, n=9
GEP-NENs
[from the small intestine]) and one published array from the ArrayExpress
database (accession
122
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
number: E-TABM-389: n=6 controls, n=3 primary midgut NENs, and n=3 GEP-NEN
metastases [METs]).
[00442] Tumor samples were clearly differentiated from controls (FIG. 21A-C)
and
differentially expressed genes were identified for each of the groups: Blood
(n=2.354), "In-
house" (n=1,976) and Public datasets (n=4,353) (FIG. 21D-F).
[00443] As expected, there was a large correlation between changes in gene
expression for the
"In-house" and Public tissue datasets (R=0.59, FIG. 22A). While the
correlation between the
Blood and "In-house" and Public datasets was low (R=-0.11 and -0.05,
respectively) (FIG.
22A,B), 157 (33%) of the 483 significantly changed genes ("In-house"/blood)
and 423 (45%) of
the 947 significantly changed genes (Public/blood), were positively
correlated.
[00444] Overall, between the Blood, "In-house", and Public datasets, 85 genes
were
correlated in blood and tissue, while 196 were inversely or anti-correlated
(FIG. 23A). The
correlated genes encoded processes such as intracellular signaling, cell
death, and regulation of
transcription (FIG. 23B) while the anti-correlated genes encoded processes
such as telomere
maintenance, neural tube development, and protein complex assembly (FIG. 23C).
[00445] Thirty-nine of the 85 (46%) concordantly expressed genes in both blood
and tissue
were up-regulated and 46 transcripts are down-regulated. An analysis of the up-
regulated genes
identified that 22 had 0-3 paralogs and were expressed at levels >3-fold.
Integration of these
genes with the blood interactome confirmed that they were highly inter-
connected (more central
in the interactome), demonstrating their "putative" biological relevance in
the context of GEP-
NENs (FIG. 24).
[00446] These approaches, including analysis and integration of tumor tissue
and circulating
peripheral blood transcripts, enabled identification of a panel of 75
candidate marker genes
associated with GEP-NENs. The utility of these genes to identify GEP-NENs was
then studied
in peripheral blood samples.
The Circulating GEP-NEW Fingerprint (51 Marker Gene Panel)
[00447] In order to develop a useable marker panel, transcript levels of each
of the 75
candidate markers in mRNA isolated from 77 blood samples (controls: n=49; GEP-
NENs: n=28)
were measured. A 2-step protocol (RNA isolation, cDNA production and PCR) was
developed
as this is more accurate than 1-step protocols. The reproducibility of 2-step
protocols is high
123
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
(Pearson's correlation >0.97; for the 2-step approach, the correlation is
0.987-0.996). In
preliminary studies, the preferred method for mRNA isolation from blood
samples was the mini
blood kit (Qiagen: RNA quality >1.8 A260:280 ratio, RIN>5.0, appropriate for
PCR applications
37) with cDNA produced using the High Capacity Reverse transcriptase kit
(Applied Biosystems:
cDNA production 2000-2500ng/up. Real-time PCR was consistently performed with
200ng/ul
of cDNA on a HT-7900 machine using 384-well plates and 16u1 of reagents/well
(Fast Universal
PCR master mix, Applied Biosystems). The limit of detection for PCR was
determined as 40
cycles (200ng/u1 cDNA positively amplified in 95.3 0.2% of cases). Increasing
the number of
cycles to 45-50 cycles identified positive expression in <1% of target
samples; the false negative
rate was calculated using a CT cut-off of 40 to be 0.8%. This cycle number is
more stringent than
the accepted European approach for leukemia detection, but is consistent with
other PCR-based
detection protocols. Primers were exon spanning to minimize genomic DNA
amplification and
were <150bprs. Commercially available Applied Biosystems primers (5'-nuclease
assay) were
used. The consistent parameters for RNA isolation, cDNA synthesis and real-
time PCR provide
a stable platform for target and house-keeping gene analysis.
[00448] 51 of the 75 candidate markers were as identified as producing
detectable product (CT
<40 cycles) in blood. This 51 gene panel included: AKAP8L, APLP2, ARAF1,
ATP6V1H,
BNIP3L, BRAF, C2lorf7, CD59, COMMD9, CTGF, ENPP4, FAM13A, FLJ10357, FZD7,
GLT8D1, HDAC9, HSF2, Ki67, KRAS, LE01, MORF4L2, NAP1L1, NOL3, NUDT3, OAZ2,
PANK2, PHF2l A, PKDl PLD3, PNMA2, PQBPl , RAF1, RNF41, RSF1, RTN2, SMARCD3,
SPATA7, SST1, SST3, SST4, SSTS, TECPR2, TPH1. TRMT112, VMAT1, VMAT2. VPS13C,
WDFY3, ZFHX3, ZXDC. ZZZ3. Thirteen of these 51 marker genes have previously
been
associated with GEP-NENs, either of the previous studies, or in those of
others.
[00449] Having defined a potentially useful marker gene panel, the GEP-NEN
transcriptornie
resources were examined to identify preferred house-keeping genes and
determine preferred
methods for normalization of the data. Identifying appropriate house-keeping
genes and applying
normalization protocols would facilitate quantification of each of the 51
candidate transcripts
and determine whether they represented a panel of GEP-NEN marker genes.
124
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
EXAMPLE 5C: Detection of circulating GEP-NEN cells and biomarker expression in
whole
blood
[00450] Using real-time PCR, flow cytometry, and florescence-activated cell
sorting (FACS )-
sorting, CD164 was identified as a marker capable of detecting circulating GEP-
NEN cells in
whole blood. Detection of CD164 transcript expression levels by real-time PCR
demonstrated
that this biomarker is consistently overexpressed (300-10,000 x) in GEP-NEN
patient samples
(29/29 GEP-NEN cells and 4 GEP-NEN cell lines) compared to normal EC cells and
leukocytes,
demonstrating CD164 is useful as a biomarker for identification of GEP-NEN
cells in human
samples, e.g.. whole blood.
[00451] Multi-parameter flow cytometry was performed on whole blood samples
obtained
from 10 GEP-NEN patients and 10 age- and sex- matched controls. A population
of GEP-NEN
cell-sized cells (FIG. 25A), which was double-positive for acridine orange
(A0)-PE-CY7 and
CD l 64-APC was detected in GEP-NEN samples, but absent in normal control
samples (FIG.
25B). Collection and immunostaining of these cells for TPH expression
confirmed they were
serotonin-positive GEP-NEN cells (FIG. 25C7 inset).
[00452] After dual labeling with AO and CD164, 3-12 GEP-NEN cells, per mL of
blood,
were sorted by FACS and collected. Real-time PCR identified elevated (>2-fold,
p<0.03)
expression levels of the 21 GEP-NEN biomarkers described above (MAGE-D2, MTAL
NAP1L1,
Ki67, Survivin, FZD7, Kiss], NRP2, X2BTB48, CXCL14, GRIA2, NKX2-3, 0R51E1,
PNMA2,
SPOCK1, HOXC6, CTGF, PTPRN2, SCG5, and Tphl), normalized to house-keeping
genes
compared to normal whole blood samples, confirming that these cells were GEP-
NEN tumor
cells. Significantly higher expression levels (3-5 fold, p<0.05) were
identified in samples
obtained from six patients with metastatic disease, as compared to four
patients with local
disease.
[00453] Expression of a thirteen GEP-NEN biomarker panel was detected by real-
time PCR
on RNA prepared directly from whole blood obtained from 12 patients. For
comparison, PCR
was performed in parallel on RNA purified from FACS-purified circulating blood
GEP-NEN
cells (as described above), and tumor mucosa from 12 patients from same study.
The expression
levels of the biomarker transcripts detected in whole blood were highly
correlated with levels
detected in purified circulating GEP-NEN cells (R2=0.6, p<0.0001) (FIG. 26A)
and in tumor
tissue (R2=0.81, p<0.0001) (FIG. 26B).
125
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
[00454] These results confirm that circulating GEP-NEN cells (CNCs) exist in
blood and can
be detected by PCR using RNA prepared from whole blood and other blood samples
for the
detection, staging, prognosis and prediction with the methods and compositions
provided herein.
EXAMPLE 5D: Detection of GEP-NEN biomarker expression and statistical analysis
using
whole blood samples
[00455] Expression levels of individual biomarker transcripts (VMA T2, NAP
11,1 , and
PNMA2), as well as the summed expression levels of a panel of thirteen (13)
GEP-NEN
biornarkers (APLP2, ARAFI, BRAF, CD59, CTGF, FZD7, Ki67, KRAS, NAP1L1, PNMA2,
RAF], TPH1, VMAT2) were determined by real-time PCR as described above, on
whole blood
samples from 3 groups of human samples obtained from: 1) a training group from
Yale New
Haven Hospital, including 55 GEP-NEN patients (all comers, including patients
with high level
disease as well as those considered disease-free) and 47 control patients, 2)
an independent test
group from Berlin (n=144 (n=120 patients, n=24 controls)) and 3) an
independent test group
from Uppsala (n=34 (n=20 patients; n=14 controls))), respectively. The primer
pair sequences
and other information about primers are listed in Tables 1A and 1B.
[00456] To facilitate representation, detected expression levels of the 13
biomarker transcripts
were vectorally summed (>+1= over-expressed genes; >-1= genes whose expression
is
decreased) and plotted.
[00457] An ROC curve strategy was employed for identification of GEP-NENs, in
group (1)
samples. Results demonstrated areas under the curve (AUCs) for the three
individual biomarker
transcripts ranging from 0.66 to 0.90 (0.92 for summed transcripts (V1:
p<0.0001)) in GEP-NEN
patient samples (FIG. 27, showing ROCs for each). The sensitivity,
specificity, positive
predictive value and negative predictive value for determining GEP-NEN disease
using the
summed transcript expression levels were 96.1%; 90.2%; 83.3%, and 97.9%,
respectively. Use
of predicted cutoffs was tested in the two independent test sets (2) and (3).
The sensitivities and
specificities for V1 ranged from 95-97% and 81-87%. It was also observed that
gender was not
associated with transcript expression (Mann Whitney score = 0.11, p=0.19)
levels detected in
blood. Storage at -80 C had no significant effect on the transcript expression
levels of the
detected 13 biomarkers (R=0.987-0.996, p<0.0001).
126
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
[00458] Housekeeping and summed GEP-NEN biomarker transcript expression levels
were
detected in 29 control and patient samples (Yale and Berlin), assayed on
separate days in two
separate PCR runs. Expression of the ALG9 housekeeping gene and the FZD7 GEP-
NEN
marker were each highly correlated when assayed on separate days: R2: 0.92-
0.97, p<0.0001 in
two separate runs (FIG. 28A-B) and no significant differences were noted
between normalized
FZD7 in controls and tumor samples on separate days (FIG. 28C-D). Intra- and
inter-assay
reproducibility was high for FDZ7 (C.V. = 2.28-3.95%) demonstrating that blood
measurements
of target genes are highly reproducible. The results demonstrate intra- and
inter-assay
reproducibility for housekeeping and GEP-NEN biomarker detection using real-
time PCR on
RNA obtained from whole blood.
[00459] These data demonstrated that detection of GEP-NEN biomarker transcript
expression
levels in whole blood can be used to identify circulating GEP-NEN cells
(CNCs), that the
detected expression levels in whole blood correlate well with tissue
expression levels and can
identify GEP-NEN patients with high sensitivity and specificity, and with high
reproducibility.
EXAMPLE 5E: Detection of lesions and treatment response
[00460] To evaluate the utility of the 51 marker gene panel both as a
technique as a
circulating GEP-NEN signature to detect these lesions as well as treatment
response, a test set of
130 samples (controls: n=67, GEP-NENs: n=63 [untreated disease, n=28, treated,
n=35]) was
established. PCR was performed on all markers, and values normalized to ALG9
(AACT), using
the control group as the population control (calibrator sample). The work-flow
for identifying
the utility of the marker panel included normalization (ANOVA identified 39 of
51 genes to be
differentially expressed in all 3 sets) and the support-machine bases
mathematical assessments of
gene expression.
[00461] Using the four algorithms, an average 88% correct call rate was
determined (FIG.
29), while the performance metrics are included in Table 10. The data of the
molecular test for
differentiating normal samples from GEP-NENs (both treated and untreated) are
as follows:
overall sensitivity (94.0%). specificity (85.7%), positive predictive value
(PPV) (87.5%) and
negative predictive value (NPV) (93.1%).
Table 10. Performance evaluation of distinguishing normal samples from GEP-
NENs.
Normal (True) GEP-NENs
127
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
(True)
Normal (Predicted) 63 9
GEP-NENs (Predicted) 4 54
[00462] Using the same gene panel, it was determined that treated and un-
treated GEP-NENs
could be distinguished with the following performance metrics (Table 11):
Sensitivity = 85.7%,
Specificity = 85.7%, PPV = 88.2% and NPV = 82.8%.
Table 11. Performance evaluation of distinguishing Treated from Untreated GEP-
NENs.
Treated GEP-NENs Untreated GEP-NENs
(True) (True)
Treated GEP-NENs 30 4
(Predicted)
Untreated GEP-NENs 5 24
(Predicted)
[00463] For overall performance as a test to differentiate NENs from controls,
the call rate
was 94%, while the ability to identify treated samples was 85%.
[00464] These results indicate that pattern recognition protocols which enable
analysis of
expression of 51 candidate markers (as a group) have utility for
differentiating between
"normal" or "GEP-NENs". This confirmed that approaches e.g., SVM used in tumor
tissue, are
applicable to peripheral blood transcript analysis and identification of
neuroendocrine tumor
disease.
EXAMPLE 5F: Evaluation of the Molecular Fingerprint as a Predictor of GEP-NENs
[00465] The efficacy of this 51 marker gene panel as a potential test was
examined in four
independent datasets to establish whether it could correctly identify GEP-NENs
versus controls.
Four independent sets were constructed: Independent set 1 included 35 GEP-NENs
and 36
controls; Independent set 2 included 33 GEP-NENs and 31 controls; Independent
set 3
included 47 GEP-NENs and 24 controls; and Independent set 4 included 89 GEP-
NENs and no
controls.
[00466] The four algorithms were assessed: SVM, LDA, KNN and Bayes for utility
in
determining whether a blood sample was a GEP-NEN or a control in each of the
independent
sets. Tabulated results identified that overall correct call rates
(identifying both GEP-NENs and
128
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
controls correctly) ranged from 56-68% in independent set 1, 53-78% in set 2,
82-92% in set 3
and 48-74% in set 4 (Table 12). The average rates over all sets were 67-69%
for SVM, LDA and
Bayes; KNN scored higher: 73%.
Table 12. Overall call rates (percentage) for each of the algorithms in each
of the independent
sets
SVM LDA KNN Bayes
Set 1 56 57 68 59
Set 2 78 77 70 53
Set 3 90 92 89 82
Set 4 48 48 65 74
AVE (%) 68 69 73 67
[00467] Further analysis of the calls identified whether the correct call
rates corresponded to
identifying controls or tumor samples (Table 13). Most consistent correct
calls for controls were
the SVM (90% overall) and LDA (91%) algorithms. The highest correct call rates
for GEP-
NENs were identified with the Bayes algorithm (85%).
Table 13. Call rates (percentage) for each of the groups, control or GEP-NENs,
in each of the
independent sets
SVM LDA KNN Bayes
CON NEN CON NEN CON NEN CON NEN
Set 1 97 14 97 17 97 37 33 86
Set 2 73 70 77 76 58 82 3 100
Set 3 100 85 100 87 100 83 88 79
Set 4 NA 48 NA 48 NA 65 NA 74
AVE 90 54 91 57 85 67 41 85
(%)
NA -= not applicable (no controls included in this set)
[00468] Sensitivities, specificities, positive predictive values and negative
predictive values
calculated for each of the algorithms in the 3 independent sets are included
in Table 14.
Table 14. Performance metrics for each of the algorithms in each of the
independent sets
SVM LDA KNN Bayes
A B C DAB C DAB C DAB C D
Set 14 97 83 54 17 97 86 54 37 97 93 61 86 33 55 70
1
129
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
Set 70 87 85 73 76 77 78 75 81 58 68 75 100 3 52 100
2
Set 85 100 100 77 87 100 100 80 83 100 100 75 79 88 93 68
3
A=sensitivity, B=specificity, C=positive predictive value, D=negative
predictive value. Set 4
had no controls.
[00469] The Bayes algorithm performed best for detecting GEP-NENs (sensitivity
= 83%),
while the SVM algorithm performed best for determining controls (specificity =
96%). The
weakness of Bayes is a high false-positive; the weakness of SVM is that it
does not perform
adequately in sample sets that exhibit a majority of well-treated (complete
remission/stable
disease) samples.
[00470] For overall performance as a test to differentiate NENs from controls.
algorithms
SVM, LDA and KNN had positive predictive values of ¨90% and negative
predictive values of
70%.
EXAMPLE 5G: 51 marker gene panel for GEP-NEN Identification
[00471] To confirm that the 51 marker gene panel was effective, correct call
rates for the
panel were compared in each of the independent sets (Table 12) and compared
this with a 13
marker and 25 marker subsets. The 13 marker subset was limited to genes
confirmed as
predictive of GEP-NEN malignancy in tissue; the 25 marker panel included these
genes as well
as an additional 12 GEP-NEN specific genes identified in FIG. 20D. Examining
correct calls in
each of the 4 independent sets identified that the 51 marker panel performed
significantly better
than either the 13 or 25 marker panel (FIG. 30).
[00472] These results indicate that a number of pattern recognition protocols
based on the 51
candidate marker genes can distinguish between control samples and GEP-NENs
with high
efficiency and sensitivity.
Example 5H: Detection of GEP-NEN-biomarker expression levels in whole blood,
for evaluation
of therapeutic responsiveness and prediction of metastases (comparison to CgA)
[00473] Detection of summed GEP-NEN biomarker transcript expression levels (13-
biorrtarker panel) in whole blood, before and following therapeutic
intervention (resection and
130
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
Octreotide LAR) was carried out, demonstrating the clinical utility of
embodiments of the
provided methods and systems. Moreover, comparison to detection of CgA
expression alone
demonstrated improved sensitivity of the provided methods in GEP-NEN
detection, risk
determination, and monitoring of therapeutic responses. CgA is an SI GEP-NEN
marker present
in 60-80% of GEP NETs, as described by Modlin IM etal., Chromogranin A-
Biological
Function and Clinical Utility in Neuro Endocrine Tumor Disease, Ann Surg
Oncol. 2010
Sep;17(9):2427-43. Epub 2010 Mar 9.
Detection of GEP-NEN biomarkers following surgical intervention:
[00474] Nine patients underwent small bowel and hepatic met resection
(resulting in an
approximately 90% reduction in tumor volume). Expression levels of the 13
summed GEP-
NEN biomarker transcripts (APLP2, ARAL], BRAF, CD59, CTGF, FZD7, Ki67, KRAS,
NAP1L1, PNMA2, RAF], TPH1, VMAT2) were determined as described above using
real-time
PCR on samples prepared from whole blood samples, taken one day before surgery
and then two
weeks post-operatively.
[00475] The results are shown in FIG. 31 (horizontal bars representing mean
expression
levels pre and post surgery). Two weeks following surgery, summed expression
levels (as
described above) of the GEP-NEN Biomarkers levels were significantly decreased
(from a mean
of 84 before surgery to a mean of 19 after surgery, greater than 75%
reduction, with p<0.02)
(FIG. 31A). As shown in FIG. 31B, detection of CgA expression levels alone did
not show a
significant decrease (20 % reduction in mean expression).
[00476] These results demonstrate that biomarker expression levels detected
with the
provided methods and systems accurately reflect tumor removal and can be used
to evaluate
responsiveness and efficacy of surgical intervention.
Detection of GEP-NEN-biomarkers following somatostatin analogue (Sandostatin
JAR
(Octreotide acetate injection)) drug therapy
[00477] Summed expression levels (as described above) for the thirteen
biomarkers also were
detected in eight patient samples by real-time PCR, before, one month after,
and two months
after treatment with Sandostatin LAR (Octreotide acetate injection), a
somatostatin analog.
The results are shown in FIG. 32.
131
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
[00478] Results showed a significant (p=0.017) reduction in expression of the
summed
biomarker transcripts one month after continuous treatment. After six months
of continued
treatment, transcript levels were reduced by an additional 50% (p=0.06 vs. 1
month) and were in
the normal range (FIG. 32A). In contrast, no significant change in CgA
expression levels,
alone, was observed at one month post-LARO treatment (FIG. 32B); in this
study, detected
levels of CgA expression alone decreased only at the 6 month time-point. These
results show
that as for surgical intervention, the provided systems and methods for
biomarker detection can
be used to monitor LARO treatment, providing a higher sensitivity as compared
to detection of a
single GEP-NEN biomarker alone. e.g. CgA.
Early detection of low volume micrometastasis and evaluation of treatment
efficacy in
individual patients
[00479] The summed 13 GEP-NEN biomarker expression levels (as described above)
were
monitored to evaluate treatment efficacy and predict risk in two individual
patients, treated with
CryoAblation and hepatic met resection, respectively.
[00480] Patient SK, a male of 63 yrs, with metastatic small intestinal (SI)
GEP-NEN was
evaluated as normal by stereotactic radiosurgery (SRS) / computed tomography
(CT), and was
considered disease free. Summed expression of transcripts in whole blood was
evaluated using
real-time PCR as described above. The results, presented in FIG. 33, showed
normal expression
levels of CgA. In contrast, summed expression levels of the panel of 13 GEP-
NEN biomarkers
(APLP2, ARAF1, BRAE, CD59, CTGF, E7D7, Ki67, KRAS, NAP111, PNMA2, RAP], TPH1,
VMAT2) ("PCR(+)") were elevated (FIG. 33). Based on this information, the
patient underwent
11C-PET-CT in Sweden, demonstrating he had a liver metastasis of approximately
0.5 cm.
Subsequently, the patient underwent cryoablation, which liberates GEP-NEN
tissue for entry
into the blood, inducing symptoms, as described by Mazzaglia PJ, et al.,
"Laparoscopic
radiofrequency ablation of neuroendocrine liver metastases: a 10-year
experience evaluating
predictors of survival." Surgery 2007;142(1):10-9.
[00481] Expression levels were monitored monthly for six months following
cryoablation, by
real-time PCR on RNA prepared from whole blood. The results demonstrated
elevated
expression levels of the biomarker panel, but not of CgA alone, after
cryoablation. Between four
and five months following cryoablation, bone micrometastases were identified;
PCR
demonstrated the appearance of these micrometastases correlated with elevated
GEP-NEN
biomarker panel transcript expression levels; CgA expression alone was
detected as normal.
132
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
Following LAR therapy (which blocks secretion and proliferation of GEP-NEN
cells),
biomarker panel expression levels in the blood were determined to normalize
(real-time PCR).
[00482] This study demonstrates that detection of GEP-NEN biomarker panels by
the
provided methods can accurately reflect acute GEP-NEN-associated events,
demonstrating the
impmroved sensitivity of the provided methods (e.g., as compared to detection
of an available
biomarker, CgA alone) and systems for detecting GEP-NEN biomarkers in
prognostic and
predictive analysis and evaluation of treatment efficacy and detection of
relapse or early-stage
disease, particularly when disease is limited to rare micrometastases.
[00483] Patient BG, a 69-year old female with metastatic small intestinal GEP-
NEN. Grade
T2N1M1, underwent surgical small bowel and hepatic met resection, followed by
9 months of
treatment with Octreotide LAW). Expression levels of CgA and the 13-GEP-NEN
biomarkers
("PCR(+)") were monitored by real-time PCR on whole blood samples as described
above, prior
to surgery, two weeks post-op, and monthly for twelve months. All symptoms
resolved after
twelve months of treatment. As shown in FIG. 34, detection of CgA expression
levels alone
revealed dramatic fluctuations, not correlating with treatment or symptom
reduction. Biomarker
panel expression levels ("PCR(+)"), by contrast, were detected as
significantly reduced
following surgical tumor excision, measured two weeks post-op, and
significantly reduced
following LAR treatment, remaining reduced out to twelve months (at which
point all
symptoms remained resolved).
[00484] This study demonstrates that detection of GEP-NEN biomarker panels by
the
provided methods can sensitively reflect disease severity and responsiveness
to treatment,
providing an improvement over available biomarker detection methods. The
provided methods
and systems are useful for monitoring treatment responsiveness and relapse,
and can detect both
the presence (FIG. 33) and the absence (FIG. 34) of GEP-NEN disease with high
fidelity. These
results demonstrate that detection of the provided biomarkers in blood
provides added diagnostic
and treatment value, for example, as surrogate markers for treatment efficacy
to monitor the
effects of surgery (removal of tumors) or targeted medical therapy (
inhibition of tumor
secretion/ proliferation). See Arnold R, et al., "Placebo-controlled, double-
blind, prospective,
randomized study of the effect of octreotide LAR in the control of tumor
growth in patients with
metastatic neuroendocrine midgut tumors: A report from the PROMID study
group," ASCO
2009, Gastrointestinal Cancers Symposium, Abstract #121. 2009.
133
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
EXAMPLE 6: Evaluation of the Molecular Fingerprint as an Indicator of
Treatment Efficacy
[00485] The efficacy of this 51 marker gene panel as a potential test was
examined in the four
independent datasets to establish whether it could differentiate between
treatment-responsive
(those that were clinically categorized as in complete remission or as
exhibiting stable disease)
and untreated (treatment-naïve) or non-responsive (clinically categorized as
"progressive")
tumors. In addition, a 13 marker subset of the 51 marker gene panel was
evaluated to determine
if it could be used to provide additional information regarding response to
therapy, specifically if
it could provide more specific information in progressive, untreated disease
compared to stable
disease.
[00486] Independent set 1 included 35 GEP-NENs. Full clinical details were
available for all
patients; 33 of the samples were considered in complete remission or had
stable disease. Sixty
percent of samples were under treatment (predominantly LAR: 96%).
[00487] Independent set 2 included 32 GEP-NENs. Full clinical details were
available for
all: 28 of the samples were considered in complete remission or had stable
disease. Eighty-four
percent of samples were under treatment (LAR: -40%, surgery -25%).
[00488] Independent set 3 included 47 NENs. Full clinical details were
available for all: 30
of the samples were considered in complete remission or had stable disease.
Fifty-six percent of
samples were under treatment (LAR: -75%).
[00489] Independent set 4 included 89 GEP-NENs. Full clinical details were
available for all
patients; 71 of the samples were considered in complete remission or had
stable disease. Forty-
six percent of samples were under treatment (predominantly LAR: 85%).
[00490] The four algorithms were assessed: SVM, LDA, KNN and Bayes for utility
in
determining whether a blood sample was associated with a "treated" phenotype
(clinically
responsive/stable disease) or could identify untreated/progressive disease.
Tumor samples that
were called "normal" or -treated" were considered to exhibit a -treated" or
clinically responsive
("responder") phenotype. Those considered "untreated" were classified as being
non-responsive
(or "non-responders"). The algorithms as a group ("voting" algorithm) were
examined for utility
and include correct call rates from best 3 of 4 algorithms.
[00491] Tabulated results indicate that overall correct call rates
(identifying both
appropriately treated and non-responsive samples) was 73-94% in independent
set 1, 81-89% in
134
CA 02828878 2013-08-30
WO 2012/119013
PCT/US2012/027351
set 2, 82-94% in set 3 and 72-94% in set 4 (Table 15). The average rates were
83-88% for each
of the algorithms. A combination, best "3 of 4", resulted in similar (88%)
correct call rate.
Table 15. Overall call rates (%) for each of the algorithms in each of the
independent sets
SVM LDA KNN Bayes Best 3 of 4
Set 1 89 89 94 94 94
Set 2 88 88 88 88 88
Set 3 83 85 89 72 88
Set 4 73 81 82 82 82
AVE (%) 83 86 88 84 88
[00492] Further analysis of the calls identified whether the correct call
rates corresponded to
identifying clinically responsive patients or samples from those individuals
that were not
responding to treatment (Table 16).
Table 16. Call rates (%) for each of the groups, clinically responsive or non-
responders, in each
of the independent sets
SVM LDA KNN Bayes
RESP NON RESP NON RESP NON RESP NON
Set 1 94 0* 94 0* 100 0* 100 0*
Set 2 100 0* 100 0* 100 0* 100 0*
Set 3 91 73 94 80 97 73 75 67
Set 4 83 83 82 77 90 50 96 39
AVE 92 78 90 79 97 62 93 53
(%)
* = excluded from analysis as only two and four patients were classified as
"non-responders" in
each of these two sets.
[00493] The most consistent correct calls for "responders" were identified
with the KNN
algorithm (-97%). The highest correct call rates for "non-responders" were the
SVM and LDA
algorithms (-80%).
[00494] Sensitivities, specificities, positive predictive values and negative
predictive values
calculated for each of the algorithms in the 3 independent sets are included
in Table 17.
Table 17. Performance metrics for each of the algorithms in each of the
independent sets
SVM LDA KNN Bayes
ABC DABCDABCDABICD
135
CA 02828878 2013-08-30
WO 2012/119013
PCT/US2012/027351
Set 1 97 0 97 0 97 0 97
0 100 0 100 0 100 0 100 0
Set 2 88 * 100 0 88 * 100 0 88 * 100 0 88 * 100 0
Set 3 100 62 86 100 96 86 94 92 97 85 94 92 89 55 76 77
Set 4 95 56 83 83 94 52 82 77 88 56 90 50 85 88 98 39
AVE 95 39 92 46 94 46 93 42 93 47 96 36 90 48 94 29
SENS = sensitivity, SPEC = specificity, PPV = positive predictive value, NPV =
negative
predictive value, * no value (cannot be calculated)
[00495] The SVM, LDA and KNN algorithms performed best for detecting patients
that were
considered to be either in complete remission or exhibiting stable disease
(sensitivity = 93-95%).
The SVM algorithm was also the most sensitive algorithm for detecting
individuals with
untreated or progressive disease (80%). The combination, best "3 of 4",
resulted in an average
99% correct call rate for determining remission/disease stability and 75% for
detecting untreated
or progressive disease.
[00496] The best algorithms to differentiate treatment-responsive samples from
those
classified as non-responders were LDA and KNN with PPVs of ¨98% and NPVs of
¨92%.
[00497] Thereafter, the association between clinical description and PCR-based
scores was
examined. To delineate individual groups, the following descriptors were used:
[00498] "Complete remission" = all investigations negative;
[00499] "Stable disease after surgery" = abnormal investigations but no change
in serial
evaluation; and
[00500] "Stable disease after surgery + LAR" = abnormal investigations but no
change in
serial evaluation.
[00501] Analysis by clinical criteria (examination, biochemistry, scanning) of
all treated
samples in the 4 independent sets as a group identified:
1) Patients considered in complete remission (i.e. following surgery for
removal of an
appendiceal tumor (n=3) or a hemicolectomy for a <1.5cm ileo-cecal MEN with no
lymph node metastases (n=8) or <2 lymph node metastases (n=2) were correctly
identified in 100% of cases by the algorithms (FIG. 35). All 13 samples were
called
"normal" by the algorithm.
2) Patients considered as stable disease (following surgery for removal of
tumor
(hemicolectomy: n=24, gastrectomy: n=1, appendectomy: n=3, hemicolectomy and
liver
resection: n=7, ileal/colonic resection: n=3, hemicolectomy, liver resection
and lymph
136
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
node dissection: n=2, embolization: n=2) were correctly called (called tumor
"treated" by
the mathematical algorithms) in 95% of cases (40/42) (FIG. 35).
3) Patients considered as stable disease following drug therapy (long-acting
somatostatin analog (ST-NENs=72, PNENs=l 3, rectal NENs=2, gastric NENs=2),
pasireotide (ST-NEN=1) or RAD001 (ST-NEN=4) were correctly called in 90% of
cases
(70/78) (FIG. 35).
EXAMPLE 7: Use of a 13 marker panel gene panel subset to evaluate disease
responses
[00502] A subset of genes that were highly correlated with untreated,
progressive disease was
evaluated to determine if they could be used to further define patient groups
and be used to
provide additional information regarding response to therapy, particularly in
patients undergoing
treatment but considered to be "progressive".
[00503] An analysis of clinical samples in the test set identified 13 genes
selectively over-
expressed in the untreated, progressive group compared to those considered to
exhibit stable
disease. The genes identified were: AKAP8L, BRAF, CD59, COMMD9, Ki67, MORF4L2,
OAZ2, RAF1. SST1, SST3, TECPR2, ZFHX3 and ZXDC. Inclusion of these genes in an
algorithm resulted in correct call rates to differentiate stable from
progressive disease in ¨73% of
cases in the test set (FIG. 36).
[00504] An analysis of this gene panel in independent sets 1-4 (as a group;
progressive
disease ¨ irrespective of treatment: n=26 [50% of patients on treatment. 50%
treatment stopped
because considered untreatable]; stable disease: n=143) identified that the
correct call rate for
samples from patients considered to exhibit "progressive" disease was 65%
(KNN). The
sensitivities, specificities, PPV and NPV for the four different algorithms in
this group were 34-
65%, 96-100%, 64-100%, and 89-94% respectively. The best algorithm to detect
"progressive"
disease was KNN; the best algorithm for detecting "stable" disease was SVM.
This indicates that
a 13 marker panel subset is useful as an adjunct to the 51 marker panel
particularly for
identifying GEP-NENs that are not responding to therapy and are considered
clinically
"progressive".
[00505] These approaches demonstrate that treatment responsiveness can be
accurately
defined by the 51 marker panel in 90-100%. Samples that are considered
clinically progressive
and therefore not responsive to therapy (e.g., LAR or everolimus) can be
identified in 65-80%.
137
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
EXAMPLE 8: Comparison of the 51-marker gene panel with plasma Chromogranin A
levels for
Disease Prediction
[00506] The utility of the PCR-based approach was compared to Chromogranin A
levels
measured in plasma for identifying GEP-NENs and differentiating between
treated and untreated
samples.
[00507] CgA is widely utilized as a generalized NEN marker and elevated levels
are generally
considered to be a sensitive, ¨70-90% accurate as a marker for GEP-NENs.
Measurements of
this peptide, however, are non-specific (10-35% specificity) as it is also
elevated in other
neoplasia e.g., pancreatic and small cell lung neoplasia and prostate
carcinomas as well as in a
variety of cardiac and inflammatory diseases, by proton pump inhibitor usage
and in renal
failure. CgA is a component of neuroendocrine cell secretion, not
proliferation, and therefore its
use as a surrogate for tumor growth has obvious significant limitations. In
general, the sensitivity
of this biomarker for predicting GEP-NENs is dependent on the degree of
differentiation of the
tumor, the location of the tumor and whether it is metastatic or not. Despite
modest correlations
between CgA levels and hepatic tumor burden, the low (<60%) sensitivity for
detecting
metastases, the absence of a standardization of measurement in the USA, as
well as that the FDA
does not accept CgA as a supportable biomarker, it is currently the only
marker "routinely" used
to evaluate treatment efficacy (surgery, liver transplantation, bio-/chemo-
therapy, chemo-
/embolization, radiofrequency ablation). CgA levels were therefor used as the
best available
equivalent of a "gold-standard" against which to assess the PCR-based test.
[00508] CgA values were measured using the DAKO ELISA kit in the initial test
set of 130
samples (controls: n=67, GEP-NENs: n=63 [untreated disease, n=28, treated,
n=35]) used to
develop the 51 marker gene panel. The DAKO kit is art recognized to detect CgA
in plasma
samples from GEP-NENs.
[00509] CgA levels were elevated (p<0.05) in both untreated (63%) and treated
GEP-NENs
(32%) using either the Student's t-test (FIG. 37A) or non-parametric tests
(FIG. 37B).
[00510] The efficacy of CgA to identify GEP-NENs compared to controls
identified a correct
call rate of 74% (Table 18). The efficacy for correctly identifying a GEP-NEN,
irrespective of
treatment, was lower at ¨45%.
Table 18: Diagnostic capacity of CgA levels to discriminate controls from all
GEP-NENs
(treated and untreated).
138
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
Normal (True) GEP-NENs
(True)
Normal (Predicted) 65 30
GEP-NENs (Predicted) 2 26
The performance metrics of this test were: Sensitivity = 97%, Specificity =
46%, PPV = 68%
and NPV = 93%.
[00511] DAKO uses a cut-off of 19 Units/L as the upper limit of normal. Using
this value, a
total of 25 (45%) of 56 GEP-NENs were considered positive compared to 1 (1.4%)
of 67
controls for performance metrics of Sensitivity = 45%, Specificity = 98%, PPV
= 96% and NPV
= 68% (FIG. 38). The correct call rate for this cut-off was 70%.
[00512] Using CgA levels with the PCR transcript expression (51 marker panel)
was next
evaluated for the ability to provide additional value to the predictions.
Inclusion of CgA levels
did not increase the prediction ability of the marker genes, and reduced the
efficacy, particularly
of the KNN classifier (FIG. 39A-B). It was conclude that inclusion of CgA
levels do not
improve the quality of the candidate marker gene panel.
[00513] These results demonstrate that quantification of a circulating multi-
transcript
molecular signature (tumor transcripts) is more sensitive than measurement of
a single,
circulating protein (CgA). Inclusion of CgA measurement in the molecular
fingerprint provides
no "added" predictive value.
EXAMPLE 9: Comparison of the 51-marker gene panel with plasma Chromogranin A
levels for
Assessment of Disease Efficacy
[00514] The utility of the PCR-based approach was directly compared to CgA
levels
measured in plasma for identifying GEP-NENs and differentiating between
treated and untreated
samples. Analyses of the efficacy of CgA to differentiate between treated and
untreated GEP-
NENs identified that the correct call rate was 66% (Table 19). The performance
metrics were:
Sensitivity = 69%, Specificity = 63%, PPV = 67% and NPV = 65%.
Table 19: Diagnostic capacity of CgA levels to discriminate between un-treated
and treated
GEP-NEN samples
Treated (True) Untreated (True)
Treated (Predicted) 20 10
Untreated (Predicted) 9 17
139
CA 02828878 2013-08-30
WO 2012/119013
PCT/US2012/027351
Illustrative Cases
[00515] To facilitate clinical usage, a scoring system was developed based on
the calls from
the mathematical algorithm. This is a "Distance" score that measures the
Euclidean distance of
an unknown sample to gene expression profiles of the different calls "Normal"
versus "Tumor",
and "Treated" versus "Untreated". A low score: 0-25 converts to "normal", 26-
50 is "tumor ¨
treated" (or stable) and 51-100 is "tumor untreated". This provides a
physician-friendly
visualization since it is clear where an individual patient value falls in the
disease spectrum
(diagnosis of "normal" versus "tumor" and clinical interpretation of "treated"
versus
"untreated"). It also provides the opportunity to graph how treatment
influences the transcript
index of the disease. An example is provided in FIG. 40. These terms and
scores are used for the
individual, illustrative cases provided below.
Index case 1: Incidentally identified Appendiceal NEN, with subsequent
development of a
mesenteric metastasis
[00516] JPP (45 yo male with hypertension and previous splenectomy [1998]),
underwent left
hemicolectomy for an abscess and perforation [12/2009]. At surgery, a well
differentiated 0.8cm
NEN was identified with lymphatic invasion and extension to the meso-appendix
[T?N1M1].
The tumor exhibited low proliferative capacity: Ki67<2% and mitotic count
<1/10HPF). A
subsequent MRI scan (1/2010) identified residual mesenteric implants and
repeat surgery
(4/2010) was undertaken for colostomy closure. At this time a mesenteric lymph
node metastasis
(<1cm) was removed (Ki67<2%).
Table 20
Surgery Dec-09 Jan-10 Mar-10 Surgery
Apr-10 Aug-10 Apr-11
Score 68 40 45
PCR test Diagnosis TUMOR TUMOR
TUMOR
Interpretation Untreated Treated
Treated
Value
6.5U/L 9.6U/L 6.7U/L 7.8U/L
CgA ELISA Call NML NML NML NML
Left hemi-
MRI: Surgery -
colectomy
Residual colectomy lcm
PROCEDURE [<1.cm well-
mesenteric LN metastasis,
differentiated
Disease Ki67<2%
NEN, Ki67<2%]
PCR score: 0-100 (0-25=normaL 26-50=treated; 51-100=untreated disease);
Diagnosis = normal or tumor, interpretation =
treated versus untreated. CgA values: Units/liter (U/L (DAKO ELISA)
ABNML = abnormal (elevated); NML = normal range
140
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
[00517] The PCR test is more sensitive than CgA for identifying residual
(untreated) disease
(diagnosis = "tumor", interpretation = "untreated", PCR score 68) and for
demonstrating surgical
removal of the metastasis (diagnosis = "tumor", interpretation = "treated",
PCR score 40). The
identification that the blood PCR test did not revert to a call of "Normal"
following surgical
excision indicates the presence of residual metastatic disease (PCR score
remains above normal:
¨40). Since the PCR value has not changed between 2010 and 2011 to an
"untreated" phenotype
it is likely that the disease is clinically "stable".
Index case 2: Si-NEN, surgical resection.
[00518] BA (65 yr female, history of coronary artery disease, type 11 diabetes
and glaucoma
exhibited with anemia in 1996 and 2006. Colonoscopy and CT scan identified a
terminal ileal
NEN [5/2009]. She underwent a right hemicolectomy (2/2010) for removal of a
lcm SI-NEN
which exhibited lymphatic invasion but no nodes were positive [T1NOM0]. Tumor
exhibited low
proliferative index: Ki67=2% and mitotic count <2/10HPF).
Table 21
May-09 Nov-09 Surgery Feb-10 Mar-10
Score 30
61 26
PCR test Diagnosis
TUMOR TUMOR TUMOR
Interpretation Untreated Treated Treated
Value
15.5U/L 7.8U/L 17U/L
CgA ELISA Call NML NML ABNML
Endoscopy Right hemi-colectomy
PROCEDURE & CT Scan: 1cm well-differentiated
!LEAL NEN lesion, Ki67<2%
PCR score: 0-100 (0-25=normal, 26-50=treated; 51-100=untreated disease);
Diagnosis = normal or tumor, interpretation =
treated versus untreated. CgA values: Units/liter (U/L (DAKO ELISA)
ABNML = abnormal (elevated); NML = normal range
[00519] The PCR test identifies a small mass, low proliferation small
intestinal NEN
(diagnosis = "tumor", interpretation = "untreated", PCR score 61). The
identification that the
blood PCR test did not revert to a call of "Normal" following surgical
excision indicates the
presence of residual disease (PCR score 26-30) suggesting incomplete resection
(non-RO).
141
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
Index case 3: Metastatic rectal NEN (endoscopically removed), pan-segmental
liver metastases,
treated with LAR
[00520] AJ (47 yo male, with an incidentally identified rectal NEN (on
endoscopy ¨ 5/2010)).
Extensive pan-segmental liver metastases were noted on follow-up (CT/MRI scan
6/2010).
Sandostatin was initiated [7/2011]. Surgical resection for residual disease
(primary and rectal
lymph node metastasis) with removal of 2 liver metastases was undertaken
[10/2010]. A 1.5cm
rectal lymph node metastasis), with a Ki67<15% was identified. The liver
metastases had a
Ki67-3% (T2N1M1). Subsequent surgery was undertaken [2/2011] to close the
ileostomy and
remove additional liver metastases. Serial MRI scans demonstrated no change in
hepatic burden.
Sandosatin continued.
Table 22
May-10 Jun-10 Jul-10 Oct-10 Jan-11 Jun-11
Score 78 36 44 26
PCR test Diagnosis TUMOR TUMOR TUMOR TUMOR
Interpretation Untreated Treated Treated
Treated
Value
9.5U/L 9U/L 10U/L 9.3U/L
CgA ELISA Call NML NML NML NML
CT/MRI
Endoscopy MRI/CT Resection Closure, liver
& Polyp Scan: for residual mets,
Ki67<3%, no
Sandostatin change
PROCEDURE removal: Extensive nitiate disease, CT/MRI
no
id in
RECTAL NEN hepatic 1.5cm LN, change in
disease
(lcm) metastasis K167-15% disease burden
burden
PCR score: 0-100 (0-25=normal, 26-50=treated; 51-100=untreated disease);
Diagnosis = normal or tumor, interpretation =
treated versus untreated. CgA values: Units/liter [U/L) (DAKO ELISA)
ABNML = abnormal (elevated); NML = normal range
[00521] The PCR test identifies liver metastases as well as residual disease
from a non-
functional (non-secreting) lesion (diagnosis = "tumor", interpretation =
"untreated", PCR score
78). The PCR test is more effective than CgA for both identifying the disease
and monitoring
treatment response. The failure of the PCR test to revert to a call of
"Normal" is consistent with
the presence of liver metastases. The values have not changed between 2010 and
2011 to an
µ`untreated" phenotype suggesting the disease is "stable". This finding (PCR
score: 26-44) is
consistent with current imaging protocols which demonstrate stable non
progressive disease.
142
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
Index case 4: Metastatic SI-NEN, pan-segmental liver metastases, treated with
hemicolectomy,
lymph node dissection and liver resection and LAR
[00522] BG (71 yo female, initially identified with hepatic nodules and ¨4cm
mesenteric
mass (positive by octreoscan) confirmed to be a well-differentiated
neuroendocrine carcinoma
(by liver biopsy) [9/2008]. Underwent an ileal and liver wedge resections
[12/2008]. An 8cm
mesenteric nodule was removed as was a 1.5cm NEN, while 6/9 lymph nodes were
positive for
metastasis. The tumor had a low proliferative capacity, mitotic count =
2/10hpf, Ki67<2%
(T2N1M1). Octreotide was initiated 2/2009 with some control of symptoms but
increasing right
upper quadrant discomfort was noted. Octreoscan [4/2010] identified several
small liver lesions
with additional lesions confirmed in 2/2011 (Octreoscan). Underwent ERCP and
sphincterotomy
[4/2011] and cholecystectomy [6/2011].
Table 23
Sep-08 Dec-08 Feb-09 Apr-10 Feb-11 Apr-
11 Jun-11
Score 70 32 27 32 35 33
Diagnosis
PCR test TUMOR TUMOR TUMOR TUMOR TUMOR
TUMOR
Interpretation Untreate
Treated Treated Treated
Treated Treated
Value
9.2U/L 10U/L 28.8U/L 9.1U/L 8U/L
8.2U/L -- 11U/L
CgA ELISA Call NML NML ABNML NML NML NML NML
Ileal and
Octreosca hepatic ERCP &
c. 0 treoscan Octreoscan
n & Liver resection, Sandostati
sphincter Chclecyst-
PROCEDURE : small liver : additional
biospy: 1.5cm n initiated
ectomy
lesions liver lesions
WD NEC NEN, otomy
Ki67=2%
PCR score: 0-100 (0-25=normal, 26-S0=treated; S1-100=untreated disease);
Diagnosis = normal or tumor, interpretation =
treated versus untreated. CgA values: Units/liter (U/L (DAKO ELISA)
ABNML = abnormal (elevated); NML = normal range
[00523] The PCR test identifies extensive disease (diagnosis = "tumor",
interpretation =
"untreated", PCR score 70). Since the blood PCR test (PCR Score 27-35) did not
revert to
"Normal" after surgery the result is consistent with residual metastases. CgA
results performed
less effectively than the PCR test for both identifying the disease and
monitoring treatment
response.
143
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
Index case 5: Recurrent liver metastasis (following hepatectomy), treated with
JAR and
embolization
[00524] SK (64 yo male, with a history of atrial fibrillation, hyperlipedemia
and kidney
stones). SI-NEN diagnosed [12/2001] after developing flushing. He underwent
resection of the
ileal tumor and hepatic mets. Subsequent surgeries included re-resection of a
mesenteric lymph
node mass [3/2005] and lymph nodes [9/2006]. In [12/2008] cryoablation for a
liver met. PET
scan [4/2009] identified small liver nodules and a bone lesion. Sandostatin
begun [6/2009],
repeat scans [PET and MRI] identify no new lesions.
Table 24
Dec-01 Mar-05 Sep-06 Jan-09 Dec-09 Apr-10
Sep-10 Mar-11
Score 63 26 49 33
35
Diagnosis TUMO
TUMO
PCR test TUMOR TUMOR R
TUMOR
Interpretation Untreate Treate
Treate
Treated d Treated
CgA ELISA Value
8.1U/L 10U/L 8.2U/L 28.8U/L 8U/L
Call
NML NML NML ABNML NML
Ileal and
PET
hepatic
Scan:
resectio
Mesenteri Lymph
Cryoablatio liver PET/M
n for node Somatostati
PROCEDURE C LN n for liver
nodules RI no
NEN resectio n initiated
Resection metastasis
and lesions
with
bone
Liver
lesion
Mets
PCR score: 0-100 (0-25=normal, 26-50=treated; 51-100=untreated disease);
Diagnosis = normal or tumor, interpretation =
treated versus untreated. CgA values: Units/liter [U/L) (DAKO ELISA)
ABNML = abnormal (elevated); NML = normal range
[00525] The PCR test identified recurrence of the disease (diagnosis =
"tumor", interpretation
= "untreated", PCR score 63), demonstrated efficacy of cryoablation and
detected residual
disease. Becuase the blood PCR test did not revert to a call of "Normal", this
result was
considered evidence of metastases which were identified by 13C-PET (PCR score
49). CgA
results were less effective than the PCR test for both identifying the disease
and monitoring
treatment response.
144
CA 02828878 2013-08-30
WO 2012/119013 PCT/US2012/027351
EXAMPLE 10: Utility of the molecular signature to differentiate GEP-NEN
subtypes (Small
Intestine versus Pancreatic NENs)
[00526] The 51 marker gene panel was used to examine the ability to
distinguish GEP-NENs
from controls and to differentiate whether a sample is from a patient
responsive to treatment
compared to a non-responder or treatment-naive individual. The marker panel
was developed
around information derived from small intestinal NEN tissue and blood
microarrays. While the
performance metrics are significantly better than for CgA ELISA, it was a goal
of this work to
establish whether the test could differentiate between GEP-NENs from two
different sites,
namely the small intestine and the pancreas. This is relevant in the case of a
tumor of unknown
primary location and is also relevant since tumors exhibit significantly
different prognoses
depending on their site of origin. The 5-year survival of a SI-NEN is ¨80% and
¨50% of the
mortality is not disease-specific. In contrast, the 5-year survival of a PNEN
is ¨40% and ¨95%
of patients die from the disease. Determining the location of an unknown
primary can therefore
be an important variable in determining therapy; somatostatin analogs have
demonstrated utility
in SI-NENs 9 while sunitinib and everolimus have efficacy in PNENs_ENREF_10.
[00527] Examination of the 51 marker gene panel identified that it exhibited a
much larger
expression variance (0.54 0.4 versus 0.38 0.14 in SI-NENs) indicating that the
genes selected
in the panel were not as specific for PNENs as for SI-NENs. Mapping expression
identified that
tumors were spatially separated (FIG. 41A).
[00528] Furthermore, the expression in the panel could differentiate with 92%
accuracy
between the two tumor sites (FIG. 41B). The test can thus accurately
differentiate between a
pancreatic and a small bowel tumor.
EXAMPLE 11: Ability of the 51-marker gene panel to discriminate between GEP-
NENs and GI
Cancers
[00529] To further evaluate the utility of this PCR-based approach, the
molecular fingerprint
in gastrointestinal adenocarcinomas, such as gastric and hepatic cancers
(esophageal: n=2,
pancreatic: n=11, gallbladder: n=3, colon: n=10, rectal: n=7 was examined.
This was undertaken
to assess whether some genes e.g., KRAS, BRAF, Ki67 over-expressed in GI
adenocarcinoma
and included in the panel, might perturb the accuracy.
145
CA 2828878
[00530] Examination of the 51 marker gene panel identified that it exhibited a
larger
expression variance (0.5+0.25 versus 0.44+0.17 in GEP-NENs) indicating that
the NEN-
specific genes selected in the panel were less specific for GI cancers than
for GEP-NENs. PCA
identified that tumors were spatially separated (FIG. 42A) and that the NEN
panel could
differentiate with 83% accuracy between GEP-NENs and GI cancers (FIG. 42B).
[00531] The test therefore has the power to differentiate between GEP-NENs and
GI cancers
and the circulating molecular signature of NENs is different to that of GI
adenocarcinomas.
The minor overlap is consistent with the observation that ¨ 40% of GI
adenocarcinomas exhibit
neuroendocrine elements.
[00532] A direct comparison of the molecular test and CgA ELISA identified
that the PCR-
based method had a significantly more accurate call rate compared to
measurement of CgA
levels (2'12.3,p<0.0005) (FIG. 43).
[00533] The sensitivities were similar for detecting a GEP-NEN (94% versus
97%) but the
specificity of the PCR test was higher than CgA (85% versus 46%). For
differentiating treated
versus untreated samples, the PCR-based test exhibited higher performance
metrics (85%
versus ¨65%).
[00534] CgA is less useful than the circulating molecular fingerprint for
defining
"treatment" in GEP-NENs. This reflects the fact that the protein (CgA) is a
constitutive
secretory product of all neuroendocrine cells and has no specific biological
relationship to
neuroendocrine tumors, their proliferation rate or their metastasis.
[00535] Throughout this application, various website data content,
publications, patent
applications and patents are referenced. (Websites are referenced by their
Uniform Resource
Locator, or URL, addresses on the World Wide Web.)
[00536] The present invention is not to be limited in scope by the embodiments
disclosed
herein, which are intended as single illustrations of individual aspects of
the invention, and any
that are functionally equivalent are within the scope of the invention.
Various modifications to
the models and methods of the invention, in addition to those described
herein, will become
apparent to those skilled in the art from the foregoing description and
teachings, and are
similarly intended to fall within the scope of the invention. Such
modifications or other
embodiments can be practiced without departing from the true scope and spirit
of the invention.
146
CA 2828878 2018-06-11