Note: Descriptions are shown in the official language in which they were submitted.
CA 02839600 2014-01-21
Attorney Docket No. 1001P039CA01
COMPUTER AIDED DIAGNOSIS FOR DETECTING ABDOMINAL BLEEDING WITH
3D ULTRASOUND IMAGING
FIELD OF THE INVENTION
The present invention relates to a diagnosis system for
analyzing 3D ultrasound images to detect the size and
location of internal organs and free fluid.
BACKGROUND
The rapid diagnosis of invisible internal injury in austere
and hostile front-line environments remains a challenge for
medical personnel. The availability of a portable real-time
3-Dimensional (3D) ultrasound imaging system with automated
diagnostic capabilities for detecting non-visible internal
abdominal bleeding, pneumothorax, hematothorax and
facilitating image guided operations is helpful, if not
essential, in supporting triage and medical decisions for
trauma patients.
In trauma patients, inner-body fluids can accumulate at the
lowest position of the concavities within the body. The site
of fluid accumulation depends on the source of the bleeding
and the position of the patient. In general, patients are
examined on the bed lying flat on the back. In such
conditions, inner-body fluids accumulate at positions between
the liver and the kidneys, along the spleen border, at the
position posterior to the bladder or uterus, and in the
pericardial space. Similarly, there are conditions akin to
internal bleeding which also cause an accumulation of fluid
in a patient's internal cavities. For these non-trauma
- 1 -
CA 02839600 2014-01-21
Attorney Docket No. 1001P039CA01
conditions, being able to detect such fluid accumulation
would also be greatly helpful.
One system developed to address the concerns of internal
bleeding and the fluid accumulation it causes is ultrasound
imaging. Ultrasound imaging maps out ultrasound reflection
points in order to build up an internal image of the target.
These systems return information about the internal structure
of a target. In general, ultrasound refers to high frequency
longitudinal mechanical waves, more commonly known as
megahertz (MHz) sound waves. Ultrasound can propagate in any
physical medium but has better propagation characteristics
when traveling through solid or liquid media. Intermolecular
coupling in solids and liquids affects the rate at which
mechanical waves propagate. A solid with strong
intermolecular coupling allows for faster ultrasound
propagation compared to a low density fluid with weak
coupling.
Currently, inner-body fluids are detected by scanning the
human body on specific characteristic locations by means of
the so-called "Focus Assessment with Sonography in Trauma"
(FAST) method. FAST is an important method used to identify
free intraperitoneal, intrathoracic, or pericardial fluid.
FAST is primarily used at the patient's bedside by emergency
physicians and trauma surgeons. The development of hand-held
ultrasound devices facilitated the introduction of FAST into
pre-hospital trauma management (p-FAST).
FAST consists of multiple, focused, ultrasonographic views of
the abdomen and the pericardium. The use of multiple views
increases the sensitivity of the FAST examination in the
detection of hemoperitoneum.
- 2 -
CA 02839600 2014-01-21
Attorney Docket No. 1001P039CA01
Currently, there exist other known imaging systems that use
3-Dimensional ultrasound systems. For instance, United
States (U.S.) patent 7,920,731 provides a method for
differentiating between a blood vessel bifurcation and a
bleeding blood vessel in an ultrasound volume. This method
uses Doppler waveform data to determine if the bifurcation is
a point of internal bleeding.
Additionally, U.S. patent 7,803,116 discloses an ultrasound-
based method for detecting and imaging vibrations in tissue.
The tissue vibrations are analyzed as a detection method for
internal bleeding.
Other examples of internal or ultrasound imaging systems and
methods include U.S. patent 8,520,947, U.S. patent 6,561,980,
U.S. patent 6,385,332, U.S. patent 6,719,696, U.S. patent
6,482,160, and U.S. patent application 13/743,490.
However, none of the above disclosures provides a portable
real-time 3D ultrasound imaging system with automated
diagnostic capabilities for detecting non-visible internal
abdominal bleeding. There is therefore a need for a non-
invasive medical detection system to address casualty care
support.
SUMMARY OF INVENTION
The present invention provides a computer aided diagnosis
system that uses 3D ultrasound imaging to detect human
internal organs such as kidneys, livers, spleens and free
fluid (i.e. fluids due to internal bleeding). The system
detects and labels internal organs to assist the user in
locating the ultrasound probe position. Second, inner body
- 3 -
CA 02839600 2014-01-21
Attorney Docket No. 1001P039CA01
fluids are detected by the system when these appear as dark
areas in 3D ultrasound images.
In a first aspect, the present invention provides a method
for detecting an area of fluid in volumetric ultrasound data
derived from an image of a mammal's body part, the method
comprising:
a) receiving volumetric data;
b) denoising said data to result in denoised
data;
c) enhancing a contrast of tissue voxels in said
denoised data;
d) determining an initial surface of a volume
presented as a dark area in said denoised data;
e) determining if an internal organ is detected
in said volume in said denoised data; and
f) isolating said volume in said denoised data to
determine a size and location of said volume.
In another aspect, the present invention provides computer
readable media having encoded thereon computer readable and
computer executable instructions which, when executed,
implements a method for detecting an area of fluid in
volumetric data derived from an image of a mammal's body
part, the method comprising:
a) receiving volumetric data;
b) denoising said data to result in denoised
data;
- 4 -
CA 02839600 2014-01-21
Attorney Docket No. 1001P039CA01
c) enhancing a contrast of tissue voxels in said
denoised data;
d) determining an initial surface of a volume
presented as a dark area in said denoised data;
e) determining if an internal organ is detected
in said volume in said denoised data; and
f) isolating said volume in said denoised data to
determine a size and location of said volume.
BRIEF DESCRIPTION OF THE DRAWINGS
The embodiments of the present invention will now be
described by reference to the following figures, in which
identical reference numerals in different figures indicate
identical elements and in which:
FIGURE 1 is a block diagram detailing the steps in a
de-speckling method used in one aspect of the present
invention;
FIGURE 2 is a block diagram detailing the steps in a
tissue volumetric pixels (voxels) intensity
enhancement method;
FIGURE 3 is a block diagram of the steps in an
initial surface selection procedure using both manual
and automatic methods;
FIGURE 4 is a block diagram illustrating the steps
for a process for detecting kidney and liver organs
in volumetric data;
- 5 -
CA 02839600 2014-01-21
Attorney Docket No. 1001P039CA01
FIGURE 5 is a table showing an internal force
relation between each surface sample point with its
neighbours as parametric coefficients derived by the
discretized Euler-Lagrange equation;
FIGURE 6 is a block diagram of the steps in a fluid
segmentation procedure using a combination of 3D
Snake and Level-Set methods;
FIGURE 7 is a block diagram of the steps in a
procedure for determining a medical condition of a
patient based on volumetric ultrasound data according
to one aspect of the invention; and
FIGURE 8 illustrates a view panel of a developed
Graphical User Interface (GUI).
The Figures are not to scale and some features may be
exaggerated or minimized to show details of particular
elements while related elements may have been eliminated to
prevent obscuring novel aspects. Therefore, specific
structural and functional details disclosed herein are not to
be interpreted as limiting but merely as a basis for the
claims and as a representative basis for teaching one skilled
in the art to variously employ the present invention.
DETAILED DESCRIPTION OF THE INVENTION
The task of object detection in 3D medical images has been
investigated by researchers, due to its implications for
medical diagnosis. The procedural steps of object detection
in 3D volumes consist of preprocessing tasks, manual
- 6 -
CA 02839600 2014-01-21
Attorney Docket No. 1001P039CA01
adjustment, 3D segmentation and classification. The aim of
the preprocessing tasks is to put more emphasis on valuable
information and to reduce the effect of unwanted interfering
signals. Some examples of such tasks are denoising, edge
refinement, contrast enhancement and volume clipping. As a
second step, user intervention can be applied to boost the
segmentation results. The 3D segmentation task can be
considered to be the main part of an anatomical organ
detection procedure. The selected 3D segmentation method for
ultrasound volumes has to be robust against intensity
variability, speckle noise and discontinuities among object
walls. Finally, the classification step labels the segmented
region as internal bleeding area, kidney, liver or other
organs of interest.
There are numerous approaches to denoising images in order to
reduce the speckle noise in ultrasound images. Some of these
approaches include Temporal Averaging, Homomorphic Wiener
Filtering, Median Filtering, Bayesian Denoising and Wavelet
Thresholding. All of these methods result in the loss of
information and the blurring of edges. The Speckle Reducing
Anisotropic Diffusion (SRAD) was proposed by Yongjian and
Action. In this method, intensities are diffused based on
the gradient magnitude edge and speckle noise level. At edge
regions, intensities become 0, whereas at homogeneous
regions, intensities become 1 and diffusion is performed. In
another method called Squeeze Box Filter (SBF), outliers are
suppressed as a local mean of their neighbourhood.
Yet another approach to de-speckle ultrasound images is to
apply the sparse representation framework to represent the
signal while suppressing noise. The double-sparsity method
- 7 -
CA 02839600 2014-01-21
Attorney Docket No. 1001P039CA01
considers a sparse representation for the learned dictionary
based on an analytical dictionary, such as a Discrete Cosine
Transform (DCT) dictionary. This method uses a dictionary for
the denoising ultrasound volume and sparsely represents the
volume to effectively reduce the noise level.
Additionally, much research has been dedicated to segment 3D
ultrasound images, such as Level-Set based approaches, region
growing segmentation, Watershed and Graph-cuts methods, and
Snake active contour models. The task of ultrasound image
segmentation is known to be highly challenging, due to innate
problems of ultrasound images including high speckle noise,
inconsistent intensity levels, and discontinuities in regions
and boundaries.
The 3D Snake approach provides a highly robust method against
boundaries' discontinuities, if its parameters are correctly
adjusted. However, the precise segmentation using the 3D
Snake method requires an extensive number of sample points,
resulting in a very high computational cost.
Region growing segmentation methods employ iterative
propagation of an initiated region into homogenous regions
that have similar intensity levels. This method is highly
sensitive to the intensity variations and fails to correctly
operate in the ultrasound volumes which are naturally varying
in intensity.
The simplicity of extending the Level-Set approach from 2-
Dimensional (2D) to 3D-segmentation contributes to this
approach being commonly used in 3D medical image processing.
It provides an accurate solution for some medical imaging
modalities such as Magnetic Resonance Imaging (MRI) and
- 8 -
CA 02839600 2014-01-21
Attorney Docket No. 1001P039CA01
Computed Tomography (CT). However, in the case of ultrasound
images, the Level-Set method can fail in the presence of
discontinuities among boundaries.
In one aspect, the present invention is a computer aided
diagnosis system that uses, as input, 3D ultrasound images to
detect human internal organs, such as kidneys, livers and
spleens, as well as free fluid, i.e. fluid due to internal
bleeding. The present invention facilitates automatic object
recognition for non-skilled users. A person skilled in the
art will understand that the method steps of the present
invention may be programmed into a handheld device, or any
other suitable data processing device.
The present invention aims to detect and label internal
organs in order to locate the ultrasound probe position.
Another aim is to detect inner body fluids presented as dark
areas in 3D ultrasound images. The inner body fluids can be
blood fluid or a fluid-containing organ, such as a bladder or
a gallbladder. One target of the present invention is to
detect medical conditions associated with internal bleeding.
In another aspect, the present invention uses the position of
the detected internal organs to decide whether the detected
fluid area is an inner body fluid-containing organ or if the
fluid area is due to internal bleeding.
It is important to note that ultrasound volumetric images
suffer several problems including: high levels of speckle
noise, contrast variation among volume data, and
discontinuities among internal organ body boundaries. To
reduce the abovementioned ultrasound 3D imaging problems, the
present invention combines two methods, noise reduction using
- 9 -
CA 02839600 2014-01-21
Attorney Docket No. 1001P039CA01
the double-sparsity dictionary learning approach and contrast
enhancement using the Mardia and Hainsworth method.
It should be noted that the double-sparsity dictionary
learning method is used to train a sparse-dictionary that
best describes the valuable signal and is not representative
for the speckle noise. Thus, the reconstructed image using
the generated dictionary in the sparse representation
framework is de-speckled.
It should further be noted that the Mardia and Hainsworth
method is a local thresholding approach that localizes the
classification of points into tissue and non-tissue regions.
To improve the contrast of the ultrasound volumetric data,
the Mardia and Hainsworth approach is applied to the de-
speckled volume to create a binary volume which has a value
of one at tissue voxels and zero at non-tissue voxels. Then,
the weighted summation of the de-speckled volume and the
binary volume provides a De-Speckled Contrast Enhanced (DSCE)
volume at the preprocessing step.
In another aspect, the present invention provides an
effective way for a user to manually select initial seed
points or to manually draw initial contours for the
segmentation method. Specifying multiple initial seeds
provides flexibility to improve segmentation results. In
addition to the capability of manually selecting initial
points, the present invention may automatically offer seed
points in the ultrasound volume. The model of human visual
attention system is used to select conspicuity points in the
ultrasound volume. The user can switch between manually
selected points/contours or automatically specified points.
- 10 -
CA 02839600 2014-01-21
Attorney Docket No. 1001P039CA01
Two segmentation strategies are preferably applied in the
present invention: (1) a deformable model based on prior
shapes to detect organs, and (2) a combination of active
contour model, Level-Set, and region growing method to detect
fluid areas. The first strategy is applied to detect an
organ, such as a kidney or liver, based on the prior
knowledge of its shape. The known prior shape is defined as
a zero Level-Set function and a deformable model is used to
fit the shape to a region in the volume. The deformable model
consists of global and local transforms. The global
transform is formed by rotation, translation and a single
scale factor with 7 parameters. The global transform finds a
possible location of the prior shape in the volume. The
local transform is then applied using the Level-Set approach
with several iterations to maintain the minimal changes in
the prior shape. The deformed shape is then analysed to
determine how well it fits in the volume in order to decide
whether the object has been detected or not. This task is
performed for prior shapes of organs in an online fashion for
the stream of volumetric data.
In ultrasound volumetric data, the fluid areas appear as wide
homogenous dark regions. The initial seed points, either
manually drawn or automatically determined, are used to
initiate the segmentation of the fluid regions. The
segmentation processing step is a hybrid approach that
consists of the 3D Snake active contour model, 3D Level-Set,
and the minimum variance region growing approach. The 3D
Snake model is robust against discontinuities among object
boundaries, while the 3D Level-Set approach provides better
accuracy. The present invention takes advantage of the
benefits of both approaches, including robustness against
- 11 -
CA 02839600 2014-01-21
Attorney Docket No. 1001P039CA01
discontinuities and the high accuracy of segmentation. The
3D Snake is initiated with a sphere and the sphere is
iteratively evolved to extract the object boundary. The
segmentation result of the 3D Snake is a 3D surface that is
used to initiate the Level-Set function. The 3D Snake
surface is first converted to a binary volume with ones
inside the surface and zeroes outside the surface. The 3D
image-filling operator is applied to fill the inside of the
surface. For this, a surface without any gap is required.
However, the 3D Snake points are discretely separated and an
interpolation is required to create a closed surface. This
is achieved by using a few iterations of the region growing
approach. Then, the image-filled morphological operator
provides a filled binary volume which can be easily used to
create the initial Level-Set function. Further iterations of
the Level-Set deformation can be used to improve the
segmentation accuracy.
The preprocessing step is added to improve the accuracy of
the segmentation task. The two purposes of the preprocessing
step are to reduce the amount of the speckle noise and to
improve the contrast of the volumetric data. The sparse
representation framework is employed to reduce the amount of
speckle noise in the volumetric ultrasound data. Also, the
volume intensities in tissue voxels are improved by adding a
weighted volumetric classification result obtained using the
Martha and Hainsworth approach.
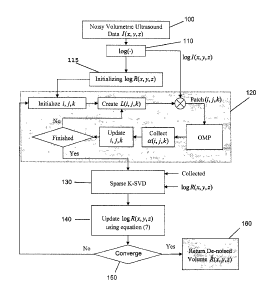
A block diagram of a denoising approach based on a sparse K-
SVD (K-Singular Value Decomposition) is shown in Figure 1.
Referring to Figure 1, the input data of the present
invention is the 3D ultrasound images, also known as the 3D
- 12 -
CA 02839600 2014-01-21
Attorney Docket No. 1001P039CA01
ultrasound volume. In this process, the first step 100 is
that of determining the volumetric data I(x,y,z) , in which
y=[1,...,Sy] and z=[1,...,Sz]. Each I(x,y,z) is a
volumetric pixel (voxel) with intensity levels in
l(x,y,z)e[0,1,2,...,255]. With the current ultrasound imaging
technology, the ultrasound volume data has a high level of
multiplicative noise, also called the speckle noise. The
general formulation for the speckle noise is represented as,
1(x, y,z)= eõ,(x, y,z)xR(x, y,z)+ Ea(x, y,z) (I)
where em(x,y,z) and ea(x,y,z) are multiplicative and additive
noise components, respectively. R(x,y,z) is the actual data.
The additive noise component is negligible and can be remove
from the above formula. In addition to the speckle noise, the
intensity level of the ultrasound volumetric data varies in
different locations in the 3D volume. Therefore, fitting a
global probability mixture distribution model is subject to
inaccuracy in ultrasound volumetric data.
De-Speckling using Sparse K-SVD
Consider 3D patches of size VT-tx.:17/4-171 taken from the
volumetric ultrasound data and reordered into a column
vector, X, E R" . Having a dictionary, DE R"<m , of m atoms,
d1ER", the sparse representation of x, is formulated as
follows,
a= arg min llot subject to !Ix-Da11
2 <e
(2)
2
a
- 13 -
CA 02839600 2014-01-21
Attorney Docket No. 1001P039CA01
where a is a sparse vector with a few number of non-zero
coefficients. The dictionary, D, can be an analytical
dictionary. If we consider the noise to have a normal
distribution with zero-mean and unit variance, the MAP
(maximum a posteriori probability) solution of the denoised
signal is attained by solving equation (2) and finding .i,=-JV
. Applying a Lagrange multiplier, equation (2) becomes,
= arg minilx-Dall: +1111allo
(3)
a
The solution to equation (3) is achieved using the Orthogonal
Matching Pursuit (OMP) method. The sparse representation task
can be better performed by using learned-based dictionaries.
In order to denoise the ultrasound volume, I(x,y,z), aiming to
achieve the noise-free volume, R(x,y,z), the second step 110
(from Figure 1) is to apply the logarithm transform on
equation (1) to convert the multiplication operator into the
summation operator as follows,
log(/(x, y, z)) = log(eõ, (x, y, z))+ log(R(x, y, z))
(4)
Note that the E(x,y,z) is removed as a negligible term in the
equation (/) (see step 115 in Figure 1). Our desired output
is to find the noise-free data which is R(x,y,z). Considering
the whole volume to be denoised, equation (3) is reformulated
as,
\\
(5)
112
Eviik X log R(x,y,z)¨ Dakm02/1ijk j,k IL
arg min i,j,k
{R(x,y,z),a,,j,k ,D}
2
+2 log R(x,y,z)¨ log I(x, y, z)112
- 14 -
CA 02839600 2014-01-21
Attorney Docket No. 1001P039CA01
is an operation that extracts a 3D patch from the
volumetric data. A is the multiplier which controls the
proximity of the logarithm of the input noisy-image,
log/(x,y,z), with the logarithm of the noise-free data,
logR(x,y,z), in the entire volume. The analytical or fixed
dictionaries can represent signals based on specific
characteristics, whereas learned-based dictionaries improve
representation accuracy sparsity level. A method developed
by M. Aheron, M. Elad, and A. Bruckstein, called the sparse
K-SVD method, considers the learned dictionary to be itself a
sparse representation of a fixed dictionary (such as DCT).
This method makes a bridge between analytical or fixed
dictionaries and learned-based dictionaries to take
advantages of both dictionaries. This method has been shown
to be more effective for image denoising applications. In
this case, the sparse dictionary is defined as D=c130,4 where
413 is a DCT dictionary and A is the matrix of sparse
coefficients. In other words, each dictionary atom, d, has
a sparse vector A to represent it based on the fixed
dictionary, 4). The sparse-dictionary is applied in equation
(5) and the modified formula is obtained as follows,
7( (6)
X log Nx, y, z) ¨ (Dila 2
i,j4112 fii,j,kliaijA 110 J
arg min
{R(x,y,z),aj ,D}
+2 Illog R(x, y, z)¨ log /(x, y, z)11:
An iterative procedure is applied to minimize equation (5) in
which three individual steps are performed in each iteration.
- 15 -
CA 02839600 2014-01-21
Attorney Docket No. 1001P039CA01
Having the initialization of logR(x,y,z)=log/(x,y,z), the first
part of the iterative procedure calculates aw, by fixing D
and logR(x,y,z). This part of the procedure is separately
performed for each akm, related to a specific 3D patch,
using the OMP method (see step 120). In the next step of the
denoising method, step 130, the dictionary, D, is updated
using the sparse K-SVD approach by fixing akm and logR(x,y,z).
Afterwards, in step 140, the minimization problem in equation
(6) is optimized with respect to logR(x,y,z). The optimization
solution is obtained using the following formula,
(
log R(x, y, z) = AI id + ELT,,Lki,,,Lk
2/(x, y, z)+ ELj,k(Noti,j,k) (7)
ijk \ ijk
where lid is an identity matrix. The first matrix on which an
inverse operation is applied is a diagonal matrix.
Therefore, the equation (7) can be separately performed for
each 3D patch in the volume. After the optimization, the
updated logR(x,y,z) is checked (step 150) with the previous
value to decide whether to proceed with another iteration, or
to stop the denoising procedure. This process results in
step 160, the return of the estimation of the noise-free
volumetric data, f?(x,y,z).
Enhancing Tissue Voxels' Intensity
A block diagram of the procedure for the tissue voxels'
intensity enhancement is illustrated in Figure 2.
- 16 -
CA 02839600 2014-01-21
Attorney Docket No. 1001P039CA01
As discussed previously, ultrasound volumetric data suffers
from inconsistent intensity level in object boundaries. More
specifically, a particular object may have brighter
boundaries in some parts of its surface and darker boundaries
in other parts. This problem highly affects the proper
functioning of the segmentation process. A new approach is
presented herein to equalize tissue voxels' intensities using
the following equation,
V (x, y, z)= a B(x, y, z)+ (1- a)h(x, y, z) ( 8 )
where a is the mixing factor and B(x, y, z) is volumetric data,
{fir if (x,y,z) is tissue ( 9 )
B(x, y,z)=
,uõ if (x, y, z) is non -tissue
where ,uT and uNT are mean values of the tissue and non-
tissue voxels, respectively. To find uT and uNT, the
expectation maximization (EM) method is applied to find
parameters of the Gaussian-Gaussian Mixture Model (GGMM) to
fit it on the input volumetric data.
The Mardia and Hainsworth approach is used to determine
tissue and non-tissue voxels in the de-speckled ultrasound
volumetric data, f?(x,y,z). In the Mardia and Hainsworth
approach, voxels in a local neighbourhood are considered an
isotropic random Gaussian process, where the cross-covariance
of two voxels are dependent on their Euclidean distance as
follows,
y,z)]= gx, y,z),Cov[h(x, y, z), .1?(x, , y N , z N)]= 2(x, .Y,z)13(11(x, .Y,z)-
(XN, Y Al, Z 0112) (10)
- 17 -
CA 02839600 2014-01-21
Attorney Docket No. 1001P039CA01
where p(dist) is an isotropic correlation function in which
p(0)=1. The correlation function may be selected as,
p(d)= expl---(41 /ç }
(//)
where 77 and are two parameters that control the decaying
of the correlation function. This definition maintains a
smooth decay of the correlation function by increasing the
distance between a voxel (x,y,z) and its neighbour (XioyN,zõ).
The linear combination of neighbour voxels is calculated to
estimate the voxel (x,y,z) as .1?-(x,y,z)= The
neNeighbors(x,y,z)
assumption of In =1 provides PT=/IT , fiNT =Jim- = Accounting
for variances provides,
= py, = (5.2NT = per2NT
(12)
where is the vector of weights. UT and UNT
are standard deviations of tissue and non-tissue voxels
obtained using the GGMM-EM method. P is the correlation
function matrix defined as follows,
P(0) p(l) p(l)
(/3)
= p(1) p(0) p(1) = = =
P
p(2) p(1) p(0) = = =
_ =
- 18 -
CA 02839600 2014-01-21
Attorney Docket No. 1001P039CA01
Having prior probabilities of tissue voxels, 137., and non-
tissue voxels,
NT 1 the spatial thresholding is obtained by
modifying the Lloyd spatial thresholding equation as follows,
t = rNT2 NTT 1 \
0.2 {\.fr."" NT
fr '" NT" T 1 iuy r +2(4
a )10g((137-0-NT) I (PNTar)
)10.5
(14)
(NT )
The local mean thresholding method is then applied to label
voxels into tissue and non-tissue classes as follows,
R(x,y,z)t label: Tissue
(15)
R(x,y,z)< t label:non¨Tissue
After labelling all voxels, the estimation of parameters
is updated with Maximum Likelihood
(ML) method. If convergence is not attained, the process is
repeated with equation (14), unless the volume intensity
enhancement process is finished and the enhanced volume,
V(x,y,z), is obtained using the equation (8). V(x,y,z) is now
ready to be used by the segmentation procedure.
From Figure 2, it can be seen that the intensity enhancement
procedure has a number of steps. The first step 200 is that
of determining the de-speckled ultrasound data. This data is
the result of the procedure illustrated in Figure 1.
Afterwards, in step 210, flT and fiNT are found by applying
the the expectation maximization (EM) method to find
parameters of the Gaussian-Gaussian Mixture Model (GGMM).
The matrix P is then found using equation (13) above (step
220). The value for spatial thresholding, t is then
calculated using equation (14) (step 230). The relevant
- 19 -
CA 02839600 2014-01-21
Attorney Docket No. 1001P039CA01
voxels are then labelled (step 240) and, using Maximum
Likelihood, the parameters W=.1Li
k. NT,uNT,PNT,IIT,crT,PT} are updated
(step 250). A check is then made to determine if the values
are converging (step 255). If there is no convergence, the
procedure returns to step 230 for another iteration. If
there is convergence, the enhanced volume data is returned as
the procedure's output (step 260).
Initial Points and Surface Selection
A block diagram of the initial surface selection procedure is
shown in Figure 3.
The segmentation task using the 3D Snake model requires an
initial surface to start the process. There are two methods
to initiate the 3D Snake method: (1) manual selection of an
initial surface, and (2) automatic selection of initial
surface based on human visual attention system. The human
visual attention system models the manual selection of points
in the ultrasound volumes.
The initial surface selection procedure in Figure 3 starts
with the intensity enhanced data that was the output from the
procedure in Figure 2 (step 300). A determination is then
made as to whether the initial surface selection is to be
made manually or automatically (step 305). If the selection
is to be performed manually (step 310), then the left
sequence in Figure 3 is followed. For an automatic selection
(step 320), the right sequence in Figure 3 is followed.
In the manual selection of the initial surface, the user
displays the volume (step 312) by moving among the volume in
different planes and deciding where to insert the initial
- 20 -
,
CA 02839600 2014-01-21
Attorney Docket No. 1001P039CA01
point (step 314) by clicking on the mouse button. Then, the
user starts to draw a sphere by moving the cursor to
determine the radius of the sphere (step 316). The manually
drawn sphere is finalized by clicking on the mouse button.
The outputs of the manually surface selection task are the
center point, C'surf=[xõyõze]T , and the surface radius, r.f.
In the automatic selection of the initial surface (step 320),
the human visual attention system model finds some points
inside the ultrasound volume to alternatively select the
center point of the initial surface. The first step of the
human visual system is to decompose the input visual field
into topographical maps. This is performed for the enhanced
ultrasound volume. V(x,y,z) is decomposed to multi-scale
intensity (step 330) and orientation-based (step 335) maps,
also called topographic maps. The topographic intensity maps
are labeled with V(x, y,z, Cr) E Rit'fr(ci"'"Al") where M(a)= S,/r.
M y (0) = S v 2 - and Mz(a)=Sz /2 . Intensity-based topographic
maps are generated for UE{0,1,2,3,4,5,6,7,8}, resulting in 9
different scales. The orientation-based topographic maps are
generated for 4 directions of OE 10,71./4,/r/2,37r/41 . Thus, there
exist 36 multi-scale orientation-based topographic maps,
0(x,y,z,a,0).
Feature maps are then generated from topographic maps (steps
340, 345). This reflects the lateral inhibition property of
the nervous system which causes the neighbours to influence
each other. Thus, only those points in topographic maps
which significantly differ from their surroundings survive in
the feature maps. To generate this feature map, the fine
resolution topographic map is subtracted with its low-passed
¨ 21 ¨
CA 02839600 2014-01-21
Attorney Docket No. 1001P039CA01
version obtained by up-sampling of coarser scales maps. This
is formulated as
V(x, y, z, C",8)=V(x, y, z, ce)(-)V(x, y, z, 8)
(16)
where ojE{2,3,4} and 8E{3,4). The (--) operation subtracts the
left side operand in 01 scale with the up-sampled version of
the right operand from of+g scale to ce scale. The result is
6 feature maps related to the image intensity property. The
same operation is applied on orientation-based topographic
maps, but in different directions as follows,
0(x, y, z, 0(x, y, z, cr', 0)(-)0(x, y, z, +8,0)
(/7)
The orientation-based features are 24 maps, created in 4
directions and 6 scales' combinations. These steps provide 30
feature maps in total.
After the feature maps are created, conspicuity maps are
created by summing together feature maps in the scale cr=4.
In step 350, 355, the intensity based features are summed to
arrive at a single intensity-based conspicuity map, according
to the following summation formula,
(x, y, z)= N(V(x,y,z, 2, 5))(+)N(V(x, y, z, 2, 6))(+)...(+)N(V(x,y,z, 4,8))
(18)
where N is the normalization operation. The purpose of using
this normalization operation is to reduce the influence of
the noisy data in a particular feature map and to increase
the importance of stand-alone pixels in other feature maps.
The result of steps 350, 355 is five conspicuity maps
- 22 -
CA 02839600 2014-01-21
Attorney Docket No. 1001P039CA01
including one intensity-based map and four orientation-based
conspicuity maps in different directions as follows,
O (x,y,z,0)= N(0(x,y,z, 2,5, 0))(+)N(0(x,y,z, 2,6, 0))(+)...(+)N(0(x,y,z, 4,
8, 0)) . (19)
Step 360 is that of summing the normalized versions of the
conspicuity maps to produce the Saliency map, S(x,y,z).
Having this saliency map, the visual attention point refers
to the location with the maximum value. From the saliency
map, the center point, asurf=[xy,,z,]T , is selected (step 370)
and the surface radius, rsurf=Rdefault is set (step 380). The Rdef.d,
can be any arbitrary value and is preferably 5.
Volumetric Ultrasound Data Segmentation
In the present invention, the segmentation task consists of
two different strategies using: (1) deformable model based on
prior shapes to detect organs, such as kidneys and livers,
and (2) a combination of active contour model, Level-Set and
region growing methods to detect fluid areas. Provided below
are the mathematical details of these strategies. In a
preferred embodiment of the present invention, the ultrasound
data is used to detect the location of the kidney or the
liver. As such, the present disclosure will focus on
analysis for the detection of kidneys and livers. However,
it will be understood by a person skilled in the art that a
similar analysis could be used to detect the location of any
internal organ.
In the first strategy, in which a deformable model based on
prior shapes is used to detect organs, the location of the
ultrasound-probe is not known to the system. Thus, the
- 23 -
CA 02839600 2014-01-21
Attorney Docket No. 1001P039CA01
kidney and liver detection strategy is applied to locate the
ultrasound-probe. This provides a possible way to
discriminate between fluid-carrying organs and free fluids in
volumetric data. One assumption is that kidney and liver have
standard shapes and can be detected based on their shape in
the volumetric data. Based on this assumption, the prior
shapes of kidney and liver are deformed to find possible fits
in each volume in the volumetric data. The deformation is
performed in both global and local fashions. The global
deformation is applied to fit the location and orientation of
the shape in the volume while the local transformation is
applied to slightly change the shape to maximize the fit of
the shape in a possible location in the volume.
Referring to Figure 4, the steps in a procedure for detecting
the liver or kidney are illustrated. The procedure begins
with the enhanced ultrasound volumetric data which was the
output of the procedure from Figure 2 (step 400). A prior
shape, S, is first selected (step 410). For this procedure,
the prior shape, S,is defined as the zero Level-Set
function. Thus, the Level-Set function is defined as
1431(k-)17V=[x,y,z]T} in which 00-1(0)=S, (130(:kon,)<0 and (1)(X,)>O,
where it70,1 and 3',õ refer to voxels outside and inside the
prior shape, respectively. An initial level-set function 0,
is then created (step 420). To find the best fit of prior
shapes in the volume, the following error function is
minimized,
Min H(4T0(X"))r(X)
(20)
xEv
- 24 -
CA 02839600 2014-01-21
Attorney Docket No. 1001P039CA01
where hr(x) is the Heaviside function and is positive for x>0
and negative elsewhere. r(X)=-1-7.00-r,,T(X) is based on
intensities of voxels and the distribution model obtained in
the enhancing tissue voxels' intensity procedure as follows,
rT(X)=¨(V(X)¨ AT)2 I (201. ) + log(pT / (V-T)
(21)
rATT (X) = )2 /(2c2NT)+ log(PNT /(V-2-crNT))
Values for r(x,y,z) for all voxels in the volume can then be
calculated (step 430).
The Level-Set function is then written as the deformation of
the prior Level-Set oo,
=210)=0,(T4')
(22)
where X'..--Wjf and T is a global transformation in the
homogenous geometry and is defined with 7 parameters as
follows,
T=7;x7;xT3 (23)
1 0 0 cx+c-
0 1 0 c = +t
v v
o o 1 cz+tz
0 0 0 1
{sin(0x) sin(0) cos(9) {cos(0,) sin(8) cos(02)
A cos(0y) cos(t9z) 0
¨ cos(0) sin(Oz )1 + sin(Ox ) sin(0z )1
{A(sin(0x) sin(0y )sin(0,) {cos(0x) sin(0y) sin(0)
T2 = COO) sin(9) 0
COS(ex) COS(61z ))} ¨sin(19,c) cos(0z )1
¨ sin(e) cos(O ) sin(9) A cos( 9 x)cos(19 x) 0
0 0 0 1
- 25 -
CA 02839600 2014-01-21
Attorney Docket No. 1001P039CA01
1 0 0 ¨c,
0 1 0
T=
3 0 0 1 --Cz
0 0 0 1
where [c,cociT is the center of the prior shape, and
j5=100000,20.,,tocl are 7 parameters of the global
transformation. These parameters can thus be initialized
(step 440). An iterative update procedure (steps 450-470)
is applied to modify these parameters to fit the shape in the
volume. The initial set for these values is {0,0,0,1,0,0,0} .
For this iterative procedure, T and partial derivatives of T
are calculated (step 450). The following equation (using the
results of step 450) is applied to iteratively modify these
parameters (step 460),
+ K1))
pider+1 = piiter
Lr(T-1 x X')x <V 4:13 0, a(T(P x(T-' x X') >
(24)
x.v api
where Ki is a constant number that controls the updating
step size.
The iterative process is repeated until a convergence is
attained. A check (step 470) determines if convergence has
been attained or not. If there is no convergence in the
results, then the logic flow returns to step 450 for another
iteration. If there is convergence, the result is a final
Level-Set function, 00G, that can be used by the Level-Set
method (step 480) to improve the fitness accuracy in the
volume. A test is then performed to determine to see if
- 26 -
CA 02839600 2014-01-21
Attorney Docket No. 1001P039CA01
EH(G(X))r(X) 490 is less than a constant threshold value,
lE V
thi (decision 490). If the result of the test is positive,
then the object sought (a kidney or liver) has been detected
(step 500). If the result is negative, then the object
sought has not been detected (step 510).
In the second strategy used to detect internal fluids, the 3D
Snake approach provides a highly robust method against
boundaries' discontinuities, if its parameters are correctly
adjusted. However, the precise segmentation using the 3D
Snake method requires an extensive number of sample points,
resulting in a very high computational cost. On the other
hand, the Level-Set approach provides accurate segmentation
with less computational cost. However, this approach is not
resistant against discontinuities among object boundaries.
In another aspect of the present invention, a novel approach
is to make a bridge between the 3D Snake and Level-Set
methods. This combined approach provides the robustness of
the 3D Snake method as well as the accuracy of Level-Set
method. The steps in this combined approach are detailed in
Figure 6.
As can be seen in Figure 6, in the 3D Snake method, a surface
is represented with two parameters, senfl and renfl, as
S(s,r)=IX(s,r),Y(s,r),Z(r,$)}. The manual or automatic initial
surface that was determined as noted above is used to define
the initial surface S(s,r) (step 610). For this implementation
of the invention, it is assumed that the surface follows the
boundary condition, S(0,r)=S(1,r), and is initialized as a
sphere. The surface evolution to segment the region of
interest is formulated as the following energy function,
- 27 -
CA 02839600 2014-01-21
Attorney Docket No. 1001P039CA01
(25)
as as
Eint = as 2 2 2 ¨ + __ 2 fisr 2 fl +
(S(s,r))ciscir
aS2 r ar2 aSar
where a, and ar are two parameters that control the surface
tension along s-axis and r-axis, respectively. A and A
determine the surface rigidity along s-axis and r-axis,
respectively. Also, Aõ prevents the surface against
twisting. To simplify the problem, one can assume that
a = as = ar and fi=16, = fir = fisr . This assumption provides an easier
parameter adjustment. The Euler-Lagrange solution to the
equation is,
a2s a4s a4s a4s (25)
+ a 2 + /1+ _____________________ + # ____ +V Eõ01(S(s,r))= 0
as2 ar as4 ar4 as2ar2
To numerically solve the Euler-Lagrange equation, the surface
model is discretized to S(lis,r/r) where rt,E0,2,...A1 and
rire11,2,,P0. This conversion maps s=0, s=1, r=0 and r=1
into ns=1, ns=111-s , nr=1 and nr=Nr, respectively. After
simplifying the difference equation obtained by discretizing
equation (25), the relation of each single surface sample,
S(i,j), and its neighbour samples is achieved, as shown in
Figure 5. The surface samples are reshaped from the 2D matrix
form, SE R' , into a vector form, S'(k) =[X(1,m),Y(1,m),Z(1,m)]
where 1=[kINr] and m=k¨lxNr. Additionally, the balloon
force is added to let the surface evolve. The image force
bounds the evolving surface to stop in desired boundaries. At
each surface sample point, the balloon force is normal to the
- 28 -
CA 02839600 2014-01-21
Attorney Docket No. 1001P039CA01
surface and inflates the surface outward. The difference
equation (25) is converted into the following matrix
representation form,
Ax X +av(s)
(26)
-Frfbxalloon(s)=0
ax
av(s)
AxY + rfbYalloon(s)=0
ax
Ax Z +av(s)
+rfbzaii.(s)=0
ax
where the matrix AER("c))4Acxk) represents the internal force,
and flallobn(S), f1
100(S) and ,Dfzalloon(S) are projections of f
,.//om n
the x-axis, y-axis and z-axis, respectively. For the next
step in the procedure in Figure 6, this matrix A is created
(step 615). After this, the balloon force fba//00,2(x,y,z) is
calculated for all the voxels (step 620) using as input the
enhanced ultrasound volumetric data lax,y,z). Equation (27)
must, therefore, now be solved. The iterative solution to
the equation (26) is
X, = (A+ 1)--1(X av(s) (27)
ax
Y, = aV(S) rflalloon(S))
ay
z1= (A+ III (Z1
, av(s) rfbzalloon(S))
- az
where t is the iteration number and IER(N,x1V0x(NvxN,)
is the unit
matrix. As Si are non-integer values, each value of V(S)
should be interpolated from the level of the surrounding 3D
image voxels. The evolution of the surface is an iterative
process (box 630) performed by repeating the equation (27).
- 29 -
CA 02839600 2014-01-21
Attorney Docket No. 1001P039CA01
The evolution stops when the 3D Snake surface finds the
object boundaries in the volume. In this iterative process,
step 623 is that of finding the balloon force for the various
surface samples by linear interpolation. The surface samples
are then updated using equation (28) (step 625). A check is
then made (step 635) to determine if the results are
converging. If there is no convergence in the results, the
logic flow returns to step 623 for another iteration. If
there is convergence, then the procedure moves to the next
step.
After the 3D Snake approximately finds the object boundaries,
the 3D surface is converted into an initial Level-Set
function. To achieve the initial Level-Set function, the
surface samples are placed in a binarized volume (step 640).
Every four neighbour points are interpolated to create a
continuous surface in the binarized volume with values equal
to 1. The surface is turned into a shape in the 3D volume.
Using the image-fill morphological operation, the shape can
be filled by values equal to 1. Then, the initial 3D Level-
Set function, 00, is created (step 645) by making voxels on
the shape surface equal to zero, making voxels inside the
shape equal to 1, and making voxels outside the shape equal
to -1. The zero-Level-Set function is pointing to the
resultant 3D Snake surface, 00(S)=0.
The Level-Set function, cri([x,y,z],t), assigns a level to each
point, [x,y,z], in the 3D grid and (1)([x,y,z],0)=43130. The volume
containing points with non-negative Level-Set values are
called the Front. The Level-Set function iteratively updates
along its normal vector fields by F([x,y,z1), which controls
the speed of Level-Set change at each time-point. The Level-
- 30 -
CA 02839600 2014-01-21
Attorney Docket No. 1001P039CA01
Set function stops changing at boundaries. The speed
function, F({x, y, z]) , has two components,
F([x, y,z]) = FA([x, y, z])+ FG([x,y,z]) , where FA([x,y,z]) is similar to
the balloon force and FG([x,y,z1) maintains the shape's
smoothness. FA([x,y,z]) is divided into FAv([x,y,z1) and FAs([x,y,z])
F AV ([x, z]) is the normal speed vector to the Level-Set
function at each point, while FAs([x,y,z]) consists of vectors
pointing toward image edges. These values are then calculated
for each of the voxels (step 650). Because the smoothness is
provided by the 3D Snake, FG([x, y, z]) is eliminated and the
evolution formula is driven as follows,
all([x,y,z],t) (28)
F AV([x, y, z]) =V(1)([x, y, z],t)+ F As([X, y, z]) =V(1)([x, y, z], t) = 0
at
The normal term of the first speed term is only effective,
and therefore the above formula is again simplified as,
ac13([x,y,z],t) (29)
F AV([x, y,zDIV4:13([x, y,z],t)1+ F As([X, y,z]) =V4)([x, y, z], t) =0
at
The speed function is now defined based on voxels'
intensities, V(x,y,z) , to reduce speed at locations with
higher intensities. Therefore,
F AV([X, Ylz]) eXPe- K 21IV * V y , z))II) (30)
where Go. is a Gaussian smoothing filter with standard
deviation, 6, and IC2 is a constant that controls the rate of
speed reduction. This definition provides a smooth reduction
- 31 -
CA 02839600 2014-01-21
Attorney Docket No. 1001P039CA01
in the speed of Level-Set change, as the front approaches
object boundaries. The term FAs(x,y,z) also is driven as
follows,
FAS([X,y1Z])= 11V(G0.*V(x, y,z)II (31)
Homogeneous regions have high values of FAvqx,y,z]) leading to
even growth of the Front while FAs(x,y,z) is small in these
regions. When the Front approaches the boundary region,
FAv([x,y,z]) decreases while FAs(x,y,z) increases, thereby
pushing the Front toward the correct boundary. Equation (32)
can thus be used to update 41)([x,y,z],t) iteratively (step 655) .
As long as the exit condition is not satisfied (step 660),
(130([x,y,z],t) is continuously updated. This update procedure is
iteratively repeated for a few iterations, ni,õ . Once the
iterative loop exits, the segmented 3D fluid region is
displayed (step 670).
Final Decision Making on Medical Condition
Figure 7 is a flowchart of the various steps in a process
according to another aspect of the invention. The overall
process of the invention is illustrated in Figure 7 with the
final step being that of deciding whether the segmented fluid
region is a fluid-carrying body organ or free blood fluid. If
the detected fluid region is free blood fluid, the medical
condition of internal bleeding is sent to the user so that
immediate action may be taken. Inputs for this final decision
step are the location of detected organs and the location and
size of the segmented free fluid region. The anatomical
knowledge, detected location of the kidney and liver, and the
- 32 -
CA 02839600 2014-01-21
Attorney Docket No. 1001P039CA01
size and location of the segmented fluid region are applied
to make the final decision.
For clarity, the process in Figure 7 starts with noisy
volumetric ultrasound data (step 700). This data is then de-
speckled or denoised using K-Singular Value Decomposition
(step 710). The resulting data set then has its contrast
enhanced (step 720). The enhanced contrast dataset is
processed to find the image or a kidney, a liver, or other
organs within the dataset (step 730). The same enhanced
contrast dataset is also sent so that a user can determine
whether to manually or automatically initial surface
selection (step 735). If the user opts for manual entry, the
user manually draws the initial surface (step 740).
Alternatively, the initial surface can be automatically
detected using the human visual attention system model (Step
750). Once the initial surface has been detected, whether
manually or automatically, the size and location of the fluid
carrying objects are determined (step 760). Once the size
and location of the fluid carrying objects have been
determined or once the kidney, liver, and other organs have
been detected, step 770 is the final decision on the medical
condition. Step 770 determines whether the area in the
ultrasound image is a fluid filled cavity or whether the area
is an internal organ.
As can be imagined, the methods and processes disclosed in
this document may be used on any ultrasound imaging data
derived from an ultrasound image of any mammal's body part.
In one implementation, the various aspects of the invention
may be used in both portable and non-portable medical
equipment designed for use under less than ideal conditions
- 33 -
CA 02839600 2014-01-21
Attorney Docket No. 1001P039CA01
such as remote locations, battlefield locations, mobile
surgical hospitals, and disaster-stricken locations. In
another aspect, the methods and processes described above may
be implemented and used in non-portable equipment or
equipment designed for a fixed installation. Such fixed
equipment may be useful for detecting fluid accumulation in
patients as well as for distinguishing internal organs from
fluid accumulation in both trauma and non-trauma patients.
It should also be noted that while the above discussion
mostly relates to 3D ultrasound imaging, the methods and
processes discussed above may also be used on equipment which
uses X-ray computed tomography (X-ray CT) or magnetic
resonance imaging (MRI) technology.
Implementation and Development of the Invented System
The system can be implemented by computer programming
software, such as MATLABTm, in conjunction with suitable
computer hardware. All described steps and methods can be
developed as individual modules and a designed GUI (graphical
user interface) can combine these modules to form a unique
toolbox to handle the detection procedure from the beginning
to the final step. The GUI provides 3D display capability
with the flexibility to move between x-plane, y-plane and z-
plane and it also displays the 3D segmented objects.
A view panel of a developed GUI according to another aspect
of the invention is shown in Figure 8. Figure 8 also shows an
automatically detected fluid region in the volumetric data.
For further reference and clarity regarding the varying
aspects of the invention, the following references may be
consulted:
- 34 -
CA 02839600 2014-01-21
Attorney Docket No. 1001P039CA01
B. J. C. and D. C., "Adaptive filtering for reduction of
speckle in ultrasound pulse-echo imagesõ" Ultrasonics,
vol. 1, pp. 41-44, 1986.
G. A, G. J., M. S. and C. K. Deepika, "De-speckling of
Medical Ultrasound Images using Wiener Filter and Wavelet
Transform," International Journal of Electronics &
Communication Technology, vol. 2, no. 3, pp. 21-24, 2011.
L. T., M. W. N. and A. P. L., "An adaptive weighted
median filter for speckle suppression in medical
ultrasonic images," IEEE Transaction on Circuits and
Systems, vol. 36, pp. 129-135, 1989.
E. P. Simoncelli and A. E. H., "Noise removal via
Bayesian wavelet coring," in Third International
Conference on Image Processing, 1996.
P. Taya, S. T. Acton and J. A. Hossack, "A wavelet
thresholding method to reduce ultrasound artifacts,"
Computerized Medical Imaging and Graphics, vol. 35, pp.
42-50, 2011.
Y. Yu and S. Acton, "Speckle reducing anisotropic
diffusion," IEEE Transactions on Image Processingõ vol.
11, no. 11, pp. 1260-1270, 2002.
S. Kalaivani and R. Wahidabanu, "Condensed anisotropic
diffusion for speckle reducton and enhancement in
ultrasonography," EURASIP Journal on Image and Video
Processing, vol. 2012, no. 12, pp. 1-17, 2012.
B. Burgeth, S. Didas, L. Florack and J. Weickert, "A
generic approach to diffusion filtering of matrix-
- 35 -
CA 02839600 2014-01-21
Attorney Docket No. 1001P039CA01
fields," Computing, vol. 81, no. 2, pp. 179-197, 2007.
P. C. Tay, C. D. Garson, S. T. Acton and J. A. Hossack,
"Ultrasound Despeckling for Contrast Enhancement," IEEE
Transactions on Image Processing, vol. 19, no. 7, pp.
1847-1860, 2010.
R. Rubinstein, M. Zibulevsky and M. Elad, "Double
sparsity: learning sparse dictionaries for sparse signal
approximation," IEEE Transactions on Signal Processing,
vol. 58, no. 3, p. 1553, 2010.
A. Belaid, D. Boukerroui, Y. Maingourd and J. F.
Lerallut, "Phase-Based Level Set Segmentation of
Ultrasound Images," Information Technology in
Biomedicine, IEEE Transactions on, vol. 5, no. 1, pp.
138-147, 2011.
E. Ukwatta, J. Awad, A. Ward, D. Buchanan, G. Parraga and
A. Fenster, "Coupled level set approach to segment
carotid arteries from 3D ultrasound images," in
Biomedical Imaging: From Nano to Macro, 2011 IEEE
International Symposium on, 2011.
M. Wahba, "An automated modified region growing technique
for prostate segmentation in trans-rectal ultrasound
images," ProQuest Dissertations and Theses, 2009.
R. J. Schneider, D. P. Perrin, N. V. Vasilyev, G. R.
Marx, P. J. d. Nido and R. D. Howe, "Mitral Annulus
Segmentation From 3D Ultrasound Using Graph Cuts," IEEE
Transactions on Medical Imaging, vol. 29, no. 9, pp.
1676-1687, 2010.
- 36 -
CA 02839600 2014-01-21
Attorney Docket No. 1001P039CA01
R. Juang, E. McVeigh, B. Hoffmann, D. Yuh and P. Burlina,
"Automatic segmentation of the left-ventricular cavity
and atrium in 3D ultrasound using graph cuts and the
radial symmetry transform," in IEEE International
Symposium on Biomedical Imaging: From Nano to Macro,
2011, 2011.
X. Yang, W. He, J. Jin, X. Zhang, M. Yuchi and M. Ding,
"A hybrid method to segment common carotid arteries from
3D ultrasound images," in IEEE-EMBS International
Conference on Biomedical and Health Informatics (axI),
2012, 2012.
R. Shekhar, R. M. Cothren, D. G. Vince, S. Chandra, J. D.
Thomas and J. F. Cornhill, "Three-dimensional
segmentation of luminal and adventitial borders in serial
intravascular ultrasound images," Computerized Medical
Imaging and Graphics, vol. 23, no. 6, pp. 299-309, 1999.
R. Whitaker, "A Level-Set Approach to 3D Reconstruction
from Range Data," International Journal of Computer
Vision, vol. 29, no. 3, pp. 203-231, 1998.
K. Mardia and T. J. Hainsworth, "A spatial thresholding
method for image segmentationõ" IEEE Transactions on
Pattern Analysis and Machine Intelligence, vol. 10, no.
6, pp. 919-927, 1988.
L. Itti, C. Koch and E. Niebur, "A Model of Saliency-
Based Visual Attention for Rapid Scene Analysis," IEEE
Transactions on Pattern Analysis and Machine
Intelligence, vol. 20, no. 11, pp. 1254-1259, 1998.
- 37 -
CA 02839600 2014-01-21
Attorney Docket No. 1001P039CA01
A. Yezzi and S. Soatto, "Deformotion: Deforming motion,
shape average and the joint registration and
approximation of structures in images," International
Journal of Computer Vision, vol. 53, pp. 153-167, 2003.
M. R., J. A. Sethian and B. C. Vemuri, "Shape modeling
with front propagation: a level set approach," IEEE
Transactions on Pattern Analysis and Machine
Intelligence, vol. 17, no. 2, pp. 158-175, 1995.
C. Revol and M. Jourlin, "A new minimum variance region
growing algorithm for image segmentation," Pattern
Recognition Letters, vol. 18, pp. 249-258, 1997.
P. Soille, Morphological Image Analysis: Principles and
Applications, 1999, pp. 173-174.
A. K. Jain, Fundamental of Digital Image Processing., NJ:
Prentice-Hall, 1989.
A. B. Watson, "Image compression using the Discrete
Cosine Transform," Mathematica Jounral, vol. 4, no. 1,
pp. 81-88, 1994.
M. Elad and M. Aheron, "Image Denoising Via Learned
Dictionaries and Sparse representation," in Proceedings
of the 2006 IEEE Computer Society Conference on Computer
Vision and Pattern Recognition (CVPR'06), 2006.
Y. C. Pati, R. R. and K. P. S., "Orthogonal Matching
Pursuit: recursive function approximation with
applications to wavelet decomposition," Conference Record
of The Twenty-Seventh Asilomar Conference on Signals,
- 38 -
CA 02839600 2014-01-21
Attorney Docket No. 1001P039CA01
Systems and Computersõ vol. 1, pp. 40-44, 1993.
D. E. Lloyd, Automatic target classification using moment
variants of iamge shapes, Franborough, UK, Rep. RAE IDN
AW126, 1985.
B. Mory, 0. Somphone, R. Prevost and R. Ardon, Template
Deformation with User Constraints for Live 3D Interactive
Surface Extraction, Toronto, Ontario, 2011.
D. Terzopoulos, A. Witkin and M. Kass, "Constrains on
deformable models: recovering 3d shape and nonrigid
motion," Artificial Intelligence, vol. 36, no. 1, pp. 91-
123, 1988.
J. Ahlberg, "Active contours in three dimensions,"
Master's thesis, Linkping University, Computer Vision,
The Institute of Technology , 1996.
M. Aheron, M. Elad and A. Bruckstein, "K-SVD: An
Algorithm for Designing Overcomplete Dictionaries for
Sparse Representationõ" IEEE Transactions on Signal
Processing, vol. 54, no. 12, pp. 4311-4322, 2006.
C. Koch and S. Ulman, "Shifts in selective visual
attention: towards the underlying neural circuitry,"
Human Neurobiology, vol. 4, pp. 219-227, 1985.
- 39 -
CA 02839600 2014-01-21
Attorney Docket No. 1001P039CA01
L. Itti, C. Koch and E. Niebur, "A Model of Saliency-
Based Visual Attention for Rapid Scene Analysis," IEEE
Transactions on Pattern Analysis and Machine
Intelligence, vol. 20, no. 11, pp. 1254-1259, 1998.
The method steps of the invention may be embodied in sets of
executable machine code stored in a variety of formats such
as object code or source code. Such code is described
generically herein as programming code, or a computer program
for simplification. Clearly, the executable machine code may
be integrated with the code of other programs, implemented as
subroutines, by external program calls or by other techniques
as known in the art.
The embodiments of the invention may be executed by a
computer processor or similar device programmed in the manner
of method steps, or may be executed by an electronic system
which is provided with means for executing these steps.
Similarly, an electronic memory means such computer
diskettes, CD-ROMs, Random Access Memory (RAM), Read Only
Memory (ROM) or similar computer software storage media known
in the art, may be programmed to execute such method steps.
As well, electronic signals representing these method steps
may also be transmitted via a communication network.
Embodiments of the invention may be implemented in any
conventional computer programming language. For example,
preferred embodiments may be implemented in a procedural
programming language (e.g."C") or an object oriented language
(e.g."C++"). Alternative embodiments of the invention may be
implemented as pre-programmed hardware elements, other
related components, or as a combination of hardware and
software components. Embodiments can be implemented as a
- 40 -
CA 02839600 2014-01-21
Attorney Docket No. 1001P039CA01
computer program product for use with a computer system. Such
implementations may include a series of computer instructions
fixed either on a tangible medium, such as a computer
readable medium (e.g., a diskette, CD-ROM, ROM, or fixed
disk) or transmittable to a computer system, via a modem or
other interface device, such as a communications adapter
connected to a network over a medium. The medium may be
either a tangible medium (e.g., optical or electrical
communications lines) or a medium implemented with wireless
techniques (e.g., microwave, infrared or other transmission
techniques). The series of computer instructions embodies all
or part of the functionality previously described herein.
Those skilled in the art should appreciate that such computer
instructions can be written in a number of programming
languages for use with many computer architectures or
operating systems. Furthermore, such instructions may be
stored in any memory device, such as semiconductor, magnetic,
optical or other memory devices, and may be transmitted using
any communications technology, such as optical, infrared,
microwave, or other transmission technologies. It is expected
that such a computer program product may be distributed as a
removable medium with accompanying printed or electronic
documentation (e.g., shrink wrapped software), preloaded with
a computer system (e.g., on system ROM or fixed disk), or
distributed from a server over the network (e.g., the
Internet or World Wide Web). Of course, some embodiments of
the invention may be implemented as a combination of both
software (e.g., a computer program product) and hardware.
Still other embodiments of the invention may be implemented
as entirely hardware, or entirely software (e.g., a computer
program product).
- 41 -
CA 02839600 2014-01-21
Attorney Docket No. 1001P039CA01
A person understanding this invention may now conceive of
alternative structures and embodiments or variations of the
above all of which are intended to fall within the scope of
the invention as defined in the claims that follow.
- 42 -