Note: Descriptions are shown in the official language in which they were submitted.
CA 02864258 2014-09-15
1
FIELD OF THE INVENTION
The invention relates to the use of Grb2 genes (for its
acronym in English, Growth factor receptor-bound protein 2) gene
or receptor-bound protein 2 growth, Gata3, immunoglobulin M
(IgM), MAVS (for its acronym in English, Mitochondrial signaling
antiviral protein) or mitocrondrial antiviral signaling protein
and Regul to determine susceptible phenotypes and resistance to
infectious salmon anemia (ISA) in Atlantic salmon (Salmo salar).
PRIOR ART
The Atlantic salmon (Salmo salar) is a widely cultivated and
highly commercial aquaculture species worldwide. However, this
species has susceptibility to various pathogens, resulting in
losses for the salmon industry. One of the most important
pathogens is the ISA virus, which can reach cumulative
mortalities between 15% to 100%. Although, it has been observed
in the field that there are families of fish that have a reduced
mortality rate against the virus (resistant families), which
could prove to be key to the development of effective treatments.
Moreover, at present, there is a clear consensus that
conventional fisheries are in crisis, since as indicated
according to scientific evidence, the quantities of catch have
declined continuously over the past 5 years (The State of world
CA 02864258 2014-09-15
2
fisheries and aquaculture, FAO (Food and Agricultural
Organization), 2012;
http://www.fao.org/docrep/016/i2727e/i2727e.pdf). Which is vital
because in the last five decades it has exceeded the population
growth of 1.7% annually, which requires the production of larger
quantities of food. Notably, in 2009, fish accounted for 16.6% of
the animal protein intake of the world's population and some
regions are almost entirely dependent on fishing.
With this scenario in between, aquaculture, which is to
cultivate various species of freshwater or saltwater under
controlled conditions, is emerging as an alternative to the
growing demand which conventional fisheries cannot meet. This
industry has presented a growth of about 7% a year (The State of
world fisheries and aquaculture, FAO,
2012;
http://www.fao.org/docrep/016/i2727e/i2727e.pdf
Among the branches of aquaculture, salmon farming is of great
economic importance to Chile, reporting an annual growth rate of
20.5% between 1990 and 2008 (Report to the National Innovation
Council for
Competitiveness,
http://biblioteca.cnic.cl/media/users/3/181868/files/18813/G Para
da ACUI final.pdf), positioning salmon in 2008 as the fourth
national export product and aquaculture as the third export
CD, 02864258 2014-09-15
3
sector after mining and forestry. Salmon production has grown
exponentially since 1990, there were 24 thousand tons worth $ 116
million through 2008, production reached 445 thousand tons
involving an income of $ 2,392 million for our country. The
consolidation of the national salmon industry over the past two
decades has led to Chile to position itself as a major salmon
producers worldwide, with 31% of production, along with Norway
(41%), United Kingdom (10 %) and Canada (7%). Salmonids with
higher production in our country are Atlantic salmon (Salmo
salar) with 46% of all salmon produced coho salmon (Oncorhynchus
kisutch) with 26% and rainbow trout (Oncorhynchus mykiss) with
25% (Sernapesca 2010 Report, SalmonChile, ProChile).
The intensive nature of the crop has favored the emergence
of a number of diseases that have caused mortalities, problems in
production and economic impact on the industry. These pathogens
in cropping systems are normally present in wild *fish
populations; However, in natural environments they rarely cause
mortalities due to the absence of stressful conditions such as
handling, transport, high population density and changes in water
quality, present in culture facilities (Toranzo A., B. and
Magarinos Romalde Jl 2005 A Review Of The Main Bacterial Diseases
In Fish Mariculture Aquaculture System. 246: 37-61). Within the
CA 02864258 2014-09-15
4
pathogens that cause major impact on this industry are bacterial:
Salmon Rickettsial Septicemia (SRS) and Bacterial Kidney Disease
(BKD), and viral: Infectious Pancreatic Necrosis (IPN) and
Infectious Salmon Anemia (ISA). In recent years and particularly
in 2007, domestic salmon industry has been severely affected by
the ISA virus, a highly contagious disease of horizontal
transmission producing a decrease in the levels of production and
exports (17% in 2008-2009), the unlinking of more than 20
thousand of the 50 thousand workers in the sector and a
corresponding to the loss in revenue of $ 883 million from
exports (Carreno A. (2010) Impacto del virus ISA en Chile.
Publicaciones Fundacion Terram 2010) Impact del virus ISA en
Chile. Publicaciones Fundacian Terram 2010).
Infectious salmon anemia (ISA) is a multisystem disease with
high mortality caused by the only virus of the genus Isavirus,
the Orthomyxoviridae family, which also owns the influenza virus
(Dannevig B, Falk K,y Namork E. Isolation of the causal virus of
infectious salmon anaemia (ISA) in a long-term cell line from
Atlantic salmon head kidney. J Gen Virol. 1995. 76:1353-1359;
Kawaoka, Y., Cox, N.J., Kaverin, N., Klenk, H.-D., Lamb, R.A.,
Mccauley, J., Nakamura,K., Palese, P., Webster, R.G., 2005.
Family Orthomyxoviridae. Disseminated around 2005, based on
CA 02864258 2014-09-15
surface glycoprotein gene sequences. Virol. J.6, 88. 3245-3259).
The first recorded outbreak of this virus dates 1984 on the coast
of southwestern Norway (Thorud K, Djupvik HO (1988) Infectious
anemia in Atlantic salmon (Salmo salar L.) Bull Eur Assoc Fish
Pathol. 8: 109-11). Subsequently, the disease has been reported
in Canada in 1996 (Mullins JE, Groman D, Wadowska D (1998)
Infectious salmon anaemia in salt water Atlantic salmon (Salmo
salar L.) in New Brunswick, Canada. Bull Eur Assoc Fish Pathol
18(4): 110-114), in Scotland in 1998 (Rodger HD, Turnbull T, Muir
F, Millar S, Richards RH (1998) Infectious salmon anaemia (ISA)
in the United Kingdom. Bull Eur Assoc Fish Pathol 18(4):115-116),
Faroe Islands in 1999 (LyngOy C (2003) Infectious salmon anaemia
in Norway and the Faeroe Islands: An industrial approach. In:
Miller 0, Cipriniano RC (eds), International response to
infectious salmon anaemia: Prevention, control and eradication,
pp 97-10), in Chile 1999 (Kibenge F.S.B., Garate 0.N., Johnson G.
Arriagada R., Kibenge M.J.T. & Wadowska D. (2001). Isolation and
identification of infectious salmon anaemia virus (ISAV) from
Coho salmon in Chile. Dis. Aquat. Org ., 45, 9-18) and in Maine,
USA, 2000 (Bouchard D.A., Brockway K., Giray C., Keleher W. &
Merrill P.L. (2001). First report of infectious salmon anemia
CD, 02864258 2014-09-15
6
(ISA) in the United States. Bull. Eur. Assoc. Fish Pathol.,
21,86-88).
This virus has a genome consisting of eight single-stranded
RNA segments of negative polarity and hemagglutinating activity,
receptor-destroying and capable of adsorptive (Dannevig B.H.,
Falk K. & Namork E. (1995). Isolation of the causal virus of
infectious salmonanemia (ISA) in a long-term cell line from
Atlantic salmon head kidney. J. Gen. Virol., 76, 1353-1359;
Eliassen T.M., Froystad M.K., Dannevig B.H., Jankowska M., Brech
A., Falk K.,Romoren K. & Gjoen T. (2000). Initial events in
infectious salmon anemia virus infection: evidence for the
requirement of a low-pH step. J. Virol., 74, 218-227; Falk K.,
Aspehaugv., VlasakR. & Endresen C. (2004). Identification and
characterization ofviral structural proteins of infectious salmon
anemia virus. J. Virol., 78, 3063-3071; Falk K, Dale OB. (2007):
Lack of receptor-destroying enzymatic activity of ISA-virus is an
important virulence factor for development of ISA in Atlantic
salmon. In Frisk Fisk Conference, Tromso: Jan 23-25th 2007;
Norway; Mjaaland, S., E. Rimstad, K. Falk, and B. H. Dannevig.
(1997). Genomiccharacterization of the virus causing infectious
salmon anemia in Atlanticsalmon (Salmo sa1ar L.): an orthomyxo-
like virus in a teleost. J. Viro1.71:7681-7686). Se ha descrito
CA 02864258 2014-09-15
7
la secuencia nucleotidica de los 8 segmentos (Clouthier, S.C.,
Rector, T., Brown, N.E., Anderson, E.D., 2002. Genomic
organization of infectious salmon anaemia virus. J. Gen. Virol.
83 (Pt. 2), 421-428) and we have identified four major structural
proteins, including a nucleoprotein (NP) of 68 kDa, a protein
matrix (MP) of 22 kDa, a hemagglutinin-esterase (HE) with
receptor-destroying activity of 42 kDa and glycoprotein of 50 kDa
with potential surface adsorption capacity (F), which are encoded
in the genome segments; 3,8,6, and 5, respectively (Falk K.,
Aspehaugv., VlasakR. & Endresen C. (2004). Identification and
characterization ofviral structural proteins of infectious salmon
anemia virus. J. Virol., 78, 3063-3071). A total of 10
polypeptides exist, presenting the remaining 6, orthology to
influenza genes (Cottet, L., Cortez-San Martin, M., Tello, M.,
Olivares, E., Rivas-Aravena, A., Vallejos, E., Sandino, A.M.,
Spencer, E., (2010). Bioinformatic analysis of the genome of
infectious salmon anemia viruses associated with outbreaks of
high mortality in Chile. J. Virol 84(22): 11916-11928). A
schematic representation of the ISA virus can be seen in the
schematic representation of the ISA virus, given Bioinformatic
analysis of the genome of infectious salmon anemia viruses
CA 02864258 2014-09-15
8
associated with outbreaks of high mortality in Chile. J. Virol
84(22): 11916-1192).
Sequence analysis of the genomic segments have revealed
differences between within isolated and defined geographical
areas (Blake, S., Bouchard, D., Keleher, W., Opitz, M.,
Nicholson, B.L., (1999). Genomic relationships of the North
American isolate of infectious salmon anemia virus (ISAV) to the
Norwegian strain of ISAV. Dis. Aquat. Organ. 35 (2), 139-144;
Dannevig BH, Falk K y Namork E. Isolation of the causal virus of
infectious salmon anaemia (ISA) in a long-term cell line from
Atlantic salmon head kidney. J Gen Virol. 1995. 76: 1353-1359;
Kibenge F.S.B., Garate 0.N., Johnson G. Arriagada R., Kibenge
M.J.T. & Wadowska D. (2001). Isolation and identification of
infectious salmon anaemia virus (ISAV) from Coho salmon in Chile.
Dis. Aquat. Org ., 45, 9-18). Accordingly, there are 2 main
genotypes of ISA virus: European (I) genotype and America (II)
genotype, which differ between 15 and 19% in their amino acid
sequence of the fragments of 5 (F) and 6 (HE). Another way of
classifying isolates lies with the ISA virus hemagglutinin-
esterase gene, specifically in a region close to the
transmembrane domain of a maximum of 35 amino acids called HPR
(Highly Polymorphic Region) which exhibits high variability in
CD, 02864258 2014-09-15
9
pathogenic isolates given by insertions or deletions (Nylund, A.,
Devoid, M., Plarre, H., Isdal, E., Aarseth, M., (2003). Emergence
and maintenance of infectious salmon anaemia virus (ISAV) in
Europe: a new hypothesis. Dis. Aquat. Organ. 56 (1), 11-24;
Mjaaland, S., Hungnes, 0., Teig, A., Dannevig, B.H., Thorud, K.,
Rimstad, E., 2002. Polymorphism in the infectious salmon anemia
virus hemagglutinin gene: importance and possible implications
for evolution and ecology of infectious salmon anemia disease.
Virology 20 (304), 379-391). To date we have identified 35 HPRs,
HPR1 and HPR2 being the most predominant, while the others are
variants of these, where amino acids are added in the carboxyl
region of hemagglutinin for HPR1 or in the amino region of HPR2.
In Chile, several types of HPRs have been found, HPR7b being the
most prevalent in years 2007-2008 and responsible for high
mortalities recorded for those years in our country (Kibenge,
F.S., Godoy, M.G., Wang, Y., Kibenge, N.J., Gherardelli, V.,
Mansilla, S.,Lisperger, A., Jarpa, M., Larroquete, G., Avendano,
F., Lara, M y Gallardo, A.. Infectious salmon anaemia virus
(ISAV) isolated from the ISA disease outbreaksin Chile diverged
from ISAV isolates from Norway around 1996 and was disseminated
around 2005, based on surface glycoprOtein gene sequences. Virol
J. 2009. 6:8-16.). A feature of this hypervariable region is the
Mk 02864258 2014-09-15
existence of a variant that contains the reasons described for
the other HPRs, of which the longest is called HPRO and is
characterized by low pathogenic and not be cultivated in standard
cell lines such as SHK-1 (Salmon Head Kidney), (McBeath, A.J.,
Bain, N., Snow, M., 2009. Surveillance for infectious salmon
anaemia virusHPRO in marine Atlantic salmon farms across
Scotland. Dis. Aquat. Organ. 87, 161 -169; Markussen, T.,
Jonassen, C.M., Numanovic, S., Braaen, S., Hjortaas, M., Nilsen,
H.,. Mjaaland, S., 2008. Evolutionary mechanisms involved in the
virulence of infectious salmon anaemia virus (ISAV), a piscine
orthomyxovirus. Virology 374, 515-527).
Endothelial cells are one of the main objectives of the
virus (Hovland T, Nylund A, Watanabe K, Endresen C. (1995).
Observation of Infectious salmon anemia virus in Atlantic salmon,
Sa1mo-salar L. J Fish Dis 17:291-6) allowing the virus to
replicate in different organs. In addition, we have detected the
presence of the virus in polymorphonuclear cells and lymphocytes
(Sommer A.I. & Mennen S. (1996). Propagation of infectious salmon
anaemia virus in Atlantic salmon, Salmo salar L., head kidney
macrophages. J. Fish Dis., 19, 179-183; Kawaoka, Y., Cox, N.J.,
Kaverin, N., Klenk, H.-D., Lamb, R.A., Mccauley, J., Nakamura,K.,
Palese, P., Webster, R.G., 2005. Family Orthomyxoviridae.
Mk 02864258 2014-09-15
11
Disseminated around 2005, based on surface glycoprotein gene
sequences. Virol. J.6, 88. 3245-3259). In erythrocytes the ISA
virus are lined in the reaction of hemagglutination by
interaction of sialic acid cell receptors and viral protein HE,
process similar to the hemagglutination of the influenza virus in
which 3 separate phenomena occurs: a) adsorption of the virus in
the erythrocyte membrane, b) elution of the virus, which is not
always complete c) virus absorption via endosome by erythrocytes
(Kawaoka, Y., Cox, N.J., Kaverin, N., Klenk, H.-D., Lamb, R.A.,
Mccauley, J., Nakamura,K., Palese, P., Webster, R.G., 2005.
Family Orthomyxoviridae. Disseminated around 2005, based on
surface glycoprotein gene sequences. Virol. J.6,88. 3245-3259;
Workenhe, S.T., Wadowska, D.W., Wright, G.M., Kibenge, N.J.,
Kibenge, F.S., (2007). Demonstration of infectious salmon anaemia
virus (ISAV) endocytosis in erythrocytes of Atlantic salmon.
Virol. J. 4, 13). For the ISA virus the elution from erythrocytes
is reported in various fish species except Atlantic salmon, which
presents elution only in mild strains. This phenomenon would be
associated with the receptor-destroying activity of the HE
protein has no effect on Atlantic salmon which would lead to
severe clinical effects suffered by this species (Falk K, Dale
OB. (2007): Lack of receptor-destroying enzymatic activity of
CA 02864258 2014-09-15
12
ISA-virus is an important virulence factor for development of ISA
in Atlantic salmon. In Frisk Fisk Conference, Tromso: Jan 23-25th
2007; Norway). A schematic representation of the virus infectious
cycle can be seen in the schematic representation of infection of
a cell by ISA virus of Cottet, L., Cortez-San Martin, M., Tello,
M., Olivares, E., Rivas-Aravena, A., Vallejos, E., Sandino, A.M.,
Spencer, E., (2010). Bioinformatic analysis of the genome of
infectious salmon anemia viruses associated with outbreaks of
high mortality in Chile. J. Virol 84(22): 11916-1192).
The disease in Atlantic salmon may occur with systemic and
lethal nature, characterized by severe anemia and hemorrhages in
various organs. The hematocrit usually is under 20% and in the
most severe cases less than 10% (normal values between 35% to
64%). The virus affects mainly five organs: liver, kidney, heart,
intestine and gills. Hepatic manifestation is characterized by
the dark color of the liver caused by hemorrhagic necrosis. In
the kidney inflammation with moderate interstitial hemorrhage and
tubular necrosis occurs. The intestine appears deep red due to
bleeding in the gut wall but not in the lumen. In the gill,
besides pallid, in some cases the accumulation of blood occurs,
particularly in the central venous sinus of these filaments. The
most striking observable symptoms in Chilean cases reported for
CA 02864258 2014-09-15
13
2007 are pale gills, eye hemorrhage, exophthalmos, ascites in the
abdominal cavity, hemorrhagic petechiae on the skin, abdominal
jaundice, external darkening, congested liver and spleen and
hemorrhages in basal fins (Godoy, M.G., Aedo, A., Kibenge, N.J.,
Groman, D.B., Yason, C.V., Grothusen, H., Lisperguer, A.,
Calbucura, M., Avendano, F., Imilan, M., Jarpa, M., Kibenge,
F.S., (2008). First detection, isolation and molecular
characterization of infectious salmon anaemia virus associated
with clinical disease in farmed Atlantic salmon (Salmo salar) in
Chile. BMC Vet. Res. 4, 28).
The main cause of transmission is direct contact and
cohabitation with infected specimens, due to natural secretions
and excretions of fish, implying that the transmission is
horizontal. Because of this the main route of infection is
through the gills, but cannot exclude the oral input (Mikalsen,
A.B., Teig, A., Helleman, A.L., Mjaaland, S., Rimstad, E.,
(2001). Detection of infectious salmon anaemia virus (ISAV) by
RT-PCR after cohabitant exposure in Atlantic salmon Salmo salar.
Dis. Aquat. Organ. 47 (3), 175-181). However, it has been
reported that vertical transmission also exist due to the
sequence similarity of strains present in Norway and Chile in
2007, so the Norwegian origin of the Chilean outbreak suggests.
CA 02864258 2014-09-15
14
Note that Chile imports large quantities of embryos from Norway
and these have been infected with the virus while bringing them
into the country. There is still controversy as to whether
vertical transmission actually occurs or whether it was due to
poor disinfection of the eggs (Nylund A, Plarre H, Karlsen M,
Fridell F, Ottem KF, Bratland A, Saether PA, (2007). Transmission
of infectious salmon anaemia virus (ISAV) in farmed populations
of Atlantic salmon (Sa1mo salar). Arch Virol 2007, 152:151-179;
Snow M., (2011). The contribution of molecular epidemiology to
the understanding and control of viral diseases of salmonid
aquaculture. Veterinary Research 2011, 42:56; Vike S., Nylund S.,
Nylund A., (2009) ISA virus in Chile: evidence of vertical
transmission. Arch Virol (2009) 154:1-8)
In experiments of viral transmission, other salmon species
such as brown trout (Salmo trutta) Rainbow trout and Coho salmon,
these show viral replication but not present clinical symptoms of
the disease, which can be reservoirs or vectors of the virus
spreading, (Nylund A, Jakobsen P (1995) Sea trout as a carrier of
infectious salmon anaemia virus. J Fish Biol 47: 174 -1 76;
Kibenge F.S.B., Gdrate 0.N., Johnson G. Arriagada R., Kibenge
M.J.T. & Wadowska D. (2001). Isolation and identification of
infectious salmon anaemia virus (ISAV) from Coho salmon in Chile.
CA 02864258 2014-09-15
Dis. Aquat. Org . 45, 9-18; MacWilliams C, Johnson G, Groman D,
Kibenge FS. Morphologic description of infectious salmon anaemia
virus (ISAV)-induced lesions in rainbow trout Oncorhynchus mykiss
compared to Atlantic salmon Salmo salar. Dis Aquat Organ. 2007;
78(1):1-12). We also detected the virus in non-salmonid species
such as cod (Gadus morhua) and saithe (Pollachius virens) (Grove
S, Hjortaas MJ, Reitan LJ, Dannevig BH. (2007). Infectious salmon
anaemia virus (ISAV) in experimentally challenged Atlantic cod
(Gadus morhua). Arch Virol.;152(10):1829-37; McClure CA, Hammell
KL, Dohoo IR, Gagne N.(2004) Lack of evidence of infectious
salmon anemia virus in pollock Pollachius virens cohabitating
with infected farmed Atlantic salmon Salmo salar. Dis Aquat
Organ.; 61(1-2):149-52).
A parasite to consider in the development of the disease and
in the salmon industry is sea lice or Caligus (Caligus
rogercresseyi). This copepod generated skin lesions and stress
which leads to a depression of the immune system resulting in a
greater susceptibility to pathogens such as ISA virus. Because of
their direct contact with the salmon they are believed to act as
a vector and may contribute to the spread of the virus (Bravo S.
(2009), "Estrategias de Manejo Integrado para el control de
Caligus en la industria del salmon en Chile").
CD, 02864258 2014-09-15
16
Finally due to the high density of fish handled by the salmon
farms (up to 30 kg / m 3) and the proximity between the farms of
southern Chile, the optimal conditions for episodes of massive
outbreaks and high mortalities are presented. According to public
account Sernapesca of November 2009 there have been 141 outbreaks
in salmon farms, and including these, a total of 228 centers have
been detecting virus, January 2009 being the highest prevalence
of the disease, ie the higher rate of affected areas in relation
to those who were populated with susceptible species (Atlantic
salmon), reaching the prevalence of positive centers in that
month to 45% (Sernapesca, 2009).
Teleost fish like salmon have an immune system, although not
equal, similar to higher vertebrates, comprising an innate immune
system and an adaptive one. *Press CM, Evensen 0. (1999). The
morphology of the immune system in teleost fishes. Fish Shellfish
Immunol 309-318; Fernandez AB, Ruiz I, de Blas I. (2002) El
sistema inmune de los teleasteos (II): Respuesta inmune
inespecifica. Revista Aquatic disponible
en:
http://www.revistaaquatic.com/aquatic/html/art1707/inmune2.htm)
The fish skin is the first line of defense against pathogens,
as well as the mucous membranes lining the gills and
gastrointestinal tract. The mucus that coats the skin is an
CA 02864258 2014-09-15
17
important defense mechanism, as it contains a variety of reactive
compounds as antimicrobial lysozymes, proteases, complement
factors, C-reactive protein, lectins, interferons, eicosanoids,
transferrin, piscidins and various carbohydrates (Buchmann K,
Lindenstrrm T, Bresciani J. (2001) Defence mechanisms against
parasites in fish and the prospect for vaccines. ActaParasitol
;46:71-81; Magnadottir B. (2006) Innate immunity of fish
(overview). Fish Shellfish Immunol 2006;20:137-151; Manning M J.
(1998) Immune defence systems. In: Black KD, Pickering AD
editors. Biology of farmed fish. Sheffield: Sheffield Academic
Press; 1998:180-221; Woo PTK. (2007) Protective immunity in fish
against protozoan diseases. Parassitologia 49:185-191). As the
complement we can mention that it is composed of a number of
serum proteins having three main defensive functions: a) coating
pathogens and particles outside the body to facilitate
recognition and destruction by phagocytic cells (opsonization),
b) initiate inflammatory responses stimulating vasodilation and
lymphocyte chemoattractant and c) lyse pathogens by drilling
their membranes (Boshra H, Li J, Sunyer JO. (2006) Recent
advances on the complement system of teleost fish. Fish Shellfish
Immunol 20:239-262; Janeway CA, Travers P, Walport M, Schlomchik
MJ. (2005) Immunobiology; the immune system in health and
CA 02864258 2014-09-15
18
disease. 6th ed. New York: Garland Science Publishing).
Eicosanoids include prostaglandins, thromboxanes
and
leukotrienes, and are potent pro-inflammatory mediators. Involved
in various physiological processes such as hemostasis, immune
regulation, increase phagocytosis and quimioatractanctes for
neutrophils (Secombes CJ. (1996) The nonspecific immune system:
cellular defenses. In: Iwama G, Nakanishi T editors. The fish
immune system: organism, pathogen and environment. London:
Academic Press Ltd.; 63-103) The nonspecific immune system:
cellular defenses. In: Iwama G, Nakanishi T editors. The fish
immune system: organism, pathogen and environment. London:
Academic Press Ltd.; 63-10.; Manning MJ. (1998) Immune defence
systems. In: Black RD, Pickering AD editors. Biology of farmed
fish. Sheffield: Sheffield Academic Press; 1998:180-221).
Interferons (IFN) are a family of proteins heterologous to confer
protection against viral infections (Secombes CJ. (1996) The
nonspecific immune system: cellular defenses. In: Iwama G,
Nakanishi T editors. The fish immune system: organism, pathogen
and environment. London: Academic Press Ltd.; 63-103). There are
the Type I, which includes the IFN and IFN-3 which are produced
by cells infected with the virus and can produce any cell type,
and type 2, which includes IFN which is a cytokine produced by T
CA 02864258 2014-09-15
19
lymphocytes (Magnadottir B. (2006) Innate immunity of fish
(overview). Fish Shellfish Immunol 2006; 20:137-151). The answer
= begins when the pathogen and host interact, leading to activation
of pathogen recognition receptors (PRR), among which the most
prominent are the family members of toll-like receptors (TLRs)
that are able to recognize pathogen-associated molecular patterns
(PAMPs) such as lipopolysaccharide (LPS) of Gram-negative
bacteria, peptidoglycan from Gram-positive bacteria, RNA of
double and single-stranded virus and Unmethylated CpG DNA (Iliev,
D., Roach, J., MacKenzie, S., Planas, J. and Goetz, FW. (2005).
Endotoxin Recognition: In fish or not in fish. FEBS Letters 579
(29):6519-6528). To this date it has been reported the existence
of at least 4 TLR, two of which have been associated with viral
infection in salmon, one is TLR-8 in mammals whose function is to
recognize single-stranded RNA. In salmon expression of TLR-8
=
messenger level is induced in cells treated with IFN- and IFN-
(Skjaeveland, D. B. Iliev, & J. B. Jorgensen (2009).
Identification and characterization of TLR8 and MyD88 homologs in
Atlantic salmon (Salmo salar.) Developmental & Comparative
Immunology. 33: 1011-1017). The other TLR-9 that recognize DNA
CpG unmethylated like in mammals (Skjaeveland, D. B. Iliev, J.
Zou T. Jorgensen & J. B. Jorgensen (2008). A TLR9 homolog that is
ak 02864258 2014-09-15
up-regulated by IFN- in Atlantic salmon (Salmon salar).
Developmental and Comparative Immunology, 32, 603-607).
Another alternative route to sense viral RNA is through
helicase RIG-I, acting through a molecule known as IPS-1 (also
called MAVS, VISA and CARDIF) activates a signaling cascade that
eventually leads to the activation of type I IFN (Onomoto K,
Yoneyama M, Fujita T. (2007). Regulation of antiviral innate
immune responses by RIG-I family of RNA helicases. Curr Top
Microbiol Immunol. 316:193-205). In salmon it is known the
existence of RIG-I and described that cells transfected with
cloned IPS-1 are able to induce an antiviral state (Biacchesi,
S., Leberre, M., Lamoureux, A., Louise, Y., Lauret, E., Boudinot,
P., and Bremont, M. (2009) MAVS Plays a Major Role in Induction
of the Fish Innate Immune Response against RNA and DNA Viruses.
J. Virol. 83 (16):7815-7827 (Journal)).
Salmonids as most non-mammalian exhibit no bone marrow or
lymph nodes, however, the thymus, kidney and spleen assume this
role. In kidney hematopoietic activity is observed, showing stem
cells that perform this function (Hanington PC, Tam J, Katzenback
BA, Hitchen SJ, Barreda DR, Belosevic M. (2009). Development of
macrophages of cyprinid fish. Dev Comp Immunol 33:411-429), in
addition together with the spleen have extensive lattice for
CA 02864258 2014-09-15
21
trapping blood hauled and presented together macrophage and
lymphocyte populations of particles capable of initiating immune
responses (Press CM, Evensen 0. (1999). The morphology of the
immune system in teleost fishes. Fish Shellfish Immunol 309-318).
It has also been reported that the kidney presents lymphoid
activity and that is the main body of B cells and antigen
production (Whyte S.K.. The innate immune response of finfish-A
review of current knowledge. Fish & Shellfish Immunology, 2007.
6:1127-1151). The adaptive immune response can be divided into
humoral and cellular immunity, dependent largely on lymphocytes B
and T respectively (Fernandez AB, Ruiz I, de Blas I. (2002)The
immune system of teleost (II): non-specific immune response.
Aquatic magazine available
at:
http://www.revistaaquatic.com/aquatic/html/art1707/inmune2.htm).
Similar to as in mammals, humoral immunity involves the
recognition and binding of antigens by B lymphocytes, which
differentiate into plasma cells and memory cells to produce
antigen-specific antibodies or immunoglobulins and generate fast
responses and pronounced in subsequent infections respectively
(Hansen JD, Landis ED, Phillips RB. (2005). Discovery of a unique
Ig heavy-chain isotype (IgT) in rainbow trout: Implications for a
distinctive B cell developmental pathway in teleost fish. Proc
CA 02864258 2014-09-15
22
Nati Acad Sci. USA. 102:6919-6924). In viral infections the
immunoglobulins can bind to the surface of viruses being the
predominant neutralizing IgM, which may be associated with B cell
membrane and act as receptor for antigen recognition, or in its
secreted form (Janeway CA, Travers P, Walport M, Schlomchik MJ.
(2005) Immunobiology; the immune system in health and disease.
6' ed. New York: Garland Science Publishing). In addition to IgM
other immunoglobulins have been described in teleosts such as
IgD, and IgZ IgT, which have been cloned in Atlantic salmon and
have sequence similarity (Randelli, E., Buonocore, F.,
Scapigliati, G. (2008). Cell markers and determinants in fish
immunology. Fish Shellfish Immunol. 25, 326-340).
On the other hand, the acquired cell mediated immunity
involves the activation of T lymphocytes mainly, which are
activates by the interaction of the major histocompatibility
complex (MHC) loaded with an antigen and the T cell receptor
(TCR) (Finelli, A., K. M. Kerksiek, S. E. Allen, N. Marshall, R.
Mercado, I. Pilip, D. H. Busch y Pamer.E.G MHC class I
restricted T cell responses to Listeria monocytogenes, an
intracellular bacterial pathogen. Immunol. Res.1999.19:211-223).
There are two types of MHC class I and II. The MHC class I is
found most cells and acts primarily against intracellular
CA 02864258 2014-09-15
23
pathogens by activating cytotoxic T lymphocytes (CD8+), whereas
MHC class II, is found only on antigen-presenting cells such as
macrophages, dendritic cells and lymphocytes B, and they lead to
the activation of T helper lymphocytes(linfocitos T helper)
(CD4+) (Nakanishi T, Ototake M. (1999). The graft-versus-host
reaction (GVHR) in the ginbuna crucian carp, Carassius auratus
langsdorfii. Dev Comp Immunol 23:15-26). The interactions between
cells are not only mediated by cell-cell contact, but also by
soluble factors such as cytokines, which account for the
different types of immune responses via differential expression
depending on the pathogen to display the individual faced.
Against intracellular pathogens, CD4 + T cells differentiate into
Thl cells by action of IL-12 and INF, which are secreted by
progenitors expressing the T-bet gene, and this gene being the
regulator key to produce Thl. Th1 cells secrete molecules finally
as IL-2, INF-and TNF-3 mainly promoting cellular response,
generating the activation of macrophages, increased antigen
presentation and differentiation of CD8 + T lymphocytes.
Extracellular pathogens against CD4 + T cells differentiate into
Th2 cells by action of IL-4, which is secreted by GATA3
progenitors expressing gene. These lymphocytes secrete IL-4, IL-
5, IL-6, IL-9, IL-10, and IL-13 that activate B cells to
CD, 02864258 2014-09-15
24
antibody-mediated response (Janeway CA, Travers P, Walport M,
Schlomchik NJ. (2005) Immunobiology; the immune system in health
and disease. 6th ed. New York: Garland Science Publishing).
The prior art, in particular the following patent documents
may be mentioned:
WO 2004024956, where it describes the identification
resistant Atlantic salmon and trout susceptible to viral diseases
(ISA, IPN), and bacterial, to determine the genotype of a salmon
kit, chip type, molecular markers typing containing gene
sequences belonging to the described English Histocompability
Main complex (MHC, its acronym in English Histocompability Main
complex) class I and MHC class II alpha/beta. The use of
technology is in the field of reproduction.
W00226784 refers to the use of nucleotide sequences encoding
the structural proteins M1 and M2 of the ISA virus for use in
vaccines, generation of antibodies against proteins or diagnostic
kits or detection of ISA virus.
W02012075597 describes a method for determining the
virulence of a virus infection by infectious pancreatic necrosis
(IPN), serotype Sp from a tissue sample of salmon. RNA sample is
extracted first, amplified by PCR the gene region corresponding
to the VP2 protein, the purified PCR product is sequenced, the
CA 02864258 2014-09-15
nucleotide sequence into an amino acid sequence results, and
finally the presence of threonine, proline or alanine amino acids
were identified at a given position, and in this case it is
determined if the IPN virus has high or medium virulence.
Furthermore, in the market the following products related to
the area of the present invention.
The product Sb + Health SalmoBreed comprising eggs of Salmo
salar (Atlantic salmon) species that are resistant to ISA and IPN
virus and furunculosis. These eggs are genetically selected to
deliver better growth rates and survival throughout the life
cycle of the salmon. This product is a selection made at a salmon
parental level, based on various parameters, including among
others, resistance to ISA.
The product AquaGen Atlantic QTL-innOva IPN/PD corresponds
to eggs of Salmo salar species originating from spawning salmon
that are highly resistant to infectious pancreatic necrosis virus
(IPN, its acronym in English, Infectious Pancreatic Necrosis) and
pancreas disease (PD, its acronym in English, Pancreas Disease),
where using the technique of marker-assisted selection is
selected directly parenting that are used in the production of
eggs that have specific genetic markers or a quantitative trait
locci, QTL (from its acronym in English, quantitative trait
CA 02864258 2014-09-15
26
locci) which are associated with resistance to a virus. The
selection is made at both family and individual levels, which
ensures high resistance to IPN and/or PD virus in the offspring
or the eggs.
The subunit vaccine against infectious salmon anemia
(injectable) Centrovet is an ISA virus vaccine for S. salar made
from this virus recombinant proteins produced in yeast cultures.
Help to prevention of infectious salmon anemia in pre-smolt
salmonid fish starting from 30 grams of weight.
Transcriptome Sequencing services, LC Science, refer to an
RNA sequencing service (RNA-seq) technology using high throughput
sequencing to determine structures Illumina gene splicing
patterns, posttranscriptional modifications, detecting new or
rare transcripts and quantification of changes at the level of
transcript expression in various organisms, and corresponds to a
generic service and not focused only on salmon.
The technical assistance services, genetic and breeding
programs in general are a service of aquaculture breeding species
of interest including Atlantic salmon. His focus has been mainly
improved the percentage of weight gain of salmon, being, between
5-8% for the Atlantic salmon.
CA 02864258 2014-09-15
27
Also, there have been developed or are being developed, a
series of investigations in the field of the invention, for
example, the following projects are listed below:
Project development and validation of a chip of SNPs to
determine the molecular basis and genetic improvement of disease
resistance in Salmo salar Aquainnovo SA 2012-2015. In this
project a system of genetic screening to identify disease
resistant Atlantic salmon produced by ISA and IPN virus, using
the technique of Single nucleotide polymorphisms, SNPs (for its
acronym in English, Single Nucleotide Polymorphism) in chip
format.
Project development and validation of genetic markers of
immune response to ISA of Fisheries and Oceans Canada, 2007-2009,
identified from a genomic point of view using DNA microchips,
immunological genetic markers involved in the response to ISA
virus. Validation of these markers in vivo provide information
for the study of disease, the resistance of the salmon and the
response thereof to vaccines.
The present invention relates to the use of Gata3, Grb2, IgM,
MAVS and Regul genes determining phenotypes and Resist
susceptible to infectious salmon anemia (ISA) in Atlantic salmon
(Salmo salar).
CA 02864258 2014-09-15
28
In the present invention, the level of expression of
selected genes specifically related to the response against ISA
virus in fish susceptible and quantified virus phenotypes and
gene expression profiles associating, to each phenotype.
They were selected according to criteria ontological and
functional genomics, genes from the group consisting in Gata3,
Grb2, IgM, MAVS y Regul and others.
To quantify the expression levels of each gene of salmon
with resistant and susceptible phenotypes, PCR was used in real-
time from tissues obtained from salmon with resistant and
susceptible phenotypes.
Generally, it resulted that the expression of selected genes
tend to be lower in the fish susceptible phenotype,and then,
Gata3, Grb2, IgM, MAVS and Regul and found useful as markers for
differentiated phenotype "resistant" to "susceptible".
BRIEF DESCRIPTION OF THE FIGURES
Figure 1 Graph of cumulative mortality of fish families
corresponding to phenotypes resistant and susceptible. Each
challenge was performed in duplicate for greater range.
CA 02864258 2014-09-15
29
Figure 2 Ribosomal bands of 18S and 28S RNAs corresponding to
susceptible groups (A), and resistant controls (B), displayed on
agarose gel at 1%.
Figure 3 Dissociation curves analyzed genes: annexin (A),
cathepsin (B), Gata 3 (C), Grb2 (D), HSP90 (E), IgM (F), IgZ (G),
Mays (H), Tbet (I), Regul (J). The presence of a single PCR
product was observed.
Figure 4 Normalized relative expression of candidate immune
genes, IgM (A), Grb2 (B), HSP 90 (C), Annexin (D) and Regul (E)
genes are shown. Bars with squares representing individuals and
susceptible to fine lines resistant individuals. Data are the
average of 9 individuals per phenotype, normalized against 18S
rRNA gene expression and adjusted for relative efficiency. (*)
Denotes significant difference from control (* p <0.05, ** p
<0.01, *** p <0.001).
Figure 5 Normalized relative expression of immune candidate
genes, genes with similar expression pattern Cathepsin (A), IgZ
(B), Mays (C), Gata3 (D) and Tbet (E) are shown. Bars with
squares representing individuals and susceptible to fine lines
CA 02864258 2014-09-15
resistant individuals. Data are the average of 9 individuals per
phenotype, normalized against 18S rRNA gene expression and
adjusted for relative efficiency. (*) Denotes significant
difference from control (* p <0.05, ** p <0.01, *** p <0.001)
DETAILED DESCRIPTION OF THE INVENTION
In general, one of the search strategies of candidate genes
is functional genomics, which aims to relate the sequence of a
gene with the function that it has. To achieve this goal
platforms like microarrays are used, which are based on the
hybridization of fluorescently labeled complementary DNA from two
different conditions, with thousands of genes present on the
microarray chip (Belbin TJ, Gaspar J, Haigentz M, Perez .. -Soler
R, Keller SM, Prystowsky MB, Childs G, Socci ND (2004) Indirect
measurements of differential gene expression with cDNA
microarrays Biotechniques 2004, 36:.. 310-314). This results in
differences in the expression of the genes studied under both
conditions. However, microarray data are subject to tests (genes)
that each platform may possess. A new "RNA sequencing" technology
allows access to the RNA sequence with differential expression in
a broad and comprehensive sense, making it an excellent tool in
the search for candidate genes (Wang Z, Gerstein M, Snyder M.
CA 02864258 2014-09-15
31
(2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev
Genet. 10(1):57-63).
Differential expression patterns of certain genes,
representative of a particular phenotype can be analyzed using
the Polymerase chain reaction (PCR). A variant of this reaction
is the real-time PCR, which uses fluorescent markers for
monitoring DNA amplification in each cycle. This is characterized
by its sensitivity and specificity, making it suitable for
detailed analysis of genes of interest (Pfaffl M.W. y Hageleit,M.
(2001) Validities of mRNA quantificationusing recombinant RNA and
recombinant DNA external calibrationcurves in real-time RT-PCR.
Biotechnol. Lett., 23, 275-282).
In the present invention, the technique "RNA sequencing"
technology that uses Illumina-Solexa sequencing, and develop an
experiment that includes resistant and susceptible Antlantic
salmon families to the ISA virus. The genetic status of families
was determined according to the level of heritability; which
corresponds to a proportion of phenotypic variation in a
population, attributable to genotypic variation among
individuals. The value of heritability can vary (for each
feature) in different populations or even in the same population
at different points in its history. For purposes of this
CA 02864258 2014-09-15
32
description, if the value is greater than or equal to 0.35 will
be classified as resistant family and less than or equal to 0.34
for susceptible family. Besides the total mortalities within each
family must be consistent with expectations (higher in
susceptible).
The experiment results indicated RNA sequencing different
candidates differentially expressed genes between the two
phenotypes. Within the main ontological categories associated
noted: a) signal transduction, b) regulation / proliferation of B
cells, c) a stress response, d) T-cell receptors, d)
differentiation of T cells, e) immune response, f) regulation of
transcription, g) regulation of endothelial cell migration h)
viral response. For each of these categories were selected
candidate genes representative. The genes, their function and
their associated biological processes are detailed in Table 1.
Table 1. Candidate genes selected for Real time with their
respective associated function and biological process.
Genes Process Function of its protein
Candidates Biological
Anexina A- Regulatory Inhibition of synthesis of
1 inflammatory prostaglandins, leukotrienes and
CA 02864258 2014-09-15
33
Response thromboxanes, anti-inflammatory
effect, inhibition of Leukocyte
chemotaxis
Cathepsin Lymphocyte Involved in proteolysis and antigen
proliferatio production to charge MHC II.
n T and B
Gata 3 T Lymphocyte Signage for synthesis of Th2
differentiat lymphocytes.
ion
Grb2 Signal Adapter protein, linked to signal
transduction transduction of tyrosine kinases
- regulation activated by lymphocyte antigen.
of the Linked to positive signaling by the
transcriptio TCR.
HSP 90 Response to Proper folding of poly peptides,
stress chaperone protein restauration.
IgM Humoral Identification and neutralization of
Response pathogens, complement activation,
precipitation, agglutination
ak 02864258 2014-09-15
34
IgZ Humoral Identification and neutralization of
Response pathogens, precipitation,
agglutination
MAVS Response to Adapter protein, involved in sensing
virus viral RNA and activation of IFN-I
Tbet T lymphocyte Signage for synthesis of Thl
differentiat lymphocytes
ion
Regul Regulation Negative regulator of G protein and
of signaling pathway, acceleration GTP-
endothelial GDP conversion, decrease of the
migration effect of chemokines and immune cell
Regulation migration
of
transcriptio
18S Ribosome Constitutive gene, used as reference
assembly gene
Al Annexin inhibits phospholipase protein 2, which releases
arachidonic acid from the cell, this finally transformed into
CA 02864258 2014-09-15
prostaglandins, leukotrienes and thromboxanes (Eicosanoids),
which are associated with inflammatory processes. Note that
prolonged inflammatory processes cause great damage to tissues
and organs, so it is important to analyze genes related to the
biosynthesis of eicosanoids (Glaser KB. (1995): Regulation of
phospholipase A2 enzymes: selective inhibitors and their
pharmacological potential. Adv Pharmacol 1995, 32:31-66).
Cathepsin S is a cysteine protease expressed in the antigen
presenting cells, which has the function to generate antigens for
loading on MHCII. It has been reported that deficiency in this
gene produces deficient mitochondrial damage and increased levels
of reactive oxygen species (ROS), as well as problems in
signaling MHCII (Costantino CM, Hang HC, Kent SC, Hafler DA,
Ploegh HL. (2008) Lysosomal cysteine and aspartic proteases are
heterogeneously expressed and act redundantly to initiate human
invariant chain degradation. J. Immunol 180:2876-2885. [PubMed:
18292509]). It is analyzed for its relation to the loading of
MHCII antigens.
Grb2 is an adapter protein linked to the activation of the
MAP kinase pathway and signal transduction in lymphocytes.
Positively regulates the signaling by the TCR (T cell receptor),
with this development activating T cells and negatively signaling
CA 02864258 2014-09-15
36
the BCR (B cell receptor, thereby inducing an immunity response
linked more cellular than humoral (Jang IK., Zhang J., Gu H.
(2009) Grb2, a simple adapter with complex roles in lymphocyte
development, function, and signaling. Inmunol Rev 232; 150-159).
HSP90is a protein involved in the correct folding of
polypeptides, protein renaturation heat damaged and as chaperone.
It is expressed mainly by either osmotic stress, by contaminants,
or immune response (Pearl LH y Prodromou C. Structure, function,
and mechanism of the Hsp90 molecular chaperone. 2001. Adv Protein
Chem 59: 157-186.). It is mainly used in this thesis as a marker
of stress. IgZ and IgM immunoglobulins are the first of these
present in greater amounts in salmon and as primary response
against pathogens (Frazer, J.K. and J.D. Capra. Immunoglobulins:
structure and function. En Fundamental Immunology, 1999. 4th ed.
W.E. Paul, editor. Lippincott-Raven, Philadelphia. 37-74.). As
IgZ not yet been described but its function is thought to be more
specific immunoglobin(Gamez-Lucia Duato E. The major
histocompatibility complex. In Manual of Veterinary Immunology
2007. Gomez-Lucia E, Blanco MM y Domenech A (eds.). Pearson-
Prentice Hall. Madrid. pp. 1/7-136).Tbet and Gata3 antagonistic
genes are involved in differentiation of T cells into Thl and
Th2, respectively. Both genes are key regulators in the fate of
CA 02864258 2014-09-15
37
CD4 + T lymphocytes and analyzed as markers of cellular and
humoral immunity. Regul is a negative regulator of G protein
signaling accelerates the conversion of GTP to GDP, reducing
signaling downstream of G-protein function This repressive effect
decreases by chemokines and immune cell migration (Tiffany Tran,
Pedro Paz, Sharlene Velichko, Jill Cifrese, Praveen Belur, Ken D.
Yamaguchi, Karin Ku, Parham Mirshahpanah, Anthony T. Reder y
Croze. E. Interferon-1b Induces the Expression of RGS1 a
Negative Regulator of G-Protein Signaling. International Journal
of Cell Biology. 2010. 7: 1-12).
Essentially, alternatives to control the infection caused by
the ISA virus in Atlantic salmon can be grouped into three: i)
vaccines, which corresponds to the currently used alternative
mainly due to the obligation of the national authority (in Chile,
Sernapesca) to vaccinate all fish in smoltification stage post
(General fisheries and Aquaculture Law N 20.632 of Chile).
There are currently, in Chile, more than 10 commercial vaccines
registered for ISA, which can be mono or polyvalent molecules
inactivated virus and primarily injectable or oral type.
Unfortunately there is no information currently published
regarding the actual coverage of vaccines against ISA, internal
reports only restricted production companies. However, it is
CD, 02864258 2014-09-15
38
widely known that the coverage of vaccines in field does not
correspond to a 100% (Mikalsen, A.B., Sindre, H., Torgersen, J. y
Rimstad, E. Protective effects of a DNA vaccine expressing the
infectious salmon anemia virus hemagglutinin-esterase in Atlantic
salmon. 2005. Vaccine 23:4895-4905; Lauscher a., KrossOy B.,
Frost P., Grove S., Kanig m., Bohlin J., Falk K., Austb0 L.,
Rimstad E. (2011). Immune responses in Atlantic salmon (Salmo
salar) following protective vaccination against Infectious salmon
anemia (ISA) and subsequent ISA virus infection. Vaccine, Volume
29, chapter 37, 6392-6401), ie not all vaccinated individuals
fail to develop immunoprotection, ii) Genetics, which corresponds
to a classical alternative selection in the context of genetic
animal model, which is mainly based on the detection and
utilization of resistant individuals with a given disease and
subsequent design of controlled crosses. To date, this
alternative has been very successful but only in certain
companies which have the advantage of the heterogeneous resistant
and iii) fish, which range from immunostimulating, antiviral and
synthetic peptides.
By improving family because controlled crosses, fish families
that have ISA virus resistance, finally resulted in lower
cumulative mortalities were obtained (less than 40% for this
CD, 02864258 2014-09-15
39
description and heritability (H) greater than or equal to 0.35)
in compared to other sites. These are called resistant families.
At the same time there are also families who have high mortality
(50% or higher for this description and H less than or equal to
0.34) against the virus, which will be classified as susceptible.
Taking these two phenotypes base (resistant and susceptible), the
expression patterns of candidate genes and their relationship to
the resistance or susceptibility to the pathogen compared.
For this, the following actions were taken:
= Quantify, evaluate and associated expression profiles of
candidate genes in resistant and susceptible fish with
ISA virus as the initial target phenotypes.
= Develop and standardize the analysis of real-time PCR
quantification of the expression of candidate genes as
the next target.
= Quantified as relative candidate genes in susceptible and
resistant phenotypes aim below.
= Analyze the feasibility of some of the selected genes may
serve as a marker of resistance to the ISA virus, as the
ultimate goal
In the initial phase, a challenge test was performed with 2
families of S. salar (SNIR and Fanad, resistant and susceptible,
CD. 02864258 2014-09-15
respectively) to determine the degree of resistance to ISA virus.
To this was used a Chilean viral isolate which was sequenced in
the hypervariable region (HPR) resulting HPR type 7b which was
inoculated intraperitoneally injected in the ventral midline at
0.2 mL inoculum per fish Trojan which contains a degree of 7.2 x
104 TCID50. Were infected only 40% of the fish pond, waiting for
the remaining 60% were infected by cohabitation, thus infection
resembles what happens in the field.
Before starting the test, the health status of fish in fish
farming was confirmed source. A visit was made in the field to
check the general condition of the fish of the selected group. At
the same time, taking 30 fish sampled for analysis in the
laboratory was performed. The analyzes cover 15 necropsies, gram
analysis, Flavo, BKD, RT-PCR IPNV and RT-PCR ISA. If positive,
the selected group of either fish pathogens analyzed, it is
discarded and a new group of fish sought.
Tanks 720 L
Salinity 32
Temperature 12 C acclimatization and
14 C defiance
Oxygen (saturation) 85-100%
CA 02864258 2014-09-15
41
Replacement rate 1.5 replacement/hour
N of fish per tank 363 trojans y 528
(acclimatization) cohabitants
N of fish per tank 110 trojans + 160
(cohabitation) cohabitants
Density 20-30 kg/m3
In the following experimental design scheme associated with
the challenge illustrated by cohabitation:
CA 02864258 2014-09-15
=
42
35 kg/m 3
Troyans
330 p FANAO > 330 Fish
Aclimacion Cohabitants:
period of 160 p 160 p 160 p SNIR > 40 Fish
14 days j F ANAD < 40 Fish
Tank
7201
25,6 kg/m)
Inoculation Trojan fish
37 kg/m 3
Experimental 110p 110p 110p Defiance
for
period 160 p 160 p 160p
Cohabitation
cohabitation:
Defiance for
cohabitation
SNIR > 40
fish
AD
35 days FAN 40
fish 720 L
Tanks
Trojans 40% FANAD> 110 Fish
Cohabitant 60% Inoculated with ISAv
CD, 02864258 2014-09-15
43
The trial looked at the reception of 528 Atlantic salmon fish
(Salmo salar) with an average weight of 115 g, corresponding to
the experimental design cohabitants previous scheme. These fish
are all marked individually with finning for identification. Also
relevant 363 Atlantic salmon fish fish contemplated "Trojans",
which were injected once the acclimation with ISA virus inoculum.
All fish were transported to a recepted Experimental Centre on 1
720 L tank for the Trojans in 3 ponds and cohabitants. The fish
were in brackish water at a salinity of 15 and then gradually
increasing the salinity within the next 7 days to finally get to
32 fish remained in the 4 ponds for a period of 14 days as
acclimatization period at a temperature of 12 C, the same
temperature of fish origin. During this period the fish were
manually fed a commercial diet, providing rations three times a
day up to 2% BW (body weight / day) .Then, a total of 480
individuals, who cohabited with 330 infected individuals were
challenged ISAv intraperitoneally with (40% of total 810 fish)
injection. It is proposed to use a total of 4 720 L tanks, 3
permanent and 1 temporary pond. In each of the 3 permanent ponds
were put 40 fish from each group (resistant or susceptible) to
challenge by cohabitation, see diagram, ie, in each of the ponds
was left with 160 cohabiting fish were individually marked with
CD, 02864258 2014-09-15
44
cuts fin , average weight 155 g. In 330 temporary pond fish
"Trojans" of the same weight, which were inoculated
intraperitoneally with virus inoculum ISA once the acclimation
were placed. This pond is only temporary and hosted the Trojans
fish during the acclimation period (14 days). Concluded the
acclimation phase, the inoculation was made by intraperitoneal
injection of the virus, to which the fish were anesthetized
Trojans benzocaine. The fish were fasted for 24 hours prior to
inoculation. These fish are adipose fin cut for easy
identification and 110 fish were placed in each of the permanent
ponds containing 160 cohabiting fish. In this way they were three
ponds with 40% of infected fish (Trojans) and 60% of cohabiting
fish finning marked to identify at all times the different groups
of fish. The cohabitation phase lasted 35 days. Fish mortality
was recorded daily. To the moribund fish were extracted samples
of kidney, spleen and heart.
Throughout the trial the fish will have a photoperiod regime
of 24 hours light to ensure complete fish smoltification.
The fish were given a 12 C since the temperature of water
in the fish breeding source is at that temperature. That
temperature was maintained for the days of acclimatization.
Throughout the period of cohabitation challenge temperature at 14
CD, 02864258 2014-09-15
C was maintained. The temperature was automatically recorded
every minute with a "logger". The oxygen was maintained between
85% and 100% during the acclimation period and cohabitation
defiance.
For the acclimation period and cohabitation challenge a
commercial diet was utilized, Golden Optima 4, of the Biomar
company in caliber 4.0 mm. Feeding was done manually, delivering
rations three times a day until completing the portion of the
day.
9 pieces of each phenotype were selected to study to which
they extracted the anterior kidney at day 15 post infection,
which was stored in "RNA later", and then stored at -80 C for
further analysis.
RNA extraction was performed from anterior kidney samples
using the RNeasy mini kit (Qiagen). Each sample was disaggregated
using a tissue homogenizer (TissueRuptor, Qiagen) in 1 mL of
Trizol LS (Invitrogen) according to what is established by the
manufacturer. The homogenate was incubated 2 minutes at room
temperature, then add 200 pL of chloroform, vortexed and
centrifuged at 12,000 g for 15 minutes at 4 C to separate the
phases. The aqueous phase to a tube containing 700 pL of 70%
ethanol and then the total volume was transferred to a mini-
CA 02864258 2014-09-15
46
column (RNeasy Mini Kit, Qiagen) and centrifuged at 9000 g for 30
seconds was transferred. 600 pL of RW1 buffer was added and
centrifuged at 9000 g for 30 seconds. Was further added a step of
adding DNase treatment 80 pL of digestion solution (10 pL of
enzyme DNase (Qiagen) and 70 1 of buffer according to the
manufacturer RDD) to each column. Then washed 2 times with 500 pL
of buffer RPE, the first wash at 9000 g for 30 seconds and the
second at 9000 g for 2 minutes to dry. Finally the column with 50
pL of nuclease free water was eluted, and aliquots of 10 1 of
extracted RNA were prepared. RNA integrity was verified by gel
electrophoresis in 1% agarose, stained with ethidium bromide. The
presence of the 28S and 18S bands (ribosomal RNA) and the level
of degradation, serve as an indicator of the integrity of
samples. Simultaneously the concentration of RNA in a
BioPhotometer (Eppendorf) was measured, which was performed by
diluting 2 L of the extracted RNA plus 58 L of nuclease free
water, and the absorbance was determined at 230, 260, 280 and 320
nm to verify the state and possible contaminations.
MMLV (Promega) enzyme and the method of Random Primers
(Promega) was used for reverse transcription. To this pre 2.5 pg
RNA incubated in water (12,875 pL final volume of the RT
reaction) at 60 C for 10 min and then quickly transferred to
CD, 02864258 2014-09-15
47
ice. RT mix comprising 5X RT buffer, dNTPs 3.125 pL of 4 pM, 100
U of ribonuclease inhibitor RNasin (Promega), 100 U of MMLV and
0.5 pg of Random Primers, in a final volume of 12,125 pL of mix
RT per reaction. RT mix in the 12,875 pL RNA was then added to
obtain a pre incubated final reaction volume of 25 pL and reverse
transcription was performed in a thermocycler with the following
program: 60 minutes at 42 C, 5 minutes at 99 C for denaturar
enzyme.
The primers of the selected genes were generated using the
program Primer3 using preset parameters based on this
Immune
Genome database developed by our laboratory (Cepeda et. a/.2011)
and multiple alignments performed with the BioEdit program to
5.09 of roughly determine the exon-exon junctions. The primers
were synthesized by IDT (Integrated DNA Technologies). They were
optimized conditions PCR primers for each set of varying
concentrations of primers(0,5-2 pM) and CDNA directly charged to
the reaction (1 to 3 pL) in order to obtain the cycle ct between
20 and 30 The real time PCR was carried out using the Green qPCR
kit TMSYBR Master Mix (Fermentas) in 96-well plates in the 7300
Real-Time PCR equipment System (Applied Biosystems). Dissociation
curve was also done in order to determine dissociation
temperatures and analyze the specificity of the amplicons. Sizes
CA 02864258 2014-09-15
48
were also confirmed by agarose gels. The conditions employed in
the real time PCR were 10 min at 95 C of pre incubation,
followed by 40 cycles at 95 C for 30s, 53-62 C for 1 min and
72 C for 30 sec. Finally dissociation curve was performed from
60 C to 95 C. The primers for each gene are listed in Table
2.
Table 2. Sequences of PCR primers for candidate genes in real
time
Primer Gene
Annexin Al F: CTCCAGGAAATTGAACACCGCGA
R: AAGGCTGCGATGAAGGACATGGT
Cathepsin S F: CGAAGG GAGGTCTGG
GAGAGGAAT
R: GCCCAGGTCATAGGTGTGCATGTC
Gata 3 F:
CCAAAAACAAGGTCATGTTCAGAAGG
R: TGGTGAGAGGTCGGTTGATATTGTG
Grb2 F: TGACTTTACTGCCACTGCTGAGGAC
R: CAGTCATCATTGGTGCCCAAGATC
HSP90 F:GAACCTCTGCAAGCTCATGAAGGA
ak 02864258 2014-09-15
49
IR: ACCAGCCTGTTTGACACAGTCACCT
IgM F: TTCTCTCCACCGGCTCATCATCA
R: ATACGGTGACCCTGACTTGCTACG
IgZ F: AGCACCAGGGACAACCACCAT
R: TTCACACTCGGTGGGTTAGAGTC
MAVS F: AGGAGGTGCTGCACAATGTTGCT
R: CGACGGCGACGGAGCCAGTA
Tbet F: GGTAACATGCCAGG GAACAGGA
R:
TGGTCTATTTTAGCTGGGTATGTCTG
Regul F: GACTCCTAACCTCCAATGCTTCGAC
R: CGAATCTCTCTCCATCAGCCCATA
18S F: CCTTAGATGTCCGGGGCT
R: CTCGGCGAAGGGTAGACA
Calibration curves were made for each gene from RNA
extractions from known concentration; to the point of maximum
concentration of 2.5 pg total RNA, which was subjected to serial
dilutions were used. Each standard curve consisting of at least 5
points in triplicate, plotting the mean Ct from each point versus
the log of the concentration of RNA. From these curves FOR
efficiency for each gene was determined to be acceptable in the
range between 90% and 110%.
CA 02864258 2014-09-15
The linear method described by Pffafl was used for relative
quantification (Pfaffl, MW (2001) A new mathematical model for
relative quantification in Real-time RT-PCR Nucleic Acids Res
29:... E45). This calculates the relative expression rate of a
gene based on the efficiency and the Ct difference in the sample
versus control. The Ct value of the gene of interest is
normalized to a reference gene, which in this case is the 18S
gene. The formula used for the relative quantification by Pfaffl,
shown below:
(E
ACT
target (control -sample)
Exchange Rate = _______________________________________________________
( e
ERf ,\ ACT Ref ( control -sample)
This equation takes into account the PCR efficiencies
obtained from calibration curves of both the target gene (Etarget)
that as the reference gene 18S (Ered, as well as the differences
of cT of the candidate genes, (Acttarget (control-sample)) between
control fish (no challenge ISA) and the sample (ISA under
challenge, belonging to one of four phenotypes challenged). The
same goes for the reference gene 18S (ACtref (control-muestra)).
The results obtained by the method were analyzed Pffafl and
plotted with the program Graphpad Prism 4 (Graphpad software,
CA 02864258 2014-09-15
51
Inc). From the results of analysis of RNA sequencing methodology
with Illumina best housekeeping gene was determined as the 18S
(which has less variation), so it was used as reference gene.
This is consistent with that reported by Pfaffl, 2004 (Pfaffl,
M.W. (2004). Quatification strategies in real time PCR. Chapter
3, pages 87-112).
The average efficiency corrected standardized expressions and
expressions relating will be determined by processing the data of
cT obtained for each condition and gene, in the program Q-gene
(Simon P. (2003). Q-Gene: processing quantitative real-time RT-
PCR data. Bioinformatics. 2003 Jul 22;19(11):1439-40).
An analysis of variance (ANOVA) one and two-way normalized
expression data was performed to find differences with this
control in the case of a road, and to analyze the effect of
factor phenotype, and their contribution to the variance of the
data in the case of two paths, that in each of the selected
genes. In addition, a posteriori Tukey test and Bonferroni were
performed to find differences between each of the groups
(phenotypes) of study.
To assess whether the genes under study can be used as
classifiers for phenotype markers and ROC curve analysis was
performed to determine genes with greater predictive power.
CD, 02864258 2014-09-15
52
Subsequently already selected the best predictors genes, a
discriminant analysis was performed with these, in order to
determine whether it is possible to classify within the
phenotypes under study, an individual was subjected to PCR
analysis in real time using using predictors genes. Also to
simplify the visualization and understanding of data, a principal
components analysis was also performed.
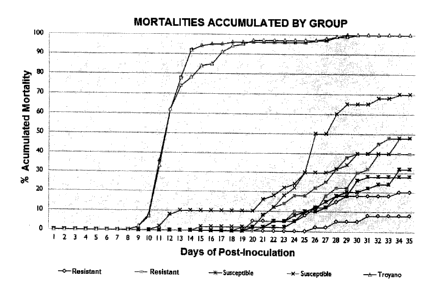
The results of the challenge test are shown in Figure 1 It
can be seen that Trojans fishes have the highest cumulative
mortality (100%) compared to the other study groups. The most
extreme values of each duplicate were selected for further
analysis by real-time PCR, which corresponds to a 70% cumulative
mortality for the susceptible phenotype and 40% for heavy duty.
The total RNA extracted from resistant and susceptible
individuals uninfected controls was
quantified
spectrophotometrically. The concentration range of the samples
varied between 0.8-3 g / 1. The purity of the sample in terms of
protein and organic solvents was determined by the ratio
A26onm/A2sonm, which ranged between 1.8 and 2.0 for protein and
reason A260runiA230 which varied between 1.95 and 2.5 for organic
solvents. Both reasons are within acceptable ranges. RNA
integrity was assessed by electrophoresis on 1% agarose gel, in
CD, 02864258 2014-09-15
53
Figure 2 the rRNA bands are seen 18S and 28S of samples
resistant, susceptible and control subjects.
In order to determine the experimental efficiency of each set
of Splitters, calibration curves were carried out from total RNA
of known concentration. The curves were prepared by plotting the
Ct versus log of the concentration of RNA in each point of the
curve (dilution). Each curve consists of a minimum of 5 points.
It was determined through the correlation coefficient points
behave linearly and that efficiencies and slopes are within
acceptable ranges (90 to 110% for efficiencies, -3.6 and -3.1 for
earrings). The results are detailed in Table 3.
Table 3. Slopes and correlation coefficient efficiencies obtained
from the calibration curve for each gene.
Gene Pending Correlation Eficiency (90-
Coef. 110%)
Annexin A-1 -3.4193 0.9839 96.09%
Cathepsin-S -3.4892 0.9695 93.65%
Gata 3 -3.4468 0.9459 95.04%
Grb2 -3.41 0.9926 96.45%
HSP90 -3.3956 0.9822 97.01%
IgM -3.3771 0.9762 97.75%
CA 02864258 2014-09-15
54
IgZ -3.5143 0.9621 92.55%
MAVS -3.3360 0.9892 99.42%
Tbet -3.1693 0.994 106.79%
Regul -3.3968 0.9985 96.97%
18S -3.5845 0.9956 90.10%
The method of real-time PCR with SYBR Green, possible to
carry out an analysis of dissociation of each of the amplicons,
allowing in turn to determine the melting temperatures of each
amplicon, and confirm the existence of a single product of PCR.
In Figure 3, the melting curves for PCR products of each of the
candidate genes, where the existence of single PCR product is
confirmed whose size was confirmed by agarose gel.
In order to evaluate the expression levels of candidate genes
and their relationship to immune response developed in resistant
and susceptible to ISA virus fish anterior kidney samples from
moribund fish or day 21 post infection were analyzed. . This
sampling time is selected based on the modulation of an immune
response and the incubation period described for the experimental
induction of ISA (Joseph, T., Kibenge, M. T. y Kibenge, F. S. B.
Antibody-mediated growth of infectious salmon anaemia virus in
macrophage-like fish cell lines. J Gen Virol. 2003. 84:1701-
.
CA 02864258 2014-09-15
1710.; Totland GK, Hjeitnes BK y Flood PR. Transmission of
infectious salmon anaemia (ISA) through natural secretions and
excretions from infected smelts of Atlantic salmon (Salmo salar)
during their presymptomatic phase. 1996. Dis Aquat Org 26:25-31).
The genes will be presented according to their expression
profile. Grb2 IgM and show a similar expression profile, wherein
in the susceptible phenotypes have low expression compared to the
control are statistically significant (p < 0,001). For the
susceptible phenotype Grb2 gene expression has a 49 fold lower
than the control, respectively (p < 0,001).
For the Annexin gene individuals from the susceptible and
resistant phenotypes significantly different from the control,
with decreases in the expression of 32 and 27 times respectively.
In the case of Regul a significant overexpression of the gene
is observed in subjects with increases of 57 times for the
susceptible phenotype and 54 times for the resistant phenotype.
As for Cathepsin, IgZ, MAVS Tbet Gata3 and genes, have a
similar expression profile comprising expression susceptible low
compared with the control phenotype of 36.5, 23, 33, 43 and 39
times respectively.
In order to explain the variance of the data and the
expression profile of each gene two-way ANOVA was performed,
CA 02864258 2014-09-15
56
taking into account factors phenotype. This analysis shows the
percentage contribution in the variability of the data. The
results are shown in Table 4, showing high rates with low
interaction between factors. Factor for the phenotype, 2 genes
are those that contribute most to the variability; Grb2 with
88.62% of a 83.02% IgM. The genes mentioned above also have a low
level of interaction (10% less), so that each factor separately
presents greater explanatory power.
Table 4. Results of the analysis ANOVA Two Way phenotype
factors. High percentages in the contribution of data
variability, attributable to a single factor is shown in bold.
(*) Denotes significant difference (*p < 0,05; **p < 0,01; ***p <
0,001).
CA 02864258 2014-09-15
57
Gene %Interaction % Phenotype
Annexin 8.44 *** 1.82 *
Cathepsin 22.16 *** 2.87 *
Gata 3 41.9 *** 8.07 ***
Grb2 5.22 ** 88.62 ***
HSP90 37.15 * 7.67 (ns)
1
IgM 8.78 *** 83.02 ***
igz 7.8 *** 0.95 *
MAVS 26.72 *** 40.64 ***
Tbet 41.31 *** 8.04 ***
Regul 1.34 * 0.01 (ns)
In order to find genes that may serve as better predictors of
phenotype, ROC curve analysis with raw data normalized relative
expression was performed, this in susceptible and resistant
phenotypes. The results are shown in Table 5 5 genes which have
100% sensitivity and specificity, resulting in the absence of
false negatives or positives are observed. These genes are Gata3,
Grb2, IgM, MAVS and Regul. These genes loom as the top candidates
CA 02864258 2014-09-15
58
to be used as predictors of phenotype and were used to perform a
discriminant analysis to test its predictive power.
Table 5. Results of ROC curve analysis. Shown in bold italics,
genes found to be the most suitable to be used as predictors of
phenotype
Cutoff Sensi- Speci- True True False False
value bility ficity
AUC (>-LOG) (%) (%) Posi Negat negat Posit
tive ive ive ive
Annexin 0.98 15.6902 100 88.89 88.9 100 11.1 0
6
Catheps 0.54 14.9695 87.5 22.22 50 66.67 50 33.33
in 2
Gata 3 1 19.814 100 100 100 100 0
0
Grb2 1 15.0971 100 100 100 100 0 0
HSP90 0.65 4.1932 100 33.33 57.1 100 42.9 0
3
IgM 1 12.5262 100 100 100 100 0 0
IgZ 0.83 12.3401 66.67 100 100 75 0 25
3
MAVS 1 21.8213 100 100 100 100 0 0
CA 02864258 2014-09-15
59
Tbet 0.61 9.1247 100 55.56 66.7 100 33.3 0
1
Regul 1 =22.2873 100 /00 /00 /00 0 0
In order to test the predictive power of the genes thrown by
the ROC curve analysis, (Gata3, Grb2, IgM, and Regul MAVS), a
discriminant analysis was performed. This one in the first
instance, was performed taking into account two phenotypes, which
could discriminate 100% resistant to susceptible individuals. The
summary of results in Table 6 are shown below.
Table 6. Discriminant analysis used Gata3, Grb2, IgM, and
Regul MAVS genes as predictors. It is observed that the 9
individuals are correctly classified with a rate corresponding to
1, which is equivalent to 100% correct classification.
Classification Table
Group Prediction by Group (Std) Correctly
classified
Susceptible Resistant
Susceptible 9 0 1.000
Resistant 0 9 1.000
Correct Global Rate 1.000
CD, 02864258 2014-09-15
This paper was able to establish that there are differences
in the expression of genes involved in innate and adaptive immune
responses. between resistant and susceptible to ISA virus
phenotypes by relative quantification of transcripts by real-time
PCR.
With respect to the technique used (real time PCR). one can
mention that has several important factors to be considered for a
reliable quantification. One factor to consider is the integrity
and quality of RNA. which has a chemical nature unstable and is
susceptible to hydrolysis by base catalysis or RNase own tissues
(Houseley JM y Tollervey D. The many pathways of RNA
degradation. 2009 Cell. 136: 763-776;; DeRose. V.J. Two decades
of RNA catalysis. 2002. Chem. Biol. 9: 961-969.). The effect of
the degradation of RNA on the real time PCR results in less
reproducible and more late Ct values (Fleige S. y Pfaffl M.W. RNA
integrity and the effect on the real-time qRT-PCR performance.
2006.Mol. Aspects Med. 27:126-139.).). As seen in Figure 2. the
RNA does not have significant degradation and can therefore be
used for further analysis without making errors in the relative
quantification of the candidate genes.
The presence of a single PCR product was verified by analysis
of the melting curves. The fluorophore SYBR Green is 25 to 100
CD, 02864258 2014-09-15
61
times more sensitive than ethidium bromide (Schneeberger C.
Speiser P. Kury F and Zeillinger R. Quantitative detection of
reverse transcriptase-PCR products by Means of a novel and
sensitive DNA stain. 1995. PCR Methods Appl 4: 234-238). and
therefore more likely to detect unspecific products by analyzing
the melting curves, which by electrophoresis on agarose gel
stained with ethidium bromide. As seen in Figure 3. the melting
curves show a single PCR product. although the products were also
analyzed by agarose gel electrophoresis to verify the occurrence
of the expected size of band.
One aspect to consider when evaluating gene expression levels
in biological samples from the same species and treatment. the
interindividual variation, which in many cases prevents
statistically significant differences are found between
experimental groups. One way to overcome the bias introduced by
heterogeneity is to increase the sample size. and thus deliver a
greater statistical weight to the results. In the case of this
thesis interindividual variations not desired can occur due to
differences in the effectiveness of the infection by the pathogen
or genetic differences in the immune response of the individual.
However, large sample size (n = 9) in order to reduce bias was
employed.
CD. 02864258 2014-09-15
62
As for the relative quantification of gene expression
profiles are observed like in some genes for the phenotypes being
studied. Two genes that show a similar expression pattern are IgM
and Grb2. In these. a decrease is observed in susceptible
phenotypes compared to resistant and control. IgM is the dominant
immunoglobulin in salmon and a first line of defense (Bengten E.
Wilson M. Miller N. Clem LW. Pilstrom L and Warr GW.
Immunoglobulin isotypes: structure. function. and genetics. Curr
Top Microbiol Immunol. 2000. 248:189-219). however this
immunoglobulin turns out to be of little specificness (Hordvik
I.. Lindstrom C.D.. Voie A.M.. Jacob A.L.J and Endresen C.
Structure and organization of the immunoglobulin M heavy chain
genes in Atlantic salmon. Salmo salar. 1997. Molecular
Immunology. 34. 631-639) and with few neutralizing antibodies.
but despite this the importance lie in the recognition and
binding to the virus for containment and complement activation.
which increases the number of neutralizing antibodies. as well as
B lymphocytes with what is promotes activation of adaptive
immunity, and therefore the effectiveness of the system to
counteract the virus (Olesen NJ. y Jsrgensen PEV. Quantification
of serum immunoglobulin in rainbow trout Salmo gairdneri under
various environmental conditions. 1986. Dis Aquat Org 1. 183-189;
CA 02864258 2014-09-15
1.
63
Boes. M. Role of natural and immune IgM antibodies in immune
responses. 2000. Molecular Immunolology 37:1141-1149). Thus. the
subject to have low levels of IgM expression individuals would
delay the activity of the immune system and present a low
activity and this reaction.
Moreover Grb2 is an adapter molecule related to the antigen-
activated signaling both T (TCR) and B lymphocytes (BCR)
receptors. This signaling both development and lymphocyte
activation is regulated both. Antigen stimulation to occur. the
activated receptor tyrosine kinases of the Src family, resulting
in phosphorylation of several substrates and multiple
signalosomas assembly. The coordination of signaling components
in signalosomas. start the corresponding signaling cascades that
control transcription and thereby cell fate. activation or other
biological processes (Samelson. L. E. Signal transduction
mediated by the T cell antigen receptor: the role of adapter
proteins. 2000. Annu. Rev. Immunol. 20:371-394). To counteract
antigen stimulation. the antigen-activated receptor as negative
regulators must recruit tyrosine kinases and phosphatases
negative in signalosomas (Shaw A yThomas ML. Coordinate
interactions of protein tyrosine kinases and protein tyrosine
phosphatases in T-cell receptor- mediated signalling. 1991.-Curr
CA 02864258 2014-09-15
64
Opin Cell Biol.;3:862-868). The balance of positive and negative
signals are essential for proper development, activation and
induction of lymphocytes. Grb2 described which is essential in
the activation of Ras-MAPK pathway and the regulation of MAPKs.
JNK and p38 which are involved in both positive and negative
selection of T cells (Houtman JC. Yamaguchi H. Barda Saad M.
Braiman A. Bowden B. Appella E. Schuck P y Samelson LE.
Oligomerization of sig- naling complexes by the multipoint
binding of GRB2 to both LAT and SOS1.2006. Nat Struct Mol Biol
13:798-805). This regulation occurs at the beginning of the
cascade triggered by TCR. It has also been described which would
be involved in the regulation of signaling for positive selection
of T cells CD4+ y CD8+ (Jang IK.. Zhang J.. Gu H. (2009) Grb2. a
simple adapter with complex roles in lymphocyte development.
function. and signaling. Inmunol Rev 232; 150-159).B lymphocytes
have been reported to both positively and negatively regulate the
signaling of the BCR and also regulate Ca2 + signaling (Stork B.
Grb2 and the non-T cell activation linker NTAL Constitute a Ca (2
+) - regulating signal circuit in B lym- phocytes.2004 Immunity.
21:. 681-691). The low expression of the Grb2 gene in individuals
susceptible phenotype would realize a reduction in the activation
of receptor-mediated immune response of antigen. generating this
CD. 02864258 2014-09-15
lower regulation in selection and development of T and B
lymphocytes. the opposite occurs in individuals with resistant
phenotype. The differential expression of this gene in
susceptible and resistant phenotypes suggests that the induction
of T and B lymphocytes would be associated with ISA virus
resistance.
Annexin while significant differences with control
individuals of both phenotypes (susceptible and resistant).
showing a lower expression of the gene. There could be the
possibility that the virus has an inhibitory effect on this gene
as described that some viruses have an immunosuppressive effect
on some genes (Ronneseth A. Wergeland HI. Pettersen EF y
Neutrophils and B-cells in Atlantic cod (Gadus morhua L.). 2007.
Fish Shellfish Immunol. 3:493-503).
Regul has a high expression of this in individuals. Regul is
a negative regulator of G protein as it accelerates the
hydrolysis of GTP to the G protein activated subunits This
inhibitory effect induced by chemokines acting on signals. which
have an important role in the movement of immune cells to sites
of inflammation . Described that this gene is highly expressed in
cells, suggesting a role in the regulation of the immune cell.
Also described is a strong relationship between the expression of
CA 02864258 2014-09-15
66
Regul and B cell migration (C. Moratz. J. R. Hayman. H. Gu. and
J. H. Kehrl. "Abnormal B-cell responses to chemokines. disturbed
plasma cell localization, and distorted immune tissue
architecture in RGS1-/- mice." Molecular and Cellular Biology.
vol. 24. no. 13. pp. 5767-5775. 2004). Inhibition by Regul
results in reducing the effects of chemokines and decreased
immune cell migration. which eventually creates a vacuum in the
immune response.
The following five genes have a similar expression pattern in
which individuals have a resistant phenotype similar to the
control with no significant differences in individual expression.
whereas susceptible individuals have low gene expression compared
to control.
Cathepsin S is a cysteine protease that plays a critical role
in antigen processing and presentation. Described that Cathepsin
S is essential for antigen presentation by MHC-II and its
inhibition leads to a reduced response of T lymphocytes CD4+
(Gupta S. Dennis J. Thurman RE. Kingston R. Stamatoyannopoulos
JA. Predicting Human Nucleosome Occupancy from Primary Sequence.
2008. PLoS Comput Biol 4:1-9). Also. its deficiency has been
linked to low mobility of antigen presenting cells and B cells
(Faure-Andre G. Vargas P. Yuseff MI. Heuze M. Diaz J. Lankar D.
ak 02864258 2014-09-15
1.
67
Steri V. Manry J. Hugues S. Vascotto F. Boulanger J. Raposo G.
Bono MR. Rosemblatt M. Piel M. Lennon-Dumenil AM. Regulation of
dendritic cell migration by CD74. the MHC class II-associated
invariant chain. 2008. Science. 322:1705-1710. The susceptible
phenotype has low expression of the gene. which could present
problems in the antigen presentation and immune cell mobility.
affecting mainly with this acquired immunity. This effect may
contribute to the susceptibility of individuals in this
phenotype.
Mays is an adapter molecule that acts after the senses
helicase RIG-I viral RNA by activating a signaling cascade
ultimately leading to the activation of expression of type I IFN
generating other signaling cascades that lead to activated
transcription of interferon (ISGs) in which genes are those
encoding proteins with antiviral function. In the case of salmon.
it may be mentioned to Mx-1 which is capable of protecting cells
by preventing the replication of IPNV. (Larsen. R.. Rokenes. T..
Robertsen. B. (2004). Inhibition of infectious pancreatic
necrosis virus replication by atlantic salmon MX1 protein. J
virol 78. 7938-7944). In this way individuals with low expression
of Mays will present less antiviral activity to be inhibited
CD. 02864258 2014-09-15
68
signaling cascade associated with IFN. which would contribute to
the susceptibility phenotype.
IgZ is an immunoglobulin recently discovered in zebrafish
which has not much information about their biological function.
It has been recently reported to be associated with mucosal
immunity and neutralizing antigens in the blood
(Ryo S. Wijdeven RH. Tyagi A. Hermsen T. Kono T. Karunasagar
I. Rombout JH. Sakai M. Verburg-van Kemenade BM y Sayan R. Common
carp have two subclasses of bonyfish specific antibody IgZ
showing differential expression in response to infection. 2010.
Dev Comp Immunol.
34:1183-1190). This immunoglobulin is
expressed in smaller amounts than IgM. but would help in
neutralizing the virus and signaling for the activation of the
immune system. Because individuals with susceptible phenotype
have low gene expression, one could infer that have lower virus
neutralization and hence further progress of the disease.
T-bet and Gata3 are antagonistic transcription factors
involved in cell fate commitment and T. which eventually lead to
the development of Thl and Th2 lymphocytes. T-bet promotes
differentiation into Thl cells. which protect against
intracellular parasites and viruses causing factors such as IFN.
IL-2 and TNF which will induce activation of macrophages and NK
CA 02864258 2014-09-15
69
cells, production of cytotoxic T lymphocytes (CTL) and complement
activation, generating a response primarily of cell type (Jenner
RG. Townsend NJ. Jackson I. Sun K. Bouwman RD. Young RA. Glimcher
LH y Lord GM. The transcription factors T-bet and GATA-3 control
alternative pathways of T-cell differentiation through a shared
set of target genes. 2009. Proc Natl Acad Sci U S A. 106:17876-
17881). Gata3 on another part promotes differentiation towards
Th2 lymphocytes. which by action of interleukins as IL-4. IL-5.
IL-6 and IL-13 produce effects such as activating antibody-
mediated immunity. eosinophil differentiation. inflammation
processes and elimination of macrophages. primarily creating a
humoral response. The effectors of both pathways have an
inhibitory effect on the other. this destination polarizing
towards the Thl or Th2 lymphocytes. IFN-y inhibits Th2
polarization while IL-4 does so with Thl (Constant SL y Bottomly
K. Induction of Thl and Th2 CD4+ T cell responses the alternative
approaches. 1997. Annu Rev Immunol. 15:297-322). Thus as can be
seen in the graphs of Figure 5. in susceptible individuals, it
was observed that both channels are decreased or inhibited which
realizes differentiation and polarization of T lymphocytes are
not occur or do so measuring less compared to resistant
CA 02864258 2014-09-15
individuals or controls. For the same reason the susceptibility
of individuals is explained.
The ROC curve analysis was employed using data from gross
normalized relative expression of resistant and susceptible
individuals with the aim of finding genes that could
differentiate the two phenotypes prediction mode and that they
possess a high level of sensitivity and specificity, resulted in
a area under the curve (AUC) equal to or close to 1 When
analyzing 5 are genes having an AUC of 1. where these Gata3.
Grb2. IgM. Mays and Regul. presenting 100% success. For this
reason these genes were selected for discriminant analysis. which
also could discriminate both phenotypes perfectly. checking the
predictive power of the ROC curve thrown genes. Comparing thrown
by the ROC curve analysis with results of two-way ANOVA genes. we
noticed that Grb2. IgM and Regul genes have significant
percentages of the variation in the data. whether this be
associated with the effect of the phenotype in the case IgM and
Grb2. because this changes only affect the susceptible phenotype
so statistically decreases their influence, resulting in smaller
percentages.