Note: Descriptions are shown in the official language in which they were submitted.
DEMANDES OU BREVETS VOLUMINEUX
LA PRESENTE PARTIE DE CETTE DEMANDE OU CE BREVETS
COMPREND PLUS D'UN TOME.
CECI EST LE TOME 1 DE 2
NOTE: Pour les tomes additionels, veillez contacter le Bureau Canadien des
Brevets.
JUMBO APPLICATIONS / PATENTS
THIS SECTION OF THE APPLICATION / PATENT CONTAINS MORE
THAN ONE VOLUME.
THIS IS VOLUME 1 OF 2
NOTE: For additional volumes please contact the Canadian Patent Office.
CA 02876864 2015-01-08
Description
METHODS FOR ELIMINATING MANNOSYLPHOS-
PHORYLATION OF GLYCANS IN TkII PRODUCTION OF
GLYCOPROTEINS
[1] (Cancelled.)
STATEMENT AS TO FEDERALLY SPONSORED RESEARCH
[2] This invention was funded, at least in part, under a grant from the
Department of
Commerce, N1ST-ATP Cooperative Agreement Number 70NANB2H3046. The
United States government may therefore have certain rights in this invention.
FIELD OF THE INVENTION
[3] The present invention relates to the elimination of mannosylphosphate
transfer on .
glycans of glycoproteins, and further relates to eliminating genes responsible
for the
addition of mannosylphosphate residues on glycans in yeast and filamentous
fungal
cells. In particular, the invention relates to engineering yeast and
filamentous fungal
host cells to produce glycans without naannosylphosphate residues.
Background of the Invention
[4] The ability to produce recoinbinant human proteins has led to major
advances in
hi-man health care and remains an active area of drug discovery. Many
therapeutic =
proteins require the cotranslational addition of glycans to specific
asparagine residues (
N-glycosylation) of the protein to ensure proper structure-function activity
and
subsequent stability in human serum. For therapeutic use in humans,
glycoproteins
require human-like N-glycosylation. Mammalian cell lines (Chinese hamster
ovary
(CHO) cells as well as human retinal cells) which can mimic human-like
glycoprotein
processing have several drawbacks including low protein titers, long
fermentation
times, heterogeneous products, and ongoing viral containment issues. Thus, the
use of
yeast and filamentous fungal expression systems having more economical
processing,
fewer safety obstacles and producing more robust heterologous protein yields
have
been heavily researched as host cells for human therapeutics.
[5] In yeast and filamentous fungus, glycoproteins are produced having
oligosaccharides which are different from those of mammalian-derived
glycoproteins.
Specifically in yeast, outer chain oligosaccharides are hypermannosylated
consisting of
30-150 mannose residues (Kukuruzinska et al., 1987, Anna. Rev. Biochem. 56:
915-944). Moreover, mannosylphosphate is often transferred to both the core
and outer
sugar chains of glycoproteins produced in yeast (Ballou, 1990, Methods
Enzymol. 185:
440-470). Of most consequence, is that these mannosylphosphorylated glycans
from
glycoproteins produced in the yeast, Saccharomyces cerevisiae, have been shown
to
CA 0287 6864 2015- 01- 08
2
=
illicit an immune response in rabbits (Rosenfeld and Ballou, 1974, JBC, 249:
2319-2321). Thus, the elimination of mannosylphosphorylation in yeast and
filamentous fungi is essential for the production of non-immunogenic
therapeutic gly-
coproteins.
[6] In .S cerevisiae there are at least two genes which participate in the
transfer of man-
nosylphosphate. The two genes, MNN4 and MNN6 have been cloned, and arinlyses
of
the gene product suggest they function in the transfer of mannosylphosphate
(for
review see Iipmi and Ofilni, 1999, Biochim. Biophys. Acta, 1426: 333-345).
MNN6
encodes a type IL membrane protein homologous to the Kre2p/Mntlp family of
proteins which has been characterized as Golgi aL1,2-mannosyl-transferases
involved
in 0-mannosylation and N-glycosylation (Lnssier et al., 1997,1BC, 272:
_ _
15527-15531). The Amnn6 mutant does nOt show a defect in the mannosylphos-
phorylation of the core glycans in vivo, but exhibits a decrease
inmannosyiphosphate
transferase activity in vitro (Wang at al, 1997, JBC, 272: 18117-18124). Mnn4p
is
also a putative type 11 membrane protein which is 33% identical to the S.
cerevisiae
Yjr061p (Odani at al., 1996, Glycobiology,6: 805-810; Bunter and Plowman,
1997,
Trends in Biochem; Sci., 22:18-22). Both the Amnn6 and Amnn4 mutants decrease
the
_
transfer of mannosylphosphate. However, the Amnn6 Amnn4 double mutant does not
further reduce this activity. These observations suggest the presence of
Pdrittional man-
nosyltransferases that add mannosylphosphate to the core glycans.
[7] Thus, despite the reduction of mannosylphosphorylation in S. cerevisiae
with the
disruption of MNN4,101N6 or both in combination, there is no evidence that
complete
*Ftlirninnfion of mannosylphosphate transferase activity is possible. Other
genes which
affect the mannosylphosphate levels have been identified in S. cerevisiae.
These genes
include P212.1, VRG4, M7N2 and MNN.5. PMR1 encodes a Golgi-localized Ca2+/Mn2+
-
AlTase required for the normal function of the Golgi apparatus (Antebi and
Fink,
1992, Mol. Biol. Cell, 3: 633-654); Vrg4p is involved in nucleotide-sugar
transport in
the Golgi (Dean at al.,1997,113C, 272: 31908-31914), and Mmap and Mnn5n are
.4,2-marmosy1transferases responsible for the initiation of branching in the
outer chain
of N.4 inlrf-d glyrans (Rayner and Munro, 1998, JBC, 273: 23836-23843). For
all four
proteins, the reduction in nazanosylphosphate groups attached to N-liniced
glycans
seems to be a consequence of Golgi malfunction or a reduction in size of the N-
linked
glycans rather than a specific defect in the transfer activity of the
mannosylphosphate
groups.
[8] Proteins expressed in the methylotrophic yeast, Pichia pastoris contPin
mamm-
sylphosphorylated glycans (Miele, etal., 1997, Biotech. Appl Biochem., 2: 79-
83).
Miula at al. reported the identification of the PND1 (Phosphorylmannosylation
of N -
finked Oligosaccharides) gene which upon disruption confers an attenuation of
manno-
sylphosphorylation on glycoproteins (WO 01/88143; Miura et al., 2004, Gene,
324:
129-137). The PNO1 gene encodes for a protein involved in the transfer of
manno-
CA 02876864 2015-01-08
3
sylphosphate to glycans in P. pastoris. Its specific function, however, is
unknown. As
mentioned, the Apnol mutant decreases but does not abolish
mannosylphosphorylation on
N-glycans relative to a P. pastoris strain having wild-type Pnolp.
[9] Currently, no methods exist to eliminate mannosylphosphorylation on
glycoproteins produced in fungal hosts. A residual amount of
mannosylphosphorylation on
glycoproteins may still be immunogenic and, thus, is undesirable for use as
human
therapeutics.
[10] What is needed, therefore, is an expression system based on yeast or
filamentous fungi that produces glycoproteins which are essentially free of
mannosylphosphorylated glycans.
SUMMARY OF THE INVENTION
[11] The present invention provides a method for eliminating
mannosylphosphate
residues on glycans of glycoproteins in a yeast or filamentous fungal host
(e.g., P. pastoris).
The present invention also provides a fungal host which normally produces
mannosylphosphorylated glycoproteins or a fraction thereof, in which the
fungal host is
modified to produce glycoproteins essentially free of mannosylphosphate
residues. In one
embodiment, the present invention provides a null mutant lacking one or more
genes
homologous to MNN4. In a preferred embodiment, the present invention provides
a host of
the genus Pichia comprising a disruption, deletion or mutation of rnnn4B and
pno 1 . The
resulting host strain is essentially free of mannosylphosphorylation on
glycans of
glycoproteins. According to another embodiment, there is provided a fungal
host comprising
a combined disruption, deletion or mutation of the MNN4B and PNOI genes
wherein the
fungal host is Pichia pastoris and is capable of producing less than 1%
mannosylphosphorylated glycoproteins of total N-glycans.
[12] The present invention further provides glycoprotein compositions that
are
essentially free of mannosylphosphorylated glycoproteins. Such glycoprotein
CA 02876864 2015-01-08
3a
compositions comprise complex N-glycans that may be used for therapeutic
applications.
[13] The present invention also provides isolated polynucleotides
comprising or
consisting of nucleic acid sequences selected from the group consisting of the
coding
sequences of the P. pastoris MNN4A, MNN4B and MNNC; nucleic acid sequences
that are
degenerate variants of these sequences; and related nucleic acid sequences and
fragments.
The invention also provides isolated polypeptides comprising or consisting of
polypeptide
sequences selected from the group consisting of sequences encoded by the P.
pastoris
MNN4A, MNN4B, MNN4C; related polypeptide sequences, fragments and fusions.
Antibodies that specifically bind to the isolated polypeptides of the
invention are also
provided.
[14] The present invention also provides host cells comprising a
disruption, deletion
or mutation of a nucleic acid sequence selected from the group consisting of
the coding
sequence of the P. pastoris MN1\T4A, MNN4B and MA/NC gene, a nucleic acid
sequence that is
a degenerate variant of the coding sequence of the P. pastoris MNN4A, MNN4B
and MNNC
gene and related nucleic acid sequences and fragments, in which the host cells
have a reduced
activity of the polypeptide encoded by the nucleic acid sequence compared to a
host cell
without the disruption, deletion or mutation.
[14a] In one aspect, the invention provides a fungal host cell
comprising a disruption,
deletion or mutation of the MNN4B (mannosyltransferase 4B) and PNO1
(phosphomarmosylation of N-linked oligosaccharides 1) genes, wherein said
disruption,
deletion or mutation reduces the activity of said MNN4B and PN01, wherein the
fungal host
cell is Pichia pastoris and is capable of producing less than 1%
mannosylphosphorylated
glycoproteins of total N-glycans, and wherein the MNN4B gene to be disrupted,
deleted or
mutated encodes a messenger RNA that corresponds to a cDNA comprising or
consisting of a
nucleic acid sequence selected from the group consisting of (a) SEQ ID NO:3;
(b) a nucleic
acid sequence that is a degenerate variant of SEQ. ID NO:3; (c) a nucleic acid
sequence at
least 90% identical to SEQ ID NO:3; (d) a nucleic acid sequence that encodes a
polypeptide
comprising the amino acid sequence of SEQ ID NO:4; and, (e) a nucleic acid
sequence that
encodes a polypeptide at least 90% identical to SEQ ID NO:4.
CA 02876864 2015-01-08
3b
[14b] In another aspect, the invention provides a method for producing
glycoprotein
compositions in the host cell as described herein comprising propagating said
host cell and
isolating the glycoprotein products.
[14c] In another aspect, the invention provides a glycoprotein produced by
the
method as described herein, wherein the glycoprotein is selected from the
group consisting of
kringle domains of human plasminogen, erythropoietin, cytokines, soluble IgE
receptor alpha-
chain, IgG, IgG fragments, IgM, urokinase, chymase, urea trypsin inhibitor,
IGF-binding
protein, epidermal growth factor, growth hormone-releasing factor, annexin V
fusion protein,
angiostatin, vascular endothelial growth factor-2, myeloid progenitor
inhibitory factor-1,
osteoprotegerin, alpha-1 antitrypsin, DNase II, alpha-fetoproteins, FSH and
peptide hormones,
wherein the glycoprotein comprises less than 1% mannosylphosphorylated
glycoproteins of
total N-glycans.
[14d] In another aspect, the invention provides a modified Pichia host cell
characterized in that the host cell does not produce the gene products
comprising the amino
acid sequence of SEQ ID NO:2, SEQ ID NO:4 and of PN01, as a result of
disruption,
deletion, or mutation of nucleic acid sequences encoding the gene products,
and thus lacks the
mannosylphosphate transferase activity of the gene products.
CA 02876864 2015-01-08
4
BRIEF DESCRIPTION OF THE DRAWINGS
[15] Figure 1. depicts the nucleic acid and amino add sequence of?.
pastoris IONN4A.
[16] Figure 2. depicts the nucleic acid and amino acid sequence of?.
pastoris Aill11N4B.
[17] Figure 3. depicts the nucleic acid and amino acid sequence of?.
pastoris M2TN4C.
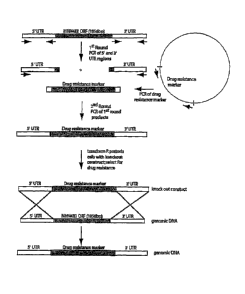
[18] Figure 4. illustrates the fusion PCR knock-out strategy of?. pastoris
2&N4B
using a drug resistance marker.
[19] Figure 5A. shows a high performance liquid chrom,gtogram for the
negative ex-
perimental control using 1120 as the sample. B. shows a high performance
liquid
chromatogram for the sample containing N-linked glycans from 1C3 purified from
P.
pastoris YSH-44 supernatant Glycans with mannosylphosphate elute between 20 -
30
-mins. C. shows a high performance liquid chromatogram for a sample containing
N -
hinkfml glycans from 1(3 purified from P. pastoris YSH-49 (Apno 1)
supernatant.
Glycans with maimosylphosphate elute between 20 - 30 rains. D. shows a high
performance liquid chromatogram for a sample,contRining N-linked glycans from
1(3
, purified from P. pastoris YAS-130 (Apno Amnn4B) supernatant. Note the
absence of
mannosylphosphorylaind glycans between 20 and 30 mins.
[20] Figure 6A. shows a highperfonnance liquid chromatogram for the sample
containing N-iinirefi glycans from 1(3 purified from P. pastoris YSH-1 (dochl)
su-
pernatant. Glycans with mannosylphospirte elite between 20 - 30 mink-B. shows
a
high performance liquid chromatogram for a sample containing N-linkfid glycans
from.
1(3 purified from P. pastoris YAS-164 (dochl dmnn4A Apno 1) supernatant.
Glycans with mannosylphosphate elute between 20 - 30 rains. C. shows a high
performance liquid chromatogram for g sample. co-ntRining N-linked glycans
from 1(3
purified from P. pastoris YAS-174 (dochl Amnn4A Apno 1 Amnn4B) supernatant.
Note the absence of marmosyiphosphorylated glycans between 20 and 30 rains.
[21] Figure 7A. shows a high performance liquid chromatogram for the
negative ex-
perimental control sample containing H20 B. shows a high performance liquid
chromatogram for the sample containing N-1inlred glycans from erythropoietin
expressed from pBK291 (His-FPO) produced in P. pastoris strain BK248 C. shows
a
high performance liquid chromatogram for the sample containing N-iinked
glycans
from His-EPO produced in P. pastoris strain BK244 I). shows a high performance
liquid chromatogram for the sample containing N-3in1red glycans from 0D40
expressed
from 111-C33 (His-CD40) produced in?. pastoris strain YJC12 E. shows a high
performance liquid chromatogram for the YAS252. Note: Glycans with manno-
sylphosphate elute between 20 - 30 mins
[22] Figure 8 A. shows a high performance liquid chromatogram for the
sample
containing N-linked glycan from invertase expressed from pPB147 produced in?.
pastoris strain YAS252.
[23] Figure 9
[24] shows an alignment of 16,7N4IPNO1 homologs in?. pastoris (Pp), S.
cerevisiae
CA 02876864 2015-01-08
(Sc), Neurospora crassa (Nc), Aspergillus nidulans (An), Candida albi cans
(Ca) and
Pichia angusta (Hansenula polYmorpha) (Pa) using Clustal W from DNAStar.
DETAILED DESCRIPTION OF Ilik: INVENTION
[25] Unless otherwise defined herein, scientific and technical terms
used in connection
with the present invention shall have the meanings that are commonly
understood by
those of ordinary skill in the art. Further, unless otherwise required by
context, singular
terms shall include the plural and plural terms shall include the singular.
Generally,
nomenclatures used in connection with, and techniques of biochemistry,
enzymology,
molecular and cellular biology, microbiology, genetics and protein and nucleic
acid
chemistry and hybridization described herein are those well known and commonly
used in the art: The methods and techniques of the present invention are
generally
performed according to conventional methods well known in the art and as
described
in various general and more specific references that are cited and discussed
throughout
the present specification unless otherwise indicated. See, e.g., Sambrook et
al.
. = ' Molecular Cloning: A Laboratory Manual, 2d ed., Cold Spring Harbor
Laboratory
Press, Cold Spring Harbor, N.Y. (1989); Ausubel et al., Current Protocols in
Molecular Biology, Greene Publishing Associates (1992, and Supplements to
2002);
. Harlow and Lane, Antibodies: A Laboratory Manual, Cold Spring Harbor
Laboratory
= Press, Cold
Spring Harbor, N.Y. (1990); Taylor and Drickamer, Introduction to Gly-
=:!==
= cobiology, Oxford Univ. Press (2003); Worthington Enzyme Manual,
Worthington
Biochemical Corp., Freehold, NJ; Handbook ofBiochemistry: Section A Proteins,
Vol =
= I, CRC Press (1976); Handbook of Biochemistry: Section A Proteins, Vol
11, CRC
Press (1976); Essentials of Glycobiology, Cold Spring Harbor Laboratory Press
(199.9). .
[26] All publications, patents and other references mentioned herein are
hereby in-
corporated-by reference in their entireties.
[27] The following terms, unless otherwise indicated, shall be
understood to have the = =
following meanings:
[28] The term tpolynucleotide or 'nucleic acid molecule' refers to a
polymeric form of
nucleotides of at least 10 bases in length. The term includes DNA molecules
(e.g.,
cDNA or genomic or synthetic DNA) and RNA molecules (e.g., mRNA or synthetic
RNA), as well as analogs of DNA or RNA containing non-natural nucleotide
analogs,
non-native intemucleoside bonds, or both. The nucleic acid can be in any
topological
conformation. For instance, the nucleic acid can be single-stranded, double-
stranded,
triple-stranded, quadruplexed, partially double-stranded, branched,
hairpinned,
circular, or in a padlocked conformation.
[29] Unless otherwise indicated, a 'nucleic acid comprising SEQ ID NO:X'
refers to a
nucleic acid, at least a portion of which has either (i) the sequence of SEQ
ID NO:X,
or (ii) a sequence complementary to SEQ ID NO:X. The choice between the two is
dictated by the context. For instance, if the nucleic acid is used as a probe,
the choice
between the two is dictated by the requirement that the probe be complementary
to the
CA 02876864 2015-01-08
6
desired target.
[30] An 'isolated' or 'substantially pure' nucleic acid or polynucleotide
(e.g., an RNA,
DNA or a mixed polymer) is one which is substantially separated from other
celhilar
components that naturally accompany the native polynucleotide in its natural
host cell,
e.g., ribosomes, polymerases and genomic sequences with which it is naturally
associated. The term embraces a nucleic acid or polynucleotide that (1) has
been
removed from its naturally occurring environment, (2) is not associated with
all or a
portion of a polynucleotide in which the 'isolated polynucleotide' is found in
nature, (3)
is operatively linked to a polynucleotide which it is not linked to in nature,
or (4) does
not occur in nature. The term 'isolated' or 'substantially pure' also can be
used in
reference to recombinant or cloned DNA isolates, chemically synthesized
polynucleotide analogs, or polynucleotide analogs that are biologically
synthesized by.
heterologous systems.
[31] However, 'isolated' does not necessarily require that the nucleic acid
or =
polynucleotide so described has itself been physically removed from its native
en-
vironment For instance, an endogenous nucleic acid sequence in the genome of
an
organism is deemed 'isolated' herein if a heterologous sequence is placed
adjacent to
= . the endogenous nucleic acid sequence, such that the expression of
this endogenous
= nucleic acid sequence is altered. In this context, a heterologous
sequence is a sequence
that is not naturally adjacent to the endogenous nucleic acid sequence,
whether or not =
the heterologous sequence is itself endogenous (originating from the same host
cell or
= progeny thereof) or exogenous (originating from a different host cell or
progeny
= thereof). By way of example, a promoter sequence can be substituted
(e.g., by
homologous recombination) for the native promoter of a gene in the genome of a
host .
cell, such that this gene has an altered expression pattern. This gene would
now
become 'isolated' because it is separated from at least some of the sequences
that
naturally flank it.
[32] A nucleic acid is also considered 'isolated' if it contains any
modifications that do
not naturally occur to the corresponding nucleic acid in a genome. For
instance, an
endogenous coding sequence is considered 'isolated' if it contains an
insertion, deletion
or a point mutation introduced artificially, e.g., by human intervention. An
'isolated
nucleic acid' also includes a nucleic acid integrated into a host cell
chromosome at a .
heterologous site and a nucleic acid construct present as an episome.
Moreover, an ,
'isolated nucleic acid' can be substantially free of other cellular material,
or sub-
stantially free of culture medium when produced by recombinant techniques, or
sub-
stantially free of chemical precursors or other chemicals when chemically
synthesized.
[33] As used herein, the phrase 'degenerate variant' of a reference nucleic
acid sequence
encompasses nucleic acid sequences that can be translated, according to the
standard
genetic code, to provide an amino acid sequence identical to that translated
from the
reference nucleic acid sequence. The term 'degenerate oligonucleotide' or
'degenerate
CA 02876864 2015-01-08
=
7
primer' is used to signify an oligonucleotide capable of hybridizing with
target nucleic
acid sequences that are not necessarily identical in sequence but that are
homologous
to one another within one or more particular segments.
[34] The term 'percent sequence identity' or 'identical' in the
context of nucleic acid
sequences refers to the residues in the two sequences which are the same when
aligned
for maximum correspondence. The length of sequence identity comparison may be
over a stretch of at least about nine nucleotides, usually at least about 20
nucleotides,
more usually at least about 24 nucleotides, typically at least about 28
nucleotides, more
typically at least about 32 nucleotides, and preferably at least about 36 or
more nu-
cleotides. There are a number of different algorithms known in the art which
can be
used to measure nucleotide sequence identity. For instance, polynucleotide
sequences
can be compared using FASTA, Gap or Bestfit, which are programs in Wisconsin
Package Version 10.0, Genetics Computer Group (GCG), Madison, Wisconsin.
FASTA provides alignments and percent sequence identity of the regions of the
best
overlap between the query and search sequences. Pearson, Methods Enzyn2oL
= 183:63-98 (1990) . For instance,
percent sequence identity between nucleic acid sequences can be determined
using
= FASTA with its defaultparameters (a word size of 6 and the NOPAM factor
for the
= scoring matrix) or using Gap with its default parameters as provided in
GCG Version
= = 6.1. Alternatively, sequences can be compared using .
the computer program, BLAST (Altschul et at., MoL Biol. 215:403-410 (1990);
Gish
. and States, Nature Genet. 3:266-272 (1993); Madden et aL, Meth. EnzymoL
=
266:131-141 (1996); Altschul et aL, Nucleic Acids Res. 25:3389-3402 (1997);
Zhang
. and Madden, Genome Res. 7:649-656 (1997)), especially blastp or tblastn
(Altschul et
aL, Nucleic Acids Res. 25:3389-3402 (1997)).
[35] The term 'substantial homology' or 'substantial similarity; when
referring to a
nucleic acid or fragment thereof, indicates that, when optimally aligned with
ap-
propriate nucleotide insertions or deletions with another nucleic acid (or its
com-
plementary strand), there is nucleotide sequence identity in at least about
50%, more
preferably 60% of the nucleotide bases, usually at least about 70%, more
usually at
least about 80%, preferably at least about 90%, and more preferably at least
about
95%, 96%, 97%, 98% or 99% of the nucleotide bases, as measured by any well-
known
algorithm of sequence identity, such as FASTA, BLAST or Gap, as discussed
above.
[36] Alternatively, substantial homology or similarity exists when a
nucleic acid or
fragment thereof hybridizes to another nucleic acid, to a strand of another
nucleic acid,
or to the complementary strand thereof, under stringent hybricii7ation
conditions.
'Stringent hybridization conditions' and 'stringent wash conditions' in the
context of
nucleic acid hybridization experiments depend upon a number of different
physical
parameters. Nucleic acid hybridization will be affected by such conditions as
salt con-
centration, temperature, solvents, the base composition of the hybridizing
species,
CA 02876864 2015-01-08
8
length of the complementRry regions, and the number of nucleotide base
mismatches
between the hybridizing nucleic acids, as will be readily appreciated by those
skilled in
the ark One having ordinary skill in the art knows how to vary these
parameters to
achieve a particular stringency of hybridization.
[37] In general, 'stringent hybridization' is performed at about 25 C
below the thermal
melting point (T.) for the specific DNA hybrid under a particular set of
conditions.
'Stringent washing' is performed at temperatures about 5 C lower than the T.
for the =.
specific DNA hybrid under a particular set of conditions. The T. is the
temperature at =
which 50% of the target sequence hybridizes to a perfectly matched probe. See
Sambrook et al., Molecular Cloning: A Laboratory Manual, 2d ed., Cold Spring
Harbor Laboratory Press, Cold Spring Harbor, N.Y. (1989), page 9.51, hereby in-
corporated by reference. For purposes herein, 'stringent conditions' are
defined for
solution phase hybridivition as aqueous hybridization (i.e., free of
forraamide) in 6X =
SSC (where 20X SSC contRins 3.0 M NaC1 and 0.3 M sodium citrate), 1% SDS at 65
C for 8-12 hours, followed by two washes in 0.2X SSC, 0.1% SDS at 65 C for 20
minutes. It will be appreciated by the skilled worker that hybridization at 65
C will =
= = occur at different rates depending on a number of factors
including the length and
percent identity of the sequences which are hybridizing. . .
.
=
[38] The nucleic acids (also referred to as polynucleotides) of this
invention may include
both sense and antisense strands of RNA, cDNA, genomic DNA, and synthetic
forms
and mixed polymers of the above. They may be modified chemically or
biochemically
or may contain non#natural or derivatized nucleotide bases, as will be readily
ap-
preciated by those of skill in the art. Such modifications include, for
example, labels, .
methylation, substitution of one or more of the naturally occurring
nucleotides with an
analog, intemucleotide modifications such as uncharged linkages (e.g., methyl
phosphonates, phosphotriesters, phosphoramidates, carbamates, etc.), charged
linkages = .=
(e.g., phosphorothioates, phosphorodithioates, etc.), pendent moieties (e.g.,
polypeptides), intercalators (e.g., acridine, psoralen, etc.), chelators,
alkylators, and
modified linkages (e.g., alpha anomeric nucleic acids, etc.) Also included are
synthetic
molecules that mimic polynucleotides in their ability to bind to a designated
sequence
via hydrogen bonding and other chemical interactions. Such molecules are known
in =
the art and include, for example, those in which peptide linkages substitute
for
phosphate linkages in the backbone of the molecule. Other modifications can
include,
for example, analogs in which the ribose ring contains a bridging moiety or
other
structure such as the modifications found in 'locked' nucleic acids.
[39] The term 'mutated' when applied to nucleic acid sequences means that
nucleotides
in a nucleic acid sequence may be inserted, deleted or changed compared to a
reference
nucleic acid sequence. A single alteration may be made at a locus (a point
mutation) or
multiple nucleotides may be inserted, deleted or changed at a single locus. In
addition,
one or more alterations may be made at any number of loci within a nucleic
acid
CA 02876864 2015-01-08
9
sequence. A nucleic acid sequence may be mutated by any method known in the
art
including but not limited to mutagenesis techniques such as 'error-prone PCR'
(a =
process for performing PCR under conditions where the copying fidelity of the
DNA
polymerase is low, such that a high rate of point mutations is obtained along
the entire
length of the PCR product; see, e.g., Leung et aL, Technique, 1:11-15 (1989)
and
Caldwell and Joyce, PCR Methods Applic. 2:28-33 (1992)); and 'oligonucleotide-
directed mutagenesis' (a process which enables the generation of site-specific
=
mutations in any cloned DNA segment of interest see, e.g., Reidhaar-Olson and
Sauer,
Science 241:53-57 (1988)).
[40] The tem]. 'vector' as used herein is intended to refer to a
nucleic acid molecule
capable of transporting another nucleic acid to which it has been linked. One
type of
vector is a 'plasmid', which refers to a circular double stranded DNA loop
into which
additional DNA segments may be ligated. Other vectors include cosmids,
bacterial
artificial chromosomes (BAC) and yeast artificial chromosomes (YAC). Another
type 4
of vector is a viral vector, wherein artditional DNA segments may be ligated
into the =
=
= = viral genome (discussed in more detail below). Certain vectors
are capable of
autonomous replication in a host cell into which they are introduced (e.g.,
vectors
having an origin of replication which functions in the host cell). Other
vectors can be -
integrated into the genome of a host cell upon introduction into the host
cell, and are
=
.
thereby replicated along with the host genome. Moreover, certain preferred
vectors are
.
= =capable of directing the expression of genes to which they are
operatively linked. Such.
= vectors are referred to herein as 'recombinant expression vectors' (or
simply,
= =
'expression vectors). =
= [41] The
term 'marker sequence' or 'marker gene' refers to a nucleic acid sequence =
capable of expressing an activity that allows either positive or negative
selection for .
the presence or absence of the sequence within a host cell. For example, the
P. pastoris
LIRAS gene is a marker gene becanse its presence can be selected for by the
ability of
cells containing the gene to grow in the absence of uracil. Its presence can
also be
selected against by the inability of cells containing the gene to grow in the
presence of "
5-F0A. Marker sequences or genes do not necessarily need to display both
positive
and negative selectability. Non-limiting examples of marker sequences or genes
from
P. pastoris include ADE1, ARG4, H1.54 and URA. 3 .
[42] 'Operatively linked' expression control sequences refers to a linkage
in which the
expression control sequence is contiguous with the gene of interest to control
the gene
of interest, as well as expression control sequences that act in trans or at a
distance to
control the gene of interest.
[43] The term 'expression control sequence' as used herein refers to
polynucleotide
sequences which are necessary to affect the expression of coding sequences to
which =
they are operatively linked. Expression control sequences are sequences which
control
the transciiption, post-transcriptional events and translation of nucleic acid
sequences.
CA 02876864 2015-01-08
Expression control sequences include appropriate transcription initiation,
termination,
promoter and enhancer sequences; efficient RNA processing sianals such as
splicing
and polyadenylation signals; sequences that stabilize cytoplasmic mRNA;
sequences
that enhance translation efficiency (e.g., ribosome binding sites); sequences
that
enhance protein stability; and when desired, sequences that enhance protein
secretion.
The nature of such control sequences differs depending upon the host organism;
in
prokaryotes, such control sequences generally include promoter, ribosomal
binding
site, and transcription termination sequence. Tho term 'control sequences' is
intended to
include, at a minimum, all components whose presence is essential for
expression, and
can also include additional components whose presence is advantageous, for
example,
leader sequences and fusion partner sequences.
[44] The term 'recombinant host cell' (or simply 'host cell), as used
herein, is intended to
refer to a cell into which a recombinant vector has been introduced. It should
be
understood that such terms are intended to refer not only to the particular
subject, cell
but to the progeny of such a cell. Because certain modifications may occur in
= = . =
succeeding generations due to either mutation or environmental influences,
such = = , .
progeny may not, in fact, be identical to the parent cell, but are still
included within the
scope of the term 'host cell' as used herein. A recombinant host cell may be
an isolated
= cell or cell line grown in culture or may be a cell which resides in a
living tissue or
= organism.
[45] = The term 'peptide' as used herein refers to a short polypeptide,
e.g., one that is
typirtally less than about 50 amino acids Jong and more typically less than
about 30 µ=
amino acids long. The term as used herein encompasses analogs and mimetics
that =,
mimic structural and thus biological function.
[46] The term polypeptide' encompasses both naturally-occurring and non-
naturally-occurring proteins, and fragments, mutants, derivatives and analogs
thereof.
A polypeptide may be monomeric or polymeric. Further, a polypeptide may
comprise
a number of different domains each of which has one or more distinct
activities.
[47] The term 'isolated protein' or 'isolated polypeptide' is a protein or
polypeptide that
by virtue of its origin or source of derivation (1) is not associated with
naturally '
associated components that accompany it in its native state, (2) exists in a
purity not
found in nature, where purity can be adjudged with respect to the presence of
other
cellular material (e.g., is free of other proteins from the same species) (3)
is expressed
by a cell from a different species, or (4) does not occur in nature (e.g., it
is a fragment
of a polypeptide found in nature or it includes amino acid analogs or
derivatives not
found in nature or linkages other than standard peptide bonds). Thus, a
polypeptide
that is chemically synthesized or synthesized in a cellular system different
from the
cell from which it naturally originates will be 'isolated' from its naturally
associated
components. A polypeptide or protein may also be rendered substantially free
of
naturally associated components by isolation, using protein purification
techniques
CA 02876864 2015-01-08
11
well known in the art. As thus defined, 'isolated' does not necessarily
require that the
protein, polypeptide, peptide or oligopeptide so described has been physically
removed
from its native environment.
[48] The term 'polypeptide fragment' as used herein refers to a
polypeptide that has a
deletion, e.g., an amino-terminal and/or carboxy-terminal deletion compared to
a full-
length polypeptide. In a preferred embodiment, the polypeptide fragment is a
contiguous sequence in which the amino acid sequence of the fragment is
identical to
the corresponding positions in the naturally-occurring sequence. Fragments
typically
are at least 5, 6,7, 8, 9 or 10 amino acids long, preferably at least 12, 14,
16 or 18
amino acids long, more preferably at least 20 amino acids long, more
preferably at
= least 25, 30, 35, 40 or 45, amino acids, even morepreferably at least 50
or 60 amino
acids long, and even more preferably at least 70 amino acids long.
[49] A 'modified derivative' refers to polypeptides or fragments thereof
that 'are sub-
stantially homologous in primary structural sequence but which include, e.g.,
in vivo or
in vitro chemical and biochemical modifications or which incorporate amino
acids that
are not found in the native polypeptide. Such modifications include, for
example,
acetylation, carboxylation, phosphorylation, glycosylation, ubiquitination,
labeling;
e.g., with radionuclides, and various enzymatic modifications, as will be
readily ap-
preciated by those skilled in the art. A variety of methods for labeling
polypeptides and
of substituents or labels useful for such purposes are well known in the art,
and include
radioactive isotopes such as 1251, 3113,35S, and ligands which
bind to labeled an-
tiligands (e.g., antibodies), finorophores, chemilumniescent agents, enzymes,
and an-
tiligands which can serve as specific binding pair members for a labeled
ligand. The
choice of label depends on the sensitivity required, ease of conjugation with
the
primer, stability requirements, and available instrumentation. Methods for
labeling
polypeptides are well known in the art. See, e.g., Ausubel et al., Current
Protocols in
Molecular Biology, Greene Publishing Associates (1992, and Supplements to
2002) .
[50] The term 'fusion protein' refers to a polypeptide comprising a polyp
eptide or
fragment coupled to heterologous amino acid sequences. Fusion proteins are
useful
because they can be constructed to contain two or more desired functional
elements
from two or more different proteins. A fusion protein comprises at least 10
contiguous
amino acids from a polypeptide of interest, more preferably at least 20 or 30
amino
acids, even more preferably at least 40, 50 or 60 amino acids, yet more
preferably at
least 75, 100 or 125 amino acids. Fusions that include the entirety of the
proteins of the
present invention have particular utility. The heterologous polypeptide
included within
the fusion protein of the present invention is at least 6 amino acids in
length, often at
least 8 amino acids in length, and usefully at least 15, 20, and 25 amino
acids in length.
Fusions that include larger polypeptides, such as an IgG Fc region, and even
entire
proteins, such as the green fluorescent protein ('GFP') chromophore-containing
CA 02876864 2015-01-08
12
proteins, have particular utility. Fusion proteins can be produced
recombinantly by
constructing a nucleic acid sequence which encodes the polypeptide or a
fragment
thereof in frame with a nucleic acid sequence encoding a different protein or
peptide
and then expressing the fusion protein. Alternatively, a fusion protein can be
produced
chemically by crosslinking the polypeptide or .a fragment thereof to another
protein.
[51] The term 'non-peptide analog' refers to a compound with properties
that are
analogous to those of a reference polypeptide. A non-peptide compound may also
be
termed a 'peptide mimetic' or alpeptidomimetic'. See, e.g., Jones, Amino Acid
and
Peptide Synthesis, Oxford University Press (1992); Jung, Combinatorial Peptide
and
Non_peptide Libraries: A Handbook, John Wiley (1997); Bodanszky et al.,
Peptide
Chemistry¨A Practical Textbook, Springer Verlag (1993); Synthetic Peptides: A
Users-
Guide, (Grant, ed., W. H. Freeman and Co., 1992); Evans at al., J. Med. Chem.
30:1229 (1987); Fauchere, J. Adv. Drug Res. 15:29 (1986); Veber and
Freidinger,
Trends Neurosci., 8:392-396 (1985); and references sited in each of the
above..
Such compounds are often developed with the
aid of computerized molecular modeling. Peptide raimetics that are
structurally similar
to-useful peptides of the invention may be used to produce an equivalent
effect and are
therefore envisioned to be part of the invention.
[52] A 'polypeptide mutant or 'mutein' refers to a polypeptide whose
sequence contains
an insertion, duplication, deletion, rearrangement or substitution of one or
more =amino
acids compared to the amino acid sequence of a native or wild-type protein. A
mutein
may have one or more amino acid point substitutions, in which a single amino
acid.at a
position has been changed to another amino ,acid, one or more insertions
and/or
deletions, in which one or more amino acids are inserted or deleted,
respectively, in the
sequence of the naturally-occurring protein, and/or truncations of the amino
acid
sequence at either or both the amino or carboxy termini. A mutein may have the
same
but preferably has a cli-fferent biological activity compared to the naturally-
occurring
protein.
[53] A mutein has at least 50% overall sequence homology to its wild-type
counterpart.
Even more preferred are muteins having at least 70%, 75%, 80%, 85% or 90%
overall
sequence homology to the wild-type protein. In an even more preferred
embodiment, a
mutein exhibits at least 95% sequence identity, even more preferably 98%, even
more
preferably 99% and even more preferably 99.9% overall sequence identity.
Sequence
homology may be measured by any common sequence analysis algorithm, such as
Gap
or Bestdt
[54] Amino acid substitutions can include those which: (1) reduce
susceptibility to
proteolysis, (2) reduce susceptibility to oxidation, (3) alter binding
affinity for forming
protein complexes, (4) alter binding affinity or enzymatic activity, and (5)
confer or
modify other physicochemical or functional properties of such ariPlogs
[55] As used herein, the twenty conventional amino acids and their
abbreviations follow
CA 02876864 2015-01-08
13
conventional usage. See Immunology ¨A Synthesis (Golub and Gren eds.,
Sinauer Associates, Sunderland, Mass., 2'd ed. 1991).
Stereoisomers (e.g., D-amino acids) of the twenty conventional
amino acids, unnatural amino acids such as ce-,a-disubstituted amino acids,
N-alkyl amino acids, and other unconventional amino acids may also be suitable
components for polypeptides of the present invention. Examples of
unconventional
amino acids include: 4-hydroxyproline, &-carboxyglutamate, E-N,N,N-
trimethyllysine,
e-N-acetyllysine, 0-phosphoserine, N-acetylserine, N-formylmethionine, 3-
methylhistidine,
5-hydroxylysine, N-methylarginine, and other similar amino acids and imino
acids
(e.g., 4-hydroxyproline). In the polypeptide notation used herein, the left-
hand end
corresponds to the amino terminal end and the right-hand end corresponds to
the
carboxy-terminal end, in accordance with standard usage and convention.
[56] A protein has 'homology' or is 'homologous to a second protein lithe
nucleic acid
sequence that encodes the protein Im a similar sequence to the nucleic acid
sequence
that encodes the second protein. Alternatively, a protein has homology to a
second
proteinif the two proteins have 'similar amino acid sequences. (Thus, the lerm
'homologous proteins' is defined to mean that the two proteins have citrular
amino acid
sequences.) In a preferred embodiment, a homologous protein is one that
exhibits at
least 65% sequence homology to the wild type protein, more preferred is at
least 70%
.sequence homology. Even more preferred are horo.ologious proteins that
exhibit at least
75%, 80%, 85% or 90% sequence homology to the wild type protein. In a yet more
=.preferred-embodiment, a homologous protein exhibits at least 95%, 98%, 99%
or
99.9% sequence identity. As used herein, homology between two regions of amino
acid sequence (especially with respect to predicted structural similarities)
is interpreted
as implying similarity in function.
[57] When 'homologous' is used in reference to proteins or peptides, it is
recogni7ed that
residue positions that are not identical often differ by conservative amino
acid sub-
- stitutions. A 'conservative amino acid substitution' is one in which an
amino acid
residue is substituted by another amino acid residue having a side chain (R
group) with
similar chemical properties (e.g., charge or hydrophobicity). In general, a
conservative
amino acid substitution will not substantially change the functional
properties of a
protein. In cases where two or more amino acid sequences differ from each
other by
conservative substitutions, the percent sequence identity or degree of
homology may
be adjusted upwards to correct for the conservative nature of the
substitution. Means
for making this adjustment are well known to those of skill in the art. See,
e.g.,
Pearson, 1994, Methods Mol. Biol. 24:307-31 and 25:365-89.
[58] The following six groups each contain amino acids that are
conservative sub-
stititions for one another: 1) Serine (S), Tlareonine (T); 2) Aspartic Acid
(D), GlutRmic
CA 02876864 2015-01-08
14
Acid (E); 3) Asparagine (N), Glutamirte (Q); 4) Arginine (R), Lysine (K); 5)
lsoleucine (I), Leucine (L), Methionine (M), Alanine (A), Valine (V), and 6)
Phenylalanine (F), Tyrosine (Y), Ttyptophan (W).
[59] Sequence homology for polypeptides, which is also referred to as
percent sequence
identity, is typically measured using sequence analysis software. See, e.g.,
the
Sequence Analysis Software Package of the Genetics Computer Group (GCG),
University of Wisconsin Biotechnology Center, 910 University Avenue, Madison,
Wisconsin 53705. Protein analysis software matches similar sequences using a
=
measure of homology assigned to various substitutions, deletions and other
modi-
fications, including conservative amino acid substitutions. For instance, GCG
contains
programs such as 'Gap' and Sestfit which can be used with default parameters
to
determine sequence homology or sequence identity between closely related
polypeptides, such as homologous polypeptides from different species of
organisms or
between a wild-type protein and a mutein thereof. See, e.g., GCG Version 6.1.
[60] A preferred
algorithm when comparing a particular polypepitde sequence to a =
= database.containing a large number of sequences from different organisms
is the = -*
.= computer program BLAST (Altschul at al., J. Mol. Biol. 215:403-410
(1990); Gish and
= States, Nature Genet. 3:266-272 (1993); Madden et at., Meth. Enzymol.
266:131-141
= (1996); Altschul at al., Nucleic Acids Res. 25:3389-3402 (1997); Zhang
and Madden, =
Genorne Res. 7:649-656 (1997)), especially blastp or.tblastn (Altschul etal.,
Nucleic
Acids Res. 25:3389-3402 (1997)).
[61] Preferred parameters for BLASTp are: =
[62] Expectation value: 10 (default); Filter. seg (default); Cost to open a
gap: 11
= (default); Cost to extend a gap: 1 (default); Max. alignments: 100
(default); Word size:
11 (default); No. of descriptions: 100 (default); Penalty Matrix: BLOWSUM62. -
[63] The length of polypeptide sequences compared for homology will
generally be at
least about 16 amino acid residues, usually at least about 20 residues, more
usually at
least about 24 residues, typically at least about 28 residues, and preferably
more than
about 35 residues. When searching a database containing sequences from a large
number of different organisms, it is preferable to compare amino acid
sequences.
Database searching using amino acid sequences can be measured by algorithms
other
than blastp known in the art. For instance, polypeptide sequences can be
compared
using FASTA, a program in GCG Version 6.1. FASTA provides alignments and
percent sequence identity of the regions of the best overlap between the query
and
search sequences. Pearson, Methods Erzzymol. 183:63-98 (1990),
For example, percent sequence identity between amino acid sequences
can be determined using FASTA with its default parameters (a word size of 2
and the
PAM250 scoring matrix), as provided in GCG Version 6.1.
[64] The term 'region' as used herein refers to a physically contiguous
portion of the
CA 02876864 2015-01-08
primary structure of a biomolecule. In the case of proteins, a region is
defmed by a
contiguous portion of the amino acid sequence of that protein.
[65] The term 'domain' as used herein refers to a structure of a
biomolecule that
contributes to a known or suspected function of the biomolecule. Domains may
be co-
extensive with regions or portions thereof, domains may also include distinct,
non-
contiguous regions of a biomolecule. Examples of protein domains include, but
are not
limited to, an Ig domain, an extracellular domain, a transmembrane domain, and
a cy-
toplasmic domain. =
[66] As used herein, the term 'molecule' means any compound, including, but
not limited
to, a small molecule, peptide, protein, sugar, nucleotide, nucleic acid,
lipid, etc., and
such a compound can be natural or synthetic.
[67] The term 'elimination' as used with respect to mannosylphosphorylation
refers to
mannosphosphorylated glycan detection levels indicating no apparent detectable
man-
nosylphosphate residues using HPLC under the stated setting.
[68] Unless otherwise defined, all technical and scientific terms used
herein have the
same meaning as commonly understood by one of ordinary skill in the art to
which this
invention pertains. Exemplary methods and materials are described below,
although
= methods and materials similar or equivalent to those described herein can
also be used
in the practice of the present invention and will be apparent to those of
skill in the art. =
In case of conflict, the present specification, including definitions, will
= control. The materials, methods, and examples are illustrative only
and not intended, to .=
be limiting.
= [69] Methods For Producing a Fungal Host Strain Lacking
Mannosylphosphorylation
on Glycoproteins
[70] The present invention provides methods for eliminating
marmosylphosphate
= transfer on glycans of glycoproteins in yeast or filamentous fungal host
cells which
= normally produce glycoproteins having mannosylphosphorylation. In one
embodiment,
the yeast or filamentous fungal host cell which normally produces
glycoproteins
having mannosylphosphorylation is engineered so that it is essentially free of
manno-
sylphosphorylation on glycans of glycoproteins. In another embodiment, the
fungal
hosts are genetically modified to have disrupted, attenuated or mutated at
least one
gene encoding a protein participating in mannosylphosphate transferase.
Preferably,
the method involves disruption, attenuation or mutation of one or more genes
selected
from MNN4A, M3IN4B, MNN4C and PNOI.
[71] Using known genes encoding mannosylphosphate transferases, novel
genes
encoding mannosylphosphate transferase in P. pastoris were isolated. The MNN4
gene
sequence from S. cerevisiae (Genbank accession # P36044) was blasted against
the
genome of P. pastoris (Integrated Genomics, Chicago, IL). This search resulted
in the
identification of three previously unknown ORFs in addition to the PNOI gene.
The
CA 02876864 2015-01-08
16
three ORFs were designated as MNN4A (SEQ ID NO: 1), MNN4B (SEQ ID NO: 3), =
andMNN4C (SEQ ID NO: 1). These ORFs were amplified and subsequently
sequenced and are shown respectively in Figs. 1-3 (Example 1). The encoded
amino
acid sequences for MNN4A (SEQ ID NO: 2),MNN4B (SEQ ID NO: 4),MNN4C (SEQ
ID NO: 6) are also set forth in Figs. 1-3.
[72] Nucleic Acid Sequences
[73] In one aspect, the present invention provides a nucleic acid molecule
comprising or
consisting of a sequence which is a variant of the P. pastoris M1VN4 A gene
having at
least 50% identity to SEQ ID NO:l. The nucleic acid sequence can preferably
have at
least 65%, 70%, 75% or 80% identity to the wild-type gene. Even more
preferably, the
nucleic acid sequence can have 85%, 90%, 95%, 98%, 99%, 99.9% or even higher
identity to the SEQ 11) NO:1. The present invention also provides polypeptide
comprising or consisting of a sequence which is a variant of the P. pastoris
MNN4A
gene having at least 50% identity to SEQ NO:2. The amino acid sequence can
preferably have at least 65%, 70%, 75% or 80% identity to the wild-type gene.
Even =
= more
preferably, the amino acid sequence can have 85%, 90%, 95%, 98%, 99%, =
=
= = = 99.9% or even higher identity to the SEQ ID NO:2.
=
= [74] = In another embodiment, the P . pastoris MNN4B gene is
particularly useful in the
= == elimination of mannosylphosphate transfer on glycans of
glycoproteins in a yeast
=
strain. The present invention provides a nucleic acid molecule comprising or
consisting = .
of a sequence which is a variant of the P. pastoris M11N4 B gene having at
least 50% -
'identity to SEQ ID NO:3. The nucleic acid sequence can preferably have at
least 65%,
70%, 75% or 80% identity to the wild-type gene. Even more preferably, the
nucleic
= acid sequence can have 85%, 90%, 95%, 98%, 99%, 99.9% or even higher
identity to -
the SEQ ID NO:3. The present invention also provides polypeptide comprising or
consisting of a sequence which is a variant of the P. pastoris MNN4B gene
having at =
least 50% identity to SEQ ID NO:4. The amino acid sequence can preferably have
at
least 65%, 70%, 75% or 80% identity to the wild-type gene. Even more
preferably, the
amino acid sequence can have 85%, 90%, 95%, 98%, 99%, 99.9% or even higher
identity to the SEQ ID NO:4.
[75] In yet another embodiment, the present invention provides a
nucleic acid molecule
comprising or consisting of a sequence which is a variant of the P. pastoris
MNN4 C
gene having at least 50% identity to SEQ ID NO:5. The nucleic acid sequence
can
preferably have at least 65%, 70%, 75% or 80% identity to the wild-type gene.
Even
more preferably, the nucleic acid sequence can have 85%, 90%, 95%, 98%, 99%,
99.9% or even higher identity to the SEQ NO:5. The present invention also
provides an polypeptide comprising or consisting of a sequence which is a
variant .of
the P. pastoris M1'sN4C gene having at least 50% identity to SEQ ID NO:6. The
amino acid sequence can preferably have at least 65%, 70%, 75% or 80% identity
to
the wild-type gene. Even more preferably, the amino acid sequence can have
85%,
CA 02876864 2015-01-08
17
90%, 95%, 98%, 99%, 99.9% or even higher identity to the SEQ ID NO:6.
P6] Also provided are vectors, incluciiii,s expression vectors and knock-
out vectors
comprising the above nucleic acid molecules of the invention. A knock-out
vector
comprising a 101N4A, MNN4B or lvINN4C may be used to disrupt the MNN4A,
MNN4B or MNN4C gene locus. Alternatively, an integration vector comprising a
drug
resistance marker or an auxotrophic marker is used to disrupt the M1VN4 gene
locus.
[77] Combination of Marmosylphosphorylation Gene Knock-huts
[78] Each of the three newly identified P. pastoris genes, MNN4A, MIVN4B,
1IVN4C , is
disrupted using the PCR overlap strategy as shown in Fig. 4 to determine the
effect on
mannosylphosphorylation. The individual Amnn4A, Amnn4B, and Amnn4C mutants
did not show a significant decrease in mannosylphosphorylation transfer
activity on
glycans of the kringle 3 domain of human plasminogen (K.3) protein, whereas
the
4no/mutant (YSH-49) displayed only an attenuation in mannosylphosphorylation
transfer-decreased to 6% (Fig. 5C) - but not to the levels described
previously in
Miura et al. (WO 01/88143). It has been postulated that different
glycoproteins may
= display varying degrees and types of glycosylation in the same host cell
(IvIontesino et
al, 1998, Prot. .Fapr. Purif 14: 197-207). In one embodiment of the present
invention,
combinations of mill mutants were constructed, one of which, the double mutant
Apno
Amnn4b in P. pastoris resulted in undetectable levels of
mannosylphosphorylation on
glycans of the K3 reporter protein (Fig. SD). Similarly, other glycoproteins
(e.g, CD40
and invertase) produced from the double mutant Apno 1 Amnn4b in P. pastoris
also
resulted in lack of mannosylphosphorylation. The double mutant, therefore,
produces
various glycoproteins of interest that are free of mannosylphosphorylation on
glycang.
Accordingly, a method is provided for disrupting a combination of genes
involved in,
the transfer of mannosylphosphate residues on glycans of glycoproteins in a
host (e.g.,
Pichia sp.). Preferably, the combination includes disruption ofMIVN4B and
PNOI.
[79] In case the disruption of the P. pastoris 1011f4B locus alone does
not, confer
elimination of mannosylphosphorylation on glycans, a combination of
mannosylphos-
phorylation genes are disrupted. In a preferred embodiment, the disruption of
the
MNN4B locus is in combination with at least a second gene involved in manno-
sylphosphate transfer, such as MNN4A, MIVN4B, MNN4C or PN01. The second gene
in this case is preferably the?. pastoris PNO1 gene (Genbank accession
#BD105434).
It is contemplated that a skilled artisan may disrupt or mutate any gene
involved in
oligosaccharide synthesis or a fragment thereof in combination with a
disrupted or
mutated .116NN4B, which would result in the elimination of mannosylphosphate
transfer
to glycans in other fungal hosts.
[80] In another embodiment, the method provides for disrupting a gene
encoding
MNN4B (SEQ ID NO: 3) in a host (e.g., P. pastoris) that already has attenuated
man-
nosylphosphate tranaferase activity. Additionally, it is contemplated that the
elimination of mannosylphosphate transfer to glycans in other Pichia species
involves
CA 02876864 2015-01-08
18
the disruption or mutation of any combination of genes having homology to
1v1117N4.4
lvINN4C, orPN01.
[81] In yet another aspect of the invention each of the three newly
identified P. pastoris
genes, 1vflYN4A, MNN4B, PINN4C , was disrupted using a fusion knock out
strategy as
described in Example 3 in order determine if any combination of gene knockouts
had
an effect on mannosylphosphorylation of glycoproteins expressed in this mutant
strain.
The individual dmnn4A, Amnn4B, and Amnn4C mutants as with the PCR overlap
knockout strategy (Fig. 4) did not show a decrease in mannosylphosphorylation
transfer activity on glycans of the kring,le 3 domain of human p3asminogen (K)
protein (data not shown). However, the K3 reporter protein expressed in a
Apnol
Amnn4b double null mutant (YAS174) is essentially free of any marmosylphos-
phorylation (Fig. 6C, compare with Fig. 6A, B). Note the absence of
mannosylphos-
phorylated glycans between 20 and 30 ruins. .
[82] Fieterologous Glycoprotein Expression System
[83] Using established techniques for expressing heterologous glycoproteins
in yeast and -
filamentous fungi, a gene encoding a therapeutic glycoprotein is expressed. A
fungal
recombinant protein expression system may typirnlly include promoters such as
AOXI, A0X2, or other inducible promoters, transcriptional term:mat= such as
CYC,
selectable markers such as URA3, URA.5, G418, ADE1, ARG4, BIS4, Zeocin and
secretion signals such as S. cerevisiae oNIF. In one embodiment, this
expression
system is modified to be at least a mnn4B mutant Preferably, the glycoproteins
are
produced in P. pastoris having at least oarinn4B.
[84] Glycoproteins of interest can be produced by anyin-nns through the use
of the
methods disclosed herein. Glycoprotein production can be provided by any
rormis in a .
host cell, including accumnlation in an intracellular compartment or secretion
from the
cell into a culture supernatant. Host cells of the present invention may be
propagated
or cultured by any method known or contemplated in the art, including but not
limited
to growth in culture tubes, flasks, roller bottles, shake flasks or
fermentors. Isolation
andfor purification of the glycoprotein products may be conducted by any means
known or contemplated in the art such as fractionation, ion exchange, gel
filtration, hy-
drophobic chromatography and Pfanity chromatography. An example of
glycoprotein
production and purification is disclosed in Example 7.
[85) The glycoproteins expressed without maminsylphosphorylated glycans
using the
methods described herein can include but are not limited to: erythropietin,
cytokines
such as interferon-a, interferon-$, interferon-6, interferon-c, TNF-ce,
granulocyte-CSF, GM-CSF, interleukin,s such as IL-lra, coagulation factors
such as
factor VIII, factor D.:, human protein C, anfithrombin III and thrombopoeitin
antibodies; IgG, IgA, IgD, IgE, IgM and fragments thereof, Pc and Fab regions,
soluble IgE receptor a -chain, urolcina.se, chymase, and urea trypsin
inhibitor, IGF-
binding protein, epidermal growth factor, growth hormone-releasing factor,
FSH,
CA 02876864 2015-01-08
19
anrien V fusion protein, angiostatin, vascular endothelial growth factor-2,
myeloid
progenitor inhibitory factor-1, osteoprotegerin, a-1 antitryp sin, DNase II, a-
feto
proteins and glucocembrosidase.
[86] Production of Complex Glvcoproteins Laclthis Mannosvlphosphoryl tion
[87] In another aspect of the invention, the present invention provides
methods for
producing complex N-linked glycans in fungi and yeast (e.g., P. pastoris) that
comprises eliminating mannosylphosphate transfer to glycans on glycoproteins.
Such
method provides a glycoprotein composition that is essentially free of manno-
sylphosphate residues on glycoproteins. In one embodiment, the invention
provides
less than I% mannosylphosphorylated glycoproteins of total N-glycans. In a
more
preferred embodiment, the invention provides less than 0.5%
marmosylphosphorylated
glycoproteins of total N-glycans.
[88] In another aspect of the present invention, the glycoprotein
compositions are es- ,
sentia fly free of mannosylphospha e residues on complex N-glycans. The method
to
produce such glycans involve disrupting the PNO1 and MNAT4B genes in a host
strain
expressing complex N-glycans (e.g., P. pastoris YSH-44 expressing K3 reporter
protein) (Hamilton et aL, 2003, Science, 301: 1244-1246) . The engineered
strain
comprisingpno/ mnn4B disruptions, designatpd as YAS-130, lacks manno-
sylphosphate residues on glycans of glycoproteins (Example 5). Although a
genetic
disruption of the PNO1 gene in YSH-44 (designated YSH-49) rerInces the mole %
of
glycans exhibiting mannosylphosphorylaticm (acidic fraction),
rammosylphosphate
residues still remain (Fig. 5C). Treatment of the glycans from YSH-44 with
mild acid
hydrolysis followed by PlicniinP phosphrtace demonstrates that the acidic
fraction is
comprised of about 5-15% of total glycans. This YSH-49 strain shows an acidic'
fraction of about 6%, which does compare favorably with the about 9% acidic
fraction
of the YSH-44 (Fig. 513).
[89] . By contrast, Fig. 5D shows elimination of mannosylphosphate transfer to
glycans in
P. pastoris YAS-130 (Apno 1 Amnn4B) in comparison to Fig. 5A control -(H20),
-Fig. 5B YSH-44 with about 9% mannosylphosphorylation, and Fig. 5C YSH-49
(Apno 1)
With about 6% roannosylphosphorylation. Herein is described for the first time
a yeast
strain engineered to be essentially free of mannosylphosphorylated glycans.
[90] It is also contempl atd that other types of yeast and filamentous
fungus can be
modified to lack mannosylphosphate transfer activity using the methods
described
herein. While Pichia pastoris is the pi ________________ craned host strain
for producing complex N-
linked glycoproteins lacking mannos-ylphosphate residues, the following host
cells may
be also engineered: Pichia finlandica, Pichia trehalophik, koclamae, Pichia
inembranaefaciens, Pichia methanolica, Pichia minuta (Ogataea minuta ,Pichia
lindnerz), Pichia opuntiae, Pichia thermotolerans, Pichi salictaria, Pichia
guercum,
Pichia pijperi, Pichia stiptis, and Pichia ang,usta (Hansenula paymorpha).
[91] Therapeutic Glycoproteins Produced in Yeast (e.g., P. pastorisl
CA 02876864 2015-01-08
[92] Different glycoproteins may display varying degrees and types of
glycosylation in
the same host cell (Montesino et a/, 1998). The present invention provides
methods for
producing various glycoproteins in a recombinant yeast strain that essentially
lack
mannosylphospborylation. Preferably, the method involves engineering
expression of a
heterologous glycoprotein in P. pastoris Apnol Amnn4B. As such, the present
=
invention demonstrates elimination of mannosylphosphorylation from glycans on
various therapeutic glycoproteins (Fig. 7A-E, Fig. 8).
[93] While the reporter protein 1C3, contains a single N-Iinked
glycosylation site, the
reporter protein His-erytbropoietin (E)0) disclosed herein contains three N-
linked gly-
cosylations sites, the reporter protein His-CD40 disclosed herein contnins two
gly-
cosylation sites, and the His-invertase protein disclosed herein contains up
to 24 gly-
cosylation sites. }Es-tagged erythropoietin (1fis-EPO) is expressed from P.
pastoris
strain expressing mannosylphosphorylation in Fig. 7B and a P. pastoris dpnol
dmn
n4B strain lselcing mannosylphosphorylation in Fig. 7C. His-tagged CD40 (His-
CD40)
is expressed from P. pastoris strain expressing marmosylphosphorylation in
Fig. 713
and P. pastoris L'.pnol Amnn4b strain lacking mannosylphosphorylation in Fig.
7E.
His-tagged invertase is expressed from P. pastoris strain lacking manno-
sylphophorylation in Fig. 8. Strain construction for each of these
glycoproteins is
disclosed in Example 6.
194] ;dentification of MININ4 omologs
[95] In another aspect of the present invention, a method is provided for
identifying the
homologs to a 1vJJN4 gene in any yeast preferably Pichia sp. or filamentous
fungi. A
= skilled artisan can perform a BLAST database search using the amino acid
sequence of
:MNN4A, MNN4B, MNN4C or PN0.1 (Genbank accession gBD105434) against the
gen:ome of any yeast, ineferably Pichia and obtain the homologs to any of
these genes.
With the identification of the MNN4/PNOI homologs in Pichia yeast, one
qlcilled in
the art can subsequently disrupt or mutate any combination of these homologous
genes. An alignment is shown in Figure 9 of ./i4NN4IPNO1 homologs in P.
pastoris, S.
cerevisiae, Neurospora crassa, Aspergillus nidulans, Candida albicans and
Pichia
angusta (Hansenula polymorpha). Upon screening for the presence of
mannosylphos-
phorylated glycans on proteins =pressed from the Pichia host (Example 7), one
slcilled in the art can determinp the gene or combination of genes, which upon
disruption confer the expression of glycoproteins from the Pichia host which
are es-
sentially free of mannosylphosphorylation.
[96] The disrupted genes or genes which encode for proteins participating
in the transfer
of mannosylphosphate to glycans of glycoproteins are preferably from a yeast
strain
belonging to the genus Pichia. Yeasts belonging to the genus Pichia according
to the
present invention include, but are not limited to: Fichte pastoris, Fichte
finiandica,
Pichia irehalophila, Fichte koclamae, Pickle membranaeaciens, Pichia
methanolica,
Pichia minutia (Ogataea mimaa ,Pichia lindnerz), Pichia opuntiae, Pichia titer-
CA 02876864 2015-01-08
21
motolerans, Pichi salictaria, Pichia guercum, Pichia pijperi, Pichia stiptis,
and Pichia
angusta (Hansenula polymorpha). Pichia pastoris is preferably used among
these.
Other yeast and filamentous fungi include Saccharomyces cerevisiae , Schizosac-
charomyces pombe, Sczccharomyce sp. Hansenula polymorpha, lauyveronzyces sp.,
Cczndida sp., Candida albicans, Aspergilhz nidulans, Aspergillus niger,
Aspergillus
oryzae, Trichoderma reesei, Cloysosporium lucknowense, Fusarium sp., Fusarium
gramineum, Fusarium venenatum and Neurospora crassa.
[97] The following are examples which illustrate the compositions and
methods of this
invention. These examples should not be construed as limiting¨the examples are
included for the purposes of illustration only.
[98] Example 1
[99] Identification and sequencing of M/V7V4A. MATIV4B. MArN4C in P.
pastoris
(Figs. 1-31
[100] The Saccharonryces cerevisiae PINN4 protein sequence (Genbank
accession #
P36044) was blasted against a Pichi pastoris genomic sequence (Integrated
Ge.n.omics, Chicago, IL) for open reading frames encoding for proteins with
homology.
This search identified three ORFs with regions of homology to 1VINN4p. These
ORFs
were designated IviNN4A, MNN4B and kiNN4C. Each of these three genes was sub-
sequently sequenced. The MNN4A gene was found to contain an open reading frame
containing 2580.nucleotide bases coding for 860 amino acids (Fig. 1). The
MNN4B
gene was found to.contain an open reading frame containing 1956 nucleotide
bases'
coding for 652 amino acids (Fig. 2), and the MMV4C gene was found to contain
an
open reading frame containing 2289 nucleotide bases coding for 763 amino acids
(Fig.
3).
[101] Example.2
[102] Construction of P. pastoris strains: YSH-44 and YS'Ef-1
[103] P. pastoris YSH-44 and YSH-1 were engineered from BK64-1, an Aochl
deletion
mutant secreting 1(3, a reporter protein with a single N-linked glycosylation
site (Choi
et al., 2003, PNAS, 100: 5022-5027; Astnihon et aL, 2003, Science, 301: 1244-
1246).
YSH-1 expresses glycopron,im having predominantly GleNAcMan5GlcliAn2 N-
glycans and YSII-44 expresses glycoproteins having predominantly GIcNA.c2Man3
GloNAc2 N-glycans.
[104] Deletion of PNO1 gene in YSH-44 strain
[105] The pnol deletion allele (pnoI::Hyg A ) in YSFI-44 was generated by
the PCR
overlap method (Davidson et al., 1999, Microbial. 148:2607-2615'). Primers
PNK.1
(5'-CATAGCCCACTGCTAAGCC-AGAATTCTAATATG-31) (SEQ ID NO:7)
paired with PNK2
(5'-GCAGCGTACGAAGCTTCAGCTAGAATTGTAAAGTG-AATTATCAAG-TCT
TTC-3') (SEQ ID NO:8), PNK3
(51-CAGATCCACTAGTGGCCTATOCAACAA-TATAGCACCTCTCAAATACAC
CA 02876864 2015-01-08
22
G'TTG-3') (SEQ ID NO:9) paired with PNK4
(5'-TCTTGAAGTAGATTTGGAGA- GCGCTATG-3') (SEQ
NO:10) were
used to amplify the 5' and 3' flanking regions of the PNOI gene from genomic
DNA
(NRRL-Y11430). Primers KAN1 (5'-AGCTGAAGCT-TCGTACGCTGC-3') (SEQ
NO:11) paired with KAN2 (5'-GCATAGGCCACTAGTGGATCTG-3 (SEQ ID
NO:12) were used to amplify the Hyg resistance marker from vector pAG32
(Goldstein atul McCusker, 1999, Yeast, 14: 1541-1553). Primers PNIC1 and PNK4
were then used in a second reaction with all three products from first round
of PCR
reactions to generate an overlap product The resulting fusion PCR product was
used to
transform strain YSH-44, an engineered P. pastoris strain expressing
predominantly.
GIcNAc2Man3G1c,NAc2. Transfurmants were selected an YPD (1% yeast extract, 2%
peptone, 2% dextrose) medium contnining 200 mg/m1 of hygromycin B. Proper in-
=
tegration of deletion allele pnol::Hyg Rwas confirmed by pca. This Apno I
strain was
desigreed YSH-49.
[106] Example 3
[1073. PNOVIIINN4B knockout strategy in P. pastoris strain YSE1-49 Mg. 41
[108] YAS-130 (Apno1 Amnn4b) double mutant strain was achieved by PCR
overlap in
YSH-49. The TAS54 (TTCAACGAGTG-ACCAATGTAGA) (SEQ ID NO: 13) and .
TAS51 (CCAT-CCAGTGTCGAAAACGAGCTGGCGAA(..ITT1 __ CTGGGTCGAAG)
(SEQ ID NO:14) primers were used to amplify the 521 bp DNA fragment 5' of the
predicted start codon from Pichia pastoris genomic DNA (NRRL-Y 11430). TAS51
= contains a 22 bp=overhang that is complimentary to the 5' end of a drug
resistance
marker. TAS49 (TGAAGACGTCCCL'i ii GAACA) (SEQ ID NO:15) and TAS52
(ACGAGGCAAGCTAAAC.AG-ATCTAGTTG'TITTTTCTATATAAAA.C) (SEQ
NO:16) were used to amplify the 503 bp DNA fragment 3' of the predicted stop
codon.
TAS52 also contaim a 22 bp overhang that is complimentary to the 3' end of the
drug
resistance marker. PCR of the drag resistance marker used pAG29 (contains pat
ORF)
as the DNA source (Goldstein and McCuster, 1999). The drug resistance marker
was
amplified using primers TAS53
riCGA.CCCAGAAAAGITCGCCAGCTCG- fill CGACACTGGATGG) (SEQ
ID NO:17) and TAS50
(G ______________________________________________________ ATATAG-
AAAAAACAACTAGATCTGTTTAGCTTGCCTCGT) (SEQ ID
NO:14). TAS53 has a22 bp overhang that is comp1immtaty to the 22 bp 5' to the
predicted PINN4B start codon. TASK hoc a 22 bp overhang that is complimentary
to
the 22 bp 3' to the predicted WOMB stop codon. The 5' MIVN4B fragment, 3'
PINN4B
fragment, and the gene that confers resistance to a selectable marker were
combined in
an equimolar ratio and used as template DNA with primers TAS54 and TA549 for
the
PCR overlap reaction.
[109] ,PNO1/AINN4B knockout strategy in P. pastoris strain YSH-1
[110) YSH-1 was transformed by electroporation with SA-digested pIN503b
(Amnn4A
CA 02876864 2015-01-08
23
Apno1::URA3) to yield the Aochl Arram4A Apnol strain YAS159. The URA3
selectable marker
was recovered in this strain by 5-FOA counterselection. The resulting strain,
YAS164 (Aochl;
Amnn4A Apnol; ura3; his4; adel ; arg4), was transformed with SfiI-digested
pAS19
(Amnn4B::URA3) giving rise to the Aochl Amnn4A Lipnol Amnn4B strain YAS170.
The
YAS170 strain was subsequently countersele,cted on 5-FOA to yield the strain
YAS174 (Aochl
Amnn4A Apnol Amnn4B; ura3; his4; adel; arg4). YAS174 thus represents a Pichia
pastoris
strain that is deficient in mannose outer chain formation and void of
mannosylphosphate on
N-linked glycans.
[111] Example 4
= [112] PCR amplification
[113] An Eppendorf*Mastercycler was used for all PCR reactions. PCR
reactions
contained template DNA, 125 mM dNTPs, 0_2 mM each of forward and reverse
primer, Ex Taq polymerase buffer (Takara Bio Inc.), and Ex Taq polymerase. The
DNA fragments 5' to the predictedfrEVN4B ORF, 3' to the predicted .tlINN4B
ORF, and
the drug resistance marker were amplified with 30 cycles of 15 sec at 97 C, 15
sec at
55 C and 90 sec at 72 C with an initial denaturation step of 2 min at 97 C and
a final
extension step of 7 min at 72 C. PCP. samples were separated by agarose gel
elec-
trophoresis and the DNA bands were extracted and purified using a Gel
Extraction Kit
from Qiagen. All DNA purifications were elated in 10 mM Tris, pH 8.0 except
for the
final PCR (overlap of all three fragments) which was elated in deionized H20.
- [114] Example 5
[115] DNA Transformations. Culture Conditions for Production of Comolex
Glvcans in
P.pastoris for marmosylphosphorylation analysis
[116] DNA for transformation was prepared by adding sodium acetate to a
frul con-
centration of 0_3 M. One hunched percent ice cold ethanol was then added to a -
R-rtal
concentration of 70% to the DNA sample. DNA was pelleted by centrifugation
(12000g x 10min) and washed twice with 70% ice cold ethanol. The DNA was dried
and then resuspended in 50 ml of 10mM Tris, pH 8Ø YSH-49 and YAS-130 were
prepared by expanding a yeast culture in BMGY (buffered minimal glycerol: 100
raM
potassium phosphate, pH 6.0; 134% yeast nitrogen base; 4x10-5% biotin; 1%
glycerol)
to an OD. of ¨2-6. The yeast were made eleutiocompeteut by washing 3 times in
1M
sorbitol and resuspending in ¨1-2 mls IM sorbitol. DNA (1-2 mg) was mixed with
106
ml of competent yeast and incubated on ice for 10 min. Yeast were then
electroporated =
with a BTX Electrocell Manipulator 600 using the following parameters; 1.5 kV,
129
ohms, and 25 mF One milliliter of YPDS (1% yeast extract, 2% peptone, 2%
dextrose,
1M sorbitol) was added to the electroporated cells. Transfomed yeast were sub-
sequently plated on selective agar plates. Cells transformed with knockout
constructs
containing the hph resistance gene were spread onto YPD (1% yeast exiiact,
2%
peptone, 2% dextrose, 1.34% yeast nitrogen base without amino acids) agar
plates
*Trade mark
CA 02876864 2015-01-08
24
containing 0.4 mg/m1 hygromycin B. Cells transformed with knockout constructs
containing the pat resistance gene were spread onto defined medium (1.34%
yeast
nitrogen base lacking amino acids and 1H4SO4, 2% dextrose, 0.1% L-proline,
4x10-5%
biotin) agar plates containing 0.6 mg/ml glufosinate. Colonies were patched
onto
another plate containing the same drug selection. DNA was isolated from these
patches
and analyzed by PCR for replacement of the wild-type MNN4B ORF with the drag
resistance marker.
[117] Screening for knockouts was performed by PCR amplification
(Example 4) of both
the 5' and 3' portions of the knockout construct. TAS81
(TAGTCCAAGTACGA-AACGACACTATCG) (SEQ ID NO:19) and TAS08
(AGCTGCGCACGTCAAGAC-TGTCAAGG) (SEQ ID NO:20) primers were used to
screen the 5' portion of the knockout construct while TAS82
(ACGACGGTGAGITCAAACAGTTTGGTT) (SEQ 13) NO:21) and TAS07
(TCGCTATACTGCTGTCGATTCGATAC) (SEQ ID NO:22) primers were used to
= screen the 3' portion of the knockout construct Observation of a PCR
product in both
screens is indicative of a successful knockout of the MNN4B ORF since primers
TAS08 and TAS07 anneal at the 5' and 3' ends of the drag resistance marker
sequence,
respectively and TAS81 and TAS82 are complimentary to sequences in the genome
that flank the 5' and 3' regions of DNA used in the knockout construct Ninety
six
transformants were screened with four testing positive as an MNN4B knockout
All
four Apno 1 Amnn4b strains expressed the 13 reporter protein without
detectable
levels of mannosylphosphate. An example of this is shown in Figure 5D.
[118] Example 6
[119] Strain construction for His-tagged EPO, CD40 and Invertase proteins
Figs.
[120] For Ths-tagged erythropoietin (EPO), the first 166 amino acids of EPO
was
amplified from a human Edney cDNA library (Clontech) and inserted into the C-
terminal 6His pPICZA (Invitrogen) plaRmid at the BcoRI and KpiaI sites. This
plasmid
(pBK291) was transformed into two P. pastoris strains, resulting in the
following
strains expressing EPO-61fis: BK248 (ura3, izis4, adel, arg4, Aochi ::UR,43)
and
BK244 [YSH44 transformed with pBKI16 and p131C284 having the pnohnnn4b (
pno 1:.Hyg A) (mnn4b: :Kan 4) knockouts as described and shown in Example 2,
Figure 4. pBK116 results from a 1551 bp A0X1 3'UTR DNA fragment isolated from
NRRL11430 (ATCC) inserted into Invitrogen pPIC6A plasrnid at the Afila site
and a
1952bp A0X1 5TUTR DNA fragment isolated from NRRL11430 inserted into the
same pPIC6A plaRmid at the Bgla and BarnHT sites with the removal of the 573
bp
Pmel/BamEl DNA fragment This pBK116 was then digested with Notl and the
resulting NotI fragments were transformed into YSH44 in order to knock out the
reporter 1(3 protein. pB1(284 results from a 3196 bp DNA fragment inclnriing
the
A0X1 promoter, A0X1 ORF and A0X1 terminator sequence isolated from
CA 02876864 2015-01-08
NRRL11430 (ATCC) and cloned into the multiple cloning site of the Invitrogen
plasmid pCR2.1-TOPO. This plasmid was then digested with MscI and Bsslil in
order
to delete the kanamycin gene. This resulted in pBK284 which was digested with
PmeI
prior to transformation into the YSH44 stain transformed with pBK116 for
integration
into the AOXI promoter locus. HPLC glycan analysis of EPO-6His in BK248 and
BK244 is shown in Fig. 7B, C. For His-tagged CD40, the human CD40 DNA was
amplified by PCR from phCD40/GemT (Pullen et aL, 1999, JBC, 274: 14246-14254)
using a 5' EcoRI primer and a 3' His10-Kpn.1 primer for cloning into pPICZ aA
resulting in pIC33. pJC33 was expressed in P. pastoris strain YJC12 (ura3,
his4,
ade I, arg4) and YAS252-2 (YAS-130 transformed with pBK116, pBK284 and
= pRCD465 containing galactosyltransferase) resulting in YAS252. =
= HPLC
glycan analysis of CD49-6His in YJC12 and YAS252 is shown in Fig. 71), E. =
For His-tagged-invertase, the full length invertase sequence was amplified by
PCR
from Kluyveromyces lactis genomic DNA, strain CBS683, purchased from Cen-
- traalbureau voor Schimmelcultures. The invertase ORF was amplified using
blunt '
ended 5' and 3' primers for insertion into pPICZA plasmid (providing the C-
terminal =
6Efis tag) at the Pm1.1.site. This pPB147 was transformed into the P. pastoris
strain =
YAS245-2 (YAS130 transformed with pBK116, pBK284, and pRCD465
= resulting in YAS253.
HPLC glycan analysis of invertase-6His in YAS253 =
is shown in Fig. 8. , =
[121] Example 7
[122] Determination of mannosylphosphorylation in P. pastoris =
[123] The extent of mannosylphosphate transfer to N-linked glycans in the
strains shown
in Figs. 5-8 was determined by secreting a His-tagged reporter protein
(It:tingle 3 =
protein in Figs. 5,6; erythropoietin protein and CD40 protein in Fig. 7 and
invertase
protein in Fig. 8) expressed under the control of the methanol inducible AOXI
promoter. Briefly, a shake flask containing BMGY was inoculated with a fresh
yeast
culture (e.g., YAS-130) and grown to an O.D. of ¨20. The culture was
centrifuged and
the cell pellet washed with BMMY (buffered minimal methanol: same as BMGY
except 0.5% methanol instead of 1% glycerol). The cell pellet was resuspended
in
BMIvIY to a volume 1/5 of the original BMGY culture and placed in a shaker for
24 h.
The secreted protein was harvested by pelleting the biomass by centrifugation
and
= transferring the culture medium to a fresh tube. The His-tagged K3, EPO,
CD40 and
invertase proteins were then purified on a Ni-affinity column and digested
with
PNGase (Choi et al., 2003). Glycan was separated from protein and then labeled
with
.2-amino-ben7an-tide (2-AB). The 2-AB-labeled glycan was lyophili7ed,
resuspended in
HPLC grade water and subjected to HPLC using a GlycoSep C column (Glyco,
Novato, CA). This analysis allows separation of neutral and acidic glycans.
These
glycans were determined to be phosphoryIated from experiments with mild acid
hydrolysis which removes the terminal mannose group, exposing the phosphate.
With
CA 02876864 2015-01-08
26
subsequent alkaline phosphatase treatment, the terminal phosphate group can be
cleaved, leaving a neutral glycan. Successive experiments showed that
phosphorylated
N-linked glycans (acidic glycans) in all strains migrated between 20 and 30
minutes.
Baseline conditions were assessed using dH20 as a blank. The percentage of
phos-
phorylation was calculated by dividing the acidic peak areas by the sum of the
neutral
and the acidic peaks. This HPLC analysis was performed under the conditions
below.
[124] HPLC Analysis
[125] The HPLC conditions are as follows: Solvent A (acetonitrile), solvent
B (500mM
ammonium acetate, 500 raM, pH 4.5) and solvent C (water). The flow rate was
0.4
mL/min for 50 min. After eluting isocratically (20% A:80% C) for 10 min a
linear
solvent gradient (20% A:0% B:80% C to 20% A:50% B: 30%C) was employed over
30 min to elute the glycaus. The column was equilibrated with solvent (20% A:
80%C)
for 20 min between runs.
Sequence List Text
[126] SEQ ID NO:1 MNN4A (Fig. 1)
[127] " SEQ ID NO:2 MNN4A AA (Fig. 1)
[128] SEQ ID NO:3 MNN4B (Fig 2)
. ,
[129]
SEQ ID NO:4 MNN4B AA (Fig. 2) =
[130] SEQ
ID NO:5 IANN4C (Fig 3) ==
[131]
SEQ ID NO:6 IVINN4C AA (Fig. 3) =
[132]
SEQ ID NO:7 PNK1: CATAGCCCACTGCTAAGCCAGAATTCTMTATG =
[133] SEQ ID NO:8 PN1C2: GCAGCGTACGAAGCTTCAGCTAGAATTGTAAAGT-
GAATTATCAAGTCTTTC =
= =
[134] SEQ ID NO:9 PNIC.3: CAGATCCACTAGTGGCCTATGCAACAATATAG- =
CACCTCTCAAATACACGTTG
[135] SEQ ID NO:10 PNK4: TCTTGAAGTAGATTTGGAGATTTTGCGCTATG
[136] SEQ ID NO:11 KAN1: AGCTGAAGCTTCGTACGCTGC
[137] SEQ ID NO:12 KAM: GCATAGGCCACTAGTGGATCTG
[138) SEQ ID NO:13 TAS54: TTCAACGAGTGACCAATGTAGA
[139] SEQ ID NO:14 TAS51: CCATCCAGTGTCGAAAACGAGCTGGC-
GAACI ff1 __________ CTGGGTCGAAG
[140] SEQ ID NO:15 TAS49: TGAAGACGTCCCCTTTGAACA
[141] SEQ ID NO:16 TAS52: ACGAGGCAAGCTAAACAGATCTAGTTG TTC-
TATATAAAAC
[142] SEQ ID NO:17 TAS53: C'1'1CGACCCAGAAAAGTTCGCCAGCTCGTTTTC-
GACACTGGATGG
[143] SEQ ID NO:18 TAS50: G1"1 I 1ATATAGAAAAAACAACTAGATCTGTT-
TAGCTTGCCTCGT
[144] SEQ ID NO:19 TAS81: TAGTCCAAGTACGAAACGACACTATCG
[145] SEQ ID NO:20 TAS08: AGCTGCGCACGTCAAGACTGTCAAGG
CA 02876864 2015-01-08
27
[146] SEQ ID NO:21 TAS82: ACGACGGTGAGTTCAAACAGTTTGGTT
[147] SEQ NO:22 TAS07: TCGCTATACTGCTGTCGATTCGATAC
=
=
= = =
=
DEMANDES OU BREVETS VOLUMINEUX
LA PRESENTE PARTIE DE CETTE DEMANDE OU CE BREVETS
COMPREND PLUS D'UN TOME.
CECI EST LE TOME 1 DE 2
NOTE: Pour les tomes additionels, veillez contacter le Bureau Canadien des
Brevets.
JUMBO APPLICATIONS / PATENTS
THIS SECTION OF THE APPLICATION / PATENT CONTAINS MORE
THAN ONE VOLUME.
THIS IS VOLUME 1 OF 2
NOTE: For additional volumes please contact the Canadian Patent Office.