Note: Descriptions are shown in the official language in which they were submitted.
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
MUTATIONS IN SOLANACEAE PLANTS THAT MODULATE SHOOT
ARCHITECTURE AND ENHANCE YIELD-RELATED PHENOTYPES
FEDERALLY SPONSORED RESEARCH OR DEVELOPMENT
This invention was made with government support under federal grant number IOS-
1237880 awarded by the National Science Foundation. The government has certain
rights in
the invention.
CROSS-REFERENCE TO RELATED APPLICATIONS
This application claims the benefit of the filing date of U.S. Provisional
Application
No. 61/728,654, filed November 20, 2012, and of the filing date of U.S.
Provisional
Application No. 61/869,052, filed August 22, 2013 and claims priority to U.S.
Application
No. 13/799,831 filed March 13, 2013. The entire contents of each of these
referenced
applications are incorporated by reference herein.
BACKGROUND
There are ongoing attempts to enhance yield and quality, as well as life span,
of food
crops and other plants, such as ornamental plants and trees, in an effort to
use resources more
efficiently and produce more food, flowers and trees. Additional approaches
for doing so are
still needed, and one of the primary targets for manipulating plant
productivity is the
flowering process and its corresponding effects on vegetative and reproductive
shoot
architecture.
SUMMARY
Described herein are novel genetic variants of Solanaceae plants, e.g., tomato
plants,
that (a) exhibit modified flowering time and shoot architecture; (b) exhibit
higher yield,
higher quality products (e.g., fruits); (c) produce products (e.g., fruits)
with different
compositions, (e.g., brix, also known as enhanced soluble solids or sugar
concentration in the
fruits,); or (d) any combination of (a) to (c), compared to corresponding
"wild-type (WT)"
Solanaceae plants (Solanaceae plants that have not been genetically altered).
In one embodiment, a genetically-altered plant, such as a genetically-altered
semi-
determinate or semi-indeterminate Solanaceae plant, comprises a mutant
suppressor of spl
- 1 -
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
(sspl) gene and a mutant single flower truss (sft) gene. Such Solanaceae
plants are
heterozygous or homozygous for mutant genes (e.g., heterozygous or homozygous
for a
mutant suppressor of spl (sspl) gene or heterozygous or homozygous for a
mutant single
flower truss (sft) gene) or heterozygous or homozygous for both a mutant
suppressor of spl
(sspl) gene and a mutant single flower truss (sft) gene (double heterozygotes
or double
homozygotes). Such plants may further comprise a mutant self pruning (sp)
gene. Such
plants may be heterozygous or homozygous for a mutant self pruning (sp) gene.
Such plants
can also be in the wild type SELF PRUNING (SP) background.
In a further embodiment, Solanaceae plants comprise a mutant self pruning (sp)
gene
and a mutant single flower truss (sft) gene. Such Solanaceae plants are
heterozygous or
homozygous for mutant genes (e.g., for a mutant self pruning (sp) gene or for
a mutant single
flower truss (sft) gene) or heterozygous or homozygous for both a mutant self
pruning (sp)
gene and a mutant single flower truss (sft) gene (double heterozygotes or
double
homozygotes).
In another embodiment, Solanaceae plants comprise a mutant self pruning (sp)
gene
and a mutant suppressor of spl (sspl) gene. Such Solanaceae plants are
heterozygous or
homozygous for mutant genes (e.g., for a mutant self pruning (sp) gene or for
a mutant
suppressor of spl (sspl) gene) or heterozygous or homozygous for both a mutant
self pruning
(sp) gene and a mutant suppressor of spl (sspl) gene (double heterozygotes or
double
homozygotes).
In yet another embodiment, Solanaceae plants comprise a mutant suppressor of
spl
(sspl) gene or a mutant single flower truss (sft) gene. Such Solanaceae plants
are
heterozygous or homozygous for a mutant suppressor of spl (sspl) gene or
heterozygous or
homozygous for a mutant single flower truss (sft) gene. Such plants may
further comprise a
mutant self pruning (sp) gene. Such plants may be heterozygous or homozygous
for a mutant
self pruning (sp) gene. Such plants can also be in the wild type SELF PRUNING
(SP)
background.
In some embodiments, the mutant sspl gene comprises a nucleic acid sequence
that
encodes a mutant sspl protein that comprises a mutant SAP motif. In some
embodiments,
the mutant sspl gene comprises a nucleic acid sequence that encodes a mutant
sspl protein
that comprises a mutant SAP motif with a sequence of SEQ ID NO: 14 or SEQ ID
NO: 15.
- 2 -
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
In some embodiments, the mutant sspl gene encodes a mutant sspl polypeptide
comprising
the sequence of SEQ ID NO: 5 or SEQ ID NO: 6. In some embodiments, the mutant
sspl
gene comprises a C to T mutation at position 641 of SEQ ID NO: 1, a C to T
mutation at
position 647 of SEQ ID NO: 1, or a C to T mutation at position 641 and 647 of
SEQ ID NO:
1. In some embodiments, the mutant sspl gene comprises a coding sequence that
comprises
the nucleic acid sequence of SEQ ID NO: 2 or SEQ ID NO: 3. In some
embodiments, the
mutant sspl gene comprises the nucleic acid sequence of SEQ ID NO: 2 or SEQ ID
NO: 3.
In some embodiments, the mutant sspl gene comprises the nucleic acid sequence
of SEQ ID
NO: 25 or SEQ ID NO: 26. In some embodiments, the mutant sspl gene is ssp-2129
or ssp-
610. In some embodiments, the mutant sp gene comprises a coding sequence that
comprises
the nucleic acid sequence of SEQ ID NO: 8. In some embodiments, the mutant sp
gene
comprises the nucleic acid sequence of SEQ ID NO: 8. In some embodiments, the
mutant sp
gene comprises the nucleic acid sequence of SEQ ID NO: 23. In some
embodiments, the
mutant sp gene comprises a nucleic acid that encodes the amino acid sequence
of mutant sp
protein (e.g., SEQ ID NO: 10). SEQ ID NO: 2 is the nucleotide sequence of the
coding
sequence of ssp-2129, a mutant allele of SSP1. SEQ ID NO: 3 is the nucleotide
sequence of
the coding sequence of ssp-610, a mutant allele of SSP1. SEQ ID NO: 5 is the
amino acid
sequence of ssp-2129 mutant protein. SEQ ID NO: 6 is the amino acid sequence
of ssp-610
mutant protein. In some embodiments, the mutant sft gene comprises a coding
sequence that
comprises the nucleic acid sequence of SEQ ID NO: 20. In some embodiments, the
mutant
sft gene comprises SEQ ID NO: 20. In some embodiments, the mutant sft gene
comprises the
nucleic acid sequence of SEQ ID NO: 17. In some embodiments, the mutant sft
gene encodes
a mutant sft polypeptide comprising the sequence of SEQ ID NO: 21. In some
embodiments,
the mutant sft gene comprises a nucleotide sequence that encodes a mutant sft
polypeptide
comprising a Val to Met mutation at position 132 of SEQ ID NO: 19. In some
embodiments,
the mutant sft gene is sft-1906.
In some embodiments, the genetically-altered Solanaceae plant is a genetically
altered
semi-determinate or semi-indeterminate Solanaceae plant, such as a tomato
(Solanum
lycopersicum) plant. In some embodiments, the genetically-altered semi-
determinate or
semi-indeterminate Solanaceae plant is isogenic. In some embodiments, the
genetically-
altered semi-determinate or semi-indeterminate Solanaceae plant is inbred. In
some
- 3 -
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
embodiments, the genetically-altered semi-determinate or semi-indeterminate
Solanaceae
plant is a hybrid.
In another aspect, the disclosure relates to a seed for producing a
genetically-altered
Solanaceae plant, such as a semi-determinate or semi-indeterminate genetically-
altered
Solanaceae plant, as described herein, e.g., a genetically-altered semi-
determinate or semi-
indeterminate Solanaceae plant comprising (e.g., heterozygous or homozygous
for) a mutant
single flower truss (sft) gene and a mutant suppressor of spl (sspl) gene, a
genetically-altered
semi-determinate or semi-indeterminate Solanaceae plant comprising (e.g.,
heterozygous or
homozygous for) a mutant single flower truss (sft) gene and a mutant self
pruning (sp) gene,
a genetically-altered semi-determinate or semi-indeterminate Solanaceae plant
comprising
(e.g., heterozygous or homozygous for) a mutant suppressor of spl (sspl) gene
and a mutant
self pruning (sp) gene, or a genetically-altered semi-determinate or semi-
indeterminate
Solanaceae plant comprising (e.g., heterozygous or homozygous for) a mutant
single flower
truss (sft) gene, a mutant suppressor of spl (sspl) gene, and a mutant self
pruning (sp) gene.
In some embodiments, the mutant sspl gene comprises a nucleic acid sequence
that encodes
a mutant sspl protein that comprises a mutant SAP motif. In some embodiments,
the mutant
sspl gene comprises a nucleic acid sequence that encodes a mutant sspl protein
that
comprises a mutant SAP motif comprising SEQ ID NO: 14 or SEQ ID NO: 15. In
some
embodiments, the mutant sspl gene encodes a mutant sspl polypeptide comprising
the
sequence of SEQ ID NO: 5 or SEQ ID NO: 6. In some embodiments, the mutant sspl
gene
comprises a C to T mutation at position 641 of SEQ ID NO: 1, a C to T mutation
at position
647 of SEQ ID NO: 1, or a C to T mutation at position 641 and 647 of SEQ ID
NO: 1. In
some embodiments, the mutant sspl gene comprises a coding sequence that
comprises the
nucleic acid sequence of SEQ ID NO: 2 or SEQ ID NO: 3. In some embodiments,
the mutant
sspl gene comprises the nucleic acid sequence of SEQ ID NO: 2 or SEQ ID NO: 3.
In some
embodiments, the mutant sspl gene comprises the nucleic acid sequence of SEQ
ID NO: 25
or SEQ ID NO: 26. In some embodiments, the mutant sp gene comprises a coding
sequence
that comprises the nucleic acid sequence of SEQ ID NO: 8. In some embodiments,
the
mutant sp gene comprises the nucleic acid sequence of SEQ ID NO: 8. In some
embodiments, the mutant sp gene comprises the nucleic acid sequence of SEQ ID
NO: 23. In
some embodiments, the mutant sp gene comprises a nucleic acid that encodes the
amino acid
- 4 -
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
sequence of mutant sp protein (e.g., SEQ ID NO: 10).
In yet another aspect, the disclosure relates to methods of producing a
genetically-
altered Solanaceae plant, such as a semi-determinate or semi-indeterminate
Solanaceae plant.
In some embodiments, the method comprises:
(a) introducing a mutant sft gene into a Solanaceae plant containing a mutant
sspl
gene (or alternatively introducing a mutant sspl gene into a Solanaceae plant
containing a
mutant sft gene), thereby producing a genetically-altered Solanaceae plant
containing a
mutant sft gene and a mutant sspl gene; and
(b) self-crossing the genetically-altered Solanaceae plant produced in (a) or
crossing
two genetically-altered Solanaceae plants produced in (a) under conditions
appropriate for
producing a genetically-altered Solanaceae plant heterozygous or homozygous
for the mutant
sft gene and heterozygous or homozygous for the mutant sspl gene. In some
embodiments,
the plant produced in (b) is heterozygous for the mutant sft gene and
heterozygous for the
mutant sspl gene. In some embodiments, the plant produced in (b) further
comprises a
mutant sp gene (e.g., is heterozygous or homozygous for a mutant sp gene).
In some embodiments, the method of producing a genetically-altered Solanaceae
plant
comprises:
(a) introducing a mutant sft gene into a Solanaceae plant part containing a
mutant
sspl gene (or alternatively introducing a mutant sspl gene into a Solanaceae
plant part
containing a mutant sft gene), thereby producing a genetically-altered
Solanaceae plant part
containing the mutant sft gene and the mutant sspl gene;
(b) maintaining the genetically-altered Solanaceae plant part containing a
mutant
sft gene and a mutant sspl gene produced in (a) under conditions and for
sufficient time for
production of a genetically-altered Solanaceae plant containing the mutant sft
gene and the
mutant sspl gene from the plant part, thereby producing a genetically-altered
Solanaceae
plant containing the mutant sft gene and the mutant sspl gene;
(c) self-crossing the genetically-altered Solanaceae plant produced in (b)
or
crossing two genetically-altered Solanaceae plants produced in (b) under
conditions
appropriate for producing a genetically-altered Solanaceae plant heterozygous
or
homozygous for the mutant sft gene and heterozygous or homozygous for the
mutant sspl
gene, thereby producing a genetically-altered Solanaceae plant. In some
embodiments, the
- 5 -
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
plant produced in (c) is heterozygous for the mutant sft gene and heterozygous
for the mutant
sspl gene. In some embodiments, the plant produced in (c) further comprises a
mutant sp
gene (e.g., is heterozygous or homozygous for a mutant sp gene).
In some embodiments, the mutant sspl gene comprises a nucleic acid sequence
that
encodes a mutant sspl protein that comprises a mutant SAP motif. In some
embodiments,
the mutant sspl gene comprises a nucleic acid sequence that encodes a mutant
sspl protein
that comprises a mutant SAP motif with a sequence of SEQ ID NO: 14 or SEQ ID
NO: 15.
In some embodiments, the mutant sspl gene encodes a mutant sspl polypeptide
comprising
the sequence of SEQ ID NO: 5 or SEQ ID NO: 6. In some embodiments, the mutant
sspl
gene comprises a C to T mutation at position 641 of SEQ ID NO: 1, a C to T
mutation at
position 647 of SEQ ID NO: 1, or a C to T mutation at position 641 and 647 of
SEQ ID NO:
1. In some embodiments, the mutant sspl gene comprises a coding sequence that
comprises
the nucleic acid sequence of SEQ ID NO: 2 or SEQ ID NO: 3. In some
embodiments, the
mutant sspl gene comprises the nucleic acid sequence of SEQ ID NO: 2 or SEQ ID
NO: 3.
In some embodiments, the mutant sspl gene comprises the nucleic acid sequence
of SEQ ID
NO: 25 or SEQ ID NO: 26. In some embodiments, the mutant sp gene comprises a
coding
sequence that comprises the nucleic acid sequence of SEQ ID NO: 8. In some
embodiments,
the mutant sp gene comprises the nucleic acid sequence of SEQ ID NO: 8. In
some
embodiments, the mutant sp gene comprises the nucleic acid sequence of SEQ ID
NO: 23. In
some embodiments, the mutant sp gene comprises a nucleic acid that encodes the
amino acid
sequence of mutant sp protein (e.g., SEQ ID NO: 10). In some embodiments, the
mutant sft
gene comprises a coding sequence that comprises SEQ ID NO: 20. In some
embodiments,
the mutant sft gene comprises SEQ ID NO: 20. In some embodiments, the mutant
sft gene
comprises SEQ ID NO: 17. In some embodiments, the mutant sft gene comprises a
nucleic
acid sequence that encodes a mutant sft polypeptide comprising the sequence of
SEQ ID NO:
21. In some embodiments, the mutant sft gene and/or mutant sspl gene is
introduced into a
plant or a plant part by a method selected from the group consisting of:
Agrobacterium-
mediated recombination, viral-vector mediated recombination, microinjection,
gene gun
bombardment/biolistic particle delivery, nuclease mediated recombination, and
electroporation. In some embodiments, the Solanaceae plant is a tomato
(Solanum
lycopersicum) plant. In some embodiments, the genetically-altered Solanaceae
plant is a
- 6 -
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
plant that suppresses sp imposed determinate growth and exhibits a sympodial
index of less
than three and either terminates (is semi-determinate), or continues to
produce sympodial
shoots (is semi-indeterminate). Accordingly, in some embodiments, the
Solanaceae plant is
semi-determinate or semi-indeterminate. In some embodiments, the Solanaceae
plant is a
semi-determinate or semi-indeterminate plant with a sympodial index of less
than 3, less than
2.5, less than 2, or less than 1.5. In some embodiments, the Solanaceae plant
is a semi-
determinate or semi-indeterminate tomato plant. In some embodiments, the
Solanaceae plant
is a semi-determinate or semi-indeterminate tomato plant with a sympodial
index of less than
3, less than 2.5, less than 2, or less than 1.5
In some embodiments, the Solanaceae plant is inbred. In some embodiments, the
genetically-altered semi-determinate Solanaceae plant is a hybrid. In another
aspect, the
disclosure relates to a genetically-altered Solanaceae plant produced by or
producible by the
methods described herein.
In another aspect, the disclosure relates to a genetically-altered Solanaceae
plant, e.g.,
a semi-determinate or semi-indeterminate Solanaceae plant, homozygous for a
mutant
suppressor of spl (sspl) gene and homozygous for a self pruning (sp) gene. In
some
embodiments, the mutant sspl gene comprises a nucleic acid sequence that
encodes a mutant
sspl protein that comprises a mutant SAP motif. In some embodiments, the
mutant sspl
gene comprises a nucleic acid sequence that encodes a mutant sspl protein that
comprises a
mutant SAP motif with a sequence of SEQ ID NO: 14 or SEQ ID NO: 15. In some
embodiments, the mutant sspl gene encodes a mutant sspl polypeptide comprising
the
sequence of SEQ ID NO: 5 or SEQ ID NO: 6. In some embodiments, the mutant sspl
gene
comprises a C to T mutation at position 641 of SEQ ID NO: 1, a C to T mutation
at 647 of
SEQ ID NO: 1, or a C to T mutation at position 641 and 647 of SEQ ID NO: 1. In
some
embodiments, the mutant sspl gene comprises a coding sequence that comprises
the nucleic
acid sequence of SEQ ID NO: 2 or SEQ ID NO: 3. In some embodiments, the mutant
sspl
gene comprises the nucleic acid sequence of SEQ ID NO: 2 or SEQ ID NO: 3. In
some
embodiments, the mutant sspl gene comprises the nucleic acid sequence of SEQ
ID NO: 25
or SEQ ID NO: 26. In some embodiments, the mutant sp gene comprises a coding
sequence
that comprises the nucleic acid sequence of SEQ ID NO: 8. In some embodiments,
the
mutant sp gene comprises the nucleic acid sequence of SEQ ID NO: 8. In some
- 7 -
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
embodiments, the mutant sp gene comprises the nucleic acid sequence of SEQ ID
NO: 23. In
some embodiments, the mutant sp gene comprises a nucleic acid that encodes the
amino acid
sequence of mutant sp protein (e.g., SEQ ID NO: 10).
In some embodiments, the genetically-altered Solanaceae plant, e.g., a semi-
determinate or semi-indeterminate Solanaceae plant, is a tomato (Solanum
lycopersicum)
plant. In some embodiments, the genetically-altered Solanaceae plant, e.g., a
semi-
determinate or semi-indeterminate Solanaceae plant, is isogenic. In some
embodiments, the
genetically-altered Solanaceae plant, e.g., a semi-determinate or semi-
indeterminate
Solanaceae plant, is inbred. In some embodiments, the genetically-altered
Solanaceae plant,
e.g., a semi-determinate or semi-indeterminate Solanaceae plant, is a hybrid.
In another aspect, the disclosure relates to a genetically-altered Solanaceae
plant
heterozygous for a mutant suppressor of spl (sspl) gene and homozygous for a
mutant self
pruning (sp) gene. In some embodiments, the mutant sspl gene comprises a
nucleic acid
sequence that encodes a mutant sspl protein that comprises a mutant SAP motif.
In some
embodiments, the mutant sspl gene comprises a nucleic acid sequence that
encodes a mutant
sspl protein that comprises a mutant SAP motif with a sequence of SEQ ID NO:
14 or SEQ
ID NO: 15. In some embodiments, the mutant sspl gene encodes a mutant sspl
polypeptide
comprising the sequence of SEQ ID NO: 5 or SEQ ID NO: 6. In some embodiments,
the
mutant sspl gene comprises a C to T mutation at position 641 of SEQ ID NO: 1,
a C to T
mutation at 647 of SEQ ID NO: 1, or a C to T mutation at position 641 and 647
of SEQ ID
NO: 1. In some embodiments, the mutant sspl gene comprises a coding sequence
that
comprises the nucleic acid sequence of SEQ ID NO: 2 or SEQ ID NO: 3. In some
embodiments, the mutant sspl gene comprises the nucleic acid sequence of SEQ
ID NO: 2 or
SEQ ID NO: 3. In some embodiments, the mutant sspl gene comprises the nucleic
acid
sequence of SEQ ID NO: 25 or SEQ ID NO: 26. In some embodiments, the mutant sp
gene
comprises a coding sequence that comprises the nucleic acid sequence of SEQ ID
NO: 8. In
some embodiments, the mutant sp gene comprises the nucleic acid sequence of
SEQ ID NO:
8. In some embodiments, the mutant sp gene comprises the nucleic acid sequence
of SEQ ID
NO: 23. In some embodiments, the mutant sp gene comprises a nucleic acid that
encodes the
amino acid sequence of mutant sp protein (e.g., SEQ ID NO: 10). In some
embodiments, the
genetically-altered Solanaceae plant is a tomato (Solanum lycopersicum) plant.
In some
- 8 -
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
embodiments, the genetically-altered Solanaceae plant is isogenic. In some
embodiments, the
genetically-altered Solanaceae plant is inbred. In some embodiments, the
genetically-altered
semi-determinate Solanaceae plant is a hybrid.
In another aspect, the disclosure relates to a genetically-altered Solanaceae
plant
homozygous for a mutant suppressor of spl (sspl) gene. In some embodiments,
the mutant
sspl gene comprises a nucleic acid sequence that encodes a mutant sspl protein
that
comprises a mutant SAP motif. In some embodiments, the mutant sspl gene
comprises a
nucleic acid sequence that encodes a mutant sspl protein that comprises a
mutant SAP motif
with a sequence of SEQ ID NO: 14 or SEQ ID NO: 15. In some embodiments, the
mutant
sspl gene encodes a mutant sspl polypeptide comprising the sequence of SEQ ID
NO: 5 or
SEQ ID NO: 6. In some embodiments, the mutant sspl gene comprises a C to T
mutation at
position 641 of SEQ ID NO: 1, a C to T mutation at 647 of SEQ ID NO: 1, or a C
to T
mutation at position 641 and 647 of SEQ ID NO: 1. In some embodiments, the
mutant sspl
gene comprises a coding sequence that comprises the nucleic acid sequence of
SEQ ID NO: 2
or SEQ ID NO: 3. In some embodiments, the mutant sspl gene comprises the
nucleic acid
sequence of SEQ ID NO: 2 or SEQ ID NO: 3. In some embodiments, the mutant sspl
gene
comprises the nucleic acid sequence of SEQ ID NO: 25 or SEQ ID NO: 26. In some
embodiments, the mutant sp gene comprises a coding sequence that comprises the
nucleic
acid sequence of SEQ ID NO: 8. In some embodiments, the mutant sp gene
comprises the
nucleic acid sequence of SEQ ID NO: 8. In some embodiments, the mutant sp gene
comprises the nucleic acid sequence of SEQ ID NO: 23. In some embodiments, the
mutant
sp gene comprises a nucleic acid that encodes the amino acid sequence of
mutant sp protein
(e.g., SEQ ID NO: 10). In some embodiments, the genetically-altered Solanaceae
plant is a
tomato (Solanum lycopersicum) plant. In some embodiments, the genetically-
altered
Solanaceae plant is isogenic. In some embodiments, the genetically-altered
Solanaceae plant
is inbred. In some embodiments, the genetically-altered semi-determinate
Solanaceae plant is
a hybrid. In some embodiments, the genetically-altered Solanaceae plant is
homozygous for
a wild-type SELF PRUNING (SP) gene.
In another aspect, the disclosure relates to a seed for producing a
genetically-altered
semi-determinate or semi-indeterminate Solanaceae plant as described herein,
e.g. a
genetically-altered semi-determinate or semi-indeterminate Solanaceae plant
homozygous for
- 9 -
CA 02892012 2015-05-20
WO 2014/081730
PCT/US2013/070825
a mutant suppressor of spl (sspl) gene and homozygous for a mutant self
pruning (sp) gene.
In some embodiments, the mutant sspl gene comprises a nucleic acid sequence
that encodes
a mutant sspl protein that comprises a mutant SAP motif. In some embodiments,
the mutant
sspl gene comprises a nucleic acid sequence that encodes a mutant sspl protein
that
comprises a mutant SAP motif with a sequence of SEQ ID NO: 14 or SEQ ID NO:
15. In
some embodiments, the mutant sspl gene encodes a mutant sspl polypeptide
comprising the
sequence of SEQ ID NO: 5 or SEQ ID NO: 6. In some embodiments, the mutant sspl
gene
comprises a C to T mutation at position 641 of SEQ ID NO: 1, a C to T mutation
at 647 of
SEQ ID NO: 1, or a C to T mutation at position 641 and 647 of SEQ ID NO: 1. In
some
embodiments, the mutant sspl gene comprises a coding sequence that comprises
the nucleic
acid sequence of SEQ ID NO: 2 or SEQ ID NO: 3. In some embodiments, the mutant
sspl
gene comprises the nucleic acid sequence of SEQ ID NO: 2 or SEQ ID NO: 3. In
some
embodiments, the mutant sspl gene comprises the nucleic acid sequence of SEQ
ID NO: 25
or SEQ ID NO: 26. In some embodiments, the mutant sp gene comprises a coding
sequence
that comprises the nucleic acid sequence of SEQ ID NO: 8. In some embodiments,
the
mutant sp gene comprises the nucleic acid sequence of SEQ ID NO: 8. In some
embodiments, the mutant sp gene comprises the nucleic acid sequence of SEQ ID
NO: 23.
In some embodiments, the mutant sp gene comprises a nucleic acid that encodes
the amino
acid sequence of mutant sp protein (e.g., SEQ ID NO: 10).
In yet another aspect, the disclosure relates to methods of producing a
genetically-
altered semi-determinate or semi-indeterminate Solanaceae plant. In some
embodiments, the
method comprises:
(a) introducing a mutant sspl gene into a Solanaceae plant containing a mutant
sp
gene, thereby producing a genetically-altered Solanaceae plant containing a
mutant sspl gene
and a mutant sp gene; and
(b) self-crossing the genetically-altered Solanaceae plant produced in (a) or
crossing
two genetically-altered Solanaceae plants produced in (a) under conditions
appropriate for
producing a genetically-altered Solanaceae plant homozygous for the mutant
sspl gene and
the mutant sp gene, thereby producing a genetically-altered Solanaceae plant
that is semi-
determinate or semi-indeterminate. In some embodiments, the method of
producing a
genetically-altered semi-determinate or semi-indeterminate Solanaceae plant
comprises:
- 10-
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
(a) introducing a mutant sspl gene into a Solanaceae plant part containing
a
mutant sp gene, thereby producing a genetically-altered Solanaceae plant part
containing the
mutant sspl gene and the mutant sp gene;
(b) maintaining the genetically-altered Solanaceae plant part containing a
mutant
sspl and a mutant sp gene produced in (a) under conditions and for sufficient
time for
production of a genetically-altered Solanaceae plant containing the mutant
sspl gene and the
mutant sp gene from the plant part, thereby producing a genetically-altered
Solanaceae plant
containing the mutant sspl gene and the mutant sp gene;
(c) self-crossing the genetically-altered Solanaceae plant produced in (b)
or
crossing two genetically-altered Solanaceae plants produced in (b) under
conditions
appropriate for producing a genetically-altered Solanaceae plant homozygous
for the mutant
sspl gene and the mutant sp gene, thereby producing a genetically-altered
Solanaceae plant
that is semi-determinate or semi-indeterminate. In some embodiments, the
mutant sspl gene
comprises a nucleic acid sequence that encodes a mutant sspl protein that
comprises a mutant
SAP motif. In some embodiments, the mutant sspl gene comprises a nucleic acid
sequence
that encodes a mutant sspl protein that comprises a mutant SAP motif with a
sequence of
SEQ ID NO: 14 or SEQ ID NO: 15. In some embodiments, the mutant sspl gene
encodes a
mutant sspl polypeptide comprising the sequence of SEQ ID NO: 5 or SEQ ID NO:
6. In
some embodiments, the mutant sspl gene comprises a C to T mutation at position
641 of
SEQ ID NO: 1, a C to T mutation at position 647 of SEQ ID NO: 1, or a C to T
mutation at
position 641 and 647 of SEQ ID NO: 1. In some embodiments, the mutant sspl
gene
comprises a coding sequence that comprises the nucleic acid sequence of SEQ ID
NO: 2 or
SEQ ID NO: 3. In some embodiments, the mutant sspl gene comprises the nucleic
acid
sequence of SEQ ID NO: 2 or SEQ ID NO: 3. In some embodiments, the mutant sspl
gene
comprises the nucleic acid sequence of SEQ ID NO: 25 or SEQ ID NO: 26. In some
embodiments, the mutant sp gene comprises a coding sequence that comprises the
nucleic
acid sequence of SEQ ID NO: 8. In some embodiments, the mutant sp gene
comprises the
nucleic acid sequence of SEQ ID NO: 8. In some embodiments, the mutant sp gene
comprises the nucleic acid sequence of SEQ ID NO: 23. In some embodiments, the
mutant
sp gene comprises a nucleic acid that encodes the amino acid sequence of
mutant sp protein
(e.g., SEQ ID NO: 10). In some embodiments, the mutant sspl gene is introduced
into a
- 11-
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
plant or a plant part by a method selected from the group consisting of:
Agrobacterium-
mediated recombination, viral-vector mediated recombination, microinjection,
gene gun
bombardment/biolistic particle delivery, nuclease mediated recombination, and
electroporation. In some embodiments, the Solanaceae plant is a tomato
(Solanum
lycopersicum) plant. In some embodiments, the Solanaceae plant is inbred. In
some
embodiments, the genetically-altered semi-determinate Solanaceae plant is a
hybrid. In
another aspect, the disclosure relates to a genetically-altered semi-
determinate Solanaceae
plant produced by or producible by the methods herein.
Other aspects of the disclosure relate to isolated polynucleotides or isolated
polypeptides. In some embodiments, the isolated polynucleotide encodes a
mutant sspl
protein having the amino acid sequence of SEQ ID NO: 5 or SEQ ID NO: 6. In
some
embodiments, the isolated polynucleotide comprises the nucleic acid sequence
of SEQ ID
NO: 2 or SEQ ID NO: 3. In some embodiments, the isolated polynucleotide
encodes a
mutant sft protein having the amino acid sequence of SEQ ID NO: 21. In some
embodiments, the isolated polynucleotide comprises the nucleic acid sequence
of SEQ ID
NO: 20.
Other aspects of the disclosure relate to plant cells and food products as
described
herein.
BRIEF DESCRIPTION OF THE DRAWINGS
Figure 1 shows the SSP1 protein and SFT protein. The bZIP domain and SAP motif
are indicated and the two mutations (ssp-2129 and ssp-610) in the SAP motif of
the SSP1
protein are indicated: Proline to Leucine (P216L) and Threonine to Isoleucine
(T214I),
respectively. Two putative ligand binding sites are indicated and the mutation
in the external
loop region of the SFT protein is indicated (se).
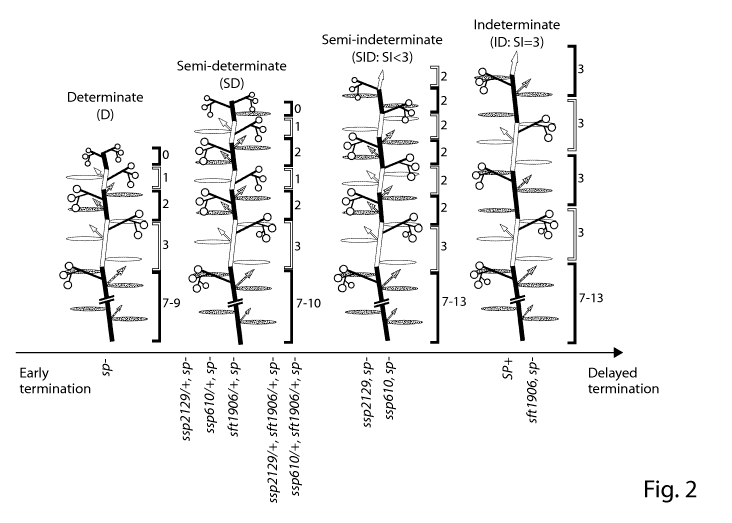
Figure 2 shows graphical representations of the suppression of sp mutant
imposed
sympodial shoot termination with the sspl and sft mutants in the sp mutant
background. Left
depicts a sp "Determinate" (D) mutant plant. Right depicts the wild-type
"Indeterminate"
(ID) plant with fully functional SP, SSP1 and SFT genes. Note the three-leaf
reiteration of
vegetative sympodial shoots (SYM) after the primary shoot produces 7-9 leaves
and
transitions to flowering. The number of leaves in each sympodial unit, known
as the
- 12-
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
Sympodial Index (SI) is consistently three. The second diagram from left
depicts the "Semi-
determinate" (SD) single heterozygote plants of ssp-2129, ssp610 or sft1906,
and double
heterozygote plants of ssp-2129, ssp610 or sft1906 in the sp mutant
background. Note that
shoot growth ends after a few more units of vegetative shoot (SYM) production
compared to
the sp determinate plants. Right middle depicts the "semi-indeterminate" ssp-
2129; sp
double mutant plant. Note the reduction in leaf number from three to two in
each SYM
(SI=2). Dots indicate flowers/fruits produced in each inflorescence. Black and
gray arrows
indicate axillary shoots on the leaf axils. The long horizontal arrow
indicates the gradual
change in determinacy due to individual and combined mutations in homozygous
and
heterozygous conditions.
Figure 3 shows internode length between inflorescences in sympodial units in
determinate (cultivar M82D), indeterminate (cultivar M82ID and sft1906; sp)
and semi-
indeterminate (ssp-2129; sp and, ssp-610; sp) plants. Note the intermediate
internode length
for the semi-indeterminate ssp-2129; sp and ssp-610; sp double mutant plants,
which reflects
a reduced sympodial index to an average of two leaves. ** P<0.01, student t-
test against
M82D, and against M82ID indicated with lines.
Figure 4 is a series of graphs showing shoot determinacy of the primary shoot,
the
inflorescence, and the sympodial shoot in M82 indeterminate (M82ID), M82
determinate
(M82D), two sspl mutant alleles (ssp-2129 and ssp-610), the sft-1906 mutant
allele, and all
genotypic combinations among mutant alleles. Figure 4A shows the numbers of
leaves
produced by the primary shoot. Figure 4B shows the numbers of flowers per
inflorescence.
Figure 4C shows the total number of inflorescences generated from each
genotype. Figure
4D shows numbers of leaves within each sympodial shoot (i.e.; sympodial index)
in M82ID,
M82D, ssp-2129 and ssp-610 and sft1906. The primary and sympodial shoot
architectures
from all genotypic combinations are shown in Figure 4E. Note the quantitative
range of
sympodial indices created by different homozygous and heterozygous mutant
combinations.
ID, indeterminate SYM growth; D, determinate SYM growth; SD, semi-determinate
SYM
growth. The font size of "D" and "ID" indicate relative values of shoot
terminations from 20
plants. ** P<0.01, students t-test against M82D indicated with asterisks above
bars, and
against M82ID indicated asterisks above lines. Standard error is shown for
Figure 4B, C, and
E, and in all other cases standard deviation.
- 13 -
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
Figure 5 is a schematic of the map-based cloning procedure used to identify
the SSP1
gene. The highlighted region is the mapping interval identified using map-
based cloning.
Arrows indicate single nucleotide polymorphisms (SNPs). The coding region of
SSP1 is
shown. The C to T mutations in the protein C-terminus of ssp-2129 and ssp-610
are
indicated.
Figure 6A and 6B is a ClustalW analysis of the nucleotide sequence of the
coding
sequence of the wild-type SSPlgene and the coding sequences of the two mutant
alleles e610
and e2129 (SEQ ID NOs: 1, 3 and 2, respectively).
Figure 7 is a ClustalW analysis of the amino acid sequence of the wild-type
SSPlgene
and the two mutant alleles e610 and e2129 (SEQ ID NOs: 4, 6 and 5,
respectively).
Figure 8A-8D are graphic representations of results of assessment of
characteristics of
plants of the genotypes indicated. Figure 8A shows brix-yield (brix x yield)
of plants of the
genotypes indicated. Figure 8B shows (soluble solids, sugar concentration in
the fruits) of
plants of the genotypes indicated. Figure 8C shows yield of red fruit for
plants of the
genotypes indicated. Figure 8D shows total yield of fruit for plants of the
genotypes
indicated. Yield was measured by taking 15 plants from each genotype in a
completely
randomized design with 1 plant/meter squared. * P<0.05, ** P<0.01, students t-
test against
M82D. Error bars are standard error.
Figure 9 is a ClustalW analysis of the nucleotide sequence of the wild-type
SFT
coding sequence and the mutant allele sft-1906 coding sequence (SEQ ID NOs: 18
and 20,
respectively).
Figure 10 is a ClustalW analysis of the amino acid sequence of the wild-type
SFT
gene and the mutant allele sft-1906 (SEQ ID NOs: 19 and 21, respectively).
SEQUENCES
SEQ ID NO: 1 is the nucleotide sequence of the coding sequence of wild-type
tomato SSP1.
SEQ ID NO: 2 is the nucleotide sequence of the coding sequence of ssp-2129, a
mutant allele
of SSP1.
SEQ ID NO: 3 is the nucleotide sequence of the coding sequence of ssp-610, a
mutant allele
of SSP1.
- 14-
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
SEQ ID NO: 4 is the amino acid sequence of wild-type tomato SSP1 protein.
SEQ ID NO: 5 is the amino acid sequence of ssp-2129 mutant protein.
SEQ ID NO: 6 is the amino acid sequence of ssp-610 mutant protein.
SEQ ID NO: 7 is the nucleotide sequence of the coding sequence of wild-type
tomato SP
gene.
SEQ ID NO: 8 is the nucleotide sequence of the coding sequence of a mutant sp
gene.
SEQ ID NO: 9 is the amino acid sequence of wild-type SP protein.
SEQ ID NO: 10 is the amino acid sequence of mutant sp protein.
SEQ ID NO: 11 is the amino acid sequence of the SAP motif of the tomato SSP1
protein.
SEQ ID NO: 12 is the nucleotide sequence of ssp-2129 that encodes the mutant
SAP motif of
ssp-2129 protein.
SEQ ID NO: 13 is the nucleotide sequence of ssp-610 that encodes the mutant
SAP motif of
ssp-610 protein.
SEQ ID NO: 14 is the amino acid sequence of the mutant SAP motif of ssp-2129
protein.
SEQ ID NO: 15 is the amino acid sequence of the mutant SAP motif of ssp-610
protein.
SEQ ID NO: 16 is the nucleic acid sequence of SFT wild-type genomic region.
SEQ ID NO: 17 is the nucleic acid sequence of sft-1906, a mutant allele of
SFT, genomic
region.
SEQ ID NO: 18 is the nucleic acid sequence of the coding sequence of wild-type
SFT DNA.
SEQ ID NO: 19 is the amino acid sequence of wild-type SFT protein.
SEQ ID NO: 20 is the nucleic acid sequence of the coding sequence of sft-1906,
a mutant of
SFT.
SEQ ID NO: 21 is the amino acid sequence of mutant sft protein.
SEQ ID NO: 22 is the nucleic acid sequence of SP wild-type genomic region.
SEQ ID NO: 23 is the nucleic acid sequence of sp mutant genomic region.
SEQ ID NO: 24 is the nucleic acid sequence of SSP wild-type genomic region.
SEQ ID NO: 25 is the nucleic acid sequence of ssp-2129 mutant genomic region.
SEQ ID NO: 26 is the nucleic acid sequence of ssp-610 mutant genomic region.
- 15 -
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
DETAILED DESCRIPTION
Tomato yield, on both a per plant basis and in the context of tons per acre,
depends
partly on fruit size, but is mainly driven by the production of dozens of
multi-flowered
inflorescences and resulting fruit clusters that develop according to the
"sympodial" growth
habit. The defining feature of sympodial plants is that the shoot apical
meristem (SAM) ends
growth by differentiating into a terminal flower after producing a set number
of leaves, and
growth then renews from a specialized axillary (sympodial) meristem (SYM)
that, in tomato,
produces just three leaves before undergoing its own flowering transition and
termination.
Indefinite reiteration of three-leaf sympodial flowering events results in an
"indeterminate"
plant that continuously produces equally spaced inflorescences. The regular
production of
leaves between inflorescences is known as the "sympodial index" (SI).
Tomato breeding goals are multifaceted and shift according to the needs and
desires
of growers (e.g. improved pest resistances) and consumers (e.g. better
quality), but one
unwavering aim is to improve yield. Indeterminate cultivars are grown
commercially to
enable continuous market delivery of "round", "roma", "cocktail", "grape", and
"cherry"
tomato types that are eaten fresh and command a premium price. Indeterminate
tomatoes are
primarily grown in greenhouses where successively ripening clusters are
harvested by hand
multiple times over an extended period, in some cases up to a year, to
maximize yield on
plants that must be pruned to one or two main shoots to enable efficient
greenhouse growth
and maintain fresh market quality. While the necessary pruning of
indeterminate tomatoes
facilitates agronomic practices that maximize quality, such as size, shape,
and flavor, it also
limits yield. In contrast, tomatoes grown for sauces, pastes, juices, or other
processed can or
jar products where fruit quality is less relevant, must be managed
agronomically to produce
maximum yields (per acre) through once-over mechanical harvests to be
economically
justified. Maximal yields for processing tomatoes are achieved by growing
determinate sp
mutants in the open field to their full potential, because sequential
sympodial shoots
transition to flowering progressively faster in sp plants, which results in a
compact bush-like
form where fruits ripen uniformly. Thus, sp varieties lend themselves to once-
over
mechanical harvesting and have therefore come to dominate the processing
tomato industry,
although determinate varieties have also been bred for fresh market
production. In a parallel
to the physical pruning of indeterminate tomatoes, one drawback of sp-imposed
determinate
- 16-
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
growth is that inflorescence and fruit production is restricted, because of a
genetic pruning
that causes sympodial cycling to stop. Thus, maximizing inflorescence and
fruit production
while simultaneously minimizing shoot production for the tomato industry has
remained a
challenging goal.
One approach to increase inflorescence production and yield is to reduce SI to
less
than three. As described herein, chemically induced mutant populations of
tomatoes in the
self pruning (sp) background were screened for mutations that suppressed sp
imposed growth
determinacy, with the goal of producing indeterminate tomatoes with reduced
SI. Three
mutants were identified. The first mutant carried a novel mutation in the gene
SINGLE
FLOWER TRUSS (SFT), the mutation referred to herein as sft-1906, which
resulted in a weak
allele. SFT is the tomato ortholog of the Arabidopsis thaliana gene FLOWERING
LOCUS T
(FT). The other two mutants were defective in a gene designated SUPPRESSOR OF
SP]
(SSP1), which encodes a protein partner of SFT. SSP1 is the tomato ortholog of
the
Arabidopsis thaliana gene FLOWERING LOCUS D (FD). These other two mutants are
referred to herein as ssp-2129 or ssp-610. Remarkably, each homozygous sft and
sspl mutant
transforms determinate tomato plants into indeterminate tomato plants with a
SI of less than
three.
As described herein, data was generated in cherry and roma tomato types that
demonstrate the utility of the new SFT and SSP1 mutations to serve as novel
germplasm for
breeding. Specifically, it was found that mixing and matching these mutations
in various
homozygous and heterozygous combinations resulted in a quantitative range of
sympodial
indices and novel determinate, semi-determinate, and indeterminate shoot
architectures that
have not been possible to achieve with existing breeding germplasm. Notably,
it has been
also found that these mutations increase the number of flowers per
inflorescence.
The SSP1 and SFT mutations were introduced into the classic mutant self-
pruning (sp)
determinate background (sp-classic, coding sequence is SEQ ID NO: 8), which is
a
prerequisite genotype for generating indeterminate plants with a SI of less
than 3. SP is the
tomato ortholog of the Arabidopsis thaliana gene TERMINAL FLOWER] (TFL1). Data
from
the roma cultivar `M82' (which is homozygous for sp-classic) showed that
homozygosity for
ssp-2129 or ssp-610 in the sp-classic background results in semi-indeterminate
plants with an
average SI of 2. Data from a determinate variety of the cherry type 'Currant'
tomato (S.
- 17 -
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
pimpinellifolium) showed that homozygosity for sft-1906 or either of the sspl
mutations
created a SI of 2. Double heterozygotes of sft-1906 and ssp-2129 or ssp-610 in
hybrids
resulted in an average SI less than 2, illustrating that combining mutations
in either
homozygous or heterozygous states can lead to subtle quantitative manipulation
of sympodial
shoot architecture and yield potential. Finally, all three mutants increase
the number of
flowers per inflorescence, with sft-1906 exhibiting the strongest effect.
Described herein are genetically-altered Solanaceae plants, such as
genetically-altered
semi-determinate (SD) or semi-indeterminate (SID) Solanaceae plants, e.g., a
tomato plant
(Solanum lycopersicum), that comprise a mutant gene or mutant genes and
exhibit
characteristics different from those of the corresponding plant that has not
been genetically
altered. The characteristics include any combination of the following:
modified flowering
time and shoot architecture; higher yield; higher quality products (e.g.,
fruits); and products
(e.g., fruits) with different compositions (e.g., brix, also known as enhanced
soluble solids or
sugar concentration in the fruits,), compared to corresponding "wild-type
(WT)" Solanaceae
plants that have not been genetically altered. In some embodiments, the
genetically-altered
Solanaceae plant has a sympodial index of less than 3, less than 2.5, less
than 2, or less than
1.5. In some embodiments, the genetically-altered Solanaceae plant has a
sympodial index of
between 1 and 3 or between 1.5 and 2.5.
In some embodiments, genetically-altered heterozygous or homozygous Solanaceae
plants, e.g., tomato plants (Solanum lycopersicum), comprise a mutant
flowering gene SSP1
(sspl) and mutant FLOWERING LOCUS gene SFT (sft). In some embodiments, the
genetically-altered heterozygous plants are semi-determinate (SD) or semi-
indeterminate
(SID). Such plants have increased yield; higher quality products (e.g.,
fruits); and products
(e.g., fruits) with different compositions (e.g., brix, also known as enhanced
soluble solids or
sugar concentration in the fruits,) relative to these characteristics of
corresponding plants that
do not comprise such mutant genes. The plants comprise a variety of
combinations of the
different mutant alleles, such as mutant sspl (e.g., ssp-2129, ssp-610) with
sft mutations (e.g.,
sft-1906). The genetically-altered plants are heterozygotes or homozygotes
and, in some
cases, are double heterozygotes or double homozygotes. In one embodiment, such
plants
comprise ssp-2129, described herein, and mutant SFT, such as sft-1906. In
another
embodiment, the plants comprise ssp-610, described herein, and mutant SFT,
such as sft-
- 18 -
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
1906.
In a specific embodiment, genetically-altered Solanaceae plants, such as
tomato plants
designated herein as sft-1906 x M82 Fl, exhibited greater yield than the
corresponding wild
type tomato plants. See, for example, Figure 8D. Similarly, other genetically-
engineered
tomato plants produced greater yields (total fruit, red fruit, for example)
than the
corresponding wild-type tomato plants. See Figure 8C and 8D, graphic
representations of
yield of fruit by heterozygous tomato plants described herein (e.g., double
heterozygotes
ssp2129 x sft1906 and ssp610 x sft1906, as well as the three heterozygotes
indicated: sft1906
x M82F1; ssf610 x M82F1; ssp2129 x M82F1).
Also described herein are semi-determinate (SD) or semi-indeterminate (SID)
Solanaceae plants, e.g., a tomato plant (Solanum lycopersicum), that comprise
a mutant of
SUPPRESSOR OF SP1 (SSP1). The SSP1 gene encodes a bZIP transcription factor
that
physically interacts with the florigen hormone, SFT, to induce the flowering
transition and
flower production. SSP1 has two domains, the bZIP domain and the SAP motif
(Figure 1).
The bZIP domain contains a basic leucine zipper capable of interacting with
DNA. The SAP
motif, "RTSTAPF" (SEQ ID NO: 11), is found in the C-terminus of SSP1 from
amino acid
position 211 to 217 in SEQ ID NO: 4. The SAP motif is similar to a
conventional 14-3-3
recognition motif.
Mutant sspl genes, such as ssp-2129 and ssp-610
In some embodiments, the mutant sspl gene comprises, for example, a coding
sequence comprising a nucleic acid (e.g., DNA) having the sequence of SEQ ID
NO: 2; a
coding sequence comprising a portion of SEQ ID NO: 2 that exhibits
substantially the same
activity (e.g., encoding the same polypeptide or substantially the same
polypeptide that has
the same activity) as a nucleic acid (e.g., DNA) having the sequence of SEQ ID
NO: 2; a
coding sequence comprising a nucleic acid (e.g., DNA) having at least 85%, at
least 90%, at
least 95%, at least 96%, at least 97%, at least 98%, or at least 99% identity
with the sequence
of SEQ ID NO: 2; a nucleic acid (e.g., DNA) having the sequence of positions
631 to 651of
SEQ ID NO: 2 (CGGACGTCAACTGCTCTATTT, SEQ ID NO: 12); a coding sequence
comprising an orthologue or homologue of the nucleic acid having the sequence
of SEQ ID
NO: 2; an orthologue or homologue of the nucleic acid sequence of positions
631 to 651 of
- 19-
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
SEQ ID NO: 2; a coding sequence comprising a nucleic acid (e.g., DNA) having
the
sequence of SEQ ID NO: 3; a coding sequence comprising a portion of SEQ ID NO:
3 that
exhibits substantially the same activity (e.g., encoding the same polypeptide
or substantially
the same polypeptide that has the same activity) as a nucleic acid (e.g., DNA)
having the
sequence of SEQ ID NO: 3; a coding sequence comprising a nucleic acid (e.g.,
DNA) having
at least 85%, at least 90%, at least 95%, at least 96%, at least 97%, at least
98%, or at least
99% identity with the sequence of SEQ ID NO: 3; a nucleic acid (e.g., DNA)
having the
sequence of positions 631 to 651of SEQ ID NO: 3 (CGGACGTCAATTGCTCCATTT, SEQ
ID NO: 13); a coding sequence comprising an orthologue or homologue of the
nucleic acid
having the sequence of SEQ ID NO: 3; or an orthologue or homologue of the
nucleic acid
sequence of positions 631 to 651of SEQ ID NO: 3. In some embodiments, the
mutant sspl
gene comprises a C to T mutation at position 641 of SEQ ID NO: 1, a C to T
mutation at
position 647 of SEQ ID NO: 1, or a C to T mutation at position 641 and
position 647 of SEQ
ID NO: 1. In some embodiments, the mutant sspl gene comprises a nucleotide
sequence that
encodes a polypeptide of SEQ ID NOs: 5 or 6 or a nucleotide sequence that
encodes a
polypeptide that comprises SEQ ID NOs: 14 or 15. In some embodiments, the
mutant sspl
gene comprises a nucleic acid sequence that encodes a mutant sspl protein that
comprises at
least one mutation in a SAP motif, wherein the at least one mutation alters
flowering time and
shoot architecture of the Solanaceae plant, e.g., by conferring semi-
determinacy. In some
embodiments, the SAP motif with the at least one mutation has the amino acid
sequence SEQ
ID NO: 14 or SEQ ID NO: 15. In some embodiments, the mutant sspl gene
comprises a
nucleic acid sequence that encodes a mutant sspl protein that comprises at
least one mutation
in a SAP motif or in the two amino acids flanking the N-terminal position of
the SAP motif
and the one amino acid flanking the C-terminal position of the SAP motif,
which includes the
phosphorylation site for Ca-dependent protein kinases (CDPKs), wherein the at
least one
mutation alters flowering time and shoot architecture of the Solanaceae plant,
e.g., by
conferring semi-determinacy. In some embodiments, the mutant sspl gene
comprises, for
example, a nucleic acid (e.g., DNA) having the sequence of SEQ ID NO: 25; a
portion of
SEQ ID NO: 25 that exhibits substantially the same activity (e.g., encoding
the same
polypeptide or substantially the same polypeptide that has the same activity)
as a nucleic acid
(e.g., DNA) having the sequence of SEQ ID NO: 25; a nucleic acid (e.g., DNA)
having at
- 20 -
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
least 85%, at least 90%, at least 95%, at least 96%, at least 97%, at least
98%, or at least 99%
identity with the sequence of SEQ ID NO: 25; an orthologue or homologue of the
nucleic
acid having the sequence of SEQ ID NO: 25; a nucleic acid (e.g., DNA) having
the sequence
of SEQ ID NO: 26; a portion of SEQ ID NO: 26 that exhibits substantially the
same activity
(e.g., encoding the same polypeptide or substantially the same polypeptide
that has the same
activity) as a nucleic acid (e.g., DNA) having the sequence of SEQ ID NO: 26;
a nucleic acid
(e.g., DNA) having at least 85%, at least 90%, at least 95%, at least 96%, at
least 97%, at
least 98%, or at least 99% identity with the sequence of SEQ ID NO: 26; or an
orthologue or
homologue of the nucleic acid having the sequence of SEQ ID NO: 26.
In some embodiments, the semi-determinate (SD) or semi-indeterminate (SID)
Solanaceae plant, e.g., a tomato plant, comprises a mutant sspl polypeptide
(e.g., a mutant
sspl protein) encoded by a mutant sspl gene. In some embodiments, the mutant
sspl
polypeptide comprises the sequence of SEQ ID NO: 5; a portion of SEQ ID NO: 5
that
exhibits substantially the same activity as a polypeptide (e.g., a protein)
having the sequence
of SEQ ID NO: 5; an amino acid sequence having at least 85%, at least 90%, at
least 95%, at
least 96%, at least 97%, at least 98%, or at least 99% identity with the
sequence of SEQ ID
NO: 5; the amino acid sequence of positions 211 to 217 in SEQ ID NO: 5
(RTSTALF, SEQ
ID NO: 14); an orthologue or homologue of the polypeptide having the sequence
of SEQ ID
NO: 5; an orthologue or homologue of the amino acid sequence of positions 211
to 217 in
SEQ ID NO: 5 (RTSTALF, SEQ ID NO: 14); a polypeptide (e.g., a protein) having
the
sequence of SEQ ID NO: 6; a portion of SEQ ID NO: 6 that exhibits
substantially the same
activity as a polypeptide (e.g., a protein) having the sequence of SEQ ID NO:
6; an amino
acid sequence having at least 85%, at least 90%, at least 95%, at least 96%,
at least 97%, at
least 98%, or at least 99% identity with the sequence of SEQ ID NO: 6; the
sequence of
positions 211 to 217 in SEQ ID NO: 6 (RTSIAPF, SEQ ID NO: 15); an orthologue
or
homologue of the polypeptide having the sequence of SEQ ID NO: 6; or an
orthologue or
homologue of the polypeptide sequence of positions 211 to 217 in SEQ ID NO: 6
(RTSIAPF,
SEQ ID NO: 15). In some embodiments, the polypeptide comprises a Thr to Ile
mutation at
position 214 of SEQ ID NO: 4 or a Pro to Leu mutation at position 216 of SEQ
ID NO: 4, or
a Thr to Ile mutation at position 214 and a Pro to Leu mutation at position
216 of SEQ ID
NO: 4. In some embodiments, the mutant sspl polypeptide comprises an amino
acid
- 21 -
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
sequence with at least one mutation in a SAP motif, wherein the at least one
mutation alters
flowering time and shoot architecture of the Solanaceae plant, e.g., by
conferring semi-
determinacy or semi-indeterminacy. In some embodiments, the SAP motif with the
at least
one mutation has the amino acid sequence SEQ ID NO: 14 or SEQ ID NO: 15. In
some
embodiments, the mutant sspl polypeptide comprises an amino acid sequence with
at least
one mutation in a SAP motif or in the two amino acids flanking the N-terminal
position of the
SAP motif and the one amino acid flanking the C-terminal position of the SAP
motif, which
includes the phosphorylation site for Ca-dependent protein kinases (CDPKs),
wherein the at
least one mutation alters flowering time and shoot architecture of the
Solanaceae plant, e.g.,
by conferring semi-determinacy or semi-indeterminacy.
Mutant sft gene, such as sft-1906
In some embodiments, the mutant sft gene comprises, for example, a coding
sequence
comprising a nucleic acid (e.g., DNA) having the sequence of SEQ ID NO: 20; a
coding
sequence comprising a portion of SEQ ID NO: 20 that exhibits substantially the
same activity
(e.g., encoding the same polypeptide or substantially the same polypeptide
that has the same
activity) as a nucleic acid (e.g., DNA) having the sequence of SEQ ID NO: 20;
a coding
sequence comprising a nucleic acid (e.g., DNA) having at least 85%, at least
90%, at least
95%, at least 96%, at least 97%, at least 98%, or at least 99% identity with
the sequence of
SEQ ID NO: 20; a coding sequence comprising an orthologue or homologue of the
nucleic
acid having the sequence of SEQ ID NO:20. In some embodiments, the mutant sft
gene
comprises a G to A mutation at position 394 of SEQ ID NO: 20. In some
embodiments, the
mutant sft gene comprises, for example, a nucleic acid (e.g., DNA) having the
sequence of
SEQ ID NO: 17; a portion of SEQ ID NO: 17 that exhibits substantially the same
activity
(e.g., encoding the same polypeptide or substantially the same polypeptide
that has the same
activity) as a nucleic acid (e.g., DNA) having the sequence of SEQ ID NO: 17;
a nucleic acid
(e.g., DNA) having at least 85%, at least 90%, at least 95%, at least 96%, at
least 97%, at
least 98%, or at least 99% identity with the sequence of SEQ ID NO: 17; or an
orthologue or
homologue of the nucleic acid having the sequence of SEQ ID NO: 17. In some
embodiments, the mutant sft gene comprises a nucleotide sequence that encodes
a mutant sft
polypeptide of SEQ ID NO: 21. In some embodiments, the mutant sft gene
comprises a
- 22 -
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
nucleotide sequence that encodes a mutant sft polypeptide that comprises a Val
to Met
mutation at position 132 of SEQ ID NO: 19.
Solanaceae plants comprising mutant genes
Flowering time and shoot architecture; higher yield, higher quality products
(e.g.,
fruits); and products (e.g., fruits) with different compositions (e.g., brix,
also known as
enhanced soluble solids or sugar concentration in the fruits,), can be
manipulated in a wide
variety of types of Solanaceae plants that comprise a mutant sspl gene or a
mutant sft gene,
or two or three mutant genes¨a mutant sspl gene and a mutant self-pruning (sp)
gene; a
.. mutant sspl and a mutant sft gene; a mutant sft gene and a mutant self-
pruning (sp) gene; or a
mutant sft gene, a mutant sspl gene, and a mutant self-pruning (sp) gene. The
mutant sspl
gene can comprise, for example, any of the sspl nucleic acids described
herein. The mutant
sft gene can comprise, for example, any of the sft nucleic acids described
herein. The mutant
sp gene can comprise, for example, any of the sp nucleic acids described
herein.
In specific embodiments, the mutant sspl gene and/or the mutant sft gene is
present
along with a mutant self-pruning (sp) gene in a double mutant background and
the mutant sp
gene (Pnueli et al. Development. 1998;125(11):1979-89) comprises, for example,
a coding
sequence comprising a nucleic acid (e.g., DNA) having the sequence of SEQ ID
NO: 8; a
coding sequence comprising a portion of SEQ ID NO: 8 that exhibits
substantially the same
.. activity (e.g., encoding the same polypeptide or substantially the same
polypeptide that has
the same activity) as a nucleic acid (e.g., DNA) having the sequence of SEQ ID
NO: 8; a
coding sequence comprising a nucleic acid (e.g., DNA) having at least 85%, at
least 90%, at
least 95%, at least 96%, at least 97%, at least 98%, or at least 99% identity
with the sequence
of SEQ ID NO: 8; or a coding sequence comprising an orthologue or homologue of
the
.. nucleic acid having the sequence of SEQ ID NO: 8. In some embodiments, the
mutant sp
gene comprises, for example, a nucleic acid (e.g., DNA) having the sequence of
SEQ ID NO:
23; a portion of SEQ ID NO: 23 that exhibits substantially the same activity
(e.g., encoding
the same polypeptide or substantially the same polypeptide that has the same
activity) as a
nucleic acid (e.g., DNA) having the sequence of SEQ ID NO: 23; a nucleic acid
(e.g., DNA)
.. having at least 85%, at least 90%, at least 95%, at least 96%, at least
97%, at least 98%, or at
least 99% identity with the sequence of SEQ ID NO: 23; or an orthologue or
homologue of
-23 -
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
the nucleic acid having the sequence of SEQ ID NO: 23. In some embodiments,
the mutant
sp gene comprises a C to T mutation at position 227 of SEQ ID NO: 7. In some
embodiments, the mutant sp gene comprises a nucleotide sequence with at least
one mutation
that reduces the activity of a sp protein encoded by the mutant sp gene. In
some
embodiments, the mutant sp gene comprises a nucleotide sequence that encodes a
polypeptide of SEQ ID NO: 10. In some embodiments, the SD or SID Solanaceae
plant
comprises a mutant sp polypeptide (e.g., a protein) encoded by a mutant sp
gene. In some
embodiments, the mutant sp polypeptide comprises, for example, the sequence of
SEQ ID
NO: 10; a portion of SEQ ID NO: 10 that exhibits substantially the same
activity as a
polypeptide (e.g., a protein) having the sequence of SEQ ID NO: 10; an amino
acid sequence
having at least 85%, at least 90%, at least 95%, at least 96%, at least 97%,
at least 98%, or at
least 99% identity with the sequence of SEQ ID NO: 10; or an orthologue or
homologue of
the polypeptide having the sequence of SEQ ID NO: 10. In some embodiments, the
mutant
sp polypeptide comprises a Pro to Leu mutation at position 76 of SEQ ID NO: 9.
In some
embodiments, the mutant sp polypeptide comprises at least one mutation that
reduces
(partially or completely) the activity of the sp polypeptide. In some
embodiments, the mutant
sp polypeptide comprises at least one mutation that reduces the activity of
the sp polypeptide,
wherein the reduced (partially or completely) activity of the sp polypeptide
can confer
determinacy.
The Solanaceae plant can be, for example, inbred, isogenic or hybrid, as long
as the
plant comprises a mutant sspl gene, a mutant sft gene, a mutant ssplgene and a
mutant sft
gene, a mutant sft gene and a mutant sp gene, a mutant sspl gene and a mutant
sp gene, or a
mutant ssplgene, a mutant sft gene, and a mutant sp gene. Plants in the
Solanaceae family
include, e.g., tomato, potato, eggplant, petunia, tobacco, and pepper. In some
embodiments,
the Solanaceae plant is a tomato plant. In some embodiments, the Solanaceae
plant, e.g.
tomato plant, is not a variety.
In some embodiments, the Solanaceae plant comprises one wild-type copy of the
SSP1 gene and one mutant copy of the sspl gene as described herein (is
heterozygous for the
mutant sspl gene). In some embodiments, the Solanaceae plant comprises two
copies of a
mutant sspl gene as described herein (is homozygous for the mutant sspl gene).
In some
embodiments, the Solanaceae plant comprises a first mutant sspl gene as
described herein
- 24 -
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
and a second mutant sspl gene as described herein, wherein the first mutant
sspl gene and
the second mutant sspl gene are different (e.g., the first mutant sspl gene
comprises a coding
sequence comprising SEQ ID NO: 2 and the second mutant sspl gene comprises a
coding
sequence comprising SEQ ID NO: 3). In some embodiments, the Solanaceae plant
comprises
one copy of a mutant sspl gene as described herein and one copy of a mutant sp
gene as
described herein (is heterozygous for the mutant sspl gene and heterozygous
for the mutant
sp gene). In some embodiments, the Solanaceae plant comprises one copy of a
mutant sspl
gene as described herein and two copies of a mutant sp gene as described
herein (is
heterozygous for the mutant sspl gene and homozygous for the mutant sp gene).
In some
embodiments, the Solanaceae plant comprises two copies of a mutant sspl gene
as described
herein and two copies of a mutant sp gene as described herein (is homozygous
for the mutant
sspl gene and homozygous for the mutant sp gene).
In some embodiments, the Solanaceae plant comprises one wild-type copy of a
SFT
gene and one mutant copy of a sft gene as described herein (is heterozygous
for the mutant sft
gene). In some embodiments, the Solanaceae plant comprises two copies of a
mutant sft gene
as described herein (is homozygous for the mutant sft gene). In some
embodiments, the
Solanaceae plant comprises one copy of a mutant sft gene as described herein
and one copy
of a mutant sp gene as described herein (is heterozygous for the mutant sft
gene and
heterozygous for the mutant sp gene). In some embodiments, the Solanaceae
plant comprises
one copy of a mutant sft gene as described herein and two copies of a mutant
sp gene as
described herein (is heterozygous for the mutant sft gene and homozygous for
the mutant sp
gene). In some embodiments, the Solanaceae plant comprises two copies of a
mutant sft gene
as described herein and two copies of a mutant sp gene as described herein (is
homozygous
for the mutant sft gene and homozygous for the mutant sp gene).
In some embodiments, the Solanaceae plant comprises one wild-type copy of a
SFT
gene and one mutant copy of a sft gene as described herein (is heterozygous
for the mutant sft
gene) and comprises one wild-type copy of the SSP1 gene and one mutant copy of
the sspl
gene as described herein (is heterozygous for the mutant sspl gene). In some
embodiments,
the Solanaceae plant comprises two copies of a mutant sft gene as described
herein (is
homozygous for the mutant sft gene) and comprises two copies of a mutant
ssplgene as
described herein (is homozygous for the mutant ssplgene). In some embodiments,
the
- 25 -
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
Solanaceae plant comprising a mutant sft gene (one or two copies) as described
herein and a
mutant sspl gene (one or two copies) further comprises one copy of a mutant sp
gene as
described herein (is heterozygous or homozygous for the mutant sft gene and
the mutant
sspl gene and heterozygous for the mutant sp gene). In some embodiments, the
Solanaceae
plant further comprises two copies of a mutant sp gene as described herein (is
homozygous
for the mutant sp gene). Exemplary, non-limiting genotype combinations
include:
Combination # SSP1 genotype SFT genotype SP genotype
1 ssp-2129I+ +/+ sp/sp
2 ssp-610I+ +/+ sp/sp
3 +1+ sft-1906I+ sp/sp
4 ssp-2129I+ sft-1906I+ sp/sp
5 ssp-610I+ sft-1906I+ sp/sp
6 ssp-2129/ssp-2129 +/+ sp/sp
7 ssp-610/ssp-610 +/+ sp/sp
8 ssp-2129/ ssp-610 +/+ sp/sp
9 +1+ sft-1906/sft-1906 sp/sp
In some embodiments, a genetically-altered Solanaceae plant (e.g., a tomato
plant)
provided herein is semi-determinate or semi-indeterminate. As used herein, a
Solanaceae
plant that is "semi-determinate" is a plant that (1) has a sympodial index
that is less than 3
and (2) undergoes termination of the sympodial meristem. As used herein, a
Solanaceae
plant that is "semi-indeterminate" is a plant that (1) has a sympodial index
that is less than 3
and (2) continuously produces sympodial shoots (the sympodial meristem does
not undergo
termination). The term "sympodial index", as used herein, refers to the number
of leaves per
sympodium (i.e., the number of leaves between successive clusters of flowers).
In some
- 26 -
CA 02892012 2015-05-20
WO 2014/081730
PCT/US2013/070825
embodiments, the genetically-altered Solanaceae plant has a sympodial index of
less than 3,
less than 2.9, less than 2.8, less than 2.7, less than 2.6, less than 2.5,
less than 2.4, less than
2.3, less than 2.2, less than 2.1, less than 2, less than 1.9, less than 1.8,
less than 1.7, less than
1.6, or less than 1.5. In some embodiments, the genetically-altered Solanaceae
plant has a
sympodial index of between 1 and 3, 1.5 and 3, 1 and 2.5, 1.5 and 2.5, 1 and
2, or 1.5 and 2.
Solanaceae plant cells are also contemplated herein. A Solanaceae plant cell
may
comprise any genotype described herein in the context of the Solanaceae plant
(e.g., a
Solanaceae plant cell heterozygous for a mutant sft gene and a mutant sspl
gene or a
Solanaceae plant cell homozygous for a mutant sspl gene and a mutant sp gene).
In some
embodiments, the Solanaceae plant cell is isolated. In some embodiments, the
Solanaceae
plant cell is a non-replicating plant cell.
In some embodiments, any of the Solanaceae plants described above have an
altered
flowering time and shoot architecture compared to a wild-type Solanaceae plant
(e.g., a
Solanaceae plant comprising two copies of a wild-type SP gene), to a
determinate Solanaceae
plant (e.g., a Solanaceae plant comprising a mutant sp gene), or to both a
wild-type
Solanaceae plant and a determinate Solanaceae plant. In some embodiments, any
of the
Solanaceae plants described above have a higher yield than a corresponding
wild-type
Solanaceae plant. In some embodiments, a Solanaceae plant comprising two
copies of a
mutant sspl gene as described herein and two copies of a wild-type SP gene as
described
herein has altered flowering time compared to a wild-type Solanaceae plant. In
some
embodiments, a Solanaceae plant comprising one copy of a mutant sspl gene as
described
herein and two copies of a mutant sp gene as described herein is semi-
determinate or semi-
indeterminate. In some embodiments, a Solanaceae plant comprising two copies
of a mutant
sspl gene as described herein and two copies of a mutant sp gene as described
herein is semi-
determinate or semi-indeterminate.
Food products are also contemplated herein. Such food products comprise a
Solanaceae plant part, such as a fruit (e.g., a tomato fruit). Non-limiting
examples of food
products include sauces (e.g., tomato sauce or ketchup), purees, pastes,
juices, canned fruits,
and soups. Food products may be produced or producible by using methods known
in the art.
Isolated polynucleotides are also described herein, including wild-type and
mutant
alleles of the SSP1 gene, and specifically, two mutant alleles designated
herein as ssp-2129
- 27 -
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
and ssp-610. Isolated polynucleotides including wild-type and mutant alleles
of the SFT gene
are also contemplated, e.g., sft-1906. Isolated polynucleotides can comprise,
for example, a
nucleic acid (e.g., DNA) having the sequence of SEQ ID NO: 2; a portion of SEQ
ID NO: 2
that exhibits substantially the same activity as a nucleic acid (e.g., DNA)
having the sequence
of SEQ ID NO: 2; a nucleic acid (e.g., DNA) having at least 85%, at least 90%,
at least 95%,
at least 96%, at least 97%, at least 98%, or at least 99% identity with the
sequence of SEQ
ID NO: 2; a nucleic acid (e.g., DNA) having the sequence of positions 631 to
651of SEQ ID
NO: 2 (CGGACGTCAACTGCTCTATTT, SEQ ID NO: 12); an orthologue or homologue of
the nucleic acid having the sequence of SEQ ID NO: 2; an orthologue or
homologue of the
nucleic acid sequence of positions 631 to 651of SEQ ID NO: 2; a nucleic acid
(e.g., DNA)
having the sequence of SEQ ID NO: 3; a portion of SEQ ID NO: 3 that exhibits
substantially
the same activity as a nucleic acid (e.g., DNA) having the sequence of SEQ ID
NO: 3; a
nucleic acid (e.g., DNA) having at least 85%, at least 90%, at least 95%, at
least 96%, at least
97%, at least 98%, or at least 99% identity with the sequence of SEQ ID NO: 3;
a nucleic
acid (e.g., DNA) having the sequence of positions 631 to 651of SEQ ID NO: 3
(CGGACGTCAATTGCTCCATTT, SEQ ID NO: 13); an orthologue or homologue of the
nucleic acid having the sequence of SEQ ID NO: 3; or an orthologue or
homologue of the
nucleic acid sequence of positions 631 to 651of SEQ ID NO: 3. In some
embodiments, the
isolated polynucleotide comprises a mutant sspl gene that includes a C to T
mutation at
position 641 of SEQ ID NO: 1, a C to T mutation at position 647 of SEQ ID NO:
1, or a C to
T mutation at position 641 and position 647 of SEQ ID NO: 1. In some
embodiments, the
isolated polynucleotide comprises a nucleotide sequence that encodes the
polypeptide of SEQ
ID NOs: 5 or 6 or a nucleotide sequence that encodes a polypeptide that
comprises SEQ ID
NOs: 14 or 15. In some embodiments, the isolated polynucleotide comprises a
nucleic acid
sequence that encodes a mutant sspl protein that comprises at least one
mutation in a SAP
motif, wherein the at least one mutation alters flowering time and shoot
architecture of the
Solanaceae plant, e.g., by conferring semi-determinacy. In some embodiments,
the SAP
motif with the at least one mutation has the amino acid sequence SEQ ID NO: 14
or SEQ ID
NO: 15. In some embodiments, the isolated polynucleotide comprises a nucleic
acid
sequence that encodes a mutant sspl protein that comprises at least one
mutation in the SAP
motif or in the two amino acids flanking the N-terminal position of the SAP
motif and the
- 28 -
CA 02892012 2015-05-20
WO 2014/081730
PCT/US2013/070825
one amino acid flanking the C-terminal position of the SAP motif, which
includes the
phosphorylation site for Ca-dependent protein kinases (CDPKs), wherein the at
least one
mutation alters flowering time and shoot architecture of the Solanaceae plant,
e.g., by
conferring semi-determinacy. In some embodiments, isolated polynucleotides can
comprise,
for example, a nucleic acid (e.g., DNA) having the sequence of SEQ ID NO:
20;_a portion of
SEQ ID NO: 20 that exhibits substantially the same activity as a nucleic acid
(e.g., DNA)
having the sequence of SEQ ID NO: 20; a nucleic acid (e.g., DNA) having at
least 70%, at
least 80%, at least 90%, at least 95%, at least 96%, at least 97%, at least
98%, or at least 99%
identity with the sequence of SEQ ID NO: 20; or an orthologue or homologue of
the nucleic
acid having the sequence of SEQ ID NO: 20. In some embodiments, the isolated
polynucleotide comprises a mutant sft gene comprising a G to A mutation at
position 394 of
SEQ ID NO: 20. In some embodiments, the isolated polynucleotide comprising a
mutant sft
gene comprises a nucleotide sequence that encodes a polypeptide of SEQ ID NO:
21. In some
embodiments, the isolated polynucleotide comprises a mutant sft gene
comprising a
nucleotide sequence that encodes a mutant sft polypeptide that comprises a Val
to Met
mutation at position 132 of SEQ ID NO: 19. In some embodiments, the isolated
polynucleotide is a cDNA. Such isolated polynucleotides can be used, for
example, in
methods of producing genetically-altered plants, such as genetically-altered
semi-determinate
or semi-indeterminate plants.
Isolated polypeptides (e.g., proteins) are also described herein, including
wild-type
and mutant sspl polypeptides, and specifically, the polypeptides encoded by
the two mutant
alleles ssp-2129 and ssp-610. Isolated polypeptides also include wild-type and
mutant sft
polypeptides, such as those encoded by the mutant allele sft-1906. In some
embodiments, the
isolated polypeptide comprises, for example, the sequence of SEQ ID NO: 5; a
portion of
SEQ ID NO: 5 that exhibits substantially the same activity as a polypeptide
(e.g., a protein)
having the sequence of SEQ ID NO: 5; an amino acid sequence having at least
85%, at least
90%, at least 95%, at least 96%, at least 97%, at least 98%, or at least 99%
identity with the
sequence of SEQ ID NO: 5; the amino acid sequence of positions 211 to 217 in
SEQ ID NO:
5 (RTSTALF, SEQ ID NO: 14); an orthologue or homologue of the polypeptide
having the
sequence of SEQ ID NO: 5; an orthologue or homologue of the amino acid
sequence of
positions 211 to 217 in SEQ ID NO: 5 (RTSTALF, SEQ ID NO: 14); a polypeptide
(e.g., a
- 29 -
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
protein) having the sequence of SEQ ID NO: 6; a portion of SEQ ID NO: 6 that
exhibits
substantially the same activity as a polypeptide (e.g., a protein) having the
sequence of SEQ
ID NO: 6; an amino acid sequence having at least 85%, at least 90%, at least
95%, at least
96%, at least 97%, at least 98%, or at least 99% identity with the sequence of
SEQ ID NO: 6;
the sequence of positions 211 to 217 in SEQ ID NO: 6 (RTSIAPF, SEQ ID NO: 15);
an
orthologue or homologue of the polypeptide having the sequence of SEQ ID NO:
6; or an
orthologue or homologue of the polypeptide sequence of positions 211 to 217 in
SEQ ID NO:
6 (RTSIAPF, SEQ ID NO: 15). In some embodiments, the isolated polypeptide
comprises a
Thr to Ile mutation at position 214 of SEQ ID NO: 4 or a Pro to Leu mutation
at position 216
of SEQ ID NO: 4, or a Thr to Ile mutation at position 214 and a Pro to Leu
mutation at
position 216 of SEQ ID NO: 4. In some embodiments, the isolated polypeptide
comprises a
mutant sspl protein comprising an amino acid sequence with at least one
mutation in a SAP
motif, wherein the at least one mutation alters flowering time and shoot
architecture of the
Solanaceae plant, e.g., by conferring semi-determinacy. In some embodiments,
the SAP
motif with the at least one mutation has the amino acid sequence SEQ ID NO: 14
or SEQ ID
NO: 15. In some embodiments, the isolated polypeptide comprises a mutant sspl
protein
comprising an amino acid sequence with at least one mutation in a SAP motif or
in the two
amino acids flanking the N-terminal position of the SAP motif and the one
amino acid
flanking the C-terminal position of the SAP motif, which includes the
phosphorylation site
for Ca-dependent protein kinases (CDPKs), wherein the at least one mutation
alters flowering
time and shoot architecture of the Solanaceae plant, e.g., by conferring semi-
determinacy.
An isolated polypeptide may also comprise the sequence of SEQ ID NO: 21; a
portion of
SEQ ID NO: 21 that exhibits substantially the same activity as a polypeptide
(e.g., a protein)
having the sequence of SEQ ID NO: 21; an amino acid sequence having at least
85%, at least
90%, at least 95%, at least 96%, at least 97%, at least 98%, or at least 99%
identity with the
sequence of SEQ ID NO: 21; an orthologue or homologue of the polypeptide
having the
sequence of SEQ ID NO: 21. Such isolated polypeptides can be used, for
example, in
methods of producing genetically-altered plants, such as genetically-altered
semi-determinate
or semi-indeterminate plants.
Also described herein are methods of modifying flowering time and shoot
architecture
in a Solanaceae plant, such as tomato, particularly suppressing sympodial
shoot termination,
- 30 -
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
which results in increased number of leaves in a Solanaceae plant compared to
the number of
leaves in corresponding determinate (e.g., a sp mutant) plants maintained
under the same
conditions, but fewer leaves than the number of leaves in corresponding
indeterminate wild-
type plants (see, e.g., Figure 2 and 4).
Methods described are methods of producing a genetically-altered Solanaceae
plant,
such as a semi-determinate (SD) or semi-indeterminate (SID) plant; and/or a
Solanaceae plant
with an altered flowering time and shoot architecture, increased yield, higher
quality products
(e.g., fruits), and/or products (e.g., fruits) with different compositions
(e.g., brix, also known
as enhanced soluble solids or sugar concentration in the fruits,) compared to
a wild-type
Solanaceae plant. In one embodiment, a method of producing a genetically-
altered semi-
determinate or semi-indeterminate Solanaceae plant comprises: (a) introducing
a mutant sspl
gene into a Solanaceae plant that comprises a mutant sp gene or producing a
mutant sspl
gene in a Solanaceae plant that comprises a mutant sp gene, thereby producing
a genetically-
altered plant that comprises the mutant sspl gene and the mutant sp gene; (b)
self-crossing
the genetically-altered Solanaceae plant produced in (a) or crossing two
genetically-altered
Solanaceae plants produced in (a) under conditions appropriate for producing a
genetically-
altered Solanaceae plant homozygous for the mutant sspl gene and homozygous
for the
mutant sp gene, thereby producing a genetically-altered Solanaceae plant that
is homozygous
for the mutant sspl gene and the mutant sp gene and is semi-determinate or
semi-
indeterminate. In another embodiment, a method of producing a genetically-
altered
Solanaceae plant with an altered flowering time and shoot architecture
compared to a wild-
type Solanaceae plant comprises: (a) introducing a mutant sspl gene into a
Solanaceae plant
that comprises a mutant sp gene or producing a mutant sspl gene in a
Solanaceae plant that
comprises a mutant sp gene, thereby producing a genetically-altered plant that
comprises the
mutant sspl gene and the mutant sp gene; (b) self-crossing the genetically-
altered Solanaceae
plant produced in (a) or crossing two genetically-altered Solanaceae plants
produced in (a)
under conditions appropriate for producing a genetically-altered Solanaceae
plant
heterozygous for the mutant sspl gene and homozygous for the mutant sp gene,
thereby
producing a genetically-altered Solanaceae plant that is heterozygous for the
mutant sspl
gene and homozygous for the mutant sp gene and has an altered flowering time
and shoot
architecture compared to a wild-type Solanaceae plant.
- 31 -
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
In other embodiments, the method comprises:
(a) introducing a mutant sft gene into a Solanaceae plant containing a mutant
sspl
gene or introducing a mutant sspl gene into a Solanaceae plant containing a
mutant sft gene,
thereby producing a genetically-altered Solanaceae plant containing a mutant
sft gene and a
mutant sspl gene; and
(b) self-crossing the genetically-altered Solanaceae plant produced in (a) or
crossing
two genetically-altered Solanaceae plants produced in (a) under conditions
appropriate for
producing a genetically-altered Solanaceae plant heterozygous or homozygous
for the mutant
sft gene and heterozygous or homozygous for the mutant sspl gene. In some
embodiments,
the plant produced in (b) is heterozygous for the mutant sft gene and
heterozygous for the
mutant sspl gene.
In specific embodiments of the method, the mutant sspl gene comprises, for
example,
a coding sequence comprising a nucleic acid (e.g., DNA) having the sequence of
SEQ ID
NO: 2; a coding sequence comprising a portion of SEQ ID NO: 2 that exhibits
substantially
the same activity (e.g., encoding the same polypeptide or substantially the
same polypeptide
that has the same activity) as a nucleic acid (e.g., DNA) having the sequence
of SEQ ID NO:
2; a coding sequence comprising a nucleic acid (e.g., DNA) having at least
85%, at least
90%, at least 95%, at least 96%, at least 97%, at least 98%, or at least 99%
identity with the
sequence of SEQ ID NO: 2; a nucleic acid (e.g., DNA) having the sequence of
positions 631
to 651of SEQ ID NO: 2 (CGGACGTCAACTGCTCTATTT, SEQ ID NO: 12); a coding
sequence comprising an orthologue or homologue of the nucleic acid having the
sequence of
SEQ ID NO: 2; an orthologue or homologue of the nucleic acid sequence of
positions 631 to
651of SEQ ID NO: 2; a coding sequence comprising a nucleic acid (e.g., DNA)
having the
sequence of SEQ ID NO: 3; a coding sequence comprising a portion of SEQ ID NO:
3 that
exhibits substantially the same activity (e.g., encoding the same polypeptide
or substantially
the same polypeptide that has the same activity) as a nucleic acid (e.g., DNA)
having the
sequence of SEQ ID NO: 3; a coding sequence comprising a nucleic acid (e.g.,
DNA) having
at least 85%, at least 90%, at least 95%, at least 96%, at least 97%, at least
98%, or at least
99% identity with the sequence of SEQ ID NO: 3; a nucleic acid (e.g., DNA)
having the
sequence of positions 631 to 651 of SEQ ID NO: 3 (CGGACGTCAATTGCTCCATTT, SEQ
ID NO: 13); a coding sequence comprising an orthologue or homologue of the
nucleic acid
- 32-
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
having the sequence of SEQ ID NO: 3; or an orthologue or homologue of the
nucleic acid
sequence of positions 631 to 651 of SEQ ID NO: 3. In some embodiments, the
mutant sspl
gene comprises, for example, a nucleic acid (e.g., DNA) having the sequence of
SEQ ID NO:
25; a portion of SEQ ID NO: 25 that exhibits substantially the same activity
(e.g., encoding
the same polypeptide or substantially the same polypeptide that has the same
activity) as a
nucleic acid (e.g., DNA) having the sequence of SEQ ID NO: 25; a nucleic acid
(e.g., DNA)
having at least 85%, at least 90%, at least 95%, at least 96%, at least 97%,
at least 98%, or at
least 99% identity with the sequence of SEQ ID NO: 25; an orthologue or
homologue of the
nucleic acid having the sequence of SEQ ID NO: 25; a nucleic acid (e.g., DNA)
having the
sequence of SEQ ID NO: 26; a portion of SEQ ID NO: 26 that exhibits
substantially the same
activity (e.g., encoding the same polypeptide or substantially the same
polypeptide that has
the same activity) as a nucleic acid (e.g., DNA) having the sequence of SEQ ID
NO: 26; a
nucleic acid (e.g., DNA) having at least 85%, at least 90%, at least 95%, at
least 96%, at least
97%, at least 98%, or at least 99% identity with the sequence of SEQ ID NO:
26; or an
orthologue or homologue of the nucleic acid having the sequence of SEQ ID NO:
26. In
some embodiments, the mutant sspl gene comprises a C to T mutation at position
641 of
SEQ ID NO: 1, a C to T mutation at position 647 of SEQ ID NO: 1, or a C to T
mutation at
position 641 and position 647 of SEQ ID NO: 1. In some embodiments, the mutant
sspl
gene comprises a nucleotide sequence that encodes a polypeptide of SEQ ID NOs:
5 or 6 or a
nucleotide sequence that encodes a polypeptide that comprises SEQ ID NOs: 14
or 15. In
some embodiments, the mutant sspl gene comprises a nucleic acid sequence that
encodes a
mutant sspl protein that comprises at least one mutation in a SAP motif,
wherein the at least
one mutation alters flowering time and shoot architecture of the Solanaceae
plant, e.g., by
conferring semi-determinacy. In some embodiments, the SAP motif with the at
least one
mutation has the amino acid sequence SEQ ID NO: 14 or SEQ ID NO: 15. In some
embodiments, the mutant sspl gene comprises a nucleic acid sequence that
encodes a mutant
sspl protein that comprises at least one mutation in a SAP motif or in the two
amino acids
flanking the N-terminal position of the SAP motif and the one amino acid
flanking the C-
terminal position of the SAP motif, which includes the phosphorylation site
for Ca-dependent
protein kinases (CDPKs), wherein the at least one mutation alters flowering
time and shoot
architecture of the Solanaceae plant, e.g., by conferring semi-determinacy.
- 33 -
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
In some embodiments, the mutant sft gene comprises, for example, a coding
sequence
comprising a nucleic acid (e.g., DNA) having the sequence of SEQ ID NO: 20;_a
coding
sequence comprising a portion of SEQ ID NO: 20 that exhibits substantially the
same activity
(e.g., encoding the same polypeptide or substantially the same polypeptide
that has the same
activity) as a nucleic acid (e.g., DNA) having the sequence of SEQ ID NO: 20;
a coding
sequence comprising a nucleic acid (e.g., DNA) having at least 85%, at least
90%, at least
95%, at least 96%, at least 97%, at least 98%, or at least 99% identity with
the sequence of
SEQ ID NO: 20; or a coding sequence comprising an orthologue or homologue of
the nucleic
acid having the sequence of SEQ ID NO:20. In some embodiments, the mutant sft
gene
comprises a G to A mutation at position 394 of SEQ ID NO: 20. In some
embodiments, the
mutant sft gene comprises a nucleotide sequence that encodes a polypeptide of
SEQ ID NO:
21. In some embodiments, the mutant sft gene comprises a nucleotide sequence
that encodes
a mutant sft polypeptide comprising a Val to Met mutation at position 132 of
SEQ ID NO:
19. In some embodiments, the mutant sft gene comprises, for example, a nucleic
acid (e.g.,
DNA) having the sequence of SEQ ID NO: 17; a portion of SEQ ID NO: 17 that
exhibits
substantially the same activity (e.g., encoding the same polypeptide or
substantially the same
polypeptide that has the same activity) as a nucleic acid (e.g., DNA) having
the sequence of
SEQ ID NO: 17; a nucleic acid (e.g., DNA) having at least 85%, at least 90%,
at least 95%, at
least 96%, at least 97%, at least 98%, or at least 99% identity with the
sequence of SEQ ID
NO: 17; or an orthologue or homologue of the nucleic acid having the sequence
of SEQ ID
NO: 17.
In specific embodiments, the sp mutant gene comprises, for example, a coding
sequence comprising a nucleic acid (e.g., DNA) having the sequence of SEQ ID
NO: 8; a
coding sequence comprising a portion of SEQ ID NO: 8 that exhibits
substantially the same
activity (e.g., encoding the same polypeptide or substantially the same
polypeptide that has
the same activity) as a nucleic acid (e.g., DNA) having the sequence of SEQ ID
NO: 8; a
coding sequence comprising a nucleic acid (e.g., DNA) having at least 85%, at
least 90%, at
least 95%, at least 96%, at least 97%, at least 98%, or at least 99% identity
with the sequence
of SEQ ID NO: 8; or a coding sequence comprising an orthologue or homologue of
the
nucleic acid having the sequence of SEQ ID NO: 8. In some embodiments, the
mutant sp
gene comprises a C to T mutation at position 227 of SEQ ID NO: 7. In some
embodiments,
- 34 -
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
the mutant sp gene comprises at least one mutation that reduces the activity
of a sp protein
encoded by the mutant sp gene. In some embodiments, the mutant sp gene
comprises at least
one mutation that reduces (partially or completely) the activity of a sp
protein encoded by the
mutant sp gene, wherein the reduced activity can confer determinacy. In some
embodiments, the mutant sp gene comprises a nucleotide sequence that encodes a
polypeptide of SEQ ID NO: 10. In some embodiments, the mutant sp gene
comprises, for
example, a nucleic acid (e.g., DNA) having the sequence of SEQ ID NO: 23; a
portion of
SEQ ID NO: 23 that exhibits substantially the same activity (e.g., encoding
the same
polypeptide or substantially the same polypeptide that has the same activity)
as a nucleic acid
(e.g., DNA) having the sequence of SEQ ID NO: 23; a nucleic acid (e.g., DNA)
having at
least 85%, at least 90%, at least 95%, at least 96%, at least 97%, at least
98%, or at least 99%
identity with the sequence of SEQ ID NO: 23; or an orthologue or homologue of
the nucleic
acid having the sequence of SEQ ID NO: 23.
Alternatively, a method of producing a genetically-altered Solanaceae plant,
such as a
genetically-altered semi-determinate or semi-indeterminate Solanaceae plant,
comprises: (a)
introducing a mutant sspl gene into a Solanaceae plant part (e.g., a cell, a
leaf or seed) that
comprises a mutant sp gene or producing a mutant sspl gene in a Solanaceae
plant part (e.g.,
a cell, a leaf or seed) that comprises a mutant sp gene, thereby producing a
genetically-altered
Solanaceae plant part that contains the mutant sspl gene and the mutant sp
gene; (b)
maintaining the genetically-altered Solanaceae plant part containing the
mutant sspl gene
produced in (a) under conditions and for sufficient time for production of a
genetically-
altered Solanaceae plant containing the mutant sspl gene and the mutant sp
gene from the
plant part, thereby producing a genetically-altered Solanaceae plant that
contains the mutant
sspl gene and a mutant sp gene; (c) self-crossing the genetically-altered
Solanaceae plant
produced in (b) or crossing two genetically-altered Solanaceae plants produced
in (b) under
conditions appropriate for producing a genetically-altered Solanaceae plant
homozygous for
the mutant sspl gene and the mutant sp gene, thereby producing a genetically-
altered
Solanaceae plant that is homozygous for the mutant sspl gene and the mutant sp
gene and is
semi-determinate or semi-indeterminate. In another embodiment, a method of
producing a
genetically-altered Solanaceae plant with an altered flowering time and shoot
architecture
compared to a wild-type Solanaceae plant comprises: (a) introducing a mutant
sspl gene into
- 35 -
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
a Solanaceae plant part (e.g., a cell, a leaf or seed) that comprises a mutant
sp gene or
producing a mutant sspl gene in a Solanaceae plant part (e.g., a cell, a leaf
or seed) that
comprises a mutant sp gene, thereby producing a genetically-altered Solanaceae
plant part
that contains the mutant sspl gene and the mutant sp gene; (b) maintaining the
genetically-
altered Solanaceae plant part containing the mutant sspl gene produced in (a)
under
conditions and for sufficient time for production of a genetically-altered
Solanaceae plant
containing the mutant sspl gene and the mutant sp gene from the plant part,
thereby
producing a genetically-altered Solanaceae plant that contains the mutant sspl
gene and a
mutant sp gene; (c) self-crossing the genetically-altered Solanaceae plant
produced in (b) or
crossing two genetically-altered Solanaceae plants produced in (b) under
conditions
appropriate for producing a genetically-altered Solanaceae plant heterozygous
for the mutant
sspl gene and homozygous for the mutant sp gene, thereby producing a
genetically-altered
Solanaceae plant that is heterozygous for the mutant sspl gene and homozygous
for the sp
gene and has an altered flowering time and shoot architecture compared to a
wild-type
Solanaceae plant. The mutant sspl gene and the mutant sp gene can be as
described above.
In some embodiments, the method of producing a genetically-altered Solanaceae
plant
comprises:
(a) introducing a mutant sft gene into a Solanaceae plant part containing a
mutant
sspl gene or introducing a mutant sspl gene into a Solanaceae plant part
containing a mutant
sft gene, thereby producing a genetically-altered Solanaceae plant part
containing the mutant
sft gene and the mutant sspl gene;
(b) maintaining the genetically-altered Solanaceae plant part containing a
mutant
sft gene and a mutant sspl gene produced in (a) under conditions and for
sufficient time for
production of a genetically-altered Solanaceae plant containing the mutant sft
gene and the
mutant sspl gene from the plant part, thereby producing a genetically-altered
Solanaceae
plant containing the mutant sft gene and the mutant sspl gene; and
(c) self-crossing the genetically-altered Solanaceae plant produced in (b)
or
crossing two genetically-altered Solanaceae plants produced in (b) under
conditions
appropriate for producing a genetically-altered Solanaceae plant heterozygous
or
homozygous for the mutant sft gene and heterozygous or homozygous for the
mutant sspl
gene, thereby producing a genetically-altered Solanaceae plant. In some
embodiments, the
- 36 -
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
plant produced in (c) is heterozygous for the mutant sft gene and heterozygous
for the mutant
ssp/ gene.
In any of the methods described herein, the mutant sspl gene or the mutant sft
gene
can be introduced into a Solanaceae plant or a plant part or produced in a
Solanaceae plant or
plant part by a method known to those of skill in the art, such as
Agrobacterium-mediated
recombination, viral-vector mediated recombination, microinjection, gene gun
bombardment/biolistic particle delivery, electroporation, mutagenesis (e.g.,
by ethyl
methanesulfonate or fast neutron irradiation), TILLING (Targeting Induced
Local Lesions in
Genomes), conventional marker-assisted introgression, and nuclease mediated
recombination
(e.g., use of custom-made restriction enzymes for targeting mutagenesis by
gene replacement,
see, e.g., CRISPR-Cas9: Genome engineering using the CRISPR-Cas9 system. Ran
FA, Hsu
PD, Wright J, Agarwala V, Scott DA, Zhang F. Nat Protoc. 2013 Nov;8(11):2281-
308;
TALEN endonucleases: Nucleic Acids Res. 2011 Jul;39(12):e82. Efficient design
and
assembly of custom TALEN and other TAL effector-based constructs for DNA
targeting.
Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia
NV,
Bogdanove AJ, Voytas DF and Plant Biotechnol J. 2012 May;10(4):373-89. Genome
modifications in plant cells by custom-made restriction enzymes. Tzfira T,
Weinthal D,
Marton I, Zeevi V, Zuker A, Vainstein A.). Genetically-altered Solanaceae
plants, such as
genetically-altered semi-determinate or semi-indeterminate Solanaceae plants,
produced by
or producible by a method described herein are also claimed.
Alternatively, a method of producing a semi-determinate or semi-indeterminate
Solanaceae plant or a Solanaceae plant having one or more other
characteristics described
herein (increased yield, brix, etc.) comprises: (a) reducing (partially or
completely) function
of a wild-type SSP1 gene comprising SEQ ID NO: 1 in a Solanaceae plant
homozygous for a
mutant sp gene, thereby producing a semi-determinate or semi-indeterminate
Solanaceae
plant. In some embodiments, reducing the function of the wild-type SSP1 gene
comprising
SEQ ID NO: 1 comprises performing any of the following methods of RNA-
interference
(e.g., administering to the Solanaceae plant a micro-RNA or a small
interfering (si)-RNA or
hairpin RNA) or translational blocking (e.g., administering to the Solanaceae
plant a
morpholino). Methods of RNA-interference and translational blocking are well-
known in
the art. Methods of producing micro-RNAs, si-RNAs, and morpholinos are well-
known in
- 37 -
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
the art and can involve use of the nucleotides sequences provided herein,
e.g., SEQ ID NO: 1.
The mutant sp gene can be any mutant sp gene described herein. In some
embodiments, the
method further comprises (b) reducing (partially or completely) function of a
wild-type SFT
gene comprising SEQ ID NO: 16 or a coding sequence comprising SEQ ID NO: 18.
In some
embodiments, reducing the function of the wild-type SFT gene comprises
performing any of
the above methods of RNA-interference or translational blocking.
Further aspects and embodiments of the disclosure are provided below.
In some embodiments, the disclosure provides a genetically-altered Solanaceae
plant
comprising a mutant suppressor of spl (sspl) gene and a mutant self pruning
(sp) gene. In
some embodiments, the mutant sspl gene comprises a nucleic acid sequence that
encodes a
mutant sspl protein that comprises a mutant SAP motif. In some embodiments,
the mutant
sspl gene comprises a nucleic acid sequence that encodes a mutant sspl protein
that
comprises a mutant SAP motif with a sequence of SEQ ID NO: 14 or SEQ ID NO:
15. In
some embodiments, the mutant sspl gene encodes a mutant sspl polypeptide
comprising the
sequence of SEQ ID NO: 5 or SEQ ID NO: 6. In some embodiments, the mutant sspl
gene
comprises a C to T mutation at position 641 of SEQ ID NO: 1, a C to T mutation
at 647 of
SEQ ID NO: 1, or a C to T mutation at position 641 and 647 of SEQ ID NO: 1. In
some
embodiments, the mutant sspl gene comprises a coding sequence having the
nucleic acid
sequence of SEQ ID NO: 2 or SEQ ID NO: 3. In some embodiments, the mutant sp
gene
encodes a mutant sp polypeptide comprising the sequence of SEQ ID NO: 10. In
some
embodiments, the mutant sp gene comprises a coding sequence having the nucleic
acid
sequence of SEQ ID NO: 8. In some embodiments, the genetically-altered
Solanaceae plant
is a tomato (Solanum lycopersicum) plant. In some embodiments, the genetically-
altered
Solanaceae plant is isogenic. In some embodiments, the genetically-altered
Solanaceae plant
is inbred. In some embodiments, the genetically-altered Solanaceae plant is
homozygous for
the mutant sspl gene and homozygous for the mutant sp gene. In some
embodiments, the
genetically-altered Solanaceae plant is semi-determinate.
Other aspects relate to a seed for producing a genetically-altered Solanaceae
plant as
described in any of the embodiments provided in the preceding paragraph.
- 38 -
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
Yet other aspects relate to a method of producing a genetically-altered
Solanaceae
plant comprising:
(a) introducing a mutant sspl gene into a Solanaceae plant containing a mutant
sp
gene, thereby producing a genetically-altered Solanaceae plant containing a
mutant sspl gene
and a mutant sp gene; and
(b) self-crossing the genetically-altered Solanaceae plant produced in (a) or
crossing
two genetically-altered Solanaceae plants produced in (a) under conditions
appropriate for
producing a genetically-altered Solanaceae plant heterozygous or homozygous
for the mutant
sspl gene and heterozygous or homozygous for the mutant sp gene, thereby
producing a
genetically-altered Solanaceae plant. In some embodiments, the method of
producing a
genetically-altered Solanaceae plant comprises:
(a) introducing a mutant sspl gene into a Solanaceae plant part
containing a
mutant sp gene, thereby producing a genetically-altered Solanaceae plant part
containing the
mutant sspl gene and the mutant sp gene;
(b) maintaining the genetically-altered Solanaceae plant part containing a
mutant
sspl and a mutant sp gene produced in (a) under conditions and for sufficient
time for
production of a genetically-altered Solanaceae plant containing the mutant
sspl gene and the
mutant sp gene from the plant part, thereby producing a genetically-altered
Solanaceae plant
containing the mutant sspl gene and the mutant sp gene;
(c) self-crossing the genetically-altered Solanaceae plant produced in (b)
or
crossing two genetically-altered Solanaceae plants produced in (b) under
conditions
appropriate for producing a genetically-altered Solanaceae plant heterozygous
or
homozygous for the mutant sspl gene and heterozygous or homozygous for the
mutant sp
gene, thereby producing a genetically-altered Solanaceae plant. In some
embodiments, the
genetically-altered Solanaceae plant is homozygous for the mutant sspl gene
and
homozygous for the mutant sp gene. In some embodiments, the mutant sspl gene
comprises
a nucleic acid sequence that encodes a mutant sspl protein that comprises a
mutant SAP
motif. In some embodiments, the mutant sspl gene comprises a nucleic acid
sequence that
encodes a mutant sspl protein that comprises a mutant SAP motif with a
sequence of SEQ ID
NO: 14 or SEQ ID NO: 15 In some embodiments, the mutant sspl gene encodes a
mutant
sspl polypeptide comprising the sequence of SEQ ID NO: 5 or SEQ ID NO: 6. In
some
- 39 -
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
embodiments, the mutant sspl gene comprises a C to T mutation at position 641
of SEQ ID
NO: 1, a C to T mutation at 647 of SEQ ID NO: 1, or a C to T mutation at
position 641 and
647 of SEQ ID NO: 1. In some embodiments, the mutant sspl gene comprises a
coding
sequence having the nucleic acid sequence of SEQ ID NO: 2 or SEQ ID NO: 3. In
some
embodiments, the mutant sp gene encodes a mutant sp polypeptide comprising the
sequence
of SEQ ID NO: 10. In some embodiments, the mutant sp gene comprises a coding
sequence
having the nucleic acid sequence of SEQ ID NO: 8. In some embodiments, in (a),
the mutant
sspl gene is introduced into a plant or a plant part by a method selected from
the group
consisting of: Agrobacterium-mediated recombination, viral-vector mediated
recombination,
microinjection, gene gun bombardment/biolistic particle delivery, nuclease
mediated
recombination, and electroporation. In some embodiments, in (a), the mutant
sspl gene is
introduced into a plant or a plant part by nuclease mediated recombination. In
some
embodiments, the genetically-altered Solanaceae plant is a tomato (Solanum
lycopersicum)
plant. In some embodiments, the genetically-altered Solanaceae plant is
inbred. In some
embodiments, the genetically-altered Solanaceae plant is semi-determinate.
Another aspect relates to a genetically-altered Solanaceae plant produced by
the
method of any of the embodiments provided in the preceding paragraph.
Other aspects relate to a genetically-altered Solanaceae plant comprising a
mutant
suppressor of spl (sspl) gene and a mutant single flower truss (sft) gene. In
some
embodiments, the genetically-altered Solanaceae plant comprises a mutant
suppressor of spl
(sspl) gene and a mutant single flower truss (sft) gene, wherein the mutant
genes are
heterozygous. In some embodiments, the sft gene comprises a coding sequence
having the
nucleic acid sequence of SEQ ID NO: 20. In some embodiments, the sft gene
comprises
SEQ ID NO: 20. In some embodiments, the mutant sspl gene comprises a coding
sequence
having a nucleic acid sequence of SEQ ID NO: 2 or SEQ ID NO: 3 or encodes a
mutant sspl
protein that comprises SEQ ID NO: 5 or SEQ ID NO: 6. In some embodiments, the
mutant
sspl gene comprises a nucleic acid sequence that comprises SEQ ID NO: 2 or SEQ
ID NO: 3
or encodes a mutant sspl protein that comprises SEQ ID NO: 5 or SEQ ID NO: 6.
In some
embodiments, the mutant sspl gene encodes a mutant sspl polypeptide that
comprises a
mutant SAP motif with a sequence of SEQ ID NO: 14 or SEQ ID NO: 15. In some
embodiments, the mutant sspl gene comprises a C to T mutation at position 641
of SEQ ID
- 40 -
CA 02892012 2015-05-20
WO 2014/081730
PCT/US2013/070825
NO: 1, a C to T mutation at position 647 of SEQ ID NO: 1, or a C to T mutation
at position
641 and 647 of SEQ ID NO: 1. In some embodiments, the mutant sspl gene
comprises a
coding sequence having the nucleic acid sequence of SEQ ID NO: 2 or SEQ ID NO:
3. In
some embodiments, the mutant sspl gene comprises the nucleic acid sequence of
SEQ ID
NO: 2 or SEQ ID NO: 3. In some embodiments, the genetically-altered Solanaceae
plant is
semi-determinate. In some embodiments, the genetically-altered semi-
determinate
Solanaceae plant is a tomato (Solanum lycopersicum) plant.
In some embodiments, the the genetically-altered semi-determinate Solanaceae
plant is
inbred. In some embodiments, the mutant sft gene is sft-1906.
Yet another aspect relates to a seed for producing a genetically-altered
Solanaceae
plant as provided in any of the embodiments in the proceding paragraph.
Other aspects relate to genetically-altered Solanaceae plant comprising a
mutant self-
pruning (sp) gene that comprises a coding sequence having the nucleic acid
sequence of SEQ
ID NO: 8. and a mutant single flower truss (sft) gene that comprises SEQ ID
NO: 20. In
some embodiments, the mutant self-pruning (sp) gene is heterozygous and the
mutant single
flower truss (sft) gene is heterozygous.
Further aspects relate to a genetically-altered Solanaceae plant comprising a
mutant
self-pruning (sp) gene that comprises the nucleic acid sequence of SEQ ID NO:
8. and a
mutant single flower truss (sft) gene that comprises SEQ ID NO: 20, wherein
the mutant
genes are heterozygous.
Another aspect relates to a method of producing a genetically-altered
Solanaceae
plant comprising:
(a) introducing a mutant sft gene into a Solanaceae plant containing a mutant
ssplgene, thereby producing a genetically-altered Solanaceae plant containing
a mutant sspl
gene and a mutant sft gene; and
(b) self-crossing the genetically-altered Solanaceae plant produced in (a) or
crossing
two genetically-altered Solanaceae plants produced in (a) under conditions
appropriate for
producing a genetically-altered Solanaceae plant homozygous for the mutant
sspl gene and
the mutant sft gene, thereby producing a genetically-altered Solanaceae plant.
In some
embodiments, the method of producing a genetically-altered Solanaceae plant
comprises:
- 41 -
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
(a) introducing a mutant sft gene into a Solanaceae plant part containing a
mutant
sspl gene, thereby producing a genetically-altered Solanaceae plant part
containing the
mutant sspl gene and the mutant sft gene;
(b) maintaining the genetically-altered Solanaceae plant part containing a
mutant
sspl and a mutant sft gene produced in (a) under conditions and for sufficient
time for
production of a genetically-altered Solanaceae plant containing the mutant
sspl gene and the
mutant sft gene from the plant part, thereby producing a genetically-altered
Solanaceae plant
containing the mutant sspl gene and the mutant sft gene;
(c) self-crossing the genetically-altered Solanaceae plant produced in (b)
or
crossing two genetically-altered Solanaceae plants produced in (b) under
conditions
appropriate for producing a genetically-altered Solanaceae plant homozygous
for the mutant
sspl gene and the mutant sft gene, thereby producing a genetically-altered
Solanaceae plant
that is semi-determinate.
Other aspects relate to a genetically-altered Solanaceae plant produced by a
method
provided in the preceding paragraph. Another aspect relates to a genetically-
altered
Solanaceae plant heterozygous for a mutant suppressor of spl (sspl) gene and
homozygous
for a mutant single flower truss (sft) gene, wherein the genetically-altered
Solanaceae plant
has an altered yield compared to the yield of a wild-type Solanaceae plant.
Other aspects relate to a seed for producing a genetically-altered Solanaceae
plant
provided in the preceding paragraph.
Yet other aspects relate to a genetically-altered semi-determinate Solanaceae
plant
homozygous for a mutant suppressor of spl (sspl) gene and homozygous for a
mutant self
pruning (sp) gene. In some embodiments, the mutant sspl gene comprises a
nucleic acid
sequence that encodes a mutant sspl protein that comprises a mutant SAP motif.
In some
embodiments, the mutant sspl gene comprises a nucleic acid sequence that
encodes a mutant
sspl protein that comprises a mutant SAP motif with a sequence of SEQ ID NO:
14 or SEQ
ID NO: 15. In some embodiments, the mutant sspl gene encodes a mutant sspl
polypeptide
comprising the sequence of SEQ ID NO: 5 or SEQ ID NO: 6. In some embodiments,
the
mutant sspl gene comprises a C to T mutation at position 641 of SEQ ID NO: 1,
a C to T
mutation at 647 of SEQ ID NO: 1, or a C to T mutation at position 641 and 647
of SEQ ID
NO: 1. In some embodiments, the mutant sspl gene comprises a coding sequence
having the
- 42 -
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
nucleic acid sequence of SEQ ID NO: 2 or SEQ ID NO: 3. In some embodiments,
the mutant
sspl gene comprises the nucleic acid sequence of SEQ ID NO: 2 or SEQ ID NO: 3.
In some
embodiments, the mutant sp gene comprises a coding sequence having the nucleic
acid
sequence of SEQ ID NO: 8. In some embodiments, the mutant sp gene comprises
the nucleic
acid sequence of SEQ ID NO: 8. In some embodiments, the genetically-altered
semi-
determinate Solanaceae plant is a tomato (Solanum lycopersicum) plant. In some
embodiments, the genetically-altered semi-determinate Solanaceae plant is
isogenic. In some
embodiments, the genetically-altered semi-determinate Solanaceae plant is
inbred.
Other aspects relate to a seed for producing a genetically-altered semi-
determinate
Solanaceae plant as described in any of the embodiments provided in the
preceding
paragraph.
Another aspect relates to a method of producing a genetically-altered semi-
determinate Solanaceae plant comprising:
(a) introducing a mutant sspl gene into a Solanaceae plant containing a mutant
sp
gene, thereby producing a genetically-altered Solanaceae plant containing a
mutant sspl gene
and a mutant sp gene; and
(b) self-crossing the genetically-altered Solanaceae plant produced in (a) or
crossing
two genetically-altered Solanaceae plants produced in (a) under conditions
appropriate for
producing a genetically-altered Solanaceae plant homozygous for the mutant
sspl gene and
the mutant sp gene, thereby producing a genetically-altered Solanaceae plant
that is semi-
determinate. In some embodiments, the method of producing a genetically-
altered semi-
determinate Solanaceae plant comprises:
(a) introducing a mutant sspl gene into a Solanaceae plant part containing
a
mutant sp gene, thereby producing a genetically-altered Solanaceae plant part
containing the
mutant sspl gene and the mutant sp gene;
(b) maintaining the genetically-altered Solanaceae plant part containing a
mutant
sspl and a mutant sp gene produced in (a) under conditions and for sufficient
time for
production of a genetically-altered Solanaceae plant containing the mutant
sspl gene and the
mutant sp gene from the plant part, thereby producing a genetically-altered
Solanaceae plant
containing the mutant sspl gene and the mutant sp gene;
- 43 -
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
(c) self-crossing the genetically-altered Solanaceae plant
produced in (b) or
crossing two genetically-altered Solanaceae plants produced in (b) under
conditions
appropriate for producing a genetically-altered Solanaceae plant homozygous
for the mutant
sspl gene and the mutant sp gene, thereby producing a genetically-altered
Solanaceae plant
that is semi-determinate. In some embodiments, the mutant sspl gene comprises
a nucleic
acid sequence that encodes a mutant sspl protein that comprises a mutant SAP
motif. In
some embodiments, the mutant sspl gene comprises a nucleic acid sequence that
encodes a
mutant sspl protein that comprises a mutant SAP motif with a sequence of SEQ
ID NO: 14
or SEQ ID NO: 15. In some embodiments, the mutant sspl gene encodes a mutant
sspl
polypeptide comprising the sequence of SEQ ID NO: 5 or SEQ ID NO: 6. In some
embodiments, the mutant sspl gene comprises a C to T mutation at position 641
of SEQ ID
NO: 1, a C to T mutation at 647 of SEQ ID NO: 1, or a C to T mutation at
position 641 and
647 of SEQ ID NO: 1. In some embodiments, the mutant sspl gene comprises a
coding
sequence having the nucleic acid sequence of SEQ ID NO: 2 or SEQ ID NO: 3. In
some
embodiments, the mutant sspl gene comprises the nucleic acid sequence of SEQ
ID NO: 2 or
SEQ ID NO: 3. In some embodiments, the mutant sp gene comprises a coding
sequence
having the nucleic acid sequence of SEQ ID NO: 8. In some embodiments, the
mutant sp
gene comprises the nucleic acid sequence of SEQ ID NO: 8. In some embodiments,
in (a),
the mutant sspl gene is introduced into a plant or a plant part by a method
selected from the
group consisting of: Agrobacterium-mediated recombination, viral-vector
mediated
recombination, microinjection, gene gun bombardment/biolistic particle
delivery, nuclease
mediated recombination, and electroporation. In some embodiments, the
Solanaceae plant
is a tomato (Solanum lycopersicum) plant. In some embodiments, the Solanaceae
plant is
inbred.
Another aspect relates to a genetically-altered semi-determinate Solanaceae
plant
produced by a method provided in the preceding paragraph.
Other aspects relate to a isolated polynucleotide encoding a mutant sspl
protein
having the amino acid sequence of SEQ ID NO: 5 or SEQ ID NO: 6. In some
embodiments,
the isolated polynucleotide comprises the nucleic acid sequence of SEQ ID NO:
2 or SEQ ID
NO: 3.
- 44 -
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
Yet another aspect relates to a genetically-altered Solanaceae plant
heterozygous for a
mutant suppressor of spl (sspl) gene and homozygous for a mutant self pruning
(sp) gene,
wherein the genetically-altered Solanaceae plant has an altered flowering time
and shoot
architecture compared to a wild-type Solanaceae plant. In some embodiments,
the mutant
sspl gene comprises a nucleic acid sequence that encodes a mutant sspl protein
that
comprises a mutant SAP motif. In some embodiments, the mutant sspl gene
comprises a
nucleic acid sequence that encodes a mutant sspl protein that comprises a
mutant SAP motif
with a sequence of SEQ ID NO: 14 or SEQ ID NO: 15. In some embodiments, the
mutant
sspl gene encodes a mutant sspl polypeptide comprising the sequence of SEQ ID
NO: 5 or
SEQ ID NO: 6. In some embodiments, the mutant sspl gene comprises a C to T
mutation at
position 641 of SEQ ID NO: 1, a C to T mutation at 647 of SEQ ID NO: 1, or a C
to T
mutation at position 641 and 647 of SEQ ID NO: 1. In some embodiments, the
mutant sspl
gene comprises a coding sequence having the nucleic acid sequence of SEQ ID
NO: 2 or
SEQ ID NO: 3. In some embodiments, the mutant sspl gene comprises the nucleic
acid
sequence of SEQ ID NO: 2 or SEQ ID NO: 3. In some embodiments, the mutant sp
gene
comprises a coding sequence having the nucleic acid sequence of SEQ ID NO: 8.
In some
embodiments, the mutant sp gene comprises the nucleic acid sequence of SEQ ID
NO: 8. In
some embodiments, the genetically-altered Solanaceae plant is a tomato
(Solanum
lycopersicum) plant. In some embodiments, the genetically-altered Solanaceae
plant is
isogenic. In some embodiments, the genetically-altered Solanaceae plant is
inbred. In some
embodiments, the genetically-altered Solanaceae plant is semi-determinate.
Other aspects relate to a seed for producing a genetically-altered Solanaceae
plant of
any of the embodiments provided in the preceding paragraph.
Yet another aspect relates to a genetically-altered Solanaceae plant
homozygous for a
mutant suppressor of spl (sspl) gene, wherein the genetically-altered
Solanaceae plant has an
altered flowering time compared to a wild-type Solanaceae plant. In some
embodiments, the
mutant sspl gene comprises a nucleic acid sequence that encodes a mutant sspl
protein that
comprises a mutant SAP motif. In some embodiments, the mutant sspl gene
comprises a
nucleic acid sequence that encodes a mutant sspl protein that comprises a
mutant SAP motif
with a sequence of SEQ ID NO: 14 or SEQ ID NO: 15. In some embodiments,
wherein the
mutant sspl gene encodes a mutant sspl polypeptide comprising the sequence of
SEQ ID
- 45 -
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
NO: 5 or SEQ ID NO: 6. In some embodiments, the mutant sspl gene comprises a C
to T
mutation at position 641 of SEQ ID NO: 1, a C to T mutation at 647 of SEQ ID
NO: 1, or a C
to T mutation at position 641 and 647 of SEQ ID NO: 1. In some embodiments,
the mutant
sspl gene comprises a coding sequence having the nucleic acid sequence of SEQ
ID NO: 2 or
SEQ ID NO: 3. In some embodiments, the mutant sspl gene comprises the nucleic
acid
sequence of SEQ ID NO: 2 or SEQ ID NO: 3. In some embodiments, the mutant sp
gene
comprises a coding sequence having the nucleic acid sequence of SEQ ID NO: 8.
In some
embodiments, the mutant sp gene comprises the nucleic acid sequence of SEQ ID
NO: 8. In
some embodiments, the genetically-altered Solanaceae plant is a tomato
(Solanum
lycopersicum) plant. In some embodiments, the genetically-altered Solanaceae
plant is
isogenic. In some embodiments, the genetically-altered Solanaceae plant is
inbred. In some
embodiments, the genetically-altered Solanaceae plant is homozygous for a wild-
type SELF
PRUNING (SP) gene.
Other aspects relate to a seed for producing a genetically-altered Solanaceae
plant of
any embodiment provided in the preceding paragraph.
EXAMPLES
Example 1: Identifying mutant plants providing semi-determinate or semi-
indeterminate shoot architecture phenotypes
The crop plant tomato (Solanum lycopersicum) was used to identify mutant
plants
with a semi-determinate phenotype. The previously generated tomato EMS
mutation library
in the determinate M82 background was used in the methods described below
(Menda N Y et
al. 2004. Plant J 38.861-72). The mutation library was previously generated by
treating seeds
with ethyl methanesulfonate (0.5 percent EMS for 12 h; LD15), producing an M1
generation.
These seeds were self-crossed to produce 13,000 M2 families with random
mutations in
unknown locations.
The M82 determinate (M82D) isogenic background is known to contain a mutation
in
the SELF PRUNING (SP) gene (Pnueli, et al. Development 1989), resulting in a
tomato plant
with a determinate (D) growth habit. In D-type plants, the sympodial shoots
produce
progressively fewer leaves until the plant terminates growth in two successive
inflorescences.
- 46 -
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
In contrast, indeterminate (ID) tomato plants generate successive sympodial
units (SYM) that
produce three leaves each, and SYMs continue to reiterate to generate an ID
shoot. The M2
EMS mutation library described above was generated in an M82D background and
self-
crossed to produce approximately six thousand independent mutant tomato plants
and many
fertile mutants which contained mutations at unidentified locations which were
carried over
into the M3 inbred generation. These lines were screened in both the M2 and M3
generations
for homozygous recessive mutant phenotypes that partially suppressed the
determinate
growth phenotype of the M82D background, resulting in a semi-determinate shoot
architecture phenotype. In particular, flowering time, flower production per
inflorescence,
and shoot architecture were examined. Single lines were identified that
contained at least one
M3 plant with a semi-determinate phenotype.
Two M2 families, e2129 and e610, were identified that had a low frequency
(less than
or equal to ¨25%) of plants with an altered flowering time and shoot
architecture resembling
a semi-determinate phenotype. Figure 2 shows that homozygosity for the e2129
mutant
(designated ssp-2129) in the sp-/- M82 mutant background suppressed sympodial
shoot
termination compared to the sp-/- mutant alone. Figure 2, left, depicts a sp-/-
determinate plant
where leaf number in each sympodial shoot gradually decreases, leading to
shoot termination
in two successive inflorescences. Figure 2, right, depicts an indeterminate
wild-type plant
where each sympodial unit produces three leaves and this process continues
indefinitely.
Figure 2, center, depicts an ssp-2129; sp-/- double mutant with semi-
determinate sympodial
shoot development, such that each sympodial shoot unit now produces two
leaves, instead of
the typical three leaves produced in wild-type indeterminate tomato plants.
Figure 3 shows
quantification of internode length among 4 inflorescences in determinate sp-/-
mutants
(M82D), semi-determinate ssp-2129; sp-/- double mutants and ssp-610; sp-/-
double mutants
(ssp-2129 and ssp-610, respectively), and wild-type plants (M82ID). The p-
value was
measured by a student's t-test against M82D and M82ID (P<0.01 for ssp-2129 and
ssp-610,
indicated by **).
Figure 4 shows quantification of leaf numbers produced by the primary shoot
meristem (PSM) indicating flowering time and sympodial units in both sp-/- and
ssp-2129;sp-/-
3 0 backgrounds. Note that ssp-2129;sp-/- double mutant plants displayed a
delayed flowering
time as indicated by the increased leaf numbers in the PSM compared to sp-/-
single mutant
- 47 -
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
plants. ssp-2129;sp-1- plants also displayed more leaves within sympodial
units compared to
sp4-
plants, but fewer leaves within sympodial units compared to wild-type
indeterminate
plants. This semi-determinacy was observed for a second mutant allele, ssp-6/0
in the sp
mutant background.
To identify the genetic mutation(s) resulting in the altered flowering time
and shoot
architecture in these two families, one family, e2129, was further analyzed
using previously
reported map-based cloning procedures (Lippman et al, PLoS Biology 2008). A
single M3
ssp-2129 mutant plant was crossed to a wild-type species, S. piminellifolium,
with known
polymorphisms throughout the genome (The Tomato Genome sequence, Nature,
2012). The
Fl hybrid was then self-crossed to produce an F2 generation of progeny plants
segregating
for both the ssp-2129 mutation and the DNA polymorphisms between M82 and S.
pimpinellifolium. Approximately 200 F2 plants were scored for altered
flowering time and
semi-determinate shoot architecture phenotypes, reflecting homozygosity of the
ssp-2129
mutation.
The homozygous mutant F2 plants (those with the altered flowering time and
shoot
architecture) were then genotyped with evenly spaced polymorphic DNA markers
spanning
all 12 tomato chromosomes using the bulk segregant mapping technique. DNA was
isolated
from at least 20 mutant plants (those with the altered flowering time and
shoot architecture
phenotype) and from 20 wild-type plants. DNA from the mutant plants was pooled
to form
pool 1 and DNA from the wild-type plants was pooled to form pool 2. A 10
centiMorgan
(cM) scan was performed to identify M82 polymorphisms that were over-
represented in pool
1 relative to pool 2, reflective of the origin of the ssp-2129 mutation in the
M82 background.
Over-representation of polymorphisms in pool 1 was found within at 2Mb mapping
interval
between PCR markers 4029 and 4230, corresponding to 40M and 42M on chromosome
2, as
shown in Figure 5.
To find the location of the mutation within this region, RNA was extracted
from wild-
type and mutant plants. This RNA was converted to cDNA and sequenced using
Illumina
sequencing. The sequencing reads were then mapped to the known gene
annotations and
only those within the 2 Mb region described above were analyzed. A large
number of C to T
mutations were observed at a single location, position 647 according to SEQ ID
NO: 2 which
corresponds to amino acid position 216 according to SEQ ID NO: 5, within the C-
terminus of
- 48 -
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
the tomato orthologue of the Arabidopsis FLOWERING LOCUS D (FD) gene. The
tomato
orthologue is referred herein as SUPPRESSOR OF SP1 (SSP1).
Following identification of the mutation present in the e2129 family, the e610
mutant,
a confirmed allele in the SSP1 gene by complementation test with the ssp-2129
mutant as
described below, was examined to see if the unknown mutation in that family
also occurred
in the SSP1 gene. Heterozygous plants with one copy of ssp-2129 and one copy
of ssp-610
phenocopied the ssp-2129 homozygous mutant and the ssp-610 homozygous mutant
plant,
indicating that these two mutations were in the same gene. Following the
complementation
test, DNA was extracted from e610 mutant plants and Sanger sequencing was
performed to
determine if the SSP1 gene locus contained a mutation. A 'C' to 'T' mutation
was identified
in SSP1 at position 641 according to SEQ ID NO: 3 which corresponds to amino
acid
position 214 according to SEQ ID NO: 6.
Figure 5 summarizes the map-based cloning of the SSP1 gene discussed above.
Figure
5A shows the SSP1 map position on chromosome 2 localized to a 2.1M interval
between
markers inde14029 (4029x10k) and inde14230 (4230 x20k) was defined by bulk
segragant
analysis. An F2 mapping population segregating for the recessive sspl mutant
was generated
by self-pollinating an S. pimpinellifolium sp- x sspl sp- (cv. M82) Fl plant.
At least four
insertion-deletion (indel) PCR markers were used for each of the 12 tomato
chromosomes on
a pool of DNA composed of 20 sspl mutant individuals compared to a pool of DNA
composed of 20 wild-type individuals. Deconvolution of the mutant pool
revealed a
recombination-defined internal of 40M-42.1M on chromosome 2. Figure 5B depicts
Illumina
RNA-sequencing (RNA-seq), which was performed on RNA isolated from
reproductive
meristems and used to identify and reconstruct DNA protein coding sequences
from
expressed genes in the mapping interval. RNA-seq reads revealed eight genes
(positions
indicated by arrows) expressed in meristems. Single nucleotide polymorphisms
(SNP) were
identified relative to the reference annotation for the eight expressed genes,
which revealed a
C-to-T DNA change in the coding sequence of 5o1yc02g083520 of the ssp-2129
mutant
allele. Sequencing of the ssp-610 allele revealed a C-to-T change at a nearby
location in the
coding sequence of 5o1yc02g083520. Each mutation causes a missense amino acid
changes
in the conserved SAP motif of the closest tomato homolog of the Arabidopsis
flowering gene
FLOWERING LOCUS D (FD), encoding a bZIP transcription factor.
- 49 -
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
A summary of the sequences of both the wild-type SSP1 gene and the two mutant
alleles, ssp-2129 and ssp-610, is provided in Figures 6 and 7, which depict
ClustalW analyses
of the nucleotide and amino acid sequences, respectively. The nucleotide and
amino acid
sequences for wild-type SSP1, mutant ssp-2129, mutant ssp-610, wild-type SP,
and mutant sp
are also listed after Example 2. Mutations present in the nucleotide and amino
acid
sequences of each mutant nucleic acid or protein are indicated as underlined
and bolded
nucleotides or amino acids.
Example 2: Identifying mutant plants providing desirable plant traits such as
brix, brix
yield, red fruit yield, and total yield
A mutant SINGLE FLOWER TRUSS (SFT) gene, sft-1906, was identified in tomato
plants using the same techniques described above in Example 1. sft-1906 was
determined to
have a mutation of Valine to Methionine at position 132. A summary of the
nucleotide and
amino acid sequences of the wild-type SFT gene and the mutant allele, sft-
1906, is provided
in Figures 9 and 10, which depict ClustalW analyses of the nucleotide and
amino acid
sequences, respectively. The nucleotide and amino acid sequences for the wild-
type SFT
gene and the mutant allele, sft-1906, are also listed below. Mutations present
in the
nucleotide and amino acid sequences of each mutant nucleic acid or protein are
indicated as
underlined and bolded nucleotides or amino acids.
Tomato plants carrying sft-1906 were crossed with tomato plants carrying
ssp2120 or
ssp610 and compared to plants having other genetic backgrounds (M82 (sp),
ssp2129 x M82
Fl, ssp610 x M82 Fl, and sft1906 x M82 F1). It was found that all of the
combined mutant
tomato plants (ssp2129 x sft1906, ssp610 x sft1906, ssp2129 x M82 Fl, ssp610 x
M82 Fl,
and sft1906 x M82 Fl) had improved Brix Yield, Brix, Red Fruit Yield, and
Total Yield
compared to M82 plants (FIG. 8). Data on red and total fruit yield were
generated from
replicated field trials and Brix data was obtained using a refractometer (see
below). Tomato
plants doubly heterozygous for both an sspl mutation and a sft mutation had
the highest
improvement in Brix Yield, Brix, Red Fruit Yield, and Total Yield (FIG. 8).
Tomato plants
were grown under wide (1 plant per m2) spacing using drip irrigation for field
trials. When
all plants have 80% or more red fruits, total fruits and total red fruits were
measured by
weight (kg) per plant. Ten red fruits collected randomly from each plant were
used for
- 50 -
CA 02892012 2015-05-20
WO 2014/081730
PCT/US2013/070825
determine Brix value, which was measured by a digital Brix refractometer
(ATAGO PAL-1).
Brix-yield was calculated by the multiplied output of Brix and total fruit
yield measured in
g/m2. Each genotype in the experiments was represented by a minimum of 15
replicates in
two locations (Riverhead, NewYork USA and Akko, Israel). All plants were
transplanted in
a completely randomized design.
Nucleic Acid and Polypeptide Sequences
Nucleic Acid
>SSP1 coding sequence
ATGTGGTCATCAAGCAGTGATAACAGGGGACTCTCTGCTTCTTCTTCTTCATCTTCATCC
TCATCTCATTCACCATTTTCTCCAAGACTCAAAACAATGGAAGAAGTGTGGAAAGATATT
AATCTTTCTTCACTTCAAGATCACACTACGAATTACTCTAGAGATCATCATCATCTTCAT
GATCATAATCATCAAGCTGCTAATTTTGGTGGAATGATTTTACAAGATTTTTTGGCAAGG
CCTTTTGCTAATGAATCTTCACCAGCAGCAGCAGCAGCAGCAGCCTCCCCTGTTTCAGCT
ACAACTATGCTGAATTTGAACTCTGTTCCTGAGCTTCATTTCTTTGATAACCCATTGAGG
CAAAACTCAATCTTGCACCAACCAAATGCAAGTGGAAGAAAAAGGGTTGTCCCTGAAACA
GAAGACAATTCTACAGGGGATAGAAGAAATCAGAGGATGATCAAGAACAGAGAGTCTGCT
GCTAGATCAAGAGCTAGAAAGCAGGCTTATATGAACGAGTTGGAATCAGAAGTGGCACAT
TTAGTTGAAGAAAATGCAAGGCTCAAGAAGCAGCAGCAACAGTTACGAGTAGATGCAGCT
AATCAAGTTCCCAAAAAGAACACTCTTTATCGGACGTCAACTGCTCCATTTTGA (SEQ ID NO: 1)
>ssp-2129 coding sequence
ATGTGGTCATCAAGCAGTGATAACAGGGGACTCTCTGCTTCTTCTTCTTCATCTTCATCC
TCATCTCATTCACCATTTTCTCCAAGACTCAAAACAATGGAAGAAGTGTGGAAAGATATT
AATCTTTCTTCACTTCAAGATCACACTACGAATTACTCTAGAGATCATCATCATCTTCAT
GATCATAATCATCAAGCTGCTAATTTTGGTGGAATGATTTTACAAGATTTTTTGGCAAGG
CCTTTTGCTAATGAATCTTCACCAGCAGCAGCAGCAGCAGCAGCCTCCCCTGTTTCAGCT
ACAACTATGCTGAATTTGAACTCTGTTCCTGAGCTTCATTTCTTTGATAACCCATTGAGG
CAAAACTCAATCTTGCACCAACCAAATGCAAGTGGAAGAAAAAGGGTTGTCCCTGAAACA
GAAGACAATTCTACAGGGGATAGAAGAAATCAGAGGATGATCAAGAACAGAGAGTCTGCT
GCTAGATCAAGAGCTAGAAAGCAGGCTTATATGAACGAGTTGGAATCAGAAGTGGCACAT
TTAGTTGAAGAAAATGCAAGGCTCAAGAAGCAGCAGCAACAGTTACGAGTAGATGCAGCT
AATCAAGTTCCCAAAAAGAACACTCTTTATCGGACGTCAACTGCTCTATTTTGA (SEQ ID NO: 2)
>ssp-610 coding sequence
ATGTGGTCATCAAGCAGTGATAACAGGGGACTCTCTGCTTCTTCTTCTTCATCTTCATCC
- 51 -
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
TCATCTCATTCACCATTTTCTCCAAGACTCAAAACAATGGAAGAAGTGTGGAAAGATATT
AATCTTTCTTCACTTCAAGATCACACTACGAATTACTCTAGAGATCATCATCATCTTCAT
GATCATAATCATCAAGCTGCTAATTTTGGTGGAATGATTTTACAAGATTTTTTGGCAAGG
CCTTTTGCTAATGAATCTTCACCAGCAGCAGCAGCAGCAGCAGCCTCCCCTGTTTCAGCT
ACAACTATGCTGAATTTGAACTCTGTTCCTGAGCTTCATTTCTTTGATAACCCATTGAGG
CAAAACTCAATCTTGCACCAACCAAATGCAAGTGGAAGAAAAAGGGTTGTCCCTGAAACA
GAAGACAATTCTACAGGGGATAGAAGAAATCAGAGGATGATCAAGAACAGAGAGTCTGCT
GCTAGATCAAGAGCTAGAAAGCAGGCTTATATGAACGAGTTGGAATCAGAAGTGGCACAT
TTAGTTGAAGAAAATGCAAGGCTCAAGAAGCAGCAGCAACAGTTACGAGTAGATGCAGCT
AATCAAGTTCCCAAAAAGAACACTCTTTATCGGACGTCAATTGCTCCATTTTGA (SEQ ID NO: 3)
Protein
>SSP
MWSSSSDNRGLS ASSSSSSSSSHSPFSPRLKTMEEVWKDINLSSLQDHTTNYSRDHHHLH
DHNHQAANFGGMILQDFLARPFANESSPAAAAAAASPVSATTMLNLNSVPELHEFDNPLR
QNSILHQPNASGRKRVVPETEDNSTGDRRNQRMIKNRES AARSRARKQAYMNELESEVAH
LVEENARLKKQQQQLRVDAANQVPKKNTLYRTSTAPF* (SEQ ID NO: 4)
>ssp-2129
MWSSSSDNRGLS ASSSSSSSSSHSPFSPRLKTMEEVWKDINLSSLQDHTTNYSRDHHHLH
DHNHQAANFGGMILQDFLARPFANESSPAAAAAAASPVSATTMLNLNSVPELHEFDNPLR
QNSILHQPNASGRKRVVPETEDNSTGDRRNQRMIKNRES AARSRARKQAYMNELESEVAH
LVEENARLKKQQQQLRVDAANQVPKKNTLYRTSTALF* (SEQ ID NO: 5)
>ssp-610
MWSSSSDNRGLS ASSSSSSSSSHSPFSPRLKTMEEVWKDINLSSLQDHTTNYSRDHHHLH
DHNHQAANFGGMILQDFLARPFANESSPAAAAAAASPVSATTMLNLNSVPELHEFDNPLR
QNSILHQPNASGRKRVVPETEDNSTGDRRNQRMIKNRES AARSRARKQAYMNELESEVAH
LVEENARLKKQQQQLRVDAANQVPKKNTLYRTSIAPF* (SEQ ID NO: 6)
Nucleic acid
>SP (wild-type) coding sequence
ATGGCTTCCAAAATGTGTGAACCCCTTGTGATTGGTAGAGTGATTGGTGAAGTTGTTGATTATTTCT
GTCCAAGTGTTAAGATGTCTGTTGTTTATAACAACAACAAACATGTCTATAATGGACATGAATTCT
TTCCTTCCTCAGTAACTTCTAAACCTAGGGTTGAAGTTCATGGTGGTGATCTCAGATCCTTCTTCAC
ACTGATCATGATAGATCCAGATGTTCCTGGTCCTAGTGATCCATATCTCAGGGAACATCTACACTG
GATTGTCACAGACATTCCAGGCACTACAGATTGCTCTTTTGGAAGAGAAGTGGTTGGGTATGAAAT
GCCAAGGCCAAATATTGGAATCCACAGGTTTGTATTTTTGCTGTTTAAGCAGAAGAAAAGGCAAA
- 52 -
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
CAATATCGAGTGCACCAGTGTCCAGAGATCAATTTAGTAGTAGAAAATTTTCAGAAGAAAATGAA
CTTGGCTCACCAGTTGCTGCTGTTTTCTTCAATTGTCAGAGGGAAACTGCCGCTAGAAGGCGTTGA
(SEQ ID NO: 7)
>sp (mutant) coding sequence
ATGGCTTCCAAAATGTGTGAACCCCTTGTGATTGGTAGAGTGATTGGTGAAGTTGTTGATTATTTCT
GTCCAAGTGTTAAGATGTCTGTTGTTTATAACAACAACAAACATGTCTATAATGGACATGAATTCT
TTCCTTCCTCAGTAACTTCTAAACCTAGGGTTGAAGTTCATGGTGGTGATCTCAGATCCTTCTTCAC
ACTGATCATGATAGATCCAGATGTTCTTGGTCCTAGTGATCCATATCTCAGGGAACATCTACACTG
GATTGTCACAGACATTCCAGGCACTACAGATTGCTCTTTTGGAAGAGAAGTGGTTGGGTATGAAAT
GCCAAGGCCAAATATTGGAATCCACAGGTTTGTATTTTTGCTGTTTAAGCAGAAGAAAAGGCAAA
CAATATCGAGTGCACCAGTGTCCAGAGATCAATTTAGTAGTAGAAAATTTTCAGAAGAAAATGAA
CTTGGCTCACCAGTTGCTGCTGTTTTCTTCAATTGTCAGAGGGAAACTGCCGCTAGAAGGCGTTGA
(SEQ ID NO: 8)
Protein
>SP (wild-type)
MASKMCEPLVIGRVIGEVVDYFCPSVKMSVVYNNNKHVYNGHEFFPSSVTSKPRVEVHGG
DLRS1-t TLIMIDPDVPGPSDPYLREHLHWIVTDIPGTTDCSFGREVVGYEMPRPNIGIHR
FVFLLFKQKKRQTISSAPVSRDQFSSRKFSEENELGSPVAAVFFNCQRETAARRR* (SEQ ID NO: 9)
>sp (mutant)
MASKMCEPLVIGRVIGEVVDYFCPSVKMSVVYNNNKHVYNGHEFFPSSVTSKPRVEVHGG
DLRSFFTLIMIDPDVLGPSDPYLREHLHWIVTDIPGTTDCSFGREVVGYEMPRPNIGIHR
FVFLLFKQKKRQTISSAPVSRDQFSSRKFSEENELGSPVAAVFFNCQRETAARRR* (SEQ ID NO: 10)
Nucleic acid
>SFT (wild-type) coding sequence
ATGCCTAGAGAACGTGATCCTCTTGTTGTTGGTCGTGTGGTAGGGGATGTATTGGACCCTTTCACA
AGAACTATTGGCCTAAGAGTTATATATAGAGATAGAGAAGTTAATAATGGATGCGAGCTTAGGCC
TTCCCAAGTTATTAACCAGCCAAGGGTTGAAGTTGGAGGAGATGACCTACGTACCTTTTTCACTTT
GGTTATGGTGGACCCTGATGCTCCAAGTCCGAGTGATCCAAATCTGAGAGAATACCTTCACTGGTT
GGTCACCGATATTCCAGCTACCACAGGTTCAAGTTTTGGGCAAGAAATAGTGAGCTATGAAAGTCC
AAGACCATCAATGGGAATACATCGATTTGTATTTGTATTATTCAGACAATTAGGTCGGCAAACAGT
GTATGCTCCAGGATGGCGTCAGAATTTCAACACAAGAGATTTTGCAGAACTTTATAATCTTGGTTT
ACCTGTTGCTGCTGTCTATTTTAATTGTCAAAGAGAGAGTGGCAGTGGTGGACGTAGAAGATCTGC
TGATTGA (SEQ ID NO: 18)
>sft (mutant) coding sequence
- 53 -
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
ATGCCTAGAGAACGTGATCCTCTTGTTGTTGGTCGTGTGGTAGGGGATGTATTGGACCCTTTCACA
AGAACTATTGGCCTAAGAGTTATATATAGAGATAGAGAAGTTAATAATGGATGCGAGCTTAGGCC
TTCCCAAGTTATTAACCAGCCAAGGGTTGAAGTTGGAGGAGATGACCTACGTACCTTTTTCACTTT
GGTTATGGTGGACCCTGATGCTCCAAGTCCGAGTGATCCAAATCTGAGAGAATACCTTCACTGGTT
GGTCACCGATATTCCAGCTACCACAGGTTCAAGTTTTGGGCAAGAAATAGTGAGCTATGAAAGTCC
AAGACCATCAATGGGAATACATCGATTTGTATTTGTATTATTCAGACAATTAGGTCGGCAAACAAT
GTATGCTCCAGGATGGCGTCAGAATTTCAACACAAGAGATTTTGCAGAACTTTATAATCTTGGTTT
ACCTGTTGCTGCTGTCTATTTTAATTGTCAAAGAGAGAGTGGCAGTGGTGGACGTAGAAGATCTGC
TGATTGA (SEQ ID NO: 20)
Protein
>SFT (wild-type)
MPRERDPLVVGRVVGDVLDPFTRTIGLRVIYRDREVNNGCELRPSQVINQPRVEVGGDDL
RT1-t TLVMVDPDAPSPSDPNLREYLHWLVTDIPATTGS SFGQEIVSYESPRPSMGIHRFV
FVLFRQLGRQTVYAPGWRQNFNTRDFAELYNLGLPVAAVYFNCQRESGSGGRRRSAD* (SEQ ID NO:
19)
>sft (mutant)
MPRERDPLVVGRVVGDVLDPFTRTIGLRVIYRDREVNNGCELRPSQVINQPRVEVGGDDL
RT1-t TLVMVDPDAPSPSDPNLREYLHWLVTDIPATTGS SFGQEIVSYESPRPSMGIHRFV
FVLFRQLGRQTMYAPGWRQNFNTRDFAELYNLGLPVAAVYFNCQRESGSGGRRRSAD* (SEQ ID NO:
21)
Genomic Sequences
* Marker : bold with underbar, * Exon: Italics and bold,
>SP (wild-type) genomic
ATGGCTTCCAAAATGTGTGAACCCCTTGTGATTGGTAGAGTGATTGGTGAAGTTGTTGATTATTTCTG
TCCAAGTGTTAAGATGTCTGTTGTTTATAACAACAACAAACATGTCTATAATGGACATGAATTCTTTC
CTTCCTCAGTAACTTCTAAACCTAGGGTTGAAGTTCATGGTGGTGATCTCAGATCCTTCTTCACACTG
GTATATATTAATCTTCAACACTTCCAATTTACTCCGTCTGTCTGTCCTAATTTATGTCACACATTTTC
TATGATATATAGTTTTAGAAATTATTCAAGACCATAACTTTTTAAAGAAAAAATCATAGACTTTCTT
AGTCAACGTCAAATAAATTGAGACGGACAAGATGACATGATTAGTACATTTATCTTCTATTATTGA
CCTCTCATTTTCTTTTATACATTATTTGACAGATCATGATAGATCCAGATGTTCCTGGTCCTAGTGAT
CCATATCTCAGGGAACATCTACACTGGTATAGACAACATATGCCTTAAAACTAACTCAGTCAATTTT
ATCTTCAATTGTTTACTTTGGAAGGGGAAATGACATGATCATTATATCATAGTACAAATTATTATGT
AATTTCTGTTCGTCTAAAAAATGTCACTTTAGAAAAAACTGATAATCATATACAATACCACAATAA
AGATAGAAGAACATGTACTAATATTGAACTTAAATAATGAGTACTAGGAGTATTATTAATTAACTT
TAAAAATGCTAGTCAATATACCTATGTTTATATGTTAAAAAATCCTTTATATTTGGAAACATGAGT
ACTCCTATACCATACAATGTTGTCGTACAGTTGATTAGACGGGCAAATTAAACAAATGTCCAATAA
TTGTACTAATTAATAACTACTTGTTCTCTTCATCTATTATTAGTTATTACCAAAAAAAGAGGACTGC
AAAATGGTGATATTATTATGTGTAACGGAAAAAAACGTACTCTATTTAATATGATAGAATCAAAGT
GACATATTTTGTTCTAGTTAGACAAATAAGTAACTGAAAAGAGGATTTGACCATCTTTACAGGAT
TGTCA CA GA CA TTCCAGGCACTA CA GATTGCTCTTTTGGTATGTATCCTTAACCCATAAATCAAAAT
AATGTACTTTCTTTTTATTTGCCATTAATATCTCTAGTACAAAAAAGAAATATTATAAAAAAAATTA
ATTTCAATTTTTATATTATAGGTTTAAGATAATAATATTAAACGATATTTTAGTCTCTACCAAATAG
- 54 -
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
ACGAGCAAATTAAAACTAAGAAAGCACTACATGTTTTCTTTATATTATTAGTATAAAAATATATTA
TAATTTGCCTGGTGGTAATAGGATCAAAGTATTGATTCTTAATTATTATTATATAATTAATAATAAT
GGTAAACAAAAAGATATAAAGTGCTTACCTCCTAATTCCCTATATGAAAAAATATACTTACTTAAT
TACTCTTTTTACACGTAAGCATGCATTTAAAAAAATATTAAAAAATTATTCCAGAGGTTATATATA
ATATGTATGGATAAAAAAAAAATTCACCTATATACATAATAATATAATTTTCGAGTGAATTGACCG
CCCTTCAGCATCATTATATAATGTTATCGATCTAGGTCTTTGTGTGAAATTAAAAGTTATTTATACG
GTTAGTACGATCGCGTAATAACGAAGGTAAAAATATTTCAGGAAGAGAAGTGGTTGGGTATGAAAT
GCCAAGGCCAAATATTGGAATCCACAGGTTTGTATTTTTGCTGTTTAAGCAGAAGAAAAGGCAAACA
ATATCGAGTGCACCAGTGTCCAGAGATCAATTTAGTAGTAGAAAATTTTCAGAAGAAAATGAACTTG
GCTCACCAGTTGCTGCTGTTTTCTTCAATTGTCAGAGGGAAACTGCCGCTAGAAGGCGTTGA (SEQ ID
NO: 22)
>sp(mutant)genomic(mutation in brackets)
ATGGCTTCCAAAATGTGTGAACCCCTTGTGATTGGTAGAGTGATTGGTGAAGTTGTTGATTATTTCTG
TCCAAGTGTTAAGATGTCTGTTGTTTATAACAACAACAAACATGTCTATAATGGACATGAATTCTTTC
CTTCCTCAGTAACTTCTAAACCTAGGGTTGAAGTTCATGGTGGTGATCTCAGATCCTTCTTCACACTG
GTATATATTAATCTTCAACACTTCCAATTTACTCCGTCTGTCTGTCCTAATTTATGTCACACATTTTC
TATGATATATAGTTTTAGAAATTATTCAAGACCATAACTTTTTAAAGAAAAAATCATAGACTTTCTT
AGTCAACGTCAAATAAATTGAGACGGACAAGATGACATGATTAGTACATTTATCTTCTATTATTGA
CCTCTCATTTTCTTTTATACATTATTTGACAGATCATGATAGATCCAGATGTTC[[T][TGGTCCTAGTG
ATCCATATCTCAGGGAACATCTACACTGGTATAGACAACATATGCCTTAAAACTAACTCAGTCAATT
TTATCTTCAATTGTTTACTTTGGAAGGGGAAATGACATGATCATTATATCATAGTACAAATTATTAT
GTAATTTCTGTTCGTCTAAAAAATGTCACTTTAGAAAAAACTGATAATCATATACAATACCACAAT
AAAGATAGAAGAACATGTACTAATATTGAACTTAAATAATGAGTACTAGGAGTATTATTAATTAA
CTTTAAAAATGCTAGTCAATATACCTATGTTTATATGTTAAAAAATCCTTTATATTTGGAAACATGA
GTACTCCTATACCATACAATGTTGTCGTACAGTTGATTAGACGGGCAAATTAAACAAATGTCCAAT
AATTGTACTAATTAATAACTACTTGTTCTCTTCATCTATTATTAGTTATTACCAAAAAAAGAGGACT
GCAAAATGGTGATATTATTATGTGTAACGGAAAAAAACGTACTCTATTTAATATGATAGAATCAAA
GTGACATATTTTGTTCTAGTTAGACAAATAAGTAACTGAAAAGAGGATTTGACCATCTTTACAGG
ATTGTCACAGACATTCCAGGCACTACAGA TTGCTCTTTTGGTATGTATCCTTAACCCATAAATCAAA
ATAATGTACTTTCTTTTTATTTGCCATTAATATCTCTAGTACAAAAAAGAAATATTATAAAAAAAAT
TAATTTCAATTTTTATATTATAGGTTTAAGATAATAATATTAAACGATATTTTAGTCTCTACCAAAT
AGACGAGCAAATTAAAACTAAGAAAGCACTACATGTTTTCTTTATATTATTAGTATAAAAATATAT
TATAATTTGCCTGGTGGTAATAGGATCAAAGTATTGATTCTTAATTATTATTATATAATTAATAATA
ATGGTAAACAAAAAGATATAAAGTGCTTACCTCCTAATTCCCTATATGAAAAAATATACTTACTTA
ATTACTCTTTTTACACGTAAGCATGCATTTAAAAAAATATTAAAAAATTATTCCAGAGGTTATATA
TAATATGTATGGATAAAAAAAAAATTCACCTATATACATAATAATATAATTTTCGAGTGAATTGAC
CGCCCTTCAGCATCATTATATAATGTTATCGATCTAGGTCTTTGTGTGAAATTAAAAGTTATTTATA
CGGTTAGTACGATCGCGTAATAACGAAGGTAAAAATATTTCAGGAAGAGAAGTGGTTGGGTATGAA
ATGCCAAGGCCAAATATTGGAATCCACAGGTTTGTATTTTTGCTGTTTAAGCAGAAGAAAAGGCAAA
CAATATCGAGTGCACCAGTGTCCAGAGATCAATTTAGTAGTAGAAAATTTTCAGAAGAAAATGAACTT
GGCTCACCAGTTGCTGCTGTTTTCTTCAATTGTCAGAGGGAAACTGCCGCTAGAAGGCGTTGA (SEQ
ID NO: 23)
* Marker: bold with underbar, * Exon: Italics and bold,
>S SP1 (wild-type) genomic
ATGTGGTCATCAAGCAGTGATAACAGGGGACTCTCTGCTTCTTCTTCTTCATCTTCATCCTCATCTCA
TTCACCATTTTCTCCAAGACTCAAAACAATGGAAGAAGTGTGGAAAGATATTAATCTTTCTTCACTTC
AAGATCACACTACGAATTACTCTAGAGATCATCATCATCTTCATGATCATAATCATCAAGCTGCTAAT
TTTGGTGGAATGATTTTACAAGATTTTTTGGCAAGGCCTTTTGCTAATGAATCTTCACCAGCAGCAGC
AGCAGCAGCAGCCTCCCCTGTTTCAGCTACAACTATGCTGAATTTGAACTCTGTTCCTGAGCTTCAT
TTCTTTGATAACCCATTGAGGCAAAACTCAATCTTGCACCAACCAAATGCAAGTGGAAGAAAAAG
GGTTGTCCCTGAAACAGAAGACAATTCTACAGGGGATAGAAGAAATCAGAGGATGATCAAGAAC
AGAGAGTCTGCTGCTAGATCAAGAGCTAGAAAGCAGGTAAGTGACACTCAACTTTGTCTTAATCCT
- 55 -
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
GTCAATTTTGTGCTTATACATCAACTATGTTCCATATTGTTACTCTTTTGCTGCTTCTATTCTTGATT
TGAACAATATGCCGAGTTACTCTGTTTGCAGGCTTA TATGAACGAGTTGGAATCAGAAGTGGCACAT
TTAGTTGAAGAAAATGCAAGGCTCAAGAAGCAGCAGCAACAGGTTCTCTTATCTTTCTTTATTTCTGT
CACTTTTAAAATTCAGTTTATAAAAAAAATGGATATAACTGATTCATAATAAATTGGTGTTTTCTTA
ATTTGTACAGTTACGAGTAGATGCAGCTAATCAAGTTCCCAAAAAGAACACTCTTTATCGGACGTCA
ACTGCTCCATTTTGAGATCTTATTATAATTTGGTTTCCTAGTGCTACATTAGTATTAAGAACAATTTC
CCATTTGGCTGTATTTTGTTTGTAATATGCACCAACTGTTGTTTTGATGGTGGCCTTGTGGGGCGAT
GAATATTCGTATGACAAAATAAGAAGGGAAAAAATAGGTTGTGAATTAAGGAAAGTGTAGGCCAT
TATTAGTACTCTATCA (SEQ ID NO: 24)
>ssp-2129 genomic (mutation in brackets)
ATGTGGTCATCAAGCAGTGATAACAGGGGACTCTCTGCTTCTTCTTCTTCATCTTCATCCTCATCTCA
TTCACCATTTTCTCCAAGACTCAAAACAATGGAAGAAGTGTGGAAAGATATTAATCTTTCTTCACTTC
AAGATCACACTACGAATTACTCTAGAGATCATCATCATCTTCATGATCATAATCATCAAGCTGCTAAT
TTTGGTGGAATGATTTTACAAGATTTTTTGGCAAGGCCTTTTGCTAATGAATCTTCACCAGCAGCAGC
AGCAGCAGCAGCCTCCCCTGTTTCAGCTACAACTATGCTGAATTTGAACTCTGTTCCTGAGCTTCAT
TTCTTTGATAACCCATTGAGGCAAAACTCAATCTTGCACCAACCAAATGCAAGTGGAAGAAAAAG
GGTTGTCCCTGAAACAGAAGACAATTCTACAGGGGATAGAAGAAATCAGAGGATGATCAAGAAC
AGAGAGTCTGCTGCTAGATCAAGAGCTAGAAAGCAGGTAAGTGACACTCAACTTTGTCTTAATCCT
GTCAATTTTGTGCTTATACATCAACTATGTTCCATATTGTTACTCTTTTGCTGCTTCTATTCTTGATT
TGAACAATATGCCGAGTTACTCTGTTTGCAGGCTTA TATGAACGAGTTGGAATCAGAAGTGGCACAT
TTAGTTGAAGAAAATGCAAGGCTCAAGAAGCAGCAGCAACAGGTTCTCTTATCTTTCTTTATTTCTGT
CACTTTTAAAATTCAGTTTATAAAAAAAATGGATATAACTGATTCATAATAAATTGGTGTTTTCTTA
ATTTGTACAGTTACGAGTAGATGCAGCTAATCAAGTTCCCAAAAAGAACACTCTTTATCGGACGTCA
ACTGCTC[[T][ATTTTGAGATCTTATTATAATTTGGTTTCCTAGTGCTACATTAGTATTAAGAACAATT
TCCCATTTGGCTGTATTTTGTTTGTAATATGCACCAACTGTTGTTTTGATGGTGGCCTTGTGGGGCG
ATGAATATTCGTATGACAAAATAAGAAGGGAAAAAATAGGTTGTGAATTAAGGAAAGTGTAGGCC
ATTATTAGTACTCTATCA (SEQ ID NO: 25)
>ssp-610 genomic (mutation in brackets)
ATGTGGTCATCAAGCAGTGATAACAGGGGACTCTCTGCTTCTTCTTCTTCATCTTCATCCTCATCTCA
TTCACCATTTTCTCCAAGACTCAAAACAATGGAAGAAGTGTGGAAAGATATTAATCTTTCTTCACTTC
AAGATCACACTACGAATTACTCTAGAGATCATCATCATCTTCATGATCATAATCATCAAGCTGCTAAT
TTTGGTGGAATGATTTTACAAGATTTTTTGGCAAGGCCTTTTGCTAATGAATCTTCACCAGCAGCAGC
AGCAGCAGCAGCCTCCCCTGTTTCAGCTACAACTATGCTGAATTTGAACTCTGTTCCTGAGCTTCAT
TTCTTTGATAACCCATTGAGGCAAAACTCAATCTTGCACCAACCAAATGCAAGTGGAAGAAAAAG
GGTTGTCCCTGAAACAGAAGACAATTCTACAGGGGATAGAAGAAATCAGAGGATGATCAAGAAC
AGAGAGTCTGCTGCTAGATCAAGAGCTAGAAAGCAGGTAAGTGACACTCAACTTTGTCTTAATCCT
GTCAATTTTGTGCTTATACATCAACTATGTTCCATATTGTTACTCTTTTGCTGCTTCTATTCTTGATT
TGAACAATATGCCGAGTTACTCTGTTTGCAGGCTTA TATGAACGAGTTGGAATCAGAAGTGGCACAT
TTAGTTGAAGAAAATGCAAGGCTCAAGAAGCAGCAGCAACAGGTTCTCTTATCTTTCTTTATTTCTGT
CACTTTTAAAATTCAGTTTATAAAAAAAATGGATATAACTGATTCATAATAAATTGGTGTTTTCTTA
ATTTGTACAGTTACGAGTAGATGCAGCTAATCAAGTTCCCAAAAAGAACACTCTTTATCGGACGTCA
A[[T][TGCTCCATTTTGAGATCTT ATT AT AATTTGGTTTCCT AGTGCT ACATT AGT ATT AAGAACAATT
TCCCATTTGGCTGTATTTTGTTTGTAATATGCACCAACTGTTGTTTTGATGGTGGCCTTGTGGGGCG
ATGAATATTCGTATGACAAAATAAGAAGGGAAAAAATAGGTTGTGAATTAAGGAAAGTGTAGGCC
ATTATTAGTACTCTATCA (SEQ ID NO: 26)
* Marker : bold with underbar, * Exon: Italics and bold, * Deletion : :
>SFT (wild-type) genomic
ATGCCTAGAGAACGTGATCCTCTTGTTGTTGGTCGTGTGGTAGGGGATGTATTGGACCCTTTCACAA
GAACTA TTGGCCTAA GAGTTA TA TA TA GA GA TA GA GAAGTTAA TAA TGGA TG
CGAGCTTAGGCCTTC
CCAAGTTATTAACCAGCCAAGGGTTGAAGTTGGAGGAGATGACCTACGTACCTTTTTCACTTTGGT A
ATATTTCTTATATTTTTTGTTTGGGAATATAGTTAAGTTGATTTTCATAAGCAAAGTAAAAAGTATT
- 56 -
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
TTTGTCTTTTTGTAAAGGTTATGGTGGACCCTGATGCTCCAAGTCCGAGTGATCCAAATCTGAGAGA
ATACCTTCACTGGTCCGTATTTTTTCCTTATTCTCTCTTCTTTTCATCTCTTTCTTTTTTGACCTTTTTA
CTTAATTATATTCTTTAGTAATAATATATGATGATATCCTTTTTAAAAATTGGAAATACGAAAAGG
AGAAATGAAGAGGAGATTTACATGTGAGGGAGCAGATGGTAGAAATATATAAATGTGAAGATAT
ATATTCTTGAACTTAAAAACAAGCTACTAAAATAAAAATGAATAAAATATTTACTCTGTCAATATT
CTGTACTATATTGGTCAATGAATATTTATATTATTCATGACTTTAAAAATAGTCAAACGAGACATA
ACGTAAAAGTCAAAATACGTTTAAGCTCATTCATATAAATGAATATTTTTAAAATTTGTTGCATCC
ATCAAAATATCTACTTTTTAAGGAATGATATTTATTTCATAATATTCATATTTGATTCGTTGATGGA
TAGATTTTATTCTTTAAAAAATTAAATAAAAAAAATAAAATTGGCCTAGTCATATCCATCTAAAAT
GGGTGAGATTCTGGTACGCTGACCGTCTTATAATTCCCAATAAAAACTTTTGGAGAAAAAAGGGA
ACACAAAAAAATGAAGTAGTGCACCAATAGAATCACTTCTCACCTCCTTATAGCTAGTACGGATTA
TTCCCTTCATGTGTGCCACAGTCATGCACAATCCATATTATAATTTCCAAAATAATTAGTTGTTCAC
GTTTGAATTGATCATAAATGATATTACCATTTATCCTTTTTACTTATTAAGTAGATAGATTAAAAAA
TTTAAGATTTTCAAAAAGTTCTACATTTTTAAAAATAATCAATTGAAGGTATAAAAAAGTTGTCCT
TCCTTAATTTCTCAAGATGGATAAGTAATTAAGAACAACTAAAAAAAAGCGAACAAATAATTAGA
GATCGAATGAATATTTATCAATCCTCATTTCACCAAGTCATTAAATTATTTTATGACCAAAATGTTT
ACTCATTTTGCTTAAATATCAAGAAAATTGTTGAATTATTTCTTATAGAAATATCACTCAACATCAG
TATCTAAGTAGTACTCATTTCGTTTCTATTTATATATCATTTTTATTAAATATAAATGTTTTCTTGAT
ATTTATTTATTTCACAAAATCAAAATTTGACTTATGATTACTAAATAATTAATTTAATTTAATTAAT
CAAAATAAATTAATTTATCTCTTTTGCAAAAGTTAACTTTAAGAGAACACTAATTAAGAATATAAT
AATAAATTTAGTTAATTTTTTTAAAAGATATAAAATCTAAATTAGTGACATATAAATAGAAAGAGG
GGAAAGTAGTAGTTTAACTCTTATGGTTTGATAAGGTGTGTGCTAAATGACAACATCTTTCTTGTCT
CGTAAAGTTAACATCTTTGTAGGTGGTGAGTAAGTGAGTGAATGCCATTGAATGAAGAGATTATTT
GTTTTTGTCACCTTTACCACTAAAGTTTTGTCTATTTTTATTCTTCGAATTCCTCCAGTACAAGATTT
TATTTTTGATATTCCTTTCTTTGGAATTCAGTGTTGGTATAAATAGGATCTATTTGGCTATCCACAT
ATATTTTTAAATAAAAATCAGTATTTAGTCATTTAAATTACATTTCATGGATTATACTCGTTAAAAA
AAATATATTTAAGCAATTAAATATTATTTGTTGAACATAGGAAAAATGATTTGAAATATATTCAAA
CTTTGATCACAATTGTGATAACAATTTCAAATTTTGGGAAGGACCTTTTACCCCTTGCACTATTTAT
AGTATATTTTAAATGTATATATATGTCAACATAAATATAATAAATATTGCATTATTATATATAGTAA
CTTGTTCACGTGGATACATATATACCTGTAAAATATACTATTAAATAATATAGGAGATAGTAGGTC
CTGCTCAAAGTTAGAGATTGTTATAGCAATTTCGATCAAAGATATATTTCAAACTATTTTTCCTAAA
AGATATAACCAAATACAATTTTATCTTTAATTTCAATATTTGCAAATAAAGTGAAAAAATATTTAT
ACCAAGTAGGATGAATTAAAAATTAAGGGTTTTTTTCCTCTTGTTATATATATAACTAATCGTCATT
TTTTTATTAATGAATCGTCGACAGGTTGGTCACCGATATTCCAGCTACCACAGGTTCAAGTTTTGGT
GAGAATCCTCTTTTTGTTAATTGTTTGTTTGTTGTCTTCCCATGTTTACATTTTTTTAAAAAAAAACA
AACTAATTTTAAAGGTAGAATTAAAAAAAAATCATTATCGTATTTAAAAATATATTTTTATAATAA
TATGGACGAATAATATGAAACTAACAGAGTAATGACAAAGGAATTTATACTGAGCGGGCAATGTT
GCGTTAAATCATGTTTGTCCTAAACTTTTAAAACCTAGGAAAGGGAATGAAATCTATTCTCAATTA
ACGTGATTAAATATTCTAAACAATTGATATCCTTTAATTATGTCCCACACTACGCCAAAAGTTCTTA
AGCATTACACTCTAAAATTTGTATGCATAACATTAAAAGATCATTACCTATTTGGCTAAAATTTTTA
CAATAAGTTTATTTTAAAAAGTGTTCCTTTTTTTCCCCTCTCAAAAACACACTTGTGTTACTCTTGAT
TTTTCTCTCAAAAGTTTAGTTAAATACTTAAGTTTTTTTAAAATAATTTTTTTATGAAAAAAGAAAA
AAAACATTTTTGGCTAACCAAACAGGTTTAGGAGACTTGCGCTCTGCCATAAGTATTTCCCCATTC
ACTTTTCTTCCATTTTTATTTATGATTTTTTTTAACATATTAAGAAAGCTATTTGTTTCATGCTCTTC
AATAATTTCTTATTCTCCAAATTAACATAGATATTGTGGTAAAACACCATAATAGTTATTGTATATT
TGTATACCTTTTCAAATATATATACTCTCTAATAAGATCACAAGATAAAAAAACATTTATTGGTGA
ATAAATTTGACATAACTTTAATTTAATTATAACACAAAATTCAAAAGTTTTATTTCTCAACTTAAAA
ATTTGGTGTCAAGTCAGAAGTAGATGTGATAATTTTTGTTTTTGAAATTGGAGGGAGTATCTTGT
TGAAAATATTGGATATGTACATAAGAAGTAGTCATTTGAAATGCATTGAAACTTGATAAAAACAT
AAGTAGCTAGCTAGTGCATGAAAGTTTGGTTGTTTATGTACTTTTAATATGTAGGGCAAGAAA TAGT
GAGCTATGAAAGTCCAAGACCATCAATGGGAATACATCGATTTGTATTTGTATTATTCAGACAATTAG
GTCGGCAAACAGTGTATGCTCCAGGATGGCGTCAGAATTTCAACACAAGAGATTTTGCAGAACTTTA
TAATCTTGGTTTACCTGTTGCTGCTGTCTATTTTAATTGTCAAAGAGAGAGTGGCAGTGGTGGACGTA
GAAGATCTGCTGATTGATCAACTCCATCTACTACAAAAAACAAAAAAACAATGATATTTTTAGCTA
ATAATAACCACCAATATCTACTACTTCTCTTACAACTTTAGTAGTATCTATAGTTATCTTTTTTAATC
TACTCTTTTACTTCTTTACTATATTGTCTTCCTCTCAATTTATTTGAATTAGTGACTTGATATCAAGT
- 57 -
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
TTCAATAAAGAAACAAAGACTGACTTTAGAATTTTGTGATTTACAATAAGTTGTACATATTTGTAT
GACTATCTTAAAAAGTTAAATCATTATTATTAAATATAAAAATATGATTAATTTAAAAGGAAGTAA
ATTATATAAAACGTTAATTTTTTTTTATAGTTTAGCTCTTAAAAAAAAATTATAACAATTAAAAGTA
TTGAATGAAAGAAGTTTGTAACTAGTCTCTGTTATTCCTCTATAAAACAGTATATTTTCTTGTTACT
TTTATAAATTTCTAAGATATGAACTTGAGT (SEQ ID NO: 16)
>sft-1906 genomic(mutation in brackets)
ATGCCTAGAGAACGTGATCCTCTTGTTGTTGGTCGTGTGGTAGGGGATGTATTGGACCCTTTCACA
AGAACTATTGGCCTAAGAGTTATATATAGAGATAGAGAAGTTAATAATGGATGCGAGCTTAGGCC
TTCCCAAGTTATTAACCAGCCAAGGGTTGAAGTTGGAGGAGATGACCTACGTACCTTTTTCACTTT
GGTAATATTTCTTATATTTTTTGTTTGGGAATATAGTTAAGTTGATTTTCATAAGCAAAGTAAAAAG
TATTTTTGTCTTTTTGTAAAGGTTATGGTGGACCCTGATGCTCCAAGTCCGAGTGATCCAAATCTGA
GAGAATACCTTCACTGGTCCGTATTTTTTCCTTATTCTCTCTTCTTTTCATCTCTTTCTTTTTTGACCT
TTTTACTTAATTATATTCTTTAGTAATAATATATGATGATATCCTTTTTAAAAATTGGAAATACGAA
AAGGAGAAATGAAGAGGAGATTTACATGTGAGGGAGCAGATGGTAGAAATATATAAATGTGAAG
ATATATATTCTTGAACTTAAAAACAAGCTACTAAAATAAAAATGAATAAAATATTTACTCTGTCAA
TATTCTGTACTATATTGGTCAATGAATATTTATATTATTCATGACTTTAAAAATAGTCAAACGAGAC
ATAACGTAAAAGTCAAAATACGTTTAAGCTCATTCATATAAATGAATATTTTTAAAATTTGTTGCA
TCCATCAAAATATCTACTTTTTAAGGAATGATATTTATTTCATAATATTCATATTTGATTCGTTGAT
GGATAGATTTTATTCTTTAAAAAATTAAATAAAAAAAATAAAATTGGCCTAGTCATATCCATCTAA
AATGGGTGAGATTCTGGTACGCTGACCGTCTTATAATTCCCAATAAAAACTTTTGGAGAAAAAAGG
GAACACAAAAAAATGAAGTAGTGCACCAATAGAATCACTTCTCACCTCCTTATAGCTAGTACGGA
TTATTCCCTTCATGTGTGCCACAGTCATGCACAATCCATATTATAATTTCCAAAATAATTAGTTGTT
CACGTTTGAATTGATCATAAATGATATTACCATTTATCCTTTTTACTTATTAAGTAGATAGATTAAA
AAATTTAAGATTTTCAAAAAGTTCTACATTTTTAAAAATAATCAATTGAAGGTATAAAAAAGTTGT
CCTTCCTTAATTTCTCAAGATGGATAAGTAATTAAGAACAACTAAAAAAAAGCGAACAAATAATT
AGAGATCGAATGAATATTTATCAATCCTCATTTCACCAAGTCATTAAATTATTTTATGACCAAAAT
GTTTACTCATTTTGCTTAAATATCAAGAAAATTGTTGAATTATTTCTTATAGAAATATCACTCAACA
TCAGTATCTAAGTAGTACTCATTTCGTTTCTATTTATATATCATTTTTATTAAATATAAATGTTTTCT
TGATATTTATTTATTTCACAAAATCAAAATTTGACTTATGATTACTAAATAATTAATTTAATTTAAT
TAATCAAAATAAATTAATTTATCTCTTTTGCAAAAGTTAACTTTAAGAGAACACTAATTAAGAATA
TAATAATAAATTTAGTTAATTTTTTTAAAAGATATAAAATCTAAATTAGTGACATATAAATAGAAA
GAGGGGAAAGTAGTAGTTTAACTCTTATGGTTTGATAAGGTGTGTGCTAAATGACAACATCTTTCT
TGTCTCGTAAAGTTAACATCTTTGTAGGTGGTGAGTAAGTGAGTGAATGCCATTGAATGAAGAGAT
TATTTGTTTTTGTCACCTTTACCACTAAAGTTTTGTCTATTTTTATTCTTCGAATTCCTCCAGTACAA
GATTTTATTTTTGATATTCCTTTCTTTGGAATTCAGTGTTGGTATAAATAGGATCTATTTGGCTATCC
ACATATATTTTTAAATAAAAATCAGTATTTAGTCATTTAAATTACATTTCATGGATTATACTCGTTA
AAAAAAATATATTTAAGCAATTAAATATTATTTGTTGAACATAGGAAAAATGATTTGAAATATATT
CAAACTTTGATCACAATTGTGATAACAATTTCAAATTTTGGGAAGGACCTTTTACCCCTTGCACTAT
TTATAGTATATTTTAAATGTATATATATGTCAACATAAATATAATAAATATTGCATTATTATATATA
GTAACTTGTTCACGTGGATACATATATACCTGTAAAATATACTATTAAATAATATAGGAGATAGTA
GGTCCTGCTCAAAGTTAGAGATTGTTATAGCAATTTCGATCAAAGATATATTTCAAACTATTTTTCC
TAAAAGATATAACCAAATACAATTTTATCTTTAATTTCAATATTTGCAAATAAAGTGAAAAAATAT
TTATACCAAGTAGGATGAATTAAAAATTAAGGGTTTTTTTCCTCTTGTTATATATATAACTAATCGT
CATTTTTTTATTAATGAATCGTCGACAGGTTGGTCACCGATATTCCAGCTACCACAGGTTCAAGTTT
TGGTGAGAATCCTCTTTTTGTTAATTGTTTGTTTGTTGTCTTCCCATGTTTACATTTTTTTAAAAAAA
AACAAACTAATTTTAAAGGTAGAATTAAAAAAAAATCATTATCGTATTTAAAAATATATTTTTATA
ATAATATGGACGAATAATATGAAACTAACAGAGTAATGACAAAGGAATTTATACTGAGCGGGCAA
TGTTGCGTTAAATCATGTTTGTCCTAAACTTTTAAAACCTAGGAAAGGGAATGAAATCTATTCTCA
ATTAACGTGATTAAATATTCTAAACAATTGATATCCTTTAATTATGTCCCACACTACGCCAAAAGTT
CTTAAGCATTACACTCTAAAATTTGTATGCATAACATTAAAAGATCATTACCTATTTGGCTAAAATT
TTTACAATAAGTTTATTTTAAAAAGTGTTCCTTTTTTTCCCCTCTCAAAAACACACTTGTGTTACTCT
TGATTTTTCTCTCAAAAGTTTAGTTAAATACTTAAGTTTTTTTAAAATAATTTTTTTATGAAAAAAG
AAAAAAAACATTTTTGGCTAACCAAACAGGTTTAGGAGACTTGCGCTCTGCCATAAGTATTTCCCC
ATTCACTTTTCTTCCATTTTTATTTATGATTTTTTTTAACATATTAAGAAAGCTATTTGTTTCATGCT
CTTCAATAATTTCTTATTCTCCAAATTAACATAGATATTGTGGTAAAACACCATAATAGTTATTGTA
- 58 -
CA 02892012 2015-05-20
WO 2014/081730 PCT/US2013/070825
TATTTGTATACCTTTTCAAATATATATACTCTCTAATAAGATCACAAGATAAAAAAACATTTATTGG
TGAATAAATTTGACATAACTTTAATTTAATTATAACACAAAATTCAAAAGTTTTATTTCTCAACTTA
AAAATTTGGTGTCAAGTCAGAAGTAGATGTGATAATTTTTGTTTTTGAAATTGGAGGGAGTATCT
TGTTGAAAATATTGGATATGTACATAAGAAGTAGTCATTTGAAATGCATTGAAACTTGATAAAAAC
ATAAGTAGCTAGCTAGTGCATGAAAGTTTGGTTGTTTATGTACTTTTAATATGTAGGGCAAGAAAT
AGTGAGCTATGAAAGTCCAAGACCATCAATGGGAATACATCGATTTGTATTTGTATTATTCAGACA
ATTAGGTCGGCAAACA[[A]]TGTATGCTCCAGGATGGCGTCAGAATTTCAACACAAGAGATTTTGCA
GAACTTTATAATCTTGGTTTACCTGTTGCTGCTGTCTATTTTAATTGTCAAAGAGAGAGTGGCAGTG
GTGGACGTAGAAGATCTGCTGATTGATCAACTCCATCTACTACAAAAAACAAAAAAACAATGATA
TTTTTAGCTAATAATAACCACCAATATCTACTACTTCTCTTACAACTTTAGTAGTATCTATAGTTAT
CTTTTTTAATCTACTCTTTTACTTCTTTACTATATTGTCTTCCTCTCAATTTATTTGAATTAGTGACTT
GATATCAAGTTTCAATAAAGAAACAAAGACTGACTTTAGAATTTTGTGATTTACAATAAGTTGTAC
ATATTTGTATGACTATCTTAAAAAGTTAAATCATTATTATTAAATATAAAAATATGATTAATTTAAA
AGGAAGTAAATTATATAAAACGTTAATTTTTTTTTATAGTTTAGCTCTTAAAAAAAAATTATAACA
ATTAAAAGTATTGAATGAAAGAAGTTTGTAACTAGTCTCTGTTATTCCTCTATAAAACAGTATATTT
TCTTGTTACTTTTATAAATTTCTAAGATATGAACTTGAGT (SEQ ID NO: 17)
Without further elaboration, it is believed that one skilled in the art can,
based on the
above description, utilize the present disclosure to its fullest extent. The
specific
embodiments are, therefore, to be construed as merely illustrative, and not
limitative of the
remainder of the disclosure in any way whatsoever. All publications cited
herein are
incorporated by reference for the purposes or subject matter referenced
herein.
From the above description, one skilled in the art can easily ascertain the
essential
characteristics of the present disclosure, and without departing from the
spirit and scope
thereof, can make various changes and modifications of the disclosure to adapt
it to various
usages and conditions. Thus, other embodiments are also within the claims.
- 59 -