Note: Descriptions are shown in the official language in which they were submitted.
CA 02913512 2015-11-25
WO 2014/190422
PCT/CA2014/000471
Title: Methods and Compositions for Detecting Progressive Hearing Loss
Related Applications
[0001] This is a Patent Cooperation Treaty Application which claims
the benefit of 35
U.S.C. 119 based on the priority of U.S. Provisional Patent Application No.
61/829,050,
filed May 30, 2013 which is incorporated herein by reference in its entirety.
Field of the Disclosure
[0002] The disclosure relates to methods and products for detecting
susceptibility of
a subject of to having and/or developing hearing loss and specifically to
methods of
detecting a mutation in FOXL1 associated with otosclerosis.
Introduction
[0003] Hearing loss due to otosclerosis is relatively common and is
estimated to
contribute to 0.3-1% of cases of late onset hearing loss ([1, 2]). Both
sporadic and hereditary
cases are currently recognized, and at least eight known loci for otosclerosis
inherited as a
dominant trait (OTSC1, OTSC2, OTSC3, OTSC4, OTSC5, OTSC7, OTSC8 and OTSC10)
and have been mapped [3-10].
[0004] Otosclerosis of the temporal bone is characterized by abnormal bone
metabolism, with focal resorption and formation of abnormal bone (greater
density, cellularity
and vascularity and eventually thick sclerotic bone) in the otic capsule.
Otosclerotic foci
occurring anterior to the oval window can cause stapes fixation and conductive
hearing loss,
and more rarely sensorineural hearing loss caused by otosclerotic foci within
the
cochlea. Certain diagnosis of otosclerosis requires surgical confirmation and
is based on the
finding of otosclerotic foci on the oval window with stapedial fixation during
surgery.
[0005] The decision to undergo surgical exploration of the middle ear
for suspected
otosclerosis is based on the clinical findings of conductive hearing loss with
a normal
tympanic membrane and absent/ abnormal stapedial reflexes. Although high
resolution CT
scan is proving to be useful for assisting with presurgical diagnosis, it is
not widely available
clinically, and the results appear to be false negatives in a significant
proportion of cases.
Most importantly, relying on currently available clinical tests (high
resolution CT, audiological
testing) means that the disease has progressed significantly before surgical
treatment is
considered and a diagnosis of otosclerosis can be confirmed.
[0006] Early detection of the disease through genetic testing, prior to
the
development of advanced clinical symptoms, will facilitate earlier treatment,
with the
potential for improved surgical outcomes and the
reduction of surgical
complications. Furthermore, insights provided by identifying a genetic
etiology may
1
CA 02913512 2015-11-25
WO 2014/190422
PCT/CA2014/000471
elucidate the underlying pathophysiological mechanisms associated with this
metabolic bone
disorder and improve understanding of the onset and natural progression of
otosclerotic
disease processes, thus facilitating identification of ways for the prevention
of this disorder
through targeted treatments.
Summary of the Disclosure
[0007] An aspect includes a method for identifying a human subject as
having or
having an increased likelihood to develop otosclerosis and/or hearing loss,
the method
comprising (a) obtaining a suitable sample from the subject; (b) assaying the
sample for the
presence or absence of a mutation in: i) a FOXL1 polynucleotide at a
nucleotide sequence
encoding one or more of amino acid residues in FOXL1 polypeptide C-terminus;
ii) a FOXL1
polynucleotide at a nucleotide sequence encoding one or more of amino acid
residues
corresponding to positions 326 to 330 of SEQ ID NO:3; iii) a FOXL1 polypeptide
at one or
more amino acid residues in FOXL1 polypeptide C-terminus; or iv) a FOXL1
polypeptide at
one or more amino acid residues corresponding to positions 326 to 330 of SEQ
ID NO:3;
and (c) identifying the subject as having or having an increased likelihood to
develop
otosclerosis and/or hearing loss if said mutation is detected.
[0008]
Another aspect includes a method of determining if a human subject is a
carrier of a mutation that increases the likelihood of developing otosclerosis
and/or hearing
loss, the method comprising (a) obtaining a suitable sample from the subject;
(b) assaying
the sample for the presence or absence of a mutation in: i) a FOXL1
polynucleotide at a
nucleotide sequence encoding one or more of amino acid residues in FOXL1
polypeptide C-
terminus; ii) a FOXL1 polynucleotide at a nucleotide sequence encoding one or
more of
amino acid residues corresponding to positions 326 to 330 of SEQ ID NO:3; iii)
a FOXL1
polypeptide at one or more amino acid residues in FOXL1 polypeptide C-
terminus; or iv) a
FOXL1 polypeptide at one or more amino acid residues corresponding to
positions 326 to
330 of SEQ ID NO:3; and (c) identifying the subject as a carrier of a mutation
that increased
the likelihood of developing otosclerosis and/or hearing loss if said mutation
is detected.
[0009]
Another aspect includes a method for identifying a human subject as having
or having an increased likelihood to develop otosclerosis and/or hearing loss,
the method
comprising (a) obtaining a suitable sample from the subject; and (b) assaying
the sample for
the presence or absence of a mutation in: i) a FOXL1 polynucleotide at a
nucleotide
sequence encoding one or more of amino acid residues in FOXL1 polypeptide C-
terminus; ii)
a FOXL1 polynucleotide at a nucleotide sequence encoding one or more of amino
acid
residues corresponding to positions 326 to 330 of SEQ ID NO:3; iii) a FOXL1
polypeptide at
one or more amino acid residues in FOXL1 polypeptide C-terminus; or iv) a
FOXL1
2
CA 02913512 2015-11-25
WO 2014/190422
PCT/CA2014/000471
polypeptide at one or more amino acid residues corresponding to positions 326
to 330 of
SEQ ID NO:3; ; wherein detection of the presence of said mutation indicates
that the subject
has, or has an increased likelihood, to develop otosclerosis and/or hearing
loss.
[0010] Yet another aspect includes an isolated polynucleotide
comprising the
sequence of SEQ ID NO: 2 or 5.
[0011] Another aspect includes an isolated polypeptide comprising the
sequence of
SEQ ID NO: 4 or 6.
[0012] Another aspect includes an isolated antibody or binding
fragment that binds
SEQ ID NO: 6 or that binds an epitope comprising amino acid residues 325 and
331 of SEQ
ID NO: 4 that does not bind an epitope comprising amino acids 326 to 330 of
SEQ ID NO: 3.
[0013] In an embodiment, the isolated polynucleotide, isolated
polypeptide or
isolated antibody or binding fragment thereof is conjugated to and/or
comprises a
heterologous moiety, optionally a detectable label.
[0014] A further aspect includes a composition comprising: i) a
polynucleotide
comprising the sequence of SEQ ID NO: 2 or 5; ii) a polypeptide comprising the
sequence of
SEQ IN NO: 4 or 6; or iii) an antibody that binds SEQ ID NO: 6 or that binds
an epitope
comprising amino acids 325 and 331 of SEQ ID NO: 3 that does not bind an
epitope
comprising amino acids 326 to 330 of SEQ ID NO: 3 and a diluent.
[0015] Other features and advantages of the present disclosure will
become
apparent from the following detailed description. It should be understood,
however, that the
detailed description and the specific examples while indicating preferred
embodiments of the
disclosure are given by way of illustration only, since various changes and
modifications
within the spirit and scope of the disclosure will become apparent to those
skilled in the art
from this detailed description.
Brief description of the drawings
[0016] An embodiment of the present disclosure will now be described in
relation to the drawings in which:
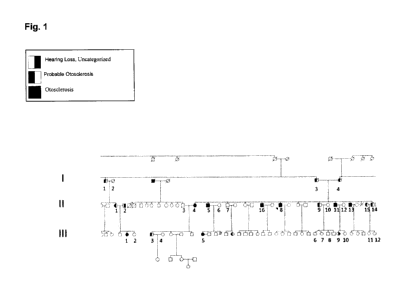
[0017] Figure 1: Partial pedigree of family 2081 segregating a form of
autosomal
dominant otosclerosis.
[0018] Figure 2: Audiological series of the family 2081 proband.
[0019] Figure 3: Partial pedigree (right hand side) of NL family 2081 co-
segregating
a 9Mb green haplotype that located 2Mb downstream of OTSC4.
3
CA 02913512 2015-11-25
WO 2014/190422
PCT/CA2014/000471
[0020] Figure 4: Electropherogram showing 15 base pair deletion causing in
frame
deletion.
[0021] Figure 5: Multiple alignment of FOXL1 showing the deletion of 5
amino acids
when compared across Eukaryotic and Prokaryotic species: XP 001231599.2
(G.gallus), NP
032050.2 (M.musculus), XP 002694802.1 (B.taurus), XP 851625.1 (C.lupus), NP
005241.1
(Homo species) and XP 511154.2 (p.troglodytes).
[0022] Figure 6: Gene structure of FOXL1.
[0023] Figure 7: Expression of FOXL1 gene from lymphoblastoid cell
line in subjects
with otosclerosis.
[0024] Figure 8: Effect of 15bp deletion on FOXL1 expression.
[0025] Figure 9: Expression of FOXL1-mutant protein in nuclear extract from
HEK
293A cell line transfected FOXL1 (WT and Mutant).
[0026] Figure 10: Transient transfection of FOXL1 (/VT and Mutant) in
osteoblast
cell line and measuring FOXL1 by real time PCR.
[0027] Figure 11: Nuclear localization of FOXL1 (WT and Mutant) in
osteoblast cell
line.
[0028] Figure 12: Transient transfection of FOXL1 (WT and Mutant) in
osteoblast
cell line and measuring protein expression.
[0029] Figure 13: Expression of predicted genes involved in
otosclerosis.
[0030] Table 1: Sequenced functional candidate genes at extended OTSC4
region.
[0031] Table 2: Variants detected from sequencing of 13 genes in extended
OTSC4.
[0032] Table 3: Sequences
Detailed description of the Disclosure
I. Definitions
[0033] The term "FOXL1" as used herein means forkhead box protein L1
and
optionally refers to the gene, and/or its gene products including nucleic acid
transcripts and
translated polypeptide as determinable from its context usage and includes all
naturally
occurring variants thereof. FOXL1 is an 345-amino acid protein with one
isoform
(NM_005250) and one known functional domain (Fork head domain). The
polynucleotide
sequence of wildtype human FOXL1 is shown in SEQ ID NO:1 and the amino-acid
sequence
of wildtype human FOXL1 is shown in SEQ ID NO: 3. The polynucleotide sequence
of
4
CA 02913512 2015-11-25
WO 2014/190422
PCT/CA2014/000471
mutant human FOXL1 is shown in SEQ ID NO:2 and the amino acid sequence of
mutant
human FOXL1 is shown in SEQ ID NO: 4. The FOXL1 gene is found at positions
86612115
86615304 of chromosome 16 (Accession number NM_005250, Genomic BUILD: hg19).
[0034] The term "c 976 990het deIGGGATCCCCTTCCTC"
. _ or
"de1976_990GGGATCCCCTTCCTC" as used herein means a nucleotide mutation in a
FOXL1 polynucleotide, the mutation being a deletion of the GGGATCCCCTTCCTC
(SEQ ID
NO: 5) nucleotides corresponding to position 976 to position 990. SEQ ID NO:2
is a nucleic
acid sequence that shows the polynucleotide sequence of FOXL1 deleted for this
sequence.
[0035] The term "p.G1y326_Leu330del" as used herein means an amino
acid
mutation in a FOXL1 polypeptide, the mutation comprising mutation of the amino
acids
corresponding to position 326 to amino acid at position 330 from Glycine (Gly)
to Leucine
(Leu) (the deleted amino acid sequence is shown in SEQ ID NO:6). SEQ ID NO:4
is a
polypeptide sequence that shows the polypeptide sequence of FOXL1deleted for
this
sequence.
[0036] The term "c.976_990het_deIGGGATCCCCTTCCTC: p.G1y326_Leu330del"
as used herein means both or either the c.976_990het_deIGGGATCCCCTTCCTC FOXL1
polynucleotide mutation or the p.G1y326_Leu330del FOXL1 polypeptide mutation.
Deletion
of the GGGATCCCCTTCCTC from position 976 to position 990 of SEQ ID NO:1
results in a
mutated FOXL1 polypeptide comprising p.G1y326_Leu330del.
[0037] The term "mutated FOXL1" as used herein refers to a FOXL1
molecule,
including a FOXL1 polynucleotide encoding a polypeptide deleted for one or
more or all
amino acids corresponding to position 326 to position 330 of SEQ ID NO:3
and/or a FOXL1
polypeptide encoded by such a polynucleotide.
[0038] The term "corresponding to" as used herein includes a residue
situated in a
different sequence position but having sequence characteristics in common,
including
identical, or substantially identical, nucleotide sequence flanking the
mutation (e.g.
substantial identity is optionally at least 100% identity over four or more
contiguous
nucleotides). For example the deleted nucleotide positions are given relative
to SEQ ID NO:
1, which is the cDNA sequence of wildtype human FOXL1 and the deleted amino
acid
positions are given relative to SEQ ID NO: 3. A person skilled in the art
would readily be
able to determine the amino acids corresponding to position 326-330 of SEQ ID
NO:3 in a
different FOXL1 polypeptide e.g. different species, or naturally occurring
variant. Similarly in
methods involving genomic DNA, a person skilled in the art would readily be
able to
determine the genomic residue corresponding to position 976-990 of SEQ ID NO:
1.
5
CA 02913512 2015-11-25
WO 2014/190422
PCT/CA2014/000471
[0039] The term "heterologous moiety" as used herein means a molecule,
optionally
a nucleotide or amino acid sequence that is not a FOXL1 sequence, such as a
targeting
moiety that targets the sequence when expressed in a cell to a particular cell
compartment
and/or a signal sequence that directs cellular localization of a fused
polypeptide. The
heterologous moiety can also be a detectable label that is conjugated to an
isolated
polypeptide or isolated polynucleotide such as a fluorescent polypeptide or
polynucleotide
encoding a fluorescent polypeptide or an enzyme etc. The detectable label can
also be
comprised in the polynucleotide or polypeptide, for example when the
detectable label is a
radioactive nucleotide or amino acid.
[0040] The term "nucleic acid" and/or "oligonucleotide" as used herein
refers to a
sequence of nucleotide or nucleoside monomers consisting of naturally
occurring bases,
sugars, and intersugar (backbone) linkages, and is intended to include DNA and
RNA which
can be either double stranded or single stranded, represent the sense or
antisense strand as
well as cDNA. The term also includes modified or substituted oligomers
comprising non-
naturally occurring monomers or portions thereof, which function similarly,
which are referred
to herein as "chemical analogues" and/or "oligonucleotide analogues" such as
"peptide
nucleic acids". Such modified or substituted nucleic acids may be preferred
over naturally
occurring forms because of properties such increased stability in the presence
of nucleases.
[0041] The term "isolated nucleic acid sequence, and/or "isolated
polynucleotide" as
used herein refers to a nucleic acid substantially free of cellular material
or culture medium
when produced by recombinant DNA techniques, or chemical precursors, or other
chemicals
when chemically synthesized.
[0042] The term "isolated DNA" as used herein means genomic or cDNA
that is
purified sufficiently for a method of detecting a mutation, for example
sufficiently purified for
PCR or other method described herein.
[0043] The term "probe" as used herein refers to a nucleic acid sequence
that will
hybridize to a nucleic acid target sequence. In one example, the probe
hybridizes to a
mutated FOXL1 polynucleotide such as a RNA or DNA FOXL1 nucleic acid deleted
or
substituted for GGGATCCCCTTCCTC (SEQ ID NO: 5) corresponding to position 976
to
position 990 of SEQ ID NO: 1 or a nucleic acid sequence complementary to said
RNA or
DNA. For example, the probe can comprise residues complementary to 5' and 3'
sequence
flanking the deletion sequence, for example comprising at least 8 contiguous
nucleotides of
SEQ ID NO: 2 , the at least 8 contiguous nucleotides including residues
corresponding to
position 975 and 976. The length of probe depends on the hybridize conditions
and the
6
CA 02913512 2015-11-25
WO 2014/190422
PCT/CA2014/000471
sequences of the probe and nucleic acid target sequence. In one embodiment,
the probe is
at least 8, 10, 12, 15, 20, 25, 50, 75, 100, 150, 200, 250, 400, 500 or more
nucleotides in
length. In an another embodiment, the probe nucleotide length is any whole
number
between 8 and 500 nucleotides.
[0044] The
term "primer" as used herein refers to a nucleic acid sequence, whether
occurring naturally as in a purified restriction digest or produced
synthetically, which is
capable of acting as a point of synthesis when placed under conditions in
which synthesis of
a primer extension product, which is complementary to a nucleic acid strand is
induced (e.g.
in the presence of nucleotides and an inducing agent such as DNA polymerase
and at a
suitable temperature and pH). The primer must be sufficiently long to prime
the synthesis of
the desired extension product in the presence of the inducing agent. The exact
length of the
primer will depend upon factors, including temperature, sequences of the
primer and the
methods used. A primer typically contains 15-25 or more nucleotides, although
it can contain
less. In an embodiment, the primer is about 12 nucleotides. The primer can be
from about
15 to about 30 nucleotides in length or any whole number in between 12 and 30
nucleotides.
The factors involved in determining the appropriate length of primer are
readily known to one
of ordinary skill in the art.
[0045] As
used herein "all or part of" of a probe or primer refers to the portion
sufficient for in the case a probe, sufficient to specifically hybridize to
the intended target and
in the case of a primer, sufficient to prime amplification of the intended
template.
[0046] The term
"hybridize" refers to the sequence specific non-covalent binding
interaction with a complementary nucleic acid. One aspect of the application
provides an
isolated nucleotide sequence, which hybridizes to a RNA product of FOXL1 or a
nucleic acid
sequence which is complementary to an RNA product of a gene of FOXL1. In one
embodiment the hybridization is conducted under at least moderately stringent
conditions. In
a preferred embodiment, the hybridization is under high stringency conditions.
Appropriate
stringency conditions which promote hybridization are known to those skilled
in the art, or
can be found in Current Protocols in Molecular Biology, John Wiley & Sons,
N.Y. (1989),
6.3.1 6.3.6. For example, 6.0 x sodium chloride/sodium citrate (SSC) at about
45 C for 15
minutes, followed by a wash of 2.0 x SSC at 50 C for 15 minutes may be
employed.
[0047] The
stringency may be selected based on the conditions used in the
wash step. For example, the salt concentration in the wash step can be
selected from a high
stringency of about 0.2 x SSC at 50 C for 15 minutes. In addition, the
temperature in the
wash step can be at high stringency conditions, at about 65 C for 15 minutes.
7
CA 02913512 2015-11-25
WO 2014/190422
PCT/CA2014/000471
[0048] By "at least moderately stringent hybridization conditions" it is
meant
that conditions are selected which promote selective hybridization between two
complementary nucleic acid molecules in solution. Hybridization may occur to
all or a portion
of a nucleic acid sequence molecule. The hybridizing portion is typically at
least 15 (e.g. 20,
25, 30, 40 or 50) nucleotides in length. Those skilled in the art will
recognize that the stability
of a nucleic acid duplex, or hybrids, is determined by the Tm, which in sodium
containing
buffers is a function of the sodium ion concentration and temperature (Tm =
81.5 C ¨ 16.6
(Log10 [Na+]) + 0.41( /0(G+C) ¨ 600/1), or similar equation). Accordingly, the
parameters in
the wash conditions that determine hybrid stability are sodium ion
concentration and
temperature. In order to identify molecules that are similar, but not
identical, to a known
nucleic acid molecule a 1% mismatch may be assumed to result in about a 1 C
decrease in
Tm, for example if nucleic acid molecules are sought that have a >95% sequence
identity,
the final wash temperature will be reduced by about 5 C. Based on these
considerations
those skilled in the art will be able to readily select appropriate
hybridization conditions. In
preferred embodiments, stringent hybridization conditions are selected. By way
of example
the following conditions may be employed to achieve stringent hybridization:
hybridization at
5x sodium chloride/sodium citrate (SSC)/5x Denhardt's solution/1.0% SDS at Tm -
5 C
based on the above equation, followed by a wash of 0.2x SSC/0.1% SDS at 60 C
for 15
minutes. Moderately stringent hybridization conditions include a washing step
in 3x SSC at
42 C for 15 minutes. It is understood, however, that equivalent stringencies
may be
achieved using alternative buffers, salts and temperatures. Additional
guidance regarding
hybridization conditions may be found in: Current Protocols in Molecular
Biology, John Wiley
& Sons, N.Y., 1989, 6.3.1-6.3.6 and in: Sambrook et al., Molecular Cloning, a
Laboratory
Manual, Cold Spring Harbor Laboratory Press, 2000, Third Edition.
[0049] The
term "sequence identity" as used herein refers to the percentage of
sequence identity between two polypeptide sequences or two nucleic acid
sequences. To
determine the percent identity of two amino acid sequences or of two nucleic
acid
sequences, the sequences are aligned for optimal comparison purposes (e.g.,
gaps can be
introduced in the sequence of a first amino acid or nucleic acid sequence for
optimal
alignment with a second amino acid or nucleic acid sequence). The amino acid
residues or
nucleotides at corresponding amino acid positions or nucleotide positions are
then
compared. When a position in the first sequence is occupied by the same amino
acid
residue or nucleotide as the corresponding position in the second sequence,
then the
molecules are identical at that position. The percent identity between the two
sequences is a
function of the number of identical positions shared by the sequences (i.e., %
8
CA 02913512 2015-11-25
WO 2014/190422
PCT/CA2014/000471
identity=nunnber of identical overlapping positions/total number of
positions×100 /0). In
one embodiment, the two sequences are the same length. The determination of
percent
identity between two sequences can also be accomplished using a mathematical
algorithm.
A preferred, non-limiting example of a mathematical algorithm utilized for the
comparison of
two sequences is the algorithm of Karlin and Altschul, 1990, Proc. Natl. Acad.
Sci. U.S.A.
87:2264-2268, modified as in Karlin and Altschul, 1993, Proc. Natl. Acad. Sci.
U.S.A.
90:5873-5877. Such an algorithm is incorporated into the NBLAST and XBLAST
programs of
Altschul et al., 1990, J. Mol. Biol. 215:403. BLAST nucleotide searches can be
performed
with the NBLAST nucleotide program parameters set, e.g., for score=100,
wordlength=12 to
obtain nucleotide sequences homologous to a nucleic acid molecules of the
present
application. BLAST protein searches can be performed with the XBLAST program
parameters set, e.g., to score-50, wordlength=3 to obtain amino acid sequences
homologous to a protein molecule described herein. To obtain gapped alignments
for
comparison purposes, Gapped BLAST can be utilized as described in Altschul et
al., 1997,
Nucleic Acids Res. 25:3389-3402. Alternatively, PSI-BLAST can be used to
perform an
iterated search which detects distant relationships between molecules (Id.).
When utilizing
BLAST, Gapped BLAST, and PSI-Blast programs, the default parameters of the
respective
programs (e.g., of XBLAST and NBLAST) can be used (see, e.g., the NCBI
website). The
percent identity between two sequences can be determined using techniques
similar to
those described above, with or without allowing gaps. In calculating percent
identity, typically
only exact matches are counted. In an
embodiment, the isolated nucleic acids are useful
as primers.
[0050] As
used herein "NASBA" refers to a sensitive isothermal transcription-based
amplification method used for example for RNA research. NASBA technology is
optionally
applied to single nucleotide polymorphism (SNP) analysis using human genomic
DNA as a
template. For example combination of DNA NASBA with multiplex hybridization of
specific
molecular beacons makes it possible to discriminate the presence of mutations
of interest
(Berard, C, Cazalis MA, Leissner P, Mougin B., DNA nucleic acid sequence-based
amplification-based genotyping for polymorphism analysis. Biotechniques. 2004,
37:680-2,
684, 686).
[0051] The term
"isolated polypeptide" as used herein refers to a proteinaceous
agent, such as a peptide, polypeptide or protein, which is substantially free
of cellular
material or culture medium when produced recombinantly, or chemical
precursors, or other
chemicals, when chemically synthesized.
9
CA 02913512 2015-11-25
WO 2014/190422
PCT/CA2014/000471
[0052] The term
"antibody" as used herein is intended to include monoclonal
antibodies, polyclonal antibodies, and chimeric antibodies. The antibody may
be from
recombinant sources and/or produced in transgenic animals. The term "antibody
fragment"
as used herein is intended to include Fab, Fab', F(ab')2, scFv, dsFv, ds-scFv,
dimers,
minibodies, diabodies, and multimers thereof and bispecific antibody
fragments. Antibodies
can be fragmented using conventional techniques. For example, F(ab')2
fragments can be
generated by treating the antibody with pepsin. The
resulting F(ab')2 fragment can be
treated to reduce disulfide bridges to produce Fab' fragments. Papain
digestion can lead to
the formation of Fab fragments. Fab, Fab' and F(ab')2, scFv, dsFv, ds-scFv,
dimers,
minibodies, diabodies, bispecific antibody fragments and other fragments can
also be
synthesized by recombinant techniques.
[0053] To
produce human monoclonal antibodies, antibody producing cells
(lymphocytes) can be harvested from a human having cancer and fused with
myeloma cells
by standard somatic cell fusion procedures thus immortalizing these cells and
yielding
hybridoma cells. Such techniques are well known in the art, (e.g. the
hybridoma technique
originally developed by Kohler and Milstein (Nature 256:495-497 (1975)) as
well as other
techniques such as the human B-cell hybridoma technique (Kozbor et al.,
lmmunol. Today
4:72 (1983)), the EBV-hybridoma technique to produce human monoclonal
antibodies (Cole
et al., Methods Enzymol, 121:140-67 (1986)), and screening of combinatorial
antibody
libraries (Huse et al., Science 246:1275 (1989)).
Hybridoma cells can be screened
immunochemically for production of antibodies specifically reactive with
mutated FOXL1
polypeptide and the monoclonal antibodies can be isolated.
[0054]
Specific antibodies, or antibody fragments, reactive against mutated
FOXL1 antigen, may also be generated by screening expression libraries
encoding
immunoglobulin genes, or portions thereof, expressed in bacteria with cell
surface
components. For example, complete Fab fragments, VH regions and FV regions can
be
expressed in bacteria using phage expression libraries (See for example Ward
et al., Nature
341:544-546 (1989); Huse et al., Science 246:1275-1281 (1989); and McCafferty
et al.,
Nature 348:552-554 (1990)).
[0055] The
term "amplifying a portion of a FOXL1 coding sequence, the portion
comprising minimally FOXL1 residues corresponding to positions 976 to
position_990 of
SEQ ID NO:1" as used herein means amplifying a portion of exon 1 of FOXL1 in
genomic
DNA or in FOXL1 cDNA, the portion including minimally residues corresponding
to positions
976 to 990 of SEQ ID NO:1, optionally all of SEQ ID NO:1. In a subject
carrying the
mutation, amplifying minimally positions 976 to 990 produces no amplified
product. The
CA 02913512 2015-11-25
WO 2014/190422
PCT/CA2014/000471
amplified FOXL1 polynucleotide if larger for example comprising an amplified
product
corresponding to positions 900 to 999, produces an amplified product of 100
nucleotides in a
wild type carrier and 85 nucleotides in a FOXL1
c.976_990het_deIGGGATCCCCTTCCTC
mutant carrier.
[0056] A "microarray" as used herein refers to an ordered set of
probes fixed to a
solid surface that permits analysis such as gene analysis of a plurality of
genes. A DNA
microarray refers to an ordered set of DNA fragments fixed to the solid
surface. For
example, in one embodiment the microarray is a gene chip. A tissue microarray
refers to an
ordered set of tissue specimens fixed to a solid surface. For example, in one
embodiment
the tissue microarray comprises a slide comprising an array of arrayed tumor
biopsy
samples in paraffin. Tissue microarray technology optionally allows multiple
specimens, such
as biopsy samples, to be analyzed in a single analysis at the DNA, RNA or
protein level.
Tissue microarrays are analyzed by a number of techniques including
immunohistochemistry, in situ hybridization, in situ PCR, RNA or DNA
expression analysis
and and/or morphological and clinical characterization or a combination of
techniques. The
specimens are optionally from the same subject or from a plurality of
subjects. Methods of
detecting gene expression using arrays are well known in the art. Such methods
are
optionally automated.
[0057] The term "subject" as used herein includes all members of the
animal
kingdom including multicellular organisms, including mammals, and preferably
means
humans.
[0058] The term "suitable sample" as used herein means a sample that
can be
assayed for FOXL1 protein and/or polynucleotide sequence. Suitable samples for
detecting
the FOXLI deletion are blood, saliva, throat swab and any DNA accessible
tissue.
[0059] As used herein, "a relative" or "blood relation" is a relative
genetically related,
or related by birth, and includes without limitation 1st, 2nd, 3rd , 4th, 5th,
6th , 7th, 8th, 9th
and 10th degree relations, for example but not limited to parents, children,
grandchildren,
grandparents, cousins and/or 2nd cousins related by blood.
[0060] In understanding the scope of the present disclosure, the term
"comprising"
and its derivatives, as used herein, are intended to be open ended terms that
specify the
presence of the stated features, elements, components, groups, integers,
and/or steps, but
do not exclude the presence of other unstated features, elements, components,
groups,
integers and/or steps. The foregoing also applies to words having similar
meanings such as
the terms, "including", "having" and their derivatives. Finally, terms of
degree such as
11
CA 02913512 2015-11-25
WO 2014/190422
PCT/CA2014/000471
"substantially", "about" and "approximately" as used herein mean a reasonable
amount of
deviation of the modified term such that the end result is not significantly
changed. These
terms of degree should be construed as including a deviation of at least 5%
of the modified
term if this deviation would not negate the meaning of the word it modifies
[0061] In understanding the scope of the present disclosure, the term
"consisting"
and its derivatives, as used herein, are intended to be close ended terms that
specify the
presence of stated features, elements, components, groups, integers, and/or
steps, and also
exclude the presence of other unstated features, elements, components, groups,
integers
and/or steps.
[0062] The recitation of numerical ranges by endpoints herein includes
all numbers
and fractions subsumed within that range (e.g. 1 to 5 includes 1, 1.5, 2,
2.75, 3, 3.90, 4, and
5). It is also to be understood that all numbers and fractions thereof are
presumed to be
modified by the term "about." Further, it is to be understood that "a," "an,"
and "the" include
plural referents unless the content clearly dictates otherwise.
[0063] Further, the definitions and embodiments described in
particular sections are
intended to be applicable to other embodiments herein described for which they
are suitable
as would be understood by a person skilled in the art. For example, in the
following
passages, different aspects of the invention are defined in more detail. Each
aspect so
defined may be combined with any other aspect or aspects unless clearly
indicated to the
contrary. In particular, any feature indicated as being preferred or
advantageous may be
combined with any other feature or features indicated as being preferred or
advantageous.
Methods
[0064] A novel 15bp deletion, c.976_990het_deIGGGATCCCCTTCCTC, in
FOXL1
deleted in subjects with otosclerosis and/or hearing loss is disclosed herein.
The deletion
causes an inframe deletion of 5 amino acids (p.Gly326_Leu330).
[0065] Accordingly, an aspect includes a method for identifying a
human subject as
having or having an increased likelihood to develop otosclerosis and/or
hearing loss, the
method comprising (a) obtaining a suitable sample from the subject; (b)
assaying the sample
for the presence or absence of a mutation in: i) a FOXL1 polynucleotide at a
nucleotide
sequence encoding one or more of amino acid residues in FOXL1 polypeptide C-
terminus; ii)
a FOXL1 polynucleotide at a nucleotide sequence encoding one or more of amino
acid
residues corresponding to positions 326 to 330 of SEQ ID NO:3; iii) a FOXL1
polypeptide at
12
CA 02913512 2015-11-25
WO 2014/190422
PCT/CA2014/000471
one or more amino acid residues in FOXL1 polypeptide C-terminus; or iv) a
FOXL1
polypeptide at one or more amino acid residues corresponding to positions 326
to 330 of
SEQ ID NO:3; and (c) identifying the subject as having or having an increased
likelihood to
develop otosclerosis and/or hearing loss if said mutation is detected.
[0066] Another aspect includes a method for identifying a human
subject as having
or having an increased likelihood to develop a otosclerosis and/or hearing
loss, the method
comprising (a) obtaining a suitable sample from the subject; (b) assaying the
sample for the
presence or absence of a mutation in: i) a FOXL1 polynucleotide encoding an
amino acid
corresponding to position 326 to position 330 of SEQ ID NO:3 or ii) a FOXL1
polypeptide
encoded by said polynucleotide; and (c) identifying the subject as having or
having an
increased likelihood to develop otosclerosis and/or hearing loss if said
mutation is detected.
[0067] A further aspect includes a method of determining if a human
subject is a
carrier of a mutation that increases the likelihood of developing otosclerosis
and/or hearing
loss, the method comprising (a) obtaining a suitable sample from the subject;
(b) assaying
the sample for the presence or absence of a mutation in: i) a FOXL1
polynucleotide at a
nucleotide sequence encoding one or more of amino acid residues in FOXL1
polypeptide C-
terminus; ii) a FOXL1 polynucleotide at a nucleotide sequence encoding one or
more of
amino acid residues corresponding to positions 326 to 330 of SEQ ID NO:3; iii)
a FOXL1
polypeptide at one or more amino acid residues in FOXL1 polypeptide C-
terminus; or iv) a
FOXL1 polypeptide at one or more amino acid residues corresponding to
positions 326 to
330 of SEQ ID NO:3; and (c) identifying the subject as a carrier of a mutation
that increased
the likelihood of developing otosclerosis and/or hearing loss if said mutation
is detected.
[0068] In another aspect the method comprises: (a) obtaining a
suitable sample from
the subject; (b) assaying the sample for the level of a FOXL1 polypeptide
wherein an
increased level of is indicative the subject is a carrier of a mutation and/or
has or has an
increased likelihood of developing otosclerosis and/or hearing loss.
[0069] In an embodiment, the presence or absence of a mutation in a
FOXL1
polynucleotide is assessed by detecting deletion of the sequence in SEQ ID NO:
5. For
example SEQ ID NO: 1 comprises the 15 nucleotide sequence in SEQ ID NO: 5
whereas
SEQ ID NO:2 is deleted for this sequence. Detecting a FOXL1 polynucleotide
sequence
deleted for SEQ ID NO: 5 is indicative that the subject has or has an
increased likelihood to
develop otosclerosis and/or hearing loss.
[0070] Deafness research has demonstrated that there are many examples
where a
single deafness gene can cause several, distinct forms of hearing loss. For
example, a
13
CA 02913512 2015-11-25
WO 2014/190422
PCT/CA2014/000471
particular gene mutation can cause both syndromic and non-syndromic forms of
hearing
loss; both dominant and recessive patterns of inheritance in families; mild
and severe forms;
early and late onset; various patterns as detected by audiogram. The detected
deletion
mutation occurs in a highly conserved region of the gene and at the c-terminus
of the FOXL1
transcription factor. The c-terminus of transcription factors is typically
involved in binding to
genomic targets (other genes) in order to regulate downstream effects.
[0071] The suitable sample can be any sample comprising DNA containing
cells,
such as a saliva sample, blood sample or DNA accessible tissue sample. In an
embodiment,
said sample is a blood sample or fraction thereof. For example the blood can
be any fraction
such as a plasma fraction or a serum fraction. In an embodiment, the sample
comprises
leukocytes. In an embodiment, the sample comprises DNA and/or is a DNA sample.
For
protein and RNA transcript related applications, the sample comprises and/or
is derived from
cells that express FOXL1, for example bone cells and/or gastric tissue cells.
In yet another
embodiment, the sample comprises protein or is a protein fraction.
[0072] An aspect includes a method for detecting the presence of a
mutation
identified as c.976_990het_deIGGGAT000CTTCCTC in FOXL1 polynucleotide at a
nucleotide sequence encoding one or more of amino acid residues corresponding
to
positions 326 to 330 of SEQ ID NO:3, in a suitable sample, the method
comprising: a)
contacting the sample with a pair of primers, wherein the oligonucleotide
primers hybridize
under stringent conditions with a region flanking the mutation, under
conditions permitting
hybridization of the primers with the DNA contained in the sample; b)
amplifying the region of
interest of the FOXL1 gene or transcript; c) detecting the mutation in the
amplification
products.
[0073] In another aspect, the method comprises a method for detecting
the presence
or absence of a mutation identified as c.976_990het_deIGGGATCCCCITCCTC in
FOXL1
polynucleotide at a nucleotide sequence encoding one or more of amino acid
residues
corresponding to positions 326 to 330 of SEQ ID NO:3, in a suitable sample,
the method
comprising: a) contacting the sample with a probe, wherein the probe
hybridizes with a
FOXL1 gene or transcript comprising a c.976_990het_deIGGGATCCCOTTCCTC deletion
mutation, and does not hybridize with a wild-type FOXL1 probe, under
conditions permitting
hybridization of the probe with the DNA contained in the sample; and b)
detecting the
presence or absence of hybrids formed between the probe and the DNA contained
in the
sample, thereby indicating the presence or absence, respectively, of the
mutation.
14
CA 02913512 2015-11-25
WO 2014/190422
PCT/CA2014/000471
[0074] Either a FOXL1
polynucleotide or FOXL1 polypeptide can be assayed for said
mutation. Methods are known for obtaining a sample from a subject that can be
assayed for
a mutated FOXL1 polynucleotide (e.g. DNA or RNA) or a mutated FOXL1
polypeptide (e.g.
protein fraction). In an embodiment the subject is pre-symptomatic. In another
embodiment,
the subject exhibits or is suspected of having hearing loss. In another
embodiment, the
subject has one or more blood relative carriers of the
c.976_990het_deIGGGATCCCCTTCCTC:p.Gly326_Leu330del mutation.
[0075] In an
embodiment, said sample or DNA or RNA isolated therefrom or DNA
amplified therefrom, is assayed for the presence or absence of said mutation
in said FOXL1
polynucleotide.
[0076] In an embodiment,
the presence or absence of said mutation can be
determined by assessing in the FOXL1 polynucleotide comprises a mutation in
one or more
of the codon that encodes glycine 326 to the codon that encodes leucine 330 of
SEQ ID
NO:3. The nucleotides that encode G326, 1327, P328, F329 and L330 are
nucleotides 976-
990of SEQ ID NO:1 (e.g. GGGATCCCCTTCCTC SEQ ID NO:5).
[0077] Said mutation in
said polynucleotide can correspond to deletion of one or
more nucleotides that encodes glycine, isoleucine, proline, phenylalanine
and/or leucine at
position 326,327,328,329 and/or 330 of SEQ ID NO:3. For example, as shown
here, the
mutation can be deletion of the nucleotides corresponding to positions 976 to
position 990 of
SEQ ID NO:1.
[0078] In an embodiment,
said mutation in said FOXL1 polynucleotide comprises
mutation of the nucleotides corresponding to position 976 to position 990 of
SEQ ID NO:1
(e.g. mutation of nucleotides in SEQ ID NO:5).
[0079] In an
embodiment, said mutation in said FOXI1 polynucleotide is
976_990het_deIGGGATCCCCTTCCTC. The wildtype sequence of FOXL1 transcript is
provided is SEQ ID NO:1 and comprises three guanine (G) at position 976, 977,
and 978,
adenine at position 979, thymine (T) at position 980, four cytosine ( C) at
position 981- 984,
two thymine (T) at position (985-986), two cytosine ( C) at position (987-
988), thymine (T) at
position 989 and thymine (T) at position 990. The corresponding polypeptide
sequence is
provided in SEQ ID NO: 3 and comprises glycine, isoleucine, proline,
phenylalanine and
leucine at position
326,327,328,329 and 330. The mutated polynucleotide sequence FOXL1
which is deleted for GGGATCCCCTTCCTC (SEQ ID NO:5) corresponding to position
976 to
position 990 as found in SEQ ID NO:1, is provided in SEQ ID NO:2. Deletion of
the
GGGATCCCCTTCCTC (SEQ ID NO:5) at position 976 to position 990 causes an in
frame
CA 02913512 2015-11-25
WO 2014/190422
PCT/CA2014/000471
deletion which is reflected in the mutant polypeptide sequence provided in SEQ
ID NO:4.
SEQ ID NO: 3 comprises a glycine, isoleucine, proline, phenylalanine and
leucine at position
326,327,328,329 and 330 consecutively and is deleted in SEQ ID NO: 4 in
comparison to
the wildtype sequence in SEQ ID NO:3.
[0080] Methods for detecting a mutation in a polynucleotide or nucleic
acid alteration
within a sample, include genotyping, microarrays, Restriction Fragment Length
Polymorphism, Southern Blots, single-strand conformation polymorphism (SSCP),
dHPLC,
single nucleotide primer extension, allele-specific hybridization, allele-
specific primer
extension, oligonucleotide ligation assay, and invasive signal amplification,
Matrix-assisted
laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry, and
Fluorescence
polarization (FP). Such methods optionally employ the isolated nucleic acid
primers, probes
compounds and compositions of the disclosure described herein.
[0081] Some methods employ polymerase chain reaction (PCR), real time
PCR,
multiplex ligation dependent probe amplification (MLPA), nucleic acid sequence
based
amplification (NASBA) and/or real time NASBA.
[0082] As an example, one assay strategy comprises gene amplification
analysis
such as polymerase chain reaction (PCR) analysis, optionally followed by
sequencing, and
comparing the amplification profile or identified sequence to a wild-type
amplification profile
or wild-type FOXL1 sequence as found for example in SEQ ID NO:1. Methods of
sequencing
are well known in the art. In an embodiment, one or more FOXI1 exons are
amplified by
PCR, and analyzed for gene mutations, for example by SSCP.
[0083] In an embodiment, DNA or RNA is isolated from and/or DNA is
amplified said
sample and the DNA or RNA is assayed for the presence or absence of said
mutation.
[0084] In an embodiment, the mutation in FOXL1 polynucleotide is
detected by a
FOR based method and/or a hybridization based method.
[0085] In an embodiment, said DNA is genomic DNA.
[0086] In another embodiment, assaying said sample for the presence or
absence of
the mutation in FOXL1 corresponding to positions 976-990 comprises: a)
amplifying a
portion of a FOXL1 coding sequence, the portion spanning minimally FOXL1
residues
corresponding to position 976 to position 990 of SEQ ID NO:1, to produce
amplified FOXL1
polynucleotide, b) sequencing the amplified FOXL1 polynucleotide and
determining if said
mutation is present.
16
CA 02913512 2015-11-25
WO 2014/190422
PCT/CA2014/000471
[0087] In an embodiment, the portion comprises at least 40 nucleotides, at
least 50
nucleotides, at least 60 nucleotides, at least 70 nucleotides, at least 80
nucleotides, at least
90 nucleotides, at least 100 nucleotides, at least 110 nucleotides, at least
120 nucleotides, at
least 130 nucleotides, at least 140 nucleotides, at least 150 nucleotides, at
least 160
nucleotides, at least 170 nucleotides, at least 1800 nucleotides, at least 200
nucleotides, at
least 220 nucleotides, at least 240 nucleotides, at least 260 nucleotides, at
least 280
nucleotides, at least 300 nucleotides, or at least 320 nucleotides.
[0088] In
another embodiment, mRNA is isolated from said sample, for example a
bone cell sample or gastric cell sample, and the mRNA is assayed directly for
the presence
or absence of said mutation and/or cDNA is prepared using said mRNA and the
cDNA is
assayed for the presence or absence of said mutation.
[0089] In an
embodiment, the assay comprises: a) isolating mRNA from said sample;
b) deriving corresponding DNA (cDNA) from said isolated mRNA; c) amplifying
all or a
portion of FOXL1 coding sequence, the portion spanning minimally FOXL1
nucleotide
residues corresponding to position 976 to position 990 of SEQ ID NO:1 to
produce amplified
FOXL1 polynucleotide, d) sequencing the amplified FOXL1 polynucleotide; e)
determining if
said mutation is present; and f) identifying the subject as having or having
an increased
likelihood to develop otosclerosis and/or hearing loss if said mutation is
detected.
[0090] The
amplification step can comprise use of one or more primers described
herein. For example, FOXL1 specific primers flanking said FOXL1 mutation are
selected, for
example primers which amplify all or part of FOXL1 coding sequence either in
genomic
sequence and/or corresponding transcripts, are used to amplify the gene region
comprising
said FOXL1 mutation.
[0091] The
amplified portion can comprise for example a 490 bp fragment as
described below. The amplified portion can be about or at least 50
nucleotides, about or at
least 100 nucleotides, about or at least 150 nucleotides, about or at least
200 nucleotides,
about or at least 250 nucleotides, about or at least 300 nucleotides, about or
at least 350
nucleotides, about or at least 400 nucleotides, or about or at least 450
nucleotides or any
whole number between for example 15 nucleotides and the full length sequence.
[0092]
Instead of sequencing the amplified FOXL1 polynucleotide, in certain
embodiments, the mutation is detected using a probe that hybridizes to the
mutated FOXL1
polynucleotide. In an embodiment, the probe comprises a probe described
herein. For
example, a probe specific for the mutant FOXL1 polynucleotide can comprise for
example
residues complementary to 5' and 3' sequence flanking the deletion sequence,
for example
17
CA 02913512 2015-11-25
WO 2014/190422
PCT/CA2014/000471
comprising at least 8 contiguous nucleotides of SEQ ID NO: 2 , the at least 8
contiguous
nucleotides including residues corresponding to positions 975 and 976. For
example the
probe specific for detecting mutant FOXL1 can comprise residues 972, 973, 974,
975, 976,
977, 978 and 979 of SEQ ID NO:2.
[0093] The probe can also be specific for the wildtype FOXL1 sequence.
A probe
specific for the wildtype FOXL1 comprises for example all or part of the
sequence of SEQ ID
NO: 5. In an embodiment, detecting mutatnt FOXL1 comprises using a wildtype
FOXL1
probe, wherein hybridization identifies the subject as not having or not
having an increased
likelihood of developing otosclerosis and/or hearing loss and lack of
hybridization identifies
the subject as having or having an increased likelihood of developing
otosclerosis.
[0094] In an embodiment, the method comprises assaying for the presence
or
absence of mutant FOXL1 and assaying for the presence or absence of wildtype
FOXL1.
[0095] In an embodiment, assaying the sample for the presence or
absence of a
mutation in a FOXL1 polynucleotide encoding an amino acid in FOXL1 polypeptide
C-
terminus comprises:
1. a) contacting the sample with a pair of primers, wherein the
oligonucleotide primers
hybridize under stringent conditions with a region flanking the C-terminus,
optionally wherein
an upstream primer binds upstream of the DNA binding forkhead domain and a
downstream
primer binds in the 3' UTR, under conditions permitting hybridization of the
primers with the
DNA contained in the sample; b) amplifying the region of interest of the FOXL1
gene or
transcript; c) detecting the mutation in the amplification products; or
2. a) contacting the sample with a Probe, wherein the probe hybridizes with a
FOXL1 gene or transcript comprising a C-terminus deletion mutation, and does
not hybridize
with a wild-type FOXL1 probe, under conditions permitting hybridization of the
probe with the
DNA or RNA contained in the sample; and b) detecting the presence or absence
of hybrids
formed between the probe and the DNA or RNA contained in the sample, thereby
indicating
the presence or absence, respectively, of the mutation.
[0096] In an embodiment, the C-terminus end is the 70, 60, 50, 40 or
30 C-terminal
coding amino acids and/or the corresponding nucleotides encoding said coding
region.
[0097] FOXL1 polypeptide can also be assayed for said mutation. A
sample can also
be assayed for the level of FOXL1 polypeptide. As demonstrated herein, the
level of FOXL1
protein is increased. In an embodiment, the mRNA level is not increased.
18
CA 02913512 2015-11-25
WO 2014/190422
PCT/CA2014/000471
[0098] Accordingly in an embodiment, a sample from the subject, for example
a
bone sample, and/or a protein fraction derived therefrom, is assayed for the
presence or
absence of said mutation in said FOXL1 polypeptide and/or the level of FOXL1
polypeptide.
[0099] In an embodiment, assaying the sample for the presence or
absence of a
mutation in a FOXL1 polynucleotide encoding an amino acid corresponding to
position 326
to position 330 of SEQ ID NO:3 comprises:
1. a) contacting the sample with a pair of primers, wherein the
oligonucleotide
primers hybridize under stringent conditions with a region flanking the
mutation, under
conditions permitting hybridization of the primers with the DNA contained in
the sample; b)
amplifying the region of interest of the FOXL1 gene or transcript; c)
detecting the mutation in
the amplification products; or
2. a) contacting the sample with a probe, wherein the probe hybridizes with a
FOXL1 gene or transcript comprising a c.976_990het_deIGGGAT000CTTCCTC deletion
mutation, and does not hybridize with a wild-type FOXL1 probe, under
conditions permitting
hybridization of the probe with the DNA or RNA contained in the sample; and b)
detecting
the presence or absence of hybrids formed between the probe and the DNA or RNA
contained in the sample, thereby indicating the presence or absence,
respectively, of the
mutation.
[00100] In an embodiment, the method comprises assaying for a level of
FOXL1
expression, optionally mRNA expression or protein expression, wherein an
increased level
of FOXL1 expression compared to a control is indicative that the subject is a
carrier of a
mutation and/or has or has an increased likelihood of developing otosclerosis
and/or hearing
loss.
[00101] In another embodiment said mutation in said FOXL1 polypeptide
is
p.G1y326_Leu330del.
[00102] Methods that can be used to assess if mutated FOXL1 is in the
sample. As
GGGATCCCCTTCCTC corresponding to position 976 to position 990 in SEQ ID NO:1
results in an encoded polypeptide that is missing five amino acids, analysis
of FOXL1
polypeptide size can indicate whether the subject has a mutated FOXL1
polypeptide. For
example, an antibody that binds specifically to the sequence in SEQ ID NO: 6
detects wild
type FOXL1, and an antibody that is specific for an epitope comprising
contiguous amino
acids 325-327 of SEQ ID NO: 4 is specific for the mutant FOXL1.
19
CA 02913512 2015-11-25
WO 2014/190422
PCT/CA2014/000471
[00103] In an embodiment, assaying the sample for the presence or absence
of
mutated FOXL1, e.g., assaying the sample for the presence or absence a
mutation in a
FOXL1 polypeptide at position 326 to position 330 of SEQ ID NO:3 comprises
contacting the
sample obtained from the subject with an antibody having specific binding
affinity for FOXL1
deleted for amino acids 326, 327, 328, 329 and/or 330, thereby forming a
complex of the
antibody and FOXL1 deleted for amino acids 326, 327, 328, 329 and/or 330.
[00104] In an embodiment, the method comprises assaying the sample for
the level of
FOXL1 compared to a control, wherein an increased protein level compared to a
control is
indicative the subject is a carrier of a mutation and/or has or has an
increased likelihood of
developing otosclerosis and/or hearing loss.
[00105] In an embodiment, the antibody is conjugated to and/or comprises a
heterologous moiety. In an embodiment, the antibody is conjugated to and/or
comprises a
detectable label. In another embodiment, the detectable signal is detectable
indirectly, for
example, using a secondary antibody. In an embodiment, the secondary antibody
is
conjugated to and/or comprises a detectable label.
[00106] The detectable label is preferably capable of producing, either
directly or
indirectly, a detectable signal. For example, the label may be radio-opaque or
a radioisotope,
such as 3H, 140, 32p, 35s, 1231, 1251 , 1311; a fluorescent (fluorophore) or
chemiluminescent
(chromophore) compound, such as fluorescein isothiocyanate, rhodamine or
luciferin; an
enzymatic label, such as alkaline phosphatase, beta-galactosidase or
peroxidase such as
horseradish peroxidase; an imaging agent; or a metal ion. In an embodiment,
the detectable
label is a hapten label such as dintrophenol, biotin or digoxigenin.
[00107] In an embodiment, the antibody contacted with the sample can be
complexed
to a solid support. In an embodiment the antibody FOXL1 complex is coupled to
a solid
support prior to separating.
[00108] In an embodiment, the method comprises separating the complex
formed
from antibody not complexed of the mutated FOXL1; detecting or quantifying a
signal from
the detectable label of the antibody, optionally the signal being proportional
to an amount of
mutated FOXL1.
[00109]
Detecting a signal proportional to the amount of mutated FOXL1 identifies the
subject as having or having an increased likelihood to develop otosclerosis
and/or hearing
loss.
CA 02913512 2015-11-25
WO 2014/190422
PCT/CA2014/000471
[00110] In an
embodiment, the method comprises detecting wildtype FOXL1
polypeptide, wherein the detection of wildtype polypeptide identifies the
subject has not
having an increased likelihood to develop otosclerosis and/or hearing loss
(e.g. mutated
FOXL1 related otosclerosis and/or hearing loss.
[00111]
Detection of mutant FOXL1 is achieved in antibody based binding methods
and/or hybridization based methods when for example the signal measured is at
least 2X, at
least 3X at least 4X assay background levels (e.g. in an assay control where
binding is not
expected). A positive control (e.g. assay test known to comprise mutant FOXL1
polypeptide,
wildtype FOXL1 polypeptide, mutant FOXL1 polynucleotide or wildtype FOXL1
polynucleotide, assayed with a mutant detecting probe or antibody (e.g. for
mutant
molecules) or assayed with a wildtype detecting probe or antibody (e.g. for
wildtype
molecules).
[00112] In an
embodiment, the detectable label comprises a peroxidase conjugate or
digoxigenin.
[00113] In an
embodiment, the antibody is coupled to a solid support and/or
conjugated to a component configured for coupling to a solid support.
[00114] In an
embodiment, the assaying comprises contacting the sample obtained
from the subject with a capture antibody having specific binding affinity for
mutated FOXL1,
thereby forming a complex of the capture antibody and the mutated FOXL1, the
capture
antibody being coupled to a solid support or comprising a component configured
for coupling
the capture antibody to a solid support; contacting the sample with a
detection antibody
having specific binding affinity for mutated FOXL1, thereby forming a complex
of the capture
antibody, mutated FOXL1, and the detection antibody, the detection antibody
having a
detectable label; contacting the complex of the capture antibody, mutated
FOXLI and the
detection antibody with a solid support, whereby the complex of the capture
antibody,
mutated and the detection antibody couples to the solid support; separating
the complex of
the capture antibody, mutated FOXL1 and the detection antibody coupled to the
solid
support from detection antibody, capture antibody and mutated FOXL1 not
coupled to the
solid support; exposing the complex of the capture antibody, mutated FOXL1 and
the
detection antibody to a substrate, thereby producing a signal from the
detectable label of the
detection antibody comprising the complex of the capture antibody, FOXL1 and
the detection
antibody which couples to the solid support in said step of contacting;
detecting or
quantifying the signal produced in said step of exposing the complex of the
capture antibody,
21
CA 02913512 2015-11-25
WO 2014/190422
PCT/CA2014/000471
FOXL1 and the detection antibody, the signal being proportional to an amount
of FOXL1 in
the sample from the subject.
[00116] In an embodiment, the second epitope is different than the
first epitope.
In an embodiment, the step of contacting the sample with the capture antibody
and the step
of contacting the sample with the detection antibody occur at substantially
the same time.
[00116] In an embodiment, the solid support is a mircrowell.
[00117] Further, in some embodiments, one or more mutations in FOXL1 in
addition
to a mutation in p.G1y326_Leu330 (and/or the corresponding GGGATCCCCTTCCTC
deletion) are assessed.
[00118] The control is for example a suitable sample from a subject
without
otosclerosis and/or hearing loss.
[00119] In an embodiment, the assay is conducted as part of a multiplex
assay. In an
embodiment, the method further comprises audiological testing, optionally
measuring
conductive and/or sensorineural hearing, and/or surgical diagnosis. In an
embodiment, a
subject identified with a FOXL1 mutation and decreased conductive and/or
sensorineural
hearing is identified as having otosclerosis.
[00120] Surgery and hearing aids are the typical treatments for
otosclerosis.
Otosclerosis is heterogeneous and can involve both conductive and
sensorineural
components. As hearing loss progresses, and particularly with significant
sensorineural
components, hearing aids may be less effective and in these cases cochlear
implantation
requiring surgery may be necessary.
[00121] A further aspect includes a method of treating a subject, the
method
comprising determining if the subject is a carrier
of
c.976_990het_deIGGGATCCCCTTCCTC: p.Gly326_Leu330del for example in FOXL1
polynucleotide and/or polypeptide, for example a mutation corresponding to
positions 326 to
position 330 of SEQ ID NO: 3 optionally p.G1y326_Leu330 del FOXL1 as described
herein;
and providing subjects determined to be carriers of the mutation, optionally
p.G1y326_Leu330 del FOXL1, with a hearing loss therapy.
[00122] In an embodiment, the hearing loss therapy comprises providing
the subject
with a suitable hearing aid, FM hearing system or cochlear implant.
III. Products and Kits
22
CA 02913512 2015-11-25
WO 2014/190422
PCT/CA2014/000471
[00123] Also provided
isolated nucleic acids, isolated polypeptides, nucleic acid
primers and probes, antibodies, compounds, compositions as well as kits
comprising said
products which can for example be used in a methods described herein.
[00124] Accordingly in
another aspect is provided an isolated polynucleotide encoding
at least 5 and a maximum 50, 60, 70 or 80 contiguous amino acids of SEQ ID
NO:3, 4 or 6,
the at least 5 contiguous amino acids comprising the amino acid residues of
SEQ ID NO:6
and/or excluding the amino acid residues in SEQ ID NO: 6. In an embodiment,
the
polynucleotide comprises at least 15, at least 20, at least 25, at least 30,
at least 35, at least
40, at least 45, or at least 50 nucleotides of SEQ ID NO: 5, 1 or 2. In an
embodiment, the
polynucleotide comprises the nucleotides of SEQ ID NO: 5, 1 or 2.
[00125] A further
aspect includes an isolated polypeptide comprising at least 5 and a
maximum 50, 60, 70 or 80 contiguous amino acids of SEQ ID NO:3, 4 or 6, the at
least 5
contiguous amino acids comprising the amino acid corresponding to position 326-
330 of
SEQ ID NO:3 or comprising the amino acids of SEQ ID NO:6. and/or excluding the
amino
acid residues in SEQ ID NO: 6. In an embodiment, the polypeptide comprises at
least 5, at
least 10, at least 15, at least 20, at least 25, at least 30, at least 35, at
least 40, at least 45 or
at lest 50 amino acids of SEQ ID NO: 3, 4 or 6.
[00126] In an
embodiment, the isolated polynucleotide encodes and/or the isolated
polypeptide comprises at least 5, at least 10, at least 12, at least 15 or at
least 20 amino
acids.
[00127] A further
aspect includes a compound comprising a polynucleotide encoding
at least 5 and a maximum 50, 60, 70 or 80 contiguous amino acids of SEQ ID
NO:3, 4 or 5,
the at least 5 contiguous amino acids comprising the amino acid corresponding
to residue
326-330 of SEQ ID NO: 3and/or excluding the nucleotides encoding the amino
acids in SEQ
ID NO: 6. In an embodiment, the compound comprises a polynucleotide comprising
at least
15, at least 20, at least 25, at least 30, at least 35, at least 40, at least
45, or at least 50
nucleotides of SEQ ID NO: 5, 1 or 2. In an embodiment, the polynucleotide
comprises the
nucleotides of SEQ ID NO: 5, 1 01 2.
[00128] In an embodiment, the FOXL1 polynucleotide encoding the at
least 5 and a
maximum of 50, 60 , 70 or 80 contiguous amino acids comprises least 15
nucleotides of
SEQ ID NO: 1, 2 or 5, optionally said at least 15 nucleotides comprising the
nucleotide
corresponding to position 976 to 990 of SEQ ID NO:1. GGGATCCCCTTCCTC.
23
CA 02913512 2015-11-25
WO 2014/190422
PCT/CA2014/000471
[00129] Yet another aspect includes a compound comprising a polypeptide
comprising at least 5 and a maximum 50, 60, 70 or 80 contiguous amino acids of
SEQ ID
NO: 3, 4 or 6, optionally the at least 5 contiguous amino acids comprising the
amino acid
corresponding to position 326 to position 330 of SEQ ID NO:3 .
[00130] In an embodiment, the polynucleotide encodes and/or the
polypeptide
comprises at least 5, at least 10 at least 12, at least 15 or at least 20
amino acids.
[00131] In a further embodiment, the isolated polynucleotide or the
isolated
polypeptide is conjugated to and/or the compound comprises a heterologous
moiety. In an
embodiment, the isolated polynucleotide or the isolated polypeptide is
conjugated and/or the
compound comprises a detectable label.
[00132] In an embodiment, the compound comprises a detectable label. For
example,
the label can be conjugated to a polynucleotide, primer, probe, polypeptide,
antibody, or
compound described herein.
[00133] Also provided in another aspect is a nucleic acid primer for
diagnosing
otosclerosis and/or hearing loss which hybridizes under stringent conditions
to a portion of
the nucleic acid sequence of SEQ ID NO: 1 or complement thereof, wherein the
primer
amplifies all or a portion of the coding sequence of FOXL1 such that one or
more
nucleotides encoding an amino acid corresponding to position 326 to position
330 in SEQ ID
NO: 3 is amplified.
[00134] The amplified portion can comprise for example a 490 bp
fragment as
described below. The amplified portion can be about or at least 50
nucleotides, about or at
least 100 nucleotides, about or at least 150 nucleotides, about or at least
200 nucleotides,
about or at least 250 nucleotides, about or at least 300 nucleotides, about or
at least 350
nucleotides, about or at least 400 nucleotides, or about or at least 450
nucleotides or any
whole number between for example 15 nucleotides and the full length sequence.
[00135] In an embodiment, the primer is at least 12 nucleotides in length,
for example
about 15 to 30 nucleotides, or any whole number in between.
[00136] In an embodiment, the primer comprises at least 12 consecutive
nucleotides
of SEQ ID NO: 7 or 8. In another embodiment, the primer consists of at least
12 consecutive
nucleotides of SEQ ID NO: 7 or 8.
[00137] In an embodiment, the primer is conjugated to and/or comprises a
heterologous moiety, optionally a detectable label.
24
CA 02913512 2015-11-25
WO 2014/190422
PCT/CA2014/000471
[00138] A further aspect includes a nucleic acid probe for diagnosing
otosclerosis
hearing disorder which hybridizes under stringent conditions to a portion of
the complement
of SEQ ID NO: 1 or 2 comprising residues encoding amino acid corresponding to
position
326 to position 330 of SEQ ID NO: 3 or a nucleic acid sequence that differs
from the SEQ
ID NO: 1 at one or more nucleotides encoding arginine at residue 326 to
position 330 of
SEQ ID NO:3, and which detects the presence or absence of a substitution or
lesion at one
or more nucleotides that encode the Glycine, lsoleucine, Proline,
phenylalanine and Leucine
at position 326,327,328,329 and 330 consequence of SEQ ID NO:3.
[00139] In an embodiment, the probe detects a substitution or lesion at
a nucleotide
corresponding to position 976 to position 990 of SEQ ID NO: 1.
[00140] In a further embodiment, the probe is at least 12 nucleotides in
length. In an
embodiment, the probe is a maximum of 50, 100 or 150 nucleotides in length. A
probe for
detecting the mutant FOXL1 polynucleotide can comprise none of the deleted
sequence. A
wildtype probe for example comprising at least 12 nucleotides of SEQ ID NO: 5,
can be used
to detect the wildtype sequence. Accordingly another aspect includes a method
using both a
probe specific for the deletion mutant and a probe specific for the wild type
allele.
[00141] In yet a further embodiment, the probe comprises at least 12
consecutive
nucleotides of SEQ ID NO:1 or 2.
[00142] In another embodiment, the probe hybridizes to or hybrids to
the complement
of at least 50, at least 100 or at least 150 consecutive nucleotides of SEQ ID
NO: 1 or 2.
[00143] In yet another embodiment, the probe comprises and/or is conjugated
to a
detectable label.
[00144] In an embodiment, the isolated polynucleotide, compound,
nucleic acid
primer, nucleic acid probe, is comprised in a composition with a suitable
diluent or carrier.
[00145] In an embodiment the diluent is a saline such as PBS, water,
such as sterile
water, or hybridization solution.
[00146] A further aspect is an immunogen comprising SEQ ID NO: 6. A
further aspect
is an immunogen comprising contiguous amino acid residues 326-327 or 325-328
of SEQ ID
NO: 4. The immunogen can comprise a tag such as KLH to improve immunogenicity,
can be
administered for example with an adjuvant. An antibody specific for SEQ ID NO:
6 detect
wild-type FOXL1. An antibody specific for amino acid residues 326-327 or 325-
328 of SEQ
ID NO: 4 would be predicted to recognize mutant FOXL1.
CA 02913512 2015-11-25
WO 2014/190422
PCT/CA2014/000471
[00147] Another aspect is an isolated antibody specific for SEQ ID N0:6 or
specific for
an epitope comprising amino acid residues 326-327 or 325-328 of SEQ ID NO: 4.
The
antibody specific for said epitope comprising amino acid residues 326-327 or
325-328 binds
said epitope with at least 2 fold, 3 fold or 4 fold specificity over SEQ ID
NO:6 and/or 3.
[00148] A further aspect includes a kit for diagnosing a subject at
risk for a
otosclerosis hearing loss, comprising: at least two nucleic acid primers which
hybridize under
stringent conditions to a nucleic acid sequence of SEQ ID NO: 1 or complement
thereof,
wherein the primers amplify all or a portion of SEQ ID NO: 1 or 2, such that
minimally one or
more nucleotides encoding an amino acid residue corresponding to position 326
to position
330 of SEQ ID NO:3 is amplified; and instructions for diagnosing a
otosclerosis hearing loss
by detecting a substitution or deletion of one or more nucleotides encoding
amino acid
residues at 326 to position 330.
[00149] In an embodiment, the kit comprises a primer that is described
herein.
[00150] In an embodiment, the kit further comprises a nucleic acid
probe which
hybridizes under stringent conditions to the complement of SEQ ID NO: 2 at one
or more
nucleotides encoding Glycine, Isoleucineõ Proline, phenylalanine and Leucine
at residue
326 to position 330 of SEQ ID NO:3 and which detects the presence or absence
of a
substitution or lesion at one or more nucleotides that encode the Glycine,
Isoleucineõ
Proline, phenylalanine and Leucine at residues 326 to position 330 of SEQ ID
NO:3.
[00151] In an embodiment, the kit comprises a probe that is described
herein.
[00152] In an embodiment, the kit comprises more than one probe.
[00153] In a further embodiment, one or more of the probes is
conjugated to and/or
comprises a heterologous moiety optionally a detectable label. In an
embodiment, one or
more of the probes is a labeled probe.
[00154] The kit can also comprise in an embodiment, one or more primers
or probes
for detecting additional mutations in FOXL1,
[00155] In an embodiment, the kit comprises a probe that comprises at
least 12
nucleotides in length.
[00156] In yet another embodiment, the kit comprises a probe that
comprises at least
12 consecutive nucleotides of SEQ ID NO:1 or 2.
[00157] In a further embodiment, the kit comprises a probe that comprises
at least 50
consecutive nucleotides of SEQ ID NO: 1 or 2.
26
CA 02913512 2015-11-25
WO 2014/190422
PCT/CA2014/000471
[00158] In an embodiment, the kit comprises a primer that is at least 12
nucleotides in
length.
[00159] In yet another embodiment, the kit comprises a primer that
comprises at least
12 consecutive nucleotides of SEQ ID NO: 7 or 8. In an embodiment, the kit
comprises a
primer wherein the primer is labeled.
[00160] In an embodiment, the kit comprises one or more of 10X Buffer,
dNTPs,
MgC12 and a DNA polymerase such as Taq polymerase.
[00161] In certain embodiments, the kit is a diagnostic kit for medical
use. In other
embodiments, the kit is a diagnostic kit for laboratory use.
[00162] In another aspect the disclosure provides a commercial package
comprising
an isolated nucleic acid or composition described herein and instructions for
use.
[00163] In an embodiment, the kit comprises a microarray, for example a
DNA
microarray such as a gene chip, or a tissue array.
[00164] For example, in an embodiment the tissue microarray comprises a
slide
comprising an array of arrayed biopsy samples in paraffin. Tissue microarray
technology
optionally allows multiple specimens, such as biopsy samples, to be analyzed
in a single
analysis at the DNA, RNA or protein level. Tissue microarrays are analyzed by
a number of
techniques including immunohistochemistry, in situ hybridization, in situ PCR,
RNA or DNA
expression analysis and and/or morphological and clinical characterization or
a combination
of techniques. The specimens are optionally from the same subject or from a
plurality of
subjects. Methods of detecting gene expression using arrays are well known in
the art. Such
methods are optionally automated. In an embodiment, the sample from the
subject is
analyzed using a tissue microarray.
[00165] In an embodiment the polypeptide, optionally the isolated
polypeptide,
polynucleotide optionally isolated polynucleotide, primer, probe, antibody or
antibody
fragment is conjugated to and/or comprises a detectable label.
[00166] The detectable label is preferably capable of producing, either
directly or
indirectly, a detectable signal. For example, the label may be radio-opaque or
a radioisotope,
such as 3H, 140, 32p, 35s, 1231, 1251, 1311; a fluorescent (fluorophore) or
chemiluminescent
(chromophore) compound, such as fluorescein isothiocyanate, rhodamine or
luciferin; an
enzyme, such as alkaline phosphatase, beta-galactosidase or horseradish
peroxidase; an
imaging agent; or a metal ion.
27
CA 02913512 2015-11-25
WO 2014/190422
PCT/CA2014/000471
[00167] In another embodiment, the detectable signal is detectable
indirectly, for
example, using a secondary antibody.
[00168] The above disclosure generally describes the present
application. A more
complete understanding can be obtained by reference to the following specific
examples.
These examples are described solely for the purpose of illustration and are
not intended to
limit the scope of the application. Changes in form and substitution of
equivalents are
contemplated as circumstances might suggest or render expedient. Although
specific terms
have been employed herein, such terms are intended in a descriptive sense and
not for
purposes of limitation.
[00169] The following non-limiting examples are illustrative of the
present disclosure:
Examples
Example 1
[00170] Otosclerosis hearing loss is a common form of hearing loss
contributing to
0.3-1% of all cases worldwide. We identified a multiplex family from
Newfoundland (Family
2081) segregating otosclerosis inherited as an autosomal dominant trait.
Haplotyping of the
9Mb region at chr16q23.1-q24.2 and sequencing positional candidate genes
revealed a 15
bp deletion (c.976_990het_deIGGGATCCCCTTCCTC) in the FOXL1 gene which encodes
a
Forkhead protein. This novel deletion causes an in frame deletion of five
amino acids:
glycine, isoleucine, proline, phenylalanine and leucine (p.Gly326_Leu330). To
investigate
the role of FOXL1 in other patients, FOXL1 was sequenced in 17 probands from
NL with
otosclerosis but did not identify a mutation in FOXL1. Further screening of
otosclerosis
families for this mutation will help in identifying other families with the
mutation.
Example 2
[00171] A multiplex, extended pedigree (Family 2081) segregating non-
syndromic
otosclerosis hearing loss from the founder population of Newfoundland, Canada
(Figure 1)
was identified. Family 2081 is a multiplex family spanning 4 generations and
includes 36
available family members. Otosclerosis in this family appears to segregate in
an autosomal
dominant pattern, both parents of the proband have a medical history of
hearing loss
(subject 1-3 and 1-4 ) (Figure 1). The diagnosis of otosclerosis in this
family is based on; the
surgical report of otosclerosis in seven affected member (subjects 11-4, 11-5,
11-8, 11-11, 11-13,
III-1) and the audiological profile of conductive hearing loss with absence of
stapedial reflex
and low compliance of the tympanic membrane in subject III-5. The hearing loss
in the all
affected family members starts around the age of 12 years as conductive
hearing loss in six
out of seven affected (subjects 11-4, 11-8, 11-11, 11-13, 111-1, 111-5) and as
a pure SNHL in the
28
CA 02913512 2015-11-25
WO 2014/190422
PCT/CA2014/000471
seventh member (11-5) (Figure 2). Genomic DNA was isolated from peripheral
leukocytes of
7 affecteds (11-4, 11-5, 11-8, 11-12, 11-13, 111-1, 111-5) and also from four
relatives with no history
of hearing loss (11-6, 11-7.111-2,111-6, 111-10 and 111-12) and three
relatives with hearing loss
uncategorized (11-9,11-14, 11-15) as previously described (Miller, et al.,
1988). Ethics approval
was obtained (Human Investigation Committee #01.186). Genotyping the 9Mb
region at
chr16q23.1-q24.2 was performed on all available DNA samples spanning two
generations.
Six microsatellite markers were amplified and run on an ABI 3130x1 or 3730 DNA
Analyzer
and analyzed using GeneMapper v4.0 (Applied Biosystems). Haplotype analysis
revealed
that all affected members (11-4, 11-5, 11-8, 11-12, 11-13, 111-1, 111-5)
inherited the green
(presumably disease) haplotype (Figure 3). This 9Mb contains 79 positional
candidate
genes. Thirteen positional candidate genes (Table 1) were sequenced based on a
biological
role in bone remodeling. All sequencing variants were tested for co-
segregation with the
otosclerosis phenotype and a 15 bp deletion in
FOXL1
(c.976_990het_deIGGGATCCCCTTCCTC) (SEQ ID NO: 5) was shown to segregate with
the disease haplotype. This in frame deletion in the coding region of FOXL1 is
absent in 116
chromosomes tested from NL population controls.
[00172] In
order to evaluate the effect of FOXL1 deletion on the nnRNA expression,
RNA was extracted from lymphoblastoid cell line of 4 deletion carriers (11-4,
11-5, 11-8 and II-
12) and compared to controls (11-1 and TA09). No amplification from the
lymphoblastoid cell
line RNA extract using RT-PCR analysis was detected, which was further
confirmed by real
time PCR. These data suggested that FOXL1 is normally not expressed in
lymphocyte
(Figure 7).
[00173] To
determine the effect of the FOXL1 deletion on the mRNA level, HEK293A
cells were transiently transfected with FOXL1 (both wild and-mutant forms).
RNAs were
isolated and the level of FOXL1 expression compared between the transfectants
by RT-
PCR. No significant difference in the level of FOXL1 expression between HEK-
FOXL1 wild
type and HEK-FOXL1-mutant was detected (Figure 8).
[00174] The
localization and expression of FOXL1 at the protein level were next
examined in HEK293-FOXL1-wildtype and HEK293 FOXL1-mutant cells to explore the
possibility of protein degradation as a result of post-transcription or post-
translational
modification. Nuclear and cytoplasmic extracts were prepared from HEK-23-FOXL1
wild-
type cells and HEK-293-FOXL1-mutantcells. FOXL1 expression was determined
using Anti-
FOXL1 rabbit polyclonal IgG and Anti-GFP mouse IgG for the FOXL1-GFP tagged
transcripts. No significant difference was detected in the FOXL1 protein level
between the
29
CA 02913512 2015-11-25
WO 2014/190422
PCT/CA2014/000471
mutant FOXL1 and wild type in the cytoplasm. However, the mutant FOXL1 did
show an up
regulation in the nuclear extract (Figure 9).
[00175]
Functional studies showed that the FOXL1 deletion up regulated the level of
the FOXL1 protein. To confirm the result from HEK293A and exclude any other
confounding
factor that related to type of cell line (hFOB 1.19), Human fetal osteoblast
femur HEK293A
cells were transiently transfected with FOXL1 wild and-mutant forms. No
significant
difference in the level of FOXL1 expression between hF0B-FOXL1 wild type and
hF0B-
FOXL1-mutant as detected by RT-PCR (Figure 10). Up regulation of the FOXL1
mutant
protein was observed using the nuclear extract of FOXL protein (Figure12). The
nuclear
localization of FOXL1 was visualized using living cells that were transfected
with mock, wild,
or mutant types FOXL1-GFP expression plasmids. Living cells were examined
under the
fluorescence microscope and showed that nuclear localization of FOXL1 was not
altered by
the mutation (Figure 11). As FOXL1 is a known transcription factor, it was
hypothesized that
FOXL1 deletion could affect the FOXL1 protein structure by interfering with
signaling of
downstream gene targets. Expression of 13 genes that have been predicted to be
involved
in bone formation (Table 4) was tested by RT-PCR, using RNA from HEK293A
transfected
with wild FOXL1 and HEK293A transected by the mutant FOXL1. No significant
change in
the expression of these genes between the wild-type and mutant FOXL1
transfected cell line
was observed (Figure 13).
[00176] A
positional mapping approach and a large extended family from NL were
used to identify a 15bp deletion in FOXL1 gene that segregates with the
otosclerosis in the
study family. Family 2081 is a large multiplex five-generation family. Linkage
analysis
excluded linkage of family 2081 to any previously mapped loci and suggest
linkage of the
family to 9Mb at chr16q23.1-q24.2. This new locus is located 2Mb telomeric to
the published
OTSC4 locus. The 9Mb region contains 79 annotated genes. Bidirectional Sanger
sequencing of the coding region and intron/exon boundaries of 13 functional
candidates
within the region identified a 15 bp deletion in
FOXL/
(c.976_990het_deIGGGATCCCCTTCCTC) that segregates with the disease haplotype
and
is absent from dbSNP and not found in 116 NL control chromosomes. The deletion
mutation
occurs in a highly conserved region of the gene and at the c-terminus of the
FOXL1
transcription factor.
[00177] A
potential NL founder effect for the 15bp deletion in FOXL1 was explored by
screening 17 NL otosclerosis families for the 15 bp deletion but none was
found. As well, the
full gene was sequenced in these other otosclerosis families and no variants
identified. The
prevalence of otosclerosis in NL is unknown.
CA 02913512 2015-11-25
WO 2014/190422
PCT/CA2014/000471
[00178] Human Forkhead (FKH)-box (FOX) gene family consist of at least 43
members. This gene family was named after the discovery of Drosophila FKH gene
in 1989.
FOX proteins act as a transcription factor and are characterized by sharing a
common
element of 110 amino acid DNA binding site, the fork head domain (FHD). FOX
genes are
important during embryogenesis and cell differentiation [11, 12] and play an
important role in
the regulation of other genes in different species from yeast to human and
have been
involved in different biological process including tumorigenesis and speech
acquisition in
human [13]. Mutations have been identified indifferent FOX genes, for example,
mutations
in FOXC1 cause a range of developmental defects associated with glaucoma, as
in
Axenfeld anamoly, Rieger anomly, iris hyperplasia and Rieger syndrome[14] [15]
[00179] FOXL1 gene (Accession number; NM_005250) consists of one coding
exon,
a small 3UTR and a huge 5UTR. FOXL1 protein as other members of FOX family has
the
conserved sequence domain FHD which is the DNA binding site. FOXL1 is highly
expressed
in otic vesicles in vertebrates and is important in their development by
acting as a negative
regulator to the SHH signaling [16] and has a role in embryonic
transcriptional regulator.
Previous in vivo studies has shown that Foxf1 and Fox11 are target genes of
HHL pathway in
developing stomach and their expression is controlled by G1i2 and G1i3
transcription factors
that harbour a binding site on the 5UTR of FOXI1 gene and this finding shows
that FOXL1
genes involve in HHL pathway [17] [18]. HHL signaling pathway is important in
regulating
many cell processes including cell proliferation, differentiation,
angiogenesis, cellular matrix
remodeling and stem cell homeostasis and disregulation of this pathway lead to
development a different forms of tumors[19]. FOXL1 has no identified role in
bone
regulation. The FOXL1 protein has been identified as one of a number of
proteins associated
with the BMR2 (bone morphogenic receptor2) receptor as seen with two-
dimensional gel
electrophoresis and mass spectrometry [20]. FOXL1 has also been implicated in
the
expression of BMP4 as has shown in the study by Madison etal [18] FOXL1 and
FOXF2
mutants show reduced expression of BMP4 in the gut. [18]. The mechanism under
development of otosclerosis as a result of a mutation in FOXL1 is not known
yet. The
deletion mutation didn't cause any change in the RNA level but did cause up
regulation of
the FOXL1 mutant protein. The identified mutation is located at the c-terminus
end of the
FOXL1 protein and the up regulation of the FOXL1 mutant protein could be as
result of
FOXL1 protein misfolding. The effect of FOXL1 mutant transcription factor on
the some
genes that known to be involved in bone metabolism was tested. The expression
of ten
genes (SCD1, BMP2, BMP4, BMP7, CDKN1A, LTB, ATP1B3, TGB1, TNFSF11 and
BMPR2) were chosen for their expression in four cell lines (hF0B-empty, hF0B-
non-
31
CA 02913512 2015-11-25
WO 2014/190422
PCT/CA2014/000471
transfected, hF0B-FOXL1 mutant and hF0B-FOXL1wild) using RT-PCR. No difference
in
expression between the hF0B- FOXL1 mutant and hF0B- FOXL1 wild type was found.
Conclusion
[00180] This study has identified a novel 15 bp deletion in FOXL1 gene
that co
segregates in a Newfoundland family with otosclerosis and support association
of FOXL1 to
the newly identified locus (extended OTSC4). The level of FOXL1 carrying the
c.976_990het_deIGGGAT0000TTCCTC mutation results in up regulated FOXL1 protein
levels. Further screening of patient with otosclerosis will help in
identifying families with this
deletion.
[00181] While the present application has been described with reference to
what are
presently considered to be the preferred examples, it is to be understood that
the application
is not limited to the disclosed examples. To the contrary, the application is
intended to cover
various modifications and equivalent arrangements included within the spirit
and scope of
the appended claims.
[00182] All publications, patents and patent applications are herein
incorporated by
reference in their entirety to the same extent as if each individual
publication, patent or
patent application was specifically and individually indicated to be
incorporated by reference
in its entirety. Specifically, the sequences associated with each accession
numbers provided
herein including for example accession numbers and/or biomarker sequences (
e.g. protein
and/or nucleic acid) provided in the Tables or elsewhere, are incorporated by
reference in its
entirely.
32
CA 02913512 2015-11-25
WO 2014/190422 PCT/CA2014/000471
Table 1 : sequenced functional candidate genes at extended OTSC4 region
Genomic
position
Genes Accession Strand Start Exons Function or expression
Number End
PLCG2 NM_002661 81812930 33 This gene encodes by protein which
is
81991899 responsible for conversion of 1-
phosphatidy1-1D-
myo-inositol 4,5-bisphosphate to 1D-myo-inositol
1,4,5-trisphosphate (IP3) and diacylglycerol
(DAG) which is important for transmitting signals
from growth factors receptors and immune
system receptors a cross the cell membranes
IRF8 NM_002163 85932774 9 It plays a negative regulatory
role in cells of the
85956211 immune system, and it is expressed
in connective
tissue.
SLC38 NM_00108044 - 84043389 10 It is a multi-pass membrane
protein and it is
A8 2 84075762 widely expressed in tissue
including immune
system, connective tissue and ear.
ZDHHC NM_00114554 - 85008067 9 It is one of zinc finger protein,
it is expressed in
7 8 85045141 connective tissue and bone.
SLC7A NM_003486 87863629 10 It is a multi-pass membrane
protein which is
5 87903100 abundantly expressed in bone marrow
and
inflammatory immune system.
HSD17 NM_002153 82660339 5 It is a single -pass type
membrane protein and it
B2 82132139 is expressed in the bone and bone
marrow.
CDH13 NM_001257.4 + 82660339 14 This gene encodes a membrane of
cadherin
83830215 superfamily and this protein acts
as a negative
inhibitors axon growth during neural different ion.
COTL1 NM 021149 84599204 4 This gene encodes one of the
numerous actin-
84651609 binding proteins which regulate
actin
cytoskeleton.
FOXF1 NM_001451 86544133 1 This gene belongs to the forkhead
family of
86548070 transcription factors . Function of
this gene has
not been determined yet. it is abundantly
expressed in the connective tissue.
FOXL1 NM 005250 86612115 1 This gene belongs to the forkhead
family of
86615304 transcription factors . Function of
this gene has
not been determined yet. it is abundantly
expressed in the connective tissue.
FOXC1 NM_001453 1610681 This gene belongs to the forkhead
family of
1614129 transcription factors. Function of
this gene has not
been determined and it has been shown a role in
regulation of embryonic and ocular development.
Mutation in this gene has been identified in
various glaucoma phenotypes.
CA5A NM_001739 87921625 7 Carbonic anhydrase are a large
family of zinc
87970112 metalloenzyme. They h have an
important role in
a variety of biological process including
calcification, bone resorption.
OSGIN NM 182980 83982672 7 It regulates the differentiation
and proliferation of
1 83999937 normal cells through apoptosis.
Also it regulate
33
inflammatory and anti inflammatory molecules.
Absence of this gene and it is protein leads to
uncontrolled proliferation and growth.
ZNF46 NM_00112746 + 88493879 2 It encodes a zinc-finger protein,
it shares a low
9 4 88507165 percent of homology with some
collagen genes.
CA 02913512 2015-11-25
WO 2014/190422 PCT/CA2014/000471
The homology between ZNF469 and collagen
genes predicts that ZN469 can function s
regulatory factor for synthesis and organization o
collagen fibers.
10
34
CA 02913512 2015-11-25
WO 2014/190422
PCT/CA2014/000471
Table 2: variants detected from sequencing of 13 genes in extended OTSC4
Gene Sequence Variants Location of Protein Effects
dbSNP Disease
Variant Segregati
on
CA5A c.-26C>T 5'UTR Not predicted Not listed No
c.-4C>T 5'UTR Not predicted Not listed No
c.11G>A Coding Ex1 p.R4K Not listed No
c.41 43deITCT Coding Ex1 Phe14del Not listed No
c.44-T>G Coding Ex1 p.L15W Not listed No
c.94C>T Coding Ex1 p.R32X Not listed No
c.120A>G Coding Ex1 p.A40A Not listed No
c.121T>C Coding EX1 p.W41R Not listed No
c.125A>G Coding EX1 p.Q42R Not listed No
c.134A>G Coding EX1 p.N45S Not listed No
c.142+23T>C Coding EX1 None Not listed No
c.142+26deIG Intron 1 None Not listed No
c.142+31G>A Intronl None Not listed No
c.142+65G>A Intron 1 None Not listed No
c.143-111C>T Intron 1 None Not listed No
c.143-9T>A Intron 1 None Not listed No
c.453C>T Intron / p.P151P rs7186698n No
c.459+84G>A Coding (EX3) None rs72816331 No
c.807A>T Coding (EX 4) None rs72816311 No
c.912G>A Coding (EX7) R304RR rs118155826 No
c.918+81C>T 3'UTR) None rs58800453 No
c.918+107C>A 3'UTR None Not listed No
c.918+128G>A 3'UTR None rs61607056 No
COTL1 c.-136T>C 5'UTR not predicted rs71390483 No
c.319-34A>G Intron 3 not predicted rs12929880 No
c."104A>G 3'UTR not predicted Not listed No
c."180T>C 3'UTR not predicted rs1046623 No
c."430A>G 3'UTR not predicted rs247861 No
c."585T>C 3'UTR not predicted rs247862 No
c."921T>C 3'UTR not predicted rs7458 No
c."1047T>A 3'UTR not predicted rs1047121 No
FOXF1 c."196T>C 3'UTR
FOXL1 c.976 990het Coding (EX1) p.Gly326 Leu33 Not listed yes
delGdGATCCCCTT Odel "(infram)
CCTC loss of 5 residue
c.873C>T Coding (EX1) Not listed No
c.45 ins AA Coding (EX1) p.G291G Not listed No
p.Pro16AsnfsX2
4
HSD17B2 c.-133T>C 5'UTR No prediction Not listed
No
IRF8 c.432C>T Coding (EX4) p.D144DD rs16939945 No
c.573G>A Coding (EX4) p.G191GG rs17444416 No
CA 02913512 2015-11-25
WO 2014/190422
PCT/CA2014/000471
OSGIN c.-95C>T 5'UTR Not predicted rs4782864
' No
c.17C>T Coding (EX1) p.P6L Not listed No
c.101A>C Coding (EX2) p.N34T rs28555129 No
c.169A>C Coding (EX2) p.M57L rs2244899 No
c.175C>G Coding (EX2) p.L59V rs2244898 No
c.218-104C>T Intron 2 not predicted no No
c.316+115T>C lntron 3 not predicted rs824396
No
c.316+116T>G Intron 3 not predicted rs862200
No
c.316-'-153_316+156 Intron 3 not predicted Not listed
No
het delATT
c.317-80G>A Intron 3 not predicted rs2665296
No
c.454-48C>A Intron4 not predicted rs2665294
No
c.454-21C>T Intron 4 not predicted Not listed
No
c.646-17G>T Intron 5 not predicted rs2136662
No
c.646-38G>A Intron 5 not predicted rs2741876
No
c.1332C>A Coding (EX7) p.L444LL rs35132222 No
c.*205T>C 3'UTR not predicted rs4782574
No
c.*41A>G 3'UTR not predicted rs77204347
i No
i
SLC7A5 c.-21G>T 5'UTR Not predicted Not listed
No
c.345C>A Coding (EX1) p.G115GG rs17853938 No
c.770+114T>A Intron 3 not predicted Not listed
No
c.770+124T>A Intron 3 not predicted Not listed
No
c.770+125G>A Intron 3 not predicted Not listed
No
c.770+127C>A Intron 3 not predicted Not listed
No
c.770+135A>G Intron 3 not predicted Not listed
No
c.770+145T>A Intron 3 not predicted Not listed
No
c.770+146G>T Intron 3 not predicted Not listed
No
c.[939+61T>G lntron 6 not predicted rs750950
No
c.[1044-35C>T Intron 6 not predicted rs2287120
No
c.[1140+85C>G Intron 7 not predicted rs3815559
No
SLC38A8 c.[189+11G>T Inton 1 not predicted rs1105355
No
c.[189+151A>T Intron1 not predicted rs13339651
No
c.[190-9T>G Intron 1 not predicted rs1876960
No
c.[190-5C>T Intron 1 not predicted rs1876962
No
c.[195G>C Coding (Ex2) p.S65SS rs1317524 No
c.[273G>A Coding (Ex2) p.V91VV Not listed No
c.[388+69G>T Intron 2 not predicted rs8057543
No
c.[388+128A>T Intron 2 not predicted rs34702590
No
c.[531-147C>T Intron 3 not predicted rs907044
No
c.[531-137T>C Intron 3 not predicted rs907043
No
c.[690+65C>T Intron 5 not predicted Not listed
No
c.[690+127G>C Intron 5 not predicted Not listed
No
c.805+53_805+54het Intron 6 not predicted Not listed
No
_delAA
c.[1162+87C>G lntron 8 not predicted rs12929392
No
c.[1214+116C>T Intron8 not predicted rs11864162
No
c.[1214+174T>C Intron8 not predicted rs11864037
No
c.[1214+194A>G Intron8 not predicted rs11149615
No
c.[1214+204C>G Intron8 not predicted rs11864124
No
36
CA 02913512 2015-11-25
WO 2014/190422
PCT/CA2014/000471
PLCG2 c.565-22deIG Intron 6 not predicted Not listed
No
c.[648+134T>C Intron 7 not predicted rs7192509
No
c.[648+171T>A Intron 7 not predicted rs7192535
No
c.[648+237A>T Intron 7 not predicted rs7187837
No
c.[648+242C>T Intron 7 not predicted rs7187163
No
c.[648+319T>C Intron 7 not predicted Not listed
No
c.[692+25C>T Intron 8 not predicted rs11865395
No
c.[692+61A>C Intron 8 not predicted rs41305761
No
c.[765+137T>A Intron 9 not predicted Not listed
No
c.[770A>T Coding EX10 His257Leu rs45443101
No
c.[802C>T Coding EX10 p.H257H rs17537869
No
c.[987-97G>C Intron 11 p.R268RW rs4889430
No
c.[1073-33G>A Intron 12 not predicted rs13333348
No
c.[1149C>T Coding EX12 not predicted rs1143688
No
c.[1188C>G Coding EX12 p.D383D rs13333716
No
c.[1467+38G>C Intron 15 p.T396TT rs4435248
No
c.[1467+45G>T Intron 15 not predicted rs11859176
No
c.[1497C>T Coding EX16 p.A499A rs1143689
No
c.[1557+65C>T Intron 16 not predicted rs71400183
No
c.[1557+87T>A Intron 16 not predicted Not listed
No
c.[2054+7G>A Intron 19 not predicted Not listed
No
c.[2054+47A>C Intron 19 not predicted rs41305765
No
c.[2055-8T>C Intron 19 not predicted rs12448130
No
c.[2236-14C>G Intron 20 not predicted rs12446127
No
c.[3093T>C Coding EX28 p.N1031NN rs1071644
No
c.[3198+74A>G Intron 28 not predicted rs1071644
No
c.[3314-55T>C Intron 29 not predicted rs11150425
No
c.[3314-23C>A Intron 29 not predicted rs4405546
No
c.[3798+35T>C 3'UTR not predicted rs45554137
No
ST3GAL2 c."25G>A] 3'UTR not predicted Not listed
No
ZDHHC7 c.[315+55G>A Intron 3 not predicted rs931713
No
c.316-366het delT Intron 4 not predicted Not listed
No
c.316-57hetileIC Intron 4 not predicted Not listed
No
c.316-50het_delT Intron 4 not predicted Not listed
No
c.375C>T Coding EX5 p.P125PP rs16975086 No
c.538-11C>T Intron 6 not predicted rs8044638
No
c.750+77T>C Intron 8 not predicted rs80330099
No
c.906T>C Coding EX9 p.G302GG rs7195377 No
37
CA 02913512 2015-11-25
WO 2014/190422
PCT/CA2014/000471
ZNF469 c.1069T>C Coding EX1 p.S357P rs11648572 No
c.1098A>C Coding EX1 p.R366S rs11640794 No
c.1529G>C Coding EX1 p.G510A rs7199961 No
c.1776A>G Coding EX1 p.P592P rs12927001 No
c.2130C>T Coding EX1 p.P71OPP rs12918876 No
c.2717C>T Coding EX1 p.P906L rs77951481 No
c.3438G>A Coding EX1 p.P1146P rs9938800 No
c.3484A>G Coding EX1 p.K116GLU rs7197071 No
c.7072G>C Coding EX2 p.G2358R rs12598474 No
c.8520C>T Coding EX2 p.R2840RR rs3812953 No
Coding EX2 p.H2848R rs1983014 No
c.8543A>G Coding EX2 E3630EQ rs904783 No
c.10888G>C Coding EX2 p.T3636A rs904783 No
c.10906A>G Coding EX2 p.S3924S rs4782362 No
c.11772C>T Coding EX2 p.P3891P rs4782301 No
c.11673A>G 5'UTR not predicted rs45504291 No
"8G>A
38
CA 02913512 2015-11-25
WO 2014/190422
PCT/CA2014/000471
Table 3: Sequences
SEQ ID NO:1 Polynucleotide sequence for wildtype FOXI1
ATGAG TCACCTCTTC GATCCCCGGC TGCCTGCCCT 250
GGCCGCCTCG CCCATGCTGT ATCTGTACGG TCCCGAGAGA CCCGGCCTCC 300
CTCTGGCCTT CGCCCCCGCG GCTGCTCTAG CTGCCTCGGG CCGGGCCGAG 350
ACCCCGCAGA AGCCTCCCTA CAGCTACATC GCGCTCATCG CCATGGCGAT 400
CCAGGACGCG CCCGAGCAGA GGGTCACGCT CAACGGCATC TACCAGTTCA 450
TCATGGACCG CTTCCCCTTC TACCACGACA ACCGGCAGGG CTGGCAGAAC 500
AGCATCCGCC ACAACCTCTC GCTCAACGAC TGCTTCGTCA AGGTGCCCCG 550
CGAGAAAGGG CGGCCGGGCA AGGGCAGCTA CTGGACGCTG GACCCCCGCT 600
GCCTGGACAT GTTTGAGAAC GGCAACTACC GGCGCCGGAA GAGGAAGCCC 650
AAGCCGGGCC CCGGGGCCCC GGAGGCCAAG AGGCCCCGCG CCGAGACGCA 700
CCAGCGCAGC GCGGAGGCGC AGCCGGAGGC GGGGAGCGGG GCAGGGGGCT 750
CGGGCCCCGC AATCTCCCGC CTGCAGGCAG CGCCCGCGGG CCCCTCGCCC 800
CTCCTGGACG GCCCCTCTCC GCCGGCGCCC CTCCACTGGC CGGGGACCGC 850
GTCCCCGAAC GAGGACGCTG GTGACGCTGC CCAGGGCGCA GCGGCCGTGG 900
CGGTCGGCCA GGCAGCGCGC ACAGGGGACG GCCCGGGGTC CCCTCTGCGC 950
CCCGCCTCCC GCAGCTCTCC GAAGAGCTCC GACAAGTCCA AGAGCTTCAG 1000
CATAGACAGC ATCCTGGCGG GAAAGCAGGG CCAGAAGCCG CCTTCAGGGG 1050
ACGAACTCCT AGGGGGTGCC AAGCCTGGGC CCGGCGGCCG TCTGGGTGCC 1100
TCGCTCCTGG CCGCCTCCTC CAGCCTCCGT CCGCCTTTCA ACGCTTCCCT 1150
GATGCTCGAC CCGCATGTCC AGGGCGGCTT TTACCAGCTC GGGATCCCCT 1200
TCCTCTCTTA TTTCCCCCTG CAGGTTCCCG ACACGGTACT CCACTTCCAG 1250
TAA
SEQ ID NO:2 Polynucleotide sequence for mutant FOXL1 comprising
c.976_990het_deIGGGATOCCOTTCCTC . Note : dashes represent the deleted 15 bp
sequence
ATGAG TCACCTCTTC GATCCCCGGC TGCCTGCCCT 250
GGCCGCCTCG CCCATGCTGT ATCTGTACGG TCCCGAGAGA CCCGGCCTCC 300
CTCTGGCCTT CGCCCCCGCG GCTGCTCTAG CTGCCTCGGG CCGGGCCGAG 350
ACCCCGCAGA AGCCTCCCTA CAGCTACATC GCGCTCATCG CCATGGCGAT 400
CCAGGACGCG CCCGAGCAGA GGGTCACGCT CAACGGCATC TACCAGTTCA 450
TCATGGACCG CTTCCCCTTC TACCACGACA ACCGGCAGGG CTGGCAGAAC 500
AGCATCCGCC ACAACCTCTC GCTCAACGAC TGCTTCGTCA AGGTGCCCCG 550
CGAGAAAGGG CGGCCGGGCA AGGGCAGCTA CTGGACGCTG GACCCCCGCT 600
GCCTGGACAT GTTTGAGAAC GGCAACTACC GGCGCCGGAA GAGGAAGCCC 650
AAGCCGGGCC CCGGGGCCCC GGAGGCCAAG AGGCCCCGCG CCGAGACGCA 700
CCAGCGCAGC GCGGAGGCGC AGCCGGAGGC GGGGAGCGGG GCAGGGGGCT 750
CGGGCCCCGC AATCTCCCGC CTGCAGGCAG CGCCCGCGGG CCCCTCGCCC 800
CTCCTGGACG GCCCCTCTCC GCCGGCGCCC CTCCACTGGC CGGGGACCGC 850
GTCCCCGAAC GAGGACGCTG GTGACGCTGC CCAGGGCGCA GCGGCCGTGG 900
CGGTCGGCCA GGCAGCGCGC ACAGGGGACG GCCCGGGGTC CCCTCTGCGC 950
CCCGCCTCCC GCAGCTCTCC GAAGAGCTCC GACAAGTCCA AGAGCTTCAG 1000
39
CA 02913512 2015-11-25
WO 2014/190422
PCT/CA2014/000471
CATAGACAGC ATCCTGGCGG GAAAGCAGGG CCAGAAGCCG CCTTCAGGGG 1050
ACGAACTCCT AGGGGGTGCC AAGCCTGGGC CCGGCGGCCG TCTGGGTGCC 1100
TCGCTCCTGG CCGCCTCCTC CAGCCTCCGT CCGCCTTTCA ACGCTTCCCT 1150
GATGCTCGAC CCGCATGTCC AGGGCGGCTT TTACCAGCTC ------------------ 1200
-------------------------------------------------------------- TCTTA
TTTCCCCCTG CAGGTTCCCG ACACGGTACT CCACTTCCAG 1250
TAA
Amino acid sequence Sequences
[00183] SEQ ID NO:3 Amino acid sequence for wildtype FOXI1
>NP 005241 length=345
MSHLFDPRLPALAASPMLYLYGPERPGLPLAFAPAAALAASGRAETPQKP
PYSYIALIAMAIQDAPEQRVTLNGIYQFIMDRFPFYHDNRQGWQNSIRHN
LSLNDCFVKVPREKGRPGKGSYWTLDPRCLDMFENGNYRRRKRKPKPGPG
APEAKRPRAETHQRSAEAQPEAGSGAGGSGPAISRLQAAPAGPSPLLDGP
SPPAPLHWPGTASPNEDAGDAAQGAAAVAVGQAARTGDGPGSPLRPASRS
SPKSSDKSKSFSIDSILAGKQGQKPPSGDELLGGAKPGPGGRLGASLLAA
SSSLRPPFNASLMLDPHVQGGFYQLGIPFLSYFPLQVPDTVLHFQ
[00184] SEQ ID NO:4 Amino acid sequence for mutant FOXL1 comprising
deletion of
5 amino acid p.Gly326_Leu330. Note: dots represent the deleted 5 amino acid
sequence
>
MSHLFDPRLPALAASPMLYLYGPERPGLPLAFAPAAALAASGRAETPQKP
PYSyIALIAMAIQDAPEQRVTLNGIYQFIMDRFPFYHDNRQGWQNSIRHN
LSLNDCFVKVPREKGRPGKGSYWTLDPRCLDMFENGNYRRRKRKPKPGPG
APEAKRPRAETHQRSAEAQPEAGSGAGGSGPAISRLQAAPAGPSPLLDGP
SPPAPLHWPGTASPNEDAGDAAQGAAAVAVGQAARTGDGPGSPLRPASRS
SPKSSDKSKSFSIDSILAGKQGQKPPSGDELLGGAKPGPGGRLGASLLAA
SSSLRPPFNASLMLDPHVQGGFYQL ------- SYFPLQVPDTVLHFQ
Deleted sequences
[00185] SEQ ID NO: 5 GGGATCCCCTTCCTC.
[00186] SEQ ID NO: 6 GIPFL
CA 02913512 2015-11-25
WO 2014/190422
PCT/CA2014/000471
[00187] Primer
sequence that detect the FOXL1 deletion.
FOXL1_F:AACGAGGACGCTGGTGAC SEQ ID NO: 7
FOXL1_R:CCCAGGCAAAGATCATTTTA _ SEQ ID NO: 8
[00188] The amplified product is about 490bp.
XP 001231599.2 275 QVHGRLYHIGIPFLSCFPFHFSE-AVFNFQ --------------------- 303
SEQ ID NO:
9
NP 032050.2 307 HVQGGFSQLGIPFLSYFPLQVPEATVLRFH ------- 336 SEQ ID
NO: 10
XP 002694802.1 318 HVQGGFYQLGIPFLSYFPLQLPE-AVLHFQ --------- 346 SEQ ID
NO: 11
XP 851625.1 328 HVQGGFYQLGIPFLSYFPLQLPD-
TVLHFQ 356 SEQ ID NO:
12
NP 005241.1 317
HVQGGFYQLGIPFLSYFPLQVPD-TVLHFQ 345 SEQ ID NO:
13
XP 511154.2 317
HVQGGFYQLGIPFLSYFPLQLPD-TVLHFQ 345 SEQ ID NO:
14
41
CA 02913512 2015-11-25
WO 2014/190422
PCT/CA2014/000471
References:
1. de Souza C, G.M., Otosclerosis and stapedectomy. Thieme, Stuttgart,
2004.
2. Somers, ID., F. Kuhweide, R. andRobillard, Th., Otosclerosis. B-ENT,3,
Suppl. 6, 3-
10, 2007.
3. Bel Hadj All, I., et al., A new locus for otosclerosis, OTSC8, maps to
the
pericentromeric region of chromosome 9. Hum Genet, 2008. 123(3): p. 267-72.
4. Ealy, M. and R.J. Smith, The Genetics of otosclerosis. Hear Res, 2010.
266(1-2): p.
70-4.
5. Chen, W., et al., Linkage of otosclerosis to a third locus (OTSC3) on
human
chromosome 6p21.3-22.3. J Med Genet, 2002. 39(7): p. 473-7.
6. Thys, M., et al., A seventh locus for otosclerosis, OTSC7, maps to
chromosome
6q13-16.1. Eur J Hum Genet, 2007. 15(3): p. 362-8.
7. Van Den Bogaert, K., et al., A second gene for otosclerosis, OTSC2, maps
to
chromosome 7q34-36. Am J Hum Genet, 2001. 68(2): p. 495-500.
8. Tomek, M.S., et al., Localization of a gene for otosclerosis to
chromosome 15q25-
q26. Hum Mol Genet, 1998. 7(2): p. 285-90.
9. Van Den Bogaert, K., et al., A fifth locus for otosclerosis, OTSC5, maps
to
chromosome 3q22-24. J Med Genet, 2004. 41(6): p. 450-3.
10. Brownstein, Z., et at., Chromosomal mapping and phenotypic
characterization of
hereditary otosclerosis linked to the OTSC4 locus. Arch Otolaryngol Head Neck
Surg, 2006. 132(4): p. 416-24.
11. Carlsson, P. and M. Mahlapuu, Forkhead transcription factors: key
players in
development and metabolism. Dev Blot, 2002. 250(1): p. 1-23.
12. Kaufmann, E. and W. Knochel, Five years on the wings of fork head. Mech
Dev,
1996. 57(1): p. 3-20.
13. Lai, CS., et al., A forkhead-domain gene is mutated in a severe speech
and
language disorder. Nature, 2001. 413(6855): p. 519-23.
14. Nishimura, D.Y., et at., The forkhead transcription factor gene FKHL7
is responsible
for glaucoma phenotypes which map to 6p25. Nat Genet, 1998. 19(2): p. 140-7.
15. Mirzayans, F., et at., Axenfeld-Rieger syndrome resulting from mutation
of the FKHL7
gene on chromosome 6p25. Eur J Hum Genet, 2000. 8(1): p. 71-4.
16. Nakada, C., et at., Transcriptional repressor fox11 regulates central
nervous system
development by suppressing shh expression in zebra fish. Mol Cell Blot, 2006.
26(19): p. 7246-57.
17. Hallikas, 0., et al., Genome-wide prediction of mammalian enhancers
based on
analysis of transcription-factor binding affinity. Cell, 2006. 124(1): p. 47-
59.
18. Madison, B.B., et at., FoxF1 and FoxL1 link hedgehog signaling and the
control of
epithelial proliferation in the developing stomach and intestine. J Blot Chem,
2009.
284(9): p. 5936-44.
19. Bale, A. E., Hedgehog signaling and human disease. Annu Rev Genomics
Hum
Genet, 2002. 3: p. 47-65.
20. Hassel, S., et al., Proteins associated with type II bone morphogenetic
protein
receptor (BMPR-II) and identified by two-dimensional gel electrophoresis and
mass
spectrometry. Proteomics, 2004. 4(5): p. 1346-58.
21. Schrauwen, I., et al., Association of bone morphogenetic proteins with
otosclerosis. J
Bone Miner Res, 2008. 23(4): p. 507-16.
22. Teng, Y.H., R.S. Aquino, and P.W. Park, Molecular functions of syndecan-
1 in
disease. Matrix Biol, 2012. 31(1): p. 3-16.
23. Perreault, N., et at., Fox11 controls the Wnt/beta-catenin pathway by
modulating the
expression of proteoglycans in the gut. J Blot Chem, 2001. 276(46): p. 43328-
33.
42