Note: Descriptions are shown in the official language in which they were submitted.
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
DESCRIPTION
NUCLEAR LOCALIZATION OF GLP-1 STIMULATES MYOCARDIAL
REGENERATION AND REVERSES HEART FAILURE
[0001]
This application claims priority to U.S. Provisional Patent Application
Serial No. 61/901,693, filed November 8, 2013, and claims priority to U.S.
Provisional Patent
Application Serial No. 62/052,141, filed September 18, 2014, both of which
applications are
incorporated by reference herein in their entirety.
TECHNICAL FIELD
[0002] The
field for the present disclosure includes at least the fields of cell
biology, molecular biology, and medicine, such as cardiac medicine.
BACKGROUND
[0003]
There are nearly 5 million Americans with congestive heart failure
(CHF) and approximately 550,000 new cases are diagnosed in the U.S. each year.
Congestive
heart failure affects people of all ages, from children and young adults to
the middle-aged and
the elderly (Roger, et al., 2012). It is very crucial to find new resource of
cardiac muscle
regeneration for CHF treatments. There are various theories about the origin
of regenerating
cardiac muscle cells. These include self-replication of pre-existing adult
cardiac muscle cells
(Senyo, etal., 2012; Eulalio, etal., 2012), differentiation of adult resident
cardiac progenitor
cells (Smart, etal., 2011; Bolli, et al., 2011), dedifferentiation and
proliferation adult cardiac
muscle cells (Beltrami, et al., 2003; Jopling, et al., 2010; Porrello, et al.,
2011), and
transdifferentiation of fibroblast cells into cardiac muscle cells (Song, et
al., 2012; Qian, et
al., 2012). However, it remains controversial whether or not cardiomyocyte
regeneration can
be sufficient to reverse established cardiomyopathy.
[0004]
Glucagon-like peptide-1 (GLP-1) is synthesized in intestinal endocrine
cells in 2 principal major molecular forms, GLP-1 (7-36) amide and GLP-1(7-37)
amide,
which have wide bioactivities on CNS satiety centers, gastrointestinal
motility, islet function
and p cell growth, and energy homeostasis (Drucker, et al., 2001; Drucker, et
al., 2002).
Recently GLP-1 was found to have cardioprotective effects independent of those
attributable
to tight glycemic control (Halbirk, et al., 2010; Timmers, et al., 2009).
Intravenous infusions
1
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
2
of GLP-1 protein to patients with myocardial infarction or chronic heart
failure improved
global LV function and the function of ischemic LV segments. So far it has
been only known
that GLP-1 indirectly acts on GLP-1 receptors distributed on the membrane of
cardiomyocytes and GLP-1R signaling to cAMP generation produces distinct
downstream
signaling events in intracellular calcium or ERK1/2 activation (Ussher, et
al., 2012).
However, no data have been published regarding the effects of GLP-1 gene
delivery to heart.
[0005]
The present disclosure satisfies a long-felt need in the art to provide
therapy for one or more cardiac-related medical conditions, including to
provide therapy for
myocardial regeneration and reversal of heart failure, for example.
BRIEF SUMMARY
[0006] Embodiments of the present disclosure are directed to methods and/or
compositions related to therapy and/or prevention of one or more cardiac-
related medical
conditions. Embodiments of the present disclosure concern regeneration of
tissue, including
muscle tissue, such as myocardial tissue. Certain embodiments relate to
reversal of a cardiac-
related medical condition (or improvement of at least one symptom thereof),
including at
least cardiac disease, cardiomyopathy, cardiotoxicity, congestive heart
failure, ischemic heart
disease, acute myocardial infarction, atrial fibrillation, and arrhythmias.
[0007] Particular aspects of the disclosure concern delivery of a
polynucleotide,
protein, peptide, or mixture thereof to a certain tissue for proliferation
and/or differentiation
of certain cells in the tissue. The tissue may be of any kind, but in specific
cases it is muscle
tissue, including cardiac tissue. In particular embodiments, methods and
compositions of the
disclosure allow for self-replication of pre-existing adult cardiac muscle
cells, differentiation
of adult resident cardiac progenitor cells, dedifferentiation and
proliferation adult cardiac
muscle cells, and/or transdifferentiation of fibroblast cells into cardiac
muscle cells.
[0008] In specific embodiments, a polynucleotide, protein, peptide, or mixture
thereof is targeted to a particular tissue of interest, including a muscle
tissue, such as cardiac
tissue, for example. The targeting may include an ultrasound targeted
microbubble
destruction (UTMD) system for delivery of the polynucleotide or protein or
peptide to the
tissue of interest, including cardiac tissue, for example. In
particular cases, the
polynucleotide, protein, peptide, or mixture thereof localizes to the nucleus
of cells in the
targeted tissue. In certain aspects, the nuclear localization occurs in the
absence of receptors
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
3
for the expression product of the polynucleotide or the protein or peptide on
one or more cells
in the targeted tissue. In specific embodiments, cardiac tissue cells lack
receptors for GLP-1
and a particular liposome composition comprising GLP-1 is provided to the
cells and uptaken
therein under certain conditions. Particular embodiments include nuclear
localization of
G LP-1 .
[0009] Embodiments of the disclosure include GLP-1 polynucleotides utilized
for a therapeutic purpose, including for therapy for a cardiac-related medical
condition. The
polynucleotide may encompass part or all of GLP-1, for example. In certain
cases, the
polynucleotide is an expression vector that may or may not include a nuclear
localization
signal (NLS).
[0010]
Embodiments of the disclosure include delivery of one or more
polynucleotides that stimulate regeneration of cells (such as muscle cells,
including
cardiomyocytes) and/or tissue (including cardiac tissue). Particular aspects
for such
embodiments result in reversal of one or more cardiac-related medical
conditions. Certain
aspects for such embodiments result in improvement of at least one symptom of
a cardiac-
related medical condition. In exemplary embodiments, the cardiac-related
medical condition
is heart failure. The heart failure may be the result of one or more causes,
including heart
failure following exposure to one or more drugs, including chemotherapy drugs,
such as
Adriamycin.
[0011]
Embodiments of the disclosure include at least one of the following:
targeted delivery of GLP-1 gene viral or plasmid vectors to the heart and/or
targeted delivery
of nuclear localization of therapeutic peptides to heart; for example, one can
utilize one or
more nuclear location signal peptides with GLP-1 peptides.
[0012]
Particular but exemplary indications of embodiments of the disclosure
include at least applications for 1) congestive heart failure; 2) prevention
from ventricular
remodeling or aneuysm of myocardial infarction; and/or 3) cardiomyopathy.
Other
indications may also include coronary artery disease, ischemic heart disease,
valvular heart
disease, arrhythmias, etc. In specific embodiments, methods and compositions
of the
disclosure provide cardiomyocyte regeneration that is sufficient to reverse
established
cardiomyopathy, congestive heart failure, and prevention from ventricular
remodeling or
aneuysm of myocardial infarction.
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
4
[0013] Aspects of the disclosure include delivery of GLP-1 gene directly to
the
heart of a mammal that has or is susceptible to heart function failure, such
as that induced by
adriamycin. Ultrasound targeted microbubble destruction (UTMD) (Chen, et at.,
2003;
Bekeredjian, et at., 2003; Korpany, et at., 2005; Chen, et at., 2012; Chen, et
at., 2013in vivo
may be employed, in embodiments. In particular aspects, UTMD is used to
deliver GLP-1
gene, for example under a piggybac transposon plasmid system (Saridey, et at.,
2009; Cary,
et at., 1989; Fraser, et at., 1995; Cadinanos, et at., 2007), to mammalian
hearts. Provided
herein is demonstration that after a single UTMD treatment, transgenic GLP-1
was
surprisingly over-expressed in nuclei of rat heart cells with evidence that
transfected cardiac
cells underwent proliferation and differentiation. However, in specific
embodiments multiple
deliveries of UTMD/GLP-1 are utilized. GLP-1 delivery to heart stimulates the
regeneration
of cardiac muscle and reversal of cardiomyopathy, for example induced by
adriamycin.
[0014] In
particular embodiments, an individual that receives methods or
compositions of the disclosure is not diabetic, has not been diagnosed as
diabetic, has no
signs of being diabetic, is not suspected of being diabetic, and/or is not at
risk of being
diabetic. In some cases, an individual happens to have diabetes but is in need
of cardiac
therapy that was not previously diagnosed; such methods of the present
disclosure may
include the step of diagnosing a need for cardiac therapy in an individual
with diabetes.
[0015] In embodiments of the disclosure, there is a method of localizing GLP-1
and/or TB4 to the nucleus of cells, comprising the steps of providing to the
cells an effective
amount of GLP-1- (and/or TB4-) comprising lipid-stabilized microbubbles; and
exposing the
microbubbles to ultrasound conditions sufficient to deliver GLP-1 and/or TB4
into the nuclei
of the cells. In particular cases, the cell membrane of the cells lacks GLP-1
receptors,
although in some cases the cell membrane of the cells has GLP-1 receptors. The
cells may be
located in vitro or in vivo. The cells may be muscle cells, neural cells,
kidney cells, brain
cells, cartilage cells, cardiac cells, cardiac progenitor cells, yellow
adipocytes, white
adipocytes, or liver cells. Particular muscle cells include cardiomyocytes,
skeletal myocytes,
or smooth muscle myocytes. In some cases, the neural cell is a central neural
cell or a
peripheral neural cell.
[0016] In
particular aspects of the disclosure, a GLP-1 is further defined as a
GLP-1 nucleic acid or a GLP-1 protein. A GLP-1 nucleic acid may be comprised
in a vector,
such as a retroviral vector, lentiviral vector, adenoviral vector, adeno-
associated vector, or
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
plasmid, including a piggyback transposon gene delivery plasmid, for example.
Expression of
a GLP-1 nucleic acid may be regulated by CMV or a tissue-specific promoter. In
some cases,
a GLP-1 nucleic acid further comprises sequence that encodes a nuclear
localization signal.
In particular aspects of the disclosure, TB4 is further defined as a TB4
nucleic acid or a TB4
protein. A TB4 nucleic acid may be comprised in a vector, such as a retroviral
vector,
lentiviral vector, adenoviral vector, adeno-associated vector, or plasmid,
including a
piggyback transposon gene delivery plasmid, for example. Expression of a TB4
nucleic acid
may be regulated by CMV or a tissue-specific promoter. In some cases, a TB4
nucleic acid
further comprises sequence that encodes a nuclear localization signal.
[0017]
Microbubbles employed in particular methods comprise albumin,
polymer shell, phospholipid, or a graphite shell, and the microbubbles may
further comprise a
gas, such as perfluoropropane, air, sulfur hexafluoride, perfluorobutane,
perfluoropentane,
and nitrogen.
[0018] The cells targeted in methods of the disclosure may be in an individual
that has a cardiac-related medical condition, such as one selected from the
group consisting
of cardiac disease, cardiomyopathy, cardiotoxicity, congestive heart failure,
myocardial
infarction, cardiac ischemia, pericarditis, cardiac systolic dysfunction, and
arryhthmia. In
cases of cardiomyopathy, the condition may be induced by a drug, such as a
chemotherapy
drug (like Adriamycin) or a monoclonal antibody. Examples of drugs include
those selected
from the group consisting of anthracyclines; taxanes; fluoropyrimidine;
cyclophosphamide;
bevacizumab; trastuzomab; lapatinib; sorafenib; and sunitinib. The
cardiomyopathy may be
ischemic or non-ischemic cardiomyopathy. The cardiomyopathy may be caused by
long-
term high blood pressure, heart valve problems, heart tissue damage from a
previous heart
attack, chronic rapid heart rate, metabolic disorders, nutritional
deficiencies, pregnancy,
alcohol abuse, drug abuse, chemotherapy drugs, viral infection,
hemochromatosis, genetic
condition, elevated cholesterol levels, or a combination thereof.
[0019] In particular aspects, an individual is provided with an additional
cardiac
disease therapy, such as an additional cardiomyopathy therapy.
[0020] In
certain embodiments, there is a method of regenerating cells at a
desired location in an individual, comprising the steps of delivering to the
location an
effective amount of GLP-1- (and/or TB4-) comprising lipid-stabilized
microbubbles; and
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
6
exposing the microbubbles to ultrasound conditions sufficient to deliver GLP-1
and/or TB4
into the nuclei of cells at the location. The cell membrane of the cells may
or may not lack
GLP-1 receptors. The cells may be muscle cells, neural cells, kidney cells,
brain cells,
cartilage cells, cardiac cells, cardiac progenitor cells, yellow adipocytes,
white adipocytes,
liver cells, and bone marrow-derived progenitor cells. In some cases, the
location is at a
region of the heart. Exemplary muscle cells include cardiomyocytes, skeletal
myocytes, or
smooth muscle myocytes. In embodiments wherein the location is the brain, the
method
steps may circumvent the skull of the individual, such as the microbubbles
being delivered to
the brain through the eye, up through the chin, or in a hole or flap in the
skull. In some cases,
the location is at a region of the spinal cord or is at a region of the
peripheral nervous system.
A delivering step may comprise injection, intravenous perfusion, intra-
coronary artery
myocardium perfusion, intra-artery organ perfusion by catheter, or coronary
sinus perfusion
catheter, for example. The individual may have a cardiac-related medical
condition, such as
cardiac disease, cardiomyopathy, cardiotoxicity, congestive heart failure,
myocardial
infarction, cardiac ischemia, pericarditis, cardiac systolic dysfunction, and
arrhythmia.
10021] Embodiments of the disclosure include methods and/or compositions for
regeneration of cardiac muscle and reversal of myocardial ischemic injury, for
example. In
particular embodiments, there are methods for stimulating proliferation of
resident adult
cardiac progenitor or cardiac muscle cells in mammalian hearts that have had a
cardiac-
related medical condition, such as acute ischemic injury, for example. In
certain
embodiments, such methods are achieved with compositions comprising GLP-1 and,
in
particular embodiments, also thymosin beta 4 (TB4); in specific embodiments,
the GLP-1
and/or TB4 includes a nuclear localization signal. In particular embodiments,
GLP-1 (with or
without a nuclear localization signal (NLS)) and TB4 efficiently stimulate
proliferation and
differentiation of adult cardiac muscle cells into three intact cardiac cell
lineages in
mammalian ischemic heart- vascular endothelial cells, coronary artery smooth
muscle cells
and cardiac muscle cells. The GLP-1 and/or TB4 may be provided to an
individual in
microbubbles as contemplated herein, although in some embodiments they are
provided
without microbubbles. The GLP-1 and/or TB4 may be provided in nucleic acid
form or in
proteinaceous form. In specific embodiments, the GLP-1 and TB4 are provided in
nucleic
acid form, and they may or may not be on the same nucleic acid molecule. In
any event, the
expression of GLP-1 and TB4 may or may not be controlled by the same
regulatory
element(s). GLP-1 and TB4 in microbubbles may be delivered locally to the
heart. In
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
7
specific embodiments, GLP-1 and/or TB4 may be associated with a piggyback
transposon
plasmid.
[0022]
Embodiments of the disclosure include myocardial regeneration using
methods and/or compositions as contemplated herein. The myocardial
regeneration may be
following any cardiac-related medical condition. In specific embodiments,
myocardial
regeneration occurs following cardiomyopathy, for example. In particular
embodiments,
myocardial regeneration after UTMD GLP-1 myocardial nuclear delivery is
mediated by
dedifferentiation and proliferation of nuclear FOX01-positive cardiac muscle
cells that, in
specific embodiments, express embryonic stem cell markers (such as OCT4,
Nanog, SOX2,
and/or c-kit) and proliferating markers (such as Ki-67, BrDU, PHH3, and/or
Aurora B).
[0023] In
embodiments of the disclosure, treatment of an individual with
UTMD comprising GLP-1 (with or without NLS) and/or TB4 resulted in
overexpression of
GLP-1 and/or TB4, respectively, in nuclei of heart cells, and the transfected
cardiac cells
undergo dedifferentiation and proliferation. Such delivery results in
myocardial regeneration
and reversal of cardiomyopathy.
[0024] In
some embodiments, there is a kit, housed in a suitable container,
comprising: a polynucleotide encoding at least part of GLP-1 and/or primers
suitable to
amplify at least part of a GLP-1 nucleic acid sequence, and; one or more
reagents suitable for
generating liposomes. Functional fragments of GLP-1 may be provided in the
kit. The GLP-
1 may be in nucleic acid form or in protein form. The kit may further comprise
an additional
therapeutic compound, such as a cardiac disease therapeutic compound. The
additional
therapeutic compound may be an ACE Inhibitor, aldosterone inhibitor,
angiotensin II
receptor blocker (ARBs); beta-blocker, calcium channel blocker, cholesterol-
lowering drug,
digoxin, diuretics, inotropic therapy, potassium, magnesium, vasodilator,
anticoagulant
medication, aspirin, or a combination thereof. In particular embodiments, the
kit comprises a
polynucleotide encoding at least part of thymosin beta 4 (TB4) and/or primers
suitable to
amplify at least part of a TB4 nucleic acid sequence. In some embodiments, the
TB4 is in
protein form and is provided in the kit. In particular embodiments, the kit
comprises a
polynucleotide that comprises both GLP-1 and TB4 nucleic acid sequence.
[0025] The
foregoing has outlined rather broadly the features and technical
advantages of the present disclosure in order that the detailed description of
the disclosure
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
8
that follows may be better understood. Additional features and advantages of
the disclosure
will be described hereinafter which form the subject of the claims of the
disclosure. It should
be appreciated by those skilled in the art that the conception and specific
embodiment
disclosed may be readily utilized as a basis for modifying or designing other
structures for
carrying out the same purposes of the present disclosure. It should also be
realized by those
skilled in the art that such equivalent constructions do not depart from the
spirit and scope of
the disclosure as set forth in the appended claims. The novel features which
are believed to
be characteristic of the disclosure, both as to its organization and method of
operation,
together with further objects and advantages will be better understood from
the following
description when considered in connection with the accompanying figures. It is
to be
expressly understood, however, that each of the figures is provided for the
purpose of
illustration and description only and is not intended as a definition of the
limits of the present
disclosure.
BRIEF DESCRIPTION OF THE DRAWINGS
[0026] For a more complete understanding of the present disclosure, reference
is now made to the following descriptions taken in conjunction with the
accompanying
drawing, in which:
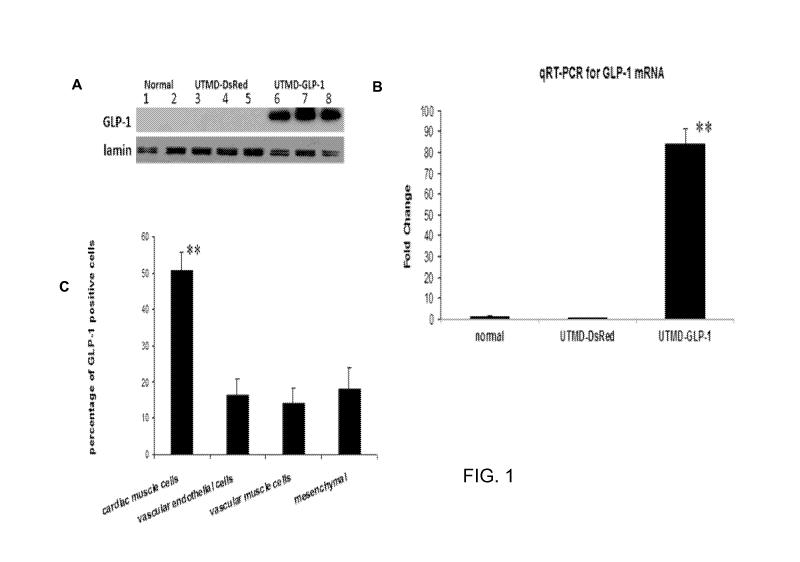
[0027] Fig 1. UTMD GLP-1NLS gene delivery to rat heart showing nuclear
localization of GLP-1 signal in cardiac cells. Panel A is western blot to
detect GLP-1 from
nuclear protein extract of heart tissue, lamin is a marker of nuclear protein.
Panel B is qRT-
PCR for GLP1 cDNA. Panel C is percentage of cell type in GLP-1 positive cells.
Values are
presented as mean SEM. n = 6 per group; **P<0.001 vs control groups.
[0028] Fig
2. Masson's trichrome staining and GLP-1 staining. Panel A is
normal rat heart. Panel B is ADM only. Panel C is ADM plus GLP peptide
treatment. Panel
D is ADM plus UTMD-GLP1 peptide delivery. Panel E is ADM plus UTMD-GLP1 gene
therapy. Panel F is ADM injection first and 14 day late UTMD-GLP1 gene
therapy. The
upper panel is Masson's trichrome staining for whole heart crossing section.
Lower panel is
lower power imaging, scale bar is 200 1..1111.
[0029] Fig
3. Echocardiography evaluated heart structure and functioning. A:
Normal Rat heart; B: ADM injection only; C: ADM injection plus GLP protein
treatment; D:
ADM injection plus UTMD-GLP1 protein delivery; E: ADM injection plus UTMD-GLP1
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
9
gene therapy; F: ADM injection first and 14 day late UTMD-GLP1 gene
therapy.the upper of
left panel is M-model images; down of left panel is 2 dimensional left
ventricle images at
short axis view. Right panels are graphics for LV mass, fraction shorten
index, LVPWd and
abdomen ascite solution. Values are presented as mean SEM; n = 6 per group;
**P<0.001 vs
control groups.
[0030] Fig 4. Therapeutic effects of GLP1 gene heart delivery on established
adriamycin cardiomyopathy. Panel A is a graphic for fractional shortening (%).
Panel B is a
graphics for left ventricular mass. Panel C is a graphics for left ventricular
post wall depths
(LVPWd). Panel D is a graphics for ascites volume. Values are presented as
mean SEM. n
= 6 per group; **P<0.001 vs control groups.
[0031] Fig
5. Dedifferentiated adult cardiac muscle cells are in proliferation.
Panels are the graphics for the percentage of anti-PHH3 (A), anti-Aurora B
(B), anti-Ki-67
(C), and anti-BrDu (D) positive cardiac muscle cells. Values are presented as
mean SEM. n
= 6 per group; **P<0.001 vs control groups.
[0032] Fig
6. Myocardial nuclear overexpression of FOX01 induced by
adriamycin. Panel A is western blot for detecting nuclear FOX01 from nuclear
protein
extracts of heart tissue, lamin is a marker of nuclear proteins. Panel B is
qRT-PCR for
FOX01 mRNA level. Values are presented as mean SEM, n = 6 per group; **P<0.001
vs
normal group and normal plus UTMD-GLP-1NLS group, # P <0.001 vs ADM only.
[0033] Fig 7. Expression of myocardial nuclear enzyme Topoisomerase II a and
13. Panel A is western blots for detecting nuclear TOP Ha and TOP 1113 from
nuclear protein
extracts of heart tissue, lamin is a marker of nuclear proteins. Panel B and C
are qRT-PCR
for TOP Ha and TOP 11f3 mRNA level. Values are presented as mean SEM, n = 6
per group;
**P<0.001 vs normal group and ADM injection only group; ##P<0.001 vs ADM
injection
only group and ADM injection plus UTMD-GLP-1NLS group.
[0034] Fig
8. Overexpression of myocardial nuclear cyclin Dl. Panel A is
western blots for detecting nuclear cyclin D1 from nuclear protein extracts of
heart tissue,
lamin is a marker of nuclear proteins. Panel B is qRT-PCR for cyclin D1 mRNA
level.
Values are presented as mean SEM. n = 6 per group: **P<0.001 vs normal group
and ADM
injection only group.
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
[0035] Fig 9. Panel A is a graph showing the percentage of anti-
NKX2.5
positive cardiac muscle cells. Panel B is a graph showing the percentage of
anti-ISL-1
positive cardiac muscle cells. Values are presented as mean SEM. n = 6 per
group;
**P<0.001 vs control groups.
[0036] Fig 10. Schematic depiction of GLP1NLS cDNA;
[0037] Fig 11. Echocardiography evaluated heart structure and functioning (2D
echo imaging at a short axis view of left ventricle and then measures with M-
model). Panel A
is graphic for LV mass. Panel B is graphic for fraction shorten index. Panel C
is graphic for
LVPWd. Panel D is graphic for abdomen ascite solution. Values are presented as
mean SEM. n = 6 per group; **P<0.001 vs control groups.
[0038] Fig 12. FOX01 inhibitor (AS1842856) inhibited the activation
of
myocardial nuclear FOX01 in ADM cardiomyopathy are associated with reversal of
heart
function failure. Panel A is a graphic for fractional shortening (%). Panel B
is a graphics for
left ventricular post wall depths (LVPWd), Values are presented as mean SEM. n
= 6 per
group; *P<0.01 vs normal and ADM plus FOX01 inhibitor groups.
[0039] Fig 13. Masson's trichromone staining images and evaluation of heart
function with echocardiography. A: Normal rat heart; B: Ligation of rat
coronary artery plus
UTMD-DsRed; C: UTMD-GLP-1NLS/TB4 gene therapy one week after ligation of rat
coronary artery; D: UTMD-GLP-1NLS/TB4 gene therapy and then quickly ligation
of rat
coronary artery; E: UTMD-GLP-1NLS/TB4 gene therapy one week and then ligation
of rat
coronary artery; F: UTMD-GLP-1NLS/TB4 gene therapy two weeks and then ligation
of rat
coronary artery; The left upper panel is Masson's trichromone staining gross
images. The left
down panel is Masson's trichromone staining microscope images, Scale bar is
1000 um. the
right panel are graphics of heart scar tissue, LV mass, Fraction shortening
and LVPWd.
Error bars represent mean SEM; Number of hearts analyzed for each group:
N=6, *P<0.05;
**P<0.001 vs control groups.
[0040] Fig 14. SMAct staining shows coronary artery distribution in in
ischemic
risk area. Illustrated is the graphic for arteriolar density A: Normal rat
heart; B: Ligation of
rat coronary artery plus UTMD-DsRed; C: UTMD-GLP-1NLS/TB4 gene therapy one
week
after ligation of rat coronary artery; D: UTMD-GLP-1NLS/TB4 gene therapy and
then
quickly ligation of rat coronary artery; E: UTMD-GLP-1NLS/TB4 gene therapy one
week
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
11
and then ligation of rat coronary artery; F: UTMD-GLP-1NLS/TB4 gene therapy
two weeks
and then ligation of rat coronary artery Error bars represent mean SEM;
Number of hearts
analyzed for each group: N=6, *P<0.05; **P<0.001 vs control group(Ligation of
rat coronary
artery plus UTMD-DsRed).
[0041] Fig 15. CD31 staining shows vascular endothelial cells density as shown
by the graphic for capillary density. A: Normal rat heart; B: Ligation of rat
coronary artery
plus UTMD-DsRed; C: UTMD-GLP-1NLS/TB4 gene therapy one week after ligation of
rat
coronary artery; D: UTMD-GLP-1NLS/TB4 gene therapy and then quickly ligation
of rat
coronary artery; E: UTMD-GLP-1NLS/TB4 gene therapy one week and then ligation
of rat
coronary artery; F: UTMD-GLP-1NLS/TB4 gene therapy two weeks and then ligation
of rat
coronary artery. Error bars represent mean SEM; Number of hearts analyzed
for each
group: N=6, *P<0.05; **P<0.001 vs control group (Ligation of rat coronary
artery plus
UTMD-DsRed).
[0042] Fig 16. Proliferation markers staining shown cardiac muscle cells are
in
proliferation after UTMD-GLP-1NLS/TB4 gene therapy. The graphics are for the
percentage
of PHH3(A) , Aurora B(B), Ki-67(C), BrDu(D) positive cardiac muscle cells.
Values are
presented as mean SEM; n = 6 per group; **P<0.001 vs control groups. In the
graphics, A:
Normal rat heart; B: Ligation of rat coronary artery plus UTMD-DsRed; C: UTMD-
GLP-
1NLS/TB4 gene therapy one week after ligation of rat coronary artery; D: UTMD-
GLP-
1NLS/TB4 gene therapy and then quickly ligation of rat coronary artery; E:
UTMD-GLP-
1NLS/TB4 gene therapy one week and then ligation of rat coronary artery; F:
UTMD-GLP-
1NLS/TB4 gene therapy two weeks and then ligation of rat coronary artery.
DETAILED DESCRIPTION
I. Exemplary Definitions
[0043] As used herein, the use of the word "a" or "an" when used
in
conjunction with the term "comprising" in the claims and/or the specification
may mean
"one," but it is also consistent with the meaning of "one or more," "at least
one," and "one or
more than one." Some embodiments of the disclosure may consist of or consist
essentially of
one or more elements, method steps, and/or methods of the disclosure. It is
contemplated that
any method or composition described herein can be implemented with respect to
any other
method or composition described herein.
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
12
[0044] The
term "cardiac-related medical condition" as used herein refers to
any medical condition that affects heart tissue, including that affects heart
function.
II. General Aspects of the Disclosure
[0045] The
present disclosure provides methods and/or compositions for
treatment and/or prevention of at least one cardiac-related medical condition.
Such methods
and compositions employ at least part of GLP-1 and a UTMD system. In
particular aspects,
UTMD directly delivers a GLP-1 polynucleotide into the nuclei of
cardiomyocytes (that lack
GLP-1 cell surface receptors). Upon translation of the polynucleotide inside
the cell, this
GLP-1 gene product concentrates in the nuclei of cardiomyocytes and does not
excrete out
into circulation. This nuclear GLP-1 stimulates myocardial regeneration via
proliferation or
self-replication of existing cardiomyocytes, in specific embodiments.
Embodiments provide
a new molecular mechanism of nuclear GLP-1 action different from routine GLP-1
peptide in
cardioprotecting effects.
[0046] In at least some cases, particular GLP-1 polynucleotides are employed,
including those that encode partial but functional GLP-1 gene products. In
particular
embodiments, TB4 polynucleotides are used in conjunction with GLP-1
polynucleotides,
such as on the same or different polynucleotide in a UTMD system. Certain
expression
vectors may be useful for harboring the GLP-1 (and/or TB4) polynucleotide(s).
Particular
UTMD components may be utilized for delivery of the GLP-1 and/or TB4
polynucleotide(s).
III. Cardiac-Related Medical Conditions and Treatment and/or Prevention
Thereof
[0047] In
specific embodiments, the cardiac-related medical condition is
selected from the group consisting of cardiac disease, cardiovascular disease,
heart disease,
cardiomyopathy, cardiotoxicity, myocardial infarction, cardiac ischemic
disease, arrhythmias,
coronary artery disease, and a combination thereof.
[0048]
Particular types of cardiovascular disease may be treated or prevented,
such as coronary artery disease (also known as coronary heart disease and
ischaemic heart
disease); cardiomyopathy (diseases of cardiac muscle); hypertensive heart
disease; heart
failure; cor pulmonale; cardiac dysrhythmias; inflammatory heart disease;
endocarditis;
inflammatory cardiomegaly; myocarditis; valvular heart disease;
cerebrovascular disease;
peripheral arterial disease; congenital heart disease; and rheumatic heart
disease.
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
13
[0049] In particular aspects of the disclosure, cardiomyopathy is
the cardiac-
related medical condition. The cardiac-related medical condition (including,
for example,
cardiomyopathy) may be caused by one or more of a variety of characteristics,
including, for
example, long-term high blood pressure; heart valve problems; heart tissue
damage (such as
from a previous heart attack); chronic rapid heart rate; metabolic disorders,
such as thyroid
disease or diabetes; nutritional deficiencies of essential vitamins or
minerals, such as thiamin
(vitamin B-1), selenium, calcium and/or magnesium; pregnancy; alcohol abuse;
drug abuse,
including of narcotics or prescription drugs, such as cocaine or
antidepressant medications,
such as tricyclic antidepressants; use of some chemotherapy drugs to treat
cancer (including
Adriamycin); certain viral infections; hemochromatosis and/or an unknown cause
or
undetected cause. The cardiac-related medical condition may be directly or
indirectly caused
by cancer therapeutics, both small molecule drugs and biologics, that are
associated with
cardiotoxicity, for example. Examples include anthracyclines; taxanes;
fluoropyrimidine;
cyclophosphamide; bevacizumab; trastuzomab; lapatinib; sorafenib; and
sunitinib. The drug
may be an immunogenic composition, such as a monoclonal antibody, such as
Herceptin, for
example.
100501 In some cases, methods and compositions of the present disclosure are
employed for prevention of one or more cardiac-related medical conditions or
delay of onset
of one or more cardiac-related medical conditions or reduction of extent of
one or more
symptoms of one or more cardiac-related medical conditions. In particular
cases, such
prevention, delay or onset, or reduction of extent of one or more symptoms,
occurs in an
individual that is at risk for a cardiac-related medical condition. Exemplary
risk factors
include one or more of the following: age, gender (male, although it occurs in
females), high
blood pressure, high serum cholesterol levels, tobacco smoking, excessive
alcohol
consumption, sugar consumption, family history, obesity, lack of physical
activity,
psychosocial factors, diabetes mellitus, overweight, genetic predisposition,
and/or exposure
to air pollution.
IV. GLP-1 and TB4 Compositions
100511 Certain embodiments of the present disclosure concern a GLP-1 and/or
TB4 nucleic acid, which also may be referred to as a GLP-1 polynucleotide
and/or TB4
polynucleotide, respectively. In certain aspects, a GLP-1 and/or TB4 nucleic
acid comprises
a wild-type or a mutant GLP-1 and/or TB4 nucleic acid. In particular aspects,
a GLP-1
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
14
and/or TB4 nucleic acid encodes for or comprises a transcribed nucleic acid.
In other
aspects, a GLP-1 and/or TB4 nucleic acid comprises a nucleic acid segment of
GLP-1 and/or
TB4, respectively, or a biologically functional equivalent thereof. In
particular aspects, a
GLP-1 and/or TB4 nucleic acid encodes a protein, polypeptide, or peptide. An
exemplary
human GLP-1 nucleic acid is at the GenBank database of National Center for
Biotechnology Information, Accession No. J04040.1 (SEQ ID NO:1), which is
incorporated
by reference herein in its entirety. An exemplary GLP-1 polypeptide is at
GenBank
Accession Number AAA52567.1 (SEQ ID NO:2), which is incorporated by reference
herein
in its entirety. The skilled artisan recognizes that the entire GLP1 gene
sequence is from 311
to 421 of proglucagon mRNA and that proglucagon mRNA includes glucagon, GLP-1
and
GLP-2. SEQ ID NO:5 is an exemplary proglucagon nucleic acid sequence, and SEQ
ID
NO:6 is an exemplary proglucagon protein sequence.
[0052] In specific embodiments, GLP-1 (7-36) amide or GLP-1(7-37) amide are
utilized in the methods. In certain embodiments, nucleic acid encoding GLP-1
(7-36) amide
and GLP-1(7-37) amide is utilized in the methods.
[0053] An
exemplary human thymosin beta 4 (TB4) polynucleotide is at
GenBank Accession Number BC139925; SEQ ID NO:9), which is incorporated by
reference herein in its entirety. An exemplary human thymosin beta 4
polypeptide is at
GenBank Accession Number AAI39926; SEQ ID NO:10), which is incorporated by
reference herein in its entirety.
[0054] In
specific embodiments, a functional fragment of GLP-1 is utilized
instead of the entire GLP-1 polynucleotide or entire GLP-1 peptide. A
functional fragment of
GLP-1 is one that is sufficient to allow regeneration of cells upon exposure
to the fragment
and upon its uptake into the nucleus of the cells, either alone or in
conjunction with TB4. In
specific embodiments, the functional fragment of GLP-1 nucleic acid encodes
(or the peptide
comprises) at least 27, 26, 25, 24, 23, 22, 21, 20, 19, 18, 17, 16, 15, 14,
13, 12, 11, or 10
amino acids of SEQ ID NO:2. In specific embodiments, the functional fragment
of GLP-1
nucleic acid encodes (or the peptide comprises) no more than 27, 26, 25, 24,
23, 22, 21, 20,
19, 18, 17, 16, 15, 14, 13, 12, 11, or 10 amino acids of SEQ ID NO:2. The
functional GLP-1
fragment may be 100%, 99%, 98%, 97%, 96%, 95%, 94%, 93%, 92%, 91%, 90%, 85%,
80%.
75%, or 70% identity to SEQ ID NO:2.
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
[0055] In
specific embodiments, a functional fragment of TB4 is utilized
instead of the entire TB4polynucleotide or entire TB4 peptide. A functional
fragment of TB4
is one that is sufficient to allow regeneration of cells upon exposure to the
fragment and upon
its uptake into the nucleus of the cells, either alone or in conjunction with
GLP-1. In specific
embodiments, the functional fragment of TB4 nucleic acid encodes (or the
peptide
comprises) at least 43, 42, 41, 40, 39, 38, 37, 36, 35, 34, 33, 32, 31, 30,
29, 28, 27, 26, 25, 24,
23, 22, 21, 20, 19, 18, 17, 16, 15, 14, 13, 12, 11, or 10 amino acids of SEQ
ID NO:10. In
specific embodiments, the functional fragment of TB4 nucleic acid encodes (or
the peptide
comprises) no more than 43, 42, 41, 40, 39, 38, 37, 36, 35, 34, 33, 32, 31,
30, 29, 28, 27, 26,
25, 24, 23, 22, 21, 20, 19, 18, 17, 16, 15, 14, 13, 12, 11, or 10 amino acids
of SEQ ID NO:10.
The functional TB4 fragment may be 100%, 99%, 98%, 97%, 96%, 95%, 94%, 93%,
92%,
91%, 90%, 85%, 80%, 75%, or 70% identity to SEQ ID NO:10.
[0056] The term "nucleic acid" is well known in the art. A "nucleic acid" as
used herein will generally refer to a molecule (i.e., a strand) of DNA, RNA or
a derivative or
analog thereof, comprising a nucleobase. A nucleobase includes, for example, a
naturally
occurring purine or pyrimidine base found in DNA (e.g., an adenine "A," a
guanine "G," a
thymine "T" or a cytosine "C") or RNA (e.g., an A, a G, an uracil "U" or a C).
The term
"nucleic acid" encompass the terms "oligonucleotide" and "polynucleotide,"
each as a
subgenus of the term "nucleic acid." The term "oligonucleotide" refers to a
molecule of
between about 3 and about 100 nucleobases in length. The term "polynucleotide"
refers to at
least one molecule of greater than about 100 nucleobases in length.
[0057]
These definitions generally refer to a single-stranded molecule, but in
specific embodiments will also encompass an additional strand that is
partially, substantially
or fully complementary to the single-stranded molecule. Thus, a nucleic acid
may encompass
a double-stranded molecule or a triple-stranded molecule that comprises one or
more
complementary strand(s) or "complement(s)" of a particular sequence comprising
a molecule.
As used herein, a single stranded nucleic acid may be denoted by the prefix
"ss," a double
stranded nucleic acid by the prefix "ds," and a triple stranded nucleic acid
by the prefix "ts."
A. Nucleobases
[0058] As used herein a "nucleobase" refers to a heterocyclic base, such as
for
example a naturally occurring nucleobase (i.e., an A, T, G, C or U) found in
at least one
naturally occurring nucleic acid (i.e., DNA and RNA), and naturally or non-
naturally
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
16
occurring derivative(s) and analogs of such a nucleobase. A nucleobase
generally can form
one or more hydrogen bonds ("anneal" or "hybridize") with at least one
naturally occurring
nucleobase in manner that may substitute for naturally occurring nucleobase
pairing (e.g., the
hydrogen bonding between A and T, G and C, and A and U).
10059]
"Purine" and/or "pyrimidine" nucleobase(s) encompass naturally
occurring purine and/or pyrimidine nucleobases and also derivative(s) and
analog(s) thereof,
including but not limited to, those a purine or pyrimidine substituted by one
or more of an
alkyl, caboxyalkyl, amino, hydroxyl, halogen (i.e., fluoro, chloro, bromo, or
iodo), thiol or
alkylthiol moiety. Preferred alkyl (e.g., alkyl, caboxyalkyl, etc.) moeities
comprise of from
about 1, about 2, about 3, about 4, about 5, to about 6 carbon atoms. Other
non-limiting
examples of a purine or pyrimidine include a deazapurine, a 2,6-diaminopurine,
a 5-
fluorouracil, a xanthine, a hypoxanthine, a 8-bromoguanine, a 8-chloroguanine,
a
bromothymine, a 8-aminoguanine, a 8-hydroxyguanine, a 8-methylguanine, a 8-
thioguanine,
an azaguanine, a 2-aminopurine, a 5-ethylcytosine, a 5-methylcyosine, a 5-
bromouracil, a 5-
ethyluracil, a 5-iodouracil, a 5-chlorouracil, a 5-propyluracil, a thiouracil,
a 2-methyladenine,
a methylthioadenine, a N,N-diemethyladenine, an azaadenines, a 8-bromoadenine,
a 8-
hydroxyadenine, a 6-hydroxyaminopurine, a 6-thiopurine, a 4-(6-
aminohexyl/cytosine), and
the like.
[0060] A nucleobase may be comprised in a nucleoside or nucleotide, using any
chemical or natural synthesis method described herein or known to one of
ordinary skill in
the art.
B. Nucleosides
[0061] As
used herein, a "nucleoside" refers to an individual chemical unit
comprising a nucleobase covalently attached to a nucleobase linker moiety. A
non-limiting
example of a "nucleobase linker moiety" is a sugar comprising 5-carbon atoms
(i.e., a "5-
carbon sugar"), including but not limited to a deoxyribose, a ribose, an
arabinose, or a
derivative or an analog of a 5-carbon sugar. Non-limiting examples of a
derivative or an
analog of a 5-carbon sugar include a 2'-fluoro-2'-deoxyribose or a carbocyclic
sugar where a
carbon is substituted for an oxygen atom in the sugar ring.
[0062]
Different types of covalent attachment(s) of a nucleobase to a
nucleobase linker moiety are known in the art. By way of non-limiting example,
a nucleoside
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
17
comprising a purine (i.e., A or G) or a 7-deazapurine nucleobase typically
covalently attaches
the 9 position of a purine or a 7-deazapurine to the 1'-position of a 5-carbon
sugar. In another
non-limiting example, a nucleoside comprising a pyrimidine nucleobase (i.e.,
C, T or U)
typically covalently attaches a 1 position of a pyrimidine to a 1 '-position
of a 5-carbon sugar
(Kornberg and Baker, 1992).
C. Nucleotides
[0063] As used herein, a "nucleotide" refers to a nucleoside further
comprising
a "backbone moiety". A backbone moiety generally covalently attaches a
nucleotide to
another molecule comprising a nucleotide, or to another nucleotide to form a
nucleic acid.
The "backbone moiety" in naturally occurring nucleotides typically comprises a
phosphorus
moiety, which is covalently attached to a 5-carbon sugar. The attachment of
the backbone
moiety typically occurs at either the 3'- or 5'-position of the 5-carbon
sugar. However, other
types of attachments are known in the art, particularly when a nucleotide
comprises
derivatives or analogs of a naturally occurring 5-carbon sugar or phosphorus
moiety.
D. Nucleic Acid Analogs
[0064] A nucleic acid may comprise, or be composed entirely of, a derivative
or
analog of a nucleobase, a nucleobase linker moiety and/or backbone moiety that
may be
present in a naturally occurring nucleic acid. As used herein a "derivative"
refers to a
chemically modified or altered form of a naturally occurring molecule, while
the terms
"mimic" or "analog" refer to a molecule that may or may not structurally
resemble a naturally
occurring molecule or moiety, but possesses similar functions. As used herein,
a "moiety"
generally refers to a smaller chemical or molecular component of a larger
chemical or
molecular structure. Nucleobase, nucleoside and nucleotide analogs or
derivatives are well
known in the art, and have been described (see for example, Scheit, 1980,
incorporated herein
by reference).
[0065] Additional non-limiting examples of nucleosides, nucleotides or nucleic
acids comprising 5-carbon sugar and/or backbone moiety derivatives or analogs,
include
those in U.S. Patent No. 5,681,947 which describes oligonucleotides comprising
purine
derivatives that form triple helixes with and/or prevent expression of dsDNA:
U.S. Patents
5,652,099 and 5,763,167 which describe nucleic acids incorporating fluorescent
analogs of
nucleosides found in DNA or RNA, particularly for use as flourescent nucleic
acids probes;
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
18
U.S. Patent 5,614,617 which describes oligonucleotide analogs with
substitutions on
pyrimidine rings that possess enhanced nuclease stability; U.S. Patents
5,670,663, 5,872,232
and 5,859,221 which describe oligonucleotide analogs with modified 5-carbon
sugars
(i.e., modified 2'-deoxyfuranosyl moieties) used in nucleic acid detection;
U.S. Patent
5,446,137 which describes oligonucleotides comprising at least one 5-carbon
sugar moiety
substituted at the 4' position with a substituent other than hydrogen that can
be used in
hybridization assays; U.S. Patent 5,886,165 which describes oligonucleotides
with both
deoxyribonucleotides with 3'-5' internucleotide linkages and ribonucleotides
with 2'-5'
internucleotide linkages; U.S. Patent 5,714,606 which describes a modified
internucleotide
linkage wherein a 3'-position oxygen of the internucleotide linkage is
replaced by a carbon to
enhance the nuclease resistance of nucleic acids; U.S. Patent 5,672,697 which
describes
oligonucleotides containing one or more 5' methylene phosphonate
internucleotide linkages
that enhance nuclease resistance; U.S. Patents 5,466,786 and 5,792,847 which
describe the
linkage of a substituent moiety which may comprise a drug or label to the 2'
carbon of an
oligonucleotide to provide enhanced nuclease stability and ability to deliver
drugs or
detection moieties; U.S. Patent 5,223,618 which describes oligonucleotide
analogs with a 2 or
3 carbon backbone linkage attaching the 4' position and 3' position of
adjacent 5-carbon sugar
moiety to enhanced cellular uptake, resistance to nucleases and hybridization
to target RNA;
U.S. Patent 5,470,967 which describes oligonucleotides comprising at least one
sulfamate or
sulfamide internucleotide linkage that are useful as nucleic acid
hybridization probe; U.S.
Patents 5,378,825, 5,777,092, 5,623,070, 5,610,289 and 5,602,240 which
describe
oligonucleotides with three or four atom linker moiety replacing
phosphodiester backbone
moiety used for improved nuclease resistance, cellular uptake and regulating
RNA
expression; U.S. Patent 5,858,988 which describes hydrophobic carrier agent
attached to the
2'-0 position of oligonuceotides to enhanced their membrane peimeability and
stability; U.S.
Patent 5,214,136 which describes olignucleotides conjugated to anthraquinone
at the 5'
terminus that possess enhanced hybridization to DNA or RNA; enhanced stability
to
nucleases; U.S. Patent 5,700,922 which describes PNA-DNA-PNA chimeras wherein
the
DNA comprises 2'-deoxy-erythro-pentofuranosyl nucleotides for enhanced
nuclease
resistance, binding affinity, and ability to activate RNase H; and U.S. Patent
5.708.154 which
describes RNA linked to a DNA to form a DNA-RNA hybrid.
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
19
E. Polyether and Peptide Nucleic Acids
[0066] In
certain embodiments, it is contemplated that a nucleic acid
comprising a derivative or analog of a nucleoside or nucleotide may be used in
the methods
and compositions of the disclosure. A non-limiting example is a "polyether
nucleic acid",
described in U.S. Patent Serial No. 5,908,845, incorporated herein by
reference. In a
polyether nucleic acid, one or more nucleobases are linked to chiral carbon
atoms in a
polyether backbone.
[0067] Another non-limiting example is a "peptide nucleic acid", also known as
a "PNA", "peptide-based nucleic acid analog" or "PENAM", described in U.S.
Patent Serial
Nos. 5,786,461, 5891,625, 5,773,571, 5,766,855, 5,736,336, 5,719,262,
5,714,331, 5,539,082,
and WO 92/20702, each of which is incorporated herein by reference. Peptide
nucleic acids
generally have enhanced sequence specificity, binding properties, and
resistance to enzymatic
degradation in comparison to molecules such as DNA and RNA (Egholm et al.,
1993;
PCT/EP/01219). A peptide nucleic acid generally comprises one or more
nucleotides or
nucleosides that comprise a nucleobase moiety, a nucleobase linker moiety that
is not a 5-
carbon sugar, and/or a backbone moiety that is not a phosphate backbone
moiety. Examples
of nucleobase linker moieties described for PNAs include aza nitrogen atoms,
amido and/or
ureido tethers (see for example, U.S. Patent No. 5,539,082). Examples of
backbone moieties
described for PNAs include an aminoethylglycine, polyamide, polyethyl,
polythioamide,
polysulfinamide or polysulfonamide backbone moiety.
[0068] In
certain embodiments, a nucleic acid analogue such as a peptide
nucleic acid may be used to inhibit nucleic acid amplification, such as in
PCR, to reduce false
positives and discriminate between single base mutants, as described in U.S.
Patent Serial
No. 5,891,625. Other modifications and uses of nucleic acid analogs are known
in the art,
and are encompassed herein. In a non-limiting example, U.S. Patent 5,786,461
describes
PNAs with amino acid side chains attached to the PNA backbone to enhance
solubility of the
molecule. In another example, the cellular uptake property of PNAs is
increased by
attachment of a lipophilic group. U.S. application Ser. No. 117.363 describes
several
alkylamino moeities used to enhance cellular uptake of a PNA. Another example
is
described in U.S. Patent Nos. 5,766,855, 5.719.262, 5,714,331 and 5,736.336,
which describe
PNAs comprising naturally and non-naturally occurring nucleobases and
alkylamine side
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
chains that provide improvements in sequence specificity, solubility and/or
binding affinity
relative to a naturally occurring nucleic acid.
F. Preparation of Nucleic Acids
[0069] A nucleic acid may be made by any technique known to one of ordinary
skill in the art, such as for example, chemical synthesis, enzymatic
production or biological
production. Non-
limiting examples of a synthetic nucleic acid (e.g., a synthetic
oligonucleotide), include a nucleic acid made by in vitro chemical synthesis
using
phosphotriester, phosphite or phosphoramidite chemistry and solid phase
techniques such as
described in EP 266,032, incorporated herein by reference, or via
deoxynucleoside H-
phosphonate intermediates as described by Froehler et al., 1986 and U.S.
Patent Serial No.
5,705,629, each incorporated herein by reference. In the methods of the
present disclosure,
one or more oligonucleotide may be used. Various different mechanisms of
oligonucleotide
synthesis have been disclosed in for example, U.S. Patents. 4,659,774,
4,816,571, 5,141,813,
5,264,566, 4,959,463, 5,428,148, 5,554,744, 5,574,146, 5,602,244, each of
which is
incorporated herein by reference.
[0070] A
non-limiting example of an enzymatically produced nucleic acid
include one produced by enzymes in amplification reactions such as PCRTM (see
for example,
U.S. Patent 4,683,202 and U.S. Patent 4,682,195, each incorporated herein by
reference), or
the synthesis of an oligonucleotide described in U.S. Patent No. 5,645,897,
incorporated
herein by reference. A non-limiting example of a biologically produced nucleic
acid includes
a recombinant nucleic acid produced (i.e., replicated) in a living cell, such
as a recombinant
DNA vector replicated in bacteria (see for example, Sambrook et al. 1989,
incorporated
herein by reference).
G. Purification of Nucleic Acids
[0071] A nucleic acid may be purified on polyacrylamide gels, cesium chloride
centrifugation gradients, or by any other means known to one of ordinary skill
in the art (see
for example, Sambrook et al., 1989, incorporated herein by reference).
[0072] In certain aspect, the present disclosure concerns a nucleic acid that
is an
isolated nucleic acid. As used herein, the term "isolated nucleic acid" refers
to a nucleic acid
molecule (e.g., an RNA or DNA molecule) that has been isolated free of, or is
otherwise free
of, the bulk of the total genomic and transcribed nucleic acids of one or more
cells. In certain
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
21
embodiments, "isolated nucleic acid" refers to a nucleic acid that has been
isolated free of, or
is otherwise free of, bulk of cellular components or in vitro reaction
components such as for
example, macromolecules such as lipids or proteins, small biological
molecules, and the like.
H. Nucleic Acid Segments
[0073] In certain embodiments, the nucleic acid is a nucleic acid segment. As
used herein, the term "nucleic acid segment," are smaller fragments of a
nucleic acid, such as
for non-limiting example, those that encode only part of the GLP-1 peptide or
polypeptide
sequence. Thus, a "nucleic acid segment" may comprise any part of a gene
sequence, of from
about 2 nucleotides to the full length of the GLP-1 peptide or polypeptide
encoding region.
[0074]
Various nucleic acid segments may be designed based on a particular
nucleic acid sequence, and may be of any length. By assigning numeric values
to a sequence,
for example, the first residue is 1, the second residue is 2, etc., an
algorithm defining all nucleic
acid segments can be created:
[0075] n to n + y
[0076] where n is an integer from 1 to the last number of the sequence and y
is the
length of the nucleic acid segment minus one, where n + y does not exceed the
last number of
the sequence. Thus, for a 10-mer, the nucleic acid segments correspond to
bases 1 to 10, 2 to 11,
3 to 12 ... and so on. For a 15-mer, the nucleic acid segments correspond to
bases 1 to 15, 2 to
16, 3 to 17 ... and so on. For a 20-mer, the nucleic segments correspond to
bases 1 to 20, 2 to 21,
3 to 22 ... and so on. In certain embodiments, the nucleic acid segment may be
a probe or
primer. As used herein, a "probe" generally refers to a nucleic acid used in a
detection method
or composition. As used herein, a "primer" generally refers to a nucleic acid
used in an
extension or amplification method or composition.
I. Nucleic Acid Complements
[0077]
The present disclosure also encompasses a nucleic acid that is
complementary to a GLP-1 nucleic acid. In particular embodiments the
disclosure
encompasses a nucleic acid or a nucleic acid segment complementary to the GLP-
1 encoding
sequence. A nucleic acid "complement(s)" or is "complementary" to another
nucleic acid
when it is capable of base-pairing with another nucleic acid according to the
standard
Watson-Crick, Hoogsteen or reverse Hoogsteen binding complementarity rules. As
used
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
22
herein "another nucleic acid" may refer to a separate molecule or a spatial
separated sequence
of the same molecule.
[0078] As
used herein, the term "complementary" or "complement(s)" also
refers to a nucleic acid comprising a sequence of consecutive nucleobases or
semiconsecutive
nucleobases (e.g., one or more nucleobase moieties are not present in the
molecule) capable
of hybridizing to another nucleic acid strand or duplex even if less than all
the nucleobases do
not base pair with a counterpart nucleobase. In certain embodiments, a
"complementary"
nucleic acid comprises a sequence in which about 70%, about 71%, about 72%,
about 73%,
about 74%, about 75%, about 76%, about 77%, about 77%, about 78%, about 79%,
about
80%, about 81%, about 82%, about 83%, about 84%, about 85%, about 86%, about
87%,
about 88%, about 89%, about 90%, about 91%, about 92%, about 93%, about 94%,
about
95%, about 96%, about 97%, about 98%, about 99%, to about 100%, and any range
derivable
therein, of the nucleobase sequence is capable of base-pairing with a single
or double
stranded nucleic acid molecule during hybridization. In certain embodiments,
the term
"complementary" refers to a nucleic acid that may hybridize to another nucleic
acid strand or
duplex in stringent conditions, as would be understood by one of ordinary
skill in the art.
[0079] In
certain embodiments, a "partly complementary" nucleic acid
comprises a sequence that may hybridize in low stringency conditions to a
single or double
stranded nucleic acid, or contains a sequence in which less than about 70% of
the nucleobase
sequence is capable of base-pairing with a single or double stranded nucleic
acid molecule
during hybridization.
J. Hybridization
[0080] As used herein, "hybridization", "hybridizes" or "capable of
hybridizing"
is understood to mean the forming of a double or triple stranded molecule or a
molecule with
partial double or triple stranded nature. The term "anneal" as used herein is
synonymous
with "hybridize." The term "hybridization", "hybridize(s)" or "capable of
hybridizing"
encompasses the terms "stringent condition(s)" or "high stringency" and the
tetins "low
stringency" or "low stringency condition(s)."
[0081] As
used herein "stringent condition(s)" or "high stringency" are those
conditions that allow hybridization between or within one or more nucleic acid
strand(s)
containing complementary sequence(s), but precludes hybridization of random
sequences.
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
23
Stringent conditions tolerate little, if any, mismatch between a nucleic acid
and a target
strand. Such conditions are well known to those of ordinary skill in the art,
and are preferred
for applications requiring high selectivity. Non-limiting applications include
isolating a
nucleic acid, such as a gene or a nucleic acid segment thereof or detecting at
least one
specific mRNA transcript or a nucleic acid segment thereof, and the like.
[0082] Stringent conditions may comprise low salt and/or high
temperature
conditions, such as provided by about 0.02 M to about 0.15 M NaCl at
temperatures of about
50 C to about 70 C. It is understood that the temperature and ionic strength
of a desired
stringency are determined in part by the length of the particular nucleic
acid(s), the length and
nucleobase content of the target sequence(s), the charge composition of the
nucleic acid(s),
and to the presence or concentration of formamide, tetramethylammonium
chloride or other
solvent(s) in a hybridization mixture.
[0083] It is also understood that these ranges, compositions and conditions
for
hybridization are mentioned by way of non-limiting examples only, and that the
desired
stringency for a particular hybridization reaction is often determined
empirically by
comparison to one or more positive or negative controls. Depending on the
application
envisioned it is preferred to employ varying conditions of hybridization to
achieve varying
degrees of selectivity of a nucleic acid towards a target sequence. In a non-
limiting example,
identification or isolation of a related target nucleic acid that does not
hybridize to a nucleic
acid under stringent conditions may be achieved by hybridization at low
temperature and/or
high ionic strength. Such conditions are termed "low stringency" or "low
stringency
conditions", and non-limiting examples of low stringency include hybridization
performed at
about 0.15 M to about 0.9 M NaCl at a temperature range of about 20 C to about
50 C. Of
course, it is within the skill of one in the art to further modify the low or
high stringency
conditions to suite a particular application.
[0084] As used herein "wild-type" refers to the naturally occurring sequence
of
a nucleic acid at a genetic locus in the genome of an organism, or a sequence
transcribed or
translated from such a nucleic acid. Thus, the term "wild-type" also may refer
to an amino
acid sequence encoded by a nucleic acid. As a genetic locus may have more than
one
sequence or alleles in a population of individuals, the term "wild-type"
encompasses all such
naturally occurring allele(s). As used herein the term "polymorphic" means
that variation
exists (i.e., two or more alleles exist) at a genetic locus in the individuals
of a population. As
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
24
used herein "mutant" refers to a change in the sequence of a nucleic acid or
its encoded
protein, polypeptide or peptide that is the result of the hand of man.
[0085] The
present disclosure also concerns the isolation or creation of a
recombinant construct or a recombinant host cell through the application of
recombinant
nucleic acid technology known to those of skill in the art or as described
herein. A
recombinant construct or host cell may comprise a GLP-1 nucleic acid, and may
express a
GLP-1 protein, peptide or peptide, or at least one biologically functional
equivalent thereof.
[0086] Herein, in certain embodiments, a "gene" refers to a nucleic acid that
is
transcribed. In
certain aspects, the gene includes regulatory sequences involved in
transcription, or message production or composition. In particular
embodiments, the gene
comprises transcribed sequences that encode for a protein, polypeptide or
peptide. As will be
understood by those in the art, this function term "gene" includes both
genomic sequences,
RNA or cDNA sequences or smaller engineered nucleic acid segments, including
nucleic
acid segments of a non-transcribed part of a gene, including but not limited
to the non-
transcribed promoter or enhancer regions of a gene. Smaller engineered gene
nucleic acid
segments may express, or may be adapted to express using nucleic acid
manipulation
technology, proteins, polypeptides, domains, peptides, fusion proteins,
mutants and/or such
like.
[0087]
"Isolated substantially away from other coding sequences" means that
the gene of interest forms the significant part of the coding region of the
nucleic acid, or that
the nucleic acid does not contain large portions of naturally-occurring coding
nucleic acids,
such as large chromosomal fragments, other functional genes, RNA or cDNA
coding regions.
Of course, this refers to the nucleic acid as originally isolated, and does
not exclude genes or
coding regions later added to the nucleic acid by the hand of man.
[0088] The nucleic acid(s) of the present disclosure, regardless of the length
of
the sequence itself, may be combined with other nucleic acid sequences,
including but not
limited to, promoters, enhancers, polyadenylation signals, restriction enzyme
sites, multiple
cloning sites, coding segments, and the like, to create one or more nucleic
acid construct(s).
As used herein, a "nucleic acid construct" is a nucleic acid engineered or
altered by the hand
of man, and generally comprises one or more nucleic acid sequences organized
by the hand
of man.
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
[0089] In a non-limiting example, one or more nucleic acid constructs may be
prepared that include a contiguous stretch of nucleotides identical to or
complementary (at
least in part) to SEQ ID NO:1 or SEQ ID NO:9. A nucleic acid construct may be
about 3,
about 5, about 8, about 10 to about 14, or about 15, about 20, about 30, about
40, about 50,
about 100, about 115, about 200, about 500. about 600, or about 650
nucleotides in length, as
well as constructs of greater size, up to and including chromosomal sizes
(including all
intermediate lengths and intermediate ranges), given the advent of nucleic
acids constructs
such as a yeast artificial chromosome are known to those of ordinary skill in
the art. It will
be readily understood that "intermediate lengths" and "intermediate ranges",
as used herein,
means any length or range including or between the quoted values (i.e., all
integers including
and between such values). Non-limiting examples of intermediate lengths
include about 11,
about 12, about 13, about 16, about 17, about 18, about 19, etc.; about 21,
about 22, about 23,
etc.; about 31, about 32, etc.; about 51, about 52, about 53, etc.; about 101,
about 102, about
103, etc.; about 151, about 152, about 153, etc.; about 600, about 601, about
605, about 610,
etc.etc., etc. Non-limiting examples of intermediate ranges include about 3 to
about 32, about
150 to about 750, etc.
[0090] In certain
embodiments, the nucleic acid construct is a recombinant
vector. In particular embodiments, the disclosure concerns one or more
recombinant
vector(s) comprising nucleic acid sequences that encode an GLP-1 protein,
polypeptide or
peptide that includes within its amino acid sequence a contiguous amino acid
sequence in
accordance with, or essentially as set forth in, SEQ ID NO:2, corresponding to
SEQ ID NO:1
nucleic acid. In particular aspects, the recombinant vectors are DNA vectors.
[0091] In certain
embodiments, the nucleic acid construct is a recombinant
vector. In particular embodiments, the disclosure concerns one or more
recombinant
vector(s) comprising nucleic acid sequences that encode TB4 protein,
polypeptide or peptide
that includes within its amino acid sequence a contiguous amino acid sequence
in accordance
with, or essentially as set forth in, SEQ ID NO:10, corresponding to SEQ ID
NO:9 nucleic
acid. In particular aspects, the recombinant vectors are DNA vectors.
[0092] The term
"biologically functional equivalent" is well understood in the
art and is further defined in detail herein. Accordingly, a sequence that has
70%, 71%, 72%,
73%, 74%, 75%, 76%, 77%, 78%, 79%, 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%,
88%,
89%, 90%, 91%, 92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99%of amino acids that
are
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
26
identical or functionally equivalent to the amino acids of SEQ ID NO:2 or SEQ
ID NO:10,
provided the biological activity of the protein, polypeptide or peptide is
maintained.
[0093] In
certain other embodiments, the disclosure concerns at least one
recombinant vector that include within its sequence a nucleic acid sequence
essentially as set
forth in SEQ ID NO:1 or SEQ ID NO:9. In particular embodiments, the
recombinant vector
comprises DNA sequences that encode protein(s), polypeptide(s) or peptide(s)
exhibiting
GLP-1 activity.
[0094] The
term "functionally equivalent codon" is used herein to refer to
codons that encode the same amino acid, such as the six codons for arginine
and serine, and
also refers to codons that encode biologically equivalent amino acids. Codon
usage for
various organisms and organelles can be found in the literature. Thus, it is
contemplated that
codon usage may be optimized for other animals, as well as other organisms
such as a
prokaryote (e.g., an eubacteria, an archaea), an eukaryote (e.g., a protist, a
plant, a fungi, an
animal), a virus and the like, as well as organelles that contain nucleic
acids, such as
mitochondria, chloroplasts and the like, based on the preferred codon usage as
would be
known to those of ordinary skill in the art.
[0095] It
will also be understood that amino acid sequences or nucleic acid
sequences may include additional residues, such as additional N- or C-terminal
amino acids
or 5' or 3' sequences, or various combinations thereof, and yet still be
essentially as set forth
in one of the sequences disclosed herein, so long as the sequence meets the
criteria set forth
above, including the maintenance of biological protein, polypeptide or peptide
activity where
expression of a proteinaceous composition is concerned. The addition of
terminal sequences
particularly applies to nucleic acid sequences that may, for example, include
various non-
coding sequences flanking either of the 5' and/or 3' portions of the coding
region or may
include various internal sequences, i.e., introns, which are known to occur
within genes.
[0096]
Excepting intronic and flanking regions, and allowing for the
degeneracy of the genetic code, nucleic acid sequences that have between about
70% and
about 79%; or more preferably, between about 80% and about 89%; or even more
particularly, between about 90% and about 99%; of nucleotides that are
identical to the
nucleotides of SEQ ID NO:1 or SEQ ID NO:9 are encompassed in the disclosure.
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
27
[0097] Encompassed in the disclosure include sequences that are 99, 98, 97,
96,
95, 94, 93, 92, 91, 90, 89, 88, 87, 86, 85, 84, 83, 82, 81, 80, 79, 78, 77,
76, 75, 74, 73, 72, 71,
70, or less percent identical to SEQ ID NO:1 or SEQ ID NO:2 (or SEQ ID NO:9 or
SEQ ID
NO:10). The sequences that are not 100 percent identical to SEQ ID NO:1 or SEQ
ID NO:2
(or SEQ ID NO:9 or SEQ ID NO:10) may share their identity therewith at any
region of the
sequence, including the N-terminal or C-terminal ends, or inbetween therof of
SEQ ID NO:2
or the 5' or 3' ends or inbetween there of SEQ ID NO:1 (or SEQ ID NO:9 or SEQ
ID NO:10
respectively).
[0098] It will also be understood that this disclosure is not
limited to the
particular nucleic acid or amino acid sequences of SEQ ID NO:1 or SEQ ID NO:2
(or SEQ
ID NO:9 or SEQ ID NO:10), respectively. Recombinant vectors and isolated
nucleic acid
segments may therefore variously include these coding regions themselves,
coding regions
bearing selected alterations or modifications in the basic coding region, and
they may encode
larger polypeptides or peptides that nevertheless include such coding regions
or may encode
biologically functional equivalent proteins, polypeptide or peptides that have
variant amino
acids sequences.
[0099] The nucleic acids of the present disclosure encompass
biologically
functional equivalent GLP-1 proteins, polypeptides, or peptides. Such
sequences may arise
as a consequence of codon redundancy or functional equivalency that are known
to occur
naturally within nucleic acid sequences or the proteins, polypeptides or
peptides thus
encoded. Alternatively, functionally equivalent proteins, polypeptides or
peptides may be
created via the application of recombinant DNA technology, in which changes in
the protein,
polypeptide or peptide structure may be engineered, based on considerations of
the properties
of the amino acids being exchanged. Changes designed by man may be introduced,
for
example, through the application of site-directed mutagenesis techniques as
discussed herein
below, e.g., to introduce improvements or alterations to the antigenicity of
the protein,
polypeptide or peptide, or to test mutants in order to examine GLP-1 protein,
polypeptide or
peptide activity at the molecular level.
V. Ultrasound Targeted Microbubble Destruction (UTMD) System
[0100] In one aspect, there are methods of delivering a bioactive
agent to a
target organ or tissue in vivo by using an ultrasound-targeted microbubble
destruction
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
28
(UTMD), using microbubbles loaded with nanosphere cationic liposomes
containing the
bioactive agent. Exemplary microbubbles comprise but are not limited to
neutrally charged
lipids, polymers, metals, or acrylic shells suitable for in vivo ultrasound-
targeted microbubble
destruction. In one embodiment, the bioactive agent is first encapsulated
within or attached to
tiny cationic liposomes of nanoparticle size (10-60 nm) (hereinafter,
nanosphere cationic
liposomes either "loaded with" or "including" the bioactive agent refers to
any bioactive
agent encapsulated within or attached to the liposomes, e.g., cationic
liposomes), and the
liposomes are then attached to neutrally charged lipid-coated or albumin-
coated microbubbles
filled with a gas suitable for ultrasound microbubble destruction techniques,
for example
perfluoropropane. The liposomes may be attached to the outer surface of the
microbubble
shell, incorporated within the microbubble shell and/or encapsulated within
the microbubble
shell. In the present disclosure, one or more bioactive agents can be
delivered either
concomitantly or subsequently by ultrasound-targeted microbubble destruction
using the
neutrally charged lipid microbubbles loaded with bioactive agent-containing
nanosphere
cationic liposomes. In another aspect, the present disclosure is a method of
treating a
mammal in need of such treatment comprising administration of an effective
amount of a
composition comprising neutrally charged lipid microbubbles loaded with
nanosphere
cationic liposomes containing a bioactive agent via ultrasound-targeted
microbubble
destruction.
[0101] Examples of bioactive agents suitable for the present disclosure
include
one or more of polynucleotides (including GLP-1 polynucleotides)
pharmaceuticals and
drugs, bioactive synthetic organic molecules, proteins, peptides,
polypeptides, vitamins,
steroids, polyanionic agents, genetic material, and diagnostic agents.
Bioactive vitamins,
steroids, proteins, peptides and polypeptides can be of natural origin or
synthetic. Exemplary
polyanionic agents include but are not limited to sulphated polysaccharides,
negatively
charged serum albumin and milk proteins, synthetic sulphated polymers,
polymerized anionic
surfactants, and polyphosphates. Suitable diagnostic agents include but are
not limited to dyes
and contrast agents for use in connection with magnetic resonance imaging,
ultrasound or
computed tomography of a patient.
[0102] Suitable genetic material includes nucleic acids,
nucleosides,
nucleotides, and polynucleotides that can be either isolated genomic,
synthetic or
recombinant material; either single or double stranded; and either in the
sense or antisense
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
29
direction, with or without modifications to bases, carbohydrate residues or
phosphodiester
linkages. Exemplary sources for the genetic material include but are not
limited to
deoxyribonucleic acids (DNA), ribonucleic acids (RNA), complementary DNA
(cDNA),
messenger RNA (mRNA), ribosomal RNA (rRNA), short interfering RNA (siRNA),
ribozymes, and mixed duplexes and triplexes of RNA and DNA.
[0103] Genetic materials are genes carried on expression vectors including but
not limited to helper viruses, plasmids, phagemids, cosmids, and yeast
artificial
chromosomes. The genetic material suitable for the present disclosure is
capable of coding
for at least a portion of a therapeutic, regulatory, and/or diagnostic
protein. Moreover, genetic
materials can preferably code for more than one type of protein. For example,
a bioactive
agent may comprise plasmid DNA comprising genetic material encoding
therapeutic protein
and a selectable or diagnostic marker to monitor the delivery of the plasmid
DNA, e.g.,
pDsRed-human insulin promoter. Such proteins include but are not limited to
histocompatibility antigens, cell adhesion molecules, growth factors,
coagulation factors,
hormones, insulin, cytokines, chemokines, antibodies, antibody fragments, cell
receptors,
intracellular enzymes, transcriptional factors, toxic peptides capable of
eliminating diseased
or malignant cells. Other genetic materials that could be delivered by this
technique included
adenovirus, adeno-associated virus, retrovirus, lentivirus, RNA, siRNA, or
chemicals that
selectively turn on or off specific genes, such as polyamides or peptide
fragments.
Modifications to wild-type proteins resulting in agonists or antagonists of
the wild type
variant fall in the scope of this disclosure. The genetic material may also
comprise a tissue-
specific promoter or expression control sequences such as a transcriptional
promoter, an
enhancer, a transcriptional terminator, an operator or other control
sequences.
[0104] Examples of other agents that may be used with the polynucleotides of
the present disclosure include one or more of the following therapeutics pre-
loaded into a
liposome and associated with microbubbles including, but are not limited to,
hormone
products such as, vasopressin and oxytocin and their derivatives, glucagon and
thyroid agents
as iodine products and anti-thyroid agents; cardiovascular products as
chelating agents and
mercurial diuretics and cardiac glycosides; respiratory products as xanthine
derivatives
(theophylline and aminophylline); anti-infectives as aminoglycosides,
antifungals (e.g.,
amphotericin), penicillin and cephalosporin antibiotics, antiviral agents
(e.g., Zidovudine.
Ribavirin, Amantadine. Vidarabine and Acyclovir), antihelmintics,
antimalarials, and
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
antituberculous drugs; biologicals such as antibodies (e.g., antitoxins and
antivenins), vaccine
antigens (e.g., bacterial vaccines, viral vaccines, toxoids); antineoplastics
(e.g., nitrosoureas,
nitrogen mustards, antimetabolites (fluorouracil, hormones, progestins and
estrogens agonists
and/or antagonists); mitotic inhibitors (e.g., Etoposide and/or Vinca
alkaloids),
radiopharmaceuticals (e.g., radioactive iodine and phosphorus products); and
Interferon,
hydroxyurea, procarbazine, Dacarbazine, Mitotane, Asparaginase and
cyclosporins, including
mixtures and combinations thereof. Other suitable therapeutics include, but
are not limited
to: thrombolytic agents such as urokinase; coagulants such as thrombin;
antineoplastic agents,
such as platinum compounds (e.g., spiroplatin, cisplatin, and carboplatin),
methotrexate,
adriamycin, taxol, mitomycin, ansamitocin, bleomycin, cytosine arabinoside,
arabinosyl
adsnine, mercaptopolylysine, vincristine, busulfan, chlorambucil, melphalan
(e.g., PAM, L-
PAM or phenylalanine mustard), mercaptopurine, mitotane, procarbazine
hydrochloride
dactinomycin (actinomycin D), daunorubicinhydrochloride, doxorubicin
hydrochloride,
mitomycin, plicamycin (mithramycin), aminoglutethimide, estramustine phosphate
sodium,
flutamide, leuprolide acetate, megestrol acetate, tamoxifen citrate,
testolactone, trilostane,
amsacrine (m-AMSA), asparaginase (L-asparaginase), Erwinaasparaginase,
etoposide (VP-
16), interferon alpha-2a, interferon alpha-2b, teniposide (VM-26), vinblastine
sulfate (VLB),
vincristine sulfate, bleomycin, bleomycin sulfate, methotrexate, adriamycin,
and arabinosyl;
blood products such as parenteral iron, hemin; biological response modifiers
such as
muramyldipeptide, muramyltripeptide, microbial cell wall components,
lymphokines (e.g.,
bacterial endotoxin such as lipopolysaccharide, macrophage activation factor),
sub-units of
bacteria (such as Mycobacteria, Corynebacteria), the synthetic dipeptide N-
acetyl-muramyl-
L-alanyl-D-isog-lutamine; anti-fungalagents such as ketoconazole, nystatin,
griseofulvin,
flucytosine (5-fc), miconazole, amphotericin B, ricin, and beta-lactam
antibiotics (e.g.,
penicillin, ampicillin, sulfazecin); hormones such as growth hormone, PDGF,
EGF, CSF,
GM-CSF, melanocyte stimulating hotmone, estradiol, beclomethasone
dipropionate,
betamethasone, betamethasone acetate and betamethasone sodium phosphate,
vetamethasonedisodiumphosphate, vetamethasone sodium phosphate, cortisone
acetate,
dexamethasone, dexamethasone acetate, dexamethasone sodium phosphate,
flunsolide,
hydrocortisone, hydrocortisone acetate, hydrocortisone cypionate,
hydrocortisone sodium
phosphate, hydrocortisone sodium succinate, methylprednisolone,
methylprednisolone
acetate, methylprednisolone sodium succinate, paramethasone acetate,
prednisolone,
prednisoloneacetate, prednisolone sodium phosphate, prednisolone rebutate,
prednisone,
triamcinolone, triamcinolone acetonide, triamcinolone diacetate, triamcinolone
hexacetonide
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
31
and fludrocortisone acetate; vitamins such vitamin C, E, A, K,
ascyanocobalamin, neinoic
acid, retinoids and derivatives such as retinolpalmitate, and alpha-
tocopherol(s); peptides
(e.g., T cell epitopes such as MAGE, GAGE, DAGE, etc.); proteins, such as
manganese super
oxide dimutase, alcohol dehydrogenase, nitric oxide synthase; enzymes such as
alkaline
phosphatase; anti-allergic agents such as amelexanox; anti-coagulation agents
such as
phenprocoumon and heparin; circulatory drugs such as propranolol; metabolic
potentiators
such asglutathione; antituberculars such as para-aminosalicylic acid,
isoniazid, capreomycin
sulfate cycloserine, ethambutol hydrochloride ethionamide, pyrazinamide,
rifampin, and
streptomycin sulfate; antivirals such as acyclovir, amantadine azidothymidine
(AZT or
Zidovudine), Ribavirin andvidarabine monohydrate (adenine arabinoside, ara-A);
antianginals such asdiltiazem, nifedipine, verapamil, erythrityl tetranitrate,
isosorbidedinitrate, nitroglycerin (glyceryl trinitrate) and
pentaerythritoltetranitrate;
anticoagulants such as phenprocoumon, heparin; antibiotics such as dapsone,
chloramphenicol, neomycin, cefaclor, cefadroxil, cephalexin, cephradine
erythromycin,
clindamycin, lincomycin, amoxicillin, ampicillin, bacampicillin,
carbenicillin, dicloxacillin,
cyclacillin, picloxacillin, hetacillin, methicillin, nafcillin, oxacillin,
penicillin G, penicillin V,
ticarcillin rifampin and tetracycline; antiinflammatories such as difunisal,
ibuprofen,
indomethacin, meclofenamate, mefenamic acid, naproxen, oxyphenbutazone,
phenylbutazone, piroxicam, sulindac, tolmetin, aspirin and salicylates;
antiprotozoans such as
chloroquine, hydroxychloroquine, metronidazole, quinine and meglumine
antimonate;
antirheumatics such as penicillamine; narcotics such as paregoric; opiates
such as codeine,
heroin, methadone, morphine and opium; cardiac glycosides such as deslanoside,
digitoxin,
digoxin, digitalin and digitalis; neuromuscular blockers such as atracurium
besylate,
gallamine triethiodide, hexafluorenium bromide, metocurine iodide, pancuronium
bromide,
succinylcholine chloride (suxamethonium chloride), tubocurarine chloride and
vecuronium
bromide; sedatives (hypnotics) such as amobarbital, amobarbital sodium,
aprobarbital,
butabarbital sodium, chloral hydrate, ethchlorvynol, ethinamate, flurazepam
hydrochloride,
glutethimi de, methotrimeprazine hydrochloride, methyprylon, midazolam
hydrochloride,
paraldehyde, pentobarbital, pentobarbital sodium, phenobarbital sodium,
secobarbital sodium,
talbutal, temazepam and triazolam; local anesthetics such as bupivacaine
hydrochloride,
chloroprocaine hydrochloride, etidocainehydrochloride, lidocaine
hydrochloride,
mepivacaine hydrochloride, procainehydrochloride and tetracaine hydrochloride;
general
anesthetics such asdroperidol, etomidate, fentanyl citrate with droperidol,
ketaminehydrochloride, methohexital sodium and thiopental sodium; and
radioactive
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
32
particles or ions such as strontium, iodide rhenium and yttrium, and
combinations and
mixtures thereof.
[0105] In addition to the polynucleotides of the disclosure,
prodrugs may be
pre-loaded into the liposomes prior to attachment to the microbubbles.
Prodrugs are well
known in the art and may include inactive drug precursors that are metabolized
to form active
drugs. The skilled artisan will recognize suitable prodrugs (and if necessary
their salt forms)
as described by, e.g., in Sinkula, et al., 1975, the relevant portions of
which are incorporated
herein by reference. Prodrugs, for example, may include inactive forms of the
active drugs
wherein a chemical group is present on the prodrug which renders it inactive
and/or confers
solubility or some other property to the drug. In this form, the prodrugs are
generally inactive,
but once the chemical group has been cleaved from the prodrug, by heat,
cavitation, pressure,
and/or by enzymes in the surrounding environment or otherwise, the active drug
is generated.
Such prodrugs are well described in the art, and comprise a wide variety of
drugs bound to
chemical groups through bonds such as esters to short, medium or long chain
aliphatic
carbonates, hemiesters of organic phosphate, pyrophosphate, sulfate, amides,
amino acids,
azo bonds, carbamate, phosphamide, glucosiduronate, N-acetylglucosamine and
beta-
glucoside. Examples of drugs with the parent molecule and the reversible
modification or
linkage are as follows: convallatoxin with ketals, hydantoin with alkyl
esters, chlorphenesin
with glycine or alanins esters, acetaminophen with caffeine complex,
acetylsalicylic acid with
THAM salt, acetylsalicylic acid with acetamidophenyl ester, naloxone with
sulfateester, 15-
methylprostaglandin F sub 2 with methyl ester, procaine with polyethylene
glycol,
erythromycin with alkyl esters, clindamycin with alkylesters or phosphate
esters, tetracycline
with betains salts, 7-acylaminocephalosporins with ring-substituted
acyloxybenzyl esters,
nandrolone with phenylproprionate decanoate esters, estradiol with enolether
acetal,
methylprednisolone with acetate esters, testosterone with n-
acetylglucosaminide
glucosiduronate (trimethylsily1) ether, cortisol or prednisolone or
dexamethasone with 21-
phosphate esters. Prodrugs may also be designed as reversible drug derivatives
and used as
modifiers to enhance drug transport to site-specific tissues. Examples of
carrier molecules
with reversible modifications or linkages to influence transport to a site
specific tissue and for
enhanced therapeutic effect include isocyanate with haloalkyl nitrosurea,
testosterone with
propionateester. methotrexate (3-5'-dichloromethotrexat-e) with dialkyl
esters, cytosine
arabinoside with 5'-acylate, nitrogen mustard (2,2'-dichloro-N-
methyldiethylamine), nitrogen
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
33
mustard with aminomethyltetracycline, nitrogen mustard with cholesterol or
estradiol
ordehydroepiandrosterone esters and nitrogen mustard with azobenzene.
[0106] The skilled artisan will recognize that a particular chemical group may
be modified in any given agent may be selected to influence the partitioning
of the drug into
either the shell or the interior of the microbubbles. The bond selected to
link the chemical
group to the drug may be selected to have the desired rate of metabolism,
e.g., hydrolysis in
the case of ester bonds in the presence of serum esterases after release from
the microbubbles.
Additionally, the particular chemical group may be selected to influence the
biodistribution
of the drug employed in the microbubbles, e.g., N,N-bis(2-chloroethyl)-
phosphorodiamidic
acid with cyclic phosphoramide. Additionally, the prodrugs employed within the
microbubbles may be designed to contain reversible derivatives that are used
as modifiers of
duration of activity to provide, prolong or depot action effects.
[0107] For
example, nicotinic acid may be modified with dextran and
carboxymethlydextran esters, streptomycin with alginic acid salt,
dihydrostreptomycin with
pamoate salt, cytarabine (ara-C) with 5'-adamantoats ester, ara-adenosine (ara-
A) with 5-
palmirate and 5'-benzoate esters, amphotericin B with methyl esters,
testosterone with 17-
beta-alkyl esters, estradiol with formate ester, prostaglandin with 2-(4-
imidazoly1) ethylamine
salt, dopamine with amino acid amides, chloramphenicol with mono- and
bis(trimethylsily1)
ethers, and cycloguanil with pamoate salt. In this form, a depot or reservoir
of long-acting
drug may be released in vivo from the prodrug bearing microbubbles. The
particular chemical
structure of the therapeutics may be selected or modified to achieve a desired
solubility such
that the therapeutic is loaded into a liposome prior to attaching or loading
in, to, at or about a
microbubble. Similarly, other therapeutics may be formulated with a
hydrophobic group
which is aromatic or sterol in structure to incorporate into the surface of
the microbubble.
[0108]
Cationic liposomes suitable for use in the present disclosure comprise
one or more monocationic or polycationic lipids, optionally combined with one
or more
neutral or helper lipids. The cationic lipids suitable for the present
disclosure can be obtained
commercially or made by methods known in the art. Cationic lipids suitable for
the formation
of cationic liposomes are well known in the art and include but are not
limited to any
phospholipid-related materials, such as lecithin, phosphatidylethanolamine,
lysolecithin,
lysophosphatidylethanolamine, phosphatidylserine, phosphatidylinositol,
sphingomyelin,
cephalin, cardiolipin, phosphatidic
acid, cerebrosides, dicetylphosphate,
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
34
dioleoylphosphatidylcholine (DOPC), dipalmitoylphosphatidylcholine
(DPPC),
dioleoylphosphatidylglycerol (DOPG),
dipalmitoylphosphatidylglycerol (DPPG),
dipalmitoylphosphatidylethanolamine 5-carboxyspermylamide
(DPPES),
dioleoylphosphatidylethanolamine (DOPE), palmitoyloleoylphosphatidylcholine
(POPC),
palmitoyloleoylphosphatidylethanolamine (POPE) and dioleoylphosphatidyl-
ethanolamine 4-
(N-maleimidomethyl)cyclohexane-1-carboxylate (DOPE-ma!). Additional non-
phosphorous
containing lipids include but are not limited to stearylamine, dodecylamine,
hexadecylamine,
acetyl palmitate, glycerol ricinoleate, hexadecyl stearate, isopropyl
myristate, amphoteric
acrylic polymers, triethanolamine-lauryl sulfate, alkyl-aryl sulfate
polyethyloxylated fatty
acid amides, dioctadecyldimethyl ammonium bromide and steroids such as
cholesterol,
ergosterol, ergosterol B 1, B2 and B3, androsterone, cholic acid, desoxycholic
acid,
chenodesoxycholic acid, lithocholic
acid, N- [1-(2,3-dioleoyloxy)propyl] -N,N,N-
trimethylammonium chloride (DOTMA), 1,2-bis(oleoyloxy)-3-3-
(trimethylammonia)propane
(DOTAP), and 5-carboxyspermylglycine dioctadecylamide (DOGS). A preferred
liposome
foi ululation comprises the
polycationic lipid 2,3-dioleyloxy-N-[2-
(sperminecarboxaido)ethy1]-N,N-dimethyl-1-propanaminu- m trifluoroacetate (DO
SPA) and
the neutral lipid dioleoyl phosphatidylethanolamine (DOPE) at (3:1, w/w), and
mixtures and
combinations thereof.
101091 In
the method of the present disclosure, the cationic liposomes are
loaded with the polynucleotide, at least. In one embodiment, a cationic lipid
formulation of
one or more lipids dissolved in one or more organic solvents is first dried or
lyophilized to
remove the organic solvent(s), resulting in a lipid film. Just prior to use,
the lipid film is
mixed with a polynucleotide suitable for the present disclosure suspended in a
suitable
aqueous medium for forming liposomes from the dried lipid film. For example,
water, an
aqueous buffer solution, or a tissue culture media can be used for rehydration
of the lipid
film. A suitable buffer is phosphate buffered saline, i.e., 10 mM potassium
phosphate having
a pH of 7.4 in 0.9% NaCl solution. In another embodiment, the dried lipid film
is rehydrated
with a suitable aqueous medium to folin liposomes before the addition of the
bioactive agent.
This method is preferred when the bioactive agent comprises genetic material.
The
incorporation of the bioactive agent into the cationic liposomes is often
performed at a
temperature within the range of about 0 to 30 C., e.g., room temperature, in
about 5, 10-20
minutes.
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
[0110] In
the methods of the present disclosure, the cationic liposomes with
attached bioactive agent(s) (such as a polynucleotide) are then loaded onto
neutrally charged
microbubbles. In a preferred embodiment, this is accomplished by adding to the
cationic
liposomes with attached bioactive agent(s) a lipid composition suitable for
making the
microbubble shell, mixing well, and then adding an appropriate gas for
encapsulation by the
microbubble shell, followed by vigorous shaking for about 5 to 60 seconds,
preferably for
about 20 seconds. In a preferred method, the lipid composition is kept at
about 0 to 30 C.
before the addition of the cationic liposomes with attached bioactive
agent(s).
[0111] To
form the microbubble shell, any biocompatible lipid of natural or
synthetic origin known to be useful in ultrasound-targeted microbubble
destruction are
contemplated as part of the present disclosure. Exemplary lipids can be found
in International
Application No. WO 2000/45856 and include but are not limited to fatty acids,
phosphatides,
glycolipids, glycosphingolipids, sphingolipids, aliphatic alcohols, aliphatic
waxes, terpenes,
sesquiterpenes, and steroids. Examples of lipids are phosphocholines,
phosphatidylcholines,
phosphatidylethanolamines, phosphatidylserines,
phosphatidylglycerols, and
phosphatidylinositol. A particular lipid is 1,2-palmitoyl-sn-glycero-3-
phosphocholine or 1,2-
palmitoyl-sn-glycero-phosphatidylethanolamine. The specific lipid is L-1,2-
palmitoyl-sn-
glycero-3-phosphocholine and L-1,2-palmitoyl-sn-glycero-
phosphatidylethanolamine.
[0112]
Gases suitable for the present disclosure are generally inert and
biocompatible, including but not limited to air; carbon dioxide; nitrogen;
oxygen; fluorine;
noble gases such as helium, neon, argon, and xenon; sulfur-based gases;
fluorinated gases;
and mixtures thereof. The gas may be a perfluoropropane, e.g.,
octafluoropropane.
[0113] As is well known to those versed in the art, targeting ligands can also
be
attached to the microbubbles to confer additional tissue specificity. Such
ligands could
include monoclonal antibodies, peptides, polypeptides, proteins,
glycoproteins, hormones or
hormone analogues, monosaccharides, polysaccharides, steroids or steroid
analogues,
vitamins, cytokines, or nucleotides.
[0114] The
delivery methods of the present disclosure comprising neutrally
charged microbubbles loaded with nanosphere cationic liposomes containing one
or more
bioactive agents provide all the advantages of an ultrasound-targeted
microbubble delivery
system combined with all the advantages of a liposome delivery system. The
ultrasound-
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
36
targeted microbubble delivery system allows for delivery of a drug/gene
bioactive agent to a
specific organ or tissue while minimizing the exposure of other organs or
tissues to the
bioactive agent. During delivery, the bioactive agent(s) remain within the
protective cationic
liposome, which shields the bioactive agent(s) from proteases, nucleases,
lipases,
carbohydrate-cleaving enzymes, free radicals, or other chemical alterations.
This method
increases the delivery of the bioactive agent and its bioavailability to the
target tissue. For
example, in the delivery of neutrally charged microbubbles loaded with
nanosphere cationic
liposomes containing plasmid DNA, the level of gene expression at the target
site is increased
over the level of expression possible with either a microbubble delivery or a
liposome
delivery of the same plasmid DNA.
[0115] In one aspect, the present disclosure is a method of treating a mammal
in
need of such treatment comprising administration of an effective amount of a
composition
comprising neutrally charged lipid microbubbles loaded with nanosphere
cationic liposomes
containing a bioactive agent (such as a GLP-1 polynucleotide) via ultrasound-
targeted
microbubble destruction. Administration of the composition comprising
neutrally charged
lipid microbubbles loaded with nanosphere cationic liposomes containing a
bioactive agent
and the ultrasound-targeted microbubble destruction of these microbubbles to
release the
bioactive agent can be accomplished by any means known in the art. Repeat
administration of
the microbubbles is possible, particularly to prolong the duration of the
therapeutic effect. For
example, repeated transfection of cardiomyocytes by ultrasound targeted
microbubble
destruction has been shown to extend the peak duration of luciferase activity
in the heart from
4 days to 12 days (Bekeredjian et al, 2003). This potentially allows for the
duration of gene or
drug delivery to be tailored to the specific biological or medical need.
VI. Nucleic Acid-Based Expression Systems
[0116] Glucagon-like peptide (GLP)-1 may be provided as a polynucleotide in
the UTMD system to a target tissue, such as the heart. In specific
embodiments, the GLP-1
polynucleotide is provided in an expression vector for use in the UTMD system.
Also, TB4
may be provided as a polynucleotide in the UTMD system to a target tissue,
such as the heart.
In specific embodiments, the TB4 polynucleotide is provided in an expression
vector for use
in the UTMD system. In certain embodiments, the GLP-1 and TB4 polynucleotides
are the
same molecule, although in some embodiments the GLP-1 and TB4 polynucleotides
are
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
37
different molecules. When GLP-1 and TB4 is expressed from the same
polynucleotide, they
may have the same or different regulatory regions for their expression.
[0117] In particular embodiments, an expression vector for use in the UTMD
system may comprise one or more suitable restriction enzyme digestion
sequences, start
codons, stop codons, nuclear localization signals, protease cutting codons,
selectable markers,
origins of replication, regulatory regions, multiple cloning sites, and a
combination thereof.
Such moieties may be positioned in the expression vector in any suitable
order.
[0118] As an exemplary embodiment, the following pattern of GLP-1 construct
may be used in an expression vector:
X hol Start codon Fuxin cutting codon GLP-1 (7-37) NLS Stop cotton Notl
[0119] NLS sequence: 5'-CCT-AAA-AAA-AAG-CGG-AAG-GTC-3' (SEQ ID
NO:3)
[0120] In one embodiment, the following GLP-1 construct may be employed in
an expression vector:
[0121] Xho--Start codon--Furin cutting codon--GLP-1(7-37) ____ NLS-
-Stop
codon--Not 1
[0122] 5 '
-AAA-CTC-GAG-ATG-CGT-CAA-CGT-CGT-CAT-GCT-GAA-
GGG-ACC-TTT-ACC-AGT-GAT-GTG-AGT-TCT-TAC-TTG-GAG-GGC-CAG-GCA-
GCA-AAG-GAA-TTC-ATT-GCT-TGG-CTG-GTG-AAA-GGC-CGA-GGA-CCT-AAA-
AAA-AAG-CGG-AAG-GTC-TAG-GCG-GCC-GCA-AAA-3' (SEQ ID NO:4)
A. Vectors
[0123] The term "vector" is used to refer to a carrier nucleic acid molecule
into
which a nucleic acid sequence can be inserted for introduction into a cell
where it can be
replicated. A nucleic acid sequence can be "exogenous," which means that it is
foreign to the
cell into which the vector is being introduced or that the sequence is
homologous to a
sequence in the cell but in a position within the host cell nucleic acid in
which the sequence is
ordinarily not found. Vectors include plasmids, cosmids, viruses
(bacteriophage, animal
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
38
viruses, and plant viruses), and artificial chromosomes (e.g., YACs). One of
skill in the art
would be well equipped to construct a vector through standard recombinant
techniques (see,
for example, Maniatis et al., 1988 and Ausubel et al., 1994, both incorporated
herein by
reference).
[0124] The term "expression vector" refers to any type of genetic
construct
comprising a nucleic acid coding for a RNA capable of being transcribed. In
some cases,
RNA molecules are then translated into a protein, polypeptide, or peptide. In
other cases,
these sequences are not translated, for example, in the production of
antisense molecules or
ribozymes. Expression vectors can contain a variety of "control sequences,"
which refer to
nucleic acid sequences necessary for the transcription and possibly
translation of an operably
linked coding sequence in a particular host cell. In addition to control
sequences that govern
transcription and translation, vectors and expression vectors may contain
nucleic acid
sequences that serve other functions as well and are described infra.
a. Promoters and Enhancers
[0125] A "promoter" is a control sequence that is a region of a
nucleic acid
sequence at which initiation and rate of transcription are controlled. It may
contain genetic
elements at which regulatory proteins and molecules may bind, such as RNA
polymerase and
other transcription factors, to initiate the specific transcription a nucleic
acid sequence. The
phrases "operatively positioned," "operatively linked," "under control," and
"under
transcriptional control" mean that a promoter is in a correct functional
location and/or
orientation in relation to a nucleic acid sequence to control transcriptional
initiation and/or
expression of that sequence.
[0126] In embodiments of the disclosure, a CMV promoter or a tissue-specific
promoter may be employed. The tissue-specific promoter may be a cardiac tissue
specific
promoter. Examples of cardiac tissue specific promoters include ventricle-
specific myosin
light chain-2 (mlc-2v); alpha-myosin heavy chain (a-MHC). Another example of a
promoter
that may be employed includes the insulin promoter, including the rat insulin
promoter.
[0127] A promoter generally comprises a sequence that functions to position
the
start site for RNA synthesis. The best known example of this is the TATA box,
but in some
promoters lacking a TATA box, such as, for example, the promoter for the
mammalian
terminal deoxynucleotidyl transferase gene and the promoter for the SV40 late
genes, a
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
39
discrete element overlying the start site itself helps to fix the place of
initiation. Additional
promoter elements regulate the frequency of transcriptional initiation.
Typically, these are
located in the region 30-110 bp upstream of the start site, although a number
of promoters
have been shown to contain functional elements downstream of the start site as
well. To
bring a coding sequence "under the control of' a promoter, one positions the
5' end of the
transcription initiation site of the transcriptional reading frame
"downstream" of (i.e., 3' of)
the chosen promoter. The "upstream" promoter stimulates transcription of the
DNA and
promotes expression of the encoded RNA.
[0128] The
spacing between promoter elements frequently is flexible, so that
promoter function is preserved when elements are inverted or moved relative to
one another.
In the tk promoter, the spacing between promoter elements can be increased to
50 bp apart
before activity begins to decline. Depending on the promoter, it appears that
individual
elements can function either cooperatively or independently to activate
transcription. A
promoter may or may not be used in conjunction with an "enhancer," which
refers to a cis-
acting regulatory sequence involved in the transcriptional activation of a
nucleic acid
sequence.
[0129] A
promoter may be one naturally associated with a nucleic acid
sequence, as may be obtained by isolating the 5' non-coding sequences located
upstream of
the coding segment and/or exon. Such a promoter can be referred to as
"endogenous."
Similarly, an enhancer may be one naturally associated with a nucleic acid
sequence, located
either downstream or upstream of that sequence. Alternatively, certain
advantages will be
gained by positioning the coding nucleic acid segment under the control of a
recombinant or
heterologous promoter, which refers to a promoter that is not normally
associated with a
nucleic acid sequence in its natural environment. A recombinant or
heterologous enhancer
refers also to an enhancer not normally associated with a nucleic acid
sequence in its natural
environment. Such promoters or enhancers may include promoters or enhancers of
other
genes, and promoters or enhancers isolated from any other virus, or
prokaryotic or eukaryotic
cell, and promoters or enhancers not "naturally occurring," i.e., containing
different elements
of different transcriptional regulatory regions, and/or mutations that alter
expression. For
example, promoters that are most commonly used in recombinant DNA construction
include
the p-lactamase (penicillinase), lactose and tryptophan (trp) promoter
systems. In addition to
producing nucleic acid sequences of promoters and enhancers synthetically,
sequences may
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
be produced using recombinant cloning and/or nucleic acid amplification
technology,
including PCRTM, in connection with the compositions disclosed herein (see
U.S. Patent Nos.
4,683,202 and 5,928,906, each incorporated herein by reference). Furthermore,
it is
contemplated the control sequences that direct transcription and/or expression
of sequences
within non-nuclear organelles such as mitochondria, chloroplasts, and the
like, can be
employed as well.
[0130] Naturally, it will be important to employ a promoter and/or
enhancer
that effectively directs the expression of the DNA segment in the organelle,
cell type, tissue,
organ, or organism chosen for expression. Those of skill in the art of
molecular biology
generally know the use of promoters, enhancers, and cell type combinations for
protein
expression, (see, for example Sambrook et al. 1989, incorporated herein by
reference). The
promoters employed may be constitutive, tissue-specific, inducible, and/or
useful under the
appropriate conditions to direct high level expression of the introduced DNA
segment, such
as is advantageous in the large-scale production of recombinant proteins
and/or peptides. The
promoter may be heterologous or endogenous.
[0131] Additionally any promoter/enhancer combination (as per, for example,
the Eukaryotic Promoter Data Base EPDB, http://www.epd.isb-sib.ch/) could also
be used to
drive expression. Use of a T3, T7 or SP6 cytoplasmic expression system is
another possible
embodiment. Eukaryotic cells can support cytoplasmic transcription from
certain bacterial
promoters if the appropriate bacterial polymerase is provided, either as part
of the delivery
complex or as an additional genetic expression construct.
b. Initiation Signals and Internal Ribosome Binding Sites
101321 A specific initiation signal also may be required for efficient
translation
of coding sequences. These signals include the ATG initiation codon or
adjacent sequences.
Exogenous translational control signals, including the ATG initiation codon,
may need to be
provided. One of ordinary skill in the art would readily be capable of
determining this and
providing the necessary signals. It is well known that the initiation codon
must be "in-frame"
with the reading frame of the desired coding sequence to ensure translation of
the entire
insert. The exogenous translational control signals and initiation codons can
be either natural
or synthetic. The efficiency of expression may be enhanced by the inclusion of
appropriate
transcription enhancer elements.
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
41
[0133] In certain embodiments of the disclosure, the use of internal
ribosome
entry sites (IRES) elements are used to create multigene, or polycistronic,
messages. IRES
elements are able to bypass the ribosome scanning model of 5' methylated Cap
dependent
translation and begin translation at internal sites (Pelletier and Sonenberg,
1988). IRES
elements from two members of the picornavirus family (polio and
encephalomyocarditis)
have been described (Pelletier and Sonenberg, 1988), as well an IRES from a
mammalian
message (Macejak and Samow, 1991). IRES elements can be linked to heterologous
open
reading frames. Multiple open reading frames can be transcribed together, each
separated by
an IRES, creating polycistronic messages. By virtue of the IRES element, each
open reading
frame is accessible to ribosomes for efficient translation. Multiple genes can
be efficiently
expressed using a single promoter/enhancer to transcribe a single message (see
U.S. Patent
Nos. 5,925,565 and 5,935,819, each herein incorporated by reference).
c. Multiple Cloning Sites
[0134] Vectors can include a multiple cloning site (MCS), which is a nucleic
acid region that contains multiple restriction enzyme sites, any of which can
be used in
conjunction with standard recombinant technology to digest the vector (see,
for example,
Carbonelli et al., 1999, Levenson et al., 1998, and Cocea, 1997, incorporated
herein by
reference.) "Restriction enzyme digestion" refers to catalytic cleavage of a
nucleic acid
molecule with an enzyme that functions only at specific locations in a nucleic
acid molecule.
Many of these restriction enzymes are commercially available. Use of such
enzymes is
widely understood by those of skill in the art. Frequently, a vector is
linearized or
fragmented using a restriction enzyme that cuts within the MCS to enable
exogenous
sequences to be ligated to the vector. "Ligation" refers to the process of
forming
phosphodiester bonds between two nucleic acid fragments, which may or may not
be
contiguous with each other. Techniques involving restriction enzymes and
ligation reactions
are well known to those of skill in the art of recombinant technology.
d. Splicing Sites
101351 Most transcribed eukaryotic RNA molecules will undergo RNA splicing
to remove introns from the primary transcripts. Vectors containing genomic
eukaryotic
sequences may require donor and/or acceptor splicing sites to ensure proper
processing of the
transcript for protein expression (see, for example, Chandler et al.. 1997,
herein incorporated
by reference.)
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
42
e. Termination Signals
[0136] The vectors or constructs of the present disclosure will
generally
comprise at least one termination signal. A "termination signal" or
"terminator" is comprised
of the DNA sequences involved in specific termination of an RNA transcript by
an RNA
polymerase. Thus, in certain embodiments a termination signal that ends the
production of an
RNA transcript is contemplated. A terminator may be necessary in vivo to
achieve desirable
message levels.
[0137] In eukaryotic systems, the terminator region may also comprise specific
DNA sequences that permit site-specific cleavage of the new transcript so as
to expose a
polyadenylation site. This signals a specialized endogenous polymerase to add
a stretch of
about 200 A residues (polyA) to the 3' end of the transcript. RNA molecules
modified with
this polyA tail appear to more stable and are translated more efficiently.
Thus, in other
embodiments involving eukaryotes, it is preferred that that terminator
comprises a signal for
the cleavage of the RNA, and it is more preferred that the terminator signal
promotes
polyadenylation of the message. The terminator and/or polyadenylation site
elements can
serve to enhance message levels and to minimize read through from the cassette
into other
sequences.
[0138] Terminators contemplated for use in the disclosure include any known
terminator of transcription described herein or known to one of ordinary skill
in the art,
including but not limited to, for example, the termination sequences of genes,
such as for
example the bovine growth hormone terminator or viral termination sequences,
such as for
example the SV40 terminator. In certain embodiments, the termination signal
may be a lack
of transcribable or translatable sequence, such as due to a sequence
truncation.
f. Polyadenylation Signals
[0139] In expression, particularly eukaryotic expression, one will
typically
include a polyadenylation signal to effect proper polyadenylation of the
transcript. The
nature of the polyadenylation signal is not believed to be crucial to the
successful practice of
the disclosure, and any such sequence may be employed. Preferred embodiments
include the
SV40 polyadenylation signal or the bovine growth hormone polyadenylation
signal,
convenient and known to function well in various target cells. Polyadenylation
may increase
the stability of the transcript or may facilitate cytoplasmic transport.
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
43
g= Origins of Replication
[0140] In order to propagate a vector in a host cell, it may contain one or
more
origins of replication sites (often termed "on"), which is a specific nucleic
acid sequence at
which replication is initiated. Alternatively an autonomously replicating
sequence (ARS) can
be employed if the host cell is yeast.
h. Selectable and Screenable Markers
[0141] In certain embodiments of the disclosure, cells containing a nucleic
acid
construct of the present disclosure may be identified in vitro or in vivo by
including a marker
in the expression vector. Such markers would confer an identifiable change to
the cell
permitting easy identification of cells containing the expression vector.
Generally, a
selectable marker is one that confers a property that allows for selection. A
positive
selectable marker is one in which the presence of the marker allows for its
selection, while a
negative selectable marker is one in which its presence prevents its
selection. An example of
a positive selectable marker is a drug resistance marker.
[0142] Usually the inclusion of a drug selection marker aids in the cloning
and
identification of transformants, for example, genes that confer resistance to
neomycin,
puromycin, hygromycin, DHFR, GPT, zeocin and histidinol are useful selectable
markers. In
addition to markers conferring a phenotype that allows for the discrimination
of
transformants based on the implementation of conditions, other types of
markers including
screenable markers such as GFP, whose basis is colorimetric analysis, are also
contemplated.
Alternatively, screenable enzymes such as herpes simplex virus thymidine
kinase (tk) or
chloramphenicol acetyltransferase (CAT) may be utilized. One of skill in the
art would also
know how to employ immunologic markers, possibly in conjunction with FACS
analysis.
The marker used is not believed to be important, so long as it is capable of
being expressed
simultaneously with the nucleic acid encoding a gene product. Further examples
of
selectable and screenable markers are well known to one of skill in the art.
i. Plasmid Vectors
[0143] In certain embodiments, a plasmid vector is contemplated for
use to
transform a host cell. In general, plasmid vectors containing replicon and
control sequences
which are derived from species compatible with the host cell are used in
connection with
these hosts. The vector ordinarily carries a replication site, as well as
marking sequences
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
44
which are capable of providing phenotypic selection in transformed cells. In a
non-limiting
example, E. coli is often transformed using derivatives of pBR322, a plasmid
derived from an
E. coli species. pBR322 contains genes for ampicillin and tetracycline
resistance and thus
provides easy means for identifying transformed cells. The pBR plasmid, or
other microbial
plasmid or phage must also contain, or be modified to contain, for example,
promoters which
can be used by the microbial organism for expression of its own proteins.
[0144] In addition, phage vectors containing replicon and control sequences
that
are compatible with the host microorganism can be used as transforming vectors
in
connection with these hosts. For example, the phage lambda GEMTm-11 may be
utilized in
making a recombinant phage vector which can be used to transform host cells,
such as, for
example, E. coli LE392.
[0145] Genomic integrated plasmids, such as piggybac or sleeping
beauty
transposon gene delivery plasmids, may be employed for long term transgenic
expression of
GLP-1 gene in heart or other organ.
[0146] Further useful plasmid vectors include pIN vectors (Inouye et al.,
1985);
and pGEX vectors, for use in generating glutathione S-transferase (GST)
soluble fusion
proteins for later purification and separation or cleavage. Other suitable
fusion proteins are
those with P-galactosidase, ubiquitin, and the like.
[0147] Bacterial host cells, for example, E. coli, comprising the
expression
vector, are grown in any of a number of suitable media, for example, LB. The
expression of
the recombinant protein in certain vectors may be induced, as would be
understood by those
of skill in the art, by contacting a host cell with an agent specific for
certain promoters,
e.g., by adding IPTG to the media or by switching incubation to a higher
temperature. After
culturing the bacteria for a further period, generally of between 2 and 24 h,
the cells are
collected by centrifugation and washed to remove residual media.
j. Viral Vectors
[0148] The ability of certain viruses to infect cells or enter
cells via
receptor-mediated endocytosis, and to integrate into host cell genome and
express viral genes
stably and efficiently have made them attractive candidates for the transfer
of foreign nucleic
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
acids into cells (e.g., mammalian cells). Non-limiting examples of virus
vectors that may be
used to deliver a nucleic acid of the present disclosure are described below.
1. Adenoviral Vectors
[0149] A particular method for delivery of the nucleic acid involves the use
of
an adenovirus expression vector. Although adenovirus vectors are known to have
a low
capacity for integration into genomic DNA, this feature is counterbalanced by
the high
efficiency of gene transfer afforded by these vectors. "Adenovirus expression
vector" is
meant to include those constructs containing adenovirus sequences sufficient
to (a) support
packaging of the construct and (b) to ultimately express a tissue or cell-
specific construct that
has been cloned therein. Knowledge of the genetic organization or adenovirus,
a 36 kb,
linear, double-stranded DNA virus, allows substitution of large pieces of
adenoviral DNA
with foreign sequences up to 7 kb (Grunhaus and Horwitz, 1992).
2. AAV Vectors
[0150]
The nucleic acid may be introduced into the cell using adenovirus
assisted transfection. Increased transfection efficiencies have been reported
in cell systems
using adenovirus coupled systems (Kelleher and Vos, 1994; Cotten et at., 1992;
Curiel,
1994). Adeno-associated virus (AAV) is an attractive vector system for use in
embodiments
of the present disclosure as it has a high frequency of integration and it can
infect nondividing
cells, thus making it useful for delivery of genes into mammalian cells, for
example, in tissue
culture (Muzyczka, 1992) or in vivo. AAV has a broad host range for
infectivity (Tratschin et
al., 1984; Laughlin et al., 1986; Lebkowski et al., 1988; McLaughlin et al.,
1988). Details
concerning the generation and use of rAAV vectors are described in U.S. Patent
Nos.
5,139,941 and 4,797,368, each incorporated herein by reference.
3. Retroviral Vectors
[0151]
Retroviruses have promise as delivery vectors due to their ability to
integrate their genes into the host genome, transferring a large amount of
foreign genetic
material, infecting a broad spectrum of species and cell types and of being
packaged in
special cell-lines (Miller, 1992).
[0152] In
order to construct a retroviral vector, a nucleic acid is inserted into
the viral genome in the place of certain viral sequences to produce a virus
that is
replication-defective. In order to produce virions, a packaging cell line
containing the gag,
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
46
poi, and env genes but without the LTR and packaging components is constructed
(Mann et
al., 1983). When a recombinant plasmid containing a cDNA, together with the
retroviral
LTR and packaging sequences is introduced into a special cell line (e.g., by
calcium
phosphate precipitation for example), the packaging sequence allows the RNA
transcript of
the recombinant plasmid to be packaged into viral particles, which are then
secreted into the
culture media (Nicolas and Rubenstein, 1988; Temin, 1986; Mann et al., 1983).
The media
containing the recombinant retroviruses is then collected, optionally
concentrated, and used
for gene transfer. Retroviral vectors are able to infect a broad variety of
cell types. However,
integration and stable expression require the division of host cells (Paskind
et al., 1975).
[0153] Lentiviruses are complex retroviruses, which, in addition to the common
retroviral genes gag, poi, and env, contain other genes with regulatory or
structural function.
Lentiviral vectors are well known in the art (see, for example, Naldini et
al., 1996; Zufferey
et al., 1997; Blomer et al., 1997; U.S. Pat. Nos. 6,013,516 and 5,994,136).
Some examples
of lentivirus include the Human Immunodeficiency Viruses: HIV-1, HIV-2 and the
Simian
Immunodeficiency Virus: SIV. Lentiviral vectors have been generated by
multiply
attenuating the HIV virulence genes, for example, the genes env, vif, vpr, vpu
and nef are
deleted making the vector biologically safe.
[0154] Recombinant lentiviral vectors are capable of infecting non-
dividing
cells and can be used for both in vivo and ex vivo gene transfer and
expression of nucleic acid
sequences. For example, recombinant lentivirus capable of infecting a non-
dividing cell
wherein a suitable host cell is transfected with two or more vectors carrying
the packaging
functions, namely gag, poi and env, as well as rev and tat is described in
U.S. Pat. No.
5,994,136, incorporated herein by reference. One may target the recombinant
virus by
linkage of the envelope protein with an antibody or a particular ligand for
targeting to a
receptor of a particular cell-type. By inserting a sequence (including a
regulatory region) of
interest into the viral vector, along with another gene which encodes the
ligand for a receptor
on a specific target cell, for example, the vector is now target-specific.
4. Other Viral Vectors
[0155] Other viral vectors may be employed as vaccine constructs in the
present
disclosure. Vectors derived from viruses such as vaccinia virus (Ridgeway.
1988; Baichwal
and Sugden, 1986; Coupar et al., 1988), sindbis virus, cytomegalovirus and
herpes simplex
virus may be employed. They offer several attractive features for various
mammalian cells
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
47
(Friedmann, 1989; Ridgeway, 1988; Baichwal and Sugden, 1986; Coupar et at.,
1988;
Horwich et at., 1990).
5. Delivery Using Modified Viruses
[0156] A nucleic acid to be delivered may be housed within an infective virus
that has been engineered to express a specific binding ligand. The virus
particle will thus
bind specifically to the cognate receptors of the target cell and deliver the
contents to the cell.
A novel approach designed to allow specific targeting of retrovirus vectors
was developed
based on the chemical modification of a retrovirus by the chemical addition of
lactose
residues to the viral envelope. This modification can permit the specific
infection of
hepatocytes via sialoglycoprotein receptors.
[0157] Another approach to targeting of recombinant retroviruses was designed
in which biotinylated antibodies against a retroviral envelope protein and
against a specific
cell receptor were used. The antibodies were coupled via the biotin components
by using
streptavidin (Roux et at., 1989). Using antibodies against major
histocompatibility complex
class I and class II antigens, they demonstrated the infection of a variety of
human cells that
bore those surface antigens with an ecotropic virus in vitro (Roux et at.,
1989).
B. Vector Delivery and Cell Transformation
[0158] Suitable methods for nucleic acid delivery for
transformation of an
organelle, a cell, a tissue or an organism for use with the current disclosure
are believed to
include virtually any method by which a nucleic acid (e.g., DNA) can be
introduced into an
organelle, a cell, a tissue or an organism, as described herein or as would be
known to one of
ordinary skill in the art. Such methods include, but are not limited to,
direct delivery of DNA
such as by ex vivo transfection (Wilson et at., 1989, Nabel et at, 1989), by
injection (U.S.
Patent Nos. 5,994,624, 5,981,274, 5,945,100, 5,780,448, 5,736,524, 5,702,932,
5,656,610,
5,589,466 and 5,580,859, each incorporated herein by reference), including
microinjection
(Harlan and Weintraub, 1985; U.S. Patent No. 5,789,215, incorporated herein by
reference);
by electroporation (U.S. Patent No. 5,384,253, incorporated herein by
reference; Tur-Kaspa
et at., 1986; Potter et at., 1984); by calcium phosphate precipitation (Graham
and Van Der
Eb, 1973; Chen and Okayama, 1987; Rippe et at., 1990); by using DEAE-dextran
followed
by polyethylene glycol (Gopal, 1985); by direct sonic loading (Fechheimer et
at., 1987): by
liposome mediated transfection (Nicolau and Sene. 1982: Fraley et at., 1979:
Nicolau et
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
48
al., 1987; Wong et al., 1980; Kaneda et al., 1989; Kato et al., 1991) and
receptor-mediated
transfection (Wu and Wu, 1987; Wu and Wu, 1988); by microprojectile
bombardment (PCT
Application Nos. WO 94/09699 and 95/06128; U.S. Patent Nos. 5,610,042;
5,322,783
5,563,055, 5,550,318, 5,538,877 and 5,538,880, and each incorporated herein by
reference);
by agitation with silicon carbide fibers (Kaeppler et al., 1990; U.S. Patent
Nos. 5,302,523 and
5,464,765, each incorporated herein by reference); by Agrobacterium-mediated
transformation (U.S. Patent Nos. 5,591,616 and 5,563,055, each incorporated
herein by
reference); by PEG-mediated transformation of protoplasts (Omirulleh et al.,
1993; U.S.
Patent Nos. 4,684,611 and 4,952,500, each incorporated herein by reference);
by
desiccation/inhibition-mediated DNA uptake (Potrykus et al., 1985), and any
combination of
such methods. Through the application of techniques such as these,
organelle(s), cell(s),
tissue(s) or organism(s) may be stably or transiently transformed.
a. Ex vivo Transformation
[0159] Methods for tranfecting vascular cells and tissues removed
from an
organism in an ex vivo setting are known to those of skill in the art. For
example, cannine
endothelial cells have been genetically altered by retrovial gene tranfer in
vitro and
transplanted into a canine (Wilson et al., 1989). In another example, yucatan
minipig
endothelial cells were tranfected by retrovirus in vitro and transplated into
an artery using a
double-ballonw catheter (Nabel et al., 1989). Thus, it is contemplated that
cells or tissues
may be removed and tranfected ex vivo using the nucleic acids of the present
disclosure. In
particular aspects, the transplanted cells or tissues may be placed into an
organism. In
preferred facets, a nucleic acid is expressed in the transplated cells or
tissues.
b. Injection
[0160] In certain embodiments, a nucleic acid may be delivered to an
organelle,
a cell, a tissue or an organism via one or more injections (i.e., a needle
injection), such as, for
example, subcutaneously, intradermally, intramuscularly, intervenously,
intraperitoneally,
etc. Methods of injection of vaccines are well known to those of ordinary
skill in the art (e.g.,
injection of a composition comprising a saline solution). Further embodiments
of the present
disclosure include the introduction of a nucleic acid by direct
microinjection. Direct
microinjection has been used to introduce nucleic acid constructs into
Xenopu.s. oocytes
(Harland and Weintraub, 1985).
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
49
c. Electroporation
[0161] In certain embodiments of the present disclosure, a nucleic acid is
introduced into an organelle, a cell, a tissue or an organism via
electroporation.
Electroporation involves the exposure of a suspension of cells and DNA to a
high-voltage
electric discharge. In some variants of this method, certain cell wall-
degrading enzymes,
such as pectin-degrading enzymes, are employed to render the target recipient
cells more
susceptible to transformation by electroporation than untreated cells (U.S.
Patent No.
5,384,253, incorporated herein by reference). Alternatively, recipient cells
can be made more
susceptible to transformation by mechanical wounding.
[0162] Transfection of eukaryotic cells using electroporation has been
quite
successful. Mouse pre-B lymphocytes have been transfected with human
kappa-immunoglobulin genes (Potter et al., 1984), and rat hepatocytes have
been transfected
with the chloramphenicol acetyltransferase gene (Tur-Kaspa et al., 1986) in
this manner.
[0163] To effect transformation by electroporation in cells such as, for
example,
plant cells, one may employ either friable tissues, such as a suspension
culture of cells or
embryogenic callus or alternatively one may transform immature embryos or
other organized
tissue directly. In this technique, one would partially degrade the cell walls
of the chosen
cells by exposing them to pectin-degrading enzymes (pectolyases) or
mechanically wounding
in a controlled manner. Examples of some species which have been transfaimed
by
electroporation of intact cells include maize (U.S. Patent No. 5,384,253;
Rhodes etal., 1995;
D'Halluin et al., 1992), wheat (Zhou et al., 1993), tomato (Hou and Lin,
1996), soybean
(Christou et al., 1987) and tobacco (Lee et al., 1989).
[0164] One also may employ protoplasts for electroporation transformation of
plant cells (Bates, 1994; Lazzeri, 1995). For example, the generation of
transgenic soybean
plants by electroporation of cotyledon-derived protoplasts is described by
Dhir and Widholm
in International Patent Application No. WO 9217598, incorporated herein by
reference.
Other examples of species for which protoplast transformation has been
described include
barley (Lazerri, 1995), sorghum (Battraw et al., 1991). maize (Bhattacharjee
et al., 1997),
wheat (He et al., 1994) and tomato (Tsukada, 1989).
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
d. Calcium Phosphate
[0165] In
other embodiments of the present disclosure, a nucleic acid is
introduced to the cells using calcium phosphate precipitation. Human KB cells
have been
transfected with adenovirus 5 DNA (Graham and Van Der Eb, 1973) using this
technique.
Also in this manner, mouse L(A9), mouse C127, CHO, CV-1, BHK, NIH3T3 and HeLa
cells
were transfected with a neomycin marker gene (Chen and Okayama, 1987), and rat
hepatocytes were transfected with a variety of marker genes (Rippe etal.,
1990).
e. DEAE-Dextran
[0166] In
another embodiment, a nucleic acid is delivered into a cell using
DEAE-dextran followed by polyethylene glycol. In this manner, reporter
plasmids were
introduced into mouse myeloma and erythroleukemia cells (Gopal, 1985).
f. Sonication Loading
[0167]
Additional embodiments of the present disclosure include the
introduction of a nucleic acid by direct sonic loading. LTK- fibroblasts have
been transfected
with the thymidine kinase gene by sonication loading (Fechheimer etal., 1987).
g. Liposome-Mediated Transfection
[0168] In
a further embodiment of the disclosure, a nucleic acid may be
entrapped in a lipid complex such as, for example, a liposome. Liposomes are
vesicular
structures characterized by a phospholipid bilayer membrane and an inner
aqueous medium.
Multilamellar liposomes have multiple lipid layers separated by aqueous
medium. They form
spontaneously when phospholipids are suspended in an excess of aqueous
solution. The lipid
components undergo self-rearrangement before the formation of closed
structures and entrap
water and dissolved solutes between the lipid bilayers (Ghosh and Bachhawat,
1991). Also
contemplated is an nucleic acid complexed with Lipofectamine (Gibco BRL) or
Superfect
(Qiagen).
[0169]
Liposome-mediated nucleic acid delivery and expression of foreign
DNA in vitro has been very successful (Nicolau and Sene. 1982: Fraley et al.,
1979;
Nicolau et al., 1987). The feasibility of liposome-mediated delivery and
expression of
foreign DNA in cultured chick embryo, HeLa and hepatoma cells has also been
demonstrated
(Wong etal., 1980).
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
51
[0170] In certain embodiments of the disclosure, a liposome may be complexed
with a hemagglutinating virus (HVJ). This has been shown to facilitate fusion
with the cell
membrane and promote cell entry of liposome-encapsulated DNA (Kaneda et al.,
1989). In
other embodiments, a liposome may be complexed or employed in conjunction with
nuclear
non-histone chromosomal proteins (HMG-1) (Kato et al., 1991). In yet further
embodiments,
a liposome may be complexed or employed in conjunction with both HVJ and HMG-
1. In
other embodiments, a delivery vehicle may comprise a ligand and a liposome.
h. Receptor Mediated Transfection
[0171]
Still further, a nucleic acid may be delivered to a target cell via
receptor-mediated delivery vehicles. These take advantage of the selective
uptake of
macromolecules by receptor-mediated endocytosis that will be occurring in a
target cell. In
view of the cell type-specific distribution of various receptors, this
delivery method adds
another degree of specificity to the present disclosure.
[0172]
Certain receptor-mediated gene targeting vehicles comprise a cell
receptor-specific ligand and a nucleic acid-binding agent.
Others comprise a cell
receptor-specific ligand to which the nucleic acid to be delivered has been
operatively
attached. Several ligands have been used for receptor-mediated gene transfer
(Wu and Wu,
1987; Wagner et al., 1990; Perales et al., 1994; Myers, EPO 0273085), which
establishes the
operability of the technique. Specific delivery in the context of another
mammalian cell type
has been described (Wu and Wu, 1993; incorporated herein by reference). In
certain aspects
of the present disclosure, a ligand will be chosen to correspond to a receptor
specifically
expressed on the target cell population.
[0173] In other embodiments, a nucleic acid delivery vehicle component of a
cell-specific nucleic acid targeting vehicle may comprise a specific binding
ligand in
combination with a liposome. The nucleic acid(s) to be delivered are housed
within the
liposome and the specific binding ligand is functionally incorporated into the
liposome
membrane. The liposome will thus specifically bind to the receptor(s) of a
target cell and
deliver the contents to a cell. Such systems have been shown to be functional
using systems
in which, for example, epidermal growth factor (EGF) is used in the receptor-
mediated
delivery of a nucleic acid to cells that exhibit upregulation of the EGF
receptor.
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
52
[0174] In still further embodiments, the nucleic acid delivery
vehicle
component of a targeted delivery vehicle may be a liposome itself, which will
preferably
comprise one or more lipids or glycoproteins that direct cell-specific
binding. For example,
lactosyl-ceramide, a galactose-terminal asialganglioside, have been
incorporated into
liposomes and observed an increase in the uptake of the insulin gene by
hepatocytes
(Nicolau et at., 1987). It is contemplated that the tissue-specific
transforming constructs of
the present disclosure can be specifically delivered into a target cell in a
similar manner.
i. Microprojectile Bombardment
[0175] Microprojectile bombardment techniques can be used to
introduce a
nucleic acid into at least one, organelle, cell, tissue or organism (U.S.
Patent No. 5,550,318;
U.S. Patent No. 5,538,880; U.S. Patent No. 5,610,042; and PCT Application WO
94/09699;
each of which is incorporated herein by reference). This method depends on the
ability to
accelerate DNA-coated microprojectiles to a high velocity allowing them to
pierce cell
membranes and enter cells without killing them (Klein et at., 1987). There are
a wide variety
of microprojectile bombardment techniques known in the art, many of which are
applicable
to the disclosure.
[0176] Microprojectile bombardment may be used to transform various cell(s),
tissue(s) or organism(s), such as for example any plant species. Examples of
species which
have been transformed by microprojectile bombardment include monocot species
such as
maize (PCT Application WO 95/06128), barley (Ritala et at., 1994; Hensgens et
at., 1993),
wheat (U.S. Patent No. 5,563,055, incorporated herein by reference), rice
(Hensgens et
at., 1993), oat (Torbet et at., 1995; Torbet et at., 1998), rye (Hensgens et
at., 1993),
sugarcane (Bower et at., 1992), and sorghum (Casas et at., 1993; Hagio et at.,
1991); as well
as a number of dicots including tobacco (Tomes et at., 1990; Buising and
Benbow, 1994),
soybean (U.S. Patent No. 5,322,783, incorporated herein by reference),
sunflower (Knittel et
at. 1994), peanut (Singsit et at., 1997), cotton (McCabe and Martinell, 1993),
tomato
(VanEck et at. 1995), and legumes in general (U.S. Patent No. 5,563,055,
incorporated herein
by reference).
[0177] In this microprojectile bombardment, one or more particles
may be
coated with at least one nucleic acid and delivered into cells by a propelling
force. Several
devices for accelerating small particles have been developed. One such device
relies on a
high voltage discharge to generate an electrical current, which in turn
provides the motive
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
53
force (Yang et at., 1990). The microprojectiles used have consisted of
biologically inert
substances such as tungsten or gold particles or beads. Exemplary particles
include those
comprised of tungsten, platinum, and preferably, gold. It is contemplated that
in some
instances DNA precipitation onto metal particles would not be necessary for
DNA delivery to
a recipient cell using microprojectile bombardment. However, it is
contemplated that
particles may contain DNA rather than be coated with DNA. DNA-coated particles
may
increase the level of DNA delivery via particle bombardment but are not, in
and of
themselves, necessary.
[0178] For the bombardment, cells in suspension are concentrated on filters or
solid culture medium. Alternatively, immature embryos or other target cells
may be arranged
on solid culture medium. The cells to be bombarded are positioned at an
appropriate distance
below the macroprojectile stopping plate.
[0179] An illustrative embodiment of a method for delivering DNA into a cell
(e.g., a plant cell) by acceleration is the Biolistics Particle Delivery
System, which can be
used to propel particles coated with DNA or cells through a screen, such as a
stainless steel or
Nytex screen, onto a filter surface covered with cells, such as for example, a
monocot plant
cells cultured in suspension. The screen disperses the particles so that they
are not delivered
to the recipient cells in large aggregates. It is believed that a screen
intervening between the
projectile apparatus and the cells to be bombarded reduces the size of
projectiles aggregate
and may contribute to a higher frequency of transformation by reducing the
damage inflicted
on the recipient cells by projectiles that are too large.
C. Host Cells
[0180] As used herein, the terms "cell," "cell line," and "cell
culture" may be
used interchangeably. All of these terms also include their progeny, which is
any and all
subsequent generations. It is understood that all progeny may not be identical
due to
deliberate or inadvertent mutations. In the context of expressing a
heterologous nucleic acid
sequence, "host cell" refers to a prokaryotic or eukaryotic cell, and it
includes any
transformable organism that is capable of replicating a vector and/or
expressing a
heterologous gene encoded by a vector. A host cell can, and has been, used as
a recipient for
vectors. A host cell may be "transfected- or "transformed," which refers to a
process by
which exogenous nucleic acid is transferred or introduced into the host cell.
A transformed
cell includes the primary subject cell and its progeny. As used herein, the
terms "engineered"
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
54
and "recombinant" cells or host cells are intended to refer to a cell into
which an exogenous
nucleic acid sequence, such as, for example, a vector, has been introduced.
Therefore,
recombinant cells are distinguishable from naturally occurring cells which do
not contain a
recombinantly introduced nucleic acid.
[0181] In certain embodiments, it is contemplated that RNAs or proteinaceous
sequences may be co-expressed with other selected RNAs or proteinaceous
sequences in the
same host cell. Co-expression may be achieved by co-transfecting the host cell
with two or
more distinct recombinant vectors. Alternatively, a single recombinant vector
may be
constructed to include multiple distinct coding regions for RNAs, which could
then be
expressed in host cells transfected with the single vector.
[0182] A tissue may
comprise a host cell or cells to be transformed with a
polynucleotide encoding part or all of GLP-1. The tissue may be part or
separated from an
organism. In certain embodiments, a tissue may comprise, but is not limited
to, myocytes,
adipocytes, alveolar, ameloblasts, axon, basal cells, blood (e.g.,
lymphocytes), blood vessel,
bone, bone marrow, brain, breast, cartilage, cervix, colon, cornea, embryonic,
endometrium,
endothelial, epithelial, esophagus, facia, fibroblast, follicular, ganglion
cells, glial cells,
goblet cells, kidney, liver, lung, lymph node, muscle, neuron, ovaries,
pancreas, peripheral
blood, prostate, skin, skin, small intestine, spleen, stem cells, stomach,
testes, anthers, ascite
tissue, cobs, ears, flowers, husks, kernels, leaves, meristematic cells,
pollen, root tips, roots,
silk, stalks, and all cancers thereof.
[0183] In certain embodiments, the host cell or tissue may be comprised in at
least one organism. In certain embodiments, the organism may be, but is not
limited to, a
prokayote (e.g., a eubacteria, an archaea) or an eukaryote, as would be
understood by one of
ordinary skill in the art (see, for
example, webpage
http://phylogeny.arizona.edu/tree/phylogeny.html).
[0184] Numerous cell lines and cultures are available for use as a host cell,
and
they can be obtained through the American Type Culture Collection (ATCC),
which is an
organization that serves as an archive for living cultures and genetic
materials
(www.atcc.org). An appropriate host can be determined by one of skill in the
art based on the
vector backbone and the desired result. A plasmid or cosmid, for example, can
be introduced
into a prokaryote host cell for replication of many vectors. Cell types
available for vector
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
replication and/or expressioninclude, but are not limited to, bacteria, such
as E. coli (e.g.,
E. coli strain RR1, E. coli LE392, E. coli B, E. coli X 1776 (ATCC No. 31537)
as well as E.
coli W3110 (F-, lambda-, prototrophic, ATCC No. 273325), DH5oc, JM109, and
KC8, bacilli
such as Bacillus subtilis; and other enterobacteriaceae such as Salmonella
typhimurium,
Serratia marcescens, various Pseudomonas specie, as well as a number of
commercially
available bacterial hosts such as SURE Competent Cells and SOLOPACKTM Gold
Cells
(STRATAGENE , La Jolla). In certain embodiments, bacterial cells such as E.
coli LE392 are
particularly contemplated as host cells for phage viruses.
[0185] Examples of eukaryotic host cells for replication and/or expression of
a
vector include, but are not limited to, HeLa, NIH3T3, Jurkat, 293, Cos, CHO,
Saos, and
PC12. Many host cells from various cell types and organisms are available and
would be
known to one of skill in the art. Similarly, a viral vector may be used in
conjunction with
either a eukaryotic or prokaryotic host cell, particularly one that is
permissive for replication
or expression of the vector.
[0186]
Some vectors may employ control sequences that allow it to be
replicated and/or expressed in both prokaryotic and eukaryotic cells. One of
skill in the art
would further understand the conditions under which to incubate all of the
above described
host cells to maintain them and to permit replication of a vector. Also
understood and known
are techniques and conditions that would allow large-scale production of
vectors, as well as
production of the nucleic acids encoded by vectors and their cognate
polypeptides, proteins,
or peptides.
D. Expression Systems
[0187] Numerous expression systems exist that comprise at least a part or all
of
the compositions discussed above. Prokaryote- and/or eukaryote-based systems
can be
employed for use with the present disclosure to produce nucleic acid
sequences, or their
cognate polypeptides, proteins and peptides. Many such systems are
commercially and
widely available.
[0188] The
insect cell/baculovirus system can produce a high level of protein
expression of a heterologous nucleic acid segment, such as described in U.S.
Patent No.
5,871,986, 4,879,236, both herein incorporated by reference, and which can be
bought, for
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
56
example, under the name MAxBAc 2.0 from INVITROGEN and BAcPAcKTM BACULOVIRUS
EXPRESSION SYSTEM FROM CLONTECH .
[0189]
Other examples of expression systems include STRATAGENE S
COMPLETE CONTROLTm Inducible Mammalian Expression System, which involves a
synthetic
ecdysone-inducible receptor, or its pET Expression System, an E. coli
expression system.
Another example of an inducible expression system is available from INVITROGEN
, which
carries the T-RExTm (tetracycline-regulated expression) System, an inducible
mammalian
expression system that uses the full-length CMV promoter. INVITROGEN also
provides a
yeast expression system called the Pichia methanolica Expression System, which
is designed
for high-level production of recombinant proteins in the methylotrophic yeast
Pichia
methanolica. One of skill in the art would know how to express a vector, such
as an
expression construct, to produce a nucleic acid sequence or its cognate
polypeptide, protein,
or peptide.
[0190] It
is contemplated that the proteins, polypeptides or peptides produced
by the methods of the disclosure may be "overexpressed", i.e., expressed in
increased levels
relative to its natural expression in cells. Such overexpression may be
assessed by a variety
of methods, including radio-labeling and/or protein purification. However,
simple and direct
methods are preferred, for example, those involving SDS/PAGE and protein
staining or
western blotting, followed by quantitative analyses, such as densitometric
scanning of the
resultant gel or blot. A specific increase in the level of the recombinant
protein, polypeptide
or peptide in comparison to the level in natural cells is indicative of
overexpression, as is a
relative abundance of the specific protein, polypeptides or peptides in
relation to the other
proteins produced by the host cell and, e.g., visible on a gel.
[0191] In some embodiments, the expressed proteinaceous sequence forms an
inclusion body in the host cell, the host cells are lysed, for example, by
disruption in a cell
homogenizer, washed and/or centrifuged to separate the dense inclusion bodies
and cell
membranes from the soluble cell components. This centrifugation can be
performed under
conditions whereby the dense inclusion bodies are selectively enriched by
incorporation of
sugars, such as sucrose, into the buffer and centrifugation at a selective
speed. Inclusion
bodies may be solubilized in solutions containing high concentrations of urea
(e.g. 8M) or
chaotropic agents such as guanidine hydrochloride in the presence of reducing
agents, such as
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
57
13-mercaptoethanol or DTT (dithiothreitol), and refolded into a more desirable
conformation,
as would be known to one of ordinary skill in the art.
E. Proteins, Polypeptides, and Peptides
[0192] In
some cases, embodiments may utilize purified GLP-1 proteins,
polypeptides, or peptides. The term "purified proteins, polypeptides, or
peptides" as used
herein, is intended to refer to an proteinaceous composition, isolatable from
mammalian cells
or recombinant host cells, wherein the at least one protein, polypeptide, or
peptide is purified
to any degree relative to its naturally-obtainable state, i.e., relative to
its purity within a
cellular extract. A purified protein, polypeptide, or peptide therefore also
refers to a
wild-type or mutant protein, polypeptide, or peptide free from the environment
in which it
naturally occurs.
[0193] The
nucleotide and protein, polypeptide and peptide sequences for
various genes have been previously disclosed, and may be found at computerized
databases
known to those of ordinary skill in the art. One such database is the National
Center for
Biotechnology Information's GenBank and GenPept0 databases. The coding
regions for
these known genes may be amplified and/or expressed using the techniques
disclosed herein
or by any technique that would be known to those of ordinary skill in the art.
Additionally,
peptide sequences may be sythesized by methods known to those of ordinary
skill in the art,
such as peptide synthesis using automated peptide synthesis machines, such as
those
available from Applied Biosystems (Foster City, CA).
[0194]
Generally, "purified" will refer to a specific protein, polypeptide, or
peptide composition that has been subjected to fractionation to remove various
other proteins,
polypeptides, or peptides, and which composition substantially retains its
activity, as may be
assessed, for example, by the protein assays, as described herein below, or as
would be
known to one of ordinary skill in the art for the desired protein, polypeptide
or peptide.
[0195]
Where the term "substantially purified" is used, this will refer to a
composition in which the specific protein, polypeptide, or peptide forms the
major
component of the composition, such as constituting about 50% of the proteins
in the
composition or more. In preferred embodiments, a substantially purified
protein will
constitute more than 60%, 70%, 80%, 90%, 95%, 99% or even more of the proteins
in the
composition.
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
58
[0196] A
peptide, polypeptide or protein that is "purified to homogeneity," as
applied to the present disclosure, means that the peptide, polypeptide or
protein has a level of
purity where the peptide, polypeptide or protein is substantially free from
other proteins and
biological components. For example, a purified peptide, polypeptide or protein
will often be
sufficiently free of other protein components so that degradative sequencing
may be
performed successfully.
[0197] Various methods for quantifying the degree of purification of proteins,
polypeptides, or peptides will be known to those of skill in the art in light
of the present
disclosure. These include, for example, determining the specific protein
activity of a fraction,
or assessing the number of polypeptides within a fraction by gel
electrophoresis.
[0198] To
purify a desired protein, polypeptide, or peptide a natural or
recombinant composition comprising at least some specific proteins,
polypeptides, or
peptides will be subjected to fractionation to remove various other components
from the
composition. In addition to those techniques described in detail herein below,
various other
techniques suitable for use in protein purification will be well known to
those of skill in the
art. These include, for example, precipitation with ammonium sulfate, PEG,
antibodies and
the like or by heat denaturation, followed by centrifugation; chromatography
steps such as
ion exchange, gel filtration, reverse phase, hydroxylapatite, lectin affinity
and other affinity
chromatography steps; isoelectric focusing; gel electrophoresis; and
combinations of such
and other techniques.
[0199] Another example is the purification of a specific fusion protein using
a
specific binding partner. Such purification methods are routine in the art. As
the present
disclosure provides DNA sequences for the specific proteins, any fusion
protein purification
method can now be practiced. This is exemplified by the generation of an
specific
protein-glutathione S-transferase fusion protein, expression in E. coli, and
isolation to
homogeneity using affinity chromatography on glutathione-agarose or the
generation of a
polyhistidine tag on the N- or C-terminus of the protein, and subsequent
purification using
Ni-affinity chromatography. However, given many DNA and proteins are known, or
may be
identified and amplified using the methods described herein, any purification
method can
now be employed.
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
59
[0200] Although preferred for use in certain embodiments, there is no general
requirement that the protein, polypeptide, or peptide always be provided in
their most
purified state. Indeed, it is contemplated that less substantially purified
protein, polypeptide
or peptide, which are nonetheless enriched in the desired protein
compositions, relative to the
natural state, will have utility in certain embodiments.
[0201] Methods exhibiting a lower degree of relative purification
may have
advantages in total recovery of protein product, or in maintaining the
activity of an expressed
protein. Inactive products also have utility in certain embodiments, such as,
e.g., in
deteimining antigenicity via antibody generation.
VII. Combination Therapy
[0202] In certain cases, the GLP-1/UTMD therapy of the present
disclosure
(which may also include TB4) is utilized in conjunction with one or more other
therapies for
a cardiac-related medical condition. GLP-1/UTMD may also be used in
combination with
other genes or gene products, including TB4 (in peptide, protein, or nucleic
acid form). The
one or more other therapies may be directly or indirectly related to the
cardiac-related
medical condition (examples of indirectly related therapies include those for
pain or
infection).
[0203] The GLP-1/UTMD therapy may precede or follow the other agent
treatment by intervals ranging from minutes to to hours to days to weeks or
months. In
embodiments where the other agent and the GLP-1/UTMD therapy are applied
separately to
the individual, one would generally ensure that a significant period of time
did not expire
between the time of each delivery, such that the agent and GLP-1/UTMD therapy
would still
be able to exert an advantageously combined effect on the cell. In such
instances, it is
contemplated that one may contact the individual with both modalities
simultaneously or
within minutes of each other or within about 1-12, 6-12, or 12-24 h of each
other. In some
situations, it may be desirable to extend the time period for treatment
significantly, however,
where several days (2, 3, 4, 5, 6 or 7) to several weeks (1, 2, 3, 4, 5, 6, 7
or 8) lapse between
the respective administrations.
[0204] Yet further, GLP-1/UTMD may also include other nucleic acids
or
peptides, for example, but not limited to TB4. In such instances, GLP-1/TB4
UTMD therapy
may also be used in combination may precede or follow the other agent
treatment by intervals
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
ranging from minutes to weeks or months. In embodiments where the other agent
and the
GLP-1/TB4/UTMD therapy are applied separately to the individual, one would
generally
ensure that a significant period of time did not expire between the time of
each delivery, such
that the agent and GLP-1/TB4/UTMD therapy would still be able to exert an
advantageously
combined effect on the cell. In such instances, it is contemplated that one
may contact the
individual with both modalities simultaneously or within minutes of each other
or within
about 1-12, 6-12, or 12-24 h of each other. In some situations, it may be
desirable to extend
the time period for treatment significantly, however, where several days (2,
3, 4, 5, 6 or 7) to
several weeks (1, 2, 3, 4, 5, 6, 7 or 8) lapse between the respective
administrations.
[0205] In specific embodiments, GLP-1/UTMD therapy and TB4/UTMD
therapy are provided at the same time or at different times. The GLP-1 and TB4
entities may
be within the same plurality of microbubbles or they may be comprised in
separate pluralities
of microbubbles. In cases wherein the GLP-1/UTMD therapy and TB4/UTMD therapy
are
provided at different times, they may be separated by any suitable range in
times, such as
minutes, hours, days, or weeks. In embodiments wherein they are provided
separately, the
order of delivery of GLP-1/UTMD therapy and TB4/UTMD therapy may be of any
suitable
order, including delivery of GLP-1/UTMD prior to or subsequent to TB4/UTMD
therapy. In
cases wherein GLP-1/UTMD therapy and TB4/UTMD therapy are provided to an
individual
in need thereof, they may be provided with yet another therapy for a cardiac-
related medical
condition.
[0206] Examples of other treatments to be employed with the GLP-1/UTMD
and/or GLP-1/TB4/UTMD therapy of the disclosure includes one or more of the
following:
ACE Inhibitors, Aldosterone Inhibitor, Angiotensin II Receptor Blocker (ARBs);
Beta-
Blockers, Calcium Channel Blockers, Cholesterol-Lowering Drugs, Digoxin,
Diuretics,
Inotropic Therapy, Potassium or Magnesium, Vasodilators, anticoagulant
medication, aspirin,
or a combination thereof
VIII. Kits of the Disclosure
[0207] Any of the compositions described herein may be comprised in a kit. In
a non-limiting example, a GLP-1 and/or TB4 polynucleotide or primers for
amplification of it
may be comprised in a kit. In specific embodiments, the kit comprises GLP-1
and/or TB4
peptides. The kit may alternatively or additionally comprise reagents for
generating
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
61
liposomes and/or microbubbles and optionally may have additional agents for
therapy of a
cardiac-related medical condition.
[0208] The components of the kits may be packaged either in aqueous media or
in lyophilized form. The container means of the kits will generally include at
least one vial,
test tube, flask, bottle, syringe or other container means, into which a
component may be
placed, and preferably, suitably aliquoted. Where there are more than one
component in the
kit, the kit also will generally contain a second, third or other additional
container into which
the additional components may be separately placed. However, various
combinations of
components may be comprised in a vial. The kits of the present disclosure also
will typically
include a means for containing the one or more compositions in close
confinement for
commercial sale. Such containers may include injection or blow-molded plastic
containers
into which the desired vials are retained.
[0209] The composition may be formulated into a syringeable composition. In
which case, the container means may itself be a syringe, pipette, and/or other
such like
apparatus, from which the foimulation may be applied to an infected area of
the body,
injected into an animal, and/or even applied to and/or mixed with the other
components of the
kit. However, the components of the kit may be provided as dried powder(s).
When reagents
and/or components are provided as a dry powder, the powder can be
reconstituted by the
addition of a suitable solvent. It is envisioned that the solvent may also be
provided in
another container means.
[0210] The kits of the present disclosure will also typically include a means
for
containing the vials in close confinement for commercial sale, such as, e.g.,
injection and/or
blow-molded plastic containers into which the desired vials are retained.
[0211] In
particular embodiments, the kit comprises reagents and/or tools for
determining that an individual has a cardiac-related medical condition. In
some
embodiments, the kit comprises one or more additional therapies for a cardiac-
related
medical condition, such as one or more of ACE Inhibitor, aldosterone
inhibitor, angiotensin II
receptor blocker (ARBs); beta-blocker, calcium channel blocker, cholesterol-
lowering drug,
digoxin, diuretics, inotropic therapy, potassium, magnesium, vasodilator,
anticoagulant
medication, aspirin, and a combination thereof.
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
62
EXAMPLES
[0212] The
following examples are included to demonstrate preferred
embodiments of the disclosure. It should be appreciated by those of skill in
the art that the
techniques disclosed in the examples which follow present techniques
discovered by the
inventors to function well in the practice of the disclosure, and thus can be
considered to
constitute preferred modes for its practice. However, those of skill in the
art should, in light
of the present disclosure, appreciate that many changes can be made in the
specific
embodiments which are disclosed and still obtain a like or similar result
without departing
from the spirit and scope of the disclosure.
EXAMPLE 1
EXEMPLARY MATERIALS AND METHODS
Animal protocols and UTMD
[0213] All animal studies were performed in accordance with National Institute
of Health (NIH) recommendations and the approval of our institutional animal
research
committee. Male Sprague¨Dawley rats (230-270g) were anesthetized with
intraperitoneal
ketamine (60 mg/kg) and xylazine (5 mg/kg), and a polyethylene tube (PE 50,
Becton
Dickinson, Franklin Lakes, TN, USA) was inserted into the right internal
jugular vein by cut-
down.
[0214]
Animal protocol-1: A total of 18 normal rats received one of 3
treatments: (1) no treatment (normal control rats, n=6); (2) UTMD with pXL-
BASII-CI-
DsRed/pCI-hyPB (n=6); (3) UTMD with pXL-BASII-CI-GLP-1/pCI-hyPB (n=6), all
were
killed at 4 weeks after UTMD. The thymidine analog, 5-bromo-2-deoxyuridine
(BrdU)
(100mg/kg, Sigma, St Louis, MO) intraperitoneal injected 6 hours prior to
killing.
[0215] Animal protocol-2: A total of 60 rats were divided into: (1) no
treatment
(normal control rats, n=12); (2) adriamycin (ADM)38 injection only, at total
dose of 15
mg/kg/ip, 2.5mg/kg/ip 6 times over 2 weeks, n=12. (3) ADM plus GLP-1 peptide
injection, at
GLP-1 peptide fragment 7-37 human (dose of 50 nmol/kg/ip/time x 6, N=12, from
Sigma, St
Louis, MO), (4) ADM injection plus UTMD-GLP1 peptide delivery (same dose of
GLP-1
peptide fragment 7-37 human mixed with microbubble for UTMD). n=12: (5) ADM
injection
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
63
plus UTMD with pXL-BASII-CI-GLP-1/pCI-hyPB, n=12. Half rats were killed at 16
hours
after last ADM injection; other half rats were killed at 4 weeks after UTMD.
The thymidine
analog, 5-bromo-2-deoxyuridine (BrdU) (100mg/kg) was injected
intraperitoneally 6 hours
prior to euthanasia.
[0216] Animal protocol-3: UTMD was performed after established adriamycin
cardiomyopathy defined as a fractional shortening < 30% by echocardiography.
There were
3 groups of rats as follows: (1) normal control rats, n=6; (2) UTMD with pXL-
BASII-CI-
DsRed/pCI-hyPB (n=6); (3) UTMD with pXL-BASII-CI-GLP-1/pCI-hyPB (n=12). All
rats
were killed 4 weeks after UTMD. The thymidine analog, 5-bromo-2-deoxyuridine
(BrdU)
(100mg/kg) was injected intraperitoneally 6 hours prior to euthanasia.
[0217] Animal protocol-4: Specific FOX01 inhibitor (AS1842856, a
small
molecular compound from Millipore) was injected into rats with ADM
cardiomyopathy to
investigate if FOX01 mediates ADM cardiomyopathy. There were 3 groups of rats
as
follows: (1) normal control rats, n=6; (2) ADM injection only group at total
dose of 15
mg/kg/ip, 2.5mg/kg/ip 6 times over 2 weeks, (n=6); (3) ADM at total dose of 15
mg/kg/ip,
2.5mg/kg/ip 6 times over 2 weeks plus FOX01 inhibitor injection (at total dose
of 24
mg/kg/ip, 4 mg/kg/ip 6 times over 2 weeks (n=6). All rats were killed 4 weeks
after ADM
injection.
[0218] Piggybac transposon donor plasmids and helper plasmids ratio
(pXL-
BASII-CI-GLP-1 /pCI-hyPB) was 5:1. Microbubble or control solutions (0.5 ml
diluted with
0.5 ml phosphate-buffered solution (PBS)) were infused over 5 min via pump
(Genie, Kent
Scientific, Torrington, CT). During the infusion, ultrasound was directed to
the heart using a
commercially available ultrasound transducer (S3, Sonos 5500, Philips
Ultrasound, Bothell,
WA). A 2D echocardiographic view of the left ventricle was obtained in a short-
axis view
and the probe was clamped in placed. Ultrasound was then applied in
ultraharmonic mode
(transmit 1.3 MHz/receive 3.6 MHz) at a mechanical index of 1.4. Four bursts
of ultrasound
were triggered to every fourth end-systole by electrocardiogram using a delay
of 45-70 ms
after the peak of the R wave. These settings have shown to be optimal for
plasmid delivery by
UTMD using this instrument. Bubble destruction was visually apparent in all
rats. After
UTMD, the jugular vein was tied off, the skin closed, and the animals allowed
to recover. All
of rats were euthanized using an overdose of sodium pentobarbital (120 mg/kg).
Hearts were
harvested for histology, western blots and qRT-PCR assay.
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
64
RNA isolation and qRT¨PCR analysis
[0219] Total RNA was isolated from 100 mg of harvested heart using the Trizol
reagent (Invitrogen), according to the manufacturer's instructions and reverse-
transcribed
using Superscript III RT (Invitrogen). Real-time RT¨qPCR analysis was
performed on an
ABI 7700 Sequence Detector (Applied Biosystems) using SYBR Green (RT2 SYBR
Green
qPCR Kit, Qiagen). Data were normalized to HKG expression (endogenous
control).
Changes in gene expression were normalized to control rat heart tissue.
Complementary
DNA PCR primer sequences information should be requested to the correspondence
author.
Immunohistochemistry
[0220] Tissue samples were fixed in 10% formalin for 24 hours and transferred
into 70% alcohol for paraffin embedding and 4% paraformaldehyde and 20%
sucrose
overnight at 4 C for frozen sections. Cryostat sections 5-8 ni in thickness
were further fixed
with acetone (-20 C) for 5 min and quenched for 5-20 min with 10 mM glycine in
PBS.
Sections were then rinsed in PBS 3 times, and permeabilized with 0.5% Triton X-
100 in PBS
for 15 min. The slides that needed further nuclear protein retrieval were
subjected to boiling
citrate buffer solution with tween 20 at pH 6.0 for 5 minutes. Sections were
blocked with
20% Aquablock solution (EastCoast Bio, North Berwick, ME) at room temperature
for 1 hr
and washed with PBS 1 time. The primary antibodies rabbit anti-NKX2.5, 1: 500
dilution,
and rabbit anti-ISL-1, 1: 500 dilution, and mouse anti-cardiac troponin T, 1:
250 dilution,
rabbit anti-phosopho-histone H3, 1: 500 dilution, rabbit anti Ki-67, 1: 500
dilution, rabbit
anti-BrDu, 1:200 dilution, rabbit anti-Aurora B, 1:200 dilution, mouse anti-
GLP-1, 1:250
dilution, rabbit anti-GLP-1,1:250 dilution, rabbit anti-topoisomerase IIa, 1:
50 dilution, rabbit
anti-topoisomerase II3, 1: 200 dilution, rabbit anti-FOX01, 1:300 dilution,
rabbit anti-cyclin
D1, 1:300 dilution, rabbit anti-c-kit, 1:300 dilution, rabbit anti-OCT4, 1:250
dilution, rabbit
anti-Nanog, 1:250 dilution, rabbit anti-S0X2, 1:500 dilution, (Abeam Inc,
Cambridge, MA),
anit-rabbit Smooth muscle actin-alpha , 1:500 dilution (Sigma, St. Louis. MO),
anti-rabbit
von Willebrand Factor, 1:200 dilution (Dako, Carpinteria, CA) (Abeam Inc,
Cambridge,
MA), were added and incubated for 2 hrs at RT. After washing with PBS three
times for 5
min, the secondary antibody (Sigma, St; Louis, MO) anti-mouse lgG conjugated
with FITC;
anti-rabbit IgG-conjugated with Texas Red, or anti-donkey lgG conjugated with
Cy5) (1:250
dilution in block solution) were added and incubated for 1 hr at RT. Sections
were rinsed
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
with PBS for 10 min, 3 times, and incubated with Dapi (Invitrogen, Carlsbad,
CA), 1: 5000
dilution for 5 min and washed 3 times with PBST, then mounted. A confocal
microscope was
used to take pictures. BrdU staining included with incubating in HC1 (IN) for
10 minutes on
ice to break open the DNA structure of the labeled cells and then followed by
HC1 (2N) for
10 minutes at room temperature before moving them to an incubator for 20
minutes at 37 C,
Immediately after the acid washes, Borate buffer (0.1M) is added to buffer the
cells for 12
minutes.
Culture of HL-1 Cardiac Muscle Cells line.
[0221] The HL-1 cell line was a generous gift from Dr. William C. Claycomb 40
in Louisiana State University Medical Center, New Orleans, LA. The cells were
maintained
in Claycomb basal medium (Sigma) supplemented with 10% fetal bovine serum, 0.1
mM
norepinephrine and 2 mM L-glutamine. HL-1 cells were treated with adriamycin
at 0 uM,
0.25 uM, 0.50 uM, or 1.00 uM for 48 hrs in complete growth medium, and were
subjected to
immunofluorescent staining.
Western blotting
[0222] Nuclear proteins extracts from cardiac tissue with a NE-PER Nuclear
and Cytoplasmic Extraction Kit (Thermo Scientific, Rockford, IL 61105, USA).
Protein
concentrations were determined using the BCA-200 Protein Assay kit (Pierce);
equal
amounts of protein were separated by SDS-PAGE. After separation and transfer
to
nitrocellulose membranes, the membranes were incubated with primary antibodies
(Abeam)
to GLP-1 (1:1,000 dilution), anti-topoisomerase Ha(1:1,000 dilution), anti-
topoisomerase
1113(1:2,000 dilution), anti-cyclin D1(1:5000 dilution), anti-Fox01(1:5000
dilution), anti-
lamin (1:5,000 dilution; Sigma-Aldrich). Horseradish peroxidase secondary
antibodies were
used, and chemiluminescence was determined using the Super Signal West Dura
detection
system (Pierce); nuclear marker (lamin) was used to confirm equal loading. All
Western blots
were performed in duplicate.
Plasmid Construction
[0223] The piggybac transposon plasmids (pXL-BSII donor plasmid)
was
provided by Dr. Fraser at University of Notre Dame (Notre Dame, IN) and mouse
piggybac
transposase helper plasmid was provided by Dr. Bradley at Wellcome Trust
Sanger Institute
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
66
(Cambridge, England). Human GLP-1 cDNA with a furin cutting site after first
ATG were
constructed. DsRed cDNA vector was purchased from Clontech (Mountain view,
CA), GLP-
1 cDNAs were cloned to pCI vector (Promega, Madison, WI), hyperative piggybac
transposase cDNA with EcoRl/NOT1 cutting was also subcloned into pCI vector
and then
GLP-1 or DsRed cDNA with CMV promoter and polyA fragment with BgL II/BamH1
cutting was subcloned to pXL-BSII vector. The plasmids digestion, ligation,
subcloning,
isolation and purification were performed by standard procedures, and once
again sequenced
to confirm that no artifactual mutations were present. GLP-1 constructs: GLP-1
cDNA(7-
37) with a furin cutting site after first ATG were constructed. A nuclear
localization signal
(NLS) fragment was fused to GLP-1 cDNA (7-37).GLP-1NLS forward primers: 5-AAA-
CTC-GAG-ATG-CGT-CAA-CGT-CGT-CAT-GCT-GAA-GGG-ACC-TTT-A-3 (SEQ ID
NO. 7); GLP-1NLS reverse primers: 5-AAA-AGC-CGC-TCA-GAC-CTT-CCG-CTT-TTT-
TTT-AGG-TCC-TCG-GCC-TTT-CAC-CAG-CCA-3 (SEQ ID NO. 8); This GLP-1NLS is
composed of a start codon, furin cut codon, GLP-1 cDNA (7-37) and a stop codon
(FIG 10).
DsRed cDNA vector was purchased from Clontech (Mountain View, CA), GLP-1NLS
cDNAs were cloned to pCI vector (Promega, Madison, WI), hyperative piggybac
transposase
cDNA with EcoRl/NOT1 cutting also subcloned into pCI vector and then GLP-1NLS
or
DsRed cDNA with CMV promoter and polyA fragment with BgL II/BamH1 cutting
subcloned to pXL-BSII vector. The plasmids digestion, ligation, subcloning,
isolation and
purification were performed by standard procedures, and once again sequenced
to confirm
that no artifactual mutations were introduced.
Manufacture of Plasmid-Containing Lipid-Stabilized Microbubble
[0224] Lipid-stabilized microbubbles were prepared using a solution of DPPC
(1,2-dipalmitoyl-sn-glycero-3-phosphatidylcholine, Sigma, St. Louis, MO) 2.5
mg/ml; DPPE
(1,2-dipalmitoyl-sn-glycero-3-phosphatidylethanolamine, Sigma, St. Louis, MO)
0.5 mg/ml;
and 10% glycerol was mixed with 2 mg of plasmids (pXL-BSII-CI-TB4 plasmid and
pCI-
mPB ratio is 5:1) dissolved in 50 ul of lipefectamine 2000 (Invitrogen,
Carlsbad, CA).
Aliquots of 0.5 ml of this phospholipid-plasmid solution were placed in 1.5 ml
clear vials; the
remaining headspace was filled with the perfluoropropane gas (Air Products,
Inc, Allentown,
PA). Each vial was incubated at 4 C for 30 min and then mechanically shaken
for 30 seconds
by a dental amalgamator (VialmixTM. Bristol-Myers Squibb Medical Imaging, N.
Billerica,
MA). The lipid-stabilized microbubbles appear as a milky white suspension
floating on the
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
67
top of a layer of liquid containing unattached plasmid DNA. The mean diameter
and
concentration of the microbubbles in the upper layer were measured by a
particle counter
(Beckman Coulter Multisizer III, Miami, FL).
Echocardiography
[0225] Echocardiographic measurements of LV fractional shortening and LV
posterior wall thickness were made from digital images acquired with a 12 MHz
broadband
transducer (S12 probe, Philips Ultrasound, Bothell, WA). Fractional shortening
was
evaluated from the following formula: FS=(LVIDd-LVIDs)/LVIDd X 100.
Data analysis
[0226] Data was analyzed with Statview software (SAS, Cary, NC, USA). The
results are expressed as mean one standard deviation. Differences were
analyzed by
repeated measures ANOVA with Fisher's post hoc test and considered significant
at P<0.05.
EXAMPLE 2
MYOCARDIAL REGENERATION IN ADRIAMYC[N CARDIOMYOPATHY BY
NUCLEAR EXPRESSION OF GLP1 USING ULTRASOUND TARGETED
MICROBUBBLE DESTRUCTION
[0227] There are nearly 5.7 million Americans with heart failure
(HF) and
approximately 670,000 new cases are diagnosed in the U.S. each year. HF
affects people of
all ages, from children and young adults to the middle-aged and the elderly
(Roger, et at.,
2012). Traditional medical therapy for HF is targeted toward relief of
symptoms and
blockade of neurohormonal activation that propagates HF. When medical therapy
fails, heart
transplant or left ventricular assist devices may be appropriate, but are
limited by donor
supply, immunosuppression, expense and complications. The ideal goal for HF
therapy is
myocardial regeneration, which has become a major goal of HF therapy since the
discovery
that cardiomyocytes are not terminally differentiated. There are various
theories about the
origin of regenerating cardiomyocytes, including self-replication of pre-
existing adult cardiac
muscle cells (Senyo, et al.. 2013; Eulalio, et al.. 2012), differentiation of
adult resident
cardiac progenitor cells (Smart, et al., 2011; Bolli, et al., 2011),
dedifferentiation and
proliferation of adult cardiac muscle cells (Beltrami, et al.. 2003; Jopling,
et al.. 2010;
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
68
Porrello, et al., 2011) and transdifferentiation of fibroblast cells into
cardiac muscle cells
(Song, et al., 2012; Qian, et al., 2012). These findings have led to numerous
clinical trials of
stem cell therapy. However, a recent meta-analysis of 49 stem cell trials for
HF identified
several flaws and internal discrepancies and no detectable benefit in left
ventricular ejection
fraction (Nowbar, et al., 2014). It remains unclear whether myocardial
regeneration in heart
failure is able to reverse established cardiomyopathy.
[0228] This disclosure evaluates the effect of gene therapy with glucagon-like
peptide-1 (GLP-1) on adriamycin-induced cardiomyopathy in a rodent model. The
adriamycin model was chosen to avoid the potential problems of decreased
myocardial blood
flow and necrosis/fibrosis associated with ischemic cardiomyopathy. GLP-1 was
selected
because it has been found to have cardioprotective effects independent of
those attributable to
tight glycemic control (Ussher, et al., 2012). Intravenous infusions of GLP-1
peptide to
patients with myocardial infarction or chronic HF improved global LV function
and the
function of ischemic LV segments (Timmers, et al., 2009; Halbirk, et al.,
2010). However,
GLP-1 acts indirectly via GLP-1 receptors distributed on the membrane of
cardiomyocytes.
GLP-1R acts via cAMP generation to produce distinct downstream signaling
events via
intracellular calcium or ERK1/2 activation (Ussher, et al., 2012). However, no
data have been
published regarding the effects of GLP-1 gene delivery to heart. It was
considered to deliver
GLP-1 gene directly to the hearts of normal rats or rats with HF induced by
adriamycin.
Ultrasound targeted microbubble destruction (UTMD) (Bekeredjian, et at., 2003;
Korpanty,
etal., 2005; Chen, et al., 2012; Chen, etal., 2013) was utilized, which has
been used to direct
gene or protein therapy to specific organs in vivo. Data showed that UTMD
directed GLP-1
to both cytoplasm and nuclei of cardiomyocytes in vivo. Because nuclear
expression of GLP-
1 gene had not been previously reported, a nuclear localizing signal was
selected to
investigate the effects of GLP-1 gene delivered specifically to the nucleus
using a piggybac
transposon plasmid system (Saridey, et al., 2009; Cary, et al., 1989;
Cadinanos, et al., 2007)
to rat hearts. After a single UTMD treatment, transgenic GLP-1 was
overexpressed in nuclei
of rat heart cells with evidence that transfected cardiac cells underwent
dedifferentiation and
proliferation. The results show that GLP-1 gene delivery to heart stimulates
myocardial
regeneration and reversal of adriamycin cardiomyopathy.
Successful overexpression of GLP-1 in nuclei of heart cells after UTMD-GLP-1
gene delivery to heart of normal rats
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
69
[0229] There is the absence of GLP-1 signal in the heart of normal control
rats
or UTMD-DsRed control rats. However, GLP-1 is seen in the hearts of rats
treated with
UTMD-GLP1NLS , notably in nuclei of heart cells. GLP-1 was not detected in the
bloodstream using GLP-1 ELISA kit. Western blot was employed to detect GLP-1
from
cardiac nuclear protein extracts and the results showed that GLP-1 signal
existed in cardiac
muscle nuclear protein extracts of UTMD-GLP1NLS group (Figure 1, panel A).
GLP1
mRNA level was further evaluated with quantitative RT-PCR. The result (Figure
1, panel B)
shows that the GLP-1 mRNA level in UTMD-GLP1NLS group was 84-fold greater than
in
normal control or UTMD-DsRed control groups (P<0.001). The percentage of GLP-1
positive cells was also counted in heart slides. GLP-1 was present in 50.7% of
cardiomyocytes, 16.7% of vascular endothelial cells, 14.3% of vascular muscle
cells and
18.3% of vimentin positive cells (Figure 1, panel C).
Reversal of adriamycin cardiomyopathy after UTMD-GLP-1 gene therapy
[0230] It was considered if there is any possible pharmacological
effects of
GLP-1NLS gene on rat hearts with established adriamycin cardiomyopathy. Figure
2 (upper
panels) shows the gross pathology of LV walls in the groups of ADM injection
only, ADM
injection plus GLP1 peptide treatment, and ADM injection plus UTMD-GLP1
peptide
delivery (B, C, and D). Rats with adriamycin cardiomyopathy (B, C) had thin LV
walls and
dilated LV cavities compared to normal controls (A). Adriamycin rats who were
treated with
UTMD-GLP-1NLS gene therapy early (E) or 14 days later (F) resembled normal
controls.
The lower panels show the results of Masson's trichrome staining for fibrosis,
which was
significantly decreased after UTMD-GLP1-NLS treatment (E and F). The nuclear
location of
GLP-1 in UTMD-GLP1-NLS gene therapy groupswas determined. Echocardiography was
utilized to evaluate heart structure and function in all groups. Figures 4 and
11 demonstrated
decreased LV mass and wall thickness in adriamycin cardiomyopathy with
restoration to
normal values by GLP-1NLS gene therapy but not GLP-1 peptide therapy. Similar
findings
were seen for LV fractional shortening and volume of abdominal ascites
(Figures 4 and 11).
Of 12 rats with established adriamycin cardiomyopathy, 10 demonstrated
complete
normalization of LV size and function after UTMD-GLP-1 gene therapy, the other
2 were not
improved significantly.
Nuclear GLP-1 inhibits the activation of nuclear Fox01
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
[0231] Recently Fox() proteins (forkhead box proteins, 0) were considered as
key factors during the development of diabetic cardiomyopathy or ischemic
cardiomyopathy.
This is the first report showing that Fox01 is involved in adriamycin-induced
cardiomyopathy. Fox01 signal is present in nuclei of cardiac muscle cells with
established
adriamycin cardiomyopathy. However, in normal rat heart slides Fox01 signal
was not seen
in nuclei of cardiac muscle cells, although it was occasionally present in
cytoplasm. After
UTMD GLP-1-NLS gene delivery, transgenic nuclear overexpression of GLP-1 gene
was
associated with disappearance of nuclear Fox01. The results (Figure 6A) of
Fox01 western
blots confirm a significant increase of Fox01 in cardiac nuclear protein
extracts of ADM
only group, and Fox01 decreased to a nearly normal level after UTMD GLP-1NLS
gene
delivery. The result (Figure 6B) shows that the Fox01 mRNA level in ADM only
group was
12.6-fold greater than in normal control or normal plus UTMD-GLP-1NLS groups
(P<0.001).
However, Fox01 mRNA level was decreased to 4.5-fold (P<0.01 vs ADM only) after
UTMD
GLP-1NLS gene delivery. It was considered to confirm this finding in an in
vitro cell
culture. Adriamycin under 0.25, 0.5, and 1.0 viM concentration significantly
activates
overexpression of nuclear Fox01 in HL-1 cardiac muscle cell line. Figure 12
showed that
specific Fox01 inhibitor (AS1842856, a small molecular compound) is able to
block the
activation of myocardial nuclear Fox01 and reverse ADM cardiomyopathy (panel A-
B). The
data taken together demonstrates that activation of myocardial nuclear Fox01
mediates ADM
cardiomyopathy.
Nuclear GLP-1 activates myocardial topoisomerase Hot to initiate
overexpression
of Cyclin D1 for Gl/S transition of cell cycle of adult cardiomyocytes.
[0232] Topoisomerase II is a nuclear enzyme that has an important
role in
topological rearrangement of DNA during replication, transcription and
resolution/separation
of daughter chromosomes at mitosis. Topo Ha which is considered a specific
marker for cell
proliferation, does not exist in adult cardiomyocytes, however Topo 1113 is
normally present in
adult mammalian cardiomyocytes. Topo 1113 exists in nuclei of cardiomyocytes
of normal
controls, but after ADM injection, is no longer seen and could not be
recovered by UTMD-
GLP-1 gene delivery. However, Topo ha was detected in the nuclei of some
cardiomyocytes
after UTMD-GLP-1 gene delivery, providing further evidence for cardiomyocyte
proliferation after GLP-1 myocardial nuclear expression. The result (Figure
7A) of TOP II
western blots also shows TOP ha expressed in cardiac nuclear protein extracts
of ADM plus
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
71
UTMD-GLP-1 group. Interestingly TOP 1113 bands significantly decreased after
adriamycin
treatment. The qRT-PCR (Figure 7B-7C) shows that the TOP Ha mRNA level in ADM
plus
UTMD-GLP-1NLS group was 21.8-fold greater than in normal control or ADM only
groups
(P<0.001), however TOP 1113 mRNA level in normal group was 18-fold greater
than in ADM
only or ADM plus UTMD-GLP-1NLS groups (P<0.001). Figure 8 addresses
overexpression
of cyclin D1 in the nucleus of cardiomyocytes after GLP-1NLS gene delivery.
The results
(Figure 8A-8B) of western blots and qRT-PCR support the overexpression of
cyclin D1 in
nuclei of cardiac muscle cells after UTMD-GLP-1NLS.
Regenerating Adult Cardiomyocytes are in Dedifferentiation.
[0233]
Dedifferentiation and proliferation of mature cardiomyocytes has only
been observed in zebrafish, amphibians (Jopling, et al., 2010) or in neonatal
mouse heart
(Porrello, et al., 2011). Although adult mammalian cardiomyocytes generally
lose the
capability of dedifferentiation and proliferation they exhibited in the fetus
stage, it was
considered if this capability can be recovered by genetic manipulation in
adriamycin
cardiomyopathy. Dedifferentiation of adult pancreatic beta cell (Talchai, et
al., 2012) was
mediated by the activation of nuclear Fox01 under diabetic condition. The data
(Figure 6)
suggests that ADM induced overexpression of nuclear Fox01. In addition,
nuclear Fox01-
positive myocardium in adriamycin-induced cardiomyopathy does not undergo
dedifferentiation but apoptosis. However, after UTMD-GLP-1NLS treatment,
embryonic
stem cell markers (OCT4, Nanog, and SOX2) were induced in nuclei of adult
cardiac muscle
cells. Further evidence to support dedifferentiation of adult rodent
cardiomyocytes is co-
expression of smooth muscle actin alpha (a marker of coronary artery muscle
cells) with
cardiac troponin T (a marker of mature cardiac muscle cell) in sarcomere
disassembly area.
von Willebrand factor (vWf), a marker of vascular endothelial cells was not
seen to co-
localize with cINT . There are some c-kit positive adult cardiac muscle cells
to support
dedifferentiation of adult cardiac muscle cells into multipotent cardiac stem-
like cells.
[0234]
There is the presence of NKX2.5-positive adult cardiomyocytes in
adriamycin cardiomyopathy rats treated with UTMD-GLP1 gene therapy early and
14 day
later but not in the controls. NKX2.5 signal was clearly seen in the nucleus
of a small
number of cardiac troponin T positive cells. Similar findings were seen with
ISL-1. The
percentage of NKX2.5 or ISL-1 positive cardiomyocytes was counted in 1000 cTnT
positive
cardiomyocytes cells in anterior wall and posterior LV walls with serial
sections through each
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
72
rat heart (n=6 each group). The percentage of NKX2.5 positive cardiomyocytes
in the gene
therapy groups (Figure 9A) was 8.7310.83% and 8.5111.45% (p<0.001 vs
controls). The
percentage of ISL-1 positive cardiomyocytes (Figure 9B) was 11.5712.31% and
11.9713.24
% in the gene therapy groups. There were GATA4 positive nuclei in these
treated groups.
NKX2.5, GATA4 and ISL-1 are considered markers of early cardiomyocyte
differentiation
(Smart, et at., 2011; Boni, et at., 2008). Sarcomere disassembly is considered
specific for
cardiomyocyte proliferation (Porrello, et at., 2011; Bersell, et at., 2009).
There were NKX2.5
and ISL-1 positive nuclei located in sarcomere disassembly structure. Thus, in
particular
embodiments gene therapy with GLP-1NLS by UTMD leads to dedifferentiation and
proliferation of nuclear Fox01 positive cardiomyocytes in the rat adramycin-
induced
cardiomyopathy model.
Dedifferentiated Adult Cardiomyocytes are in Proliferation.
[0235] Two proliferation markers (anti-BrDu and anti-Ki-67) and two mitotic
markers (anti-phospho-histone H3 (Serl 0) (PHH3) and anti-aurora B) were used
to
demonstrate if dedifferentiated cardiac muscle cells were proliferating. The
percentage of
BrDu, PHH3 or Ki-67 or aurora B positive cardiomyocytes was calculated by
counting
stained nuclei from 1000 cInT positive cardiomyocytes cells in the anterior
and posterior LV
walls using serial sections through each rat heart (n=6 each group). BrDu
signal was observed
within the nucleus of cTnT positive cardiomyocytes by confocal microscopy
(Fig. 5). The
percentage of BrDu positive cardiac muscle cells in the rats treated with UTMD-
GLP1NLS
gene therapy early and 14 days later was 8.7310.83% and 8.5111.45% (p<0.001 vs
controls).
Similarly, the percentage of Ki-67 positive cardiac muscle cells was
6.5711.12% and
7.3711.35% (p<0.001 vs controls) (Figure 5). Aurora B staining was clearly
seen in nuclei of
cardiomyocytes treated with GLP-1 gene therapy by UTMD. The percentage of
aurora B
positive cardiac muscle cells in the two gene therapy groups was 8.9310.90%
and
8.11+1.62% (p<0.001 vs controls). PHH3 signal was observed within the nucleus
of cInT
positive cardiomyocytes by confocal microscopy. The percentage of PHH3
positive cardiac
muscle cells in the UTMD-GLP1NLS gene therapy groups was 7.7311.20% and
8.1011.70%
(p<0.001 vs controls) (Fig. 5).
Significance of Certain Embodiments
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
73
102361
After systemic administration of GLP-1 peptide, GLP-1 binds with its
receptor on the cell membrane and activates adenylate cyclase, leading to
increased
intracellular cAMP levels.
This triggers a wide range of cardioactive effects including
increased heart rate and blood pressure, increased p38 MAPK activity,
Akt/ERK1/2
phosphorylation, GLUT4, glucose intake, and decreased apoptosis in the heart
(Ussher et al.,
2012). However, it was recently reported (Kim et al., 2013) that GLP-lr exists
only in atrial,
but not in ventricular cardiomyocytes, and that excited GLP-1 r signal
activates atrial ANP
and mediates "so called GLP-1 cardioprotection". The data indicate that GLP-1
targeted to
the cell nucleus of ventricular cardiomyocytes triggers sufficient myocardial
regeneration to
reverse established adriamycin cardiomyopathy in rats. This is a novel finding
that has
important clinical implications for treatment of HF. Established adriamycin
cardiomyopathy
is a lethal disease. When HF develops, mortality is approximately 50% in a
year. Moreover,
treatment of established adriamycin cardiomyopathy is directly only to HF
symptoms. There
is no known therapy for reversing the underlying cardiomyopathy (Chatterjee et
al., 2010).
Heart transplant is often not an option in these patients for fear that the
requisite
immunosuppressive therapy could activate whatever malignancy necessitated
adriamycin
therapy. Zhang et al. (2012) reported that adriamycin-induced cardiomyopathy
was mediated
by topoisomerase-11f3. Adriamycin binds with topoisomerase-11f3 and formed a
complex,
activated topoisomerase-II13 to cut double strands of DNA and leaded to
mitochondia
biogenesis, and caused cardiac muscle cell death. Topoisomerase-lla, which
does not exist in
adult cardiomyocytes, is a specific marker of cell proliferation. The data
shows a novel
molecular mechanism of GLP-1 that stimulates cardiomyocyte proliferation
associated with
activation of topoisomerase-Ha, when GLP- 1 is targeted to the cell nucleus by
UTMD and a
nuclear localizing signal. This was not seen with systemic administration of
GLP-1 peptide
that is known to bind to the cell membrane. Further studies are needed to
decipher how
nuclear GLP-1 activates topoisomerase-lla and cyclinD1 in adult mammlian
cardiomyocytes.
102371 Battiprolu et al. (2012) reported that metabolic stress¨induced
activation
of Fox01 in nuclei of cardiac muscle cells is central to the development of
diabetic
cardiomyopathy in diabetic mice, and the knockout Fox01 blocked the formation
of diabetic
cardiomyopathy induced by high fat diet. The present studies included in vivo
and in vitro
data shows that adriamycin cardiomyopathy is also associated with, and
possibly mediated
by, overexpression of Fox01 in the nuclei of cardiac muscle cells.
Interestingly, nuclear
localization of GLP-1 in cardiac muscle cells or specific Fox01 inhibitor
(AS1842856)
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
74
appears to inhibit Fox01 and restores nearly normal cardiac function and
morphology after
UTMD GLP-1 gene delivery. Abundant evidence now suggests that 3 members of the
Fox()
subfamily, Fox01, Fox03, and Fox04, are critical for maintenance of cardiac
function and
cardiac stress responsiveness (Skurk et at., 2005; Ni et at., 2006; Sengupta
et at., 2009; Stahl
et at., 2002; Kops et at., 2002; Puigserver et at., 2003; Medema et at., 2000;
Schmidt et at.,
2002). Taken together, these data indicate that Fox() factors are an important
pathway for the
development of cardiomyopathy and an important therapeutic target.
[0238] As shown herein, nuclear expression of GLP-1 by UTMD
promotes
adult myocardial regeneration sufficient to reverse adriamycin cardiomyopathy
by inhibition
of nuclear Fox01 and by activation of nuclear topoisomerase Ha. This appears
to mediate
cell cycle re-entry by the activation of cyclin D1 in adult cardiomyocytes.
The data indicate
that the process of myocardial regeneration is accompanied by
dedifferentiation of nuclear
Fox01 positive adult cardiac muscle cells into cardiac progenitor-like cells
expressing OCT4,
Nanog, SOX2 and c-kit, with subsequent redifferentiation into mature cardiac
muscle cells.
This phenomenon is not known to occur in adult mammalian cardiac muscle cells
under
physiological conditions. One can characterize by standard means in the art
why nuclear
Fox01 positive adult cardiomyocytes in cardiomyopathy are able to recover the
capability of
dedifferentiation and proliferation after intranuclear delivery of GLP-1.
EXAMPLE 3
GLP-1 PLUS THYMOSIN BETA 4 STIMULATES REGENERATION OF ADULT RAT
CARDIAC MUSCLE AND REVERSAL OF MYOCARDIAL ISCHEMIC INJURY
[0239] It is estimated that acute myocardial infarction (MI) afflicts 1.3
million
Americans every year. Approximately 10% of those patients will die from the
MI, typically
from ventricular arrhythmias, pump failure, or myocardial rupture. In
survivors, infarcted
myocardium is replaced by fibrous scar tissue, leading to progressive left
ventricular (LV)
remodeling and congestive heart failure. Although prompt revascularization of
the occluded
coronary artery by percutaneous coronary intervention has been shown to reduce
mortality,
the myocardial microcirculation may not be completely reperfused due to the
"no-reflow"
phenomenon, and some degree of myocardial necrosis typically remains.
Therefore, a major
goal of therapy for acute MI has been to develop strategies for regeneration
of
cardiomyocytes and blood vessels in the damaged area of the heart. Novel
strategies for
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
myocardial regeneration include cell therapy with embryonic stem cells, iPS
cells and bone
marrow stem cells. Recently, resident cardiac progenitor cells were discovered
in the adult
mammalian heart. Although sparse in number, these cells are self-renewing,
clonogenic, and
multipotent. In particular embodiments, they can differentiate into all three
major cardiac
lineages¨vascular smooth muscle cells, endothelial cells and cardiac muscle
cells. However,
it remains controversial whether adult cardiac muscle cells can be formed from
these resident
progenitor cells in vivo.
[0240] As
demonstrated herein there is delivery of thymogen beta-4 (TB4)
/GLP-1NLS genes directly to the ischemic heart of rats, in order to stimulate
resident cardiac
progenitor cells.
[0241]
Objectives: To determine whether gene therapy with GLP-1NLS/
thymosin beta 4 (TB4) stimulates proliferation of resident adult cardiac
progenitor or cardiac
muscle cells in rat heart with acute ischemic injury.
[0242] Background: It has been proposed that TB4 protein delivery stimulates
proliferation and differentiation of resident adult WTI positive cardiac
progenitor cells, but
with very low efficiency and GLP-1 has cardioprotective effects.
[0243]
Methods: Ultrasound targeted microbubble destruction (UTMD) was
used to deliver the GLP-1NLS/ TB4 genes under a piggybac transposon plasmid to
rat
ischemic heart. The rat hearts were assayed by quantitative RT-PCR and
immunohistology
with a confocal microscope at 2 weeks post UTMD.
[0244]
Results: Fig. 13 demonstrates Masson's trichromone staining images
and evaluation of heart function with echocardiography. Fig 14 demonstrates
that SMAa
staining shows coronary artery distribution in ischemic risk area. Fig. 15
provides CD31
staining that shows vascular endothelial cells density. Fig. 16 demonstrates
proliferation
marker staining showing that cardiac muscle cells are in proliferation after
UTMD-GLP-
1NLS/TB4 gene therapy.
[0245] Thus, GLP-1NLS/ TB4 genes stimulation resulted in the ischemic area
reduction in ischemic myocardium.
GLP-1NLS/ TB4 stimulated angiogenesis and
arteriogenesis. One month after GLP-1/TB4 gene therapy by UTMD, the percentage
of
NKX2.5 positive cardiomyocytes was 5.5 1.0%, and NKX2.5 mRNA was 24-fold
higher
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
76
than in the control groups (p<0.001). Similar results were found for ISL-1,
BrDu, Ki-67,
PHH3 and aurora B (p<0.001).
[0246] Conclusions: GLP-1NLS/TB4 genes efficiently stimulates proliferation
and differentiation of adult cardiac muscle cells into three intact cardiac
cell lineages-
vascular endothelial cells, coronary artery smooth muscle cells and cardiac
muscle cells in rat
ischemic heart.
REFERENCES
[0247] All patents and publications mentioned in this
specification are
indicative of the level of those skilled in the art to which the disclosure
pertains. All patents
and publications herein are incorporated by reference to the same extent as if
each individual
publication was specifically and individually indicated to be incorporated by
reference in
their entirety.
Battiprolu, etal., J Clin Invest. 2012; 122:1109-1118.
Bekeredjian, etal., Circulation. 2003; 108(8): 1022-26
Beltrami, etal., Cell. 2003; 114:763-776.
Bersell, etal., Cell. 2009; 138:257-270.
Bolli, etal., Lancet. 2011; 378: 1847-1857.
Boni, etal., Proc Nat! Acad Sci USA. 2008; 105:15529-15534.
Cadifianos, etal., Nucleic Acids Res. 2007; 35:e87.
Cary, etal., Virology 1989; 172:156-69.
Chatterjee, etal., Cardiology . 2010; 115:155-162.
Chen, etal., Gene Ther. 2013; 20:225-33.
Chen, etal., Cell Cycle. 2012; 11:695-705.
Chen, etal., J Am Coll Cardiol. 2003; 42: 301-8.
Claycomb, etal., Proc Nat! Acad Sci USA.1998; 95:2979-2984.
Drucker, et al., Gastroenterology. 2002;122:531-44.
Drucker, et al., Endocrinology. 2001;142:521-7
Eulalio, etal., Nature. 2012; 492:376-81.
Fraser, etal., Virology 1995; 211: 397-407.
Halbirk, etal., Am J Physiol Heart Circ Physiol. 2010; 298:H1096-102.
Iliskovic, etal., American Journal of Pathology. 1997; 150:727-734.
Jopling, etal., Nature. 2010; 464: 606-609.
Kim, etal., Nat Med. 2013; 19:567-575.
Kops, etal., Nature. 2002; 419:316-321.
Korpanty, etal., Gene Therapy. 2005; 12: 1305-12
Medema, etal., Nature. 2000; 404:782-787.
Ni, etal., Circulation. 2006; 114:1159-1168.
CA 02929555 2016-05-03
WO 2015/070050 PCT/US2014/064606
77
Nowbar, et al., BMJ. 2014; 348:g2688.
Porrello, etal., Science. 2011; 331:1078-1080.
Puigserver, etal., Nature. 2003; 423:550-555.
Qian, etal., Nature. 2012; 485:593-598.
Roger, etal., Circulation. 2012; 125:e2-e220.
Saridey, etal., Mol Ther. 2009; 17: 2115-20.
Schmidt, etal., Mol Cell Biol. 2002; 22:7842-7852.
Sengupta, et al., J Biol Chem. 2009; 284:28319-28331.
Senyo, et al., Nature 2012; 493,433-436.
Sinkula, etal., J Pharm Sci. 64:181-210,1975.
Skurk, etal., J Biol Chem. 2005; 280:20814-20823.
Smart, et al., Nature. 2011; 474: 640-644.
Song, et al., Nature 2012; 485:599-604.
Stahl, etal., J Immunol. 2002; 168:5024-5031.
Talchai, etal., Cell. 2012; 150:1223-1234.
Timmers, etal., J Am Coll Cardiol. 2009; 53:501-10.
Ussher, et al., Endocr Rev. 2012; 33:187-215.
Yusa, et aL, Proc Natl Acad Sci U S A. 2011; 108:1531-1536.
Zhang, etal., Nat Med. 2012; 18:1639-1642.
[0248] Although the present disclosure and its advantages have been described
in detail, it should be understood that various changes, substitutions and
alterations can be
made herein without departing from the spirit and scope of the disclosure as
defined by the
appended claims. Moreover, the scope of the present application is not
intended to be limited
to the particular embodiments of the process, machine, manufacture,
composition of matter,
means, methods and steps described in the specification. As one of ordinary
skill in the art
will readily appreciate from the disclosure of the present disclosure,
processes, machines,
manufacture, compositions of matter, means, methods, or steps, presently
existing or later to
be developed that perform substantially the same function or achieve
substantially the same
result as the corresponding embodiments described herein may be utilized
according to the
present disclosure. Accordingly, the appended claims are intended to include
within their
scope such processes, machines, manufacture, compositions of matter, means,
methods, or
steps.