Note: Descriptions are shown in the official language in which they were submitted.
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
IDENTIFICATION OF PREDICTIVE BIOMARKERS ASSOCIATED WITH WNT
PATHWAY INHIBITORS
CROSS-REFERENCE TO RELATED APPLICATONS
[0001] This application claims priority benefit of U.S. Provisional
Application No. 61/910,663, filed
December 2, 2013, and U.S. Provisional Application No. 61/975,339, filed April
4, 2014, each of
which are hereby incorporated by reference herein in their entirety.
FIELD OF INVENTION
[0002] The present invention relates to the field of cancer treatment. More
particularly, the invention
provides methods for identifying tumors that are likely to be responsive or
non-responsive to
treatment with a Wnt pathway inhibitor. In addition, the invention provides
methods for identifying,
selecting, and/or treating patients with cancer who are likely to respond to
treatment with a Wnt
pathway inhibitor, either alone or in combination with other therapeutic
agents.
BACKGROUND OF THE INVENTION
[0003] Cancer is one of the leading causes of death in the developed world,
with approximately 1.6
million people diagnosed with cancer and over 550,000 deaths per year in the
United States alone.
Overall it is estimated that more than 1 in 3 people will develop some form of
cancer during their
lifetime. There are more than 200 different types of cancer, four of which -
breast, lung, colorectal,
and prostate¨account for almost half of all new cases in the United States
(Siegel et al., 2012, CA: A
Cancer J. for Clin., 62:10-29).
[0004] Signaling pathways normally connect extracellular signals to the
nucleus leading to the
expression of genes that directly or indirectly control cell growth,
differentiation, survival, and death.
However, in a wide variety of cancers signaling pathways are dysregulated and
may be linked to
tumor initiation and/or progression. Signaling pathways implicated in human
oncogenesis include,
but are not limited to, the Wnt pathway, the Ras-Raf-MEK-ERK or MAPK pathway,
the PI3K-AKT
pathway, the CDKN2A/CDK4 pathway, the Bc1-2/TP53 pathway, and the NOTCH
pathway.
[0005] The Wnt signaling pathway is one of several critical regulators of
embryonic pattern
formation, post-embryonic tissue maintenance, and stem cell biology. More
specifically, Wnt
signaling plays an important role in the generation of cell polarity and cell
fate specification including
self-renewal by stem cell populations. Unregulated activation of the Wnt
pathway is associated with
numerous human cancers where it is believed the activation can alter the
developmental fate of cells.
It is believed that the activation of the Wnt pathway may maintain tumor cells
in an undifferentiated
state and/or lead to uncontrolled proliferation. This may allow carcinogenesis
to proceed by
1
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
overtaking homeostatic mechanisms which control normal development and tissue
repair (reviewed in
Reya & Clevers, 2005, Nature, 434:843-50; Beachy et al., 2004, Nature, 432:324-
31).
[0006] The Wnt signaling pathway was first elucidated in the Drosophila
developmental mutant
wingless (wg) and from the murine proto-oncogene int-1, now Wntl (Nusse &
Varmus, 1982, Cell,
31:99-109; Van Ooyen & Nusse, 1984, Cell, 39:233-40; Cabrera et al., 1987,
Cell, 50:659-63;
Rijsewijk et al., 1987, Cell, 50:649-57). Wnt genes encode lipid-modified
glycoproteins which are
secreted and 19 different Wnt proteins have been identified in mammals. These
secreted ligands
activate a receptor complex consisting of a Frizzled (FZD) receptor family
member and low-density
lipoprotein (LDL) receptor-related protein 5 or 6 (LRP5/6). The FZD receptors
are members of the
G-protein coupled receptor (GPCR) superfamily and contain seven transmembrane
domains and a
large extracellular N-terminal ligand binding domain. The N-terminal ligand
binding domain contains
conserved cysteines and is known as a cysteine-rich domain (CRD) or a "Fri
domain". There are
ten human FZD receptors, FZD1, FZD2, FZD3, FZD4, FZD5, FZD6, FZD7, FZD8, FZD9,
and
FZD10. Different FZD CRDs have different binding affinities for specific Wnt
proteins (Wu &
Nusse, 2002, J. Biol. Chem., 277:41762-9). In addition, FZD receptors may be
grouped into those
that activate the canonical f3-catenin pathway and those that activate non-
canonical pathways (Miller
et al., 1999, Oncogene, 18:7860-72).
[0007] A role for Wnt signaling in cancer was first uncovered with the
identification of Wntl
(originally intl) as an oncogene in mammary tumors transformed by the nearby
insertion of a murine
virus (Nusse & Varmus, 1982, Cell, 31:99-109). Since these early observations
additional evidence
for the role of Wnt signaling in breast cancer has continued to accumulate.
For example, over-
expression of I3-catenin in the mammary glands of transgenic mice results in
hyperplasias and
adenocarcinomas (Imbert et al., 2001, J. Cell Biol., 153:555-68; Michaelson &
Leder, 2001,
Oncogene, 20:5093-9) whereas loss of Wnt signaling disrupts normal mammary
gland development
(Tepera et al., 2003, J. Cell Sci., 116:1137-49; Hatsell et al., 2003, J.
Mammal.); Gland Biol.
Neoplasia, 8:145-58). In human breast cancer, [3-catenin accumulation
implicates activated Wnt
signaling in over 50% of carcinomas, and though specific mutations have not
been identified, up-
regulation of Frizzled receptor expression has been observed (Brennan & Brown,
2004, J. Mammal.);
Gland Biol. Neoplasia, 9:119-31; Malovanovic et al., 2004, Int. J. Oncol.,
25:1337-42).
[0008] Activation of the Wnt pathway is also associated with colorectal
cancer, lung cancer,
pancreatic cancer, and melanoma. Approximately 5-10% of all colorectal cancers
are hereditary with
one of the main cancer types being familial adenomatous polyposis (FAP). FAP
is an autosomal
dominant disease in which about 80% of affected individuals contain a germline
mutation in the
adenomatous polyposis coli (APC) gene. Mutations have also been identified in
other Wnt pathway
components including Axin and 13-catenin. Individual adenomas are clonal
outgrowths of epithelial
cells containing a second inactivated allele, and the large number of FAP
adenomas inevitably results
in the development of adenocarcinomas through additional mutations in
oncogenes and/or tumor
2
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
suppressor genes. Furthermore, activation of the Wnt signaling pathway,
including loss-of-function
mutations in APC and stabilizing mutations in P-catenin, can induce
hyperplastic development and
tumor growth in mouse models (Oshima et al., 1997, Cancer Res., 57:1644-9;
Harada et al., 1999,
EMBO J., 18:5931-42).
[0009] Thus the Wnt pathway has been identified as a target for cancer therapy
and treatment. As
drug discovery and development advances, especially in the cancer field, the
"one drug fits all"
approach is shifting to a "personalized medicine" strategy. Personalized
medicine strategies may
include treatment regimens that are based upon cancer biomarkers, including
prognostic markers,
pharmacodynamic markers, and predictive markers. In general, predictive
biomarkers assess the
likelihood that a tumor or cancer will be responsive to or sensitive to a
specific therapeutic agent, and
may allow for the identification and/or the selection of patients most likely
to benefit from the use of
that agent.
[0010] The invention provides the identification of predictive biomarkers
associated with the use of
Wnt pathway inhibitors in the treatment of cancer. Also provided are methods
of using the predictive
biomarkers for identifying, selecting, and/or classifying tumors and/or
patients with cancer as likely to
be responsive or non-responsive to treatment with a Wnt pathway inhibitor.
Methods for treating
patients with a Wnt inhibitor that are predicted to be responsive to treatment
are also provided.
SUMMARY OF THE INVENTION
[0011] Provided are biomarkers for identifying patients likely to respond to
treatment with Wnt
pathway inhibitors. Additionally provided are methods for identifying tumors
and/or patients that are
likely to be responsive or non-responsive to treatment with a Wnt pathway
inhibitor. Further
provided are methods of treating cancer in a patient with a Wnt pathway
inhibitor, wherein the patient
is predicted to be or has been identified as likely to be responsive to the
Wnt pathway inhibitor.
[0012] In one aspect, the invention provides a method of identifying a human
tumor that is likely to
be responsive or non-responsive to treatment with a Wnt pathway inhibitor, the
method comprising:
(a) obtaining a sample of the human tumor; (b) measuring the expression level
of each biomarker of a
biomarker signature in the sample, wherein the biomarker signature comprises
one or more of the
biomarkers FBXW2, CCND2, RHOU, CRBP2, WIF1, and DKK1; and (c) identifying the
tumor as
likely to be responsive or non-responsive to treatment based upon the
expression level of the
biomarkers. In some embodiments, a method of identifying a human tumor that is
likely to be
responsive or non-responsive to treatment with a Wnt pathway inhibitor
comprises: (a) obtaining a
sample of the human tumor; (b) measuring the expression level of each
biomarker of a biomarker
signature in the sample, wherein the biomarker signature comprises one or more
of the biomarkers
FBXW2, CCND2, RHOU, CRBP2, WIF1, and DKK1; and (c) calculating a decision
value based
upon the standardized expression of the biomarkers in the biomarker signature;
wherein a positive
decision value indicates the tumor is predicted to be responsive to the Wnt
pathway inhibitor and a
3
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
negative decision value indicates the tumor is predicted to be non-responsive
to the Wnt pathway
inhibitor. As used herein, "standardized" and "normalized" may be used
interchangeably. In some
embodiments, the method comprises identifying a human tumor that is likely to
be responsive or non-
responsive to treatment with a Wnt pathway inhibitor in combination with
paclitaxel.
[0013] In another aspect, the invention provides a method of classifying a
human tumor as likely to
be responsive or non-responsive to treatment with a Wnt pathway inhibitor, the
method comprising:
(a) obtaining a sample of the human tumor; (b) measuring the expression level
of each biomarker of a
biomarker signature in the sample, wherein the biomarker signature comprises
one or more of the
biomarkers FBXW2, CCND2, RHOU, CTBP2, WIF1, and DKK1; and (c) classifying the
tumor as
likely to be responsive or non-responsive to treatment based upon the
expression level of the
biomarkers. In some embodiments, a method of classifying a human tumor as
likely to be responsive
or non-responsive to treatment with a Wnt pathway inhibitor comprises: (a)
obtaining a sample of the
human tumor; (b) measuring the expression level of each biomarker of a
biomarker signature in the
sample, wherein the biomarker signature comprises one or more of the
biomarkers FBXW2, CCND2,
RHOU, CTBP2, WIF1, and DKK1; and (c) calculating a decision value based upon
the standardized
expression of the biomarkers in the biomarker signature; wherein a positive
decision value indicates
the tumor is predicted to be responsive to the Wnt pathway inhibitor and a
negative decision value
indicates the tumor is predicted to be non-responsive to the Wnt pathway
inhibitor. In some
embodiments, the method comprises classifying a human tumor as likely to be
responsive or non-
responsive to treatment with a Wnt pathway inhibitor in combination with
paclitaxel.
[0014] In another aspect, the invention provides a method of determining the
responsiveness (or
sensitivity) of a human tumor to treatment with a Wnt pathway inhibitor, the
method comprising: (a)
obtaining a sample of the human tumor; (b) measuring the expression level of
each biomarker of a
biomarker signature in the sample, wherein the biomarker signature comprises
one or more of the
genes FBXW2, CCND2, RHOU, CTBP2, WIF1, and DKK1; and (c) determining the
responsiveness
of the tumor to treatment based upon the expression level of the biomarkers.
In some embodiments, a
method of determining the responsiveness or sensitivity of a human tumor to
treatment with a Wnt
pathway inhibitor comprises: (a) obtaining a sample of the human tumor; (b)
measuring the
expression level of each biomarker of a biomarker signature in the sample,
wherein the biomarker
signature comprises one or more of the genes FBXW2, CCND2, RHOU, CTBP2, WIF1,
and DKK1;
and (c) calculating a decision value based upon the standardized expression of
the biomarkers in the
biomarker signature; wherein a positive decision value indicates the tumor is
predicted to be
responsive to or sensitive to the Wnt pathway inhibitor. In some embodiments,
the method comprises
determining the responsiveness or sensitivity of a human tumor to treatment
with a Wnt pathway
inhibitor in combination with paclitaxel.
[0015] In another aspect, the invention provides a method of identifying a
patient with cancer who is
likely to respond to treatment with a Wnt pathway inhibitor, the method
comprising: (a) obtaining a
4
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
sample of the patient's tumor; (b) measuring the expression level of each
biomarker of a biomarker
signature in the sample, wherein the biomarker signature comprises one or more
of the biomarkers
FBXW2, CCND2, RHOU, CTBP2, WIF1, and DKK1; and (c) identifying the patient who
is likely to
respond to treatment based upon the expression level of the biomarkers. In
some embodiments, a
method of identifying a patient with cancer who is likely to respond to
treatment with a Wnt pathway
inhibitor comprises: (a) obtaining a sample of the patient's tumor; (b)
measuring the expression level
of each biomarker of a biomarker signature in the sample, wherein the
biomarker signature comprises
one or more of the biomarkers FBXW2, CCND2, RHOU, CTBP2, WIF1, and DKK1; and
(c)
calculating a decision value based upon the standardized expression of the
biomarkers in the
biomarker signature; wherein a positive decision value indicates that the
patient is predicted to
respond to treatment with the Wnt pathway inhibitor. In some embodiments, the
method comprises
identifying a patient with cancer who is likely to respond to treatment with a
Wnt pathway inhibitor in
combination with paclitaxel.
[0016] In another aspect, the invention provides a method of selecting a
patient with cancer for
treatment with a Wnt pathway inhibitor, the method comprising: (a) obtaining a
sample of the
patient's tumor; (b) measuring the expression level of each biomarker of a
biomarker signature in the
sample, wherein the biomarker signature comprises one or more of the
biomarkers FBXW2, CCND2,
RHOU, CTBP2, WIF1, and DKK1; (c) selecting the patient for treatment based
upon the expression
level of the biomarkers. In some embodiments, a method of selecting a patient
with cancer for
treatment with a Wnt pathway inhibitor comprises: (a) obtaining a sample of
the patient's tumor; (b)
measuring the expression level of each biomarker of a biomarker signature in
the sample, wherein the
biomarker signature comprises one or more of the biomarkers FBXW2, CCND2,
RHOU, CTBP2,
WIF1, and DKK1; (c) calculating a decision value based upon the standardized
expression of the
biomarkers in the biomarker signature; and (d) selecting the patient for
treatment when their tumor
sample has a positive decision value. In some embodiments, the method
comprises selecting a patient
with cancer for treatment with a Wnt pathway inhibitor in combination with
paclitaxel.
[0017] In another aspect, the invention provides a method of treating cancer
in a patient, comprising:
(a) identifying if the patient is likely to respond to treatment with a Wnt
pathway inhibitor, wherein
the identification comprises: (i) obtaining a sample of the patient's cancer;
(ii) measuring the
expression level of each biomarker of a biomarker signature in the sample,
wherein the biomarker
signature comprises one or more of the biomarkers FBXW2, CCND2, RHOU, CTBP2,
WIF1, and
DKK1; and (iii) identifying the patient who is likely to respond to treatment
based upon the
expression level of the biomarkers; and (b) administering to the patient who
is likely to response to
treatment an effective amount of the Wnt pathway inhibitor. In some
embodiments, a method of
treating cancer in a patient comprises: (a) identifying if the patient is
likely to respond to treatment
with a Wnt pathway inhibitor, wherein the identification comprises: (i)
obtaining a sample of the
patient's cancer; (ii) measuring the expression level of each biomarker of a
biomarker signature in the
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
sample, wherein the biomarker signature comprises one or more of the
biomarkers FBXW2, CCND2,
RHOU, CTBP2, WIF1, and DKK1; and (iii) calculating a decision value based upon
the standardized
expression of the biomarkers in the signature; wherein a positive decision
value indicates that a
patient is predicted to respond to treatment; and (b) administering to the
patient who is predicted to
response to treatment an effective amount of the Wnt pathway inhibitor. In
some embodiments, the
method comprises identifying if the patient is likely to respond to treatment
with a Wnt pathway
inhibitor in combination with paclitaxel. In some embodiments, the method
comprises administering
to the patient the Wnt pathway inhibitor in combination with paclitaxel.
[0018] In another aspect, the invention provides a method of treating cancer
in a patient, comprising:
administering an effective amount of a Wnt pathway inhibitor to the patient;
wherein the patient is
predicted to respond to treatment with a Wnt inhibitor based upon expression
levels of a biomarker
signature in a patient tumor sample, wherein the signature comprises one or
more of the biomarkers
FBXW2, CCND2, RHOU, CTBP2, WIF1, and DKK1. In some embodiments, a method of
treating
cancer in a patient comprises: administering an effective amount of a Wnt
pathway inhibitor to the
patient; wherein the patient is predicted to respond to treatment based upon a
positive decision value
calculated from the weighted sum of the standardized expression of biomarkers
in a biomarker
signature in a patient tumor sample, wherein the set of biomarkers comprises
one or more of the
biomarkers FBXW2, CCND2, RHOU, CTBP2, WIF1, and DKK1. In some embodiments, the
patient
is predicted to respond to treatment with a Wnt pathway inhibitor in
combination with paclitaxel. In
some embodiments, the method comprises administering to the patient the Wnt
pathway inhibitor in
combination with paclitaxel.
[0019] In another aspect, the invention provides a method for increasing the
likelihood of effective
treatment with a Wnt pathway inhibitor, comprising: (a) identifying if a
patient has a tumor that is
likely to respond to treatment with a Wnt pathway inhibitor, wherein the
identification comprises: (i)
obtaining a sample of the patient's cancer; (ii) measuring the expression
level of each biomarker of a
biomarker signature in the sample, wherein the biomarker signature comprises
one or more of the
biomarkers FBXW2, CCND2, RHOU, CTBP2, WIF1, and DKK1; and (iii) identifying
the patient
who is likely to respond to treatment based upon the expression level of the
biomarkers; and (b)
administering an effective amount of the Wnt pathway inhibitor to the patient.
In some embodiments,
a method for increasing the likelihood of effective treatment with a Wnt
pathway inhibitor comprises:
(a) identifying if a patient has a tumor that is likely to respond to
treatment with a Wnt pathway
inhibitor, wherein the identification comprises: (i) obtaining a sample of the
patient's cancer; (ii)
measuring the expression level of each biomarker of a biomarker signature in
the sample, wherein the
biomarker signature comprises one or more of the biomarkers FBXW2, CCND2,
RHOU, CTBP2,
WIF1, and DKK1; and (iii) calculating a decision value based upon the
standardized expression of the
biomarkers in the biomarker signature; wherein a positive decision value
indicates that a patient is
predicted to respond to treatment; and (b) administering an effective amount
of the Wnt pathway
6
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
inhibitor to the patient whose tumor has a positive decision value. In some
embodiments, the method
comprises identifying if a patient has a tumor that is likely to respond to
treatment with a Wnt
pathway inhibitor in combination with paclitaxel. In some embodiments, the
method comprises
administering to the patient the Wnt pathway inhibitor in combination with
paclitaxel.
[0020] In another aspect, the invention provides a method for increasing the
likelihood of effective
treatment with a Wnt pathway inhibitor, comprising: administering an effective
amount of a Wnt
pathway inhibitor to a patient; wherein the patient is identified as likely to
respond to treatment with a
Wnt inhibitor based upon expression levels of a biomarker signature in a
patient tumor sample,
wherein the signature comprises one or more of the biomarkers FBXW2, CCND2,
RHOU, CTBP2,
WIF1, and DKK1. In some embodiments, a method for increasing the likelihood of
effective
treatment with a Wnt pathway inhibitor comprises: administering an effective
amount of a Wnt
pathway inhibitor to a patient; wherein the patient is identified as likely to
respond to treatment based
upon a positive decision value calculated from the weighted sum of the
standardized expression of
biomarkers in a biomarker signature in a patient tumor sample, wherein the set
of biomarkers
comprises one or more of the biomarkers FBXW2, CCND2, RHOU, CTBP2, WIF1, and
DKK1. In
some embodiments, the patient is identified as likely to respond to treatment
with a Wnt pathway
inhibitor in combination with paclitaxel. In some embodiments, the method
comprises administering
to the patient the Wnt pathway inhibitor in combination with paclitaxel.
[0021] In certain embodiments of each of the aforementioned aspects, as well
as other aspects and/or
embodiments described elsewhere herein, the biomarker signature comprises one
or more of the
biomarkers FBXW2, CCND2, RHOU, CTBP2, WIF1, DKK1, EP300, and CTBP1. In some
embodiments, the biomarker signature comprises one or more of the biomarkers
FBXW2, CCND2,
RHOU, CTBP2, WIF1, DKK1, EP300, CTBP1, WNT6, WNT3, FZD2, APC, TLE2, DVL2,
PITX2,
WISP1, GSK3B, WNT9A, FZD7, and LEF1. In some embodiments, the biomarker
signature
comprises one or more of the biomarkers FBXW2, CCND2, RHOU, CTBP2, WIF1, and
DKK1, and
at least one additional biomarker from Table 2.
[0022] In certain embodiments of each of the aforementioned aspects, as well
as other aspects and/or
embodiments described elsewhere herein, the Wnt pathway inhibitor is an
antibody. In some
embodiments, the Wnt pathway inhibitor is an antibody that specifically binds
at least one Frizzled
(FZD) protein or fragment thereof In some embodiments, the Wnt pathway
inhibitor is an antibody
that specifically binds at least one FZD protein selected from the group
consisting of: FZD1, FZD2,
FZD3, FZD4, FZD5, FZD6, FZD7, FZD8, FZD9, and FZD10. In some embodiments, the
Wnt
pathway inhibitor is an antibody that specifically binds at least one FZD
protein selected from the
group consisting of: FZD1, FZD2, FZD5, FZD7, and FZD8. In certain embodiments,
the Wnt
pathway inhibitor is an antibody which comprises: (a) a heavy chain CDR1
comprising
GFTFSHYTLS (SEQ ID NO:1), a heavy chain CDR2 comprising VISGDGSYTYYADSVKG (SEQ
ID NO:2), and a heavy chain CDR3 comprising NFIKYVFAN (SEQ ID NO:3), and (b) a
light chain
7
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
CDR1 comprising SGDNIGSFYVH (SEQ ID NO:4), a light chain CDR2 comprising
DKSNRPSG
(SEQ ID NO:5), and a light chain CDR3 comprising QSYANTLSL (SEQ ID NO:6).
[0023] In certain embodiments, the Wnt pathway inhibitor is an antibody which
comprises a heavy
chain variable region comprising SEQ ID NO:7 and a light chain variable region
comprising SEQ ID
NO: 8. In certain embodiments, the Wnt pathway inhibitor is an antibody which
comprises a heavy
chain variable region and a light chain variable region encoded by the plasmid
deposited with ATCC
as PTA-9541. In certain embodiments, the Wnt pathway inhibitor is an antibody
which comprises a
heavy chain and a light chain encoded by the plasmid deposited with ATCC as
PTA-9541. In some
embodiments, the Wnt pathway inhibitor is antibody OMP-18R5.
[0024] In certain embodiments of each of the aforementioned aspects, as well
as other aspects and/or
embodiments described elsewhere herein, the Wnt pathway inhibitor is a soluble
receptor. In some
embodiments, the Wnt pathway inhibitor comprises the extracellular domain of a
FZD receptor
protein. In some embodiments, the Wnt pathway inhibitor comprises a Fri domain
of a FZD protein.
In some embodiments, the Wnt pathway inhibitor comprises the Fri domain of
FZD8. In certain
embodiments, the Wnt pathway inhibitor comprises the Fri domain of FZD8 and a
human Fc domain.
In some embodiments, the Wnt pathway inhibitor is the soluble receptor OMP-
54F28.
[0025] In some embodiments, the tumor is selected from the group consisting of
a breast tumor, lung
tumor, a colon tumor, glioma, a gastrointestinal tumor, a renal tumor, an
ovarian tumor, a liver tumor,
a colorectal tumor, an endometrial tumor, a kidney tumor, a prostate tumor, a
thyroid tumor, a
neuroblastoma, a pancreatic tumor, a glioblastoma multiforme, a cervical
tumor, a stomach tumor, a
bladder tumor, a hepatoma, melanoma, and a head and neck tumor. In some
embodiments, the tumor
is a breast tumor.
[0026] In some embodiments, the cancer is selected from the group consisting
of a breast cancer,
lung cancer, a colon cancer, glioma, a gastrointestinal cancer, a renal
cancer, an ovarian cancer, a liver
cancer, a colorectal cancer, an endometrial cancer, a kidney cancer, a
prostate cancer, a thyroid
cancer, a neuroblastoma, a pancreatic cancer, a glioblastoma multiforme, a
cervical cancer, a stomach
cancer, a bladder cancer, a hepatoma, melanoma, and a head and neck cancer. In
some embodiments,
the cancer is breast cancer.
[0027] In some embodiments, the method further comprises administering a
second therapeutic agent
to the patient. In some embodiments, the second therapeutic agent is a
chemotherapeutic agent. In
some embodiments, the second therapeutic agent is paclitaxel.
[0028] In certain embodiments of each of the aforementioned aspects, as well
as other aspects and/or
embodiments described elsewhere herein, the sample includes, but is not
limited to, any clinically
relevant tissue sample, such as a tumor biopsy, a core biopsy tissue sample, a
fine needle aspirate, a
hair follicle, or a sample of bodily fluid, such as blood, plasma, serum,
lymph, ascitic fluid, cystic
fluid, or urine. In some embodiments, the sample is taken from a patient
having a tumor or cancer. In
some embodiments, the sample is a primary tumor. In some embodiments, the
sample is a metastasis.
8
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
In some embodiments, the sample is a tissue sample. In some embodiments, the
sample is a tumor
sample. In some embodiments, the sample is a fresh frozen (FF) tissue sample.
In some
embodiments, the sample is a formalin-fixed paraffin embedded (FFPE) tissue
sample. In some
embodiments, the sample is whole blood, plasma, or serum. In some embodiments,
the sample is
cells. In some embodiments, the sample is circulating tumor cells (CTCs).
[0029] In certain embodiments of each of the aforementioned aspects, as well
as other aspects and/or
embodiments described elsewhere herein, the expression level of a biomarker is
determined using
PCR-based methods, such as but not limited to, reverse transcription PCR (RT-
PCR), quantitative
RT-PCR (qPCR), TaqManTm, or TaqManTm low density array (TLDA). In some
embodiments, the
expression level of a biomarker is determined using a microarray.
[0030] In certain embodiments of each of the aforementioned aspects, as well
as other aspects and/or
embodiments described elsewhere herein, the standardized expression of each
biomarker is
determined by measuring an expression level for each biomarker and multiplying
it by a
corresponding weight, wherein the weight for each biomarker is determined by
the biomarker
expression. In certain embodiments, the decision value is calculated according
to the equation:
0.4560427*FBXW2 + 0.3378467*CCND2 - 0.4809354*RHOU + 0.409029*CTBP2 +
0.3291529*WIF1 + 0.2926374*DKK1 + 0.04662682.
[0031] In some embodiments, the expression level of a biomarker is measured or
determined by a
PCR-based assay. In some embodiments, the expression levels of FBXW2, CCND2,
RHOU, CTBP2,
WIF1, and DKK1 are measured using polynucleotides selected from the group
consisting of SEQ ID
NOs:62-79. In some embodiments, the expression levels of FBXW2, CCND2, RHOU,
CTBP2,
WIF1, and DKK1 are measured using (a) a forward primer of SEQ ID NO:62, a
reverse primer of
SEQ ID NO:63, and a probe comprising SEQ ID NO:64; (b) a forward primer of SEQ
ID NO:65, a
reverse primer of SEQ ID NO:66, and a probe comprising SEQ ID NO:67; (c) a
forward primer of
SEQ ID NO:68, a reverse primer of SEQ ID NO:69, and a probe comprising SEQ ID
NO:70; (d) a
forward primer of SEQ ID NO:71, a reverse primer of SEQ ID NO:72, and a probe
comprising SEQ
ID NO:73; (e) a forward primer of SEQ ID NO:74, a reverse primer of SEQ ID
NO:75, and a probe
comprising SEQ ID NO:76; and (f) a forward primer of SEQ ID NO:77, a reverse
primer of SEQ ID
NO:78, and a probe comprising SEQ ID NO:79.
[0032] In some embodiments, the expression level of a biomarker is measured or
determined by
multi-analyte profile testing, radioimmunoassay (RIA), Western blot assay,
immunofluorescent assay,
enzyme immunoassay, enzyme linked immunosorbent assay (ELISA),
immunoprecipitation assay,
chemiluminescent assay, immunohistochemical assay, dot blot assay, or slot
blot assay. In some
embodiments wherein the assay uses an antibody, the antibody is detectably
labeled. In some
embodiments, the label is selected from the group consisting of an
immunofluorescent label, a
chemiluminescent label, a phosphorescent label, an enzyme label, a radiolabel,
an avidin/biotin label,
colloidal gold particles, colored particles, and magnetic particles.
9
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
[0033] The invention also provides a kit comprising a container, wherein the
container contains at
least one reagent for specifically detecting the expression of at least one
biomarker of the invention.
In certain embodiments, the reagent is an antibody or nucleic acid probe that
binds a biomarker of the
invention.
[0034] In some embodiments, a kit comprises polynucleotides selected from the
group consisting of
SEQ ID NOs:62-79. In some embodiments, a kit comprises (a) a forward primer of
SEQ ID NO:62, a
reverse primer of SEQ ID NO:63, and a probe comprising SEQ ID NO:64; (b) a
forward primer of
SEQ ID NO:65, a reverse primer of SEQ ID NO:66, and a probe comprising SEQ ID
NO:67; (c) a
forward primer of SEQ ID NO:68, a reverse primer of SEQ ID NO:69, and a probe
comprising SEQ
ID NO:70; (d) a forward primer of SEQ ID NO:71, a reverse primer of SEQ ID
NO:72, and a probe
comprising SEQ ID NO:73; (e) a forward primer of SEQ ID NO:74, a reverse
primer of SEQ ID
NO:75, and a probe comprising SEQ ID NO:76; and (f) a forward primer of SEQ ID
NO:77, a reverse
primer of SEQ ID NO:78, and a probe comprising SEQ ID NO:79.
[0035] Where aspects or embodiments of the invention are described in terms of
a Markush group or
other grouping of alternatives, the present invention encompasses not only the
entire group listed as a
whole, but also each member of the group individually and all possible
subgroups of the main group,
and also the main group absent one or more of the group members. The present
invention also
envisages the explicit exclusion of one or more of any of the group members in
the claimed invention.
BRIEF DESCRIPTIONS OF THE DRAWINGS
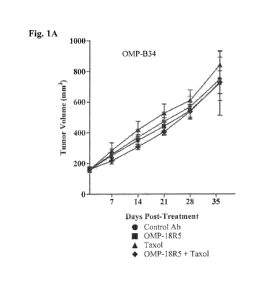
[0036] Figures 1A-1H. Classification of responsive or non-responsive breast
tumors. Figure 1A.
Breast tumor OMP-B34 cells were injected subcutaneously into NOD/SCID mice.
Figure 1B. Breast
tumor OMP-B39 cells were injected subcutaneously into NOD/SCID mice. Figure
1C. Breast tumor
OMP-B44 cells were injected subcutaneously into NOD/SCID mice. Figure 1D.
Breast tumor OMP-
B59 cells were injected subcutaneously into NOD/SCID mice. Figure 1E. Breast
tumor OMP-B60
cells were injected subcutaneously into NOD/SCID mice. Figure 1F. Breast tumor
UM-T01 cells
were injected subcutaneously into NOD/SCID mice. Figure 1G. Breast tumor UM-
T03 cells were
injected subcutaneously into NOD/SCID mice. Figure 1H. Breast tumor UM-PE13
cells were
injected subcutaneously into NOD/SCID mice. For each experiment, mice were
treated with OMP-
18R5 antibody(-.-), taxol (-=-), a combination of OMP-18R5 and taxol (- V -),
or a control antibody
(-=-). Data is shown as tumor volume (mm3) over days post-treatment.
[0037] Figure 2. Performance curve for the top 20 ranked genes.
[0038] Figure 3. PCA plot of 6 selected genes.
[0039] Figure 4. Correlation of the 6-gene biomarker signature with ratio of
tumor volume.
[0040] Figure 5. Prediction of tumor responsiveness based upon classification
probability analysis.
T = tumor used in training set for establishment of 6-gene signature.
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
[0041] Figures 6A-6F. In vivo validation of predictive biomarkers. Figure 6A.
Breast tumor OMP-
B29 cells were injected subcutaneously into NOD/SCID mice. Figure 6B. Breast
tumor OMP-B71
cells were injected subcutaneously into NOD/SCID mice. Figure 6C. Breast tumor
OMP-B84 cells
were injected subcutaneously into NOD/SCID mice. Figure 6D. Breast tumor OMP-
B90 cells were
injected subcutaneously into NOD/SCID mice. Figure 6E. Breast tumor UM-T02
cells were injected
subcutaneously into NOD/SCID mice. Figure 6F. Breast tumor UM-T06 cells were
injected
subcutaneously into NOD/SCID mice. For each experiment, mice were treated with
OMP-18R5
antibody (-0-), taxol (-=-), a combination of OMP-18R5 and taxol (- V -), or a
control antibody (-=-).
Data is shown as tumor volume (mm3) over days post-treatment.
[0042] Figure 7. Population prevalence estimation of the 6-gene biomarker
signature using three
public datasets.
DETAILED DESCRIPTION OF THE INVENTION
I. Definitions
[0043] To facilitate an understanding of the present invention, a number of
terms and phrases are
defined below.
[0044] The term "biomarker" as used herein may include but is not limited to,
nucleic acids and
proteins, and variants and fragments thereof A biomarker may include DNA
comprising the entire or
partial nucleic acid sequence encoding the biomarker, or the complement of
such a sequence.
Biomarker nucleic acids useful in the invention are considered to include both
DNA and RNA
comprising the entire or partial sequence of any of the nucleic acid sequences
of interest. Biomarker
proteins are considered to comprise the entire or partial amino acid sequence
of any of the biomarker
proteins or polypeptides.
[0045] The term "antibody" as used herein refers to an immunoglobulin molecule
that recognizes and
specifically binds a target, such as a protein, polypeptide, peptide,
carbohydrate, polynucleotide, lipid,
or combinations of the foregoing, through at least one antigen-binding site
within the variable region
of the immunoglobulin molecule. As used herein, the term encompasses intact
polyclonal antibodies,
intact monoclonal antibodies, single chain antibodies, antibody fragments
(such as Fab, Fab', F(ab')2,
and Fv fragments), single chain Fv (scFv) antibodies, multispecific antibodies
such as bispecific
antibodies, monospecific antibodies, monovalent antibodies, chimeric
antibodies, humanized
antibodies, human antibodies, fusion proteins comprising an antigen-binding
site of an antibody, and
any other modified immunoglobulin molecule comprising an antigen-binding site
as long as the
antibodies exhibit the desired biological activity. An antibody can be any of
the five major classes of
immunoglobulins: IgA, IgD, IgE, IgG, and IgM, or subclasses (isotypes) thereof
(e.g., IgGl, IgG2,
IgG3, IgG4, IgAl, and IgA2), based on the identity of their heavy chain
constant domains referred to
as alpha, delta, epsilon, gamma, and mu, respectively. The different classes
of immunoglobulins have
11
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
different and well-known subunit structures and three-dimensional
configurations. Antibodies can be
naked or conjugated to other molecules, including but not limited to, toxins
and radioisotopes.
[0046] The term "antibody fragment" refers to a portion of an intact antibody
and refers to the
antigenic determining variable regions of an intact antibody. Examples of
antibody fragments
include, but are not limited to, Fab, Fab', F(ab')2, and Fv fragments, linear
antibodies, single chain
antibodies, and multispecific antibodies formed from antibody fragments.
"Antibody fragment" as
used herein comprises at least one antigen-binding site or epitope-binding
site.
[0047] The term "variable region" of an antibody refers to the variable region
of an antibody light
chain, or the variable region of an antibody heavy chain, either alone or in
combination. The variable
region of a heavy chain or a light chain generally consists of four framework
regions (FR) connected
by three complementarity determining regions (CDRs), also known as
"hypervariable regions". The
CDRs in each chain are held together in close proximity by the framework
regions and contribute to
the formation of the antigen-binding site(s) of the antibody. There are at
least two techniques for
determining CDRs: (1) an approach based on cross-species sequence variability
(i.e., Kabat et al.,
1991, Sequences of Proteins of Immunological Interest, 5th Edition, National
Institutes of Health,
Bethesda, MD), and (2) an approach based on crystallographic studies of
antigen-antibody complexes
(Al-Lazikani et al., 1997, J. MoL Biol., 273:927-948). In addition,
combinations of these two
approaches are sometimes used in the art to determine CDRs.
[0048] The term "monoclonal antibody" as used herein refers to a homogeneous
antibody population
involved in the highly specific recognition and binding of a single antigenic
determinant or epitope.
This is in contrast to polyclonal antibodies that typically include a mixture
of different antibodies
directed against a variety of different antigenic determinants. The term
"monoclonal antibody"
encompasses both intact and full-length monoclonal antibodies as well as
antibody fragments (e.g.,
Fab, Fab', F(ab')2, Fv), single chain (scFv) antibodies, fusion proteins
comprising an antibody portion,
and any other modified immunoglobulin molecule comprising an antigen-binding
site. Furthermore,
"monoclonal antibody" refers to such antibodies made by any number of
techniques, including but not
limited to, hybridoma production, phage selection, recombinant expression, and
transgenic animals.
[0049] The term "humanized antibody" as used herein refers to antibodies that
are specific
immunoglobulin chains, chimeric immunoglobulins, or fragments thereof that
contain minimal non-
human sequences. Methods used to generate humanized antibodies are well known
in the art.
[0050] The term "human antibody" as used herein refers to an antibody produced
by a human or an
antibody having an amino acid sequence corresponding to an antibody produced
by a human. A
human antibody may be made using any of the techniques known in the art.
[0051] The term "chimeric antibody" as used herein refers to an antibody
wherein the amino acid
sequence of the immunoglobulin molecule is derived from two or more species.
Typically, the
variable regions of the light chain and the heavy chain correspond to the
variable regions of an
antibody derived from one species of mammals (e.g., mouse, rat, rabbit, etc.)
with the desired
12
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
specificity, affinity, and/or binding capability, while the constant regions
correspond to sequences
from an antibody derived from another species (usually human).
[0052] The term "affinity-matured antibody" as used herein refers to an
antibody with one or more
alterations in one or more CDRs thereof that result in an improvement in the
affinity of the antibody
for antigen, compared to a parent antibody that does not possess those
alterations(s). The definition
also includes alterations in non-CDR residues made in conjunction with
alterations to CDR residues.
Preferred affinity-matured antibodies will have nanomolar or even picomolar
affinities for the target
antigen. Affinity-matured antibodies are produced by procedures known in the
art. For example,
techniques may include affinity maturation by VH and VL domain shuffling,
random mutagenesis of
CDR and/or framework residues, and site-directed mutagenesis.
[0053] The terms "epitope" and "antigenic determinant" are used
interchangeably herein and refer to
that portion of an antigen capable of being recognized and specifically bound
by a particular antibody.
When the antigen is a polypeptide, epitopes can be formed both from contiguous
amino acids and
noncontiguous amino acids juxtaposed by tertiary folding of a protein.
Epitopes formed from
contiguous amino acids (also referred to as linear epitopes) are typically
retained upon protein
denaturing, whereas epitopes formed by tertiary folding (also referred to as
conformational epitopes)
are typically lost upon protein denaturing. An epitope typically includes at
least 3, and more usually,
at least 5 or 8-10 amino acids in a unique spatial conformation.
[0054] The terms "selectively binds" or "specifically binds" mean that a
binding agent or an antibody
reacts or associates more frequently, more rapidly, with greater duration,
with greater affinity, or with
some combination of the above to the epitope, protein, or target molecule than
with alternative
substances, including unrelated or related proteins. In certain embodiments
"specifically binds"
means, for instance, that an antibody binds a protein with a KD of about 0.1mM
or less, but more
usually less than about l[tM. In certain embodiments, "specifically binds"
means that an antibody
binds a target at times with a KD of at least about 0.1[tM or less, at other
times at least about 0.01[tM
or less, and at other times at least about 1nM or less. Because of the
sequence identity between
homologous proteins in different species, specific binding can include an
antibody that recognizes a
protein in more than one species (e.g., human FZD and mouse FZD). Likewise,
because of homology
within certain regions of polypeptide sequences of different proteins,
specific binding can include an
antibody (or other polypeptide or binding agent) that recognizes more than one
protein (e.g., human
FZD1 and human FZD7). It is understood that, in certain embodiments, an
antibody or binding agent
that specifically binds a first target may or may not specifically bind a
second target. As such,
"specific binding" does not necessarily require (although it can include)
exclusive binding, i.e.
binding to a single target. Thus, a binding agent may, in certain embodiments,
specifically bind more
than one target. In certain embodiments, multiple targets may be bound by the
same binding site on
the agent or antibody. For example, an antibody may, in certain instances,
comprise two identical
antigen-binding sites, each of which specifically binds the same epitope on
two or more proteins. In
13
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
certain alternative embodiments, an antibody may be bispecific or
multispecific and comprise at least
two antigen-binding sites with differing specificities. By way of non-limiting
example, a bispecific
agent may comprise one binding site that recognizes a target on one protein
(e.g., human FZD) and
further comprise a second, different binding site that recognizes a different
target on a second protein
(e.g., a human WNT protein). Generally, but not necessarily, reference to
binding means specific
binding.
[0055] The terms "polypeptide" and "peptide" and "protein" are used
interchangeably herein and
refer to polymers of amino acids of any length. The polymer may be linear or
branched, it may
comprise modified amino acids, and it may be interrupted by non-amino acids.
The terms also
encompass an amino acid polymer that has been modified naturally or by
intervention; for example,
disulfide bond formation, glycosylation, lipidation, acetylation,
phosphorylation, or any other
manipulation or modification, such as conjugation with a labeling component.
Also included within
the definition are, for example, polypeptides containing one or more analogs
of an amino acid
(including, for example, unnatural amino acids), as well as other
modifications known in the art. It is
understood that, because the polypeptides of this invention may be based upon
antibodies, in certain
embodiments, the polypeptides can occur as single chains or associated chains
(e.g., dimers).
[0056] The terms "polynucleotide" and "nucleic acid" are used interchangeably
herein and refer to
polymers of nucleotides of any length, and include DNA and RNA. The
nucleotides can be
deoxyribonucleotides, ribonucleotides, modified nucleotides or bases, and/or
their analogs, or any
substrate that can be incorporated into a polymer by DNA or RNA polymerase.
[0057] "Conditions of high stringency" may be identified by conditions that:
(1) employ low ionic
strength and high temperature for washing, for example 15mM sodium
chloride/1.5mM sodium
citrate/0.1% sodium dodecyl sulfate at 50 C; (2) employ during hybridization a
denaturing agent, such
as formamide, for example, 50% (v/v) formamide with 0.1% bovine serum
albumin/0.1% Fico11/0.1%
polyvinylpyrrolidone/50mM sodium phosphate buffer at pH 6.5 in 5x SSC (0.75M
NaC1, 75mM
sodium citrate) at 42 C; or (3) employ during hybridization 50% formamide in
5x SSC, 50mM
sodium phosphate (pH 6.8), 0.1% sodium pyrophosphate, 5x Denhardt's solution,
sonicated salmon
sperm DNA (50Kg/m1), 0.1% SDS, and 10% dextran sulfate at 42 C, with washes at
42 C in 0.2x
SSC and 50% formamide, followed by a wash consisting of 0.1x SSC containing
EDTA at 55 C.
[0058] The terms "identical" or percent "identity" in the context of two or
more nucleic acids or
polypeptides, refer to two or more sequences or subsequences that are the same
or have a specified
percentage of nucleotides or amino acid residues that are the same, when
compared and aligned
(introducing gaps, if necessary) for maximum correspondence, not considering
any conservative
amino acid substitutions as part of the sequence identity. The percent
identity may be measured using
sequence comparison software or algorithms or by visual inspection. Various
algorithms and
software that may be used to obtain alignments of amino acid or nucleotide
sequences are well-known
in the art. These include, but are not limited to, BLAST, ALIGN, Megalign,
BestFit, GCG Wisconsin
14
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
Package, and variations thereof In some embodiments, two nucleic acids or
polypeptides of the
invention are substantially identical, meaning they have at least 70%, at
least 75%, at least 80%, at
least 85%, at least 90%, and in some embodiments at least 95%, 96%, 97%, 98%,
99% nucleotide or
amino acid residue identity, when compared and aligned for maximum
correspondence, as measured
using a sequence comparison algorithm or by visual inspection. In some
embodiments, identity exists
over a region of the sequences that is at least about 10, at least about 20,
at least about 40-60 residues,
at least about 60-80 residues in length or any integral value therebetween. In
some embodiments,
identity exists over a longer region than 60-80 residues, such as at least
about 80-100 residues, and in
some embodiments the sequences are substantially identical over the full
length of the sequences
being compared, such as the coding region of a nucleotide sequence.
[0059] A "conservative amino acid substitution" is one in which one amino acid
residue is replaced
with another amino acid residue having a similar side chain. Families of amino
acid residues having
similar side chains have been defined in the art, including basic side chains
(e.g., lysine, arginine,
histidine), acidic side chains (e.g., aspartic acid, glutamic acid), uncharged
polar side chains (e.g.,
glycine, asparagine, glutamine, serine, threonine, tyrosine, cysteine), non-
polar side chains (e.g.,
alanine, valine, leucine, isoleucine, proline, phenylalanine, methionine,
tryptophan), beta-branched
side chains (e.g., threonine, valine, isoleucine) and aromatic side chains
(e.g., tyrosine, phenylalanine,
tryptophan, histidine). For example, substitution of a phenylalanine for a
tyrosine is a conservative
substitution. Preferably, conservative substitutions in the sequences of the
polypeptides and
antibodies of the invention do not abrogate the binding of the polypeptide or
antibody containing the
amino acid sequence, to the antigen to which the polypeptide or antibody
binds. Methods of
identifying nucleotide and amino acid conservative substitutions which do not
eliminate antigen
binding are well-known in the art.
[0060] The term "vector" as used herein means a construct, which is capable of
delivering, and
usually expressing, one or more gene(s) or sequence(s) of interest in a host
cell. Examples of vectors
include, but are not limited to, viral vectors, naked DNA or RNA expression
vectors, plasmid, cosmid,
or phage vectors, DNA or RNA expression vectors associated with cationic
condensing agents, and
DNA or RNA expression vectors encapsulated in liposomes.
[0061] As used herein the term "soluble receptor" refers to an extracellular
domain (or a fragment
thereof) of a receptor protein preceding the first transmembrane domain of the
receptor that can be
secreted from a cell in soluble form. Generally this is the N-terminal portion
of the receptor protein.
[0062] As used herein the term "FZD soluble receptor" or "soluble FZD
receptor" refers to an N-
terminal extracellular fragment of a FZD receptor protein preceding the first
transmembrane domain
of the receptor that can be secreted from a cell in soluble form. FZD soluble
receptors comprising the
entire N-terminal extracellular domain (ECD) as well as smaller fragments are
encompassed by the
term. Thus, FZD soluble receptors comprising a FZD Fri domain are also
included in this term.
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
[0063] A polypeptide, antibody, polynucleotide, vector, cell, or composition
which is "isolated" is a
polypeptide, antibody, polynucleotide, vector, cell, or composition which is
in a form not found in
nature. Isolated polypeptides, antibodies, polynucleotides, vectors, cells, or
compositions include
those which have been purified to a degree that they are no longer in a form
in which they are found
in nature. In some embodiments, a polypeptide, antibody, polynucleotide,
vector, cell, or composition
which is isolated is substantially pure.
[0064] The term "substantially pure" as used herein refers to material which
is at least 50% pure (i.e.,
free from contaminants), at least 90% pure, at least 95% pure, at least 98%
pure, or at least 99% pure.
[0065] The terms "cancer" and "cancerous" as used herein refer to or describe
the physiological
condition in mammals in which a population of cells are characterized by
unregulated cell growth.
Examples of cancer include, but are not limited to, carcinoma, blastoma,
sarcoma, and hematologic
cancers such as lymphoma and leukemia.
[0066] The terms "tumor" and "neoplasm" as used herein refer to any mass of
tissue that results from
excessive cell growth or proliferation, either benign (non-cancerous) or
malignant (cancerous)
including pre-cancerous lesions.
[0067] The term "metastasis" as used herein refers to the process by which a
cancer spreads or
transfers from the site of origin to other regions of the body with the
development of a similar
cancerous lesion at a new location. A "metastatic" or "metastasizing" cell is
one that loses adhesive
contacts with neighboring cells and migrates (e.g., via the bloodstream or
lymph) from the primary
site of disease to secondary sites.
[0068] The terms "cancer stem cell" and "CSC" and "tumor stem cell" and "tumor
initiating cell" are
used interchangeably herein and refer to cells from a cancer or tumor that:
(1) have extensive
proliferative capacity; 2) are capable of asymmetric cell division to generate
one or more types of
differentiated cell progeny wherein the differentiated cells have reduced
and/or limited proliferative or
developmental potential; and (3) are capable of symmetric cell divisions for
self-renewal or self-
maintenance. These properties confer on the cancer stem cells the ability to
form or establish a tumor
or cancer upon serial transplantation into an immunocompromised host (e.g., a
mouse) compared to
the majority of tumor cells that fail to form tumors. Cancer stem cells
undergo self-renewal versus
differentiation in a chaotic manner to form tumors with abnormal cell types
that can change over time
as mutations occur.
[0069] The terms "cancer cell" and "tumor cell" refer to the total population
of cells derived from a
cancer or tumor or pre-cancerous lesion, including both non-tumorigenic cells,
which comprise the
bulk of the cancer cell population, and tumorigenic stem cells (cancer stem
cells). As used herein, the
terms "cancer cell" or "tumor cell" will be modified by the term "non-
tumorigenic" when referring
solely to those cells lacking the capacity to renew and differentiate to
distinguish those tumor cells
from cancer stem cells.
16
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
[0070] The term "tumorigenic" as used herein refers to the functional features
of a cancer stem cell
including the properties of self-renewal (giving rise to additional
tumorigenic cancer stem cells) and
proliferation to generate all other tumor cells (giving rise to differentiated
and thus non-tumorigenic
tumor cells).
[0071] The term "tumorigenicity" as used herein refers to the ability of a
random sample of cells
from the tumor to form palpable tumors upon serial transplantation into
immunocompromised hosts
(e.g., mice). This definition also includes enriched and/or isolated
populations of cancer stem cells
that form palpable tumors upon serial transplantation into immunocompromised
hosts (e.g., mice).
[0072] The term "patient" refers to any animal (e.g., a mammal), including,
but not limited to,
humans, non-human primates, canines, felines, rodents, and the like, which is
to be the recipient of a
particular treatment. Typically, the terms "patient" and "subject" are used
interchangeably herein in
reference to a human patient.
[0073] The term "pharmaceutically acceptable" refers to a product or compound
approved (or
approvable) by a regulatory agency of the Federal government or a state
government or listed in the
U.S. Pharmacopeia or other generally recognized pharmacopeia for use in
animals, including humans.
[0074] The terms "pharmaceutically acceptable excipient, carrier or adjuvant"
or "acceptable
pharmaceutical carrier" refer to an excipient, carrier, or adjuvant that can
be administered to a subject,
together with at least one agent (e.g., an antibody) of the present
disclosure, and which does not
destroy the activity of the agent. The excipient, carrier, or adjuvant should
be non-toxic when
administered with an agent in doses sufficient to deliver a therapeutic
effect.
[0075] The terms "effective amount" or "therapeutically effective amount" or
"therapeutic effect"
refer to an amount of a binding agent, an antibody, polypeptide,
polynucleotide, small organic
molecule, or other drug effective to "treat" a disease or disorder in a
subject or mammal. In the case
of cancer, the therapeutically effective amount of a drug (e.g., an antibody)
has a therapeutic effect
and as such can reduce the number of cancer cells; decrease tumorigenicity,
tumorigenic frequency, or
tumorigenic capacity; reduce the number or frequency of cancer stem cells;
reduce the tumor size;
reduce the cancer cell population; inhibit and/or stop cancer cell
infiltration into peripheral organs
including, for example, the spread of cancer into soft tissue and bone;
inhibit and/or stop tumor or
cancer cell metastasis; inhibit and/or stop tumor or cancer cell growth;
relieve to some extent one or
more of the symptoms associated with the cancer; reduce morbidity and
mortality; improve quality of
life; or a combination of such effects. To the extent the agent, for example
an antibody, prevents
growth and/or kills existing cancer cells, it can be referred to as cytostatic
and/or cytotoxic.
[0076] The terms "treating" or "treatment" or "to treat" or "alleviating" or
"to alleviate" refer to both
1) therapeutic measures that cure, slow down, lessen symptoms of, and/or halt
progression of a
diagnosed pathologic condition or disorder and 2) prophylactic or preventative
measures that prevent
or slow the development of a targeted pathologic condition or disorder. Thus
those in need of
treatment include those already diagnosed with the disorder; those prone to
have the disorder; and
17
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
those in whom the disorder is to be prevented. In some embodiments, a subject
is successfully
"treated" according to the methods of the present invention if the patient
shows one or more of the
following: a reduction in the number of and/or complete absence of cancer
cells; a reduction in the
tumor size; an inhibition of tumor growth; inhibition of and/or an absence of
cancer cell infiltration
into peripheral organs including the spread of cancer cells into soft tissue
and bone; inhibition of
and/or an absence of tumor or cancer cell metastasis; inhibition and/or an
absence of cancer growth;
relief of one or more symptoms associated with the specific cancer; reduced
morbidity and mortality;
improvement in quality of life; reduction in tumorigenicity; reduction in the
number or frequency of
cancer stem cells; or some combination of such effects.
[0077] As used in the present disclosure and claims, the singular forms "a",
"an" and "the" include
plural forms unless the context clearly dictates otherwise.
[0078] It is understood that wherever embodiments are described herein with
the language
"comprising" otherwise analogous embodiments described in terms of "consisting
of' and/or
"consisting essentially of' are also provided. It is also understood that
wherever embodiments are
described herein with the language "consisting essentially of' otherwise
analogous embodiments
described in terms of "consisting of' are also provided.
[0079] The term "and/or" as used in a phrase such as "A and/or B" herein is
intended to include both
A and B; A or B; A (alone); and B (alone). Likewise, the term "and/or" as used
in a phrase such as
"A, B, and/or C" is intended to encompass each of the following embodiments:
A, B, and C; A, B, or
C; A or C; A or B; B or C; A and C; A and B; B and C; A (alone); B (alone);
and C (alone).
II. Methods of use of predictive biomarkers
[0080] Provided herein are methods for identifying, classifying, and/or
selecting tumors and/or
patients with cancer that are likely to be responsive ("sensitive") or non-
responsive ("resistant") to
treatment with a Wnt pathway inhibitor. In addition, provided are methods for
treating patients with
cancer who are likely to respond to treatment, are predicted to respond to
treatment, and/or have been
identified to respond to treatment with a Wnt pathway inhibitor.
[0081] Provided herein is a method of identifying a human tumor that is likely
to be responsive or
non-responsive to treatment with a Wnt pathway inhibitor, the method
comprising: (a) obtaining a
sample of the human tumor; (b) measuring the expression level of each
biomarker of a biomarker
signature in the sample, wherein the biomarker signature comprises one or more
of the biomarkers
FBXW2, CCND2, RHOU, CRBP2, WIF1, and DKK1; and (c) identifying the tumor as
likely to be
responsive or non-responsive to treatment based upon the expression level of
the biomarkers. In some
embodiments, a method of identifying a human tumor that is likely to be
responsive or non-responsive
to treatment with a Wnt pathway inhibitor comprises: (a) obtaining a sample of
the human tumor; (b)
measuring the expression level of each biomarker of a biomarker signature in
the sample, wherein the
biomarker signature comprises one or more of the biomarkers FBXW2, CCND2,
RHOU, CRBP2,
18
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
WIF1, and DKK1; and (c) calculating a decision value based upon the
standardized expression of the
biomarkers in the biomarker signature; wherein a positive decision value
indicates the tumor is
predicted to be responsive to the Wnt pathway inhibitor and a negative
decision value indicates the
tumor is predicted to be non-responsive to the Wnt pathway inhibitor.
[0082] Provided herein is a method of classifying a human tumor as likely to
be responsive or non-
responsive to treatment with a Wnt pathway inhibitor, the method comprising:
(a) obtaining a sample
of the human tumor; (b) measuring the expression level of each biomarker of a
biomarker signature in
the sample, wherein the biomarker signature comprises one or more of the
biomarkers FBXW2,
CCND2, RHOU, CTBP2, WIF1, and DKK1; and (c) classifying the tumor as likely to
be responsive
or non-responsive to treatment based upon the expression level of the
biomarkers. In some
embodiments, a method of classifying a human tumor as likely to be responsive
or non-responsive to
treatment with a Wnt pathway inhibitor comprises: (a) obtaining a sample of
the human tumor; (b)
measuring the expression level of each biomarker of a biomarker signature in
the sample, wherein the
biomarker signature comprises one or more of the biomarkers FBXW2, CCND2,
RHOU, CTBP2,
WIF1, and DKK1; and (c) calculating a decision value based upon the
standardized expression of the
biomarkers in the biomarker signature; wherein a positive decision value
indicates the tumor is
predicted to be responsive to the Wnt pathway inhibitor and a negative
decision value indicates the
tumor is predicted to be non-responsive to the Wnt pathway inhibitor.
[0083] Provided herein is a method of determining the responsiveness (or
sensitivity) of a human
tumor to treatment with a Wnt pathway inhibitor, the method comprising: (a)
obtaining a sample of
the human tumor; (b) measuring the expression level of each biomarker of a
biomarker signature in
the sample, wherein the biomarker signature comprises one or more of the genes
FBXW2, CCND2,
RHOU, CTBP2, WIF1, and DKK1; and (c) determining the responsiveness of the
tumor to treatment
based upon the expression level of the biomarkers. In some embodiments, a
method of determining
the responsiveness or sensitivity of a human tumor to treatment with a Wnt
pathway inhibitor
comprises: (a) obtaining a sample of the human tumor; (b) measuring the
expression level of each
biomarker of a biomarker signature in the sample, wherein the biomarker
signature comprises one or
more of the genes FBXW2, CCND2, RHOU, CTBP2, WIF1, and DKK1; and (c)
calculating a
decision value based upon the standardized expression of the biomarkers in the
biomarker signature;
wherein a positive decision value indicates the tumor is predicted to be
responsive to the Wnt pathway
inhibitor.
[0084] Provided herein is a method of identifying a patient with cancer who is
likely to respond to
treatment with a Wnt pathway inhibitor, the method comprising: (a) obtaining a
sample of the
patient's tumor; (b) measuring the expression level of each biomarker of a
biomarker signature in the
sample, wherein the biomarker signature comprises one or more of the
biomarkers FBXW2, CCND2,
RHOU, CTBP2, WIF1, and DKK1; and (c) identifying the patient who is likely to
respond to
treatment based upon the expression level of the biomarkers. In some
embodiments, a method of
19
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
identifying a patient with cancer who is likely to respond to treatment with a
Wnt pathway inhibitor
comprises: (a) obtaining a sample of the patient's tumor; (b) measuring the
expression level of each
biomarker of a biomarker signature in the sample, wherein the biomarker
signature comprises one or
more of the biomarkers FBXW2, CCND2, RHOU, CTBP2, WIF1, and DKK1; and (c)
calculating a
decision value based upon the standardized expression of the biomarkers in the
biomarker signature;
wherein a positive decision value indicates that the patient is predicted to
respond to treatment with
the Wnt pathway inhibitor. In some embodiments, the method further comprises
selecting the patient
for treatment when their tumor sample has a positive decision value. In some
embodiments, the
method further comprises administering a therapeutically effective amount of
the Wnt pathway
inhibitor to the patient.
[0085] Provided herein is a method of selecting a patient with cancer for
treatment with a Wnt
pathway inhibitor, the method comprising: (a) obtaining a sample of the
patient's tumor; (b)
measuring the expression level of each biomarker of a biomarker signature in
the sample, wherein the
biomarker signature comprises one or more of the biomarkers FBXW2, CCND2,
RHOU, CTBP2,
WIF1, and DKK1; (c) selecting the patient for treatment based upon the
expression level of the
biomarkers. In some embodiments, a method of selecting a patient with cancer
for treatment with a
Wnt pathway inhibitor comprises: (a) obtaining a sample of the patient's
tumor; (b) measuring the
expression level of each biomarker of a biomarker signature in the sample,
wherein the biomarker
signature comprises one or more of the biomarkers FBXW2, CCND2, RHOU, CTBP2,
WIF1, and
DKK1; (c) calculating a decision value based upon the standardized expression
of the biomarkers in
the biomarker signature; and (d) selecting the patient for treatment when
their tumor sample has a
positive decision value. In some embodiments, the method further comprises
administering a
therapeutically effective amount of the Wnt pathway inhibitor to the patient.
[0086] Provided herein is a method of treating cancer in a patient,
comprising: (a) identifying if the
patient is likely to respond to treatment with a Wnt pathway inhibitor,
wherein the identification
comprises: (i) obtaining a sample of the patient's cancer; (ii) measuring the
expression level of each
biomarker of a biomarker signature in the sample, wherein the biomarker
signature comprises one or
more of the biomarkers FBXW2, CCND2, RHOU, CTBP2, WIF1, and DKK1; and (iii)
identifying
the patient who is likely to respond to treatment based upon the expression
level of the biomarkers;
and (b) administering to the patient who is likely to response to treatment an
effective amount of the
Wnt pathway inhibitor. In some embodiments, a method of treating cancer in a
patient comprises: (a)
identifying if the patient is likely to respond to treatment with a Wnt
pathway inhibitor, wherein the
identification comprises: (i) obtaining a sample of the patient's cancer; (ii)
measuring the expression
level of each biomarker of a biomarker signature in the sample, wherein the
biomarker signature
comprises one or more of the biomarkers FBXW2, CCND2, RHOU, CTBP2, WIF1, and
DKK1; and
(iii) calculating a decision value based upon the standardized expression of
the biomarkers in the
signature; wherein a positive decision value indicates that the patient is
predicted to respond to
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
treatment; and (b) administering to the patient who is predicted to response
to treatment an effective
amount of the Wnt pathway inhibitor.
[0087] In another aspect, the invention provides a method of treating cancer
in a patient, comprising:
administering an effective amount of a Wnt pathway inhibitor to the patient;
wherein the patient is
predicted to respond to treatment with a Wnt inhibitor based upon expression
levels of a biomarker
signature in a patient tumor sample, wherein the signature comprises one or
more of the biomarkers
FBXW2, CCND2, RHOU, CTBP2, WIF1, and DKK1. In some embodiments, a method of
treating
cancer in a patient comprises: administering an effective amount of a Wnt
pathway inhibitor to the
patient; wherein the patient is predicted to respond to treatment based upon a
positive decision value
calculated from the weighted sum of the standardized expression of biomarkers
in a biomarker
signature in a patient tumor sample, wherein the set of biomarkers comprises
one or more of the
biomarkers FBXW2, CCND2, RHOU, CTBP2, WIF1, and DKK1.
[0088] Provided herein is a method for increasing the likelihood of effective
treatment with a Wnt
pathway inhibitor, comprising: (a) identifying if a patient has a tumor that
is likely to respond to
treatment with a Wnt pathway inhibitor, wherein the identification comprises:
(i) obtaining a sample
of the patient's cancer; (ii) measuring the expression level of each biomarker
of a biomarker signature
in the sample, wherein the biomarker signature comprises one or more of the
biomarkers FBXW2,
CCND2, RHOU, CTBP2, WIF1, and DKK1; and (iii) identifying the patient who is
likely to respond
to treatment based upon the expression level of the biomarkers; and (b)
administering an effective
amount of the Wnt pathway inhibitor to the patient. In some embodiments, a
method for increasing
the likelihood of effective treatment with a Wnt pathway inhibitor comprises:
(a) identifying if a
patient has a tumor that is likely to respond to treatment with a Wnt pathway
inhibitor, wherein the
identification comprises: (i) obtaining a sample of the patient's cancer; (ii)
measuring the expression
level of each biomarker of a biomarker signature in the sample, wherein the
biomarker signature
comprises one or more of the biomarkers FBXW2, CCND2, RHOU, CTBP2, WIF1, and
DKK1; and
(iii) calculating a decision value based upon the standardized expression of
the biomarkers in the
biomarker signature; wherein a positive decision value indicates that the
patient is predicted to
respond to treatment; and (b) administering an effective amount of the Wnt
pathway inhibitor to the
patient whose tumor has a positive decision value.
[0089] In another aspect, the invention provides a method for increasing the
likelihood of effective
treatment with a Wnt pathway inhibitor, comprising: administering an effective
amount of a Wnt
pathway inhibitor to a patient; wherein the patient is identified as likely to
respond to treatment with a
Wnt inhibitor based upon expression levels of a biomarker signature in a
patient tumor sample,
wherein the signature comprises one or more of the biomarkers FBXW2, CCND2,
RHOU, CTBP2,
WIF1, and DKK1. In some embodiments, a method for increasing the likelihood of
effective
treatment with a Wnt pathway inhibitor comprises: administering an effective
amount of a Wnt
pathway inhibitor to a patient; wherein the patient is identified as likely to
respond to treatment based
21
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
upon a positive decision value calculated from the weighted sum of the
standardized expression of
biomarkers in a biomarker signature in a patient tumor sample, wherein the set
of biomarkers
comprises one or more of the biomarkers FBXW2, CCND2, RHOU, CTBP2, WIF1, and
DKK1. In
some embodiments, the patient is identified as likely to respond to treatment
with a Wnt pathway
inhibitor in combination with paclitaxel. In some embodiments, the method
comprises administering
to the patient the Wnt pathway inhibitor in combination with paclitaxel.
[0090] Provided herein is a use for identifying a human tumor that is likely
to be responsive or non-
responsive to treatment with a Wnt pathway inhibitor, wherein the use
comprises (a) obtaining a
sample of the human tumor; (b) measuring the expression level of each
biomarker of a biomarker
signature in the sample, wherein the biomarker signature comprises one or more
of the biomarkers
FBXW2, CCND2, RHOU, CRBP2, WIF1, and DKK1; and (c) calculating a decision
value based
upon the standardized expression of the biomarkers in the biomarker signature;
wherein a positive
decision value indicates the tumor is predicted to be responsive to the Wnt
pathway inhibitor and a
negative decision value indicates the tumor is predicted to be non-responsive
to the Wnt pathway
inhibitor.
[0091] Provided herein is a use for classifying a human tumor as likely to be
responsive or non-
responsive to treatment with a Wnt pathway inhibitor, wherein the use
comprises (a) obtaining a
sample of the human tumor; (b) measuring the expression level of each
biomarker of a biomarker
signature in the sample, wherein the biomarker signature comprises one or more
of the biomarkers
FBXW2, CCND2, RHOU, CTBP2, WIF1, and DKK1; and (c) calculating a decision
value based
upon the standardized expression of the biomarkers in the biomarker signature;
wherein a positive
decision value indicates the tumor is predicted to be responsive to the Wnt
pathway inhibitor and a
negative decision value indicates the tumor is predicted to be non-responsive
to the Wnt pathway
inhibitor.
[0092] Provided herein is a use for determining the sensitivity of a human
tumor to treatment with a
Wnt pathway inhibitor, wherein the use comprises (a) obtaining a sample of the
human tumor; (b)
measuring the expression level of each biomarker of a biomarker signature in
the sample, wherein the
biomarker signature comprises one or more of the genes FBXW2, CCND2, RHOU,
CTBP2, WIF1,
and DKK1; and (c) calculating a decision value based upon the standardized
expression of the
biomarkers in the biomarker signature; wherein a positive decision value
indicates the tumor is
predicted to be responsive to the Wnt pathway inhibitor.
[0093] Provided herein is a use for identifying a patient with cancer who is
likely to respond to
treatment with a Wnt pathway inhibitor, wherein the use comprises (a)
obtaining a sample of the
patient's tumor; (b) measuring the expression level of each biomarker of a
biomarker signature in the
sample, wherein the biomarker signature comprises one or more of the
biomarkers FBXW2, CCND2,
RHOU, CTBP2, WIF1, and DKK1; and (c) calculating a decision value based upon
the standardized
expression of the biomarkers in the biomarker signature; wherein a positive
decision value indicates
22
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
that the patient is predicted to respond to treatment with the Wnt pathway
inhibitor. In some
embodiments, the use further comprises selecting the patient for treatment
when their tumor sample
has a positive decision value. In some embodiments, the use further comprises
administering a
therapeutically effective amount of the Wnt pathway inhibitor to the patient.
[0094] Provided herein is a use for selecting a patient with cancer for
treatment with a Wnt pathway
inhibitor, wherein the use comprises (a) obtaining a sample of the patient's
tumor; (b) measuring the
expression level of each biomarker of a biomarker signature in the sample,
wherein the biomarker
signature comprises one or more of the biomarkers FBXW2, CCND2, RHOU, CTBP2,
WIF1, and
DKK1; (c) calculating a decision value based upon the standardized expression
of the biomarkers in
the biomarker signature; and (d) selecting the patient for treatment when
their tumor sample has a
positive decision value. In some embodiments, the use further comprises
administering a
therapeutically effective amount of the Wnt pathway inhibitor to the patient.
[0095] Provided herein is a Wnt pathway inhibitor for use in treating cancer
in a patient, the use
comprising: (a) identifying if the patient is likely to respond to treatment
with a Wnt pathway
inhibitor, wherein the identification comprises: (i) obtaining a sample of the
patient's cancer; (ii)
measuring the expression level of each biomarker of a biomarker signature in
the sample, wherein the
biomarker signature comprises one or more of the biomarkers FBXW2, CCND2,
RHOU, CTBP2,
WIF1, and DKK1; and (iii) calculating a decision value based upon the
standardized expression of the
biomarkers in the signature; wherein a positive decision value indicates that
the patient is predicted to
respond to treatment; and (b) administering to the patient who is predicted to
response to treatment an
effective amount of the Wnt pathway inhibitor.
[0096] Provided herein is a use for increasing the likelihood of effective
treatment with a Wnt
pathway inhibitor, the use comprising: (a) identifying if a patient has a
tumor that is likely to respond
to treatment with a Wnt pathway inhibitor, wherein the identification
comprises: (i) obtaining a
sample of the patient's cancer; (ii) measuring the expression level of each
biomarker of a biomarker
signature in the sample, wherein the biomarker signature comprises one or more
of the biomarkers
FBXW2, CCND2, RHOU, CTBP2, WIF1, and DKK1; and (iii) calculating a decision
value based
upon the standardized expression of the biomarkers in the biomarker signature;
wherein a positive
decision value indicates that the patient is predicted to respond to
treatment; and (b) administering an
effective amount of the Wnt pathway inhibitor to the patient whose tumor has a
positive decision
value.
[0097] Provided herein is a Wnt pathway inhibitor for use in treating cancer
in a patient identified to
likely to respond to treatment with a Wnt pathway inhibitor wherein the
identification of the patient
comprises: (i) measuring the expression level of each biomarker of a biomarker
signature in the
cancer sample obtained from the patient, wherein the biomarker signature
comprises one or more of
the biomarkers FBXW2, CCND2, RHOU, CTBP2, WIF1, and DKK1; and (ii) calculating
a decision
23
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
value based upon the standardized expression of the biomarkers in the
signature; wherein a positive
decision value indicates that the patient is predicted to respond to
treatment.
[0098] Provided herein is a Wnt pathway inhibitor for use in treating cancer
in a patient, wherein the
patient is one for whom a positive decision value is calculated based upon the
standardized expression
of each biomarker of the biomarker signature in a cancer sample of the
patient, wherein the biomarker
signature comprises one or more of the biomarkers FBXW2, CCND2, RHOU, CTBP2,
WIF1, and
DKK1.
[0099] In some embodiments of the methods described herein, the biomarker
signature comprises
two or more of the biomarkers FBXW2, CCND2, RHOU, CTBP2, WIF1, and DKK1. In
some
embodiments, the biomarker signature comprises three or more of the biomarkers
FBXW2, CCND2,
RHOU, CTBP2, WIF1, and DKK1. In some embodiments, the biomarker signature
comprises four or
more of the biomarkers FBXW2, CCND2, RHOU, CTBP2, WIF1, and DKK1. In some
embodiments, the biomarker signature comprises five or more of the biomarkers
FBXW2, CCND2,
RHOU, CTBP2, WIF1, and DKK1. In some embodiments, the biomarker signature
comprises
FBXW2, CCND2, RHOU, CTBP2, WIF1, and DKK1. In some embodiments, the biomarker
signature consists of FBXW2, CCND2, RHOU, CTBP2, WIF1, and DKK1.
[00100] In some embodiments, the biomarker signature comprises one or more
additional biomarkers,
in addition to at least one of the biomarkers FBXW2, CCND2, RHOU, CTBP2, WIF1,
and DKK1. In
some embodiments, the biomarker signature comprises one or more additional
biomarkers selected
from the genes listed in Table 2, in addition to at least one of the
biomarkers FBXW2, CCND2,
RHOU, CTBP2, WIF1, and DKK1. In some embodiments, the biomarker signature
comprises one or
more of the biomarkers EP300, CTBP1, WNT6, WNT9A, SNT3, FZD2, FZD7, APC, TLE2,
DVL2,
PITX2, WISP1, GSK3B, and LEF1, in addition to at least one of the biomarkers
FBXW2, CCND2,
RHOU, CTBP2, WIF1, and DKK1. In some embodiments, the biomarker signature
comprises one or
more of the biomarkers FBXW2, CCND2, RHOU, CTBP2, WIF1, DKK1, EP300, and
CTBP1. In
some embodiments, the biomarker signature comprises one or more of the
biomarkers FBXW2,
CCND2, RHOU, CTBP2, WIF1, DKK1, EP300, CTBP1, WNT6, WNT3, FZD2, APC, TLE2,
DVL2,
PITX2, WISP1, GSK3B, WNT9A, FZD7, and LEF1.
[00101] In some embodiments of the methods described herein, the biomarker
signature comprises
FBXW2. In some embodiments, the biomarker signature comprises CCND2. In some
embodiments,
the biomarker signature comprises RHOU. In some embodiments, the biomarker
signature comprises
CTBP2. In some embodiments, the biomarker signature comprises WIF1. In some
embodiments, the
biomarker signature comprises DKK1.
[00102] In some embodiments of the methods described herein, the biomarker
signature comprises
FBXW2 and CCND2. In some embodiments, the biomarker signature comprises FBXW2
and
RHOU. In some embodiments, the biomarker signature comprises FBXW2 and CTBP2.
In some
embodiments, the biomarker signature comprises FBXW2 and WIF1. In some
embodiments, the
24
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
biomarker signature comprises FBXW2 and DKK1. In some embodiments, the
biomarker signature
comprises CCND2 and RHOU. In some embodiments, the biomarker signature
comprises CCND2
and CTBP2. In some embodiments, the biomarker signature comprises CCND2 and
WIF1. In some
embodiments, the biomarker signature comprises CCND2 and DKK1. In some
embodiments, the
biomarker signature comprises RHOU and CTBP2. In some embodiments, the
biomarker signature
comprises RHOU and WIF1. In some embodiments, the biomarker signature
comprises RHOU and
DKK1. In some embodiments, the biomarker signature comprises CTBP2 and WIF1.
In some
embodiments, the biomarker signature comprises CTBP2 and DKK1. In some
embodiments, the
biomarker signature comprises WIF1 and DKK1.
[00103] In some embodiments of the methods described herein, the biomarker
signature comprises
FBXW2, CCND2, and RHOU. In some embodiments, the biomarker signature comprises
FBXW2,
CCND2, and CTBP2. In some embodiments, the biomarker signature comprises
FBXW2, CCND2,
and WIF1. In some embodiments, the biomarker signature comprises FBXW2, CCND2,
and DKK1.
In some embodiments, the biomarker signature comprises FBXW2, RHOU, and CTBP2.
In some
embodiments, the biomarker signature comprises FBXW2, RHOU, and WIF1. In some
embodiments,
the biomarker signature comprises FBXW2, RHOU, and DKK1. In some embodiments,
the
biomarker signature comprises FBXW2, CTBP2, and WIF1. In some embodiments, the
biomarker
signature comprises FBXW2, CTBP2, and DKK1. In some embodiments, the biomarker
signature
comprises FBXW2, WIF1, and DKK1. In some embodiments, the biomarker signature
comprises
CCND2, RHOU, and CTBP2. In some embodiments, the biomarker signature comprises
CCND2,
RHOU, and WIF1. In some embodiments, the biomarker signature comprises CCND2,
RHOU, and
DKK1. In some embodiments, the biomarker signature comprises CCND2, CTBP2, and
WIF1. In
some embodiments, the biomarker signature comprises CCND2, CTBP2, and DKK1. In
some
embodiments, the biomarker signature comprises CCND2, WIF1, and DKK1. In some
embodiments,
the biomarker signature comprises RHOU, CTBP2, and WIF1. In some embodiments,
the biomarker
signature comprises RHOU, CTBP2, and DKK1. In some embodiments, the biomarker
signature
comprises RHOU, WIF1, and DKK1. In some embodiments, the biomarker signature
comprises
CTBP2, WIF1, and DKK1.
[00104] In some embodiments of the methods described herein, the biomarker
signature comprises
FBXW2, CCND2, RHOU, and CTBP2. In some embodiments, the biomarker signature
comprises
FBXW2, CCND2, RHOU, and WIF1. In some embodiments, the biomarker signature
comprises
FBXW2, CCND2, RHOU, and DKK1. In some embodiments, the biomarker signature
comprises
FBXW2, RHOU, CTBP2, and WIF1. In some embodiments, the biomarker signature
comprises
FBXW2, RHOU, CTBP2, and DKK1. In some embodiments, the biomarker signature
comprises
FBXW2, CTBP2, WIF1, and DKK1. In some embodiments, the biomarker signature
comprises
CCND2, RHOU, CTBP2, and WIF1. In some embodiments, the biomarker signature
comprises
CCND2, RHOU, CTBP2, and DKK1. In some embodiments, the biomarker signature
comprises
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
CCND2, CTBP2, WIF1, and DKK1. In some embodiments, the biomarker signature
comprises
RHOU, CTBP2, WIF1, and DKK1. In some embodiments, any of these signatures may
comprise one
or more additional biomarkers.
[00105] In some embodiments of the methods described herein, the biomarker
signature comprises
FBXW2, CCND2, RHOU, CTBP2, and WIF1. In some embodiments, the biomarker
signature
comprises FBXW2, CCND2, RHOU, CTBP2, and DKK1. In some embodiments, the
biomarker
signature comprises FBXW2, CCND2, CTBP2, WIF1, and DKK1. In some embodiments,
the
biomarker signature comprises FBXW2, CCND2, RHOU, WIF1, and DKK1. In some
embodiments,
the biomarker signature comprises FBXW2, RHOU, CTBP2, WIF1, and DKK1. In some
embodiments, the biomarker signature comprises CCND2, RHOU, CTBP2, WIF1, and
DKK1.
[00106] In some embodiments, the sample includes, but is not limited to, any
clinically relevant tissue
sample, such as a tumor biopsy, a core biopsy tissue sample, a fine needle
aspirate, a hair follicle, or a
sample of bodily fluid, such as blood, plasma, serum, lymph, ascitic fluid,
cystic fluid, or urine. In
some embodiments, the sample is taken from a patient having a tumor or cancer.
In some
embodiments, the sample is a primary tumor. In some embodiments, the sample is
a metastasis. The
sample may be taken from a human, or from non-human mammals such as, mice,
rats, non-human
primates, canines, felines, ruminants, swine, or sheep. In some embodiments,
samples are taken from
a subject at multiple time points, for example, before treatment, during
treatment, and/or after
treatment. In some embodiments, samples are taken from different locations in
the subject, for
example, a sample from a primary tumor and a sample from a metastasis in a
distant location.
[00107] In some embodiments, the sample is a paraffin-embedded fixed tissue
sample. In some
embodiments, the sample is a formalin-fixed paraffin embedded (FFPE) tissue
sample. In some
embodiments, the sample is a fresh tissue (e.g., tumor) sample. In some
embodiments, the sample is a
frozen tissue sample. In some embodiments, the sample is a fresh frozen (FF)
tissue (e.g., tumor)
sample. In some embodiments, the sample is a cell isolated from a fluid. In
some embodiments, the
sample comprises circulating tumor cells (CTCs). In some embodiments, the
sample is an archival
tissue sample. In some embodiments, the sample is an archival tissue sample
with known diagnosis,
treatment, and/or outcome history. In some embodiments, the sample is a block
of tissue. In some
embodiments, the sample is dispersed cells. In some embodiments, the sample
size is from about 1
cell to about 1 x 106 cells or more. In some embodiments, the sample size is
about 10 cells to about 1
x 105 cells. In some embodiments, the sample size is about 10 cells to about
10,000 cells. In some
embodiments, the sample size is about 10 cells to about 1,000 cells. In some
embodiments, the
sample size is about 10 cells to about 100 cells. In some embodiments, the
sample size is about 1 cell
to about 10 cells. In some embodiments, the sample size is a single cell.
[00108] In some embodiments, the sample is processed to DNA or RNA. In some
embodiments,
RNA is isolated from the sample. In some embodiments, mRNA is isolated from
the sample. In
some embodiments, RNA is isolated from cells by procedures that involve cell
lysis and denaturation
26
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
of the proteins contained therein. In some embodiments, DNase is added to
remove DNA. In some
embodiments, RNase inhibitors are added to the lysis buffer. In some
embodiments, a protein
denaturation/digestion step is added to the protocol. Methods for preparing
total and mRNA are well
known in the art and RNA isolation kits are commercially available (e.g.,
RNeasy mini kit, Qiagen,
USA). In some embodiments, the RNA is amplified by PCR-based techniques.
[00109] Determination of biomarker expression levels may be performed by any
suitable method
including, but are not limited to, methods based on analyses of polynucleotide
expression, sequencing
of polynucleotides, and/or analyses of protein expression. For example,
determination of biomarker
expression levels may be performed by detecting the expression of mRNA
expressed from the genes
of interest, and/or by detecting the expression of a polypeptide encoded by
the genes.
[00110] Commonly used methods for the analysis of polynucleotides, include
Southern blot analysis,
Northern blot analysis, and in situ hybridization, RNAse protection assays,
and polymerase chain
reaction (PCR)-based methods, such as reverse transcription polymerase chain
reaction (RT-PCR),
quantitative PCR (qPCR) as known as real-time PCR, TaqManTm, TaqManTm low
density array
(TLDA), anchored PCR, competitive PCR, rapid amplification of cDNA ends
(RACE), and
microarray analyses. RT-PCR is a quantitative method that can be used to
compare mRNA levels in
different samples to examine gene expression profiles. A variation of RT-PCR
is real time
quantitative PCR, which measures PCR product accumulation through a dual-
labeled fluorigenic
probe (e.g., TaqManTm probe). There are many other PCR-based techniques known
to one of skill in
the art, including but not limited to, differential display, amplified
fragment length polymorphism,
BeadArrayTM technology, high coverage expression profiling (HiCEP) and digital
PCR.
Representative methods for sequencing-based gene expression analyses include
Serial Analysis of
Gene Expression (SAGE), Massively Parallel Signature Sequencing (MPSS), and
NexGen sequencing
analysis, including mRNA sequencing.
[00111] In certain embodiments, the biomarker expression is determined using a
qPCR assay. For
example, total RNA is extracted from a fresh frozen (FF) tissue sample or
total RNA is extracted from
a macro-dissected formalin-fixed paraffin embedded (FFPE) tissue sample. The
quantity and quality
of the total RNA is assessed by standard spectrophotometry and/or any other
appropriate method (e.g.,
an Agilent Bioanalyzer). Following RNA extraction, the RNA sample is reverse
transcribed using
standard methods and/or a commercially available cDNA synthesis kit (e.g.,
Roche Transcriptor First
Strand cDNA synthesis kit). The resultant cDNA is pre-amplified using, for
example, an ABI pre-
amplification kit. Expression of the biomarker(s) (e.g., FBXW2, CCND2, RHOU,
CTBP2, WIF1,
and/or DKK1) are assessed on, for example, a Roche Lightcycler 480 system
(Roche Diagnostics)
using an ABI TaqMan Gene Expression Mastermix. qPCR reactions are performed in
triplicate. For
each assay a subset of the samples is run without reverse transcription (the
RT-neg control), as well
as, control samples run without template. A universal human reference RNA
sample is included on
each plate to act as a positive control. Suitable reference genes are
identified from a standard panel of
27
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
reference genes. Candidate reference genes are selected with different
cellular functions to eliminate
risk of co-regulation. The most suitable reference genes are evaluated and
selected using specific
software and algorithms (e.g., Genex software; GeNorm and Normfinder
algorithms). The expression
level of each biomarker is normalized using the selected optimum reference
genes. In some
embodiments, these normalized (or standardized) expression values for each
biomarker are used to
calculate the decision value of the sample. In some embodiments, these
normalized (or standardized)
expression values for each biomarker are used to calculate an expression
level.
[00112] In some embodiments, biomarker expression is determined using a PCR-
based assay
comprising specific primers and/or probes for each biomarker (e.g., FBXW2,
CCND2, RHOU,
CTBP2, WIF1, and/or DKK1). As used herein, the term "probe" refers to any
molecule that is
capable of selectively binding a specifically intended target biomolecule.
Probes can be synthesized
by one of skill in the art using known techniques, or derived from biological
preparations. Probes
may include but are not limited to, RNA, DNA, proteins, peptides, aptamers,
antibodies, and organic
molecules. The term "primer" or "probe" encompasses oligonucleotides that have
a sequence of a
specific SEQ ID NO or oligonucleotides that have a sequence complementary to a
specific SEQ ID
NO. In some embodiments, the probe is modified. In some embodiments, the probe
is modified with
a quencher. In some embodiments, the probe is labeled. Labels can include, but
are not limited to,
colorimetric, fluorescent, chemiluminescent, or bioluminescent labels.
[00113] In some embodiments, biomarker expression of each biomarker is
determined using a specific
primer set and probe. In some embodiments, a specific primer set consists of a
forward primer and a
reverse primer. In some embodiments, CCND2 expression is determined using a
polynucleotide
comprising the sequence of GCTGTCTCTGATCCGCAAGC (SEQ ID NO:62), a
polynucleotide
comprising the sequence of GACGGTGGGTACATGGCAAAC (SEQ ID NO:63), and a
polynucleotide comprising the sequence of CCTTCATTGCTCTGTGTGCCACCGAC (SEQ ID
NO: 64), or complements thereof In some embodiments, CCND2 expression is
determined using a
forward primer of sequence GCTGTCTCTGATCCGCAAGC (SEQ ID NO:62) and a reverse
primer
of sequence GACGGTGGGTACATGGCAAAC (SEQ ID NO:63). In some embodiments, CCND2
expression is determined using a probe of sequence CCTTCATTGCTCTGTGTGCCACCGAC
(SEQ
ID NO:64).
[00114] In some embodiments, CTBP2 expression is determined using isolated a
polynucleotide
comprising the sequence of ATCCGTGGGGAGACGCTG (SEQ ID NO:65), a polynucleotide
comprising the sequence of CTCGAACTGCAACCGCCTG (SEQ ID NO:66), and a
polynucleotide
comprising the sequence of CCCGTGCGACCAAAGCCAATGAGG (SEQ ID NO:67), or
complements thereof In some embodiments, CTBP2 expression is determined using
a forward
primer of sequence ATCCGTGGGGAGACGCTG (SEQ ID NO:65) and a reverse primer of
sequence
of CTCGAACTGCAACCGCCTG (SEQ ID NO:66). In some embodiments, CTBP2 expression
is
determined using a probe of sequence CCCGTGCGACCAAAGCCAATGAGG (SEQ ID NO:67).
28
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
[00115] In some embodiments, DKK1 expression is determined using isolated a
polynucleotide
comprising the sequence of GACCATTGACAACTACCAGCCGTA (SEQ ID NO:68), a
polynucleotide comprising the sequence of TGGGACTAGCGCAGTACTCATC (SEQ ID
NO:69),
and a polynucleotide comprising the sequence of TGCCGCACTCCTCGTCCTCTG (SEQ ID
NO:70), or complements thereof In some embodiments, DKK1 expression is
determined using a
forward primer of sequence GACCATTGACAACTACCAGCCGTA (SEQ ID NO:68) and a
reverse
primer of sequence of TGGGACTAGCGCAGTACTCATC (SEQ ID NO:69). In some
embodiments,
DKK1 expression is determined using a probe of sequence TGCCGCACTCCTCGTCCTCTG
(SEQ
ID NO:70).
[00116] In some embodiments, FBXW2 expression is determined using a
polynucleotide comprising
the sequence of GCCAGTTATGATATTCTCAGGGTCA (SEQ ID NO:71), a polynucleotide
comprising the sequence of AGCAGGGCAAAGATATCTCCAAA (SEQ ID NO:72), and a
polynucleotide comprising the sequence of AGACTCCTGAGATAGCAAACTTGGCCT (SEQ ID
NO: 73), or complements thereof In some embodiments, FBXW2 expression is
determined using a
forward primer of sequence GCCAGTTATGATATTCTCAGGGTCA (SEQ ID NO:71) and a
reverse
primer of sequence AGCAGGGCAAAGATATCTCCAAA (SEQ ID NO:72). In some
embodiments,
FBXW2 expression is determined using a probe of sequence
AGACTCCTGAGATAGCAAACTTGGCCT (SEQ ID NO:73).
[00117] In some embodiments, RHOU1 expression is determined using a
polynucleotide comprising
the sequence of CCCACCGAGTACATCCCTACTG (SEQ ID NO:74), a polynucleotide
comprising
the sequence of CAGTGTCACAGAGTTGGAGTCTCA (SEQ ID NO:75), and a polynucleotide
comprising the sequence of CGCCCATCCACAGACACCACCG (SEQ ID NO:76), or
complements
thereof In some embodiments, RHOU1 expression is determined using a forward
primer of sequence
CCCACCGAGTACATCCCTACTG (SEQ ID NO:74) and a reverse primer of sequence
CAGTGTCACAGAGTTGGAGTCTCA (SEQ ID NO:75). In some embodiments, RHOU1
expression is determined using a probe of sequence CGCCCATCCACAGACACCACCG (SEQ
ID
NO:76).
10011811n some embodiments, WIF1 expression is determined using a
polynucleotide comprising the
sequence of GTTCCAAAGGTTACCAGGGAGAC (SEQ ID NO:77), a polynucleotide
comprising
the sequence of GTTGGGTTCATGGCAGGTTCC (SEQ ID NO:78), and a polynucleotide
comprising the sequence of CCAGGCTCGCAGACAGGCTTTGAAC (SEQ ID NO:79), or
complements thereof In some embodiments, WIF1 expression is determined using a
forward primer
of sequence GTTCCAAAGGTTACCAGGGAGAC (SEQ ID NO:77) and a reverse primer of
sequence GTTGGGTTCATGGCAGGTTCC (SEQ ID NO:78). In some embodiments, WIF1
expression is determined using a probe of sequence CCAGGCTCGCAGACAGGCTTTGAAC
(SEQ
ID NO:79).
29
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
[00119] In some embodiments of any of the methods described herein, the
expression levels of
FBXW2, CCND2, RHOU, CTBP2, WIF1, and DKK1 are measured using polynucleotides
selected
from the group consisting of SEQ ID NOs:62-79. In some embodiments of any of
the methods
described herein, the expression levels of FBXW2, CCND2, RHOU, CTBP2, WIF1,
and DKK1 are
measured using (a) a forward primer of SEQ ID NO:62, a reverse primer of SEQ
ID NO:63, and a
probe comprising SEQ ID NO:64; (b) a forward primer of SEQ ID NO:65, a reverse
primer of SEQ
ID NO:66, and a probe comprising SEQ ID NO:67; (c) a forward primer of SEQ ID
NO:68, a reverse
primer of SEQ ID NO:69, and a probe comprising SEQ ID NO:70; (d) a forward
primer of SEQ ID
NO:71, a reverse primer of SEQ ID NO:72, and a probe comprising SEQ ID NO:73;
(e) a forward
primer of SEQ ID NO:74, a reverse primer of SEQ ID NO:75, and a probe
comprising SEQ ID
NO:76; and (f) a forward primer of SEQ ID NO:77, a reverse primer of SEQ ID
NO:78, and a probe
comprising SEQ ID NO:79.
[00120] In some embodiments, the expression level of each biomarker (e.g.,
FBXW2, CCND2,
RHOU, CTBP2, WIF1, and/or DKK1) is determined in a separate assay (e.g., 6
assays). In some
embodiments, the reference gene(s) and normalization methods for each assay
are the same for all 6
assays. In some embodiments, the expression levels of several biomarkers
(e.g., FBXW2, CCND2,
RHOU, CTBP2, WIF1, and/or DKK1) are detected in a single multiplex assay.
[00121] Alternatively, biomarker expression levels may be determined by
amplifying complementary
DNA (cDNA) or complementary RNA (cRNA) produced from mRNA and analyzing it
using a
microarray. Microarray technology allows for simultaneous analysis of the
expression of thousands
of genes. A number of different array configurations and methods for their
production are known to
those skilled in the art. In addition, microarrays are commercially available
(e.g., Affymetrix
GeneChips) or can be custom-produced. Microarrays currently in wide use
include cDNA arrays and
oligonucleotide arrays. In general, polynucleotides of interest (e.g., probes
or probe sets) are plated,
or arrayed, on a microchip substrate. In some embodiments, probes to at least
10, 25, 50, 100, 500,
1000, 5000, 10,000, 20,000, or 25,000 or more genes are immobilized on an
array substrate. The
substrate may be a porous or nonporous support, such as a glass, plastic or
gel surface. The probes
can include DNA, RNA, copolymer sequences of DNA and RNA, DNA and/or RNA
analogues, or
combinations thereof In some embodiments, a microarray includes a support with
an ordered array
of binding sites for each individual gene. The microarrays can be addressable
arrays or positionally
addressable arrays, e.g., each probe of the array is located at a known,
predetermined position on the
solid support such that the identity of each probe can be determined from its
position of the array.
[00122] Each probe on the microarray can be between 10-50,000 nucleotides in
length. In some
embodiments, the probes of the microarray can consist of nucleotide sequences
with lengths of less
than about 1,000 nucleotides, less than about 750 nucleotides, less than about
500 nucleotides, less
than about 250 nucleotides, less than about 100 nucleotides, or less than
about 50 nucleotides in
length. Generally, an array includes positive control probes and negative
control probes.
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
[00123] In certain embodiments, the biomarker expression is determined using a
microarray. For
example, total RNA is extracted from a fresh frozen (FF) tissue sample or
total RNA is extracted from
a macro-dissected formalin-fixed paraffin embedded (FFPE) tissue sample. The
quantity and quality
of the total RNA is assessed by standard spectrophotometry and/or any other
appropriate technology
(e.g., an Agilent Bioanalyzer). Following RNA extraction, the RNA sample is
amplified using
standard methods and/or a commercially available amplification system (e.g.,
NuGEN Ovation RNA
Amplification System V2). The amplified cDNA is fragmented, labeled, and
hybridized to a
microarray (e.g., using NuGEN Encore Biotin Module and Affymetrix GeneChip
array) following
standard procedures. The array is washed, stained, and scanned in accordance
with the instructions
for the microarray. The microarray data is pre-processed, the probe-level
intensity measurements are
background corrected, normalized, and summarized as expression measurements
using the Robust
Multichip algorithm (RMA). The probe level data is summarized to get the
expression level of each
biomarker (e.g., FBXW2, CCND2, RHOU, CTBP2, WIF1, and/or DKK1). A combination
of quality
parameter threshold and data reduction techniques (e.g., principal component
analysis) is applied to
the data set to establish profile quality and identify potential outlying
samples. These normalized (or
standardized) expression values for each biomarker are used to calculate the
decision value of the
sample.
[00124] In some embodiments, biomarker expression is analyzed by studying the
protein expression
of the gene or genes of interest. Commonly used methods for the analysis of
protein expression,
include but are not limited to, immunohistochemistry (IHC)-based, antibody-
based, and mass
spectrometry-based methods. Antibodies, generally monoclonal antibodies, may
be used to detect
expression of a gene product (e.g., protein). In some embodiments, the
antibodies can be detected by
direct labeling of the antibodies themselves. In other embodiments, an
unlabeled primary antibody is
used in conjunction with a labeled secondary antibody. Immunohistochemistry
methods and/or kits
are well known in the art and are commercially available.
[00125] In some embodiments, biomarker expression is determined by an assay
known to those of
skill in the art, including but not limited to, multi-analyte profile test,
enzyme-linked immunosorbent
assay (ELISA), radioimmunoassay, Western blot assay, immunofluorescent assay,
enzyme
immunoassay, immunoprecipitation assay, chemiluminescent assay,
immunohistochemical assay, dot
blot assay or slot blot assay. In some embodiments, wherein an antibody is
used in the assay the
antibody is detectably labeled. The antibody labels may include, but are not
limited to,
immunofluorescent label, chemiluminescent label, phosphorescent label, enzyme
label, radiolabel,
avidin/biotin, colloidal gold particles, colored particles and magnetic
particles.
[00126] Other suitable methods for analyzing biomarker expression include
proteomics-based
methods. Proteomics includes, among other things, study of the global changes
of protein expression
in a sample. In some embodiments, a proteomic method comprises the following
steps: (1) separation
of individual proteins in a sample by 2-D electrophoresis (2-D PAGE), (2)
identification of individual
31
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
proteins recovered from the gel (e.g., by mass spectrometry or N-terminal
sequencing), and (3)
analysis of the data using bioinformatics. In some embodiments, a proteomic
method comprises using
a tissue microarray (TMA). Tissue arrays may be constructed according to a
variety of techniques
known to one of skill in the art. In certain embodiments, a manual tissue
arrayer is used to remove a
"core" from a paraffin block prepared from a tissue sample. The core is then
inserted into a separate
paraffin block in a designated location on a grid. Cores from as many as about
400 samples can be
inserted into a single recipient block. The resulting tissue array may be
processed into thin sections
for analysis. In some embodiments, a proteomic method comprises an antibody
microarray. In some
embodiments, a proteomic method comprises using mass spectrometry, including
but not limited to,
SELDI, MALDI, electro spray, and surface plasmon resonance methods. In some
embodiments, a
proteomic method comprises bead-based technology, including but not limited
to, antibodies on beads
in an array format. In some embodiments, the proteomic method comprises a
reverse phase protein
microarray (RPPM). In some embodiments, the proteomic method comprises
multiplexed protein
profiling, including but not limited to, the Global Proteome Survey (GPS)
method.
[00127] In some embodiments, the biomarker signature is identified by
differential gene expression
between two samples. In some embodiments, the biomarker signature is
identified by differential
gene expression between two samples which comprise genes differentially
expressed in cancer cells
as compared to normal cells. In some embodiments, the biomarker signature
comprises genes
differentially expressed in tumorigenic cancer stem cells as compared to non-
tumorigenic cancer cells.
In some embodiments, the biomarker signature comprises genes differentially
expressed in cells from
a tumor which is responsive to a specific treatment as compared to cells from
a tumor which is non-
responsive to the same treatment.
[00128] In some embodiments, expression profiles are determined using
microarray analysis. The
microarray data identifies gene profiles comprising similarly and
differentially expressed genes
between two samples. In some embodiments, the expression profiles are refined,
filtered, and/or
subdivided into biomarker signatures based on fold expression change. In some
embodiments, all
genes above a certain fold expression change are included in the biomarker
signature. The fold
expression change may be elevated, reduced or both elevated and reduced. In
some embodiments, all
genes with a 2-fold or more expression change are included in the biomarker
signature. In some
embodiments, all genes with a 2.5-fold or more expression change are included
in the biomarker
signature. In some embodiments, all genes with a 3-fold or more expression
change are included in
the biomarker signature. In some embodiments, all genes with a 3.5-fold or
more expression change
are included in the biomarker signature. In some embodiments, all genes with a
4-fold or more
expression change are included in the biomarker signature.
[00129] In some embodiments, the gene expression profiles are refined,
filtered, and/or subdivided
into biomarker signatures based on statistical analyses. The statistical
methods may include, but are
not limited to, cluster analysis, supported vector machines (SVM) analysis,
supported vector machines
32
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
- recursive feature elimination (SVM-RFE) analysis, Platt scaling, neural
networks, and other
algorithms. In some embodiments, the gene expression profiles are analyzed
using a t-test analysis.
In some embodiments, the gene expression profiles are analyzed using paired-
sample empirical
Baysian analysis. In some embodiments, a combination of statistical analyses
is used. In some
embodiments, SVM models are used to obtain decision values based on the
training data. In some
embodiments, the decision values are calculated by a weighted sum of the
standardized expression of
a set of biomarkers. In some embodiments, a positive decision value indicates
a tumor predicted to be
a responder while a negative decision value indicates a tumor predicted to be
a non-responder. In
some embodiments, classification probabilities for responders and non-
responders are obtained using
Platt scaling (Platt, 1999, Advances in Large Margin Classifiers, pp. 61-74,
MIT Press). Platt scaling
may comprise fitting a logistic distribution using maximum likelihood to
decision values obtained, for
example, by SVM models. In some embodiments, tumors associated with
probabilities higher than
0.5 would be predicted to be a responder while tumors with probabilities lower
than 0.5 would be
predicted to be a non-responder.
10013011n some embodiments of any of the methods or uses described herein,
classification
probabilities of a tumor (in regard to responder or non-responder status) are
obtained based on the
decision values. In some embodiments, the probabilities are obtained by
fitting a logistic regression
on the decision values. In some embodiments, tumors associated with
probabilities higher than 0.5
are predicted to be a responder while tumors with probabilities lower than 0.5
are predicted to be a
non-responder.
10013111n some embodiments, a biomarker signature is obtained by a series of
analytical steps. For
example, expression data from a training set of samples are obtained from
microarray analyses. The
data are preprocessed to get an expression matrix with specific genes. Genes
with near zero variance
are removed, as are genes with expression values below a pre-determined level.
The remaining genes
are ranked using SVM-RFE analysis. Leave-one-out cross-validation (LOOCV)
methods are used to
identify and select the best predictive genes and also to measure positive
predictive value (PPV),
negative predictive value (NPV), sensitivity, and specificity.
[00132] In some embodiments, all genes with elevated expression, reduced
expression, or both, with a
P value across samples of 0.01 or less are included in the biomarker
signature. In some embodiments,
all genes with elevated expression, reduced expression or both, with a P value
across samples of 0.005
or less are included in the biomarker signature. In some embodiments, all
genes with elevated
expression, reduced expression or both, with a P value across samples of 0.001
or less are included in
the biomarker signature. In some embodiments, all genes with elevated
expression, reduced
expression or both, with a FDR (False Discovery Rate) of 0.25 or less are
included in the biomarker
signature. In some embodiments, all genes with elevated expression, reduced
expression or both, with
a FDR of 0.1 or less, 0.01 or less, or 0.001 or less are included in the
biomarker signature.
33
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
[00133] In some embodiments, the gene expression profiles and/or biomarker
signatures are refined,
filtered, and/or subdivided based on statistical models. In some embodiments,
the gene expression
profiles and/or biomarker signatures are refined, filtered, and/or subdivided
based on survival analysis
models. These models may include, but are not limited to, Kaplan-Meier
survival models, Cox
proportional models, Cox proportional hazard models, chi-square analysis,
univariate logistic
regression models, multivariate competing risk models, linear discriminate
analysis models,
parametric regression models and correlation analysis models.
[00134] In some embodiments, the gene expression profiles and/or biomarker
signatures are refined,
filtered, subdivided and/or tested using gene expression array datasets that
have associated clinical
outcomes. There are several databases that contain datasets that are available
to the public, for
example, Gene Expression Omnibus (GEO) and ArrayExpress.
[00135] In some embodiments, the gene expression profiles and/or biomarker
signatures are refined
using biological function parameters, and/or gene sets. For example, in some
embodiments, gene
expression profiles, and/or biomarker signatures are refined using Gene Set
Enrichment Analysis
(GSEA) (Subramanian et al., 2005, PNAS, 102: 15545-15550). In some
embodiments, the gene
expression profiles are refined based on their ability to predict clinical
outcome.
[00136] In some of the embodiments of the methods described herein, the Wnt
pathway inhibitor is an
anti-FZD antibody as described herein. In some of the embodiments of the
methods described herein,
the Wnt pathway inhibitor is an antibody that specifically binds at least one
Frizzled (FZD) protein or
portion thereof In some embodiments, the anti-FZD antibody specifically binds
at least one FZD
protein selected from the group consisting of: FZD1, FZD2, FZD5, FZD7, and
FZD8. In other
embodiments, the anti-FZD antibody comprises: (a) a heavy chain CDR1
comprising GFTFSHYTLS
(SEQ ID NO:1), a heavy chain CDR2 comprising VISGDGSYTYYADSVKG (SEQ ID NO:2),
and a
heavy chain CDR3 comprising NFIKYVFAN (SEQ ID NO:3), and (b) a light chain
CDR1
comprising SGDNIGSFYVH (SEQ ID NO:4), a light chain CDR2 comprising DKSNRPSG
(SEQ ID
NO:5), and a light chain CDR3 comprising QSYANTLSL (SEQ ID NO:6). In some
embodiments,
the anti-FZD antibody comprises a heavy chain variable region comprising the
amino acids of SEQ
ID NO:7. In some embodiments, the anti-FZD antibody comprises a light chain
variable region
comprising the amino acids of SEQ ID NO:8. In some embodiments, the anti-FZD
antibody
comprises a heavy chain variable region comprising the amino acids of SEQ ID
NO :7 and a light
chain variable region comprising the amino acids of SEQ ID NO:8. In some
embodiments, the anti-
FZD antibody is antibody OMP-18R5. In some embodiments, the anti-FZD antibody
is encoded by
the plasmid having ATCC deposit no. PTA-9541. In other embodiments, the anti-
FZD antibody
competes for specific binding to at least one human FZD protein with an
antibody encoded by the
plasmid deposited with ATCC having deposit no. PTA-9541.
[00137] In some embodiments of the methods described herein, the tumor is
selected from the group
consisting of a breast tumor, lung tumor, a colon tumor, glioma, a
gastrointestinal tumor, a renal
34
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
tumor, an ovarian tumor, a liver tumor, a colorectal tumor, an endometrial
tumor, a kidney tumor, a
prostate tumor, a thyroid tumor, a neuroblastoma, a pancreatic tumor, a
glioblastoma multiforme, a
cervical tumor, a stomach tumor, a bladder tumor, a hepatoma, melanoma, and a
head and neck tumor.
In some embodiments, the tumor is a breast tumor. In some embodiments, the
tumor is a HER2-
negative breast tumor. In some embodiments, the tumor is a triple negative
breast cancer (TNBC)
tumor.
[00138] In some embodiments of the methods described herein, the cancer is
selected from the group
consisting of a breast cancer, lung cancer, a colon cancer, glioma, a
gastrointestinal cancer, a renal
cancer, an ovarian cancer, a liver cancer, a colorectal cancer, an endometrial
cancer, a kidney cancer,
a prostate cancer, a thyroid cancer, a neuroblastoma, a pancreatic cancer, a
glioblastoma multiforme, a
cervical cancer, a stomach cancer, a bladder cancer, a hepatoma, melanoma, and
a head and neck
cancer. In some embodiments, the cancer is breast cancer. In some embodiments,
the cancer is a
HER2-negative breast cancer. In some embodiments, the cancer is a triple
negative breast cancer
(TNBC).
[00139] In some of the embodiments of the methods described herein, the method
comprises treating a
patient with a Wnt pathway inhibitor described herein (e.g., an anti-FZD
antibody), particularly after
the patient has been identified as being responsive to treatment with the Wnt
pathway inhibitor. In
some embodiments, the treatment comprises administering at least one
additional therapeutic agent in
combination with the Wnt pathway inhibitor. An additional therapeutic agent
can be administered
prior to, concurrently with, and/or subsequently to, administration of the Wnt
pathway inhibitor. In
some embodiments, the at least one additional therapeutic agent comprises 1,
2, 3, or more additional
therapeutic agents.
[00140] Useful classes of therapeutic agents include, for example, antitubulin
agents, auristatins, DNA
minor groove binders, DNA replication inhibitors, alkylating agents (e.g.,
platinum complexes such as
cisplatin, mono(platinum), bis(platinum) and tri-nuclear platinum complexes
and carboplatin),
anthracyclines, antibiotics, antifolates, antimetabolites, chemotherapy
sensitizers, duocarmycins,
etoposides, fluorinated pyrimidines, ionophores, lexitropsins, nitrosoureas,
platinols, purine
antimetabolites, puromycins, radiation sensitizers, steroids, taxanes,
topoisomerase inhibitors, vinca
alkaloids, or the like. In certain embodiments, the second therapeutic agent
is an alkylating agent, an
antimetabolite, an antimitotic, a topoisomerase inhibitor, or an angiogenesis
inhibitor.
[00141] Therapeutic agents that may be administered in combination with the
Wnt pathway inhibitors
include chemotherapeutic agents. Thus, in some embodiments, the method or
treatment involves the
administration of a Wnt pathway inhibitor of the present invention in
combination with a
chemotherapeutic agent or cocktail of multiple different chemotherapeutic
agents. Treatment with a
Wnt pathway inhibitor (e.g, an anti-FZD antibody) can occur prior to,
concurrently with, or
subsequent to administration of chemotherapies. Combined administration can
include co-
administration, either in a single pharmaceutical formulation or using
separate formulations, or
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
consecutive administration in either order but generally within a time period
such that all active agents
can exert their biological activities simultaneously. Preparation and dosing
schedules for such
chemotherapeutic agents can be used according to manufacturers' instructions
or as determined
empirically by the skilled practitioner. Preparation and dosing schedules for
such chemotherapy are
also described in The Chemotherapy Source Book, 4th Edition, 2008, M. C.
Perry, Editor, Lippincott,
Williams & Wilkins, Philadelphia, PA.
[00142] Chemotherapeutic agents useful in the instant invention include, but
are not limited to,
alkylating agents such as thiotepa and cyclosphosphamide (CYTOXAN); alkyl
sulfonates such as
busulfan, improsulfan and piposulfan; aziridines such as benzodopa,
carboquone, meturedopa, and
uredopa; ethylenimines and methylamelamines including altretamine,
triethylenemelamine,
trietylenephosphoramide, triethylenethiophosphaoramide and
trimethylolomelamime; nitrogen
mustards such as chlorambucil, chlornaphazine, cholophosphamide, estramustine,
ifosfamide,
mechlorethamine, mechlorethamine oxide hydrochloride, melphalan, novembichin,
phenesterine,
prednimustine, trofosfamide, uracil mustard; nitrosureas such as carmustine,
chlorozotocin,
fotemustine, lomustine, nimustine, ranimustine; antibiotics such as
aclacinomysins, actinomycin,
authramycin, azaserine, bleomycins, cactinomycin, calicheamicin, carabicin,
caminomycin,
carzinophilin, chromomycins, dactinomycin, daunorubicin, detorubicin, 6-diazo-
5-oxo-L-norleucine,
doxorubicin, epirubicin, esorubicin, idarubicin, marcellomycin, mitomycins,
mycophenolic acid,
nogalamycin, olivomycins, peplomycin, potfiromycin, puromycin, quelamycin,
rodorubicin,
streptonigrin, streptozocin, tubercidin, ubenimex, zinostatin, zorubicin; anti-
metabolites such as
methotrexate and 5-fluorouracil (5-FU); folic acid analogues such as
denopterin, methotrexate,
pteropterin, trimetrexate; purine analogs such as fludarabine, 6-
mercaptopurine, thiamiprine,
thioguanine; pyrimidine analogs such as ancitabine, azacitidine, 6-azauridine,
carmofur, cytosine
arabinoside, dideoxyuridine, doxifluridine, enocitabine, floxuridine, 5-FU;
androgens such as
calusterone, dromostanolone propionate, epitiostanol, mepitiostane,
testolactone; anti-adrenals such as
aminoglutethimide, mitotane, trilostane; folic acid replenishers such as
folinic acid; aceglatone;
aldophosphamide glycoside; aminolevulinic acid; amsacrine; bestrabucil;
bisantrene; edatraxate;
defofamine; demecolcine; diaziquone; elformithine; elliptinium acetate;
etoglucid; gallium nitrate;
hydroxyurea; lentinan; lonidamine; mitoguazone; mitoxantrone; mopidamol;
nitracrine; pentostatin;
phenamet; pirarubicin; podophyllinic acid; 2-ethylhydrazide; procarbazine;
PSK; razoxane; sizofuran;
spirogermanium; tenuazonic acid; triaziquone; 2,2',2"-trichlorotriethylamine;
urethan; vindesine;
dacarbazine; mannomustine; mitobronitol; mitolactol; pipobroman; gacytosine;
arabinoside (Ara-C);
taxoids, e.g. paclitaxel (TAXOL) and docetaxel (TAXOTERE); chlorambucil;
gemcitabine; 6-
thioguanine; mercaptopurine; platinum analogs such as cisplatin and
carboplatin; vinblastine;
platinum; etoposide (VP-16); ifosfamide; mitomycin C; mitoxantrone;
vincristine; vinorelbine;
navelbine; novantrone; teniposide; daunomycin; aminopterin; ibandronate;
CPT11; topoisomerase
inhibitor RFS 2000; difluoromethylornithine (DMF0); retinoic acid;
esperamicins; capecitabine
36
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
(XELODA); and pharmaceutically acceptable salts, acids or derivatives of any
of the above.
Chemotherapeutic agents also include anti-hormonal agents that act to regulate
or inhibit hormone
action on tumors such as anti-estrogens including for example tamoxifen,
raloxifene, aromatase
inhibiting 4(5)-imidazoles, 4-hydroxytamoxifen, trioxifene, keoxifene,
LY117018, onapristone, and
toremifene (FARESTON); and anti-androgens such as flutamide, nilutamide,
bicalutamide,
leuprolide, and goserelin; and pharmaceutically acceptable salts, acids or
derivatives of any of the
above. In certain embodiments, the additional therapeutic agent is paclitaxel
(taxol).
[00143] In certain embodiments, the chemotherapeutic agent is a topoisomerase
inhibitor.
Topoisomerase inhibitors are chemotherapy agents that interfere with the
action of a topoisomerase
enzyme (e.g., topoisomerase I or II). Topoisomerase inhibitors include, but
are not limited to,
doxorubicin HC1, daunorubicin citrate, mitoxantrone HC1, actinomycin D,
etoposide, topotecan HC1,
teniposide (VM-26), and irinotecan, as well as pharmaceutically acceptable
salts, acids, or derivatives
of any of these.
[00144] In certain embodiments, the chemotherapeutic agent is an anti-
metabolite. An anti-metabolite
is a chemical with a structure that is similar to a metabolite required for
normal biochemical reactions,
yet different enough to interfere with one or more normal functions of cells,
such as cell division.
Anti-metabolites include, but are not limited to, gemcitabine, fluorouracil,
capecitabine, methotrexate
sodium, ralitrexed, pemetrexed, tegafur, cytosine arabinoside, thioguanine, 5-
azacytidine, 6-
mercaptopurine, azathioprine, 6-thioguanine, pentostatin, fludarabine
phosphate, and cladribine, as
well as pharmaceutically acceptable salts, acids, or derivatives of any of
these.
[00145] In certain embodiments, the chemotherapeutic agent is an antimitotic
agent, including, but not
limited to, agents that bind tubulin. In some embodiments, the agent is a
taxane. In certain
embodiments, the agent is paclitaxel or docetaxel, or a pharmaceutically
acceptable salt, acid, or
derivative of paclitaxel or docetaxel. In certain embodiments, the agent is
paclitaxel (TAXOL),
docetaxel (TAXOTERE), albumin-bound paclitaxel (nab-paclitaxel; ABRAXANE), DHA-
paclitaxel,
or PG-paclitaxel. In certain alternative embodiments, the antimitotic agent
comprises a vinca
alkaloid, such as vincristine, binblastine, vinorelbine, or vindesine, or
pharmaceutically acceptable
salts, acids, or derivatives thereof In some embodiments, the antimitotic
agent is an inhibitor of
kinesin Eg5 or an inhibitor of a mitotic kinase such as Aurora A or Plkl. In
certain embodiments,
where the chemotherapeutic agent administered in combination with a Wnt
pathway inhibitor is an
anti-mitotic agent, the cancer or tumor being treated is breast cancer or a
breast tumor. In certain
embodiments, the additional therapeutic agent is paclitaxel (taxol) or albumin-
bound paclitaxel.
[00146] In some embodiments, an additional therapeutic agent comprises an
agent such as a small
molecule. For example, treatment can involve the combined administration of a
Wnt pathway
inhibitor of the present invention with a small molecule that acts as an
inhibitor against additional
tumor-associated antigens including, but not limited to, EGFR, ErbB2, HER2,
and/or VEGF. In
certain embodiments, the additional therapeutic agent is a small molecule that
inhibits a cancer stem
37
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
cell pathway. In some embodiments, the additional therapeutic agent is an
inhibitor of the Notch
pathway. In some embodiments, the additional therapeutic agent is an inhibitor
of the Wnt pathway.
In some embodiments, the additional therapeutic agent is an inhibitor of the
BMP pathway.
[00147] Certain embodiments of the present invention comprise a method of
identifying a human
breast tumor that is likely to be responsive to or non-responsive to treatment
with an antibody that
specifically binds at least one human frizzled (FZD) selected from the group
consisting of FZD1,
FZD2, FZD5, FZD7, and FZD8, the method comprising (a) obtaining a sample of
the human breast
tumor; (b) measuring the expression level of each biomarker of a biomarker
signature in the sample,
wherein the biomarker signature comprises one or more of the biomarkers FBXW2,
CCND2, RHOU,
CTBP2, WIF1, and DKK1; and (c) calculating a decision value based upon the
standardized
expression of the biomarkers in the biomarker signature; wherein a positive
decision value indicates
the breast tumor is predicted to be responsive to treatment with the antibody
and a negative decision
value indicates the tumor is predicted to be non-responsive to treatment with
the antibody. Some
embodiments comprise a method of identifying a patient with breast cancer that
is likely to be
responsive to treatment with an antibody that specifically binds at least one
human frizzled (FZD)
selected from the group consisting of FZD1, FZD2, FZD5, FZD7, and FZD8, the
method comprising:
(a) obtaining a sample of the breast cancer; (b) measuring the expression
level of each biomarker of a
biomarker signature in the sample, wherein the biomarker signature comprises
one or more of the
biomarkers FBXW2, CCND2, RHOU, CTBP2, WIF1, and DKK1; and (c) calculating a
decision
value based upon the standardized expression of the biomarkers in the
biomarker signature; wherein a
positive decision value indicates the breast cancer is predicted to be
responsive to treatment with the
antibody. Some embodiments comprise a method of selecting a patient with
breast cancer for
treatment with an antibody that specifically binds at least one human frizzled
(FZD) selected from the
group consisting of FZD1, FZD2, FZD5, FZD7, and FZD8, the method comprising:
(a) obtaining a
sample of the breast cancer; (b) measuring the expression level of each
biomarker of a biomarker
signature in the sample, wherein the biomarker signature comprises one or more
of the biomarkers
FBXW2, CCND2, RHOU, CTBP2, WIF1, and DKK1; (c) calculating a decision value
based upon the
standardized expression of the biomarkers in the biomarker signature; wherein
a positive decision
value indicates the breast cancer is predicted to be responsive to treatment
with the antibody; and
selecting the patient for treatment when their tumor sample has a positive
decision value.
[00148] Some embodiments of the present invention comprise a method of
treating breast cancer in a
patient, comprising: (a) identifying if the patient is likely to respond to
treatment with an antibody that
specifically binds at least one human frizzled (FZD) selected from the group
consisting of FZD1,
FZD2, FZD5, FZD7, and FZD8, wherein the identification comprises: (i)
obtaining a sample of the
patient's breast cancer; (ii) measuring the expression level of each biomarker
of a biomarker signature
in the sample, wherein the biomarker signature comprises one or more of the
biomarkers FBXW2,
CCND2, RHOU, CTBP2, WIF1, and DKK1; and (iii) calculating a decision value
based upon the
38
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
standardized expression of the biomarkers in the signature; wherein a positive
decision value indicates
that the patient is predicted to respond to treatment; and (b) administering
to the patient who is
predicted to response to treatment an effective amount of the antibody.
[00149] Certain embodiments of the present invention comprise a method of
identifying a human
breast tumor that is likely to be responsive to or non-responsive to treatment
with anti-FZD antibody
OMP-18R5 in combination with paclitaxel, the method comprising (a) obtaining a
sample of the
human breast tumor; (b) measuring the expression level of each biomarker of a
biomarker signature in
the sample, wherein the biomarker signature comprises the biomarkers FBXW2,
CCND2, RHOU,
CTBP2, WIF1, and DKK1; and (c) calculating a decision value based upon the
standardized
expression of the biomarkers in the biomarker signature; wherein a positive
decision value indicates
the breast tumor is predicted to be responsive to treatment and a negative
decision value indicates the
tumor is predicted to be non-responsive to treatment. Some embodiments
comprise a method of
identifying a patient with breast cancer that is likely to be responsive to
treatment with the anti-FZD
antibody OMP-18R5 in combination with paclitaxel, the method comprising: (a)
obtaining a sample
of the breast cancer; (b) measuring the expression level of each biomarker of
a biomarker signature in
the sample, wherein the biomarker signature comprises the biomarkers FBXW2,
CCND2, RHOU,
CTBP2, WIF1, and DKK1; and (c) calculating a decision value based upon the
standardized
expression of the biomarkers in the biomarker signature; wherein a positive
decision value indicates
the breast cancer is predicted to be responsive to treatment. Some embodiments
comprise a method of
selecting a patient with breast cancer for treatment with the anti-FZD
antibody OMP-18R5 in
combination with paclitaxel, the method comprising: (a) obtaining a sample of
the breast cancer; (b)
measuring the expression level of each biomarker of a biomarker signature in
the sample, wherein the
biomarker signature comprises the biomarkers FBXW2, CCND2, RHOU, CTBP2, WIF1,
and DKK1;
(c) calculating a decision value based upon the standardized expression of the
biomarkers in the
biomarker signature; wherein a positive decision value indicates the breast
cancer is predicted to be
responsive to treatment; and selecting the patient for treatment when their
tumor sample has a positive
decision value.
[00150] Some embodiments of the present invention comprise a method of
treating breast cancer in a
patient, comprising: (a) identifying if the patient is likely to respond to
treatment with the anti-FZD
antibody OMP-18R5 in combination with paclitaxel, wherein the identification
comprises: (i)
obtaining a sample of the patient's breast cancer; (ii) measuring the
expression level of each
biomarker of a biomarker signature in the sample, wherein the biomarker
signature comprises the
biomarkers FBXW2, CCND2, RHOU, CTBP2, WIF1, and DKK1; and (iii) calculating a
decision
value based upon the standardized expression of the biomarkers in the
signature; wherein a positive
decision value indicates that the patient is predicted to respond to
treatment; and (b) administering to
the patient who is predicted to response to treatment an effective amount of
the antibody and
paclitaxel.
39
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
III. Wnt pathway inhibitors
[00151] The present invention provides methods for identifying tumors and/or
patients with cancer
that are likely to be responsive to or sensitive to treatment with Wnt pathway
inhibitors. As used
herein "Wnt pathway inhibitor" includes, but is not limited to, Frizzled (FZD)
binding agents and
Wnt-binding agents. FZD-binding agents may include antibodies that
specifically bind to FZD
proteins. Wnt-binding agents may include antibodies that specifically bind to
Wnt proteins as well as
soluble FZD receptors that bind to Wnt proteins.
[00152] In certain embodiments, the Wnt pathway inhibitors are agents that
bind one or more human
FZD proteins. In some embodiments, the FZD-binding agents specifically bind
one, two, three, four,
five, six, seven, eight, nine, or ten FZD proteins. In some embodiments, the
FZD-binding agent binds
one or more FZD proteins selected from the group consisting of FZD1, FZD2,
FZD3, FZD4, FZD5,
FZD6, FZD7, FZD8, FZD9, and FZD10. In some embodiments, FZD-binding agent
binds one or
more FZD proteins comprising FZD1, FZD2, FZD5, FZD7, and/or FZD8. In certain
embodiments,
FZD-binding agent binds FZD7. In certain embodiments, FZD-binding agent binds
FZD5 and/or
FZD8. In certain embodiments, the FZD-binding agent specifically binds FZD1,
FZD2, FZD5,
FZD7, and FZD8. Non-limiting examples of FZD-binding agents can be found in
U.S. Patent No.
7,982,013.
[00153] In certain embodiments, the FZD-binding agent is a FZD antagonist. In
certain embodiments,
the FZD-binding agent is a Wnt pathway antagonist. In certain embodiments, the
FZD-binding agent
inhibits Wnt signaling. In some embodiments, the FZD-binding agent inhibits
canonical Wnt
signaling.
[00154] In some embodiments, the FZD-binding agents are antibodies. In some
embodiments, the
FZD-binding agents are polypeptides. In certain embodiments, the FZD-binding
agent is an antibody
or a polypeptide comprising an antigen-binding site. In certain embodiments,
an antigen-binding site
of a FZD-binding antibody or polypeptide described herein is capable of
binding (or binds) one, two,
three, four, five, or more human FZD proteins. In certain embodiments, an
antigen-binding site of the
FZD-binding antibody or polypeptide is capable of specifically binding one,
two, three, four, or five
human FZD proteins selected from the group consisting of FZD1, FZD2, FZD3,
FZD4, FZD5, FZD6,
FZD7, FZD8, FZD9 and FZD10. In some embodiments, when the FZD-binding agent is
an antibody
that binds more than one FZD protein, it may be referred to as a "pan-FZD
antibody".
[00155] In certain embodiments, the FZD-binding agent (e.g., antibody)
specifically binds the
extracellular domain (ECD) of the one or more human FZD proteins to which it
binds. In certain
embodiments, the FZD-binding agent specifically binds within the Fri domain
(also known as the
cysteine-rich domain (CRD)) of the human FZD protein to which it binds.
Sequences of the Fri
domain of each of the human FZD proteins are known in the art and are provided
as SEQ ID NO:13
(FZD1), SEQ ID NO:14 (FZD2), SEQ ID NO:15 (FZD3), SEQ ID NO:16 (FZD4), SEQ ID
NO:17
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
(FZD5), SEQ ID NO:18 (FZD6), SEQ ID NO:19 (FZD7), SEQ ID NO:20 (FZD), SEQ ID
NO:21
(FZD9), and SEQ ID NO:22 (FZD10).
[00156] In certain embodiments, the FZD-binding agent binds one, two, three,
four, five, or more FZD
proteins. In some embodiments, the FZD-binding agent specifically binds one,
two, three, four, or
five FZD proteins selected from the group consisting of FZD1, FZD2, FZD5,
FZD7, and FZD8. In
some embodiments, the FZD-binding agent specifically binds at least FZD5 and
FZD8.
[00157] In some embodiments, the FZD-binding agent binds at least one human
FZD protein with a
dissociation constant (KD) of about 1 M or less, about 100nM or less, about
40nM or less, about
20nM or less, about lOnM or less, about 1nM or less, or about 0.1nM or less.
In some embodiments,
a FZD-binding agent binds at least one FZD protein with a KD of about lOnM or
less. In some
embodiments, a FZD-binding agent binds at least one FZD protein with a KD of
about 1nM or less. In
some embodiments, a FZD-binding agent binds at least one FZD protein with a KD
of about 0.1nM or
less. In certain embodiments, a FZD-binding agent binds each of one or more
(e.g., 1, 2, 3, 4, or 5) of
FZD1, FZD2, FZD5, FZD7, and FZD8 with a KD of about 40nM or less. In certain
embodiments, the
FZD-binding agent binds to each of one or more of FZD1, FZD2, FZD5, FZD7, and
FZD8 with a KD
of about lOnM or less. In certain embodiments, the FZD-binding agent binds
each of FZD1, FZD2,
FZD5, FZD7, and FZD8 with a KD of about lOnM. In some embodiments, the KD of
the binding
agent (e.g., an antibody) to a FZD protein is the KD determined using a FZD-Fc
fusion protein
comprising at least a portion of the FZD extracellular domain or FZD-Fri
domain immobilized on a
Biacore chip.
[00158] In certain embodiments, the FZD-binding agent binds one or more (for
example, two or more,
three or more, or four or more) human FZD proteins with an EC50 of about 1 M
or less, about 100nM
or less, about 40nM or less, about 20nM or less, about lOnM or less, or about
1nM or less. In certain
embodiments, a FZD-binding agent binds to more than one FZD protein with an
EC50 of about 40nM
or less, about 20nM or less, or about lOnM or less. In certain embodiments,
the FZD-binding agent
has an EC50 of about 20nM or less with respect to one or more (e.g., 1, 2, 3,
4, or 5) of the following
FZD proteins: FZD1, FZD2, FZD5, FZD7, and FZD8. In certain embodiments, the
FZD-binding
agent has an EC50 of about lOnM or less with respect to one or more (e.g., 1,
2, 3, 4, or 5) of the
following FZD proteins: FZD1, FZD2, FZD5, FZD7, and FZD8. In certain
embodiments, the FZD-
binding agent has an EC50 of about 40nM or less or 20nM or less with respect
to binding of FZD5
and/or FZD8.
[00159] In certain embodiments, the Wnt pathway inhibitor is a FZD-binding
agent which is an
antibody. In some embodiments, the antibody is a recombinant antibody. In some
embodiments, the
antibody is a monoclonal antibody. In some embodiments, the antibody is a
chimeric antibody. In
some embodiments, the antibody is a humanized antibody. In some embodiments,
the antibody is a
human antibody. In certain embodiments, the antibody is an IgG1 antibody. In
certain embodiments,
the antibody is an IgG2 antibody. In certain embodiments, the antibody is an
antibody fragment
41
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
comprising an antigen-binding site. In some embodiments, the antibody is
monovalent, monospecific,
or bivalent. In some embodiments, the antibody is a bispecific antibody or a
multispecific antibody.
In some embodiments, the antibody is conjugated to a cytotoxic moiety. In some
embodiments, the
antibody is isolated. In some embodiments, the antibody is substantially pure.
[00160] The FZD-binding agents (e.g., antibodies) of the present invention can
be assayed for specific
binding by any method known in the art. The immunoassays which can be used
include, but are not
limited to, competitive and non-competitive assay systems using techniques
such as Biacore analysis,
FACS analysis, immunofluorescence, immunocytochemistry, Western blot analysis,
radioimmunoassays, ELISA, "sandwich" immunoassays, immunoprecipitation assays,
precipitation
reactions, gel diffusion precipitin reactions, immunodiffusion assays,
agglutination assays,
complement-fixation assays, immunoradiometric assays, fluorescent
immunoassays, and protein A
immunoassays. Such assays are routine and well-known in the art (see, e.g.,
Ausubel et al., Editors,
1994-present, Current Protocols in Molecular Biology, John Wiley & Sons, Inc.,
New York, NY).
[00161] In certain embodiments, the invention provides a Wnt pathway inhibitor
which is a FZD-
binding agent (e.g., an antibody) that comprises a heavy chain CDR1 comprising
GFTFSHYTLS
(SEQ ID NO:1), a heavy chain CDR2 comprising VISGDGSYTYYADSVKG (SEQ ID NO:2),
and a
heavy chain CDR3 comprising NFIKYVFAN (SEQ ID NO:3). In some embodiments, the
FZD-
binding agent further comprises a light chain CDR1 comprising SGDNIGSFYVH (SEQ
ID NO:4), a
light chain CDR2 comprising DKSNRPSG (SEQ ID NO:5), and a light chain CDR3
comprising
QSYANTLSL (SEQ ID NO:6). In some embodiments, the FZD-binding agent comprises
a light
chain CDR1 comprising SGDNIGSFYVH (SEQ ID NO:4), a light chain CDR2 comprising
DKSNRPSG (SEQ ID NO:5), and a light chain CDR3 comprising QSYANTLSL (SEQ ID
NO:6). In
certain embodiments, the FZD-binding agent comprises: (a) a heavy chain CDR1
comprising
GFTFSHYTLS (SEQ ID NO:1), a heavy chain CDR2 comprising VISGDGSYTYYADSVKG (SEQ
ID NO:2), and a heavy chain CDR3 comprising NFIKYVFAN (SEQ ID NO:3), and (b) a
light chain
CDR1 comprising SGDNIGSFYVH (SEQ ID NO:4), a light chain CDR2 comprising
DKSNRPSG
(SEQ ID NO:5), and a light chain CDR3 comprising QSYANTLSL (SEQ ID NO:6).
[00162] In certain embodiments, the invention provides a FZD-binding agent
(e.g., an antibody) that
comprises: (a) a heavy chain CDR1 comprising GFTFSHYTLS (SEQ ID NO:1), or a
variant thereof
comprising 1, 2, 3, or 4 amino acid substitutions; (b) a heavy chain CDR2
comprising
VISGDGSYTYYADSVKG (SEQ ID NO:2), or a variant thereof comprising 1, 2, 3, or 4
amino acid
substitutions; (c) a heavy chain CDR3 comprising NFIKYVFAN (SEQ ID NO:3), or a
variant thereof
comprising 1, 2, 3, or 4 amino acid substitutions; (d) a light chain CDR1
comprising
SGDNIGSFYVH (SEQ ID NO:4), or a variant thereof comprising 1, 2, 3, or 4 amino
acid
substitutions; (e) a light chain CDR2 comprising DKSNRPSG (SEQ ID NO:5), or a
variant thereof
comprising 1, 2, 3, or 4 amino acid substitutions; and (f) a light chain CDR3
comprising
42
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
QSYANTLSL (SEQ ID NO:6), or a variant thereof comprising 1, 2, 3, or 4 amino
acid substitutions.
In certain embodiments, the amino acid substitutions are conservative
substitutions.
[00163] In certain embodiments, the invention provides a FZD-binding agent
(e.g., an antibody) that
comprises a heavy chain variable region having at least about 80% sequence
identity to SEQ ID
NO:7, and/or a light chain variable region having at least 80% sequence
identity to SEQ ID NO:8. In
certain embodiments, the FZD-binding agent comprises a heavy chain variable
region having at least
about 85%, at least about 90%, at least about 95%, at least about 97%, or at
least about 99% sequence
identity to SEQ ID NO:7. In certain embodiments, the FZD-binding agent
comprises a light chain
variable region having at least about 85%, at least about 90%, at least about
95%, at least about 97%,
or at least about 99% sequence identity to SEQ ID NO:8. In certain
embodiments, the FZD-binding
agent comprises a heavy chain variable region having at least about 95%
sequence identity to SEQ ID
NO:7, and/or a light chain variable region having at least about 95% sequence
identity to SEQ ID
NO:8. In certain embodiments, the FZD-binding agent comprises a heavy chain
variable region
comprising SEQ ID NO:7 and/or a light chain variable region comprising SEQ ID
NO:8. In certain
embodiments, the FZD-binding agent comprises a heavy chain variable region
comprising SEQ ID
NO:7 and a light chain variable region comprising SEQ ID NO:8. In certain
embodiments, the FZD-
binding agent comprises a heavy chain variable region consisting essentially
of SEQ ID NO:7 and a
light chain variable region consisting essentially of SEQ ID NO:8.
[00164] In certain embodiments, the invention provides a FZD-binding agent
(e.g., an antibody) that
comprises: (a) a heavy chain having at least 90% sequence identity to SEQ ID
NO:9 (with or without
the signal sequence) or SEQ ID NO:11; and/or (b) a light chain having at least
90% sequence identity
to SEQ ID NO:10 (with or without the signal sequence) or SEQ ID NO:12. In some
embodiments,
the FZD-binding agent comprises: (a) a heavy chain having at least 95%
sequence identity to SEQ ID
NO:9 (with or without the signal sequence) or SEQ ID NO:11; and/or (b) a light
chain having at least
95% sequence identity to SEQ ID NO:10 (with or without the signal sequence) or
SEQ ID NO:12. In
some embodiments, the FZD-binding agent comprises a heavy chain comprising SEQ
ID NO:9 (with
or without the signal sequence) or SEQ ID NO:11, and/or a light chain
comprising SEQ ID NO:10
(with or without the signal sequence) or SEQ ID NO:12. In some embodiments,
the FZD-binding
agent comprises a heavy chain comprising SEQ ID NO:11 and a light chain
comprising SEQ ID
NO:12. In some embodiments, the FZD-binding agent comprises a heavy chain
consisting essentially
of amino acids 20-463 of SEQ ID NO:9 and a light chain consisting essentially
of amino acids 20-232
of SEQ ID NO:10. In some embodiments, the FZD-binding agent comprises a heavy
chain consisting
essentially of SEQ ID NO:11 and a light chain consisting essentially of SEQ ID
NO:12.
[00165] In certain embodiments, the invention provides a Wnt pathway inhibitor
which is a FZD-
binding agent (e.g., an antibody) that specifically binds at least one of
FZD1, FZD2, FZD5, FZD7,
and/or FZD8, wherein the FZD-binding agent (e.g., an antibody) comprises one,
two, three, four, five,
and/or six of the CDRs of antibody OMP-18R5. Antibody OMP-18R5 (also known as
18R5 and
43
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
vantictumab), as well as other FZD-binding agents, has been previously
described in U.S. Patent No.
7,982,013. DNA encoding the heavy chain and light chain of the OMP-18R5 IgG2
antibody was
deposited with the ATCC, under the conditions of the Budapest Treaty on
September 29, 2008, and
assigned ATCC deposit designation number PTA-9541. In some embodiments, the
FZD-binding
agent comprises one or more of the CDRs of OMP-18R5, two or more of the CDRs
of OMP-18R5,
three or more of the CDRs of OMP-18R5, four or more of the CDRs of OMP-18R5,
five or more of
the CDRs of OMP-18R5, or all six of the CDRs of OMP-18R5.
[00166] The invention provides polypeptides which are Wnt pathway inhibitors.
The polypeptides
include, but are not limited to, antibodies that specifically bind human FZD
proteins. In some
embodiments, a polypeptide binds one or more FZD proteins selected from the
group consisting of
FZD1, FZD2, FZD3, FZD4, FZD5, FZD6, FZD7, FZD8, FZD9, and FZD10. In some
embodiments,
a polypeptide binds FZD1, FZD2, FZD5, FZD7, and/or FZD8. In some embodiments,
a polypeptide
binds FZD1, FZD2, FZD5, FZD7, and FZD8.
10016711n certain embodiments, a polypeptide comprises one, two, three, four,
five, and/or six of the
CDRs of antibody OMP-18R5. In some embodiments, a polypeptide comprises CDRs
with up to four
(i.e., 0, 1, 2, 3, or 4) amino acid substitutions per CDR. In certain
embodiments, the heavy chain
CDR(s) are contained within a heavy chain variable region. In certain
embodiments, the light chain
CDR(s) are contained within a light chain variable region.
[00168] In some embodiments, the invention provides a polypeptide that
specifically binds one or
more human FZD proteins, wherein the polypeptide comprises an amino acid
sequence having at least
about 80% sequence identity to SEQ ID NO:7, and/or an amino acid sequence
having at least about
80% sequence identity to SEQ ID NO:8. In certain embodiments, the polypeptide
comprises an
amino acid sequence having at least about 85%, at least about 90%, at least
about 95%, at least about
97%, or at least about 99% sequence identity to SEQ ID NO:7. In certain
embodiments, the
polypeptide comprises an amino acid sequence having at least about 85%, at
least about 90%, at least
about 95%, at least about 97%, or at least about 99% sequence identity to SEQ
ID NO:8. In certain
embodiments, the polypeptide comprises an amino acid sequence having at least
about 95% sequence
identity to SEQ ID NO:7, and/or an amino acid sequence having at least about
95% sequence identity
to SEQ ID NO:8. In certain embodiments, the polypeptide comprises an amino
acid sequence
comprising SEQ ID NO:7, and/or an amino acid sequence comprising SEQ ID NO:8.
[00169] In some embodiments, a FZD-binding agent comprises a polypeptide
comprising a sequence
selected from the group consisting of: SEQ ID NO:7, SEQ ID NO:8, SEQ ID NO:9,
SEQ ID NO:10,
SEQ ID NO:11, and SEQ ID NO:12.
[00170] In certain embodiments, a FZD-binding agent comprises the heavy chain
variable region and
light chain variable region of the OMP-18R5 antibody. In certain embodiments,
a FZD-binding agent
comprises the heavy chain and light chain of the OMP-18R5 antibody (with or
without the leader
sequence).
44
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
[00171] In certain embodiments, a FZD-binding agent comprises, consists
essentially of, or consists
of, the antibody OMP-18R5.
[00172] In certain embodiments, a FZD-binding agent (e.g., antibody) competes
for specific binding
to one or more human FZD proteins with an antibody that comprises a heavy
chain variable region
comprising SEQ ID NO:7 and a light chain variable region comprising SEQ ID
NO:8. In certain
embodiments, a FZD-binding agent (e.g., antibody) competes for specific
binding to one or more
human FZD proteins with an antibody that comprises a heavy chain comprising
SEQ ID NO:9 (with
or without the signal sequence) and a light chain comprising SEQ ID NO:10
(with or without the
signal sequence). In certain embodiments, a FZD-binding agent (e.g., antibody)
competes for specific
binding to one or more human FZD proteins with an antibody that comprises a
heavy chain
comprising SEQ ID NO:11 and a light chain comprising SEQ ID NO:12. In certain
embodiments, a
FZD-binding agent competes with antibody OMP-18R5 for specific binding to one
or more human
FZD proteins. In some embodiments, a FZD-binding agent or antibody competes
for specific binding
to one or more human FZD proteins in an in vitro competitive binding assay.
[00173] In certain embodiments, a FZD-binding agent (e.g., an antibody) binds
the same epitope, or
essentially the same epitope, on one or more human FZD proteins as an antibody
of the invention. In
another embodiment, a FZD-binding agent is an antibody that binds an epitope
on one or more human
FZD proteins that overlaps with the epitope on a FZD protein bound by an
antibody of the invention.
In certain embodiments, a FZD-binding agent (e.g., an antibody) binds the same
epitope, or
essentially the same epitope, on one or more FZD proteins as antibody OMP-
18R5. In another
embodiment, the FZD-binding agent is an antibody that binds an epitope on one
or more human FZD
proteins that overlaps with the epitope on a FZD protein bound by antibody OMP-
18R5.
[00174] In certain embodiments, the Wnt pathway inhibitors are agents that
bind one or more human
Wnt proteins. In certain embodiments, the agents specifically bind one, two,
three, four, five, six,
seven, eight, nine, ten, or more Wnt proteins. In some embodiments, the Wnt-
binding agents bind one
or more human Wnt proteins selected from the group consisting of Wntl, Wnt2,
Wnt2b, Wnt3,
Wnt3a, Wnt4, Wnt5a, Wnt5b, Wnt6, Wnt7a, Wnt7b, Wnt8a, Wnt8b, Wnt9a, Wnt9b,
Wntl Oa,
Wntl0b, Wntll, and Wnt16. In certain embodiments, a Wnt-binding agent binds
one or more (or two
or more, three or more, four or more, five or more, etc.) Wnt proteins
selected from the group
consisting of Wntl, Wnt2, Wnt2b, Wnt3, Wnt3a, Wnt7a, Wnt7b, Wnt8a, Wnt8b, Wntl
Oa, and
Wntl Ob. In certain embodiments, the one or more (or two or more, three or
more, four or more, five
or more, etc.) Wnt proteins are selected from the group consisting of Wntl,
Wnt2, Wnt2b, Wnt3,
Wnt3a, Wnt8a, Wnt8b, Wntl Oa, and Wntl Ob.
[00175] In certain embodiments, the Wnt-binding agent is a Wnt antagonist. In
certain embodiments,
the Wnt-binding agent is a Wnt pathway antagonist. In certain embodiments, the
Wnt-binding agent
inhibits Wnt signaling. In some embodiments, the Wnt-binding agent inhibits
canonical Wnt
signaling.
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
[00176] In some embodiments, the Wnt-binding agent is an antibody. In some
embodiments, the
Wnt-binding agent is a polypeptide. In certain embodiments, the Wnt-binding
agent is an antibody or
a polypeptide comprising an antigen-binding site. In certain embodiments, an
antigen-binding site of
a Wnt-binding antibody or polypeptide described herein is capable of binding
(or binds) one, two,
three, four, five, or more human Wnt proteins. In certain embodiments, an
antigen-binding site of the
Wnt-binding antibody or polypeptide is capable of specifically binding one,
two, three, four, or five
human Wnt proteins selected from the group consisting of Wntl, Wnt2, Wnt2b,
Wnt3, Wnt3a, Wnt7a,
Wnt7b, Wnt8a, Wnt8b, Wntl0a, and Wntl Ob. Non-limiting examples of Wnt-binding
agents can be
found in International Publication WO 2011/088127.
[00177] In certain embodiments, a Wnt-binding agent binds to the C-terminal
cysteine rich domain of
one or more human Wnt proteins. In certain embodiments, the Wnt-binding agent
binds a domain
within the one or more Wnt proteins selected from the group consisting of: SEQ
ID NO:46 (Wntl),
SEQ ID NO:47 (Wnt2), SEQ ID NO:48 (Wnt2b), SEQ ID NO:49 (Wnt3), SEQ ID NO:50
(Wnt3a),
SEQ ID NO:51 (Wnt7a), SEQ ID NO:52 (Wnt7b), SEQ ID NO:53 (Wnt8a), SEQ ID NO:54
(Wnt8b),
SEQ ID NO:55 (Wntl Oa), and SEQ ID NO:56 (Wntl Ob).
[00178] In certain embodiments, the Wnt-binding agent binds one or more (e.g.,
two or more, three or
more, or four or more) Wnt proteins with a KD of about 1 M or less, about
100nM or less, about
40nM or less, about 20nM or less, or about lOnM or less. For example, in
certain embodiments, a
Wnt-binding agent described herein that binds more than one Wnt protein, binds
those Wnt proteins
with a KD of about 100nM or less, about 20nM or less, or about lOnM or less.
In certain
embodiments, the Wnt-binding agent binds each of one or more (e.g., 1, 2, 3,
4, or 5) Wnt proteins
with a KD of about 40nM or less, wherein the Wnt proteins are selected from
the group consisting of:
Wntl, Wnt2, Wnt2b, Wnt3, Wnt3a, Wnt7a, Wnt7b, Wnt8a, Wnt8b, Wntl Oa, and Wntl
Ob. In some
embodiments, the KD of the binding agent (e.g., an antibody) to a Wnt protein
is the KD determined
using a Wnt fusion protein comprising at least a portion of the Wnt C-terminal
cysteine rich domain
immobilized on a Biacore chip.
[00179] In certain embodiments, the Wnt-binding agent binds one or more (for
example, two or more,
three or more, or four or more) human Wnt proteins with an EC50 of about 111M
or less, about 100nM
or less, about 40nM or less, about 20nM or less, about lOnM or less, or about
1nM or less. In certain
embodiments, a Wnt-binding agent binds to more than one Wnt with an EC50 of
about 40nM or less,
about 20nM or less, or about lOnM or less. In certain embodiments, the Wnt-
binding agent has an
EC50 of about 20nM or less with respect to one or more (e.g., 1, 2, 3, 4, or
5) of Wnt proteins Wntl,
Wnt2, Wnt2b, Wnt3, Wnt3a, Wnt4, Wnt5a, Wnt5b, Wnt6, Wnt7a, Wnt7b, Wnt8a,
Wnt8b, Wnt9a,
Wnt9b, Wntl0a, Wntl0b, Wntll, and/or Wnt16. In certain embodiments, the Wnt-
binding agent has
an EC50 of about lOnM or less with respect to one or more (e.g., 1, 2, 3, 4,
or 5) of the following Wnt
proteins Wntl, Wnt2, Wnt2b, Wnt3, Wnt3a, Wnt8a, Wnt8b, Wntl0a, and/or Wntl0b.
46
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
[00180] In certain embodiments, the Wnt pathway inhibitor is a Wnt-binding
agent which is an
antibody. In some embodiments, the antibody is a recombinant antibody. In some
embodiments, the
antibody is a monoclonal antibody. In some embodiments, the antibody is a
chimeric antibody. In
some embodiments, the antibody is a humanized antibody. In some embodiments,
the antibody is a
human antibody. In certain embodiments, the antibody is an IgG1 antibody. In
certain embodiments,
the antibody is an IgG2 antibody. In certain embodiments, the antibody is an
antibody fragment
comprising an antigen-binding site. In some embodiments, the antibody is
monovalent, monospecific,
or bivalent. In some embodiments, the antibody is a bispecific antibody or a
multispecific antibody.
In some embodiments, the antibody is conjugated to a cytotoxic moiety. In some
embodiments, the
antibody is isolated. In some embodiments, the antibody is substantially pure.
[00181] The Wnt-binding agents (e.g., antibodies) of the present invention can
be assayed for specific
binding by any method known in the art as described herein for FZD-binding
agents.
[00182] In certain embodiments, the Wnt-binding agent is a soluble receptor.
In certain embodiments,
the Wnt-binding agent comprises the extracellular domain of a FZD receptor
protein. In some
embodiments, the Wnt-binding agent comprises a Fri domain of a FZD protein. In
some
embodiments, a soluble receptor comprising a FZD Fri domain can demonstrate
altered biological
activity (e.g., increased protein half-life) compared to a soluble receptor
comprising the entire FZD
ECD. Protein half-life can be further modified (i.e., increased) by covalent
modification with
polyethylene glycol (PEG) or polyethylene oxide (PEO). In certain embodiments,
the FZD protein is
a human FZD protein. In certain embodiments, the human FZD protein is FZD1,
FZD2, FZD3,
FZD4, FZD5, FZD6, FZD7, FZD8, FZD9, or FZD10. Non-limiting examples of soluble
FZD
receptors can be found in U.S. Patent Nos. 7,723,477 and 7,947,277 and U.S.
Patent Publication No.
2013/0034551.
[00183] The predicted Fri domains for each of the human FZD1-10 proteins are
provided as SEQ ID
NOs:13-22. The predicted minimal Fri domains for each of the human FZD1-10
proteins are
provided as SEQ ID NOs:23-32. Those of skill in the art may differ in their
understanding of the
exact amino acids corresponding to the various Fri domains. Thus, the N-
terminus and/or C-terminus
of the domains outlined above and herein may extend or be shortened by 1, 2,
3, 4, 5, 6, 7, 8, 9, or
even 10 amino acids.
[00184] In certain embodiments, the Wnt-binding agent comprises a Fri domain
of a human FZD
protein, or a fragment or variant of the Fri domain that binds one or more
human Wnt proteins. In
certain embodiments, the human FZD protein is FZD1, FZD2, FZD3, FZD4, FZD5,
FZD6, FZD7,
FZD8, FZD9, or FZD10. In certain embodiments, the human FZD protein is FZD4.
In certain
embodiments, the human FZD protein is FZD5. In certain embodiments, the human
FZD protein is
FZD8. In certain embodiments, the human FZD protein is FZD10. In certain
embodiments, the FZD
protein is FZD4 and the Wnt-binding agent comprises SEQ ID NO:16. In certain
embodiments, the
FZD protein is FZD5 and the Wnt-binding agent comprises SEQ ID NO:17. In
certain embodiments,
47
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
the FZD protein is FZD7 and the Wnt-binding agent comprises SEQ ID NO:19. In
certain
embodiments, the FZD protein is FZD8 and the Wnt-binding agent comprises SEQ
ID NO:20. In
certain embodiments, the FZD protein is FZD10 and the Wnt-binding agent
comprises SEQ ID
NO:22. In certain embodiments, the FZD protein is FZD8 and the Wnt-binding
agent comprises SEQ
ID NO:33.
[00185] In some embodiments, the Wnt-binding agent comprises a Fri domain
comprising the
minimal Fri domain of FZD1 (SEQ ID NO:23), the minimal Fri domain of FZD2 (SEQ
ID NO:24),
the minimal Fri domain of FZD3 (SEQ ID NO:25), the minimal Fri domain of FZD4
(SEQ ID
NO:26), the minimal Fri domain of FZD5 (SEQ ID NO:27), the minimal Fri domain
of FZD6 (SEQ
ID NO:28), the minimal Fri domain of FZD7 (SEQ ID NO:29), the minimal Fri
domain of FZD8
(SEQ ID NO:30), the minimal Fri domain of FZD9 (SEQ ID NO:31), or the minimal
Fri domain of
FZD10 (SEQ ID NO:32). In some embodiments, the Wnt-binding agent comprises a
Fri domain
comprising the minimal Fri domain of FZD8 (SEQ ID NO:30).
[00186] In some embodiments, the Wnt-binding agent comprises a Fri domain
consisting essentially
of the Fri domain of FZD1, the Fri domain of FZD2, the Fri domain of FZD3, the
Fri domain of
FZD4, the Fri domain of FZD5, the Fri domain of FZD6, the Fri domain of FZD7,
the Fri domain of
FZD8, the Fri domain of FZD9, or the Fri domain of FZD10. In some embodiments,
the Wnt-binding
agent comprises a Fri domain consisting essentially of the Fri domain of FZD8.
[00187] In some embodiments, the Wnt-binding agent comprises a sequence
selected from the group
consisting of: SEQ ID NO:13, SEQ ID NO:14, SEQ ID NO:15, SEQ ID NO:16, SEQ ID
NO:17, SEQ
ID NO:18, SEQ ID NO:19, SEQ ID NO:20, SEQ ID NO:21, SEQ ID NO:22, SEQ ID
NO:23, SEQ
ID NO:24, SEQ ID NO:25, SEQ ID NO:26, SEQ ID NO:27, SEQ ID NO:28, SEQ ID
NO:29, SEQ
ID NO:30, SEQ ID NO:31, SEQ ID NO:32, and SEQ ID NO:33. In some embodiments,
the Wnt-
binding agent comprises a Fri domain consisting essentially of SEQ ID NO:20.
In some
embodiments, the Wnt-binding agent comprises a Fri domain consisting
essentially of SEQ ID
NO:33.
[00188] In certain embodiments, the Wnt-binding agent comprises a variant of
any one of the
aforementioned FZD Fri domain sequences that comprises one or more (e.g., one,
two, three, four,
five, six, seven, eight, nine, ten, etc.) conservative substitutions and is
capable of binding Wnt
protein(s).
[00189] In certain embodiments, a Wnt-binding agent, such as an agent
comprising a Fri domain of a
human FZD receptor, further comprises a non-FZD polypeptide. In some
embodiments, a FZD
soluble receptor may include FZD ECD or Fri domains linked to other non-FZD
functional and
structural polypeptides including, but not limited to, a human Fc region,
protein tags (e.g., myc,
FLAG, GST), other endogenous proteins or protein fragments, or any other
useful protein sequence
including any linker region between a FZD ECD or Fri domain and a second
polypeptide. In certain
embodiments, the non-FZD polypeptide comprises a human Fc region. The Fc
region can be obtained
48
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
from any of the classes of immunoglobulin, IgG, IgA, IgM, IgD and IgE. In some
embodiments, the
Fc region is a human IgG1 Fc region. In some embodiments, the Fc region is a
human IgG2 Fc
region. In some embodiments, the Fc region is a wild-type Fc region. In some
embodiments, the Fc
region is a mutated Fc region. In some embodiments, the Fc region is truncated
at the N-terminal end
by 1, 2, 3, 4, 5, 6, 7, 8, 9, or 10 amino acids, (e.g., in the hinge domain).
In some embodiments, an
amino acid in the hinge domain is changed to hinder undesirable disulfide bond
formation. In some
embodiments, a cysteine is replaced with a serine to hinder or block
undesirable disulfide bond
formation. In some embodiments, the Fc region is truncated at the C-terminal
end by 1, 2, 3, or more
amino acids. In some embodiments, the Fc region is truncated at the C-terminal
end by 1 amino acid.
In certain embodiments, the non-FZD polypeptide comprises SEQ ID NO:34, SEQ ID
NO:35, SEQ
ID NO:36, SEQ ID NO:37, or SEQ ID NO:38. In certain embodiments, the non-FZD
polypeptide
consists essentially of SEQ ID NO:34, SEQ ID NO:35, SEQ ID NO:36, SEQ ID
NO:37, or SEQ ID
NO:38. In certain embodiments, the non-FZD polypeptide consists essentially of
SEQ ID NO:36 or
SEQ ID NO:37.
[00190] In certain embodiments, a Wnt-binding agent is a fusion protein
comprising at least a minimal
Fri domain of a FZD receptor and a Fc region. As used herein, a "fusion
protein" is a hybrid protein
expressed by a nucleic acid molecule comprising nucleotide sequences of at
least two genes. In some
embodiments, the C-terminus of the first polypeptide is linked to the N-
terminus of the
immunoglobulin Fc region. In some embodiments, the first polypeptide (e.g., a
FZD Fri domain) is
directly linked to the Fc region (i.e. without an intervening linker). In some
embodiments, the first
polypeptide is linked to the Fc region via a linker.
[00191] As used herein, the term "linker" refers to a linker inserted between
a first polypeptide (e.g., a
FZD component) and a second polypeptide (e.g., a Fc region). In some
embodiments, the linker is a
peptide linker. Linkers should not adversely affect the expression, secretion,
or bioactivity of the
polypeptide. Linkers should not be antigenic and should not elicit an immune
response. Suitable
linkers are known to those of skill in the art and often include mixtures of
glycine and serine residues
and often include amino acids that are sterically unhindered. Other amino
acids that can be
incorporated into useful linkers include threonine and alanine residues.
Linkers can range in length,
for example from 1-50 amino acids in length, 1-22 amino acids in length, 1-10
amino acids in length,
1-5 amino acids in length, or 1-3 amino acids in length. Linkers may include,
but are not limited to,
SerGly, GGSG, GSGS, GGGS, S(GGS)n where n is 1-7, GRA, poly(Gly), poly(Ala),
ESGGGGVT
(SEQ ID NO:57), LESGGGGVT (SEQ ID NO:58), GRAQVT (SEQ ID NO:59), WRAQVT (SEQ
ID
NO:60), and ARGRAQVT (SEQ ID NO:61). As used herein, a "linker" is an
intervening peptide
sequence that does not include amino acid residues from either the C-terminus
of the first polypeptide
(e.g., a FZD Fri domain) or the N-terminus of the second polypeptide (e.g.,
the Fc region).
[00192] In some embodiments, the Wnt-binding agent comprises a FZD Fri domain,
a Fc region, and
a linker connecting the FZD Fri domain to the Fc region. In some embodiments,
the FZD Fri domain
49
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
comprises SEQ ID NO:20, SEQ ID NO:30, or SEQ ID NO:33. In some embodiments,
the linker
comprises ESGGGGVT (SEQ ID NO:57) or LESGGGGVT (SEQ ID NO:58).
[00193] In some embodiments, the Wnt-binding agent comprises a first
polypeptide comprising SEQ
ID NO:13, SEQ ID NO:14, SEQ ID NO:15, SEQ ID NO:16, SEQ ID NO:17, SEQ ID
NO:18, SEQ
ID NO:19, SEQ ID NO:20, SEQ ID NO:21, SEQ ID NO:22, SEQ ID NO:23, SEQ ID
NO:24, SEQ
ID NO:25, SEQ ID NO:26, SEQ ID NO:27, SEQ ID NO:28, SEQ ID NO:29, SEQ ID
NO:30, SEQ
ID NO:31, SEQ ID NO:32, or SEQ ID NO:33; and a second polypeptide comprising
SEQ ID NO:34,
SEQ ID NO:35, SEQ ID NO:36, SEQ ID NO:37, or SEQ ID NO:38, wherein the first
polypeptide is
directly linked to the second polypeptide. In some embodiments, the Wnt-
binding agent comprises a
first polypeptide comprising SEQ ID NO:20 and a second polypeptide comprising
SEQ ID NO:34,
SEQ ID NO:35, SEQ ID NO:36, SEQ ID NO:37, or SEQ ID NO:38. In some
embodiments, the Wnt-
binding agent comprises a first polypeptide comprising SEQ ID NO:20 and a
second polypeptide
comprising SEQ ID NO:36 or SEQ ID NO:37. In some embodiments, the Wnt-binding
agent
comprises a first polypeptide consisting essentially of SEQ ID NO:20 and a
second polypeptide
consisting essentially of SEQ ID NO:36 or SEQ ID NO:37. In some embodiments,
the Wnt-binding
agent comprises a first polypeptide comprising SEQ ID NO:30 and a second
polypeptide comprising
SEQ ID NO:34, SEQ ID NO:35, SEQ ID NO:36, SEQ ID NO:37, or SEQ ID NO:38. In
some
embodiments, the Wnt-binding agent comprises a first polypeptide comprising
SEQ ID NO and a
second polypeptide comprising SEQ ID NO:36 or SEQ ID NO:37. In some
embodiments, the Wnt-
binding agent comprises a first polypeptide comprising SEQ ID NO:33 and a
second polypeptide
comprising SEQ ID NO:34, SEQ ID NO:35, SEQ ID NO:36, SEQ ID NO:37, or SEQ ID
NO:38. In
some embodiments, the Wnt-binding agent comprises a first polypeptide
comprising SEQ ID NO:33
and a second polypeptide comprising SEQ ID NO:36, SEQ ID NO:37, or SEQ ID
NO:35. In some
embodiments, the Wnt-binding agent comprises a first polypeptide consisting
essentially of SEQ ID
NO:33 and a second polypeptide consisting essentially of SEQ ID NO:36, SEQ ID
NO:37, or SEQ ID
NO:35.
[00194] In some embodiments, the Wnt-binding agent comprises a first
polypeptide comprising SEQ
ID NO:13, SEQ ID NO:14, SEQ ID NO:15, SEQ ID NO:16, SEQ ID NO:17, SEQ ID
NO:18, SEQ
ID NO:19, SEQ ID NO:20, SEQ ID NO:21, SEQ ID NO:22, SEQ ID NO:23, SEQ ID
NO:24, SEQ
ID NO:25, SEQ ID NO:26, SEQ ID NO:27, SEQ ID NO:28, SEQ ID NO:29, SEQ ID
NO:30, SEQ
ID NO:31, SEQ ID NO:32, or SEQ ID NO:33; and a second polypeptide comprising
SEQ ID NO:34,
SEQ ID NO:35, SEQ ID NO:36, SEQ ID NO:37, or SEQ ID NO:38, wherein the first
polypeptide is
connected to the second polypeptide by a linker. In some embodiments, the Wnt-
binding agent
comprises a first polypeptide comprising SEQ ID NO:20 and a second polypeptide
comprising SEQ
ID NO:34, SEQ ID NO:35, SEQ ID NO:36, SEQ ID NO:37, or SEQ ID NO:38. In some
embodiments, the Wnt-binding agent comprises a first polypeptide comprising
SEQ ID NO:20 and a
second polypeptide comprising SEQ ID NO:36 or SEQ ID NO:37. In some
embodiments, the Wnt-
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
binding agent comprises a first polypeptide consisting essentially of SEQ ID
NO:20 and a second
polypeptide consisting essentially of SEQ ID NO:36 or SEQ ID NO:37. In some
embodiments, the
Wnt-binding agent comprises a first polypeptide comprising SEQ ID NO:30 and a
second polypeptide
comprising SEQ ID NO:34, SEQ ID NO:35, SEQ ID NO:36, SEQ ID NO:37, or SEQ ID
NO:38. In
some embodiments, the Wnt-binding agent comprises a first polypeptide
comprising SEQ ID NO:33
and a second polypeptide comprising SEQ ID NO:34, SEQ ID NO:35, SEQ ID NO:36,
SEQ ID
NO:37, or SEQ ID NO:38. In some embodiments, the Wnt-binding agent comprises a
first
polypeptide comprising SEQ ID NO:33 and a second polypeptide comprising SEQ ID
NO:36, SEQ
ID NO:37, or SEQ ID NO:35. In some embodiments, the Wnt-binding agent
comprises a first
polypeptide consisting essentially of SEQ ID NO:33 and a second polypeptide
consisting essentially
of SEQ ID NO:36, SEQ ID NO:37, or SEQ ID NO:35.
[00195] In some embodiments, the Wnt-binding agent comprises a first
polypeptide that is at least
95% identical to SEQ ID NO:13, SEQ ID NO:14, SEQ ID NO:15, SEQ ID NO:16, SEQ
ID NO:17,
SEQ ID NO:18, SEQ ID NO:19, SEQ ID NO:20, SEQ ID NO:21, SEQ ID NO:22, SEQ ID
NO:23,
SEQ ID NO:24, SEQ ID NO:25, SEQ ID NO:26, SEQ ID NO:27, SEQ ID NO:28, SEQ ID
NO:29,
SEQ ID NO:30, SEQ ID NO:31, SEQ ID NO:32, or SEQ ID NO:33; and a second
polypeptide
comprising SEQ ID NO:34, SEQ ID NO:35, SEQ ID NO:36, SEQ ID NO:37, or SEQ ID
NO:38,
wherein the first polypeptide is directly linked to the second polypeptide. In
some embodiments, the
Wnt-binding agent comprises a first polypeptide that is at least 95% identical
to SEQ ID NO:20 and a
second polypeptide comprising SEQ ID NO:34, SEQ ID NO:35, SEQ ID NO:36, SEQ ID
NO:37, or
SEQ ID NO:38. In some embodiments, the Wnt-binding agent comprises a first
polypeptide that is at
least 95% identical to SEQ ID NO:30 and a second polypeptide comprising SEQ ID
NO:34, SEQ ID
NO:35, SEQ ID NO:36, SEQ ID NO:37, or SEQ ID NO:38. In some embodiments, the
Wnt-binding
agent comprises a first polypeptide that is at least 95% identical to SEQ ID
NO:33 and a second
polypeptide comprising SEQ ID NO:34, SEQ ID NO:35, SEQ ID NO:36, SEQ ID NO:37,
or SEQ ID
NO:38.
[00196] In some embodiments, the Wnt-binding agent comprises a first
polypeptide that is at least
95% identical to SEQ ID NO:13, SEQ ID NO:14, SEQ ID NO:15, SEQ ID NO:16, SEQ
ID NO:17,
SEQ ID NO:18, SEQ ID NO:19, SEQ ID NO:20, SEQ ID NO:21, SEQ ID NO:22, SEQ ID
NO:23,
SEQ ID NO:24, SEQ ID NO:25, SEQ ID NO:26, SEQ ID NO:27, SEQ ID NO:28, SEQ ID
NO:29,
SEQ ID NO:30, SEQ ID NO:31, SEQ ID NO:32, or SEQ ID NO:33; and a second
polypeptide
comprising SEQ ID NO:34, SEQ ID NO:35, SEQ ID NO:36, SEQ ID NO:37, or SEQ ID
NO:38,
wherein the first polypeptide is connected to the second polypeptide by a
linker. In some
embodiments, the Wnt-binding agent comprises a first polypeptide that is at
least 95% identical to
SEQ ID NO:20 and a second polypeptide comprising SEQ ID NO:34, SEQ ID NO:35,
SEQ ID
NO:36, SEQ ID NO:37, or SEQ ID NO:38. In some embodiments, the Wnt-binding
agent comprises
a first polypeptide that is at least 95% identical to SEQ ID NO:30 and a
second polypeptide
51
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
comprising SEQ ID NO:34, SEQ ID NO:35, SEQ ID NO:36, SEQ ID NO:37, or SEQ ID
NO:38. In
some embodiments, the Wnt-binding agent comprises a first polypeptide that is
at least 95% identical
to SEQ ID NO:33 and a second polypeptide comprising SEQ ID NO:34, SEQ ID
NO:35, SEQ ID
NO:36, SEQ ID NO:37, or SEQ ID NO:38.
[00197] FZD proteins contain a signal sequence that directs the transport of
the proteins. Signal
sequences (also referred to as signal peptides or leader sequences) are
located at the N-terminus of
nascent polypeptides. They target the polypeptide to the endoplasmic reticulum
and the proteins are
sorted to their destinations, for example, to the inner space of an organelle,
to an interior membrane,
to the cell outer membrane, or to the cell exterior via secretion. Most signal
sequences are cleaved
from the protein by a signal peptidase after the proteins are transported to
the endoplasmic reticulum.
The cleavage of the signal sequence from the polypeptide usually occurs at a
specific site in the amino
acid sequence and is dependent upon amino acid residues within the signal
sequence. Although there
is usually one specific cleavage site, more than one cleavage site may be
recognized and/or used by a
signal peptidase resulting in a non-homogenous N-terminus of the polypeptide.
For example, the use
of different cleavage sites within a signal sequence can result in a
polypeptide expressed with
different N-terminal amino acids. Accordingly, in some embodiments, the
polypeptides described
herein may comprise a mixture of polypeptides with different N-termini. In
some embodiments, the
N-termini differ in length by 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more amino
acids. In some embodiments,
the N-termini differ in length by 1, 2, 3, 4, or 5 amino acids. In some
embodiments, the polypeptide is
substantially homogeneous, i.e., the polypeptides have the same N-terminus. In
some embodiments,
the signal sequence of the polypeptide comprises one or more (e.g., one, two,
three, four, five, six,
seven, eight, nine, ten, etc.) amino acid substitutions and/or deletions. In
some embodiments, the
signal sequence of the polypeptide comprises amino acid substitutions and/or
deletions that allow one
cleavage site to be dominant, thereby resulting in a substantially homogeneous
polypeptide with one
N-terminus.
[00198] In some embodiments, the Wnt-binding agent comprises an amino acid
sequence selected
from the group consisting of: SEQ ID NO:39, SEQ ID NO:40, SEQ ID NO:41, SEQ ID
NO:42, SEQ
ID NO:43, SEQ ID NO:44, and SEQ ID NO:45.
[00199] In certain embodiments, the Wnt-binding agent comprises the sequence
of SEQ ID NO:39. In
certain embodiments, the agent comprises the sequence of SEQ ID NO:39,
comprising one or more
(e.g., one, two, three, four, five, six, seven, eight, nine, ten, etc.)
conservative substitutions. In certain
embodiments, the agent comprises a sequence having at least about 90%, about
95%, or about 98%
sequence identity with SEQ ID NO:39. In certain embodiments, the variants of
SEQ ID NO:39
maintain the ability to bind one or more human Wnt proteins.
[00200] In certain embodiments, the Wnt-binding agent comprises the sequence
of SEQ ID NO:40. In
some embodiments, the Wnt-binding agent is SEQ ID NO:40. In certain
alternative embodiments, the
agent comprises the sequence of SEQ ID NO:40, comprising one or more (e.g.,
one, two, three, four,
52
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
five, six, seven, eight, nine, ten, etc.) conservative substitutions. In
certain embodiments, the agent
comprises a sequence having at least about 90%, about 95%, or about 98%
sequence identity with
SEQ ID NO:40. In certain embodiments, the variants of SEQ ID NO:40 maintain
the ability to bind
one or more human Wnt proteins.
[00201] In certain embodiments, the Wnt-binding agent comprises the sequence
of SEQ ID NO:41. In
some embodiments, the Wnt-binding agent is SEQ ID NO:41. In certain
alternative embodiments, the
agent comprises the sequence of SEQ ID NO:41, comprising one or more (e.g.,
one, two, three, four,
five, six, seven, eight, nine, ten, etc.) conservative substitutions. In
certain embodiments, the agent
comprises a sequence having at least about 90%, about 95%, or about 98%
sequence identity with
SEQ ID NO:41. In certain embodiments, the variants of SEQ ID NO:41 maintain
the ability to bind
one or more human Wnt proteins.
[00202] In some embodiments, the Wnt-binding agent is OMP-54F28.
[00203] In certain embodiments, a Wnt-binding agent is a polypeptide
comprising an amino acid
sequence selected from the group consisting of: SEQ ID NO:39, SEQ ID NO:40,
SEQ ID NO:41,
SEQ ID NO:42, SEQ ID NO:43, SEQ ID NO:44, and SEQ ID NO:45. In certain
embodiments, the
polypeptide comprises an amino acid sequence selected from the group
consisting of SEQ ID NO:39,
SEQ ID NO:40, and SEQ ID NO:41. In some embodiments, a polypeptide consists
essentially of an
amino acid sequence selected from the group consisting of: SEQ ID NO:39, SEQ
ID NO:40, and SEQ
ID NO:41. In certain embodiments, the polypeptide comprises the amino acid
sequence of SEQ ID
NO:39. In some embodiments, the polypeptide comprises the amino acid sequence
of SEQ ID
NO:40. In certain embodiments, the polypeptide comprises the amino acid
sequence of SEQ ID
NO:41. In certain embodiments, the polypeptide comprises the amino acid
sequence of SEQ ID
NO:42. In certain embodiments, the polypeptide comprises the amino acid
sequence of SEQ ID
NO:43. In certain embodiments, the polypeptide comprises the amino acid
sequence of SEQ ID
NO:44. In certain embodiments, the polypeptide comprises the amino acid
sequence of SEQ ID
NO:45.
[00204] In some embodiments, the polypeptide is a substantially purified
polypeptide comprising an
amino acid sequence selected from the group consisting of SEQ ID NO:39, SEQ ID
NO:40, and SEQ
ID NO:41. In some embodiments, the polypeptide is a substantially purified
polypeptide comprising
SEQ ID NO:41. In certain embodiments, the substantially purified polypeptide
consists of at least
90% of a polypeptide that has an N-terminal sequence of ASA. In some
embodiments, the nascent
polypeptide comprises a signal sequence that results in a substantially
homogeneous polypeptide
product with one N-terminal sequence.
[00205] In certain embodiments, a Wnt-binding agent comprises a Fc region of
an immunoglobulin.
Those skilled in the art will appreciate that some of the binding agents of
this invention will comprise
fusion proteins in which at least a portion of the Fc region has been deleted
or otherwise altered so as
to provide desired biochemical characteristics, such as increased cancer cell
localization, increased
53
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
tumor penetration, reduced serum half-life, or increased serum half-life, when
compared with a fusion
protein of approximately the same immunogenicity comprising a native or
unaltered constant region.
Modifications to the Fc region may include additions, deletions, or
substitutions of one or more amino
acids in one or more domains. The modified fusion proteins disclosed herein
may comprise
alterations or modifications to one or more of the two heavy chain constant
domains (CH2 or CH3) or
to the hinge region. In other embodiments, the entire CH2 domain may be
removed (ACH2
constructs). In some embodiments, the omitted constant region domain is
replaced by a short amino
acid spacer (e.g., 10 aa residues) that provides some of the molecular
flexibility typically imparted by
the absent constant region domain.
[00206] In some embodiments, the modified fusion proteins are engineered to
link the CH3 domain
directly to the hinge region. In other embodiments, a peptide spacer is
inserted between the hinge
region and the modified CH2 and/or CH3 domains. For example, constructs may be
expressed
wherein the CH2 domain has been deleted and the remaining CH3 domain (modified
or unmodified)
is joined to the hinge region with a 5-20 amino acid spacer. Such a spacer may
be added to ensure
that the regulatory elements of the constant domain remain free and accessible
or that the hinge region
remains flexible. However, it should be noted that amino acid spacers may, in
some cases, prove to
be immunogenic and elicit an unwanted immune response against the construct.
Accordingly, in
certain embodiments, any spacer added to the construct will be relatively non-
immunogenic so as to
maintain the desired biological qualities of the fusion protein.
[00207] In some embodiments, the modified fusion proteins may have only a
partial deletion of a
constant domain or substitution of a few or even a single amino acid. For
example, the mutation of a
single amino acid in selected areas of the CH2 domain may be enough to
substantially reduce Fc
binding and thereby increase cancer cell localization and/or tumor
penetration. Similarly, it may be
desirable to simply delete that part of one or more constant region domains
that control a specific
effector function (e.g., complement Clq binding). Such partial deletions of
the constant regions may
improve selected characteristics of the binding agent (e.g., serum half-life)
while leaving other
desirable functions associated with the subject constant region domain intact.
Moreover, as alluded to
above, the constant regions of the disclosed fusion proteins may be modified
through the mutation or
substitution of one or more amino acids that enhances the profile of the
resulting construct. In this
respect it may be possible to disrupt the activity provided by a conserved
binding site (e.g., Fc
binding) while substantially maintaining the configuration and immunogenic
profile of the modified
fusion protein. In certain embodiments, the modified fusion proteins comprise
the addition of one or
more amino acids to the constant region to enhance desirable characteristics
such as decreasing or
increasing effector function, or provide for more cytotoxin or carbohydrate
attachment sites.
[00208] It is known in the art that the constant region mediates several
effector functions. For
example, binding of the Cl component of complement to the Fc region of IgG or
IgM antibodies
(bound to antigen) activates the complement system. Activation of complement
is important in the
54
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
opsonization and lysis of cell pathogens. The activation of complement also
stimulates the
inflammatory response and can also be involved in autoimmune hypersensitivity.
In addition, the Fc
region of an immunoglobulin can bind to a cell expressing a Fc receptor (FcR).
There are a number
of Fc receptors which are specific for different classes of antibody,
including IgG (gamma receptors),
IgE (epsilon receptors), IgA (alpha receptors) and IgM (mu receptors). Binding
of antibody to Fc
receptors on cell surfaces triggers a number of important and diverse
biological responses including
engulfment and destruction of antibody-coated particles, clearance of immune
complexes, lysis of
antibody-coated target cells by killer cells, release of inflammatory
mediators, placental transfer, and
control of immunoglobulin production.
[00209] In some embodiments, the modified fusion proteins provide for altered
effector functions that,
in turn, affect the biological profile of the administered agent. For example,
in some embodiments,
the deletion or inactivation (through point mutations or other means) of a
constant region domain may
reduce Fc receptor binding of the circulating modified agent, thereby
increasing cancer cell
localization and/or tumor penetration. In other embodiments, the constant
region modifications
increase or reduce the serum half-life of the agent. In some embodiments, the
constant region is
modified to eliminate disulfide linkages or oligosaccharide moieties.
[00210] In certain embodiments, a modified fusion protein does not have one or
more effector
functions normally associated with an Fc region. In some embodiments, the
agent has no antibody-
dependent cell-mediated cytotoxicity (ADCC) activity, and/or no complement-
dependent cytotoxicity
(CDC) activity. In certain embodiments, the agent does not bind to the Fc
receptor and/or
complement factors. In certain embodiments, the agent has no effector
function.
[00211] In some embodiments, the Wnt-binding agent (e.g., a soluble receptor)
described herein is
modified to reduce immunogenicity. In general, immune responses against
completely normal human
proteins are rare when these proteins are used as therapeutics. However,
although many fusion
proteins comprise polypeptides sequences that are the same as the sequences
found in nature, several
therapeutic fusion proteins have been shown to be immunogenic in mammals. In
some studies, a
fusion protein comprising a linker has been found to be more immunogenic than
a fusion protein that
does not contain a linker. Accordingly, in some embodiments, the polypeptides
of the invention are
analyzed by computation methods to predict immunogenicity. In some
embodiments, the
polypeptides are analyzed for the presence of T-cell and/or B-cell epitopes.
If any T-cell or B-cell
epitopes are identified and/or predicted, modifications to these regions
(e.g., amino acid substitutions)
may be made to disrupt or destroy the epitopes. Various algorithms and
software that can be used to
predict T-cell and/or B-cell epitopes are known in the art. For example, the
software programs
SYFPEITHI, HLA Bind, PEPVAC, RANKPEP, DiscoTope, ElliPro, and Antibody Epitope
Prediction are all publicly available.
[00212] In some embodiments, a cell producing any of the Wnt-binding agents
(e.g., soluble
receptors) or polypeptides described herein is provided. In some embodiments,
a composition
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
comprising any of the Wnt-binding agents (e.g., soluble receptors) or
polypeptides described herein is
provided. In some embodiments, the composition comprises a polypeptide wherein
at least 80%,
90%, 95%, 97%, 98%, or 99% of the polypeptide has an N-terminal sequence of
ASA. In some
embodiments, the composition comprises a polypeptide wherein 100% of the
polypeptide has an N-
terminal sequence of ASA. In some embodiments, the composition comprises a
polypeptide wherein
at least 80% of the polypeptide has an N-terminal sequence of ASA. In some
embodiments, the
composition comprises a polypeptide wherein at least 90% of the polypeptide
has an N-terminal
sequence of ASA. In some embodiments, the composition comprises a polypeptide
wherein at least
95% of the polypeptide has an N-terminal sequence of ASA.
[00213] The polypeptides described herein can be recombinant polypeptides,
natural polypeptides, or
synthetic polypeptides. It will be recognized in the art that some amino acid
sequences of the
invention can be varied without significant effect on the structure or
function of the protein. If such
differences in sequence are contemplated, it should be remembered that there
will be critical areas on
the protein which determine activity. Thus, the invention further includes
variations of the
polypeptides which show substantial activity or which include regions of FZD
proteins, such as the
protein portions discussed herein. Such mutants include deletions, insertions,
inversions, repeats, and
type substitutions.
[00214] Of course, the number of amino acid substitutions a skilled artisan
would make depends on
many factors, including those described above. In certain embodiments, the
number of substitutions
for any given soluble receptor polypeptide will not be more than 50, 40, 30,
25, 20, 15, 10, 5 or 3.
[00215] Fragments or portions of the polypeptides of the present invention can
be employed for
producing the corresponding full-length polypeptide by peptide synthesis;
therefore, the fragments
can be employed as intermediates for producing the full-length polypeptides.
These fragments or
portion of the polypeptides can also be referred to as "protein fragments" or
"polypeptide fragments".
[00216] A "protein fragment" of this invention is a portion or all of a
protein which is capable of
binding to one or more human Wnt proteins or one or more human FZD proteins.
In some
embodiments, the fragment has a high affinity for one or more human Wnt
proteins. In some
embodiments, the fragment has a high affinity for one or more human FZD
proteins. Some fragments
of Wnt-binding agents described herein are protein fragments comprising at
least part of the
extracellular portion of a FZD protein linked to at least part of a constant
region of an
immunoglobulin (e.g., a Fc region). The binding affinity of the protein
fragment can be in the range
of about 10-11 to 10-12M, although the affinity can vary considerably with
fragments of different sizes,
ranging from 10-7 to 10-13M. In some embodiments, the fragment is about 100 to
about 200 amino
acids in length and comprises a binding domain linked to at least part of a
constant region of an
immunoglobulin.
[00217] In some embodiments, the Wnt pathway inhibitors are polyclonal
antibodies. Polyclonal
antibodies can be prepared by any known method. In some embodiments,
polyclonal antibodies are
56
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
raised by immunizing an animal (e.g., a rabbit, rat, mouse, goat, donkey) by
multiple subcutaneous or
intraperitoneal injections of an antigen of interest (e.g., a purified peptide
fragment, full-length
recombinant protein, or fusion protein). The antigen can be optionally
conjugated to a carrier such as
keyhole limpet hemocyanin (KLH) or serum albumin. The antigen (with or without
a carrier protein)
is diluted in sterile saline and usually combined with an adjuvant (e.g.,
Complete or Incomplete
Freund's Adjuvant) to form a stable emulsion. After a sufficient period of
time, polyclonal antibodies
are recovered from blood and/or ascites of the immunized animal. The
polyclonal antibodies can be
purified from serum or ascites according to standard methods in the art
including, but not limited to,
affinity chromatography, ion-exchange chromatography, gel electrophoresis, and
dialysis.
[00218] In some embodiments, the Wnt pathway inhibitors are monoclonal
antibodies. Monoclonal
antibodies can be prepared using hybridoma methods known to one of skill in
the art (see e.g., Kohler
and Milstein, 1975, Nature, 256:495-497). In some embodiments, using the
hybridoma method, a
mouse, hamster, or other appropriate host animal, is immunized as described
above to elicit from
lymphocytes the production of antibodies that will specifically bind the
immunizing antigen. In some
embodiments, lymphocytes can be immunized in vitro. In some embodiments, the
immunizing
antigen can be a human protein or a portion thereof In some embodiments, the
immunizing antigen
can be a mouse protein or a portion thereof
[00219] Following immunization, lymphocytes are isolated and fused with a
suitable myeloma cell
line using, for example, polyethylene glycol, to form hybridoma cells that can
then be selected away
from unfused lymphocytes and myeloma cells. Hybridomas that produce monoclonal
antibodies
directed specifically against a chosen antigen may be identified by a variety
of methods including, but
not limited to, immunoprecipitation, immunoblotting, and in vitro binding
assay (e.g., flow cytometry,
FACS, ELISA, and radioimmunoassay). The hybridomas can be propagated either in
in vitro culture
using standard methods (J.W. Goding, 1996, Monoclonal Antibodies: Principles
and Practice, 3rd
Edition, Academic Press, San Diego, CA) or in vivo as ascites tumors in an
animal. The monoclonal
antibodies can be purified from the culture medium or ascites fluid according
to standard methods in
the art including, but not limited to, affinity chromatography, ion-exchange
chromatography, gel
electrophoresis, and dialysis.
[00220] In certain embodiments, monoclonal antibodies can be made using
recombinant DNA
techniques as known to one skilled in the art. The polynucleotides encoding a
monoclonal antibody
are isolated from mature B-cells or hybridoma cells, such as by RT-PCR using
oligonucleotide
primers that specifically amplify the genes encoding the heavy and light
chains of the antibody, and
their sequence is determined using conventional techniques. The isolated
polynucleotides encoding
the heavy and light chains are then cloned into suitable expression vectors
which produce the
monoclonal antibodies when transfected into host cells such as E. coli, simian
COS cells, Chinese
hamster ovary (CHO) cells, or myeloma cells that do not otherwise produce
immunoglobulin proteins.
57
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
In other embodiments, recombinant monoclonal antibodies, or fragments thereof,
can be isolated from
phage display libraries.
[00221] The polynucleotide(s) encoding a monoclonal antibody can further be
modified in a number
of different manners using recombinant DNA technology to generate alternative
antibodies. In some
embodiments, the constant domains of the light and heavy chains of, for
example, a mouse
monoclonal antibody can be substituted for those regions of, for example, a
human antibody to
generate a chimeric antibody, or for a non-immunoglobulin polypeptide to
generate a fusion antibody.
In some embodiments, the constant regions are truncated or removed to generate
the desired antibody
fragment of a monoclonal antibody. Site-directed or high-density mutagenesis
of the variable region
can be used to optimize specificity, affinity, etc. of a monoclonal antibody.
[00222] In some embodiments, the Wnt pathway inhibitor is a humanized
antibody. Typically,
humanized antibodies are human immunoglobulins in which amino acid residues of
the CDRs are
replaced by amino acid residues of a CDR from an immunoglobulin of a non-human
species (e.g.,
mouse, rat, rabbit, hamster, etc.) that have the desired specificity,
affinity, and/or binding capability
using methods known to one skilled in the art. In some embodiments, Fv
framework region amino
acid residues of a human immunoglobulin are replaced with corresponding amino
acid residues from
an antibody of a non-human species that has the desired specificity, affinity,
and/or binding capability.
In some embodiments, the humanized antibody can be further modified by the
substitution of
additional amino acid residues either in the Fv framework region and/or within
the replaced non-
human amino acid residues to refine and optimize antibody specificity,
affinity, and/or capability. In
general, the humanized antibody will comprise substantially all of at least
one, and typically two,
variable domain regions containing all, or substantially all, of the CDRs that
correspond to the non-
human immunoglobulin whereas all, or substantially all, of the framework
regions are those of a
human immunoglobulin sequence. In some embodiments, the humanized antibody can
also comprise
at least a portion of an immunoglobulin constant region or domain (Fc),
typically that of a human
immunoglobulin. In certain embodiments, such humanized antibodies are used
therapeutically
because they may reduce antigenicity and HAMA (human anti-mouse antibody)
responses when
administered to a human subject. Methods used to generate humanized antibodies
are well known in
the art.
[00223] In certain embodiments, the Wnt pathway inhibitor is a human antibody.
Human antibodies
can be directly prepared using various techniques known in the art. In some
embodiments,
immortalized human B lymphocytes immunized in vitro or isolated from an
immunized individual
that produces an antibody directed against a target antigen can be generated.
In some embodiments,
the human antibody can be selected from a phage library, where that phage
library expresses human
antibodies. Alternatively, phage display technology can be used to produce
human antibodies and
antibody fragments in vitro, from immunoglobulin variable domain gene
repertoires from
unimmunized donors. Techniques for the generation and use of antibody phage
libraries are well-
58
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
known in the art. Affinity maturation strategies including, but not limited
to, chain shuffling (Marks
et al., 1992, Bio/Technology, 10:779-783) and site-directed mutagenesis, are
known in the art and may
be employed to generate high affinity human antibodies.
[00224] In some embodiments, human antibodies can be made in transgenic mice
that contain human
immunoglobulin loci. These mice are capable, upon immunization, of producing
the full repertoire of
human antibodies in the absence of endogenous immunoglobulin production. This
approach is
described in U.S. Patent Nos. 5,545,807; 5,545,806; 5,569,825; 5,625,126;
5,633,425; and 5,661,016.
[00225] This invention also encompasses bispecific antibodies that
specifically recognize at least one
human FZD protein or at least one Wnt protein. Bispecific antibodies are
capable of specifically
recognizing and binding at least two different epitopes. The different
epitopes can either be within the
same molecule (e.g., two different epitopes on human FZD5) or on different
molecules (e.g., one
epitope on FZD5 and a different epitope on a second protein). In some
embodiments, the bispecific
antibodies are monoclonal human or humanized antibodies. In some embodiments,
the antibodies can
specifically recognize and bind a first antigen target, (e.g., a FZD protein)
as well as a second antigen
target, such as an effector molecule on a leukocyte (e.g., CD2, CD3, CD28,
CD80, or CD86) or a Fc
receptor (e.g., CD64, CD32, or CD16) so as to focus cellular defense
mechanisms to the cell
expressing the first antigen target. In some embodiments, the antibodies can
be used to direct
cytotoxic agents to cells which express a particular target antigen. These
antibodies possess an
antigen-binding arm and an arm which binds a cytotoxic agent or a radionuclide
chelator, such as
EOTUBE, DPTA, DOTA, or TETA.
[00226] Bispecific antibodies can be intact antibodies or antibody fragments.
Antibodies with more
than two valencies are also contemplated. For example, trispecific antibodies
can be prepared (Tutt et
al., 1991, J. Immunol., 147:60). Thus, in certain embodiments the antibodies
are multispecific.
Techniques for making bispecific and multispecific antibodies are known by
those skilled in the art.
[00227] In certain embodiments, the antibodies (or other polypeptides)
described herein may be
monospecific. For example, in certain embodiments, each of the one or more
antigen-binding sites
that an antibody contains is capable of binding (or binds) a homologous
epitope on different proteins.
In certain embodiments, an antigen-binding site of a monospecific antibody
described herein is
capable of binding (or binds), for example, FZD5 and FZD7 (i.e., the same
epitope is found on both
FZD5 and FZD7 proteins).
[00228] In certain embodiments, the Wnt pathway inhibitor is an antibody
fragment comprising an
antigen-binding site. Antibody fragments may have different functions or
capabilities than intact
antibodies; for example, antibody fragments can have increased tumor
penetration. Various
techniques are known for the production of antibody fragments including, but
not limited to,
proteolytic digestion of intact antibodies. In some embodiments, antibody
fragments include a F(ab')2
fragment produced by pepsin digestion of an antibody molecule. In some
embodiments, antibody
fragments include a Fab fragment generated by reducing the disulfide bridges
of an F(ab')2 fragment.
59
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
In other embodiments, antibody fragments include a Fab fragment generated by
the treatment of the
antibody molecule with papain and a reducing agent. In certain embodiments,
antibody fragments are
produced recombinantly. In some embodiments, antibody fragments include Fv or
single chain Fv
(scFv) fragments. Fab, Fv, and scFv antibody fragments can be expressed in and
secreted from E. coli
or other host cells, allowing for the production of large amounts of these
fragments. In some
embodiments, antibody fragments are isolated from antibody phage libraries as
discussed herein. For
example, methods can be used for the construction of Fab expression libraries
to allow rapid and
effective identification of monoclonal Fab fragments with the desired
specificity for a FZD or Wnt
protein or derivatives, fragments, analogs or homologs thereof In some
embodiments, antibody
fragments are linear antibody fragments. In certain embodiments, antibody
fragments are
monospecific or bispecific. In certain embodiments, the Wnt pathway inhibitor
is a scFv. Various
techniques can be used for the production of single-chain antibodies specific
to one or more human
FZD proteins or one or more human Wnt proteins.
[00229] It can further be desirable, especially in the case of antibody
fragments, to modify an antibody
in order to increase its serum half-life. This can be achieved, for example,
by incorporation of a
salvage receptor binding epitope into the antibody fragment by mutation of the
appropriate region in
the antibody fragment or by incorporating the epitope into a peptide tag that
is then fused to the
antibody fragment at either end or in the middle (e.g., by DNA or peptide
synthesis). In some
embodiments, an antibody is modified to decrease its serum half-life.
[00230] Heteroconjugate antibodies are also within the scope of the present
invention.
Heteroconjugate antibodies are composed of two covalently joined antibodies.
Such antibodies have,
for example, been proposed to target immune cells to unwanted cells. It is
also contemplated that the
heteroconjugate antibodies can be prepared in vitro using known methods in
synthetic protein
chemistry, including those involving crosslinking agents. For example,
immunotoxins can be
constructed using a disulfide exchange reaction or by forming a thioether
bond. Examples of suitable
reagents for this purpose include iminothiolate and methyl-4-
mercaptobutyrimidate.
[00231] For the purposes of the present invention, it should be appreciated
that modified antibodies
can comprise any type of variable region that provides for the association of
the antibody with the
target (i.e., a human FZD protein or a human Wnt protein). In this regard, the
variable region may
comprise or be derived from any type of mammal that can be induced to mount a
humoral response
and generate immunoglobulins against the desired tumor-associated antigen. As
such, the variable
region of the modified antibodies can be, for example, of human, murine, non-
human primate (e.g.
cynomolgus monkeys, macaques, etc.) or rabbit origin. In some embodiments,
both the variable and
constant regions of the modified immunoglobulins are human. In other
embodiments, the variable
regions of compatible antibodies (usually derived from a non-human source) can
be engineered or
specifically tailored to improve the binding properties or reduce the
immunogenicity of the molecule.
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
In this respect, variable regions useful in the present invention can be
humanized or otherwise altered
through the inclusion of imported amino acid sequences.
[00232] In certain embodiments, the variable domains in both the heavy and
light chains are altered by
at least partial replacement of one or more CDRs and, if necessary, by partial
framework region
replacement and sequence modification and/or alteration. Although the CDRs may
be derived from
an antibody of the same class or even subclass as the antibody from which the
framework regions are
derived, it is envisaged that the CDRs will be derived preferably from an
antibody from a different
species. It may not be necessary to replace all of the CDRs with all of the
CDRs from the donor
variable region to transfer the antigen binding capacity of one variable
domain to another. Rather, it
may only be necessary to transfer those residues that are necessary to
maintain the activity of the
antigen-binding site.
[00233] Alterations to the variable region notwithstanding, those skilled in
the art will appreciate that
the modified antibodies of this invention will comprise antibodies (e.g., full-
length antibodies or
immunoreactive fragments thereof) in which at least a fraction of one or more
of the constant region
domains has been deleted or otherwise altered so as to provide desired
biochemical characteristics
such as increased tumor localization and/or increased serum half-life when
compared with an
antibody of approximately the same immunogenicity comprising a native or
unaltered constant region.
In some embodiments, the constant region of the modified antibodies will
comprise a human constant
region. Modifications to the constant region compatible with this invention
comprise additions,
deletions or substitutions of one or more amino acids in one or more domains.
The modified
antibodies disclosed herein may comprise alterations or modifications to one
or more of the three
heavy chain constant domains (CH1, CH2 or CH3) and/or to the light chain
constant domain (CL). In
some embodiments, one or more domains are partially or entirely deleted from
the constant regions of
the modified antibodies. In some embodiments, the modified antibodies will
comprise domain
deleted constructs or variants wherein the entire CH2 domain has been removed
(ACH2 constructs).
In some embodiments, the omitted constant region domain is replaced by a short
amino acid spacer
(e.g., 10 amino acid residues) that provides some of the molecular flexibility
typically imparted by the
absent constant region.
[00234] In some embodiments, the modified antibodies are engineered to fuse
the CH3 domain
directly to the hinge region of the antibody. In other embodiments, a peptide
spacer is inserted
between the hinge region and the modified CH2 and/or CH3 domains. For example,
constructs may
be expressed wherein the CH2 domain has been deleted and the remaining CH3
domain (modified or
unmodified) is joined to the hinge region with a 5-20 amino acid spacer. Such
a spacer may be added
to ensure that the regulatory elements of the constant domain remain free and
accessible or that the
hinge region remains flexible. However, it should be noted that amino acid
spacers may, in some
cases, prove to be immunogenic and elicit an unwanted immune response against
the construct.
61
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
Accordingly, in certain embodiments, any spacer added to the construct will be
relatively non-
immunogenic so as to maintain the desired biological qualities of the modified
antibodies.
[00235] In some embodiments, the modified antibodies may have only a partial
deletion of a constant
domain or substitution of a few or even a single amino acid. For example, the
mutation of a single
amino acid in selected areas of the CH2 domain may be enough to substantially
reduce Fc binding and
thereby increase cancer cell localization and/or tumor penetration. Similarly,
it may be desirable to
simply delete the part of one or more constant region domains that control a
specific effector function
(e.g. complement Clq binding). Such partial deletions of the constant regions
may improve selected
characteristics of the antibody (serum half-life) while leaving other
desirable functions associated
with the subject constant region domain intact. Moreover, as alluded to above,
the constant regions of
the disclosed antibodies may be modified through the mutation or substitution
of one or more amino
acids that enhances the profile of the resulting construct. In this respect it
may be possible to disrupt
the activity provided by a conserved binding site (e.g., Fc binding) while
substantially maintaining the
configuration and immunogenic profile of the modified antibody. In certain
embodiments, the
modified antibodies comprise the addition of one or more amino acids to the
constant region to
enhance desirable characteristics such as decreasing or increasing effector
function or provide for
more cytotoxin or carbohydrate attachment sites.
[00236] It is known in the art that the constant region mediates several
effector functions. For
example, binding of the Cl component of complement to the Fc region of IgG or
IgM antibodies
(bound to antigen) activates the complement system. Activation of complement
is important in the
opsonization and lysis of cell pathogens. The activation of complement also
stimulates the
inflammatory response and can also be involved in autoimmune hypersensitivity.
In addition, the Fc
region of an antibody can bind a cell expressing a Fc receptor (FcR). There
are a number of Fc
receptors which are specific for different classes of antibody, including IgG
(gamma receptors), IgE
(epsilon receptors), IgA (alpha receptors) and IgM (mu receptors). Binding of
antibody to Fc
receptors on cell surfaces triggers a number of important and diverse
biological responses including
engulfment and destruction of antibody-coated particles, clearance of immune
complexes, lysis of
antibody-coated target cells by killer cells, release of inflammatory
mediators, placental transfer, and
control of immunoglobulin production.
[00237] In certain embodiments, the Wnt pathway inhibitors are antibodies that
provide for altered
effector functions. These altered effector functions may affect the biological
profile of the
administered antibody. For example, in some embodiments, the deletion or
inactivation (through
point mutations or other means) of a constant region domain may reduce Fc
receptor binding of the
circulating modified antibody (e.g., anti-FZD antibody) thereby increasing
cancer cell localization
and/or tumor penetration. In other embodiments, the constant region
modifications increase or reduce
the serum half-life of the antibody. In some embodiments, the constant region
is modified to
eliminate disulfide linkages or oligosaccharide moieties. Modifications to the
constant region in
62
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
accordance with this invention may easily be made using well known biochemical
or molecular
engineering techniques well within the purview of the skilled artisan.
[00238] In certain embodiments, a Wnt pathway inhibitor is an antibody does
not have one or more
effector functions. For instance, in some embodiments, the antibody has no
ADCC activity, and/or no
CDC activity. In certain embodiments, the antibody does not bind an Fc
receptor, and/or complement
factors. In certain embodiments, the antibody has no effector function.
[00239] The present invention further embraces variants and equivalents which
are substantially
homologous to the chimeric, humanized, and human antibodies, or antibody
fragments thereof, set
forth herein. These can contain, for example, conservative substitution
mutations, i.e. the substitution
of one or more amino acids by similar amino acids. For example, conservative
substitution refers to
the substitution of an amino acid with another within the same general class
such as, for example, one
acidic amino acid with another acidic amino acid, one basic amino acid with
another basic amino acid
or one neutral amino acid by another neutral amino acid. What is intended by a
conservative amino
acid substitution is well known in the art and described herein.
[00240] In certain embodiments, the antibodies described herein are isolated.
In certain embodiments,
the antibodies described herein are substantially pure.
[00241] In some embodiments of the present invention, the Wnt pathway
inhibitors are polypeptides.
The polypeptides can be recombinant polypeptides, natural polypeptides, or
synthetic polypeptides
comprising an antibody, or fragment thereof, that bind at least one human FZD
protein or at least one
Wnt protein. It will be recognized in the art that some amino acid sequences
of the invention can be
varied without significant effect on the structure or function of the protein.
Thus, the invention
further includes variations of the polypeptides which show substantial
activity or which include
regions of an antibody, or fragment thereof, against a human FZD protein or a
Wnt protein. In some
embodiments, amino acid sequence variations of FZD-binding polypeptides or Wnt-
binding
polypeptides include deletions, insertions, inversions, repeats, and/or other
types of substitutions.
[00242] The polypeptides, analogs and variants thereof, can be further
modified to contain additional
chemical moieties not normally part of the polypeptide. The derivatized
moieties can improve the
solubility, the biological half-life, and/or absorption of the polypeptide.
The moieties can also reduce
or eliminate any undesirable side effects of the polypeptides and variants. An
overview for chemical
moieties can be found in Remington: The Science and Practice of Pharmacy, 22st
Edition, 2012,
Pharmaceutical Press, London.
[00243] The isolated polypeptides that can be used in the methods described
herein can be produced
by any suitable method known in the art. Such methods range from direct
protein synthesis methods
to constructing a DNA sequence encoding polypeptide sequences and expressing
those sequences in a
suitable host. In some embodiments, a DNA sequence is constructed using
recombinant technology
by isolating or synthesizing a DNA sequence encoding a wild-type protein of
interest. Optionally, the
sequence can be mutagenized by site-specific mutagenesis to provide functional
analogs thereof
63
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
[00244] In some embodiments, a DNA sequence encoding a polypeptide of interest
may be
constructed by chemical synthesis using an oligonucleotide synthesizer.
Oligonucleotides can be
designed based on the amino acid sequence of the desired polypeptide and
selecting those codons that
are favored in the host cell in which the recombinant polypeptide of interest
will be produced.
Standard methods can be applied to synthesize a polynucleotide sequence
encoding an isolated
polypeptide of interest. For example, a complete amino acid sequence can be
used to construct a
back-translated gene. Further, a DNA oligomer containing a nucleotide sequence
coding for the
particular isolated polypeptide can be synthesized. For example, several small
oligonucleotides
coding for portions of the desired polypeptide can be synthesized and then
ligated. The individual
oligonucleotides typically contain 5' or 3' overhangs for complementary
assembly.
[00245] Once assembled (by synthesis, site-directed mutagenesis, or another
method), the
polynucleotide sequences encoding a particular polypeptide of interest can be
inserted into an
expression vector and operatively linked to an expression control sequence
appropriate for expression
of the protein in a desired host. Proper assembly can be confirmed by
nucleotide sequencing,
restriction enzyme mapping, and/or expression of a biologically active
polypeptide in a suitable host.
As is well-known in the art, in order to obtain high expression levels of a
transfected gene in a host,
the gene must be operatively linked to transcriptional and translational
expression control sequences
that are functional in the chosen expression host.
[00246] In certain embodiments, recombinant expression vectors are used to
amplify and express
DNA encoding binding agents (e.g., antibodies or soluble receptors), or
fragments thereof, against a
human FZD protein or a Wnt protein. For example, recombinant expression
vectors can be replicable
DNA constructs which have synthetic or cDNA-derived DNA fragments encoding a
polypeptide
chain of a FZD-binding agent, a Wnt-binding agent, an anti-FZD antibody or
fragment thereof, an
anti-Wnt antibody or fragment thereof, or a FZD-Fc soluble receptor
operatively linked to suitable
transcriptional and/or translational regulatory elements derived from
mammalian, microbial, viral or
insect genes. A transcriptional unit generally comprises an assembly of (1) a
genetic element or
elements having a regulatory role in gene expression, for example,
transcriptional promoters or
enhancers, (2) a structural or coding sequence which is transcribed into mRNA
and translated into
protein, and (3) appropriate transcription and translation initiation and
termination sequences.
Regulatory elements can include an operator sequence to control transcription.
The ability to replicate
in a host, usually conferred by an origin of replication, and a selection gene
to facilitate recognition of
transformants can additionally be incorporated. DNA regions are "operatively
linked" when they are
functionally related to each other. For example, DNA for a signal peptide
(secretory leader) is
operatively linked to DNA for a polypeptide if it is expressed as a precursor
which participates in the
secretion of the polypeptide; a promoter is operatively linked to a coding
sequence if it controls the
transcription of the sequence; or a ribosome binding site is operatively
linked to a coding sequence if
it is positioned so as to permit translation. In some embodiments, structural
elements intended for use
64
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
in yeast expression systems include a leader sequence enabling extracellular
secretion of translated
protein by a host cell. In other embodiments, where recombinant protein is
expressed without a leader
or transport sequence, it can include an N-terminal methionine residue. This
residue can optionally be
subsequently cleaved from the expressed recombinant protein to provide a final
product.
[00247] The choice of an expression control sequence and an expression vector
depends upon the
choice of host. A wide variety of expression host/vector combinations can be
employed. Useful
expression vectors for eukaryotic hosts include, for example, vectors
comprising expression control
sequences from SV40, bovine papilloma virus, adenovirus, and cytomegalovirus.
Useful expression
vectors for bacterial hosts include known bacterial plasmids, such as plasmids
from E. coli, including
pCR1, pBR322, pMB9 and their derivatives, and wider host range plasmids, such
as M13 and other
filamentous single-stranded DNA phages.
[00248] Suitable host cells for expression of a FZD-binding or Wnt-binding
agent (or a protein to use
as an antigen) include prokaryotes, yeast cells, insect cells, or higher
eukaryotic cells under the
control of appropriate promoters. Prokaryotes include gram-negative or gram-
positive organisms, for
example E. coli or Bacillus. Higher eukaryotic cells include established cell
lines of mammalian
origin as described below. Cell-free translation systems may also be employed.
Appropriate cloning
and expression vectors for use with bacterial, fungal, yeast, and mammalian
cellular hosts are well-
known in the art. Additional information regarding methods of protein
production, including antibody
production, can be found, e.g., in U.S. Patent Publication No. 2008/0187954,
U.S. Patent Nos.
6,413,746 and 6,660,501, and International Patent Publication No. WO
2004/009823.
[00249] Various mammalian culture systems are used to express recombinant
polypeptides.
Expression of recombinant proteins in mammalian cells may be preferred because
such proteins are
generally correctly folded, appropriately modified, and biologically
functional. Examples of suitable
mammalian host cell lines include COS-7 (monkey kidney-derived), L-929 (murine
fibroblast-
derived), C127 (murine mammary tumor-derived), 3T3 (murine fibroblast-
derived), CHO (Chinese
hamster ovary-derived), HeLa (human cervical cancer-derived), BHK (hamster
kidney fibroblast-
derived), HEK-293 (human embryonic kidney-derived) cell lines and variants
thereof Mammalian
expression vectors can comprise non-transcribed elements such as an origin of
replication, a suitable
promoter and enhancer linked to the gene to be expressed, and other 5' or 3'
flanking non-transcribed
sequences, and 5' or 3' non-translated sequences, such as necessary ribosome
binding sites, a
polyadenylation site, splice donor and acceptor sites, and transcriptional
termination sequences.
[00250] Expression of recombinant proteins in insect cell culture systems
(e.g., baculovirus) also
offers a robust method for producing correctly folded and biologically
functional proteins.
Baculovirus systems for production of heterologous proteins in insect cells
are well-known to those of
skill in the art (see, e.g., Luckow and Summers, 1988, Bio/Technology, 6:47).
[00251] Thus, the present invention provides cells comprising the FZD-binding
agents or the Wnt-
binding agents described herein. In some embodiments, the cells produce the
binding agents (e.g.,
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
antibodies or soluble receptors) described herein. In certain embodiments, the
cells produce an
antibody. In certain embodiments, the cells produce antibody OMP-18R5. In some
embodiments, the
cells produce a soluble receptor. In some embodiments, the cells produce a FZD-
Fc soluble receptor.
In some embodiments, the cells produce a FZD8-Fc soluble receptor. In some
embodiments, the cells
produce FZD8-Fc soluble receptor 54F28.
[00252] The proteins produced by a transformed host can be purified according
to any suitable
method. Standard methods include chromatography (e.g., ion exchange, affinity,
and sizing column
chromatography), centrifugation, differential solubility, or by any other
standard technique for protein
purification. Affinity tags such as hexa-histidine, maltose binding domain,
influenza coat sequence,
and glutathione-S-transferase can be attached to the protein to allow easy
purification by passage over
an appropriate affinity column. Isolated proteins can also be physically
characterized using such
techniques as proteolysis, mass spectrometry (MS), nuclear magnetic resonance
(NMR), high
performance liquid chromatography (HPLC), and x-ray crystallography.
[00253] In some embodiments, supernatants from expression systems which
secrete recombinant
protein into culture media can be first concentrated using a commercially
available protein
concentration filter, for example, an Amicon or Millipore Pellicon
ultrafiltration unit. Following the
concentration step, the concentrate can be applied to a suitable purification
matrix. In some
embodiments, an anion exchange resin can be employed, for example, a matrix or
substrate having
pendant diethylaminoethyl (DEAE) groups. The matrices can be acrylamide,
agarose, dextran,
cellulose, or other types commonly employed in protein purification. In some
embodiments, a cation
exchange step can be employed. Suitable cation exchangers include various
insoluble matrices
comprising sulfopropyl or carboxymethyl groups. In some embodiments, a
hydroxyapatite media can
be employed, including but not limited to, ceramic hydroxyapatite (CHT). In
certain embodiments,
one or more reverse-phase HPLC steps employing hydrophobic RP-HPLC media,
e.g., silica gel
having pendant methyl or other aliphatic groups, can be employed to further
purify a binding agent.
Some or all of the foregoing purification steps, in various combinations, can
also be employed to
provide a homogeneous recombinant protein.
[00254] In some embodiments, recombinant protein produced in bacterial culture
can be isolated, for
example, by initial extraction from cell pellets, followed by one or more
concentration, salting-out,
aqueous ion exchange, or size exclusion chromatography steps. HPLC can be
employed for final
purification steps. Microbial cells employed in expression of a recombinant
protein can be disrupted
by any convenient method, including freeze-thaw cycling, sonication,
mechanical disruption, or use of
cell lysing agents.
[00255] Methods known in the art for purifying antibodies and other proteins
also include, for
example, those described in U.S. Patent Publication Nos. 2008/0312425,
2008/0177048, and
2009/0187005.
66
CA 02931975 2016-05-27
WO 2015/084808
PCT/US2014/068097
[00256] In certain embodiments, the Wnt-binding agent or the FZD-binding agent
is a polypeptide
that is not an antibody. A variety of methods for identifying and producing
non-antibody
polypeptides that bind with high affinity to a protein target are known in the
art. See, e.g., Skerra,
2007, Curr. Opin. Biotechnol., 18:295-304; Hosse et al., 2006, Protein
Science, 15:14-27; Gill et al.,
2006, Curr. Opin. Biotechnol., 17:653-658; Nygren, 2008, FEBS J., 275:2668-76;
and Skerra, 2008,
FEBS J., 275:2677-83. In certain embodiments, phage display technology may be
used to produce
and/or identify a FZD-binding or Wnt-binding polypeptide. In certain
embodiments, the polypeptide
comprises a protein scaffold of a type selected from the group consisting of
protein A, protein G, a
lipocalin, a fibronectin domain, an ankyrin consensus repeat domain, and
thioredoxin.
[00257] In certain embodiments, the binding agents can be used in any one of a
number of conjugated
(i.e. an immunoconjugate or radioconjugate) or non-conjugated forms. In
certain embodiments,
antibodies can be used in a non-conjugated form to harness the subject's
natural defense mechanisms
including complement-dependent cytotoxicity and antibody dependent cellular
toxicity to eliminate
the malignant or cancer cells.
[00258] In some embodiments, the binding agent is conjugated to a cytotoxic
agent. In some
embodiments, the cytotoxic agent is a chemotherapeutic agent including, but
not limited to,
methotrexate, adriamicin, doxorubicin, melphalan, mitomycin C, chlorambucil,
daunorubicin or other
intercalating agents. In some embodiments, the cytotoxic agent is an
enzymatically active toxin of
bacterial, fungal, plant, or animal origin, or fragments thereof, including,
but not limited to, diphtheria
A chain, nonbinding active fragments of diphtheria toxin, exotoxin A chain,
ricin A chain, abrin A
chain, modeccin A chain, alpha-sarcin, Aleurites fordii proteins, dianthin
proteins, Phytolaca
americana proteins (PAPI, PAPII, and PAP-S), Momordica charantia inhibitor,
curcin, crotin,
Sapaonaria officinalis inhibitor, gelonin, mitogellin, restrictocin,
phenomycin, enomycin, and the
tricothecenes. In some embodiments, the cytotoxic agent is a radioisotope to
produce a
radioconjugate or a radioconjugated antibody. A variety of radionuclides are
available for the
production of radioconjugated antibodies including, but not limited to, 90Y,
1251, 1311, 1231, 1111n, 1311n,
105Rh, 1535m, 67ch, 67Ga, 166-Th,
ti 177LU, 186Re, 188Re and 212Bi. In some embodiments,
conjugates of an
antibody and one or more small molecule toxins, such as a calicheamicin,
maytansinoids, a
trichothene, and CC1065, and the derivatives of these toxins that have toxin
activity, can be produced.
In certain embodiments, conjugates of an antibody and a cytotoxic agent are
made using a variety of
bifunctional protein-coupling agents such as N-succinimidy1-3-(2-
pyridyidithiol) propionate (SPDP),
iminothiolane (IT), bifunctional derivatives of imidoesters (such as dimethyl
adipimidate HCL),
active esters (such as disuccinimidyl suberate), aldehydes (such as
glutareldehyde), bis-azido
compounds (such as bis(p-azidobenzoyl) hexanediamine), bis-diazonium
derivatives (such as bis-(p-
diazoniumbenzoy1)-ethylenediamine), diisocyanates (such as toluene 2,6-
diisocyanate), and bis-active
fluorine compounds (such as 1,5-difluoro-2,4-dinitrobenzene).
67
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
[00259] In certain embodiments, the Wnt pathway inhibitor (e.g., antibody or
soluble receptor) is an
antagonist of at least one Wnt protein (i.e., 1, 2, 3, 4, 5, 6, 7, 8, 9, or 10
Wnt proteins). In certain
embodiments, the Wnt pathway inhibitor inhibits activity of the Wnt protein(s)
to which it binds. In
certain embodiments, the Wnt pathway inhibitor inhibits at least about 10%, at
least about 20%, at
least about 30%, at least about 50%, at least about 75%, at least about 90%,
or about 100% of the
activity of the human Wnt protein(s) to which it binds.
[00260] In certain embodiments, the Wnt pathway inhibitor (e.g., antibody or
soluble receptor)
inhibits binding of at least one human Wnt to an appropriate receptor. In
certain embodiments, the
Wnt pathway inhibitor inhibits binding of at least one human Wnt protein to
one or more human FZD
proteins. In some embodiments, the at least one Wnt protein is selected from
the group consisting of:
Wntl, Wnt2, Wnt2b/13, Wnt3, Wnt3a, Wnt4, Wnt5a, Wnt5b, Wnt6, Wnt7a, Wnt7b,
Wnt8a, Wnt8b,
Wnt9a, Wnt9b, Wntl0a, Wntl0b, Wntl 1, and Wnt16. In some embodiments, the one
or more human
FZD proteins are selected from the group consisting of: FZD1, FZD2, FZD3,
FZD4, FZD5, FZD6,
FZD7, FZD8, FZD9, and FZD10. In certain embodiments, the Wnt pathway inhibitor
inhibits binding
of one or more Wnt proteins to FZD1, FZD2, FZD4, FZD5, FZD7, and/or FZD8. In
certain
embodiments, the Wnt pathway inhibitor inhibits binding of one or more Wnt
proteins to FZD8. In
certain embodiments, the inhibition of binding of a particular Wnt to a FZD
protein by a Wnt pathway
inhibitor is at least about 10%, at least about 25%, at least about 50%, at
least about 75%, at least
about 90%, or at least about 95%. In certain embodiments, an agent that
inhibits binding of a Wnt to
a FZD protein, also inhibits Wnt pathway signaling. In certain embodiments, a
Wnt pathway inhibitor
that inhibits human Wnt pathway signaling is an antibody. In certain
embodiments, a Wnt pathway
inhibitor that inhibits human Wnt pathway signaling is a FZD-Fc soluble
receptor. In certain
embodiments, a Wnt pathway inhibitor that inhibits human Wnt pathway signaling
is a FZD8-Fc
soluble receptor. In certain embodiments, a Wnt pathway inhibitor that
inhibits human Wnt pathway
signaling is soluble receptor 54F28.
[00261] In certain embodiments, the Wnt pathway inhibitors (e.g., antibody or
soluble receptor)
described herein are antagonists of at least one human Wnt protein and inhibit
Wnt activity. In certain
embodiments, the Wnt pathway inhibitor inhibits Wnt activity by at least about
10%, at least about
20%, at least about 30%, at least about 50%, at least about 75%, at least
about 90%, or about 100%.
In some embodiments, the Wnt pathway inhibitor inhibits activity of one, two,
three, four, five or
more Wnt proteins. In some embodiments, the Wnt pathway inhibitor inhibits
activity of at least one
human Wnt protein selected from the group consisting of: Wntl, Wnt2, Wnt2b,
Wnt3, Wnt3a, Wnt4,
Wnt5a, Wnt5b, Wnt6, Wnt7a, Wnt7b, Wnt8a, Wnt8b, Wnt9a, Wnt9b, Wntl Oa, Wntl
Ob, Wntl 1, and
Wntl 6. In some embodiments, the Wnt-binding agent binds at least one Wnt
protein selected from
the group consisting of Wntl, Wnt2, Wnt2b, Wnt3, Wnt3a, Wnt7a, Wnt7b, Wnt8a,
Wnt8b, Wntl Oa,
and Wntl Ob. In certain embodiments, the at least one Wnt protein is selected
from the group
consisting of Wntl, Wnt2, Wnt2b, Wnt3, Wnt3a, Wnt8a, Wnt8b, Wntl0a, and
Wntl0b. In certain
68
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
embodiments, a Wnt pathway inhibitor that inhibits human Wnt activity is an
antibody. In certain
embodiments, a Wnt pathway inhibitor that inhibits human Wnt activity is a FZD-
Fc soluble receptor.
In certain embodiments, a Wnt pathway inhibitor that inhibits human Wnt
activity is a FZD8-Fc
soluble receptor. In certain embodiments, a Wnt pathway inhibitor that
inhibits human Wnt activity is
soluble receptor 54F28.
[00262] In certain embodiments, the Wnt pathway inhibitor described herein is
an antagonist of at
least one human FZD protein and inhibits FZD activity. In certain embodiments,
the Wnt pathway
inhibitor inhibits FZD activity by at least about 10%, at least about 20%, at
least about 30%, at least
about 50%, at least about 75%, at least about 90%, or about 100%. In some
embodiments, the Wnt
pathway inhibitor inhibits activity of one, two, three, four, five or more FZD
proteins. In some
embodiments, the Wnt pathway inhibitor inhibits activity of at least one human
FZD protein selected
from the group consisting of: FZD1, FZD2, FZD3, FZD4, FZD5, FZD6, FZD7, FZD8,
FZD9, and
FZD10. In certain embodiments, the Wnt pathway inhibitor inhibits activity of
FZD1, FZD2, FZD4,
FZD5, FZD7, and/or FZD8. In certain embodiments, the Wnt pathway inhibitor
inhibits activity of
FZD8. In some embodiments, the Wnt pathway inhibitor is an anti-FZD antibody.
In certain
embodiments, the Wnt pathway inhibitor is anti-FZD antibody OMP-18R5.
[002631ln certain embodiments, the Wnt pathway inhibitor described herein is
an antagonist of at
least one human Wnt protein and inhibits Wnt signaling. In certain
embodiments, the Wnt pathway
inhibitor inhibits Wnt signaling by at least about 10%, at least about 20%, at
least about 30%, at least
about 50%, at least about 75%, at least about 90%, or about 100%. In some
embodiments, the Wnt
pathway inhibitor inhibits signaling by one, two, three, four, five or more
Wnt proteins. In some
embodiments, the Wnt pathway inhibitor inhibits signaling of at least one Wnt
protein selected from
the group consisting of Wntl, Wnt2, Wnt2b, Wnt3, Wnt3a, Wnt7a, Wnt7b, Wnt8a,
Wnt8b, Wntl Oa,
and Wntl Ob. In certain embodiments, a Wnt pathway inhibitor that inhibits Wnt
signaling is an
antibody. In certain embodiments, a Wnt pathway inhibitor that inhibits Wnt
signaling is a soluble
receptor. In certain embodiments, a Wnt pathway inhibitor that inhibits Wnt
signaling is a FZD-Fc
soluble receptor. In certain embodiments, a Wnt pathway inhibitor that
inhibits Wnt signaling is a
FZD8-Fc soluble receptor. In certain embodiments, a Wnt pathway inhibitor that
inhibits Wnt
signaling is soluble receptor 54F28.
[00264] In certain embodiments, a Wnt pathway inhibitor described herein is an
antagonist ofj3-
catenin signaling. In certain embodiments, the Wnt pathway inhibitor inhibits
P-catenin signaling by
at least about 10%, at least about 20%, at least about 30%, at least about
50%, at least about 75%, at
least about 90%, or about 100%. In certain embodiments, a Wnt pathway
inhibitor that inhibits (3-
catenin signaling is an antibody. In certain embodiments, a Wnt pathway
inhibitor that inhibits [3-
catenin signaling is an anti-FZD antibody. In certain embodiments, a Wnt
pathway inhibitor that
inhibits I3-catenin signaling is antibody OMP-18R5. In certain embodiments, a
Wnt pathway inhibitor
that inhibits I3-catenin signaling is a soluble receptor. In certain
embodiments, a Wnt pathway
69
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
inhibitor that inhibits f3-catenin signaling is a FZD-Fc soluble receptor. In
certain embodiments, a
Wnt pathway inhibitor that inhibits f3-catenin signaling is a FZD8-Fc soluble
receptor.
[00265] In certain embodiments, the Wnt pathway inhibitor described herein
inhibits binding of at
least one Wnt protein to a receptor. In certain embodiments, the Wnt pathway
inhibitor inhibits
binding of at least one human Wnt protein to one or more of its receptors. In
some embodiments, the
Wnt pathway inhibitor inhibits binding of at least one Wnt protein to at least
one FZD protein. In
some embodiments, the Wnt-binding agent inhibits binding of at least one Wnt
protein to FZD1,
FZD2, FZD3, FZD4, FDZ5, FDZ6, FDZ7, FDZ8, FDZ9, and/or FZD10. In certain
embodiments, the
inhibition of binding of at least one Wnt to at least one FZD protein is at
least about 10%, at least
about 25%, at least about 50%, at least about 75%, at least about 90%, or at
least about 95%. In
certain embodiments, a Wnt pathway inhibitor that inhibits binding of at least
one Wnt to at least one
FZD protein further inhibits Wnt pathway signaling and/or P-catenin signaling.
In certain
embodiments, a Wnt pathway inhibitor that inhibits binding of at least one
human Wnt to at least one
FZD protein is an antibody. In certain embodiments, a Wnt pathway inhibitor
that inhibits binding of
at least one human Wnt to at least one FZD protein is an anti-FZD antibody. In
certain embodiments,
a Wnt pathway inhibitor that inhibits binding of at least one human Wnt to at
least one FZD protein is
antibody OMP-18R5. In certain embodiments, a Wnt pathway inhibitor that
inhibits binding of at
least one human Wnt to at least one FZD protein is a soluble receptor. In
certain embodiments, a Wnt
pathway inhibitor that inhibits binding of at least one human Wnt to at least
one FZD protein is a
FZD-Fc soluble receptor. In certain embodiments, a Wnt pathway inhibitor that
inhibits binding of at
least one human Wnt to at least one FZD protein is a FZD8-Fc soluble receptor.
In certain
embodiments, a Wnt pathway inhibitor that inhibits binding of at least one
human Wnt to at least one
FZD protein is FZD8-Fc soluble receptor 54F28.
[00266] In certain embodiments, the Wnt pathway inhibitor described herein
blocks binding of at least
one Wnt to a receptor. In certain embodiments, the Wnt pathway inhibitor
blocks binding of at least
one human Wnt protein to one or more of its receptors. In some embodiments,
the Wnt pathway
inhibitor blocks binding of at least one Wnt to at least one FZD protein. In
some embodiments, the
Wnt pathway inhibitor blocks binding of at least one Wnt protein to FZD1,
FZD2, FZD3, FZD4,
FDZ5, FDZ6, FDZ7, FDZ8, FDZ9, and/or FZD10. In certain embodiments, the
blocking of binding
of at least one Wnt to at least one FZD protein is at least about 10%, at
least about 25%, at least about
50%, at least about 75%, at least about 90%, or at least about 95%. In certain
embodiments, a Wnt
pathway inhibitor that blocks binding of at least one Wnt protein to at least
one FZD protein further
inhibits Wnt pathway signaling and/or P-catenin signaling. In certain
embodiments, a Wnt pathway
inhibitor that blocks binding of at least one human Wnt to at least one FZD
protein is an antibody. In
certain embodiments, a Wnt pathway inhibitor that blocks binding of at least
one human Wnt to at
least one FZD protein is an anti-FZD antibody. In certain embodiments, a Wnt
pathway inhibitor that
blocks binding of at least one human Wnt to at least one FZD protein is
antibody OMP-18R5. In
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
certain embodiments, a Wnt pathway inhibitor that blocks binding of at least
one human Wnt to at
least one FZD protein is a soluble receptor. In certain embodiments, a Wnt
pathway inhibitor that
blocks binding of at least one human Wnt to at least one FZD protein is a FZD-
Fc soluble receptor. In
certain embodiments, a Wnt pathway inhibitor that blocks binding of at least
one human Wnt to at
least one FZD protein is a FZD8-Fc soluble receptor. In certain embodiments, a
Wnt pathway
inhibitor that blocks binding of at least one human Wnt to at least one FZD
protein is soluble receptor
54F28.
[00267] In certain embodiments, the Wnt pathway inhibitor described herein
inhibits Wnt pathway
signaling. It is understood that a Wnt pathway inhibitor that inhibits Wnt
pathway signaling may, in
certain embodiments, inhibit signaling by one or more receptors in the Wnt
signaling pathway but not
necessarily inhibit signaling by all receptors. In certain alternative
embodiments, Wnt pathway
signaling by all human receptors may be inhibited. In certain embodiments, Wnt
pathway signaling
by one or more receptors selected from the group consisting of FZD1, FZD2,
FZD3, FZD4, FDZ5,
FDZ6, FDZ7, FDZ8, FDZ9, and FZD10 is inhibited. In certain embodiments, the
inhibition of Wnt
pathway signaling by a Wnt pathway inhibitor is a reduction in the level of
Wnt pathway signaling of
at least about 10%, at least about 25%, at least about 50%, at least about
75%, at least about 90%, or
at least about 95%. In some embodiments, a Wnt pathway inhibitor that inhibits
Wnt pathway
signaling is an antibody. In some embodiments, a Wnt pathway inhibitor that
inhibits Wnt pathway
signaling is an anti-FZD antibody. In some embodiments, a Wnt pathway
inhibitor that inhibits Wnt
pathway signaling is antibody OMP-18R5. In some embodiments, a Wnt pathway
inhibitor that
inhibits Wnt pathway signaling is a soluble receptor. In some embodiments, a
Wnt pathway inhibitor
that inhibits Wnt pathway signaling is a FZD-Fc soluble receptor. In some
embodiments, a Wnt
pathway inhibitor that inhibits Wnt pathway signaling is a FZD8-Fc soluble
receptor. In some
embodiments, a Wnt pathway inhibitor that inhibits Wnt pathway signaling is
soluble receptor 54F28.
[00268] In certain embodiments, the Wnt pathway inhibitor described herein
inhibits activation of 13-
catenin. It is understood that a Wnt pathway inhibitor that inhibits
activation of f3-catenin may, in
certain embodiments, inhibit activation ofj3-catenin by one or more receptors,
but not necessarily
inhibit activation of [3-catenin by all receptors. In certain alternative
embodiments, activation of f3-
catenin by all human receptors may be inhibited. In certain embodiments,
activation of f3-catenin by
one or more receptors selected from the group consisting of FZD1, FZD2, FZD3,
FZD4, FDZ5,
FDZ6, FDZ7, FDZ8, FDZ9, and FZD10 is inhibited. In certain embodiments, the
inhibition of
activation of f3-catenin by a Wnt-binding agent is a reduction in the level of
activation of [3-catenin of
at least about 10%, at least about 25%, at least about 50%, at least about
75%, at least about 90%, or
at least about 95%. In some embodiments, a Wnt pathway inhibitor that inhibits
activation off3-
catenin is an antibody. In some embodiments, a Wnt pathway inhibitor that
inhibits activation of f3-
catenin is an anti-FZD antibody. In some embodiments, a Wnt pathway inhibitor
that inhibits
activation of13-catenin is antibody OMP-18R5. In some embodiments, a Wnt
pathway inhibitor that
71
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
inhibits activation ofj3-catenin is a soluble receptor. In some embodiments, a
Wnt pathway inhibitor
that inhibits activation of [3-catenin is a FZD-Fc soluble receptor. In some
embodiments, a Wnt
pathway inhibitor that inhibits activation ofj3-catenin is a FZD8-Fc soluble
receptor. In some
embodiments, a Wnt pathway inhibitor that inhibits activation of I3-catenin is
soluble receptor 54F28.
[00269] In vivo and in vitro assays for determining whether a Wnt pathway
inhibitor inhibits [3-catenin
signaling are known in the art. For example, cell-based, luciferase reporter
assays utilizing a
TCF/Luc reporter vector containing multiple copies of the TCF-binding domain
upstream of a firefly
luciferase reporter gene may be used to measure f3-catenin signaling levels in
vitro (Gazit et al., 1999,
Oncogene, 18; 5959-66; TOPflash, Millipore, Billerica MA). The level of f3-
catenin signaling in the
presence of one or more Wnt proteins (e.g., Wnt(s) expressed by transfected
cells or provided by Wnt-
conditioned media) in the presence of a binding agent is compared to the level
of signaling without
the binding agent present. In addition to the TCF/Luc reporter assay, the
effect of a binding agent (or
candidate agent) on I3-catenin signaling may be measured in vitro or in vivo
by measuring the effect of
the agent on the level of expression of I3-catenin-regulated genes, such as c-
myc (He et al., 1998,
Science, 281:1509-12), cyclin D1 (Tetsu et al., 1999, Nature, 398:422-6),
and/or fibronectin (Gradl et
al. 1999, Mol. Cell Biol., 19:5576-87). In certain embodiments, the effect of
a binding agent on 13-
catenin signaling may also be assessed by measuring the effect of the agent on
the phosphorylation
state of Dishevelled-1, Dishevelled-2, Dishevelled-3, LRP5, LRP6, and/or I3-
catenin.
[00270] In certain embodiments, a Wnt pathway inhibitor has one or more of the
following effects:
inhibit proliferation of tumor cells, inhibit tumor growth, reduce the
frequency of cancer stem cells in
a tumor, reduce the tumorigenicity of a tumor, reduce the tumorigenicity of a
tumor by reducing the
frequency of cancer stem cells in the tumor, trigger cell death of tumor
cells, induce cells in a tumor to
differentiate, differentiate tumorigenic cells to a non-tumorigenic state,
induce expression of
differentiation markers in the tumor cells, prevent metastasis of tumor cells,
or decrease survival of
tumor cells.
[00271] In certain embodiments, a Wnt pathway inhibitor is capable of
inhibiting tumor growth. In
certain embodiments, a Wnt pathway inhibitor is capable of inhibiting tumor
growth in vivo (e.g., in a
xenograft mouse model, and/or in a human having cancer). In some embodiments,
the tumor is a
tumor selected from the group consisting of colorectal tumor, colon tumor,
pancreatic tumor, lung
tumor, ovarian tumor, liver tumor, breast tumor, kidney tumor, prostate tumor,
gastrointestinal tumor,
melanoma, cervical tumor, bladder tumor, glioblastoma, and head and neck
tumor. In certain
embodiments, the tumor is melanoma. In certain embodiments, the tumor is a
colorectal tumor. In
certain embodiments, the tumor is a pancreatic tumor. In certain embodiments,
the tumor is a breast
tumor. In certain embodiments, the tumor is a Wnt-dependent tumor.
[00272] In certain embodiments, a Wnt pathway inhibitor is capable of reducing
the tumorigenicity of
a tumor. In certain embodiments, a Wnt pathway inhibitor is capable of
reducing the tumorigenicity
of a tumor comprising cancer stem cells in an animal model, such as a mouse
xenograft model. In
72
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
certain embodiments, the number or frequency of cancer stem cells in a tumor
is reduced by at least
about two-fold, about three-fold, about five-fold, about ten-fold, about 50-
fold, about 100-fold, or
about 1000-fold. In certain embodiments, the reduction in the number or
frequency of cancer stem
cells is determined by limiting dilution assay using an animal model.
Additional examples and
guidance regarding the use of limiting dilution assays to determine a
reduction in the number or
frequency of cancer stem cells in a tumor can be found, e.g., in International
Publication No. WO
2008/042236, and U.S. Patent Publication Nos. 2008/0064049 and 2008/0178305.
[00273] In certain embodiments, the Wnt pathway inhibitors described herein
are active in vivo for at
least 1 hour, at least about 2 hours, at least about 5 hours, at least about
10 hours, at least about 24
hours, at least about 2 days, at least about 3 days, at least about 1 week, or
at least about 2 weeks. In
certain embodiments, the Wnt pathway inhibitor is an IgG (e.g., IgG1 or IgG2)
antibody that is active
in vivo for at least 1 hour, at least about 2 hours, at least about 5 hours,
at least about 10 hours, at least
about 24 hours, at least about 2 days, at least about 3 days, at least about 1
week, or at least about 2
weeks. In certain embodiments, the Wnt pathway inhibitor is a fusion protein
that is active in vivo for
at least 1 hour, at least about 2 hours, at least about 5 hours, at least
about 10 hours, at least about 24
hours, at least about 2 days, at least about 3 days, at least about 1 week, or
at least about 2 weeks.
[00274] In certain embodiments, the Wnt pathway inhibitors described herein
have a circulating half-
life in mice, cynomolgus monkeys, or humans of at least about 5 hours, at
least about 10 hours, at
least about 24 hours, at least about 2 days, at least about 3 days, at least
about 1 week, or at least about
2 weeks. In certain embodiments, the Wnt pathway inhibitor is an IgG (e.g.,
IgG1 or IgG2) antibody
that has a circulating half-life in mice, cynomolgus monkeys, or humans of at
least about 5 hours, at
least about 10 hours, at least about 24 hours, at least about 2 days, at least
about 3 days, at least about
1 week, or at least about 2 weeks. In certain embodiments, the Wnt pathway
inhibitor is a fusion
protein that has a circulating half-life in mice, cynomolgus monkeys, or
humans of at least about 5
hours, at least about 10 hours, at least about 24 hours, at least about 2
days, at least about 3 days, at
least about 1 week, or at least about 2 weeks. Methods of increasing (or
decreasing) the half-life of
agents such as polypeptides and antibodies are known in the art. For example,
known methods of
increasing the circulating half-life of IgG antibodies include the
introduction of mutations in the Fc
region which increase the pH-dependent binding of the antibody to the neonatal
Fc receptor (FcRn) at
pH 6.0 (see, e.g., U.S. Patent Publication Nos. 2005/0276799, 2007/0148164,
and 2007/0122403).
Known methods of increasing the circulating half-life of antibody fragments
lacking the Fc region
include such techniques as PEGylation.
IV. Kits
[00275] Kits for practicing the methods of the invention are further provided.
By "kit" is intended any
manufacture (e.g., a package or a container) comprising at least one reagent,
e.g., an antibody, a
nucleic acid probe, etc. for specifically detecting the expression of at least
one biomarker of the
73
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
invention. The kit may be promoted, distributed, and/or sold as a unit for
performing the methods of
the present invention. Additionally, the kits may contain a package insert
describing the kit and
including instructional material for its use.
[00276] In some embodiments, a kit comprises reagents for practicing the
methods of the invention
using microarray technology. In some embodiments, a kit comprises reagents for
practicing the
methods of the invention using qPCR assays. Positive and/or negative controls
may be included in
the kits to validate the activity and correct usage of reagents employed in
accordance with the
invention. Controls may include samples known to be either positive or
negative for the presence of
the biomarker of interest, or other samples comprising the biomarkers of
interest. The design and use
of controls is standard and well within the routine capabilities of those in
the art.
[00277] In some embodiments, a kit comprises polynucleotides selected from the
group consisting of
SEQ ID NOs:62-79. In some embodiments, a kit comprises (a) a forward primer of
SEQ ID NO:62, a
reverse primer of SEQ ID NO:63, and a probe comprising SEQ ID NO:64; (b) a
forward primer of
SEQ ID NO:65, a reverse primer of SEQ ID NO:66, and a probe comprising SEQ ID
NO:67; (c) a
forward primer of SEQ ID NO:68, a reverse primer of SEQ ID NO:69, and a probe
comprising SEQ
ID NO:70; (d) a forward primer of SEQ ID NO:71, a reverse primer of SEQ ID
NO:72, and a probe
comprising SEQ ID NO:73; (e) a forward primer of SEQ ID NO:74, a reverse
primer of SEQ ID
NO:75, and a probe comprising SEQ ID NO:76; and (f) a forward primer of SEQ ID
NO:77, a reverse
primer of SEQ ID NO:78, and a probe comprising SEQ ID NO:79.
[00278] It will be further appreciated that any or all steps in the methods of
the invention could be
implemented by personnel or, alternatively, performed in an automated fashion.
Thus, the steps of
sample preparation, detection of biomarker expression, etc. may be automated.
[00279] Embodiments of the present disclosure can be further defined by
reference to the following
non-limiting examples, which describe in detail preparation of certain
antibodies of the present
disclosure and methods for using antibodies of the present disclosure. It will
be apparent to those
skilled in the art that many modifications, both to materials and methods, may
be practiced without
departing from the scope of the present disclosure.
EXAMPLES
Example 1
Identification of tumors responsive to treatment with a combination of OMP-
18R5 and taxol
[00280] The breast tumor xenograft models OMP-B34, OMP-B39, OMP-B44, OMP-B59,
OMP-B60,
UM-T01, UM-T03, and UM-PE13 were established at OncoMed Pharmaceuticals or the
University of
Michigan from minimally passaged, patient-derived tumor specimens. Six- to 8-
week-old
NOD/SCID mice were subcutaneously injected with 2-4 x 104 cells of OMP-B34,
OMP-B39, OMP-
B44, OMP-B59, OMP-B60, UM-T01, UM-T03, or UM-PE13 tumors. Tumors were allowed
to grow
74
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
until they reached an average volume of 100 to 150mm3. Tumor-bearing mice were
randomized into
four groups (n = 10 per group) and treated with control antibody 1B711
(15mg/kg), anti-FZD
antibody OMP-18R5 (15mg/kg), taxol (10mg/kg), or OMP-18R5 (15mg/kg) in
combination with
taxol (10mg/kg). Treatment with antibodies and/or taxol was administered on a
weekly basis. Tumor
growth was monitored and tumor volumes were measured with electronic calipers
at the indicated
time points. Data are expressed as mean S.E.M.
[00281] To determine if a tumor was responsive to anti-FZD antibody OMP-18R5,
single agent tumor
volume data was compared with the control while combination treatment with OMP-
18R5 and taxol
was compared with taxol as a single agent. For this study a "responder" tumor
was defined as a tumor
showing significantly greater tumor growth inhibition with the combination of
OMP-18R5 and taxol
as compared to tumor growth inhibition with taxol as single agent.
[00282] The results for each xenograft model are shown in Figures 1A-H. T-
tests were conducted at
each time point. Multiple comparisons used 2-way repeated measurement ANOVA
followed by
Bonferroni corrections. The t-tests and 2-way repeated measurement ANOVA were
performed using
GraphPad Prism5 (GraphPad Software Inc.). The tumors OMP-B59, OMP-B60, UM-T03,
and UM-
PE13 were shown to be responders, while tumors OMP-B34, OMP-B39, OMP-B44, and
UM-T01
were shown to be non-responders. The results are summarized in Table 1.
Table 1
Tumor Tumor Subtype Classification
OMP-B34 TNBC Non-Responder
OMP-B39 TNBC Non-Responder
OMP-B44 TNBC Non-Responder
OMP-B59 TNBC Responder
OMP-B60 TNBC Responder
UM-T01 TNBC Non-Responder
UM-T03 ER+PR+HER2+ Responder
UM-PE13 TNBC Responder
Example 2
Identification of predictive biomarkers
[00283] Microarray analyses were performed on untreated breast tumors OMP-B34,
OMP-B39, OMP-
B44 which did not respond to treatment with a combination of OMP-18R5 and
taxol, ("non-
responders"), and UM-T01 and untreated tumors OMP-B59, OMP-B60, UM-T03, and UM-
PE13
which did respond to treatment with a combination of OMP-18R5 and taxol
("responders"). RNA
was isolated from each tumor using a RNeasy Fibrous Tissue Mini Kit (Qiagen,
Valencia CA) with
DNAse treatment following the manufacturer's instructions. Samples were stored
at -80 C. RNA
CA 02931975 2016-05-27
WO 2015/084808
PCT/US2014/068097
was visualized on an Agilent 2100 Bioanalyzer and integrity was confirmed by
the presence of intact
28S and 18S ribosomal peaks. All RNA samples had 260/280 ratios > 1.8. Total
RNA isolated from
each tumor was amplified using the Ovation RNA Amplification System V2 (NuGEN,
San Carlos,
CA). Amplified, anti-sense single stranded-cDNA was fragmented and
biotinylated using the FL-
Ovation cDNA Biotin Module V2 (NuGEN). The quality of the cDNA and the
fragmented cDNA
was assessed by a spectrophotometer and a Bioanalyzer before hybridization to
the array. The
processed RNA was hybridized to Affymetrix HG-U133 plus 2.0 microarrays
(Affymetrix, Santa
Clara, CA) as outlined in the manufacturer's technical manuals. After
hybridization, the microarrays
were washed, scanned, and analyzed. Microarray data were processed to probe
set level data by using
GeneChip-RMA (Wu et al., 2004, J. Amer. Stat. Assn., 99:909-917). Probe sets
that were likely to
cross-hybridize with murine markers were removed. To summarize the data to
gene level and make
sure the probe set with the strongest signals were chosen, maximum expression
was used across all
probe sets mapping to one gene. Genes with low expression (< 5 on log2 scale)
or near-zero variance
(< 0.01) were removed. Genes were standardized to N(0,1) by subtracting the
log2 scale expression
from the mean and dividing by the standard deviation of each gene.
[00284] Analyses were performed using genes from several signaling pathways
including canonical,
planar cell polarity, Wnt/Ca+2, Wnt signaling negative regulation, cell fate,
tissue polarity, cell
growth and proliferation, cell migration, cell cycle, and cellular homeostasis
(see Table 2).
Table 2
Gene Symbol Protein Name
AES Amino-terminal enhancer of split
APC Adenomatous polyposis coli protein
AXIN1 Axin-1
BCL9 B-cell CLL/Iymphoma 9 protein
BTRC F-box/WD repeat-containing protein lA
CCND1 Gl/S-specific cyclin-Dl
CCND2 Gl/S-specific cyclin-D2
CCND3 Gl/S-specific cyclin-D3
CSNK1A1 Casein kinase I isoform alpha
CSNK1D Casein kinase I isoform delta
CSNK1G1 Casein kinase I isoform gamma-1
CSNK2A1 Casein kinase II subunit alpha
CTBP1 C-terminal-binding protein 1
CTBP2 C-terminal-binding protein 2
CTNNB 1 Catenin beta-1
CTNNBIP1 Beta-catenin-interacting protein 1
CXXC4 CXXC-type zinc finger protein 4
DAAM1 Disheveled-associated activator of morphogenesis 1
DIXDC1 Dixin
DKK1 Dickkopf-related protein 1
DVL1 Segment polarity protein disheveled homolog DVL-1
DVL2 Segment polarity protein disheveled homolog DVL-2
EP300 Histone acetyltransferase p300
FBXW11 F-box/WD repeat-containing protein 11
76
CA 02931975 2016-05-27
WO 2015/084808
PCT/US2014/068097
FBXW2 F-box/WD repeat-containing protein 2
FBXW4 F-box/WD repeat-containing protein 4
FGF4 Fibroblast growth factor 4
FOSL1 Fos-related antigen 1
FOXN1 Forkhead box protein N1
FRAT1 Proto-oncogene FRAT1
FRZB Secreted frizzled-related protein 3
FSHB Follitropin subunit beta
FZD1 Frizzled-1
FZD2 Frizzled-2
FZD3 Frizzled-3
FZD4 Frizzled-4
FZD5 Frizzled-5
FZD6 Frizzled-6
FZD7 Frizzled-7
FZD8 Frizzled-8
GSK3A Glycogen synthase kinase-3 alpha
GSK3B Glycogen synthase kinase-4 alpha
JUN Transcription factor AP-1
KREMEN1 Kremen protein 1
LEF1 Lymphoid enhancer-binding factor 1
LRP5 Low-density lipoprotein receptor-related protein 5
LRP6 Low-density lipoprotein receptor-related protein 6
MYC Myc proto-oncogene protein
NKD1 Protein naked cuticle homolog
NLK Serine/threonine-protein kinase NLK
PITX2 Pituitary homeobox 2
PORCN Protein-cysteine N-palmitoyl transferase porcupine
PPP2CA Serine/threonine-protein phosphatase 2A catalytic subunit alpha
isoform
PPP2R1A Serine/threonine-protein phosphatase 2A 65 kDa regulatory
subunit A alpha
isoform
PYGO1 Pygopus homolog 1
RHOU Rho-related GTP-binding protein RhoU
SENP2 Sentrin-specific protease 2
SFRP1 Secreted frizzled-related protein 1
SFRP4 Secreted frizzled-related protein 4
SLC9A3R1 Na(+)/H(+) exchange regulatory cofactor NHE-RF1
50X17 Transcription factor SOX-17
T Brachyury protein
TCF7 Transcription factor 7
TCF7L1 Transcription factor 7-like 1
TLE1 Transducin-like enhancer protein 1
TLE2 Transducin-like enhancer protein 2
WIF1 Wnt inhibitory factor 1
WISP1 WNT1-inducible signaling pathway protein 1
WNT1 Proto-oncogene Wnt-1
WNT2 Protein Wnt-2
WNT2B Protein Wnt-2B
WNT3 Protein Wnt-3
WNT3A Protein Wnt-3a
WNT4 Protein Wnt-4
WNT5A Protein Wnt-5a
WNT5B Protein Wnt-5b
77
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
WNT6 Protein Wnt-6
WNT7A Protein Wnt-7a
WNT7B Protein Wnt-7b
WNT8A Protein Wnt-8a
WNT9A Protein Wnt-9a
WNT10A Protein Wnt-10 a
WNT11 Protein Wnt-11
WNT16 Protein Wnt-16
[00285] Support Vector Machines - Recursive Feature Elimination (SVM-RFE)
methods (Guyon et al,
2002, Machine Learning, 46:389-422) were used to identify genes that could
distinguish between the
responder and non-responder tumors and Support Vector Machine (SVM) methods
(Cortes and
Vapnik, 1995, Machine Learning, 20:273-297) were used for classification. A
leave-one-out cross-
validation (LOOCV) method was used to select the number of genes and also to
measure positive
predictive value (PPV), negative predictive value (NPV), sensitivity, and
specificity of the models. A
biomarker signature comprising FBXW2, CCND2, RHOU, CTBP2, WIF1, and DKK1
achieved the
best performance with PPV=NPV=sensitivity=specificity=100% using the 8 breast
tumors (see Figure
2). As shown in Figure 3, principal component analysis (PCA) illustrated that
the 6-gene biomarker
signature resulted in a near perfect separation of the 8 breast tumors. In
addition, strong correlation
was observed between the 6-gene biomarker signature and the ratio of tumor
volume (RTV) from the
in vivo experiments described in Example 1 (correlation = 0.95, p-value =
0.0003; cross-validated
correlation = 0.89, p-value = 0.00027; Figure 4).
[00286] Decision values were determined from the SVM model based on the
training data. For the 6-
gene biomarker signature, decision values can be calculated by a weighted sum
of the standardized
expression of the 6 genes: 0.4560427*FBXW2 + 0.3378467*CCND2 ¨ 0.4809354*RHOU
+
0.409029*CTBP2 + 0.3291529*WIF1 + 02926374*DKK1 + 0.04662682. A positive
decision value
indicated a tumor predicted to be a responder while a negative decision value
indicated a tumor
predicted to be a non-responder. In addition, classification probabilities can
be obtained by fitting a
logistic regression on the decision values. Tumors associated with
probabilities higher than 0.5 would
be predicted to be a responder while tumors with probabilities lower than 0.5
would be predicted to be
a non-responder.
Example 3
In vivo validation of predictive biomarkers
[00287] Six additional breast cancer tumors were selected from the OncoMed
Tumor Bank and
microarray analyses were performed as described in Example 1. The six breast
cancer tumors were
OMP-B29, OMP-B71, OMP-B84, OMP-B90, UM-T02, and UM-T06. As described herein,
classification probability analysis was used with the 6-gene biomarker
signature to predict the
response of each of these tumors to treatment with anti-FZD antibody OMP-18R5
in combination
78
CA 02931975 2016-05-27
WO 2015/084808
PCT/US2014/068097
with taxol (see Figure 5). In parallel the six tumors were evaluated in in
vivo xenograft models as
described in Example 1 (see Figures 6A-F). As described in Example 1 a
"responder" in the in vivo
models is a tumor showing significantly greater tumor growth inhibition with
the combination of
OMP-18R5 and taxol as compared to tumor growth inhibition with taxol as single
agent. The
predictions based on classification probabilities were compared to the results
of the in vivo xenograft
models. The results are summarized in Table 3.
Table 3
Classification DecisionIn vivo
Tumor Tumor subtype Prediction
Probability Value
Response
OMP-B29 ER+PR+HER2- 0.3344 -0.5928 Non- Non-
responder
responder
OMP-B71 ER+PR+HER2- 0.9897 1.6789 Responder Responder
OMP-B84 ER+PR+HER2- 0.4324 -0.4002 Non- Non-
responder
responder
OMP-B90 TNBC 0.8152 0.492 Responder
Responder
UM-T02 TNBC 0.4387 -0.3972 Non- Non-
responder
responder
UM-T06 ER+PR+HER2- 0.1385 -1.0778 Non- Non-
responder
responder
[00288] As shown in Table 3, the response of each of the six breast cancer
tumors was accurately
predicted by the 6-gene biomarker signature using the decision values and the
classification
probabilities.
Example 4
Prevalence Estimation of the 6-gene biomarker signature
[00289] Prevalence of a biomarker signature can be defined as the proportion
of a population
predicted to be a responder based upon the biomarker signature. The prevalence
of the 6-gene
biomarker signature in HER2 negative (HER2-) and triple negative breast cancer
(TNBC) populations
was estimated by applying the 6-gene biomarker signature to three publicly
available breast cancer
microarray data sets. The Cremoux2001 dataset was compiled from Affymetrix
U133p1us2
microarrays with 226 patients, including 145 HER2- and 81 HER2+, where 51 TNBC
were included
within the HER2- group. The Wang2011 dataset was compiled from Affymetrix
U133plus2
microarrays with 115 patients, including 79 HER2- and 36 HER2+, where 28 TNBC
were included
within the HER2- group. The Prat2010 dataset was compiled from Agilent Human
lA microarrays
with 333 patients, including 215 HER2- and 118 HER2+, where 57 TNBC were
included within the
HER2- group. Pre-processing of the public data included downloading the data,
extracting the probe
79
CA 02931975 2016-05-27
WO 2015/084808 PCT/US2014/068097
sets mapping to the six genes, and collapsing the probe sets to the six genes.
Gene level expression
data was further standardized by subtracting the mean and dividing by the
standard deviation of each
gene in the public data. The SVM model built upon the training data was used
to classify the public
data. Classification probabilities were obtained and the proportion of
predicted responders
(probability > 0.5) was calculated based on the 6-gene biomarker signature.
[00290] As shown in Figure 7, the predicted prevalence of the 6-gene biomarker
signature within the 3
datasets was very similar (approximately 60%). This prediction would suggest
that there is a large
population of breast cancer patients that would be responsive to therapy with
the anti-FZD antibody
OMP-18R5 in combination with taxol.
Example 5
qPCR assays for 6-gene biomarker signature
[00291] qPCR assays were developed to determine the expression levels of
FBXW2, CCND2, RHOU,
CTBP2, WIF1, and DKK1 in a tumor sample. Primers and probes were designed
using publicly
available mRNA sequences. The primers and probes were generated and used in
optimization and
validation tests using human fresh frozen (FF) and formalin-fixed paraffin-
embedded (FFPE) human
tissue samples. The specific primers and probes are listed in Table 4 (all
sequences in 5' to 3'
direction). Four reference genes were used for normalization including TOP1
(topoisomerase 1),
GUSB (beta-glucuronidase), SDHA (succinate dehydrogenase), and PUM1 (pumilio
homolog 1).
Table 4
Gene Primer/Probe Sequence SEQ ID NO
Forward Primer GCTGTCTCTGATCCGCAAGC SEQ ID NO:62
CCND2 Reverse Primer GACGGTGGGTACATGGCAAAC SEQ ID NO:63
Probe CCTTCATTGCTCTGTGTGCCACCGAC SEQ ID NO:64
Forward Primer ATCCGTGGGGAGACGCTG SEQ ID NO:65
CTBP2 Reverse Primer CTCGAACTGCAACCGCCTG SEQ ID NO:66
Probe CCCGTGCGACCAAAGCCAATGAGG SEQ ID NO:67
Forward Primer GACCATTGACAACTACCAGCCGTA SEQ ID NO:68
DKK1 Reverse Primer TGGGACTAGCGCAGTACTCATC SEQ ID NO:69
Probe TGCCGCACTCCTCGTCCTCTG SEQ ID NO:70
Forward Primer GCCAGTTATGATATTCTCAGGGTCA SEQ ID NO:71
FBXW2 Reverse Primer AGCAGGGCAAAGATATCTCCAAA SEQ ID NO:72
Probe AGACTCCTGAGATAGCAAACTTGGCCT SEQ ID NO:73
Forward Primer CCCACCGAGTACATCCCTACTG SEQ ID NO:74
RHOU1 Reverse Primer CAGTGTCACAGAGTTGGAGTCTCA SEQ ID NO:75
Probe CGCCCATCCACAGACACCACCG SEQ ID NO:76
WIF1 Forward Primer GTTCCAAAGGTTACCAGGGAGAC SEQ ID NO:77
CA 02931975 2016-05-27
WO 2015/084808
PCT/US2014/068097
Reverse Primer GT TGGGT TCATGGCAGGT TCC SEQ ID NO:78
Probe CCAGGCTCGCAGACAGGCTTTGAAC
SEQ ID NO:79
[00292] qPCR was performed on total RNA obtained from 18 xenograft breast
tumors. Tumor
specimens were harvested and immediately snap frozen and stored at -80 C prior
to RNA isolation.
Total RNA was extracted using the RNeasy Fibrous Mini Kit (Qiagen, Valencia
CA, PN#74704) with
TissueLyzer homogenization and DNase I treatment according to the
manufacturer's protocol. RNAs
were visualized on a Bioanalyzer 2100 (Agilent, Santa Clara, CA) and verified
to be intact with RIN
values > 6Ø All RNAs had A260/A280 ratios > 1.8.
[00293] qPCR was performed in a two-step manner. First, cDNA was synthesized
from total RNA
using random hexamers as described in Applied Biosystems User Bulletin 2.
TaqMan Universal PCR
Master Mix (Applied Biosystems, Foster City, CA. Cat # 4304437 and 4326708)
was used in
subsequent qPCR reactions according to the manufacturer's protocol. Quantities
of gene expression
were determined using a Ct (cycle threshold) method from triplicate reactions.
Cycle threshold is
generally considered to be the number of cycles required for a signal to cross
the detection threshold.
Ct levels are inversely proportional to the amount of target nucleic acid in a
sample. Ct of the six
genes are normalized using the Ct levels of the four reference genes.
Normalized Ct of the 6-gene
signature for the 18 xenograft samples is shown in Table 5.
Table 5
FBXW2 CCND2 RHOU CTBP2 WIF1 DKK1
OMP-B84 0.8425 12.5125 4.6775
1.0775 16.4025 5.2575
OMP-B71 0.98375 14.52375 6.46875
0.08875 4.56875 1.14375
OMP-B59 0.83875 2.67375 6.43875 -
0.6012 4.90375 10.7888
OMP-B86 2.4725 11.5125 2.5425
1.3275 -0.8825 1.1125
OMP-B39 1.03 12.54 1.44 2.225 2.045 6.365
OMP-B90
1.175 6.955 6.87 1.535 17.535 10.87
pl
OMP-B94 1.67375 1.52875 5.95375
1.56875 9.34875 4.37875
OMP-B40 1.03 16.455 6.775 0.73 16.455
14.985
OMP-B29 1.445 13.63 6.425 0.695 13.63 4.185
OMP-B60 1.6725 14.7775 6.9825
0.4775 -0.8025 8.6775
OMP-B90
0.75875 14.18375 5.78875 0.13875 15.54375
10.7388
p2
UM-T06 1.19875 11.51875 4.27875
1.34375 8.87375 6.26375
OMP-B44 1.61 11.765 4.755 0.505 7.225 9.61
UM-T02 2.255 13.215 4.195 1.075 16.125 4.225
UM-T3 1.67625 12.58625 5.83625
0.20125 16.21625 4.17125
OMP-B34 0.08625 0.58625 6.21125
0.53125 9.02125 0.06125
81
CA 02931975 2016-05-27
WO 2015/084808
PCT/US2014/068097
UM-PE13 0.925 15.185 4.055 -0.7 15.185 6.62
UM-T01 2.20375 15.11375 6.44375 -0.1062 15.11375 15.1138
[00294] Decision values can be calculated by a weighted sum of the normalized
expression of the 6
genes from data generated from the qPCR assays. These decision values are
different than the
decision values generated from the analysis based on microarray data, however
the predictive
capabilities of the two models are very similar.
[00295] It is understood that the examples and embodiments described herein
are for illustrative
purposes only and that various modifications or changes in light thereof will
be suggested to persons
skilled in the art and are to be included within the spirit and purview of
this application.
[00296] All publications, patents, and patent applications cited herein are
hereby incorporated by
reference in their entirety for all purposes to the same extent as if each
individual publication, patent
or patent application were specifically and individually indicated to be so
incorporated by reference.
[00297] Following are the sequences disclosed in the application:
OMP-18R5 Heavy chain CDR1 (SEQ ID NO:1)
GFTFSHYTLS
OMP-18R5 Heavy chain CDR2 (SEQ ID NO:2)
VISGDGSYTYYADSVKG
OMP-18R5 Heavy chain CDR3 (SEQ ID NO:3)
NFIKYVFAN
OMP-18R5 Light chain CDR1 (SEQ ID NO:4)
SGDNIGSFYVH
OMP-18R5 Light chain CDR2 (SEQ ID NO:5)
DKSNRPSG
OMP-18R5 Light chain CDR3 (SEQ ID NO:6)
QSYANTLSL
OMP-18R5 Heavy chain variable region amino acid sequence (SEQ ID NO:7)
EVQLVESGGGLVQPGGSLRLSCAASGFTFSHYTLSWVRQAPGKGLEWVSVISGDGSYTYY
ADSVKGRFTISSDNSKNTLYLQMNSLRAEDTAVYYCARNFIKYVFANWGQGTLVTVSS
OMP-18R5 Light chain variable region amino acid sequence (SEQ ID NO:8)
DIELTQPPSVSVAPGQTARISCSGDNIGSFYVHWYQQKPGQAPVLVIYDKSNRPSGIPER
FSGSNSGNTATLTISGTQAEDEADYYCQSYANTLSLVFGGGTKLTVLG
OMP-18R5 Heavy chain amino acid sequence with predicted signal sequence
underlined (SEQ ID
NO:9)
MKHLWFFLLLVAAPRWVLSEVQLVESGGGLVQPGGSLRLSCAASGFTFSHYTLSWVRQAP
GKGLEWVSVISGDGSYTYYADSVKGRFTISSDNSKNTLYLQMNSLRAEDTAVYYCARNFI
KYVFANWGQGTLVTVSSASTKGPSVFPLAPCSRSTSESTAALGCLVKDYFPEPVTVSWNS
82
CA 02931975 2016-05-27
WO 2015/084808
PCT/US2014/068097
GALT S GVHT FPAVLQS SGLYSLSSVVTVPSSNFGTQTYTCNVDHKPSNTKVDKTVERKCC
VECPPCPAPPVAGPSVFLFPPKPKDTLMI SRT PEVTCVVVDVS HE DPEVQ FNWYVDGVEV
HNAKTKPREEQFNS T FRVVSVLTVVHQDWLNGKEYKCKVSNKGLPAP IEKT I SKTKGQPR
E PQVYTL PPSREEMTKNQVS LT CLVKGFYPS DIAVEWESNGQPENNYKTTPPMLDS DGS F
FLYSKLTVDKSRWQQGNVFS CSVMHEALHNHYTQKSL S LS PGK
OMP-18R5 Light chain amino acid sequence with predicted signal sequence
underlined (SEQ ID
NO:10)
MAWALLLLTLLTQGTGSWADIELTQPPSVSVAPGQTARI S CS GDNI GS FYVHWYQQKPGQ
APVLVIY DKSNRPS GI PERFSGSNSGNTATLT I SGTQAEDEADYYCQSYANTLSLVFGGG
TKLTVLGQPKAAPSVTLFPPSSEELQANKATLVCL I S DFYPGAVTVAWKADS SPVKAGVE
TT T PSKQ SNNKYAAS S YL SLT PEQWKS HRSY S CQVTHEGS TVEKTVAPTECS
OMP-18R5 Heavy chain amino acid sequence without predicted signal sequence
(SEQ ID NO:11)
EVQLVES GGGLVQPGGSLRL SCAAS GFT FSHYTLSWVRQAPGKGLEWVSVI SGDGSYTYY
ADSVKGRFT I S S DNSKNT LYLQMNS LRAE DTAVYYCARNFI KYVFANWGQGT LVTVS SAS
TKGPSVFPLAPCSRST SE S TAALGCLVKDYFPEPVTVSWNS GALT S GVHT FPAVLQ S SGL
YS LS SVVTVPS SNFGTQTYTCNVDHKPSNTKVDKTVERKCCVECPPCPAPPVAGPSVFLF
PPKPKDTLMI S RT PEVTCVVVDVS HE DPEVQ FNWYVDGVEVHNAKTKPREEQ FNS TFRVV
SVLTVVHQDWLNGKEYKCKVSNKGL PAP I EKT I SKTKGQPREPQVYTLPPSREEMTKNQV
SLTCLVKGFYP S DIAVEWESNGQPENNYKTT P PML DS DGSFFLYSKLTVDKSRWQQGNVF
SCSVMHEALHNHYTQKSLSLSPGK
OMP-18R5 Light chain amino acid sequence without predicted signal sequence
(SEQ ID NO:12)
DI ELTQP PSVSVAPGQTARI SCSGDNI GS FYVHWYQQKPGQAPVLVI YDKSNRPS GI PER
FS GSNSGNTAT LT I SGTQAEDEADYYCQSYANTLSLVFGGGTKLTVLGQPKAAPSVTLFP
PS SEELQANKATLVCL I S DFYPGAVTVAWKADS S PVKAGVE TT T PSKQSNNKYAAS SYLS
LT PEQWKSHRS YSCQVTHEGS TVEKTVAPTECS
Human FZD1 Fri domain amino acid sequence without predicted signal sequence
(SEQ ID NO:13)
QQPPPPPQQQQSGQQYNGERGI SVP DHGYCQP I S I PLCT DIAYNQT IMPNLLGHTNQEDA
GLEVHQFYPLVKVQCSAELKFFLCSMYAPVCTVLEQALPPCRSLCERARQGCEALMNKFG
FQWPDTLKCEKFPVHGAGELCVGQNTS DKGT
Human FZD2 Fri domain amino acid sequence without predicted signal sequence
(SEQ ID NO:14)
QFHGEKGI S I P DHGFCQP I S I PLCT DIAYNQT IMPNLLGHTNQEDAGLEVHQFYPLVKVQ
CS PELRFFLCSMYAPVCTVLEQAIPPCRS I CERARQGCEALMNKFGFQWPERLRCEHFPR
HGAEQ I CVGQNHSE DG
Human FZD3 Fri domain amino acid sequence without predicted signal sequence
(SEQ ID NO:15)
HS LFS CE P I TLRMCQDLPYNTTFMPNLLNHYDQQTAALAMEPFHPMVNLDCSRDF
RP FLCALYAP I CMEYGRVTLPCRRLCQRAYSECSKLMEMFGVPWPEDMECSRFPDCDEPY
PRLVDL
Human FZD4 Fri domain amino acid sequence without predicted signal sequence
(SEQ ID NO:16)
FGDEEERRCDPIRI SMCQNLGYNVTKMPNLVGHELQT DAELQLTT FT PL I QYGCS SQLQF
FLCSVYVPMCTEKINI PI GPCGGMCLSVKRRCEPVLKEFGFAWPESLNCSKFPPQNDHNH
MCMEGPGDEEV
Human FZD5 Fri domain amino acid sequence without predicted signal sequence
(SEQ ID NO:17)
AS KAPVCQE I TVPMCRGI GYNLTHMPNQFNH DTQDEAGLEVHQ FWPLVE I QC S PDLRFFL
CSMYT P I CLPDYHKPLPPCRSVCERAKAGCS PLMRQYGFAWPERMSCDRLPVLGRDAEVL
CMDYNRSEATT
Human FZD6 Fri domain amino acid sequence without predicted signal sequence
(SEQ ID NO:18)
83
CA 02931975 2016-05-27
WO 2015/084808
PCT/US2014/068097
H S LFT CE P I TVPRCMKMAYNMT FFPNLMGHYDQS I AAVEME HFLPLANLE CS PNIETFLC
KAFVPTC IEQ I HVVPPCRKLCEKVYSDCKKL I DT FGI RWPEELECDRLQYCDETVPVT FD
PHTEFLG
Human FZD7 Fri domain amino acid sequence without predicted signal sequence
(SEQ ID NO:19)
QPYHGEKGI SVPDHGFCQP I S I PLCTDIAYNQT I L PNLLGHTNQEDAGLEVHQFY PLVKV
QCSPELRFFLCSMYAPVCTVLDQAI PPCRSLCERARQGCEALMNKFGFQWPERLRCENFP
VHGAGE I CVGQNTS DGSG
Human FZD8 Fri domain amino acid sequence without predicted signal sequence
(SEQ ID NO:20)
ASAKELACQE I TVPLCKGI GYNYTYMPNQFNHDTQDEAGLEVHQFWPLVE IQCSPDLKFF
LCSMYTP I CLE DYKKPLPPCRSVCERAKAGCAPLMRQYGFAWP DRMRCDRLPEQGNPDT L
CMDYNRT DLTT
Human FZD9 Fri domain amino acid sequence without predicted signal sequence
(SEQ ID NO:21)
LE I GRFDPERGRGAAPCQAVE I PMCRGIGYNLTRMPNLLGHTSQGEAAAELAEFAPLVQY
GCHSHLRFFLCSLYAPMCT DQVSTP I PACRPMCEQARLRCAPIMEQFNFGWPDSLDCARL
PT RNDPHALCMEAPENA
Human FZD10 Fri domain amino acid sequence without predicted signal sequence
(SEQ ID NO:22)
I S SMDMERPGDGKCQP IE I PMCKDI GYNMTRMPNLMGHENQREAAIQLHEFAPLVEYGCH
GHLRFFLCSLYAPMCTEQVS TP I PACRVMCEQARLKCS PIMEQFNFKWPDSLDCRKLPNK
NDPNYLCMEAPNNG
Human FZD1 amino acids 116-227 (SEQ ID NO:23)
CQ PIS I PLCT DIAYNQT IMPNLLGHTNQEDAGLEVHQFYPLVKVQCSAELKFFLCSMYAP
VCTVLEQALP PCRS LCERARQGCEALMNKFGFQWP DT LKCEKFPVHGAGE LC
Human FZD2 amino acids 39-150 (SEQ ID NO:24)
CQ PIS I PLCT DIAYNQT IMPNLLGHTNQEDAGLEVHQFYPLVKVQCS PELRFFLCSMYAP
VCTVLEQAI PPCRS I CERARQGCEALMNKFGFQWPERLRCE HFPRHGAEQ I C
Human FZD3 amino acids 28-133 (SEQ ID NO:25)
CE P I T LRMCQ DLPYNT T FMPNLLNHYDQQTAALAME P FHPMVNLDC S RDFRP FLCALYAP
I CMEYGRVTLPCRRLCQRAYSECSKLMEMFGVPWPEDMECSRFPDC
Human FZD4 amino acids 48-161 (SEQ ID NO:26)
CDPIRI SMCQNLGYNVTKMPNLVGHELQT DAELQLTT FT PL IQYGCS SQLQFFLCSVYVP
MCTEKINI PI GPCGGMCLSVKRRCEPVLKEFGFAWPESLNCSKFPPQNDHNHMC
Human FZD5 amino acids 33-147 (SEQ ID NO:27)
CQE I TVPMCRG I GYNLTHMPNQ FNH DTQDEAGLEVHQ FWPLVE I QC S PDLRFFLCSMYT P
I CLPDYHKPLPPCRSVCERAKAGCS PLMRQYGFAWPERMSCDRLPVLGRDAEVLC
Human FZD6 amino acids 24-129 (SEQ ID NO:28)
CE P I TVPRCMKMAYNMT FFPNLMGHYDQS IAAVEMEHFLPLANLECS PNI ET FLCKAFVP
T C IEQ I HVVP PCRKLCEKVY S DCKKL I DT FGIRWPEELECDRLQYC
Human FZD7 amino acids 49-160 (SEQ ID NO:28)
CQ PIS I PLCT DIAYNQT I L PNLLGHTNQE DAGLEVHQ FYPLVKVQCS PELRFFLCSMYAP
VCTVLDQAI PPCRSLCERARQGCEALMNKFGFQWPERLRCENFPVHGAGE IC
Human FZD8 amino acids 35-148 (SEQ ID NO:30)
CQE I TVPLCKGI GYNYTYMPNQ FNHDTQDEAGLEVHQ FWPLVE IQCS PDLKFFLCSMYT P
I CLE DYKKPL P PCRSVCERAKAGCAPLMRQYGFAWPDRMRC DRLPEQGNP DT LC
84
CA 02931975 2016-05-27
WO 2015/084808
PCT/US2014/068097
Human FZD9 amino acids 39-152 (SEQ ID NO:31)
CQAVE I PMCRG I GYNLTRMPNLLGHT S QGEAAAELAE FAPLVQYGCH S HLRFFLC S LYAP
MCTDQVS T P I PACRPMCEQARLRCAPIMEQFNFGWPDSLDCARLPTRNDPHALC
Human FZD10 amino acids 34-147 (SEQ ID NO:32)
CQPIE I PMCKDIGYNMTRMPNLMGHENQREAAIQLHEFAPLVEYGCHGHLRFFLCSLYAP
MCTEQVS T P I PACRVMCEQARLKCS PIMEQFNFKWPDSLDCRKLPNKNDPNYLC
Human FZD8 Fri domain amino acid sequence without predicted signal sequence
(variant) (SEQ ID
NO:33)
ASAKELACQE I TVPLCKGI GYNYTYMPNQFNHDTQDEAGLEVHQFWPLVE IQCSPDLKFF
LCSMYT P I CLE DYKKPLPPCRSVCERAKAGCAPLMRQYGFAWP DRMRCDRLPEQGNPDT L
CMDYNRT DL
Human IgGi Fc region (SEQ ID NO:34)
DKTHT CP PCPAPELLGGPSVFL FPPKPKDTLMI SRTPEVTCVVVDVSHEDPEVKFNWYVD
GVEVHNAKTKPREEQYNS TYRVVSVLTVLHQ DWLNGKEYKCKVSNKALPAP I EKT I SKAK
GQ PRE PQVYT L PPSRDELTKNQVSLTCLVKGFYPS DIAVEWESNGQPENNYKTTPPVLDS
DGSFFLYSKLTVDKSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGK
Human IgGi Fc region (variant) (SEQ ID NO:35)
DKTHT CP PCPAPELLGGPSVFL FPPKPKDTLMI SRTPEVTCVVVDVSHEDPEVKFNWYVD
GVEVHNAKTKPREEQYNS TYRVVSVLTVLHQ DWLNGKEYKCKVSNKALPAP I EKT I SKAK
GQ PRE PQVYT L PPSREEMTKNQVSLTCLVKGFYPS DIAVEWESNGQPENNYKTTPPVLDS
DGSFFLYSKLTVDKSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGK
Human IgGi Fc region (SEQ ID NO:36)
KS S DKTHTCP PCPAPELLGGPSVFL FP PKPKDTLMI SRT PEVT CVVVDVS HE DPEVKFNW
YVDGVEVHNAKTKPREEQYNS TYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAP I EKT I S
KAKGQ PREPQVYTL PP SRDELTKNQVS LT CLVKGFYP S DIAVEWESNGQPENNYKTTPPV
L DS DGS FFLY SKLTVDKSRWQQGNVFS CSVMHEALHNHYTQKS LSL S PGK
Human IgGi Fc region (SEQ ID NO:37)
EPKSS DKTHT CPPCPAPELLGGPSVFL FP PKPKDT LMI SRT PEVTCVVVDVSHEDPEVKF
NWYVDGVEVHNAKTKPREEQYNS TYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAP I EKT
I SKAKGQ PRE PQVYTL PPSRDELTKNQVS LT CLVKGFY PS DIAVEWE SNGQPENNYKTT P
PVLDS DGS FFLYSKLTVDKSRWQQGNVFS CSVMHEALHNHYTQKSL S LS PGK
Human IgG2Fc region (SEQ ID NO:38)
CVECP PC PAP PVAGPSVFL FPPKPKDT LMI S RT PEVT CVVVDVS HE DPEVQFNWYVDGVE
VHNAKTKPREEQFNST FRVVSVLTVVHQDWLNGKEYKCKVSNKGLPAPIEKT I SKTKGQP
RE PQVYT LPP SREEMTKNQVSLTCLVKGFYP S DIAVEWESNGQPENNYKTT PPML DS DGS
FFLYSKLTVDKSRWQQGNVFSCSVMHEALHNHYTQKSLSLS PGK
FZD8-Fc variant 54F03 amino acid sequence (without predicted signal sequence)
(SEQ ID NO:39)
ASAKELACQE I TVPLCKGI GYNYTYMPNQFNHDTQDEAGLEVHQFWPLVE IQCSPDLKFF
LCSMYT P I CLE DYKKPLPPCRSVCERAKAGCAPLMRQYGFAWP DRMRCDRLPEQGNPDT L
CMDYNRT DLTTGRADKTHTCPPCPAPELLGGPSVFLFPPKPKDTLMI SRT PEVTCVVVDV
S HE DPEVKFNWYVDGVEVHNAKTKPREEQYNS TYRVVSVLTVLHQDWLNGKEYKCKVSNK
AL PAP IEKT I SKAKGQ PRE PQVYTL PP SRDELTKNQVS LTCLVKGFY PS DIAVEWE SNGQ
PENNYKTTPPVLDS DGS FFLYSKLTVDKSRWQQGNVFSCSVMHEALHNHYTQKSL S LS PG
K
CA 02931975 2016-05-27
WO 2015/084808
PCT/US2014/068097
FZD8-Fc variant 54F16, 54F17, 54F18, 54F23, 54F25, 54F27, 54F29, 54F31, and
54F34 amino acid
sequence (without predicted signal sequence) (SEQ ID NO:40)
ASAKELACQE I TVPLCKGI GYNYTYMPNQFNHDTQDEAGLEVHQFWPLVE IQCSPDLKFF
LCSMYT P I CLE DYKKPLPPCRSVCERAKAGCAPLMRQYGFAWP DRMRCDRLPEQGNPDT L
CMDYNRT DLTTKSS DKTHTCPPCPAPELLGGPSVFLFPPKPKDTLMI SRT PEVTCVVVDV
S HE DPEVKFNWYVDGVEVHNAKTKPREEQYNS TYRVVSVLTVLHQDWLNGKEYKCKVSNK
AL PAP IEKT I SKAKGQ PRE PQVYTL PP SRDELTKNQVS LT CLVKGFY PS DIAVEWE SNGQ
PENNYKTTPPVLDS DGS FFLYSKLTVDKSRWQQGNVFS CSVMHEALHNHYTQKSL S LS PG
K
FZD8-Fc variant 54F19, 54F20, 54F24, 54F26, 54F28, 54F30, 54F32, 54F34 and
54F35 amino acid
sequence (without predicted signal sequence) (SEQ ID NO:41)
ASAKELACQE I TVPLCKGI GYNYTYMPNQFNHDTQDEAGLEVHQFWPLVE IQCSPDLKFF
LCSMYT P I CLE DYKKPLPPCRSVCERAKAGCAPLMRQYGFAWP DRMRCDRLPEQGNPDT L
CMDYNRT DLTTEPKSS DKTHTCPPCPAPELLGGPSVFLFPPKPKDTLMI SRT PEVTCVVV
DVS HE DPEVKFNWYVDGVEVHNAKTKPREEQYNS TYRVVSVLTVLHQ DWLNGKEYKCKVS
NKALPAPIEKT I SKAKGQPREPQVYTL PP SRDELTKNQVSLTCLVKGFYP S DIAVEWESN
GQPENNYKTT P PVL DS DGS FFLYSKLTVDKSRWQQGNVFSCSVMHEALHNHYTQKSLSLS
PGK
FZD8-Fc variant 54F03 amino acid sequence with signal sequence (SEQ ID NO:42)
MEWGYLLEVT SLLAALALLQRS SGAAAASAKELACQE I TVPLCKGI GYNYTYMPNQFNHD
TQ DEAGLEVHQFWPLVE IQCS P DLKFFLCSMYT P I CLEDYKKPLPPCRSVCERAKAGCAP
LMRQYGFAWPDRMRCDRLPEQGNPDTLCMDYNRTDLTTGRADKTHTCPPCPAPELLGGPS
VFLFPPKPKDTLMI SRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNS T
YRVVSVLTVLHQDWLNGKEYKCKVSNKAL PAP IEKT I SKAKGQ PRE PQVYTL PPSRDELT
KNQVS LT CLVKGFY PS DIAVEWESNGQPENNYKTT PPVLDS DGSFFLYSKLTVDKSRWQQ
GNVFSCSVMHEALHNHYTQKSLSLS PGK
FZD8-Fc variant 54F16 amino acid sequence with signal sequence (SEQ ID NO:43)
MEWGYLLEVT SLLAALALLQRS S GAAAASAKELACQE I TVPLCKGI GYNYTYMPNQFNHD
TQ DEAGLEVHQFWPLVE IQCS P DLKFFLCSMYT P I CLE DYKKPLP PCRSVCERAKAGCAP
LMRQYGFAWPDRMRCDRLPEQGNPDTLCMDYNRTDLTTKSS DKTHTCPPCPAPELLGGPS
VFLFPPKPKDTLMI SRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNS T
YRVVSVLTVLHQDWLNGKEYKCKVSNKAL PAP IEKT I SKAKGQ PRE PQVYTL PPSRDELT
KNQVS LT CLVKGFY PS DIAVEWESNGQPENNYKTT PPVLDSDGSFFLYSKLTVDKSRWQQ
GNVFSCSVMHEALHNHYTQKSLSLS PGK
FZD8-Fc variant 54F26 with signal sequence (SEQ ID NO:44)
MEWGYLLEVTSLLAALFLLQRS PIVHAASAKELACQE I TVPLCKGI GYNYTYMPNQFNHD
TQ DEAGLEVHQFWPLVE I QCS P DLKFFLCSMYT P I CLEDYKKPLPPCRSVCERAKAGCAP
LMRQYGFAWPDRMRCDRLPEQGNPDTLCMDYNRT DLTTEPKS SDKTHTCPPCPAPELLGG
PSVFL FP PKPKDTLMI SRT PEVTCVVVDVS HE DPEVKFNWYVDGVEVHNAKTKPREEQYN
S TYRVVSVLTVLHQ DWLNGKEYKCKVSNKAL PAP I EKT I SKAKGQPREPQVYTLPPSRDE
LTKNQVS LT CLVKGFY PS DIAVEWESNGQPENNYKTT P PVL DS DGS FFLYSKLTVDKSRW
QQGNVFS CSVMHEALHNHYTQKSLS LS PGK
FZD8-Fc variant 54F28 with signal sequence (SEQ ID NO:45)
MEWGYLLEVTSLLAALLLLQRS PFVHAASAKELACQE I TVPLCKGI GYNYTYMPNQFNHD
TQ DEAGLEVHQFWPLVE I QCS P DLKFFLCSMYT P I CLEDYKKPLPPCRSVCERAKAGCAP
LMRQYGFAWPDRMRCDRLPEQGNPDTLCMDYNRTDLTTEPKSS DKTHTCPPCPAPELLGG
PSVFL FP PKPKDTLMI SRT PEVTCVVVDVS HE DPEVKFNWYVDGVEVHNAKTKPREEQYN
S TYRVVSVLTVLHQ DWLNGKEYKCKVSNKAL PAP I EKT I SKAKGQPREPQVYTLPPSRDE
LTKNQVS LT CLVKGFY PS DIAVEWESNGQPENNYKTT P PVL DS DGS FFLYSKLTVDKSRW
QQGNVFS CSVMHEALHNHYTQKSLS LS PGK
86
CA 02931975 2016-05-27
WO 2015/084808
PCT/US2014/068097
Human Wntl C-terminal cysteine rich domain (aa 288-370) (SEQ ID NO:46)
DLVYFEKSPNFCTYSGRLGTAGTAGRACNSS S PAL DGCELLCCGRGHRTRTQRVTERCNC
T FHWCCHVSCRNCTHTRVLHECL
Human Wnt2 C-terminal cysteine rich domain (aa 267-360) (SEQ ID NO:47)
DLVYFENSPDYCIRDREAGSLGTAGRVCNLT SRGMDS CEVMCCGRGY DT S HVTRMTKCGC
KFHWCCAVRCQ DCLEALDVHTCKAPKNADWT TAT
Human Wnt2b C-terminal cysteine rich domain (aa 298-391) (SEQ ID NO:48)
DLVYFDNSPDYCVLDKAAGSLGTAGRVCSKT SKGT DGCEIMCCGRGYDTTRVTRVTQCEC
KFHWCCAVRCKECRNTVDVHTCKAPKKAEWLDQT
Human Wnt3 C-terminal cysteine rich domain (aa 273-355) (SEQ ID NO:49)
DLVYYENSPNFCEPNPETGS FGTRDRTCNVT SHGI DGCDLLCCGRGHNTRTEKRKEKCHC
I FHWCCYVSCQECI RI YDVHTCK
Human Wnt3a C-terminal cysteine rich domain (aa 270-352) (SEQ ID NO:50)
DLVYYEASPNFCEPNPETGS FGTRDRTCNVS SHGI DGCDLLCCGRGHNARAERRREKCRC
VFHWCCYVSCQECTRVYDVHTCK
Human Wnt7a C-terminal cysteine rich domain (aa 267-359) (SEQ ID NO:51)
DLVY I EKS PNYCEE DPVTGSVGTQGRACNKTAPQAS GC DLMCCGRGYNTHQYARVWQCNC
KFHWCCYVKCNTCSERTEMYTCK
Human Wnt7b C-terminal cysteine rich domain (aa 267-349) (SEQ ID NO:52)
DLVY I EKS PNYCEE DAATGSVGTQGRLCNRT S PGADGCDTMCCGRGYNTHQYTKVWQCNC
KFHWCCFVKCNTCSERTEVFTCK
Human Wnt8a C-terminal cysteine rich domain (aa 248-355) (SEQ ID NO:53)
EL I FLEE S PDYCTCNS SLGIYGTEGRECLQNSHNT SRWERRSCGRLCTECGLQVEERKTE
VI SSCNCKFQWCCTVKCDQCRHVVSKYYCARS PGSAQ S LGRVWFGVY I
Human Wnt8b C-terminal cysteine rich domain (aa 245-351) (SEQ ID NO:54)
ELVHLEDSPDYCLENKTLGLLGTEGRECLRRGRALGRWELRSCRRLCGDCGLAVEERRAE
TVS S CNCKFHWCCAVRCEQCRRRVTKY FC SRAERPRGGAAHKPGRKP
Human WntlOa C-terminal cysteine rich domain (aa 335-417) (SEQ ID NO:55)
DLVYFEKSPDFCEREPRLDSAGTVGRLCNKS SAGS DGCGSMCCGRGHNI LRQTRSERCHC
RFHWCCFVVCEECRITEWVSVCK
Human Wntl Ob C-terminal cysteine rich domain (aa 307-389) (SEQ ID NO:56)
ELVYFEKSPDFCERDPTMGS PGTRGRACNKT SRLLDGCGSLCCGRGHNVLRQTRVERCHC
RFHWCCYVLCDECKVTEWVNVCK
Linker (SEQ ID NO:57)
ESGGGGVT
Linker (SEQ ID NO:58)
LE S GGGGVT
Linker (SEQ ID NO:59)
GRAQVT
87
CA 02931975 2016-05-27
WO 2015/084808
PCT/US2014/068097
Linker (SEQ ID NO:60)
WRAQVT
Linker (SEQ ID NO:61)
ARGRAQVT
CCND2 Forward Primer (SEQ ID NO:62)
GCTGTCTCTGATCCGCAAGC
CCND2 Reverse Primer (SEQ ID NO:63)
GACGGTGGGTACATGGCAAAC
CCND2 Probe (SEQ ID NO:64)
CCTTCATTGCTCTGTGTGCCACCGAC
CTBP2 Forward Primer (SEQ ID NO:65)
ATCCGTGGGGAGACGCTG
CTBP2 Reverse Primer (SEQ ID NO:66)
CTCGAACTGCAACCGCCTG
CTBP2 Probe (SEQ ID NO:67)
CCCGTGCGACCAAAGCCAATGAGG
DKK1 Forward Primer (SEQ ID NO:68)
GACCATTGACAACTACCAGCCGTA
DKK1 Reverse Primer (SEQ ID NO:69)
TGGGACTAGCGCAGTACTCATC
DKK1 Probe (SEQ ID NO:70)
TGCCGCACTCCTCGTCCTCTG
FBXW2 Forward Primer (SEQ ID NO:71)
GCCAGTTATGATATTCTCAGGGTCA
FBXW2 Reverse Primer (SEQ ID NO:72)
AGCAGGGCAAAGATATCTCCAAA
FBXW2 Probe (SEQ ID NO:73)
AGACTCCTGAGATAGCAAACTTGGCCT
RHOU1 Forward Primer (SEQ ID NO:74)
CCCACCGAGTACATCCCTACTG
RHOU1 Reverse Primer (SEQ ID NO:75)
CAGTGTCACAGAGTTGGAGTCTCA
RHOU1 Probe (SEQ ID NO:76)
CGCCCATCCACAGACACCACCG
WIF1 Forward Primer (SEQ ID NO:77)
GTTCCAAAGGTTACCAGGGAGAC
WIF1 Reverse Primer (SEQ ID NO:78)
88
CA 02931975 2016-05-27
WO 2015/084808
PCT/US2014/068097
GTTGGGTTCATGGCAGGTTCC
WIF1 Probe (SEQ ID NO:79)
CCAGGCTCGCAGACAGGCTTTGAAC
89