Note: Descriptions are shown in the official language in which they were submitted.
CA 02946317 2016-10-18
WO 2015/168699 PCT/US2015/029101
METHODS OF PREDICTING MEDICALLY REFRACTIVE ULCERATIVE
COLITIS (mrUC) REQUIRING COLECTOMY
GOVERNMENT RIGHTS
This invention was made with government support under Contract Nos. DK046763
and DK062413 awarded by the National Institutes of Health. The government has
certain
rights in the invention.
FIELD OF THE INVENTION
The invention relates generally to the fields of genetics and inflammatory
disease,
specifically medically refractive-UC (mrUC).
BACKGROUND
All publications herein are incorporated by reference to the same extent as if
each
individual publication or patent application was specifically and individually
indicated to
be incorporated by reference. The following description includes information
that may be
useful in understanding the present invention. It is not an admission that any
of the
information provided herein is prior art or relevant to the presently claimed
invention, or
that any publication specifically or implicitly referenced is prior art.
Crohn's disease (CD) and ulcerative colitis (UC), the two common forms of
idiopathic inflammatory bowel disease (IBD), are chronic, relapsing
inflammatory
disorders of the gastrointestinal tract. Each has a peak age of onset in the
second to fourth
decades of life and prevalences in European ancestry populations that average
approximately 100-150 per 100,000 (D.K. Podolsky, N Engl J Med 347, 417
(2002); E.V.
Loftus, Jr., Gastroenterology 126, 1504 (2004)). Although the precise etiology
of IBD
remains to be elucidated, a widely accepted hypothesis is that ubiquitous,
commensal
intestinal bacteria trigger an inappropriate, overactive, and ongoing mucosal
immune
response that mediates intestinal tissue damage in genetically susceptible
individuals
(D.K. Podolsky, N Engl J Med 347, 417 (2002)). Genetic factors play an
important role
in IBD pathogenesis, as evidenced by the increased rates of IBD in Ashkenazi
Jews,
familial aggregation of IBD, and increased concordance for IBD in monozygotic
compared to dizygotic twin pairs (S. Vermeire, P. Rutgeerts, Genes Immun 6,
637
(2005)). Moreover, genetic analyses have linked IBD to specific genetic
variants,
especially CARD15 variants on chromosome 16q12 and the IBD5 haplotype
(spanning
the organic cation transporters, 5LC22A4 and SLC22A5, and other genes) on
1
CA 02946317 2016-10-18
WO 2015/168699 PCT/US2015/029101
chromosome 5q31 (S. Vermeire, P. Rutgeerts, Genes Immun 6, 637 (2005); J.P.
Hugot et
al., Nature 411, 599 (2001); Y. Ogura et al., Nature 411, 603 (2001); J.D.
Rioux et al., Nat
Genet 29, 223 (2001); V.D. Peltekova et al., Nat Genet 36, 471 (2004)). CD and
UC are
thought to be related disorders that share some genetic susceptibility loci
but differ at
others.
Thus, there is a need in the art to identify genes, allelic variants and/or
haplotypes
that may assist in determining the need for colectomy, diagnosing
susceptibility or
treatment for medically refractive ulcerative colitis (mrUC).
SUMMARY OF THE INVENTION
Various embodiments of the present invention provide for a method of
determining the need for colectomy in a subject with mrUC comprising:
obtaining a
sample from the subject; assaying the sample to detect the presence or absence
of mrUC
genetic risk variants, wherein the mrUC genetic risk variants are selected
from the group
consisting of SEQ ID NOs: 1-99; calculating a genetic risk score based on the
detection of
the mrUC genetic risk variants; determining that the subject has an increased
likelihood of
needing colectomy if the calculated genetic risk score is at the high end of
the observed
range and determining that the subject has a decreased likelihood of needing
colectomy if
the calculated genetic risk score is at the low end of the observed range. In
various
embodiments, the genetic risk score is obtained by calculating a total number
of risk
alleles for all the mrUC genetic risk variants assayed, wherein the risk
allele for each
mrUC genetic risk variant assayed is 0, 1 or 2.
Various other embodiments, further comprise obtaining a theoretical range and
an observed range based on the genetic risk score, wherein the theoretical
range consists
of the minimum and maximum number of risk alleles possible based on the number
of
mrUC genetic risk variants assayed and wherein the observed range consists of
the actual
minimum and maximum number of risk alleles detected. In various embodiments,
the
number of mrUC genetic risk variants assayed is 46, the theoretical range is 0-
92 and the
observed range is 28-60. In various embodiments, the number of mrUC genetic
risk
variants assayed is 36, the theoretical range is 0-72 and the observed range
is 16-38.
Various other embodiments further comprise prescribing colectomy to subjects
having a
genetic risk score at the high end of the observed range. In various
embodiments, time to
colectomy is lower in a subject with a genetic risk score at the high end of
the observed
2
CA 02946317 2016-10-18
WO 2015/168699 PCT/US2015/029101
range and time to colectomy is higher in a subject with a genetic risk score
at the low end
of the observed range. In various embodiments, the time to colectomy is 10 to
70 months
from detection.
Various embodiments of the present invention provide for a method of
diagnosing susceptibility to mrUC in a subject, comprising: obtaining a sample
from the
subject; assaying the sample to detect the presence or absence of mrUC genetic
risk
variants, wherein the mrUC genetic risk variants are selected from the group
consisting of
SEQ ID NOs: 1-99; calculating a genetic risk score based on the detection of
the mrUC
genetic risk variants; and diagnosing susceptibility to mrUC based on the
calculated risk
score, wherein a subject has an increased susceptibility to mrUC if the
calculated genetic
risk score is at the high end of the observed range and a subject has a
decreased
susceptibility to mrUC if the calculated genetic risk score is at the low end
of the
observed range.
In various embodiments, the genetic risk score is obtained by calculating a
total
number of risk alleles for all the mrUC genetic risk variants assayed, wherein
the risk
allele for each mrUC genetic risk variant assayed is 0, 1 or 2.
Various other embodiments further comprise obtaining a theoretical range and
an
observed range based on the genetic risk score, wherein the theoretical range
consists of
the minimum and maximum number of risk alleles possible based on the number of
mrUC genetic risk variants assayed and wherein the observed range consists of
the actual
minimum and maximum number of risk alleles detected. In various embodiments,
an
increase in the number of risk alleles detected signifies an increase in
susceptibility to
mrUC. In various embodiments, the number of mrUC genetic risk variants assayed
is 46,
the theoretical range is 0-92 and the observed range is 28-60. In various
embodiments, the
number of mrUC genetic risk variants assayed is 36, the theoretical range is 0-
72 and the
observed range is 16-38.
Various other embodiments further comprise prescribing colectomy to subjects
diagnosed with a susceptibility for mrUC and have a genetic risk score at the
high end of
the observed range. In various embodiments, the time to colectomy is lower in
a subject
with a genetic risk score at the high end of the observed range and the time
to colectomy
is higher in a subject with a genetic risk score at the low end of the
observed range. In
various embodiments, the time to colectomy is 10 to 70 months from detection.
In various other embodiments of the present invention provides for a method of
treating mrUC in a subject, comprising: obtaining a sample from the subject;
assaying the
3
CA 02946317 2016-10-18
WO 2015/168699 PCT/US2015/029101
sample to detect the presence or absence of mrUC genetic risk variants,
wherein the
mrUC genetic risk variants are selected from the group consisting of SEQ ID
NOs: 1-99;
calculating a genetic risk score based on the detection of the mrUC genetic
risk variants;
diagnosing susceptibility to mrUC based on the calculated risk score, wherein
a subject
has an increased susceptibility to mrUC if the calculated genetic risk score
is high and a
subject has a decreased susceptibility to mrUC if the calculated genetic risk
score is low;
and prescribing colectomy to the subject with an increased susceptibility to
mrUC.
In various embodiments, the genetic risk score is obtained by calculating a
total
number of risk alleles for all the mrUC genetic risk variants assayed, wherein
the risk
allele for each mrUC genetic risk variant assayed is 0, 1 or 2.
Various other embodiments further comprise obtaining a theoretical range and
an
observed range based on the genetic risk score, wherein the theoretical range
consists of
the minimum and maximum number of risk alleles possible based on the number of
mrUC genetic risk variants assayed and wherein the observed range consists of
the actual
minimum and maximum number of risk alleles detected. In various embodiments,
an
increase in the number of risk alleles detected signifies an increase in
susceptibility to
mrUC. In various embodiments, the number of mrUC genetic risk variants assayed
is 46,
the theoretical range is 0-92 and the observed range is 28-60. In various
embodiments, the
number of mrUC genetic risk variants assayed is 36, the theoretical range is 0-
72 and the
observed range is 16-38.
In various embodiments, the treatment is colectomy and is prescribed to
subjects
diagnosed with a susceptibility for mrUC and have a genetic risk score at the
high end of
the observed range. In various other embodiments, the time to colectomy is
lower in a
subject with a genetic risk score at the high end of the observed range and
the time to
colectomy is higher in a subject with a genetic risk score at the low end of
the observed
range. In various embodiments, the time to colectomy is 10 to 70 months from
detection.
Various embodiments of the present invention provide for a kit for prognostic
use, comprising: a single prognostic panel comprising one or more medically
refractive
ulcerative colitis (mrUC) genetic risk variants described in SEQ ID NOs: 1-99.
BRIEF DESCRIPTION OF THE FIGURES
Exemplary embodiments are illustrated in referenced figures. It is intended
that the
embodiments and figures disclosed herein are to be considered illustrative
rather than
restrictive.
4
CA 02946317 2016-10-18
WO 2015/168699 PCT/US2015/029101
Figure 1 depicts, in accordance with an embodiment herein, a schematic
describing
mrUC vs. non-mrUC survival analysis and risk modeling.
Figure 2 depicts, in accordance with an embodiment herein, A) Higher risk
score
categories are associated with mrUC (x2 test for trend p <2.2x10-16). Risk
score
(observed range: 28-60) was divided into quarters: scores 28-38 (risk-A);
scores 39-45
(risk-B); scores 46-52 (risk-C); and scores 53-60 (risk-D). Percentage of mrUC
is noted,
along with the total number of UC subjects in each risk category. B) Higher
risk score
categories are associated with an earlier progression to colectomy at 24 and
60 months.
Risk score was divided into quarters: scores 28-38 (risk-A); scores 39-45
(risk-B); scores
46-52 (risk-C); and scores 53-60 (risk-D). At 24 months, risk of colectomy was
3.1%,
19.1% and 62% for risk-B, -C, and -D, respectively. Risk of colectomy at 60
months
increased to 8.3%, 48.4%, 84% for risk-B, -C, and -D, respectively. Total
number of UC
subjects in each risk category is given.
Figure 3 depicts, in accordance with an embodiment herein, serology data
demonstrating an association of mrUC with Cbir 1 , ASCA, OmpC and 12 antibody
quartile sum in mrUC and non-mrUC subjects.
Figure 4 depicts, in accordance with an embodiment herein, single SNP
association tested with logistic regression analysis in mrUC and non-mrUC
subjects.
Figure 5 depicts, in accordance with an embodiment herein, a schematic
describing
mr UC vs. Non-mrUC survival analysis and risk modeling for mrUC.
Figure 6 depicts, in accordance with an embodiment herein, a chart with the
top 36
associated SNPs from Analysis I and II, referenced herein.
Figure 7 depicts, in accordance with an embodiment herein, higher risk score
association with mrUC.
Figure 8 depicts, in accordance with an embodiment herein, higher risk score
association with earlier progression to colectomy.
Figure 9 depicts, in accordance with an embodiment herein, higher risk score
exhibits a shorter overall median time to colectomy.
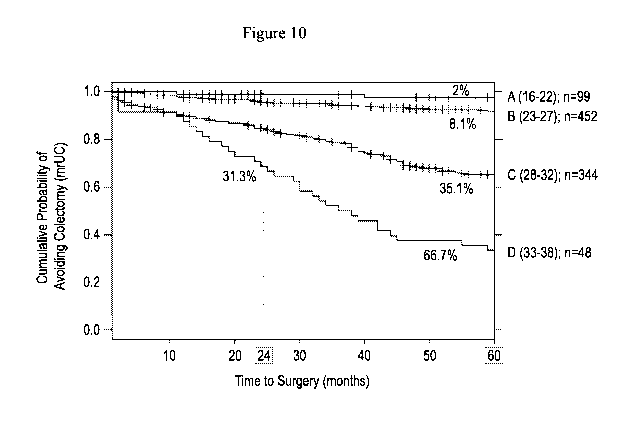
Figure 10 depicts, in accordance with an embodiment herein, potential clinical
utility of the association of a higher risk score with earlier progression to
colectomy.
Figure 11 depicts, in accordance with an embodiment herein, role for major
histocompatibility (MHC) in UC severity in mrUC versus controls.
Figure 12 depicts, in accordance with an embodiment herein, single SNP
association tested with regression analysis in mrUC versus controls.
5
CA 02946317 2016-10-18
WO 2015/168699 PCT/US2015/029101
DESCRIPTION OF THE INVENTION
All references cited herein are incorporated by reference in their entirety as
though
fully set forth. Unless defined otherwise, technical and scientific terms used
herein have
the same meaning as commonly understood by one of ordinary skill in the art to
which this
invention belongs. Singleton et at., Dictionary of Microbiology and Molecular
Biology
3rd ed., J. Wiley & Sons (New York, NY 2001); March, Advanced Organic
Chemistry
Reactions, Mechanisms and Structure 5th ed., J. Wiley & Sons (New York, NY
2001);
and Sambrook and Russel, Molecular Cloning: A Laboratory Manual 3rd ed., Cold
Spring Harbor Laboratory Press (Cold Spring Harbor, NY 2001), provide one
skilled in
the art with a general guide to many of the terms used in the present
application.
One skilled in the art will recognize many methods and materials similar or
equivalent to those described herein, which could be used in the practice of
the present
invention. Indeed, the present invention is in no way limited to the methods
and materials
described.
"IBD" as used herein is an abbreviation of inflammatory bowel disease.
"CD" as used herein is an abbreviation of Crohn's Disease.
"UC" as used herein is an abbreviation of ulcerative colitis.
"GWAS" as used herein is an abbreviation of genome wide association
study.
"mrUC" as used herein is defined as ulcerative colitis with symptoms
uncontrolled
by medical therapy. Also referred to as mr-UC.
As used herein, the term "mrUC genetic risk variant" refers to genetic
variants, or
SNPs, that have an association with the mrUC, or ulcerative colitis requiring
colectomy,
phenotype.
As used herein, the term "biological sample" means any biological material
from
which nucleic acid molecules can be prepared. As non-limiting examples, the
term
material encompasses whole blood, plasma, saliva, cheek swab, or other bodily
fluid or
tissue that contains nucleic acid.
A "Risk Score" as used herein is a calculated number, obtained by
adding/totaling
the total number of risk alleles for all the mrUC genetic risk variants
assayed. The risk
allele for each mrUC genetic risk variant assayed is 0, 1 or 2. For example,
when
analyzing a patient for 5 mrUC genetic risk variants, the detected risk
alleles may be 1, 0,
6
CA 02946317 2016-10-18
WO 2015/168699 PCT/US2015/029101
1, 2, and 1, which when added will give the patient a risk score of 5
(1+0+1+2+1 = 5).
The risk score, based on analyzed mrUC genetic risk variants, is calculated in
other
patients and the cumulative risk scores for all patients analyzed provide an
observed
range as discussed below.
"Risk Group" as used herein refers to a subset of patients who fall within the
same
category for colectomy risk based on the detected mrUC risk variants in the
subject's
biological sample.
"Treatment", as used herein refers to both therapeutic treatment and
prophylactic
or preventative measures, wherein the object is to prevent or slow down
(lessen) the
targeted pathologic condition, prevent the pathologic condition, pursue or
obtain good
overall survival, or lower the chances of the individual developing the
condition even if
the treatment is ultimately unsuccessful. Those in need of treatment include
those already
with the condition as well as those prone to have the condition or those in
whom the
condition is to be prevented. Examples of mrUC treatment include, but are not
limited to,
active surveillance, observation, surgical intervention (such as colectomy),
drug therapy
(anti-inflammatory and/or immune system suppressor drugs), targeted therapy to
genes
known to be involved in mrUC, such as, but not limited to those referenced
herein and/or
a combination thereof
"Time to colectomy" as used herein refers to the amount of time between the
determination that a subject had an increased likelihood of needing colectomy
and
actually undergoing colectomy. In one embodiments, the subject has a reduced
time to
colectomy (for example: 0-6 months, 6 months ¨ 1 year, 1 ¨ 2 years or 2-3
years) if the
subject has a high risk score. In another embodiment, the subject has an
increased time to
colectomy (for example, 3-4 years, 4-5 years or more) if the subject has a low
risk score.
"Theoretical range" as used herein refers to the minimum and maximum number
of risk alleles possible based on the number of mrUC genetic risk variants
assayed. For
example, if 46 genetic risk variants are analyzed, the theoretical range is 0-
92, where 0 is
the minimum number of risk alleles and 92 (46 x 2 alleles) is the maximum
number of
risk alleles.
"Observed range" as used herein refers to the minimum and maximum risk score,
which is based on the risk alleles detected for the patient cohort, as
described above. For
example, an observed range of 28-60, obtained when analyzing the 46 genetic
risk
variants, results in a minimum of 28 and a maximum of 60.
7
CA 02946317 2016-10-18
WO 2015/168699 PCT/US2015/029101
"High end" of an observed range as used herein refers to a genetic risk score
that
is within for example, 10 ¨ 15 points of the maximum observed range.
"Low end" of an observed range as used herein refers to a genetic score that
is
within for example, 10 ¨ 15 points of the minimum observed range.
Acute severe ulcerative colitis (UC) remains a significant clinical challenge
and the
ability to predict, at an early stage, those individuals at risk of colectomy
for medically
refractory UC (mrUC) would be a major clinical advance. As disclosed herein,
the
inventors used a genome-wide association study (GWAS) in a well characterized
cohort of
UC patients to identify genetic variation that contributes to mrUC. A GWAS
comparing
324 mrUC patients with 537 Non-mrUC patients was analyzed using logistic
regression
and Cox proportional hazards methods. In addition, the mrUC patients were
compared
with 2601 healthy controls.
As further disclosed herein, mrUC was associated with more extensive
disease (p= 2.7x10-6) and a positive family history of UC (p= 0.004). A risk
score based
on the combination of 46 SNPs associated with mrUC explained 48% of the
variance for
colectomy risk in the cohort. Risk scores divided into quarters showed the
risk of
colectomy to be 0%, 17%, 74% and 100% in the four groups. Comparison of the
mrUC
subjects with healthy controls confirmed the contribution of the major
histocompatibility
complex to severe UC (peak association: rs17207986, p= 1.4x10-16) and provided
genome-wide suggestive association at the TNFSF15 (TL1A) locus (peak
association:
rs11554257, p= 1.4x10-6). A SNP-based risk scoring system, identified herein
by GWAS
analyses, can provide a useful adjunct to clinical parameters for predicting
natural history
in UC. Furthermore, discovery of genetic processes underlying disease severity
can help
to identify pathways for novel therapeutic intervention in severe UC.
Determining the need for colectomy
Various embodiments of the present invention provide for a method of
determining the need for colectomy in a subject with mrUC comprising:
obtaining a
sample from the subject; assaying the sample to detect the presence or absence
of mrUC
genetic risk variants, wherein the mrUC genetic risk variants are selected
from the group
consisting of SEQ ID NOs: 1-99; calculating a genetic risk score based on the
detection of
the mrUC genetic risk variants; determining that the subject has an increased
likelihood of
needing colectomy if the calculated genetic risk score is at the high end of
the observed
range and determining that the subject has a decreased likelihood of needing
colectomy if
8
CA 02946317 2016-10-18
WO 2015/168699 PCT/US2015/029101
the calculated genetic risk score is at the low end of the observed range. In
various
embodiments, the genetic risk score is obtained by calculating a total number
of risk
alleles for all the mrUC genetic risk variants assayed, wherein the risk
allele for each
mrUC genetic risk variant assayed is 0, 1 or 2.
Various other embodiments, further comprise obtaining a theoretical range and
an observed range based on the genetic risk score, wherein the theoretical
range consists
of the minimum and maximum number of risk alleles possible based on the number
of
mrUC genetic risk variants assayed and wherein the observed range consists of
the actual
minimum and maximum number of risk alleles detected. In various embodiments,
the
number of mrUC genetic risk variants assayed is 46, the theoretical range is 0-
92 and the
observed range is 28-60. In various embodiments, the number of mrUC genetic
risk
variants assayed is 36, the theoretical range is 0-72 and the observed range
is 16-38.
Various other embodiments further comprise prescribing colectomy to subjects
having a genetic risk score at the high end of the observed range. In various
embodiments, time to colectomy is lower in a subject with a genetic risk score
at the high
end of the observed range and time to colectomy is higher in a subject with a
genetic risk
score at the low end of the observed range. In various embodiments, the time
to
colectomy is 10 to 70 months from detection.
Diagnosing susceptibility
Various embodiments of the present invention provide for a method of
diagnosing susceptibility to mrUC in a subject, comprising: obtaining a sample
from the
subject; assaying the sample to detect the presence or absence of mrUC genetic
risk
variants, wherein the mrUC genetic risk variants are selected from the group
consisting of
SEQ ID NOs: 1-99; calculating a genetic risk score based on the detection of
the mrUC
genetic risk variants; and diagnosing susceptibility to mrUC based on the
calculated risk
score, wherein a subject has an increased susceptibility to mrUC if the
calculated genetic
risk score is at the high end of the observed range and a subject has a
decreased
susceptibility to mrUC if the calculated genetic risk score is at the low end
of the
observed range.
In various embodiments, the genetic risk score is obtained by calculating a
total
number of risk alleles for all the mrUC genetic risk variants assayed, wherein
the risk
allele for each mrUC genetic risk variant assayed is 0, 1 or 2
Various other embodiments further comprise obtaining a theoretical range and
an
9
CA 02946317 2016-10-18
WO 2015/168699 PCT/US2015/029101
observed range based on the genetic risk score, wherein the theoretical range
consists of
the minimum and maximum number of risk alleles possible based on the number of
mrUC genetic risk variants assayed and wherein the observed range consists of
the actual
minimum and maximum number of risk alleles detected. . In various embodiments,
an
increase in the number of risk alleles detected signifies an increase in
susceptibility to
mrUC. In various embodiments, the number of mrUC genetic risk variants assayed
is 46,
the theoretical range is 0-92 and the observed range is 28-60. In various
embodiments, the
number of mrUC genetic risk variants assayed is 36, the theoretical range is 0-
72 and the
observed range is 16-38.
Various other embodiments further comprise prescribing colectomy to subjects
diagnosed with a susceptibility for mrUC and have a genetic risk score at the
high end of
the observed range. In various embodiments, the time to colectomy is lower in
a subject
with a genetic risk score at the high end of the observed range and the time
to colectomy
is higher in a subject with a genetic risk score at the low end of the
observed range. In
various embodiments, the time to colectomy is 10 to 70 months from detection.
Treatment
In various other embodiments of the present invention provides for a method of
treating mrUC in a subject, comprising: obtaining a sample from the subject;
assaying the
sample to detect the presence or absence of mrUC genetic risk variants,
wherein the
mrUC genetic risk variants are selected from the group consisting of SEQ ID
NOs: 1-99;
calculating a genetic risk score based on the detection of the mrUC genetic
risk variants;
diagnosing susceptibility to mrUC based on the calculated risk score, wherein
a subject
has an increased susceptibility to mrUC if the calculated genetic risk score
is high and a
subject has a decreased susceptibility to mrUC if the calculated genetic risk
score is low;
and prescribing colectomy to the subject with an increased susceptibility to
mrUC.
In various embodiments, the genetic risk score is obtained by calculating a
total
number of risk alleles for all the mrUC genetic risk variants assayed, wherein
the risk
allele for each mrUC genetic risk variant assayed is 0, 1 or 2.
Various other embodiments further comprise obtaining a theoretical range and
an
observed range based on the genetic risk score, wherein the theoretical range
consists of
the minimum and maximum number of risk alleles possible based on the number of
mrUC genetic risk variants assayed and wherein the observed range consists of
the actual
minimum and maximum number of risk alleles detected. In various embodiments,
an
CA 02946317 2016-10-18
WO 2015/168699 PCT/US2015/029101
increase in the number of risk alleles detected signifies an increase in
susceptibility to
mrUC. In various embodiments, the number of mrUC genetic risk variants assayed
is 46,
the theoretical range is 0-92 and the observed range is 28-60. In various
embodiments, the
number of mrUC genetic risk variants assayed is 36, the theoretical range is 0-
72 and the
observed range is 16-38.
In various embodiments, the treatment is colectomy and is prescribed to
subjects
diagnosed with a susceptibility for mrUC and have a genetic risk score at the
high end of
the observed range. In various other embodiments, the time to colectomy is
lower in a
subject with a genetic risk score at the high end of the observed range and
the time to
colectomy is higher in a subject with a genetic risk score at the low end of
the observed
range. In various embodiments, the time to colectomy is 10 to 70 months from
detection.
Those in need of treatment include those already with the condition as well as
those prone to have the condition or those in whom the condition is to be
prevented.
Examples of mrUC treatment include, but are not limited to, active
surveillance,
observation, surgical intervention (such as colectomy), drug therapy (anti-
inflammatory
and/or immune system suppressor drugs), and targeted therapy, directed to
genes known
to be involved in IBD, such as, but not limited to those referenced herein
and/or a
combination thereof. Targeted therapy can consist of administering a
composition(s) that
will modify gene regulation by inhibiting or inducing the target gene
expression and/or
activity of the gene.
Kits
Various embodiments of the present invention provide for a kit for prognostic
use, comprising: a single prognostic panel comprising one or more medically
refractive
ulcerative colitis (mrUC) genetic risk variants described in SEQ ID NOs: 1-99.
The present invention is directed to a kit to predict the risk for colectomy,
susceptibility to mrUC and/or treatment of mrUC. The kit is useful for
practicing the
inventive method of determining risk for colectomy in a subject, diagnosing
susceptibility
to mrUC in a subject and/or treatment of a subject. The kit is an assemblage
of materials
or components, including at least one of the inventive compositions. In
various
embodiments, the kit contains a composition including a drug that targets
genes known to
be involved in mrUC, such as the mrUC genetic risk variants, for treatment of
mrUC, as
described above. Thus, in some embodiments the kit contains a composition
including
11
CA 02946317 2016-10-18
WO 2015/168699 PCT/US2015/029101
primers and probes to genetic risk alleles and/or drugs useful in targeting
those genetic
risk alleles.
The exact nature of the components configured in the inventive kit depends on
its
intended purpose. For example, some embodiments are configured for the purpose
of
__ treating mrUC. In one embodiment, the kit is configured particularly for
the purpose of
treating mammalian subjects. In another embodiment, the kit is configured
particularly
for the purpose of treating human subjects. In further embodiments, the kit is
configured
for veterinary applications, treating subjects such as, but not limited to,
farm animals,
domestic animals, and laboratory animals.
Instructions for use may be included in the kit. "Instructions for use"
typically
include a tangible expression describing the technique to be employed in using
the
components of the kit to effect a desired outcome. Optionally, the kit also
contains other
useful components, such as, primers, diluents, buffers, pharmaceutically
acceptable
carriers, syringes, catheters, applicators, pipetting or measuring tools,
bandaging materials
__ or other useful paraphernalia as will be readily recognized by those of
skill in the art.
The materials or components assembled in the kit can be provided to the
practitioner stored in any convenient and suitable ways that preserve their
operability and
utility. For example the components can be in dissolved, dehydrated, or
lyophilized form;
they can be provided at room, refrigerated or frozen temperatures. The
components are
__ typically contained in suitable packaging material(s). As employed herein,
the phrase
"packaging material" refers to one or more physical structures used to house
the contents
of the kit, such as inventive compositions and the like. The packaging
material is
constructed by well-known methods, preferably to provide a sterile,
contaminant-free
environment. As used herein, the term "package" refers to a suitable solid
matrix or
__ material such as glass, plastic, paper, foil, and the like, capable of
holding the individual
kit components. The packaging material generally has an external label which
indicates
the contents and/or purpose of the kit and/or its components.
A variety of methods can be used to determine the presence or absence of an
mrUC
genetic risk variant allele or haplotype. As an example, enzymatic
amplification of
__ nucleic acid from an individual may be used to obtain nucleic acid for
subsequent analysis.
The presence or absence of a variant allele or haplotype may also be
determined directly
from the individual's nucleic acid without enzymatic amplification.
Analysis of the nucleic acid from an individual, whether amplified or not, may
be
performed using any of various techniques. Useful techniques include, without
limitation,
12
CA 02946317 2016-10-18
WO 2015/168699 PCT/US2015/029101
polymerase chain reaction based analysis, sequence analysis and
electrophoretic analysis.
As used herein, the term "nucleic acid" means a polynucleotide such as a
single or double-
stranded DNA or RNA molecule including, for example, genomic DNA, cDNA and
mRNA. The term nucleic acid encompasses nucleic acid molecules of both natural
and
synthetic origin as well as molecules of linear, circular or branched
configuration
representing either the sense or antisense strand, or both, of a native
nucleic acid
molecule.
The presence or absence of a variant allele or haplotype may involve
amplification
of an individual's nucleic acid by the polymerase chain reaction. Use of the
polymerase
chain reaction for the amplification of nucleic acids is well known in the art
(see, for
example, Mullis et al. (Eds.), The Polymerase Chain Reaction, Birkhauser,
Boston,
(1994)).
A TaqmanB allelic discrimination assay available from Applied Biosystems may
be useful for determining the presence or absence of a variant allele. In a
TaqmanB allelic
discrimination assay, a specific, fluorescent, dye-labeled probe for each
allele is
constructed. The probes contain different fluorescent reporter dyes such as
FAM and
VICTM to differentiate the amplification of each allele. In addition, each
probe has a
quencher dye at one end which quenches fluorescence by fluorescence resonant
energy
transfer (FRET). During PCR, each probe anneals specifically to complementary
sequences in the nucleic acid from the individual. The 5' nuclease activity of
Taq
polymerase is used to cleave only probe that hybridize to the allele. Cleavage
separates the
reporter dye from the quencher dye, resulting in increased fluorescence by the
reporter
dye. Thus, the fluorescence signal generated by PCR amplification indicates
which alleles
are present in the sample. Mismatches between a probe and allele reduce the
efficiency of
both probe hybridization and cleavage by Taq polymerase, resulting in little
to no
fluorescent signal. Improved specificity in allelic discrimination assays can
be achieved by
conjugating a DNA minor grove binder (MGB) group to a DNA probe as described,
for
example, in Kutyavin et al., "3'-minor groove binder-DNA probes increase
sequence
specificity at PCR extension temperature, "Nucleic Acids Research 28:655-661
(2000)).
Minor grove binders include, but are not limited to, compounds such as
dihydrocyclopyrroloindole tripeptide (DPI).
Sequence analysis also may also be useful for determining the presence or
absence of a variant allele or haplotype.
Restriction fragment length polymorphism (RFLP) analysis may also be useful
for
13
CA 02946317 2016-10-18
WO 2015/168699 PCT/US2015/029101
determining the presence or absence of a particular allele (Jarcho et al. in
Dracopoli et al.,
Current Protocols in Human Genetics pages 2.7.1-2.7.5, John Wiley & Sons, New
York;
Innis et al.,(Ed.), PCR Protocols, San Diego: Academic Press, Inc. (1990)). As
used
herein, restriction fragment length polymorphism analysis is any method for
distinguishing
genetic polymorphisms using a restriction enzyme, which is an endonuclease
that
catalyzes the degradation of nucleic acid and recognizes a specific base
sequence,
generally a palindrome or inverted repeat. One skilled in the art understands
that the use of
RFLP analysis depends upon an enzyme that can differentiate two alleles at a
polymorphic
site.
Allele-specific oligonucleotide hybridization may also be used to detect a
disease-
predisposing allele. Allele-specific oligonucleotide hybridization is based on
the use of a
labeled oligonucleotide probe having a sequence perfectly complementary, for
example, to
the sequence encompassing a disease-predisposing allele. Under appropriate
conditions,
the allele-specific probe hybridizes to a nucleic acid containing the disease-
predisposing
allele but does not hybridize to the one or more other alleles, which have one
or more
nucleotide mismatches as compared to the probe. If desired, a second allele-
specific
oligonucleotide probe that matches an alternate allele also can be used.
Similarly, the
technique of allele-specific oligonucleotide amplification can be used to
selectively
amplify, for example, a disease-predisposing allele by using an allele-
specific
oligonucleotide primer that is perfectly complementary to the nucleotide
sequence of the
disease-predisposing allele but which has one or more mismatches as compared
to other
alleles (Mullis et al., supra, (1994)). One skilled in the art understands
that the one or more
nucleotide mismatches that distinguish between the disease-predisposing allele
and one or
more other alleles are preferably located in the center of an allele-specific
oligonucleotide
primer to be used in allele-specific oligonucleotide hybridization. In
contrast, an allele-
specific oligonucleotide primer to be used in PCR amplification preferably
contains the
one or more nucleotide mismatches that distinguish between the disease-
associated and
other alleles at the 3' end of the primer.
A heteroduplex mobility assay (HMA) is another well-known assay that may be
used to detect a SNP or a haplotype. HMA is useful for detecting the presence
of a
polymorphic sequence since a DNA duplex carrying a mismatch has reduced
mobility in a
polyacrylamide gel compared to the mobility of a perfectly base-paired duplex
(Delwart et
al., Science 262:1257-1261 (1993); White et al., Genomics 12:301-306 (1992)).
The technique of single strand conformational, polymorphism (SSCP) also may be
14
CA 02946317 2016-10-18
WO 2015/168699 PCT/US2015/029101
used to detect the presence or absence of a SNP and/or a haplotype (see
Hayashi, K.,
Methods Applic.1:34-38 (1991)). This technique can be used to detect mutations
based on
differences in the secondary structure of single-strand DNA that produce an
altered
electrophoretic mobility upon non-denaturing gel electrophoresis. Polymorphic
fragments
are detected by comparison of the electrophoretic pattern of the test fragment
to
corresponding standard fragments containing known alleles.
Denaturing gradient gel electrophoresis (DGGE) also may be used to detect a
SNP
and/or a haplotype. In DGGE, double-stranded DNA is electrophoresed in a gel
containing an increasing concentration of denaturant; double-stranded
fragments made up
of mismatched alleles have segments that melt more rapidly, causing such
fragments to
migrate differently as compared to perfectly complementary sequences
(Sheffield et al.,
"Identifying DNA Polymorphisms by Denaturing Gradient Gel Electrophoresis" in
Innis
et al., supra, 1990).
Other molecular methods useful for determining the presence or absence of a
SNP
and/or a haplotype are known in the art and useful in the methods of the
invention. Other
well-known approaches for determining the presence or absence of a SNP and/or
a
haplotype include automated sequencing and RNAase mismatch techniques (Winter
et al.,
Proc. Natl. Acad. Sci. 82:7575-7579 (1985)). Furthermore, one skilled in the
art
understands that, where the presence or absence of multiple alleles or
haplotype(s) is to be
determined, individual alleles can be detected by any combination of molecular
methods.
See, in general, Birren et al. (Eds.) Genome Analysis: A Laboratory Manual
Volume 1
(Analyzing DNA) New York, Cold Spring Harbor Laboratory Press (1997). In
addition,
one skilled in the art understands that multiple alleles can be detected in
individual
reactions or in a single reaction (a "multiplex" assay). In view of the above,
one skilled in
the art realizes that the methods of the present invention may be practiced
using one or any
combination of the well-known assays described above or another art-
recognized genetic
assay.
One skilled in the art will recognize many methods and materials similar or
equivalent to those described herein, which could be used in the practice of
the present
invention. Indeed, the present invention is in no way limited to the methods
and materials
described. For purposes of the present invention, the following terms are
defined below.
CA 02946317 2016-10-18
WO 2015/168699 PCT/US2015/029101
EXAMPLE S
The following examples are provided to better illustrate the claimed invention
and
are not to be interpreted as limiting the scope of the invention. To the
extent that specific
materials are mentioned, it is merely for purposes of illustration and is not
intended to
limit the invention. One skilled in the art may develop equivalent means or
reactants
without the exercise of inventive capacity and without departing from the
scope of the
invention.
Example]
Overall
Acute severe ulcerative colitis (UC) remains a significant clinical challenge
and the ability to predict, at an early stage, those individuals at risk of
colectomy for
medically refractory UC (mrUC) would be a major clinical advance. As disclosed
herein,
the inventors used a genome-wide association study (GWAS) in a well
characterized
cohort of UC patients to identify genetic variation that contributes to mrUC.
A GWAS
comparing 324 mrUC patients with 537 Non-mrUC patients was analyzed using
logistic
regression and Cox proportional hazards methods. In addition, the mrUC
patients were
compared with 2601 healthy controls.
As further disclosed herein, mrUC was associated with more extensive disease
(p=
2.7x10-6) and a positive family history of UC (p= 0.004). A risk score based
on the
combination of 46 SNPs associated with mrUC explained 48% of the variance for
colectomy risk in the cohort. Risk scores divided into quarters showed the
risk of
colectomy to be 0%, 17%, 74% and 100% in the four groups. Comparison of the
mrUC
subjects with healthy controls confirmed the contribution of the major
histocompatibility
complex to severe UC (peak association: rs17207986 (SEQ ID NO: 47), p= 1.4x10-
16)
and provided genome-wide suggestive association at the TNFSF15 (TL1A) locus
(peak
association: rs11554257 (SEQ ID NO: 48), p= 1.4x10-6). A SNP-based risk
scoring
system, identified herein by GWAS analyses, can provide a useful adjunct to
clinical
parameters for predicting natural history in UC. Furthermore, discovery of
genetic
processes underlying disease severity can identify pathways for novel
therapeutic
intervention in severe UC.
16
CA 02946317 2016-10-18
WO 2015/168699 PCT/US2015/029101
Example 2
UC Cases
Ulcerative Colitis (UC) subjects (n= 929) were recruited at Cedars Sinai-
Medical
Center Inflammatory Bowel Disease Center following informed consent after
approval by
the Institutional Review Board. UC diagnosis was based on standard criteria
31. UC
subjects requiring colectomy for severe disease refractory to medical
therapies (including
intravenous corticosteroids, cyclosporine, and biologic therapies) were
classified as
medically refractory UC (mrUC). Subjects requiring colectomy where the
indication was
for treatment of cancer/dysplasia, in addition to subjects not requiring
colectomy, were
classified as Non-mrUC. Subjects who required colectomy for mrUC and were
subsequently found to have evidence of dysplasia or carcinoma in the resected
colon were
classified as mrUC (n= 3). For the mrUC cohort, time from diagnosis to date of
colectomy was collected; time from diagnosis to last follow-up visit was
obtained for the
Non-mrUC cohort. Samples which did not genotype successfully (n= 16),
exhibited
gender mismatch (n= 9) or cryptic relatedness (n= 13), or were considered
outliers by
principal components analysis (n= 30) were excluded. Following these measures,
861 UC
subjects (mrUC n= 324; Non-mrUC n= 537) were included in the analyses.
Example 3
Non-IBD Controls
Controls were obtained from the Cardiovascular Health Study (CHS), a
population-based cohort study of risk factors for cardiovascular disease and
stroke in
adults 65 years of age or older, recruited at four field centers. 5,201
predominantly
Caucasian individuals were recruited in 1989-1990 from random samples of
Medicare
eligibility lists, followed by an additional 687 African-Americans recruited
in 1992-1993
(total n= 5,888). CHS was approved by the Institutional Review Board at each
recruitment
site, and subjects provided informed consent for the use of their genetic
information. A
total of 2,601 Caucasian non-IBD control subjects who underwent GWAS were
included
in these analyses. African-American CHS participants were excluded from
analysis due to
insufficient number of ethnically-matched cases.
17
CA 02946317 2016-10-18
WO 2015/168699 PCT/US2015/029101
Example 4
Genotyping
All genotyping was performed at the Medical Genetics Institute at Cedars-Sinai
Medical Center using Infinium technology (IIlumina, San Diego, CA). UC cases
were
genotyped with either the HumanCNV370-Quad or Human610-Quad platform; controls
were genotyped with the HumanCNV370-Duo platform. Identity-by-descent was used
to
exclude related individuals (Pi-hat scores >0.5; PLINK). Average genotyping
rate among
cases and controls retained in the analysis was >99.8% and >99.2%,
respectively. Single
nucleotide polymorphisms (SNPs) were excluded based on: test of Hardy-
Weinberg
Equilibrium p <10-3; SNP failure rate >10%; MAF <3%; SNPs not found in dbSNP
Build
129. 313,720 SNPs passed quality control measures and were common in all data
sets.
Example 5
Population Stratification
Principal components analysis (Eigenstrat as implemented in Helix Tree)
(Golden Helix, Bozeman, MT) was conducted to examine population
stratification.
Extreme outliers, defined as subjects with more than two standard deviations
(SD) away
from the distribution of the rest of the samples for any component, were
removed. All
African-American participants identified by principal components analysis were
excluded
from these analyses. Genetic heterogeneity following correction for population
sub-
structure was low, with estimated genomic inflation factors (kGC) of 1.04 and
1.06 for
mrUC vs. Non-mrUC, and mrUC cases vs. Non-IBD controls analyses, respectively.
Example 6
mrUC vs. Non-mrUC: Survival Analysis and Risk Modeling
Single marker association analysis of mrUC vs. Non-mrUC (analysis-I) was
performed using a logistic regression model correcting for population
stratification using
20 principal components as covariates (PLINK v1.06). Association between
medically
refractory disease (mrUC) and the top 100 SNPs together (as determined by the
lowest
corrected p-values) from analysis-I were tested using a stepwise logistic
regression model.
SNPs were further analyzed by Cox proportional hazards regression utilizing
time-to
information, as described for UC cases (using the step and glm, and coxph
functions,
respectively, in R v2.9.0). 37 SNPs identified with logistic regression p<0.05
and Cox
18
CA 02946317 2016-10-18
WO 2015/168699 PCT/US2015/029101
proportional hazards p <0.1 were retained in the risk model. The 100 SNPs (p
<3x10-4)
evaluated from analysis-I are listed herein (Table 1). A genome-wide Cox
proportional
hazards regression analysis (analysis-II) was then performed on a subset of
the UC cohort
(mrUC subjects with colectomy <60 months, n= 187; Non-mrUC followed up >60
months, n= 328) correcting for population stratification using two principal
components as
covariates (PLINK). The top 65 SNPs (8 of which overlap with the 100 SNPs from
analysis-I above) were tested together (using coxph function in R). The 65
SNPs (p
<1x10-4) from analysis-II are listed herein (Table 2). From these 65 SNPs, 9
SNPs were
identified (p <3x10-4) and combined with the 37 SNPs from analysis-I to
identify a final
risk model consisting of 46 SNPs (see Figure 1 for schematic; Table 3). A
genetic risk
score was calculated from the total number of risk alleles (0, 1, or 2) across
all 46 risk
SNPs (theoretical range: 0-92). Risk score (observed range: 28-60) was divided
into
quarters: scores 28-38 (risk-A); scores 39-45 (risk-B); scores 46-52 (risk-C);
and scores
53-60 (risk-D). Receiver operating characteristic (ROC) curve and area under
the ROC
curve (AUC) were calculated using R software v2.9.0, including packages
survival and
survivalROC 39-41. Sensitivity and specificity curves, positive and negative
predictive
values, positive (sensitivity/ 1- specificity) and negative likelihood ratio
(1-
sensitivity/specificity) were all calculated using the R package ROCR 42. 1000-
fold
replication of 10-fold cross-validation was implemented to validate the fitted
logistic
regression model. Mean sensitivity and specificity were then re-calculated
using the 1000
replicated samples. Bootstrap method with 1000-fold replication was utilized
for
estimating variability of hazard ratio estimated from the Cox regression
model. The hazard
ratio in survival analysis is the effect of an explanatory variable on the
hazard or risk of an
event.
Table 1: Top 100 SNPs from Analysis I
Chr* SNP Position* Minor P-value Odds Stat Loci**
allele Ratio
1 rs260970 39323829 G 2.38E- 1.594 3.675
MACF116439101
04
1 rs6697447 54219515 A 2.06E- 0.4481 -3.712 HSPB111YIPF11
04 Clorf831
1 rs746503 54842574 A 2.27E- 1.475 3.687 ACOT111FAM151A1
04 Clorf175
1645442 1
1 rs2275612 95140004 A 1.87E- 1.838 3.736 CNN31SLC44A31
04 64689617299701
1 rs4847368 95149626 G 7.75E- 1.976 3.952 CNN316468961729970
19
CA 02946317 2016-10-18
WO 2015/168699 PCT/US2015/029101
Chr* SNP Position* Minor P-value Odds Stat Loci**
allele Ratio
05 1
1 rs2298162 95221621 G 2.85E- 0.6668 -3.628 CNN31ALG141
04
1 rs7550055 157045388 C 1.35E- 1.571 3.818 MNDAI0R6N2 1
04 OR1OAA1P1OR6K4P1
OR6N110R6K3 1
OR6K5P16463771
1 rs7367845 224512151 A 2.55E- 1.475 3.657 ACBD3 ILIN91
04
1 rs9286999 224561138 A 1.61E- 1.491 3.773 LIN911001288321
04
2 rs892878 137588330 A 2.78E- 0.6726 -3.635 THSD7B1
04
2 rs1560579 137592445 A 2.67E- 0.6326 -3.646 THSD7B1
04
2 rs9287461 137593668 G 8.64E- 0.6156 -3.926 THSD7B1
05
2 rs958323 137606935 G 1.85E- 0.6408 -3.738 THSD7B1
04
2 rs1483148 142036240 C 1.59E- 0.6225 -3.777 LRP1B1
04
2 rs1448901 206961885 G 2.13E- 1.673 4.251 ADAM231
05
2 rs7565690 224105705 A 2.10E- 0.5414 -3.706
04
2 rs4487082 229432205 G 2.03E- 0.4091 -3.716
04
3 rs403961 1575422 G 2.09E- 1.495 3.708
04
3 rs924022 65824936 G 1.98E- 0.5251 -3.721 MAGIll
04
3 rs10511119 79943297 G 2.87E- 0.6796 -3.627
04
3 rs9682694 114378369 G 1.47E- 1.549 3.797 BOC1
04
3 rs4839637 144422638 A 2.15E- 1.501 3.701
04
4 rs2286461 15572771 G 1.86E- 1.487 3.738 PROM11FGFBP11
04 FGFBP2 11001300671
4 rs12650313 41401850 A 2.57E- 1.877 3.655 LIMCH111001286541
04
4 rs1546318 79168396 C 9.31E- 1.932 3.908 FRAS113916701
05 1001282971
4 rs1393644 79175731 C 9.19E- 1.93 3.911 FRAS113916701
05 1001282971
4 rs1399403 108639264 A 1.60E- 1.538 3.775
04
4 rs11098020 110122894 A 1.90E- 1.777 3.732 COL25A1l
04
4 rs7675371 116049368 A 2.64E- 0.58 -3.648 NDST41
04
4 rs6821443 122566710 C 2.90E- 0.669 -3.624 QRFPR13916921
CA 02946317 2016-10-18
WO 2015/168699 PCT/US2015/029101
Chr* SNP Position* Minor P-value Odds Stat Loci**
allele Ratio
04 7291091729112 1
rs3846599 10308821 A 1.06E- 1.517 3.877 MARCH61CCT5 1
04 FAM173B1MIR3781
5 rs12652447 15727635 A 3.86E- 1.524 4.115 FBXL71
05
5 rs6596684 105972832 G 1.13E- 1.53 3.861 3455711
04
5 rs6870711 126446993 A 1.98E- 2.404 3.722 MARCH3
14012071
04
6 rs1536242 6876009 A 2.71E- 0.62 -3.642
04
6 rs17207986 32187545 G 4.71E- 2.297 4.069 ATF6B
IRNF51PPT21
05 EGFL81653033 1
6 rs3734263 34946407 G 2.37E- 0.4474 -3.676 TAF111ANKS1A1
04 UHRF1BP11
6 rs9470224 36248614 A 1.70E- 1.843 3.759 BRPF3IPNPLA1l
04
6 rs777649 68925053 A 1.10E- 1.618 4.396 6429021
05
6 rs3777505 75937343 A 1.04E- 2.526 3.881 COL12A1l
04
6 rs6908055 107015887 G 1.13E- 1.67 3.861 AIM11
04
6 rs9400010 107027893 G 1.44E- 1.56 3.802 AIM11
04
7 rs11760555 12563060 G 2.92E- 0.6713 -3.622 SCIN1
04
7 rs4722456 25338225 A 9.50E- 0.6644 -3.903 1001310161
05
7 rs13244827 131735522 G 1.68E- 0.3415 -3.763 PLXNA41
04
7 rs851685 147125736 A 2.25E- 1.546 3.69 CNTNAP2 1
04
8 rs2978310 2701133 A 2.68E- 1.496 3.644
04
8 rs1471474 76216530 A 1.12E- 0.6541 -3.864
04
8 rs6994721 76220268 G 7.63E- 0.6521 -3.956
05
8 rs4734754 105347978 C 1.57E- 0.6496 -3.78 TM7SF41
04
8 rs4734757 105355266 A 1.95E- 0.6537 -3.725 TM7SF41
04
8 rs263241 131931602 A 2.44E- 1.456 3.669 ADCY81
04
9 rs7861972 6759692 G 2.34E- 0.473 -3.68 JMJD2C1SNRPEL11
04
9 rs11265961 91638884 A 2.12E- 1.586 4.252
05
9 rs2145929 116621761 G 2.44E- 1.634 3.668 TNFSF1516452661
04 100129633 1
9 rs10817934 118589872 A 1.39E- 0.6364 -3.811 ASTN2 1
21
CA 02946317 2016-10-18
WO 2015/168699 PCT/US2015/029101
Chr* SNP Position* Minor P-value Odds Stat Loci**
allele Ratio
04
rs3793792 50520169 A 2.94E- 1.481 3.62 CHATIClOorf531
04
10 rs518525 129520863 A 1.65E- 1.488 3.767 PTPRE13877201
04
11 rs2403456 11134390 A 1.08E- 2.413 3.872
04
11 rs4356200 37489093 A 6.49E- 0.6231 -3.994 100132895 1
05
11 rs1461898 37546808 A 5.46E- 0.5881 -4.035
05
11 rs1075025 37582318 G 2.38E- 0.6141 -3.675
04
11 rs767289 37624038 G 3.99E- 0.6274 -4.108 1001326311
05
11 rs10837504 40775682 A 1.50E- 1.649 3.792
04
11 rs6591765 62674829 A 7.84E- 0.6468 -3.949 SLC22A241
05
11 rs7949840 62741273 G 6.75E- 0.6511 -3.985 SLC22A241SLC22A25
05 1SLC22A101
11 rs11231409 62741444 G 7.46E- 0.6527 -3.961 SLC22A241SLC22A25
05 1SLC22A101
12 rs887357 3344906 C 2.16E- 0.5947 -3.699 64311917282301
04 100128253 1
12 rs970063 13424516 A 1.87E- 1.48 3.736 C12orf361
04
12 rs12581840 19725418 G 1.05E- 0.6566 -3.88
04
12 rs526058 24326688 A 6.51E- 0.6009 -3.994
05
12 rs1144720 32157518 G 2.82E- 1.571 4.188 BICD117294571
05
12 rs1613650 32169080 G 2.65E- 1.507 3.647 BICD117294571
04
12 rs2683471 32171607 A 2.14E- 1.518 3.702 BICD117294571
04
14 rs1956388 28202628 A 1.32E- 0.672 -3.822
04
14 rs11156667 30906111 A 1.75E- 0.6296 -4.294 HEATR5A17288521
05
14 rs9323262 53863964 A 2.69E- 0.5639 -3.644 CDKN31
04
14 rs10133064 85844148 A 2.03E- 0.4618 -3.715
04
14 rs35795554 85854768 C 2.56E- 0.4743 -3.656
04
rs7172534 24855745 G 2.52E- 1.503 3.661 GABRG3 1
04
15 rs965355 57847116 G 1.04E- 1.584 3.881 BNIP2
11001301071
04
15 rs965353 57847498 G 1.38E- 1.569 3.811 BNIP2
11001301071
22
CA 02946317 2016-10-18
WO 2015/168699 PCT/US2015/029101
Chr* SNP Position* Minor P-value Odds Stat Loci**
allele Ratio
04
15 rs10519111 59169989 G 1.08E- 1.719 3.872 RORA1
04
15 rs990422 98377291 A 2.97E- 1.533 3.618 ADAMTS171
04
15 rs1585933 98403238 G 6.14E- 1.605 4.007 ADAMTS171
05
16 rs305087 84539747 G 4.72E- 0.5364 -4.069 100131952 1
05
17 rs759258 52483547 A 2.39E- 1.561 3.674 AKAP11
04
18 rs3848490 2326366 A 2.97E- 1.48 3.618
04
18 rs8088744 64685792 A 1.72E- 0.6688 -3.757 CCDC102B 1
04
19 rs2967682 8644532 A 9.96E- 0.6538 -3.892 MY01F1ADAMTS101
05 OR2Z113908801
19 rs11085825 12868458 A 6.05E- 0.6309 -4.011
05
19 rs2293683 12900284 A 8.35E- 0.636 -3.934
05
19 rs1010222 12909608 A 1.52E- 0.6469 -3.788 CALR1DNASE21
04 FARSA1NFIX1
RAD23A1KLF11
DAND5 1SYCE2 1
19 rs4808408 15881376 G 1.47E- 0.628 -4.334 CYP4F21CYP4F111
05 OR10H414405111
6465961729645 1
7296541
19 rs12459140 15882888 G 1.52E- 0.6287 -4.326 CYP4F21CYP4F111
05 OR10H414405111
6465961729645 1
7296541
20 rs6034134 15182479 A 1.00E- 0.6518 -3.89 MACROD21
04
20 rs10485594 19772393 A 6.91E- 2.126 3.979 RIN216442981
05
20 rs6059101 31182314 A 6.54E- 0.601 -3.993 C20orf711C20orf701
05 3177161391242 1
20 rs6059104 31185354 A 1.55E- 0.6545 -3.784 C20orf711C20orf701
04 3177161391242 1
22 rs909502 47050966 A 2.65E- 1.483 3.647
04
Table 2: Top 65 SNPs from Analysis II
Chr SNP Position129 Minor P_value Loci
_allele
1 rs1392127 55788503 A 5.25E-05 4007541
1 rs2298162 95221621 G 1.58E-05 CNN31ALG14 1
23
CA 02946317 2016-10-18
WO 2015/168699 PCT/US2015/029101
Chr SNP Position129 Minor P_value Loci
_allele
2 rs1448901 206961885 G 5.08E-05 ADAM231
2 rs3791994 207718164 A 2.66E-05 KLF71
3 rs900569 41834977 G 1.91E-05 ULK41
3 rs6796430 73950170 A 5.77E-05
3 rs9843732 135505746 G 6.00E-05 RYK1
4 rs1013300 13204657 G 9.52E-05 NKX3-21BOD1L 12855481
4 rs1491262 13301398 A 9.20E-05 BOD1L16448681
4 rs17476066 15461202 G 4.87E-05 CD381
4 rs2608816 39103204 G 9.90E-05 RFC11LIAS 1KLB 1642885 1
4 rs6811556 180521808 A 9.46E-06
rs6892546 5873530 G 7.03E-05
5 rs4571457 107862146 G 7.40E-06
6 rs9468256 28003483 A 8.81E-05 HIST1H1B IHIST1H2AK1
HIST1H2AMIHIST1H3I1
HIST1H3JIHIST1H4K1HIST1H4L
10R2B610R2W6P1OR2W4P1
OR2B2 1
6 rs2116984 28040741 A 4.39E-05 HIST1H1B IHIST1H2AM1
HIST1H3IIHIST1H3J1HIST1H4L 1
0R2B610R2W4P 1 OR2W2P 1
OR2B7P10R2B21
6 rs1012411 30440534 C 8.56E-05 HCG18164649111001291921
6 rs9501030 30907378 A 4.27E-06 DDR11GTF2H4 IIER31FLOT11
VARS21646553 IMIR5881
6 rs9295930 30957801 A 9.38E-05 DDR11GTF2H4 1VARS2 1DPCR11
64655316465701M1R58817297781
6 rs10947114 31010160 A 7.87E-05 VARS2 1DPCR11 SFTA21MUC211
64656316465701M1R58817297781
7297921
6 rs537160 32024379 A 1.44E-05 CFB 1C21C4A1C4B INEUll
SKIV2L1RDBPISTK19IEHMT21
SLC44A4 1ZBTB12 1
6 rs4151657 32025519 G 5.17E-05 CFB 1C21C4A1C4B INEUll
SKIV2L1RDBPISTK19IEHMT21
SLC44A4 1ZBTB12 1
6 rs630379 32030233 T 3.11E-05 CFB 1C21C4A1C4B INEUll
SKIV2L1RDBPISTK19IEHMT21
SLC44A4 1ZBTB12 1
6 rs9267845 32301676 T 4.04E-05
6 rs6910071 32390832 G 1.84E-05 NOTCH4 1C6orf101
6 rs2894253 32453518 C 1.95E-05 HLA-DRA1C6orf1016466681
6 rs34330585 32498681 G 4.30E-05 HLA-DRA1C6orflOIBTNL21
6466681
7 rs2158767 17019311 G 7.51E-05
7 rs1178163 18754088 A 5.55E-06 HDAC91
7 rs11764116 18766938 A 2.26E-06 HDAC9
7 rs2389992 18903915 G 1.30E-05 HDAC9
7 rs929351 81695829 C 8.33E-06 CACNA2D11
8 rs2980654 6480608 G 6.62E-05
8 rs6474026 56956047 G 7.63E-05 LYN1
8 rs2383847 73166848 G 8.52E-05 TRPAll
24
CA 02946317 2016-10-18
WO 2015/168699 PCT/US2015/029101
Chr SNP Position129 Minor P_value Loci
_allele
8 rs2954870 75995015 G 8.76E-05 CRISPLD11
9 rs3118292 25133480 G 2.83E-05
9 rs1331501 92432152 G 9.55E-05 DIRAS21340515 1
11 rs1783983 57177356 A 8.95E-05
11 rs1031232 57628857 G 6.93E-05
11 rs6591765 62674829 A 4.62E-05 SLC22A24 1
11 rs7949840 62741273 G 6.67E-05 SLC22A24 1SLC22A25
1SLC22A10
1
11 rs11231409 62741444 G 8.18E-05 SLC22A241SLC22A25 1SLC22A10
1
12 rs2098102 5026839 A 9.72E-05 KCNA5 13902821
12 rs906724 126243850 A 5.15E-05 1001325641
13 rs4769736 28876751 G 7.36E-05 K1AA0774 1
13 rs10507842 75481600 G 8.61E-05
13 rs7319358 78448935 A 3.34E-06
14 rs1956388 28202628 A 2.23E-06
14 rs2179891 28215603 A 6.90E-05 FOXG11C140rf23 1
14 rs8020281 94436179 A 7.28E-05
16 rs1421069 51755435 G 8.00E-05 CHD91
16 rs2388011 51770920 G 9.60E-05 CHD91
16 rs3815548 51879235 G 9.94E-05 CHD9144177011001328751
16 rs1424203 59355718 A 3.55E-05
17 rs9898519 24865310 G 3.62E-05 TAOK11TP53113 1ANKRD13B1
6459421
17 rs3744624 24885455 G 1.48E-05 TAOK11TP53113 1ANKRD13B1
6459421
18 rs669924 38725419 A 7.79E-05 RIT21
19 rs2116941 10195443 A 3.25E-05
20 rs755171 31176251 G 9.70E-05 C20orf711C20orf7013177161
3912421
20 rs6059101 31182314 A 3.71E-06 C20orf711C20orf7013177161
3912421
20 rs6059104 31185354 A 9.73E-05 C20orf711C20orf7013177161
3912421
21 rs2831462 28370367 A 4.17E-05
22 rs916234 46165555 G 5.77E-05
22 rs2051594 47259952 A 5.43E-05 6432661643325 1
Table 3: 46 SNPs associated with the risk model for mrUC
Chr SNP Position SEQ ID NO Risk_ Loci
129 allele
1 rs746503 54842574 1 A ACOT111FAM151A1C1orf175 1
6454421
1 rs2275612 95140004 2 A CNN31SLC44A3164689617299701
1 rs7550055 157045388 3 C MNDA1 0R6N2 10R2AQ1P 1
OR1OAA1P1OR6K4P1OR6N1
10R6K3 10R6K5P16463771
1 rs7367845 224512151 4 A ACBD3 IMIXHILIN91100128832 1
CA 02946317 2016-10-18
WO 2015/168699 PCT/US2015/029101
Chr SNP Position SEQ ID NO Risk_ Loci
129 allele
2 rs1448901 206961885 5 G ADAM2311001328491
2 rs4487082 229432205 6 A 2q36.3
3 rs900569 41834977 7 G ULK41
3 rs924022 65824936 8 A MAGI11
3 rs9843732 135505746 9 G RYK1
4 rs2286461 15572771 10 G PROM11FGFBP11FGFBP21
1001300671
4 rs12650313 41401850 11 A LIMCH111001286541
4 rs1399403 108639264 12 A 4q25
4 rs7675371 116049368 13 G NDST41
rs3846599 10308821 14 A MARCH61CCT5 1FAM173B1M1R378
1
5 rs6596684 105972832 15 G 3455711
6 rs1536242 6876009 16 G 6p25.1
6 rs17207986 32187545 17 G ATF6B1RNF5 1PPT2 1EGFL81653033
1
6 rs777649 68925053 18 A 6429021
7 rs11764116 18766938 19 A HDAC91
7 rs4722456 25338225 20 G 1001310161
7 rs929351 81695829 21 A CACNA2D11
8 rs2980654 6480608 22 G ANGPT2 1AGPAT5 1MCPH11
100131112 11001323011
8 rs6994721 76220268 23 A 8q21.11
8 rs4734754 105347978 24 A RIMS21TM7SF41
9 rs7861972 6759692 25 A JMJD2C1SNRPEL11
9 rs3118292 25133480 26 G 9p21.3
9 rs10817934 118589872 27 G ASTN2 1
11 rs2403456 11134390 28 A 11p15.3
11 rs1461898 37546808 29 G 100132895 11001326311
11 rs6591765 62674829 30 G SLC22A24 SLC22A251SLC22A101
12 rs887357 3344906 31 A 64311917282301100128253 1
12 rs526058 24326688 32 G 12p12.1
13 rs7319358 78448935 33 A 13q31.1
14 rs1956388 28202628 34 G 14q12
14 rs11156667 30906111 35 G GPR331HEATR5A1NUBPL1
C14orf1261728852 1
14 rs10133064 85844148 36 C 14q31.3
14 rs8020281 94436179 37 A 14q32.13
rs965353 57847498 38 G BNIP2 11001301071GTF2A2 1
16 rs305087 84539747 39 A 100131952 1
17 rs759258 52483547 40 A AKAP11
19 rs2967682 8644532 41 C MY01F1ADAMTS1010R2Z11
3908801
19 rs2293683 12900284 42 G
rs6034134 15182479 43 C MACROD21
20 rs10485594 19772393 44 A RIN216442981
20 rs6059104 31185354 45 G PL1JNC1C20orf711C20orf701
C20orf186131771613912421
21 rs2831462 28370367 46 A 21q21.3
26
CA 02946317 2016-10-18
WO 2015/168699 PCT/US2015/029101
Example 7
mrUC vs. Non-IBD Controls: Regression Analysis
Single marker analysis of genome-wide data for mrUC cases vs. Non-IBD
Caucasian controls from CHS (analysis-III) was performed as before, using
logistic
regression correcting for 20 principal components (PLINK).
Example 8
UC subject demographics
Complete temporal data was available on 861 UC subjects (mrUC n= 324; Non-
mrUC n= 537). The demographic data of the cohort is summarized herein. The
inventors
observed no differences in gender, median age of onset of disease, and smoking
status
between the medically refractory and Non-mrUC subjects. There was a
significant
difference in our median disease duration (p= 7.4x10-9), with the time from
diagnosis to
last follow-up in the Non-mrUC cohort nearly double the time from diagnosis to
colectomy in our mrUC subjects. Additionally, there was a significantly higher
incidence
of disease that extended proximal to the splenic flexure (p= 2.7x10-6) in the
mrUC group
when compared to Non-mrUC, consistent with previously published data. The
inventors
identified a novel association between a family history (first or second
degree relative) of
UC and the development of mrUC (p= 0.004).
Example 9
Forty-six SNP risk model is associated with mrUC and predicts earlier
progression to
colectomy
The inventors performed a GWAS on 324 mrUC and 537 Non-mrUC subjects.
Results of this analysis (analysis-I) are given herein and discussed below.
Following
identification of single markers associated with mrUC, the inventors proceeded
to a
multivariate approach. Beginning with the top 100 results from analysis-I (p
<3x10-4),
the inventors performed a stepwise logistic regression and identified 64 SNPs
(p <0.05)
that together were associated with medically refractory disease (mrUC) and
were carried
forward to survival analysis. Of these 64 SNPs, 37 SNPs remained (Cox
proportional
hazards regression p <0.1; OR 1.2- 1.8), which explained 40% of the variance
for mrUC.
In order to elucidate the maximum discrimination, i.e. greatest percentage of
the variance,
the inventors further performed a genome-wide Cox proportional hazards
regression
27
CA 02946317 2016-10-18
WO 2015/168699 PCT/US2015/029101
analysis (analysis-II) on a subset of the UC cohort to identify SNPs involved
in earlier
progression to colectomy. Testing together the top 65 SNPs from this analysis
(p <1x10-
4), the inventors identified nine SNPs with Cox proportional hazards p <3x10-4
(individual OR ranged from 1.4-1.6), explaining 17% of the variance. Beginning
with the
previously identified 37 risk SNP model, these 9 SNPs were added sequentially
to the
model. This analysis resulted in the final risk model of 46 SNPs (OR for MR-
UC for each
individual SNP ranged from 1.2-1.9), which explained 48% of the variance for
colectomy
in the mrUC cohort.
The inventors calculated a genetic risk score from the total number of risk
alleles
across all 46 risk SNPs (theoretical range: 0-92). The observed risk score
ranged from 28-
60, and was significantly associated with mrUC (logistic regression and Cox
proportional
hazards pvalues <10-16). An ROC curve using this risk score gave an AUC of
0.91. The
sensitivity of the fitted model for mrUC was 0.793, with a specificity of
0.858. Using
1000 replicates of the 10-fold cross-validation data, they obtained a mean
sensitivity of
0.789 (SD= 0.0067) and mean specificity of 0.859 (SD= 0.002). This indicates
that the
fitted model was robust and only ¨0.4% over-fitting was observed. The hazard
ratio was
estimated to be 1.313 from the Cox regression model. 1000 replicates of
bootstrapped
samples gave an estimated hazard ratio of 1.314 (SD= 0.017) (Table 4).
Table 4
Sensitivity/Specificity Sensitivity 0.793
(cut-off=.5) Specificity 0.858
Hazard Ratio: 1.313
1000 times of 10 fold Cross-Validation data sets with logistic regression
Variable N Mean Std Dev Minimum Maximum
Sensitivity 1000 0.789 0.0067 0.758 0.793
Specificity 1000 0.859 0.0021 0.858 0.87
1000 fold Bootstrapping:
Mean Std Dev Minimum Maximum
1000 1.314 0.017 1.269 1.372
Based on the genetic risk scores, the inventors grouped the UC cohort into
four
risk categories; less than 1% of cases in the lowest risk category (risk-A)
were mrUC and
the percentage of mrUC increased to ¨17%, ¨74% and 100% in risk-B, -C and ¨D
groups,
28
CA 02946317 2016-10-18
WO 2015/168699 PCT/US2015/029101
respectively (x2 test for trend p <2.2x10-16; Figure 2A). The median time to
colectomy for
risk-C and -D categories was 72 months and 23 months, respectively.
Progression to
colectomy within 2 and 5 years of diagnosis may be more clinically relevant
and while no
individuals in the risk-A category had undergone colectomy at either 2 or 5
years after
diagnosis, the respective incidence of mrUC at 2 years for risk groups -B, -C
and -D was
3.1%, 19.1%, and 62%, respectively, and at 5 years was 8.3%, 50%, and 80%,
respectively (Figure 2B). At five years from diagnosis, either the total risk
score (AUC
0.86) or the risk category (AUC 0.82) are able to predict patients that will
require surgery.
The operating characteristics of the risk score system are shown herein. A
score of 44 and
47 can be used to generate a test with a sensitivity (to exclude a diagnosis
of colectomy)
and specificity (to include a diagnosis) of over 90%, respectively. Loci
corresponding to
the 46 SNPs in the risk model include several compelling candidate genes for
UC severity
and suggest potential biological pathways for further avenues of study. As
each risk SNP
contributes modestly to the overall risk of mrUC (OR 1.2-1.9), this work
supports the
paradigm that a group of SNPs, identified by GWAS and combined together may
account
for a large proportion of the genetic contribution to a complex phenotype (48%
of the
variance for risk in this study) to provide a risk score with clinical
utility.
Example 10
MHC region and TLIA (TNFSF15) contribute to UC severity.
Association analyses between 324 UC subjects with mrUC and 2,601 population
matched controls confirmed a major contribution of the major
histocompatibility (MHC)
on chromosome 6p to the development of severe UC (analysis-III; Table 5). Ten
SNPs in
MHC reached a priori defined level of genome-wide significance (p <5x10-7; 87
SNPs
with p <1x10-3), with peak association at rs17207986 (SEQ ID NO: 47; p= 1.4x10-
16).
Three SNPs on chromosome 9q, a locus which contains the known IBD
susceptibility
gene TNFSF15 (TL1A), achieved genome-wide suggestive significance (p <5x10-5),
with
the most significant association seen at rs11554257 (SEQ ID NO: 48; p=1.4x10-
6).
Table 5: MHC region associated SNPs
Chr SNP Position* Minor P- Odds Stat Loci**
* Allele value Ratio
6 rs3132679 30183822 A 9.40E- 0.5327 -3.906 TRIM311RNF391
05
TRIM15 1TRIM401
6 rs9468692 30227869 A 6.64E- 1.767 3.404 TRIM10 TRIM31
29
CA 02946317 2016-10-18
WO 2015/168699 PCT/US2015/029101
Chr SNP Position* Minor P- Odds Stat Loci**
* Allele value Ratio
04 RNF391TRIM151
TRIM401
6 rs1012411 30440534 C 2.98E- 1.526 3.617 HCG1816464911
04 100129192 1
6 rs2040450 30442318 A 3.54E- 1.65 3.572 HCG1816464911
04 100129192 1100129772
1
6 rs2524211 30458639 A 4.75E- 1.612 3.495 HCG1816464911
04 64652011001291921
100129772 1
6 rs9261761 30480966 A 2.28E- 0.5646 -3.685 HLA-E1MICC1HCG18
04 16465201100129192 1
100129772 1
6 rs9261817 30486580 C 2.33E- 0.565 -3.68 HLA-E1MICC1HCG18
04 16465201100129192 1
100129772 1
6 rs9261821 30487053 G 2.46E- 0.5663 -3.667 HLA-E1MICC1HCG18
04 16465201100129192 1
100129772 1
6 rs9261846 30490419 G 2.42E- 0.5662 -3.671 HLA-E1MICC1HCG18
04 16465201100129192 1
100129772 1
6 rs9261847 30490626 C 4.44E- 0.5793 -3.512 HLA-E1MICC1HCG18
04 16465201100129192 1
100129772 1
6 rs9261860 30492482 A 4.17E- 0.5742 -3.529 HLA-E1MICC1HCG18
04 16465201100129192 1
100129772 1
6 rs9261862 30492717 G 2.17E- 0.5636 -3.698 HLA-E1MICC1HCG18
04 16465201100129192 1
100129772 1
6 rs9261871 30493873 G 2.27E- 0.5646 -3.687 HLA-E1MICC1HCG18
04 16465201100129192 1
100129772 1
6 rs9261919 30499702 A 2.34E- 0.5654 -3.679 HLA-E1MICC1HCG18
04 16465201100129192 1
100129772 1
6 rs9261923 30500139 A 2.17E- 0.5636 -3.698 HLA-E1MICC1HCG18
04 16465201100129192 1
100129772 1
6 rs9261926 30500385 A 2.47E- 0.5665 -3.665 HLA-E1MICC1HCG18
04 16465201100129192 1
100129772 1
6 rs9261947 30502607 A 2.60E- 0.5601 -3.652 HLA-E1MICC16465201
04 100129192 1100129772
1
6 rs9501447 30505819 G 3.36E- 0.5737 -3.586 HLA-E1MICC16465201
04 100129772 1
6 rs1079541 30514735 A 2.13E- 0.5634 -3.703 HLA-E1MICC16465201
04 100129772 1
6 rs9501467 30516949 A 4.48E- 0.5935 -3.51 HLA-E1MICC16465201
04 100129772 1
6 rs9295871 30519068 G 2.91E- 0.5704 -3.623 HLA-E1MICC16465201
04 100129772 1
CA 02946317 2016-10-18
WO 2015/168699 PCT/US2015/029101
Chr SNP Position* Minor P- Odds Stat Loci**
* Allele value Ratio
6 rs9295873 30522214 G 2.18E- 0.5638 -3.698 HLA-E MICC1646520
I
04 100129772 1
6 rs9918306 30527750 A 2.30E- 0.5732 -3.683 HLA-E MICC1646520
I
04 100129772 1
6 rs9295886 30529376 A 3.91E- 0.5751 -3.546 HLA-E MICC1646520
I
04 100129772 1
6 rs3540751 30531202 G 2.13E- 0.5634 -3.703 HLA-E MICC1646520
I
04 100129772 1
6 rs3410187 30531337 A 1.96E- 0.5615 -3.724 HLA-E MICC1646520
I
5 04 100129772 1
6 rs3398639 30532053 G 2.16E- 0.5637 -3.7 HLA-E MICC1646520 I
3 04 100129772 1
6 rs9501336 30535489 A 2.17E- 0.5639 -3.698 HLA-E PRR3IMICC1
04 646520 1001297721
6 rs1196661 30537012 C 2.49E- 0.5668 -3.663 HLA-E PRR3IMICC1
9 04 646520 1001297721
6 rs3579261 30538854 C 6.53E- 0.5875 -3.409 HLA-E PRR31MICC I
1 04 646520 1001297721
6 rs3132585 30795593 G 2.04E- 2.972 3.715 DHX161MDC11FLOT1
04 I NRMIKIAA1949 I
TUBB 1
6 rs3132583 30796554 C 3.00E- 2.915 3.616 DHX161MDC11FLOT1
04 I NRMIKIAA1949 I
TUBB 1
6 rs2230365 31633427 A 1.54E- 1.575 3.785 AIF11ATP6V1G21LTA
04 INFKBIL1ITNFIBAT2
IBAT11LST11APOMI
SNORA381SNORD841
SNORD1171
6 rs2229092 31648736 C 7.30E- 1.882 3.378 AIF11ATP6V1G21
04 CSNK2B1LTAILTB 1
TNFIBAT2IBAT11
LST11APOMILY6G5B
ISNORA381SNORD841
SNORD1171
6 rs537160 32024379 A 1.55E- 1.943 4.805 CFB 1C21C4A1C4B 1
06 NEU1ISKIV2LIRDBP1
STK191EHMT21
SLC44A41ZBTB121
6 rs2072633 32027557 A 3.04E- 1.513 3.612 CFB 1C21C4A1C4B 1
04 NEU1ISKIV2LIRDBP1
STK191EHMT21
SLC44A41ZBTB121
6 rs437179 32036993 A 3.37E- 2.336 5.912 CFB 1C4A1C4B 1
09 DOM3ZINEU11
SKIV2LIRDBPISTK19
1EHMT2 1SLC44A4 1
ZBTB121
6 rs386480 32054816 G 2.52E- 2.355 5.96 C4A1C4BIDOM3Z1
09 SKIV2LIRDBPISTK19
IEHMT21ZBTB121
6 rs389883 32055439 C 2.66E- 2.359 5.951 C4A1C4BIDOM3Z1
31
CA 02946317 2016-10-18
WO 2015/168699 PCT/US2015/029101
Chr SNP Position* Minor P- Odds Stat Loci**
* Allele value Ratio
09 SKIV2L1RDBPISTK19
IEHMT21ZBTB121
6 rs2856448 32122553 A 1.87E- 1.515 3.736 DOM3ZIRDBP1
04
6 rs185819 32158045 A 2.73E- 1.498 3.64 RNF5 1PPT2 1EGFL81
04 653033 1
6 rs1720798 32187545 G 1.36E- 3.953 8.268 ATF6B1RNF51PPT21
6 16 EGFL816530331
6 rs1053924 32228693 A 9.32E- 1.45 3.31 ATF6B1RNF5 1PPT2 1
04 FKBPLIPRRT11
EGFL816530331
1001305361
6 rs2269425 32231617 A 4.85E- 1.649 3.489 ATF6B1RNF51PPT21
04 FKBPLIPRRT11
EGFL81401252 1653033
11001305361
6 rs2269423 32253685 A 1.68E- 1.492 3.762 AGERIATF6B1PBX21
04 RNF51AGPAT11
FKBPLIPRRT11
EGFL81401252 1653033
11001305361
6 rs443198 32298384 G 5.76E- 0.689 -3.443 AGERIATF6B1
04 NOTCH4 1PBX2 1
AGPAT11GPSM3 1
FKBPLIPRRT11
401252 11001305361
6 rs2894252 32453421 A 2.67E- 0.6137 -3.646 HLA-DRA1C6orf101
04 6466681
6 rs2894253 32453518 C 1.45E- 1.965 4.337 HLA-DRA1C6orf101
05 6466681
6 rs9405094 32454386 A 2.66E- 0.6136 -3.647 HLA-DRA1C6orf101
04 6466681
6 rs2395157 32456123 G 2.65E- 0.6136 -3.648 HLA-DRA1C6orf101
04 6466681
6 rs9268454 32457689 G 2.67E- 0.6137 -3.645 HLA-DRA1C6orf101
04 6466681
6 rs9268456 32457924 A 2.92E- 0.6155 -3.622 HLA-DRA1C6orf101
04 6466681
6 rs9268461 32459879 A 2.66E- 0.6136 -3.647 HLA-DRA1C6orf101
04 6466681
6 rs1742364 32465111 A 3.07E- 1.695 3.609 HLA-DRA1C6orf101
9 04 BTNL216466681
6 rs1252904 32465693 A 3.15E- 1.693 3.603 HLA-DRA1C6orf101
9 04 BTNL216466681
6 rs1687012 32467438 A 3.37E- 1.687 3.585 HLA-DRA1C6orf101
3 04 BTNL216466681
6 rs2076524 32478662 G 2.64E- 0.6135 -3.648 HLA-DRA1C6orf101
04 BTNL216466681
6 rs2076522 32479157 G 2.64E- 0.6135 -3.648 HLA-DRA1C6orf101
04 BTNL216466681
6 rs3806156 32481676 A 6.24E- 0.6757 -3.421 HLA-DRA1C6orf101
04 BTNL216466681
32
CA 02946317 2016-10-18
WO 2015/168699 PCT/US2015/029101
Chr SNP Position* Minor P- Odds Stat Loci**
* Allele value Ratio
6 rs9268491 32482109 G 2.64E- 0.6135 -3.648 HLA-DRA1C6orf101
04 BTNL216466681
6 rs2395163 32495787 G 3.17E- 0.6026 -3.601 HLA-DRA1C6orf101
04 BTNL216466681
6 rs3433058 32498681 G 3.00E- 1.74 3.615 HLA-DRA1C6orf101
04 BTNL216466681
6 rs9268905 32540055 G 5.94E- 0.5461 -4.993 C6orflOIBTNL21
07
6 rs2395185 32541145 A 1.16E- 0.5558 -4.863 C6orf10IBTNL21
06
6 rs9368726 32546520 G 7.45E- 0.5503 -4.949 C6orflOIBTNL21
07
6 rs9405108 32546626 A 5.94E- 0.5462 -4.993 C6orflOIBTNL21
07
6 rs2877272 32617335 A 1.34E- 0.4848 -5.681 HLA-DQA1
4 08
6 rs2853064 32635057 C 3.55E- 1.763 4.135 HLA-DQA1
8 05
6 rs2836629 32668837 C 2.72E- 0.4843 -5.559 HLA-DQA1
8 08
6 rs3526569 32669312 G 6.31E- 0.5203 -4.001 HLA-DQA1
8 05
6 rs2860540 32677665 G 1.42E- 1.967 4.822 HLA-DQA1
4 06
6 rs2516049 32678378 G 9.52E- 0.503 -5.336 HLA-DQA1
08
6 rs9270856 32678817 A 1.30E- 1.79 4.84 HLA-DQA1
06
6 rs9271100 32684456 A 1.45E- 1.784 4.818 HLA-DQA1
06
6 rs660895 32685358 G 8.98E- 0.5616 -3.917 HLA-DQA1
05
6 rs9271170 32685867 A 1.45E- 1.784 4.818 HLA-DQA1
06
6 rs9271488 32696978 A 3.08E- 0.4876 -5.537 HLA-DQA1
08
6 rs9272105 32707977 G 4.38E- 0.6447 -4.086 HLA-DQA1
05
6 rs9272143 32708781 A 2.55E- 0.6415 -4.211 HLA-DQA1
05
6 rs3427636 32722015 A 6.62E- 0.4968 -5.401 HLA-DQA11HLA-
9 08 DQA21
6 rs2647025 32743927 A 8.19E- 0.5545 -4.46 HLA-DQA21HLA-
06 DQB116466861
6 rs2858331 32789255 G 1.25E- 1.505 3.837 HLA-DQA21HLA-
04 DQB116466861
Example 11
Utilizing a GWAS approach of a well-characterized UC cohort and a large
healthy
control group, the inventors confirmed the contribution of the MHC to severe
UC at a
33
CA 02946317 2016-10-18
WO 2015/168699 PCT/US2015/029101
genome- wide level of significance and observed more than one 'signal' from
this locus.
The inventors also implicated TNFSF15 (TEM) in UC severity, with potential
therapeutic
implications. It was confirmed an association between extensive disease and
colectomy,
and also demonstrated, for the first time, that a family history of UC is
associated with the
need for surgery. These observations support the concept that genetic
variation contributes
to the natural history of UC. The regression model of 46 SNPs presented herein
discriminates patients at risk of mrUC and explains approximately 50% of the
genetic
contribution to the risk of surgery in the cohort. When the risk score was
divided into four
categories, higher risk score categories had a higher percentage of mrUC
subjects (p
<2.2x10-16) and predicted earlier colectomy.
The predictive power of diagnostic tests can be evaluated by the area under
the
curve (AUC), an ROC summary index, which evaluates the probability that one's
test
correctly identifies a diseased subject from a pair of affected and unaffected
individuals. A
perfect test has an AUC of 1.0, while random chance gives an AUC of 0.5.
Screening
programs attempting to identify high-risk groups generally have an AUC of
¨0.80 48. The
genetic risk score reported herein yielded an AUC of 0.91.
The inventors calculated operating characteristics in an attempt to determine
whether a prognostic test based on these genetic data would be clinically
useful. The score
of 44 and 47 (out of a possible score of 60) can be used to generate a test
with a sensitivity
and specificity of over 90%, respectively. The fitted model was robust, given
the
comparable mean sensitivity and specificity following cross-validation. In
addition,
likelihood ratios can be used with differing pre-test probabilities to
calculate relevant post-
test probabilities and are therefore much more generalizable. The Cochrane
collaboration
has suggested that positive likelihood ratios of greater than 10 and negative
likelihood
ratios of less than 0.1 are likely to make a significant impact on health
care. As can be seen
from the data presented herein, these ratios are met with a risk score of 47
and 43,
respectively. For example, in a newly diagnosed patient with ulcerative
colitis, if the pre-
test probability of colectomy was approximately 20% (based on epidemiological
and
clinical data) and the patient had a genetic risk score of 47 (positive
likelihood ratio of
approximately 10), then utilizing Bayesian principles, this equates to a post-
test probability
of colectomy of approximately 75%. If patients at high risk for colectomy
could be
identified early in their course of disease, then this could have significant
consequences for
clinicians. Clinicians may suggest earlier introduction of more potent
medication for the
high risk patients and choose to clinically and endoscopically monitor these
patients more
34
CA 02946317 2016-10-18
WO 2015/168699 PCT/US2015/029101
intensively. Stressing the importance of compliance with therapy and even
monitoring
compliance in high-risk patients may also be considered by clinicians.
The inventors have confirmed the association with the MHC and disease severity
in UC and the data shows that there may be more than one 'signal' from this
locus.
Furthermore, the inventors have also implicated a realistic therapeutic target
and known
IBD locus, TNFSF15 (TL1A), suggesting that interference with this pathway is
important
in severe UC. In addition, the inventors have demonstrated the utility of a
model based on
GWAS data for predicting the need for surgery in UC. These data demonstrate
that the
effect of these variants cumulatively they may provide adequate discriminatory
power for
clinical use. These findings allow a more tailored approach to the management
of UC
patients and also identify additional targets for early therapeutic
intervention in more
aggressive UC.
Example 12
Medically refractory UC (mrUC) requiring colectomy for failure to respond
to medical therapy occurs in up to 30% UC patients and remains a significant
clinical
challenge. The inventors have shown genetic associations with mrUC, which
allows for
the timely identification of patients at risk for surgery and supports early
introduction of
more intensive therapy. Genetic loci have been identified as contributing to
mrUC using
immune-specific Immunochip arrays. These genetic associations also identify
novel
therapeutic targets for the treatment of severe UC.
Example 13
Table 6: Demographic data
mrUC non-mrUC
FACTORS P-
value
(n=323) (n=639)
Gender (F%) 43% 49% NS
Median Age of Onset - yrs (IQR) 26 (17-37) 27 (18-39) NS
Smoking (%) 8% 8% NS
Median Disease Duration - months (IQR) 47 (23-128) 109
(47-208) 9.5x10-9
Extraintestinal Manifestations (%) 14% 6% 6.1x10-
5
Extensive Disease (%) 82% 64% 1.3x10-
7
Family History of UC (%) 26% 18% 0.006
Family History of IBD (%) 32% 24% 0.006
CA 02946317 2016-10-18
WO 2015/168699 PCT/US2015/029101
Example 14
Serological associations with mrUC and Cbirl, ASCA, OmpC and 12 antibody
quartile sums calculated within UC, were observed (Figure 3). The inventors
performed a
GWAS on 323 mrUC and 639 Non-mrUC subjects. The demographic data of the cohort
is
summarized herein (Table 6).Following identification of single markers
associated with
mrUC, the inventors proceeded to a multivariate approach, as performed above
to identify
the 46 SNPs. The inventors performed a stepwise logistic regression and
identified 33
SNPs (Analysis I ¨ Logistic regression: mrUC versus non-mrUC; Figure 4) and 8
SNPs
(Analysis II ¨ Cox proportional hazards regression) that together were
associated with
mrUC (logistic regression and Cox proportional hazards; analysis schematic see
Figure 5).
This analysis resulted in the final risk model of 36 SNPs, which explained
34.7% of risk
for colectomy in mrUC (Figure 6; Table 7).
The combination of risk alleles (genetic "burden") may be useful to identify
UC
patients at high risk for colectomy. SNPs identified together explain a large
proportion of
risk: 36 SNPs: 35% risk for colectomy in the mrUC cohort. The inventors
calculated a
genetic risk score was calculated from the total number of risk alleles (0, 1,
or 2) across all
36 risk SNPs (theoretical range: 0-72; observed range: 16-38). Based on the
genetic risk
scores, the inventors grouped the UC cohort into four risk categories, scores
16-22 (risk-
A); scores 23-27 (risk-B); scores 28-32 (risk-C); and scores 33-38 (risk-D). A
higher risk
score was associated with mrUC, earlier progression to colectomy and shorter
overall
time to colectomy (Figures 7 - 10). This further supports the paradigm that a
group of
SNPs, identified by GWAS and combined together may account for a large
proportion of
the genetic contribution to a complex phenotype to provide a risk score with
clinical
utility.
Table 7: 36 SNPs associated with the risk model for mrUC
Chr SNP SEQ ID NO Gene(s) of Interest
12 rs79122070 49 CACNA1C
1 rs226476 50 TNFRSF9
1 rs2275612 51 CNN3
22 rs9610486 52 MYH9
12 rs1798613 53 BICD1
36
CA 02946317 2016-10-18
WO 2015/168699
PCT/US2015/029101
Chr SNP SEQ ID NO Gene(s) of Interest
2 rs726357 54 PFTK2 1FZD7
22 rs4823779 55 FLJ462571FAM19A5
17 rs7222857 56 RPL38
13 rs1351832 57 AKAP111TNFSF11
1 rs76505423 58 CRB1
12 rs526058 59 SOX5
3 rs17026843 60 CADM21VGLL3
6 rs17708487 61 BACH2
rs10795186 62
13 rs17612850 63 DIAPH3
12 rs216865 64 VWF
13 rs813841 65 RFC31NBEA
1 rs12025913 66 RGS211RGS1
8 rs56384685 67 XKR6
13 rs912425 68 AKAP111TNFSF11
6 rs7757174 69 TEAD3
2 rs10931144 70 ZNF804A
5 rs10060659 71 HMP19
14 rs1956388 72 FOXG1
10 rs56065922 73 PRKCQ
4 rs1032147 74 GBA3
6 rs2269423 75 AGPAT1
2 rs114855708 76 ADAM23
6 rs2296337 77 ITPR3
2 rs3024861 78 STAT4
13 rs1410434 79 GPR12
6 rs9258253 80 IFITM4P1HCG4
1 rs10875260 81 FRRS11AGI
1 rs72717025 82 FCGR2A
14 rs9323816 83 GPR65
10 rs1199075 84 ZWINT1IPMK
37
CA 02946317 2016-10-18
WO 2015/168699 PCT/US2015/029101
Example 15
mrUC Network Analysis
Analysis of 962 subjects (323 mrUC and 639 non-mrUC) resulted in 6573
candidate SNPs (logistic regression analysis (p < 0.05) > 1742 genes. A
calculated gene-
based logistic regression score was used to obtain genes with a maximal
AUC>0.56
selected for network construction. The network was constructed using pairwise
Pearson
correlation coefficient (p<10-7) between gene scores and protein-protein
interaction
database (STRING). Pathways associated with mrUC networks revealed cytokine-
cytokine receptor interactions (p=1.5x10-5), T-cell receptor signaling pathway
interactions (p=0.0001) and Rheumatoid arthritis (p=0.0015). This analysis
identified
relevant pathways for further investigation of potential new therapeutic
targets for mrUC.
Example /6
Role for MHC in UC severity
Stringent sample and SNP quality control of 323 Caucasian mrUC subjects and
5190 controls was performed to test single-SNP associations with regression
analysis
corrected for 4 principal components. Results demonstrated the association of
MHC with
UC severity (Figure 11 ¨ 12; Table 8).
Table 8
Chr SNP SEQ ID NO Gene(s) of Interest
6 rs4151651 85 CFB
6 rs9268923 86 HLA-DRAIHLA-DRB5
2 rs75412898 87 AFF3
5 rs3846599 88 CCT5
1 rs12567149 89 Clorf53
17 rs12150079 90 ORMDL3
2 rs4143571 91 ACTR2 1 SPRED2
2 rs114709725 92 DYTN
12 rs12318183 93 IFNG
6 rs16896780 94 ANKS1A 1 UHRF1BP1
2 rs3732151 95 HS1BP3
6 rs6908055 96 ATG5
38
CA 02946317 2016-10-18
WO 2015/168699 PCT/US2015/029101
12 rs74912794 97 MPHOSPH9
1 rs2281852 98 TNFRSF14
1 rs2281852 99 TNFRSF14
Example 17
Additional summary and conclusions
Cross-validation and bootstrapping was performed to validate the fitted
logistic
regression model. The model was able to identify a dataset for independent
replication. A
multivariate model will be built by integrating clinical, serological, and
genetic
associations. A truncated genetic analysis can then identify a patient
population at risk for
colectomy that would benefit from early intervention and identify therapeutic
targets
(Table 9), which would address an unmet medical need.
Example 18
Table 9: Potential Therapeutic Targets
Genes Pathways
ORMDL3, CCT5 Protein folding & ER stress/UPR
MYH9, ADAM23, CADM2 Cell adhesion & cell-cell interaction
TNFRSF14, IFNG,
TNFRSF9, STAT4, PRKCQ, T-cell mediated immune response
TNFSF11, BACH2
ATG5, PRKCQ Autophagy
TNFRSF9, IL6R,
TNFRSF18/TNFRSF4,
CCL21, TNFSF15, TNFSF11, Cytokine-cytokine receptor interaction
TNFRSF13B, CCL2/CCL7,
TNFRSF6B
While the description above refers to particular embodiments of the present
invention, it should be readily apparent to people of ordinary skill in the
art that a number
of modifications may be made without departing from the spirit thereof The
presently
disclosed embodiments are, therefore, to be considered in all respects as
illustrative and
not restrictive.
Various embodiments of the invention are described above in the Detailed
Description. While these descriptions directly describe the above embodiments,
it is
understood that those skilled in the art may conceive modifications and/or
variations to the
39
CA 02946317 2016-10-18
WO 2015/168699 PCT/US2015/029101
specific embodiments shown and described herein. Any such modifications or
variations
that fall within the purview of this description are intended to be included
therein as well.
Unless specifically noted, it is the intention of the inventor that the words
and phrases in
the specification and claims be given the ordinary and accustomed meanings to
those of
ordinary skill in the applicable art(s).
The foregoing description of various embodiments of the invention known to the
applicant at this time of filing the application has been presented and is
intended for the
purposes of illustration and description. The present description is not
intended to be
exhaustive nor limit the invention to the precise form disclosed and many
modifications
and variations are possible in the light of the above teachings. The
embodiments
described serve to explain the principles of the invention and its practical
application and
to enable others skilled in the art to utilize the invention in various
embodiments and with
various modifications as are suited to the particular use contemplated.
Therefore, it is
intended that the invention not be limited to the particular embodiments
disclosed for
carrying out the invention.
While particular embodiments of the present invention have been shown and
described, it will be obvious to those skilled in the art that, based upon the
teachings
herein, changes and modifications may be made without departing from this
invention and
its broader aspects and, therefore, the appended claims are to encompass
within their scope
all such changes and modifications as are within the true spirit and scope of
this invention.
Furthermore, it is to be understood that the invention is solely defined by
the appended
claims. It will be understood by those within the art that, in general, terms
used herein,
and especially in the appended claims (e.g., bodies of the appended claims)
are generally
intended as "open" terms (e.g., the term "including" should be interpreted as
"including
but not limited to," the term "having" should be interpreted as "having at
least," the term
"includes" should be interpreted as "includes but is not limited to," etc.).
It will be further
understood by those within the art that if a specific number of an introduced
claim
recitation is intended, such an intent will be explicitly recited in the
claim, and in the
absence of such recitation no such intent is present. For example, as an aid
to
understanding, the following appended claims may contain usage of the
introductory
phrases "at and "one or more" to introduce claim recitations. However, the use
of such
phrases should not be construed to imply that the introduction of a claim
recitation by the
indefinite articles "a" or "an" limits any particular claim containing such
introduced claim
recitation to inventions containing only one such recitation, even when the
same claim
CA 02946317 2016-10-18
WO 2015/168699 PCT/US2015/029101
includes the introductory phrases "one or more" or "at least one" and
indefinite articles
such as "a" or "an" (e.g., "a" and/or "an" should typically be interpreted to
mean "at least
one" or "one or more"); the same holds true for the use of definite articles
used to
introduce claim recitations. In addition, even if a specific number of an
introduced claim
recitation is explicitly recited, those skilled in the art will recognize that
such recitation
should typically be interpreted to mean at least the recited number (e.g., the
bare recitation
of "two recitations," without other modifiers, typically means at least two
recitations, or
two or more recitations).
Accordingly, the invention is not limited except as by the appended claims.
41