Note: Descriptions are shown in the official language in which they were submitted.
BIOMARKERS FOR RESPONSE TO PI3K INHIBITORS
CROSS-REFERENCE TO RELATED APPLICATIONS
This application claims priority to U.S. Provisional Patent Application
Serial No. 61/991,165, filed May 9, 2014, U.S. Provisional Patent Application
Serial
No. 61/992,173, filed May 12, 2014, and U.S. Provisional Patent Application
Serial
No. 62/004,518, filed May 29, 2014.
1. INTRODUCTION
This present invention relates to biomarkers which may be used to
evaluate whether a PI3K inhibitor would produce an anti-cancer effect in a
subject
during the course of treatment with the PI3K inhibitor. As such, these
biomarkers
may be used in methods of treating cancer patients.
2. BACKGROUND OF THE INVENTION
Phosphatidylinositol 3-kinases (PI3Ks) are lipid kinases that are
important in controlling signaling pathways involved in cell proliferation,
motility,
death and invasion, as well as in insulin signaling. Upon activation, PI3K
catalyzes
the phosphorylation of the cell membrane-embedded phosphatidylinositol 4,5-
bisphosphate (PIP2) to phosphatidylinositol 3,4,5-triphosphate (PIP3). In
turn, PIP3, a
critical phospholipid second messenger, acts as a docking site for signaling
proteins,
such as PDK1, to effect downstream cellular pathways critical for cell growth
and
survival. Class I PI3Ks contains four isoforms, p110a, p110f3, p1106 and
p110y,
which carry out non-redundant signaling functions. Mutations in
phosphatidylinositol-4, 5-bisphosphate 3-kinase catalytic subunit alpha
isoform
(PIK3CA), the gene encoding p110a, are frequently found in multiple human
tumor
types, such as colorectal cancer, breast cancer, ovarian cancer, endometrial
carcinoma
and hepatocellular carcinoma, suggesting that p1 10a is a key isoform for
promoting
tumor growth. In particular, in many tumors, the PI3K signaling pathway is
constitutively activated. This is thought to be a critical step in mediating
the
transforming potential and growth stimulating activity of various oncogenes
(i.e.,
HER2, EGFR, IGF1R).
1
Date Recue/Date Received 2021-08-23
CA 02948351 2016-11-07
WO 2015/172085 PCT/US2015/029995
The current standard of care in monitoring cancer patients undergoing
treatment is the use of medical imaging and invasive techniques, such as
biopsies.
However, in certain circumstances, such biopsies can be deemed unsafe,
impractical
or not feasible. Therefore, there remains a need in the art for minimally
invasive
techniques that can allow real-time monitoring of the effectiveness of a
particular
cancer treatment within a patient. The discovery of circulating DNA, also
called cell-
free DNA, in the blood of a subject provides the opportunity to analyze
genetic
material originating from normal and/or tumor cells without the risks
associated with
invasive sampling methods. Cell free DNA fragments containing tumor-specific
sequence alterations can be found in the cell free fraction of blood.
Accordingly, there is a need in the art for methods and/or biomarkers
useful for monitoring the effectiveness of anti-cancer treatments, such as
PI3K
inhibitors, in a subject.
3. SUMMARY
The present disclosure relates to the use of one or more biomarkers to
evaluate whether an anti-cancer effect is likely to be produced in a subject
during the
course of treatment with a PI3K inhibitor. It is based, at least in part, on
the discovery
that certain nucleotides, e.g., genomic DNA in free circulation, isolated from
the
serum of subjects undergoing cancer treatment, could be used as biomarkers to
indicate the effectiveness or ineffectiveness, of P13Ka treatment on cancer
growth.
Accordingly, in non-limiting embodiments, the present subject matter
provides for methods and kits for determining the presence and/or level of one
or
more biomarkers, e.g., mutant alleles of P/K3CA in cell free DNA, and methods
of
using such determinations in selecting and/or modifying a therapeutic regimen
for a
cancer patient.
In certain embodiments, the method for determining whether an anti-
cancer effect is likely to be produced in a cancer of a subject by a PI3Ka
inhibitor
comprises determining the presence and/or level of one or more PIK3CA
biomarkers
in one or more body fluid samples obtained from a subject during PI3Ka
inhibitor
treatment, where if the presence and/or level of a PIK3CA biomarker increases,
it is
less likely that a P13Ka inhibitor is having an anti-cancer effect on the
cancer. In
certain non-limiting embodiments, the method fur determining whether an anti-
cancer
CA 02948351 2016-11-07
WO 2015/172085 PCT/US2015/029995
effect is likely to be produced in a cancer of a subject by an inhibitor of
PI31(13,
P131(?, or P13K6 comprises determining the presence and/or level of one or
more
PIK3CB, PIK3CC, or PIK3CD biomarkers, respectively, in one or more body fluid
samples obtained during inhibitor treatment, where if the presence and/or
level of a
biomarker increases, it is less likely that an inhibitor is having an anti-
cancer effect on
the cancer. This information may be advantageously used to avoid ineffective
treatment and to redirect a subject to a more effective therapeutic regimen.
In certain embodiments, the method for determining whether an anti-
cancer effect is likely to be produced in a cancer of a subject by a PI3Ka
inhibitor
comprises determining the presence and/or level of one or more PIK3CA
biomarkers
in one or more body fluid samples obtained during P13 Ku inhibitor treatment,
where
if the presence and/or level of a PIK3CA biomarker decreases, it is likely
that a
PI3Ka inhibitor is having an anti-cancer effect on the cancer. In certain non-
limiting
embodiments, the method for determining whether an anti-cancer effect is
likely to be
produced in a cancer of a subject by an inhibitor of PI3K13, PI3Ky, or P13K6
comprises determining the presence and/or level of one or more PIK3CB, PIK3CC,
or
PIK3CD biomarkers, respectively, in one or more body fluid samples obtained
during
inhibitor treatment, where if the presence and/or level of a biomarker
decreases, it is
more likely that an inhibitor is having an anti-cancer effect on the cancer.
The present disclosure further provides methods for producing an anti-
cancer effect in a subject comprising determining the presence and/or level of
a
PIK3CA biomarker in one or more body fluid samples of the subject obtained
during
PI3Ka inhibitor treatment, where if the presence and/or level of a PIK3CA
biomarker
is reduced, then continuing treatment of the subject with a therapeutically
effective
amount of a PI3Ka inhibitor, but if the presence and/or level of a PIK3CA
biomarker
increases, then discontinuing treatment with the P13Ka inhibitor and
optionally
pursuing treatment of the subject with a therapeutically effective amount of
another
anti-cancer agent. Analogous methods can be performed using inhibitors that
act on
other PI3K isofonns and corresponding biomarkers.
In certain embodiments, the cancer can be liver cancer, brain cancer,
cervical cancer, colorectal cancer, breast cancer, endometrial carcinoma,
gastric
cancer, cancers of the head and neck, bladder cancer, lung cancer, ovarian
cancer,
biliary tree cancer and hepatocellular carcinoma.
3
CA 02948351 2016-11-07
WO 2015/172085
PCT/US2015/029995
In certain embodiments, the one or more samples are plasma samples.
The one or more samples can be obtained serially after initiation of PI3K
inhibitor
treatment. In certain embodiments, the one or more samples arc serially
obtained
from the patient about every four weeks after initiation of treatment.
Alternatively or
-- additionally, the one or more samples are obtained serially from the
patient about
every two weeks after initiation of treatment.
In certain embodiments, the one or more PIK3CA biomarkers are
mutations in the C2 domain, kinase domain and/or helical domain of PIK3CA
which
increases PIK3CA activity. In certain non-limiting embodiments, the one or
more
biomarkers are selected from the group consisting of the PIK3CA HI 047R
biomarker,
PIK3CA E545K biomarker, PIK3CA E542K biomarker, PIK3CA E545G biomarker,
PIK3CA E545Q biomarker, PIK3CA E545A biomarker, PIK3CA E545D biomarker,
PIK3CA E545V biomarker, PIK3CA H1047L biomarker, PIK3CA H1047Y
biomarker, PIK3CA E542Q biomarker, PIK3CA E542G biomarker, PIK3CA P539R
biomarker, PIK3CA N345K biomarker, PIK3CA C420R biomarker, PIK3CA
G1 049R biomarker, PIK3CA E726K biomarker, PIK3CA R88Q biomarker, PIK3CA
Q546K biomarker, P1K3CA Q546P biomarker, PIK3CA Q546R biomarker, PIK3CA
Q546L biomarker or combinations thereof
The present disclosure further provides kits for dctciiiiining whether an
anti-cancer effect is likely being produced in a cancer by a Pl3Kct inhibitor,
comprising a means for detecting a P1K3CA biomarker. In certain embodiments,
the
means for detecting a P1K3CA biomarker includes one or more sets of primers or
probes specific for the PIK3CA biomarker.
4. BRIEF DESCRIPTION OF FIGURES
FIGURE 1 shows the plasma concentrations of total cell free DNA
(ng/ml) and tumor-specific mutant allele fractions in cell free DNA of PIK3CA
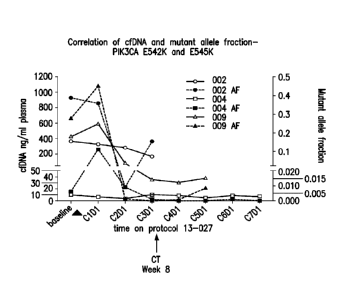
E542K and E545K in patients being treated with a PI3Kot inhibitor.
FIGURE 2 shows the plasma concentrations of total cell free DNA
(ng/ml) and tumor-specific mutant allele fractions in cell free DNA of PIK3CA
H1047R in patients being treated with a P131(ct inhibitor.
FIGURE 3 shows a representation of a serial PIK3CA E542K mutation
analysis by ddPCR from cell free DNA.
4
CA 02948351 2016-11-07
WO 2015/172085 PCT/US2015/029995
FIGURE 4 shows the measurements of the cell free PIK3CA mutant
allele fraction at day 28 for 7 patients being treated with a P13Ku inhibitor.
FIGURE 5 shows that following treatment with BYL719 and letrozole,
the cell free PIK3CA mutant allele fraction decreased markedly in patient 009.
FIGURE 6 shows that ddPCR is sensitive for detecting low levels of
cell free PIK3CA mutant allele fractions.
FIGURE 7 shows that low total cfDNA concentrations were
informative for determining and following cell free PIK3CA mutant allele
fraction.
FIGURE 8 shows that increases in the cell free PIK3CA mutant allele
fraction may precede clinical progression by I to 3 months.
FIGURE 9 shows that increases in the cell free PIK3CA mutant allele
fraction may be used to determine early resistance to a PI3Ka inhibitor.
5. DETAILED DESCRIPTION
For clarity and not by way of limitation the detailed description of the
invention is divided into the following subsections:
(i) P1K3CA as a biomarker;
(ii) PI3Kot inhibitors;
(iii) cancer targets;
(iv) biomarker detection;
(v) methods of use; and
(vi) kits.
5.1 PIK3CA AS A BIOMARKER
The present disclosure provides biomarkcrs for determining, during the
course of treatment in a subject with a P13 Ku inhibitor, whether the P13 Ku
inhibitor is
likely effective at providing an anti-cancer effect on the cancer in the
subject.
The term "biomarker" as used herein, includes nucleic acids of
phosphatidylinosito1-4, 5-hisphosphate 3-kinase catalytic subunit alpha
isoform,
denoted herein as PIK3CA. In certain embodiments of the present disclosure,
the
biomarker is a nucleic acid that is related to the activity level of PIK3CA.
A "patient" or "subject,- as used interchangeably herein, refers to a
human or a non-human subject. Non-limiting examples of non-human subjects
5
CA 02948351 2016-11-07
WO 2015/172085
PCT/US2015/029995
include non-human primates, dogs, cats, mice, rats, guinea pigs, rabbits,
pigs, fowl,
horses, cows, goats, sheep, cetaceans, etc.
In certain embodiments, the nucleic acids are present in a body fluid of
the subject. For example, but not way of limitation, the biomarker of the
present
disclosure can be a nucleic acid, e.g., DNA, that is released into vascular
system,
present in circulation, e.g., blood or plasma, present in body fluid, e.g.,
plasma, serum,
urine or pleural effusion or is extracellular, e.g., outside of (not located
within) any
cell, bound or unbound to the cell surface.
The biomarkers of the present disclosure can be obtained from a
"biological sample" or "sample." A "biological sample" or "sample," as used
interchangeably herein, refers to a sample of biological material obtained
from a
subject, including a biological fluid and/or body fluid, e.g., blood, plasma,
serum,
stool, urine, lymphatic fluid, ascites, ductal lavage, nipple aspirate,
saliva, broncho-
alveolar lavage, tears and cerebrospinal fluid. In certain non-limiting
embodiments,
the levels and/or presence of one or more biomarkers of the present disclosure
are
determined in one or more samples obtained from a subject, e.g., plasma
samples.
In certain non-limiting embodiments, a biomarker is an allelic variant
or mutation of the PIK3CA gene that results in the activation, constitutive
activation
and/or overactivation of PIK3CA, e.g., gain of function mutations. In certain
limiting
embodiments, a biomarker is an allelic variant or mutation of the PIK3CA gene
or
protein that results in the loss of functional PIK3CA protein.
In certain non-limiting embodiments, a PIK3CA biomarker is a nucleic
acid having one or more inser lions, deletions or substitutions relative to
a reference
PIK3CA gene described below. Such insertions, deletions or substitutions may
result
in a nonsense mutation, a frameshift mutation, a missense mutation or a
termination
relative to the reference PIK3CA gene and/or protein.
In a specific, non-limiting embodiment, a "reference," "reference
control" or "control," as used interchangeably herein, may be a human PIK3CA
nucleic acid having the sequence as set forth in NCBI database accession no.
NG 012113.2, or a nucleic acid encoding a PIK3CA protein molecule that has the
amino acid sequence set forth in NCBI database accession no. GI:126302584.
Reference PIK3CA nucleic acids for non-human species are known or
can be determined according to methods known in the art, for example, where
the
sequence is the allele represented in the majority of the population of that
species.
6
Where comparisons to a reference control level, quantity, expression
and/or presence are referred to herein, the biomarker is assessed relative to
the
reference control level, quantity, expression and/or presence within the same
species.
For example, a human PIK3CA biomarker level and/or presence is compared with a
human PIK3CA reference control level and/or presence.
In certain non-limiting embodiments, a PIK3CA biomarker for a
human subject is the PIK3CA H1047R mutation.
In certain non-limiting embodiments, a PIK3CA biomarker for a
human subject is the PIK3CA H1047Y mutation.
In certain non-limiting embodiments, a PIK3CA biomarker for a
human subject is the PIK3CA H1047L mutation.
In certain non-limiting embodiments, a PIK3CA biomarker for a
human subject is the PIK3CA E545K mutation.
In certain non-limiting embodiments, a PIK3CA biomarker for a
human subject is the PIK3CA E545G mutation.
In certain non-limiting embodiments, a PIK3CA biomarker for a
human subject is the PIK3CA E542K mutation.
In certain non-limiting embodiments, a PIK3CA biomarker comprises
one or more mutations in the helical, kinase domain, adaptor-binding, Ras-
binding,
C2 domains or combinations thereof.
Additional non-limiting examples of PIK3CA biomarkers are disclosed
in Samuels et al., Science (2004) 304(5670):554; Campbell et al., Cancer Res.
(2004)
64(21):7678-7681; Lee et al., Oncogene (2005) 24:1477-1480; Cizkova et al,
Breast
Cancer Res. (2012) 14(1):R28; Bachman et al., Cancer Biol. Ther. (2004)
3(8):772-
775; and Karakas et al., British Journal of Cancer (2006) 94:455-459.
In certain non-limiting embodiments, a PIK3CA biomarker comprises
one or more mutations in the helical domain of PIK3CA, which, for example,
increases PIK3CA catalytic activity. In certain non-limiting embodiments, a
PIK3CA
biomarker comprises one or more mutations in the kinase domain of PIK3CA,
which,
for example, increases PIK3CA catalytic activity. In certain non-limiting
embodiments, a PIK3CA biomarker comprises one or more mutations in the C2
domain of PIK3CA, which, for example, increases PIK3CA catalytic activity. In
certain non-limiting embodiments, the mutated PIK3CA nucleotide sequence is at
7
Date Recue/Date Received 2021-08-23
least 95 or at least 98 or at least 99 or at least 99.5 percent homologous to
the
reference control, described above. Sequence homology can be determined, for
example, by software such as BLAST or FASTA.
In certain non-limiting embodiments, a PIK3CA biomarker comprises
one or more mutations including the PIK3CA H1047R mutation, the PIK3CA E545K
mutation, the PIK3CA E542K mutation, the PIK3CA E545G mutation, the PIK3CA
E545Q mutation, the PIK3CA E545A mutation, the PIK3CA E545D mutation, the
PIK3CA E545V mutation, the PIK3CA H1047L mutation, the PIK3CA H1047Y
mutation, the PIK3CA E542Q mutation, the PIK3CA E542G mutation, the PIK3CA
P539R mutation, the PIK3CA N345K mutation, the PIK3CA C420R mutation, the
PIK3CA G1049R mutation, the PIK3CA E726K mutation, the PIK3CA R88Q
mutation, the PIK3CA Q546K mutation, the PIK3CA Q546P mutation, the PIK3CA
Q546R mutation, the PIK3CA Q546L mutation or combinations thereof.
5.2 PI3Ka INHIBITORS
Non-limiting examples of PI3Ka inhibitors include compounds,
molecules, chemicals, polypeptides and proteins that inhibit and/or reduce the
expression and/or activity of PI3Ka. Additional non-limiting examples of PI3Ka
inhibitors include ATP-competitive inhibitors of PI3Ka. In particular non-
limiting
embodiments, the PI3Ka inhibitor is derived from imidazopyridine or 2-
aminothiazole compounds. Further non-limiting examples include BYL719, INK-
1114, INK-1117, NVP-BYL719, 5RX2523, LY294002, PIK-75, PKI-587, A66,
CH5132799 and GDC-0032 (taselisib).
Further non-limiting examples of PI3Ka inhibitors are disclosed in
Schmidt-Kittler et al., Oncotarget (2010) 1(5):339-348; Wu et al., Med. Chem.
Comm. (2012) 3:659-662; Hayakawa et al., Bioorg. Med. Chem. (2007) 15(17):
5837-5844; and PCT Patent Application Nos. W02013/049581 and W02012/052745.
Further non-limiting examples of PI3Ka inhibitors include ribozymes,
antisense oligonucleotides, shRNA molecules and siRNA molecules that
specifically
inhibit and/or reduce the expression or activity of PI3Ka. One non-limiting
example
of a PI3Ka inhibitor comprises an antisense, shRNA, or siRNA nucleic acid
sequence
homologous to at least a portion of a PI3Ka nucleic acid sequence, e.g., the
nucleic
acid sequence of a PI3Ka subunit such as PIK3CA, wherein the homology of the
8
Date Recue/Date Received 2021-08-23
CA 02948351 2016-11-07
WO 2015/172085
PCT/US2015/029995
portion relative to the PI3Kot sequence is at least about 75 or at least about
80 or at
least about 85 or at least about 90 or at least about 95 or at least about 98
percent,
where percent homology can be determined by, for example, BLAST or FASTA
software. In certain non-limiting embodiments, the complementary portion may
constitute at least 10 nucleotides or at least 15 nucleotides or at least 20
nucleotides or
at least 25 nucleotides or at least 30 nucleotides and the antisensc nucleic
acid,
shRNA or siRNA molecules may be up to 15 or up to 20 or up to 25 or up to 30
or up
to 35 or up to 40 or up to 45 or up to 50 or up to 75 or up to 100 nucleotides
in length.
Antisense, shRNA, or siRNA molecules may comprise DNA or atypical or non-
naturally occurring residues, for example, but not limited to,
phosphorothioate
residues.
In certain non-limiting embodiments, the PI3Ka inhibitor can be used
alone or in combination with one or more anti-cancer agents. An "anti-cancer
agent,"
as used herein, can be any molecule, compound, chemical or composition that
has an
anti-cancer effect. Anti-cancer agents include, but are not limited to,
chemotherapeutic agents, radiotherapeutic agents, cytokines, anti-angiogenic
agents,
apoptosis-inducing agents, anti-cancer antibodies, anti-cyclin-dependent
kinase
agents, and/or agents which promote the activity of the immune system
including but
not limited to cytokines such as but not limited to interleukin 2, interferon,
anti-
CTLA4 antibody, anti-PD-1 antibody, and/or anti-PD-L1 antibody. For example,
but
not by way of limitation, a P13 Ku inhibitor can be used in combination with
letrozolc
or exemestane. "In combination with," as used herein, means that the PI3Ka
inhibitor
and the one or more anti-cancer agents are administered to a subject as part
of a
treatment regimen or plan. In certain embodiments, being used in combination
does
not require that the PI3Ka inhibitor and one or more anti-cancer agents are
physically
combined prior to administration or that they be administered over the same
time
frame.
5.3 CANCER TARGETS
Non-limiting examples of cancers that may be subject to the presently
disclosed subject matter include liver cancer, brain cancer, cervical cancer,
colorectal
cancer, breast cancer, endometrial carcinoma, gastric cancer, cancers of the
head and
neck, bladder cancer, lung cancer, ovarian cancer, biliary tree cancer and
hepatocellular carcinoma.
9
CA 02948351 2016-11-07
WO 2015/172085 PCT/US2015/029995
5.4 BIOMARKER DETECTION
A biomarker of the present disclosure can be isolated from a subject by
any means known in the art or described herein. For example, but not by way of
limitation, a biomarker, e.g., a cell free mutant allele of a PIK3C'4 nucleic
acid, can be
isolated from a biological sample obtained from a subject, such as a plasma
sample, or
other biological fluid, as described above.
There are several platforms that are known in the art and currently
available to isolate cell free nucleic acids from biological samples. In
certain
embodiments, isolation of DNA from a biological samples is based on extraction
methods using organic solvents such as a mixture of phenol and chloroform,
followed
by precipitation with ethanol (see, for example, J. Sambrook et al.,
"Molecular
Cloning: A Laboratory Manual'', 1989, 2nd Ed., Cold Spring Harbour Laboratory
Press: New York, N.Y.). Additional non-limiting examples include salting out
DNA
extraction (see, for example, P. Sunnucks et al., Genetics, 1996, 144: 747-
756; and S.
M. Aljanabi and I. Martinez, Nucl. Acids Res. 1997, 25: 4692-4693), the
trimethylammonium bromide salts DNA extraction method (see, for example, S.
Gustincich et al., BioTechniques, 1991, 11: 298-302) and the guanidinium
thiocyanate
DNA extraction method (see, for example, J. B. W. Hammond et al.,
Biochemistry,
1996, 240: 298-300).
There are also numerous different and versatile kits that can be used to
extract DNA from bodily fluids and that are commercially available from, for
example, BD Biosciences Clontech (Palo Alto, Calif.), Epicentre Technologies
(Madison, Wis.), Gentra Systems, Inc. (Minneapolis, Minn.), MicroProbe Corp.
(Bothell, Wash.), Organon Teknika (Durham, N.C.), and Qiagen Inc. (Valencia,
Calif.). Sensitivity, processing time and cost may be different from one kit
to another.
One of ordinary skill in the art can easily select the kit(s) most appropriate
for the
particular sample to be analyzed.
The presently disclosed subject matter further provides methods for
detecting and/or determining the presence and/or level of a nucleic acid
PIK3CA
biomarker. For example, but not by way of limitation, such methods include
polymerase chain reaction (PCR), including real-time PCR, quantitative PCR,
fluorescent PCR, RT-MSP (RT methylation specific polymerase chain reaction and
digital PCR, in situ hybridization, fluorescent in situ hybridization
("FISH"), gel
CA 02948351 2016-11-07
WO 2015/172085
PCT/US2015/029995
electrophoresis, radioimmunoassay, direct radio-labeling of DNA, sequencing
and
sequence analysis, microarray analysis and other techniques known in the art.
In certain embodiments, the presence and/or level of a PIK3CA
biomarker, can be detected through the use of DROPLET DIGITALT" PCR
(ddPCRT"), which is a method for performing digital PCR based on water-oil
emulsion droplet technology. Alternatively or additionally, a biomarker
disclosed
herein can be detected through direct plasma sequencing by means of tagged-
amplicon deep sequencing (see, for example, Forshew et al., Sci. Transl. Med.
(2012)
4136, p. 136).
In certain embodiments, the level and/or presence of one or more
biomarkers in one or more samples of a patient are determined by sequencing,
e.g.,
next generation sequencing. In certain embodiments, the level and/or presence
of one
or more biomarkers are determined using an microarray. In certain embodiments,
the
level and/or presence of one or more biomarkers are determined using an assay
that
.. comprises an amplification reaction, such as a polymerase chain reaction
(PCR).
5.5 METHODS OF USE
The presently disclosure provides methods for monitoring patients
undergoing anti-cancer treatment with a PI3Ka inhibitor. Early detection of an
increase in the tumor burden and/or tumor size, for example, by the detection
of an
increase in one or more of the presently disclosed biomarkers in one or more
biological samples of the subject, would allow modification of the current
treatment,
e.g., change in the anti-cancer agent being administered to the subject, to
avoid
ineffective treatment and improve the subject's possibility of survival.
Accordingly, the present disclosure provides methods for determining,
during the course of treatment of a subject having cancer with a PI3Ka
inhibitor,
whether an anti-cancer effect is likely being produced by the PI3Ka inhibitor.
In
certain embodiments, the methods include determining the presence and/or level
of a
biomarker in one or more serially collected samples of a patient, wherein if
the
presence and/or level of the biomarker decreases in the later collected
samples, there
is an increased likelihood that the PI3Ku inhibitor is having an anti-cancer
effect.
PIK3CA biomarkers are described in section 5.1 above. PI3Ku
inhibitors are described in section 5.2 above. Cancers suitable for treatment
are
CA 02948351 2016-11-07
WO 2015/172085
PCT/US2015/029995
described above in section 5.3. Methods of detecting a presently disclosed
biomarker
are described above in section 5.4.
In certain embodiments, the one or more samples are collected serially
during the course of treatment with a PI3Ka inhibitor. For example, but not by
way
of limitation, the level and/or presence of a biomarker in a patient can be
analysed
during a course of treatment with a P13 Ku inhibitor through serial sampling.
In
certain embodiments, one or more samples can be obtained from a patient
undergoing
treatment about every 5 weeks, about every 4 weeks, about every 3 weeks, about
every 2 weeks, about every week, about every 6 days, about every 5 days, about
every
4 days, about every 3 days or about every 2 days. In certain non-limiting
embodiments, one or more samples can be obtained from a patient about every 2
weeks. In certain embodiments, one or more samples can be obtained from a
patient
about every 28 days. In certain embodiments, one or more samples can be
obtained
from a patient on day 1 and on the last day, e.g., day 28, of one or more
treatment
cycles.
In certain embodiments, the one or more biological samples can be
obtained prior to treatment with a PI3Ka inhibitor, after initiation of
treatment and
during treatment with the same or different P13 Ku inhibitor. For example, and
not by
way of limitation, one or more samples obtained during the course of treatment
can be
compared to a sample obtained prior to treatment or during initiation of
treatment to
determine the presence and/or change in the mutant allele fraction of a
biomarker.
In certain non-limiting embodiments, the present disclosure provides
for a method of determining whether an anti-cancer effect is likely being
produced in
a cancer by a P13 Ku inhibitor, comprising obtaining one or more samples
sequentially
from a patient undergoing treatment with a PI3Ka inhibitor, and determining,
in each
sample, the presence and/or level of a PIK3CA biomarker, where if the presence
and/or level of the biomarker decreases, it is likely that the PI3Ku inhibitor
is
producing an anti-cancer effect on the cancer.
In certain non-limiting embodiments, a decrease in the mutant allele
fraction of a PIK3CA biotnarker, which indicates that a P13 Ku inhibitor is
more likely
producing an anti-cancer effect on the cancer, can be greater than about 70%,
greater
than about 80%, greater than about 85%, greater than about 86%, greater than
about
87%, greater than about 88%, greater than about 89%, greater than about 90%,
greater
than about 91%, greater than about 92%, greater than about 93%, greater than
about
12
CA 02948351 2016-11-07
WO 2015/172085 PCT/US2015/029995
94%, greater than about 95%, greater than about 96%, greater than about 97%,
greater
than about 98% or greater than about 99%.
In certain non-limiting embodiments, a decrease in the mutant allele
fraction of a PIK3CA biomarker of less than about 25%, less than about 20%,
less
.. than about 15%, less than about 10%, less than about 9%, less than about
8%, less
than about 7%, less than about 6%, less than about 5%, less than about 4%,
less than
about 3%, less than about 2% or less than about 1% may indicate that it is
less likely
that a P13 Ku inhibitor is producing an anti-cancer effect on the cancer.
In certain embodiments, a decrease in the mutant allele fraction of a
.. PIK3CA biomarker of greater than about 90% in one or more samples obtained
after
treatment, as compared to a sample obtained prior to treatment, may indicate
that a
PI3Ka inhibitor is more likely producing an anti-cancer effect.
In certain embodiments, a decrease in the mutant allele fraction of a
PIK3CA biomarker of greater than about 90% for two or more samples obtained
.. during P13 Ku inhibitor treatment may indicate that a P13 Ku inhibitor is
more likely
producing an anti-cancer effect. In certain embodiments, a sample can be
obtained on
day 1 of a treatment cycle and a sample can be obtained on the last day, e.g.,
day 28,
of the treatment cycle.
In certain embodiments, a decrease in the mutant allele fraction of a
PIK3CA biomarker of greater than about 90% for two or more samples obtained
after
initiation of treatment, as compared to a sample obtained prior to treatment,
may
indicate that a PI3Ka inhibitor is more likely producing a continued anti-
cancer effect.
In certain embodiments, a decrease in the mutant allele fraction of a
PIK3CA biomarker of less than about 25% in one or more samples obtained after
initiation of treatment, as compared to a sample obtained prior to treatment,
may
indicate that a PI3Ka inhibitor is less likely producing an anti-cancer
effect.
An "anti-cancer effect," as used herein, refers to one or more of a
reduction in aggregate cancer cell mass, a reduction in cancer cell growth
rate, a
reduction in cancer cell proliferation, a reduction in tumor mass, a reduction
in tumor
volume, a reduction in tumor cell proliferation, a reduction in tumor growth
rate,
and/or a reduction in tumor metastasis.
In certain non-limiting embodiments, the present invention provides
for a method of determining whether an anti-cancer effect is likely being
produced in
a cancer by a PI3Ka inhibitor, comprising obtaining one or more samples
sequentially
13
CA 02948351 2016-11-07
WO 2015/172085
PCT/US2015/029995
from a patient undergoing treatment with a PI31(a inhibitor, and determining,
in each
sample, the presence and/or level of a PIK3CA biomarker, where if the presence
and/or level of the biomarker increases, it is more likely that the PI3Ka
inhibitor is
not producing an anti-cancer effect on the cancer.
In certain embodiments, if the presence and/or level of a PIK3CA
biomarker decreases over the course of treatment with a P13 Ku inhibitor, the
method
can further include resuming and/or continuing treatment of the subject with a
therapeutically effective amount of a PI3Ka inhibitor. For example, and not by
way
of limitation, if the presence and/or level of a PIK3CA biornarker decreases
greater
than about 90%, the method can further include resuming and/or continuing
treatment
of the subject with a therapeutically effective amount of a P13 Ku inhibitor.
In certain
non-limiting embodiments, the P13 Ku can be the same or different from the P13
Ku
inhibitor administered during the determination of the change in PIK3CA
biomarker
presence and/or levels in the subject. A therapeutically effective amount is
an amount
1 5 that is able to achieve one or more of an anticancer effect,
prolongation of survival
and/or prolongation of period until relapse.
In certain non-limiting embodiments, the PI3Ku inhibitor used to treat
the subject after the detection of a decrease in the presence and/or levels of
a
biomarker may be of the same or different chemical class than the PI3Ka
inhibitor
administered during the determination of the biomarker change in the subject.
In
certain non-limiting embodiments, the PI3Ka inhibitor used to treat the
subject after
the detection of a decrease in the presence and/or levels of a biomarker may
function
by a similar or different mechanism than the PI3Ka inhibitor administered
during the
determination of the biomarker change in the subject.
In certain embodiments, if the presence and/or level of a PIK3CA
biomarker increases over the course of treatment with a P13 Ku inhibitor, the
method
can further include initiating treatment with another modality, for example,
one or
more alternative chemotherapeutic agents; radiotherapeutic agent; anti-
angiogenic
agent; apoptosis-inducing agent; anti-cancer antibody; agents which promote
the
activity of the immune system including but not limited to a cytokine such as
but not
limited to interleukin 2 or interferon, anti-CTLA4 antibody, anti-PD-1
antibody,
and/or anti-PD-L I antibody, letrozole or exemestane.
As used herein, "determining the presence and/or level- of biomarker
refers to quantitative measurements as well as detecting the presence or
absence of the
14
CA 02948351 2016-11-07
WO 2015/172085 PCT/US2015/029995
biomarker. In certain embodiments, the level of a biomarker in sample can
refer to
the level of the biomarker compared to the reference control, as a percentage
or a
fraction (referred to herein as the mutant allele fraction). For example, and
not by
way of limitation, a mutant allele fraction can be the ratio of the level of a
biomarker,
e.g., mutant allele of PIK3CA, to the level of a reference control in the same
sample.
As described above in section 5.1, in certain embodiments, the reference
control can
be the allele of PIK3CA present in the majority of the population.
Alternatively or additionally, the level of a biomarker in a sample can
refer to the total amount of the biomarker in the sample. For example, and not
by way
of limitation, the level of a biomarker can be the number of molecules of the
biomarker (e.g., number of copies of the nucleic acid biomarker molecules) or
the
concentration of the biomarker in terms of weight per volume of biological
sample,
e.g., plasma.
5.6 KITS
In certain non-limiting embodiments, the present invention provides
for kits for deten-nining the effectiveness of treatment with a PI3Ka
inhibitor in a
subject having cancer, comprising a means for detecting one or more biomarkers
set
forth in section 5.1.
Types of kits include, but are not limited to, packaged biomarker-
specific probe and primer sets (e.g., TaqMan probe/primer sets),
arrays/microarrays,
which further contain one or more probes, primers, biomarker-specific beads or
other
reagents for detecting one or more biomarkers of the present invention.
In a specific, non-limiting embodiment, a kit may comprise a pair of
oligonucleotide primers, suitable for polymerase chain reaction (PCR) or
nucleic acid
sequencing, for detecting the biomarker(s) to be identified. A pair of
printers may
comprise nucleotide sequences complementary to a biomarker set forth above,
and be
of sufficient length to selectively hybridize with said biomarker.
Alternatively, the
complementary nucleotides may selectively hybridize to a specific region in
close
.. enough proximity 5' and/or 3' to the biomarker position to perform PCR
and/or
sequencing. Multiple biomarker-specific primers may be included in the kit to
simultaneously assay large number of biomarkers.
The kit may also comprise one or more polymerases, reverse
transcriptase, and nucleotide bases, wherein the nucleotide bases can be
further
CA 02948351 2016-11-07
WO 2015/172085 PCT/US2015/029995
detectably labeled. For example, in certain embodiments, the kits may comprise
containers (including microliter plates suitable for use in an automated
implementation of the method), each with one or more of the various reagents
(typically in concentrated form) utilized in the methods, including, for
example, pre-
fabricated microarrays, buffers, the appropriate nucleotide triphosphates
(e.g., dATP,
dCTP, dGTP and dTTP, or rATP, rCTP, rGTP and UTP), reverse transcriptase, DNA
polymerase, RNA polymerase, and one or more probes and primers of the present
invention (e.g., appropriate length poly(T) or random primers linked to a
promoter
reactive with the RNA polymerase).
I 0 In certain non-limiting embodiments, a primer may be at least about
10
nucleotides or at least about 15 nucleotides or at least about 20 nucleotides
in length
and/or up to about 200 nucleotides or up to about 150 nucleotides or up to
about 100
nucleotides or up to about 75 nucleotides or up to about 50 nucleotides in
length.
In a further non-limiting embodiment, the oligonucleotide primers may
be immobilized on a solid surface or support, for example, on a microarray,
wherein
the position of each oligonucleotide primer bound to the solid surface or
support is
known and identifiable. The terms "arrays," "microarrays," and "DNA chips" are
used herein interchangeably to refer to an array of distinct polynucleotides
affixed to a
substrate, such as glass, plastic, paper, nylon or other type of membrane,
filter, chip,
.. bead, or any other suitable solid support. The polynucleotides can be
synthesized
directly on the substrate, or synthesized separate from the substrate and then
affixed to
the substrate. The arrays are prepared using known methods.
In a specific, non-limiting embodiment, a kit may comprise at least one
nucleic acid probe, suitable for in situ hybridization or fluorescent in situ
.. hybridization, for detecting the biomarker(s) to be identified. Such kits
will generally
comprise one or more oligonucleotide probes that have specificity for various
biomarkers. Means for testing multiple biomarkers may optionally be comprised
in a
single kit.
In one specific non-limiting embodiment, a kit may comprise one or
more pairs of primers, probes or microarrays suitable for detecting one or
more
PIK3CA biomarkers including the PIK3CA H 1047R biomarker, PIK3CA E545K
biomarker, PIK3CA E542K biomarker, the PIK3CA E545G biomarker, PIK3CA
E545Q biomarker, PIK3CA E545A biomarker, PIK3CA E545D biomarker, PIK3CA
E545V biomarker, PIK3CA H1047L biomarker, PIK3CA H1047Y biomarker,
16
CA 02948351 2016-11-07
WO 2015/172085 PCT/US2015/029995
PIK3CA E542Q biomarker, PIK3CA E542G biomarker, P1K3CA P539R biomarker,
P1K3CA N345K biomarker, PIK3CA C420R biomarker, PIK3CA G1049R
biomarker, PIK3CA E726K biomarker, PIK3CA R88Q biomarker, PIK3CA Q546K
biomarker, PIK3CA Q546P biomarker, PIK3CA Q546R biomarker, P1K3CA Q546L
biomarker or combinations thereof.
In one specific non-limiting embodiment, a kit may comprise a pair of
primers, a probe or microarray suitable for detecting the PIK3CA E545K
mutation
biomarker.
In one specific non-limiting embodiment, a kit may comprise a pair of
primers, a probe or microarray suitable for detecting the PIK3CA E545G
mutation
biomarker.
In one specific non-limiting embodiment, a kit may comprise a pair of
primers, a probe or microarray suitable for detecting the PIK3CA E542K
mutation
biomarker.
In one specific non-limiting embodiment, a kit may comprise a pair of
primers, a probe or microarray suitable for detecting the PIK3CA H1047Y
mutation
biomarker.
In one specific non-limiting embodiment, a kit may comprise a pair of
primers, a probe or microarray suitable for detecting the PIK3CA H1047R
mutation
biomarker.
In one specific non-limiting embodiment, a kit may comprise a pair of
primers, a probe or microarray suitable for detecting the P1K3CA H1047L
mutation
biomarker.
In certain non-limiting embodiments, where the measurement means in
the kit employs an array, the one or more biomarkers set forth above may
constitute at
least 10 percent or at least 20 percent or at least 30 percent or at least 40
percent or at
least 50 percent or at least 60 percent or at least 70 percent or at least 80
percent of the
species of markers represented on the microarray.
In certain non-limiting embodiments, a biomarker detection kit may
comprise one or more one or more probes, primers, microarrays/arrays, beads,
detection reagents and other components (e.g., a buffer, enzymes such as DNA
polymerases or ligases, chain extension nucleotides such as deoxynueleotide
triphosphates, and in the case of Sanger-type DNA sequencing reactions, chain
terminating nucleotides, and the like) to detect the presence and/or level of
a reference
17
CA 02948351 2016-11-07
WO 2015/172085
PCT/US2015/029995
control. Non-limiting examples of a reference control are described above in
section
5.1.
In certain non-limiting embodiments, a kit can further include
instructions for using the kit to detect the biomarker of interest. For
example, the
instructions can describe that a decrease in the level and/or presence of a
biomarker,
set forth herein, in serial samples from a patient undergoing treatment with a
PI3K
inhibitor, is indicative of a likelihood of an anti-cancer effect in a cancer
by a PI3K
inhibitor.
Alternatively or additionally, the instructions can further describe that
an increase in the level and/or presence of a biomarker, set forth herein, in
serial
samples from a patient undergoing treatment with a PI3K inhibitor, is
indicative of an
increased possibility of an anti-cancer effect in a cancer by the PI3K
inhibitor.
The following examples are offered to more fully illustrate the
disclosure, but are not to be construed as limiting the scope thereof.
6. EXAMPLE 1: Monitoring PIK3CA mutant allele fraction of cell free DNA
in metastatic breast cancer patients treated with a P13 Ku¨inhibitor, in
combination
with letrozole or exemestane.
In this example, results of a correlative aim assessing serial mutant
allele fraction in a phase I study of a PI3Ka-inhibitor, BYL719, with
letrozole (L) or
exemestane (E) in metastatic breast cancer patients are reported. Tumor-
derived cell
free DNA (et-DNA) was extracted from the plasma patient with tumors that
harbored
activating mutations and serially quantified by droplet digital PCR (ddPCR).
These
results indicate that mutant allele fractions may be predictive of target-
directed
therapy for initial response and evolving resistance.
6.1. MATERIALS AND METHODS
PIK3C4 status of tumors within patients was determined by molecular
analysis of the tumor. It was determined that 8 patients were mutant for
PIK3CA, 5
patients were wildtype for PIK3CA and 1 patient was unknown. Plasma samples
were collected at baseline and on day 1 of each 28 day cycle while on the
protocol.
Cell free DNA was extracted using QIAamp Circulating nucleic acid kit (Qiagen)
and
quantified by KAPA human genomic DNA qPCR (Kapa Biosystems). Allele specific
assays for PIK3CA E542K, E545K, H1047R and H1047L mutations were designed
for quantification on BioRad QX200 DROPLET DlGITALIm PCR System. For
18
CA 02948351 2016-11-07
WO 2015/172085
PCT/US2015/029995
example, to detect and quantify the PIK3CA HI 047R mutation, a forward primer
(GAGCAAGAGGCTITGGAGTAT) and reverse primer
(GCTGTTTAATTGTGTGGAAGATCC) were used for thennocycling in conjunction
with a TAQMAN probe (VIC-AATGATGCACaTCAT- MGBNFQ) for the wild
type PIK3CA 3140A PIK3CA sequence and a TAQMAN probe (6FAM-
TGAATGATGCACgTCAT- MGBNFQ) for the PIK3CA 3140G mutation. Mutant
allele fraction was determined from the counts for mutant as compared to wild-
type in
the sample. The detection limit for each assay was calculated from the number
of
events detected.
6.2. RESULTS
5 out of 8 patients with PIK3CA mutation were evaluated by ddPCR
having received treatment for at least 2 cycles with the P13Ku-inhibitor. In
the plasma
of 4 patients, we identified a brisk decrease in PIK3CA mutant allele fraction
at cycle
2, day 1 (C2D1) ranging from 91.8- 99.6% decrease from baseline (see, for
example,
.. FIGURE 1; 002 AF, 004 AF and 009 AF). All 4 patients had stable or
responding
disease as best response. In 1 patient, later determined to have disease
progression,
there was a small change (19% decrease) in PIK3CA mutant allele fraction at
C2D1
with a 16X increase in mutant allele fraction at cycle 3 (FIGURE 2, 008 AF).
Ongoing responses result in a persistent low (>98% decrease) or undetectable
mutant
alleles (see, for example, FIGURE 1; 004AF and 009 AF). In 1 patient, with a
transient response, the marked decrease in mutant allele fraction at C2D1 was
followed by an increase at cycle 3 predicting the patient's clinical
progression
(FIGURE 1, 002 AF).
6.3. DISCUSSION
Targeted therapies directed towards specific oncogene mutations may
be assessed serially by ddPCR to confirm mutant target sensitivity. Assessment
fur
early tumor resistance may allow a more rapid treatment change with real-time
monitoring of target mutant alleles. Additionally, the detection of tumor
derived
fragments of DNA in a blood sample provides the opportunity for relatively non-
.. invasive serial assessment of the genetic alterations harbored in the
tumor, which can
be helpful in the exploration of the more effective and better tolerated
dosing
schedules. Further, the analysis of cfDNA can be used to determine tumor
mutation
status for patients in whom a biopsy is not feasible and serial assessment for
new
genetic alterations that occur during tumor progression and therapeutic
resistance.
19
CA 02948351 2016-11-07
WO 2015/172085 PCT/US2015/029995
7. EXAMPLE 2: Additional monitoring of the PIK3CA mutant allele fraction
of cell free DNA in metastatic breast cancer patients treated with a PI3Ka-
inhibitor, in
combination with letrozole or exemestane.
7.1. MATERIALS AND METHODS
Peripheral blood samples were processed by separating the plasma
from the cellular fraction in a clinical hematology lab by centrifugation at
2000 x g
for 10 minutes. Plasma underwent a second centrifugation at 16,000 x g for 10
minutes and stored frozen at -80 degrees until further processing.
Cell free DNA (cfDNA) was extracted from 2-5m1 of plasma using the
QIAamp Circulating Nucleic Acid Kit (Qiagen). The concentration and integrity
of
cfDNA was determined by qPCR using the KAPA Human Genomic DNA
quantification and QC kit (KAPAbiosystems). This method generated standard
curves
based on the amplification of a 41 bp, 129 bp and 305 bp fragment of a
conserved
single copy human gene. The ratio of the amplified 129 bp quantity to the 41
bp
quantity was used as a measure of DNA quality.
Allele specific assays for PIK3CA E542K, E545K, H1047R and
H1047L mutations were designed for quantification on BioRad QX200 Droplet
Digital PCR System. The Droplet digital PCR method included the generation of
droplets with cfDNA templates, reactions primers and TAQMAN probes, followed
by
PCR. Droplets are then counted and scored for fluorescent wild-type or mutant
probes. Mutant allele fraction was determined from the counts for mutant as
compared to wild-type alleles. A representative example of serial PIK3CA E542K
mutation analysis by ddPCR from cfCDNA is shown in FIGURE 3. The lower limit
of detection for each assay is calculated from the number of events detected.
7.2. RESULTS
In all patients with tumors that were mutated for PIK3CA, a cfP1K3CA
mutant allele was detected in the patient's plasma sample (TABLE 1). For
example,
in patients 002 and 013, the PIK3CA mutant allele E545K was detected by ddPCR.
In patients 007 and 008 that had tumors with mutations in PIK3CA, the PIK3CA
141047R mutant allele was detected by ddPCR of circulating DNA, indicating
that
PIK3CA mutant allele fractions in cell free DNA can be used to monitor
therapeutic
treatments.
As shown in TABLE 2, cIPIK_3CA mutant allele fractions
demonstrated a brisk decrease at day 28 in 5 out of 6 patients with stable and
CA 02948351 2016-11-07
WO 2015/172085 PCT/US2015/029995
responding disease (FIGURE 4). In patients 007 and 004 which demonstrated
stable
disease while on the study, exhibited greater than 90% decrease in mutant
allele
fraction by day 28. In patient 002, the cfPIK3CA E545K mutant allele fraction
in
response to treatment with BYL71 9 and letrozole decreased by 97.4% at day 28
(C2D1), which was followed by a 17 fold increase at C3D1 (FIGURES 1 and 4).
Clinically, the patient had a 29% decrease in tumor volume by REC1ST but then
had
rapid disease progression.
In patient 009, who has been treated with 8 chemotherapy and 4 anti-
estrogen regimens, displayed a significant decrease in cfPIK3CA E542K mutant
allele
fraction in response to treatment with BYL719 and letrozole. At day 28, ddPCR
quantifies a 99.7% decrease in the cfPIK3CA mutant allele fraction with no
mutation
detected at C3 and C4, despite a dose reduction (FIGURE 5). The mutant allele
fraction of PIK3CA E542K in patient 009 decreased from 0.4507 at day 1 to
0.0014 at
day 28 (TABLE 2 and FIGURE 5). Although an upward trend in mutant allele
fraction has been identified since C5, the patient's response to treatment has
continued. Next-generation sequencing has identified 2 activating PIK3CA
mutations
(E542K and E543K) in patient 009. CT scan of the patient prior to treatment
displayed extensive hepatic tumor burden. At C3D1, C5D1 and C8D1, patient
exhibited a partial response (PR) by REC1ST criteria by CT scan (FIGURE 5). By
contrast, patient 008 which exhibited a 19.6% decrease in mutant allele
fraction
between day 1 and day 28 displayed a progression of the disease (TABLE 2). In
patient 008, the marked 12.5 fold increase in H1047R mutant allele fraction at
week 8
(C3D1) correlated with tumor progression, which was confirmed by CT imaging
(FIGURES 2 and 4). These results are also displayed in FIGURE 4 and indicate
that
the measurement of the cfPIK3CA mutant allele fraction at day 28 of treatment
is
predictive for clinical benefit with the P13Ka inhibitor BYL719 and aromatase
inhibition.
As shown in TABLE 2 and FIGURE 6, ddPCR was sensitive for
detecting low levels of PIK3CA mutant alleles. Mutant allele fractions as low
as
0.00068 and 0.00107 were detected (FIGURE 6). Despite low concentrations of
total
efDNA, similar to levels identified in patients without cancer, serial
cfPIK3CA
mutant allele fraction measurements were informative. For example, in patient
004,
the total cfDNA was measured to be ¨6ng/m1 plasma before treatment, yet 11% of
detected PIK3CA alleles are mutant (FIGURE 7). A 91.8% decrease at C2D1 and
CA 02948351 2016-11-07
WO 2015/172085
PCT/US2015/029995
98.9% at C3 in cfPIK3CA mutant allele fraction was observed following
treatment
and these significant decreases in cfPIK3CA mutant allele fraction correlated
with the
patient's ongoing stable disease. After cycle 11, the cfPIK3CA mutant allele
fraction
continued to be >99% decreased or undetectable (FIGURE 7).
Table 1.
- -..,. cfONA ddPC-R:,:4
13.0,27 . = = Tumor . ; ' µ,..-. ' ',, - - , ,µ :,-
.1:191m1 PtP1K3C,A
Patient . Sites of metastatic genotyping PIK364 ,,:l.
40:illa'Ot ,-," '-.):::-ilastri.a mutant
'...,,,10 ' ''-. , . ' '., itliSOAS,e method gene ?'*d:.-.
' allele,. ; " daY,1- -, k e ...i.,¶:, j.li',;,'
al - -1-. - .. netiOhl- -.,=ild*-
type `.- '' '--' v-7--' -; -.- :'-',i
' 002 bone, liver sequenom mutation E545K 328.9 Yes
- 003 . ' liver:LN sequenom wild-type
/ 004 bone sequenom mutation E542K 4.1
Yes
' 005 tone not assessed .
'' 41.0
006 bone, liver sequenom wild-type 16.5
/ 007 bone, liµer, LN, breast sequenom mutation
H1047R 20.1 Yes
' 008 bone, liver, lung. brain, LN NGS mutation
H1047R 112.3 Yes
= 009 bone, liver, LN sequenom
mutation E542K 595,4 Yes
/ 012 bone, INer, lung, LN sequenom
mutation E542K 3908.5 Yes
F 013 solitary bone sequenom mutation E545K 15.4 Yes
' 014 bone sequenom wild-
type 8.9
' 016 bene,.1iYer, lung, LN, breast NOS ' wild-
type 45.3 -
' 017 bone, her, lung, LN sequenom
mutation H1047L 61.7 Yes ,
*Patient 012 was on the study for 7 days and only the day 1 sample was
assessed by ddPCR.
Table 2.
'....','..P.Vi.n...'iT',,,,. :!!.474'.',..r,WIK., -7,..rt,41.:-'=
Best
- ' ' - Ili utanl AF 1 = =
..,µ .,,,.. ., ../., -, , : -i .. , = , . ,,n9lnit,, .KSqA.:.;1-PIK3CA =
Clinical. Days
1,t1din .,-,..:,81tess9-ftlle,Aktali=_. '.4",ilutatif ., .,-
pJi,sfro..M.I'iatgit,- rilutant AF 1% decreasej Respon. -Ve Off
f4.. , ' ':. :',4i074::'., ttioi 'idpii- ..al '
1 , -. ,..day 28 .-4a 28 i ..se, . -sluor .stuoY
,650 - bone her, LN E542K 695.4 ' -6.4507 '
0:0014 99.7 - , - PR . Ii3o '
pcw bone, I.,.er LN. breast H1047R 20.1 .
0.0705 0.0013 98.2 SD 199 POD
= , ,
002 bone, liver E545K 328.9 0.3590 0.0092 97.4 -
Mct " 88 POD
r 004 bore E542K 4.1 0 1110 0.0091 91.8 SD
289
,
DV bone, User, lung, LN H10471 61,7 0.5854
0.1290 77.9 SD 146 POD
008 bone her, lung, brain, LN H1047R 112.3 0.1052 0.0850
19.6 POD 58 POD
- . - .
613* solitary bone E545K 18.4 0.0007 Ø0007 0 SD
172
*Changes in the mutant allele fraction at or near the lower limit of detection
for the assay may not be
reliable for interpretation.
In patient 017, the cfPIK3CA mutant allele fraction decreased at day
28 (C2D1); however, the decrease was 78% as compared to other responders where
the initial decrease was >90% (FIGURE 8). This early decrease was followed by
a
22
CA 02948351 2016-11-07
WO 2015/172085 PCT/US2015/029995
progressive two-fold increase in the H1047L mutant allele fraction, although
the
patient had stable disease clinically and by imaging until progression of
disease at
cycle 6 (FIGURE 8). These results suggest that increases in the cfPIK3CA
mutant
allele fraction may precede clinical progression by 1 to 3 months. Patient 017
harbored a tumor with a PIK3CA HI 047L mutation, which is less activating than
the
H1047R hotspot mutation. The identification of such increases in mutant allele
fraction may also be used to detect early resistance of the tumor to treatment
by
analysis of cfPIK3CA mutant alleles and can be used to determine the timing of
tumor
biopsies to perform further analysis of the tumor sample, e.g., by next-
generation
DNA sequencing (FIGURE 9).
Further analysis of an additional 40 patients with tumors mutated for
PIK3CA showed that in patients that exhibited a clinical benefit (e.g.,
exhibited six
months of disease stability) to the PI3Ku inhibitor, an average decrease of
96.9% in
the cfPIK3CA mutant allele fraction was observed during the first cycle of
treatment.
This average decrease in the cfPIK3CA mutant allele fraction was observed by
day 7,
day 14 or day 28 of treatment. In patients with a progression of disease, a
transient
decrease, no decrease or an increase was observed in the cfPIK3CA mutant
allele
fraction by ddPCR during the first cycle of treatment. In patients with a
progression
of disease, an average decrease of 25% in the cfPIK3CA mutant allele fraction
was
observed.
7.3. DISCUSSION
Analysis of cfDNA in this Example, has demonstrated an early (day
28) and large (average 93%) decrease in etPIK3CA mutant allele fraction at the
first
post treatment assessment in patients with stable and responsive disease. A
patient
with progressive disease exhibited a small (approximately 20%) and transient
decrease in cfPIK3CA mutant allele fraction. A persistent disease in the
cfPIK3CA
mutant allele fraction associated with continued response. An increase in
cfPIK3CA
mutant allele fraction predicts for tumor resistance. Without being limited to
a
particular theory, the increase in cfPIK3CA mutant allele fraction may precede
clinical progression by 1 to 3 months.
Incorporating cfPIK3CA mutant allele fraction assessment may be
considered to identify early tumor response, identify evolving resistance
prior to
clinical progression, allow time tissue biopsy to assess samples arising
resistance
mechanisms. Dynamic changes in cfPIK3CA mutant allele fraction can be
23
informative for developing personalized dosing strategies that limit toxicity
and deter
therapeutic resistance.
24
Date Recue/Date Received 2021-08-23