Note: Descriptions are shown in the official language in which they were submitted.
DEMANDE OU BREVET VOLUMINEUX
LA PRESENTE PARTIE DE CETTE DEMANDE OU CE BREVET COMPREND
PLUS D'UN TOME.
CECI EST LE TOME 1 DE 2
CONTENANT LES PAGES 1 A 293
NOTE : Pour les tomes additionels, veuillez contacter le Bureau canadien des
brevets
JUMBO APPLICATIONS/PATENTS
THIS SECTION OF THE APPLICATION/PATENT CONTAINS MORE THAN ONE
VOLUME
THIS IS VOLUME 1 OF 2
CONTAINING PAGES 1 TO 293
NOTE: For additional volumes, please contact the Canadian Patent Office
NOM DU FICHIER / FILE NAME:
NOTE POUR LE TOME / VOLUME NOTE:
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
IMMUNE SYSTEM MODULATORS AND COMPOSITIONS
RELATED APPLICATIONS
[0001]
This application claims the benefit of US Provisional App. No. 62/129669
filed March 6, 2015, which is hereby incorporated by reference in its
entirety. This
application is related to PCT Application No. PCT/U52014/054612 filed
September 8, 2014,
and U.S. Provisional Application Ser. No. 61/875598, filed September 9, 2013,
each of
which is hereby incorporated by reference in its entirety.
SEQUENCE IN ELECTRONIC FORMAT
[0002] The
present application is being filed along with a Sequence Listing in
electronic format. The
Sequence Listing is provided as a file entitled
CANIG006WOSEQUENCE.TXT, created and last saved on March 1, 2016, which is
162,388 bytes in size. The information in the electronic format of the
Sequence Listing is
incorporated herein by reference in its entirety.
FIELD OF THE INVENTION
[0003]
Aspects of the present invention generally relate to compositions that
interact with molecules, which suppress the immune system.
More specifically,
embodiments described herein concern the discovery, manufacture, and use of
compositions
that modulate the immune system.
BACKGROUND OF THE INVENTION
[0004] The
immune system is finely tuned to detect and eradicate foreign
molecules and, at the same time, avoid over reactivity, which could result in
destruction of
normal tissues resulting in autoimmune or chronic inflammatory diseases. The
initiation of a
specific immune response is a well-orchestrated chain of events culminating in
the activation
of effector functions, such as the release of cytokines, production of
specific antibodies
and/or cellular cytotoxic activity.
1
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
[0005] The role of the immune system in human cancer has been under
debate for
several years. It has been puzzling, for example, that an increased incidence
of malignant
tumors is not observed in immunocompromised animals, such as nude mice. It is,
however,
now clarified that these animal models were in reality not profoundly
immunocompromised,
but were still able to mount a significant anti-tumour immune reactivity. When
severely
immunocompromised transgenic mice of the Stat 1 -/-, IF'NyR -/-, or RAG2 -/-
genotypes
were studied, the tumor incidence and the immunogenicity of cancers growing in
these
animals strongly supported the existence of an immune mediated anti-cancer
reactivity with
the capacity to control cancer development. Based on these results, the
immunoediting
model was developed (Dunn and Schreiber, Immunity, 21:137-148 (2004)).
[0006] Similarly, the modest increase in cancer incidence in
therapeutically
immunosuppressed, allo-organ transplanted patients seems to be explained by
the early
appearance of immunosuppression in epithelial cancers (Schiile J, et al.,
Breast Cancer Res
Treat. 2002; 74:33-40; Wolfram RM, et al., Int J Cancer. 2000; 88:239-44.,
Petersen RP, et
al., Cancer. 2006; 107:2866-72). The occurrence of spontaneous immune-mediated
tumor
regression, the correlation between tumor-infiltrating lymphocytes and
prognosis, the
occurrence of tumor specific cytotoxic T-lymphocytes and antibodies and the
efficacy of
immunostimulatory treatment all support a significant role of the immune
system in the
control or regulation of cancer progression.
[0007] These observations are also consistent with the results of
Clinchy et al.
(Clinchy B, et al., Cancer. 2007; 109:1742-9), showing that dysregulation of
the immune
system in cancer, with an enhanced capacity to produce IL-6, correlate to poor
prognosis in
radically resected colorectal cancer patients. Not even in the group of high
risk patients with
locally advance tumors, T3N1-2, did patients die from their cancer if their
immune cells
exhibited a normal production of IL-6. Similarly, Galon et al. (Galon J, et
al., Science. 2006;
313:1960-4, Mlecnik B, et al., J Clin Oncol. 2011, 29:610-8) have shown that T-
cell immune
parameters strongly correlate to the prognosis in these patients.
[0008] The majority of human cancers of different origin induce immune
mediated anti-tumor reactivity, but immunosuppressor mechanisms often
appearing at an
early stage, compromise the immune system. The existence of regional
immunosuppression
2
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
in the absence of systemic suppression (concomitant immunity), indicates a
regional,
systemic gradient of immunosuppression (Gorelik E., et al., Adv Cancer Res.
1983;39:71-
120). For instance, the function of immune cells can be more impaired near the
tumor than in
peripheral blood (Vose BM, et al., Int J Cancer 1977 20:895-902). Several
factors may
mediate this suppression (Menetrier-Caux C, et al., Br J Cancer 1999 79: 119-
130., Heimdal
JH, et al., Scand J Immunol 2000 51: 271-278., Heimdal JH, et al., Scand J
Immunol 2001
53: 162-170), but no fundamental mechanism has been identified (Kim R, et al.,
Cancer Res.
2006 Jun 1;66(11):5527-36, Mocellin S, et al., J Immunother 2001 24:392-407).
The impact
of the hostile intra-tumoral milieu has been described by several groups
(Perdrizet GA, et al.,
J Exp Med. 1990;171:1205-20, Yu P, et al., J Exp Med. 2005 201:779-91.) Immune
reactivity against cancer can be suppressed at various levels, e.g.,
initiation, recruitment of
effector cells to the tumor and migration of these cells within the tumor and
their cytotoxic
activity. Effector mechanisms present at the tumor site can also provide
immune mediated
cancer control.
[0009] Although data indicate that the immune system is of major
importance for
cancer control (Dunn GP, et al., Immunity. 2004 21:137-48., Galon J, et al.,
Science. 2006
313:1960-4., Koebel CM, et al., Nature. 2007 450:903-7, Clinchy B, et al.,
Cancer. 2007
109:1742-9, Teng MW, et al., J Leukoc Biol. 2008 84:988-93) malignant tumors
continue to
grow and the efficacy of immunotherapy is rather poor with an objective
remission rate of 10-
20%. There can be several reasons for this apparent paradox, e.g., tumors
avoid recognition
by the immune system due to tumor antigens being weak self-antigens, poor
antigen
presentation due to down-regulation of TAP and MHC I and II) or induction of
tolerance or
cancer related immunosuppression. The impact of an hostile intra-tumoral
milieu is
demonstrated by results from animal experiments (Perdrizet GA, et al., J Exp
Med.
1990;171:1205-20., Yu P, et al., J Exp Med. 2005 201:779-91.) and human tumors
(Gajewski
TF, et al., J Immunother. 2006 29:233-40, Whiteside TL, Oncogene. 2008 27:5904-
12).
[0010] Different types of immunosuppressor cells, regulatory T-cells,
immature
dendritic cells (iDC), tumor associated macrophages (TAM) and myeloid derived
suppressor
cells (MDSC), can function substantially in cancer related immunosuppression.
The immune
balance is generally skewed to a Th2 dominance characterized by cytokines,
such as IL-4, IL-
3
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
and PGE2. Additionally, other immunosuppressor mechanisms, such as serum
blocking
factors, circulating immune complexes, enhanced IL-1Ra production and enhanced
intra-
tumoral proteolytic activity can function in cancer related immunosuppression.
[0011] While investigating mechanisms for induction of interleukin-6
(IL-6) in
cancer patients, immunoregulatory peptide sequences derived from serum albumin
were
found (see e.g., US Patent Nos. 7,960,126; 8,110,347; and 8,110,347; as well
as, US
Publication No. 2010/0323370, each of which is hereby expressly incorporated
by reference
in their entireties. Interleukin-2 (IL-2) plays a major role in initiation and
activation of the
immune response and its capacity to induce lymphokine activated killer cells
(LAK-cells), T-
cell proliferation and cytotoxicity. Several reports have shown that
peripheral blood
mononuclear cells (PBMC) from cancer patients have a diminished capacity to
both
synthesize (Wanebo HJ, et al., Cancer. 1986 57:656-62, Mantovani, G., et al.,
Diagn. Clin.
Immunol. 1987 5: 104-111, Lauerova L, et al., Neoplasma 1999 46: 141-149) and
respond to
IL-2 (Tsubono M, et al., J Clin Lab Immunol 1990 33:107-115, Pellegrini P, et
al., Cancer
Immunol Immunother 1996 42:1-8). Soluble products from tumor explants or serum
from
cancer patients can inhibit cytokine production, inhibit IL-2 receptor
expression (Botti C, et
al., Intl J Biol Markers 1998 13:51-69, Lauerova L, et al., Neoplasma 1999
46:141-149)
and/or reduce the proliferative capacity in normal T lymphocytes (Botti C, et
al., Intl J Biol
Markers 1998 13:51-69).
[0012] Integrins are a superfamily of transmembrane glycoproteins,
found
predominantly on leukocytes that mediate cell-cell and cell substratum
interactions. Integrins
play an important role in immune regulation, as well, in particular aLf32,
(Leukocyte
Function Associated molecule-1, LFA-1) is of pivotal importance for the
initiation and
regulation of an immune response, tissue recruitment and migration of
inflammatory cells
and cytotoxic activity of lymphocytes (Hogg N, et al., J Cell Sci. 2003
116:4695-705, Giblin
PA, et al., Curr Pharm Des. 2006 12:2771-95, Evans R, et al., Cell Sci. 2009
122:215-25). In
addition, LFA-1 is involved in the proliferative response to interleukin-2
(Vyth-Dreese FA,
Eur J Immunol. 1993 12:3292-9) and some fragments of albumin bind to LFA-1
and/or the
IL-2 receptor thereby modulating the functional properties mediated through
these receptors
including immune cell proliferation (see U.S. Publication No. 2011/0262470,
which is hereby
4
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
expressly incorporated by reference in its entirety). Despite these
advancements, the need for
more compositions to modulate the immune system, especially in individuals
that have a
compromised immune system and/or cancer, is manifest.
BRIEF SUMMARY OF THE INVENTION
[0013] A number of Alternatives are provided herein:
[0014] In Alternative 1, a method of treating, inhibiting, or
ameliorating a cancer,
such as a metastatic cancer is provided, the cancer comprising a first tumor
and a second
tumor in a subject. The method can comprise administering a composition
comprising an
isolated peptide comprising the amino acid sequence FFVKLS (SEQ ID NO: 62) to
the
subject, in which (a) the composition is administered intratumorally or
peritumorally to the
first tumor, but not the second tumor, or (b) the composition is administered
at a site in the
subject that is neither intratumoral or peritumoral to either of the first
tumor or the second
tumor, such as systemically, thus ameliorating, inhibiting, or eliminating the
first tumor and
ameliorating, inhibiting, or eliminating the second tumor.
[0015] Alternative 2 comprises the method of Alternative 1, in which
the first
tumor comprises a prostate tumor, a melanoma, a lung carcinoma, a colon
cancer, an
Apocrine gland carcinoma, a testis tumor, a mast cell tumor, a mammary tumor,
a mucinous
carcinoma, or a histicytoma, and wherein the second tumor is a same or
different type of
tumor as the first tumor.
[0016] Alternative 3 comprises the method of Alternative 2, in which
the
mammary tumor comprises a malignant mammary tumor, or the mammary tumor
comprises a
mixed mammary tumor (for example a malignant mixed mammary tumor).
[0017] Alternative 4 comprises the method of Alternative 3, in which
the
mucinous carcinoma comprises a mammary gland mucinous carcinoma.
[0018] Alternative 5 comprises the method of any one of Alternatives 1-
4, in
which, wherein the second tumor is the same type of tumor as the first tumor.
[0019] Alternative 6 comprises the method of any one of Alternatives 1-
4, in
which the second tumor is a different type of tumor from the first tumor.
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
[0020] Alternative 7 comprises the method of Alternative 6, in which
the second
tumor comprises a prostate tumor, a melanoma, a lung carcinoma, a colon
cancer, an
Apocrine gland carcinoma, a testis tumor, a mast cell tumor, a mammary tumor,
a mucinous
carcinoma, or a histicytoma.
[0021] Alternative 8 comprises the method of any one of Alternatives 1-
7, in
which the first tumor is a primary tumor and the second tumor comprises a
metastatic tumor
tumor.
[0022] Alternative 9 comprises the method of any one of Alternatives 1-
7, in
which the first tumor is a metastatic tumor and the second tumor is a
metastatic tumor.
[0023] Alternative 10 comprises the method of any one of Alternatives
1-7, in
which wherein the first tumor is a primary tumor and the second tumor is a
primary tumor.
[0024] Alternative 11 comprises the method of any one of Alternatives
1-10, in
which (a) said composition is administered intratumorally or peritumorally to
the first tumor,
but not the second tumor.
[0025] Alternative 12 comprises the method of any one of Alternatives
1-10, in
which (b) said composition is administered a site in the subject that is
neither intratumoral or
peritumoral to either of the first tumor or the second tumor
[0026] Alternative 13 comprises the method of Alternative 12, in which
said
composition is administered systemically to the subject.
[0027] Alternative 14 comprises the method of Alternative 13, in which
said
systemic administration comprises enteral administration or parenteral
administration.
[0028] Alternative 15 comprises the method of Alternative 13, in which
said
systemic administration comprises at least one of subcutaneous, intravenous,
intraperitoneal,
or intramuscular administration.
[0029] Alternative 16 comprises the method of any one of Alternatives
1-15, in
which the administering further induces regressive changes in the first tumor,
thereby
ameliorating, inhibiting, or eliminating the first tumor.
[0030] Alternative 17 comprises the method of any one of Alternatives
1-16, in
which the administering further induces immune cell infiltration of the first
tumor, thereby
ameliorating, inhibiting, or eliminating the first tumor.
6
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
[0031] Alternative 18 comprises the method of any one of Alternatives
1-17, in
which the administering further induces eradication of cells of the first
tumor, thereby
ameliorating, inhibiting, or eliminating the first tumor.
[0032] Alternative 19 comprises the method of any one of Alternatives
1-18, in
which the administering further induces eradication of the first tumor,
thereby eliminating the
first tumor.
[0033] Alternative 20 comprises the method of any one of Alternatives
1-19, in
which the administering further induces regressive changes in the second
tumor, thereby
ameliorating, inhibiting, or eliminating the second tumor.
[0034] Alternative 21 comprises the method of any one of Alternatives
1-20, in
which the administering further induces immune cell infiltration of the second
tumor, thereby
ameliorating, inhibiting, or eliminating the second tumor.
[0035] Alternative 22 comprises the method of any one of Alternatives
1-21, in
which the administering further induces eradication of cells of the second
tumor, thereby
ameliorating, inhibiting, or eliminating the second tumor.
[0036] Alternative 23 comprises the method of any one of Alternatives
1-22, in
which the administering further induces eradication of the second tumor,
thereby eliminating
the second tumor.
[0037] Alternative 24 comprises the method of any one of Alternatives
1-23, in
which the isolated peptide is administered to the subject at a dose of at
least about 1 ng/kg.
[0038] Alternative 25 comprises a method of ameliorating, inhibiting,
or treating
a cancer comprising administering an isolated peptide comprising the amino
acid sequence
FFVKLS (SEQ ID NO: 62) to a subject having a first tumor, wherein said
isolated peptide is
administered to a site in the subject other than the first tumor, but is not
administered
intratumorally to the first tumor and is not administered peritumorally to the
first tumor,
thereby ameliorating or eliminating the first tumor.
[0039] Alternative 26 comprises the method of Alternative 25, in which
the
subject further comprises a second tumor in a different location than the
first tumor and
different from the site of administration, and wherein said second tumor is
further
ameliorated, inhibited, or eliminated.
7
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
[0040] Alternative 27 comprises the method of Alternative 25, in which
the
subject further comprises a second tumor in a different location than the
first tumor, wherein
said second tumor comprises the site of administration, and wherein said
second tumor is
further ameliorated, inhibited, or eliminated.
[0041] Alternative 28 comprises the method of Alternative 25, in which
the
isolated peptide is administered intratumorally or peritumroally to the second
tumor.
[0042] Alternative 28 comprises the method of any one of Alternatives
26-28, in
which said subject has metastatic cancer, said metastatic cancer comprising
said first and
second tumors, and wherein said metastatic cancer is ameliorated, inhibited,
or eliminated.
[0043] Alternative 30 comprises the method of any one of Alternatives
25-27 or
29, in which said isolated peptide is administered systemically to the
subject.
[0044] Alternative 31 comprises the method of Alternative 30, in which
said
systemic administration comprises enteral administration or parenteral
administration.
[0045] Alternative 32 comprises the method of Alternative 60, in which
said
systemic administration comprises at least one of subcutaneous, intravenous,
intraperitoneal,
or intramuscular administration.
[0046] Alternative 33 comprises the method of any one of Alternatives
25-32, in
which the administering further induces regressive changes in the first tumor,
thereby
ameliorating, inhibiting or eliminating the first tumor.
[0047] Alternative 34 comprises the method of any one of Alternatives
25-33, in
which the administering further induces immune cell infiltration of the first
tumor, thereby
ameliorating, inhibiting or eliminating the first tumor.
[0048] Alternative 35 comprises the method of any one of Alternatives
25-34, in
which the administering further induces eradication of cells of the first
tumor, thereby
ameliorating, inhibiting or eliminating the first tumor.
[0049] Alternative 36 comprises the method of any one of Alternatives
25-35, in
which the administering further induces eradication of the first tumor,
thereby ameliorating,
inhibiting or eliminating the first tumor.
8
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
[0050] Alternative 37 comprises the method of any one of Alternatives
26-36, in
which the administering further induces regressive changes in the second
tumor, thereby
ameliorating, inhibiting or eliminating the second tumor.
[0051] Alternative 38 comprises the method of any one of Alternatives
26-37, in
which the administering further induces immune cell infiltration of the second
tumor, thereby
ameliorating, inhibiting or eliminating the second tumor.
[0052] Alternative 39 comprises the method of any one of Alternatives
26-38, in
which the administering further induces eradication of cells of the second
tumor, thereby
eliminating the second tumor.
[0053] Alternative 40 comprises the method of any one of Alternatives
26-39, in
which the administering further induces eradication of the second tumor,
thereby eliminating
the second tumor.
[0054] Alternative 41 comprises the method of any one of Alternatives
1-40, in
which said isolated peptide comprises no more than 30 amino acid residues.
[0055] Alternative 42 comprises the method of any one of Alternatives
1-40, in
which said isolated peptide comprises no more than 20 amino acid residues.
[0056] Alternative 43 comprises the method of any one of Alternatives
1-40, in
which said isolated peptide comprises no more than 16 amino acid residues.
[0057] Alternative 44 comprises the method of any one of Alternatives
1-40, in
which said isolated peptide consists of the amino acid sequence FFVKLS (SEQ ID
NO: 62).
[0058] Alternative 45 comprises the method of any one of Alternatives
1-40, in
which said isolated peptide comprises the amino acid sequence KKLDTFFVKLSLFTER
(SEQ ID NO: 2).
[0059] Alternative 46 comprises the method of any one of Alternatives
1-40, in
which said isolated peptide consists of the amino acid sequence
KKLDTFFVKLSLFTER
(SEQ ID NO: 2).
[0060] Alternative 47 comprises the method of any one of Alternatives
1-42, in
which said isolated peptide comprises the amino acid sequence
RKLDTFFVKLSLFTERRR
(SEQ ID NO: 586).
9
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
[0061] Alternative 48 comprises the method of any one of Alternatives
1-40, in
which said isolated peptide consists of the amino acid sequence
RKLDTFFVKLSLFTERRR
(SEQ ID NO: 586).
[0062] Alternative 49 comprises an isolated peptide for use in
treating, inhibiting
or ameliorating a cancer, such as metastatic cancer, said cancer comprising a
first tumor and a
metastatic tumor in a subject, said isolated and said use comprising: (a)
intratumoral
administration or petitumoral administration of the isolated peptide to the
first tumor, but not
the metastatic tumor; or (b) administration of the isolated peptide to a site
in the subject that
is neither intratumoral nor peritumoral to either of the first tumor or the
metastatic tumor,
such as systemically, thus ameliorating, inhibiting or eliminating the first
tumor, and
ameliorating or eliminating the metastatic tumor.
[0063] Alternative 50 comprises the isolated peptide for use according
to
Alternative 49, in which the first tumor comprises a prostate tumor, a
melanoma, a lung
carcinoma, a colon cancer, an Apocrine gland carcinoma, a testis tumor, a mast
cell tumor, a
mammary tumor, a mucinous carcinoma, or a histicytoma, and wherein the
metastatic tumor
is a same or different type of tumor as the first tumor.
[0064] Alternative 51 comprises the isolated peptide for use according
to
Alternative 50, in which the mammary tumor comprises a malignant mammary
tumor, or the
mammary tumor comprises a mixed mammary tumor (for example a malignant mixed
mammary tumor), or wherein the mammay tumor comprises a mucinous carcinoma
comprising a mammary gland mucinous carcinoma.
[0065] Alternative 52 comprises the isolated peptide for use according
to any one
of Alternatives 49-51, in which the metastatic tumor is the same type of tumor
as the first
tumor.
[0066] Alternative 53 comprises the isolated peptide for use according
to any one
of Alternatives 49-51, in which the metastatic tumor is a different type of
tumor from the first
tumor.
[0067] Alternative 54 comprises the isolated peptide for use according
to
Alternative 50, in which the metastatic tumor comprises a prostate tumor, a
melanoma, a
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
colon cancer, a lung carcinoma, an Apocrine gland carcinoma, a testis tumor, a
mast cell
tumor, a mammary tumor, a mucinous carcinoma, or a histicytoma.
[0068] Alternative 55 comprises the isolated peptide for use according
to any one
of Alternatives 49-54, in which said use comprises administering said
composition
intratumorally or peritumorally to the first tumor.
[0069] Alternative 56 comprises the isolated peptide for use according
to any one
of Alternatives 49-54, in which said use comprises administering the
composition to a site in
the subject that is neither intratumoral or peritumoral to either of the first
tumor or the
metastatic tumor
[0070] Alternative 57 comprises the isolated peptide for use according
to any one
of Alternatives 49-56, in which said isolated peptide is administered
systemically to the
subject.
[0071] Alternative 58 comprises the isolated peptide for use according
to
Alternative 57, in which said composition is administered systemically via at
least one of
subcutaneous, intravenous, intraperitoneal, or intramuscular administration.
[0072] Alternative 59 comprises the isolated peptide for use according
to any one
of Alternatives 49-58, in which the administering further induces regressive
changes in the
first tumor, thereby ameliorating or eliminating the first tumor.
[0073] Alternative 60 comprises the isolated peptide for use according
to any one
of Alternatives 49-59, in which the administering further induces immune cell
infiltration of
the first tumor, thereby ameliorating, inhibiting or eliminating the first
tumor.
[0074] Alternative 61 comprises the isolated peptide for use according
to any one
of Alternatives 49-60, in which the administering further induces eradication
of cells of the
first tumor, thereby ameliorating, inhibiting or eliminating the first tumor.
[0075] Alternative 62 comprises the isolated peptide for use according
to any one
of Alternatives 49-61, in which the administering further induces eradication
of the first
tumor, thereby eliminating the first tumor.
[0076] Alternative 63 comprises the isolated peptide for use according
to any one
of Alternatives 49-62, in which the administering further induces regressive
changes in the
metastatic tumor, thereby ameliorating, inhibiting or eliminating the
metastatic tumor.
11
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
[0077] Alternative 64 comprises the isolated peptide for use according
to any one
of Alternatives 49-63, in which the administering further induces immune cell
infiltration of
the metastatic tumor, thereby ameliorating, inhibiting or eliminating the
metastatic tumor.
[0078] Alternative 65 comprises the isolated peptide for use according
to any one
of Alternatives 49-64, in which the administering further induces eradication
of cells of the
metastatic tumor, thereby ameliorating, inhibiting or eliminating the
metastatic tumor.
[0079] Alternative 66 comprises the isolated peptide for use according
to any one
of Alternatives 49-65, in which the administering further induces eradication
of the
metastatic tumor, thereby eliminating the metastatic tumor.
[0080] Alternative 67 comprises the isolated peptide for use according
to any one
of Alternatives 49-66, in which the first tumor is a primary tumor.
[0081] Alternative 68 comprises the isolated peptide for use according
to any one
of Alternatives 49-66, in which the first tumor is an other metastatic tumor.
[0082] Alternative 69 comprises the isolated peptide for use according
to any one
of Alternatives 49-68, in which said isolated peptide comprises no more than
30 amino acid
residues.
[0083] Alternative 70 comprises the isolated peptide for use according
to any one
of Alternatives 49-68, in which said isolated peptide comprises no more than
20 amino acid
residues.
[0084] Alternative 71 comprises the isolated peptide for use according
to any one
of Alternatives 49-68, in which said isolated peptide comprises no more than
16 amino acid
residues.
[0085] Alternative 72 comprises the isolated peptide for use according
to any one
of Alternatives 49-68, in which said isolated peptide consists of the amino
acid sequence
FFVKLS (SEQ ID NO: 62).
[0086] Alternative 73 comprises the isolated peptide for use according
to any one
of Alternatives 49-70, in which said isolated peptide comprises the amino acid
sequence
KKLDTFFVKLSLFTER (SEQ ID NO: 2).
12
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
[0087] Alternative 74 comprises the isolated peptide for use according
to any one
of Alternatives 49-68, in which said isolated peptide consists of the amino
acid sequence
KKLDTFFVKLSLFTER (SEQ ID NO: 2)
[0088] Alternative 75 comprises the isolated peptide for use according
to any one
of Alternatives 49-70, in which said isolated peptide comprises the amino acid
sequence
RKLDTFFVKLSLFTERRR (SEQ ID NO: 586).
[0089] Alternative 76 comprises the isolated peptide for use according
to any one
of Alternatives 49-68, in which said isolated peptide consists of the amino
acid sequence
RKLDTFFVKLSLFTERRR (SEQ ID NO: 586).
[0090] Alternative 77 comprises the isolated peptide for use according
to any one
of Alternatives 49-76, in which the composition is for use in administering
the isolated
peptide to the subject at a dose of at least about 1 ng/kg.
[0091] Alternative 78 comprises a composition comprising an isolated
peptide
comprising the amino acid sequence FFVKLS (SEQ ID NO: 62), and a support, such
as a
nanoparticle, in which the isolated peptide is immobilized on the
nanoparticle.
[0092] Alternative 79 comprises the composition of Alternative 78, in
which the
support comprises the nanoparticule.
[0093] Alternative 80 comprises the composition of Alternative 78, in
which the
support comprises the nanoparticule comprising at least one of: a polymer
(such as PLGA,
glycerol, chitosan, DNA, a hydrogel, or an acrylamide), a dendrimer (such as
PAMAM), a
quantum dot (such as CdSe, CuInSe, or CdTe), a gold nanoparticle (such as a
sphere, rod, or
shell), a silica nanoparticle (such as a a sphere, a shell, or a mesoporous
structure), a
magnetic particle (such as an iron oxide, a cobalt-based material, a magnetic
sphere, an
aggregate in dextran or silica, or a Dynal bead), a carbon-based material
(such as a carbon
nanotube, a buckminsterfullerene, a graphene, or an activated carbon), a
carbohydrate, a
nucleic acid, a polypeptide (such as an albumin or an albumin fragment), or a
lipid.
[0094] Alternative 81 comprises the composition of any one of
Alternatives 78-
80, in which said isolated peptide comprises no more than 30 amino acid
residues.
[0095] Alternative 82 comprises the composition of any one of
Alternatives 78-
80, in which said isolated peptide comprises no more than 20 amino acid
residues.
13
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
[0096] Alternative 83 comprises the composition of any one of
Alternatives 78-
80, in which said isolated peptide comprises no more than 16 amino acid
residues.
[0097] Alternative 84 comprises the composition of any one of
Alternatives 78-
80, in which said isolated peptide consists of the amino acid sequence FFVKLS
(SEQ ID
NO: 62).
[0098] Alternative 85 comprises the composition of any one of
Alternatives 78-
82, in which said isolated peptide comprises the amino acid sequence
KKLDTFFVKLSLFTER (SEQ ID NO: 2).
[0099] Alternative 86 comprises the composition of any one of
Alternatives 78-
80, in which said isolated peptide consists of the amino acid sequence
KKLDTFFVKLSLFTER (SEQ ID NO: 2)
[0100] Alternative 87 comprises the composition of any one of
Alternatives 78-
82, in which said isolated peptide comprises the amino acid sequence
RKLDTFFVKLSLFTERRR (SEQ ID NO: 586).
[0101] Alternative 88 comprises the composition of any one of
Alternatives 78-
80, in which said isolated peptide consists of the amino acid sequence
RKLDTFFVKLSLFTERRR (SEQ ID NO: 586).
[0102] Alternative 89 comprises the composition of any one of
Alternatives 78-88
for use in treating, inhibiting or ameliorating a cancer, such as metastatic
cancer, said cancer
comprising a first tumor and a metastatic tumor in a subject, in which the use
comprises: (a)
intratumoral administration or petitumoral administration of the composition
to the first
tumor, but not the metastatic tumor; or (b) administration of the composition
to a site in the
subject that is neither intratumoral nor peritumoral to either of the first
tumor or the
metastatic tumor, such as systemically, thus ameliorating, inhibiting or
eliminating the first
tumor, and ameliorating or eliminating the metastatic tumor.
[0103] Alternative 90 comprises the composition for use according to
Alternative
89, in which the first tumor comprises a prostate tumor, a melanoma, a lung
carcinoma, a
colon cancer, an Apocrine gland carcinoma, a testis tumor, a mast cell tumor,
a mammary
tumor, a mucinous carcinoma, or a histicytoma, and wherein the metastatic
tumor is a same
or different type of tumor as the first tumor.
14
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
[0104] Alternative 91 comprises the composition for use according to
Alternative
90, in which the mammary tumor comprises a malignant mammary tumor, or the
mammary
tumor comprises a mixed mammary tumor (for example a malignant mixed mammary
tumor), or wherein the mammay tumor comprises a mucinous carcinoma comprising
a
mammary gland mucinous carcinoma.
[0105] Alternative 92 comprises the composition for use according to
any one of
Alternatives 89-91, in which the metastatic tumor is the same type of tumor as
the first tumor.
[0106] Alternative 93 comprises the composition for use according to
any one of
Alternatives 89-91, in which the metastatic tumor is a different type of tumor
from the first
tumor.
[0107] Alternative 94 comprises the composition for use according to
Alternative
93, in which the metastatic tumor comprises a prostate tumor, a melanoma, a
colon cancer, a
lung carcinoma, an Apocrine gland carcinoma, a testis tumor, a mast cell
tumor, a mammary
tumor, a mucinous carcinoma, or a histicytoma.
[0108] Alternative 95 comprises the composition for use according to
any one of
Alternatives 89-94, in which said use comprises administering said composition
intratumorally or peritumorally to the first tumor.
[0109] Alternative 96 comprises the composition for use according to
any one of
Alternatives 89-94, in which said use comprises administering the composition
to a site in the
subject that is neither intratumoral or peritumoral to either of the first
tumor or the metastatic
tumor
[0110] Alternative 97 comprises the composition for use according to
Alternative
96, in which said composition is administered systemically to the subject.
[0111] Alternative 98 comprises the composition for use according to
Alternative
97, in which said composition is administered systemically via at least one of
subcutaneous,
intravenous, intraperitoneal, or intramuscular administration.
BRIEF DESCRIPTION OF THE DRAWINGS
[0112] Figure 1 illustrates immunohistochemical staining of a
malignant
melanoma metastases using affinity purified rabbit antibodies directed to the
P3028 epitope.
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
[0113] Figure 2 illustrates Western blot performed on tumor extracts
using
antibodies directed against the 3028-structure.
[0114] Figure 3 illustrates Sandwich ELISA detecting albumin exposing
the
P3028 epitope in serum; competition with the P3028 peptide.
[0115] Figure 4 illustrates IL-2 induced proliferation by PBMCs from
healthy
control samples and human immune cells (PBMC) from renal cell carcinoma
patients (RCC)
cultured in 10% autologous sera.
[0116] Figure 5 illustrates a Kaplan Meyer analysis of renal cell
carcinoma
patients according to proliferative response to IL-2.
[0117] Figure 6 illustrates analysis of the effect of four different
peptides on IL-2
induced proliferation of PBMCs from healthy control samples.
[0118] Figure 7 illustrates inhibition of the proliferative response
to IL-2 by
P3028 in the human ex vivo model using cancer patient PBMCs.
[0119] Figure 8 illustrates effect of P3028 on TCR stimulated
lymphocyte
proliferation of PBMCs from four healthy persons.
[0120] Figures 9A-9B illustrates effect of albumin peptides on NK-cell
cytotoxicity. Figure 9A depicts effects for K5 and K6. Figure 9B depicts
effects for K12
and K13.
[0121] Figure 10 illustrates effect of P3028 on the spreading on
peripheral blood
leukocytes.
[0122] Figure 11 illustrates effect of P3028 on migration of PBMCs
studied
using the Boyden chamber technique.
[0123] Figure 12 illustrates effect of the C- (3218) and N-terminal
(3325) parts of
P3028 on 11-2 induced proliferation in comparison with the effect of the full
length P3028.
[0124] Figure 13 illustrates the inhibitory effect of P3028 on IL-2
induced
proliferation is not neutralized by the C- (3218) and N-terminal (3325) parts
of P3028 alone
or in combination.
[0125] Figure 14 illustrates inhibition of the binding of anti-LFA-1
antibody
directed to CD1 la by incubation of normal PBMCs with patient sera.
16
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
[0126] Figure 15 illustrates inhibition of the binding of an anti-LFA-
1, mAb, to
mononuclear blood cells by P3028.
[0127] Figure 16 illustrates staining of LFA-1 on PBMCs from a healthy
control
sample (A), and a cancer patient before (B) and after (C) treatment with an
antibody directed
against the inhibitory P3028.
[0128] Figure 17 illustrates staining of mononuclear blood cells by an
anti-LFA-1
antibody (A) is blocked by P3028 (B) or cancer patient serum (C).
[0129] Figures 18A and 18B illustrates ELISA analysis showing that the
binding
of biotinylated IL-2 to rhulL-2R. Figure 18B is a contrast-enhanced image of
Figure 18A,
so as to depict the binding data for non biotinylated IL-2 (triangles;
[0130] Figure 19 illustrates the a-chain of the IL-2 receptor (CD25)
binding
P3028 (A) at the IL-2 binding site (B).
[0131] Figure 20 illustrates antisera from rabbits immunized with
P3028 binds to
P3028.
[0132] Figure 21 illustrates inhibition of the binding of rabbit-anti
3028 serum L
to wells coated with the P3028 in an ELISA by albumin peptides
[0133] Figure 22 illustrates effect of affinity purified antibodies
directed against
P3028 on the proliferative response to IL-2 of PBMCs from immunosuppressed
cancer
patients (Figure 22A) and normal control samples (Figure 22B).
[0134] Figure 23 illustrates identification of P3028 inhibitors in
solution. Based
on previous analyses potential binders of P3028 were synthesized on a chip.
Figure 23A
illustrates results for assays 1-14. Figure 23B illustrates results for assays
15-28. Figures
23A and 23B represent the left and right sides, respectively, of a single
graph that was
enlarged to show the text more clearly. The Y axis has been reproduced in
Figure 23B for
reference.
[0135] Figure 24 illustrates stimulatory activity of P28R on
suppressed
proliferative response to IL-2. Figures 24A, 24B, 24C, and 24D respectively
illustrate
stimulatory activity for four different cancer patients.
[0136] Figure 25 illustrates binding of biotinylated P28R to a fresh
frozen breast
cancer tumor.
17
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
[0137] Figure 26 illustrates breast cancer tissue incubated with
buffer (Figure
26A) or P28R (Figure 26B) stained by an antibody directed against LFA-1.
[0138] Figure 27 illustrates rampo scores for binding of P3028 to
peptides having
single amino acid substitutions of each position of P28R.
[0139] Figure 28 illustrates single amino acid substitutions of
peptide P28R
having rampo scores greater than 500.
[0140] Figure 29 illustrates rampo scores for binding of P3028 to P28R
and N-
terminal and/or C-terminal truncations of peptide P28R.
[0141] Figure 30 illustrates rampo scores for binding of P3028 to
internal
deletion mutants, and single amino acid substitution mutants of peptide P28R.
Figures 30A
and 30B represent the left and right sides, respectively, of a single graph
that was enlarged to
show the text more clearly. For reference, the Y axis has been reproduced in
Figure 30B.
[0142] Figure 31 illustrates favorable electrostatic interactions and
hydrophobic
interactions between peptide 3028 and peptide KKL15.
[0143] Figure 32 illustrates alignments of cyclic peptides identified
as binding to
P3028 in positional scan experiments (SEQ ID NOs: 265-267) and linear peptides
identified
as binding to P3028 (SEQ ID NOs: 2, and 268-293).
[0144] Figures 33A and 33B illustrate effects of various
concentrations of
peptide P28R on MTS bioreduction in (Figure 33A) PBMC's from healthy control
samples,
and (Figure 33B) PBMC's from cancer patients.
[0145] Figure 34 illustrates effect of P28R (aka "SCF 28W') (N=9) and
P27 (aka
"SCF 27") (N=8) on PBMCs from cancer patients, MTS measurements, day 7.
[0146] Figure 35 illustrates response to IL-2 in cancer patients
cells, measured by
BrdU incorporation.
[0147] Figure 36 illustrates effect of P28R (aka "P28") on IL-2
induced
proliferation in cells of (Figure 36A) high responders, and (Figure 36B) low
responders.
[0148] Figure 37 illustrates effect of P28R (aka "SCF 28R") and P27
(aka "SCF
27") on IL-2 stimulation of PBMCs from cancer patients, based on BrdU
incorporation.
[0149] Figure 38 illustrates effect of P28R (aka "SCF 28R") and P27
(aka "SCF
27") on IL-2-indueced proliferation based on BrdU incorporation (Figures 38A,
38C) and
18
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
MTS incorporation (Figures 38B, 38D). Shown are cells of two different
patients, (Figures
38A, 38B) and (Figures 38C, 38D) respectively.
[0150] Figure 39 illustrates enzyme linked immunosorbent spot assays
of cells
with (bottom row) and without (top row) P3028 peptide.
[0151] Figure 40 illustrates data from enzyme linked immunosorbent
spot assays
of cells with and without P3028 peptide.
[0152] Figure 41 is a series of graphs illustrating effects of
modified peptides on
activation of PBMCs from healthy control person. PBMCs were incubated with the
peptides
(401.tg/mL) for 24 hours in RPMI plus 10% human AB serum. Activation is
determined as
percentage of cells with enhanced marker CD69 using flow cytometry. Figure 41A
illustrates results of two experiments (410 and 412) performed for each
peptide. Figure 41B
illustrates results of two experiments (414 and 416) performed for each
peptide.
[0153] Figure 42 is a series of graphs illustrating effects of the
full length peptide
P28R and the 6 amino acid central sequence (32230, FFVKLS, SEQ ID NO: 62) in
culture
medium containing normal human AB serum. Activation is determined as
percentage of
cells with enhanced marker CD69 or CD71 using flow cytometry. PBMCs were
incubated
with the peptides (40 g/mL) for 24 hours in RPMI plus 10% human AB serum.
Figure 42A
illustrates the results of two experiments (420 and 422) performed for each
peptide. Figure
42B illustrates the results of two experiments (424 and 426) performed for
each peptide.
[0154] Figure 43 is a graph illustrating a comparison of the full
length peptide
P28R and the 6 amino acid central sequence (32230, FFVKLS, SEQ ID NO: 62) in
culture
medium containing sera from two different cancer patients ("human ca serum 1"
430 and
("human ca serum 2" 432).
[0155] Figure 44 is a series of microscope images illustrating P28R
treatment of
human prostate cancer ,PC3, in a xenograft model in nude mice. Tumor was
injected intra-
tumorally with P28R (Figure 44A) and only the drug solvent (Figure 44B).
Staining for
Caspase 3 440 (demonstrating induction of apoptosis) and an absence of
staining 442 are
depicted.
[0156] Figure 45 is a series of microscope images illustrating intra-
tumoral
treatment of B16 melanoma with P28R. The inflammatory infiltrate was
demonstrated after
19
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
3 days of treatment using a polyclonal rabbit antibody directed against CD45
(Figure 45A),
and control sections were incubated with rabbit IgG at the same concentration
(Figure 45B).
Staining 450 and an absence of staining 452 are depicted.
[0157] Figure 46 is a series of graphs illustrating Effect of modified
peptides on
activation of PBMCs from healthy control person. Activation is determined as
percentage of
cells with enhanced marker CD69 (Figure 46A, showing results of two
experiments, exp 1
460 and exp 2 462) or CD71 (Figure 46B, showing results of two experiments,
exp 1 464
and exp 2 466) using flow cytometry. PBMCs were incubated with the peptides
(40m/mL)
for 48 hours in RPMI plus 10% human AB serum.
[0158] Figure 47 is a series of microscope images illustrating
occurrence of the
immunoinhibitory 3028 structure in two areas (Figure 47A and Figure 47B,
respectively) of
a human breast cancer. Immunohistochemical staining (470) using biotinylated
P28R is
depicted. An absence of staining 472 is observed in Figure 47A.
[0159] Figure 48 is a series of microscope images illustrating that
tumor cells can
generate P3028 structures in accordance with some embodiments herein. Figure
48A depicts
human prostate cells cultured in the absence of serum proteins, and
immunostained with
rabbit antibodies against P3028 structures (depicted as 480). Substantially
low levels of
staining are noted as 482. Figure 48B depicts human prostate cells fed human
serum
albumin for 2 hours, and immunostained with rabbit antibodies against P3028
structures.
Substantial staining 480 is observed.
[0160] Figure 49 is a series of microscope images illustrating that
administration
of immunoregulatory peptide inhibitors immobilized on nanoparticles in
accordance with
some embodiments herein can remove bound dHSA from immune cells. Figure 49A
depicts
control PBMCs immunostained for dHSA (shown as 490). Figure 49B depicts PBMCs
incubated with magnetic DynabeadTM beads bound to P28 core peptide (SEQ ID NO:
62),
and immunostained for dHSA (shown as 490). Levels bound dHSA are substantially
lower
in the cells incubated with DynabeadTM beads bound to P28 core peptide (shown
as 492).
[0161] Figure 50 is a series of microscope images illustrating
immunohistochemical (MC) staining of Xenograft of a human prostate cancer,
PC3, in nude
mice. A first tumour biopsy (Figure 50A) and a second tumour biopsy (Figure
50B) are
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
shown. IHC staining of tumour biopsies for the P3028-structure using
oligoclonal rabbit
antibodies. The expression of the epitope showed considerable heterogeneity
with strongly
stained areas.
[0162] Figure 51 is a series of microscope images illustrating
cultured human
prostate carcinoma cells, starved for proteins for 18 hours in accordance with
some
embodiments herein. A first image of such cells (Figure 51A) and a second
image of such cells
(Figure 51B) are shown. IHC staining for the P3028-structure using oligoclonal
rabbit
antibodies.
[0163] Figure 52 is a microscope image illustrating cultured human
prostate
carcinoma cells, starved for proteins for 18 hours and then incubated with
human serum
albumin for 2 hours in accordance with some embodiments herein. IHC staining
for the
P3028-structure using oligoclonal rabbit antibodies.
[0164] Figure 53 is a series of microscope images illustrating intra-
tumoral
treatment of B16 melanoma P28R in accordance with some embodiments herein. The
inflammatory infiltrate was demonstrated after 3 days of treatment using a
polyclonal rabbit
antibody directed against CD45 (Figure 53A), control sections were incubated
with rabbit
IgG at the same concentration (Figure 53B).
[0165] Figure 54 is a series of microscope images illustrating a B16
melanoma in
accordance with some embodiments herein. Contralateral tumour injected with
vehicle one
day after treatment and with hematoxylin staining is shown.
[0166] Figure 55 is a series of microscope images illustrating B16
melanoma 5
days after intra-tumoral injection of P28R (Figures 55A and 55C) and the
contralateral
uninjected tumour (Figure 55B and 55D) in accordance with some embodiments
herein. MC
staining for CD45+ inflammatory cells. Extensive tumour regressive changes and
heavy
infiltration of CD45+ cells are seen in both treated (Figures 55A and 55C) and
untreated
tumours (Figure 55B and 55D).
[0167] Figure 56 is a microscope image illustrating Lewis lung
carcinoma grown
in C57B1 mice in accordance with some embodiments herein. Untreated tumour
stained with
hematoxylin.
21
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
[0168] Figure 57 is a series of microscope images illustrating
injection of P28R
into the tumours resulted in extensive tumour regressive changes in a Lewis
lung carcinoma
in accordance with some embodiments herein. The Lewis lung carcinoma was
treated
intratumorally with P28R (Figure 57A) in accordance with some embodiments
herein. A
similar anti-tumour effect is seen also in contralateral untreated tumours
(Figure 57B).
[0169] Figure 58 is a series of microscope images illustrating
spontaneous breast
tumour in a dog staining of a regional metastatic lesion showing infiltration
of CD45+
inflammatory cells in tumour areas with various degrees of regressive changes
in accordance
with some embodiments herein. Four different images of the tumor (Figure 58A,
Figure
58B, Figure 58C, Figure 58D) are shown.
[0170] Figure 59 is a microscope image illustrating H&E staining of a
dog anal
adenocarcinoma in accordance with some embodiments herein.
[0171] Figure 60 is a microscope image illustrating H&E staining of a
major
tumour nodule of a dog, apparently with two cell types, in accordance with
some
embodiments herein.
[0172] Figure 61 is a series of microscope images illustrating H&E
staining of
dog tumors in accordance with some embodiments herein. Figure 61A illustrates
staining of
the central part of the injection site with some destruction. Figure 61B
illustrates staining of
an area with an inflammatory infiltrate and some tumour regressive changes.
[0173] Figure 62 is a series of microscope images illustrating a dog
Apocrine
gland carcinoma stained for CD45+ inflammatory cells in accordance with some
embodiments herein. Figure 62A shows CD45+ inflammatory cells surrounding
tumour
nodules. Figure 62B shows CD45+ inflammatory cells infiltrating a thin lesion.
[0174] Figure 63 is a series of microscope images illustrating only
few scattered
CD3+ or CD8+ cells were found after treatment in this dog tumour in accordance
with some
embodiments herein. Two images of the tumor, Figure 63A and Figure 63B are
shown.
[0175] Figure 64 is a series of microscope images illustrating
inflammatory cells
infiltrating into a dog tumour nodule in accordance with some embodiments
herein. The
inflammatory cells infiltrating into a tumour nodule are stained for CD56
(Figure 64A) and
NCR1 (Figure 64B).
22
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
[0176] Figure 65 is a series of microscope images illustrating CD56+
cells are
found both infiltrating tumour nodules and stromal area of a dog in accordance
with some
embodiments herein. Both of Figure 65A and Figure 65B are pictures from the
same section.
It is noted that the staining of the tumour nodule infiltrating cells is much
weaker and there
seems to be a gradient with more strongly stained cells at the periphery of
the nodules.
[0177] Figure 66 is a series of microscope images illustrating
biopsies of a dog
testis tumor in accordance with some embodiments herein. Figure 66A
illustrates H&E
stained section showing the ordinary histopathological picture of a testis
tumour. Figure 66B
illustrates the low degree of infiltration of inflammatory cells was
demonstrated by IHC
staining for CD45.
[0178] Figure 67 is a series of microscope images illustrating H&E
stained
sections of a dog mastocytoma with pronounced tumour regressive changes
(Figure 67A and
Figure 67B represent different sections of the mastocytoma, in accordance with
some
embodiments herein.
[0179] Figure 68 is a series of microscope images illustrating a dog
mastocytoma
after intra-tumoral treatment with P28R in accordance with some embodiments
herein.
Staining for CD3+ (Figure 68A) and CD8+ T-lymphocytes (Figure 68B) show very
low
infiltration of these cells,with a highly variable, usually very faint,
staining intensity
[0180] Figure 69 is a series of microscope images illustrating a dog
mastocytoma
after intra-tumoral treatment with P28R in accordance with some embodiments
herein.
[0181] Figure 70 is a series of microscope images illustrating a dog
mastocytoma
after intra-tumoral treatment with P28R in accordance with some embodiments
herein.
Figure 70A, Figure 70B, Figure 70C, and Figure 70D represent different images
of the
mastocytoma. A massive tumour destruction and an extensive infiltration of
CD56+
inflammatory cells are shown.
[0182] Figure 71 is a series of microscope images illustrating H&E
stained dog
breast tumor sections indicating regressive changes at the injection site in
accordance with
some embodiments herein, presumably with some toxic effects. Two different
sites of
injection are shown (Figure 71A and Figure 71B).
23
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
[0183] Figure 72 is a series of microscope images illustrating H&E
staining of
the central slice of the dog breast tumour showing infiltration of
inflammatory cells with
various degrees of tumour regressive changes from well preserved glandular
structures to
scattered tumour cells surrounded by inflammatory cells in accordance with
some
embodiments herein. Figures 72A, Figure 72B, Figure 72C, and Figure 72D are
images
from the lesion showing various degrees of tumour regressive changes.
[0184] Figure 73 is a series of microscope images illustrating H&E
staining of a
regional metastatic lesion of dog breast tumor showing infiltration of
inflammatory cells with
various degrees of tumour regressive changes, from well preserved glandular
structures to
scattered tumour cells surrounded by inflammatory cells in accordance with
some
embodiments herein. Figures 73A, Figure 73B, Figure 73C, and Figure 73D are
images
from the lesion showing various degrees of tumour regressive changes.
[0185] Figure 74A and Figure 74B are is a series of microscope images
illustrating H&E staining of a distant metastasis / new primary tumour of a
dog breast tumor
showing infiltration of inflammatory cells with various degrees of tumour
regressive changes
from well preserved glandular structures to scattered tumour cells surrounded
by
inflammatory cells in accordance with some embodiments herein.
[0186] Figure 75 is a series of microscope images illustrating
staining of a
regional metastatic lesion of a dog breast tumor showing infiltration of CD45+
inflammatory
cells in tumour areas with various degrees of regressive changes in accordance
with some
embodiments herein. Figures 75A, Figure 75B, Figure 75C, and Figure 75D are
images
from the lesion showing various degrees of tumour regressive changes.
[0187] Figure 76 is a microscope image illustrating distant metastases
/ new
primary tumour of a dog breast tumor showing infiltration of CD45+ cells into
the tumour cell
areas in accordance with some embodiments herein.
[0188] Figure 77 is a series of microscope images illustrating
staining of a
regional metastatic lesion showing infiltration of CD45+ inflammatory cells in
dog breast
tumour areas with various degrees of regressive changes in accordance with
some
embodiments herein. Figures 77A, Figure 77B, Figure 77C, and Figure 77D are
images
from the lesion showing various degrees of tumour regressive changes.
24
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
[0189] Figure 78 is a series of microscope images illustrating a dog
breast tumour
after intra-tumoral injection of vehicle in accordance with some embodiments
herein. The
section was stained with hematoxylin showing a fairly well preserved glandular
structure
(Figure 78A). The inflammatory infiltrate was visualized by staining for CD45+
cells
(Figure 78B).
[0190] Figure 79A and Figure 79B are a series of microscope images
illustrating
hematoxylin stained sections of histiocytoma after intra-tumoral treatment
with P28R in
accordance with some embodiments herein. Extensive regressive changes of the
tumour were
observed.
[0191] Figure 80 is a series of microscope images illustrating
infiltration of
CD56+ cells (Figure 80A) and NCR1+ (Figure 80B) cells in a histiocytoma with
extensive
tumour cell destruction in accordance with some embodiments herein.
[0192] Figure 81 is a microscope image illustrating an overview of H&E
staining
of breast tumour treated with P28R for 5 days in accordance with some
embodiments herein.
A heavy inflammatory infiltrate is demonstrated.
[0193] Figure 82 is a series of microscope images illustrating H&E
staining of a
breast tumour in accordance with some embodiments herein. An intense
inflammatory
infiltration with extensive destruction of tumour glands (Figure 82A, Figure
82B, Figure
82C, and Figure 82D provide different images of the breast tumor). Figure 82D
also
demonstrates the occurrence of macrophages with hemosiderin (yellow, arrow).
[0194] Figure 83 is a microscope image illustrating canine breast
tumour stained
for CD8 treated intra-tumorally with 40nmol P28R in accordance with some
embodiments
herein. Apparently, the lymphocytes in the stroma have an increased staining
intensity
compared to some faintly stained cells infiltrating the tumour cell areas.
[0195] Figures 84A-B are a series of microscope images illustrating
one canine
breast tumour treated with 40nmol P28R in accordance with some embodiments
herein. The
inflammatory infiltrate was evaluated after 5 days treatment by staining of
parallel sections
for CD3+ cells with standard antibody concentration, diluted 1:50 (Figure 84A)
or an
increased concentration, diluted 1:25 (Figure 84B). The large differences in
expression of
CD3 are observed, as a large number of lymphocytes are completely negative
when stained
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
with a "standard" antibody concentration but are actually found to express
this marker when
an increased" concentration of the antibody is used.
[0196] Figure 85 is a microscope image illustrating Canine breast
tumour treated
with 40nmol P28R intra-tumourally in accordance with some embodiments herein.
Regressive changes of a large number of tumour cells are demonstrated as cells
with irregular
shaped nuclei and often disrupted nuclear membrane, positive in TUNELTm
staining
(arrows). This section is counterstained using methylgreen pyronin.
[0197] Figures 86A-B are a series of microscope images illustrating
untreated
(Figure 86A) and treated (Figure 86B) canine breast tumours, in accordance
with some
embodiments herein. The tumour cell density is significantly reduced in the
treated tumour
and at the same time, a large number of "damaged" tumour cells can still be
found.
[0198] Figures 87A-D are a series of microscope images illustrating
four
different untreated canine breast tumours showing a high density of unaffected
tumour cells
and few degenerative cells. The number of lymphocytes is low except for tumour
C, but even
with this degree of inflammatory cells in an untreated tumour the number of
degenerated
tumour cells is low.
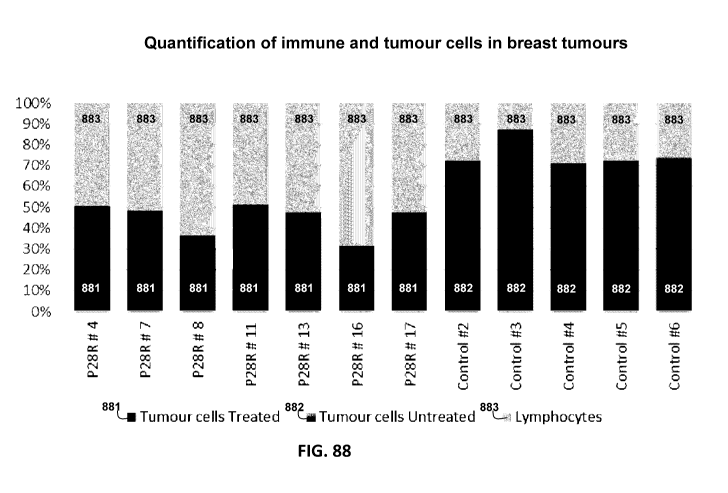
[0199] Figure 88 is a graph showing evaluation of P28R treatment in 7
dogs with
breast tumours compared with 5 untreated control dogs, in accordance with some
embodiments herein. In representative pictures (n=1-5), the total number of
tumour cells
from treated (dark bars 881 in P28R #4, #7, #8, #11, #13, #16, and #17) and
from control
tumours (dark bars 882 in Controls #2, #3, #4, #5, and #6) was counted and
compared with
the number of inflammatory cells (light grey bars 883).
[0200] Figures 89A-D. are a series of microscope images illustrating
four
examples of formalin fixed and paraffin embedded canine breast tumours with
very sparse
infiltration of inflammatory cells close to the tumour cells. The inflammatory
cells are
mainly located in the stromal areas.
[0201] Figures 90A-D are a series of microscope images illustrating
two formalin
fixed and paraffin embedded canine breast tumours with very small areas of
inflammatory
cells infiltrating into the tumour cell areas close to the tumour cells. These
areas were located
at the very periphery of the tumour section (Figures 90A and 90C). In the main
area of the
26
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
tumours, the infiltration of inflammatory cells close to tumour cells was very
sparse as seen
in the same section (Figures 90B and 90D) even if the stroma of the tumour is
heavily
infiltrated (Figure 90D), indicating immunosuppression with blockade of cell
migration at
this level.
[0202] Figures 91A-D are a series of microscope images illustrating a
comparison of inflammatory response and occurrence of degenerative tumour
cells in directly
injected breast tumors (Figures 91A and 91C) and uninjected breast tumors
(Figures 91B
and 91D) in two dogs treated with P28R in accordance with some embodiments
herein. A
large number of inflammatory cells and degenerated tumour cells were found
also in the
tumors that were not directly injected with P28R. That is, when each of the
two dog was
treated with P28R, degenerative tumor cells were observed in the tumors that
were directly
injected with P28R, and also in other tumors in the same animal, even though
these tumors
were not directly injected with P28R.
[0203] Figures 92A-B are a series of microscope images illustrating
tumours in a
CT26 colon cancer model. Apoptotic tumour cells are identified using the
TUNELTm
staining technique. The tumours are counterstained using methyl-green pyronin.
Figure 92A
shows an untreated tumour in an untreated control mouse. Figure 92B shows a
tumours
treated with 12 microgram P28R, twice weekly for two weeks.
[0204] Figures 93A-B are a series of microscope images illustrating
show
haematoxylin staining of uninjected tumours on the contralateral side of
tumours that were
injected in CT26 colon cancers in Balb/c mice. Figure 93A illustrates a saline
control, and
Figure 93B illustrates a mouse injected with oligoclonal rabbit antibody
against P3028.
DETAILED DESCRIPTION OF THE INVENTION
[0205] Several immunoregulatory peptide inhibitors, which interact
with
immunoregulatory peptides that cause immunosuppression in a human (e.g., a
human having
cancer, enduring or chronic infectious or inflammatory disease), have been
developed.
Preferred immunoregulatory peptide inhibitors bind to proteins or peptides
that comprise the
P3028 structure and/or the P3028 sequence (SEQ ID NO: 185). With reference to
some
embodiments and description herein, the P3028 structure refers to
polypeptides, such as
27
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
peptides, proteins, and the like that include the P3028 sequence (SEQ ID NO:
185). The
P3028 structure can include macromolecules such as peptides, proteins, and the
like that are
recognized by antibodies that bind specifically to P3028 structures (see
Example 1 and
Figure 2). For example, aggregates of albumin, denatured albumin and other
damaged
albumins can include the P3028 structure. In some contexts in the present
application, the
P3028 structure, P3028 sequence, and P3028 are terms used interchangeably.
Molecules
having the P3028 structure interact with receptors on immune cells, such as
the IL-2 receptor
and the LFA-1 receptor, causing immunosuppression. As such, it is contemplated
herein that
peptides, proteins, albumin fragments, damaged albumin (e.g. denature albumin)
and albumin
aggregates can include the P3028 structure, and can interact with immune cell
receptors such
as the IL-2 receptor and LFA-1 receptor. Immunosuppression can be
characterized by a
reduced immune cell proliferation, spreading and migration, as well as, NK-
cell cytotoxicity.
In the presence of an immunoregulatory peptide inhibitor, as described herein;
however, the
immunosuppression mediated by the P3028 structure can be altered (e.g.,
reduced,
ameliorated, eliminated, or removed altogether). In some experiments, for
example, it was
found that an immunoregulatory peptide inhibitor can remove a molecule
including a P3028
structure from the LFA-1 receptor thereby altering the immunosuppression
mediated by
P3028 structure. Accordingly, the description that follows provides details on
many different
classes of immunoregulatory peptide inhibitors including, but not limited to,
antibody or
antibody fragment based immunoregulatory peptide inhibitors, peptide based
immunoregulatory peptide inhibitors, peptidomimetic immunoregulatory peptide
inhibitors,
modified immunoregulatory peptide inhibitors (e.g., containing a D amino acid,
N-terminal
acetyl, or C terminal amide group), cyclic peptides inhibitors, and aptamer
based
immunoregulatory peptide inhibitors, as well as compositions comprising
immunoregulatory
inhibitors, for example compositions comprising immunoregulatory peptide
inhibitors.
Methods of using compositions (as described herein) to reduce
immunosuppression or an
aspect thereof (e.g., reducing a P3028-mediated inhibition of immune cell
proliferation,
spreading, migration, or NK-cell cytotoxicity), as well as, approaches to
inhibit, reduce, or
alter the progression of cancer (e.g. inducing immune cell infiltration of
tumors, inducing
regressive changes in tumors, and/or inducing eradiation or some or all of a
tumor) or
28
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
inflammatory disease are provided. The composition can comprise, consist of,
or consist
essentially of an immunoregulatory peptide inhibitor as described herein.
Accordingly,
compositions comprising immunoregulatory peptide inhibitors as described
herein can be
useful for ameliorating, reducing the symptoms of, reducing the severity of,
and/or treating
immuno suppres s ion.
[0206] Immunoregulatory peptide inhibitors as described herein
interact with or
bind to proteins or peptides that comprise at least one of sequence SEQ ID
NOs: 183-185 or
188-246. Such peptides can have immunoregulatory properties similar to P3028
sequences
and structures (see Examples 17 to 26).
[0207] With reference to some embodiments in the following disclosure,
amino
acids, or amino acid residues can be referred to by either a three-letter or a
one-letter
code. Twenty amino acids are typically encoded by the genetic code, and can be
referred to
using the following codes or abbreviations herein: Arginine ("Arg" or "R"),
Histidine ("His"
or "H"), Lysine ("Lys" or "K"), Aspartic Acid ("Asp" or "D"), Glutamic Acid
("Glu" or
"E"), Serine ("Ser" or "S"), Threonine ("Thr" or "T"), Asparagine ("Asp" or
"N"), Glutamine
("Gln" or "Q"), Cysteine ("Cys" or "C"), Glycine ("Gly" or "G"), Proline
("Pro" or "P"),
Alanine ("Ala" or "A"), Valine ("Val" or "V"), Isoleucine ("Be" or "I"),
Leucine ("Leu" or
"L"), Methionine ("Met" or "M"), Phenylalanine ("Phe" or "F"), Tyrosine ("Tyr"
or "Y"),
Tryptophan ("Trp" or "W").
[0208] With reference to some embodiments in the following disclosure
by
"peptide" is meant a protein and/or a fragment of a protein, which may have
several different
lengths (e.g., at least or equal to 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14
, 15, 16, 17, 18, 19, 20,
21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36,37, 38, 39, 40,
41, 42, 43, 44, 45,
46, 47, 48, 49, 50, 60, 70, 80, 90, 100, 120, 140, 160, 180, 200, 240, 260,
300, 350, 400, 450,
500, 600, 700, 800, or 1000 amino acids or a range defined by any number in
between these
numbers).
[0209] With reference to some embodiments in the following disclosure,
amino
acids (and their residues) can be categorized according to various
characteristics of the side
chains of the alpha carbon of the amino acid. It is noted that the twenty
naturally occurring
amino acids encoded by the genetic code, and also synthetic amino acids are
contemplated
29
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
herein. As used herein "hydrophobic amino acid" (including pluralaizations and
variations of
this root term) refer to naturally occurring or synthetic amino acids having a
hydrophobic side
chain, for example A, V, I, L, M, F, Y, or W. As used herein, "positively
charged amino
acid" (including pluralaizations and variations of this root term) refer to
naturally occurring
or synthetic amino acids having a positively charged side chain, for example,
R, H, or K. As
used herein, "negatively charged amino acid" (including pluralaizations and
variations of this
root term) refer to naturally occurring or synthetic amino acids having a
negatively charged
side chain, for example, D or E. As used herein, "hydrophobic non-aromatic
carbon chain
amino acid" (including pluralaizations and variations of this root term) refer
to naturally
occurring or synthetic amino acids having a hydrophobic non-aromatic carbon
side chain, for
example, A, V, I, or L. As used herein, "polar uncharged amino acid"
(including
pluralaizations and variations of this root term) refer to naturally occurring
or synthetic amino
acids having a polar uncharged side chain, for example, S, T, N, or Q.
[0210] With reference to some embodiments and description herein, the
bases of
nucleic acids, such as DNA, RNA, and the like can be referred to by either the
name of the
base or a one letter code. One skilled in the art will appreciate that the
genetic code is
degenerate, in that for some amino acid residues, two or more three-base
codons can encode
the same amino acid. Thus, some one letter codes, and described herein, can
represent one of
two or more bases, for example to describe two or more possible nucleic acids
that can
encode a single amino acid. One-letter codes used herein include: "A"
(adenine), "G"
(guanine), "C" (cytosine), "T" (thymine), "R" (one of adenine or guanine), "Y"
(one of
cytosine or thymine), "M" (one of adenine or cytosine), "K" (one of guanine or
thymine), "S"
(one of cytosine or guanine), "W" (one of adenine or thymine), "H" (one of
adenine, cytosine,
or thymine), "B" (one of cytosine, guanine, or thymine), "V" (one of adenine,
cytosine, or
guanine), "D" (one of adenine, guanine, or thymine), and "N" (one of adenine,
guanine,
cytosine, or thymine).
[0211] The terms "de-blocking" and "unblocking" as used herein
(including
pluralization and variations of this root term) refers to displacing a bound
immunoregulatory
peptide or P3028 structure from a receptor. As such, de-blocking or unblocking
a receptor
shifts the equilibrium between receptor-bound and non-receptor-bound
immunoregulatory
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
peptide towards the "non-receptor-bound" category. For example, an LFA-1
receptor or IL-2
receptor can be de-blocked in accordance with embodiments herein by displacing
a bound
peptide P3028 from the LFA-1 receptor of IL-2 receptor. For example, an LFA-1
receptor or
IL-2 receptor can be de-blocked in accordance with embodiments herein by
displacing any
immunoregulatory peptide comprising one or more sequences for Tables 1-4 from
the LFA-1
receptor or IL-2 receptor.
[0212] The term "immune cell activation" as used herein, and
pluralizations and
variations of this root term (including such as "activating an immune cell"),
refers to immune
cell proliferation, activating or enhancing expression of CD69 and/or CD71,
induction of
secretion of a signal substance (e.g. IFNy or IL-12), induction of secretion
of a cytolytic
molecule (e.g. perforin or granzyme B), enhanced cytotoxicity, cytokine
production, cell
migration, cell proliferation, or two or more of these listed items. By way of
example,
immune cell activation in accordance with some embodiments herein can occur if
an immune
cell proliferates, or if an immune cell begins to express detectable CD69, or
if an immune cell
increases its expression of CD71, or if an immune cell secretes IFNy, IL-12,
or IFNy and IL-
12.
[0213] Available data support a major role of the immune system in
cancer
control sample. Malignant tumors, however, can exploit a large number of
immunoregulatory
mechanisms to suppress immune mediated anti-tumor reactivity. Based on the
observation
that an increased serum concentration of interleukin-6 (IL-6) often is
correlated to a poor
prognosis in cancer patients of various diagnoses, the origin and induction of
this cytokine
was explored. It was found that proteolytic fragmentation or denaturation of
normal serum
albumin generated neo-structures, which exhibit immunoregulatory activity by
binding to
immune cells. Accordingly, a new class of immunoregulatory substances was
discovered.
[0214] The existence of albumin sequences having neo-structures that
bind to
immune cells was identified using a human ex vivo model based on affinity
chromatography
over an "Artificial Cell Surface Column" (ACS). The effect of different
albumin fragments
on IL-2 induced proliferation of human immune cells (PBMCs) was analyzed in
the ACS
system (see Example 9). Briefly, PBMCs were cultured for seven days in the
presence of IL-
2 and the various synthetically prepared albumin fragments. Proliferation was
measured as
31
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
incorporation of 3H thymidine during the final 18 hours. One of the peptides,
P3028 (also
referred to as "peptide 3028" and having the amino acid sequence
VFDEFKPLVEEPQNLIK
¨ SEQ ID NO: 185) regularly inhibited IL-2 induced proliferation, but none of
the other
peptides identified by their binding to the artificial cell surface showed as
much inhibitory
activity as the P3028 sequence/structure (see Figure 6). Accordingly, the
immune cell
proliferative response induced by LFA-1 or IL-2 could be inhibited by P3028,
indicating that
P3028 sequence/structure may be acting through at least the LFA-1 or IL-2
receptor.
[0215] The enhanced incorporation of 3HTdR can be the result of an
enhanced
specific activity of the intracellular thymidine pools and thereby an enhanced
specific activity
of DNA, thus, not necessarily mirroring an increase in the number of cells. It
was therefore
considered of be of importance to explore a different mode of stimulation of
lymphocyte
proliferation and to measure the response using a different method, the MTS
technique (see
Example 3). Accordingly, T-cells were stimulated in cultures on plates pre-
coated with a
monoclonal antibody directed against CD3 and the number of metabolically
active cells was
determined using MTS staining after 3 to 7 days of culture (see Figure 8). As
shown, P3028
sequence/structure had an inhibitory effect. It can be argued that the reduced
MTS staining
caused by P3028 sequence/structure might be due to a reduced cell metabolism;
however,
taken together the results from both models of lymphocyte proliferation, a
reduced
metabolism should reasonably reduce the endogenous thymidine pools and thereby
result in
an increased uptake of exogenous thymidine/ specific activity of the thymidine
pools, which
then should be erroneously registered as an enhanced proliferation. The 3H-TdR
was actually
reduced in these experiments, indicating inhibition of proliferation.
Accordingly, it was
confirmed that peptides comprising the 3028 sequence effectively inhibited IL-
2 mediated
immune cell proliferation.
[0216] Peptide fragments encompassing the C-and N-terminal parts of
P3028
were then synthesized and the ability of these peptides (separately and in
combination) to
inhibit IL-2 induced proliferation of immune cells was analyzed (see Example
6). An N-
terminal fragment of P3028 (i.e., P3325 having the amino acid sequence
VFDEFKPLVE
(SEQ ID NO: 186)) and a C-terminal fragment of P3028 (i.e., P3218 having the
amino acid
sequence EPQNLIK) (SEQ ID NO: 187)) were synthesized. It was determined that
the
32
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
inhibitory activity of these two fragments of P3028 alone or in combination
was weaker than
P3028 (see Figure 12) and the peptide fragments of 3028 do not inhibit the
effect of P3028
on IL-2 induced proliferation (see Figure 13).
[0217] It was then determined that peptides comprising the P3028
sequence/structure sequence not only interacted with the IL-2 receptor but
also interacted
with the LFA-1 receptor. In a first set of experiments, it was found that the
P3028 peptide
has the capacity to modulate the binding of an LFA-1 specific monoclonal
antibody to the
LFA-1 receptor on human immune cells (see Example 7). This LFA-1 specific
monoclonal
antibody is a potent inhibitor of IL-2 induced immune cell proliferation (see
Vyth-Dreese et
al., Eur. J. Immunol. 12:3292-3299 (1993)). A standard immunohistochemical
staining
procedure was employed in the presence and absence of the P3028 peptide.
Briefly, immune
cells (PBMCs) from healthy individuals and cancer patients were compared. The
cells were
fixed utilizing acetone, blocked with 10% human AB-serum with or without
P3028, and
incubated with a monoclonal anti-LFA-1 antibody and a secondary antibody
followed by
color development using Fast Red. As shown in Figure 16A, a clear membrane
staining 3
was found on PBMCs from healthy control samples in contrast to PBMCs from a
patient with
advanced cancer, which exhibited weak staining 5. However, when the PBMCs from
this
cancer patient were incubated with an antibody specific for the 3028 structure
for 24 hours
the membrane staining 3 appeared, indicating that the antibody bound the 3028-
structure and
thereby unblocked LFA-1 (see Figure 16C) and the discussion infra.
[0218] Since P3028 sequence/structure significantly inhibited the
proliferative
response of immune cells to IL-2, the effect of P3028 sequence/structure on
the binding of
IL-2 to CD25 was studied. The fusion protein of CD25 and the Fc-part of IgG
was bound to
protein G coated micro-plates / ELISA plates and the plates were incubated
with biotinylated
IL-2 with or without the presence of P3028. Surprisingly, the binding of IL-2
to CD25 was
enhanced by the presence of P3028, providing evidence of a three-part
interaction between
1L-2, CD25 and P3028 (see Figure 18A-B). Even if the binding of IL-2 to CD25
is
enhanced, the proper assembly of the high affinity receptor and/or signal
transduction is
blocked as the P3028 sequence/structure is a potent inhibitor of IL-2 induced
proliferation.
Using computer-assisted molecular modeling, it was determined that the P3028
33
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
sequence/structure binds to CD25 at the IL-2 binding site (see Figure 19).
These results
provide greater evidence that the P3028 sequence/structure has at least a dual
immunoregulatory capacity since it binds to both the LFA-1 receptor and the IL-
2 receptor.
[0219] The ability of specific albumin fragments to impact NK-cell
cytotoxicity
was also evaluated. In these experiments, synthetic peptides corresponding to
albumin
fragments (P3028, P3026, and P3027) (SEQ ID NOs: 185, 183, and 184,
respectively) were
prepared and the amount of lysis of K562 target cells was assessed (see
Example 4).
Inhibition was not seen in the presence of the control sample peptide P3027
but P3028 and to
a lesser degree P3026 caused a reduction in NK-cell cytotoxicity (see Figures
9A-B).
Accordingly, peptides having the sequence of P3028 effectively inhibit NK-cell
cytotoxicity.
[0220] The ability of specific albumin fragments to inhibit leukocyte
spreading
and immune cell migration was also analyzed. Briefly, buffy coat cells were
prepared from
heparinized blood by Dextran assisted sedimentation. These cells were then
washed twice in
PBS and transferred to clean slides. The cells strongly adhered to the glass
surface and
spread out; however, pre-treatment of these cells with P3028 at a
concentration of 10i.tg/m1
for 15 minutes efficiently inhibited the immune cell spreading (see Example
5). Similarly,
the impact of P3028 on PBMC migration was studied using the Boyden chamber
technique
(see Example 5). As shown in Figure 11, P3028 is a potent inhibitor of immune
cell
migration (p<0.002).
[0221] Antibodies specific for proteins having the P3028
sequence/structure were
prepared, purified, and characterized (see Example 9). Polyclonal antibodies
specific for
P3028 were generated in rabbits or goats. Briefly, rabbits were immunized with
P3028 and
specific antibodies were affinity purified using P3028. These antibodies were
found to bind
to P3325 (the N-terminal fragment SEQ ID NO: 186) but not P3218 (the C-
terminal
fragment (SEQ ID NO: 187) of P3028.
[0222] In a next series of experiments, the expression of P3028 in
malignant
tumors (e.g., malignant melanoma, renal cell carcinoma, and colorectal cancer)
was identified
by immunohistochemical staining using affinity purified rabbit anti-3028
antibodies (see
Example 9). The immunohistochemical staining of malignant melanoma, renal cell
carcinoma, and colorectal cancer tissue slices showed that the P3028 sequence
containing
34
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
molecules are widely expressed and/or localized on cancer cells. These results
were further
supported by the demonstration of 3028-structures in tumor extracts prepared
from malignant
melanoma metastases using a Western technique (see Example 1). Appreciable
3028
structures (approximately, slightly larger than 66kD) were identified by the
Western blot but
the 3028 sequence was also detected in full size albumin and larger molecules
(see Figure 2).
These results provide evidence that molecules comprising the 3028 structure
are generated
not only by proteolytic fragmentation but also by denaturation. Accordingly,
it was
determined that P3028 sequence and/or molecules that comprise this sequence
are present in
and/or localized to tumor tissue.
[0223] An ELISA technique was then used to confirm that proteins and
peptides
comprising the 3028 sequence were present in human serum. Briefly, a sandwich
assay was
employed, wherein affinity purified anti-3028 antibodies were coated onto high
protein
binding ELISA microwells (capture antibody), and a 1% solution of heat-
inactivated serum,
spiked with increasing concentrations of P3028, was then added to the wells.
After washing,
a biotinylated mouse anti-human albumin monoclonal antibody was added and the
amount of
bound antibody was detected with HRP-conjugated streptavidin and TMB chromogen
substrate (see Example 1). The serum concentration was found to be in the
range of 1.2-1.6
i.t.g/m1 P3028 equivalents in one serum pool from 5 healthy control samples, 1
healthy control
sample serum and 2 sera obtained from cancer patients. The amount of 3028
containing
molecules was determined as the amount of P3028, which inhibits 50% of the
binding of
3028 structures in the serum to the capture antibody (directed against the
3028 epitope) in the
sandwich ELISA (see Figure 3). The amount of these 3028-substances in serum
may be
considerably more as the molecular weight of albumin is about 35 times more
than that of
P3028, but their epitope specific reactivity is accurately determined using
the method
described above.
[0224] Experiments were then performed using a first class of
inhibitors that are
specific for the P3028 sequence/structure. The proliferative response of human
immune cells
from healthy individuals and cancer patients after IL-2 induction were
analyzed in the
presence and absence of antibodies specific for the P3028 sequence/structure
(see Example
9). That is, the proliferative response of PBMCs from a patient having renal
cell carcinoma
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
and a patient having malignant melanoma were compared to the proliferative
response of
PBMCs obtained from a healthy individual in the presence and absence of
antibodies specific
for the P3028 sequence/structure. It was determined that in the presence of
the antibodies
that are specific for the P3028 sequence/structure, enhanced proliferation of
the PBMCs after
IL-2 induction was seen. That is, the antibody inhibitor for the P3028
sequence/structure was
able to remove the blockade on IL-2-induce proliferation of the immune cells
mediated by the
P3028 sequence/structure. These results demonstrate that a binding partner for
the P3028
sequence/structure (e.g., an antibody or binding fragment thereof specific for
P3028), can
reduce the immune suppression mediated by the P3028 sequence/structure.
[0225] The P3028 sequence/structure is a potent physiological
inhibitor of the
immune system, and is a possible a target for therapeutic compositions that
can modulate
immune activity. Antibodies directed against the P3028 sequence/structure
reversed cancer-
related immunosuppression, which was modeled as reduced proliferative response
of PBMCs
to IL-2 in a human ex vivo model (see Example 9). Moreover, the outcome in
this model
correlated to over-all survival of the cancer patients (see Example 2).
Therefore, it was
contemplated that additional binding partners for the P3028 sequence/structure
(e.g.,
peptides, cyclic peptides, peptidomimetics, antibodies and portions thereof)
may be useful for
inhibiting the P3028 sequence/structure-mediated immune suppression.
[0226] Three peptide-based binding partners for the P3028
sequence/structure
were initially developed and the binding capacity of these inhibitors with
P3028 in solution
was tested, as shown in Figure 23 (see Example 10). Only one molecule, SCF28,
had a
solubility sufficient to allow testing in biological human ex vivo models.
Based on this
structure, a first drug candidate, P28R (SEQ ID NO: 2), was developed.
[0227] Since P28R strongly bound to P3028, the ability of P28R to
inhibit the
function of the P3028 sequence/structure was tested. As described above, the
02-integrins
plays a major role in the normal function of the immune system. However, the
binding of the
P3028 sequence/structure, to the 02-integrin LFA-1 has a substantial
immunosuppressive
effect. As demonstrated above (see Example 7), in assays staining for LFA-1,
the membrane
staining of PBMCs from cancer patients is markedly decreased compared to
normal control
samples. The exposure of LFA-1 could, however, be enhanced by incubating PBMCs
from
36
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
cancer patients with an antibody directed against the inhibitory P3028
sequence/structure (see
Example 7 and Figure 16C).
[0228] Without being limited by any theory, the occurrence of tumour
infiltrating
lymphocytes in primary tumours generally indicates good prognosis. However, in
many
tumors, tumor-infiltrating lymphocytes can have reduced function or a lack of
function, and
rather than migrate into nodules of tumour cells, can "get stuck" in the
stromal areas of the
tumour. It is observed that incubation of fresh frozen tumour sections with
peptide P28R
(SEQ ID NO: 2) de-blocks LFA-1 of tumour infiltrating lymphocytes (i.e.
displaces a bound
immunoregulatory peptides or P3028 strucutres from the LFA-1 receptors),
resulting in an
enhanced binding of the anti-CD1 la antibody (Figure 26). These results showed
that the
LFA-1 receptor was unblocked by removal of the P3028 structure by the
antibody. To test
the ability of P28R to inhibit the P3028 structure, fresh frozen tumor
sections without
fixation were incubated for 4-20 hours in the presence of the drug candidate,
P28R before
staining for LFA-1 (see Example 15). For comparison, tumor sections were
incubated with
phosphate buffered saline only. As shown in Figure 26, P28R effectively
unblocked the
LFA-1 receptor (e.g. displaced bound immunoregulatory peptides or 3028
structures from the
LFA-1 receptor) and thereby markedly enhanced the functional expression of LFA-
1 enabling
migration and cytotoxic activity of these cells. Accordingly, P28R decreases
the binding of
P3028 to LFA-1 and effectively inhibits the immune suppression mediated by
P3028. It is
contemplated that incubation with P28 core (SEQ ID NO: 62) in accordance with
some
embodiments herein also de-blocks LFA-1 (e.g. displaces bound immunoregulatory
peptides
or 3028 structures from the LFA-1 receptor).
[0229] As such, the receptors of P3028 include LFA-1 and the alpha
chain of the
IL-2 receptor (CD25). Binding of a monoclonal antibody to CD1 la (the alpha
chain of LFA-
1) was used to study the possible occurrence of a physiological blocker of LFA-
1 and the de-
blocking activity of P28R and antibodies directed to P3028. Accordingly, it is
further
contemplated that, similar to the LFA-1 receptor, the IL-2 receptor can be de-
blocked by
immunoregulatory peptide inhibitors as described herein (e.g. bound
immunoregulatory
peptides or 3028 structures can be displaced from the IL-2 receptor).. As
such, in some
embodiments, an immunoregulatory peptide inhibitor as described herein
deblocks an IL-2
37
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
receptor, for example an IL-2 receptor that has been blocked by any one or
more of the
peptides listed in Tables 1-4 (e.g. a peptide comprising SEQ ID NO: 185).
[0230] Incubation of PBMCs from healthy controls with P3028 (Figures
15 and
17) or cancer patient sera (Figure 17) blocks the binding of the anti-CD11 a
antibody to LFA-
1. Furthermore, incubation of PBMCs from advanced cancer patients with an
antibody
directed against P3028 restitutes the binding of the anti-CD1 la antibody to
LFA-1 (Figure
16). P3028 can bind to PBMCs (see Figure 15A depicting no peptide added, and
Figure
15B, depicting preincubation with peptide 3028; anti-LFA-1 mAb HIM was
inhibited by
preincubation with peptide 3028, indicating binding to mononuclear blood cells
by peptide
3028).
[0231] Since P28R unblocks LFA-1 receptors that are suppressed by the
P3028
sequence/structure (e.g. displaces bound immunoregulatory peptides or 3028
structures from
the LFA-1 receptor), the ability of P28R to enhance immune stimulation was
tested in human
ex vivo models. The stimulatory activity of P28R on PBMCs was measured using
the MTS
or CFSE techniques in 7 healthy control samples and 7 cancer patients of
various diagnoses
(see Example 13). Even in the absence of other types of stimulation, P28R has
a significant
stimulatory activity in 6 out of 7 cancer patients; whereas PBMCs from control
samples
showed only weak or no stimulation (see Example 13). Similar to the studies on
the efficacy
of antibodies directed against P3028 to reverse cancer related
immunosuppression above (see
Example 9; see Figure 22), the ability of the P28R inhibitor to unblock the IL-
2 receptor and
thereby induce immune cell proliferation was investigated. Cultures of PBMCs
from four
different treatment naïve patients were each treated with P28R, and
proliferation of PBMCs
was measured. While PBMC' s that had high proliferative activity before P28R
treatment
were largely unaffected by the drug (see Figure 24C and Figure 24D), PBMCs
with a low
initial proliferation were markedly stimulated (see Figure 24A and Figure 23B;
see
Example 13). Thus, the P28R inhibitor effectively induces immune cell
proliferation when
the immune cells are bound and suppressed by the P3028 sequence/structure,
even in the
absence of additional stimulation.
[0232] Since cancer cells have been shown to be enriched for P3028
structures
(see Example 1 and Figures 1-2), the ability of P28R to specifically bind
cancer cells was
38
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
investigated. The binding of biotinylated P28R to tumors was studied. Three
breast cancers,
two renal cell carcinomas and four malignant melanomas were analyzed. Notably,
all of the
different types of tumors analyzed in the experiments bound P28R. The stained
breast cancer
section, shown in Figure 25, for example, exhibits a strong positive signal,
indicating the
presence of the inhibitory P3028-structure in this tumor, and ability of P28R
to bind to this
tumor (see Example 14).
[0233] Since the P3028-structure inhibits lymphocyte migration, as
well as,
cytotoxic activity (see Examples 4 and 5), an immune system mediated attack
against
positively-staining tumor areas is expected to be efficiently suppressed so
long as the a
P3028-containing structure is present and not sequestered by a binding partner
for the P3028
sequence/structure (e.g., an antibody, binding fragment thereof, and/or an
inhibitory peptide,
such as P28R, or a peptidomimetic corresponding to the P28R structure).
Consistent with the
observation that P3028 strongly binds the LFA-1 receptor, lymphocytes were not
stained by
this procedure since the P3028 structure was blocked by binding to LFA-1 on
these cells.
[0234] Based on the ability of P28R to bind the P3028
sequence/structure,
unblock the LFA-1 receptor, and ameliorate the P3028 sequence/structure-
dependent
immunosuppression, P28R was used as a template compound to identify additional
compounds that bind to and sequester P3028. Variants of the P28R structure
were
synthesized, and tested for the ability to bind P3028 using PEPSCAN technology
(see
Example 12). A library of peptides that include each genetically-coded amino
acid
substitution at each amino acid position of P28R (i.e., 19 substitutions for
each position) was
synthesized. Each peptide was affixed to a support pin, and the peptide
library was incubated
with P3028. The binding of the candidate inhibitors to P3028 was detected by a
sandwich
ELISA, where a rabbit anti-mouse peroxidase (rampo) secondary antibody was
employed
(see Example 12). The binding of each peptide was then assigned a rampo score
(see Figure
27). Peptide P28R had rampo values ranging between about 262 and 460 with a
mean value
of 370. In some embodiments, the immunoregulatory peptide inhibitor as
disclosed herein, is
selected for a desired P3028 binding rampo score. In some embodiments, the
desired P3028
binding rampo score is greater than or equal to the rampo score of P28R. It is
also
contemplated that some peptides that bind to P3028 with less affinity than
P28R have
39
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
therapeutic application. Some peptides with binding affinities that are less
than P28R, for
example, may modulate signal transduction events differently than P28R by
virtue of the fact
that the affinity to P3028 is less. Accordingly, embodiments also include any
peptide that
binds to P3028, wherein said peptides have a rampo score that is less than
that exhibited by
P28R. Accordingly, contemplated embodiments include peptides that bind with
any affinity
to P3028 (e.g., any one or more of the peptides provided in Table 5.1,
preferably peptides
that modulate the immune system (e.g., modulate, upregulate or down regulate a
marker of
the immune system or immunosuppression, such as reducing a P3028-mediated
inhibition of
immune cell proliferation, spreading, migration, or NK-cell cytotoxicity).
[0235] A total of 31 substitutions of peptide P28R (SEQ ID NOs: 3-33)
had
rampo values greater than 500 (see Figure 28), indicating that these 31
peptides (strong
binding partners for P3028) can be used to efficiently bind and sequester
P3028 and thereby
reduce P3028-mediated immunosuppression. Table 6.1 lists these 31 peptides
that were
evaluated in assays and shown to have appreciable binding to P3028.
Additionally, the
binding strength of substituted peptides at each position (based on rampo
score) was
compared to the binding strength of a P28R (SEQ ID NO: 2) control sample for
the same
position (see Example 12). Peptides that bound with a rampo score
substantially equal to or
greater than that of the P28R control sample (i.e., at peptides that bound to
P3028 with at
least 98% of the rampo score of the P28R control sample) were identified (SEQ
ID NOs:
268-393). Table 6.2 lists these 126 peptides that were shown to have
appreciable binding to
P3028. It is noted that these 126 peptides include the 31 peptides of Table
6.1. Accordingly,
126 different binding partners for P3028 were identified by this initial
screen and these
molecules or variants thereof (e.g., variants having D amino acids, N-terminal
amides, and/or
C terminal acetyl groups or peptidomimetics or aptamers corresponding to these
binding
partners) can be used to inhibit the binding of the P3028 sequence/structure
to an immune
cell and thereby alleviate, or reduce P3028-dependent immunosuppression. One
variant of
P28R, Peptide KKL15 (SEQ ID NO: 1), which lacks only a C-terminal arginine, is
thought
to bind to the P3028 sequence/structure through both charged and hydrophobic
interactions.
As shown in Figure 31, positively charged amino acids of KKL15 interact with
negatively
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
charged amino acids on P3028 and hydrophobic amino acids generate hydrophobic
contacts
enhancing the interaction.
[0236] To further map the P3028 binding domain of P28R, deletions, and
truncations of P28R were synthesized, and tested for binding to P3028 using
the PEPSCAN
assay. This approach led to the development of many more binding partners for
P3028.
While deletion of residues 6-9 ("FFVK" ¨ SEQ ID NO: 182) and the C-terminal
amino acids
tended to reduce the binding of peptides to P3028 based on rampo score (see
Example 12
and Figure 30), several deletions and truncations of peptide P28R have a rampo
score
comparable to, or higher than peptide P28R (see, e.g., SEQ ID NOs: 34, 64-66,
68, and 76).
Additionally, peptides deleted up to at least 8 amino acids from the N-
terminus of P28R (see,
e.g., SEQ ID NOs: 46-53) retained a high affinity to P3028, as measured by
rampo score,
providing evidence that inhibitors that are smaller than P28R can be useful
for binding to and
sequestering P3028, preventing the interaction of P3028 with immune cell
receptors, such as
the IL-2 or LFA-1 receptors, thereby reducing P3028-induced immunosuppression.
[0237] Because P28R was shown to have a modulatory effect on IL-2
stimulation
of immune cell proliferation (see Example 2), it was further investigated
whether P28R
would have a modulatory effect on other aspects of IL-2 stimulation of immune
cells.
PBMC's from eight healthy control samples and nine cancer patients with
various diagnoses
were cultured in a modified version of the ex vivo model of Example 2 for
seven days in the
presence of various doses of P28R (either "no P28R" control samples, or
5[tg/mL, 10[tg/ml,
or 20[tg/m1 of P28R). A dose dependent stimulation of the mitochondrial
metabolism
measured as conversion of MTS was observed in 5/8 (see Figure 33A) control
samples and
9/9 cancer patients (see Figure 33B). Similar results were obtained when the
PBMCs were
cultured for only three days (see Example 28).
[0238] To identify the effectiveness of other inhibitors of
immunomodulatory
peptides, the effect of P28R (SEQ ID NO: 2) on mitochondrial metabolism based
on MTS
converstion was compared to the effect of a closely related peptide P27. P27
has the sequence
KKLDTFFKKLSLFTER (SEQ ID NO: 264), and is a variant of P28R that differs in
that V8
of P28R is substituted to K8 in P27. P28R binds to P3028 more efficiently than
P27 (P27 binds
P3028 with a rampo score of 253, while a P28R control sample binds P3028 with
a rampo score
41
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
of 308; see Example 12). The concentrations were either untreated control
samples, 5pg/mL
("SCF28-R5" and "SCF275"), 10pg/m1 ("SCF28-R10" and "SCF2710"), 20pg/m1("SCF28-
R20"
and "SCF2720"), or 40pg/m1("SCF28-R40" and "SCF2740"). The results are shown
in Figure
34. While P28R stimulated the cells of cancer patients in a dose-dependent
manner, P27 had no
effect (see Example 29).
[0239] The effect of P28R (SEQ ID NO: 2) on IL-2 induced proliferation
was also
measured in a BrdU incorporation assay. PBMCs from six healthy control samples
and ten
cancer were harvested in a modified version of the ex vivo model. Four out of
six control
samples had a high proliferative response to IL-2 compared to four out of ten
cancer patient
samples (see Figure 35). These differences in proliferative response to IL-2
in PBMCs
demonstrated the difference existence of high and low responders to IL-2
stimulation (see
Example 30).
[0240] The response of high responders and low responders to various
doses of
P28R was compared. Cells from either high responders or low responders were
cultured with
various doses of P28R (see Figures 36A and 36B). IL-2-induced proliferation
was measured
as BrdU incorporation. While P28R had no stimulatory effect in cells from
patients with a
high response to IL-2 (N=4) (see Figure 36A), P28R had a stimulatory effect on
cells from
patients with a low response to IL-2 (N=6) (see Figure 36B). Accordingly
effects of P28R
on binding to and blocking immunoinhibitory activity of P3028 were
demonstrated in in the
ex vivo model, as addition of P28R to the cultures had no effect on
proliferation when added
to PBMCs from healthy controls and cancer patients with a normal proliferative
rate, but the
proliferation of PBMCs from immunosuppressed cancer patients were
significantly
stimulated by P28R. Without being limited by any theory, in some embodiments
P28R (SEQ
ID NO: 2) or P28 core (SEQ ID NO: 62) binds to a blocker of immune cell
proliferation, and
induces immune cell proliferation.
[0241] The effect of P27 (SEQ ID NO: 264) was then compared to the
effect of
P28R (SEQ ID NO: 2) on IL-2 induced proliferation as measured by BrdU
Incorporation.
PBMCs from low responder cancer patients were with various concentrations of
either P28R of
P27, ranging from no peptide ("untreated cells"), to 5pg/mL, 10pg/ml, or
20pg/ml. As shown in
Figure 37, both P28R and P27 enhanced the proliferative rate of PBMC's induced
by IL-2 as
measured by BrdU incorporation. When comparing the results shown in Figure 37
to those of
42
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
Figure 34, P27 was observed to enhance IL-2 stimulation of cell proliferation
as measured by
BrdU incorporation, but not mitochondrial metabolism as measured by MTS
conversion. On
the other hand, P28R was observed to enhance both MTS conversion and BrdU
incorporation
in response to IL-2 stimulation (see Example 31).
[0242] The different effects of different inhibitors of
immunoregulatory peptides
on BrdU incorporation and MTS conversion were further investigated. The
effects of P28R
on IL-2 stimulation of immune cell proliferation differed significantly,
depending on which
assay was used (see Figure 38). Peptide P28R had a stimulatory activity of MTS
conversion
in seven day cultures of PBMCs in 100% of cancer patients examined (N=9) and
in 63% of
healthy control samples examined (N=8). In contrast, P28R stimulated
incorporation of
BrdU in seven day cultures of PBMCs from only 17% (N=6) and 20% of (N=10)
patients.
P28R stimulated IL-2 induced proliferation, measured as incorporation of BrdU,
in PBMC
cultures from cancer patients with a low proliferative response to IL-2. On
the other hand,
PBMCs from 67% of healthy control samples examined (N=3) and 50% of cancer
patients
(N=4) were not stimulated by IL-2 when the effect was measured as MTS
conversion (see
Example 32 and Figure 38). However, PBMCs from all these persons ("non-
responders")
who did not respond when measured with MTS were significantly stimulated by IL-
2 when
the effect was measured as incorporation of BrdU (see Figure 38). In two
patients, the
response to IL-2, measured as BrdU incorporation, was enhanced by P28R (see
Figure 38A
and 38C), but this effect of P28R was only observed in one of these patients
when MTS
conversion was used (see Figure 38B). Thus, while in one patient (see Figures
38A and
38B) the stimulatory activity of IL-2 was registered using both BrdU and MTS,
in the other
patient, the stimulatory activity of IL-2 was registered using BrDU only (see
Figure 38C)
(see Example 32). Based on these observations, it was contemplated that
effects on the
metabolic activity measured as MTS conversion does not always correlate with
DNA
synthesis measured as incorporation of BrdU, and different populations of
patients can
respond differently to inhibitors of immunoregulatory peptides.
[0243] It was contemplated that other molecules that bind to P3028
could be
identified. These additional binding molecules could also potentially block
P3028. Looped
6-mere peptides were synthesized, and 6-meres that demonstrated appreciable
binding to
43
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
P3028 were identified (see Table 12, SEQ ID NOs: 265-267) (see Example 19). It
was
observed that two of the 6-meres with the strongest binding to P3028 based on
rampo score
possessed homology to linear peptides that bind 3028 (see Figure 32).
[0244] In addition to P3028, several other albumin fragments and
synthetic
peptides were found to bind to the immune cells. Some of these fragments can
have
immunomodulatory activity similar to P3028, can bind to immune cells similar
to P3028,
and/or can bind to immunomodulatory antibodies that recognized P3028. In a
first set of
experiments, albumin fragments were generated by trypsin digestion and the
tryptic
fragments were found to bind to immune cells in the ACS system described
herein (see
Example 17). Table 1 provides a listing of trypsin-generated fragments of
albumin, which
bind to immune cells in the ACS system, as detected by MALDI-TOF analysis.
Table 1: Trypsin-generated albumin fragments that bind to ACS
SEQ ID NO: Percent Sequence Albumin
Absorbed Positions
194 71% KYLYEIAR 161-168
195 64% KVPQVSTPTLVEVSR 438-452
196 60% VFDEFKPLVEEPQNLIK 397-413
197 59% VPQVSTPTLVEVSR 439-452
198 42% RPCFSALEVDETYVPK 509-524
199 41% FQNALLVR 427-434
200 36% SLHTLFGDK 89-97
201 36% LKECCEKPLLEK 299-310
202 35% LCTVATLR 98-105
203 34% YLYEIAR 162-168
204 32% CCAAADPHECYAK 384-396
205 29% AAFTECCQAADK 187-198
206 26% CCTESLVNR 500-508
207 25% QEPERNECFLQHK 118-130
208 23% AVMDDFAAFVEK 570-581
209 22% NECFLQHK 123-130
210 20% ONCELFEQLGEYK 414-426
211 18% QEPERNECFLQHK 118-130
212 13% VHTECCHGDLLECADDR 265-281
213 8% FKDLGEENFK 35-44
214 3% YICENQDSISSK 287-298
215 2% LDELRDEGK 206-214
216 1% DDNPNLPR 131-138
44
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
[0245] In a second set of experiments, denatured human serum albumin
was
degraded by asparaginase (ASN-N), and the ability of these proteolytic
fragments to bind
with immune cells was evaluated in the ACS system. Again, the immune cell
binding
peptides were identified by comparing adsorbed and unadsorbed peptide
solutions using the
MALDI TOF technique. These peptides are shown in Table 2.
Table 2: Asp-N-generated albumin fragments that bind to ACS
SEQ ID Percent Sequence Albumin
NO: Absorbed Positions
217 100% DHVKLVNEVTEFAKTCVA 62-79
218 100% DDKETCFAEEGKKLVAASQAALGL 586-609
219 87% DRVTKCCTESLVNRRPCFSALEV 495-517
220 86% DETYVPKEFNAETFTHA 518-535
221 65% DSISSKLKECCEKPLLEKSHCIAEVEN 293-319
222 65% DKLCTVATLRETYGEM 96-112
223 100% YSVVLLLRLAKTYETTLEKCCAAADPHEC 364-398
YAKVF
224 100% KLCTVATLRETYGEMADCCAKQEPERNEC 96-130
FLQHK
225 100% ICTLSEKERQIKKQTALVELVKHKPKATKE 536-572
QLKAVM
226 100% LAKYICENQDSISSKLKECCEKPLLEKHCIA 283-319
EVEN
227 100% VFLGMFLYEYARRHPDYSVVLLLRLAKTY 348-388
ETT LEKCCAAA
228 100% LGEENFKALVLIAFAQYLQQCPFEDHVKLV 37-79
NEVTEFAKTCVA
229 100% RVTKCCTESLVNRRPCFSALEVDETYVPKE 495-535
FNAETFTFHA
230 37% YLSVVLNQLCVLHEKTPVSDRVTKCCCTES 475-517
LVNRRPFSALEV
[0246] Additionally, several synthetic peptides were synthesized, as
shown in
Table 3, and the binding of these molecules to immune cells using the ACS
system was
evaluated.
Table 3: Synthetic albumin peptides
SEQ ID NO: Peptide Name Sequence Albumin
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
Positions
183 3026 NEETFLKKYLYEIARRHPYFYAP 153-176
184 3027 ELFEQLGEYKFQNALLVR 417-434
188 3029 KVPQVSTPTLVEVSR 438-452
189 2604 KLVNEVTEFAKT 65-76
190 2605 NEETFLKKYLYE 153-168
191 2606 LDELRDEGKAS 205-217
192 2607 EMADCCAKQEPE 110-122
193 2608 ELFEQLGEYKF 417-427
[0247] Additionally, several albumin fragment peptides bind
specifically to an
dHSA-specific antibody with immunomodulatory effects (mAb A) (see Example 18).
These
peptides are shown in Table 4.
Table 4: Albumin peptides that bind to monoclonal antibody mAb A
SEQ ID NO: Sequence Albumin Positions
231 LVNEVTEFAK 066-075
232 SLHTLFGDK 089-097
233 LCTVATLR 098-105
234 ETYGEMADCCAK 106-117
235 YLYEIAR 162-168
236 LDELRDEGK 206-214
237 YICENQDSISSK 287-298
238 LKECCEKPLLEK 299-310
239 HPDYSVVLLLR 362-372
240 CCAAADPHECYAK 384-396
241 QNCELFEQLGEYK 414-426
242 FQNALLVR 427-434
243 CCTESLVNR 500-508
244 AVMDDFAAFVEK 570-581
245 LS QRFPK 243-249
246 DDNPNLPR 131-138
[0248] It is contemplated that inhibitors to any one or more of the
peptides listed
in Tables 1-4 can be generated in much the same way that inhibitors to P3028
were
generated. In brief, polyclonal and monoclonal antibodies that are specific
for any one or
more of the peptides in Tables 1-4 can be easily generated using conventional
techniques in
immunology. Antibody binding fragments can also be prepared and isolated using
46
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
conventional techniques in immunology. These antibodies or antibody fragments
can be
human, or humanized, as described herein. Using an approach similar to that
described supra
and in Examples 9 and 10, these peptide inhibitors can be evaluated on a chip
based assay
and biochemical assays, such as immune cell proliferation in the presence and
absence of the
peptide inhibitors, can be evaluated. The section below provides more
information on the
development of immunoregulatory peptide inhibitors, preferably inhibitors of
P3028.
[0249] It is contemplated that inhibitors of any one or more of the
peptides listed
in Tables 1-4 can comprise modifications of the P28R (SEQ ID NO: 2) or P28
core (SEQ
ID NO: 62) sequence, and further can be useful for reducing inhibition of the
LFA-1
receptor, or for stimulating immune cells. To identify modification to
inhibitor peptides in
accordance with some embodiments herein, positional scan data was used to
study the
influence of substitution of different types of amino acids in each position
of P28R (SEQ ID
NO: 2) on the binding of P3028 (SEQ ID NO: 185). Each amino acid in the
peptide
sequence of P28R (SEQ ID NO: 2) was exchanged with all of the naturally
occurring amino
acids, and binding of P3028 (SEQ ID NO: 185) to each peptide on a solid phase
chip was
assessed (see, e.g. Example 36). A number of optional modifications to P28R in
accordance
with embodiments herein are summarized in Tables 5.3, 5.4, 5.5, 5.6, and 13.
Optionally, an
inhibitor peptide in accordance with some embodiments herein can comprise one
or more of
the modifications of Table 5.3 or Table 13. Optionally, an inhibitor peptide
comprises a
central core of positions 2, 5-11, and 15 as provided in Table 5.3, and the
remaining position
are omitted or substituted with substantially any amino acid. Optionally, an
inhibitor peptide
comprises a central core of positions K2, T5-S11, and EIS of SEQ ID NO: 2, and
the
remaining position are omitted or substituted with substantially any amino
acid.
[0250] From the positional scan data it is also noted that a "core
peptide" can be
identified, FFVKLS (SEQ ID NO: 62) (referred to herein as "P28 core"). In some
embodiments, a peptide comprising, consisting of, or consisting essentially of
P28 core (SEQ
ID NO: 62) is provided. The peptide can comprise no more than about 30 amino
acid
residues, for example no more than about 30, 29, 28, 27, 26, 25, 24, 23, 22,
21, 20, 19, 18,
17, 16, 15, 14, 13, 12, 11, 10, 9, 8, 7, or 6 amino acid residues. In some
embodiments, the
core peptide de-blocks an LFA-1 receptor (e.g. displaces bound
immunoregulatory peptides
47
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
or 3028 structures from the LFA-1 receptor) that has been bound by one or more
immunoregulatory peptides of Tables 1-4.
[0251] Based on the positional scan data, it is contemplated that
substitutions of
SEQ ID NO: 2 can be useful in accordance with some embodiments herein for
binding
P3028, de-blocking the LFA-1 receptor from P3028-mediated inhibition (e.g.
displacing
bound P3028 peptide and P3028-structure containing molecules from the LFA-1
receptor),
and/or stimulating immune cells. The activity of peptide P28R (SEQ ID NO: 2)
and
modifications of P28R was studied in a human ex vivo model using PBMCs from a
healthy
control human in short term cultures, and with PBMC activation measured as a
percentage of
cells with enhanced CD69 (see Example 37). It was observed that P28R (SEQ ID
NO: 2)
and peptide 31135 (KKLDTFFVYLSLFTER)(SEQ ID NO: 589) directly stimulate
healthy
PBMC's in this ex vivo model, but peptides 30677 (KKLDTFFVKLSLMTER)(SEQ ID NO:
583), 30678 (KKLDTFFVKLQLFTER)(SEQ ID NO: 584), 30680
(KKLDTVMVKLQLMTER)(SEQ ID NO: 585), 30864 (KSLDTFFVKLSLFTER)(SEQ ID
NO: 587); 30685 (KKLDTFFVKLSLFTFR)(SEQ ID NO: 588); and 31136
(KKLDTFFVNLSLFTER)(SEQ ID NO: 590), and 31138 (KKLDTFFVDLSLFTER)(SEQ
ID NO: 591) did not stimulate the healthy PBMC's in this ex vivo model (see
Figures 41A
and 41B). As such, in some embodiments, a composition comprising, consisting
essentially
of, or consisting of P28R (SEQ ID NO: 2), peptide 31135 (SEQ ID NO: 589), or a
combination of P28R and peptide 31135 is provided to directly stimulate immune
cells. As
such, in some embodiments, a composition comprising, consisting essentially of
a peptide of
SEQ ID NO: 2, SEQ ID NO: 62, or any of SEQ ID NOs: 583-586 or 587-595, or a
combination of these peptides is provided.
[0252] It is noted that peptide 31135 comprises a Y at the position
corresponding
to position 9 of SEQ ID NO: 2 and position 4 of SEQ ID NO: 62. (see Tables 5.3
and 5.5).
In some embodiments, a composition comprising, consisting essentially of, or
consisting of a
modified peptide comprising a modification of P28R comprising a Y at position
9 of SEQ ID
NO: 2 is provided. Optionally, the immune cells can comprise healthy immune
cells.
Optionally, the immune cells can comprise immune cells in cancer patient
serum, for
example cancer patient immune cells. In some embodiments, a composition
comprising,
48
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
consisting essentially of, or consisting of a modified peptide comprising a
modification of
P28 core comprising a Y at position 4 of SEQ ID NO: 62 is provided.
Optionally, the
immune cells can comprise healthy immune cells. Optionally, the immune cells
can
comprise immune cells in cancer patient serum, for example cancer patient
immune cells.
[0253] As P28R (SEQ ID NO: 2) can bind to P3028 and stimulate PBMC's
from
healthy controls in short term cultures, for example when in a culture medium
comprising
RPMI plus 10% normal human AB serum (see Example 37), it is contemplated that
truncations of P28R in accordance with some embodiments herein can be useful
for binding
to inhibitors of any one or more of the peptides listed in Tables 1-4.
Truncations of P28R
were assessed for their ability to activate PBMC's (see Example 38). PBMCs
were
incubated with the peptides (40i.tg/mL) for 24 hours in RPMI plus 10% human AB
serum.
PBMC activation was measured as percent cells with enhanced expression of
either CD69
(Figure 42A) or CD71 (Figure 42B) using flow cytometry. As shown in Figures
42A and
42B, peptide P28R (SEQ ID NO: 2) effectively activated healthy PBMC's in this
ex vivo
model, but peptide 32251 (SEQ ID NO: 592) and peptide 32230 ("P28
core")(FFVKLS)(SEQ ID NO: 62) did not. However, PBMCs were also incubated with
the
peptides in cancer sera from dogs, or in caner sera from human cancer patients
(see Figure
43). It was observed that full length peptide P28R (SEQ ID NO: 2) and the P28
core peptide
(peptide 32230)(SEQ ID NO: 62) activated PBMCs in the presence of cancer
serum. As
such, it is contemplated that in accordance with some embodiments herein,
P28R, P28 core,
or combinations of these peptides are useful for stimulating immune cells in
the serum of a
subject that has cancer.
[0254] In some embodiments, a peptide comprising, consisting of, or
consisting
essentially of P28 core (SEQ ID NO: 62) is provided. Optionally, the peptide
comprising,
consisting of, or consisting essentially of P28 core (SEQ ID NO: 62) can bind
to P3028
peptide. It was observed that P28 core peptide (SEQ ID NO: 62) can bind the
3028 peptide
as efficiently as the full length peptide P28R, and can induce activation
(e.g. proliferation,
enhanced expression of CD69 and/or CD71, secretion of IL-12 or IFNy, or
secretion of
perforin or granzyme B, enhanced cytotoxicity, cell migration, or cytokine
production) of
PBMC's in cancer serum (see Example 38 and Figure 43), but that in an ex vivo
model
49
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
comprising short term cultures of PBMC's, the P28 core peptide (SEQ ID NO: 62)
not
stimulate PBMC activation (CD69 and CD71) as the P28R peptide does (see
Figures 42A
and 42B). Accordingly, in some embodiments, a peptide comprising, consisting
of, or
consisting essentially of P28 core (SEQ ID NO: 62) binds to P3028 peptide as
efficiently or
substantially as efficiently as P28R (SEQ ID NO: 2). In some embodiments, P28R
(SEQ ID
NO: 2 is provided to bind to P3028 and de-block cellular receptors (e.g.
displaces bound
immunoregulatory peptides or 3028 structures from the cellular receptors).
Optionally P28R
can further have a direct stimulatory activity on immune cells. In some
embodiments, P28
core (SEQ ID NO: 62) is provided to bind to P3028 and de-block cellular
receptors (e.g.
displaces bound P3028 peptides or 3028 structures from the cellular
receptors).
[0255] It has also been observed that, biotinylated P28R has been
shown to bind
directly to PBMCs as demonstrated by immunocytochemistry or rosetting of P28R
coated
beads (binding of beads to the cells). Accordingly, in some embodiments, P28R
is provided
to bind directly to PBMCs. In some embodiments, P28R comprising a detectable
moiety is
provided to bind to PBMCs. In some embodiments, P28R comprising a toxin is
provided to
bind to PBMCs. In some embodiments, peptide 31135 comprising a toxin or a
detectable
moiety is provided.
[0256] The effect of P28R (SEQ ID NO:2) on cancer cells was further
studied in
in vivo models in nude and immunocompetent mice. P28R was injected intra-
tumorally into
human pancreas cancer in a xenograft model in nude mice, and induced tumor
cell apoptosis
after one day (see Example 39). P28R induced Caspase 3, a marker of ongoing
apoptosis,
while treatment of tumors with the drug solvent only did not induce Caspase 3
(see Figures
44A and 44B). In some embodiments, P28R (SEQ ID NO: 2) has a direct cytotoxic
action
on tumor cells, for example, prostate cancer cells. In some embodiments, a
peptide of Table
5.3, or a modified P28R peptide comprising at least one modification of Table
5.2 has a
direct cytotoxic action on tumor cells, for example prostate cancer cells.
[0257] As it was observed that P28R has an immunostimulatory effect
(see, e.g.
Example 37), the capacity of P28R (SEQ ID NO: 2) to activate the immune system
was also
evaluated. P28R, 40 microgram in 100 microliter was injected intra-tumorally
into B16
melanoma in B16 melanoma-inoculated immunocompetent mice, C57B1 (see Example
40).
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
Tumors were taken out after 3 days, and sections were immunohistochemically
stained using
a polyclonal rabbit anti-CD45 antibody. The dominating cells in the tumors
after P28R
treatment were inflammatory cells, as indicated by CD45 immunostaining 450
(see Figure
45A). The staining was not observed 452 in a control tumor section incubated
with rabbit
IgG at the same concentration (Figure 45B). It is contemplated that in some
embodiments
P28R (SEQ ID NO: 2), P28 core (SEQ ID NO: 62), a peptide of SEQ ID NO: 586 or
589,
or a modified P28R peptide comprising at least one modification of Table 5.2
can activate
the immune system, for example to direct an immune response against tumor
cells. In some
embodiments, one or more of the listed peptides is administered at or near a
tumor. In some
embodiments, one or more of the listed peptides is administered peri-
tumorally. In some
embodiments, one or more of the listed peptides is administered systemically.
[0258] As
it is contemplated that modifications of P28R can be useful for immune
cell stimulation, the influence of various amino acid substitutions and
additions to P28R on
the immunostimulatory effect was further studied. Effects of modified peptides
on the
activation of PBMCs from a healthy control person were assessed (see Example
41).
PBMCs were incubated with the peptides (40i.tg/mL) for 48 hours in RPMI plus
10% human
AB serum, and PBMC activation was determined by flow cytometry based on the
percentage
of cells with enhanced marker CD69 or CD71. Peptides P28R (SEQ ID NO: 2), P28
core
(peptide 32230)(SEQ ID NO: 62), 32251 (KKLDTFFPKLSLFTER)(SEQ ID NO: 592),
32814 (RKLDTFFVKLSLFTERRR)(SEQ ID NO: 586),
32815
(KKLDQFFVKLSQHNER)(SEQ ID NO: 595), 32665 (KKLDTFMVKLSQHTER)(SEQ ID
NO: 593), and 32819 (KKLDTFFVKLSLFTER(C(PEG24)))(SEQ ID NO: 594) were tested.
As shown in Figure 46, peptide 32814 (SEQ ID NO: 586), had a stimulatory
effect in short
term cultures similar to that of P28R (SEQ ID NO: 2) (batch CS8040) for both
CD69
enhancement (see Figure 46A) and CD71 enhancement (see Figure 46B).
Accordingly, it is
contemplated herein that
[0259] In
addition to therapeutic applications, diagnostic applications of P28R
and truncations and modifications thereof were also contemplated. For example,
information
about patients systemic and local (intra-tumoural) immune status can be
obtained using
reagents comprising P28R, or a truncation or modification thereof.
51
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
[0260] It is contemplated that the occurrence of immunoinhibitory 3028-
structures in tumors can be identified by immunohistochemical staining using
either an
antibody directed against P3028 or using labeled P28R (SEQ ID NO: 2) or P28
core (SEQ
ID NO: 62), for example biotinylated P28R or P28 core. Figure 47 shows two
areas of a
human breast cancer stained using biotinylated P28R. Staining 470 is observed
in Figure
47B. Staining is not observed in Figure 47A. An absence of staining is
indicated 472.
[0261] As such, areas of tumors comprising P3028 structures (as well
as areas not
comprising these structures) can be identified using labeled peptides in
accordance with
embodiments herein. In some embodiments, a peptide of SEQ ID NO: 2, SEQ ID NO:
62,
SEQ ID NO: 584, a peptide listed in Table 5.4, or a modified P28R or P28 core
peptide
comprising one or more modifications listed in Table 5.3 or Table 13 is
provided, and
further comprises a detectable moiety. The peptide comprising the detectable
moiety can
bind to one or more immunoregulatory peptides of Tables 1-4, for example P3028
(SEQ ID
NO: 185).
[0262] Additionally, human prostate cancer cells were cultured in the
absence of
serum proteins, and exhibited minimal immunostaining for P3028 strucutres,
based on
detection by rabbit antibodies (Figure 48A). However, when human prostate
cancer cells
were fed human serum albumin for 2 hours, and were stained for the presence of
P3028
structures using rabbit antibodies, substantial immunostaining was observed
(Figure 48B).
Accordingly it is contemplated that tumors can generate P3028 strucutres.
Moreover, it is
contemplated that immunoregulatory peptides inhibitors in accordance with some
embodiments herein can be administered to tumor cells to counteract
immunoinhibitory
effects of P3028 structures. In some embodiments, a composition comprising a
peptide of
SEQ ID NO: 2, SEQ ID NO: 62, SEQ ID NO: 584, a peptide listed in Table 5.4, or
a
modified P28R or P28 core peptide comprising one or more modifications listed
in Table 5.3
or Table 13, is administered to a tumor cell, and can bind to one or more
P3028 structures so
as to de-block an LFA-1 and/or IL-2 receptor and enhance immune cell
stimulation.
[0263] It was observed that compositions comprising immunoregulatory
peptide
inhibitors immobilized on nanoparticles in accordance with some embodiments
herein can
displace bound dHSA from immune cells. Magnetic DynabeadTM beads were bound to
P28
52
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
core peptide (FFVKLS)(SEQ ID NO: 62). As shown in Figure 49A, control PBMC's
cultured with dHSA exhibited substantial levels of bound dHSA. However, as
shown in
Figure 49B, PBMC's cultured with dHSA and incubated for 24 hours with the
DynabeadTM
bead-P28 core peptide composition exhibited substantially lower levels of
bound dHSA.
Accordingly, it is contemplated that compositions comprising immunoregulatory
peptide
inhibitors immobilized on nanoparticles in accordance with some embodiments
herein can be
useful for enhancing immune cell stimulation, for example by providing
immunoregulatory
peptide inhibitors to immune cells (e.g. lymphocyte, monocyte, macrophage, or
NK-cell)
bound to P3028. In some embodiments, a composition comprising a peptide of SEQ
ID NO:
2, SEQ ID NO: 62, SEQ ID NO: 584, a peptide listed in Table 5.4, or a modified
P28R or
P28 core peptide comprising one or more modifications listed in Table 5.3 or
Table 13 is
provided. The composition can comprise a nanoparticle, and the
immunoregulatory peptide
inhibitor (e.g. a peptide of SEQ ID NO: 62) can be immobilized on the
nanoparticle.
Optionally, the composition can be administered to a patient in need of immune
cell
stimulation. Optionally, stimulation of immune cells of the subject can be
detected, for
example, as enhanced expression of CD69 and/or CD71, secretion of IL-12 or
IFNy, or
secretion of perforin or granzyme B, enhanced cytotoxicity, cytokine
production, cell
migration, and/or cell proliferation. Optionally, the patient in need of
immune cell
stimulation is suffering from a tumor, for example a prostate tumor, a
melanoma, a colon
cancer, a lung carcinoma, an Apocrine gland carcinoma, a testis tumor, a mast
cell tumor, a
mammary tumor (e.g. a benign mammary tumor or a malignant mammary tumor, for
example
a mixed mammary tumor such as a benign mixed mammary tumor or a malignant
mixed
mammary tumor), a mucinous carcinoma (e.g. a mammary gland mucinous
carcinoma), or a
histicytoma. Optionally, administration of the composition to the patient
induces regressive
changes in the tumor, and/or eradicates or contributes to the eradication of
the tumor.
[0264] It has been observed that compositions comprising
immunoregulatory
peptide inhibitors in accordance with some embodiments herein can induce
immune cell
infiltration of tumors in mammal models. For example, administration of a
composition
comprising P28R (SEQ ID NO: 2) directly to a B16 melanoma in C57B1 mice
induced
regressive changes in the tumor and permeation of the tumour by CD45+
inflammatory cells,
53
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
and it was observed that a systemic immune activation against the tumor was
achieved (see
Figures 53A-53B). Moreover, contralateral tumors of the mouse that did not
receive the
direct administration of the P28R composition also underwent regressive
changes, and were
also infiltrated by immune cells (Figures 55A-D). It was further observed that
direct
injection of P28R composition into a Lewis lung carcinoma model in B7B 1 mice
induced
regressive changes in both the tumor that received the P28R composition, and
in contralateral
tumors that were not directly injected (Figures 57A and 57B). It was further
observed that
intra-tumoral administration of a composition comprising P28R to a breast
tumor of a dog
induced regressive changes in the tumor and infiltration of the tumor by CD45+
inflammatory cells (see Figure 58A-D). It was further observed that intra-
tumoral injection
of a composition comprising P28R to an apocrine gland carcinoma of a dog
induced
infiltration of the tumor by CD45+ inflammatory cells (Figure 62A-B) and NK
cells (e.g.
CD56+ cells and/or NCR1+ cells)(Figures 64a-B and 65A-B), and also induced
regressive
changed in the tumor. It was further observed that intra-tumoral injection of
a control
composition (vehicle that did not comprise P28R) to a testis tumor of a dog
did not induce
infiltration by immune cells (Figures 66A-B). It was further observed that
intra-tumoral
injection of a composition comprising P28R to a mast cell tumor of a dog
induced massive
tumor destruction and infiltration by CD45+ inflammatory cells (see Figure 69)
and
extensive infiltration by CD56+ inflammatory cells (see Figure 70A-D). It was
further
observed that intra-tumoral injection of a composition comprising P28R to a
benign mixed
mammary tumor of a dog induced regressive changes at the injection site
(Figure 71A-B)
and infiltration of the tumor by inflammatory cells (Figure 72A-D, Figure 73A-
D, and
Figure 74A-B), for example CD45+ cells (Figure 75A-D). Distant metastases of
the tumor
were also infiltrated by CD45+ cells (Figure 76 and Figure 77A-D). It was
further observed
that intra-tumoral injection of a composition comprising P28R to a mammary
gland mucous
carcinoma of a dog induced infiltration of the tumor by CD45+ inflammatory
cells (Figure
78A-B) and extensive regressive changes of the tumor. It was further observed
that intra-
tumoral injection of a composition comprising P28R to a histiocytoma of a dog
induced
extensive regressive changes of the tumor (Figure 79A-B) and infiltration of
the tumor by
CD56+ cells (Figure 80A) and NCR1+ cells (Figure 80B). It was further observed
that
54
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
intra-tumoral injection of a composition comprising P28R to a intraductal
papillary adenoma
of a dog induced intense inflammatory infiltration of the tumor (Figure 81 and
Figure 82A-
D), and extensive eradication of the tumor cells. It was further observed that
of CD3 (T
cells), CD8 (T cells), and CD45 (leukocytes) were down-regulated or lost when
inflammatory
cells infiltrated into the tumour-cell-areas, so that a number of lymphocytes
infiltrating tumor
cell areas appear to have lower staining intensity compared to the stroma (see
Figure 83).
The use or morphological criteria, however, confirmed that a number of
lymphocystes close
to the tumour cells were present, but unstained. The presence of lymphocytes
with reduced
CD3, CD8 and CD45 was futher confirmed using more intense immunostaining,
which
yielded increased background staining, but did not yield any more specific
staining (see
Figure 84). Tumor cells in dogs treated with P28R also exhibited faintly-
stained, often
irregularly-shaped nuclei and disruption of the nuclear membrane, and were
confirmed to be
apoptotic, as demonstrated by TUNELTm staining (Figure 85). Accordingly,
administration
of immunoregulatory peptide inhibitor comprising, consisting of, or consisting
essentially of
SEQ ID NO: 2, SEQ ID NO: 62, SEQ ID NO: 584, a peptide listed in Table 5.4, or
a
modified P28R or P28 core peptide comprising one or more modifications listed
in Table 5.3
or Table 13 to a subject with a tumor in accordance with some embodiments
herein can
induce apoptosis of the tumor cells. The ratio between normal and "damaged"
tumour cells
was observed, and treatment with P28R was confirmed to result in a
significantly lower
density of tumour cells in treated tumours (see Figure 86B) compared with
tumours from
untreated control dogs (see Figure 86A and Figures 87A-D). The inflammatory
infiltrate in
tissues of was further quantified and compared with total tumor cell number in
breast tumors
for both P28R-treated dogs and untreated dogs. It was observed that treated
tumours
contained more than a 3-fold higher ratio between inflammatory cells and
tumour cells
compared with untreated tumours (see Figure 88). In addition, as a control,
the inflammatory
infiltrate was evaluated in ten formalin fixed and paraffin embedded ("FFPE")
tumours, and
generally the infiltration of inflammatory cells was very low (see Figure 89).
Accordingly,
administration of immunoregulatory peptide inhibitor comprising, consisting
of, or consisting
essentially of SEQ ID NO: 2, SEQ ID NO: 62, SEQ ID NO: 584, a peptide listed
in Table
5.4, or a modified P28R or P28 core peptide comprising one or more
modifications listed in
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
Table 5.3 or Table 13 in accordance with some embodiments herein induces
infiltration of
tumour cells by inflammatory cells, and decreases the number of tumour cells
realative to
inflammatory cells.
[0265] The effects of P28R on tumors at locations other than the site
of
administration were evaluated. In P28R-treated dogs, tumours that were not
directly injected
with P28R exhibited regressive changes, providing strong evidence of treatment
of these
uninjected, remote, or peripheral tumours. Enhanced inflammatory infiltrate
and an enhanced
amount of "damaged" tumour cells were observed in large remote or peripheral
tumours after
injection of only 200 [IL of P28R, indicating a distant effect of P28R (see
Figures 91B and
91D). Accordingly, this experiment provides strong evidence that
administration of an
immunoregulatory peptide inhibitor comprising, consisting of, or consisting
essentially of
SEQ ID NO: 2, SEQ ID NO: 62, SEQ ID NO: 584, a peptide listed in Table 5.4,
P28R or a
modified P28R or P28 core peptide comprising one or more modifications listed
in Table 5.3
or Table 13 is effective for inhibiting, ameliorating, or treating cancer
cells and/or tumors
that are distant from a primary tumor, such as metastasis. In another
experiment, mice with
inoculate CT26 colon cancer were treated with 12 microgram P28R, twice weekly
for two
weeks. Apoptosis, identified using the TUNELTm staining technique, was induced
in the
majority of tumour cells (see Figures 92A-B). In another experiment, P28R was
administered subcutaneously twice weekely to two weeks to BALBc mice with
inoculate
CT26 colon cancer cells. Two dose levels, 4 (D10) or 12 mg (D30) per injection
were
compared with injection of the vehicle, and apoptotic tumour cells were
identified by staining
using the TUNELTm staining technique. The systemic administration of P28R
decreased the
number of viable tumour cells, and increased the number of apoptotic tumour
cells at both
D10 and D30 (see Table 16). Accordingly, both low and high doses of P28R,
administered
systemically in accordance with some embodiments herein (e.g. enterally, or
example orally;
or parenterally, for example subcutaneously, intravenously, intraperitoneally,
or via
implantation), systemically induces apoptosis in cancer cells and tumours. In
some
embodiments, an immunoregulatory peptide inhibitor as described herein (or a
composition
comrpsing the immunoregulatory peptide inhibitor) is administered intra-
tumorally or peri-
tumorally to a subset of tumors in a subject having multiple tumors (for
example, metastatic
56
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
cancer), so as to treat, inhibit, or ameliorate cancer cells, tumors, and/or
metastasis, even
those cancer cells and tumors that did not intra-tumorally or peri-tumorally
receive the
immunoregulatory peptide inhibitor. In some embodiments, an immunoregulatory
peptide
inhibitor as described herein is administered intra-tumorally or peri-
tumorally to one tumor in
a subject having metastatic cancer, but is not administered intra-tumorally or
peri-tumorally
to another tumor in the subject, so as to treat, ameliorate, destroy, and/or
eliminate both
tumors and/or cancer cells that originated from said primary tumor, such as in
the case of
metastasis.
[0266] Systemic effects of immunoregulatory peptide inhibitors were
also
observed in CT26 colon cancers in Balb/c mice. The Balb/c mice were injected
with an
oligoclonal rabbit antibody (oligoclonal antibody "R") against the human
albumin fragment
P3028. The oligoclonal rabbit antibody was 100 micrograms in 100 microliters,
or with the
same volume of saline as a control ("A"), and eradication of the tumor cells
was observed in
the antibody-injected mice after five days. The number of tumour cells was
substantially
reduced in the oligoclonal antibody-injected mice (see Figure 93B) compared to
saline
controls (Figure 93A). As shown in Figure 93B, and summarized numerically in
Table 17,
the tumour cell density is reduced by antibody treatment. Thus, an abscopal
and/or systemic
effect in an uninjected tumour is observed in a mouse treated with an
immunoregulatory
peptide inhibitor. As such, systemic effects of immunoregulatory peptide
inhibitors are
contemplated in accordance with some embodiments herein. In some embodiments,
an
immunoregulatory peptide inhibitor is administered intratumorally to a tumor,
or
peritumorally to a tumor in a subject having multiple tumors, so as to
ameliorate, inhibit or
eliminate at least one tumor that did not intratumorally or peritumorally
receive the
immunoregulatory peptide inhibitor. In some embodiments, the immunoregulatory
peptide
inhibitor comprises an antibody against any of the peptides of SEQ ID NOs: 183-
185 or 188-
246, for example P3028 (SEQ ID NO: 185). Optionally, the antibody binds
specifically to
P3028 (SEQ ID NO: 185). Optionally, the antibody comprises a polyclonal
antibody.
Optionally, the antibody comprises an oliogoclonal antibody. Optionally, the
antibody
comprises a monoclonal antibody. Optionally, the antibody comprises a full-
length
monoclonal antibody. Optionally, the antibody comprises a binding fragment of
a
57
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
monoclonal antibody. In some embodiments, immunoregulatory peptide inhibitor
induces
regressive changes in the tumor(s) that did not intratumorally or
peritumorally receive the
immunoregulatory peptide inhibitor. In some embodiments, immunoregulatory
peptide
inhibitor induces immune cell infiltration in the tumor(s) that did not
intratumorally or
peritumorally receive the immunoregulatory peptide inhibitor.
[0267] is contemplated that compositions comprising immunoregulatory
peptide
inhibitors as described herein can treat, ameliorate, eliminate, inhibit,
and/or eradicate
multiple tumors or cancer cells in a subject, even if the compositions are not
directly
administered intratumorally or peri-tumorally to each of the tumors or cancer
cells.
Moreover, it is contemplated that compositions comprising immunoregulatory
peptide
inhibitors as described herein can treat, ameliorate, inhibit, eliminate,
and/or eradicate
metstatic cancer without being intratumorally or peri-tumorally administered
to each any
every tumor of the metastatic cancer. As such, in some embodiments a
composition
comprising an immunoregulatory peptide inhibitor comprising, consisting of, or
consisting
essentially of SEQ ID NO: 2, SEQ ID NO: 62, SEQ ID NO: 584, a peptide listed
in Table
5.4, P28R, or a modified P28R or P28 core peptide comprising one or more
modifications
listed in Table 5.3 or Table 13 is administred systemically (e.g., enterally,
or example orally;
or parenterally, for example subcutaneously, intravenously, intraperitoneally,
or via
implantation), and induces apoptosis of tumour and/or cancer cells throughout
the subject
including in a subject having metastasis. In some embodiments, the composition
comprising
the immunoregulatory peptide inhibitor is administered intratumorally or
peritumorally to
some, but not all, of the tumors and/or cancer cells in a metatstatic cancer
in a subject having
metastasis, for example at least 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13,
14, 15, 16, 17, 18, 19,
or 20 tumors of the subject (including ranges between any two fo the listed
values), for use in
treating, inhibiting, or ameliorating the metatstatic tumors and/or cancer
cells of the subject
(e.g., the composition is for treating or inhibiting at least one more tumor
than the number of
tumors that receives an intratumoral or peritumoral administration of the
aforementioned
compositions).
[0268] Accordingly, it is contemplated that in some embodiments, a
composition
comprising an immunoregulatory peptide inhibitor of SEQ ID NO: 2, SEQ ID NO:
62, SEQ
58
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
ID NO: 584, a peptide listed in Table 5.4, or a modified P28R or P28 core
peptide
comprising one or more modifications listed in Table 5.3 or Table 13 is
provided. The
composition can comprise a nanoparticle, and the immunoregulatory peptide
inhibitor (e.g.
SEQ ID NO: 2, SEQ ID NO: 62, SEQ ID NO: 584, a peptide listed in Table 5.4, or
a
modified P28R or P28 core peptide comprising one or more modifications listed
in Table 5.3
or Table 13) can be immobilized on the nanoparticle. Optionally, the
composition is for use
in direct administration to a tumor, or is directly administered to a tumor.
Optionally, the
tumor can be part of a metastatic cancer. Optionally, the composition induces
regressive
changes of the tumor. Optionally, the composition induces eradication of some
or all of the
tumor or inhibits proliferation of tumor cells and/or metastasis. Optionally,
the composition
is for use in treating, ameliorating, inducing regressive changes in, inducing
eradiation of
some of, or inducing eradication of all of a prostate tumor, a melanoma, a
colon cancer, a
lung carcinoma, an Apocrine gland carcinoma, a testis tumor, a mast cell
tumor, a mammary
tumor (e.g. a benign mammary tumor or a malignant mammary tumor, for example a
mixed
mammary tumor such as a benign mixed mammary tumor or a malignant mixed
mammary
tumor), a mucinous carcinoma (e.g. a mammary gland mucinous carcinoma), a
histicytoma,
or an adenoma (e.g. an intraductal papillary adenoma) or inhibits
proliferation of cells and/or
metastasis associated with the aforementioned cancers. As such, the
composition can be
administered to a subject having a cancer or tumor or at subject at risk for
metastasis, for
example prostate tumor, a melanoma, a colon cancer, a lung carcinoma, an
Apocrine gland
carcinoma, a testis tumor, a mast cell tumor, a mammary tumor (e.g. a benign
mammary
tumor or a malignant mammary tumor, for example a mixed mammary tumor such as
a
benign mixed mammary tumor or a malignant mixed mammary tumor), a mucinous
carcinoma (e.g. a mammary gland mucinous carcinoma), a histicytoma, or an
adenoma (e.g.
an intraductal papillary adenoma). The subject can be in need of treatment of
the cancer or
tumor. Optionally, the composition induces immune cell infiltration of the
tumor to which
the composition was directly administered, for example infiltration by CD45+
and/or NK
cells. Optionally, the composition induces immune cell infiltration of a tumor
of the subject
that it was not directly administered, such as a metastatic tumor or a
contralateral tumor (e.g.
a second, metastatic tumor and/or contralateral tumor, if the composition is
directly
59
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
administered to a first tumor). Optionally, the composition induces a systemic
immune
response.
Ameliorating immunosuppression
[0269] As the inhibitors of immunoregulatory peptides described herein
can be
useful for removing immunosuppression, some embodiments herein comprise
methods of
ameliorating, reducing the symptoms of, reducing, or treating
immunosuppression. In some
embodiments a subject suffering from immunosuppression is identified. The
subject can
comprise a human, or a non-human mammal. A composition comprising at least one
of the
inhibitors of immunoregulatory peptides described herein can be administered
to the patient.
The composition can comprise at least one peptide comprising, consisting of,
or consisting
essentially of any one of SEQ ID NOs: 1-33, 34, 46-53, 62, 64-66, 68, 76, 94-
96 ,98, 265-
393, 583-586, 587-595, or a modified P28R or P28 core peptide comprising one
or more of
the modifications of Table 5.3 or Table 13. The peptide can have length is
less than or equal
to 1100 amino acids, for example, less than or equal to 4, 5, 6, 7, 8, 9, 10,
11, 12, 13, 14, 15,
16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34,
35, 36, 37, 38, 39, 40,
41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59,
60, 61, 62, 63, 64, 65,
66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84,
85, 86, 87, 88, 89, 90,
91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105, 106, 107,
108, 109, 110, 111,
112, 113, 114, 115, 116, 117, 118, 119, 120, 121, 122, 123, 124, 125, 126,
127, 128, 129,
130, 131, 132, 133, 134, 135, 136, 137, 138, 139, 140, 150, 160, 170, 180,
190, 200, 210,
220, 230, 240, 250, 260, 270, 280, 290, 300, 320, 340, 360, 380, 400, 450,
500, 550, 600,
650, 700, 750, 800, 850, 900, 950, 1000, 1050, or 1100 amino acids or a length
defined by a
range between any two of these numbers. Optionally, the composition can
further comprise a
buffer as described herein, for example, Trizma, Bicine, Tricine, MOPS, MOPSO,
MOBS,
Tris, Hepes, HEPBS, MES, phosphate, carbonate, acetate, citrate, glycolate,
lactate, borate,
ACES, ADA, tartrate, AMP, AMPD, AMPSO, BES, CABS, cacodylate, CHES, DIPSO,
EPPS, ethanolamine, glycine, HEPPSO, imidazole, imidazolelactic acid, PIPES,
SSC, SSPE,
POPSO, TAPS, TABS, TAPSO or TES. Optionally, the composition can further
comprise a
degradable particle as described herein. The composition can be administered
to the subject
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
via a variety of routes, for example, systemically, at the site of
immunosuppression (e.g. if
there is local immunosuppression by a tumor), or near the site of
immunosuppression, for
example within 10cm 9cm, 8cm, 7cm, 6cm, 5cm, 4cm, 3cm, 2cm, lcm, or 0.5cm of
the site
of immunosuppression. Optionally a second therapeutic agent can be
administered in
addition to the composition, for example prior to, concurrently with, or
subsequent to the
administration of the composition. For example, the second therapeutic agent
can comprise
an immunostimulatory agent. Optionally, activation of immune cells (e.g.
enhanced
expression of CD69 and/or CD71, secretion of IL-12 or IFNy, or secretion of
perforin or
granzyme B, enhanced cytotoxicity, cytokine production, cell migration, and/or
cell
proliferation) of the subject can be detected. For example, activation of
immune cells can be
detected as enhanced expression of one or more markers of immune cells, for
example CD69,
CD71, and the like. Activation of immune cells (e.g. enhanced expression of
CD69 and/or
CD71, secretion of IL-12 or IFNy, or secretion of perforin or granzyme B,
enhanced
cytotoxicity, cytokine production, cell migration, and/or cell proliferation)
can be detected by
a number of techniques known to the skilled artisan, for example flow
cytometry,
immunohistochemistry, ELISA, western blotting, immunoblotting, quantitative
PCR,
detection of BUdR incorporation to measure proliferation, and the like.
Without being
limited by any theory, different types of immunosuppressor cells, regulatory T-
cells,
immature dendritic cells (iDC), tumor associated macrophages (TAM) and myeloid
derived
suppressor cells (MDSC), can function immunosuppression, and further, other
immunosuppressor mechanisms, such as serum blocking factors, circulating
immune
complexes, enhanced IL-1Ra production and enhanced intra-tumoral proteolytic
activity can
function in cancer related immunosuppression. As such, in some embodiments,
treatment,
amelioration, reduction, or reduction of the symptoms of immunosuppression can
be
determined by a change in activity, phenotype, or proliferation of an
immunosuppressive cell,
or a change in expression level or localization of an immunosuppressive
factor.
Inhibitors of Immunoregulatory Peptides
[0270] Some embodiments include inhibitors of immunoregulatory
peptides such
as P3028 and/or one or more of the immunoregulatory peptides listed in Tables
1-4 (SEQ ID
61
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
NOs: 183-184, and 188-246), also referred to as blockers of albumin derived
immunoregulatory peptides, binding partners for immunoregulatory peptides, or
immunoregulatory peptide inhibitors. The immunoregulatory peptide inhibitors
can include,
but are not limited to: peptides, cyclic peptides, peptidomimetics, proteins,
nucleic acids,
antibodies; antibody fragments, nucleic acid aptamers; peptide aptamers; and
small
molecules. The following section provides more details on antibody or antibody
fragment-
based immunoregulatory peptide inhibitors.
Antibody or antibody fragment-based immunoregulatory peptide inhibitors
[0271] Some embodiments include antibody or antibody fragment based
immunoregulatory peptide inhibitors. Methods that use these immunoregulatory
peptide
inhibitors to inhibit immunosuppression in a subject (e.g., a subject having
cancer or a
pathogenic infection such as a bacterial or viral infection) are also
contemplated. The core
antibody structural unit is known to comprise a tetramer. Each tetramer is
composed of two
identical pairs of polypeptide chains, each pair having one "light" chain
(about 25 kDa) and
one "heavy" chain (about 50-70 kDa). The amino-terminal portion of each chain
includes a
variable region of about 100 to 110 or more amino acids primarily responsible
for antigen
recognition. The carboxy-terminal portion of each chain defines a constant
region primarily
responsible for effector function. Heavy chains are classified as mu, delta,
gamma, alpha, or
epsilon, and define the antibody's isotype as IgM, IgD, IgG, IgA, and IgE,
respectively. An
additional isotope, IgY is found in avian hosts. The chains all exhibit the
same general
structure of relatively conserved framework regions (FR) joined by three hyper
variable
regions, also called complementarity determining regions or CDRs. The CDRs
from the two
chains of each pair are aligned by the framework regions, enabling binding to
a specific
epitope. From N-terminal to C-terminal, both light and heavy chains comprise
the domains
FR1, CDR1, FR2, CDR2, FR3, CDR3 and FR4. The assignment of amino acids to each
domain is in accordance with the definitions of Kabat, Sequences of Proteins
of
Immunological Interest (National Institutes of Health, Bethesda, Md. (1987 and
1991)), or
Chothia & Lesk J. Mol. Biol. 196:901-917 (1987); Chothia et al., Nature
342:878-883
(1989).
62
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
[0272] Accordingly, some embodiments include a composition that
comprises,
consists of, or consists essentially of an immunoregulatory peptide inhibitor
that comprises an
antibody or antibody fragment comprising a domain, which binds to one or more
regions of
an immunoregulatory peptide, such as P3028 or one or more of the
immunoregulatory
peptides provided in Tables 1-4 (SEQ ID NOs: 183-184 and 188-246). In some
embodiments, the antibody or antibody fragment is from a mouse, rabbit, rat,
hamster, guinea
pig, goat, donkey, bovine, horse, camel, cow, chicken, or human host. In some
embodiments,
the antibody or fragment is of isotype IgG, IgM, IgA, IgD, IgE, or IgY. In
some
embodiments, the antibody or fragment is part of a collection of polyclonal
antibodies. In
some embodiments, the antibody is monoclonal. In some embodiments, the
antibody or
fragment is chimeric. In some embodiments, the antibody or fragment includes
at least one
region form a human host, which can be at least one of the following Fc; Fab;
light chain
variable region; light chain CDR1, CDR2, or CDR3; heavy chain variable region;
heavy
chain CDR1, CDR2, or CDR3; light chain framework region; light chain FR1, FR2,
FR3, or
FR4; heavy chain framework region; heavy chain FR1, FR2, FR3, or FR4. In some
embodiments, the antibody includes at least one CDR or FR of a non-human host.
In some
embodiments, the antibody regions are in accordance with the definition of
Kabat. In some
embodiments, the antibody regions are in accordance with the definition of
Chothia. In some
embodiments, the antibody regions are in accordance with a combination of the
definition of
Kabat and Chothia. In some embodiments, the antibody or antibody fragment
mimics one or
more of the peptides described in Table 5.1, Table 5.4, Table 5.5, or Table
5.6.
[0273] Antibodies can be readily produced using conventional
techniques in
immunology, for example techniques described in US Pat Nos (8,142,784 and
7,628,986).
Antibodies generated in non-human hosts can be humanized, for example by
substituting at
least one variable region of the antibody of the non-human host into a human
antibody.
Moreover, human antibodies can be generated, for example in a transgenic host
animal.
Transgenic animals (e.g., mouse, such as XENOMOUSE) can be engineered, upon
immunization, to produce a full repertoire of human antibodies in the absence
of endogenous
immunoglobulin production (Jakobovits et al. (1993) Proc. Natl. Acad. Sci.
USA, 90:2551;
Jakobovits et al. (1993) Nature 362:255-258; Bruggermann et al. (1993) Year in
Immuno.
63
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
7:33; and U.S. Pat. No. 5,591,669; U.S. Pat. No. 5,589,369; U.S. Pat. No.
5,545,807).
Moreover, phage display technology (McCafferty et al. (1990) Nature 348:552-
553) can be
used to produce human antibodies and antibody fragments in vitro, from
immunoglobulin
variable (V) domain gene repertoires from unimmunized donors (Johnson, Kevin
S. and
Chiswell, David J. (1993) Current Opinion in Structural Biology 3:564-571). A
repertoire of
V genes from unimmunized human donors can be constructed and antibodies to a
diverse
array of antigens (including self-antigens) can be isolated essentially (Marks
et al. (1991) J.
Mol. Biol. 222:581-597; Griffith et al. (1993) EMBO J. 12:725-734; U.S. Pat.
No. 5,565,332;
U.S. Pat. No. 5,573,905). Many phage display libraries are known, or can be
generated, for
example those of (US Pat. No. 7,985,840). Human antibodies may also be
generated by in
vitro activated B cells (U.S. Pat. No. 5,567,610; U.S. Pat. No. 5,229,275).
Thus, some
embodiments include generating antibodies that bind to P3028 (SEQ ID NO: 185)
and/or the
peptides of Tables 1-4 (SEQ ID NOs: 183-184 and 188-246). In some embodiments,
the
antibodies are humanized antibodies that include at least one variable region
of a non-human
host antibody. In some embodiments, the antibodies are human antibodies
generated in a
non-human host, for example a transgenic animal. In some embodiments, the
transgenic
animal is a transgenic mouse. In some embodiments, the antibodies are
generated in vitro. In
some embodiments, the antibodies are generated using phage display technology.
In some
embodiments, the antibodies are generated in activated B cells in vitro.
[0274] Antibodies and antibody fragments can be configured to deliver
cytotoxic
compounds to a target site. Thus, some embodiments include antibodies and/or
antibody
fragments bound to cytotoxic compounds as described herein. In some
embodiments, the
antibodies or antibody fragments are bound to the cytotoxic compounds via a
cleavable linker
as described herein.
[0275] Some embodiments include a composition that comprises, consists
of, or
consists essentially of an immunoregulatory peptide inhibitor that comprises
antibodies or a
binding fragment thereof, which specifically binds to P3028 (SEQ ID NO: 185).
Some
embodiments include antibodies or fragments thereof, which specifically bind
to a fragment
of P3028 (SEQ ID NOs: 186 and 187). Exemplary antibodies that bind to P3028
are
described in Example 9.
64
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
[0276] In some embodiments, the antibody or fragment thereof described
above
can be used to inhibit or sequester P3028. In some embodiments, the antibody
or fragment
thereof specific for P3028 can be administered to a patient having at least
one immune cell
bound to P3028 so as to unblock at least one of the patient's LFA-1 or IL-2
receptors. In
some embodiments, the antibody or fragment thereof can be administered to a
patient in need
of treatment immunosuppression, as described herein, thereby stimulating or
enhancing an
immune response of said patient. For example, the antibody or fragment thereof
can be
provided to a patient in need of an inhibition of immunosuppression (e.g., a
subject that has
cancer or a pathogenic infection such as a bacterial or viral infection).
After providing the
antibody or fragment thereof the patient can be evaluated for an inhibition of
immunosupression, which can be accomplished by determining immune cell
infiltration of a
tumor or a reduction in a bacterial or viral infection, for example, or an
improved immune
response by the PBMCs of said subject.
[0277] In other embodiments, the antibody or fragment thereof can be
used to
detect the presence of P3028, for example, in a biological sample. The
antibody or fragment
thereof can be used to detect the formation of a complex, for example when an
immunoregulatory peptide inhibitor (e.g., a peptide SEQ ID NOs: 1-33, 34, 46-
53, 64-66,
68, 76, 94-96, 98 or 264-393) is attached to a support, and the antibody is
used as a primary
antibody or fragment thereof is used to detect the presence of P3028 bound to
the inhibitor.
[0278] Some embodiments include an antibody or fragment thereof that
specifically binds to an immunoregulatory peptide inhibitor of P3028 (e.g., an
antibody or
fragment thereof that mimics or has at least 70%, 75%, 80%, 85%, 90%, 95%, or
98%
identity to one or more of the peptides of Table 5.1). The antibody or
fragment thereof can
specifically bind to a peptide that includes at least one of SEQ ID NOs: 1-33,
34, 46-53, 64-
66, 68, 76, 94-96, 98 or 264-393. In some embodiments, the antibody or
fragment thereof
specific for an immunoregulatory peptide inhibitor of P3028 can be used to
detect the
presence of an immunoregulatory peptide inhibitor of P3028 in a biological
sample. The
antibody or fragment thereof specific for an immunoregulatory peptide
inhibitor of P3028 can
also be used to detect the formation of a complex, for example, if P3028 is
attached to a
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
support, and the antibody or fragment thereof is used as a primary antibody to
detect the
presence of an immunoregulatory peptide inhibitor bound to P3028.
[0279] In some embodiments, the antibody or fragment thereof specific
for an
immunoregulatory peptide inhibitor of P3028 can be used to isolate or identify
the presence
of an inhibitor of P3028. For example, the antibody or fragment thereof can be
used to purify
an inhibitor to be used for stimulating an immune cell of a human, and/or for
binding to the
cancer cell of a human. For example, the antibody or fragment thereof, such as
a binding
fragment, can be used to purify an inhibitor to be used for stimulating an
immune cell of a
non-human mammal, and/or for binding to the cancer cell of a non-human mammal.
[0280] In some embodiments, the antibody or fragment thereof specific
for an
immunoregulatory peptide inhibitor of P3028 can be used to detect the presence
of P3028.
For example, the antibody or fragment thereof specific for an immunoregulatory
peptide
inhibitor of P3028 can be used for immunohistochemical staining of a
biological sample to
detect the presence of a cancer cell that has been contacted with an
immunoregulatory peptide
inhibitor. For example, the antibody specific for an immunoregulatory peptide
inhibitor of
P3028 can be used in flow cytometry to detect and/or isolate immune or cancer
cells that are
bound to an immunoregulatory peptide inhibitor. The following section provides
more
details on peptide-based immunoregulatory peptide inhibitors.
Peptide-based immunoregulatory peptide inhibitors
[0281] In some embodiments, an isolated peptide that comprises a
domain, which
binds to one or more regions of an immunoregulatory peptide, such as P3028, is
provided.
The term "isolated" requires that the material be removed from its original
environment (e.g.,
the natural environment if it is naturally occurring). For example, a
naturally-occurring
polynucleotide present in a living animal is not isolated, but the same
polynucleotide,
separated from some or all of the coexisting materials in the natural system,
is isolated. It is
also advantageous that the sequences be in purified form. The term "purified"
does not
require absolute purity; rather, it is intended as a relative definition.
Isolated proteins have
been conventionally purified to electrophoretic homogeneity by Coomassie
staining, for
example. Purification of starting material or natural material to at least one
order of
66
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
magnitude, preferably two or three orders, and more preferably four or five
orders of
magnitude is expressly contemplated. An isolated peptide can exist, for
example, in a
substantially salt form, crystal form, lyophilized form, in solution (for
example aqueous
solution which can include buffer), and/or in a pharmaceutically carrier or
diluent. An
isolated peptide can exist in a substantially pure form, for example a
composition that
includes at least or equal to about 1% of the peptide by weight, for example
at least or equal
to about 1%, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19,
20, 21, 22, 23, 24, 25,
26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44,
45, 46, 47, 48, 49, 50,
51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69,
70, 71, 72, 73, 74, 75,
76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94,
95, 96, 97, 98, 98.5,
99, 99.5, 99.9, 99.99, or 99.999% peptide by weight.
[0282] In some embodiments, the isolated immunoregulatory peptide
inhibitors
described herein (e.g., a peptide comprising, consisting of, or consisting
essentially of any
one of SEQ ID NOs: 1-33, 34, 46-53, 62, 64-66, 68, 76, 94-96 ,98, 265-393, 583-
586, 587-
595, or a modified P28R or P28 core peptide comprising one or more of the
modifications of
Table 5.3 or Table 13 have lengths that are less than or equal to 1100 amino
acids, for
example, less than or equal to 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16,
17, 18, 19, 20, 21, 22,
23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41,
42, 43, 44, 45, 46, 47,
48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66,
67, 68, 69, 70, 71, 72,
73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91,
92, 93, 94, 95, 96, 97,
98, 99, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113,
114, 115, 116,
117, 118, 119, 120, 121, 122, 123, 124, 125, 126, 127, 128, 129, 130, 131,
132, 133, 134,
135, 136, 137, 138, 139, 140, 150, 160, 170, 180, 190, 200, 210, 220, 230,
240, 250, 260,
270, 280, 290, 300, 320, 340, 360, 380, 400, 450, 500, 550, 600, 650, 700,
750, 800, 850,
900, 950, 1000, 1050, or 1100 amino acids, including ranges between any two of
the listed
values. For example, an immunoregulatory peptide inhibitor consisting of the
sequence
(FVKL) can bind to P3028 with a comparable rampo score to immunoregulatory
peptide
inhibitors, which comprise FVKL, that are 6 to 16 amino acids in length (see
Figure 29 and
Example 12). Additionally, amino acids sequences near an N terminal, C
terminal, or
exposed loop of a peptide are more likely to be accessible to potential
binding targets rather
67
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
than incorporated into a higher-order peptide structure, thus permitting a
peptide of 1100
amino acids or less to bind P3028. Therefore, some embodiments of the
invention concern
compositions and methods of use thereof (e.g., a method of binding P3028 or a
method of
reducing P3028-mediated immunosuppression), which comprise, consist of, or
consist
essentially of any one or more of immunoregulatory peptide inhibitors
described herein (e.g.,
any one or more of the peptides provided in Table 5.1, 5.4, 5.5, or 5.6).
Desirably these
peptides (e.g., any one or more of the peptides of Table 5.1, 5.4, 5.5, or
5.6) have lengths
that are less than or equal to 1100 amino acids, for example, less than or
equal to 4, 5, 6, 7, 8,
9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28,
29, 30, 31, 32, 33,
34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52,
53, 54, 55, 56, 57, 58,
59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77,
78, 79, 80, 81, 82, 83,
84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102,
103, 104, 105,
106, 107, 108, 109, 110, 111, 112, 113, 114, 115, 116, 117, 118, 119, 120,
121, 122, 123,
124, 125, 126, 127, 128, 129, 130, 131, 132, 133, 134, 135, 136, 137, 138,
139, 140, 150,
160, 170, 180, 190, 200, 210, 220, 230, 240, 250, 260, 270, 280, 290, 300,
320, 340, 360,
380, 400, 450, 500, 550, 600, 650, 700, 750, 800, 850, 900, 950, 1000, 1050,
or 1100 amino
acids, including ranges between any two of the listed values.
Table 5.1: Sequences and Corresponding Rampo Scores
RAM P0 10 KKLDTFSVKLSLFTER 700
SEQ ID Sequence Score 369 KKLDTFFVKLSLPTER 696
367 KKLDTFFVKLSLMTER 1190 26 KKLDTFFVKLSLPTER 696
22 KKLDTFFVKLSLMTER 1190 343 KKLDTFFVKVSLFTER 658
370 KKLDTFFVKLSLQTER 1144 14 KKLDTFFVKVSLFTER 658
23 KKLDTFFVKLSLQTER 1144 355 KKLDTFFVKLSQFTER 651
364 KKLDTFFVKLSLHTER 1046 19 KKLDTFFVKLSQFTER 651
24 KKLDTFFVKLSLHTER 1046 372 KKLDTFFVKLSLSTER 635
368 KKLDTFFVKLSLNTER 862 27 KKLDTFFVKLSLSTER 635
25 KKLDTFFVKLSLNTER 862 382 KKLDTFFVKLSLFNER 599
348 KKLDTFFVKLQLFTER 768 31 KKLDTFFVKLSLFNER 599
15 KKLDTFFVKLQLFTER 768 313 KKLDTAFVKLSLFTER 575
346 KKLDTFFVKLMLFTER 744 7 KKLDTAFVKLSLFTER 575
16 KKLDTFFVKLMLFTER 744 287 KKGDTFFVKLSLFTER 563
321 KKLDTFMVKLSLFTER 712 94 KKGDTFFVKLSLFTER 563
9 KKLDTFMVKLSLFTER 712 4 KKGDTFFVKLSLFTER 563
323 KKLDTFSVKLSLFTER 700 383 KKLDTFFVKLSLFPER 551
68
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
32 KKLDTF FVKLSLFP ER 551 268 AKLDTFFVKLSLFTER 466
319 KKLDTVFVKLSLFTER 547 378 KKLDTFFVKLSLFH ER 463
8 KKLDTVFVKLSLFTER 547 354 KKLDTFFVKLSNFTER 462
359 KKLDTFFVKLSVFTER 545 350 KKLDTFFVKLSAFTER 462
20 KKLDTFFVKLSVFTER 545 396 KKLDTFFVKLSLFTER 460
345 KKLDTFFVKLHLFTER 535 351 KKLDTFFVKLSHFTER 460
308 KKLDQFFVKLSLFTER 535 336 KKLDTFFVKMSLFTER 460
18 KKLDTFFVKLHLFTER 535 291 KKMDTFFVKLSLFTER 460
6 KKLDQFFVKLSLFTER 535 310 KKLDSFFVKLSLFTER 458
363 KKLDTFFVKLSLGTER 531 275 MKLDTFFVKLSLFTER 457
100 KKLDTFFVKLSLGTER 531 352 KKLDTFFVKLSIFTER 456
28 KKLDTFFVKLSLGTER 531 329 KKLDTFFPKLSLFTER 456
285 KKEDTFFVKLSLFTER 528 278 QKLDTFFVKLSLFTER 455
KKEDTFFVKLSLFTER 528 289 KKI DTFFVKLSLFTER 454
325 KKLDTFVVKLSLFTER 527 347 KKLDTFFVKLNLFTER 451
11 KKLDTFVVKLSLFTER 527 296 KKTDTFFVKLSLFTER 451
361 KKLDTFFVKLSLATER 525 304 KKLDCFFVKLSLFTER 449
29 KKLDTFFVKLSLATER 525 274 LKLDTFFVKLSLFTER 449
279 RKLDTFFVKLSLFTER 523 366 KKLDTFFVKLSLLTER 448
3 RKLDTFFVKLSLFTER 523 397 KKLDTFIVKLSLFTER 446
349 KKLDTFFVKLTLFTER 520 374 KKLDTFFVKLSLVTER 446
17 KKLDTFFVKLTLFTER 520 316 KKLDTNFVKLSLFTER 446
324 KKLDTFTVKLSLFTER 517 398 KKLDTFFVKLSLFTER 445
320 KKLDTFLVKLSLFTER 517 276 NKLDTFFVKLSLFTER 445
13 KKLDTFLVKLSLFTER 517 302 KKLWTFFVKLSLFTER 443
12 KKLDTFTVKLSLFTER 517 399 KKLDTFFVKLSLFTER 442
322 KKLDTFQVKLSLFTER 511 281 VKLDTFFVKLSLFTER 442
371 KKLDTFFVKLSLRTER 502 340 KKLDTFFVKRSLFTER 439
30 KKLDTFFVKLSLRTER 502 400 KKLDTFFVKLSLFTER 437
381 KKLDTFFVKLSLFMER 501 358 KKLDTFFVKLSTFTER 437
353 KKLDTFFVKLSMFTER 499 338 KKLDTFFVKPSLFTER 436
21 KKLDTFFVKLSMFTER 499 306 KKLDNFFVKLSLFTER 436
317 KKLDTPFVKLSLFTER 497 401 KKLDTSFVKLSLFTER 432
334 KKLDTFFVKGSLFTER 495 402 KNLDTFFVKLSLFTER 432
373 KKLDTFFVKLSLTTER 494 283 KKCDTFFVKLSLFTER 432
298 KKLATFFVKLSLFTER 494 375 KKLDTFFVKLSLWTER 430
280 TKLDTFFVKLSLFTER 493 309 KKLDRFFVKLSLFTER 430
284 KKDDTFFVKLSLFTER 492 300 KKLITFFVKLSLFTER 430
356 KKLDTFFVKLSRFTER 483 403 KKLDTFFVKLSLFTER 428
273 IKLDTFFVKLSLFTER 483 272 HKLDTFFVKLSLFTER 428
318 KKLDTTFVKLSLFTER 481 307 KKLDPFFVKLSLFTER 427
357 KKLDTFFVKLSSFTER 478 282 KKADTFFVKLSLFTER 427
288 KKHDTFFVKLSLFTER 477 404 KKLDTFAVKLSLFTER 426
305 KKLDMFFVKLSLFTER 475 332 KKLDTFFVKASLFTER 426
293 KKQDTFFVKLSLFTER 473 405 KPLDTFFVKLSLFTER 425
339 KKLDTFFVKQSLFTER 470 312 KKLDYFFVKLSLFTER 425
365 KKLDTFFVKLSLITER 468 406 KKLDTFFVKLSLFTER 424
315 KKLDTMFVKLSLFTER 467 303 KKLYTFFVKLSLFTER 422
314 KKLDTIFVKLSLFTER 466 311 KKLDW FFVKLSLFTER 418
69
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
407 KRLDTFFVKLSLFTER 417 432 KKLDTFRVKLSLFTER 369
299 KKLETFFVKLSLFTER 417 384 KKLDTFFVKLSLFSER 369
335 KKLDTFFVKISLFTER 415 271 GKLDTFFVKLSLFTER 367
408 KKLDTFFVKLSLFTER 414 93 GKLDTFFVKLSLFTER 367
409 KKLDTFCVKLSLFTER 411 391 KKLDTFFVKLSLFTER 366
328 KKLDTFFLKLSLFTER 411 337 KKLDTFFVKNSLFTER 365
410 KKLDTQFVKLSLFTER 410 330 KKLDTFFRKLSLFTER 365
360 KKLDTFFVKLSW FTER 409 433 KKLDTFHVKLSLFTER 364
411 KKLDTLFVKLSLFTER 408 434 KKLDTYFVKLSLFTER 364
412 KG LDTFFVKLSLFTER 405 435 KKLPTFFVKLSLFTER 364
413 KKLTTFFVKLSLFTER 405 436 KKPDTFFVKLSLFTER 361
387 KKLDTFFVKLSLFTDR 404 380 KKLDTFFVKLSLFLER 360
333 KKLDTFFVKFSLFTER 403 326 KKLDTFFFKLSLFTER 358
414 KKLDTFFVKLSLFTER 402 437 KKLDTFPVKLSLFTER 356
415 KKLDTFFVKLYLFTER 402 438 KKLDTFFVKLSKFTER 355
416 KKLDTFFIKLSLFTER 401 439 KKLDTFFVKLSLFTPR 351
417 KMLDTFFVKLSLFTER 400 341 KKLDTFFVKSSLFTER 351
362 KKLDTFFVKLSLCTER 400 440 KQLDTFFVKLSLFTER 350
342 KKLDTFFVKTSLFTER 399 441 KELDTFFVKLSLFTER 349
270 EKLDTFFVKLSLFTER 396 442 KKLDTFFVKLSLFTER 348
418 KHLDTFFVKLSLFTER 394 443 KKLDTFNVKLSLFTER 348
295 KKSDTFFVKLSLFTER 393 444 KKLDTW FVKLSLFTER 348
286 KKFDTFFVKLSLFTER 393 376 KKLDTFFVKLSLF FE R 348
419 KKLDTFFVKLVLFTER 392 445 KKLDTFFVTLSLFTER 347
420 KKLDHFFVKLSLFTER 391 446 KKLDTG FVKLSLFTER 347
421 KFLDTFFVKLSLFTER 390 96 KKLDTFGVKLSLFTER 347
422 KKLDTFFVKLSFFTER 389 447 KKLDAFFVKLSLFTER 346
277 PKLDTFFVKLSLFTER 387 448 KKLQTFFVKLSLFTER 345
290 KKKDTFFVKLSLFTER 386 449 KKLCTFFVKLSLFTER 344
95 KKLDGFFVKLSLFTER 386 450 KKLDTFFVKLSLFTQR 344
423 KKLMTFFVKLSLFTER 384 451 KKLSTFFVKLSLFTER 344
344 KKLDTFFVKYSLFTER 382 452 KKYDTFFVKLSLFTER 344
424 KKLDTFEVKLSLFTER 381 453 SKLDTFFVKLSLFTER 344
425 KKLDTFWVKLSLFTER 380 454 KLLDTFFVKLSLFTER 343
426 KKLFTFFVKLSLFTER 380 377 KKLDTFFVKLSLFG ER 343
385 KKLDTFFVKLSLFVER 380 455 KKLDTFFVKLSCFTER 342
327 KKLDTFFGKLSLFTER 379 456 KKLDEFFVKLSLFTER 341
427 KKLDTFFVKLSLFTER 377 457 KKLDTFFVKLCLFTER 341
297 KKVDTFFVKLSLFTER 377 458 KKW DTFFVKLSLFTER 341
428 KKLDTFFVKLSLFTER 375 459 KKLDTFFVKLSLFTYR 340
379 KKLDTFFVKLSLFI ER 375 460 KKLDTKFVKLSLFTER 337
429 KKLDVFFVKLSLFTER 374 461 KDLDTFFVKLSLFTER 335
386 KKLDTFFVKLSLFW ER 374 462 KKLDTCFVKLSLFTER 335
331 KKLDTFFVRLSLFTER 374 463 KKLDTFYVKLSLFTER 334
292 KKNDTFFVKLSLFTER 374 464 KKLDTFFVKLRLFTER 333
269 DKLDTFFVKLSLFTER 373 465 FKLDTFFVKLSLFTER 332
430 KKLDTFFVKLSLFTER 371 466 KKLDTHFVKLSLFTER 332
431 KKLDTFFVKLSGFTER 370 467 KILDTFFVKLSLFTER 331
294 KKRDTFFVKLSLFTER 370 468 KTLDTFFVKLSLFTER 331
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
469 KKLDTFFVQLSLFTER 330 512 KKLKTFFVKLSLFTER 274
470 KKLDTFFVKLPLFTER 328 513 KKLDTFFQKLSLFTER 271
471 KKLDTFFVKLSLFTKR 324 514 KKLDTFFVKLSLFYER 270
472 KKLDTFFVKLWLFTER 324 515 KKLGTFFVKLSLFTER 264
473 KKLDTFFVKLKLFTER 323 33 KKLDTFFVKLSLFRER 264
474 KKLDTFFVKLDLFTER 322 516 KKLDTFFVKLSLFTER 260
475 KKLDTFFVKLSYFTER 320 517 KKLDTFFVKLSLFKER 259
476 KKLDTFFVKLSLFTER 319 518 KKLDTFFVNLSLFTER 256
477 KKLDTFFVKLALFTER 318 519 KKLDTFFCKLSLFTER 254
478 KKLDTFFVKLSLFTHR 318 520 KKLDTFFVKLSLFCER 254
479 KKLHTFFVKLSLFTER 317 521 KKLDTFFVKLSLFTEV 254
480 KKLRTFFVKLSLFTER 317 264 KKLDTFFKKLSLFTER 253
481 KVLDTFFVKLSLFTER 317 522 KKLDTFFVKLFLFTER 250
482 KKLDTFFVKWSLFTER 316 523 KKLDTFFVVLSLFTER 248
483 YKLDTFFVKLSLFTER 315 524 KKLDTFFVKLSLFTMR 247
484 KKLDLFFVKLSLFTER 311 525 KKLDTFFVKLSLFTLR 246
393 KKLDTFFVKLSLFTEY 311 526 KKLDTFFVWLSLFTER 245
390 KKLDTFFVKLSLFTEN 311 527 KKLDTFFVELSLFTER 240
485 KALDTFFVKLSLFTER 309 528 KKLDTFFVKLSLFTEH 239
486 KKLDTRFVKLSLFTER 309 529 KKLDTFFVKLSLFTEM 238
487 KKLDTFFVKLSLFTER 308 530 KKLDKFFVKLSLFTER 237
488 KKLDTFFVHLSLFTER 306 531 KKLDTFFVKLSLFTRR 237
489 KKLDTFFVKLSLFAER 305 532 KKLDTFFVKLELFTER 234
490 KWLDTFFVKLSLFTER 304 533 KKLDTFFVKLSLFTEP 234
491 KKLLTFFVKLSLFTER 303 534 KKLDTFFVPLSLFTER 233
492 KKLDTFDVKLSLFTER 301 101 KKLDTFFVKLSLFTGR 233
493 KKLDTFFVKLSLFQER 301 535 KKLDTFKVKLSLFTER 232
494 KYLDTFFVKLSLFTER 301 536 KKLDTEFVKLSLFTER 229
495 KKLDTFFAKLSLFTER 299 537 KKLDTFFWKLSLFTER 228
496 KKLDTFFTKLSLFTER 298 538 KKLDTFFVKLSLFTEA 226
497 KKLDTFFVKLSPFTER 297 539 KKLDTFFVKLSLFTWR 226
388 KKLDTFFVKLSLFTEF 297 540 KKLDTFFMKLSLFTER 221
498 KKLNTFFVKLSLFTER 296 541 KKLDTFFVCLSLFTER 220
499 KCLDTFFVKLSLFTER 295 542 KKLDTFFVKLSLKTER 220
500 KKLDDFFVKLSLFTER 295 543 KKLDTFFVKLSLFTEG 218
501 KKLDIFFVKLSLFTER 293 544 KKLDTFFVKLSLFTEL 217
502 KKLDTFFVKHSLFTER 293 545 KKLDTFFSKLSLFTER 216
392 KKLDTFFVKLSLFTET 292 546 CKLDTFFVKLSLFTER 215
503 KKLDTFFVKLSLYTER 291 547 KKLDTFFHKLSLFTER 213
389 KKLDTFFVKLSLFTEK 291 548 KKLDTFFVKLLLFTER 213
504 KKLDFFFVKLSLFTER 290 549 KKLDTFFYKLSLFTER 211
505 KKLDTFFVKLILFTER 289 550 KKLDTFFNKLSLFTER 203
99 KKLDTFFVKLGLFTER 288 551 KKLDTFFVKLSLFTEW 202
506 KKLDTFFVKKSLFTER 285 552 KKLDTFFVYLSLFTER 198
507 WKLDTFFVKLSLFTER 284 553 KKLDTDFVKLSLFTER 193
508 KKLDTFFVKCSLFTER 283 554 KKLDTFFVALSLFTER 191
509 KKLDTFFVMLSLFTER 283 555 KKLDTFFVILSLFTER 190
510 KSLDTFFVKLSLFTER 281 98 KKLDTFFVGLSLFTER 188
511 KKLDTFFVSLSLFTER 274 97 KKLDTFFVGLSLFTER 188
71
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
556 KKLDTFFVKLSLFTCR 185
557 KKLDTFFVKLSLFTES 184
558 KKLDTFFVKLSLFTEI 176
559 KKLDTFFVKLSLFTEC 175
560 KKLDTFFVFLSLFTER 174
561 KKLDTFFVKLSLFTAR 174
562 KKLDTFFVLLSLFTER 166
563 KKLDTFFVKLSLFTSR 165
564 KKLDTFFVKLSLFTIR 163
565 KKLDTFFVKLSLFTVR 163
566 KKLDTFFVKLSLFTNR 161
567 KKLDTFFVKLSLFDER 159
568 KKLDTFFVKLSLFTTR 152
569 KKLDTFFVDLSLFTER 149
570 KKLDTFFEKLSLFTER 139
571 KKLDTFFVKLSLFTFR 137
572 KKLDTFFVKLSLFTED 133
573 KKLDTFFVKLSLFTEQ 133
574 KKLDTFFDKLSLFTER 122
575 KKLDTFFVKLSLDTER 112
576 KKLDTFFVKLSLFEER 110
577 KKLDTFFVKLSLFTEE 107
578 KKLDTFFVKDSLFTER 102
579 KKLDTFFVKLSLETER 98
580 KKLDTFFVKLSDFTER 89
581 KKLDTFFVKLSEFTER 82
582 KKLDTFFVKESLFTER 81
72
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
[0283] As shown in Example 12, at least 31 single amino acid
substitutions of
P28R shown in Table 6.1 (SEQ ID NOs: 3-34) bind to P3028 with a higher rampo
score
than P28R. Additionally at least 4 single substitutions of glycine residues
for residues of
P28R (SEQ ID NOs: 94-96 and 98) bind to P3028 with a rampo scores at least
comparable to P28R, for example a rampo score greater than about 500.
Additionally at
least 129 single amino acid substitutions bind to P3028 with a rampo score at
least
substantially equal to (i.e., at least 98% of) P28R, as shown in Table 6.2
(SEQ ID NOs:
268-393). Additionally, truncations of at least the N terminal arginine of
P28R (SEQ ID
NO: 34), and up to the first 8 C terminal amino acids of P28R (SEQ ID NOs: 46-
53)
provide peptides with rampo scores at least comparable to P28R. Additionally,
at least
some internal amino acid residue deletions of P28 (SEQ ID NOs: 64-66, 68, 76)
provide
peptides with ramp scores at least comparable to P28R. Thus, contemplated
herein are
peptides that include substitutions of P28R that include combinations of two
or more of
the substitutions of SEQ ID NOs: 3-34. Moreover, contemplated herein are
peptides that
include at least one deletion of P28R as in SEQ ID NOs: 34, 46-53, 64-66, 68,
and/or 74,
and at least one substitution (of a non-deleted residue) of P28R as in SEQ ID
NOs: 3-34,
94-96, 98 and/or 268-393.
[0284] Accordingly, some embodiments concern compositions that
comprise,
consist of, or consist essentially of an immunoregulatory peptide inhibitor
that comprises,
consists of, or consists essentially of Formula (I):
Formula (I):
[0285] XX1VKX2X3X4 (SEQ ID NO: 166).
[0286] wherein X is an optional sequence, and can be KKLDT (SEQ ID
NO:
167), RKLDT (SEQ ID NO: 168), KKGDT (SEQ ID NO: 169), KKEDT (SEQ ID NO:
170), KKLDQ (SEQ ID NO: 171), KKGDQ (SEQ ID NO: 252), KKEDQ (SEQ ID
NO: 253), RKLDQ (SEQ ID NO: 254), RKGDQ (SEQ ID NO: 255), RKEDQ (SEQ ID
NO: 256), RKGTD (SEQ ID NO: 257), RKEDT (SEQ ID NO: 258), KLDT (SEQ ID
NO: 172), KGDT (SEQ ID NO: 259), KEDT (SEQ ID NO: 260), KLDQ (SEQ ID NO:
261), KGDQ (SEQ ID NO: 262), KEDQ (SEQ ID NO: 263), LDT, LDQ, GDT, GDQ,
EDT, EDQ, DT, DQ, T, or Q, or absent.
73
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
[0287] Xi can be one of 1-1-, FM, FS, FV, FT, FL, AF, AM, AS, AV, AT,
AL,
VF, VM, VS, VV, VT, or VL.
[0288] X2 can be one of LS, LQ, LM, LT, LH, VS, VQ, VM, VT, or VH.
[0289] X3 can be one of LFT, LMT, LQT, LHT, LNT, LPT, LST, LGT, LAT,
LRT, QFT, QMT, QQT, QHT, QNT, QPT, QST, QGT, QAT, QRT, VFT, VMT, VQT,
VHT, VNT, VPT, VST, VGT, VAT, VRT, MFT, MMT, MQT, MHT, MNT, MPT, MST,
MGT, MAT, MRT, LFN, LMN, LQN, LHN, LNN, LPN, LSN, LGN, LAN, LRN, QFN,
QMN, QQN, QHN, QNN, QPN, QSN, QGN, QAN, QRN, VFN, VMN, VQN, VHN,
VNN, VPN, VSN, VGN, VAN, VRN, MFN, MMN, MQN, MHN, MNN, MPN, MSN,
MGN, MAN, MRN, LFP, LMP, LQP, LHP, LNP, LPP, LSP, LGP, LAP, LRP, QFP,
QMP, QQP, QHP, QNP, QPP, QSP, QGP, QAP, QRP, VFP, VMP, VQP, VHP, VNP,
VPP, VSP, VGP, VAP, VRP, MFP, MMP, MQP, MHP, MNP, MPP, MSP, MGP, MAP,
MRPR, LFR, LMR, LQR, LHR, LNR, LPR, LSR, LGR, LAR, LRR, QFR, QMR, QQR,
QHR, QNR, QPR, QSR, QGR, QAR, QRR, VFR, VMR, VQR, VHR, VNR, VPR, VSR,
VGR, VAR, VRR, MFR, MMR, MQR, MHR, MNR, MPR, MSR, MGR, MAR, or MRR.
[0290] X4 is an optional sequence, and can be ER, or E, or absent.
[0291] In some embodiments, if X is absent, Xi is FF, and X2 is LS.
[0292] In some embodiments, the isolated peptide comprising Formula
(I) has
a length that is less than or equal to 1100 amino acids, for example, less
than or equal to
4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24,
25, 26, 27, 28, 29,
30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48,
49, 50, 51, 52, 53,
54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72,
73, 74, 75, 76, 77,
78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96,
97, 98, 99, 100,
101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115,
116, 117, 118,
119, 120, 121, 122, 123, 124, 125, 126, 127, 128, 129, 130, 131, 132, 133,
134, 135, 136,
137, 138, 139, 140, 150, 160, 170, 180, 190, 200, 210, 220, 230, 240, 250,
260, 270, 280,
290, 300, 320, 340, 360, 380, 400, 450, 500, 550, 600, 650, 700, 750, 800,
850, 900, 950,
1000, 1050, or 1100 amino acids, including ranges between any two of the
listed values.
[0293] Some embodiments concern compositions that comprise, consist
of, or
consist essentially of an immunoregulatory peptide inhibitor that comprises,
consists of,
or consists essentially of Formula (11):
Formula (11):
[0294] X20TFFVKLSX21X22 (SEQ ID NO: 173)
74
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
[0295] wherein X20 is an optional sequence, and can be KKLD (SEQ ID
NO:
174), RKLD (SEQ ID NO: 175), KKGD (SEQ ID NO: 176), KKED (SEQ ID NO:
177), KLD, LD, or D, or absent.
[0296] X21 is an optional sequence, and can be LFT, LMT, LQT, LHT,
LNT,
LPT, LST, LGT, LAT, LRT, QFT, QMT, QQT, QHT, QNT, QPT, QST, QGT, QAT,
QRT, VFT, VMT, VQT, VHT, VNT, VPT, VST, VGT, VAT, VRT, MFT, MMT, MQT,
MHT, MNT, MPT, MST, MGT, MAT, MRT, LFN, LMN, LQN, LHN, LNN, LPN, LSN,
LGN, LAN, LRN, QFN, QMN, QQN, QHN, QNN, QPN, QSN, QGN, QAN, QRN, VFN,
VMN, VQN, VHN, VNN, VPN, VSN, VGN, VAN, VRN, MFN, MMN, MQN, MHN,
MNN, MPN, MSN, MGN, MAN, MRN, LFP, LMP, LQP, LHP, LNP, LPP, LSP, LGP,
LAP, LRP, QFP, QMP, QQP, QHP, QNP, QPP, QSP, QGP, QAP, QRP, VFP, VMP,
VQP, VHP, VNP, VPP, VSP, VGP, VAP, VRP, MFP, MMP, MQP, MHP, MNP, MPP,
MSP, MGP, MAP, MRPR, LFR, LMR, LQR, LHR, LNR, LPR, LSR, LGR, LAR, LRR,
QMR, QQR, QHR, QNR, QPR, QSR, QGR, QAR, QRR, VFR, VMR, VQR, VHR,
VNR, VPR, VSR, VGR, VAR, VRR, MFR, MMR, MQR, MHR, MNR, MPR, MSR,
MGR, MAR, or MRR, or absent.
[0297] X22 is an optional sequence, and can be ER, or E, or absent.
[0298] In some embodiments, the isolated peptide comprising Formula
(II) has
a length that is less than or equal to 1100 amino acids, for example, less
than or equal to
4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24,
25, 26, 27, 28, 29,
30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48,
49, 50, 51, 52, 53,
54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72,
73, 74, 75, 76, 77,
78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96,
97, 98, 99, 100,
101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115,
116, 117, 118,
119, 120, 121, 122, 123, 124, 125, 126, 127, 128, 129, 130, 131, 132, 133,
134, 135, 136,
137, 138, 139, 140, 150, 160, 170, 180, 190, 200, 210, 220, 230, 240, 250,
260, 270, 280,
290, 300, 320, 340, 360, 380, 400, 450, 500, 550, 600, 650, 700, 750, 800,
850, 900, 950,
1000, 1050, or 1100 amino acids, including ranges between any two of the
listed values.
[0299] Some embodiments concern compositions that comprise, consist
of, or
consist essentially of an immunoregulatory peptide inhibitor that comprises,
consists of,
or consists essentially of Formula (III):
Formula (III):
[0300] X30X31VKLX32LX33TEX34 (SEQ ID NO: 178)
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
[0301] wherein X30 is an optional sequence, and can be KKLDTF (SEQ ID
NO: 179), KLDTF (SEQ ID NO: 180), LDTF (SEQ ID NO: 181), DTF, TF, or F, or
absent.
[0302] X31 is an optional sequence, and can be F, S, M, V, T, or L,
or absent.
In some embodiments, X31 is F.
[0303] X32 can be S, Q, M, T, or H. In some embodiments, X32 is S.
[0304] X33 can be F, M, Q, H, N, P, S, G, A, or R. In some
embodiments, X34
is F.
[0305] X34 is an optional sequence, and can be R, or absent.
[0306] In some embodiments, the isolated peptide comprising Formula
(III)
has a length that is less than or equal to 1100 amino acids, for example, less
than or equal
to 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23,
24, 25, 26, 27, 28,
29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47,
48, 49, 50, 51, 52,
53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71,
72, 73, 74, 75, 76,
77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95,
96, 97, 98, 99,
100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114,
115, 116, 117,
118, 119, 120, 121, 122, 123, 124, 125, 126, 127, 128, 129, 130, 131, 132,
133, 134, 135,
136, 137, 138, 139, 140, 150, 160, 170, 180, 190, 200, 210, 220, 230, 240,
250, 260, 270,
280, 290, 300, 320, 340, 360, 380, 400, 450, 500, 550, 600, 650, 700, 750,
800, 850, 900,
950, 1000, 1050, or 1100 amino acids, including ranges between any two of the
listed
values.
[0307] Some embodiments concern compositions that comprise, consist
of, or
consist essentially of an immunoregulatory peptide inhibitor that comprises,
consists of,
or consists essentially of Formula (VIE):
Formula (VIE):
[0308] X700K X701X702X703 X704X705X706K X707 X708 X709 X710 X711E
X712
(SEQ ID NO: 394),
[0309] wherein X700 is an optional sequence, and can be
K,A,D,E,G,H,I,L,M,N,P,Q,R,T,or V, or absent.
[0310] X701 is an optional sequence, and can be
L,A,C,D,E,F,G,H,I,K,M,N,Q,R,S,T,or V, or absent.
[0311] X702 is an optional sequence, and can be D,A,E,I,V,W, or Y, or
absent.
76
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
[0312] X703 is an optional sequence, and can be T,C,M,N,P,Q,R,S,W,
orY, or
absent.
[0313] X704 is an optional sequence, and can be F,A,I,M,N,P,T, or V,
or
absent.
[0314] X705 is an optional sequence, and can be F,L,M,Q,S,T or V, or
absent.
[0315] X706 is an optional sequence, and can be V,F,G,L,P, or R, or
absent.
[0316] X707 is an optional sequence, and can be
L,A,F,G,I,M,N,P,Q,R,S,T,V,
or Y, or absent.
[0317] X708 is an optional sequence, and can be S,H,M,N,Q, or T, or
absent.
[0318] X709 is an optional sequence, and can be
L,A,H,I,M,N,Q,R,S,T,V, or
W, or absent.
[0319] X710 is an optional sequence, and can be
F,A,C,G,H,I,L,M,N,P,Q,R,S,T,V, or W, or absent.
[0320] X711 is an optional sequence, and can be
T,F,G,H,I,L,M,N,P,S,V, or W,
or absent.
[0321] X712 is an optional sequence, and can be R,F,K,N,R,T, orY, or
absent.
[0322] In some embodiments, the isolated peptide comprising Formula
(VII)
has a length that is less than or equal to 1100 amino acids, for example, less
than or equal
to 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23,
24, 25, 26, 27, 28,
29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47,
48, 49, 50, 51, 52,
53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71,
72, 73, 74, 75, 76,
77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95,
96, 97, 98, 99,
100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114,
115, 116, 117,
118, 119, 120, 121, 122, 123, 124, 125, 126, 127, 128, 129, 130, 131, 132,
133, 134, 135,
136, 137, 138, 139, 140, 150, 160, 170, 180, 190, 200, 210, 220, 230, 240,
250, 260, 270,
280, 290, 300, 320, 340, 360, 380, 400, 450, 500, 550, 600, 650, 700, 750,
800, 850, 900,
950, 1000, 1050, or 1100 amino acids, including ranges between any two of the
listed
values.
[0323] Some embodiments concern compositions that comprise, consist
of, or
consist essentially of an immunoregulatory peptide inhibitor that comprises,
consists of,
or consists essentially of Formula (VIII):
Formula (VIII):
[0324] X8001( X8011( X802E X803 (SEQ ID NO: 395)
77
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
[0325] wherein X800 is K, A, D, E, G, H, I, L, M, N, P, Q, R, T, V,
or K, or
absent.
[0326] X801 is LDTFFV, GDTFFV, EDTFFV, LDQI-1-V, LDTAFV,
LDTVFV, LDTFMV, LDTFSV, LDTFVV, LDTFTV, LDTFLV, LDGFFV, LDTFGV,
LDTFFK, ADTFFV, CDTH-V, DDTI-1-V, FDTI-1-V, HDTFFV, IDTFFV, KDTI-1-V,
MDTFFV, NDTFFV, QDTFFV, RDTFFV, SDTFFV, TDTFFV, VDTFFV, LATH-AT,
LETFFV, LITFFV, LVTFFV, LWTFFV, LYTH-V, LDCFFV, LDMFFV, LDNI-1-V,
LDPFFV, LDRFFV, LDSFFV, LDWFFV, LDYFFV, LDTlFV, LDTMFV, LDTNFV,
LDTPFV, LDTTFV, LDTFQV, LDTFFF, LDTFFG, LDTFFL, LDTFFP, LDTFFR,
LDTFIV, LDTSFV, LDTFAV, LDTFCV, LDTQFV, LDTLFV, LTTFFV, LDTFFI,
LDHFFV, LMTFFV, LDTFEV, LDTFWV, LFTFFV, LDVFFV, LDTFRV, LDTFHV,
LDTYFV, LPTI-1-V, PDTFFV, LDTFPV, LDTFNV, LDTWFV, LDTGFV, LDAI-1-V,
LQTFFV, LCTI-1-V, LSTFFV, YDTFFV, LDEFFV, WDTFFV, LDTKFV, LDTCFV,
LDTFYV, LDTHFV, LHTFFV, LRTFFV, LDLFFV, LDTRFV, LLTFFV, LDTFDV,
LDTFFA, LDTFFT, LNTFFV, LDDFFV, LUFFY, LDI-1-FV, LKTI-1-V, LDTI-1-Q,
LGTFFV, LDTFFC, LDKFFV, LDTFKV, LDTEFV, LDTFFW, LDTFFM, LDTFFS,
LDTFFH, LDTFFY, LDTFFN, LDTDFV, LDTFFE, LDTI-1-D, LTFFV, LDTI-1-, TH-V,
LDF, LDTE, FFV, LDV, LV, or L, or absent;
[0327] wherein X802 is LSLFT, VSLFT, LQLFT, LMLFT, LTLFT, LHLFT,
LSQFT, LSVFT, LSMFT, LSLMT, LSLQT, LSLHT, LSLNT, LSLPT, LSLST, LSLGT,
LSLAT, LSLRT, LSLFN, LSLFP, LSLFR, LGLFT, ASLFT, FSLFT, GSLFT, ISLFT,
MSLFT, NSLFT, PSLFT, QSLFT, RSLFT, SSLFT, TSLFT, YSLFT, LNLFT, LSAFT,
LSHFT, LSIFT, LSNFT, LSRFT, LSSFT, LSTFT, LSWFT, LSLCT, LSLIT, LSLLT,
LSLTT, LSLVT, LSLWT, LSLFF, LSLFG, LSLFH, LSLFI, LSLFL, LSLFM, LSLFS,
LSLFV, LSLFW, LYLFT, LVLFT, LSFFT, LSGFT, LSKFT, LSCFT, LCLFT, LRLFT,
LPLFT, LWLFT, LKLFT, LDLFT, LSYFT, LALFT, WSLFT, LSLFA, LSLFQ, LSPFT,
HSLFT, LSLYT, LILFT, KSLFT, CSLFT, LSLFY, LSLFK, LSLFC, LFLFT, LELFT,
LSLKT, LLLFT, LSLI-D, LSLDT, LSLFE, DSLFT, LSLET, LSDFT, LSEFT, ESLFT,
SLFT, LSFT, LFT, LSL, LT, or T, or absent; and
[0328] wherein X803 is R, F, K, N, R, T, or Y, or absent.
[0329] In some embodiments, the isolated peptide comprising Formula
(VIII)
has a length that is less than or equal to 1100 amino acids, for example, less
than or equal
to 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23,
24, 25, 26, 27, 28,
78
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47,
48, 49, 50, 51, 52,
53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71,
72, 73, 74, 75, 76,
77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95,
96, 97, 98, 99,
100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114,
115, 116, 117,
118, 119, 120, 121, 122, 123, 124, 125, 126, 127, 128, 129, 130, 131, 132,
133, 134, 135,
136, 137, 138, 139, 140, 150, 160, 170, 180, 190, 200, 210, 220, 230, 240,
250, 260, 270,
280, 290, 300, 320, 340, 360, 380, 400, 450, 500, 550, 600, 650, 700, 750,
800, 850, 900,
950, 1000, 1050, or 1100 amino acids, including ranges between any two of the
listed
values.
[0330] Some
embodiments concern compositions that comprise, consist of, or
consist essentially of an immunoregulatory peptide inhibitor that comprises,
consists of,
or consists essentially of any one or more of the peptides set forth in Table
5.1. In some
embodiments, the isolated peptide from Table 5.1 used in these compositions
has a length
that is less than or equal to 1100 amino acids, for example, less than or
equal to 4, 5, 6, 7,
8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27,
28, 29, 30, 31,
32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50,
51, 52, 53, 54, 55,
56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74,
75, 76, 77, 78, 79,
80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98,
99, 100, 101, 102,
103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115, 116, 117,
118, 119, 120,
121, 122, 123, 124, 125, 126, 127, 128, 129, 130, 131, 132, 133, 134, 135,
136, 137, 138,
139, 140, 150, 160, 170, 180, 190, 200, 210, 220, 230, 240, 250, 260, 270,
280, 290, 300,
320, 340, 360, 380, 400, 450, 500, 550, 600, 650, 700, 750, 800, 850, 900,
950, 1000,
1050, or 1100 amino acids, including ranges between any two of the listed
values.
[0331] In some
embodiments, the peptide comprises one of SEQ ID NOs: 1-
33, 34, 46-53, 64-66, 68, 76, 94-96 or 98. Again, this isolated peptide can
have a length
that is less than or equal to 1100 amino acids, for example, less than or
equal to 4, 5, 6, 7,
8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27,
28, 29, 30, 31,
32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50,
51, 52, 53, 54, 55,
56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74,
75, 76, 77, 78, 79,
80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98,
99, 100, 101, 102,
103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115, 116, 117,
118, 119, 120,
121, 122, 123, 124, 125, 126, 127, 128, 129, 130, 131, 132, 133, 134, 135,
136, 137, 138,
139, 140, 150, 160, 170, 180, 190, 200, 210, 220, 230, 240, 250, 260, 270,
280, 290, 300,
79
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
320, 340, 360, 380, 400, 450, 500, 550, 600, 650, 700, 750, 800, 850, 900,
950, 1000,
1050, or 1100 amino acids, including ranges between any two of the listed
values.
[0332]
Embodiments of the invention also include immunoregulatory peptide
inhibitors that have a specific affinity to P3028 sequences or structures. In
some
embodiments, the immunoregulatory peptide inhibitors have specific affinity to
P3028
sequences or structures as measured by a rampo assay in which the
immunoregulatory
peptide inhibitors are affixed to a solid phase, P3028 is added, and the
enzymatic activity
of a rampo secondary antibody is measured so as to detect binding (see Example
12). In
some embodiments, the immunoregulatory peptide inhibitors bind to P3028
structures or
sequences with a rampo score that is at least substantially equal to the rampo
score of
P28R (see Example 12, Table 6.2). Preferably, the immunoregulatory peptide
inhibitors
have a specific affinity to P3028 by this rampo assay of at least or equal to
about 300
rampo units, for example, at least or equal to about 300, 310, 320, 330, 340,
350, 360,
370, 380, 390, 400, 410, 420, 430, 440, 450, 460, 470, 480, 490, 500, 510,
520, 530, 540,
550, 560, 570, 580, 590, 600, 610, 620, 630, 640, 650, 660, 670, 680, 690,
700, 710, 720,
730, 740, 750, 760, 770, 780, 790, 800, 820, 840, 860, 880, 900, 920, 940,
960, 980,
1000, 1020, or 1040 rampo units, including ranges between any two of the
listed values.
In some embodiments, the immunoregulatory peptide inhibitors bind to P3028
structures
or sequences with a rampo score of at 500 (see Example 12, Table 6.1).
Exemplary
peptides with affinity to P3028 are provided in Example 12 (see Tables 6.1,
6.2, and
Figures 29-30).
[0333]
Similarly, embodiments include isolated immunoregulatory peptide
inhibitors that have an affinity to any one or more of the immunoregulatory
peptides listed
in Tables 1-4 (SEQ ID NOs: 183-184 and 188-246). In some embodiments, the
immunoregulatory peptide inhibitors have specific affinity to any one or more
of the
immunoregulatory peptides listed in Tables 1-4 (SEQ ID NOs: 183-184 and 188-
246), as
measured by a rampo assay in which the immunoregulatory peptide inhibitors are
affixed
to a solid phase, any one or more of the immunoregulatory peptides listed in
Tables 1-4
(SEQ ID NOs: 183-184 and 188-246) is added, and the enzymatic activity of a
rampo
secondary antibody is measured so as to detect binding. For example, aspects
of the
invention include any peptide provided in Table 5.1 and any of the methods
described
herein can be practiced using one or more of the peptides described in Table
5.1.
Preferably, the immunoregulatory peptide inhibitors have a specific affinity
to any one or
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
more of the immunoregulatory peptides listed in Tables 1-4 (SEQ ID NOs: 183-
184 and
188-246) by this rampo assay of at least or equal to about 300 rampo units,
for example,
at least or equal to about 300, 310, 320, 330, 340, 350, 360, 370, 380, 390,
400, 410, 420,
430, 440, 450, 460, 470, 480, 490, 500, 510, 520, 530, 540, 550, 560, 570,
580, 590, 600,
610, 620, 630, 640, 650, 660, 670, 680, 690, 700, 710, 720, 730, 740, 750,
760, 770, 780,
790, 800, 820, 840, 860, 880, 900, 920, 940, 960, 980, 1000, 1020, or 1040
rampo units,
including ranges between any two of the listed values.
Peptide Sequence Variations
[0334] A number
of sequence variations to the immunoregulatory peptide
inhibitor P28R (KKLDTFFVKLSLFTER; SEQ ID NO: 2) have been shown to have
immunostimulatory activity and/or cytotoxicity to tumor cells (see Examples 37-
40).
Without being limited by any theory, SEQ ID NO: 2 and variations of SEQ ID NO:
2 as
described in Table 5.3 for example, one or more of the peptides of Table 5.4
can be
useful for binding peptide 3028 (SEQ ID NO: 185), binding a peptide or albumin
fragment that comprises SEQ ID NO: 185, binding any one or more of the
peptides listed
in Tables 1-4, directly stimulating immune cells, and/or killing tumor cells
in accordance
with some embodiments herein (see Examples 36-40). As such, in some
embodiments, a
immunoregulatory peptide inhibitor peptide comprises, consists of, or consists
essentially
of an amino acid sequence with one or more of the modifications to SEQ ID NO:
2 as
shown in Table 5.3, for example, 1, 2, 3, 4, 5, 6, 7, 8, 9, or 10
modifications, for example,
1-2, 1-3, 1-4, 1-5, 1-6, 1-7, 1-8, 1-9, 1-10, 2-3, 2-4, 2-5, 2-6, 2-7, 2-8, 2-
9, 2-10, 3-4, 3-5,
3-6, 3-7, 3-8, 3-9, 3-10, 4-5, 4-6, 4-7, 4-8, 4-9, 4-10, 5-6, 5-7, 5-8, 5-9, 5-
10, 6-7, 6-8, 6-9,
6-10, 7-8, 7-9, 7-10, 8-9, 8-10, or 9-10 variations. The inhibitor peptide can
further
comprise a further variation at one or more of positions 1, 3-4, 12-14, or 16
in SEQ ID
NO: 2, wherein the further variation comprises any amino acid or the absence
of an amino
acid, for example, 1, 2, 3, 4, 5, 6, or 7 further variations:
Table 5.3
Position in Type of Variation Exemplary Amino
KKLDTFFVKLSLFTER Acids for Variations
(SEQ ID NO: 2)
K1 Any type of amino acid Any amino acid or
absent
K2 Positive charged amino acid R, H, K
81
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
L3 Any type of amino acid Any amino acid or
absent
D4 Any type of amino acid Any amino acid or
absent
T5 Polar uncharged amino acid S, T, N, Q
F6 Hydrophobic or uncharged polar A, V, I, L, F, Y,
W, S,
amino acid T, N, Q
F7 Hydrophobic or uncharged polar A, V, I, L, F, Y,
W, S,
amino acid T, N, Q
V8 Hydrophobic, non-aromatic A, V, I, L
carbon chain amino acids that are
not M
K9 Positively charged amino acids, R, H, K, T, Q, Y
T, Q, or Y
L10 Any type of amino acid except R, H, K, S, T, N, Q,
C,
negatively charged U, G, P, A, V, I, L, M,
F, Y, W
Sll Polar uncharged amino acids S, T, N, Q
L12 Any type of amino acid except R, H, K, S, T, N, Q,
C,
negatively charged U, G, P, A, V, I, L, M,
F, Y, W
F13 Any type of amino acid except R, H, K, S, T, N, Q,
C,
negatively charged U, G, P, A, V, I, L, M,
F, Y, W
T14 Any type of amino acid except R, H, K, S, T, N, Q,
C,
negatively charged U, G, P, A, V, I, L, M,
F, Y, W
El5 Negatively charged amino acids D, E
[0335] In some
embodiments, the varied peptide does not comprise a M at
position 8. In some embodiments, the varied peptide does not comprise a M at
position 9.
In some embodiments, the varied peptide does not comprise a M at position 15.
In some
embodiments, the modified peptide does not comprise a M at any of positions 8,
9, or 15.
[0336]
Accordingly, in some embodiments, the peptide inhibitor comprising a
variation of P28R comprises, consists essentially of, or consists of a peptide
of Formula
(IX):
Formula (IX)
[0337]
X901X902X903X904X905X906X907X908X909X910X911X912X913X914X915X916X
917,
wherein X901 is any amino acid or absent,
X902 is a positively charged amino acid, F, or N,
82
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
X903 is any amino acid,
X904 is any amino acid,
X905 is a polar uncharged amino acid, R, Y, or W,
X906 is a hydrophobic or uncharged polar amino acid,
X907 is a hydrophobic or uncharged polar amino acid,
X908 is a hydrophobic, non-aromatic carbon chain amino acid that is not M
or F,
X909 is a positively charged amino acid, T, Q, or Y,
X910 is any amino acid that is not negatively charged,
X911 is a polar uncharged amino acid or H,
X912 is any amino acid that is not negatively charged,
X913 is any amino acid that is not negatively charged,
X914 is any amino acid that is not negatively charged,
X915 is a negatively charged amino acid, Y, or Q,
X916 is any amino acid that is not negatively charged, and
X917 is one or more positively charged amino acids or is absent.
Optionally, X901 comprises a positively charged amino acid. Optionally, X901
is an R or
K. Optional, X917 comprises or consists of RR.
[0338] A number
of peptide inhibitors based on variation of peptides
described herein have been shown to stimulate immune cells (see Example 36).
Exemplary varied peptides are shown in Table 5.4. Accordingly, in some
embodiments,
the peptide inhibitor comprises, consists of, or consists essentially of a
peptide of Table
5.4. Additional exemplary varied peptides shown to have low binding to P3028
(see
Example 36) or low stimulation of healthy PBMC's in healthy serum (see Example
37)
are shown in Tables 5.5 and 5.6. In some embodiments, a peptide comprising,
consisting
of, or consisting essentially of a peptide of Table 5.4, 5.5, or 5.6 is
provided.
Table 5.4: Peptides with "high" binding to P3028 based on positional scans
SE Q ID Amino Acid Sequence May also be referred to
NO: (variation(s) to SEQ ID NO: 2 are as:
underlined)
583 KKLDTFFVKLSLMTER 30677
584 KKLDTFFVKLQLFTER 30678
585 KKLDTVMVKLQLMTER 30680
83
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
586 RKLDTFFVKLSLFTERRR 32814
Table 5.5: Peptides with "low" binding to P3028 based on positional scans
SEQ ID Amino Acid Sequence May also be referred to as:
NO: (variation(s) to SEQ ID NO: 2 are
underlined)
587 KS LDTFFVKLS LFTER 30684
588 KKLDTFFVKLSLFTFR 30685
589 KKLDTFFVYLSLFTER 31135
590 KKLDTFFVNLSLFTER 31136
591 KKLDTFFVDLSLFTER 31138
Table 5.6: Additional modification of P28R
SEQ ID Amino Acid Sequence May also be referred to
NO: (variation(s) to SEQ ID NO: 2 are as:
underlined)
592 KKLDTFFPKLSLFTER 32251
593 KKLDTFMVKLS QHTER 32665
594 KKLDTFFVKLSLFTER(C(PEG24)) 32819
595 KKLDQFFVKLS QHNER 32815
[0339]
Embodiments of the invention also include peptides and proteins with
identity to an isolated immunoregulatory peptide inhibitor described herein.
The term
"identity" is meant to include nucleic acid or protein sequence homology or
three-
dimensional homology. Several techniques exist to determine nucleic acid or
peptide
sequence homology and/or three-dimensional homology to peptides. These methods
are
routinely employed to discover the extent of identity that one sequence,
domain, or model
has to a target sequence, domain, or model. A vast range of functional
immunoregulatory
peptide inhibitors (e.g., an immunoregulatory peptide inhibitor for P3028
sequence or
structures) can incorporate features of peptide inhibitors disclosed herein,
thus providing
for a vast degree of identity to the immunoregulatory peptide inhibitors of
SEQ ID NOs:
1-33, 34, 46-53, 64-66, 68, 76, 94-96, 98, 264-393, 583-586, or 589. For
example, a
fusion protein having a small region of an inhibitor can exhibit a low degree
of overall
identity to an immunoregulatory peptide inhibitor of SEQ ID NOs: 1-33, 34, 46-
53, 64-
66, 68, 76, 94-96, 98, 264-393, 583-586, or 589, yet retain the ability to
function as
inhibitor (e.g., an inhibitor of P3028, such as a molecule that binds to
P3028), or to
enhance immune cell stimulation via the LFA-1 and/or IL-2 receptor (e.g.,
modulate,
upregulate or down regulate a marker of the immune system or
immunosuppression, such
84
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
as reducing a P3028-mediated inhibition of immune cell proliferation,
spreading,
migration, or NK-cell cytotoxicity), or to enhance immune cell stimulation.
Thus,
embodiments of the invention can have from 1% identity to 100% identity to the
sequences of SEQ ID NOs: 1-33, 34, 46-53, 62, 64-66, 68, 76, 94-96, 98, 264-
393, 583-
586, or 589. That is, embodiments can have at least or equal to about, 1%, 2%,
3%, 4%,
5%, 6%, 7%, 8%, 9%, 10%, 11%, 12%, 13%, 14%, 15%, 16%, 17%, 18%, 19%, 20%,
21%, 22%, 23%, 24%, 25%, 26%, 27%, 28%, 29%, 30%, 31%, 32%, 33%, 34%, 35%,
36%, 37%, 38%, 39%, 40%, 41%, 42%, 43%, 44%, 45%, 46%, 47%, 48%, 49%, 50%,
51%, 52%, 53%, 54%, 55%, 56%, 57%, 58%, 59%, 60%, 61%, 62%, 63%, 64%, 65%,
66%, 67%, 68%, 69%, 70%, 71%, 72%, 73%, 74%, 75%, 76%, 77%, 78%, 79%, 80%,
81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%, 92%, 93%, 94%, 95%,
96%, 97%, 98%, 99%, or 100% identity to any one of SEQ ID NOs: 1-33, 34, 46-
53, 64-
66, 68, 76, 94-96, 98, 264-393, 583-586, or 589. Preferably, these peptides or
modified
peptides also retain the ability to modulate the immune system (e.g.,
modulate, upregulate
or down regulate a marker of the immune system or immunosuppression, such as
reducing a P3028-mediated inhibition of immune cell proliferation, spreading,
migration,
or NK-cell cytotoxicity).
[0340]
Embodiments also include compositions that comprise multimers of
isolated immunoregulatory peptide inhibitors and/or isolated immunoregulatory
peptide
inhibitors bound to a support. Some embodiments include compositions that
comprise
multimers of immunoregulatory peptide inhibitors that include multiple copies
of a single
immunoregulatory peptide inhibitor. Some embodiments include compositions that
comprise multimers that include two or more different immunoregulatory peptide
inhibitors. Some multimers include at least or equal to two immunoregulatory
peptide
inhibitors, for example 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16,
17, 18, 19, 20, 21,
22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40,
45, 50, 55, 60, 65,
70, 75, 80, 85, 90, 95, 99, 100, or 101 immunoregulatory peptide inhibitors.
In some
embodiments, the multimers are of the same immunoregulatory peptide inhibitor
and in
other embodiments, the multimers are of different immunoregulatory peptide
inhibitors.
Accordingly, some embodiments concern compositions that comprise one or more
immunoregulatory peptide inhibitors and in some embodiments, the one or more
immunoregulatory peptide inhibitor are multimers of the same molecule.
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
Methods of making peptide-based immunoregulatory peptide inhibitors
[0341] Many
methods of making peptides are known in the art. Examples of
methods of making peptides can be found in US Patent No: 6,495,674, hereby
expressly
incorporated by reference in its entirety. In some embodiments, peptide
inhibitors are
chemically synthesized. Chemical synthesis of peptides is also well-known. For
example, solid phase chemical synthesis can be used to produce peptides of up
to at least
about 100 amino acids in length. Accordingly, in some embodiments, the
immunoregulatory peptide inhibitor is a synthetic peptide.
[0342] In other
embodiments, immunoregulatory peptide inhibitors are
prepared by recombinant DNA technology using techniques well known in the art.
Such
methods can be used to construct expression vectors containing nucleotide
sequences
encoding an immunoregulatory peptide inhibitor, for example, and appropriate
transcriptional and translational control signals. These methods can include,
for example,
in vitro recombinant DNA techniques, synthetic techniques, and in vivo genetic
recombination. Alternatively, RNA capable of encoding a peptide inhibitor can
be
chemically synthesized using, for example, synthesizers. See, for example, the
techniques
described in Oligonucleotide Synthesis, 1984, Gait, M. J. ed., lRL Press,
Oxford, which is
incorporated by reference herein in its entirety. Alternatively, a DNA or RNA
encoding a
peptide or protein substantially longer that the peptide inhibitor can be
provided, in which
the peptide inhibitor is flanked by protease target sites, thus producing the
peptide
inhibitor from a larger peptide or protein. Exemplary proteases include
thrombin, trypsin,
chymotrypsin, LysC, GluC, and AspN. Alternatively, a DNA or RNA encoding two
or
more copies of the peptide inhibitor can be provided, in which the peptide
inhibitors are
flanked by protease target sites, thus producing the peptide inhibitor from a
larger peptide
or protein. Thus, in some embodiments, the peptide inhibitor of P3028 is
produced by a
ribosome.
[0343] In
several embodiments, the immunoregulatory peptide inhibitors are
expressed in a cell line. For example, some cells are provided a nucleic acid
encoding
one or more immunoregulatory peptide inhibitors, said cells are made to
express the
peptides of SEQ ID NOs: 1-33, 34, 46-53, 64-66, 68, 76, 94-96, 98, 264-393,
583-586,
or 589 or any one or more of the peptides provided in Table 5.1 and the
immunoregulatory peptide inhibitors are isolated and/or purified. Exemplary
nucleic
acids are listed in Table 5.2, SEQ ID NOs: 102-165.
86
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
[0344] A
variety of host-expression vector systems can be utilized to express
inhibitor peptides of some embodiments of the invention. Where the
immunoregulatory
peptide inhibitor is a soluble peptide, it can be recovered from the culture,
i.e., from the
host cell in cases where the peptide or polypeptide is not secreted, and from
the culture
media in cases where the peptide or polypeptide is secreted by the cells.
However, the
expression systems also encompass engineered host cells that express the
peptide or
functional equivalents in situ, i.e., anchored in the cell membrane.
Purification or
enrichment of the peptide from such expression systems can be accomplished
using
appropriate detergents and lipid micelles and methods well known to those
skilled in the
art. However, such engineered host cells themselves can be used in situations
where it is
important not only to retain the structural and functional characteristics of
the peptide, but
to assess biological activity, e.g., in drug screening assays.
[0345] The
expression systems that can be used for purposes of the invention
include, but are not limited to, microorganisms such as bacteria (e.g., E.
coli or B. subtilis)
transformed with recombinant bacteriophage DNA, plasmid DNA or cosmid DNA
expression vectors containing nucleotide sequences encoding inhibitor
peptides; yeast
(e.g., Saccharomyces, Pichia) transformed with recombinant yeast expression
vectors
containing the nucleotide sequences encoding inhibitor peptides; insect cell
systems
infected with recombinant virus expression vectors (e.g., baculovirus)
containing
sequences encoding inhibitor peptides; plant cell systems infected with
recombinant virus
expression vectors (e.g., cauliflower mosaic virus, CaMV; tobacco mosaic
virus, TMV) or
transformed with recombinant plasmid expression vectors (e.g., Ti plasmid)
containing
nucleotide sequences encoding inhibitor peptides; mammalian cell systems
(e.g., COS,
CHO, BHK, 293, 3T3) harboring recombinant expression constructs containing
promoters
derived from the genome of mammalian cells (e.g., metallothionein promoter) or
from
mammalian viruses (e.g., the adenovirus late promoter; the vaccinia virus 7.5K
promoter);
or cell-free expression systems, which can include cell lysates or fractions
thereof, and
nucleic acids encoding the inhibitor peptides.
[0346] In
bacterial systems, a number of expression vectors can be
advantageously selected depending upon the use intended for the peptide being
produced.
For example, when a large quantity of such a peptide is to be produced, for
the generation
of pharmaceutical compositions or for raising antibodies to the peptide, for
example,
vectors which direct the expression of high levels of fusion protein products
that are
87
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
readily purified can be desirable. Such vectors include, but are not limited,
to the E. coli
expression vector pUR278 (Ruther et al., EMBO J., 2:1791 (1983), in which the
inhibitor
peptide coding sequence can be ligated individually into the vector in frame
with the lacZ
coding region so that a fusion protein is produced; pliN vectors (Inouye &
Inouye, Nucleic
Acids Res., 13:3101-3109 (1985); Van Heeke & Schuster, J. Biol. Chem.,
264:5503-5509
(1989)); and the like. pGEX vectors can also be used to express foreign
polypeptides as
fusion proteins with glutathione S-transferase (GST). In general, such fusion
proteins are
soluble and can be purified from lysed cells by adsorption to glutathione-
agarose beads
followed by elution in the presence of free glutathione. The PGEX vectors are
designed
to include thrombin or factor Xa protease cleavage sites so that the cloned
target gene
product can be released from the GST moiety.
[0347] In an
insect system, Auto grapha californica nuclear polyhedrosis virus
(AcNPV) is used as a vector to express foreign genes. The virus grows in
Spodoptera
frugiperda cells. The peptide coding sequence can be cloned individually into
non-
essential regions (for example the polyhedrin gene) of the virus and placed
under control
of an AcNPV promoter (for example the polyhedrin promoter). Successful
insertion of
peptide coding sequence will result in inactivation of the polyhedrin gene and
production
of non-occluded recombinant virus, (i.e., virus lacking the proteinaceous coat
coded for
by the polyhedrin gene). These recombinant viruses are then used to infect
Spodoptera
frugiperda cells in which the inserted gene is expressed. (E.g., see Smith et
al., J. Virol.
46: 584 (1983); and Smith, U.S. Pat. No. 4,215,051).
[0348] In
mammalian host cells, a number of viral-based expression systems
can be utilized. In cases where an adenovirus is used as an expression vector,
the
nucleotide sequence of interest can be ligated to an adenovirus
transcription/translation
control complex, e.g., the late promoter and tripartite leader sequence. This
chimeric
gene can then be inserted in the adenovirus genome by in vitro or in vivo
recombination.
Insertion in a non-essential region of the viral genome (e.g., region El or
E3) will result in
a recombinant virus that is viable and capable of expressing the peptide in
infected hosts.
(E.g., see Logan & Shenk, Proc. Natl. Acad. Sci. USA 81:3655-3659 (1984)).
Specific
initiation signals can also be required for efficient translation of inserted
nucleotide
sequences encoding peptides. These signals include the ATG initiation codon
and
adjacent sequences.
88
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
[0349] In cell
free systems, cellular extracts, or fractions thereof are provided
for the translation of nucleic acids into polypeptides in vitro. Cell free
systems can
include, for example e coli extracts, yeast extracts. The extracts can be
lysates. The
extracts can be purified, for example, to enrich for ribosomes and/or to
remove undesired
materials such as debris or host genomic DNA. Nucleic
acids encoding
immunoregulatory peptide inhibitors in cell-free systems can include plasmid
DNA, linear
DNA, or RNA.
[0350] In some
embodiments, immunoregulatory peptide inhibitors are
isolated or purified after expression. Isolation or purification can include
affinity
purification. In some embodiments, the peptide product of the expression
system includes
an affinity tag, for example GST separated by a cleavable linker, for example
a thrombin
or factor Xa protease cleavage site. After affinity purification, the affinity
tag can be
cleaved, producing a substantially pure peptide that does not have an affinity
tag or
cleavage site. In some embodiments, purification results in a composition that
is at least
or equal to about 1,2, 3, 4, 5, 6, 7, 8,9, 10, 11, 12, 13, 14, 15, 16, 17, 18,
19, 20, 21, 22,
23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41,
42, 43, 44, 45, 46,
47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65,
66, 67, 68, 69, 70,
71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89,
90, 91, 92, 93, 94,
95, 96, 97, 98, 98.5, 99, 99.5, 99.9, 99.99, or 99.999% peptide by weight. The
section
below provides more information on pharmaceutically acceptable carriers and
diluents
that can be used with the embodiments described herein.
D amino acids and non-natural amino acids
[0351] Some
embodiments include compositions that comprise, consist, or
consist essentially of one or more immunoregulatory peptide inhibitors that
include at
least one D amino acid. With the exception of glycine, the chiral carbon of an
amino acid
can exist as the D or the L isomer. Typically, amino acids synthesized by
ribosomes are
in the L configuration. However, peptides that include D amino acids, or a
combination
of D and L amino acids can have activity, for example as ligands or
inhibitors. For
example, a peptide including at least one D amino acid can bind to the P3028
sequence/structure and inhibit the ability of the P3028 sequence/structure to
bind to the
LFA-1 receptor and/or the IL-2 receptor.
89
CA 02978617 2017-09-01
WO 2016/144650 PCT/US2016/020510
[0352]
Accordingly, some embodiments include immunoregulatory peptide
inhibitors that comprise at least one non-natural amino acid. Non-natural
amino acids
include amino acids having R groups other than the R group of the 20 amino
acids
encoded by the standard genetic code. Non-natural amino acids can exist in the
L or D
configuration. Thus, some embodiments include peptides having non-natural
amino acids
in the D configuration and/or the L configuration. Exemplary non-natural amino
acids are
described in US Pat Nos: 8,153,758, 7,888,533, 6,344,483, each of which is
expressly
incorporated by reference in its entirety herein. Some embodiments concern a
composition that comprises, consists of, or consists essentially of one or
more of the
immunoregulatory peptide inhibitors described herein (e.g., an
immunoregulatory peptide
inhibitor of the P3028 sequence/structure, such as one or more of the
immunoregulatory
peptide inhibitors provided by of SEQ ID NOs: 1-33, 34, 46-53, 64-66, 68, 76,
94-96,
98, 264-393, 583-586, or 589 or any one or more of the peptides provided in
Table 5.1,
5.4, 5.5, 5.6, or any variation or combination of variations of P28R or P28
core as
provided in Tables 5.3 and 13, wherein said immunoregulatory peptide inhibitor
comprises at least one D amino acid. Similarly, some embodiments concern a
composition comprising immunoregulatory peptide inhibitor of the P3028
sequence/structure, wherein said immunoregulatory peptide inhibitors (e.g.,
any one or
more of the immunoregulatory peptide inhibitors provided by of SEQ ID NOs: 1-
33, 34,
46-53, 64-66, 68, 76, 94-96, 98, 264-393, 583-586, or 589 or any one or more
of the
peptides provided in Table 5.1, 5.4, 5.5, 5.6, or any variation or combination
of variations
of P28R or P28 core as provided in Tables 5.3 and 13 comprises at least one
non-natural
amino acid. Further embodiments
include a composition comprising an
immunoregulatory peptide inhibitor (e.g., any one or more of the
immunoregulatory
peptide inhibitors provided by SEQ ID NOs: 1-33, 34, 46-53, 64-66, 68, 76, 94-
96, 98,
264-393, 583-586, or 589 or any one or more of the peptides provided in Table
5.1, 5.4,
5.5, 5.6, or any variation or combination of variations of P28R or P28 core as
provided in
Tables 5.3 and 13, wherein each non-glycine amino acid of the immunoregulatory
peptide inhibitor is a D amino acid.
[0353] The
crystal structure of the IL-2 receptor (CD25) has been solved, and
computer modeling of P3028 binding to the IL-2 binding site of the IL-2
receptor has
been performed (see Figure 19). Moreover, the crystal structure of the ligand
binding
domain of IL-2 is known (see Qu, A and Leahy, DJ, Proc. Nall. Acad. Sci. USA
1995, 92:
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
10277-10281, which is expressly incorporated by reference in its entirety).
Moreover,
favorable interactions between P3028 and at least one immunoregulatory peptide
inhibitor
can facilitate the selection of additional amino acid residues, D amino acid
residues,
and/or non-natural amino acid residues to maintain favorable interactions.
[0354] In some
embodiments, at least some of these immunoregulatory
peptide inhibitors include D amino acids positions that are selected using
rational design
or P3028 sequence/structure inhibitors. As noted in U.S. Pat No. 7,957,912,
rational
design of peptides can start with a protein backbone structure and designs the
amino acid
sequence to modify the protein's properties, while maintaining its three
dimensional
folding properties. In some embodiments, large numbers of sequences can be
manipulated using computer modeling, allowing for the design of protein
structures
(sequences, subsequences, etc.). Aspects of rational design are described in a
number of
publications, including, e.g., Malakauskas and Mayo (1998) "Design, Structure
and
Stability of a Hyperthermophilic Protein Variant" Nature Struc. Biol. 5:470;
Dahiyat and
Mayo (1997) "De Novo Protein Design: Fully Automated Sequence Selection"
Science,
278, 82-87. DeGrado, (1997) "Proteins from Scratch" Science, 278:80-81;
Dahiyat,
Sarisky and Mayo (1997) "De Novo Protein Design: Towards Fully Automated
Sequence
Selection" J. Mol. Biol. 273:789-796; Dahiyat and Mayo (1997) "Probing the
Role of
Pachng Specificity in Protein Design" Proc. Natl. Acad. Sci. USA, 94:10172-
10177;
Hellinga (1997) "Rational Protein Design--Combining Theory and Experiment"
Proc.
Natl. Acad. Sci. USA, 94: 10015-10017; Su and Mayo (1997 j "Coupling Backbone
Flexibility and Amino Acid Sequence Selection in Protein Design" Prot. Sci.
6:1701-
1707; Dahiyat, Gordon and Mayo (1997) "Automated Design of the Surface
Positions of
Protein Helices" Prot. Sci., 6:1333-1337; Dahiyat and Mayo (1996) "Protein
Design
Automation" Prot. Sci., 5:895-903.
[0355] In some
embodiments, a library of variant of immunoregulatory
peptide inhibitors of the P3028 sequence/structure containing one or more D
amino acids
and/or non-natural amino acids is screened for binding to the P3028
sequence/structure.
In some embodiments, the library is screened for binding to P3028 (see
Examples 10 and
12). In some embodiments, the library is screened for inhibiting binding of
the P3028
sequence/structure to the LFA-1 receptor (see Example 15).
[0356] In some
embodiments, a lead molecule is used as a template for
directed drug design. A lead peptide, for example, can include, but is not
limited to one
91
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
or more of the exemplary immunoregulatory peptide inhibitors that bind to the
P3028
sequence/structure provided herein, such as in Example 12, (e.g., any one or
more of the
immunoregulatory peptide inhibitors provided by SEQ ID NOs: 1-33, 34, 46-53,
64-66,
68, 76, 94-96, 98, 264-393, 583-586, or 589 or any one or more of the peptides
provided
in Table 5.1, 5.4, 5.5, 5.6, or any variation or combination of variations of
P28R or P28
core as provided in Tables 5.3 and 13). The lead peptide can be synthesized to
include at
least one D amino acid and/or at least one non-natural amino acid. In some
embodiments,
the binding activity of the first generation artificial immunoregulatory
peptide inhibitor is
then detected, for example by evaluating the binding affinity for the P3028
sequence/structure, as described herein. Additionally, the immunostimulatory
activity of
the first generation artificial immunoregulatory peptide inhibitor can be
detected, for
example by evaluating the stimulation of an LFA-1 and/or IL-2 dependent
response in a
cell having a LFA-1 receptor or IL-2 receptor, which can be inhibited by the
P3028
sequence/structure. Once the binding and/or immunostimulatory activity of the
first
generation artificial immunoregulatory peptide inhibitor is obtained, at least
one
additional modification is made to the lead peptide and this second generation
immunoregulatory peptide inhibitor is evaluated for binding to the P3028
sequence/structure and immunostimulatory activity. The additional modification
can
include, but is not limited to the addition or substitution, of at least one
additional D
amino acid and/or a non-natural amino acid. By iteratively conducting this
screening and
modification procedure, more immunoregulatory peptide inhibitors can be made.
[0357]
Additionally, any one or more of the immunoregulatory peptide
inhibitors described herein (e.g., any one or more of the immunoregulatory
peptide
inhibitors provided by SEQ ID NOs: 1-33, 34, 46-53, 64-66, 68, 76, 94-96, 98,
264-393,
583-586, or 589 or any one or more of the peptides provided in Table 5.1, 5.4,
5.5, 5.6, or
any variation or combination of variations of P28R or P28 core as provided in
Tables 5.3
and 13 can comprise an N-terminal acetyl group and/or a C-terminal amide
group.
Furthermore, any one or more of the immunoregulatory peptide inhibitors
described
herein that comprise at least one D amino acid and/or at least one non-natural
amino acid
(e.g., any one or more of the immunoregulatory peptide inhibitors provided by
SEQ ID
NOs: 1-33, 34, 46-53, 64-66, 68, 76, 94-96, 98, 264-393, 583-586, or 589 or
any one or
more of the peptides provided in Table 5.1, 5.4, 5.5, 5.6, or any variation or
combination
92
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
of variations of P28R or P28 core as provided in Tables 5.3 and 13 can be
prepared to
comprise an N-terminal acetyl group and/or a C-terminal amide group).
Pep tidomimetics
[0358] Some
embodiments include compositions that comprise, consist of, or
consist essentially of peptidomimetic-based immunoregulatory peptide
inhibitors.
Peptidomimetics can include, but are not limited to small-molecule compounds
having at
least one biochemical interaction that a peptide also has. Some
peptidomimetics can
include a small molecule backbone. Some peptidomimetics can include at least
one R
group of a naturally-occurring amino acid covalently bonded to a small
molecule
backbone. Some peptidomimetics are substituted into at least one position of a
known
peptide sequence. Accordingly, some embodiments include a composition that
comprises, consists of, or consists essentially of one or more of the
exemplary
immunoregulatory peptide inhibitors that bind to the P3028 sequence/structure
provided
herein (e.g., any one or more of the immunoregulatory peptide inhibitors
provided by
SEQ ID NOs: 1-33, 34, 46-53, 64-66, 68, 76, 94-96, 98, 264-393, 583-586, or
589 or any
one or more of the peptides provided in Table 5.1, 5.4, 5.5, 5.6, or any
variation or
combination of variations of P28R or P28 core as provided in Tables 5.3 and
13),
wherein said immunoregulatory peptide inhibitor comprises at least one
peptidomimetic
substitution (e.g., a non-peptide bond, a small molecule backbone, or an
artificial peptide
linkage).
[0359] Some
embodiments include a composition that comprises, consists of,
or consists essentially of one or more of the exemplary isolated
immunoregulatory peptide
inhibitors that bind to the P3028 sequence/structure provided herein (e.g.,
any one or
more of the immunoregulatory peptide inhibitors provided by SEQ ID NOs: 1-33,
34, 46-
53, 64-66, 68, 76, 94-96, 98, 264-393, 583-586, or 589 or any one or more of
the peptides
provided in Table 5.1, 5.4, 5.5, 5.6, or any variation or combination of
variations of P28R
or P28 core as provided in Tables 5.3 and 13, wherein said immunoregulatory
inhibitors
comprise a peptidomimetic substitution, which includes two or more monomers,
wherein
each monomer comprises a small molecule backbone covalently bound to at least
one R
group. More embodiments, include a composition that comprises, consists of, or
consists
essentially of one or more of the exemplary immunoregulatory peptide
inhibitors that bind
to the P3028 sequence/structure provided herein (e.g., any one or more of the
93
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
immunoregulatory peptide inhibitors provided by SEQ ID NOs: 1-33, 34, 46-53,
64-66,
68, 76, 94-96, 98, 264-393, 583-586, or 589 or any one or more of the peptides
provided
in Table 5.1, 5.4, 5.5, 5.6, or any variation or combination of variations of
P28R or P28
core as provided in Tables 5.3 and 13, wherein said immunoregulatory
inhibitors
comprise at least one peptidomimetic small molecule backbone, wherein each
backbone
molecule includes one of an aryl group, for example a benzene, pyrrole, furan,
thiophene,
imidazole, oxazole, thiazole, triazole, pyrazole, pyridine, pyrazine,
pyridazine and
pyrimidine, and the like; a cycloalkane or heterocycloalkane; a cycloalkene or
heterocycloalkene; or a combination of two or more of the listed molecules.
Each R
group can be the R group of a naturally occurring amino acid, or optionally
can be a
synthetic molecule. Each R group can be different, but two or more R groups
can be the
same. Some peptidomimetics include a first monomer that binds to a first
position of
P3028, for example, and a second monomer that binds to a second position of
P3028, in
which the first and second monomers are covalently bonded (see, for example,
the
approach of Chen et al., ACS Chemical Biology 2009; 4(9): 769-81, hereby
expressly
incorporated by reference in its entirety). The peptidomimetic backbone that
is
incorporated into one or more of the exemplary immunoregulatory peptide
inhibitors that
bind to the P3028 sequence/structure provided herein (e.g., any one or more of
the
immunoregulatory peptide inhibitors provided by SEQ ID NOs: 1-33, 34, 46-53,
64-66,
68, 76, 94-96, 98, 264-393, 583-586, or 589 or any one or more of the peptides
provided
in Table 5.1, 5.4, 5.5, 5.6, or any variation or combination of variations of
P28R or P28
core as provided in Tables 5.3 and 13, can include a derivative of a 13-turn
peptidomimetic cyclic compound of formula (IV), as taught by U.S. Pat No.
6,881,719,
hereby expressly incorporated by reference in its entirety:
94
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
[0360] Formula (IV)
/ R4
CO
_________________________________________ 0
0
RS"- N HN
0
1 INKER
[0361] In some embodiments, R1 and R3 of the above Formula (IV)
include R
groups of natural and/or synthetic amino acids. Some embodiments include a
composition that comprises, consists of, or consists essentially of one or
more of the
exemplary immunoregulatory peptide inhibitors that bind to the P3028
sequence/structure
provided herein (e.g., any one or more of the immunoregulatory peptide
inhibitors
provided by SEQ ID NOs: 1-33, 34, 46-53, 64-66, 68, 76, 94-96, 98, 264-393,
583-586,
or 589 or any one or more of the peptides provided in Table 5.1, 5.4, 5.5,
5.6, or any
variation or combination of variations of P28R or P28 core as provided in
Tables 5.3 and
13), wherein said immunoregulatory inhibitors comprise a peptidomimetic
substitution
that includes a polymer of two or more derivatives of Formula (IV). In some
embodiments, individual peptidomimetic monomers or dimers derived from Formula
(IV)
are selected for their ability to bind the P3028 sequence/structure, and are
then assembled
into polymers, thus producing a peptidomimetic polymer that specifically binds
the P3028
sequence/structure.
[0362] As described in US Pat No. 7,816,324, peptidomimetics of
either
Formula (V) or Formula (VI) can be modified to mimic alpha-helix motifs that
bind to
peptides.
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
[0363] Formula (V)
A
X
X
i
Y X
.4hr X
Y X
001 X s,õ
[0364] Formula (VI)
1110 R
X
Y X
Olt R2
X
411 RA
X
Y X
96
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
[0365]
Accordingly, aspects of the invention include a composition that
comprises, consists of, or consists essentially of one or more of the
exemplary
immunoregulatory peptide inhibitors that bind to the P3028 sequence/structure
provided
herein (e.g., any one or more of the immunoregulatory peptide inhibitors
provided by
SEQ ID NOs: 1-33, 34, 46-53, 64-66, 68, 76, 94-96, 98, 264-393, 583-586, or
589 or any
one or more of the peptides provided in Table 5.1, 5.4, 5.5, 5.6, or any
variation or
combination of variations of P28R or P28 core as provided in Tables 5.3 and
13),
wherein said immunoregulatory inhibitors comprise a peptidomimetic
substitution that
incorporates the scaffold of formula V or formula VI, which provide a rigid
structure and
places and orients substituents as an alpha-helix does. Substitution on the
rigid iris-
benzamide, for instance, can allow placement of three functional groups (R1-
R3)
corresponding to the side chains of amino acids found at the i, i+4, and i+7
positions of an
ideal alpha-helix, bound by the peptide. As shown in Figure 19, P3028 is
modeled to
bind to alpha helix-containing regions of the IL-2 receptor. Thus, some
embodiments
include a composition that comprises, consists of, or consists essentially of
one or more of
the exemplary immunoregulatory peptide inhibitors that bind to P3028 provided
herein
(e.g., any one or more of the immunoregulatory peptide inhibitors provided by
SEQ ID
NOs: 1-33, 34, 46-53, 64-66, 68, 76, 94-96, 98, 264-393, 583-586, or 589 or
any one or
more of the peptides provided in Table 5.1, 5.4, 5.5, 5.6, or any variation or
combination
of variations of P28R or P28 core as provided in Tables 5.3 and 13), wherein
said
immunoregulatory inhibitors comprise a peptidomimetic substitution that
incorporates a
peptidomimetic of formula V or formula VI, wherein R1 - R3 are selected from
positions
on a known binding partner of P3028, for example the alpha subunit of the IL-2
receptor
(CD25) (SEQ ID NO: 247), the LFA-1 receptor (CD 11 a - SEQ ID NO: 248 and CD18
- SEQ ID NO: 249), or a peptide of SEQ ID NOs: 1-33, 34, 46-53, 64-66, 68, 76,
94-96,
98, 264-393, 583-586, or 589 or any one or more of the peptides provided in
Table 5.1,
5.4, 5.5, 5.6, or any variation or combination of variations of P28R or P28
core as
provided in Tables 5.3 and 13.
[0366]
Embodiments also include a library of peptidomimetics. In some
embodiments, the library of peptidomimetics is selected and/or synthesized
using a
rational design approach. As disclosed in U.S. Pat No. 7,816,324, hereby
expressly
incorporated by reference in its entirety, a peptidomimetic library can be
developed based
on based on a structural knowledge of the interface of protein complexes.
Thus, in some
97
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
embodiments, peptidomimetic compounds are based on the structure of P3028, and
its
interactions with known binding partners, for example the IL-2 receptor for
which the
crystal structure is known (see Figure 19), the LFA-1 receptor, for which the
crystal
structure is known, the KKL15 peptide (see Example 11), and known inhibitors
of the
P3028 sequence/structure (e.g., SEQ ID NOs: 1-33, 34, 46-53, 64-66, 68, 76, 94-
96, 98,
or 264-393 or any one or more of the peptides provided in Table 5.1). In some
embodiments, alpha.-helix mimetics may be used to modulate protein-protein or
protein-
peptide interaction. Thus, synthetic scaffolds that mimic key elements found
in the
interface between the P3028 sequence/structure and its binding partners is
contemplated
for the development of small molecule immunoregulatory protein inhibitors. In
some
embodiments, the molecules of the peptidomimetic library are attached to a
support, chip,
surface, or substrate, for example a microarray, as in U.S. Pat No. 7,153,68,
hereby
expressly incorporated by reference in its entirety. The section below
provides more
details on aptamer-based immunoregulatory peptide inhibitors.
Cyclic peptides
[0367] Some embodiments include at least one cyclic peptide
immunoregulatory peptide inhibitor. Cyclic peptides, sometimes referred to as
"looped
peptides" are known in the art, and can be chemically synthesized (see, e.g.,
U.S. Pat. No.
7,589,170, hereby expressly incorporated by reference in its entirety herein),
or
synthesized in vivo (see, e.g., U.S. Pat. No. 7,252,952, hereby expressly
incorporated by
reference in its entirety herein). As taught in U.S. Pat. No. 7,589,170,
cyclisation can be
accomplished, for example by disulfide bond formation between two side chain
functional
groups, amide or ester bond formation between one side chain functional group
and the
backbone alpha-amino or carboxyl function, amide or ester bond formation
between two
side chain functional groups, amide bond formation between the backbone alpha-
amino
and carboxyl functions, or via a linker connecting two or more positions of
the peptide.
[0368] A
portion of a peptide can be cyclized, or optionally, the entire peptide
can be cyclized, thereby forming a cyclic peptide. Thus, in some embodiments,
the N
terminus of the peptide is bonded to the C terminus of the peptide, thereby
cyclizing the
entire peptide. In some embodiments, the N terminus is bonded to the C
terminus via an
alpha-amide linkage. In some embodiments, the N terminus is bonded to the C
terminus
via a non-alpha-amide linkage, for example a bond between the side chain of a
Ser (S) or
98
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
Thr(T) and the C-terminal carboxyl group, a disulfide bond between two Cys (C)
residues, or a thioether between a Trp (W) and Cys (C) residue, or a synthetic
linker
molecule. In some embodiments, the C terminus is bonded to an internal amino
acid via a
non-alpha-amide linkage, for example, a bond between the side chain of a Ser
(S) or
Thr(T) and the C-terminal carboxyl group, or a synthetic linker molecule. In
some
embodiments, the N terminus or the C terminus is bonded to an internal amino
acid, or
two internal amino acids are bonded to each other via a non-alpha-amide
linkage, for
example a disulfide bond between two Cys (C) residues, or a thioether between
a Trp (W)
and Cys (C) residue.
[0369] In some
embodiments, a cyclic peptide immunoregulatory peptide
inhibitor includes a single cyclic polypeptide structure. In some embodiments,
a cyclic
peptide immunoregulatory peptide inhibitor includes two or more cyclic
polypeptide
structures, for example 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, or 12 cyclic
polypeptide structures.
Each cyclic polypeptide structure can include at least two amino acid
residues, for
example, about 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19,
20, 25, 26, 27,
28, 29, 30, 35, or 40 amino acid residues or a range that is defined by any
two of these
numbers.
[0370] In some
embodiments, a library of cyclic peptides is screened for cyclic
peptides that bind to albumin-derived immunoregulatory peptides, for example,
the
peptides of Tables 1-4 or 5.4 (SEQ ID NOs: 183-184, 188-246). Screening of
cyclic
peptides libraries is described in PCT Publication WO 95/09344, hereby
incorporated by
reference in its entirety. In some embodiments, a library of cyclic peptides
is synthesized.
In some embodiments, each looped peptide in the library has the same length,
for example
5-meres, 6-meres, 7-meres, 8-meres, 9-meres, 10-meres, 11-meres, or 12-meres.
In some
embodiments, the library includes cyclic peptides of two or more lengths. As
shown in
Example 12, a library of 6-meres was synthesized and was screened for peptides
that bind
to P3038. Positional scans (i.e., single amino acid substitutions at each
position) of a lead
cyclic peptide (SEQ ID NO: 265) identified as exhibiting appreciable binding
to P3028
were performed to identify additional cyclic 6-meres that bind to P3028. It
was observed
that the two 6-meres that bound to P3028 with the highest affinity (SEQ ID
NOs: 266-
267) had homology to linear peptides that bind to P3028 (see Figure 32). Thus,
it is
contemplated herein that aspects of linear peptides that bind to albumin-
derived
99
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
immunoregulatory peptides can be incorporated into cyclic peptides, thus
producing
cyclic peptides that bind albumin-derived immunoregulatory peptides.
[0371] In some embodiments, inhibitors of albumin-derived
immunoregulatory peptides or structures, or a portion thereof is cyclized. In
some
embodiments, a peptide of any of SEQ ID NOs: 1-33, 34, 46-53, 64-66, 68, 76,
94-96,
98, 264-393, 583-586, or 589 or any one or more of the peptides provided in
Table 5.1,
5.4, 5.5, 5.6, or any variation or combination of variations of P28R or P28
core as
provided in Tables 5.3 and 13, or a portion thereof is modified to facilitate
cyclization. In
some embodiments, amino residues containing side chains that can for cyclic
structures,
for example Cysteine, are added to the N terminus, C terminus, and/or internal
positions
of any of the peptide of SEQ ID NOs: 1-33, 34, 46-53, 64-66, 68, 76, 94-96,
98, 264-
393, 583-586, or 589 or any one or more of the peptides provided in Table 5.1,
5.4, 5.5,
5.6, or any variation or combination of variations of P28R or P28 core as
provided in
Tables 5.3 and 13.
Aptamers
[0372] Aptamers
are small molecules that specifically bind to a target
molecule. Aptamers can include oligonucleotide aptamers, for example DNA, RNA,
or
synthetic oligonucleotides. In some embodiments, oligonucleotide aptamers
include
oligonucleotides with a synthetic backbone, for example morpholinos. Aptamers
can also
include peptide aptamers. Aspects of the invention include a composition that
comprises,
consists of, or consists essentially of an aptamer (e.g., nucleic acid based
or peptide
based), wherein said aptamer corresponds or mimics one or more of the
exemplary
immunoregulatory peptide inhibitors that bind to the P3028 sequence/structure
provided
herein (e.g., any one or more of the immunoregulatory peptide inhibitors
provided by of
SEQ ID NOs: 1-33, 34, 46-53, 64-66, 68, 76, 94-96, 98, 264-393, 583-586, or
589 or any
one or more of the peptides provided in Table 5.1, 5.4, 5.5, 5.6, or any
variation or
combination of variations of P28R or P28 core as provided in Tables 5.3 and
13). Some
embodiments of the invention include aptamers that bind specifically to the
P3028
sequence/structure.
[0373] Some
embodiments include a library of oligonucleotide aptamers.
Oligonucleotide aptamers that bind to the P3028 sequence/structure can be
readily
developed given the teachings described herein. As described in US Pat No:
7,745,607,
100
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
which is hereby expressly incorporated by reference in its entirety herein, an
aptamer that
binds specifically to a target, for example the P3028 sequence/structure can
be identified
by interacting an antisense oligonucleotide with a library oligonucleotide
having a
complementary antisense binding domain to form a double stranded duplex, said
library
oligonucleotide further having a random nucleotide domain; ii) immobilizing
the duplex
structure on a solid support; iii) incubating the duplex structure in the
presence of the
P3028 sequence/structure; and iv) collecting library oligonucleotides that
dissociate from
the duplex structure and bind to the P3028 sequence/structure. Alternatively,
a library of
oligonucleotides can be provided in which the library oligonucleotide is
hybridized to a
biotinylated antisense oligonucleotide to form a duplex molecule. The duplex
molecules
are immobilized on a surface, for example avidin-coated beads. A target, such
as P3028
is provided and contacted with the oligonucleotides. Oligonucleotides which
have bound
to the target, are collected and amplified. Similar screening approaches can
be used to
identify peptide-based aptamers that bind to the P3028 sequence/structure.
Peptide based
aptamers that bind to the P3028 sequence/structure, can mimic the
immunoregulatory
peptide inhibitors described herein (e.g., any one or more of SEQ ID NOs: 1-
33, 34, 46-
53, 64-66, 68, 76, 94-96, 98, 264-393, 583-586, or 589 or any one or more of
the peptides
provided in Table 5.1, 5.4, 5.5, 5.6, or any variation or combination of
variations of P28R
or P28 core as provided in Tables 5.3 and 13), and variants thereof. The
section below
discusses many of the modifications that can be incorporated in an
immunoregulatory
peptide inhibitor described herein.
Modifications
[0374]
Embodiments described herein also include a composition that
comprises, consists of, or consists essentially of one or more of the
exemplary isolated
immunoregulatory peptide inhibitors that bind to the P3028 sequence/structure
provided
herein (e.g., any one or more of the immunoregulatory peptide inhibitors
provided by
(SEQ ID NOs: 1-33, 34, 46-53, 64-66, 68, 76, 94-96, 98, 264-393, 583-586, or
589 or
any one or more of the peptides provided in Table 5.1, 5.4, 5.5, 5.6, or any
variation or
combination of variations of P28R or P28 core as provided in Tables 5.3 and
13),
wherein said immunoregulatory inhibitors comprise at least one modification
(e.g.,
glycosylation, nitrosylation, a cytotoxin, a detectable moiety, or a
radionuclide).
Glycosylation can include the addition of polyethylene glycol (PEG). The
addition of
101
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
PEG can increase the solubility of one or more of the immunoregulatory peptide
inhibitors described herein in aqueous solution, protect the molecule from
attack by a
host's immune system, and/or increase the half-life of the molecule in the
host.
[0375] In some
embodiments, the immunoregulatory peptide inhibitors are
directly bound to a cytotoxin. In some embodiments, a peptide consisting of,
consisting
essentially of, or comprising one of SEQ ID NOs: 1-33, 34, 46-53, 64-66, 68,
76, 94-96,
98, 264-393, 583-586, or 589 or any one or more of the peptides provided in
Table 5.1,
5.4, 5.5, 5.6, or any variation or combination of variations of P28R or P28
core as
provided in Tables 5.3 and 13 is covalently bound to a cytotoxin. In some
embodiments,
the immunoregulatory peptide inhibitor is attached to the toxin via a linker.
In some
embodiments, a peptide consisting of, consisting essentially of, or comprising
one of SEQ
ID NOs: 1-33, 34, 46-53, 64-66, 68, 76, 94-96, 98, 264-393, 583-586, or 589 or
any one
or more of the peptides provided in Table 5.1, 5.4, 5.5, 5.6, or any variation
or
combination of variations of P28R or P28 core as provided in Tables 5.3 and 13
is
attached to a cytotoxin via a linker. A wide array of linker technologies can
be
employed. Linkers can be cleavable or non-cleavable. It is known that in many
cases, the
full cytotoxic potential of a drug can be observed when the cytotoxic
molecules are
released from a conjugates, for example an inhibitor of an immunoregulatory
peptide, in
unmodified form at the target site. One of the cleavable linkers that has been
employed
for the preparation of cytotoxin conjugates is an acid-labile linker based on
cis-aconitic
acid that takes advantage of the acidic environment of different intracellular
compartments such as the endosomes encountered during receptor mediated
endocytosis
and the lysosomes. Shen and Ryser introduced this method for the preparation
of
conjugates of daunorubicin with macromolecular carriers (Biochem. Biophys.
Res.
Commun. 102:1048-1054 (1981)). Yang and Reisfeld used the same technique to
conjugate daunorubicin to an anti-melanoma antibody (J. Natl. Canc. Inst.
80:1154-1159
(1988)). Recently, Dillman et al. also used an acid-labile linker in a similar
fashion to
prepare conjugates of daunorubicin with an anti-T cell antibody (Cancer Res.
48:6097-
6102 (1988)). An alternative approach, explored by Trouet et al. involved
linking
daunorubicin to a targeting molecule via a peptide spacer arm (Proc. Natl.
Acad. Sci.
79:626-629 (1982)). This was done under the premise that free drug could be
released
from such a conjugate by the action of lysosomal peptidases. One skilled in
the art will
appreciate that cleavable linker approaches employed for conjugating
cytotoxins to
102
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
antibodies can also be employed to conjugate a peptide, for example one of SEQ
ID
NOs: 1-33, 34, 46-53, 64-66, 68, 76, 94-96, 98, 264-393, 583-586, or 589 or
any one or
more of the peptides provided in Table 5.1, 5.4, 5.5, 5.6, or any variation or
combination
of variations of P28R or P28 core as provided in Tables 5.3 and 13 to a
cytotoxin.
[0376]
Exemplary cytotoxins that can be incorporated into one or more of the
exemplary immunoregulatory peptide inhibitors that bind to the P3028
sequence/structure
provided herein (e.g., any one or more of the immunoregulatory peptide
inhibitors
provided by SEQ ID NOs: 1-33, 34, 46-53, 64-66, 68, 76, 94-96, 98, 264-393,
583-586,
or 589 or any one or more of the peptides provided in Table 5.1, 5.4, 5.5,
5.6, or any
variation or combination of variations of P28R or P28 core as provided in
Tables 5.3 and
13) include: radiotoxins, monomethylauristatin-E, monomethylauristatin-F,
aplidin,
azaribine, anastrozole, azacytidine, bleomycin, bortezomib, bryostatin-1,
busulfan,
calicheamycin, camptothecin, 10-hydroxycamptothecin,
c armus tine, celebrex,
chlorambucil, cisplatin, irinotec an (CPT-11), S N-38, carboplatin,
cladribine,
cyclophosphamide, cytarabine, dacarbazine, docetaxel, dactinomycin, daunomycin
glucuronide, daunorubicin, dexamethasone, diethylstilbestrol, doxorubicin,
doxorubicin
glucuronide, epirubicin glucuronide, ethinyl estradiol, estramustine,
etoposide, etoposide
glucuronide, etoposide phosphate, floxuridine (FUdR), 3',5'--0-dioleoyl-FudR
(FUdR-
d0), fludarabine, flutamide, fluorouracil, fluoxymesterone, gemcitabine,
hydroxyprogesterone caproate, hydroxyurea, idarubicin, ifosfamide, L-
asparaginase,
leucovorin, lomustine, mechlorethamine, medroprogesterone acetate, megestrol
acetate,
melphalan, mercaptopurine, 6-mercaptopurine, methotrexate, mitoxantrone,
mithramycin,
mitomycin, mitotane, phenyl butyrate, prednisone, procarbazine, paclitaxel,
pentostatin,
PSI-341, saporin, semustine streptozocin, tamoxifen, taxanes, testosterone
propionate,
thalidomide, thioguanine, thiotepa, teniposide, topotecan, uracil mustard,
velcade,
vinblastine, vinorelbine, vincristine, ricin, for example ricin A chain,
abrin, ribonuclease,
onconase, rapLR1, DNase I, Staphylococcal enterotoxin-A, pokeweed antiviral
protein,
gelonin, diphtheria toxin, Pseudomonas exotoxin, and Pseudomonas endotoxin.
Optionally, the cytotoxin is conjugated to the immunoregulatory peptide
inhibitor via a
linker (e.g. a cleavable or non-cleavable linker) or non-covalent
immobilization as
described herein. Optionally, a composition comprising the immunoregulatory
peptide
inhibitor immobilized on a nanoparticle is provided as described herein, and
the cytotoxin
is immobilized on the nanoparticle (e.g. each of the immunoregulatory peptide
inhibitor
103
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
immobilized and cytotoxin can be immobilized on the nanoparticle, but not
necessarily
connected to each other).
Optionally, the immunoregulatory peptide inhibitor
immobilized on the nanoparticle by a first linker, and the cytotoxin is
immobilized on the
nanoparticle via a second linker that can be the same as or different from the
first
linker. [0435]
Exemplary detectable moieties (which may also be referred to herein as
"detectable labels" that can be incorporated into one or more of the exemplary
immunoregulatory peptide inhibitors that bind to the P3028 sequence/structure
provided
herein (e.g., any one or more of the immunoregulatory peptide inhibitors
provided by
SEQ ID NOs: 1-33, 34, 46-53, 64-66, 68, 76, 94-96, 98, 264-393, 583-586, or
589 or any
one or more of the peptides provided in Table 5.1, 5.4, 5.5, 5.6, or any
variation or
combination of variations of P28R or P28 core as provided in Tables 5.3 and
13) or to an
antibody that binds specifically to P3028 include: a radiolabel, a
fluorophore, biotin, a
fluorescent protein, colloidal gold, and/or a coenzyme. Radiolabels can
include 3H and
14C. Fluorophores can include Alexa-Fluor dyes, Pacific Blue, Pacific Orange,
Cascade
Blue, Cascade Yellow and R-phycoerythrin, fluorescein (FITC), rhodamine, Texas
red,
BODIPY family dyes, Cy2, Cy3, C5, and Cy7. Fluorescent proteins can include
Blue,
Cyan, Green, Yellow, and Red fluorescent proteins. In some embodiments,
fluorescent
labels include a FRET pair. For example, a single peptide can be attached to a
FRET
donor and FRET acceptor, which are configured so that the FRET acceptor is
substantially within a FRET radius of the FRET donor when the peptide is in a
first
configuration (for example, bound to target), but not when the peptide is in a
second
configuration (for example, unbound to target). For example, a first peptide
can be
attached to a FRET acceptor, and a second peptide can be attached to a FRET
donor, so
that the FRET acceptor is substantially within a FRET radius of the FRET donor
when the
first peptide and second peptide are each bound to a target, for example a
target cell, but
not when at least one peptide is unbound to the target. In some embodiments,
fluorescent
label includes a fluorophore and a quencher. The fluorophore and quencher can
each be
attached to the peptide so that the quench absorbs electromagnetic radiation
emitted by
the fluorophore when the peptide is in a first configuration (for example,
bound to target),
but not when the peptide is in a second configuration (for example, unbound to
target).
Coenzymes can include vitamins such as biotin.
[0377]
Exemplary radionuclides that can be incorporated into one or more of
the exemplary immunoregulatory peptide inhibitors that bind to the P3028
104
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
sequence/structure provided herein (e.g., any one or more of the
immunoregulatory
peptide inhibitors provided by SEQ ID NOs: 1-33, 34, 46-53, 64-66, 68, 76, 94-
96, 98,
264-393, 583-586, or 589 or any one or more of the peptides provided in Table
5.1, 5.4,
5.5, 5.6, or any variation or combination of variations of P28R or P28 core as
provided in
Tables 5.3 and 13) or an antibody that binds specifically to P3028 include:
1111, 1311, 90Y,
67Cu, "6Re, issRe, 212Bi or 211At. Preferable radiolabeled immunoregulatory
peptide
inhibitors are able to deliver more than 6000 rads to a tumor, for example,
and have
sufficient affinity so that the patient's bone marrow is not exposed to more
than 300 rads.
The section that follows describes in greater detail some of the embodiments,
which
encompass protein complexes comprising an immunoregulatory peptide inhibitor
described herein.
[0378] In some
embodiments, a diagnostic kit is provided. The kit can include
any one or more of the immunoregulatory peptide inhibitors provided by SEQ ID
NOs:
1-33, 34, 46-53, 64-66, 68, 76, 94-96, 98, 264-393, 583-586, or 589 or any one
or more of
the peptides provided in Table 5.1, 5.4, 5.5, 5.6, or any variation or
combination of
variations of P28R or P28 core as provided in Tables 5.3 and 13 or an antibody
that
binds specifically to any of the peptides of SEQ ID NOs: 183-185 or 188-246,
for
example P3028 (SEQ ID NO: 185). The kit can also include a detectable moiety
as
described herein. In some embodiments, the peptide inhibitor or antibody of
the kit is
biotinylated.
Carrier molecules
[0379] Some
embodiments include a carrier molecule. Carrier molecules, can
for example, increase the stability or half-life, increase the solubility,
increase the
absorption, target the peptide to an appropriate cell, organ or tissue, and/or
minimize an
immune response against a therapeutic molecule.
[0380]
Exemplary carrier molecules include human serum albumin; a polymer
having a plurality of acid moieties (see PCT Pub. No. WO 01/93911); anionic
group-
containing amphiphilic block copolymers that, when used as a drug carrier for
a cationic
therapeutic molecule can improve stability of that molecule (see PCT Pub. No.
WO
03/00778); cyclodextrin and acids for improving the properties of basic
therapeutic
molecules (European Pat. No. 0 681 481); lipids as carriers for hydrophobic
therapeutic
molecules (see PCT Pub. No. WO 04/064731); immunoglobulins; and Fc fragments
as
105
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
carriers for improving half-life and minimizing immune response (see U.S. Pat.
No.
7,736,653). In some embodiments, an immunoregulatory peptide inhibitor (e.g.,
a peptide
comprising, consisting of, or consisting essentially of SEQ ID NOs: 1-33, 34,
46-53, 64-
66, 68, 76, 94-96, 98, 264-393, 583-586, or 589 or any one or more of the
peptides
provided in Table 5.1, 5.4, 5.5, 5.6, or any variation or combination of
variations of P28R
or P28 core as provided in Tables 5.3 and 13) includes or is joined to a
carrier. In some
embodiments, an immunoregulatory peptide inhibitor (e.g. a peptide comprising,
consisting of, or consisting essentially of SEQ ID NOs: 1-33, 34, 46-53, 64-
66, 68, 76,
94-96, 98, 264-393, 583-586, or 589 or any one or more of the peptides
provided in Table
5.1, 5.4, 5.5, 5.6, or any variation or combination of variations of P28R or
P28 core as
provided in Tables 5.3 and 13) includes two or more carriers.
[0381] In some
embodiments, an immunoregulatory peptide inhibitor is
provided with a degradable particle. Without being limited by any theory, it
is
contemplated that a degradable particle can permit an immunoregulatory
particle to be
soluble and exert its activity for a controlled period of time in the systemic
circulation.
Accordingly, in some embodiments, a degradable particle comprising an
immunoregulatory peptide inhibitor (for example, a peptide comprising,
consisting of, or
consisting essentially of SEQ ID NOs: 1-33, 34, 46-53, 64-66, 68, 76, 94-96,
98, 264-
393, 583-586, or 589 or any one or more of the peptides provided in Table 5.1,
5.4, 5.5,
5.6, or any variation or combination of variations of P28R or P28 core as
provided in
Tables 5.3 and 13) is provided. In some embodiments the degradable particle
comprising
the immunoregulatory peptide inhibitor is administered to a subject in need.
Optionally,
the degradable particle can be administered systemically. Optionally, the
degradable
particle can be administered locally, for example at or near a site of
immunosuppression
(e.g. within 10cm, 9cm, 8cm 7cm, 6c, 5cm, 4cm, 3cm, 2cm, 1 cm, or 0.5cm of the
site of
immunosuppression or a range defined by any two of these numbers). In some
embodiments, the subject suffers from LFA-1 receptor blockage by an
immunoregulatory
peptide sequence of any of Tables 1-4. Optionally, the degradable particle can
be
coadministered with one or more additional therapeutic agents. For example, if
a the
immunoregulatory peptide inhibitor is useful for de-blocking an LFA-1 receptor
(e.g.
displaces bound immunoregulatory peptides or P3028 structures from the LFA-1
receptor), a therapeutic agent that stimulates an immune response, for example
via an
LFA-1 receptor can be useful for co-administering with the immunoregulatory
peptide
106
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
inhibitor and degradable particle. In some embodiments, the additional
therapeutic agent
is administered at the same time as the immunoregulatory peptide inhibitor,
for example
as part of the degradable particle. In some embodiments, the additional
therapeutic agent
is administered after the immunoregulatory peptide inhibitor, for example at
least about 1,
2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22,
23, 24 hours
afterwards, or about 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17,
18, 19, 20, 21, 22,
23, 24, 25, 26, 27, 28, 29, 30, or 31 days afterwards or a range defined by
any two of
aforementioned times. A variety of suitable degradable particles can be used
in
accordance with embodiments herein. In some embodiments, the degradable
particle
comprises a sphere, for example a microsphere. In some embodiments, the
degradable
particle comprises a nanoparticle. In some embodiments, the degradable
particle
comprises a starch or sugar. In some embodiments, the degradable particle
comprises an
organic polymer or a combination of organic polymers, for example, polyesters,
polyphosphate esters, polyphosphazenes, polyorthoesters, polyanhydrides,
polycarbonates,
polyamides, poly-lactic acid, a poly-glycoloyic acid, or a combination of two
or more
polymers, for example two or more of the listed polymers.
Protein Complexes
[0382] Some
embodiments include a composition comprising an isolated
protein complex that comprises an immunoregulatory peptide inhibitor. The
isolated
protein complex can include an immunoregulatory peptide, for example P3028
(SEQ ID
NO: 185) or any one or more of the immunoregulatory peptides described in
Tables 1-4
(SEQ ID NOs: 183-184 and 188-246) and at least one immunoregulatory peptide
inhibitor (e.g., any one or more of the peptides provided in Table 5.1). In
some
embodiments, the isolated protein complex includes peptide 3028 (SEQ ID NO:
185) and
an inhibitor peptide that includes the sequence of SEQ ID NOs: 1-33, 34, 46-
53, 64-66,
68, 76, 94-96 or 98 or any one or more of the peptides provided in Table 5.1.
Exemplary
protein complexes that include each of the peptides SEQ ID NOs: 1-33, 34, 46-
53, 64-66,
68, 76, 94-96, 98, 264-393, 583-586, or 589 or any one or more of the peptides
provided
in Table 5.1, 5.4, 5.5, 5.6, or any variation or combination of variations of
P28R or P28
core as provided in Tables 5.3 and 13 bound to the P3028 sequence/structure
are
provided in Examples 10, 11 and 12 and Table 5.1. The protein complex can
include at
least one favorable electrostatic interaction between an amino acid residue of
P3028 or a
107
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
variant thereof, and an amino acid of an inhibitor peptide or peptide mimetic.
The protein
complex can include at least one favorable hydrophobic interaction between an
amino
acid residue of P3028 or a variant thereof, and an amino acid of an inhibitor
peptide or
peptide mimetic (see Example 11). In some embodiments, the protein complex
includes
a variant of P3028 having at least about 80% identity to P3028, for example
greater than
or equal to about 80%, 81%, 82%, 83%, 84%, 85%, 86%, 87%, 88%, 89%, 90%, 91%,
92%, 93%, 94%, 95%, 96%, 97%, 98%, or 99% identity to P3028. The protein
complex
can further include at least one protein bound to a cancer cell, for example a
surface
protein. Thus, in some embodiments, the isolated protein complex can localize
to the
surface of a cancer cell.
[0383]
Accordingly, some embodiments include a method of making a protein
complex that comprises one or more of the immunoregulatory peptide inhibitors
described herein. The methods can be practiced, for example, by binding an
immunoregulatory peptide inhibitor, as described herein to P3028, or a variant
or
fragment thereof. The method can optionally include detecting the presence of
the
complex, which can be accomplished by rampo studies, as described herein.
[0384] Some
embodiments include methods of binding a peptide comprising,
consisting or, or consisting essentially of SEQ ID NOs: 1-33, 34, 46-53, 64-
66, 68, 76,
94-96, 98, 264-393, 583-586, or 589 or any one or more of the peptides
provided in Table
5.1, 5.4, 5.5, 5.6, or any variation or combination of variations of P28R or
P28 core as
provided in Tables 5.3 and 13 to a molecule that comprises the P3028
sequence/structure
(SEQ ID NO: 185). Some embodiments include methods of binding a peptide
comprising, consisting of, or consisting essentially of at least one of SEQ ID
NOs: 1-33,
34, 46-53, 64-66, 68, 76, 94-96, 98, 264-393, 583-586, or 589 or any one or
more of the
peptides provided in Table 5.1, 5.4, 5.5, 5.6, or any variation or combination
of variations
of P28R or P28 core as provided in Tables 5.3 and 13 to a molecule comprising
a variant
of the P3028 sequence/structure (SEQ ID NO: 185). Some embodiments include
methods of binding a peptide including at least one of SEQ ID NOs: 1-33, 34,
46-53, 64-
66, 68, 76, 94-96, 98, 264-393, 583-586, or 589 or any one or more of the
peptides
provided in Table 5.1, 5.4, 5.5, 5.6, or any variation or combination of
variations of P28R
or P28 core as provided in Tables 5.3 and 13 to a protein that comprises the
P3028
sequence/structure or a fragment of P3028 (SEQ ID NO: 185), wherein the
fragment of
108
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
P3028 has a length of at least about 10 amino acids, more preferably 11 amino
acids,
more preferable 12 amino acids, more preferably 13 amino acids, more
preferably 14
amino acids, more preferably 15 amino acids, more preferably 16 amino acids,
or more
preferably 17 amino acids. In some embodiments, the binding includes favorable
hydrophilic and/or electrostatic interactions between members of the protein
complex. In
some embodiments, the binding includes covalent bonds between members of the
protein
complex, for example through crosslinking. Crosslinking can be induced
chemically,
and/or via electromagnetic radiation, for example electromagnetic radiation in
the
ultraviolet spectrum.
[0385] In some
embodiments, the peptide comprises at least one of SEQ ID
NOs: 1-33, 34, 46-53, 64-66, 68, 76, 94-96, 98, 583-586, or 589 or any one or
more of the
peptides provided in Table 5.1, 5.4, 5.5, 5.6, or any variation or combination
of variations
of P28R or P28 core as provided in Tables 5.3 and 13. Exemplary supports
include a pin,
bead, surface, matrix, artificial cell surface, or cell surface. For example,
the peptide can
be affixed via an affinity tag to a support. In some embodiments, P3028, or a
variant or
fragment thereof is affixed to a support. In some embodiments, the peptide
including at
least one of SEQ ID NOs: 1-33, 34, 46-53, 64-66, 68, 76, 94-96, 98, 583-586,
or 589 or
any one or more of the peptides provided in Table 5.1, 5.4, 5.5, 5.6, or any
variation or
combination of variations of P28R or P28 core as provided in Tables 5.3 and 13
is
affixed to a support, and P3028 or a variant or fragment thereof is dissolved
in a solvent.
In some embodiments, the peptide including at least one of SEQ ID NOs: 1-33,
34, 46-
53, 64-66, 68, 76, 94-96, 98, 583-586, or 589 or any one or more of the
peptides provided
in Table 5.1, 5.4, 5.5, 5.6, or any variation or combination of variations of
P28R or P28
core as provided in Tables 5.3 and 13 is dissolved in a solvent, and P3028, or
a variant or
fragment thereof is affixed to a support. In some embodiments, the peptide
including at
least one of SEQ ID NOs: 1-33, 34, 46-53, 64-66, 68, 76, 94-96, 98, 583-586,
or 589 or
any one or more of the peptides provided in Table 5.1, 5.4, 5.5, 5.6, or any
variation or
combination of variations of P28R or P28 core as provided in Tables 5.3 and 13
and
P3028 are each dissolved in a solvent, for example serum.
[0386] In some
embodiments, the binding occurs in an organism, for example
in extracellular matrix, and/or serum or in a biological sample obtained from
an organism,
such as a human or a non-human mammal. Biological samples can include at least
one
cell, tissue, or extracellular composition of an organism, include extracts,
purified
109
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
extracts, and/or fractions thereof. Exemplary biological samples include whole
blood,
serum, bone marrow, isolated immune cells, and tumor biopsies. Isolated immune
cells
can include leukocytes, and peripheral blood mononuclear cells (PBMC's), for
example
lymphocytes, monocytes, or macrophages. The method can include delivering at
least one
member of the complex, for example a peptide including at least one of SEQ ID
NOs: 1-
33, 34, 46-53, 64-66, 68, 76, 94-96, 98, 583-586, or 589 or any one or more of
the
peptides provided in Table 5.1, 5.4, 5.5, 5.6, or any variation or combination
of variations
of P28R or P28 core as provided in Tables 5.3 and 13, to the organism. In some
embodiments, the binding occurs in vitro, for example in a buffer solution or
in a
biological sample. The method can include adding at least one member of the
complex,
for example a peptide including at least one of SEQ ID NOs: 1-33, 34, 46-53,
64-66, 68,
76, 94-96, 98, 583-586, or 589 or any one or more of the peptides provided in
Table 5.1,
5.4, 5.5, 5.6, or any variation or combination of variations of P28R or P28
core as
provided in Tables 5.3 and 13, to a solution that contains the remaining
members of the
complex. Alternatively, the method can include adding two or more members of
the
complex to a solution for example a peptide including at least one of SEQ ID
NOs: 1-33,
34, 46-53, 64-66, 68, 76, 94-96, 98õ 583-586, or 589 or any one or more of the
peptides
provided in Table 5.1, 5.4, 5.5, 5.6, or any variation or combination of
variations of P28R
or P28 core as provided in Tables 5.3 and 13 and P3028 or a fragment or
variant thereof.
In some embodiments, a peptide including at least one of SEQ ID NOs: 1-33, 34,
46-53,
64-66, 68, 76, 94-96, 98, 583-586, or 589 or any one or more of the peptides
provided in
Table 5.1, 5.4, 5.5, 5.6, or any variation or combination of variations of
P28R or P28 core
as provided in Tables 5.3 and 13 is added to a biological sample.
[0387] Some
embodiments include detecting the presence of the complex.
Some embodiments include detecting the presence of the P3028
sequence/structure bound
to a peptide that is affixed to a support (see Example 12), for example by
ELISA. Some
embodiments include detecting the presence of a complex by FRET. For example a
FRET donor fluorophore can be attached to a first member of the complex, and a
FRET
acceptor fluorophore can be attached to a second member of the complex, so
that FRET
transfer occurs only when the complex is formed. Some embodiments include
detecting
the presence of a complex by cessation of quenching. For example a member of
the
complex can be attached to a fluorophore and a quencher for electromagnetic
radiation
110
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
emitted by the fluorophore, so that when the complex member is unbound, the
fluorophore is substantially within the quencher radius, and the quencher
absorbs
electromagnetic radiation emitted by the fluorophore (e.g., a quencher can be
attached to
the N terminal and a fluorophore attached to the C terminal, or a quencher can
be attached
to the C terminal, and a fluorophore attached to the N terminal). Upon complex
formation, the fluorophore can be outside of the quencher radius, thus
permitting
detection of electromagnetic radiation emitted by the fluorophore.
[0388] Some
embodiments include detecting the presence of the complex by
detecting of complex function. For example, an immune cell in which peptide
3028 is
bound to the LFA-1 and/or IL-2 receptor can exhibit reduced IL-2-induced
proliferation,
T cell receptor stimulation, leukocyte spreading, immune cell migration,
and/or NK cell
cytotoxicity (see Examples 2-6). Direct or indirect detection of increased IL-
2-induced
proliferation, T cell receptor stimulation, leukocyte spreading, immune cell
migration,
and/or NK cell cytotoxicity, for example increase in comparison to an
untreated control
sample in which at least one member of the complex was not added, can detect
complex
formation. For example, as shown in Example 13, the formation of a complex
between
the P3028 sequence/structure and an immunoregulatory peptide inhibitor can
increase
lymphocyte stimulation. For example, as shown in Example 1, the formation of a
complex can unblock theLFA-1 receptor. Thus, some embodiments include
detecting
complex formation indirectly by, for example, detecting increased lymphocyte
stimulation, detecting unblocked LFA-1 receptor, and/or detecting immune cell
stimulation via an unblock LFA-1 receptor, as compared to a control sample
that is
known to lack complex formation.
[0389] Some
embodiments include detecting the presence of the complex by
detecting localization of complex members. In some embodiments, detecting the
presence of the complex includes detecting the presence of an immunoregulatory
peptide
inhibitor including at least one of SEQ ID NOs: 1-33, 34, 46-53, 64-66, 68,
76, 94-96,
98, 264-393, 583-586, or 589 or any one or more of the peptides provided in
Table 5.1,
5.4, 5.5, 5.6, or any variation or combination of variations of P28R or P28
core as
provided in Tables 5.3 and 13, or a peptidomimetic that binds specifically to
the P3028
sequence/structure on tumor cells. As shown in Example 1, the P3028
sequence/structure
can bind to tumor cells. As shown in Example 14, an inhibitor of the P3028
sequence/structure can bind to tumor cells, for example by binding to the
P3028
111
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
sequence/structure. Thus, in some embodiments, the presence of an inhibitor of
the
P3028 sequence/structure, for example, at least one of SEQ ID NOs: 1-33, 34,
46-53, 64-
66, 68, 76, 94-96, 98, 264-393, 583-586, or 589 or any one or more of the
peptides
provided in Table 5.1, 5.4, 5.5, 5.6, or any variation or combination of
variations of P28R
or P28 core as provided in Tables 5.3 and 13 on a tumor cell can indicate
complex
formation. Thus, complex formation can be detected by colocalization of an
inhibitor
with at least one marker of a tumor cell. Colocalization can be detected, for
example by
immunohistochemistry or flow cytometry. In some embodiments, the inhibitor is
labeled,
for example with a fluorophore or radiolabel. In some embodiments, the
inhibitor is
detected, for example with a primary antibody that specifically binds to the
inhibitor. The
section that follows describes in greater detail some of the nucleic acid
embodiments,
which encode an immunoregulatory peptide inhibitor.
Nucleic acids encoding inhibitor peptides
[0390] Some
embodiments include isolated nucleic acids encoding an
immunoregulatory peptide inhibitor. One skilled in the art will appreciate
that for a given
peptide sequence, a nucleic acid sequence encoding that peptide sequence can
readily be
determined, and due to the degeneracy of the genetic code, more than one
nucleic acid
sequence can encode any one peptide. A nucleic acid sequence encoding a
peptide can be
incorporated into an expression vector using known techniques, as well.
Expression
vectors can be used to produce the peptide in an expression system, for
example a host
cell, a host organism, a cell-free expression system, and the like. Expression
vectors can
also be used to produce a peptide in an organism, for example a patient in
need of
blocking of immunosuppression, as described herein. Exemplary expression
vectors
include plasmid DNA, such as a pVAX construct, bacteriophage DNA, cosmid DNA,
artificial chromosomes such as BACs and YACs, retrovirus systems, for example
lentivirus, DNA virus systems, for example adenovirus or vaccinia virus (e.g.,
MVA).
For peptides that do not have an N-terminal amino acid that corresponds to a
translation
start codon (typically Met corresponding to ATG), expression vectors can
include an in-
frame translation start codon. Such an amino acid can be separated from the N-
terminal
of the peptide by a cleavable linker, for example a peptide sequence that is
cleaved by a
protease. Expression vectors can include transcriptional regulatory sequences,
for
example core promoters, transcriptional enhancers, and/or insulator sequences.
Such
112
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
sequences can facilitate the assembly of transcriptional machinery (for
example RNA
Polymerase III), and the subsequent production of a transcript encoding the
peptide (for
example, by facilitating a heterochromatic environment that is favorable to
transcription).
[0391] In some
embodiments, an expression vector encodes two or more
copies of a peptide, and/or two or more unique peptides. In some embodiments,
an
expression vector encodes two or more peptides, and each peptide is under the
control of
a unique transcription unit (e.g., promoter, transcriptional enhancers, and/or
transcription
terminator). In some embodiments, a nucleic acid encoding two or more peptides
is under
the control of a single transcription unit. In such embodiments, a sequence
encoding an
individual peptide can be under the control of an individual translation start
site, for
example an Internal Ribosome Entry Site (IRES). In such embodiments, a single
nucleic
acid can encode a protein or polypeptide encoding two or more peptides, which
are
separated by at least one protease target site.
[0392] One
skilled in the art will appreciate that polynucleotides encoding
peptides, such as peptide inhibitors, can be readily constructed based upon
the sequence
of the peptide. Exemplary polynucleotides encoding the sequences of
immunoregulatory
peptide inhibitor peptides of (SEQ ID NOs: 2-33) are provided in Table 5.2.
One skilled
in the art will appreciate that due to the degeneracy of the genetic code, a
given
polypeptide can be encoded by more than one polynucleotide may encode. Thus,
provided herein, for example in Table 5.2, are consensus polynucleotides that
account for
typical degeneracy of the genetic code, as well as exemplary polynucleotides.
The
polynucleotides of Table 5.2 are provided by way of example, and include SEQ
ID NOs:
102-165. On skilled in the art will further appreciate that additional
polynucleotides can
encode peptide inhibitors such as the peptide inhibitors disclosed herein
(e.g.,
polynucleotides encoding any one or more of the peptides provided in Table 5.1
are
embodiments). For example, polynucleotides can be modified post-
transcriptionally, for
example by alternative splicing, and/or by enzymes such as RNA-specific
adenosine
deaminase (ADAR) that can modify the bases of polynucleotides.
Table 5.2: Polynucleotides encoding peptide inhibitors of the P3028
sequence/structure
Seq ID Description
NO
102 Consensus polynucleotide encoding P28R (SEQ ID NO: 2)
103 Exemplary NT encoding P28R (SEQ ID NO: 2)
113
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
104 Consensus polynucleotide encoding SEQ ID NO: 3
105 Exemplary NT encoding SEQ ID NO: 3
106 Consensus polynucleotide encoding SEQ ID NO: 4
107 Exemplary NT encoding SEQ ID NO: 4
108 Consensus polynucleotide encoding SEQ ID NO: 5
109 Exemplary NT encoding SEQ ID NO: 5
110 Consensus polynucleotide encoding SEQ ID NO: 6
111 Exemplary NT encoding SEQ ID NO: 6
112 Consensus polynucleotide encoding SEQ ID NO: 7
113 Exemplary NT encoding SEQ ID NO: 7
114 Consensus polynucleotide encoding SEQ ID NO: 8
115 Exemplary NT encoding SEQ ID NO: 8
116 Consensus polynucleotide encoding SEQ ID NO: 9
117 Exemplary NT encoding SEQ ID NO: 9
118 Consensus polynucleotide encoding SEQ ID NO: 10
119 Exemplary NT encoding SEQ ID NO: 10
120 Consensus polynucleotide encoding SEQ ID NO: 11
121 Exemplary NT encoding SEQ ID NO:11
122 Consensus polynucleotide encoding SEQ ID NO: 12
123 Exemplary NT encoding SEQ ID NO: 12
124 Consensus polynucleotide encoding SEQ ID NO: 13
125 Exemplary NT encoding SEQ ID NO: 13
126 Consensus polynucleotide encoding SEQ ID NO: 14
127 Exemplary NT encoding SEQ ID NO: 14
128 Consensus polynucleotide encoding SEQ ID NO: 15
129 Exemplary NT encoding SEQ ID NO: 15
130 Consensus polynucleotide encoding SEQ ID NO: 16
131 Exemplary NT encoding SEQ ID NO: 16
132 Consensus polynucleotide encoding SEQ ID NO: 17
133 Exemplary NT encoding SEQ ID NO: 17
134 Consensus polynucleotide encoding SEQ ID NO: 18
135 Exemplary NT encoding SEQ ID NO: 18
136 Consensus polynucleotide encoding SEQ ID NO: 19
137 Exemplary NT encoding SEQ ID NO: 19
138 Consensus polynucleotide encoding SEQ ID NO: 20
139 Exemplary NT encoding SEQ ID NO: 20
140 Consensus polynucleotide encoding SEQ ID NO: 21
141 Exemplary NT encoding SEQ ID NO: 21
142 Consensus polynucleotide encoding SEQ ID NO: 22
143 Exemplary NT encoding SEQ ID NO: 22
144 Consensus polynucleotide encoding SEQ ID NO: 23
145 Exemplary NT encoding SEQ ID NO: 23
146 Consensus polynucleotide encoding SEQ ID NO: 24
147 Exemplary NT encoding SEQ ID NO: 24
148 Consensus polynucleotide encoding SEQ ID NO: 25
149 Exemplary NT encoding SEQ ID NO: 25
150 Consensus polynucleotide encoding SEQ ID NO: 26
114
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
151 Exemplary NT encoding SEQ ID NO: 26
152 Consensus polynucleotide encoding SEQ ID NO: 27
153 Exemplary NT encoding SEQ ID NO: 27
154 Consensus polynucleotide encoding SEQ ID NO: 28
155 Exemplary NT encoding SEQ ID NO: 28
156 Consensus polynucleotide encoding SEQ ID NO: 29
157 Exemplary NT encoding SEQ ID NO: 29
158 Consensus polynucleotide encoding SEQ ID NO: 30
159 Exemplary NT encoding SEQ ID NO: 30
160 Consensus polynucleotide encoding SEQ ID NO: 31
161 Exemplary NT encoding SEQ ID NO: 31
162 Consensus polynucleotide encoding SEQ ID NO: 32
163 Exemplary NT encoding SEQ ID NO: 32
164 Consensus polynucleotide encoding SEQ ID NO: 33
165 Exemplary NT encoding SEQ ID NO: 33
[0393]
Accordingly, embodiments described herein also include a composition
that comprises, consists of, or consists essentially of an isolated nucleic
acid or
polynucleotide that encodes one or more of the exemplary immunoregulatory
peptide
inhibitors that bind to the P3028 sequence/structure provided herein (e.g.,
any one or
more of the immunoregulatory peptide inhibitors provided by SEQ ID NOs: 1-33,
34, 46-
53, 64-66, 68, 76, 94-96 or 98 or any one or more of the peptides provided in
Table 5.1).
Vectors, constructs, and plasmids comprising the aforementioned nucleic acids
or
polynucleotides are also embodiments. The following section discusses
additional
components that may be included in one or more of the compositions described
herein.
Pharmaceutical compositions
[0394] In some
embodiments, a pharmaceutical composition comprising,
consisting essentially of or consisting of a peptide inhibitor (e.g., any one
or more of the
peptides provided in Table 5.1) is provided. The pharmaceutical composition
can include
a peptide inhibitor as described herein and a pharmaceutically acceptable
ingredient as
described herein. Exemplary pharmaceutically acceptable ingredients include
diluents,
carriers, excipients and/or buffers. In some embodiments, the peptide
inhibitor
comprises, consists of, or consists essentially of a peptide inhibitor as
described herein.
For example, a composition can comprise, consist of, or consist essentially of
a peptide
inhibitor that comprises, consists of, or consists essentially of the peptide
of Formula (I),
XX1VKX2X3X4 (SEQ ID NO: 166). In some embodiments, X is an optional sequence,
115
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
and can be KKLDT (SEQ ID NO: 167), RKLDT (SEQ ID NO: 168), KKGDT (SEQ ID
NO: 169), KKEDT (SEQ ID NO: 170), KKLDQ (SEQ ID NO: 171), KKGDQ (SEQ ID
NO: 252), KKEDQ (SEQ ID NO: 253), RKLDQ (SEQ ID NO: 254), RKGDQ (SEQ ID
NO: 255), RKEDQ (SEQ ID NO: 256), RKGTD (SEQ ID NO: 257), RKEDT (SEQ ID
NO: 258), KLDT (SEQ ID NO: 172), KGDT (SEQ ID NO: 259), KEDT (SEQ ID NO:
260), KLDQ (SEQ ID NO: 261), KGDQ (SEQ ID NO: 262), KEDQ (SEQ ID NO:
263), LDT, LDQ, GDT, GDQ, EDT, EDQ, DT, DQ, T, or Q, or absent. In some
embodiments, Xi is one of FF, FM, FS, FV, FT, FL, AF, AM, AS, AV, AT, AL, VF,
VM,
VS, VV, VT, or VL. In some embodiments, X2 is one of LS, LQ, LM, LT, LH, VS,
VQ,
VM, VT, or VH. In some embodiments, X3 is one of LFT, LMT, LQT, LHT, LNT, LPT,
LST, LGT, LAT, LRT, QFT, QMT, QQT, QHT, QNT, QPT, QST, QGT, QAT, QRT,
VFT, VMT, VQT, VHT, VNT, VPT, VST, VGT, VAT, VRT, MFT, MMT, MQT, MHT,
MNT, MPT, MST, MGT, MAT, MRT, LFN, LMN, LQN, LHN, LNN, LPN, LSN, LGN,
LAN, LRN, QFN, QMN, QQN, QHN, QNN, QPN, QSN, QGN, QAN, QRN, VFN,
VMN, VQN, VHN, VNN, VPN, VSN, VGN, VAN, VRN, MFN, MMN, MQN, MHN,
MNN, MPN, MSN, MGN, MAN, MRN, LFP, LMP, LQP, LHP, LNP, LPP, LSP, LGP,
LAP, LRP, QFP, QMP, QQP, QHP, QNP, QPP, QSP, QGP, QAP, QRP, VFP, VMP,
VQP, VHP, VNP, VPP, VSP, VGP, VAP, VRP, MFP, MMP, MQP, MHP, MNP, MPP,
MSP, MGP, MAP, MRPR, LFR, LMR, LQR, LHR, LNR, LPR, LSR, LGR, LAR, LRR,
QMR, QQR, QHR, QNR, QPR, QSR, QGR, QAR, QRR, VFR, VMR, VQR, VHR,
VNR, VPR, VSR, VGR, VAR, VRR, MFR, MMR, MQR, MHR, MNR, MPR, MSR,
MGR, MAR, or MRR. In some embodiments, X4 is an optional sequence, and can be
ER,
or E, or absent. In some embodiments, if X is absent, Xi is FF, and X2 is LS.
In some
embodiments, the isolated peptides comprising Formula (I). have a length that
is less than
or equal to 1100 amino acids, for example, less than or equal to 4, 5, 6, 7,
8, 9, 10, 11, 12,
13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31,
32, 33, 34, 35, 36,
37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55,
56, 57, 58, 59, 60,
61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79,
80, 81, 82, 83, 84,
85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102,
103, 104, 105,
106, 107, 108, 109, 110, 111, 112, 113, 114, 115, 116, 117, 118, 119, 120,
121, 122, 123,
124, 125, 126, 127, 128, 129, 130, 131, 132, 133, 134, 135, 136, 137, 138,
139, 140, 150,
160, 170, 180, 190, 200, 210, 220, 230, 240, 250, 260, 270, 280, 290, 300,
320, 340, 360,
116
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
380, 400, 450, 500, 550, 600, 650, 700, 750, 800, 850, 900, 950, 1000, 1050,
or 1100
amino acids, including ranges between any two of the listed values.
[0395]
Additionally, a composition can comprise, consist of, or consist
essentially of a peptide inhibitor that comprises, consists of, or consists
essentially of a
peptide of Formula (II) X20TFFVKLSX21X22, (SEQ ID NO: 173). In some
embodiments, X20 is an optional sequence, and can be KKLD (SEQ ID NO: 174),
RKLD
(SEQ ID NO: 175), KKGD (SEQ ID NO: 176), KKED (SEQ ID NO: 177), KLD, LD,
or D, or absent. X21 is an optional sequence, and can be LFT, LMT, LQT, LHT,
LNT,
LPT, LST, LGT, LAT, LRT, QFT, QMT, QQT, QHT, QNT, QPT, QST, QGT, QAT,
QRT, VFT, VMT, VQT, VHT, VNT, VPT, VST, VGT, VAT, VRT, MFT, MMT, MQT,
MHT, MNT, MPT, MST, MGT, MAT, MRT, LFN, LMN, LQN, LHN, LNN, LPN, LSN,
LGN, LAN, LRN, QFN, QMN, QQN, QHN, QNN, QPN, QSN, QGN, QAN, QRN, VFN,
VMN, VQN, VHN, VNN, VPN, VSN, VGN, VAN, VRN, MFN, MMN, MQN, MHN,
MNN, MPN, MSN, MGN, MAN, MRN, LFP, LMP, LQP, LHP, LNP, LPP, LSP, LGP,
LAP, LRP, QFP, QMP, QQP, QHP, QNP, QPP, QSP, QGP, QAP, QRP, VFP, VMP,
VQP, VHP, VNP, VPP, VSP, VGP, VAP, VRP, MFP, MMP, MQP, MHP, MNP, MPP,
MSP, MGP, MAP, MRPR, LFR, LMR, LQR, LHR, LNR, LPR, LSR, LGR, LAR, LRR,
QMR, QQR, QHR, QNR, QPR, QSR, QGR, QAR, QRR, VFR, VMR, VQR, VHR,
VNR, VPR, VSR, VGR, VAR, VRR, MFR, MMR, MQR, MHR, MNR, MPR, MSR,
MGR, MAR, or MRR, or absent. In some embodiments, X22 is an optional sequence,
and can be ER, or E, or absent. In some embodiments, the isolated peptides
comprising
Formula (II) have a length that is less than or equal to 1100 amino acids, for
example, less
than or equal to 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20,
21, 22, 23, 24,
25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43,
44, 45, 46, 47, 48,
49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67,
68, 69, 70, 71, 72,
73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91,
92, 93, 94, 95, 96,
97, 98, 99, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112,
113, 114,
115, 116, 117, 118, 119, 120, 121, 122, 123, 124, 125, 126, 127, 128, 129,
130, 131, 132,
133, 134, 135, 136, 137, 138, 139, 140, 150, 160, 170, 180, 190, 200, 210,
220, 230, 240,
250, 260, 270, 280, 290, 300, 320, 340, 360, 380, 400, 450, 500, 550, 600,
650, 700, 750,
800, 850, 900, 950, 1000, 1050, or 1100 amino acids, including ranges between
any two
of the listed values.
117
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
[0396] Additionally, a
composition can comprise, consist of, or consist
essentially of a peptide inhibitor that comprises, consists of, or consists
essentially of a
peptide of Formula (III) X30X31VKLX32LX33TEX34 (SEQ ID NO: 178), or of SEQ ID
NOs: 1-33, 34, 46-53, 64-66, 68, 76, 94-96 or 98. In some embodiments, X31 is
an
optional sequence, and can be F, S, M, V, T, or L, or absent. In some
embodiments, X31
is F. In some embodiments, X32 can be S, Q, M, T, or H. In some embodiments,
X32 is S.
X33 can be F, M, Q, H, N, P, S, G, A, or R. In some embodiments, X34 is F. X34
is an
optional sequence, and can be R, or absent. In some embodiments, X30 is an
optional
sequence, and can be KKLDTF (SEQ ID NO: 179), KLDTF (SEQ ID NO: 180), LDTF
(SEQ ID NO: 181), DTF, TF, or F, or absent. In some embodiments, the isolated
peptides comprising Formula (Ill) have a length that is less than or equal to
1100 amino
acids, for example, less than or equal to 4, 5, 6, 7, 8, 9, 10, 11, 12, 13,
14, 15, 16, 17, 18,
19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37,
38, 39, 40, 41, 42,
43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61,
62, 63, 64, 65, 66,
67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85,
86, 87, 88, 89, 90,
91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105, 106, 107,
108, 109, 110,
111, 112, 113, 114, 115, 116, 117, 118, 119, 120, 121, 122, 123, 124, 125,
126, 127, 128,
129, 130, 131, 132, 133, 134, 135, 136, 137, 138, 139, 140, 150, 160, 170,
180, 190, 200,
210, 220, 230, 240, 250, 260, 270, 280, 290, 300, 320, 340, 360, 380, 400,
450, 500, 550,
600, 650, 700, 750, 800, 850, 900, 950, 1000, 1050, or 1100 amino acids,
including
ranges between any two of the listed values.
[0397] Additionally, a
composition can comprise, consist of, or consist
essentially of a peptide inhibitor that comprises, consists of, or consists
essentially of a
peptide of Formula (VII), X7001( X701X702X703 X704X705X706K X707 X708 X709
X710 X711E
X712 (SEQ ID NO: 394), as described herein. In some embodiments, X700 is an
optional
sequence, and can be K,A,D,E,G,H,I,L,M,N,P,Q,R,T, or V, or absent. In some
embodiments, X701 is an optional sequence, and
can be
L,A,C,D,E,F,G,H,I,K,M,N,Q,R,S,T, or V, or absent. In some embodiments, X702 is
an
optional sequence, and can be D,A,E,I,V,W, or Y, or absent. In some
embodiments, X703
is an optional sequence, and can be T,C,M,N,P,Q,R,S,W, or Y, or absent. In
some
embodiments, X704 is an optional sequence, and can be F,A,I,M,N,P,T, or V, or
absent. In
some embodiments, X705 is an optional sequence, and can be F,L,M,Q,S,T or V,
or
absent. In some embodiments, X706 is an optional sequence, and can be
V,F,G,L,P, or R,
118
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
or absent. In some embodiments, X707 is an optional sequence, and can be
L,A,F,G,I,M,N,P,Q,R,S,T,V, or Y, or absent. In some embodiments, X708 is an
optional
sequence, and can be S,H,M,N,Q, or T, or absent. In some embodiments, X709 is
an
optional sequence, and can be L,A,H,I,M,N,Q,R,S,T,V, or W, or absent. In some
embodiments, X710 is an optional sequence, and can be
F,A,C,G,H,I,L,M,N,P,Q,R,S,T,V,
or W, or absent. In some embodiments, X711 is an optional sequence, and can be
T,F,G,H,I,L,M,N,P,S,V, or W, or absent. In some embodiments, X712 is an
optional
sequence, and can be R,F,K,N,R,T, or Y, or absent. In some embodiments, the
isolated
peptide comprising Formula (VII) has a length that is less than or equal to
1100 amino
acids, for example, less than or equal to 4, 5, 6, 7, 8, 9, 10, 11, 12, 13,
14, 15, 16, 17, 18,
19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37,
38, 39, 40, 41, 42,
43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61,
62, 63, 64, 65, 66,
67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85,
86, 87, 88, 89, 90,
91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105, 106, 107,
108, 109, 110,
111, 112, 113, 114, 115, 116, 117, 118, 119, 120, 121, 122, 123, 124, 125,
126, 127, 128,
129, 130, 131, 132, 133, 134, 135, 136, 137, 138, 139, 140, 150, 160, 170,
180, 190, 200,
210, 220, 230, 240, 250, 260, 270, 280, 290, 300, 320, 340, 360, 380, 400,
450, 500, 550,
600, 650, 700, 750, 800, 850, 900, 950, 1000, 1050, or 1100 amino acids,
including
ranges between any two of the listed values.
[0398]
Additionally, a composition can comprise, consist of, or consist
essentially of a peptide inhibitor that comprises, consists of, or consists
essentially of a
peptide of Formula (VIII), X800K XstiiK X802E X803 (SEQ ID NO: 395), as
described
herein. In some embodiments, X800 is an optional sequence, and can be K, A, D,
E, G, H,
I, L, M, N, P, Q, R, T, V, or K, or absent. In some embodiments, X801 is an
optional
sequence, and can be LDTFFV, GDTH-V, EDTFFV, LDQFFV, LDTAFV, LDTVFV,
LDTFMV, LDTFSV, LDTFVV, LDTFTV, LDTFLV, LDGFFV, LDTFGV, LDTH-K,
ADTFFV, CDTFFV, DDTFFV, FDTFFV, HDTFFV, IDTFFV, KDTFFV, MDTH-V,
NDTFFV, QDTH-V, RDTFFV, SDTFFV, TDTFFV, VDTH-V, LATFFV, LETH-V,
LITFFV, LVTFFV, LWTFFV, LYTH-V, LDCH-V, LDMFFV, LDNFFV, LDPH-V,
LDRFFV, LDSFFV, LDW141-V, LDYFFV, LDTlFV, LDTMFV, LDTNFV, LDTPFV,
LDTTFV, LDTFQV, LDTF141-, LDTFFG, LDTFFL, LDTFFP, LDTFFR, LDTFIV,
LDTSFV, LDTFAV, LDTFCV, LDTQFV, LDTLFV, LTTFFV, LDTFFI, LDH141-V,
LMTFFV, LDTFEV, LDTFWV, LFTFFV, LDVFFV, LDTFRV, LDTFHV, LDTYFV,
119
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
LPTFFV, PDTH-V, LDTFPV, LDTFNV, LDTWFV, LDTGFV, LDAH-V, LQT1-1-V,
LCTFFV, LSTFFV, YDTFFV, LDEFFV, WDTFFV, LDTKFV, LDTCFV, LDTFYV,
LDTHFV, LHTFFV, LRTFFV, LDLFFV, LDTRFV, LLTFFV, LDTFDV, LDTH-A,
LDTFFT, LNTFFV, LDDFFV, LUFFY, LD1-1-FV, LKTFFV, LDTH-Q, LGTH-V,
LDTFFC, LDKFFV, LDTFKV, LDTEFV, LDTFFW, LDTFFM, LDTFFS, LDT1-141,
LDTFFY, LDTFFN, LDTDFV, LDTFFE, LDTF1-D, LTH-V, LDTFF, TFFV, LDF,
LDTE, FFV, LDV, LV, or L, or absent. In some embodiments, X802 is an optional
sequence, and can be LSLFT, VSLFT, LQLFT, LMLFT, LTLFT, LHLFT, LSQFT,
LSVFT, LSMFT, LSLMT, LSLQT, LSLHT, LSLNT, LSLPT, LSLST, LSLGT, LSLAT,
LSLRT, LSLFN, LSLFP, LSLFR, LGLFT, ASLFT, FSLFT, GSLFT, ISLFT, MSLFT,
NSLFT, PSLFT, QSLFT, RSLFT, SSLFT, TSLFT, YSLFT, LNLFT, LSAFT, LSHFT,
LSIFT, LSNFT, LSRFT, LSSFT, LSTFT, LSWFT, LSLCT, LSLIT, LSLLT, LSLTT,
LSLVT, LSLWT, LSLFF, LSLFG, LSLFH, LSLFI, LSLFL, LSLFM, LSLFS, LSLFV,
LSLFW, LYLFT, LVLFT, LSFFT, LSGFT, LSKFT, LSCFT, LCLFT, LRLFT, LPLFT,
LWLFT, LKLFT, LDLFT, LSYFT, LALFT, WSLFT, LSLFA, LSLFQ, LSPFT, HSLFT,
LSLYT, LlLFT, KSLFT, CSLFT, LSLFY, LSLFK, LSLFC, LFLFT, LELFT, LSLKT,
LLLFT, LSLFD, LSLDT, LSLFE, DSLFT, LSLET, LSDFT, LSEFT, ESLFT, SLFT,
LSFT, LFT, LSL, LT, or T, or absent. In some embodiments, X803 is an optional
sequence, and can be R, F, K, N, R, T, or Y, or absent. In some embodiments,
the
isolated peptide comprising Formula (VIII) has a length that is less than or
equal to 1100
amino acids, for example, less than or equal to 4, 5, 6, 7, 8, 9, 10, 11, 12,
13, 14, 15, 16,
17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35,
36, 37, 38, 39, 40,
41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59,
60, 61, 62, 63, 64,
65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83,
84, 85, 86, 87, 88,
89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105, 106,
107, 108,
109, 110, 111, 112, 113, 114, 115, 116, 117, 118, 119, 120, 121, 122, 123,
124, 125, 126,
127, 128, 129, 130, 131, 132, 133, 134, 135, 136, 137, 138, 139, 140, 150,
160, 170, 180,
190, 200, 210, 220, 230, 240, 250, 260, 270, 280, 290, 300, 320, 340, 360,
380, 400, 450,
500, 550, 600, 650, 700, 750, 800, 850, 900, 950, 1000, 1050, or 1100 amino
acids,
including ranges between any two of the listed values. Additionally, a
composition can
comprise, consist of, or consist essentially of a peptide inhibitor that
comprises, consists
of, or consists essentially of any one or more of the peptides set forth in
Table 5.1. In
some embodiments, the isolated peptide from Table 5.1 used in these
compositions has a
120
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
length that is less than or equal to 1100 amino acids, for example, less than
or equal to 4,
5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25,
26, 27, 28, 29,
30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48,
49, 50, 51, 52, 53,
54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72,
73, 74, 75, 76, 77,
78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96,
97, 98, 99, 100,
101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115,
116, 117, 118,
119, 120, 121, 122, 123, 124, 125, 126, 127, 128, 129, 130, 131, 132, 133,
134, 135, 136,
137, 138, 139, 140, 150, 160, 170, 180, 190, 200, 210, 220, 230, 240, 250,
260, 270, 280,
290, 300, 320, 340, 360, 380, 400, 450, 500, 550, 600, 650, 700, 750, 800,
850, 900, 950,
1000, 1050, or 1100 amino acids, including ranges between any two of the
listed values.
[0399] The
pharmaceutical composition can comprise one or more other
pharmaceutical acceptable pharmaceutical ingredients, such as a
pharmaceutically
acceptable diluent, carrier, excipient and/or buffer. "Pharmaceutically
acceptable" means
a non-toxic compound that does not decrease the effectiveness of the
biological activity of
the active ingredients. Such pharmaceutically acceptable additives, diluents
buffers,
carriers or excipients are well-known in the art (see Remington's
Pharmaceutical
Sciences, 18th edition, A.R Gennaro, Ed., Mack Publishing Company (1990) and
handbook of Pharmaceutical Excipients, 3rd edition, A. Kibbe, Ed.,
Pharmaceutical Press
(2000).
[0400] The
pharmaceutical composition can include a buffer. The term
"buffer" is intended to refer to an aqueous solution containing an acid-base
mixture with
the purpose of stabilizing pH. Examples of buffering agents are magnesium
hydroxide
and aluminum hydroxide; alginic acid; pyrogen-free water; isotonic saline;
Ringer's
solution; ethyl alcohol; pH buffered solutions; polyesters, polycarbonates
and/or
polyanhydrides; and other non-toxic compatible substances employed in
pharmaceutical
formulations. Other examples of buffers are Trizma, Bicine, Tricine, MOPS,
MOPSO,
MOBS, Tris, Hepes, HEPBS, MES, phosphate, carbonate, acetate, citrate,
glycolate,
lactate, borate, ACES, ADA, tartrate, AMP, AMPD, AMPSO, BES, CABS, cacodylate,
CHES, DlPSO, EPPS, ethanolamine, glycine, HEPPSO, imidazole, imidazolelactic
acid,
PIPES, SSC, SSPE, POPSO, TAPS, TABS, TAPSO and TES.
[0401] The
pharmaceutical composition can include a diluent. The term
"diluent" is intended to refer to an aqueous or non-aqueous solution with the
purpose of
121
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
diluting the compounds in the pharmaceutical preparation. The diluent may be
one or
more of saline, water, polyethylene glycol, propylene glycol or ethanol.
[0402] The
pharmaceutical composition can include an excipient. The
excipient can be one or more of carbohydrates, surfactants, polymers, lipids
and minerals.
Examples of carbohydrates include lactose, sucrose, mannitol, and
cyclodextrines, which
are added to the composition, e.g., for facilitating lyophilisation. Examples
of polymers
are starch, cellulose ethers, cellulose carboxymethylcellulose,
hydroxypropylmethyl
cellulose, hydroxyethyl cellulose, ethylhydroxyethyl cellulose, alginates,
carageenans,
hyaluronic acid and derivatives thereof, polyacrylic acid, polysulphonate,
polyethylenglycol/polyethylene oxide, polyethyleneoxide/polypropylene oxide
copolymers, polyvinylalcohol/polyvinylacetate of different degree of
hydrolysis, and
polyvinylpyrrolidone, all of different molecular weight, which are added to
the
composition, e.g., for viscosity control, for achieving bioadhesion, or for
protecting the
lipid from chemical and proteolytic degradation. Examples of lipids are fatty
acids,
phospholipids, mono-, di-, and triglycerides, ceramides, sphingolipids and
glycolipids, all
of different acyl chain length and saturation, egg lecithin, soy lecithin,
hydrogenated egg
and soy lecithin, which are added to the composition for reasons similar to
those for
polymers. Examples of minerals are talc, magnesium oxide, zinc oxide and
titanium
oxide, which are added to the composition to obtain benefits such as reduction
of liquid
accumulation or advantageous pigment properties.
[0403] The
pharmaceutical composition can include a carrier. In some
embodiments, the carrier is a non-aqueous carrier. Examples of suitable
aqueous and
nonaqueous carriers which can be employed in the pharmaceutical compositions
of the
invention include water, ethanol, polyols (such as glycerol, propylene glycol,
polyethylene
glycol, and the like), and suitable mixtures thereof, vegetable oils, such as
olive oil, and
injectable organic esters, such as ethyl oleate. Proper fluidity can be
maintained, for
example, by the use of coating materials, such as lecithin, by the maintenance
of the
required particle size in the case of dispersions, and by the use of
surfactants.
[0404] These
compositions can contain adjuvants such as preservatives,
wetting agents, emulsifying agents and dispersing agents. Prevention of the
action of
microorganisms upon the subject compounds may be ensured by the inclusion of
various
antibacterial and antifungal agents, for example, paraben, chlorobutanol,
phenol sorbic
acid, and the like. It may also be desirable to include isotonic agents, such
as sugars,
122
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
sodium chloride, and the like into the compositions. In addition, prolonged
absorption of
the injectable pharmaceutical form may be brought about by the inclusion of
agents which
delay absorption such as aluminum monostearate and gelatin.
[0405] The
pharmaceutical composition can be formulated for a extended
release. In some embodiments, the pharmaceutical compositon is formulated as a
gel or
gel-like substance for extended release. The gel or gel-like substance can
remain stable
under physiological conditions for about 3 days, 4, 5, 6, 7, 8, 9, 10, 11, 12,
13, or 14 days,
3-4 days, 3-5, 3-6, 3-7, 3-8, 3-9, 3-10, 3-11, 3-12, 3-13, 3-14, 4-5, 4-6, 4-
7, 4-8, 4-9, 4-10,
4-11, 4-12, 4-13, 4-14, 5-6, 5-7, 5-8, 5-9, 5-10, 5-11, 5-12, 5-13, 5-14, 6-7,
6-8, 6-9, 6-10,
6-11, 6-12, 6-13, 6-14, 7-8, 7-9, 7-10, 7-11, 7-12, 7-13, 7-14, 8-14, 9-14, or
10-14 days.
In some embodiments, the gel comprises an inhibitor peptide comprising,
consisting of, or
consisting essentially of any of SEQ ID NOs: 1-33, 34, 46-53, 64-66, 68, 76,
94-96, 98,
264-393, or 583-586, in which the inhibitor peptide is not water soluble and a
buffer or
adjuvant selected to formulate a gel when combined with the inhibitor peptide.
Without
being limited by any theory, in accordance with some embodiments herien, gels
can be
suitable for slow release of the inhibitor peptide.
[0406] The
pharmaceutical composition can be formulated for solubility in
aqueous solution. By way of example, an inhibitor peptide consisting of or
consisting
essentially of SEQ ID NO: 589 has been shown to be soluble in aqueous
solution. As
such, in some emboduments, a pharmaceutical composition comprises an inhibitor
peptide consisting of or consisting essentially of SEQ ID NO: 589 solubleized
or partially
solubleized in an aqueous solution. Optionally, the aqueous solution can be
provided as
an adjuvant.
Compositions comprising nanoparticles
[0407] It is
contemplated that nanoparticles can be useful in formulations
comprising immunoregulatory peptide inhibitors in accordance with some
embodiments
herein. Without being limited by any theory, nanoparticles can be useful in
solubilizing
immunoregulatory peptide inhibitors, minimizing aggregates of immunoregulatory
peptide inhibitors, and/or delivering immunoregulatory peptide inhibitors to
tumors in
accordance with some embodiments herein. For example, nanoparticles have been
observed to exhibit enhanced permeation (EPR) effects, and it is contemplated
that EPR's
123
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
can be useful for facilitating permeation of tumors by immunoregulatory
peptide
inhibitors in accordance with some embodiments herein.
[0408] As used
herein, "nanoparticle" refers to a particle having a diameter of
0.1 to 9000 nm, for example at least or equal to 0.1 nm, mm, 5nm, lOnm, 20nm,
50nm,
90nm, 100nm, 200nm, 300nm 500nm, 900nm, 1000nm, 2000nm, 3000 nm, 4000 nm,
5000nm, and/or, for example, less than or equal to 9000 nm, 5000 nm, 2000 nm,
1000
nm, 500 nm, 300nm, 200 nm, 100 nm, 90 nm, 50 nm, 30 nm, 20 nm, 10 nm, 5 nm, 1
nm,
or within a range defined by any two of the aforementioned diameters, for
example, 0.1
nm to lOnm, 0.1nm to 20nm, 0.1nm to 50nm, 0.1nm to 90nm, 0.1nm to 100nm, 0.1nm
to
200nm, 0.1nm to 300nm, 0.1nm to 500nm, 0.1nm to 900nm, 0.1nm to 1000nm, 0.1nm
to
1500nm, 0.1nm to 2000nm, 0.1nm to 3000nm, 0.1nm to 5000nm, 0.1nm to 9000nm, 1
nm to lOnm, mm to 20nm, mm to 50nm, mm to 90nm, mm to 100nm, mm to 200nm,
mm to 300nm, mm to 500nm, mm to 900nm, mm to 1000nm, mm to 1500nm, mm to
2000nm, mm to 3000nm, mm to 5000nm, mm to 9000nm, 5 nm to lOnm, 5nm to 20nm,
5nm to 50nm, 5nm to 90nm, 5nm to 100nm, 5nm to 200nm, 5nm to 300nm, 5nm to
500nm, 5nm to 900nm, 5nm to 1000nm, 5nm to 1500nm, 5nm to 2000nm, 5nm to
3000nm, 5nm to 5000nm, 5nm to 9000nm, lOnm to 20nm, lOnm to 50nm, lOnm to
90nm, lOnm to 100nm, lOnm to 200nm, lOnm to 300nm, lOnm to 500nm, lOnm to
900nm, lOnm to 1000nm, lOnm to 1500nm, lOnm to 2000nm, lOnm to 3000nm, lOnm to
5000nm, lOnm to 9000nm, 20nm to 50nm, 20nm to 90nm, 20nm to 100nm, 20nm to
200nm, 20nm to 300nm, 20nm to 500nm, 20nm to 900nm, 20nm to 1000nm, 20nm to
1500nm, 20nm to 2000nm, 20nm to 3000nm, 20nm to 5000nm, 20nm to 9000nm, 50nm
to 90nm, 50nm to 100nm, 50nm to 200nm, 50nm to 300nm, 50nm to 500nm, 50nm to
900nm, 50nm to 1000nm, 50nm to 1500nm, 50nm to 2000nm, 50nm to 3000nm, 50nm to
5000nm, 50nm to 9000nm, 100nm to 200nm, 100nm to 300nm, 100nm to 500nm, 100nm
to 900nm, 100nm to 1000nm, 100nm to 1500nm, 100nm to 2000nm, 100nm to 3000nm,
100nm to 5000nm, 100nm to 9000nm, 500nm to 1000nm, 500nm to 2000nm, 500nm to
3000nm, 500nm to 5000nm, or 500nm to 9000nm, 1000nm to 2000nm, 1000nm to
3000nm, 1000nm to 5000nm, or 1000nm to 9000nm or within a range defined by any
two
of the aforementioned diameters. Optionally, the nanoparticle comprises a
degradable
particle and/or a non-degradable particle. Optionally, the nanoparticle
comprises a non-
degradable particle. Optionally, the nanoparticle consists of or consists
essentially of a
124
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
non-degradable particle. Optionally, the nanoparticle comprises a degradable
particle.
Optionally, the nanoparticle consists of or consists essentially of a
degradable particle.
[0409] A number
of nanoparticle materials are suitable for nanoparticles in
compositions, uses, and methods in accordance with some embodiments herein.
The
nanoparticles can comprise various properties, for example biocompatibility,
ability to
carry hydrophobic cargo, and the like. Examples of materials for suitable
nanoparticles in
accordance with some embodiments herein include semiconductors (e.g. quantum
dots)
(Gao,X.H. et al.,(2004). "In vivo cancer targeting and imaging with
semiconductor
quantum dots". Nat. Biotechnol. 22, 969-976; Medintz,I.L., et al. (2005).
"Quantum dot
bioconjugates for imaging, labelling and sensing". Nat. Mater. 4, 435-446;
Michalet, X.,
et al. (2005). "Quantum dots for live cells, in vivo imaging, and
diagnostics". Science
307, 538-544; Park et al. (2011). "CuInSe/ZnS Core/Shell NlR quantum dots for
biomedical imaging". Small 7, 3148-3152; Chen et al., (2012)
"Pharmacokinetics,
dosimetry and comparative efficacy of Re-188-liposome and 5-FU inaCT26-luclung-
metastaticmicemodel". Nucl. Med. Biol. 39, 35-43; Petryayeva et al., (2013),
"Quantum
dots in bioanalysis: a review of applications across various platforms for
fluorescence
spectroscopy and imaging". Appl. Spectrosc. 67, 215-252), each of which is
hereby
incorporated by reference in its entirety. Examples of materials for suitable
nanoparticles
in accordance with some embodiments herein include silica (Vanblaaderen et
al., (1992)
"Synthesis and characterization of colloidal dispersions of fluorescent,
monodispersesilicaspheres". Langmuir 8, 2921-2931; Giri,S.,et al.(2007).
"Mesoporous
silica nanomaterial-b as ed biotechnological and biomedical delivery systems".
Nanomedicine 2; Gin i et al., (2005) "Stimuli-responsive controlled-release
delivery
system based on mesoporous silica nanorods capped with magnetic
nanoparticles".
Angew. Chem. Int. Ed. Engl. 44, 5038-504; Burns et al., (2006) "Fluorescent
core-shell
silica nanoparticles: towards "LabonaParticle"architectures for
nanobiotechnology".
Chem. Soc. Rev. 35, 1028-1042), each of which is hereby incorporated by
reference in its
entirety. Examples of materials for suitable nanoparticles in accordance with
some
embodiments herein include gold (e.g. gold spheres, gold rods, gold
shells)(Boisselier E.,
et al. (2009) "Gold nanoparticles in nanomedicine: preparations, imaging,
diagnostics,
therapies and toxicity". Chem. Soc. Rev. 38, 1759-1782; Arvizo et al., (2010),
"Gold
nanoparticles: opportunities and challenges in nanomedicine". Expert Opin.
Drug Deliv.
7, 753-763), each of which is hereby incorporated by reference in its
entirety. Examples
125
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
of materials for suitable nanoparticles in accordance with some embodiments
herein
include magnetic materials (e.g. magnetic DynabeadTM (Invitrogen)) (Arruebo et
al.,
(2007) "Magnetic nanoparticles for drug delivery". Nano Today 2, 22-32;
Banerjee et al.,
(2010) "Nanomedicine: magnetic nanoparticles and their biomedical
applications". Curr.
Med. Chem. 17, 3120-3141; Haun et al., (2010) "Magnetic nanoparticle
biosensors".
Wiley Interdiscip. Rev. Nanome. Nanobiotechnol. 2, 291-304), each of which is
hereby
incorporated by reference in its entirety. Examples of materials for suitable
nanoparticles
in accordance with some embodiments herein include carbon-based materials
(e.g. a
carbon nanotube, an activated carbon, buckminsterfullerene, or graphene)
(Prato et al.,
(2008) "Functionalized carbon nanotubes in drug design and discovery". Acc.
Chem. Res.
41, 60-68; JaM, (2012) "Advances in use of functionalized carbon nanotubes for
drug
design and discovery". Expert Opin. Drug Discov. 7, 1029-1037; Ye et al.,
(2015)
"Targeted delivery of docetaxel to the metastatic lymph nodes: A comparison
study
between nanoliposomes and activated carbon nanoparticles". Asian Journal of
Pharmaceutical Sciences 10: pp. 64-72), each of which is hereby expressly
incorporated
by reference in its entirety.
[0410] In some
embodiments, a composition (or method or use thereof)
comprising an immunoregulatory peptide inhibitor comprises a nanoparticle
comprising at
least one of a polymer (e.g. PLGA, glycerol, chitosan, DNA, a hydrogel, an
acrylamide,
and the like), a dendrimer (e.g. PAMAM and the like), a quantum dot (e.g.
CdSe,
CuInSe, CdTe, and the like), a gold nanoparticle (e.g. a sphere, rod, or
shell), a silica
nanoparticle (e.g. a sphere, shell, mesoporous structure, and the like), a
magnetic particle
(e.g. iron oxide, cobalt-based material, a magnetic sphere, an aggregate in
dextran or
silica, a Dynal bead, and the like), a carbon-based material (e.g. a carbon
nanotube,
buckminsterfullerene, graphene, or an activated carbon), a carbohydrate, a
nucleic acid, a
polypeptide (e.g. an albumin or an albumin fragment), or a lipid. Optionally,
the
nanoparticle comprises two or more of the listed substances. Optionally, the
nanoparticle
is PEGylated. Optionally, the nanoparticle is coated with PEGylated lipid.
[0411] It is
contemplated that in accordance with compositions, uses, and
methods in accordance with embodiments herein, the shape of a nanoparticle can
be
tuned, for example to affect solubility, delivery, or release of an
immunoregulatory
peptide inhibitor. Examples of nanoparticles suitable for use in accordance
with some
embodiments herein include beads, tubes, spheres, shells, rods, mesoporous
structures,
126
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
hydrogels, aggregates, carbon-based materials, polymers, fullerenes, and the
like. In some
embodiments, the nanoparticle comprises a cage that can contain (and
subsequently
release, or make accessible) the immunoregulatory peptide, for example an
albumin or
nucleic acid cage (see, e.g. Andersen, et al (2009). Nature. 459: 73-6, hereby
incorporated
by reference in its entirety), or a porous metal nanocage such as gold (see,
e.g. Yavuz, et
al. (2009), Nat Mater. 8:935-9, hereby incorporated by reference in its
entirety). It is
contemplated that engineered viruses or portions thereof, such as engineered
plant viruses
(e.g. cowpea mosaic virus) can provide nanoparticles with readily scalable
production. In
some embodiments, the nanoparticle comprises a viral capsid, or portion
thereof (see, e.g.
Steinmetz (2013). Mo/ Pharm. 10: 1-2, hereby incorporated by reference in its
entirety).
In some embodiments, the nanoparticle comprises a lipid capsule or liposome.
It is
contemplated that that lipid capsules such as liposomes can permit a
relatively high level
of inhibitory peptide-to-carrier ratio, and minimize escape of inhibitor
peptide during
circulation. It is contemplated that polymers, such as polyacrylamide or
chitosan, can
faciliatate sustained release (e.g. slow release) of the immunoregulatory
peptide inhibitors
(see, e.g., Kashyap et al., Hydrogels for Pharmaceutical and Biomedical
Applications,
(2005) Critical Reviewsin Therapeutic Drug Carrier Systems, 22(2):107-150,
hereby
incorporated by reference in its entirety). It is contemplated that active
carbon supports,
for example activated carbon nanoparticles can facilitate delivery to lymph
nodes, for
example delivery of the immunoregulatory to lymph nodes in metastatic cancer
(see, e.g.
Ye et al., (2015) "Targeted delivery of docetaxel to the metastatic lymph
nodes: A
comparison study between nanoliposomes and activated carbon nanoparticles".
Asian
Journal of Pharmaceutical Sciences 10: pp. 64-72, hereby incorporated by
reference in its
entirety).
[0412] In some
compositions, methods, and uses in accordance with some
embodiments herein, a nanoparticle as described herein is conjugated to an
immunoregulatory peptide inhibitor. As used herein "conjugate" (and variations
of this
root term) of immunoregulatory peptide inhibitors refers broadly to covalent,
non-
covalent, or a combination of covalent and non-covalent associations between
an
immunoregulatory peptide inhibitor and another particle, such as a
nanoparticle. As such,
an immunoregulatory peptide inhibitor can be immobilized on the surface of a
nanoparticle by conjugation to the nanoparticle. In some
embodiments, the
immunoregulatory peptide inhibitor is conjugated to a nanoparticle covalently.
Examples
127
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
of covalent linkers are reviewed in Trail (2013), Antibodies 2: 113-129,
hereby
incorporated by reference in its entirety. In some embodiments, the
immunoregulatory
peptide inhibitor is conjugated to a nanoparticle non-covalently, for example
via
hydrophobic or electrostatic interactions. Without being limited by any
theory, it is
contemplated that it can be useful for immunoregulatory peptide inhibitors in
accordance
with some embodiments herein to subsequently dissociate from the
nanoparticles.
Accordingly, in some embodiments, the immunoregulatory peptide inhibitor is
noncovalently conjugated to the nanoparticle, or is covalently conjugated to
the
nanoparticle via a cleavable or reversible covalent linker, for example a
cleavable linker
or a pH-sensitive linker.
[0413] Examples
of non-covalent interactions in accordance with some
embodiments herein include van der Waals, steric, hydrogen bonding,
hydrophobic and
electrostatic interactions. In some embodiments, non-covalent associations
between
immunoregulatory peptide inhibitors and nanoparticles include hydrophobic
interactions,
electrostatic interactions, hydrogen bonding, and steric immobilization, or
combinations
of two or more of these. Examples of hydrophobic interactions between
nanoparticles
and therapeutic moieties are described in Cheng et al. (2008) J Am Chem Soc.
130:
10643-10647, hereby incorporated by reference in its entirety. Examples of
electrostatic
interactions between nanoparticles and therapeutic moieties are described in
Manju et al.
(2011),. Langmuir. 27: 14489-14496, hereby incorporated by reference in its
entirety.
Examples of hydrogen bonding between nanoparticles and therapeutic moieties
are
described in Kim et al. (2008), ACS Nano. 2: 386-392, hereby incorporated by
reference
in its entirety. Examples of steric immobilization interactions between
nanoparticles and
therapeutic moieties are described in Kester et al. (2008), Nano Lett. 8: 4116-
4121,
hereby incorporated by reference in its entirety. Optionally, an
immunoregulatory peptide
inhibitor and nanoparticle are associated by one or more non-covalent
interaction, and one
or more covalent interaction (for example a cleavable linker, or a non-
cleavable linker), as
described herein.
[0414] Examples
of cleavable linkers suitable for some embodiments herein
include peptides comprising protease target sites, such as targets of
cathepsin, matrix
metalloproteinases, furin, pepsin, trypsin, and the like. By way of example,
it has been
observed that some solid tumors express proteases such as matrix
metalloproteinases. As
128
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
such, in some embodiments, a linker comprises a matrix metalloproteinase
target site such
as MMP-sensitive peptide (GPQGIAGQ, SEQ ID NO: 596).
[0415] By way
of example, it has been observed that some solid tumors
comprise a slightly acidic pH. Accordingly, in some embodiments, the linker
comprises a
pH-sensitive linker that permits dissociation of the immunoregulatory peptide
inhibitor
from the nanoparticle at an acidic pH. Example pH-sensitive linkers include
acid-labile
hydrazones, disulfide, thioethers, silyl linkers and peptide. An example acid-
labile
hydrazone is provided in Formula (IX) below (with the site of acid-sensitive
separation
depicted as a dashed line).
Formula (IX):
0
N R2
1\1"
R1
H
[0416] Silyl
linkers have also been shown to be cleaved at acidic pH's (see,
e.g. Finniss et al. (2014), MedChemComm DOI: 10.1039/c4md00150h). An example
acid-labile silyl linker is provided in Formula (X) below:
Formula (X):
/R
[0417] Without
being limited by any theory, it is contemplated that some
receptors that are un-inhibited (e.g. de-blocked) by immunoregulatory peptide
inhibitors
in accordance with various embodiments herein can be internalized. For
example, the
LFA-1 receptor has been shown to undergo internalization (see, Yusuf-
Makagiansar et al.
(2001), Pharm Res. 18: 329-35.). For example, the IL-2 receptor has been shown
to
undergo internalization (see, Hemar et al. (1994), J. Cell Biology 129: 55-
64).
129
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
Accordingly, in some embodiments, the cleavable linker is cleaved upon
internalization,
for example, a citrulline linker (e.g. dipeptide valine-citrulline) or a
cathepsin B target
(e.g. Arg-Arg).
[0418] In accordance with some embodiments herein, a nanoparticle as
described herein is conjugated to a immunoregulatory peptide inhibitor. In
some
embodiments, the nanoparticle is conjugated to an isolated P28R peptide (SEQ
ID NO:
2), or a peptide comprising SEQ ID NO: 2. In some embodiments, the
nanoparticle is
conjugated to an isolated P28 core peptide (SEQ ID NO: 62) or a peptide
comprising
SEQ ID NO: 62. Without being limited by any theory, it is contemplated that a
composition comprising P28 core peptide (SEQ ID NO: 62) immobilized on a
nanoparticle can be useful for facilitating stimulation or activation of
immune cells (for
example, by removing immunoregulatory peptides from blocking LFA-1 and/or IL-2
receptors), without having a direct immunostimulatory effect itself. In
some
embodiments, the nanoparticle is conjugated to an isolated peptide comprising
Formula
VII, wherein Formula VII is:
[0419] X7001KX701 X702X703X704X705X706KX707X708X709X710X711EX712 (SEQ
ID
NO: 394)
[0420] wherein X700 is K,A,D,E,G,H,I,L,M,N,P,Q,R,T,V, or K, or
absent;
[0421] wherein X701 is L,A,C,D,E,F,G,H,I,K,M,N,Q,R,S,T, or V, or
absent;
[0422] wherein X702 is D,A,E,I,V,W, or Y, or absent;
[0423] wherein X703 is T,C,M,N,P,Q,R,S,W, or Y, or absent;
[0424] wherein X704 is F,A,I,M,N,P,T, or V, or absent;
[0425] wherein X705 is F,L,M,Q,S,T or V, or absent;
[0426] wherein X706 is V,F,G,L,P, or R, or absent;
[0427] wherein X707 is L,A,F,G,I,M,N,P,Q,R,S,T,V, or Y, or absent;
[0428] wherein X708 is S,H,M,N,Q, or T, or absent;
[0429] wherein X709 is L,A,H,I,M,N,Q,R,S,T,V, or W, or absent;
[0430] wherein X710 is F,A,C,G,H,I,L,M,NP,Q,R,S,T,V, or W, or absent;
[0431] wherein X711 is T,F,G,H,I,L,M,N,P,S,V, or W, or absent; and
[0432] wherein X712 is R,F,K,N,R,T, or Y, or absent.
[0433] Said formula VII may be one of SEQ ID NO: 1-101, 167-172, 174-
177, 179-393, 396-581, or 582.
130
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
[0434] Some embodiments of the invention include compositions that
comprise an isolated peptide comprising Formula VIII, wherein Formula VIII is:
[0435] X800K X8011( X802E X803 (SEQ ID NO: 395)
[0436] wherein X800 is K, A, D, E, G, H, I, L, M, N, P, Q, R, T, V,
or K, or
absent;
[0437] wherein X801 is LDTFFV, GDTFFV, EDTFFV, LDQFFV, LDTAFV,
LDTVFV, LDTFMV, LDTFSV, LDTFVV, LDTFTV, LDTFLV, LDGFFV, LDTFGV,
LDTFFK, ADTFFV, CDTH-V, DDTH-V, FDTH-V, HDTFFV, IDTFFV, KDTFFV,
MDTFFV, NDTFFV, QDTFFV, RDTFFV, SDTFFV, TDTFFV, VDTH-V, LATFFV,
LETFFV, LITFFV, LVT1-1-V, LWTFFV, LYTFFV, LDCFFV, LDMFFV, LDNFFV,
LDPFFV, LDR1-1-V, LDSFFV, LDWFFV, LDYFFV, LDTIFV, LDTMFV, LDTNFV,
LDTPFV, LDTTFV, LDTFQV, LDTFFF, LDTFFG, LDTFFL, LDTFFP, LDTH-R,
LDTFIV, LDTSFV, LDTFAV, LDTFCV, LDTQFV, LDTLFV, LTTFFV, LDT1-1-4,
LDH1-1-V, LMTFFV, LDTFEV, LDTFWV, LFTFFV, LDVFFV, LDTFRV, LDTFHV,
LDTYFV, LPTFFV, PDTFFV, LDTFPV, LDTFNV, LDTWFV, LDTGFV, LDAFFV,
LQTFFV, LCTFFV, LSTFFV, YDTFFV, LDEFFV, WDTFFV, LDTKFV, LDTCFV,
LDTFYV, LDTHFV, LHTFFV, LRTFFV, LDLFFV, LDTRFV, LLTFFV, LDTFDV,
LDTFFA, LDTH-T, LNTFFV, LDDFFV, LDIFFV, LD1-1-FV, LKT1-1-V, LDTFFQ,
LGTFFV, LDTFFC, LDKFFV, LDTFKV, LDTEFV, LDTFFW, LDTFFM, LDTFFS,
LDTFFH, LDTFFY, LDTFFN, LDTDFV, LDTFFE, LDTH-D, LTFFV, LDT1-1-, TFFV,
LDF, LDTE, FFV, LDV, LV, or L, or absent;
[0438] wherein X802 is LSLFT, VSLFT, LQLFT, LMLFT, LTLFT, LHLFT,
LSQFT, LSVFT, LSMFT, LSLMT, LSLQT, LSLHT, LSLNT, LSLPT, LSLST, LSLGT,
LSLAT, LSLRT, LSLFN, LSLFP, LSLFR, LGLFT, ASLFT, FSLFT, GSLFT, ISLFT,
MSLFT, NSLFT, PSLFT, QSLFT, RSLFT, SSLFT, TSLFT, YSLFT, LNLFT, LSAFT,
LSHFT, LSIFT, LSNFT, LSRFT, LSSFT, LSTFT, LSWFT, LSLCT, LSLIT, LSLLT,
LSLTT, LSLVT, LSLWT, LSLFF, LSLFG, LSLFH, LSLFI, LSLFL, LSLFM, LSLFS,
LSLFV, LSLFW, LYLFT, LVLFT, LSFFT, LSGFT, LSKFT, LSCFT, LCLFT, LRLFT,
LPLFT, LWLFT, LKLFT, LDLFT, LSYFT, LALFT, WSLFT, LSLFA, LSLFQ, LSPFT,
HSLFT, LSLYT, LILFT, KSLFT, CSLFT, LSLFY, LSLFK, LSLFC, LFLFT, LELFT,
LSLKT, LLLFT, LSLFD, LSLDT, LSLFE, DSLFT, LSLET, LSDFT, LSEFT, ESLFT,
SLFT, LSFT, LFT, LSL, LT, or T, or absent; and
[0439] wherein X803 is R, F, K, N, R, T, or Y, or absent.
131
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
[0440] Said
formula VIII may be one of SEQ ID NOs: 1-34, 64-68, 70-72,
74-77, 80, 83, 86, 89, 92-96, 99-100, 264, 268-269, 270-386, 388-393, 396-401,
403,
404, 406, 408-411, 413-416, 419-420, 422-438, 442-444, 446-449, 451-453, 455-
458,
460, 462-466, 470, 472-477, 479-480, 482-484, 486, 487, 489, 491-493, 495-498,
500-
508, 512-517, 519-522, 528-530, 532, 533, 535-538, 540, 542-551, 553, 557-559,
567,
570, 572-581, or 582.
[0441] In some
embodiments of the invention, the nanoparticle is conjugated
to an isolated peptide comprising Formula I, wherein Formula I is:
[0442] XX1VKX2X3X4 (SEQ ID NO: 166)
[0443] wherein X is KKLDT (SEQ ID NO: 167), RKLDT (SEQ ID NO:
168), KKGDT (SEQ ID NO: 169), KKEDT (SEQ ID NO: 170), KKLDQ (SEQ ID NO:
171), KKGDQ (SEQ ID NO: 252), KKEDQ (SEQ ID NO: 253), RKLDQ (SEQ ID
NO: 254), RKGDQ (SEQ ID NO: 255), RKEDQ (SEQ ID NO: 256), RKGTD (SEQ
ID NO: 257), RKEDT (SEQ ID NO: 258), KLDT (SEQ ID NO: 172), KGDT (SEQ ID
NO: 259), KEDT (SEQ ID NO: 260), KLDQ (SEQ ID NO: 261), KGDQ (SEQ ID NO:
262), KEDQ (SEQ ID NO: 263), LDT, LDQ, GDT, GDQ, EDT, EDQ, DT, DQ, T, Q, or
absent.
[0444] wherein
Xi is 1-1-, FM, FS, FV, FT, FL, AF, AM, AS, AV, AT, AL,
VF, VM, VS, VV, VT, or VL, or absent;
[0445] wherein
X2 is LS, LQ, LM, LT, LH, VS, VQ, VM, VT, or VH, or
absent;
[0446] wherein
X3 is LFT, LMT, LQT, LHT, LNT, LPT, LST, LGT, LAT,
LRT, QFT, QMT, QQT, QHT, QNT, QPT, QST, QGT, QAT, QRT, VFT, VMT, VQT,
VHT, VNT, VPT, VST, VGT, VAT, VRT, MFT, MMT, MQT, MHT, MNT, MPT,
MGT, MAT, MRT, LFN, LMN, LQN, LHN, LNN, LPN, LSN, LGN, LAN, LRN, QFN,
QMN, QQN, QHN, QNN, QPN, QSN, QGN, QAN, QRN, VFN, VMN, VQN, VHN,
VNN, VPN, VSN, VGN, VAN, VRN, MFN, MMN, MQN, MHN, MNN, MPN, MSN,
MGN, MAN, MRN, LFP, LMP, LQP, LHP, LNP, LPP, LSP, LGP, LAP, LRP, QFP,
QMP, QQP, QHP, QNP, QPP, QSP, QGP, QAP, QRP, VFP, VMP, VQP, VHP, VNP,
VPP, VSP, VGP, VAP, VRP, MFP, MMP, MQP, MHP, MNP, MPP, MSP, MGP, MAP,
MRPR, LFR, LMR, LQR, LHR, LNR, LPR, LSR, LGR, LAR, LRR, QFR, QMR, QQR,
QHR, QNR, QPR, QSR, QGR, QAR, QRR, VFR, VMR, VQR, VHR, VNR, VPR, VSR,
132
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
VGR, VAR, VRR, MFR, MMR, MQR, MHR, MNR, MPR, MSR, MGR, MAR, or
and
[0447] wherein X4 is ER, E, or absent.
[0448] Said formula I may be one of SEQ ID NOs: 2-40, 46-52, 58-65,
67-
71, 74-77, 80-83, 86-88, 92-96, 99-101, 166, 173, 178, 182, 268-325, 332-392-
393, 396-
415, 417-444, 446-468, 470-487, 489-494, 497-508, 510, 512, 514-517, 520-522,
524-
525, 528-533, 535-536, 538-539, 542-544, 546, 548, 551, 553, 556-559, 561, 563-
568,
571-573, 575-581 or 582, such as said formula I may be one of SEQ ID NOs: 2 to
33.
[0449] In some embodiments of the invention, the nanoparticle is
conjugated
to an isolated peptide comprising formula II, wherein formula II is
XTFFVKLSX1X2
(SEQ ID NO: 173),
[0450] wherein X is KKLD (SEQ ID NO: 174), RKLD (SEQ ID NO: 175),
KKGD (SEQ ID NO: 176), KKED (SEQ ID NO: 177), KLD, LD, D, or absent
[0451] wherein X1 is LFT, LMT, LQT, LHT, LNT, LPT, LST, LGT, LAT,
LRT, QFT, QMT, QQT, QHT, QNT, QPT, QST, QGT, QAT, QRT, VFT, VMT, VQT,
VHT, VNT, VPT, VST, VGT, VAT, VRT, MFT, MMT, MQT, MHT, MNT, MPT,
MST, MGT, MAT, MRT, LFN, LMN, LQN, LHN, LNN, LPN, LSN, LGN, LAN, LRN,
QFN, QMN, QQN, QHN, QNN, QPN, QSN, QGN, QAN, QRN, VFN, VMN, VQN,
VHN, VNN, VPN, VSN, VGN, VAN, VRN, MFN, MMN, MQN, MHN, MNN, MPN,
MSN, MGN, MAN, MRN, LFP, LMP, LQP, LHP, LNP, LPP, LSP, LGP, LAP, LRP,
QFP, QMP, QQP, QHP, QNP, QPP, QSP, QGP, QAP, QRP, VFP, VMP, VQP, VHP,
VNP, VPP, VSP, VGP, VAP, VRP, MFP, MMP, MQP, MHP, MNP, MPP, MSP, MGP,
MAP, MRPR, LFR, LMR, LQR, LHR, LNR, LPR, LSR, LGR, LAR, LRR, QFR, QMR,
QQR, QHR, QNR, QPR, QSR, QGR, QAR, QRR, VFR, VMR, VQR, VHR, VNR, VPR,
VSR, VGR, VAR, VRR, MFR, MMR, MQR, MHR, MNR, MPR, MSR, MGR, MAR, or
MRR, or absent; and
[0452] wherein X2 is ER, or E, or absent, such as said formula II may
be one
of SEQ ID NO: 2-5, 19-38, 46-49, 58-61, 64, 68-70, 75, 81, 87, 93, 94, 100,
101, 173,
268-303, 350-393, 396, 398, 399, 400, 402, 403, 405, 406-408, 412-414, 417,
418, 421-
423, 426-428, 430, 431, 435, 436, 438, 439, 440-442, 448-455, 458, 459, 461,
465, 467,
468, 471, 475, 476, 478-481, 483, 485, 487, 489-491, 493, 494, 497-499, 503,
507, 510,
512, 514-517, 520, 521, 524, 525, 528, 529, 531, 533, 538, 539, 542-, 544,
546, 551,
559, 561, 563-568, 571-573, 575-577, 579, 580, or 581. Other examples includes
an
133
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
isolated peptide, wherein X is KKLD (SEQ ID NO: 174) or wherein X2 is ER or
wherein
said formula is TFFVKLSLFTER (SEQ ID NO: 49) or TFFVKLSLFTE (SEQ ID NO:
250) or wherein said formula is KKLDTFFVKLSLFTER (SEQ ID NO: 2) or
KKLDTFFVKLSLFTE (SEQ ID NO: 34).
[0453] In some embodiments of the invention, the nanoparticle is
conjugated
to an isolated peptide comprising Formula III, wherein Formula III is:
[0454] XX1VKLX2LX3TEX4 (SEQ ID NO: 178)
[0455] wherein X is KKLDTF (SEQ ID NO: 179), KLDTF (SEQ ID NO:
180), LDTF (SEQ ID NO: 181), DTF, TF, or F, or absent;
[0456] wherein Xi is F, M, S, V, T, or L, or absent;
[0457] wherein X2 is S, Q, M, T, or H, or absent;
[0458] wherein X3 is F, M, Q, H, N, P, S, G, A, or R, or absent; and
[0459] wherein X4 is R or absent.
[0460] Said formula III may be one of SEQ ID NO: 2-13, 15-18, 22-30,
34,
46-52, 58, 64, 65, 70, 71, 76, 77, 82, 83, 88, 93-96, 99, 100, 178, 268-325.
Examples
includes wherein X is KKLDTF (SEQ ID NO: 178) or wherein X4 is R or wherein
said
formula is VKLSLFTER (SEQ ID NO: 52) or VKLSLFTE (SEQ ID NO: 251) or
wherein said formula is KKLDTFFVKLSLFTER (SEQ ID NO: 2) or
KKLDTFFVKLSLFTE (SEQ ID NO: 34).
[0461] Other examples includes isolated peptides comprising at least
one of
SEQ ID NOs: 1-101, 167-172, 174-177, 179-393, 396-581 and 582 or at least one
of
SEQ ID NOs: 1-32, 34, 64-66, 68, 76, 94-96, 98, and 264-393 or at least one of
the
sequences of Table 5.1.
[0462] The above mentioned isolated peptides conjugatged to the
nanoparticle, may have at least one amino acid being a D amino acid,
artificial amino
acid, or chemically modified amino acid and/or comprise an N-terminal acetyl
group
and/or comprise a C-terminal amide group and/or be glycosylated or
nitrosylated.
[0463] The above mentioned isolated peptides conjugated to the
nanoparticle
may be joined to at least one of polyethylene glycol, a fatty acid, or a
pharmacokinetic
modifier and/or comprises a cyclic peptide.
[0464] The above mentioned isolated peptides conjugated to the
nanoparticle
may comprise at least one modification, for example at least one of a D amino
acid
134
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
a N-terminal acetyl group and/or a C-terminal amide group and/or glycosylation
and/or
nitrosylation and/or carbonylation and/or oxidation and/or a linked
pharmacokinetic
modifier and/or a linked polyethylene glycol or any combination thereof.
[0465] The
above mentioned isolated peptides conjugated to the nanoparticle
can be less than or equal to 1100 amino acids in length, such as between 7
amino acids
and 20 amino acids in length.
[0466] The
above mentioned isolated peptides conjugated to the nanoparticle
may be multimerized.
[0467] The
above defined peptides conjugated to the nanoparticle may be less
than or equal to 50, 49, 48, 47, 46, 45, 44, 43, 42, 41, 40, 39, 38, 37, 36,
35, 34 , 33, 32,
31, 30, 29, 28, 27, 26, 25, 24, 23, 22, 21, 20, 19, 16, 15, 14, 13, 12, 11,
10, 9, 8, 7, 6 or 5
amino acids in length or any length in between any of these numbers.
Administration form
[0468] The pharmaceutical formulations described herein (e.g.
immunoregulatory peptide inhibitors, and/or immunoregulatory peptide
inhibitors
immobilized on nanoparticles as described herein) may be administered locally
or
systemically. Routes of administration include topical, ocular, nasal,
pulmonar, buccal,
parenteral (intravenous, subcutaneous, and intramuscular), oral, vaginal and
rectal. Most
commonly used being oral administration.
[0469] In some
embodiments, for example if immune cell invasion of a
cytotoxicity of a tumor, or deblocking of a an immune cell receptor of a tumor
is desired,
the pharmaceutical formulation is administered at or near a tumor. For
example, the
pharmaceutical formulation can be administered peri-tumorally, or within 10cm
of the
tumor, for example within 10cm, 9, 8, 7, 6, 5, 4, 3, 2, 1, or 0.5cm of the
tumor or a range
defined by any two of these distances. Optionally, the pharmaceutical
formulation is
administered directly to a tumor, and induces regressive changes in the tumor.
Optionally, the pharmaceutical formulation is administered to a subject, and
induces
regressive changes of a tumor to which the composition is not directly
administered.
Optionally, the pharmaceutical formulation is administered directly to a
tumor, and
induces regressive changes in the tumor, and further induces regressive
changes in a
second tumor to which the formulation was not directly administered (e.g. a
metastatic or
contralateral tumor). Optionally, the pharmaceutical formulation is
administered directly
to a tumor, and induces eradication of the tumor. Optionally, the
pharmaceutical
135
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
formulation is administered to a subject, and induces eradication of a tumor
to which the
composition is not directly administered. Optionally, the pharmaceutical
formulation is
administered directly to a tumor, and induces eradication of the tumor, and
further
eradication of a second tumor to which the formulation was not directly
administered
a metastatic or contralateral tumor). Optionally, the pharmaceutical
formulation is
administered directly to a tumor, and induces immune cell infiltration of the
tumor.
Optionally, the pharmaceutical formulation is administered directly to a
tumor, and
induces immune cell infiltration of the tumor, and further induces immune cell
of a second tumor to which the formulation was not directly administered (e.g.
a
metastatic or contralateral tumor). Optionally, the pharmaceutical formulation
is
administered to a subject, and induces immune cell infiltration of a tumor to
which the
composition is not directly administered. Example tumors to which the
pharmaceutical
composition can be directly or indirect administered include a prostate tumor,
a
melanoma, a colon cancer, a lung carcinoma, an Apocrine gland carcinoma, a
testis
tumor, a mast cell tumor, a mammary tumor (e.g. a benign mammary tumor or a
malignant mammary tumor, for example a mixed mammary tumor such as a benign
mixed mammary tumor or a malignant mixed mammary tumor), a mucinous carcinoma
(e.g. a mammary gland mucinous carcinoma), or a histicytoma
[0470] The
pharmaceutical compositions will be administered to a patient in a
therapeutically effective amount or dose. A therapeutically effective amount
includes a
dose of pharmaceutical composition sufficient to at least partially arrest a
symptom of a
disorder from which a patient is suffering. The exact dose is dependent on the
manner of
administration, the nature and severity of the disorder. Depending on the
general health,
sex, age and body weight of the patient different doses may be needed. The
administration of the dose can be carried out both by single administration in
the form of
an individual dose unit or else several smaller dose units and also by
multiple
administration of subdivided doses at specific intervals, for example daily
intervals (e.g.,
1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 19, 20, 21, 22, 23,
24, 25, 26, 27,
29, or 30 days between doses, including ranges between any two of the listed
values).
Exemplary dosing can comprise doses in the milligram, microgram, or nanogram-
range,
for example milligrams, micrograms, or nanograms per kg of body weight of the
subject.
The active compounds or substances may also be administered together or
separately
depending on the administration form. Exemplary dosing regiments in accordance
with
136
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
some embodiments herein include "prime boost" approaches in which a first dose
of
compound or substance is administered in a first administration, and second
dose of
compound or substance is administered in second administration. Optionally,
additional
subsequent administrations (e.g. third, fourth, fifth, sixth, seventh, eighth,
ninth, or tenth)
are performed. Optionally, the first dose is greater than a subsequent dose
(e.g. the
second dose, or if performed, third, fourth, fifth, sixth, seventh, eighth,
ninth, or tenth),
for example at least 1.1x, 1.2x, 1.5x, 2x, 3x, 4x, 5x, 10x, 20x, 30x, 40x,
50x, 100x, 200x,
500x, 1000x, 2000x, 5000x, or 10000x of the subsequent dose. Optionally, the
subsequent dose (e.g. second, third, fourth, fifth, sixth, seventh, eighth,
ninth, or tenth) is
greater than the first dose, for example at least 1.1x, 1.2x, 1.5x, 2x, 3x,
4x, 5x, 10x, 20x,
30x, 40x, 50x, 100x, 200x, 500x, 1000x, 2000x, 5000x, or 10000x of the first
dose. In
some embodiments a subsequent dose (e.g. second dose after first dose, third
dose after
second dose, if performed, fourth dose after fifth dose, if performed) is
administered at
least one day after the preceding dose, for example, 1, 2, 3, 4, 5, 6, 7, 8,
9, 10, 11, 12, 13,
14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 60, 90, or
100 days after,
including ranges between any two of the listed values.
[0471] In some
embodiments, the immunoregualtory peptide inhibitor as
described herein is administered systemically to the subject (e.g., a subject
suffering
a cancer having more than one tumour, such as metastisis) so as to inhibit or
prevent
cancer (e.g., metastatic cancer) at a does of at least about 1 ng/kg, for
example at least
about 1 ng/kg, 2 ng/kg, 3 ng/kg, 4 ng/kg, 5 ng/kg, 6 ng/kg, 7 ng/kg, 8 ng/kg,
9 ng/kg, 10
ng/kg, 15 ng/kg, 20 ng/kg, 25 ng/kg 30 ng/kg, 35 ng/kg, 40 ng/kg, 45 ng/kg, 50
ng/kg, 60
ng/kg, 70 ng/kg, 80 ng/kg, 90 ng/kg, 100 ng/kg, 110 ng/kg, 120 ng/kg, 130
ng/kg, 140
ng/kg, 150 ng/kg, 160 ng/kg, 170 ng/kg, 180 ng/kg, 190 ng/kg, 200 ng/kg, 210
ng/kg, 220
ng/kg, 230 ng/kg, 240 ng/kg, 250 ng/kg, 260 ng/kg, 270, ng/kg, 280 ng/kg, 290
ng/kg,
ng/kg, 310 ng/kg, 320 ng/kg, 330 ng/kg, 340 ng/kg, 350 ng/kg, 360 ng/kg, 370,
ng/kg,
ng/kg, 390 ng/kg, 400 ng/kg, 410 ng/kg, 420 ng/kg, 430 ng/kg, 440 ng/kg, 450
ng/kg, 460
ng/kg, 470, ng/kg, 480 ng/kg, 490 ng/kg, 200 ng/kg, 510 ng/kg, 520 ng/kg, 530
ng/kg,
ng/kg, 550 ng/kg, 560 ng/kg, 570, ng/kg, 580 ng/kg, 590 ng/kg, 200 ng/kg, 610
ng/kg,
ng/kg, 630 ng/kg, 640 ng/kg, 650 ng/kg, 660 ng/kg, 670, ng/kg, 680 ng/kg, 690
ng/kg,
ng/kg, 710 ng/kg, 720 ng/kg, 730 ng/kg, 740 ng/kg, 750 ng/kg, 760 ng/kg, 770,
ng/kg,
ng/kg, 790 ng/kg, 800 ng/kg, 810 ng/kg, 820 ng/kg, 830 ng/kg, 840 ng/kg, 850
ng/kg, 860
ng/kg, 870, ng/kg, 880 ng/kg, 890 ng/kg, 900 ng/kg, 910 ng/kg, 920 ng/kg, 930
ng/kg,
137
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
ng/kg, 950 ng/kg, 960 ng/kg, 970, ng/kg, 980 ng/kg, 990 ng/kg, 1000 ng/kg, 2
pg/kg, 3
pg/kg, 4 pg/kg, 5 pg/kg, 6 pg/kg, 7 pg/kg, 8 pg/kg, 9 pg/kg, 10 pg/kg, 15
pg/kg, 20
25 pg/kg, 30 pg/kg, 40 pg/kg, 50 pg/kg, 60 pg/kg, 70 pg/kg, 80 pg/kg, 90
pg/kg, 100
pg/kg, 110 pg/kg, 120 pg/kg, 130 pg/kg, 140 pg/kg, 150 pg/kg, 160 pg/kg, 170
pg/kg,
180 pg/kg, 190 pg/kg, 200 pg/kg, 250 pg/kg, 300 pg/kg, 350 pg/kg, 400 pg/kg,
450
pg/kg, 500 pg/kg, 600 pg/kg, 700 pg/kg, 800 pg/kg, 900 pg/kg, 1000 pg/kg, 2000
pg/kg,
3000 pg/kg, 4000 pg/kg, or 5000 pg/kg, including ranges between any two of the
listed
values, for example 1 ng/kg ¨ 100 ng/kg, 1 ng/kg ¨ 200 ng/kg, 1 ng/kg ¨ 300
ng/kg, 1
ng/kg ¨ 400 ng/kg, 1 ng/kg ¨ 500 ng/kg, 1 ng/kg ¨ 600 ng/kg, 1 ng/kg ¨ 700
ng/kg, 1
ng/kg ¨ 800 ng/kg, 1 ng/kg ¨ 900 ng/kg, 1 ng/kg ¨ 1000 ng/kg, 1 ng/kg ¨ 2
pg/kg, 1 ng/kg
¨ 3 pg/kg, 1 ng/kg ¨ 5 pg/kg, 1 ng/kg ¨ 10 pg/kg, 1 ng/kg ¨ 20 pg/kg, 1
ng/kg ¨ 50 pg/kg,
1 ng/kg ¨ 100 pg/kg, 1 ng/kg ¨ 200 pg/kg, 1 ng/kg ¨ 500 pg/kg, 1 ng/kg ¨ 1000
pg/kg, 5
ng /kg ¨ 100 ng/kg, 5 ng /kg ¨ 200 ng/kg, 5 ng /kg ¨ 300 ng/kg, 5 ng /kg ¨ 400
ng/kg, 5
/kg ¨ 500 ng/kg, 5 ng /kg ¨ 600 ng/kg, 5 ng /kg ¨ 700 ng/kg, 5 ng /kg ¨ 800
ng/kg, 5 ng
/kg ¨ 900 ng/kg, 5 ng /kg ¨ 1000 ng/kg, 5 ng/kg ¨ 2 pg/kg, 5 ng/kg ¨ 3 pg/kg,
5 ng/kg ¨ 5
pg/kg, 5 ng/kg ¨ 10 pg/kg, 5 ng/kg ¨ 20 pg/kg, 5 ng/kg ¨ 50 pg/kg, 5 ng/kg ¨
100 pg/kg,
ng/kg ¨ 200 pg/kg, 5 ng/kg ¨ 500 pg/kg, 5 ng/kg ¨ 1000 pg/kg, 10 ng /kg ¨ 100
ng/kg, 10
ng /kg ¨ 200 ng/kg, 10 ng /kg ¨ 300 ng/kg, 10 ng /kg ¨ 400 ng/kg, 10 ng /kg ¨
500 ng/kg,
ng /kg ¨ 600 ng/kg, 10 ng /kg ¨ 700 ng/kg, 10 ng /kg ¨ 800 ng/kg, 10 ng /kg ¨
900
ng/kg, 10 ng /kg ¨ 1000 ng/kg, 10 ng/kg ¨ 2 pg/kg, 10 ng/kg ¨ 3 pg/kg, 10
ng/kg ¨ 5
pg/kg, 10 ng/kg ¨ 10 pg/kg, 10 ng/kg ¨ 20 pg/kg, 10 ng/kg ¨ 50 pg/kg, 10 ng/kg
¨ 100
pg/kg, 10 ng/kg ¨ 200 pg/kg, 10 ng/kg ¨ 500 pg/kg, 10 ng/kg ¨ 1000 pg/kg, 20
ng /kg ¨
100 ng/kg, 20 ng /kg ¨ 200 ng/kg, 20 ng /kg ¨ 300 ng/kg, 20 ng /kg ¨ 400
ng/kg, 20 ng
¨ 500 ng/kg, 20 ng /kg ¨ 600 ng/kg, 20 ng /kg ¨ 700 ng/kg, 20 ng /kg ¨ 800
ng/kg, 20 ng
/kg ¨ 900 ng/kg, 20 ng /kg ¨ 1000 ng/kg, 20 ng/kg ¨ 2 pg/kg, 20 ng/kg ¨ 3
pg/kg, 20
ng/kg ¨ 5 pg/kg, 20 ng/kg ¨ 10 pg/kg, 20 ng/kg ¨ 20 pg/kg, 20 ng/kg ¨ 50
pg/kg, 20
¨ 100 pg/kg, 20 ng/kg ¨ 200 pg/kg, 20 ng/kg ¨ 500 pg/kg, 20 ng/kg ¨ 1000
pg/kg, 30 ng
/kg ¨ 100 ng/kg, 30 ng /kg ¨ 200 ng/kg, 30 ng /kg ¨ 300 ng/kg, 30 ng /kg ¨ 400
ng/kg, 30
ng /kg ¨ 500 ng/kg, 30 ng /kg ¨ 600 ng/kg, 30 ng /kg ¨ 700 ng/kg, 30 ng /kg ¨
800 ng/kg,
30 ng /kg ¨ 900 ng/kg, 30 ng /kg ¨ 1000 ng/kg, 30 ng/kg ¨ 2 pg/kg, 30 ng/kg ¨
3 pg/kg,
30 ng/kg ¨ 5 pg/kg, 30 ng/kg ¨ 10 pg/kg, 30 ng/kg ¨ 20 pg/kg, 30 ng/kg ¨ 50
pg/kg, 30
ng/kg ¨ 100 pg/kg, 30 ng/kg ¨ 200 pg/kg, 30 ng/kg ¨ 500 pg/kg, 30 ng/kg ¨ 1000
pg/kg,
40 ng /kg ¨ 100 ng/kg, 40 ng /kg ¨ 200 ng/kg, 40 ng /kg ¨ 300 ng/kg, 40 ng /kg
¨ 400
138
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
ng/kg, 40 ng /kg ¨ 500 ng/kg, 40 ng /kg ¨ 600 ng/kg, 40 ng /kg ¨ 700 ng/kg, 40
ng /kg ¨
800 ng/kg, 40 ng /kg ¨ 900 ng/kg, 40 ng /kg ¨ 1000 ng/kg, 40 ng/kg ¨ 2 pg/kg,
40 ng/kg ¨
3 pg/kg, 40 ng/kg ¨ 5 pg/kg, 40 ng/kg ¨ 10 pg/kg, 40 ng/kg ¨ 20 pg/kg, 40
ng/kg ¨ 50
pg/kg, 40 ng/kg ¨ 100 pg/kg, 40 ng/kg ¨ 200 pg/kg, 40 ng/kg ¨ 500 pg/kg, 40
ng/kg ¨
1000 pg/kg, 50 ng /kg ¨ 100 ng/kg, 50 ng /kg ¨ 200 ng/kg, 50 ng /kg ¨ 300
ng/kg, 50 ng
/kg ¨ 400 ng/kg, 50 ng /kg ¨ 500 ng/kg, 50 ng /kg ¨ 600 ng/kg, 50 ng /kg ¨ 700
ng/kg, 50
ng /kg ¨ 800 ng/kg, 50 ng /kg ¨ 900 ng/kg, 50 ng /kg ¨ 1000 ng/kg, 50 ng/kg ¨
2 pg/kg,
50 ng/kg ¨ 3 pg/kg, 50 ng/kg ¨ 5 pg/kg, 50 ng/kg ¨ 10 pg/kg, 50 ng/kg ¨ 20
pg/kg, 50
ng/kg ¨ 50 pg/kg, 50 ng/kg ¨ 100 pg/kg, 50 ng/kg ¨ 200 pg/kg, 50 ng/kg ¨ 500
pg/kg, 50
ng/kg ¨ 1000 pg/kg, 100 ng /kg ¨ 200 ng/kg, 100 ng /kg ¨ 300 ng/kg, 100 ng /kg
¨ 400
ng/kg, 100 ng /kg ¨ 500 ng/kg, 100 ng /kg ¨ 600 ng/kg, 100 ng /kg ¨ 700 ng/kg,
100 ng
/kg ¨ 800 ng/kg, 100 ng /kg ¨ 900 ng/kg, 100 ng /kg ¨ 1000 ng/kg, 100 ng/kg ¨
2 pg/kg,
100 ng/kg ¨ 3 pg/kg, 100 ng/kg ¨ 5 pg/kg, 100 ng/kg ¨ 10 pg/kg, 100 ng/kg ¨ 20
pg/kg,
100 ng/kg ¨ 50 pg/kg, 100 ng/kg ¨ 100 pg/kg, 100 ng/kg ¨ 200 pg/kg, 100 ng/kg
¨ 500
pg/kg, 100 ng/kg ¨ 1000 pg/kg, 200 ng /kg ¨ 300 ng/kg, 200 ng /kg ¨ 400 ng/kg,
200 ng
/kg ¨ 500 ng/kg, 200 ng /kg ¨ 600 ng/kg, 200 ng /kg ¨ 700 ng/kg, 200 ng /kg ¨
800 ng/kg,
200 ng /kg ¨ 900 ng/kg, 200 ng /kg ¨ 1000 ng/kg, 200 ng/kg ¨ 2 pg/kg, 200
ng/kg ¨ 3
pg/kg, 200 ng/kg ¨ 5 pg/kg, 200 ng/kg ¨ 10 pg/kg, 200 ng/kg ¨ 20 pg/kg, 200
ng/kg ¨ 50
pg/kg, 200 ng/kg ¨ 100 pg/kg, 200 ng/kg ¨ 200 pg/kg, 200 ng/kg ¨ 500 pg/kg,
200 ng/kg
¨ 1000 pg/kg, 500 ng /kg ¨ 600 ng/kg, 500 ng /kg ¨ 700 ng/kg, 500 ng /kg ¨ 800
ng/kg,
500 ng /kg ¨ 900 ng/kg, 500 ng /kg ¨ 1000 ng/kg, 500 ng/kg ¨ 2 pg/kg, 500
ng/kg ¨ 3
pg/kg, 500 ng/kg ¨ 5 pg/kg, 500 ng/kg ¨ 10 pg/kg, 500 ng/kg ¨ 20 pg/kg, 500
ng/kg ¨ 50
pg/kg, 500 ng/kg ¨ 100 pg/kg, 500 ng/kg ¨ 200 pg/kg, 500 ng/kg ¨ 500 pg/kg, or
500
ng/kg ¨ 1000 pg/kg. In some embodiments, the dosage is administered in a
suitable
volume of solution. The exact volume is dependent on the dosage, manner of
administration, and the nature and severity of the disorder. For example,
suitable
can include about 10 pi, 20 pi, 30 pi, 40 pi, 50 pi, 60 pi, 70 pi, 80 pi, 90
pi, 100 pi, 110
pi, 120 pi, 130 pi, 140 pi, 150 pi, 160 pi, 170 pi, 180 pi, 190 pi, 200 pi,
250 pi, 300 pi,
250 pi, 400 pi, 450 pi, 500 pi, 550 pi, 600 pi, 650 p1,700 pi, 750 pi, 800 pi,
850 pi, 900
pi, 950 pi, 1000 pi, 1100 p1,1200 pi, 1300 pi, 1400 pi, 1500 pi, 1600 pi, 1700
p1,1800
pi, 1900 pi, 2000 pi, 2500 pi, 3000 pi, 3500 pi, 4000 pi, 4500 pi, 5000 pi,
6000 pi, 7000
pi, 8000 pi, 9000 pi, and 10000 pi, including ranges between any of two of the
listed
values, for example, about 10 pi ¨ 1000 pi, 10 pi ¨ 5000 pi, 10 pi - 10000 pi,
50 tl ¨
139
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
1000 Ill, 50 tl ¨ 5000 Ill, 50 tl - 10000 Ill, 100 Ill -1000 Ill, 100 Ill -
5000 Ill, 100 Ill -
10000 Ill, 500 tl ¨ 1000 Ill, 500 tl ¨ 5000 Ill, or 500 tl ¨ 10000 1.11. A
number of
manners of systemic administration are contemplated. For example, the systemic
administration can be performed subcutaneously, intravenously, orally,
intraperitoneally,
or peritumorally or intratumorally (contemplating that the intratumoral
administration
also ameliorate and/or eliminate tumors other than the one(s) at or near the
site of
administration).
[0472] Suitable
preparation forms are, for example granules, powders, tablets,
coated tablets, (micro) capsules, microgranulates effervescent powders or
granules,
suppositories, injectable solution in ampule form and also preparations with
protracted
release of active compounds, in whose preparation excipients, diluents or
carriers are
customarily used as described above. Other preparations may be those which
give rise to
different release profiles of the active ingredients which are well-known for
a person
skilled in the art. Examples include sustained-release, sustained-action,
extended-
time-release or timed-release, controlled-release, modified release, or
continuous-release.
The advantages of sustained-release tablets or capsules are that they can
often be taken
less frequently than immediate-release formulations of the same drug, and that
they keep
steadier levels of the drug in the bloodstream. Today, many time-release drugs
are
formulated so that the active ingredient is embedded in a matrix of insoluble
substance(s)
(for example some acrylics, or chitin) such that the dissolving drug must find
its way out
through the holes in the matrix. Some drugs are enclosed in polymer-based
tablets with a
laser-drilled hole on one side and a porous membrane on the other side.
Stomach acids
push through the porous membrane, thereby pushing the drug out through the
laser-
hole. In time, the entire drug dose releases into the system while the polymer
container
remains intact, to be excreted later through normal digestion. In some
formulations, the
drug dissolves into the matrix, and the matrix physically swells to form a
gel, allowing
drug to exit through the gel's outer surface. Micro-encapsulation is also
regarded as a
more complete technology to produce complex dissolution profiles. Through
coating an
active pharmaceutical ingredient around an inert core, and layering it with
insoluble
substances to form a microsphere it is possible to obtain more consistent and
replicable
dissolution rates. In some embodiments, the composition comprises at least
about at
0.1% of the immunoregulatory peptide inhibitor by weight, for example, at
least 0.1%,
0.2%, 0.5%, 1%, 2%, 3%, 4%, 5%, 6%, 7%, 8%, 9%, 10%, 20%, or 30% of the
140
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
immunoregulatory peptide inhibitor by weight, including ranges between any two
of the
listed values. All of those being well-known for a person skilled in the art.
Method of treating, preventing, or inhibiting multiple tumors, such as in
metastatic
cancer
[0473] Many
conditions and diseases are associated with immunosuppression,
for example, many types of cancer, infection, and inflammatory disease are
associated
with immunosuppression. Thus,
exemplary conditions associated with
immunosuppression that can be treated, prevented, or inhibited using one or
more of the
immunoregulatory peptide inhibitors described herein include many types of
cancer, such
as colorectal cancer, colon cancer, renal cancer, breast cancer, skin cancer,
ovarian
cancer, prostate cancer, pancreatic cancer, lung cancer, or hematopoietic cell
cancer.
Additional examples include a prostate tumor, a melanoma, a lung carcinoma, an
Apocrine gland carcinoma, a testis tumor, a mast cell tumor, a mammary tumor
(e.g. a
benign mammary tumor or a malignant mammary tumor, for example a mixed mammary
tumor such as a benign mixed mammary tumor or a malignant mixed mammary
tumor),
mucinous carcinoma (e.g. a mammary gland mucinous carcinoma), or a
histicytoma. In
particular, it is contemplated that cancers comprising two or more tumors, for
example
metastatic cancer, or two or more tumors (in which the tumors can be of the
same or
different type of cancer, for example any of the cancers listed herein, or any
two
cancers listed here) can be treated, prevented, inhibited, or ameliorated by
the
immunoregulatory peptide inhibitors described herein.
Optionally, the
peptide inhibitors or compositions comprising the immunoregulatory peptide
inhibitors
described herein can be administerd to a subset of one or more tumors in a
subject (but
not all of the tumors), so as to treat, prevent, inhibit, or ameliorate at
least one tumor that
was not at or near the site of administration. Optionally, the
immunoregualtory peptide
inhibitors are administered to a primary tumor (but not a remore tumor) of the
metastasis
so as to treat primary and metastatic tumors. Optionally, the immunoregualtory
peptide
inhibitors are administered to a metastatic tumor (but not a primary tumor) of
the
metastasis so as to treat primary and metastatic tumors.
Optionally, the
immunoregualtory peptide inhibitors are administered to some, but not all
primary and
metastatic tumors of the metastasis so as to treat primary and metastatic
tumors.
Optionally, the immunoregualtory peptide inhibitors are administered to
primary and
141
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
metastatic tumors of the metastasis so as to treat these primary and
metastatic tumors.
Optionally, the immunoregulatory peptide inhibitors or compositions comprising
the
immunoregulatory peptide inhibitors as described herein can be administerd
systemically
to a subject having two or more tumors, so as to treat, prevent, inhibit, or
ameliorate two
or more tumors in the subject, even if at least one tumor was not at or near
the site of
administration. Optionally, the immunoregulatory peptide inhibitors or
compositions
comprising the immunoregulatory peptide inhibitors can induce regressive
changes in,
immune cell infiltration of, and/or eradication of the tumor, even if the
tumor itself was
not at or near the site of administration of the immunoregulaotyr peptide
inhibitor.
Exemplary conditions associated with immunosuppression that can be treated,
prevented,
or inhibited by using one or more of the immunoregulatory peptide inhibitors
described
herein further include hormonal imbalances, such as increased and/or ectopic
cortisol
activity.
[0474]
Accordingly, some embodiments include methods of treating,
preventing, or reducing immunosuppression or one or more of the aforementioned
infections or diseases in a human, in which the diseases comprise two or more
tumors,
example metastatic cancer, or two or more tumors at two or more different
sites in the
human (in which any two of the tumors can be of the same type of cancer or a
different
type of cancer). In some embodiments, the method includes identifying a
patient having
acondition associated with immunosuppression and comprising two or more
tumors, for
example metastatic cancer, and/or a first tumor at a first site and a
metastatic tumor at a
second site (in which the first and second tumors can be of the same type of
cancer, or of
different types of cancer). By way of example, the metatstatic cancer can
comprise one
more primary tumors, and one or more remote tumors. Such an identification
step can be
accomplished by clinical evaluation (e.g., CT, MRI, or PET scan) or diagnostic
assay.
The method further includes administering to the identified or selected
patient a
composition comprising, consisting of, or consisting essentially of an
immunoregulatory
peptide inhibitor sequence (for example, a composition comprising an
immunoregulatory
peptide inhibitor immobilized on a nanoparticle as described herein), or a
nucleic acid
encoding such a molecule as described herein. For example, the composition
consisting of, or consisting essentially of an immunoregulatory peptide
inhibitor can
include any one of SEQ ID NOs: 1-33, 34, 46-53, 64-66, 68, 76, 94-96, 98, 264-
393,
583-586, or 589 or any one or more of the peptides provided in Table 5.1, 5.4,
5.5, 5.6,
142
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
any variation or combination of variations of P28R or P28 core as provided in
Tables 5.3
and 13. In some embodiments, the composition is administered peri-tumorally,
or near a
tumor, for example within 10, 9, 8, 7, 6, 5, 4, 3, 2, 1, or 0.5cm of a first
tumor in the
subject, but is not administered peri-tumorally or near a second tumor in the
subject (so
that the composition is not administered directly to the second tumor). Both
the first and
second tumors can undergo regressive changes, so as to treat ameliorate both
the first and
second tumor. Optionally, the first tumor is a primary tumor, and the second
tumor is a
metastatic tumor. Optionally, the first tumor is a metastatic tumor, and the
second tumor
is a primary tumor Optionally, the first tumor and secondary tumors are both
primary
tumors. Optionally, the first tumor and secondary tumors are both metastatic
tumors.
Optionally, the first and second tumor are of the same type of cancer.
Optionally, the
second tumor is of a different type of cancer than the first tumor.
Optionally, the first
tumor and second tumor are part of a metastatic cancer. Optionally, the first
tumor and
second tumor are in different tissues of the subject. Optionally, the first
tumor and
tumor are in the same tissue, but the tumors are at least 1 mm apart from each
other, for
example, at least 1 mm, 2 mm, 3 mm, 4 mm, 5 mm, 6 mm, 7 mm, 8 mm, 9 mm, 10 mm,
11 mm, 12 mm, 13 mm, 14 mm, 15 mm, 16 mm, 17 mm, 18 mm, 19 mm, 20 mm, 25
mm, 30 mm, 40 mm, 45 mm, 50 mm 55 mm, 60 mm, 65 mm, 70 mm, 80 mm, 90 mm,
100 mm, 200 mm, 300 mm, 400 mm, or 500 mm apart. Some embodiments include
methods of treating, preventing, or reducing immunosuppression or one or more
of the
aforementioned infections or diseases in a non-human mammal, in which the
diseases
comprise two or more tumors, for example metastatic cancer, or two or more
tumors at
two or more different sites in the non-human mammal (in which any two of the
tumors
can be of the same type of cancer or a different type of cancer).
[0475] In some
embodiments, the composition is administered systemically.
In some embodiments, the composition is administered in conjunction with a
second
therapeutic agent, for example a therapeutic agent selected to stimulate an
immune cell
after an LFA-1 receptor of the immune cell has been de-blocked (e.g. bound
immunoregulatory peptides or 3028 structures have been displaced from the LFA-
1
receptor). In some embodiments, these isolated peptides used in these methods
have a
length that is less than or equal to 1100 amino acids, for example, less than
or equal to 4,
5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25,
26, 27, 28, 29,
30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48,
49, 50, 51, 52,
143
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72,
73, 74, 75, 76,
78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96,
97, 98, 99, 100,
101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115,
116, 117, 118,
119, 120, 121, 122, 123, 124, 125, 126, 127, 128, 129, 130, 131, 132, 133,
134, 135, 136,
137, 138, 139, 140, 150, 160, 170, 180, 190, 200, 210, 220, 230, 240, 250,
260, 270, 280,
290, 300, 320, 340, 360, 380, 400, 450, 500, 550, 600, 650, 700, 750, 800,
850, 900, 950,
1000, 1050, or 1100 amino acids, including ranges between any two of the
listed values.
Optionally, the composition is administered directly to a tumor, and induces
regressive
changes in the tumor. Optionally, the composition is administered to a
subject, and
induces regressive changes of a tumor to which the composition is not directly
administered. Optionally, the composition is administered directly to a tumor,
and
induces regressive changes in the tumor, and further induces regressive
changes in a
second tumor to which the formulation was not directly administered (e.g. a
metastatic or
contralateral tumor). Optionally, the composition is administered directly to
a tumor, and
induces eradication of the tumor. Optionally, the composition is administered
to a
subject, and induces eradication of a tumor to which the composition is not
directly
administered. Optionally, the composition is administered directly to a tumor,
and
induces eradication of the tumor, and further induces eradication of a second
tumor to
which the formulation was not directly administered (e.g. a metastatic or
contralateral
tumor). Optionally, the composition is administered directly to a tumor, and
induces
immune cell infiltration of the tumor. Optionally, the composition is
administered
directly to a tumor, and induces immune cell infiltration of the tumor, and
further
immune cell infiltration of a second tumor to which the formulation was not
directly
administered (e.g. a metastatic or contralateral tumor). Optionally, the
composition is
administered to a subject, and induces immune cell infiltration of a tumor to
which the
composition is not directly administered. Example tumors to which the
pharmaceutical
composition can be directly or indirect administered include a prostate tumor,
a
melanoma, a colon cancer, a lung carcinoma, an Apocrine gland carcinoma, a
testis
tumor, a mast cell tumor, a mammary tumor (e.g. a malignant mammary tumor, for
example a mixed mammary tumor such as a malignant mixed mammary tumor), a
mucinous carcinoma (e.g. a mammary gland mucinous carcinoma), or a
histicytoma. As
shown in Figures 55-92 and Table 16, administration of compositions comprising
144
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
immunoregulatory peptide inhibitors as described herein induced regressive
changes,
immune cell infiltration of, and/or eradication of tumors.
[0476]
Additionally, the composition comprising, consisting of, or consisting
essentially of the immunoregulatory peptide inhibitor used in these methods
can
comprise, consist of, or consist essentially of a peptide as described herein,
or a nucleic
acid encoding such a molecule. For example, the peptide inhibitor used in
these methods
can comprise, consist of, or consist essentially of Formula (I), XX1VKX2X3X4
(SEQ ID
NO: 166) as described herein. In some embodiments, X is an optional sequence,
and can
be KKLDT (SEQ ID NO: 167), RKLDT (SEQ ID NO: 168), KKGDT (SEQ ID NO:
169), KKEDT (SEQ ID NO: 170), KKLDQ (SEQ ID NO: 171), KKGDQ (SEQ ID
252), KKEDQ (SEQ ID NO: 253), RKLDQ (SEQ ID NO: 254), RKGDQ (SEQ ID
255), RKEDQ (SEQ ID NO: 256), RKGTD (SEQ ID NO: 257), RKEDT (SEQ ID NO:
258), KLDT (SEQ ID NO: 172), KGDT (SEQ ID NO: 259), KEDT (SEQ ID NO:
KLDQ (SEQ ID NO: 261), KGDQ (SEQ ID NO: 262), KEDQ (SEQ ID NO: 263),
LDT, LDQ, GDT, GDQ, EDT, EDQ, DT, DQ, T, or Q, or absent. In some
Xi is one of FF, FM, FS, FV, FT, FL, AF, AM, AS, AV, AT, AL, VF, VM, VS, VV,
VT,
or VL. In some embodiments, X2 is one of LS, LQ, LM, LT, LH, VS, VQ, VM, VT,
or
VH. In some embodiments, X3 is one of LFT, LMT, LQT, LHT, LNT, LPT, LST, LGT,
LAT, LRT, QFT, QMT, QQT, QHT, QNT, QPT, QST, QGT, QAT, QRT, VFT, VMT,
VQT, VHT, VNT, VPT, VST, VGT, VAT, VRT, MFT, MMT, MQT, MHT, MNT,
MST, MGT, MAT, MRT, LFN, LMN, LQN, LHN, LNN, LPN, LSN, LGN, LAN, LRN,
QFN, QMN, QQN, QHN, QNN, QPN, QSN, QGN, QAN, QRN, VFN, VMN, VQN,
VHN, VNN, VPN, VSN, VGN, VAN, VRN, MFN, MMN, MQN, MHN, MNN, MPN,
MSN, MGN, MAN, MRN, LFP, LMP, LQP, LHP, LNP, LPP, LSP, LGP, LAP, LRP,
QFP, QMP, QQP, QHP, QNP, QPP, QSP, QGP, QAP, QRP, VFP, VMP, VQP, VHP,
VNP, VPP, VSP, VGP, VAP, VRP, MFP, MMP, MQP, MHP, MNP, MPP, MSP, MGP,
MAP, MRPR, LFR, LMR, LQR, LHR, LNR, LPR, LSR, LGR, LAR, LRR, QFR, QMR,
QQR, QHR, QNR, QPR, QSR, QGR, QAR, QRR, VFR, VMR, VQR, VHR, VNR, VPR,
VSR, VGR, VAR, VRR, MFR, MMR, MQR, MHR, MNR, MPR, MSR, MGR, MAR, or
MRR. In some embodiments, X4 is an optional sequence, and can be ER, or E, or
In some embodiments, if X is absent, X1 is FF, and X2 is LS. In some
embodiments, the
isolated peptides that comprise Formula (I) used in these methods have a
length that is
less than or equal to 1100 amino acids, for example, less than or equal to 4,
5, 6, 7, 8, 9,
145
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28,
29, 30, 31, 32,
34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52,
53, 54, 55, 56,
58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76,
77, 78, 79, 80,
82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100,
101, 102, 103,
104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115, 116, 117, 118,
119, 120, 121,
122, 123, 124, 125, 126, 127, 128, 129, 130, 131, 132, 133, 134, 135, 136,
137, 138, 139,
140, 150, 160, 170, 180, 190, 200, 210, 220, 230, 240, 250, 260, 270, 280,
290, 300, 320,
340, 360, 380, 400, 450, 500, 550, 600, 650, 700, 750, 800, 850, 900, 950,
1000, 1050,
1100 amino acids, including ranges between any two of the listed values.
[0477]
Additionally, the peptide inhibitor used in these methods can
consist of, or consist essentially of Formula (II), X20TFFVKLSX21X22 (SEQ ID
NO:
173), as described herein. In some embodiments, X20 is an optional sequence,
and can be
KKLD (SEQ ID NO: 174), RKLD (SEQ ID NO: 175), KKGD (SEQ ID NO: 176),
KKED (SEQ ID NO: 177), KLD, LD, D, or absent. X21 is an optional sequence, and
be LFT, LMT, LQT, LHT, LNT, LPT, LST, LGT, LAT, LRT, QFT, QMT, QQT, QHT,
QNT, QPT, QST, QGT, QAT, QRT, VFT, VMT, VQT, VHT, VNT, VPT, VST, VGT,
VAT, VRT, MFT, MMT, MQT, MHT, MNT, MPT, MST, MGT, MAT, MRT, LFN,
LMN, LQN, LHN, LNN, LPN, LSN, LGN, LAN, LRN, QFN, QMN, QQN, QHN, QNN,
QPN, QSN, QGN, QAN, QRN, VFN, VMN, VQN, VHN, VNN, VPN, VSN, VGN,
VAN, VRN, MFN, MMN, MQN, MHN, MNN, MPN, MSN, MGN, MAN, MRN, LFP,
LMP, LQP, LHP, LNP, LPP, LSP, LGP, LAP, LRP, QFP, QMP, QQP, QHP, QNP, QPP,
QSP, QGP, QAP, QRP, VFP, VMP, VQP, VHP, VNP, VPP, VSP, VGP, VAP, VRP,
MFP, MMP, MQP, MHP, MNP, MPP, MSP, MGP, MAP, MRPR, LFR, LMR, LQR,
LHR, LNR, LPR, LSR, LGR, LAR, LRR, QFR, QMR, QQR, QHR, QNR, QPR, QSR,
QGR, QAR, QRR, VFR, VMR, VQR, VHR, VNR, VPR, VSR, VGR, VAR, VRR,
MMR, MQR, MHR, MNR, MPR, MSR, MGR, MAR, or MRR, or absent. In some
embodiments, X22 is an optional sequence, and can be ER, or E, or absent. In
some
embodiments, the isolated peptides that comprise Formula (II) used in these
methods
a length that is less than or equal to 1100 amino acids, for example, less
than or equal to
4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24,
25, 26, 27, 28,
30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48,
49, 50, 51, 52,
54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72,
73, 74, 75, 76,
78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96,
97, 98, 99, 100,
146
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115,
116, 117, 118,
119, 120, 121, 122, 123, 124, 125, 126, 127, 128, 129, 130, 131, 132, 133,
134, 135, 136,
137, 138, 139, 140, 150, 160, 170, 180, 190, 200, 210, 220, 230, 240, 250,
260, 270, 280,
290, 300, 320, 340, 360, 380, 400, 450, 500, 550, 600, 650, 700, 750, 800,
850, 900, 950,
1000, 1050, or 1100 amino acids, including ranges between any two of the
listed values.
[0478]
Additionally, the peptide inhibitor used in these methods can
comprise, consist of, or consist essentially of Formula (III),
X30X31VKLX32LX33TEX34
(SEQ ID NO: 178). In some embodiments, X30 is an optional sequence, and can be
KKLDTF (SEQ ID NO: 179), KLDTF (SEQ ID NO: 180), LDTF (SEQ ID NO: 181),
DTF, TF, or F, or absent. In some embodiments, X31 is an optional sequence,
and can be
F, S, M, V, T, or L, or absent. In some embodiments, X31 is F. In some
embodiments,
X32 can be S, Q, M, T, or H. In some embodiments, X32 is S. X33 can be F, M,
Q, H, N,
P, S, G, A, or R. In some embodiments, X34 is F. X34 is an optional sequence,
and can
be R or absent. In some embodiments, the isolated peptides that comprise
Formula (III)
used in these methods have a length that is less than or equal to 1100 amino
acids, for
example, less than or equal to 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16,
17, 18, 19, 20,
21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39,
40, 41, 42, 43,
44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62,
63, 64, 65, 66,
67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85,
86, 87, 88, 89,
90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105, 106,
107, 108, 109,
110, 111, 112, 113, 114, 115, 116, 117, 118, 119, 120, 121, 122, 123, 124,
125, 126, 127,
128, 129, 130, 131, 132, 133, 134, 135, 136, 137, 138, 139, 140, 150, 160,
170, 180, 190,
200, 210, 220, 230, 240, 250, 260, 270, 280, 290, 300, 320, 340, 360, 380,
400, 450, 500,
550, 600, 650, 700, 750, 800, 850, 900, 950, 1000, 1050, or 1100 amino acids,
including
ranges between any two of the listed values.
[0479]
Additionally, the peptide inhibitor used in these methods can
consist of, or consist essentially of Formula (VII), X700K X701X702X703
X704X705X706K
X707 X708 X700 X710 X711E X712 (SEQ ID NO: 394), as described herein. In some
embodiments, X700 is an optional sequence, and can be
K,A,D,E,G,H,I,L,M,N,P,Q,R,T,
V, or absent. In some embodiments, X701 is an optional sequence, and can be
L,A,C,D,E,F,G,H,I,K,M,N,Q,R,S,T, or V, or absent. In some embodiments, X702 is
an
optional sequence, and can be D,A,E,I,V,W, or Y, or absent. In some
embodiments,
is an optional sequence, and can be T,C,M,N,P,Q,R,S,W, or Y, or absent. In
some
147
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
embodiments, X704 is an optional sequence, and can be F,A,I,M,N,P,T, or V, or
absent.
some embodiments, X705 is an optional sequence, and can be F,L,M,Q,S,T, or V,
or
absent. In some embodiments, X706 is an optional sequence, and can be
V,F,G,L,P, or R,
or absent. In some embodiments, X707 is an optional sequence, and can be
L,A,F,G,I,M,N,P,Q,R,S,T,V, or Y, or absent. In some embodiments, X708 is an
optional
sequence, and can be S,H,M,N,Q, or T, or absent. In some embodiments, X709 is
an
optional sequence, and can be L,A,H,I,M,N,Q,R,S,T,V, or W, or absent. In some
embodiments, X710 is an optional sequence, and can be
F,A,C,G,H,I,L,M,N,P,Q,R,S,T,V,
or W, or absent. In some embodiments, X711 is an optional sequence, and can be
T,F,G,H,I,L,M,N,P,S,V, or W, or absent. In some embodiments, X712 is an
optional
sequence, and can be R,F,K,N,R,T, or Y, or absent. In some embodiments, the
isolated
peptide comprising Formula (VII) has a length that is less than or equal to
1100 amino
acids, for example, less than or equal to 4, 5, 6, 7, 8, 9, 10, 11, 12, 13,
14, 15, 16, 17, 18,
19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37,
38, 39, 40, 41,
43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61,
62, 63, 64, 65,
67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85,
86, 87, 88, 89,
91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105, 106, 107,
108, 109, 110,
111, 112, 113, 114, 115, 116, 117, 118, 119, 120, 121, 122, 123, 124, 125,
126, 127, 128,
129, 130, 131, 132, 133, 134, 135, 136, 137, 138, 139, 140, 150, 160, 170,
180, 190, 200,
210, 220, 230, 240, 250, 260, 270, 280, 290, 300, 320, 340, 360, 380, 400,
450, 500, 550,
600, 650, 700, 750, 800, 850, 900, 950, 1000, 1050, or 1100 amino acids,
including
ranges between any two of the listed values.
[0480]
Additionally, the peptide inhibitor used in these methods can
consist of, or consist essentially of Formula (VIII), X800K X801K X802E X803
(SEQ ID
NO: 395), as described herein. In some embodiments, X800 is an optional
sequence, and
can be K, A, D, E, G, H, I, L, M, N, P, Q, R, T, V, or K, or absent. In some
X801 is an optional sequence, and can be LDTFFV, GDTH-V, EDTH-V, LDQFFV,
LDTAFV, LDTVFV, LDTFMV, LDTFSV, LDTFVV, LDTFTV, LDTFLV, LDGFFV,
LDTFGV, LDTFFK, ADTFFV, CDTFFV, DDTFFV, FDTFFV, HDTFFV, IDTFFV,
KDTH-V, MDTFFV, NDTFFV, QDTFFV, RDTFFV, SDTFFV, TDTFFV, VDTFFV,
LATFFV, LETH-V, LITFFV, LVTFFV, LWTFFV, LYTH-V, LDCFFV, LDMFFV,
LDN1-1-V, LDPFFV, LDR1-1-V, LDSFFV, LDWFFV, LDYFFV, LDTIFV, LDTMFV,
LDTNFV, LDTPFV, LDTTFV, LDTFQV, LDTH-F, LDTFFG, LDTFFL, LDTFFP,
148
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
LDTFFR, LDTFIV, LDTSFV, LDTFAV, LDTFCV, LDTQFV, LDTLFV, LTTFFV,
LDTFFI, LDHFFV, LMTFFV, LDTFEV, LDTFWV, LFTFFV, LDVFFV, LDTPRV,
LDTFHV, LDTYFV, LPTFFV, PDTFFV, LDTFPV, LDTFNV, LDTWFV, LDTGFV,
LDAH-V, LQTFFV, LCTFFV, LSTFFV, YDTFFV, LDEFFV, WDTFFV, LDTKFV,
LDTCFV, LDTFYV, LDTHFV, LHTFFV, LRTFFV, LDLFFV, LDTRFV, LLTFFV,
LDTFDV, LDT1-1-A, LDTFFT, LNTFFV, LDD1-1-V, LDIFFV, LDFFFV, LKTFFV,
LDTFFQ, LGTH-V, LDTH-C, LDKFFV, LDTFKV, LDTEFV, LDTFFW, LDTFFM,
LDTFFS, LDTFFH, LDTFFY, LDTH-N, LDTDFV, LDTFFE, LDTFFD, LTFFV,
LDTFF, TFFV, LDF, LDTE, FFV, LDV, LV, or L, or absent. In some embodiments,
is an optional sequence, and can be LSLFT, VSLFT, LQLFT, LMLFT, LTLFT, LHLFT,
LSQFT, LSVFT, LSMFT, LSLMT, LSLQT, LSLHT, LSLNT, LSLPT, LSLST, LSLGT,
LSLAT, LSLRT, LSLFN, LSLFP, LSLFR, LGLFT, ASLFT, FSLFT, GSLFT, ISLFT,
MSLFT, NSLFT, PSLFT, QSLFT, RSLFT, SSLFT, TSLFT, YSLFT, LNLFT, LSAFT,
LSHFT, LSIFT, LSNFT, LSRFT, LSSFT, LSTFT, LSWFT, LSLCT, LSLIT, LSLLT,
LSLTT, LSLVT, LSLWT, LSLFF, LSLFG, LSLFH, LSLFI, LSLFL, LSLFM, LSLFS,
LSLFV, LSLFW, LYLFT, LVLFT, LSFFT, LSGFT, LSKFT, LSCFT, LCLFT, LRLFT,
LPLFT, LWLFT, LKLFT, LDLFT, LSYFT, LALFT, WSLFT, LSLFA, LSLFQ, LSPFT,
HSLFT, LSLYT, LILFT, KSLFT, CSLFT, LSLFY, LSLFK, LSLFC, LFLFT, LELFT,
LSLKT, LLLFT, LSLFD, LSLDT, LSLFE, DSLFT, LSLET, LSDFT, LSEFT, ESLFT,
SLFT, LSFT, LFT, LSL, LT, or T, or absent. In some embodiments, X803 is an
optional
sequence, and can be R, F, K, N, R, T, or Y, or absent. In some embodiments,
the
isolated peptide comprising Formula (VIII) has a length that is less than or
equal to 1100
amino acids, for example, less than or equal to 4, 5, 6, 7, 8, 9, 10, 11, 12,
13, 14, 15, 16,
17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35,
36, 37, 38, 39,
41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59,
60, 61, 62, 63,
65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83,
84, 85, 86, 87,
89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105, 106,
107, 108,
109, 110, 111, 112, 113, 114, 115, 116, 117, 118, 119, 120, 121, 122, 123,
124, 125, 126,
127, 128, 129, 130, 131, 132, 133, 134, 135, 136, 137, 138, 139, 140, 150,
160, 170, 180,
190, 200, 210, 220, 230, 240, 250, 260, 270, 280, 290, 300, 320, 340, 360,
380, 400, 450,
500, 550, 600, 650, 700, 750, 800, 850, 900, 950, 1000, 1050, or 1100 amino
acids,
including ranges between any two of the listed values.
149
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
[0481]
Additionally, the peptide inhibitor can comprise, consist of, or consist
essentially of Formula(IX). Accordingly, in some embodiments, the peptide
inhibitor
comprises a peptide of Formula (IX):
X901X902X903X904X905X906X907X908X909X910X911X912X913X914X915X916X917, wherein
X901
is any amino acid or absent; X902 is a positively charged amino acid, F, or N;
X903 is
any amino acid; X904 is any amino acid; X905 is a polar uncharged amino acid,
R, Y, or
W; X906 is a hydrophobic or uncharged polar amino acid; X907 is a hydrophobic
or
uncharged polar amino acid; X908 is a hydrophobic, non-aromatic carbon chain
amino
acid that is not M or F; X909 is a positively charged amino acid, T, Q, or Y;
X910 is any
amino acid that is not negatively charged; X911 is a polar uncharged amino
acid or H;
X912 is any amino acid that is not negatively charged; X913 is any amino acid
that is not
negatively charged; X914 is any amino acid that is not negatively charged;
X915 is a
negatively charged amino acid, Y, or Q; X916 is any amino acid that is not
negatively
charged; and X917 is one or more positively charged amino acids or is absent.
Optionally,
X901 comprises a positively charged amino acid. Optionally X901 is an R or K.
Optionally X917 is RR. In some embodiments, the isolated peptide comprising
Formula
(IX) has a length that is less than or equal to 1100 amino acids, for example,
less than or
equal to 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22,
23, 24, 25, 26,
27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45,
46, 47, 48, 49,
50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68,
69, 70, 71, 72,
73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91,
92, 93, 94, 95,
96, 97, 98, 99, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111,
112, 113,
114, 115, 116, 117, 118, 119, 120, 121, 122, 123, 124, 125, 126, 127, 128,
129, 130, 131,
132, 133, 134, 135, 136, 137, 138, 139, 140, 150, 160, 170, 180, 190, 200,
210, 220, 230,
240, 250, 260, 270, 280, 290, 300, 320, 340, 360, 380, 400, 450, 500, 550,
600, 650, 700,
750, 800, 850, 900, 950, 1000, 1050, or 1100 amino acids, including ranges
between any
two of Wailed vatittltlitionally, the peptide inhibitor used in these methods
can
consist of, or consist essentially of and/or SEQ ID NOs: 1-33, 34, 46-53, 64-
66, 68, 76,
94-96, 98, 264-393, 583-586, or 589 or any one or more of the peptides
provided in
5.1, 5.4, 5.5, 5.6, or any variation or combination of variations of P28R or
P28 core as
provided in Tables 5.3 and 13. In some embodiments, these isolated peptides
used in
these methods have a length that is less than or equal to 1100 amino acids,
for example,
less than or equal to 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18,
19, 20, 21, 22, 23,
150
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42,
43, 44, 45, 46,
48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66,
67, 68, 69, 70,
72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90,
91, 92, 93, 94,
96, 97, 98, 99, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111,
112, 113,
115, 116, 117, 118, 119, 120, 121, 122, 123, 124, 125, 126, 127, 128, 129,
130, 131, 132,
133, 134, 135, 136, 137, 138, 139, 140, 150, 160, 170, 180, 190, 200, 210,
220, 230, 240,
250, 260, 270, 280, 290, 300, 320, 340, 360, 380, 400, 450, 500, 550, 600,
650, 700, 750,
800, 850, 900, 950, 1000, 1050, or 1100 amino acids, including ranges between
any two
of the listed values.
[0483]
Additionally, the peptide inhibitor used in these methods can
comprise, consist of, or consist essentially of a peptide inhibitor that
comprises, consists
of, or consists essentially of any one or more of the peptides set forth in
SEQ ID NOs: 1-
33, 34, 46-53, 64-66, 68, 76, 94-96, 98, 264-393, 583-586, or 589 or any one
or more of
the peptides provided in Table 5.1, 5.4, 5.5, 5.6, or any variation or
combination of
variations of P28R or P28 core as provided in Tables 5.3 and 13. In some
embodiments,
the isolated peptide used in these methods has a length that is less than or
equal to 1100
amino acids, for example, less than or equal to 4, 5, 6, 7, 8, 9, 10, 11, 12,
13, 14, 15, 16,
17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35,
36, 37, 38, 39,
40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58,
59, 60, 61, 62,
63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81,
82, 83, 84, 85,
86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103,
104, 105, 106,
107, 108, 109, 110, 111, 112, 113, 114, 115, 116, 117, 118, 119, 120, 121,
122, 123, 124,
125, 126, 127, 128, 129, 130, 131, 132, 133, 134, 135, 136, 137, 138, 139,
140, 150, 160,
170, 180, 190, 200, 210, 220, 230, 240, 250, 260, 270, 280, 290, 300, 320,
340, 360, 380,
400, 450, 500, 550, 600, 650, 700, 750, 800, 850, 900, 950, 1000, 1050, or
1100 amino
acids, including ranges between any two of the listed values.
[0484] In some
embodiments, a nucleic acid encoding such a peptide
can be provided, for example a nucleic acid of SEQ ID NOs: 102-165.
Preferably, the
immunoregulatory peptide inhibitor used in the aforementioned methods is P28R,
a
derivative thereof, or a nucleic acid encoding such a molecule (e.g., any one
or more of
the immunoregulatory peptide inhibitors comprise, consist of, or consist
essentially of a
peptide as described herein. For example, the peptide inhibitor can comprise,
consist of,
or consist essentially of Formula (I), XX1VKX2X3X4 (SEQ ID NO: 166) as
described
151
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
herein. In some embodiments, X is an optional sequence, and can be KKLDT (SEQ
ID
NO: 167), RKLDT (SEQ ID NO: 168), KKGDT (SEQ ID NO: 169), KKEDT (SEQ ID
NO: 170), KKLDQ (SEQ ID NO: 171), KKGDQ (SEQ ID NO: 252), KKEDQ (SEQ
ID NO: 253), RKLDQ (SEQ ID NO: 254), RKGDQ (SEQ ID NO: 255), RKEDQ (SEQ
ID NO: 256), RKGTD (SEQ ID NO: 257), RKEDT (SEQ ID NO: 258), KLDT (SEQ
ID NO: 172), KGDT (SEQ ID NO: 259), KEDT (SEQ ID NO: 260), KLDQ (SEQ ID
NO: 261), KGDQ (SEQ ID NO: 262), KEDQ (SEQ ID NO: 263), LDT, LDQ, GDT,
GDQ, EDT, EDQ, DT, DQ, T, or Q, or absent. In some embodiments, Xi is one of
FF,
FM, FS, FV, FT, FL, AF, AM, AS, AV, AT, AL, VF, VM, VS, VV, VT, or VL. In some
embodiments, X2 is one of LS, LQ, LM, LT, LH, VS, VQ, VM, VT, or VH. In some
embodiments, X3 is one of LFT, LMT, LQT, LHT, LNT, LPT, LST, LGT, LAT, LRT,
QFT, QMT, QQT, QHT, QNT, QPT, QST, QGT, QAT, QRT, VFT, VMT, VQT, VHT,
VNT, VPT, VST, VGT, VAT, VRT, MFT, MMT, MQT, MHT, MNT, MPT, MST,
MGT, MAT, MRT, LFN, LMN, LQN, LHN, LNN, LPN, LSN, LGN, LAN, LRN, QFN,
QMN, QQN, QHN, QNN, QPN, QSN, QGN, QAN, QRN, VFN, VMN, VQN, VHN,
VNN, VPN, VSN, VGN, VAN, VRN, MFN, MMN, MQN, MHN, MNN, MPN, MSN,
MGN, MAN, MRN, LFP, LMP, LQP, LHP, LNP, LPP, LSP, LGP, LAP, LRP, QFP,
QMP, QQP, QHP, QNP, QPP, QSP, QGP, QAP, QRP, VFP, VMP, VQP, VHP, VNP,
VPP, VSP, VGP, VAP, VRP, MFP, MMP, MQP, MHP, MNP, MPP, MSP, MGP, MAP,
MRPR, LFR, LMR, LQR, LHR, LNR, LPR, LSR, LGR, LAR, LRR, QFR, QMR, QQR,
QHR, QNR, QPR, QSR, QGR, QAR, QRR, VFR, VMR, VQR, VHR, VNR, VPR, VSR,
VGR, VAR, VRR, MFR, MMR, MQR, MHR, MNR, MPR, MSR, MGR, MAR, or
In some embodiments, X4 is an optional sequence, and can be ER, or E, or
absent. In
some embodiments, if X is absent, Xi is FF, and X2 is LS. In some embodiments,
the
isolated peptides that comprise Formula (I) have a length that is less than or
equal to
amino acids, for example, less than or equal to 4, 5, 6, 7, 8, 9, 10, 11, 12,
13, 14, 15, 16,
17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35,
36, 37, 38, 39,
41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59,
60, 61, 62, 63,
65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83,
84, 85, 86, 87,
89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105, 106,
107, 108,
109, 110, 111, 112, 113, 114, 115, 116, 117, 118, 119, 120, 121, 122, 123,
124, 125, 126,
127, 128, 129, 130, 131, 132, 133, 134, 135, 136, 137, 138, 139, 140, 150,
160, 170, 180,
190, 200, 210, 220, 230, 240, 250, 260, 270, 280, 290, 300, 320, 340, 360,
380, 400, 450,
152
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
500, 550, 600, 650, 700, 750, 800, 850, 900, 950, 1000, 1050, or 1100 amino
acids,
including ranges between any two of the listed values.
[0485]
Additionally, the peptide inhibitor can comprise, consist of, or consist
essentially of Formula (II), X20TFFVKLSX21X22 (SEQ ID NO: 173). In some
embodiments, X20 is an optional sequence, and can be KKLD (SEQ ID NO: 174),
RKLD (SEQ ID NO: 175), KKGD (SEQ ID NO: 176), KKED (SEQ ID NO: 177),
KLD, LD, or D, or absent. X21 is an optional sequence, and can be LFT, LMT,
LQT,
LHT, LNT, LPT, LST, LGT, LAT, LRT, QFT, QMT, QQT, QHT, QNT, QPT, QST,
QGT, QAT, QRT, VFT, VMT, VQT, VHT, VNT, VPT, VST, VGT, VAT, VRT, MFT,
MMT, MQT, MHT, MNT, MPT, MST, MGT, MAT, MRT, LFN, LMN, LQN, LHN,
LNN, LPN, LSN, LGN, LAN, LRN, QFN, QMN, QQN, QHN, QNN, QPN, QSN, QGN,
QAN, QRN, VFN, VMN, VQN, VHN, VNN, VPN, VSN, VGN, VAN, VRN, MFN,
MMN, MQN, MHN, MNN, MPN, MSN, MGN, MAN, MRN, LFP, LMP, LQP, LHP,
LNP, LPP, LSP, LGP, LAP, LRP, QFP, QMP, QQP, QHP, QNP, QPP, QSP, QGP, QAP,
QRP, VFP, VMP, VQP, VHP, VNP, VPP, VSP, VGP, VAP, VRP, MFP, MMP, MQP,
MHP, MNP, MPP, MSP, MGP, MAP, MRPR, LFR, LMR, LQR, LHR, LNR, LPR, LSR,
LGR, LAR, LRR, QFR, QMR, QQR, QHR, QNR, QPR, QSR, QGR, QAR, QRR, VFR,
VMR, VQR, VHR, VNR, VPR, VSR, VGR, VAR, VRR, MFR, MMR, MQR, MHR,
MNR, MPR, MSR, MGR, MAR, or MRR, or absent. In some embodiments, X22 is an
optional sequence, and can be ER, or E, or absent. In some embodiments, the
isolated
peptides that comprise Formula (II) have a length that is less than or equal
to 1100 amino
acids, for example, less than or equal to 4, 5, 6, 7, 8, 9, 10, 11, 12, 13,
14, 15, 16, 17, 18,
19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37,
38, 39, 40, 41,
42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60,
61, 62, 63, 64,
65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83,
84, 85, 86, 87,
88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105,
106, 107,
108, 109, 110, 111, 112, 113, 114, 115, 116, 117, 118, 119, 120, 121, 122,
123, 124, 125,
126, 127, 128, 129, 130, 131, 132, 133, 134, 135, 136, 137, 138, 139, 140,
150, 160, 170,
180, 190, 200, 210, 220, 230, 240, 250, 260, 270, 280, 290, 300, 320, 340,
360, 380, 400,
450, 500, 550, 600, 650, 700, 750, 800, 850, 900, 950, 1000, 1050, or 1100
amino acids,
including ranges between any two of the listed values.
[0486]
Additionally, the peptide inhibitor can comprise, consist of, or consist
essentially of Formula (III), X30X31VKLX32LX33TEX34 (SEQ ID NO: 178). In some
153
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
embodiments, X30 is an optional sequence, and can be KKLDTF (SEQ ID NO: 179),
KLDTF (SEQ ID NO: 180), LDTF (SEQ ID NO: 181), DTF, TF, F, or absent. In some
embodiments, X31 is an optional sequence, and can be F, S, M, V, T, or L, or
absent. In
some embodiments, X31 is F. In some embodiments, X32 can be S, Q, M, T, or H.
In
some embodiments, X32 is S. X33 can be F, M, Q, H, N, P, S, G, A, or R. In
some
embodiments, X34 is F. X34 is an optional sequence, and can be R or absent. In
some
embodiments, the isolated peptides that comprise Formula (III) have a length
that is less
than or equal to 1100 amino acids, for example, less than or equal to 4, 5, 6,
7, 8, 9, 10,
11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29,
30, 31, 32, 33,
35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53,
54, 55, 56, 57,
59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77,
78, 79, 80, 81,
83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101,
102, 103, 104,
105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115, 116, 117, 118, 119,
120, 121, 122,
123, 124, 125, 126, 127, 128, 129, 130, 131, 132, 133, 134, 135, 136, 137,
138, 139, 140,
150, 160, 170, 180, 190, 200, 210, 220, 230, 240, 250, 260, 270, 280, 290,
300, 320, 340,
360, 380, 400, 450, 500, 550, 600, 650, 700, 750, 800, 850, 900, 950, 1000,
1050, or
amino acids, including ranges between any two of the listed values.
[0487]
Additionally, the peptide inhibitor can comprise, consist of, or consist
essentially of Formula (VII), ,C700K ,c7o1)(702,C7o3 ,C704)(705,C706K )(707
,C708 ,C709 )(710
X711E X712 (SEQ ID NO: 394), as described herein. In some embodiments, X700 is
an
optional sequence, and can be K,A,D,E,G,H,I,L,M,N,P,Q,R,T, or V, or absent. In
some
embodiments, X701 is an optional sequence, and can
be
L,A,C,D,E,F,G,H,I,K,M,N,Q,R,S,T, or V, or absent. In some embodiments, X702 is
an
optional sequence, and can be D,A,E,I,V,W, or Y, or absent. In some
embodiments,
is an optional sequence, and can be T,C,M,N,P,Q,R,S,W, or Y, or absent. In
some
embodiments, X704 is an optional sequence, and can be F,A,I,M,N,P,T, or V, or
absent.
some embodiments, X705 is an optional sequence, and can be F,L,M,Q,S,TV, or
absent.
In some embodiments, X706 is an optional sequence, and can be V,F,G,L,P, or R,
or
absent. In some
embodiments, X707 is an optional sequence, and can be
L,A,F,G,I,M,N,P,Q,R,S,T,V, or Y, or absent. In some embodiments, X708 is an
optional
sequence, and can be S,H,M,N,Q, or T, or absent. In some embodiments, X709 is
an
optional sequence, and can be L,A,H,I,M,N,Q,R,S,T,V, or W, or absent. In some
embodiments, X710 is an optional sequence, and can be
F,A,C,G,H,I,L,M,N,P,Q,R,S,T,V,
154
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
or W, or absent. In some embodiments, X711 is an optional sequence, and can be
T,F,G,H,I,L,M,N,P,S,V, or W, or absent. In some embodiments, X712 is an
optional
sequence, and can be R,F,K,N,R,T,+ or Y, or absent. In some embodiments, the
isolated
peptide comprising Formula (VII) has a length that is less than or equal to
1100 amino
acids, for example, less than or equal to 4, 5, 6, 7, 8, 9, 10, 11, 12, 13,
14, 15, 16, 17, 18,
19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37,
38, 39, 40, 41,
43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61,
62, 63, 64, 65,
67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85,
86, 87, 88, 89,
91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105, 106, 107,
108, 109, 110,
111, 112, 113, 114, 115, 116, 117, 118, 119, 120, 121, 122, 123, 124, 125,
126, 127, 128,
129, 130, 131, 132, 133, 134, 135, 136, 137, 138, 139, 140, 150, 160, 170,
180, 190, 200,
210, 220, 230, 240, 250, 260, 270, 280, 290, 300, 320, 340, 360, 380, 400,
450, 500, 550,
600, 650, 700, 750, 800, 850, 900, 950, 1000, 1050, or 1100 amino acids,
including
ranges between any two of the listed values.
[0488]
Additionally, the peptide inhibitor can comprise, consist of, or consist
essentially of Formula (VIII), X800K X801K X802E X803 (SEQ ID NO: 395), as
described
herein. In some embodiments, X800 is an optional sequence, and can be K, A, D,
E, G,
I, L, M, N, P, Q, R, T, V, or K, or absent. In some embodiments, X801 is an
optional
sequence, and can be LDTFFV, GDTH-V, EDTFFV, LDQFFV, LDTAFV, LDTVFV,
LDTFMV, LDTFSV, LDTFVV, LDTFTV, LDTFLV, LDGFFV, LDTFGV, LDT141-K,
ADTFFV, CDTFFV, DDTFFV, FDTH-V, HDTFFV, IDTFFV, KDTH-V, MDTFFV,
NDTFFV, QDTH-V, RDTFFV, SDTFFV, TDTH-V, VDTH-V, LATFFV, LETFFV,
LITFFV, LVTFFV, LWTH-V, LYTFFV, LDCFFV, LDMFFV, LDNFFV, LDPFFV,
LDRFFV, LDSFFV, LDW141-V, LDYFFV, LDTIFV, LDTMFV, LDTNFV, LDTPFV,
LDTTFV, LDTFQV, LDTFFF, LDTFFG, LDTFFL, LDTFFP, LDTFFR, LDTFIV,
LDTSFV, LDTFAV, LDTFCV, LDTQFV, LDTLFV, LTTFFV, LDTFFI, LDHFFV,
LMTFFV, LDTFEV, LDTFWV, LFTFFV, LDVFFV, LDTFRV, LDTFHV, LDTYFV,
LPTFFV, PDTH-V, LDTFPV, LDTFNV, LDTWFV, LDTGFV, LDAFFV, LQTFFV,
LCTFFV, LSTFFV, YDTFFV, LDEFFV, WDTFFV, LDTKFV, LDTCFV, LDTFYV,
LDTHFV, LHTFFV, LRTFFV, LDLFFV, LDTRFV, LLTFFV, LDTFDV, LDTFFA,
LDTFFT, LNTFFV, LDDH-V, LDI141-V, LDFFFV, LKTFFV, LDTH-Q, LGTFFV,
LDTFFC, LDKFFV, LDTFKV, LDTEFV, LDTFFW, LDTFFM, LDTFFS, LDTFFH,
LDTFFY, LDTFFN, LDTDFV, LDTFFEõ LDTFFD, LTH-V, LDTFF, TFFV, LDF,
155
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
LDTE, FFV, LDV, LV, or L, or absent. In some embodiments, X802 is an optional
sequence, and can be LSLFT, VSLFT, LQLFT, LMLFT, LTLFT, LHLFT, LSQFT,
LSVFT, LSMFT, LSLMT, LSLQT, LSLHT, LSLNT, LSLPT, LSLST, LSLGT, LSLAT,
LSLRT, LSLFN, LSLFP, LSLFR, LGLFT, ASLFT, FSLFT, GSLFT, ISLFT, MSLFT,
NSLFT, PSLFT, QSLFT, RSLFT, SSLFT, TSLFT, YSLFT, LNLFT, LSAFT, LSHFT,
LSIFT, LSNFT, LSRFT, LSSFT, LSTFT, LSWFT, LSLCT, LSLIT, LSLLT, LSLTT,
LSLVT, LSLWT, LSLFF, LSLFG, LSLFH, LSLFI, LSLFL, LSLFM, LSLFS, LSLFV,
LSLFW, LYLFT, LVLFT, LSH-T, LSGFT, LSKFT, LSCFT, LCLFT, LRLFT, LPLFT,
LWLFT, LKLFT, LDLFT, LSYFT, LALFT, WSLFT, LSLFA, LSLFQ, LSPFT, HSLFT,
LSLYT, LILFT, KSLFT, CSLFT, LSLFY, LSLFK, LSLFC, LFLFT, LELFT, LSLKT,
LLLFT, LSLFD, LSLDT, LSLFEõ DSLFT, LSLET, LSDFT, LSEFT, ESLFT, SLFT,
LSFT, LFT, LSL, LT, or T, or absent. In some embodiments, X803 is an optional
sequence, and can be R, F, K, N, R, T, or Y, or absent. In some embodiments,
the
isolated peptide comprising Formula (VIII) has a length that is less than or
equal to 1100
amino acids, for example, less than or equal to 4, 5, 6, 7, 8, 9, 10, 11, 12,
13, 14, 15, 16,
17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35,
36, 37, 38, 39,
41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59,
60, 61, 62, 63,
65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83,
84, 85, 86, 87,
89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105, 106,
107, 108,
109, 110, 111, 112, 113, 114, 115, 116, 117, 118, 119, 120, 121, 122, 123,
124, 125, 126,
127, 128, 129, 130, 131, 132, 133, 134, 135, 136, 137, 138, 139, 140, 150,
160, 170, 180,
190, 200, 210, 220, 230, 240, 250, 260, 270, 280, 290, 300, 320, 340, 360,
380, 400, 450,
500, 550, 600, 650, 700, 750, 800, 850, 900, 950, 1000, 1050, or 1100 amino
acids,
including ranges between any two of the listed values.
[0489]
Additionally, the peptide inhibitor can comprise, consist of, or consist
essentially of Formula(IX). Accordingly, in some embodiments, the peptide
inhibitor
comprises a peptide of Formula (IX):
X901X902X903X904X905X906X907X908X909X910X911X912X913X914X915X916X917, wherein
X901
is any amino acid or absent; X902 is a positively charged amino acid, F, or N;
X903 is
any amino acid; X904 is any amino acid; X905 is a polar uncharged amino acid,
R, Y, or
X906 is a hydrophobic or uncharged polar amino acid; X907 is a hydrophobic or
uncharged
polar amino acid; X908 is a hydrophobic, non-aromatic carbon chain amino acid
that is
M or F; X909 is a positively charged amino acid, T, Q, or Y; X910 is any amino
acid that is
156
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
not negatively charged; X911 is a polar uncharged amino acid or H; X912 is any
amino
that is not negatively charged; X913 is any amino acid that is not negatively
charged; X914
is any amino acid that is not negatively charged; X915 is a negatively charged
amino acid,
Y, or Q; X916 is any amino acid that is not negatively charged; and X917 is
one or more
positively charged amino acids or is absent. Optionally, X901 comprises a
positively
charged amino acid. Optionally X901 is an R or K. Optionally X917 is RR. In
some
embodiments, the isolated peptide comprising Formula (IX) has a length that is
less than
or equal to 1100 amino acids, for example, less than or equal to 4, 5, 6, 7,
8, 9, 10, 11,
13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31,
32, 33, 34, 35,
37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55,
56, 57, 58, 59,
61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79,
80, 81, 82, 83,
85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102,
103, 104, 105,
106, 107, 108, 109, 110, 111, 112, 113, 114, 115, 116, 117, 118, 119, 120,
121, 122, 123,
124, 125, 126, 127, 128, 129, 130, 131, 132, 133, 134, 135, 136, 137, 138,
139, 140, 150,
160, 170, 180, 190, 200, 210, 220, 230, 240, 250, 260, 270, 280, 290, 300,
320, 340, 360,
380, 400, 450, 500, 550, 600, 650, 700, 750, 800, 850, 900, 950, 1000, 1050,
or 1100
amino acids, including ranges between any two of the listed values.
[0490]
Additionally, the peptide inhibitor can comprise, consist of, or consist
essentially of and/or SEQ ID NOs: 1-33, 34, 46-53, 64-66, 68, 76, 94-96, 98,
264-393,
583-586, or 589 or any one or more of the peptides provided in Table 5.1, 5.4,
5.5, 5.6,
or any variation or combination of variations of P28R or P28 core as provided
in Tables
5.3 and 13, as described herein. In some embodiments, the isolated peptides
have a
length that is less than or equal to 1100 amino acids, for example, less than
or equal to 4,
5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25,
26, 27, 28, 29,
30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48,
49, 50, 51, 52,
53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71,
72, 73, 74, 75,
76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94,
95, 96, 97, 98,
99, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114,
115, 116,
117, 118, 119, 120, 121, 122, 123, 124, 125, 126, 127, 128, 129, 130, 131,
132, 133, 134,
135, 136, 137, 138, 139, 140, 150, 160, 170, 180, 190, 200, 210, 220, 230,
240, 250, 260,
270, 280, 290, 300, 320, 340, 360, 380, 400, 450, 500, 550, 600, 650, 700,
750, 800, 850,
900, 950, 1000, 1050, or 1100 amino acids, including ranges between any two of
the
listed values.
157
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
[0491]
Additionally, the peptide inhibitor can comprise, consist of, or
consist essentially of a peptide inhibitor that comprises, consists of, or
consists
essentially of any one or more of the peptides set forth in Table 5.1, 5.4,
5.5, or 5.6 or
any variation or combination of variations of P28R or P28 core as provided in
Tables 5.3
and 13 In some embodiments, the isolated peptide from Table 5.1, 5.4, 5.5, or
5.6 or
any variation or combination of variations of P28R or P28 core as provided in
Tables 5.3
and 13 has a length that is less than or equal to 1100 amino acids, for
example, less than
or equal to 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21,
22, 23, 24, 25,
26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44,
45, 46, 47, 48,
49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67,
68, 69, 70, 71,
72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90,
91, 92, 93, 94,
95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110,
111, 112, 113,
114, 115, 116, 117, 118, 119, 120, 121, 122, 123, 124, 125, 126, 127, 128,
129, 130, 131,
132, 133, 134, 135, 136, 137, 138, 139, 140, 150, 160, 170, 180, 190, 200,
210, 220, 230,
240, 250, 260, 270, 280, 290, 300, 320, 340, 360, 380, 400, 450, 500, 550,
600, 650, 700,
750, 800, 850, 900, 950, 1000, 1050, or 1100 amino acids, including ranges
between any
two of the listed values. For example, a nucleic acid encoding such a peptide
inhibitor
can be provided, by SEQ ID NOs: 102-165.
[0492] The
immunoregulatory peptide inhibitors used in the aforementioned
methods can comprise at least one D amino acid, at least one non-natural amino
acid, an
N-terminal acetyl group, or a C terminal amide group and said immunoregulatory
peptide
inhibitors can be glycosylated or joined to PEG, a cytotoxin, or radionuclide.
The
can be administered to at least one cell of the patient. The administration
can be
performed in vivo, for example therapeutically. The administration can be
performed ex
vivo, for example as a diagnostic tool, or as an ex vivo therapy to stimulate
immune cells
of the patient before the immune cells are administered to the patient.
Administration of
an immunoregulatory peptide inhibitor comprising, consisting, or consisting
essentially
a peptide inhibitor as described herein, or a nucleic acid encoding such a
molecule to
human immune cells, and detection of immune cell stimulation is described in
Example
13). For example, the peptide inhibitor used in these methods can comprise,
consist of,
consist essentially of Formula (I), XX1VKX2X3X4 (SEQ ID NO: 166) as described
herein. In some embodiments, X is an optional sequence, and can be KKLDT (SEQ
ID
NO: 167), RKLDT (SEQ ID NO: 168), KKGDT (SEQ ID NO: 169), KKEDT (SEQ ID
158
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
NO: 170), KKLDQ (SEQ ID NO: 171), KKGDQ (SEQ ID NO: 252), KKEDQ (SEQ
ID NO: 253), RKLDQ (SEQ ID NO: 254), RKGDQ (SEQ ID NO: 255), RKEDQ (SEQ
ID NO: 256), RKGTD (SEQ ID NO: 257), RKEDT (SEQ ID NO: 258), KLDT (SEQ
ID NO: 172), KGDT (SEQ ID NO: 259), KEDT (SEQ ID NO: 260), KLDQ (SEQ ID
NO: 261), KGDQ (SEQ ID NO: 262), KEDQ (SEQ ID NO: 263), LDT, LDQ, GDT,
GDQ, EDT, EDQ, DT, DQ, T, or Q, or absent. In some embodiments, Xi is be one
of
FM, FS, FV, FT, FL, AF, AM, AS, AV, AT, AL, VF, VM, VS, VV, VT, or VL. In some
embodiments, X2 is one of LS, LQ, LM, LT, LH, VS, VQ, VM, VT, or VH. In some
embodiments, X3 is be one of LFT, LMT, LQT, LHT, LNT, LPT, LST, LGT, LAT, LRT,
QFT, QMT, QQT, QHT, QNT, QPT, QST, QGT, QAT, QRT, VFT, VMT, VQT, VHT,
VNT, VPT, VST, VGT, VAT, VRT, MFT, MMT, MQT, MHT, MNT, MPT, MST,
MGT, MAT, MRT, LFN, LMN, LQN, LHN, LNN, LPN, LSN, LGN, LAN, LRN, QFN,
QMN, QQN, QHN, QNN, QPN, QSN, QGN, QAN, QRN, VFN, VMN, VQN, VHN,
VNN, VPN, VSN, VGN, VAN, VRN, MFN, MMN, MQN, MHN, MNN, MPN, MSN,
MGN, MAN, MRN, LFP, LMP, LQP, LHP, LNP, LPP, LSP, LGP, LAP, LRP, QFP,
QMP, QQP, QHP, QNP, QPP, QSP, QGP, QAP, QRP, VFP, VMP, VQP, VHP, VNP,
VPP, VSP, VGP, VAP, VRP, MFP, MMP, MQP, MHP, MNP, MPP, MSP, MGP, MAP,
MRPR, LFR, LMR, LQR, LHR, LNR, LPR, LSR, LGR, LAR, LRR, QFR, QMR, QQR,
QHR, QNR, QPR, QSR, QGR, QAR, QRR, VFR, VMR, VQR, VHR, VNR, VPR, VSR,
VGR, VAR, VRR, MFR, MMR, MQR, MHR, MNR, MPR, MSR, MGR, MAR, or
In some embodiments, X4 is an optional sequence, and can be ER, or E, or
absent. In
some embodiments, if X is absent, X1 is FF, and X2 is LS. In some embodiments,
the
isolated peptides that comprise Formula (I) used in these methods have a
length that is
less than or equal to 1100 amino acids, for example, less than or equal to 4,
5, 6, 7, 8, 9,
10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28,
29, 30, 31, 32,
34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52,
53, 54, 55, 56,
58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76,
77, 78, 79, 80,
82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100,
101, 102, 103,
104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115, 116, 117, 118,
119, 120, 121,
122, 123, 124, 125, 126, 127, 128, 129, 130, 131, 132, 133, 134, 135, 136,
137, 138, 139,
140, 150, 160, 170, 180, 190, 200, 210, 220, 230, 240, 250, 260, 270, 280,
290, 300, 320,
340, 360, 380, 400, 450, 500, 550, 600, 650, 700, 750, 800, 850, 900, 950,
1000, 1050,
1100 amino acids, including ranges between any two of the listed values.
159
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
[0493]
Additionally, the peptide inhibitor used in these methods can
comprise, consist of, or consist essentially of Formula (II), X20TFFVKLSX21X22
(SEQ
ID NO: 173). In some embodiments, X20 is an optional sequence, and can be KKLD
(SEQ ID NO: 174), RKLD (SEQ ID NO: 175), KKGD (SEQ ID NO: 176), KKED
(SEQ ID NO: 177), KLD, LD, or D, or absent. X21 is an optional sequence, and
can be
LFT, LMT, LQT, LHT, LNT, LPT, LST, LGT, LAT, LRT, QFT, QMT, QQT, QHT,
QNT, QPT, QST, QGT, QAT, QRT, VFT, VMT, VQT, VHT, VNT, VPT, VST, VGT,
VAT, VRT, MFT, MMT, MQT, MHT, MNT, MPT, MST, MGT, MAT, MRT, LFN,
LMN, LQN, LHN, LNN, LPN, LSN, LGN, LAN, LRN, QFN, QMN, QQN, QHN, QNN,
QPN, QSN, QGN, QAN, QRN, VFN, VMN, VQN, VHN, VNN, VPN, VSN, VGN,
VAN, VRN, MFN, MMN, MQN, MHN, MNN, MPN, MSN, MGN, MAN, MRN, LFP,
LMP, LQP, LHP, LNP, LPP, LSP, LGP, LAP, LRP, QFP, QMP, QQP, QHP, QNP, QPP,
QSP, QGP, QAP, QRP, VFP, VMP, VQP, VHP, VNP, VPP, VSP, VGP, VAP, VRP,
MFP, MMP, MQP, MHP, MNP, MPP, MSP, MGP, MAP, MRPR, LFR, LMR, LQR,
LHR, LNR, LPR, LSR, LGR, LAR, LRR, QFR, QMR, QQR, QHR, QNR, QPR, QSR,
QGR, QAR, QRR, VFR, VMR, VQR, VHR, VNR, VPR, VSR, VGR, VAR, VRR,
MFR, MMR, MQR, MHR, MNR, MPR, MSR, MGR, MAR, or MRR, or absent. In
some embodiments, X22 is an optional sequence, and can be ER, or E, or absent.
In
some embodiments, the isolated peptides that comprise Formula (II) used in
these
methods have a length that is less than or equal to 1100 amino acids, for
example, less
than or equal to 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20,
21, 22, 23, 24,
25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43,
44, 45, 46, 47,
48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66,
67, 68, 69, 70,
71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89,
90, 91, 92, 93,
94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110,
111, 112,
113, 114, 115, 116, 117, 118, 119, 120, 121, 122, 123, 124, 125, 126, 127,
128, 129, 130,
131, 132, 133, 134, 135, 136, 137, 138, 139, 140, 150, 160, 170, 180, 190,
200, 210, 220,
230, 240, 250, 260, 270, 280, 290, 300, 320, 340, 360, 380, 400, 450, 500,
550, 600, 650,
700, 750, 800, 850, 900, 950, 1000, 1050, or 1100 amino acids, including
ranges
betweeiI.0494wo olsithlitiishtilly41tteit. peptide inhibitor used in these
methods can
consist of, or consist essentially of Formula (III), X30X31VKLX32LX33TEX34
(SEQ ID
NO: 178). In some embodiments, X30 is an optional sequence, and can be KKLDTF
(SEQ ID NO: 179), KLDTF (SEQ ID NO: 180), LDTF (SEQ ID NO: 181), DTF, TF,
160
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
F, or absent. In some embodiments, X31 is an optional sequence, and can be F,
S, M, V,
T, or L, or absent. In some embodiments, X31 is F. In some embodiments, X32
can be S,
Q, M, T, or H. In some embodiments, X32 is S. X33 can be F, M, Q, H, N, P, S,
G, A, or
R. In some embodiments, X34 is F. X34 is an optional sequence, and can be R or
absent.
In some embodiments, the isolated peptides that comprise Formula (III) used in
these
methods have a length that is less than or equal to 1100 amino acids, for
example, less
than or equal to 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20,
21, 22, 23, 24,
25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43,
44, 45, 46, 47,
49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67,
68, 69, 70, 71,
73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91,
92, 93, 94, 95,
97, 98, 99, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112,
113, 114,
115, 116, 117, 118, 119, 120, 121, 122, 123, 124, 125, 126, 127, 128, 129,
130, 131, 132,
133, 134, 135, 136, 137, 138, 139, 140, 150, 160, 170, 180, 190, 200, 210,
220, 230, 240,
250, 260, 270, 280, 290, 300, 320, 340, 360, 380, 400, 450, 500, 550, 600,
650, 700, 750,
800, 850, 900, 950, 1000, 1050, or 1100 amino acids, including ranges between
any two
of the listed values.
[0495]
Additionally, the peptide inhibitor used in these methods can
consist of, or consist essentially of Formula (VII), X700K X701X702X703
X704X705X706K
X707 X708 X700 X710 X711E X712 (SEQ ID NO: 394), as described herein. In some
embodiments, X700 is an optional sequence, and can be
K,A,D,E,G,H,I,L,M,N,P,Q,R,T,
V, or absent. In some embodiments, X701 is an optional sequence, and can be
L,A,C,D,E,F,G,H,I,K,M,N,Q,R,S,T, or V, or absent. In some embodiments, X702 is
an
optional sequence, and can be D,A,E,I,V,W,Y, or absent. In some embodiments,
X703 is
an optional sequence, and can be T,C,M,N,P,Q,R,S,W,Y, or absent. In some
embodiments, X704 is an optional sequence, and can be F,A,I,M,N,P,T, or V, or
absent.
some embodiments, X705 is an optional sequence, and can be F,L,M,Q,S,T, or V,
or
absent. In some embodiments, X706 is an optional sequence, and can be
V,F,G,L,P, or R,
or absent. In some embodiments, X707 is an optional sequence, and can be
L,A,F,G,I,M,N,P,Q,R,S,T,V, or Y, or absent. In some embodiments, X708 is an
optional
sequence, and can be S,H,M,N,Q, or T, or absent. In some embodiments, X709 is
an
optional sequence, and can be L,A,H,I,M,N,Q,R,S,T,V, or W, or absent. In some
embodiments, X710 is an optional sequence, and can be
F,A,C,G,H,I,L,M,N,P,Q,R,S,T,V,
or W, or absent. In some embodiments, X711 is an optional sequence, and can be
161
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
T,F,G,H,I,L,M,N,P,S,V, or W, or absent. In some embodiments, X712 is an
optional
sequence, and can be R,F,K,N,R,T, or Y, or absent. In some embodiments, the
isolated
peptide comprising Formula (VII) has a length that is less than or equal to
1100 amino
acids, for example, less than or equal to 4, 5, 6, 7, 8, 9, 10, 11, 12, 13,
14, 15, 16, 17, 18,
19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37,
38, 39, 40, 41,
43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61,
62, 63, 64, 65,
67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85,
86, 87, 88, 89,
91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105, 106, 107,
108, 109, 110,
111, 112, 113, 114, 115, 116, 117, 118, 119, 120, 121, 122, 123, 124, 125,
126, 127, 128,
129, 130, 131, 132, 133, 134, 135, 136, 137, 138, 139, 140, 150, 160, 170,
180, 190, 200,
210, 220, 230, 240, 250, 260, 270, 280, 290, 300, 320, 340, 360, 380, 400,
450, 500, 550,
600, 650, 700, 750, 800, 850, 900, 950, 1000, 1050, or 1100 amino acids,
including
ranges between any two of the listed values.
[0496]
Additionally, the peptide inhibitor used in these methods can
consist of, or consist essentially of Formula (VIII), X800K X801K X802E X803
(SEQ ID
NO: 395), as described herein. In some embodiments, X800 is an optional
sequence, and
can be K, A, D, E, G, H, I, L, M, N, P, Q, R, T, V, or K, or absent. In some
X801 is an optional sequence, and can be LDTFFV, GDTH-V, EDTH-V, LDQFFV,
LDTAFV, LDTVFV, LDTFMV, LDTFSV, LDTFVV, LDTFTV, LDTFLV, LDGFFV,
LDTFGV, LDTFFK, ADTFFV, CDTFFV, DDTFFV, FDTFFV, HDTH-V, IDTFFV,
KDTH-V, MDTFFV, NDTFFV, QDTFFV, RDTFFV, SDTFFV, TDTFFV, VDTFFV,
LATFFV, LETH-V, LITFFV, LVTFFV, LWTFFV, LYTH-V, LDCFFV, LDMFFV,
LDNIA-V, LDPFFV, LDRIA-V, LDSFFV, LDWFFV, LDYFFV, LDTIFV, LDTMFV,
LDTNFV, LDTPFV, LDTTFV, LDTFQV, LDTH-F, LDTFFG, LDTFFL, LDTFFP,
LDTFFR, LDTFIV, LDTSFV, LDTFAV, LDTFCV, LDTQFV, LDTLFV, LTTFFV,
LDTFFI, LDHFFV, LMTFFV, LDTFEV, LDTFWV, LFTFFV, LDVFFV, LDTI-RV,
LDTFHV, LDTYFV, LPTH-V, PDTFFV, LDTFPV, LDTFNV, LDTWFV, LDTGFV,
LDAFFV, LQTFFV, LCTFFV, LSTFFV, YDTFFV, LDEFFV, WDTFFV, LDTKFV,
LDTCFV, LDTFYV, LDTHFV, LHTFFV, LRTFFV, LDLFFV, LDTRFV, LLTFFV,
LDTFDV, LDTH-A, LDTFFT, LNTFFV, LDDIA-V, LDIFFV, LDFFFV, LKTFFV,
LDTFFQ, LGTH-V, LDTH-C, LDKFFV, LDTFKV, LDTEFV, LDTFFW, LDTFFM,
LDTFFS, LDTFFH, LDTFFY, LDTH-N, LDTDFV, LDTFFE, LDTFFD, LTFFV,
LDTFF, TFFV, LDF, LDTE, FFV, LDV, LV, or L, or absent. In some embodiments,
162
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
is an optional sequence, and can be LSLFT, VSLFT, LQLFT, LMLFT, LTLFT, LHLFT,
LSQFT, LSVFT, LSMFT, LSLMT, LSLQT, LSLHT, LSLNT, LSLPT, LSLST, LSLGT,
LSLAT, LSLRT, LSLFN, LSLFP, LSLFR, LGLFT, ASLFT, FSLFT, GSLFT, ISLFT,
MSLFT, NSLFT, PSLFT, QSLFT, RSLFT, SSLFT, TSLFT, YSLFT, LNLFT, LSAFT,
LSHFT, LSIFT, LSNFT, LSRFT, LSSFT, LSTFT, LSWFT, LSLCT, LSLIT, LSLLT,
LSLTT, LSLVT, LSLWT, LSLFF, LSLFG, LSLFH, LSLFI, LSLFL, LSLFM, LSLFS,
LSLFV, LSLFW, LYLFT, LVLFT, LSFFT, LSGFT, LSKFT, LSCFT, LCLFT, LRLFT,
LPLFT, LWLFT, LKLFT, LDLFT, LSYFT, LALFT, WSLFT, LSLFA, LSLFQ, LSPFT,
HSLFT, LSLYT, LILFT, KSLFT, CSLFT, LSLFY, LSLFK, LSLFC, LFLFT, LELFT,
LSLKT, LLLFT, LSLFD, LSLDT, LSLFE, DSLFT, LSLET, LSDFT, LSEFT, ESLFT,
SLFT, LSFT, LFT, LSL, LT, or T, or absent. In some embodiments, X803 is an
optional
sequence, and can be R, F, K, N, R, T, or Y, or absent. In some embodiments,
the
isolated peptide comprising Formula (VIII) has a length that is less than or
equal to 1100
amino acids, for example, less than or equal to 4, 5, 6, 7, 8, 9, 10, 11, 12,
13, 14, 15, 16,
17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35,
36, 37, 38, 39,
41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59,
60, 61, 62, 63,
65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83,
84, 85, 86, 87,
89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105, 106,
107, 108,
109, 110, 111, 112, 113, 114, 115, 116, 117, 118, 119, 120, 121, 122, 123,
124, 125, 126,
127, 128, 129, 130, 131, 132, 133, 134, 135, 136, 137, 138, 139, 140, 150,
160, 170, 180,
190, 200, 210, 220, 230, 240, 250, 260, 270, 280, 290, 300, 320, 340, 360,
380, 400, 450,
500, 550, 600, 650, 700, 750, 800, 850, 900, 950, 1000, 1050, or 1100 amino
acids,
including ranges between any two of the listed values.
[0497]
Additionally, the peptide inhibitor can comprise, consist of, or consist
essentially of and/or SEQ ID NOs: 1-33, 34, 46-53, 64-66, 68, 76, 94-96, 98,
264-393,
583-586, or 589 or any one or more of the peptides provided in Table 5.1, 5.4,
5.5, 5.6,
any variation or combination of variations of P28R or P28 core as provided in
Tables 5.3
and 13, as described herein. In some embodiments, these isolated peptides used
in these
methods have a length that is less than or equal to 1100 amino acids, for
example, less
than or equal to 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20,
21, 22, 23, 24,
25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43,
44, 45, 46, 47,
49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67,
68, 69, 70, 71,
73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91,
92, 93, 94, 95,
163
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
97, 98, 99, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112,
113, 114,
115, 116, 117, 118, 119, 120, 121, 122, 123, 124, 125, 126, 127, 128, 129,
130, 131, 132,
133, 134, 135, 136, 137, 138, 139, 140, 150, 160, 170, 180, 190, 200, 210,
220, 230, 240,
250, 260, 270, 280, 290, 300, 320, 340, 360, 380, 400, 450, 500, 550, 600,
650, 700, 750,
800, 850, 900, 950, 1000, 1050, or 1100 amino acids, including ranges between
any two
of the listed values.
[0498] Additionally, the
peptide inhibitor used in these methods can
comprise, consist of, or consist essentially of a peptide inhibitor that
comprises, consists
of, or consists essentially of any one or more of the peptides set forth in
Table 5.1. In
some embodiments, the isolated peptide from Table 5.1 used in these methods
has a
length that is less than or equal to 1100 amino acids, for example, less than
or equal to 4,
5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25,
26, 27, 28, 29,
30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48,
49, 50, 51, 52,
53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71,
72, 73, 74, 75,
76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94,
95, 96, 97, 98,
99, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114,
115, 116,
117, 118, 119, 120, 121, 122, 123, 124, 125, 126, 127, 128, 129, 130, 131,
132, 133, 134,
135, 136, 137, 138, 139, 140, 150, 160, 170, 180, 190, 200, 210, 220, 230,
240, 250, 260,
270, 280, 290, 300, 320, 340, 360, 380, 400, 450, 500, 550, 600, 650, 700,
750, 800, 850,
900, 950, 1000, 1050, or 1100 amino acids, including ranges between any two of
the
listed v4U99] A nucleic acid
encoding such a peptide inhibitor can be provided, for
example a nucleic acid of SEQ ID NOs: 102-165. Following administration of the
immunoregulatory peptide inhibitor, stimulation of human immune cells of the
human
be detected (e.g., an increase in immune cell proliferation, migration of NK
cell
cytotoxicity). Once the immunoregulatory peptide inhibitor has been
administered, these
methods can, optionally, include measuring or observing a reduction in
immunosuppression in the patient (e.g., an increase in immune cell
proliferation,
migration, or spreading or NK-cell cytotoxicity can be evaluated or detecting
activation
or stimulation of an immune cell, as evidenced by an increase in CD69 or CD71
expression, induction of the secretion of a signal substance, as evidenced by
interferon
gamma or IL-12 production, or stimulation of the release of a cytolytic
substance, as
evidenced by the release of granzyme B or perforM, enhanced cytotoxicity,
cytokine
production, cell migration, and/or cell proliferation).
164
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
[0500] As
mentioned above, some embodiments include a step of identifying
a patient suffering from immunosuppression. This analysis can include
generally
determining the immune cell activity of the patient, for example determining
the quantity
of at least one immune cell type, for example leukocytes, PBMC's, lymphocytes,
monocytes, macrophages in a biological sample of the patient. The presence of
the
P3028 sequence/structure in the serum of a patient, and/or on a cancer cell of
a patient
(an evaluation that can be accomplished using a labeled immunoregulatory
peptide
inhibitor) is also indicative of suppression of the immune system of the
patient.
Accordingly, some embodiments of the invention include detecting the presence
of the
P3028 sequence/structure in a biological sample of a patient, for example a
sample that
includes blood, plasma, serum, or a cancer cell biopsy. Examples, methods, and
compositions for detecting the presence of Peptide 3028 in a biological sample
of a
patient can be found in US Pat Nos. 7960126, 8133688, 8110347, and US
Publication
Nos. 2010/0323370 and 2011/0262470, each of which is hereby expressly
incorporated
by reference in its entirety. The P3028 sequence/structure can be detected,
for example,
by immunoassays, a blotting technique, ELISA, ELISpot, flow cytometry,
cytometric
bead assay, proteomics, and/or immunohistochemistry of a biological sample,
using at
least one antibody that binds to the P3028 sequence/structure. The
P3028
sequence/structure can also be detected, for example, by mass spectrometry of
a
biological sample of a patient or a fraction thereof. The P3028
sequence/structure can
further be detected by direct detection of a labeled peptide inhibitor of the
P3028
sequence/structure as described herein, for example by histological staining,
fluorescent
microscopy, immunohistochemistry, or colorimetric enzymatic assays (see
Example 14).
The P3028 sequence/structure can also be detected, for example, functionally,
by
comparing an immune cell contacted by a patient's serum to an immune cell
contacted
by control sample serum known not to contain the P3028 sequence/structure. In
some
embodiments, the serum is denatured. Exemplary immune cells include PBMCs. In
some embodiments, the serum is not denatured. The immune cells can be
optionally
stimulated, for example, by IL-2 or lipopolysaccharide (LPS). In some
embodiments, the
immun41a0111are ahttlyarnefeAlodiprektbsctioipatient suffering from
immunosuppression
can be identified by diagnosing the patient with cancer, for example
metastatic cancer,
and/or two or more tumors in different locations. In some embodiments, cancer
cells can
be identified, and the patient can thus be identified, by detecting the
binding of cells of
165
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
patient to the P3028 sequence/structure (see Example 7) or an inhibitor of the
P3028
sequence/structure (see Example 14). For example, binding of P3028 to cells or
tissues
from multiple locations in the patient can indicate the presence of multiple
tumors, such
as primary and remote tumors in metasatis. Exemplary cancers that can be
identified,
that are associated with immunosuppression include breast cancer, renal cell
carcinoma,
and malignant melanoma. Additional examples include a prostate tumor, a
melanoma, a
colon cancer, a lung carcinoma, an Apocrine gland carcinoma, a testis tumor, a
mast cell
tumor, a mammary tumor (e.g. a benign mammary tumor or a malignant mammary
for example a mixed mammary tumor such as a benign mixed mammary tumor or a
malignant mixed mammary tumor), a mucinous carcinoma (e.g. a mammary gland
mucinous carcinoma), or a histicytoma
[0502] The
administration of the immunoregulatory peptide inhibitor (or a
composition comprising the immunoregulatory peptide inhibitor, for example an
immunoregulatory peptide inhibitor immobilized on a nanoparticle as described
herein)
the patient can be accomplished by a variety of approaches. In some
embodiments, the
immunoregulatory peptide inhibitor is administered directly to the patient.
The
immunoregulatory peptide inhibitor can be administered intravenously,
intraperitoneally,
subcutaneousously, intramuscularly, topically, transdermally, orally, and/or
peri-
tumorally. In some
embodiments, the immunoregulatory peptide inhibitor is
at the site of a tumor, for example via direct injection. In some embodiments,
the
immunoregulatory peptide inhibitor is administered near a tumor, for example
within
10cm, 9cm, 8cm, 7cm, 6cm, 5cm, 4cm, 3cm, 2cm, 1 cm, or 0.5cm of the tumor or a
range
defined by any tow of the aforementioned distances. In some
embodiments, the
immunoregulatory peptide inhibitor is administered with a pharmaceutically
acceptable
diluent or carrier, as described herein. In some embodiments, the
immunoregulatory
peptide inhibitor is administered ex vivo. Immune cells of the patient can be
isolated
the patient, contacted with the inhibitor, and returned to the patient, for
example.
Examples 13 and 14 describe contacting immune cells of a patient with an
inhibitor of
the P3028 sequence/structure.
Optionally, the pharmaceutical formulation is
directly to a tumor, and induces regressive changes in the tumor. Optionally,
the
pharmaceutical formulation is administered to a subject, and induces
regressive changes
of a tumor to which the composition is not directly administered. Optionally,
the
pharmaceutical formulation is administered directly to a tumor, and induces
regressive
166
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
changes in the tumor, and further induces regressive changes in a second tumor
to which
the formulation was not directly administered (e.g. a metastatic or
contralateral tumor).
Optionally, the pharmaceutical formulation is administered directly to a
tumor, and
induces eradication of the tumor. Optionally, the pharmaceutical formulation
is
administered to a subject, and induces eradication of a tumor to which the
composition is
not directly administered. Optionally, the pharmaceutical formulation is
administered
directly to a tumor, and induces eradication of the tumor, and further induces
eradication
of a second tumor to which the formulation was not directly administered (e.g.
a
metastatic or contralateral tumor). Optionally, the pharmaceutical formulation
is
administered directly to a tumor, and induces immune cell infiltration of the
tumor.
Optionally, the pharmaceutical formulation is administered directly to a
tumor, and
induces immune cell infiltration of the tumor, and further induces immune cell
of a second tumor to which the formulation was not directly administered (e.g.
a
metastatic or contralateral tumor). Optionally, the pharmaceutical formulation
is
administered to a subject, and induces immune cell infiltration of a tumor to
which the
composition is not directly administered. Example tumors to which the
pharmaceutical
composition can be directly or indirect administered include a prostate tumor,
a
melanoma, a colon cancer, a lung carcinoma, an Apocrine gland carcinoma, a
testis
tumor, a mast cell tumor, a mammary tumor (e.g. a benign mammary tumor or a
malignant mammary tumor, for example a mixed mammary tumor such as a benign
mixed mammary tumor or a malignant mixed mammary tumor), a mucinous carcinoma
(e.g. a mammary gland mucinous carcinoma), or a histicytoma. As shown in
Figures
82, administration of compositions comprising immunoregulatory peptide
inhibitors as
described herein induced regressive changes, immune cell infiltration of,
and/or
eradication of tumors.
[0503] Any one
or more of the immunoregulatory peptide inhibitors described
herein can be employed with one or more of the aforementioned methods. In some
embodiments, the immunoregulatory peptide inhibitor comprises at least one of
SEQ ID
NOs: 1-33, 34, 46-53, 64-66, 68, 76, 94-96, 98, 264-393, 583-586, or 589 or
any one or
more of the peptides provided in Table 5.1, 5.4, 5.5, 5.6, or any variation or
combination
of variations of P28R or P28 core as provided in Tables 5.3 and 13. In some
embodiments, the immunoregulatory peptide inhibitor includes at least one
peptidomimetic inhibitor of the P3028 sequence/structure corresponding to any
one or
167
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
more of SEQ ID NOs: 1-33, 34, 46-53, 64-66, 68, 76, 94-96, 98, 264-393, 583-
586, or
589 or any one or more of the peptides provided in Table 5.1, 5.4, 5.5, 5.6,
or any
variation or combination of variations of P28R or P28 core as provided in
Tables 5.3 and
13. In some embodiments, the immunoregulatory peptide inhibitor is a small
molecule
inhibitor of Peptide 3028 corresponding to any one or more of SEQ ID NOs: 1-
33, 34,
46-53, 64-66, 68, 76, 94-96, 98, 264-393, 583-586, or 589 or any one or more
of the
peptides provided in Table 5.1, 5.4, 5.5, 5.6, or any variation or combination
of
of P28R or P28 core as provided in Tables 5.3 and 13. In some embodiments, the
immunoregulatory peptide inhibitor includes an antibody or fragment thereof
that
specifically binds to the P3028 sequence/structure. Antibodies that inhibit
the P3028
sequence/structure are described in Example 9.
[0504] In some
of the aforementioned methods, the immunoregulatory
inhibitor of the P3028 sequence/structure comprises a nucleic acid encoding an
immunoregulatory peptide inhibitor, such as a peptide described herein. For
example,
peptide inhibitor encoded by the nucleic acid can comprise, consist of, or
consist
essentially of Formula (I), XX1VKX2X3X4 (SEQ ID NO: 166) as described herein.
In
some embodiments, X is an optional sequence, and can be KKLDT (SEQ ID NO:
167),
RKLDT (SEQ ID NO: 168), KKGDT (SEQ ID NO: 169), KKEDT (SEQ ID NO: 170),
KKLDQ (SEQ ID NO: 171), KKGDQ (SEQ ID NO: 252), KKEDQ (SEQ ID NO:
253), RKLDQ (SEQ ID NO: 254), RKGDQ (SEQ ID NO: 255), RKEDQ (SEQ ID NO:
256), RKGTD (SEQ ID NO: 257), RKEDT (SEQ ID NO: 258), KLDT (SEQ ID NO:
172), KGDT (SEQ ID NO: 259), KEDT (SEQ ID NO: 260), KLDQ (SEQ ID NO:
KGDQ (SEQ ID NO: 262), KEDQ (SEQ ID NO: 263), LDT, LDQ, GDT, GDQ, EDT,
EDQ, DT, DQ, T, or Q, or absent. In some embodiments, Xi is one of FF, FM, FS,
FV,
FT, FL, AF, AM, AS, AV, AT, AL, VF, VM, VS, VV, VT, or VL. In some
embodiments, X2 is one of LS, LQ, LM, LT, LH, VS, VQ, VM, VT, or VH. In some
embodiments, X3 is one of LFT, LMT, LQT, LHT, LNT, LPT, LST, LGT, LAT, LRT,
QFT, QMT, QQT, QHT, QNT, QPT, QST, QGT, QAT, QRT, VFT, VMT, VQT, VHT,
VNT, VPT, VST, VGT, VAT, VRT, MFT, MMT, MQT, MHT, MNT, MPT, MST,
MGT, MAT, MRT, LFN, LMN, LQN, LHN, LNN, LPN, LSN, LGN, LAN, LRN, QFN,
QMN, QQN, QHN, QNN, QPN, QSN, QGN, QAN, QRN, VFN, VMN, VQN, VHN,
VNN, VPN, VSN, VGN, VAN, VRN, MFN, MMN, MQN, MHN, MNN, MPN, MSN,
168
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
MGN, MAN, MRN, LFP, LMP, LQP, LHP, LNP, LPP, LSP, LGP, LAP, LRP, QFP,
QMP, QQP, QHP, QNP, QPP, QSP, QGP, QAP, QRP, VFP, VMP, VQP, VHP, VNP,
VPP, VSP, VGP, YAP, VRP, MFP, MMP, MQP, MHP, MNP, MPP, MSP, MGP, MAP,
MRPR, LFR, LMR, LQR, LHR, LNR, LPR, LSR, LGR, LAR, LRR, QFR, QMR, QQR,
QHR, QNR, QPR, QSR, QGR, QAR, QRR, VFR, VMR, VQR, VHR, VNR, VPR, VSR,
VGR, VAR, VRR, MFR, MMR, MQR, MHR, MNR, MPR, MSR, MGR, MAR, or
In some embodiments, X4 is an optional sequence, and can be ER, or E, or
absent. In
some embodiments, if X is absent, Xi is FF, and X2 is LS. In some embodiments,
the
isolated peptides that comprise Formula (I) encoded by the nucleic acids used
in these
methods have a length that is less than or equal to 1100 amino acids, for
example, less
than or equal to 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20,
21, 22, 23, 24,
25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43,
44, 45, 46, 47,
49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67,
68, 69, 70, 71,
73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91,
92, 93, 94, 95,
97, 98, 99, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112,
113, 114,
115, 116, 117, 118, 119, 120, 121, 122, 123, 124, 125, 126, 127, 128, 129,
130, 131, 132,
133, 134, 135, 136, 137, 138, 139, 140, 150, 160, 170, 180, 190, 200, 210,
220, 230, 240,
250, 260, 270, 280, 290, 300, 320, 340, 360, 380, 400, 450, 500, 550, 600,
650, 700, 750,
800, 850, 900, 950, 1000, 1050, or 1100 amino acids, including ranges between
any two
of the listed values.
[0505]
Additionally, the peptide inhibitor encoded by the nucleic acids can
comprise, consist of, or consist essentially of Formula (II), X20TFFVKLSX21X22
(SEQ
NO: 173). In some embodiments, X20 is an optional sequence, and can be KKLD
(SEQ
ID NO: 174), RKLD (SEQ ID NO: 175), KKGD (SEQ ID NO: 176), KKED (SEQ ID
NO: 177), KLD, LD, or D, or absent. X21 is an optional sequence, and can be
LFT,
LQT, LHT, LNT, LPT, LST, LGT, LAT, LRT, QFT, QMT, QQT, QHT, QNT, QPT,
QST, QGT, QAT, QRT, VFT, VMT, VQT, VHT, VNT, VPT, VST, VGT, VAT, VRT,
MFT, MMT, MQT, MHT, MNT, MPT, MST, MGT, MAT, MRT, LFN, LMN, LQN,
LHN, LNN, LPN, LSN, LGN, LAN, LRN, QFN, QMN, QQN, QHN, QNN, QPN, QSN,
QGN, QAN, QRN, VFN, VMN, VQN, VHN, VNN, VPN, VSN, VGN, VAN, VRN,
MFN, MMN, MQN, MHN, MNN, MPN, MSN, MGN, MAN, MRN, LFP, LMP, LQP,
LHP, LNP, LPP, LSP, LGP, LAP, LRP, QFP, QMP, QQP, QHP, QNP, QPP, QSP, QGP,
QAP, QRP, VFP, VMP, VQP, VHP, VNP, VPP, VSP, VGP, YAP, VRP, MFP, MMP,
169
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
MQP, MHP, MNP, MPP, MSP, MGP, MAP, MRPR, LFR, LMR, LQR, LHR, LNR,
LPR, LSR, LGR, LAR, LRR, QFR, QMR, QQR, QHR, QNR, QPR, QSR, QGR, QAR,
QRR, VFR, VMR, VQR, VHR, VNR, VPR, VSR, VGR, VAR, VRR, MFR, MMR,
MQR, MHR, MNR, MPR, MSR, MGR, MAR, or MRR, or absent. In some
embodiments, X22 is an optional sequence, and can be ER, E, or absent. In some
embodiments, the isolated peptides that comprise Formula (II) encoded by the
nucleic
acids used in these methods have a length that is less than or equal to 1100
amino acids,
for example, less than or equal to 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15,
16, 17, 18, 19,
21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39,
40, 41, 42, 43,
45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63,
64, 65, 66, 67,
69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87,
88, 89, 90, 91,
93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109,
110, 111,
112, 113, 114, 115, 116, 117, 118, 119, 120, 121, 122, 123, 124, 125, 126,
127, 128, 129,
130, 131, 132, 133, 134, 135, 136, 137, 138, 139, 140, 150, 160, 170, 180,
190, 200, 210,
220, 230, 240, 250, 260, 270, 280, 290, 300, 320, 340, 360, 380, 400, 450,
500, 550, 600,
650, 700, 750, 800, 850, 900, 950, 1000, 1050, or 1100 amino acids, including
ranges
between any two of the listed values.
[0506]
Additionally, the peptide inhibitor encoded by the nucleic acids can
comprise, consist of, or consist essentially of Formula (III),
X30X31VKLX32LX33TEX34
(SEQ ID NO: 178). In some embodiments, X30 is an optional sequence, and can be
KKLDTF (SEQ ID NO: 179), KLDTF (SEQ ID NO: 180), LDTF (SEQ ID NO: 181),
DTF, TF, or F, or absent. In some embodiments, X31 is an optional sequence,
and can be
F, S, M, V, T, or L, or absent. In some embodiments, X31 is F. In some
embodiments,
X32 can be S, Q, M, T, or H. In some embodiments, X32 is S. X33 can be F, M,
Q, H, N,
P, S, G, A, or R. In some embodiments, X34 is F. X34 is an optional sequence,
and can
R or absent. In some embodiments, the isolated peptides that comprise Formula
(III)
encoded by the nucleic acids used in these methods have a length that is less
than or
to 1100 amino acids, for example, less than or equal to 4, 5, 6, 7, 8, 9, 10,
11, 12, 13, 14,
15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33,
34, 35, 36, 37,
39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57,
58, 59, 60, 61,
63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81,
82, 83, 84, 85,
87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104,
105, 106, 107,
108, 109, 110, 111, 112, 113, 114, 115, 116, 117, 118, 119, 120, 121, 122,
123, 124, 125,
170
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
126, 127, 128, 129, 130, 131, 132, 133, 134, 135, 136, 137, 138, 139, 140,
150, 160, 170,
180, 190, 200, 210, 220, 230, 240, 250, 260, 270, 280, 290, 300, 320, 340,
360, 380, 400,
450, 500, 550, 600, 650, 700, 750, 800, 850, 900, 950, 1000, 1050, or 1100
amino acids,
including ranges between any two of the listed values.
[0507]
Additionally, the peptide inhibitor encoded by the nucleic acids can
comprise, consist of, or consist essentially of Formula (VII), X700K
X70X702X703
X704X705X706K X707 X708 X709 X710 X711E X712 (SEQ ID NO: 394), as described
herein.
In some embodiments, X700 is an optional sequence, and can be
K,A,D,E,G,H,I,L,M,N,P,Q,R,T, or V, or absent. In some embodiments, X701 is an
optional sequence, and can be L,A,C,D,E,F,G,H,I,K,M,N,Q,R,S,T, or V, or
absent. In
some embodiments, X702 is an optional sequence, and can be D,A,E,I,V,W, or Y,
or
absent. In some
embodiments, X703 is an optional sequence, and can be
T,C,M,N,P,Q,R,S,W, or Y, or absent. In some embodiments, X704 is an optional
sequence, and can be F,A,I,M,N,P,T, or V, or absent. In some embodiments, X705
is an
optional sequence, and can be F,L,M,Q,S,T, or V, or absent. In some
embodiments, X706
is an optional sequence, and can be V,F,G,L,P, or R, or absent. In some
embodiments,
X707 is an optional sequence, and can be L,A,F,G,I,M,N,P,Q,R,S,T,V, or Y, or
absent. In
some embodiments, X708 is an optional sequence, and can be S,H,M,N,Q, or T, or
absent.
In some embodiments, X709 is an optional sequence, and can be
L,A,H,I,M,N,Q,R,S,T,V,
or W, or absent. In some embodiments, X710 is an optional sequence, and can be
F,A,C,G,H,I,L,M,N,P,Q,R,S,T,V, or W, or absent. In some embodiments, X711 is
an
optional sequence, and can be T,F,G,H,I,L,M,N,P,S,V, or W, or absent. In some
embodiments, X712 is an optional sequence, and can be R,F,K,N,R,T, or Y, or
absent. In
some embodiments, the isolated peptide comprising Formula (VII) has a length
that is
than or equal to 1100 amino acids, for example, less than or equal to 4, 5, 6,
7, 8, 9, 10,
11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29,
30, 31, 32, 33,
35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53,
54, 55, 56, 57,
59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77,
78, 79, 80, 81,
83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101,
102, 103, 104,
105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115, 116, 117, 118, 119,
120, 121, 122,
123, 124, 125, 126, 127, 128, 129, 130, 131, 132, 133, 134, 135, 136, 137,
138, 139, 140,
150, 160, 170, 180, 190, 200, 210, 220, 230, 240, 250, 260, 270, 280, 290,
300, 320, 340,
171
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
360, 380, 400, 450, 500, 550, 600, 650, 700, 750, 800, 850, 900, 950, 1000,
1050, or
amino acids, including ranges between any two of the listed values.
[0508]
Additionally, the peptide inhibitor encoded by the nucleic acids can
comprise, consist of, or consist essentially of Formula (VIII), X800K X801K
X802E X803
(SEQ ID NO: 395), as described herein. In some embodiments, X800 is an
optional
sequence, and can be K, A, D, E, G, H, I, L, M, N, P, Q, R, T, V, or K, or
absent. In
embodiments, X801 is an optional sequence, and can be LDTFFV, GDTFFV, EDTFFV,
LDQFPV, LDTAFV, LDTVFV, LDTFMV, LDTFSV, LDTFVV, LDTFTV, LDTFLV,
LDGEEN, LDTFGV, LDTH-K, ADTFFV, CDTFFV, DDTFFV, FDTH-V, HDTFFV,
IDTFFV, KDTFFV, MDTFFV, NDTFFV, QDTH-V, RDTFFV, SDTFFV, TDTFFV,
VDTFFV, LATFFV, LETFFV, LITFFV, LVTFFV, LWTFFV, LYTH-V, LDCFFV,
LDMEEN, LDNFFV, LDPFFV, LDRFFV, LDSFFV, LDWFFV, LDYFFV, LDTIFV,
LDTMFV, LDTNFV, LDTPFV, LDTTFV, LDTFQV, LDTFFF, LDTFFG, LDTFFL,
LDTFFP, LDTH-R, LDTFIV, LDTSFV, LDTFAV, LDTFCV, LDTQFV, LDTLFV,
LTTFFV, LDT141-4, LDHFFV, LMTFFV, LDTFEV, LDTFWV, LFTFFV, LDVFFV,
LDTFRV, LDTFHV, LDTYFV, LPTFFV, PDTFFV, LDTFPV, LDTFNV, LDTWFV,
LDTGFV, LDAFFV, LQT141-N, LCTFFV, LSTFFV, YDTFFV, LDEFFV, WDTFFV,
LDTKFV, LDTCFV, LDTFYV, LDTHFV, LHTFFV, LRTFFV, LDLFFV, LDTRFV,
LLTFFV, LDTFDV, LDTFFA, LDTFFT, LNTFFV, LDDFFV, LDIFFV, LDFFFV,
LKTFFV, LDTFFQ, LGTFFV, LDTFFC, LDKFFV, LDTFKV, LDTEFV, LDTFFW,
LDTFFM, LDTFFS, LDTFFH, LDTFFY, LDTH-N, LDTDFV, LDTH-E, LDTFFD,
LTH-V, LDTFF, TFFV, LDF, LDTE, FFV, LDV, LV, or L, or absent. In some
embodiments, X802 is an optional sequence, and can be LSLFT, VSLFT, LQLFT,
LMLFT, LTLFT, LHLFT, LSQFT, LSVFT, LSMFT, LSLMT, LSLQT, LSLHT, LSLNT,
LSLPT, LSLST, LSLGT, LSLAT, LSLRT, LSLFN, LSLFP, LSLFR, LGLFT, ASLFT,
FSLFT, GSLFT, ISLFT, MSLFT, NSLFT, PSLFT, QSLFT, RSLFT, SSLFT, TSLFT,
YSLFT, LNLFT, LSAFT, LSHFT, LSIFT, LSNFT, LSRFT, LSSFT, LSTFT, LSWFT,
LSLCT, LSLIT, LSLLT, LSLTT, LSLVT, LSLWT, LSLFF, LSLFG, LSLFH, LSLFI,
LSLFL, LSLFM, LSLFS, LSLFV, LSLFW, LYLFT, LVLFT, LSFFT, LSGFT, LSKFT,
LSCFT, LCLFT, LRLFT, LPLFT, LWLFT, LKLFT, LDLFT, LSYFT, LALFT, WSLFT,
LSLFA, LSLFQ, LSPFT, HSLFT, LSLYT, LILFT, KSLFT, CSLFT, LSLFY, LSLFK,
LSLFC, LFLFT, LELFT, LSLKT, LLLFT, LSLFD, LSLDT, LSLFE, DSLFT, LSLET,
LSDFT, LSEFT, ESLFT, SLFT, LSFT, LFT, LSL, LT, or T, or absent. In some
172
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
embodiments, X803 is an optional sequence, and can be R, F, K, N, R, T, or Y,
or absent.
In some embodiments, the isolated peptide comprising Formula (VIII) has a
length that is
less than or equal to 1100 amino acids, for example, less than or equal to 4,
5, 6, 7, 8, 9,
10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28,
29, 30, 31, 32,
34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52,
53, 54, 55, 56,
58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76,
77, 78, 79, 80,
82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100,
101, 102, 103,
104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115, 116, 117, 118,
119, 120, 121,
122, 123, 124, 125, 126, 127, 128, 129, 130, 131, 132, 133, 134, 135, 136,
137, 138, 139,
140, 150, 160, 170, 180, 190, 200, 210, 220, 230, 240, 250, 260, 270, 280,
290, 300, 320,
340, 360, 380, 400, 450, 500, 550, 600, 650, 700, 750, 800, 850, 900, 950,
1000, 1050,
1100 amino acids, including ranges between any two of the listed values.
[0509]
Additionally, the peptide inhibitor encoded by the nucleic acid used in
these methods can comprise, consist of, or consist essentially of Formula
(IX).
Accordingly, in some embodiments, the peptide inhibitor comprises a peptide of
(IX): X901X902X903X904X905X906X907X908X909X910X911X912X913X914X915X916X917,
wherein
X901 is any amino acid or absent; X902 is a positively charged amino acid, F,
or N;X903 is
any amino acid; X904 is any amino acid; X905 is a polar uncharged amino acid,
R, Y, or
X906 is a hydrophobic or uncharged polar amino acid; X907 is a hydrophobic or
uncharged
polar amino acid; X908 is a hydrophobic, non-aromatic carbon chain amino acid
that is
M or F; X909 is a positively charged amino acid, T, Q, or Y; X910 is any amino
acid that is
not negatively charged; X911 is a polar uncharged amino acid or H; X912 is any
amino
that is not negatively charged; X913 is any amino acid that is not negatively
charged; X914
is any amino acid that is not negatively charged; X915 is a negatively charged
amino acid,
Y, or Q; X916 is any amino acid that is not negatively charged; and X917 is
one or more
positively charged amino acids or is absent. Optionally, X901 comprises a
positively
charged amino acid. Optionally X901 is an R or K. Optionally X917 is RR. In
some
embodiments, the isolated peptide comprising Formula (IX) has a length that is
less than
or equal to 1100 amino acids, for example, less than or equal to 4, 5, 6, 7,
8, 9, 10, 11,
13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31,
32, 33, 34, 35,
37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55,
56, 57, 58, 59,
61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79,
80, 81, 82, 83,
85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102,
103, 104, 105,
173
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
106, 107, 108, 109, 110, 111, 112, 113, 114, 115, 116, 117, 118, 119, 120,
121, 122, 123,
124, 125, 126, 127, 128, 129, 130, 131, 132, 133, 134, 135, 136, 137, 138,
139, 140, 150,
160, 170, 180, 190, 200, 210, 220, 230, 240, 250, 260, 270, 280, 290, 300,
320, 340, 360,
380, 400, 450, 500, 550, 600, 650, 700, 750, 800, 850, 900, 950, 1000, 1050,
or 1100
amino acids, including ranges between any two of the listed values.
[0510]
Additionally, the peptide inhibitor encoded by the nucleic acid used in
these methods can comprise, consist of, or consist essentially of and/or SEQ
ID NOs: 1-
33, 34, 46-53, 64-66, 68, 76, 94-96, 98, 264-393, 583-586, or 589 or any one
or more of
the peptides provided in Table 5.1, 5.4, 5.5, 5.6, or any variation or
combination of
variations of P28R or P28 core as provided in Tables 5.3 and 13, as described
herein. In
some embodiments, these isolated peptides encoded by the nucleic acids used in
these
methods have a length that is less than or equal to 1100 amino acids, for
example, less
than or equal to 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20,
21, 22, 23, 24,
25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43,
44, 45, 46, 47,
48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66,
67, 68, 69, 70,
71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89,
90, 91, 92, 93,
94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110,
111, 112,
113, 114, 115, 116, 117, 118, 119, 120, 121, 122, 123, 124, 125, 126, 127,
128, 129, 130,
131, 132, 133, 134, 135, 136, 137, 138, 139, 140, 150, 160, 170, 180, 190,
200, 210, 220,
230, 240, 250, 260, 270, 280, 290, 300, 320, 340, 360, 380, 400, 450, 500,
550, 600, 650,
700, 750, 800, 850, 900, 950, 1000, 1050, or 1100 amino acids, including
ranges
between any two of the listed values.
[0511]
Additionally, the peptide inhibitor encoded by the nucleic acid used
in these methods can comprise, consist of, or consist essentially of a peptide
inhibitor
comprises, consists of, or consists essentially of any one or more of the
peptides set forth
in Table 5.1, 5.4, 5.5, 5.6, or any variation or combination of variations of
P28R or P28
core as provided in Tables 5.3 and 13. In some embodiments, the isolated
peptide from
Table 5.1 5.4, 5.5, 5.6, or any variation or combination of variations of P28R
or P28 core
as provided in Tables 5.3 and 13, which is encoded by the nucleic acid used in
these
methods has a length that is less than or equal to 1100 amino acids, for
example, less
or equal to 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21,
22, 23, 24, 25,
27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45,
46, 47, 48, 49,
51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69,
70, 71, 72, 73,
174
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93,
94, 95, 96, 97,
99, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114,
115, 116,
117, 118, 119, 120, 121, 122, 123, 124, 125, 126, 127, 128, 129, 130, 131,
132, 133, 134,
135, 136, 137, 138, 139, 140, 150, 160, 170, 180, 190, 200, 210, 220, 230,
240, 250, 260,
270, 280, 290, 300, 320, 340, 360, 380, 400, 450, 500, 550, 600, 650, 700,
750, 800, 850,
900, 950, 1000, 1050, or 1100 amino acids, including ranges between any two of
the
listed values.
[0512] For
example, a nucleic acid encoding such a peptide inhibitor can be
provided, for example a nucleic acid of SEQ ID NOs: 102-165. The nucleic acid
can be
provided in an expression vector as described herein. The nucleic acid can be
provided
to the human by directly administering an expression vector comprising the
nucleic acid
that encodes the immunoregulatory peptide inhibitor to the human, for example
via a
retroviral or adenoviral vector or expression plasmid used in genetic
immunization (e.g.,
pVAX). The expression vector can be provided to cells of the human ex vivo,
and the
cells can be returned to the human or in vivo using electroporation
technology. Methods
of delivering nucleic acids to a host cell via viral vectors are described in
US Pat No.
7,572,906, which is expressly incorporated by reference in its entirety
herein. Methods
of transducing immune cells with an adenovirus ex vivo and returning them to a
patient
are described in US Pat No. 8.012,468, which is expressly incorporated by
reference in
its entirety herein. In some embodiments, a host cell, is contacted with a
vector encoding
the immunoregulatory peptide inhibitor of P3028. The vector can replicates in
the host
cell. In some embodiments, the host cell is also contacted with a "helper-
expression
vector," i.e., a viral genome that promotes the replication of the vector in
an uninfected
host. In some embodiments, the inhibitor is administered as in Example 16. In
some
embodiments, the cell is contacted ex vivo. In some embodiments, the cell is
an immune
cell. In some embodiments, the cell is one of a lymphocyte, a PBMC, or a
leukocyte. In
some embodiments, the inhibitor is administered as in Example 13. In some
embodiments, the nucleic acid encoding the peptide inhibitor is administered
to a non-
human mammal, for treatment of immunosuppression or cancer (for example
metastatic
cancer) in the non-human mammal.
[0513] Preferably, a therapeutic ally
effective amount of the
peptide inhibitor is provided. For a patient already suffering from P3028-
dependent
immunosuppression, a therapeutically effective amount of inhibitor may include
a dose
175
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
immunoregulatory peptide inhibitor sufficient to at least partially arrest a
symptom of
immunosuppression (e.g., an amount sufficient to improve proliferation or
migration of
immune cells). In some embodiments, a therapeutically effective amount
includes at
about 1 nanogram of substantially pure immunoregulatory peptide inhibitor, for
example,
at least or equal to about 1 nanogram, 2, 3, 4, 5, 6, 7, 8, 9, 10, 15, 20, 25,
30, 40, 50, 60,
70, 80, 90, 100, 120, 140, 160, 180, 200, 250, 300, 350, 400, 450, 500, 550,
600, 650,
700, 750, 800, 850, 900, 950, 1000 nanograms, 1 microgram, 2, 3, 4, 5, 6, 7,
8, 9, 10, 15,
20, 25, 30, 40, 50, 60, 70, 80, 90, 100, 120, 140, 160, 180, 200, 250, 300,
350, 400, 450,
500, 550, 600, 650, 700, 750, 800, 850, 900, 950, 1000 micrograms, about 1
milligram,
3, 4, 5, 6, 7, 8, 9, 10, 15, 20, 25, 30, 40, 50, 60, 70, 80, 90, 100, 120,
140, 160, 180, 200,
250, 300, 350, 400, 450, 500, 550, 600, 650, 700, 750, 800, 850, 900, 950,
1000
milligrams, or 1.1 gram, 1.2, 1.3, 1.4, 1.5, 1.6, 1.7, 1.8, 1.9, 2, 2., 3,
3.5, 4, 4.5, 5, 5.5, 6,
6.5, 7, 7.5, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24,
25, 26, 27, 28,
29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47,
48, 49, 50, 55,
65, 70, 75, 80, 85, 90, 95, 99, 100, 105, 110, 120, 130, 140, 150, 160, 170,
180, 190,
250, 300, 350, 400, 450, or 500 grams, including ranges between any two of the
listed
values can be provided to a patient in need.
[0514] In some
embodiments, a therapeutically effective amount can be
provided according to a schedule that includes one, or more than one
administration of a
therapeutically effective amount of inhibitor, for example at least or equal
to about 1, 2,
3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23,
24, 25, 26, 27, 28,
29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47,
48, 49, 50, 55,
60, 65, 70, 75, 80, 85, 90, 95, 99, 100, 105, 110, 120, 130, 140, 150, 160,
170, 180, 190,
200, 250, 300, 350, 400, 450, or 500 administrations. An administration can be
provided
hourly or less, for example no more than once every 1, 2, 3, 4, 5, 6, 7, 8, 9,
10, 11, 12,
13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24 hours, or no more than once
every 1 day,
1.5, 2, 2.5, 3, 3.5, 4, 4.5, 5, 5.5, 6, 6.5, 7, 7.5, 8, 9, 10, 11, 12, 13,15,
16, 17, 18, 19, 20,
21, 22, 23, 24, 25, 26, 27, 28, 29, 30, or 31 days.
[0515] By some
methods, after administration of the immunoregulatory
peptide inhibitor, a reduction in immunosuppression is measured, detected, or
observed.
In some embodiments, a reduction in immunosuppression is detected, measured,
or
observed by obtaining a biological sample from the patient that received the
immunoregulatory peptide inhibitor and detecting a reduction in immune cell
receptor
176
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
binding to P3028 and/or a detecting immune cell proliferation after IL-2
induction of the
immune cells present in the biological sample. In some embodiments, the
analysis of the
biological sample obtained from the patient above is compared to the same
analysis (e.g.,
determining the amount of immune cell receptor binding to the P3028
sequence/structure
or IL-2 induced immune cell proliferation) conducted on a control biological
sample, for
example, a biological sample from the same patient taken prior to
administration of the
immunoregulatory peptide inhibitor or a biological sample taken from a healthy
human.
Examples 9 and 13 describe detection of a reduction of immunosuppression in
cells
contacted by serum as compared to a control sample. In some embodiments, the
reduction in immunosuppression is observed as immune cell infiltration of a
tumor,
regressive changes in a tumor, and/or eradication of a tumor. As shown in
Figures 55-82,
administration of compositions comprising immunoregulatory peptide inhibitors
as
described herein induced regressive changes, immune cell infiltration of,
and/or
eradication of tumors.
[0516] As
mentioned above, a reduction in immunosuppression can be
detected as an increase in immune cell stimulation, for example immune cell
proliferation or immune cell cytotoxicity. A
reduction in P3028-induced
immunosuppression, which can be measured in the methods described supra, can
include: increased T-Cell receptor stimulation (see Example 3); increased NK-
Cell
cytotoxicity (see Example 4); increased leukocyte spreading (see Example 5);
increased
immune cell migration (see Example 5); and/or IL-2 Induced Proliferation (see
Example
6). Decreased IL-6 production can also an improvided prognosis for cancer
patients, for
example cancer patients suffering from immunosuppression (see US Pat No.
8,110,347,
herein expressly incorporated by reference in its entirety). Desirably, a
reduction in
immunosuppression is detected by an increased proliferative response of PBMC's
to IL-
2, as shown in Example 9, or by detecting activation or stimulation of an
immune cell,
as evidenced by an increase in CD69 or CD71 expression, induction of the
secretion of a
signal substance, as evidenced by interferon gamma or IL-12 production, or
stimulation
of the release of a cytolytic substance, as evidenced by the release of
granzyme B or
perforM, enhanced cytotoxicity, cytokine production, cell migration, and/or
cell
proliferriOlg] In some
embodiments, the reduction in immunosuppression is detected
by detecting the presence or quantity of markers from immune cells and/or
serum and/or
albumin collected from a patient. In some embodiments, the detection includes
177
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
patient serum, blood, and/or patient albumin, and contacting the patient
serum, plasma,
blood, or albumin with an immune cell ex vivo. In some embodiments, the immune
cell
also contacted with IL-2. The proliferative response of the immune cell to IL-
2 can be
used to detect a decrease in immunosuppression. The immune cell can be a
patient cell,
or a cell from another human, or a cell from cell culture. In some
embodiments, the
reduction in immunosuppression can be detected by detecting effects of
increased
immune system activity, for example reduction in cancer cell number, a
reduction in
tumor size, or a reduction or inhibition of cancer cell proliferation. In some
embodiments, cancer cells can be identified, and cancer cells can thus be
quantified, by
detecting cells that bind to the P3028 sequence/structure (see Example 7) or
an inhibitor
of the P3028 sequence/structure (see Example 14).
Methods of binding cancer cells with an immunoregulatory peptide inhibitor
[0518]
Embodiments also include methods of binding cancer cells in two or
more different tumors with an immunoregulatory peptide inhibitor (e.g., an
immunoregulatory peptide inhibitor having a cytotoxin, radionuclide, or
detectable label)
of two or more tumors, in which an immunoregualtory peptide inhibitor is
neither
intratumorally nor peri-tumorally administered to at least one of the tumors.
For
for a patient with metastatic cancer, an immunoregualtory peptide inhibitor
can bind to
both primary tumor cells and remote tumor cells. These methods are practiced
by
contacting cancer cells (e.g., in vitro or in vivo) with a composition that
comprises,
consists of, or consists essentially of any one or more of the
immunoregulatory peptide
inhibitors described herein. For example, the peptide inhibitor used in these
methods can
comprise, consist of, or consist essentially of Formula (I), XX1VKX2X3X4 (SEQ
ID NO:
166) as described herein. In some embodiments, X is an optional sequence, and
can be
KKLDT (SEQ ID NO: 167), RKLDT (SEQ ID NO: 168), KKGDT (SEQ ID NO: 169),
KKEDT (SEQ ID NO: 170), KKLDQ (SEQ ID NO: 171), KKGDQ (SEQ ID NO:
KKEDQ (SEQ ID NO: 253), RKLDQ (SEQ ID NO: 254), RKGDQ (SEQ ID NO:
RKEDQ (SEQ ID NO: 256), RKGTD (SEQ ID NO: 257), RKEDT (SEQ ID NO: 258),
KLDT (SEQ ID NO: 172), KGDT (SEQ ID NO: 259), KEDT (SEQ ID NO: 260),
KLDQ (SEQ ID NO: 261), KGDQ (SEQ ID NO: 262), KEDQ (SEQ ID NO: 263),
LDT, LDQ, GDT, GDQ, EDT, EDQ, DT, DQ, T, or Q, or absent. In some
Xi is one of FF, FM, FS, FV, FT, FL, AF, AM, AS, AV, AT, AL, VF, VM, VS, VV,
VT,
178
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
or VL. In some embodiments, X2 is one of LS, LQ, LM, LT, LH, VS, VQ, VM, VT,
or
VH. In some embodiments, X3 is one of LFT, LMT, LQT, LHT, LNT, LPT, LST, LGT,
LAT, LRT, QFT, QMT, QQT, QHT, QNT, QPT, QST, QGT, QAT, QRT, VFT, VMT,
VQT, VHT, VNT, VPT, VST, VGT, VAT, VRT, MFT, MMT, MQT, MHT, MNT,
MST, MGT, MAT, MRT, LFN, LMN, LQN, LHN, LNN, LPN, LSN, LGN, LAN, LRN,
QFN, QMN, QQN, QHN, QNN, QPN, QSN, QGN, QAN, QRN, VFN, VMN, VQN,
VHN, VNN, VPN, VSN, VGN, VAN, VRN, MFN, MMN, MQN, MHN, MNN, MPN,
MSN, MGN, MAN, MRN, LFP, LMP, LQP, LHP, LNP, LPP, LSP, LGP, LAP, LRP,
QFP, QMP, QQP, QHP, QNP, QPP, QSP, QGP, QAP, QRP, VFP, VMP, VQP, VHP,
VNP, VPP, VSP, VGP, VAP, VRP, MFP, MMP, MQP, MHP, MNP, MPP, MSP, MGP,
MAP, MRPR, LFR, LMR, LQR, LHR, LNR, LPR, LSR, LGR, LAR, LRR, QFR, QMR,
QQR, QHR, QNR, QPR, QSR, QGR, QAR, QRR, VFR, VMR, VQR, VHR, VNR, VPR,
VSR, VGR, VAR, VRR, MFR, MMR, MQR, MHR, MNR, MPR, MSR, MGR, MAR, or
MRR. In some embodiments, X4 is an optional sequence, and can be ER, or E, or
In some embodiments, if X is absent, Xi is FF, and X2 is LS. In some
embodiments, the
isolated peptides that comprise Formula (I) used in these methods have a
length that is
less than or equal to 1100 amino acids, for example, less than or equal to 4,
5, 6, 7, 8, 9,
10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28,
29, 30, 31, 32,
34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52,
53, 54, 55, 56,
58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76,
77, 78, 79, 80,
82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100,
101, 102, 103,
104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115, 116, 117, 118,
119, 120, 121,
122, 123, 124, 125, 126, 127, 128, 129, 130, 131, 132, 133, 134, 135, 136,
137, 138, 139,
140, 150, 160, 170, 180, 190, 200, 210, 220, 230, 240, 250, 260, 270, 280,
290, 300, 320,
340, 360, 380, 400, 450, 500, 550, 600, 650, 700, 750, 800, 850, 900, 950,
1000, 1050,
1100 amino acids, including ranges between any two of the listed values.
[0519]
Additionally, the peptide inhibitor used in these methods can
comprise, consist of, or consist essentially of Formula (II), X20TFFVKLSX21X22
(SEQ
NO: 173). In some embodiments, X20 is an optional sequence, and can be KKLD
(SEQ
ID NO: 174), RKLD (SEQ ID NO: 175), KKGD (SEQ ID NO: 176), KKED (SEQ ID
NO: 177), KLD, LD, D, or absent. X21 is an optional sequence, and can be LFT,
LMT,
LQT, LHT, LNT, LPT, LST, LGT, LAT, LRT, QFT, QMT, QQT, QHT, QNT, QPT,
QST, QGT, QAT, QRT, VFT, VMT, VQT, VHT, VNT, VPT, VST, VGT, VAT, VRT,
179
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
MFT, MMT, MQT, MHT, MNT, MPT, MST, MGT, MAT, MRT, LFN, LMN, LQN,
LHN, LNN, LPN, LSN, LGN, LAN, LRN, QFN, QMN, QQN, QHN, QNN, QPN, QSN,
QGN, QAN, QRN, VFN, VMN, VQN, VHN, VNN, VPN, VSN, VGN, VAN, VRN,
MFN, MMN, MQN, MHN, MNN, MPN, MSN, MGN, MAN, MRN, LFP, LMP, LQP,
LHP, LNP, LPP, LSP, LGP, LAP, LRP, QFP, QMP, QQP, QHP, QNP, QPP, QSP, QGP,
QAP, QRP, VFP, VMP, VQP, VHP, VNP, VPP, VSP, VGP, VAP, VRP, MFP, MMP,
MQP, MHP, MNP, MPP, MSP, MGP, MAP, MRPR, LFR, LMR, LQR, LHR, LNR,
LPR, LSR, LGR, LAR, LRR, QFR, QMR, QQR, QHR, QNR, QPR, QSR, QGR, QAR,
QRR, VFR, VMR, VQR, VHR, VNR, VPR, VSR, VGR, VAR, VRR, MFR, MMR,
MQR, MHR, MNR, MPR, MSR, MGR, MAR, or MRR, or absent. In some
embodiments, X22 is an optional sequence, and can be ER, or E, or absent. In
some
embodiments, the isolated peptides that comprise Formula (II) used in these
methods
a length that is less than or equal to 1100 amino acids, for example, less
than or equal to
4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24,
25, 26, 27, 28,
30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48,
49, 50, 51, 52,
54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72,
73, 74, 75, 76,
78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96,
97, 98, 99, 100,
101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115,
116, 117, 118,
119, 120, 121, 122, 123, 124, 125, 126, 127, 128, 129, 130, 131, 132, 133,
134, 135, 136,
137, 138, 139, 140, 150, 160, 170, 180, 190, 200, 210, 220, 230, 240, 250,
260, 270, 280,
290, 300, 320, 340, 360, 380, 400, 450, 500, 550, 600, 650, 700, 750, 800,
850, 900, 950,
1000, 1050, or 1100 amino acids, including ranges between any two of the
listed values.
[0520]
Additionally, the peptide inhibitor used in these methods can
consist of, or consist essentially of Formula (III), X30X31VKLX32LX33TEX34
(SEQ ID
NO: 178). In some embodiments, X30 is an optional sequence, and can be KKLDTF
(SEQ ID NO: 179), KLDTF (SEQ ID NO: 180), LDTF (SEQ ID NO: 181), DTF, TF,
F, or absent. In some embodiments, X31 is an optional sequence, and can be F,
S, M, V,
T, or L, or absent. In some embodiments, X31 is F. In some embodiments, X32
can be S,
Q, M, T, or H. In some embodiments, X32 is S. X33 can be F, M, Q, H, N, P, S,
G, A, or
R. In some embodiments, X34 is F. X34 is an optional sequence, and can be R or
absent.
In some embodiments, the isolated peptides that comprise Formula (III) used in
these
methods have a length that is less than or equal to 1100 amino acids, for
example, less
than or equal to 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20,
21, 22, 23, 24,
180
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43,
44, 45, 46, 47,
49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67,
68, 69, 70, 71,
73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91,
92, 93, 94, 95,
97, 98, 99, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112,
113, 114,
115, 116, 117, 118, 119, 120, 121, 122, 123, 124, 125, 126, 127, 128, 129,
130, 131, 132,
133, 134, 135, 136, 137, 138, 139, 140, 150, 160, 170, 180, 190, 200, 210,
220, 230, 240,
250, 260, 270, 280, 290, 300, 320, 340, 360, 380, 400, 450, 500, 550, 600,
650, 700, 750,
800, 850, 900, 950, 1000, 1050, or 1100 amino acids, including ranges between
any two
of the listed values.
[0521]
Additionally, the peptide inhibitor used in these methods can
comprise, consist of, or consist essentially of Formula (VII), X7001(
X701X702X703
X704X705X706K X707 X708 X709 X710 X711E X712 (SEQ ID NO: 394), as described
herein.
In some embodiments, X700 is an optional sequence, and can be
K,A,D,E,G,H,I,L,M,N,P,Q,R,T, or V, or absent. In some embodiments, X701 is an
optional sequence, and can be L,A,C,D,E,F,G,H,I,K,M,N,Q,R,S,T, or V, or
absent. In
some embodiments, X702 is an optional sequence, and can be D,A,E,I,V,W, or Y,
or
absent. In some
embodiments, X703 is an optional sequence, and can be
T,C,M,N,P,Q,R,S,W, or Y, or absent. In some embodiments, X704 is an optional
sequence, and can be F,A,I,M,N,P,T, or V, or absent. In some embodiments, X705
is an
optional sequence, and can be F,L,M,Q,S,T, or V, or absent. In some
embodiments, X706
is an optional sequence, and can be V,F,G,L,P, or R, or absent. In some
embodiments,
X707 is an optional sequence, and can be L,A,F,G,I,M,N,P,Q,R,S,T,V, or Y, or
absent. In
some embodiments, X708 is an optional sequence, and can be S,H,M,N,Q, or T, or
absent.
In some embodiments, X709 is an optional sequence, and can be
L,A,H,I,M,N,Q,R,S,T,V,
or W, or absent. In some embodiments, X710 is an optional sequence, and can be
F,A,C,G,H,I,L,M,N,P,Q,R,S,T,V, or W, or absent. In some embodiments, X711 is
an
optional sequence, and can be T,F,G,H,I,L,M,N,P,S,V, or W, or absent. In some
embodiments, X712 is an optional sequence, and can be R,F,K,N,R,T, or Y, or
absent. In
some embodiments, the isolated peptide comprising Formula (VII) has a length
that is
than or equal to 1100 amino acids, for example, less than or equal to 4, 5, 6,
7, 8, 9, 10,
11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29,
30, 31, 32, 33,
35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53,
54, 55, 56, 57,
59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77,
78, 79, 80, 81,
181
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101,
102, 103, 104,
105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115, 116, 117, 118, 119,
120, 121, 122,
123, 124, 125, 126, 127, 128, 129, 130, 131, 132, 133, 134, 135, 136, 137,
138, 139, 140,
150, 160, 170, 180, 190, 200, 210, 220, 230, 240, 250, 260, 270, 280, 290,
300, 320, 340,
360, 380, 400, 450, 500, 550, 600, 650, 700, 750, 800, 850, 900, 950, 1000,
1050, or
amino acids, including ranges between any two of the listed values.
[0522]
Additionally, the peptide inhibitor used in these methods can
consist of, or consist essentially of Formula(VIII), X800K X801K X802E X803
(SEQ ID
395), as described herein. In some embodiments, X800 is an optional sequence,
and can
K, A, D, E, G, H, I, L, M, N, P, Q, R, T, V, or K, or absent. In some
embodiments, X801
is an optional sequence, and can be LDTFFV, GDTFFV, EDTFFV, LDQFFV, LDTAFV,
LDTVFV, LDTFMV, LDTFSV, LDTFVV, LDTFTV, LDTFLV, LDGFFV, LDTFGV,
LDTFFK, ADTFFV, CDTH-V, DDTH-V, FDTH-V, HDTFFV, IDTFFV, KDTFFV,
MDTFFV, NDTFFV, QDTFFV, RDTFFV, SDTFFV, TDTFFV, VDTH-V, LATFFV,
LETFFV, LITFFV, LVT1-1-V, LWTFFV, LYTFFV, LDCFFV, LDMFFV, LDNFFV,
LDPFFV, LDR1-1-V, LDSFFV, LDWFFV, LDYFFV, LDTIFV, LDTMFV, LDTNFV,
LDTPFV, LDTTFV, LDTFQV, LDTFFF, LDTFFG, LDTFFL, LDTFFP, LDTH-R,
LDTFIV, LDTSFV, LDTFAV, LDTFCV, LDTQFV, LDTLFV, LTTFFV, LDT1-1-4,
LDH1-1-V, LMTFFV, LDTFEV, LDTFWV, LFTFFV, LDVFFV, LDTFRV, LDTFHV,
LDTYFV, LPTFFV, PDTFFV, LDTFPV, LDTFNV, LDTWFV, LDTGFV, LDAFFV,
LQTFFV, LCTFFV, LSTFFV, YDTFFV, LDEFFV, WDTFFV, LDTKFV, LDTCFV,
LDTFYV, LDTHFV, LHTFFV, LRTFFV, LDLFFV, LDTRFV, LLTFFV, LDTFDV,
LDTFFA, LDTH-T, LNTFFV, LDDFFV, LDIFFV, LD1-1-FV, LKT1-1-V, LDTFFQ,
LGTFFV, LDTFFC, LDKFFV, LDTFKV, LDTEFV, LDTFFW, LDTFFM, LDTFFS,
LDTFFH, LDTFFY, LDTFFN, LDTDFV, LDTFFE, LDTH-D, LTFFV, LDT1-1-, TFFV,
LDF, LDTE, FFV, LDV, LV, or L, or absent. In some embodiments, X802 is an
optional
sequence, and can be LSLFT, VSLFT, LQLFT, LMLFT, LTLFT, LHLFT, LSQFT,
LSVFT, LSMFT, LSLMT, LSLQT, LSLHT, LSLNT, LSLPT, LSLST, LSLGT, LSLAT,
LSLRT, LSLFN, LSLFP, LSLFR, LGLFT, ASLFT, FSLFT, GSLFT, ISLFT, MSLFT,
NSLFT, PSLFT, QSLFT, RSLFT, SSLFT, TSLFT, YSLFT, LNLFT, LSAFT, LSHFT,
LSIFT, LSNFT, LSRFT, LSSFT, LSTFT, LSWFT, LSLCT, LSLIT, LSLLT, LSLTT,
LSLVT, LSLWT, LSLFF, LSLFG, LSLFH, LSLFI, LSLFL, LSLFM, LSLFS, LSLFV,
LSLFW, LYLFT, LVLFT, LSH-T, LSGFT, LSKFT, LSCFT, LCLFT, LRLFT, LPLFT,
182
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
LWLFT, LKLFT, LDLFT, LSYFT, LALFT, WSLFT, LSLFA, LSLFQ, LSPFT, HSLFT,
LSLYT, LILFT, KSLFT, CSLFT, LSLFY, LSLFK, LSLFC, LFLFT, LELFT, LSLKT,
LLLFT, LSLFD, LSLDT, LSLFEõ DSLFT, LSLET, LSDFT, LSEFT, ESLFT, SLFT,
LSFT, LFT, LSL, LT, or T, or absent. In some embodiments, X803 is an optional
sequence, and can be R, F, K, N, R, T, or Y, or absent. In some embodiments,
the
isolated peptide comprising Formula (VIII) has a length that is less than or
equal to 1100
amino acids, for example, less than or equal to 4, 5, 6, 7, 8, 9, 10, 11, 12,
13, 14, 15, 16,
17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35,
36, 37, 38, 39,
41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59,
60, 61, 62, 63,
65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83,
84, 85, 86, 87,
89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105, 106,
107, 108,
109, 110, 111, 112, 113, 114, 115, 116, 117, 118, 119, 120, 121, 122, 123,
124, 125, 126,
127, 128, 129, 130, 131, 132, 133, 134, 135, 136, 137, 138, 139, 140, 150,
160, 170, 180,
190, 200, 210, 220, 230, 240, 250, 260, 270, 280, 290, 300, 320, 340, 360,
380, 400, 450,
500, 550, 600, 650, 700, 750, 800, 850, 900, 950, 1000, 1050, or 1100 amino
acids,
including ranges between any two of the listed values.
[0523]
Additionally, the peptide inhibitor used in these methods can
comprise, consist of, or consist essentially of and/or SEQ ID NOs: 1-33, 34,
46-53, 64-
66, 68, 76, 94-96, 98, 264-393, 583-586, or 589 or any one or more of the
peptides
provided in Table 5.1, 5.4, 5.5, 5.6, or any variation or combination of
variations of
P28R or P28 core as provided in Tables 5.3 and 13 as described herein. In some
embodiments, these isolated peptides used in these methods have a length that
is less
than or equal to 1100 amino acids, for example, less than or equal to 4, 5, 6,
7, 8, 9, 10,
11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29,
30, 31, 32, 33,
34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52,
53, 54, 55, 56,
57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75,
76, 77, 78, 79,
80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98,
99, 100, 101,
102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115, 116,
117, 118, 119,
120, 121, 122, 123, 124, 125, 126, 127, 128, 129, 130, 131, 132, 133, 134,
135, 136, 137,
138, 139, 140, 150, 160, 170, 180, 190, 200, 210, 220, 230, 240, 250, 260,
270, 280, 290,
300, 320, 340, 360, 380, 400, 450, 500, 550, 600, 650, 700, 750, 800, 850,
900, 950,
1000, 1050, or 1100 amino acids, including ranges between any two of the
listed values.
183
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
[0524]
Additionally, the peptide inhibitor used in these methods can
comprise, consist of, or consist essentially of a peptide inhibitor that
comprises, consists
of, or consists essentially of any one or more of the peptides set forth in
Table 5.1. In
some embodiments, the isolated peptide from Table 5.1 used in these methods
has a
length that is less than or equal to 1100 amino acids, for example, less than
or equal to 4,
5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25,
26, 27, 28, 29,
30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48,
49, 50, 51, 52,
53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71,
72, 73, 74, 75,
76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94,
95, 96, 97, 98,
99, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114,
115, 116,
117, 118, 119, 120, 121, 122, 123, 124, 125, 126, 127, 128, 129, 130, 131,
132, 133, 134,
135, 136, 137, 138, 139, 140, 150, 160, 170, 180, 190, 200, 210, 220, 230,
240, 250, 260,
270, 280, 290, 300, 320, 340, 360, 380, 400, 450, 500, 550, 600, 650, 700,
750, 800, 850,
900, 950, 1000, 1050, or 1100 amino acids, including ranges between any two of
the
listed values. Additionally, a nucleic acid encoding such a peptide inhibitor
can be
provided, for example a nucleic acid of SEQ ID NOs: 102-165.
[0525]
Preferably, the immunoregulatory peptide inhibitor used in the
aforementioned methods is P28R, P28 core, a derivative thereof, or a nucleic
acid
encoding such a molecule (e.g., any one or more of the immunoregulatory
peptide
inhibitors provided by SEQ ID NOs: 1-33, 34, 46-53, 64-66, 68, 76, 94-96, 98,
264-393,
583-586, or 589 or any one or more of the peptides provided in Table 5.1, 5.4,
5.5, 5.6,
or any variation or combination of variations of P28R or P28 core as provided
in Tables
5.3 and 13, or a nucleic acid encoding such a molecule (e.g., SEQ ID NOs: 102-
165)).
The immunoregulatory peptide inhibitors used in the aforementioned methods can
comprise at least one D amino acid, at least one non-natural amino acid, an N-
terminal
acetyl group, or a C terminal amide group and said immunoregulatory peptide
inhibitors
can be glycosylated or joined to PEG, a cytotoxin, or radionuclide.
[0526] Once the
immunoregulatory peptide inhibitor or antibody that binds
specifically to any immunoregulatory peptide of Tables 1-4 is bound to the
cancer cell, it
can be detected. That is, optionally, the method above includes a detecting
step whereby
the binding of the immunoregulatory peptide inhibitor is determined directly
or
In some embodiments, the binding of the immunoregulatory peptide inhibitor is
directly
detected as in Example 14. In some embodiments, the binding of the
immunoregulatory
184
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
peptide inhibitor is indirectly detected. As described herein, the presence of
P3028 on
cancer cells can locally suppress an immune response. Thus, in some
embodiments,
detecting the binding of an immunoregulatory peptide inhibitor to a cancer
cell can also
include a step of detecting a reversal of immunosuppression, as described in
Example
Reversal of immunosuppression can be determined, for example as a reversal of
PBMC proliferation (see Examples 2 and 13), reversal of T cell receptor
stimulation (see
Example 3), reversal of decreased NK cell cytotoxicity (see Example 4),
reversal of
decreased leukocyte spreading (see Example 5) or decreased immune cell
migration (see
Example 6), or increased IL-2 induced proliferation (see Examples 6 and 9). In
some
embodiments, cancer cells are bound to an immunoregulatory peptide inhibitor
in vivo.
Example 16 describes delivery of an inhibitor of P3028 to cancer cells in
vivo. Example
42 describes detection of an inhibitor of P3028 on cancer cells.
[0527] In some embodiments,
the detection of an immunoregulatory peptide
inhibitor can occur on tissue biopsies obtained from a human. In some
embodiments, the
tissue biopsies can include putative cancer cells, or the biopsies can be
screened for
cancer cells. By these methods, the
tissue biopsies are contacted with an
immunoregulatory peptide inhibitor, as described herein.
Preferably, the
immunoregulatory peptide inhibitor comprises a detectable label, as described
herein. In
some embodiments, live cells are contacted with the immunoregulatory peptide
inhibitor
(see Example 14). In some embodiments, histological sections are bound with
the
immunoregulatory peptide inhibitor. The detectable label is then detected,
thus
permitting identification of cancer cells which cannot be attacked by the
immune system.
The detectable label can be detected through methods known in the art, for
example by
immunoassays, a blotting technique, ELISA, ELISpot, flow cytometry, cytometric
bead
assay, proteomics, and/or immunohistochemistry.
Methods of inhibiting the proliferation of cancer cells
[0528] Some embodiments of
the invention include methods of inhibiting the
proliferation of cancer cells of two or more tumors, in which an
immunoregualtory
peptide inhibitor is neither intratumorally nor peri-tumorally administered to
at least one
of the tumors. For example, the two or more tumors can comprise tumors of a
such as a primary tumor and a remote tumore, and/or two or more primary
tumors, and/or
two or more remote tumors. The method can include identifying a human cancer
patient.
185
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
The patient can be suffering from one or more cancers, for example colorectal
cancer,
renal cancer, breast cancer, skin cancer, ovarian cancer, prostate cancer,
pancreatic
lung cancer, malignant melanoma, small cell lung cancer, non-small lung cancer
(adenocarcinoma), squamous cell carcinoma, bladder cancer, osteosarcoma,
bronchial
cancer, hematopoietic cell cancer, and/or may have a tumor, for example a
prostate
a melanoma, a colon cancer, a lung carcinoma, an Apocrine gland carcinoma, a
testis
tumor, a mast cell tumor, a mammary tumor (e.g. a benign mammary tumor or a
malignant mammary tumor, for example a mixed mammary tumor such as a benign
mixed mammary tumor or a malignant mixed mammary tumor), a mucinous carcinoma
(e.g. a mammary gland mucinous carcinoma), or a histicytoma. The method can
include
contacting immune cells of the human by an immunoregulatory peptide inhibitor.
In
some embodiments, contacting the immune cells comprises intra-tumoral
administration,
or administration near a tumor, for example within 10, 9, 8, 7, 6, 5,4, 3, 2,
1, or 0.5 cm of
the tumor. Optionally, the immunoregulatory peptide inhibitor (or a
composition
comprising the immunoregulatory peptide inhibitor immobilized on a
nanoparticle as
described herein) is administered directly to a tumor, and induces regressive
changes in
the tumor. Optionally, the immunoregulatory peptide inhibitor (or a
composition
comprising the immunoregulatory peptide inhibitor immobilized on a
nanoparticle as
described herein) is administered to a subject, and induces regressive changes
of a tumor
to which the composition is not directly administered. Optionally, the
immunoregulatory
peptide inhibitor (or a composition comprising the immunoregulatory peptide
inhibitor
immobilized on a nanoparticle as described herein)is administered directly to
a tumor,
induces regressive changes in the tumor, and further induces regressive
changes in a
second tumor to which the formulation was not directly administered (e.g. a
metastatic or
contralateral tumor).
Optionally, the immunoregulatory peptide inhibitor (or a
composition comprising the immunoregulatory peptide inhibitor immobilized on a
nanoparticle as described herein)is administered directly to a tumor, and
induces
eradication of the tumor. Optionally, the immunoregulatory peptide inhibitor
(or a
composition comprising the immunoregulatory peptide inhibitor immobilized on a
nanoparticle as described herein)is administered to a subject, and induces
eradication of a
tumor to which the composition is not directly administered.
Optionally, the
immunoregulatory peptide inhibitor (or a composition comprising the
immunoregulatory
peptide inhibitor immobilized on a nanoparticle as described herein)is
administered
186
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
directly to a tumor, and induces eradication of the tumor, and further induces
eradication
of a second tumor to which the formulation was not directly administered (e.g.
a
metastatic or contralateral tumor). Optionally, the immunoregulatory peptide
inhibitor
a composition comprising the immunoregulatory peptide inhibitor immobilized on
a
nanoparticle as described herein)is administered directly to a tumor, and
induces immune
cell infiltration of the tumor. Optionally, the immunoregulatory peptide
inhibitor (or a
composition comprising the immunoregulatory peptide inhibitor immobilized on a
nanoparticle as described herein) is administered directly to a tumor, and
induces
cell infiltration of the tumor, and further induces immune cell infiltration
of a second
tumor to which the formulation was not directly administered (e.g. a
metastatic or
contralateral tumor). Optionally, the immunoregulatory peptide inhibitor (or a
composition comprising the immunoregulatory peptide inhibitor immobilized on a
nanoparticle as described herein)is administered to a subject, and induces
immune cell
infiltration of a tumor to which the composition is not directly administered.
Example
tumors to which the pharmaceutical composition can be directly or indirect
administered
include a prostate tumor, a melanoma, a colon cancer, a lung carcinoma, an
Apocrine
gland carcinoma, a testis tumor, a mast cell tumor, a mammary tumor (e.g. a
benign
mammary tumor or a malignant mammary tumor, for example a mixed mammary tumor
such as a benign mixed mammary tumor or a malignant mixed mammary tumor), a
mucinous carcinoma (e.g. a mammary gland mucinous carcinoma), or a
histicytoma. As
shown in Figures 55-82, administration of compositions comprising
immunoregulatory
peptide inhibitors as described herein induced regressive changes, immune cell
of, and/or eradication of tumors. In some embodiments, the method of
inhibiting the
proliferation of cancer cells of two or more tumors is applied to a non-human
mammal.
[0529] In some
embodiments, the immunoregulatory peptide inhibitor
comprises, consists of or consists essentially of a peptide as described
herein. For
example, the peptide inhibitor used in these methods can comprise, consist of,
or consist
essentially of Formula (I), XX1VKX2X3X4 (SEQ ID NO: 166) as described herein.
In
some embodiments, X is an optional sequence, and can be KKLDT (SEQ ID NO:
167),
RKLDT (SEQ ID NO: 168), KKGDT (SEQ ID NO: 169), KKEDT (SEQ ID NO: 170),
KKLDQ (SEQ ID NO: 171), KKGDQ (SEQ ID NO: 252), KKEDQ (SEQ ID NO:
253), RKLDQ (SEQ ID NO: 254), RKGDQ (SEQ ID NO: 255), RKEDQ (SEQ ID NO:
256), RKGTD (SEQ ID NO: 257), RKEDT (SEQ ID NO: 258), KLDT (SEQ ID NO:
187
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
172), KGDT (SEQ ID NO: 259), KEDT (SEQ ID NO: 260), KLDQ (SEQ ID NO:
KGDQ (SEQ ID NO: 262), KEDQ (SEQ ID NO: 263), LDT, LDQ, GDT, GDQ, EDT,
EDQ, DT, DQ, T, or Q, or absent. In some embodiments, Xi is one of FF, FM, FS,
FV,
FT, FL, AF, AM, AS, AV, AT, AL, VF, VM, VS, VV, VT, or VL. In some
embodiments, X2 is one of LS, LQ, LM, LT, LH, VS, VQ, VM, VT, or VH. In some
embodiments, X3 is one of LFT, LMT, LQT, LHT, LNT, LPT, LST, LGT, LAT, LRT,
QFT, QMT, QQT, QHT, QNT, QPT, QST, QGT, QAT, QRT, VFT, VMT, VQT, VHT,
VNT, VPT, VST, VGT, VAT, VRT, MFT, MMT, MQT, MHT, MNT, MPT, MST,
MGT, MAT, MRT, LFN, LMN, LQN, LHN, LNN, LPN, LSN, LGN, LAN, LRN, QFN,
QMN, QQN, QHN, QNN, QPN, QSN, QGN, QAN, QRN, VFN, VMN, VQN, VHN,
VNN, VPN, VSN, VGN, VAN, VRN, MFN, MMN, MQN, MHN, MNN, MPN, MSN,
MGN, MAN, MRN, LFP, LMP, LQP, LHP, LNP, LPP, LSP, LGP, LAP, LRP, QFP,
QMP, QQP, QHP, QNP, QPP, QSP, QGP, QAP, QRP, VFP, VMP, VQP, VHP, VNP,
VPP, VSP, VGP, VAP, VRP, MFP, MMP, MQP, MHP, MNP, MPP, MSP, MGP, MAP,
MRPR, LFR, LMR, LQR, LHR, LNR, LPR, LSR, LGR, LAR, LRR, QFR, QMR, QQR,
QHR, QNR, QPR, QSR, QGR, QAR, QRR, VFR, VMR, VQR, VHR, VNR, VPR, VSR,
VGR, VAR, VRR, MFR, MMR, MQR, MHR, MNR, MPR, MSR, MGR, MAR, or
In some embodiments, X4 is an optional sequence, and can be ER, or E, or
absent. In
some embodiments, if X is absent, X1 is FF, and X2 is LS. In some embodiments,
the
isolated peptides that comprise Formula (I) used in these methods have a
length that is
less than or equal to 1100 amino acids, for example, less than or equal to 4,
5, 6, 7, 8, 9,
10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28,
29, 30, 31, 32,
34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52,
53, 54, 55, 56,
58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76,
77, 78, 79, 80,
82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100,
101, 102, 103,
104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115, 116, 117, 118,
119, 120, 121,
122, 123, 124, 125, 126, 127, 128, 129, 130, 131, 132, 133, 134, 135, 136,
137, 138, 139,
140, 150, 160, 170, 180, 190, 200, 210, 220, 230, 240, 250, 260, 270, 280,
290, 300, 320,
340, 360, 380, 400, 450, 500, 550, 600, 650, 700, 750, 800, 850, 900, 950,
1000, 1050,
1100 amino acids, including ranges between any two of the listed values.
[0530]
Additionally, the peptide inhibitor used in these methods can
consist of, or consist essentially of Formula (II), X20TFFVKLSX21X22 (SEQ ID
NO:
173), as described herein. In some embodiments, X20 is an optional sequence,
and can
188
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
KKLD (SEQ ID NO: 174), RKLD (SEQ ID NO: 175), KKGD (SEQ ID NO: 176),
KKED (SEQ ID NO: 177), KLD, LD, or D, or absent. X21 is an optional sequence,
and
can be LFT, LMT, LQT, LHT, LNT, LPT, LST, LGT, LAT, LRT, QFT, QMT, QQT,
QHT, QNT, QPT, QST, QGT, QAT, QRT, VFT, VMT, VQT, VHT, VNT, VPT, VST,
VGT, VAT, VRT, MFT, MMT, MQT, MHT, MNT, MPT, MST, MGT, MAT, MRT,
LFN, LMN, LQN, LHN, LNN, LPN, LSN, LGN, LAN, LRN, QFN, QMN, QQN, QHN,
QNN, QPN, QSN, QGN, QAN, QRN, VFN, VMN, VQN, VHN, VNN, VPN, VSN,
VGN, VAN, VRN, MFN, MMN, MQN, MHN, MNN, MPN, MSN, MGN, MAN, MRN,
LFP, LMP, LQP, LHP, LNP, LPP, LSP, LGP, LAP, LRP, QFP, QMP, QQP, QHP, QNP,
QPP, QSP, QGP, QAP, QRP, VFP, VMP, VQP, VHP, VNP, VPP, VSP, VGP, VAP,
VRP, MFP, MMP, MQP, MHP, MNP, MPP, MSP, MGP, MAP, MRPR, LFR, LMR,
LQR, LHR, LNR, LPR, LSR, LGR, LAR, LRR, QFR, QMR, QQR, QHR, QNR, QPR,
QSR, QGR, QAR, QRR, VFR, VMR, VQR, VHR, VNR, VPR, VSR, VGR, VAR, VRR,
MFR, MMR, MQR, MHR, MNR, MPR, MSR, MGR, MAR, or MRR, or absent. In
some embodiments, X22 is an optional sequence, and can be ER, E, or absent. In
some
embodiments, the isolated peptides that comprise Formula (II) used in these
methods
a length that is less than or equal to 1100 amino acids, for example, less
than or equal to
4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24,
25, 26, 27, 28,
30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48,
49, 50, 51, 52,
54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72,
73, 74, 75, 76,
78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96,
97, 98, 99, 100,
101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115,
116, 117, 118,
119, 120, 121, 122, 123, 124, 125, 126, 127, 128, 129, 130, 131, 132, 133,
134, 135, 136,
137, 138, 139, 140, 150, 160, 170, 180, 190, 200, 210, 220, 230, 240, 250,
260, 270, 280,
290, 300, 320, 340, 360, 380, 400, 450, 500, 550, 600, 650, 700, 750, 800,
850, 900, 950,
1000, 1050, or 1100 amino acids, including ranges between any two of the
listed values.
[0531]
Additionally, the peptide inhibitor used in these methods can
consist of, or consist essentially of Formula (III), X30X31VKLX32LX33TEX34
(SEQ ID
NO: 178). In some embodiments, X30 is an optional sequence, and can be KKLDTF
(SEQ ID NO: 179), KLDTF (SEQ ID NO: 180), LDTF (SEQ ID NO: 181), DTF, TF,
F, or absent. In some embodiments, X31 is an optional sequence, and can be F,
S, M, V,
T, or L, or absent. In some embodiments, X31 is F. In some embodiments, X32
can be S,
Q, M, T, or H. In some embodiments, X32 is S. X33 can be F, M, Q, H, N, P, S,
G, A, or
189
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
R. In some embodiments, X34 is F. X34 is an optional sequence, and can be R or
absent.
In some embodiments, the isolated peptides that comprise Formula (III) used in
these
methods have a length that is less than or equal to 1100 amino acids, for
example, less
than or equal to 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20,
21, 22, 23, 24,
25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43,
44, 45, 46, 47,
49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67,
68, 69, 70, 71,
73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91,
92, 93, 94, 95,
97, 98, 99, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112,
113, 114,
115, 116, 117, 118, 119, 120, 121, 122, 123, 124, 125, 126, 127, 128, 129,
130, 131, 132,
133, 134, 135, 136, 137, 138, 139, 140, 150, 160, 170, 180, 190, 200, 210,
220, 230, 240,
250, 260, 270, 280, 290, 300, 320, 340, 360, 380, 400, 450, 500, 550, 600,
650, 700, 750,
800, 850, 900, 950, 1000, 1050, or 1100 amino acids, including ranges between
any two
of the listed values.
[0532]
Additionally, the peptide inhibitor used in these methods can
consist of, or consist essentially of Formula (VII), X700K X701X702X703
X704X705X706K
X707 X708 X700 X710 X711E X712 (SEQ ID NO: 394), as described herein. In some
embodiments, X700 is an optional sequence, and can be
K,A,D,E,G,H,I,L,M,N,P,Q,R,T,
V, or absent. In some embodiments, X701 is an optional sequence, and can be
L,A,C,D,E,F,G,H,I,K,M,N,Q,R,S,T, or V, or absent. In some embodiments, X702 is
an
optional sequence, and can be D,A,E,I,V,W, or Y, or absent. In some
embodiments,
is an optional sequence, and can be T,C,M,N,P,Q,R,S,W, or Y, or absent. In
some
embodiments, X704 is an optional sequence, and can be F,A,I,M,N,P,T, or V, or
absent.
some embodiments, X705 is an optional sequence, and can be F,L,M,Q,S,T, or V,
or
absent. In some embodiments, X706 is an optional sequence, and can be
V,F,G,L,P, or R,
or absent. In some embodiments, X707 is an optional sequence, and can be
L,A,F,G,I,M,N,P,Q,R,S,T,V, or Y, or absent. In some embodiments, X708 is an
optional
sequence, and can be S,H,M,N,Q, or T, or absent. In some embodiments, X700 is
an
optional sequence, and can be L,A,H,I,M,N,Q,R,S,T,V, or W, or absent. In some
embodiments, X710 is an optional sequence, and can be
F,A,C,G,H,I,L,M,N,P,Q,R,S,T,V,
or W, or absent. In some embodiments, X711 is an optional sequence, and can be
T,F,G,H,I,L,M,N,P,S,V, or W, or absent. In some embodiments, X712 is an
optional
sequence, and can be R,F,K,N,R,T, or Y, or absent. In some embodiments, the
isolated
peptide comprising Formula (VII) has a length that is less than or equal to
1100 amino
190
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
acids, for example, less than or equal to 4, 5, 6, 7, 8, 9, 10, 11, 12, 13,
14, 15, 16, 17, 18,
19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37,
38, 39, 40, 41,
43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61,
62, 63, 64, 65,
67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85,
86, 87, 88, 89,
91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105, 106, 107,
108, 109, 110,
111, 112, 113, 114, 115, 116, 117, 118, 119, 120, 121, 122, 123, 124, 125,
126, 127, 128,
129, 130, 131, 132, 133, 134, 135, 136, 137, 138, 139, 140, 150, 160, 170,
180, 190, 200,
210, 220, 230, 240, 250, 260, 270, 280, 290, 300, 320, 340, 360, 380, 400,
450, 500, 550,
600, 650, 700, 750, 800, 850, 900, 950, 1000, 1050, or 1100 amino acids,
including
ranges between any two of the listed values.
[0533]
Additionally, the peptide inhibitor used in these methods can
consist of, or consist essentially of Formula (VIII), X800K X801K X802E X803
(SEQ ID
NO: 395), as described herein. In some embodiments, X800 is an optional
sequence, and
can be K, A, D, E, G, H, I, L, M, N, P, Q, R, T, V, or K, or absent. In some
X801 is an optional sequence, and can be LDTFFV, GDTH-V, EDTH-V, LDQFFV,
LDTAFV, LDTVFV, LDTFMV, LDTFSV, LDTFVV, LDTFTV, LDTFLV, LDGFFV,
LDTFGV, LDTFFK, ADTFFV, CDTFFV, DDTFFV, FDTFFV, HDTFFV, IDTFFV,
KDTH-V, MDTFFV, NDTFFV, QDTFFV, RDTFFV, SDTFFV, TDTFFV, VDTFFV,
LATFFV, LETH-V, LITFFV, LVTFFV, LWTFFV, LYTH-V, LDCFFV, LDMFFV,
LDNIA-V, LDPFFV, LDRIA-V, LDSFFV, LDWFFV, LDYFFV, LDTIFV, LDTMFV,
LDTNFV, LDTPFV, LDTTFV, LDTFQV, LDTH-F, LDTFFG, LDTFFL, LDTFFP,
LDTFFR, LDTFIV, LDTSFV, LDTFAV, LDTFCV, LDTQFV, LDTLFV, LTTFFV,
LDTFFI, LDHFFV, LMTFFV, LDTFEV, LDTFWV, LFTFFV, LDVFFV, LDTI-RV,
LDTFHV, LDTYFV, LPTH-V, PDTFFV, LDTFPV, LDTFNV, LDTWFV, LDTGFV,
LDAFFV, LQTFFV, LCTFFV, LSTFFV, YDTFFV, LDEFFV, WDTFFV, LDTKFV,
LDTCFV, LDTFYV, LDTHFV, LHTFFV, LRTFFV, LDLFFV, LDTRFV, LLTFFV,
LDTFDV, LDTH-A, LDTFFT, LNTFFV, LDDIA-V, LDIFFV, LDFFFV, LKTFFV,
LDTFFQ, LGTH-V, LDTH-C, LDKFFV, LDTFKV, LDTEFV, LDTFFW, LDTFFM,
LDTFFS, LDTFFH, LDTFFY, LDTH-N, LDTDFV, LDTFFE, LDTFFD, LTFFV,
LDTFF, TFFV, LDF, LDTE, FFV, LDV, LV, or L, or absent. In some embodiments,
is an optional sequence, and can be LSLFT, VSLFT, LQLFT, LMLFT, LTLFT, LHLFT,
LSQFT, LSVFT, LSMFT, LSLMT, LSLQT, LSLHT, LSLNT, LSLPT, LSLST, LSLGT,
LSLAT, LSLRT, LSLFN, LSLFP, LSLFR, LGLFT, ASLFT, FSLFT, GSLFT, ISLFT,
191
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
MSLFT, NSLFT, PSLFT, QSLFT, RSLFT, SSLFT, TSLFT, YSLFT, LNLFT, LSAFT,
LSHFT, LSIFT, LSNFT, LSRFT, LSSFT, LSTFT, LSWFT, LSLCT, LSLIT, LSLLT,
LSLTT, LSLVT, LSLWT, LSLFF, LSLFG, LSLFH, LSLFI, LSLFL, LSLFM, LSLFS,
LSLFV, LSLFW, LYLFT, LVLFT, LSFFT, LSGFT, LSKFT, LSCFT, LCLFT, LRLFT,
LPLFT, LWLFT, LKLFT, LDLFT, LSYFT, LALFT, WSLFT, LSLFA, LSLFQ, LSPFT,
HSLFT, LSLYT, LILFT, KSLFT, CSLFT, LSLFY, LSLFK, LSLFC, LFLFT, LELFT,
LSLKT, LLLFT, LSLFD, LSLDT, LSLFE, DSLFT, LSLET, LSDFT, LSEFT, ESLFT,
SLFT, LSFT, LFT, LSL, LT, or T, or absent. In some embodiments, X803 is an
optional
sequence, and can be R, F, K, N, R, T, or Y, or absent. In some embodiments,
the
isolated peptide comprising Formula (VIII) has a length that is less than or
equal to 1100
amino acids, for example, less than or equal to 4, 5, 6, 7, 8, 9, 10, 11, 12,
13, 14, 15, 16,
17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35,
36, 37, 38, 39,
41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59,
60, 61, 62, 63,
65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83,
84, 85, 86, 87,
89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104, 105, 106,
107, 108,
109, 110, 111, 112, 113, 114, 115, 116, 117, 118, 119, 120, 121, 122, 123,
124, 125, 126,
127, 128, 129, 130, 131, 132, 133, 134, 135, 136, 137, 138, 139, 140, 150,
160, 170, 180,
190, 200, 210, 220, 230, 240, 250, 260, 270, 280, 290, 300, 320, 340, 360,
380, 400, 450,
500, 550, 600, 650, 700, 750, 800, 850, 900, 950, 1000, 1050, or 1100 amino
acids,
including ranges between any two of the listed values.
[0534]
Additionally, the peptide inhibitor used in these methods can
consist of, or consist essentially of Formula(IX). Accordingly, in some
embodiments,
peptide inhibitor comprises a peptide of Formula
(IX):
X901X902X903X904X905X906X907X908X909X910X911X912X913X914X915X916X917, wherein
X901
is any amino acid or absent; X902 is a positively charged amino acid, F, or N;
X903 is
any amino acid; X904 is any amino acid; X905 is a polar uncharged amino acid,
R, Y, or
X906 is a hydrophobic or uncharged polar amino acid; X907 is a hydrophobic or
uncharged
polar amino acid; X908 is a hydrophobic, non-aromatic carbon chain amino acid
that is
M or F; X909 is a positively charged amino acid, T, Q, or Y; X910 is any amino
acid that is
not negatively charged; X911 is a polar uncharged amino acid or H; X912 is any
amino
that is not negatively charged; X913 is any amino acid that is not negatively
charged; X914
is any amino acid that is not negatively charged; X915 is a negatively charged
amino acid,
Y, or Q; X916 is any amino acid that is not negatively charged; and X917 is
one or more
192
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
positively charged amino acids or is absent. Optionally, X901 comprises a
positively
charged amino acid. Optionally X901 is an R or K. Optionally X917 is RR. In
some
embodiments, the isolated peptide comprising Formula (IX) has a length that is
less than
or equal to 1100 amino acids, for example, less than or equal to 4, 5, 6, 7,
8, 9, 10, 11,
13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31,
32, 33, 34, 35,
37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55,
56, 57, 58, 59,
61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79,
80, 81, 82, 83,
85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102,
103, 104, 105,
106, 107, 108, 109, 110, 111, 112, 113, 114, 115, 116, 117, 118, 119, 120,
121, 122, 123,
124, 125, 126, 127, 128, 129, 130, 131, 132, 133, 134, 135, 136, 137, 138,
139, 140, 150,
160, 170, 180, 190, 200, 210, 220, 230, 240, 250, 260, 270, 280, 290, 300,
320, 340, 360,
380, 400, 450, 500, 550, 600, 650, 700, 750, 800, 850, 900, 950, 1000, 1050,
or 1100
amino acids, including ranges between any two of the listed values.
[0535]
Additionally, the peptide inhibitor used in these methods can
comprise, consist of, or consist essentially of and/or SEQ ID NOs: 1-33, 34,
46-53, 64-
66, 68, 76, 94-96, 98, 264-393, 583-586, or 589 or any one or more of the
peptides
provided in Table 5.1, 5.4, 5.5, 5.6, or any variation or combination of
variations of
P28R or P28 core as provided in Tables 5.3 and 13 as described herein. In some
embodiments, these isolated peptides have a length that is less than or equal
to 1100
amino acids, for example, less than or equal to 4, 5, 6, 7, 8, 9, 10, 11, 12,
13, 14, 15, 16,
17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35,
36, 37, 38, 39,
40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58,
59, 60, 61, 62,
63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81,
82, 83, 84, 85,
86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103,
104, 105, 106,
107, 108, 109, 110, 111, 112, 113, 114, 115, 116, 117, 118, 119, 120, 121,
122, 123, 124,
125, 126, 127, 128, 129, 130, 131, 132, 133, 134, 135, 136, 137, 138, 139,
140, 150, 160,
170, 180, 190, 200, 210, 220, 230, 240, 250, 260, 270, 280, 290, 300, 320,
340, 360, 380,
400, 450, 500, 550, 600, 650, 700, 750, 800, 850, 900, 950, 1000, 1050, or
1100 amino
acids, including ranges between any two of the listed values.
[0536]
Additionally, the peptide inhibitor used in these methods can
consist of, or consist essentially of a peptide inhibitor that comprises,
consists of, or
consists essentially of any one or more of the peptides set forth in Table
5.1, 5.4, 5.5,
or any variation or combination of variations of P28R or P28 core as provided
in Tables
193
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
5.3 and 13. In some embodiments, the isolated peptide from Table 5.1 used in
these
methods has a length that is less than or equal to 1100 amino acids, for
example, less
or equal to 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21,
22, 23, 24, 25,
27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45,
46, 47, 48, 49,
51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69,
70, 71, 72, 73,
75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93,
94, 95, 96, 97,
99, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114,
115, 116,
117, 118, 119, 120, 121, 122, 123, 124, 125, 126, 127, 128, 129, 130, 131,
132, 133, 134,
135, 136, 137, 138, 139, 140, 150, 160, 170, 180, 190, 200, 210, 220, 230,
240, 250, 260,
270, 280, 290, 300, 320, 340, 360, 380, 400, 450, 500, 550, 600, 650, 700,
750, 800, 850,
900, 950, 1000, 1050, or 1100 amino acids, including ranges between any two of
the
listed values.
[0537] In some
embodiments, the method includes providing to the human a
polynucleotide encoding such a peptide inhibitor (e.g., any one or more of the
peptides
provided in Table 5.1, 5.4, 5.5, 5.6, or any variation or combination of
variations of
P28R or P28 core as provided in Tables 5.3 and 13). For example, a
polynucleotide
encoding such a peptide inhibitor can be provided, for example a nucleic acid
of SEQ ID
NOs: 102-165.
[0538]
Reduction of cancer-associated immunosuppression can induce and/or
enhance an immune response against cancer cells. An immune response against
cancer
cells can reduce cancer cell proliferation, and/or cause cancer cells to
undergo cell death
or apoptosis. Thus, the method can include detecting an inhibition in the
proliferation of
cancer cells of the patient. The method can include detecting an induction of
cell death
or apoptosis of cancer cells of the patient. The method can include detecting
an
inhibition in the proliferation of cancer cells of the patient, and an
induction of cell death
or apoptosis of cancer cells of the patient. Apoptosis can be identified as
known in the
art, for example by neutral red assay, by trypan blue exclusion of dead cells,
by acridine
orange staining, by TUNEL staining, and/or by detection of cleaved PARP,
and/or
cleaved caspases.
Methods of identifying a patient in need
[0539] It is
contemplated herein that different populations of patients can
different albumin-derived immunoregulatory peptides, and that a given albumin-
derived
194
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
immunoregulatory peptide can have different effects in different individual
patients. As
shown in Example 30, some cancer patients have immune cells with a high
proliferative
response to IL-2, while other cancer patients have immune cells with a low
proliferative
response to IL-2. As shown in Examples 31 and 32, different populations of
patients can
respond differently to the same inhibitor of immunoregulatory peptides.
Additionally, a
given inhibitor can modulate the immune system in some patients, but not in
other
patients. Thus, some embodiments of the invention include methods of
identifying a
patient in need. A patient in need can include a patient having albumin-
derived
immunoregulatory peptides bound to at least some of his or her immune cells. A
patient
in need can include a patient that is likely to respond to an inhibitor of an
immunoregulatory peptide. In some embodiments, immune cells of a patient can
be
isolated. The presence of immunoregulatory structures on the immune cells can
be
detected. The effect of an inhibitor of an immunoregulatory peptide on the
immune cells
can be detected. If an immunoregulatory structure is present and/or if immune
cell
function is modulated by the inhibitor, the patient can be classified as a
patient in need.
Optionally, an effective dose of the inhibitor can be determined. A
therapeutically
effective dose of the inhibitor can be administered to the patient in need.
[0540] Some
embodiments include methods of detecting the presence of
immunoregulatory peptides in an in vitro assay. In vitro methods of detecting
the
presence of albumin-derived immunoregulatory peptides bound to immune cells,
immunoregulatory sequences and structures, and in vitro methods of detecting
the effects
of albumin-derived immunoregulatory peptides on immune cell activity are
provided in
U.S. Pat. No. 8,182,983, hereby expressly incorporated by reference in its
entirety herein;
U.S. Pat. No. 7,960,126, hereby expressly incorporated by reference in its
entirety herein;
U.S. Pat. No. 8,133,688 hereby expressly incorporated by reference in its
entirety herein;
U.S. Pat. No. 8,110,347, hereby expressly incorporated by reference in its
entirety herein;
and U.S. Pub. No. 2011/0262470, hereby expressly incorporated by reference in
its
entirety herein.
[0541] Some
embodiments include detecting the response of inhibited
immune cells to an inhibitor of immunoregulatory peptides. In some
embodiments,
immune cells are isolated from a patient. In some embodiments, the immune
cells
PBMCs. In some embodiments, the immune cells are contacted with an inhibitor
of
immunoregulatory peptides.
195
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
[0542] In some
embodiments, the immune cells are contacted with an
inhibitor that comprises a peptide comprising, consisting of or consisting
essentially of at
least one peptide of SEQ ID NOs: 1-33, 34, 46-53, 64-66, 68, 76, 94-96, 98,
264-393,
583-586, or 589 or any one or more of the peptides provided in Table 5.1, 5.4,
5.5, 5.6,
6.1, 6.2, or 12 or any variation or combination of variations of P28R or P28
core as
provided in Tables 5.3 and 13.
[0543] In some
embodiments, the immune cells are contacted with an
that comprises a peptide comprising, consisting of or consisting essentially
of Formula
XX1VKX2X3X4 (SEQ ID NO: 166). In some embodiments, X is an optional sequence,
and can be KKLDT (SEQ ID NO: 167), RKLDT (SEQ ID NO: 168), KKGDT (SEQ ID
NO: 169), KKEDT (SEQ ID NO: 170), KKLDQ (SEQ ID NO: 171), KKGDQ (SEQ
NO: 252), KKEDQ (SEQ ID NO: 253), RKLDQ (SEQ ID NO: 254), RKGDQ (SEQ
NO: 255), RKEDQ (SEQ ID NO: 256), RKGTD (SEQ ID NO: 257), RKEDT (SEQ ID
NO: 258), KLDT (SEQ ID NO: 172), KGDT (SEQ ID NO: 259), KEDT (SEQ ID NO:
260), KLDQ (SEQ ID NO: 261), KGDQ (SEQ ID NO: 262), KEDQ (SEQ ID NO:
263), LDT, LDQ, GDT, GDQ, EDT, EDQ, DT, DQ, T, or Q, or absent. In some
embodiments, X1 is one of FF, FM, FS, FV, FT, FL, AF, AM, AS, AV, AT, AL, VF,
VM, VS, VV, VT, or VL. In some embodiments, X2 can be one of LS, LQ, LM, LT,
VS, VQ, VM, VT, or VH. In some embodiments, X3 can be one of LFT, LMT, LQT,
LHT, LNT, LPT, LST, LGT, LAT, LRT, QFT, QMT, QQT, QHT, QNT, QPT, QST,
QGT, QAT, QRT, VFT, VMT, VQT, VHT, VNT, VPT, VST, VGT, VAT, VRT, MFT,
MMT, MQT, MHT, MNT, MPT, MST, MGT, MAT, MRT, LFN, LMN, LQN, LHN,
LNN, LPN, LSN, LGN, LAN, LRN, QFN, QMN, QQN, QHN, QNN, QPN, QSN, QGN,
QAN, QRN, VFN, VMN, VQN, VHN, VNN, VPN, VSN, VGN, VAN, VRN, MFN,
MMN, MQN, MHN, MNN, MPN, MSN, MGN, MAN, MRN, LFP, LMP, LQP, LHP,
LNP, LPP, LSP, LGP, LAP, LRP, QFP, QMP, QQP, QHP, QNP, QPP, QSP, QGP, QAP,
QRP, VFP, VMP, VQP, VHP, VNP, VPP, VSP, VGP, VAP, VRP, MFP, MMP, MQP,
MHP, MNP, MPP, MSP, MGP, MAP, MRPR, LFR, LMR, LQR, LHR, LNR, LPR, LSR,
LGR, LAR, LRR, QFR, QMR, QQR, QHR, QNR, QPR, QSR, QGR, QAR, QRR, VFR,
VMR, VQR, VHR, VNR, VPR, VSR, VGR, VAR, VRR, MFR, MMR, MQR, MHR,
MNR, MPR, MSR, MGR, MAR, or MRR. In some embodiments, X4 is an optional
sequence, and can be ER, or E, or absent. In some embodiments, if X is absent,
X1 is
and X2 is LS. In some embodiments, the peptide comprises one of SEQ ID NOs: 2-
33.
196
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
In some embodiments, the isolated peptides that comprise Formula (I) have a
length that
is less than or equal to 1100 amino acids, for example, less than or equal to
4, 5, 6, 7, 8,
10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28,
29, 30, 31, 32,
34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52,
53, 54, 55, 56,
58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76,
77, 78, 79, 80,
82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100,
101, 102, 103,
104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115, 116, 117, 118,
119, 120, 121,
122, 123, 124, 125, 126, 127, 128, 129, 130, 131, 132, 133, 134, 135, 136,
137, 138, 139,
140, 150, 160, 170, 180, 190, 200, 210, 220, 230, 240, 250, 260, 270, 280,
290, 300, 320,
340, 360, 380, 400, 450, 500, 550, 600, 650, 700, 750, 800, 850, 900, 950,
1000, 1050,
1100 amino acids, including ranges between any two of the listed values.
[0544] In some
embodiments, the immune cells are contacted with an
that comprises a peptide comprising, consisting of or consisting essentially
of Formula
(II), X20TFFVKLSX21X22 (SEQ ID NO: 173). In some embodiments, X20 is an
optional
sequence, and can be KKLD (SEQ ID NO: 174), RKLD (SEQ ID NO: 175), KKGD
(SEQ ID NO: 176), KKED (SEQ ID NO: 177), KLD, LD, or D, or absent. X21 is an
optional sequence, and can be LFT, LMT, LQT, LHT, LNT, LPT, LST, LGT, LAT,
QFT, QMT, QQT, QHT, QNT, QPT, QST, QGT, QAT, QRT, VFT, VMT, VQT, VHT,
VNT, VPT, VST, VGT, VAT, VRT, MFT, MMT, MQT, MHT, MNT, MPT, MST,
MGT, MAT, MRT, LFN, LMN, LQN, LHN, LNN, LPN, LSN, LGN, LAN, LRN, QFN,
QMN, QQN, QHN, QNN, QPN, QSN, QGN, QAN, QRN, VFN, VMN, VQN, VHN,
VNN, VPN, VSN, VGN, VAN, VRN, MFN, MMN, MQN, MHN, MNN, MPN, MSN,
MGN, MAN, MRN, LFP, LMP, LQP, LHP, LNP, LPP, LSP, LGP, LAP, LRP, QFP,
QMP, QQP, QHP, QNP, QPP, QSP, QGP, QAP, QRP, VFP, VMP, VQP, VHP, VNP,
VPP, VSP, VGP, VAP, VRP, MFP, MMP, MQP, MHP, MNP, MPP, MSP, MGP, MAP,
MRPR, LFR, LMR, LQR, LHR, LNR, LPR, LSR, LGR, LAR, LRR, QFR, QMR, QQR,
QHR, QNR, QPR, QSR, QGR, QAR, QRR, VFR, VMR, VQR, VHR, VNR, VPR, VSR,
VGR, VAR, VRR, MFR, MMR, MQR, MHR, MNR, MPR, MSR, MGR, MAR, or
or absent. In some embodiments, X22 is an optional sequence, and can be ER, or
E, or
absent. In some embodiments, the isolated peptides that comprise Formula (II)
have a
length that is less than or equal to 1100 amino acids, for example, less than
or equal to 4,
5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25,
26, 27, 28, 29,
30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48,
49, 50, 51, 52,
197
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72,
73, 74, 75, 76,
78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96,
97, 98, 99, 100,
101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115,
116, 117, 118,
119, 120, 121, 122, 123, 124, 125, 126, 127, 128, 129, 130, 131, 132, 133,
134, 135, 136,
137, 138, 139, 140, 150, 160, 170, 180, 190, 200, 210, 220, 230, 240, 250,
260, 270, 280,
290, 300, 320, 340, 360, 380, 400, 450, 500, 550, 600, 650, 700, 750, 800,
850, 900, 950,
1000, 1050, or 1100 amino acids, including ranges between any two of the
listed values.
[0545] In some
embodiments, the immune cells are contacted with an
inhibitor that comprises a peptide comprising, consisting of or consisting
essentially of
Formula (III), X30X31VKLX32LX33TEX34 (SEQ ID NO: 178). In some embodiments,
X30 is an optional sequence, and can be KKLDTF (SEQ ID NO: 179), KLDTF (SEQ ID
NO: 180), LDTF (SEQ ID NO: 181), DTF, TF, or F, or absent. In some
embodiments,
X31 is an optional sequence, and can be F, S, M, V, T, or L, or absent. In
some
embodiments, X31 is F. In some embodiments, X32 can be S, Q, M, T, or H. In
some
embodiments, X32 is S. X33 can be F, M, Q, H, N, P, S, G, A, or R. In some
embodiments, X34 is F. X34 is an optional sequence, and can be R, or absent.
In some
embodiments, the isolated peptides that comprise Formula (III) used in these
methods
have a length that is less than or equal to 1100 amino acids, for example,
less than or
equal to 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22,
23, 24, 25, 26,
27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45,
46, 47, 48, 49,
50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68,
69, 70, 71, 72,
73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91,
92, 93, 94, 95,
96, 97, 98, 99, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111,
112, 113,
114, 115, 116, 117, 118, 119, 120, 121, 122, 123, 124, 125, 126, 127, 128,
129, 130, 131,
132, 133, 134, 135, 136, 137, 138, 139, 140, 150, 160, 170, 180, 190, 200,
210, 220, 230,
240, 250, 260, 270, 280, 290, 300, 320, 340, 360, 380, 400, 450, 500, 550,
600, 650, 700,
750, 800, 850, 900, 950, 1000, 1050, or 1100 amino acids, including ranges
between any
two of te546led vailesome embodiments, the immune cells are contacted with an
that comprises a peptide comprising, consisting of or consisting essentially
of Formula
X7001( X701X702X703 X704X705X706K X707 X708 X709 X710 X711E X712 (SEQ ID NO:
394), as described herein. In some embodiments, X700 is an optional sequence,
and can
K,A,D,E,G,H,I,L,M,N,P,Q,R,T, or V, or absent. In some embodiments, X701 is an
optional sequence, and can be L,A,C,D,E,F,G,H,I,K,M,N,Q,R,S,T, or V, or
absent. In
198
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
some embodiments, X702 is an optional sequence, and can be D,A,E,I,V,W, or Y,
or
absent. In some
embodiments, X703 is an optional sequence, and can be
T,C,M,N,P,Q,R,S,W, or Y, or absent. In some embodiments, X704 is an optional
sequence, and can be F,A,I,M,N,P,T, or V, or absent. In some embodiments, X705
is an
optional sequence, and can be F,L,M,Q,S,T, or V, or absent. In some
embodiments, X706
is an optional sequence, and can be V,F,G,L,P, or R, or absent. In some
embodiments,
X707 is an optional sequence, and can be L,A,F,G,I,M,N,P,Q,R,S,T,V, or Y, or
absent. In
some embodiments, X708 is an optional sequence, and can be S,H,M,N,Q, or T, or
absent.
In some embodiments, X709 is an optional sequence, and can be
L,A,H,I,M,N,Q,R,S,T,V,
or W, or absent. In some embodiments, X710 is an optional sequence, and can be
F,A,C,G,H,I,L,M,N,P,Q,R,S,T,V, or W, or absent. In some embodiments, X711 is
an
optional sequence, and can be T,F,G,H,I,L,M,N,P,S,V, or W, or absent. In some
embodiments, X712 is an optional sequence, and can be R,F,K,N,R,T, or Y, or
absent. In
some embodiments, the isolated peptide comprising Formula (VII) has a length
that is
than or equal to 1100 amino acids, for example, less than or equal to 4, 5, 6,
7, 8, 9, 10,
11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29,
30, 31, 32, 33,
35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53,
54, 55, 56, 57,
59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77,
78, 79, 80, 81,
83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101,
102, 103, 104,
105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115, 116, 117, 118, 119,
120, 121, 122,
123, 124, 125, 126, 127, 128, 129, 130, 131, 132, 133, 134, 135, 136, 137,
138, 139, 140,
150, 160, 170, 180, 190, 200, 210, 220, 230, 240, 250, 260, 270, 280, 290,
300, 320, 340,
360, 380, 400, 450, 500, 550, 600, 650, 700, 750, 800, 850, 900, 950, 1000,
1050, or
amino acids, including ranges between any two of the listed values.
[0547] In some
embodiments, the immune cells are contacted with an
that comprises a peptide comprising, consisting of or consisting essentially
of Formula
(VIII), X800K X801K X802E X803 (SEQ ID NO: 395), as described herein. In some
embodiments, X800 is an optional sequence, and can be K, A, D, E, G, H, I, L,
M, N, P,
R, T, V, or K, or absent. In some embodiments, X801 is an optional sequence,
and can be
LDTFFV, GDTFFV, EDTFFV, LDQ141-V, LDTAFV, LDTVFV, LDTFMV, LDTFSV,
LDTFVV, LDTFTV, LDTFLV, LDGFFV, LDTFGV, LDT141-K, ADTFFV, CDTFFV,
DDTFFV, FDTFFV, HDTFFV, IDTFFV, KDTFFV, MDTFFV, NDTFFV, QDTFFV,
RDTFFV, SDTFFV, TDTFFV, VDTFFV, LATFFV, LETFFV, LITFFV, LVTFFV,
199
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
LWTFFV, LYTFFV, LDCFFV, LDMFFV, LDNFFV, LDPFFV, LDRFFV, LDSFFV,
LDWFFV, LDYFFV, LDTIFV, LDTMFV, LDTNFV, LDTPFV, LDTTFV, LDTFQV,
LDTFFF, LDTFFG, LDTFFL, LDTFFP, LDTFFR, LDTFIV, LDTSFV, LDTFAV,
LDTFCV, LDTQFV, LDTLFV, LTTFFV, LDT1-1-4, LDHFFV, LMTFFV, LDTFEV,
LDTFWV, LFTFFV, LDVFFV, LDTFRV, LDTFHV, LDTYFV, LPTFFV, PDTFFV,
LDTFPV, LDTFNV, LDTWFV, LDTGFV, LDAFFV, LQTFFV, LCTFFV, LSTFFV,
YDTFFV, LDEFFV, WDTFFV, LDTKFV, LDTCFV, LDTFYV, LDTHFV, LHTFFV,
LRTFFV, LDLFFV, LDTRFV, LLTFFV, LDTPDV, LDTFFA, LDTFFT, LNTFFV,
LDFFFV, LKTFFV, LDTFFQ, LGTFFV, LDTFFC, LDKFFV,
LDTFKV, LDTEFV, LDTFFW, LDTFFM, LDTFFS, LDTFFH, LDTFFY, LDTFFN,
LDTDFV, LDTFFE, LDTH-D, LTFFV, LDTFF, TFFV, LDF, LDTE, FFV, LDV, LV, or
L, or absent. In some embodiments, X802 is an optional sequence, and can be
LSLFT,
VSLFT, LQLFT, LMLFT, LTLFT, LHLFT, LSQFT, LSVFT, LSMFT, LSLMT, LSLQT,
LSLHT, LSLNT, LSLPT, LSLST, LSLGT, LSLAT, LSLRT, LSLFN, LSLFP, LSLFR,
LGLFT, ASLFT, FSLFT, GSLFT, ISLFT, MSLFT, NSLFT, PSLFT, QSLFT, RSLFT,
SSLFT, TSLFT, YSLFT, LNLFT, LSAFT, LSHFT, LSIFT, LSNFT, LSRFT, LSSFT,
LSTFT, LSWFT, LSLCT, LSLIT, LSLLT, LSLTT, LSLVT, LSLWT, LSLFF, LSLFG,
LSLFH, LSLFI, LSLFL, LSLFM, LSLFS, LSLFV, LSLFW, LYLFT, LVLFT, LSFFT,
LSGFT, LSKFT, LSCFT, LCLFT, LRLFT, LPLFT, LWLFT, LKLFT, LDLFT, LSYFT,
LALFT, WSLFT, LSLFA, LSLFQ, LSPFT, HSLFT, LSLYT, LILFT, KSLFT, CSLFT,
LSLFY, LSLFK, LSLFC, LFLFT, LELFT, LSLKT, LLLFT, LSLFD, LSLDT, LSLFE,
DSLFT, LSLET, LSDFT, LSEFT, ESLFT, SLFT, LSFT, LFT, LSL, LT, T, or absent. In
some embodiments, X803 is an optional sequence, and can be R, F, K, N, R, T,
or Y, or
absent. In some embodiments, the isolated peptide comprising Formula (VIII)
has a
length that is less than or equal to 1100 amino acids, for example, less than
or equal to 4,
5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25,
26, 27, 28, 29,
30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48,
49, 50, 51, 52,
54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72,
73, 74, 75, 76,
78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96,
97, 98, 99, 100,
101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115,
116, 117, 118,
119, 120, 121, 122, 123, 124, 125, 126, 127, 128, 129, 130, 131, 132, 133,
134, 135, 136,
137, 138, 139, 140, 150, 160, 170, 180, 190, 200, 210, 220, 230, 240, 250,
260, 270, 280,
200
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
290, 300, 320, 340, 360, 380, 400, 450, 500, 550, 600, 650, 700, 750, 800,
850, 900, 950,
1000, 1050, or 1100 amino acids, including ranges between any two of the
listed values.
[0548] In some
embodiments, the immune cells are contacted with an
inhibitor that comprises a peptide comprising, consisting of or consisting
essentially of
Formula (IX). Accordingly, in some embodiments, the peptide inhibitor
comprises a
peptide of Formula (IX):
X901X902X903X904X905X906X907X908X909X910X911X912X913X914X915X916X917, wherein
X901
is any amino acid or absent; X902 is a positively charged amino acid, F, or N;
X903 is
any amino acid; X904 is any amino acid; X905 is a polar uncharged amino acid,
R, Y, or
W; X906 is a hydrophobic or uncharged polar amino acid; X907 is a hydrophobic
or
uncharged polar amino acid; X908 is a hydrophobic, non-aromatic carbon chain
amino
acid that is not M or F; X909 is a positively charged amino acid, T, Q, or Y;
X910 is any
amino acid that is not negatively charged; X911 is a polar uncharged amino
acid or H;
X912 is any amino acid that is not negatively charged; X913 is any amino acid
that is not
negatively charged; X914 is any amino acid that is not negatively charged;
X915 is a
negatively charged amino acid, Y, or Q; X916 is any amino acid that is not
negatively
charged; and X917 is one or more positively charged amino acids or is absent.
Optionally,
X901 comprises a positively charged amino acid. Optionally X901 is an R or K.
Optionally X917 is RR. In some embodiments, the isolated peptide comprising
Formula
(IX) has a length that is less than or equal to 1100 amino acids, for example,
less than or
equal to 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22,
23, 24, 25, 26,
27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45,
46, 47, 48, 49,
50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68,
69, 70, 71, 72,
73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91,
92, 93, 94, 95,
96, 97, 98, 99, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111,
112, 113,
114, 115, 116, 117, 118, 119, 120, 121, 122, 123, 124, 125, 126, 127, 128,
129, 130, 131,
132, 133, 134, 135, 136, 137, 138, 139, 140, 150, 160, 170, 180, 190, 200,
210, 220, 230,
240, 250, 260, 270, 280, 290, 300, 320, 340, 360, 380, 400, 450, 500, 550,
600, 650, 700,
750, 800, 850, 900, 950, 1000, 1050, or 1100 amino acids, including ranges
between any
two of 1054191ed vdkesome embodiments, the immune cells are contacted with an
that comprises, consists of, or consists essentially of a peptide inhibitor
that comprises,
consists of, or consists essentially of any one or more of the peptides set
forth in Table
5.1. In some embodiments, the isolated peptide from Table 5.1 used in these
methods
201
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
a length that is less than or equal to 1100 amino acids, for example, less
than or equal to
4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24,
25, 26, 27, 28,
30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48,
49, 50, 51, 52,
54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72,
73, 74, 75, 76,
78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96,
97, 98, 99, 100,
101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114, 115,
116, 117, 118,
119, 120, 121, 122, 123, 124, 125, 126, 127, 128, 129, 130, 131, 132, 133,
134, 135, 136,
137, 138, 139, 140, 150, 160, 170, 180, 190, 200, 210, 220, 230, 240, 250,
260, 270, 280,
290, 300, 320, 340, 360, 380, 400, 450, 500, 550, 600, 650, 700, 750, 800,
850, 900, 950,
1000, 1050, or 1100 amino acids, including ranges between any two of the
listed values.
In some embodiments, the response of the immune cells is detected. In some
embodiments, the response to IL-2 stimulation is detected (see Example 2). In
some
embodiments, T cell stimulation is detected (see Example 3). In some
embodiments,
NK-Cell cytotoxicity is assayed (see Example 4). In some embodiments,
leukocyte
spreading is detected (see Example 5). In some embodiments, unblocking of the
LFA-1
receptor is detected (see Example 6). In some embodiments, binding of P28R to
the
tumor can be demonstrated. In some embodiments, binding of P3028 (SEQ ID NO:
to the IL-2 receptor is detected (see Example 8). In some embodiments, MTS
by the immune cells is detected, for example in response to immune cell
stimulation (see
Examples 31-32). In some embodiments, BrdU incorporation by the immune cells
is
detected, for example in response to immune cell stimulation (see Examples 31-
32). It
contemplated herein that some patients will exhibit some immune cell responses
in
response to the inhibitor, but will not exhibit other immune cells responses
in response to
that same inhibitor (see Example 31-32, and Figures 34, 37, and 38, showing,
among
other results, that P28R enhanced the IL-2 induced stimulation of BrdU uptake
and MTS
conversion in one patient, but enhanced BrdU updated and not MTS conversion in
another patient). Thus, some embodiments include detecting two or more immune
cell
responses described herein. Detection of two or more immune cell responses can
allow
the identification of a patient that is likely to elicit a first response, but
not a second
response, and can be useful in guiding clinical decisions such as which
inhibitors or
combinations of inhibitors to apply, and whether to apply additional therapies
to the
patient in need. In some embodiments, detecting activation or stimulation of
an immune
cell, as evidenced by an increase in CD69 or CD71 expression, induction of the
secretion
202
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
of a signal substance, as evidenced by interferon gamma or IL-12 production,
or
stimulation of the release of a cytolytic substance, as evidenced by the
release of
granzyme B or perforM is performed. In some embodiments, detecting activation
or
stimulation of an immune cell includes detecting one or more of enhanced
cytotoxicity,
cytokine production, cell migration, and/or cell proliferation
[0550] In some
embodiments, optionally, an effective dose of the inhibitor for
the patient in need is determined. In some embodiments, cells of the patient
are
contacted in vitro with two or more doses of the inhibitor, and an immune
response. As
shown in Figures 33A, 33B, and 34, P28R can have dose-dependent
immunomodulatory
effects, for example on mitochondrial metabolism (see Example 28 and 29).
[0551] As shown
in Figure 34, increasing doses of P28R (SEQ ID NO: 2)
were provided to the immune cells of cancer patients in vitro. A dose of
20pg/m1 of
P28R resulted in significantly higher MTS conversion than a dose of 40 pg/ml
of P28R.
Thus, one skilled in the art will appreciate that some embodiments include
determining
an effective dose of an inhibitor for the cells of a patient in vitro, and
then providing an
appropriate dose of the inhibitor to the patient.
Additional alternative embodiments
[0552]
Alternative 1001 includes a composition comprising an isolated
peptide comprising the amino acid sequence FFVKLS (SEQ ID NO: 62), in which
the
isolated peptide comprises no more than 30 amino acid residues; and a
nanoparticle, in
which the isolated peptide is immobilized on the nanoparticle. Alternative
1002 includes
the composition of Alternative 1, in which the nanoparticle comprises at least
one of: a
polymer, a dendrimer, a quantum dot, a gold nanoparticle, a silica
nanoparticle, a
magnetic particle, a carbon-based material, a carbohydrate, a nucleic acid, a
polypeptide,
or a lipid. Alternative 1003 includes the composition of Alternative 1001 or
Alternative
1002, in which the nanoparticle comprises a polymer comprising at least one of
PLGA,
glycerol, chitosan, DNA, or a hydrogel. Alternative 1004 includes the
composition of
one of Alternatives 1001-1003, in which the nanoparticle comprises a gold
nanoparticle
comprising at least one of a sphere, rod, or shell. Alternative 1005 includes
the
composition of any one of alternatives 1001-1004, in which the nanoparticle
comprises a
dendrimer comprising PAMAM. Alternative 1006 includes the composition of any
one
Alternatives 1001-1005, in which the nanoparticle comprises a silica
nanoparticle
203
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
comprising at least one of a sphere, shell, or mesoporous structure.
Alternative 1007
includes the composition of any one of Alternatives 1001-1006, in which the
comprises a quantum dot comprising at least one of CdSe, CuInSe, or CdTe.
Alternative
1008 includes the composition of any one of Alternatives 1001-1007, in which
the
nanoparticle comprises a magnetic particle comprising at least one of iron
oxide, cobalt-
based material, a magnetic sphere, an aggregate in dextran or silica, or a
Dynal bead.
Alternative 1009 includes the composition of any one of Alternatives 1001-
1008, in
which the nanoparticle comprises a magnetic comprising a carbon-based material
comprising at least one of a carbon nanotube, buckminsterfullerene, or
graphene.
Alternative 1010 includes the composition of any one of Alternatives 1001-
1009, in
which the nanoparticle comprises a polypeptide comprising an albumin or an
albumin
fragment. Alternative 1011 includes the composition of any one of Alternatives
1001-
1010, in which the nanoparticle comprises a lipid comprising a lipid capsule
or liposome.
Alternative 1012 includes the composition of any one of Alternatives 1001-
1011, in
which the nanoparticle is PEGylated. Alternative 1013 includes the composition
of any
one of Alternatives 1001-1012, in which the nanoparticle comprises a non-
degradable
particle. Alternative 1014 includes the composition of any one of Alternatives
1001-
1012, in which the nanoparticle comprises a degradable particle. Alternative
1015
includes the composition of any one of Alternatives 1001-1014, in which the
comprises a structure selected from the group consisting of: a sphere, a rod,
a shell, a
mesoporous structure, a bead, a hydrogel, an aggregate, a fullerene, a cage, a
porous
nanocage, a viral capsid, a viral capsid fragment, or a lipid capsule.
Alternative 1016
includes the composition of any one of Alternatives 1001-1015, in which the
isolated
peptide is non-covalently immobilized on the nanoparticle. Alternative 1017
includes the
composition of Alternative 1016, in which the nanoparticle is non-covalently
on the nanoparticle by at least one of a van der Waals interaction, steric
interaction,
hydrogen bonding interaction, hydrophobic interaction or electrostatic
interaction.
Alternative 1018 includes the composition of any one of Alternatives 1001-
1017, in
which the isolated peptide is immobilized on the nanoparticle covalently.
Alternative
1019 includes the composition of Alternative 1018, in which the isolated
peptide is
immobilized on the nanoparticle via a cleavable linker or a non-cleavable
linker.
Alternative 1020 includes the composition of Alternative 1019, in which the
cleavable
linker comprises one of an acid-labile linker, a matrix metalloproteinase
target site, or a
204
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
cathepsin target site. Alternative 1021 includes the composition of any one of
Alternatives 1001-1020, in which the nanoparticle has a diameter of at least
10 nm.
Alternative 1022 includes the composition of any one of Alternatives 1001-
1021, in
which the nanoparticle has a diameter of no more than 5000 nm. Alternative
1023
includes the composition of any one of Alternatives 1001-1022, in which the
isolated
peptide comprises no more than 16 amino acid residues. Alternative 1024
includes the
composition of any one of Alternatives 1001-1022, in which the isolated
peptide
comprises no more than 8 amino acid residues.
Alternative 1025 includes the
composition of any one of Alternatives 1001-1022, in which the isolated
peptide consists
of the amino acid sequence1-1-VKLS (SEQ ID NO: 62).
[0553]
Alternative 1026 includes a composition comprising an isolated
peptide comprising the amino acid sequence KKLDTFFVKLSLFTER (SEQ ID NO: 2);
and a nanoparticle, in which the isolated peptide is immobilized on the
nanoparticle.
Alternative 1027 includes the composition of Alternative 1026, in which the
nanoparticle
comprises at least one of: a polymer, a dendrimer, a quantum dot, a gold
nanoparticle, a
silica nanoparticle, a magnetic particle, a carbon-based material, a
carbohydrate, a
acid, a polypeptide, or a lipid. Alternative 1028 includes the composition of
Alternative
1026 or Alternative 1027, in which the nanoparticle comprises a polymer
comprising at
least one of PLGA, glycerol, chitosan, DNA, or a hydrogel. Alternative 1029
includes
composition of any one of Alternatives 1026-1028, in which the nanoparticle
comprises a
gold nanoparticle comprising at least one of a sphere, rod, or shell.
Alternative 1030
includes the
composition of any one of Alternatives 1026-1029, in which the
nanoparticle comprises a dendrimer comprising PAMAM. Alternative 1031 includes
the
composition of any one of Alternatives 1026-1030, in which the nanoparticle
comprises a
silica nanoparticle comprising at least one of a sphere, shell, or mesoporous
structure.
Alternative 1032 includes the composition of any one of Alternatives 1026-
1031, in
which the nanoparticle comprises a quantum dot comprising at least one of
CdSe,
or CdTe. Alternative 1030 includes the composition of any one of Alternatives
1026-
1032, in which the nanoparticle comprises a magnetic particle comprising at
least one of
iron oxide, cobalt-based material, a magnetic sphere, an aggregate in dextran
or silica, or
Dynal bead. Alternative 1002 includes the composition of any one of
Alternatives 1026-
1033, in which the nanoparticle comprises a magnetic comprising a carbon-based
comprising at least one of a carbon nanotube, buckminsterfullerene, or
graphene.
205
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
Alternative 1035 includes the composition of any one of Alternatives 1026-
1034, in
which the nanoparticle comprises a polypeptide comprising an albumin or an
albumin
fragment. Alternative 1036 includes the composition of any one of Alternatives
1026-
1035, in which the nanoparticle comprises a lipid comprising a lipid capsule
or liposome.
Alternative 1037 includes the composition of any one of Alternatives 1026-
1036, in
which the nanoparticle is PEGylated. Alternative 1038 includes the composition
of any
one of Alternatives 1026-1037, in which the nanoparticle comprises a non-
degradable
particle. Alternative 1039 includes the composition of any one of Alternatives
1026-
1037, in which the nanoparticle comprises a degradable particle. Alternative
1040
includes the composition of any one of Alternatives 1026-1039, in which the
comprises a structure selected from the group consisting of: a sphere, a rod,
a shell, a
mesoporous structure, a bead, a hydrogel, an aggregate, a fullerene, a cage, a
porous
nanocage, a viral capsid, a viral capsid fragment, or a lipid capsule.
Alternative 1041
includes the composition of any one of Alternatives 1026-1040, in which the
isolated
peptide is non-covalently immobilized on the nanoparticle. Alternative 1042
includes the
composition of Alternative 1041, in which the nanoparticle is non-covalently
on the nanoparticle by at least one of a van der Waals interaction, steric
interaction,
hydrogen bonding interaction, hydrophobic interaction or electrostatic
interaction.
Alternative 1043 includes the composition of any one of Alternatives 1026-
1042, in
which the isolated peptide is immobilized on the nanoparticle covalently.
Alternative
1044 includes the composition of Alternative 1043, in which the isolated
peptide is
immobilized on the nanoparticle via a cleavable linker or a non-cleavable
linker.
Alternative 1045 includes the The composition of Alternative 1044, in which
the
cleavable linker comprises one of an acid-labile linker, a matrix
metalloproteinase target
site, or a cathepsin target site. Alternative 1046 includes the composition of
any one of
Alternatives 1026-1045, in which the nanoparticle has a diameter of at least
10 nm.
Alternative 1047 includes the composition of any one of Alternatives 1026-
1046, in
which the nanoparticle has a diameter of no more than 5000 nm. Alternative
1048
includes the composition of any one of Alternatives 1026-1047, in which the
isolated
peptide comprises no more than 100 amino acid residues. Alternative 1049
includes the
composition of any one of Alternatives 1026-1047, in which the isolated
peptide
comprises no more than 30 amino acid residues. Alternative 1050 includes the
206
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
composition of any one of Alternatives 1026-1047, in which the isolated
peptide consists
of the amino acid sequence of SEQ ID NO: 2.
[0554]
Alternative 1051 includes a composition comprising an isolated
peptide comprising the amino acid sequence RKLDTFFVKLSLFTERRR (SEQ ID NO:
586); and a nanoparticle, in which the isolated peptide is immobilized on the
Alternative 1052 includes the composition of Alternative 1051, in which the
nanoparticle
comprises at least one of: a polymer, a dendrimer, a quantum dot, a gold
nanoparticle, a
silica nanoparticle, a magnetic particle, a carbon-based material, a
carbohydrate, a
acid, a polypeptide, or a lipid. Alternative 1053 includes the composition of
Alternative
1051 or Alternative 1052, in which the nanoparticle comprises a polymer
comprising at
least one of PLGA, glycerol, chitosan, DNA, or a hydrogel. Alternative 1054
includes
composition of any one of Alternatives 1051-1053, in which the nanoparticle
comprises a
gold nanoparticle comprising at least one of a sphere, rod, or shell.
Alternative 1055
includes the composition of any one of Alternatives 1051-1054, in which the
comprises a dendrimer comprising PAMAM. Alternative 1056 includes the
composition
of any one of Alternatives 1051-1055, in which the nanoparticle comprises a
silica
nanoparticle comprising at least one of a sphere, shell, or mesoporous
structure.
Alternative 1057 includes the composition of any one of Alternatives 1051-
1056, in
which the nanoparticle comprises a quantum dot comprising at least one of
CdSe,
or CdTe. Alternative 1058 includes the composition of any one of Alternatives
1051-
1057, in which the nanoparticle comprises a magnetic particle comprising at
least one of
iron oxide, cobalt-based material, a magnetic sphere, an aggregate in dextran
or silica, or
Dynal bead. Alternative 1059 includes the composition of any one of
Alternatives
1051-1058, in which the nanoparticle comprises a magnetic comprising a carbon-
based
material comprising at least one of a carbon nanotube, buckminsterfullerene,
or
Alternative 1060 includes the composition of any one of Alternatives 1051-
1059, in
which the nanoparticle comprises a polypeptide comprising an albumin or an
albumin
fragment. Alternative 1061 includes the composition of any one of Alternatives
1051-
1060, in which the nanoparticle comprises a lipid comprising a lipid capsule
or liposome.
Alternative 1062 includes the composition of any one of Alternatives 1051-
1061, in
which the nanoparticle is PEGylated. Alternative 1063 includes the composition
of any
one of Alternatives 1051-1062, in which the nanoparticle comprises a non-
degradable
particle. Alternative 1064 includes the composition of any one of Alternatives
1051-
207
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
1062, in which the nanoparticle comprises a degradable particle. Alternative
1065
includes the composition of any one of Alternatives 1051-1054, in which the
comprises a structure selected from the group consisting of: a sphere, a rod,
a shell, a
mesoporous structure, a bead, a hydrogel, an aggregate, a fullerene, a cage, a
porous
nanocage, a viral capsid, a viral capsid fragment, or a lipid capsule.
Alternative 1066
includes the composition of any one of Alternatives 1051-1055, in which the
isolated
peptide is non-covalently immobilized on the nanoparticle. Alternative 1067
includes the
composition of Alternative 1066, in which the nanoparticle is non-covalently
on the nanoparticle by at least one of a van der Waals interaction, steric
interaction,
hydrogen bonding interaction, hydrophobic interaction or electrostatic
interaction.
Alternative 1068 includes the composition of any one of Alternatives 1051-
1067, in
which the isolated peptide is immobilized on the nanoparticle covalently.
Alternative
1069 includes the composition of Alternative 1058, in which the isolated
peptide is
immobilized on the nanoparticle via a cleavable linker or a non-cleavable
linker.
Alternative 1070 includes the composition of Alternative 1059, in which the
cleavable
linker comprises one of an acid-labile linker, a matrix metalloproteinase
target site, or a
cathepsin target site. Alternative 1071 includes the composition of any one of
Alternatives 1051-1070, in which the nanoparticle has a diameter of at least
10 nm.
Alternative 1072 includes the composition of any one of Alternatives 1051-
1071, in
which the nanoparticle has a diameter of no more than 5000 nm. Alternative
1073
includes the composition of any one of Alternatives 1051-1072, in which the
isolated
peptide comprises no more than 16 amino acid residues. Alternative 1074
includes the
composition of any one of Alternatives 1051-1073, in which the isolated
peptide
comprises no more than 100 amino acid residues. Alternative 1075 includes the
composition of any one of Alternatives 1051-1074, in which the isolated
peptide
comprises no more than 30 amino acid residues. Alternative 1076 includes the
composition any one of Alternatives 1051-1075, in which the isolated peptide
consists of
the amino acid sequence of SEQ ID NO: 586.
[0555]
Alternative 1077 includes a composition comprising: an isolated
peptide comprising the formula XiX2X3X4X5X6X7X8X9XioXi iXi2X13X14X15X16X17, in
which Xi is any amino acid or is absent; X2 is a positively charged amino
acid, F, or N;
X3 is any amino acid; X4 is any amino acid; X5 is a polar uncharged amino
acid, R, Y, or
W; X6 is a hydrophobic or uncharged polar amino acid; X7 is a hydrophobic or
208
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
polar amino acid; X8 is a hydrophobic, non-aromatic carbon chain amino acid
that is not
M or F; X9 is a positively charged amino acid, T, Q, or Y; Xio is any amino
acid that is
not negatively charged; Xii is a polar uncharged amino acid or H; X12 is any
amino acid
that is not negatively charged; Xi 3 is any amino acid that is not negatively
charged; X14 is
any amino acid that is not negatively charged; X15 is a negatively charged
amino acid, Y,
or Q; X16 is any amino acid that is not negatively charged; X17 is one or more
positively
charged amino acids or is absent; and a nanoparticle, in which the isolated
peptide is
immobilized on the nanoparticle. Alternative 1078 includes the composition of
Alternative 1077, in which the nanoparticle comprises at least one of: a
polymer, a
dendrimer, a quantum dot, a gold nanoparticle, a silica nanoparticle, a
magnetic particle,
carbon-based material, a carbohydrate, a nucleic acid, a polypeptide, or a
lipid.
Alternative 1079 includes the composition of Alternative 1077 or Alternative
1078, in
which the nanoparticle comprises a polymer comprising at least one of PLGA,
glycerol,
chitosan, DNA, or a hydrogel. Alternative 1080 includes the composition of any
one of
Alternatives 1077-1079, in which the nanoparticle comprises a gold
nanoparticle
comprising at least one of a sphere, rod, or shell. Alternative 1081 includes
the
composition of any one of Alternatives 1077-1080, in which the nanoparticle
comprises a
dendrimer comprising PAMAM. Alternative 1082 includes the composition of any
one
of Alternatives 1077-1081, in which the nanoparticle comprises a silica
nanoparticle
comprising at least one of a sphere, shell, or mesoporous structure.
Alternative 1083
includes the composition of any one of Alternatives 1077-1082, in which the
comprises a quantum dot comprising at least one of CdSe, CuInSe, or CdTe.
Alternative
1084 includes the composition of any one of Alternatives 1077-1083, in which
the
nanoparticle comprises a magnetic particle comprising at least one of iron
oxide, cobalt-
based material, a magnetic sphere, an aggregate in dextran or silica, or a
Dynal bead.
Alternative 1085 includes the composition of any one of Alternatives 1077-
1084, in
which the nanoparticle comprises a magnetic comprising a carbon-based material
comprising at least one of a carbon nanotube, buckminsterfullerene, or
graphene.
Alternative 1086 includes the composition of any one of Alternatives 1077-
1085, in
which the nanoparticle comprises a polypeptide comprising an albumin or an
albumin
fragment. Alternative 1087 includes the composition of any one of Alternatives
1077-
1086, in which the nanoparticle comprises a lipid comprising a lipid capsule
or liposome.
Alternative 1088 includes the composition of any one of Alternatives 1077-
1087, in
209
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
which the nanoparticle is PEGylated. Alternative 1089 includes the composition
of any
one of Alternatives 1077-1088, in which the nanoparticle comprises a non-
degradable
particle. Alternative 1090 includes the composition of any one of Alternatives
1077-
1088, in which the nanoparticle comprises a degradable particle. Alternative
1091
includes the composition of any one of Alternatives 1077-1090, in which the
comprises a structure selected from the group consisting of: a sphere, a rod,
a shell, a
mesoporous structure, a bead, a hydrogel, an aggregate, a fullerene, a cage, a
porous
nanocage, a viral capsid, a viral capsid fragment, or a lipid capsule.
Alternative 1092
includes the composition of any one of Alternatives 1077-1091, in which the
isolated
peptide is non-covalently immobilized on the nanoparticle. Alternative 1093
includes the
composition of Alternative 1092, in which the nanoparticle is non-covalently
on the nanoparticle by at least one of a van der Waals interaction, steric
interaction,
hydrogen bonding interaction, hydrophobic interaction or electrostatic
interaction.
Alternative 1094 includes the composition of any one of Alternatives 1077-
1093, in
which the isolated peptide is immobilized on the nanoparticle covalently.
Alternative
1095 includes the composition of Alternative 1094, in which the isolated
peptide is
immobilized on the nanoparticle via a cleavable linker or a non-cleavable
linker.
Alternative 1096 includes the composition of Alternative 1095, in which the
cleavable
linker comprises one of an acid-labile linker, a matrix metalloproteinase
target site, or a
cathepsin target site. Alternative 1097 includes the composition of any one of
Alternatives 1077-1096, in which the nanoparticle has a diameter of at least
10 nm.
Alternative 1098 includes the composition of any one of Alternatives 1077-
0197, in
which the nanoparticle has a diameter of no more than 5000 nm. Alternative
1099
includes the composition of any one of Alternatives 1077-1098, in which Xi
comprises at
least one positively charged amino acid. Alternative 1100 includes the
composition of
any one of Alternatives 1077-1099, in which Xi comprises R and X17 comprises
RR.
Alternative 1101 includes the composition of any one of Alternatives 1077-
1100, in
which the peptide is soluble in an aqueous solution. Alternative 1102 includes
the
composition of any one of Alternatives 1077-1101, in which at least one of:Xi
is K; X2 is
K; X3 is L; X4 is D; X5 is T; X6 is F; X7 is F; X8 is V;X9 is K;X10 is L; X11
is S; X12 is L;
Xi 3 is F; X14 is T; Xi 5 is E; or X16 is R. Alternative 1103 includes the
composition of
one of Alternatives 1077-1102, in which the isolated peptide has a length of
30 amino
acid residues or less. Alternative 1104 includes the composition of any one of
210
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
Alternatives 1077-1102, in which the isolated peptide consists of the formula
XiX2X3X4X5X6X7X8XoXioXiiXi2X13X14X15X16X17. Alternative 1105 includes the
composition of any one of Alternatives 1077-1104, in which the isolated
peptide
comprises the amino acid sequence KKLDTFFVKLSLFTER (SEQ ID NO: 2).
Alternative 1106 includes the composition of any one of Alternatives 1077-
1104, in
which the isolated peptide comprises the amino acid sequence
RKLDTFFVKLSLFTERRR (SEQ ID NO: 586). Alternative 1107 includes a
composition comprising: an isolated synthetic peptide consisting of the amino
acid
sequence FFVKLS (SEQ ID NO: 62); and a nanoparticle, in which the isolated
synthetic
peptide is immobilized on the nanoparticle. Alternative 1108 includes the
composition
Alternative 1107, in which the nanoparticle comprises at least one of: a
polymer, a
dendrimer, a quantum dot, a gold nanoparticle, a silica nanoparticle, a
magnetic particle,
carbon-based material, a carbohydrate, a nucleic acid, a polypeptide, or a
lipid.
Alternative 1109 includes the composition of Alternative 1107 or Alternative
1108, in
which the nanoparticle comprises a polymer comprising at least one of PLGA,
glycerol,
chitosan, DNA, or a hydrogel. Alternative 1110 includes the composition of any
one of
Alternatives 1107-1109, in which the nanoparticle comprises a gold
nanoparticle
comprising at least one of a sphere, rod, or shell. Alternative 1111 includes
the
composition of any one of Alternatives 1107-1110, in which the nanoparticle
comprises a
dendrimer comprising PAMAM. Alternative 1112 includes the composition of any
one
of Alternatives 1107-1111, in which the nanoparticle comprises a silica
nanoparticle
comprising at least one of a sphere, shell, or mesoporous structure.
Alternative 1113
includes the composition of any one of Alternatives 1107-1112, in which the
comprises a quantum dot comprising at least one of CdSe, CuInSe, or CdTe.
Alternative
1114 includes the composition of any one of Alternatives 1107-1113, in which
the
nanoparticle comprises a magnetic particle comprising at least one of iron
oxide, cobalt-
based material, a magnetic sphere, an aggregate in dextran or silica, or a
Dynal bead.
Alternative 1115 includes the composition of any one of Alternatives 1107-
1114, in
which the nanoparticle comprises a magnetic comprising a carbon-based material
comprising at least one of a carbon nanotube, buckminsterfullerene, or
graphene.
Alternative 1116 includes the composition of any one of Alternatives 1107-
1115, in
which the nanoparticle comprises a polypeptide comprising an albumin or an
albumin
fragment. Alternative 1117 includes the composition of any one of Alternatives
1107-
211
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
1116, in which the nanoparticle comprises a lipid comprising a lipid capsule
or liposome.
Alternative 1118 includes the composition of any one of Alternatives 1107-
1117, in
which the nanoparticle is PEGylated. Alternative 1119 includes the composition
of any
one of Alternatives 1107-1118, in which the nanoparticle comprises a non-
degradable
particle. Alternative 1120 includes the composition of any one of Alternatives
1107-
1118, in which the nanoparticle comprises a degradable particle. Alternative
1121
includes the composition of any one of Alternatives 1107-1120, in which the
comprises a structure selected from the group consisting of: a sphere, a rod,
a shell, a
mesoporous structure, a bead, a hydrogel, an aggregate, a fullerene, a cage, a
porous
nanocage, a viral capsid, a viral capsid fragment, or a lipid capsule.
Alternative 1122
includes the composition of any one of Alternatives 1107-1121, in which the
isolated
peptide is non-covalently immobilized on the nanoparticle. Alternative 1123
includes the
composition of Alternative 1122, in which the nanoparticle is non-covalently
on the nanoparticle by at least one of a van der Waals interaction, steric
interaction,
hydrogen bonding interaction, hydrophobic interaction or electrostatic
interaction.
Alternative 1124 includes the composition of any one of Alternatives 1107-
1123, in
which the isolated peptide is immobilized on the nanoparticle covalently.
Alternative
1125 includes the composition of Alternative 1124, in which the isolated
peptide is
immobilized on the nanoparticle via a cleavable linker or a non-cleavable
linker.
Alternative 1126 includes the composition of Alternative 1125, in which the
cleavable
linker comprises one of an acid-labile linker, a matrix metalloproteinase
target site, or a
cathepsin target site. Alternative 1127 includes the composition of any one of
Alternatives 1107-1126, in which the nanoparticle has a diameter of at least
10 nm.
Alternative 1128 includes the composition of any one of Alternatives 1107-
1127, in
which the nanoparticle has a diameter of no more than 5000 nm. Alternative
1129
includes the composition of any one of Alternatives 1001-1128, in which the
isolated
peptide comprises a modification comprising at least one of a D amino acid, an
N-
terminal acetyl group, a C-terminal amide group, glycosylation, nitrosylation,
carbonylation, oxidation, a linked pharmacokinetic modifier, and a linked
polyethylene
glycol or any combination thereof. Alternative 1130 includes the composition
of any one
of Alternatives 1001-1129 in which the isolated peptide activates an immune
cell.
Alternative 1131 includes the composition of any one of Alternatives 1001-1130
in
the isolated peptide activates an immune cell, if a solution comprising the
immune cell
212
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
comprises a second peptide comprising, consisting essentially of, or
consisting of the
sequence VFDEFKPLVEEPQNLIK (SEQ ID NO: 185), or if an LFA-1 receptor of the
immune cell is bound to the second peptide. Alternative 1132 includes the
composition
of any one of Alternatives 1001-1131, in which, if the isolated peptide is
contacted with
second peptide comprising, consisting essentially of, or consisting of the
amino acid
sequence VFDEFKPLVEEPQNLIK (SEQ ID NO: 185), the isolated peptide specifically
binds to the second peptide. Alternative 1133 includes the composition of any
one of
Alternatives 1001-1132, in which, if the isolated peptide is contacted with an
immune
comprising an LFA-1 receptor and a second peptide comprising, consisting
essentially of,
or consisting of the amino acid sequence VFDEFKPLVEEPQNLIK (SEQ ID NO: 185),
the isolated peptide inhibits binding of the second peptide to the LFA-1
receptor.
Alternative 1134 includes the composition of any one of Alternatives 1001-
1133, further
comprising a pharmaceutically acceptable carrier or diluent. Alternative 1135
includes
the composition of Alternative 1134, in which the pharmaceutically acceptable
carrier or
diluent comprises a degradable particle. Alternative 1136 includes the
composition of
one of Alternatives 1001-1135, in which the composition comprises at least
about 10pg
the isolatedpeptide.
Alternative 1137 includes the composition of any one of
1134-1136, comprising a buffer selected from the group consisting of: Trizma,
Bicine,
Tricine, MOPS, MOPSO, MOBS, Tris, Hepes, HEPBS, MES, phosphate, carbonate,
acetate, citrate, glycolate, lactate, borate, ACES, ADA, tartrate, AMP, AMPD,
AMPSO,
BES, CABS, cacodylate, CHES, DIPSO, EPPS, ethanolamine, glycine, HEPPSO,
imidazole, imidazolelactic acid, PIPES, SSC, SSPE, POPSO, TAPS, TABS, TAPSO
and
TES. Alternative 1138 includes the composition of any one of Alternatives 1001-
1137,
which if contacted with a cancer cell, the composition induces cytotoxicity of
the cancer
cell. Alternative 1139 includes the composition of Alternative 1038, in which
the cancer
cell comprises a prostate cancer cell. Alternative 1140 includes the
composition of any
one of Alternatives 1001-1139, in which the composition comprises a gel.
Alternative
1141 includes the composition of Alternative 1140 in which the composition
will remain
in a gel format for at least 72 hours under physiological conditions.
[0556]
Alternative 1142 includes a method comprising administering to an
individual having a cancer, and in need of treatment therefor, an effective
amount of the
composition of any of Alternatives 1001-1141, thereby inducing at least one of
the
following: (a) activation of an immune cell; (b) inhibition of binding of a
damaged
213
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
albumin, an aggregate of albumins, an albumin fragment, or a second peptide to
an LFA-
receptor or IL-2 receptor, in which the second peptide or albumin fragment, if
present,
comprises, consists of, or consists essentially of at least one of SEQ ID NOs:
183-246;
or(c)
cytotoxicity to the tumor cell. Alternative 1143 includes the method
Alternative 1142, in which (a) and (b) are induced. Alternative 1144 includes
the
of Alternative 1142, in which (a), (b), and (c) are induced. Alternative 1145
includes the
method of any one of Alternatives 1142-1144, in which the individual having a
cancer
a tumor. Alternative 1146 includes the method of Alternative 1145, in which
the tumor
comprises at least one of a prostate tumor, a melanoma, a colon cancer, a lung
an Apocrine gland carcinoma, a testis tumor, a mast cell tumor, a mammary
tumor, a
mucinous carcinoma, or a histicytoma. Alternative 1147 includes the method of
Alternative 1146, in which the mammary tumor comprises a benign mammary tumor
or a
malignant mammary tumor, or the mammary tumor comprises a mixed mammary tumor
(for example a benign mixed mammary tumor, or a malignant mixed mammary
tumor).
Alternative 1148 includes the method of Alternative 1146, in which the
mucinous
carcinoma comprises a mammary gland mucinous carcinoma. Alternative 1149
includes
the method of any one of Alternatives 1142-1148, in which the administering
further
induces regressive changes in the cancer. Alternative 1150 includes the method
of any
one of Alternatives 1145-1149, in which the administering further induces
immune cell
infiltration of the tumor. Alternative 1151 includes the method of any one of
1145-1150, in which the administering further induces eradication of cells of
the tumor.
Alternative 1152 includes the method of any one of Alternatives 1145-1151, in
which the
administering further induces eradication of the tumor. Alternative 1153
includes the
method of any one of Alternatives 1145-1152, in which the composition is
administered
directly to the tumor in the subject. Alternative 1154 includes the method of
any one of
Alternatives 1145-1152, in which the composition induces regressive changes in
a tumor
to which the composition is not directly administered. Alternative 1155
includes the
method of any one of Alternatives 1145-1153, in which the composition induces
eradication of a tumor to which the composition is not directly administered.
Alternative
1156 includes the method of any one of Alternatives 1154-1155, in which the
tumor to
which the composition was not administered comprises a contralateral or
metastatic
different from a tumor to which the composition is directly administered.
Alternative
1157 includes the method of any one of Alternatives 1142-1156, in which the
albumin
214
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
fragment or second peptide comprises no more than 100 amino acid residues.
1158 includes the method of any one of Alternatives 1142-1157, in which the
albumin
fragment or second peptide comprises SEQ ID NO: 185. Alternative 1159 includes
the
method of any one of Alternatives 1142-1156 in which the albumin fragment or
second
peptide consists of or consists essentially of SEQ ID NO: 185. Alternative
1160 includes
the method of any one of Alternatives 1142-1159, in which the LFA-1 receptor
is
available for stimulation following inhibition of binding of the albumin,
albumin
fragment, or second peptide. Alternative 1161 includes the method of any one
of
Alternatives 1142-1160, in which the immune cell is stimulated following
inhibition of
binding of the albumin, albumin fragment, or second peptide. Alternative 1162
includes
the method of Alternative 1161, in which the immune cell is stimulated by a
second
therapeutic agent. Alternative 1163 includes the method of Alternative 1162,
in which
the second therapeutic agent is administered concurrently with the
composition.
Alternative 1164 includes the method of Alternative 1162, in which the
composition
comprises the second therapeutic agent. Alternative 1165 includes the method
of
Alternative 1162, in which the second therapeutic agent is administered prior
to
administering the composition. Alternative 1166 includes the method of
Alternative
1162, in which the second therapeutic agent is administered subsequent to
administering
the composition. Alternative 1167 includes the method of any one of
Alternatives 1142-
1166, in which the peptide of the composition is administered to the
individual at a dose
of at least about 0.1 mg/kg. Alternative 1168 includes the method of any one
of
Alternatives 1142-1167, in which the peptide of the composition is
administered in at
least a first administration and a second administration at least five days
after the first
administration. Alternative 1169 includes the method of any one of
Alternatives 1142-
1168, in which the peptide is administered to a tissue within about 10cm of a
tumor of
cancer. Alternative 1170 includes the method of any one of Alternatives 1142-
1169, in
which the peptide is administered peri-tumorally to a tumor of the cancer.
Alternative
1171 includes the method of any one of Alternatives 1142-1170, in which the
cancer
comprises at least one of colorectal cancer, renal cancer, breast cancer, skin
cancer,
ovarian cancer, prostate cancer, pancreatic cancer, lung cancer, malignant
melanoma,
small cell lung cancer, non-small lung cancer (adenocarcinoma), squamous cell
carcinoma, bladder cancer, osteosarcoma, bronchial cancer, or hematopoietic
cell cancer.
Alternative 1172 includes the method of any one of Alternatives 1142-1171, in
which the
215
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
individual comprises serum comprising a damaged albumin, an aggregate of
albumins,
albumin fragment, or a second peptide, in which the albumin fragment or second
peptide
comprises at least one of SEQ ID NOs: 183-246. Alternative 1173 includes the
method
Alternative 1172, in which the second peptide or albumin fragment comprises
the amino
acid sequence VFDEFKPLVEEPQNLIK (SEQ ID NO: 185). Alternative 1174 includes
the method of Alternative 1172, in which the second peptide or albumin
fragment
comprises no more than 100 amino acid residues.
[0557]
Alternative 1175 includes a method of activating an immune cell in a
cancer patient, the method comprising contacting the immune cell with a
composition
comprising: an isolated peptide comprising the amino acid sequence FFVKLS (SEQ
ID
NO: 62), in which the peptide consists of about six to thirty amino acids; and
a
nanoparticle, in which the isolated peptide is immobilized on the
nanoparticle.
Alternative 1176 includes the method of Alternative 1175, in which the cancer
patient
a tumor. Alternative 1177 includes the method of Alternative 1176, in which
the tumor
comprises at least one of a prostate tumor, a melanoma, a colon cancer, a lung
an Apocrine gland carcinoma, a testis tumor, a mast cell tumor, a mammary
tumor, a
mucinous carcinoma, or a histicytoma. Alternative 1178 includes the method of
Alternative 1177, in which the mammary tumor comprises a benign mammary tumor
or a
malignant mammary tumor, or the mammary tumor comprises a mixed mammary tumor
(for example a benign mixed mammary tumor, or a malignant mixed mammary
tumor).
Alternative 1179 includes the method of Alternative 1177, in which the
mucinous
carcinoma comprises a mammary gland mucinous carcinoma. Alternative 1180
includes
the method of any one of Alternatives 1175-0179, in which the contacting
further
regressive changes in the cancer. Alternative 1181 includes the method of any
one of
Alternatives 1175-1180, in which the contacting further induces immune cell
infiltration
of the tumor. Alternative 1182 includes the method of any one of Alternatives
1175-
1181, in which the contacting further induces eradication of cells of the
tumor.
Alternative 1183 includes the method of any one of Alternatives 1175-1182, in
which the
contacting further induces eradication of the tumor. Alternative 1184 includes
the
of any one of Alternatives 1175-1183, in which the composition is administered
directly
to the tumor in the cancer patient. Alternative 1185 includes the method of
any one of
Alternatives 1175-1184, in which the composition induces regressive changes in
a tumor
to which the composition is not directly administered. Alternative 1186
includes the
216
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
method of any one of Alternatives 1175-1185, in which the composition induces
eradication of a tumor to which the composition is not directly administered.
Alternative
1187 includes the method of any one of Alternatives 1175-1186, in which the
tumor to
which the composition is not directly administered comprises a contralateral
or
tumor different from a tumor to which the composition is directly
administered.
Alternative 1188 includes the method of any one of Alternatives 1175-1187, in
which the
nanoparticle comprises at least one of: a polymer, a dendrimer, a quantum dot,
a gold
nanoparticle, a silica nanoparticle, a magnetic particle, a carbon-based
material, a
carbohydrate, a nucleic acid, a polypeptide, or a lipid. Alternative 1189
includes the
method of any one of Alternatives 1175-1188, in which the nanoparticle
comprises a
polymer comprising at least one of PLGA, glycerol, chitosan, DNA, or a
hydrogel.
Alternative 1190 includes the method of any one of Alternatives 1175-1189, in
which the
nanoparticle comprises a gold nanoparticle comprising at least one of a
sphere, rod, or
shell. Alternative 1191 includes the method of any one of Alternatives 1175-
1190, in
which the nanoparticle comprises a dendrimer comprising PAMAM. Alternative
1192
includes the method of any one of Alternatives 1175-1191, in which the
nanoparticle
comprises a silica nanoparticle comprising at least one of a sphere, shell, or
mesoporous
structure. Alternative 1193 includes the method of any one of Alternatives
1175-1192, in
which the nanoparticle comprises a quantum dot comprising at least one of
CdSe,
or CdTe. Alternative 1194 includes the method of any one of Alternatives 1175-
1193, in
which the nanoparticle comprises a magnetic particle comprising at least one
of iron
oxide, cobalt-based material, a magnetic sphere, an aggregate in dextran or
silica, or a
Dynal bead. Alternative 1195 includes the method of any one of Alternatives
1175-
in which the nanoparticle comprises a magnetic comprising a carbon-based
material
comprising at least one of a carbon nanotube, buckminsterfullerene, or
graphene.
Alternative 1196 includes the method of any one of Alternatives 1175-1195, in
which the
nanoparticle comprises a polypeptide comprising an albumin or an albumin
fragment.
Alternative 1197 includes the method of any one of Alternatives 1175-1196, in
which the
nanoparticle comprises a lipid comprising a lipid capsule or liposome.
Alternative 1198
includes the method of any one of Alternatives 1175-1197, in which the
nanoparticle is
PEGylated. Alternative 1199 includes the method of any one of Alternatives
1175-1198,
in which the nanoparticle comprises a non-degradable particle. Alternative
1200
the method of any one of Alternatives 1175-1199, in which the nanoparticle
comprises a
217
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
degradable particle. Alternative 1201 includes the method of any one of
Alternatives
1175-1200, in which the nanoparticle comprises a structure selected from the
group
consisting of: a sphere, a rod, a shell, a mesoporous structure, a bead, a
hydrogel, an
aggregate, a fullerene, a cage, a porous nanocage, a viral capsid, a viral
capsid fragment,
or a lipid capsule. Alternative 1202 includes the method of any one of
Alternatives
1201, in which the isolated peptide is non-covalently immobilized on the
nanoparticle.
Alternative 1203 includes the method of Alternative 1202, in which the
nanoparticle is
non-covalently immobilized on the nanoparticle by at least one of a van der
Waals
interaction, steric interaction, hydrogen bonding interaction, hydrophobic
interaction or
electrostatic interaction.
Alternative 1204 includes the method of any one of
1175-1203, in which the isolated peptide is immobilized on the nanoparticle
covalently.
Alternative 1205 includes the method of Alternative 1204, in which the
isolated peptide
immobilized on the nanoparticle via a cleavable linker or a non-cleavable
linker.
Alternative 1206 includes the method of Alternative 1205, in which the
cleavable linker
comprises one of an acid-labile linker, a matrix metalloproteinase target
site, or a
cathepsin target site. Alternative 1207 includes the method of any one of
Alternatives
1175-1206, in which the nanoparticle has a diameter of at least 10 nm.
Alternative 1208
includes the method of any one of Alternatives 1175-1207, in which the
nanoparticle has
a diameter of no more than 5000 nm. Alternative 1209 includes the method of
any one
Alternatives 1175-1208, in which the isolated peptide comprises no more than
16 amino
acid residues. Alternative 1210 includes the method of any one of Alternatives
1175-
1209, in which the isolated peptide comprises no more than 8 amino acid
residues.
Alternative 1211 includes the method of any one of Alternatives 1175-1210, in
which the
isolated peptide consists of the amino acid sequence FFVKLS (SEQ ID NO: 62).
Alternative 1212 includes the method of any one of Alternatives 1175-1211, in
which
contacting the immune cell with the composition inhibits binding of a damaged
albumin,
an aggregate of albumins, an albumin fragment, or a second peptide to an LFA-1
in which the albumin fragment or second peptide comprises at least one of SEQ
ID NOs:
183-246. Alternative 1213 includes the method of Alternative 1212, in which
the
albumin fragment or second peptide comprises no more than 100 amino acids.
Alternative 1214 includes the method of Alternative 1212 or 1213, in which the
albumin
fragment or second peptide comprises SEQ ID NO: 185. Alternative 1215 includes
the
method of Alternative 1214, in which the albumin fragment or second peptide
consists of
218
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
or consists essentially of SEQ ID NO: 185. Alternative 1216 includes the
method of any
one of Alternatives 1212-1215, in which the LFA-1 receptor is available for
stimulation
following inhibition of binding of the albumin, albumin fragment, or second
peptide.
Alternative 1217 includes the method of any one of Alternatives 1215-1216, in
which the
LFA-1 receptor is stimulated following inhibition of binding of the albumin,
albumin
fragment, or second peptide. Alternative 1218 includes the method of
Alternative 1217,
in which the immune cells are stimulated by a second therapeutic agent.
Alternative
includes the method of Alternative 1218, in which the second therapeutic agent
is
administered concurrently with the composition. Alternative 1220 includes the
method
Alternative 1218, in which the composition comprises the second therapeutic
agent.
Alternative 1221 includes the method of Alternative 1218, in which the second
therapeutic agent is administered prior to administration of the composition.
Alternative
1222 includes the method of Alternative 1218, in which the second therapeutic
agent is
administered subsequent to administration of the composition.
[0558]
Alternative 1223 includes a method of binding cancer cells with a
peptide, the method comprising: contacting a cancer cell with the composition
of any one
of Alternatives 1001-1141;and detecting the binding of said peptide to said
cancer cell.
Alternative 1224 includes the method of Alternative 1223, in which the peptide
a detectable moiety. Alternative 1225 includes the method of Alternative 1224,
in which
the detectable moiety comprises a biotinylated label, a radioactive label, a
fluorescent
label, an enzyme, or a colloidal gold label. Alternative 1226 includes the
method of any
one of Alternatives1 1123-1225, in which the cancer cell is a colorectal
cancer cell, a
renal cancer cell, a breast cancer cell, a skin cancer cell, an ovarian cancer
cell, a
cancer cell, a pancreatic cancer cell, a lung cancer cell, a malignant
melanoma cell, a
small cell lung cancer cell, a non-small lung cancer (adenocarcinoma) cell, a
squamous
cell carcinoma cell, a bladder cancer cell, an osteosarcoma cell, a bronchial
cancer cell,
a hematopoietic cell cancer cell. Alternative 1227 includes the method of any
one of
Alternatives 1223-1226, in which the cancer cell comprises a prostate tumor
cell, a
melanoma cell, a colon cancer cell, a lung carcinoma cell, an Apocrine gland
carcinoma
cell, a testis tumor cell, a mast cell tumor cell, a mammary tumor cell, a
mucinous
carcinoma cell, or a histicytoma cell. Alternative 1228 includes the method of
1227, in which the mammary tumor comprises a benign mammary tumor or a
malignant
mammary tumor, or the mammary tumor comprises a mixed mammary tumor (for
219
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
example a benign mixed mammary tumor, or a malignant mixed mammary tumor).
Alternative 1229 includes the method of Alternative 1227, in which the
mucinous
carcinoma comprises a mammary gland mucinous carcinoma. Alternative 1230
includes
the method of any one of Alternatives 1223-1229, in which said peptide
comprises an
antibody or antibody fragment.
[0559] Alternative 1231 includes a method of ameliorating
immunosuppression in a subject in need thereof, the method comprising
administering to
the subject an effective amount of the composition of any of Alternatives 1001-
10141,
thereby inducing at least one of the following: (a) activation of an immune
cell; or (b)
inhibition of binding of a damaged albumin, an aggregate of albumins, an
albumin
fragment, or a second peptide to an LFA-1 receptor, in which the second
peptide or
albumin fragment, if present, comprises at least one of SEQ ID NOs: 183-246.
Alternative 1232 includes the method of Alternative 1231, in which the albumin
fragment or second peptide comprises no more than 100 amino acid residues.
Alternative 1233 includes the method of Alternative 1231 or 1232, in which the
albumin
fragment or second peptide comprises SEQ ID NO: 185. Alternative 1234 includes
the
method of any one of Alternatives 1231-1233, in which the albumin fragment or
second
peptide consists of or consists essentially of SEQ ID NO: 185. Alternative
1235 includes
the method of any one of Alternatives 1231-1234, in which the LFA-1 receptor
is
available for stimulation following inhibition of binding of the albumin,
albumin
fragment, or second peptide. Alternative 1236 includes a kit comprising: the
composition
of any one of Alternatives 1001-1141; and a detectable moiety. Alternative
1237
includes a kit of Alternative 1236, in which the detectable moiety comprises a
biotinylated label, a radioactive label, a fluorescent label, an enzyme, or a
colloidal gold
label. Alternative 1238 includes use of the composition of any one of
Alternatives 1001-
1141 for the treatment of cancer.
[0560]
Alternative 1238 includes use of the composition of any one of
Alternatives 1001-1141 for stimulating an immune cell in a cancer patient.
Alternative
1240 includes the use of any of Alternatives 1238-1239, in which the cancer
comprises at
least one of colorectal cancer, renal cancer, breast cancer, skin cancer,
ovarian cancer,
prostate cancer, pancreatic cancer, lung cancer, melanoma, malignant melanoma,
small
cell lung cancer, non-small lung cancer (adenocarcinoma), lung carcinoma,
squamous
carcinoma, bladder cancer, osteosarcoma, bronchial cancer, hematopoietic cell
cancer,
220
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
mammary tumor, mucinous carcinoma, or histicytoma. Alternative 1241 includes
the
of any of Alternatives 1238-1240, in which the cancer comprises a tumor
comprising at
least one of a prostate tumor, a melanoma, a colon cancer, a lung carcinoma,
an
gland carcinoma, a testis tumor, a mast cell tumor, a mammary tumor, a
mucinous
carcinoma, or a histicytoma. Alternative 1242 includes the use of Alternative
1241, in
which the mammary tumor comprises a benign mammary tumor or a malignant
tumor, or the mammary tumor comprises a mixed mammary tumor (for example a
benign
mixed mammary tumor, or a malignant mixed mammary tumor). Alternative 1243
includes the use of Alternative 1241, in which the mucinous carcinoma
comprises a
mammary gland mucinous carcinoma. Alternative 1244 includes the use of any of
Alternatives 1238-1243, in which the composition is further for use in
inducing
changes in the cancer. Alternative 1245 includes the use of any of
Alternatives 1238-
1244, in which the composition is further for use in inducing immune cell
infiltration of a
tumor of the cancer. Alternative 1246 includes the use of any of Alternatives
1238-1245,
in which the composition is further for use in eradicating cells of a tumor of
the cancer.
Alternative 1247 includes the use of any of Alternatives 1238-1246, in which
the
composition is further for use in eradicating a tumor of the cancer.
Alternative 1248
includes the use of any of Alternatives 1238-1247, in which the composition is
for
administration directly to a tumor in a cancer patient. Alternative 1249
includes the use
of any one of Alternatives 1238-1248, in which the composition is for use in
inducing
regressive changes in a tumor to which the composition is not directly
administered.
Alternative 1250 includes the use of any one of Alternatives 1238-1249, in
which the
composition is for use in eradicating a tumor to which the composition is not
directly
administered. Alternative 1251 includes the use of any one of Alternatives
1238-1250, in
which the composition is for use in direct administration to a first tumor,
and is further
use in inducing regressive changes in a second tumor to which the composition
was not
administered. Alternative 1252 includes the use of Alternative 1251, in which
the
tumor comprises a contralateral or metastatic tumor different from the first
tumor.
Additional supporting information
[0561] We have
previously noted that immunoregulating neo-structures from
albumin can be generated by proteolytic fragmentation (see W003099312A1,
hereby
incorporated by reference in its entirety herein) or denaturation (see
W006043891A1,
221
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
hereby incorporated by reference in its entirety herein). In addition to this
observation,
without being limited by any theory, it is contemplated that tumour cells
under stress can
have the capacity for albumin uptake via endosomes in order to increase their
energy
supply and also provide them with amino acids.
[0562] It is
contemplated that compositions comprising immunoregulatory
inhibitors in accordance with some embodiments herein can be administered to
subjects
with cancers, especially tumors, and can facilitate an immune response (for
example, by
permiting or enhancing) against the cancer and/or tumor. Without being limited
by any
theory, it is contemplated that some immunomodulators, for example
immunoregulatory
inhibitors, in accordance with some embodiments herein can facilitate an
immune
response against cancer and/or tumors via two modes of action: (1)
counteracting P3028
structures, for example by binding and/or displacing P3028 so as to remove
P3028-
mediated inhibition of immune cells receptors such as the LFA-1 receptor
and/or the IL-2
receptor; and (2) immunostimulatory activity of the immunoregulatory inhibitor
itself.
For example, P28R has been observed to exhibit both modes of action.
[0563] As such,
in some embodiments, a composition comprising a peptide of
SEQ ID NO: 2, SEQ ID NO: 62, SEQ ID NO: 584, a peptide listed in Table 5.4, or
a
modified P28R or P28 core peptide comprising one or more modifications listed
in Table
5.3 or Table 13 is provided. Optionally, the peptide is immobilized on a
nanoparticle as
described herein. Optionally, the composition has effects in counteracting
P3028
structures and immunostimulatory activity. Optionally, the composition has
effects in
counteracting P3028 structures, but does not have direct immunostimulatory
activity.
Optionally the composition is administered to, or is for use in administering
to, a subject
suffering from a cancer and/or a tumor, for example a prostate tumor, a
melanoma, a
colon cancer, a lung carcinoma, an Apocrine gland carcinoma, a testis tumor, a
mast cell
tumor, a mammary tumor (e.g. a benign mammary tumor or a malignant mammary
for example a mixed mammary tumor such as a benign mixed mammary tumor or a
malignant mixed mammary tumor), a mucinous carcinoma (e.g. a mammary gland
mucinous carcinoma), or a histicytoma and/or cancer cells associated
therewith.
Optionally, the composition is directly administered to the cancer
cells/tumor, for
example via intra-tumoral injection, implantation of a capsule or infuser
comprising the
composition, or topically. Optionally, administration of the composition
induced
cell infiltration of the tumor, for example, infiltration by CD45+
inflammatory cells
222
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
and/or CD56+ NCR1+ NK-cells. Optionally, if the composition is administered
directly
to the cancer cells and/tumor, the composition further induces immune cell
infiltration of
at least on additional tumor of the subject, for example a contralateral tumor
and/or
cells at locations that are different or removed from the site of
administration.
[0564] As
outlined in Table 14, without being limited by any theory, it is
contemplated that the Beta2-integrin, LFA-1, can be involved in multiple
immune
functions.
Table 14: Involvement of the Beta2-integrin LFA-1 in multiple immune functions
Function Comments Ref, e.g.
Th1-Th2 Immunostimulation without LFA-1 Smits et al. J Immunol. 2002
Feb
balance function results in Th2 polarization 15;168(4):1710-6; Salomon
et al., J
and reasonably also in macrophage Immunol. 1998 Nov 15;161(10):5138-
M2 polarization 42; Varga et al., J Invest
Dermatol.
2010;130(4):1005-12.
Initiation of an LFA-1 plays a pivotal role in the Jo et al., J Cell
Biochem.
immune immune synapse between antigen 2010;111(5):1125-37; Zheng et
al., J
response presenting cells and T--cells Biol Chem. 2009;284(32):21280-
7;
Graf et al., J Immunol.
2007;179(3):1616-24.; Marwali et al.,
J Immunol. 2004;173(5):2960-7.
Recruitment of Binding LFA-1 to ICAM-1 on Shulman et al., Immunity. 2009;
inflammatory vascular endothelium is a 30(3):384-96; Borthwick et al.,
Clin
cells to prerequisite for transmigration of Exp Immunol. 2003;
134(2):246-52;
tumours inflammatory cells into tissues/ Ding et al., J Leukoc Biol.
2001;
tumours. 69(3):458-66.
Cell migration Migration of inflammatory cells Verma et al., J Cell
Physiol. 2011;
depends on the function of 226(6):1489-98; Smith et al.,
Immunol
adhesion molecules, LFA-1 plays a Rev. 2007; 218:135-46.
major role in this context
Cytotoxic LFA-1 is required for binding of Perez et al., Blood.
2004;104(4):1083-
activity of T- effector cells to target cells in order 93; Suttman et al.,
Urol Res.
cells and NK- to achieve tumour cell lysis 2002;30(4):233-9.; Luo et
al., J
223
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
cells Hepatol. 1999; 31(1):110-6.
[0565] Table
15, below, summarizes features of modified peptides in
accordance with some embodiments herien, for example modifications of P28R.
The
indicated "changed positions" are with reference to P28R (SEQ ID NO: 2).
Table 15: Features of Modified peptides
Peptide designation Changed Peptide designation Changed
(see, e.g. Figure 41A) position (see, e.g. Figure 41B) position
30677 F13 --- M13 30684 K2 --- S2
30678 Sll --- Q11 30685 El5 --- F15
30680 F6 --- V6 31135 K9 --- Y9
F7 --- M7 31136 K9 --- N9
S11 - Q11 31138 K9 --- D9
F13 --- M13
Summary of additional supporting information
[0566] Five
spontaneous tumours have been treated intra-tumorally with
P28R in accordance with some embodiments herein. In all of these, a strong
inflammatory infiltrate was observed, mainly characterized as CD45+ cells and
NK cells
stained by antibodies directed against CD56 and NCR1. Extensive tumour
regressive
changes were found in three of these and in one, the apocrine gland carcinoma,
with
thick tumour nodules, regressive changes were seen at least in thin lesions
and at the
periphery of the tumour nodules. The thick tumour nodules were, however,
heavily
infiltrated by NK-cells. Interestingly, in a breast tumour with regional
metastases, also
these lesions were heavily infiltrated with inflammatory cells and showed
extensive
tumour regressive changes. Two tumours were injected with the vehicle, in one
of these,
a breast tumour, a spontaneous inflammatory infiltrate was found. The other, a
testis
tumour, did not show any inflammatory reaction.
[0567] So far
nine dogs have been treated with P28R in accordance with
embodiments herein, 4 in the toxicological study (CiToxLab, Denmark) with
200nM
administered in 1 mL subcutaneously and 5 dogs in the treatment study reported
here
224
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
40nM in 200 microliters intra tumourally. None of these dogs showed any
systemic side
effects.
[0568] A class
of immunoregulatory substances is provided in accordance
with some embodiments herein.
[0569] A
general mechanism, whereby this class of immunoregulatory
fragments are produced, has been identified in accordance with some
embodiments
herein.
[0570] The
enhanced proteolytic activity or enhance capacity to denature
proteins in malignant tumours generates neostructures of normally occurring
serum
proteins such as albumin and immunoglobulin in accordance with some
embodiments
herein.
[0571]
Neostructures with both stimulatory and inhibitory immunoregulatory
activity have been found in accordance with some embodiments herein.
[0572] This
class of immunomodulatory substances in accordance with some
embodiments herein can comprise targets for immunomodulation in cancer and
inflammatory diseases.
[0573] A potent
inhibitory peptide, P3028, blocking the proliferative response
to IL-2, NK-cell cytotoxicity, T-cell receptor stimulation, leukocyte
spreading and
lymphocyte migration was developed in accordance with some embodiments herein.
[0574] The
structure of P3028 has been characterized in accordance with
some embodiments herein.
[0575] P3028
binds to LFA-1 and CD25 in accordance with some
embodiments herein.
[0576] Affinity
purified antibodies to P3028 in accordance with some
embodiments herein reverse the suppressed proliferative response to IL-2 in a
culture
model where the response to IL-2 has been shown to correlate to overall
survival.
[0577] A low
molecular weight immunoregulatory inhibitor peptide of P3028
has been developed, P28R, in accordance with some embodiments herein.
[0578] The
capacity of the P28R immunoregulatory inhibitor peptide of
P3028 to reverse suppressed IL-2 induced proliferation of PBMCs from cancer
patients
in accordance with some embodiments herein was demonstrated.
[0579] The
distribution of P3028 and binding of P28R in tumour tissue have
been studied in accordance with some embodiments herein.
225
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
[0580] The
capacity of P28R to unblock LFA-1 was demonstrated in
accordance with some embodiments herein.
[0581] The P28R
has a strong immunostimulatory activity in accordance with
some embodiments herein as shown, for example, in a human ex vivo model.
[0582] In vivo
administration of P28R intra-tumorally in immunocompetent
mouse models elicits in an extensive inflammatory reaction resulting in tumour
cell
eradication in accordance with some embodiments herein.
[0583] In vivo
administration of P28R subcutaneously in immunocompetent
mouse models elicits in an extensive inflammatory reaction resulting in tumour
cell
eradication in accordance with some embodiments herein.
[0584] In vivo
administration of P28R intra-tumorally in spontaneous tumours
in dogs elicits in an extensive inflammatory reaction resulting in tumour cell
eradication
in accordance with some embodiments herein.
[0585]
Systemic, SC, administration of P28R, in accordance with some
embodiments herein, is as efficient as intra-tumoural administration.
Materials and methods
[0586] Except
when stated otherwise, the following materials and or methods
were used as appropriate in the Examples provided below.
Human serum
[0587] Human
serum was collected in serum collection tubes without
additives (Vacutainer, Becton Dickinson, Franklin Lakes, NJ) at the same time
as blood
samples for isolation of PBMC. The sera were heat-inactivated at 56EC for 30
minutes.
Isolation of PBMC's
[0588] To
isolate PBMC' s, venous blood was drawn from healthy volunteers
or from cancer patients in glass vacuum tubes with acid dextrose citrate
solution A as
anti-coagulant (Vacutainer, Becton Dickinson, Franklin Lakes, NJ).
Erythrocytes were
removed by sedimentation on 2% dextran T500 solution (Amersham Pharmacia
Biotech
AB, Uppsala, Sweden) in 0.9% NaC1 (this step was omitted for cultures with PHA-
stimulation ¨ see below). PBMC were then isolated by Ficoll-Paque Plus (GE
Healthcare
Bio-Sciences AB, Uppsala, Sweden) density gradient centrifugation after which
the cells
226
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
were washed twice in RPMI 1640 Dutch's modification (Gibco, InVitrogen AB,
Stockholm, Sweden) with 2% human serum albumin (HSA) (Pharmacia & Upjohn,
Stockholm, Sweden) (RPMI/2%HSA). For cell cultures with PHA-stimulation, PBMC
were washed in Hank's Balanced Salt Solution (HBSS) with 10% autologous plasma
instead of RPMI/2%HSA. Cell viability was assessed by exclusion of 0.05%
Trypan
and was always above 95%. The cell suspension was stained with Ttirk's
solution and
number of lymphocytes and monocytes in the PBMC preparation were counted in a
hemocytometer. PBMCs were suspended in RPMI/2%HSA and the cell concentration
adjusted to 5 x 105 lymphocytes/ml.
IL-2 induced proliferation of PBMC in uncoated and coated culture plates
[0589] Pre-
coating of culture plates with HSA and HSA/IgG. Round-
bottomed, 96-well tissue culture plates (Costar, Corning Inc. NY, US) were pre-
coated
with HSA only or HSA and pooled human IgG for intravenous injection
(Gammagard,
Baxter AS, DK). HSA was diluted in RPMI1640 without supplements to a
concentration
of 10 mg/ml. In some experiments, 1 mg/ml IgG was mixed into a solution of 9
mg/ml
HSA in RPMI (HSA/IgG). 200 pi of HSA or HSA/IgG were then added to each well
of
the plate. The plates were incubated at 4 C for 30 minutes after which the
wells were
washed twice with 200 pl of RPMI1640. The coated plates were used immediately.
[0590] 100 pl
of RPMI1640 supplemented with 200 IU/ml penicillin, 200
pl/ml streptomycin, 4 mM L-glutamine (all from Sigma Chemical Co. MO, US) and
20%
heat-inactivated human serum (autologous or from cancer patients) were added
to
uncoated, HSA or HSA/IgG coated tissue culture microtiter plates. PBMC,
isolated from
healthy individuals or patients with metastatic renal cell carcinoma, were
diluted in
RPMI/2%HSA at a concentration of 5 x 105/m1 and 100p1 were added to the
microtiter
wells. Interleukin-2 (IL-2, Proleukin, Chiron, NL), at a final concentration
of 120
IU/well, was added to some wells. Cells were cultured for 7 days in a
humidified, 5%
CO2-atmosphere at 37 C. Proliferation was assayed by incorporation of 1.6
pCi/well of
31-11-thymidine (Amersham Int., UK) during the last 18 hrs. Mean values of dpm
(disintegrations per minute) of triplicates were used for the calculations.
227
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
Interleukin-2 (IL-2) induced proliferation of PBMC in the presence of albumin
peptides
[0591] Cultures
for IL-2 induced proliferation was set up with PBMC from
healthy donors and autologous serum as described above with the exception that
PBMC
were first pre-incubated for 30 mm at room temperature with the indicated
albumin
peptides at a concentration of 10 pg/ml.
Interleukin-2 (IL-2) induced proliferation of PBMC in the presence of albumin
peptides
in coated and uncoated tissue culture plates
[0592] Round-
bottomed, 96-well tissue culture plates (Costar, Corning Inc.
NY, US) were pre-coated with HSA only or HSA and pooled human IgG for
intravenous
injection (Gammagard, Baxter AS, DK) as follows; HSA was diluted in RPMI1640
without supplements to a concentration of 10 mg/ml. A mixture of 1 mg/ml IgG
in a
solution of 9 mg/ml HSA in RPMI (HSA/IgG) was also prepared. 200 pl of HSA or
HSA/IgG were then added to each well of the plate. The plates were incubated
at 4 C
for 30 minutes after which the wells were washed twice with 200 pl of
RPMI1640. The
coated plates were used immediately. 100 pl of RPMI1640 supplemented with 200
IU/ml penicillin, 200 p1/ml streptomycin, 4 mM L-glutamine (all from Sigma
Chemical
Co. MO, US) and 20% heat-inactivated human serum (autologous) were added to
the
HSA or HSA/IgG coated tissue culture microtiter wells. PBMC, isolated from
healthy
individuals, were diluted in RPMI/2%HSA and peptides were added directly to
the cell
suspension at a concentration of 10 pg/ml. One hundred pl of this cell
suspension (5x104
lymphocytes) was then added per well providing a final concentration of 5
pg/ml peptide
per well. IL-2 (Proleukin, Chiron, NL), at a final concentration of 120
IU/well, was
added to the wells. Cells were cultured for 7 days in a humidified, 5% CO2-
atmosphere
at 37 C. Proliferation was assayed by incorporation of 1.6 pCi/well of 113H1-
thymidine
(Amersham Int., UK) during the last 18 hrs. Mean values of dpm
(disintegrations per
minute) of triplicates were used for the calculations.
Albumin peptides
[0593]
Synthetic albumin peptides were custom prepared by CSBio Co,
Park, CA. Peptides were > 95% pure as confirmed by HPLC. Peptides were kept
freeze
dried at minus 20 C. Peptides were reconstituted in sterile H20 (Sigma) for
use in
ELISA or in RPMI1640 (GIBCO) for use in cell culture experiments. Peptides
were
228
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
sterile filtered through a 0.22 pm syringe filter (Millipore Co) before use in
cell culture
experiments.
ELISA for the detection of murine antibodies binding to human albumin
[0594]
Duplicate wells in Hibinding microtitre plates (Costar 2592, Corning
Inc, NY, USA) were coated with 100 1 of dHSA diluted in PBS at various
concentrations or, alternatively, control albumin sample at the same
concentrations. The
plates were incubated at room temperature overnight. The wells where then
washed with
wash buffer consisting of 0.05% Tween-20 in PBS (Sigma) followed by blocking
for 1 h
at 25 C with 200 1 0.1% gelatin prepared from bovine skin (Sigma) in PBS
followed by
washing in wash buffer. Either of two murine monoclonal antibodies (IgG1) with
specificity for denatured, human albumin (anti-dAbc1h040801 or anti-
dAlbc1h040809)
was added at 1 lig/m1 in ELISA reagent diluent (0.01% gelatin (Sigma) and
0.05%
Tween-20 (Sigma) in 20 mM Tris-buffered saline (TBS, Sigma)). The antibodies
were
incubated for 1.5 h at 25 C followed by washing. Envision-HRP (DakoCytomation
Norden A/S, Glostrup, Denmark) was added diluted 1/5 to 1/10 in ELISA reagent
diluent
and incubated for 30 mm at 25 C followed by washing. Finally, a substrate
solution
consisting of H202 and tetramethylbenzidine (R&D Systems Europe, Ltd,
Abingdon,
UK) was added. The reaction was stopped with 1M H2504 and the optical density
measured as absorbance (Abs) at dual wavelengths, 450 nm and 570 nm, with a
Multiscan EX microplate reader (Labsystems).
ELISA with rabbit-anti 3028 antiserum
[0595]
Duplicate wells in Hi-binding microtitre plates (Costar 2592, Corning
Inc, NY, USA) were coated with 100 pl of P3028 (10 ug/ml), denatured HSA
(denHSA,
4.5 ug/ml) or control HSA sample (4.5 ug/ml). All coating reagent were diluted
in PBS
and incubated at room temperature overnight. The wells where then washed with
wash
buffer consisting of 0.05% Tween-20 in PBS (Sigma) followed by blocking for 1
hr at
25 C with 200 pl 0.5% gelatin prepared from bovine skin (Sigma) in PBS
followed by
washing in wash buffer. Rabbit preimmune sera or anti-3028 sera, diluted
1/1000 000 in
ELISA reagent diluent (0.01% gelatin and 0.05% Tween-20 in PBS), were added
and
incubated for 1 h at 25 C followed by washing. Biotinylated horse anti-
rabbit/mouse IgG
(Vectastain ELITE, Vetor Laboratories Inc, CA, USA) diluted 1/5 in ELISA
reagent
229
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
diluent was then added and the plates incubated for 1 h at 25 C followed by
washing.
Next, HRP-conjugated strreptavidine (R&Dsystems Europe, Ltd, UK) was added.
Finally, after washing in wash buffer, substrate solution consisting of H202
and
tetramethylbenzidine (R&D Systems) was added. The reaction was stopped with 1M
H2504 and the optical density measured as absorbance (A) at dual wavelengths,
450 nm
and 570 nm, with a Multiscan EX microplate reader (Labsystems).
Statistical considerations
[0596]
Comparisons of the means of different patient groups or different test
occasions were performed using an unpaired t-test. Time to progression and
survival was
analyzed using the Kaplan-Meier method and Logrank test.
[0597]
Comparisons between the proliferative response to PHA in different
groups or at different test occasions were done on logarithmated mean values
of dpm of
triplicates using unpaired t-test. For the determination of the effect of
addition of CHL
on the proliferative response of PHA-stimulated PBMCs, a modulation index (MI)
was
calculated according to the following formula: MI = log (dpm PHA + drug/dpm
PHA).
Example 1: Serum peptides with immune inhibitory activities
Identification of immunoregulatory peptides
[0598] An
artificial cell surface (ACS) was prepared by selectively
biotinylating cell surface structures of PBMCs and after lysing the cells
binding the
biotinylated proteins to streptavidin columns (see Example 17 for further
description of
the ACS). The mixture of peptides obtained after trypsination was adsorbed by
ACS and
the binding peptides were identified by comparing adsorbed and unabsorbed
peptide
solutions using the MALDI TOF ms technique. Based on their degree of binding
and
their spatial relation to previously identified immunoregulatory structures,
four new
peptides were selected to be synthesized and investigated for their
immunoregulatory
activity, primarily the effect on the proliferative response to IL-2. One of
these peptides,
P3028 (SEQ ID NO: 185) was found to have multiple immunoinhibitory activities.
Expression of the P3028 epitope in malignant tumors
[0599] Rabbit
polyclonal antibodies against P3028 were generated and affinity
purified (see Example 9). To determine the localization of P3028 in tumor
cells, sections of
230
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
malignant metastases were immunostained using the anti-P3028 rabbit polyclonal
antibodies.
Tissue sections were prepared from formalin fixed biopsies from cancer
patients. Sections
were de-paraffinased and blocked with 10% normal, human AB-serum in Hank's
balanced
salt solution supplemented with 0.01 M Hepes (BSS, GIBCO BRL) for one hour
prior to
staining. Sections were then stained with 10 ug/ml affinity purified rabbit
anti-P3028 diluted
in BSS with 2% AB-serum and 0.1 g/ml saponin for 30 min. After washing in BSS
with 0.1
g/ml saponin, Ultravison One alkaline phosphatase polymer specific for mouse
and rabbit Ig
(Lab Vision Co., CA, USA) was added. Excess polymer was then washed from the
sections
with BSS with 0.1 g/ml saponin. Bound polymer complex was the detected by
naphthol
phosphate substrate and liquid Fast Red chromogen (Lab Vision Corp.) The
sections were
counter stained in Mayer's haematoxylin and mounted in Glycergel. As shown in
Figure 1,
structures 1 to which anti-P3028 antibodies bind are widely expressed in human
malignant
tumors, e.g., malignant melanoma, renal cell carcinoma and colorectal cancer.
[0600] Western
blotting was performed on extracts of malignant melanoma
metastases to detect the presence of P3028 structures. Western blotting was
performed
using standard techniques, and P3028 structures were detected using affinity
purified
Rabbit polyclonal antibodies against P3028 (see Example 9). P3028 structures
in tumor
extracts from malignant melanoma metastases were identified in the extracts of
7 out of
7 mestases from 4 patients that were screened (see Figure 2). The P3028
peptide was
present in all patients. Additionally, the P3028 structure was present in full-
length
albumin. In addition this structure was found in larger molecules. These
results are
compatible with the P3028 structure being generated not only by proteolytic
fragmentation but also by denaturation.
Occurrence of P3028 structures in serum
[0601]
Substances exposing the structure of P3028 were determined in human
serum by using affinity purified antibodies in a sandwich ELISA. That is, the
ability to
detect P3028 structures in human serum was confirmed.
[0602] A
sandwich ELISA was performed to detect albumin exposing the
P3028 epitope in serum as follows: An affinity polyclonal purified rabbit
antisera,
specific for human albumin P3028, was coated onto high protein binding ELISA
microwells (capture antibody; see Example 9). A 1% solution of heat-
inactivated serum
(from a serum pool of 5 healthy control samples, 1 healthy control serum
sample and 2
231
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
sera obtained from cancer patients), spiked with increasing concentrations of
P3028, was
then added to the wells. After washing, a biotinylated mouse anti-human
albumin
monoclonal antibody was added and the amount of bound antibody was detected
with
HRP-conjugated streptaviddin and TMB chromogen substrate. (One representative
experiment out of two is shown Figure 3).
[0603] The
amount of P3028 structures were determined as the amount of
P3028, which inhibits 50% of the binding of P3028 structures in the serum to
the capture
antibody (see Figure 3). The serum concentration was determined to be in the
range of
1.2-1.6pg/m1 P3028 equivalents in one serum pool from 5 healthy control
samples, 1
healthy control serum sample and 2 sera obtained from cancer patients. The
amount of
these P3028-substances in serum can be considerably more as the molecular
weight of
albumin is about 35 times more than that of P3028. The epitope specific
reactivity of
P3028-substances was accurately determined using the methods of this Example.
Example 2: Effect of ACS-identified peptides on IL-2 induced proliferation
Human ex vivo model for immunosuppression in cancer patients
[0604]
Interleukin-2 (IL-2) plays a major role in initiation and activation of an
immune response and its capacity to induce lymphokine activated killer cells
(LAK-
cells), T-cell proliferation and cytotoxicity. Accordingly, a human ex vivo
model of IL-2
stimulation of immune cells was developed. This model was useful for studying
the
effects of immune system modulators, such as P3028, and inhibitors thereof.
[0605] The
model included PBMCs isolated from venous blood samples from
healthy blood donors (control samples) or cancer patients. One hundred pI of
culture
medium (RPMI 1640 Dutch's modification (Gibco, InVitrogenAB, Stockholm,
Sweden)
supplemented with 200 IV/ml penicillin, 200 ug/ml streptomycin, 4 mM L-
glutamine (all
from Sigma Chemical Co. MO, US) and 20% heat-inactivated human serum) were
added
to roundbottomed, 96-well tissue culture plates (Costar, Corning Inc. NY, US).
One
hundred ul of PBMCs in RPMI/2% HSA (5x104 lymphocytes) was then added per well
followed by IL-2 (Proleukin, Chiron, NL) at a final concentration of 120
IU/well.
sample wells without IL-2 was set up in parallel. Cells were cultured for 7
days in a
humidified, 5% CO2- atmosphere at 37 C. Cell proliferation was assayed by
incorporation of 1.6 pCi/well of 113H1-thymidine (Amersham Int., UK) during
the last 18-
232
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
24 h hrs. Mean values of dpm (disintegrations per minute) of triplicate wells
were used
for the calculations.
[0606] IL-2
induced proliferation by PBMC from healthy control samples and
PBMC from renal cell carcinoma patients (RCC) cultured in 10% autologous sera
was
studied using this model. Results of the study are shown in Figure 4. IL-2
induced
proliferation was significantly reduced (p <0.0002) for PBMC's cultured in
serum of a
renal carcinoma patient as compared to a healthy control sample.
Correlation between IL-2 response in ex vivo model and overall survival of
renal
cell carcinoma patients
[0607] The
response to IL-2 in this model was demonstrated to correlate to
overall survival of renal cell carcinoma patients. Patients, included in the
analyses of over-all
survival according to proliferative response of PBMCs to interleukin-2, were
diagnosed with
systemic metastatic renal cell carcinoma. They were previously untreated and
scheduled for
Interleukin-2 treatment (Proleukin, Chiron, NL). Blood samples were taken
prior to initiation
of treatment. Survival curves were plotted using the method of Kaplan and
Meier and time to
progression and survival comparisons between subgroups were performed using
the log rank
test. In addition, the prognostic significance of the level of LPS-stimulated
IL-6 production
was also calculated using Cox regression.
[0608] Figure 5
illustrates a Kaplan Meyer analysis of renal cell carcinoma
patients according to proliferative response to IL-2. Patients were classified
as having a
proliferative response of > 30,000 dpm 52, 15,000-30,000 dpm 54, or <15,000
dpm 56. A log
rank analysis we performed, and overall patient survival correlated with
proliferative
response (p=0.0042). As illustrated in Figure 5, patients with the lowest IL-2
induced
proliferation of PBMCs in autologous serum in the ex vivo model 56 also had
the lowest
overall survival time. Thus, a low proliferative rate indicates a poor
survival.
Effect of different peptides on IL-2 induced proliferation
[0609] The
effect of different peptides on IL-2 induced proliferation was
analyzed in the human ex vivo model, using PBMCs from healthy control samples.
PBMCs
were cultured for 7 days in the presence of IL-2 (20 U/ml) and the peptides. A
control
sample was also performed in which no peptide was added ("None").
Proliferation was
measured as incorporation of 3H-thymidine during the final 18 hours. The
peptides
233
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
included P3026 (SEQ ID NO: 183), P3027 (SEQ ID NO: 184), P3028 (SEQ ID NO:
185),
and P3029 (SEQ ID NO: 186). One of the peptides, P3028, regularly inhibited IL-
2 induced
proliferation (p < 0.0006, as compared to control sample; n=17), but none of
the other
peptides identified by their binding to the artificial cell surface had any
inhibitory activity
(For P3026, P3027, P3029 n= 4 or 5). Figure 6 illustrates the analysis of the
effect of
four different peptides.
[0610] The
inhibition of the proliferative response to IL-2 by P3028 was also
observed for cancer patient PBMCs studied in the human ex vivo model. The ex
vivo model
of IL-2 stimulation was constituted using the PBMCs of a cancer patient, and
IL-2 stimulation
was compared in the presence and absence of P3028. As illustrated in Figure 7,
the
inhibitory activity of P3028 on IL-2 induced proliferation can be demonstrated
also in
cultures with cancer patient PBMCs, even if the response to IL-2 was already
suppressed
(see Figure 7).
Example 3: Effect of P3028 on T-Cell Receptor Stimulation
[0611] To
examine the effects of P3028 on T cell receptor stimulation, Blood
for PBMC isolation was provided from healthy control samples in 50m1
transfusion bags
with acid dextrose citrate solution A. .Whole blood was diluted 1:1 in PBS
containing
2mM EDTA. PBMCs were then isolated by Ficoll-paque Plus (GE Healthcare Bio-
Sciences AB, Sweden) density gradient centrifugation after which the cells
were washed
first in PBS with 2mM EDTA and second in lymphocyte culture media. Cell
viability
was assessed by exclusion of 0.02% Trypan Blue and was always above 95%. The
cell
suspension was counted in a haemocytometer. PBMCs were suspended in the
culture
medium without sera and the cell concentration adjusted to 1x106
lymphocytes/ml for
proliferation assays and 6.4x105 for migration assays respectively. The
lymphocyte
culture medium RPMI 1640 (Invitrogen, Sweden) was complemented with 1%
Penicillin/Streptomycin (Invitrogen, Sweden) and 4mM Gluta-Max (Invitrogen,
Sweden).
For CD3 induced proliferation the plates were coated with purified anti-human
CD3
antibodies (BD Pharmingen, Sweden). Therefore 50 Ill of 2.51.1g/m1 antibody
PBS
solution were pipetted into each well incubated for 1 hour. Cells were
cultured for 4, 5
7 days in a humidified, 5% CO2-atmosphere at 37 C. Cell proliferation was
assayed by
the mitochondrial activity test CellTiter 96 AQueous Non-Radioactive Cell
Assay (MTS, Promega, Sweden) during the last 4 hours. To each well 10111 of
the MTS
234
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
solution was added and measured after 4 hours of incubation at 37 C. The
measured
values of the reference dye were subtracted of each well. The peptide
solutions were
prepared by dissolving peptides 3028, SCF28R, 28209 and SCF27 (Schafer-N,
Copenhagen, Denmark) in lymphocyte media to a concentration of 25 g/ml. The
final
concentration in the cultures was 5 or lOug/ml.
[0612] T cells
were stimulated in cultures on plates pre-coated with a
monoclonal antibody directed against CD3 and the number of metabolically
active cells
(i.e., cell proliferation) was determined using MTS staining after 3 to 7 days
of culture.
Detection of solid phase CD3 monoclonal antibody was used as a measurement of
T cell
proliferation. Figure 8 illustrates the effect of P3028 on TCR stimulated
lymphocyte
proliferation of PBMCs from four healthy persons. For each person,
proliferation of
lymphocytes was measured in the absence of stimulation 82, IL-2 stimulation
84,
treatment with P3028 alone 86, and IL-2 stimulation plus P3028 88. Bars of the
bar
graph of Figure 8 are in the same order for each person.
[0613] As can
be seen in Figure 8, P3028 had an inhibitory effect in at least
three out of four experiments (p<0.001). It is unlikely that reduced MTS
staining caused
by P3028 was be due to a reduced cell metabolism. Taken together, the results
from both
models of lymphocyte proliferation, a reduced metabolism should reasonably
reduce the
endogenous thymidine pools and thereby result in an increased uptake of
exogenous
thymidine/ specific activity of the thymidine pools, which then should be
erroneously
registered as an enhanced proliferation. The 3H-TdR was actually reduced in
these
experiments, indicating inhibition of proliferation.
Example 4: Effect of P3028 on NK-Cell Cytotoxicity
[0614] The NK-
cell cytotoxic activity of blood mononuclear cells from four
healthy donors was tested. Mononuclear cells were separated by standard Ficoll-
paque
Plus (Pharmacia AB, Sweden) density gradient centrifugation from heparinized
blood
obtained from healthy donors. NK cell cytotoxic activity of the mononuclear
cells was
then tested using a commercial kit (NKTEST, Orpegen Pharma GmblI, Heidelberg,
Germany) following the manufacturers protocol. Briefly, the kit contains
cryopreserved,
NK-sensitive target cells (K562) labeled with a lipophilic green fluorescent
membrane
dye, which enables discrimination of effector and target cells. After
incubation with
effector cells, killed target cells are identified by a DNA-stain, which
penetrates and
235
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
specifically stain the nuclei of dead target cells. This way the percentage of
killed targets
can be determined by flow cytometry. The mononuclear cells were preincubated
for 30
min at 37 C with the indicated peptides (peptides have been described
previously) at 10
ug/ml. Target cells were then added, giving an effector:target ratio of 40: 1,
and the cell
mixture incubated at 37 C for 3-4 hours. Samples were analysed on a
FACSCalibur (BD
Biosciences, San Jose, Calif.).
[0615] Figures
9A-B illustrate the effect of albumin peptides on NK-cell
cytotoxicity (p = 0.015, paired t-test, normal transformation log-values). As
shown in
Figure 9A-B, the presence of P3028 and, to a lesser degree, peptide 3026
reduced the
per cent specific lysis of K562 target cells by all four donors. Inhibition
was not seen in
the presence of the control sample peptide 3027 with no structural
relationship with
P3028. Inhibition of NK-cell cytotoxicity, in this model, was not due to an
effect of
P3028 on the activity of IL-2 as no IL-2 was added to the short-term cultures.
Example 5: Effect of P3028 on Leukocyte Spreading and Immune Cell Migration
[0616] In
properly functioning immune systems, immune cells are recruited to
tissues, and migrate within tissues. The effect of P3028 in two functional
tests,
leukocyte spreading and immune cell migration was investigated.
Leukocyte spreading
[0617] To
analyze the effect of P3028 on leukocyte spreading, buffy coat
cells were prepared from heparinized blood by Dextran assisted sedimentation.
These
cells were then washed twice in PBS and transferred to slides washed in 70%
and 96%
ethanol. The cell suspension was dropped onto the slides and incubated for 15
mm in a
moist chamber with or without P3028, 10pg/ml, the solution was carefully
drained off,
the slides were air dried and stained in May Grilnewals Giemsa for 1 minute.
As shown
in Figure 10A, the cells strongly adhered to the glass surface and spread out.
Pre-
treatment of these cells with P3028 efficiently inhibited the spreading (see
Figure 10B).
Immune cell migration
[0618] Blood
for PBMC isolation was provided from healthy control samples
in 50m1 transfusion bags with acid dextrose citrate solution A. Whole blood
was diluted
1:1 in PBS containing 2mM EDTA. PBMCs were then isolated by Ficoll-paque Plus
236
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
Healthcare Bio-Sciences AB, Sweden) density gradient centrifugation after
which the
cells were washed first in PBS with 2mM EDTA and second in lymphocyte culture
media. Cell viability was assessed by exclusion of 0.02% Trypan Blue and was
always
above 95%. The cell suspension was counted in a haemocytometer. PBMCs were
suspended in the culture medium without sera and the cell concentration
adjusted to
lymphocytes/ml for proliferation assays and 6.4x105 for migration assays
respectively.
The lymphocyte culture medium RPMI 1640 (Invitrogen, Sweden) was complemented
with 1% Penicillin/Streptomycin (Invitrogen, Sweden) and 4mM Gluta-Max
(Invitrogen,
Sweden). For CD3 induced proliferation the plates were coated with purified
anti-human
CD3 antibodies (BD Pharmingen, Sweden). Therefore 50 ul of 2.5 g/m1 antibody
PBS
solution were pipetted into each well incubated for 1 hour. Cells were
cultured for 4, 5
7 days in a humidified, 5% CO2-atmosphere at 37 C. Cell proliferation was
assayed by
the mitochondrial activity test CellTiter 96 AQueous Non-Radioactive Cell
Proliferation
Assay (Promega, Sweden) during the last 4 hours. To each well 10111 of the MTS
was added and measured after 4 hours of incubation at 37 C. The measured
values of
reference dye were subtracted of each well. The peptide solutions were
prepared by
dissolving peptides 3028, SCF28R, 28209 and 5CF27 (Schafer-N, Copenhagen,
Denmark) in lymphocyte media to a concentration of 25 g/ml. The final
concentration
the cultures was 5 or lOug/ml.
[0619] 50u1 of
the prepared 6.4x105 PBMC dilution were pipetted into
Eppendorfs tubes and centrifuged for 5 minutes at 400g, then the prepared
dilutions of
blank, P3028 and the inhibitors were added. The PBMCs were incubated at the 37
C
with the test substances for one hour. Meanwhile the Boyden Chamber was
prepared by
pipetting 25111 of either media without fMLP or media containing 1x10-8M fMLP
to the
lower wells. Then 50u1 of the PBMCs final concentration, 3,2x104, were
transferred to
the upper wells of the chamber. The PBMCs were allowed to migrate for one hour
at the
37 C. The filters were removed and stored in 70% ethanol overnight. Thereafter
the
filters were dehydrated in increasing alcohol concentration and finally placed
in Xylene.
Subsequently they were placed on slides, mounted and counted with a
microscope,
containing a um scale. Each test was done in duplicates and migration was
calculated as
percentage of the mean of the blank duplicates without fMLP. As shown in
Figure 11,
P3028 is a potent inhibitor of immune cell migration across the membrane of
the Boyden
chamber (p<0.002). Migration for healthy control samples (N=6) is illustrated
in Figure
237
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
11 using dark bars (left), while cancer patients (N= 3) are shown as light
bars (right). In
Figure 11, Error bars: 95% CI. P3028 reduced the migration of PBMCs of both
healthy
cells and cancer patients.
Example 6: Further Characterization of the Effect of P3028 on IL-2 Induced
Proliferation
[0620] The C
and N-terminal parts of P3028 were synthesized and analyzed
separately and in combination. The inhibitory activity of these two parts of
P3028 alone
or in combination is much weaker (see Figure 12) and they do not inhibit the
effect of
P3028 on IL-2 induced proliferation (see Figure 13) in the ex vivo human
model. Figure
12 illustrates effects of the C- (P3218) (SEQ ID NO: 187) and N-terminal
(P3325) (SEQ
ID NO: 186) parts of P3028 on 11-2 induced proliferation in comparison with
the effect
of the full length P3028. One representative experiment is shown. Figure 13
illustrates
that the inhibitory effect of P3028 on IL-2 induced proliferation is not
neutralized by the
C- (P3218) and N-terminal (P3325) parts of P3028 alone or in combination.
Example 7: Binding of P3028 to LFA-1
[0621] The
presence of 02-integrins on PBMCs was demonstrated by
immunocytochemical staining. The occurrence of factors interfering with the
binding of
monoclonal antibodies directed against 02-integrins in cancer patient sera was
analysed
staining of 02-integrins on PBMCs. A standard immunohistochemical staining
procedure
using acetone fixation, 10% human AB-serum for blocking, incubation with anti-
LFA-1
antibody. PBMCs were separated as described above and immediately spun down on
cleaned microscope slides in a Shandon Cytospin (Shandon Scientific Ltd, UK)
at 1000
RPM for 7 mm at 5 x 104 cells per slide. The slides were left to dry at room
temperature
over night, after which they were wrapped in parafilm and stored at 70 C.
Immediately
before use, the cytospins were thawed and fixed with acetone for 5 mm at room
temperature. The cytospins were first blocked with 10% normal human AB-serum
with
and without albumin peptides (40 pg/ml) or serum from cancer patients for 1 h
before
staining. Primary antibody, consisting of a monoclonal mouse anti-human CD1 la
(BD
Biosciences) diluted in Tris buffered saline (TBS, pH 7.6) at 1 pg/ml (PBMC),
was
added. The slides were incubated for 30 mm and then washed in TBS followed by
Envision-Alkaline Phosphatase (Dako Norden A/S, Denmark) or, alternatively,
Ultravision-Alkaline Phosphatase (Lab Vision Co) for 30 mm. After additional
washing
238
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
in TBS, the slides were incubated in alkaline phosphatase substrate consisting
of Fast
TR salt (Sigma), naphtol AS-MX (Sigma) and 5 mM levamisol (Sigma) to block
endogenous alkaline phosphatase activity, for 20 mm followed by washing in
TBS. They
were then counterstained in Mayer's haematoxylin for 1 minute and mounted in
(Dako Norden A/S). Monoclonal mouse IgG1 against an irrelevant antigen
(Aspergillus
niger glukosoxidase, Dako Norden A/S) was used as a negative control sample.
All
incubations were performed at room temperature in a moist chamber.
[0622] Pre-
incubation with peptides added to the AB serum was either no
peptide added (see Figure 15A), or P3028 added (see Figure 15B). Notably, the
anti-
LFA-1 antibody used in these experiments was a potent inhibitor of IL-2
induced
proliferation.
[0623] As shown
in Figure 14, the presence of 02-integrin blocking factors
was then demonstrated as a reduced stainability 5 of these cells after
incubation with
cancer patient sera (see Figure 14B), compared to preparations pre-incubated
with
control serum sample (see Figure 14A) which showed strong staining 3 for LFA-
1.
[0624] As shown
in Figure 15, similar to the results described for cancer
patient sera, treatment with P3028 can modulate the binding of the LFA-1
antibody to
LFA-1 of mononuclear blood cells, Figure 15 illustrates inhibition of the
binding of an
anti-LFA-1, mAb, to mononuclear blood cells by P3028. Strong staining 3 for
LFA-1
was observed in cells in which no peptide was added (see Figure 15A), while
weak
staining 5 for LFA-1 was observed in cells in which P3028 was added (see
Figure 15B).
[0625] In order
to further demonstrate the blockade of LFA-1 by the P3028
structure, the staining of this integrin on PBMCs from healthy control samples
and
cancer patients was compared. Figure 16 illustrates staining of LFA-1 on PBMCs
from
a healthy control sample (see Figure 16A), and a cancer patient before (see
Figure 16B)
and after (see Figure 16C) treatment with an antibody directed against P3028.
As shown
in Figure 16A, a clear membrane staining 3 is found on PBMCs from healthy
control
samples in contrast to PBMCs from a patient with advanced cancer, which
exhibited
weak staining 5. However, when the PBMCs from this patient were incubated with
an
antibody directed towards the P3028 structure for 24 hours the membrane
staining
appeared 3, indicating that the antibody bound the P3028-structure and thereby
unblocked LFA-1 (see Figure 16C).
239
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
[0626]
Similarly, as shown in Figure 17, incubation of PBMCs from a
healthy control sample with either P3028 or serum from a cancer patient
blocked the
membrane staining of LFA-1. Figure 17 illustrates staining 3 of mononuclear
blood
cells by an anti-LFA-1 antibody (A) is blocked 5 by P3028 (B) or cancer
patient serum
(C).
Example 8: Binding of P3028 to the cc-chain (CD25) of the IL-2 receptor
[0627] Because
P3028 significantly inhibits the proliferative response to IL-2,
the effect of P3028 on the binding of IL-2 to its receptor, CD25 was studied.
The fusion
protein of CD25 and the Fc-part of IgG was bound to protein G coated micro-
plates /
ELISA plates and the plates were incubated with biotinylated IL-2 with or
without P3028
present. Figures 18-B illustrate the results of this ELISA analysis for
dilution of
biotinylated IL-2 that were as follows: (diamond #) 1:300, (square.) 1:600,
(triangle 1;
see Figure 18B) no biotinylated IL-2. The binding of biotinylated IL-2 to
rhuIL-2R
alpha was increased by increasing amounts of P3028. Surprisingly, the binding
of IL-2
to CD25 was enhanced by P3028, indicating a three-part interaction between IL-
2, CD25
and P3028 (see Figure 18-B). Even if the binding of IL-2 to CD25 is enhanced
the
proper assembly of the high affinity receptor and/or signal transduction is
blocked as
P3028 is a potent inhibitor of IL-2 induced proliferation.
[0628] It was
demonstrated using computer assisted molecular modeling that
P3028 binds to CD25 at the IL-2 binding site (see Figure 19). The crystal
structure of
the IL-2 receptor bound to IL-2 is known in the art (see Wang et al., Science
2005,
310(5751): 1159-1163, and Stauber et al, Proc. Natl. Acad. Sci. USA 2006,
103(8):
2788-2793, each of which is hereby incorporated by reference in its entirety),
and
binding of P3028 was modeled according. In Figure 19, the cc-chain 190 of the
IL-2
receptor (CD25) binding P3028 192 (A) at the IL-2 binding site 194 (B) is
depicted. IL-2
196 is also shown.
Example 9: Antibodies that bind to P3028
[0629] Rabbit
antisera directed against the albumin P3028 were generated.
P3028 was synthesized with a cysteine added to the N-terminus end and then
conjugated
with keyhole limpet hemocyanin (KLH) as a carrier protein. Polyclonal antisera
were
generated by repeated immunizations of rabbits with KLH-conjugated P3028 and
adjuvants. For some experiments, the antisera were affinity purified by
chromatography
240
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
on P3028-conjugated Ultralink Iodoacetyl gels (Pierce Biotechnology Inc.). For
cell
culture experiments, buffer exchange to RPMI 1640 Dutch's modification (Gibco,
InVitrogen AB, Stockholm, Sweden) was performed by passage over PD-10 sephadex
columns (Amersham Biosciences, Uppsala, Sweden) followed by filter
sterilization on
0.22 pm Millex syringe filters (Millipore Co., MA, USA). Rabbit immunizations
and
purification of antisera were carried out by Agrisera AB, Sweden.
[0630] Two
antisera, R and L, from two different rabbits were tested for their
ability to bind human serum and denatured Human Serum Albumin (dHSA). Human
serum albumin commercially available for therapeutic purposes was tested,
heated 10
times in order to be virus free. Wells were coated with the P3028, dHSA, or
control
sample treated (not denatured, but heated 10 times) HSA, which has been
prepared just
as the denatured HSA except for the denaturation procedure. As shown in Figure
20,
antisera, but not preimmune sera, from two rabbits immunized with the albumin
P3028
bind to plates coated with the P3028 204, dHSA 206 and, to a lesser extent, to
control
sample treated HSA 208. No substantial binding was detected for wells with no
coat
202. Thus, rabbit antisera directed against the albumin P3028 binds to dHSA
and to a
lesser extent to control sample HSA.
[0631] The
binding of the rabbit anti-P3028 serum to P3028 fragments was
assayed using competition ELISA assay. Rabbit antisera, diluted 1/1000 000 in
ELISA
reagent diluent, was pre-incubated for 1 hr at room temperature with the
indicated
concentrations of the peptides. 100 pl
of the monoclonal antibody alone, or,
alternatively, the monoclonal antibody mixed with peptides, was then added to
P3028
coated wells and the ELISA carried out. Inhibition of the binding of rabbit
anti-P3028
serum L to wells coated with the P3028 was determined for albumin peptides
2607 (SEQ
ID NO: 192), 3218 (C terminal of P3028) (SEQ ID NO: 187), 3325 (N terminal of
P3028) (SEQ ID NO: 186), and full-length P3028 (SEQ ID NO: 185). Peptide 2607,
containing the E5K structure, was used as a negative control sample. As shown
in
Figure 21, these serum antibodies bound preferentially to the 3325 but not to
the 3218
fragment of P3028. Similar results are also obtained with the affinity
purified antibodies.
[0632] The
effects of affinity purified antibodies directed against P3028 on
proliferative response to IL-2 were studied in the ex vivo model, using PBMCs
from
immunosuppressed cancer patients and normal control samples. Cultures to test
the
immunomodulatory effect of affinity purified rabbit antibodies specific for
3028 were
241
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
performed as described above for IL-2 induced proliferation with the following
exceptions; 2% HSA was omitted from the washing medium and from the PBMC
suspension medium. Serum containing culture medium (100 p1/well) was pre-
incubated
with 20 pg/ml of rabbit antibodies for 30 mm at room temperature before the
addition of
100 pl PBMC suspension to the culture wells.
[0633] P21 had
renal cell carcinoma and p26, p28 and p29 had malignant
melanoma. As shown in Figure 22, affinity-purified rabbit antibodies against
P3028
overcame inhibition of the proliferative response to IL-2 in immunosuppressed
cancer
patients (Figure 22A). In normal control samples with normal proliferative
response to
IL-2, no effect of addition of these antibodies was seen (see Figure 22B)
(antibody: R.,
cancer patients, p = 0.0002, paired t-test, normal transformation log-values).
In normal
control samples with down-regulation of the immune reactivity having a
proliferative rate
of less than 100,000 dpm, the proliferative rate was stimulated similar to the
situation in
cultures from cancer patients.
[0634]
Polyclonal rabbit IgG was added to control sample cultures in order to
make sure that the effect of the affinity purified antibodies was not due to
an unspecific
activity of rabbit IgG in this model. Rabbit IgG had only minimal activity.
The
specificity of the anti-P3028 antibodies was further demonstrated as the
stimulatory
effect of these antibodies was neutralized by a small amount of P3028 having
no
inhibitory activity per se. Similar to the results in the autologous ex vivo
model, the
immunosuppressor activity of sera from persons with a low proliferative
response to IL-2
was over-come by addition of the anti-P3028 antibodies to the cultures.
Example 10: Peptides that bind to P3028
[0635] The
information obtained by studying the effect of cancer patient sera
and the synthetic peptide P3028, on staining of the cc-chain, CD11a, of LFA-1
on
was used in order to design the structure of a potential binder / inhibitor of
the
immunomodulatory peptide P3028. The epitope of the particular monoclonal mouse
antibody used, HI 111, was mapped to residues 249-300 of CD1 1 a (Ma Q, et
al., J Biol
Chem. 2002;277:10638-41). Based on complementarity of charged and hydrophobic
amino acid sequences the first candidate binding to the P3028 peptide was
designed.
sequence was then optimized by synthesizing and testing the binding efficacy
of
peptides where each amino acid was substituted for all 19 L-amino acids.
242
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
[0636] Three
candidate peptide inhibitors of P3028 sequences/structures were
identified and their blocking capacity in solution was tested. Potential
peptide inhibitors
of P3028 were synthesized on a chip. The linear and/or CLIPS peptides were
synthesized based on the amino acid sequence of the target protein using
standard Fmoc-
chemistry and deprotected using trifluoric acid with scavengers. The
constrained
peptides were synthesized on chemical scaffolds in order to reconstruct
conformational
epitopes, using Chemically Linked Peptides on Scaffolds (CLIPS) technology
(Timmerman et al. (2007)). For example, the single looped peptides were
synthesized
containing a dicysteine, which was cyclized by treating with alpha, alpha'-
dibromoxylene and the size of the loop is varied by introducing cysteine
residues at
variable spacing. If other cysteines besides the newly introduced cysteines
are present,
they were replaced by alanine. The side-chains of the multiple cysteines in
the peptides
are coupled to CLIPS templates by reacting onto credit-card format
polypropylene
PEPSCAN cards (455 peptide formats/card) with a 0.5 mM solution of CLIPS
template
such as 1,3-bis (bromomethyl) benzene in ammonium bicarbonate (20 mM, pH
7.9)/acetonitrile (1:1(v/v)). The cards were gently shaken in the solution for
30 to 60
minutes while completely covered in solution. Finally, the cards are washed
extensively
with excess of H20 and sonicated in disrupt-buffer containing 1 percent
SDS/0.1 percent
beta-mercaptoethanol in PBS (pH 7.2) at 70 C for 30 minutes, followed by
sonication in
H20 for another 45 minutes. The binding of His-tagged P3028 to each peptide
was
tested in a PEPSCAN-based ELISA. The 455-well credit card format polypropylene
cards containing the covalently linked peptides are incubated with peptide
solution for
example consisting of 1 micrograms/mL diluted in blocking solution, for
example 4%
horse serum, 5% ovalbumin (w/v) in PBS/1% Tween. After washing, the peptides
were
incubated with a monoclonal mouse anti-his tag antibody (1/1000, Novagen,
70796-3)
and subsequently after washing with a rabbit-anti-mouse antibody peroxidase
conjugate
(1/1000, Southern Biotech, 6175-05) for one hour at 25 C. After washing, the
peroxidase substrate 2,2' -azino-di-3-ethylbenzthiazoline sulfonate (ABTS) and
2
microlitres of 3 percent H202 were added. After one hour, the color
development was
measured. The color development was quantified with a charge coupled device
(CCD) ¨
camera R6134 ima'gthpmues
sgttesn.Data: Optical density, Arbitrary OD units) are
optical values obtained by a CCD-camera. The values mostly range from 0 to
3000, a
scale similar to 1 to 3 of a standard 96-well plate ELISA-reader. First the
CCD-camera
243
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
makes a picture of the card before peroxidase coloring and then again a
picture after the
peroxidase coloring. These two pictures are substracted from each other which
results in
the data which is called raw-data. This is copied into the PeplabTM database.
Then the
values are copied to excel and this file is labeled as raw-data file. One
follow-up
manipulation is allowed. Sometimes a well contains an air-bubble resulting in
a false-
positive value, the cards are manually inspected and any values caused by an
air-bubble
are scored as 0.
[0638] As shown
in Position 17, 22 and 26 contained good binders of P3028
(PGE73 = His-tag-P3028). As shown in the diagram, peptide 5CF28 and 5CF29
efficiently block the binding of P3028 (PGE73) but 5CF27 does not. Peptide
5CF28
(SEQ ID NO: 1), had a solubility good enough to allow testing in biological
human ex
vivo models. Based on this structure, peptide P28R (SEQ ID NO: 2) was
developed.
For each position, shown are data for no peptide added assay in PBS buffer
230, 5CF027
assay in PBS buffer 232, 5CF029 assay in PBS buffer + 10% DMSO 234, no peptide
added in PBS buffer + 10% DMSO 236, and 5CF028 assay in PBS buffer + 10 DMSO
238. In the bar graph of Figure 23, bars representing each assay were in the
same left-
to-right order for each position. Each peptide, when present in an assay, was
at a
concentration of 0.5 mg/mL.
Example 11: Peptide interactions with P3028
[0639] The
information obtained by studying the effect of cancer patient sera
and the synthetic peptide P3028, on staining of the alpha-chain, CD1 la (SEQ
ID NO:
248), of LFA-1 on PBMCs was used in order to design the structure of a
potential binder
/ inhibitor of the immunoinhibitory peptide P3028. The epitope of the
particular
monoclonal mouse antibody used, HIM, was mapped to residues 274-325 of CD11a,
(SEQ ID NO: 248) (UniProt accession code P20701; Ma Q, et al., J Biol Chem.
2002;
277: 10638-41). Based on complementarity of charged and hydrophobic amino acid
sequences (see Figure 31) the first candidate binding to the P3028 peptide was
designed
using the sequence comprising 312-326 of CD1 la. This resulted in the peptide
KKL15
(SEQ ID NO: 1).
[0640] Peptide
KKL15 (SEQ ID NO: 1), for example appears to be
complementary to P3028. As shown in Figure 31, positively charged amino acids
244
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
interact with negatively charged amino acids of P3028 and hydrophobic amino
acids
make hydrophobic contacts enhancing the interaction.
Example 12: Peptides that bind to P3028
[0641] Based on
the structure of peptide P28R, additional peptides were
identified that bind to P3028. The additional binders included deletions,
truncations, and
or amino acid substitutions of peptide P28R. Binding of peptides to P3028 was
assayed
using PEPSCAN technology. PEPSCAN technology, or "rampo" assays are
biochemical
binding assays, details of which are provided below:
[0642] A
peptide microarray screening technology was used to measure
binding of P28R (SEQ ID NO: 2) and variants of P28R to P3028 (SEQ ID NO: 185).
In
this technology libraries of synthetic peptides are synthesized and covalently
linked onto
polypropylene microarray chips. The linear peptides were synthesized onto
credit-card
format polypropylene cards (455 peptide formats/card) as described by
(Timmerman et
al., 2004) using standard Fmoc-chemistry using hexamethylenediamine (HMDA) as
linker and deprotection using trifluoroacetic acid (TFA) with scavengers.
[0643] The
binding of His-tagged P3028 to each peptide on the card was
tested in an ELISA assay. The 455-well credit card format polypropylene cards
containing the covalently linked peptides were incubated with His-tagged P3028
peptide
(PGE73) solution consisting of 0.5 pg/mL diluted in blocking solution (4%
horse serum,
5% ovalbumin (w/v) in PBS/1% Tween). After washing, the peptides were
incubated
with a monoclonal mouse anti-His-tag antibody (Novagen, 70796-3, diluted
1/1000 in the
incubation buffer) and subsequently after washing with a rabbit-anti-mouse
antibody
peroxidase (Rampo) conjugate (Southern Biotech, 6175-05, diluted 1/1000,), for
one
hour at 25 C. After
washing, the peroxidase substrate 2,2' -azine-di-3-
ethylbenzthiazoline sulfonate (ABTS) and 2 pL of 3% H202 were added. The
binding
capacity of the mAb was measured as a color development at 405 nm (optical
density,
0D405). The color development was quantified with a charge-coupled device
(CCD) ¨
camera and an image processing system.
[0644] The
0D405-values obtained by a CCD-camera was considered as raw
data values ("rampo values," "rampo units," or "rampo scores"). The values
mostly
ranged from 0 to 3000, a log scale similar to 1 to 3 of a standard 96-well
plate ELISA-
reader. First the CCD-camera made a picture of the card before peroxidase
coloring and
245
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
then again a picture after the peroxidase coloring. These two pictures were
subtracted
from each other, which resulted in the data which was considered raw data.
These values
were copied into an excel file and labeled as a raw data file. One follow-up
was allowed. Sometimes a well can contain an air-bubble resulting in a false-
positive
value. If manual inspection of the cards detect an air-bubble the value are
set to 0 for
well.
[0645] A
library of peptides tested for binding to peptide P3028 included all
substitutions for each position of the peptide P28R (SEQ ID NO: 2) (i.e., 19
substitutions for each position). The results of the binding experiments are
shown in
Figures 27, 28, 29 and 30 and Table 5.1. Rampo scores ranged between 102 and
1190
for all substitutions in each of the 16 positions of P28R. P28R had rampo
values ranging
between 262 and 460 with a mean value of 370. As shown in Figure 28, 31 single-
amino acid substitutions of the peptide P28R (SEQ ID NO: 2) had a rampo score
above
500. These 31 substituted peptides include SEQ ID NOs: 3-31, and are shown in
Table
6.1. Significant higher values were observed for the substitutions M, Q, H, N
in position
13 (SEQ ID NOs: 22 to 25, respectively), all with values above 800. In
addition, M and
S in position 7 (SEQ ID NOs: 9 and 10, respectively), and Q and M in position
11 (SEQ
ID NOs: 15 and 16, respectively) all have rampo values over 700.
l07401 Table 6.1: Peptides that bind to P3028 with a rampo score above 500
SEQ ID NO Sequence
3 RKLDTFFVKLSLFTER
4 KKGDTFFVKLSLFTER
KKEDTFFVKLSLFTER
6 KKLD Q1-1- V KL S LFTER
7 KKLDTAFVKLSLFTER
8 KKLDTVFVKLSLFTER
9 KKLDTFMVKLSLFTER
KKLDTFSVKLSLFTER
11 KKLDTFVVKLSLFTER
12 KKLDTFTVKLSLFTER
13 KKLDTFLVKLSLFTER
14 KKLDTFFVKVSLFTER
246
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
15 KKLDTFFVKLQLFTER
16 KKLDTFFVKLMLFTER
17 KKLDTFFVKLTLFTER
18 KKLDTFFVKLHLFTER
19 KKLDTFFVKLS QFTER
20 KKLDTFFVKLSVFTER
21 KKLDTFFVKLSMFTER
22 KKLDTFFVKLSLMTER
23 KKLDTFFVKLSLQTER
24 KKLDTFFVKLSLHTER
25 KKLDTFFVKLSLNTER
26 KKLDTFFVKLSLPTER
27 KKLDTFFVKLSLSTER
28 KKLDTFFVKLSLGTER
29 KKLDTFFVKLSLATER
30 KKLDTFFVKLSLRTER
31 KKLDTFFVKLSLFNER
32 KKLDTFFVKLSLFPER
33 KKLDTFFVKLSLFRER
[0646] For each
position of P28R, the rampo scores of the group of 19
different peptides containing an L-amino acid substitution were compared to
the rampo
score of a control sample P28R peptide (SEQ ID NO: 2) for that group. Single-
amino
acid substitutions having a rampo score greater than or substantially
equivalent to P28R
were identified. As used herein, a rampo score "substantially equivalent to
P28R" is a
rampo score that is at least 98% of the rampo score of P28R. Thus, variants of
P28R
having equivalent or better binding to P3028 were identified.
[0647] For
example, at position 8 of P28R (SEQ ID NO: 2) is a V. The
control sample P28R peptide had a rampo score of 308, and peptides having an
F, G, L,
or R at position 8 (SEQ ID NOs: 326-330, respectively) each had a rampo score
greater
than or equal to 302 (98% of 308). The single amino acid substitutions of P28R
having a
247
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
score greater than or equal to that of the P28R control sample peptide for
that group are
shown in Table 6.2.
Table 6.2: Peptides that bind to a rampo score greater than or substantially
equivalent to
that of P28R
SEQ ID Position Sequence Rampo Score Rampo score of
NO P28R control
sample
268 1 AKLDTFFVKLSLFTER 466 308
269 1 DKLDTFFVKLSLFTER 373 308
270 1 EKLDTFFVKLSLFTER 396 308
271 1 GKLDTFFVKLSLFTER 367 308
272 1 HKLDTFFVKLSLFTER 428 308
273 1 IKLDTFFVKLSLFTER 483 308
274 1 LKLDTFFVKLSLFTER 449 308
275 1 MKLDTFFVKLSLFTER 457 308
276 1 NKLDTFFVKLSLFTER 445 308
277 1 PKLDTFFVKLSLFTER 387 308
278 1 QKLDTFFVKLSLFTER 455 308
279 1 RKLDTFFVKLSLFTER 523 308
280 1 TKLDTFFVKLSLFTER 493 308
281 1 VKLDTFFVKLSLFTER 442 308
282 3 KKADTFFVKLSLFTER 427 375
283 3 KKCDTFFVKLSLFTER 432 375
284 3 KKDDTFFVKLSLFTER 492 375
285 3 KKEDTFFVKLSLFTER 528 375
286 3 KKFDTFFVKLSLFTER 393 375
287 3 KKGDTFFVKLSLFTER 563 375
288 3 KKHDTFFVKLSLFTER 477 375
289 3 KKIDTFFVKLSLFTER 454 375
290 3 KKKDTFFVKLSLFTER 386 375
291 3 KKMDTFFVKLSLFTER 460 375
248
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
292 3 KKNDTFFVKLSLFTER 374 375
293 3 KKQDTFFVKLSLFTER 473 375
294 3 KKRDTFFVKLSLFTER 370 375
295 3 KKSDTFFVKLSLFTER 393 375
296 3 KKTDTFFVKLSLFTER 451 375
297 3 KKVDTFFVKLSLFTER 377 375
298 4 KKLATFFVKLSLFTER 494 414
299 4 KKLETFFVKLSLFTER 417 414
300 4 KKL I TFFVKLSLFTER 430 414
301 4 KKLVTFFVKLSLFTER 424 414
302 4 KKLWTFFVKLSLFTER 443 414
303 4 KKLYTFFVKLSLFTER 422 414
304 5 KKLDCFFVKLSLFTER 449 424
305 5 KKLDMFFVKLSLFTER 475 424
306 5 KKLDNFFVKLSLFTER 436 424
307 5 KKLDPFFVKLSLFTER 427 424
308 5 KKLDQFFVKLSLFTER 535 424
309 5 KKLDRFFVKLSLFTER 430 424
310 5 KKLDSFFVKLSLFTER 458 424
311 5 KKLDWFFVKLSLFTER 418 424
312 5 KKLDYFFVKLSLFTER 425 424
313 6 KKLDTAFVKLSLFTER 575 437
314 6 KKLDT IFVKLSLFTER 466 437
315 6 KKLDTMFVKLSLFTER 467 437
316 6 KKLDTNFVKLSLFTER 446 437
317 6 KKLDTPFVKLSLFTER 497 437
318 6 KKLDTTFVKLSLFTER 481 437
319 6 KKLDTVFVKLSLFTER 547 437
320 7 KKLDTFLVKLSLFTER 517 460
321 7 KKLDTFMVKLSLFTER 712 460
322 7 KKLDTFQVKLSLFTER 511 460
249
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
323 7 KKLDTFSVKLSLFTER 700 460
324 7 KKLDTFTVKLSLFTER 517 460
325 7 KKLDTFVVKLSLFTER 527 460
326 8 KKLDTFFFKLSLFTER 358 308
327 8 KKLDTFFGKLSLFTER 379 308
328 8 KKLDTFFLKLSLFTER 411 308
329 8 KKLDTFFPKLSLFTER 456 308
330 8 KKLDTFFRKLSLFTER 365 308
331 9 KKLDTFFVRLSLFTER 374 377
332 10 KKLDTFFVKASLFTER 426 348
333 10 KKLDTFFVKFSLFTER 403 348
334 10 KKLDTFFVKGSLFTER 495 348
335 10 KKLDTFFVKISLFTER 415 348
336 10 KKLDTFFVKMSLFTER 460 348
337 10 KKLDTFFVKNSLFTER 365 348
338 10 KKLDTFFVKPSLFTER 436 348
339 10 KKLDTFFVKQSLFTER 470 348
340 10 KKLDTFFVKRSLFTER 439 348
341 10 KKLDTFFVKSSLFTER 351 348
342 10 KKLDTFFVKTSLFTER 399 348
343 10 KKLDTFFVKVSLFTER 658 348
344 10 KKLDTFFVKYSLFTER 382 348
345 11 KKLDTFFVKLHLFTER 535 442
346 11 KKLDTFFVKLMLFTER 744 442
347 11 KKLDTFFVKLNLFTER 451 442
348 11 KKLDTFFVKLQLFTER 768 442
349 11 KKLDTFFVKLTLFTER 520 442
350 12 KKLDTFFVKLSAFTER 462 428
351 12 KKLDTFFVKLSHFTER 460 428
352 12 KKLDTFFVKLSIFTER 456 428
353 12 KKLDTFFVKLSMFTER 499 428
250
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
354 12 KKLDTFFVKLSNFTER 462 428
355 12 KKLDTFFVKLSQFTER 651 428
356 12 KKLDTFFVKLSRFTER 483 428
357 12 KKLDTFFVKLSSFTER 478 428
358 12 KKLDTFFVKLSTFTER 437 428
359 12 KKLDTFFVKLSVFTER 545 428
360 12 KKLDTFFVKLSWFTER 409 428
361 13 KKLDTFFVKLSLATER 525 402
362 13 KKLDTFFVKLSLCTER 400 402
363 13 KKLDTFFVKLSLGTER 531 402
364 13 KKLDTFFVKLSLHTER 1046 402
365 13 KKLDTFFVKLSLITER 468 402
366 13 KKLDTFFVKLSLLTER 448 402
367 13 KKLDTFFVKLSLMTER 1190 402
368 13 KKLDTFFVKLSLNTER 862 402
369 13 KKLDTFFVKLSLPTER 696 402
370 13 KKLDTFFVKLSLQTER 1144 402
371 13 KKLDTFFVKLSLRTER 502 402
372 13 KKLDTFFVKLSLSTER 635 402
373 13 KKLDTFFVKLSLTTER 494 402
374 13 KKLDTFFVKLSLVTER 446 402
375 13 KKLDTFFVKLSLWTER 430 402
376 14 KKLDTFFVKLSLFFER 348 319
377 14 KKLDTFFVKLSLFGER 343 319
378 14 KKLDTFFVKLSLFHER 463 319
379 14 KKLDTFFVKLSLFIER 375 319
380 14 KKLDTFFVKLSLFLER 360 319
381 14 KKLDTFFVKLSLFMER 501 319
382 14 KKLDTFFVKLSLFNER 599 319
383 14 KKLDTFFVKLSLFPER 551 319
384 14 KKLDTFFVKLSLFSER 369 319
251
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
385 14 KKLDTFFVKLSLFVER 380 319
386 14 KKLDTFFVKLSLFWER 374 319
387 15 KKLDTFFVKLSLFTDR 404 371
388 16 KKLDTFFVKLSLFTEF 297 260
389 16 KKLDTFFVKLSLFTEK 291 260
390 16 KKLDTFFVKLSLFTEN 311 260
391 16 KKLDTFFVKLSLFTER 260 260
392 16 KKLDTFFVKLSLFTET 292 260
393 16 KKLDTFFVKLSLFTEY 311 260
[0648] The
positional substitutions of P28R in Table 6.2, (SEQ ID NOs:
268-393) are summarized in Figure 32. It is noted that positions 2 (K), 9 (K)
and 15 (E)
tolerate relatively few substitutions while still binding to P3028.
Substitution of the
residue at positions 2, 9, and/or 15 of P28R can result in binding to P3028
(as measured
by rampo scores) substantially lower than unsubstituted P28R. Thus, it is
contemplated
herein that these 3 positions appear to modulate signal transduction. One
skilled in the
art will appreciate that signal transduction modulatory activity of these
positions can be
useful in designing inhibitors of immunomodulatory peptides.
[0649] PEPSCAN
analysis was also performed on truncations and internal
deletions of peptide P28R. Shown in Figure 29 are rampo scores for peptides
having the
sequences KKLDTFFVKLSLFTER (SEQ ID NO: 2); KKLDTFFVKLSLFTE (SEQ ID
NO 34); KKLDTFFVKLSLFT (SEQ ID NO: 35); KKLDTH-VKLSLF (SEQ ID NO
36); KKLDTH-VKLSL (SEQ ID NO: 37); KKLDTFFVKLS (SEQ ID NO: 38);
KKLDTFFVKL (SEQ ID NO: 39); KKLDTFFVK (SEQ ID NO: 40); KKLDTFFV
(SEQ ID NO: 41); KKLDTFF (SEQ ID NO: 42); KKLDTF (SEQ ID NO: 43); KKLDT
(SEQ ID NO: 44); KKLD (SEQ ID NO: 45); KLDTH-VKLSLFTER (SEQ ID NO:
LDTFFVKLSLFTER (SEQ ID NO: 47); DTFFVKLSLFTER (SEQ ID NO: 48);
TFFVKLSLFTER (SEQ ID NO: 49); FFVKLSLFTER (SEQ ID NO: 50);
FVKLSLFTER (SEQ ID NO:51); VKLSLFTER (SEQ ID NO: 52); KLSLFTER (SEQ
ID NO: 53); LSLFTER (SEQ ID NO: 54); SLFTER (SEQ ID NO: 55); LFTER (SEQ
ID NO: 56); FTER (SEQ ID NO: 57); KLDTFFVKLSLFTE (SEQ ID NO: 58);
252
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
LDTFFVKLSLFT (SEQ ID NO: 59); DTFFVKLSLF (SEQ ID NO: 60); TFFVKLSL
(SEQ ID NO: 61); FFVKLS (SEQ ID NO: 62); FVKL (SEQ ID NO: 63).
[0650] Shown in
Figure 30 are rampo scores for peptides having the
sequences KKLDTFFVKLSLFTER (SEQ ID NO: 2); KLDTFFVKLSLFTER (SEQ ID
NO: 46); KKLTFFVKLSLFTER (SEQ ID NO: 64); KKLDTFVKLSLFTER (SEQ ID
NO: 65); KKLDTFFKLSLFTER (SEQ ID NO: 66); KKLDTFFVKSLFTER (SEQ ID
NO: 67); KKLDTFFVKLSFTER (SEQ ID NO: 68); KLDTFFVKLSLFER (SEQ ID
NO: 69); KLDTH-VKLSLFTE (SEQ ID NO: 58); LDTFFVKLSLFTER (SEQ ID NO:
47); KKTFFVKLSLFTER (SEQ ID NO: 70); KKLDFVKLSLFTER (SEQ ID NO: 71);
KKLDTEKLSLFTER (SEQ ID NO: 72); KKLDTFFVSLFTER (SEQ ID NO:73);
KKLDTFFVKLFTER (SEQ ID NO: 74); KKLDTFFVKLSLER (SEQ ID NO: 75);
LDTFFVKLSLFT (SEQ ID NO: 59); DTFFVKLSLFTER (SEQ ID NO: 48);
KKH-VKLSLFTER (SEQ ID NO: 76); KKLDVKLSLFTER (SEQ ID NO: 77);
KKLDTFLSLFTER (SEQ ID NO: 78); KKLDTFFVLFTER (SEQ ID NO: 79);
KKLDTFFVKLTER (SEQ ID NO: 80); KKLDTFFVKLSLR (SEQ ID NO: 81);
KFFVKLSLFTER (SEQ ID NO: 82); KKLVKLSLFTER (SEQ ID NO: 83);
KKLDTLSLFTER (SEQ ID NO: 84); KKLDTFFLFTER (SEQ ID NO: 85);
KKLDTFFVKTER (SEQ ID NO: 86); KKLDTFFVKLSR (SEQ ID NO: 87);
KFVKLSLFTER (SEQ ID NO: 88); KKLKLSLFTER (SEQ ID NO: 89);
KKLDTSLFTER (SEQ ID NO:90); KKLDTFFFTER (SEQ ID NO: 91);
KKLDTFFVKER (SEQ ID NO: 92); KKLDTFFVKLS (SEQ ID NO: 38);
GKLDTFFVKLSLFTER (SEQ ID NO: 93); KKGDTH-VKLSLFTER (SEQ ID NO:
94); KKLDGFFVKLSLFTER (SEQ ID NO: 95); KKLDTFGVKLSLFTER (SEQ ID
NO: 96); KKLDTFFVGLSLFTER (SEQ ID NO: 97); KKLDTFFVGLSLFTER (SEQ
ID NO: 98); KKLDTFFVKLGLFTER (SEQ ID NO: 99); KKLDTFFVKLSLGTER
(SEQ ID NO: 100); KKLDTFFVKLSLFTGR (SEQ ID NO: 101).
[0651] As shown
in Figure 30, several deletions and truncations of peptide
P28R have a rampo score comparable to, or higher than peptide P28R, including
peptides
of the sequences SEQ ID NOs: 64, 65, 68, and 76. Additionally several glycine
substitutions had rampo scores comparable to P28R, including peptides of SEQ
ID
94, 95, 96, 98, and 99. Deleting up to at least 8 amino acids from the N
terminal of
253
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
(SEQ ID NOs: 46to 53) retained a high affinity to P3028 as measured by rampo
score.
Deleting the C terminal R of P28R (SEQ ID NO: 34) retained a high affinity to
P 3028.
Example 13: Effect of a low molecular weight inhibitor of P3028 on lymphocyte
activation
[0652] Analyses
of the inhibitor of P3028, P28R, were performed in human
ex vivo models. The stimulatory activity on PBMCs, measured using the MTS or
CFSE
techniques, were studied in 7 healthy control samples and 7 cancer patients of
various
diagnoses. Interestingly, even in the absence of other types of stimulation
P28R has a
significant stimulatory activity in 6 out of 7 cancer patients whereas PBMCs
from control
samples showed only a weak or no stimulation.
[0653] As shown
in Figure 24, stimulatory activity of P28R on suppressed
proliferative response to IL-2. PBMCs were cultured for 7 days with IL-2 and
the
proliferative rate was determined as incorporation of BrdU. Each bar
represents mean
value of triplets. Similar to the studies on the efficacy of antibodies (see
Figure 22)
directed against P3028 to reverse cancer related immunosuppression determined
as a
poor proliferative response of PBMCs from cancer patients to IL-2, the
efficacy of the
low molecular weight inhibitor P28R on reversal of suppressed IL-2 induced
proliferation was investigated. The results of cultures of PBMCs from four
different
treatment naïve patients are shown in Figure 24. For each quantity of added
P28R, IL-2
stimulated cells 240 are shown in the left, and unstimulated 242 are shown on
the right.
PBMCs with a low initial proliferation (see Figures 24A and 24B) were markedly
stimulated by P28R whereas a high initial proliferation was essentially
unaffected by the
drug (see Figures 24C and 24D). As expected, systemic immunosuppression was
not
present in all patients and only those with immunosuppression were stimulated.
Example 14: Binding of a low molecular weight inhibitor of P3028 to tumor
cells
[0654] As
demonstrated herein, P3028 structures are present in tumors. A
biotinylated inhibitor of P3028, P28R, was used to further study the
distribution of 3028
structures and the binding of the inhibitor in tumor tissue. Three breast
cancers, two
cell carcinomas and four malignant melanomas were analyzed. All investigated
tumors
bound the inhibitor. An example of a stained breast cancer is shown in Figure
25, and a
strong positive reaction 7 is seen indicating the presence of the inhibitory
3028-structure
254
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
in this tumor. As the P3028-structure inhibits lymphocyte migration as well as
cytotoxic
activity (described above), an immune mediated attack against positively
staining tumor
areas can be efficiently suppressed as long as exposed P3028 is not blocked by
binding
P28R. However, lymphocytes were not stained by this procedure since the P3028
structure was blocked by binding to LFA-1 on these cells.
Example 15: Unblocking the LFA-1 receptor by P28R
[0655] As
described herein, 02-integrins play a role in the normal function of
the immune system. Also described herein are immunosuppressor mechanisms based
on
the binding of an endogenous inhibitor, P3028, to the 02-integrin LFA-1. As
described
in Example 7, the membrane staining of PBMCs from cancer patients is markedly
decreased compared to normal control samples. The exposure of LFA-1 could,
however,
be enhanced by incubating PBMCs from cancer patients with an antibody directed
against the inhibitor P3028 (see Example 7 and Figure 16). Staining for LFA-1
was
performed using anti-LFA-1 antibody of Example 7 and a secondary antibody
(Ultravision) followed by development with Fast Red. Fresh frozen tumor
sections
without any fixation were incubated for 4-20 hours with the drug candidate,
P28R before
staining for LFA-1 (see Figure 26B). For comparison, control sample tumor
sections
were incubated with phosphate buffered saline only (see Figure 26A).
[0656] As is
shown in Figure 26, P28R unblocked LFA-1, and thereby
markedly enhanced the functional expression of LFA-1 enabling migration and
cytotoxic
activity of these cells. Strong LFA-1 staining 3 in P28R-treated cells is
contrasted with
weak LFA-1 staining 5 in untreated cells. These results show that LFA-1 was
unblocked
by removal of the P3028 structure by the P28R.
Example 16: Delivery of immunoregulatory peptide inhibitors via nanodosing to
cancer
patients
[0657] Cancer
patients with immunosuppression due to the presence of P3028
structures and having subcutaneous melanoma metastases are selected. A micro-
dialysis
catheter is inserted into one of these metastases after the inflammatory
infiltrate has been
determined using a fine needle biopsy. The base line: inflammatory infiltrate,
cytokine
profile and concentration of P3028 structures are determined before infusion
of the
P3028-specific immunoregulatory peptide inhibitor. Changes of the cytokine
profile and
255
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
concentration of P3028 structures are then determined during and after the
infusion. The
infusion will continue for 24 or 48 hours and the area supplied by the micro-
dialysis
catheter will be excised immediately after the infusion and then after one and
two weeks
in order to study the inflammatory infiltrate and tumor regressive changes. It
is expected
that the administration of the immunoregulatory peptide inhibitor will reduce
the
immunosuppression of the cancer patient, as measured, for example, by de-
blocking
1, binding P3028 structures, and/or enhancing immune cell recruitment.
Example 17: Albumin peptide binders of cell surface molecules
Albumin fragments that bind to cell surface molecules
[0658] As
taught in US Publication No: 2011/0262470 (hereby expressly
incorporated by reference in its entirety) some albumin fragments can bind to
cell surface
molecules. U.S. Publication No: 2011/0262470 reports the identification of
serum
peptides that bind to Artificial Cell Surface (ACS) columns. The ACS columns
were
prepared as follows:
[0659] First,
biotinylated cell surface proteins were prepared. Buffy coats
generated from 450 ml blood each were collected from 4 healthy donors.
Erythrocytes
were removed by sedimentation on 2% dextran T500 solution (Amersham Pharmacia
Biotech AB, Uppsala Sweden) in 0.9% NaCl. Mononuclear cells (PBMC) were then
isolated by Ficoll-Paque Plus (GE Healthcare BioscienceAB Sweden) density
gradient
centrifugation. The PBMCs were then suspended in phosphate buffered saline
(PBS)
containing Ca and Mg (GIBCO) at a concentration of 10 x 106 /ml. EZ Link Sulfo-
NHS-
biotin (Pierce USA) was added at a final concentration of 0.2 mg/ml and the
mixture
incubated on a shaker at room temperature for 10 mM. Excess biotin was then
removed
by washing the PBMC in PBS. Biotinylated PBMC were then lysed by adding 1.0 ml
ice-cold lysing buffer (50 mM Tris-HCL, pH 7.5, with 0.15 MNaCI, 5mM MgCl2
containing 100 mM Octyl glucoside and 1 mM Phenylmethylsulfonyl fluoride) per
2 x
107 pelleted cells with gentle shaking, then incubated for 30 mM. on ice.
Debris was
removed by centrifugation at 5000xg at 4 C. for 10 mM and the supernatants
were
collected and pooled from all four donors. The lysate was then stored at -70
C. in
polypropylene plastic tubes.
[0660] To study
the absorptions by trypsin-fragment dHSA, affinity columns
with biotinylated cell surface proteins from mononuclear cells coupled to
streptavidin-
256
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
sepharose were prepared as follows: 18 ml biotinylated cell lysate in lysate
buffer was
diluted 1/10 in binding buffer (20 mM NaH2PO4, 0.15 M NaCI, pH 7.5). This
amount of
lysate corresponds to 36x107 mononuclear cells. It was added to a 1 ml Hitrap
Streptavidin HP affinity column (Amersham Biosciences). To block possible
remaining
free biotin, 5 ml of 0.1 M glycine (Sigma) was added to the column.
Unsaturated
streptavidin on the column was then reacted with 150 ug biotin (Sigma) in
binding
The column was carefully washed with PBS and stored in PBS with 0.1% NaN3 at 4
C.
until use.
[0661] To study
the absorptions by of ASP-N fragmented dHSA, affinity
columns with biotinylated cell surface proteins from mononuclear cells coupled
to
streptavidin-sepharose were prepared as follows: Biotinylated cell lys ate in
lys ate buffer
underwent buffer exchange by dialysis with Spectrapore 4 dialysis tubing
(Spectrum
Europe, Breda, The Netherlands) in binding buffer (20 mM NaH2PO4, 0.15 MNaCI
pH
7.5). 27 ml biotinylated cell lysate in binding buffer (corresponding to 54 x
107
mononuclear cells) was added to 1.5 ml washed Streptavidin Sepharose HP
(Amersham
Biosciences). To block possible remaining free biotin, 25 ml of 0.1 M glycine
(Sigma)
was added to the Streptavidin Sepharose. Unsaturated streptavidin was then
reacted with
225 ug biotin (Sigma) in binding buffer. The Streptavidin Sepharose was
carefully
washed in PBS. One ml of the biotinylated cell lysate coupled Streptavidin
Sepharose
was then packed in an empty column (Tricorn Empty High Performance Column,
Amersham Bioscience) and washed with phosphate buffered saline (PBS)
containing
Ca2+ and Mg2+ (GIBCO).
[0662]
Digestion with trypsin or ASP-N was performed as follows. Freeze
dried dHSA (0.5 mg) was reconstituted in 25 mM NH4HCO3, pH 8, containing 10 mg
sequencing grade modified trypsin (Promega Corporation, WI) or 2 mg
Endoproteinase
ASP-N (Sigma) and incubated at 37 C overnight. To remove unfragmented albumin
and
enzyme, the sample was ultra filtered through an Amicon Ultra 4 (mw cut-off of
5000) or
a Centriplus (mw cut-off 10000) centrifugal filter (Millipore AB, Solna,
Sweden). The
filtrate, containing fragmented dHSA without enzymes, was collected and
diluted with
PBS with Ca and Mg (GIBCO).
[0663] dHSA was
trypsinated, and the mixture of peptides obtained after
trypsination was adsorbed by ACS. Two ml of enzyme-fragmented dHSA in PBS,
corresponding to a total of 0.2 mg protein, was passaged over the ACS column.
The
257
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
flow-through was collected with consideration taken to void volume and
dilution of
adsorbed sample by collecting in small portions of 0.2 ml. Thirty microliters
of each
sample, including a control sample that has not been adsorbed, were dried in a
Speed-
centrifuge. The binding peptides were identified by comparing adsorbed and
unadsorbed
peptide solutions using the MALDI TOF mass spectrometry technique. Dried
samples
were reconstituted in 10 ul of 0.1 % TFA. Zip Tip pipette tips (Millipore,
USA)
containing C18 reversed-phase media were used for desalting reconstituted
samples. For
analysis of samples in the mass range 700 ¨ 3600 Da, one ul of each Zip Tip
eluted
sample was mixed with 1111 of a saturated solution of oc-cyano-4-
hydroxycinamic acid
(0.02mg/m1) in 70%acetonitrile/0.3% trifluoro acetic acid. For the analysis of
samples in
the mass range 1500 ¨ 9000 Da, one ul of each Zip Tip eluted sample was mixed
with
of sinapinic acid (3-methoxy-4-hydroxycinnamic acid). 1111 of the mixture was
spotted on
the MALDI plate and analysed using MALDI-TOF MS (Voyager-DE PRO, Applied
Biosystems, CA, US). Mass identity search of resulting spectra was performed
in the
SwissProt or NCBI databases using MS-Fit.
[0664] These peptides are shown in Table 7.
Table 7: Trypsin-generated albumin fragments that bind to ACS
SEQ ID NO: Percent Sequence Albumin
Absorbed Positions
194 71% KYLYEIAR 161-168
195 64% KVPQVSTPTLVEVSR 438-452
196 60% VFDEFKPLVEEPQNLIK 397-413
197 59% VPQVSTPTLVEVSR 439-452
198 42% RPCFSALEVDETYVPK 509-524
199 41% FQNALLVR 427-434
200 36% SLHTLFGDK 89-97
201 36% LKECCEKPLLEK 299-310
202 35% LCTVATLR 98-105
203 34% YLYEIAR 162-168
204 32% CCAAADPHECYAK 384-396
205 29% AAFTECCQAADK 187-198
258
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
206 26% CCTESLVNR 500-508
207 25% QEPERNECFLQHK 118-130
208 23% AVMDDFAAFVEK 570-581
209 22% NECFLQHK 123-130
210 20% ONCELFEQLGEYK 414-426
211 18% QEPERNECFLQHK 118-130
212 13% VHTECCHGDLLECADDR 265-281
213 8% FKDLGEENFK 35-44
214 3% YICENQDSISSK 287-298
215 2% LDELRDEGK 206-214
216 1% DDNPNLPR 131-138
[0665] Because
the full peptide sequence of albumin is not recovered using
the MALDI-TOF technique after trypsin degradation, and because some sequences
with
the capacity to bind to cell surface receptors of immune cells, might have
been degraded
by trypsin treatment, dHSA was also degraded by asparaginase (ASN-N), and the
mixture of peptides obtained after degradation was adsorbed by ACS. The
binding
peptides were identified by comparing adsorbed and unadsorbed peptide
solutions using
the MALDI TOF ms technique. These peptides are shown in Table 8.
Table 8: Asp-N-generated albumin fragments that bind to ACS
SEQ ID Percent Sequence Albumin
NO: Absorbed Positions
217 100% DHVKLVNEVTEFAKTCVA 62-79
218 100% DDKETCFAEEGKKLVAASQAALGL 586-609
219 87% DRVTKCCTESLVNRRPCFSALEV 495-517
220 86% DETYVPKEFNAETFTHA 518-535
221 65% DSISSKLKECCEKPLLEKSHCIAEVEN 293-319
222 65% DKLCTVATLRETYGEM 96-112
223 100% YSVVLLLRLAKTYETTLEKCCAAADPHEC 364-398
YAKVF
224 100% KLCTVATLRETYGEMADCCAKQEPERNEC 96-130
259
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
FLQHK
225 100% ICTLSEKERQIKKQTALVELVKHKPKATKE 536-572
QLKAVM
226 100% LAKYICENQDSISSKLKECCEKPLLEKHCIA 283-319
EVEN
227 100% VFLGMFLYEYARRHPDYSVVLLLRLAKTY 348-388
ETT LEKCCAAA
228 100% LGEENFKALVLIAFAQYLQQCPFEDHVKLV 37-79
NEVTEFAKTCVA
229 100% RVTKCCTESLVNRRPCFSALEVDETYVPKE 495-535
FNAETFTFHA
230 37% YLSVVLNQLCVLHEKTPVSDRVTKCCCTES 475-517
LVNRRPFSALEV
[0666]
Additionally, nine synthetic albumin peptides were synthesized, as
shown in Table 9.
Table 9: Synthetic albumin peptides
SEQ ID NO: Peptide Name Sequence Albumin
Positions
183 3026 NEETFLKKYLYEIARRHPYFYAP 153-176
184 3027 ELFEQLGEYKFQNALLVR 417-434
185 3028 VFDEFKPLVEEPQNLIK 397-413
188 3029 KVPQVSTPTLVEVSR 438-452
189 2604 KLVNEVTEFAKT 65-76
190 2605 NEETFLKKYLYE 153-168
191 2606 LDELRDEGKAS 205-217
192 2607 EMADCCAKQEPE 110-122
193 2608 ELFEQLGEYKF 417-427
260
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
Example 18: Albumin peptide binders of cell surface molecules
[0667]
Monoclonal antibody mAb A was shown to have immunomodulatory
activity. Structures of the epitope bound by mAb A were further investigated.
Briefly,
albumin fragments were incubated with antibody, and Matrix-Assisted Laser
Desorption/Ionisation Time-of-Flight mass spectrometry (MALDI-TOF ms) were
used in
order to define the possible site or sites on human serum albumin to which a
mouse
monoclonal antibody specific for denatured albumin binds. One approach took
advantage of the fact that some tryptic peptides to which an antibody is bound
will not
generate characteristic mass spectra in MALDI as they are "hidden" from the
analysis.
Another approach takes advantage of the fact that sites on a protein where an
antibody
has bound are protected from proteolysis.
[0668] Purified
human serum albumin (HSA) was denatured with urea,
reduced with DTT and alkylated. The denatured HSA was then subjected to
trypsin
treatment with a low concentration (0.02-2 ng/ml) of trypsin. However, the
spectra
obtained with MALDI were unsatisfactory, as the peptides masses typical for
albumin
were not found. Based on gel electrophoresis this preparation (digested by
0.02 ng/ml of
trypsin) was found to contain substantial amounts of undigested albumin.
Therefore,
trypsin digestion was continued, at a higher concentration (5ug/m1) in order
to obtain the
mass spectra usually used for identification of proteins by MALDI.
[0669] To
identify albumin fragments bound by mAb A, some of the now
completely cleaved albumin solution was incubated with the mAb A. MALDI-TOF ms
was performed and spectra of enzyme-treated denatured albumin obtained in the
presence or absence of mAb A were compared. Fourteen albumin (SEQ ID NOs: 231-
244) massed were absent or reduced after incubation with mAb A. The amino acid
sequence of these peptides is shown in Table 10. The spectra represent
multiple areas
encompassing residues 66 to 508 of the albumin molecule.
[0670] In order
to further confirm these results the monoclonal antibody mAb
A was allowed to bind to the denatured albumin (previously digested by trypsin
at a
concentration of 0.02 ng/ml) in order to protect the peptide sequences of the
epitope.
complex was then again treated with trypsin. MALDI-TOF ms was then performed
and
the peptide mass spectra generated from albumin were compared with spectra
generated
from denatured albumin trypsin-treated in the absence of antibody. The same
fourteen
masses out of 39 albumin masses disappeared completely or were significantly
reduced
261
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
the sample were the mAb was present during trypsin treatment (see Table 10,
Column
Multiple readings were taken to verify the results.
Table 10: Albumin peptides that bind to monoclonal antibody mAb A
SEQ Sequence Albumin Peak area Peak area Peak area
ID Positions before after trypsiniated
NO: Ab incub. Ab incub. Albumin+Ab
(2 spectra) (5 spectra) (6 spectra)
231 LVNEVTEFAK 066-075 1970, 4092 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0,
232 SLHTLFGDK 089-097 1695, 5089 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0,
233 LCTVATLR 098-105 1862, 4869 0, 0, 132, 0, 0 0, 0, 0,
0, 0, 0,
234 ETYGEMADCCAK 106-117 809, 1010 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0,
235 YLYEIAR 162-168 6036, 13066 504, 118, 473, 448, 895,
216,
281, 288 724, 2346, 1571
236 LDELRDEGK 206-214 3064, 7917 0, 0, 0, 0, 0 0, 0, 0, 0,
0, 0
237 YICENQDSISSK 287-298 583, 1394 0, 0, 0, 0, 0, 0, 0, 53,
0, 0, 0,
238 LKECCEKPLLEK 299-310 2283, 4675 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0,
239 HPDYSVVLLLR 362-372 1036, 1482 0, 0, 0, 0, 0, 0, 0, 51,
0, 407
(1312),
226(1312)
240 CCAAADPHECYAK 384-396 2186, 3327 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
241 QNCELFE,QLGEYK 414-426 2519, 2978 0, 0, 0, 0, 0, 0, 0, 0, 0,
0,212(1656.64)
242 FQNALLVR 427-434 15276, 267, 315, 931, 591, 1284, 199,
32846 494, 309 1015, 2963,
1998
243 CCTESLVNR 500-508 1360, 4659 0, 0, 0, 0, 0, 0, 258, 0,
0,
0,204 (1139)
244 AVMDDFAAFVEK 570-581 2720, 3758 0, 0, 0, 0, 0 0, 0, 0, 0,
0, 0
[0671] Some
peptide fragments of albumin might not be identified by binding
an antibody to trypsinated fragments of albumin because of the possibility
that the mAb
262
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
binding epitope of albumin is cleaved by trypsin, resulting in fragments of
the epitope
with too low binding affinity to bind to the mAb. Therefore, an additional
method was
used to identify fragments bound by the antibody. MALDI epitope mapping of mAb
A
based on antibody protection of proteolysis was repeated. This time a slightly
different
approach was used. Denatured HSA was incubated with mAb A. Albumin not bound
by
the antibody, was removed from the sample by size exclusion on an ultra
filter. The
remaining free mAbs and the complexes of mAb-albumin was then digested with
trypsin
(sequences of the albumin molecule to which mAb is bound should resist the
trypsin
digestion). Small cleaved fragments of mAb and unprotected albumin was then
removed
from the sample by ultrafiltration (30 IcD). The complexes of mAb and bound
albumin
fragments were dissociated by lowering the pH to 2.7. Again ultrafiltration at
301(D was
performed to separate whole mAb from albumin fragments smaller than 30kD.
MALDI
TOF analysis of these fragments did not identify spectra typical for albumin.
because the fragments containing the epitope of mAb A were still too large.
This filtrate
(< 30 kD) was then further digested with trypsin (for cleavage of sites
previously
protected by the mAb) in order to generate peptide masses suitable for
analysis with
MALDI TOF ms.
[0672] After
this second trypsin treatment, eight of 32 masses detected by
MALDI TOF ms matched to albumin (see Table 11). Thus, these new amino acid
sequences represent a part of the epitope, which also contains sequences on
the other side
of the trypsin cleavage point. Six of the eight peptide masses ((SEQ ID NOs:
231, 233,
235, 236, 242, and 243) were peptide masses that also disappeared when
analysed
previously when completely cleaved albumin was incubated with the mAb A before
the
MALDI-TOF analysis (see Table 10). Two of the eight peptides (SEQ ID NOs: 245
and 346) had not been identified in the binding assays with completely cleaved
albumin.
The epitope/s of this antibody was thus established. It is important to note
that multiple
such structures are present in the albumin molecule, which can then cause
cross-linking
of the receptors to which they are bound. However, multiple epitope sites for
mAb A
can indeed exist on albumin.
Table 11: Albumin peptides that bind to monoclonal antibody mAb A
SEQ ID NO: Sequence Albumin Positions
263
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
245 LS QRFPK 243-249
246 DDNPNLPR 131-138
235 YLYEIAR 162-168
233 LCTVATLR 98-105
242 FQNALLVR 427-434
236 LDELRDEGK 206-214
243 CCTESLVNR 500-508
231 LVNEVTEFAK 66-75
Example 19: Cyclic Peptides that Bind P3028
[0673] In order
to identify cyclic peptides that bind to P3028, all possible
variants of di- and tripeptides were synthesized on chips and the binding of
the His-tag
labeled P 3028 was analyzed using the ELISA-technique. Based on the identified
binding motifs, looped 6-meres were produced and tested. These results
together enable
the construction of a lead cyclic peptide CLALNVMCG (SEQ ID NO: 264).
Positional
scans were performed in each position of the lead cyclic peptide was replaced
with each
of the other 19 L-amino acids. Binding of each of the substituted peptides was
tested,
and peptide sequences with even better binding capacity than that of the lead
peptide
were identified. The two peptides with the highest affinity were CLRLNVFCG
(SEQ ID
NO: 265) and CLRLIVMCG (SEQ ID NO: 266). The two best looped peptides that
bind to P3028 based on the positional scan binding assay are summarized in
Table 12.
Table 12: Cyclic peptides that bind to P3028
SEQ ID NO: SEQUENCE
264 CLALNVMCG
265 CLRLNVFCG
266 CLRLIVMCG
[0674]
Substitutable amino acid residues in the lead looped peptide that were
identified in the positional scans as providing improved binding to P3028 (SEQ
ID NO:
185) are summarized in Figure 33 (i.e., SEQ ID NOs: 264 to 266). Positional
substitutions of P28R that result in equivalent or better binding to P28R to
P3028 that
264
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
were identified as providing higher or substantially equal binding to P3028
(see Tables
6.1 and 6.2) are also summarized in Figure 33. It was observed that there was
very good
homology between looped peptide sequences that bind to P3028 based on the scan
data
(SEQ ID NOs: 264-266), and sequences of linear peptides that identified as
binding to
P3028 (SEQ ID NOs: 2-31 and 268-393) (see Figure 33). It is noted that the N-
terminal
C residues and C-terminal CG residues of the cyclic peptides are involved in
cyclization
of the peptide. Thus, as shown by shaded boxes in Figure 33, there is strong
homology
between 6-mere cyclic peptides identified as binders of P3028 (SEQ ID NOs: 264-
266)
and either the N terminus of C-terminus of P28R-related peptide (SEQ ID NOs: 2-
31
268-393). It is contemplated that additional cyclic peptides that bind to and
inhibit
albumin-derived immunoregulatory peptides can be identified.
Example 20: Effect of albumin peptides on IL-2 induced proliferation
[0675] The
effect of albumin peptides including at least one of SEQ ID NOs:
183-185 or 188-246 is determined using the ex vivo human model as described in
Example 2.
[0676] PBMCs
are isolated from venous blood samples from healthy blood
donors (control samples) or cancer patients. One hundred pI of culture medium
(RPMI
1640 Dutch's modification (Gibco, InVitrogenAB, Stockholm, Sweden)
supplemented
with 200 IV/ml penicillin, 200 ul/rnl streptomycin, 4 mM L-glutamine (all from
Sigma
Chemical Co. MO, US) and 20% heat-inactivated human serum) is added to
roundbottomed, 96-well tissue culture plates (Costar, Corning Inc. NY, US).
For
experimental cultures, the culture medium of each well is supplemented with a
peptide of
SEQ ID NOs: 183-185 or 188-246. One hundred pI of PBMCs in RPMI/2% HAS
(5x104 lymphocytes) is then added per well followed by IL-2 (Proleukin,
Chiron, NL) at
a final concentration of 120 IU/well. Control wells without IL-2 are set up in
parallel.
Cells are cultured for 7 days in a humidified, 5% CO2- atmosphere at 37 C.
Cell
proliferation is assayed by incorporation of 1.6 pCi/well of 113H1-thymidine
(Amersham
Int., UK) during the last 18-24 h hrs. Mean values of dpm (disintegrations per
minute) of
triplicate wells are used for the calculations.
[0677] Thus,
albumin peptides that inhibit IL-2 stimulation of PBMC's are
identified.
265
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
Example 21: Effect of albumin peptides on T cell receptor stimulation
[0678] The effect of albumin peptides including at least one of SEQ
ID NOs:
183-185 or 188-246 on T Cell receptor stimulation is determined as in Example
3. Cells
are stimulated in cultures on plates pre-coated with a monoclonal antibody
directed
against CD3 and the number of metabolically active cells (i.e., cell
proliferation) is
determined using MTS staining after 3 to 7 days of culture. Detection of solid
phase
CD3 monoclonal antibody is used as a measurement of T cell proliferation.
[0679] Thus, albumin peptides that inhibit T cell receptor
stimulation are
identified.
Example 22: Effect of albumin peptides on NK cell cytotoxicity
[0680] The effect of albumin peptides including at least one of SEQ
ID
NOs: 183-185 or 188-246 on NK cell cytotoxicity is determined as in Example 4.
[0681] Mononuclear cells are separated by standard Ficoll-paque Plus
(Pharmacia AB, Sweden) density gradient centrifugation from heparinized blood
obtained from healthy donors. NK cell cytotoxic activity of the mononuclear
cells is
then tested using a commercial kit (NKTEST, Orpegen Pharma GmblI, Heidelberg,
Germany) following the manufacturers protocol. Briefly, the kit contains
cryopreserved,
NK-sensitive target cells (K562) labeled with a lipophilic green fluorescent
membrane
dye, which enables discrimination of effector and target cells. After
incubation with
effector cells, killed target cells are identified by a DNA-stain, which
penetrates and
specifically stain the nuclei of dead target cells. This way the percentage of
killed targets
can be determined by flow cytometry. The mononuclear cells were preincubated
for 30
min at 37 C with the indicated peptides (peptides have been described
previously) at 10
ug/ml. Target cells were then added, giving an effector:target ratio of 40: 1,
and the cell
mixture incubated at 37 C for 3-4 hours. Samples are analysed on a FACSCalibur
(BD
Biosciences, San Jose, Calif.).
[0682] Thus, albumin peptides that inhibit NK cell cytotoxicity are
identified.
Example 23: Effect of albumin peptides on leukocyte spreading
[0683] The effect of albumin peptides including at least one SEQ ID
NOs:
183-185 or 188-246 on leukocyte spreading is determined as in Example 5. Buffy
coat
cells are prepared from heparinized blood by Dextran assisted sedimentation.
To test the
266
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
effects of each peptide, a samples of cells are treated with of one of the
peptides of (SEQ
ID NOs: 183-185 or 188-246 at a concentration of 10pg/m1 for 15 minutes
efficiently
inhibited the spreading. These cells are then washed twice in PBS and
transferred to
clean slides. Cells adherance to the glass surface and spreading is detected.
[0684] Thus, albumin peptides that inhibit leukocyte spreading are
identified.
Example 24: Effect of albumin peptides on immune cell migration
[0685] The effect of albumin peptides including at least one of SEQ
ID NOs:
183-185 or 188-246 on immune cell migration is determined as in Example 5.
PBMC
migration is studied using the Boyden chamber technique. Migration for PBMCs
of
healthy control samples and cancer patients is assessed in both the presence
and absence
of each of the peptides of SEQ ID NOs: 183-185 or 188-246. Thus, albumin
peptides
that inhibit immune cells migration are identified.
Example 25: Binding of albumin peptides to LFA-1
[0686] The binding of albumin peptides including at least one of SEQ
ID
NOs: 183-185 or 188-246 to LFA-1 is determined as in Example 7. A standard
immunohistochemical staining procedure is performed using acetone fixation,
10%
human AB-serum for blocking, incubation with anti-LFA-1 antibody and a
secondary
antibody (Ultravision) followed by development with Fast Red. Pre-incubation
with
peptides added to the AB serum is either no peptide added, or a peptide of SEQ
ID NOs:
183-185 or 188-246 is added.
[0687] Peptides that bind to LFA-1 prevent the binding of the
antibody, thus
decreasing the amount of Fast Red staining in antibody-treated cells as
compared to
untreated control samples.
Example 26: Antibodies that bind albumin peptides
[0688] Antibodies that specifically bind to peptides including at
least one of
SEQ ID NOs: 183-185 or 188-246 are generated as in Example 9. Rabbit antisera
directed against each of the peptides of SEQ ID NOs: 183-185 or 188-246 are
generated.
Each peptide of SEQ ID NOs: 183-185 or 188-246 is synthesized with a cysteine
added
to the N-terminus end and then conjugated with keyhole limpet hemocyanin (KLH)
as a
carrier protein. Polyclonal antisera is generated by repeated immunizations of
rabbits
267
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
with KLH-conjugated P3028 and Freund's adjuvants. The antisera are affinity
purified
chromatography on P3028-conjugated Ultralink Iodoacetyl gels (Pierce
Biotechnology
Inc.).
[0689] The antisera are
tested for their ability to bind human serum and
dHSA. Human serum commercially available for therapeutic purposes is tested,
heated
times in order to be virus free. Thus, rabbit antisera that specifically binds
the
albumin peptide binds to dHSA and/or control sample HSA.
[0690] The binding of the
rabbit antiserum to peptides of SEQ ID NOs: 183-
185 or 188-246 is assayed using competition ELISA assay.
[0691] Effects of affinity
purified antibodies directed against of SEQ ID
NOs: 183-185 or 188-246 on the proliferative response to IL-2 are examined the
ex vivo
model, using PBMCs from immunosuppressed cancer patients and normal control
sample[O692] Thus, antibodies that
bind peptides of SEQ ID NOs: 183-185 or 188-
246 are identified.
Example 27: Peptides that bind to albumin-derived peptides
[0693] Peptides that bind to
peptides including at least one of SEQ ID NOs:
183-185 or 188-246 are identified as in Example 10. Potential binders of the
peptides
are synthesized. For each peptide of SEQ ID NOs: 183-185 or 188-246 a His-
tagged
peptide is contacted with the potential binders in solution, and then isolated
from solution
using the His tag. Binders of each peptide are isolated along with the
peptide, and
subsequently identified.
[0694] Additionally,
substitutions, truncations, and deletions of peptides that
bind to each of the albumin peptides are identified as in Example 12.
Substitutions,
truncations, and deletions are synthesized on a chip, and contacted with the
albumin
peptide of one of SEQ ID NOs: 183-185 or 188-246 to determine binding. The
amount
of bound peptide is quantified using a rampo assay as in Example 12. The
binders with
the highest rampo scores are isolated.
[0695] The highest-score
binders of each peptide are assessed for their ability
to reduce immunosuppression, as in Examples 13 and 15. Each binder is assessed
for its
ability to induce lymphocyte activation, and unblock the LFA-1 receptor.
Additionally,
each binder is assessed to bind to tumor cells, as in Example 14.
268
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
Example 28: Effect of P28R on mitochondrial metabolism and conversion of MTS
[0696] PBMCs
from eight healthy control samples and nine cancer patients
with various diagnoses (including renal cell cancer, malignant melanoma,
rectal cancer,
small cell lung cancer, non-small cell lung cancer (adenocarcinoma), squamous
cell
carcinoma, bladder cancer, osteosarcoma, pancreatic cancer, or bronchial
cancer) were
cultured in a modified version of the ex vivo model of Example 2 for seven
days in the
presence of various quantities of P28R (SEQ ID NO: 2), and control samples
were
untreated with P28R. As shown in Figures 33A and 33B, the cells were cultured
in
either no P28R 322, 5pg/mL 324, 10pg/m1 326, or 20pg/m1 328 of P28R. A dose
dependent stimulation of the mitochondrial metabolism measured as conversion
of MTS
was observed in 5/8 (see Figure 33A) control samples and 9/9 cancer patients
(see
Figure 33B). Similar results were obtained when the PBMCs were cultured for
only
three days.
Example 29: Effects of inhibitors of immunoregulatory peptides on
mitochondrial
metabolism and conversion of MTS
[0697] The
effect of P28R (SEQ ID NO: 2) on mitochondrial metabolism based
on MTS conversion was compared to the effect of a closely related peptide P27.
P27 (aka
"SCF 27") has the sequence KKLDTFFKKLSLFTER (SEQ ID NO: 264), and is a variant
of P28R that differs in that V8 of P28R is substituted to K8 in P27. P28R
binds to P3028
more efficiently than P27 (P27 binds P3028 with a rampo score of 253, while a
P28R control
sample binds P3028 with a rampo score of 308; see Example 12).
[0698] PBMCs
from cancer patients with various diagnoses were cultured in
a modified version of the ex vivo model of Example 2 with various
concentrations of
P28R or P27 (N=9 for P28R: N=8 for P27). The concentrations were either
untreated
control samples, 5pg/mL ("5CF28-R5" and "5CF275"), 10pg/m1 ("5CF28-R10" and
"SCF2710"), 20pg/m1("SCF28-R20" and "5CF2720"), or 40pg/m1("SCF28-R40" and
"5CF2740"). The results are shown in Figure 34. While P28R stimulated the
cells of
cancer patients in a dose-dependent manner, P27 had no effect.
Example 30: Effect of P28R on IL-2 induced proliferation (BrdU incorporation)
[0699] The
effect of P28R (SEQ ID NO: 2) on IL-2 induced proliferation was
measured in a BrdU incorporation assay. PBMCs from six healthy control samples
and ten
cancer patients (including renal cell cancer, malignant melanoma, rectal
cancer, small
269
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
lung cancer, non-small cell lung cancer (adenocarcinoma), squamous cell
carcinoma,
bladder cancer, osteosarcoma, pancreatic cancer, or bronchial cancer) were
harvested in
modified version of the ex vivo model of Example 2. One hundred pI of culture
medium
(RPMI 1640 Dutch's modification (Gibco, InVitrogenAB, Stockholm, Sweden)
supplemented with 200 IV/ml penicillin, 200 ul/rnl streptomycin, 4 mM L-
glutamine (all
from Sigma Chemical Co. MO, US) and 20% heat-inactivated human serum) were
added
to roundbottomed, 96-well tissue culture plates (Costar, Corning Inc. NY, US).
One
hundred pI of PBMCs in RPMI/2% HAS (5x104 lymphocytes) was then added per well
followed by IL-2 (Proleukin, Chiron, NL) at a final concentration of 120
IU/well.
sample wells without IL-2 was set up in parallel. Cells were cultured for 7
days in a
humidified, 5% CO2- atmosphere at 37 C. Cell proliferation was assayed by
incorporation of BrdU.
[0700] As shown
in Figure 35, four out of six control samples had a high
proliferative response to IL-2 compared to four out of ten cancer patients.
These
differences in proliferative response to IL-2 in PBMCs demonstrated the
difference
existence of high and low responders to IL-2.
[0701] The
response of high responders and low responders to various doses
of P28R was compared. Cells from either high responders or low responders were
cultured for 7 days with either no P28R, 5pg/mL, 10pg/m1, or 20pg/m1 of P28R.
IL-2-
induced proliferation was measured as BrdU incorporation, as in the above
example, and
the results are shown for high responders in Figure 36A, and low responders in
Figure
36B. While P28R had no stimulatory effect in cells from patients with a high
response to
IL-2 (N=4) (see Figure 36A), P28R had a stimulatory effect on cells from
patients with a
low response to IL-2 (N=6) (see Figure 36B).
Example 31: Effects of inhibitors of immunoregulatory peptides on IL-2 induced
proliferation (BrdU incorporation and MTS conversion)
[0702] The
effect of P27, a peptide related to P28R was compared to the
of P28R on I1-2induced proliferation as measured by BrdU Incorporation. P27
(aka "SCF
27") has the sequence KKLDTFFKKLSLFTER (SEQ ID NO: 264), and is a variant of
P28R that differs in that V8 of P28R is substituted to K8 in P27. P28R binds
to P3028 more
efficiently than P27 (P27 binds P3028 with a rampo score of 253, while a P28R
control
sample binds P3028 with a rampo score of 308; see Example 12).
270
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
[0703] PBMCs
from low responder cancer patients of Example 30 were
cultured as in Example 30, except that some samples were cultured with various
concentrations P28R (aka "SCF28-R"), and others were cultured with various
concentrations of P27 (aka "SCF27"). The concentrations were either no peptide
("untreated cells"), 5pg/mL, 10pg/ml, or 20pg/ml. BrdU incorporation was
measured as
in Example 30. As shown in Figure 37, both P28R and P27 enhanced the
proliferative
rate of PBMC's induced by IL-2. A comparison can be drawn to the data of
Example 29
and Figure 34, in which P28R, but not P27 enhanced IL-2 stimulation of
mitochondrial
metabolism, as measured by MTS conversion. P27 was observed to enhance IL-2
stimulation of cell proliferation as measured by BrdU incorporation, but not
mitochondrial metabolism as measured by MTS conversion. On the other hand,
P28R
was observed to enhance both parameters. The inhibitory peptide P3028 binds to
different receptors, including CD25 (see Example 8 and Figures 18-19) and LFA-
1 (see
Example 7 and Figures 15-16), as described herein. It is contemplated that the
more
efficient binder of P3028, P28R, is capable of removing P3028 from LFA-1
and/or
unblocking CD25. However, it is contemplated that P27 with a lower /weaker
binding to
P3028, does not have the capacity to unblock LFA-1 but can unblock CD25. Thus,
it is
contemplated that different populations of patients may be affected in
different ways by
immunoregulatory peptides such as P3028. Moreover, it is contemplated that
different
inhibitors of immunoregulatory peptides can modulate the activity of different
receptors,
and/or different signal transduction pathways.
Example 32: Comparison of MTS and BrdU assays
[0704] The two
cell proliferation assays in this study are both widely used in
order to measure cell proliferation. Peptide P28R had a stimulatory activity
of MTS
conversion in seven day cultures of PBMCs in 9/9 patients and in 5/8 healthy
control
samples. In contrast, P28R stimulated incorporation of BrdU in seven day
cultures of
PBMCs from only 1/6 and 2/10 patients.
[0705] IL-2
induced proliferation, measured as incorporation of BrdU, was
stimulated by P28R in PBMC cultures from cancer patients with a low
proliferative
response to IL-2 (experimental conditions were as described in Example 30).
PBMCs
from 2/3 healthy control samples and 2/4 cancer patients were not stimulated
by IL-2
when the effect was measured as MTS conversion (experimental conditions were
as
271
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
described in Example 28). However, PBMCs from all these persons ("non-
responders")
who did not respond when measured with MTS were significantly stimulated by IL-
2
when the effect was measured as incorporation of BrdU.
[0706] The
above results are illustrated in Figure 38. PBMC cultures from
two different patients (A, B) and (C, D), with IL-2 382 (bars on left) or
without IL-2 384
(bars on right). The effect of IL-2 and the peptides P28R (aka "SCR28R") and
P27 (aka
"SCF27") were measured at concentrations of either no peptide ("untreated
cells"),
5pg/mL, 10pg/m1, or 20pg/m1 of peptide.
[0707] In two
patients, the response to IL-2, measured as BrdU incorporation,
was enhanced by P28R (see Figures 38A and 38C), but this effect of P28R was
only
observed in one of these patients when MTS conversion was used (see Figure
38B).
Thus, while in one patient (see Figures 38A and 38B) the stimulatory activity
of IL-2
was registered using both BrdU and MTS, in the other patient, the stimulatory
activity of
IL-2 was registered using BrdU only (see Figure 38C). Based on these
observations, it
is concluded that effects on the metabolic activity measured as MTS conversion
does not
always correlate with DNA synthesis measured as incorporation of BrdU.
[0708]
Additionally, P28R enhanced the effect of IL-2 measured both with
BrdU and MTS, but the stimulatory effect of SCF27 was observed only when BrdU
incorporation is measured. In the patient shown in C the results are very
similar to those
shown in A, but in D no stimulatory effect is seen when the effect is
determined using
MTS conversion.
[0709] These
results indicate that albumin-derived immunomodulatory
structures such as P3028 appear to modulate signal transduction through
different
mechanisms. Thus, different patient populations can respond differently to
inhibitors of
immunomodulatory peptides. It is contemplated that in vitro diagnostic assays
can be
helpful in identifying which patients have albumin-derived immunomodulatory
structures, and can be further helpful in identifying which patients will
respond to certain
inhibitors (or combinations of inhibitors) of immunomodulatory structures.
Example 33: Effects of binders of immunoregulatory peptides on lymphocyte
activation
[0710] Binders
of immunoregulatory peptides, for example the peptides of
Tables 5.1, 6.1, 6.2, or 12 (SEQ ID NOs: 1-32, 265-393), or SEQ ID NOs: 34, 46-
53,
64-66, 68, 76, 94-96, 98, or 264, are assayed for effects on lymphocyte
activation, as in
272
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
Example 13. Analyses of these peptides are performed in human ex vivo models.
The
stimulatory activity on PBMCs, measured using the MTS or CFSE techniques, are
in 7 healthy control samples and 7 cancer patients of various diagnoses. The
peptides are
assayed for stimulatory activity even in the absence of other types of
stimulation, and are
compared to untreated control samples.
[0711]
Stimulatory activity of the peptides of Tables 5.1, 6.1, 6.2, or 12
(SEQ ID NOs: 1-32, 265-393), or SEQ ID NOs: 34, 46-53, 64-66, 68, 76, 94-96,
98, or
264 on a proliferative response to IL-2 suppressed by a P3028 sequence or
structure.
PBMCs are cultured for 7 days with IL-2 and the proliferative rate is
determined as
incorporation of BrdU. Each set of conditions is assayed in triplicate.
Initial
proliferation of PBMCs is compared to proliferation of PBMCs from the same
donor
after treatment with each peptide.
Example 34: Binding of inhibitors of immunoregulatory peptides to tumor cells
[0712] A
biotinylated version of each of the P28R peptides of Tables 5.1,
6.1, 6.2, or 12 (SEQ ID NOs: 1-32, 265-393), or SEQ ID NOs: 34, 46-53, 64-66,
68, 76,
94-96, 98, or 264, each of which has been shown to bind to P3028, is used to
assay
binding of the peptide to tumor cells. Five breast cancers, two renal cell
carcinomas and
four malignant melanomas are analyzed, as in Example 14.
Example 35: Unblocking the LFA-1 receptor by inhibitors of immunoregulatory
peptides
[0713] As
described herein, 02-integrins play a role in the normal function of
the immune system. Also described herein are immunosuppressor mechanisms based
on
the binding of an endogenous inhibitor, P3028, to the 02-integrin LFA-1. As
described
in Example 7, the membrane staining of PBMCs from cancer patients is markedly
decreased compared to normal control samples. The exposure of LFA-1 could,
however,
be enhanced by incubating PBMCs from cancer patients with an antibody directed
against the inhibitor P3028 (see Example 7 and Figure 16).
[0714] Staining
for LFA-1 is performed with the anti-LFA-1 antibody of
Example 7 and a secondary antibody (Ultravision) followed by development with
Fast
Red. Fresh frozen tumor sections without any fixation are incubated for 4-20
hours with
each of the P28R peptides of Tables 5.1, 6.1, 6.2, or 12 (SEQ ID NOs: 1-32,
265-393),
or SEQ ID NOs: 34, 46-53, 64-66, 68, 76, 94-96, 98, or 264, each of which has
been
273
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
shown to bind to P3028, before staining for LFA-1. For comparison, control
sample
tumor sections were incubated with phosphate buffered saline only. The amount
of anti-
LFA-1 antibody staining is observed, and used to determine the amount of
blocking, if
any, of the LFA-1 receptor. Migration and cytotoxic activity of treated cells
is also
ongoing.
Example 36: Positional scans of amino acid residues in SEQ ID NO: 2
[0715]
Positional scan data was used to study the influence of substitution of
different types of amino acids in each position of P28R (SEQ ID NO: 2) on the
binding
of P3028 (SEQ ID NO: 185). Each amino acid in the peptide sequence of P28R
(SEQ ID
NO: 2) was exchanged with all of the naturally occurring amino acids, and
immobilized
on a solid phase chip. The binding of P3028 to these "mutated" P28 R peptides
synthesized on a chip was determined using the ELISA technique. The results
are
summarized in Table 13. In view of the results, Table 13 includes a column
identifying
optional substitutions at each position that can maintain binding to P3028.
Table 13: Analysis of P3028 Binding to Solid Phase P28R Variants
Position Substitution ELISA signal Avg Optional
Category Substitutions
that maintain
3028 binding
K1 RHK 523 428 366 439 any type of
DE 373 396 385 amino acid
AVIL 466 442 483 449 460 possible
457 457
FYVV 332 315 284 310
STNQ 344 493 445 455 434
K2 RHK 417 394 445 419 positively
DE 335 349 342 charged amino
AVIL 309 317 331 343 325 acids
preferable,
274
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
400 400 F and N
FYW 390 301 304 332 possible*
STNQ 281 331 432 350 349
L3 RHK 370 477 386 411 any type of
DE 492 528 510 amino acid
AVIL 427 377 454 375 408 possible
460 460
FYW 393 344 341 359
STNQ 393 451 374 473 423
D4 RHK 317 317 274 303 any type of
DE 414 417 416 amino acid
AVIL 494 424 430 303 413 possible
384 384
FYW 380 422 443 415
STNQ 344 405 296 345 348
T5 RHK 430 391 237 353 polar
uncharged
DE 295 341 318 amino acids
AVIL 346 374 293 311 331
preferable, R, Y
475 475 and W are
FYW 290 425 418 378 possible*
STNQ 458 424 436 535 463
F6 RHK 309 332 309 317
hydrophobic and
DE 193 229 211
uncharged polar
AVIL 575 547 466 408 499 amino
acids are
467 467
preferable; avoid
FYW 437 364 348 383 positively and
STNQ 432 481 446 410 442 negatively
charged
F7 RHK 369 364 232 322
hydrophobic and
DE 301 381 341
uncharged polar
AVIL 426 527 446 517 479 amino
acids are
712 712 preferable; avoid
275
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
FYW 460 334 380 391 positively and
STNQ 700 517 348 511 519 negatively
charged
V8 RHK 365 213 253 277 hydrophobic
DE 122 139 131 non-aromatic
AVIL 299 308 401 411 355 carbon chain
221 221 amino acids are
FYW 358 211 228 266 preferable,
STNQ 216 298 203 271 247 F possible, avoid
negatively
charged
K9 RHK 374 306 377 352 positively
DE 149 240 195 charged amino
AVIL 191 248 190 166 199 acids preferable,
283 283 polar uncharged
FYW 174 198 245 206 T and Q possible
STNQ 274 347 256 330 302
L10 RHK 439 293 285 339 any type of
DE 102 81 92 amino acid
AVIL 426 658 415 348 462 except negatively
460 460 charged are
FYW 403 382 316 367 possible
STNQ 351 399 365 470 396
Sll RHK 333 535 323 397 polar uncharged
DE 322 234 278 amino acids are
AVIL 318 392 289 213 303 preferable, H is
744 744 possible*
FYW 250 402 324 325
STNQ 442 520 451 768 545
L12 RHK 483 460 355 433 any type of
DE 89 82 86 amino acid
AVIL 462 545 456 428 473 except negatively
276
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
499 499 charged
FYW 389 320 409 373
STNQ 478 437 462 651 507
F13 RHK 502 1046 220 589 any type of
DE 112 98 105 amino acid
AVIL 525 446 468 448 472 except negatively
1190 1190 charged*
FYW 402 291 430 374
STNQ 635 494 862 1144 784
T14 RHK 264 463 259 329 any type of
DE 159 110 135 amino acid
AVIL 305 380 375 360 355 except negatively
501 501 charged
FYW 348 270 374 331
STNQ 369 319 599 301 397
El5 RHK 237 318 324 293 negatively
DE 404 371 388 charged amino
AVIL 174 163 163 246 187 acids preferable,
247 247 possibly Y or Q
FYW 137 340 226 234
STNQ 165 152 161 344 206
R16 RHK 260 239 291 263 any type of
DE 133 107 120 amino acid
AVIL 226 254 176 217 218 except negatively
238 238 charged
FYW 297 311 202 270
STNQ 184 292 311 133 230
[0716] *It is
noted that M has a sulfur atom in the side chain, and without
being limited by any theory, it is contemplated that substitution of M only at
positions 8,
9, and/or 15 can result in reduced binding of the inhibitor peptide to P3028.
277
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
[0717] It was noted that the following categories of amino acid
residues at the
following positions are likely to be involved in binding of P3028 to P28R
(some
additional "possible" substitutions are noted in Table 13):
[0718] K2 positive charged amino acids
[0719] T5 polar uncharged amino acids
[0720] F6 hydrophobic and uncharged polar
[0721] F7 hydrophobic and uncharged polar
[0722] V8 hydrophobic, non-aromatic carbon chain amino acids
[0723] K9 positively charged amino acids
[0724] Sll polar uncharged amino acids
[0725] EIS negatively charged amino acids
[0726] Thus, in some embodiments, a central core, T5-S11, and two
additional amino acids, K2 and EIS, are identified to be involved in the
binding of the
peptide P3028.
[0727] From the positional scan data it is also noted that a "core
peptide" can
be identified, FFVKLS (SEQ ID NO: 62) (also referred to herein as "P28 core"),
bind the
3028 peptide as efficiently as the full length peptide P28R. However, the P28
core
peptide does not stimulate PBMC activation (CD69 and CD71) in short term
cultures of
this model, while the P28R peptide does stimulate PBMC activation in short
term
cultures of this model.
[0728] However, in cultures with human and dog cancer sera, P28 core
has a
stimulatory activity. As such, without being limited by any theory, it is
contemplated
that P28 core can be useful in de-blocking inhibitory effects of P3028 (e.g.
displacing
bound 3028 structures from the cellular receptors). For example, in some
embodiments,
P28 core can be useful in de-blocking P3028-mediated inhibition of the LFA-1
receptor.
[0729] Based on the positional scan data, it is contemplated that
substitutions
of SEQ ID NO: 2 could be useful in binding P3028, de-blocking the LFA-1
receptor
from P3028-mediated inhibition, and/or stimulating immune cells.
Example 37: Effect of modified peptides on PBMC activation
[0730] The activity of peptide P28R (SEQ ID NO: 2) and modifications
of
P28R was studied in a human ex vivo model using PBMCs in short term cultures,
24 or
48 hours. Effects of P28R and modifications of P28R on PBMC's from a healthy
control
278
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
person were studied. Activation was measured as percentage of cells with
enhanced
marker CD69 using flow cytometry. PBMCs were incubated with the peptides
(40 g/mL) for 24 hours in RPMI plus 10% human AB serum.
[0731] The
influence of various amino acid substitutions on the stimulatory
effect (measured as expression of CD69) in this ex vivo model was studied.
Stimulatory
effects of P28R and amino acid substitutions that exhibit a good binding
capacity
according to the positional scan were assessed. P28R (KKLDTFFVKLSLFTER)(SEQ
ID NO: 2), peptide 30677 (KKLDTFFVKLSLMTER)(SEQ ID NO: 583), peptide 30678
(KKLDTFFVKLQLFTER)(SEQ ID NO: 584), and peptide 30680
(KKLDTVMVKLQLMTER)(SEQ ID NO: 585) were examined (see Figure 41A).
Figure 41A illustrates the results of two experiments (410 and 412) for each
peptide. All
four peptides induced activation of PBMCs from the healthy control person.
[0732] P28R
(SEQ ID NO: 2), peptide 30864 (KSLDTFFVKLSLFTER, SEQ
ID NO: 586); peptide 30685 (KKLDTFFVKLSLFTFR, SEQ ID NO: 587); peptide 31135
(KKLDTFFVYLSLFTER)(SEQ ID NO: 588); peptide 31136
(KKLDTFFVNLSLFTER)(SEQ ID NO: 589), and peptide 31138
(KKLDTFFVDLSLFTER)(SEQ ID NO: 590) were examined (see Figure 41B). Figure
41B shows two experiments (414 and 416) for each peptide. Peptide 31135 also
stimulated immune cells. Accordingly, in addition to the analysis of Table 13,
tyrosine
may also be substituted in position 9 of SEQ ID NO: 2 in accordance with some
embodiments herein.
[0733] These
results show general agreement with the data from the analysis
based on the positional scan in Table 13 (see Example 36). Without being
limited by
any theory, some differences between the position scan data and immune cell
stimulation
data are not inconsistent with the disclosure herein. It is noted that Table
13 relates to
ability to bind to P3028 in an ELISA assay, while Figures 41A-B relates to an
assay for
PBMC activation. In some embodiments, a peptide comprising, consisting
essentially of,
or consisting of SEQ ID NO 2 or 583-585 stimulates healthy immune cells, for
example
PBMC' s.
Example 38: Effect of P28 core peptide on PBMC activation
[0734] As
observed in Example 37, P28R (SEQ ID NO: 2) can stimulate
PBMC's from healthy controls in short term cultures when RPMI plus 10% normal
279
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
human AB serum is used as culture medium. Truncations of P28R were also
assessed
their ability to activate PBMC's. PBMCs were incubated with the peptides (40
g/mL)
24 hours in RPMI plus 10% human AB serum. PBMC activation was measured as
percent cells with enhanced expression of either CD69 (Figure 42A) or CD71
(Figure
42B) using flow cytometry. Two experiments were performed for each peptide.
[0735] As shown
in Figures 42A and 42B, peptide P28R (SEQ ID NO: 2)
effectively activated healthy PBMC's in this model, but peptide 32251 (SEQ ID
NO:
592) and peptide 32230 ("P28 core")(FFVKLS)(SEQ ID NO: 62) did not activate
healthy
PBMC's in this model.
[0736] However,
in PBMC cultures where normal human AB-serum in the
culture medium was substituted for by sera from dogs with cancer or human
patients
with cancer, P28R (SEQ ID NO: 2) and P28 core (peptide 32230(FFVKLS)(SEQ ID
NO:
62) each activated PBMCs, measured as enhanced expression of CD69 (see Figure
43).
Figure 43 shows a comparison between the full length peptide P28R (SEQ ID NO:
2)
and the 6 amino acid P28 core sequence (peptide 32230)(FFVKLS)(SEQ ID NO: 62)
in
culture medium containing sera from two different cancer patients (human ca
serum 1
430 and human ca serum 2 432). Both P28R (SEQ ID NO: 2) and P28 core (SEQ ID
NO: 62) activated PBMCs in the presence of cancer serum.
[0737] In
addition, biotinylated P28R has been shown to bind directly to
PBMCs as demonstrated by immunocytochemistry or rosetting of P28R coated beads
(binding of beads to the cells).
[0738] Taken
together, these results show that P28R (SEQ ID NO: 2) can
bind to P3028 and de-block cellular receptors and can also have a direct
stimulatory
activity on immune cells. Additionally, P28 core (SEQ ID NO: 62) can bind to
P3028
and de-block cellular receptors.
Example 39: Cytotoxic activity of P28R
[0739] The
effect of P28R (SEQ ID NO:2) was further studied in in vivo
models in nude and immunocompetent mice. Injection of P28R intra-tumorally
into
human pancreas cancer in a xenograft model in nude mice demonstrated a
capacity to
induce tumor cell apoptosis after one day. Figures 44A and 44B shows
immunohistochemical staining for Caspase 3 (440), indicating an ongoing
apoptosis)
a significantly enhanced activation of this enzyme in P28R treated tumors
(Figure 44A)
280
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
compared to tumors which were treated with the drug solvent only (Figure 44B).
An
absence of staining is also indicated 442. It is noted that the results shown
were obtained
only one day after administration of P28R in animals with no capacity to form
an
reactivity to the tumor.
[0740] As such,
intra-tumoral administration of P28R a can have a cytotoxic
action on tumor cells in accordance with some embodiments herein. In some
embodiments, P28R has a direct cytotoxic action on tumor cells.
Example 40: Therapeutic activity of P28R
[0741] The
capacity of P28R (SEQ ID NO: 2) to activate the immune system
and thereby induce tumor cell-lysis was studied in immunocompetent mice,
C57B1, with
inoculated B16 melanoma. P28R, 40 microgram in 100 microliter, was injected
intra-
tumorally and the tumors were taken out after 3 days. As shown in Figure 45,
the
dominating cells in the tumors after this treatment are inflammatory cells,
which were
identified by immunohistochemical staining 450 using a polyclonal rabbit anti-
CD45
antibody (Figure 45A). For comparison a control tumor section was incubated
with
rabbit IgG at the same concentration (Figure 45B). An absence of staining is
also
indicated 452.
[0742]
Accordingly, it was demonstrated that P28R can induce infiltration of
a B16 melanoma tumor by inflammatory cells. In accordance with some
embodiments
herein, P28R can induce infiltration of tumors, for example melanomas, by
immune
cells.
Example 41: Effects of modified peptides on immune cell stimulation
[0743] The
influence of various amino acid substitutions and additions on the
immunostimulatory effect was studied. Effects of modified peptides on
activation of
PBMCs from healthy control person were assessed. Activation was determined as
percentage of cells with enhanced marker CD69 or CD71 using flow cytometry.
PBMCs
were incubated with the peptides (40 g/mL) for 48 hours in RPMI plus 10% human
AB
serum. Two experiments (460 and 462 in Figure 46A; 464 and 466 in Figure 46B,
respectively) were performed for each peptide. Peptides P28R (SEQ ID NO: 2),
P28
core (peptide 32230)(SEQ ID NO: 62), 32251 (KKLDTFFPKLSLFTER)(SEQ ID NO:
592), 32814 (RKLDTFFVKLSLFTERRR)(SEQ ID NO: 591), 32815
281
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
(KKLDQFFVKLSQHNER)(SEQ ID NO: 595), 32665 (SEQ ID NO: 593), and 32819
(SEQ ID NO: 594) were tested.
[0744] As shown
in Figure 46, peptide 32814 (SEQ ID NO: 591), had a
stimulatory effect in short term cultures similar to that of P28R (SEQ ID NO:
2) (batch
CS8040). Accordingly, peptide 32814 (SEQ ID NO: 591) activated healthy PBMCs
as
indicated by enhanced CD69 (Figure 46A) and also by enhanced CD71 (Figure
46B).
Example 42: Diagnostic uses
[0745] In
addition to therapeutic applications, diagnostic applications of P28R
and truncations and modifications thereof were also contemplated. For example,
information about patients systemic and local (intra-tumoural) immune status
can be
obtained using reagents comprising P28R, or a truncation or modification
thereof.
[0746] It is
contemplated that the occurrence of immunoinhibitory 3028-
structures in tumors can be identified by immunohistochemical staining using
either an
antibody directed against P3028 or using labeled P28R (SEQ ID NO: 2) or P28
core
(SEQ ID NO: 62), for example biotinylated P28R or P28 core. Figure 47 shows
two
areas of a human breast cancer stained using biotinylated P28R. Staining 470
is
observed in Figure 47B. Staining is not observed in Figure 47A. An absence of
staining is indicated 472.
[0747] As such,
areas of tumors comprising P3028 structures (as well as areas
not comprising these structures) can be identified using labeled peptides in
accordance
with embodiments herein.
Example 43: Treatment of a tumor using a P28 peptide inhibitor
[0748] A
patient having a melanoma is identified. A pharmaceutical
composition comprising 40pg/100m1 of a peptide consisting of the amino acid
sequence
SEQ ID NO: 2 and a PBS buffer formulated as a gel-like substance is injected
peri-
tumorally in the patient once a week for three weeks. Tumor cytotoxicity is
observed.
Immune cell invasion of the tumor is observed.
Example 44: Treatment of a tumor using a P28 core peptide inhibitor
[0749] A
patient having breast cancer is identified. A pharmaceutical
composition comprising a 80pg/100m1 of a peptide consisting the amino acid
sequence
282
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
SEQ ID NO: 62 and a tris buffer formulated as a gel-like substance is injected
peri-
tumorally in the patient. Immune cell invasion of the tumor is observed.
Example 45: Treatment of a tumor using a P28R-modification peptide inhibitor
[0750] A
patient having prostate cancer is identified. A pharmaceutical
composition comprising lmg/kg of a peptide consisting of the amino acid
sequence SEQ
ID NO: 586 dissolved in an aqueous buffer is administered systemically to the
patient
once every two days for five total administrations. Tumor cytotoxicity is
observed.
Immune cell invasion of the tumor is observed.
Example 46: Generation of immunoinhibitory P3028 structures by cancer cells
[0751] Human
prostate cancer cells were cultured in the absence of serum
proteins, and exhibited minimal immunostaining for P3028 strucutres, based on
detection
by rabbit antibodies (Figure 48A). The human prostate cancer cells were fed
human
serum albumin for 2 hours, and were stained for the presence of P3028
strucutres using
rabbit antibodies (Figure 48B). The albumin-fed cancer cells exhibited
substantially
higher levels of P3028 strucutres (as depicted by red staining 480 in Figure
48B) as
compared to the non-albumin-fed cells (as indicated by substantially lower
levels 482 of
red staining 480 in Figure 48A).
[0752] As such,
it has been shown that immunoinhibitory structures such as
3028 structures can be generated by cancer cells. It is contemplated that
inhibitors of
immunoregulatory proteins in accordance with some embodiments herein can be
useful
for countering the effects of such immunohibitory structures on cancer cells.
Example 47: Nanoparticle-inhibitors of albumin-derived immunoregulatory
peptide
compositions
[0753] Magnetic
DynabeadTM beads were bound to P28 core peptide
(FFVKLS)(SEQ ID NO: 62). The DynabeadsTM coated with P28 core peptide were
incubated with PBMC's for 24 hours. As shown in Figure 49A-B, untreated
control
PBMC's had substantial amounts of bound dHSA (shown as red staining 490 in
Figure
49A). After incubation with the DynabeadTm-P28 core particles, the PBMC's had
significantly reduced bound dHSA 492 (as indicated by substantially lower
levels of
staining 490 in Figure 49B) in comparison to the untreated PBMCs.
283
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
[0754]
Accordingly, it has been shown that nanoparticles associated with
immunoregulatory peptide inhibitors such as P28 core peptide in accordance
with some
embodiments herein can be delivered to immune cells bound by immunoregulatory
peptides, and further can reduce inhibition of immune cell receptors by
immunoregulatory peptides.
Example 48: Expression of P3028 epitopes
[0755] IHC
staining of P3028 of human tumours using a rabbit oligo-clonal
anti-P3028 antibody (Rimbo) generally shows high expression of this epitope.
Some
tumours even show a high expression intra-tumorally, in the cytoplasm. This
phenomenon has been further investigated in a mouse xenograft model of
prostate
cancer, PC3. As shown in Figure 50A, the cytoplasm was strongly stained in
certain
areas, however, a remarkable heterogeneity in the expression of this epitope
was seen
with extensive faintly staining areas (Figure 50B). The P3028 epitope has also
been
observed to be expressed by damaged HSA, and unexpectedly, this tumour (showin
in
Figures 50A-B) shows a high expression of the P3028 epitope although it is
grown in a
mouse with no HSA present. Without being limited by any theory, there can be
several
explanations, e.g. cross reactivity of the antibody with unknown prostate
cancer
structures / epitopes, uptake of mouse albumin by the tumour cells followed by
generation of the 3028 epitope also in mouse albumin (fragmentation or
denaturation).
[0756] In order
to further analyze then latter possibility (uptake of albumin)
the PC3 cell line was set up in cell culture. Figure 51A-B clearly shows that
when the
cells were "starved" (cultured only medium F12 without proteins) for 18 hours
the
stainability was markedly reduced, in particular in confluent areas but also
in
peripherally growing tumour cells, compared to sections of the same tumour.
[0757] The
cultures were then supplemented with HSA, MSA or BFS for 30
or 120 minutes. Figure 52 shows cultured human prostate carcinoma cells,
starved for
proteins for 18 hours and then incubated with human serum albumin for 2 hours.
IHC
staining for the 3028-structure using oligoclonal rabbit antibodies. Strong
expression of
the P3028 epitope was observed (Figure 52). Based on these observations it is
highly
unlikely that the antibody directed against the P3028 epitope cross-reacts
with some
unknown prostate cancer structures, reasonably in tumour sections from the
prostate
284
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
cancer mouse albumin has been transformed to expose some epitope binding the
anti-
P3028 antibody.
[0758] The
P3028 structure has been demonstrated to be a very potent
immunoinhibitor, and now has been shown to be efficiently produced by the
tumour cells
themselves. This reasonably means that these types of tumours have the
capacity to
efficiently inhibit immune mediated anti-tumour reactivity. Human breast
cancers were
stained using a biotinylated peptide binding to the P3028 structure.
Interestingly, great
differences in the staining patterns were obtained where the malignant cells
in some
tumours did not show any cytoplamatic expression of the P3028 structure. It is
contemplated that in accordance with some embodiments herein, the intra-
tumoural
expression of this structure can be of prognostic or predictive value.
[0759] The
production or immunoregulatory effect of this immunosuppressor
can to be blocked in accordance with some embodiments herein. Considering the
rapid
and extensive production of the P3028 structure by tumour cells, it is
contemplated that
conventional approaches might be difficult to supply substances blocking the
P3028
structure (antibodies or low molecular weight blockers) in sufficient amounts
in order to
maintain control of this type of immunosuppression long enough to achieve a
therapeutic
anti-tumour effect. There are then at least two alternatives: Block the uptake
and
generation of the P3028 structure by the tumour cells or use binders to the
P3028
structure to target toxins to the tumour cells.
Example 49: Activation of the immune system by P28R in immunocompetent mice
[0760] The
capacity of P28R to activate the immune system and thereby
induce tumor cell-lysis was studied in immunocompetent mice, C57B1, with
inoculated
B16 melanoma. P28R, 20 nM in 100 microliter, was injected intra-tumorally and
the
tumors were taken out after 3-5 days. As shown in Figure 53, the tumors were
permeated
by CD45+ inflammatory cells after this treatment (Figure 53A). For comparison
a
control tumor section was incubated with rabbit IgG at the same concentration
(Figure
53B). It is noted that in animals treated intra-tumorally with P28R a regional
lymph
node reaction was regularly found. Accordingly, these findings provide
evidence of an
inhibition of the cancer and such a systemic immune activation against the
cancer, a
vaccination effect, was achieved.
285
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
[0761] In this
tumour model bilateral tumours were inoculated, one in each
flank. P28R was injected into one of these tumours and tumour regressive
changes were
studied in these tumours as well as in the contralateral tumours either left
uninjected or
injected with the vehicle only. Figure 54 shows a tumour injected with the
vehicle after
one day with only minor tumour regressive changes.
[0762] It is
noted that intra-tumoral injection of P28R resulted in remarkable
tumour regressive changes not only in P28R treated tumours but also in
uninjected
contralateral tumour (Figure 55A-D) or tumours injected with saline only. The
effect in
the untreated distant / contralateral tumours increased with time after
injection of P28R
into the treated tumour.
[0763] Similar
results were obtained in a Lewis lung carcinoma model in
B57B1 mice. Compared to tumours in animals treated with P28R, tumours in
untreated
animals showed a predominance of tumour cells with only minor tumour
regressive
changes (Figure 56).
[0764]
Injection of P28R into the Lewis lung carcinoma tumours resulted in
extensive tumour regressive changes; both in the treated tumour (Figure 57A)
and in the
untreated contralateral tumour (Figure 57B). Effects of intra-tumoural
treatment of
spontaneous tumours in dogs with P28R in accordance with some embodiments
herein
are summarized below.
[0765]
Accordingly, it is shown that administration of immunoregulatory
peptide inhibitors in accordance with some embodiments herein can induce
regressive
changes in tumors, including tumors that receive the the immunoregulatory
peptide
inhibitor intratumorally, as well as tumors in other parts of the subject
(e.g. tumors
contralateral to the tumor that received the immunoregulatory peptide
inhibitor).
Examples 50-60: Intratumoral treatment of spontaneous tumors in dogs with P28R
[0766]
Seventeen spontaneous canine tumours, of variable histology have been
treated by intra-tumoral injection of 40nmol P28R in 200 microliters. The
tumours were
resected 3-5 days later, immediately snap frozen and stored at -80 C until
further processed.
A detailed description of treatment of the tumors is provided below (see
Examples 50-60). In
these examples, 15 control dogs with untreated tumours (5 snap frozen and 10
FFPE) were
also examined.
286
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
[0767] A
summary is provided for 7 dogs with treated mammary tumours and 5
untreated control dogs (see Figure 88). Tumour
cells (881 for treated dogs, 882 for
untreated control dogs) and lymphocytes (883 for both treated dogs and
untreated control
dogs) were counted at 400x magnification. The ratio between these cells was
used as a
measure on the inflammatory infiltrate (Figure 88). The reproducibility and
the influence of
tumour heterogeneity on cell counts were evaluated by counting several areas
from each
tumour section.
[0768] The
therapeutic efficacy was evaluated as the presence of an
inflammatory infiltrate and the occurrence of tumour cell regressive changes.
As migration of
effector cells close to tumour cells is one important, but often inhibited
function of these
cells, the effect of P28R was specifically evaluated as the presence of
inflammatory cells
infiltrating into the tumour cell areas close to the tumour cells. Antibodies
against the
following markers of inflammatory cells have been used: CD3, CD8, CD45 and
CD68.
[0769] [0865] As
outlined in Examples 50-54, five spontaneous tumours
were treated intra-tumorally with P28R, in all of these a strong inflammatory
infiltrate
was observed, mainly characterized as CD45+ cells and NK cells stained by
antibodies
directed against CD56 and NCR1 (see Figure 78A-B). Extensive tumour regressive
changes were found in three of these and in one, the apocrine gland carcinoma,
with
thick tumour nodules, regressive changes were seen at least in thin lesions
and at the
periphery of the tumour nodules. The thick tumour nodules were, however,
heavily
infiltrated by NK-cells. Interestingly, in a breast tumour with regional
metastases, also
these lesions were heavily infiltrated with inflammatory cells and showed
extensive
tumour regressive changes (Figure 58). Two tumours were injected with the
vehicle, in
one of these, a breast tumour, a spontaneous inflammatory infiltrate was
found. The
other, a testis tumour, did not show any inflammatory reaction.
[0770] More
than 20 dogs have been treated with P28R, including 4 in the
toxicological study (CiToxLab, Denmark) with 200nM administered in 1 mL
subcutaneously and 17 dogs in the treatment study reported here with 40nM in
200
microliters intra-tumourally. None of these dogs showed any systemic side
effects.
[0771]
Treatment schedule / strategy: The drug, P28R, is injected intra-
tumorally, 40 nM in 200 pl. The tumours are then resected within 3-5 days and
representative biopsis are immediately "snap frozen" and stored frozen / at -
80 C until
further investigated.
287
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
[0772] Objective of the treatments, performed in accordance with some
embodiments herein, include:
[0773] 1. Evaluate infiltration of inflammatory cells in H&E stained
sections
[0774] 2. Characterize inflammatory cells immunohistochemically.
(Based on
the pronounced stimulation CD3+ lymphocytes 48 h after exposure to P28R in a
human
ex vivo model, staining for T-cells was included in the analysis of the
inflammatory
infiltrate,)
[0775] 3. Analyse occurrence of tumour regressive changes,
morphologically
and immunohistochemic ally.
[0776] Dog tumor tissues were removed, snap-frozen in isopentane in
dry ice
and kept at -80 C until further processed. Frozen tissues were embedded in OCT
cryomount (Histolab, Goteborg, Sweden) and sectioned with 5 -7 pm thickness
using
Cryostat (CM3050S, Leica, Sweden) and the tissue sections were kept at -80 C
until
Tissue sections were fixed either in aceton or with 4% formalin for 10 mm
followed by
rinsing in distilled water. Tissue sections were then washed with PBS for 5
min for three
times and blocked with lx animal free blocker (VECTOR, Goteborg, Sweden) and
10%
serum (human AB serum (Sigma-Aldrich, Schnelldorf, Germany) or normal dog
serum
(Abcam, Cambridge, UK) for 30 min followed by incubation with primary antibody
for
lh at room temperature and then washed three times with PBS for 5 mm.
Immunohistochemistry staining was visualized with alkaline phosphatase (AP) or
diaminobenzidine (DAB). After primary antibody incubation and washing, AP
staining
was carried out using EXPOSE Mouse and Rabbit specific AP (Abcam, Cambridge,
UK)
by applying AP-conjugate (Expose Mouse and Rabbit specific AP, Abcam,
Cambridge,
Cambridge, UK) on the tissue sections and incubating for 30 mm, followed by
washing
four times with PBS for 5 mm. Thereafter tissue sections were incubated with
Enhancer
(Expose Mouse and Rabbit specific AP, Abcam, Cambridge, UK) for 4 mm. Without
rinsing off the Enhancer, mixture of equal volume of Naphtol Phosphate (Expose
Mouse
and Rabbit specific AP, abcam, Cambridge, UK) and Fast Red (Expose Mouse and
specific AP, abcam, Cambridge, UK) (1:1) and levamisole (DAKO, carpinteria,
USA) (1
drop/mi) were applied on tissue sections and incubated for 8 mm at room
temperature.
Tissues were then washed with PBS four times. After rinsing in distilled water
counter-
staining was carried out with Mayers hematoxylin (Histolab, Goteborg, Sweden)
for 10
min, following washing with running tap water for 10 mm and rinsing in
distilled water
288
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
before mounting tissue sections with aqueous mounting medium (AQUA PERTEX,
Histolab, Goteborg, Sweden). For immunohistochemistry with DAB, staining was
performed using DAKO Autostainer plus (DAKO, Glostrup, Denmark) using the
EnVision Flex High pH-kit (DAKO, Glostrup, Denmark) according to the company's
instruction. The stained sections were mounted with CYTOSEAL XYL (Thermo
Scientific, Cheshire, UK).
[0777]
Antibodies that have been used in this study were the following.
Rabbit active pro caspase 3 (ab13847, abcam, Cambridge, UK), mouse pan-
cytokeratin
(C1801, Sigma-Aldrich, Schnelldorf, Germany), Rabbit mouse NCR1 (bs10027R,
BioSite, Sweden), Rabbit IgG (ab27478, abcam, Cambridge, UK), mouse CD3
(ab699,
abcam, Cambridge, UK), mouse CD8a (ab34105, abcam, Cambridge, UK), mouse CD45
(ab34126, abcam, Cambridge, UK), goat anti-Rabbit IgG Envision FLEX/HRP (DAKO,
Glostrup, Denmark), goat anti-Rabbit and mouse IgG Envision FLEX/HRP (DAKO,
Glostrup, Denmark).
[0778] Accordingly, intra-tumoral administration of immunoregulatory
peptide inhibitors to subjects in accordance with some embodiments herein can
treat,
ameliorate, eliminate, and/or destroy tumors.
Example 50: D DOG TUMOUR 1
Clinical data
[0779] Breed of
dog: Cross breed. Weight: 15.1 kg. Sex: Spayed. Age: 85
months. Type of tumour: Apocrine gland carcinoma of the anal sac (primary
tumour).
Size of tumour: 25 mm. Clinical observations immediately after injection of
the P28R
and during the time interval until resection: No systemic adverse events were
observed.
Biopsies obtained:
[0780] A large
biopsy was taken from the site of injection in direction of drug
injection from the periphery towards the centre of the tumour. This was
divided into two
equal parts, biopsy 1 and 2. At the "bottom" of first biopsy a small
haemorrhage, 2-3 mm
in diameter was observed. Another biopsy was cut out, injection site 1, and
still at the
bottom of this third biopsy the haemorrhage continued. A fourth biopsy was
obtained
from this area, injection site 2.
Histopathological examination.
289
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
[0781] H&E
stained sections were obtained from all biopsies described
above. These were investigated for occurrence of tumour growth, infiltration
of
inflammatory cells and various degrees / types of tumour regressive changes.
H&E stained sections
[0782] This
tumour grew in large bulky tumour nodules, sometimes with
central necroses, surrounded by stromal tissue. Tumour regressive areas were
only
observed at the periphery of such nodules or in areas of thin tumour growth
(see Figure
59).
[0783] A
careful examination of such tumour nodules revealed the existence
of at least two types of cells, large cells with faintly staining nuclei and
small cell with
dense nuclei (Figures 60-61).
IHC characterization of the inflammatory infiltrate
[0784]
Characterization of the inflammatory cells has mainly focused on the
innate immune response as the tumours were resected 3 days after intra-tumoral
injection
of the drug. Staining with an antibody directed against CD45 is shown in
Figure 62.
[0785] As shown
in Figure 62, the CD45+ inflammatory cells are mainly
localized in the stromal areas surrounding the tumour nodules, which are not
infiltrated
by these cells. Tumour regressive changes, immune mediated tumour cell lysis,
is only
observed at the periphery of the tumour nodules or in "thin" tumour areas.
[0786] As shown
in Figure 63, only few scattered CD3+ or CD8+ cells were
found after treatment in this tumour.
Staining for NK-cells, CD56 and NCR]
[0787] As the
tumour was resected three days after intra-tumoural treatment
and only minor T-cell recruitment was found, the innate response was
investigated by
staining for NK-cells and macrophages (see Figures 64-65).
[0788] As shown
in Figures 64-65, The more intense staining of CD56+ cells
in the stromal areas and the gradient of staining intensity of these cells in
the tumour
nodules indicate that the CD56 marker might be lost from the cells (shedding)
possibly
due to high proteolytic activity in these areas.
[0789]
Accordingly, it has been shown that apocrine gland carcinoma of the
anal sac, resected 3 days after intra-tmoural treatment with P28R in
accordance with
embodiments herein. A strong mainly stromal infiltration of CD45+ cells was
found with
infiltration close to tumour cells with tumour regressive changes found in
"thin" tumour
290
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
areas and the peripheral parts of some tumour nodules. CD56+ cells were found
to
permeate the tumour nodules.
[0790] Accordingly, intra-tumoral administration of immunoregulatory
peptide inhibitors to subjects in accordance with some embodiments herein can
cause
regressive changes in carcinoma tumors of the anal sac, and can treat,
ameliorate,
eliminate, and/or destroy such tumors.
Example 51: D DOG TUMOUR 2
Clinical data
[0791] Breed of
dog: N/A. Weight: 36.0 kg. Age: 106 months. Sex: Male.
Type of tumour: Testis tumour (primary tumour). Size of tumour: 50 mm.
Clinical
observations immediately after injection of the vehicle and during the time
interval until
resection: No systemic adverse events were observed.
Biopsies obtained:
[0792] One
large biopsy was taken from the site of injection in direction of
drug injection from the periphery towards the centre of the tumour.
Histopathological examination.
[0793] H&E
stained sections were investigated for occurrence of tumour
growth, infiltration of inflammatory cells and various degrees / types of
tumour
regressive changes.
H&E stained sections
[0794] All
three biopsies were carefully analysed and showed the same result,
a testicular tumour with no signs of tissue destruction and only a weak
inflammatory
reaction of the degree usually found in these tumours (Figure 66A). The degree
of
inflammatory reaction was confirmed using staining for CD45 (Figure 66B).
[0795]
Accordingly, it has been shown that no inflammatory reaction or
tumour regressive changes were observed in this tumour after intra-tumoral
injection of
the vehicle of the drug formulation in accordance with some embodiments
herein.
Example 52: D DOG TUMOUR 3
Clinical data
[0796] Breed of
dog: Pug. Weight: 12.5 kg. Sex: Spayed. Age: 88 months.
Type of tumour: Mast cell tumour (primary tumour). Size of tumour: 10 mm.
Clinical
291
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
observations immediately after injection of the P28R and during the time
interval until
resection: No systemic adverse events were observed.
Biopsies obtained:
[0797] A large
biopsy was taken from the site of injection in direction of drug
injection from the periphery towards the centre of the tumour.
Histopathological examination.
[0798] H&E
stained sections were investigated for occurrence of tumour
growth, infiltration of inflammatory cells and various degrees / types of
tumour
regressive changes (see Figures 67A-B).
[0799] Only few
scattered CD3+ or CD8+ cells were found after treatment in
this tumour (see Figures 68A-68B).
[0800] In
Figure 69, a mastocytoma after intra-tumoral treatment with P28R
is shown. A massive tumour destruction and as extensive infiltration of CD45+
inflammatory cells are shown.
[0801] Staining
for NK-cells, CD56 and NCR1 was performed (see Figures
70A-D). As the tumour was resected three days after intra-tumoural treatment
and only
minor T-cell recruitment was found, the innate response was investigated by
staining for
NK-cells and macrophages.
[0802] As such,
mastocytoma after intra-tumoral injection of P28R in
accordance with some embodiments herein was observed. The tumour, resected
after 3
days, is permeated with CD56+ cells and shows extensive tumour regressive
changes.
[0803] Accordingly, intra-tumoral administration of immunoregulatory
peptide inhibitors to subjects in accordance with some embodiments herein can
cause
regressive changes in mast cell tumors, and can treat, ameliorate, eliminate,
and/or
destroy such tumors.
Example 53: D DOG TUMOUR 4
Clinical data
[0804] Breed of
dog: Farm dog. Weight: 8.6 kg. Sex: Female. Age: 86
months. Type of tumour: Benign mixed mammary tumour (primary tumour and
metastases). Size of tumour: 25 mm. Clinical observations immediately after
injection of
the P28R and during the time interval until resection: No systemic adverse
events were
observed.
292
CA 02978617 2017-09-01
WO 2016/144650
PCT/US2016/020510
Biopsies obtained:
[0805] The primary tumour was localized in a distal breast gland
close to the
groin:
1. A central slice, 5-7 mm thick, of the primary tumour corresponding to
the injection site was cut out.
2. A peripheral biopsy close to the central slice
3. In the peripheral slice a small haemorrhage was observed, probably the
centre of the injection, called injection site.
4. A separate metastasis close to the primary tumour, about 3cm in
diameter
5. A second separate metastasis close to the primary tumour
6. An additional tumour localized in the upper most breast gland close to
the axilla.
Histopathological examination.
[0806] H&E stained sections were obtained from all biopsies described
above. These were investigated for occurrence of tumour growth, infiltration
of
inflammatory cells and various degrees / types of tumour regressive changes.
[0807] All areas of all biopsies were infiltrated by inflammatory
cells. Only
small areas completely without infiltration of such cells were observed in any
of the
biopsies. In general, the vast majority of the tumour cell areas had a marked
infiltration
often with strongly distorted morphology of the tumour growth. The following
types of
tumour regressive areas were identified.
[0808] 1. Regressive changes related to the injection site
[0809] 2. Clear glandular structure with inflammatory infiltrate
[0810] 3. Highly distorted glandular structure with inflammatory
infiltrate
[0811] 4. Diffuse, confluent growth of tumour cells with pronounced
tumour
regressive changes and often a strong inflammatory infiltrate
[0812] H&E stained sections from different parts of the primary
tumour, a
regional metastasis and a distant tumour were analysed, The results are shown
Figures
71-74.
[0813] The characterization of the inflammatory infiltrate of this
tumour has
so far been performed using antibodies directed against CD45 and CD8 (see,
e.g. Figure
75).
293
DEMANDE OU BREVET VOLUMINEUX
LA PRESENTE PARTIE DE CETTE DEMANDE OU CE BREVET COMPREND
PLUS D'UN TOME.
CECI EST LE TOME 1 DE 2
CONTENANT LES PAGES 1 A 293
NOTE : Pour les tomes additionels, veuillez contacter le Bureau canadien des
brevets
JUMBO APPLICATIONS/PATENTS
THIS SECTION OF THE APPLICATION/PATENT CONTAINS MORE THAN ONE
VOLUME
THIS IS VOLUME 1 OF 2
CONTAINING PAGES 1 TO 293
NOTE: For additional volumes, please contact the Canadian Patent Office
NOM DU FICHIER / FILE NAME:
NOTE POUR LE TOME / VOLUME NOTE: