Note: Descriptions are shown in the official language in which they were submitted.
METHODS AND SYSTEMS FOR CONSTRUCTION OF NORMALIZED NUCLEIC
ACID LIBRARIES
RELATED APPLICATIONS
[0001] This application claims priority to U.S. Provisional Patent
Application
Serial No. 62/319,746, filed on April 7, 2016, and U.S. Provisional Patent
Application Serial
No. 62/348,766, filed on June 10, 2016.
FIELD
[0002] This disclosure relates generally to the field of production
and
normalization of nucleic acid libraries, and particularly relates to devices,
systems and
methods for preparation of normalized nucleic acid libraries suitable for
various downstream
analytical applications such as nucleic acid sequencing, particularly next
generation
sequencing.
BACKGROUND
[0003] The materials described in this section are not admitted to
be prior art by
inclusion in this section.
[0004] As common practice, biological samples used to prepare
nucleic acid
libraries for downstream analyses are homogeneously processed according to
standardized
assay protocols without regard to customized procedures for each sample. An
important
quality control (QC) measurement is nucleic acid concentration of nucleic acid
libraries since
performance of many modern nucleic acid analysis technologies is dependent on
the nucleic
acid concentration of input nucleic acids. Since such downstream analyses
typically use
expensive reagents and incur significant costs to perform, samples not meeting
nucleic acid
concentration requirements and/or other QC measurements are simply discarded
on the
assumption that the sample preparation was intrinsically unsuitable for the
desired
application.
[0005] For example, in next-generation sequencing (NGS)
applications, also
known as high-throughput sequencing, it is often desirable but logistically
challenging to
-1-
CA 3016221 2019-11-29
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
prepare nucleic acid libraries with substantially uniform DNA molar
concentrations. This
problem often arises in the preparation of nucleic acid libraries constructed
from a plurality
of heterogeneous biological samples, particularly when a desirable source
biological sample
is disproportionately underrepresented. This problem additionally arises when
a target
nucleic acid molecule is a relatively rarer and/or more unstable nucleic acid
species when
compared to even other nucleic acids that are derived from the same biological
sample.
SUMMARY
[0006] This section provides a general summary of the disclosure, and is
not
comprehensive of its full scope or all of its features.
[0007] The present disclosure generally relates to systems and methods
for
amplification and/or normalization of nucleic acids that can be performed on
individual
nucleic acid samples or libraries in order to generate a master pool of
nucleic acid libraries
having customized molar concentrations of target nucleic acid molecules. In
some
embodiments, the disclosed methods allow for generating nucleic acid libraries
having
substantially uniform concentrations across multiple nucleic acid libraries.
In some
embodiments, the disclosed systems and methods allow for preparation of
targeted amplicon
samples or libraries on a droplet actuator using a digital fluidic, e.g.
microfluidic, procedure.
[0008] A droplet actuator as described herein typically includes one or
more
substrates configured to form a surface or gap for conducting droplet
operations. The one or
more substrates establish a droplet operations surface or gap for conducting
droplet
operations and may also include electrodes arranged to conduct the droplet
operations. The
droplet operation substrate or the gap between the substrates may be coated or
filled with a
filler fluid that is immiscible with the liquid that forms the droplets.
[0009] Droplet actuators are used in a variety of applications in which
precise and
accurate handling of small volumes of liquid samples and reagents are required
for achieving
reliable analytical results. For example, droplet actuators are used to
conduct a variety of
molecular protocols, such as amplification of nucleic acids (e.g., polymerase
chain reaction
(PCR)). In one exemplary application, PCR techniques are used in the
preparation of targeted
amplicon samples or libraries for sequencing. On-bench protocols (e.g., plate-
based
protocols) have been developed for preparation of targeted amplicon samples or
libraries.
However, on-bench protocols typically involve multiple labor and time
intensive steps (e.g.,
-2-
pipetting, reagent preparation, etc.) and relatively large sample input
amounts. In addition,
the quality of sequence data and subsequent data analysis depends on the
quality of amplicon
sample or library construction. Therefore, there is a need for a flexible,
automated platform
for construction of nucleic acid samples or libraries that provides high
yield, uniformity, and
specificity across multiple samples and/or libraries from relatively low DNA
inputs, with less
reagent consumption, and allows a user to operate over a range of multiplexed
operations.
[0010] In
one aspect, disclosed herein are embodiments of methods for nucleic
acid amplification that include (a) providing a nucleic acid sample including
target nucleic
acid molecules; (b) contacting the nucleic acid sample with a reaction mixture
comprising a
solid phase and a liquid phase, where the solid phase includes a plurality of
first
amplification primers immobilized on a solid support, the first amplification
primers capable
of specifically hybridizing to a first sequence of the target nucleic acid
molecules, and where
the liquid phase includes a plurality of second amplification primers in
solution, the plurality
of second primers capable of specifically hybridizing to a second sequence of
the target
nucleic acid molecules; and (c) amplifying the target nucleic acid molecules
under isothermal
conditions such that substantially all of the first amplification primers are
incorporated into
amplification products, wherein the plurality of first primers is provided in
an amount which
limits the yield of amplification products to a predefined amount, and the
plurality of second
amplification primers is provided in an amount that exceeds the amount of the
first
amplification primers.[0010a] In
some embodiments, the present disclosure provides a
method for nucleic acid amplification comprising:
a) providing an input sample comprising target nucleic acid molecules;
b) contacting the sample with a reaction mixture comprising a solid phase and
a liquid
phase, wherein:
i. the solid phase comprises a plurality of first amplification primers
immobilized
on a solid support, the first amplification primers capable of specifically
hybridizing to a first sequence of the target nucleic acid molecules; and
ii. the liquid phase comprises a plurality of second amplification primers in
solution, the plurality of second primers capable of specifically hybridizing
to
a second sequence of the target nucleic acid molecules;
-3-
CA 3016221 2019-11-29
c) amplifying the target nucleic acid molecules under isothermal conditions,
wherein
the plurality of first amplification primers is provided in an amount which
limits the
yield of amplification products to a predefined amount; and
the plurality of second amplification primers is provided in an amount that
exceeds the
amount of the first amplification primers.
[0011]
Implementations of embodiments of the methods according to this aspect
and other aspects of the disclosure can include one or more of the following
features. In some
embodiments, the plurality of second amplification primers is provided in an
amount within
an order of magnitude of the amount of first amplification primer. In some
embodiments, the
plurality of second amplification primers is provided in an amount that
exceeds the amount
of the first amplification primers by at least 100%. In some embodiments, the
methods
further include separating the amplification products from the solid support.
In some
embodiments, the solid support includes a plurality of beads. In some
embodiments, the solid
support includes beads selected from magnetic beads, paramagnetic beads,
plastic beads,
polystyrene beads, glass beads, agarose beads, flow cytometry microbeads,
polystyrene
microparticles, polystyrene nanoparticles, functionalized polystyrene
microparticles,
-3a-
CA 3016221 2019-11-29
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
functionalized polystyrene nanoparticles, coated polystyrene microparticles,
coated
polystyrene nanoparticles, silica microbeads, fluorescent microspheres,
fluorescent
nanospheres, functionalized fluorescent microspheres, functionalized
fluorescent
nanospheres, coated fluorescent microspheres, coated fluorescent nanospheres,
color dyed
microparticles, color dyed nanoparticles, magnetic microparticles, magnetic
nanoparticles,
superparamagnetic microparticles, superparamagnetic nanoparticles, and
combinations
thereof In some embodiments, the beads are in an aqueous reaction buffer. In
some
embodiments, the beads are in fluid communication with each other. In some
embodiments,
the beads include streptavidin beads onto which the first amplification
primers are affixed
through conjugated biotin. In some embodiments, the beads are monoclonal. In
some
embodiments, the beads are polyclonal. In some embodiments, the solid support
is a surface
of a reaction site. In some embodiments, the surface of a reaction site
includes a bottom
portion of an inner surface of a well, a groove, a flow cell, a reaction
chamber or channel.
[0012] Some embodiments disclosed herein relate to methods for nucleic
acid
amplification in which the nucleic acid sample includes single-stranded
nucleic acid
molecules. Some embodiments disclosed herein relate to methods for nucleic
acid
amplification in which the nucleic acid sample includes double-stranded
nucleic acid
molecules.
[0013] In some embodiments of the methods for nucleic acid amplification
disclosed herein, the first amplification primers and the second amplification
primers
includes sequences complementary to known nucleotide sequences within the
target nucleic
acid molecules. In some embodiments, the known nucleotide sequences correspond
to the
first ends and second ends of the target nucleic acid molecules. In some
embodiments, the
first ends and second ends of the target nucleic acid molecules include
universal sequencing
tail-adaptors that have been added to the target nucleic acid molecules. In
some
embodiments, each of the first and/or second amplification primers further
includes an
indexing portion. In some embodiments, at least a portion of the first
amplification primers
further includes a capture portion having sequence complementarity to a region
of the target
nucleic acid molecules in addition to a known sequence of the target nucleic
acid molecules.
In some embodiments, the capture portion is generated by hybridizing a capture
oligonucleotide to a first amplification primer immobilized onto the solid
support and
-4-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
extending the immobilized amplification primer to generate an extended
amplification primer
having sequence complementarity to the capture oligonucleotide.
[0014] In some embodiments of the methods for nucleic amplification
disclosed
herein, the amplification step is performed on a plurality of nucleic acid
samples. In some
embodiments, the amount of each of the plurality of nucleic acid samples is
not normalized
across the plurality of nucleic acid samples. In some embodiments, the
plurality of nucleic
acid samples is combined before the amplification step. In some embodiments,
the
amplification products from the plurality of nucleic acid samples are combined
to form a
nucleic acid pooled nucleic acid library. In some embodiments, the
amplification products
from the plurality of nucleic acid samples are combined before being separated
from the
respective solid support. In some embodiments, the amplification products from
the plurality
of nucleic acid samples are combined after being separated from the respective
solid support.
[0015] Some embodiments of the methods disclosed herein further include
a step
of obtaining a nucleotide sequence of the amplification products. In some
embodiments, the
nucleotide sequence of the amplification products is obtained by nucleic acid
sequencing. In
In some embodiments, the nucleic acid sequencing includes high-throughput
sequencing, e.g.
next generation sequencing (NGS).
[0016] In some embodiments, the present disclosure provides methods for
nucleic
acid amplification that include (a) providing a nucleic acid sample including
target nucleic
acid molecules; (b) contacting the nucleic acid sample with a reaction mixture
comprising a
solid phase and a liquid phase, where the solid phase includes a plurality of
first
amplification primers immobilized on a solid support, the first amplification
primers capable
of specifically hybridizing to a first sequence of the target nucleic acid
molecules, and where
the liquid phase includes a plurality of second amplification primers in
solution, the plurality
of second primers capable of specifically hybridizing to a second sequence of
the target
nucleic acid molecules; and (c) amplifying the target nucleic acid molecules
under isothermal
conditions such that substantially all of the first amplification primers are
incorporated into
amplification products, wherein the plurality of first primers is provided in
an amount which
limits the yield of amplification products to a predefined amount, and where
at least a portion
of the first amplification primers further comprises a capture portion having
sequence
complementarity to a region of the target nucleic acid molecules in addition
to a known
-5-
sequences of the target nucleic acid molecules, and the capture portion is
generated by
hybridizing a capture oligonucleotide to a first amplification primer
immobilized onto the
solid support and extending the immobilized amplification primer to generate
an extended
amplification primer having sequence complementarity to the capture
oligonucleotide. In
some embodiments of the methods disclosed herein, the reaction mixture further
comprises
one or more of a recombinase, a single-strand DNA-binding protein, a helicase,
and a strand-
displacing polymerase. Further provided are compositions that include
amplification products
produced by a method according to this aspect and other aspects of the
disclosure.
[0016a] In some embodiments, the present disclosure provides a method for
nucleic acid amplification comprising:
a) providing an input sample comprising target nucleic acid molecules;
b) contacting the sample with a reaction mixture comprising a solid phase and
a liquid
phase, wherein:
i. the solid phase comprises a plurality of first amplification primers
immobilized
on a solid support, the first amplification primers capable of specifically
hybridizing to a first sequence of the target nucleic acid molecules; and
ii. the 'liquid phase comprises a plurality of second amplification primers in
solution, the plurality of second primers capable of specifically hybridizing
to
a second sequence of the target nucleic acid molecules;
c) amplifying the target nucleic acid molecules under isothermal conditions,
wherein:
the plurality of first amplification primers is provided in an amount which
limits the
yield of amplification products to a predefined amount,
at least a portion of the first amplification primers further comprises a
capture portion
having sequence complementarity to a region of the target nucleic acid
molecules in addition
to a known sequences of the target nucleic acid molecules, and the capture
portion is
generated by hybridizing a capture oligonucleotide to a first amplification
primer
immobilized onto the solid support and extending the immobilized amplification
primer to
generate an extended amplification primer having sequence complementarity to
the capture
oligonucleotide.
[0017] In
one aspect, disclosed herein are embodiments of methods for nucleic
acid amplification and/or normalization that include (a) providing an input
nucleic acid
-6-
CA 3016221 2019-11-29
sample including target nucleic acid molecules; (b) contacting the input
nucleic acid sample
with a reaction mixture comprising a plurality of first normalization primers
and a plurality
of second normalization primers, where the plurality of first normalization
primers
immobilized on a solid support, the first normalization primers capable of
specifically
hybridizing to a first sequence of the target nucleic acid molecules, and
where the plurality of
second normalization primers in solution, the plurality of second primers
capable of
specifically hybridizing to a second sequence of the target nucleic acid
molecules; and (c)
amplifying the target nucleic acid molecules under isothermal conditions such
that
substantially all of the first normalization primers are incorporated into
amplification
products, wherein the plurality of first primers is provided in an amount
which limits the
yield of amplification products to a predefined amount, and the plurality of
second
normalization primers is provided in an amount that exceeds the amount of the
first
normalization primers.
10017a1 In some embodiments, the present disclosure provides a method for
nucleic acid amplification comprising:
a) providing an input sample comprising target nucleic acid molecules;
b) contacting the input sample with a reaction mixture comprising a plurality
of first
normalization primers and a plurality of second normalization primers,
wherein:
i. the plurality of first normalization primers is immobilized on a solid
support, the
first normalization primers being capable of specifically hybridizing to a
first
sequence of the target nucleic acid molecules; and
ii. the plurality of second normalization primers is in solution, the
plurality of
second primers being capable of specifically hybridizing to a second sequence
of the target nucleic acid molecules;
c) amplifying the target nucleic acid molecules under isothermal conditions,
wherein
the plurality of first normalization primers is provided in an amount which
limits the
yield of amplification products to a predefined amount; and
the plurality of second normalization primers is provided in an amount that
exceeds the
amount of the first normalization primers.
[0018]
Implementations of embodiments of the methods according to the
disclosure can include one or more of the following features. In some
embodiments, the
-6a-
CA 3016221 2019-11-29
plurality of second normalization primers is provided in an amount within an
order of
magnitude of the amount of first normalization primer. In some embodiments,
the plurality of
second normalization primers is provided in an amount that exceeds the amount
of the first
normalization primers by at least 100%. In some embodiments, the methods
further include
separating the amplification products from the solid support. In some
embodiments, the
plurality of first normalization primers is hybridized with the target nucleic
acid molecules
-6b-
CA 3016221 2019-11-29
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
prior to being immobilized on the solid support. In some embodiments, the
plurality of first
normalization primers is immobilized on the solid support prior to being
hybridized with the
target nucleic acid molecules. In some embodiments, the solid support includes
a plurality of
beads. In some embodiments, the solid support includes beads selected from
magnetic beads,
paramagnetic beads, magnetically responsive beads, plastic beads, polystyrene
beads, glass
beads, agarose beads, and combinations thereof In some embodiments, the solid
support
includes beads selected from magnetic beads, paramagnetic beads, plastic
beads, polystyrene
beads, glass beads, agarose beads, flow cytometry microbeads, polystyrene
microparticles,
polystyrene nanoparticles, functionalized polystyrene microparticles,
functionalized
polystyrene nanoparticles, coated polystyrene microparticles, coated
polystyrene
nanoparticles, silica microbeads, fluorescent microspheres, fluorescent
nanospheres,
functionalized fluorescent microspheres, functionalized fluorescent
nanospheres, coated
fluorescent microspheres, coated fluorescent nanospheres, color dyed
microparticles, color
dyed nanoparticles, magnetic microparticles, magnetic nanoparticles,
superparamagnetic
microparticles, superparamagnetic nanoparticles, and combinations thereof. In
some
embodiments, the beads are in an aqueous reaction buffer. In some embodiments,
the beads
are in fluid communication with each other. In some embodiments, the beads
include
streptavidin beads onto which the first normalization primers are affixed
through conjugated
biotin. In some embodiments, the beads are monoclonal. In some embodiments,
the beads are
polyclonal. In some embodiments, the solid support is a surface of a reaction
site. In some
embodiments, the surface of a reaction site includes a bottom portion of an
inner surface of a
well, a groove, a flow cell, a reaction chamber or channel.
[0019] Some embodiments disclosed herein relate to methods for nucleic
acid
amplification and/or normalization in which the input nucleic acid sample
includes single-
stranded nucleic acid molecules. Some embodiments disclosed herein relate to
methods for
nucleic acid amplification in which the input nucleic acid sample includes
double-stranded
nucleic acid molecules. In some embodiments, the input nucleic acid sample
includes a
mixture of single-stranded nucleic acids and double stranded nucleic acids.
[0020] In some embodiments of the methods for nucleic acid amplification
and/or
normalization disclosed herein, at least one of the first normalization
primers and/or the
second normalization primers includes a region having sequence complementarity
to known
-7-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
nucleotide sequences within the target nucleic acid molecules. In some
embodiments, the
known nucleotide sequences correspond to the first ends and second ends of the
target
nucleic acid molecules. In some embodiments, the first ends and second ends of
the target
nucleic acid molecules include universal primer regions that have been added
to the target
nucleic acid molecules. In some embodiments, the universal primer regions
include a
sequencing-by-synthesis (SBS) primer sequence. In some embodiments, at least
one of the
first and/or second normalization primers further includes an indexing
portion. In some
embodiments, at least one of the first and/or second normalization primers
further includes a
region having sequence complementarity to the universal primer regions added
to the target
nucleic acid molecules. In some embodiments, at least a portion of the first
normalization
primers further comprises a capture portion having sequence complementarity to
a cognate
region of the target nucleic acid molecules in addition of a known sequence of
the target
nucleic acid molecules. In some embodiments, the capture portion is generated
by
hybridizing a capture oligonucleotide to a first normalization primer and
extending the first
normalization primer to generate an extended normalization primer having
sequence
complementarity to the capture oligonucleotide.
[0021] In some embodiments of the methods for nucleic acid amplification
and/or
normalization disclosed herein, the normalizing amplification step is
performed on a plurality
of input nucleic acid samples. In some embodiments, the amount of each of the
input nucleic
acid samples is not equalized across the plurality of input nucleic acid
samples. In some
embodiments, the plurality of input nucleic acid samples is combined before
the
amplification step. In some embodiments, the amplification products from the
plurality of
input nucleic acid samples are combined to form a nucleic acid pooled nucleic
acid library. In
some embodiments, the amplification products from the plurality of input
nucleic acid
samples are combined before being separated from the respective solid support.
In some
embodiments, the amplification products from the plurality of input nucleic
acid samples are
combined after being separated from the respective solid support.
[0022] Some embodiments of the methods disclosed herein further include
a step
of obtaining a nucleotide sequence of the amplification products. In some
embodiments, the
nucleotide sequence of the amplification products is obtained by nucleic acid
sequencing. In
-8-
In some embodiments, the nucleic acid sequencing includes high-throughput
sequencing,
e.g., next generation sequencing (NGS).
[0023] In
some embodiments, the present disclosure provides methods for nucleic
acid amplification that include (a) providing an input nucleic acid sample
including target
nucleic acid molecules; (b) contacting the input nucleic acid sample with a
reaction mixture
comprising plurality of first normalization primers and a plurality of second
normalization
primers, where the plurality of first normalization primers is immobilized on
a solid support,
the first normalization primers capable of specifically hybridizing to a first
sequence of the
target nucleic acid molecules, and where the plurality of second normalization
primers is in
solution, the plurality of second primers capable of specifically hybridizing
to a second
sequence of the target nucleic acid molecules; and (c) amplifying the target
nucleic acid
molecules under isothermal conditions such that substantially all of the first
normalization
primers are incorporated into amplification products, wherein the plurality of
first primers is
provided in an amount which limits the yield of amplification products to a
predefined
amount, and where at least a portion of the first normalization primers
further comprises a
capture portion having sequence complementarity to a region of the target
nucleic acid
molecules in addition to a known sequences of the target nucleic acid
molecules, and the
capture portion is generated by hybridizing a capture oligonucleotide to a
first normalization
primer and extending the first normalization primer to generate an extended
normalization
primer having sequence complementarity to the capture oligonucleotide.
[0023a] In some embodiments, the present disclosure provides a method for
nucleic acid amplification comprising:
a) providing an input sample comprising target nucleic acid molecules;
b) contacting the input sample with a reaction mixture comprising a plurality
of first
normalization primers and a plurality of second normalization primers,
wherein:
i. the plurality of first normalization primers is immobilized on a solid
support, the
first normalization primers capable of specifically hybridizing to a first
sequence of the target nucleic acid molecules; and
ii. the plurality of second normalization primers is in solution, the
plurality of
second primers capable of specifically hybridizing to a second sequence of the
target nucleic acid molecules;
-9-
CA 3016221 2019-11-29
amplifying the target nucleic acid molecules under isothermal conditions,
wherein the
plurality of first normalization primers is provided in an amount which limits
the yield of
amplification products to a predefined amount, at least a portion of the first
normalization
primers further comprises a capture portion having sequence complementarity to
a region of
the target nucleic acid molecules in addition to a known sequences of the
target nucleic acid
molecules, and the capture portion is generated by hybridizing a capture
oligonucleotide to a
first normalization primer and extending the first normalization primer to
generate an
extended normalization primer having sequence complementarity to the capture
ol igonucleotide.
[0024] In some embodiments of the methods disclosed herein, the
reaction
mixture further comprises one or more of a recombinase, a single-strand DNA-
binding
protein, a helicase, and a strand-displacing polymerase.
[0025] In some embodiments of the methods according to this aspect
and other
aspects of the present disclosure, prior to step (a), the target nucleic acid
molecules in the
input nucleic acid sample are subjected to a first enrichment amplification
reaction which
includes a first target-specific primer and a second target-specific primer.
In some
embodiments, each of the first target-specific primer and the second target-
specific primer
includes a region having sequence complementarity to known sequences of the
target nucleic
acid molecules. In some embodiments, each of the first target-specific primer
and the second
target-specific primer includes a universal primer region. In some
embodiments, the
universal primer region of the first target-specific primer includes a
sequencing-by-synthesis
(SBS) primer sequence.
[0026] In some embodiments of the methods disclosed herein, prior
to step (a),
the target nucleic acid molecules in the input nucleic acid sample are
subjected to a second
enrichment amplification reaction which includes a first universal primer and
a second
universal primer, wherein the first universal primer includes a region having
sequence
complementarity to the universal primer region of the first target-specific
primer, and the
second universal primer includes a region having sequence complementarity to
the universal
primer region of the second target-specific primer. In some embodiments, each
of the first
and/or second universal primers further includes an indexing portion.
-10-
CA 3016221 2019-11-29
[0027] In some embodiments, the method for nucleic acid
amplification as
disclosed herein is performed in multiplexed format. In some embodiments, the
method
disclosed herein is performed in multiplexed format on a droplet actuator.
[0028] In on aspect, some embodiments disclosed herein relate to a
method for
multiplexed amplification of nucleic acid samples on a droplet actuator, the
method
including: (a) providing a plurality of input nucleic acid samples including
target nucleic acid
molecules; (b) loading the plurality of input nucleic acid samples onto a
droplet operations
surface of the droplet actuator having droplet operations electrodes arranged
thereon; (c)
dispending a normalization reagent droplet comprising a plurality of first
normalization
primers and a plurality of second normalization primers, wherein (i) the
plurality of first
normalization primers is immobilized on a solid support and capable of
specifically
hybridizing to a first sequence of the target nucleic acid molecules, (ii) the
plurality of second
normalization primers is in solution and capable of specifically hybridizing
to a second
sequence of the target nucleic acid molecules; and (d) amplifying the target
nucleic acid
molecules under isothermal conditions such that substantially all of the first
normalization
primers are incorporated into amplification products. In some embodiments, the
method of
this aspect and other aspects further includes, prior to step (c), dispensing
a first enrichment
PCR reagent droplet onto a droplet operations surface of the droplet actuator,
wherein the
first enrichment PCR reagent droplet including a first target-specific primer
and a second
target-specific primer; and thermally cycling the target nucleic acid
molecules in the plurality
of input nucleic acid samples to form enriched nucleic acid samples. In some
embodiments,
each of the first target-specific primer and/or the second target-specific
primer includes a
region having sequence complementarity to known sequences of the target
nucleic acid
molecules. In some embodiments, each of the first target-specific primer
and/or the second
target-specific primer further includes a universal primer region. In some
embodiments, the
universal primer region of the first target-specific primer includes a
sequencing-by-synthesis
(SBS) primer sequence. In some embodiments, at least one of the first and/or
second target-
specific primers further includes an indexing portion.
[0028a] In some embodiments, the present disclosure provides a method for
multiplexed amplification of samples on a droplet actuator, comprising:
a) providing a plurality of input samples comprising target nucleic acid
molecules;
-11-
CA 3016221 2019-11-29
b) loading the plurality of input samples onto a droplet operations surface of
the
droplet actuator having droplet operations electrodes arranged thereon;
c) dispending a normalization reagent droplet comprising a plurality of first
normalization primers and a plurality of second normalization primers,
wherein:
i. the plurality of first normalization primers is immobilized on a solid
support,
the first normalization primers being capable of specifically hybridizing to a
first sequence of the target nucleic acid molecules; and
ii. the plurality of second normalization primers is in solution, the
plurality of
second primers being capable of specifically hybridizing to a second
sequence of the target nucleic acid molecules;
d) amplifying the target nucleic acid molecules under isothermal conditions.
[0029] In some embodiments, the method of this aspect and other
aspects further
includes, prior to step (c), dispensing a second enrichment PCR reagent
droplet onto a droplet
operations surface of the droplet actuator, wherein the second enrichment PCR
reagent
droplet comprising a first universal primer and a second universal primer; and
thermally
cycling the target nucleic acid molecules in the plurality of input nucleic
acid samples to
form enriched nucleic acid samples. In some embodiments, at least one of the
first and/or
second universal primers further includes a primer sequence region. In some
embodiments, at
least one of the first and/or second universal primers further includes an
indexing portion.
[0030] In some embodiments, the method of this aspect and other
aspects further
includes, prior to step (c), dispensing a first enrichment PCR reagent droplet
onto a droplet
operations surface of the droplet actuator, wherein the first enrichment PCR
reagent droplet
including a first target-specific primer and a second target-specific primer;
dispensing a
second enrichment PCR reagent droplet onto a droplet operations surface of the
droplet
actuator, wherein the second enrichment PCR reagent droplet including a first
universal
primer and a second universal primer; combining the second enrichment PCR
reagent droplet
with the first enrichment PCR reagent droplet using droplet operations to form
a combined
enrichment PCR reagent droplet; and thermally cycling the target nucleic acid
molecules in
the plurality of input nucleic acid samples to form enriched nucleic acid
samples.
[0031] In one aspect, some embodiments disclosed herein relate to a
microfluidic
system for performing multiplexed amplification of nucleic acid samples on a
droplet
-12-
CA 3016221 2019-11-29
actuator, in which the microfluidic system includes a processor for executing
code, a memory
communicatively coupled to the processor, and a program code stored in the
memory that
causes the processor to execute a method of multiplexed amplification of
nucleic acid
samples, wherein the method comprising (a) loading a plurality of input
nucleic acid samples
onto a droplet operations surface of the droplet actuator having droplet
operations electrodes
arranged thereon, each of the plurality of input nucleic acid samples
comprising target
nucleic molecules; (b) dispending a normalization reagent droplet comprising a
plurality of
first normalization primers and a plurality of second normalization primers,
wherein the
plurality of first normalization primers is immobilized on a solid support and
capable of
specifically hybridizing to a first sequence of the target nucleic acid
molecules, and the
plurality of second normalization primers is in solution and capable of
specifically
hybridizing to a second sequence of the target nucleic acid molecules; and (c)
amplifying the
target nucleic acid molecules under isothermal conditions such that
substantially all of the
first normalization primers are incorporated into amplification products.
[0031a] In some embodiments, the present disclosure provides a microfluidic
system for performing multiplexed amplification of nucleic acid samples on a
droplet
actuator, comprising a processor for executing code, a memory communicatively
coupled to
the processor, and a program code stored in the memory that causes the
processor to execute
a method of multiplexed amplification of nucleic acid samples, according to
the method as
defined herein comprising:
e)
loading a plurality of input samples onto a droplet operations surface of the
droplet
actuator having droplet operations electrodes arranged thereon, each of the
plurality of input
samples comprising target nucleic molecules;
0 dispensing a normalization reagent droplet comprising a plurality of first
normalization primers and a plurality of second normalization primers,
wherein:
i. the plurality of first normalization primers is immobilized on a solid
support,
the first normalization primers being capable of specifically hybridizing to a
first sequence of the target nucleic acid molecules; and
ii. the plurality of second normalization primers is in solution, the
plurality of
second primers being capable of specifically hybridizing to a second
sequence of the target nucleic acid molecules;
-12a-
CA 3016221 2019-11-29
g) amplifying the target nucleic acid molecules under isothermal conditions.
[0032] In various embodiments of the disclosure, implementations of
the
microfluidic system disclosed herein can include one or more of the following
components:
(a) a heating device; (b) a droplet actuator thermally coupled to the heating
device; (c) a
detector optically coupled to the droplet actuator; (d) an impedance sensing
module; (e) a
disruption device for lysing a biomaterial comprising nucleic acids; and a
controller that is
electronically coupled to one or more of the components of (a) to (d). In some
embodiments,
the controller of the microfluidic system disclosed herein includes a program
code, a
processor for executing the program code, and a local memory in communication
with the
processor, wherein the program code causes the processor to execute a method
of
multiplexed amplification of nucleic acid samples, the method including ( a)
loading a
plurality of input nucleic acid samples onto a droplet operations surface of
the droplet
actuator having droplet operations electrodes arranged thereon, each of the
plurality of input
nucleic acid samples comprising target nucleic molecules; (b) dispensing a
normalization
reagent droplet including a plurality of first normalization primers and a
plurality of second
normalization primers, wherein the plurality of first normalization primers is
immobilized on
a solid support and capable of specifically hybridizing to a first sequence of
the target nucleic
acid molecules, the plurality of second normalization primers is in solution
and capable of
specifically hybridizing to a second sequence of the target nucleic acid
molecules; (c)
amplifying the target nucleic acid molecules under isothermal conditions such
that
substantially all of the first normalization primers are incorporated into
amplification
products. _______________________________________________________________
-12b-
CA 3016221 2019-11-29
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
[0033] In some embodiments, the program code of the controller causes
the
processor to execute a method of multiplexed amplification of nucleic acid
samples that
further includes, prior to step (a), dispensing a first enrichment PCR reagent
droplet onto a
droplet operations surface of the droplet actuator, wherein the first
enrichment PCR reagent
droplet including a first target-specific primer and a second target-specific
primer; and
thermally cycling the target nucleic acid molecules in the plurality of input
nucleic acid
samples to form enriched nucleic acid samples. In some embodiments, the
program code
causes the processor to execute a method of multiplexed amplification of
nucleic acid
samples that further includes, prior to step (a), dispensing a second
enrichment PCR reagent
droplet onto a droplet operations surface of the droplet actuator, wherein the
second
enrichment PCR reagent droplet comprising a first universal primer and a
second universal
primer, and thermally cycling the target nucleic acid molecules in the
plurality of input
nucleic acid samples to form enriched nucleic acid samples. In some
embodiments, the
program code causes the processor to execute a method of multiplexed
amplification of
nucleic acid samples that further includes, prior to step (a), dispensing a
first enrichment PCR
reagent droplet onto a droplet operations surface of the droplet actuator,
wherein the first
enrichment PCR reagent droplet including a first target-specific primer and a
second target-
specific primer; dispensing a second enrichment PCR reagent droplet onto a
droplet
operations surface of the droplet actuator, wherein the second enrichment PCR
reagent
droplet including a first universal primer and a second universal primer;
combining the
second enrichment PCR reagent droplet with the first enrichment PCR reagent
droplet using
droplet operations to form a combined enrichment PCR reagent droplet; and
thermally
cycling the target nucleic acid molecules in the plurality of input nucleic
acid samples to
form enriched nucleic acid samples. In some embodiments, the microfluidic
system
disclosed herein further includes one or more magnets movable from and into
proximity to
one or more of the fluid reservoirs, wherein the positions of magnets are
optionally
controlled by a motor. In some embodiments, the microfluidic system disclosed
herein
further includes one or more heating devices to providing thermal control
thereof In some
embodiments, the program code is partially or entirely stored in a local
memory of the
controller or on a remote computing device. In some embodiments, the program
code is
locally and/or remotely executed.
-13-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
[0034] In some embodiments, the droplet actuator of the microfluidic
systems
disclosed herein includes (a) a bottom substrate and a top substrate separated
by a droplet
operations gap, wherein either one or both of the bottom and the top substrate
including
electrodes configured for conducting droplet operations in the gap; (b) an
electrode
arrangement including one or more of paths, reaction lanes, and an array of
droplet
operations electrodes; (c) a plurality of fluid reservoirs interconnected
through the electrode
arrangement configured for dispensing separated fluids along the electrodes;
and (d) a
plurality of temperature control zones. In some embodiments, the droplet
operations gap is
filled with a filler fluid. In some embodiments, the filler fluid is a
silicone oil fluid or a
hexadecane filler fluid. In some embodiments, the plurality of fluid
reservoirs includes one
or more reagent reservoirs, one or more sample reservoirs, one or more index
reservoirs, one
or more waste reservoirs, or a combination thereof. In some embodiments, the
droplet
actuator further includes one or more biochemical reaction zones for
performing certain
processing steps for each nucleic acid amplification reaction. In some
embodiments, at least
one of the fluid reservoirs includes an input port for loading fluids therein.
In some
embodiments, the droplet actuator disclosed herein further includes one or
more magnets
movable from and into proximity to one or more of the droplet operations
electrodes. In some
embodiments, the magnets are permanent magnets. In some embodiments, the
magnets are
electromagnets. In some embodiments, the temperature control zones include
differing
temperature from one another. In some embodiments, the temperature control
zones include
essentially the same temperature. In some embodiments, the electrode
arrangement in the
droplet actuator disclosed herein includes one or more of dispensing operation
electrodes,
transporting operation electrodes, merging operation electrodes, incubating
operation
electrodes, splitting operation electrodes, mixing operation electrodes, or
combinations
thereof
[0035] In one aspect, some embodiments disclosed herein relate to a
computer
readable medium storing processor executable instructions for performing a
method of
multiplexed nucleic acid amplification on a droplet actuator, the method
including ( a)
loading the plurality of input nucleic acid samples onto a droplet operations
surface of the
droplet actuator having droplet operations electrodes arranged thereon, each
of the plurality
of input nucleic acid samples including target nucleic molecules; (b)
dispensing a
-14-
normalization reagent droplet including a plurality of first normalization
primers and a
plurality of second normalization primers, wherein the plurality of first
normalization primers
is immobilized on a solid support and capable of specifically hybridizing to a
first sequence
of the target nucleic acid molecules, the plurality of second normalization
primers is in
solution and capable of specifically hybridizing to a second sequence of the
target nucleic
acid molecules; (c) amplifying the target nucleic acid molecules under
isothermal conditions
such that substantially all of the first normalization primers are
incorporated into
amplification products.
[0035a] In some embodiments, the present disclosure provides a computer
readable medium storing processor executable instructions for performing a
method of
multiplexed nucleic acid amplification on a droplet actuator, the method
comprising:
h) loading a plurality of input samples onto a droplet operations surface
of the droplet
actuator having droplet operations electrodes arranged thereon, each of the
plurality of input
samples comprising target nucleic molecules;
i) dispensing a normalization reagent droplet comprising a plurality of first
normalization primers and a plurality of second normalization primers,
wherein:
i. the plurality of first normalization primers is immobilized on a solid
support,
the first normalization primers being capable of specifically hybridizing to a
first sequence of the target nucleic acid molecules; and
ii. the plurality of second normalization primers is in solution, the
plurality of
second primers being capable of specifically hybridizing to a second
sequence of the target nucleic acid molecules;
amplifying the target nucleic acid molecules under isothermal conditions.
[0036] Also provided, in one aspect of the disclosure, are
compositions that
include amplification products produced by a method or a system disclosed
herein.
[0037] The foregoing summary is illustrative only and is not
intended to be in any
way limiting. In addition to the illustrative embodiments and features
described herein,
further aspects, embodiments, objects and features of the disclosure will
become fully
apparent from the drawings and the detailed description and the claims.
-15-
CA 3016221 2019-11-29
BRIEF DESCRIPTION OF THE DRAWINGS
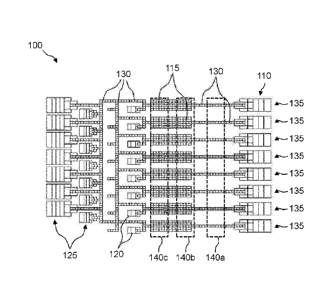
[0038] Figure 1 illustrates a top view of an example of, an
electrode arrangement
of a droplet actuator suitable for use in conducting a multiplexed targeted
amplicon sample
preparation protocol.
[0039] Figure 2 illustrates a flow diagram of a non-limiting
example of a method
of nucleic acid amplification and normalization in accordance with some
embodiments of the
disclosure. The hybridization between target nucleic acid molecules and the
first
amplification or normalization primers takes place on a solid support. In this
example, the
solid support is a plurality of capture beads. Alternatively, the solid
support can also be a
surface of a reaction site.
[0040] Figure 3 shows pictorially the steps of the method of Figure
2.
[0041] Figure 4 depicts a flow diagram of another non-limiting
example of a
method of nucleic acid amplification and normalization as described herein,
wherein the
hybridization between target nucleic acid molecules and the first
amplification or
normalization primers takes place in solution phase.
[0042] Figure 5 shows pictorially the steps of the method of Figure
4.
-15a-
CA 3016221 2019-11-29
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
[0043] Figure 6 illustrates a flow diagram of another non-limiting
example of a
method of preparing an amplified sample and/or a normalized sample for
downstream
analytical applications such as, for example, sequencing.
[0044] Figure 7A and 7B show pictorially the steps of the method of
Figure 6.
[0045] Figure 8 depicts a flow diagram of another non-limiting example
of a
method of nucleic acid amplification and normalization in accordance with some
embodiments of the disclosure.
[0046] Figure 9A and 9B show pictorially the steps of the method of
Figure 8.
[0047] Figure 10 shows a plot of the fragment size distributions in
three targeted
amplicon samples prepared using three different protocols, one being the two-
stage on-
actuator amplification reaction of the method of Figure 6.
[0048] Figures 11A and 11B show a bar graph of PCR efficiency per cycle
and a
bar graph of the uniformity of amplicons in each library of Figure 10. Figure
11A: PCR
efficiency per PCR cycle. Figure 11B: Uniformity improvements.
[0049] Figure 12A and 12B show a three dimensional plot of the predicted
extended intermediate product yield and a plot of the predicted yield vs the
actual yield based
on PCR product and capture probe input.
[0050] Figure 13 shows a plot of the relative library output as a
function of PCR
product input in libraries prepared using the method of Figure 6.
[0051] Figures 14A and 14B show a plot of library uniformity and fold
amplification as a function of PCR product input (input dilution) and a plot
of amplification
bias, respectively, in libraries prepared using the method of Figure 6.
[0052] Figures 15A and 15B show a three dimensional plot of ExAmp yield
and
a plot of predicted vs. actual ExAmp yield as a function of P5 primer and P7-
biotin primer
concentrations.
[0053] Figure 16 shows a plot of per target sample reads obtained from
samples
eluted from streptavidin beads using heat denaturation on a droplet actuator
and samples that
has been additionally denatured by NaOH treatment on bench.
[0054] Figure 17 shows a plot of library uniformity as a function of
genomic
DNA input for 5 different samples.
-16-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
[0055] Figures 18A and 18B show a plot of TP (true positive) variant
calling
accuracy in the NA20mix samples and a plot of TP variant calling accuracy in
the HD200
sample, respectively, of Figure 17.
[0056] Figure 19A shows a plot of the TP and FP (false positive) variant
frequency as a function of chromosome and position in genomic libraries
prepared using an
on-bench protocol and on-actuator using the method of Figure 6.
[0057] Figure 19B shows a plot of the average number of FP variant calls
in the
on-bench samples and the on-actuator samples of Figure 19A.
[0058] Figure 20 shows a plot of library uniformity as a function of
genomic
DNA and primer pool complexity.
[0059] Figures 21A, 21C, and 21B show a plot of target coverage in the
192-plex
Pool 1 library, a plot of 192-plex Pool 1 target coverage in the 384-plex Pool
Mix 1-2 library,
and a plot of 192-plex Pool 1 target coverage in the 580-plex Pool Mix 1-3
library described
in Figure 20.
[0060] Figure 22 illustrates a functional block diagram of a non-
limiting example
of a microfluidics system that includes a droplet actuator, which is one
example of a fluidics
cartridge.
[0061] Figure 23 graphically illustrates a non-limiting example of a
method of
using Ex-Amp reagents for nucleic acid amplification and normalization in
accordance with
some embodiments of the disclosure.
[0062] Figure 24 illustrates another non-limiting example of a method of
using
Ex-Amp reagents for nucleic acid amplification and normalization in accordance
with some
embodiments of the disclosure. Depending on the specific workflow, extendable
primers
used in this exemplary method can be amplification primers or normalization
primers. First
amplification or normalization primers P7 immobilized on a solid phase support
offer ability
to normalize the nucleic acid samples and ease of purification, while second
amplification or
normalization primers P5 in solution phase offer fast kinetics.
[0063] Figure 25 graphically summarizes net effects of a non-limiting
example of
a method of nucleic acid amplification in accordance with some embodiments of
the
disclosure.
-17-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
[0064] Figure 26 illustrates a non-limiting example of a method of
nucleic acid
amplification and normalization in accordance with some embodiments of the
disclosure, in
which the input sample contains double-stranded nucleic acids. In this
experiment, double-
stranded DNA (dsDNA) enters the reaction core by strand invasion, either by
first
amplification or normalization primer PS in solution phase or by second
amplification or
normalization primer P7 immobilized to bead surface (indicated by a boxed
bead).
[0065] Figure 27 illustrates a non-limiting example of a method of
nucleic acid
amplification and normalization in accordance with some embodiments of the
disclosure, in
which the input nucleic acid sample contains single-stranded nucleic acids
such as, for
example, single-stranded DNA (ssDNA). Single-stranded DNA enters either by
hybridization
to primer P7 immobilized to bead surface or by being converted into dsDNA by
primer PS in
solution phase (indicated by boxed beads).
[0066] Figure 28 illustrates another non-limiting example of a method of
nucleic
acid amplification and normalization in accordance with some embodiments of
the
disclosure. The input nucleic acid sample, which contains nucleic acids
already affixed onto
beads (boxed), enters the reaction core by hybridizing to primer PS in
solution phase.
[0067] Figure 29 graphically summarizes the results from experiments
performed
to illustrate a non-limiting example of a method of library normalization in
accordance with
some embodiments of the disclosure. 29A: Dilution factors in input nucleic
acid libraries
29B: Library yields obtained from a standard library preparation procedure;
29C: Library
yields obtained from a library preparation procedure that was performed
according to a non-
limiting example of a method of nucleic acid amplification and normalization
of the
disclosure.
DETAILED DESCRIPTION
[0068] The present disclosure generally relates to nucleic acid
amplification
devices, systems, and methods including novel approaches suitable for
construction of
nucleic acid samples, including construction of amplified/normalized nucleic
acid samples
and libraries, optionally in customized fashion, for downstream analytical
applications,
including sequencing applications utilizing techniques such as next-generation
sequencing
(NGS) and related methodologies such as genotyping-by-sequencing (GBS). In
some
embodiments, the method includes contacting a plurality of input nucleic acid
samples with a
-18-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
reaction mixture including first primers and second primers, wherein the first
and second
primers can be amplification primers or normalization primers depending on
specific
workflows. In some embodiments, the first amplification or normalization
primers are
immobilized on a solid support and the second amplification or normalization
primers are in
solution phase, and the amplification or normalization of the input nucleic
acid samples is
performed under conditions such that the amounts of nucleic acids in the
resultant
amplified/normalized samples are in substantially similar concentrations
relative to one
another regardless of the amounts of input samples. In some embodiments,
substantially all
of the first amplification or normalization primers are incorporated into
amplification
products. For example, in some embodiments at least 80%, at least 85%, at
least 90%, at least
95%, at least 96%, at least 97%, at least 98%, at least 99%, 100%, or a range
defined by any
two of the preceding values, of the first amplification or normalization
primers are
incorporated into amplification products. Further provided are systems and
droplet actuator
devices that are configured to carry out the methods disclosed herein. Nucleic
acid libraries
prepared in accordance with the disclosed methods are also provided.
[0069] In one aspect, some embodiments of the disclosure relate to
droplet
actuator devices for and methods of preparing targeted amplicon samples or
libraries for
downstream analytical applications such as, for example, sequencing
applications. Using
digital microfluidics technology, the droplet actuator devices and methods of
the disclosure
provide automated liquid handling for amplification and selection of targeted
regions of
genomic DNA for processing into amplicon samples or libraries for downstream
analytical
applications, e.g. sequencing. In various embodiments, library construction
parameters, such
as DNA input (e.g., 1 ng or less), library yield and uniformity, time-to-
result, and reagent
consumption, are substantially improved over the existing state of the art.
[0070] In one aspect, some embodiments of the methods disclosed herein
use a 2-
stage amplification reaction, wherein a first amplification reaction is driven
by target-specific
primers and a second amplification reaction is driven by universal primers.
The 2-stage
amplification reaction allows for selection of reaction conditions (e.g.,
annealing and
extension temperatures, incubation times, and number of PCR cycles) specific
for each type
of amplification, e.g., target-specific primer amplification or universal
primer amplification,
-19-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
and provides for more efficient targeted amplification (e.g., improved library
yield and
uniformity).
[0071] In some embodiments, the targeted amplicons in the resulting
nucleic acid
samples or libraries are then enriched in a solution-based hybridization
reaction using target-
specific capture probes and then immobilized on capture beads for subsequent
processing
steps. In some embodiments of the methods disclosed herein, a solution-based
hybridization
reaction is generally more kinetically favorable compared to a solid-phase
hybridization
reaction, thereby creating a more robust system and providing for improved
target capture, as
well as improved uniformity and specificity across multiple samples and/or
libraries.
[0072] In one aspect, some embodiments of the methods disclosed herein
employ
at least one set of first amplification or normalization primers in solution
phase and at least
another set of second amplification or normalization primers immobilized on a
solid phase
support during amplification or normalization of the nucleic acid library.
This feature of this
aspect of the disclosure advantageously differs from current library
preparation procedures in
which both primers are either in a solution phase or both primers are affixed
onto a solid
phase. Additionally, in some particular implementations of the methods
disclosed herein, the
set of first primers immobilized on the solid support is provided in an amount
which limits
the yield of amplification products to a predefined amount, and the set of
second primers in
solution is provided in an amount that exceeds the amount of the first
primers. Advantages
achieved by this specific primer configuration is twofold: (1) It allows
conveniently washing
away of any amplification products that is found in solution after
amplification, resulting in
an amplified/normalized amount of amplification products remained immobilized
on the
solid support which can be subsequently isolated, such that a predefined
amount of
amplification products can be generated. In contrast, existing library
preparation methods
typically produce excess amounts of amplification products, which are
measured, and then
subjected to a dilution step to obtain desired amounts is required; (2) in
some particular
implementations, the methods disclosed herein allow construction of a
plurality of nucleic
acid libraries in which the output DNA amounts are normalized to substantially
uniform
concentrations across the libraries regardless of the amounts of input DNA
samples.
[0073] In one aspect, the methods of the disclosure use a kinetic
exclusion
amplification procedure, also referred to as exclusion amplification (ExAmp)
reaction to
-20-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
equalize sample quantities and adjust the concentration of amplicon DNA for
subsequent
sequencing applications. In some embodiments, the ExAmp library normalization
reaction is
an isothermal amplification reaction that uses a first primer sequence (e.g.,
primer including a
P7 primer sequence) immobilized on the capture beads and a second primer
sequence (e.g.,
primer including a 135 primer sequence) in solution as normalization primers
for library
normalization. Generally, a process of library normalization can be performed
over a wide
range of PCR product (amplicon) inputs (e.g., those having over at least two
orders of
magnitude change of input). In addition, the reaction conditions (e.g.,
incubation time, P7
and/or P5 primer concentration, and reaction components) can be selected such
that all
available primers immobilized on the capture beads are extended into
amplicons, e.g., the
reaction is allowed to run to saturation.
[0074] In general, the flexibility and programmability of a droplet
actuator device
provides for precise control over the various biochemical reactions performed
during
construction of a targeted amplicon sample or library.
[0075] In one exemplified application, the droplet actuators and methods
of the
disclosure are used for preparation of a targeted amplicon sample or library
for identification
of genetic variants, e.g., single nucleotide polymorphisms (SNPs).
[0076] In the following detailed description, reference is made to the
accompanying Figures, which form a part hereof The illustrative embodiments
described in
the detailed description, Figures, and claims are not meant to be limiting.
Other embodiments
may be used, and other changes may be made, without departing from the spirit
or scope of
the subject matter presented here. It will be readily understood that the
embodiments of the
present disclosure, as generally described herein, and illustrated in the
Figures, can be
arranged, substituted, combined, and designed in a wide variety of different
configurations,
all of which are explicitly contemplated and make part of this disclosure.
[0077] Unless expressly defined otherwise, all terms of art, notations
and other
scientific terms or terminology used herein are intended to have the meanings
commonly
understood by those of skill in the art to which this disclosure pertains when
read in light of
this disclosure.
-21-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
SOME DEFINITIONS
[0078] Unless otherwise defined, all terms of art, notations and other
scientific
terms or terminology used herein are intended to have the meanings commonly
understood
by those of skill in the art to which this disclosure pertains when read in
light of this
disclosure. In some cases, terms with commonly understood meanings are defined
herein for
clarity and/or for ready reference, and the inclusion of such definitions
herein should not
necessarily be construed to represent a substantial difference over what is
generally
understood in the art. Many of the techniques and procedures described or
referenced herein
are well understood and commonly employed using conventional methodology by
those
skilled in the art.
[0079] The singular form "a", "an", and "the" include plural references
unless the
context clearly dictates otherwise. For example, the term "a molecule"
includes one or more
molecules, including mixtures thereof. As used in this disclosure and the
appended claims,
the term "and/or" can be singular or inclusive. For example, "A and/or B" is
used herein to
include all of the following alternatives: "A", "B", and "A and B".
[0080] The term "about", as used herein, has its ordinary meaning of
approximately. If the degree of approximation is not otherwise clear from the
context,
"about" means either within plus or minus 10% of the provided value, or
rounded to the
nearest significant figure, in all cases inclusive of the provided value.
Where ranges are
provided, they are inclusive of the boundary values.
[0081] The term "activate," as used herein with reference to one or more
electrodes, means affecting a change in the electrical state of the one or
more electrodes
which, in the presence of a droplet, results in a droplet operation.
Activation of an electrode
can be accomplished using alternating current (AC) or direct current (DC). Any
suitable
voltage may be used. For example, an electrode may be activated using a
voltage which is
greater than about 150 V, or greater than about 200 V, or greater than about
250 V, or from
about 275 V to about 1000 V, or about 300 V. Where an AC signal is used, any
suitable
frequency may be employed. For example, an electrode may be activated using an
AC signal
having a frequency from about 1 Hz to about 10 MHz, or from about 10 Hz to
about 60 Hz,
or from about 20 Hz to about 40 Hz, or about 30 Hz.
-22-
[0082] As used herein, the term "droplet" means a volume of liquid
on a droplet
actuator. Typically, a droplet is at least partially bounded by a filler
fluid. For example, a
droplet may be completely surrounded by a filler fluid or may be bounded by
filler fluid and
one or more surfaces of the droplet actuator. As another example, a droplet
may be bounded
by filler fluid, one or more surfaces of the droplet actuator, and/or the
atmosphere. As yet
another example, a droplet may be bounded by filler fluid and the atmosphere.
Droplets may,
for example, be aqueous or non-aqueous or may be mixtures or emulsions
including aqueous
and non-aqueous components. Droplets may take a wide variety of shapes; non-
limiting
examples include generally disc shaped, slug shaped, truncated sphere,
ellipsoid, spherical,
partially compressed sphere, hemispherical, ovoid, cylindrical, combinations
of such shapes,
and various shapes formed during droplet operations, such as merging or
splitting or formed
as a result of contact of such shapes with one or more surfaces of a droplet
actuator. For
examples of droplet fluids that may be subjected to droplet operations using
the approach of
the present disclosure, see Eckhardt et al., International Patent Pub. No.
WO/2007/120241,
entitled, "Droplet-Based Biochemistry," published on October 25, 2007.
[0083] In various embodiments of the disclosure, a droplet may
include a
biological sample, such as whole blood, lymphatic fluid, serum, plasma, sweat,
tear, saliva,
sputum, cerebrospinal fluid, amniotic fluid, seminal fluid, vaginal excretion,
serous fluid,
synovial fluid, pericardial fluid, peritoneal fluid, pleural fluid,
transudates, exudates, cystic
fluid, bile, urine, gastric fluid, intestinal fluid, fecal samples, liquids
containing single or
multiple cells, liquids containing organelles, fluidized tissues, fluidized
organisms, liquids
containing multi-celled organisms, biological swabs and biological washes.
Moreover, a
droplet may include a reagent, such as water, deionized water, saline
solutions, acidic
solutions, basic solutions, detergent solutions and/or buffers. A droplet can
include nucleic
acids, such as DNA, genomic DNA, RNA, mRNA or analogs thereof; nucleotides
such as
deoxyribonucleotides, ribonucleotides or analogs thereof such as analogs
having terminator
moieties such as those described in Bentley et cd., Nature 456:53-59 (2008);
Gormley et al.,
International Patent Pub. No. WO/2013/131962, entitled, "Improved Methods of
Nucleic
Acid Sequencing," published on September 12, 2013; Barnes et al., U.S. Patent
No.
7,057,026, entitled "Labelled Nucleotides," issued on June 6, 2006; Kozlov et
al.,
International Patent Pub. No. WO/2008/042067, entitled, "Compositions and
Methods for
-23-
CA 3016221 2019-11-29
Nucleotide Sequencing," published on April 10, 2008; Rigatti et al.,
International Patent Pub.
No. WO/2013/117595, entitled, "Targeted Enrichment and Amplification of
Nucleic Acids
on a Support," published on August 15, 2013; Hardin et al., U.S. Patent No.
7,329,492,
entitled "Methods for Real-Time Single Molecule Sequence Determination,"
issued on
February 12, 2008; Hardin et al., U.S. Patent No. 7,211,414, entitled
"Enzymatic Nucleic
Acid Synthesis: Compositions and Methods for Altering Monomer Incorporation
Fidelity,"
issued on May 1, 2007; Turner etal., U.S. Patent No. 7,315,019, entitled
"Arrays of Optical
Confinements and Uses Thereof," issued on January 1, 2008; Xu et al., U.S.
Patent No.
7,405,281, entitled "Fluorescent Nucleotide Analogs and Uses Therefor," issued
on July 29,
2008; and Ranket al., U.S. Patent Pub. No. 20080108082, entitled "Polymerase
Enzymes and
Reagents for Enhanced Nucleic Acid Sequencing," published on May 8, 2008;
enzymes such
as polymerases, ligases, recombinases, or transposases; binding partners such
as antibodies,
epitopes, streptavidin, avidin, biotin, lectins or carbohydrates; or other
biochemically active
molecules. Other examples of droplet contents include reagents, such as a
reagent for a
biochemical protocol, such as a nucleic acid amplification protocol, an
affinity-based assay
protocol, an enzymatic assay protocol, a sequencing protocol, and/or a
protocol for analyses
of biological fluids. According to some of embodiments disclosed herein, a
droplet may
include one or more beads depending on specific workflows and/or downstream
applications.
[0084] The
term "droplet actuator," as used herein, means a device for
manipulating droplets. For examples of droplet actuators, see Pamula et al.,
U.S. Patent No.
6,911,132, entitled "Apparatus for Manipulating Droplets by Electrowetting-
Based
Techniques," issued on June 28, 2005; Pamula et al., U.S. Patent Pub. No.
20060194331,
entitled "Apparatuses and Methods for Manipulating Droplets on a Printed
Circuit Board,"
published on August 31, 2006; Pollack et al., International Patent Pub. No.
WO/2007/120241, entitled "Droplet-Based Biochemistry," published on October
25, 2007;
Shenderov, U.S. Patent No. 6,773,566, entitled "Electrostatic Actuators for
Microfluidics and
Methods for Using Same," issued on August 10, 2004; Shenderov, U.S. Patent No.
6,565,727, entitled "Actuators for Microfluidics Without Moving Parts," issued
on May 20,
2003; Kim et al., U.S. Patent Pub. No. 20030205632, entitled "Electrowetting-
driven
Micropumping," published on November 6, 2003; Kim et al., U.S. Patent Pub. No.
20060164490, entitled "Method and Apparatus for Promoting the Complete
Transfer of
-24-
CA 3016221 2019-11-29
Liquid Drops from a Nozzle," published on July 27, 2006; Kim et al., U.S.
Patent Pub. No.
20070023292, entitled "Small Object Moving on Printed Circuit Board,"
published on
February 1, 2007; Shah et al., U.S. Patent Pub. No. 20090283407, entitled
"Method for
Using Magnetic Particles in Droplet Microfluidics," published on November 19,
2009; Kim
et al., U.S. Patent Pub. No. 20100096266, entitled "Method and Apparatus for
Real-time
Feedback Control of Electrical Manipulation of Droplets on Chip," published on
April 22,
2010; Velev, U.S. Patent No. 7,547,380, entitled "Droplet Transportation
Devices and
Methods Having a Fluid Surface," issued on June 16, 2009; Sterling et al.,
U.S. Patent No.
7,163,612, entitled "Method, Apparatus and Article for Microfluidic Control
via
Electrowetting, for Chemical, Biochemical and Biological Assays and the Like,"
issued on
January 16, 2007; Becker et aL, U.S. Patent No. 7,641,779, entitled "Method
and Apparatus
for Programmable Fluidic Processing," issued on January 5, 2010; Becker et
al., U.S. Patent
No. 6,977,033, entitled "Method and Apparatus for Programmable Fluidic
Processing,"
issued on December 20, 2005; Decre et al., U.S. Patent No. 7,328,979, entitled
"System for
Manipulation of a Body of Fluid," issued on February 12, 2008; Yamakawa et
al., U.S.
Patent Pub. No. 20060039823, entitled "Chemical Analysis Apparatus," published
on
February 23, 2006; Wu, U.S. Patent Pub. No. 20110048951, entitled "Digital
Microfluidics
Based Apparatus for Heat-exchanging Chemical Processes," published on March 3,
2011;
Fouillet et al., U.S. Patent Pub. No. 20090192044, entitled "Electrode
Addressing Method,"
published on July 30, 2009; Fouillet et al., U.S. Patent No. 7,052,244,
entitled "Device for
Displacement of Small Liquid Volumes Along a Micro-catenary Line by
Electrostatic
Forces," issued on May 30, 2006; Marchand et al., U.S. Patent Pub. No.
20080124252,
entitled "Droplet Microreactor," published on May 29, 2008; Adachi et al.,
U.S. Patent Pub.
No. 20090321262, entitled "Liquid Transfer Device," published on December 31,
2009;
Roux et al., U.S. Patent Pub. No. 20050179746, entitled "Device for
Controlling the
Displacement of a Drop Between Two or Several Solid Substrates," published on
August 18,
2005; and Dhindsa et al., "Virtual Electrowetting Channels: Electronic Liquid
Transport with
Continuous Channel Functionality," Lab Chip, 10:832-836 (2010).
-25-
CA 3016221 2019-11-29
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
[0085] In some embodiments disclosed herein, certain droplet actuators
will
include one or more substrates arranged with a droplet operations gap
therebetween and
electrodes associated with (e.g., layered on, attached to, and/or embedded in)
the one or more
substrates and arranged to conduct one or more droplet operations. For
example, certain
droplet actuators will include a base (or bottom) substrate, droplet
operations electrodes
associated with the substrate, one or more dielectric layers atop the
substrate and/or
electrodes, and optionally one or more hydrophobic layers atop the substrate,
dielectric layers
and/or the electrodes forming a droplet operations surface. A top substrate
may also be
provided, which is separated from the droplet operations surface by a gap,
commonly
referred to as a droplet operations gap. Various electrode arrangements on the
top and/or
bottom substrates are discussed in the patents and applications referenced
herein and certain
novel electrode arrangements are discussed in the description of the present
disclosure.
During droplet operations it is preferred that droplets remain in continuous
contact or
frequent contact with a ground or reference electrode. A ground or reference
electrode may
be associated with the top substrate facing the gap, the bottom substrate
facing the gap, in the
gap. Where electrodes are provided on both substrates, electrical contacts for
coupling the
electrodes to a droplet actuator instrument for controlling or monitoring the
electrodes may
be associated with one or both plates. In some cases, electrodes on one
substrate are
electrically coupled to the other substrate so that only one substrate is in
contact with the
droplet actuator. In some embodiments, a conductive material (e.g., an epoxy,
such as
MASTER BONDTM Polymer System EP79, available from Master Bond, Inc.,
Hackensack,
NJ) provides the electrical connection between electrodes on one substrate and
electrical
paths on the other substrates, e.g, a ground electrode on a top substrate may
be coupled to an
electrical path on a bottom substrate by such a conductive material. In some
embodiments
disclosed herein where multiple substrates are used, a spacer may be provided
between the
substrates to determine the height of the gap therebetween and define on-
actuator dispensing
reservoirs. The spacer height may, for example, be at least about 5 gm, 100
gm, 200 gm, 250
gm, 275 gm or more. Alternatively or additionally the spacer height may be at
most about
600 gm, 400 gm, 350 gm, 300 gm, or less. The spacer may, for example, be
formed of a
layer of projections form the top or bottom substrates, and/or a material
inserted between the
top and bottom substrates. One or more openings may be provided in the one or
more
-26-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
substrates for forming a fluid path through which liquid may be delivered into
the droplet
operations gap. The one or more openings may in some cases be aligned for
interaction with
one or more electrodes, e.g., aligned such that liquid flowed through the
opening will come
into sufficient proximity with one or more droplet operations electrodes to
permit a droplet
operation to be effected by the droplet operations electrodes using the
liquid. The base (or
bottom) and top substrates may in some cases be formed as one integral
component One or
more reference electrodes may be provided on the base (or bottom) and/or top
substrates
and/or in the gap. Examples of reference electrode arrangements are provided
in the patents
and patent applications referenced herein. In various embodiments, the
manipulation of
droplets by a droplet actuator may be electrode mediated, e.g., electrowetting
mediated or
dielectrophoresis mediated or Coulombic force mediated.
[0086] Examples of other techniques for controlling droplet operations
that may
be used in the droplet actuators of the present disclosure include using
devices that induce
hydrodynamic fluidic pressure, such as those that operate on the basis of
mechanical
principles (e.g., external syringe pumps, pneumatic membrane pumps, vibrating
membrane
pumps, vacuum devices, centrifugal forces, piezoelectric/ultrasonic pumps and
acoustic
forces); electrical or magnetic principles (e.g., electroosmotic flow,
electrokinetic pumps,
ferrofluidic plugs, electrohydrodynamic pumps, attraction or repulsion using
magnetic forces
and magnetohydrodynamic pumps); thermodynamic principles (e.g., gas bubble
generation/phase-change-induced volume expansion); other kinds of surface-
wetting
principles (e.g., el ectrowetting, and opto el ectrowetting, as well as
chemically, thermally,
structurally and radioactively induced surface-tension gradients); gravity;
surface tension
(e.g., capillary action); electrostatic forces (e.g., electroosmotic flow);
centrifugal flow
(substrate disposed on a compact disc and rotated); magnetic forces (e.g.,
oscillating ions
causes flow); magnetohydrodynamic forces; and vacuum or pressure differential.
In certain
embodiments, combinations of two or more of the foregoing techniques may be
employed to
conduct a droplet operation in a droplet actuator of the present disclosure.
Similarly, one or
more of the foregoing may be used to deliver liquid into a droplet operations
gap, e.g., from a
reservoir in another device or from an external reservoir of the droplet
actuator (e.g., a
reservoir associated with a droplet actuator substrate and a flow path from
the reservoir into
the droplet operations gap).
-27-
100871
Droplet operations surfaces of certain droplet actuators of the present
disclosure may be made from hydrophobic materials or may be coated or treated
to make
them hydrophobic. For example, in some cases some portion or all of the
droplet operations
surfaces may be derivatized with low surface-energy materials or chemistries,
e.g., by
deposition or using in situ synthesis using compounds such as poly- or per-
fluorinated
compounds in solution or polymerizable monomers. Examples include TEFLON AF
(available from DuPont, Wilmington, DE), members of the cytop family of
materials,
coatings in the FLUOROPEL family of hydrophobic and superhydrophobic coatings
(available from Cytonix Corporation, Beltsville, MD), silane coatings,
fluorosilane coatings,
hydrophobic phosphonate derivatives (e.g., those sold by Aculon, Inc.), and
NOVECTM
electronic coatings (available from 3M Company, St. Paul, MN), other
fluorinated monomers
for plasma-enhanced chemical vapor deposition (PECVD), and organosiloxane
(e.g., Si0C)
for PECVD. In some cases, the droplet operations surface may include a
hydrophobic coating
having a thickness ranging from about 10 nm to about 1,000 nm. Moreover, in
some
embodiments, the top substrate of the droplet actuator includes an
electrically conducting
organic polymer, which is then coated with a hydrophobic coating or otherwise
treated to
make the droplet operations surface hydrophobic. For example, the electrically
conducting
organic polymer that is deposited onto a plastic substrate may be poly(3,4-
ethylenedioxythiophene) poly(styrenesulfonate) (PEDOT:PSS). Other examples of
electrically conducting organic polymers and alternative conductive layers are
described in
Pollack et al., International Patent Pub. No. WO/2011/002957, entitled
"Droplet Actuator
Devices and Methods," published on January 6, 2011. In some embodiments
disclosed
herein, one or both substrates may be fabricated using a printed circuit board
(PCB), glass,
indium tin oxide (ITO)-coated glass, and/or semiconductor materials as the
substrate. When
the substrate is ITO-coated glass, the ITO coating is, for example, a
thickness of at least
about 20 nm, 50 nm, 75 nm, 100 nm or more. Alternatively or additionally, the
thickness can
be at most about 200 nm, 150 nm, 125 nm or less. In some cases, the top and/or
bottom
substrate includes a PCB substrate that is coated with a dielectric, such as a
polyimide
dielectric, which may in some cases also be coated or otherwise treated to
make the droplet
operations surface hydrophobic. When the substrate includes a PCB, the
following materials
are examples of
suitable
-28-
CA 3016221 2019-11-29
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
materials: MITSUITm BN-300 (available from MITSUI Chemicals America, Inc., San
Jose
CA); ARLONTM 11N (available from Arlon, Inc., Santa Ana, CA).; NELCO N4000-6
and
N5000-30/32 (available from Park Electrochemical Corp., Melville, NY); ISOLATM
FR406
(available from Isola Group, Chandler, AZ), especially IS620; fluoropolymer
family (suitable
for fluorescence detection since it has low background fluorescence);
polyimide family;
polyester; polyethylene naphthalate; polycarbonate; polyetheretherketone;
liquid crystal
polymer; cyclo-olefin copolymer (COC); cyclo-olefin polymer (COP); aramid;
THER1VIOUNT nonwoven aramid reinforcement (available from DuPont, Wilmington,
DE); NOMEXI3 brand fiber (available from DuPont, Wilmington, DE); and paper.
Various
materials are also suitable for use as the dielectric component of the
substrate. Examples
include: vapor deposited dielectric, such as PARYLENETM C (especially on
glass),
PARYLENETM N, and PARYLENETM HT (for high temperature, ¨300 C) (available from
Parylene Coating Services, Inc., Katy, TX); TEFLON AF coatings; cytop;
soldermasks,
such as liquid photoimageable soldermasks (e.g., on PCB) like TAIYOTm PSR4000
series,
TAIYOTm PSR and AUS series (available from Taiyo America, Inc. Carson City,
NV) (good
thermal characteristics for applications involving thermal control), and
PROBIMERTm 8165
(good thermal characteristics for applications involving thermal control
(available from
Huntsman Advanced Materials Americas Inc., Los Angeles, CA); dry film
soldermask, such
as those in the VACREL dry film soldermask line (available from DuPont,
Wilmington,
DE); film dielectrics, such as polyimide film (e.g., KAPTON polyimide film,
available
from DuPont, Wilmington, DE), polyethylene, and fluoropolymers (e.g., FEP),
polytetrafluoroethylene; polyester; polyethylene naphthalate; cyclo-olefin
copolymer (COC);
cyclo-olefin polymer (COP); any other PCB substrate material listed above;
black matrix
resin; polypropylene; and black flexible circuit materials, such as DuPontTM
Pyralux HXC
and DuPontTM Kapton MBC (available from DuPont, Wilmington, DE).
[0088] Droplet transport voltage and frequency may be selected for
performance
with reagents used in specific assay protocols. Design parameters may be
varied, e.g.,
number and placement of on-actuator reservoirs, number of independent
electrode
connections, size (volume) of different reservoirs, placement of magnets/bead
washing
zones, electrode size, inter-electrode pitch, and gap height (between top and
bottom
substrates) may be varied for use with specific reagents, protocols, droplet
volumes, etc. In
-29-
some cases, a substrate of the present disclosure may be derivatized with low
surface-energy
materials or chemistries, e.g., using deposition or in situ synthesis using
poly- or per-
fluorinated compounds in solution or polymerizable monomers. Examples include
TEFLON AF coatings and FLUOROPEL coatings for dip or spray coating, other
fluorinated monomers for plasma-enhanced chemical vapor deposition (PECVD),
and
organosiloxane (e.g., Si0C) for PECVD. Additionally, in some cases, some
portion or all of
the droplet operations surface may be coated with a substance for reducing
background noise,
such as background fluorescence from a PCB substrate. For example, the noise-
reducing
coating may include a black matrix resin, such as the black matrix resins
available from
Toray industries, Inc., Japan.
[0089] Electrodes of a droplet actuator are typically controlled by
a controller or a
processor, which is itself provided as part of a system, which may include
processing
functions as well as data and software storage and input and output
capabilities. Reagents
may be provided on the droplet actuator in the droplet operations gap or in a
reservoir fluidly
coupled to the droplet operations gap. The reagents may be in liquid form,
e.g., droplets, or
they may be provided in a reconstitutable form in the droplet operations gap
or in a reservoir
fluidly coupled to the droplet operations gap. Reconstitutable reagents may
typically be
combined with liquids for reconstitution. An example of reconstitutable
reagents suitable for
use with the methods and apparatus set forth herein includes those described
in Meathrel et
al., U.S. Patent No. 7,727,466, entitled "Disintegratable Films for Diagnostic
Devices,"
issued on June 1, 2010.
[0090] "Droplet operation", as used herein, means any manipulation
of a droplet
on a droplet actuator. A droplet operation may, for example, include: loading
a droplet into
the droplet actuator; dispensing one or more droplets from a source droplet;
splitting,
separating or dividing a droplet into two or more droplets; transporting a
droplet from one
location to another in any direction; merging or combining two or more
droplets into a single
droplet; diluting a droplet; mixing a droplet; agitating a droplet; deforming
a droplet;
retaining a droplet in position; incubating a droplet; heating a droplet;
vaporizing a droplet;
cooling a droplet; disposing of a droplet; transporting a droplet out of a
droplet actuator;
other droplet operations described herein; and/or any combination of the
foregoing. The
terms "merge," "merging," "combine," "combining" and the like are used to
describe the
-30-
CA 3016221 2019-11-29
creation of one droplet from two or more droplets. It should be understood
that when such a
term is used in reference to two or more droplets, any combination of droplet
operations that
are sufficient to result in the combination of the two or more droplets into
one droplet may be
used. For example, "merging droplet A with droplet B," can be achieved by
transporting
droplet A into contact with a stationary droplet B, transporting droplet B
into contact with a
stationary droplet A, or transporting droplets A and B into contact with each
other. The terms
"splitting," "separating" and "dividing" are not intended to imply any
particular outcome
with respect to volume of the resulting droplets (i.e., the volume of the
resulting droplets can
be the same or different) or number of resulting droplets (the number of
resulting droplets
may be 2, 3, 4, 5 or more). The term "mixing" refers to droplet operations
which result in
more homogenous distribution of one or more components within a droplet
Examples of
"loading" droplet operations include microdialysis loading, pressure assisted
loading, robotic
loading, passive loading, and pipette loading. Droplet operations may be
electrode-mediated.
In some cases, droplet operations are further facilitated by the use of
hydrophilic and/or
hydrophobic regions on surfaces and/or by physical obstacles. For non-limiting
examples of
droplet operations, see the patents and patent applications cited above under
the definition of
"droplet actuator." Impedance or capacitance sensing or imaging techniques may
sometimes
be used to determine or confirm the outcome of a droplet operation. Examples
of such
techniques are described in Sturmer et al., U.S. Patent Pub. No. 20100194408,
entitled
"Capacitance Detection in a Droplet Actuator," published on Aug. 5, 2010.
Generally
speaking, the sensing or imaging techniques may be used to confirm the
presence or absence
of a droplet at a specific electrode. For example, the presence of a dispensed
droplet at the
destination electrode following a droplet dispensing operation confirms that
the droplet
dispensing operation was effective. Similarly, the presence of a droplet at a
detection spot at
an appropriate step in an assay protocol may confirm that a previous set of
droplet operations
has successfully produced a droplet for detection. Droplet transport time can
be quite fast
For example, in various embodiments, transport of a droplet from one electrode
to the next
may exceed about 1 sec, or about 0.1 sec, or about 0.01 sec, or about 0.001
sec. In some
embodiments disclosed herein, the electrode is operated in AC mode but is
switched to DC
mode for imaging. It is helpful for conducting droplet operations for the
footprint area of
droplet to be similar to electrowetting area; in other words, lx-, 2x- 3x-
droplets are usefully
-31-
CA 3016221 2019-11-29
controlled operated using 1, 2, and 3 electrodes, respectively. If the droplet
footprint is
greater than number of electrodes available for conducting a droplet operation
at a given
time, the difference between the droplet size and the number of electrodes
should typically
not be greater than 1; in other words, a 2x droplet is usefully controlled
using 1 electrode and
a 3x droplet is usefully controlled using 2 electrodes. When droplets include
beads, it is
useful for droplet size to be equal to the number of electrodes controlling
the droplet, e.g.,
transporting the droplet.
[0091] "Filler fluid" means a fluid associated with a droplet
operations substrate
of a droplet actuator, which fluid is sufficiently immiscible with a droplet
phase to render the
droplet phase subject to electrode-mediated droplet operations. For example,
the droplet
operations gap of a droplet actuator is typically filled with a filler fluid.
The filler fluid may,
for example, be or include a low-viscosity oil, such as silicone oil or
hexadecane filler fluid.
The filler fluid may be or include a halogenated oil, such as a fluorinated or
perfluorinated
oil. The filler fluid may fill the entire gap of the droplet actuator or may
coat one or more
surfaces of the droplet actuator. Filler fluids may be conductive or non-
conductive. Filler
fluids may be selected to improve droplet operations and/or reduce loss of
reagent or target
substances from droplets, improve formation of microdroplets, reduce cross
contamination
between droplets, reduce contamination of droplet actuator surfaces, reduce
degradation of
droplet actuator materials, etc. For example, filler fluids may be selected
for compatibility
with droplet actuator materials. As an example, fluorinated filler fluids may
be usefully
employed with fluorinated surface coatings. Fluorinated filler fluids are
useful to reduce loss
of lipophilic compounds, such as umbelliferone substrates like 6-
hexadecanoylamido-4-
methylumbelliferone substrates (e.g., for use in Krabbe, Niemann-Pick, or
other assays);
other umbelliferone substrates are described in Winger et al., U.S. Patent
Pub. No.
20110118132, entitled "Enzymatic Assays Using Umbelliferone Substrates with
Cyclodextrins in Droplets of Oil," published on May 19, 2011. Examples of
suitable
fluorinated oils include those in the Galden line, such as Galden HT170 (bp =
170 C,
viscosity = 1.8 cSt, density = 1.77), Galden HT200 (bp = 200 C, viscosity =
2.4 cSt, d =
1.79), Galden HT230 (bp = 230 C, viscosity = 4.4 cSt, d = 1.82) (all from
Solvay Solexis);
those in the Novec line, such as Novec 7500 (bp
-32-
CA 3016221 2019-11-29
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
= 128 C, viscosity = 0.8 cSt, d = 1.61), Fluorinert FC-40 (bp = 155 C,
viscosity = 1.8 cSt, d
= 1.85), Fluorinert FC-43 (bp = 174 C, viscosity = 2.5 cSt, d = 1.86) (both
from 3M). In
general, selection of perfluorinated filler fluids is based on kinematic
viscosity (< 7 cSt is
preferred, but not required), and on boiling point (> 150 C is preferred, but
not required, for
use in DNA/RNA-based applications (PCR, etc.)). Filler fluids may, for
example, be doped
with surfactants or other additives. For example, additives may be selected to
improve
droplet operations and/or reduce loss of reagent or target substances from
droplets, formation
of microdroplets, cross contamination between droplets, contamination of
droplet actuator
surfaces, degradation of droplet actuator materials, etc. Composition of the
filler fluid,
including surfactant doping, may be selected for performance with reagents
used in the
specific assay protocols and effective interaction or non-interaction with
droplet actuator
materials. Examples of filler fluids and filler fluid formulations suitable
for use with the
methods and apparatus set forth herein are provided in Srinivasan et al,
International Patent
Pub. No. WO/2010/027894, entitled "Droplet Actuators, Modified Fluids and
Methods,"
published on June 3, 2010; Srinivasan et al, International Patent Pub. No.
WO/2009/021173,
entitled "Use of Additives for Enhancing Droplet Operations," published on
February 12,
2009; Sista etal., International Patent Pub. No. WO/2008/098236, entitled
"Droplet Actuator
Devices and Methods Employing Magnetic Beads," published on January 15, 2009;
and
Monroe et al., U.S. Patent Pub. No. 20080283414, entitled "Electrowetting
Devices,"
published on November 20, 2008, the entire disclosures of which are
incorporated herein by
reference, as well as the other patents and patent applications cited herein.
Fluorinated oils
may in some cases be doped with fluorinated surfactants, e.g., Zonyl FSO-100
(Sigma-
Aldrich) and/or others. A filler fluid is typically a liquid. In some
embodiments, a filler gas
can be used instead of a liquid.
[0092] The term "hybridization", as used herein, refers generally to the
ability of
nucleic acid molecules to join via complementary base strand pairing. Such
hybridization
may occur when nucleic acid molecules are contacted under appropriate
conditions and/or
circumstances. As used herein, two nucleic acid molecules are said to be
capable of
specifically hybridizing to one another if the two molecules are capable of
forming an anti-
parallel, double-stranded nucleic acid structure. A nucleic acid molecule is
said to be the
"complement" of another nucleic acid molecule if they exhibit complete
complementarity. As
-33-
used herein, nucleic acid molecules are said to exhibit "complete
complementarity" when
every nucleotide of one of the molecules is complementary to its base pairing
partner
nucleotide of the other. Two molecules are said to be "minimally
complementary" if they can
hybridize to one another with sufficient stability to permit them to remain
annealed to one
another under at least conventional "low-stringency" conditions. In some
instances, the
molecules are said to be "complementary" if they can hybridize to one another
with sufficient
stability to permit them to remain annealed to one another under conventional
"high-
stringency" conditions. Nucleic acid molecules that hybridize to other nucleic
acid
molecules, e.g., at least under low stringency conditions are said to be
"hybridizable
cognates" of the other nucleic acid molecules. Conventional stringency
conditions are
described by Sambrook et al., Molecular Cloning, A Laboratory Handbook, Cold
Spring
Harbor Laboratory Press, 1989), and by Haymes et al. In: Nucleic Acid
Hybridization, A
Practical Approach, IRL Press, Washington, D.C. (1985), and for the disclosure
discussed
herein. Departures from complete complementarity are therefore permissible in
some
embodiments, as long as such departures do not completely preclude the
capacity of the
molecules to form a double-stranded structure. Thus, in order for a nucleic
acid molecule or
fragment thereof of the present disclosure to serve as a primer or probe in
some embodiments
it needs only be sufficiently complementary in sequence to be able to form a
stable double-
stranded structure under the particular solvent and salt concentrations
employed. In other
embodiments, the nucleic acids disclosed herein are fully complementary to
their targets.
[0093] The
term "immobilize", as used herein with respect to magnetically
responsive beads, means that the beads are substantially restrained in
position in a droplet or
in filler fluid on a droplet actuator. For example, in some embodiments
disclosed herein,
immobilized beads are sufficiently restrained in position in a droplet to
permit execution of a
droplet splitting operation, yielding one droplet with substantially all of
the beads and one
droplet substantially lacking in the beads. In reference to nucleic acid
molecule, e.g. a primer
or oligonucleotide, the term "immobilize," and its derivatives, as used herein
refers to the
attachment of a nucleic acid molecule directly to a solid support through at
least one
intermediate component such as, for example, biotin. As used herein, the terms
"attach" and
"affix" and their respective derivatives include adsorption, such as,
physisorption or
-34-
CA 3016221 2019-11-29
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
chemisorption, ligand/receptor interaction, covalent bonding, hydrogen
bonding, or ionic
bonding of a polymeric substance or a nucleic acid molecule to a solid
support.
[0094] As used herein, the term "magnetically responsive" means
responsive to a
magnetic field. "Magnetically responsive beads" include or are composed of
magnetically
responsive materials. Examples of magnetically responsive materials include
paramagnetic
materials, ferromagnetic materials, ferrimagnetic materials, and metamagnetic
materials.
Examples of suitable paramagnetic materials include iron, nickel, and cobalt,
as well as metal
oxides, such as Fe304, BaFe12019, CoO, NiO, Mm03, Cr2O3, and CoMnP.
[0095] The terms "nucleic acid molecule and "polynucleotide" are used
interchangeably herein, and refer to both RNA and DNA molecules, including
nucleic acid
molecules comprising cDNA, genomic DNA, synthetic DNA, and DNA or RNA
molecules
containing nucleic acid analogs. Nucleic acid molecules can have any three-
dimensional
structure. A nucleic acid molecule can be double-stranded or single-stranded
(e.g., a sense
strand or an antisense strand). Non-limiting examples of nucleic acid
molecules include
genes, gene fragments, exons, introns, messenger RNA (mRNA), transfer RNA,
ribosomal
RNA, siRNA, micro-RNA, tracrRNAs, crRNAs, guide RNAs, ribozymes, cDNA,
recombinant polynucleotides, branched polynucleotides, nucleic acid probes and
nucleic acid
primers. A nucleic acid molecule may contain unconventional or modified
nucleotides. The
terms "polynucleotide sequence" and "nucleic acid sequence" as used herein
interchangeably
refer to the sequence of a polynucleotide molecule. The nomenclature for
nucleotide bases as
set forth in 37 CFR 1.822 is used herein.
[0096] "Reservoir" means an enclosure or partial enclosure configured
for
holding, storing, or supplying liquid. In some embodiments, a droplet actuator
system of the
present disclosure may include on-cartridge reservoirs and/or off-cartridge
reservoirs. On-
cartridge reservoirs may be (1) on-actuator reservoirs, which are reservoirs
in the droplet
operations gap or on the droplet operations surface; (2) off-actuator
reservoirs, which are
reservoirs on the droplet actuator cartridge, but outside the droplet
operations gap, and not in
contact with the droplet operations surface; or (3) hybrid reservoirs which
have on-actuator
regions and off-actuator regions. An example of an off-actuator reservoir is a
reservoir in the
top substrate. An off-actuator reservoir is typically in fluid communication
with an opening
or flow path arranged for flowing liquid from the off-actuator reservoir into
the droplet
-35-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
operations gap, such as into an on-actuator reservoir. An off-cartridge
reservoir may be a
reservoir that is not part of the droplet actuator cartridge at all, but which
flows liquid to
some portion of the droplet actuator cartridge. For example, an off-cartridge
reservoir may be
part of a system or docking station to which the droplet actuator cartridge is
coupled during
operation. Similarly, an off-cartridge reservoir may be a reagent storage
container or syringe
which is used to force fluid into an on-cartridge reservoir or into a droplet
operations gap. In
some embodiments, a system using an off-cartridge reservoir will typically
include a fluid
passage means whereby liquid may be transferred from the off-cartridge
reservoir into an on-
cartridge reservoir or into a droplet operations gap.
[0097] As used herein, the term a "nucleic acid sample" refers to a
collection of
nucleic acid molecules. In some embodiments, the nucleic acid sample is from a
single
biological source, e.g. one individual or one tissue sample, and in other
embodiments the
nucleic acid sample is a pooled sample, e.g., containing nucleic acids from
more than one
organism, individual or tissue.
[0098] The term nucleic acid sample encompasses "nucleic acid library"
which,
as used herein, includes a nucleic acid library that has been prepared by any
method known
in the art. In some embodiments, providing the nucleic acid library includes
the steps
required for preparing the library, for example, including the process of
incorporating one or
more nucleic acid samples into a vector-based collection, such as by ligation
into a vector
and transformation of a host. In some embodiments, providing a nucleic acid
library includes
the process of incorporating a nucleic acid sample into a non-vector-based
collection, such as
by ligation to adaptors. In some embodiments, the adaptors can anneal to PCR
primers to
facilitate amplification by PCR or can be universal primer regions such as,
for example,
sequencing tail adaptors. In some embodiments, the adaptors can be universal
sequencing
adaptors.
[0099] The term "substantially" as used herein has its ordinary meaning
as read in
light of the specification, and can mean, for example, at least 80%, at least
85%, at least 90%,
at least 95%, at least 96%, at least 97%, at least 98%, at least 99%.
[0100] "Transporting into the magnetic field of a magnet," "transporting
towards
a magnet," and the like, as used herein to refer to droplets and/or
magnetically responsive
beads within droplets, is intended to refer to transporting into a region of a
magnetic field
-36-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
capable of substantially attracting magnetically responsive beads in the
droplet. Similarly,
"transporting away from a magnet or magnetic field," "transporting out of the
magnetic field
of a magnet," and the like, as used herein to refer to droplets and/or
magnetically responsive
beads within droplets, is intended to refer to transporting away from a region
of a magnetic
field capable of substantially attracting magnetically responsive beads in the
droplet, whether
or not the droplet or magnetically responsive beads is completely removed from
the magnetic
field. It will be appreciated that in any of such cases described herein, the
droplet may be
transported towards or away from the desired region of the magnetic field,
and/or the desired
region of the magnetic field may be moved towards or away from the droplet
Reference to
an electrode, a droplet, or magnetically responsive beads being "within" or
"in" a magnetic
field, or the like, is intended to describe a situation in which the electrode
is situated in a
manner which permits the electrode to transport a droplet into and/or away
from a desired
region of a magnetic field, or the droplet or magnetically responsive beads
is/are situated in a
desired region of the magnetic field, in each case where the magnetic field in
the desired
region is capable of substantially attracting any magnetically responsive
beads in the droplet
Similarly, reference to an electrode, a droplet, or magnetically responsive
beads being
"outside of' or "away from" a magnetic field, and the like, is intended to
describe a situation
in which the electrode is situated in a manner which permits the electrode to
transport a
droplet away from a certain region of a magnetic field, or the droplet or
magnetically
responsive beads is/are situated away from a certain region of the magnetic
field, in each case
where the magnetic field in such region is not capable of substantially
attracting any
magnetically responsive beads in the droplet or in which any remaining
attraction does not
eliminate the effectiveness of droplet operations conducted in the region. In
various aspects
of the present disclosure, a system, a droplet actuator, or another component
of a system may
include a magnet, such as one or more permanent magnets (e.g., a single
cylindrical or bar
magnet or an array of such magnets, such as a Halbach array) or an
electromagnet or array of
electromagnets, to form a magnetic field for interacting with magnetically
responsive beads
or other components on chip. Such interactions may, for example, include
substantially
immobilizing or restraining movement or flow of magnetically responsive beads
during
storage or in a droplet during a droplet operation or pulling magnetically
responsive beads
out of a droplet.
-37-
[0101] As used herein, the term "universal sequence" refers to a
region of
sequence that is common to two or more nucleic acid molecules where the
molecules also
have regions of sequence that differ from each other. A universal sequence
that is present in
different members of a collection of molecules can allow capture of multiple
different
nucleic acids using a population of universal capture nucleic acids that are
complementary to
the universal sequence. Similarly, a universal sequence present in different
members of a
collection of molecules can allow the replication or amplification of multiple
different
nucleic acids using a population of universal primers that are complementary
to the universal
sequence. Thus, a universal capture nucleic acid or a universal primer such
as, a universal
sequencing tail-adaptor, includes a sequence that can hybridize specifically
to a universal
sequence. In some embodiments of the methods disclosed herein, target nucleic
acid
molecules may be modified to attach universal adaptors, for example, at one or
both ends of
the different target sequences, for example by ligation or by amplification
using primer-
directed amplification.
[0102] "Washing" with respect to washing a bead means reducing the
amount
and/or concentration of one or more substances in contact with the bead or
exposed to the
bead from a droplet in contact with the bead. The reduction in the amount
and/or
concentration of the substance may be partial, substantially complete, or even
complete. The
substance may be any of a wide variety of substances; examples include target
substances for
further analysis, and unwanted substances, such as components of a sample,
contaminants,
and/or excess reagent In some embodiments, a washing operation begins with a
starting
droplet in contact with a magnetically responsive bead, where the droplet
includes an initial
amount and initial concentration of a substance. The washing operation may
proceed using a
variety of droplet operations. The washing operation may yield a droplet
including the
magnetically responsive bead, where the droplet has a total amount and/or
concentration of
the substance which is less than the initial amount and/or concentration of
the substance.
Examples of suitable washing techniques are described in Pamula et al., U.S.
Patent No.
7,439,014, entitled "Droplet-Based Surface Modification and Washing," issued
on October
21, 2008.
[0103] The terms "top," "bottom," "over," "under," and "on" are
used throughout
the description with reference to the relative positions of components of the
droplet actuator,
-38-
CA 3016221 2019-11-29
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
such as relative positions of top and bottom substrates of the droplet
actuator. It will be
appreciated that the droplet actuator is functional regardless of its
orientation in space.
[0104] When a liquid in any form (e.g., a droplet or a continuous body,
whether
moving or stationary) is described as being "on", "at", or "over" an
electrode, array, matrix
or surface, such liquid could be either in direct contact with the
electrode/array/matrix/surface, or could be in contact with one or more layers
or films that
are interposed between the liquid and the electrode/array/matrix/surface. In
one example,
filler fluid can be considered as a film between such liquid and the
electrode/array/matrix/surface.
[0105] When a droplet is described as being "on" or "loaded on" a
droplet
actuator, it should be understood that the droplet is arranged on the droplet
actuator in a
manner which facilitates using the droplet actuator to conduct one or more
droplet operations
on the droplet, the droplet is arranged on the droplet actuator in a manner
which facilitates
sensing of a property of or a signal from the droplet, and/or the droplet has
been subjected to
a droplet operation on the droplet actuator.
[0106] The terms "fluidics cartridge," "digital fluidics cartridge,"
"droplet
actuator," and "droplet actuator cartridge" as used throughout the description
can be
synonymous.
[0107] As will be understood by one having ordinary skill in the art,
for any and
all purposes, such as in terms of providing a written description, all ranges
disclosed herein
also encompass any and all possible sub-ranges and combinations of sub-ranges
thereof. Any
listed range can be easily recognized as sufficiently describing and enabling
the same range
being broken down into at least equal halves, thirds, quarters, fifths,
tenths, etc. As a non-
limiting example, each range discussed herein can be readily broken down into
a lower third,
middle third and upper third, etc. As will also be understood by one skilled
in the art all
language such as "up to," "at least," "greater than," "less than," and the
like include the
number recited and refer to ranges which can be subsequently broken down into
sub-ranges
as discussed above. Finally, as will be understood by one skilled in the art,
a range includes
each individual member. Thus, for example, a group having 1-3 articles refers
to groups
having 1, 2, or 3 articles. Similarly, a group having 1-5 articles refers to
groups having 1, 2,
3, 4, or 5 articles, and so forth.
-39-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
[0108] In some embodiments of the methods or processes described herein,
the
steps can be carried out in any order, except when a temporal or operational
sequence is
explicitly recited. Furthermore, in some embodiments, the specified steps can
be carried out
concurrently unless explicit claim language recites that they be carried out
separately. For
example, in some embodiments a claimed step of doing X and a claimed step of
doing Y can
be conducted simultaneously within a single operation, and the resulting
process will fall
within the literal scope of the claimed process.
[0109] As used herein, "comprising" is synonymous with "including,"
"containing," or "characterized by," and is inclusive or open-ended and does
not exclude
additional, unrecited elements or method steps. As used herein, "consisting
of' excludes any
elements, steps, or ingredients not specified in the claimed composition or
method. As used
herein, "consisting essentially of' does not exclude materials or steps that
do not materially
affect the basic and novel characteristics of the claimed composition or
method. Any
recitation herein of the term "comprising", particularly in a description of
components of a
composition or in a description of steps of a method, is understood to
encompass those
compositions and methods consisting essentially of and consisting of the
recited components
or steps.
[0110] Headings, e.g., (a), (b), (i) etc., are presented merely for ease
of reading
the specification and claims, and do not limit in any way the scope of the
disclosure or its
alternatives. The use of headings in the specification or claims does not
require the steps or
elements be performed in alphabetical or numerical order or the order in which
they are
presented.
I. METHODS FOR NORMALIZING NUCLEIC ACID SAMPLES
[0111] Various embodiments of the disclosure generally relate to systems
and
methods for construction of nucleic acid samples, including construction of
amplified/normalized nucleic acid samples and nucleic acid libraries for
downstream
analytical applications. In one aspect, some embodiments of the methods
disclosed herein
employ at least one set of first amplification or normalization primers
immobilized on a solid
phase support and at least another set of second amplification or
normalization primers in
solution phase during normalization of the nucleic acid samples or nucleic
acid libraries. As
discussed above, this biphasic feature of the disclosed methods differs from
current library
-40-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
preparation procedures in which both primers are either in a solution phase or
both primers
are affixed onto a solid phase. The first amplification or normalization
primers in solution
phase offer fast kinetics, while the second amplification or normalization
primers
immobilized on a solid phase support offer ability to normalize the nucleic
acid samples and
ease of purification. Additionally, in some particular implementations of the
methods
disclosed herein, the set of first primers immobilized on the solid support is
provided in an
amount which limits the yield of amplification products to a predefined
amount, and the set
of second primers in solution is provided in an amount that exceeds the amount
of the first
primers. In some embodiments, the primer configuration can provide one or more
of the
following advantages: (1) convenient washing away of any amplification
products that is
found in solution after amplification, resulting in an amplified/normalized
amount of
amplification products remained immobilized on the solid support which can be
subsequently
isolated, such that a predefined amount of amplification products can be
generated; (2)
construction of a plurality of nucleic acid samples or nucleic acid libraries
in which the DNA
amounts are normalized to substantially uniform concentrations across the
nucleic acid
libraries regardless of the amounts of input DNA in the original samples.
[0112] In one aspect, the present disclosure provides a method for
nucleic acid
amplification that includes providing a nucleic acid sample including target
nucleic acid
molecules; contacting the nucleic acid sample with a reaction mixture
comprising a solid
phase and a liquid phase, the solid phase includes a plurality of first
amplification primers
immobilized on a solid support, the first amplification primers capable of
specifically
hybridizing to a first sequence of the target nucleic acid molecules, and the
liquid phase
includes a plurality of second amplification primers in solution, the
plurality of second
primers capable of specifically hybridizing to a second sequence of the target
nucleic acid
molecules; and amplifying the target nucleic acid molecules under isothermal
conditions
such that substantially all of the first amplification primers are
incorporated into
amplification products, wherein the plurality of first primers is provided in
an amount which
limits the yield of amplification products to a predefined amount, and the
plurality of second
amplification primers is provided in an amount that exceeds the amount of the
first
amplification primers.
-41-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
[0113] Figure 2 illustrates a flow diagram of a non-limiting example of
a method
of amplifying and/or normalizing nucleic acid sample according to some
embodiments of the
disclosure, in which the amount of target nucleic acid molecules in a nucleic
acid sample is
amplified/normalized in a reaction mixture that includes a solid phase and a
liquid phase.
Depending on specific workflows and downstream applications, extendable
primers used in
this and other exemplary methods of the present disclosure can be
amplification primers or
normalization primers. Method 200 may include, but is not limited to, the
following steps.
[0114] At a step 210, an input nucleic acid sample such as, for example,
a
genomic DNA sample, comprising target nucleic acid molecules is provided. This
step can
be achieved, for example, by loading the nucleic acid sample into a sample
reservoir of a
droplet actuator. At a step 230, the target nucleic acid molecules are brought
into contact
with a reaction mixture comprising a solid phase and a liquid phase. The solid
phase of the
reaction mixture includes a plurality of first amplification or normalization
primers (e.g.,
primers including a P7 primer sequence) capable of specifically hybridizing to
a first
sequence of the target nucleic acid molecules where the first amplification or
normalization
primers are immobilized on a solid support such as, for instance, capture
beads. The liquid
phase of the reaction mixture includes a plurality of second amplification or
normalization
primers in solution, the plurality of second primers capable of specifically
hybridizing to a
second sequence of the target nucleic acid molecules (e.g., primers including
a P5 primer
sequence). At a step 235, the first amplification or normalization primer
hybridized to
targeted nucleic acid sequences is extended to form an immobilized
complementary DNA
strand and, at a step 245, the extended target nucleic acid molecules are
amplified under
isothermal conditions such that substantially all of the first amplification
or normalization
primers are incorporated into amplification products. In some embodiments of
the disclosed
methods, the plurality of first immobilized primers is provided in an amount
which limits the
yield of amplification products to a predefined amount, and the plurality of
second
amplification or normalization primers is provided in an amount that exceeds
the amount of
the first amplification or normalization primers. At an optional step 250, the
amplified/normalized nucleic acid samples are eluted from the capture beads
for downstream
analytical applications such as, for example, high-throughput sequencing.
-42-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
[0115] Figure 3 shows pictorially the steps of method 200 of Figure 2.
Namely,
an input nucleic acid sample (not shown) includes a target nucleic acid
molecule 310. First
amplification or normalization primer (e.g., reverse primer) includes a target-
specific region
340. Second amplification or normalization primer (e.g., forward primer)
includes a target-
specific region 330. In some embodiments, the target-specific regions 330 and
340 flank a
region of interest on the target nucleic acid molecule 310. In some
embodiments, the target-
specific regions can have sequence complementarity to adaptors or universal
primer
sequences in the target nucleic acids, which can be added to the target
nucleic acids in a
sequence specific manner. In some embodiments, as disclosed in more detail
below,
adaptors or universal primer sequences in the target nucleic acids can be
added to the target
nucleic acids in a sequence independent manner. An amplicon 350 is then
synthesized using
the forward primer and reverse primer. In the reaction, the first
amplification or
normalization primer (e.g., reverse primer) is immobilized on the solid
support. For
example, the reverse amplification or normalization primer is conjugated to a
biotin label 380
and is immobilized on a streptavidin (SA) coated capture bead 385.
[0116] In some embodiments of the methods according to this and other
aspects
of the disclosure, the plurality of first amplification or normalization
primers (e.g., reverse
primers) is hybridized with the target nucleic acid molecules prior to being
immobilized on
the solid support. A flow diagram of a non-limiting example of a method
according to these
embodiments of the disclosure is shown at Figures 4 and 5. At a step 425, the
first
amplification or normalization primers 340 are hybridized to a target nucleic
acid molecule
310 of the nucleic acid sample to form target molecule\normalization-primer
duplexes. This
hybridization step is carried out in solution phase. At a step 430, capture
beads such as, for
example, streptavidin-coated capture beads, are added to the hybridization
reaction for
capture of hybridized target molecule\primer duplexes. Hybridized
molecule\normalization-
primer duplexes and un-hybridized primers are immobilized on the capture beads
by
formation of a biotin-streptavidin binding complex. The remaining steps 435,
445, and 450
are carried out similarly to the corresponding steps 235, 245, and 250 of the
exemplary
methods described in Figures 2 and 3.
[0117] According to some embodiments of the methods disclosed herein, a
sample or a library of target nucleic acids can have an average strand length
that is desired or
-43-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
appropriate for a particular application of the methods or compositions set
forth herein. For
example, the average strand length can be less than about 100,000 nucleotides,
50,000
nucleotides, 10,000 nucleotides, 5,000 nucleotides, 1,000 nucleotides, 500
nucleotides, 100
nucleotides, or 50 nucleotides. Alternatively or additionally, the average
strand length can be
greater than about 10 nucleotides, 50 nucleotides, 100 nucleotides, 500
nucleotides, 1,000
nucleotides, 5,000 nucleotides, 10,000 nucleotides, 50,000 nucleotides, or
100,000
nucleotides. The average strand length for populations of target nucleic acids
as disclosed
herein can be in a range between a maximum and minimum value set forth above.
Solid support
[0118] In some embodiments, the amplification and/or normalization
reaction
mixture of the methods disclosed herein includes one or more solid supports.
Solid supports
suitable for the methods disclosed herein can generally be of any convenient
size and
fabricated from any number of known materials. Preferably, the solid support
used in the
methods disclosed herein can be of any suitable type that provides a known
binding capacity,
resulting in a substantially unfluctuating amount of bound nucleic acids per
fixed amount of
solid support. Example of such materials include: inorganics, natural
polymers, and synthetic
polymers. Specific examples of these materials include: cellulose, cellulose
derivatives,
acrylic resins, glass, silica gels, gelatin, polystyrene, polyvinyl
pyrrolidone, co-polymers of
vinyl and acrylamide, polystyrene cross-linked with divinylbenzene or the
like,
polyacrylamides, latex gels, silicon, plastics, nitrocellulose, polystyrene,
dextran, rubber,
natural sponges, silica gels, control pore glass, metals, cross-linked
dextrans (e.g.,
SephadexTM) agarose gel (SepharoseTm), and other solid supports known to those
of skill in
the art.
[0119] In some embodiments, the solid phase supports can include
synthetic
polymer supports, such as polystyrene, polypropylene, substituted polystyrene
(e.g.,
carboxylated or aminated polystyrene), polyamides, polyacrylamides,
polyvinylchloride, and
the like, or any material useful in nucleic acid affinity chromatography.
[0120] In some embodiments of the methods disclosed herein, the solid
support
can include beads. The term "bead," as used herein with respect to beads on a
droplet
actuator, means any bead or particle that is capable of interacting with a
droplet on or in
proximity with a droplet actuator. Beads may be any of a wide variety of
shapes, such as
-44-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
spherical, generally spherical, egg shaped, disc shaped, cubical, amorphous
and other three
dimensional shapes. The bead may, for example, be capable of being subjected
to a droplet
operation in a droplet on a droplet actuator or otherwise configured with
respect to a droplet
actuator in a manner which permits a droplet on the droplet actuator to be
brought into
contact with the bead on the droplet actuator and/or off the droplet actuator.
Beads may be
provided in a droplet, in a droplet operations gap, or on a droplet operations
surface. Beads
may be provided in a reservoir that is external to a droplet operations gap or
situated apart
from a droplet operations surface, and the reservoir may be associated with a
flow path that
permits a droplet including the beads to be brought into a droplet operations
gap or into
contact with a droplet operations surface. Beads may be manufactured using a
wide variety
of materials, including for example, resins, and polymers. The beads may be
any suitable
size, including for example, microbeads, microparticles, nanobeads and
nanoparticles. In
some cases, beads are magnetically responsive; in other cases beads are not
significantly
magnetically responsive. For magnetically responsive beads, the magnetically
responsive
material may constitute substantially all of a bead, a portion of a bead, or
only one
component of a bead. The remainder of the bead may include, among other
things, polymeric
material, coatings, and moieties which permit attachment of an assay reagent
Examples of
suitable beads include flow cytometry microbeads, polystyrene microparticles
and
nanoparticles, functionalized polystyrene microparticles and nanoparticles,
coated
polystyrene microparticles and nanoparticles, silica microbeads, fluorescent
microspheres
and nanospheres, functionalized fluorescent microspheres and nanospheres,
coated
fluorescent microspheres and nanospheres, color dyed microparticles and
nanoparticles,
magnetic microparticles and nanoparticles, superparamagnetic microparticles
and
nanoparticles (e.g., DYNABEADS particles, available from Invitrogen Group,
Carlsbad,
CA), fluorescent microparticles and nanoparticles, coated magnetic
microparticles and
nanoparticles, ferromagnetic microparticles and nanoparticles, coated
ferromagnetic
microparticles and nanoparticles, and those described in Watkins etal., U.S.
Patent Pub. No.
20050260686, entitled "Multiplex Flow Assays Preferably with Magnetic
Particles as Solid
Phase," published on November 24, 2005; Chandler., U.S. Patent Pub. No.
20030132538,
entitled "Encapsulation of Discrete Quanta of Fluorescent Particles,"
published on July 17,
2003; Chandler et al., U.S. Patent Pub. No. 20050118574, entitled "Multiplexed
Analysis of
-45-
Clinical Specimens Apparatus and Method," published on June 2, 2005; Chandler
et
al., U.S. Patent Pub. No. 20050277197, entitled "Microparticles with Multiple
Fluorescent
Signals and Methods of Using Same," published on December 15, 2005; and
Chandler et al.,
U.S. Patent Pub. No. 20060159962, entitled "Magnetic Microspheres for use in
Fluorescence-based Applications," published on July 20, 2006, for their
teaching concerning
beads and magnetically responsive materials and beads. Beads may be pre-
coupled with a
biomolecule or other substance that is able to bind to and form a complex with
a
biomolecule. Beads may be pre-coupled with an antibody, protein or antigen,
DNA/RNA
probe or any other molecule with an affinity for a desired target. Examples of
droplet
actuator techniques for immobilizing magnetically responsive beads and/or non-
magnetically
responsive beads and/or conducting droplet operation protocols using beads are
described in
Pollack et al., U.S. Patent Pub. No. 20080053205, entitled "Droplet-Based
Particle Sorting,"
published on March 6, 2008; U.S. Patent App. No. 61/039,183, entitled
"Multiplexing Bead
Detection in a Single Droplet," filed on March 25, 2008; Pamula et al., U.S.
Patent App. No.
61/047,789, entitled "Droplet Actuator Devices and Droplet Operations Using
Beads," filed
on April 25, 2008; U.S. Patent App. No. 61/086,183, entitled "Droplet Actuator
Devices and
Methods for Manipulating Beads," filed on August 5, 2008; Eckhardt et al.,
International
Patent Pub. No. WO/2008/098236, entitled "Droplet Actuator Devices and Methods
Employing Magnetic Beads," published on August 14, 2008; Grichko et al.,
International
Patent Pub. No. WO/2008/134153, entitled "Bead-based Multiplexed Analytical
Methods
and Instrumentation," published on November 6, 2008; Eckhardt et al.,
International Patent
Pub. No. WO/2008/116221, "Bead Sorting on a Droplet Actuator," published on
September
25, 2008; and Eckhardt et al., International Patent Pub. No. WO/2007/120241,
entitled
"Droplet-based Biochemistry," published on October 25, 2007. Bead
characteristics may be
employed in the multiplexing aspects of the present disclosure. Examples of
beads having
characteristics suitable for multiplexing, as well as methods of detecting and
analyzing
signals emitted from such beads, may be found in Whitman et al., U.S. Patent
Pub. No.
20080305481, entitled "Systems and Methods for Multiplex Analysis of PCR in
Real Time,"
published on December 11, 2008; Roth, U.S. Patent Pub. No. 20080151240,
"Methods and
Systems for Dynamic Range Expansion," published on June 26, 2008; Sorensen et
al., U.S.
Patent Pub. No. 20070207513, entitled "Methods, Products, and Kits for
Identifying an
-46-
CA 3016221 2019-11-29
Analyte in a Sample," published on September 6, 2007; Roth, U.S. Patent Pub.
No.
20070064990, entitled "Methods and Systems for Image Data Processing,"
published on
March 22, 2007; Chandler et al., U.S. Patent Pub. No. 20060159962, entitled
"Magnetic
Microspheres for use in Fluorescence-based Applications," published on July
20, 2006;
Chandler et al., U.S. Patent Pub. No. 20050277197, entitled "Microparticles
with Multiple
Fluorescent Signals and Methods of Using Same," published on December 15,
2005; and
Chandler et al., U.S. Patent Publication No. 20050118574, entitled
"Multiplexed Analysis of
Clinical Specimens Apparatus and Method," published on June 2, 2005.
[0121] Accordingly, in some preferred embodiments, the beads
suitable for the
methods disclosed herein can be of any convenient size and fabricated from any
number of
known materials. In some embodiments, the bead can be, for example, magnetic
beads,
paramagnetic beads, plastic beads, polystyrene beads, glass beads, agarose
beads, and
combinations thereof. In some embodiments, the beads are beads approximately 2
to 100 gm
in diameter, or 10 to 80 gm in diameter, or 20 to 40 gm in diameter. In some
embodiments,
the beads can be provided in solution. In some embodiments, the beads can be
immobilized
on a solid support. In some embodiments, the beads can be provided both in
solution and in
an immobilized state on a solid support.
[0122] In some preferred embodiment, the solid support can include
streptavidin.
In some embodiments, the solid support is, or can include, streptavidin-coated
beads. In some
embodiments, the solid support is or can include streptavidin-coated magnetic
beads. In some
embodiments where the solid phase support includes streptavidin, the nucleic
acids
molecules can be biotinylated to facilitate binding of the nucleic acids to
the solid support.
[0123] In some embodiments of the methods disclosed herein, the
solid phase
support includes a surface of a reaction site. For example, a glass side can
be treated to have
nucleic acids bound at specific locations on the solid support, e.g., as a
high density array. In
some embodiments, the reaction site can include a bottom portion of an inner
surface of a
well, groove, flow cell, reaction chamber or channel. In some embodiments, the
site can
include a reaction chamber or well. In some embodiments, the reaction site can
be part of an
-47-
CA 3016221 2019-11-29
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
array of similar or identical sites. For example, the solid support can be the
bottom and/or
sides of a well on a microtiter plate.
[0124] In some embodiments of the methods disclosed herein, the beads or
reactions sites are in an aqueous phase such as, for example, in aqueous
reaction buffer. In
some embodiments, the beads or reactions sites are in continuous aqueous
phase. In some
embodiments, the amplifying is performed without sealing the beads or reaction
sites from
each other. For example, the beads or reaction sites can remain in fluid
communication with
each other during the amplifying. Accordingly, in some embodiments of the
methods
disclosed herein, the beads or reaction sites are in fluid communication with
each other
during the amplifying. In other embodiments, the solid support or beads can be
located in
discrete locations that are separated from each other and not in fluid
communication, e.g.,
wells of a microtiter plate, or beads located in wells of a microtiter plate.
[0125] In some embodiments, the amplification or normalization reaction
mixture
of the methods disclosed herein include one or more solid supports with
amplification or
normalization primers affixed thereon. Amplification or normalization primers
can be
attached to the solid support by methods well known to those of skill in the
art. At least one
of the supports can include one or more instances of a first primer including
a first primer
sequence. In some embodiments, at least one polynucleotide template in the
reaction mixture
(e.g., a member of a nucleic acid sample or nucleic acid library) includes a
first primer
binding sequence. The first primer binding sequence can be fully or
substantially identical, or
fully or substantially complementary, to the first primer sequence. In some
embodiments, at
least one, some or all of the solid supports include a plurality of first
primers that are
identical to each other. In some embodiments, all of the primers on the solid
supports are
identical to each other, or all include an identical first primer sequence. In
a preferred
embodiment, the solid support is a plurality of beads, wherein each bead in
the plurality has a
plurality of identical primers attached.
[0126] In various embodiments of the disclosure, a second amplification
or
normalization primer is provided in solution phase, which can be optionally
exposed to the
immobilized first amplification or normalization primer. In some embodiments,
the amount
of the second amplification or normalization primers in solution phase is
greater than the
amount of the first amplification or normalization primers immobilized on a
solid phase
-48-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
support. By providing an excess amount of the second primer in solution, the
amount of first
primer immobilized on the solid support will determine the amount of
amplification product
produced on the solid support. Consequently the amplification results in a
substantially
constant amount of amplification products per fixed amount of solid phase
support. In some
embodiments, the plurality of second amplification or normalization primers is
provided in
an amount within an order of magnitude of the amount of first amplification or
normalization
primer. In some embodiments, the plurality of second amplification or
normalization primers
is provided in an amount that exceeds the amount of the first amplification or
normalization
primers by at least, or at least about, 100%. In other embodiments, the amount
of the second
primer exceeds the amount of the first primer by at least, or at least about,
150%, at least, or
at least about 200%, at least, or at least about 300%, at least, or at least
about 400%, or at
least, or at least about 1000%, or a range of any two of the proceeding
values, for example
from about 100% to about 1000%.
Output amplified/normalized samples and libraries
[0127] In some embodiments, the amount of amplification products
obtained
from each nucleic acid sample can be substantially uniformly represented in a
pooled nucleic
acid library. In some other embodiments, the amounts of amplification products
obtained
from the input nucleic acid samples can be present in a pooled library in
different,
predetermined concentrations by obtaining the amplification products from
different,
predetermined amounts of solid support or by pooling different amounts of the
amplification
products.
[0128] Accordingly, in some embodiments, the methods disclosed herein
allow
for generating nucleic acid libraries having normalized amounts and which
amounts are
substantially uniform across multiple nucleic acid samples and/or libraries.
In some
embodiments, the nucleic acid amounts in the normalized nucleic acid libraries
vary by less
than 10%, 5%, 3%, 2%, or 1%. In some embodiments, the methods disclosed herein
allow
for generating pooled nucleic acid libraries in which the amount of
constituent nucleic acids
in the resultant pooled nucleic acid libraries are at substantially similar
amounts regardless of
the amount of input DNA samples. In some embodiments, the amounts of
constituent nucleic
acids in the pooled nucleic acid libraries vary by less than 10%, 5%, 3%, 2%,
or 1%.
-49-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
[0129] In some embodiments, the nucleic acid libraries or pooled
libraries
generated by the methods described herein can be suitable for downstream
analytical
applications, including sequencing application utilizing techniques such as
next-generation
sequencing (NGS) and related methodologies such as genotyping-by-sequencing
(GBS).
[0130] In some embodiments, the nucleic acid amplification methods
disclosed
herein can be automated and/or can be performed in a multiplexed format, for
example the
methods can be performed by a droplet actuator or liquid handling robot.
[0131] In some embodiments of the disclosed methods and systems, the
beads as
described herein can be monoclonal, that is, they can include a single
population of
amplification or normalization primers that are identical to each other.
[0132] In some embodiments, the beads can be polyclonal, that is, they
can
include a pool of a plurality of monoclonal beads, wherein the pooled
monoclonal beads
include amplification or normalization primers comprising more than one
capture portion
having sequence similarity to a cognate region of a target nucleic acid. In
some
embodiments, the beads can include individual polyclonal beads, that is, they
can include
amplification or normalization primers comprising more than one capture
portion per bead.
[0133] In some embodiments, at least one solid support includes two or
more
different primers affixed thereto. In some embodiments, the at least one
support can include
at least 2, 3, 4, 5, 6, 7, 8, 9, or 10 different amplification or
normalization primers having
different nucleic acid sequences. In some embodiments, the solid support has a
plurality of
discrete locations, each location having a plurality of primers having the
same sequence. In
some embodiments, the sequences at each of the plurality of locations is the
same, in other
embodiments the sequences at one or more of the plurality of locations differ
from that of
one or more other locations.
[0134] In some embodiments, the nucleic acid amplification methods
disclosed
herein further include a step of separating the amplification products from
the solid support.
Generally, any suitable method for removing nucleic acids from the solid
support can be
used. In some embodiments, the amplification products are separated from the
solid support
by elution. In some embodiments, the amplification products are eluted in a
heated buffer. In
some embodiments where streptavidin is included in solid phase support and the
nucleic
acids are biotinylated to facilitate binding of the nucleic acids to the solid
support, the nucleic
-50-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
acid amplification products can be separated from the solid support via by
heat avidin-biotin
cleavage.
Input nucleic acid samples
[0135] In some embodiments of the methods and systems disclosed herein,
the
input nucleic acid sample includes single-stranded nucleic acid molecules. In
some
embodiments, at least one of the target nucleic acid molecules in the input
nucleic acid
sample is double-stranded, or is rendered at least partly double-stranded
using appropriate
procedures prior to amplification. In some embodiments, the input nucleic acid
sample
includes a mixture of single-stranded nucleic acid molecules and double-
stranded nucleic
acid molecules. In some embodiments, the target nucleic acid molecules can be
linear.
Alternatively, the target nucleic acid molecules can be circular, or include a
combination of
linear and circular regions.
[0136] In some embodiments, the double stranded target nucleic acid
molecules
can include a forward strand. In some embodiments, the double stranded target
nucleic acid
molecules can further include a reverse strand. The forward strand, in some
embodiments of
the disclosed methods, can include a first primer binding site. The reverse
strand, in some
embodiments of the disclosed methods, can include a second primer binding
site.
[0137] In various embodiments of the disclosure, the amount of input
nucleic acid
can be from about 0.01 ng to 100 ng. In some embodiments, the amount of input
nucleic
acid is about 0.1 ng, 0.2 ng, 0.3 ng, 0.4 ng, 0.5 ng, 0.6 ng, 0.7 ng, 0.8 ng,
0.9 ng, 1 ng, 1.1 ng,
1.2 ng, 1.3 ng, 1.5 ng, 2.0 ng, 2.5 ng, 3.0 ng, 3.5 ng, 4.0 ng, 4.5 ng, 5.0
ng, or within a range
defined by any two of the aforementioned values. In some embodiments, the
amount of
input nucleic acid is about 5.5 ng, 6.0 ng, 6.5 ng, 7.0 ng, 7.5 ng, 8.0 ng,
8.5 ng, 9.0 ng, 9.5 ng,
10.0 ng, 11.0 ng, 11.5 ng, 12.0 ng, 12.5 ng, 13.0 ng, 13.5 ng, 14.0 ng, 15.0
ng, 15.5 ng, 16.0
ng, 16.5 ng, 17.0 ng, 18.0 ng, 18.5 ng, 19.0 ng, 19.5 ng, 20.0 ng, or within a
range defined by
any two of the aforementioned values. In some embodiments, the amount of input
nucleic
acid is about 21.0 ng, 22.0 ng, 22.5 ng, 23.0 ng, 23.5 ng, 24.0 ng, 24.5 ng,
25.0 ng, 26.0 ng,
27.0 ng, 28.0 ng, 29.0 ng, 30.0 ng, 32.5 ng, 35.0 ng, 37.5 ng, 40.0 ng, 42.5
ng, 45.0 ng, 47.5
ng, 50.0 ng, 52.5 ng, 55.0 ng, 60.0 ng, 65.0 ng, 70.0 ng, 75.0 ng, 80.0 ng,
85.0 ng, 90.0 ng, or
within a range defined by any two of the aforementioned values. In some
embodiments, the
amount of input nucleic acid is about 0.08, 0.4, 2.0, 10.0, or 50.0 ng.
-51-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
[0138] In some embodiments of the methods disclosed herein, the target
nucleic
acid molecules already include a first and/or second primer binding site prior
to the
amplification step(s). Alternatively, the target nucleic acid molecules do not
originally
include a primer binding site, and the disclosed methods optionally include
attaching or
introducing a primer binding site to the target nucleic acid molecules prior
to the amplifying.
For example, in some embodiments, the disclosed methods can optionally include
ligating or
otherwise introducing (e.g., by primer directed amplification, including PCR)
an adaptor
containing a primer binding site to, or into, the target nucleic acid
molecules. The adapter can
be ligated or otherwise introduced to an end of a linear target nucleic acid
molecule, or
within the body of a linear or circular nucleic acid molecule. In some
embodiments, the
target nucleic acid molecule can be circularized after the adapter is ligated
or introduced. In
some embodiments, a first adapter can be ligated or introduced at a first end
of a linear target
nucleic acid molecule, and a second adaptor can be ligated or introduced at a
second end of
the target nucleic acid molecule.
[0139] In some embodiments, the first amplification or normalization
primers and
the second amplification or normalization primers are complementary to binding
sites with
known nucleotide sequences within the target nucleic acid molecules. In some
embodiments,
the primer binding sites with known nucleotide sequences correspond to the
first ends and
second ends of the target nucleic acid molecules.
[0140] In some embodiments of the nucleic acid amplification methods
disclosed
herein, the first ends and second ends of the target nucleic acid molecules
include universal
primer regions such as, for example, universal sequencing tail adaptors, that
have been added
to the target nucleic acid molecules.
[0141] In some embodiments, at least one of the first and/or second
amplification
or normalization primers further includes an indexing portion. The indexing
portion can be
used to identify the source of the target nucleic acids, e.g., the individual
or biological
sample, such that if amplification products are pooled, the source of the
target nucleic acids
can later be determined. Alternatively, the indexing portion can be added to
the target
nucleic acids prior to the normalization step, for example, when adaptors or
universal primer
sequences are added to the target nucleic acids.
-52-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
[0142] In some embodiments, at least a portion of the first
amplification or
normalization primers further includes a capture portion having sequence
complementarity to
a cognate region of the target nucleic acid molecules in addition to a known
sequences of the
target nucleic acid molecules. For example, if adaptors and/or universal
primers are added to
the nucleic acid molecules of the input sample, the adaptors and/or universal
primers can
constitute the "known sequences," while the "capture portion" is complementary
to a
sequence native to the nucleic acids in the library. In this way, only those
portions of the
library having the capture sequence will be amplified even though the adaptors
and/or
universal primers are present on most or all of nucleic acids of the library.
In a preferred
embodiment, the use of a capture portion permits selective amplification of
nucleic acids in
the nucleic acid sample, e.g., the nucleic acid library. Embodiments using
capture probes are
illustrated in Figures 6-9, described in more detail below.
[0143] In some embodiments, the capture portion is generated by
hybridizing a
capture oligonucleotide to a first amplification or normalization primer
immobilized onto the
solid support and extending the immobilized amplification or normalization
primer to
generate an extended amplification or normalization primer having sequence
complementarity to the capture oligonucleotide.
[0144] Applicant has demonstrated that the method disclosed herein can
be
advantageously applied to a single nucleic acid sample or a plurality of
nucleic acid samples
by using a plurality of solid phase supports. Accordingly, in some embodiments
of methods
disclosed herein, the amplification step is performed on a plurality of
nucleic acid libraries.
In some embodiments, the amplification products from the plurality of nucleic
acid libraries
are combined to form a combined pooled nucleic acid library. In some
embodiments, the
amplification products derived from the plurality of nucleic acid libraries
are combined after
being removed from the solid phase support. In some embodiments, the
amplification
products from the plurality of nucleic acid libraries are combined before
being removed from
the solid phase support. In some embodiments, the plurality of input nucleic
acid samples is
combined before the amplification step. In some embodiments, the amount of
each input
nucleic acid sample is not normalized across the plurality of nucleic acid
samples.
-53-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
[0145] In some embodiments, the plurality of input nucleic acid samples
comprises at least 2,4, 8, 12, 24, 48, 96, 200, 384, 400, 500, 100, 1500, or a
number of input
nucleic acid samples within a range defined by any two of the aforementioned
numbers.
[0146] In some embodiments, the relative representation of each
population of
constituent amplification products can be advantageously adjusted in the
pooled nucleic acid
library. By "advantageously adjusted", as used herein, is meant that the
amount of each
constituent amplification products in the pooled nucleic acid library can be
controlled or
predetermined. In some preferred embodiments, the amount of each constituent
amplification
products is substantially uniformly represented in the pooled nucleic acid
library.
Alternatively, the plurality of constituent amplification products can be
present in the pooled
nucleic acid library in different, predetermined concentrations. This may be
achieved by
assembling different amounts of the solid phase supports with amplification
products
remaining affixed thereon or by pooling different amounts of the plurality of
constituent
amplification products after being recovered from the solid phase support.
Stated differently,
the advantageous adjustment can include selectively adjusting both the
proportional
representation and the population number of constituent amplification products
in the pooled
nucleic acid library. In yet some further embodiments, an advantageous
adjustment may
include subjecting a sample of constituent amplification products to at least
one processing
step in addition to recovering amplification products from the solid phase
support.
Normalization step
[0147] In some embodiments of the methods disclosed herein, nucleic acid
amplification is performed under substantially isothermal conditions. The
optimal
temperature for amplification varies and may for example depend on primer
characteristics,
such as sequence length, melting temperature as described elsewhere herein,
and choice of
polymerase. In some embodiments of the present disclosure, the amplification
temperature is
lower than 60 degrees Celsius, lower than 50 degrees Celsius, lower than 45
degrees Celsius,
or lower than 42, 38, 35, 30, 25, or 20 degrees Celsius. In some preferred
embodiments, the
amplification temperature is 38 degrees Celsius.
[0148] In one aspect, the methods of the disclosure include isothermal
amplification which can be performed by using, for example, kinetic exclusion
amplification
(IKEA), also referred to as exclusion amplification (Ex-Amp). In some
embodiments, the
-54-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
methods of the disclosure use an ExAmp normalization reaction to equalize
sample quantities
and adjust the concentration of amplicon DNA for subsequent sequencing
applications. In
some embodiments, the ExAmp library normalization reaction is an isothermal
amplification
reaction that uses a first normalization primer sequence (e.g., P7 primer
sequence)
immobilized on the capture beads and a second normalization primer sequence
(e.g., P5
primer sequence) in solution as normalization primers for library
normalization. Library
normalization can be performed over a wide range of PCR product (amplicon)
input (e.g.,
over at least two orders of magnitude change of input). The reaction
conditions (e.g.,
incubation time, P7 and/or P5 primer concentration, and reaction components)
can be
selected such that all available primers on the capture beads are extended
into amplicons,
e.g., the reaction is run to saturation.
[0149] Accordingly, an amplified/normalized nucleic acid library of the
present
disclosure can be constructed using a method that includes a step of reacting
an amplification
reagent to produce a plurality of amplification sites that each includes a
substantially clonal
population of amplicons from an individual target nucleic acid that has seeded
the site. In
some embodiments the amplification reaction proceeds until a sufficient number
of
amplicons are generated to fill the capacity of the respective amplification
site. Filling an
already seeded site to capacity in this way inhibits target nucleic acids from
landing and
amplifying at the site thereby producing a clonal population of amplicons at
the site. In some
embodiments, apparent clonality can be achieved even if an amplification site
is not filled to
capacity prior to a second target nucleic acid arriving at the site. Under
some conditions,
amplification of a first target nucleic acid can proceed to a point that a
sufficient number of
copies are made to effectively outcompete or overwhelm production of copies
from a second
target nucleic acid that is transported to the site. For example in an
embodiment that uses a
bridge amplification process on a circular feature (e.g., bead) that is
smaller than 500 nm in
diameter, it has been determined that after 14 cycles of exponential
amplification for a first
target nucleic acid, contamination from a second target nucleic acid at the
same site will
produce an insufficient number of contaminating amplicons to adversely impact
sequencing-
by-synthesis analysis on an Illumina sequencing platform.
[0150] In some embodiments, kinetic exclusion can occur when a process
occurs
at a sufficiently rapid rate to effectively exclude another event or process
from occurring.
-55-
array of similar or identical sites. For example, the solid support can be the
bottom and/or
sides of a well on a microtiter plate.
[0124] In some embodiments of the methods disclosed herein, the
beads or
reactions sites are in an aqueous phase such as, for example, in aqueous
reaction buffer. In
some embodiments, the beads or reactions sites are in continuous aqueous
phase. In some
embodiments, the amplifying is performed without sealing the beads or reaction
sites from
each other. For example, the beads or reaction sites can remain in fluid
communication with
each other during the amplifying. Accordingly, in some embodiments of the
methods
disclosed herein, the beads or reaction sites are in fluid communication with
each other
during the amplifying. In other embodiments, the solid support or beads can be
located in
discrete locations that are separated from each other and not in fluid
communication, e.g.,
wells of a microtiter plate, or beads located in wells of a microtiter plate.
[0125] In some embodiments, the amplification or normalization
reaction mixture
of the methods disclosed herein include one or more solid supports with
amplification or
normalization primers affixed thereon. Amplification or normalization primers
can be
attached to the solid support by methods well known to those of skill in the
art. At least one
of the supports can include one or more instances of a first primer including
a first primer
sequence. In some embodiments, at least one polynucleotide template in the
reaction mixture
(e.g., a member of a nucleic acid sample or nucleic acid library) includes a
first primer
binding sequence. The first primer binding sequence can be fully or
substantially identical, or
fully or substantially complementary, to the first primer sequence. In some.
embodiments, at
least one, some or all of the solid supports include a plurality of first
primers that are
identical to each other. In some embodiments, all of the primers on the solid
supports are
identical to each other, or all include an identical first primer sequence. In
a preferred
embodiment, the solid support is a plurality of beads, wherein each bead in
the plurality has a
plurality of identical primers attached.
[0126] In various embodiments of the disclosure, a second
amplification or
normalization primer is provided in solution phase, which can be optionally
exposed to the
immobilized first amplification or normalization primer. In some embodiments,
the amount
of the second amplification or normalization primers in solution phase is
greater than the
amount of the first amplification or normalization primers immobilized on a
solid phase
-56-
CA 3016221 2019-11-29
Take for example a solution of beads having universal primers where the beads
are
randomly seeded with target nucleic acids in a solution and copies of the
target nucleic acid
are generated in an amplification process to fill each of the beads to
capacity. In accordance
with the kinetic exclusion methods of the present disclosure, the seeding and
amplification
processes can proceed simultaneously under conditions where the amplification
rate exceeds
the seeding rate. As such, the relatively rapid rate at which copies are made
on a particular
bead that has been seeded by a first target nucleic acid will effectively
exclude a second
nucleic acid from seeding that particular bead for amplification. Kinetic
exclusion
amplification methods can be performed as described in detail in the
disclosure of U.S.
Application Pub. No. 2013/0338042.
[0151] Kinetic exclusion can exploit a relatively slow rate for
initiating
amplification (e.g., a slow rate of making a first copy of a target nucleic
acid) vs. a relatively
rapid rate for making subsequent copies of the target nucleic acid (or of the
first copy of the
target nucleic acid). In the example of the previous paragraph, kinetic
exclusion occurs due to
the relatively slow rate of target nucleic acid seeding (e.g., relatively slow
diffusion or
transport) vs. the relatively rapid rate at which amplification occurs to fill
the site (e.g., bead
or other site on a solid substrate (e.g., reaction site or well)) with copies
of the nucleic acid
seed. In another exemplary embodiment, kinetic exclusion can occur due to a
delay in the
formation of a first copy of a target nucleic acid that has seeded a site
(e.g., delayed or slow
activation) vs. the relatively rapid rate at which subsequent copies are made
to fill the site. In
this example, an individual site may have been seeded with several different
target nucleic
acids (e.g., several target nucleic acids can be present at each site prior to
amplification).
However, first copy formation for any given target nucleic acid can be
activated randomly
such that the average rate of first copy formation is relatively slow compared
td the rate at
which subsequent copies are generated. In this case, although an individual
site (e.g., bead)
may have been seeded with several different target nucleic acids, kinetic
exclusion will allow
only one of those target nucleic acids to be amplified. More specifically,
once a first target
nucleic acid has been activated for amplification, the site will rapidly fill
to capacity with its
copies, thereby preventing copies of a second target nucleic acid from being
made at the site.
[0152] An amplification reagent can include further components that
facilitate
amplicon formation and in some cases increase the rate of amplicon formation.
An example
-57-
CA 3016221 2019-11-29
is a recombinase. Recombinase can facilitate amplicon formation by allowing
repeated
invasion/extension. More specifically, recombinase can facilitate invasion of
a target nucleic
acid by the polymerase and extension of a primer by the polymerase using the
target nucleic
acid as a template for amplicon formation. This process can be repeated as a
chain reaction
where amplicons produced from each round of invasion/extension serve as
templates in a
subsequent round. The process can occur more rapidly than standard PCR since a
denaturation cycle (e.g., via heating or chemical denaturation) is not
required. As such,
recombinase-facilitated amplification can be carried out isothermally. It is
generally desirable
to include ATP, or other nucleotides (or in some cases non-hydrolyzable
analogs thereof) in a
recombinase-facilitated amplification reagent to facilitate amplification. A
mixture of
recombinase and single stranded binding (SSB) protein is particularly useful
as SSB can
further facilitate amplification. Exemplary formulations for recombinase-
facilitated
amplification include those sold commercially as TwistAmp kits by TwistDx
(Cambridge,
UK). Useful components of recombinase-facilitated amplification reagent and
reaction
conditions are set forth in US 5,223,414 and US 7,399,590.
[0153] Another example of a component that can be included in an
amplification
reagent to facilitate amplicon formation and in some cases to increase the
rate of amplicon
formation is a helicase. Helicase can facilitate amplicon formation by
allowing a chain
reaction of amplicon formation. The process can occur more rapidly than
standard PCR since
a denaturation cycle (e.g., via heating or chemical denaturation) is not
required. As such,
helicase-facilitated amplification can be carried out isothermally. A mixture
of helicase and
single stranded binding (SSB) protein is particularly useful as SSB can
further facilitate
amplification. Exemplary formulations for helicase-facilitated amplification
include those
sold commercially as IsoAmp kits from Biohelix (Beverly, MA). Further,
examples of useful
formulations that include a helicase protein are described in US 7,399,590 and
US 7,829,284.
101541 In some embodiments of the methods disclosed herein, the
amplification
reagents used in the methods disclosed herein can further include one or more
origin binding
proteins. Without being bound by any particular theory, the inclusion of an
origin binding
-58-
CA 3016221 2019-11-29
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
droplets, including temperature adjustments as needed, may also be performed
on a droplet
actuator.
[0158] In some embodiments, certain protocol steps may be conducted
outside of
a droplet actuator and certain protocol steps may be conducted on a droplet
actuator. For
example, in some embodiments, samples and reagents may be prepared outside the
droplet
actuator and combined and incubated on the droplet actuator. Reagent
preparation (e.g.,
buffers, PCR master mix solutions, and normalization solutions) may also be
prepared using
on-bench protocols prior to loading on a droplet actuator. In another
embodiment reagent
and/or samples may be prepared in reservoirs associated with the droplet
actuator then
flowed to different operations gaps, and/or prepared in the droplet operations
gap.
[0159] Fi2ure 6 illustrates a flow diagram of a non-limiting example of
a method
600 of preparing a targeted amplicon library, for example on a droplet
actuator, for
downstream analytical applications such as, for example, sequencing in
accordance with
some embodiments of the disclosure. Method 600 may include, but is not limited
to, the
following steps.
[0160] At a step 610, a nucleic acid sample such as, for example, a
genomic DNA
sample (e.g., from about 1 ng to about 10 ng) is provided, for example by
loading into a
sample reservoir of a droplet actuator. In some embodiments, a bead-based
protocol
performed on the droplet actuator is used to concentrate and purify a genomic
DNA sample
prior to subsequent sample processing steps. In one example, the bead-based
protocol uses
magnetically responsive SPRI beads (e.g, Solid Phase Reversible Immobilization
beads,
Agencourt AMPureXP available from Beckman Coulter) to concentrate and purify
the
genomic DNA sample. For example, the genomic DNA is immobilized on
magnetically
responsive beads (e.g., SPRI beads) and a magnet and a series of washes are
used to
concentrate and purify the DNA prior to subsequent processing steps.
[0161] At a step 615, target nucleic acid sequences are amplified in a
multiplex
PCR amplification reaction using target-specific primer pairs that flank
regions of interest.
The target-specific primer pairs include, for example, (1) a forward primer
that comprises a
target-specific region and optionally a sequencing-by-synthesis (SBS) primer
sequence, and
(2) a reverse primer that comprises a target-specific sequence and optionally
a universal
sequence. The forward and reverse primers in each primer pair flank a region
of interest in
-59-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
the target nucleic acid molecule. In general, any number of primer pairs can
be used. In
some examples, 200 primer pairs are used in a multiplex amplification format
(e.g., 200-
plex) to target 200 DNA sequences of interest. The number of PCR cycles can
generally be
any number of cycles and can be, for example, from 2 to about 100 cycles,
about 5 to about
60, about 10 to about 40, about 15 to about 30 PCR cycles. In some examples of
the methods
disclosed herein, the number of PCR cycles can be from 4 to about 10 cycles,
about 6 to
about 20, about 8 to about 15, about 10 to about 30 PCR cycles. In some
embodiments, from
4 to 6 PCR cycles can be used to amplify the targeted DNA sequences.
[0162] At a step 620, a second amplification reaction is performed using
a
universal primer pair. The universal primer pair includes a forward primer and
optionally a
reverse primer. In some embodiments, the forward primer includes an SBS
complementary
sequence, optionally a unique index sequence, and a primer sequence, for
example P5 primer
sequence. The reverse primer includes a complementary universal primer
sequence. The
number of PCR cycles can generally be any number of cycles and can be, for
example, from
4 to about 10 cycles, about 6 to about 20, about 8 to about 15, about 10 to
about 30 PCR
cycles. In one example, 14 PCR cycles are used in the second amplification
reaction.
[0163] At a step 625, amplicons are hybridized to capture probes in a
solution-
based hybridization reaction. The capture probe includes, for example, a
target-specific
capture sequence, a primer sequence (e.g., P7 primer sequence), and a biotin
label. The
target-specific capture sequence has sequence complementarity to a sequence in
a targeted
region of interest in the nucleic acid sample. In one example, 200 capture
probes with
different capture sequences are used in a hybridization reaction to target 200
DNA
sequences.
[0164] At a step 630, capture beads such as, for example, magnetically-
responsive streptavidin-coated capture beads (SA capture beads) are added to
the
hybridization reaction for capture of hybridized amplicon\capture probe
duplexes.
Hybridized amplicon\capture probe duplexes and un-hybridized capture probes
are
immobilized on the SA capture beads by formation of a biotin-streptavidin
binding complex.
[0165] At a step 635, the target-specific capture sequence of capture
probes
hybridized to targeted DNA sequences is extended to form an immobilized
complementary
DNA strand.
-60-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
[0166] At a step 640, a quantity of capture probes such as, for example,
P7-biotin
primers are captured onto the SA capture beads with extended amplicon/capture
probe
duplexes thereon. The P7-biotin primers include a P7 primer sequence and a
biotin label. In
this example, the P7-biotin primers are immobilized on the SA capture beads by
formation of
a biotin-streptavidin binding complex. The SA capture beads with extended
amplicon/capture
probe duplexes thereon now include a quantity of immobilized P7-biotin
primers. The P7-
biotin primers are used in subsequent exclusion amplification reaction for
library
normalization at step 645 as described below.
[0167] At a step 645, the nucleic acid sample is normalized using an
exclusion
amplification (ExAmp) reaction. Sample (ExAmp) normalization is performed to
equalize
sample quantities and adjust the concentration of DNA for subsequent
sequencing. In some
embodiments, preparation of the ExAmp reaction solution for library
normalization is
prepared on-actuator. In some embodiments, the ExAmp reaction solution for
library
normalization is prepared on-bench and subsequently loaded into a reagent
dispensing
reservoir of a droplet actuator. The ExAmp reaction solution includes reaction
reagents and
normalization primers (e.g., P5 primer sequences). The ExAmp library
normalization
reaction is an isothermal amplification reaction that uses a first set of
normalization primers
(e.g., P7-biotin primers) immobilized on the SA beads and a second set of
normalization
primers (e.g., P5 primers) in solution as normalization primers for library
normalization. The
reaction conditions (e.g., incubation time, P7 and/or P5 primer concentration,
and reaction
components) can be selected such that all P7-primer sites on the SA beads are
converted,
e.g., the reaction is run to saturation.
[0168] At a step 650, the library amplicons are eluted from the capture
beads for
sequencing. In one example, illustrated in Figure 7B, amplicons 390 are eluted
from SA
beads 385 and denatured by heating at 95 C for 4 minutes.
[0169] Figures 7A and 71B show pictorially the steps of method 600 of
Figure 6.
Namely, a genomic DNA sample (not shown) includes a target nucleic acid
molecule 310.
The target nucleic acid molecule 310 includes a capture region 315. At step
615, target
nucleic acid molecule 310 is amplified in a first enrichment amplification
reaction, which is
optionally carried out in multiplex format using a target-specific primer
pair. The target-
specific primer pair includes, for example, a target-specific forward primer
320 and a target-
-61-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
specific reverse primer 325. Forward primer 320 includes a target-specific
region 330 and
optionally a universal sequence. In some embodiments, the universal sequence
includes a
SBS primer sequence 335 (e.g., SBS3). Reverse primer 325 includes a target-
specific region
340 and optionally a universal primer region 345. Target-specific regions 330
and 340 flank
a region of interest in the target nucleic acid molecule 310. In general, any
number of target-
specific primer pairs can be used. In some examples, 200 target-specific
primer pairs can be
used in a multiplex amplification format (e.g., 200-plex) to target 200 DNA
sequences of
interest. The number of PCR cycles can generally be any number of cycles and
can be, for
example, from 2 to about 100 cycles, about 5 to about 60, about 10 to about
40, about 15 to
about 30 PCR cycles. In some examples of the methods disclosed herein, the
number of PCR
cycles can be from 4 to about 10 cycles, about 6 to about 20, about 8 to about
15, about 10 to
about 30 PCR cycles. In some embodiments, from 4 to 6 PCR cycles can be used
to amplify
the targeted DNA sequences. In some embodiments, 4 PCR cycles can be used to
amplify
the targeted DNA sequences. An amplicon 350 synthesized using forward primer
320 and
reverse primer 325 now includes SBS primer region 335 and universal primer
region 345.
[0170] At step 620, an optional second enrichment amplification reaction
(e.g., 14
PCR cycles) is performed using a universal primer pair. The universal primer
pair includes,
for example, a universal forward primer 355 and a universal reverse primer
345a. In some
embodiments, the universal forward primer 355 includes a region having
sequence
complementarity to the universal sequence of the target-specific forward
primer 320 at step
615. In some embodiments, the universal forward primer 355 includes an SBS
complementary region 335a that is complementary to SBS primer region 335. In
some
embodiments, universal forward primer 355 includes an index region 360, and
optionally a
primer region such as, for example a P5 primer region 365. The universal
reverse primer
345a includes a sequence having sequence complementarity to the universal
primer region
345 of the target-specific reverse primer 325 described at step 615 above. In
general, any
number of universal primer pairs can be used. The number of PCR cycles can
generally be
any number of cycles and can be, for example, from 2 to about 100 cycles,
about 5 to about
60, about 10 to about 40, about 15 to about 30 PCR cycles. In some examples of
the methods
disclosed herein, the number of PCR cycles can be from 4 to about 10 cycles,
about 6 to
about 20, about 8 to about 15, about 10 to about 30 PCR cycles. In some
embodiments, from
-62-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
to 20 PCR cycles can be used to amplify the targeted DNA sequences. In some
embodiments, 14 PCR cycles can be used to amplify the targeted DNA sequences.
Amplicon
350 now includes P5 primer region 365, index region 360, SBS primer region 335
and
universal primer region 345.
[0171] At step 625, amplicon 350 is hybridized to a capture probe 370 in
a
solution-based hybridization reaction. In some embodiments, the capture probe
370 includes
a capture complementary region 315a that is complementary to capture region
315 in
amplicon 350. In some embodiments, the capture probe 370 further includes a
primer region
such as, for example a P7 primer region 375, and optionally a labeling reagent
such as, for
example, a biotin label 380.
[0172] At step 630, a quantity of capture beads such as, for example, SA
capture
beads 385 are added to the hybridization reaction for capture of hybridized
amplicon
350 \capture probe 370 duplexes and optionally unhybridized capture probes
370. Hybridized
amplicon\capture probe duplexes and un-hybridized capture probes are
immobilized on the
SA capture beads by formation of a biotin-streptavidin binding complex.
[0173] At step 635, the capture complementary region 315a of capture
probe 370
is extended to form an immobilized DNA strand 390. DNA strand 390 immobilized
on
capture bead 385 now includes biotin label 380, P7 primer region 375, SBS
primer region
335, index region 360, and 135 primer region 365.
[0174] At optional step 640, a quantity of modified capture probe 395,
which is
essentially the capture probe 370 devoid of the capture region the capture
region 315, are
added to the SA capture bead reaction. In this exemplary embodiment, the
modified capture
probe 395 includes P7 primer region 375 attached to biotin label 380. Modified
capture probe
395 (e.g., P7-biotin primers) are immobilized on SA capture beads 385 by
formation of a
biotin-streptavidin binding complex. SA capture beads 385 with amplicon
390/capture probe
370 duplexes thereon now include a quantity of immobilized P7-biotin primers
395.
[0175] At step 645, a normalizing amplification is carried out, for
example, under
isothermal amplification procedure such as, for example, in an ExAmp reaction
using
immobilized P7-biotin primer 395 and P5 primers in the reaction solution (not
shown) as
normalization primers. The reaction conditions (e.g., incubation time, P7
and/or P5 primer
concentration, and reaction components) can be selected such that all P7-
primer sites on the
-63-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
SA beads are converted, e.g, the reaction is run to saturation. The amount of
P7 primers
immobilized on the solid substrate is used to control the amount of
amplification product,
thereby normalizes the amounts of end products across multiple nucleic acid
samples or
libraries.
[0176] At optional step 650, amplicons 390 are eluted from SA beads 385
for
sequencing. In one example, amplicons 390 are eluted from SA beads 385 and
denatured by
heating at 95 C for 4 minutes.
[0177] In addition or alternatively, in some embodiments of the methods
according to this and other aspects of the disclosure, the hybridization of
capture probes 370
with amplicon 350 can be carried out in solid phase, e.g., on capture beads.
As illustrated in
the flow diagrams shown at Figures 8 and 9A-9B, at step 830, a quantity of
capture beads
385 with capture probes 370 affixed thereon are added to the hybridization
reaction for
capture of amplicons 350. The hybridization step 825 is then carried out,
wherein the capture
probes immobilized on the capture beads are hybridized with the amplicons 350
to form
amplicon 350\capture probe 370 duplexes that are immobilized on the solid
phase, e.g.,
capture beads. The remaining steps 830, 835, 840, 845, and 850 are carried out
similarly to
the corresponding steps 630, 635, 640, 645, and 650 of the alternative methods
described in
Figures 6 and 7A-7B.
III. DROPLET ACTUATOR CONFIGURED FOR GENOMIC DNA INPUT TO TARGETED AMPLICON
SAMPLE OUTPUT
[0178] In one aspect, some embodiments disclosed herein relate to
certain droplet
actuated molecular techniques. In some embodiments, a droplet actuator may,
for example,
include a bottom substrate and a top substrate that are separated by a droplet
operations gap.
The droplet operations gap contains filler fluid, such as silicone oil or
hexadecane filler fluid.
The bottom substrate can be, for example, a printed circuit board (PCB) that
may include an
arrangement of droplet operations electrodes (e.g., electrowetting
electrodes). The top
substrate can be, for example, a plastic or glass substrate. The top substrate
may include a
ground reference plane or electrode.
[0179] In one example, a droplet actuator may be adapted for use in
conducting a
multiplexed targeted amplicon sample preparation protocol. For example, the
composition of
the filler fluid may be selected for performance with reagents used in a
particular protocol.
-64-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
Droplet transport voltage (e.g., electrowetting voltage) and frequency may
also be selected
for performance with reagents used in a particular protocol. Design parameters
may be
varied, e.g., number and placement of on-actuator reservoirs, number of
independent
electrode connections, size (volume) of different reservoirs, placement of
magnets/bead
washing zones, electrode size, inter-electrode pitch, and height (between top
and bottom
substrates) of the droplet operations gap.
[0180] The droplet actuator may be designed to fit onto an instrument
deck that
houses additional-droplet actuator features, such as one or more magnets for
immobilization
of magnetically responsive beads and one or more heater assemblies for
controlling the
temperature within certain reaction and/or washing zones.
[0181] Manipulation of droplets on a droplet actuator includes droplet
operations
such as dispensing, transporting, merging, incubating, splitting, and mixing.
The size of a
droplet can vary depending on the droplet operation used in a protocol step.
In one example,
a unit sized droplet, e.g., "droplet unit" (DU), is about 0.34 III, and can be
described as a lx
droplet. Typical protocol reactions use a range of droplet sizes from about 1
DU (e.g., a 1 x
droplet) to about a 6 DU droplet (e.g., a 6x droplet).
[0182] Figure 1 illustrates a top view of an example of an electrode
arrangement
100 of a droplet actuator suitable for use in conducting a multiplexed
targeted amplicon
library preparation protocol in accordance with some exemplary embodiments of
the
disclosure. Electrode arrangement 100 is configured for multiplexed processing
of multiple
genomic DNA samples for construction of one or more targeted amplicon
libraries. Droplet
operations are conducted atop electrode arrangement 100 on a droplet
operations surface. In
this example, electrode arrangement 100 is configured for processing up to 8
different
samples in parallel in dedicated reaction regions for construction of 8
different targeted
amplicon sequencing libraries.
[0183] Electrode arrangement 100 includes 8 sample reservoir electrodes
110
(hereafter called sample reservoir electrodes 110a through 110h) for inputting
and dispensing
sample solutions (e.g., a genomic DNA sample). Electrode arrangement 100 also
includes 8
PCR/biochemical reaction zones 115 (hereafter called PCR/biochemical reaction
zones 115a
through 115h) for performing certain processing steps for construction of each
targeted
amplicon library. Each of the PCR/biochemical reaction zones 115 includes a
cluster or
-65-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
arrangement of multiple electrodes for conducting droplet operations. The
processing steps
include, for example, PCR amplification, capture probe hybridization, bead
capture, primer
extension, library normalization, and library elution. Electrode arrangement
100 also
includes 8 index reservoir electrodes 120 (hereafter called index reservoir
electrodes 120a
through 120h) for dispensing 8 unique indexing oligonucleotide solutions for
indexing each
targeted amplicon library. Electrode arrangement 100 also includes 14 reagent
reservoir
electrodes 125 (hereafter called reagent reservoir electrodes 125a through
125n) configured
for dispensing different reagent liquids (e.g., wash buffers, PCR master mix
solutions,
magnetically responsive capture beads, primer extension reagents, library
normalization
reagents, and elution/denaturation buffer solutions). In some implementations,
reservoir
electrodes 110 and index reservoir electrodes 120 are used as waste reservoir.
In some
implementations, reagent reservoir electrodes 125 are not used for waste.
[0184] Generally, the sample reservoir electrodes 110, PCR/biochemical
zones
115, index reservoir electrodes 120, and reagent reservoir electrodes 125 are
interconnected
through an arrangement, such as a path or array, of droplet operations
electrodes 130.
[0185] In electrode arrangement 100, sample reservoir electrode 110a
corresponds to PCR/biochemical reaction zone 115a, which corresponds to index
reservoir
electrode 120a; sample reservoir electrode 110b corresponds to PCR/biochemical
reaction
zone 115b, which corresponds to index reservoir electrode 120b; and so on
through sample
reservoir electrode 110h corresponding to PCR/biochemical reaction zone 115h,
which
corresponds to index reservoir electrode 120h. The 8 arrangements of
corresponding sample
reservoir electrodes 110, PCR/biochemical reaction zones 115, and index
reservoir electrodes
120 form 8 dedicated reaction lanes 135 (hereafter called reaction lanes 135a
through 135h)
for processing each sample input. The use of dedicated lanes for sample
droplets minimizes
cross-contamination among different genomic DNAs.
[0186] One or more magnets (not shown) may be located in proximity to
certain
droplet operations electrodes 130 for retaining a quantity of magnetically
responsive beads.
The magnet may, for example, be a permanent magnet or an electromagnet. In one
example,
the magnet may be a movable magnet that may be moved into proximity of and
away from
its respective droplet operations electrode 130. Each magnet is positioned in
a manner which
ensures spatial immobilization of nucleic acid-attached beads during certain
processing steps
-66-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
(e.g., bead washing, library capture, enzymatic reactions, and bead removal
following
elution/denaturation of processed genomic DNA).
[0187] Electrode arrangement 100 may include one or more temperature
control
zones 140. In one example, three temperature control zones 140 may be used
(e.g.,
temperature control zones 140a, 140b, and 140c). Temperature control elements
(not shown)
control the temperature of filler fluid (not shown) in vicinity of temperature
control zones
140. Each temperature control zone 140 may be independently controlled to a
certain
temperature(s) sufficient for the different processing steps in a library
construction protocol.
For example, temperature control zone 140a may be heated to about 98 C, which
is a
temperature sufficient for denaturation of DNA, while temperature control zone
140b may be
heated to from about 60 C to about 72 C, which is a temperature range
suitable for
annealing and extension reactions. While three temperature control zones 140
are shown,
any number of temperature control zones 140 may be associated with electrode
arrangement
100.
[0188] Electrode arrangement 100 is an example of an electrode
arrangement on a
droplet actuator that can be used to facilitate the methods in accordance with
some
embodiments of the disclosure; namely, to facilitate automated liquid handling
for
amplification and selection of targeted regions of genomic DNA for processing
into amplicon
libraries for sequencing, as described herein.
IV. GENERAL DIGITAL MICROFLUDIC PROTOCOL FOR PREPARATION OF A TARGETED
AlvIPLICON LIBRARY
[0189] In another example, methods 200, 400, 600, and 800 of Figures 2,
4, 6,
and 8, respectively, can be described as step-by-step, droplet-based protocols
for preparation
of a targeted amplicon library. An example of a droplet-based protocol for
preparation of a
targeted amplicon library includes, but is not limited to, the following
droplet movements.
[0190] In another example of step 610 of method 600 of Figure 6, a
genomic
DNA sample comprising a quantity of magnetically responsive SPRI beads is
loaded into a
sample reservoir of a droplet actuator. In one example, the genomic DNA sample
includes
about I ng of genomic DNA, a DNA binding buffer solution, and a quantity of
magnetically
responsive SPRI beads (e.g., from about 20 pi, to about 50 L). The genomic
DNA sample
with SPRI beads therein is incubated for a period of time sufficient for
binding of the DNA
-67-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
onto the beads. Sample solution with SPRI beads is transported using droplet
operations to an
area of sample reservoir within the magnetic field of a magnet. In some
examples, the entire
volume of the sample (20-50 1) is used rather than a few DUs are used. The
magnetically
responsive SPRI beads with genomic DNA thereon are immobilized by the magnetic
field of
the magnet. In one example, the magnet is a moveable magnet that can be moved
into
proximity of the certain droplet operations electrode and away from the
certain droplet
operations electrode. A supernatant is pulled back from the immobilized beads
and returned
using droplet operations to the sample reservoir for disposal. In some
examples, the entire
supernatant volume (20-50 1) is pulled back. A 2x bead wash buffer droplet is
dispensed
from a reagent reservoir and transported using droplet operations to the
immobilized SPRI
beads with genomic DNA thereon to form a 2x wash buffer/SPRI bead droplet. The
SPRI
beads with genomic DNA thereon are re-suspended and washed using a bead
washing
protocol. The magnet is positioned in proximity of the 2x wash buffer/ SPRI
bead droplet
such that the magnetically responsive SPRI beads are immobilized by the
magnetic field of
the magnet. A 2x supernatant droplet is pulled off and transported using
droplet operations to
the sample reservoir for disposal. A lx elution buffer droplet is transported
from a reagent
reservoir to the immobilized magnetically responsive SPRI beads with genomic
DNA
thereon to form a Ix elution buffer/SPRI bead droplet. The 1 x elution
buffer/SPRI bead
droplet is incubated at about 55 C for about 2 minutes to elute the DNA from
the SPRI
beads. The magnetically responsive SPRI beads are then immobilized by the
magnetic field
of the magnet and a 1 x DNA sample droplet is transported using droplet
operations away
from the immobilized magnetically responsive beads to a dedicated
PCR/biochemical
reaction zone. A 1>< bead wash buffer droplet is transported from a reagent
dispensing
reservoir and merged with the immobilized magnetically responsive SPRI beads
(now devoid
of genomic DNA) to form a 1x SPRI bead/wash buffer droplet. The 1x SPRI
bead/wash
buffer droplet is transported using droplet operations to the sample reservoir
for disposal.
[0191] In another example of step 615 of method 600 of Figure 6, a 1 x
PCR
reagent droplet is dispensed from a reagent dispensing reservoir. The 1 x PCR
reagent droplet
includes, for example, buffer, polymerase, and dNTPs. The 1 x PCR reagent
droplet is
combined using droplet operations with the 1x DNA sample droplet to yield a 2x
amplification droplet. A 1x target-specific primer droplet is dispensed from a
reagent
-68-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
dispensing reservoir and combined using droplet operations with the 2x
amplification droplet
to yield a 3x target-specific amplification droplet. In one example, the 1><
target-specific
primer droplet includes forward (e.g., forward primer 320 of Figures 7A-7B)
and reverse
primer (reverse primer 325 of Figures 7A-7B) pairs for a 200-plex
amplification reaction.
PCR cycling (e.g., 4 cycles) is performed in a flow-through format where for
each cycle the
3x target-specific amplification droplet is cyclically transported using
droplet operations
between different temperature control zones on the droplet actuator. In one
example, the
amplification reaction is an initial incubation at 98 C for 2 minutes, then 4
PCR cycles of
the following sequence: 98 C for 20 seconds, 70 C for 20 seconds, 56 C for
60 seconds, 72
C for 75s; followed by a final incubation at 72 C for 60 seconds.
[0192] In another example of step 620 of method 600 of Figure 6, a 2x
index
primer droplet (e.g., forward primer 355 of Figures 7A-7B) and a 1>< PCR
reagent droplet
are dispensed from reagent dispensing reservoirs and combined using droplet
operations to
yield a 3x universal primer droplet. The 1x PCR reagent droplet includes, for
example,
buffer, polymerase, dNTPs, and a universal reverse primer (e.g., reverse
primer 345a of
Figure 7A. In one example, the universal reverse primer is part of the 2x
index primer
droplet. The 3x universal primer droplet is split using droplet operations to
yield two 1.5x
universal primer droplets. One 1.5x universal primer droplet is combined using
droplet
operations with the 3x target-specific amplification droplet to yield a 4.5x
universal
amplification droplet. The second 1.5x universal primer droplet is transported
using droplet
operations to a waste collection reservoir. In one example, the waste
collection reservoir is
the sample port used as waste at this point of the workflow. PCR cycling
(e.g., 14 cycles) is
performed in a flow-through format where for each cycle the 4.5x universal
amplification
droplet is cyclically transported using droplet operations between different
temperature
control zones on the droplet actuator. In one example, the amplification
reaction is an initial
incubation at 98 C for 30 seconds, then 14 PCR cycles of the following
sequence: 98 C for
20 seconds, 70 C for 20 seconds, 60 C for 60 seconds, 72 C for 60s;
followed by a final
incubation at 72 C for 120 seconds.
[0193] In another example of step 625 of method 600 of Figure 6, a 1 x
capture
probes droplet (e.g., a plurality of different capture probe 370 of Figures 7A-
7B) and two
capture buffer droplets (e.g., a 2x and a 1 x capture buffer droplet
comprising 20x SSC) are
-69-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
dispensed from reagent dispensing reservoirs and combined using droplet
operations to yield
a 4x capture probes droplet. The 4x capture probes droplet is split using
droplet operations
into two 2x capture probes droplets. One 2x capture probes droplet is combined
using droplet
operations with the 4.5x universal amplification reaction droplet to yield a
6.5x
amplicon/capture probe droplet. The second 2x capture probes droplet is
transported using
droplet operations to a waste collection reservoir. In one example, capture
probe
hybridization is performed by incubating the amplicon/capture probe
hybridization droplet at
98 C for 3 minutes, then 75 C for 30 seconds, then 60 C for 10 minutes, and
then 40 C
for 5 minutes.
[0194] In another example of step 630 of method 600 of Figure 6,
streptavidin
(SA)-coated magnetically responsive beads are prepared for capture of the
hybridized
amplicon/probe duplexes. For example, a 1 x SA bead droplet is dispensed from
a reagent
dispensing reservoir and transported using droplet operations to a certain
droplet operations
electrode within the magnetic field of a magnet. The magnetically responsive
beads within
the 1x SA bead droplet are immobilized by the magnetic field of the magnet and
a 1x
supernatant droplet is split off and transported using droplet operations to a
waste reservoir.
A 2x PR2 wash buffer droplet is dispensed from a reagent reservoir and
transported using
droplet operations to the immobilized SA beads to form a 2x SA bead PR2 wash
droplet. The
beads are re-suspended and washed at room temperature for about 1 minute using
a bead-
wash protocol. A magnet is positioned in proximity of the 2x SA bead PR2 wash
droplet
such that the magnetically responsive SA beads are immobilized by the magnetic
field of the
magnet. A 2x supernatant droplet is pulled off and transported using droplet
operations to a
waste reservoir for disposal. The bead-washing protocol is repeated once using
a 2x HT1
wash buffer droplet. At the end of the bead-washing protocol, a lx HT1
resuspension droplet
is dispensed and transported using droplet operations to the immobilized SA
beads and the
beads are re-suspended to form a 1x washed SA bead droplet. The lx washed SA
bead
droplet is transported using droplet operations and merged with the 6.5x
amplicon/capture
probe droplet to yield a 7.5x library capture droplet. The 7.5x library
capture droplet is
incubated at room temperature for about 6 min for capture of amplicon/capture
probe
duplexes onto the SA beads via formation of a biotin-streptavidin binding
complex. At the
end of the incubation period, the magnetically responsive SA beads with
amplicon/capture
-70-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
probe duplexes thereon are immobilized by the magnetic field of a magnet and a
7.5x
supernatant droplet is transported using droplet operations away from the
magnetic field to a
waste collection reservoir. A 2x PR2 wash buffer droplet is dispensed from a
reagent
reservoir and transported using droplet operations to the immobilized SA beads
with
amplicon/capture probe duplexes thereon to form a 2x bead/amplicon wash
droplet. The
beads are re-suspended and washed at room temperature for about 1 minute using
a bead-
wash protocol. At the end of the incubation period, the magnetically
responsive SA beads
with amplicon/capture probe duplexes thereon are immobilized by the magnetic
field of a
magnet and a 2x supernatant droplet is transported using droplet operations
away from the
magnetic field to a waste collection reservoir.
[0195] In another example of step 635 of method 600 of Figure 6, two 2x
extension buffer droplets are dispensed from a reagent dispensing reservoir
and transported
using droplet operations to the immobilized SA beads with amplicon/capture
probe duplexes
thereon to form a 4x extension reaction droplet. The extension buffer
includes, for example,
buffer, polymerase, and dNTPs for synthesis of a complementary DNA strand. The
4x
extension reaction droplet is incubated at about 60 C for about 5 minutes to
for extension of
the target-specific capture sequence of capture probes hybridized to targeted
DNA sequences.
At the end of the incubation period, the magnetically responsive SA beads with
extended
amplicon/capture probe duplexes thereon are immobilized by the magnetic field
of a magnet
and a 4x supernatant droplet is transported using droplet operations away from
the magnetic
field to a waste collection reservoir. A 2x PR2 wash buffer droplet is
dispensed from a
reagent reservoir and transported using droplet operations to the immobilized
SA beads with
extended amplicon/capture probe duplexes thereon to form a 2x bead/amplicon
wash droplet.
The beads are re-suspended and washed at room temperature for about 1 minute
using a
bead-wash protocol. At the end of the incubation period, the magnetically
responsive SA
beads with extended amplicon/capture probe duplexes thereon are immobilized by
the
magnetic field of a magnet and a 2x supernatant droplet is transported using
droplet
operations away from the magnetic field to a waste collection reservoir.
[0196] In another example of step 640 of method 600 of Figure 6, a 2x P7-
biotin
primer droplet is dispensed from a reagent dispensing reservoir and
transported using droplet
operations to the immobilized SA beads with extended amplicon/capture probe
duplexes
-71-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
thereon to form a 2>< library normalization droplet. The SA beads with
extended
amplicon/capture probe duplexes thereon are re-suspended and incubated at room
temperature for about 6 minutes for capture of P7-biotin primers onto the SA
capture beads.
At the end of the incubation period, the magnetically responsive SA beads with
extended
amplicon/capture probe duplexes and P7-biotin primers thereon are immobilized
by the
magnetic field of a magnet and a 2x supernatant droplet is transported using
droplet
operations away from the magnetic field to a waste collection reservoir.
[0197] In another example of step 645 of method 600 of Figure 6, the
library is
normalized using an exclusion amplification (ExAmp) reaction. To prepare an
ExAmp
reaction solution, two 2x ExAmpl reagent droplets, one 1 x ExAmp2 reagent
droplet, two 2x
ExAmp3-P5 reagent droplets, and one ExAmp3-P5 reagent droplet are dispensed
from
reagent dispensing reservoirs and combined using droplet operations to yield a
10x ExAmp
reaction solution droplet. The 10x ExAmp reaction solution droplet is split
using droplet
operations into five 2x ExAmp reaction solution droplets. Three 2x ExAmp
reaction solution
droplets are transported using droplet operations to a waste collection
reservoir. Two 2x
ExAmp reaction solution droplet are transported using droplet operations to
the immobilized
SA beads with extended amplicon/capture probe duplexes and P7-biotin primers
thereon to
form a 4x normalization reaction droplet. The 4x normalization reaction
droplet is incubated
at 38 C for about 20 minutes. At the end of the incubation period, the
magnetically
responsive SA beads with normalized amplicon library thereon are immobilized
by the
magnetic field of a magnet and a 4x supernatant droplet is transported using
droplet
operations away from the magnetic field to a waste collection reservoir.
[0198] In another example of step 650 of method 600 of Figure 6, two 2x
elution
buffer droplets (e.g., TE buffer droplets) are dispensed from a reagent
dispensing reservoir
and transported using droplet operations to the immobilized SA beads with
library amplicons
thereon to form a 4x library elution droplet. The 4x library elution droplet
is incubated at
about 95 C for about 4 for elution and denaturation of amplicons from the SA
beads. At the
end of the incubation period, the SA beads are immobilized by the magnetic
field of the
magnet and a 4x amplicon library droplet is transported using droplet
operations away from
the immobilized magnetically responsive SA beads for collection and subsequent
sequencing.
-72-
[0199] [0200] No admission is made that any reference cited herein
constitutes prior art. The discussion of the references states what their
authors assert, and the
applicant reserves the right to challenge the accuracy and pertinence of the
cited documents.
It will be clearly understood that, although a number of information sources,
including
scientific journal articles, patent documents, and textbooks, are referred to
herein, this
reference does not constitute an admission that any of these documents forms
part of the
common general knowledge in the art.
[0201] The discussion of the general methods given herein is intended for
illustrative purposes only. Other alternative methods and alternatives will be
apparent to
those of skill in the art upon review of this disclosure, and are to be
included within the spirit
and purview of this application.
EXAMPLES
[0202] Additional embodiments are disclosed in further detail in the
following
examples, which are not in any way intended to limit the scope of this
disclosure or the
claims.
EXAMPLE 1
Reaction Efficiency and Library Uniformity
[0203] To evaluate the reaction efficiency and uniformity of
amplicons prepared
using method 600 of Figure 6, three different targeted amplicon libraries were
prepared. A
first targeted amplicon library was prepared using an on-bench amplification
protocol,
wherein target-specific primer pairs and a universal primer pair are combined
in a single PCR
reaction (e.g., a one-stage reaction). A second targeted amplicon library was
prepared on a
droplet actuator using a one-stage digital fluidic protocol, wherein target-
specific primer
pairs and a universal primer pair are combined in a single PCR reaction. A
third targeted
amplicon library was prepared on a droplet actuator using the two-stage
amplification (e.g.,
steps 615 and 620) of method 600 of Figure 6. For each library, amplicons were
prepared
using 1 ng of genomic DNA and a 161-plex, target-specific primer pool.
-73-
CA 3016221 2019-11-29
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
[0204] Figure 10 shows a plot 1000 of the fragment size distributions in
three
targeted amplicon libraries prepared using three different protocols in
accordance with some
embodiments of the disclosure. Namely, one targeted amplicon library prepared
using a one-
stage on-bench amplification reaction, another targeted amplicon library
prepared using a
one-stage on-actuator amplification reaction, and yet another targeted
amplicon library
prepared using the two-stage on-actuator amplification reaction of method 600
of Figure 6.
Plot 1000 shows (1) a line 1010 of the fragment size distribution in the
amplicon library
prepared on-bench using a one-stage PCR reaction, (2) a line 1015 of the
fragment size
distribution in the amplicon library prepared on-actuator using a one-stage
PCR reaction, and
(3) a line 1020 of the fragment size distribution in the amplicon library
prepared on-actuator
using the two-stage amplification method 600 of Figure 6. The data presented
in Figure 10
show that in all three amplicon libraries (lines 1010, 1015, and 1020) there
was a bimodal
distribution of desirable PCR products (indicated by arrows) and a peak of
unwanted by-
product (e.g., primer dimers). In the on-bench amplicon library prepared using
a one-stage
PCR reaction (line 1010), there was a substantial peak of primer dimers (a
reaction by-
product) and relatively low yield of desirable product. In the amplicon
library prepared using
an on-actuator one-stage PCR reaction (line 1015), there was a substantial
reduction in the
amount of primer dimers and an increase in the amount of desirable PCR
product; that is the
ratio of by-product to product is shifted toward desirable product and the
formation of by-
product (e.g., primer dimers) is minimized. Finally, in the amplicon library
prepared using
the two-stage on-actuator amplification reaction (line 1020) of method 600 of
Figure 6,
formation of primer dimers was essentially eliminated and the amount of
desirable PCR
product was further increased. In addition, the two-stage on-actuator
amplification reaction
generally offers faster cycling in second stage (which saves time) and more
modular system
(which is better for development).
[0205] Figures 11A and 11B respectively show a bar graph 1100
illustrating
PCR efficiency per cycle and a bar graph 1110 illustrating the uniformity of
amplicons in
each library of Figure 10. Referring now to Figure 11A, the data show that the
efficiency of
the PCR reaction performed on-actuator (about 92% per PCR cycle) is
substantially
improved using the two-stage amplification reaction of method 600 of Figure 6
("DF 2
stage') compared to the efficiency (about 85% per PCR cycle) of the one-stage
amplification
-74-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
reaction ("DF 1 stage") performed on a droplet actuator. Regarding the two
samples labelled
"Digitized 1 stage" and "Digitized 2 stage", the term "digitized" refers to a
type of
experiment where reactions were run on bench but under conditions "simulating"
DF. These
conditions were designed to match different DF aspects as closely as possible
but using
regular lab equipment. In specific experiments shown in FIG. 11A as well as
FIG. 11B
described below, the PCR reactions were performed with the same reagent ratios
as in the
corresponding DF reactions, e.g. "DF 1 stage" and "DF 2 stage" (those
digitized conditions
were different than "Original bench" conditions) and under DF oil to simulate
DF conditions.
However, these digitized reactions were performed in regular thermocycler.
[0206] Referring now to Figure 11B, the data show that distribution (or
variability) of amplicons (about 89.5%) within the library generated using the
two-stage
amplification reaction of method 600 of Figure 6 ("DF 2 stage") is
substantially improved
compared to the uniformity of amplicons (about 83%) within the library
generated using the
one-stage amplification reaction ("DF 1 stage") performed on a droplet
actuator. Uniformity
is defined as the distribution/variability of amplicons within the library. A
higher uniformity
means a more even distribution of amplicons in the library and provides for
more efficient
library coverage in sequencing.
[0207] Table 1 below shows a summary of an example of changes that were
made
to adapt an on-bench targeted amplification protocol to a digital fluidic
format.
TABLE 1
Comparison of on-bench and digital fluidic targeted amplification protocols
Changes On-bench Digital fluidic Observed Effect
Yield and uniformity
Protocol One-stage PCR Two-stage PCR
improvements
Saving reagents and improving
Reaction volume 50 !IL 2 pi
performance
gDNA concentration 0.02 ng/tit 0.5 ng/tiL Yield improvement
Yield and robustness
Phusion polymerase 0.04 U411_, 0.12 U/ 1_,
improvements
Uniformity and yield
Target-specific primers 10 nM each 20 nM each
improvements
Universal reverse primer 0.2 p.M 1.5 tiM Yield
improvements
Annealing 60 C 56 C Uniformity improvements
Thermal profile 0.2 C/s cool <0.4 C/s cool Slower cycling
-75-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
EXAMPLE 2
Solution-based Hybridization and Library Uniformity
[0208] This Example summarizes the results of experiments performed to
evaluate the effect of a solution-based hybridization reaction (step 625 of
method 600 of
Figure 6) on uniformity. To this end, three different libraries were prepared.
One library,
referred to here as "bead-based hybridization library", was prepared on-bench
using
traditional procedure where library capture probes are first immobilized on
streptavidin
coated magnetic beads through biotin streptavidin interaction and PCR amplicon
is then
hybridized to capture probes. In another library, referred to here as
"Solution-based
hybridization library and produced on-bench", PCR amplicon was first
hybridized in solution
to biotinylated capture probes and then immobilized through biotin
streptavidin interaction
on streptavidin coated magnetic beads. Finally, a third library referred to
here as "Solution-
based hybridization DF library" was prepared using the same sequence of steps
as Solution-
based hybridization library but was performed on-actuator.
[0209] Table 2 below shows the uniformity of the libraries prepared by
these
three different protocols. In some experiments, a fourth library was prepared
based on a
Bead-based hybridization performed on-actuator (DF). It was observed that the
uniformity of
Solution-based hybridization library was substantially improved over
traditional Bead-based
hybridization library. Uniformity of Solution-based hybridization in DF
library prepared on-
actuator was found to be improved even further.
TABLE 2
Comparison of bead- and solution-based hybridization reactions
Condition Uniformity
Bead-based hybridization 82.5%
Solution-based hybridization 87.5%
Solution-based hybridization in DF 90.0%
EXAMPLE 3
Mathematical Model for Predicting Library Output
[0210] This Example summarizes experimental results illustrating that
the
flexibility and programmability of a droplet actuator device provides for fine
control over the
various biochemical reactions performed during construction of a targeted
amplicon library.
-76-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
Because of the precise control of the biochemical reactions performed on a
droplet actuator,
mathematical models can be used to predict certain process outcomes. For
example, product
yield of amplicon hybridization and capture probes extension can be predicted
from PCR
product input (amplicon input) and capture probe input used in the
hybridization and
extension steps of method 600 of Figure 6 using the following equation:
[0211] Log(yield[pM]) = ¨0.703 + 0.846*log(probes conc.[pM each] +
0.741*log(input conc. [pM each])
[0212] Figure 12A shows a three dimensional plot 1200 of the
hybridization and
extension product yield. Figure 12B shows a plot 1210 of the predicted yield
vs the actual
yield based on PCR product and capture probe input. Referring now to Figure
12A, the data
show dependency of hybridization and extension product yield on capture probes
and PCR
amplicon input concentration. Referring now to Figure 12B, the plot shows
correlation
between actual yield (Y-axis) and yield predicted by mathematical equation
described above.
Good correlation between the two yields as shown in Figure 12B is
representative of good
quality of the mathematical model and indicative of predictable and
reproducible nature of
the biochemical processes involved.
EXAMPLE 4
Library (ExAmp) Normalization
[0213] This Example summarizes experimental results illustrating that
the process
of library normalization (step 645 of method 600 of Figure 6) can be performed
over a wide
range of PCR product (amplicon) input by using a suitable amplification
procedure such as
kinetic exclusion amplification (KEA), also referred to as exclusion
amplification (Ex-Amp).
In this example, an ExAmp normalization reaction was used to substantially
equalize sample
quantities and adjust the concentration of PCR product (amplicon) input for
subsequent
sequencing applications.
[0214] Figure 13 shows a plot 1300 of the relative library output as a
function of
PCR product input in libraries prepared using method 600 of Figure 6. The data
show that
the change in library output is relatively small compared to the change in PCR
product input,
e.g., there is about a 2-fold change in library output over a broad range of
PCR product input
concentrations (e.g., about a 128-fold change in PCR product input). The data
also show that
even beyond the 128-fold change in PCR product input that decrease in library
output is
-77-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
relatively small relative to the change in PCR product input (e.g., about a 10-
fold decrease in
library output at about a 100,000-fold dilution in PCR product input).
[0215] Figures 14A and 14B show a plot 1400 of library uniformity and
fold
amplification as a function of PCR product input (input dilution) and a plot
1410 of
amplification bias, respectively, in libraries prepared using method 600 of
Figure 6.
Referring to plot 1400 of Figure 14A, the data show that as PCR product input
decreases,
there is a slight decrease in library uniformity (bar graph). The data also
show that as the fold
amplification (line) increases, library uniformity decreases. By minimizing
the fold
amplification during library normalization (e.g., targeting from about 10 to
about 20 fold
amplification), the effect on library uniformity can be substantially avoided.
[0216] Referring to plot 1410 of Figure 14B, the data show correlation
of library
members coverage for two different DNA input quantities. Several library
members that
deviate from diagonal correlation are highlighted in dashed ellipse. This is
indicative of
amplification bias where these particular library members are not efficiently
amplified.
Overlaying amplicon GC content information, defined as fraction of amplicon
DNA
sequence consisting of dG or dC bases and shown as dot shade, shows all
highlighted library
members with amplification bias has high amplicon GC content.
[0217] Figures 15A and 15B show a three dimensional plot 1500 of ExAmp
yield and a plot 1510 of predicted vs. actual ExAmp yield as a function of P5
primer and P7-
biotin primer concentrations. Referring to Figure 15A, plot 1500 shows
dependency of
ExAmp amplification product yield on concentration of P5 and biotin-P7
primers. The data
shows that yield can be modulated by changing both P5 and biotin-P7
concentrations.
[0218] Referring to Figure 15B, plot 1510 shows correlation between
actual
ExAmp normalization yield and yield predicted by mathematical equation derived
from the
data on plot 1500. The predicted ExAmp normalization yield can be described as
a function
of P5 and P7-biotin primer concentrations by the following equation:
[0219] log(yield[plVI]) = 3.7974 + 1.0340*log(P5 concUMB + 1.0014*log(P7-
biotin conc. [hiN4]).
-78-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
EXAMPLE 5
Library Elution Efficiency
[0220] Table 3 below shows the effect of buffer composition,
temperature, and
incubation time on the efficiency of library elution from streptavidin beads
(step 650 of
method 600 of Figure 6). The highest elution efficiency was achieved using a
TE + Tween
elution buffer at a temperature of 95 C and an incubation time of 3 minutes.
The data
presented in Table 3 also show that the elution was robust both to times
shorter than 3 min
and temperature lower than 95 C.
TABLE 3
Streptavidin beads biotin elution efficiency
TE with Tween lmin 2min 3min 4min 5min
95C 106% 105% 110% 102% 99%
90.2C 97% 98% 101% 94% 93%
84.4C 91% 93% 95% 90% 89%
79.1C 84% 86% 89% 85% 85%
75.1C 76% 82% 84% 81% 82%
70C 67% 71% 76% 76% 77%
Water lmin 2min 3min 4min 5min
95C 100% 103% 102% 99% 90%
90.2C 97% 101% 101% 97% 90%
84.4C 94% 99% 98% 96% 90%
79.1C 89% 96% 94% 92% 89%
75.1C 80% 91% 90% 90% 86%
70C 70% 77% 82% 84% 81%
HT1 lmin 2min 3min 4min 5min
95C 99% 105% 101% 98% 75%
90.2C 73% 86% 84% 86% 69%
84.4C 38% 53% 54% 57% 47%
79.1C 11% 16% 21% 21% 18%
75.1C 4% 5% 9% 7% 9%
70C 2% 2% 4% 3% 4%
[0221] Figure 16 shows a plot 1600 of correlation between the library-
per-target
coverage obtained from libraries eluted from streptavidin beads using heat
denaturation on a
droplet actuator and the same libraries subsequently denatured by standard
NaOH treatment.
-79-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
Plot 1600 shows good correlation, demonstrating that subsequent NaOH
denaturation was
not needed and that library was fully and efficiently denatured by head on-
actuator.
EXAMPLE 6
Library Uniformity versus Genomic Input
[0222] To evaluate library uniformity as a function of genomic DNA
input, 5
libraries were prepared using method 600 of Figure 6 and 5 different genomic
DNA
samples.
[0223] Figure 17 shows a plot 1700 of library uniformity as a function
of
genomic DNA input for 5 different libraries prepared in accordance with some
embodiments
of the disclosure. The libraries were prepared using 0.2, 1, and 10 ng of 5
different genomic
DNA samples (HD200, HD701, HD729, NA12878, and NA20Mix genomic DNA samples)
and MJ191-plex target-specific primer pool. Typically, genomic DNA sample
input of 1 ng
corresponds to about 300 genomes, 0.2 ng corresponds to about 60 genomes, and
10 ng
corresponds to about 3000 genomes. The data presented at Figure 17 show good
uniformity
(> 90% for most libraries) across all genomic DNA samples and input ranges
examined.
Specificity across all genomic DNA samples was > 95%. In this experiment, the
specificity
was defined as the percentage of filtered reads corresponding to the intended
targets.
EXAMPLE 7
Variant Calling Accuracy
[0224] To evaluate the accuracy of variant calling in libraries prepared
using
method 600 of Figure 6, the correlation between expected and observed variant
frequencies
for certain alleles was determined for the NA20mix and HD200 libraries of
Figure 17.
[0225] Figures 18A and 18B show a plot 1800 of TP (true positive)
variant
calling accuracy in the NA20mix library described in Figure 17 and a plot 1810
of TP
variant calling accuracy in the HD200 library described in Figure 17,
respectively. The data
show, for example, that for 1 ng (e.g., about 300 genome equivalents) input,
there is a good
correlation between the expected variant frequency and observed variant
frequency for TP
variant calling in both the NA20mix and HD200 libraries. Referring to Figures
18A and 18B,
the boxed areas represent about less than 30 copies of the genome for lng of
gDNA input
(e.g., less than 30 copies of those specific variants/mutations). Some of the
boxed variants
-80-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
correspond to as low as 7 or 3 genome copies per 1 ng gDNA input and
demonstrate the
sensitivity of the digital fluidic system and method for accurate variant
calling using
relatively low genomic DNA input. Circled area in Figure 18A corresponds to
particular
variant that was detected at frequency higher than expected. This is
attributed to low reads
coverage on this particular target resulting in increased noise.
[0226] Figure 19A shows a plot 1900 of the TP and FP (false positive)
variant
frequency as a function of chromosome and position in genomic libraries
prepared using an
on-bench protocol and on-actuator using method 600 of Figure 6. Libraries
(e.g., 19 digital
fluidic (on-actuator) samples and 9 on-bench samples) were prepared using 1 ng
input
NA12878 genomic DNA and the MJ191-plex target-specific primer pool. In the
NA12878
genome, there are 7 true positive variants. The data show that all TP variants
were accurately
called in both the on-bench and on-actuator samples. The data also show the
frequency of
random and persistent (indicated by an asterisk) FP variants that were called
in either all the
on-bench or all the on-actuator samples. The frequency of the FP variant calls
is substantially
lower than the frequency of TP variant calls. Additionally, sequencing base
quality is often,
but not always, substantially lower for FP variants relative to TP variants
indicative of
sequencing noise. FP variants can be removed, for example, during analysis by
filtering using
both these criteria. It is apparent that on-actuator method results in fewer
FP variants than on-
bench method.
[0227] Figure 19B shows a plot 1910 of the average number of FP variant
calls
in the on-bench samples and the on-actuator samples of plot 1900 of Figure
19A. The data
show that on average, there were about 12 FP variants called in the on-bench
samples
compared to about 2 FP variants called in the on-actuator samples. The lower
FP variant call
rate in the samples prepared on-actuator compared to the samples prepared on-
bench may be
due, in part, to the differences in the PCR step(s) of the library preparation
protocols. For
example, the on-bench library preparation protocol used a combined target-
specific and
universal PCR reaction of about 30 cycles for amplification of the input
genomic DNA. The
on-actuator library preparation protocol used a first target-specific
amplification of about 4 to
about 6 cycles (step 615 of method 600 of Figure 6) and a second universal
amplification
(step 620 of method 600 of Figure 6) of about 14 cycles for amplification of
the input
genomic DNA (e.g., total PCR of about 20 cycles or less). Lower total number
of PCR cycles
-81-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
is possible to achieve in on-actuator protocol due to improved PCR and
biochemistry
efficiency.
EXAMPLE 8
Amplicon Scalability
[0228] To evaluate library uniformity as a function of the complexity of
target-
specific primer pools, targeted amplicon libraries were prepared using method
600 of Figure
6 and 5 different multiplexed primer pools. Libraries were prepared using 1
and 5 ng of
NA12878 and NA20Mix genomic DNA and a 192-plex primer pool (Pool 1; 192
targets), or
a second 192-plex primer pool (Pool 2; 192 targets), or a 196-plex primer pool
(Pool 3; 193
targets), or a 384-plex combined primer pool (Pool Mix I to 2; 384 targets),
or a 580-plex
combined primer pool (Pool Mix 1 to 3; 580 targets).
[0229] Figure 20_shows a plot 2000 of library uniformity as a function
of
genomic DNA and primer pool complexity. In plot 2000, the data show good
uniformity
(most libraries > 90%) for both genomic DNA samples and input amounts. The
data also
show that as the complexity of the primer pool increased from a lower
complexity (e.g., 192-
or 196-plex) to a higher complexity (e.g., 384- or 580-plex), there was
relatively little change
in uniformity.
[0230] Uniformity for samples prepared on-bench (data not shown) was
about
70% for pool mix 1 to 3. The higher uniformity in libraries prepared on-
actuator (about 90%)
compared to on-bench uniformity (about 70%) provides for more efficient
library coverage in
sequencing (e.g., decreases sequencing depth by about an order of magnitude).
Specificity for
all genomic DNA samples and primer pools was about 95%.
[0231] Figures 21A, 21B, and 21C show a plot 2100 of target coverage in
the
192-plex Pool 1 library, a plot 2110 of 192-plex Pool 1 target coverage in the
384-plex Pool
Mix 1-2 library, and a plot 2120 of 192-plex Pool 1 target coverage in the 580-
plex Pool Mix
1-3 library described in plot 2000 of Figure 20.
[0232] The data show that when the complexity of a library increases
(e.g.,
increased target-specific primer pairs and genomic targets in Pool Mix 1-2 and
Pool Mix 1-3
libraries), there is a relatively small effect on the coverage of the 192-plex
Pool 1 library
targets.
-82-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
EXAMPLE 9
Systems
[0233] Figure 22 illustrates a functional block diagram of an example of
a
microfluidics system 2200 that includes a droplet actuator 2205, which is one
example of a
fluidics cartridge. Digital microfluidic technology conducts droplet
operations on discrete
droplets in a droplet actuator, such as droplet actuator 2205, by electrical
control of their
surface tension (electrowetting). The droplets may be sandwiched between two
substrates of
droplet actuator 2205, a bottom substrate and a top substrate separated by a
droplet
operations gap. The bottom substrate may include an arrangement of
electrically addressable
electrodes. The top substrate may include a reference electrode plane made,
for example,
from conductive ink or indium tin oxide (ITO). The bottom substrate and the
top substrate
may be coated with a hydrophobic material. Droplet operations are conducted in
the droplet
operations gap. The space around the droplets (e.g., the gap between bottom
and top
substrates) may be filled with an immiscible inert fluid, such as silicone
oil, to prevent
evaporation of the droplets and to facilitate their transport within the
device. Other droplet
operations may be effected by varying the patterns of voltage activation;
examples include
merging, splitting, mixing, and dispensing of droplets.
[0234] Droplet actuator 2205 may be designed to fit onto an instrument
deck (not
shown) of microfluidics system 2200. The instrument deck may hold droplet
actuator 2205
and house other droplet actuator features, such as, but not limited to, one or
more magnets
and one or more heating devices. For example, the instrument deck may house
one or more
magnets 2210, which may be permanent magnets. Optionally, the instrument deck
may house
one or more electromagnets 2215. Magnets 2210 and/or electromagnets 2215 are
positioned
in relation to droplet actuator 2205 for immobilization of magnetically
responsive beads.
Optionally, the positions of magnets 2210 and/or electromagnets 2215 may be
controlled by
a motor 2220. Additionally, the instrument deck may house one or more heating
devices
2225 for controlling the temperature within, for example, certain reaction
and/or washing
zones of droplet actuator 2205. In one example, heating devices 2225 may be
heater bars that
are positioned in relation to droplet actuator 2205 for providing thermal
control thereof.
[0235] A controller 2230 of microfluidics system 2200 is electrically
coupled to
various hardware components of the apparatus set forth herein, such as droplet
actuator 2205,
-83-
electromagnets 2215, motor 2220, and heating devices 2225, as well as to a
detector
2235, an impedance sensing system 1640, and any other input and/or output
devices (not
shown). Controller 2230 controls the overall operation of microfluidics system
2200.
Controller 2230 may, for example, be a general purpose computer, special
purpose computer,
personal computer, or other programmable data processing apparatus. Controller
2230 serves
to provide processing capabilities, such as storing, interpreting, and/or
executing software
instructions, as well as controlling the overall operation of the system.
Controller 2230 may
be configured and programmed to control data and/or power aspects of these
devices. For
example, in one aspect, with respect to droplet actuator 2205, controller 2230
controls droplet
manipulation by activating/deactivating electrodes.
[0236] In one example, detector 2235 may be an imaging system that
is
positioned in relation to droplet actuator 2205. In one example, the imaging
system may
include one or more light-emitting diodes (LEDs) (e.g., an illumination
source) and a digital
image capture device, such as a charge-coupled device (CCD) camera. Detection
can be
carried out using an apparatus suited to a particular reagent or label in use.
For example, an
optical detector such as a fluorescence detector, absorbance detector,
luminescence detector
or the like can be used to detect appropriate optical labels. Systems designed
for array-based
detection are particularly useful. For example, optical systems for use with
the methods set
forth herein may be constructed to include various components and assemblies
as described
in Banerjee et al., U.S. Patent No. 8,241,573, entitled "Systems and Devices
for Sequence by
Synthesis Analysis," issued on August 14, 2012; Feng et al., U.S. Patent No.
7,329,860,
entitled "Confocal Imaging Methods and Apparatus," issued on February 12,
2008; Feng et
al., U.S. Patent No. 8,039,817, entitled "Compensator for Multiple Surface
Imaging," issued
on October 18, 2011; Feng et al., U.S. Patent Pub. No. 20090272914, entitled
"Compensator
for Multiple Surface Imaging," published on November 5, 2009; and Reed et al.,
U.S. Patent
Pub. No. 20120270305, entitled "Systems, Methods, and Apparatuses to Image a
Sample for
Biological or Chemical Analysis," published on October 25, 2012. Such
detection systems
are particularly useful for nucleic acid sequencing embodiments.
102371 Impedance sensing system 2240 may be any circuitry for
detecting
impedance at a specific electrode of droplet actuator 2205. In one example,
impedance
sensing system 2240 may be an impedance spectrometer. Impedance sensing system
2240
-84-
CA 3016221 2019-11-29
may be used to monitor the capacitive loading of any electrode, such as any
droplet
operations electrode, with or without a droplet thereon. For examples of
suitable capacitance
detection techniques, see Sturmer et al., International Patent Pub. No.
WO/2008/101194,
entitled "Capacitance Detection in a Droplet Actuator," published on December
30, 2009;
and Kale et al., International Patent Pub. No. WO/2002/080822, entitled
"System and
Method for Dispensing Liquids," published on February 26, 2004.
[0238] Droplet actuator 2205 may include disruption device 2245.
Disruption
device 2245 may include any device that promotes disruption (lysis) of
materials, such as
tissues, cells and spores in a droplet actuator. Disruption device 2245 may,
for example, be a
sonication mechanism, a heating mechanism, a mechanical shearing mechanism, a
bead
beating mechanism, physical features incorporated into the droplet actuator
2205, an electric
field generating mechanism, armal cycling mechanism, and any combinations
thereof.
Disruption device 2245 may be controlled by controller 2230.
[0239] It will be appreciated that various aspects of the present
disclosure may be
embodied as a method, system, computer readable medium, and/or computer
program
product. Aspects of the present disclosure may take the form of hardware
embodiments,
software embodiments (including firmware, resident software, micro-code,
etc.), or
embodiments combining software and hardware aspects that may all generally be
referred to
herein as a "circuit," "module," or "system." Furthermore, the methods of the
present
disclosure may take the form of a computer program product on a computer-
usable storage
medium having computer-usable program code embodied in the medium.
[0240] Any suitable computer useable medium may be utilized for
software
aspects of the present disclosure. The computer-usable or computer-readable
medium may
be, for example but not limited to, an electronic, magnetic, optical,
electromagnetic, infrared,
or semiconductor system, apparatus, device, or propagation medium. The
computer readable
medium may include transitory embodiments. More specific examples (a non-
exhaustive list)
of the computer-readable medium would include some or all of the following: an
electrical
connection having one or more wires, a portable computer diskette, a hard
disk, a random
access memory (RAM), a read-only memory (ROM), an erasable programmable read-
only
-85-
CA 3016221 2019-11-29
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
memory (EPROM or Flash memory), an optical fiber, a portable compact disc read-
only
memory (CD-ROM), an optical storage device, a transmission medium such as
those
supporting the Internet or an intranet, or a magnetic storage device. Note
that the computer-
usable or computer-readable medium could even be paper or another suitable
medium upon
which the program is printed, as the program can be electronically captured,
via, for instance,
optical scanning of the paper or other medium, then compiled, interpreted, or
otherwise
processed in a suitable manner, if necessary, and then stored in a computer
memory. In the
context of this document, a computer-usable or computer-readable medium may be
any
medium that can contain, store, communicate, propagate, or transport the
program for use by
or in connection with the instruction execution system, apparatus, or device.
[0241] Program code for carrying out operations of the methods and
apparatus set
forth herein may be written in an object oriented programming language such as
Java,
Smalltalk, C++ or the like. However, the program code for carrying out
operations of the
methods and apparatus set forth herein may also be written in conventional
procedural
programming languages, such as the "C" programming language or similar
programming
languages. The program code may be executed by a processor, application
specific integrated
circuit (ASIC), or other component that executes the program code. The program
code may
be simply referred to as a software application that is stored in memory (such
as the computer
readable medium discussed herein). The program code may cause the processor
(or any
processor-controlled device) to produce a graphical user interface ("GUI").
The graphical
user interface may be visually produced on a display device, yet the graphical
user interface
may also have audible features. The program code, however, may operate in any
processor-
controlled device, such as a computer, server, personal digital assistant,
phone, television, or
any processor-controlled device utilizing the processor and/or a digital
signal processor.
[0242] The program code may locally and/or remotely execute. The program
code, for example, may be entirely or partially stored in local memory of the
processor-
controlled device. The program code, however, may also be at least partially
remotely stored,
accessed, and downloaded to the processor-controlled device. A user's
computer, for
example, may entirely execute the program code or only partly execute the
program code.
The program code may be a stand-alone software package that is at least partly
on the user's
computer and/or partly executed on a remote computer or entirely on a remote
computer or
-86-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
server. In the latter scenario, the remote computer may be connected to the
user's computer
through a communications network.
[0243] The methods and apparatus set forth herein may be applied
regardless of
networking environment. The communications network may be a cable network
operating in
the radio-frequency domain and/or the Internet Protocol (IP) domain. The
communications
network, however, may also include a distributed computing network, such as
the Internet
(sometimes alternatively known as the "World Wide Web"), an intranet, a local-
area network
(LAN), and/or a wide-area network (WAN). The communications network may
include
coaxial cables, copper wires, fiber optic lines, and/or hybrid-coaxial lines.
The
communications network may even include wireless portions utilizing any
portion of the
electromagnetic spectrum and any signaling standard (such as the IEEE 802
family of
standards, GSM/CDMA/TDMA or any cellular standard, and/or the ISM band). The
communications network may even include powerline portions, in which signals
are
communicated via electrical wiring. The methods and apparatus set forth herein
may be
applied to any wireless/wireline communications network, regardless of
physical
componentry, physical configuration, or communications standard(s).
[0244] Certain aspects of present disclosure are described with
reference to
various methods and method steps. It will be understood that each method step
can be
implemented by the program code and/or by machine instructions. The program
code and/or
the machine instructions may create means for implementing the functions/acts
specified in
the methods.
[0245] The program code may also be stored in a computer-readable memory
that
can direct the processor, computer, or other programmable data processing
apparatus to
function in a particular manner, such that the program code stored in the
computer-readable
memory produce or transform an article of manufacture including instruction
means which
implement various aspects of the method steps.
[0246] The program code may also be loaded onto a computer or other
programmable data processing apparatus to cause a series of operational steps
to be
performed to produce a processor/computer implemented process such that the
program code
provides steps for implementing various functions/acts specified in the
methods of the
present disclosure.
-87-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
EXAMPLE 10
Indexed Library Preparation
[0247] Indexed multiplex PCR was performed with the OTMI buffer
(I1lumina
Part# 15059758), 161-plex SBS3-GS-F primer (gene specific forward) and GS-R
primer
(gene specific reverse) mix in betaine, universal forward primer (UF-4) with
different
indexes, universal reverse primer (UR) and human genomic DNA. Primer
concentrations
used were 10 nM each of GS-F and GS-R, 300 n1\4 of UF-4, and 200 nM of UR
primer in
final PCR reaction. Coriell NA12878 genomic DNA was used at 0.02ng/RL final as
input for
PCR. The DNA polymerase used was 0.04U/t1 Phusion HSII DNA polymerase (Thermo
Fisher Scientific). The thermocycling parameters used in this experiment are
shown in Table
4.
TABLE 4: Parameters used in thermocycling reactions
98 C 2 min
98 C 20"
30 cycles 70 C 30"
60 C 60" (from 70 C ramp to 60 C, 0.2 C/sec)
72 C 75" (from 60 C ramp to 72 C, 0.2 C/sec)
72 C 2 min
C hold
[0248] The resulting PCR product had adaptors on the ends that were
complementary to the P7 and P5 primers used in Example 12 below.
EXAMPLE 11
Preparation of capture beads for Ex-Amp normalization
[0249] Streptavidin beads (I1lumina Patt#11118442) were washed with a
buffer
containing 5mM Tris-HC1 pH 7.5, 500nM EDTA, and 1M NaCl (BW buffer) and re-
suspended in 75 n1 Biotin-P7-Index-5B5491' in BW-Buffer to a final
concentration of 10
RM. The beads were vortexed for 15min at room temperature, washed in 1 x BW
buffer and
re-suspended in 37.5 pi or 0.25nM each Capture Probe Template oligo, in HT1
buffer (as in
MiSeq V3 150 cycle kit; Illumina catalog# MS-102-3001). The beads were
incubated for 5
minutes at 60 C followed by 5 minutes at 40 C, then washed and re-suspended in
AMS-6
buffer (as in HiSeq PE Cluster Kit V4 for cBot (I1lumina Catalog#PE-401-4001).
After
incubation at 40 C for 5 minutes, the beads were re-suspended in 100 I, of
0.1N NaOH and
-88-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
incubated for additional 5 minutes at room temperature. The beads were then
washed at least
once in 100 uL HT1 buffer and re-suspended in 150 L HT1 buffer. The resulting
beads
contained bound biotin-P7 amplification or normalization primer.
EXAMPLE 12
On-bead extension and Ex-Amp normalization
[0250] Ten-fold serial dilutions of the indexed PCR product of Example
10 were
prepared in 1 x HT1 buffer and denatured in 20 x SSC by heating at 98 C for 3
minutes then
keep on ice. Equal volume of capture beads of Example 11 was added to
denatured PCR
product and incubated at 75 C for 30 sec, then at 65 C for 10 min, then 40 C
for 5 mm,
followed by washing in PR2 buffer (in MiSeq V3 150 cycle kit, Illumina
catalog# MS-102-
3001).
[0251] On bead Ex-Amp normalization was performed as follows. Briefly,
each
sample was incubated at room temperature for 8 min with 1 L of 100 M P5
primer, 4 pIL of
I120 and 5 L of 0.1N NaOH. Prepared EPX mix following the manufacturer's
recommendations (I1lumina PN 15067046) and 35 IA, of the EPX mix was added to
15 L of
the denatured P5 primer. PR2 buffer was removed from the beads (on magnet) and
50 L of
the EPX+P5 cocktail was added to the beads, and then re-suspended and
incubated at 38 C
for 20 min followed by removal of supernatant. The libraries were denatured
with 0.1N
NaOH, washed and re-suspended in PR2 buffer.
[0252] The libraries were quantitated using SYBR Green qPCR library
quantification kits (KAPA Biosystems, Part number KK4824). As shown in Figure
29A, the
output library yields were normalized regardless of the diverse input sample
concentrations.
EXAMPLE 13
High-throughput sequencing (HiSeq)
[0253] Samples from Example 12 were normalized based on the reported
qPCR
yield. Total library concentration used was 10 pM. In this Example, normalized
samples from
Example 12 were sequenced on Illumina's HiSeq2000 using HiSeq V4 chemistry
(Illumina
Catalog numbers: PE-401-4001, FC-401-4002 and FC-401-4003), following standard
single
index settings as per manufacturer's recommendations. The top level sequencing
metrics are
shown in Table 5.
-89-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
TABLE 5: Summary of sequencing metrics for individual nucleic acid samples
whose
DNA concentration have been normalized as described in Example 12.
Sample Name Total PF On Target Mean Specificity
Uniformity CV Span95
S1-30Cyc-Di1-1-
77760589 77169353 482308 0.992 0.90625 0.713 22
ExAmp_S1
S2-30Cyc-Di1-10-
78706859 78295323 489346 0.995 0.85 0.965 49
ExAmp S2
S3-30Cyc-Dil-
75718469 75305133 470657 0.995 0.825 1.089 74
100-ExAmp_S3
S4-30Cyc-Dil-
73640185 72479744 452998 0.984 0.8125 1.112 79
1000-ExAmp_S4
[0254] As shown in Table 5, it was observed that even with the aggressive
dilution of the input PCR product, the library specificity remained high
(>0.98). The
uniformity dropped as expected with further dilution of the library however,
even at 1000
fold diluted PCR product input, the uniformity is still at acceptable level
(>0.80). Thus, as
shown in Table 5, the sequencing metrics for the libraries of Example 12
resulting from the
method described in the present disclosure are comparable to the standard
library preparation
method.
EXAMPLE 14
On-bead extension and Ex-Amp normalization
[0255] In another experiment, the experimental procedure outlined above in
Example 12 was followed except that the template DNA used was genomic DNA from
formalin-fixed paraffin-embedded (FFPE) samples (HorizonDX HD751). Input
samples
containing 50, 10, 2, 0.4 and 0.08ng of genomic DNA was tested using the
standard library
prep protocol using TrueSeq library preparation kit (Illumina FC-121-4001) as
per
manufacturer's recommendations or using the method described in the present
disclosure.
The library yields were quantitated using KAPA library quantitation kit. The
data shown in
Figure 29B was generated by using the standard library preparation method
where the output
library yields were approximately 659, 216, 67, 18 and 5.3 pM for the input
DNA range 50,
10, 2, 0.4 and 0.08ng respectively. In comparison, using the method described
in the present
disclosure, the output library yields were consistent across the input range
resulting in
approximately 51500, 46250, 47628, 45220 and 43238 pM, respectively, as shown
in Figure
29C.
-90-
CA 03016221 2018-08-29
WO 2017/176896 PCT/US2017/026169
EXAMPLE 15
On-bead extension and Ex-Amp normalization
[0256] In this Example, the
samples were normalized based on the reported qPCR
yield. The yield from the standard library preparation using 0.4 ng and 0.08ng
of DNA,
(samples SOS and 506 in the table below) were too low to be normalized. Total
normalized
library concentration was 20 pM. The libraries were sequenced on Illumina's
MiSeq
sequencer using MiSeq V3 150 cycle kit (IIlumina Catalog# MS-102-3001),
following
standard dual index settings. The top level sequencing metrics are given in
Table 6. Samples
502-S06 are the standard library preparations shown in Figure 29B, while S08-
5012 are the
samples from Figure 29C. Samples 501 and 507 are controls.
TABLE 6: Summary of sequencing metrics for individual nucleic acid samples
whose
DNA concentrations have been normalized as described in Example14.
Sample Name Total PF On Target Mean Specificity
Uniformity CV Span95
SO 1-AXE- 75263 22117 138 0.294 0.388 6.699 462
NTC Si
S02-AXE- 4622638 4590000 28688 0.993 0.906 0.905 26
50ngHD751_S2
S03-AXE- 5624965 5592099 34951 0.994 0.844 0.99 49
10ngHD751 53
SO4-AXE- 4387777 4359280 27246 0.994 0.756 1.286
138
2ngHD751_54
S05-AXE- 2890741 2854572 17841 0.987 0.681 1.471
604
o4ngHD751_S5
S06-AXE- 615260 593796 3711 0.965 0.544 1.699 inf
o08ngHD751_S6
507-ExAmp- 1215577 422982 2644 0.348 0.031 8.812 435
NTC_S7
S08-ExAmp- 4042970 4009345 25058 0.992 0.863 1.031 38
50ngHD751_S8
509-ExAmp- 3993705 3967177 24795 0.993 0.794 1.132 70
10ngHD751 S9
S10-ExAmp- 3897994 3869129 24182 0.993 0.688 1.449
210
2ngHD751_S10
S11-ExAmp- 3825284 3787323 23671 0.99 0.65 1.689 833
o4ngHD751 S 11
512-ExAmp- 3284845 3177453 19859 0.967 0.512 1.829
200862
o08ngHD751 S12
[0257] As shown in Table 6,
the sequencing metrics for the libraries resulting
from the amplification and normalization method described in the present
disclosure (508-
512) are comparable to the standard library preparation method (S02-506).
-91-
102581 Thus, as Figures 29A and 29C demonstrate, the amplification
and
normalization methods disclosed herein provide a simple method of providing a
normalized
amount of output DNA libraries across a wide range of concentrations of input
DNA
samples, where no further dilution or concentration of the resulting output
libraries is
required before pooling the libraries for subsequent sequencing. In
particular, the
experimental data presented in Tables 5 and 6 show that the quality of the
amplified product
is comparable to existing library preparation methods across a wide range of
input DNA
concentrations.
***
In some aspects, embodiments of the present invention as described herein
include
the following items:
1. A method for nucleic acid amplification comprising:
a) providing an input sample comprising target nucleic acid molecules;
b) contacting the sample with a reaction mixture comprising a solid phase and
a liquid
phase, wherein:
i. the solid phase comprises a plurality of first amplification primers
immobilized
on a solid support, the first amplification primers capable of specifically
hybridizing to a first sequence of the target nucleic acid molecules; and
ii. the liquid phase comprises a plurality of second amplification primers in
solution, the plurality of second primers capable of specifically hybridizing
to
a second sequence of the target nucleic acid molecules;
and
c) amplifying the target nucleic acid molecules under isothermal conditions,
wherein
the plurality of first amplification primers is provided in an amount which
limits the yield of
amplification products to a predefined amount; and
the plurality of second amplification primers is provided in an amount that
exceeds the
amount of the first amplification primers.
2. The method of item 0, further comprising separating the amplification
products from
the solid support.
3. The method of item 0 or 2, wherein the solid support comprises a
plurality of beads.
-92-
Date Recue/Date Received 2020-10-05
4. The method of any one of items 0 to 3, wherein the solid support
comprises beads
selected from magnetic beads, paramagnetic beads, plastic beads, polystyrene
beads, glass
beads, agarose beads, flow cytometry microbeads, polystyrene microparticles,
polystyrene
nanoparticles, functionalized polystyrene microparticles, functionalized
polystyrene
nanoparticles, coated polystyrene microparticles, coated polystyrene
microparticles
nanoparticles, silica microbeads, fluorescent microspheres, fluorescent
microspheres
nanospheres, functionalized fluorescent microspheres, functionalized
fluorescent
nanospheres, coated fluorescent microspheres, coated fluorescent microspheres
nanospheres,
color dyed microparticles, color dyed microparticles nanoparticles, magnetic
microparticles,
magnetic nanoparticles, superparamagnetic microparticles, superparamagnetic
nanoparticles,
and combinations thereof.
5. The method of item 3 or 4, wherein the beads are in an aqueous reaction
buffer.
6. The method of any one of items 3 to 5, wherein the beads are in fluid
communication
with each other.
7. The method of any one of items 3 to 6, wherein the beads comprise
streptavidin beads
onto which the first amplification primers are affixed through conjugated
biotin.
8. The method of any one of items 3 to 7, wherein the beads are monoclonal.
9. The method of any one of items 3 to 7, wherein the beads are polyclonal.
10. The method of any one of items 0 to 9, wherein the solid support is a
surface of a
reaction site.
11. The method of item 10, wherein the surface of a reaction site comprises a
bottom
portion of an inner surface of a well, a groove, a flow cell, a reaction
chamber or channel.
12. The method of any one of items 0 to 11, wherein the sample comprises
single-stranded
nucleic acid molecules.
13. The method of any one of items 0 to 11, wherein the sample comprises
double-stranded
nucleic acid molecules.
14. The method of any one of items 0 to 13, wherein the first amplification
primers and the
second amplification primers comprises sequences complementary to known
nucleotide
sequences within the target nucleic acid molecules.
15. The method of item 14, wherein the known nucleotide sequences
correspond to the first
end and second end of the target nucleic acid molecules.
-93-
Date Recue/Date Received 2020-10-05
16. The method of item 15, wherein the first ends and second ends of the
target nucleic acid
molecules comprise either universal sequencing tail-adaptors or universal
primer regions that
have been added to the target nucleic acid molecules.
17. The method of item 16 wherein
i. the universal primer regions comprise a sequencing-by-synthesis (SBS)
primer
sequence; and
ii. wherein at least one of the first and/or second primers further comprises
a
region having sequence complementarity to the universal primer regions
added to the target nucleic acid molecules.
18. The method of any one of items 0 to 17, wherein each of the first and/or
second
amplification primers further comprises an indexing portion.
19. The method of any one of items 0 to 18, wherein at least a portion of the
first
amplification primers further comprises a capture portion having sequence
complementarity
to a cognate region of the target nucleic acid molecules as well as to a known
sequence of the
target nucleic acid molecules.
20. The method of any one of items 0 to 19, wherein the capture portion is
generated by
hybridizing a capture oligonucleotide to a first amplification primer
immobilized onto the
solid support and extending the immobilized amplification primer to generate
an extended
amplification primer having sequence complementarity to the capture
oligonucleotide.
21. The method of any one of items 0 to 20, wherein the amplification step
(c) is performed
on a plurality of samples.
22. The method of item 21, wherein the amount of each input sample is
varies across the
plurality of samples.
23. The method of item 21 or 22, wherein the plurality of input samples is
combined before
the amplification step.
24. The method of any one of items 21 to 23, wherein amplification products
from the
plurality of samples are combined to form a pooled nucleic acid library.
25. The method of any one of items 21 to 24, wherein the amplification
products from the
plurality of samples are combined before being separated from the respective
solid support.
26. The method of any one of items 21 to 24, wherein the amplification
products from the
plurality of samples are combined after being separated from the respective
solid support.
-94-
Date Recue/Date Received 2020-10-05
27. The method of any one of items 0 to 26, further comprising obtaining a
nucleotide
sequence of the amplification products.
28. A method for nucleic acid amplification comprising:
a) providing an input sample comprising target nucleic acid molecules;
b) contacting the sample with a reaction mixture comprising a solid phase and
a liquid
phase, wherein:
i. the solid phase comprises a plurality of first amplification primers
immobilized
on a solid support, the first amplification primers capable of specifically
hybridizing to a first sequence of the target nucleic acid molecules; and
ii. the liquid phase comprises a plurality of second amplification primers in
solution, the plurality of second primers capable of specifically hybridizing
to
a second sequence of the target nucleic acid molecules;
and
c) amplifying the target nucleic acid molecules under isothermal conditions,
wherein:
the plurality of first amplification primers is provided in an amount which
limits the
yield of amplification products to a predefined amount,
at least a portion of the first amplification primers further comprises a
capture portion
having sequence complementarity to a region of the target nucleic acid
molecules in addition
to a known sequences of the target nucleic acid molecules, and the capture
portion is
generated by hybridizing a capture oligonucleotide to a first amplification
primer
immobilized onto the solid support and extending the immobilized amplification
primer to
generate an extended amplification primer having sequence complementarity to
the capture
oligonucleotide.
29. The method of item 28, wherein the plurality of second amplification
primers is
provided in an amount that exceeds the amount of the first amplification
primers.
30. The method of any one of items 0 to 29, wherein the plurality of second
amplification
primers is provided in an amount within an order of magnitude of the amount of
first
amplification primers.
31. The method of any one of items 0 to 29, wherein the plurality of second
amplification
primers is provided in an amount that exceeds the amount of the first
amplification primers
by at least 100%.
-95-
Date Recue/Date Received 2020-10-05
32. The method of any one of items 0 to 31, wherein the reaction mixture
further comprises
one or more of a recombinase, a single-strand DNA-binding protein, a helicase,
and a strand-
displacing polymerase.
33. A method for nucleic acid amplification comprising:
a) providing an input sample comprising target nucleic acid molecules;
b) contacting the input sample with a reaction mixture comprising a plurality
of first
normalization primers and a plurality of second normalization primers,
wherein:
i. the plurality of first normalization primers is immobilized on a solid
support, the
first normalization primers being capable of specifically hybridizing to a
first
sequence of the target nucleic acid molecules; and
ii. the plurality of second normalization primers is in solution, the
plurality of
second primers being capable of specifically hybridizing to a second sequence
of the target nucleic acid molecules;
and
c) amplifying the target nucleic acid molecules under isothermal conditions,
wherein
the plurality of first normalization primers is provided in an amount which
limits the yield of
amplification products to a predefined amount; and
the plurality of second normalization primers is provided in an amount that
exceeds the
amount of the first normalization primers.
34. The method of item 0, wherein the plurality of first normalization
primers is hybridized
with the target nucleic acid molecules prior to being immobilized on the solid
support.
35. The method of item 0, wherein the plurality of first normalization primers
is
immobilized on the solid support prior to being hybridized with the target
nucleic acid
molecules.
36. The method of any one of items 0 to 35, further comprising separating
the amplification
products from the solid support.
37. The method of any one of items 0 to 35, wherein the solid support
comprises a plurality
of beads.
38. The method of any one of items 0 to 36, wherein the solid support
comprises beads
selected from magnetic beads, paramagnetic beads, magnetically responsive
beads, plastic
beads, polystyrene beads, glass beads, agarose beads, and combinations thereof
-96-
Date Recue/Date Received 2020-10-05
39. The method of any one of items 0 to 38, wherein the solid support
comprises beads
selected from magnetic beads, paramagnetic beads, plastic beads, polystyrene
beads, glass
beads, agarose beads, flow cytometry microbeads, polystyrene microparticles,
polystyrene
nanoparticles, functionalized polystyrene microparticl es, functionalized
polystyrene
nanoparticles, coated polystyrene microparticles, coated polystyrene
nanoparticles, silica
microbeads, fluorescent microspheres, fluorescent nanospheres, functionalized
fluorescent
microspheres, functionalized fluorescent nanospheres, coated fluorescent
microspheres,
coated fluorescent nanospheres, color dyed microparticles, color dyed
nanoparticles,
magnetic microparticl es, magnetic nanoparticles, superparamagnetic
microparticles,
superparamagnetic nanoparti cies, and combinations thereof
40. The method of item 39, wherein the beads are in an aqueous reaction
buffer.
41. The method of item 39 or 40, wherein the beads are in fluid
communication with each
other.
42. The method of any one of items 39 to 41, wherein the beads comprise
streptavidin
beads onto which the first normalization primers are affixed through
conjugated biotin.
43. The method of any one of items 39 to 42, wherein the beads are
monoclonal.
44. The method of any one of items 39 to 42, wherein the beads are
polyclonal.
45. The method of any one of items 39 to 44, wherein the solid support is a
surface of a
reaction site.
46. The method of item 45, wherein the surface of a reaction site comprises a
bottom
portion of an inner surface of a well, a groove, a flow cell, a reaction
chamber or channel.
47. The method of any one of items 0 to 46, wherein the input sample
comprises single-
stranded nucleic acid molecules.
48. The method of any one of items 0 to 46, wherein the input sample
comprises double-
stranded nucleic acid molecules.
49. The method of any one of items 0 to 48, wherein at least one of the
first normalization
primers and/or the second normalization primers comprises a region having
sequence
complementarity to known nucleotide sequences within the target nucleic acid
molecules.
50. The method of item 49, wherein the known nucleotide sequences
correspond to the first
end and second end of the target nucleic acid molecules.
-97-
Date Recue/Date Received 2020-10-05
51. The method of item 50, wherein the first ends and second ends of the
target nucleic acid
molecules comprise universal primer regions that have been added to the target
nucleic acid
molecules.
52. The method of item 51, wherein the universal primer regions comprise a
sequencing-
by-synthesis (SBS) primer sequence.
53. The method of any one of items 0 to 52, wherein at least one of the
first and/or second
normalization primers further comprises an indexing portion.
54. The method of any one of items 51 to 53, wherein at least one of the
first and/or second
normalization primers further comprises a region having sequence
complementarity to the
universal primer regions added to the target nucleic acid molecules.
55. The method of any one of items 0 to 54, wherein at least a portion of the
first
normalization primers further comprises a capture portion having sequence
complementarity
to a cognate region of the target nucleic acid molecules as well as to a known
sequence of the
target nucleic acid molecules.
56. The method of any one of items 0 to 55, wherein a capture portion is
generated by
hybridizing a capture oligonucleotide to a first normalization primer and
extending the first
normalization primer to generate an extended normalization primer having
sequence
complementarity to the capture oligonucleotide.
57. The method of any one of items 0 to 56, wherein the amplification step
is performed on
a plurality of input samples.
58. The method of any one of items 0 to 57, wherein the amount of each
input sample is not
equalized across the plurality of input samples.
59. The method of item 57 or 58, wherein the plurality of input samples is
combined before
the amplification step.
60. The method of item 57 or 58, wherein the amplification products from
the plurality of
input samples are combined to form a pooled nucleic acid library.
61. The method of any one of items 57 to 60, wherein the amplification
products from the
plurality of input samples are combined before being separated from the
respective solid
support.
-98-
Date Recue/Date Received 2020-10-05
62. The method of any one of items 57 to 61, wherein the amplification
products from the
plurality of input samples are combined after being separated from the
respective solid
support.
63. The method of any one of items 0 to 62, further comprising obtaining a
nucleotide
sequence of the amplification products.
64. A method for nucleic acid amplification comprising:
a) providing an input sample comprising target nucleic acid molecules;
b) contacting the input sample with a reaction mixture comprising a plurality
of first
normalization primers and a plurality of second normalization primers,
wherein:
i. the plurality of first normalization primers is immobilized on a solid
support, the
first normalization primers capable of specifically hybridizing to a first
sequence of the target nucleic acid molecules; and
ii. the plurality of second normalization primers is in solution, the
plurality of
second primers capable of specifically hybridizing to a second sequence of the
target nucleic acid molecules;
and
c) amplifying the target nucleic acid molecules under isothermal conditions,
wherein
the plurality of first normalization primers is provided in an amount which
limits the yield of
amplification products to a predefined amount, at least a portion of the first
normalization
primers further comprises a capture portion having sequence complementarity to
a region of
the target nucleic acid molecules in addition to a known sequences of the
target nucleic acid
molecules, and the capture portion is generated by hybridizing a capture
oligonucleotide to a
first normalization primer and extending the first normalization primer to
generate an
extended normalization primer having sequence complementarity to the capture
oligonucleotide.
66. The method of item 0, wherein the plurality of second normalization
primers is
provided in an amount that exceeds the amount of the first normalization
primers.
67. The method of item 64 or 66, wherein the plurality of second
normalization primers is
provided in an amount within an order of magnitude of the amount of first
normalization
primers.
-99-
Date Recue/Date Received 2020-10-05
68. The method of item 64 or 66, wherein the plurality of second
normalization primers is
provided in an amount that exceeds the amount of the first normalization
primers by at least
100%.
69. The method of any one of items 64 to 68, wherein the reaction mixture
further
comprises one or more of a recombinase, a single-strand DNA-binding protein, a
helicase,
and a strand-displacing polymerase.
70. The method of any one of items 64 to 68, wherein the first and second
amplification
primers are first and second normalization primers wherein:
a) the plurality of first normalization primers is hybridized with the target
nucleic acid
molecules prior to being immobilized on the solid support; or
b) the plurality of first normalization primers is immobilized on the solid
support prior
to being hybridized with the target nucleic acid molecules.
71. The method of any one of items 64 to 69, wherein prior to step (a), the
target nucleic
acid molecules in the input sample are subjected to a first enrichment
amplification reaction
comprising a first target-specific primer and a second target-specific primer.
72. The method of item 70, wherein each of the first target-specific primer
and/or the
second target-specific primer comprises a region having sequence
complementarity to known
sequences of the target nucleic acid molecules.
73. The method of item 70 or 71, wherein each of the first target-specific
primer and/or the
second target-specific primer further comprises a universal primer region.
74. The method of item 72, wherein the universal primer region of the first
target-specific
primer comprises a sequencing-by-synthesis (SBS) primer sequence.
75. The method of items 73 or 73, wherein prior to step (a), the target
nucleic acid
molecules in the input sample are subjected to a second enrichment
amplification reaction
comprising a first universal primer and a second universal primer, wherein:
a) the first universal primer comprises a region having sequence
complementarity to
the universal primer region of the first target-specific primer; and
b) the second universal primer comprises a region having sequence
complementarity
to the universal primer region of the second target-specific primer.
76. The method of item 75, wherein at least one of the first and/or second
universal primers
further comprises an indexing portion.
-100-
Date Recue/Date Received 2020-10-05
77. The method of any one of items 64 to 76, wherein the method is performed
in
multiplexed format using a droplet actuator.
78. A method for multiplexed amplification of samples on a droplet
actuator, comprising:
a) providing a plurality of input samples comprising target nucleic acid
molecules;
b) loading the plurality of input samples onto a droplet operations surface of
the
droplet actuator having droplet operations electrodes arranged thereon;
c) dispensing a normalization reagent droplet comprising a plurality of first
normalization primers and a plurality of second normalization primers,
wherein:
i. the plurality of first normalization primers is immobilized on a solid
support,
the first normalization primers being capable of specifically hybridizing to a
first sequence of the target nucleic acid molecules; and
ii. the plurality of second normalization primers is in solution, the
plurality of
second primers being capable of specifically hybridizing to a second
sequence of the target nucleic acid molecules;
and
d) amplifying the target nucleic acid molecules under isothermal conditions,
wherein
the plurality of first normalization primers is provided in an amount which
limits the yield of
amplification products to a predefined amount.
79. The method of item 0, wherein prior to step (c) further comprising:
dispensing a first enrichment PCR reagent droplet onto a droplet operations
surface of
the droplet actuator, wherein the first enrichment PCR reagent droplet
comprising
a first target-specific primer and a second target-specific primer; and
thermally cycling the target nucleic acid molecules in the plurality of input
samples to
form enriched nucleic acid samples.
80. The method of item 0 or 79, wherein each of the first target-specific
primer and/or the
second target-specific primer comprises a region having sequence
complementarity to known
sequences of the target nucleic acid molecules.
81. The method of item 80, wherein each of the first target-specific primer
and/or the
second target-specific primer further comprises a universal primer region.
82. The method of item 80, wherein the universal primer region of the first
target-specific
primer comprises a sequencing-by-synthesis (SBS) primer sequence.
-101-
Date Recue/Date Received 2020-10-05
83. The method of any one of items 0 to 82, wherein at least one of the
first and/or second
target-specific primers further comprises an indexing portion.
84. The method of item 0, wherein prior to step (c) further comprising:
dispensing a second enrichment PCR reagent droplet onto a droplet operations
surface of the droplet actuator, wherein the second enrichment PCR reagent
droplet comprising a first universal primer and a second universal primer; and
thermally cycling the target nucleic acid molecules in the plurality of input
samples to
form enriched nucleic acid samples.
85. The method of item 84, wherein at least one of the first and/or second
universal primers
further comprises a primer sequence region.
86. The method of item 84 or 85, wherein at least one of the first and/or
second universal
primers further comprises an indexing portion.
87. The method of item 0, wherein prior to step (c) further comprising:
dispensing a first enrichment PCR reagent droplet onto a droplet operations
surface of
the droplet actuator, wherein the first enrichment PCR reagent droplet
comprising
a first target-specific primer and a second target-specific primer;
dispensing a second enrichment PCR reagent droplet onto a droplet operations
surface of the droplet actuator, wherein the second enrichment PCR reagent
droplet comprising a first universal primer and a second universal primer;
combining the second enrichment PCR reagent droplet with the first enrichment
PCR
reagent droplet using droplet operations to form a combined enrichment PCR
reagent droplet; and
thermally cycling the target nucleic acid molecules in the plurality of input
samples to
form enriched nucleic acid samples.
88. A mi croflui di c system for performing multiplexed amplification of
nucleic acid
samples on a droplet actuator, comprising a processor for executing code, a
memory
communicatively coupled to the processor, and a program code stored in the
memory that
causes the processor to execute the method of multiplexed amplification of
nucleic acid
samples, as defined in the method of any one of items 77 to 86 comprising:
-102-
Date Recue/Date Received 2020-10-05
a) loading a plurality of input samples onto a droplet operations surface of
the droplet
actuator having droplet operations electrodes arranged thereon, each of the
plurality of input
samples comprising target nucleic molecules;
b) dispensing a normalization reagent droplet comprising a plurality of first
normalization primers and a plurality of second normalization primers,
wherein:
i. the plurality of first normalization primers is immobilized on a solid
support,
the first normalization primers being capable of specifically hybridizing to a
first sequence of the target nucleic acid molecules; and
ii. the plurality of second normalization primers is in solution, the
plurality of
second primers being capable of specifically hybridizing to a second
sequence of the target nucleic acid molecules;
and
c) amplifying the target nucleic acid molecules under isothermal conditions,
wherein
the plurality of first normalization primers is provided in an amount which
limits the yield of
amplification products to a predefined amount.
89. The microfluidic system of item 87, further comprising one or more of
the following
components:
a) a heating device;
b) a detector optically coupled to the droplet actuator;
c) an impedance sensing module;
d) a disruption device for lysing a biomaterial comprising nucleic acids; and
e) a controller electronically coupled to one or more of the components of
(a) to (c).
90. The microfluidic system of item 87 or 88, wherein the droplet actuator
comprises one
or more of the following:
i. a bottom substrate and a top substrate separated by a droplet operations
gap;
ii. an electrode arrangement comprising one or more of paths, reaction
lanes, and
an array of droplet operations electrodes;
iii. a plurality of fluid reservoirs interconnected through the electrode
arrangement
configured for dispensing separated fluids along the electrodes,;
iv. a plurality of temperature control zones;
-103-
Date Recue/Date Received 2020-10-05
v. one or more biochemical reaction zones for performing certain processing
steps
for each nucleic acid amplification reaction; or
vi. one or more magnets movable from and into proximity to one or more of
the
droplet operations electrodes.
91. The microfluidic system of item 89, wherein the droplet actuator
comprises the bottom
substrate and the top substrate separated by the droplet operations gap,
wherein either one or both of the bottom and the top substrate comprising
electrodes
configured for conducting droplet operations in the gap; and/or
wherein the droplet operations gap is filled with a filler fluid or a filler
gas.
92. The microfluidic system of item 90, wherein the filler fluid is
selected from the group
consisting of a silicone oil, a hexadecane filler fluid, a halogenated oil, a
fluorinated, and a
perfluorinated oil.
93. The microfluidic system of item 89, wherein the plurality of fluid
reservoirs
interconnected through the electrode arrangement configured for dispensing
separated fluids
along the electrodes, comprises one or more reagent reservoirs, one or more
sample
reservoirs, one or more index reservoirs, one or more waste reservoirs, or a
combination
thereof.
94. The microfluidic system of item 89, wherein the one or more magnets
movable from
and into proximity to one or more of the droplet operations electrodes, are
permanent
magnets or electromagnets.
95. The microfluidic system of any one of items 88 to 93, wherein the
controller comprises
a program code, a processor for executing the program code, and a local memory
in
communication with the processor, wherein the program code causes the
processor to
execute a method of multiplexed amplification of nucleic acid samples, the
method
comprising:
a) loading a plurality of input samples onto a droplet operations surface of
the droplet
actuator having droplet operations electrodes arranged thereon, each of the
plurality of input
samples comprising target nucleic molecules;
b) dispensing a normalization reagent droplet comprising a plurality of first
normalization primers and a plurality of second amplification primers,
wherein:
-104-
Date Recue/Date Received 2020-10-05
i. the plurality of first normalization primers is immobilized on a solid
support,
the first normalization primers being capable of specifically hybridizing to a
first sequence of the target nucleic acid molecules, and
ii. the plurality of second normalization primers is in solution, the
plurality of
second primers being capable of specifically hybridizing to a second
sequence of the target nucleic acid molecules;
and
c) amplifying the target nucleic acid molecules under isothermal conditions,
wherein
the plurality of first normalization primers is provided in an amount which
limits the yield of
amplification products to a predefined amount.
96. The microfluidic system of any one of items 87 to 94, wherein the method
of
multiplexed amplification of nucleic acid samples further comprising prior to
step (a):
dispensing a first enrichment PCR reagent droplet onto a droplet operations
surface of
the droplet actuator, wherein the first enrichment PCR reagent droplet
comprising
a first target-specific primer and a second target-specific primer; and
thermally cycling the target nucleic acid molecules in the plurality of input
samples to
form enriched nucleic acid samples.
97. The microfluidic system of any one of items 87 to 95, wherein the method
of
multiplexed amplification of nucleic acid samples further comprising prior to
step (a):
dispensing a second enrichment PCR reagent droplet onto a droplet operations
surface of the droplet actuator, wherein the second enrichment PCR reagent
droplet comprising a first universal primer and a second universal primer; and
thermally cycling the target nucleic acid molecules in the plurality of input
samples to
form enriched nucleic acid samples.
98. The microfluidic system of any one of items 87 to 96, wherein the method
of
multiplexed amplification of nucleic acid samples further comprising prior to
step (a):
dispensing a first enrichment PCR reagent droplet onto a droplet operations
surface of
the droplet actuator, wherein the first enrichment PCR reagent droplet
comprising
a first target-specific primer and a second target-specific primer;
-105-
Date Recue/Date Received 2020-10-05
dispensing a second enrichment PCR reagent droplet onto a droplet operations
surface of the droplet actuator, wherein the second enrichment PCR reagent
droplet comprising a first universal primer and a second universal primer;
combining the second enrichment PCR reagent droplet with the first enrichment
PCR
reagent droplet using droplet operations to form a combined enrichment PCR
reagent droplet; and
thermally cycling the target nucleic acid molecules in the plurality of input
samples to
form enriched nucleic acid samples.
99. The microfluidic system of any one of items 87 to 97, further
comprising one or more
magnets movable from and into proximity to one or more of the fluid
reservoirs.
100. The microfluidic system of item 98, wherein the positions of magnets are
controlled by
a motor.
101. The microfluidic system of any one of items 87 to 99, further comprising
one or more
heating devices to providing thermal control thereof.
102. The microfluidic system of any one of items 87 to 100, wherein the
program code is
partially or entirely stored in a local memory of the controller or on a
remote computing
device.
103. The microfluidic system of any one of items 87 to 101, wherein the
program code is
locally and/or remotely executed.
104. The microfluidic system of any one of items 87 to 102, wherein the
droplet actuator
comprising:
a) a bottom substrate and a top substrate separated by a droplet operations
gap,
wherein either one or both of the bottom and the top substrate comprising
electrodes
configured for conducting droplet operations in the gap;
b) an electrode arrangement comprising one or more of paths, reaction lanes,
and an
array of droplet operations electrodes;
c) a plurality of fluid reservoirs interconnected through the electrode
arrangement
configured for dispensing separated fluids along the electrodes; and
d) a plurality of temperature control zones.
105. The microfluidic system of item 103, wherein the droplet operations gap
is filled with a
filler fluid or a filler gas.
-106-
Date Recue/Date Received 2020-10-05
106. The microfluidic system of item 104, wherein the filler fluid is selected
from the group
consisting of a silicone oil, a hexadecane filler fluid, a halogenated oil, a
fluorinated, and a
perfluorinated oil.
107. The microfluidic system of any one of items 103 to 105, wherein the
plurality of fluid
reservoirs comprises one or more reagent reservoirs, one or more sample
reservoirs, one or
more index reservoirs, one or more waste reservoirs, or a combination thereof
108. The microfluidic system of any one of items 103 to 106, wherein the
droplet actuator
further comprising one or more biochemical reaction zones for performing
certain processing
steps for each nucleic acid amplification reaction.
109. The microfluidic system of any one of items 103 to 107, wherein at least
one of the
fluid reservoirs comprises an input port for loading fluids therein.
110. The microfluidic system of any one of items 103 to 108, the droplet
actuator further
comprising one or more magnets movable from and into proximity to one or more
of the
droplet operations electrodes.
111. The microfluidic system of any one of items 103 to 109, wherein the
magnets are
permanent magnets or electromagnets.
112. The microfluidic system of any one of items 103 to 110, wherein the
temperature
control zones comprise differing temperature from one another.
113. The microfluidic system of any one of items 103 to 111, wherein the
temperature
control zones include essentially the same temperature.
114. The microfluidic system of any one of items 103 to 112, wherein the
electrode
arrangement comprises one or more of dispensing, transporting, merging,
incubating,
splitting, mixing operation electrodes, or combinations thereof.
115. A computer readable medium storing processor executable instructions for
performing
a method of multiplexed nucleic acid amplification on a droplet actuator, the
method
comprising:
a)
loading a plurality of input samples onto a droplet operations surface of the
droplet
actuator having droplet operations electrodes arranged thereon, each of the
plurality of input
samples comprising target nucleic molecules;
b) dispensing a normalization reagent droplet comprising a plurality of first
normalization primers and a plurality of second normalization primers,
wherein:
-107-
Date Recue/Date Received 2020-10-05
i. the plurality of first normalization primers is immobilized on a solid
support,
the first normalization primers being capable of specifically hybridizing to a
first sequence of the target nucleic acid molecules, and
ii. the plurality of second normalization primers is in solution, the
plurality of
second primers being capable of specifically hybridizing to a second
sequence of the target nucleic acid molecules;
and
c) amplifying the target nucleic acid molecules under isothermal conditions,
wherein
the plurality of first normalization primers is provided in an amount which
limits the yield of
amplification products to a predefined amount.
-108-
Date Recue/Date Received 2020-10-05