Note: Descriptions are shown in the official language in which they were submitted.
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
HUMAN CYTOMEGALOVIRUS GB POLYPEPTIDE
CROSS REFERENCE TO RELATED APPLICATIONS
This application claims priority to United States Provisional Patent
Application
.. Serial No. 62/307,423 filed on March 11, 2016, the contents of which is
incorporated
in its entirety by reference herein.
FIELD OF THE INVENTION
The present invention relates to human cytomegalovirus (HCMV)
compositions and methods thereof.
BACKGROUND
Human cytomegalovirus (HCMV) is a double stranded DNA virus of the 13-
herpesvirus family. HCMV is the leading cause of congenital and neonatal
hearing
loss resulting from vertical virus transmission following infection or
reactivation of
latent virus in pregnant women. In addition, HCMV is a common opportunistic
pathogen affecting immunosuppressed transplant patients, such as solid organ /
stem cell transplant patients, AIDS patients, etc. Though development of a
vaccine
against HCMV has been listed as a top priority by the Institute of Medicine,
none has
been licensed to date.
The HCMV genome encodes several envelope glycoproteins, one of which is
glycoprotein B (gB). Glycoprotein B is an important surface target for
neutralizing
antibody (nAb) response in natural infection and is required for virus entry
into cells
by functioning as a fusogen.
HCMV subunit vaccines incorporating gB have been under development.
Studies have suggested that gB subunit vaccines were safe and immunogenic,
though further improvements in potency and durability of protection were
desirable.
Accordingly, safe and effective immunogenic compositions against
cytomegalovirus infections, as well diagnostic reagents capable of detecting
immunogenic stimuli resulting from CMV infections, guiding the design of gB-
based
HCMV vaccines, and/or supporting the development of therapeutic antibodies
against this medically relevant human pathogen are needed.
1
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
SUMMARY OF THE INVENTION
To meet these and other needs, in one aspect, the present invention relates
to a polypeptide that includes at least one mutation in the fusion loop 1 (F
Li) region
.. of an HCMV gB polypeptide.
In one aspect, the invention relates to a polypeptide that includes at least
one
mutation in the fusion loop 2 (FL2) region of an HCMV gB polypeptide.
In one aspect, the invention relates to a polypeptide that includes at least
one
mutation in the fusion loop 1 (FL1) region and the fusion loop 2 (FL2) region
of an
HCMV gB polypeptide.
In one aspect, the invention relates to a polypeptide that includes at least
one
mutation in the furin-like cleavage site of an HCMV gB polypeptide.
In one aspect, the invention relates to a polypeptide that includes at least
two
mutations in the fusion loop 2 (FL2) region of an HCMV gB polypeptide.
In one aspect, the invention relates to a polypeptide that includes at least
two
mutations in the fusion loop 1 (FL1) region and the fusion loop 2 (FL2) region
of an
HCMV gB polypeptide.
In one aspect, the invention relates to a polypeptide that includes at least
two
mutations in the furin-like cleavage site of an HCMV gB polypeptide.
In one aspect, the invention relates to a polypeptide that includes a mutation
at position Y155, as compared to SEQ ID NO: 6.
In one aspect, the invention relates to a polypeptide that includes a mutation
at positions Y155 and 1156, as compared to SEQ ID NO: 6.
In one aspect, the invention relates to a polypeptide that includes a mutation
at positions Y155, 1156, and H157, as compared to SEQ ID NO: 6.
In one aspect, the invention relates to a polypeptide that includes a mutation
at positions 1156 and H157, as compared to SEQ ID NO: 6.
2
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
In one aspect, the invention relates to a polypeptide that includes a mutation
at positions Y155, 1156, H157, and W240, as compared to SEQ ID NO: 6.
In one aspect, the invention relates to a polypeptide that includes a mutation
at positions Y155 and W240, as compared to SEQ ID NO: 6.
In one aspect, the invention relates to a polypeptide that includes a mutation
at positions Y155, H157, and W240, as compared to SEQ ID NO: 6.
In one aspect, the invention relates to a polypeptide that includes a mutation
at positions Y155 and H157, as compared to SEQ ID NO: 6.
In one aspect, the invention relates to a polypeptide that includes the
mutation
Y155G, as compared to SEQ ID NO: 6.
In one aspect, the invention relates to a polypeptide that includes the
mutations Y155G and I156H, as compared to SEQ ID NO: 6.
In one aspect, the invention relates to a polypeptide that includes the
mutations Y155G, I156H, and H157R, as compared to SEQ ID NO: 6.
In one aspect, the invention relates to a polypeptide that includes the
mutations I156H and H157R, as compared to SEQ ID NO: 6.
In one aspect, the invention relates to a polypeptide that includes the
mutations Y155G, I156H, H157R, and W240A, as compared to SEQ ID NO: 6.
In one aspect, the invention relates to a polypeptide that includes the
mutations Y155G and W240A, as compared to SEQ ID NO: 6.
In one aspect, the invention relates to a polypeptide that includes the
mutations Y155G, H157R, and W240A, as compared to SEQ ID NO: 6.
In one aspect, the invention relates to a polypeptide that includes the
mutations Y155G and H157R, as compared to SEQ ID NO: 6.
3
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
In one aspect, the invention relates to a polypeptide that includes an amino
acid sequence that is at least about 60% identical to SEQ ID NO: 1.
In one aspect, the invention relates to a polypeptide that includes an amino
acid sequence that is at least about 60% identical to SEQ ID NO: 2.
In one aspect, the invention relates to a polypeptide that includes an amino
acid sequence that is at least about 60% identical to SEQ ID NO: 3.
In one aspect, the invention relates to a polypeptide that includes an amino
acid sequence that is at least about 60% identical to SEQ ID NO: 5.
In one aspect, the invention relates to a polypeptide that includes an amino
acid sequence that is at least about 60% identical to SEQ ID NO: 7.
In one aspect, the invention relates to a polypeptide that includes an amino
acid sequence that is at least about 60% identical to SEQ ID NO: 8.
In one aspect, the invention relates to a polypeptide that includes the amino
acid sequence set forth in SEQ ID NO: 1.
In one aspect, the invention relates to a polypeptide that includes the amino
acid sequence set forth in SEQ ID NO: 2.
In one aspect, the invention relates to a polypeptide that includes the amino
acid sequence set forth in SEQ ID NO: 3.
In one aspect, the invention relates to a polypeptide that includes the amino
acid sequence set forth in SEQ ID NO: 5.
In one aspect, the invention relates to a polypeptide that includes the amino
acid sequence set forth in SEQ ID NO: 7.
In one aspect, the invention relates to a polypeptide that includes the amino
acid sequence set forth in SEQ ID NO: 8.
4
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
In one embodiment, the polypeptide does not include a mutation at any one of
the following positions: (i) R236, (ii) G237, (iii) T158; (iv) Y242. In one
embodiment,
the polypeptide does not include any one of the following mutations: (i)
R236N, (ii)
G237N, (iii) T158N; (iv) Y242T; (v) Y242S; (vi) Y2420. In one embodiment, the
amino acid sequence SEQ ID NO: 9 is a part of the polypeptide sequence. In one
embodiment, the amino acid sequence SEQ ID NO: 10 is a part of the polypeptide
sequence. In one embodiment, the amino acid sequences of SEQ ID NO: 9 and
SEQ ID NO: 10 is a part of the polypeptide sequence. In one embodiment, the
polypeptide does not include a protease cleavage site. In one embodiment, the
polypeptide does not include a wild-type CMV protease cleavage site. In one
embodiment, the polypeptide does not include a non-naturally occurring
protease
cleavage site that replaces the wild-type CMV protease cleavage site. In one
embodiment, the polypeptide does not include an N- glycosylation site that
includes
N-X-S/T/C motif, wherein X is any amino acid residue. In one embodiment, the
polypeptide does not include a modified amino acid sequence that introduces an
0-
linked glycosylation site. In one embodiment, the polypeptide does not include
a
deletion or substitution of any one of the amino acid residues selected from
the
group consisting of 154, 158, 159, 160, 230, 231, 232, 233, 234, 235, 236,
237, 238,
239, 241, and 242, according to the numbering of SEQ ID NO: 6. In one
embodiment, the polypeptide does not include a mutation of any one of the
amino
acid residues: Y160, R236, S238, T239, and Y242, according to the numbering of
SEQ ID NO: 6. In one embodiment, the polypeptide does not include the
cytoplasmic tail of HCMV gB.
5
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
In one embodiment, the polypeptide does not contain an insect cell pattern of
glycosylation. In one embodiment, the polypeptide is contacted with
ethylenediaminetetraacetic acid (EDTA).
In one aspect, the invention relates to a composition that includes the
.. polypeptide described herein, and a diluent.
In one aspect, the invention relates to a composition that includes the
polypeptide described herein, and an adjuvant. In one embodiment, the
composition
is immunogenic. In one embodiment, the composition is for use in inducing an
immune response against cytomegalovirus.
In one aspect, the invention relates to a recombinant nucleic acid molecule
encoding the polypeptide described herein, wherein the polypeptide undergoes a
structural conformation change in response to a pH change. In one embodiment,
said recombinant nucleic acid (a) is not a self-replicating RNA molecule; (b)
is not an
alphavirus replicon; (c) does not encode any alphavirus nonstructural
proteins, such
as NSP1, NSP2, NSP3 and NSP4; (d) does not contain: an Internal Ribosomal
Entry
Site (IRES), such as EMCV or EV71; and/or (e) does not contain a viral 2A
site, such
as FMDV.
In one aspect, the invention relates to a method for raising antibodies using
the polypeptide described herein. In one embodiment, said antibody is for use
in a
diagnostic assay. In one embodiment, said antibody is labelled directly or
indirectly.
In one embodiment, said antibody is for use in therapy.
In one aspect, the invention relates to a method of eliciting an immune
response in a mammal, the method that includes administering to the mammal an
effective amount of the polypeptide described herein.
6
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
BRIEF DESCRIPTION OF THE DRAWINGS
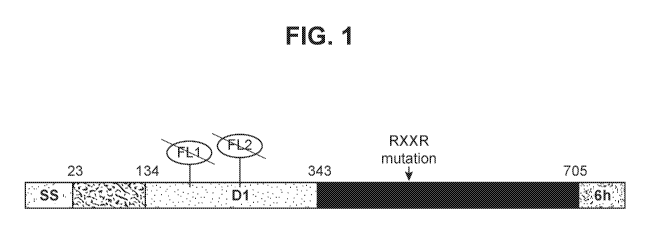
FIG. 1- illustration of HCMV gB705(SEQ ID NO: 1). SS represents a signal
sequence; D1 represents the Domain I; FL represents a fusion loop; RxxR
mutation
represents knock-out mutations of the furin-like cleavage site; 6h represents
an
optional 6xHis tag.
FIG. 2 - sequence of HCMV gB705 (SEQ ID NO: 2).
FIG. 3 - The gB705 protein was transiently expressed in Expi293 and 293T cells
after transfection respectively. The target protein was secreted into culture
medium,
precipitated by an affinity tag, and resolved on SDS-PAGE under reducing
condition.
The presence of the gB705 protein was visualized by a Western blot analysis
using
poly-clonal anti-gB serum. As shown, gB705 exhibited a single band with an
apparent molecular weight about 130kDa, consistent with the design of the
furin-
cleavage site being knocked out by mutations.
FIG. 4A-B - FIG. 4A Binding of human anti-HCMV gB monoclonal antibodies to
gB705 protein; FIG. 4B Binding of human anti-HCMV gB monoclonal antibodies to
Sino gB protein (gB from Sino Biologicals, Inc.) (ELN# 00710043-0177).
FIG. 5 - HCMV gB705 protein (SEQ ID NO: 2) was analyzed on SDS-PAGE and
exhibited a single band under a fully reduced condition and showed one
additional
high molecular weight band under partially reduced conditions, representing a
trimer.
FIG. 6- Sedimentation Velocity analysis of gB705 (ELN# 00708337-0110).
FIG. 7 - Velocity sedimentation analysis of 10 mM EDTA-treated gB705 (ELN#
00708337-0110).
FIG. 8A-C- Sedimentation velocity analysis of EDTA-treated gB705 at pH 5.2
(FIG.
8A) and pH 8.7 (FIG. 8B); Cryo electron microscopy analysis of EDTA-treated
gB705
proteins pH 5.2 (left) or pH 8.7 (right) (FIG. 8C)
FIG. 9A-B - FIG. 9A Far UV CD analysis; FIG. 9B intrinsic fluorescence
spectroscopy (ELN# 00708337-0115, 0117) of gB705 at pH 5.2 and pH 8.7.
7
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
FIG. 10- Near UV CD spectroscopy analysis of gB705 at pH 5.2 and pH 8.7.
Buffer
1, 50 mM citrate, 100 mM phosphate; Buffer 2, 50 mM Na0Ac; Buffer 3, 100 mM Na
carbonate/bicarbonate; Buffer 4, 50 mM Na borate (ELN# 00708337-0115, 0117).
FIG. 11A-B - DSC analysis of gB705 at (FIG. 11A) pH 5.2 and (FIG. 11B) pH 8.7.
FIG. 12A-C - (A) ANS binding of gB705 at pH 5.2, pH 7.4 and pH 8.7 (ELN#
00708337-0115, 0117); (B) ANS binding of gB705 at pH 8.7 and 5.2 and shifted
from
pH 8.7 to pH 5.2; (C) ANS binding of gB705 at pH 8.7 and 5.2 and shifted from
pH
5.2 to pH 8.7 (ELN# 00708329-0153)
FIG. 13- illustration of RhCMV gB674 (SEQ ID NO: 7). SS represents a signal
sequence; FL represents a fusion loop; 6h represents a 6xHis tag
FIG. 14- Graph showing anti-gB binding titers from six vaccines on week 9
after
three doses of vaccination
FIG. 15A-C - Six RhCMV(-) monkeys were immunized with placebo (A), recombinant
RhCMV pentamer (B), or pentamer + RhCMV gB674 (C) intramuscularly in QS-21
adjuvant at week 0, 4 & 8. The animals were then challenged with live RhCMV
(UCDE52 strain) orally for five times. Virus shedding in saliva samples were
measured using qPCR (low limit of quantitation (LLOQ) is 125 DNA copy per ml)
after virus challenge. Week 9 was one week after the third vaccination and one
week prior to the first oral challenge. Week 15 was first sample at one week
post the
fifth virus challenge.
FIG. 16- TEM image of negative staining of gB705 (62000X magnification) (ELN#
00702423-0124)
FIG. 17A-B - (A) SDS-PAGE analysis of rhesus gB674, demonstrating the lack of
protease cleavage as designed and >95% purity, NR = non-reducing; R =
reducing;
(B) Size Exclusion chromatography of rhesus gB674 protein showing elution as a
major peak (MW -313 kDa), consistent with a homogenous individual trimer
8
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
SEQUENCE IDENTIFIERS
SEQ ID NO: 1 sets forth the amino acid sequence for HCMV VR1814 gB705.
SEQ ID NO: 2 sets forth the amino acid sequence for HCMV VR1814 gB705, without
an initial methionine. SEQ ID NO: 2 is identical to SEQ ID NO: 1 but for the
absence
of the initial methionine.
SEQ ID NO: 3 sets forth the amino acid sequence for a fragment of HCMV VR1814
gB705 (residues 25-705 OF SEQ ID NO: 1).
SEQ ID NO: 4 sets forth the amino acid sequence for the signal sequence of an
HCMV VR1814 gB.
SEQ ID NO: 5 sets forth the amino acid sequence for HCMV VR1814 gB705 with
linker
SEQ ID NO: 6 sets forth the amino acid sequence for GenBank Accession #
A0Z79977.1 for envelope glycoprotein B of strain VR1814 [Human beta-
herpesvirus
5].
SEQ ID NO: 7 sets forth the amino acid sequence for an analogous construct of
HCMV gB705 construct, named as RhCMV gB674, which was designed and tested
in RhCMV(-) monkeys
SEQ ID NO: 8 sets forth the amino acid sequence for an analogous construct of
HCMV gB705 construct, named as RhCMV gB674, which was designed and tested
in RhCMV(-) monkeys, without an initial methionine. SEQ ID NO: 8 is identical
to
SEQ ID NO: 7 but for the absence of the initial methionine.
SEQ ID NO: 9 sets forth the amino acid sequence for VVDPLPP705, a part of the
membrane proximal region (MPR) of HCMV gB.
SEQ ID NO: 10 sets forth the amino acid sequence for RA457TKA4595.
SEQ ID NO: 11 sets forth the amino acid sequence for GenBank Accession #
GU552457.1 for macacine herpesvirus 3 isolate 21252 glycoprotein B (RhUL55).
9
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
SEQ ID NO: 12 sets forth the amino acid sequence for the predicted fusion loop
1
region of the envelope glycoprotein B of strain VR1814 [Human beta-herpesvirus
5],
from GenBank Accession # ACZ79977.1.
SEQ ID NO: 13 sets forth the amino acid sequence for the predicted fusion loop
2
region of the envelope glycoprotein B of strain VR1814 [Human beta-herpesvirus
5],
based on GenBank Accession # ACZ79977.1.
SEQ ID NO: 14 sets forth the amino acid sequence for the furin-like cleavage
site of
the envelope glycoprotein B of strain VR1814 [Human beta-herpesvirus 5], based
on
GenBank Accession # ACZ79977.1.
SEQ ID NO: 15: sets forth the amino acid sequence for the protease cleavage
site of
the envelope glycoprotein B of strain VR1814 [Human beta-herpesvirus 5], based
on
GenBank Accession # ACZ79977.1.
10
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
DETAILED DESCRIPTION
The inventors surprisingly discovered mutations that can be introduced into a
cytomegalovirus (CMV) gB polypeptide, which can, among other things, greatly
facilitate the production and subsequent purification of a stable gB antigen;
significantly improve the efficiency of production of a gB polypeptide;
maintain and/or
increase antigenicity of a gB polypeptide, as compared to the wild-type gB
polypeptide; facilitate a focused immune response to gB; and reduce and/or
eliminate steric occlusion of neutralizing epitopes of gB. The modified gB
polypeptide also surprisingly demonstrated strong structural integrity and a
potential
to undergo structural conformational changes in tertiary structure in response
to
significant pH changes. The structural changes were strictly pH-dependent.
Without
being bound by theory, the potential to undergo structural conformational
changes in
response to significant pH changes may be a newly recognized property of the
HCMV gB ectodomain.
gB is an envelope glycoprotein B having numerous roles, one of which is the
involvement in the fusion of the cytomegalovirus with host cells. It is
encoded by the
UL55 gene of HCMV genome.
In one aspect, the invention relates to a modified HCMV gB that is based on
the VR1814 gB sequence (GenBank# ACZ79977.1; SEQ ID NO: 6). However, the
.. present invention is applicable to gB proteins originating from any CMV
strain.
Unless otherwise stated, references to the numbering of amino acid residue
positions of a CMV gB polypeptide as used herein are in relation to the amino
acid
sequence of the gB protein of SEQ ID NO: 6, from the clinical isolate VR1814
strain.
Comparable amino acid positions in a gB protein of any other CMV strains can
be
.. determined by those of ordinary skill in the art. Accordingly, the term
"CMV gB
protein" or "HCMV gB protein" as used herein is to be understood as a HCMV gB
protein from any human HCMV strain (not limited to the VR1814 strain). The
actual
residue position number may need to be adjusted for gB proteins from other
human
CMV strains depending on the actual sequence alignment.
In one embodiment, the modified HCMV gB polypeptide includes amino acids
1-705 of SEQ ID NO: 6. In another embodiment, the modified HCMV gB polypeptide
includes at least one mutation in the fusion loop 1 region of an HCMV gB
polypeptide. The precise boundaries of the fusion loop 1 region of a wild-type
VR1814 gB polypeptide are currently not fully defined, however, the predicted
11
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
sequence of the fusion loop 1 region of a wild-type VR1814 gB polypeptide is
Y153AYIHT158 (SEQ ID NO: 12).
In another embodiment, the modified HCMV gB polypeptide includes at least
one mutation in the fusion loop 2 region of an HCMV gB polypeptide. The
precise
.. boundaries of the fusion loop 2 region of a wild-type VR1814 gB polypeptide
are
currently not fully defined, however, the predicted sequence of the fusion
loop 2
region of a wild-type VR1814 gB polypeptide is G237STWLYRE244 (SEQ ID NO: 13).
In a preferred embodiment, the modified HCMV gB polypeptide includes at
least one mutation in the fusion loop 1 region and in the fusion loop 2 region
of an
.. HCMV gB polypeptide. In another preferred embodiment, the modified HCMV gB
polypeptide includes a total of at most four mutations in the fusion loop 1
region and
in the fusion loop 2 region of an HCMV gB polypeptide.
In another embodiment, the modified HCMV gB polypeptide includes at least
one mutation in the furin-like cleavage site of an HCMV gB polypeptide. In a
preferred embodiment, the modified HCMV gB polypeptide includes at most two
mutations in the furin-like cleavage site of an HCMV gB polypeptide.
In another preferred embodiment, the modified HCMV gB polypeptide
includes at least one mutation in the fusion loop 1 region, at least one
mutation in the
fusion loop 2 region, and at least one mutation in the furin-like cleavage
site of an
HCMV gB polypeptide. The sequence of the furin-like cleavage site of a wild-
type
VR1814 gB polypeptide is R458TKR459 (SEQ ID NO: 14).
In another preferred embodiment, the modified HCMV gB polypeptide
includes at least two mutations in the fusion loop 1 region, at least one
mutation in
the fusion loop 2 region, and at least one mutation in the furin-like cleavage
site of an
HCMV gB polypeptide.
In one aspect, the invention relates to a CMV gB polypeptide or immunogenic
fragment thereof having a mutation at position Y155, as compared to SEQ ID NO:
6.
In one aspect, the invention relates to a CMV gB polypeptide or immunogenic
fragment thereof having a mutation at positions Y155 and 1156, as compared to
SEQ
ID NO: 6.
In one aspect, the invention relates to a CMV gB polypeptide or immunogenic
fragment thereof having a mutation at positions Y155, 1156, and H157, as
compared
to SEQ ID NO: 6.
12
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
In one aspect, the invention relates to a CMV gB polypeptide or immunogenic
fragment thereof having a mutation at positions 1156 and H157, as compared to
SEQ
ID NO: 6.
In one aspect, the invention relates to a CMV gB polypeptide or immunogenic
fragment thereof having a mutation at positions Y155,1156, H157, and W240, as
compared to SEQ ID NO: 6.
In one aspect, the invention relates to a CMV gB polypeptide or immunogenic
fragment thereof having a mutation at positions Y155 and W240, as compared to
SEQ ID NO: 6.
In one aspect, the invention relates to a CMV gB polypeptide or immunogenic
fragment thereof having a mutation at positions Y155, H157, and W240, as
compared to SEQ ID NO: 6.
In one aspect, the invention relates to a CMV gB polypeptide or immunogenic
fragment thereof having a mutation at positions Y155 and H157, as compared to
SEQ ID NO: 6.
In one aspect, the invention relates to a CMV gB polypeptide or immunogenic
fragment thereof having the mutation Y155G, as compared to SEQ ID NO: 6.
In one aspect, the invention relates to a CMV gB polypeptide or immunogenic
fragment thereof having the mutations Y155G and I156H, as compared to SEQ ID
NO: 6.
In one aspect, the invention relates to a CMV gB polypeptide or immunogenic
fragment thereof having the mutations Y155G, I156H, and H157R, as compared to
SEQ ID NO: 6.
In one aspect, the invention relates to a CMV gB polypeptide or immunogenic
fragment thereof having the mutations I156H and H157R, as compared to SEQ ID
NO: 6.
In one aspect, the invention relates to a CMV gB polypeptide or immunogenic
fragment thereof having the mutations Y155G, I156H, H157R, and W240A, as
compared to SEQ ID NO: 6.
In one aspect, the invention relates to a CMV gB polypeptide or immunogenic
fragment thereof having the mutations Y155G and W240A, as compared to SEQ ID
NO: 6.
13
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
In one aspect, the invention relates to a CMV gB polypeptide or immunogenic
fragment thereof having the mutations Y155G, H157R, and W240A, as compared to
SEQ ID NO: 6.
In one aspect, the invention relates to a CMV gB polypeptide or immunogenic
fragment thereof having the mutations Y155G and H157R, as compared to SEQ ID
NO: 6.
In one embodiment, the gB polypeptide does not include a mutation at any
one of the following positions, individually or in combination: (i) R236, (ii)
G237, (iii)
T158; (iv) Y242.
For example, in one embodiment, the gB polypeptide does not include any
one of the following mutations, individually or in combination: (i) R236N,
(ii) G237N,
(iii) T158N; (iv) Y242T; (v) Y2425; (vi) Y242C.
14
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
Exemplary Modified Human CMV gB polypeptides
In one aspect, the invention relates to a polypeptide having the amino acid
sequence that is at least about 60%, 65%, 70%, 75%, 80%, 81%, 82%, 83%, 84%.
85%, 86%, 87%, 88%, 89%, 90%, 91 %, 92%, 93%, 94%, 95%, 96%, 97%, 98%,
99%, or 100% identical to SEQ ID NO: 1.
In one embodiment, the polypeptide having at least about 60%, 65%, 70%,
75%, 80%, 81%, 82%, 83%, 84%. 85%, 86%, 87%, 88%, 89%, 90%, 91 %, 92%,
93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity to SEQ ID NO:
1, includes the amino acid sequence VVDPLPP705 (SEQ ID NO: 9) as a part of the
polypeptide sequence.
Without being bound by theory, the sequence set forth in SEQ ID NO: 9 may
be highly hydrophobic and structurally stiff, which facilitates immunogenicity
of the
polypeptide. Accordingly, in one embodiment, the sequence set forth in SEQ ID
NO:
9 is hydrophobic.
In another embodiment, the polypeptide having at least about 60%, 65%,
70%, 75%, 80%, 81%, 82%, 83%, 84%. 85%, 86%, 87%, 88%, 89%, 90%, 91 %,
92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity to SEQ ID
NO: 1, includes the amino acid sequence RA457TKA4595 (SEQ ID NO: 10) as a part
of the polypeptide sequence.
Without being bound by theory, the sequence set forth in SEQ ID NO: 10 may
prevent cleavage of the gB polypeptide by cellular proteases. Preferably, the
polypeptide of the invention does not include a protease cleavage site. For
example,
in one embodiment, the polypeptide does not include a wild-type, natural CMV
protease cleavage site, e.g., RTKR459 (SEQ ID NO: 15) for the envelope
glycoprotein
B of strain VR1814 [Human beta-herpesvirus 5], based on GenBank Accession #
A0Z79977.1. In another embodiment, the polypeptide does not include a non-
naturally occurring protease cleavage site. In yet another embodiment, the
polypeptide does not include a non-naturally occurring protease cleavage site
that
replaces the wild-type, natural CMV protease cleavage site. In one embodiment,
the
polypeptide does not include a protease cleavage site at or encompassing
residue
R459 at position 459 of a human CMV gB, according to the numbering of SEQ ID
NO:
6, wherein the protease cleavage site includes at most 20, 19, 18, 17, 16, 15,
14, 13,
12, 11, 10, 9, 8, 7, 6, 5, 4, 0r3 amino acid residues in length. For example,
the
polypeptide does not include a protease cleavage site encompassing residue
R459 at
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
position 459 of a human CMV gB, according to the numbering of SEQ ID NO: 6,
wherein the protease cleavage site includes at most 6, more preferably at most
5,
and most preferably at most 4 amino acid residues in length.
In another embodiment, the polypeptide having at least about 60%, 65%,
70%, 75%, 80%, 81%, 82%, 83%, 84%. 85%, 86%, 87%, 88%, 89%, 90%, 91 %,
92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity to SEQ ID
NO: 1, includes both the amino acid sequence VVDPLPID705 (SEQ ID NO: 9) and
the
amino acid sequence RA457TKA4595 (SEQ ID NO: 10) as a part of the polypeptide
sequence.
16
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
In one aspect, the invention relates to a polypeptide having the amino acid
sequence that is at least about 60%, 65%, 70%, 75%, 80%, 81%, 82%, 83%, 84%.
85%, 86%, 87%, 88%, 89%, 90%, 91 %, 92%, 93%, 94%, 95%, 96%, 97%, 98%,
99%, or 100% identical to SEQ ID NO: 2.
In one embodiment, the polypeptide having at least about 60%, 65%, 70%,
75%, 80%, 81%, 82%, 83%, 84%. 85%, 86%, 87%, 88%, 89%, 90%, 91 %, 92%,
93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity to SEQ ID NO:
2, includes the amino acid sequence VVDPLPP705 (SEQ ID NO: 9) as a part of the
polypeptide sequence.
In another embodiment, the polypeptide having at least about 60%, 65%,
70%, 75%, 80%, 81%, 82%, 83%, 84%. 85%, 86%, 87%, 88%, 89%, 90%, 91 %,
92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity to SEQ ID
NO: 2, includes the amino acid sequence RA457TKA4595 (SEQ ID NO: 10) as a part
of the polypeptide sequence.
Preferably, the polypeptide does not include a protease cleavage site. For
example, in one embodiment, the polypeptide does not include a wild-type,
natural
CMV protease cleavage site, e.g., RTKR459 (SEQ ID NO: 15) for the envelope
glycoprotein B of strain VR1814 [Human beta-herpesvirus 5], based on GenBank
Accession # A0Z79977.1. In another embodiment, the polypeptide does not
include
a non-naturally occurring protease cleavage site. In yet another embodiment,
the
polypeptide does not include a non-naturally occurring protease cleavage site
that
replaces the wild-type, natural CMV protease cleavage site.
In another embodiment, the polypeptide having at least about 60%, 65%,
70%, 75%, 80%, 81%, 82%, 83%, 84%. 85%, 86%, 87%, 88%, 89%, 90%, 91 %,
92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity to SEQ ID
NO: 2, includes both the amino acid sequence VVDPLPP705 (SEQ ID NO: 9) and the
amino acid sequence RA457TKA4595 (SEQ ID NO: 10) as a part of the polypeptide
sequence.
17
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
In one aspect, the invention relates to a polypeptide having the amino acid
sequence that is at least about 60%, 65%, 70%, 75%, 80%, 81%, 82%, 83%, 84%.
85%, 86%, 87%, 88%, 89%, 90%, 91 %, 92%, 93%, 94%, 95%, 96%, 97%, 98%,
99%, or 100% identical to SEQ ID NO: 3.
In one embodiment, the polypeptide having at least about 60%, 65%, 70%,
75%, 80%, 81%, 82%, 83%, 84%. 85%, 86%, 87%, 88%, 89%, 90%, 91 %, 92%,
93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity to SEQ ID NO:
3, includes the amino acid sequence VVDPLPP705 (SEQ ID NO: 9) as a part of the
polypeptide sequence.
In another embodiment, the polypeptide having at least about 60%, 65%,
70%, 75%, 80%, 81%, 82%, 83%, 84%. 85%, 86%, 87%, 88%, 89%, 90%, 91 %,
92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity to SEQ ID
NO: 3, includes the amino acid sequence RA457TKA4595 (SEQ ID NO: 10) as a part
of the polypeptide sequence.
Preferably, the polypeptide does not include a protease cleavage site. For
example, in one embodiment, the polypeptide does not include a wild-type,
natural
CMV protease cleavage site, e.g., RTKR459 (SEQ ID NO: 15) for the envelope
glycoprotein B of strain VR1814 [Human beta-herpesvirus 5], based on GenBank
Accession # A0Z79977.1. In another embodiment, the polypeptide does not
include
.. a non-naturally occurring protease cleavage site. In yet another
embodiment, the
polypeptide does not include a non-naturally occurring protease cleavage site
that
replaces the wild-type, natural CMV protease cleavage site.
In another embodiment, the polypeptide having at least about 60%, 65%,
70%, 75%, 80%, 81%, 82%, 83%, 84%. 85%, 86%, 87%, 88%, 89%, 90%, 91 %,
92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity to SEQ ID
NO: 3, includes both the amino acid sequence VVDPLPP705 (SEQ ID NO: 9) and the
amino acid sequence RA457TKA4595 (SEQ ID NO: 10) as a part of the polypeptide
sequence.
18
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
In one aspect, the invention relates to a polypeptide having the amino acid
sequence that is at least about 60%, 65%, 70%, 75%, 80%, 81%, 82%, 83%, 84%.
85%, 86%, 87%, 88%, 89%, 90%, 91 %, 92%, 93%, 94%, 95%, 96%, 97%, 98%,
99%, or 100% identical to SEQ ID NO: 5.
In one embodiment, the polypeptide having at least about 60%, 65%, 70%,
75%, 80%, 81%, 82%, 83%, 84%. 85%, 86%, 87%, 88%, 89%, 90%, 91 %, 92%,
93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity to SEQ ID NO:
5, includes the amino acid sequence VVDPLPP705 (SEQ ID NO: 9) as a part of the
polypeptide sequence.
In another embodiment, the polypeptide having at least about 60%, 65%,
70%, 75%, 80%, 81%, 82%, 83%, 84%. 85%, 86%, 87%, 88%, 89%, 90%, 91 %,
92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity to SEQ ID
NO: 5, includes the amino acid sequence RA457TKA4595 (SEQ ID NO: 10) as a part
of the polypeptide sequence.
Preferably, the polypeptide does not include a protease cleavage site. For
example, in one embodiment, the polypeptide does not include a wild-type,
natural
CMV protease cleavage site, e.g., RTKR459 (SEQ ID NO: 15) for the envelope
glycoprotein B of strain VR1814 [Human beta-herpesvirus 5], based on GenBank
Accession # A0Z79977.1. In another embodiment, the polypeptide does not
include
a non-naturally occurring protease cleavage site. In yet another embodiment,
the
polypeptide does not include a non-naturally occurring protease cleavage site
that
replaces the wild-type, natural CMV protease cleavage site.
In another embodiment, the polypeptide having at least about 60%, 65%,
70%, 75%, 80%, 81%, 82%, 83%, 84%. 85%, 86%, 87%, 88%, 89%, 90%, 91 %,
92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity to SEQ ID
NO: 5, includes both the amino acid sequence VVDPLPP705 (SEQ ID NO: 9) and the
amino acid sequence RA457TKA4595 (SEQ ID NO: 10) as a part of the polypeptide
sequence.
19
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
Exemplary Modified Rhesus CMV gB polypeptides
In one aspect, the invention relates to a polypeptide having the amino acid
sequence that is at least about 60%, 65%, 70%, 75%, 80%, 81%, 82%, 83%, 84%.
85%, 86%, 87%, 88%, 89%, 90%, 91 %, 92%, 93%, 94%, 95%, 96%, 97%, 98%,
99%, or 100% identical to SEQ ID NO: 7.
In one embodiment, the polypeptide having at least about 60%, 65%, 70%,
75%, 80%, 81%, 82%, 83%, 84%. 85%, 86%, 87%, 88%, 89%, 90%, 91 %, 92%,
93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity to SEQ ID NO:
7, includes the amino acid sequence VVDPLPP (SEQ ID NO: 9) as a part of the
polypeptide sequence.
In another embodiment, the polypeptide having at least about 60%, 65%,
70%, 75%, 80%, 81%, 82%, 83%, 84%. 85%, 86%, 87%, 88%, 89%, 90%, 91 %,
92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity to SEQ ID
NO: 7, includes the amino acid sequence RATKAS (SEQ ID NO: 10) as a part of
the
polypeptide sequence. See, for example, residues 428-433 of SEQ ID NO: 7.
Preferably, the polypeptide does not include a protease cleavage site. For
example, in one embodiment, the polypeptide does not include a wild-type,
natural
CMV protease cleavage site. In another embodiment, the polypeptide does not
include a non-naturally occurring protease cleavage site. In yet another
embodiment, the polypeptide does not include a non-naturally occurring
protease
cleavage site that replaces the wild-type, natural CMV protease cleavage site.
In another embodiment, the polypeptide having at least about 60%, 65%,
70%, 75%, 80%, 81%, 82%, 83%, 84%. 85%, 86%, 87%, 88%, 89%, 90%, 91 %,
92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity to SEQ ID
NO: 7, includes both the amino acid sequence VVDPLPP (SEQ ID NO: 9) and the
amino acid sequence RATKAS (SEQ ID NO: 10) as a part of the polypeptide
sequence.
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
In one aspect, the invention relates to a polypeptide having the amino acid
sequence that is at least about 60%, 65%, 70%, 75%, 80%, 81%, 82%, 83%, 84%.
85%, 86%, 87%, 88%, 89%, 90%, 91 %, 92%, 93%, 94%, 95%, 96%, 97%, 98%,
99%, or 100% identical to SEQ ID NO: 8.
In one embodiment, the polypeptide having at least about 60%, 65%, 70%,
75%, 80%, 81%, 82%, 83%, 84%. 85%, 86%, 87%, 88%, 89%, 90%, 91 %, 92%,
93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity to SEQ ID NO:
8, includes the amino acid sequence VVDPLPP (SEQ ID NO: 9) as a part of the
polypeptide sequence.
In another embodiment, the polypeptide having at least about 60%, 65%,
70%, 75%, 80%, 81%, 82%, 83%, 84%. 85%, 86%, 87%, 88%, 89%, 90%, 91 %,
92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity to SEQ ID
NO: 8, includes the amino acid sequence RATKAS (SEQ ID NO: 10) as a part of
the
polypeptide sequence.
Preferably, the polypeptide does not include a protease cleavage site. For
example, in one embodiment, the polypeptide does not include a wild-type,
natural
CMV protease cleavage site. In another embodiment, the polypeptide does not
include a non-naturally occurring protease cleavage site. In yet another
embodiment, the polypeptide does not include a non-naturally occurring
protease
cleavage site that replaces the wild-type, natural CMV protease cleavage site.
In another embodiment, the polypeptide having at least about 60%, 65%,
70%, 75%, 80%, 81%, 82%, 83%, 84%. 85%, 86%, 87%, 88%, 89%, 90%, 91 %,
92%, 93%, 94%, 95%, 96%, 97%, 98%, 99%, or 100% sequence identity to SEQ ID
NO: 8, includes both the amino acid sequence VVDPLPP (SEQ ID NO: 9) and the
amino acid sequence RATKAS (SEQ ID NO: 10) as a part of the polypeptide
sequence.
21
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
GLYCOSYLATION
Although CMV gB may be referred to as a glycoprotein, this nomenclature
should not be taken to mean that the polypeptides described herein must be
glycosylated when used with the invention. In some embodiments of the
invention,
the modified gB polypeptide is not glycosylated.
In one preferred embodiment, the gB polypeptide does not include a
glycosylation site in the fusion loop regions of the gB polypeptide. That is,
the gB
polypeptide does not include a glycan moiety attached to the gB polypeptide
within
the fusion loop 1 region or within the fusion loop 2 region. The fusion loop 1
region
of the VR1814 (SEQ ID NO: 6) is located at residues 153-158. The fusion loop 2
region of the VR1814 (SEQ ID NO: 6) is located at residues 237-244.
In one preferred embodiment, the gB polypeptide does not include a modified
amino acid sequence that introduces an N-linked glycosylation site. For
example,
the modified gB polypeptide does not include an N- glycosylation site
comprising N-
X-S/T/C motif, wherein X is any amino acid residue.
In another preferred embodiment, the modified gB polypeptide does not
include a modified amino acid sequence that introduces an 0-linked
glycosylation
site. For example, the modified gB polypeptide does not include a carbohydrate
moiety linked to the hydroxyl oxygen of serine and threonine. As another
example,
the modified gB polypeptide does not include an 0- linked glycosylation at
tyrosine,
5-hydroxylysine, or 4-hydroxyproline.
22
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
UNMODIFIED HYDROPHOBIC SURFACE RESIDUES
In one preferred embodiment, the modified gB polypeptide does not include a
deletion or substitution of any one of the amino acid residues selected from
the
group consisting of 154, 158, 159, 160, 230, 231, 232, 233, 234, 235, 236,
237, 238,
239, 241, and 242, according to the numbering of SEQ ID NO: 6, or any
combination
thereof. For example, the gB polypeptide does not include a mutation of any
one of
the amino acid residues selected from the group consisting of Y160, R236,
S238,
T239, and Y242, according to the numbering of SEQ ID NO: 6, or any combination
thereof.
DELETION OF C-TERMINAL CYTOPLASMIC DOMAIN
The inventors further discovered that eliminating the cytoplasmic tail of HCMV
gB may help to focus the mammal's immune response to the ectodomains of gB.
Additionally, deletion of the cytoplasmic tail may reduce steric occlusion and
facilitate
access of antibodies to important neutralizing epitopes on gB.
Accordingly, in one embodiment, the native C-terminal cytoplasmic domain of
the gB polypeptide (e.g., 100% of the amino acids of the cytoplasmic domain)
is
deleted. For example, the native C-terminal cytoplasmic domain is not deleted
to a
varying extent. That is, no less than 100% of the native C-terminal
cytoplasmic
domain is deleted. As used herein, the cytoplasmic tail of HCMV gB refers the
amino acid sequence located at positions 771-905 of SEQ ID NO: 6.
23
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
EXPRESSION OF GB
In one aspect, the invention relates to nucleic acids that encode a gB
polypeptide of the invention. Preferably, the recombinant nucleic acid
molecule: (a)
is not a self- replicating RNA molecule; (b) is not an alphavirus replicon;
(c) does not
.. encode any alphavirus nonstructural proteins, such as NSP 1 , NSP2, NSP3
and
NSP4; (d) does not contain: an Internal Ribosomal Entry Site (IRES), such as
EIV1CV or EV71 ; and/or (e) does not contain a viral 2A site, such as FM DV.
In one embodiment, the invention relates to a nucleic acid sequence that
encodes an amino acid sequence that is at least about 60%, 65%, 70%, 75%, 80%,
81%, 82%, 83%, 84%. 85%, 86%, 87%, 88%, 89%, 90%, 91 %, 92%, 93%, 94%,
95%, 96%, 97%, 98%, 99%, or 100% identical to SEQ ID NO: 1.
In one embodiment, the invention relates to a nucleic acid sequence that
encodes an amino acid sequence that is at least about 60%, 65%, 70%, 75%, 80%,
81%, 82%, 83%, 84%. 85%, 86%, 87%, 88%, 89%, 90%, 91 %, 92%, 93%, 94%,
.. 95%, 96%, 97%, 98%, 99%, or 100% identical to SEQ ID NO: 2.
In one embodiment, the invention relates to a nucleic acid sequence that
encodes an amino acid sequence that is at least about 60%, 65%, 70%, 75%, 80%,
81%, 82%, 83%, 84%. 85%, 86%, 87%, 88%, 89%, 90%, 91 %, 92%, 93%, 94%,
95%, 96%, 97%, 98%, 99%, or 100% identical to SEQ ID NO: 3.
In one embodiment, the invention relates to a nucleic acid sequence that
encodes an amino acid sequence that is at least about 60%, 65%, 70%, 75%, 80%,
81%, 82%, 83%, 84%. 85%, 86%, 87%, 88%, 89%, 90%, 91 %, 92%, 93%, 94%,
95%, 96%, 97%, 98%, 99%, or 100% identical to SEQ ID NO: 5.
In one embodiment, the invention relates to a nucleic acid sequence that
encodes an amino acid sequence that is at least about 60%, 65%, 70%, 75%, 80%,
81%, 82%, 83%, 84%. 85%, 86%, 87%, 88%, 89%, 90%, 91 %, 92%, 93%, 94%,
95%, 96%, 97%, 98%, 99%, or 100% identical to SEQ ID NO: 7.
In one embodiment, the invention relates to a nucleic acid sequence that
encodes an amino acid sequence that is at least about 60%, 65%, 70%, 75%, 80%,
81%, 82%, 83%, 84%. 85%, 86%, 87%, 88%, 89%, 90%, 91 %, 92%, 93%, 94%,
95%, 96%, 97%, 98%, 99%, or 100% identical to SEQ ID NO: 8.
The invention also provides a host cell comprising the nucleic acids described
herein. When the host cell is cultured under a suitable condition, the nucleic
acids
24
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
can express a gB polypeptide. Preferably, said gB polypeptide forms a
monodispersed trimer. For example, the gB polypeptide of the invention does
not
form a dimeric association of two trimers. In another exemplary embodiment,
the gB
polypeptide of the invention does not form a higher order association of
trimers.
Preferably, the monodispersed trimer can be secreted from the host cell.
Most preferably, the host cells are mammalian cells (e.g., human, non-human
primate, horse, cow, sheep, dog, cat, and rodent (e.g., hamster). Suitable
mammalian cells include, for example, Chinese hamster ovary (CHO) cells, human
embryonic kidney cells (HEK-293 cells, typically transformed by sheared
adenovirus
type 5 DNA), NIH-3T3 cells, 293-T cells, Vera cells, HeLa cells, PERC.6 cells
(ECACC deposit number 96022940), Hep G2 cells, MRC-5 (ATCC CCL-171), WI-38
(ATCC CCL-75), fetal rhesus lung cells (ATCC CL-160), Madin-Darby bovine
kidney
("MDBK") cells, Madin-Darby canine kidney ("MDCK") cells (e.g., MDCK (NBL2),
ATCC CCL34; or MDCK 33016, DSM ACC 2219), baby hamster kidney (BHK) cells,
such as BHK21 -F, HKCC cells, and the like.
In a most preferred embodiment, the host cell is EXPI293FTM (ThermoFisher
Scientific) human cells, derived from the 293 cell line.
In certain embodiments, the host cell is a CHO cell. In certain embodiments,
the polynucleotide encoding the gB polypeptide described herein is stably
integrated
into the genome of the CHO cell.
Various CHO cell lines are also available from European Collection of Cell
Cultures (ECACC), or American Type Culture Collection (ATCC), such as CHO cell
lines hCBE1 1 (ATCC PTA-3357Tm), E77.4 (ATCC PTA-3765Tm), hLT-B: R-hG1
CHO #14 (ATCC CRL-1 1965Tm), MOR-CHO- MORAb-003-RCB (ATCC PTA-
7552Tm), AQ.C2 clone 11 B (ATCC PTA-3274Tm), AQ.C2 clone 11 B (ATCC
PTA-3274Tm), hsAQC2 in CHO-DG44 (ATCC PTA-3356Tm), xr55 (ATCC CRL-
2348Tm), CHO-K1 (ATCC CCL-61TM), Led [originally named Pro-5WgaRI3C]
(ATCCO CRL-1735Tm), Pro-5 (ATCCO CRL-1781Tm), ACY1 -E (ATCC 65421TM),
ACY1 -E (ATCC 65420Tm), pgsE-606 (ATCC CRL-2246Tm), CHO-CD36 (ATCC
CRL-2092Tm), pgsC- 605 (ATCC CRL-2245Tm), MC2/3 (ATCC CRL-2143Tm),
CHO-ICAM-1 (ATCC CRL-2093Tm), and pgsB-618 (ATCC CRL-2241Tm). Any one
of these CHO cell lines may be used.
Other commercially available CHO cell lines include, e.g., FREESTYLETm
CHO-S Cells and Flp-InTm-CHO Cell Line from Life Technologies.
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
Alternative suitable host cells may also include, for example, insect cells
(e.g.,
Aedes aegypti, Auto grapha califomica, Bombyx mori, Drosophila melanogaster,
Spodoptera frugiperda, and Trichoplusia ni), mammalian cells (e.g., human, non-
human primate, horse, cow, sheep, dog, cat, and rodent (e.g., hamster)), avian
cells
(e.g., chicken, duck, and geese), bacteria (e.g., E. coli, Bacillus subtilis,
and
Streptococcus spp.), yeast cells (e.g., Saccharomyces cerevisiae, Candida
albicans,
Candida maltosa, Hansenual polymorpha, Kluyveromyces fragilis, Kluyveromyces
lactis, Pichia guillerimondii, Pichia pastoris, Schizosaccharomyces pombe and
Yarrowia lipolytica), Tetrahymena cells (e.g., Tetrahymena thermophila) or
combinations thereof.
Suitable insect cell expression systems, such as baculovirus systems, are
known to those of skill in the art. Materials and methods for baculo
virus/insect cell
expression systems are commercially available. Suitable insect cells include,
for
example, Sf9 cells, Sf21 cells, Tn5 cells, Schneider S2 cells, and High Five
cells
(Invitrogen)).
In a most preferred embodiment, the host cell is a mammalian cell, not an
insect cell. Accordingly in a preferred embodiment, the polypeptide does not
have
an insect cell pattern of glycosylation.
In certain embodiments, the recombinant nucleic acids are codon optimized
for expression in a selected prokaryotic or eukaryotic host cell.
To facilitate replication and expression, the nucleic acids can be
incorporated
into a vector, such as a prokaryotic or a eukaryotic expression vector.
Exemplary
vectors include plasmids that are able to replicate autonomously or to be
replicated
in a host cell. Typical expression vectors contain suitable promoters,
enhancers, and
terminators that are useful for regulation of the expression of the coding
sequence(s)
in the expression construct. The vectors may also comprise selection markers
to
provide a phenotypic trait for selection of transformed host cells (such as
conferring
resistance to antibiotics such as ampicillin or neomycin).
Also provided herein is a process of producing cytomegalovirus (CMV) gB
polypeptide comprising: (i) culturing the host cell described herein under a
suitable
condition, thereby expressing said gB polypeptide, or immunogenic fragment
thereof;
26
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
and (ii) harvesting said gB polypeptide, or immunogenic fragment thereof, from
the
culture.
In one embodiment, the gB polypeptide described herein is purified. The gB
polypeptide can be purified using any suitable methods, such as HPLC, various
types of chromatography (such as hydrophobic interaction, ion exchange,
affinity,
chelating, and size exclusion), electrophoresis, density gradient
centrifugation,
solvent extraction, or the like. As appropriate, the gB polypeptide may be
further
purified, as required, so as to remove substantially any polypeptides which
are also
secreted in the medium or result from lysis of host cells, so as to provide a
product
which is at least substantially free of host debris, e.g., polypeptides,
lipids and
polysaccharides.
As used herein, a "purified" protein or polypeptide is a protein or
polypeptide
which is recombinantly or synthetically produced, or produced by its natural
host,
and has been isolated from other components of the recombinant or synthetic
production system or natural host such that the amount of the protein relative
to
other macromolecular components present in a composition is substantially
higher
than that present in a crude preparation. In general, a purified protein will
be at least
about 50% homogeneous and more preferably at least about 75%, at least about
80%, at least about 85%, at least about 90%, at least about 95% or
substantially
homogeneous.
In another embodiment, the process of purifying the modified gB polypeptide
of the invention allows for production of the polypeptide at a purity of >85%,
>86%,
>87%, >88%, >89%, >90%, >91%, >92%, >93%, >94% or >95% of total protein by
mass, as determined by gel electrophoresis. These high levels of purity make
the
modified gB polypeptide suitable for use as an immunogen in diagnostic
applications
or as an antigen in immunogenic compositions.
27
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
PREPARATION OF ANTIBODIES AGAINST CMV EPITOPES
The immunogenic polypeptides prepared as described above may be used to
produce antibodies, both polyclonal and monoclonal. If polyclonal antibodies
are
desired, a selected mammal (e.g., mouse, rabbit, goat, guinea pig, horse,
etc.) is
.. immunized with an immunogenic polypeptide bearing a CMV epitope(s). Serum
from the immunized animal is collected and treated according to known
procedures.
If serum containing polyclonal antibodies to a CMV epitope contains antibodies
to
other antigens, the polyclonal antibodies can be purified by immunoaffinity
chromatography. Techniques for producing and processing polyclonal antisera
are
known in the art.
Monoclonal antibodies directed against CMV epitopes can also be readily
produced by one skilled in the art. The general methodology for making
monoclonal
antibodies by hybridomas is known. Immortal antibody-producing cell lines can
be
created by cell fusion, and also by other techniques such as direct
transformation of
B lymphocytes with oncogenic DNA, or transfection with Epstein-Barr virus.
Panels
of monoclonal antibodies produced against CMV epitopes can be screened for
various properties; i.e., for isotype, epitope affinity, etc.
Antibodies, both monoclonal and polyclonal, which are directed against CMV
epitopes are particularly useful in diagnosis, and those which are
neutralizing are
useful in passive immunotherapy. Monoclonal antibodies, in particular, may be
used
to raise anti-idiotype antibodies.
IMMUNOASSAY AND DIAGNOSTIC KITS
Both the recombinant polypeptides which react immunologically with serum
containing CMV antibodies, and the antibodies raised against these recombinant
polypeptides, are useful in immunoassays to detect the presence of CMV
antibodies,
or the presence of the virus, in biological samples, including for example,
blood or
serum samples. Design of the immunoassays is subject to a great deal of
variation,
and a variety of these are known in the art. For example, the immunoassay may
utilize the polypeptide having the sequence set forth in SEQ ID NO: 2.
Alternatively,
the immunoassay may use a combination of viral antigens derived from the gB
polypeptides described herein. It may use, for example, a monoclonal antibody
directed towards one modified gB polypeptides described herein, a combination
of
monoclonal antibodies directed towards the modified gB polypeptides described
28
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
herein, monoclonal antibodies directed towards different viral antigens,
polyclonal
antibodies directed towards the modified gB polypeptides described herein, or
polyclonal antibodies directed towards different viral antigens. Protocols may
be
based, for example, upon competition, or direct reaction, or may be sandwich
type
assays. Protocols may also, for example, use solid supports, or may be by
immunoprecipitation. Most assays involve the use of labeled antibody or
polypeptide;
the labels may be, for example, fluorescent, chemiluminescent, radioactive, or
dye
molecules. Assays which amplify the signals from the probe are also known;
examples of which are assays which utilize biotin and avidin, and enzyme-
labeled
and mediated immunoassays, such as ELISA assays.
Kits suitable for immunodiagnosis and containing the appropriate labeled
reagents are constructed by packaging the appropriate materials, including the
recombinant polypeptides of the invention containing CMV epitopes or
antibodies
directed against epitopes in suitable containers, along with the remaining
reagents
and materials required for the conduct of the assay, as well as a suitable set
of
assay instructions.
The polynucleotide probes can also be packaged into diagnostic kits.
Diagnostic kits include the probe DNA, which may be labeled; alternatively,
the
probe DNA may be unlabeled and the ingredients for labeling may be included in
the
kit. The kit may also contain other suitably packaged reagents and materials
needed
for the particular hybridization protocol, for example, standards, as well as
instructions for conducting the test.
29
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
COMPOSITIONS AND METHODS OF TREATMENT
The invention relates to compositions and methods of treatment using the
cytomegalovirus gB polypeptide described herein, or a nucleic acid encoding
such
gB polypeptide described herein. For example, the polypeptide of the invention
can
be delivered directly as a component of an immunogenic composition.
Alternatively,
nucleic acids that encode the gB polypeptide of the invention can be
administered to
produce the CMV protein or immunogenic fragment in vivo. Certain preferred
embodiments, such as protein formulations, recombinant nucleic acids (e.g.,
DNA,
RNA, self-replicating RNA, or any variation thereof) and viral vectors (e.g.,
live,
single-round, non-replicative assembled virions, or otherwise virus-like
particles, or
alphavirus VRP) that contain sequences encoding gB polypeptides are further
described herein and may be included in the composition.
In one aspect, the invention provides an immunogenic composition
comprising the recombinant CMV gB polypeptide described herein. The
immunogenic composition can include additional CMV proteins, such as gO, gH,
gL,
pUL128, pUL130, pUL131 , an immunogenic fragment thereof, or a combination
thereof. For example, the gB polypeptide can be combined with CMV pentameric
complex comprising: gH or a pentamer-forming fragment thereof, gL or a
pentamer-
forming fragment thereof, pUL128 or a pentamer-forming fragment thereof,
pUL130
or a pentamer-forming fragment thereof, and pUL131 or a pentamer-forming
fragment thereof. The gB polypeptide of the invention can also be combined
with
CMV trimeric complex comprising: gH or a trimer-forming fragment thereof, gL
or a
trimer-forming fragment thereof, and g0 or a trimer- forming fragment thereof.
The immunogenic composition may include an adjuvant. Exemplary adjuvants
to enhance effectiveness of the composition include: (1) aluminum salts
(alum), such
as aluminum hydroxide, aluminum phosphate, aluminum sulfate, etc.; (2) oil-in-
water
emulsion formulations (with or without other specific adjuvants such as
muramyl
peptides (see below) or bacterial cell wall components), such as for example
(a)
MF59 (PCT Publ. No. WO 90/14837), containing 5% Squalene, 0.5% TWEEN 80,
and 0.5% Span 85 formulated into submicron particles using a microfluidizer,
(b)
SAF, containing 10% Squalane, 0.4% Tween 80, 5% pluronic-blocked polymer L121
, and thr-MDP either microfluidized into a submicron emulsion or vortexed to
generate a larger particle size emulsion, and (c) RIBI TM adjuvant system
(RAS), (Ribi
lmmunochem, Hamilton, Mont.) containing 2% Squalene, 0.2% Tween 80, and one
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
or more bacterial cell wall components from the group consisting of
monophosphorylipid A (MPL), trehalose dimycolate (TDM), and cell wall skeleton
(CWS), preferably MPL+CWS (DetoxTm); (3) saponin adjuvants, such as QS-21,
STIMU LON TM (Cambridge Bioscience, Worcester, Mass.), which may be used or
particles generated therefrom such as ISCOMs (immunostimulating complexes);
(4)
Complete Freunds Adjuvant (CFA) and Incomplete Freunds Adjuvant (I FA); (5)
cytokines, such as interleukins (IL-1 , IL-2, etc.), macrophage colony
stimulating
factor (M-CSF), tumor necrosis factor (TN F), etc.; and (6) other substances
that act
as adjuvants to enhance the effectiveness of the composition. In a preferred
embodiment, the adjuvant is a saponin adjuvant, namely QS-21.
Each of the immunogenic compositions discussed herein may be used alone
or in combination with one or more other antigens, the latter either from the
same
viral pathogen or from another pathogenic source or sources. These
compositions
may be used for prophylactic (to prevent infection) or therapeutic (to treat
disease
after infection) purposes.
In one embodiment, the composition may include a "pharmaceutically
acceptable carrier," which includes any carrier that does not itself induce
the
production of antibodies harmful to the individual receiving the composition.
Suitable
carriers are typically large, slowly metabolized macromolecules such as
proteins,
polysaccharides, polylactic acids, polyglycolic acids, polymeric amino acids,
amino
acid copolymers, lipid aggregates (such as oil droplets or liposomes), and
inactive
virus particles. Such carriers are well known to those of ordinary skill in
the art.
Additionally, these carriers may function as adjuvants. Furthermore, the
antigen may
be conjugated to a bacterial toxoid, such as a toxoid from diphtheria,
tetanus,
.. cholera, H. pylori, and etc. pathogens.
In one embodiment, the composition includes a diluent, such as water, saline,
glycerol, ethanol, etc. Additionally, auxiliary substances, such as wetting or
emulsifying agents, pH buffering substances, and the like, may be present in
such
vehicles.
The compositions described herein may include an immunologically effective
amount of the polypeptide, as well as any other of the above-mentioned
components, as needed. By "immunologically effective amount," it is meant that
the
administration of that amount to an individual, either in a single dose or as
part of a
series, is effective for eliciting an immune response. The immune response
elicited
31
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
may be sufficient, for example, for treatment and/or prevention and/or
reduction in
incidence of illness, infection or disease. This amount varies depending upon
the
health and physical condition of the individual to be treated, the taxonomic
group of
individual to be treated (e.g., nonhuman primate, primate, etc.), the capacity
of the
individual's immune system to synthesize antibodies, the degree of protection
desired, the formulation of the vaccine, the treating doctor's assessment of
the
medical situation, and other relevant factors. It is expected that the amount
will fall in
a relatively broad range that can be determined through routine trials.
The composition may be administered parenterally, e.g., by injection, either
subcutaneously or intramuscularly. Additional formulations suitable for other
modes
of administration include oral and pulmonary formulations, suppositories, and
transdermal applications. Oral formulations may be preferred for certain viral
proteins. Dosage treatment may be a single dose schedule or a multiple dose
schedule. The immunogenic composition may be administered in conjunction with
.. other immunoregulatory agents.
In another aspect, the invention provides a method of eliciting an immune
response against cytomegalovirus (CMV), comprising administering to a subject
in
need thereof an immunologically effective amount of a modified CMV gB
polypeptide
and/or an immunogenic composition described herein, which comprises the
proteins,
DNA molecules, RNA molecules (e.g., self-replicating RNA molecules), or VRPs
as
described above. In certain embodiments, the immune response comprises the
production of neutralizing antibodies against CMV.
The immune response can comprise a humoral immune response, a cell-
mediated immune response, or both. In some embodiments an immune response is
.. induced against each delivered CMV protein. A cell-mediated immune response
can
comprise a Helper T-cell (Th) response, a CD8+ cytotoxic T-cell (CTL)
response, or
both. In some embodiments the immune response comprises a humoral immune
response, and the antibodies are neutralizing antibodies.
Neutralizing antibodies block viral infection of cells. CMV infects epithelial
cells and also fibroblast cells. In some embodiments the immune response
reduces
or prevents infection of both cell types. Neutralizing antibody responses can
be
complement-dependent or complement- independent. In some embodiments the
neutralizing antibody response is complement- independent. In some embodiments
the neutralizing antibody response is cross-neutralizing; i.e., an antibody
generated
32
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
against an administered composition neutralizes a CMV virus of a strain other
than
the strain used in the composition.
The modified CMV gB polypeptide and/or immunogenic composition
described herein may also elicit an effective immune response to reduce the
likelihood of a CMV infection of a non-infected mammal, or to reduce symptoms
in
an infected mammal, e.g., reduce the number of outbreaks, CMV shedding, and
risk
of spreading the virus to other mammals.
For example in one aspect, the modified CMV gB polypeptide and/or
immunogenic composition described herein reduces viral shedding in a mammal.
The term "viral shedding" is used herein according to its plain ordinary
meaning in
medicine and virology and refers to the production and release of virus from
an
infected cell. In some embodiments, the virus is released from a cell of a
mammal.
In some embodiments, virus is released into the environment from an infected
mammal. In some embodiments the virus is released from a cell within a mammal.
In one aspect, the invention relates to a method for reducing CMV viral
shedding in a mammal. The method includes administering the modified CMV gB
polypeptide and/or immunogenic composition described herein to the mammal that
is
infected with or is at risk of a CMV infection. In one embodiment, the
reduction in
CMV viral shedding in a mammal is as compared to the viral shedding in mammals
that were not administered the modified CMV gB. In another embodiment, the
reduction in CMV viral shedding in a mammal is as compared to the viral
shedding
following an administration of a CMV pentamer alone or following an
administration
of a CMV pentamer in the absence of the modified CMV gB.
In one embodiment, the challenge cytomegalovirus strain is a human CMV
strain. In one embodiment, the challenge cytomegalovirus strain is homologous
to
the CMV strain from which the modified CMV gB polypeptide is derived. In
another
embodiment, the challenge cytomegalovirus strain is homologous to the VR1814
CMV strain.
In one embodiment, the challenge cytomegalovirus strain is a human CMV
strain that is heterologous to the CMV strain from which the modified CMV gB
polypeptide is derived. In another embodiment, the challenge cytomegalovirus
strain
is a human CMV strain that is heterologous to the VR1814 CMV strain. In
another
embodiment, the challenge cytomegalovirus strain is the VR1814 CMV strain.
33
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
In another embodiment, the challenge cytomegalovirus strain is a rhesus
CMV strain homologous to the macacine herpesvirus 3 isolate 21252 CMV strain.
In
another embodiment, the challenge cytomegalovirus strain is the macacine
herpesvirus 3 isolate 21252 CMV strain.
A useful measure of antibody potency in the art is "50% neutralization titer."
Another useful measure of antibody potency is any one of the following: a "60%
neutralization titer"; a "70% neutralization titer"; a "80% neutralization
titer"; and a
"90% neutralization titer." To determine, for example, a 50% neutralizing
titer,
serum from immunized animals is diluted to assess how dilute serum can be yet
retain the ability to block entry of 50% of infectious viruses into cells. For
example, a
titer of 700 means that serum retained the ability to neutralize 50% of
infectious virus
after being diluted 700-fold. Thus, higher titers indicate more potent
neutralizing
antibody responses. In some embodiments, this titer is in a range having a
lower
limit of about 200, about 400, about 600, about 800, about 1000, about 1500,
about
2000, about 2500, about 3000, about 3500, about 4000, about 4500, about 5000,
about 5500, about 6000, about 6500, or about 7000. The 50%, 60%, 70%, 80%, or
90% neutralization titer range can have an upper limit of about 400, about
600, about
800, about 1000, about 1500, about 2000, about 2500, about 3000, about 3500,
about 4000, about 4500, about 5000, about 5500, about 6000, about 6500, about
7000, about 8000, about 9000, about 10000, about 11000, about 12000, about
13000, about 14000, about 15000, about 16000, about 17000, about 18000, about
19000, about 20000, about 21000, about 22000, about 23000, about 24000, about
25000, about 26000, about 27000, about 28000, about 29000, or about 30000. For
example, the 50% neutralization titer can be about 3000 to about 6500. "About"
means plus or minus 10% of the recited value. Neutralization titer can be
measured
as described in the specific examples, below.
An immune response can be stimulated by administering proteins, DNA
molecules, RNA molecules (e.g., self-replicating RNA molecules), or VRPs to an
individual, typically a mammal, including a human. In some embodiments the
immune response induced is a protective immune response, i.e., the response
reduces the risk or severity of or clinical consequences of a CMV infection.
Stimulating a protective immune response is particularly desirable in some
populations particularly at risk from CMV infection and disease. For example,
at-risk
populations include solid organ transplant (SOT) patients, bone marrow
transplant
34
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
patients, and hematopoietic stem cell transplant (HSCT) patients. VRPs can be
administered to a transplant donor pre-transplant, or a transplant recipient
pre-
and/or post-transplant. Because vertical transmission from mother to child is
a
common source of infecting infants, administering VRPs to a woman who is
pregnant
or can become pregnant is particularly useful.
Any suitable route of administration can be used. For example, a composition
can be administered intramuscularly, intraperitoneally, subcutaneously, or
transdermally. Some embodiments will be administered through an intra-mucosal
route such as intra-orally, intra- nasally, intra-vaginally, and intra-
rectally.
Compositions can be administered according to any suitable schedule.
Also provided herein is a method of inhibiting cytomegalovirus (CMV) entry
into a cell, comprising contacting the cell with the immunogenic composition
described herein.
The invention will be further described by reference to the following, non-
limiting, examples.
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
EXAMPLES
The following Examples illustrate embodiments of the invention.
EXAMPLE 1: Description of the gB705 expression construct
Pfizer's expression construct of HCMV gB705 (SEQ ID NO: 1) is based on the
VR1814 gB sequence (GenBank# A0Z79977.1) that contains amino acids 1-705. A
6xHis affinity tag is optionally included at the C-terminus in a pLH115 vector
(Pfizer).
Pfizer's gB705 contains four mutations ((Y155G/I156H/H157R/VV240A) as well as
mutations (R456A/R459A) in the furin-like cleavage site as illustrated in FIG.
1,
wherein SS represents a signal sequence; D1 represents the Domain I; FL
represents a fusion loop; 6h represents a 6xHis tag. FIG. 2 shows the sequence
of
HCMV gB705 (SEQ ID NO: 2)
EXAMPLE 2: Expression of gB705
The initial assessment of gB705 expression was carried out by transfecting
the construct DNA into HEK293T or Expi293F cells using Effectene Transfection
Reagent (Qiagen) or Expifetamin 293 Transfection kit (Gibco), respectively.
The
culture supernatants were harvested -48 hours after transfection. The gB705
protein
was pulled down from the culture supernatant of transfected cells using a
MAGNEHISTM Protein Purification System (Promega) and was analyzed on 4-12%
Bis-Tris protein gel (NUPAGEO NOVEXO) under reducing condition. Presence of
the target proteins was detected by Western blot using anti-HCMV polyclonal
antibody (CytoGam). The result showed that recombinant gB705 existed as a
single
protein band with molecular weight about 120 kDa (FIG. 3). This apparent
molecular
weight is higher than that predicted from the peptide sequence of residues 25-
705 [of
SEQ ID NO: 1] from the mature gB705 polypeptide, indicating that the gB705
protein
is heavily glycosylated. The gB705 protein is expressed efficiently as a
homogeneous protein in transfection of HEK293 or Expi293F cells.
36
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
EXAMPLE 3: Recombinant gB705 contains intact neutralizing epitopes
recognized by human anti-HCMV gB monoclonal antibodies (mAbs)
To evaluate the structural integrity of recombinant gB, a panel of five
neutralizing human anti-HCMV gB mAbs were tested for the binding of gB705 (SEQ
ID NO: 2). Briefly, the culture supernatants from gB705 transfected Expi293F
cells
were added on a pre-blocked HISGRABTM Nickel Coated Plate (Pierce) and
incubated for 1 hour at room temperature. After wash, serial diluted antibody
solutions were added to the plate and incubated for 1 hour at room
temperature,
followed by addition of HRP-conjugated anti-human IgG secondary antibody.
After 1
hour incubation, the plate was washed and the peroxidase substrate TMB was
added to be read by a plate reader when the color was developed. The results
showed all five human anti-gB mAbs bound efficiently to gB705 (FIG. 4A).
Recombinant gB705 binds to three anti-AD5 mAbs including 2B11, 5F1 and
4H9 (each described in Table 1 of US patent publication no. 20160280770 and
Table
1 of WIPO publication no. W02010007533), whereas recombinant gB proteins from
a commercial source (Sino gB, from Sino Biologicals, Inc.) did not (FIG. 4B).
Similarly, these three mAbs did not bind to a recombinant gB protein expressed
in
insect cells from RedBiotech, Inc. (data not shown). The recombinant gB
protein
from the commercial source and from RedBiotec do not include any one of the
following mutations: Y155G, I156H, H157R, and W240A, according to the
numbering
of SEQ ID NO: 6. Without being bound by theory, the inventive combination of
mutations Y155G, I156H, H157R, and W240A include deletions of the cytoplasmic
tail and mutations in hydrophobic fusion loops of gB. These mutations appear
to
relieve steric hindrance that potentially would have otherwise blocked
antibody
access to these sites in an unmodified gB protein or in a protein that does
not
include the inventive combination of mutations.
The 2B11 mAb has a sequence set forth in SEQ ID NO: 367 for the heavy
chain and SEQ ID NO: 368 for the light chain; 5F1 mAb has a sequence set forth
in
SEQ ID NO: 290 for the heavy chain and SEQ ID NO: 291 for the light chain; and
4H9 mAb has a sequence set forth in SEQ ID NO: 308 for the heavy chain and SEQ
ID NO: 309 for the light chain, as disclosed in US patent publication no.
20160280770 and WIPO publication no. W02010007533, each sequence of which is
incorporated by reference herein).
37
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
FIG. 4A Binding of human anti-HCMV gB monoclonal antibodies to gB705
protein. Culture supernatants from gB705 transfected Expi293F cells were
captured
on a pre-blocked HISGRABTM Nickel Coated Plate (Pierce) and incubated serially
diluted monoclonal antibodies followed by detection with HRP conjugated anti-
s human Ig secondary antibody (ELN# 00710043-0177). FIG. 4B Binding of
human
anti-HCMV gB monoclonal antibodies to Sino gB protein (gB from Sino
Biologicals,
Inc.) (ELN# 00710043-0177).
38
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
EXAMPLE 4: Scale-up production and purification of gB705
The gB705 construct was transiently transfected into 1.8 liters of Expi293F
cells and
about 95 mg of protein was purified from the conditioned media collected on
day 4
through a series of processes of diafiltration, Ni-sepharose and size
exclusion
chromatography. The protein was analyzed on SDS-PAGE and exhibited a single
band under reducing condition, as detected by Coomassie blue staining (FIG.
5).
Under non-reducing conditions, there were higher molecular weight bands that
may
be corresponding to a trimeric form of the protein. FIG. 5 shows an SDS-PAGE
of
gB705 under reducing and non-reducing conditions (ELN#00709755-0086).
39
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
EXAMPLE 5: Structural characterization of recombinant gB705 expressed in
Expi293F cells: Oligomer formation and stability
Postfusion gB protein is in a trimer form. To characterize whether the gB705
forms
higher order of oligomers, a series of experiments were carried out as
summarized
.. below.
Singular and oligomeric trimers of gB705 with characteristics of post-fusion
gB observed by transmission electron microscopy
Recombinant gB705 protein was subjected to electron microscopy analysis after
negative staining. The image (FIG. 16) shows that the protein forms a singular
trimer
as well as larger oligomeric complexes (that is, associations between
trimers). The
singular trimer of gB705 exhibited strong similarity in general morphology
with the
postfusion gB structure as published in Burke et al., PLoS Pathog. 2015 Oct;
11(10).
In contrast to the singular trimer formed by the inventive gB705, a modified
gB
protein, termed "gB706," which does not include the inventive combination of
fusion
.. loop mutations, forms a dimer and trimer of trimers, as discussed in in
Sharma et al.,
Virology. 2013 Jan 20;435(2):239-49, namely on page 243 and in Figure 20. More
specifically, the gB706 protein of Sharma et al. is described as encoding the
mature
ectodomain of HCMV (strain AD169) gB, residues 25-706, with a signal sequence.
Sharma et al. further describe gB706 as lacking the hydrophobic membrane-
proximal region, the transmembrane region, and the cytoplasmic domain of gB.
Multiple species of gB705 observed in velocity analytical ultracentrifugation
analysis
The purified gB705 protein (SEQ ID NO: 2) was also subjected to velocity
analytical
ultracentrifugation analysis and the result showed that the protein formed
multiple
peaks corresponding to the sizes of singular trimer or larger / high-order
complexes
(FIG. 6). FIG. 6 velocity analytical ultracentrifugation analysis of gB705
(ELN#
00708337-0110).
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
Transforming recombinant gB705 to homogeneous singular trimer by
treatment of ethylenediaminetetraacetic acid (EDTA)
To assess the stability of the gB705 higher order oligomers, the gB705 protein
was subjected to various treatment including surfactants, chaotropes, heat,
reductant
and EDTA (data not shown). Among these conditions, it was observed that EDTA
can break down the high molecular weight oligomers to homogenous monodispersed
trimers. This was confirmed by velocity sedimentation analysis as shown in FIG
7.
This homogenous material establishes a strong base entity for studying its
structural
characteristics and its potential for change under biologically relevant
conditions.
FIG. 7 shows Velocity sedimentation analysis of 10 mM EDTA-treated gB705 (
ELN#
00708337-0110).
In the structure of a postfusion HCMV gB protein, a calcium ion (Ca++) was
observed in the core of the central coiled coil, coordinated by residue D508.
To
evaluate whether Ca++ ion has an effect on the overall shape of the gB705
after
EDTA treatment, sedimentation velocity analysis was performed on the EDTA-
treated gB705 at pH 5.2 with and without adding back 10 mM Ca++ ion. The
result
showed that adding back Ca++ ion had no impact on hydrodynamic properties of
gB705 at pH 5.2 (data not shown). Furthermore, adding back Ca++ ion did not
cause
the protein to form high molecular weight oligomers seen in the non-EDTA-
treated
gB705 protein. In contrast, adding back Ni+ or Cu++ ions led to formation of
high-
order oligomeric complexes of gB705 trimer (data not shown). Taken together,
oligomer formation of gB705 trimer may be a result of the existence of the
6xHis
affinity tag, rather than due to some unknown intrinsic properties of gB705
trimer.
41
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
EXAMPLE 6: Structural characterization of gB705 trimer at different pH
conditions:
To evaluate the effect of pH on the HCMV gB705 protein, EDTA-treated gB705
protein was dialyzed into buffers at pH 5.2 and pH 8.7 and subjected to a
series of
.. biophysical characterization experiments to determine the changes, if any,
in the
secondary, tertiary and quaternary structure or hydrodynamic properties.
Sedimentation velocity analysis
Sedimentation velocity analysis was performed on EDTA-treated gB705 at pH 5.2
and pH 8.7 (Fig. 8). No significant difference in migration or apparent
molecular
weight of the protein at different pHs was observed. FIG. 8A-B Sedimentation
velocity analysis of EDTA-treated gB705 at pH 5.2 (A) and pH 8.7 (B).
Cryo electron microscopy analysis
Cryo electron microscopy analysis was performed on EDTA-treated gB705
proteins. The two
images were collected on a Tecnia F20 electron microscope at the magnification
of x50,000
.. using low dose beam condition on a cryo frozen grid. The images showed that
the gB705
was in monodispersed trimer form at either pH5.2 or pH 8.7 (FIG. 8C).
Far UV CD and intrinsic fluorescence spectroscopy
Both the Far UV CD (FIG. 9A) and intrinsic fluorescence spectroscopy (FIG. 9B)
analyses suggest little change in the secondary structure of gB705 at pH 5.2
and pH
.. 8.7. Fig. 9A Far UV CD analysis; FIG. 9B intrinsic fluorescence
spectroscopy (ELN#
00708337-0115, 0117)
Change in tertiary structure of gB705 trimer at different pHs detected by near-
UV CD spectroscopy
Near-UV CD analysis was also performed to determine whether there is any
difference in the tertiary structure of gB705 trimer at pH 5.2 and pH 8.7. The
gB705
trimer exhibited differences in the spectroscopy profile under the two pH
conditions
(Fig. 10). This difference was strictly pH-dependent, because it was
consistently
observed when two different buffers were used at both pH 5.2 and pH 8.7. The
lower
fluorescence of gB705 in the normalized near-UV CD spectrum at pH 5.2 may be
indicative of looser packing of the aromatic residues in gB705 trimer under
acidic
conditions. In short, change in environmental pH condition has a noticeable
influence
on the tertiary structure of gB705 trimer. FIG. 10 Near UV CD spectroscopy
analysis
of gB705 at pH 5.2 and pH 8.7. Buffer 1, 50 mM citrate, 100 mM phosphate;
Buffer
42
CA 03016867 2018-09-06
WO 2017/153954 PCT/IB2017/051401
2, 50 mM Na0Ac; Buffer 3, 100 mM Na carbonate/bicarbonate; Buffer 4, 50 mM Na
borate (ELN# 00708337-0115, 0117).
43
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
Difference in thermodynamic melting at high and low pH conditions
DSC analysis was performed to study the thermodynamic properties of gB705 at
pH
5.2 and pH 8.7 (Fig. 11). The gB705 protein underwent a two-phase transition
at pH
5.2, with a transitional melting temperature of 81.0 C. In stark contrast, the
same
protein prep demonstrated a three-phase transition at pH 8.7, with two
transitional
melting points at 72 C and 78 C respectively. The observed difference here
indicates
that there is significant structural difference, in terms of biophysical
stability, in the
gB705 trimer at pH 5.2 versus pH 8.7. FIG. 11 A-B: DSC analysis of gB705 at
(FIG.
11A) pH 5.2 and (FIG. 11B) pH 8.7. At pH 8.7, the observed melting curve (in
block
line) was fit to the 4-state unfolding model and resolved to a three-phase
transition
pattern (in red lines) using the software provided by the DSC manufacturer.
(ELN#
00708337-0115, 0117).
Increase in exposure of hydrophobic elements in gB705 trimer at acidic
condition.
Difference in ANS staining of gB705 at different pH
ANS is a fluorescent dye that binds to hydrophobic surfaces of protein,
resulting in
increased fluorescence intensity and shift in the emission maximum towards
lower
wavelengths. It was observed that ANS fluorescence intensity of gB705 was
increased and its maximal emission was shifted to shorter wavelength of UV at
pH
5.2, relative to those at pH 8.7 or pH 7.4 (FIG. 12A). These data are
consistent with
that hydrophobic surfaces of gB705 trimer are more exposed under acidic pH
than
under neutral or basic pH conditions.
The exposure of hydrophobic surfaces of gB705 trimer at different pH
conditions is
reversible
To assess whether the difference in ANS binding ability of gB705 at different
pHs is
reversible or not, the gB705 protein at pH 5.2 was dialyzed into pH 8,7 buffer
and
vice versa, and then subjected to ANS analysis. The results showed that the
protein
gained ANS binding when shifted from high pH to low pH (FIG. 12B) and lost ANS
binding when shifted from low pH to high pH (FIG. 12C). Therefore, the
observed
difference in ANS binding to gB705 are reversible.
FIG. 12A-C: (A) ANS binding of gB705 at pH 5.2, pH 7.4 and pH 8.7 (ELN#
00708337-0115, 0117); (B) ANS binding of gB705 at pH 8.7 and 5.2 and shifted
from
44
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
pH 8.7 to pH 5.2; (C) ANS binding of gB705 at pH 8.7 and 5.2 and shifted from
pH
5.2 to pH 8.7 (ELN# 00708329-0153)
Accordingly, a HCMV gB705 protein was produced. The protein contains the
ectodomain of the VR1814 gB and mutations in the fusion loops and furin
cleavage
site. This protein may be in the postfusion conformation under neutral pH. The
protein forms various oligomers after purification but can be transformed into
homogenous singular trimer upon EDTA treatment.
No significant difference was observed in the secondary structure of EDTA-
treated
gB705 at pH 5.2 and pH 8.7, based on assays of far-UV CD spectroscopy,
intrinsic
fluorescence as well as sedimentation velocity analysis. However, significant
differences in conformation of gB705 trimer are observed with various assays.
Data
from near-UV spectroscopy revealed significant difference in tertiary
structure in
gB705 trimer between pH 5.2 and pH 8.7. Data on extrinsic fluorescence after
ANS
staining suggest that the protein exhibits more exposed hydrophobic surfaces
at pH
5.2, compared with at pH 8.7 and pH 7.2. The thermodynamic melting pattern of
gB705 was significantly different between pH 5.2 and pH 8.7. Finally, the ANS
staining analysis demonstrated that the conformational states of gB705 trimer
exist
in a reversible equilibrium, a property similar to VSV-G as a prototypic type
Ill viral
fusogen.
The gB705 protein exhibits a pH-dependent transition of conformation, which is
a
unique property and is biologically important. A non-postfusion conformation
of gB
(for example, gB705 at pH 8.7) may be a presented in a specific
chemical/physical
condition or by stabilizing its conformation with treatment of crosslinking
agents.
Such an immunogen may assume conformation state(s) that have a potential to
elicit
more robust nAb response than for example, recombinant gB protein in the
postfusion conformation. Moreover, the pH-dependent transition of tertiary
conformation of the gB705 trimer may be useful for improving solubility of the
gB
immunogen in an immunogenic composition, for improving stability of the
immunogen, for improving binding of the immunogen to aluminum or other
adjuvants, and/or for improving immunization of a vaccine comprising the
immunogen, as compared, for example, to a recombinant gB protein that does not
exhibit a pH-dependent transition of tertiary conformation, and/or as compared
to a
CA 03016867 2018-09-06
WO 2017/153954 PCT/IB2017/051401
recombinant gB protein that does not include the following mutations Y155G,
I156H,
H157R, and W240A.
46
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
>HCMV VR1814 gB705
M ESR IWC LVVCVN LC I VCLGAVVSSSSTSHATSSA H NGS HTSRTTSAQTRSVSSQ H
VTSSEAVSH RAN ETIYNTTLKYG DVVGVNTTKYPYRVCSMAQGTDLI RFERNIVCTP
MKPI NEDLDEGIMVVYKRNIVAHTFKVRVYQKVLTFRRSYAGH RTTYLLGSNTEYVA
PPMWEIHHINRHSQCYSSYSRVIAGTVFVAYHRDSYEN KTMQLM LDDYSNTHSTR
YVTVKDQWHSRGSTALYRETCN LNCMVTITTARSKYPYHFFATSTGDVVDISPFYN
GTNRNTSYFGENADKFFI FPNYTIVSDFGRANSAPETHRLVAFLERADSVISWDIQD
EKNVTCQLTFWEASERTI RSEAEDSYHFSSAKMTATFLSKKQEVNMSDPVLDCVR
DQALNKLQQI FNASYNQTYEKYG NVSVFETTGGLVVFWQG I KQKSLLELERLANSS
GVNSTRATKASTGNTTTLSLESESVRNVLYAQLQFTYDTLRSYI NRALAQIAEAWCV
DQRRTLEVFKELSKI NPSAI LSAIYNKPIAARFMGDVLGLASCVTI NQTSVKVLRDMN
VKESPGRCYSRPVVI FN FVNSSYVQYGQ LGEDN El LLGNHRTEECQFPSLKI FIAGN
SAYEYVDYLFKRMI DLSSISTVDSMIALDI DPLENTDFRVLELYSQKELRSSNVFDLE
El MR EFNSYKQRVKYVEDKVVDPLPP (SEQ ID NO: 1)
>HCMV VR1814 gB705 (w/o Met)
ESR IWC LVVCVN LC I VC LGAVVSSSSTSHATSSA H NGSH TSRTTSAQT RSVSSQ HV
TSSEAVSH RAN ETIYNTTLKYG DVVGVNTTKYPYRVCSMAQGTD LI RFERNIVCTPM
KPI NEDLDEGIMVVYKRN IVAHTFKVRVYQKVLTFRRSYAGHRTTYLLGSNTEYVAP
PMWEI HHINRHSQCYSSYSRVIAGTVFVAYHRDSYENKTMQLMLDDYSNTHSTRY
VTVKDQWHSRGSTALYRETCNLNCMVTITTARSKYPYHFFATSTGDVVDISPFYNG
TN RNTSYFGENADKF Fl FPNYTIVSDFGRANSAPETHRLVAFLERADSVISWDIQDE
KNVTCQLTFWEASERTI RSEAEDSYHFSSAKMTATFLSKKQEVNMSDPVLDCVRD
QALNKLQQI FNASYN QTYEKYGNVSVFETTGG LVVFWQG I KQKSLLELERLANSSG
VNSTRATKASTGNTTTLSLESESVRNVLYAQLQFTYDTLRSYI NRALAQIAEAWCVD
QRRTLEVFKELSKINPSAILSAIYNKPIAARFMGDVLGLASCVTI NQTSVKVLRDM NV
KESPGRCYSRPVVI FNFVNSSYVQYGQLGEDN El LLGNHRTEECQFPSLKI FIAGNS
AYEYVDYLFKRMI DLSSISTVDSMIALDI DPLENTDFRVLELYSQKELRSSNVFDLEEI
MREFNSYKQRVKYVEDKVVDPLPP (SEQ ID NO: 2)
47
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
>HCMV VR1814 gB705 (residues 25-705 OF SEQ ID NO: 1)
SSSTSHATSSAHNGSHTSRTTSAQTRSVSSQHVTSSEAVSH RAN ETIYNTTLKYGD
VVGVNTTKYPYRVCSMAQGTD LI RFE RN IVCTPM KPI N EDLDEG I MVVYKRN IVAHT
FKVRVYQKVLTFRRSYAGHRTTYLLGSNTEYVAPPMWEIHHIN RHSQCYSSYSRVI
AGTVFVAYHRDSYENKTMQLMLDDYSNTHSTRYVTVKDQWHSRGSTALYRETCN
LNCMVTITTARSKYPYHFFATSTGDVVDISPFYNGTNRNTSYFGENADKFFI FPNYTI
VSDFGRANSAPETH RLVAFLERADSVISWDIQDEKN VTCQLTFWEASERTI RSEAE
DSYHFSSAKMTATFLSKKQEVNMSDPVLDCVRDQALNKLQQI FNASYNQTYEKYG
NVSVFETTGGLVVFWQGI KQKSLLELERLANSSGVNSTRATKASTGNTTTLSLESE
SVRNVLYAQLQFTYDTLRSYIN RALAQIAEAWCVDQRRTLEVFKELSKI NPSAI LSAI
YNKPIAARFMGDVLGLASCVTINQTSVKVLRDMNVKESPGRCYSRPVVI FNFVNSS
YVQYGQLGEDN El LLGNH RTEECQFPSLKI FIAGNSAYEYVDYLFKRM I DLSSISTVD
SMIALDI DPLENTDFRVLELYSQKELRSSNVFDLEEI M REFNSYKQRVKYVEDKVVD
PLPP (SEQ ID NO: 3)
> signal sequence
MESRIWCLVVCVNLCIVCLGAV (SEQ ID NO: 4)
>HCMV VR1814 gB705 with linker
VSSSSTSHATSSAH NGSHTSRTTSAQTRSVSSQHVTSSEAVSH RAN ETIYNTTLKY
GDVVGVNTTKYPYRVCSMAQGTDLI RFER NI VCTPMKPI N EDLDEGI MVVYKRN IVA
HTFKVRVYQKVLTFRRSYAGHRTTYLLGSNTEYVAPPMWEI HHINRHSQCYSSYSR
VIAGTVFVAYH R DSYEN KTMQLM LDDYSNTHSTRYVTVKDQWHSRGSTALYRETC
NLNCMVTITTARSKYPYHFFATSTGDVVDISPFYNGTNRNTSYFGENADKFFI FPNY
TIVSDFGRANSAPETH RLVAFLERADSVISWDIQDEKNVTCQLTFWEASERTI RSEA
EDSYHFSSAKMTATFLSKKQEVNMSDPVLDCVRDQALNKLQQI FNASYNQTYEKY
G NVSVFETTGG LVVFWQGI KQKSLLELERLANSSGVNSTRATKASTG NTTTLSLES
ESVRNVLYAQLQFTYDTLRSYI NRALAQIAEAWCVDQRRTLEVFKELSKI NPSAI LSA
IYNKPIAARFMGDVLGLASCVTI NQTSVKVLRDMNVKESPGRCYSRPVVI FNFVNSS
YVQYGQLGEDN El LLGNH RTEECQFPSLKI FIAGNSAYEYVDYLFKRM I DLSSISTVD
SMIALDI DPLENTDFRVLELYSQKELRSSNVFDLEEI M REFNSYKQRVKYVEDKVVD
PLPPGSG (SEQ ID NO: 5)
48
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
>ACZ79977.1 envelope glycoprotein B [Human betaherpesvirus 5]
MESRIWCLVVCVNLCIVCLGAVVSSSSTSHATSSAHNGSHTSRTTSAQTRSVSSQH
VTSSEAVSHRANETIYNTTLKYGDVVGVNTTKYPYRVCSMAQGTDLIRFERNIVCTP
MKPINEDLDEGIMVVYKRNIVAHTFKVRVYQKVLTFRRSYAYIHTTYLLGSNTEYVAP
PMWEIHHINRHSQCYSSYSRVIAGTVFVAYHRDSYENKTMQLMLDDYSNTHSTRY
VTVKDQWHSRGSTWLYRETCNLNCMVTITTARSKYPYHFFATSTGDVVDISPFYNG
TNRNTSYFGENADKFFIFPNYTIVSDFGRANSAPETHRLVAFLERADSVISWDIQDE
KNVTCQLTFWEASERTIRSEAEDSYHFSSAKMTATFLSKKQEVNMSDPVLDCVRD
QALNKLQQIFNASYNQTYEKYGNVSVFETTGGLVVFWQGIKQKSLLELERLANSSG
VNSTRRTKRSTGNTTTLSLESESVRNVLYAQLQFTYDTLRSYINRALAQIAEAWCVD
QRRTLEVFKELSKINPSAILSAIYNKPIAARFMGDVLGLASCVTINQTSVKVLRDMNV
KESPGRCYSRPVVIFNFVNSSYVQYGQLGEDNEILLGNHRTEECQFPSLKIFIAGNS
AYEYVDYLFKRMIDLSSISTVDSMIALDIDPLENTDFRVLELYSQKELRSSNVFDLEEI
MREFNSYKQRVKYVEDKVVDPLPPYLKGLDDLMSGLGAAGKAVGVAIGAVGGAVA
SVVEGVATFLKNPFGAFTIILVAIAVVIIIYLIYTRQRRLCMQPLQNLFPYLVSADGTTV
TSGNTKDTSLQAPPSYEESVYNSGRKGPGPPSSDASTAAPPYTNEQAYQMLLALA
RLDAEQRAQQNGTDSLDGQTGTQDKGQKPNLLDRLRHRKNGYRHLKDSDEEENV
(SEQ ID NO: 6)
49
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
EXAMPLE 7:
CMV is a species-specific virus. Just has humans are infected by HCMV,
rhesus macaques are infected by rhCMV, which has a biology similar to HCMV.
This provides an animal model system for testing CMV vaccine inventions and
technologies in ways that may be impractical or unethical in humans, such as
certain
infectious challenge experiments. Therefore, rhCMV analogues of HCMV antigens
may be made to allow experimental investigation. RhCMV has a gB with a
structure
and function very similar to those of HCMV gB. A satisfactory correlation
exists
between effects in rhesus macaques relating to CMV and effects ultimately
observed
in humans. Accordingly, data from testing in rhesus macaques is reasonably
predictive of the response in humans. Following this line reasoning, an
analogous
construct of HCMV gB705 construct, named as RhCMV gB674, was designed and
tested in RhCMV(-) monkeys.
RhCMV gB674 Design
Following the same design of HCMV gB705, Pfizer's expression construct of
RhCMV
gB674 (SEQ ID NO: 7) was based on gB sequence of RhCMV strain UCD52, which
contains amino acids 1-674 of the ectodomains and a 6xHis affinity tag at the
C-
terminus. In addition, the gB674 protein contains three mutations (F128G,
I129H,
W213A) in the predicted fusion loops, because a fourth change in HCMV gB705 is
pre-existing in the natural UCD52 sequence. Similarly, mutations
(R429A/R430T/R432A) were introduced to totally inactivate the furin-like
cleavage
site, as illustrated.
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
Amino Acid Sequence:
> RhCMV gB674
1 msknwfpllc asylvvyvai assstgtasa vtpaptentt geiianttlr thevfrvnms
61 kfpyrvcsma qgtdllrfeq nircdsfkpt kedfdegimv vykrdikpyt fkvhiyqkil
.. 121 tfrqsysGHr enhlIgfsqe rlavpmwevn yinrInrcyn svvrnvagvt yvnyhkdsyv
181 netmhliedd ysnthsaryv tvkelwhkpg stAlyttscn vncmvtvtta rskypydffv
241 tsggevvdis pfyngsnneh fgenrdkfyi rrnysmvesy grdnaplvah elvafferpd
301 mlmswdivde anntceytfw eqsertirse adetyhftsh smtatfltlk kelnesdsdf
361 dcirdeaner lekifnttyn etyvksgnvs vyetsggliv fwlpvkekai wemqklateh
421 anntnatrAT kAstnsgnst kevlqnvvya qlqftydtlr nyinralrqi aeawckdqkr
481 tlevlkelsk inpsamlsai ydkpiaarfv gdvislarcv evdqnsvqvl rdmhtkekgl
541 cysrpvvlyt fvnsshvqyg qlgedneill grhrteaces pslkifiagn ssyeyvdyly
601 krmipldsis tvdtmisldi dplentdfra lelysrdelr ssnvfdledi mrefntykqr
661 mvhvegkvfd nvpa gsghhhhhh (SEQ ID NO: 7)
> RhCMV gB674
sknwfplIcasylvvyvaiassstgtasavtpaptenttgeiianttIrthevfrvnmskfpyrvcsmaqgtdllrfeq
nirc
dsfkptkedfdegimvvykrdikpytfkvhiyqkiltfrqsysGHrenhlIgfsgerlavpmwevnyinrInrcynsvv
r
nvagytyvnyhkdsyvnetmhlieddysnthsaryvtvkelwhkpgstAlyttscnvncmvtvttarskypydffvts
ggevvdispfyngsnnehfgenrdkfyi rrnysmvesygrdnaplvahelvafferpdm
Imswdivdeanntceytf
weqsertirseadetyhftshsmtatfltIkkelnesdsdfdcirdeanerlekifnttynetyvksgnvsvyetsggl
ivfw
I pvkekaiwemqklatehanntnatrATkAstnsgnstkevlqnvvyaql qftydtl rnyi nral rq
iaeawckdqkrtl
evIkelskinpsamIsaiydkpiaarfvgdvislarcvevdqnsvqvIrdmhtkekgIcysrpvvlytfvnsshvqygq
1
gednei Ilgrhrteacespslkifiagnssyeyvdylykrmi pldsistvdtm
isldidplentdfralelysrdelrssnvfdl
.. edimrefntykqrmvhvegkvfdnvpagsghhhhhh (SEQ ID NO: 8)
51
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
> RhCMV gB
MSKNWFPLLCASVLVVYVAIASSSTGTASAVTPAPTENTTGEIIANTTLRTHEVFRVN
MSKF PYRVCSMAQGTDLLR F EQ N I RCDSF KPTKED F DEG I MVVYKR DI KPYTFKVH I
YQKI LTF RQSYSF I R EN H LLGFSQERLAVPMWEVNYI N RLN RCYNSVVRNVAGVTY
.. VNYH KDSYVN ETM H LI EDDYSNTHSARYVTVKELWH KPGSTWLYTTSCNVNCMVT
VTTARSKYPYDFFVTSGGEVVDISPFYNGSN N EH FG EN RDKFYI RRNYSMVESYGR
DNA P LVA H ELVAF F ERP DM LMSWDI VD EAN NTCEYTFWEQSERTI RSEADETYH FT
SHSMTATF LTLKKELN ESDSDF DCI R DEAN ER LEKI FNTTYN ETYVKSGNVSVYETS
GGLIVFWLPVKEKAIWEMQKLATEHANNTNATRRRKRSTNSGNSTKEVLQNVVYA
QLQFTYDTLRNYI N RA LRQ IAEAWCKDQ KRTLEVLKELSKI N PSAM LSA IYDKP IAA R
FVGDVISLARCVEVDQNSVQVLRDM HTKEKGLCYSRPVVLYTFVNSSHVQYGQLG
EDN El LLGRH RTEACESPSLKI FIAGNSSYEYVDYLYKR M I PLDSI STVDTM I SLD I DP L
ENTDFRALELYSRDELRSSNVFDLEDIM REFNTYKQRMVHVEGKVFDNVPAYLRGL
DDM MSGLGSAGKA LGVA I GAVGGAVASFVEGVVG Fl EN PFGSFTVI LFLLAVLGVIY
LI YM RQ KRAYEKP F EH F F PYVVPPTTVKEA PPSYEQSQYEN I KEKAASATKEFSLEE
AYQM LLALQKLDQEKRRKAEADDEDFASNGQSAGFLDRLRN RR RGGYQ KI QN EYE
V (SEQ ID NO: 11)
52
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
EXAMPLE 8: Analytics of RhCMV gB674
1. SDS-PAGE analysis of gB674 (ELN 00707788-0011)
The gB674 protein was transiently expressed in Expi293 cells and purified via
the
C-terminal 6xHis tag. On SDS-PAGE, the protein exhibited a single band under
.. reducing (R) conditions, demonstrating the lack of protease cleavage as
designed
and >98% purity. Some oligomeric forms were shown under a non-reducing
condition. See FIG. 17A.
2. Size Exclusion chromatography (ELN 00707788-0011)
The gB674 protein was treated with EDTA after purification and analyzed on
Superose-6 column. As observed with HCMV gB705, gB674 protein was eluted as a
major peak, consistent with a homogenous individual trimer. See FIG. 17B.
Immunogenicity testing:
RhCMV(-) rhesus monkeys were screened and proven as seronegative for anti-
gB by an ELISA test (data not shown). Six animals were immunized
intramuscularly
with 100 ug of gB674 per dose and 50 pg of RhCMV pentamer in 50 ug of QS-21
adjuvant at week 0, 4 and 8. A group of six animals were injected with a
placebo
regimen lacking only the gB protein, as controls.
At week 9, anti-gB response was detected in an ELISA using recombinant
RhCMV gB lacking a C-terminal 6xHis tag. In so doing, the ELISA avoids
detection
.. of antibody responses to the 6His tags on the gB and pentamer antigens in
the
vaccine. Sera from all six animals in the control group (animals C1-C6 in FIG.
14)
were negative of anti-gB at a 1:100 dilution on week 9, as expected. The anti-
gB
binding titers from the six vaccines (animals V1-V6 in FIG. 14) on week 9
after three
doses of vaccination were detected. Thus, recombinant gB674 of RhCMV UCD52 is
highly immunogenic. This provides supporting evidence of recombinant gB705 of
HCMV as a vaccine antigen.
53
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
EXAMPLE 9: Efficacy testing of the Rhesus equivalent of HCMV gB705
Utility of the new design of HCMV post-fusion gB705 was tested by evaluating
the protective efficacy of immunization with RhCMV gB674 protein (SEQ ID NO:
7) in
an oral RhCMV infectious challenge model in RhCMV(-) rhesus monkeys. The
candidate monkeys were screened and proven seronegative for gB by an ELISA
test
(data not shown). Six animals in each of three experimental groups were
immunized
intramuscularly at weeks 0, 4 and 8. All formulations, including the control
formulation, had a pH of 7.7 and contained 50 lig of the QS-21 adjuvuant. Six
animals in one group of six animals were injected with a placebo formulation,
containing no antigen, as negative controls. Six animals in a second group
were
immunized with 50 lig each of a recombinant RhCMV pentamer complex (containing
the gH, gL, pUL128, pUL130, and pUL131 RhCMV antigens). Six animals in a third
group were each immunized with 100 lig of gB674 + 50 lig of recombinant RhCMV
pentamer. Two weeks after the third vaccination, all animals were challenged
with
8x105 plaque-forming units (PFU) of UCD52 RhCMV viral stock by oral
inoculation,
and the challenge was repeated weekly for a total of 5 times. This study was
designed to demonstrate whether recombinant gB674 would add extra protective
efficacy to a vaccine formulation containing recombinant RhCMV pentamer as the
sole antigen. Virus infection was monitored by the presence of viremia and by
virus
shedding in saliva and urine compartments. The presence of virus in these
target
samples was quantified by a qPCR assay with a limit of quantitation of 125 DNA
copies/ml. As shown in Fig 15 panel A, five oral challenges led to virus
shedding in
saliva in 5 of 6 animals in the control group. Week 9 and week 15 are one week
prior to the first virus challenge and one week after the fifth virus
challenge,
.. respectively. The same five control animals had viremia and high level of
virus
shedding in urine (data not shown). Five of six animals in both the pentamer
and the
pentamer + gB groups of animals also acquired virus infection, but with
modestly
reduced levels of viremia and urine virus shedding (data not shown).
Vaccination
with pentamer alone led to some reduction of virus shedding in saliva, Fig 15
panel
.. B. Importantly, inclusion of gB674 in combination with the pentamer
resulted in
significant delay and reduction in saliva virus shedding, relative to
immunization with
the pentamer alone, Fig 15 panel C. This result indicates that adding the
gB674 to
the pentamer reduced viral shedding in the urine. This likely reflects
limitation of
54
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
virus infection of the kidneys of the monkeys that received the combination of
pentamer and gB antigens. This result demonstrates the utility of the rhCMV
gB674
antigen as a protective vaccine antigen, and by extension, of the analogous
HCMV
gB705 antigen.
55
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
The following clauses describe additional embodiments of the invention:
Cl. A polypeptide comprising at least one mutation in the fusion loop 1
(FL1)
region of an HCMV gB polypeptide.
02. A polypeptide comprising at least one mutation in the fusion loop 2
(FL2)
region of an HCMV gB polypeptide.
03. A polypeptide comprising at least one mutation in the fusion loop 1
(FL1)
region and the fusion loop 2 (FL2) region of an HCMV gB polypeptide.
04. A polypeptide comprising at least one mutation in the furin-like
cleavage site
of an HCMV gB polypeptide.
05. A polypeptide comprising at least two mutations in the fusion loop 2
(FL2)
region of an HCMV gB polypeptide.
06. A polypeptide comprising at least two mutations in the fusion loop 1
(FL1)
region and the fusion loop 2 (FL2) region of an HCMV gB polypeptide.
07. A polypeptide comprising at least two mutations in the furin-like
cleavage site
of an HCMV gB polypeptide.
08. A polypeptide comprising a mutation at position Y155, as compared to
SEQ
ID NO: 6.
09. A polypeptide comprising a mutation at positions Y155 and 1156, as
compared
to SEQ ID NO: 6.
010. A polypeptide comprising a mutation at positions Y155, 1156, and H157, as
compared to SEQ ID NO: 6.
C11. A polypeptide comprising a mutation at positions 1156 and H157, as
compared to SEQ ID NO: 6.
012. A polypeptide comprising a mutation at positions Y155, 1156, H157, and
W240, as compared to SEQ ID NO: 6.
56
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
013. A polypeptide comprising a mutation at positions Y155 and W240, as
compared to SEQ ID NO: 6.
014. A polypeptide comprising a mutation at positions Y155, H157, and W240, as
compared to SEQ ID NO: 6.
015. A polypeptide comprising a mutation at positions Y155 and H157, as
compared to SEQ ID NO: 6.
016. A polypeptide comprising the mutation Y155G, as compared to SEQ ID NO:
6.
017. A polypeptide comprising the mutations Y155G and I156H, as compared to
SEQ ID NO: 6.
018. A polypeptide comprising the mutations Y155G, I156H, and H157R, as
compared to SEQ ID NO: 6.
019. A polypeptide comprising the mutations I156H and H157R, as compared to
SEQ ID NO: 6.
020. A polypeptide comprising the mutations Y155G, I156H, H157R, and W240A,
as compared to SEQ ID NO: 6.
021. A polypeptide comprising the mutations Y155G and W240A, as compared to
SEQ ID NO: 6.
022. A polypeptide comprising the mutations Y155G, H157R, and W240A, as
compared to SEQ ID NO: 6.
023. A polypeptide comprising the mutations Y155G and H157R, as compared to
SEQ ID NO: 6.
024. A polypeptide comprising an amino acid sequence that is at least about
60%
identical to SEQ ID NO: 1.
57
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
025. A polypeptide comprising an amino acid sequence that is at least about
60%
identical to SEQ ID NO: 2.
026. A polypeptide comprising an amino acid sequence that is at least about
60%
identical to SEQ ID NO: 3.
.. 027. A polypeptide comprising an amino acid sequence that is at least about
60%
identical to SEQ ID NO: 5.
028. A polypeptide comprising an amino acid sequence that is at least about
60%
identical to SEQ ID NO: 7.
029. A polypeptide comprising an amino acid sequence that is at least about
60%
identical to SEQ ID NO: 8.
030. A polypeptide comprising the amino acid sequence set forth in SEQ ID NO:
1.
031. A polypeptide comprising the amino acid sequence set forth in SEQ ID NO:
2.
032. A polypeptide comprising the amino acid sequence set forth in SEQ ID NO:
3.
033. A polypeptide comprising the amino acid sequence set forth in SEQ ID NO:
5.
034. A polypeptide comprising the amino acid sequence set forth in SEQ ID NO:
7.
035. A polypeptide comprising the amino acid sequence set forth in SEQ ID NO:
8.
036. The polypeptide according to any one of clauses 01-035, wherein the
polypeptide does not comprise a mutation at any one of the following
positions: (i)
R236, (ii) G237, (iii) T158; (iv) Y242.
037. The polypeptide according to any one of clauses 01-035, wherein the
polypeptide does not comprise any one of the following mutations: (i) R236N,
(ii)
G237N, (iii) T158N; (iv) Y242T; (v) Y2425; (vi) Y2420.
038. The polypeptide according to any one of clauses 01-037, wherein the amino
acid sequence SEQ ID NO: 9 is a part of the polypeptide sequence.
58
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
039. The polypeptide according to any one of clauses 01-037, wherein the amino
acid sequence SEQ ID NO: 10 is a part of the polypeptide sequence.
040. The polypeptide according to any one of clauses 01-037, wherein the amino
acid sequences of SEQ ID NO: 9 and SEQ ID NO: 10 is a part of the polypeptide
sequence.
041. The polypeptide according to any one of clauses 01-040, wherein the
polypeptide does not comprise a protease cleavage site.
042. The polypeptide according to any one of clauses 01-040, wherein the
polypeptide does not comprise a wild-type CMV protease cleavage site.
043. The polypeptide according to any one of clauses 01-040, wherein the
polypeptide does not comprise a non-naturally occurring protease cleavage site
that replaces the wild-type CMV protease cleavage site.
044. The polypeptide according to any one of clauses 01-040, wherein the
polypeptide does not comprise an N- glycosylation site comprising N-X-S/T/C
motif, wherein X is any amino acid residue.
045. The polypeptide according to any one of clauses 01-040, wherein the
polypeptide does not comprise a modified amino acid sequence that introduces
an 0-linked glycosylation site.
046. The polypeptide according to any one of clauses 01-045, wherein the
polypeptide does not include a deletion or substitution of any one of the
amino
acid residues selected from the group consisting of 154, 158, 159, 160, 230,
231,
232, 233, 234, 235, 236, 237, 238, 239, 241, and 242, according to the
numbering of SEQ ID NO: 6.
047. The polypeptide according to any one of clauses 01-046, wherein the
polypeptide does not include a mutation of any one of the amino acid residues:
59
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
Y160, R236, S238, T239, and Y242, according to the numbering of SEQ ID NO:
6.
048. The polypeptide according to any one of clauses 01-047, wherein the
polypeptide does not include the cytoplasmic tail of HCMV gB.
049. The polypeptide according to any one of clauses 01-048, wherein the
polypeptide does not contain an insect cell pattern of glycosylation.
050. The polypeptide according to any one of clauses 01-049, wherein the
polypeptide is contacted with ethylenediaminetetraacetic acid (EDTA).
051. The polypeptide according to any one of clauses 01-050, wherein the
polypeptide undergoes a structural conformation change in response to a pH
change.
052. The polypeptide according to clause 051, wherein the polypeptide exhibits
improved solubility or stability, as compared to a recombinant gB protein that
does not include the following mutations Y155G, I156H, H157R, and W240A.
053. A composition comprising the polypeptide according to any one of clauses
01-051, and a diluent.
054. A composition comprising the polypeptide according to any one of clauses
01-051, and an adjuvant.
055. The composition according to any one of clauses 053-054, wherein the
composition is immunogenic.
056. The composition of clause 055, for use in inducing an immune response
against cytomegalovirus.
057. A recombinant nucleic acid molecule encoding the polypeptide according to
any one of clauses 01-051, wherein the polypeptide undergoes a structural
conformation change in response to a pH change.
CA 03016867 2018-09-06
WO 2017/153954
PCT/IB2017/051401
058. The recombinant nucleic acid molecule according to clause 057, wherein
said
recombinant nucleic acid (a) is not a self-replicating RNA molecule; (b) is
not an
alphavirus replicon; (c) does not encode any alphavirus nonstructural
proteins,
such as NSP1, NSP2, NSP3 and NSP4; (d) does not contain: an Internal
Ribosomal Entry Site (I RES), such as EMCV or EV71 ; and/or (e) does not
contain a viral 2A site, such as FMDV.
059. A method for raising antibodies using the polypeptide according to any
one of
clauses 01-051.
060. The antibody according to clause 059, wherein said antibody is for use in
a
diagnostic assay.
061. The antibody according to clause 059, wherein said antibody is labelled
directly or indirectly.
062. The antibody according to clause 059, wherein said antibody is for use in
therapy.
063. A method of eliciting an immune response in a mammal, the method
comprising administering to the mammal an effective amount of the polypeptide
according to any one of clauses 01-051.
064. A method for reducing cytomegalovirus viral shedding in a mammal, the
method comprising administering to the mammal an effective amount of the
polypeptide according to any one of clauses 01-051.
065. The method according to clause 064, wherein the reduction in viral
shedding
is as compared to the viral shedding following an administration of a CMV
pentamer.
066. The method according to clause 064, wherein the challenge virus is
homologous to the VR1814 CMV strain.
067. The method according to clause 064, wherein the challenge virus is
homologous to the macacine herpesvirus 3 isolate 21252 CMV strain.
61