Note: Descriptions are shown in the official language in which they were submitted.
CA 03026340 20112-133
- 1 -
Description
[Title of Invention] FUSION PROTEIN FOR IMPROVING PROTEIN
EXPRESSION LEVEL FROM TARGET mRNA
[Technical Field]
[0001]
The present invention relates to fusion proteins for
improving protein expression levels from target mRNAs.
[Background Art]
[0002]
Techniques of binding nucleic acid-binding protein
factors revealed by a variety of analyses to sequences of
interest are established and used in recent years. Use
of this sequence-specific binding enables removal of a
target DNA sequence or regulation (activation or
inactivation) of expression of a protein coding gene
present downstream of the target DNA sequence in some
extent.
[0003]
While zinc finger nuclease (ZFN), TAL effector
nuclease (TALEN), Crispr-ca59, and the like are known as
techniques using protein factors which act on DNA, the
development of techniques using protein factors which act
specifically to RNA is still limited.
[0004]
CA 03026340 2018-12-03
- 2 -
The present inventors have proposed a method of
designing a protein which can specifically bind to a
target RNA sequence using the properties of PPR proteins
(protein having one or more pentatricopeptide repeat
(PPR) motifs), which are proteins mainly found in plants
(Patent Literature 1).
[Citation List]
[Patent Literature]
[0005]
[Patent Literature 1]
W02013/058404
[Summary of Invention]
[Technical Problem]
[0006]
In the disclosure according to Patent Literature 1,
the amino acids which function when a PPR motif
demonstrates RNA-binding properties were identified, and
the relation between the structure of the PPR motif and
the target base was revealed, thereby enabling the
construction of proteins which have one or more PPR
motifs and can bind to RNAs having any sequence and
length. However, no method has ever been found which
actually regulates target RNAs using the techniques
according to Patent Literature 1.
CA 03026340 2018-12-03
- 3 -
[Solution to Problem]
[0007]
As a result of extensive research on a method of
improving a protein expression level from a target mRNA
using a PPR protein, the present inventors have found
that a fusion protein of a predetermined functional
domain and a PPR protein improves the protein expression
level from the target mRNA, and have completed the
present invention.
[0008]
Specifically, an embodiment of the present invention
relates to a fusion protein for improving a protein
expression level from a target mRNA, the fusion protein
comprising:
(A) one or more functional domains which improve a
protein expression level from an mRNA; and
(B) a polypeptide moiety which can bind to a target
mRNA in an RNA base-selective or RNA base sequence-
specific manner,
wherein polypeptide moiety (B) is a polypeptide
moiety comprising one or more PPR motifs, each PPR motif
comprising a polypeptide consisting of 30 to 38 amino
acids in length and being represented by Formula 1:
[Formula 1]
(Helix A)-X-(Helix B)-!. (Formula 1)
where
CA 03026340 2018-12-03
- 4 -
Helix A is a moiety which consists of 12 amino acids
in length and can form an a-helix structure, and is
represented by Formula 2:
[Formula 2]
A1-A2-A3-A4-A6-A8-k-A8-A94110-Ai1-Al2 (Formula 2)
where Ai to Al2 each independently represent an amino
acid;
X is not present, or is a moiety consisting of 1 to
9 amino acids in length;
Helix B is a moiety which consists of 11 to 13 amino
acids in length and can form an a-helix structure;
L is a moiety consisting of 2 to 7 amino acids in
length and represented by Formula 3:
[Formula 3]
LVIrLVis-LwatiV"Liiie*Lirti (Formula 3)
where the amino acids are numbered from the C-
terminal as "i" (-1), "ii" (-2), ... and
Li,, to Lvi, may not be present, and
a combination of three amino acids Ai, A4, and Li, or
a combination of two amino acids A4 and Lii corresponds to
a base or base sequence of the target mRNA.
[0009]
In an embodiment according to the present invention,
polypeptide moiety (B) comprises 2 to 30 PPR motifs, and
CA 03026340 2018-12-03
- 5 -
the plurality of PPR motifs is arranged so as to
specifically bind to the base sequence of the target mRNA.
[0010]
Moreover, in an embodiment according to the present
invention, polypeptide moiety (B) comprises 5 to 25 PPR
motifs.
[0011]
Moreover, in an embodiment according to the present
invention, one or more functional domains (A) each bind
to an N-terminal side and/or a C-terminal side of
polypeptide moiety (B).
[0012]
Moreover, in an embodiment according to the present
invention, one or more functional domains (A) are
selected from the group consisting of a domain which
guides ribosome to the mRNA, a domain associated with
initiation or promotion of translation of the mRNA, a
domain associated with nuclear export of the mRNA, a
domain associated with binding to an endoplasmic
reticulum membrane, a domain containing an endoplasmic
reticulum retention signal (ER retention signal) sequence,
and a domain containing an endoplasmic reticulum signal
sequence.
[0013]
Moreover, in an embodiment according to the present
invention, the domain which guides ribosome to the mRNA
is a domain containing all or functional part of a
CA 03026340 2018-12-03
- 6 -
polypeptide selected from the group consisting of DENR
(Density-regulated protein), MCT-1 (Malignant T-cell
amplified sequence 1), TPT1 (Translationally-controlled
tumor protein), and Lerepo4 (Zinc finger CCCH-domain),
the domain associated with initiation or promotion
of translation of the mRNA is a domain containing all or
functional part of a polypeptide selected from the group
consisting of eIF4E and eIF4G,
the domain associated with nuclear export of the
mRNA is a domain containing all or functional part of
SLBP (Stem-loop binding protein),
the domain associated with binding to an endoplasmic
reticulum membrane is a domain containing all or
functional part of a polypeptide selected from the group
consisting of SEC61B, TRAP-alpha (Translocon associated
protein alpha), SR-alpha, Dial (Cytochrome b5 reductase
3), and p180,
the endoplasmic reticulum retention signal (ER
retention signal) sequence is a signal sequence
containing a KDEL (KEEL) sequence, or
the endoplasmic reticulum signal sequence is a
signal sequence containing MGWSCIILFLVATATGAHS (SEQ ID
NO: 22).
[0014]
Moreover, in an embodiment according to the present
invention, the combination of the three amino acids AI,
A4, and Li, in each of the PPR motifs is:
CA 03026340 2018-12-03
- 7 -
(valine, threonine, asparagine), (phenylalanine,
serine, asparagine), (phenylalanine, threonine,
asparagine), (isoleucine, asparagine, aspartic acid), or
(threonine, threonine, asparagine) in order of (Au A4,
Lli) if a target base for the PPR motif is A (adenine);
(glutamic acid, glycine, aspartic acid), (valine,
threonine, aspartic acid), (lysine, threonine, aspartic
acid), or (leucine, threonine, aspartic acid) in order of
(Au AA, Lil) if the target base for the PPR motif is G
(guanine);
(valine, asparagine, aspartic acid), (isoleucine,
asparagine, asparagine), (isoleucine, asparagine,
aspartic acid), (isoleucine, methionine, aspartic acid),
(phenylalanine, proline, aspartic acid), or (tyrosine,
proline, aspartic acid) in order of (Au ALI, L11) if the
target base for the PPR motif is U (uracil); or
(valine, asparagine, asparagine), (isoleucine,
asparagine, asparagine), (valine, asparagine, serine), or
(isoleucine, methionine, aspartic acid) in order of (Au
if the target base for the PPR motif is C
(cytosine).
[0015]
Moreover, in an embodiment according to the present
invention, the combination of the two amino acids A4 and
in each of the PPR motifs is:
CA 03026340 2018-12-03
- 8 -
(threonine, asparagine), (serine, asparagine), or
(glycine, asparagine) in order of (A4, Lii) if a target
base for the PPR motif is A (adenine);
(threonine, aspartic acid) or (glycine, aspartic
acid) in order of (A4, Lid if the target base for the PPR
motif is G (guanine);
(asparagine, aspartic acid), (proline, aspartic
acid), (methionine, aspartic acid), or (valine,
threonine) in order of (A4, Lii) if the target base for
the PPR motif is U (uracil); or
(asparagine, asparagine), (asparagine, serine), or
(leucine, aspartic acid) in order of (A4, Lid if the
target base for the PPR motif is C (cytosine).
[0016]
Another embodiment according to the present
invention relates to a nucleic acid which encodes the
fusion protein according to the present invention.
[0017]
Still another embodiment according to the present
invention relates to a vector (preferably an expression
vector) comprising the nucleic acid according to the
present invention.
[0018]
Further still another embodiment according to the
present invention relates to a method of improving a
protein expression level from a target mRNA within a cell,
the method comprising:
CA 03026340 2018-12-03
- 9 -
a step of providing the fusion protein according to
the present invention or the vector according to the
present invention; and
a step of introducing the fusion protein or the
vector into the cell.
[0019]
Moreover, in an embodiment according to the present
invention, the cell is a eukaryotic cell.
[0020]
Moreover, in an embodiment according to the present
invention, the cell is an animal cell.
[0021]
Moreover, in an embodiment according to the present
invention, the animal cell is a human cell.
[0022]
Inventions having any combination of one or more
features of the present invention described above are
also included in the scope of the present invention.
[Brief Description of Drawings]
[0023]
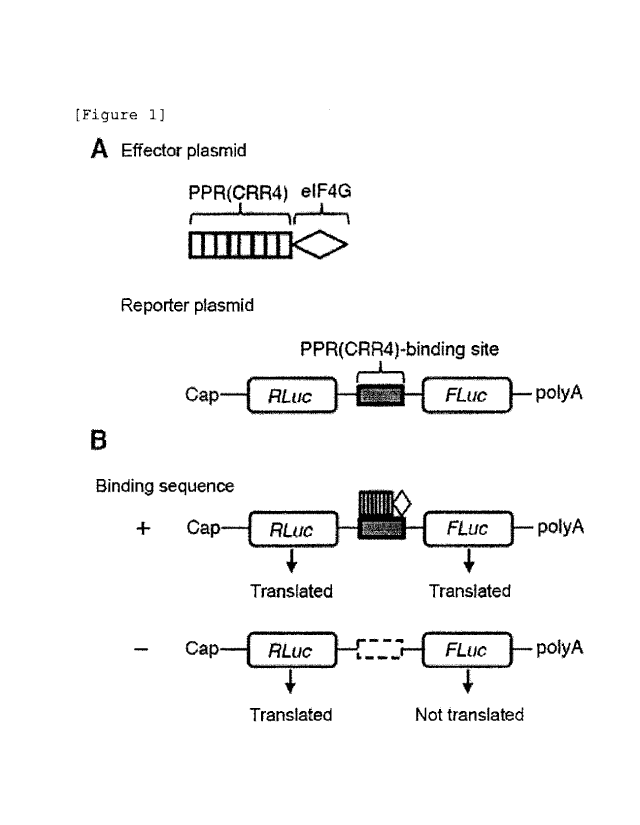
[Figure 1] Figure 1 illustrates a schematic view of
an effector plasmid and a reporter plasmid used in
Examples, and a schematic view of an experimental outline.
Figure 1A illustrates a schematic view of the effector
plasmid and the reporter plasmid used in Examples. A
fusion protein of PPR motifs and eIF4G expresses from the
CA 03026340 2018-12-03
- 10 -
effector plasmid. In Examples, a CRR4 protein was used,
whose target sequence is well researched. From the
reporter plasmid, renilla luciferase (RLuc) and firefly
luciferase (FLuc) are transcribed in the form of a
dicistronic mRNA. A PPR-binding sequence (here, CRR4-
binding sequence) was inserted into a site on the 5' end
of FLuc. Figure 1B illustrates a schematic view of an
experimental outline of Examples. Irrespective of the
presence/absence of the PPR-binding sequence, RLuc is
translated at a similar level. For this reason, the
activity value of RLuc can be treated as a control in
transfection in this reporter system. The translation of
Fluc is started only when PPR-eIF4G binds to the PPR-
binding sequence and translation factors can be attracted
by the effects of eIF4G. In contrast, the translation of
FLuc remains at a low level if the PPR-binding sequence
is not present and thus, PPR-eIF4G cannot bind to the
PPR-binding sequence.
[Figure 2] Figure 2 illustrates an experimental
procedure of a reporter assay using HEK293T cells.
[Figure 3] Figure 3 shows the experimental results
of Example 1. The activation of sequence-specific
translation depends on CRR4-eIF4G and the PPR-binding
sequence. This experiment was performed using an
effector plasmid, into which CRR4-Flag (without
translation activating factor, in white) or CRR4-eIF4G
(with translation activating factor, in gray) was
CA 03026340 2018-12-03
- 11 -
inserted, and a reporter vector with or without an
inserted PPR-binding sequence. From the results, it was
verified that specific translation activity increased
2.75 times in the presence of both PPR-eIF4G and the PPR-
binding sequence. The value represents the average and
the standard deviation (N = 3).
[Figure 4] Figure 4 illustrates an outline of the
experiment in Example 2.
[Figure 5] Figure 5 illustrates the experimental
results in Example 2 and the functions of the domains.
[Figure 6] Figure 6 illustrates the experimental
results in Example 2 and the functions of the domains.
[Description of Embodiment]
[0024]
[PPR motifs and PPR proteins]
Unless otherwise specified, the term "PPR motif"
used in the present invention indicates a polypeptide
which is composed of 30 to 38 amino acids and has an
amino acid sequence having an E value equal to or less
than a predetermined value (desirably E-03), the E value
being obtained at PF01535 in Pfam and PS51375 at Prosite
during the analysis of the amino acid sequence with a
protein domain search program on the Web. The position
number of an amino acid forming the PPR motif defined in
the present invention is substantially as defined as
PF01535 while it corresponds to the number obtained by
CA 03026340 2018-12-03
- 12 -
subtracting 2 from the location of the amino acid in
PS51375 (for example, position 1 in the present invention
corresponds to position 3 in PS51375). Note that the
term "ii" (-2)-th amino acid refers to the second amino
acid from the tail end (C-terminal side) of the amino
acids forming one PPR motif or the amino acid close to
the N-terminal by two amino acids from the first amino
acid of the next PPR motif (that is, -2 amino acid). If
the next PPR motif is not clearly identified, the forward
amino acid by two amino acids from the first amino acid
of the next helix structure is defined as "ii". See
http://pfam.sanger.ac.uk/ for Pfam and
http://www.expasy.org/prosite/ for Prosite.
[0025]
Although the conserved amino acid sequence of the
PPR motif has low conservation properties at the amino
acid level, two a-helices are well conserved on the
secondary structure. Although a typical PPR motif is
composed of 35 amino acids, its length is variable from
30 to 38 amino acids.
[0026]
More specifically, the term PPR motif used in the
present invention is composed of a polypeptide having 30
to 38 amino acids in length and being represented by
Formula 1:
[0027]
CA 03026340 2018-12-03
- 13 -
[Formula 411
(Helix A)-X-(Helix B)-1_ (Formula 1)
where
Helix A is a moiety which consists of 12 amino acids
in length and can form an a-helix structure, and is
represented by Formula 2:
[0028]
[Formula 511
ArA2-k-A4-A5-A6-A7-A8-A9AIVA11-Al2 (Formula 2)
where Ai to Al2 each independently represent an amino
acid;
X is not present, or is a moiety consisting of 1 to
9 amino acids in length;
Helix B is a moiety which consists of 11 to 13 amino
acids in length and can form the a-helix structure; and
L is a moiety consisting of 2 to 7 amino acids in
length and represented by Formula 3:
[0029]
[Formula 6]
(Formula 3)
where the amino acids are numbered from the C-
terminal side as "i" (-1), "ii" (-2), ... and
Lill to Lvii may not be present.
[0030]
CA 03026340 2018-12-03
- 14 -
Unless otherwise specified, the term "PPR protein"
used in the present invention indicates a PPR protein
comprising one or more PPR motifs described above,
preferably two or more PPR motifs described above.
Unless otherwise specified, the term "protein" used
herein generally indicates substances consisting of
polypeptides (chains of several amino acids bound through
peptide bond), also including those consisting of
relatively low molecular weight polypeptides. The term
"amino acid" used in the present invention may indicates
a usual amino acid molecule, or otherwise may indicate an
amino acid residue forming a peptide chain in some cases.
Persons skilled in the art clearly understand from
contexts which case the term indicates.
[0031]
Unless otherwise specified, the "selective" used in
the present invention about the binding properties of the
PPR motif to the RNA bases indicates that the binding
activity of a PPR motif to one of the RNA bases is higher
than the binding activity thereof to other bases.
Persons skilled in the art can plan the experiment for
this selectivity and verify it, and can also determine
through calculation.
[0032]
Unless otherwise specified, the term "RNA base" used
in the present invention indicates a base of a
ribonucleotide forming an RNA, specifically adenine (A),
CA 03026340 2018-12-03
- 15 -
guanine (G), cytosine (C), or uracil (U). Note that
although the PPR protein can have selectivity to the base
in the RNA, it does not bind to a nucleic acid monomer.
[0033]
PPR protein is present in many plants, and 500
proteins, about 5000 motifs can be found in Arabidopsis
thaliana. PPR motifs and PPR proteins having a variety
of amino acid sequences are also present in many land
plants such as Oryza, Populus, and Selaginella
tamariscina. In the present invention, PPR motifs and
PPR proteins present in the natural world may be used, or
PPR motifs and PPR proteins designed based on the method
disclosed in W02013/058404, for example, may be used.
Specifically, desired PPR motifs and PPR proteins can be
designed based on the following information disclosed in
W02013/058404.
[0034]
(I) Information on the position of the amino acid
essential for selective binding
The combination (Al, A4, Lii) of three, i.e., 1st,
4th, and "ii" (-1)-th amino acids of a PPR motif or the
combination (A4, Lii) of two, i.e., 4th and "ii" (-1)-th
amino acids is essential for selective binding to the RNA
base, and the target RNA base for binding can be
determined by these combinations.
[0035]
CA 03026340 2018-12-03
- 16 -
The present invention can use the findings about the
combination of three amino acids Al, A4, and Lii, and/or
the combination of two amino acids A4 and Lii disclosed
in W02013/058404.
[0036]
(II) Information about the correspondence of the
combination of three amino acids Al, A4, and Lii to RNA
bases
(3-1) If the combination of three amino acids Al, A4, and
Lii is valine, asparagine, and aspartic acid in this
order, the PPR motif has a selective RNA base binding
ability as follows: binding to U is the strongest, and
binding to C is the second strongest, followed by binding
to A or G.
(3-2) If the combination of three amino acids Al, A4, and
Lii is valine, threonine, and asparagine in this order,
the PPR motif has a selective RNA base binding ability as
follows: binding to A is the strongest, and binding to G
is the second strongest, followed by binding to C without
binding to U.
(3-3) If the combination of three amino acids Al, A4, and
Lii is valine, asparagine, and asparagine in this order,
the PPR motif has a selective RNA base binding ability as
follows: binding to C is the strongest and binding to A
or U is the second strongest, without binding to G.
(3-4) If the combination of three amino acids Al, A4, and
Lii is glutamic acid, glycine, and aspartic acid in this
CA 03026340 2018-12-03
- 17 -
order, the PPR motif has a selective RNA base binding
ability as follows: binding to G is strong, without
binding to A, U, or C.
(3-5) If the combination of three amino acids Al, A4, and
Lii is isoleucine, asparagine, and asparagine in this
order, the PPR motif has a selective RNA base binding
ability as follows: binding to C is the strongest, and
binding to U is the second strongest, followed by binding
to A, without binding to G.
(3-6) If the combination of three amino acids Al, A4, and
Lii is valine, threonine, and aspartic acid in this order,
the PPR motif has a selective RNA base binding ability as
follows: binding to G is the strongest and binding to U
is the second strongest, without binding to A or C.
(3-7) If the combination of three amino acids Al, A4, and
Lii is lysine, threonine, and aspartic acid in this order,
the PPR motif has a selective RNA base binding ability as
follows: binding to G is the strongest and binding to A
is the second strongest, without binding to U or C.
(3-8) If the combination of three amino acids Al, A4, and
Lii is phenylalanine, serine, and asparagine in this
order, the PPR motif has a selective RNA base binding
ability as follows: binding to A is the strongest, and
binding to C is the second strongest, followed by binding
to G and U.
(3-9) If the combination of three amino acids Al, A4, and
Lii is valine, asparagine, and serine in this order, the
CA 03026340 2018-12-03
- 18 -
PPR motif has a selective RNA base binding ability as
follows: binding to C is the strongest and binding to U
is the second strongest, without binding to A or G.
(3-10) If the combination of three amino acids Al, A4,
and Lii is phenylalanine, threonine, and asparagine in
this order, the PPR motif has a selective RNA base
binding ability as follows: binding to A is strong,
without binding to G, U, or C.
(3-11) If the combination of three amino acids Al, A4,
and Lii is isoleucine, asparagine, aspartic acid in this
order, the PPR motif has a selective RNA base binding
ability as follows: binding to U is the strongest and
binding to A is the second strongest, without binding to
G or C.
(3-12) If the combination of three amino acids Al, A4,
and Lii is threonine, threonine, and asparagine in this
order, the PPR motif has a selective RNA base binding
ability as follows: binding to A is strong, without
binding to G, U, or C.
(3-13) If the combination of three amino acids Al, A4,
and Lii is isoleucine, methionine, and aspartic acid in
this order, the PPR motif has a selective RNA base
binding ability as follows: binding to U is the strongest
and binding to C is the second strongest, without binding
to A or G.
(3-14) If the combination of three amino acids Al, A4,
and Lii is phenylalanine, proline, and aspartic acid in
CA 03026340 2018-12-03
- 19 -
this order, the PPR motif has a selective RNA base
binding ability as follows: binding to U is the strongest
and binding to C is the second strongest, without binding
to A or G.
(3-15) If the combination of three amino acids Al, A4,
and Lii is tyrosine, proline, and aspartic acid in this
order, the PPR motif has a selective RNA base binding
ability as follows: binding to U is strong, without
binding to A, G, or C.
(3-16) If the combination of three amino acids Al, A4,
and Lii is leucine, threonine, and aspartic acid in this
order, the PPR motif has a selective RNA base binding
ability as follows: binding to G is strong, without
binding to A, U, or C.
[0037]
(II) Information about the correspondence of the
combination of two amino acids A4 and Lii to the RNA
bases
(2-1) If A4 and Lii in this order are asparagine and
aspartic acid, the PPR motif has a selective RNA base
binding ability as follows: binding to U is the strongest,
and binding to C is the second strongest, followed by
binding to A and G.
(2-2) If A4 and Lii in this order are asparagine and
asparagine, the PPR motif has a selective RNA base
binding ability as follows: binding to C is the strongest,
CA 03026340 2018-12-03
- 20 -
binding to U is the second strongest, followed by binding
to A and G.
(2-3) If A4 and Lii in this order are threonine and
asparagine, the PPR motif has a selective RNA base
binding ability with strong binding to A and weak binding
to G, U, and C.
(2-4) If A4 and Lii in this order are threonine and
aspartic acid, the PPR motif has a selective RNA base
binding ability with strong binding to G and weak binding
to A, U, and C.
(2-5) If A4 and Lii in this order are serine and
asparagine, the PPR motif has a selective RNA base
binding ability as follows: binding to A is the strongest
and binding to G, U, and C is the second strongest.
(2-6) If A4 and Lii in this order are glycine and
aspartic acid, the PPR motif has a selective RNA base
binding ability as follows: binding to G is the strongest,
and binding to U is the second strongest, followed by
binding to A, without binding to C.
(2-7) If A4 and Lii in this order are asparagine and
serine, the PPR motif has a selective RNA base binding
ability as follows: binding to C is the strongest, and
binding to U is the second strongest, followed by binding
to A and G.
(2-8) If A4 and Lii in this order are proline and
aspartic acid, the PPR motif has a selective RNA base
binding ability as follows: binding to U is the strongest,
CA 03026340 2018-12-03
- 21 -
and binding to G, C, and C is the second strongest,
without binding to A.
(2-9) If A4 and Lii in this order are glycine and
asparagine, the PPR motif has a selective RNA base
binding ability as follows: binding to A is the strongest,
and binding to G is the second strongest, without binding
to C or U.
(2-10) If A4 and Lii in this order are methionine and
aspartic acid, the PPR motif has a selective RNA base
binding ability with strong binding to U and weak binding
to A, G, and C.
(2-11) If A4 and Lii in this order are leucine and
aspartic acid, the PPR motif has a selective RNA base
binding ability as follows: binding to C is the strongest,
and binding to U is the second strongest, without binding
to A or G.
(2-12) If A4 and Lii in this order are valine and
threonine, the PPR motif has a selective RNA base binding
ability as follows: binding to U is the strongest, and
binding to A is the second strongest, without binding to
G or C.
[0038]
[Use of PPR motifs and PPR proteins]
Identification and design:
One PPR motif can recognize a specific base of an
RNA. According to the present invention, PPR motifs
selective to A, U, G, or C can be selected or designed by
CA 03026340 2018-12-03
- 22 -
disposing appropriate amino acids in specific positions
of a PPR motif. Furthermore, a protein containing an
appropriate series of such PPR motifs can recognize its
corresponding specific sequence. Moreover, according to
the findings described above, a PPR motif which can
selectively bind to a desired RNA base and a protein
having a plurality of PPR motifs which can sequence-
specifically bind to a desired RNA can be designed. In
design, the sequence information of a naturally occurring
PPR motif may be referred with respect to moieties other
than the amino acids disposed in the important positions
of the PPR motif. Alternatively, a PPR motif may be
designed by using a naturally occurring PPR motif as a
whole and replacing only the amino acids in the important
positions with other amino acids. The repetition number
of the PPR motif can be appropriately determined
according to the target sequence; for example, the
repetition number can be 2 or more, or 2 to 30.
[0039]
The PPR motif or PPR protein thus designed can be
prepared by a method well known to persons skilled in the
art. For example, a nucleic acid sequence encoding an
amino acid sequence of the designed PPR motif or PPR
protein can be determined from the amino acid sequence,
and may be cloned to prepare a transformant (such as an
expression vector) which produces a desired PPR motif or
PPR protein.
CA 03026340 2018-12-03
- 23 -
[0040]
Preparation and use of fusion protein:
The present invention relates to a fusion protein of
the PPR motif or PPR protein described above (i.e., a
polypeptide which can bind RNA base-selectively or RNA
base sequence-specifically to the target mRNA) and one or
more functional domains which improve a protein
expression level from an mRNA.
[0041]
The "functional domain which improves a protein
expression level from an mRNA" which can be used in the
present invention may be all or functional part of a
functional domain of a known protein which directly or
indirectly promotes the translation of the mRNA, for
example. More specifically, the functional domain which
can be used in the present invention may be a domain
which guides ribosome to the mRNA, a domain associated
with initiation or promotion of translation of the mRNA,
a domain associated with nuclear export of the mRNA, a
domain associated with binding to an endoplasmic
reticulum membrane, a domain containing an endoplasmic
reticulum retention signal (ER retention signal) sequence,
or a domain containing an endoplasmic reticulum signal
sequence, for example.
[0042]
More specifically, the domain which guides ribosome
to the mRNA may be a domain containing all or functional
CA 03026340 2018-12-03
- 24 -
part of a polypeptide selected from the group consisting
of DENR (Density-regulated protein), NOT-1 (Malignant T-
cell amplified sequence 1), TPT1 (Translationally-
controlled tumor protein), and Lerepo4 (Zinc finger CCCH-
domain). The domain associated with initiation or
promotion of translation of the mRNA may be a domain
containing all or functional part of a polypeptide
selected from the group consisting of eIF4E and eIF4G.
The domain associated with nuclear export of the mRNA may
be a domain containing all or functional part of SLBP
(Stem-loop binding protein). The domain associated with
binding to an endoplasmic reticulum membrane may be a
domain containing all or functional part of a polypeptide
selected from the group consisting of SE061B, TRAP-alpha
(Translocon associated protein alpha), SR-alpha, Dial
(Cytochrome b5 reductase 3), and p180. The endoplasmic
reticulum retention signal (ER retention signal) sequence
may be a signal sequence containing a KDEL (KEEL)
sequence. The endoplasmic reticulum signal sequence may
be a signal sequence containing MGWSCIILFLVATATGAHS (SEQ
ID NO: 22).
[0043]
In the fusion protein according to the present
invention, the functional domain may be fused to the N-
terminal side of the PPR protein, may be fused to the C-
terminal side of the PPR protein, or may be fused to both
of the N-terminal side and the C-terminal side thereof.
CA 03026340 20112-133
- 25 -
Moreover, the fusion protein according to the present
invention may include several functional domains (for
example, 2 to 5 functional domains). Furthermore, in the
fusion protein according to the present invention, the
functional domain and the PPR protein may be indirectly
fused via a linker, for example.
[0044]
The present invention also relates to a nucleic acid
encoding the fusion protein described above, and a vector
(such as an expression vector) comprising the nucleic
acid. The expression vector herein refers to, for
example, a vector comprising a DNA having a promoter
sequence, a DNA encoding a desired protein, and a DNA
having a terminator sequence, in this order from upstream.
The expression vector may not have these DNAs in this
order as long as it demonstrates desired functions. A
variety of expression vectors which can be usually used
by persons skilled in the art can be used in the present
invention.
[0045]
Because the fusion protein according to the present
invention uses the RNA translation mechanism of
eukaryotes, it can function in cells of eukaryotes (such
as animals, plants, microorganisms (e.g., yeasts), and
protists). The fusion protein according to the present
invention can function within animal cells (in vitro or
in vivo) in particular. Examples of animal cells into
CA 03026340 2018-12-03
- 26 -
which the fusion protein according to the present
invention or a vector which expresses the fusion protein
according to the present invention can be introduced can
include cells derived from human, monkey, pig, cow, horse,
dog, cat, mouse, and rat. Examples of cultured cells
into which the fusion protein according to the present
invention or a vector which expresses the fusion protein
according to the present invention can be introduced can
include, but should not be limited to, Chinese hamster
ovarian (CHO) cells, COS-1 cells, COS-7 cells, VERO (ATCC
CCL-81) cells, BHK cells, dog kidney-derived MDCK cells,
hamster AV-12-664 cells, HeLa cells, WI38 cells, 293
cells, 293T cells, and PER.C6 cells.
[0046]
The terms used herein excluding those particularly
defined are used for illustration of the specific
embodiments, and are not intended to be limitative to the
invention.
[0047]
The term "comprise" used herein, unless contexts
clearly require different understandings, is intended to
express that a described entry (such as a member, a step,
a component, or a number) is present, and is intended not
to exclude the presence of other entries (such as a
member, a step, a component, or a number).
[0048]
CA 03026340 2018-12-03
- 27 -
Unless otherwise defined, all the terms used herein
(including technical terms and scientific terms) have the
same meanings as those broadly understood by persons
skilled in the art to which the present invention belongs.
Unless otherwise clearly defined, the terms used herein
should be interpreted as having the meanings consistent
to those herein and its related technical field, and
should not be interpreted as idealized or excessively
formal meanings.
[0049]
Hereinafter, the present invention will be described
more in detail with reference to Examples. However, the
present invention can be implemented with a variety of
aspects, and should not be construed as limitative to
Examples described below.
[Examples]
[0050]
Example 1: Improvement in protein expression level from
target mRNA by fusion protein of PPR motif and eIF4G
[0051]
Materials
(Equipment)
- Basic facility for molecular biological experiment
(for construction of plasmids, for example)
- Inverted microscope (DM IL S40, Leica Microsystems,
Wetzlar, Germany)
CA 03026340 2018-12-03
- 28 -
- CO2 incubator (KM-CC17RH2, Panasonic Healthcare,
Tokyo, Japan)
- Clean bench (MHE-S1300A2, Panasonic Healthcare,
Tokyo, Japan)
- Aspirator (SP-30, Air Liquide Medical Systems,
Bovezzo BS, Italy)
- Centrifuge (swing rotor) (LC-200, Tomy Seiko,
Tokyo, Japan)
- Ultra-low temperature freezer (-80 C) (MDF-C8V,
Panasonic Healthcare, Tokyo, Japan)
- plate reader (EnSight Kaleido, PerkinElmer,
Waltham, MA, USA)
[0052]
(Cell culturing)
- HEK293T cell line (see note 1)
- Dulbecco's modified Eagle's culture medium (DMEM,
glucose-rich) (see note 2)
- 100x penicillin-streptomycin solution
- Fetal bovine serum (FBS) (see note 3)
- EDTA-NaCl solution: 10 mM EDTA and 0.85% (w/v)
NaCl, pH adjusted to 7.2 to 7.4, autoclave sterilized,
stored at room temperature
- 100 x 20 mm cell culture petri dish (Greiner bio
one, Frickenhausen, Germany)
- 10 mL disposable sterilized pipette
- 15 mL and 50 mL plastic centrifuge tubes
CA 03026340 2018-12-03
- 29 -
- 1.8 mL cryotube (Nunc; Thermo Fisher Scientific,
Waltham, MA, USA)
- Freeze container (Nalgene; Thermo Fisher
Scientific, Waltham, MA, USA)
- Bambanker (Lymphotec, Tokyo, Japan)
[0053]
(Transfection)
- Effector plasmid: pcDNA3.1 (Thermo Fisher
Scientific, Waltham, MA, USA) was used as a basic vector.
A fusion gene of PPR and eIF4G is inserted into an
expression cassette (100 ng/ L) (see note 4).
- Reporter plasmid: pcDNA3.1 (Thermo Fisher
Scientific, Waltham, MA, USA) was used as a basic vector.
Luciferase genes are inserted into an expression cassette,
and a PPR-binding sequence is inserted into its 5'-UTR
(100 ng/ L).
- 96-well plate coated with poly-L-lysine (AGC
Techno glass, Shizuoka, Japan)
- lx phosphate-buffered saline, PBS(-): 1.47 mM
KH2PO4, 8.1 mM Na2HPO4, 137 mM NaCl, and 2.7 mM KC1. pH
adjusted to 7.4, autoclave sterilized, stored at room
temperature
- Hemocytometer (for counting the number of cells)
(Improved Neubauer Type Cell counter plate, Watson, Hyogo,
Japan)
- Transfection reagent (HilyMax, Dojindo Molecular
Technologies, Kumamoto, Japan)
CA 03026340 2018-12-03
- 30 -
[0054]
(Luciferase assay)
- Dual-Glo Luciferase Assay System (Promega, Madison,
WI, USA.)
- 96-well luminometer plate (PerkinElmer, Waltham,
MA, USA).
[0055]
Experimental method
(Construction of vector)
The reporter assay requires an effector plasmid and
a reporter plasmid. These two plasmids both are
constructed based on pcDNA3.1. The effector plasmid
includes a fusion gene encoding a PPR protein and a
partial domain of human eIF4G (SEQ ID NO: 1) (Figure 1A).
The PPR protein moiety used was CRR4 (SEQ ID NO: 2). The
reporter plasmid includes two open reading frames (ORFs),
specifically, renilla luciferase (RLuc) and firefly
luciferase (FLuc), which are dicistronically transcribed
(Figure 1A). The RLuc gene is located on the side of the
5'-end of the FLuc gene, and was used as a control of
gene expression. The PPR-binding region is inserted into
the 5'-UTR of the ORE of FLuc, and consists of three
repetitions of a CRR4-recognizing sequence (5V-
UAUCUUGUCUUUA-3') (SEQ ID NO: 3) interrupted with four-
base sequences (ATCG and GATC). To express both of the
fused effector gene and the reporter gene, a
cytomegalovirus promoter (CMV) and a bovine growth
CA 03026340 2018-12-03
- 31 -
hormone gene-derived polyadenylation signal were used.
For a control experiment, an effector plasmid having no
eIF4G was constructed by fusing a FLAG epitope tag to the
PPR. A control reporter plasmid without a PPR-binding
region was also constructed.
[0056]
The outline of the procedures from cell culturing to
the reporter assay in Examples is shown in Figure 2.
[0057]
(Cell culturing from frozen stock)
This step is aseptically performed. All the tools
are preliminarily antisepticized with 70% ethanol.
1. A 9 mL DMEM culture medium is placed into a 15 mL
centrifuge tube (sterilized).
2. 1 mL of frozen HEK293T cells in a cryotube is
incubated within a water bath at 37 C to quickly melt the
cells.
3. The cells are placed into the 15 mL centrifuge
tube containing 9 mL DMEM.
4. The centrifuge tube is centrifuged at room
temperature and 1100 xg for two minutes, and the
supernatant is removed.
5. The cells are resuspended in 10 mL DMEM (FBS is
added such that the final concentration is 10%).
6. The suspended cells are transferred into a 100 mm
petri dish. The petri dish was left to stand in an
incubator at 37 C and under a 5% CO2 condition. If the
CA 03026340 2018-12-03
- 32 -
culturing was started from the frozen stock, the cultured
cells were subcultured after 24 hours.
[0058]
To keep the cells healthy (see note 5), the cell
density on the surface of the petri dish is maintained
between 10% and 80%. The passage is basically performed
every three days (two times a week), or is performed
according to the growth rate of the cells. Furthermore,
to keep the number of passages small, cells are freshly
cultured from the frozen stock once a month. Keeping the
number of passages small and thus keeping the cells
healthy are important for efficient DNA transfection.
[0059]
(Passages to maintain cells)
1. New 100 mm petri dishes are provided as required.
8 mL DMEM and 1 mL FBS are preliminarily placed onto each
of the petri dishes.
2. The culture medium on a petri dish containing the
cultured cells is removed with an aspirator (see note 6).
3. 2 mL EDTA-NaCl solution is gently added onto
adhering cells on the surface of the petri dish so as not
to peel off the cells. The petri dish is turned around
to evenly distribute the solution across the entire
surface of the petri dish. The EDTA-NaCl solution is
removed with an aspirator. The petri dish is tapped to
peel off the cells.
CA 03026340 2018-12-03
- 33 -
4. 10 mL DMEM is added to the cells in the petri
dish, and the cells are suspended by gently pipetting.
5. 1 mL suspended cells (10% cultured cells) are
added to the petri dishes preliminarily provided and each
containing 9 mL culture medium. Each of the petri dishes
is turned around to distribute the cells across the
entire surface thereof.
[0060]
(Freeze storage of cells)
A frozen stock is prepared with Bambanker reagent
and cultured cells in a logarithmic growth phase at a
cell density up to 50%. Use of Bambanker provides a high
recovery rate and facilitates long-term storage.
1. The cells on the second day since the passage are
peeled off according to the procedure for passage. 5 to
mL DMEM is added, and the cells are recovered in a 50
mL centrifuge tube.
2. The centrifuge tube is centrifuged at room
temperature and 1100 xg for two minutes, and the
supernatant is removed.
3. 1 mL Bambanker per petri dish is added to suspend
the cells.
4. The suspended cells are quickly dispensed into
cryotubes, and the cryotubes are covered with their lids.
5. The cryotubes are placed in a dedicated freeze
container, and are left to stand at -80 C for 12 hours
(see note 7).
CA 03026340 2018-12-03
- 34 -
6. The cryotubes are transferred into a standard
sample box, and are stored at -80 C or in liquid nitrogen.
[0061]
(Transient gene introduction (transfection))
1. Before starting transfection, petri dishes each
containing the cells on the second day since the passage
are provided as required, and the cells are checked
whether they are healthy (normal) or not (see note 8).
About 96 assays can be performed with one petri dish as
an estimate.
2. The cells on the second day since the passage are
peeled off according to the procedure for passage, and
the suspended cells are transferred into a 50 mL
centrifuge tube.
3. The centrifuge tube is centrifuged at room
temperature and 1100 xg for two minutes, and the
supernatant is removed.
4. Cell clusters are completely dispersed in 10 mL
DMEM (FBS is added such that the final concentration is
10%).
5. The number of cells is counted with a
hemocytometer and an inverted microscope. The cells are
suspended in an appropriate amount of DMEM (FBS is added
such that the final concentration is 10%) such that the
number of cells is 1 to 2 x 105 cells/mL.
6. A 96-well plate is provided. 200 L (2 to 4 x 104
cells/mL) per well of suspended cultured cells is placed
CA 03026340 2018-12-03
- 35 -
into each well, and the plate is left to stand overnight
in an incubator at 37 C under a 5% CO2 condition. One
well is used for one assay.
7. On the next day, the culture medium is carefully
removed from each well, and is replaced with 100 L of
new DMEM (FBS is added such that the final concentration
is 10%).
8. 400 ng effector plasmid (4 L of 100 ng/ L) and
100 ng reporter plasmid (1 L of 100 ng/ L) are placed
into a single well on a new 96-well PCR plate (or a 0.2
mL tube).
9. For one assay, 1 L HilyMAX is diluted with 10 L
serum-free DMEM.
10. 11 L diluted solution is placed into each of
the wells containing the plasmids. The solution is well
mixed with the plasmids by pipetting.
11. The solution is left to stand at room
temperature for 15 minutes. The total amount of the
mixture is placed into the wells containing the cultured
cells. The plate is left to stand in an incubator at
37 C under a 5% CO2 condition for 24 hours.
[0062]
(Luciferase assay)
The dual luciferase assay is performed using Dual-
Glo Luciferase Assay System according to the usage
instruction from the manufacturer except for a few
modifications.
CA 03026340 2018-12-03
- 36 -
1. After 24 hours from the transfection, the culture
medium of each well is replaced with 40 L lx PBS(-).
2. 40 L of Dual-Glo luciferase reagent is placed
into each well, and is well mixed with the culture medium
by pipetting.
3. The mixture is left to stand at room temperature
for 10 minutes, and the total amount thereof is
transferred into a 96-well luminometer plate.
4. The light emission from firefly luciferase
related with expression of FLuc gene is measured with a
plate reader.
5. A Stop & Glo substrate is 100-fold diluted with a
Dual-Glo Stop & Glo buffer. 40 L of the diluted
solution is added into each well.
6. The plate is left to stand at least at room
temperature for 10 minutes, and then the light emission
from renilla luciferase related with expression of RLuc
gene is measured.
[0063]
(Data analysis)
1. The value of FLuc/RLuc is calculated to correct a
difference in transfection efficiency between the assays
or experimental errors.
2. An increase in activity of reporter gene
expression is determined in the presence of the PPR-
binding region and in the absence thereof by dividing an
experimental value obtained using the plasmid according
CA 03026340 2018-12-03
- 37 -
to the present invention (plasmid encoding a fusion
protein of CRR4 and a translation activation domain
eIF4G) by an experimental value obtained using a control
plasmid (plasmid encoding a fusion protein of CRR4 and
FLAG-tag).
[0064]
Experimental results
The results of the luciferase assay are shown in
Figure 3. As shown in Figure 3, 2.75-fold translation
activity was specifically verified in the presence of
both of the PPR-eIF4G and the PPR-binding sequence. That
is, it is demonstrated that the fusion protein of the PER
protein and the functional domain which improve a protein
expression level from an mRNA improves the protein
expression level from the target mRNA.
[0065]
Notes
(Note 1) HEK293T is a human fetus-derived kidney cell
line which expresses an SV40 large T antigen. The cell
line is readily cultured, and can be transfected with
high efficiency by a variety of methods. HEK293T cells
are available from RIKEN BRC (ja.brc.riken.jp) or ATCC
(www.atcc.org).
(Note 2) lx penicillin-streptomycin solution is added to
DMEM to avoid contamination with microorganisms.
(Note 3) Before use, FBS is inactivated at 56 C for 30
minutes, and is stored at 4 C.
CA 03026340 2018-12-03
- 38 -
(Note 4) The purity of the plasmid is significantly
important to the transfection efficiency. The plasmid
should be isolated using a kit of a transfection grade.
(Note 5) A daily growth rate is an index indicating that
the cells are healthy. To avoid suppression of cell
growth, the cells should be always cultured in a
sufficient space under a sufficient nutritional condition.
(Note 6) HEK293T cells should be gently treated when the
culture medium is replaced because the cells are readily
peeled off from the culturing petri dish.
(Note 7) The dedicated freeze container is a box whose
freezing speed can be adjusted (about -1 C per minute at
-80 C), and enables the cells to be freeze stored in a
non-programmable -80 C freezer.
(Note 8) In transfection, cells are used at a culture
density of 50 to 80%. However, an appropriate cell
density depends on the transfection reagent.
Additionally, the ratio of the transfection reagent ( L)
to the plasmid DNA ( g) should be also optimized
according to the usage instructions from the manufacturer.
The procedure described herein is optimized for a
condition where a 96-well plate, HEK293T cells, and
HilyMAX as a transfection reagent are used.
[0066]
Example 2: Improvement in protein expression level from
target mRNA by fusion protein of PPR and another
functional domain
CA 03026340 2018-12-03
- 39 -
[0067]
In the case where useful substances are produced
using cells, the amounts of protein synthesized by
endogenous genes and exogenous genes should be precisely
controlled. The final amount of the synthesized protein
is determined by the insertion positions of genes, the
mRNA transcription amount, post-transcriptional
regulation (regulation at an RNA level), post-
translational modification, and the like. For these
reasons, the present inventors have devised a method of
enhancing the translation of mRNAs taking advantage of
the fact that a PPR protein sequence-specifically binds
to a target RNA molecule (Figure 4). In the translation
of mRNAs in eukaryotes, an mRNA undergoes action of a
translation initiation factor (eukaryotic initiation
factor; eIF). As a result, the ribosome is recruited
near the translation starting point, and then the
translation of the mRNA is started. In other words, the
present inventors have considered that the translation of
the mRNA can be artificially enhanced if the ribosome can
only be recruited onto the mRNA. Moreover, the
translation of an mRNA into a protein is usually
performed in the ER. For this reason, the present
inventors have considered that the translation of the
mRNA can be enhanced by intentionally localizing the
target mRNA into the ER.
[0068]
CA 03026340 20112-133
- 40 -
Verification by experiment
To verify the idea above, a reporter assay system
using animal cultured cells (HEK293T) was prepared (the
experiment was performed by the same method as that in
Example 1 except that different functional domains were
used). The system was constructed using CRR4 protein
(one of Arabidopsis thaliana PPR proteins), which is
known to bind to a specific RNA sequence (UAUCUUGUCUUUA)
(SEQ ID NO: 3). First, a fusion protein expression
vector (effector plasmid) of CRR4 and a candidate protein
functional domain was prepared. The selected candidate
domains were (a) eIF proteins (eIF4E and eIF4G), (b)
ribosome-bound proteins (DENR, MCT-1, TPT1, and Lerepo4),
(c) translational regulation factors (SLBPs) of Histone
which promote transport of the transcribed mRNA from the
nucleus to the cytoplasm, (d) ER anchor proteins (SEC61B,
TRAP-alpha, SR-alpha, Dial, and p180), (e) ER retention
signal (KDEL), and (f) ER signal peptide. The fusion
proteins were cloned so as to express in the form of HA-
CRR4-XX or XX-CRR4-HA (HA: epitope tag (SEQ ID NO: 4);
XX: candidate domain).
[0069]
The reporter plasmid included an expression cassette
where renilla luciferase (RLuc) and firefly luciferase
(FLuc) are transcribed in the form of a dicistronic mRNA
under the control of a CMV promoter. Three PPR-binding
CA 03026340 2018-12-03
- 41 -
sequences (UAUCUUGUCUUUA) (SEQ ID NO: 3) are inserted
into a site on the 5'-end of Fluc.
[0070]
The effector plasmid and the reporter plasmid were
transfected into HEK293T cells, and the intensities of
light emission from RLUC and FLUC were measured. The
intensity of light emission from RLUC was treated as a
transfection control, and the value of the intensity of
light emission from FLUC/the intensity of light emission
from RLUC was treated as a translation activity amount.
[0071]
Results
The results shown in Figures 5 and 6 were examined
using the following indices (A) and (B).
(A) Comparison between the absence of and the presence of
the target
The comparison shows an amount of sequence-specific
change in translation.
(B) Comparison to the presence of the target and the
absence of the effector (empty) (black dashed line)
The comparison shows an amount of change in
translation caused by addition of the domain.
[0072]
1. eIF4E was fused to the C-terminal side of CRR4.
(A) 2.7 times
(B) 1.6 times
2. eIF4G was fused to the C-terminal side of CRR4.
CA 03026340 20112-133
- 42 -
(A) 4.5 times
(B) 3.3 times
3. DENR was fused to the N-terminal side of CRR4.
(A) 1.7 times
(B) 1.3 times
4. DENR was fused to the C-terminal side of CRR4.
(A) 2.4 times
(B) 1.7 times
5. MCT-1 was fused to the N-terminal side of CRR4.
(A) 1.3 times
(B) 1.0 time
6. MCT-1 was fused to the C-terminal side of CRR4.
(A) 2.0 times
(B) 1.2 times
7. TPT-1 was fused to the N-terminal side of CRR4.
(A) 1.4 times
(B) 1.0 time
8. TPT-1 was fused to the C-terminal side of CRR4.
(A) 2.4 times
(B) 1.9 times
9. Lerepo4 was fused to the N-terminal side of CRR4.
(A) 3.0 times
(B) 1.8 times
10. Lerepo4 was fused to the C-terminal side of CRR4.
(A) 3.3 times
(B) 2.6 times
11. SLBP was fused to the C-terminal side of CRR4.
CA 03026340 2018-12-03
- 43 -
(A) 4.1 times
(B) 3.3 times
12. Sec61B was fused to the C-terminal side of CRR4.
(A) 1.6 times
(B) 1.6 times
13. Sec61BTM was fused to the C-terminal side of CRR4.
(A) 2.4 times
(B) 1.9 times
14. TRAP-alpha was fused to the C-terminal side of CRR4.
(A) 3.5 times
(B) 4.5 times
15. TRAPTM was fused to the C-terminal side of CRR4.
(A) 2.3 times
(B) 1.6 times
16. SR-alpha was fused to the N-terminal side of CRR4.
(A) 1.7 times
(B) 1.5 times
17. Dia1TM was fused to the N-terminal side of CRR4.
(A) 1.8 times
(B) 1.2 times
18. P180TM2R was fused to the N-terminal side of CRR4.
(A) 2.1 times
(B) 1.5 times
19. P180TMH was fused to the N-terminal side of CRR4.
(A) 2.3 times
(B) 2.5 times
20. P180TM2 was fused to the N-terminal side of CRR4.
CA 03026340 2018-12-03
- 44 -
(A) 3.0 times
(B) 2.1 times
21. KDEL was fused to the C-terminal side of CRR4.
(A) 1.8 times
(B) 1.4 times
22. KEEL was fused to the C-terminal side of CRR4.
(A) 2.3 times
(B) 2.1 times
23. Signal peptide (SP) was fused to the N-terminal side
of CRR4.
(A) 1.4 times
(B) 2.0 times
[0073]
As shown above, an increase in translation was found
in all the functional domains in both of the indices (A)
and the targets (B). Namely, it was clearly shown that
the fusion protein according to the present invention can
enhance the translation of the target mRNA.
[0074]
The amino acid sequences of the functional domains
used in Examples are listed below:
CA 03026340 2018-12-03
- 45 -
[Table 1-1]
Domain Sequence
eIF4E
MATVEPETTPTPNETTTEEEKTESNOEVANPEHYIKHPLORWALW
FTKNDKSKTWQANLRLISKFDTVEDFWALYNHIQLSSNLMPGCDYS
LEKDGIEMLEDEKNKRGGRWLITLNKOORRSDLDRFSILETLE.CLI
GESFDDYSDDVC,GAVVNVRAKGDKIAIWTTECENREAVTHIGRVYK
ERLGLPPKIVIGYQSHADTATKSGSTIKNRFINGRI (SEQ ID NO: 5)
eING
EEKKRYDREFLLGFQFIFASMOKPEGLPHISDVVLDKANKTPLRPL
DPTRLQGINCGPDFTPSFANLGRTTLSTRGPPRGGPGGELPRGPQA
GLGPRRSQQGPRKEPRKIIATVLMTEDIKLNKAEKAWKPSSKRTAA
DKDRGEEDADGSKTQDLFRRVRSILNKLTPQMFQQLMKQVTQLAID
TEERLKGVIDLIFEKAISEPNFSVAYANMCRCLMALKVPTTEKPTV
TVNFRKLLLNRCQKEFEKDYMDDEVFEKKQKEMDEAATAEERGFILK
EELEEARDIARRRSLGNIKFIGELFKLKMLTEAIMHDCVVKLLKNH
DEESLECLCRLLTTIGKDLDFEKAKPRMDQYFNQMEKIIKEKKTSS
RIRFMLODVLDLRGSNWVPRRGDQGPKTIDOIHKEAEMEEHREHIK
VQQLMAKGSDKRRGGPPGPPISRGLPLVDDGGWNTVPISKGSRPID
TSRLTKITKPGSIDSNNQLFAPGGRLSWGKGSSGGSGAKPSDAASE
AARPATSTLNRFSALQQAVPTESTDNRRVVQRSSLSRERGEKAGDR
GDRLERSERGGDRGDRLDRARTPATKRSFSKEVEERSRERPSQPEG
LRKAASLTEDRDRGRDAVKREAALPPVSPLKAALSEEELEKKSKAI
TEMAILNIDMKEAVQCVQELASPSLIZIFVRHGVESTLERSATARE
HMGQLLHQLLCAGHLSTAQYYQGLYEILELAEDMEIDIPHVWLYLA
ELVTPILOEGGVPMGELFREITKPLRPLGKAASULEILGLLCKSM
GPKKVGTLWREAGLSWKEFLPEGQDIGAFVAEQKVEYTLGEESEAP
GQRALPSEELNRQLEKLLKEGSSNQRVFDWIEANLSEQQIVSNTLV
RALMTAVCYSAIIFETPLRVDVAVLKARAKLLOKYLCDEOKELOAL
YALQALVVTLEQPPNLLRMFFDALYDEDVVKEDAFYSWESSKDPAE
QQGKGVALKSVTAFFKWLREAEEESDH (SEQ ID NO: I)
DENR MAADISESSGADCKGDPRNSAKLDADYPLRVLYCGVCSLPTEYCEY
MPDVAKCRQWLEKNFPNEFAKLTVENSPKQEAGISEGQGTAGEEEE
KKKQKRGGRGQIKQKKKTVPQKVTIAKIPRAKKKYVTRVCGLATFE
IDLKEAQRFFAQKFSCGASVTGEDEIIIQGDFTDDIIDVIQEKWPE
,VDDDSIEDLGEVKK (Sc ID NO: 6)
MCT-1, MFKKFDEKENVSNCIQLKTSVIKGIKNQLIEQFPGIEPWLNQIMPK
KDPVKIVRCHEHIEILTVNGELLFFRQREGPFYPTLRLLHKYFFIL
PHQQVDKGAIKFVLSGANIMCPGLTSPGAKLYPAAVDTIVAIMAEG
CA 03026340 2018-12-03
- 46 -
[Table 1-2]
KONA LCVGVMKNSAEDIEKVNKGIGI ENI HYLNDGLWIIMKTYK
(SEQ ID NO: 7)
T PT- 1 MI I YRDL I SHDEMFSDIYKI REIADGLC LEVEGKMVSRTEGN I DDS
LIGGNASAEGPEGEGTESTVI TGVD I VIINHH WETS FTKEAYKICY I
KDYMKSI KGKLEEQR PERVKPFMTGAAEQ I ICH I LANFKNYQFPIGE
NMNPDGMVALLDYREDGVTPYNI FFKDGLEMEKC ( SEC ID NO: B)
Le repo 4 P PKKQAQAGGS ICKAKKKKEIC I I EDKT FGL KNIUCGAKQQKF I KAVT
HQVKFGQQNPRQVAQS EAEIU(LKKDDICKKELQELNE LFKPVVAAQK
I SKGADPKSVVCAFFKQGQCTKGDK.CKFSHDLTLERKCEKRSVYID
ARDEELEKDTMDNWDEKICLEEVVNKKHGEAEICIU(PKTQIVCKHFLE
A I ENNKYGWFWVC PGGGDI CMYRHALP PG FVLKKDKKKEEKEDE I S
LEDLI ERERSALGPNVTKITLESPLAWKICRKRQEKIDKLEQDMERR
KADF KAG KALV I SGREVF EF'R PELVNDDDE EADDTRYTQGTGGDEV
DDS VSVNDI DLS LY I PRD'VDETGITVASLERESTYTSDICDENKLSE
ASGGRAENGERSDLEEDNEREGTENGA I DAVPVDEKS FHWRGFG
(SEC ID NO: 9)
SL BP ACRPRSP PRHQSRCDGDAS PPSPARWS LGR ICRRADGRRWRPEDAEE
AEHRGAERRPESFTTPEGPKPRSRCSDWASAVEEDEMRTRVNKEMA
RYK.RKLL I NDFGRER KSSSGSSDSKESMSTVPADPETDESVLMRRQ
KQ I NYG KNT I AYDRY I KEV PRIILRQPG I HP KTPNKFKKYS RRSWDQ
Q I KLW KVALH PIMP PAEEGCDLQE I HPVDLESAESSSEPQTSSQDD
FDVYSGTPTKVRIIMDSQVEDEFDLEACLTE PLRDFSAMS
(SEQ ID NO: 10)
Sec6 1B PG PT PSGTNVGS SCRS PSKAVAARAAGSTVRORKNAS CGTR SAGRT
TSAGTGGMVIR PYTEDS PGLKVGPVPVLVMSLLF I ASVPMLH IWGKY
TRS (SEQ ID NO: 11)
Sec6113-TM VGPVPVLVNSLLFIASVFICH I W (SEQ ID NO: 12)
TRAP-alpha RLLPRLLLLLLLVFPATVLFRGGPRGLLAVAQDLTEDEETVEDS II
EDEDDEPLEVEEDEPTDLVEDKEEEDVSGEPEA.SPSADTT I LPVKGE
DPPANNIVKFLVGPTNICGTEDFIVESLDASPRYPQDYQPYIQNP'rA
LPLNTVVPPQRQATFEYSP I PAEPMGGRPPGLVINLNYKDLNGNVF
()DAV FNQTVTV I EREDGLDG ET I FMYMFLAGLGLLV I VGLHQLLES
RKR,KRPIQKVENGTSSQNDVDMSW I PQEI'LNQ I NKASPRRLPRKRA
QKRSVGSDE (SEC ID NO: 13)
TRAP-TM T I FMYMPLAGLGLLVIVGLHQLL ( SEQ ID NO: 14)
SR-alpha LDFFT I FS KGG LVLWCFQGVSDS CTG PVNAL I RS VL LQVG FQKI LT
CA 03026340 2018-12-03
- 47 -
[Table 1-3]
L'I'YvDKLIDDVERLFRDKYRTErQQQSALELLNGTFDFQNDFLELL
REAEESEKI RAPITMKKFEDS EKAXXIIVREM ETRGEKPKEKAKNS
KKKGAKKEGSDOPLATS KPVPAEKSOLPVGPENGvELSKEEL I PAK
REEPIQKHGRGMEK$NKETKEDAPKEKGKKAPRINELGOCANKEVL
DYSTPTTNGTPEAALSEDINLX RGTOSGGOLQDLDCSSEDDEGAAQ
NETKPSATXGTLMMFGMLKGLVGEKELSREDMESVLDKMRDEL
KNVAADIAVQLCEEVANKLEGKVMGTFSTVTETVKQALQBELVQIL
QPQRRVDMLEDIMDAORRORPYVVTFCGVNGVGKSTNLAKISPWLL
ENGFEVLIAACDTFRAGAVEQLRTHTERLSALEPPEKHGGRTMVQL
PEKGYOKDAA0 TAMA IAPARNOPDVVLVDTAGEMQDNAPLMTAL
AKLITIMTPDLVLFVGEALVONEAvDOLVKFIMALADEEMAQTPRL
IDGIVLICKFDTIDDKVGAAXEMTYITSKPIVFVGTGOTYCDLESLN
AXAVVAALMKA ( SEQ ID NO: 15)
Diant STLGIMLETVWFLYSLL ( SEQ ID NO: 16)
M0TKR2 DI YDIVTLGVVVPGGFMVVSAIGX FLVSTPSWETEYEEALANQRK
EmAKTEHQKVEKXXXEKTVEKKOKIKKKEEKPNGKI PDXDPAPNyr
VLLREPVEAPAVAVAPTPVQPII I VAPVATVPAMKEKLASEPKDK
KICKEKKVAKVEPAVESVVNE IQVLTEKAA LETA PKEGRETDVAQE
PEAPKQEAPAKKILEGSIUUCGP PDADGPLYLPYKTLVSTVGSMVPNE
GEAQRL I E I LSEICAGI IQDTWEKATQKGDPV ( SEG ID NO: 17
P 8 Onsti LaVVVFGOPMVVEAIGIFLVETP(sEQ ID NO: 18)
P1801142 DIYDTOTLGVWPGGFMWSAIGIFINSTF ( SEQ ID NO: 1))
KDEL KDEL (SEQ ID NO: 20)
KEEL KEEL (SEQ ID NO: 21)
Ert signal peptide MGWECIILFLVATATGAHS(sE4 ID NO: 22)