Note: Descriptions are shown in the official language in which they were submitted.
RECOMBINASE POLYMERASE AMPLIFICATION
=
BACKGROUND OF THE INVENTION
The ability to amplify DNA lies at the heart of modem biological and medical
research.
This is because most molecular biology techniques rely on samples containing
many identical
molecules to increase the sensitivity of an assay or to prepare enough
material for further
processing. Among the various nucleic acid amplification techniques,
polymerase chain
reaction (PCR) is the most common because of its sensitivity and efficiency at
amplifying short
nucleic acid sequences.
While PCR is of great utility, it is also limited in a number of ways. The
first limitation
of PCR is that it relies on multiple cycles of thermal melting (denaturing) at
high temperatures
followed by hybridization and elongation at a reduced temperature. To maximize
efficiency and
to minimize noise, complex temperature control of multiple reactions is
required. This
necessitates the use of a thermocycler controllable rapid heating/cooling
block made with exotic
material (e.g., gold plated silver blocks), or a robotic mechanism to move
samples between
temperature-controlled zones. Because of the high-temperature required to melt
DNA in
physiological salt conditions, PCR technology requires either the addition of
fresh polymerase
per cycle or the use of thermostable polyrnerases. The approach of adding
fresh polymerase has
not been automated and is thus labor intensive and prone to errors (e.g.,
contamination, dropped
tubes, labeling errors). Furthermore, the need to add enzymes and to mix each
reaction
individually presents serious drawbacks that have limited adaptation of enzyme-
addition PCR
methods to the small scale.
Compared to methods involving the addition of fresh polymerase, the use of
thermostable polymerases in PCR is the most widely practiced. This approach
suffers from the
fact that thermostable polymerases are found in a limited number of organisms,
and the
replication mechanisms used by thermophilic organisms are poorly understood.
The available
repertoire of thermostable polymerases is limited to single polypeptide
polymerase enzymes
involved in DNA repair, and/or lagging strand synthesis. DNA repair and/or
lagging strand
1
CA 3041471 2019-04-26
polymerases are poor choices for DNA amplification because they exhibit poor
processivity
(distributive synthesis). In part as a consequence of using repair and/or
lagging strand
polymerases (e.g. Taq, Pfu, Vent polymerases), and due to the formation of
inhibitory secondary
or tertiary nucleic acid structures following thermal melting, current PCR
protocols do not
readily amplify sequences longer than several thousands of base pairs.
Reliable synthesis (and
amplification) of longer templates will rely on polymerases and auxiliary
enzymatic complexes
collectively exhibiting much higher levels of processivity, strand
displacement, and secondary
structure resolution, as well as limiting the formation of inhibitory higher
order nucleic acid
structures that may form on cooling heat-denatured DNA.
A second limitation of PCR is that it relies on solution hybridization between
oligonucleotides (PCR primers) and denatured template DNA (i.e., the DNA to be
amplified) in
an aqueous environment. To be effective, PCR reactions are performed in a
short time because
the therniostable polymerases have a rapidly declining activity at PCR
temperatures. Further,
for effective hybridization in a short time, a feature critical to rapid
turnaround, it is necessary to
perform PCR in an environment with high concentrations of oligonucleotides.
The high
oligonucleotide concentration also ensures rapid interaction of target
sequences with the
oligonucleotides in competition with the heat-denatured complementary strand
still present in
solution. High oligonucleotide primer concentrations can cause problems,
particularly when the
copy number of the target sequence is low and present in a complex mixture of
DNA molecules.
This would be the case, for example, in a PCR of a genome to determine the
genetic
polymorphism in one locus.
One problem with using high oligonucleotide concentrations is that it enhances
the
degree of false priming at only partly matched sequences in the complex DNA
mixture. False
priming refers to the hybridization of a primer to a template DNA in PCR even
when the primer
sequence is not completely complementary to the template nucleic acid, which
can lead to non-
specific amplification of nucleic acids. Noise, due to false priming,
increases with the
oligonucleotide concentration and the complexity of total starting DNA. In
addition, the
possibility of false priming increases as the copy number of target sequences
decreases. Where
the conditions for false priming are favorable (i.e., high oligonucleotide
concentration, high
2
CA 3041471 2019-04-26
complexity, low copy number), errant amplified sequences can become a dominant
reaction
product. Consequently it can be difficult to identify conditions, and
oligonucleotides, for clean
amplification of target sequences from a sample DNA without an excess of false
priming
background. Thus a further disadvantage of using PCR is the limited success at
cleanly
amplifying rare target DNAs from complex sequences mixtures.
One solution to the problems of specificity and template-melting problem
incurred by
PCR is to employ methods that rely on the biological properties of the
bacterial RecA
recombinase protein, or its prokaryotic and eukaryotic relatives. These
proteins coat single-
stranded DNA (ssDNA) to form filaments, which then scan double-stranded DNA
(dsDNA) for
regions of sequence homology. When homologous sequences are located, the
nucleoprotein
filament strand invades the dsDNA creating a short hybrid and a displaced
strand bubble known
as a D-loop. The free 3'-end of the filament strand in the D-loop can be
extended by DNA
polymerases to synthesize a new complementary strand. The complementary strand
displaces
the originally paired strand as it elongates. By utilizing pairs of
oligonucleotides in a manner
similar to that used in PCR it should be possible to amplify target DNA
sequences in an
analogous fashion but without any requirement for thermal melting
(thermocycling). This has
the advantage both of allowing the use of heat labile polymerases previously
unusable in PCR,
and increasing the fidelity and sensitivity by template scanning and strand
invasion instead of
hybridization.
Although the use of RecA and its homologues for in vitro amplification of
nucleic acids
has been previously described (U.S. Patent 5,223,414 to Zarling et al.,
referred to herein as
"Zarling"), the method and results are limited. Zarling's method has critical
failings that limit
its ability to achieve exponential amplification of double-stranded DNA. The
failure of the
Zarling method to achieve exponential amplification may be due to its
specification for the use
of ATPyS rather than ATP. The Zarling method urges the use of ATPyS, instead
of ATP, in the
assembly of RecA nucleoprotein filaments because it results in a more stable
RecA/ssDNA
filament structure. Normally, filaments are assembled in a 5' to 3' direction
and will
spontaneously disassemble in the same 5' to 3' direction as RecA hydrolyzes
ATP. This process
is dynamic in that assembly and disassembly occurs at the same time and the
amount of
3
CA 3041471 2019-04-26
assembled filaments is at equilibrium. If the non-hydolyzable ATP analog,
ATPyS, is used,
hydrolysis of the ATPyS and the 5' to 3' disassembly of the filaments are
inhibited. The great
stability of RecA/ATPyS filaments, both before and after strand exchange,
while helpful in the
method of targeting (i.e., the Zarling method) is detrimental and unpractical
for DNA
amplification.
In the Zarling method, RecA protein involved in strand invasion will remain
associated
with the double-stranded portion of the exchanged material after strand
exchange. This
interaction occurs because the newly formed duplex is bound in the high-
affinity site of RecA.
The displaced strand occupies a different low-affinity site, unless it is
bound to another single-
stranded DNA binding protein (SSB), such as E. coil SSB. If ATP had been
utilized to generate
the exchange structure, spontaneous 5' to 3' disassembly might occur, although
the exchange
complex can be quite stable and may require additional factors to stimulate
ATP-dependent
disassembly. Regardless of whether spontaneous or stimulated, in the presence
of ATPyS, 5' to
3' disassembly of the RecA filament is inhibited (Paulus, B. F. and Bryant, F.
R. (1997).
Biochemistry 36, 7832-8; Rosselli, W. and Stasialc, A. (1990). J Mol Biol 216,
335-52; Shan, Q.
et al., (1997). J Mol Biol 265, 519-40).
These RecA/dsDNA complexes are precisely the sites targeted by the RecA/ssDNA
primer complexes used to initiate subsequent rounds of invasion and synthesis.
Indeed, with the
RecA bound, the intermediate may not be accessible to polymerase, and
certainly the dsDNAs
can no longer be invaded by RecA/ssDNA primer complexes and are therefore not
amplifiable
from this point. Further synthesis from these templates might occur if
initiated at the other end
of the template, which is free of RecA, and this might eventually lead to
physical displacement
of the bound RecA. It is not clear, however, whether many polymerases can
displace RecA in
this manner. Moreover, the initiation site for that synthetic round will now
be 'blocked' instead.
In such a situation, amplification is only linear with time, and will
predominately generate
single-stranded DNA amplification products.
Thus, the described Zarling method, at best, is likely to generate little more
than small
quantities of ssDNA copies from each template. The linear amplification
potentially given by
4
CA 3041471 2019-04-26
the Zarling method will only occur in the presence of SSB, since the displaced
strand will
continue to bind to the second interaction site on RecA, and single-stranded
DNA will not be
released (Mazin, A. V. and Kowalczykowski, S. C. (1998). EMBO J 17, 1161-8).
This probably
explains why the Zarling method observed additional faster-migrating fragments
when they
included SSB. These additional fragments were most likely displaced single-
stranded
fragments. Hence, in the Zarling method only linear amplification of single-
stranded DNA will
occur at best. There is, therefore, a need in the art for an improved
recombinase-dependent
DNA amplification method.
This invention utilizes two new amplification strategies that avoid any
requirement for
thermal melting of DNA or thermostable components. These strategies also
overcome the
inefficiencies of the Zarling method. As with the Zarling strategy, these
methods rely on the
biological properties of the bacterial RecA protein, or its prokaryotic and
eukaryotic relatives, in
particular, the phage T4 uvsX protein. However, in contrast to the Zarling
method, these
methods are devised to achieve exponential amplification of dsDNA. They
achieve this by
permitting rapid regeneration of targetable sequences in the target nucleic
acid in the presence of
dynamic recombinase/DNA filaments, rather than ATP-y-S loaded non-dynamic
filaments, and
in an environment that concomitantly succeeds in maintaining high
recombination activity.
Furthermore, and critically, while the concept of elongating from
recombination intermediates
has been visited earlier in concept, and limited practice, both in the Zarling
approach, and also in
the Alberts laboratory (Formosa and Alberts, 1996; Morrical and Alberts, 1990;
Morrical,
Wong, and Alberts 1991) and elsewhere (Salinas, Jiang, and Kodadek, 1995;
Morel, Cherney,
Ehrlich, and Cassuto, 1997; International patent application WO 02/086167,
Benkovic and
Salinas), none of the descriptions to date teaches a practical method to allow
exquisitely specific,
sensitive exponential DNA amplification with a capacity for amplification up
to 10 to the power
of 12 fold. This is because establishing this necessary environment which
supports high
recombinase/filament activity, but in the presence of large quantities of the
necessary single-
stranded DNA binding proteins in an in vitro environment has proved extremely
challenging,
and this environment is entirely dependant on a strict combination of
components. This includes,
most critically and unexpectedly, very specific crowding agents which alter
the behaviour of the
CA 3041471 2019-04-26
in vitro system in a remarkable, and essentially unpredictable way. This
remarkable and largely
unpredictable alteration of system behaviour with specific volume-occupying
agents presumably
reflects their capacity to engender fractal-like kinetics, phase separation
effects, or other
additional properties on the biochemical system. By identifying such precise
conditions to
enable rapid and highly geometric DNA amplification, as well as conditions for
driving high
persistent and dynamic recombination activity in vitro for other uses, this
invention enables a
new generation of in vitro molecular techniques. We refer to the described
amplification method
performed under these enabling conditions as Recombinase Polymerase
Amplification (RPA).
We envision herein yet further methods based upon this high activity,
persistent, yet dynamic
recombination environment, which will likely become practiced in due course.
This invention
enables this new generation of approaches, and should be contrasted to the
current circumstance
in which, despite decades of research, no other widely used application of
recombinases for in
vitro technology has appeared apart from a very limited number dependant on
the use of ATP-y-
S.
In this invention we go further and demonstrate that RPA reactions can be
fully
integrated with dynamic detection of reaction products. This validates that
RPA reactions
achieve two general criteria for real-time analysis. First a biochemical
sensor, such as sensing
dye like SYBR green or 'third' probe, is compatible with the RPA reaction
environment. Such
compatibility is not a trivial assumption because RPA employs saturating
quantities of DNA
binding proteins, which might interfere with dye or probe binding behaviour.
Conversely the
binding of dyes or probes to nucleic acids might have interfered with the
activity of the DNA
binding proteins. Secondly to be employed in real-time quantitative
applications RPA would
need to demonstrate exponential DNA amplification of target DNA over a
significant range of
starting template quantities, and be able to maintain exponential
amplification up to
concentrations easily within the detection range of the overall sensor system.
Also in this invention we disclose approaches to control, and potentially
synchronise
aspects of, RPA reactions. In current configurations of RPA there is no
temporal separation
between the DNA targeting and DNA synthesis phases. For RPA, it is difficult
to ensure that
all reactions in RPA are initiated at exactly the same moment unless a rate-
limiting reagent is
6
CA 3041471 2019-04-26
supplied to all samples simultaneously, or the reactions are assembled at a
non-permissive
temperature. We suggest approaches by which RPA reactions may be initiated,
and individual
'rounds' of priming activity may be regulated, by limiting invasion to well-
spaced short bursts.
Such approaches to limit recombinase activity to short limited bursts could
improve
amplification. One way to control DNA invasion in RPA may be by regulating the
concentration
of free ATP. In the absence of sufficient ATP, or an excess of ADP,
recombinase/DNA
filaments disassemble and recombination halts. Caged ATP does not support recA
loading, but
subsequently uncaged material does [Butler BC, Hanchett RH, Rafailov
MacDonald G
(2002) Investigating Structural Changes Induced By Nucleotide Binding to RecA
Using
Difference FTIR. Biophys 3 82(4): 2198-2210]. Thus the use of caged ATP
analogues in RPA
reactions, which can be deprotected in pulses by light thus permitting bursts
of recombinase
activity, should be an effective means to control the invasion phase of an RPA
reaction.
Alternatively ATP concentration could be cyclically controlled by alternative
methods such as
periodic addition of ATP to the reaction from an external source, or by
establishing a
biochemical oscillator capable of generating periodic increases of ATP in the
reaction.
In this invention we extend the knowledge of how to attain ideal
recombinase/ssDNA
loading by virtue of 5' sequence design, and widen the repertoire of contexts
in which this key
stable dynamic recombination environment can be employed in addition to DNA
amplification
reactions. We ,describe how this unique composition may be used to replace
classical
hybraisation steps in any process that otherwise would require thermal or
chemical melting, or
other duplex targeting approach, in a variety of molecular applications. In
particular the use of
stable dynamic recombination environments in the presence of synthetic
oligonucleotides will be
useful in combination with other enzyme systems than the polymerase systems
previously
described, due to the lack of need for thermal or chemical melting, and the
concomitant capacity
to employ a wider range of enzymes and avoid thermal cycling equipment.
CA 3041471 2019-04-26
SUMMARY OF THE INVENTION
Aspects of the invention include:
RecA / Primer Loading Prior to the addition of template DNA and/or Polymerase.
RecA and SSB will compete for binding to single-stranded oligonucleotide
primers. In the
presence of a RecR and RecO, RecA is selectively stabilized onto the single-
stranded primers
forming RecA nucleoprotein filaments in a complex with Rec0 and RecR. This
complex is
competent to invade double-stranded DNA to form a D-loop at sites homologous
to the
oligonucleotide primers. Alternatively, RecA, Rec0 and RecR can be pre-loaded
onto
oligonucleotide primers prior to the introduction of SSB to the reaction
mixture.
Leading strand Recombinase - Polymerase Amplification (IsRPA) RecA/primer
nucleoprotein filaments invade double stranded template DNA preferentially
associating with
homologous target sites. As D-loops are formed and synthesis proceeds
displacedssingle
stranded DNA becomes coated with SSB. RecA release from double-stranded DNA
can occur via
ATP hydrolysis in a 5'-3' direction or as a result of helicaseiresolvase or
polymerase activity.
. Leading strand Recombinase - Polymerase Amplification (IsRPA) As synthesis
continues, polymerases encounter SSB bound, displaced, single-stranded
template. Double-
stranded target site are re-invaded by RecA/primer nucleoprotein filaments.
Subsequent rounds of
IsRPA proceed from re-invaded sites.
Leading/Lagging Strand Recombinase - Polymerase Amplification: Initiation
First, the primosome loads onto the D-loop formed by RecA nucleoprotein
filament invasion. The
primosome synthesizes a stretch of RNA primer. Finally, pdmosome recruits the
clamp loader,
which recruits both the sliding clamp dimer and the asymmetric DNA polymerase
core.
Leading/Lagging Strand Recombinase - Polymerase Amplification: Synthesis
Synthesis occurs simultaneously in both the leading and lagging directions.
Eventually lagging
strand synthesis stops and the lagging strand clamp is unloaded. Synthesis of
the leading strand -
continues until a new site of lagging stand synthesis is formed.
Leading/Lagging Strand Recombinase Polymerase Amplification While leading
strand synthesis continues, a new site of lagging stand synthesis is formed.
Lagging strand
synthesis continues back to the previous Okazaki fragment where the lagging
strand clamp is
unloaded. Leading/Lagging Strand Recombinase - Polymerase Amplification DNA
Polymerase I removes the RNA primer, and fills in the gap while DNA ligase
connects the two
Okazaki fragments forming a continuous lagging strand.
According to another aspect, the invention provides a method of DNA
amplification, termed RPA, which comprises the following steps. First, a
recombinase agent is contacted with a first and a second nucleic
7.1
CA 3041471 2019-04-26
acid primer to form a first and a second nucleoprotein primer. Second, the
first and second
nucleoprotein primers are contacted to a double stranded target sequence to
form a first double
stranded structure at a first portion of said first strand and form a double
stranded structure at a
second portion of said second strand so the 3' ends of said first nucleic acid
primer and said
second nucleic acid primer are oriented towards each other on a given template
DNA molecule.
Third, the 3' end of said first and second nucleoprotein primers are extended
by DNA
polymeraSes to generate first and second double stranded nucleic acids, and
first and second
displaced strands of nucleic acid. Finally, the second and third steps are
repeated until a desired
degree of amplification is reached.
The invention also provides for a method of nested RPAs. In a nested RPA, a
first
region of nucleic acid is amplified by RPA to form a first amplified region.
Then a second
region of nucleic acid that is completely within the first amplified region is
amplified using RPA
to form a second amplified region. This process may be repeated as often as
necessary. For
example, a third region of nucleic acid, which is completely within the second
region, may be
amplified from the second amplified region by RPA. hi addition to the one, two
and three
rounds of RPA discussed above, the invention contemplates at least 4, and
preferably at least 5
rounds of nested RPAs also.
The invention also provides for methods of detecting a genotype using RPA.
This
method is useful for genotypirig, for detecting a normal or diseased
condition, a predisposition,
or a lack of a disposition for a diseased condition. Further, RPA can be used
for detecting the
presence of a genome, such as for example, a genome of a pathogen. In this
use, the method is
useful for diagnosis and detection.
The invention also details the nature and concentrations of recombinases,
single-stranded
binding proteins, polymerases, and nucleotides necessary to establish an
effective amplification
reaction. The invention further provides detailed enablement on the nature of
the target DNA,
the length, and composition of targeting oligonucleotides, and the inter-
oligonucleotide length
optimal for amplification under various conditions. The invention provides for
the inclusion of
additional components, or the use of modified components, that contribute to
establishing a
8
CA 3041471 2019-04-26
recombination-polymerase amplification system that is sensitive, robust, and
with optimal
signal-to-noise properties. In particular the use of more than one species of
recombinase is
demonstrated, and the utility of engineered and modified analogues of the
recombinases E. coli
recA and T4 bacteriophage uvsX, of polymerases including the E. coli DNA
polymerase I
Klenow fragment, Bst polymerase, Phi-29 polymerase, Bacillus subtilis Pot I
(Bsu), as well as
single-stranded DNA binding proteins from E. coli and T4 (the gp32 protein) is
detailed.
The utility of forms of gp32 with altered cooperativity and/or strand
assimilation
properties is demonstrated. Also shown is the use of 14 uvsY protein, and most
particularly of
molecular crowding agents especially PEG compound (also known as Carbowax
20M), to aid in
establishing an optimal reaction environment. Further the present invention
details effects and
the possible use of other enzymes involved in DNA metabolism including
toposiomerases,
helicases and nucleases, in order to improve the amplification behaviour. The
present invention
also includes the use of optimised conditions for repeated invasion/extension
of a primer
targeted to a supercoiled or linear template to generate a linear
amplification, and the use of this
method for DNA sequencing. The present invention also describes the use of a
recombinase in
detection of a specific amplified product of a reaction by directing
oligonucleotides labeled in
some manner to the specific product species and measuring a change in the
appearance or
property of the reactants as a consequence.
This invention also provides data and approaches to improve* the
implementation of the
RPA method, notably for diagnostic applications. Careful design of
oligonucleotide length, base
composition, and use of modified backbone sugar residues underpin strategies
for high
sensitivity and specificity tests. We also disclose approaches to combine
oligonucleotides with
distinct activities as nucleoprotein filaments to improve signal-to-noise
ratios. Also disclosed
are methods of product detection that obviate gel electrophoresis, in some
cases employing
'third' specific oligonucleotides. We disclose the constitution of an active
lyophilizate that can
be stored at ambient temperature for at least 10 days and retain amplification
activity when
reconstituted with buffered sample only.
This invention also discloses enabling data to permit the use of the RPA
method in
9
CA 3041471 2019-04-26
quantitative real-time applications. We show that appropriate dilutions of
SYBR green or
SYBR gold fluorescent nucleic acid binding dyes are compatible with RPA
reactions and permit
the monitoring of the accumulation of products. Products continue to
accumulate in an
apparently exponential fashion for sufficient time to permit quantification
after threshold
detection levels are achieved. We show experimentally that using this approach
RPA is
quantitative over at least four or five orders of magnitude of starting
template quantity.
Simultaneous initiation of many parallel reactions is achieved by establishing
reaction mixes on
ice then simultaneously shifting the samples to reaction temperatures (33-39
C). Alternatively
parallel RPA reactions might be simultaneously initiated by other means, such
as light-driven
uncaging of ATP, or of caged oligonucleotide primers. We detail other product-
specific real-
time monitoring approaches that may be compatible with the RPA system. We also
describe the
overall composition of a real-time RPA device composed of low power solid
state components
which could enable cheap portable implementation in both laboratory and non-
laboratory
contexts.
This invention also discloses method to control RPA reactions, achieved by
controlling
the presence of the necessary supporting nucleoside triphosphate cofactors
such as ATP, to
limited periods during the reaction. If chemically caged nucleotide
triphosphates are used,
pulses of free ATP may be generated by illumination with a defined burst of
light corresponding
to the uncaging wavelength of the photoprotecting group. The released ATP will
then, allow the
binding of recombinase proteins to single-stranded DNA (ssDNA), and the
subsequent
homology searching and strand exchange activity of the recombinase-ssDNA
complexes.
Alternatively, ATP may be periodically added to the reaction. Over time, the
concentration of
ATP will decrease as a consequence of hydrolysis, either recombinase
hydrolysis or hydrolysis
by other reaction components specifically added to hydrolyse excess ATP, and
ADP.
Consequently after a period of time defined by the decreases in ATP
concentration and / or an
increase in the ADP concentration, recombinase molecules will cease to
function and dissociate
from DNA. Subsequent pulses of light at the uncaging wavelength can be
delivered to release
fresh ATP, or fresh ATP can be added to re-initiate recombinase activity. In
this manner a series
of controlled periods of homology searching and priming are enabled so that
initiation of
CA 3041471 2019-04-26
elongation can be phased.
As a second method to control the invasion phase of RPA reactions, we describe
an
approach to separate the activity driven by one primer from the other. This
method utilises
recombinase-mediated invasion at one primer target site and the completion of
synthesis from
that primer. This will generate a single-stranded displaced DNA, which can
then be used as a
template for a second facing primer that is modified and thus unable to
support recombinase-
mediated initiation of synthesis. This will possibly avoid conflicts arising
through collision of
polymerases.
This invention also discloses approaches to assess the polymorphic nature of
amplified
products without a need for size fractionation. Amplification reaction
products are permitted to
form duplex hybrids with immobilized probes of either initially single or
double stranded
character by the action of recombinases and/or single-stranded DNA binding
proteins and other
accessory molecules. Methods to destabilize imperfect hybrids formed between
products and
probes are described, occurring in a dynamic environment of recombinase
action, and supportive
of the activity of a wide variety of additional enzymatic components.
Productive hybrids are
detected by one of many standard approaches used to reveal the presence of
absence of a
molecular interaction.
This invention also describes the combination of the determined in vitro
conditions
which support a stable persistent dynamic recombination environment with other
enzymatic
activities, thus permitting strand invasion and pairing between ssDNAs and
duplexes to occur
continuously and in the presence of other metabolic enzymes, especially those
non-thermophilic
enzymes that would be necessitated in conventional approaches, as well as
other processes that
are not equivalent, or attainable, in a system employing thermal or chemical
melting. We also
describe findings that improve the design of oligonucleotides to permit high
activity in stable
dynamic recombination systems by virtue of including optimised sequences
within, and
particularly at the 5' end of desired sequences.
Other embodiments, objects, aspects, features, and advantages of the invention
will be
apparent from the accompanying description and claims.
11
CA 3041471 2019-04-26
BRIEF DESCRIPTION OF THE DRAWINGS
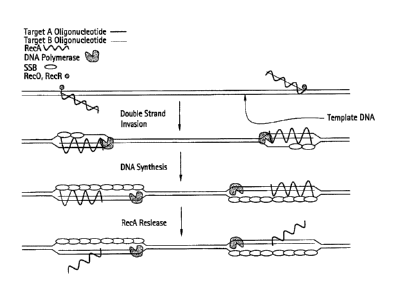
Figure 1 depicts a schematic representation of RecA/Primer Loading.
Figures 2A-2B depict a schematic of succeeding steps, shown in panels (A) and
(B), of Leading
Strand Recombinase Polymerase Amplification (1sRPA).
Figures 3A-3D depict a schematic of succeeding steps, shown in panels (A),
(B), (C), and (D),
of Leading and Lagging Strand Recombinase Polymerase Amplification.
Figure 4 depicts an example of nested primers chosen for nested RPA.
Figure 5 depicts examples of suitable double stranded template nucleic
acids.
Figures 6A-6B depict in panels (a) and (b) the various orientations of the RPA
primer pairs in
hybridization with the target nucleic acid.
Figures 7A-7C in panels (A), (B), and (C) depict a schematic representation of
an RPA reaction
in progress.
Figures 8A-8C depict (A) examples of double stranded primers; (B) double
stranded primers
after elongation and after annealing of the second member of a primer pair;
(C)
after the elongation of the second member of a primer pair with the non-
invading
strand displaced.
Figure 9 depicts investigation into the nature of double-stranded DNA
targets and
targeting oligonucleotides. Experiments using either supercoiled templates or
linearized DNAs suggest that recA catalyses the formation of intermediates
capable of supporting polymerase elongation most readily on supercoiled DNA or
at the ends of linearized DNA. Tester3bio oligonucleotide (SEQ ID NO:19) is
shown.
Figure 10 depicts backfire synthesis. Backfire synthesis occurs when a
recA-coated
targeting oligonucleotide possessing a 5' overhang invades a duplex DNA end in
the presence of a suitable polymerase and dNTPs. This new duplex region is
stable to subsequent branch migration and can be utilised as a platform for
other
12
CA 3041471 2019-04-26
activities. Forward fire, that is the elongation from the invading
oligonucleotide,
can also occur.
Figure 11 depicts uses of backfire synthesis. Backfire synthesis can be
useful because it
generates a branch migration resistant structure that can be used for
application
other than normal oligonucleotide priming. Some examples are shown here
including introduction of a nicking enzyme target site, introduction of an RNA
polymerase promoter, and the linear generation of short dsDNA fragments
through successive invasion/synthesis/cleavage events.
Figure 12 depicts single stranded binding proteins facilitate recombinase
invasion and
primer extension. Both E. coli SSB and T4 gp32 with an N-terminal His tag
(gp32(N)) stimulate recA-mediated invasion/elongation on a linear DNA
template.
Figure 13 depicts the requirement for a minimal oligonucleotide length or
overhang for
invasion and elongation during end targeting of linear templates. When the
invading oligonucleotide is targeted at ends of a linearized template a
minimal
oligonucleotide length, or an overhang is needed for invasion/elongation to
occur.
Figure 14 depicts paranemic and pletonemic joints. Schematic description of
the formation
of paranemic and pIectonemic joints by recombination events involving DNA
ends or embedded sequences.
Figure 15 depicts the effect of crowding agents. Crowding agents can alter
the reaction
behaviour. In the presence of polyethylene glycols, gp32(N) and recA
recombinase, multiple invasion events can be stimulated on single templates
without the requirement for 5' overhang in the targeting oligonucleotide.
Figure 16 depicts end targeted amplification using leading strand RPA.
Amplification of a
target DNA using end-directed oligonucleotides, recA(C) protein, and the
Klenow fragment of E. coli DNA polymerase 1.
Figure 17 depicts leading strand RPA and limits of Klenow processivity.
Limited
13
CA 3041471 2019-04-26
amplification of an approximately 300 base pair fragment when only 0.5
ffiioles
of starting template is utilised with the recA protein, gp32(N), and the
Klenow
fragment of E. coil. Strong accumulation of shorter products suggests that the
poor processivity of Klenow (10-50 nucleotides) may underlie the template
concentration dependence of the reaction.
Figure 18 depicts spacing dependence of RPA primers. There is an
optimal inter-
oligonucleotide length for RPA when using the Klenow fragment of E. coil DNA
polymerase I. The template (SEQ ID NO:1) and EcoRI overhang (SEQ ID
NO:124) sequences are shown.
Figure 19 depicts RPA products that are largely double stranded. RPA
reaction can
generate double-stranded DNA products as evidenced by agarose gel
electrophoresis and restriction enzyme cleavage.
Figure 20 depicts activity of a recA C-terminal truncation mutant.
Mutant recA protein
with a deletion of the C-terminal acidic peptide (recA(C)delta) can promote
strand exchange and extension in a linear template run-on assay.
Figure 21 depicts modified gp32 proteins. Shown is a schematic
representation of the
bacteriophage T4 gp32 proteins used in this study and the position of various
modification and mutations.
Figure 22 depicts activity of gp32 protein. Modified gp32 proteins show
a variety of
activities in linear invasion/run-on assays.
Figure 23 depicts invasion and extension using uvsX. Modified uvsX
protein with a C
terminal His tag (uvsX(C)), or an additional deletion of the C-terminal acidic
peptide, stimulates invasion/extension in a linear template run-on assay.
Figure 24 depicts RPA using uvsX(C). The modified recombinase uvsX(C)
can support
DNA amplification in the presence of gp32(N), the Klenow fragment of E. coli
DNA polymerase I and polyethylene glycol (PEG).
Figure 25 depicts wild-type versus modified gp32. The modified version
of gp32, gp32(C)
14
CA 3041471 2019-04-26
is qualitatively different to wild-type untagged gp32.
Figure 26 depicts titration of gp32 and effect of uvsY. Titration of gp32
reveals a
requirement for a minimal quantity of gp32, and a requirement for uvsY(N)
protein when untagged gp32 is employed.
Figure 27 depicts factors affecting reaction rate and noise. Shown is a
schematic
representation of the factors affecting reaction behaviour, particularly
reaction
rate and noise. The predicted effects and interactions of gp32, uvsX, UvsY and
PEG are suggested, with the conclusion that an optimal balance between
reaction
rate and noise must be struck.
Figure 28 depicts effects of PEG. PEG can reduce the average length of
linear
invasion/run-on products in an uvsX-mediated linear run-on experiment in the
presence of gp32(C).
Figure 29 depicts DNA end directed invasion. Shown is a schematic
depicting possible
outcomes of end-directed invasion events.
Figure 30 depicts RPA in a complex sample. The amplification of specific
DNA targets
from human genomic DNA.
Figure 31 depicts RPA sensitivity. The sensitivity of amplification of
specific DNA targets
from human genomic DNA.
Figure 32 depicts RPA sensitivity and template independent artifacts. The
sensitivity of
amplification of specific DNA targets from human genomic DNA, and the
existence of competing template-independent primer artifacts.
Figure 33 depicts how primer artifacts may arise. Shown is a schematic
representation of
the possible mechanism by which primer artifacts may arise.
Figure 34 depicts primer artifact suppression. Shown in schematic are
methods to suppress
primer artifacts.
Figure 35 depicts the use of hairpin oligonucleotides to stimulate self-
priming of displaced
CA 3041471 2019-04-26
strands. Shown is a schematic diagram of an amplification using self-
complementary hairpin oligonucleotides deliberately to stimulate self-priming
of
displaced strands.
Figure 36 depicts conditions that enable highly efficient noise-free
amplification from
complex DNA sources. The sensitivity of amplification of specific DNA targets
from human genomic DNA under optimised conditions that reduce or eliminate
primer artifacts.
Figure 37 depicts a schematic representation of RPA method as shown in
(A), (B), and (C).
Figure 38 depicts (A) STR markers from two individuals (1 and 2, father
and son) amplified
with primer pairs for seven independent markers using RPA conditions C4; (B)
Titration of reaction components to determine concentrations that support in
vitro
amplification.
Figure 39 depicts (A) Size limits of RPA reactions; (B) Elongation
efficiencies from
embedded or end sequences; (C) Sensitivity of RPA reactions; (D) Human DNA
of the indicated copy number amplified with primers ApoB4 and Apo300
generating a 300 bp fragment using conditions C2; (E,F) Human DNA from
single individuals amplified with primers D18S51 5' and 3'. Conditions
employed were C2 in (E), and C4 in (F).
Figure 40 depicts specificity of RPA reactions for: (A) Primers BsA3 and
BsB3, which
amplify a 380 bp fragment using conditions C3. An asterisk indicates the
position of the expected reaction product, and an arrow indicates the position
of
the genomic DNA; (B,C) Oligonucleotides Apo600bio and Apo300rev which
generate a 345bp fragment using conditions C4; (D) Mixtures of reaction
components assembled in the absence of the indicated components, PEG and
buffer. Primers used are indicated. Target DNA was human male genomic DNA
at 150 copies/Al; (E) Oligonucleotides targeting three independent loci in
human
genomic DNA incubated with overlapping primer pairs of 25, 28, or 32 bases as
indicated.
16
CA 3041471 2019-04-26
Figure 41 Primer noise at low target copy number. The consequences of
reducing the
starting copy density of a target in a typical RPA reaction are shown.
Figure 42 Selection of optimal primers by combining a selection of
candidate forward and
reverse primers and testing the outcome at very low start copy densities.
(SEQ.
ID NO. 123 IS SHOWN).
Figure 43 Theoretical consideration of how primer noise initiates.
Figure 44 Oligonucleotide improvement strategies ¨ overview of three
general schemes.
Figure 45 Consequences of reducing primer length ¨ short primers give
less product, but
can still retain activity at less than 30 residues.
Figure 46 Locked nucleic acids can function in RPA and exhibit significant
differences in
product accumulation, nOise accumulation, and polymerase concentration
requirement.
Figure 47 Adding homopolymeric stretches to the 5 ends of primers may
alter
nucleoprotein activity.
Figure 48 Betaine reduces levels of product and noise.
Figure 49 Combination strategies in which nucleoprotein filaments with
differential
activities are combined.
Figure 50 Detection formats incorporating a 'third' primer for product
enrichment
Figure 51 Bead capture I. Schematic of experimental strategy in which
a third probe
enriches bona fide products derived from the target.
Figure 52 Bead capture II. Experimental results demonstrating that a
low activity
nucleoprotein filament immobilized on a solid support can participate as a
third
primer and separate target amplicons from primer noise.
Figure 53 Trehalose stabilizes lyophilizates to permit all components
except buffered
sample to remain active for at least 10 days at room temperature.
Figure 54 Minor groove binding dyes SYBR green and SYBR gold can be
included in RPA
17
CA 3041471 2019-04-26
reactions and will sense the accumulation of DNA reaction products.
Figure 55 Using SYBR green dye for quantitative assessment of Bacillus
subtilis genomic
DNA copy number can be made over four order of magnitude.
Figure 56 Using SYBR green dye quantitative assessment of Bacillus subtilis
genomic
DNA copy number can be made over at least five orders of magnitude.
Figure 57 Quantitative real-time RPA responds excellently to titration of
human genomic
DNA.
Figure 58 The presence of single-strand bubbles in imperfect probe:target
hybrids leads to
enhanced overall duplex disruption in a dynamic recombination environment
Figure 59 Hybridization of amplicons to an array of potential length
polymorphism probes
in a dynamic recombination environment leads to enrichment of hybrids of
perfect or near-perfect match as compared to classical hybridization, or non-
dynamic recombination environment.
Figure 60 Single tube reverse transcription RPA (RT-RPA) shows high
sensitivity without
significant optimisation
Figure 61 dUTP can be included in RPA reactions to wholly or partially
replace dTTP thus
providing a mechanism to develop effective carry-over contamination control.
Figure 62 A strategy for controlling carry-over contamination in RPA
reactions.
Figure 63 Format of a stand-alone RPA assay to determine the presence of a
specific
nucleic acid comprising disposable amplification pouch and lateral flow strip
to
detect successful amplification.
Figure 64 Compatibility of RPA with crude lysates of human blood indicate
the possibility
that sophisticated sample preparations may be obviated for many samples.
Figure 65 Utility of real-time RPA analysis in optimising reaction
environment; salt and
PEG concentrations.
Figure 66 Utility of real-time RPA analysis in optimising reaction
environment; Magnesium
18
CA 3041471 2019-04-26
concentrations.
Figure 67 Primer sequences affect amplification rate behaviour ¨
preliminary analysis
suggests that 'fast' primers may be associated with low G content, and high C
or
C/T content.
Figure 68 Utility of real-time RPA analysis in optimising reaction
environment;
heterologous 5' sequences appended to oligonucleotides influence reaction
behaviour.
Figure 69 Utility of real-time RPA analysis in optimising reaction
environment;
heterologous 5' sequences appended to oligonucleotides influence reaction
behaviour ¨ not all pyrimidine-rich 5' sequences drive excellent kinetic
behaviour.
Figure 70 Utility of real-time RPA analysis in optimising reaction
environment;
heterologous 5' sequences appended to oligonucleotides influence reaction
behaviour¨ not all pyrimidine-rich 5' sequences drive excellent kinetic
behaviour.
Figure 71 So-called 'third probes' may be employed to monitor RPA
reactions and increase
specificity. Candidate enzymes for processing specific duplexes containing the
target and probe.
Figure 72 Enzymes that recognise abnormal features such as helix
distortions and damaged
bases/abasic sites may be employed to process probe/target hybrids.
Figure 73 Fluorescent probes demonstrate significantly different
properties in the RPA
environment compared to standard environments used in other amplification
reactions.
Figure 74 A probe containing a tetrahydrofuranyl residue in efficiently
cut and extended in
an RPA environment containing amplified target, the E.coli Nfo enzyme, and
polymerase.
Figure 75 A probe containing a tetrahydrofuranyl residue in efficiently,
rapidly, and
specifically cut and extended in an RPA environment containing amplified
target,
19
CA 3041471 2019-04-26
the E.coli Nfo enzyme, and polymerase.
Figure 76 Examples of probe design for real-time RPA studies using the
E.coli Nfo
enzyme.
DETAILED DESCRIPTION OF THE INVENTION
The present invention provides for Recombinase-Polymerase Amplification (RPA) -
- a
method for the amplification of target nucleic acid polymers. It also provides
for a general in
vitro environment in which high recombinase activity is maintained in a highly
dynamic
recombination environment, supported by ATP. One benefit of RPA is that it may
be
performed without the need for thermal melting of double-stranded templates.
Therefore, the
need for expensive thermocyclers is also eliminated. The present invention
describes two
related strategies by which RPA can be configured to permit exponential
amplification of target
nucleic acid polymers.
Throughout this specification, various patents, published patent applications
and scientific
references are cited to describe the state and content of the art.
Leading strand recombinase-polymerase amplification (1sRPA)
In leading strand Recombinase-polymerase Amplification (IsRPA) single-
stranded, or
partially single-stranded, nucleic acid primers are targeted to homologous
double-stranded, or
partially double-stranded, sequences using recombinase agents, which would
form D-loop
structures. The invading single-stranded primers, which are part of the D-
loops, are used to
initiate polymerase synthesis reactions. A single primer species will amplify
a target nucleic acid
sequence through multiple rounds of double-stranded invasion followed by
synthesis. If two
opposing primers are used, amplification of a fragment -- the target sequence -
- can be achieved.
LsRPA is described briefly in Figures 1 and 2.
The target sequence to be amplified, in any of the methods of the invention,
is preferably
a double stranded DNA. However, the methods of the invention are not limited
to double
stranded DNA because other nucleic acid molecules, such as a single stranded
DNA or RNA can
CA 3041471 2019-04-26
be turned into double stranded DNA by one of skill in the arts using known
methods. Suitable
double stranded target DNA may be a genomic DNA or a cDNA. An RPA of the
invention may
amplify a target nucleic acid at least 10 fold, preferably at least 100 fold,
more preferably at least
1,000 fold, even more preferably at least 10,000 fold, and most preferably at
least 1,000,000
fold.
The target sequence is amplified with the help of recombinase agents. A
recombinase
agent is an enzyme that can coat single-stranded DNA (ssDNA) to form
filaments, which can
then scan double-stranded DNA (dsDNA) for regions of sequence homology. When
homologous sequences are located, the nucleoprotein filament (comprising the
recombinase
agent) strand invades the dsDNA creating a short hybrid and a displaced strand
bubble known as
a D-loop. Suitable recombinase agents include the E. coli RecA protein, the T4
uvsX protein, or
any homologous protein or protein complex from any phyla. Eukaryotic RecA
homologues are
generally named Rad51 after the first member of this group to be identified.
Other non-
homologous recombinase agents may be utilized in place of RecA, for example as
RecT or
RecO. Recombinase agents generally require the presence of ATP, ATPTS, or
other nucleoside
triphosphates and their analogs. It is preferred that recombinase agents are
used in a reaction
environment in which regeneration of targeting sites can occur shortly
following a round of D-
loop stimulated synthesis. Completed recombination events involving
recombinase disassembly
will avoid a stalling of amplification or very inefficient linear
amplification of ssDNA caused by
oscillating single-sided synthesis from one end to the other.
Naturally, any derivatives and functional analogs of the recombinase agent
above may
also function itself as a recombinase agent and these derivatives and analogs
are also
contemplated as embodiments of the invention. For example, a small peptide
from recA, which
has been shown to retain some aspects of the recombination properties of recA,
may be used.
This peptide comprises residues 193 to 212 of E. coli recA and can mediate
pairing of single
stranded oligos (Oleg N. Voloshin, Lijang Wang, R. Daniel Camerini-Otero,
Homologous DNA
pairing Promoted by a 20-amino Acid Peptide Derived from RecA. Science Vol.272
10 May
1996).
21
CA 3041471 2019-04-26
Since the use of ATPyS results in the formation of stable Recombinase-
agent/dsDNA
complexes that are likely incompatible with efficient amplification, it is
preferable to use ATP
and auxiliary enzymes to load and maintain the Recombinase-agent/ssDNA primer
complexes.
Alternatively, the limitations of the use of ATPyS may be overcome by the use
of additional
reaction components capable of stripping recA bound to ATPyS from exchange
complexes.
This role might be played by helicases such as the RuvA/RuvB complex.
The terms 'nucleic acid polymer' or 'nucleic acids' as used in this
description can be
interpreted broadly and include DNA and RNA as well as other hybridizing
nucleic-acid-like
molecules such as those with substituted backbones e.g. peptide nucleic acids
(PNAs),
morpholino backboned nucleic acids, locked nucleic acid or other nucleic acids
with modified
bases and sugars.
Structurally similar to RNA, LNA monomers are bicyclic compounds that bear a
methylene linker that connects the nucleotide sugar ring's 2'-oxygen to its 4'-
carbon. LNA
polymers obey standard base-pairing rules, but their physical properties make
them suitable for
mismatch discrimination applications. LNA are available from Exiqon (Denmark)
or Proligo
(USA, Colorado).
One embodiment of the invention is directed to a method of performing RPA. The
method comprises two steps. In the first step, the following reagents are
combined in a reaction:
(1) at least one recombinase; (2) at least one single stranded DNA binding
protein; (3) at least
one DNA polymerase; (4) dNTPs or a mixture of dNTPs and ddNTPs; (5) a crowding
agent; (6)
a buffer; (7) a reducing agent; (8) ATP or ATP analog; (9) at least one
recombinase loading
protein; (10) a first primer and optionally a second primer; and (11) a target
nucleic acid
molecule. In the second step, the reagents are incubated until a desired
degree of amplification
is achieved.
The recombinase may be uvsX, recA or a combination of both. The recombinase
may
also comprise a C terminal deletion of acidic residues to improve its
activity. While any
recombinase concentration disclosed in the specification may be used,
preferred recombinase
concentrations may be, for example, in the range of 0.2-12 uM, 0.2-1 M, 1-4
0/1, 4-6 01, and
22
CA 3041471 2019-04-26
6-12 M.
The single stranded DNA binding protein may be the E. cob SSB or the T4 gp32
or a
derivative or a combination of these proteins. gp32 derivative may include, at
least, gp32(N),
gp32(C), gp32(C)K3A, gp32(C)R4Q, gp32(C)R4T, gp32K3A, gp32R4Q, gp32R4T and a
combination thereof (See Figures 13). The DNA binding protein may be present
at a
concentration of between 1 uM and 30 uM.
The DNA polymerase may be a eukaryotic polymerase. Examples of eukaryotic
polymerases include poi-a, pol-P, pol-8, pol-e and derivatives and
combinations thereof.
Examples of prokaryotic polymerase include E. coli DNA polymerase I Klenow
fragment,
bacteriophage 14 gp43 DNA polymerase, Bacillus stearothermophilus polymerase I
large
fragment, Phi-29 DNA polymerase, T7 DNA polymerase, Bacillus subtilis Pol I,
E. coli DNA
polymerase I, E. coli DNA polymerase II, E. coli DNA polymerase III, E. cob
DNA polymerase
IV, E. coli DNA polymerase V and derivatives and combinations thereof. In a
preferred
embodiment, the DNA polymerase is at a concentration of between 10,000
units/ml to 10
units/ml, such as between 5000 units/ml to 500 units/ml. In another preferred
embodiment, the
DNA polymerase lacks 3'-5' exonuclease activity. In yet another preferred
embodiment, the
DNA polymerase contains strand displacing properties.
Any of the proteins mentioned in the methods of the invention is understood to
also
include its derivative. These proteins includes at least the following:
recombinases, polymerase,
recombinase loading protein, single stranded DNA binding protein, accessory
agents,
RecA/ssDNA nucleoprotein filaments stabilizing agent and the like. The
derivative of these
proteins include, at least, a.fusion protein comprising a C terminus tag, N
terminus tag, or C and
N terminus tags. Non-limiting examples of suitable sequence tags include 6-
histidine (6X-His;
111111111114; SEQ ID NO:82), c-myc epitope (EQKLISEEDL; SEQ ID NO:83), FLAG
octapeptide (DYKDDDDK; SEQ ID NO:84), Protein C (EDQVDPRLIDGK; SEQ ID NO:85),
Tag-100 (EETARFQPGYRS; SEQ ID NO:86), V5 epitope (GKPIPNPLLGLDST; SEQ ID
NO:87), VSV-G (YTDIEMNRLGK; SEQ ID NO:88), Xpress (DLYDDDDK; SEQ ID NO:89),
and hemagglutinin (YPYDVPDYA; SEQ ID NO:90). Non-limiting examples of suitable
protein
23
CA 3041471 2019-04-26
tags include 13-galactosidase, thioredoxin, His-patch thioredoxin, IgG-binding
domain, intein-chitin
binding domain, T7 gene 10, glutathione-S-transferase (GST), green fluorescent
protein (GFP), and
maltose binding protein (MBP). It will be understood by those in the art that
sequence tags and
protein tags can be used interchangeably, e.g., for purification and/or
identification purposes.
Accordingly, as used herein, the terms "His tag" and -hexahistidine tag"
encompass all suitable
sequence tags and protein tags as known in the art and indicated in this
paragraph.
The dNTPs includes, for example, dATP, dGTP, dCTP, and dTTP. In leading and
lagging
strand RPA, ATP, GTP, CTP, and UTP may also be included for synthesis of RNA
primers. In
addition, ddNTPs (ddATP, ddTTP, ddGTP and ddGTP) may be used to generate
fragment ladders.
The dNTP may be used at a concentration of between 1 M to 200 LM of each NIP
species. A
mixture of dNTP and ddNTP may be used with ddNTP concentrations at 1/100 to
1/1000 of that of
the dNTP (luM to 2001aM).
The crowding agents used in the RPA include polyethylene glycol (PEG), dextran
and
FicollTM. The crowding agent may be at a concentration of between 1% to 12% by
volume or 1% to
12% by weight of the reaction. While all PEGs are useful, preferred PEGs
include PEG1450,
PEG3000, PEG8000, PEG10000, PEG compound molecular weight 15000-to 20,000
(also known
as Carbowax 20M), and a combination thereof.
The buffer solution in a an RPA may be a Tris-HCI buffer, a Tris-Acetate
buffer, or a
combination thereof. The buffers may be present at a concentration of between
10 to 100 mM. The
buffered pH may be between 6.5 to 9Ø The buffer may further contain Mg ions
(e.g., in the form
of Mg Acetate) at a concentration between 1 to 100 mM with a concentration of
between 5 to 15
mM being preferred. One preferred Mg concentration is 10 mM (Mg concentration
or Mg Acetate
concentration).
Reducing agents to be used include DTT. The DTT concentration may be between I
mM
and 10 mM.
The ATP or ATP analog may be any of ATP, ATP-y-S, ATP-f3-S, ddATP or a
combination
thereof. A preferred ATP or ATP analog concentration is between 1 and 10 mM.
24
CA 3041471 2019-04-26
Recombinase loading protein may include, for example, T4uvsY, E. coli ma), E.
coli
recR and derivatives and combinations of these proteins. One preferred
concentration of these
proteins is between 0.2 and 812M.
The primers used may be made from DNA, RNA, PNA, LNA, morpholino backbone
nucleic acid, phosphorothiorate backbone nucleic acid and a combination
thereof. Combinations
thereof in this can refers to a single nucleic acid molecules which may
contain one or more of
one base connected to one of more of another base. Preferred concentration of
these molecules
may be in the range of between 25 nM to 1000 nM. In one preferred embodiment,
the primers
may contain a non-phosphate linkage between the two bases at its 3' end and is
resistant to 3' to
5' nuclease activity. In another embodiment, the primers may contain a locked
nucleic acid at
its 3' ultimate base or 3' penultimate base. For example, in a nucleic acid of
the sequence 5'-
AGT-3', the T is the 3' ultimate base and the G is the 3' penultimate base.
The primers may be
at least 20 bases in length or at least 30 bases in length. In one preferred
embodiment, the
primers are between 20 to 50 bases in length. In another preferred embodiment,
the primers are
between 20 to 40 bases in length such as between 30 to 40 bases in length.
The primers may contain additional 5' sequence that is not complementary to
the target
nucleic acid. These 5' sequence may contain, for example a restriction
endonuclease
recognition site. The primers may be partly double stranded with a single
stranded 3' end.
In addition, any nucleic acid of any of the methods of the invention may be
labeled with
a detectable label. A detectable label include, for example, a fluorochrome,
an enzyme, a
fluorescence quencher, an enzyme inhibitor, a radioactive label and a
combination thereof.
The target nucleic acid may be single stranded or double stranded. It is known
that
single stranded nucleic acids would be converted to double stranded nucleic
acid in the methods
of the invention. The target nucleic acid may be supercoiled or linear. The
sequence to be
amplified (target nucleic acid) may be in between other sequences. The
sequence to be
amplified may also be at one end of a linear nucleic acid. In one embodiment,
the target nucleic
acid is linear and not connected to non-target nucleic acids. In other words,
where the target
nucleic acid is linear, it can be in any of the following formats:
CA 3041471 2019-04-26
1. [non-target nucleic acid]-[target nucleic acid]-[non-target nucleic
acid]
2. [non-target nucleic acidittarget nucleic acid]
3. [target nucleic acid]-[non-target nucleic acid]
4. [target nucleic acid]
It should be noted that the arrangement above is intended to represent both
single
stranded nucleic acids and double stranded nucleic acids. "1" may be described
as a target
nucleic acid molecule which is linear with two ends and wherein both ends are
linked to a non-
target nucleic acid molecule. "2" may be described as a target nucleic acid
molecule which is
linear with two ends and wherein one end is linked to a non-target nucleic
acid molecule. "3"
may be described as a target nucleic acid molecule which is a linear nucleic
acid molecule (with
no non-target nucleic acid).
In another embodiment, the target nucleic acid may be a single-stranded
nucleic acid
which is converted to a double stranded nucleic acid by a polymerase or a
double stranded
nucleic acid denatured by the action of heat or chemical treatment.
The target nucleic acid may be of any concentration such as less than 10,000
copies, less
than 1000 copies, less than 100 copies, less than 10 copies or 1 copy in a
reaction. A reaction
volume may be 5 I, 10 I, 20 I, 30 I, 50 1, 75 I, 100 1, 300 I, I ml, 3
ml, 10ml, 30 ml, 50
ml or 100 ml.
The reaction may be incubated between 5 minutes and 16 hours, such as between
15
minutes and 3 hours or between 30 minutes and 2 hours. The incubation may be
performed until
a desired degree of amplification is achieved. The desired degree of
amplification may be 10
fold, 100 fold, 1000 fold, 10,000 fold, 100,000 fold or 1000000 fold
amplification. Incubation
temperature may be between 20 C and 50 C, between 20 C and 40 C, such as
between 20 C and
30 C. One advantage of the methods of the invention is that the temperature is
not critical and
precise control, while preferred, is not absolutely necessary. For example, in
a field
environment, it is sufficient to keep the RPA at room temperature, or close to
body temperature
(35oC to 38oC) by placing the sample in a body crevice. Furthermore, the RPA
may be
26
CA 3041471 2019-04-26
performed without temperature induced melting of the template nucleic acid.
In another embodiment of the invention, the RPA further comprise accessory
agents.
These accessory agents includes helicase, topoisomerase, resolvase and a
combination thereof
which possess unwinding, relaxing, and resolving activities respectively on
DNA. The
accessory agents may also include RuvA, RuvB, RuvC, RecG, PriA, PriB, PriC,
DnaT, DnaB,
DnaC, DnaG, DnaX clamp loader, polymerase core complex, DNA ligase and a
sliding clamp
and a combination thereof. The sliding claim may be E. coli I3-dimer sliding
clamp, the
eukaryotic PCNA.sliding clamp, or the T4 sliding clamp gp45 and a combination
thereof. The
accessory agents may include, in addition, DNA Polymerase III holoenzyme
complex consisting
of 0-Clamp, DnaX Clamp Loader, and the Polymerase Core Complex. These latter
accessory
agents would allow the performance of leading and lagging RPA.
In another embodiment, the RPA may be performed in the presence of a
RecA/ssDNA
nucleoprotein filaments stabilizing agent. Examples of such stabilizing
include RecR, RecO,
RecF and a combination thereof. These stabilizing agents may be present at a
concentration of
between 0.01 M to 20 M. Other examples of stabilizing agents incolude the T4
uvsY protein
which stabilizes uvsX/ssDNA nucleoproteiun complexes.
Other components of RPA include a system for ATP regeneration (convert ADP to
ATP). Such system may be, for example, phosphocreatine and creatine kinase.
The RPA reaction may also include a system to regenerate ADP from AMP and a to
convert pyrophosphate to phosphate (pyrophosphate).
In one preferred embodiment, the RPA reaction as listed above are performed
with E.
coli components completely by using recA, SSB, red), recR and E coli
polymerase.
In another preferred embodiment, the RPA reaction is performed with T4
components by
using uvsX, gp32, uvxY, and T4 polymerase.
In one preferred embodiment, RPA may be performed by combining the following
reagents: (1) a uvsX recombinase at a concentration of between 0.2 to 12 M;
(2) a gp32 single
stranded DNA binding protein at a concentration between 1 to 30 M; (3) a
Bacillus subtilis
27
CA 3041471 2019-04-26
DNA polymerase I large fragment (Bsu polymerase) at a concentration between
500 to 5000
units per ml; (4) dNTPs or a mixture of dNTPs and ddNTPs at a concentration of
between 1-300
M; (5) polyethylene glycol at a concentration of between 1% to 12 % by weight
or by volume;
(6) Tris-acetate buffer at a concentration of between 1 mM to 60 mM; (7) DTT
at a
concentration of between 1 mM -10 mM; (8) ATP at a concentration of between 1
mM - 10 mM;
(9) uvsY at a concentration of between 0.2 i.tM -8 1..tM; (10) a first primer
and optionally a
second primer, wherein said primers are at a concentration of between 50 nM to
1 itM; and (11)
a target nucleic acid molecule of at least one copy. After the reaction is
assembled, it is
incubated until a desired degree of amplification is achieved. This is usually
within 2 hours,
preferably within 1 hour, such as, for example, in 50 minutes.
One advantage of the invention is that the reagents for RPA, with the possible
exception
of the crowding agent and buffer, may be freeze dried (i.e., lyophilzed)
before use. Freezed
dried reagent offer the advantage of not requiring refrigeration to maintain
activity. For
example, a tube of RPA reagents may be stored at room temperature. This
advantage is
especially useful in field conditions where access to refrigeration is
limited.
In one embodiment, the RPA reagents may be freeze dried onto the bottom of a
tube, or
on a bead (or another type of solid support). To perform an RPA reaction, the
reagents are
reconstituted in a buffer solution and with a crowding reagent, or simply a
buffered solution or
water, dependant on the composition of the freeze-dried reagents. Then a
target nucleic acid, or
a sample suspected to contain a target nucleic acid is added. The
reconstitution liquid may also
contain the sample DNA. The reconstituted reaction is incubated for a period
of time and the
amplified nucleic acid, if present, is detected.
Detection may be performed using any method, such as, for example, using
electrophoresis on an agarose or PAGE gel followed by ethidium bromide
staining.
In any of the methods of the invention, the reagents that can be freeze dried
before use
would include, at least, the recombinase, the single stranded DNA binding
protein, the DNA
polymerase, the dNTPs or the mixture of dNTPs and ddNTPs, the reducing agent,
the ATP or
ATP analog, the recombinase loading protein, and the first primer and
optionally a second
28
CA 3041471 2019-04-26
primer or a combination of any of these.
In one preferred embodiment, the reagents are assembled by combining the
reagents such
that when constituted, they will have the following concentrations: (1) a uvsX
recombinase at a
concentration of between 0.2 to 12 M; (2) a gp32 single stranded DNA binding
protein at a
concentration between 1 to 30 M; (3) a 14 gp43 DNA polyrnerase or Bsu
polymerase at a
concentration between 500 to 5000 units per ml; (4) dNTPs or a mixture of
dNTPs and ddNTPs
at a concentration of between 1-300 M; (5) DTT at a concentration of between
1 mM -10 mM;
(6) ATP at a concentration of between 1 mM - 10 mM; (7) uvsY at a
concentration of between
0.2 M -8 M. Optionally, a first primer and optionally a second prime may be
added where
their concentration would be between 50 nM to 1 AM when reconstituted. The
reagents are
freeze dried before use. Stabilizing agents such as trehalose sugar may be
included in the freeze
dried mixture, for example at 20mM to 200mM and most optimally 40inM to 80mM
in the
reconstituted reaction, in order to improve freeze-drying performance and
shelf life. If desired,
the freeze dried reagents may be stored for 1 day, 1 week, 1 month or 1 year
or more before use.
In use, the reagents are reconstituted with buffer (a) Iris-acetate buffer at
a concentration
of between 1 mM to 60 mM; and (b) polyethylene glycol at a concentration of
between 1% to 12
% by weight or by volume, or (c) with water. If the primers were not added
before freeze
drying, they can be added at this stage. Finally, a target nucleic acid, or a
sample suspected of
containing a target nucleic acid is added to begin the reaction. The target,
or sample, nucleic acid
may be contained within the reconstitution buffer as a consequence of earlier
extraction or
processing steps. The reaction is incubated until a desired degree of
amplification is achieved.
Any of the RPA reaction conditions discussed anywhere in this specification
may be
freeze dried. For example, the following reagents can be assembled by
combining each reagent
such that when constituted, they will have the following concentrations: (1)
100-200 ng,4t1 uvsX
recombinase; (2) 600 ng/ I gp32; (3) 20 ng/ I Bsu polymerase or 14 polymerase;
(4) 200 M
dNTPs; (5) 1 mM DTT (6) 3 mM ATP or an Al? analog; (7) 16 ng/ I to 60ng/ 1
uvsY; (8)
50nM to 300 nM of a first primer and 50nM to 300 nM of a second primer; (9) 80
mM
Potassium acetate; (10) 10 mM Magnesium acetate; (11) 20 mM Phosphocreatine;
(12) 50ng/ 1
29
CA 3041471 2019-04-26
to 100 ng/ I Creatine kinase. The reagents may be freeze dried onto the bottom
of a tube or in a
well of a multi-well container. The reagent may be dried or attached onto a
mobile solid support
such as a bead or a strip, or a well.
In use, the tube with the reagent may be reconstituted with (1) Tris-acetate
buffer at a
concentration of between 1 mM to 60 triM and polyethylene glycol at a
concentration of
between 1% to 12 % by weight or by volume. If the reagents were dried or
attached to a mobile
solid support, the support may be dropped in a tube and reconstituted. As
discussed above, the
primers may be dried as part of the reagent or added after reconstitution.
Finally, a target
nucleic acid, or a sample suspected of containing a target nucleic acid is
added to begin the
reaction. The reaction is incubated until a desired degree of amplification is
achieved.
As another example, the following reagents can be assembled by combining each
reagent
such that when constituted, they will have the following concentrations: (1)
100-200 ng/ I uvsX
recombinase; (2) 300-1000 ngtul gp32; (3) 10-50 ng/ 1 Bsu polymerase or T4
polymerase; (4)
50-500 uM dNTPs; (5) 0.1 to 10 mM DTI; (6) 3 mM ATP or an ATP analog; (7)
16ng/u1 to
60ng4t1 uvsY; (8) 50nM to1000 nM of a first primer and 50nM to 1000 nM of a
second primer;
(9) 40mM to160 mM Potassium acetate; (10) 5mM to20 mM Magnesium acetate; (11)
lOmM
to40 mM Phosphocreatine; (12) 50 ng/ I to 200 ng4i1 Creatine kinase. These
reagents are
freeze dried and stored. In use, the reagents are reconstituted with Tris-
acetate buffer at a
concentration of between 1 mM to 60 iriM and polyethylene glycol at a
concentration of
between 1% to 12 % by weight or by volume. The primers, item 8 above, may be
omitted
before freeze drying and added after reconstitution. To initiate the RPA, a
target nucleic acid, or
a sample suspected of containing a target nucleic acid is added. The reaction
is incubated until a
desired degree of amplification is achieved.
Another embodiment of the invention comprises a kit for performing RPA. The
kit may
comprise any of the reagents discussed above for RPA in the concentrations
described above.
The reagents of the kit may be freeze dried. For example, the kit may contain
(1) 100-200 ng/u1
uvsX recombinase; (2) 300 ng/ 1 to 1000 ng/1.11 gp32; (3) 10 ng/u1 to 50 ng/u1
Bsu polymerase
or T4 polymerase; (4) 50 uM to 500 p.M dNTPs; (5) 0.1 to 10 mM DTT; (6) 1mM to
5mM ATP
CA 3041471 2019-04-26
or an ATP analog; (7) 16ng,/ 1 to 60ng41 uvsY; (8) 50nM to 1000 nM of a first
primer and
50nM to 1000 riM of a second primer (optional); (9) 40mM to 160 mM Potassium
acetate; (10)
5mM to 20 mM Magnesium acetate; (11) 10mM to 40 mM Phosphocreatine; (12) 50
ng,4t1 to
200 ng/ 1 Creatine kinase.
In a preferred embodiment, RPA is performed with several auxiliary enzymes
that can
promote efficient disassembly of Recombinase-agent/dsDNA complexes after DNA
synthesis
initiation. These auxiliary enzymes include those that are capable of
stimulating 3' to 5'
disassembly and those capable of supporting 5' to 3' disassembly.
Auxiliary enzymes include several polymerases that can displace RecA in the 3'
to 5'
direction and can stimulate 3' to 5' disassembly of Recombinase-agent/dsDNA
complexes
(Pham et al., 2001). These DNA polymerase include E. coil PolV and homologous
polymerase
of other species. Normally in the life of E. coil, displacement of RecA in the
3' to 5' direction
occurs as part of SOS-lesion-targeted synthesis in concert with SSB, sliding
clamps and DNA
polymerase. The polymerase essential for this activity in E. coil is PolV, a
member of the
recently discovered superfamily of polymerases including UmuC, DinB, Rad30,
and Rev I,
whose function in vivo is to copy DNA lesion templates. Critical to RPA, the
in vitro 3' to 5'
disassembly of RecA filaments cannot be catalyzed by Poll, PolIII, or PolIV
alone. Only PolV,
in concert with SSB, has measurable ATP-independent 3' to 5' RecA/dsDNA
disassembly
activity. In effect, PolV pushes and removes RecA from DNA in a 3' to 5'
direction ahead of
the polymerase (Pham et al., 2001; Tang et al., 2000). Inclusion of PolV or a
functional
homologue may improve the amplification efficiency.
Other auxiliary enzymes include a class of enzymes called helicases that can
be used to
promote the disassembly of RecA from dsDNA. These promote disassembly in both
the 5' to 3'
and 3' to 5' directions. Helicases are essential components of the
recombination process in vivo
and function to move the branch points of recombination intermediates from one
place to
another, to separate strands, and to disassemble and recycle components bound
to DNA. After
the first round of invasion/synthesis has occurred in RPA, two new DNA
duplexes are "marked"
by the presence of RecA bound over the site to which primers must bind for
additional rounds of
31
CA 3041471 2019-04-26
synthesis. In such a situation dsDNA tends to occupy the high affinity site in
RecA, or
homologs, until it is actively displaced, either by ATP hydrolysis-dependent
dissociation in the
5' to 3' direction, which may be limiting, or by 3' to 5' dissociation by some
other active
process. An ideal helicase complex for stimulating disassembly of RecA from
intermediates
consists of the E. coil proteins RuvA and RuvB. The RuvAB complex promotes
branch
migration, and dissociates the RecA protein, allowing RecA to be recycled
(Adams et at, 1994).
Normally, the RuvAB complex is targeted to recombination intermediates,
particularly Holliday
junction-like structures. As it works the RuvAB complex encircles DNA and
forces RecA from
the DNA in an ATP-driven translocation (Cromie and Leach, 2000; Eggleston and
West, 2000).
This RecA dissociation activity has been demonstrated using supercoiled dsDNA
bound with
RecA, which does not even possess Holliday junctions (Adams et al., PNAS
1994). The RuvAB
complex can recognize branched structures within the RecA coated DNA.
Incorporation of
RuvAB into the RPA mixture will promote the dissociation of RecA from dsDNA
following
strand exchange and displacement, allowing renewed synthesis of the duplicated
template from
the same site. Additionally, the RuvAB complex can act in concert with RuvC,
which finally
cuts and resolves Holliday junctions. With RuvC added to the RPA reaction
mixture,
complicated structures such as Holliday junctions formed at invasion sites,
can be resolved.
Resolvase activity, such as that provided by RuvC, is particularly important
when the targeting
oligonucleotides are partially double-stranded. In such situations reverse
branch migration can
generate Holliday junctions, which can then be resolved by the RuvABC complex,
to generate
clean separated amplification products.
Still other auxiliary enzymes include the E. coli RecG protein. RecG can
stimulate
disassembly of branch structures. In vivo this protein functions to reverse
replication forks at
sites of DNA damage by unwinding both leading and lagging strands driving the
replication fork
back to generate a 4-way junction (Cox et al., 2000; Dillingham and
Kowalczykowski, 2001;
Singleton et al., 2001). In vivo such junctions function as substrates for
strand switching to
allow lesion bypass. In vitro RecG will bind to D-loops, and will lead to a
decrease in D-loop
structures by driving reverse branch migration. RecG prefers a junction with
double-stranded
elements on either side, hence partly double-stranded targeting
oligonucleotides, homologous to
32
CA 3041471 2019-04-26
the targeting site in both single-stranded and double-stranded regions, would
be ideal. This
would stimulate reverse branch migration and formation of a Holliday junction,
which can be
resolved by the RuvABC complex. In vivo RecG and RuvAB may compete to give
different
outcomes of recombination since branch migration will be driven in both
directions (McGlynn
and Lloyd, 1999; McGlynn et al., 2000). In both cases the proteins target
junction DNA coated
with RecA, and disassemble it in an active manner.
Other auxiliary enzymes useful in an RPA reaction mixture are those that allow
continual
generation of RecA nucleoprotein filaments in the presence of ATP and SSB. In
order to allow
removal of RecA at the appropriate moments, it is preferred to use ATP rather
than ATPyS in an
RPA reaction. Unfortunately RecA/ssDNA filaments formed with ATP spontaneously
depolymerize in the 5' to 3' direction, and in the presence of SSB, as
required here,
repolymerization will not occur at significant rates. The solution to this
problem is the use of the
RecO, RecR, and possibly RecF proteins. Alternatively the uvsY protein may be
employed to
stabilize the T4 uvsX nucleoprotein filaments in a similar manner. In the
presence of SSB and
ATP, RecA/ssDNA filaments dissociate (Bork et al., 2001; Webb et al., 1995;
Webb et al.,
1997; Webb et al., /999). If RecAissDNA is incubated in the presence of RecO
and RecR
proteins this dissociation does not occur. Indeed the RecR protein remains
associated with the
filament and stabilizes the structure indefinitely. Even if ssDNA is bound by
SSB, in the
presence of RecR and RecO, filaments of RecA can reassemble displacing SSB. In
the T4
phage system similar properties are attributed to the uvsY protein. Thus it is
possible to obviate
the use of ATPyS, if necessary, by using ATP in the presence of RecO and RecR
to maintain
ReckssDNA filament integrity, or uvsY to maintain the uvsX/ssDNA filament
integrity. The
RecF protein interacts with the RecO and RecR system in a seemingly opposing
manner. RecF
competes with RecR tending to drive filament disassembly in vitro. It is
likely that all three
components in vivo function together to control the generation of invading
structures, while
limiting the extent of RecA coating of ssDNA. In another preferred embodiment,
RecF is
included in RPA reactions at an appropriate concentration to re-capitulate the
dynamics of the in
vivo processes. In addition, RecF may facilitate dissociation of RecA-coated
intermediates after
invasion has occurred.
33
CA 3041471 2019-04-26
As described, the use of ATP rather than ATPyS, and/or the use of displacing
polymerases and helicases (e.g. the RuvA/RuvB complex), RecO, RecR and RecF,
or
alternatively the T4 uvsX recombinase with the uvsY protein, should permit
exponential
amplification of double-stranded DNA by driving continual regeneration of
targeting sites. This
method, however, remains responsive to differences in initiation rate that
might occur at the two
opposing targeting sites. Such differences may lead to a decrease in
amplification efficiency,
and to the production of some single-stranded DNA. The PCR method largely
avoids these
complications because temperature cycling leads to coordinated synthesis from
either side. In
another embodiment, a situation analogous to the PCR condition just described
may be induced
by using temperature sensitive (ts) mutants of RecA that are non-functional at
42 C, but function
at lower temperatures in the range 25 to 37 C (Alexseyev et al., 1996; Hickson
et al., 1981). In
this case, synthesis from either end can be synchronized by periodic lowering
to the permissive
temperature and then raising the reaction to a temperature non-permissive for
the mutant RecA
protein function, but permissive for the other components. By performing RPA
with tsRecA
mutants in combination with cycling of reaction temperatures, the number of
molecules of DNA
produced can be controlled. While this will require some mechanism to provide
temperature
cycling, the temperatures are well below those that would require the use of
thermophile-derived
proteins. Indeed, a simple chemical-based or portable low-power temperature-
cycling device
may be sufficient to control such reaction cycles.
RPA, as all other present-day nucleic acid amplification methods, employs
polymerases
to generate copies of template nucleic acid molecules. It is a necessity of
most nucleic acid
polymerases that incorporation requires a free 3'-hydroxyl moiety on the
terminal sugar of a
short stretch of double-stranded nucleic acid adjacent to the site of new
synthesis. This stretch
of double-stranded nucleic acid is typically formed on a template by a short
complementary
sequence, called a primer, which serves as an initiation site for the
polymerase synthesis
reaction. In some cases a 3' modification, such as a sulfydryl, may utilized
to prime the
synthesis reaction. The primer nucleic acid, which is base-paired with the
template and
extended by the polymerase, can be RNA or DNA. In vivo during genomic DNA
replication,
RNA primer sequences are synthesized de novo onto template DNA by primase
enzymes.
34
CA 3041471 2019-04-26
Typically, for in vitro reactions the primer is supplied as a short, often
chemically synthesized,
single-stranded DNA (or modified DNA or RNA), and is usually referred to as an
oligonucleotide primer. The primer is often of a specific sequence, although
random primers
can also be used. The primer is targeted to complementary sequences by virtue
of its specific
base-pairing capacity. Formation of hybrids between the oligonucleotide primer
and target
nucleic acid are typically formed by incubation of the two in solution under
conditions of salt,
pH, and temperature that allow spontaneous annealing.
In the case of the PCR the oligonucleotide primer is usually in vast excess
for two main
reasons. First, the high concentration will drive rapid annealing. Second, as
the reaction
proceeds through rounds of melting, annealing and extension the primer is
consumed and
becomes limiting. PCR targeted nucleic acids are often initially
double¨stranded in character,
and if not, become double stranded following the first synthetic cycle. Such
double-stranded
molecules cannot anneal new oligonucleotides at temperature and solvent
conditions appropriate
for the catalytic activity and stability of most prokaryotic and eukaryotic
proteins.
Consequently, in order to allow cycles of amplification the original template
and the newly
synthesized strands must be first separated before annealing can occur once
again. In practice
this is achieved by thermal melting. For PCR, temperatures of at least 80 C
are required for
thermal melting of most double-stranded nucleic acid molecules of lengths
greater than 100 base
pairs. In most PCR protocols a temperature of 90 to 100 C is applied to melt
the DNA. Such
temperatures allow only rare thermostable enzymes to be used. These
polymerases are typically
derived from thermophilic prokaryotes.
The advantage of RPA is that it allows the formation of short stretches of
double-
stranded nucleic acids bearing a free 3' ¨OH for extension from double-
stranded templates
without thermal melting. This is achieved by using the RecA protein from E.
coli (or a RecA
relative from other phyla including the 14 uvsX protein). In the presence of
ATP, dATP,
ddATP, UTP, ATP7S, and possibly other types of nucleoside triphosphates and
their analogs,
RecA or uvsX will form a nucleoprotein filament around single-stranded DNA.
This filament
will then scan double-stranded DNA. When homologous sequences are located the
recombinase
CA 3041471 2019-04-26
will catalyze a strand invasion reaction and pairing of the oligonucleotide
with the homologous
strand of the target DNA. The original pairing strand is displaced by strand
invasion leaving a
bubble of single stranded DNA in the region.
RecA protein can be obtained from commercial sources. Alternatively it can be
purified
according to standard protocols e.g. (Cox et al., 1981; Kuramitsu et al.,
1981). RecA
homologues have been purified from therrnophilic organisms including
Thermococcus
kodakaraensis (Rashid et at., 2001), Thermotoga maritima (Wetmur et at.,
1994), Aquifex
pyrophilus (Wetmur et al., 1994), Pyrococcus furiosus (Komori et at., 2000),
Thermus aquaticus
(Wetmur et at., 1994), Pyrobaculum islandicum (Spies et at., 2000), and Therm
us thermophilus
(Kato and Kuramitsu, 1993). RecA has also been purified from other prokaryotes
e.g.
Salmonella typhimurium (Pierre and Paoletti, 1983), Bacillus subtilis (Lovett
and Roberts,
1985), Streptococcus pneumoniae (Steffen and Bryant, 2000), Bacteroides
fragilis (Goodman et
al., 1987), Proteus mirabilis (West et at., 1983), Rhizobium meliloti (Better
and Helinski, 1983),
Pseudomonas aeruginosa (Kurumizaka et al., 1994), from eukaryotes e.g.
Saccharomyces
cerevisiae (Heyer and Kolodner, 1989), Ustilago maydis (Bennett and Holloman,
2001),
including vertebrates e.g. Human Rad51 (Baumann et al., 1997) and Xenopus
laevis (Maeshima
et al., 1996), as well as plants including broccoli (Tissier et at., 1995). We
here also show that
E. coil recA, and T4 uvsX protein, can be purified from overexpression
cultures using a
hexahistidine tag at the C terminus, and remain biologically active. This is
of great utility for
the production of recombinant protein.
For clarity of description, leading strand Recombinase-Polymerase
Amplification
method (lsRPA) can be divided into four phases.
1) Sequence targeting
RPA is initiated by targeting sequences using synthetic oligonucleotides
coated with
RecA, or a functional homologue such as the T4 uvsX protein. In order to
permit exponential
amplification two such synthetic oligonucleotides would be employed in a
manner such that
their free 3'-ends are orientated toward one another. Nucleoprotein filaments
comprising these
oligonucleotides and recombinase protein will identify targets in complex DNA
rapidly and
36
CA 3041471 2019-04-26
specifically. Once targeted the recombinase protein catalyses strand exchange
such that D-loop
structures are formed. It may be necessary to use ATP rather than ATPyS in the
procedure for
efficient amplification. If ATP is used, RecO, RecR, and/or RecF, molecules
may prove
essential for efficient amplification, or uvsY protein if uvsX recombinase is
employed.
2) Initiation of DNA synthesis
DNA polymerases will detect and bind to the hybrid between the invading
oligonucleotides and the template DNA and initiate DNA synthesis from the free
3'-hydroxyl
exposed in the hybrid. Exposure of this 3'-hydroxyl, and subsequent DNA
synthesis, will likely
require disassembly of recombinase protein from the double-stranded hybrid
formed by strand
exchange. To attain this disassembly it will probably be necessary to employ
ATP, which can
support spontaneous disassembly of recombinase from invasion complexes.
Additionally
disassembly can be stimulated/enhanced by the use of other proteins contained
within the
reaction mixture such as RuvA, RuvB, RuvC, recG, other helicases, or other
stimulatory
components, which can act to strip recombinase from the strand exchange
product.
3) Strand displacement DNA synthesis and replicon separation.
As the DNA polymerases synthesize complementary copies of template DNAs using
the
free 3 '-hydroxyls of invading oligonucleotides, or their partly extended
products, the
polymerases displace single-stranded DNAs, which may be coated with single
strand binding
proteins (SSB) included in the reaction. In an ideal configuration, invasion
of oligonucleotides
at both ends of the target nucleic acid sequence will occur in similar
timeframes, such that two
polymerases on the same template nucleic acid will initially progress toward
one another. When
these extending complexes meet one another, the original template should
simply fall apart, and
the polymerases will continue to synthesize without a need for strand
displacement, now
copying SSB-bound ssDNA template. Because of steric hinderance, polymerases
may become
dissociated from the template temporarily when the polymerases meet to permit
the separation
of the two template strands
37
CA 3041471 2019-04-26
4) Completion of synthesis and re-invasion.
Once the template strands have separated, polymerases can complete the
extension to the
end of the template (or past the sequence acting as the second, facing,
targeting site if the initial
template is longer than the desired product). To permit exponential
amplification it is necessary
for new products to be targeted and replicated in a manner similar to the
original templates, that
is from both targeted ends. The newly synthesized targeted site will be freely
available to
targeting recombinase/oligonucleotide filaments. The site initially used to
prime synthesis
should also have been freed as a consequence of the use of conditions in the
reaction that favor
disassembly of recombinase from strand exchange products. Providing the re-
invasion at this
latter site occurs in less time than it takes the polymerase to synthesize
past the second targeting
site, be primed at that second site, and return to the first site, then single-
stranded DNA will not
be the primary product and exponential amplification will occur. Having
multiple synthetic
complexes operating on the same template raises the possibility that very
short amplification
times can be achieved.
Recombinase-Polymerase Amplification (RPA) using simultaneous leading and
lagging
strand synthesis
In our description of (leading strand RPA) IsRPA we detail a multi-component
system
with the capacity to regenerate targeting sequences thus permitting
exponential amplification of
double-stranded DNA. Unlike the Zarling method, IsRPA avoids the linear
production of
single-stranded DNA. There is another approach to solving this problem that
completely avoids
the possibility of single-stranded products and a requirement for simultaneous
end initiation.
This method necessarily involves a more complex reaction mixture. Nevertheless
all of the
required components are now well understood and should be amenable to assembly
into a single
system. This system will recapitulate events occurring during the normal
replication cycle of
cells to permit coupled leading and lagging strand synthesis. This method,
leading/lagging
strand RPA is described briefly in Figures 1 and 3.
During normal replication in vivo, double-stranded DNA is simultaneously
separated into
2 strands and both are copied to give 2 new molecules of double-stranded DNA
by a replication
38
CA 3041471 2019-04-26
machine. This 'machine' couples conventional 5' to 3' leading strand synthesis
with lagging
strand synthesis, in which short RNA primers are synthesized onto template
nucleic acids by
primase enzymes. During lagging strand synthesis, short fragments of DNA are
produced,
called Okazaki fragments, which are ligated together to form contiguous
lagging strands. This
simultaneous leading-strand/lagging-strand synthesis is responsible for
duplication of the entire
genome of prokaryotic and eukaryotic organisms alike. The essential components
of this system
have been identified and characterized biochemically. The components can be
assembled in
vitro to achieve a more efficient amplification than possible using only
leading-strand synthesis.
The essential components of the replication 'machine' are now well
characterized for E.
coli and certain other organisms such as T4 phage. This machine comprises the
PolIII
holoenzyme (Glover and McHenry, 2001; Kelman and O'Donnell, 1995) and the
primosome
(Benkovic et al., 2001; Marians, 1999). The Poll!! holoenzyme is made up of
ten polypeptide
components. Each holoenzyme contains two, asymmetrically oriented, core
structures, each
consisting of a polymerase (a subunit) and two additional core components the
c subunit, which
possesses 3' to 5' exonuclease activity, and the 0 subunit. In addition to the
core complex
another set of polypeptides provide the holoenzyme with processivity and
couple leading and
lagging strand synthesis. The f3-dimer sliding clamp encircles the template
DNA affixing the
complex to the template with extremely high affinity. The sliding clamp loaded
onto DNA by
the DnaX clamp loader comprising the 12y68'xtii polypeptide subunits.
For clarity of description, the RPA method can be divided into four phases. In
reality all
phases will occur simultaneously in a single reaction.
1) Sequence targeting
RPA is initiated by targeting sequences using synthetic oligonucleotides
coated with
RecA, or T4 uvsX, or a functional homologue. Such nucleoprotein filaments will
identify
targets in complex DNA rapidly and specifically. Once targeted, the RecA or
uvsX protein
catalyses strand exchange such that a D-loop structure is formed. It may be
necessary to use--.
ATP rather than ATPyS in the procedure for efficient amplification. The
linkage of leading and
lagging strand syntheses however may obviate the requirement for very rapid
recombinase
39
CA 3041471 2019-04-26
stripping after initiation of synthesis. If ATP is used, RecO, RecR, and RecF
may need to be
employed with bacterial recA recombinase, or the T4 uvsY , proteins may prove
essential for
efficient amplification with T4 uvsX protein.
2) Primosome assembly
Primosomes can be assembled at D-loops. Normally, in E .coli, D-loop
structures are
formed by RecA as part of the mechanism to rescue damaged DNA in vivo, or
during other
forms of recombination. The purpose of the combined action of RecA-mediated
strand
exchange and primosome assembly is to generate a replication fork. A
replication fork is the
nucleoprotein structure comprising the separated template DNA strands and the
replisome. The
replisome consists of the polyrnerase holoenzyme complex, the primosome, and
other
components needed to simultaneously replicate both strands of template DNA.
Primosomes
provide both the DNA unwinding and the Okazaki fragment priming functions
required for
replication fork progression. Similar primosome assembly occurs at
recombination
intermediates in T4 phage directed by gp59 and gp41 protein.
Primosome assembly has been studied intensively through genetic and
biochemical
analysis in E. coil. The minimal set of polypeptides required for this process
is well known and
exist as purified components. The primosome assembly proteins are PriA, PriB,
PriC, Dual,
DnaC, Dna% and DnaG. These proteins have been shown sufficient to assemble a
primosome
complex on bacteriophage (DX174 DNA in vitro (Komberg and Baker, 1992;
Marians, 1992).
PriA binds to the primosome assembly site (PAS) on the (1)X174 chromosome.
Then PriB,
DnaT, and PriC bind sequentially to the PriA-DNA complex. PriB appears to
stabilize PriA at
the PAS and facilitate the binding of DnaT (Liu et al., 1996). PriC is only
partially required for
the full assembly reaction. Omission of PriC from the reaction will lower
priming 3 to 4 fold
(Ng and Mariam, 1996a; Ng and Mariam, 1996b). The function of PriC in the
bacterium is
genetically redundant to PriB. DnaC then loads DnaB into the complex in an ATP-
dependent
fashion. This PriABC-DnaBT complex is competent to translocate along the
chromosome. The
DnaG primase can interact transiently with the complex to synthesize RNA
primers.
During replication in E. coil. DnaB and DnaG function as a helicase and
primase
CA 3041471 2019-04-26
respectively. These two components are continually required in association
with the PolIII
holoenzyme to synthesize primers for the Okazaki fragments. Hence, DnaB and
DnaG are the
core components of the mobile primosome associated with the replication fork.
The other
primosome components described are essential for assembly of the primosome
onto DNA, and
for associating a dimeric polymerase. The primosome assembly proteins are
required for the re-
establishment of replication forks at recombination intermediates formed by
RecA and strand
exchange. PriA can initiate assembly of a replisome, competent for DNA
synthesis, on
recombination intermediates. It is possible to target D-loops in vitro with a
mixture of PriA,
PriB, and DnaT, which are then competent to incorporate DnaB and DnaC. Once a
primosome
has been formed at the D-loop, all that remains to initiate replication is to
load a holoenzyme
complex to the site. Alternatively in the phage T4 system the gp59 helicase
loader protein
recruits and assembles the gp41 replicative helicase to D-loop structures
3) Fork assembly and initiation of DNA synthesis
Replication forks will assemble at the site of primosome assembly. hi E. coli
the
presence of a free 3'-end on the invading strand of the D-loop stimulates the
DnaX clamp loader
complex detailed earlier to assemble a fl-dimer at this site to act as a
sliding clamp. The
holoenzyme and 2 core units are joined together by the scaffold 2 subunit. The
r subunit also
has interaction surfaces for the p-dimer, for the clamp loader, and for the
DnaB helicase
component of the primosome. These multiple interactions are necessary to
coordinate synthesis
of both leading and lagging strands using the 2 asymmetrically joined core
polymerase
complexes. In T4 phage the gp59/41 proteins with uvsY and gp32 proteins, and
with other
components coordinate assembly of the sliding clamp gp45 aided by gp44 and
gp62 proteins
initiates replisome assembly.
In E. coil the primosomal primase, DnaG, synthesizes a short RNA primer onto
the
unwound lagging strand DNA template. In the presence of the holoenzyme, the
clamp loader
recognizes the RNAJDNA duplex and loads a second 13-dimer clamp onto this
site. The
presence of an active primosome and the interaction of the r subunit with
'DnaB are critical to
ensure simultaneous leading/lagging strand synthesis. Without this interaction
the polymerase
41
CA 3041471 2019-04-26
will move away from the primosome site without coupling.
A replication fork is now assembled. Synthesis of both leading and lagging
strand will
now occur simultaneously, and the DnaB helicase will separate template strands
ahead of the
oncoming holoenzyme. The lagging strand holoenzyme core will generate Okazaki
fragments
of I to 2 kilobases in length. Once the lagging strand polymerase encounters
the previous RNA
primer, it dissociates from the 13-clamp and synthesis is initiated from a
newly assembled clamp
loaded in the vicinity of the front of the leading strand. The same lagging
strand holoenzyme
core will be re-used since it is physically tethered to leading strand core.
There is a dynamic interaction between I3-dimer clamps, core subunits, and
clamp
loaders. Their affinities can switch depending upon the physical
circumstances. The fl-dimer
that has been 'abandoned' at the end of the Okazaki fragments may be recycled
via active
removal by clamp loaders, or excess 8 subunit that may be present.
The RNA primers at the ends of Okazaki fragments are removed by the 5' to 3'
exonuclease activity of DNA polymerase I. DNA ligase then joins the Okazaki
fragments
together forming a continuous lagging strand.
4) Fork meeting and termination
In RPA, replication is initiated at two distant sites and the replication
forks are oriented
toward each other. As replication forks converge the two original template
strands will
dissociate from one another as they become separated entirely both behind, and
in front, of each
fork. The leading strand core of each fork will then complete synthesis, the
remaining RNA
primers will be processed, and the final products will be two double-stranded
molecules. We
can reasonably expect to amplify DNA's on the order of several Megabases (Mb)
by such an
approach. In this disclosure, megabase also encompasses megabasepairs. Based
on the known
synthetic rate of the PolIII holoenzyme we can expect the replication forks to
proceed at a rate of
approximately 1 Mb / 1000 seconds, i.e., approximately 15 to 20 minutes per
cycle for a 1 Mb
fragment.
The final consideration is the mechanism by which rapid exponential
amplification of
42
CA 3041471 2019-04-26
DNA will be achieved. The key to this process will be to allow efficient
reinvasion of the
targeting sites by the use of mixtures of helicases, resolvases and the RecO,
RecR, and RecF
proteins. Under appropriate conditions reinvasion and primosome assembly
should be possible
shortly after a holoenzyme has moved away from the fork-assembly site.
Continual invasions
should present no problems since the DNA will simply become branched at many
points. Each
branch will naturally resolve as it encounters the oncoming fork. Under these
conditions it may
be possible to achieve enormous amplification in times similar to the time
taken to replicate the
DNA only once. It may be critical however to limit the concentrations of
targeting
oligonucleotides to avoid nucleotide depletion prior to the completion of
synthesis.
In addition to the holoenzyme complex, the replication machine employs another
complex known as the primosome, which synthesizes the lagging strand and moves
the
replication fork forwards. The primosome complex comprises a helicase encoded
by DnaB and
a primase encoded by DnaG. Finally, in addition to the proteins of the
holoenzyme and
primosome, replication requires the activity of single-stranded DNA binding
protein (SSB), E.
coli DNA polynterase I and DNA ligase. These latter two components are
required to process
Okazaki fragments.
Nested RPA
In another embodiment, RPA amplification may be performed in a process
referred to
herein as "nested RPA." A difficulty in detecting a rare sequence is that
there can be a high ratio
of non-target to target sequence. The ability of a RPA to discriminate between
target and non-
target DNA and amplify only target sequences is a key aspect of improved
sensitivity.
Discrimination between non-target and target is a reflection of the
specificity of the primers and
reaction conditions. The more specific a reaction is the greater the relative
amount of the
specific target sequence that is produced and the easier that product is to
detect. An increase in
specificity can, therefore, increase sensitivity as well.
The need for improved sensitivity and specificity can be addressed by using
nested RPA.
The nested RPA involves a first RPA of a first region of DNA. Then the
reaction mixture is
diluted, for example, by 10, 20, 30, 40, 50, 75, or 100 fold or more to reduce
the concentration
43
CA 3041471 2019-04-26
of the first primer pair, and a second primer pair is introduced into the
reaction mixture and RPA
repeated. According to one embodiment of the invention, the second primer pair
is designed to
be internal to the first primer pair to amplify a subsequence of the first RPA
product. The
method increases specific amplification, i.e., reduces non-specific background
amplification
products and therefore increases sensitivity. Such non-specific amplification
products, although
they arise by virtue of fortuitous partial homology to the flanking primers,
are unlikely to also
have sufficient homology to the nested primers to continue to amplify.
Detection and specificity
of RPA may be further improved by labeling one or both of the second primer
pair such that
only primers amplified with one or both of the second primer pair is detected.
Nested RPA is not limited to the use of two sets of primer. Naturally, more
sets of
primers may be used to increase specificity or sensitivity. Thus, three, four,
or five pairs of
primers may be used. Furthermore, the different sets of primers, as another
embodiment of the
invention, may share common primers as illustrated in Figure 4.
In Figure 4, the primer sets are designed to be used sequentially. For
example, a first
RPA is performed with primer set I, a second RPA using the amplified product
of the first RPA
is performed with a primer set 2, a third RPA using the amplified product of
the second RPA is
performed with a primer set 3, a fourth RPA using the amplified sequence of
the third RPA is
performed with a primer set 4, and finally, a fifth RPA is performed using the
amplified product
of the fourth RPA is performed with a primer set 5. In this case, primer set
1, 2, and 3, share a
common primer ¨ primer (a). Primer 3, 4, and 5 share a common primer ¨ primer
(b).
Nested RPA may be performed using any of the two RPA methods described as well
as a
combination of the two methods in any particular order. That is, RPA may be
performed solely
by leading strand RPA, solely by leading and lagging strand RPA, or a
combination of leading
strand RPA and leading and lagging strand RPA in any particular order.
One benefit of any of the RPA methods of the invention is the size of the
amplified
product. While current methods of amplification such as PCR are limited to an
upper limit of
about 10 Kb, RPA methods are capable of amplifying regions of nucleic acids of
up to hundreds
of megabases. For leading/lagging strand RPA, the sizes of a target sequence
to be amplified
44
CA 3041471 2019-04-26
can be hundreds of megabases, such as, for example, less than 500 megabases,
less than 300
megabase, less than 100 megabase, less than 70 megabase, less than 50
megabase, less than 25
megabase, less than 10 megabase, less than 5 megabase, less than 2 megabases,
less than one
megabase, less than 500 kb, less than 200 kb, less than 100 kb, less than 50
kb, less than 25 kb,
or less than 10 kb, less than 5 kb, less than 2 kb, less than 1 kb. For lsRPA,
the sizes of a target
sequence can be in the megabase range such as, less than 5 megabase, less than
2 megabases,
less than one megabase, less than 500 kb, less than 200 kb, less than 100 kb,
less than 50 kb, less
than 25 kb, or less than 10 kb, less than 5 kb, less than 2 kb, less than 1
kb.
General considerations for reconstituting and enabling recombinase-mediated
amplification reactions
Both IsRPA and leading/lagging RPA rely on similar use of recombinase proteins
to
target oligonucleotide primers, however they differ in the mode by which the
new daughter
duplexes are formed during amplification. In leading/lagging RPA a full
replication fork is
established that simultaneously synthesises leading and lagging strands so
that two new
duplexes are concomitantly formed. In leading strand RPA (IsRPA), only leading
strand
synthesis occurs so that synthesis generates one duplex and one displaced
single-stranded DNA
as products.
In RPA DNA synthesis initiated after strand exchange is accomplished by a
polymerase.
During extension of the newly synthesised strand the polymerase must be able
to displace the
outgoing strand, either alone or in combination with a helicase capable of
mediating outgoing
strand displacement. Extension of the invading primer results eventually in
the release of the
outgoing strand as a single-stranded DNA. To ensure that geometric
amplification occurs, and
that the reaction produces a vast majority of double-stranded DNA, it is
necessary for this
displaced single-stranded DNA to serve as template for DNA synthesis from the
opposite
direction. This is a central consideration for amplification using IsRPA. Two
other central
considerations are the polymerase species used, and the existence of a stable,
dynamic,
recombinase system that functions efficiently in the presence saturating
levels of single-strand
binding proteins. These considerations are important for both the
leading/lagging RPA and
CA 3041471 2019-04-26
lsRPA.
A) Ensuring generation of double-stranded DNA from the displaced strand
The generation of the second strand of DNA in lsRPA can be achieved in one of
several
ways:
1) The displaced single-stranded DNA can simply hybridise to the complementary
strand which has been displaced from invasion and extension of a second
'facing' targeting
oligonucleotide. Alternatively the displaced single-stranded DNA can hybridise
directly with
the second 'facing' oligonucleotide. Such hybridisation events may occur
spontaneously, or may
be mediated by the strand assimilating activities of DNA binding proteins such
as recombinases
or single-stranded DNA binding proteins. Following hybridisation a polymerase
will extend
from the free 3'end to generate a double-stranded product. Note that for this
to occur efficiently
the reaction environment must enable hybridisation of complementary single-
stranded DNAs, a
situation not always compatible with the other aspects of the RPA reaction. In
some
circumstances a hybridising oligonucleotide with a modified backbone, unable
to interact with
most DNA binding protein, could be used.
2) If
strand-displacement synthesis begins simultaneously from opposing
oligonucleotide primer on the same template then the two converging
replication complexes will
eventually meet somewhere in the middle of the template. Provided that these
converging
complexes are able to pass one another the template strands will separate and
each complex will
complete replication by copying a single-stranded rather than double-stranded
template with no
further need for strand displacement.
3) If the outgoing strand possesses the capacity to form a hairpin then self-
priming
second strand synthesis may occur. This activity would result in a covalently
linked duplex with
a hairpin at one end, which could become a target for further
invasion/extension reactions. This
situation is not ideal for many applications, as it will generate products
with variable lengths and
structures. This may, however, be acceptable for detection assays, such as
some diagnostic tests.
Furthermore it may be possible to engineer primers such that after the first
few rounds of
invasion/extension most outgoing strands are capable of self-priming. This
mode of duplex
46
CA 3041471 2019-04-26
DNA formation may be very efficient.
Which of these three general processes dominate in an lsRPA reaction will
depend on
many factors. The most important factors are the distance separating the two
oligonucleotides
primers in the target, the invasion rate, and the sequence of the
oligonucleotides and template.
In the second general format of RPA, leading/lagging RPA, the generation of
substantial
single-stranded DNA is avoided by establishing a full replication fork at the
invasion site. A full
replication fork permits the simultaneous copying of both leading and lagging
strands (which
would be equivalent to the outgoing strand). Leading/lagging RPA is elegant in
its avoidance of
the generation of single-stranded DNA, however a larger number of distinct
proteins is required
to generate full replication forks. Nevertheless most aspects of optimisation
for RPA reactions
apply to both lsRPA and leading/lagging RPA.
B) Choice of polymerase, or polymerase/helicase system
The lsRPA method is similar in some respects to PCR. Both processes use of
pairs of
oligonucleotide primers orientated with 3' ends pointed toward one another in
the target DNA
and both methods achieve geometric amplification through the use of reaction
products as
targets for subsequent rounds of DNA synthesis. There are, however,
fundamental differences
in the reaction configurations. For example, in RPA target DNA is double-
stranded prior to
synthesis, whereas in PCR it is single-stranded after thermal separation of
strands. In RPA,
DNA synthesis must necessarily use DNA polymerases or polymerase complexes
capable of
strand displacement. Furthermore, because partially copied strands are
physically associated
with displaced strand through the template, there is a risk that if the
polymerase dissociates
temporarily from the template the 3' end of the new strand, and eventually the
whole new strand,
will be lost by the action of branch migration or another phenomenon known as
bubble
migration. This suggests that ideally processive polymerases will be used for
RPA reactions. It
is also important to consider that if converging replication complexes cannot
readily pass one
another without polymerase dissociation then some processive polymerases may
inhibit the RPA
reaction on some templates. In summary the ideal choice of polymerase will
depend on the
precise format and objective of the particular RPA reaction, in particular the
size of the product
47
CA 3041471 2019-04-26
to be amplified.
C) Establishment of a stable persistent active recombinase activity in a noise-
suppressed
environment
A third consideration is how to establish a stable, but dynamic, recombinase
activity,
while silencing the noise generated by aberrant primer annealing seen at low
temperatures. This
means establishing a reaction environment that balances several seemingly
incompatible
requirements. For efficient RPA recombinase proteins must remain active while
assembled into
oligonucleotide/recombinase filaments scanning for target double-stranded
DNAs. These
complexes must also disassemble after completing strand exchanges to permit
DNA synthesis.
This must happen in an environment rich in single-stranded DNA proteins. These
proteins are
needed to stimulate recombination and DNA synthesis while preventing aberrant
oligonucleotide
behaviour by melting secondary structures. Fundamentally, the recombinases and
single-
stranded binding proteins are in competition for oligonucleotide binding.
While the single-
strand binding proteins are necessary to enable efficient strand displacement
during synthesis,
they suppress recombination activity because they have a higher affinity for
single-stranded
DNA and bind with more cooperativity than do recombinases.
Establishment of a functional recombinase/replication reaction environment
requires
nucleotide cofactors. RecA, and other recombinases, require nucleotide co-
factors, such as
ATP, to assemble filaments onto single-stranded DNA, perform homology
searches, and
complete strand exchange. We have surmised that non-hydrolysable analogues
such as ATP-y-S
would be incompatible with RPA because the extremely high stability of the 3-
stranded
DNA/recA intermediate formed in the presence of with ATP-y-S would prevent
reinvasion at
primer targets and would thus prevent efficient amplification. They may even
prevent any
useful access of polymerase to the recombination intermediate. Earlier
attempts to amplify
DNA using E. coli recA (Zarling et al) were probably limited by the ATP-y-S in
the described
reactions.
The requirement for ATP in the reaction and the fact that recombinase-
complexes will be
dynamically forming and disassembling introduces additional complexities,
primarily due to
48
CA 3041471 2019-04-26
complex interactions and competition between key reaction components. In
particular single-
stranded binding proteins, such as the E. coil single-stranded binding
proteins, such as E. colt
SSB or 14 phage gp32 protein, are necessary to stimulate recombination by recA
and
homologues, due both to their capacity to collect the outgoing strand, and to
melt secondary
structures in single-stranded DNAs thus enhancing recombinase loading. In RPA
it is likely that
single-stranded binding proteins will further stimulate DNA synthesis by
binding and stabilising
the displaced DNA strand, preventing undesirable branch migration.
Despite the clear requirement for single-stranded binding proteins, these
proteins
generally have a considerably higher affinity for single-stranded DNA than
recombinases such
as recA, or uvsX, and can inhibit nucleation of recombinaseiDNA filaments.
Moreover, as
filaments formed in the presence of ATP undergo end-dependant disassembly
(Bork, Cox and
Inman J Biol Chem. 2001 Dec 7;276(49):45740-3), such filaments are likely to
be rapidly
saturated with single-stranded binding proteins and inactivated soon after
initiation of the
reaction. Thus for efficient RPA conditions that prevent inactivation of the
reaction components
are key in establishing robust amplification.
We have predicted a potentially stable reaction composition using E. coil recA
protein in
the presence of ATP and the E. coli single-stranded binding protein SSB, or
the T4 uvsX protein
in the presence of gp32. We suggested the presence of recO, recR, and possibly
recF proteins
(Bork, Cox and Inman EMBO J. 2001 Dec 17;20(24):7313-22), could lead to an
environment in '-
which pre-loaded recA filaments were stabilised, and in which recA could
nucleate successfully
onto SSB-bound oligonucleotides. A similar recombinase loading system has been
described in
other organisms including the recombination/replication/repair system of
bacteriophage T4. The
T4 recombinase uvsX can be loaded onto single-stranded DNA coated with the T4
single-
stranded DNA binding protein gp32 by the action of a third component, the uvsY
protein
(Morrical and Alberts J Biol Chem. 1990 Sep 5;265(25):15096-103).
Interestingly a principal
role of this recombination system in vivo is to permit recombination-dependant
DNA synthesis
by assembling replication components at recombination intermediates such as D-
loops (Formosa
and Alberts Cell. 1986 Dec 5;47(5):793-806). This process is similar to what
should happen in
RPA driven from D-loops made by the invasion of synthetic oligonucleotides. In
addition to
49
CA 3041471 2019-04-26
interactions between the three components uvsX, uvsY, and gp32, there are also
interactions
between these components and the replication machinery such as the polymerase,
clamp loader,
primase/helicase, and dda helicase (Reddy, Weitzel and Von Hippel, Proc Natl
Acad Sci U S A.
1993 Apr 15;90(8):3211-5., Hacker and Alberts, J Biol Chem. 1992 Oct
15;267(29):20674-81).
Taken together these facts suggest that the components of the T4
recombination/replication
machinery would perhaps be even more ideal for RPA than the E. coli
equivalents.
In addition to the use of recombinase loading proteins, such as rec0 and recR,
or uvsY,
there are other ways to create an appropriate balance between recombinase
activity and the
activity of single-stranded DNA binding proteins. The DNA binding and/or
cooperativity
behaviour of recombinases and single-stranded DNA binding proteins can be
modulated by
mutation. In addition, recombinases from different sources have distinct
properties (Eggler,
Lusetti and Cox, J Biol Chem. 2003 May 2;278(18):16389-96. Epub 2003 Feb 20,
Villemain et
al., J Biol Chem. 2000 Oct 6;275(40):31496-504). This suggests that a range of
recombinase
and single-strand DNA binding protein activities could be explored. The use of
mutated
proteins or proteins from different species, in a set of optimisation
experiments could lead to the
identification of an optimal ratio of the competing recombinase and single-
stranded binding
activities. Ultimately the activities would be balanced such that DNA
association/dissociation
for the two DNA-binding species permits sufficient recombinase activity
together with sufficient
DNA melting, activity of the single-strand DNA binding protein to perform its
.necessary
functions also. In addition, reduction of noise due to mispriming may be
achieved through the
optimisation of such parameters as oligonucleotide sequence design, reaction
buffer, the use of
partly modified oligonucleotides, the use of part duplex oligonucleotides or
the addition of other
specific reaction components detailed below.
Here we provide the results of experiments that validate the RPA method. In
particular,
we provide a description and demonstration of reaction compositions capable of
supporting
DNA amplification. We demonstrate that relatively short synthetic
oligonucleotides can be used
to target specific sequences and support initiation of DNA synthesis. We
describe the
requirements for particular types and concentrations of certain recombinases,
single-stranded
binding proteins, ATP, and oligonucleotide concentrations. We further describe
the optimisation
CA 3041471 2019-04-26
and modulation of the reaction environment, which supports an active and
dynamic
recombination system with desired rate behaviour, through the inclusion of
crowding agents
(such as polyethylene glycols), recombinase loading factors and/or mutated
proteins with altered
biochemical activities. We establish that in the presence of distributive
polymerases at least
(e.g. the E. coli DNA polymerase I Klenow fragment), there are substantial
improvements in
amplification efficiency when the distance between the amplification priming
sites is optimised.
We establish that a balance between polymerase exonuclease activity and
oligonucleotide
protecting agents must be employed to avoid non-specific degradation of
oligonucleotide
primers. We show that amplification of sequences embedded within linear (or
relaxed) DNA
substrates is relatively inefficient (at least with very distributive
polymerases such as Klenow),
whereas amplification reactions directed toward the ends of linear DNA
substrates are most
effective. We provide methods to prepare target DNA to be more efficiently
amplified in an
lsRPA reaction, including the methods of thermal or chemical melting or
restriction enzyme
digestion. We also provide evidence that the nature of the single-stranded
binding protein is
critical to establish efficient RPA reactions, and provide a rationale for
this. Furthermore we
suggest improvements and novel approaches to reduce noise and optimise
amplification
reactions performed at relatively low, or ambient, temperatures by the use of
part-double-
stranded oligonucleotides, or oligonucleotide wholly or partly lacking a
phosphate backbone.
We also provide evidence that other enzymes and proteins involved in DNA
metabolism can
influence RPA reactions, and some may be configured to improve reaction
efficiency and
specificity. These include topoisomerases, which can relax recombination /
replication
intermediates and may aid targeting of embedded sequences, as well as
helicases such as T4 dda
helicase or 14 gp41 which can improve the polymerase initiation and elongation
efficiency,
particularly if non-strand displacing polymerases are used. Finally we show
that priA and the
ruvA/B helicases have activities that might be used to optimise amplification
efficiency.
Selection of RPA reagents and reaction parameters
The details of leading strand RPA, leading and lagging strand RPA, and nested
RPA
were listed above. This section will describe the selection of reagents and
parameter for any of
51
CA 3041471 2019-04-26
the three methods discussed above.
One benefit of RPA is that the amplified product of RPA is double stranded DNA
that
could be used for other molecular biology procedures. Thus, RPA may be
combined with other
methods in molecular biology. For example, the starting material for RPA may
be a PCR
amplified fragment. Alternatively, the product of an RPA may be used for PCR.
If necessary, the RPA products in any of the methods of the invention may be
purified.
For example, in the nested RPA method, the amplified product may be purified
after each RPA
step before a subsequent RPA step. Methods of purification of nucleic acids
are known in the
art and would include, at least, phenol extraction, nucleic acid precipitation
(e.g., with salt and
ethanol), column chromatography (e.g., size exclusion, ionic column, affinity
column and the
like) or any combination of these techniques.
As discussed, the primers used in RPA may be "double stranded" or "capable of
forming
double stranded structures." These terms refer to DNA molecules that exist in
a double
stranded condition in a reaction solution such as a RPA reaction solution or a
PCR reaction
solution. The composition of a PCR solution is known. The composition of a RPA
reaction is
listed in this detailed description section and in the Examples.
The primers may have a single stranded region for hybridization to the target
DNA in the
presence of a recombinase agent. The single stranded region may be, for
example, about 10
bases about 15 bases, about 20 bases, about 25 bases, about 30 bases, about 40
bases, and about
50 bases. Even longer regions such as about 75 bases, about 100 bases, about
150 bases or more
may be used but it is not necessary. The choice of single stranded regions
will depend on the
complexity of the starting nucleic acid so that for example, a human genome
may require a
longer primer while a plasmid may require a much shorter primer.
The two strands of nucleic acid in a double stranded DNA need not be
completely
complementary. For example, the double-stranded region of a double-stranded
DNA may differ
by up to 1% in sequence. That is, the sequence of two strands of nucleic acid
may differ by one
base in one hundred bases and would still exist in a double stranded condition
in solution.
Nucleic acids with 1% difference in their complementary sequence are
contemplated as double
52
CA 3041471 2019-04-26
stranded DNA for the purposes of this disclosure.
In addition, the target nucleic acid (i.e., the nucleic acid to be amplified
by the RPA
methods of the invention) may be partially double stranded and partially
single stranded. For
example, nucleic acid in any of the configuration of Figure 5 would be
suitable as a target
nucleic acid of the invention. As discussed, the target nucleic acid may be
RNA. RNA can be
converted to double-stranded cDNA using known methods and the double-stranded
cDNA may
be used as the target nucleic acid. As shown if Figure 5, the template nucleic
acid may have any
combination of ends selected from 3' overhang, 5' overhang, or blunt ends.
The IsRPA method of the invention comprises at least the following steps.
First, a
recombinase agent is contacted to two nucleic acid primers (referred to herein
as a first and a
second primer) to form two nucleoprotein primers (referred to herein as a
first nucleoprotein
primer and a second nucleoprotein primer).
Second, the first and second nucleoprotein primers are contacted to the
template nucleic
acid to form a double stranded structure at a first portion of the first
strand and a second double
stranded structure at a second portion of the second strand. The two primers
are designed so that
when hybridized, they are pointed at each other as illustrated in Figure 6A.
Alternatively,
primer 1 and primer 2 may hybridize different target nucleic acids as
illustrated in Figure 6B.
Third, the nucleoprotein primers are extended at their 3' ends to generate a
first and a
second double stranded nucleic acid (Figure 7A). Where the primers are
hybridized to different
target nucleic acids, the elongation of the primers will generate displaced
strands (Figure 7B).
In this case, the two displaced strands that result from primer elongation may
hybridize and form
a new double stranded template nucleic acid (Figure 7C).
Step two and three are repeated until the desired degree of amplification is
reached. The
process is a dynamic process in that primer hybridization to the target
nucleic acid and
elongation are allowed to proceed continuously. One advantage of this
invention is that the
amplification is performed continuously without the need for temperature
cycling or enzyme
addition after initiation of the reaction.
In an embodiment, steps two and three are repeated at least 5 times.
Preferably, it is
53
CA 3041471 2019-04-26
repeated at least 10 times. More preferably, it is repeated at least 20 times,
such as at least 30
times. Most preferably, the two steps are repeated at least 50 times. For
multiple repetitions of
the amplification step (e.g., step 2 and 3) a RPA of the invention is
preferably started with a
primer to target nucleic acid ration of at least 100 to 1, preferably at least
300 to 1, and most
preferably at least 1000 to 1. That is, there are at least 100, 300 or 1000
copies of the primer per
copy of a target nucleic acid.
In an optional step, after a sufficient round of amplification, additional
components may
be added to the reaction after a period of time to enhance the overall
amplification efficiency. In
one embodiment, the additional components may be one or more of the following:
recombinase
agents, one or more primers, polymerase, and one or more of the additional
agents (discussed in
a separate section below).
In a preferred embodiment, a small fraction of a first RPA reaction is used as
a supply of
template DNA for subsequent rounds or RPA amplification. In this method, a
first RPA
amplification reaction is performed on a target nucleic acid. After the first
RPA reaction, a
small fraction of the total reaction is used as a substitute of the target
nucleic acid for a
subsequent round of RPA reaction. The fraction may be, for example, less than
about 10% of
the first reaction. Preferably, the fraction may be less than about 5% of the
first reaction. More
preferably, the fraction may be less than 2% of the first reaction. Most
preferably, the fraction
may be less than 1% of the initial reaction.
The primer used in RPA is preferably DNA although PNA, and RNA are also
suitable
for use as primers. It is noted that in fact, in DNA replication, DNA
polymerases elongate
genomic DNA from RNA primers.
Synthetic oligonucleotides may serve as DNA primer and can be used as
substrates for
formation of nucleoprotein filaments with RecA or its homologues. Sequences as
short as 15
nucleotides are capable of targeting double-stranded DNA (Hsieh et al., 1992).
Such
oligonucleotides can be synthesized according to standard phophoroamidate
chemistry, or
otherwise. Modified bases and/or linker backbone chemistries may be desirable
and functional
in some cases. Additionally oligonucleotides may be modified at their ends,
either 5' or 3', with
54
CA 3041471 2019-04-26
groups that serve various purposes e.g. fluorescent groups, quenchers,
protecting (blocking)
groups (reversible or not), magnetic tags, proteins etc. In some cases single-
stranded
oligonucleotides may be used for strand invasion, in others only partly single
stranded nucleic
acids may be used, the 5' stretch of sequence of an invading nucleic acid
being already
hybridized to an oligonucleotide.
In another embodiment of the invention, the primers may comprise a 5' region
that is not
homologous to the target nucleic acid. It should be noted that the processes
of the invention
should be functional even if the primers are not completely complementary to
the target nucleic
acid. The primers may be noncomplementary by having additional sequences at
their 5' end.
These additional sequences may be, for example, the sequence for a restriction
endonuclease
recognition site or the sequence that is complementary to a sequencing primer.
The restriction
endonuclease recognition site may be useful for subsequent cleavage of the
amplified sequence.
The use of restriction endonuclease that cleaves nucleic acid outside the
restriction endonuclease
recognition site is also contemplated. The sequence that is complementary for
a sequencing
primer may allow rapid DNA sequencing of the amplified product using
commercially available
primers or commercially available sequencing apparatus.
Formation of nucleoprotein filaments can be performed by incubation of the
primer
(oligonucleotides) with RecA protein or its homologues in the presence of ATP,
and auxiliary
proteins such as RecO, RecR and RecF, or uvsY in the case 01 14 proteins. When
incubated at
37 C in RecA buffer (20 mM Tris-HC1 pH 7.5, 10 mM MgCl2, 2mM ATP, 2 mM DTT and
100
n/m1 Bovine Serum Albumin), RecA will form helical filaments on ssDNA with 6
protomers
per turn. The DNA is located within the interior of the protein helix. In the
presence of dsDNA,
the RecA/ssDNA nucleoprotein filament can scan DNA at rates of at least 107 bp
per hour. The
mode of scanning is unclear but it is at a speed (>103 bp per second) that it
may involve only the
initial few base pairs that can be easily accessed along one face of the major
groove. Successful
binding may result in a transition to a triple-helical intermediate, which is
then followed by
strand invasion and displacement to form a D-loop. Such joint molecules can be
formed under
similar condition to those described above for formation of helical filaments,
and hence in the
presence of ssDNA, the homologous dsDNA, RecA, ATP, auxiliary proteins and
suitable buffer
CA 3041471 2019-04-26
and temperature conditions, joint molecules will form spontaneously. If ATP is
used the
assembly is reversible and will reach equilibrium, but RecA/ssDNA filaments
can be stabilized,
even in the presence of SSB, by the auxiliary proteins Rec0 and RecR.
Alternatively the 14
uvsX protein may be stabilized in the presence of uvsY protein. In the case of
thermostable
proteins the temperature of incubation can be higher. If a renewable supply of
ATP is required a
standard ATP regeneration system can be included in the reaction.
DNA polymerases can use the free 3'-hydroxyl of the invading strand to
catalyze DNA
synthesis by incorporation of new nucleotides. A number of polymerases can use
the 3'-
hydroxyl of the invading strand to catalyze synthesis and simultaneously
displace the other
strand as synthesis occurs. For example E. coli polymerase II or III can be
used to extend
invaded D-loops (Morel et al., 1997). In addition, E. coli polymerase V
normally used in SOS-
lesion-targeted mutations in E. coli can be used (Pham et al., 2001). All of
these polymerases
can be rendered highly processive through their interactions and co-operation
with the f3-dimer
clamp, as well as single stranded DNA binding protein (SSB) and other
components. Other
polymerases from prokaryotes, viruses, and eukaryotes can also be used to
extend the invading
strand.
In another embodiment of the invention, the primer may be partially double
stranded,
partially single stranded and with at least one single stranded 3' overhang.
In this embodiment,
the primer may comprise a invading strand and a non-invading strand as shown
in Figure 8A. In
this case, after the invading strand is hybridized to the target DNA and
elongated, it serves as a
target nucleic acid for a second primer as shown in Figure 8B. The elongation
of the second
primer would displace the noninvading strand as shown in Figure 8C. In this
embodiment, as
the target nucleic acid is amplified, the non-invading strand of primer 1 is
displaced. If both
primer one and primer two are partly double stranded primers, then the non-
invading strands of
both primer one and primer two will accumulate in solution as the target
nucleic acid is
amplified.
In one embodiment of the invention, at least two of the primers in a RPA
reaction are
partially double stranded and partially single stranded each generated by the
hybridization of an
56
CA 3041471 2019-04-26
invading strand and a non-invading oligonucleotide strand, which possess
sequences of
sufficiently complementary that they form a double stranded region.
Preferably, the two
oligonucleotide strands are sufficiently complementary over the relevant
region that they can
form a double stranded structure in RPA reaction conditions.
In an embodiment of the invention, the primers, including single-stranded and
partially
double-stranded primers, are labeled with a detectable label. It should be
noted that a
fluorescence quencher is also considered a detectable label. For example, the
fluorescence
quencher may be contacted to a fluorescent dye and the amount of quenching is
detected. The
detectable label should be such that it does not interfere with an elongation
reaction. Where the
primer is partially double stranded with an invading strand and a non-invading
strand, the
detectable label should be attached in such a way so it would not interfere
with the elongation
reaction of the invading strand. The non-invading strand of a partially double
stranded primer is
not elongated so there are no limitations on the labeling of the non-invading
strand with the sole
exception being that the label on the non-invading strand should not interfere
with the elongation
reaction of the invading strand. Labeled primers offer the advantage of a more
rapid detection
of amplified product. In addition, the detection of unincorporated label, that
is, labeled
oligonucleotides that have not been extended, will allow the monitoring of the
status of the
reaction.
Monitoring a RPA reaction may involve, for example, removing a fraction of an
RPA
reaction, isolating the unincorporated fraction, and detecting the
unincorporated primer. Since
the size of an unincorporated primer may be less than 50 bp, less than 40 bp,
less than 30 bp or
less than 25 bp, and the size of the amplified product may be greater than
1Kb, greater than 2
Kb, greater than 5 Kb, or greater than 10 Kb, there is a great size difference
between the
incorporated and unincorporated primer. The isolation of the unincorporated
primer may be
performed rapidly using size exclusion chromatography such as, for example, a
spin column. If
a primer is labeled, a monitor procedure comprising a spin column and a
measurement (e.g.,
fluorescence or radioactivity) can be performed in less than one minute.
Another alternative for
separating elongated primers from unelongated primers involve the use of PAGE.
For example,
the elongated primer may be separated from the unelongated primer by gel
electrophoresis in
57
CA 3041471 2019-04-26
less than 5 minutes. Yet another alternative for separating elongated primers
involves the use of
immobilized oligonucleotides. For example oligonucleotides homologous to
sequences found
uniquely within the amplified DNA sequence can be used to capture nucleic
acids produced by
primer elongation specifically. These capturing oligonucleotides can be
immobilized on a chip,
or other substrate. Capture of the elongated oligonucleotides by the capturing
oligonucleotides
can be performed by RecA protein mediated methods, or by traditional solution
hybridizations if
necessary.
In another embodiment of the invention, a double stranded primer may be
labeled such
that the separation of the two strands of the primer may be detected. As
discussed above, after
multiple rounds of elongation, the invading strand and the noninvading strands
of a partially
double stranded primer is separated. After this separation, the non-invading
strand does not
participate in the RPA reaction. This characteristic may be used to detect and
monitor a RPA
reaction in a number of ways.
In this application, the detectable label may be a fluorescent label or an
enzyme and the
label quencher (also referred to as the label inhibitor) may be a fluorescence
quencher or an
enzyme inhibitor. In these cases, the label is detected by fluorescence or
enzyme inhibition.
The delectability of the label would be the fluorescence if a fluorescent
label is used or enzyme
activity if an enzyme is used.
In the first method, the invading strand may be labeled with a label and the
non-invading
strand may be labeled with a detectable label quencher. The label, in the
proximity of the label
quencher (label inhibitor) on the partially double stranded primer would not
be highly
detectable. After RPA, the invading strand would be separated from the
noninvading strand and
thus, the label and the label quencher would be separated. The separation
would cause the label
to be more detectable. Thus, measuring the increases in the amount of
detectable label may
monitor RPA reactions.
The second method is similar to the first method except that the invading
strand is
modified with a label quencher while the noninvading strand is modified with a
label. Then
RPA is allowed to proceed with the result (same as method 1) of the label
being separated from
58
CA 3041471 2019-04-26
the label quencher. Thus, the overall delectability of the label would
increase.
The third method involves labeling the noninvading strand of one double
stranded primer
with a label. In addition, the noninvading strand of a second double stranded
primer is labeled
with a label quencher. The two non-invading stands are designed to be
complementary to each
other. In this configuration, the RPA reaction is initially fluorescent. As
the RPA reaction
progresses, the two noninvading strands are displaced into solution and they
hybridize to each
other because they are designed to be complementary. As they hybridize, the
label and the label
quencher are brought into proximity to each other and the fluorescence of the
reaction is
decreased. The progress of the RPA reaction may be measured by monitoring the
decrease in
label detectability.
In a fourth method, the noninvading strands of a first and second double
stranded primers
are labeled with a first label and a second label. The two noninvading strands
are also designed
to be complementary to each other. As in the third method, after RPA, the two
noninvading
strands are hybridized to each other and the proximity of the two labels will
be a reflection of the
progress of the RPA reaction. The proximity of the two labels may be
determined, for example,
by direct observation or by isolation of the non-invading strands. As
discussed above, isolation
of primers and other small nucleic acids can be accomplished by size exclusion
columns
(including spin columns) or by gel electrophoresis.
In another embodiment of the invention, the non-invading strand of one or both
of the
primers is homologous to a second region of nucleic acid such that the primer
can hybridize to
and primer DNA synthesis at the second region of nucleic acid. Using this
method, a second
RPA reaction using the noninvading stand from the primer of a first RPA may be
started. The
product of the second RPA may be monitored to determine the progress of the
first RPA.
In yet another embodiment of the invention, the non-invading strand is
detected by a
biosensor specific for the sequence of the non-invading strand. For example,
the biosensor may
be a surface with a nucleic acid sequence complementary to the non-invading
strand. The
biosensor may monitor a characteristic that results from the binding of the
non-invading strand.
The characteristic may be a detectable label.
59
CA 3041471 2019-04-26
Suitable detectable labels for any of the methods of the invention include
enzymes,
enzyme substrates, coenzymes, enzyme inhibitors, fluorescent markers,
chromophores,
luminescent markers, radioisotopes (including radionucleotides), and one
member of a binding
pair. More specific examples include fluorescein, phycobiliprotein, tetraethyl
rhodamine, and
beta-gal. Bind
pairs may include biotin/avidin, biotin/strepavidin, antigen/antibody,
ligand/receptor, and analogs and mutants of the binding pairs.
The recombinase agent of the invention may be RecA, uvsX, RadA, RadB, Rad 51
or a
functional analog or homologues of these proteins. If desired, the recombinase
may be a
temperature-sensitive (referred to herein as "ts") recombinase agent. If a ts
recombinase is used,
the RPA reaction may be started at one temperature (the permissive
temperature) and terminated
at another temperature (the non permissive temperature). Combinations of
permissive
temperatures may be, for example 25 C/30 C, 30 C/37 C, 37 C/42 C and the like.
In a
preferred embodiment, the ts protein is reversible. A reversible ts protein's
activity is restored
when it is shifted from the nonpermissive temperature to the permissive
temperature.
In a preferred embodiment, the RPA is performed in the presences of ATP, an
ATP
analog, or another nucleoside triphosphate. The ATP analog may be, for
example, ATPyS,
dATP, ddATP, or another nucleoside triphosphate analog such as UTP.
Other useful reagents that may be added to an RPA reaction include nucleotide
triphosphates (i.e., dNTPs such as dATP, dTTP, dCTP, dGTP and derivatives and
analogs
thereof) and a DNA polymerase. Other useful reagents useful for
leading/lagging RPA include
NTPs (ATP, GTP, CTP, UTP and derivatives and analogs thereof). One advantage
of the RPA
reaction is that there is no limit on the type of polymerase used. For
example, both eukaryotic
and prokaryotic polyrnerases can be used. Prokaryotic polymerase include, at
least, E. coil poll,
E. coli pol II, E. coli poi III, E. coli pol IV and E. coil polV. Eukaryotic
polymerases include,
for example, multiprotein polymerase complexes selected from the group
consisting of pol-a,
pol-S, and pol-c.
In another embodiment of the invention, the RPA process is performed in the
presence of
an accessory component to improve polymerase processivity or fidelity. Both
eukaryotic and
CA 3041471 2019-04-26
prokaryotic accessory components may be used. Preferably, the accessory
component is an
accessory protein is from E. coli. Useful accessory proteins include single-
strand binding
protein, helicase, topoisomerase, and resolvase. Other useful accessory
proteins include a
sliding clamp selected from the group consisting of an E. coli -dimer
sliding clamp, a
eukaryotic PCNA sliding clamp and a T4 sliding clamp gp45. Other accessory
components
include a DNA Polymerase III holoenzyme complex consisting of 13-Clamp, DnaX
Clamp
Loader, and the Polymerase Core Complex. Still other accessory components
include RuvA,
RuvB, RuvC, and RecG. The properties endowed by the use of additional
components will
likely enable the amplification of large DNAs not previously successfully
targeted by current
methods such as PCR.
In another embodiment, the RPA is performed in the presence of agents used to
stabilize
recombinase/ssDNA nucleoprotein filaments. For example, the agent may be RecR,
RecO,
RecF, or a combination of these proteins, or T4 uvsY protein if T4 components
are used.
Molecular crowding agents may also be employed to modulate biochemical
interactions in a
favourable manner. Other useful agents include PriA, PriB, DnaT, DnaB, DnaC,
and DnaG.
One benefit of the present invention is that the RPA reaction may be performed
at
reduced temperatures compared to a PCR reaction. For example, the RPA process
may be
performed between 20 C and 50 C. Preferably, the RPA process is performed at
less than 45 C.
More preferably, the RPA process may. be performed at less than 40 C. Even
more preferably,
the RPA process may be performed at less than 35 C. Most preferably, the RPA
process may be
performed at less than 30 C. One of the reasons that the RPA process can be
performed at these
reduced temperatures is because RPA may be performed without temperature
induced melting of
the template nucleic acid. Further, unlike PCR, absolute temperature control
is not required and
the temperature can fluctuate without adversely affecting RPA. For example,
the amount of
fluctuation may be anywhere within the temperatures specified above. The
temperature
necessary for melting of double stranded DNA also contribute to premature
enzyme inactivation,
a disadvantage absent in the methods of this invention.
RPA may be performed to test for the presences or absences of a genotype. The
61
CA 3041471 2019-04-26
genotype tested may be associated with a disease or a predisposition to a
disease. Alternatively,
the genotype may be associated with a normal phenotype or a phenotype that
confers special
resistance to a disease. The genotype as disclosed above may be any standard
genetic variant
such as a point mutation, a deletion, an insertion, an inversion, a frameshift
mutation, a
crossover event, or the presence or absences of multiple copies of a genetic
sequence (e.g., the
presences of mini chromosomes).
One method of detecting a genotype is to detect the distance between a primer
pair in an
RPA reaction. The distance between a primer pair is reflected by the size of
the amplified
sequence. In that method, the two primers are selected such that it spans a
target region such as,
for example, a gene. Then RPA is performed using the primer pair and the RPA
product is
analyzed. The analysis may involve determining the size or sequence of the
amplified product.
Methods of determining the size of a DNA sequence, including at least
techniques such as
agarose gels, PAGE gels, mass spectroscopy, pulsed field gels, gene chips,
sucrose
sedimentation and the like are known. There are many DNA sequencing methods
and their
variants, such as the Sanger sequencing using dideoxy termination and
denaturing gel
electrophoresis (Sanger, F., Nichlen, S. & Coulson, A. R. Proc. Natl. Acad.
Sci. U.S.A. 75,
5463-5467 (1977)), Maxam-Gilber sequencing using chemical cleavage and
denaturing gel
electrophoresis (Maxam, A. M. & Gilbert, W. Proc Natl Acad Sci USA 74, 560-564
(1977)),
pyrosequencing detection pyrophosphate (PPi) released during the DNA
polymerase reaction
(Ronag,hi, M., Uhlen, M. & Nyren, P. Science 281, 363, 365 (1998)), and
sequencing by
hybridization (SBH) using oligonucleotides (Lysov, I., Florentiev, V. L.,
Khorlin, A.A.,
Khrapko, K. R. & Shik, V. V. Dokl Akad Nauk SSSR 303, 1508-1511 (1988); Bains
W. &
Smith G. C. J. Theor. Biol 135, 303-307(1988); Drnanac, R., Labat, I.,
Brulcner, 1. &
Crkvenjakov, R. Genomics 4, 114-128 (1989); Khrapko, K. R., Lysov, Y.,
Khorlyn, A. A.,
Shick, V. V., Florentiev, V. L. & Mirzabekov, A. D. FEBS Lett 256. 118-122
(1989); Pevzner P.
A. J Biomol Struct Dyn 7, 63-73 (1989); Southern, E. M., Maskos, U. & Elder,
J. K. Genomics
13, 1008-1017(1992)).
One method of detecting a genotype is to use primers that are specific for a
particular
genotype. For example, a primer may be designed to efficiently amplified one
genotype but
62
CA 3041471 2019-04-26
inefficiently or not amplify another genotype at all. In an embodiment, the
primer may comprise
a 3' sequence that is complementary to one genotype (e.g., a genetic disease
genotype) but not to
another genotype (e.g., a normal genotype).
The genotype to be determined may be indicative of a disease such as, for
example, the
presence of an activated oncogene; the presence of the gene for Huntington's
disease or the
absence of an anti--oncogene.
The 3' bases of the primers are especially important in determining the
specificity and
efficiency of an RPA reaction. A primer may be designed so that the 3' base is
complementary
to one genotype and not complementary to another genotype. This will allow
efficient RPA of
one genotype and an inefficient RPA (if any) of the second genotype. It is
noted that the method
is effective if only one primer of the primer pair can differentiate between
different phenotypes
(by having different efficiencies of amplification). In a preferred
embodiment, both primers in
an RPA reaction can differentiate between different genotypes. In this above
example, the
primers are complementary to one genotype and are not complementary to a
second genotype by
one base at its 3' end. In a preferred embodiment, the primer is not
complementary to the
second genotype by at least one base at its 3' end. Preferably, the primer is
not complementary
to the second genotype by at least 2, 3, 4, 5, 6, 7, 8, 9, or 10 bases at its
3' end. Most preferably,
the primer is completely non-complementary or cannot hybridize to the second
genotype while it
can hybridize to the first genotype. =
In some of the methods discussed, the presence or absence of an amplified
product
provides the indication of the presence or absence of a genotype. In these
cases, the RPA
reaction may be monitored by the methods discussed throughout the
specification.
In a preferred embodiment, an RPA reaction for genotyping will amplify a
sequence
regardless of the genotype of the patient. However, the genotype of a patient
will alter a
characteristic of the amplified sequence. For example, the amplified sequence
may be a
different size, or sequence for one genotype than for another genotype. In
that way, the RPA
reaction will contain an internal control to indicate that the amplification
reaction was performed
successfully. Naturally, a method of RPA, which includes one or more
additional pairs of
63
CA 3041471 2019-04-26
primers as controls for the performance of the RPA reaction, is also
envisioned.
In another embodiment, an RPA reaction may be used to determine the presence
or
absences of a nucleic acid molecule. The nucleic acid molecule may be from any
organism. For
example, the microbial composition of a sample may be determined by using a
battery of RPA
reactions directed to the nucleic acid of different microbes. RPA is
especially useful for the
detection of microbes. In one embodiment, the pathogens are selected from
viruses, bacteria,
parasites, and fungi. In further embodiments, the pathogens are viruses
selected from influenza,
rubella, varicella-zoster, hepatitis A, hepatitis B, other hepatitis viruses,
herpes simplex, polio,
smallpox, human immunodeficiency virus, vaccinia, rabies, Epstein Barr,
retroviruses, and
rhinoviruses. In another embodiment, the pathogens are bacteria selected from
Escherichia coli,
Mycobacterium tuberculosis, Salmonella, Chlamydia and Streptococcus. In yet a
further
embodiment, the pathogens are parasites selected from Plasmodium, Trypanosoma,
Toxoplasma
gondii, and Onchocerca. However, it is not intended that the present invention
be limited to the
specific genera and/or species listed above.
Here we present data that help to define reaction conditions that permit
efficient
amplification of DNA by RPA.
Single-stranded DNA binding protein
Single-stranded DNA binding proteins are required for RPA reactions. These
proteins
=
bind to single-stranded DNA, melt secondary structure, facilitate outgoing
strand displacement,
and suppress branch migration. In RPA their activity is required during
several distinct phases.
We have investigated the activities of two single-stranded DNA binding
proteins, E. coli SSB
and bacteriophages T4 gp32. The T4 gp32 has proven to be most useful in our
hands.
Furthermore we have generated a number of distinct forms of this protein by
including
hexahistidine (His) peptide tags at the N or C termini, as well as
investigating several previously
described point mutations. Activities of gp32 variants are depicted
schematically in Figure 21.
Variant forms of gp32
14 gp32 protein possesses several features that are of potential utility to
RPA reactions.
64
CA 3041471 2019-04-26
Foremost gp32 has a relatively small DNA binding site (8-10 nucleotides),
displays similar
binding properties under a wide range of salt concentrations, and displays
high (unlimited)
cooperativity between monomers (Scheerhagen et al., J Biomol Struct Dyn. 1986
Apr;3(5):887-
98; Kuil et al., Biophys Chem. 1988 Dec;32(2-3):211-27). In contrast, E. coli
SSB protein has
several distinct DNA binding modes that vary with salt concentration all of
which possess
relatively large DNA binding sites (32, 56 or 65 nucleotides)(Ferrari et al.,
J Mol Biol. 1994 Feb
11;236(1):106-23) and there is complex cooperativity behaviour (Lohman and
Ferrari, Annu
Rev Biochem. 1994;63:527-70). Because the initial size of the outgoing strand
is small when
synthetic oligonucleotides are employed, we reasoned that the properties of
the gp32 protein
would be optimal for RPA. We expressed and purified gp32 possessing an N-
terminal His tag
(gp32(N)). In initial experiments we found gp32(N) to function at least as
well as the E. coli
SSB protein, even when combined in a heterologous system with the E. coli recA
recombinase
(Figure 12). This was surprising result as gp32 is reported to display
extremely high
cooperativity between monomeric subunits and it seemed unlikely that recA
would be able to
compete effectively for oligonucleotide binding in its presence. When we
compared the
behaviour of gp32(N) to untagged gp32, however, we discovered that the two
proteins did not
behave equivalently. As the N-terminal His tag is directly adjacent to the 'B'
domain of the gp32
protein, which is required for cooperativity between monomers, we reasoned
that gp32(N) must
have attenuated cooperativity. We therefore generated a gp32 protein
possessing a C-terminal
His tag (gp32(C)), as well as point mutant forms of gp32(C) in accordance with
previously
published mutants having a lysine to alanine change at position 3 (K3 to A),
or an arginine either
glutamine (R4 to Q) or threonine at position 4 (R4 to T) (Figure 21). These
three point mutant
proteins exhibit progressively less cooperativity (Figure 22)(Villemain, et
al. J Biol Chem. 2000
Oct 6;275(40):31496-504). We tested the capacity of these proteins at two
different
concentrations to support invasion / extension reactions on a linearized
template in combination
with the bacteriophage T4 uvsX protein and the Klenow fragment of E. coli DNA
polymerase I
(Figure 22). Firstly we note that gp32(C) yields much less product than other
gp32 variants at
either concentration (Figure 22 compare with gp32(N)) and that the products
are almost
exclusively full-length, in contrast to gp32(N). When we compare these results
to those
CA 3041471 2019-04-26
obtained with the point mutant allelic series we note that the gp32(N) protein
most closely
resembles the profile obtained with gp32(C) R4 to T, which has been reported
to be significantly
attenuated in cooperativity. This suggests that the N terminal His tag of
gp32(N) interferes with
the function of the B domain in a manner similar to the point mutations.
There are several other relevant observations. Firstly, the proteins believed
to
demonstrate the highest cooperativity, i.e. gp32(C) > gp32(C)K3A seem to
produce less product.
Secondly, for gp32(C) and gp32(C)IC3A, more amplified product is generated
when less single-
stranded binding protein is used, in contrast to gp32(N) and gp32(C)R4T which
generate more
product in run-on assays when more proteins is used. Taken together these
observations suggest
an explanation. If the gp32 species is progressively more cooperative then it
will form
progressively more stable filaments on the oligonucleotides and make it
progressively more
difficult for recombinase to load. Consequently when the most cooperative gp32
species are
employed there is a significant limitation on the availability of recombinase-
loaded filaments. If
the concentration of gp32 is raised, recombinase loading is further suppressed
by gp32 single-
stranded DNA binding activity. Consistent with this, as the cooperativity of
the gp32 is
progressively decreased the amount of amplified product increases. This is
consistent with a
substantial increase in recombinase-coated filament formation. As relatively
un-cooperative
gp32 monomers are less likely to coat oligonucleotides and permit the
recombinase, uvsX, to
seed instead. We note that in the case of gp32(N) and gp32(C)R4T, more product
is generated
with increased gp32(N) or gp32(C)R4T in DNA run-on assays. This contrasts
results using the
more cooperative gp32 variants. One possibility is that during and following
the strand
exchange reaction gp32 is required to stabilise the recombination and
synthesis intermediates.
This may happen because gp32 cooperativity is so attenuated that it may no
longer participate in
those aspects of the reaction, or because the recombinase out-titrates gp32
and halts the reaction.
The contrasting results of amplification versus run-on assays suggest that the
optimal amount of
cooperativity may be less than that possessed by the most cooperative gp32
variants. Thus for
RPA mutant gp32 variants, with attenuated cooperativity, may be the most
appropriate as these
will permit higher levels of recombinase loading. Attenuation of gp32
cooperativity must,
however, be balanced against noise generated by mispriming events as may be
encountered in
66
CA 3041471 2019-04-26
environments with less gp32 activity.
Finally, there is a substantial difference in the quantity of product
generated in RPA
using 7.5 pg versus 15 1.ig of gp32(C)K3A. The 7.5 pg level more closely
resembles results
with more weakly cooperative mutants. Most likely in some cases during the
course of the
reaction, which was held at 37 C for 2 hours, the amount of single-stranded
run-on product
increases to the point that gp32 no longer effectively saturates single-
stranded DNA in the
reaction. In such a situation two things would occur. First there would be a
rapid increase in the
number of homology-searching filaments leading to a significant rate increase
in invasion. Then
the lack of gp32 to stabilise outgoing strands would lead to many partial
extensions by Klenow
not being stabilised, and possibly a greater rate of bubble migration
separating the new strand
from the template. Hence many more synthesised strands would not achieve full
length.
- A model of gp32 function in multiple invasion/extension reactions
We had earlier noted that in a heterologous system involving recA and gp32(N),
polyethylene glycol was required to permit more than one cycle of invasion and
extension from
a given template. Here we describe a model to rationalise these observations
and explain why
targeting DNA ends multiple times requires specific types of single-stranded
DNA binding
protein. It is clear gp32 activity is required to stabilise the outgoing
strand during the strand
exchange process. If this does not occur then as .the recombinase disassembles
the outgoing
strand re-hybridises with its complementary strand and displaces the invading
oligonucleotide.
Following strand exchange the outgoing strand can exist in one of two states,
which likely place
different demands on the single-stranded DNA binding protein. These two states
arise as a
consequence of the relationship between the single-stranded searching DNA and
the duplex
target DNA. If the recombination event can extend to an end of a linear duplex
DNA, and it is
possible for the outgoing strand to be completely removed from its complement
at one end, then
the outgoing strand is un-constrained at one end. Under this un-constrained
condition the new
duplex involving the incoming DNA strand and its complement is able to rewind
to form B form
DNA. This is necessary because during the pairing reaction the target DNAs are
under wound
by the activity of the invading recombinase filament. Un-constrained outgoing
strands can
67
CA 3041471 2019-04-26
readily bind single-stranded DNA binding proteins, which will prevent branch
migration from
occurring and allowing the invading strand to be removed.
Alternatively if the recombination event does not extend to the end of the
target duplex,
as would occur if an embedded sequence were targeted, then the outgoing strand
is topologically
constrained because it is physically joined to the complementary strand
upstream and
downstream of the recombining region. Such intermediates are highly unstable
because the
newly formed duplex cannot rewind without making the outgoing strand also wind
around it.
And since the outgoing strand is constrained at both ends this is
energetically unstable, and the
new hybrid is placed under considerable strain. Consequently a far higher
demand is placed on
the activity of single-stranded DNA binding proteins in this context because
there is substantial
drive to eject the incoming strand and rewind the original duplex (Figure 6).
In our efforts to establish effective conditions for repeated strand invasion
and extension
of linear DNA targets we have noted that only oligonucleotides targeted to the
ends of linear
sequences are readily extended to full template length, at least when using a
distributive
polymerase such as the Klenow fragment of E. Coli DNA polymerase 1. We have
found that
under certain conditions, for example when using recA with gp32(N) or E. coil
SSB, only one
round of invasion and extension can readily occur on each target template.
These observations
may be understood by considering the outcome of invasion events occurring
under several
different circumstances leading to either fully released outgoing strands
(plectonemic joints) or
topologically constrained intermediates (paranemic joints). Finally, we have
identified certain
other conditions that are permissive for multiple rounds of invasion and
extension from a single
template, a situation ideally suited to the RPA reaction. We propose a model
based on our own
data to consolidate these observations, and forms a framework around which
optimisation of the
RPA reaction can be designed. This model takes into account the behaviour of
gp32 protein
under different reaction environments and justifies gp32 behaviours with the
effects of other
reaction components (Figure 29).
The model schematically describes the nature and outcome of recombination
events
between the end of a target duplex and an invading oligonucleotide that
initially possesses a 5'
68
CA 3041471 2019-04-26
overhang relative to the target. This situation typifies the experimental
circumstance we have
studied. Despite
this starting situation all but the initial cycle involves the invading
oligonucleotides having their 5' extreme flush to the 5' extreme of the target
duplex DNA. This
is the case because the target DNA complementary strand possesses a 3'-end
that can be
extended during the first round to copy the overhang region of the invading
primer, which we
term backfire synthesis. Experiments performed with gp32(N) protein (in the
absence of PEG),
suggest that once the target is flush at the 5'-end to the oligonucleotide
i.e. after the first round,
then subsequent invasion / extension cycles are very inefficient or do not
occur at all. Early
experiments showed only a roughly equimolar quantity of run-on product to
start template. How
does this occur?
We believe that this block to re-invasion / extension arises because invading
oligonucleotides are rarely fully coated by recombinase. Thus complete
exchange to the 5' flush
end of a target will also occur very rarely and corresponding outgoing strands
will remain in a
constrained state (Figure 13). The exchange is initially incomplete and the
intermediate is
unstable due to an inability to allow the new duplex to relax into a B-DNA
helix in the presence
of the topologically constrained outgoing strand. Such unstable intermediates
will have a
tendency to rewind the original duplex and eject the invading strand as the
recombinase
disassembles.
We found, however, that if a Crowding agent such as polyethylene glycol is
included in
the presence of gp32(N), subsequent invasion / extension occurs. One
possibility is that under
these conditions unstable intermediates are temporarily stabilised by gp32(N),
such that
elongation can occur. It is possible that polyethylene glycol acts to partly
rescue the poor
cooperativity of the compromised gp32(N) allowing it to stabilise these
otherwise unstable
intermediates. This conclusion is supported both by our data showing that N-
terminally his-
tagged gp32 is cooperatively attenuated, and the known capacity of crowding
agents to enhance
the effectiveness of interactions between molecules, thus partly making up for
the poorer
interaction between monomers. We initially believed that this experiment might
be best
explained by suggesting that recA filaments were more abundant in the presence
of PEG, as
previously reported elsewhere (Lavery PE, Kowalczykowski SC. J Biol Chem. 1992
May
69
CA 3041471 2019-04-26
5;267(13):9307-14), however we feel that this does not adequately account for
the observed
switch from a dead-end only one round of invasion to productive multiple
rounds of invasion /
extension.
The difference between PEG stimulation of full-length multiple run-ons at end-
directed
targets versus the far poorer activity at truly embedded targets (Figure 15),
suggests that the
temporary stabilisation of an intermediate, as suggested above, is
insufficient alone to generate
substantial elongation. If this were the case for run-on assays at embedded
targets it should
work as well in reinvasion at end-directed targets, but this is not the case
(Figure 15).
Alternatively the difference between efficiencies may be explained by
supposing that gp32
temporarily stabilises an intermediate in which the 5' extent of the incoming
oligonucleotide is
not paired but free, and possibly even coated with gp32 dependent on the
length. Provided that
this unexchanged segment is not coated with gp32, or that the gp32 can readily
dissociate, as
would be the case without cooperativity, then the 5'-most portion of the
oligonucleotide could
become paired via rapid branch migration (Fig 29, scenario 1). In fact there
is a difference
between truly embedded sequences and re-invasion at DNA ends (Figure 15). For
embedded
targets, secondary branch migration leading to unconstrained release of the
outgoing strand can
never occur because the ends are too distant.
We would therefore conclude that the small DNA binding size, and high
cooperativity
between subunits permits gp32 Proteins permits multiple invasion and extension
reactions by
stabilising the constrained paranemic joint structures for a sufficient time.
There are various
possible outcomes (Figure 29). Either the last small section of
oligonucleotide at the 5' end that
was not initially exchanged becomes hybridised through a branch migration
event, or
disassembly of gp32 on the outgoing strand permits the invading / extending
oligonucleotide to
be lost through branch migration. Alternatively, loading of recombinase onto
the outgoing
strand and re-invasion ejects the incoming strand (bubble migration). If
hybridisation via branch
migration occurs then an unconstrained structure arises that can be readily
stabilised and
extended. If either gp32 disassembly or bubble migration occur then there is a
substantial risk
that the new extending strand will be lost before it is fully extended. If a
single stranded DNA
binding protein with a large binding site, like E. coli SSB, or one with poor
cooperativity, like.
CA 3041471 2019-04-26
gp32(N), is used in the absence of PEG, then no temporary stabilisation occurs
and the invading
oligonucleotide is ejected without being extended.
Consequently based on this model, and the earlier conclusions about the
frequency of
recombinase-loaded filaments in the presence of various g32 forms, we conclude
that a balance
between the activities of recombinases and gp32 molecules must be struck that
best meets the
various requirements of the amplification reactions. We can summarise the
needs and effects of
single-stranded DNA binding proteins in different phases of the RPA reaction
as follows.
Phase 1
Single-stranded DNA binding proteins help to prepare single-stranded DNAs for
recombinase loading by melting secondary structure so that recombinase loading
can occur
consistently. Thus the melting activity of single-stranded DNA binding
proteins is desirable and
also plays a part in silencing non-specific annealing of primers. Despite
this, however,
excessive levels of protein and excessive cooperativity can significantly
reduce number of
recombinase-loaded filaments available for invasion.
Phase 2
Single-stranded DNA binding proteins collect the outgoing strand and prevent
spontaneous ejection of the incoming oligonucleotide as the recombinase
disassembles. The
instability of paranemic joints means that invasions occurring on embedded
sequences, including
the case that oligonucleotide ends are flush to the duplex ends (as would
occur during most
cycles of an amplification), means that significant cooperative activity may
be required for many
situations. In general this phase of the reaction will benefit from a surplus
of highly cooperative
single-stranded DNA binding proteins.
Phase 3
Single-stranded DNA binding proteins bind to the displaced strand that forms
during
DNA synthesis. As in phase 2 this displaced strand may be unconstrained, or
topologically
constrained, and these two circumstances place different demands on the single-
stranded DNA
71
CA 3041471 2019-04-26
binding protein.
Phase 4
In certain configuration of the EPA reaction the displaced single-stranded
outgoing
strand must hybridise to a partner oligonucleotide to permit subsequent
generation of a new
duplex. Many single-stranded DNA binding proteins prevent complementary
strands from
annealing, however T4 gp32 protein aids re-annealing of complementary DNA.
Therefore
bacteriophage T4 gp32 protein is an ideal protein for this phase.
Oligonucleotide length:
There is little published evidence to support how effectively recA, or other
recombinases,
might be used with relatively the short synthetic oligonucleotide primers used
for RPA. It is
also unclear whether a stable reaction environment can be generated in which
such short DNA
oligonucleotides remain actively loaded with recombinase. Most studies
performed with recA
utilise comparatively large substrates such as single-stranded and double-
stranded forms of the
bacteriophage M13 genome (many thousands of residues) as donor and acceptor
DNAs.
Experimental assays for recombinase activity often consist of the formation of
intermediate, or
completed, recombination events measured by electrophoretic migration or
electron microscopy
(Harris LD, Griffith J. J Biol Chem. 1987 Jul 5;262(19):9285-92). A few
experiments have been
described using short oligonucleotides. Sequences as short as 15 nucleotides
have been shown
to assemble functional homology-searching complexes with recA in the presence
of the non-
hydrolysable cofactor analogue ATP-y-S, but investigations combining short
oligonucleotides
and ATP are ambiguous (Hsieh P, Camerini-Otero CS, Camerini-Otero RD. Proe
Nati Acad Sci
U S A. 1992 Jul l5;89(14):6492-). The homology-searching function of
recombinases is not
necessarily sufficient to complete strand exchange and to release from the
invasion complex to
allow access by other DNA metabolising proteins such as polymerases. Indeed,
studies have
shown that recA shows transitions between low ATP hydrolysis rate (a useful
indicator of
combined DNA binding activity and functional recombinase activity) and high
hydrolysis rate at
oligonucleotide lengths substantially longer than the 15 nucleotides required
for searching.
Furthermore the type of nucleotide cofactor seems to influence on the length
at which such
72
CA 3041471 2019-04-26
hydrolysis transitions occur (Katz FS, Bryant FR. Biochemistry. 2001 Sep
18;40(37):11082-9).
The bacteriophage 14 recombinase uvsX has also been shown to exhibit variable
properties on
short oligonucleotides, and shows some sensitivity to the base composition
(Formosa T, Alberts
BM. J Biol Chem. 1986 May 5;261(13):6107-18). Despite this, both uvsX and recA
are capable
of performing recombination events with single-stranded substrates of roughly
30 base pairs or
more in the presence of hydrolysable nucleotides like ATP, suggesting that the
use of such short
synthetic targeting oligonucleotides is reasonable (Salinas F, Jiang H,
Kodadek T. J Biol Chem.
1995 Mar 10;270(10):5181-6; Formosa T, Alberts BM. J Biol Chem, 1986 May
5;261(13):6107-18).
An oligonucleotide bearing 33 residues of homology with the end of a
linearized DNA
target can form a pairing intermediate capable of elongation by the Klenow
fragment of E. coil
(Figure 9). This experiment and others demonstrate that homology lengths as
short as 33
nucleotides are sufficient to direct recombinase/ssDNA filaments to
appropriate targets in the
presence of ATP and to permit complete strand exchange. Similar results are
found when using
the bacteriophage 14 uvsX protein.
In addition to a requirement for a minimal oligonucleotide length there may be
a
progressive loss of invasion / extension efficiency if the oligonucleotide is
extended significantly
beyond the minimal length required for recombination, at least when
distributive polymerases
are used. One possibility lies in the nature of recombinase/ssDNA filaments.
Filaments coated
with recombinases have varying 5' limits to their coating, as recombinases
seed randomly and
then extend the filament in a 5'-3' direction, there will be a distribution of
5' extents of coating.
If less than roughly 25-30 nucleotides are coated then little recombination
can occur because
there is insufficient nucleoprotein filament for strand exchange. If more than
this is coated it is
potentially beneficial from the point of view of recombination, but if greater
than 10-20 residues
is added beyond the minimal length required for exchange there is a
possibility that
progressively more active filaments will posses recombinase of a sufficient
length of DNA to
permit exchange, but retain sufficient 5' uncoated DNA for gp32 to bind, which
through
cooperative binding of the 5' extreme could inhibit the branch migration phase
and prevent the
outgoing strand from being un-constrained. Consistent with this notion, we
note that stimulation
73
CA 3041471 2019-04-26
of multiple invasion events was less apparent with the oligonucleotide the
longer oligonucleotide
primer Testerl bio compared to Tester3 bio (Figure 15). The only difference
between these
oligonucleotides is that the first has 25 additional overhanging nucleotides
beyond the initial 33
residues of homology, while the second has only 15 additional residues.
Regardless of the
explanation this experimental observation argues that an optimal maximal
length may exist.
There are other reasons to suspect shorter oligonucleotides will be best for
efficient RPA.
At the relatively low temperatures used in RPA reactions there is a
substantial increase in stable
secondary structures of oligonucleotides as well as a greater probability of
inappropriate
hybridisation between primer pairs. Despite an overall excess of single-
stranded DNA binding
proteins, the dynamic nature of the reaction suggested by the instability of
nucleoprotein
filaments formed with recombinases in the presence of ATP means that there is
likely to be a
constant cycling of proteins on and off oligonucleotides, and a steady-state
concentration of
uncoated, unprotected, oligonucleotides. Consequently the use of short
oligonucleotides should
reduce the likelihood of undesirable intra- and intermolecular interactions.
Our data indicate that optimal length of oligonucleotide lay between 30
nucleotides and
50 nucleotides, and that progressively larger oligonucleotides can decrease
the rate of invasion /
extension. It may, however, be desirable to extend the length of the
oligonucleotide to
accommodate a duplex region in the 3' or 5' region of the searching
oligonucleotide. Thus, in
one aspect of the invention, the preferred primer length is between about 30
to about 50 bases.
Examples of primers sizes that would fit at least one of these criteria
includes primers of
between 30 to 45 bases, between 30 to 40 bases, between 30 to 35 bases,
between 35 to 40
bases, between 40 to 45 bases, and between 45 to 50 bases. While the above-
referenced primer
sizes are preferred, a recombinase and/or single-stranded binding protein with
an optimum
primer length of less than 30 bases is also possible and envisioned.
Oligonucleotide composition, sequence and single-stranded/duplex character:
1) Composition and sequence:
Software to design oligonucleotides for use in vitro DNA synthesis reactions
is well
74
CA 3041471 2019-04-26
established, particularly for use in PCR. The considerations for the RPA
method are similar and
include the optimisation of the melting temperature of the oligonucleotide,
avoidance of hairpin
formation within an oligonucleotide and selection against complementarity with
other
oligonucleotides present in a given reaction. As RPA is performed at
relatively low
temperatures such considerations are potentially more important. We have
observed the
accumulation of extended primer products in RPA reactions, which are
apparently template-
independent and dependent on the combination of primers used. The sizes of the
aberrant
products generated suggest they are primer dimers, or the consequence of self-
priming of a
single oligonucleotide. These undesirable primer artifacts are well known for
other methods
such as PCR. It is therefore important to design oligonucleotide primer pairs
to avoid
undesirable side reactions. We have observed that oligonucleotides capable of
forming hairpins
can erroneously self-prime in an RPA reaction.
Besides optimising oligonucleotide sequence design there are additional
approaches to
reduce or eliminate primer dimer formation. We have observed that reaction
noise can be
significantly reduced by utilising polymerases lacking 3'-5' exonuclease
activity. This suggests
mispriming may result from oligonucleotides that have been shortened by the 3'-
5' exonuclease
activity of polymerases. Consequently 3'-5' exonuclease editing activity,
pyrophosphorylysis, or
any other similar editing activity can be a source of noise. In addition to
using polymerases
lacking exonuclease activity and the removal of pyrophosphate with
pyrophosphatase, use of
synthetic oligonucleotides with a non-hydrolysable backbone at the ultimate
and/or penultimate
link may be beneficial to reduce reaction noise. Alternative backbones could
be selected from
the considerable range of chemistries available such as phosphorothiorate,
morpholino, locked
nucleic acid, or peptide nucleic acid.
2) Single-stranded/duplex character:
Deterring aberrant extension of oligonucleotide 3' ends may also be achieved
by
designing, and including, short competitor oligonucleotides that can
efficiently compete for the
formation of hybrids with the 3' region of the targeting oligonucleotides.
Such an approach
could be configured in various ways.
CA 3041471 2019-04-26
1) A short independent oligonucleotide comprising a perfect complement to the
most 3'
residues of the targeting oligonucleotide could be employed. This
oligonucleotide would be
sufficiently long that at the reaction temperature it would be likely to form
a hybrid with the
targeting oligonucleotide moderately efficiently. This might be between 6 and
15 residues in
length. The short oligonucleotide will be non-extendable by having a blocking
group at its 3'
terminus, such as a dideoxy sugar, or other 3' blocking group. This
oligonucleotide also may
require a non-phosphate linkage at the ultimate and/or penultimate backbone
link to deter
removal of bases in the 3'-5' direction by editing activities. Although a
large proportion of non
protein-coated targeting oligonucleotides may form duplexes with this short
oligonucleotide this
should not significantly decrease the rate and efficiency of the RPA reaction.
Firstly, because at
the low RPA reaction temperature there is likely to be an equilibrium between
hybridised and
unhybridised oligonucleotides there will always be a pool of free melted
targeting
oligonucleotides available. As the oligonucleotide is a better competitor than
other random
sequences in the reaction, it will be favoured against other transient
interactions. Secondly,
single-stranded DNA binding proteins such as recA, uvsX, gp32, and E. coli
SSB, tend to melt
duplex DNA and thus even if hybrids are relatively stable at the reaction
temperature when no
proteins are bound, when they bind to the single-stranded part of the
oligonucleotide and extend
cooperatively to the region of duplex they are likely to enhance its melting,
thus the duplex state
= will tend only to exist on naked oligonucleotides. Finally, recombinases
have the capacity to
extend strand exchange initiated between a single-stranded DNA region and
duplex target into
regions in which both DNAs are duplex. This appears to be an ATP-dependant
branch
migration activity. Taken together these considerations suggest that the short
duplex region
should not significantly reduce the rate of the RPA reaction, but instead act
to suppress the
formation of primer dimer or other artifacts generated from non protein-coated
oligonucleotides
by being a better competitor for binding to the 3' region of the targeting
primer than other
oligonucleotide sequences available in the reaction. If exonuclease deficient
polymerases are
used, it may be optimal to design this oligonucleotide to have its 5' most
base pairing to the
penultimate, rather than ultimate, 3' nucleotide as many polymerases tend to
add an additional
base to a perfectly blunt end.
76
CA 3041471 2019-04-26
, ¨
2) In a second approach, the targeting oligonucleotide possesses a 5' overhang
not
present in the initial target DNA, and this overhang is the precise reverse
complement to the
sequence to the 3' end of the same targeting oligonucleotide, perhaps with the
most 3' base only
unpaired (Figure 35 part D). The length of this complementary oligonucleotide
should be
relatively short, but long enough to be a far better competitor than other
oligonucleotide
sequences present in the reaction. For example between 6 and 10 nucleotides
might be optimal.
As described for the first approach this arrangement is likely to lead to any
uncoated
oligonucleotides forming hairpin structures to themselves far more efficiently
than to any other
sequences. As the design will place the 3' base, or penultimate 3' base, of
the oligonucleotide in
a perfect base-pairing environment, but at the 5' end of the targeting
oligonucleotide, it cannot be
extended, apart from the addition of the single residue often catalysed by
many polymerases if
the end is blunt. In this context it may be fine to leave the editing activity
of polymerases intact.
Such hairpin forming oligonucleotides may suppress erroneous activity of naked
oligonucleotides without deterring the activity of protein-loaded filaments.
There are however some important considerations to taking this approach.
Firstly, if the
recombinase loading is initiated directly on the partly duplex naked
oligonucleotide without
initial melting of the duplex section, recombinase may not extend as close to
the 5' end of the
targeting oligonucleotide as might be optimal. Secondly, as the amplification
reaction continues
beyond the first round there will be active displacement of products generated
from previous
rounds of synthesis and if a complete displacement occurs then the very 3' end
of a displaced
strand, which is complementary to immediately adjacent sequences, may be able
to hairpin and
rapidly self-primer. Such a rapid self-priming event would result in DNA
synthesis and
formation of a novel double-stranded DNA with both strands joined by a hairpin
at one end.
This could be a substrate for further rounds of invasion / synthesis and
result in formation of
dimer-like products, arid possibly more complex products (see figure 35). We
anticipate that
this situation may be perfectly acceptable for diagnostic tests. As the
amplified sequences are all
dependant upon the presence of bona fide target DNA, and will contain the
unique inter-
oligonucleotide sequences, and because the self-priming event may be
engineered to function
efficiently, then this may prove an ideal format for diagnostic assays. We
have already
77
CA 3041471 2019-04-26
experienced the generation of greater than unit length amplified DNA fragment
apparently
generated from specific targets and suspect that this mechanism may operate in
the absence of
specific oligonucleotide design. A similar activity has been described
although in this case the
activity was initiated in a totally different manner using a single very large
single-stranded DNA
(Morrical SW, Wong ML, Alberts BM. J Biol Chem. 1991 Jul 25;266(21):14031-8).
3) In a final approach separate short oligonucleotides with blocked 3'-ends
would be
employed as described in approach I. In this case however a linkage is
engineered between the
5' end of the targeting oligonucleotide and the 5' or 3' end of the short
competitor
oligonucleotide (Figure 35 parts B and C). This approach is similar to
approach 1, except that
by tethering the competitor oligonucleotide in the close vicinity of the
targeting oligonucleotide
one ensures efficient competition of this oligonucleotide with any other
sequences in the
reaction.
Polymerase choice:
There are many DNA polymerases that might be used for RPA. There are, however,
a
number of criteria that should be considered when designing the optimal RPA
format for a given
application. We have identified a number of different polymerases with
activity in RPA
reactions, and deduced which properties confer specific advantages for
different circumstances.
One exciting conclusion is that polymerases from heterologous systems can be
used effectively.
We discuss below the polymerase activities most relevant to RPA.
Polymerase processivity
Polymerase processivity is measured as the typical number of incorporation
events
catalysed on each individual interaction with a DNA template. Many of the
polymerase
enzymes used in molecular biology applications are not highly processive,
often because they
are functional analogues of the of E. coli DNA polymerase I whose primary role
is DNA repair
and Okazaki fragment processing. Processivity is a more critical consideration
for RPA than for
PCR. Because RPA uses a double-stranded template, distributive polymerases
will produce
partially copied strands possessing a joint, which could be ejected by branch
migration. In
addition, we have evidence to suggest that bubble migration may occur in a
variety of RPA
78
CA 3041471 2019-04-26
configurations. In bubble migration parent strands of the template DNA re-
hybridises shortly
behind the replication complex leading to ejection of the newly synthesised
strand, which is
freed as a single-stranded DNA instead of the outgoing strand of the original
recombination
event. This re-hybridisation has been previously described in the 14
recombination/DNA
synthesis system and thought to involve coating of the outgoing strand with
the uvsX
recombinase and subsequent re-invasion. Alternatively, in the presence of
uncooperative single-
stranded DNA binding protein, monomers may be lost progressively from one end
of the
outgoing strand and may lead to progressive branch migration running in a 5'-
3' direction,
chasing the replication complex.
Thus if the polymerase dissociates prematurely from the template, bubble
migration or
branch migration may result in the newly synthesised strand being separated
from the template
as an incomplete single strand. This could be catastrophic for RPA reactions,
because if such
truncated products are too short to form productive hybrids with similar
products generated from
the opposing side, these undesired short products will accumulate linearly.
The T4 single-
stranded DNA binding protein gp32 can alleviate undesirable branch migration.
However RPA
urges, where possible, the use of relatively processive polymerases, or
polymerase complexes, to
efficiently amplify larger DNA fragments. Several now commonly used simple
polymerases
such as Phi-29 DNA polymerase, Bst DNA polymerase and T7 DNA polymerase in
complex
with thioredoxin are known to be processive. However not all of the known
classes of relatively
processive polymerases may possess the combined additional properties suitable
for RPA; Phi-
29 polymerase has not proven able to support geometric RPA amplifications to
date, possibly
due to an inability to efficiently load onto the recombination intermediates.
17 DNA polymerase
lacks sufficient strand displacing activity to be effective. In contrast we
surmised that related Pot
I enzymes from the Bacilli, such as Bacillus subtilis Poll (Bsu), would likely
demonstrate
optimal characteristics by virtue of sharing with Bst polymerase relative
processivity and
exonuclease minus status, but retaining optimal solubility and activity
profiles at temperatures in
the 30-37 C range. Additionally, multi-subunit replication complexes
incorporating sliding
clamps could be used such as that from bacteriophage T4, E. coli, and others,
however
assembling all of these components in effective in vitro reactions that
maintain the excellent
79
CA 3041471 2019-04-26
signal:noise and kinetic properties of current RPA reactions remains
challenging. We purified
the Polymerase I from B. subtilis which is related to Bst polymerase. This
polymerase is readily
overproduced and purified from E. coli with an N-terminal hexhistidine tag,
and appears to
possess ideal biochemical attributes for RPA.
Finally, there are some situations where a distributive polymerase may be more
appropriate for RPA. Because in many configurations DNA synthesis is initiated
from opposing
ends and replication complexes move toward one another, there is the
possibility of a collision
between complexes resulting in a stalemate in which neither progresses
further. This requires
either that one polymerase temporarily dissociates from the template, or that
the polymerases
used are able to pass one another effectively without dissociation. In
consequence of the varying
requirements for an ideal polymerase for RPA we suggest that experimental
evidence will be the
best guide, and to date this implies that Pol I class enzymes from eubacteria,
in particular from
Bacilli, currently show the best properties.
3'-5' exonuclease activity present associated with the DNA polymerase:
Many DNA polymerases possess 3'-5 exonuclease activity, and some also possess
5'-3'
exonuclease activity, which is probably undesirable in RPA as it results in
digestion of one DNA
strand progressively as the polymerase moves forward, rather than
displacement. The 3'-5'
exonuclease has potential advantages as well as its,pbvious disadvantages. On
the one hand 3'-5'
exonuclease activity increases the fidelity of the replication reaction, and
can also prevent
stalling of polymerases at points of misincorporation. High fidelity
amplification is desirable for
many DNA applications. The 3'-5' exonuclease activity may also be appropriate
for
amplification of larger DNA fragments where stalling due to misincorporation
could inhibit
effective amplification.
Despite these clear advantages of 3'-5' exonuclease activity there are some
disadvantages. We have observed that the free oligonucleotides can be subject
to end-dependant
degradation when polymerases possessing 3'-5' exonuclease are employed. This
can be
suppressed to a large extent by using saturating amounts of relatively
cooperative gp32 protein
with some polymerases such as the Klenow fragment, but with enzymes possessing
aggressive
CA 3041471 2019-04-26
exonucleases such as T4 DNA polymerase or Phi-29 DNA polymerase, gp32 appears
to be
insufficient and the oligonucleotides appear to be completely degraded. These
data argue that it
is advantageous to at least limit to some extent the efficacy of an
exonuclease activity in the
reaction.
We have found that 3'-5' exonuclease activity of some polymerases may
contribute
substantially to noise in the reaction. At the relatively low temperatures
used in RPA reactions
there is a significant tendency for uncoated single-stranded DNA molecules to
form
inappropriate hybrids, at low complementarity, with other DNAs in the
reaction. Such hybrids
will prime DNA polymerases elongation. For extension to occur the last base or
two must be
paired correctly with its complement. While poorly complementing segments
within or between
oligonucleotides can form weak hybrids at low temperatures these will rarely
be combined with
a good hybrid match at the very 3' end. Regardless, in the presence of a 3'-5'
exonuclease
activity the unpaired 3' most bases will be excised until a correctly paired
3' end is formed, as
happens normally when an incorrect base is inserted by the polymerase. Our
data suggest that
partly extended strands that have been displaced by branch migration or bubble
migration can
fold back onto themselves leaving an unpaired 3' end, which is trimmed thus
promoting
inappropriate polymerase elongation. There are therefore good reasons to limit
or remove
exonuclease activities from polymerases used in RPA. There are other methods
to inhibit
oligonucleotide degradation that may also be used.
Access to 3' ends
The recombination intermediate formed after invasion must be accessible to the
DNA
polymerase. The structure near the 3'-end of the targeting oligonucleotide, is
not equivalent to
the 3'-end of an oligonucleotide hybridised to otherwise single-stranded DNA,
which is the
situation in PCR. Instead, the outgoing strand, which is hybridised to the
template strand
immediately, or shortly, downstream of the 3'-end of the invading
oligonucleotide may block
polymerase loading. Moreover, whether of the outgoing strand is constrained or
unconstrained
may affect the capacity of certain polymerases to load successfully. Whether a
particular
polymerase can function effectively in these situations must be addressed
experimentally. We
81
CA 3041471 2019-04-26
find that the Klenow fragment of E. coli DNA polymerase I, as well as the Bst
DNA polymerase
purified from Bacillus stearothermophilus, can load onto and extend such
recombination
intermediates. Helicases such as the T4 dda helicase and T4 gp41 helicase may
also function to
process recombination intermediates and separate the template and outgoing
strands downstream
of the exchange event permitting other polymerases to be used, but these
helicases may also
interfere with other aspects of RPA (see below). Finally, it may be beneficial
to use mixtures of
polymerases, acting synergistically in the RPA reaction, for example one
polyrnerase efficient at
accessing the 3' ends of recombination intermediates, and the other possessing
processive,
strand-displacing synthetic activity.
Cooperative component interactions
We have demonstrated that enzymatic components from heterologous systems can
be
combined together effectively in various RPA formats. For example, both the
Klenow fragment
of E. coli DNA polymerase I, the Bst DNA polymerase of Bacillus
stearothermophilus, and the
large fragment of Bacillus subtilis polymerase I,can extend recombination
products generated
with the uvsX recombinase of bacteriophage T4 in the presence of the
bacteriophage 14 gp32
protein. This suggests that the class of polymerases similar to E.coli Poll
are generally effective
in RPA reactions, and this may reflect their distinctive properties as strand
displacing enzymes,
combined with access to recombination intermediates needed for their repair
activities.
Furthermore, that elongation of recombination intermediates can easily be
mediated by
polymerases from widely different structure groups is uncertain. Despite early
indications from
end-directed recombination intermediates stabilized by backfire synthesis, we
have been unable
to drive RPA reactions using Phi-29 polymerase, and this may reflect that this
enzyme cannot
readily load onto recombination intermediates. There may be additional
synergistic effects when
proteins from the same organism are employed together. For example there are
known to be
physical interactions between the bacteriophage T4 components such as uvsY and
uvsX, as well
as gp32. This might be extended to other components, foe example the 14
polymerase
functionally interacts with the gp41 helicase, and physically with gp32
(Formosa and Alberts
PNAS 80, 2442-2446, 1983). However despite the seeming attractiveness of using
components
82
CA 3041471 2019-04-26
from the same organism to enhance RPA efficiency, we have found that one
helicase employed
for processive DNA synthesis by T4 polymerase may not combine easily with the
effective RPA
environment as established here. Early experiments suggested that inclusion of
T4 dda helicase
may disrupt the coating of oligonucleotides in the RPA environment and lead to
excessive
primer noise preventing sensitive RPA reactions. This is consistent with an
earlier published
report [insert reference JBC. 1991 May 25;266(15):9712-8 Inhibition of protein-
mediated
homologous pairing byb a DNA helicase. Kodadek T.
Resolution of replication complex collisions
In some formats of RPA, such as when a large DNA product is desired, a
processive
polymerase would be the optimal choice. Under these conditions, however, there
is significant
likelihood that replication complexes will converge with one another on the
same template.
There is a danger that replication complexes will become locked head-to head
so that neither can
pass. Most useful in this situation are polymerases that are both processive
and able to resolve
collisions as has been demonstrated for Phi29 DNA polymerase (Elias-Arnanz M,
Salas M.
EMBO J. 1997 Sep 15;16(18):5775-83). Alternatively a fine but critical balance
of an ideal level
of processivity to permit useful replicative runs combined with a distributive
capacity to
dissociate fairly frequently may solve this issue.
Recombinases
We have assayed both E. coli recA protein, and bacteriophage 14 uvsX protein
in RPA
experiments. Both these proteins share some limited protein sequence homology
and are
believed to have evolved from a common progenitor. Crystallographic and
electron microscope
studies of nucleoprotein filaments of these proteins, which show a conserved
filament structure,
in terms of the pitch of the helices formed in both ATP and ADP bound states,
suggest a
remarkable similarity in their mechanism of action. Furthermore, all
prokaryotes possess
proteins highly homologous to recA, which suggests that the principle
activities of recombinases
have been conserved throughout evolution. Hence, what is learned from one
recombinase may
be applied to, or substituted by, another.
83
CA 3041471 2019-04-26
In addition to their similarities, however, there are differences between recA
and uvsX
relevant to RPA and as additional components of recombination/replication
machinery are used
in RPA reactions organism-specific protein-protein interactions may have a
significant effect on
reaction efficiency. The nucleotide hydrolysis rate of uvsX is 10-20 times
higher than that of
recA, suggesting that it might perform recombination reactions at an
accelerated rate (Formosa
T, Alberts BM. J Biol Chem. 1986 May 5;261(13):6107-18.). An increased
hydrolysis rate
could be beneficial for RPA reactions in several ways.
1) More dynamic turnover of uvsX on oligonucleotides could increase the
overall
regeneration of nucleoprotein filaments leading to near complete recombinase
coverage to the
most 5' end of invading oligonucleotides.
2) More rapid completion and disassembly of recombinase from successful
recombination events will permit more efficient polymerase access
3) A more active recombinase will produce a more flexible nucleoprotein
filament.
Another major difference between uvsX and recA is that uvsX hydrolyses ATP to
ADP
plus phosphate, and to AMP plus pyrophosphate whereas recA and other
recombinases do not
(Formosa T, Alberts BM. I Biol Chem. 1986 May 5;261(13):6107-18.). The
biological
significance of this difference is not known but the activity might affect RPA
efficiency. For
instance, the pitch of nucleoprotein filaments formed with ATP and ADP is
different and the
hydrolysis of ATP to ADP is associated with overall filament flexibility. It
may be that AMP-
bound uvsX can adopt a different pitch and possess a distinct flexibility.
The bacteriophage T4 recombinase uvsX stimulates a mode of DNA displacement
following synthesis known as bubble migration. In bubble migration uvsX
assembles onto the
outgoing strand and mediates a re-invasion of the outgoing strand thus
displacing the newly
synthesised strand. This mode may have biological significance because by
displacing the
newly synthesised strand, the length of region with topological constraint is
limited. This
process has not been described for recA, although it may occur. Nevertheless
several aspects of
bubble migration suggest real differences between uvsX and recA. For example,
the bubble
migration model suggests that it is possible that uvsX bound DNA extends to
the end of the
84
CA 3041471 2019-04-26
invading or partially extended DNA, but that this structure is still
accessible by polymerases for
elongation (Formosa T, Alberts BM. Cell. 1986 Dec 5;47(5):793-806). This has
certainly not
been observed for recA, and if anything there is evidence that may be to the
contrary (Xu L,
Mariam KJ. J Biol Chem. 2002 Apr 19;277(16):14321-8). It is unclear how
similar uvsX and
recA filaments are in their respective abilities to promote polymerase loading
at 3' ends with or
without recombinase dissociation. We have found differences between uvsX(C)
and recA(C)
consistent with the notion that their differences have can affect RPA
efficiency. Contrary to our
findings with recA, uvsX can mediate multiple invasions to an end structure
target even when N-
terminally tagged gp32 is used in the absence of polyethylene glycol (Figure
23). Thus uvsX
may be more optimal for use in RPA.
The bacteriophage T4 uvsX recombinase has a well-characterised interaction
with its
partner loading protein uvsY. Despite reports suggesting that E. coil rec0 and
recR may be
functional analogues of uvsY we have not observed significant improvement for
RPA reactions.
This may be due to problems with our protein preparations, or due to the use
of the heterologous
gp32 rather than E. coli SSB.
Finally uvsX is likely to behave better with gp32, which we find to be an
optimal single-
stranded DNA binding protein, because they have evolved to function in concert
and may have
relevant interactions. Indeed uvsX and other components of the
bacteriophage T4
recombination dependant replication machinery, such as the dda helicase, have
known protein-
protein interactions that may useful for establishing an optimal RPA reaction
(Hacker KJ,
Alberts BM. 3 Biol Chem. 1992 Oct 15;267(29):20674-81).
Despite the apparent advantages of uvsX for RPA, there are features of E. coli
recA that
may be useful. It has been reported that recA nucleoprotein filaments are more
stable, which
could be of utility in some circumstances. The lower ATP hydrolysis rate
places less strain on
establishing a durable ATP regeneration system, and the lack of generation of
AMP and
pyrophosphate obviates the need to regenerate and mop-up these side-products.
It may also be
the case that other recA homologues possess activities that are optimal or
RPA.
CA 3041471 2019-04-26
Establishment of a dynamic recombination system:
To make RPA robust, it is critical to configure the reaction to provide a
sufficient
number of active, coated, homology-searching recombinase/DNA filaments. In
addition,
following completion of homology-searching, filaments must efficiently
disassemble, or be
otherwise processed, to permit loading DNA polymerase and other components. It
is also
essential that a sufficient quantity of single-stranded DNA binding protein be
present both to
facilitate oligonucleotide melting and to collect displaced outgoing strands.
And finally, robust
RPA requires will require processive strand-displacing DNA synthesis.
Underlying these
requirements is a competition between two the recombinase and the single-
stranded DNA
binding protein.
It is widely known that of recombinase-loaded DNA filaments are unstable in
the
presence of single-stranded DNA binding proteins. Coupled to the finding that
nucleotide
cofactor hydrolysis is not strictly required for homology searching, led to
the use of non-
hydrolysable nucleotide analogues of nucleotides such as ATP-y-S, to load recA
onto filaments
and produce stable homology-searching complexes. A recA-mediated amplification
method
using ATP-y-S has been described (Zarling, et al.), however, has not been
widely used. We
previously identified a flaw in the method, which we can now observe in our
experimental
results. The Zarling, et al. method probably fails because recombinase-loaded
filaments needs to
be dynamic and capable of disassembly as well as other ATP-hydrolysis
dependant events to
complete strand exchange and permit loading of DNA polymerase other components
to the 3'
end of the invading oligonucleotide. The use of ATP-y-S, as well as other
modifications such as
removing acidic sequences from the C terminus of recombinases, leads to a
constant general
high affinity for DNA that likely prevents strand exchange and dissociation
from the invasion
complex. Thus ATP-y-S -loaded recA filaments become effectively locked onto
the target site
in the recombination event for an abnormally long time. Consequently non-
hydrolysable
nucleotide analogues are not generally permissive for recombinase-mediated
replication and
amplification. Instead,
ATP, or other hydrolysable nucleotides capable of supporting
recombinase loading, must be employed. In ATP the recombinases are constantly
associating
with and dissociating from oligonucleotide and are in competition with single-
stranded DNA
86
CA 3041471 2019-04-26
binding proteins. We have addressed the problem this competition poses in two
general ways;
first by including a recombinase loading protein specific for uvsX, the uvsY
protein, and
secondly by modulating the cooperative behaviour of gp32, and the recombinases
uvsX and
recA, by mutation and/or inclusion of crowding agents. It is possible however
that limited
quantities of non-hydrolysable analogues such as ATP-y-S, or of non-
phosphorylatable
analogues such as ADP-13-S, may be included to modulate the global
loading/unloading activity
of the recombinase.
Additional reaction components
= A number of specific reaction components have a significant influence on
RPA reaction
efficacy.
Polyethylene glycol
Polyethylene glycols (PEGs) have a profound effect on recombination/DNA
synthesis.
Firstly, we find that PEGs influence the number of multiple invasion/extension
cycles that occur,
for example when recA is combined with gp32(N). We have also found that PEGs
stimulate
amplification reactions configured in several different ways (Figure 15,
Example 3). We also
know that in some configurations PEGs alter the length distribution of
products formed (Figure
28). In summary polyethylene glycols, and presumably other similar crowding
agents may
affect the 'Ciioperativity of gp32 and recombinases, affect polymerase
processivity and affect the
hybridisation rate and behaviour of oligonucleotides in solution. They may
imbue cell-like
environments by phase partitioning the reactants, and/or causing the
development of fractal-like
kinetics. Furthermore the chain length of the polyethylene glycol appears to
be critical. We find,
that of those tested, PEGs of average molecular weight 1450 and 15-20,000 (PEG
'compound')
produce the best results. The PEGs in aiding gp32 function, particularly gp32
variants with
attenuated cooperativity, has been detailed above. PEG is also likely to
increase the stability of
recombinase-loaded filaments and the increased persistence may increase RPA
efficacy. Most
importantly we have been completely unable to establish amplification
conditions capable of
amplifying from trace amounts of sample to detectable levels without employing
polyethylene
glycols (poorly with PEG 1450, very efficiently with PEG compound). Presumably
other agents
87
CA 3041471 2019-04-26
may ' also stimulate RPA reactions, however this effect was not evident in
preliminary
experiments with polyvinyl alcohol. Thus specific features of polyethylene
glycols, and more
particularly specific variants of PEGs, may be essential.
In what manner PEG compound (carbowax 20M) achieves such a significant
stimulatory
effect is mysterious. Kinetic models of the manner in which volume exclusion
may affect
reaction rates would place significant emphasis on the molecular weights of
the substrates and
enzymes under study in comparison to the crowding component. However the next-
most
effective agent that we tested, PEG 1450, was also the smallest and
stimulation by polyethylene
glycols intermediate between PEG1450 and PEG compound were less effective.
Consequently
we suspect that properties additional to simply average chain length are
critical to the effects of
these agents. For example specific phase transitions may occur at variant
temperatures for
different PEGs. It is also known that the alcohol groups of PEGs can lead to
additional
interactions, such as with proteins, or influencing the ionic environment. We
note (data not
shown) that the addition of 5% PEG compound (while developing a pH below 7.0
during
storage at room temperature) to Iris-buffered solutions used in RPA causes a
sharp rise in pH.
As a consequence there is currently no formulaic manner to determine which
volume-
excluding agents will have the corrent properties to stimulate high-level
dynamic recombination
environments, and RPA in particular, except through experimental validation.
ATP regeneration system components
An ATP regeneration system is crucial to permit persistent recombination
reactions as
recombinases have an extremely high rate of ATP hydrolysis when bound to
nucleic acids. In
particular, the uvsX protein has a hydrolysis rate 10-20 times higher than
recA and can consume
200 molecules of ATP per minute per monomer. A number of systems are available
and we
have routinely used the creatine kinase/phosphocreatine system. When uvsX is
employed the
AMP that is produced must be converted into ATP. We have used chicken
myokinase, which
converts a molecule of AMP and one of ATP to two molecules of ADP. ADP is then
converted
to ATP using the creatine kinase/phosphocreatine system. Poor regeneration of
ATP will reduce
the reaction rate and likely stop the reaction before detectable levels of
product have
88
CA 3041471 2019-04-26
accumulated.
Pyrophosphatase
Pyrophosphate (PPi) accumulates during DNA synthesis and when uvsX is
employed, as
it hydryolyses ATP to AMP + PPi. PPi accumulation in the reaction can have
several
detrimental consequences. Significant pyrophosphate accumulation will permit
an unacceptable
rate of pyrophosphorylsis, whereby the synthetic reaction of a polymerase is
driven into reverse
and removes the 3'-most nucleotides from duplex templates. This could lead to
unacceptable
levels of primer noise, or enhanced levels of undesired self-priming of
outgoing strands, because
editing activities of the polymerase tend to trim 3' -ends back until a
suitable duplex region is
revealed to permit rapid elongation. Additionally pyrophosphatase accumulation
will lead to
inhibition of the recombinase, and to a slowing of the polymerase synthetic
reaction.
Improved signal:noise by design of RPA reactions
RPA displays high of sensitivity and specificity. However samples containing
no target
at all often generate products derived only from the primers. Such phenomena
are not
uncommon in other methods. For example the PCR method will ultimately generate
non-
specific products such as so-called 'primer dimers'. RPA is somewhat prone to
such primer-
related artifacts because there is no cycle control allowing small amplicons
to achieve many
doublings within the period of the incubation. Electrophoresis of reaction
products permits easy
separation of signal and noise, however in simple non-laboratory diagnostic
products other
approaches must be taken. Here we disclose approaches to make such non-gel
diagnostic tests
function with adequate signal to noise, and to easily carry out detection. We
also disclose the
composition of a reaction lyophilizate that can be stored for many days at
ambient temperatures,
a prerequisite for easy non-laboratory use.
Signal to noise ratios in the RPA system
DNA amplification systems generally contend with the fact that amplification
of DNA
may occur which does not initiate from the bona fide target. This 'noise',
particularly apparent
under low target conditions, may impose restrictions on the permitted
sensitivity, and methods to
89
CA 3041471 2019-04-26
assess amplification. While initial configurations of RPA offer exquisite
sensitivity we believe
novel properties of the recombinase-based system lend themselves to previously
untapped
approaches to improve specificity, and to developing novel product detection
schema.
RPA amplification reactions, like PCR, are generally established by combining
two
oligonucleotides, which flank the desired target DNA. Doubling events occur
and, as in
PCR, exponential DNA amplification ensues, permitting as much as 1012-fold
amplification
for fragments of -450-400 bp.
When primers of 30-35 residues and acceptable to standard design algorithms
are tested,
a significant proportion amplify target with high sensitivity and specificity
(Fig. 41). For
example, such 'good' primers can successfully amplify targets from starting
target
concentrations of 2-3 copies per microliter to generate significant levels of
the correct amplicon.
However below this copy density even good primers tend to generate noise
(smear, laddering,
and/or well staining), as assayed by gel electrophoresis of all reaction
products (see Figure 41),
these products apparently of purely primer origin (although contaminating
E.coli DNA is likely
present and could in theory be specifying highly inefficient spurious
amplicons).
In some cases target DNA may not be particularly rare, for example if one
wishes to
determine a limited SNP profile of a human individual and a modest blood
sample can easily be
obtained. On the other hand, some tests for pathogens may need to detect just
a few copies to be
of great practical utility, and in particular distinguish this situation
clearly from complete
absence of target.
Our efforts to determine the level and nature of noise generated in very low,
or no, copy
number experiments, as well as other experiments investigating primer length
and composition,
has revealed a number of key observations, which we believe will aid the
development of ideal
highly sensitive portable diagnostic systems using RPA.
Firstly we have found that oligonucleotides of less than approximately 30
residues are
less effective at RPA, and notably become less 'noisy', generating no visible
noise at all when
reduced to 25-26 (see Figures 45 and 52). Nevertheless shorter
oligonucleotides are still capable
of, at least, hybridization based elongation (see Fig. 52), and their
'activity' in (recombinase-
CA 3041471 2019-04-26
mediated) RPA events is probably not zero, instead decreasing progressively
with shortening
(Fig. 45). This permits some degree of activity/noise tuning.
Secondly, we note that primers which are identical except for additional small
number of
residues at the 5' extent show significant variability in noisiness (even when
all are >30
residues), implying that the exact nature of the 5'-most base(s) plays a
significant role in the
mechanism and likelihood of noise (see Figs 42,45 and 47).
Thirdly, primers possessing a locked nucleic acid (LNA) sugar at the 3'-most
end
reduce/eliminate noise as determined by gel electrophoresis. They also reduce
the quantity of
product slightly if only one is employed, and significantly if both are used
at 'standard'
polymerase concentrations, but remain fully active if polymerase concentration
is increased, and
under the elevated polymerase concentrations noise still appears suppressed.
Finally low-target concentration noise appears to derive mostly/solely from
primers, not
sample DNA, and we thus refer to this noise as 'primer gymnastics'. This is
clear from
experiments in which sample DNA is decreased to zero. Consequently it is
possible to focus
attention on the limited mechanisms by which primers may interact with and
between
themselves to find solutions to this problem. How noise may arise is discussed
in detail in the
following section. An important conclusion of this analysis is that lowering
nucleoprotein
filament recombination/priming activity may impinge more severely on primer
noise generation
than bona fide amplicon generation.
Analysis of how primer noise may originate
Unlike PCR, or other methods involving conventional hybridization, RPA's
reliance on
the formation of extended nucleoprotein filaments undergoing ATP hydrolysis as
active
homology-searching complexes likely impinges on the parameters determining
oligonucleotide
activity in ways previously unencountered for in vitro amplification
reactions. We suggest that
the rapid drop in oligonucleotide activity below 30 residues described above
reflects
recombination rate slowing, and that most likely this parallels observations
made elsewhere on
the length-dependance of ATP hydrolysis for the E.coli recA recombinase
(Bianco PR and
Weinstock GM). In particular the length and composition of short sequences was
shown to
91
CA 3041471 2019-04-26
influence ATP hydrolysis. Roughly 30 residues or more are required for maximal
ATP
hydrolysis (moles ATP per mole DNA-bound recA) when recA is bound to synthetic
oligonucleotides, shorter sequences displaying marked drops in hydrolysis rate
until it was
absent by about 15 residues. We connect the observation of low amplification
behaviour with
previous data on ATP hydrolysis because there is a clear rationale to think
that nucleoprotein
recombination activity will be influenced by ATP hydrolysis rate. In a sense
the slowest
oligonucleotides we have observed are those bound with unhydrolyzable ATP-y-S
in which the
rate of completed exchange (formation of resolved plectonemic joints) is near
zero (Riddles PW
and Lehman IR) We have also proven that ATP-y-S effectively poisons RPA
reactions
(Piepenburg et al.). We suggest that high rates of ATP hydrolysis in a
nucleoprotein filament
underpins the activity of 'fast' primers, and low rates 'slow' primers, who
differ principally by
the average number of complete recombination events completed in a unit time.
Interpreted
simply, ATP-hydrolysis is required for dynamic activity of filaments.
Oligonucleotides of less
than 30 residues become more stable and undynamic, eventually resembling the
locked state
generated when ATP-y-S is employed, and are unable to complete exchange
('slow'
oligonucleotides may locate sequences as readily but be unable to disssassmble
readily to permit
completion and polymerase access).
A possible difference in dependence on nucleoprotein filament activity for
bona fide
product and noise
To understand why there might be any advantage to selecting less active
'slower'
oligonucleotides we need to explore how primer 'gymnastic' noise might arise.
Why might
shortened oligonucleotide reduce noise faster than the specific signal? Unlike
the geometric
phase of amplification seen for both bona fide targets and noise products, the
early formation
phase of noisy targets from starting primers is probably more complex, and is
presumably rather
slow. In Fig. 43 we have detailed 2 general mechanisms by which individual
primers can
become converted to structures with the capacity for geometric amplification.
Both strategies
diagrammed illustrate the formation of amp licons from a single primer
species. In both cases the
first step involves self-priming of the oligonucleotide by formation of a
short hairpin at the 3'-
92
CA 3041471 2019-04-26
end. In strategy I the resulting hairpin will not easily amplify in geometric
phase because of
significant mismatches. Instead to become geometric phase amplicons they would
need to
proceed through a step more akin to that described in strategy 2. In this case
the first self-
priming event is followed by a second involving the reverse complement to the
5' end of the
original oligonucleotide. There are several simple remarks from such modeling.
First is that 2
types of event have to occur; there must be elongation from the 3'-end of an
oligonucleotide,
and then a second similar 3' hairpin and extension which involves the reverse
complement to the
5'-end of the original oligo. Note that the very rare and unstable nature of
the first events
preceding geometric phase are going to be most strongly inhibited if most
oligos are coated with
either an SSB or recombinase, which have DNA melting activites when bound to
ssDNAs.
Conversely dynamic recombinase filaments which disassemble spontaneously
create time
windows in which all, or parts, of an oligonucleotide are uncoated. This very
rate limiting step
will therefore likely be sensitive to primer ATP hydrolysis rate, dynamicity,
and by
extrapolation oligonucleotide length. Consequently shortening oligonucleotides
may be
particularly effective and stemming noise, sufficiently so to justify slowing
bona fide target
amplification rate in order to achieve it.
These final geometric phase amplicons are inverted repeats. This means that an
invasion/elongation originating from one side will displace a strand that
immediately folds back
on itself to form a long hairpin. This molecule must be targeted by
recombinase action in order
to be converted back to one similar to the parent, while bona fide targets can
employ solution
hybridization additionally. This imposes additional necessity for the primer
artifacts on
recombinase-based invasion to achieve geometric amplification, and
consequently they will
probably be suppressed more rapidly by lower recombination rate. Further the
presence of
internal repeats means that even larger and more diverse versions of these
amplicons are likely
to arise quickly, and this is consistent with the common phenotype of a smear,
or ladder, running
most of the way up a gel.
A third remark is that the time taken to double a given species of DNA during
exponential amplification will be a compound of recombination rate and time
taken to synthesise
down the template by polymerase. As primer noise products (at least initially)
are very small, the
93
CA 3041471 2019-04-26
time taken for DNA synthesis is very low and so the principle rate limitation
may be the
recombination rate. Thus decreasing nucleoprotein recombination rate will,
while slowing
amplification in general, impinge more heavily on short amplicons than long
ones, thus
generally benefiting bona fide amplicons.
We draw on the above observations to devise ways to combine primers of
different
length, composition and concentration, to give highly sensitive and specific
reactions.
Primary oligonucleotide design:
The four observations described above lend themselves to some straightforward
oligo
design principals.
Oligonucleotide length
Optimal primer length may be around 30 residues and in some cases a few
residues
shorter will yield an excellent signal-to-noise ratio. As the average primer
length is decreased,
the noise generation decreases and seems absent altogether when 26-28 mers are
employed
(Figure 45). Although the amplification of the target also decreases, we
suspect that noise
decreases more quickly than signal. In the previous analysis of how noise may
arise, we
conclude there are several reasons why primer noise might be more affected by
decreased
recombination rate than correct amplicons.
5'-most sequence optimization
By comparing the activity of oligonucleotides possessing identical sequences
except for
the extent of the 5'-most residues we have noted that significant variation in
oligonucleotide
noisiness is observed (see Figs 42, 45, 47). One possibility is that this
purely reflects the effects
of length variation on nucleoprotein activity as described above. This may not
be, however, the
only reason if primers being compared are all 30 residues or longer. For
example in Figure 42,
the oligonucleotides NEST-1 and J1 are identical except that 31 contains
several extra 5'
residues. Instead of being noisier, this oligonucleotide is less noisy, while
amplifying very well.
Based on this and other cases, we propose two other likely mechanisms. First,
that an
oligonucleotide to become exponentially amplifiable may require a priming
event by the reverse
94
CA 3041471 2019-04-26
complement of the 5'-most sequences, this event critically depends on the
exact sequence of
bases at the 5'-end. This means that varying the 5'-most bases may lead to
significant variation
in noise generation. Second, that the base composition of even 5' sequences,
even when the
oligonucleotide is great than 30 bases, may affect the recombination activity
of primers.
Evidence to support this comes from an experiment in which 5'tags consisting
of
homopolymeric stretches were added to primers, and the consequences observed.
Figure 47
shows an experiment in which a run of C residues, or G residues, were appended
to the
oligonucleotides ii and K2, and subsequently amplification reaction were
established containing
all possible modified and unmodified oligonucleotides alone, or in the
possible combinations.
Surprisingly these oligonucleotides, already determined to be very sensitive
and specific, were
not improved by adding these particular homopolymeric stretches. Instead it
was observed that
if a run of Cytosines was added to the 5' end this lead to increased
production of non-specific
products when incubated alone, while a stretch of Guanosines had the reverse
effect, making the
oligonucleotides very quiet when left alone. Also, apart from the combination
of the original
parent unmodified oligonucleotides, the only pair to apparently successfully
generate a correct
product was the combination of the 2 Cytosine-tailed oligonucleotides (i.e the
most noisy
oligonucleotides). One explanation that can justify these observations is that
a stretch of
Cytosines at the 5' end makes the consequent nucleoprotein filament more
active, while
Guanosines make it less so. In summary oligonucleotide composition, like
length, may affect
rate behavior, and additional and unrelated sequences at the 5' end can have a
significant
influence in this respect. Optimal primers may be found by a rational approach
to 5' sequence
design. Several different 5'-most bases might be tried, and the best selected
for employment.
Additionally it is likely that tags can be placed at the 5' end whose sequence
is unrelated to the
target, but confer resistance to snapback hairpin priming from the reverse
complement.
Determination of optimal sequences that work on many oligonucleotides may be
possible.
Measurement of ATP hydrolysis rate of oligonucleotides in the presence of
reaction components
may also prove of utility in designing optimal primer pairs, and permit one to
create primer
mixes which all share identical amplification rate behavior. Like length,
these observations and
improvements would not be obvious in other systems because they depend on
specific features
CA 3041471 2019-04-26
of a recombinase-driven amplification system and the biochemistry that
underlies it.
Locked nucleic acid, ribose, or other sugar modifications
Locked nucleic acids (LNAs) are nucleotides where a linkage has been
engineered
between the 2' and 4' Carbons of the sugar ring of ribose. Oligonucleotides
containing such
sugars, most notably at the very 3' position, have been successfully used in
PCR assays, and
shown to significantly improve the discrimination of correct 3' base pairing
(see Vester B. and
Wengel J.). We anticipated that LNA residues might be tolerated by the RPA
system providing
that they were not present at too many position within the oligonucleotides.
As they are known
to increase specificity in PCR amplifications they might well improve signal-
noise in the RPA
system.
Earlier experiments employing phosphorothiorate linkages at the most 3'
positions
proved unable to function properly in RPA, and were notably more noisy (data
not shown). We
interpreted this as a reflection that both uvsX and gp32, which interact with
the sugar-phosphate
backbone, would not bind these backbones leading to seriously anomalous
reaction behavior.
However gp32 binding is less dependant on the sugar as evidenced by its
ability to bind to RNA
as well as DNA (albeit with altered less cooperativity) (Kowakzykowski SC, et
al.). Thus we
anticipated that LNA sugars, which retain a normal phosphate group might
remain functional
with gp32.
We have performed experiments in which oligonucleotides possessing a locked
nucleic
acid sugar at the most 3' position have been used in amplification reactions.
Comparison has
been made between reactions with identical oligonucleotides with and without
the sugar
modification, and reactions involving one modified and one unmodified
oligonucleotide. Based
on experiment it appears RPA can amplify DNA when one or both oligonucleotides
possess a
locked nucleic acid residue at the 3' position. Notably the presence of a LNA
nucleotide at the 3'
end significantly reduced product accumulation under 'standard' polymerase
concentration, and
apparently eliminated associated noise. Very low noise, and reduced product,
may arise because
of lower successful recombination frequency, reduced polymerase takeoff, or a
combination.
Whatever the reason it is reasonable based on the findings that using locked
nucleic acids in one
96
CA 3041471 2019-04-26
or both primers will significantly reduce or eliminate noise, while retaining
an acceptable
amplification rate for bona fide product. We found that product levels can be
returned to high
levels if the polym erase concentration is increased several-fold (from about
30-40ng/p.1 to 150
ng/p1), but that noise levels do not rise at an equivalent rate.
Similarly other sugar-modified residues may display improved signal to noise
behaviour
when present at some frequency in oligonucleotides. For example ribose, 2%0-
methyl
modifications, acyclic sugars, or otherwise may prove of utility.
Approaches that improve signal to noise by combining nucleoprotein filaments
with
differential activities
The ability to control the rate of recombination and/or elongation from
primers on the
basis of their length, base composition and distribution, and use of modified
sugars (or possibly
other backbone modifications) suggests mechanisms to design amplification
reactions with
optimal signal to noise properties. There are two general ways in which we
envision
oligonucleotide combination strategies to be employed to benefit. First are
strategies in which
single tube nesting occurs, and second are the employment of a non-noisy
oligonucleotide paired
with a more conventional oligonucleotide in such a way that bona fide
amplicons are readily
formed, but noisy amplicons involve only single oligonucleotides.
Single tube nesting
Nesting is the process by which improved sensitivity and signal to noise has
been
enabled by carrying out a first amplification with an outer pair of primers,
and then perform a
second amplification with an inner pair of primers. In its most familiar
context this approach has
been employed in conjunction with the PCR method. In principal the first round
amplifies the
target more efficiently than random DNA, such that after the first
amplification even if it was
insufficiently clean to give a clear amplicon against background, it still is
so relatively enriched
that a second amplification with primers internal to for the first will
readily give a very clear and
clean result. In practice this normally relies on performing one
amplification, then removing a
small portion to a new tube, and performing a second amplification with
internal 'nested'
primers.
97
CA 3041471 2019-04-26
While the concept of nesting is visited elsewhere our results suggest an
attractive
alternative nesting approach in which all the participating primers are
combined in a single
reaction. The rationale being that the inner and outer oligonucleotide pair
are differentially
active in recombination and/or elongation rate, and be present at different
concentration. For
example the outer pair would be configured to have a fast
recombination/elongation rate, and the
inner pair a slower recombination/elongation rate. Also the outer pair would
be present at lower
concentration (but still sufficiently high concentration for acceptable
activity) and the inner pair
would be present at high concentration. What would then happen? In isolation
the inner pair
would amplify cleanly but rather slowly so that they might not achieve an
acceptable degree of
amplification within a desirable timeframe. In contrast, in isolation the
outer pair would amplify
quickly, albeit with some primer noise, but would stop at a rather low
concentration due to
exhaustion. In combination however these two behaviours would potentially
complement nicely.
In practice outer primer pairs would generally be fast, over 30 residues in
length, unmodified at
the 3'-end, and possibly containing additional 5' residues that promote
'fastness'. Inner primers
would be slower, as potentially conferred by making them short, modifying the
3' end with LNA
or otherwise, adding slowness sequences at the 5' end, and so on.
Non-noisy/noisy nucleoprotein pairs
Experimental data that we have obtained suggests that much of the primer-
derived
smears or ladders contain sequences from one primer species only (see Fig.
46). If true this fact
alone suggests a simple approach to distinguishing signal from noise merely by
determining
whether or not the two primers used in the reaction become physically
associated in products.
Here we suggest that this can be further improved by combining very quiet
oligonucleotides
with faster (noisier) oligonucleotides. Quiet oligonucleotides, for example
short oligonucleotide,
may still function efficiently if present with noisier ones providing that
they can function in
hybridization reactions (see Fig.52). However if they only engage in
amplification via normal
hybridization whose kinetics are very different to lecombinase-promoted noise
amplification
they may remain uninvolved in the noise that the faster oligonucleotide
engages in alone.
Third primer and detection protocols
98
CA 3041471 2019-04-26
The possibility that primers might principally indulge in gymnastics alone, as
mentioned
above, suggests simple approaches to determining whether or not amplification
has occurred
efficiently. One simple non-gel detection format would employ one
oligonucleotide labeled with
a dye or enzyme, or other easily detectable group, and the other labeled with
an immobilizable
group e.g. biotin (see Fig.50 Part 1). Thus before, during, or after the main
reaction phase the
immobilizable oligonucleotide would be immobilized to a surface, and at
reaction end this
surface would be washed with an appropriate buffer. If the other labeled
primer co-associates
with the immobilizable primer, which can be easily determined, then
amplification of the
product has occurred.
Beyond this we imagine additional ways in which additional levels of
stringency on bona
fide product amplification might be generated. A simple additional approach
involves a 'third'
primer which functions to capture onto a surface liquid phase amplicons. This
primer would be
deterred from generating noise itself either by being engineered as quiet
(e.g. a short oligo as
described above), or by being added to the system only at a late phase of the
reaction. This third
primer might target novel internal sequences, but also could immobilize the
targets via 'backfire'
synthesis described elsewhere to avoid disruptive branch migration (Fig.50,
Parts 2 and 3).
Lyophilization of the RPA reaction
In order to configure non-laboratory, or near-patient, diagnostic and forensic
tests it will
be necessary to provide ambient-stable reagents. One obvious way to achieve
this is by
lyophilizing the reactions, omitting only the sample DNA, and possibly any
other component
which is stable separately and can be added with the sample such as buffer
used to dissolve the
sample nucleic acid.
While lyophilization is a well-established process there is no guarantee that
all
components of a reaction system will successfully by co-lyophilized and
reconstituted under the
same conditions. We have attempted to lyophilize RPA reactions with and
without various of the
final reaction components. Fig. 53 shows that we have successfully managed to
lyophilize RPA
reactions containing everything except the sample DNA and some buffer
component (which can
be stored stably at ambient temperatures and used to reconstitute sample DNA).
The
99
CA 3041471 2019-04-26
disaccharide sugar trehalose proves in these experiments to be required to
stabilize the
lyophilizate, permitting room temperature storage for at least 10 days.
Real-time kinetic analysis of RPA reactions
The ability to perform kinetic analysis of DNA amplification reactions
provides
enormous utility compared with purely end-point analysis of similar reactions.
For one thing
this allows the determination of the number of DNA, or RNA, copies present in
a sample and
has many uses in research, clinical, and environmental testing applications.
In addition to
straightforward quantification applications, dynamic reaction product
detection offer excellent
solutions to other problems, such as permitting the discrimination of single
nucleotide
polymorphic alleles by virtue of altered accumulation kinetics, and a
mechanism to assess
presence versus absence of targets in a non gel-based format.
RPA offers an excellent alternative to PCR to amplify specific DNA targets,
but by
obviating thermal cycling, RPA requires less expensive instrumentation and is
easier to
implement in non-laboratory settings.
Currently real-time analysis of DNA accumulation is most widely employed in
combination with the PCR. PCR shows exponential amplification of DNA up to
several cycles
after reaching a product detection threshold and can be implemented using
relatively
.inexpensive optics when various fluorescence-based sensors are used.
Consequently assessing
the cycle number a given sample crosses the detectability threshold, in
combination with
equivalent results from control samples, permits assignment of the number of
copies present in
anonymous samples. A number of sensor approaches have been described, and many
are
currently employed. Fluorescent minor groove binding dyes can be employed
which develop
strong fluorescence once bound to DNA. Examples of such dyes include SYBR gold
and SYBR
green (Wittwer et al. [1)). Additional other approaches have been successfully
implemented, and
these gain from adding target specificity to the sensing permitting greater
sensitivity and
specificity, particularly at low target levels in the initial sample. One
simple format comprises
two oligonucleotide primers recognising adjacent internal sequences of the
amplicon, each
labelled with a fluorophore. If the target is present the two fluorophores
become closely situated
100
CA 3041471 2019-04-26
and fluorescence resonance energy transfer (FRET) occurs, which can be
detected with suitable
excitation and emission filters (Wittwer et al. 1997 [2]). Other preferred
systems employ probes
combining the presence of both a fluorophore and a quencher. For some methods
hybridisation
to the target amplicon causes separation of hairpin-associated fluorophore and
quencher and
hence an increase in fluorescence. In another method hybridisation of the
probe is assessed via
the action of an oncoming polymerase possessing a 5'-3' exonuclease which
attacks the probe
and leads to permanent separation of the fluorophore and quencher (so called
`Taqinan' probes)
[Heid CA, Stevens J, Livak KJ, Williams PM., Real time quantitative PCR.
Genome Res. (1996)
6:986-943. Implementation of these approaches and others is widely documented.
During PCR reactions several distinct and temporally independent phases can be
identified. All DNA is single-stranded at the high temperature (e.g. 94 C)
used to melt all DNA
strands. Subsequently amplification primers may be annealed at a second low
temperature, such
as 50-65 C. Finally elongation of primers to generate principally double-
stranded products is
performed at, typically, 70-75 C. Due to the controlled cyclic nature of PCR
it is desired and
necessary to assess the accumulated level of DNA at specific distinct points
in the cycle. This
may occur at the end of the synthesis stage. Alternatively it may be desirable
to measure
fluorescence at some other point, and at another (fourth) temperature
depending on the approach
employed.
Unlike PCR, RPA reactions are principally configured to operate at a single
temperature.
Current configurations of RPA are unphased and therefore a complete variety of
reaction
'stages' will be simultaneously present in a sample (phasing may be attained
in theory by
approaches, for example, such as controlled uncaging of ATP). As a consequence
of this lack of
phasing at any given moment there is likely to be a mixture of double-stranded
DNA, single-
stranded DNA (such as displaced strands, as well as oligonucleotides), and
intermediates of
heterogeneous nature (such as triplex intermediates and/or homology-searching
complexes).
Despite the heterogeneity of the reaction mixture it will generally be the
case that the steady
state levels of double-stranded DNA, and possibly single-stranded DNA, will
increase during the
reaction as product accumulates. On this basis it seemed reasonable that
straightforward
approaches to measuring product accumulation might be readily employed.
101
CA 3041471 2019-04-26
Here we describe quantities of SYBR green and SYBR gold dyes that are
compatible
with RPA and permit real-time detection of accumulating reaction products. It
is also the case
that sequence-specific monitoring protocols will be compatible with RPA. Such
approaches
would include the employment of two fluorescently labelled oligonucleotides,
which on
hybridisation undergo fluorescence resonance energy transfer, or the use of
dual-labeled probes
such as those that are quenched until hybridisation results in alterations in
FRET, or they are
separated by associated nuclease activity. We provide evidence that the
'ragman' approach
which relies on the so-called 5'-3' exonuclease activity of certain
polymerases cannot be used in
RPA reactions, as these nucleases are actually structure-specific FLAP
endonucleases which
inhibit RPA reactions. Furthermore certain approaches, such as molecular
beacons, in which
natural hairpin-forming properties of the probe are essential, may be less
successful in RPA due
to likelihood of the probe to be in a melted state in the present of single-
stranded DNA binding
proteins and recombinases.
Figure 54 shows the results of experiments to determine whether SYBR gold and
SYBR
green stains are compatible with RPA. An initial experiment with SYBR gold in
which various
dilutions of the SYBR gold dye were made form the supplied stock (described as
a 10,000x
stock from molecular probes in DMS0) was performed using primers to amplify a
fragment of
the human apoliporotein B locus (Figure 1 A,B). The reaction is clearly
inhibited if the final
concentration was lx (1:10,000 from stock) or 0.4x (1:25,000 from stock), .but
was not
significantly observed at 0.2x (1:50,000 from stock). This concentration of
0.2x was then
employed as the highest concentration in a dilution series in 50 microliter
reactions established
with the same two human-specific primers, at two (low) target concentrations
of total human
genomic DNA (2 or 20 copies per microliters starting copy density)(Figure 54
C). A master mix
was assembled on ice and aliquoted into wells in a 96-well microplate cooled
to ice temperature.
Once mixed the plate we transferred to a fluorescence microplate reader with a
stage set to 37 C.
The reader was set to collect fluorescence readings at excitation 485nrn,
emission at 528nm, at 1
minute intervals over a period of 1 hour. In the same run SYBR green dye was
diluted to the
same degree (D) and concurrently assayed in similar samples. This experiment
revealed several
key facts relevant for configuring real-time RPA reactions with SYBR gold or
SYBR green.
102
CA 3041471 2019-04-26
First, direct comparison of detection times within the SYBR gold experiment
shows that
apparently even 1:50,000 fold dilutions slow the reaction relative to higher
dilutions. Indeed
1:100,000 or 1:80,000 gave significantly faster reaction kinetics, and thus
would appear to be a
better quantity of SYBR gold to use than the 1:50,000, which appeared
acceptable by earlier
end-point analysis. However, the relative maximal fluorescence signals
obtained with the higher
dilutions were significantly less than with the lower, suggesting that the dye
were becoming
limiting. Also, even at the highest dilution of SYBR gold fluorescence
increases over
background was detected later than the samples in the SYBR green experiment,
and the overall
fluorescence signals for all SYBR gold concentrations were much less than for
SYBR green. In
contrast the SYBR green samples were in all cases detected earlier than the
SYBR gold
experiments. In conclusion we suggest that while both dyes can be implemented
in real-time
RPA, SYBR green appears the more robust for standard analyses. It appears to
work well at
final dilutions of between 1:50,000 and 1:100,000, but the more concentrated
samples (1:50,000)
gave higher overall fluorescence and longer detectable exponential phase
suggesting that
1:50,000 dilution was the best in these experiments. We have also assayed with
higher SYBR
green concentrations successfully, but there is some indication that
inhibition may eventually
occur. We suggest that between 1:50,000 and 1:25,000 is the optimal SYBR green
concentration for dynamic real-time RPA assays.
Figure 55 shows an example of two experiments to determine the, capacity of
the system
to distinguish between different numbers of copies of a DNA target (present in
B.subtilis
genomic DNA). Starting number of template molecules of 500,000, 50,000, 5,000,
500, 50 or
zero copies of B.subtilis DNA were present in the various samples and
amplification was
performed with B.subtilis Spo0B locus-specific primers BsJ1 and BsK2. In many
independent
experiments the system successfully generated profiles similar to the
representative experiment
shown in Figure 56. Note that the spacing between the exponential phase curves
is similar
between 10-fold copy number dilutions as required for a quantitative system.
We conclude that
RPA is quantitative over at least 4 orders of magnitude of starting template
number, but
quantitative results over a larger range may be possible. Note that in (Figure
55C), the second
B.subtilis titration experiment, the highest target concentration trailed off
unexpectedly early.
103
CA 3041471 2019-04-26
Later experiments indicate that this may arise sporadically as a consequence
of the relatively low
levels of gp32 and uvsX employed in these experiments compared to later
experiments with
roughly double the levels (Figure 56).
We also investigated similarly the ability of the SYBR green RPA system to
monitor a
range of starting template copies of human DNA by amplifying with a number of
dilutions of
human DNA. Figure 57 shows the results of such analyses. As expected the data
fit the
expected profile indicating that RPA behaves in a quantitative manner and can
easily be assayed
with SYBR green. A similar trailing effect was observed for the highest target
samples, but was
corrected in later experiments in which higher amounts of gp32 and uvsX were
employed (see
Figure 57 C versus B, and the concomitant increase in overall end-point
product levels).
We show that using fluorescent dyes it is possible to monitor the kinetics of
RPA
reactions, and that this in turn can be used to indicate the quantity of
specific targets in a sample.
The simplest interpretation is that double-stranded DNA accumulates in an
exponential fashion
in RPA reactions until a mass point above the detection limits of SYBR green
fluorescence. It is
possible, however, that the fluorescence profile of SYBR green RPA reactions
reflects additional
activities. For example RPA products may under some circumstances engage in
recombination
reactions with other products leading to complex intermediates whose SYBR
green binding
behaviour is not well understood. Also the kinetic profile of product
accumulation may alter as
a consequence of reaction ageing phenomena only partly related to the starting
copy number.
This may underlie the spurious early fluorescence trailing observed for the
highest copy number
samples in some of these early experiments, although later experiments with
increased uvsX and
gp32 levels avoided this phenomenon.
We observed in a number of cases that the water control appeared to begin
rising in a
similar timeframe to the lowest concentration of sample (typically 1 copy per
microliter start
density, or less), however the kinetics of the water control product
accumulation seemed distinct
from the target-containing samples, having a shallower exponential phase
indicative of slower
doubling times. This observation is probably due to structural properties the
amplicons possess
(e.g. internal repeats) that reduce the rate of synthesis in the RPA system.
We commented
104
CA 3041471 2019-04-26
earlier on the possibility that primer-derived amplicons would tend to contain
inverted repeat
structures that would often require 2 rounds of invasion/synthesis per duplex
doubling, rather
than the single one that could suffice for bona fide target amplicons.
Regardless of the origin of
this phenomenon we speculate that with sophisticated data analysis software
and suitable
experimental internal controls, this noise might be identified and
distinguished from the kinetic
behaviour of true target amplification. Furthermore we observed some distinct
variation in the
time taken to reach detection, and the slopes of the curves, between distinct
amplicons. This
would be expected as variation in the activity of different primers, and the
variant lengths and
sequence composition of different amplicons, suggests that average doubling
times would vary
between amplicons. The slopes of the fluorescence profiles by this criteria
alone are likely to
vary between different types of amplicons, and this may be usefully employed
in analysis.
The experiments performed here suggest that kinetic RPA reactions can be used
to
decrease the time taken to assess the presence of target DNA in samples
compared to later gel
electrophoresis. The experimental set-up we have used here is far from ideal
as the 96-well
plates used are made of thick plastic, and the heated plate on the fluorometer
is not in direct
contact with the sample wells. We estimate that it takes up to 5 minutes for a
501.11 volume
reaction to attain temperature under these conditions, and perhaps 8 minutes
for a 100141 volume.
In an optimised device these long lag times would not exist. Consequently we
estimate that for a
clinically relevant quantity of human DNA (say 1000-3000 copies, ¨3ng-9ng)
amplification
could be assessed readily in roughly 30 minutes with the appropriate equipment
using typical
conditions shown here. Despite the limitations of these experiments using 96-
well plates and a
conventional fluorometer, these pilot experiments are highly encouraging and
indicate that
kinetic monitoring of RPA reactions offers a tractable approach to
quantification, and that this is
quantification may be practically implemented to assess DNA levels over at
least 5 orders of
magnitude.
RPA reaction control mediated by ATP concentration
RPA is a versatile method, but it can be improved by incorporation of features
to control
precisely when in the reaction recombinases are active. Such control can be
gained through the
105
CA 3041471 2019-04-26
periodic release of ATP by photolysis of caged-ATP. Alternatively, the
reaction concentration
ATP or other mibleoside triphosphates, can be cyclically regulated by repeated
manual addition
or the use of a biochemical oscillator.
Caged ATP cannot supporting either the DNA binding and recombinase function of
E.coli recA protein [Butler BC, Hanchett RI!, Rafailov H, MacDonald G (2002)
Investigating
Structural Changes Induced By Nucleotide Binding to RecA Using Difference
FTIR.. Biophys J
82(4): 2198-2210]. Following photolysis, however, released ATP enables
recombinase
function. All prokaryotic recombinases so far studied are direct homologues of
the recA protein
with primary sequence homology, and structural homology. Moreover, all recA
homologues
studied, including the eukaryotic homologues, require binding to ATP to enable
recombinase
function. We expect that similar phenomena with therefore be mediated with T4
uvsX and other
recombinases accordingly.
The role of nucleotides in regulating recombinase action is relatively well
documented.
In the case of prokaryotic recombinases, e.g. E.coli recA and 14 phage uvsX,
the recombinases
hydrolyse ATP to ADP (and AMP in the case of uvsX). Hydrolysis is occurring
with high
activity as long as the recombinases are bound to DNA. The ADP-bound state has
lower affinity
for DNA and is generally associated with filament disassembly from DNA (and
altered
nucleoprotein filament pitch). Under typical in vitro conditions ATP is
maintained at a
relatively high concentratidn in excess to ADP, to ensure that rapid of
exchange of ADP for
ATP in nucleoprotein filaments deters premature disassembly. Consequently, the
recombinases
in a reaction respond to the ATP:ADP ratio and to some approximation net
disassembly of the
nucleoprotein filament occurs when the ADP concentration exceeds the ATP
concentration.
RPA reactions rely on the action of recombinases to load onto synthetic single-
stranded
oligonucleotides and carry out homology-searching activity. As described the
activity of the
recombinases is dependant on the presence of nucleotide triphosphates, most
obviously ATP.
When recombinases are DNA bound they hydrolyse the nucleotide triphosphate at
a high rate,
for example the 14 uvsX protein is known to hydrolyse 200 molecules of ATP per
protein
molecule per minute at 37 C on single-stranded DNA (although rates on shorter
106
CA 3041471 2019-04-26
oligonucleotides may be more variable). Consequently there is a need for a
large supply of ATP
to maintain an active recombination reaction in which a large proportion of
oligonucleotides are
associated with recombinase filaments, particularly when the oligonucleotides
are present at near
micromolar concentrations.
Consider the flux of reaction nucleotide levels that might occur during a
typical RPA
reaction. If a pair of oligonucleotides is employed in a reaction at a
concentration oft i.iM each,
the oligonucleotides being 35 residues in length, then at 10% saturation of
the oligonucleotide
with recombinase, there is roughly a bound-recombinase concentration of 2.8
1.1M (a value of
10% saturation in 3-5% PEG compound is roughly consistent with the results of
experiments in
our hands, but could be slightly higher or lower). This would convert a 0.56
mM solution of
ATP to the equivalent amount of Al)? every minute based on the published
hydrolysis rate.
Consequently if a 3mM solution of ATP was used to initiate the reaction, and
no AT?
regeneration system were present, then after only 3 minutes the ADP
concentration would rise to
levels equal to the ATP and the recombinase would become inactive.
We have routinely used a total oligonucleotide concentration of 0.6 1.1M, and
a uvsX
concentration of roughly 3 M. Should all of this uvsX be bound to
oligonucleotides it would
consume roughly a 0.6niM solution of ATP in 3 minutes, given a reaction rate
200 molar
quantities per minute on oligonucleotides. Based on our data limited
hydrolysis data, however,
it is unlikely that complete binding of uvsX molecules to single-stranded DNA
is ever achieved
under our typical RPA conditions. Although it is formally possible that the
per monomer
hydrolysis rate is lower for the length of oligonucleotides we employ. These
alternatives are
suggested by the fact that in some cases we have been able to amplify a target
DNA to levels
detectable by ethidium bromide staining levels without the presence of a
regeneration system.
Other experiments suggest that is takes roughly 30 minutes to achieve this
level of amplification,
and by deduction we calculate that a maximum of only 0.15mM ATP could possibly
have been
consumed in each 3 minute period, a quarter of the predicted level).
Analysis of effective concentrations of uvsY in reactions is consistent with
the expected
stoichiometries. We have routinely used roughly a half to equi-molar
concentrations of uvsY
107
CA 3041471 2019-04-26
compared to uvsX protein, and published data suggests that uvsY protein
probably functions as a
hexamer. If this is so and one hexamer is required to load, and stabilise,
each loaded filament,
then whereas there would be a need for roughly 12-14 uvsX molecules per
oligonucleotide (30-
35 mer) there would need to be 6 uvsY molecules per oligonucleotide, i.e.
roughly half the
molar concentration. This experimentally determined optimum satisfies nicely
the theoretical
prediction that half to equi-molar concentrations of uvsY to uvsX are
required.
In summary, the conversion of ATP to ADP in a typical uvsX-supported RPA
reaction
with 0.61iM oligonucleotides, 3 M uvsX, and 1-3 WI uvsY is about 50 M per
minute and no
more than that. Roughly 16 molecules of ATP are hydrolysed per uvsX molecule
per minutes in
the reaction. This figure is six-fold less than that predicted if all uvsX
molecules were bound to
ssDNA and hydrolysing ATP at 200 molecules per uvsX per minute, and presumably
reflects
either that only a fraction of the uvsX is bound at any one time, that
hydrolysis rates are lower
on short oligonucleotides, and/or that in reactions with no regeneration the
hydrolysis rate falls
off significantly as the ADP levels rise. We suggest that while the last
factor may become
important eventually, during most of the reaction this is not the primary
factor.
Having deduced experimentally the consumption rate of ATP in a typical RPA
reaction,
it now remains to estimate what size pulse of ATP concentration we would need
to use to
stimulate suitable bursts of recombinase activity. To do this we need some
estimate of the Km
for a particular recombinase.
The Km of ATP hydrolysis by recA is reported to be 20 M. Consequently
relatively
little free ATP needs to be released into a recombinase system to promote
activity. Assuming
this is also true for uvsX, a pulse of ATP that changes the reaction
concentration from zero to
501.LM would amply support homology searching until such time that the ADP
level accumulates
to a 1:1 ratio. Under standard conditions this would be roughly 30 seconds and
would
eventually produce 25 M ADP and 251.tM ATP. A 30 second burst of recombinase
activity is
likely to be longer than what is required for around of invasions to occur.
Additional pulses of
ATP can readily generate additional bursts of recombinase activity. For
example a further pulse
of 501tM rise would raise the ATP level to 751.iM compared to 25 M ADP. After
30 seconds
108
CA 3041471 2019-04-26
the ATP level will equilibrate to 5011M ATP and 5012M ADP and the reaction
will again halt A
further pulse will raise the ATP to 100 M and the ADP to 5011M, and after 30
seconds further
75 M of each will be equilibrated. Thus 30 second bursts of excess ATP could
be released in
504M bursts to support burst of recombinase activity. Of course the changing
overall absolute
concentrations of ADP and ATP are likely to affect the reaction behaviour
necessitating
adjustments and possibly slightly larger pulses of ATP may need to be release
at each round.
Nevertheless it is apparent that with a starting concentration of caged-ATP of
3mM (similar to
that used in our earlier experiments) and pulses of 50 M, it is possible to
support 60
independent pulses. Even if the burst size needs to increase progressively to
account for the
overall increase in ADP concentration it is fairly likely that 30 cycles can
be accommodated.
Furthermore the ADP may be removed from the reaction by inclusion of ADP
metabolising
enzymes in the reaction. These could include hydrolysis of ADP, or
alternatively regeneration
of ADP to ATP and a constant source of alternative consuming ATP activity such
as NADH
generation.
Caged ATP is readily commercially available, and recently cheap low power
devices that
might be used to drive uncaging have become available. Advances in light-
emitting diodes
(LEDs) have lead to the development of small cheap low power sources to
generate light of
wavelength 365 urn. We envision integration of such devices into low power
heated cells,
which are portable and battery-:operated. Alternatively brief periods of
recombinase activity
could be generated simply by repeated addition of small volumes of additional
ATP, or by using
an oscillator system, such as the phosophofructokinase oscillator.
RPA reaction control mediated by use of asymmetric primers
One-sided RPA reactions can be driven from a single primer target site. In the
absence
of a facing primer, such reactions will generate single-stranded DNA. We have
found that
oligonucleotides possessing a 3' locked nucleic acid (LNA) [Di Giusto DA, King
GC (2004)
Strong positional preference in the interaction of LNA oligonucleotides with
DNA polymerase
and proofreading exonuclease activities: implications for genotyping assays.
Nucl Acids Res
32(3): e32] nucleotide, cannot server as primers for recombinase-mediated
elongation by certain
109
CA 3041471 2019-04-26
polymerases, such as the Bacillus stearothermophilus and Bacillus subtilis
polymerase L but do
serve as primers for other polymerase such as the E.coli Klenow fragment
consistent with prior
data showing that such 3'LNA capped primers can serve as primers in polymerase
extension
reactions when they hybridise to target single-stranded DNAs. We have also
found that not all
polymerases seem to be able to initiate from recombination intermediates,
perhaps reflecting that
for some polymerases longer stretches of single-stranded DNA with bound
primers are required.
For example we have found that phi-29 polymerase is unable to initiate
synthesis from a
recombination intermediate. We also have evidence that it can however
synthesise from a
3'LNA-capped primer. Finally consistent with published reports, we have found
that for a given
recombinase, there is a minimal primer length required for recombinase-
assisted strand-
exchange to occur efficiently. Specifically oligonucleotides shorter than 27-
30 base pairs are
poor substrates for uvsX. Nevertheless primers as short as 20-27 nucleotides
may be perfectly
adequate to support hybridisation-mediated priming. Thus by combining a longer
and a shorter
primer one can bias RPA reactions to use one primer for invasion-driven
elongation and the
other for hybridisation-driven elongation.
Taken together these facts suggest a variety of configurations in which
reactions can be
assembled in a way that synthesis initiates from one side, and only when it
has passed the
second site does opposing synthesis begin. For example, an RPA reaction may be
configured
.õ such that one primer is a normal oligonucleotide and the opposing primer is
a 3'LNA-capped
oligonucleotide. In the same reaction a polymerase that cannot use 3'LNA-
capped
oligonucleotides, but works from invasion structures, is mixed with one that
can use 3'LNA-
capped oligonucleotides but works only from hybridisation structures.
Recombinase-mediated
invasion and extension from the normal primer will generate single-stranded
DNA molecules,
which can then serve as templates for synthesis by the second polymerase when
a 3'LNA-
capped opposing primer hybridises to its target site. Alternatively a short
second primer that
functions only in hybridisation would also ensure asymmetric primer use. Other
configurations
of oligonucleotide length and nature, with different polymerases, may be used
to generate the
desired effect. Thus the use of asymmetric primers should resolve any
interference from
replication fork collision; however, it may be necessary also to control
recombinase activity to
110
CA 3041471 2019-04-26
avoid re-invasion of single-stranded DNAs displaced by synthesis from the
normal primer.
The combination of asymmetric primers and control of ATP levels may provide
conditions sufficient to amplify long (>10kb) DNAs. One other factor that may
affect the
efficiency of amplification of long DNAs is interference from other template
DNAs, other non-
template DNA, and product of the RPA reaction itself such as displaced single-
stranded DNAs.
In addition to the possibility that recombinase may associate with a displaced
single stranded
DNA and mediate an invasion reaction onto template DNA, there is also the
possibility that
single stranded DNAs and other non-template DNAs from the sample may hybridise
inappropriately with target template DNAs and interfere with efficient
replication. To avoid
some of these difficulties, at least for the first few rounds of replication
it would be helpful to
spatially fix template DNA and prevent association with other long DNAs. One
convenient
means to achieve this is to assemble RPA reactions in a gel matrix, such as a
polyacrylamide gel
adjusted with an average pore size that will allow free mobility of small RPA
components, such
as the enzymes and primers, but not allow free motility of long DNAs. Sample
DNAs at low
starting concentration will be physically separated, unable to associate with
one another, while
smaller RPA components will remain relatively free to associate with template
molecules.
Church and colleagues have described using polyacrylamide gels in this way for
PCR and
colleagues to produce spatially isolated amplicons, resolvable in two
dimensions, known as
polymerase colonies, or polonies [Mitra R, Church G (1999) In situ localized
amplification and
contact replication of many individual DNA molecules. Nucl Acids Res 27(24):
e34i-vi.j.
Typically, polonies are used to image amplicons from distinct templates and so
are generated on
microscope slides. For amplification of long DNAs, however, it would not be
necessary to
resolve individual polonies, thus reactions could be assembled in any
appropriate vessel.
The polony assay itself may benefit from using RPA instead of PCR. There are
at least
two difficulties with the use of PCR. Firstly, because polonies are usually
generated on
microscope slides, and glass is a poor conductor of heat, the times required
for PCRs can be
significantly longer than for normal bulk phase PCR. This means that diffusion
of amplification
product leads to fairly large polony sizes, incompatible with high density,
high throughput
assays. Secondly, because PCR requires the thermal melting of template DNA,
temperatures in
111
CA 3041471 2019-04-26
excess of 90 C are required for extended periods. These high temperatures will
increase
diffusion rates further increasing the average polony size. RPA with its low
constant
temperature will overcome both of these problems.
Using the EPA dynamic recombination environment to permit identification of
the
polymorphic status of a given amplicon
It is widely demonstrated that the presence or absence of a defined nucleic
acid sequence
can be determined by forming a hybrid between a sample nucleic acid with a
nucleic acid probe
of previously determined nature, followed by an appropriate method of
detecting such an
interaction. For example, microarrays are now widely used in which DNA, RNA or
other
backbone oligonucleotides are spatially separated and immobilized to a
support. The presence
or absence of a homologous sequence in a sample is then determined by co-
incubating the
samples under appropriate buffer and temperature conditions that allows
hybrids to form
between the immobilized probe and sample nucleic acids. In this case both
sample and probe
are provided in a completely or partially melted status so that they are able
to hybridize.
Provided a label is incorporated into the sample then the interaction can be
subsequently
quantified.
Alternative approaches to forming sequence-specific hybrids have been
described in
which one of the participating nucleic acids is double-stranded at the outset,
and a three-stranded
protein-containing hybrid is formed by the action of recombinase enzymes such
as E. coil recA
in the presence of non-hydrolysable nucleotide triphosphate analogs (see US
patent number
5,460,941 , and US patent 5,223,414). Also described are alternative methods
that employ
recombinases but seek to stabilize protein-free recombination intermediates by
forming four-
stranded structures (see US patent number 5,273,881). These approaches however
differ from
those described herein both in method and outcome as described further below.
In many cases the process of hybrid formation is of sufficient fidelity to
discriminate
between the presence and absence of the target, and further to determine
whether it is a perfect
match or not. For example, short primers (e.g. 7 to 18 nucleotides in length)
can discriminate
between perfect and imperfect complements if hybridization conditions are
stringently
112
CA 3041471 2019-04-26
controlled. However, if the nucleic acid is longer and there are only small
variations between a
perfect hybrid and an imperfect hybrid, for example a few nucleotides over a
region of say one
hundred or more, then it unlikely that a sufficiently great difference in
efficiency of
hybridization will exist between such a variant and a perfect match.
This invention concerns determining the polymorphic state of a sample nucleic
acid by
forming hybrids between the products of an amplification reaction and
previously synthesized
and immobilized probe nucleic acids present at defined locations, and each
individual location
containing a pure population of fragments representing one of the known repeat
lengths present
in the population.
DNA sequences amplified by the recombination polymerase amplification (RPA)
method are ideal targets for such a hybrid-formation based assay for the
following reasons.
Firstly, it is possible to configure the RPA method to generate mostly double-
stranded DNA
product, or mostly single-stranded DNA product, for example by altering the
ratios of
amplification primers. Secondly, RPA reactions contain all of the necessary
components to
permit the association of hybrids between initially duplex DNA and single-
stranded DNAs with
no requirement to thermally melt the double-stranded DNA. Thus little
additional sample
handling is required.
Other methods have been described which employ recombinases and the use of non-
hydrolysable analogs of ATP, such as ATP-y-S, to stabilize hybrids of related
complementary
nucleic acids in a poorly defined 'triplex' intermediate involving the
continued presence of the
recombinase. Such an approach, however, will not work in this the context
described here,
because these highly stable structures are not dynamic and present significant
resistance of
reaction products to access by other agents. As a consequence use of
recombinases with non-
hydrolysable ATP analogs leads to formation of extremely stable hybrids
between DNAs
containing a significant degree of mis-match, thus not allowing efficiently
discrimination
between sequences. In contrast, the approach described here employs a dynamic
system
utilizing ATP or other hydrolysable analogs in which hybrids are readily
destabilized as well as
generated. The net result is a large enrichment for interactions of perfect
complementary.
113
CA 3041471 2019-04-26
In a preferred embodiment, following the completion of an RPA reaction or
concomitant
with it, the reaction mixture is contacted to an array of probe molecules such
that each amplified
polymorphic DNA, will in the first instance locate the correct immobilised
probe nucleic acid
containing exactly the same, or complementary, sequence. Should this initial
hybrid-forming
reaction not occur with sufficient efficiency or fidelity, as is likely with
very similar sequences,
then the dynamic nature of the recombinase-driven reaction should resolve
mismatches. This is
because imperfect hybrids will contain bubbles and non-duplex features, which
act to aid the
reloading of recombinase onto these hybrids and cause an increased rate of
hybrid disruption
when compared to perfect matches. New duplex-forming events are permitted to
occur and only
if perfect hybrids are formed do they become relatively resistant to further
reaction. In practice,
if all of the possible number of an STR repeat were arrayed in order of repeat
number, then at
the end there may result a gradient of hybrids formed which peaks at exact
matches, is weaker
on the direct flanks and absent further way. As the mis-match bubbles increase
in size, there is a
progressively greater tendency for recombinase to reload onto the single-
stranded bubble region.
(Figs.58 and 59).
Additionally, if the dynamic recombinase system is not sufficiently specific
to discern
between perfect and imperfect hybrids, additional hybrid-disrupting components
can be included
in the reaction. Such agents include, but are not limited to, helicases,
nucleases, recombinases,
polymerases, and other DNA-binding agents. Various helicases and nucleases
exist which
selectively target specific forms of DNA, or structures, such that they
interact with and resolve
mismatches or bubbles, but not on perfect hybrids. For example, the PriA
helicase of E.coli
interacts with regions in which single-stranded DNA is exposed adjacent to
double-stranded
DNA, as would occur in a mismatch in repeat number that could occur with STR
hybridizations,
and subsequently can act to unwind DNA in a 3' to 5' direction. Such
destabilization would
permit recombinase to re-load onto these separated strands allowing a new
alternative hybrid to
form instead. Over time such a mechanism would enrich for hybrids only between
the correct
target and probe. Similarly, the replication helicase of E.coli, DnaB, will
load onto single-
stranded DNA and unwind duplex DNA, in a 5'-3' direction, opposite of that of
PriA. The dda
helicase of bacteriophage T4 loads onto single-stranded DNA. The dda helicase
is so powerful
114
CA 3041471 2019-04-26
it can displace other DNA bound proteins in its path. The dda helicase has
been shown to
disrupt very high affinity streptavidin-biotin interactions (when the biotin
is on the 3' end of the
oligonucleotide), thus dda helicase could be used to= destabilize immobilized
probe complexes on
a surface (Byrd and Raney, 2004)(Morris and Raney, 1999).
Alternatively the E. coli RuvA and RuvB gene products form a helicase that can
encircle
double-stranded DNA and drive branchpoints along DNA. In this manner a bubble
could be
'pushed' to the end of the template causing sufficient destabilization to
permit recombinase
loading as well as other event. Nucleases that would target imperfections in
duplex character
would include, for example, S1 nuclease, which can nick double-stranded DNA at
a mismatch or
bubble. Other such nucleases with structure-specific characteristics exist.
Such nicks can serve
to initiate strand-displacing polymerase elongation. Alternatively if the
nuclease cleaves the
probe DNA then it is possible to have a chemical or enzymatic group released
from the site of
immobilization so that over time a signal generated later remains only at the
point at which
perfect hybrids formed.
Thus by combining the necessary components it should be possible to create an
environment in which perfect hybrids significantly dominate and imperfect
sites are depleted, or
are rendered undetectable. Key to this approach is the establishment of a
dynamic environment
in which hybrids can be formed, and disrupted such that the stability of
perfect and imperfect
hybrids allows sensitive discrimihation of reaction products with differing
polymorphic features.
This environment is ideally provided by the dynamic/stable recombinase system
comprising
gp32, uvsX, uvsY, PEG compound, and an ATP regenerating system as described.
The presence or absence of a productive hybrid, or the loss of a label from an
immobilization site (e.g. see nuclease approach above) can be measured by
standard methods.
These include incubation of the reaction at the end-point with a substrate for
an enzyme
immobilized on the DNA target or DNA probe. Other detection approaches are
possible and
described widely elsewhere.
The dynamic persistent recombinase environment for broad use in molecular
techniques
We set about establishing the perfect environment for dynamic recombinase
activity with
115
CA 3041471 2019-04-26
a mind to permitting massively geometric amplification of nucleic acids at low
constant
temperature. However the established environment might obviously be employed
in
combination with other enzymes, or indeed without them, in a variety of
contexts to replace
classical hybridisation reactions in general. For example molecular cloning
procedures might
take advantage of the possibility of recombining large tracts of DNA with one
another much
more effectively than by thermal melting and annealing. Also many other
enzymes that might be
employed for in vitro molecular processes could benefit from the low
temperature environment
compatible with most mesophilic enzymes. Such enzymes would include, in
addition to
polymerases, helix-distortion recognising nucleases (e.g. SI nuclease), FLAP
endonucleases,
restriction endonucleases, base modifying or removing enzymes, lyases,
helicases,
topoisomerases, ligases, reverse transcriptases, RNase H activity, resolvases,
RNA polymerases,
and any other enzyme that acts on, or interacts with, nucleic acids. They
could also involve other
enzymes pertinent to the in vitro system in addition to DNA metabolising
enzymes, such as
alkaline phosphatase or horseradish peroxidase used in detection protocols.
The combination of
the unique properties of the stable dynamic recombination system with other
enzymatic
activities enables a potentially very large number of new methods and
applications.
In addition to recognising the important impact of combining wholly low-
temperature
dynamic enzymatic hybridisation systems with other enzyme systems, we have
established
sequence knowledge for the optimal assembly of highly active recombination
using short
oligonucleotide primers on the basis of experiments using RPA. We have noted
that very active
primers were characterised by a particularly rich distribution of pyrimidines.
Guanosine
residues, in contrast, appeared poorly represented in the most active
oligonucleotides that we
have analysed, and earlier results suggested that when appended to the 5' end
of
oligonucleotides they might lower their activity. In this disclosure we report
the experimental
results of dynamic monitoring of the reaction to study the affects of
appending DNA sequences
to the 5' ends of oligonucleotides. Appending sequences to the very 5' extent
of otherwise
identical oligonucleotides has a significant effect on their activity in RPA.
We suggest that these
observations are best interpreted as reflecting the loading behaviour of the
recombinase on
single-stranded, and possibly duplex, DNA. They may also have roles in
controlling defined
116
CA 3041471 2019-04-26
phasing of recombinases on ssDNAs.
As a starting point, we had come to note through kinetic studies (non real-
time), and
general observations, that some primer pairs were capable of mediating more
rapid DNA
amplification than others. Particularly rapid primers were identified for the
human SIR marker
CSF1P0, which appeared to have an average doubling time of roughly 30 seconds
(as estimated
as average doublings per unit time from start to detectable product
accumulation), and capable
of generating detectable levels of product within 15 minutes starting with a
few thousand copies
of target (see Figure 67). Other primer pairs typically took, taking 20-35
minutes to achieve
similar results. We sought to learn more about the source of this variability
(see Figure 67, and
data not shown).
Analysis of the DNA sequence of the CSFIPO primers revealed that they were
relatively
rich in pyrimidines, and also were rather low in guanosine. This observation
correlated with a
piece of data arising from an earlier experiment in which we had investigated
how appending a
stretch of cytosine or guanosine residues to the 5' end of oligonucleotides
affected their
behaviour in RPA. In Figure 68 a real-time amplification experiment is shown
in which primers
specific for the Bacillus subtilis sporulation locus SpOB, referred to as J1
and K2, were used to
amplify a fragment from B. subtilis genomic DNA. In te experiment shown in
figure 67 we had
noted that a stretch of guanosines had 'quietened' the overall reaction, while
a stretch of
appended cytosines had `noisened' amplification reactions. We repeated this
experiment and
monitored the reaction in real-time, the findings shown in Figure 68. The
results were consistent
with the notion that appended bases affected, at the very least, the rate
behaviour and
presumably activity of primers. Primers Jl and K2, in which all bases match
genomic target,
generate a product which is first detectable with SYBR green dye at a little
over 30 minutes in
this experiment (faster kinetics were observed in later experiments, and
variation may have
arisen due to non-optimal temperature ramping when using a conventional
microplate reader, as
occurred in these experiments). A sample with target DNA, and one without,
were
distinguishable in this experiment by virtue of a small delay in product
accumulation, and
variant accumulation kinetics. Primers appended with additional cytosine
residues, J1(C) and
IC2(C), were clearly capable of amplifying DNA significantly more quickly. We
note however
117
CA 3041471 2019-04-26
that the accumulation of (primer-derived) artefacts occurs on a timescale
similar to that with the
target DNA, distinctly less-well separated than in the unappended primers. We
deduce that C-
tailed primers are simply faster primers (although 'noisier' also in this
case). Conversely
accumulation kinetics for the primers appended with G residues was very poor,
and no clearly
identifiable expected end-product was observed on gel electrophoresis even at
90 minutes.
Based on our results, and integrating these findings with published work, lead
us to
speculate that sequence-composition might affect recombinase-loading onto
primers, and that
pyrimidines might promote this loading process. Published work is conflicting
in this area; in
some studies a case has been made for preference of recA binding to
recombination processes
for G and T residues which are enriched in E.coli recombination hotspots
(Tracy and
Kowalczykowski). Conversely, studies of recA loading dynamics performed using
fluorescence
anisotropy drew startlingly different conclusions (Bar-Ziv and Libchaber). In
this case the
barrier to recA nucleation (the slow phase of nucleoprotein filament
formation) was highly
sensitive to composition, showing a strong favourable bias towards
pyrimidines. These latter
observations would be more consistent with our observations. Indeed, as RPA
tends to employ
rather short oligonucleotides, it becomes easy to see how important the
recombinase nucleation
event is in giving effective amplification behaviour. Not only is rapid
nucleation highly desirable
to permit acceptable levels of filament loading, but additionally it will be
critical that this occurs
preferentially towards the very 5' end of the filament. If the 5' end is a
poor substrate for
nucleation, while an internal sequence is rather good, there is a likelihood
that most filaments
will be only partially loaded, and as such fail to function properly in RPA.
Such partially loaded
filaments may be insufficiently loaded to hydrolyse ATP properly, unable to
efficiently undergo
strand exchange, and be effectively drawn away from an active pool of
filaments. Even worse,
they may 'poison' the reaction by forming homology-searching complexes with
DNA, but,
being unable to undergo the normal dynamic behaviour (if they are below
necessary length to
promote hydrolysis), lock up target DNA in unproductive complexes, as occurs
when ATP-y-S
is employed.
Despite our early results being consistent with the notion that pyrimidines
are good
nucleation sites, and the attractive possibility that this alone accounts for
the observed variation,
118
CA 3041471 2019-04-26
we must point out that neither of the above assays is truly equivalent to the
experimental system
that we are employing. Filament loading, unloading, hydrolysis behaviour and
so on, are all
relevant in concert in the amplification situation, and multiple factors could
be at play. Also, it
likely that recombinase phasing could play a role in determining the activity
of primers, and that
this established by preferential nucleation at particular locations, a
phenomenon reported earlier
for recA (Volodin et al.; Volodin and Camerini-Otero). In this case nucleation
would occur
preferentially at the 5' end, establish a committed phase to the entire
filament, and the filament
length would need to have a perfect length to place the last protein over the
3' end in the most
desirable manned. In addition to the further complexities associated with the
amplification
situation as described, these earlier studies were all performed with E. coli
RecA protein, and not
T4 UvsX protein, and as these proteins are significantly diverged at the
primary sequence level,
it may not be possible to extrapolate conclusions from RecA studies to the
UvsX protein.
Nonetheless, realising the importance of sequence composition, and in
particular 5' sequence
composition, we have begun to analyse in more depth which sequences are most
effective for
appending to the very 5' end, i.e. we seek to determine idealised 'landing
sites' for the
recombinase in this system, and/or those that are associated with other
beneficial properties for
amplification and in vitro processes in general. Yet further, we anticipate
that phasing of
recombinases may play a significant role in overall activity, and that this
will be influenced by
composition and precise length of the oligonucleotide.
We investigated whether appending a stretch of Thymine residues, or a mixture
of
cytosine and thymines (pyrimidines) would be as effective as a stretch of
cytosine residues in
stimulating greater primary activity. The results of this analysis are shown
in figure 69.
Interestingly, appending a run of Thymines to the very 5' end of the
oligonucleotides did not, in
this experiment, improve the amplification rate behaviour. In fact rather to
the contrary
combining primers with appended thyrnines gave very poor amplification
behaviour in this
experiment. We also tested primers in which the appended sequence was a
mixture of cytosine
and thymine, as indicated. The behaviour of these primers in various
combinations was rather
more variable and complex. For example we noted the presence of additional
bands consistent,
but not proven to be, single-stranded equivalents of the product. Careful
analysis shows that in
119
CA 3041471 2019-04-26
different combinations the migration behaviour of this possible single-
stranded band was either
of one type, or a second type, and that this correlated with the primer used.
In brief, we suspect
that in these reactions one, or other, primer was more active leading to
development of
asymmetry in the reaction and single-stranded DNA accumulation not seen with
the parent,
unappended, oligonucleotides. It was difficult to rationalise the results, but
we can conclude that
in this experiment stretches of thymine were not effective, nor were mixed
polymers very
effective despite a slight tendency to appear more active if more cytosines
were present.
In our next experiment we appended yet more residues to the end of
oligonucleotides and
compared their behaviour in a real-time analysis. Once again there was
variability between
primers, and in this experiment we included as a control each primer incubated
on its own.
Interestingly we noted that, at the very low concentrations of target used
here (1 copy per
microliter start density), noise generated by single primers appeared in a
timeframe similar to the
accumulation of product DNA observed when primer pairs were used. Furthermore,
this is most
obvious for the oligonucleotide 1(2(C), studied previously, and responsible
for very rapid
amplification when combined with J1 oligonucleotides. This suggests that
monitoring
oligonucleotides individually for their rate of noise generation is of utility
in determining their
overall activity. Finally, and puzzlingly, an oligonucleotide just one base
pair shorter than
1(2(C), oligonucleotide 9 in the series in Figure 70, lacked this strong and
rapid amplification
behaviour. While the oligonucleotides used in this study were not HPLC
purified, we
nonetheless assume that most of the oligonucleotides are the full length form
(based on gel
pictures supplied by the manufacturer). Taken at face value, this surprising
observation must be
taken to reflect that either the number of C residues, changing from 5 to 6,
at the 5' end is a
point of significant transition for some structural feature of this
oligonucleotide, or it could be
taken to reflect the additional influence of some phasing behaviour. In this
latter case we would
speculate that 5' sequences strongly influence the phasing and deposition of
the first UvsX
monomer, and that this phasing is maintained all the way down to the 3' end.
Presumably
whether or not the most 3' UvsX monomer sits perfectly over the end of the
oligonucleotide, or
there is slightly too many or too few base pairs to permit each UvsX monomer
to bind to its
maximal number of backbone residues, may well influence the likelihood that
recombination
120
CA 3041471 2019-04-26
proceeds efficiently to completion. It may also influence the background, as
if there are a few
'spare' bases popping out of the 3' end this might well promoter enhanced
primer noise.
In conclusion, we have shown here how critical the length and sequence
composition of
oligonucleotides may be to elicit the best performance in RPA, and other
recombinase-driven
processes. Further, we formally demonstrate that non-target sequences can be
appended to the 5'
ends of oligonucleotides to effect regulation of their activity in RPA.
In addition to DNA amplification, a recombinase system including hydrolysable
ATP
analogs, and components to ensure high loading of DNA molecules in this
situation, could be
employed as a replacement for other described hybridisation approaches.
Contamination control in RPA reactions
As RPA is a very sensitive detection method we have encountered problems with
carry-
over contamination seen with other ultra-sensitive amplification protocols
such as PCR. We
show here that dUTP can be used to partially or wholly replace dTTP in RPA
reactions, thus
offering a simple way to distinguish the products of previous RPA reactions
from bona fide
sample targets. As heat treating RPA reactions at their initiation with a mind
to inactivating
dUTP deglycosylase (as occurs in PCR protocols with contamination control) is
not desirable,
we suggest as alternative approach in which deglycosylase inhibitors are mixed
with reactions to
permit initiation (see Figures 61 and 62).
Reverse-transcription RPA
In circumstances in which detection of the presence of specific RNA molecules
is
desired it is possible to use RPA to detect them providing that the RNA is
first converted to a
DNA form using reverse transcriptase activity. Most convenient would be if
reverse
transcription and RPA could all be performed in a single homogeneous
environement. We show
that reverse transcription RPA reactions (RT-RPA) can be performed in a single
tube
environment by including reverse transcriptase in the reaction environment,
and with other
minor modifications (see figure 60).
Straightforward detection and assessment of amplification reactions using
lateral flow
membranes. =
Features of RPA make it ideally suited to configuring portable easy-access
diagnostic
121
CA 3041471 2019-04-26
intergrated products. In one set of cases assessment of whether a specific DNA
amplification
reaction has occurred or not, with only a moderate need for quantitative
analysis, can be
performed by simple formats to assess whether two labelled primers have become
physically
associated within an amplicon. We here show that this simple idea is
effective, and in particular
that the widely-employed technology of lateral-flow strip systems are ideal to
perform this role.
RPA reactions can be mixed directly with sample running buffer for these
systems, and the
presence of amplicon determined within several minutes (see figure 63).
Compatibility of crude preparations of biological samples with RPA
We here show that simple lysates of blood can be used directly in RPA
reactions. This
offers the possibility that the format of diagnostic products containing the
RPA system may
require only trivial treatment of samples prior to direct addition to RPA
reactions (see figure
64).
Behaviour of fluorescent probes in the stable, persistent, dynamic
recombination environment
We here show that the presence of the dynamic recombination environment used
in RPA
alters the behaviour of fluorescent probes relative to equivalent environments
in other techniques
such as PCR. Most particularly if dual-labeled probes containing fluorophores
and quenchers are
to be used, the number of bases separating the two groups in the
oligonucleotide must be less,
presumably because the saturating DNA binding proteins stretch the probes
relative to both
their state in B-form duplex DNA, as well as the random coil that exists for
oligonucleotides in
free solution (figure 73). Furthermore we have identified key enzymes that may
be employed to
specifically process duplex hybrids between such probes and their target
DNA's. These
approaches teach the approach by which real-time 'third' probe strategies
musts be configured
with RPA, and how that differs to the well-established approaches in PCR such
as the Taqman'
approach, molecular beacons, and the like. In particular we have determined
that the `Taqinan'
approach cannot be employed in RPA presumably because the 5' nuclease
associated with E.coli
Poll like enzymes has FLAP endonuclease activity which inhibits RPA reactions.
REFERENCES:
Adams, D. E., Tsaneva, I. R. and West, S. C. (1994). Dissociation of RecA
filaments
from duplex DNA by the RuvA and RuvB DNA repair proteins. Proc Natl Acad Sci U
S A 91,
122
CA 3041471 2019-04-26
9901-5.
Alexseyev, A. A., Bakhlanova, I. V., Zaitsev, E. N. and Lanzov, V. A. (1996).
Genetic
characteristics of new RecA mutants of Escherichia coli K-12. J Bacteriol 178,
2018-24.
Bar-Ziv R, Libchaber A. (2001). Effects of DNA sequence and structure on
binding of
RecA to single-stranded DNA. PNAS USA Jul 31;98(16):9068-73
Baumann, P., Benson, F. E., Hajibagheri, N. and West, S. C. (1997).
Purification of
human Rad51 protein by selective spermidine precipitation. Mutat Res 384, 65-
72.
Benkovie, S. J., Valentine, A. M. and Salinas, F. (2001). Replisome-mediated
DNA
replication. Annu Rev Biochem 70, 181-208.
Bennett, R. L. and Holloman, W. K. (2001). A RecA homologue in Ustilago maydis
that
is distinct and evolutionarily distant from Rad51 actively promotes DNA
pairing reactions in the
absence of auxiliary factors. Biochemistry 40, 2942-53.
Better, M. and Helinski, D. R. (1983). Isolation and characterization of the
RecA gene of
Rhizobium meliloti. J Bacteriol 155, 311-6.
Bianco, P.R. and Weinstock, G.M. (1996). Interaction of the RecA protein of
Escherichia
colli with single-stranded oligodeoxyribonucleotides. Nucleic Acids Research
Dec
15;24(24):4933-9
Bork, J. M., Cox, M. M. and Inman, R. B. (2001). The RecOR proteins modulate
RecA
protein function at 5' ends of single- stranded DNA. EMBO J 20, 7313-22.
Bork, Cox and Inman J Biol Chem. 2001 Dec 7;276(49):45740-3
Butler BC, Hanchett RH, Rafailov H, MacDonald G (2002) Investigating
Structural
Changes Induced By Nucleotide Binding to RecA Using Difference FTIR. Biophys J
82(4):
2198-2210
Byrd AKõ Raney /CD (2004). Protein displacement by an assembly of helicase
molecules
aligned along single-stranded DNA. Nat Struct Mol Biol. Jun;11(6):531-8
Cox, M. M., Goodman, M. F., ICreuzer, K. N., Sherratt, D. J., Sandler, S. J.
and Marians,
123
CA 3041471 2019-04-26
K. J. (2000). The importance of repairing stalled replication forks. Nature
404, 37-41.
Cox, M. M., McEntee, K. and Lehman, I. R. (1981). A simple and rapid procedure
for
the large scale purification of the RecA protein of Escherichia coil. J Biol
Chem 256, 4676-8.
Cromie, G. A. and Leach, D. R. (2000). Control of crossing over. Mot Cell 6,
815-26.
Dillingham, M. S. and Kowalczykowski, S. C. (2001). A step backward in
advancing
DNA replication: rescue of stalled replication forks by RecG. Mol Cell 8, 734-
6.
Eggler, Lusetti and Cox, J Biol Chem. 2003 May 2;278(18):16389-96
Eggleston, A. K. and West, S. C. (2000). Cleavage of holiday junctions by the
Escherichia coil RuvABC complex. J Blot Chem 275, 26467-76.
Elias-Arnanz M, Salas M. EMBO J. 1997 Sep 15;16(18):5775-83
Ferrari et al., J Mol Biol. 1994 Feb 11;236(1):106-23
Formosa T, Alberts BM. J Biol Chem. 1986 May 5;261(13):6107-18
Formosa T, Alberts BM. Cell. 1986 Dec 5;47(5):793-806. DNA synthesis dependant
on
genetic recombination: characterization of a reaction catalyzed by purified
bacteriophage 14
proteins.
Glover, B. P. and McHenry, C. S. (2001). The DNA polymerase III holoenzyrne:
an
asymmetric dimeric replicative complex with leading and lagging strand
polymerases. Cell 105,
925-34.
Goodman, H. J., Parker, J. R., Southern, J. A. and Woods, D. R. (1987).
Cloning and
expression in Escherichia coli of a RecA-like gene from Bacteroides fragilis.
Gene 58, 265-71.
Hacker KJ, Alberts BM. J Biol Chem. 1992 Oct 15;267(29):20674-81
Harris LD, Griffith J. J Biol Chem. 1987 Jul 5;262(l 9):9285-92
Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome
Res.
1996 Oct;6(10):986-94.
Heyer, W. D. and Kolodner, R. D. (1989). Purification and characterization of
a protein
124
CA 3041471 2019-04-26
from Saccharomyces cerevisiae that binds tightly to single-stranded DNA and
stimulates a
cognate strand exchange protein. Biochemistry 28, 2856-62.
Hickson, I. D., Gordon, R. L., TomIcinson, A. E. and Emmerson, P. T. (1981). A
temperature sensitive RecA protein of Escherichia co/i. Mol Gen Genet 184, 68-
72.
Hsieh, P., Camerini-Otero, C. S. and Camerini-Otero, R. D. (1992). The
synapsis event
in the homologous pairing of DNAs: RecA recognizes and pairs less than one
helical repeat of
DNA. Proc Natl Acad Sci U S A 89, 6492-6.
Hsieh P. Camerini-Otero CS, Camerini-Otero RD. Proc Natl Acad Sci U S A. 1992
Jul
15;89(14):6492-6
Kato, R. and Kuratnitsu, S. (1993). RecA protein from an extremely
thermophilic
bacterium, Thermus thermophilus HB8. J Biochem (Tokyo) 114, 926-9.
Katz FS, Bryant FR. Biochemistry. 2001 Sep 18;40(37):11082-9
Kelman, Z. and O'Donnell, M. (1995). DNA polymerase III holoenzyme: structure
and
function of a chromosomal replicating machine. Annu Rev Biochem 64, 171-200.
Komori, K., Miyata, T., DiRuggiero, J., Holley-Shanks, R., Hayashi, I., Cann,
I. K.,
Mayanagi, K., Shinagawa, H. and Ishino, Y. (2000). Both RadA and RadB are
involved in
homologous recombination in Pyrococcus furiosus. J Biol Chem 275, 33782-90.
Komberg, A. and Baker, T. A. (1992). DNA Replication. New York: W. H. Freeman
and
Company.Kuil et al., Biophys Chem. 1988 Dec;32(2-3):2I1 -27
Kowalczykowski SC, Lonberg N, Newport JW, Paul LS, von Hippel PH., Biophys J.
1980 Oct;32(1):403-18.
Kuramitsu, S., Hamaguchi, K., Ogawa, T. and Ogawa, H. (1981). A large-scale
preparation and some physicochemical properties of RecA protein. J Biochem
(Tokyo) 90,
1033-45.
Kurumizaka, H., Ikawa, S., Ikeya, T., Ogawa, T. and Shibata, T. (1994). A
chimeric
RecA protein exhibits altered double-stranded DNA binding. J Biol Chem 269,
3068-75.
125
CA 3041471 2019-04-26
Lavery PE, Kowalczykowski SC. J Biol Chem. 1992 May 5;267(13):9307-14
Liu, J., Nurse, P. and Marians, K. J. (1996). The ordered assembly of the
phiX174-type
primosome. 111. PriB facilitates complex formation between PriA and DnaT. J
Biol Chem 271,
15656-61.
Lohman and Ferrari, Annu Rev Biochem. 1994;63:527-70
Lovett, C. M., Jr. and Roberts, J. W. (1985). Purification of a RecA protein
analogue
from Bacillus subtilis. J Biol Chem 260, 3305-13.
Maeshima, K., Morirnatsu, K. and Horii, T. (1996). Purification and
characterization of
XRad51.1 protein, Xenopus RAD51 homologue: recombinant XRad51.1 promotes
strand
exchange reaction. Genes Cells 1, 1057-68.
Marians, K. J. (1992). Prokaryotic DNA replication. Annu Rev Biochem 61, 673-
719.
Marians, K. J. (1999). PriA: at the crossroads of DNA replication and
recombination.
Prog Nucleic Acid Res Mol Biol 63, 39-67.
Mazin, A. V. and Kowalczykowski, S. C. (1998). The function of the secondary
DNA-
binding site of RecA protein during DNA strand exchange, EMBO J 17, 1161-8.
McGlynn, P. and Lloyd, R. G. (1999). RecG helicase activity at three- and four-
strand
DNA structures. Nucleic Acids Res 27, 3049-56.
McGlynn, P., Mandi, A. A. and Lloyd, R. G. (2000). Characterisation of the
catalytically
active form of RecG helicase. Nucleic Acids Res 28, 2324-32.
Mitra, R., Church, G. (1999). In situ localized amplification and contact
replication of
many individual DNA molecules. Nucl Acids Res 27(24): e341-vi
Morel, P., Chemy, D., Ehrlich, S. D. and Cassuto, E. (1997). Recombination-
dependent
repair of DNA double-strand breaks with purified proteins from Escherichia
coli. J Biol Chem
272, 17091-6.
Morrical SW, Wong ML, Alberts BM. J Biol Chem. 1991 Jul 25;266(21):14031-8.
Amplification of snapback DNA synthesis reactions by the uvsX recombinase of
bacteriophage
126
CA 3041471 2019-04-26
T4.
Morrical and Alberts J Biol Chem. 1990 Sep 5;265(25):15096-103. The UvsY
protein of
bacteriophage T4 modulates recombination-dependant DNA synthesis in vitro.
Morris PD, and Raney 10. (1999). DNA helicases displace streptavidin from
biotin-
labeled oligonucleotides. Biochemistry Apr 20;38(16):5164-71
Ng, J. Y. and Marians, K. J. (1996a). The ordered assembly of the phiX I74-
type
primosome. I. Isolation and identification of intermediate protein-DNA
complexes. J Biol Chem
271, 15642-8.
Ng, J. Y. and Marians, K. J. (1996b). The ordered assembly of the phiXI 74-
type
primosome. H. Preservation of primosome composition from assembly through
replication. J
Biol Chem 271, 15649-55.
Paulus, B. F. and Bryant, F. R. (1997). Time-dependent inhibition of RecA
protein-
catalyzed ATP hydrolysis by ATP-gammaS: evidence for a rate-determining
isomerization of
the RecA- ssDNA complex. Biochemistry 36, 7832-8.
Pharn, P., Bertram, J. G., O'Donnell, M., Woodgate, R. and Goodman, M. F.
(2001). A
model for SOS-lesion-targeted mutations in Escherichia coil. Nature 409, 366-
70.
Pierre, A. and Paoletti, C. (1983). Purification and characterization of RecA
protein from
salmonella typhimurium. J Biol Chem 258, 2870-4.
Rashid, N., Morikawa, M., Kanaya, S., Atomi, H. and Imanaka, T. (2001).
RecA/Rad5 I
homolog from Thermococcus kodakaraensis KODI. Methods Enzymol 334, 261-70.
Reddy, Weitzel and Von Hippel, Proc Natl Acad Sci U S A. 1993 Apr
15;90(8):3211-5
Riddles PW, Lehman W. J Biol Chem. 1985 Jan 10;260(1):170-3
Rosselli, W. and Stasiak, A. (1990). Energetics of RecA-mediated recombination
reactions. Without ATP hydrolysis RecA can mediate polar strand exchange but
is unable to
recycle. J Mol Biol 216, 335-52.
Salinas F, Jiang H, Kodadek T. J Biol Chem. 1995 Mar 10;270(10):5181-6.
Homology
127
CA 3041471 2019-04-26
dependence of UvsX protein-catalyzed joint molecule formation.
Scheerhagen et al., J Biomol Struct Dyn. 1986 Apr;3(5):887-98
Shan, Q., Bork, J. M., Webb, B. L., Inman, R. B. and Cox, M. M. (1997). RecA
protein
filaments: end-dependent dissociation from ssDNA and stabilization by Rec0 and
RecR
proteins. J Mol Biol 265, 519-40.
Singleton, M. R., Scaife, S. and Wigley, D. B. (2001). Structural analysis of
DNA
replication fork reversal by RecG. Cell 107, 79-89.
Spies, M., Kil, Y., Masui, R., Kato, R., Kujo, C., Ohshima, T., Kuramitsu, S.
and
Lanzov, V. (2000). The RadA protein from a hyperthermophilic archaeon
Pyrobaculum
islandicum is a DNA-dependent ATPase that exhibits two disparate catalytic
modes, with a
transition temperature at 75 degrees C. Eur J Biochem 267, 1125-37.
Steffen, S. E. and Bryant, F. R. (2000). Purification and characterization of
the RecA
protein from Streptococcus pneumoniae. Arch Biochem Biophys 382, 303-9.
Tang, M., Pham, P., Shen, X., Taylor, J. S., Donnell, M., Woodgate, R. and
Goodman,
M. F. (2000). Roles of E. coli DNA polymerases IV and V in lesion-targeted and
untargeted
SOS mutagenesis. Nature 404, 1014-8.
Tissier, A. F., Lopez, M. F. and Signer, E. R. (1995). Purification and
characterization of
a DNA strand transferase from broccoli. Plant Physiol 108, 379-86.
Tracy RB, Kowalczykowski SC (1996). In vitro selection of preferred DNA
pairing
sequences by the Escherichia coli recA protein. Genes Dev. Aug 1;10(15):1890-
903.
Vester B, Wengel J., Biochemistry. 2004 Oct 26;43(42):13233-41Villemain, et
al. J Biol
Chem. 2000 Oct 6;275(4431496-504
Volodin AA, Camerini-Otero RD (2002). Influence of DNA sequence on the
positioning
of RecA monomers in RecA-DNA cofilaments. J Biol Chem. Jan 11;277(2):1614-8.
Volodin AA, Smirnova HA, Bocharova TN, Camerini-Otero RD (2003). Phasing of
RecA monomers on quasi-random DNA sequence. FEBS Lett. Jul 10;546(2-3):203-8
128
CA 3041471 2019-04-26
Webb, B. L., Cox, M. M. and Inman, R. B. (1995). An interaction between the
Escherichia coli RecF and RecR proteins dependent on ATP and double-stranded
DNA. J Biol
Chem 270, 31397-404.
Webb, B. L., Cox, M. M. and Inman, R. B. (1997). Recombinational DNA repair:
the
RecF and RecR proteins limit the extension of RecA filaments beyond single-
strand DNA gaps.
Cell 91, 347-56.
Webb, B. L., Cox, M. M. and Intnan, R. B. (1999). ATP hydrolysis and DNA
binding by
the Escherichia coli RecF protein. J Biol Chem 274, 15367-74.
West, S. C., Countryman, J. K. and Howard-Flanders, P. (1983). Purification
and
properties of the RecA protein of Proteus mirabilis. Comparison with
Escherichia coli RecA
protein; specificity of interaction with single strand binding protein. J Biol
Chem 258, 4648-54.
Wetmur, J. G., Wong, D. M., Ortiz, B., Tong, J., Reichert, F. and Gelfand, D.
H. (1994).
Cloning, sequencing, and expression of RecA proteins from three distantly
related thermophilic
eubacteria. J Biol Chem 269, 25928-35.
Wittwer CT, Hermann M., Moss AA, and Rasmussen RP. Continuous fluorescence
monitoring of rapid cycle DNA amplification. Biotechniques. 1997 Jan;22(1):130-
1, 134-8
Wittwer C.T., Ririe KM, Andrew RV, David DA, Gundry RA, Balis UJ. The
LightCycler: a microvolume multisample fluorimeter with rapid temperature
control.
Biotechniques. 1997 Jan;22(1):176-81Xu L, Marians KJ. J Biol Chem. 2002 Apr
19;277(16):14321-8.
EXAMPLES
As shown herein, we have developed an in vitro DNA amplification system that
couples
recombinase-driven sequence targeting with strand-displacement synthesis. This
permits DNA
amplification without global thermal, chemical, or enzymatic template melting.
Reactions are
sensitive, specific and operate at 37 C with no pre-treatment of sample DNA.
As much as 1012-
fold amplification is observed within 1-11/2 hours. Less than 10 copies of a
given target DNA
can be detected in a complex sample with a simple single-step reaction. This
method is an ideal
129
CA 3041471 2019-04-26
alternative to PCR for a variety of applications and will enable highly
portable DNA diagnostic
systems.
The examples are presented in order to more fully illustrate the preferred
embodiments
of the invention. These examples should in no way be construed as limiting the
scope of the
invention, as encompassed by the appended claims.
Example I: An example of a leading strand recombinase-polymerase amplification
(IsRPA)
DNA sequences can be amplified using leading strand synthesis according to the
Recombinase-Polymerase amplification (RPA) method depicted in Figure 1. Figure
1 shows
RecA/primer loading. Prior to the addition of template DNA and/or Polymerase,
RecA and SSB
will compete for binding to single-stranded oligonucleotide primers. In the
presence of a RecR
and RecO, RecA is selectively stabilized onto the single-stranded primers
forming RecA
nucleoprotein filaments in a complex with Rec0 and RecR. This complex is
competent to
invade double-stranded DNA to form a D-loop at sites homologous to the
oligonucleotide
primers. Alternatively, RecA, Rec0 and RecR can be pre-loaded onto
oligonucleotide primers
prior to the introduction of SSB to the reaction mixture.
The following details the likely composition of an RPA reaction assembled with
E.coli
recA and E.coli red) and recR stabilizing agents: =
D-LOOP FORMATION/RESOLUTION COMPONENTS:
Component Concentration
RecA 20 A1
Single-stranded oligonucleotide 0.25 01
primers
ATP 3 mM
RecF 0.1 11.1sA
Red) 0.13 p.M
RecR 0.5 1.IM
Single-stranded Binding protein 1 to 10
(SSB)
DNA polymerase V 5 units
130
CA 3041471 2019-04-26
POLYMERASE/HELICASE/RESOLVASE Mix:
Component Concentration
DNA Po lymerase 5 units
Ruv A 0.5 1.11µ4
RuvB 0.5 piM
RuvC 0.51.1M
RecG 10 nM
REACTION BUFFER:
Component Concentration
MgCl2 2 to 10mM
TrisHC1 pH 7.2 50 mM
DTT 0 to 10 mM
KC1 0 to 50 mM
Deoxyribonucleotide triphosphates 0.2 niM
Bovine serum albumin (BSA) 0 to 10 pg per ml
The reaction is assembled so that the final concentration satisfies the D-Loop
Formation/Resolution Components, Polymerase/Helicase/Resolvase Mix, and
Reaction Buffer
with the DNA polymerase and/or template added last if necessary. For example,
a 2X
concentrated solution of D-Loop Formation/Resolution Components and of the
Polymerase/Helicase/Resolvase Mix may be made in 1 X reaction buffer. The
reaction may be
initiated by mixing an equal volume of each of the two components (each in Ix
reaction buffer).
Optionally, and as stated above, the DNA polymerase or template (target DNA)
may be added
last. The reaction is incubated for a sufficient time of until the reactants
are exhausted. Typical
incubation times would range from 1 hour, 2 hours, 3 hours, 5 hours, 10 hours
or overnight
(about 16 hours). Unlike PCR, which requires small volumes for rapid
temperature change,
there is no limit to the reaction volume of RPA. Reaction volumes of 25 IA, 50
Ill, 100 iii, 1 ml,
ml and 100 ml or larger may be performed in one vessel. Incubation temperature
may be a
typical laboratory temperature such as 25 C, 30 C, or 37 C.
Prior to the addition of template DNA and/or Polyrnerase, recombinase and SSB
will
compete for binding to single-stranded oligonucleotide primers. In the
presence of a RecR and
131
CA 3041471 2019-04-26
RecO, RecA is selectively stabilized onto the single-stranded primers forming
RecA
nucleoprotein filaments in a complex with RecO and RecR. This complex is
competent to
invade double-stranded DNA to form a D-loop at sites homologous to the
oligonucleotide
primers. Alternatively, RecA, RecO, and RecR can be pre-loaded onto
oligonucleotide primers
prior to the introduction of SSB to the reaction mixture (Figure 1).
The invading strands will be extended by the polymerase in a 5' to 3'
direction. As D-
loops are formed and synthesis proceeds, displaced single stranded DNA becomes
coated with
SSB. RecA release from double-stranded DNA can occur via ATP hydrolysis in a
5' to 3'
direction or as a result of helicase/resolvase or polymerase activity (Figure
2A, B). New rounds
of invasion/synthesis will continuously occur. The third round of strand-
invasion/synthesis will
release discrete products released whose ends correspond to the two facing
primer sites. These
fragments will soon become the dominant reaction product and will accumulate
to high levels.
As each synthetic complex processes to the end of the template RecA protein is
displaced either
by polymerase activity or by the activity of helicases, such as RuvAB or
resolvases, such as
RuvC. Once primers, ATP, deoxynucleoside triphosphates, or any other limiting
component is
exhausted, the reaction will stop.
The inclusion of temperature-sensitive recombinase mutants will allow the
controlled
initiation of DNA synthesis. In such a situation, the initiation reaction is
performed at 25 to
37 C permitting the formation of D-loops. Elongation reactions are performed
at 42 C, which is
non-permissive for RecA mediated double-strand invasion. The number of cycles
will
determine the amount of reaction product. Extended elongation phases will
permit the
amplification of extremely long DNAs without interference of re-invasion.
Example 2: Nested RPA
The RPA reaction is performed as described in Example 1. A fraction of one
tenth
(1/10) and one hundredth (1/100) of the reaction is removed and used in place
of the DNA
template in a second round of RPA. LsRPA, leading/lagging RPA, and
combinations thereof
may be used for nested RPA.
132
CA 3041471 2019-04-26
Example 3: Simultaneous leading and lagging strand recombinase-polymerase
amplification
DNA sequences can be amplified using simultaneous leading and lagging strand
synthesis according to the Recombinase-Polymerase amplification (RPA) method
depicted in
Figure 2. This figure specifically illustrates lsRPA. Figure 2A shows that
RecA/primer
nucleoprotein filaments invade double stranded template DNA preferentially
associating with
homologous target sites. As D-loops are formed and synthesis proceeds,
displaced single
stranded DNA becomes coated with SSB (Figure 2A). RecA release from double-
stranded DNA
can occur via ATP hydrolysis in a 5'-3' direction or as a result of
helicase/resolvase or
polymerase activity (Figure 2A). As synthesis continues (Figure 23),
polymerases encounter
SSB bound, displaced single-stranded template. Double-stranded target sites
are re-invaded by
RecA/primer nucleoprotein filaments. Subsequent rounds of lsRPA proceed from
re-invaded
sites (Figure 2B).
The following details likely components of a replisome-mediated amplification
utilizing
components from E. coil. A reaction is assembled with the following
composition:
D-LOOP FORMATION/RESOLUTION COMPONENTS
Component Concentration
RecA 20 OA
Single-stranded oligonucleotide 0.25 1.11µ4
primers
ATP 3 mM
RecF 0.1 RM
Rec0 0.13 uM
RecR 0.5 uM
Single-stranded Binding protein 1 to 10 tilkA
(SSB)
DNA polymerase V 5 units
HELICASE/RESOLVASE MIX
Component Concentration
RuvA 0.5 uM
RuvB 0.5 uM
133
CA 3041471 2019-04-26
RuvC 0.5 NI
RecG 10 nM
PRIMOSOME COMPLEX
Component Concentration
PriA 20 nM
PriB 20 nM
DnaT 100 nM
DnaB 100 nM
DnaC 200 nM
DnaG 200 nM
DNA POLYMERASE III HOLOENZYME COMPLEX
Component Concentration
0-C1amp 2 M
DnaX Clamp Loader 500 nM
Polymerase Core Complex 500 nM
LAGGING STRAND MIX
Component Concentration
DNA polymerase I 5 units
DNA ligase 2 units
REAMON BUFFER
Component Concentration
MgCl2 2 to 10 mM
TrisHC1 pH 7.2 10 to 60 mM
DTT 0 to 10 mM
KCI 0 to 50 mM
Deoxyribonucleotide triphosphates 0.2 to 0.4 mM
Bovine serum albumin (BSA) 0 to 101.1g per ml
The reaction is assembled so that the final concentration of all the reagents
is as listed
above. Thus, for example, a 5 fold concentrated solution of each of the
components (D-loop
Formation/Resolution Components, Helicase/Resolvase Mix, Primosome Complex,
DNA
Polymerase III holoenzyme Complex, Lagging Strand Mix) is made in 1 X reaction
buffer.
134
CA 3041471 2019-04-26
Then, the five solutions are mixed together in equal volumes to initiate the
reaction. The
reaction is incubated for a sufficient time of until the reactants are
exhausted. Typical
incubation times would range from 1 hour, 2 hours, 3 hours, 5 hours, 10 hours
or overnight
(about 16 hours). As stated above, there is no limit to the reaction volume of
RPA. Reaction
volumes of 25 isl, 50 41, 100 1.11, 1 ml, 10 ml and 100 ml or larger may be
performed in one
vessel. Incubation temperature may be a typical laboratory temperature such as
25 C, 30 C, or
37 C.
Figure 3 shows initiation (Figure 3A), synthesis (Figure 3B), and polymerase
amplification (Figure 3C-3D). First, the primosome loads onto the D-loop
formed by RecA
nucleoprotein filament invasion (Figure 3A). The primosome synthesizes a
stretch of RNA
primer. Finally, the primosome recruits the clamp loader, which recruits both
the sliding clamp
dimer and the asymmetric DNA polymerase core (Figure 3A). Synthesis occurs
simultaneously
in both the leading and lagging directions. Eventually lagging strand
synthesis stops and the
lagging strand clamp is unloaded (Figure 3B). Synthesis of the leading strand
continues until a
new site of lagging stand synthesis is formed (Figure 3B). While leading
strand synthesis
continues, a new site of lagging stand synthesis is formed. Lagging strand
synthesis continues
back to the previous Okazaki fragment where the lagging strand clamp is
unloaded (Figure 3C).
DNA Polymerase I removes the RNA primer, and fills in the gap while DNA ligase
connects the
two Okazaki fragments forming a continuous lagging strand (Figure 3D).
Example 4: Establishment of an amplification environment using the assembly of
heterologous components E. coli recA(C) and T4 gp32(N)
Figure 18 shows the results of an experiment in which recA(C) has been
combined with
gp32(N) in the presence of pairs of oligonucleotides, Tester3bio (possessing a
5' biotin label)
and Sizerl, Sizer2, Sizer3, Sizer4 or Tester2. These latter unbiotinylated
oligonucleotides were
positioned progressively further away from the common Tester3bio
oligonucleotide. The
template was a linear DNA fragment, approximately 300 bp, released from a
plasmid.
Tester3bio was designed to be complementary to one end of this fragment and
included a 5'
overhang relative to this sequence.
135
CA 3041471 2019-04-26
The reaction buffer included Magnesium acetate at 10 mM, required to support
recA
binding to DNA, and 3 mM ATP. Also included was an ATP regeneration system,
comprising
phosphocreatine and creatine kinase, as well as dNTPS at 200 M, and the
Klenow fragment of
E. coli DNA polymerase I. PEG compound was employed as shown. Double stranded
template
DNA (0.5 fmoles), derived from a plasmid carrying the E. coli ruvB gene, was
used as a starting
target. The Sizerl, Sizer2, Sizer3, and Sizer4 oligonucleotides did not
recognise the other end of
the template. Instead, these oligonucleotides were positioned to face
Tester3bio with increasing
distance between their relative 3' ends.
After an incubation of 2 hours at 37 C, there was a substantial amplification
of specific
fragments of the correct size when Tester2, 3, and 4 were used. In the best
conditions (with
Sizer2), we estimated that the= amplification product were 104 fold greater
than the starting
template.
Example 5: The nature of amplification products and the sensitivity of the
reaction using
a heterologous assembly of E. coli recA(C) and bacteriophage T4 gp32(N)
Figure 19 shows the results of an experiment in which recA(C) has been
combined with
gp32(N) in the presence of the pair of oligonucleotides, Tester3bio
(possessing a 5' biotin label)
and Sizer2, under conditions similar to those used in Example 1. PEG compound
or PEG 1450
were employed as shown and 0.5 fmoles of template was used as a starting
template amount.,...In
this example, progressive dilution of the template was investigated.
Alternatively we explored
the use of linearised starting template possessing no end that overlaps the
primer (by using a
ClaI digest of the E. coli ruvB gene carrying plasmid), and dilution of the
Klenow fragment.
Amplification of correctly sized fragments occurred in all lanes and was
strongest in the case of
0.5 frnoles-starting template in the presence of PEG compound.
When the products of these optimal reactions were electrophoresed on agarose
gels and
stained with ethidium bromide, a clean band of double-stranded DNA of the
correct size was
observed. When this sample was treated with BbvC1 restriction enzyme prior to
electrophoresis
the expected increase in gel mobility occurs consistent with a single cut as
expected.
Amplification of a product of the correct size was observed with starting
template dilutions of
136
CA 3041471 2019-04-26
100-fold, or greater, although the product was less abundant and includes a
ladder of shorter
products below the main band. A similar pattern was observed when uncut
template is
employed or when no template is employed. We reasoned that the proteins used
in these studies
were significantly contaminated with E. coil genomic DNA (naturally carrying
the ruvB gene) as
they were purified in single column purifications without the use of
nucleases. Consequently we
believe this test system generates false positives when the sensitivity is
high enough.
Example 6: Establishment of an amplification environment using an assembly of
gp32(N) and uvsX(C)
Figure 24 shows results of an experiment in which uvsX(C) has been combined
with
gp32(N) in the presence of the oligonucleotides Tester3bio and Sizer2. The
template DNA in
this experiment was an EcoRV digestion of the E. coil ruvB gene carrying
plasmid used in
Examples 1 and 2. Tester3bio recognised one end of an approximately 300 base
pair fragment
and included a 5' overhang relative to the end of the target sequence. Sizer2
recognised the
other strand of this template. This oligonucleotide was directed toward
embedded sequences
such that its 3' end was about three and a half helical turns from the end of
Tester3bio.
In the presence of PEG1450, we observed the amplification of the expected
fragment
within the 2 hours of the reaction. In the cases where amplification has
occurred, almost the
entire of population of oligonucleotides was consumed indicating an
amplification of 3-5 X 104.
The reaction components are indicated on Figure 24. Included in some samples
were additional
components. We found that 200 AM ADP-P-S included in this reaction slightly
increased the
amount of product formed under these conditions. Conversely, under the
conditions used here,
inclusion of E. coil toposiomerase I was inhibitory to DNA amplification.
Under the conditions
used, we detected no amplification with uvsX(C)delta protein. However, no
PEG1450 was
included in these samples and uvsX(C) also failed to amplify under these
conditions without
PEG1450.
137
CA 3041471 2019-04-26
Example 7: Amplification of a target from human genomic DNA using T4
recombination proteins
Figure 30 shows the results of an experiment in which several pairs of primers
were
employed to amplify a specific DNA fragment from human genomic DNA. The
reaction
included bacteriophage 14 gp32(C)K3A, uvsX(C) and uvsY(N) proteins, as well as
exonuclease
deficient Klenow fragment, and proteins comprising the ATP regeneration system
to convert
ADP and AMP. To detect the specific DNA fragment, we transferred the
electrophoretically
separated reaction products to nylon membrane, then hybridised a biotinylated
probe, which
recognised a unique non-primer internal sequence.
Three primer pairs were employed, and in each case a comparison was made
between no
input genomic DNA, 10,000 copies of uncut human genomic DNA, and 10,000 copies
of HpaII
cut genomic DNA (which generates at least one end for the primer pairs). In
all cases, specific
amplification of the desired DNA sequence occurred, while the efficiency
showed variation
between primer pairs, and between uncut and cut DNAs. In all cases, prior
Hpall digestion of
the DNA sample was not absolutely required, but improved the efficiency of
amplification. In
all cases, input genomic DNA was important. In the best amplification (shown
in lane 4), we
estimated at least 1011 molecules, indicating an amplification of the
approximate order 107.
. Example 8: Sensitivity of lsRPA when targeting a complex DNA - human
genomic
DNA
Figure 31 shows the results of an experiment in which several pairs of primer
were
employed to amplify a specific DNA fragment from human genomic DNA. The
reaction
included bacteriophage T4 gp32(C)K3A, uvsX(C) and uvsY(N) proteins, as well as
an
exonuclease deficient Klenow fragment, and comprising the ATP regeneration
system to convert
ADP and AMP. To detect the specific DNA fragment, we transferred the
electrophoretically
separated reaction products to nylon membrane. Then we hybridised a
biotinylated probe,
which recognises a unique non-primer internal sequence.
Three primer pairs were employed and in each case a comparison is made between
no
starting template and approximately 10, 100, 1000, 3000, and 10,000 copies of
the genomic
138
CA 3041471 2019-04-26
target. In all cases, clear amplification was detected when at least 1000
copies of the genomic
target were used (a weak signal is seen with the best primer pair at 100
copies). We concluded
that during that lsRPA reactions configured in this way were capable of
amplifying DNA from
very complex targets with a sensitivity of at least 1000 copies, and
potentially higher.
Example 9: Competition between the accumulation of bona fide product and
primer
(template-independent) artifacts during reactions
Figure 32 shows the results of an experiment in which a pair of primers was
employed to
amplify a specific DNA fragment from human genomic DNA. Employed in the
reaction were
bacteriophage T4 gp32(C), uvsX(C) and uvsY(N) proteins, as well as an
exonuclease deficient
Klenow fragment, and proteins comprising the ATP regeneration system to
convert ADP and
AMP. PEG 1450 was included at 10%w/v. One of the oligonucleotides included a
5'-biotin so
that all reaction products could be observed at the end of the amplification.
Samples were taken
at 1, 2 and 3 hours to observe how the reaction progressed. In one sample,
when a minimal
amount of uvsY(N) was employed (50 ng,/ 1), amplification of the correct
fragment was
observed (see arrow in lane 4). This fragment was cleaved by BstXI to the
expected size
fragment, indicating it was principally double-stranded. However the fragment
was less
abundant than apparently template-independent bands that also accumulated
during the reaction.
The size and template-independent nature of these bands suggested that they
were primer
artifacts, e.g., primer dimers and/or snapback synthesis products. The absence
of amplification
of the specific fragment suggested that, at uvsY(N) concentrations greater
than 50 ng/}.1.1, the
reaction occurred suboptimally. This was borne out by later experiments.
Example 10: Optimisation of reaction composition to severely limit or
eliminate primer
artifacts and enable sensitive noise-free amplification from complex templates
Figure 36 shows the results of an experiment in which a pair of primers was
employed to
amplify a specific DNA fragment from human genomic DNA. Employed in the
reaction were
bacteriophage T4 gp32(C), uvsX(C) and uvsY(N) proteins, as well as an
exonuclease deficient
Klenow fragment, or Bst polymerase, and proteins comprising the ATP
regeneration system to
139
CA 3041471 2019-04-26
convert ADP and AMP. One of the oligonucleotides included a 5'-biotin so that
all reaction
products could be observed at the end of the amplification. Amplified
fragments were visualised
following separation of fragments by size by running a small sample of the
reaction on an
acry1amide gel. In this experiment, uncut human genomic DNA was titrated from
zero copies,
45 copies, and then doublings in target copy number up to 2880. Slightly
different conditions
were employed in this experiment for each of the two polymerase species with
regard to both
buffer and temperature. The reaction with the Klenow fragment was performed at
37 C, while
that with Bst polymerase was performed at 42 C. The details of the buffer
composition are
given in the figure description.
Of note, and important to the efficiency of reactions under these optimised
conditions,
PEG compound was included at 5% final weight to volume in both cases. Both
polymerases
have effectively amplified the correct fragment, and in some cases, utilised
most of the available
primers. Under the conditions used for the Klenow fragment, the sensitivity
was so great that a
weak signal was observed even in the zero copies lane presumably reflecting
contamination with
a quantity of human DNA representing less than the 45 copies present in the
lane immediately
adjacent. At the level of sensitivity that is demonstrated here, it was
difficult to eliminate trace
levels of contamination from the equipment that was used leading to signals in
the negative
controls. Routine employment of conditions similar to those utilised in the
Klenow-mediated
amplification proved effective for noise-free amplification of numerous primer
pairs in later
experiments. This suggested that these conditions were close to one optimum
for reactions
involving this set of protein components.
Example 11: Experimental methods for production of clones and proteins
All clones have been constructed by cloning PCR amplified products from E.
colt, 14
phage, B. subtilis, or Phi-29 phage. All stock organisms used for
amplification were obtained
from a public source at the DSMZ. Cloned DNA's used for protein expression
have in general
been cloned into pET vectors (e.g., pET-21) with the insertion of a
hexahistidine peptide tag at
either the N or C terminus during the PCR amplification of the fragment, or
into pQE vectors
(e.g., pQE31) in the case of Pol I from B. subtilis (Bsu polymerase). In this
disclosure all
140
CA 3041471 2019-04-26
proteins containing an N terminal tag are referred to as the protein name
followed by (N), e.g.
gp32(N), or if containing the tag at the C terminus the name is followed by
(C), e.g. gp32(C).
Additionally we have constructed several clones to produce otherwise modified
proteins. These
include a recA(C) with a deletion of the last 17 amino acid residues of the
native protein,
referred to as recA(C)de1ta17. A similar form of the T4 UvsX(C) protein has
been generated
and is referred to as UvsX(C)delta 21. We have also constructed mutant forms
of gp32, which
modify either lysine 3 or arginine4.
All proteins were overexpressed in E. coli and purified using conventional
protocols.
Proteins have generally been purified by standard procedures on Nickel resin
in 1 M NaC1 and
phosphate buffer. Proteins were eluted with 250 mM imidazole and dialysed into
appropriate
buffers. Proteins produced from clones generated in-house include: E. coli
recA(C), E. coli
SSB(N), E. coli PriA(N), E. coli PriB, E. coli PriC, E. coli DnaB, E. coli
DnaC, E. coli
DnaC810, E. coli DnaT, E. coli RuvA, E. coli RuvB, T4 phage UvsX(C), 14 phage
UvsX(N),
T4 gp32(N), T4 gp32(C), 14 gp32(C)K3A, T4 phage gp32(C)R4Q, T4 phage
gp32(C)R4T, T4
phage gp32, T4 phage gp32 K3A, 14 phage gp32R4Q, 14 phage gp32R4T, T4 phage
UvsY(N),
T4 phage UvsY(C), T4 phage gp43, T4 phage gp43(exo-), E. coif Klenow fragment,
E. coli
Klenow exo-. Untagged gp32 proteins were purified by a 2-column procedure
involving DEAE
sepharose anionic exchange followed by binding to single-stranded DNA
cellulose matrix.
DNAs used in RPA reactions.
We have employed several different target DNAs in this study, and a number of
oligonucleotides. The sequence of the relevant section of the templates, and
the sequence of the
oligonucleotides is given below.
The E. coli RuvB gene target
The sequence of the EcoRV fragment of the RuvB gene is given below.
ATCATGATTGGTGAAGGTCCGGCGGC ACGCTCCATTAAAATTGATTTGCCGC
CGTTTACCCTGATTGGTGCAACCACGCGCGCAGGTTCGCTGACATCACCGTTGCGCG
ACCGTTTTGGTATTGTGCAACGTCTGGAGTT7TATCAGGTGCCGGATCTGCAATATA
141
CA 3041471 2019-04-26
TCGTCAGTCGCAGCGCACGCTTTATGGGGCTTGAGATGAGTGATGACGGCGCGCTG
GAAGTTGCTCGTCGCGCTCGCGGTACGCCGCGCATTGCCAACCGTCTGCTGCGTCGA
GTGCGTGATTTCGCCGAAGTGAAGCACGATGGCACCATCTCGGCAGAT (SEQ ID
NO:1)
The sequence of oligonucleotides targeting this template mentioned in this
study are
given below:
Tester2
CTAGCGATGGTGCCATCGTACAGAATTCCCTCAGCATCTGCCGA (SEQ ID NO:2)
Tester3
CTCACTATACCTCAGCATCATGATTGGTGAAGGTCCGGCGGCAC (SEQ ID NO:3)
Tester 1 bio
'-biotin-GCTAATACGACTCACTATACCTCAGCATCATGATTGGTGAAGGTC CGGCGGCAC
(SEQ ID NO:4)
Tester3bio
5'-biotin-CTCACTATACCTCAGCATCATGATTGGTGAAGGTCCGGCGGCAC
(SEQ ID NO:5)
Sizer 1
CTATGCGAATTCAGCGAACCTGCGCGCGTGGTTGCACCAATCAGGG (SEQ ID:6)
Sizer2
CTATGCGAATTCGGTGATGTCAGCGAACCTGCGCGCGTGGTTGCA (SEQ ID NO:7)
Sizer3
CTATGCGAATTCTCCAGACGTTGCACAATACCAAAACGGTCGCGC (SEQ ID NO:8)
Sizer4
CTATGCGAATTCCGTGCGCTGCGACTGACGATATATTGCAGATCC (SEQ ID NO:9)
Gen2bio
5'-biotin-ATCTGCCGAGATGGTGCC (SEQ ID NO:10)
142
CA 3041471 2019-04-26
The sequence of part of the human angiotensin converting enzyme targeted in
this study
is shown below:
AACCAACTCCGCCCCGGGCCACGGCCTCGCTCTGCTCCAGGTACTTTGTCAG
CTTCATCATCCAGTTCCAGTTCCACGAGGCACTGTGCCAGGCAGCTGGCCACACGG
GCCCCCTGCACAAGTGTGACATCTACCAGTCCAAGGAGGCCGGGCAGCGC (SEQ ID
NO:11)
Underlined are HpaIl restriction sites that have been targeted with Hpall in
the
preparation of some DNAs in some experiments.
The sequence of oligonucleotides used to target part of the human ACE gene are
shown
below:
Up3
ATTCGTCAGCCTCGCTCTGCTCCAGGTACTTTGTCAGCTTCATC (SEQ ID NO:12)
Downl
GCCTCCTTGGACTGGTAGATGTCACACTTGTGC (SEQ ID NO:13)
Down2
GCGCTGCCCGGCCTCCTTGGACTGGTAGATGTCACACTTGTGC (SEQ ID NO:14)
Down3
TATGCGAATTGCCTCCTTGGACTGGTAGATGTCACACTTGTGC (SEQ ID NO:15)
Angiolbio
5'-biotin-GCCTCCTTGGACTGGTAGATGTCACACTTGTG (SEQ ID NO:16)
Angio3
GGCCACGGCCTCGCTCTGCTCCAGGTACTTTGTCAGCTTCATC (SEQ ID NO:17)
Example 12: Experimental Results and Analysis
Figure 9 shows the results from investigations into the nature of double-
stranded DNA
targets and targeting oligonucleotides. Experiments using either supercoiled
templates or
linearised DNAs suggested that recA catalyses the formation of intermediates
capable of
143
CA 3041471 2019-04-26
supporting polymerase elongation most readily on supercoiled DNA, or the ends
of linearised
DNA. Shown are the results of an experiment in which the biotinylated
oligonucleotide,
Tester3bio, has been incubated with either supercoiled target DNA, or a target
template
linearized with EcoRV, or ClaI. This generated an end that overlapped with the
oligonucleotide
or embedded sequences respectively. The reaction solution included 20 mM Tris-
acetate pH
7.9, 10 mM Mg-acetate, 13 !As rec A, 1 pg E. coil SSB, 27 mM phosphocreatine,
I U creatine
kinase, 0.2 0.4 Tester3bio, 3 mM ATP, 200 1./M dG, dC, and dT; 1 mM dA, 50 U
Klenow, 0.5
pmoles template, 120 ng red), 120 ng recR, 0.5 RM dnaB, and 0.5 1.1M dnaC810.
E. coil Tea)
and recR proteins, as well as dnaB and dnaC810 proteins, were included in this
experiment
although they did not significantly affect the results. After 2 hours of
reaction at 37 C, the
reaction was precipitated and run on a 6% denaturing gel, transferred to nylon
membrane, and
incubated with streptavidin-HRP prior to performing ECL to detect reactive
material. In each
reaction, 0.5 pmoles of template was used. Included on the gel as a control
for size and amount
was 0.5 pmoles of biotinylated PCR fragment (labeled CON). Other reaction
components and
conditions are indicated on the figure.
Figure 10 shows backfire synthesis. Backfire synthesis occurs when a
recombinase-
coated targeting oligonucleotide possessing a 5' overhang invades a duplex DNA
end in the
presence of a suitable polymerase and dNTPs. This new duplex region is stable
to subsequent
branch migration and can be utilised as a platform for other applications.
Forward fire is the
elongation of the invading oligonucleotide, which also occurs in these
reactions. Shown are the
results of experiments to detect the activity of polymerases on intermediates
formed when the
oligonucleotide Tester3, possessing a 5' overhang relative to the end of a
linearized target DNA,
is incubated with various templates.
In part A, the template used is a double-stranded PCR product generated such
that the
product has a biotin label at the 5' end of the strand complementary to the
targeting
oligonucleotide. This fragment is otherwise similar to the EcoRV fragment
released from a
plasmid carrying the E. coil RuvB gene used elsewhere in this study, and which
is a target for
the Tester3bio oligonucleotide. The reaction solution included 10 mM Mg-
acetate, 7.5 zg recA,
144
CA 3041471 2019-04-26
1 p.g SSB, 27 iriM phosphocreatine, 1 U creatine kinase, 0.3 M Tester3bio, 3
mM ATP, 200
M dNTPs, 50 U Klenow, 0.5 pmoles biotinylated template. Optionally, we
included 0.5 pAif
ruvA and 0.5 M ruvB; or 1 M ruvA and 1 M ruvB; or 1.5 M ruvA and 1.5 M
ruvB. The
final volume was 30 p.l. Incubation was carried out for 1 hour at 37 C. In the
presence of recA,
the biotinylated strand of the target was extended by 16 bases, as would be
expected if a
recombination intermediate were accessible by a polymerase to copy the
overhang region of the
invading oligonucleotide.
In part B, the reaction is configured in a similar manner except that the
template is not
biotinylated, and the invading oligonuleotide is biotinylated. Several
polymerases were
investigated in this experiment, and only unmodified Klenow fragment gave a
significant
production of product. In this
experiment, we also investigated including a small
oligonucleotide designed to recognise the target directly downstream of the
Tester3 targeting
site. The reaction solution included 10 mM Mg-acetate, 10 g recA, 1 ig SSB,
27 mM
phosphocreatine, 1 U creatine kinase, 0.3 M Tester3bio, 3 mM ATP, 200 l.LM
dNTPs, 50 U
Klenow, 0.5 pmoles unbiotinylated template. Optionally, we included 5 I
preloaded stable
ATM oligonucleotide. The final volume was 30 p.l. Incubation was carried out
for 1 hour at
37 C. We pre-incubated with recA in the presence of ATP-y-S in an effort to
load recombinase
stably onto it. Pre-load solution included 10 mM Mg-acetate, 2.5 fig recA, 50
AM ATM, and
0.15 p.iM oligonucleotide. The pre-
load solution was added into the Tester3bio
invasion/extension mixture. In all cases, the yield of product was decreased
by inclusion of this
premixed material. Based on our data, we believe that the presence of ATP-y-S
(final
concentration ¨8 p.M) in the reaction was mildly inhibitory. The purpose of
this experiment was
to address whether the presence of a stable 3-stranded hybrid formed
immediately downstream
of the Tester3 targeting site would stabilise these invasions to branch
migration.
Figure 11 shows uses of backfire synthesis. Backfire synthesis can be useful
because it
generates a branch migration resistant platform that can be employed in
applications other than
straightforward forward fire. Some examples are shown here, including
introduction of a
nicking enzyme target site, introduction of an RNA polyrnerase promoter, and
the linear
145
CA 3041471 2019-04-26
generation of short dsDNA fragments through successive
invasion/synthesis/cleavage events. If
a restriction enzyme site is included in the additional overhang sequence such
that after targeting
a suitable linearized fragment, backfire synthesis will generate the duplex
target for the
restriction enzyme. The enzyme can then cut the sequence releasing a short
double-stranded
DNA, and a longer double-stranded DNA, which is a target for further invasion
events.
In Figure 11B, the 5' overhang of a targeting oligonucleotide is designed such
that should
backfire synthesis occur, a target for a nicking endonuclease is generated. In
the presence of the
nicking endonuclease, for example BbvCla orb, a suitable polymerase, for
example the Klenow
fragment, can extend from the nick and displace a DNA strand. Multiple strands
may be run-off
by successive nicking and elongation from a single template. In Figure 11C,
the 5' overhand
that is converted to duplex by backfire synthesis contains the sequence of an
RNA polymerase
promoter, such as the phage T7 RNA polymerase gene. In the presence of the
necessary
polymerase and suitable nucleoside triphosphates, transcription can initiate
downstream of the
promoter to generate an RNA as shown. The presence of a break in the non-
template strand is
not predicted to prevent successful elongation. RNA products might be used in
some form of
amplification reaction, or for other purposes.
Figure 12 shows that single stranded binding proteins facilitate recombinase
invasion and
primer extension. Both E. coli SSB and bacteriophage T4 gp32 with an N-
terminal His tag
(gp32(N)) are able to stimulate recA-mediated invasion/elongation on a linear
DNA template.
The results of an experiment are shown in which 0.5 pmoles of target template
(the EcoRV
fragment released from a plasmid carrying the E. coli ruvB gene) was incubated
with the
Tester3bio oligonucleotide that overlaps one end of the template. Either the
E. coli SSB protein,
or the 14 gp32(N) protein was included to stimulate the reaction. The reaction
solution included
mM Mg-acetate, 6 lig rec A, 8.8 lAg gp32 or 1 SSB, 27 mM
phosphocreatine, 1 U creatine
kinase, 0.3 mM Tester3bio, 3 mM ATP, 200 1AM dNTPs, 50 U Klenow, 0.5 pmoles
template.
Optionally, we included 120 ng red) and 120 ng recR. The final volume was 30
ul. Incubation
was carried out for I hour at 37 C. Other reaction components and conditions
are indicated in
the figure. The figure also shows the general relationship of the primer and
target DNA. In the
146
CA 3041471 2019-04-26
reactions where E. coli red) and recR proteins were included, little effect
was seen from their
addition under these conditions. Invasion and elongation appeared to have
proceeded in all
cases, and the gp32(N) appeared to have stimulated synthesis even better than
E. coli SSB,
although it was used at higher concentration in this experiment.
Figure 13 shows the requirement for a minimal oligonucleotide length or
overhang for
invasion and elongation during end targeting of linear templates. The results
of an experiment
are shown in which 0.5 pmoles of target template (the EcoRV fragment released
from a plasmid
carrying the E. coli ruvB gene) was incubated with the either the Tester3bio
oligonucleotide.
This oligonucleotide overlaps one end of the template, or the Gen2bio
oligonucleotide, which is
flush to the other end of the template and is only 18 residues long. The
reaction solution
included 10 mM Mg-acetate, 27 rriM phosphocreatine, 1 U creatine kinase, 0.2
M Tester3bio
or Gen2bio, 10 mM dATP, 3 mM ATP, 200 M dNTP mixture, 50 U Klenow or Phi29
polymerase, 13 g recA(C), I g E. coil SSB, and 0.5 pmoles template. The
final volume was
30 I. Incubation was carried out for 2 hours at 37 C, and 2 j.tl or the
reaction was loaded in
each lane of the gel. Other reaction components, conditions, and the general
relationship of the
primers and target DNA are indicated on the figure. Invasion and elongation
appeared to have
proceeded efficiently in the presence of the Klenow fragment, less efficiently
with the Phi29
polymerase, and less well with the Gen2bio primer and the Klenow fragment. We
concluded
that a minimal primer length and/or an overhand relative to the template was
required to
stimulate efficient invasion and elongation.
Figure 14 shows paranemic (A-E) and pletonemic (F-H) joints. For paranemic
joints, the
interaction of the recombinase filament with DNA stimulates unwinding (Figure
14A). The
unwound region moves with the homology search (Figure 14B). The homology is
found (Figure
14C). The recombinase dissociates and a new duplex attempts to rewind (Figure
14D). Because
it topologically restrained, the 'outgoing' strand is forced to rewind around
the new duplex
(Figure 14E). This state is highly unfavorable and unstable, and cannot always
be managed by
SSBs (Figure 14E). For plectonemic joints, the interaction of the recombinase
filament with
DNA stimulates unwinding (Figure 14F). If strand exchange overlaps with a DNA
end, the
147
CA 3041471 2019-04-26
'outgoing' strand is freed and can relax as the incoming oligo and its
complement rewind as the
recombinase dissociates (Figure 14G). This forms a deconstrained product, and
a single-strand
DNA binding protein inhibits branch migration (Figure 14H).
This figure compares the likely events that occur when a nucleoprotein
filament initiates
strand exchange with a homologous sequence located at the end of a linearized
duplex (right
side of figure), or within a duplex which lacks homology on either side (left
side of figure).
Starting with the left side, once the nucleoprotein filament has located the
correct sequence it
will pair the searching DNA to its complement, and one strand of the original
duplex becomes
unpaired. In fact, the exchange complex consists of 3 strands, which are
relatively under-wound
and stabilised by the recombinase. As the recombinase begins to disassemble in
a 5' to 3'
direction, the under-wound 3-stranded intermediate becomes unstable. For the
new duplex to
regain the normal conformation of relaxed DNA, it must rotate. However, in
doing so, it must
co-rotate the outgoing strand, as it is linked upstream and downstream to its
original partner.
This results in over-winding the outgoing strand, as it has to make the same
number of turns but
take a longer path around the new duplex, and is energetically unfavourable.
Consequently there
is a requirement for single-stranded binding proteins with very stable DNA
interactions to
permit such structures to exist for any significant time. Alternatively the
right side of the
diagram indicates that should the exchange include an end of the duplex then
exchange can
cause the complete release of the outgoing strand at one end and thus permit,
it to rotate freely
unconstrained by the other strands involved in recombination. This leads to a
stable situation in
which the new duplex is free to rewind after recombinase disassembly, and
single-stranded DNA
binding proteins need only deter spontaneous branch migration.
Figure 15 shows the effect of crowding agents. In the presence of polyethylene
glycols,
gp32(N) and recA recombinase can mediate multiple invasion events on single
templates
without a requirement for regeneration of the template ends that would permit
5' overhangs in
the targeting oligonucleotide. Figure 15A shows the results of an experiment
in which either the
Tester] bio, or Tester3bio oligonucleotides (which differ in the length of 5'
overhang relative to
the template) were incubated with the EcoRV fragment released from a plasmid
carrying the E.
coil ruvB gene, or the ClaI digest of the plasmid, in the presence or absence
of 10% PEG 8000.
148
CA 3041471 2019-04-26
The reaction solution included 10 mM Mg-acetate, 10.6 pig recA, 8.8 g gp32,
27 mM
phosphocreatine, 1 U creatine lcinase, 0.3 mM Tester3bio or TesterIbio, 3 mM
ATP, 200 M
dNTP mixture, 50 U Klenow, and 0.5 pmoles template (species indicated in
figure). Optionally,
we included 120 ng rec0 and 120 ng recR. PEG8000 was included as shown. The
final volume
was 30 1. Incubation was carried out for 1 hour at 37 C.
The diagram shown represents the relationship of the oligonucleotides to the
two
possible templates. In particular, both oligonucleotides recognised an
embedded sequence
within the ClaI fragment. In each case, 0.5 pmoles of template was used, other
conditions were
carried out as indicated. Both primers stimulated invasion/elongation on the
EcoRV template.
Based on signal intensity, approximately one elongation occurred per target
template. However,
in the presence of 10% PEG 8000, the intensity of the fully elongated fragment
was significantly
greater than in its absence and stronger than the 0.5 pmoles of control
biotinylated PCR product.
The strongest signal was seen with the Tester3bio oligonucleotide. In that
case, we estimated at
least 10 invasion/run-ons occurred per template.
In Figure 15B, we compared the stimulation of invasion/elongation in 10% w/v
of
various commercially available polyethylene glycols. The reaction solution
included 10 mM
Mg-acetate, 10.6 g recA, 8.8 tig gp32, 27 mM phosphocreatine, 1 U creatine
kinase, 0.3 mM
Tester3bio or Testerlbio, 3 mM ATP, 200 AM dNTPs, 50 U Klenow, and 0.5 pmoles
RV
=
template. PEG species were included as shown. There was significant variation
observed in the
degree of stimulation. PEG compound (MW=15,000 to 20,000) appeared to be the
most
effective, followed by PEG1450.
Figure 16 shows the effect of end targeted amplification using leading strand
RPA.
Amplification comprising several rounds of invasion and extension was
demonstrated, achieving
at least a 10 fold amplification from 0.05 pmoles of template. In this
experiment, we have
employed pairs of oligonucleotide primers to establish an amplification
reaction. Shown
schematically is the relationship of the oligonucleotides used to the EcoRV
fragment from a
plasmid carrying the E. coli nivB gene, which was used as template. The
reaction solution
included 10 mM Mg-acetate, 6 pig recA, 8.8 1.ig gp32(N), 27 mM
phosphocreatine, 1 U creatine
149
CA 3041471 2019-04-26
kinase, 0.3 mM Tester3bio, 0.3 mM variable oligonucleotide, 3 mM ATP, 200 uM
dNTPs, 10%
PEG compound, SO U Klenow, and 0.5 pmoles template. Additional proteins were
used as
indicated.
Tester3bio included a 16-nucleotide overhang relative to the starting
template, while
Tester2 included a 21-nucleotide overhang and was targeted to the other end of
the template.
Phosphol was used as an oligonucleotide with a phosphorothiorate backbone.
This
oligonucleotide was 15 residues long, and was flush to the target end. Phospho-
1 was predicted
not to interact with recombinase or single-stranded DNA binding protein as it
lacked a
phosphate backbone. However, it was predicted to function in straightforward
solution
hybridisation. A control fragment of biotinylated PCR product was employed to
demonstrate
the signal intensity of 0.5 pmoles of DNA, and was also the precise size of
the starting template.
The reaction products were run on a 6% denaturing gel, transferred to nylon,
and bound with
streptavidin-HRP prior to performing enhanced chemiluminescence to reveal the
biotinylated
products of the reactions.
In all cases, successful invasion and elongation with the biotinylated
Tester3bio has
occurred as seen by presence of fully elongated products. The products were
slightly slower
mobility that the control, due to the presence of overhangs on the
oligonucleotides.
Furthermore, there was evidence for several rounds of invasion/nm-ons as the
signal intensity
was at least as great as the 0.5 pmoles control (we initiated the reaction
with only 0.05 pmoles).
There was a significant accumulation of a product roughly 37 nucleotides
larger than the control.
This was predicted to arise from Tester3bio elongating on a strand previously
copied from, and
including the overhang from, the opposing primer. Two exposures of the same
gel are shown.
The inclusion of various different proteins, which are normally involved in
DNA
metabolism, had varying effects. DNA gyrase, and toposiomerase I (human)
decreased the yield
of amplification product, and the topoisomerase profoundly reduced the
generation of shorter
elongation products. Inclusion of E. coli ruvA and ruvB also lead to a general
reduction in
product formation. E. coli priA increased the amount of product formed, and
significantly
increased the number of shorter products formed. Inclusion of E. coil dnaB and
dnaC810
150
CA 3041471 2019-04-26
protein slightly increased the amount of product formed. Note that a
significantly stronger
signal detected in reactions containing Phospho-1 oligonucleotide in
comparison to Tester3bio
alone. This suggested that Phospho-1 was able to hybridise with displaced
strands and lead to
formation of duplex DNA.
Figure 17 shows leading strand RPA and Klenow processivity. In this
experiment, we
have employed pairs of oligonucleotide primers in an effort to establish an
amplification
reaction in a manner similar to that shown in Figure16, except using a further
100-fold dilution
of the start template. The reaction solution included 10 mM Mg-acetate, 6 or
12 pg recA, 8.8 or
14.3 tig gp32, 27 mM phosphocreatine, 1 U creatine kinase, 0.3 or 0.9 01
Tester3bio, 0.3 or 0.9
tiM Tester2, 3 mM ATP, 200 1.1M dNTPs, 10% PEG compound, 50 U Klenow, and 0.5
pmoles
template. The final volume was 30 1. Incubation was carried out for 2 hours
at 37 C. Shown
schematically is the relationship of the oligonucleotides used to an EcoRV
fragment from a
plasmid carrying the E. coil ruvB gene, which is used as target template.
Tester3bio included a
16-nucleotide overhang relative to the starting template, while Tester2
included a 21-nucleotide
overhang, is targeted to the other end of the template, and encodes an EcoRI
site within the
overhang. A control fragment of biotinylated PCR product was employed to
demonstrate the
signal intensity of 0.5 pmoles of DNA, and was also the precise size of the
starting template.
The reaction products were run on a 6% denaturing gel, transferred to nylon,
and bound with
streptavidin-HRP prior to performing enhanced chemiltuninescence to reveal the
biotinylated
products of the reactions.
The concentration of the oligonucleotides, gp32(N) and the recA(C) were
varied, and we
have also investigated whether the inclusion of EcoRI restriction enzyme,
which can cleave part
of the additional sequence incorporated by the Tester2 overhang, has any
effect on the reaction.
In most cases, there was evidence of some limited degree of amplification of
the expected size
of fragment, but there was principally generation of shorter DNA fragments. We
deduced that
the relatively poor accumulation of bona fide full length product may occur at
these more dilute
template concentrations because the poor processivity of the E. coli DNA
polyrnerase I Klenow
fragment (10-50 nucleotides) results in most interactions generating short
fragments, which is
151
CA 3041471 2019-04-26
more significant at low target template concentrations.
Figure 18 shows spacing dependence of RPA primers. As a consequence of earlier
results, we attempted to establish whether decreasing the distance between
primer pairs would
result in an increase in amplification efficiency. To test this, we employed a
series of
oligonucleotides, Sizerl , 2, 3, and 4, which were positioned at increasing
distances away from
the 3' end of the Tester3bio oligonucleotide. All Sizer oligonucleotides
included the EcoRI
overhang indicated in the bottom right side of the figure. The sequence of the
target DNA, an
EcoRV fragment from a plasmid carrying the E. coli ruvB gene, and the position
of the
oligonucleotides used are shown. The reaction solution included 10 mM Mg-
acetate, 6 ttg recA,
8.8 j.ig gp32, 27 mM phosphocreatine, 1 U creatine kinase, 0.3 1.1M
Tester3bio, 0.3 uM variable
oligonucleotide, 3 mM ATP, 200 1AM dNTPs, 10% PEG compound, 50 U Klenow, 5 U
EcoRI,
and and 0.5 pmoles template. The final volume was 30 IA The reaction solution
was incubated
for 2 hours at 37 C. Other reactions conditions are indicated on the figure.
A control fragment of biotinylated PCR product was employed to demonstrate the
signal
intensity of 0.5 pmoles of DNA, and was also the precise size of the starting
template. The
reaction products were run on a 6% denaturing gel, transferred to nylon, and
bound with
streptavidin-HRP prior to performing enhanced chemiluminescence to reveal the
biotinylated
products of the reactions. Specific fragments of the expected lengths were
efficiently amplified
from 0.5 fmoles of starting template when Sizer2, 3, and 4 were employed. The
yield of product
decreased somewhat however as the length of the product increased. Little or
no product of the
expected size is produced when Sizerl was used. Lane 4 was estimated to
contain ¨4 X 104 fold
amplification. This primer included the shortest inter-oligonucleotide
distance of only 25
nucleotides. This suggested there is a minimal required distance to separate
the ends of the
oligonucleotides, although other explanations such as poor primer behaviour
could also explain
the result. Included in the experiment were several samples containing no
template DNA. In the
case of Sizer2, a faint band of approximately the expected size is observed
even in the absence
of exogenous DNA. Based on a variety of data we believe that this arises from
contamination of
our protein preparations with significant quantities of E. coil genomic DNA.
152
CA 3041471 2019-04-26
Figure 19 shows that RPA products are largely double stranded. RPA reaction
can
generate double-stranded DNA products as evidenced by agarose gel
electrophoresis and
restriction enzyme cleavage. However, under the conditions used here, there
were significant
decreases in reaction efficiency if the starting template dropped
significantly below 0.5 fmoles.
Furthermore, signals observed in the water-only control suggested significant
genomic
contamination of E. coli DNA. In this experiment we have incubated 0.5 fmoles,
or more dilute
quantities, of the fragment detailed in Figure 10 with Tester3bio and Sizer2
under the conditions
indicated. The reaction solution included 10 rnM Mg-acetate, 6 g recA, 8.8 gg
gp32, 27 mM
phosphocreatine, 1 U creatine kinase, 0.3 M Tester3bio, 0.3 M Sizer2, 3 niM
ATP, 200 M
dNTPs, 10% PEG compound, 50 U Klenow, and 0.5 pmoles template or dilution
indicated in
figure. The final volume was 30 I.
We have included a no DNA control, progressive dilution of the template, and
investigated initiating the reaction on embedded template (the ClaI fragment
detailed on Figure
1 and 7), of using PEG 1450, and diluting the Klenow fragment. A fraction
(1/10) of the
reaction products were run on a 6% denaturing gel, transferred to nylon, and
bound with
streptavidin-HRP prior to performing enhanced chemiluminescence to reveal the
biotinylated
products of the reactions (Figure 19A). A further fraction (3/10) was phenol
extracted,
precipitated, and run on a 2% agarose gel and stained with ethidium bromide
(Figure 19B). A
final fraction (3/10) was cut with BbvC1 and electrophoresed on a 2% agarose
gel alongside
equivalent uncut DNA (Figure 19C).
Lane 2 (Figure 19A) was estimated to contain 5 X 104 fold amplification, which
corresponded to 10" molecules of final product. In the presence of 0.5 fmoles
of starting
template, an extremely robust amplification of the expected size fragment was
seen as evidenced
by denaturing gel electrophoresis. Furthermore, when part of the sample was
electrophoresed on
agarose a strong clean band of precisely the correct size for a double-
stranded DNA product was
observed. This product could be cut by BbvC1 to yield a slightly smaller
fragment of the
expected size. Dilution of the template by 100-fold or more resulted in a
significantly less
intense band, and a much larger quantity of fragments shorter than the
expected length. This
153
CA 3041471 2019-04-26
was determined by denaturing gel electrophoresis and by agarose gel
electrophoresis. A similar
pattern was observed when ClaI cut DNA was used, or if no DNA was used. We
believe that
when less than a threshold quantity of DNA is used under these conditions,
there is a suboptimal
amplification reaction, which leads to heterogeneous products, and furthermore
that our samples
are highly contaminated with E. coli genomic DNA from the single-step
purification procedures
used in generating our recombinant proteins used here.
Figure 20 shows activity of a recA C-terminal truncation mutant. RecA proteins
with a
deletion of the C-terminal acidic peptide (recA(C)A) are active in promoting
strand exchange
and extension in a linear template run-on assay. However, there was some
suggestion that the
optimal protein concentration was lower than that with the recA(C) protein.
This experiment
addresses whether a C terminal truncated form of the E. coli recA protein,
described elsewhere,
could be used successfully in invasion/extension reactions. Shown is the
result of a single-sided
run-on assay using either the E. coli recA(C) protein or the E. coli recA
17(C) protein, which
lacks the 17 C-terminal acidic residues. The relationship between primers and
template and
other experimental conditions are indicated. The reaction solution included 10
mM Mg-acetate,
27 mM phosphocreatine, 1 U creatine lcinase, 0.3 pM Tester3bio, 3 mM ATP, 200
pM dNTPs,
50 U Klenow, 0.5 pmoles RV template, and recA species and amount as indicated
in figure.
PEG was included as shown. The final volume was 30 ul. The solution was
incubated for 2
hours at 37 C. Reactions were performed with or without 10% PEG1450, and with
the indicated
quantities of the respective recombinase. Both
recombinases successfully supported
invasion/extension, although under the conditions used here there appears to
be a different
optimum for the amount of protein required.
Figure 21 shows Modified gp32 proteins. Shown is a schematic representation of
T4
gp32 proteins used in this study and the position of various modification and
mutations.
Figure 22 shows activity of gp32 proteins. Significant variations confirm that
gp32
cooperativity has a substantial effect on the rate of invasion/extension
reactions, and further
confirms that gp32(N) displays a significant decrease in cooperativity
consistent with
interference with the function of the N-terminal B domain. Shown are the
results of an
154
CA 3041471 2019-04-26
experiment in which linear run-ons were generated using as template the EcoRV
fragment of a
plasmid carrying the E. coli RuvB gene and the Tester3bio oligonucleotide. The
reaction
solution included 10 mM Mg-acetate, 27 mM phosphocreatine, 100 ng/ 1 creatine
kinase, 400
nM Tester3bio, 3 mM ATP, 200 M dNTPs, 10 U chicken myolcinase, 8 g C-tag
uvsX, 7.5 or
15 g gp32 (each species), 50 U Klenow, and 0.5 pmoles template; no PEG was
included. The
final volume was 30 I. The solution was incubated for 2 hours at 37 C.
Reactions contained
uvsX(C) and various gp32 forms as indicated. Two concentrations of each gp32
form were used
in this experiment. To analyze the reaction products, 2 I of the total volume
(30 I) was loaded
onto the gel.
In all cases, elongated products of the oligonucleotide were generated that
extend up to a
length consistent with full length run-ons. We note that in this experiment
there was a gel
artifact which we occasionally observed. We observed a slower mobility shadow
of the bands.
This was seen in the PCR control fragment lane, also. The smallest quantity of
product was
formed when using the gp32(C) protein, which was consistent with it permitting
only a low level
of recombinase-loaded filaments to be present in the reaction. A smaller
quantity of product was
formed with 15 ug compared with 7.5 g, consistent with the notion that higher
concentrations
decreased the efficiency of recombinase loading.
The gp32(C)K3A protein was predicted to be the next most cooperative form.
Consistent with this, it produced a limited number of full-length products
when 15 g of protein
is used. However, the number of run-ons was greater than that observed with
either quantity of
gp32(C), indicating that there were more recombinase-loaded filaments in the
reaction and a
higher rate of invasion/elongation. When 7.5 mg of gp32(C)K3A was used, there
was a
dramatic change in the quantity of product formed. One explanation is that an
increased rate of
invasion/elongation in this reaction could lead to the out-titration of the
gp32(C)K3A by single-
stranded DNA run-offs. Under these conditions, most of the oligonucleotide
would become
coated with uvsX(C), leading to a high invasion rate and to an inability to
stabilise the outgoing
strand and coat it with gp32. This would result in shorter truncated products,
some of which
would be folded back on themselves, self-primed, and formed into a variety of
other such
155
CA 3041471 2019-04-26
products. This suggested that the rate of invasion/elongation of gp32(C)K3A
was notably
higher for this protein than gp32(C).
By comparing the intensities of the products produced when using 15 ).i.g of
each protein,
we estimated that reactions containing gp32(C)K3A have an invasion/elongation
rate of
approximately 10 times that of gp32(C). All of the other gp32 proteins tested
in this expoiment
produced a pattern similar to that seen when gp32(C)K3A was employed and large
amounts of
product. This was the case even when 15 n of the relevant protein was
employed, suggesting
that they all exhibited lower cooperativity than either gp32(C) or gp32(C)K3A.
Notably,
however, both gp32(N) and gp32(C)R4T produced significantly less product when
only 7.5 In
of protein was used when compared with 15 pg. This contrasted to the situation
with the other
proteins. On this basis, we suggest that gp32(N) and gp32(C)R4T possess a
similar degree of
cooperativity. An earlier study has suggested that gp32K3A and gp32R4Q are of
similar
cooperativity. However, our data would suggest that gp32(C)R4Q lies somewhere
between
gp32(C)K3A and gp32(C)R4T with regard to its behaviour in supporting
invasion/synthesis.
Figure 23 shows invasion and extension using uvsX. This experiment addresses
whether
a C terminal truncated form of the bacteriophage 14 uvsX protein could be used
successfully in
invasion/extension reactions. Shown are the results of single-sided run-on
assays using either
the uvsX(C) protein or the uvsX421(C) protein which lacks the 21 C-terminal
acidic residues.
The reaction solution included 10 mM Mg-acetate, 27 mM phosphocreatine, I U
creatine kinase,
0.4 1.1M Tester3bio, 3 mM ATP, 200 AM dNTPs, 50 U Klenow, 1 U chicken
myokinase,
uvsX(C) or uvsXA21(C), 8.8 jig gp32(N), and 0.5 pmoles RV template. The final
volume was
30 j.tl. The solution was incubated for 2 hours at 37 C, and 2 ill or reaction
mixture was loaded
onto each lane of the gel. The relationship of the template and
oligonucleotides and other
experimental conditions are indicated in the figure. Reactions were performed
with the
indicated quantities of the respective recombinase.
Both recombinases successfully supported invasion/extension. However, under
the
conditions used here, there appears to be a different optimum for the amount
of protein required.
In the case of uvsX(C), the rate of invasion/extension increased progressively
with the
156
CA 3041471 2019-04-26
concentration of protein within the range tested. However for the uvsX621(C)
protein, the rate
was inhibited at higher concentration and the overall level of product
production was lower
under these conditions. In contrast to recA-mediated invasion/extension in
similar reactions,
uvsX(C), appeared to stimulate multiple invasion/extension events without the
need for the
addition of polyethylene glycol.
Figure 24 shows RPA using uvsX(C). In this experiment, uvsX(C) has been
combined
with gp32(N) in the presence of the oligonucleotides Tester3bio and Sizer2.
The template DNA
in this experiment was the EcoRV digested plasmid carrying the E. coli ruvB
gene used in
Example 1. The reaction solution included 10 mM Mg-acetate, 27 mM
phosphocreatine, 100
ng/ 1 creatine kinase, 400 nM Tester3bio, 400 nM Sizer2, 3 mM ATP, 200 M
dNTPs, 50 U
Klenow fragment, 10 U chicken myokinase, 10 g (IX) or 20 g (2X) C-tag uvsX,
8.8 g
gp32(N). Optionally, we included 0.2 mM ADP13S, 10 g E. coli topoisomerase I,
10% PEG
1450, and 10 tig uvsXA21(C). Tester3bio recognised one end of an approximately
300 base pair
fragment and included a 5' overhang relative to the end of the target. Sizer2
recognised the other
strand of this template and was directed toward an embedded sequence such that
its 3' end is
about three and a half !wheal turns from the end of Tester3bio.
In the presence of PEG1450, we observed the amplification of the expected
fragment
within 2 hours. In the cases where amplification occurred, almost the entire
of population of
oligonucleotides was consumed indicating an amplification of 3-5 X 104. The
reaction
components are indicated. Included in some samples are additional components.
We found that
200 M ADP-13-S slightly increased the amount of product generated under these
conditions.
Conversely under the conditions used here, addition of E. coli toposiomerase I
inhibited
amplification. Under the conditions used, we detected no amplification with
uvsX.6.21(C)
protein. However, no PEG1450 was included in these samples, and uvsX(C) did
not amplify
either under these conditions without PEG1450.
Figure 25 shows wild-type versus modified gp32. The variant uvsX(C) protein
was
determined to be qualitatively different to native untagged gp32. The reaction
solution included
mM Mg-acetate, 27 tnM phosphocreatine, 100 ng/ 1 creatine kinase, 300 nM
Tester3bio, 300
157
CA 3041471 2019-04-26
nM Sizer2, 3 mM ATP, 200 1.11s4 dNTPs, 50 U Klenow fragment, 10 U chicken
myokinase, 300
ng/p.1 uvsX(C), 300 ng/ml gp32 (untagged) or 100, 200, 300, 400, 500, or 600
ng/ml gp32(C),
and PEG as indicated. The reaction was incubated for 2 hours at 37 C. A
comparison is shown
between amplification reactions performed in the presence of untagged gp32 and
a titration of
gp32(C), either in the presence or absence of PEG1450. We observed that PEG
was required in
the reaction for gp32(C) to function, while this is not the case for untagged
gp32. However,
PEG significantly increased the quantity of product formed during the reaction
period in either
case. Even in the presence of PEG, untagged gp32 consistently appeared to
generate slightly
more product than gp32(C) at each point on the titration curve.
Figure 26 shows titration of gp32 and effect of uvsY. Titration of gp32
revealed a
requirement for a minimal quantity of gp32, and a requirement for uvsY(N)
protein when
untagged gp32 was employed. In Figure 26A, the results of an experiment are
shown
demonstrating that when untagged gp32 was used, uvsY(N) protein was required
for
amplification. The reaction solution included 10 mM Mg-acetate, 27 mM
phosphocreatine, 100
ng/ 1 creatine kinase, 300 nM Tester3bio, 300 nM Sizer2, 3 mM ATP, 200 [tM
dNTPs, 10%
PEG1450, 50 U Klenow fragment, 10 U chicken myokinase, 300 ng/iAl uvsX(C), 300
ng/vil gp32
(untagged), and uvsY(N) as indicated in the figure. The solution was incubated
for 2 hours at
37 C. The uvs(Y) protein operated over a range of concentrations shown here
(50 to 300 ng/ 1).
Other experiments demonstrated that higher quantities inhibited the reaction.
Thus, an optimum
must be established for any given reaction (probably between 5 and 50
ng/}.11).
Figure 26B shows the results of titrating the untagged gp32 protein in the
presence or
absence of uvsY(N). The reaction solution included 10 mM Mg-acetate, 27 mM
phosphocreatine, 100 ng/i.t1 creatine kinase, 300 nM Tester3bio, 300 nM
Sizer2, 3 mM ATP, 200
1.1.M dNTPs, 10% PEG1450, 10 U chicken myokinase, 750 ng/111 uvsX(C), 300
gp32
(untagged), and 300 ng/p.1 uvsY(N). The solution was incubated for 2 hours at
37 C. Once
again, there is a requirement for uvsY(N) to achieve amplification.
Additionally, this
experiment demonstrates a need for a minimal amount of gp32. In this
experiment, varying the
gp32 concentrations between 80 and 160 ng/111 gp32 resulted in a sharp
transition from no
158
CA 3041471 2019-04-26
amplification to efficient amplification. A simple analysis of known binding
site size for gp32,
the length of the oligonucleotides, and their concentration, suggested that
this rise in
concentration would represent a transition from substoichiometric levels of
gp32 for the primer,
to saturating levels. Consequently, the simplest interpretation is that gp32
saturates the
oligonucleotides in order to have excess gp32 in the reaction to collect and
stabilise the outgoing
strand.
Figure 27 shows factors affecting reaction rate and noise. High gp32
cooperativity
inhibits recombinase filament formation (Figure 27A). In addition, uvsY acts
to increase
recombinase loading in an unfavorable gp32 environment (Figure 27B). PEG aids
the function
of cooperatively-disabled gp32 (Figure 27C). The reaction rate is influenced
by both
recombinase activity and effectiveness of gp32 (Figure 27D). Post-invasion
phases are
enhanced in the presence of cooperative gp32 (Figure 27E). Cooperative gp32 is
more effective
at silencing reaction noise (Figure 27F).
The predicted effects and interactions of gp32, uvsX, uvsY, and PEG were
suggested,
with the conclusion that an optimal balance between reaction rate and noise
must be reached.
The degree of cooperativity of gp32 is indicated across the top of the figure.
High cooperativity
favoured efficient binding to single-stranded DNA, which prevented significant
reaction noise
by inhibiting undesirable priming behaviour. High cooperativity also favoured
stabilisation of
the outgoing strand during recombination and DNA synthesis. Conversely highly
cooperative
gp32 reduced the abundance of recombinase-loaded searching filaments, and can
affect reaction
rate considerably.
This behaviour could be partly overcome by including uvsY in the reaction.
However,
whether this could achieve as high a loading rate as desired is yet to be
determined. One could
employ modified gp32 proteins that are less cooperative. Also, the
cooperativity of gp32
proteins and uvsX proteins can be affected by the inclusion of PEGs. PEG may
also have
beneficial, or sometimes detrimental, consequences on other components of the
reaction such as
DNA hybridisation behaviour and polymerase processivity. Optimal rate may be
acquired at
some position away from either extreme, as indicated, as a balance between
recombinase
159
CA 3041471 2019-04-26
loading and gp32 function may need to be reached.
Figure 28 shows the effects of PEG. PEG was able to reduce the average length
of linear
invasion/run-on products in uvsX-mediated linear run-on experiment in the
presence of gp32(C).
Shown are the results of a linear run-on experiment utilising the Tester3bio
oliognucleotide
targeted to the end of the approximately 300 base pair EcoRV fragment of E.
coil RuvB. This
fragment was used throughout the disclosed experiments and is depicted
schematically on the
right of the figure. Reaction were reconstituted with 8 mg of UvsX(C) in a
final reaction volume
of 30 ml, in the presence of gp32(C), and in some cases, varying amounts of
UvsY(N) or
UvsY(C). The reaction solution included 10 mM Mg-acetate, 27 inM
phosphocreatine, 1 U
creatine kinase, 0.4 M Tester3bio, 3 InM ATP, 200 M dNTPs, 50 U Klenow
fragment, 1 U
chicken myokinase, 8 pg uvsX(C), 7.5 g gp32(C) or 8.8 g gp32(N), and 0.5
pmoles template.
The final volume was 30 I. The solution was incubated for 2 hours at 37 C,
and 2 I of the
solution was loaded in each lane.
In the absence of PEG, multiple invasion/run-on cycles appear to have occurred
on each
template, and there was generation of a significant amount full-length
product. However, an
even greater amount of slightly shorter fragment was produced, which we
believe constitute
slightly shorter run-ons which may have folded back on themselves and
synthesised a short
hairpin. We interpreted all bands not full-length as resulting from some form
of bona fide
invasion/extension reaction that has not ac' hieved full extension. Both
UvsY(N) and UvsY(C)
stimulate the quantity of product formed to some extent. In the case of no
PEG, UvsY(C) seems
to be more effective than UvsY(C). This is in contrast to other geometric
amplification data we
generated, suggesting that only UvsY(N) supported efficient geometric
amplification.
Most notably, the inclusion of PEG, in contradiction to findings with E. colt
recA,
seemed to decrease the overall amount of product on this gel. It also
decreased the average
length distribution of products. In order to explain this, we suggest that
under these conditions
the cooperativity of gp32(C), perhaps already at a maximum, cannot be
increased by PEG whilst
that of UvsX can be. Thus, relatively hyperactive UvsX behaviour results in
rapid loading onto
the outgoing stand, reinvasion, and efficient 'bubble migration' which chases
the newly
160
CA 3041471 2019-04-26
synthesised strand and displaces it more readily. Consequently, the average
product length is
significantly reduced.
Figure 29 shows DNA end directed invasion. The first round of
invasion/synthesis using
end-targeting and oligonucleotide overhang is illustrated (Figure 29A).
Additional ¨10 to 15
residues (5' end) and ¨30 residues (3' end) approximate the minimal
requirement for strand
exchange (Figure 29B). Invasion occurs, followed by release of the
deconstrained outgoing
strand, and backfire synthesis (Figure 29C). Most nucleoprotein filaments that
are coated
enough to complete strand exchange will catalyze complete and deconstrained
release of the
outgoing strand (Figure 29D). Subsequent rounds of invasion/synthesis occur
(Figure 29E).
Few or no nucleoprotein filaments can exchange to the end of the target
(Figure 29F). One or no
gp32 molecules are provided (Figure 29G). Following this is recombinase
loading (Figure 29H)
and branch migration (Figure 291).
This figure describes how targeting oligonucleotides initially possessing an
overhang
relative to a linear target template might behave during the first and then
subsequent attempts to
carry out strand exchange. The purpose of this model is to rationalise data
suggesting that when
such a situation in reconstituted experimentally, there is a significant
difference between the first
and subsequent invasions. The top of the figure depicts recombinase loaded
oligonucleotide
filaments, displaying different 5' extents of coverage. The 5 to 3'
directional assembly means
that most should have coating to the very 3' end. As depicted in the figure,
the oligonucleotides
all possess a 5' overhang relative to the initial target.
The first invasion event is likely to result in complete release of the
outgoing strand as
there is a significant likelihood that recombinase will coat the searching
oligonucleotide to a
further 5' extent than the sequence that will be involved in the strand
exchange. Once the
outgoing strand is freed, it is topologically unconstrained and can be easily
stabilised by single
stranded DNA binding proteins, presumably even those with relatively poor
cooperativity.
Furthermore, stability is also generated by polymerases extending the 3' end
of the duplex DNA
to generate the complement to the very 5' end of the targeting
oligonucleotide. We refer to this
synthesis as backfire synthesis. As a consequence of backfire synthesis any
subsequent invasion
161
CA 3041471 2019-04-26
will be flush with the extended template.
Under these circumstances, most oligonucleotides are not completely coated
with
recombinase to the their very 5' ends. In some cases, there may be one or more
gp32 molecules
coating the 5' part of the oligonucleotide. When these oligonucleotides
perform strand exchange
on the now extended target, the outgoing strand is unlikely to be immediately
freed. As a
consequence, the event initially resembles the topologically constrained event
already depicted
in Figure 6. The model suggests that only if, the cooperativity of the single-
stranded DNA
binding protein is sufficient will these strained unstable intermediates be
able to exist for some
limited period. In the bottom part of the figure, we explore what might occur
to these unstable
intermediates.
In scenario 1, the unexchanged 5' extent of the oligonucleotide undergoes
branch
migration with the equivalent duplex portion of the target. This could easily
lead to complete
dissociation of this part of the outgoing strand which would then rapidly
rotate to release any
stress and be stabilised by single-stranded DNA binding proteins as occurred
in the first
invasion. These now stable substrates will be ideal and relatively stable
assemblies for
polymerase elongation. Alternatively, in scenario 2, the single-stranded DNA
binding protein
disassembles from the outgoing strand and branch migration proceeds in the
opposite direction
to that in scenario 1, so that the invading DNA is ejected. In scenario 3, the
outgoing strand
becomes coated with recombinase and re-invades leading to ejection of the
oligonucleotide.
This process resembles a process described elsewhere as bubble migration. If
recombinase loads
onto the freed outgoing strand in scenario 1 then bubble migration could also
occur. We have
experimental data that is most easily reconciled by considering the existence
of bubble migration
as shown in Figure 28.
Figure 30 shows RPA in a complex sample. In this experiment we have examined
the
sensitivity of RPA, and need for a DNA-end, in an RPA reaction on a complex
DNA target. A
schematic representation is given of the DNA sequence, which corresponds to
part of the human
angiotensin converting enzyme (ACE) gene. Three different combinations of
primers were
used. The experiment used a mixture of uvsX(C), uvsY(N), and gp32(C)K3A. The
reaction
162
CA 3041471 2019-04-26
solution included 10 mM Mg-acetate, 27 triM phosphocreatine, 100 ng/i.il
creatine kinase; 300
nM 1Jp3 primer; 300 nM Downl, 2, or 3 primer; 3 mM ATP, 200 KM dNTPs, 10%
PEG1450,
50 U Klenow fragment, 10 U chicken myokinase, 300 nevi uvsX(C), 300 ng41
gp32(C)K3A,
and 50 ng/ 1 uvsY(N). The final volume was 30 Ill. The solution was incubated
for 5 hours at
37 C. Reaction products were electrophoretically separated on an acrylamide
gel, transferred to
a nylon membrane, and probed with a biotinylated oligonucleotide recognising a
unique internal
sequence.
For the RPA reaction, uncut template (genomic DNA) and cut template were
compared,
and primer pairs were compared. A fragment of the expected size was detected.
In all cases,
there was no specific product when no genomic DNA is added to the reaction,
but a specific
product was generated when DNA (equivalent to roughly 10,000 copies of any
sequence) was
added. Digestion of the DNA prior to RPA with Hpall resulted in the generation
of at least one
end overlapping with one of the oligonucleotides, and an increase in signal
strength. However,
there was not an absolute requirement for HpaII digestion for RPA to occur.
Figure 31 shows RPA sensitivity. In this experiment, we have examined the
sensitivity
in an RPA reaction on a complex DNA target. A schematic representation is
given of the DNA
sequence, which corresponds to part of the human angiotensin converting enzyme
(ACE) gene.
Three different combinations of primers were used. The experiment used a
mixture of uvsX(C),
uvsY(N), and gp32(C)K3A. The reaction solution included 10 mM Mg-acetate, 27
mM
phosphocreatine, 100 ng/ 1 creatine kinase; 300 nM Up3 primer; 300 nM Down 1,
2, or 3
primer; 3 mM ATP, 200 iilsA dNTPs, 10% PEG1450, 50 U Klenow fragment, 10 U
chicken
myokinase, 300 ng/ 1 uvsX(C), 300 ng/ 1 gp32(C)K3A, and 50 ng,/ 1 uvsY(N). The
final
volume was 30 pi. The solution was incubated for 5 hours at 37 C, and probe
ACE-hyb was
used. Reaction products were electrophoretically separated on an acrylamide
gel, transferred to
a nylon membrane, and probed with a biotinylated oligonucleotide recognising a
unique internal
sequence. A fragment of the expected size was detected. In all cases, there
was no specific
product when no genomic DNA is added to the reaction, but a specific product
was generated
when sufficient DNA was added. In all cases, 1000 copies were sufficient to
generate a
163
CA 3041471 2019-04-26
significant signal, and in one case we could detect a very faint signal at 100
copies.
Figure 32 shows RPA sensitivity and template independent artifacts. The
results of an
experiment are shown in which we have investigated the sensitivity in an RPA
reaction on a
complex DNA target. A schematic representation is given of the DNA sequence,
which
corresponds to part of the human angiotensin converting enzyme (ACE) gene. A
time course of
the amplification was performed taking reaction samples at 1, 2 and 3 hours.
Reaction products
were detected by virtue of the presence of a biotin residues attached to the
5' end of one of the
oligonucleotides used in the amplification. In this way, it was possible to
visualise all the
reaction products involving this oligonucleotide, including any artifacts that
might arise. We
tested several different concentrations of the uvsY(N) protein. The reaction
solution included 10
triM Mg-acetate, 27 mM phosphocreatine, 100 ng4t1 creatine kinase; 300 nM Up3
primer; 300
nM Downl primer; 3 mM ATP, 200 [t/vI dNTPs, 10% PEG1450, 50 U Klenow fragment,
IOU
chicken myokinase, 300 ng/til uysX(C), 300 ng/pil gp32(C), and 50 ng/p.1
uvsY(N). The final
volume was 30 pl. The solution was incubated for 5 hours at 37 C. At an
uvsY(N)
concentration of 50 ng/pl, we detected the correct product directly, albeit
faintly, after 3 hours of
incubation. During this period there was an accumulation of strong bands of
roughly twice the
length of the oligonucleotide, which accumulated similarly in the template
minus sample. These
were most likely to be primer artifacts.
Figure 33 shows how primer artifacts may arise. Primer artifacts likely
initiate by
erroneous self-priming events as depicted here. Primers may form hairpins, as
occurs in Figure
33A, or hybridise to a second primer, as occurs in Figure 33B. If a polymerase
can extend such
a hairpin a significant stretch of double-stranded DNA may be formed, as seen
in A* and B*.
These structures might become targets for other recombinase-loaded filaments,
titrate active
filaments from bona fide targets, and possibly enter into geometric forms of
amplification
themselves.
Figure 34 shows primer artifact suppression. Depicted schematically are
several
strategies to suppress primer artifact noise. In Figure 34A, a second short,
3'-blocked
oligonucleotide complementary to the 3' sequences of the targeting
oligonucleotide is included
164
CA 3041471 2019-04-26
in the reaction to compete for the formation of secondary structure formation
that might result in
erroneous priming. In Figure 348 and 34C, a similar short blocked
oligonucleotide is employed
as in (A), but in this case a covalent bridge is engineered between the 5' end
of the targeting
oligonucleotide and the 5', or 3', end of the competing short primer. In this
way, the blocked
nucleotide is tethered at its 5'end to the 5' end of the targeting
oligonucleotide (Figure 34A-B)
In Figure 34D, a short sequence complementary to the 3' region of the
targeting oligonucleotide
is added to the 5' region of the targeting oligonucleotide. It competes
efficiently with secondary
structure formation.
Figure 35 shows use of hairpin oligonucleotides to stimulate self-priming of
displaced
strands. Depicted is a scheme showing how oligonucleotides whose design
includes a 5' section
with perfect complementarity to the 3' section might be used to stimulate
amplification through
self-priming. At the top of the diagram is shown a target DNA, designated A,
which has distinct
ends which are targets for one of the two targeting oligonucleotides shown in
the upper left and
right of the figure. Both of these oligonucleotides possess complementarity
between their 5' and
3' regions as indicated by short arrows. The target, A, would likely have been
generated by
earlier invasion/elongation events involving these oligonucleotides and an
initial target lacking
the 5'-most regions of these oligonucleotides. On the left or right side of
the figure we follow
the outcome of invasion/elongation events initiated by targeting by the left
or right primers
respectively. The outcome is similar in both cases, albeit the final products
are arranged slightly
differently.
Focusing on the left side of the figure we observe that when target A is
subject to
invasion and elongation with the left primer the result is formation of a new
duplex identical to
A and a single-stranded DNA equivalent to the top strand of the initial
target, designated B. Due
to the presence of the complementarity between the very 3' region and adjacent
sequences, B is
capable of forming a hairpin which will prime DNA synthesis to generate a
largely double-
stranded product C. The product C can readily be targeted once again by the
left
oligonucleotide. However, in this case, no single-stranded displaced strand is
formed. Instead,
product D is formed with a length that is roughly twice that of the original
target. This product
is an inverted repeat and contains two sequences that are targets for the left
oligonucleotide, and
165
CA 3041471 2019-04-26
one for the right oligonucleotide located in the middle.
Subsequent invasion/elongation events, and the possible occurrence of
hybridisation
events between displaced strands, could easily lead to such `dimeric' species
becoming further
enlarged, and the formation generally of more complex products. A similar
course of events is
shown on the right side of the figure, this time initiated by
invasion/elongation by the right
primer. The final dimeric product, D', is not equivalent to D as the two end
sequences are
targets for the right primer, and the central region is a target for the left
primer. Processes
similar or identical to those shown here are likely to occur with some
frequency under some
conditions even in the absence of the deliberate design of oligonucleotides to
promote it, as there
is often some limited capacity for self-priming of single-stranded DNAs.
Figure 36 shows conditions that support the amplification of DNA with little
or no
primer artifacts. The results of an experiment are shown in which we have
investigated the
sensitivity of an RPA reaction on a complex DNA target. A schematic
representation is given of
the DNA sequence, which corresponds to part of the human angiotensin
converting enzyme
(ACE) gene. Oligonucleotides used are the biotinylated Angiolbio primer
and the
unbiotinylated Angio3 primer whose sequence is given in the experimental
methods. These
primers amplified a 132 bp double-stranded DNA fragment. Uncut human genomic
DNA was
titrated from 45 copies up to 2880 copies. Reaction products were detected by
virtue of the
presence of a biotin residue attached to the 5' end of one of the
Vligonucleotides used in* the
amplification. In this way, it was possible to visualise all the reaction
products involving this
oligonucleotide, including any artifacts that might arise.
The reaction was incubated for 2 hours at 37 C in the case of the Klenow exo-,
and 2
hours at 42 C in the case of the Bst polymerase. The reaction included the
following: 50 ng/p.1
uvsY(N), 300 ng/p.1 gp32(C), 100 ng/ 1 uvsX(C), 20 mM phosphocreatine, 3 mM
ATP, 25
milliunits/ 1 myokinase, 100 ng/ill creatine kinase, 200 p.M dNTPs, 5% w/v PEG
compound,
300 nM Angiolbio primer, 300 nM Angio3 primer, 800 ng/ 1 Klenow exo- or 1.2
units/1.d Bst
polymerase. The Klenow-mediated amplification was performed in U2 buffer
comprising a
final composition of 20 mM Tris acetate pH 7.9, 8 mM magnesium acetate, 120 mM
potassium
166
CA 3041471 2019-04-26
acetate. The Bst polymerase-mediated amplification was performed in Ul buffer
comprising a
final composition of 20 tnM Iris acetate pH 7.5, 6 mM magnesium acetate, 100
xnM potassium
acetate.
Example 13: DNA amplification for point-of-use applications
Clones and proteins were produced as described in Example 11, above.
DNAs used in RPA reactions
We have employed several different target DNAs in this study, and a number of
oligonucleotides. The sequence of the oligonucleotides is given below, and the
template target
in the experiment shown in Figure 39B. The E. coli RuvB gene target was used
for linear run-on
assay (Figure 398). Identical quantities of plasmid template containing this
fragment were cut
either with EcoRV, releasing a roughly 300 bp fragment, or with ClaI which
linearises the DNA.
Equal molar quantities of template were used in the run-on experiments (20 nM
each template).
The sequence of a Kpnl/ClaI fragment of this template is given below. The
EcoRV fragment is
embedded within this sequence, and the sites are highlighted.
GGTACCACTTTGCCGGAAGATGTAGCAGATCGCGCCATTCGCCCCAAATTAC
TGGAAGAGTATGTTGGTCAGCCGAGGTTCGTTCACAGATGGAGATTTTCATCAAAG
CAGCGAAACTGCGCGGCGATGCCCTCGATCATTTGTTGATTTTTGGTCCTCCGGGGT
TGGGTAAAACTACGCTTGCCAACATTGTCGCCAATGAAATGGGCGTTAATTTACGC
ACGACTTCTGGTCCGGTGCTGGAAAAGGCGGGCGATTTGGCTGCGATGCTCACTAA
CCTTGAACCGCATGACGTGCTGTTTATTGATGAGATCCACCGTCTATCGCCAGTTGT
TGAAGAAGTGCTGTACCCGGCAATGGAAGACTACCAACTGGATATCATGATTGGTG
AAGGICCGGCGGCACGCTCCATTAAAATTGATTTGCCGCCGTTTACCCTGATTGGTG
CAACCACGCGCGCAGGTTCGCTGACATCACCGTTGCGCGACCGTTTTGGTATTGTGC
AACGTCTGGAGTTTTATCAGGTGCCGGATCTGCAATATATCGTCAGTCGCAGCGCAC
GCTTTATGGGGCTTGAGATGAGTGATGACGGCGCGCTGGAAGTTGCTCGTCGCGCT
CGCGGTACGCCGCGCATTGCCAACCGTCTGCTGCGTCGAGTGCGTGATTTCGCCGAA
GTGAAGCACGATGGCACCATCTCGGCAGATATCGCTGCTCAGGCGCTGGATATGTT
GAATGTCGATGCTGAAGGTTTCGAT'TATATGGACCGCAAATTGTTGCTGGCGGTAAT
CGAT (SEQ ID NO:18)
Oligonucleotide Tester3bio sequence. Bases homologous to the target are in
bold.
5'biotin-CTCACTATACCICAGCATCATGATTGGTGAAGGTCCGGCGGCAC (SEQ ID
NO:19).
Human DNAs
167
CA 3041471 2019-04-26
We have used human genomic DNAs from several sources. A mixed population male
genomic DNA from Promega was utilised. Also, DNA from individual male samples
was
studied in Figure 38A. Individual 1 and 2 were father and son. Individual 2
DNA was used in
the experiment in Figure 39E, while DNA for the experiment in Figure 39E was
another male
individual. Sequences of oligonucleotides used for amplifying human and B.
subtilis sequences
are as follows:
Corresponding to the human ApolipoproteinB locus:
ApoB4 5' CAGTGTATCTGGAAAGCCTACAGGACACCAAAA 3' (SEQ ID NO:20)
Apo300 5' TGCTTTCATACGTTTAGCCCAATCTTGGATAG 3' (SEQ ID NO:21)
Apo700 5' TGGTAAACGGAAGTCTGGCAGGGTGATTCTCG 3' (SEQ ID NO:22)
Apo800 5' CAATTGTGTGTGAGATGTGGGGAAGCTGGAAT 3' (SEQ ID NO:23)
Apo900 5' GAGGTGGTTCCATTCCCTATGTCAGCATTTGC 3' (SEQ ID NO:24)
Apol000 5' GGGTTTGAGAGTTGTGCATTTGCTTGAAAATC 3' (SEQ ID NO:25)
Apo 1500 5' TTGAAITTCAAGTTTAGAAAAGTTGAGGGAGCCAG 3' (SEQ ID
NO:26)
Corresponding to the human SRY locus:
SRY3 5' AAAGCTGTAACTCTAAGTATCAGTGTGAAAC 3' (SEQ ID NO:27)
SRY4 5' GTTGTCCAGTTGCACTTCGCTGCAGAGTACC 3'(SEQ ID NO:28)
Corresponding to B. subtilis genomic DNA:
BSA1 5' TTGGGCACTTGGATATGATGGAACTGGCAC 3' (SEQ ID NO:29)
BSA3 5' ACAGAAAGCTATTAAAGCAACTGACGGTGTGG 3'(SEQ ID NO:30)
BSB3 5' CCATCTTCAGAGAACGCITTAACAGCAATCC l'(SEQ ID NO:31)
Human STR marker primers:
CSF1P0 5' GTTGCTAACCACCCTGTGTCTCAGITI-ICCTAC (SEQ ID NO:32)
CSF1P0 3' AGACTCTTCCACACACCACTGGCCATCTTCAGC (SEQ ID NO:33)
D7S820 5' GAACACTTGTCATAGTTTAGAACGAACTAACG (SEQ ID NO:34)
168
CA 3041471 2019-04-26
D7S820 3' GAA'TTATAACGATTCCACATTTATCCTCATTGAC (SEQ ID NO:35)
D13S317 5' TTGCTGGACATGGTATCACAGAAGTCTGGGATG (SEQ ID NO:36)
D13S317 3' CCATAGGCAGCCCAAAAAGACAGACAGAAAGA (SEQ ID NO:37)
D 16S539 5' AAACAAAGGCAGATCCCAAGCTCTTCCTCTTCC (SEQ ID NO: 38)
D16S539 5' ATACCATTTACGTTTGTGTGTGCATCTGTAAGC (SEQ ID NO:39)
D l8S51 5' GGTGGACATGTTGGCTTCTCTCTGTTCTTAAC (SEQ ID NO:40)
Dl 3' GGTGGCACGTGCCTGTAGTCTCAGCTACTTGC (SEQ ID NO:41)
TH01 5' TACACAGGGCTTCCGGTGCAGGTCACAGGGA (SEQ ID NO:42)
THOI 3' CCTTCCCAGGCTCTAGCAGCAGCTCATGGTGG (SEQ ID NO:43)
TPDX 5' ACTGGCACAGAACAGGCACTTAGGGAACCC (SEQ ID NO:44)
TPDX 3' GGACrGAACTGGGAACCACACAGGITAATTA (SEQ ID NO:45)
Timecourse experiment:
APOB600 GCTCACTGTTCTGCATCTGGTCAATGGTTCTG (SEQ ID NO:46)
APOB300REV CTATCCAAGA'TTGGGCTAAACGTATGAAAGCA (SEQ ID NO:47)
Shorter oligonucleotide experiment:
APOB500 ATGGTAAATTCTGGTGTGGAAAACCTGGATGG (SEQ TD NO:48)
AP0500-28 TAAATTCTGGIGTGGAAAACCTGGATGG (SEQ ID NO:49)
AP0500-25 ATTCTGGTGTGGAAAACCTGGATGG (SEQ ID NO:50)
APOB300REV CTATCCAAGATTGGGCTAAACGTATGAAAGCA (SEQ ID NO:51)
APOB300REV-28 CCAAGATTGGGCTAAACGTATGAAAGCA (SEQ ID NO:52)
APOB300REV-25 AGATTGGGCTAAACGTATGAAAGCA (SEQ ID NO:53)
D18S51 5' GGIGGACATGTTGGCTICTCTCTGTTCTTAAC (SEQ ID NO:54)
D18S51 5-28 GACATGTTGGCTTCTCTCTGTTCTTAAC (SEQ ID NO:55)
D18S51 5'-25 ATGTTGGCTTCTCTCTGTTCTTAAC (SEQ ID NO:56)
169
CA 3041471 2019-04-26
D18S51 3' GGTGGCACGTGCCTGTAGTCTCAGCTACTTGC (SEQ ID NO:57)
D18S51 3'-28 GCACGTGCCTGTAGTCTCAGCTACTTGC (SEQ ID NO:58)
D18S51 3'-25 CGTGCCTGTAGTCTCAGCTACTTGC (SEQ ID NO:59)
SRY3 AAAGCTGTAACTCTAAGTATCAGTGTGAAAC (SEQ ID NO:60)
SRY3-28 GCTGTAACTCTAAGTATCAGTGTGAAAC (SEQ ID NO:61)
SRY3-25 GTAACTCTAAGTATCAGTGTGAAAC (SEQ ID NO:62)
SRY4 GTTGTCCAGTTGCACTTCGCTGCAGAGTACC (SEQ ID NO:63)
SRY4-28 GTCCAGTTGCACTTCGCTGCAGAGTACC (SEQ ID NO:64)
SRY4-25 CAGTT'GCACTTCGCTGCAGAGTACC (SEQ ID NO:65)
Conditions of standard RPA reactions included: 50 mM Iris pH 8.4, 80 mM
Potassium
acetate, 10 mM Magnesium acetate, 1 mM DTT, 5% PEG compound (Carbowax-20 M), 3
mM
ATP, 20 mM Phosphocreatine, 100 ng/ 1 Creatine kinase, 600 ng/p1 gp32; 109
ng/pl, or 125
ng/pl, or 200 ng/ 1 uvsX; 16 ng,/ 1, or 25 ng,/ 1, or 40 ng/p.1, or 60 ng/ 1
uvsY; 20 ng41 Bsu
polymerase, 200 pM dNTPs, and 300 nM each oligonucleotide. Reaction conditions
Cl-C4 are
as above with: Cl = 109 ng/ 1 uvsX, 16ng/p1 usvY; C2 = 125 ng/pl uvsX, 25
ng/pl uvsY; C3 =
200 ng/pl uvsX, 40 ng/pl uvsY; C4 = 200 ng/ml uvsX, 60 ng,/ 1 uvsY.
Experimental Results
Figure 37 shows a schematic representation of RPA method. In Figure 37A(i),
Recombinase protein uvsX binds cooperatively to single-stranded
oligonucleotides in the
presence of ATP. Nucleoprotein filaments actively hydrolyse ATP to ADP.
Spontaneous
disassembly can lead to competitive binding of single stranded binding protein
gp32, this being
deterred and reloading aided by uvsY protein and polyethylene glycol. In
Figure 37A(ii),
recombinase filaments catalyse strand exchange if homologous DNA is detected.
In Figure
37A(iii), strand exchange matches the searching strand with its complement and
displaces a
strand then bound by gp32. Recombinase disassembles. In Figure 37A(iv),
polymerases access
the structure and extend the oligonucleotide, displacing more of the original
strand. In Figure
378(i), two opposing targeting nucleoprotein complexes recombine with their
respective targets
170
CA 3041471 2019-04-26
and DNA synthesis is initiated. In Figure 37B(ii), polymerase complexes
encounter one another
and one of the polymerases dissociates. In Figure 378(iii), the remaining
polymerase continues
synthesis freeing the two parent strands, a polymerase re-binds to the free 3'-
end thus replication
of both strands occurs. In Figure 37B(iv), new targeting events occur. In the
second round one
targeting primer will displace a free end. In Figure 37C, comparison of the
products of strand
exchange at a DNA end or at an embedded DNA sequence.
Figure 38A shows the results of amplification of STR markers from two
individuals (1
and 2, father and son) using primer pairs for seven independent markers. RPA
conditions C4
were employed (see above). Figure 38B.,shows titration of reaction components
to determine
concentrations that support in vitro amplification. Reactions included the
primers SRY3 and
SRY4 at 0.3 pM (targeting the SRY gene), 80 mM potassium acetate, 50 mM TrisC1
pH 8.4, 2
mM DTT, 5% Carbowax-20M, 200 ng4t1 uvsX, 60 ng/p1 uvsY, 600 ng/pl gp32, 20
ng/pl Bsu
polymerase, and 50 copies4t1 Y chromosomal DNA, except when a given component
was that
under investigation. Optimal quantities of gp32, ATP, uvsX, uvsY, PEG, and Bsu
polymerase
for effective amplification of this particular product were determined. ATP-y-
S and ADP--S
inhibited the reactions.
Figure 39A shows the size limits of RPA reactions. Primer ApoB4 was combined
with
opposing primers capable of generating amplified products of the indicated
sizes. Conditions of
125 ng/pluvsX and 25 ng/ luvsY (C2) were employed except Cl where 109 ng/ I
uvsX and 16
ng/ 1 uvsY were used; 15 copies/ 1 human DNA were used (30 I reactions).
Under conditions
C2, some hairpin-mediated product duplication occurred converting some of the
300 bp
amplicon to 2x and 3x unit length (*) (L. D. Harris, J. D. Griffith, J Mol
Biol 206, 19-27 (Mar 5,
1989)).
Figure 39B shows elongation efficiencies from embedded or end sequences. A
biotinylated primer was incubated with linearized plasmid DNA. Equal
quantities (20 nM final)
of templates linearized with either ClaI (lane 3) or EcoRV (lanes 1 and 2)
were used, the primer
either overlapping the cut end, or the target site being embedded (lane 3).
Incubation with
recombinase targeting components with (lanes 2 and 3) or without (lane 1)
Klenow reveals
171
CA 3041471 2019-04-26
limited elongation from the embedded site (product 1*), and abundant
elongation from the end
site (product 2*). Electrophoresed products were transferred to nylon and
biotin detected by
chemiluminescence. The weak common band (-300 bp, lanes 1 and 3) was an
artifact arising
from this particular protocol.
Figure 39C shows the sensitivity of RPA reactions. The indicated copy number
of B.
subtilis genomes was amplified with oligonucleotides BsAl and BsB3, which
amplify a 200 bp
fragment. Conditions Cl were employed. Figure 39D shows human DNA of the
indicated copy
number amplified with primers ApoB4 and Apo300 to generate a 300bp fragment.
Conditions
C2 were employed. Figure 39E, F show results from human DNA from single
individuals. The
DNA was diluted and samples theoretically containing the indicated copy number
were
amplified with primers D18S51 5' and 3' which amplify an STR of size ¨300-360
bp. At
predicted copy numbers of 2 or 3, a number of samples amplified single alleles
(*). Conditions
employed were C2 in (E), and C4 in (F).
Figure 40 shows specificity of RPA reactions. Primers BsA3 and BsB3, which
amplify a
380 bp fragment from B. subtilis genomic DNA were incubated with 1 1.ig of
human DNA, with
(+) or without (-) addition of 100 copies of B. subtilis DNA (Figure 40A). An
asterisk indicates
the position of the expected reaction product, and an arrow indicated the
position of the genomic
DNA. Conditions C3 were employed. To investigate how long it takes RPA to
generate
detectable reaction products a series of amplification reactions were
established with =
oligonucleotides Apo600bio and Apo300rev generating a 345bp fragment. A copy
number of
60 copies/pi (Figure 40B) or 6 copies/Ill (Figure 40C) were used. Individual
reactions were
stopped at the indicated number of minutes and analysed on a gel. Conditions
C4 were
employed (Figure 40D).
For long-term storage of reaction components we lyophilised RPA reactions.
Mixtures
of reaction components were assembled in the absence of the indicated
components, PEG and
buffer. The material was freeze-dried, and then reconstituted with PEG and
buffer plus the
additional omitted components. Primers used are indicated. All components
apart from PEG
and buffer could be lyophilised and successfully reconstituted in a functional
reaction. Target
172
CA 3041471 2019-04-26
DNA was human male genomic DNA at 150 copies/pi (Figure 40E). Oligonucleotides
targeting
three independent loci in human genomic DNA were incubated with overlapping
primer pairs of
25, 28, or 32 bases as indicated. Only the primers of 32 bases in length
successfully amplified
targets. Other experiments show that primers of 30 residues are also effective
at amplifying
target DNAs.
Figure 41 shows primer noise at low target copy number.
Two primers, BsA I and BsB3 (BsA 1
5 'TTGGGCACTTGGA TATGATGGAACTGGCAC3' (SEQ ID NO:29), BsB3
¨5'CCATCTICAGAGAACGCTTTAACAGCAATCC3' (SEQ ID NO:31)), which flank a
roughly 300bp fragment of the Bacillus subtilis genome were incubated with
B.subtilis genomic
DNA serially diluted with water. Conditions used were 80mM Potassium acetate,
50mM Tris.CI
pH 8.4, 2mM DTT, 5% Carbowax 20M, 200pM dNTPS, 3mM ATP, 20mM Phosphocreatine,
=
50ng/p.1 creatine kinase, 300nM each oligonucleotide, 800ng41 gp32, 120ng4I
uvsX, 25ng/p.I
uvsY, and 28ng/ 1 Bsu polymerase. The reaction was incubated for 90 minutes at
37 C.
Products were separated on a 2.5% agarose gel and stained with ethidium
bromide. Arrows
indicate the expected position of the correct amplicon.
Figure 42 details the selection of optimal primers by combining a selection of
candidate
forward and reverse primers and testing the outcome at very low start copy
densities.
Several primer pairs were designed to sequences in the Bacillus subtilis
sporulation locus
Spo0B. The position and orientation of these primers is indicated in (B) as
the following:
J1 ACGGCATTAACAAACGAACTGATTCATCTGCTTGG ¨ SEQ ID NO:66
J2 ATAACCATTTCTTCAATCATTTCAAAGACACGGTC ¨ SEQ ID NO:67
K1 TATAGACGCAAAGCACGAATCAAAGCTCTCAAACC ¨ SEQ ID NO:68
K2 CCTTAATTICTCCGAGAACTTCATAT ¨ SEQ ID NO:69
LI ATATGAAGTTCTCGGAGAAATTAAGGA I __ 1 I GTCGG ¨ SEQ ID NO:70
L2 AATCAGCTGTCTGTCAGGATGATCCGTTTGAAGCG ¨ SEQ ID NO:71
173
CA 3041471 2019-04-26
NEST I CATTAACAAACGAACTGATTCATCTGCTTGG ¨ SEQ ID NO:72
NEST2 ACAGATGAAACAGCTTTCTCATCAGTTTCG ¨ SEQ ID NO:73.
These primers were combined as indicated in (A), and incubated for 90 minutes
under
the following conditions: 80mM Potassium acetate, 50mM Tris.C1 pH 8.4, 2mM
DTT, 5%
Carbowax 20M, 2001.1.M dNTPS, 3mM ATP, 20mM Phosphocreatine, creatine
kinase,
300nM each oligonucleotide, 800nthil gp32, 120ng/p1 uvsX, 25ng/ 1 uvsY, and
28ng/111 Bsu
polymerase, ¨2copies/ 1 B.subtilis genomic DNA (estimated by serial dilution),
37 C. Arrows
indicated the position of products determined to be the expected fragment
based on size. Some
primer pairs give product, but it was accompanied by smearing, or some
additional bands. A few
combinations fail to give product of the expected size. The primer JI and K2
give a significant
amount of the product of the expected size, and rather cleanly. Interestingly
when NEST I
primer, lacking the 4 5'-most bases present in JI, replaces .11 in the
equivalent reaction, the
reaction is not improved indicating that adding as well as removing bases may
improve primer
performance even when all tested are over 30 residues in length.
Figure 43 shows a theoretical consideration of how primer noise initiates.
(A) An oligonucleotide is shown, and above it is an arrow, the head indicating
the 3'-
end of the oligonucleotide. The sequence was generated arbitrarily, although 4
of the 5 most 3'
bases form a palindrome, which might well be avoided in practice. (B) It is
assumed that most
initial events consist of the 3' end folding transiently onto more 5'
sequences to transiently form
a Watson-Crick base pair. (C) Occasionally a polymerase successfully elongates
this structure to
give a long double-stranded hairpin with a turn at one end (labeled H). The
sequence and 5'-3'
direction of the reverse complement of the original is indicated by a pale
arrow. (D) In strategy 1
it is assumed that a second recombinase-coated oligonucleotide of identical
sequence targets the
homologous double-stranded body of the hairpin, thus unraveling it. Mismatches
occur at the 3'
region of the invading oligonucleotide because the complement to this section
was not correctly
generated in step C. If a 4 base pair palindrome had not fortuitously been
generated the
mismatches would be even more severe than shown here. (E) A polymerase, very
occasionally,
might extend the mismatched 3' region of this intermediate to generate an
inverted repeat with
174
CA 3041471 2019-04-26
mismatches in the middle. This structure would also resemble the product if
two identical
primers had formed a brief 3' overlap resulting in formation of a primer
dimer. These structures
cannot readily enter geometric phase amplification. (F) In strategy 2 the long
hairpin formed in
(C) is briefly unzipped at one end. This step might be enhanced by recombinase
binding to
dsDNA because some reports indicate that recA may bind to dsDNA and promote
melting of
short duplexes. (G) The transiently melted 3'-end (reverse complement to the
original 5' end of
the oligonucleotide) folds back in a similar manner to (B) and forms a
structure capable of
elongation. (H) Polymerase elongation creates a large hairpin. In this case
one end contains an
identical duplex sequence to the parent oligonucleotide. (I) Another parent
oligonucleotide
invades the perfect duplex target. (J) Polymerase elongation of (I) generates
a large inverted
repeat with the indicated structure. Both ends contain perfect primer target
sites and so this
structure can amplify exponentially. Interestingly, the inverted repeat
structure means that if an
invasion and elongation occurs from one end, the displaced strand will rapidly
tend to fold back
onto itself to form the structure shown in (H). This product will require
another recombination
event to become structure (J), and thus it may be that higher recombination
activity particularly
benefits noise amplification, and is strongly impeded by lower recombination
activity.
Figure 44 shows several oligonucleotide design strategies to improve signal to
noise in
RPA reactions
Shown are three general strategies to improve signal to noise ratios in RPA
reactions by '
facets of oligonucleotide design. (1) Progressively shortened oligonucleotides
may be tested.
The assumption is that shortening of an oligonucleotide progressively from the
5' end leads to a
drop in activity of the respective nucleoprotein filament, and that this
influences the doubling
time of noisy artifacts more rapidly than that of target. Low activity
nucleoprotein filaments may
also be combined effectively with more active ones in some cases. (2) Locked
nucleic acids, or
other modified sugars, may be included in the primers with consequent effect
on amplification
behaviour. The structure of a locked nucleic acid (LNA) sugar is indicated.
(3) Oligonucleotides
behaviour may be improved by the addition of 5' residues, possibly unrelated
to the target,
which suppress noise via mechanisms such as deterring snapback priming, or by
altering
nucleoprotein recombination activity. Sequences may be homopolymer stretches,
or otherwise.
175
CA 3041471 2019-04-26
Ideal sequences may be determined empirically.
Figure 45 shows the consequences of reducing primer length
(A) Schematic description of experimental design. Pairs of oligonucleotides
were
synthesized to three human targets, and sets of primers were made in which the
5'-most residues
were progressively deleted (full length primers have been described above:
D18S51 5'
GGIGGACATGTTGGCTTCTCTCTGITCTTAAC (SEQ ID NO:54), Dl 3'
GGTGGCACGTGCCTGTAGTCTCAGCTACTTGC (SEQ ID NO:57), SRY3 5'
AAAGCTGTAACTCTAAGTATCAGTGTGAAAC 3' (SEQ ID NO:27), SRY4 5'
GTTGTCCAGTTGCACITCGCTGCAGAGTACC 3 '(SEQ ID NO:28), APOB 500
ATGGTAAATTCTGGTGTGGAAAACCTGGATGG (SEQ ID NO:48), APOB300REV
CTATCCAAGATTGGGCTAAACGTATGAAAGCA (SEQ ID NO:51). Shorter primers were
made by deleting 5'-most bases to give the final indicated length). Primer
pairs of identical
length were combined in amplification reactions in the fashion indicated. (B)
Amplification
reactions were performed on human genomic DNA, present in the start reaction
at 50 copies/pl.
The three targets, an SIR marker D18S51II, part of the human SRY gene, and the
human
Apolipoprotein B locus, were amplified with pairs of oligonucleotides. The
oligonucleotides
were progressively truncated from the 5' end in the manner as described in
(A), and the length of
both primers in the primer pairs used is indicated above the respective lane.
It is evident that
there is a fairly precipitous drop in amplification behaviour as
oligonucleotides are shortened
from just over to just under 30 residues. Consequently comparing
oligonucleotides with length
variation of just over to just under 30 residues is generally likely to reveal
optimal length to get
sufficient specific amplification , but very little or no background. For
example two 29-mers
were sufficient to amplify the D18S51 II locus, and do so cleanly.
Amplification conditions
were: 50mM Tris.C1 pH 8.3, 90mM Magnesium acetate, 2mM DTT, 80mM Potassium
acetate,
200 M dNTPs, 600ng/I.11 gp32, 200ng/u1 uvsX, 60ng/ 1 uvsY, 300nM primers,
20ng/u1 Bsu
polymerase, 50 copies/u1 genomic DNA, 90 minutes at 37 C.
Figure 46 shows how a 3' LNA residues alter amplification behavior in RPA
Amplification of a part of the human ACE locus is shown using combination of
regular
176
CA 3041471 2019-04-26
oligonucleotides and counterparts in which the 3'-most residue has an LNA
sugar. (A)
Schematic representation of the relationship of oligonucleotide primers to one
another. LNA1
and DNAI are identical except that LNAI contains a LNA sugar at the most 3'
position.
Likewise LNA2 and DNA2 differ similarly. (sequence of LNAJDNA I
is:GCTCCTTGGACTGGTAGATGTCACACTTGTG ¨ SEQ ID 74, sequence of LNA/DNA2
is GCCTTGGCTCTGCTGTGCGCATGTGACTTAGC ¨ SEQ ID 75). (B) RPA reactions were
assembled with the indicated oligonucleotide primer combinations. Product
levels are
significantly reduced when both oligonucleotides contain a 3' LNA under
standard reaction
conditions. However substitution of only one oligonucleotide does not suppress
the reaction too
greatly (lanes 2 and 3). Note that the background pattern in lane 3 resembles
that in lane 4, while
in 2 it is largely absent. This suggests that the background derives
principally from the activities
of a single primer species. Amplification conditions were: 50mM Tris.C1 pH
8.3, 90mM
Magnesium acetate, 2mM DTT, 80mM Potassium acetate, 200 M dNTPs, 800ng/ 1
gp32,
200ng/ I uvsX, 60ng/ 1 uvsY, 300nM primers, 20ng/ 1 Bsu polymerase, 50 copies/
1 genomic
DNA, 120 minutes at 37 C. (C) Increasing the levels of polymerase result in
restoration of
robust activity when two LNA-modified oligonucleotides are used. LNA1 and
LNA2, or DNAI
and DNA2, were combined in the presence of the indicated concentrations of
polymerase.
Notably the DNA primers displayed high activity at the lowest used
concentration, and if
anything the products looked less good as polymerase concentration was
increased. Short
products, 50-100 base pairs, are notably also seen. Conversely the LNA primers
were only
optimally active at higher polymerase concentrations. Less small products were
seen, and those
seen other than the expected size are likely product related (see below).
Amplification conditions
were: 50mM Tris.C1 pH 8.3, 90mM Magnesium acetate, 2mM DTT, 80mM Potassium
acetate,
20011M dNTPs, 800ng/ 1 gp32, 200ng/ 1 uvsX, 60ng4t1 uvsY, 300nM primers, 43 to
1400ng/ 1
Bsu polymerase, 100 copies41 genomic DNA, 120 minutes at 37 C. (D) LNA-
modified
oligonucleotides give little, if any, background when no target is present,
but efficiently amplify
bona fide product to high levels when reaction conditions are optimized. A
polymerase
concentration of 200ng/ 1 Bsu polymerase was employed. Reactions were carried
out with DNA
primers pairs, or LNA primer pairs, in the presence or absence of target. A
faint band of the
177
CA 3041471 2019-04-26
expected size was found in the negative controls indicating a contamination
problem.
Nevertheless both primer pairs amplified efficiently in the presence of
target, but in its absence
the DNA pair gave significant noisy smear, while the LNA pair gave no other
product than the
slight contamination. This indicates that the LNA primer pair may be able to
cleanly distinguish
between the presence and absence of target in the sample, while the DNA
primers generate
primer-derived products in the absence of target. Amplification conditions
were: 50mM Tris.CI
pH 8.3, 90tnM Magnesium acetate, 2mM DTT, 80mM Potassium acetate, 2001.tM
dNTPs,
800ng/pl gp32, 200ng/p1 uvsX, 60ng/p1 uvsY, 300nM primers, 200ng/p1 Bsu
polymerase, 50
copies4t1 genomic DNA, 120 minutes at 37 C.
Figure 47 shows that adding homopolymeric stretches to the 5' ends of primers
may alter
nucleoprotein activity.
(A) The sequence of six oligonucleotides used in the experiment is shown. The
.11 and
K2 oligonucleotide target the B.subtilis Spo0B locus (SEQ IDs 66 and 69
respectively), and the
other listed oligonucleotides are derivatives of them, carrying a
homopolymeric stretch of
cytosines, or guanosines as indicated. These oligonucleotides can be incubated
in a pairwise
fashion to assess which pair most effectively amplifies the target. (B) The
results of incubating
the six primers alone, or in all possible combinations, is shown. Reactions
were incubated at
37 C for 2 hours. Reaction conditions were 50mM Tris.CI pH 8.4, 2mM DTT, 5%
Carbowax
20M, 80mM Potassium acetate, 200pM dNTPS, 3mM ATP, 20mM Phosphocreatine, 50ng/
1
creatine kinase, 300nM each relevant oligonucleotide, 800ng/111 gp32, 120ng/p1
uvsX, 25ng/ 1
uvsY, and 28ng/ 1 Bsu polymerase, ¨1 copy/p1 B.subtilis genomic DNA (estimated
by serial
dilution)(20p.1 reaction volume). Optimal primers pairs were J1+K2, and then
J1C+K2C. in
general a homopolymeric Cytosine stretch appeared to make nucleoprotein
filaments more
active, and a Guanosine stretch less active.
Figure 48 shows the effects of betaine on RPA reactions
RPA reactions were established in which an increasing concentration of betaine
was
present in each reaction. Betaine concentration of 0.5M, or greater, in this
experiment reduced
the degree of amplification. At 0.75M the expected product band was still
clearly visible,
178
CA 3041471 2019-04-26
however the most of the smear present in some samples had been prevented. We
believe that the
reduction in smear far outweighs the reduction in product consistent with the
possibility that
levels of betaine can be employed that improve signal to noise ratios.
Amplification conditions
were: SOmM Tris.C1 pH 8.3, 90mM Magnesium acetate, 2mM DTT, 80mM Potassium
acetate,
200 M dNTPs, 600ng4t1 gp32, 200ng/111 uvsX, 60ng/1.11 uvsY, 300nM of primers
ApoB4 and
Apo300 (SEG IDs 20 and 21), 40ng/i.11 Bsu polymerase, 2.5 copies/A1 genomic
DNA, 120
minutes at 37 C.
Figure 49 shows combination strategies involving oligonucleotides with
different
nucleoprotein activities
(A) Single reaction nesting strategies are outlined. An outer pair of active
nucleoprotein
filaments rapidly amplifies DNA, although these primers may also be fairly
noisy. The outer
primer pair may be maintained at relatively low concentration, low enough to
be unable to
achieve gel-detectable levels of product. The inner oligonucleotides are less
active, but much
cleaner. These inner oligonucleotides are maintained at high levels. Activity
may be tuned by
altering length, composition, and backbone character as detailed in this
disclosure. The outer
primers quickly enrich the target in the sample, for example through a million-
fold
amplification. The inner primers, being slow but clean, generate little/no
noise, but are less
sensitive. The enrichment of target by the outer primers is sufficient to
permit robust product
accumulation by action of the slow inner primers in an appropriate timeframe.
(B) Amplification
schemes are shown in which an active primer is combined with a less active
primer. While the
active primer may engage in noise at low target concentrations, the slower
oligonucleotide does
not engage in artifacts, and so if association of the primers is tested at the
end of the reaction this
can cleanly determine whether target was presence or absent in the sample.
Figure 50 describes several detection protocols. Three simple strategies are
shown
schematically by which amplification of a target might be assessed without a
need for gel
electrophoresis. In all cases a primer is utilized that can be attached to a
solid phase to permit its
separation from bulk reactants, and which can be washed. In the figure a
biotin residue on one of
the primers represents this immobilizable primer, however other attachement
chemistries could
179
CA 3041471 2019-04-26
be used. (1) One primer is immobilizable, and the other contains a label of
some description, for
example an enzyme. (2) One of the amplification primers is labeled in some
way, and the
immobilizable primer is a third primer, which recognizes sequences within the
main amplicons.
This third primer may be present in the reaction environment during the main
amplification, but
may be for example designed to be quiet (e.g. by being short), or could be
added only at the end
of the reaction. (3) A third primer is employed. In this case however the
immobilizable probe is
related to one of the amplification primers, but contains additional 5'
residues that permit it to
stably capture amplicons at the end of the reaction, as polymerase extension
of the 3'-end of the
complement in the amplicon (i.e. 'backfire' synthesis) creates a stable duplex
not subject to
branch migration if reaction proteins are washed away.
Figure 51 shows a third probe enrichment strategy: Bead capture Part I.
Schematic of
experimental strategy in which a third probe enriches bona fide products
derived from the target
(A) Schematic representation of the relationship between primers. A PstI
restriction
enzyme site is indicated, located in the middle of NEST3-28
(GGATGAATAAGCTGCAGCTGATTAAAGG ¨ SEQ ID 76) and NEST3-26
(ATGAATAAGCTGCAGCTGATTAAAGG ¨ SEQ ID 77) primers which are homologous to
an amplified sequence generated by primer ii and K2 (SEQ IDs 66 and 69
respectively above).
(B) Illustration of the immobilization of a 5'-biotinylated version of N3-26
which is bound to
streptavidin-coated magnetic particles. (C) Scheme for capture experiment. An
amplification
reaction is established in which oligonucloetides ii and K2 are combined in
solution phase with
the N3-26 primer immobilized on a bead. The reaction is incubated for 90
minutes, and every 20
minutes the beads are dispersed by flicking (they settle only very slowly in
the 5% PEG reaction
solution). At the end of the incubation period the beads are concentrated
using a magnet, and the
supernatant is removed for analysis. The beads are washed twice quickly with
water, and then
incubated for 30 minutes at 37 C with excess PstI enzyme in appropriate
buffer. Once again the
supernatant is removed and analysed.
Figure 52 shows a third probe enrichment strategy: Bead capture 11.
Experimental results
demonstrating that a low activity nucleoprotein filament immobilized on a
solid support can
180
CA 3041471 2019-04-26
participate as a third primer and separate target amplicons from primer noise.
(A) Schematic representation of the relationship between primers. A PstI
restriction
enzyme site is indicated, located in the middle of N3-28 and N3-26 primers.
(B) Amplification
of B.subtilis DNA has been performed in the presence (lower panel) or absence
(upper panel) of
target DNA at 2 copies/Al. Lane 1 contains size ladder. The primers J1 and K2
efficiently
amplify their target in the presence of few copies of target genomic DNA
(Lower panel, lane 2),
however in the absence of target in the sample generate a smear as evidenced
in the upper panel
lane 2. Oligonucleotides N3-28 and N3-26 are homologous to sequences slightly
3', and the
same sense, to JI. N3-28 is a 28-mer, and N3-26 is a 26-mer lacking the two 5'-
most bases from
N3-28. Co-incubation of N3-28 or N3-26 with J1 and K2 primers results in the
generation of
two main products corresponding to the J1/K2 product, and the expected N3-
28/K2 or N3-26/K2
product (Lower panel, lanes 3 and 4). When N3-28 is incubated alone it
generates some smear,
but N3-26 incubated alone does not. We deduce that N3-26 is a 'quiet'
olignucleotide and may
rely significantly on hybridization to displaced strands from the elongation
of more active
primers in order to generate products. (C) The results of an experiment
carried out as detailed
schematically in Figure 51, and using start target density of 30 copies/ 1.
Lane 1, size marker.
Lane 2 and 3, products of an entirely liquid phase reaction (no immobilization
of N3-26)
perfomed without (lane 2) or with (lane 3) target. This is effectively
equivalent to lanes 4 of the
upper and lower panels of (A). Lanes 4 and 5 contain the first supernatant
removed after the
amplification incubation, without and with target respectively. As expected
there is product in
the target containing reaction, and the smaller product derived from use of
the N3-26 primer
(immobilized on the bead) is not seen. Also there is a smear, as expected, in
the targetless
sample. Lanes 6 and 7 contain the supernatant following PstI digestion of the
washed beads.
Interestingly the smear in the non-target sample is not present, however in
the target sample
there is DNA. The products released from the beads appear to comprise the
expected N3-26/K2
product, slightly faster mobility than in lane 3 due to clipping by PstI, as
well uncut .11 /K2
product that has co-purified. It is perhaps unsurprising that some J1/K.2
product co-purifies
because hybrids may well have formed between such products and the bead-
immobilized
products and oligonucleotides. It is, however, surprising that this material
has not apparently
181
CA 3041471 2019-04-26
been cut by PstI enzyme. The reason for this strange observation is not known
, but could
indicate that these products were present in hybrids that were resistant to
digestion, or lead to
nicking of only one strand of these products.
Figure 53 shows that Trehalose stabilizes lyophilizates to permit all
components except
; buffered sample to remain active for at least 10 days at room
temperature.
RPA reactions were assembled using primers specific for the human
Apolipoprotein B
locus. In general final reaction composition was 55mM Tris.C1 pH 8.4, 1mM DTT,
80mM
Potassium acaetate, 5% Carbowax 20M, 200 M dNTPS, 3mM ATP, 20mM
Phosphocreatine,
10Ong4t1 creatine lcinase, 300nM ApoB4 and Apo300 oligonucle,otide (SEQ ID 20
and 21
respectively) , 600ng/ 1 gp32, 200ng/ 1 uvsX, 60ng/ 1 uvsY, and 17ng/ 1. Bsu
polymerase,
¨160copies/ 1 Human genomic DNA. However, the 55mM Tris.C1 pH 8.4 and 80mM
Potassium acetate were not included in the lyophilizate, but added back with
the DNA sample
during reconstitution. Furthermore certain components were also omitted from
the lyophilizates
detailed as follows. Condition A, no trehalose used, Carbowax added with
sample DNA (not in
lyophilizate), polymerase added with sample DNA. Condition B, 50mM Trehalose
included in
lyophilizate, Carbowax added with sample DNA, polymerase added with sample
DNA.
Condition C, 50mM trehalose in lyophilizate, polymerase added with sample.
Condition D,
50mM trehalose in lyophilizate, only buffered sample DNA added for
reconstitution. After 3, 6,
or 10 days 'storage on the bench at room temperature, and with no special
storage conditions
(e.g. use of dessicants etc.) the activity of the lyophilizates were tested by
adding the missing
reagents to the dry pellet in a final volume of 50 1. Reaction condition D in
each case lacked
only the sample DNA in reaction buffer (55mM Tris.CI and 80mM Potassium
acetate). Only if
trehalose was present in the lyophilizate were reactions stable, but in those
cases the reactions
remained stable for at least 10 days. Longer periods were not investigated due
to running out of
test material, but may be possible.
Example 14: RPA reactions can be monitored in real time to allow assessment of
start
target copy number
Figure 54 shows how SYBR gold and SYBR green fluorescent dyes were assessed
for
182
CA 3041471 2019-04-26
their compatibilities in RPA reactions, and their reaction-monitoring
properties. (A)
Arrangement of the primers ApoB4 and Apo300 (SEQ IDs 20 and 21 respectively)
which flank
a fragment of the human Apolipoprotein B locus. (B) SYBR gold can be included
in RPA
reactions at dilutions of 1:50,000 or greater without complete inhibition of
the reaction.
Reactions were composed of 50mM Tris pH 8.4, 80mM Potassium acetate, 200 M
dATP,
dCTP, dGTP, and dTTP, 50 ng/ I creatine kinase, 1.5 mM ATP, lOrnM Magnesium
acetate,
2mM DTT, 30mM phosphocreatine, 300nM ApoB4 primer, 300nM ApoB300 primer, 5%
Carbowax 20M, 360 ng4t1 gp32, 86 ng/i.il uvsX, 15ng/ 1 uvsY, 35ng/p.lBsu
polymerase. SYBR
gold at the indicated dilution was included in the reaction. (C) RPA reactions
were established
in a 96-well plate, each with a final volume of 50 1. A mastennix of
components lacking only
the target DNA and the SYBR gold dye was established on ice. Five different
dilutions of SYBR
gold' were tested, with final dilutions from stock of 1:50,000, 1:60,000,
1:70,000, 1:80,000, and
1:100,000. For each dilution starting target copy densities of 2 and 20 copies
per microliter were
investigated (human genomic DNA diluted in TE to the expected copy density).
Once reactions
had been thoroughly mixed on ice, the plate was transferred to the pre-warmed
plate (37 C) of a
BIO-TEK FLX 800 fluorescent microplate reader. Readings were collected every 1
minute, and
300ng of human genomic DNA diluted in 50 1 of water was used as a sensitivity
standard for
the device. Data was transferred to Microsoft Excel and relative fluorescence
(arbitrary) was
plotted against incubation time for each reaction. (D) An experiment was
established as in (C)
except that SYBR green rather than SYBR gold Was assayed.
Figure 55 shows that real time RPA demonstrates quantitative behaviour over
four order
of magnitude using SYBR green. Figure 55 (A) Arrangment of primers used in
this analysis. (B)
RPA reactions were established in 96-well microwell plates, with a final
volume of 50 1.1.1 per
reaction volume, using primers specific or the B.subtilis Spo0B locus J1 and
K2 (J1 ¨ 5'-
ACGGCATTAACAAACGAACTGATTCATCTGCTTGG-3' SEQ ID 66, 1(2 ¨ 5' ¨
CCTTAATTTCTCCGAGAACTTCATATTCAAGCGTC-3' SEQ ID 69). A defined number of
copies of Bacillus subtilis genomic DNA was pipetted into wells in to the 96-
well plate, and
stored on ice. A reaction mastermix was mixed on ice, and then aliquoted in
the wells containing
DNA 'and mixed well. The reactions were then transferred to the pre-warmed
plate of a 1310-
183
CA 3041471 2019-04-26
TEK FLX 800 fluorescent microplate reader (set at 38 C). The reaction
composition was 50mM
Iris pH 8.4, 80mM Potassium acetate, 10mM Magnesium acetate, 2mM DTT, 200p.M
dATP,
dCTP, dGTP, and dTTP, 50 creatine lcinase, 1.5 InM ATP, 30mM
phosphocreatine,
300nM BOI primer, 300nM BsK2 primer, 5% Carbowax 20M, 360 ng/p.1 gp32, 86 ng/
I uvsX,
15ng/ 1 uvsY, 35ng/ 1 Bsu polymerase, and 1:50,000 dilution of the DMSO stock
of SYBR
green from molecular probes. The results of two equivalent experiments is
shown in (B) and (C).
After reactions had finished 7 11 of the reactions was diluted with lx sucrose
loading buffer and
separated on a 2% agarose gel (D, E). Note that when the highest concentration
of target was
used fluorescence became detectable earliest as expected, but failed to show
an enduring
exponential rise. This may arise due to consumption of reaction reagents (gp32
was used at
lower than typical concentrations in this experiment), or to an inhibitor
build-up, or otherwise.
Later experiments suggest this may arise due to gp32/uvsX undertitration. See
below. Also,
there some evidence of product contamination in (B) suggested by the faint
presence of the
expected band in the endpoint analysis gel (D).
Figure 56 shows that RPA reactions can kinetically assess of B.subtilis
genomic DNA
copy number of at least 5 orders of magnitude. RPA reactions was established
using Bacillus
subtilis primers 51 and K2 as in figure 55. However the quantity of protein
reagents was
increased as follows: gp32 was used at 600ng4i1, uvsX was used at 14Ong/ 1,
uvsY was used at
35ng/111, and ATP was at 3mM. Reaction volumes were increased to 10041 final
volume, and a
greater range of genomic DNA concentrations was used (from 0.1 to 100,000
copies/p1). No
early trailing of higher copy number samples was observed, and we believe this
reflects that
earlier experiments had somewhat undertitrated gp32 and uvsX. After reactions
had finished 7
of the reactions was diluted with lx sucrose loading buffer and separated on a
2% agarose
gel.
Figure 57 shows that real time RPA demonstrates quantitative response to
variation in
human genomic DNA copy number. Figure 57 (A) Arrangement of ApoB4 and Apo300
primers
which amplify a human DNA sequence. (ApoB4
5'- =
CAGTGTATCTGGAAAGCCTACAGGACACCAAAA3' SED 1D 20, Apo300 ¨ 5'-
184
CA 3041471 2019-04-26
TGC1TTCATACGTTTAGCCCAATCTTGGATAG3' SEQ ID 21). (B) RPA reactions were
established in 96-well microwell plates, with a final volume of 100 I per
reaction volume. A
defined number of copies of human genomic DNA was pipetted into wells in to
the 96-well
plate, and stored on ice. A reaction mastermix was mixed on ice, and then
aliquoted in the wells
containing DNA and mixed well. The reactions were then transferred to the pre-
warnted plate of
BIOTEK FLX 800 fluorescent microplate reader. The reaction composition was
50mM Tris pH
8.4, 80mM Potassium acetate, 10mM Magnesium acetate, 2mM DTT, 2001LM dATP,
dCTP,
dGTP, and dTTP, 50 ng4t1 creatine kinase, 1.5 mM ATP, 30mM phosphocreatine,
300nM
ApoB4 primer, 300nM Apo300 primer, 5% Carbowax 20M, 360 ng/u1 gp32, 86 ng4i1
uvsX,
15ng4i1 uvsY, 35ng/t.il Bsu polymerase, and 1:50,000 dilution of the DMSO
stock of SYBR
green from molecular probes. Note that, as in Example 2 (C), the highest
concentration of target
generated a curve which trailed off unexpectedly early. As with Figure 55
these experiments
were performed under conditions of uvsX, uvsY, gp32, ATP and dNTP
concentrations that may
require finessing to ensure robust detectable exponential phase at higher
target concentrations.
This is supported by (C), in which a similar experiment was performed to that
in (B), except that
concentrations of certain reagents were as follows: gp32 was used at 600ng/ul,
uvsX was used at
140ng/ 1, uvsY was used at 35ng/ 1, ATP was at 3mM. After reactions had
finished 7 ul of the
reactions was diluted with lx sucrose loading buffer and separated on a 2%
agarose gel.
" Example 15: Asymmetric primer RPA using a 3'LNA-capped primer
As an example of amplification of a target DNA using the asymmetric primer RPA
method, standard RPA conditions are employed with two differences. First, one
primer is a
3'LNA-capped primer. Secondly, an additional polymerase, phi29, is used, which
is unable to
initiate synthesis from recombinase intermediates.
Thus amplification conditions are as follows:
50mM Tris.CI pH 7.85
10mM Magnesium acetate
2mM Dithiothreitol
80mM Potassium acetate
5% PEG compound (Carbowax 20M)
15mM Phosphocreatine
185
CA 3041471 2019-04-26
3mM ATP
2001.LM dNTPs
12 ng/u1 creatine kinase
0.3 uM oligonucleotide 1, 3'LNA-capped
0.3 04 oligonucleotide 2
350 ng4i1 T4 gp32
10Ong/ 1 T4 uvsX
20ng/p1 T4 uvsY
20ng/ 1 Bsu polymerase
20n8/ 1 ph129 polymerase (exo-, or exo-attenuated)
Incubation at 33-40 C for 30 minutes to 2 hours
Example 16: Initiation of real-time RPA with deprotection of caged ATP
As an example of the use of ATP pulsed RPA to quantify input template DNA
reactions
are assembled as standard RPA reactions with several differences. Firstly,
caged-ATP is used
instead of ATP. Secondly, a double-stranded DNA detection reagent, SYBR green,
is used to
quantify RPA products. Primers are designed such that short (<200bp) reaction
products are
generated, to ensure complete synthesis with each pulse of ATP.
Thus amplification conditions are as follows:
50mM Tris.C1 pH 7.85
10mM Magnesium acetate
2mM Dithiothreitol
80mM Potassium acetate
5% PEG compound (Carbowax 20M)
3mM caged-ATP
1:50,000 dilution of SYBR-green I commercial stock (Molecular Probes)
200uM dNTPs
0.3 p.M oligonucleotide 1
0.3 OA oligonucleotide 2
600 ng/u.I T4 gp32
150ng/ 1 T4 uvsX
25ng/pil T4 uvsY
20ng,/ 1Bsu polymerase
Incubation at 33-40 C
Prior to each cycle of ATP uncaging, SYBR-green I fluorescence (494nm
Excitation
Max ¨ 521m Emission Max) is measured in the sample. The detected fluorescence
will be the
basis of quantification. Every minute a short pulse of 365nm light is applied
to uncage ATP,
186
CA 3041471 2019-04-26
leading to a burst of recombinase activity. Fluorescence measured at each
cycle is then
compared to an input template standard and the quantity of input sample
template can be
determined.
Example 17 describes the use of caged ATP to control the initiation of RPA in
a
Polony analysis
As an example of RPA-based polony analysis polyacylamide gels are generated on
microscope slides as described by Church and colleagues [Mitra R, Church G
(1999) In situ
localized amplification and contact replication of many individual DNA
molecules. Nucl Acids
Res 27(24): e34i-vi.]. In this analysis distinct polymorphic products are
detected after an initial
amplification by single-base extension using fluorescent nucleotides and
polymorphic-target
specific primers. One difference between this configuration and normal polony
analysis is that it
may not be necessary to use the 5'acrydite modified primers, as RPA performed
at a low
constant temperature and for significantly shorter times (normally PCR-based
polony reactions
would take 3-7 hours). In this example, both 5'acrydite modified primers as
well as normal
primers are used.
In place of the PCR "diffuse-in" mix, an RPA "diffuse-in" mix is used and
comprises the
following:
50mM Tris.CI pH 7.85
10mM Magnesium acetate
2mM Dithiothreitol
80mM Potassium acetate
5% PEG compound (Carbowax 20M)
15mM Phosphocreatine (included depending upon configuration)
200 M dNTPs
* 3niM caged-ATP (included depending upon configuration)
12 ngiul creatine kinase (included depending upon configuration)
0.3 1.1A4 oligonucleotide 1
0.3 g.i.M oligonucleotide 2
350 ng/AI 14 gp32
10Ong/p.1 T4 uvsX
20ng,/ 1 T4 uvsY
20ng/11 Bsu polymerase
The reaction is then initiated either by diffusing in ATP, or activating caged-
ATP
187
CA 3041471 2019-04-26
included in the original "diffuse-in" mix. Polonies are generated after
incubation at 33-40 C for
30 minutes to 2 hours
After amplification, a new polymorphic-target primer is diffused in along with
fluorescently labelled nucleotides, and the reaction is re-started with an
additional pulse of ATP,
either applied manually, or by photolysis. Incorporated fluorescence can then
be quantified
using fluorescence microscopy.
Example 18: Non gel-based determination of polymorphic repeat number in an
amplicon
using recombinase-mediated hybrid formation between single-stranded amplified
DNA and
duplex probes
In this example an Slit is amplified by flanking oligonucleotide primers. The
STR is
the marker known as TPDX and comm-only used in both US and European standard
STR marker
sets used for forensic analysis. The marker is amplified by flanking primers
with the sequence
for primer I of 5'-ACTGGCACAGAACAGGCACTTAGGGAACCC-3 (SEQ. ID. NO: 92),
and primer 2 5"-GGAGGAACTGGGAACCACACAGGTTAATTA-3' (SEQ. ID NO: 93). The
marker has a repeat of 4 nucleotides, AATG, which vary between 5 and 14
repeats.
Amplification conditions are as follows:
50mM Tris.C1 pH 7.85
10mM Magnesium acetate
2mM Dithiothreitol
80mM Potassium acetate
5% PEG compound (Carbowax 20M)
i 5mM Phosphocreatine
3mM ATP
2001,jM dNTPs
12 ng/ 1creatine kinase
0.3 M oligonucleotide I, 5'-fluorescein-labeled
0.05 M oligonucleotide 2
600 ng/ I 14 gp32
188
CA 3041471 2019-04-26
150ng,/ 1 T4 uvsX
35ng/ 1 T4 uvsY
20ng/ 1Bsu polymerase
Incubation at 33-40 C for 30 minutes to 2 hours
The amplification reaction generates a quantity of single-stranded DNA due to
primer
imbalance, and this single-stranded DNA is fluorescently labeled. At the end
of the
amplification reaction the reaction mixture is contacted directly to an array
of spatially
immobilized double-stranded probes. Each probe corresponds to one of the known
polymorphic
forms of the DNA under study, and is the same length as the amplicon that
would be made if
this form exists in the sample.
Recombinase loads onto the amplified single-stranded sample DNA to form
homology-
searching filaments. These filaments will be able to target the double-
stranded probes
immobilized on the array support surface. When a recombination event occurs
between the
recombinase-coated amplification product and the probes it will lead to the
amplification
product becoming productively Watson-Crick base-paired with its complement in
the probe,
while the originally paired equivalent strand is displaced and freed into
solution. Ideally
completion of such recombination events will be inhibited if the event
initiates between a sample
DNA and an imperfectly matched probe leading to enrichment of only perfect
hybrids.
The reaction is dynamic such that hybrids formed between probe and sample may
themselves become targets for other targeting DNAs. This dynamic behavior
coupled to a lower
efficiency of completion of recombination of imperfectly formed hybrids should
lead to an
enrichment of the perfect hybrids in the population of probe/sample mixtures.
This is further
enhanced by the addition of either a helicase such as E.coli PriA (2-20ng4t1),
E.coli DnaB (5-
50ng/ 1), E. coil RuvAB (5-50ng/ 1), T4 phage dda helicase (5-50ng,/ I), T4
phage gp41 (5-
50ng/ 1) or an appropriate nuclease. These agents utilize, or enhance,
disruptions in the helix,
which exist due to mismatches and lead to such imperfect hybrids being less
stable than perfect
hybrids.
After a suitable incubation period, such as 10-30 minutes at 28-40 C, the
reaction is
189
CA 3041471 2019-04-26
terminated if necessary, for example by the addition of EDTA or chaotropic
agents such as urea
or guanidine hydrochloride, although no such termination may be necessary.
Productive
interactions between probe and sample are then measured by exposing the array
to light of an
appropriate wavelength, and recording fluorescence using appropriate filters
and camera.
Example 19 shows a non gel-based determination of polymorphic repeat number in
an
amplicon using recombinase-mediated hybrid formation between duplex amplified
DNA and
single-stranded probes
As above this example would involve amplification of the TPDX STR marker used
in
forensics. The amplification conditions would be the same as for Example 18
except that the
ratio of amplification primers would be 1:1, with both used at a concentration
of 0.3 114. The
amplification reaction is then contacted to an array of sinRle-stranded probes
corresponding to
the known possible polymorphic forms of the DNA under study. With this format
the
recombinase loads preferentially onto the probes which then search and form
hybrids with the
double-stranded sample DNAs. As in Example 18 hybrids will ideally become
enriched if they
are perfect, and similarly the inclusion of agents such as helicases (e.g.
PriA, recG, DnaB,
RuvAB, gp41, or dda) or appropriate nucleases can accelerate and improve this
enrichment.
After a suitable incubation period, with or without arrest of the reaction as
necessary,
interactions are visualized as described in Example 18.
Example 20 shows a non gel-based determination of polymorphic repeat number in
an
amplicon using recombinase-mediated hybrid formation and nuclease processing
of heterologies
to release a label from an immobilization point
In this example the amplification reaction and hybrid formation between probe
and
sample are performed as described in Example 1, except that the amplification
primers need not
contain a label. The probe instead of the sample is labeled at one end. After
an appropriate
incubation period, as described for Examples 18 and 19, arrays are treated
with an appropriate
nuclease, which will cleave imperfect DNA duplexes. As such cuts are made in
the backbone of
imperfect hybrids and the label is thus freed from an immobilization point.
Additionally a DNA
polymerase may be included such that if a nick is introduced by the nuclease,
and the
190
CA 3041471 2019-04-26
polymerase is strand displacing then it acts to synthesize in a strand-
displacing manner and thus
remove the labeled strand accordingly. The array can be then quantified as
described in
Example 18.
Example 21 shows a non gel-based determination of polymorphic repeat number in
an
amplicon using recombinase-mediated hybrid formation and nuclease processing
of heterologies
followed by DNA synthesis labeling
In this example amplification and hybrid formation are as for either Example
18 or
Example 19 except that the amplification primers are not necessarily labeled.
During the hybrid
formation phase a nuclease is included that nicks at bubbles or helix
disturbances and the
inclusion in the reaction of labeled nucleotides and a strand displacing
polymerase leads to
imperfect hybrids being processed to a perfect hybrid form which includes
modified labeled
bases that can then be visualized.
Example 22 shows the use of a helicase to disrupt imperfect hybrids in a
dynamic
recombination system environment
In this example amplification and hybrid formation is done as for either
Example 18 or
Example 19 except that the amplification primers are not necessarily labeled.
The probe or
sample DNA should however contain a label. The probe or sample DNA is
immobilized at one
end to a solid support or bead through a high affinity non-covalent
interaction such as a biotin-
streptavidin interaction. During the hybrid-forming phase a helicase such as
the T4 dda helicase,
or combination of helicases such as PriA and dda helicases, or other mixtures
are include to
target imperfect hybrids and unwind them, effectively accelerating the
dissociation of high
affinity non-covalent interaction by physical disruption. After a suitable
incubation period the
association between the labeled nucleic acid and the solid support is measured
positive signal
indicating that perfect hybrids have formed.
Figure 58 shows in principal how imperfect hybrids will possess bubbles which
permit
enhanced overall duplex disruption in a dynamic recombination environment
A double-stranded amplicon shown in A containing 8 repeats of a short tandem
repeat
and flanked by unique sequences used for amplfication is targeted in B by
single-stranded probe
191
CA 3041471 2019-04-26
oligonucleotides containing varying numbers of the repeat unit. The
hybridization is driven by
recombinase (not shown here). In C the resultant double-stranded hybrids are
shown, some of
which contain bubbles caused by non-identical repeat numbers between hybrids.
These bubbles
are larger as the number of repeats different grows in number, and thus such
hybrids become
better targets for recombinase, or other DNA processing enzymes.
Figure 59 shows the possible outcome of a hybridization reaction as described
in
Example 18, of figure 58, is shown. A sample DNA containing 8 repeats is
incubated with an
array of probes corresponding to different repeat numbers. Over time hybrids
are enriched for
perfect repeat numbers, and less so but to a small extent those that differ
only by one or two
repeats.
Figure 60 shows how easily RPA reactions can be performed to assess the
presence of
specific RNA species by including an enzyme capable of reverse transcription
in the reaction.
Defined copy numbers of viral MS2 RNA (from a commercial source) were
incubated in
RPA reactions which included reverse transcriptase as indicated. Two primers
were employed
which recognised the MS2 sequence as indicated, more specifically these
primers were:
MS2up2 TTCCGACTGCGAGCTTATTGTTAAGGCAATG - SEQ ID78
MS2down2 CTTAAGTAAGCAATTGCTGTAAAGTCGTCAC - SEQ ID79
Reaction conditions were standard except for increased concentration of dNTPs
(50011M), an increase in DTT, the inclusion of RNase inhibitor, and the
inclusion of reverse
transcriptase where indicated. Conditions were:
50mM Tris.C1 pH 8.5
10mM Magnesium acetate
10mM Dithiothreitol
80inM Potassium acetate
5% PEG compound (Carbowax 20M)
15mM Phosphocreatine
3mM ATP
50004 dNTPs
100 ng/ 1 creatine kinase
0.3RM MS2UP
0.3 M MS2DOWN
192
CA 3041471 2019-04-26
800 ng/ I T4 gp32
120ng4t1 T4 uvsX
30ng/ 1 T4 uvsY
70ng/ 1Bsu polyrnerase
0.13 Units/ 1RNase inhibitor
MuMLV (RNase H containing) reverse transcriptase 10 units/ I (Promega)
Reactions products were phenol extracted, precipitated and separated on a non-
denaturing acrylamide gel before staining with SYBR-gold. RNA concentrations
as low as 100
copies per microliter can be readily detected in this format without further
optimization.
Figure 61 shows that dUTP can be used to partially or completely replace dTTP
in RPA
reactions, thus offering a strategy to control carry-over contamination.
RPA reactions were established using the following conditions:
50mM Tris.C1 pH 8.5
10mM Magnesium acetate
2mM Dithiothreitol
100mM Potassium acetate
5% PEG compound (Carbowax 20M)
15mM Phosphocreatine
3mM ATP
200 M dNTPs (dA, dC & dG)
100 ng/ I creatine kinase
0.3 M J1 primer for B.subtilis DNA (SEQ ID 66)
0.3 M K2 primer for B.subtilis DNA (SEQ ID 69)
630 ng/ I T4 gp32
140ng/ 1 14 uvsX
35ng/ 1 T4 uvsY
20ng/ 1 Bsu polymerase
100 copies/p.1 B.subtilis genomic DNA
The product of reactions was separated on a 2% agarose gel and stained with
ethidium
bromide. As indicated in the left-most panels, dUTP or dTTP was employed in
different
reactions. More specifically the upper left-most lane contained 200 M dTTP,
and was therefore
typical of a standard RPA reaction. The following six lanes also contained 200
M dTTP, but in
this case dUTP was also present at the indicated concentrations. The last six
lanes of the lower
left-most panel contain the products of RPA reactions in which no dTTP was
included, and the
indicated concentration (50-800 M dUTP) of dUTP was used. In all cases
amplification of the
193
CA 3041471 2019-04-26
expected fragment occurred, but notably the presence of dUTP decreased the
quantity of
amplicon generated, and inhibited the 'doubling-up' phenomenon often seen in
which snap-back
synthesis has occurred.
The right-most panel indicates the results of using as starting material
products generated
from the first reactions in the left-most panel following processing as
indicated in the schematic.
More specifically the reactions shown as A, B, C, D and E in the left-most
panels were
processed as follows. Based on staining intensity and estimation of DNA
content, an estimated
106 molecules of each product were incubated for 20 minutes with 411 of a
commercial
preparation (Roche) of heat-labile dUTP-deglycosylase, a similar amount was
left untreated, and
then finally all samples were heated to 94 C for 10 minutes. This enzyme was
quality-controlled
by the manufacturer to effectively combat carry-over contamination from 105
molecules, thus
we imposed a stringent challenge in this case. The samples were then used to
seed an RPA
reaction configured under normal RPA conditions with 20011M of each dATP,
dCTP, dGTP, and
dTTP (see conditions above). As indicated samples untreated with dUTP-
deglycosylase were
excellent start templates, while those that had been treated were extremely
poor unless no dUTP
had been initially employed. In particular even the mixed blends were
effective.
Figure 62 shows how one might combat carry-over contamination in the RPA
system.
The capacity to employ dUTP in RPA reactions could be used to prevent carry-
over
contamination according to the depicted scheme. RPA reactions would be
performed with pure
dUTP, or mixes of dTTP and dUTP as shown in figure 61. Prior to, or at the
initiation of an RPA
reaction, E.coli Uracil deglycosylase (UDG) or similar would be incubated with
the sample and
would attack carry-over material. Then after several minutes, or at the start
of the reaction, an
UDG inhibitor would be added. A specific inhibitor of E.coli UDG is known as
is referred to as
Uracil Gygosylase Inhibitor (UGI) and is a 9.5 1(13 peptide from Bacillus
subtilis, and is
commercially available.
Figure 63 shows how a simple integrated disposable system can be configured to
permit
rapid point-of-use testing for the presence of specific DNA sequences in a
sample. In (A) a
schematic description is given of a disposable RPA reaction/lateral flow strip
that might be
194
CA 3041471 2019-04-26
placed into a cheap heater than can deliver roughly 30-39 C in the vicinity of
a reaction pouch
containing dried RPA reagents. It is assumed that processed/lysed sample would
be used to
rehydrate the contents of the pouch in an appropriate volume, and then the
pouch would be
incubated at appropriate temperature for 15-60 minutes, as necessary.
Subsequently part or all of
the sample would be transferred to the sample pad of a lateral flow strip,
configured in a manner
that only if the correct amplicons have formed will a visible line form at a
specific position on
the strip. In (B) is shown the arrangement of two oligonucleotide primers used
for an RPA
reaction performed on male and female DNA respectively. The oligonucleotides
SRY3 and
SRY4 were used, and one contained a 5'-fluorescein moiety, and the other a 5'-
biotin moiety. At
the end of the reaction 1/500 of a 50 I reaction was mixed with 'running
buffer' (Milenia,
gennany) and applied to a commercially-available lateral flow strip (Milenia,
germany) such
that only products in which the fluorescein and biotin were co-associated
(i.e. in an amplification
product) would lead to the accumulation of visible gold particles on the
detection line. In (C) the
result of this experiment is shown. Only the reaction performed in male DNA
generates the
signal line, and the efficacy of the amplification reaction was separately
validated on agarose
gels (data not shown).
Figure 64 shows that RPA reactions are not inhibited by the presence of
materials in
blood providing appropriate ratios of reagents are employed.
The ability to amplify a genomic DNA fragment from whole and lysed blood added
directly to RPA reactions was investigated. RPA reactions were configured as
follows:
50mM Tris.C1 pH 8.5
10mM Magnesium acetate
2mM Dithiothreitol
80mM Potassium acetate
5% PEG compound (Carbowax 20M)
25mM Phosphocreatine
3mM ATP
200 M dNTPs
100 ng,4t1 creatine kinase
0.3 JAM oligonucleotide ApoB4 (SEQ ID 20)
0.3 M oligonucleotide ApoB300 (SEQ ID 21)
420 ng/ 1 14 gp32
195
CA 3041471 2019-04-26
140ng/ 1 T4 uvsX
35ng/ I T4 uvsY
20ng/ 1Bsu polymerase
In (A), either 1 I of water or 1 I of fresh whole human blood was added to
an RPA
reaction. Products were separated on a 2% agarose gel, and an arrow indicates
the position of the
expected amplicon. Non-specific 'primer' artefacts were present in water
control, and in the
sample, and presumably arise as a consequence of absent or very low copies of
target. We
deduce that very few copies of genomic DNA were available for amplification in
the blood
sample amplification. In (B) samples were analysed in which 1 I of blood had
first been mixed
with either 1 1, 2 1, 3 1, .4411, or 5121 of a lysis solution comprising
10m1'I Iris, 1mM EDTA,
120mM NaOH, 0.1%SDS (as indicated). In each case 1 1 of the respective lysate
was used to
start an RPA reaction (thus those lysed with more buffer contained less blood
sample per
amplification). Also shown are samples from the same experiment in which water
only (lane 1),
or purified DNA equivalent to 500 copies of target (Promega)(lane 7) were used
to seed the
amplification reaction. When 4 1 or Sul of lysis buffer were used, lysis of
the red blood cells
was clearly visible and the solution became viscous. We note that these
samples became
excellent start materials for RPA reactions, presumably releasing most of the
DNA as accessible
templates.
Figure 65 shows how real-time monitoring of RPA reactions can be employed to
optimise reactions.
In this case RPA reactions were established using the primer pair J1 and 1C2
mentioned
above, and template consisting of Bacillus subtilis genomic DNA (start density
1 copy per
microliter). Either Potassium acetate concentration, or PEG compound
concentration, was
varied. Other conditions were, unless specified otherwise:
50mM Tris.C1 pH 8.5
10mM Magnesium acetate
2mM Dithiothreitol
80mM Potassium acetate (or as indicated)
5% PEG compound (Carbowax 20M)(or as indicated)
25mM Phosphocreatine
196
CA 3041471 2019-04-26
3mM ATP
200 AM dNTPs
100 ng/ 1creatine kinase
= 0.3 AM oligonucleotide J1 primer (SEQ ID 66)
0.3 AM oligonucleotide K2 (SEQ ID 69)
620 ng/ 1 14 gp32
14Ong/ 1 T4 uvsX
35ng,/ 1 T4 uvsY
1:50,000 dilution form stock of SYBR green (lnvitrogen)
20ng/ 1 Bsu polymerase. At the end of the reaction a portion of the reactants
were mixed
with sucrose loading buffer and separated on an agarose gel, and then stained
with ethidium
bromide. We note that high salt slows the reaction, but there is a broad range
of salt
concentrations that work well. We also note that increasing PEG concentrations
from 4% to 9%
resulted in ever faster reaction behaviour, however the appearance of products
in the gel system
was adversely affected by higher PEG concentrations. We surmise that the
smeary appearance of
high PEG concentration samples may arise from ongoing networks of DNA
generated by very
high recombination activity. While this may be undesirable for gel analysis,
it may be perfectly
adequate for real-time analysis using fluorescence.
Figure 66 shows how RPA reactions are affected by Magnesium concentration.
Real-time RPA reactions were established as follows:
50mM Tris.C1 pH 8.5
6, 10, or 16mM Magnesium acetate as indicated
2mM Dithiothreitol
80mM Potassium acetate
5% PEG compound (Carbowax 20M)
25mM Phosphocreatine
3mM ATP
200 M dNTPs
100 ng/ 1 creatine kinase
0.3 M oligonucleotide J1 primer (SEQ II) 66)
0.3 AM oligonucleotide IC2 primer (SEQ PD 69)
600 ng/ 1 T4 gp32
120 ng/ul T4 uvsX
30 ng/ 1 T4 uvsY
20rtgAtlBsu polymerase
1:50,000 dilution from stock of SYBR green (Invitrogen)
197
CA 3041471 2019-04-26
The start density of target DNA, Bacillus subtilis genomic DNA, in copies per
microliter
is indicated. In the 6rnM and 16mM Magnesium acetate samples some carry-over
contamination
was evident when samples at end-point were checked on agarose gels, and this
may in part
explain the poor resolution between zero and 1 copy per microliter start
density. Note that RPA
is effective at all tested Magnesium concentrations, but is much faster as the
Magnesium
concentration rises. End-products from these higher Magnesium samples still
appear of high
quality (data not shown). This indicates that in order to achieve maximal
reaction rates,
Magnesium concentrations significantly higher than lOrnM (used as a standard
through most of
this document) may be usefully employed.
Figure 67 shows that different primer pairs/amplicons show different
amplification
kinetics.Reaction time dependence varies between oligonucleotide primers, and
analysis
indicates a possible sequence-bias underlying this phenomenon.
Two different timecourses were performed using different primer pairs.
Conditions were
not identical between these reactions, nevertheless the level of variation
indicated is typical of
the range of behaviours that we have observed between primer pairs in our
hands and is included
as just one typical example of rates that are observed (CSF primers used at
125ng,4t1 UvsX,
45ng/ 1 uvsY; Apo primers used at 200ng/u1 UvsXõ 60ng,/ 1 uvsY). Primer
sequences are
shown. Primers CSF5'(SEQ ID 32) and CSF3' (SEQ ID 33)(targeting the human STR
locus
CSF1P0) are really an exceptional primer pair insofar as they demonstrated
particularly rapid
product accumulation kinetics, the fastest we have as yet determined. Primers
ApoB600 (SEQ
ID 46) and ApoB300rev (SEQ ID 47), targeting part of the human Apoliprotein B
locus,
demonstrate slower amplification kinetics. In this experiment this latter pair
generated a similar
amount of material after 40 minutes as occurred after only 20 minutes with the
CSF primers.
Based on this, and other, data we conclude that typical doubling times vary,
on the average,
between about 30 seconds and 1 minute for most primer pairs of 30-35 residues
in RPA. CSF
primers were also notably dominant in multiplex experiments (data not shown),
consistent with
the notion that they were particularly efficient and fast. Composition
analysis of the primers,
comparing both CSF primers with, in this case, the Apo primers used here,
highlights that
guanosine content is low in both CSF primers, while average in the Apo
primers, and cytosine
198
CA 3041471 2019-04-26
content is one third or more for both CSF primers, while one quarter or less
for the Apo primers.
This suggested to us a possible composition origin for rate variability.
Reactions were performed
under similar conditions apart from the aforementioned differences in UvsX and
UvsY, and
slightly variant dNTP concentrations:
50mM Tris pH 8.4
80mM Potassium acetate
10mM Magnesium acetate
2mM DTT
5% PEG compound (Carbowax-20M)
3mM ATP
20mM Phosphocreatine
10Ong/01 creatine kinase
600ng/m1 gp32
200 OM dNTPs for Apo primer, 100 OM dNTPs for CSF primers
300nM each oligonucleotide
Figure 68 shows that appended 5' homopolymeric stretches influence reaction
behaviour
¨ cytosine increases activity and guanosine decreases it.
Primers were as indicated. JI and K.2 (SEQ IDs 66 and 69) are homologous to
part of the
Bacillus subtilis Spo0B locus. Reaction conditions were:
50mM Tris pH 8.4
80inM Potassium acetate
10mM Magnesium acetate
2mM DTT
5% PEG compound (Carbowax-20M)
3mM ATP
30mM Phosphocreatine
100ng/plcreatine kinase
420ng/ 1 gp32
UvsX
35ng/I.J.1UvsY
200 p.M dNTPs
300nM each oligonucleotide
35ng/plBsu polymerase
25 copies/pi B.subtilis genomic DNA
1:50,000 dilution from stock of SYBR green (Molecular probes)
Reaction volumes 501.1.1, 37 C
Primers J1(C), K(C), J1(G), and K2(G) were equivalent to JI and K2, but
possessing
199
CA 3041471 2019-04-26
appended stretches of cytosine or guanosine, as indicated. Reactions were
established on ice in a
microtitre plate, and then transferred to the heated stage of a fluorometer
(FLX800). After 90
minutes the reactions were diluted with lx sucrose loading buffer to twice the
original reaction
volume, and then 201.tI of this was run directly on a 2% agarose gel.
Reactions either contained
target DNA, or no target. We observed that appending cytosine residues to the
primers resulted
in faster amplification rate, while appending guanosines resulted in defective
amplification.
Cytosine-appended primers also amplified 'noise' very rapidly.
Figure 69 shows that appended 5' sequences consisting of Thymine and Cytosine
residues demonstrate significant variation in amplification behaviour.
Primers were as indicated. J1 and K2 (SEQ IDs 66 and 69) are homologous to
part of the
Bacillus subtilis Spo0B locus. Reaction conditions were:
50mM Tris pH 8.4
80mM Potassium acetate
10mM Magnesium acetate
2mM DTT
5% PEG compound (Carbowax-20M)
3mM ATP
30mM Phosphocreatine
10Ong,/ 1 creatine kinase
420ng/ 1 gp32
140ng/ 1UvsX
= = 35ng/ 1UvsY
200 M dNTPs
300nM each oligonucleotide
35ng/ 1 Bsu polymerase
20 copies4t1B.subtilis genomic DNA
1:50,000 dilution from stock of SYBR green (Molecular probes)
Reaction volumes 50u1, 37 C
Amplification reactions were assessed in real-time employing SYBR green dye as
described in Figure 66. Conditions were similar, except that only 20 copies/ 1
of start target
template were employed, and the primer sequences varied. Endpoint analysis was
made by
running products generated at 60 minutes on a 2% agarose gel after dilution as
described for
Figure 68. Reactions containing homopolymeric thymidine stretches, or very
thymidine-rich
stretches, amplified somewhat poorly. Typically less DNA was generated, and
there was some
200
CA 3041471 2019-04-26
indication that asymmetry of amplification was occurring substantially. In
general this data was
consistent with cytosine residues being preferred in the 5' sequences, and
that other hard-to-
determine phenomena were occurring, possibly some effects of preferred
recombinase 'phasing'.
Figure 70 shows more detailed study of the effects of 5' appended sequences.
We investigated yet more primers derived from the .1.1 and K2 primers (SEQ IDs
66 and
69), mostly containing additional pyrimidine residues, but one pair containing
a very 5'
guanosine residue. Some primers (6 and 12 in this series) contained 5'
pyrimidines, but were
shortened to a total of 33 residues by removing bases at the 5' (J1) or 3'
ends (K2) respectively.
These primers thus shared less than 30 residues of homology with the genomic
target. Primers
were incubated under similar conditions to those used in Figures 69 and 70.
The main
differences were that 63Ong/m1 of gp32 were employed, and only 1 copy per
microliter of target
genomic DNA was used. Furthermore, in these experiments we also examined the
rate of
amplification occurring in reactions containing only single primers. In those
reactions containing
primer pairs, pairs were examined containing one oligonucleotide that is a J1
derivative, and one
that is a K2 derivative. Pairs were used in which derivatives were containing
similar 5'
modifications. Notably single primer amplification rates were quite similar to
two primer
amplification rates at this low target density, indicating that primer noise
tends to initiate as a
competitive effect at about this very low target density. Oddly, primers 8 and
9, differing in only
one cytosine residues, showed profoundly different amplification behaviour.
This may indicate a
that a minimal stretch of cytosines is required for high rate activity, or
that some specific
phasing effect is in evidence.
Figure 71 shows several strategies by which the specificity of sequence
detection might
be improved by the use of 'third probes', and how they might be configured to
function with
fluorophore/quencher pairs.
The presence of a specific DNA sequence can be detected by use of a third
oligonucleotide probe which is blocked at the 3' end, and hence does not
engage in noisy
amplification (at least without subsequent processing). Such third probes may
form hybrids with
the amplicon specifically, and in such a duplex environment become the
substrates for DNA
201
CA 3041471 2019-04-26
processing enzymes which cleave the probe, thus separating the fluorophore and
quencher.
Given the nature of the recombinase environment, fluorophore and quencher will
be separated
by no more than about 10-12 nucleotides. A selection of candidate nuclease is
listed, and
modified bases that would be included in the probes, or the backbone nature of
the probe for
each nuclease, are indicated.
Figure 72 shows how a helix-distorting or base-specific nuclease can be
employed to
detect specific sequenced including polymorphisms.
A single-stranded DNA is shown which has been coated with recombinase to form
a
homology-searching nucleoprotein filament. Arrows indicate the interaction of
such a structure
will homologous duplexes, either identical to the probe with regard to a
single nucleotide
polymorphism (SNP), or different (A instead of G). In both cases a strand
exchange occurs
successfully, and the outgoing strand is stabilised by single-stranded DNA
binding protein. The
resulting new duplexes are either perfectly complementary, or contain a base
mismatch, which
will cause a local distortion of the helix. In the presence of a suitable
structure-specific nuclease
the probe/template containing the distortion is specifically cleaved, either
as nicks possibly on
either strand, or as a double-strand break, depending on the nuclease. In
other formats a similar
approach could be used to detect the presence or absence of a specific
sequence, regardless or
not of polymorphic state, by including modified bases in the probe, such as 8-
oxoguanine. In
this case a base-removing and abasic-site-cleaving enzyme specific for 8-
oxoguanine and duplex
environments could be employed, such as E.coli Fpg protein (8-oxoguanine DNA
glycosylase).
In either case probe molecules might contain both a fluorophore and a quencher
which are
separated on cleavage.
Figure 73 shows how dual-labeled fluorescent oligonueleotides show
differential
properties in an RPA environment compared to an environment lacking saturating
recombinase
and single-stranded DNA binding proteins.
Upper left, three probes are shown which were used in an experiment to
investigate the
fluorescent properties of those oligonucleotides in the presence or absence of
proteins used in
RPA environments. Conditions were as follows:
202
CA 3041471 2019-04-26
50mM Tris pH 8.4
80mM Potassium acetate
10mM Magnesium acetate
2mM DTT
5% PEG compound (Carbowax-20M)
3mM ATP
30mM Phosphocreatine
100ng/ 1 creatine kinase
200 1.tM dNTPs
120nM each oligonucleotide as indicated
Optional:
630ng/iAl gp32
140ng/p1 UvsX
35ng/ 1 UvsY
We observe that the FAM only containing oligonucleotide is slightly quenched
in the
presence of DNA binding proteins, likely indicating some quenching of
fluorescence by protein
bound close to the fluorophore. In marked contrast, however, the
oligonucleotides containing
both fluorophore and quencher show a reverse phenomenon, and this is markedly
different
depending on whether they are 10 or 15 residues in length. More specifically
both latter probes
were significantly quenched in the absence of proteins, consistent with the
idea that formation of
a random coil would ensure efficient quenching regardless of length. However
in the presence of
proteins this quenching is reduced. In particular the quenching for the 15-mer
is almost
abolished, and it assumes a fluorescence close to that level of the PAM only
containing
oligonucleotide in the presence of proteins. Conversely the 10-mer is little-
affeC-fed- and remains
highly quenched. This reduction in quenching for the 15-met is most easily
explained by the
suggestion that in the presence of DNA binding proteins the probe is stretched
out into a fairly
rigid rod. For example 1.JvsX and recA stretch single and double-stranded DNA
to about 1.5
times the equivalent length of B-form DNA, and similar extension is described
for gp32. Thus,
and in contrast to free solution behaviour, fluorophore and quencher are
highly separated in an
RPA environment, even more so than in a DNA duplex, and are likely to become
more
quenched on hybridization (although not investigated in this experiment). A
schematic is
provided on the right-hand side indicating what the states of the 10-mer and
15-mer probe may
be in the absence or presence of a recombinase environment, and potentially
when hybridized to
203
CA 3041471 2019-04-26
duplex DNA. Fluorophore and quencher are shown with a larger 'excitation'
radius, which when
overlapping would lead to efficient fluorescence resonance energy transfer,
and a concomitant
decrease in fluorescence emission.
Figure 74 shows that an amplicon-probe containing a tetrahydrofuranyl residue
becomes
a substrate for the E.coli Nfo enzyme in an RPA amplification reaction, is
cleaved, and that the
cleavage product can be elongated.
RPA reactions were established using the B. subtilis genomic DNA specific
primers J1
and K2 (SEQ 1D NO:66 and SEQ ID NO:69), the nested primer NEST26 (SEQ BD
NO:77) and
the biotin and Digoxygenin labeled probe THF-Probe 1 (5%BIOTIN-
CATGATTGGATGAATAAGCTGCAG-tetrahydrofuran-TGATTAAAGGAAAC-DIG-3' SEQ
ID NO:80) , which also contains a Tetrahydrofuranyl residue within the probe
body. This probe
is specific for the fragment amplified by .11 and K2 primers, and overlaps the
sequence of the
NEST26 primer (all these primers are discussed elsewhere). NEST26, a primer
which generates
no noise on its own, was included so that a smaller nested product might be
generated one of
whose ends would be the target for the probe, just in case the probe was
highly unstable in the
topologically cosnstrained environment which will result from probe invasion
into duplex J1/K2
amplicon. Reactions had the following conditions:
50mM Tris pH 7.9
80mM Potassium acetate
lOrnM Magnesium acetate
2mM DTT
5% PEG compound (Carbowax-20M)
3mM ATP
25mM Phosphocreatine
10Ong/ 1 creatine kinase
600ng/ 1 gp32
120nthil UvsX
30ng4i1UvsY
10011M dNTPs
200nM of J1 primer, K2 primer, and NEST26 primer
100nM Probe
30ng/ 1 Bsu polymerase
1000 copies/1A B.subtilis genomic DNA, or water controls as indicated
204
CA 3041471 2019-04-26
Amplification reactions were phenol extracted, precipitated, and separated on
a 16.5%
denaturing Urea-PAGE. Separated products were transferred to nylon membrane
and products
were detected using a Streptavidin-HRP conjugate. As the 5' end of the probe
was biotinylated,
this permitted the detection of uncut, cut and elongated probes. (A) Detection
of free, and
processed probes. Note that in the presence of an RPA environment capable of
amplifying the
target fragment, the probe was elongated preferentially elongated. The size of
the elongated
probe is consistent with elongation to the end of the J1/K2 amplification
product, and this can
occur because Nfo cleavage activity generates a 3'-hydroxyl, thus 'unblocking'
the probe and
permitting synthetic extension. More elongation was seen when a higher
concentration of Nfo
enzyme was employed. In (B) the structure of the probe used in this experiment
is shown. It
comprises a sequence homologous to part of the amplicons generated by Jl and
K2 primers, but
contains a tetrahydrofuranyl residue and is labeled at the 5' end with biotin,
and at the 3' end
(and thus blocked) with digoxygenin. In (C) a schematic representation is
given of the sequences
of events thought to underly the generation of elongated product via
interaction of the probe
with the desired amplicon, cleavage and subsequent polyrnerase elongation of
the free 3' end. In
this experiment apparent activity in reactions lacking start template is
believed to arise from
carry-over contamination of the amplicon from previous laboratory experiments,
a phenomenon
that has become problematic in our laboratory due to the very high sensitivity
of RPA.
Supporting evidence is provided in the subsequent figure.
Figure 75 shows that an amplicon-probe containing a tetrahydrofuranyl residue
becomes
a substrate for the E.coli Nfo enzyme in an RPA amplification reaction, is
cleaved, and that the
cleavage product can be elongated
In these experiments evidence is presented that the cleavage and extension of
a
tetrahydrofuranyl residue-containing amplicon-specific probe is specific and
dependant upon the
presence of that target sequences in the reaction, and also that having a free
end overlapping the
probe target (to prevent topological strain of the probe/target recombination
intermediate) is not
necessary. In (A) the reaction setup is depicted schematically. The Bacillus
subtilis locus
targeted by the primers J1, K2, L2, NEST26, (SEQ ID'S 66, 69, 71 AND 77) and
the probe are
depicted (the actual sequence is shown in figure 42B). Usually the primers J1
and K2 are
205
CA 3041471 2019-04-26
routinely used to amplify this locus and thus the principal laboratory
contaminant is the fragment
generated by these two primers. Thus we have also combined J1 with L2 in some
experiments,
this fragment not being subject to amplification from carry-over contamination
from the former
amplicon. In (B) are shown the results of an experiment in which RPA reactions
were
configured to amplify either the J1/K2 product, or the J1/L2 product, in the
presence of added
start template (genomic DNA) or without. Included in the reaction are the
probe and the E.coli
Nfo protein. In more detail the following conditions were used:
50mM Tris pH 7.9
80mM Potassium acetate
10mM Magnesium acetate
2mM DTT
5% PEG compound (Carbowax-20M)
3rnM ATP
25mM Phosphocreatine
10Ong/ 1 creatine kinase
600ng/ 1 gp32
120ng/ 1UvsX
30ng/ 1UvsY
100 M dNTPs
200nM of J1 primer, 200nM K2 OR L2 primer, and 200mM NEST26 primer (lanes 1-
4), in
lanes 5 and 6 JI and L2 primers were at 300nM and NEST26 was not used
120nM Probe
30ng/ 1Bsu polymerase
1000 copies/ I B.subtilis genomic DNA, or water controls as indicated
Amplification reactions were phenol extracted, precipitated, and separated on
a 16.5%
denaturing Urea-PAGE gel. Separated products were transferred to nylon
membrane and
products were detected using a Streptavidin-HRP conjugate. As the 5' end of
the probe was
biotinylated, this permitted the detection of uncut, cut and elongated probes.
(A) Detection of
probes, free and processed. Note that in the presence of an RPA environment
capable of
amplifying the target fragment, the probe was elongated preferentially
elongated. Also note that
while significant elongation occurs in the absence of the added target for the
J1/K2 primer pair
(due we believe to contamination), when the Jl /L2 primers pair is used this
does not occur (a
very faint band is, we believe, a sample cross-loading artifact). Consequently
we deduce that
probe cutting/elongation is a specific event. Furthermore the significant
signals in the no-
206
CA 3041471 2019-04-26
template J1/K2 samples suggests that the system is highly responsive to even
very small
numbers of targets, as is presumably arising from the carry-over
contamination. In (C) is shown
the results of a similar experiment in which the J1 and L2 primers were used
in combination
with the probe on either no target DNA, or on a sample with target DNA at 1000
copies per
microliter (genomic DNA). Oligonucleotide concentrations are as follows: J1
and L2 at 240nM,
probe at just 12nM, and ¨1000ng per microliter of Nfo enzyme were used. Other
conditions are
as described for the experiment in part (B). Samples were removed at the
indicated times and
stopped. Samples were phenol extracted, precipitated, and run on a denaturing
gel as already
described, transferred to nylon membrane, and detected as described above. We
note that
cleavage/elongation products can be detected by 30 minutes even when the probe
is as low as
12nM. This suggests that the kinetics of Nfo action under these conditions is
sufficiently fast to
monitor reactions excellently in real-time.
Figure 76 shows the general structure of several possible sorts of probe that
might be
used for real-time analysis of RPA reactions.
By combining the data suggesting that dual-labeled probes for use in RPA would
require
relatively short separation distances to permit quenching with our knowledge
of the efficient
action of the E.coli Nfo protein on processing intermediates between THF-
residue containing
probes and duplex targets, we suggest several probe structures. In (A) and (B)
we show probes
in which both a fluorophore and quencher are present internally (through base
modifications),
and between them is positioned a THF residue. The separation of the
fluorophore and quencher
is less than 10-12 residues to ensure efficient FRET between the groups. The
probe is blocked at
the 3' end with a suitable group. Cleavage by the Nfo enzyme at the THF
residue will eliminate
covalent association of the fluorophore and quencher, ultimately leading to an
increase in
solution fluorescence. In (C) and (D) and alternative arrangement is depicted.
In this case either
fluorophore or quencher is positioned at the 3'terminus of the probe, thus
blocking elongation.
The other light-absorbing moiety is attached via and internal linkage slightly
more 5'. The THF
residue is positioned between these 2 groups, and the 2 light-absorbing groups
are separated by
no more than 10-12 residues to ensure good FRET even when stretched by DNA
binding
proteins.
207
CA 3041471 2019-04-26
Discussion
There is a long-standing need for in vitro methods to amplify specific DNA
sequences.
Since the late 1980's the polymerase chain reaction (PCR) method has
principally met this need
(R. K. Saiki et al., Science 239, 487-91 (Jan 29, 1988)). The requirement for
thermal cycling
equipment, however, poses a significant barrier to the use of PCR outside of a
laboratory setting.
As described herein, we have developed a method called RPA, which obviates the
need for
thermal melting of template DNAs. RPA combines components of the bacteriophage
T4
recombination/replication system in vitro under critical defined conditions to
mediate the
hybridisation of oligonucleotide primers to template DNAs. Specifically, the
bacteriophage T4
recombinase uvsX, single-stranded DNA binding (SSB) protein gp32, and
recombinase loading
factor uvsY together with molecular crowding agents, allow high fidelity in
vitro recombinase-
mediated DNA targeting. When this targeting system is combined with strand
displacement
DNA synthesis mediated by enzymes of the E.coli or B.subtilis Poll class,
efficient exponential
DNA amplification is achieved.
Any oligonucleotide sequence may be coated by recombinase to form homology
searching filaments (Figure 37) giving RPA a broad utility allowing
amplification of virtually
any DNA sequence. This feature has been one of the major advantages of PCR
over other in
vitro DNA amplification methods (G. T. Walker, M. C. Little, J. G. Nadeau, D.
D. Shank, Proc
' Natl Acad Sci U SA 89, 392-6 (Jan 1, 1992); D. Y. Zhang, M. Brandwein, T.
Hsuih, H. B. Li,
Mol Diagn 6, 141-50 (Jun, 2001); M. Vincent, Y. Xu, H. Kong, EMBO Rep 5, 795-
800 (Aug,
2004); J. Compton, Nature 350, 91-2 (Mar 7, 1991)). Resembling their in vivo
role, homology-
searching filaments scan duplex DNA for sequences complementary to that of the
oligonucleotide (T. Yonesaki, Y. Ryo, T. Minagawa, H. Takahashi, Eur J Biochem
148, 127-34
(Apr 1, 1985); T. Shibata, C. DasGupta, R. P. Cunningham, C. M. Radding, Proc
Natl Acad Sci
U S A 76, 1638-42 (Apr, 1979)). On finding a match, the recombinase catalyses
several
reactions: the primer is paired with its complement, the similar 'outgoing'
strand is displaced,
and the recombinase dissociates. This establishes a 'D-loop' structure
accessible to other
reaction components. Exchange events occurring away from a free DNA end
generate
topologically strained joints, as the outgoing strand is attached on both
sides of the exchanged
208
CA 3041471 2019-04-26
region (Figure 37C).
Embedded sequences generate topologically constrained intermediates that are
unstable.
Joints formed at DNA ends permit free rotation of the displaced strand (P. W.
Riddles, I. R.
Lehman, J Biol Chem 260, 165-9 (Jan 10, 1985)). Because these two structures
have different
stabilities elongation of initial strand invasion events are less efficient
than subsequent ones
(Figure 39B). The free 3' end of the oligonucleotide primes synthesis by a
strand-displacing
DNA polymerase such as the Klenow fragment of E. coli, or the Bacillus
subtilis DNA
polymerase I (Bsu). The synthetic and strand-displacing activities of the
polymerase result in
the production of a double-stranded DNA and a displaced single strand. This
displaced strand is
replicated either by direct hybridisation and elongation of the second
oligonucleotide, or by
strand displacement synthesis if an invasion event had already occurred from
the opposite end.
The generation of two complete daughter duplexes completes one round of RPA.
Invasions
continue to act on products of previous synthesis reactions with the
endtargeted products
eventually dominating the reaction.
In developing the method of the invention, we have found that several
important
conditions are important for optimal RPA to occur. First, there needs to be
saturating quantities
of nucleic acid melting proteins present in the reaction especially a SSB such
as gp32. Second,
there needs to be a sufficient quantity of recombinase-loaded primer to
achieve an acceptable
invasion/strand-exchange rate. Finally, the recombinase/single-stranded DNA
primer filaments
need to be dynamic and capable of disassembly. There are competing biochemical
activities of
the reaction components. For example, in a typical in vitro situation
recombinases are usually
out-competed by saturating amounts of Sais such as gp32.
To overcome this problem, others have used nonhydrolysable ATP analogues such
as
ATP-y¨S, which stabilises the recombinase/single-stranded primer DNA
interaction (S. C.
Kowalczykowski, J. Clow, R. Somani, A. Varghese, J Mol Biol 193, 81-95 (Jan 5,
1987); A. L.
Eggler, S. L. Lusetti, M. M. Cox, J Biol Chem 278, 16389-96 (May 2, 2003); T.
Shibata, C.
DasGupta, R. P. Cunningham, C. M. Radding, Proc Nat! Acad Sci U S A 77, 2606-
10 (May,
1980)). Non-hydrolysable ATP analogues are, however, incompatible with the
dynamic activity
209
CA 3041471 2019-04-26
of recombinase/single-stranded DNA primer filaments needed to complete strand-
exchange
reactions and allow polymerases access to the D-loops (L. Xu, K. J. Marians, J
Biol Chem 277,
14321-8 (Apr 19, 2002); P. W. Riddles, L R. Lehman, J Biol Chem 260, 170-3
(Jan 10, 1985);
N. Armes, D. Stemple, in patent application PCT WO 03/072805 (ASM Scientific,
Inc., USA,
2003)) (Figure 38).
There are other ways the recombinase/single-stranded primer DNA interaction
could be
stabilised. For example, another bacteriophage T4 protein called uvsY is known
to aid loading
of uvsX onto gp32-coated DNA (L. D. Harris, J. D. Griffith, I Mol Biol 206, 19-
27 (Mar 5,
1989)). In addition, molecular crowding agents are also known to facilitate
loading and
stabilisation of the E. coli recA recombinase protein (P. E. Lavery, S. C.
Kowalczykowski, J
Biol Chem 267, 9307-14 (May 5, 1992)). We therefore tested whether uvsY and
molecular
crowding agents might alleviate the unfavourable competition between uvsX and
gp32 in a
dynamic, ATP-dependent system.
Titration of reaction components reveals that defined quantities of T4 gp32,
T4 uvsX, T4
uvsY, ATP, and PEG are required for DNA amplification (Figure 38). Indeed the
T4 uvsY
recombinase mediator protein and PEG-compound (Carbowax 20M) are required to
achieve
detectable amplification. At reduced concentrations of gp32, amplification
efficiency is
impaired generating smearing and laddering of reaction products. The
recombinase uvsX
protein is important for RPA and the reaction rate is accelerated at higher
concentrations,
although this can also increase artifacts (Figure 39A). ATP is important for
the reaction, and we
have found that an ATP regeneration system is needed to reach detectable
levels of product for
most reactions. Conversely, ATP-y-S is a powerful inhibitor of amplification.
To be a useful tool for routine DNA amplification applications such as
diagnostic testing,
RPA should be sensitive and specific, applicable to diverse sequence targets
and be capable of
amplifying fragments of sufficient size. We first investigated the size of
products that could be
generated. Amplified products up to 1000 base pairs in size could be amplified
using standard
reaction conditions (Figure 38A). Larger amplified products may also be
generated using RPA.
We tested the sensitivity of RPA under the most stringent conditions, using a
single-step
210
CA 3041471 2019-04-26
RPA reaction, without nesting, and detecting products by conventional ethidium
bromide
staining of agarose gels. With several independent primer/target sets we
routinely detected less
than 10 copies of starting duplex template. We observed variability between
experiments at
lowest detectable copy number but all attempts were successful at detecting
less than 10 copies.
With proper sample handling, RPA may be used to amplify from single molecules
to detectable
levels in a single reaction. To explore this possibility, we amplified a
polymorphic simple
tandem repeat (STR) marker from human DNA diluted until only a few copies
should remain.
We generated a number of amplified products corresponding to both possible
separate alleles
present in the sample DNA (Figure 39E,F). This allele separation effect
suggests that RPA has
single molecule sensitivity and thus surpasses the sensitivity of many other
DNA amplification
methods.
Analysis of the amount of amplified product shows that RPA will routinely
amplify
DNA samples by 1011-12-fold from small quantities of starting template. Final
product levels are
typically in the range of 10-250 nM, generating more than sufficient
quantities of DNA for even
the least sensitive detection protocols. To assess specificity of
amplification reactions we have
analysed many primer pairs, most directed to human DNA sequences. For every
primer/template set, we have tested the predicted product sizes, restriction
enzyme digestion
patterns, or product DNA sequence to show that amplification is specific. We
have not observed
amplification ofnon-target sequences from sample DNA to the best
interpretation of our, data.
Artifacts we have observed are product or primer related (Figure 39A).
Often in diagnostic settings, specificity problems arise from the large
amounts of
nontarget DNA present in a sample. For example, in pathogen detection, human
DNA from
blood samples can interfere with detection of pathogen DNA. We therefore
sought to detect
trace quantities of target DNA in a large mass of unrelated DNA. We found that
RPA was able
to amplify a target to detectable levels from 100 copies of Bacillus subtilis
DNA in the presence
of I tig of human DNA (i.e., I 08-fold less B. subtilis DNA than human DNA by
mass). With
such a large mass of sample DNA we found we had to increase levels of uvsX and
uvsY
compared to equivalent reactions without excess competitor DNA to achieve an
acceptable
reaction rate. This is perhaps due to out-titration of the homology-searching
components.
211
CA 3041471 2019-04-26
In time-course experiments with several different primer/template sets, we
found that
amplification rate was partially dependent on product length. For fragments in
the range 150-
400 base pairs, however, fairly similar rates were observed, allowing
amplification from
hundreds to thousands of starting copies to gel detectable levels (-1012
copies) in as little as 20
minutes). We estimate that we have been able to reduce average 'cycle' times
to as little as 30
seconds on average for such fragments. Using optimally short target sequences
and sensitive
detection method, we expect that a diagnostic amplification/detection assay
could be performed
well within an hour. Our studies show that for a large number of arbitrary DNA
targets in
complex samples high-quality primer pairs can be easily designed. We have
addressed minimal
oligonucleotide length and found that while oligonucleotides of less than 30
nucleotides do not
amplify DNA effectively, those of 30-35 nucleotides in length are excellent
primers and are
short enough for easy synthesis (Figure 40).
As demonstrated herein, RPA is an excellent general method to amplify specific
DNA
sequences. We have shown that RPA reactions can be monitored in real-time
using minor
groove binding dyes and demonstrate an excellent capacity to assess start
target copy number.
Furthermore multiple targets can be co-amplified, and sequence-specific real-
time sensors
('third probes') can be readily included in the reaction environment. We have
identified and
demonstrated that DNA repair enzymes such as fn, Nth and particularly Nfo
operate in RPA
reactions .and can be combined with suitable probes to permit real-time
assessment of reaction
behaviour in a sequence-specific manner. We show that fluorescence-based probe
systems show
markedly different properties in the RPA environment to conventional PCR
reaction
environments, or similar. Finally our initial experiments indicate that it is
easy to lyophilise the
components of the RPA reaction for convenient storage and reconstitution
(Figure 40). This
method, which operates robustly at constant low temperature, can be
lyophilised for easy storage
and requires no thermal cycling or melting for high sensitivity, nor other
complex handling, and
can be readily monitored in real-time, offers a significant breakthrough in
the development of
DNA diagnostic, forensic and other point-of-use applications. Once integrated
with portable
sample DNA extraction and product detection systems, RPA should enable easy-to-
use clinical
or domestic testing kits for a variety of pathogens (e.g., Clamydia or MRSA)
as well as field kits
212
CA 3041471 2019-04-26
for other applications.
The details of one or more embodiments of the invention have been set forth in
the
accompanying description above. Although any methods and materials similar or
equivalent to
those described herein can be used in the practice or testing of the present
invention, the preferred
methods and materials are now described. Other features, objects, and
advantages of the
invention will be apparent from the description and from the claims.
In the specification and the appended claims, the singular forms include
plural referents
unless the context clearly dictates otherwise. Unless defined otherwise, all
technical and
scientific terms used herein have the same meaning as commonly understood by
one of
ordinary skill in the art to which this invention belongs. Unless expressly
stated otherwise, the
techniques employed or contemplated herein are standard methodologies well
known to one of
ordinary skill in the art.
213
CA 3041471 2019-04-26