Note: Descriptions are shown in the official language in which they were submitted.
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
ANTI-GLYCO-MUC1 ANTIBODIES AND THEIR USES
1. SEQUENCE LISTING
[0001] The instant application contains a Sequence Listing which has been
submitted
electronically in ASCII format and is hereby incorporated by reference in its
entirety. Said ASCII
copy, created on October 24, 2017 is named GOT-001WO_Sequence_Listing.txt and
is 33,265
bytes in size.
2. BACKGROUND
[0002] The human mucin MUC1 is a polymorphic transmembrane glycoprotein
expressed on
the apical surfaces of simple and glandular epithelia (Taylor-Papadimitriou
etal., 1999). MUC1
is highly overexpressed and aberrantly 0-glycosylated in adenocarcinomas. The
extracellular
domain of the mucin contains variable number of tandem repeats (TRs) (25-125)
of 20 amino
acid residues with five potential sites for 0-glycosylation. 0-Glycans are
incompletely
processed in cancer cells resulting in the expression of the pancarcinoma
carbohydrate
antigens Tn (GaINAca1-0-Ser/Thr) (Springer, 1984). Simple mucin-type 0-
glycans, Tn, are
widely expressed in adenocarcinomas (including breast and ovarian cancers) and
show limited
distribution in normal adult tissues (Springer, 1984). The expression of these
0-glycans in
cancer correlates with poor prognosis and natural antibodies to these
carbohydrate haptens
increases in cancer patients (Miles etal., 1995; Soares etal., 1996; Werther
etal., 1996).
There is a need in the art for therapeutic modalities that utilize glyco-MUC1
epitopes that are
overexpressed in cancer cells.
3. SUMMARY
[0003] The disclosure captures the tumor specificity of glycopeptide variants
by providing
therapeutic and diagnostic agents based on antibodies and antigen binding
fragments that are
selective for cancer-specific epitopes of glyco-MUC1.
[0004] The present disclosure provides anti-glyco-MUC1 antibodies and antigen
binding
fragments thereof that bind to a cancer-specific glycosylation variant of
MUC1. The present
disclosure further provides fusion proteins and antibody-drug conjugates
comprising anti-glyco-
MUC1 antibodies and antigen binding fragments, and nucleic acids encoding the
anti-glyco-
MUC1 antibodies, antigen binding fragments and fusion proteins.
[0005] The present disclosure further provides methods of using the anti-glyco-
MUC1
antibodies, antigen-binding fragments, fusion proteins, antibody-drug
conjugates and nucleic
acids for cancer therapy.
[0006] In certain aspects, the disclosure provides bispecific and other
multispecific anti-glyco-
MUC1 antibodies and antigen binding fragments that bind to a cancer-specific
glycosylation
variant of MUC1 and to a second epitope. The second epitope can either be on
MUC1 itself, on
1
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
another protein co-expressed on cancer cells with MUC1, or on another protein
presented on a
different cell, such as an activated T cell. Further, also disclosed are
nucleic acids encoding
such antibodies, including nucleic acids comprising codon-optimized coding
regions and nucleic
acids comprising coding regions that are not codon-optimized for expression in
a particular host
cell.
[0007] The anti-glyco-MUC1 antibodies and binding fragments can be in the form
of fusion
proteins containing a fusion partner. The fusion partner can be useful to
provide a second
function, such as a signaling function of the signaling domain of a T cell
signaling protein, a
peptide modulator of T cell activation or an enzymatic component of a labeling
system.
Exemplary T cell signaling proteins include 4-1BB, 0030, and fusion peptides,
e.g., 0D28-
CD3-zeta and 4-IBB-CD3-zeta. 4-1BB, or CD137, is a co-stimulatory receptor of
T cells; CD3-
zeta is a signal-transduction component of the T-cell antigen receptor. The
moiety providing a
second function can be a modulator of T cell activation, such as IL-15, IL-
15Ra, or an IL-15/1L-
15Ra fusion, or it can encode a label or an enzymatic component of a labeling
system useful in
monitoring the extent and/or location of binding in vivo or in vitro.
Constructs encoding these
prophylactically and therapeutically active biomolecules placed in the context
of T cells, such as
autologous T cells, provide a powerful platform for recruiting adoptively
transferred T cells to
prevent or treat a variety of cancers in some embodiments of the disclosure.
[0008] In certain aspects, an anti-glyco-MUC1 antibody or antigen-binding
fragment of the
disclosure comprises heavy and/or light chain variable sequences (or encoded
by the
nucleotide sequences) set forth in Table 1. For clarity, when the term "anti-
glyco-MUC1
antibody" is used in this document, it is intended to include monospecific and
multi-specific
(including bispecific) anti-glyco-MUC1 antibodies, antigen-binding fragments
of the
monospecific and multi-specific antibodies, and fusion proteins and conjugates
containing the
antibodies and their antigen-binding fragments, unless the context dictates
otherwise. Likewise,
when the term when the term "anti-glyco-MUC1 antibody or antigen-binding
fragment" is used,
it is also intended to include monospecific and multi-specific (including
bispecific) anti-glyco-
MUC1 antibodies and their antigen-binding fragments, together with fusion
proteins and
conjugates containing such antibodies and antigen-binding fragments, unless
the context
dictates otherwise.
[0009] In other aspects, an anti-glyco-MUC1 antibody or antigen-binding
fragment of the
disclosure comprises heavy and/or light chain CDR sequences (or encoded by the
nucleotide
sequences) set forth in Tables 1-3. The CDR sequences set forth in Table 1
include CDR
sequences defined according to the IMGT (Lefranc et al., 2003, Dev Comparat
Immunol 27:55-
77, Kabat (Kabat etal., 1991, Sequences of Proteins of Immunological Interest,
5th Ed. Public
Health Service, National Institutes of Health, Bethesda, Md.), and Chothia (Al-
Lazikani etal.,
1997, J. Mol. Biol 273:927-948) schemes for defining CDR boundaries. The CDR
sequences
2
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
set forth in Table 2 are the combined regions of overlap for the CDR sequences
shown in Table
1, with the IMGT, Kabat and Chothia sequences shown in underlined bold text.
The CDR
sequences set forth in Table 3 are the common regions of overlap for the CDR
sequences
shown in Table 1. The framework sequences for such anti-glyco-M UC1 antibody
and antigen-
binding fragment can be the native murine framework sequences in Table 1 or
can be non-
native (e.g., humanized or human) framework sequences.
Table 1
Description Sequence SEQ ID NO:
VH amino acid MGWSGIFLFFLSVTTGVHSQVQLQQSDAELVKPGASVKI 1
sequence (incl.
signal SCKAS GYTFTDHAIHVVVKQRPEQGLEWIGYFSPGNDDI
sequence) HYNEKFEGKATLTADKSSSTAYMQLNSLTSEDSAVYFC
KRSYDKDFDCWGQGTTLTVSS
VL amino acid MVLILLLLVVVSGTCGDIVMSQSPSSLGVSVGEKVTMSCK 2
sequence (incl.
signal SSQSLLYSTNQKNYQSLLYSTNQKNYLAVVYQQKPGQSP
sequence) KLLIYVVVSNRKSGVPDRFTGSGSGTDFTLTISSVKAEDL
AVYYC QQYYRYPLTFGAGTKLELK
VH amino acid QVQLQQSDAELVKPGASVKISCKASGYTFTDHAIHVVVK 3
sequence
(predicted QRPEQGLEWIGYFSPGNDDIHYNEKFEGKATLTADKSS
mature) STAYMQLNSLTSEDSAVYFCKRSYDKDFDCWGQGTTLT
VSS
VL amino acid DIVMSQSPSSLGVSVGEKVTMSCKSSQSLLYSTNQKNY 4
sequence
(predicted QSLLYSTNQKNYLAVVYQQKPGQSPKLLIYVVVSNRKSGV
mature) PDRFTGSGSGTDFTLTISSVKAEDLAVYYC
QQYYRYPLTFGAGTKLELK
CDR-H1 amino GYTFTDHA 5
acid sequence
(IMGT
definition)
CDR-H2 amino FSPGNDDI 6
acid sequence
(IMGT
definition)
CDR-H3 amino KRSYDKDFDC 7
acid sequence
(IMGT
definition)
CDR-L1 amino QSLLYSTNQKNY 8
acid sequence
(IMGT
definition)
CDR-L2 amino VVVS 9
acid sequence
3
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
Table 1
Description Sequence SEQ
ID NO:
(IMGT
definition)
CDR-L3 amino QQYYRYPLT 10
acid sequence
(IMGT
definition)
VH nucleotide ATGGGATGGAGCGGGATCTTTCTCTTCTTCCTGTCAG 11
sequence (incl.
signal TAACTACAGGTGTCCACTCCCAGGTTCAGCTGCAGCA
sequence) GTCTGACGCGGAGTTGGTGAAACCTGGGGCTTCAGT
GAAGATATCCTGCAAGGCTTCTGGCTACACTTTCACT
GACCATGCTATTCACTGGGTGAAGCAGAGGCCTGAAC
AGGGCCTGGAATGGATTGGATATTTTTCTCCCGGAAA
TGATGACATTCACTACAATGAGAAGTTCGAGGGCAAG
GCCACACTGACTGCAGACAAATCCTCCAGCACTGCCT
ACATGCAGCTCAACAGCCTGACATCTGAAGATTCTGC
AGTGTATTTCTGTAAAAGATCTTACGACAAGGACTTTG
ACTGCTGGGGCCAAGGCACCACTCTCACAGTCTCCTC
A
VL nucleotide ATGGTTCTTATCTTACTGCTGCTATGGGTATCTGGTAC 12
sequence (incl.
signal CTGTGGGGACATTGTGATGTCACAGTCTCCATCCTCC
sequence) CTAGGTGTGTCAGTTGGAGAGAAGGTTACTATGAGCT
GCAAGTCCAGTCAGAGCCTTTTATACAGTACCAATCAA
AAGAACTACCTGGCCTGGTACCAGCAGAAACCAGGG
CAGTCTCCTAAGTTGCTGATTTACTGGGTATCTAATAG
GAAATCTGGGGTCCCTGATCGCTTCACAGGCAGTGGA
TCTGGGACAGATTTCACTCTCACCATCAGTAGTGTGA
AGGCTGAAGACCTGGCAGTTTATTACTGTCAGCAATA
TTATAGGTATCCGCTCACGTTCGGTGCTGGGACCAAG
CTGGAGCTGAAA
VH nucleotide CAGGTTCAGCTGCAGCAGTCTGACGCGGAGTTGGTG 13
sequence (excl.
signal AAACCTGGGGCTTCAGTGAAGATATCCTGCAAGGCTT
sequence) CTGGCTACACTTTCACTGACCATGCTATTCACTGGGT
GAAGCAGAGGCCTGAACAGGGCCTGGAATGGATTGG
ATATTTTTCTCCCGGAAATGATGACATTCACTACAATG
AGAAGTTCGAGGGCAAGGCCACACTGACTGCAGACA
AATCCTCCAGCACTGCCTACATGCAGCTCAACAGCCT
GACATCTGAAGATTCTGCAGTGTATTTCTGTAAAAGAT
CTTACGACAAGGACTTTGACTGCTGGGGCCAAGGCAC
4
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
Table 1
Description Sequence SEQ ID NO:
CACTCTCACAGTCTCCTCA
VL nucleotide GACATTGTGATGTCACAGTCTCCATCCTCCCTAGGTG 14
sequence (excl.
signal TGTCAGTTGGAGAGAAGGTTACTATGAGCTGCAAGTC
sequence) CAGTCAGAGCCTTTTATACAGTACCAATCAAAAGAACT
ACCTGGCCTGGTACCAGCAGAAACCAGGGCAGTCTC
CTAAGTTGCTGATTTACTGGGTATCTAATAGGAAATCT
GGGGTCCCTGATCGCTTCACAGGCAGTGGATCTGGG
ACAGATTTCACTCTCACCATCAGTAGTGTGAAGGCTG
AAGACCTGGCAGTTTATTACTGTCAGCAATATTATAGG
TATCCGCTCACGTTCGGTGCTGGGACCAAGCTGGAG
CTGAAA
FR-H1
QVQLQQSDAELVKPGASVKISCKAS 15
FR-H2 IHVVVKQRPEQGLEWIGY 16
FR-H3
HYNEKFEGKATLTADKSSSTAYMQLNSLTSEDSAVYFC 17
FR-H4 WGQGTTLTVSS 18
FR-L1 DIVMSQSPSSLGVSVGEKVTMSCKSS 19
FR-L2 LAVVYQQKPGQSPKLLIY 20
FR-L3 NRKSGVPDRFTGSGSGTDFTLTISSVKAEDLAVYYC 21
FR-L4 FGAGTKLELK 22
CDR-H1 amino DHAIH 23
acid sequence
(Kabat
definition)
CDR-H2 amino YFSPGNDDIHYNEKFEG 24
acid sequence
(Kabat
definition)
CDR-H3 amino SYDKDFDC 25
acid sequence
(Kabat
definition)
CDR-L1 amino KSSQSLLYSTNQKNYLA 26
acid sequence
(Kabat
definition)
CDR-L2 amino VVVSNRKS 27
acid sequence
(Kabat
definition)
CDR-L3 amino QQYYRYPLT 10
acid sequence
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
Table 1
Description Sequence SEQ
ID NO:
(Kabat
definition)
CDR-H1 amino GYTFTDH 28
acid sequence
(Chothia
definition)
CDR-H2 amino SPGNDD 29
acid sequence
(Chothia
definition)
CDR-H3 amino SYDKDFDC 25
acid sequence
(Chothia
definition)
CDR-L1 amino SQSLLYSTNQKNY 30
acid sequence
(Chothia
definition)
CDR-L2 amino VVVS 9
acid sequence
(Chothia
definition)
CDR-L3 amino YYRYPLT 31
acid sequence
(Chothia
definition)
Table 2
Description Sequence SEQ
ID NO:
CDR-H1 amino GYTFTDHAIH ( IMG ) 32
acid sequence
(combined GYTFTDHAIH (Kabat )
overlap) GYTFTDHAIH (Chothia)
CDR-H2 amino YFSPGNDDIHYNEK-EEG (LMGT) 24
acid sequence
(combined YFSPGNDDIHYNEKFEG (Kabat )
overlap)
IFSPGNDDIHYNEKFEG (Chothia.)
CDR-H3 amino Kp
,SYDKDFDC ( ) 7
acid sequence -
(combined KRSYDKDFDC (Kabat)
overlap)
KRSYDKDFDC (Chotbia)
CDR-L1 amino KS SQSLLYS TNQKNYLA ( IMGT ) 26
acid sequence
(combined KSSQSLLYSTNQKNYLA (Kabat)
overlap)
NSSULLYSTNQKNYLA (Chothia)
6
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
Table 2
Description Sequence SEQ ID NO:
CDR-L2 amino wysi\TREs 27
acid sequence ¨
(combined WVSNRKS (Kabat)
overlap)
WVSNRKS ( C b 0 t. .1_ a)
CDR-L3 amino QQYYRYPLT (IMGT) 10
acid sequence
(combined QQYYRYPLT (Kabat)
overlap)
QQYYRYPLT (Chothia)
Table 3
Description Sequence SEQ ID NO:
CDR-H1 amino DH 33
acid sequence
(common
sequence)
CDR-H2 amino s,,DGN-Dr-) 29
acid sequence
(common
sequence)
CDR-H3 amino ¨
S 25
acid sequence
(common
sequence)
CDR-L1 amino QS LLYS TNQKNY 8
acid sequence
(common
sequence)
CDR-L2 amino 9
acid sequence
(common
sequence)
CDR-L3 amino YyRYPL T 31
acid sequence
(common
sequence)
[0010] In certain aspects, the disclosure provides an anti-glyco-MUC1 antibody
or antigen-
binding fragment of the disclosure comprises CDRs comprising the amino acid
sequences of
any of the CDR combinations set forth in numbered embodiments 3 to 17. Thus,
in certain
embodiments, an anti-glyco-MUC1 antibody or antigen-binding fragment of the
disclosure
comprises a CDR-H1 comprising the amino acid sequence of SEQ ID NO:33, a CDR-
H2
comprising the amino acid sequence of SEQ ID NO:29, a CDR-H3 comprising the
amino acid
sequence of SEQ ID NO:25, a CDR-L1 comprising the amino acid sequence of SEQ
ID NO: 8,
a CDR-L2 comprising the amino acid sequence of SEQ ID NO:9, and a CDR-L3
comprising the
7
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
amino acid sequence of SEQ ID NO:31. In some embodiments, CDR-H1 comprises the
amino
acid sequence of SEQ ID NO: 5, 23, 28, or 32. In some embodiments, CDR-H2
comprises the
amino acid sequence of SEQ ID NO: 6 or 24. In some embodiments, CDR-H3
comprises the
amino acid sequence of SEQ ID NO: 7. In some embodiments, CDR-L1 comprises the
amino
acid sequence of SEQ ID NO:30 or 26. In some embodiments, CDR-L2 comprises the
amino
acid sequence of SEQ ID NO:27. In some embodiments, CDR-L3 comprises the amino
acid
sequence of SEQ ID NO:10. In other aspects, an anti-glyco-MUC1 antibody or
antigen-binding
fragment of the disclosure comprises heavy chain CDRs of SEQ ID NOS: 5-7 and
light chain
CDRs of SEQ ID NOS: 8-10. In other aspects, an anti-glyco-MUC1 antibody or
antigen-binding
fragment of the disclosure comprises heavy chain CDRs of SEQ ID NOS: 23-25 and
light chain
CDRs of SEQ ID NOS: 26, 27, and 10. In other aspects, an anti-glyco-MUC1
antibody or
antigen-binding fragment of the disclosure comprises heavy chain CDRs of SEQ
ID NOS: 28,
29, and 25 and light chain CDRs of SEQ ID NOS: 30, 9, and 31. In other
aspects, an anti-glyco-
MUC1 antibody or antigen-binding fragment of the disclosure comprises heavy
chain CDRs of
SEQ ID NOS: 32, 24, and 7 and light chain CDRs of SEQ ID NOS: 26, 27, and 10.
In other
aspects, an anti-glyco-MUC1 antibody or antigen-binding fragment of the
disclosure comprises
heavy chain CDRs of SEQ ID NOS: 33, 29, and 25 and light chain CDRs of SEQ ID
NOS: 8, 9,
and 31. The antibody or antigen-binding fragment can be murine, chimeric,
humanized or
human.
[0011] In further aspects, an anti-glyco-M UC1 antibody or antigen binding
fragment of the
disclosure competes with an antibody or antigen binding fragment comprising
heavy and light
chain variable regions of SEQ ID NOS: 3 and 4, respectively. In yet other
aspects, the
disclosure provides an anti-M UC1 antibody or antigen binding fragment having
heavy and light
chain variable regions having at least 95%, 98%, 99%, or 99.5% sequence
identity of SEQ ID
NOS: 3 and 4, respectively.
[0012] In yet other aspects, an anti-glyco-MUC1 antibody or antigen-binding
fragment of the
disclosure is a single-chain variable fragment (scFv). An exemplary scFv
comprises the heavy
chain variable fragment N-terminal to the light chain variable fragment. In
some embodiments,
the scFv heavy chain variable fragment and light chain variable fragment are
covalently bound
to a linker sequence of 4-15 amino acids. The scFv can be in the form of a bi-
specific T-cell
engager or within a chimeric antigen receptor (CAR).
[0013] The anti-glyco-MUC1 antibodies and antigen-binding fragments can be in
the form of a
multimer of a single-chain variable fragment, a bispecific single-chain
variable fragment and a
multimer of a bispecific single-chain variable fragment. In some embodiments,
the multimer of a
single chain variable fragment is selected a divalent single-chain variable
fragment, a tribody or
a tetrabody. In some of these embodiments, the multimer of a bispecific single-
chain variable
fragment is a bispecific T-cell engager.
8
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
[0014] Other aspects of the disclosure are drawn to nucleic acids encoding the
anti-glyco-
MUC1 antibodies and antibody-binding fragments of the disclosure. In some
embodiments, the
portion of the nucleic acid nucleic acid encoding an anti-glyco-MUC1 antibody
or antigen-
binding fragment is codon-optimized for expression in a human cell. In certain
aspects, the
disclosure provides an anti-glyco-MUC1 antibody or antigen binding fragment
having heavy and
light chain variable regions encoded by a heavy chain nucleotide sequence
having at least
95%, 98%, 99%, or 99.5% sequence identity to SEQ ID NO:11 or SEQ ID NO:13 and
a light
chain nucleotide sequence having at least 95%, 98%, 99%, or 99.5% sequence
identity to SEQ
ID NO:12 or SEQ ID NO:14. Vectors (e.g., a viral vector such as a lentiviral
vector) and host
cells comprising the nucleic acids are also within the scope of the
disclosure. The heavy and
light chains coding sequences can be present on a single vector or on separate
vectors.
[0015] Yet another aspect of the disclosure is a pharmaceutical composition
comprising an
anti-glyco-M UC1 antibody, antigen-binding fragment, nucleic acid (or pair of
nucleic acids),
vector (or pair or vectors) or host cell according to the disclosure, and a
physiologically suitable
buffer, adjuvant or diluent.
[0016] Still another aspect of the disclosure is a method of making a chimeric
antigen receptor
comprising incubating a cell comprising a nucleic acid or a vector according
to the disclosure,
under conditions suitable for expression of the coding region and collecting
the chimeric antigen
receptor.
[0017] Another aspect of the disclosure is a method of detecting cancer
comprising contacting
a cell or tissue sample with an anti-glyco-MUC1 antibody or antigen-binding
fragment of the
disclosure and detecting whether the antibody is bound to the cell or tissue
sample.
[0018] Yet another aspect of the disclosure is a method of treating cancer
comprising
administering a prophylactically or therapeutically effective amount of an
anti-glyco-MUC1
antibody, antigen-binding fragment, nucleic acid, vector, host cell or
pharmaceutical
composition according to the disclosure to a subject in need thereof.
4. BRIEF DESCRIPTION OF THE FIGURES
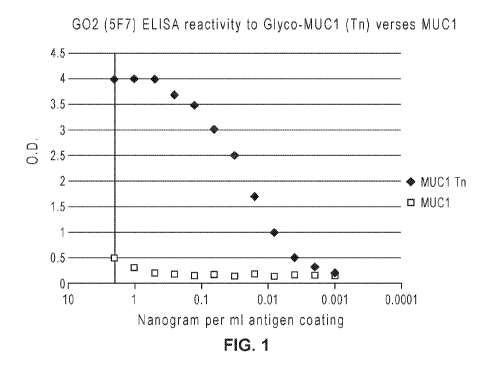
[0019] FIG. 1: Results of ELISA assay showing specificity of binding of G02 to
glyco-M UC1
relative to MUC1.
[0020] FIG. 2: Binding of G02 to colon cancer tissue. lmmunohistochemistry
labeling of
invasive colon carcinoma tissue and adjacent healthy tissue using mAbs G02.
mAb G02
shows distinct binding to colon cancer tissue with high reactivity with both
intracellular and
surface structures on cancer cells. In contrast no reactivity is seen to
surface structures on
healthy colon cells.
9
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
[0021] FIG.3: Binding of G02 to pancreatic cancer tissue. lmmunohistochemistry
labeling of
pancreatic cancer tissue using mAbs G02. mAb G02 show distinct binding to
pancreatic
cancer cells. In contrast no or limited reactivity is seen to surrounding
healthy tissue.
[0022] FIG. 4: Binding of G02 to breast cancer tissue. lmmunohistochemistry
labeling of breast
cancer tissue using mAbs G02. mAb G02 showed distinct binding to invasive
breast cancer
cells.
[0023] FIG.5: Results of an antibody dependent cellular cytotoxicity assay
with antibody G02
and a secondary antibody conjugated to the antitubulin agent monomethyl
auristatin F (MMAF).
[0024] FIG. 6: Results of an ELISA assay quantifying circulating tumor cells
using G02. X-axis
shows number of cells and Y-axis shows 0D450 values.
[0025] FIGS. 7A-E: Representative images of MUC1 positive TMA tumor cores.
FIG. 7A:
breast cancer; FIG. 7B: non-small cell lung cancer; FIG. 70: ovarian cancer;
FIG. 7D: colorectal
cancer; FIG. 7E: prostate cancer.
[0026] FIG. 8: Schematic of an exemplary anti-glyco-M UC1 and anti-CD3 T-cell
bispecific
antibody (TCB).
[0027] FIGS. 9A-B: Jurkat-N FAT activation assay with undigested patient-
derived tumor
samples (malignant neoplasm of bronchus and lung: middle lobe, bronchus or
lung, squamous
cell carcinoma) and different TCBs at 50 nM (FIG. 9A) or 5 nM (FIG. 9B).
[0028] FIG. 10: Jurkat-N FAT activation assay with undigested patient-derived
tumor samples
(malignant neoplasm of bronchus and lung: lower lobe, bronchus or lung, non-
keratinizing
squamous cell carcinoma) and different TCBs at 50 nM.
[0029] FIG. 11: Jurkat-N FAT activation assay with undigested patient-derived
tumor samples
(malignant neoplasm of bronchus and lung: upper lobe, bronchus or lung,
adenocarcinoma with
acinar type) and different TCBs at 50 nM.
[0030] FIGS. 12A-12B: Binding of G02 TCB to MUC1 expressed on MCF7 cs (FIG.
12A) and
T3M4 pzfv (FIG. 12B) cells measured by flow cytometry.
[0031] FIGS. 13A-X: Induction of tumor cell killing and T cell activation
measured by
upregulation of 0D25 and 0D69 on CD4 T cells and CD8 T cells as well as
release of IL6, IL8,
IL10, IFNy, TNFa and Granzyme B with G02 TCB on T3M4 pzfv in the presence of
PBMCs
from two healthy donors (donor 1 FIG. 13A-13L; donor 2 FIG. 13M-13X). Same
legend for each
of FIGS. 13A-13X.
[0032] FIGS. 14A-14F: Induction of tumor cell killing (FIGS. 14A-14B) and T
cell activation
measured by upregulation of 0D25 and 0D69 on CD8 T cells and CD4 T cells
(FIGS. 140-14F,
respectively) with G02 TCB on MCF7 cs in the presence of PBMCs. Same legend
for each of
FIGS. 14A-14F.
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
[0033] FIG. 15A-B: Binding of G02 TCB and HMFG1 TCB to MCF10A (human non-
tumorigenic mammary epithelial cell line) (FIG. 15A) and HBEpiC (human
bronchial epithelial
cells) (FIG. 15B).
[0034] FIG. 16A-C: Induction of tumor cell killing (FIG. 16A) and T cell
activation measured by
upregulation of 0D25 on CD4 T cells (FIG. 16B) and CD8 T cells (FIG. 160) with
G02 TCB and
HMFG1 TCB on MCF10A cells in the presence of PBMCs.
[0035] FIG. 17: Illustration of G02 and G02 TCB flowing through a flow cell
having coupled
glycopeptides.
[0036] FIG. 18A-B: Sensorgrams showing binding of G02 (FIG. 18A) and G02 TCB
(FIG.
18B) to human and cynomolgous glycopeptides.
[0037] FIG. 19A-D: Binding (avidity) of G02 antibody (FIG. 19A-19B) and G02
TCB (FIG.
19C-19D) to human and cynomolgus glycopeptides, and estimate of the "apparent"
KD.
5. DETAILED DESCRIPTION
5.1 Antibodies
[0038] The inventor has developed novel antibodies that are directed to a
glycoform of MUC1
present on tumor cells. These are exemplified by the antibody 5F7, referred to
herein as "G02".
G02 was identified in a screen for antibodies that bind to a glycosylated 60-
mer representing 3
copies of one of the tandem repeats present in MUC1, VTSAPDTRPAPGSTAPPAHG (SEQ
ID
NO:50), glycosylated with purified recombinant human glycosyltransferases
polypeptides
GaINAc-T2, GaINAc-T4, and GaINAc-T1 so as to mimic the glycosylation pattern
of MUC1
present on tumor cells.
[0039] The anti-glyco-MUC1 antibodies of the disclosure, exemplified by
antibody G02, are
useful as tools in cancer diagnosis and therapy.
[0040] Thus, in certain aspects, the disclosure provides antibodies and
antigen binding
fragments that bind to a glycoform of MUC1 present on tumor cells (referred to
herein as
"glyco-M UC1"), and preferably to the 60-mer peptide (VTSAPDTRPAPGSTAPPAHG)3
(SEQ ID
NO:47) glycosylated with GaINAc-T2, GaINAc-T4, and GaINAc-T1 as described in
US Patent
No. 6,465,220.
[0041] The anti-glyco-MUC1 antibodies of the disclosure may be polyclonal,
monoclonal,
genetically engineered, and/or otherwise modified in nature, including but not
limited to chimeric
antibodies, humanized antibodies, human antibodies, primatized antibodies,
single chain
antibodies, bispecific antibodies, dual-variable domain antibodies, etc. In
various embodiments,
the antibodies comprise all or a portion of a constant region of an antibody.
In some
embodiments, the constant region is an isotype selected from: IgA (e.g., IgAi
or IgA2), IgD, IgE,
11
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
IgG (e.g., IgGi, IgG2, IgG3 or !gat), and IgM. In specific embodiments, the
anti-glyco-MUC1
antibodies of the disclosure comprise an IgGi constant region isotyope.
[0042] The term "monoclonal antibody" as used herein is not limited to
antibodies produced
through hybridoma technology. A monoclonal antibody is derived from a single
clone, including
any eukaryotic, prokaryotic, or phage clone, by any means available or known
in the art.
Monoclonal antibodies useful with the present disclosure can be prepared using
a wide variety
of techniques known in the art including the use of hybridoma, recombinant,
and phage display
technologies, or a combination thereof. In many uses of the present
disclosure, including in vivo
use of the anti-glyco-MUC1 antibodies in humans, chimeric, primatized,
humanized, or human
antibodies can suitably be used.
[0043] The term "chimeric" antibody as used herein refers to an antibody
having variable
sequences derived from a non-human immunoglobulin, such as a rat or a mouse
antibody, and
human immunoglobulin constant regions, typically chosen from a human
immunoglobulin
template. Methods for producing chimeric antibodies are known in the art. See,
e.g., Morrison,
1985, Science 229(4719):1202-7; Oi etal., 1986, BioTechniques 4:214-221;
Gillies etal., 1985,
J. lmmunol. Methods 125:191-202; U.S. Pat. Nos. 5,807,715; 4,816,567; and
4,816397, which
are incorporated herein by reference in their entireties.
[0044] "Humanized" forms of non-human (e.g., murine) antibodies are chimeric
immunoglobulins that contain minimal sequences derived from non-human
immunoglobulin. In
general, a humanized antibody will comprise substantially all of at least one,
and typically two,
variable domains, in which all or substantially all of the CDR regions
correspond to those of a
non-human immunoglobulin and all or substantially all of the FR regions are
those of a human
immunoglobulin sequence. The humanized antibody can also comprise at least a
portion of an
immunoglobulin constant region (Fc), typically that of a human immunoglobulin
consensus
sequence. Methods of antibody humanization are known in the art. See, e.g.,
Riechmann etal.,
1988, Nature 332:323-7; U.S. Pat. Nos. 5,530,101; 5,585,089; 5,693,761;
5,693,762; and
6,180,370 to Queen etal.; EP239400; PCT publication WO 91/09967; U.S. Pat. No.
5,225,539;
EP592106; EP519596; Padlan, 1991, Mol. Immunol., 28:489-498; Studnicka etal.,
1994, Prot.
Eng. 7:805-814; Roguska etal., 1994, Proc. Natl. Acad. Sci. 91:969-973; and
U.S. Pat. No.
5,565,332, all of which are hereby incorporated by reference in their
entireties.
[0045] "Human antibodies" include antibodies having the amino acid sequence of
a human
immunoglobulin and include antibodies isolated from human immunoglobulin
libraries or from
animals transgenic for one or more human immunoglobulin and that do not
express
endogenous immunoglobulins. Human antibodies can be made by a variety of
methods known
in the art including phage display methods using antibody libraries derived
from human
immunoglobulin sequences. See U.S. Pat. Nos. 4,444,887 and 4,716,111; and PCT
publications WO 98/46645; WO 98/50433; WO 98/24893; WO 98/16654; WO 96/34096;
WO
12
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
96/33735; and WO 91/10741, each of which is incorporated herein by reference
in its entirety.
Human antibodies can also be produced using transgenic mice which are
incapable of
expressing functional endogenous immunoglobulins but which can express human
immunoglobulin genes. See, e.g., PCT publications WO 98/24893; WO 92/01047; WO
96/34096; WO 96/33735; U.S. Pat. Nos. 5,413,923; 5,625,126; 5,633,425;
5,569,825;
5,661,016; 5,545,806; 5,814,318; 5,885,793; 5,916,771; and 5,939,598, which
are incorporated
by reference herein in their entireties. Fully human antibodies that recognize
a selected epitope
can be generated using a technique referred to as "guided selection." In this
approach, a
selected non-human monoclonal antibody, e.g., a mouse antibody, is used to
guide the
selection of a completely human antibody recognizing the same epitope (see,
Jespers etal.,
1988, Biotechnology 12:899-903).
[0046] "Primatized antibodies" comprise monkey variable regions and human
constant regions.
Methods for producing primatized antibodies are known in the art. See, e.g.,
U.S. Pat. Nos.
5,658,570; 5,681,722; and 5,693,780, which are incorporated herein by
reference in their
entireties.
[0047] Anti-glyco-MUC1 antibodies of the disclosure include both full-length
(intact) antibody
molecules, as well as antigen-binding fragments that are capable of binding
glyco-M UC1.
Examples of antigen-binding fragments include by way of example and not
limitation, Fab, Fab',
F (ab')2, Fv fragments, single chain Fv fragments and single domain fragments.
[0048] A Fab fragment contains the constant domain of the light chain (CL) and
the first
constant domain (CH1) of the heavy chain. Fab' fragments differ from Fab
fragments by the
addition of a few residues at the carboxyl terminus of the heavy chain CH1
domain including
one or more cysteines from the antibody hinge region. F(ab') fragments are
produced by
cleavage of the disulfide bond at the hinge cysteines of the F(ab')2 pepsin
digestion product.
Additional chemical couplings of antibody fragments are known to those of
ordinary skill in the
art. Fab and F(ab')i fragments lack the Fc fragment of intact antibody, clear
more rapidly from
the circulation of animals, and may have less non-specific tissue binding than
an intact antibody
(see, e.g., Wahl etal., 1983, J. Nucl. Med. 24:316).
[0049] An "Fv" fragment is the minimum fragment of an antibody that contains a
complete
target recognition and binding site. This region consists of a dimer of one
heavy and one light
chain variable domain in a tight, non-covalent association (VH-VL dimer). It
is in this
configuration that the three CDRs of each variable domain interact to define a
target binding
site on the surface of the VH-VL dimer. Often, the six CDRs confer target
binding specificity to
the antibody. However, in some instances even a single variable domain (or
half of an Fv
comprising only three CDRs specific for a target) can have the ability to
recognize and bind
target, although at a lower affinity than the entire binding site.
13
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
[0050] "Single-chain Fv" or "scFv" antigen-binding fragments comprise the VH
and VL domains
of an antibody, where these domains are present in a single polypeptide chain.
Generally, the
Fv polypeptide further comprises a polypeptide linker between the VH and VL
domains which
enables the scFv to form the desired structure for target binding.
[0051] "Single domain antibodies" are composed of single VH or VL domains
which exhibit
sufficient affinity to glyco-MUC1. In a specific embodiment, the single domain
antibody is a
camelized antibody (See, e.g., Riechmann, 1999, Journal of Immunological
Methods 231:25-
38).
[0052] The anti-glyco-MUC1 antibodies of the disclosure may also be bispecific
and other
multiple specific antibodies. Bispecific antibodies are monoclonal, often
human or humanized,
antibodies that have binding specificities for two different epitopes on the
same or different
antigen. In the present disclosure, one of the binding specificities can be
directed towards
glyco-MUC1, the other can be for any other antigen, e.g., for a cell-surface
protein, receptor,
receptor subunit, tissue-specific antigen, virally derived protein, virally
encoded envelope
protein, bacterially derived protein, or bacterial surface protein, etc. In
certain preferred
embodiments, the bispecific and other multispecific anti-glyco-MUC1 antibodies
and antigen
binding fragments that specifically bind to a second MUC1 epitope, an epitope
on another
protein co-expressed on cancer cells with MUC1, or an epitope on another
protein presented
on a different cell, such as an activated T cell. Bispecific antibodies of the
disclosure include
IgG format bispecific antibodies and single chain-based bispecific antibodies.
[0053] IgG format bispecific antibodies of the disclosure can be any of the
various types of IgG
format bispecific antibodies known in the art, such as quadroma bispecific
antibodies, "knobs-
in-holes" bispecific antibodies, CrossMab bispecific antibodies, charge paired
bispecific
antibodies, common light chain bispecific antibodies, one-arm single-chain Fab-
immunoglobulin
gamma bispecific antibodies, disulfide stabilized Fv bispecific antibodies,
DuetMabs, controlled
Fab-arm exchange bispecific antibodies, strand-exchange engineered domain body
bispecific
antibodies, two-arm leucine zipper heterodimeric monoclonal bispecific
antibodies, KA-body
bispecific antibodies, dual variable domain bispecific antibodies, and cross-
over dual variable
domain bispecific antibodies. See, e.g., KOhler and Milstein, 1975, Nature
256:495-497; Milstein
and Cuello, 1983, Nature 305:537-40; Ridgway etal., 1996, Protein Eng. 9:617-
621; Schaefer
etal., 2011, Proc Natl Acad Sci USA 108:11187-92; Gunasekaran et al., 2010, J
Biol Chem
285:19637-46; Fischer etal., 2015 Nature Commun 6:6113; Schanzer etal., 2014,
J Biol Chem
289:18693-706; Metz etal., 2012 Protein Eng Des Sel 25:571-80; Mazor etal.,
2015 MAbs
7:377-89; Labrijn etal., 2013 Proc Natl Acad Sci USA 110:5145-50; Davis etal.,
2010 Protein
Eng Des Sel 23:195-202; Wranik etal., 2012, J Biol Chem 287:43331-9; Gu et
al., 2015, PLoS
One 10(5):e0124135; Steinmetz et al., 2016, MAbs 8(5):867-78; Klein etal.,
2016, mAbs,
14
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
8(6):1010-1020; Liu etal., 2017, Front. lmmunol. 8:38; and Yang etal., 2017,
Int. J. Mol. Sci.
18:48, which are incorporated herein by reference in their entireties.
[0054] In some embodiments, the bispecific antibodies of the disclosure are
CrossMabs. The
CrossMab technology is described in detail in WO 2009/080251, WO 2009/080252,
WO
2009/080253, WO 2009/080254, WO 2013/026833, WO 2016/020309, and Schaefer
etal.,
2011, Proc Natl Acad Sci USA 108:11187-92, which are incorporated herein by
reference in
their entireties. Briefly, the CrossMab technology is based on a domain
crossover between
heavy and light chains within one Fab-arm of a bispecific IgG, which promotes
correct chain
association. A CrossMab bispecific antibody of the disclosure can be a
"CrossMab" antibody,
in which the heavy and light chains of the Fab portion of one arm of a
bispecific IgG antibody
are exchanged. In other embodiments, a CrossMab bispecific antibody of the
disclosure can be
a "CrossMabv"-v1-" antibody, in which the only the variable domains of the
heavy and light
chains of the Fab portion of one arm of a bispecific IgG antibody are
exchanged. In yet other
embodiments, a CrossMab bispecific antibody of the disclosure can be a
"CrossMab'-"
antibody, in which only the constant domains of the heavy and light chains of
the Fab portion of
one arm of a bispecific IgG antibody are exchanged. CrossMabCH1-CL antibodies,
in contrast to
CrossMabFAB and CrossMabv"-vL, do not have predicted side products and,
therefore, in some
embodiments CrossMabCH1-CL bispecific antibodies are preferred. See, Klein et
al., 2016, mAbs,
8(6):1010-1020. Further embodiments of CrossMabs of the disclosure are
described below in
Section 5.2.
[0055] In some embodiments, the bispecific antibodies of the disclosure are
controlled Fab-arm
exchange bispecific antibodies. Methods for making Fab-arm exchange bispecific
antibodies
are described in PCT Publication No. W02011/131746 and Labrijn etal., 2014 Nat
Protoc.
9(10):2450-63, incorporated herein by reference in their entireties. Briefly,
controlled Fab-arm
exchange bispecific antibodies can be made by separately expressing two
parental IgG1s
containing single matching point mutations in the CH3 domain, mixing the
parental IgG1s under
redox conditions in vitro to enable recombination of half-molecules, and
removing the reductant
to allow reoxidation of interchain disulfide bonds, thereby forming the
bispecific antibodies.
[0056] Bispecific antibodies of the disclosure can comprise an Fc domain
composed of a first
and a second subunit. In one embodiment, the Fc domain is an IgG Fc domain. In
a particular
embodiment, the Fc domain is an IgGi Fc domain. In another embodiment the Fc
domain is an
IgG4 Fc domain. In a more specific embodiment, the Fc domain is an IgG4 Fc
domain
comprising an amino acid substitution at position S228 (Kabat EU index
numbering),
particularly the amino acid substitution 5228P. This amino acid substitution
reduces in vivo Fab
arm exchange of IgG4 antibodies (see Stubenrauch etal., 2010, Drug Metabolism
and
Disposition 38:84-91). In a further particular embodiment, the Fc domain is a
human Fc domain.
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
In an even more particular embodiment, the Fc domain is a human IgGi Fc
domain. An
exemplary sequence of a human IgGi Fc region is given in SEQ ID NO:42.
[0057] In particular embodiments, the Fc domain comprises a modification
promoting the
association of the first and the second subunit of the Fc domain. The site of
most extensive
protein-protein interaction between the two subunits of a human IgG Fc domain
is in the CH3
domain. Thus, in one embodiment said modification is in the CH3 domain of the
Fc domain.
[0058] In a specific embodiment said modification promoting the association of
the first and the
second subunit of the Fc domain is a so-called "knob-into-hole" modification,
comprising a
"knob" modification in one of the two subunits of the Fc domain and a "whole"
modification in
the other one of the two subunits of the Fc domain. The knob-into-hole
technology is described
e.g. in US 5,731,168; US 7,695,936; Ridgway etal., 1996, Prot Eng 9:617-621,
and Carter, J,
2001, Immunol Meth 248:7-15. Generally, the method involves introducing a
protuberance
("knob") at the interface of a first polypeptide and a corresponding cavity
("hole") in the interface
of a second polypeptide, such that the protuberance can be positioned in the
cavity so as to
promote heterodimer formation and hinder homodimer formation. Protuberances
are
constructed by replacing small amino acid side chains from the interface of
the first polypeptide
with larger side chains (e.g. tyrosine or tryptophan). Compensatory cavities
of identical or
similar size to the protuberances are created in the interface of the second
polypeptide by
replacing large amino acid side chains with smaller ones (e.g. alanine or
threonine).
[0059] Accordingly, in some embodiments, an amino acid residue in the CH3
domain of the first
subunit of the Fc domain is replaced with an amino acid residue having a
larger side chain
volume, thereby generating a protuberance within the CH3 domain of the first
subunit which is
positionable in a cavity within the CH3 domain of the second subunit, and an
amino acid
residue in the CH3 domain of the second subunit of the Fc domain is replaced
with an amino
acid residue having a smaller side chain volume, thereby generating a cavity
within the CH3
domain of the second subunit within which the protuberance within the CH3
domain of the first
subunit is positionable. Preferably said amino acid residue having a larger
side chain volume is
selected from the group consisting of arginine (R), phenylalanine (F),
tyrosine (Y), and
tryptophan (\A/). Preferably said amino acid residue having a smaller side
chain volume is
selected from the group consisting of alanine (A), serine (S), threonine (T),
and valine (V). The
protuberance and cavity can be made by altering the nucleic acid encoding the
polypeptides,
e.g. by site-specific mutagenesis, or by peptide synthesis. An exemplary
substitution is Y470T.
[0060] In a specific such embodiment, in the first subunit of the Fc domain
the threonine
residue at position 366 is replaced with a tryptophan residue (T366VV), and in
the second
subunit of the Fc domain the tyrosine residue at position 407 is replaced with
a valine residue
(Y407V) and optionally the threonine residue at position 366 is replaced with
a serine residue
(T3665) and the leucine residue at position 368 is replaced with an alanine
residue (L368A)
16
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
(numbering according to Kabat EU index). In a further embodiment, in the first
subunit of the Fc
domain additionally the serine residue at position 354 is replaced with a
cysteine residue
(S3540) or the glutamic acid residue at position 356 is replaced with a
cysteine residue
(E3560) (particularly the serine residue at position 354 is replaced with a
cysteine residue), and
in the second subunit of the Fc domain additionally the tyrosine residue at
position 349 is
replaced by a cysteine residue (Y3490) (numbering according to Kabat EU
index). In a
particular embodiment, the first subunit of the Fc domain comprises the amino
acid
substitutions S3540 and T366W, and the second subunit of the Fc domain
comprises the
amino acid substitutions Y3490, T366S, L368A and Y407V (numbering according to
Kabat EU
index).
[0061] In some embodiments, electrostatic steering (e.g., as described in
Gunasekaran etal.,
2010, J Biol Chem 285(25):19637-46) can be used to promote the association of
the first and
the second subunit of the Fc domain.
[0062] In some embodiments, the Fc domain comprises one or more amino acid
substitutions
that reduces binding to an Fc receptor and/or effector function.
[0063] In a particular embodiment the Fc receptor is an Fey receptor. In one
embodiment the
Fc receptor is a human Fc receptor. In one embodiment the Fc receptor is an
activating Fc
receptor. In a specific embodiment the Fc receptor is an activating human Fey
receptor, more
specifically human FeyRIlla, FeyRI or FeyRIla, most specifically human
FeyRIlla. In one
embodiment the effector function is one or more selected from the group of
complement
dependent cytotoxicity (CDC), antibody-dependent cell-mediated cytotoxicity
(ADCC), antibody-
dependent cellular phagocytosis (ADCP), and cytokine secretion. In a
particular embodiment,
the effector function is ADCC.
[0064] Typically, the same one or more amino acid substitution is present in
each of the two
subunits of the Fc domain. In one embodiment, the one or more amino acid
substitution
reduces the binding affinity of the Fc domain to an Fc receptor. In one
embodiment, the one or
more amino acid substitution reduces the binding affinity of the Fc domain to
an Fc receptor by
at least 2-fold, at least 5-fold, or at least 10-fold.
[0065] In one embodiment, the Fc domain comprises an amino acid substitution
at a position
selected from the group of E233, L234, L235, N297, P331 and P329 (numberings
according to
Kabat EU index). In a more specific embodiment, the Fc domain comprises an
amino acid
substitution at a position selected from the group of L234, L235 and P329
(numberings
according to Kabat EU index). In some embodiments, the Fc domain comprises the
amino acid
substitutions L234A and L235A (numberings according to Kabat EU index). In one
such
embodiment, the Fc domain is an IgGi Fc domain, particularly a human IgGi Fc
domain. In one
embodiment, the Fc domain comprises an amino acid substitution at position
P329. In a more
specific embodiment, the amino acid substitution is P329A or P329G,
particularly P329G
17
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
(numberings according to Kabat EU index). In one embodiment, the Fc domain
comprises an
amino acid substitution at position P329 and a further amino acid substitution
at a position
selected from E233, L234, L235, N297 and P331 (numberings according to Kabat
EU index). In
a more specific embodiment, the further amino acid substitution is E233P,
L234A, L235A,
L235E, N297A, N297D or P331S. In particular embodiments, the Fc domain
comprises amino
acid substitutions at positions P329, L234 and L235 (numberings according to
Kabat EU index).
In more particular embodiments, the Fc domain comprises the amino acid
mutations L234A,
L235A and P329G ("P329G LALA", "PGLALA" or "LALAPG"). Specifically, in
particular
embodiments, each subunit of the Fc domain comprises the amino acid
substitutions L234A,
L235A and P329G (Kabat EU index numbering), i.e. in each of the first and the
second subunit
of the Fc domain the leucine residue at position 234 is replaced with an
alanine residue
(L234A), the leucine residue at position 235 is replaced with an alanine
residue (L235A) and
the proline residue at position 329 is replaced by a glycine residue (P329G)
(numbering
according to Kabat EU index). In one such embodiment, the Fc domain is an IgGi
Fc domain,
particularly a human IgGi Fc domain.
[0066] Single chain-based bispecific antibodies of the disclosure can be any
of the various
types of single chain-based bispecific antibodies known in the art, such as
bispecific T-cell
engagers (BiTEs), diabodies, tandam diabodies (tandabs), dual-affinity
retargeting molecules
(DARTs), and bispecific killer cell engagers. See, e.g., LOffier etal., 2000,
Blood 95:2098-103;
Holliger etal., 1993, Proc Natl Acad Sci USA, 90:6444-8; Kipriyanov etal.,
1999, Mol Biol
293:41-56; Johnson etal., 2010, Mol Biol 399:436-49; VViernik et al., 2013,
Olin Cancer Res
19:3844-55; Liu etal., 2017, Front. lmmunol. 8:38; and Yang etal., 2017, Int.
J. Mol. Sci.
18:48, which are incorporated herein by reference in their entireties.
[0067] In some embodiments, the bispecific antibodies of the disclosure are
bispecific T-cell
engagers (BiTEs). BiTEs are single polypeptide chain molecules that having two
antigen-
binding domains, one of which binds to a T-cell antigen and the second of
which binds to an
antigen present on the surface of a target (See, PCT Publication WO 05/061547;
Baeuerle et
al., 2008, Drugs of the Future 33: 137-147; Bargou, etal., 2008, Science
321:974-977,
incorporated herein by reference in their entireties). Thus, the BiTEs of the
disclosure have an
antigen binding domain that binds to a T-cell antigen, and a second antigen
binding domain that
is directed towards glyco-MUC1.
[0068] In some embodiments, the bispecific antibodies of the disclosure are
dual-affinity
retargeting molecules (DARTs). DARTs comprise at least two polypeptide chains
that associate
(especially through a covalent interaction) to form at least two epitope
binding sites, which may
recognize the same or different epitopes. Each of the polypeptide chains of a
DART comprise
an immunoglobulin light chain variable region and an immunoglobulin heavy
chain variable
region, but these regions do not interact to form an epitope binding site.
Rather, the
18
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
immunoglobulin heavy chain variable region of one (e.g., the first) of the
DART polypeptide
chains interacts with the immunoglobulin light chain variable region of a
different (e.g., the
second) DARTTm polypeptide chain to form an epitope binding site. Similarly,
the
immunoglobulin light chain variable region of one (e.g., the first) of the
DART polypeptide
chains interacts with the immunoglobulin heavy chain variable region of a
different (e.g., the
second) DART polypeptide chain to form an epitope binding site. DARTs may be
monospecific,
bispecific, trispecific, etc., thus being able to simultaneously bind one,
two, three or more
different epitopes (which may be of the same or of different antigens). DARTs
may additionally
be monovalent, bivalent, trivalent, tetravalent, pentavalent, hexavalent,
etc., thus being able to
simultaneously bind one, two, three, four, five, six or more molecules. These
two attributes of
DARTs (i.e., degree of specificity and valency may be combined, for example to
produce
bispecific antibodies (i.e., capable of binding two epitopes) that are
tetravalent (i.e., capable of
binding four sets of epitopes), etc. DART molecules are disclosed in PCT
Publications WO
2006/113665, WO 2008/157379, and WO 2010/080538, which are incorporated herein
by
reference in their entireties.
[0069] In some embodiments of the bispecific antibodies of the disclosure, one
of the binding
specificities is directed towards glyco-MUC1, and the other is directed to an
antigen expressed
on immune effector cells. The term "immune effector cell" or "effector cell"
as used herein refers
to a cell within the natural repertoire of cells in the mammalian immune
system which can be
activated to affect the viability of a target cell. Immune effector cells
include cells of the
lymphoid lineage such as natural killer (NK) cells, T cells including
cytotoxic T cells, or B cells,
but also cells of the myeloid lineage can be regarded as immune effector
cells, such as
monocytes or macrophages, dendritic cells and neutrophilic granulocytes.
Hence, said effector
cell is preferably an NK cell, a T cell, a B cell, a monocyte, a macrophage, a
dendritic cell or a
neutrophilic granulocyte. Recruitment of effector cells to aberrant cells
means that immune
effector cells are brought in close vicinity to the aberrant target cells such
that the effector cells
can directly kill, or indirectly initiate the killing of the aberrant cells
that they are recruited to. In
order to avoid non specific interactions it is preferred that the bispecific
antibodies of the
disclosure specifically recognize antigens on immune effector cells that are
at least over-
expressed by these immune effector cells compared to other cells in the body.
Target antigens
present on immune effector cells may include CD3, CD8, CD16, 0D25, 0D28, 0D64,
0D89,
NKG2D and NKp46. Preferably, the antigen on immune effector cells is CD3
expressed on T
cells.
[0070] As used herein, "CD3" refers to any native CD3 from any vertebrate
source, including
mammals such as primates (e.g. humans), non-human primates (e.g. cynomolgus
monkeys)
and rodents (e.g. mice and rats), unless otherwise indicated. The term
encompasses "full-
length," unprocessed CD3 as well as any form of CD3 that results from
processing in the cell.
19
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
The term also encompasses naturally occurring variants of CD3, e.g., splice
variants or allelic
variants. The most preferred antigen on an immune effector cell is the CD3
epsilon chain. This
antigen has been shown to be very effective in recruiting T cells to aberrant
cells. Hence, a
bispecific antibody of the disclosure preferably specifically recognizes CD3
epsilon. The amino
acid sequence of human CD3 epsilon is shown in UniProt (www.uniprot.org)
accession no.
P07766 (version 144), or NCB! (www.ncbi.nlm.nih.gov/) RefSeq NP_000724.1. The
amino acid
sequence of cynomolgus [Macaca fascicularis] CD3 epsilon is shown in NCB!
GenBank no.
BAB71849.1. For human therapeutic use, bispecific antibodies in which the CD3-
binding
domain specifically binds to human CD3 (e.g., the human CD3 epsilon chain) are
used. For
preclinical testing in non-human animals and cell lines, bispecific antibodies
in which the CD3-
binding domain specifically binds to the CD3 in the species utilized for the
preclinical testing
(e.g., cynomolgus CD3 for primate testing) can be used.
[0071] As used herein, a binding domain that "specifically binds to" or
"specifically recognizes"
a target antigen from a particular species does not preclude the binding to or
recognition of the
antigen from other species, and thus encompasses antibodies in which one or
more of the
binding domains have inter-species cross-reactivity. For example, a CD3-
binding domain that
"specifically binds to" or "specifically recognizes" human CD3 may also bind
to or recognize
cyomolgus CD3, and vice versa.
[0072] In some embodiments, a bispecific antibody of the disclosure can
compete with
monoclonal antibody H2C (described in PCT publication no. W02008/119567) for
binding an
epitope of CD3. In other embodiments, a bispecific antibody of the disclosure
can compete with
monoclonal antibody V9 (described in Rodrigues etal., 1992, Int J Cancer Suppl
7:45-50 and
U.S. Pat. No. 6,054,297) for binding an epitope of CD3. In yet other
embodiments, a bispecific
antibody of the disclosure can compete with monoclonal antibody FN18
(described in Nooij et
al., 1986, Eur J Immunol 19:981-984) for binding an epitope of CD3. In yet
other embodiments,
a bispecific antibody of the disclosure can compete with monoclonal antibody
5P34 (described
in Pessano etal., 1985, EMBO J 4:337-340) for binding an epitope of CD3.
[0073] The anti-glyco-MUC1 antibodies of the disclosure include derivatized
antibodies. For
example, but not by way of limitation, derivatized antibodies are typically
modified by
glycosylation, acetylation, pegylation, phosphorylation, amidation,
derivatization by known
protecting/blocking groups, proteolytic cleavage, linkage to a cellular ligand
or other protein.
Any of numerous chemical modifications can be carried out by known techniques,
including, but
not limited to, specific chemical cleavage, acetylation, formylation,
metabolic synthesis of
tunicamycin, etc. Additionally, the derivative can contain one or more non-
natural amino acids,
e.g., using ambrx technology (See, e.g., Wolfson, 2006, Chem. Biol.
13(10):1011-2).
[0074] The anti-glyco-MUC1 antibodies or binding fragments may be antibodies
or fragments
whose sequences have been modified to alter at least one constant region-
mediated biological
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
effector function. For example, in some embodiments, an anti-glyco-MUC1
antibody may be
modified to reduce at least one constant region-mediated biological effector
function relative to
the unmodified antibody, e.g., reduced binding to the Fc receptor (FcyR). FcyR
binding can be
reduced by mutating the immunoglobulin constant region segment of the antibody
at particular
regions necessary for FcyR interactions (See, e.g., Canfield and Morrison,
1991, J. Exp. Med.
173:1483-1491; and Lund etal., 1991, J. lmmunol. 147:2657-2662). Reduction in
FcyR binding
ability of the antibody can also reduce other effector functions which rely on
FcyR interactions,
such as opsonization, phagocytosis and antigen-dependent cellular cytotoxicity
("ADCC").
[0075] The anti-glyco-MUC1 antibody or binding fragments described herein
include antibodies
and/or binding fragments that have been modified to acquire or improve at
least one constant
region-mediated biological effector function relative to an unmodified
antibody, e.g., to enhance
FcyR interactions (See, e.g., US 2006/0134709). For example, an anti-glyco-
MUC1 antibody of
the disclosure can have a constant region that binds FcyRIIA, FcyRIIB and/or
FcyRIIIA with
greater affinity than the corresponding wild type constant region.
[0076] Thus, antibodies of the disclosure may have alterations in biological
activity that result in
increased or decreased opsonization, phagocytosis, or ADCC. Such alterations
are known in
the art. For example, modifications in antibodies that reduce ADCC activity
are described in
U.S. Pat. No. 5,834,597. An exemplary ADCC lowering variant corresponds to
"mutant 3"
(shown in FIG. 4 of U.S. Pat. No. 5,834,597) in which residue 236 is deleted
and residues 234,
235 and 237 (using EU numbering) are substituted with alanines.
[0077] In some embodiments, the anti-glyco-MUC1 antibodies of the disclosure
have low levels
of, or lack, fucose. Antibodies lacking fucose have been correlated with
enhanced ADCC
activity, especially at low doses of antibody. See Shields etal., 2002, J.
Biol. Chem. 277:26733-
26740; Shinkawa etal., 2003, J. Biol. Chem. 278:3466-73. Methods of preparing
fucose-less
antibodies include growth in rat myeloma YB2/0 cells (ATCC CRL 1662). YB2/0
cells express
low levels of FUT8 mRNA, which encodes a-1, 6-fucosyltransferase, an enzyme
necessary for
fucosylation of polypeptides.
[0078] In yet another aspect, the anti-glyco-MUC1 antibodies or binding
fragments include
modifications that increase or decrease their binding affinities to the fetal
Fc receptor, FcRn, for
example, by mutating the immunoglobulin constant region segment at particular
regions
involved in FcRn interactions (see, e.g., WO 2005/123780). In particular
embodiments, an anti-
glyco-MUC1 antibody of the IgG class is mutated such that at least one of
amino acid residues
250, 314, and 428 of the heavy chain constant region is substituted alone, or
in any
combinations thereof, such as at positions 250 and 428, or at positions 250
and 314, or at
positions 314 and 428, or at positions 250, 314, and 428, with positions 250
and 428 a specific
combination. For position 250, the substituting amino acid residue can be any
amino acid
residue other than threonine, including, but not limited to, alanine,
cysteine, aspartic acid,
21
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
glutamic acid, phenylalanine, glycine, histidine, isoleucine, lysine, leucine,
methionine,
asparagine, proline, glutamine, arginine, serine, valine, tryptophan, or
tyrosine. For position
314, the substituting amino acid residue can be any amino acid residue other
than leucine,
including, but not limited to, alanine, cysteine, aspartic acid, glutamic
acid, phenylalanine,
glycine, histidine, isoleucine, lysine, methionine, asparagine, proline,
glutamine, arginine,
serine, threonine, valine, tryptophan, or tyrosine. For position 428, the
substituting amino acid
residues can be any amino acid residue other than methionine, including, but
not limited to,
alanine, cysteine, aspartic acid, glutamic acid, phenylalanine, glycine,
histidine, isoleucine,
lysine, leucine, asparagine, proline, glutamine, arginine, serine, threonine,
valine, tryptophan, or
tyrosine. Specific combinations of suitable amino acid substitutions are
identified in Table 1 of
U.S. Pat. No. 7,217,797, which is incorporated herein by reference. Such
mutations increase
binding to FcRn, which protects the antibody from degradation and increases
its half-life.
[0079] In yet other aspects, an anti-glyco-MUC1 antibody of antigen-binding
fragment of the
disclosure has one or more amino acids inserted into one or more of its
hypervariable regions,
for example as described in Jung and Pluckthun, 1997, Protein Engineering
10:9, 959-966;
Yazaki etal., 2004, Protein Eng. Des Sel. 17(5):481-9. Epub 2004 Aug. 17; and
U.S. Pat. App.
No. 2007/0280931.
[0080] In yet other aspects, particularly useful for diagnostic applications,
an anti-glyco-MUC1
antibody of antigen-binding fragment of the disclosure is attached to a
detectable moiety.
Detectably moieties include a radioactive moiety, a colorimetric molecule, a
fluorescent moiety,
a chemiluminescent moiety, an antigen, an enzyme, a detectable bead (such as a
magnetic or
electrodense (e.g., gold) bead), or a molecule that binds to another molecule
(e.g., biotin or
streptavidin)).
[0081] Radioisotopes or radionuclides may include 3H, 140, 15N, 355, 90y, 99-
rc, 111in, 1251, 1311.
[0082] Fluorescent labels may include rhodamine, lanthanide phosphors,
fluorescein and its
derivatives, fluorochrome, GFP (GFP for "Green Fluorescent Protein"), dansyl,
umbelliferone,
phycoerythrin, phycocyanin, allophycocyanin, o-phthaldehyde, and
fluorescamine.
[0083] Enzymatic labels may include horseradish peroxidase, 13 galactosidase,
luciferase,
alkaline phosphatase, glucose-6-phosphate dehydrogenase ("G6PDH"), alpha-D-
galactosidase,
glucose oxydase, glucose amylase, carbonic anhydrase, acetylcholinesterase,
lysozyme,
malate dehydrogenase and peroxidase.
[0084] Chemiluminescent labels or chemiluminescers, such as isoluminol,
luminol and the
dioxetanes
[0085] Other detectable moieties include molecules such as biotin, digoxygenin
or 5-
bromodeoxyuridine.
22
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
[0086] In certain aspects, an anti-glyco-MUC1 antibody or antigen binding
fragment of the
disclosure competes with G02 or an antibody or antigen binding fragment
comprising heavy
and light chain variable regions of G02 (SEQ ID NOS:3 and 4, respectively).
[0087] The competition can be assayed on cells that express the glyco-MUC1
epitope bound
by G02 or on a glycosylated MUC1 peptide containing the epitope bound by G02,
e.g., the 60-
mer peptide (VTSAPDTRPAPGSTAPPAHG)3 glycosylated with GaINAc-T2, GaINAc-T4,
and
GaINAc-T1 as described in US Patent No. 6,465,220. Cells that do not express
the epitope or
unglycosylated peptides can be used as controls.
[0088] Cells on which a competition assay can be carried out include but are
not limited to the
breast cancer cell lines MCF7 or T47D and recombinant cells that are
engineered to express
the glyco-MUC1 epitope. In one non-limiting example, CHO IdID cells, which
lack the UDP-
Gal/GaINAc epimerase and are deficient in GaINAc 0-glycosylation and
galactosylation in the
absence of exogenous addition of GaINAc and Gal, respectively, are engineered
to express
MUC1 and grown in the absence or presence of GaINAc, the latter yielding cells
expressing the
Tn glycoform of MUC1 to which G02 binds. Cells expressing the unglycosylated
form of MUC1
can be used as a negative control.
[0089] Assays for competition include, but are not limited to, a radioactive
material labeled
immunoassay (RIA), an enzyme-linked immunosorbent assay (ELISA), a sandwich
ELISA
fluorescence activated cell sorting (FACS) assays and Biacore assays.
[0090] In conducting an antibody competition assay between a reference
antibody and a test
antibody (irrespective of species or isotype), one may first label the
reference with a detectable
label, such as a fluorophore, biotin or an enzymatic (or even radioactive)
label to enable
subsequent identification. In this case, cells expressing glyco-MUC1 are
incubated with
unlabeled test antibody, labeled reference antibody is added, and the
intensity of the bound
label is measured. If the test antibody competes with the labeled reference
antibody by binding
to an overlapping epitope, the intensity will be decreased relative to a
control reaction carried
out without test antibody.
[0091] In a specific embodiment of this assay, the concentration of labeled
reference antibody
that yields 80% of maximal binding ("conc80%") under the assay conditions
(e.g., a specified
density of cells) is first determined, and a competition assay carried out
with 10 x conc80% of
unlabeled test antibody and conc80% of labeled reference antibody.
[0092] The inhibition can be expressed as an inhibition constant, or K, which
is calculated
according to the following formula:
[0093] K=IC50/(1+[reference Ab concentration]/Kd),
[0094] where IC50 is the concentration of test antibody that yields a 50%
reduction in binding of
the reference antibody and Kd is the dissociation constant of the reference
antibody, a measure
23
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
of its affinity for glyco-MUC1. Antibodies that compete with anti-glyco-MUC1
antibodies
disclosed herein can have a K, from 10 pM to 10 nM under assay conditions
described herein.
[0095] In various embodiments, a test antibody is considered to compete with a
reference
antibody if it decreases binding of the reference antibody by at least about
20% or more, for
example, by at least about 20%, 30%, 40%, 50%, 60%, 70%, 80%, 90%, 95% or even
more, or
by a percentage ranging between any of the foregoing values, at a reference
antibody
concentration that is 80% of maximal binding under the specific assay
conditions used, and a
test antibody concentration that is 10-fold higher than the reference antibody
concentration.
[0096] In one example of a competition assay, the glycosylated MUC1 60-mer
peptide is
adhered onto a solid surface, e.g., a microwell plate, by contacting the plate
with a solution of
the peptide (e.g., at a concentration of 1 pg/mL in PBS over night at 4 C).
The plate is washed
(e.g., 0.1% Tween 20 in PBS) and blocked (e.g., in Superblock, Thermo
Scientific, Rockford,
IL). A mixture of sub-saturating amount of biotinylated G02 (e.g., at a
concentration of 80
ng/mL) and unlabeled G02 (the "reference" antibody) or competing anti-glyco-M
UC1 antibody
(the "test" antibody) antibody in serial dilution (e.g., at a concentration of
2.8 pg/mL, 8.3 pg/mL,
or 25 pg/mL) in ELISA buffer (e.g., 1% BSA and 0.1% Tween 20 in PBS) is added
to wells and
plates are incubated for 1 hour with gentle shaking. The plate is washed, 1
pg/mL HRP-
conjugated Streptavidin diluted in ELISA buffer is added to each well and the
plates incubated
for 1 hour. Plates are washed and bound antibodies were detected by addition
of substrate
(e.g., TMB, Biofx Laboratories Inc., Owings Mills, MD). The reaction is
terminated by addition of
stop buffer (e.g., Bio FX Stop Reagents, Biofx Laboratories Inc., Owings
Mills, MD) and the
absorbance is measured at 650 nm using microplate reader (e.g., VERSAmax,
Molecular
Devices, Sunnyvale, CA).
[0097] Variations on this competition assay can also be used to test
competition between G02
and another anti-glyco-MUC1 antibody. For example, in certain aspects, the
anti-glyco-MUC1
antibody is used as a reference antibody and G02 is used as a test antibody.
Additionally,
instead of glycosylated MUC1 60-mer peptide, membrane-bound glyco-M UC1
expressed on
cell surface (for example on the surface of one of the cell types mentioned
above) in culture can
be used. Generally, about 104 to 106 transfectants, e.g., about 105
transfectants, are used.
Other formats for competition assays are known in the art and can be employed.
[0098] In various embodiments, an anti-glyco-MUC1 antibody of the disclosure
reduces the
binding of labeled G02 by at least 40%, by at least 50%, by at least 60%, by
at least 70%, by at
least 80%, by at least 90%, or by a percentage ranging between any of the
foregoing values
(e.g., an anti-glyco-M UC1 antibody of the disclosure reduces the binding of
labeled G02 by
50% to 70%) when the anti-glyco-MUC1 antibody is used at a concentration of
0.08 pg/mL, 0.4
pg/mL, 2 pg/mL, 10 pg/mL, 50 pg/mL, 100 pg/mL or at a concentration ranging
between any of
the foregoing values (e.g., at a concentration ranging from 2 pg/mL to 10
pg/mL).
24
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
[0099] In other embodiments, G02 reduces the binding of a labeled anti-glyco-M
UC1 antibody
of the disclosure by at least 40%, by at least 50%, by at least 60%, by at
least 70%, by at least
80%, by at least 90%, or by a percentage ranging between any of the foregoing
values (e.g.,
G02 reduces the binding of a labeled an anti-glyco-MUC1 antibody of the
disclosure by 50% to
70%) when G02 is used at a concentration of 0.4 pg/mL, 2 pg/mL, 10 pg/mL, 50
pg/mL, 250
pg/mL or at a concentration ranging between any of the foregoing values (e.g.,
at a
concentration ranging from 2 pg/mL to 10 pg/mL).
[0100] In the foregoing assays, the G02 antibody can be replaced by any
antibody or antigen-
binding fragment comprising the CDRs or the heavy and light chain variable
regions of G02,
such as a humanized or chimeric counterpart of G02.
[0101] In certain aspects, an anti-glyco-MUC1 antibody or antigen-binding
fragment of the
disclosure comprises heavy and/or light chain variable sequences (or encoded
by the
nucleotide sequences) set forth in Table 1. In other aspects, an anti-glyco-
MUC1 antibody or
antigen-binding fragment of the disclosure comprises heavy and/or light chain
CDR sequences
(or encoded by the nucleotide sequences) set forth in Table 1. The framework
sequences for
such anti-glyco-MUC1 antibody and antigen-binding fragment can be the native
murine
framework sequences in Table 1 or can be non-native (e.g., humanized or human)
framework
sequences.
[0102] In yet other aspects, the disclosure provides an anti-MUC1 antibody or
antigen binding
fragment having heavy and light chain variable regions having at least 95%,
98%, 99%, or
99.5% sequence identity of SEQ ID NOS: 3 and 4, respectively.
[0103] In yet other aspects, an anti-glyco-MUC1 antibody or antigen-binding
fragment of the
disclosure is a single-chain variable fragment (scFv). An exemplary scFv
comprises the heavy
chain variable fragment N-terminal to the light chain variable fragment. In
some embodiments,
the scFv heavy chain variable fragment and light chain variable fragment are
covalently bound
to a linker sequence of 4-15 amino acids. The scFv can be in the form of a bi-
specific T-cell
engager or within a chimeric antigen receptor (CAR).
5.2 Anti-glyco-MUC1 and Anti-CD3 Bispecific Antibodies
[0104] In some aspects, bispecific antibodies of the disclosure can comprise a
first antigen
binding domain that specifically binds to CD3 (e.g., which comprises the CDRs
or VH and VL
set forth in Table 4), and a second antigen binding domain that specifically
binds to glyco-
MUC1. The second antigen binding domain may comprise, singly or in
combination, the
features described for the glyco-M UC1 antibodies hereinabove (e.g., comprise
a combination of
CDRs identified in Tables 1-3, for example CDRs comprising the amino acid
sequences of any
of the CDR combinations set forth in numbered embodiments 3 to 17, infra, or
the VH and VL
sequences identified in Table 1).
CA 03078812 2020-04-08
WO 2019/083506
PCT/US2017/058036
Table 4
Description Sequence SEQ ID
NO:
CD3 CDR- TYAMN 34
H1 (Kabat)
CD3 CDR- RIRSKYNNYATYYADSVKG 35
H2 (Kabat)
CD3 CDR- HGNFGNSYVSWFAY 36
H3 (Kabat)
CD3 CDR-L1 GSSTGAVTTSNYAN 37
(Kabat)
CD3 CDR-L2 GTNKRAP 38
(Kabat)
CD3 CDR-L3 ALVVYSNLVVV 39
(Kabat)
CD3 VH EVQLLESGGGLVQPGGSLRLSCAASGFTFSTYAMNVVVRQAPG 40
KGLEVVVSRIRSKYNNYATYYADSVKGRFTISRDDSKNTLYLQM
NSLRAEDTAVYYCVRHGNFGNSYVSWFAYWGQGTLVTVSS
CD3 VL QAVVTQEPSLTVSPGGTVTLTCGSSTGAVTTSNYANVVVQEKP 41
GQAFRGLIGGTNKRAPGTPARFSGSLLGGKAALTLSGAQPEDE
AEYYCALVVYSNLVVVFGGGTKLTVL
hIgG1 Fc DKTHTCPPCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVV 42
region DVSHEDPEVKFNVVYVDGVEVHNAKTKPREEQYNSTYRVVSVL
TVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYT
LPPSRDELTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKT
TPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEALHNH
YTQKSLSLSP
MUC1 VL- DIVMSQSPSSLGVSVGEKVTMSCKSSQSLLYSTNQKNYQSLLY 43
CL(RK) STNQKNYLAVVYQQKPGQSPKLLIYVVVSNRKSGVPDRFTGSGS
GTDFTLTISSVKAEDLAVYYC QQYYRYPLTFGAGTKLELK
RTVAAPSVFIFPPSDRKLKSGTASVVCLLNNFYPREAKVQWKVD
NALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYAC
EVTHQGLSSPVTKSFNRGEC
CD3 VH-CL EVQLLESGGGLVQPGGSLRLSCAASGFTFSTYAMNVVVRQAPG 44
KGLEVVVSRIRSKYNNYATYYADSVKGRFTISRDDSKNTLYLQM
NSLRAEDTAVYYCVRHGNFGNSYVSWFAYWGQGTLVTVSSAS
VAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDN
ALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACE
VTHQGLSSPVTKSFNRGEC
MUC1 VH- QVQLQQSDAELVKPGASVKISCKASGYTFTDHAIHVVVKQRPEQ 45
CH1(EE)-Fc GLEWIGYFSPGNDDIHYNEKFEGKATLTADKSSSTAYMQLNSLT
(hole, P329G SEDSAVYFCKRSYDKDFDCWGQGTTLTVSSASTKGPSVFPLAP
LALA) SSKSTSGGTAALGCLVEDYFPEPVTVSWNSGALTSGVHTFPAV
LQSSGLYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDEKVEP
KSCDKTHTCPPCPAPEAAGGPSVFLFPPKPKDTLMISRTPEVTC
VVVDVSHEDPEVKFNVVYVDGVEVHNAKTKPREEQYNSTYRVV
SVLTVLHQDWLNGKEYKCKVSNKALGAPIEKTISKAKGQPREPQ
VCTLPPSRDELTKNQVSLSCAVKGFYPSDIAVEWESNGQPENN
YKTTPPVLDSDGSFFLVSKLTVDKSRWQQGNVFSCSVMHEALH
NHYTQKSLSLSPGK
MUC1 VH- QVQLQQSDAELVKPGASVKISCKASGYTFTDHAIHVVVKQRPEQ 46
CH 1(EE)- GLEWIGYFSPGNDDI HYN EKFEGKATLTADKSSSTAYMQLNSLT
0D3 VL- SEDSAVYFCKRSYDKDFDCWGQGTTLTVSSASTKGPSVFPLAP
CH1-Fc SSKSTSGGTAALGCLVEDYFPEPVTVSWNSGALTSGVHTFPAV
(knob, LQSSGLYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDEKVEP
P329G KSCDGGGGSGGGGSQAVVTQEPSLTVSPGGTVTLTCGSSTGA
26
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
LA LA) VTTSNYANVVVQEKPGQAFRGLIGGTNKRAPGTPARFSGSLLG
GKAALTLSGAQPEDEAEYYCALVVYSNLVVVFGGGTKLTVLSSAS
TKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGA
LTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTQTYICNVNHKP
SNTKVDKKVEPKSCDKTHTCPPCPAPEAAGGPSVFLFPPKPKD
TLMISRTPEVTCVVVDVSHEDPEVKFNVVYVDGVEVHNAKTKPR
EEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKALGAPIEKTI
SKAKGQPREPQVYTLPPCRDELTKNQVSLWCLVKGFYPSDIAV
EWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGN
VFSCSVMHEALHNHYTQKSLSLSPGK
[0105] In some embodiments, the first antigen binding domain comprises a heavy
chain
variable region comprising the heavy chain CDR-H1 of SEQ ID NO:34, the CDR-H2
of SEQ ID
NO:35, and the CDR-H3 of SEQ ID NO:36; and a light chain variable region
comprising the
light chain CDR-L1 of SEQ ID NO:37, the CDR-L2 of SEQ ID NO:38 and the CDR-L3
of SEQ
ID NO:39.
[0106] In some embodiments, the second antigen binding domain comprises for
example
CDRs comprising the amino acid sequences of any of the CDR combinations set
forth in
numbered embodiments 3 to 17, for example (i) a heavy chain variable region
comprising the
heavy chain CDR-H1 of SEQ ID NO: 5, the CDR-H2 of SEQ ID NO: 6, and the CDR-H3
of SEQ
ID NO: 7; and a light chain variable region comprising the light chain CDR
(CDR-L) 1 of SEQ ID
NO: 8, the CDR-L2 of SEQ ID NO: 9 and the CDR-L3 of SEQ ID NO:10.
[0107] In a particular embodiment, the bispecific antibody comprises
(i) a first antigen binding domain that specifically binds to CD3 and
comprises a heavy
chain variable region comprising a CDR-H1 comprising the amino acid sequence
of SEQ ID
NO:34, a CDR-H2 comprising the amino acid sequence of SEQ ID NO:35, and a CDR-
H3
comprising the amino acid sequence of SEQ ID NO:36; and a light chain variable
region
comprising a CDR-L1 comprising the amino acid sequence of SEQ ID NO:37, a CDR-
L2
comprising the amino acid sequence of SEQ ID NO:38, and a CDR-L3 comprising
the amino
acid sequence of SEQ ID NO:39; and
(ii) a second antigen binding domain that specifically binds to glyco-MUC1 and
comprises (i) a heavy chain variable region comprising a CDR-H1 comprising the
amino acid
sequence of SEQ ID NO:33, more preferably a CDR-H1 comprising the amino acid
sequence
of SEQ ID NO:5, a CDR-H2 comprising the amino acid sequence of SEQ ID NO:29,
more
preferably a CDR-H1 comprising the amino acid sequence of SEQ ID NO:6, and a
CDR-H3
comprising the amino acid sequence of SEQ ID NO:25, more preferably a CDR-H3
comprising
the amino acid sequence of SEQ ID NO:7; and a light chain variable region
comprising aCDR-
L1 comprising the amino acid sequence of of SEQ ID NO:8, a CDR-L2 comprising
the amino
acid sequence of SEQ ID NO:9 and a CDR-L3 comprising the amino acid sequence
of SEQ ID
NO:31, more preferably a CDR-L3 comprising the amino acid sequence of SEQ ID
NO:10.
27
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
[0108] In some embodiments, the first antigen binding domain comprises a heavy
chain
variable region sequence that is at least about 95%, 96%, 97%, 98%, 99% or
100% identical to
the amino acid sequence of SEQ ID NO:40 and a light chain variable region
sequence that is at
least about 95%, 96%, 97%, 98%, 99% or 100% identical to the amino acid
sequence of SEQ
ID NO:41.
[0109] In some embodiments, the first antigen binding domain comprises the
heavy chain
variable region sequence of SEQ ID NO:40 and the light chain variable region
sequence of
SEQ ID NO:41.
[0110] In some embodiments, the second antigen binding domain comprises a
heavy chain
variable region sequence that is at least about 95%, 96%, 97%, 98%, 99% or
100% identical to
the amino acid sequence of SEQ ID NO:3 and a light chain variable region
sequence that is at
least about 95%, 96%, 97%, 98%, 99% or 100% identical to the amino acid
sequence of SEQ
ID NO:4.
[0111] In some embodiments, the second antigen binding domain comprises the
heavy chain
variable region sequence of SEQ ID NO:3 and the light chain variable region
sequence of SEQ
ID NO:4.
[0112] In some embodiments, the first and/or the second antigen binding domain
is a Fab
molecule. In some embodiments, the first antigen binding domain is a crossover
Fab molecule
wherein either the variable or the constant regions of the Fab light chain and
the Fab heavy
chain are exchanged. In such embodiments, the second antigen binding domain
preferably is a
conventional Fab molecule.
[0113] In some embodiments wherein the first and the second antigen binding
domain of the
bispecific antibody are both Fab molecules, and in one of the antigen binding
domains
(particularly the first antigen binding domain) the variable domains VL and VH
of the Fab light
chain and the Fab heavy chain are replaced by each other,
i) in the constant domain CL of the first antigen binding domain the amino
acid at
position 124 is substituted by a positively charged amino acid (numbering
according to Kabat),
and wherein in the constant domain CH1 of the first antigen binding domain the
amino acid at
position 147 or the amino acid at position 213 is substituted by a negatively
charged amino acid
(numbering according to Kabat EU index); or
ii) in the constant domain CL of the second antigen binding domain the amino
acid at
position 124 is substituted by a positively charged amino acid (numbering
according to Kabat),
and wherein in the constant domain CH1 of the second antigen binding domain
the amino acid
at position 147 or the amino acid at position 213 is substituted by a
negatively charged amino
acid (numbering according to Kabat EU index).
28
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
[0114] The bispecific antibody does not comprise both modifications mentioned
under i) and ii).
The constant domains CL and CH1 of the antigen binding domain having the VHA/L
exchange
are not replaced by each other (i.e., they remain unexchanged).
[0115] In a more specific embodiment,
i) in the constant domain CL of the first antigen binding domain the amino
acid at
position 124 is substituted independently by lysine (K), arginine (R) or
histidine (H) (numbering
according to Kabat), and in the constant domain CH1 of the first antigen
binding domain the
amino acid at position 147 or the amino acid at position 213 is substituted
independently by
glutamic acid (E), or aspartic acid (D) (numbering according to Kabat EU
index); or
ii) in the constant domain CL of the second antigen binding domain the amino
acid at
position 124 is substituted independently by lysine (K), arginine (R) or
histidine (H) (numbering
according to Kabat), and in the constant domain CH1 of the second antigen
binding domain the
amino acid at position 147 or the amino acid at position 213 is substituted
independently by
glutamic acid (E), or aspartic acid (D) (numbering according to Kabat EU
index).
[0116] In one such embodiment, in the constant domain CL of the second antigen
binding
domain the amino acid at position 124 is substituted independently by lysine
(K), arginine (R) or
histidine (H) (numbering according to Kabat), and in the constant domain CH1
of the second
antigen binding domain the amino acid at position 147 or the amino acid at
position 213 is
substituted independently by glutamic acid (E), or aspartic acid (D)
(numbering according to
Kabat EU index).
[0117] In a further embodiment, in the constant domain CL of the second
antigen binding
domain the amino acid at position 124 is substituted independently by lysine
(K), arginine (R) or
histidine (H) (numbering according to Kabat), and in the constant domain CH1
of the second
antigen binding domain the amino acid at position 147 is substituted
independently by glutamic
acid (E), or aspartic acid (D) (numbering according to Kabat EU index).
[0118] In a particular embodiment, in the constant domain CL of the second
antigen binding
domain the amino acid at position 124 is substituted independently by lysine
(K), arginine (R) or
histidine (H) (numbering according to Kabat) and the amino acid at position
123 is substituted
independently by lysine (K), arginine (R) or histidine (H) (numbering
according to Kabat), and in
the constant domain CH1 of the second antigen binding domain the amino acid at
position 147
is substituted independently by glutamic acid (E), or aspartic acid (D)
(numbering according to
Kabat EU index) and the amino acid at position 213 is substituted
independently by glutamic
acid (E), or aspartic acid (D) (numbering according to Kabat EU index).
[0119] In a more particular embodiment, in the constant domain CL of the
second antigen
binding domain the amino acid at position 124 is substituted by lysine (K)
(numbering according
to Kabat) and the amino acid at position 123 is substituted by lysine (K)
(numbering according
29
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
to Kabat), and in the constant domain CH1 of the second antigen binding domain
the amino
acid at position 147 is substituted by glutamic acid (E) (numbering according
to Kabat EU
index) and the amino acid at position 213 is substituted by glutamic acid (E)
(numbering
according to Kabat EU index).
[0120] In an even more particular embodiment, in the constant domain CL of the
second
antigen binding domain the amino acid at position 124 is substituted by lysine
(K) (numbering
according to Kabat) and the amino acid at position 123 is substituted by
arginine (R)
(numbering according to Kabat), and in the constant domain CH1 of the second
antigen binding
domain the amino acid at position 147 is substituted by glutamic acid (E)
(numbering according
to Kabat EU index) and the amino acid at position 213 is substituted by
glutamic acid (E)
(numbering according to Kabat EU index).
[0121] In particular embodiments, if amino acid substitutions according to the
above
embodiments are made in the constant domain CL and the constant domain CH1 of
the second
antigen binding domain, the constant domain CL of the second antigen binding
domain is of
kappa isotype.
[0122] In some embodiments, the first and the second antigen binding domain
are fused to
each other, optionally via a peptide linker.
[0123] In some embodiments, the first and the second antigen binding domain
are each a Fab
molecule and either (i) the second antigen binding domain is fused at the C-
terminus of the Fab
heavy chain to the N-terminus of the Fab heavy chain of the first antigen
binding domain, or (ii)
the first antigen binding domain is fused at the C-terminus of the Fab heavy
chain to the N-
terminus of the Fab heavy chain of the second antigen binding domain.
[0124] In some embodiments, the bispecific antibody provides monovalent
binding to CD3.
[0125] In particular embodiments, the bispecific antibody comprises a single
antigen binding
domain that specifically binds to CD3, and two antigen binding domains that
specifically bind to
glyco-MUC1. Thus, in some embodiments, the bispecific antibody comprises a
third antigen
binding domain that specifically binds to glyco-MUC1. In some embodiments, the
third antigen
moiety is identical to the first antigen binding domain (e.g. is also a Fab
molecule and
comprises the same amino acid sequences).
[0126] In particular embodiments, the bispecific antibody further comprises an
Fc domain
composed of a first and a second subunit. In one embodiment, the Fc domain is
an IgG Fc
domain. In a particular embodiment, the Fc domain is an IgGi Fc domain. In
another
embodiment the Fc domain is an IgG4 Fc domain. In a more specific embodiment,
the Fc
domain is an IgG4 Fc domain comprising an amino acid substitution at position
S228 (Kabat EU
index numbering), particularly the amino acid substitution S228P. In a further
particular
embodiment, the Fc domain is a human Fc domain. In an even more particular
embodiment,
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
the Fc domain is a human IgGi Fc domain. An exemplary sequence of a human IgGi
Fc region
is given in SEQ ID NO: 42.
[0127] In some embodiments wherein the first, the second and, where present,
the third
antigen binding domain are each a Fab molecule, (a) either (i) the second
antigen binding
domain is fused at the C-terminus of the Fab heavy chain to the N-terminus of
the Fab heavy
chain of the first antigen binding domain and the first antigen binding domain
is fused at the C-
terminus of the Fab heavy chain to the N-terminus of the first subunit of the
Fc domain, or (ii)
the first antigen binding domain is fused at the C-terminus of the Fab heavy
chain to the N-
terminus of the Fab heavy chain of the second antigen binding domain and the
second antigen
binding domain is fused at the C-terminus of the Fab heavy chain to the N-
terminus of the first
subunit of the Fc domain; and (b) the third antigen binding domain, where
present, is fused at
the C-terminus of the Fab heavy chain to the N-terminus of the second subunit
of the Fc
domain.
[0128] In particular embodiments, the Fc domain comprises a modification
promoting the
association of the first and the second subunit of the Fc domain, for example,
as described in
Section 5.1.
[0129] In some embodiments, the Fc domain comprises one or more amino acid
substitutions
that reduces binding to an Fc receptor and/or effector function, for example
as described in
Section 5.1.
[0130] In a particular embodiment the bispecific antibody comprises
(i) a first antigen binding domain that specifically binds to CD3, wherein the
first antigen
binding domain is a crossover Fab molecule wherein either the variable or the
constant regions,
particularly the variable regions, of the Fab light chain and the Fab heavy
chain are exchanged;
(ii) a second and a third antigen binding domain that specifically bind to
glyco-M UC1,
comprising a heavy chain variable region comprising the heavy chain CDR-H1 of
SEQ ID NO:
5, the CDR-H2 of SEQ ID NO: 6, and the CDR-H3 of SEQ ID NO: 7; and a light
chain variable
region comprising the light chain CDR-L1 of SEQ ID NO: 8, the CDR-L2 of SEQ ID
NO: 9 and
the CDR-L3 of SEQ ID NO:10, wherein the second and third antigen binding
domain are each a
Fab molecule, particularly a conventional Fab molecule;
(iii) an Fc domain composed of a first and a second subunit capable of stable
association,
wherein the second antigen binding domain is fused at the C-terminus of the
Fab heavy
chain to the N-terminus of the Fab heavy chain of the first antigen binding
domain, and the first
antigen binding domain is fused at the C-terminus of the Fab heavy chain to
the N-terminus of
the first subunit of the Fc domain, and wherein the third antigen binding
domain is fused at the
C-terminus of the Fab heavy chain to the N-terminus of the second subunit of
the Fc domain.
31
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
[0131] In one embodiment the first antigen binding domain comprises a heavy
chain variable
region comprising the heavy chain CDR-H1 of SEQ ID NO:34, the CDR-H2 of SEQ ID
NO:35,
and the CDR-H3 of SEQ ID NO:36; and a light chain variable region comprising
the light chain
CDR-L1 of SEQ ID NO:37, the CDR-L2 of SEQ ID NO:38 and the CDR-L3 of SEQ ID
NO:39.
[0132] In one embodiment, the first antigen binding domain comprises a heavy
chain variable
region sequence that is at least about 95%, 96%, 97%, 98%, 99% or 100%
identical to the
amino acid sequence of SEQ ID NO:40 and a light chain variable region sequence
that is at
least about 95%, 96%, 97%, 98%, 99% or 100% identical to the amino acid
sequence of SEQ
ID NO:41.
[0133] In one embodiment, the first antigen binding domain comprises the heavy
chain variable
region sequence of SEQ ID NO:40 and the light chain variable region sequence
of SEQ ID
NO:41.
[0134] In one embodiment, the second and third antigen binding domain comprise
a heavy
chain variable region sequence that is at least about 95%, 96%, 97%, 98%, 99%
or 100%
identical to the amino acid sequence of SEQ ID NO:3 and a light chain variable
region
sequence that is at least about 95%, 96%, 97%, 98%, 99% or 100% identical to
the amino acid
sequence of SEQ ID NO:4. Preferably, the antigen binding domain comprises CDRs
comprising
the amino acid sequences of any of the CDR combinations set forth in numbered
embodiments
3 to 17. In one embodiment, the second and third antigen binding domains
comprise the heavy
chain variable region of SEQ ID NO:3 and the light chain variable region of
SEQ ID NO:4.
[0135] The Fc domain according to the above embodiments may incorporate,
singly or in
combination, all of the features described hereinabove in relation to Fc
domains.
[0136] In some embodiments, the antigen binding domains and the Fc region are
fused to each
other by peptide linkers, for example by peptide linkers as in SEQ ID NO:45
and SEQ ID
NO:46.
[0137] In one embodiment, in the constant domain CL of the second and the
third Fab
molecule under (ii) the amino acid at position 124 is substituted by lysine
(K) (numbering
according to Kabat) and the amino acid at position 123 is substituted by
lysine (K) or arginine
(R), particularly by arginine (R) (numbering according to Kabat), and in the
constant domain
CH1 of the second and the third Fab molecule under (ii) the amino acid at
position 147 is
substituted by glutamic acid (E) (numbering according to Kabat EU index) and
the amino acid
at position 213 is substituted by glutamic acid (E) (numbering according to
Kabat EU index).
[0138] In one embodiment, the bispecific antibody comprises a polypeptide
comprising a
sequence that is at least 80%, 85%, 90%, 95%, 96%, 97%, 98%, or 99% identical
to the
sequence of SEQ ID NO:43 (and preferably comprises a CDR-L1 comprising the
amino acid
sequence of SEQ ID NO: 8, a CDR-L2 comprising the amino acid sequence of SEQ
ID NO:9,
32
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
and a CDR-L3 comprising the amino acid sequence of SEQ ID NO:31), a
polypeptide
comprising a sequence that is at least 80%, 85%, 90%, 95%, 96%, 97%, 98%, or
99% identical
to the sequence of SEQ ID NO:44 (and preferably comprises the CD3 heavy and
light chain
CDR sequences set forth in Table 4), a polypeptide comprising a sequence that
is at least 80%,
85%, 90%, 95%, 96%, 97%, 98%, or 99% identical to the sequence of SEQ ID NO:45
(and
preferably comprises a CDR-H1 comprising the amino acid sequence of SEQ ID
NO:33, a
CDR-H2 comprising the amino acid sequence of SEQ ID NO:29, a CDR-H3 comprising
the
amino acid sequence of SEQ ID NO:25), and a polypeptide comprising a sequence
that is at
least 80%, 85%, 90%, 95%, 96%, 97%, 98%, or 99% identical to the sequence of
SEQ ID
NO:46 (and preferably comprises a CDR-H1 comprising the amino acid sequence of
SEQ ID
NO:33, a CDR-H2 comprising the amino acid sequence of SEQ ID NO:29, a CDR-H3
comprising the amino acid sequence of SEQ ID NO:25, a CDR-L1 comprising the
amino acid
sequence of SEQ ID NO:37, a CDR-L2 comprising the amino acid sequence of SEQ
ID NO:38,
and a CDR-L3 comprising the amino acid sequence of SEQ ID NO:39).
[0139] In one embodiment, the bispecific antibody comprises a polypeptide
(particularly two
polypeptides) comprising the sequence of SEQ ID NO:43, a polypeptide
comprising the
sequence of SEQ ID NO:44, a polypeptide comprising the sequence of SEQ ID
NO:45, and a
polypeptide comprising the sequence of SEQ ID NO:46.
5.3 Antibody-Drug Conjugates
[0140] Another aspect of the disclosure concerns antibody drug conjugates
(ADCs) including
the anti-glyco-MUC1 antibodies and antigen-binding fragments of the
disclosure. The ADCs
generally comprise an anti-glyco-M UC1 antibody and/or binding fragment as
described herein
having one or more cytotoxic and/or cytostatic agents linked thereto by way of
one or more
linkers. In specific embodiments, the ADCs are compounds according to
structural formula (I):
[D-L-XY]n-Ab
or salts thereof, where each "D" represents, independently of the others, a
cytotoxic and/or
cytostatic agent ("drug"); each "L" represents, independently of the others, a
linker; "Ab"
represents an anti-glyco-MUC1 antigen binding domain, such as an anti-glyco-
MUC1 antibody
or binding fragment described herein; each "XY" represents a linkage formed
between a
functional group Rx on the linker and a "complementary" functional group RY on
the antibody,
and n represents the number of drugs linked to, or drug-to-antibody ratio
(DAR), of the ADC.
[0141] Specific embodiments of the various antibodies (Ab) that can comprise
the ADCs
include the various embodiments of anti-glyco-MUC1 antibodies and/or binding
fragments
described above.
[0142] In some specific embodiments of the ADCs and/or salts of structural
formula (I), each D
is the same and/or each L is the same.
33
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
[0143] Specific embodiments of cytotoxic and/or cytostatic agents (D) and
linkers (L) that can
comprise the anti-glyco-M UC1 ADCs of the disclosure, as well as the number of
cytotoxic
and/or cytostatic agents linked to the ADCs, are described in more detail
below.
5.3.1. Cytotoxic and/or Cytostatic Agents
[0144] The cytotoxic and/or cytostatic agents may be any agents known to
inhibit the growth
and/or replication of and/or kill cells, and in particular cancer and/or tumor
cells. Numerous
agents having cytotoxic and/or cytostatic properties are known in the
literature. Non-limiting
examples of classes of cytotoxic and/or cytostatic agents include, by way of
example and not
limitation, radionuclides, alkylating agents, topoisomerase I inhibitors,
topoisomerase II
inhibitors, DNA intercalating agents (e.g., groove binding agents such as
minor groove binders),
RNA/DNA antimetabolites, cell cycle modulators, kinase inhibitors, protein
synthesis inhibitors,
histone deacetylase inhibitors, mitochondria inhibitors, and antimitotic
agents.
[0145] Specific non-limiting examples of agents within certain of these
various classes are
provided below.
[0146] Alkylating Agents: asaley ((L-Leucine, N-[N-acetyl-4-[bis-(2-
chloroethyl)amino]-DL-
phenylalany1]-, ethylester; NSC 167780; CAS Registry No. 3577897)); AZQ ((1,4-
cyclohexadiene-1,4-dicarbamic acid, 2,5-bis(1-aziridinyI)-3,6-dioxo-, diethyl
ester; NSC 182986;
CAS Registry No. 57998682)); BCNU ((N,N'-Bis(2-chloroethyl)-N-nitrosourea; NSC
409962;
CAS Registry No. 154938)); busulfan (1,4-butanediol dimethanesulfonate; NSC
750; CAS
Registry No. 55981); (carboxyphthalato)platinum (NSC 27164; CAS Registry No.
65296813);
CBDCA ((cis-(1,1-cyclobutanedicarboxylato)diammineplatinum(II)); NSC 241240;
CAS Registry
No. 41575944)); CCNU ((N-(2-chloroethyl)-N'-cyclohexyl-N-nitrosourea; NSC
79037; CAS
Registry No. 13010474)); CHIP (iproplatin; NSC 256927); chlorambucil (NSC
3088; CAS
Registry No. 305033); chlorozotocin ((2-[[[(2-chloroethyl)
nitrosoamino]carbonyl]amino]-2-
deoxy-D-glucopyranose; NSC 178248; CAS Registry No. 54749905)); cis-platinum
(cisplatin;
NSC 119875; CAS Registry No. 15663271); clomesone (NSC 338947; CAS Registry
No.
88343720); cyanomorpholinodoxorubicin (NCS 357704; CAS Registry No. 88254073);
cyclodisone (NSC 348948; CAS Registry No. 99591738); dianhydrogalactitol (5,6-
diepoxydulcitol; NSC 132313; CAS Registry No. 23261203); fluorodopan ((5-[(2-
chloroethyl)-(2-
fluoroethyl)amino]-6-methyl-uracil; NSC 73754; CAS Registry No. 834913);
hepsulfam (NSC
329680; CAS Registry No. 96892578); hycanthone (NSC 142982; CAS Registry No.
23255938); melphalan (NSC 8806; CAS Registry No. 3223072); methyl CCNU ((1-(2-
chloroethyl)-3-(trans-4-methylcyclohexane)-1-nitrosourea; NSC 95441;
13909096); mitomycin
C (NSC 26980; CAS Registry No. 50077); mitozolamide (NSC 353451; CAS Registry
No.
85622953); nitrogen mustard ((bis(2-chloroethyl)methylamine hydrochloride; NSC
762; CAS
Registry No. 55867); PCNU ((1-(2-chloroethyl)-3-(2,6-dioxo-3-piperidy1)-1-
nitrosourea; NSC
95466; CAS Registry No. 13909029)); piperazine alkylator ((1-(2-chloroethyl)-4-
(3-
34
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
chloropropyI)-piperazine dihydrochloride; NSC 344007)); piperazinedione (NSC
135758; CAS
Registry No. 41109802); pipobroman ((N,N-bis(3-bromopropionyl) piperazine; NSC
25154;
CAS Registry No. 54911)); porfiromycin (N-methylmitomycin C; NSC 56410; CAS
Registry No.
801525); spirohydantoin mustard (NSC 172112; CAS Registry No. 56605164);
teroxirone
(triglycidylisocyanurate; NSC 296934; CAS Registry No. 2451629); tetraplatin
(NSC 363812;
CAS Registry No. 62816982); thio-tepa (N,N',N"-tri-1,2-ethanediyIthio
phosphoramide; NSC
6396; CAS Registry No. 52244); triethylenemelamine (NSC 9706; CAS Registry No.
51183);
uracil nitrogen mustard (desmethyldopan; NSC 34462; CAS Registry No. 66751);
Yoshi-864
((bis(3-mesyloxy propyl)amine hydrochloride; NSC 102627; CAS Registry No.
3458228).
[0147] Topoisomerase 1 Inhibitors: camptothecin (NSC 94600; CAS Registry No.
7689-03-4);
various camptothecin derivatives and analogs (for example, NSC 100880, NSC
603071, NSC
107124, NSC 643833, NSC 629971, NSC 295500, NSC 249910, NSC 606985, NSC 74028,
NSC 176323, NSC 295501, NSC 606172, NSC 606173, NSC 610458, NSC 618939, NSC
610457, NSC 610459, NSC 606499, NSC 610456, NSC 364830, and NSC 606497);
morpholinisoxorubicin (NSC 354646; CAS Registry No. 89196043); SN-38 (NSC
673596; CAS
Registry No. 86639-52-3).
[0148] Topoisomerase 11 Inhibitors: doxorubicin (NSC 123127; CAS Registry No.
25316409);
amonafide (benzisoquinolinedione; NSC 308847; CAS Registry No. 69408817); m-
AMSA ((4'-
(9-acridinylamino)-3'-methoxymethanesulfonanilide; NSC 249992; CAS Registry
No.
51264143)); anthrapyrazole derivative ((NSC 355644); etoposide (VP-16; NSC
141540; CAS
Registry No. 33419420); pyrazoloacridine ((pyrazolo[3,4,5-kl]acridine-2(6H)-
propanamine, 9-
methoxy-N, N-dimethy1-5-nitro-, monomethanesulfonate; NSC 366140; CAS Registry
No.
99009219); bisantrene hydrochloride (NSC 337766; CAS Registry No. 71439684);
daunorubicin (NSC 821151; CAS Registry No. 23541506); deoxydoxorubicin (NSC
267469;
CAS Registry No. 63950061); mitoxantrone (NSC 301739; CAS Registry No.
70476823);
menogaril (NSC 269148; CAS Registry No. 71628961); N,N-dibenzyl daunomycin
(NSC
268242; CAS Registry No. 70878512); oxanthrazole (NSC 349174; CAS Registry No.
105118125); rubidazone (NSC 164011; CAS Registry No. 36508711); teniposide (VM-
26; NSC
122819; CAS Registry No. 29767202).
[0149] DNA Intercalating Agents: anthramycin (CAS Registry No. 4803274);
chicamycin A
(CAS Registry No. 89675376); tomaymycin (CAS Registry No. 35050556); DC-81
(CAS
Registry No. 81307246); sibiromycin (CAS Registry No. 12684332);
pyrrolobenzodiazepine
derivative (CAS Registry No. 945490095); SGD-1882 ((S)-2-(4-aminophenyI)-7-
methoxy-8-(3-
4(S)-7-methoxy-2-(4-methoxypheny1)-- 5-oxo-5,11a-dihydro-1H-
benzo[e]pyrrolo[1,2-
a][1,4]diazepin-8-yl)oxy)propox- y)-1H-benzo[e]pyrrolo[1,2-a][1,4]diazepin-
5(11aH)-one);
SG2000 (SJG-136; (11aS,11a'S)-8,8'-(propane-1,3-diyIbis(oxy))bis(7-methoxy-2-
methylene-
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
2,3- -dihydro-1H-benzo[e]pyrrolo[1,2-a][1,4]diazepin-5(11aH)-one); NSC 694501;
CAS Registry
No. 232931576).
[0150] RNA/DNA Antimetabolites: L-alanosine (NSC 153353; CAS Registry No.
59163416); 5-
azacytidine (NSC 102816; CAS Registry No. 320672); 5-fluorouracil (NSC 19893;
CAS
Registry No. 51218); acivicin (NSC 163501; CAS Registry No. 42228922);
aminopterin
derivative N-[2-chloro-5-[[(2,4-diamino-5-methy1-6-
quinazolinyl)methyl]amino]benzoyl- ]L-
aspartic acid (NSC 132483); aminopterin derivative N-[4-[[(2,4-diamino-5-ethy1-
6-
quinazolinyl)methyl]amino]benzoyl]L-asparti- c acid (NSC 184692); aminopterin
derivative N-[2-
chloro-4-[[(2,4-diamino-6-pteridinyl)methyl]amino]benzoyl]L-aspartic acid
monohydrate (NSC
134033); an antifo ((N -(4-amino-4-deoxypteroyI)-N7-hemiphthaloyl-L-ornithin-
e; NSC
623017)); Baker's soluble antifol (NSC 139105; CAS Registry No. 41191042);
dichlorallyl
lawsone ((2-(3,3-dichloroallyI)-3-hydroxy-1,4-naphthoquinone; NSC 126771; CAS
Registry No.
36417160); brequinar (NSC 368390; CAS Registry No. 96201886); ftorafur ((pro-
drug; 5-fluoro-
1-(tetrahydro-2-fury1)-uracil; NSC 148958; CAS Registry No. 37076689); 5,6-
dihydro-5-
azacytidine (NSC 264880; CAS Registry No. 62402317); methotrexate (NSC 740;
CAS
Registry No. 59052); methotrexate derivative (N-[[4-[[(2,4-diamino-6-
pteridinyl)methyl]methylamino]-1-naphthalenyl]car- bonyl]L-glutamic acid; NSC
174121); PALA
((N-(phosphonoacetyI)-L-aspartate; NSC 224131; CAS Registry No. 603425565);
pyrazofurin
(NSC 143095; CAS Registry No. 30868305); trimetrexate (NSC 352122; CAS
Registry No.
82952645).
[0151] DNA Antimetabolites: 3-HP (NSC 95678; CAS Registry No. 3814797); 2'-
deoxy-5-
fluorouridine (NSC 27640; CAS Registry No. 50919); 5-HP (NSC 107392; CAS
Registry No.
19494894); a-TGDR (a-2'-deoxy-6-thioguanosine; NSC 71851 CAS Registry No.
2133815);
aphidicolin glycinate (NSC 303812; CAS Registry No. 92802822); ara C (cytosine
arabinoside;
NSC 63878; CAS Registry No. 69749); 5-aza-2'-deoxycytidine (NSC 127716; CAS
Registry No.
2353335); 13-TGDR ([3-2'-deoxy-6-thioguanosine; NSC 71261; CAS Registry No.
789617);
cyclocytidine (NSC 145668; CAS Registry No. 10212256); guanazole (NSC 1895;
CAS
Registry No. 1455772); hydroxyurea (NSC 32065; CAS Registry No. 127071);
inosine
glycodialdehyde (NSC 118994; CAS Registry No. 23590990); macbecin 11 (NSC
330500; CAS
Registry No. 73341738); pyrazoloimidazole (NSC 51143; CAS Registry No.
6714290);
thioguanine (NSC 752; CAS Registry No. 154427); thiopurine (NSC 755; CAS
Registry No.
50442).
[0152] Cell Cycle Modulators: silibinin (CAS Registry No. 22888-70-6);
epigallocatechin gallate
(EGCG; CAS Registry No. 989515); procyanidin derivatives (e.g., procyanidin Al
[CAS
Registry No. 103883030], procyanidin B1 [CAS Registry No. 20315257],
procyanidin B4 [CAS
Registry No. 29106512], arecatannin B1 [CAS Registry No. 79763283]);
isoflavones (e.g.,
genistein [4%5,7-trihydroxyisoflavone; CAS Registry No. 446720], daidzein [4,7-
36
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
dihydroxyisoflavone, CAS Registry No. 486668]; indole-3-carbinol (CAS Registry
No. 700061);
quercetin (NSC 9219; CAS Registry No. 117395); estramustine (NSC 89201; CAS
Registry No.
2998574); nocodazole (CAS Registry No. 31430189); podophyllotoxin (CAS
Registry No.
518285); vinorelbine tartrate (NSC 608210; CAS Registry No. 125317397);
cryptophycin (NSC
667642; CAS Registry No. 124689652).
[0153] Kinase Inhibitors: afatinib (CAS Registry No. 850140726); axitinib (CAS
Registry No.
319460850); ARRY-438162 (binimetinib) (CAS Registry No. 606143899); bosutinib
(CAS
Registry No. 380843754); cabozantinib (CAS Registry No. 1140909483); ceritinib
(CAS
Registry No. 1032900256); crizotinib (CAS Registry No. 877399525); dabrafenib
(CAS Registry
No. 1195765457); dasatinib (NSC 732517; CAS Registry No. 302962498); erlotinib
(NSC
718781; CAS Registry No. 183319699); everolimus (NSC 733504; CAS Registry No.
159351696); fostamatinib (NSC 745942; CAS Registry No. 901119355); gefitinib
(NSC 715055;
CAS Registry No. 184475352); ibrutinib (CAS Registry No. 936563961); imatinib
(NSC 716051;
CAS Registry No. 220127571); lapatinib (CAS Registry No. 388082788);
lenvatinib (CAS
Registry No. 857890392); mubritinib (CAS 366017096); nilotinib (CAS Registry
No.
923288953); nintedanib (CAS Registry No. 656247175); palbociclib (CAS Registry
No.
571190302); pazopanib (NSC 737754; CAS Registry No. 635702646); pegaptanib
(CAS
Registry No. 222716861); ponatinib (CAS Registry No. 1114544318); rapamycin
(NSC 226080;
CAS Registry No. 53123889); regorafenib (CAS Registry No. 755037037); AP 23573
(ridaforolimus) (CAS Registry No. 572924540); INCB018424 (ruxolitinib) (CAS
Registry No.
1092939177); ARRY-142886 (selumetinib) (NSC 741078; CAS Registry No. 606143-52-
6);
sirolimus (NSC 226080; CAS Registry No. 53123889); sorafenib (NSC 724772; CAS
Registry
No. 475207591); sunitinib (NSC 736511; CAS Registry No. 341031547);
tofacitinib (CAS
Registry No. 477600752); temsirolimus (NSC 683864; CAS Registry No.
163635043);
trametinib (CAS Registry No. 871700173); vandetanib (CAS Registry No.
443913733);
vemurafenib (CAS Registry No. 918504651); SU6656 (CAS Registry No. 330161870);
CEP-
701 (lesaurtinib) (CAS Registry No. 111358884); XL019 (CAS Registry No.
945755566); PD-
325901 (CAS Registry No. 391210109); PD-98059 (CAS Registry No. 167869218);
ATP-
competitive TORC1/TORC2 inhibitors including PI-103 (CAS Registry No.
371935749), PP242
(CAS Registry No. 1092351671), PP30 (CAS Registry No. 1092788094), Torin 1
(CAS Registry
No. 1222998368), LY294002 (CAS Registry No. 154447366), XL-147 (CAS Registry
No.
934526893), CAL-120 (CAS Registry No. 870281348), ETP-45658 (CAS Registry No.
1198357797), PX 866 (CAS Registry No. 502632668), GDC-0941 (CAS Registry No.
957054307), BGT226 (CAS Registry No. 1245537681), BEZ235 (CAS Registry No.
915019657), XL-765 (CAS Registry No. 934493762).
[0154] Protein Synthesis Inhibitors: acriflavine (CAS Registry No. 65589700);
amikacin (NSC
177001; CAS Registry No. 39831555); arbekacin (CAS Registry No. 51025855);
astromicin
37
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
(CAS Registry No. 55779061); azithromycin (NSC 643732; CAS Registry No.
83905015);
bekanamycin (CAS Registry No. 4696768); chlortetracycline (NSC 13252; CAS
Registry No.
64722); clarithromycin (NSC 643733; CAS Registry No. 81103119); clindamycin
(CAS Registry
No. 18323449); clomocycline (CAS Registry No. 1181540); cycloheximide (CAS
Registry No.
66819); dactinomycin (NSC 3053; CAS Registry No. 50760); dalfopristin (CAS
Registry No.
112362502); demeclocycline (CAS Registry No. 127333); dibekacin (CAS Registry
No.
34493986); dihydrostreptomycin (CAS Registry No. 128461); dirithromycin (CAS
Registry No.
62013041); doxycycline (CAS Registry No. 17086281); emetine (NSC 33669; CAS
Registry No.
483181); erythromycin (NSC 55929; CAS Registry No. 114078); flurithromycin
(CAS Registry
No. 83664208); framycetin (neomycin B; CAS Registry No. 119040); gentamycin
(NSC 82261;
CAS Registry No. 1403663); glycylcyclines, such as tigecycline (CAS Registry
No. 220620097);
hygromycin B (CAS Registry No. 31282049); isepamicin (CAS Registry No.
67814760);
josamycin (NSC 122223; CAS Registry No. 16846245); kanamycin (CAS Registry No.
8063078); ketolides such as telithromycin (CAS Registry No. 191114484),
cethromycin (CAS
Registry No. 205110481), and solithromycin (CAS Registry No. 760981837);
lincomycin (CAS
Registry No. 154212); lymecycline (CAS Registry No. 992212); meclocycline (NSC
78502; CAS
Registry No. 2013583); metacycline (rondomycin; NSC 356463; CAS Registry No.
914001);
midecamycin (CAS Registry No. 35457808); minocycline (NSC 141993; CAS Registry
No.
10118908); miocamycin (CAS Registry No. 55881077); neomycin (CAS Registry No.
119040);
netilmicin (CAS Registry No. 56391561); oleandomycin (CAS Registry No.
3922905);
oxazolidinones, such as eperezolid (CAS Registry No. 165800044), linezolid
(CAS Registry No.
165800033), posizolid (CAS Registry No. 252260029), radezolid (CAS Registry
No.
869884786), ranbezolid (CAS Registry No. 392659380), sutezolid (CAS Registry
No.
168828588), tedizolid (CAS Registry No. 856867555); oxytetracycline (NSC 9169;
CAS
Registry No. 2058460); paromomycin (CAS Registry No. 7542372); penimepicycline
(CAS
Registry No. 4599604); peptidyl transferase inhibitors, e.g., chloramphenicol
(NSC 3069; CAS
Registry No. 56757) and derivatives such as azidamfenicol (CAS Registry No.
13838089),
florfenicol (CAS Registry No. 73231342), and thiamphenicol (CAS Registry No.
15318453), and
pleuromutilins such as retapamulin (CAS Registry No. 224452668), tiamulin (CAS
Registry No.
55297955), valnemulin (CAS Registry No. 101312929); pirlimycin (CAS Registry
No.
79548735); puromycin (NSC 3055; CAS Registry No. 53792); quinupristin (CAS
Registry No.
120138503); ribostamycin (CAS Registry No. 53797356); rokitamycin (CAS
Registry No.
74014510); rolitetracycline (CAS Registry No. 751973); roxithromycin (CAS
Registry No.
80214831); sisomicin (CAS Registry No. 32385118); spectinomycin (CAS Registry
No.
1695778); spiramycin (CAS Registry No. 8025818); streptogramins such as
pristinamycin (CAS
Registry No. 270076603), quinupristin/dalfopristin (CAS Registry No.
126602899), and
virginiamycin (CAS Registry No. 11006761); streptomycin (CAS Registry No.
57921);
tetracycline (NSC 108579; CAS Registry No. 60548); tobramycin (CAS Registry
No.
38
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
32986564); troleandomycin (CAS Registry No. 2751099); tylosin (CAS Registry
No. 1401690);
verdamicin (CAS Registry No. 49863481).
[0155] Histone Deacetylase Inhibitors: abexinostat (CAS Registry No.
783355602); belinostat
(NSC 726630; CAS Registry No. 414864009); chidamide (CAS Registry No.
743420022);
entinostat (CAS Registry No. 209783802); givinostat (CAS Registry No.
732302997);
mocetinostat (CAS Registry No. 726169739); panobinostat (CAS Registry No.
404950807);
quisinostat (CAS Registry No. 875320299); resminostat (CAS Registry No.
864814880);
romidepsin (CAS Registry No. 128517077); sulforaphane (CAS Registry No.
4478937);
thioureidobutyronitrile (Kevetrin TM ; CAS Registry No. 6659890); valproic
acid (NSC 93819; CAS
Registry No. 99661); vorinostat (NSC 701852; CAS Registry No. 149647789); ACY-
1215
(rocilinostat; CAS Registry No. 1316214524); CUDC-101 (CAS Registry No.
1012054599);
CHR-2845 (tefinostat; CAS Registry No. 914382608); CHR-3996 (CAS Registry No.
1235859138); 4SC-202 (CAS Registry No. 910462430); 0G200745 (CAS Registry No.
936221339); SB939 (pracinostat; CAS Registry No. 929016966).
[0156] Mitochondria Inhibitors: pancratistatin (NSC 349156; CAS Registry No.
96281311);
rhodamine-123 (CAS Registry No. 63669709); edelfosine (NSC 324368; CAS
Registry No.
70641519); d-alpha-tocopherol succinate (NSC 173849; CAS Registry No.
4345033);
compound 1113 (CAS Registry No. 865070377); aspirin (NSC 406186; CAS Registry
No.
50782); ellipticine (CAS Registry No. 519233); berberine (CAS Registry No.
633658); cerulenin
(CAS Registry No. 17397896); GX015-070 (Obatoclax0; 1H-Indole, 2-(24(3,5-
dimethy1-1H-
pyrrol-2-Amethylene)-3-methoxy-2H-pyrrol-5-y1)-; NSC 729280; CAS Registry No.
803712676);
celastrol (tripterine; CAS Registry No. 34157830); metformin (NSC 91485; CAS
Registry No.
1115704); Brilliant green (NSC 5011; CAS Registry No. 633034); ME-344 (CAS
Registry No.
1374524556).
[0157] Antimitotic Agents: allocolchicine (NSC 406042); auristatins, such as
MMAE
(monomethyl auristatin E; CAS Registry No. 474645-27-7) and MMAF (monomethyl
auristatin
F; CAS Registry No. 745017-94-1; halichondrin B (NSC 609395); colchicine (NSC
757; CAS
Registry No. 64868); cholchicine derivative (N-benzoyl-deacetyl benzamide; NSC
33410; CAS
Registry No. 63989753); dolastatin 10 (NSC 376128; CAS Registry No 110417-88-
4);
maytansine (NSC 153858; CAS Registry No. 35846-53-8); rhozoxin (NSC 332598;
CAS
Registry No. 90996546); taxol (NSC 125973; CAS Registry No. 33069624); taxol
derivative ((2'-
N43-(dimethylamino)propyl]glutaramate taxol; NSC 608832); thiocolchicine (3-
demethylthiocolchicine; NSC 361792); trityl cysteine (NSC 49842; CAS Registry
No. 2799077);
vinblastine sulfate (NSC 49842; CAS Registry No. 143679); vincristine sulfate
(NSC 67574;
CAS Registry No. 2068782).
[0158] Any of these agents that include or that may be modified to include a
site of attachment
to an antibody may be included in the ADCs disclosed herein.
39
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
[0159] In a specific embodiment, the cytotoxic and/or cytostatic agent is an
antimitotic agent.
[0160] In another specific embodiment, the cytotoxic and/or cytostatic agent
is an auristatin, for
example, monomethyl auristatin E ("MMAE") or monomethyl auristatin F ("MMAF").
5.3.2. Linkers
[0161] In the anti-glyco-MUC1 ADCs of the disclosure, the cytotoxic and/or
cytostatic agents
are linked to the antibody by way of linkers. The linker linking a cytotoxic
and/or cytostatic agent
to the antibody of an ADC may be short, long, hydrophobic, hydrophilic,
flexible or rigid, or may
be composed of segments that each independently have one or more of the above-
mentioned
properties such that the linker may include segments having different
properties. The linkers
may be polyvalent such that they covalently link more than one agent to a
single site on the
antibody, or monovalent such that covalently they link a single agent to a
single site on the
antibody.
[0162] As will be appreciated by skilled artisans, the linkers link cytotoxic
and/or cytostatic
agents to the antibody by forming a covalent linkage to the cytotoxic and/or
cytostatic agent at
one location and a covalent linkage to antibody at another. The covalent
linkages are formed by
reaction between functional groups on the linker and functional groups on the
agents and
antibody. As used herein, the expression "linker" is intended to include (i)
unconjugated forms
of the linker that include a functional group capable of covalently linking
the linker to a cytotoxic
and/or cytostatic agent and a functional group capable of covalently linking
the linker to an
antibody; (ii) partially conjugated forms of the linker that includes a
functional group capable of
covalently linking the linker to an antibody and that is covalently linked to
a cytotoxic and/or
cytostatic agent, or vice versa; and (iii) fully conjugated forms of the
linker that is covalently
linked to both a cytotoxic and/or cytostatic agent and an antibody. In some
specific
embodiments of linkers and anti-glyco-M UC1 ADCs of the disclosure, as well as
synthons used
to conjugate linker-agents to antibodies, moieties comprising the functional
groups on the linker
and covalent linkages formed between the linker and antibody are specifically
illustrated as R,
and XY, respectively.
[0163] The linkers are preferably, but need not be, chemically stable to
conditions outside the
cell, and may be designed to cleave, immolate and/or otherwise specifically
degrade inside the
cell. Alternatively, linkers that are not designed to specifically cleave or
degrade inside the cell
may be used. Choice of stable versus unstable linker may depend upon the
toxicity of the
cytotoxic and/or cytostatic agent. For agents that are toxic to normal cells,
stable linkers are
preferred. Agents that are selective or targeted and have lower toxicity to
normal cells may
utilize, chemical stability of the linker to the extracellular milieu is less
important. A wide variety
of linkers useful for linking drugs to antibodies in the context of ADCs are
known in the art. Any
of these linkers, as well as other linkers, may be used to link the cytotoxic
and/or cytostatic
agents to the antibody of the anti-glyco-MUC1 ADCs of the disclosure.
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
[0164] Exemplary polyvalent linkers that may be used to link many cytotoxic
and/or cytostatic
agents to a single antibody molecule are described, for example, in WO
2009/073445; WO
2010/068795; WO 2010/138719; WO 2011/120053; WO 2011/171020; WO 2013/096901;
WO
2014/008375; WO 2014/093379; WO 2014/093394; WO 2014/093640, the content of
which are
incorporated herein by reference in their entireties. For example, the
Fleximer linker technology
developed by Mersana et al. has the potential to enable high-DAR ADCs with
good
physicochemical properties. As shown below, the Mersana technology is based on
incorporating drug molecules into a solubilizing poly-acetal backbone via a
sequence of ester
bonds. The methodology renders highly-loaded ADCs (DAR up to 20) while
maintaining good
physicochemical properties.
[0165] Additional examples of dendritic type linkers can be found in US
2006/116422; US
2005/271615; de Groot etal. (2003) Angew. Chem. Int. Ed. 42:4490-4494; Amir
etal. (2003)
Angew. Chem. Int. Ed. 42:4494-4499; Shamis et al.(2004) J. Am. Chem. Soc.
126:1726-1731;
Sun et al.(2002) Bioorganic & Medicinal Chemistry Letters 12:2213-2215; Sun et
al.(2003)
Bioorganic & Medicinal Chemistry 11:1761-1768; King et al.(2002) Tetrahedron
Letters
43:1987-1990, each of which is incorporated herein by reference.
[0166] Exemplary monovalent linkers that may be used are described, for
example, in Nolting,
2013, Antibody-Drug Conjugates, Methods in Molecular Biology 1045:71-100;
Kitson etal.,
2013, CROs/CM0s--Chemica Oggi¨Chemistry Today 31(4):30-38; Ducry etal., 2010,
Bioconjugate Chem. 21:5-13; Zhao etal., 2011, J. Med. Chem. 54:3606-3623; U.S.
Pat. No.
7,223,837; U.S. Pat. No. 8,568,728; U.S. Pat. No. 8,535,678; and W02004010957,
each of
which is incorporated herein by reference.
[0167] By way of example and not limitation, some cleavable and noncleavable
linkers that
may be included in the anti-glyco-M UC1 ADCs of the disclosure are described
below.
5.3.3. Cleavable Linkers
[0168] In certain embodiments, the linker selected is cleavable in vivo.
Cleavable linkers may
include chemically or enzymatically unstable or degradable linkages. Cleavable
linkers
generally rely on processes inside the cell to liberate the drug, such as
reduction in the
cytoplasm, exposure to acidic conditions in the lysosome, or cleavage by
specific proteases or
other enzymes within the cell. Cleavable linkers generally incorporate one or
more chemical
bonds that are either chemically or enzymatically cleavable while the
remainder of the linker is
noncleavable. In certain embodiments, a linker comprises a chemically labile
group such as
hydrazone and/or disulfide groups. Linkers comprising chemically labile groups
exploit
differential properties between the plasma and some cytoplasmic compartments.
The
intracellular conditions to facilitate drug release for hydrazone containing
linkers are the acidic
environment of endosomes and lysosomes, while the disulfide containing linkers
are reduced in
the cytosol, which contains high thiol concentrations, e.g., glutathione. In
certain embodiments,
41
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
the plasma stability of a linker comprising a chemically labile group may be
increased by
introducing steric hindrance using substituents near the chemically labile
group.
[0169] Acid-labile groups, such as hydrazone, remain intact during systemic
circulation in the
blood's neutral pH environment (pH 7.3-7.5) and undergo hydrolysis and release
the drug once
the ADC is internalized into mildly acidic endosomal (pH 5.0-6.5) and
lysosomal (pH 4.5-5.0)
compartments of the cell. This pH dependent release mechanism has been
associated with
nonspecific release of the drug. To increase the stability of the hydrazone
group of the linker,
the linker may be varied by chemical modification, e.g., substitution,
allowing tuning to achieve
more efficient release in the lysosome with a minimized loss in circulation.
[0170] Hydrazone-containing linkers may contain additional cleavage sites,
such as additional
acid-labile cleavage sites and/or enzymatically labile cleavage sites. ADCs
including exemplary
hydrazone-containing linkers include the following structures:
0 (Ig)
N N Ab
0
-n
0 (Ih)
N N
S ____________________________________________________________ Ab
0
0 - n
(Ti)
DZ
H 3C N¨Ab
- n
0
wherein D and Ab represent the cytotoxic and/or cytostatic agent (drug) and
Ab, respectively,
and n represents the number of drug-linkers linked to the antibody. In certain
linkers such as
linker (Ig), the linker comprises two cleavable groups--a disulfide and a
hydrazone moiety. For
such linkers, effective release of the unmodified free drug requires acidic pH
or disulfide
reduction and acidic pH. Linkers such as (1h) and (Ii) have been shown to be
effective with a
single hydrazone cleavage site.
42
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
[0171] Additional linkers which remain intact during systemic circulation and
undergo hydrolysis
and release the drug when the ADC is internalized into acidic cellular
compartments include
carbonates. Such linkers can be useful in cases where the cytotoxic and/or
cytostatic agent can
be covalently attached through an oxygen.
[0172] Other acid-labile groups that may be included in linkers include cis-
aconityl-containing
linkers. cis-Aconityl chemistry uses a carboxylic acid juxtaposed to an amide
bond to accelerate
amide hydrolysis under acidic conditions.
[0173] Cleavable linkers may also include a disulfide group. Disulfides are
thermodynamically
stable at physiological pH and are designed to release the drug upon
internalization inside
cells, wherein the cytosol provides a significantly more reducing environment
compared to the
extracellular environment. Scission of disulfide bonds generally requires the
presence of a
cytoplasmic thiol cofactor, such as (reduced) glutathione (GSH), such that
disulfide-containing
linkers are reasonably stable in circulation, selectively releasing the drug
in the cytosol. The
intracellular enzyme protein disulfide isomerase, or similar enzymes capable
of cleaving
disulfide bonds, may also contribute to the preferential cleavage of disulfide
bonds inside cells.
GSH is reported to be present in cells in the concentration range of 0.5-10 mM
compared with a
significantly lower concentration of GSH or cysteine, the most abundant low-
molecular weight
thiol, in circulation at approximately 5 Tumor cells, where irregular blood
flow leads to a hypoxic
state, result in enhanced activity of reductive enzymes and therefore even
higher glutathione
concentrations. In certain embodiments, the in vivo stability of a disulfide-
containing linker may
be enhanced by chemical modification of the linker, e.g., use of steric
hindrance adjacent to the
disulfide bond.
[0174] ADCs including exemplary disulfide-containing linkers include the
following structures:
43
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
R R
DS N¨Ab
R R 0 - n
(Ik)
__________________________________________________________ Ab
(I1)
R R
DS
S¨Ab
- n
wherein D and Ab represent the drug and antibody, respectively, n represents
the number of
drug-linkers linked to the antibody and R is independently selected at each
occurrence from
hydrogen or alkyl, for example. In certain embodiments, increasing steric
hindrance adjacent to
the disulfide bond increases the stability of the linker. Structures such as
(ID and (II) show
increased in vivo stability when one or more R groups is selected from a lower
alkyl such as
methyl.
[0175] Another type of cleavable linker that may be used is a linker that is
specifically cleaved
by an enzyme. Such linkers are typically peptide-based or include peptidic
regions that act as
substrates for enzymes. Peptide based linkers tend to be more stable in plasma
and
extracellular milieu than chemically labile linkers. Peptide bonds generally
have good serum
stability, as lysosomal proteolytic enzymes have very low activity in blood
due to endogenous
inhibitors and the unfavorably high pH value of blood compared to lysosomes.
Release of a
drug from an antibody occurs specifically due to the action of lysosomal
proteases, e.g.,
cathepsin and plasmin. These proteases may be present at elevated levels in
certain tumor
cells.
[0176] In exemplary embodiments, the cleavable peptide is selected from
tetrapeptides such as
Gly-Phe-Leu-Gly (SEQ ID NO:128), Ala-Leu-Ala-Leu (SEQ ID NO:129) or dipeptides
such as
Val-Cit, Val-Ala, Met-(D)Lys, Asn-(D)Lys, Val-(D)Asp, Phe-Lys, Ile-Val, Asp-
Val, His-Val,
NorVal-(D)Asp, Ala-(D)Asp 5, Met-Lys, Asn-Lys, Ile-Pro, Me3Lys-Pro, PhenylGly-
(D)Lys, Met-
(D)Lys, Asn-(D)Lys, Pro-(D)Lys, Met-(D)Lys, Asn-(D)Lys, AM Met-(D)Lys, Asn-
(D)Lys, AW Met-
44
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
(D)Lys, and Asn-(D)Lys. In certain embodiments, dipeptides are preferred over
longer
polypeptides due to hydrophobicity of the longer peptides.
[0177] A variety of dipeptide-based cleavable linkers useful for linking drugs
such as
doxorubicin, mitomycin, camptothecin, pyrrolobenzodiazepine, tallysomycin and
auristatin/auristatin family members to antibodies have been described (see,
Dubowchik etal.,
1998, J. Org. Chem. 67:1866-1872; Dubowchik etal., 1998, Bioorg. Med. Chem.
Lett.
8(21):3341-3346; Walker etal., 2002, Bioorg. Med. Chem. Lett. 12:217-219;
Walker etal.,
2004, Bioorg. Med. Chem. Lett. 14:4323-4327; Sutherland etal., 2013, Blood
122: 1455-1463;
and Francisco etal., 2003, Blood 102:1458-1465, of each of which is
incorporated herein by
reference). All of these dipeptide linkers, or modified versions of these
dipeptide linkers, may be
used in the anti-glyco-MUC1 ADCs of the disclosure. Other dipeptide linkers
that may be used
include those found in ADCs such as Seattle Genetics' Brentuximab Vendotin SGN-
35
(AdcetrisTm), Seattle Genetics SG N-75 (anti-CD-70, Val-Cit-monomethyl
auristatin F(MMAF),
Seattle Genetics SGN-CD33A (anti-CD-33, Val-Ala-(SGD-1882)), Celldex
Therapeutics
glembatumumab (CDX-011) (anti-NMB, Val-Cit-monomethyl auristatin E (MMAE), and
Cytogen
PSMA-ADC (PSMA-ADC-1301) (anti-PSMA, Val-Cit-MMAE).
[0178] Enzymatically cleavable linkers may include a self-immolative spacer to
spatially
separate the drug from the site of enzymatic cleavage. The direct attachment
of a drug to a
peptide linker can result in proteolytic release of an amino acid adduct of
the drug, thereby
impairing its activity. The use of a self-immolative spacer allows for the
elimination of the fully
active, chemically unmodified drug upon amide bond hydrolysis.
[0179] One self-immolative spacer is the bifunctional para-aminobenzyl alcohol
group, which is
linked to the peptide through the amino group, forming an amide bond, while
amine containing
drugs may be attached through carbamate functionalities to the benzylic
hydroxyl group of the
linker (PABC). The resulting prodrugs are activated upon protease-mediated
cleavage, leading
to a 1,6-elimination reaction releasing the unmodified drug, carbon dioxide,
and remnants of the
linker group. The following scheme depicts the fragmentation of p-amidobenzyl
ether and
release of the drug:
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
0
0
protease
0 X
peptide
0
1,6-elimination
0
________________________________________________________________ 7/0
H2N X¨D
+CO2
HN
wherein X-D represents the unmodified drug.
[0180] Heterocyclic variants of this self-immolative group have also been
described. See for
example, U.S. Pat. No. 7,989,434, incorporated herein by reference.
[0181] In some embodiments, the enzymatically cleavable linker is a p-
glucuronic acid-based
linker. Facile release of the drug may be realized through cleavage of the p-
glucuronide
glycosidic bond by the lysosomal enzyme p-glucuronidase. This enzyme is
present abundantly
within lysosomes and is overexpressed in some tumor types, while the enzyme
activity outside
cells is low. 13-Glucuronic acid-based linkers may be used to circumvent the
tendency of an
ADC to undergo aggregation due to the hydrophilic nature of p-glucuronides. In
some
embodiments, p-glucuronic acid-based linkers are preferred as linkers for ADCs
linked to
hydrophobic drugs. The following scheme depicts the release of the drug from
and ADC
containing a p-glucuronic acid-based linker:
46
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
HO
0
HO 13-glucuronidase
HO:)
HO 0
0 0
HO
HN
Ab 0
HO
0OH
OH
0
ID 1,6-elimination
j
HO 0
HN
Ab
0
101
0 +CO2
HN
Ab
0
[0182] A variety of cleavable p-glucuronic acid-based linkers useful for
linking drugs such as
auristatins, camptothecin and doxorubicin analogues, CBI minor-groove binders,
and
psymberin to antibodies have been described (see, see Nolting, Chapter 5
"Linker Technology
in Antibody-Drug Conjugates," In: Antibody-Drug Conjugates: Methods in
Molecular Biology,
vol. 1045, pp. 71-100, Laurent Ducry (Ed.), Springer Science & Business
Medica, LLC, 2013;
Jeffrey etal., 2006, Bioconjug. Chem. 17:831-840; Jeffrey etal., 2007, Bioorg.
Med. Chem.
Lett. 17:2278-2280; and Jiang etal., 2005, J. Am. Chem. Soc. 127:11254-11255,
each of which
is incorporated herein by reference). All of these p-glucuronic acid-based
linkers may be used
in the anti-glyco-MUC1 ADCs of the disclosure.
[0183] Additionally, cytotoxic and/or cytostatic agents containing a phenol
group can be
covalently bonded to a linker through the phenolic oxygen. One such linker,
described in WO
2007/089149, relies on a methodology in which a diamino-ethane "SpaceLink" is
used in
conjunction with traditional "PABO"-based self-immolative groups to deliver
phenols. The
47
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
cleavage of the linker is depicted schematically below, where D represents a
cytotoxic and/or
cytostatic agent having a phenolic hydroxyl group.
representative
HO 0 linker with
PABO unit
0 "SpaceLink"
HO 0 o lysosomal
enzyme
OH
N \/C)D
0
0
0
to mAb
________________________________________ )110. HO¨D
HN D
0
SpaceLink's ultimate
> _______________________________________ 0 fate is a cyclic urea
[0184] Cleavable linkers may include noncleavable portions or segments, and/or
cleavable
segments or portions may be included in an otherwise non-cleavable linker to
render it
cleavable. By way of example only, polyethylene glycol (PEG) and related
polymers may
include cleavable groups in the polymer backbone. For example, a polyethylene
glycol or
polymer linker may include one or more cleavable groups such as a disulfide, a
hydrazone or a
dipeptide.
[0185] Other degradable linkages that may be included in linkers include ester
linkages formed
by the reaction of PEG carboxylic acids or activated PEG carboxylic acids with
alcohol groups
on a biologically active agent, wherein such ester groups generally hydrolyze
under
physiological conditions to release the biologically active agent.
Hydrolytically degradable
linkages include, but are not limited to, carbonate linkages; imine linkages
resulting from
reaction of an amine and an aldehyde; phosphate ester linkages formed by
reacting an alcohol
with a phosphate group; acetal linkages that are the reaction product of an
aldehyde and an
alcohol; orthoester linkages that are the reaction product of a formate and an
alcohol; and
48
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
oligonucleotide linkages formed by a phosphoramidite group, including but not
limited to, at the
end of a polymer, and a 5' hydroxyl group of an oligonucleotide.
[0186] In certain embodiments, the linker comprises an enzymatically cleavable
peptide moiety,
for example, a linker comprising structural formula (IVa) or (IVb):
0
(IVa)
_ _
)(0
0
Ra
N peptide _ _ x
0
_ Y
0
(IVb)
)(0
0
N peptide
Ra
or a salt thereof, wherein: peptide represents a peptide (illustrated C¨>N1
and not showing the
carboxy and amino "termini") cleavable by a lysosomal enzyme; T represents a
polymer
comprising one or more ethylene glycol units or an alkylene chain, or
combinations thereof; Ra
is selected from hydrogen, alkyl, sulfonate and methyl sulfonate; p is an
integer ranging from 0
to 5; q is 0 or 1; x is 0 or 1; y is 0 or 1;
represents the point of attachment of the linker to a
cytotoxic and/or cytostatic agent; and * represents the point of attachment to
the remainder of
the linker.
[0187] In certain embodiments, the peptide is selected from a tripeptide or a
dipeptide. In
particular embodiments, the dipeptide is selected from: Val-Cit; Cit-Val; Ala-
Ala; Ala-Cit; Cit-Ala;
Asn-Cit; Cit-Asn; Cit-Cit; Val-Glu; Glu-Val; Ser-Cit; Cit-Ser; Lys-Cit; Cit-
Lys; Asp-Cit; Cit-Asp;
Ala-Val; Val-Ala; Phe-Lys; Val-Lys; Ala-Lys; Phe-Cit; Leu-Cit; Ile-Cit; Phe-
Arg; and Trp-Cit. In
certain embodiments, the dipeptide is selected from: Cit-Val; and Ala-Val.
[0188] Specific exemplary embodiments of linkers according to structural
formula (IVa) that
may be included in the anti-glyco-MUC1 ADCs of the disclosure include the
linkers illustrated
below (as illustrated, the linkers include a group suitable for covalently
linking the linker to an
antibody):
49
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
(IVa.1)
0
o 0 0 0
. H
NN 0 ONNN 1.1
\
H H
0 H
0
HN/
H2 N /.0
(IVa.2)
0
0 0 0 0
. H
NN 0 0NNyN 101
\
&
H H
0 H
0
\..----
(IVa.3)
0
0 0 0 C)
H H
___NCNNNCN
\ 0 H
0 H
0 SO3
(IVa.4)
0
o 0 0 (7)
H
N
H H H
0
0
(IVa.5)
0
0 0 o
I H
Cl.,õ,.....,......-^.......v=-...,..,......,N..,-",...,.....,,,N N
H H H
0
NH2
N
H
(IVa.6)
0 0
0
0
H H
BrNNNCN
0 H 0 H
NH
,õ===..zz.
H2N 0
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
(IVa.7)
o
o o
_ o o))'
E"r
N
H H H
0
NH2
N0
H
[0189] Specific exemplary embodiments of linkers according to structural
formula (IVb) that
may be included in the anti-glyco-MUC1 ADCs of the disclosure include the
linkers illustrated
below (as illustrated, the linkers include a group suitable for covalently
linking the linker to an
antibody):
(IVb.1)
o
0 0 0 0)'
H
___NCNNN
\ H
0 7.....,, H
0
NH
0 NH2
0
(IVb.2)
0
0
V 0 0
. H
N,.....,......õ..--..õ.............õ.õ,-..õ,.N.,A,..N
N
H H
0 0
HN
H2N 0
(IVb.3)
o
0
c-- o _
H
N,..,..,...õ.õ--...,,,-..............N,,,;,õ,.........,.N...,..c
N
H H
0 0
(IVb.4)
o
o 0 0 0
H
____I\NXrNIN
\ H H
0
0
NH
=-"N",
0 NH2
51
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
(IVb.5)
NH2 0
0 0 xo
0 0.'-=-=-=.-.X
H
\ H _ H
0
0
NH
0NH2
(IVb.6)
0
0 0 0 0
H
NC=N N.,,_,,,N
\ H E H
0
0
H2N, 0
HN
(IVb.7)
0
0 0 cp
1 NN,)EN
C.L H i H
0
0
NH
0%NH2
(IVb.8)
0
0
cr 0 0
H 0
N,,,,,,,.,,,,,s,,,.--,,,,.,,,.--,,, .F1
0 0
0 OH
(IVb.9)
0
0 OH
0 f 0 0
H
cr1N Nõ,.s,õ-^=õ,N
H i H
-
0 0
NH
0%NH2
52
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
NH2
(IVb.10)
0
0
O''''...A
V
(FIN.,....õ.õ.... N
H A H
0 0
-.....õ
NH
0 NH2
(IVb.11)
0
0
0
0 H
NCNN 1 H E H
HO-g=0 0
ll
0
-..,,
NH
0 NH2
(IVb.12)
0
0
0 H
yjrNN
H
HO-S=0 0
ll
0
-.õ...
NH
0 NH2
(IVb.13)
OH 0
0
H
H E
i H
0 0
NH
0 NH2
53
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
(IVb.14)
0
0
0 H 0
V 0
N.,,,,,,,,,,,,,N
N
H H
0 0
HN/
H2N/c)
\/
(IVb.15)
o
p 0 E 0 0
Sif ? H
/1
N
0 H H
0
NH
0 NH2
\/
(IVb.16)
0
0 0 0 0
\___LICN
H H
0 ''..(N
0
0 SO3
NH
0N H2
(IVb.17)
0
0
c---1( 0 0 c)
0 N N
H ' H
0 0
NH
0 NH2
54
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
NH2
(IVb.18)
0
0 0
N
10rH N NH
0 /7
0 el
0
0
(IVb.19)
0
0
c-
0!.V NfXr Fc(
_
0 0 E
[0190] In certain embodiments, the linker comprises an enzymatically cleavable
peptide moiety,
for example, a linker comprising structural formula (IVc) or (IVd):
(IVc)
_ _
0 0 Ra
T' N
peptide _ _x
0
-y
0 (IVd)
?.22(peptide
Ra
or a salt thereof, wherein: peptide represents a peptide (illustrated C¨>N1
and not showing the
carboxy and amino "termini") cleavable by a lysosomal enzyme; T represents a
polymer
comprising one or more ethylene glycol units or an alkylene chain, or
combinations thereof; Ra
is selected from hydrogen, alkyl, sulfonate and methyl sulfonate; p is an
integer ranging from 0
to 5; q is 0 or 1; x is 0 or 1; y is 0 or 1; .x represents the point of
attachment of the linker to a
cytotoxic and/or cytostatic agent; and * represents the point of attachment to
the remainder of
the linker.
[0191] Specific exemplary embodiments of linkers according to structural
formula (IVc) that
may be included in the anti-glyco-M UC1 ADCs of the disclosure include the
linkers illustrated
below (as illustrated, the linkers include a group suitable for covalently
linking the linker to an
antibody):
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
(IVC. 1)
o o o 0
H
\
H H
0
0
HN/
H 2N 0
(IVc.2)
0 0 0 0
\
H H
0 =
0
(IVc.3)
(IVc.4)
0 0 0 0 0
H H
H
...,i(.....-........................N _ NX....,,,Nj../
Cl.........õ),......... ,.........................õ......j.õ
\ 0 .,...,2 H 0 E H H
0 E
0 SO3
(IVc.5) (IVc.6)
0 0 0 )CH 0 0
Cl......,......,L H
N
H H Br N
H
NH2 0 0
N 0
H NH
0 NH2
(IVc.7)
0 0 0
H
I N .NXN1)4
H H
0 . . _. . . . . .
. = 7
NH2
'`,, N . = - = ' "\\,0
H
[0192] Specific exemplary embodiments of linkers according to structural
formula (IVd) that
may be included in the anti-glyco-MUC1 ADCs of the disclosure include the
linkers illustrated
below (as illustrated, the linkers include a group suitable for covalently
linking the linker to an
antibody):
(IVd.1) c(jo 0 (IVd.2)
0 0 0 0
_...NcINII N
\ H
0 7..,.... 0 H
0 ,...,..õ
0
H/
NH N
..---.
0 NH2 H2N 0
56
CA 03078812 2020-04-08
WO 2019/083506
PCT/US2017/058036
(IVd.4)
(IVd.3) 0 0
0 0 0 0
c( NX
__.NC/;'1
H
H
0 0 0 \
0
NH
0 NH2
NH2
( (IVd.5)
IVd.6)
0 0 0 õ.......0 0 0 0
NH
N
\ H 0 0 0 0(,1.
NH
N H2
(IVd.8)
H2N (IVd.7)
0 0
0 0
.....,,,
c( H
HN N \NXN
H
0 0
0
1 H N
N
0 OH
HE
0
0
NH
NH2
(IVd.9)
(IVd.10)
0 r,,OH 0 0
0 0 0
cr,,..õ----.c.../ .µ"=.-/.---.../ c(../ \.----"\/ N '''(.='-' NH '''''''---
)4
H
H
0 0 0 z0
N
NH H
57
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
o
(IVd.11) c(0
(IVd.12)
c(N
O 0 0
0 r LIsi
0
YI\X)\114
N
1 H
HO -g=0 0
HO-S=0 H
0
ll II
0 0
NH
NH
ONH2
0 NH2
0
0 (IVd.13) 0
(IVd.14)
0
V 0 OH 0
H
V H 0
N,...................õN N..............,...)4 NN v. '
N
H
0 0 0 0
H/
NH N
0,.NH2 /.
H2N 0
(IVd.15)
(IVd.16)
0 0
8 N
....c...................õ...,...........õN........(1.,.N.......).............õ,
N.....õ
0 H
O \ 0
0 SO3-
NH NH
0N H2 0 NH2
0
(IVd.17)
c"( 0
H 0
Nõ,......,,...õ,-........0õ,..---0,...........X.,.....õ,,N,......)4
H
0 0
NH
0NH2
[0193] In certain embodiments, the linker comprising structural formula (IVa),
(IVb), (IVc), or
(IVd) further comprises a carbonate moiety cleavable by exposure to an acidic
medium. In
particular embodiments, the linker is attached through an oxygen to a
cytotoxic and/or
cytostatic agent.
5.3.4. Non-Cleavable Linkers
[0194] Although cleavable linkers may provide certain advantages, the linkers
comprising the
anti-glyco-MUC1 ADC of the disclosure need not be cleavable. For noncleavable
linkers, the
release of drug does not depend on the differential properties between the
plasma and some
cytoplasmic compartments. The release of the drug is postulated to occur after
internalization of
the ADC via antigen-mediated endocytosis and delivery to lysosomal
compartment, where the
58
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
antibody is degraded to the level of amino acids through intracellular
proteolytic degradation.
This process releases a drug derivative, which is formed by the drug, the
linker, and the amino
acid residue to which the linker was covalently attached. The amino acid drug
metabolites from
conjugates with noncleavable linkers are more hydrophilic and generally less
membrane
permeable, which leads to less bystander effects and less nonspecific
toxicities compared to
conjugates with a cleavable linker. In general, ADCs with noncleavable linkers
have greater
stability in circulation than ADCs with cleavable linkers. Non-cleavable
linkers may be alkylene
chains, or maybe polymeric in natures, such as, for example, based upon
polyalkylene glycol
polymers, amide polymers, or may include segments of alkylene chains,
polyalkylene glocols
and/or amide polymers.
[0195] A variety of non-cleavable linkers used to link drugs to antibodies
have been described.
See, Jeffrey etal., 2006, Bioconjug. Chem. 17; 831-840; Jeffrey etal., 2007,
Bioorg. Med.
Chem. Lett. 17:2278-2280; and Jiang etal., 2005, J. Am. Chem. Soc. 127:11254-
11255, each
of which is incorporated herein by reference. All of these linkers may be
included in the anti-
glyco-MUC1 ADCs of the disclosure.
[0196] In certain embodiments, the linker is non-cleavable in vivo, for
example a linker
according to structural formula (Via), (Vlb), (Vic) or (VId) (as illustrated,
the linkers include a
group suitable for covalently linking the linker to an antibody:
(Via)
0 0
0-7
0 0 (Vic) 0
(VId)
Rx
Ra
or salts thereof, wherein: Ra is selected from hydrogen, alkyl, sulfonate and
methyl sulfonate; Rx
is a moiety including a functional group capable of covalently linking the
linker to an antibody;
and represents the point of attachment of the linker to a cytotoxic and/or
cytostatic agent.
[0197] Specific exemplary embodiments of linkers according to structural
formula (V1a)-(VId)
that may be included in the anti-glyco-MUC1 ADCs of the disclosure include the
linkers
59
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
illustrated below (as illustrated, the linkers include a group suitable for
covalently linking the
linker to an antibody, and ' represents the point of attachment to a cytotoxic
and/or cytostatic
agent):
(Via)
0 0
0-7
0 0
(V1a.1)
0
(Vl
0 N N
0
c.1
(VIc.2)
0 0
CI
0 0
(VId.1)
(VId.2)
N
0
SO3H 0
id .3)
0
0
%
S,
0
5.3.5. Groups Used to Attach Linkers to Antibodies
[0198] A variety of groups may be used to attach linker-drug synthons to
antibodies to yield
ADCs. Attachment groups can be electrophilic in nature and include: maleimide
groups,
activated disulfides, active esters such as NHS esters and HOBt esters,
haloformates, acid
halides, alkyl and benzyl halides such as haloacetamides. As discussed below,
there are also
emerging technologies related to "self-stabilizing" maleimides and "bridging
disulfides" that can
be used in accordance with the disclosure. The specific group used will
depend, in part, on the
site of attachment to the antibody.
[0199] One example of a "self-stabilizing" maleimide group that hydrolyzes
spontaneously
under antibody conjugation conditions to give an ADC species with improved
stability is
depicted in the schematic below. See U520130309256 Al; also Lyon etal., Nature
Biotech
published online, doi:10.1038/nbt.2968.
CA 03078812 2020-04-08
WO 2019/083506
PCT/US2017/058036
Normal system:
\ > __ NH
\ /
TAb
/ ________________________ :71n%Fi
S 0
mAb /
I 0
/ ______________ /
------ plasma
facile
protein
Ow-
0
N_....k_
-------
)
0 NH
0
______________________________________________ /
-----(
......."1 N /
0
N
mAb H
0
S /
N ____________
-------
0
0
/ > ____ NH
------N
0
Pro .... ej( /
0
Leads to "DAR loss" over time
61
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
SGN MaIDPR (maleimido dipropylamino) system:
/ 0
mAb
NH
mAb-SH 0 0\ L711õ
NH
spontaneous at
pH 7.4
OA-
__________________________________________________________________________ OP-
0 H2N
II-1)
US20130309256A1
mAb
0 0
NH
stable in plasma
4 HN _____________________________ (retro hetero-Michael
reaction shown above slow)
OH H2N
[0200] Polytherics has disclosed a method for bridging a pair of sulfhydryl
groups derived from
reduction of a native hinge disulfide bond. See, Badescu etal., 2014,
Bioconjugate Chem.
25:1124-1136. The reaction is depicted in the schematic below. An advantage of
this
methodology is the ability to synthesize enriched DAR4 ADCs by full reduction
of IgGs (to give
4 pairs of sulfhydryls) followed by reaction with 4 equivalents of the
alkylating agent. ADCs
containing "bridged disulfides" are also claimed to have increased stability.
02s NA
in situ
elimination
SO2 0
62
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
... ......... ....
.== .
0¨s¨s-
0
reduce
disulfide c
- 0¨SH HS-
() ;
i
. SH -
0
9
N)1-,
H
N)11.
ArO2S H
0
0
_ -
: s
NX.
,
I
, H
41(t S
"bridged disulfide"
[0201] Similarly, as depicted below, a maleimide derivative (1, below) that is
capable of
bridging a pair of sulfhydryl groups has been developed. See W02013/085925.
9 õ
0
\s
s_r_i(N
0
_)õ...
s- 1 s ,
N5 0
---....õ
5.3.6. Linker Selection Considerations
[0202] As is known by skilled artisans, the linker selected for a particular
ADC may be
influenced by a variety of factors, including but not limited to, the site of
attachment to the
antibody (e.g., lys, cys or other amino acid residues), structural constraints
of the drug
pharmacophore and the lipophilicity of the drug. The specific linker selected
for an ADC should
seek to balance these different factors for the specific antibody/drug
combination. For a review
of the factors that are influenced by choice of linkers in ADCs, see Nolting,
Chapter 5 "Linker
Technology in Antibody-Drug Conjugates," In: Antibody-Drug Conjugates: Methods
in Molecular
Biology, vol. 1045, pp. 71-100, Laurent Ducry (Ed.), Springer Science &
Business Medica, LLC,
2013.
63
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
[0203] For example, ADCs have been observed to effect killing of bystander
antigen-negative
cells present in the vicinity of the antigen-positive tumor cells. The
mechanism of bystander cell
killing by ADCs has indicated that metabolic products formed during
intracellular processing of
the ADCs may play a role. Neutral cytotoxic metabolites generated by
metabolism of the ADCs
in antigen-positive cells appear to play a role in bystander cell killing
while charged metabolites
may be prevented from diffusing across the membrane into the medium and
therefore cannot
affect bystander killing. In certain embodiments, the linker is selected to
attenuate the
bystander killing effect caused by cellular metabolites of the ADC. In certain
embodiments, the
linker is selected to increase the bystander killing effect.
[0204] The properties of the linker may also impact aggregation of the ADC
under conditions of
use and/or storage. Typically, ADCs reported in the literature contain no more
than 3-4 drug
molecules per antibody molecule (see, e.g., Chari, 2008, Acc Chem Res 41:98-
107). Attempts
to obtain higher drug-to-antibody ratios ("DAR") often failed, particularly if
both the drug and the
linker were hydrophobic, due to aggregation of the ADC (King etal., 2002, J
Med Chem
45:4336-4343; Hollander etal., 2008, Bioconjugate Chem 19:358-361; Burke
etal., 2009
Bioconjugate Chem 20:1242-1250). In many instances, DARs higher than 3-4 could
be
beneficial as a means of increasing potency. In instances where the cytotoxic
and/or cytostatic
agent is hydrophobic in nature, it may be desirable to select linkers that are
relatively
hydrophilic as a means of reducing ADC aggregation, especially in instances
where DARS
greater than 3-4 are desired. Thus, in certain embodiments, the linker
incorporates chemical
moieties that reduce aggregation of the ADCs during storage and/or use. A
linker may
incorporate polar or hydrophilic groups such as charged groups or groups that
become charged
under physiological pH to reduce the aggregation of the ADCs. For example, a
linker may
incorporate charged groups such as salts or groups that deprotonate, e.g.,
carboxylates, or
protonate, e.g., amines, at physiological pH.
[0205] Exemplary polyvalent linkers that have been reported to yield DARs as
high as 20 that
may be used to link numerous cytotoxic and/or cytostatic agents to an antibody
are described in
WO 2009/073445; WO 2010/068795; WO 2010/138719; WO 2011/120053; WO
2011/171020;
WO 2013/096901; WO 2014/008375; WO 2014/093379; WO 2014/093394; WO
2014/093640,
the content of which are incorporated herein by reference in their entireties.
[0206] In particular embodiments, the aggregation of the ADCs during storage
or use is less
than about 10% as determined by size-exclusion chromatography (SEC). In
particular
embodiments, the aggregation of the ADCs during storage or use is less than
10%, such as
less than about 5%, less than about 4%, less than about 3%, less than about
2%, less than
about 1%, less than about 0.5%, less than about 0.1%, or even lower, as
determined by size-
exclusion chromatography (SEC).
64
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
5.3.7. METHODS OF MAKING ANTI-GLYCO-MUC1 ADCs
[0207] The anti-glyco-MUC1 ADCs of the disclosure may be synthesized using
chemistries that
are well-known. The chemistries selected will depend upon, among other things,
the identity of
the cytotoxic and/or cytostatic agent(s), the linker and the groups used to
attach linker to the
antibody. Generally, ADCs according to formula (I) may be prepared according
to the following
scheme:
D-L-Rx+Ab-RY-41D-L-XY]n-Ab (I)
[0208] where D, L, Ab, XY and n are as previously defined, and Rx and RY
represent
complementary groups capable of forming a covalent linkages with one another,
as discussed
above.
[0209] The identities of groups Rx and RY will depend upon the chemistry used
to link synthon
D-L- Rx to the antibody. Generally, the chemistry used should not alter the
integrity of the
antibody, for example its ability to bind its target. Preferably, the binding
properties of the
conjugated antibody will closely resemble those of the unconjugated antibody.
A variety of
chemistries and techniques for conjugating molecules to biological molecules
such as
antibodies are known in the art and in particular to antibodies, are well-
known. See, e.g., Amon
et al., "Monoclonal Antibodies For lmmunotargeting Of Drugs In Cancer
Therapy," in:
Monoclonal Antibodies And Cancer Therapy, Reisfeld etal. Eds., Alan R. Liss,
Inc., 1985;
Hellstrom etal., "Antibodies For Drug Delivery," in: Controlled Drug Delivery,
Robinson etal.
Eds., Marcel Dekker, Inc., 2nd Ed. 1987; Thorpe, "Antibody Carriers Of
Cytotoxic Agents In
Cancer Therapy: A Review," in: Monoclonal Antibodies '84: Biological And
Clinical Applications,
Pinchera etal., Eds., 1985; "Analysis, Results, and Future Prospective of the
Therapeutic Use
of Radiolabeled Antibody In Cancer Therapy," in: Monoclonal Antibodies For
Cancer Detection
And Therapy, Baldwin etal., Eds., Academic Press, 1985; Thorpe etal., 1982,
lmmunol. Rev.
62:119-58; PCT publication WO 89/12624. Any of these chemistries may be used
to link the
synthons to an antibody.
[0210] A number of functional groups Rx and chemistries useful for linking
synthons to
accessible lysine residues are known, and include by way of example and not
limitation NHS-
esters and isothiocyanates.
[0211] A number of functional groups Rx and chemistries useful for linking
synthons to
accessible free sulfhydryl groups of cysteine residues are known, and include
by way of
example and not limitation haloacetyls and maleimides.
[0212] However, conjugation chemistries are not limited to available side
chain groups. Side
chains such as amines may be converted to other useful groups, such as
hydroxyls, by linking
an appropriate small molecule to the amine. This strategy can be used to
increase the number
of available linking sites on the antibody by conjugating multifunctional
small molecules to side
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
chains of accessible amino acid residues of the antibody. Functional groups Rx
suitable for
covalently linking the synthons to these "converted" functional groups are
then included in the
synthons.
[0213] The antibody may also be engineered to include amino acid residues for
conjugation.
An approach for engineering antibodies to include non-genetically encoded
amino acid residues
useful for conjugating drugs in the context of ADCs is described by Axup
etal., 2012, Proc Natl
Acad Sci USA. 109(40):16101-16106, as are chemistries and functional group
useful for linking
synthons to the non-encoded amino acids.
[0214] Typically, the synthons are linked to the side chains of amino acid
residues of the
antibody, including, for example, the primary amino group of accessible lysine
residues or the
sulfhydryl group of accessible cysteine residues. Free sulfhydryl groups may
be obtained by
reducing interchain disulfide bonds.
[0215] For linkages where RY is a sulfhydryl group (for example, when Rx is a
maleimide), the
antibody is generally first fully or partially reduced to disrupt interchain
disulfide bridges
between cysteine residues.
[0216] Cysteine residues that do not participate in disulfide bridges may
engineered into an
antibody by mutation of one or more codons. Reducing these unpaired cysteines
yields a
sulfhydryl group suitable for conjugation. Preferred positions for
incorporating engineered
cysteines include, by way of example and not limitation, positions S1120,
S1130, A1140,
S1150, A1760, 51800, S2520, V2860, V2920, S3570, A3590, S3980, S4280 (Kabat
numbering) on the human IgGi heavy chain and positions V1100, S1140, S1210,
S1270,
S1680, V2050 (Kabat numbering) on the human Ig kappa light chain (see, e.g.,
U.S. Pat. No.
7,521,541, U.S. Pat. No. 7,855,275 and U.S. Pat. No. 8,455,622).
[0217] As will appreciated by skilled artisans, the number of cytotoxic and/or
cytostatic agents
linked to an antibody molecule may vary, such that a collection of ADCs may be
heterogeneous
in nature, where some antibodies contain one linked agent, some two, some
three, etc. (and
some none). The degree of heterogeneity will depend upon, among other things,
the
chemistries used for linking the cytotoxic and/or cytostatic agents. For
example, where the
antibodies are reduced to yield sulfhydryl groups for attachment,
heterogeneous mixtures of
antibodies having zero, 2, 4, 6 or 8 linked agents per molecule are often
produced.
Furthermore, by limiting the molar ratio of attachment compound, antibodies
having zero, 1, 2,
3, 4, 5, 6, 7 or 8 linked agents per molecule are often produced. Thus, it
will be understood that
depending upon context, stated DARs may be averages for a collection of
antibodies. For
example, "DAR4" can refer to an ADC preparation that has not been subjected to
purification to
isolate specific DAR peaks and can comprise a heterogeneous mixture of ADC
molecules
having different numbers of cytostatic and/or cytotoxic agents attached per
antibody (e.g., 0, 2,
4, 6, 8 agents per antibody), but has an average drug-to-antibody ratio of 4.
Similarly, in some
66
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
embodiments, "DAR2" refers to a heterogeneous ADC preparation in which the
average drug-
to-antibody ratio is 2.
[0218] When enriched preparations are desired, antibodies having defined
numbers of linked
cytotoxic and/or cytostatic agents may be obtained via purification of
heterogeneous mixtures,
for example, via column chromatography, e.g., hydrophobic interaction
chromatography.
[0219] Purity may be assessed by a variety of methods, as is known in the art.
As a specific
example, an ADC preparation may be analyzed via HPLC or other chromatography
and the
purity assessed by analyzing areas under the curves of the resultant peaks.
5.4 Chimeric Antigen Receptors
[0220] The present disclosure provides chimeric antigen receptors (CARs)
comprising the anti-
glyco-MUC1 antibodies or antigen-binding fragments described herein.
[0221] The CARs of the disclosure typically comprise an extracellular domain
operably linked
to a transmembrane domain which is in turn operably linked to an intracellular
domain for
signaling.
[0222] The extracellular domains of the CARs of the disclosure comprise the
sequence of an
anti-glyco-M UC1 antibody or antigen-binding fragment (e.g., as described in
Section 5.1 or
embodiments 1 to 90).
[0223] Exemplary transmembrane domain sequence and intracellular domain
sequences are
described in Section 5.4.1 and 5.4.2, respectively.
[0224] Several fusion proteins described herein (e.g., embodiments 92 and 94-
96) are CARs,
and the CAR-related disclosures apply to such fusion proteins.
5.4.1. Transmembrane Domain
[0225] VVith respect to the transmembrane domain, the CAR can be designed to
comprise a
transmembrane domain that is operably linked (e.g., fused) to the
extracellular domain of the
CAR.
[0226] The transmembrane domain may be derived either from a natural or from a
synthetic
source. Where the source is natural, the domain may be derived from any
membrane-bound or
transmembrane protein. Transmembrane regions of particular use in this
disclosure may be
derived from (i.e., comprise at least the transmembrane region(s) of) the
alpha, beta or zeta
chain of the T-cell receptor, 0D28, 0D3 epsilon, 0D45, 0D4, 0D5, 0D8, 0D9,
0D16, 0D22,
0D33, 0D37, 0D64, 0D80, 0D86, 0D134, 0D137, 0D154. In some instances, a
variety of
human hinges can be employed as well including the human Ig (immunoglobulin)
hinge.
[0227] In one embodiment, the transmembrane domain is synthetic (i.e., non-
naturally
occurring). Examples of synthetic transmembrane domains are peptides
comprising
predominantly hydrophobic residues such as leucine and valine. Preferably a
triplet of
67
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
phenylalanine, tryptophan and valine will be found at each end of a synthetic
transmembrane
domain. Optionally, a short oligo- or polypeptide linker, preferably between 2
and 10 amino
acids in length may form the linkage between the transmembrane domain and the
cytoplasmic
signaling domain of the CAR. A glycine-serine doublet provides a particularly
suitable linker.
[0228] In one embodiment, the transmembrane domain in the CAR of the
disclosure is the CD8
transmembrane domain. In one embodiment, the CD8 transmembrane domain
comprises the
amino acid sequence YLHLGALGRDLWGPSPVTGYHPLL.
[0229] In one embodiment, the transmembrane domain in the CAR of the
disclosure is the
0D28 transmembrane domain. In one embodiment, the 0D28 transmembrane domain
comprises the amino acid sequence FVVVLVVVGGVLACYSLLVTVAFIIFVVV.
[0230] In some instances, the transmembrane domain of the CAR of the
disclosure comprises
the CD8a hinge domain. In one embodiment, the CD8a hinge domain comprises the
amino acid
sequence TTTPAPRPPTPAPTIASQPLSLRPEACRPAAGGAVHTRGLDFAC.
5.4.2. Intracellular Domain
[0231] The intracellular signaling domain of the CAR of the disclosure is
responsible for
activation of at least one of the normal effector functions of the immune cell
in which the CAR is
expressed. The term "effector function" refers to a specialized function of a
cell. Effector
function of a T cell, for example, may be cytolytic activity or helper
activity including the
secretion of cytokines. Thus the term "intracellular signaling domain" refers
to the portion of a
protein which transduces the effector function signal and directs the cell to
perform a
specialized function. While usually the entire intracellular signaling domain
can be employed, in
many cases it is not necessary to use the entire chain. To the extent that a
truncated portion of
the intracellular signaling domain is used, such truncated portion may be used
in place of the
intact chain as long as it transduces the effector function signal. The term
intracellular signaling
domain is thus meant to include any truncated portion of the intracellular
signaling domain
sufficient to transduce the effector function signal.
[0232] Preferred examples of intracellular signaling domains for use in the
CAR of the
disclosure include the cytoplasmic sequences of the T cell receptor (TCR) and
co-receptors
that act in concert to initiate signal transduction following antigen receptor
engagement, as well
as any derivative or variant of these sequences and any synthetic sequence
that has the same
functional capability.
[0233] Signals generated through the TCR alone may be insufficient for full
activation of the T
cell and a secondary or co-stimulatory signal is also required. Thus, T cell
activation can be
said to be mediated by two distinct classes of cytoplasmic signaling sequence:
those that
initiate antigen-dependent primary activation through the TCR (primary
cytoplasmic signaling
68
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
sequences) and those that act in an antigen-independent manner to provide a
secondary or co-
stimulatory signal (secondary cytoplasmic signaling sequences).
[0234] Primary cytoplasmic signaling sequences regulate primary activation of
the TCR
complex either in a stimulatory way, or in an inhibitory way. Primary
cytoplasmic signaling
sequences that act in a stimulatory manner may contain signaling motifs which
are known as
immunoreceptor tyrosine-based activation motifs or ITAMs.
[0235] Examples of ITAM containing primary cytoplasmic signaling sequences
that are of
particular use in the CARs of the disclosure include those derived from TCR
zeta, FcR gamma,
FcR beta, CD3 gamma, CD3 delta, CD3 epsilon, CD5, 0D22, CD79a, CD79b, and
CD66d. It is
particularly preferred that cytoplasmic signaling molecule in the CAR of the
disclosure
comprises a cytoplasmic signaling sequence from CD3-zeta.
[0236] In a preferred embodiment, the cytoplasmic domain of the CAR is
designed to include
an ITAM containing primary cytoplasmic signaling sequences domain (e.g., that
of CD3-zeta)
by itself or combined with any other desired cytoplasmic domain(s) useful in
the context of the
CAR of the disclosure. For example, the cytoplasmic domain of the CAR can
include a CD3
zeta chain portion and a costimulatory signaling region.
[0237] The costimulatory signaling region refers to a portion of the CAR
comprising the
intracellular domain of a costimulatory molecule. A costimulatory molecule is
a cell surface
molecule other than an antigen receptor or its ligands that is required for an
efficient response
of lymphocytes to an antigen. Examples of such molecules include 0D27, 0D28, 4-
i BB
(0D137), 0X40, CD30, CD40, PD-1, ICOS, lymphocyte function-associated antigen-
1 (LFA-1),
CD2, CD7, LIGHT, NKG2C, B7-H3, and a ligand that specifically binds with 0D83,
and the like.
[0238] The cytoplasmic signaling sequences within the cytoplasmic signaling
portion of the
CAR of the disclosure may be linked to each other in a random or specified
order. Optionally, a
short oligo- or polypeptide linker, preferably between 2 and 10 amino acids in
length may form
the linkage. A glycine-serine doublet provides a particularly suitable linker.
[0239] In one embodiment, the cytoplasmic domain comprises the signaling
domain of CD3-
zeta and the signaling domain of 0D28. In another embodiment, the cytoplasmic
domain
comprises the signaling domain of CD3-zeta and the signaling domain of 4-i BB.
5.5 Nucleic Acids, Recombinant Vectors and Host Cells
[0240] The present disclosure encompasses nucleic acid molecules encoding
immunoglobulin
light and heavy chain genes for anti-glyco-MUC1 antibodies, vectors comprising
such nucleic
acids, and host cells capable of producing the anti-glyco-M UC1 antibodies of
the disclosure. In
certain aspects, the nucleic acid molecules encode, and the host cells are
capable of
expressing, the anti-glyco-MUC1 antibodies and antibody-binding fragments of
the disclosure
(e.g., as described in Section 5.1 and embodiments 1 to 90) as well as fusion
proteins (e.g., as
69
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
described in embodiments 91-96) and chimeric antigen receptors (e.g., as
described in Section
5.4 and embodiments 97-98) containing them. Exemplary vectors of the
disclosure are
described in embodiments 111-113 and exemplary host cells are described in
embodiments
114-117.
[0241] An anti-glyco-MUC1 antibody of the disclosure can be prepared by
recombinant
expression of immunoglobulin light and heavy chain genes in a host cell. To
express an
antibody recombinantly, a host cell is transfected with one or more
recombinant expression
vectors carrying DNA fragments encoding the immunoglobulin light and heavy
chains of the
antibody such that the light and heavy chains are expressed in the host cell
and, optionally,
secreted into the medium in which the host cells are cultured, from which
medium the
antibodies can be recovered. Standard recombinant DNA methodologies are used
to obtain
antibody heavy and light chain genes, incorporate these genes into recombinant
expression
vectors and introduce the vectors into host cells, such as those described in
Molecular Cloning;
A Laboratory Manual, Second Edition (Sambrook, Fritsch and Maniatis (eds),
Cold Spring
Harbor, N.Y., 1989), Current Protocols in Molecular Biology (Ausubel, F. M.
etal., eds.,
Greene Publishing Associates, 1989) and in U.S. Pat. No. 4,816,397.
[0242] To generate nucleic acids encoding such anti-glyco-M UC1 antibodies,
DNA fragments
encoding the light and heavy chain variable regions are first obtained. These
DNAs can be
obtained by amplification and modification of germline DNA or cDNA encoding
light and heavy
chain variable sequences, for example using the polymerase chain reaction
(PCR). Germline
DNA sequences for human heavy and light chain variable region genes are known
in the art
(See, e.g., the "VBASE" human germline sequence database; see also Kabat
etal., 1991,
Sequences of Proteins of Immunological Interest, Fifth Edition, U.S.
Department of Health and
Human Services, NIH Publication No. 91-3242; Tomlinson etal., 1992, J. Mol.
Biol. 22T:116-
198; and Cox etal., 1994, Eur. J. lmmunol. 24:827-836; the contents of each of
which are
incorporated herein by reference).
[0243] Once DNA fragments encoding anti-glyco-MUC1 antibody-related VH and VL
segments
are obtained, these DNA fragments can be further manipulated by standard
recombinant DNA
techniques, for example to convert the variable region genes to full-length
antibody chain
genes, to Fab fragment genes or to a scFv gene. In these manipulations, a VH-
or VL -encoding
DNA fragment is operatively linked to another DNA fragment encoding another
protein, such as
an antibody constant region or a flexible linker. The term "operatively
linked," as used in this
context, is intended to mean that the two DNA fragments are joined such that
the amino acid
sequences encoded by the two DNA fragments remain in-frame.
[0244] The isolated DNA encoding the VH region can be converted to a full-
length heavy chain
gene by operatively linking the VH-encoding DNA to another DNA molecule
encoding heavy
chain constant regions (CHi, CH2, CH3 and, optionally, CH4). The sequences of
human heavy
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
chain constant region genes are known in the art (See, e.g., Kabat etal.,
1991, Sequences of
Proteins of Immunological Interest, Fifth Edition, U.S. Department of Health
and Human
Services, NI H Publication No. 91-3242) and DNA fragments encompassing these
regions can
be obtained by standard PCR amplification. The heavy chain constant region can
be an IgGi,
IgG2, IgG3, IgG4, IgA, IgE, IgM or IgD constant region, but in certain
embodiments is an IgGi or
IgG4 constant region. For a Fab fragment heavy chain gene, the VH-encoding DNA
can be
operatively linked to another DNA molecule encoding only the heavy chain CH1
constant
region.
[0245] The isolated DNA encoding the VL region can be converted to a full-
length light chain
gene (as well as a Fab light chain gene) by operatively linking the VL-
encoding DNA to another
DNA molecule encoding the light chain constant region, CL. The sequences of
human light
chain constant region genes are known in the art (See, e.g., Kabat etal.,
1991, Sequences of
Proteins of Immunological Interest, Fifth Edition, U.S. Department of Health
and Human
Services, NI H Publication No. 91-3242) and DNA fragments encompassing these
regions can
be obtained by standard PCR amplification. The light chain constant region can
be a kappa or
lambda constant region, but in certain embodiments is a kappa constant region.
[0246] To create a scFv gene, the VH- and VL-encoding DNA fragments can be
operatively
linked to another fragment encoding a flexible linker, e.g., encoding the
amino acid sequence
(GlyeSer)3 , such that the VH and VL sequences can be expressed as a
contiguous single-
chain protein, with the VH and VL regions joined by the flexible linker (See,
e.g., Bird etal.,
1988, Science 242:423-426; Huston etal., 1988, Proc. Natl. Acad. Sci. USA
85:5879-5883;
McCafferty etal., 1990, Nature 348:552-554).
[0247] To express the anti-glyco-M UC1 antibodies of the disclosure, DNAs
encoding partial or
full-length light and heavy chains, obtained as described above, are inserted
into expression
vectors such that the genes are operatively linked to transcriptional and
translational control
sequences. In this context, the term "operatively linked" is intended to mean
that an antibody
gene is ligated into a vector such that transcriptional and translational
control sequences within
the vector serve their intended function of regulating the transcription and
translation of the
antibody gene. The expression vector and expression control sequences are
chosen to be
compatible with the expression host cell used. The antibody light chain gene
and the antibody
heavy chain gene can be inserted into separate vectors or, more typically,
both genes are
inserted into the same expression vector.
[0248] The antibody genes are inserted into the expression vector by standard
methods (e.g.,
ligation of complementary restriction sites on the antibody gene fragment and
vector, or blunt
end ligation if no restriction sites are present). Prior to insertion of the
anti-glyco-MUC1
antibody-related light or heavy chain sequences, the expression vector can
already carry
antibody constant region sequences. For example, one approach to converting
the anti-glyco-
71
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
MUC1 monoclonal antibody-related VH and VL sequences to full-length antibody
genes is to
insert them into expression vectors already encoding heavy chain constant and
light chain
constant regions, respectively, such that the VH segment is operatively linked
to the CH
segment(s) within the vector and the VL segment is operatively linked to the
CL segment within
the vector. Additionally or alternatively, the recombinant expression vector
can encode a signal
peptide that facilitates secretion of the antibody chain from a host cell. The
antibody chain gene
can be cloned into the vector such that the signal peptide is linked in-frame
to the amino
terminus of the antibody chain gene. The signal peptide can be an
immunoglobulin signal
peptide or a heterologous signal peptide (i.e., a signal peptide from a non-
immunoglobulin
protein).
[0249] In addition to the antibody chain genes, the recombinant expression
vectors of the
disclosure carry regulatory sequences that control the expression of the
antibody chain genes
in a host cell. The term "regulatory sequence" is intended to include
promoters, enhancers and
other expression control elements (e.g., polyadenylation signals) that control
the transcription or
translation of the antibody chain genes. Such regulatory sequences are
described, for example,
in Goeddel, Gene Expression Technology: Methods in Enzymology 185, Academic
Press, San
Diego, Calif., 1990. It will be appreciated by those skilled in the art that
the design of the
expression vector, including the selection of regulatory sequences may depend
on such factors
as the choice of the host cell to be transformed, the level of expression of
protein desired, etc.
Suitable regulatory sequences for mammalian host cell expression include viral
elements that
direct high levels of protein expression in mammalian cells, such as promoters
and/or
enhancers derived from cytomegalovirus (CMV) (such as the CMV
promoter/enhancer), Simian
Virus 40 (5V40) (such as the 5V40 promoter/enhancer), adenovirus, (e.g., the
adenovirus
major late promoter (AdMLP)) and polyoma. For further description of viral
regulatory elements,
and sequences thereof, see, e.g., U.S. Pat. No. 5,168,062 by Stinski, U.S.
Pat. No. 4,510,245
by Bell etal., and U.S. Pat. No. 4,968,615 by Schaffner etal.
[0250] In addition to the antibody chain genes and regulatory sequences, the
recombinant
expression vectors of the disclosure can carry additional sequences, such as
sequences that
regulate replication of the vector in host cells (e.g., origins of
replication) and selectable marker
genes. The selectable marker gene facilitates selection of host cells into
which the vector has
been introduced (See, e.g., U.S. Pat. Nos. 4,399,216, 4,634,665 and 5,179,017,
all by Axel et
al.). For example, typically the selectable marker gene confers resistance to
drugs, such as
G418, hygromycin or methotrexate, on a host cell into which the vector has
been introduced.
Suitable selectable marker genes include the dihydrofolate reductase (DHFR)
gene (for use in
DHFR- host cells with methotrexate selection/amplification) and the neo gene
(for G418
selection). For expression of the light and heavy chains, the expression
vector(s) encoding the
heavy and light chains is transfected into a host cell by standard techniques.
The various forms
72
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
of the term "transfection" are intended to encompass a wide variety of
techniques commonly
used for the introduction of exogenous DNA into a prokaryotic or eukaryotic
host cell, e.g.,
electroporation, lipofection, calcium-phosphate precipitation, DEAE--dextran
transfection and
the like.
[0251] It is possible to express the antibodies of the disclosure in either
prokaryotic or
eukaryotic host cells. In certain embodiments, expression of antibodies is
performed in
eukaryotic cells, e.g., mammalian host cells, of optimal secretion of a
properly folded and
immunologically active antibody. Exemplary mammalian host cells for expressing
the
recombinant antibodies of the disclosure include Chinese Hamster Ovary (CHO
cells) (including
DHFR- CHO cells, described in Urlaub and Chasin, 1980, Proc. Natl. Acad. Sci.
USA 77:4216-
4220, used with a DHFR selectable marker, e.g., as described in Kaufman and
Sharp, 1982,
Mol. Biol. 159:601-621), NSO myeloma cells, COS cells and 5P2 cells. When
recombinant
expression vectors encoding antibody genes are introduced into mammalian host
cells, the
antibodies are produced by culturing the host cells for a period of time
sufficient to allow for
expression of the antibody in the host cells or secretion of the antibody into
the culture medium
in which the host cells are grown. Antibodies can be recovered from the
culture medium using
standard protein purification methods. Host cells can also be used to produce
portions of intact
antibodies, such as Fab fragments or scFv molecules. It is understood that
variations on the
above procedure are within the scope of the present disclosure. For example,
it can be
desirable to transfect a host cell with DNA encoding either the light chain or
the heavy chain
(but not both) of an anti-glyco-MUC1 antibody of this disclosure.
[0252] For expression of a CAR of the disclosure, for example as described in
Section 5.4 and
in embodiments 97 and 98, it is preferably that the host cell is a T cell,
preferably a human T
cell. In some embodiments, the host cell exhibits an anti-tumor immunity when
the cell is cross-
linked with MUC1 on a tumor cell. Detailed methods for producing the T cells
of the disclosure
are described in Section 5.5.1
[0253] Recombinant DNA technology can also be used to remove some or all of
the DNA
encoding either or both of the light and heavy chains that is not necessary
for binding to glyco-
MUC1. The molecules expressed from such truncated DNA molecules are also
encompassed
by the antibodies of the disclosure.
[0254] For recombinant expression of an anti-glyco-M UC1 antibody of the
disclosure, the host
cell can be co-transfected with two expression vectors of the disclosure, the
first vector
encoding a heavy chain derived polypeptide and the second vector encoding a
light chain
derived polypeptide. The two vectors can contain identical selectable markers,
or they can each
contain a separate selectable marker. Alternatively, a single vector can be
used which encodes
both heavy and light chain polypeptides.
73
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
[0255] Once a nucleic acid encoding one or more portions of an anti-glyco-MUC1
antibody,
further alterations or mutations can be introduced into the coding sequence,
for example to
generate nucleic acids encoding antibodies with different CDR sequences,
antibodies with
reduced affinity to the Fc receptor, or antibodies of different subclasses.
[0256] The anti-glyco-MUC1 antibodies of the disclosure can also be produced
by chemical
synthesis (e.g., by the methods described in Solid Phase Peptide Synthesis,
2nd ed., 1984 The
Pierce Chemical Co., Rockford, Ill.). Variant antibodies can also be generated
using a cell-free
platform (See, e.g., Chu etal., Biochemia No. 2, 2001 (Roche Molecular
Biologicals) and
Murray etal., 2013, Current Opinion in Chemical Biology, 17:420-426).
[0257] Once an anti-glyco-MUC1 antibody of the disclosure has been produced by
recombinant expression, it can be purified by any method known in the art for
purification of an
immunoglobulin molecule, for example, by chromatography (e.g., ion exchange,
affinity, and
sizing column chromatography), centrifugation, differential solubility, or by
any other standard
technique for the purification of proteins. Further, the anti-glyco-MUC1
antibodies of the present
disclosure and/or binding fragments can be fused to heterologous polypeptide
sequences
described herein or otherwise known in the art to facilitate purification.
[0258] Once isolated, the anti-glyco-MUC1 antibody can, if desired, be further
purified, e.g., by
high performance liquid chromatography (see, e.g., Fisher, Laboratory
Techniques In
Biochemistry And Molecular Biology, Work and Burdon, eds., Elsevier, 1980), or
by gel filtration
chromatography on a SuperdexTM 75 column (Pharmacia Biotech AB, Uppsala,
Sweden).
5.5.1. Recombinant Production of CARs in T Cells
[0259] In some embodiments, nucleic acids encoding the anti-glyco-MUC1 CARs of
the
disclosure are delivered into cells using a retroviral or lentiviral vector.
CAR-expressing
retroviral and lentiviral vectors can be delivered into different types of
eukaryotic cells as well as
into tissues and whole organisms using transduced cells as carriers or cell-
free local or
systemic delivery of encapsulated, bound or naked vectors. The method used can
be for any
purpose where stable expression is required or sufficient.
[0260] In other embodiments, the CAR sequences are delivered into cells using
in vitro
transcribed mRNA. In vitro transcribed mRNA CAR can be delivered into
different types of
eukaryotic cells as well as into tissues and whole organisms using transfected
cells as carriers
or cell-free local or systemic delivery of encapsulated, bound or naked mRNA.
The method
used can be for any purpose where transient expression is required or
sufficient.
[0261] In another embodiment, the desired CAR can be expressed in the cells by
way of
transponsons.
[0262] One advantage of RNA transfection methods of the disclosure is that RNA
transfection
is essentially transient and a vector-free: an RNA transgene can be delivered
to a lymphocyte
74
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
and expressed therein following a brief in vitro cell activation, as a minimal
expressing cassette
without the need for any additional viral sequences. Under these conditions,
integration of the
transgene into the host cell genome is unlikely. Cloning of cells is not
necessary because of the
efficiency of transfection of the RNA and its ability to uniformly modify the
entire lymphocyte
population.
[0263] Genetic modification of T cells with in vitro-transcribed RNA (IVT-RNA)
makes use of
two different strategies both of which have been successively tested in
various animal models.
Cells are transfected with in vitro-transcribed RNA by means of lipofection or
electroporation.
Preferably, it is desirable to stabilize IVT-RNA using various modifications
in order to achieve
prolonged expression of transferred IVT-RNA.
[0264] Some IVT vectors are known in the literature which are utilized in a
standardized
manner as template for in vitro transcription and which have been genetically
modified in such a
way that stabilized RNA transcripts are produced. Currently protocols used in
the art are based
on a plasmid vector with the following structure: a 5' RNA polymerase promoter
enabling RNA
transcription, followed by a gene of interest which is flanked either 3'
and/or 5' by untranslated
regions (UTR), and a 3' polyadenyl cassette containing 50-70 A nucleotides.
Prior to in vitro
transcription, the circular plasmid is linearized downstream of the polyadenyl
cassette by type II
restriction enzymes (recognition sequence corresponds to cleavage site). The
polyadenyl
cassette thus corresponds to the later poly(A) sequence in the transcript. As
a result of this
procedure, some nucleotides remain as part of the enzyme cleavage site after
linearization and
extend or mask the poly (A) sequence at the 3' end. It is not clear, whether
this
nonphysiological overhang affects the amount of protein produced
intracellularly from such a
construct.
[0265] RNA has several advantages over more traditional plasmid or viral
approaches. Gene
expression from an RNA source does not require transcription and the protein
product is
produced rapidly after the transfection. Further, since the RNA has to only
gain access to the
cytoplasm, rather than the nucleus, and therefore typical transfection methods
result in an
extremely high rate of transfection. In addition, plasmid based approaches
require that the
promoter driving the expression of the gene of interest be active in the cells
under study.
[0266] In another aspect, the RNA construct can be delivered into the cells by
electroporation.
See, e.g., the formulations and methodology of electroporation of nucleic acid
constructs into
mammalian cells as taught in US 2004/0014645, US 2005/0052630A1, US
2005/0070841A1,
US 2004/0059285A1, US 2004/0092907A1. The various parameters including
electric field
strength required for electroporation of any known cell type are generally
known in the relevant
research literature as well as numerous patents and applications in the field.
See e.g., U.S. Pat.
No. 6,678,556, U.S. Pat. No. 7,171,264, and U.S. Pat. No. 7,173,116. Apparatus
for therapeutic
application of electroporation are available commercially, e.g., the
MedPulserTM DNA
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
Electroporation Therapy System (lnovio/Genetronics, San Diego, Calif.), and
are described in
patents such as U.S. Pat. No. 6,567,694; U.S. Pat. No. 6,516,223, U.S. Pat.
No. 5,993,434,
U.S. Pat. No. 6,181,964, U.S. Pat. No. 6,241,701, and U.S. Pat. No. 6,233,482;
electroporation
may also be used for transfection of cells in vitro as described e.g. in
U520070128708A1.
Electroporation may also be utilized to deliver nucleic acids into cells in
vitro. Accordingly,
electroporation-mediated administration into cells of nucleic acids including
expression
constructs utilizing any of the many available devices and electroporation
systems known to
those of skill in the art presents an exciting new means for delivering an RNA
of interest to a
target cell.
5.5.1.1 Sources of T Cells
[0267] Prior to expansion and genetic modification, a source of T cells is
obtained from a
subject. The term "subject" is intended to include living organisms in which
an immune
response can be elicited (e.g., mammals). Examples of subjects include humans,
dogs, cats,
mice, rats, and transgenic species thereof. Preferably, subjects are human.
[0268] T cells can be obtained from a number of sources, including peripheral
blood
mononuclear cells, bone marrow, lymph node tissue, cord blood, thymus tissue,
tissue from a
site of infection, ascites, pleural effusion, spleen tissue, and tumors. In
certain embodiments of
the present disclosure, any number of T cell lines available in the art, may
be used. In certain
embodiments of the present disclosure, T cells can be obtained from a unit of
blood collected
from a subject using any number of techniques known to the skilled artisan,
such as FicollTM
separation. In one preferred embodiment, cells from the circulating blood of
an individual are
obtained by apheresis. The apheresis product typically contains lymphocytes,
including T cells,
monocytes, granulocytes, B cells, other nucleated white blood cells, red blood
cells, and
platelets. In one embodiment, the cells collected by apheresis may be washed
to remove the
plasma fraction and to place the cells in an appropriate buffer or media for
subsequent
processing steps. In one embodiment of the disclosure, the cells are washed
with phosphate
buffered saline (PBS). In an alternative embodiment, the wash solution lacks
calcium and may
lack magnesium or may lack many if not all divalent cations. Again,
surprisingly, initial activation
steps in the absence of calcium lead to magnified activation. As those of
ordinary skill in the art
would readily appreciate a washing step may be accomplished by methods known
to those in
the art, such as by using a semi-automated "flow-through" centrifuge (for
example, the Cobe
2991 cell processor, the Baxter CytoMate, or the Haemonetics Cell Saver 5)
according to the
manufacturer's instructions. After washing, the cells may be resuspended in a
variety of
biocompatible buffers, such as, for example, Ca-free, Mg-free PBS, PlasmaLyte
A, or other
saline solution with or without buffer. Alternatively, the undesirable
components of the
apheresis sample may be removed and the cells directly resuspended in culture
media.
76
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
[0269] In another embodiment, T cells are isolated from peripheral blood
lymphocytes by lysing
the red blood cells and depleting the monocytes, for example, by
centrifugation through a
PERCOLLTM gradient or by counterflow centrifugal elutriation. A specific
subpopulation of T
cells, such as CD3+, 0D28', CD4+, CD8+, CD45RA+ and CD45R0+ T cells, can be
further
isolated by positive or negative selection techniques. For example, in one
embodiment, T cells
are isolated by incubation with anti-CD3/anti-0D28 (i.e., 3 x 28)-conjugated
beads, such as
DYNABEADSO M-450 CD3/0D28 T, for a time period sufficient for positive
selection of the
desired T cells. In one embodiment, the time period is about 30 minutes. In a
further
embodiment, the time period ranges from 30 minutes to 36 hours or longer and
all integer
values there between. In a further embodiment, the time period is at least 1,
2, 3, 4, 5, or 6
hours. In yet another preferred embodiment, the time period is 10 to 24 hours.
In one preferred
embodiment, the incubation time period is 24 hours. For isolation of T cells
from patients with
leukemia, use of longer incubation times, such as 24 hours, can increase cell
yield. Longer
incubation times may be used to isolate T cells in any situation where there
are few T cells as
compared to other cell types, such in isolating tumor infiltrating lymphocytes
(TIL) from tumor
tissue or from immunocompromised individuals. Further, use of longer
incubation times can
increase the efficiency of capture of CD8+ T cells. Thus, by simply shortening
or, lengthening
the time T cells are allowed to bind to the CD3/0D28 beads and/or by
increasing or decreasing
the ratio of beads to T cells (as described further herein), subpopulations of
T cells can be
preferentially selected for or against at culture initiation or at other time
points during the
process. Additionally, by increasing or decreasing the ratio of anti-CD3
and/or anti-0D28
antibodies on the beads or other surface, subpopulations of T cells can be
preferentially
selected for or against at culture initiation or at other desired time points.
The skilled artisan
would recognize that multiple rounds of selection can also be used in the
context of this
disclosure. In certain embodiments, it may be desirable to perform the
selection procedure and
use the "unselected" cells in the activation and expansion process.
"Unselected" cells can also
be subjected to further rounds of selection.
[0270] Enrichment of a T cell population by negative selection can be
accomplished with a
combination of antibodies directed to surface markers unique to the negatively
selected cells.
One method is cell sorting and/or selection via negative magnetic
immunoadherence or flow
cytometry that uses a cocktail of monoclonal antibodies directed to cell
surface markers present
on the cells negatively selected. For example, to enrich for CD4+ cells by
negative selection, a
monoclonal antibody cocktail typically includes antibodies to CD14, CD20, CD11
b, CD16, HLA-
DR, and CD8. In certain embodiments, it may be desirable to enrich for or
positively select for
regulatory T cells which typically express CD4+, CD25+, CD62Lhi, GITR+, and
FoxP3+.
Alternatively, in certain embodiments, T regulatory cells are depleted by anti-
025 conjugated
beads or other similar method of selection.
77
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
[0271] For isolation of a desired population of cells by positive or negative
selection, the
concentration of cells and surface (e.g., particles such as beads) can be
varied. In certain
embodiments, it may be desirable to significantly decrease the volume in which
beads and cells
are mixed together (i.e., increase the concentration of cells), to ensure
maximum contact of
cells and beads. For example, in one embodiment, a concentration of 2 billion
cells/ml is used.
In one embodiment, a concentration of 1 billion cells/ml is used. In a further
embodiment,
greater than 100 million cells/ml is used. In a further embodiment, a
concentration of cells of 10,
15, 20, 25, 30, 35, 40, 45, or 50 million cells/ml is used. In yet another
embodiment, a
concentration of cells from 75, 80, 85, 90, 95, or 100 million cells/ml is
used. In further
embodiments, concentrations of 125 or 150 million cells/ml can be used. Using
high
concentrations can result in increased cell yield, cell activation, and cell
expansion. Further, use
of high cell concentrations allows more efficient capture of cells that may
weakly express target
antigens of interest, such as 0D28-negative T cells, or from samples where
there are many
tumor cells present (i.e., leukemic blood, tumor tissue, etc.). Such
populations of cells may
have therapeutic value and would be desirable to obtain. For example, using
high concentration
of cells allows more efficient selection of CD8+ T cells that normally have
weaker 0D28
expression.
[0272] In a related embodiment, it may be desirable to use lower
concentrations of cells. By
significantly diluting the mixture of T cells and surface (e.g., particles
such as beads),
interactions between the particles and cells is minimized. This selects for
cells that express high
amounts of desired antigens to be bound to the particles. For example, CD4+ T
cells express
higher levels of 0D28 and are more efficiently captured than CD8+ T cells in
dilute
concentrations. In one embodiment, the concentration of cells used is 5 x
106/ml. In other
embodiments, the concentration used can be from about 1 x 105/mIto 1 x 106/ml,
and any
integer value in between.
[0273] In other embodiments, the cells may be incubated on a rotator for
varying lengths of
time at varying speeds at either 2-10 C. or at room temperature.
[0274] T cells for stimulation can also be frozen after a washing step.
VVishing not to be bound
by theory, the freeze and subsequent thaw step provides a more uniform product
by removing
granulocytes and to some extent monocytes in the cell population. After the
washing step that
removes plasma and platelets, the cells may be suspended in a freezing
solution. While many
freezing solutions and parameters are known in the art and will be useful in
this context, one
method involves using PBS containing 20% DMSO and 8% human serum albumin, or
culture
media containing 10% Dextran 40 and 5% Dextrose, 20% Human Serum Albumin and
7.5%
DMSO, or 31.25% Plasmalyte-A, 31.25% Dextrose 5%, 0.45% NaCI, 10% Dextran 40
and 5%
Dextrose, 20% Human Serum Albumin, and 7.5% DMSO or other suitable cell
freezing media
containing for example, Hespan and PlasmaLyte A, the cells then are frozen to -
80 C. at a rate
78
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
of 10 per minute and stored in the vapor phase of a liquid nitrogen storage
tank. Other methods
of controlled freezing may be used as well as uncontrolled freezing
immediately at -20 C. or in
liquid nitrogen.
[0275] In certain embodiments, cryopreserved cells are thawed and washed as
described
herein and allowed to rest for one hour at room temperature prior to
activation using the
methods of the present disclosure.
[0276] Also contemplated in the context of the disclosure is the collection of
blood samples or
apheresis product from a subject at a time period prior to when the expanded
cells as described
herein might be needed. As such, the source of the cells to be expanded can be
collected at
any time point necessary, and desired cells, such as T cells, isolated and
frozen for later use in
T cell therapy for any number of diseases or conditions that would benefit
from T cell therapy,
such as those described herein. In one embodiment a blood sample or an
apheresis is taken
from a generally healthy subject. In certain embodiments, a blood sample or an
apheresis is
taken from a generally healthy subject who is at risk of developing a disease,
but who has not
yet developed a disease, and the cells of interest are isolated and frozen for
later use. In certain
embodiments, the T cells may be expanded, frozen, and used at a later time. In
certain
embodiments, samples are collected from a patient shortly after diagnosis of a
particular
disease as described herein but prior to any treatments. In a further
embodiment, the cells are
isolated from a blood sample or an apheresis from a subject prior to any
number of relevant
treatment modalities, including but not limited to treatment with agents such
as natalizumab,
efalizumab, antiviral agents, chemotherapy, radiation, immunosuppressive
agents, such as
cyclosporin, azathioprine, methotrexate, mycophenolate, and FK506, antibodies,
or other
immunoablative agents such as CAM PATH, anti-CD3 antibodies, cytoxan,
fludarabine,
cyclosporin, FK506, rapamycin, mycophenolic acid, steroids, FR901228, and
irradiation. These
drugs inhibit either the calcium dependent phosphatase calcineurin
(cyclosporine and FK506)
or inhibit the p70S6 kinase that is important for growth factor induced
signaling (rapamycin).
(Liu etal., Cell 66:807-815, 1991; Henderson etal., lmmun. 73:316-321, 1991;
Bierer etal.,
Curr. Opin. lmmun. 5:763-773, 1993). In a further embodiment, the cells are
isolated for a
patient and frozen for later use in conjunction with (e.g., before,
simultaneously or following)
bone marrow or stem cell transplantation or T cell ablative therapy using
either chemotherapy
agents such as, fludarabine, external-beam radiation therapy (XRT),
cyclophosphamide.
[0277] In a further embodiment of the present disclosure, T cells are obtained
from a patient
directly following treatment. In this regard, it has been observed that
following certain cancer
treatments, in particular treatments with drugs that damage the immune system,
shortly after
treatment during the period when patients would normally be recovering from
the treatment, the
quality of T cells obtained may be optimal or improved for their ability to
expand ex vivo.
Likewise, following ex vivo manipulation using the methods described herein,
these cells may
79
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
be in a preferred state for enhanced engraftment and in vivo expansion. Thus,
it is
contemplated within the context of the present disclosure to collect blood
cells, including T
cells, dendritic cells, or other cells of the hematopoietic lineage, during
this recovery phase.
Further, in certain embodiments, mobilization (for example, mobilization with
GM-CSF) and
conditioning regimens can be used to create a condition in a subject wherein
repopulation,
recirculation, regeneration, and/or expansion of particular cell types is
favored, especially
during a defined window of time following therapy. Illustrative cell types
include T cells, B cells,
dendritic cells, and other cells of the immune system.
5.5.1.2 Activation and Expansion of T Cells
[0278] T cells are activated and expanded generally using methods as
described, for example,
in U.S. Pat. Nos. 6,352,694; 6,534,055; 6,905,680; 6,692,964; 5,858,358;
6,887,466;
6,905,681; 7,144,575; 7,067,318; 7,172,869; 7,232,566; 7,175,843; 5,883,223;
6,905,874;
6,797,514; 6,867,041; and U.S. Patent Application Publication No. 20060121005.
[0279] Generally, the T cells of the disclosure are expanded by contact with a
surface having
attached thereto an agent that stimulates a CD3/TCR complex associated signal
and a ligand
that stimulates a co-stimulatory molecule on the surface of the T cells. In
particular, T cell
populations may be stimulated as described herein, such as by contact with an
anti-CD3
antibody, or antigen-binding fragment thereof, or an anti-CD2 antibody
immobilized on a
surface, or by contact with a protein kinase C activator (e.g., bryostatin) in
conjunction with a
calcium ionophore. For co-stimulation of an accessory molecule on the surface
of the T cells, a
ligand that binds the accessory molecule is used. For example, a population of
T cells can be
contacted with an anti-CD3 antibody and an anti-0D28 antibody, under
conditions appropriate
for stimulating proliferation of the T cells. To stimulate proliferation of
either CD4+ T cells or
CD8+ T cells, an anti-CD3 antibody and an anti-0D28 antibody. Examples of an
anti-0D28
antibody include 9.3, B-T3, XR-0D28 (Diaclone, Besancon, France) can be used
as can other
methods commonly known in the art (Berg etal., Transplant Proc. 30(8):3975-
3977, 1998;
Haanen etal., J. Exp. Med. 190(9):13191328, 1999; Garland etal., J. Immunol
Meth. 227(1-
2):53-63, 1999).
[0280] In certain embodiments, the primary stimulatory signal and the co-
stimulatory signal for
the T cell may be provided by different protocols. For example, the agents
providing each signal
may be in solution or coupled to a surface. When coupled to a surface, the
agents may be
coupled to the same surface (i.e., in "cis" formation) or to separate surfaces
(i.e., in "trans"
formation). Alternatively, one agent may be coupled to a surface and the other
agent in
solution. In one embodiment, the agent providing the co-stimulatory signal is
bound to a cell
surface and the agent providing the primary activation signal is in solution
or coupled to a
surface. In certain embodiments, both agents can be in solution. In another
embodiment, the
agents may be in soluble form, and then cross-linked to a surface, such as a
cell expressing Fc
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
receptors or an antibody or other binding agent which will bind to the agents.
In this regard, see
for example, U.S. Patent Application Publication Nos. 20040101519 and
20060034810 for
artificial antigen presenting cells (aAPCs) that are contemplated for use in
activating and
expanding T cells in the present disclosure.
[0281] In one embodiment, the two agents are immobilized on beads, either on
the same bead,
i.e., "cis," or to separate beads, i.e., "trans." By way of example, the agent
providing the primary
activation signal is an anti-CD3 antibody or an antigen-binding fragment
thereof and the agent
providing the co-stimulatory signal is an anti-0D28 antibody or antigen-
binding fragment
thereof; and both agents are co-immobilized to the same bead in equivalent
molecular
amounts. In one embodiment, a 1:1 ratio of each antibody bound to the beads
for CD4+ T cell
expansion and T cell growth is used. In certain aspects of the present
disclosure, a ratio of anti
CD3:0D28 antibodies bound to the beads is used such that an increase in T cell
expansion is
observed as compared to the expansion observed using a ratio of 1:1. In one
particular
embodiment an increase of from about 1 to about 3 fold is observed as compared
to the
expansion observed using a ratio of 1:1. In one embodiment, the ratio of
CD3:0D28 antibody
bound to the beads ranges from 100:1 to 1:100 and all integer values there
between. In one
aspect of the present disclosure, more anti-0D28 antibody is bound to the
particles than anti-
CD3 antibody, i.e., the ratio of CD3:0D28 is less than one. In certain
embodiments of the
disclosure, the ratio of anti 0D28 antibody to anti CD3 antibody bound to the
beads is greater
than 2:1. In one particular embodiment, a 1:100 CD3:0D28 ratio of antibody
bound to beads is
used. In another embodiment, a 1:75 CD3:0D28 ratio of antibody bound to beads
is used. In a
further embodiment, a 1:50 CD3:0D28 ratio of antibody bound to beads is used.
In another
embodiment, a 1:30 CD3:0D28 ratio of antibody bound to beads is used. In one
preferred
embodiment, a 1:10 CD3:0D28 ratio of antibody bound to beads is used. In
another
embodiment, a 1:3 CD3:0D28 ratio of antibody bound to the beads is used. In
yet another
embodiment, a 3:1 CD3:0D28 ratio of antibody bound to the beads is used.
[0282] Ratios of particles to cells from 1:500 to 500:1 and any integer values
in between may
be used to stimulate T cells or other target cells. As those of ordinary skill
in the art can readily
appreciate, the ratio of particles to cells may depend on particle size
relative to the target cell.
For example, small sized beads could only bind a few cells, while larger beads
could bind
many. In certain embodiments the ratio of cells to particles ranges from 1:100
to 100:1 and any
integer values in-between and in further embodiments the ratio comprises 1:9
to 9:1 and any
integer values in between, can also be used to stimulate T cells. The ratio of
anti-CD3- and
anti-0D28-coupled particles to T cells that result in T cell stimulation can
vary as noted above,
however certain preferred values include 1:100, 1:50, 1:40, 1:30, 1:20, 1:10,
1:9, 1:8, 1:7, 1:6,
1:5, 1:4, 1:3, 1:2, 1:1, 2:1, 3:1, 4:1, 5:1, 6:1, 7:1, 8:1, 9:1, 10:1, and
15:1 with one preferred ratio
being at least 1:1 particles per T cell. In one embodiment, a ratio of
particles to cells of 1:1 or
81
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
less is used. In one particular embodiment, a preferred particle: cell ratio
is 1:5. In further
embodiments, the ratio of particles to cells can be varied depending on the
day of stimulation.
For example, in one embodiment, the ratio of particles to cells is from 1:1 to
10:1 on the first
day and additional particles are added to the cells every day or every other
day thereafter for up
to 10 days, at final ratios of from 1:1 to 1:10 (based on cell counts on the
day of addition). In
one particular embodiment, the ratio of particles to cells is 1:1 on the first
day of stimulation and
adjusted to 1:5 on the third and fifth days of stimulation. In another
embodiment, particles are
added on a daily or every other day basis to a final ratio of 1:1 on the first
day, and 1:5 on the
third and fifth days of stimulation. In another embodiment, the ratio of
particles to cells is 2:1 on
the first day of stimulation and adjusted to 1:10 on the third and fifth days
of stimulation. In
another embodiment, particles are added on a daily or every other day basis to
a final ratio of
1:1 on the first day, and 1:10 on the third and fifth days of stimulation. One
of skill in the art will
appreciate that a variety of other ratios may be suitable for use in the
present disclosure. In
particular, ratios will vary depending on particle size and on cell size and
type.
[0283] In further embodiments of the present disclosure, the cells, such as T
cells, are
combined with agent-coated beads, the beads and the cells are subsequently
separated, and
then the cells are cultured. In an alternative embodiment, prior to culture,
the agent-coated
beads and cells are not separated but are cultured together. In a further
embodiment, the
beads and cells are first concentrated by application of a force, such as a
magnetic force,
resulting in increased ligation of cell surface markers, thereby inducing cell
stimulation.
[0284] By way of example, cell surface proteins may be ligated by allowing
paramagnetic
beads to which anti-CD3 and anti-0D28 are attached (3 x 28 beads) to contact
the T cells. In
one embodiment the cells (for example, 104 to 109 T cells) and beads (for
example,
DYNABEADSO M-450 CD3/0D28 T paramagnetic beads at a ratio of 1:1) are combined
in a
buffer, preferably PBS (without divalent cations such as, calcium and
magnesium). Again, those
of ordinary skill in the art can readily appreciate any cell concentration may
be used. For
example, the target cell may be very rare in the sample and comprise only
0.01% of the sample
or the entire sample (i.e., 100%) may comprise the target cell of interest.
Accordingly, any cell
number is within the context of the present disclosure. In certain
embodiments, it may be
desirable to significantly decrease the volume in which particles and cells
are mixed together
(i.e., increase the concentration of cells), to ensure maximum contact of
cells and particles. For
example, in one embodiment, a concentration of about 2 billion cells/ml is
used. In another
embodiment, greater than 100 million cells/ml is used. In a further
embodiment, a concentration
of cells of 10, 15, 20, 25, 30, 35, 40, 45, or 50 million cells/ml is used. In
yet another
embodiment, a concentration of cells from 75, 80, 85, 90, 95, or 100 million
cells/ml is used. In
further embodiments, concentrations of 125 or 150 million cells/ml can be
used. Using high
concentrations can result in increased cell yield, cell activation, and cell
expansion. Further, use
82
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
of high cell concentrations allows more efficient capture of cells that may
weakly express target
antigens of interest, such as 0D28-negative T cells. Such populations of cells
may have
therapeutic value and would be desirable to obtain in certain embodiments. For
example, using
high concentration of cells allows more efficient selection of CD8+ T cells
that normally have
weaker 0D28 expression.
[0285] In one embodiment of the present disclosure, the mixture may be
cultured for several
hours (about 3 hours) to about 14 days or any hourly integer value in between.
In another
embodiment, the mixture may be cultured for 21 days. In one embodiment of the
disclosure the
beads and the T cells are cultured together for about eight days. In another
embodiment, the
beads and T cells are cultured together for 2-3 days. Several cycles of
stimulation may also be
desired such that culture time of T cells can be 60 days or more. Conditions
appropriate for T
cell culture include an appropriate media (e.g., Minimal Essential Media or
RPM! Media 1640
or, X-vivo 15, (Lonza)) that may contain factors necessary for proliferation
and viability,
including serum (e.g., fetal bovine or human serum), interleukin-2 (IL-2),
insulin, IFN-y, IL-4, IL-
7, GM-CSF, IL-10, IL-12, IL-15, TGFO, and TNF-a or any other additives for the
growth of cells
known to the skilled artisan. Other additives for the growth of cells include,
but are not limited
to, surfactant, plasmanate, and reducing agents such as N-acetyl-cysteine and
2-
mercaptoethanol. Media can include RPM! 1640, AIM-V, DMEM, MEM, a-MEM, F-12, X-
Vivo
15, and X-Vivo 20, Optimizer, with added amino acids, sodium pyruvate, and
vitamins, either
serum-free or supplemented with an appropriate amount of serum (or plasma) or
a defined set
of hormones, and/or an amount of cytokine(s) sufficient for the growth and
expansion of T cells.
Antibiotics, e.g., penicillin and streptomycin, are included only in
experimental cultures, not in
cultures of cells that are to be infused into a subject. The target cells are
maintained under
conditions necessary to support growth, for example, an appropriate
temperature (e.g., 37 C.)
and atmosphere (e.g., air plus 5% CO2).
[0286] T cells that have been exposed to varied stimulation times may exhibit
different
characteristics. For example, typical blood or apheresed peripheral blood
mononuclear cell
products have a helper T cell population (TH, CD4+) that is greater than the
cytotoxic or
suppressor T cell population (Tc, CD8+). Ex vivo expansion of T cells by
stimulating CD3 and
CD28 receptors produces a population of T cells that prior to about days 8-9
consists
predominately of TH cells, while after about days 8-9, the population of T
cells comprises an
increasingly greater population of Tc cells. Accordingly, depending on the
purpose of treatment,
infusing a subject with a T cell population comprising predominately of TH
cells may be
advantageous. Similarly, if an antigen-specific subset of Tc cells has been
isolated it may be
beneficial to expand this subset to a greater degree.
[0287] Further, in addition to CD4 and CD8 markers, other phenotypic markers
vary
significantly, but in large part, reproducibly during the course of the cell
expansion process.
83
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
Thus, such reproducibility enables the ability to tailor an activated T cell
product for specific
purposes.
5.6 COMPOSITIONS
[0288] The anti-glyco-MUC1 antibodies and/or anti-glyco-MUC1 ADCs of the
disclosure may
be in the form of compositions comprising the anti-glyco-MUC1 antibody and/or
ADC and one
or more carriers, excipients and/or diluents. The compositions may be
formulated for specific
uses, such as for veterinary uses or pharmaceutical uses in humans. The form
of the
composition (e.g., dry powder, liquid formulation, etc.) and the excipients,
diluents and/or
carriers used will depend upon the intended uses of the antibody and/or ADC
and, for
therapeutic uses, the mode of administration.
[0289] For therapeutic uses, the compositions may be supplied as part of a
sterile,
pharmaceutical composition that includes a pharmaceutically acceptable
carrier. This
composition can be in any suitable form (depending upon the desired method of
administering it
to a patient). The pharmaceutical composition can be administered to a patient
by a variety of
routes such as orally, transdermally, subcutaneously, intranasally,
intravenously,
intramuscularly, intratumorally, intrathecally, topically or locally. The most
suitable route for
administration in any given case will depend on the particular antibody and/or
ADC, the subject,
and the nature and severity of the disease and the physical condition of the
subject. Typically,
the pharmaceutical composition will be administered intravenously or
subcutaneously.
[0290] Pharmaceutical compositions can be conveniently presented in unit
dosage forms
containing a predetermined amount of an anti-glyco-M UC1 antibody and/or anti-
glyco-MUC1
ADC of the disclosure per dose. The quantity of antibody and/or ADC included
in a unit dose
will depend on the disease being treated, as well as other factors as are well
known in the art.
Such unit dosages may be in the form of a lyophilized dry powder containing an
amount of
antibody and/or ADC suitable for a single administration, or in the form of a
liquid. Dry powder
unit dosage forms may be packaged in a kit with a syringe, a suitable quantity
of diluent and/or
other components useful for administration. Unit dosages in liquid form may be
conveniently
supplied in the form of a syringe pre-filled with a quantity of antibody
and/or ADC suitable for a
single administration.
[0291] The pharmaceutical compositions may also be supplied in bulk from
containing
quantities of ADC suitable for multiple administrations.
[0292] Pharmaceutical compositions may be prepared for storage as lyophilized
formulations or
aqueous solutions by mixing an antibody and/or ADC having the desired degree
of purity with
optional pharmaceutically-acceptable carriers, excipients or stabilizers
typically employed in the
art (all of which are referred to herein as "carriers"), i.e., buffering
agents, stabilizing agents,
preservatives, isotonifiers, non-ionic detergents, antioxidants, and other
miscellaneous
84
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
additives. See, Remington's Pharmaceutical Sciences, 16th edition (Osol, ed.
1980). Such
additives should be nontoxic to the recipients at the dosages and
concentrations employed.
[0293] Buffering agents help to maintain the pH in the range which
approximates physiological
conditions. They may be present at a wide variety of concentrations, but will
typically be present
in concentrations ranging from about 2 mM to about 50 mM. Suitable buffering
agents for use
with the present disclosure include both organic and inorganic acids and salts
thereof such as
citrate buffers (e.g., monosodium citrate-disodium citrate mixture, citric
acid-trisodium citrate
mixture, citric acid-monosodium citrate mixture, etc.), succinate buffers
(e.g., succinic acid-
monosodium succinate mixture, succinic acid-sodium hydroxide mixture, succinic
acid-disodium
succinate mixture, etc.), tartrate buffers (e.g., tartaric acid-sodium
tartrate mixture, tartaric acid-
potassium tartrate mixture, tartaric acid-sodium hydroxide mixture, etc.),
fumarate buffers (e.g.,
fumaric acid-monosodium fumarate mixture, fumaric acid-disodium fumarate
mixture,
monosodium fumarate-disodium fumarate mixture, etc.), gluconate buffers (e.g.,
gluconic acid-
sodium glyconate mixture, gluconic acid-sodium hydroxide mixture, gluconic
acid-potassium
glyuconate mixture, etc.), oxalate buffer (e.g., oxalic acid-sodium oxalate
mixture, oxalic acid-
sodium hydroxide mixture, oxalic acid-potassium oxalate mixture, etc.),
lactate buffers (e.g.,
lactic acid-sodium lactate mixture, lactic acid-sodium hydroxide mixture,
lactic acid-potassium
lactate mixture, etc.) and acetate buffers (e.g., acetic acid-sodium acetate
mixture, acetic acid-
sodium hydroxide mixture, etc.). Additionally, phosphate buffers, histidine
buffers and
trimethylamine salts such as Tris can be used.
[0294] Preservatives may be added to retard microbial growth, and can be added
in amounts
ranging from about 0.2%-1% (w/v). Suitable preservatives for use with the
present disclosure
include phenol, benzyl alcohol, meta-cresol, methyl paraben, propyl paraben,
octadecyldimethylbenzyl ammonium chloride, benzalconium halides (e.g.,
chloride, bromide,
and iodide), hexamethonium chloride, and alkyl parabens such as methyl or
propyl paraben,
catechol, resorcinol, cyclohexanol, and 3-pentanol. lsotonicifiers sometimes
known as
"stabilizers" can be added to ensure isotonicity of liquid compositions of the
present disclosure
and include polyhydric sugar alcohols, for example trihydric or higher sugar
alcohols, such as
glycerin, erythritol, arabitol, xylitol, sorbitol and mannitol. Stabilizers
refer to a broad category of
excipients which can range in function from a bulking agent to an additive
which solubilizes the
therapeutic agent or helps to prevent denaturation or adherence to the
container wall. Typical
stabilizers can be polyhydric sugar alcohols (enumerated above); amino acids
such as arginine,
lysine, glycine, glutamine, asparagine, histidine, alanine, ornithine, L-
leucine, 2-phenylalanine,
glutamic acid, threonine, etc., organic sugars or sugar alcohols, such as
lactose, trehalose,
stachyose, mannitol, sorbitol, xylitol, ribitol, myoinisitol, galactitol,
glycerol and the like, including
cyclitols such as inositol; polyethylene glycol; amino acid polymers; sulfur
containing reducing
agents, such as urea, glutathione, thioctic acid, sodium thioglycolate,
thioglycerol, a-
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
monothioglycerol and sodium thio sulfate; low molecular weight polypeptides
(e.g., peptides of
residues or fewer); proteins such as human serum albumin, bovine serum
albumin, gelatin
or immunoglobulins; hydrophylic polymers, such as polyvinylpyrrolidone
monosaccharides,
such as xylose, mannose, fructose, glucose; disaccharides such as lactose,
maltose, sucrose
and trehalose; and trisaccacharides such as raffinose; and polysaccharides
such as dextran.
Stabilizers may be present in amounts ranging from 0.5 to 10 wt % per wt of
ADC.
[0295] Non-ionic surfactants or detergents (also known as "wetting agents")
may be added to
help solubilize the glycoprotein as well as to protect the glycoprotein
against agitation-induced
aggregation, which also permits the formulation to be exposed to shear surface
stressed
without causing denaturation of the protein. Suitable non-ionic surfactants
include polysorbates
(20, 80, etc.), polyoxamers (184, 188 etc.), and pluronic polyols. Non-ionic
surfactants may be
present in a range of about 0.05 mg/mL to about 1.0 mg/mL, for example about
0.07 mg/mL to
about 0.2 mg/mL.
[0296] Additional miscellaneous excipients include bulking agents (e.g.,
starch), chelating
agents (e.g., EDTA), antioxidants (e.g., ascorbic acid, methionine, vitamin
E), and cosolvents.
5.7 Methods of Use
[0297] The anti-glyco-MUC1 antibody or binding fragments described herein can
be used in
various diagnostic assays. For example, the antibodies and binding fragments
can be
employed in immunoassays, such as competitive binding assays, direct and
indirect sandwich
assays, and immunoprecipitation assays, including immunohistochemistry, enzyme-
linked
immunosorbent assay (ELISA), fluorescence-activated cell sorting (FACS), and
Western blots.
[0298] The anti-glyco-MUC1 antibody or binding fragments described herein also
are useful for
radiographic in vivo imaging, wherein an antibody labeled with a detectable
moiety such as a
radio-opaque agent or radioisotope is administered to a subject, preferably
into the
bloodstream, and the presence and location of the labeled antibody in the host
is assayed. This
imaging technique is useful in the staging and treatment of malignancies.
[0299] The anti-glyco-MUC1 antibody or binding fragments, ADCs and CARs
described herein
are useful for treatment of glyco-MUC1 expressing cancers, particularly
epithelial cancers such
as breast cancer, ovarian cancer, pancreatic cancer, and lung cancer.
[0300] When using the CARs of the disclosure for therapy, the therapeutic
methods of the
disclosure comprising administering to a subject with a glyco-MUC1-expressing
tumor an
effective amount of a genetically modified cell engineered to express a CAR of
the disclosure,
for example as described in Section 5.4 or in embodiment 97 or embodiment 98.
Methods of
modifying cells, particularly T cells, to express a CAR, are described in
Section 5.5.1.
86
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
6. EXAMPLES
6.1 Example 1: Identification Of Anti-Glyco-Muc1 Antibodies
6.1.1. Overview
[0301] Chemoenzymatic synthesis of multiple-repeat MUC1 glycopeptides with
different 0-
glycan density and Tn (GaINAca1-0-Ser/Thr) glycoforms was developed using
recombinant
glycosyltransferases. Different polypeptide GaINAc-transferase isoforms were
used to direct
sites of 0-glycan occupancy (Bennett etal., 1998). The optimal vaccine design
was found to be
Tn glycoforms with high 0-glycan density, and glycopeptides conjugated to KLH
were found to
overcome tolerance. In wild-type Balb/c mice, the glycopeptides with complete
0-glycan
occupancy elicited the strongest antibody response reacting with MUC1
expressed in breast
cancer cell lines, thus representing the most effective vaccine design. The
elicited humoral
immune response showed remarkable specificity for cancer cells.
6.1.2. Materials and Methods
6.1.2.1 Chemoenzymatic synthesis of multimeric Tn MUC1
glycopeptides
[0302] MUC1 60-mer (VTSAPDTRPAPGSTAPPAHG)n = 3 (SEQ ID NO:47) peptide was
synthesized, as originally reported by Fontenot etal., 1993. Control peptides
used were derived
from tandem repeats (TRs) of MUC2 (PTTTPISTTTMVTPTPTPTC) (SEQ ID NO:51) and
MUC4
(CPLPVTDTSSASTGHATPLPV) (SEQ ID NO:52). Peptides were glycosylated in vitro
using
purified recombinant human glycosyltransferases polypeptides GaINAc-T2, GaINAc-
T4, and
GaINAc-T1 (Bennett etal., 1998; Schwientek etal., 2002) as described in US
Patent No.
6,465,220. GaINAc glycosylation of the peptides was performed in a reaction
mixture (1 mg
peptide/mL) containing 25 mM cacodylate buffer (pH 7.4), 10 mM MnCl2, 0.25%
Triton X-100,
and 2 mM UDP-GaINAc. Glycosylation of 1 mg 60-mer peptide with two GaINAc per
TR
(MUC160Tn6) was obtained using GaINAc-T1. Incorporation of three GaINAc per TR
(MUC160Tn9) was obtained using GaINAc-T2. Substitution of all five putative 0-
glycosylation
sites in the MUC1 TR (MUC160Tn15) was performed using MUC160Tn9 as substrate
in a
reaction with GaINAc-T4. Glycosylation was monitored using nano-scale reversed-
phase
columns (Poros R3, PerSeptive Biosystems, Framingham, MA) and MALDI-TOF mass
spectrometry. The glycopeptides were purified by high-performance liquid
chromatography
(H PLC) on a Zorbax 3005B-C3 column (9.4 mm x 25 cm) (Agilent Technologies,
Palo Alto, CA)
in an 1100 Hewlett Packard system (Avondale, PA) using 0.1% trifluoroacetic
acid (TFA) and a
gradient of 0-80% acetonitrile. Quantification and estimation of yields of
glycosylation reactions
were performed by comparison of HPLC peaks by UV 210 absorbance using 10 pg
weighed
peptide as standard. GaINAc glycosylation of peptides generally yielded 80-90%
recovery.
Purified glycopeptides were characterized by MALDI-TOF mass spectrometry on a
Voyager DE
or Voyager DE Pro MALDI- TOF mass spectrometer (PerSeptive Biosystems)
equipped with
87
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
delayed extraction. The MALDI matrix was 2,5-dihydroxybenzoic acid 10 g/L
(Aldrich,
Milwaukee, WI) dissolved in 2:1 mixture of 0.1% TFA in 30% aqueous
acetonitrile. Samples
dissolved in 0.1% TFA to a concentration of ¨1 pmol/uL were prepared for
analysis by placing
1pL of sample solution on a probe tip followed by luL of matrix. All mass
spectra were obtained
in the linear mode. Data processing was carried out using GRAMS/386 software
(Galactic
Industries, Salem, NH).
6.1.2.2 Immunization Protocol
[0303] Glycopeptides were coupled to KLH (Pierce, Rockford, IL) using
glutaraldehyde.
Efficiency of conjugation was assessed by analyzing the reaction by size
exclusion
chromatography on a PD-10 column using anti-MUC1 ELISA of fractions.
Essentially all
reactivity was found with the excluded fraction and insignificant reactivity
in the included
fractions expected to contain peptides. Further evaluation included
comparative titration
analysis of the KLH conjugate with the corresponding glycopeptide in ELISA.
Both analyses
indicated that the conjugation was near complete, which should result in a KLH
to glycopeptide
ratio of 1:300. Female Balb/c wild-type mice were injected subcutaneously with
10 or 15 pg of
(glyco)peptide in a total volume of 200 uL (1:1 mix with Freunds adjuvant,
Sigma). Mice
received four immunizations 14 days apart, and blood samples were obtained by
tail or eye
bleeding 1 week following the third and fourth immunization.
6.1.2.3 Generation of mouse MAb anti-Tn-MUC1
[0304] A MAb was produced, from a wild-type Balb/c mouse immunized with the
fully GaINAc-
glycosylated 60-mer MUC1 glycopeptide coupled to KLH. Screening was based on
glycopeptide ELISA followed by immunocytology with breast cancer cell lines
(MCF7 and
T47D) and immunohistology with cancer tissues. Selection was based on
reactivity pattern
similar to total sera of the same mouse.
6.1.2.4 ELISA
[0305] ELISA were performed using 96-well MaxiSorp plates (Nunc, Denmark).
Plates were
coated overnight at 4 C with 1 pg/mL of glycopeptides in bicarbonate¨carbonate
buffer (pH
9.6), blocked with 5% bovine serum albumin (BSA) in phosphate-buffered saline
(PBS),
followed by incubation with sera (diluted in PBS) or MAbs for 2 h at room
temperature. Bound
antibodies were detected with peroxidase-conjugated rabbit anti-mouse
immunoglobulins
(DakoCytomation, Glostrup, Denmark) or isotype-specific antibodies peroxidase-
conjugated
goat anti-mouse IgM, IgG1, IgG2a, IgG2b, or IgG3 (Southern Biotechnology
Associates,
Birmingham, AL). Plates were developed with 0-phenylenediamine tablets
(DakoCytomation)
and read at 492 nm. Control antibodies included anti-MUC1 antibodies HMFG2 and
5M3
(Burchell etal., 1987) and anticarbohydrate antibodies 5F4 (Tn) and 3F1 (STn)
(Mandel etal.,
1991). Control sera included mice immunized with MUC4 mucin peptide linked to
KLH.
88
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
6.1.3. Results
[0306] Glycopeptide specific mAbs were produced to GaINAc-MUC1 using GaINAc-
MUC1 60-
mer glycopeptide conjugated to KLH as immunogen. Using an ELISA assay, the
generated
mAb G02 (5F7) reacted specifically with the MUC1 tandem repeat
(VTSAPDTRPAPGSTAPPAHG)3 (SEQ ID NO:47) that has been glycosylated in vitro
using
purified recombinant human glycosyltransferases GaINAc-T1, GaINAc-T2, and
GaINAc-T4, with
no reaction with the corresponding MUC1 peptide without GaINAc-residues or
irrelevant
glycopeptides with the same type of Tn glycoform. Results of the ELISA assay
are shown in
Figure 1.
6.2 Example 2: Characterization Of Anti-Glyco-Muc1 Antibodies
6.2.1. Overview
[0307] Monoclonal antibody G02 (5F7) was characterized for the specificity of
its binding to the
Tn glycoforms of MUC1 associated with cancer cells.
6.2.2. Materials and Methods
6.2.2.1 Immunocytochemistry
[0308] Cell lines were fixed for 10 min in ice-cold acetone or in
methanol:acetone. Fixed cells
were incubated overnight at 5 C with mouse sera (1:200/1:400/1:800) or MAbs,
followed by
incubation for 45 min at room temperature with fluorescein isothiocyanate
(FITC)-conjugated
rabbit anti-mouse immunoglobulins (DakoCytomation). Slides were mounted in
glycerol
containing p-phenylenediamine and examined in a Zeiss fluorescence microscope
(FluoresScience, Hallbergmoos, Germany).
6.2.2.2 Immunohistochemistry
[0309] Formalin fixed, paraffin wax embedded tissues of breast carcinoma were
obtained. All
cases were conventionally classified by histological type. The avidin¨biotin
peroxidase complex
method was used for immunostaining. Paraffin sections were dewaxed,
rehydrated, and treated
with 0.5% H202 in methanol for 30 min. Sections were rinsed in TBS and
incubated for 20 min
with rabbit non-immune serum. Sections were rinsed and incubated overnight at
5 C with
primary antibody. Sections were rinsed and incubated with biotin-labeled
rabbit anti-mouse
serum (DakoCytomation) diluted 1:200 in TBS for 30 min, rinsed with TBS, and
incubated for 1
h with avidin¨biotin peroxidase complex (DakoCyto- mation). Sections were
rinsed with TBS
and developed with 0.05% 3,3'-diaminobenzidine tetrahydrochloride freshly
prepared in 0.05 M
TBS containing 0.1% H202. Sections were stained with hematoxylin, dehydrated,
and mounted.
6.2.3. Results
[0310] lmmunohistochemistry of colorectal carcinoma, pancreatic carcinoma, and
invasive
breast adenocarcimas were perfromed with G02. Staining of colorectal cancer
tissue (Figure 2)
demonstrated strong labeling of intracellular and surface structures on a
large proportion of the
89
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
cancer cells. In contrast no or significantly lower reactivity was seen to
healthy columnar
epithelial cells. The labeling in healthy cells was restricted to
intracellular structures, which is
expected due to the presence of large amounts of biosynthetic intermediates
(GaINAc-modified
glycoproteins) in cells with high secretory capacity such as colonic cumnar
epithelia. A similar
pattern was observed with pancreatic (Figure 3) and breast cancer tissue
(Figure 4) with strong
reactivity with cancer cells and none or limited reactivity with intracellular
structures in
surrounding healthy epithelia or connective tissue cells.
6.3 Example 3: Sequence Analysis of Anti-Glyco-Muc1 Antibodies
[0311] mRNA from the hybridoma producing monoclonal antibody G02 (5F7) was
prepared,
reverse transcribed and sequenced.
[0312] The nucleotide sequences encoding the heavy and light chain variable
regions with their
signal sequences are set forth in SEQ ID NO:11 and SEQ ID NO:12, respectively.
The heavy
and light chain variable regions encoded by SEQ ID NO:11 and SEQ ID NO:12 are
set forth in
SEQ ID NO:1 and SEQ ID NO:2, respectively. The predicted mature variable
region sequences
(following truncation of the signal peptide) are set forth in SEQ ID NO:3 and
SEQ ID NO:4,
respectively, and are encoded by SEQ ID NO:13 and SEQ ID NO:14, respectively.
The
predicted heavy chain CDR sequences (IMGT definition) are set forth in SEQ ID
NOs:5-7,
respectively, and the predicted light chain CDR sequences (IMGT definition)
are set forth in
SEQ ID NOs:8-10, respectively.
6.4 Example 4: Drug Delivery to Cancer Cells with Anti-Glyco-Muc1
Antibodies
6.4.1. Overview
[0313] Monoclonal antibody G02 (5F7) was tested for its ability to deliver a
cytotoxic agent to
target cells.
6.4.2. Materials and Methods
[0314] OVCar human ovarian carcinoma cells were added to a 24-well cell
culture plate at
1,000 cells/well. Monoclonal antibody G02 and a secondary antibody conjugated
to the
antitubulin agent monomethyl auristatin F (MMAF) (anti-mFc-NC-MMAF) (Moradec
catalog no.
AM-101-AF) were added to the plate to give the following concentrations (in
pg/ml) of G02 and
ADC:
Table 2
Column
1 2 3 4 5 6
Row
G02: 5 G02: 1 G02: 0.2 G02: 0.04 G02: 0.008 G02: 0
A
ADC: 2.0 ADC: 2.0 ADC: 2.0 ADC: 2.0 ADC: 2.0 ADC: 2.0
G02: 5 G02: 1 G02: 0.2 G02: 0.04 G02: 0.008 G02: 0
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
Table 2
ADC: 0.6 ADC: 0.6 ADC: 0.6 ADC: 0.6 ADC: 0.6 ADC: 0.6
G02: 5 G02: 1 G02: 0.2 G02: 0.04 G02: 0.008 G02: 0
ADC: 0.2 ADC: 0.2 ADC: 0.2 ADC: 0.2 ADC: 0.2 ADC: 0.2
G02: 5 G02: 1 G02: 0.2 G02: 0.04 G02: 0.008 G02: 0
ADC: 0 ADC: 0 ADC: 0 ADC: 0 ADC: 0 ADC: 0
[0315] The plate was incubated at 37 C for 48 hours. After the 48 hour
incubation,
AlamarBlue (Invitrogen) was added to each well, and fluorescence at 600 nm
measured.
6.4.3. Results
[0316] Results are shown in Figure 5. The results show that the cellular
toxicity is dependent
on primary antibody (G02) concentration and presence of the antibody, and
secondary ADC
conjugated antibody concentration and presence. In other words, G02 induces
cellular toxicity
of this cancer cell line when coupled with a secondary antibody that carries a
cytotoxic agent
MMAF.
6.5 Example 5: Circulating Tumor Cell Quantification with Anti-Mud1
Antibodies
6.5.1. Overview
[0317] Monoclonal antibody G02 (5F7) was tested for its ability to be used to
quantify
circulating tumor cells.
6.5.2. Materials and Methods
[0318] G02 was conjugated to a magnetic separation bead and allowed to
interact with
samples of different concentrations of tumor cells. Cells and beads were
pulled out of solution
with a magnet and washed several times to remove unbound material. G02 that
was
conjugated to horseradish peroxidase was then applied to the magnetic
separation beads
containing bound cancer cells, incubated, and then unbound conjugated G02 was
washed
away. A colorimetric reaction was performed using TNB as a substrate. The
reaction was
terminated with sulfuric acid and then OD 450 readings were taken on the
samples.
6.5.3. Results
[0319] Results are shown in Figure 6. The results of the assay demonstrated
G02 binding of
tumor cells.
6.6 Example 6: Immunohistochemical staining of tumor tissue using Anti-
Glyco-Muc1 Antibodies
6.6.1. Materials and Methods
[0320] Sections from six formalin fixed, paraffin embedded (FFPE) tissue micro
arrays (TMAs)
were cut at 2.5 pm thickness. TMAs from breast cancer (BC), colorectal cancer
(CRC), ovarian
91
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
cancer (OVC), non-small cell lung cancer (NSCLC) and prostate cancer (PrC)
tumors were
used in the study. 25-47 tumor tissue cores per TMA were available for
evaluation. Core size
was 1 mm, 2 mm or 3mm, depending on the TMA. Each tissue core represented one
patient.
[0321] Staining was performed on a Discovery XT autostainer (Ventana Medical
Systems).
Following antigen retrieval with cell condition 1 (CC1) solution (Ventana
Medical Systems), the
G02 was applied at a concentration of 1 pg/ mL in Dako green medium antibody
diluent and
incubated for 60 min at 37 C. Binding of G02 to tumor cells was detected using
the Optiview
DAB IHC detection kit (Ventana Medical Systems) visualized in DAB (brown
precipitate).
6.6.2. Results
[0322] Representative images of MUC1 positive TMA tumor cores are shown in
Figure 7. In BC
and OVC TMAs, the majority of spots (> 90%) showed moderate or strong binding
of G02 to
tumor cells. 70% and 51% of NSCLC and CRC cases showed moderate and strong
binding of
the antibody to tumor cells, respectively. In prostate cancer, the antigen
appeared to be less
expressed. Only 28% of the spots in the TMA 1 revealed a moderate or strong
staining intensity
when applying GO-2. Staining patterns were always cytoplasmic and in many
cases
membrane-bound. An apical membrane staining pattern was observed in few cores.
6.7 Example 7: Production and purification of an anti-MUC1 antibody in
T-cell
bispecific (TCB) format
6.7.1. Materials and Methods
6.7.1.1 Expression vector production
[0323] The G02 antibody was converted into TCB format, including knob-into-
holes and
P329G/L234A/L235A ("PGLALA") mutations in the Fc region and charged residues
in the
MUC1 CH1 (147E/213E; "EE") and CL (123R/124K; "RK") regions (see SEQ ID NOs:
43-46).
The TCB antibody is illustrated in Figure 8. Briefly, the variable heavy and
variable light chains
of G02 mAb were synthesized (Geneart, Regensburg, Germany) and inserted into
suitable
expression vectors in which they are fused to the appropriate human constant
heavy or human
constant light chains. The expression cassettes in these vectors consist of
the CMV promoter,
lntron A with 5' UTR and a BGH polyadenylation site. In addition, the plasmids
contain the oriP
region from the Epstein Barr virus for the stable maintenance in HEK293 cells
harboring the
EBV nuclear antigen (EBNA).
6.7.1.2 .. Transient transfection and production
[0324] The antibodies were transiently produced in HEK293 EBNA cells using a
PEI mediated
transfection procedure as follows. HEK293 EBNA cells are cultivated in
suspension serum free
in Excell culture medium, containing 6 mM L-Glutamine. For the production of
antibodies in a
500 ml shake flask, 300 million HEK293 EBNA cells are seeded 24 hours before
transfection
(for alternative scales all amounts were adjusted accordingly). For
transfection, cells are
92
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
centrifuged for 10 min at 210 x g and the supernatant is replaced by pre-
warmed 20 ml CD
CHO medium. Expression vectors are mixed in 20 ml CD CHO medium to a final
amount of
200 pg DNA. After addition of 540 pl PEI, solution is vortexed for 15 s and
subsequently
incubated for 10 min at room temperature. Afterwards cells are mixed with the
DNA/PEI
solution, transferred to a 500 ml shake flask and incubated for 3 hours by 37
C in an incubator
with a 5 % CO2 atmosphere. After incubation, 160 ml Ex-cell medium (Sigma-
Aldrich),
containing 6mM glutamine, 1.25 mM valproic acid and 12.5% Pepsoy, is added and
cells are
cultivated for 24 hours. One day after transfection, 12% Feed 7 (48 mL) + 3
g/L glucose is
added. After 7 days, cultivation supernatant is collected for purification by
centrifugation for 45
min at 3000 x g. The solution is sterile filtered (0.22 pm filter) and sodium
azide in a final
concentration of 0.01 % w/v is added. The solution is then stored at 4 C.
6.7.1.3 Antibody purification
[0325] Secreted proteins were purified by affinity chromatography using
Protein A, followed by
size exclusion chromatography. For affinity chromatography, the supernatant
was loaded on a
Protein A MabSelect SuRe column (GE Healthcare) equilibrated with 20 mM sodium
phosphate, 20 mM sodium citrate pH 7.5. Unbound protein was removed by washing
with
equilibration buffer. The bound protein was eluted using either a step
(standard IgG) or a
gradient (bispecific antibody) elution created with 20 mM sodium citrate, 100
mM sodium
chloride, 100 mM glycine, pH 3Ø The pH of collected fractions was adjusted
by adding 1/10
(v/v) of 0.5 M sodium phosphate pH8Ø The protein was concentrated and
filtered prior to
loading on a HiLoad Superdex 200 column (GE Healthcare) equilibrated with 20
mM histidine,
140 mM sodium chloride, pH 6Ø
[0326] The aggregate content of eluted fractions was analyzed by analytical
size exclusion
chromatography. Therefore, 30 pl of each fraction was applied to a TSK G3000
SVVXL
column (TOSOH, 7.8 mm x 30 cm) equilibrated with 200 mM Arginine, 25 mM K2PO4,
125 mM
Sodium chloride, 0.02% NaN3, pH 6.7. Fractions containing less than 2 %
oligomers were
pooled and concentrated to final concentration of 1 - 1.5 mg/ml using
centrifugal concentrators
(Millipore, Amicone ULTRA ¨ 15, 30k MWCO). Purified proteins were stored at -
80 C.
6.7.2. Results
[0327] Production yield and quality of G02 TCB antibodies are shown in Table
5.
Table 5
Molecule Yield Monomer HMW [%] LMW [%] Purity Ng
[mg/L] Ng SEC CE-SOS
G02 TCB 0.51 94.16 0.00 5.84 85.61
93
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
6.8 Example 8: Jurkat-NFAT reporter assay to monitor target expression
ex
vivo in undigested patient-derived tumor samples
6.8.1. Overview
[0328] A Jurkat NFAT reporter assay was used to monitor target expression
(glyco-MUC1) ex
vivo in undigested primary human tumor samples using a G02 TCB.
6.8.2. Materials and Methods
6.8.2.1 Materials
= G02 TCB (see Example 7)
= DP47 TCB (non-targeted, negative control)
= Matrigel (Item no. 734-1101, Corning/VWR, Switzerland)
= Corning Costar Ultra-Low attachment multiwell plates (Item no. CL57007-
24EA,
Sigma)
= Cell culture microplate, 96 well (Item no. 655098, Greiner Bio-one,
Switzerland)
= RPM 11640 Medium (Item no. 42401-018, FisherScientific, Schweiz)
= Jurkat Medium: RPMI1640 Medium with 2 g/I D-Glucose, 2 g/I NaHCO3, 10 %
FCS, 25
mM HEPES, 2 mM L-Glutamine, 1 x NEAA, 1 x Sodium-Pyruvate, 200 pg / ml
Hygromycine B
= Jurkat NFAT luciferase reporter cells (Promega)
= Tumor samples received from I ndivumed GmbH, Germany. Samples were
shipped over
night in transport medium. About 24 h after surgery the samples were cut in
small
pieces.
6.8.2.2 Methods
[0329] 96-well cell culture microplates were prepared by adding 17 pl of cold
matrigel. The
plate was incubated for 2 min at 37 C before tumor pieces were added
(triplicates). 33 pl of
cold matrigel was added per well and the plate was incubated again for 2 min
at 37 C. 100 pl
(50 nM or 5 nM) of TCB antibody dilution (diluted in Jurkat medium without
Hygromycine but
with 2X Penicillin/Streptomycine) was added per well. Jurkat-NFAT reporter
cells were
harvested and viability was assessed using ViCell. Cells were centrifuged at
350 x g for 7 min
before they were resuspended in Jurkat medium without Hygromycine. 50 pl of
the cell
suspension was added per well (50,000 cells / well). The plate was incubated
for 4 to 5 h at 37
C in a humidified incubator before it was taken out for luminescence read out.
50 pl of ONE-
Glo solution was added to each well and incubated for 10 min at room
temperature in the dark.
Luminescence was detected using WALLAC Victor3 ELISA reader (PerkinElmer2030),
with a 5
sec/well detection time.
94
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
6.8.3. Results
[0330] Results from tumor samples from three patients are shown in Figures 9-
11. The results
shown in Figure 9 are from tumor samples obtained from a patient having a
malignant
neoplasm of bronchus and lung: middle lobe, bronchus or lung, squamous cell
carcinoma. The
results shown in Figure 10 are from tumor samples obtained from a patient
having a malignant
neoplasm of bronchus and lung: lower lobe, bronchus or lung, non-keratinizing
squamous cell
carcinoma. The results shown in Figure 11 are from tumor samples obtained from
a patient
having a malignant neoplasm of bronchus and lung: upper lobe, bronchus or
lung,
adenocarcinoma with acinar type. Each bar in Figures 9-11 represents the mean
of triplicates.
Standard error is indicated by error bars. The dotted line indicates
luminescence for Jurkat
NFAT cells incubated with tumor samples without any TCB. Two-tailed, unpaired
t-test was
used for statistical analysis. P-values below 0.05 were considered as
significant and were
indicated with stars (* P 0.05; ** P 0.001; *** P 0.001). In each of Figures 9-
11, tumor
samples incubated with G02 TCB displayed significantly more luminescence than
samples
incubated with DP47 negative control TCB.
6.9 Example 9: In vitro characterization of G02 TCB
6.9.1. Overview
[0331] G02 TCB (Example 7) recognizing the tumor-specific aberrantly
glycosylated MUC1
was functionally characterized on tumor cells expressing MUC1.
6.9.2. Materials and Methods
6.9.2.1 Cell lines and PBMCs
[0332] T3M4 pfzv and MCF7 es engineered tumor cell lines were cultured in DMEM
with 10%
FCS and 2 mM Glutamine. MCF10A is a human non-tumorigenic mammary epithelial
cell line
(ATCCO CRL-10317). HBEpiC are human bronchial epithelial cells (Science!!
#3210). PBMCs
were isolated by gradient centrifugation using whole blood from healthy
volunteers.
6.9.2.2 Target binding by flow cytometry
[0333] Target cells as indicated were harvested with Cell Dissociation Buffer,
washed with PBS
and resuspended in FACS buffer. The antibody staining was performed in a 96-
well round
bottom plate. 200,000 cells were seeded per well. The plate was centrifuged
for 4 min at 400g
and the supernatant was removed. The test antibodies were diluted in FACS
buffer and 30 pl of
the antibody solution was added to the cells for 30 min at 4 C. To remove
unbound antibody
the cells were washed twice with FACS buffer before addition of the diluted
secondary antibody
(PE-conjugated AffiniPure F(ab')2 Fragment goat anti-human IgG FCy Fragment
Specific,
Jackson ImmunoResearch #109-116-170). After 30 min incubation at 4 C unbound
secondary
antibody was washed away. Before measurement the cells were resuspended in 200
pl FACS
buffer and analyzed by flow cytometry using BD Fortessa. Assays were performed
in triplicates.
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
6.9.2.3 T cell mediated tumor cell killing and T cell activation
[0334] Target cells were harvested with Trypsin/EDTA, counted and viability
was checked. The
cells were resuspended in their respective medium with a final concentration
of 300,000 cells
per ml. Then 100 pl of the target cell suspension was transferred into each
well of a 96-flat
bottom plate. The plate was incubated overnight at 37 C in the incubator to
allow adherence of
the cells to the plate. On the next day PBMCs were isolated from whole blood.
The blood was
diluted 2:1 with PBS and overlayed on 15 ml Histopaque-1077 (# 10771, Sigma-
Aldrich) in
Leucosep tubes and centrifuged for 30 min at 450g without brake. After
centrifugation, the band
containing the cells was collected with a 10 ml pipette and transferred into
50 ml tubes. The
tubes were filled up with PBS until 50 ml and centrifuged (400g, 10 min, room
temperature).
The supernatant was removed and the pellet resuspended in PBS. After
centrifugation (300g,
min, room temperature), supernatants were discarded, 2 tubes were pooled and
the
washing step was repeated (this time centrifugation 350g, 10 min, room
temperature).
Afterwards, the cells were resuspended and the pellets pooled in 50 ml PBS for
cell counting.
After counting cells were centrifuged (350g, 10 min, room temperature) and
resuspended at 6
million cells per ml in RPM! with 2 % FCS and 2 nM Glutamine. Medium was
removed from
plated target cells and the test antibodies diluted in RPM! with 2% FCS and 2
nM Glutamine
were added. 300,000 cells of the effector cell solution were transferred to
each well resulting in
a E:T ratio of 10:1. To determine the maximal release target cells were lysed
with Triton X-100.
LDH release was determined after 24 h and 48 h using Cytotoxicity Detection
Kit (1644793,
Roche Applied Science). Activation marker upregulation on T cells after tumor
cell killing was
measured by flow cytometry. Briefly, PBMCs were harvested, transferred into a
96-well round
bottom plate and stained with CD4 APC (300514, BioLegend), CD8 FITC (344704,
BioLegend),
CD25 BV421 (302630, BioLegend), CD69 PE (310906, BioLegend) antibodies diluted
in FACS
buffer. After 30 min incubation at 4 C the cells were washed twice with FACS
buffer. Before
measuring the fluorescence using BD Fortessa II the cells were resuspended in
200 pl FACS
buffer. Assays were performed in triplicates.
6.9.2.4 Cytokine/chemokine release by cytometric bead array
[0335] Cytokine/chemokine secretion in the supernatant was measured by flow
cytometry,
using the cytometric bead array (CBA), according to the manufacturer's
guidelines.
Supernatants from T cell mediated killing assays were collected and stored at -
20 C.
Supernatants were subsequently thawed and tested according to manufacturer's
instructions.
The following CBA kits (BD Biosciences) were used: CBA human interferon gamma
(IFNy) Flex
Set (E7), CBA human Granzyme B Flex Set (D7), CBA human 1L6 Flex Set (A7), CBA
human
1L8 Flex Set (A9), CBA human I L10 Flex Set (B7) and CBA human tumor necrosis
factor (TN F)
Flex Set (D9). Samples were measured using the BD FACS Canto II and analyses
were
performed using the Diva Software (BD Biosciences). Assays were performed in
triplicates.
96
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
6.9.3. Results
[0336] Binding of G02 TCB to the breast cancer cell line MCF7 and the
pancreatic cancer cell
line T3M4, both engineered to express the aberrantly glycosylated MUC1, was
confirmed
(Figure 12). Subsequently, activity of G02 TCB was tested on both tumor cell
lines, MCF7 and
T3M4, using freshly insolated PBMCs (Figure 13 and Figure 14). Tumor cell
killing of both cell
lines was detected after 24 h and even stronger after 48 h. This was
accompanied by strong
activation of CD4 T cells and CD8 T cells determined by upregulation of the
two activation
markers 0D25 and 0D69 and the release of IL6, IL8, IL10, IFNy, TNFa and
Granzyme B into
the supernatant. As negative control the respective untargeted TCB was
included.
[0337] To prove that G02 TCB does not bind to normally glycosylated MUC1 on
epithelial cells,
binding to MCF10A, which is a human non-tumorigenic mammary epithelial cell
line, and to
HBEpiC, which are primary human bronchial epithelial cells, was tested. As a
positive control
the HMFG1 TCB, which does not discriminate between MUC1 expressed on normal
and on
tumor cells, was included. HFMG1 TCB was found to bind to both tested cells,
confirming the
expression of MUC1, but G02 TCB was not able to bind to these cells (Figure
15). In addition,
G02 TCB was tested to see if it would induce killing or T cell activation in
the presence of
normal epithelial cells expressing MUC1. This was tested on MCF10A cells and
there was no
killing or T cell activation detectable with G02 TCB, whereas HMFG1 TCB
induced killing as
well as T cell activation (Figure 16).
6.10 Example 10: Functional characterization of G02 and G02 TCB antibodies
by surface plasmon resonance
6.10.1. Overview
[0338] G02 and G02 TCB (Example 7) were characterized by surface plasmon
resonance.
6.10.2. Materials and Methods
6.10.2.1 Binding of G02 and G02 TCB to immobilized
glycopeptides
[0339] Binding of the G02 antibody and G02 TCB to human and cynomolgus
glycopeptides
(Table 6) was assessed by surface plasmon resonance (SPR). All SPR experiments
were
performed on a Biacore T200 at 25 C with HBS-EP as running buffer (0.01 M
HEPES pH 7.4,
0.15 M NaCI, 3 mM EDTA, 0.005% Surfactant P20, Biacore, Freiburg/Germany).
Table 6
Glycopeptide Sequence Concentration in PBS
Human peptide PDTSAAPGSTAPPAHVVTSAP 0.9 mg/ml
(SEQ ID NO:48)
Cynomolgus peptide PDTSAAPGSTGPPAHVVTSAP 1.8 mg/ml
(SEQ ID NO:49)
97
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
[0340] The biotinylated glycopeptides were dissolved in PBS and the final
concentration was
between 0.9 and 1.8 mg/ml (Table 6). Biotinylated glycopeptides were directly
coupled to a flow
cell of a streptavidin (SA) sensor chip. Immobilization levels up to 880
resonance units (RU)
were used. The G02 antibody or the G02 TCB were injected with a flow of 30
pliminute
through the flow cells, over 240 seconds and at a concentration of 1000 nM
(Figure 17). The
dissociation was monitored for 500 sec. Bulk refractive index differences were
corrected for by
subtracting the response obtained in a reference flow cell, where no protein
was immobilized.
6.10.2.2 Avidity of G02 and G02 TCB to immobilized
glycopeptides
[0341] The avidity of G02 and G02 TCB was assessed by surface plasmon
resonance (SPR).
All SPR experiments were performed on a Biacore T200 at 25 C with HBS-EP as
running
buffer (0.01 M HEPES pH 7.4, 0.15 M NaCI, 3 mM EDTA, 0.005% Surfactant P20,
Biacore,
Freiburg/Germany). Biotinylated glycopeptides (Table 6) were directly coupled
to a flow cell of a
streptavidin (SA) sensor chip. Immobilization levels up to 200 resonance units
(RU) were used.
[0342] The G02 antibody or the G02 TCB were injected with a flow of 30
pliminute through
the flow cells over 120 seconds and at a concentration range from 3.9 to 1000
nM (1:2 dilution).
The dissociation was monitored for 400 sec. Bulk refractive index differences
were corrected for
by subtracting the response obtained in a reference flow cell, where no
protein was
immobilized. The KDs were derived, despite the bivalency of the interaction,
by fitting the curve
to a 1:1 Langmuir binding using the Biaeval software (GE Healthcare). The
"apparent" KD can
therefore be used for comparison purposes only.
6.10.3. Results
[0343] As can be seen in the sensorgrams of Figure 18, G02 antibody (Figure
18A) and G02
TCB (Figure 18B) bind both human and cynomolgous glycopeptides. G02 antibody
and G02
TCB bind with higher avidity to cynomolgus than to human glycopeptide.
[0344] As can be seen in Figure 19, binding of a bivalent G02 binder (IgG,
TCB) to human
glycopeptide is in the three-digit nanomolar (Figure 19A and 190), whereas
binding to
cynomolgus glycopeptide is in the two-digit nanomolar (Figure 19B and 19D).
6.11 Example 11: Exploratory single dose pharmacokinetic and tolerability
study of G02 TCB in cynomolgus monkeys
6.11.1. Overview
[0345] The objectives of this study are to determine the pharmacokinetics and
tolerability of
the G02 TCB described in Example 7, when given by a single intravenous
injection to
cynomolgus monkeys.
98
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
6.11.2. Materials and Methods
6.11.2.1 Preparation of G02 TCB
[0346] Thawing of the frozen stock solution of G02 TCB (2.12 mg/mL) and
formulation buffer
(20 mM Histidine, 140 mM NaCI, 0.01% Tween 20; pH 6.0) is done overnight in a
fridge set to
maintain 4 C. Test item dosing formulations are prepared under sterile
conditions at
appropriate concentrations to meet dose level requirements by dilution with
formulation buffer.
[0347] The dosing formulation is prepared within 2 hours before injection and
stored at room
temperature until use. Polypropylene containers are used for preparation and
storage of dosing
formulation to prevent adsorption. Dosing formulations should are not
filtered, nor stirred or
shaken. Any mixing is done either by gentle pipetting or gentle swinging of
the container.
6.11.2.2 Animals
[0348] Cynomolgus monkeys (Macaca fascicularis) 2-4 years of age and weighing
less
than 4 kg are used in the study. The animals are allowed to acclimate to the
test facility
primate toxicology accommodation for at least 6 weeks before the commencement
of
dosing.
[0349] During the week before the commencement of dosing, the animals are
approved for
entry into the experiment on the basis of a satisfactory veterinary
examination (performed
shortly after arrival), clinical observation records, body weight profile and
clinical pathology
investigations.
[0350] Animals selected for the study are randomly allocated to cages based on
supplied group
compatibility information and then allotted individual study numbers. The
animals are allocated
a cage in groups of up to 5.
6.11.2.3 Husbandry
[0351] Animals are socially housed where possible, in groups of up to 5 by sex
in two storey
gang pens measuring 1.61 x 1.66 x 2.5 m on the lower storey and 1.61 x 1.66 x
2.03 m on the
upper. Bedding material is wood shavings. There are no known contaminants in
the bedding
that would interfere with the objectives of the study.
[0352] The targeted conditions for animal room environment are be as follows:
Temperature: 18 - 24 C
Humidity: 40 - 70%
Ventilation: a minimum of 10 air changes per hour
Light Cycle: 12 hours light and 12 hours dark (except when interrupted by
study
procedures/activities).
[0353] There is automatic control of temperature and humidity which is
continuously monitored
and recorded. There is automatic control of light cycle.
99
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
[0354] Special Diets Services (SDS) MP(E) Short SQC Diet is provided as a
daily ration
throughout the study. Approximately 200 gram ration of feed per animal is
provided once daily.
There are no known contaminants in the feed that would interfere with the
objectives of the
study. The animals have access to water ad libitum from the public supply.
There are no
known contaminants in the water that would interfere with the objectives of
the study.
[0355] The animal's home environment is enriched to promote social
interaction, play and
exploration. The animals have perches and materials such as plastic toys,
balls, climbing
frames and stainless steel mirrors. These are exchanged frequently to reduce
familiarity. Prior
to exchange, all toys and climbing frames are thoroughly cleaned to avoid
cross-contamination.
The animals are also offered a range of other treats such as forage mix,
vegetables, nuts,
biscuits and fruits normally on a daily basis.
[0356] Veterinary care is available throughout the course of the study and
animals are
examined by the veterinary staff as warranted by clinical signs or other
changes.
6.11.2.4 Experimental design
[0357] One male and one female animal are administered G02 TCB
Table 7
Group No. Dose Level (pg/kg) Dose Volume Dose Concentration
(mL/kg) (pg/kg)
1 (1 male and 1 100 1 100
female)
2 (1 male and 1 300 1 300
female)
[0358] G02 TCB is administered to the appropriate animals by a single
intravenous slow
bolus injection (1-2 min) in the saphenous vein or tail vein at least 8 days
apart. is
staggered so that only one male and one female receive a new dose level on any
single
day. Based on the observations from the previous dose level (including
clinical pathology
data), the doses are increased or decreased for the next dose group. Naïve
animals are
used for each dose level. The doses are given using a syringe with attached
Vygon infusion
needle.
[0359] Animals are necropsied ca 72 hours after dosing (after the last
scheduled sample
has been taken). For all animals which have to be terminated prior to the
scheduled date
due to severe clinical symptoms, complete panel of clinical pathology
(additional sampling
prior to termination, if feasible) and histopathology are analyzed.
[0360] The intravenous injection route of administration has been selected for
this study as this
route is a possible route of clinical application. The low dose and the high
dose levels were
chosen to cover a clinically relevant dose range and to minimize the potential
harm to the
100
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
animals. The low dose was selected based on the experience in the cynomolgus
monkey with
similar T-cell bispecific antibodies of similar potency and the high-dose
represents a 3-fold
increment thereof.
6.11.3. Results
[0361] G02 TCB is tolerated at the tested doses.
7. SPECIFIC EMBODIMENTS, CITATION OF REFERENCES
[0362] While various specific embodiments have been illustrated and described,
it will be
appreciated that various changes can be made without departing from the spirit
and scope of
the disclosure(s). The present disclosure is exemplified by the numbered
embodiments set forth
below.
1. An anti-glyco-MUC1 antibody or antigen binding fragment that:
a. preferentially binds to a glyco-MUC1 epitope that is overexpressed on
cancer
cells as compared to normal cells; and
b. competes with an antibody or antigen binding fragment comprising a heavy
chain variable (VH) sequence of SEQ ID NO:3 and a light chain variable (VL)
sequence of SEQ ID NO:4 for binding to the breast cancer cell line MCF7 or
T47D.
2. An anti-Glyco-MUC1 antibody or antigen binding fragment that
a. binds to the MUC1 tandem repeat (VTSAPDTRPAPGSTAPPAHG)3 that has
been glycosylated in vitro using purified recombinant human
glycosyltransferases GaINAc-T1, GaINAc-T2, and GaINAc-T4, and (referred to
hereinafter as the "first epitope"); and
b. competes with an antibody or antigen binding fragment comprising a heavy
chain variable (VH) sequence of SEQ ID NO:3 and a light chain variable (VL)
sequence of SEQ ID NO:4 for binding to the breast cancer cell line MCF7 or
T47D.
3. The anti-glyco-MUC1 antibody or antigen-binding fragment of
embodiment 1 or
embodiment 2 comprising a complementarity determining region (CDR) H1
comprising the
amino acid sequence of SEQ ID NO:33, a CDR-H2 comprising the amino acid
sequence of
SEQ ID NO:29, a CDR-H3 comprising the amino acid sequence of SEQ ID NO:25, a
CDR-
L1 comprising the amino acid sequence of SEQ ID NO: 8, a CDR-L2 comprising the
amino
acid sequence of SEQ ID NO:9, and a CDR-L3 comprising the amino acid sequence
of
SEQ ID NO:31.
101
CA 03078812 2020-04-08
WO 2019/083506
PCT/US2017/058036
4. The anti-glyco-MUC1 antibody or antigen-binding fragment of
embodiment 3,
wherein CDR-H1 comprises the amino acid sequence of SEQ ID NO:5.
5. The anti-glyco-MUC1 antibody or antigen-binding fragment of
embodiment 3,
wherein CDR-H1 comprises the amino acid sequence of SEQ ID NO:23.
6. The anti-glyco-MUC1 antibody or antigen-binding fragment of
embodiment 3,
wherein CDR-H1 comprises the amino acid sequence of SEQ ID NO:28.
7. The anti-glyco-MUC1 antibody or antigen-binding fragment of
embodiment 3,
wherein CDR-H1 comprises the amino acid sequence of SEQ ID NO:32.
8. The anti-glyco-MUC1 antibody or antigen-binding fragment of any one
of
embodiments 3 to 7, wherein CDR-H2 comprises the amino acid sequence of SEQ ID
NO:6.
9. The anti-glyco-MUC1 antibody or antigen-binding fragment of any one
of
embodiments 3 to 7, wherein CDR-H2 comprises the amino acid sequence of SEQ ID
NO:24.
10. The anti-glyco-MUC1 antibody or antigen-binding fragment of any one
of
embodiments 3 to 9, wherein CDR-H3 comprises the amino acid sequence of SEQ ID
NO:7.
11. The anti-glyco-MUC1 antibody or antigen-binding fragment of any one
of
embodiments 3 to 10, wherein CDR-L1 comprises the amino acid sequence of SEQ
ID
NO:30.
12. The anti-glyco-MUC1 antibody or antigen-binding fragment of any one
of
embodiments 3 to 10, wherein CDR-L1 comprises the amino acid sequence of SEQ
ID
NO:26.
13. The anti-glyco-MUC1 antibody or antigen-binding fragment of any one
of
embodiments 3 to 12, wherein CDR-L2 comprises the amino acid sequence of SEQ
ID
NO:27.
14. The anti-glyco-MUC1 antibody or antigen-binding fragment of any one
of
embodiments 3 to 13, wherein CDR-L3 comprises the amino acid sequence of SEQ
ID
NO:10.
102
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
15. The anti-glyco-MUC1 antibody or antigen-binding fragment of embodiment
1 or
embodiment 2 in which the VH comprises complementarity determining regions
(CDRs) of
SEQ ID NOS:5-7 and the VL comprises CDRs of SEQ ID NOS:8-10.
16. The anti-glyco-MUC1 antibody or antigen-binding fragment of embodiment
1 or
embodiment 2 in which the VH comprises complementarity determining regions
(CDRs) of
SEQ ID NOS:23-25 and the VL comprises CDRs of SEQ ID NOS:26, 27, and 10.
17. The anti-glyco-MUC1 antibody or antigen-binding fragment of embodiment
1 or
embodiment 2 in which the VH comprises complementarity determining regions
(CDRs) of
SEQ ID NOS:28, 29, and 25 and the VL comprises CDRs of SEQ ID NOS:30, 9, and
31.
18. The anti-glyco-MUC1 antibody or antigen-binding fragment of any one of
embodiments 1 to 17 which is a chimeric or humanized antibody.
19. The anti-glyco-MUC1 antibody or antigen-binding fragment of any one of
embodiments 1 to 18 in which the VH comprises an amino acid sequence having at
least
95% sequence identity to SEQ ID NO:3 and the VL comprises an amino acid
sequence
having at least 95% sequence identity to SEQ ID NO:4.
20. The anti-glyco-MUC1 antibody or antigen-binding fragment of any one of
embodiments 1 to 18 in which the VH comprises an amino acid sequence having at
least
97% sequence identity to SEQ ID NO:3 and the VL comprises an amino acid
sequence
having at least 97% sequence identity to SEQ ID NO:4.
21. The anti-glyco-MUC1 antibody or antigen-binding fragment of any one of
embodiments 1 to 18 in which the VH comprises an amino acid sequence having at
least
99% sequence identity to SEQ ID NO:3 and the VL comprises an amino acid
sequence
having at least 99% sequence identity to SEQ ID NO:4.
22. The anti-glyco-MUC1 antibody or antigen-binding fragment of embodiment
1 or
embodiment 2 in which the VH comprises the amino acid sequence of SEQ ID NO:3
and
the VL comprises the amino acid sequence of SEQ ID NO:4.
23. The anti-glyco-MUC1 antibody or antigen-binding fragment of any of
embodiments 1 to 22 which is multivalent.
24. The anti-glyco-MUC1 antibody or antigen-binding fragment of any of
embodiments 1 to 22 which is in the form of a single-chain variable fragment
(scFv).
103
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
25. The anti-glyco-MUC1 antibody or antigen-binding fragment of embodiment
24
wherein the scFv comprises the heavy chain variable fragment N-terminal to the
light chain
variable fragment.
26. The anti-glyco-MUC1 antibody or antigen-binding fragment of embodiment
24
wherein the scFv heavy chain variable fragment and light chain variable
fragment are
covalently bound to a linker sequence of 4-15 amino acids.
27. The anti-glyco-MUC1 antibody or antigen-binding fragment of any of
embodiments 1 to 22 which is in the form of a multispecific antibody.
28. The anti-glyco-MUC1 antibody or antigen-binding fragment of embodiment
27
wherein the multispecific antibody is a bispecific antibody that binds to a
second epitope
that is different from the first epitope.
29. The anti-glyco-MUC1 antibody or antigen-binding fragment of embodiment
28,
wherein the bispecific antibody is a CrossMab, a Fab-arm exchange antibody, a
bispecific
T-cell engager (BiTE), or a dual-affinity retargeting molecule (DART).
30. The anti-glyco-MUC1 antibody or antigen-binding fragment of embodiment
29,
wherein the bispecific antibody is a CrossMab.
31. The anti-glyco-MUC1 antibody or antigen-binding fragment of embodiment
30,
wherein the bispecific antibody is a CrossMabFAB.
32. The anti-glyco-MUC1 antibody or antigen-binding fragment of embodiment
30,
wherein the bispecific antibody is a CrossMabv"-vL.
33. The anti-glyco-MUC1 antibody or antigen-binding fragment of embodiment
30,
wherein the bispecific antibody is a CrossMabcHl-CL.
34. The anti-glyco-MUC1 antibody or antigen-binding fragment of embodiment
29,
wherein the bispecific antibody is a Fab-arm exchange antibody.
35. The anti-glyco-MUC1 antibody or antigen-binding fragment of embodiment
29,
wherein the bispecific antibody is a dual-affinity retargeting molecule
(DART).
36. The anti-glyco-MUC1 antibody or antigen-binding fragment of embodiment
29,
wherein the bispecific antibody is a bispecific T-cell engager (BiTE).
37. The anti-glyco-MUC1 antibody or antigen-binding fragment of any one of
embodiments 28 to 35, wherein the second epitope is a MUC1 epitope.
104
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
38. The anti-glyco-MUC1 antibody of antigen-binding fragment of any one of
embodiments 28 to 35, wherein the second epitope is a MUC1 epitope that is
overexpressed on cancer cells as compared to normal cells.
39. The anti-glyco-MUC1 antibody or antigen-binding fragment of any one of
embodiments 28 to 36, wherein the second epitope is a T-cell epitope.
40. The anti-glyco-MUC1 antibody or antigen-binding fragment of embodiment
39,
wherein the T-cell epitope comprises a CD3 epitope, a CD8 epitope, a CD 16
epitope, a
0D25 epitope, a 0D28 epitope, or an NKG2D epitope.
41. The anti-glyco-MUC1 antibody or antigen-binding fragment of embodiment
40,
wherein the T-cell epitope comprises a CD3 epitope, which is optionally an
epitope present
in human CD3.
42. The anti-glyco-MUC1 antibody or antigen-binding fragment of embodiment
41,
wherein the CD3 epitope comprises a CD3 gamma epitope, a CD3 delta epitope, a
CD3
epsilon epitope, or a CD3 zeta epitope.
43. The anti-glyco-MUC1 antibody or antigen-binding fragment of any one of
embodiments 1 to 42 which is conjugated to a detectable moiety.
44. The anti-glyco-MUC1 antibody or antigen binding fragment of embodiment
43 in
which the detectable marker is an enzyme, a radioisotope, or a fluorescent
label.
45. A bispecific antibody comprising a first antigen binding domain that
binds to CD3
(optionally human CD3) and a second antigen binding domain that binds to glyco-
MUC1,
wherein the bispecific antibody competes with an antibody or antigen binding
fragment
comprising a heavy chain variable (VH) sequence of SEQ ID NO:3 and a light
chain
variable (VL) sequence of SEQ ID NO:4 for binding to the breast cancer cell
line MCF7 or
T47D, and wherein the first antigen binding domain comprises a heavy chain
variable
region comprising the heavy chain CDR-H1 of SEQ ID NO:34, the CDR-H2 of SEQ ID
NO:35, and the CDR-H3 of SEQ ID NO:36; and a light chain variable region
comprising the
light chain CDR-L1 of SEQ ID NO:37, the CDR-L2 of SEQ ID NO:38 and the CDR-L3
of
SEQ ID NO:39.
46. The bispecific antibody of embodiment 45, wherein the second antigen
binding
domain comprises (i) a heavy chain variable region comprising the CDR-H1 of
SEQ ID NO:
5, the CDR-H2 of SEQ ID NO: 6, and the CDR-H3 of SEQ ID NO: 7; and a light
chain
variable region comprising the light chain CDR-L1 of SEQ ID NO: 8, the CDR-L2
of SEQ ID
NO: 9 and the CDR-L3 of SEQ ID NO:10.
105
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
47. The bispecific antibody of embodiment 45 or embodiment 46, wherein the
first
antigen binding domain comprises a heavy chain variable region sequence that
is at least
about 95%, 96%, 97%, 98%, 99% or 100% identical to the amino acid sequence of
SEQ ID
NO:40 and a light chain variable region sequence that is at least about 95%,
96%, 97%,
98%, 99% or 100% identical to the amino acid sequence of SEQ ID NO:41.
48. The bispecific antibody of embodiment 47, the first antigen binding
domain
comprises the heavy chain variable region sequence of SEQ ID NO:40 and the
light chain
variable region sequence of SEQ ID NO:41.
49. The bispecific antibody of any one of embodiments 45 to 48, wherein the
second
antigen binding domain comprises a heavy chain variable region sequence that
is at least
about 95%, 96%, 97%, 98%, 99% or 100% identical to the amino acid sequence of
SEQ ID
NO:3 and a light chain variable region sequence that is at least about 95%,
96%, 97%,
98%, 99% or 100% identical to the amino acid sequence of SEQ ID NO:4.
50. The bispecific antibody of embodiment 49, wherein the second antigen
binding
domain comprises the heavy chain variable region sequence of SEQ ID NO:3 and
the light
chain variable region sequence of SEQ ID NO:4.
51. The bispecific antibody of any one of embodiments 45 to 50, wherein the
first
and/or the second antigen binding domain is a Fab molecule.
52. The bispecific antibody of embodiment 51, wherein the first antigen
binding
domain is a crossover Fab molecule, wherein either the variable or the
constant regions of
the Fab light chain and the Fab heavy chain are exchanged.
53. The bispecific antibody of embodiment 52, wherein the first and the
second
antigen binding domain of the bispecific antibody are both Fab molecules, and
in one of the
antigen binding domains (particularly the first antigen binding domain) the
variable domains
VL and VH of the Fab light chain and the Fab heavy chain are replaced by each
other,
wherein
a. in the constant domain CL of the first antigen binding domain the amino
acid at
position 124 is substituted by a positively charged amino acid (numbering
according to Kabat), and wherein in the constant domain CH1 of the first
antigen
binding domain the amino acid at position 147 or the amino acid at position
213
is substituted by a negatively charged amino acid (numbering according to
Kabat
EU index); or
106
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
b. in the constant domain CL of the second antigen binding domain the amino
acid
at position 124 is substituted by a positively charged amino acid (numbering
according to Kabat), and wherein in the constant domain CH1 of the second
antigen binding domain the amino acid at position 147 or the amino acid at
position 213 is substituted by a negatively charged amino acid (numbering
according to Kabat EU index),
wherein the constant domains CL and CH1 of the antigen binding domain having
the
VHA/L exchange are not replaced by each other.
54. The bispecific antibody of embodiment 53, wherein
a. in the constant domain CL of the first antigen binding domain the amino
acid at
position 124 is substituted independently by lysine (K), arginine (R) or
histidine
(H) (numbering according to Kabat), and in the constant domain CH1 of the
first
antigen binding domain the amino acid at position 147 or the amino acid at
position 213 is substituted independently by glutamic acid (E), or aspartic
acid
(D) (numbering according to Kabat EU index); or
b. in the constant domain CL of the second antigen binding domain the amino
acid
at position 124 is substituted independently by lysine (K), arginine (R) or
histidine
(H) (numbering according to Kabat), and in the constant domain CH1 of the
second antigen binding domain the amino acid at position 147 or the amino acid
at position 213 is substituted independently by glutamic acid (E), or aspartic
acid
(D) (numbering according to Kabat EU index).
55. The bispecific antibody of embodiment 54, wherein in the constant
domain CL of
the second antigen binding domain the amino acid at position 124 is
substituted
independently by lysine (K), arginine (R) or histidine (H) (numbering
according to Kabat),
and in the constant domain CH1 of the second antigen binding domain the amino
acid at
position 147 or the amino acid at position 213 is substituted independently by
glutamic acid
(E), or aspartic acid (D) (numbering according to Kabat EU index).
56. The bispecific antibody of embodiment 55, wherein in the constant
domain CL of
the second antigen binding domain the amino acid at position 124 is
substituted
independently by lysine (K), arginine (R) or histidine (H) (numbering
according to Kabat),
and in the constant domain CH1 of the second antigen binding domain the amino
acid at
position 147 is substituted independently by glutamic acid (E), or aspartic
acid (D)
(numbering according to Kabat EU index).
107
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
57. The bispecific antibody of embodiment 55, in the constant domain CL of
the
second antigen binding domain the amino acid at position 124 is substituted
independently
by lysine (K), arginine (R) or histidine (H) (numbering according to Kabat)
and the amino
acid at position 123 is substituted independently by lysine (K), arginine (R)
or histidine (H)
(numbering according to Kabat), and in the constant domain CH1 of the second
antigen
binding domain the amino acid at position 147 is substituted independently by
glutamic acid
(E), or aspartic acid (D) (numbering according to Kabat EU index) and the
amino acid at
position 213 is substituted independently by glutamic acid (E), or aspartic
acid (D)
(numbering according to Kabat EU index).
58. The bispecific antibody of embodiment 57, wherein in the constant
domain CL of
the second antigen binding domain the amino acid at position 124 is
substituted by lysine
(K) (numbering according to Kabat) and the amino acid at position 123 is
substituted by
lysine (K) (numbering according to Kabat), and in the constant domain CH1 of
the second
antigen binding domain the amino acid at position 147 is substituted by
glutamic acid (E)
(numbering according to Kabat EU index) and the amino acid at position 213 is
substituted
by glutamic acid (E) (numbering according to Kabat EU index).
59. The bispecific antibody of embodiment 57, wherein in the constant
domain CL of
the second antigen binding domain the amino acid at position 124 is
substituted by lysine
(K) (numbering according to Kabat) and the amino acid at position 123 is
substituted by
arginine (R) (numbering according to Kabat), and in the constant domain CH1 of
the second
antigen binding domain the amino acid at position 147 is substituted by
glutamic acid (E)
(numbering according to Kabat EU index) and the amino acid at position 213 is
substituted
by glutamic acid (E) (numbering according to Kabat EU index).
60. The bispecific antibody of any one of embodiments 53 to 59, wherein the
constant domain CL of the second antigen binding domain is of kappa isotype.
61. The bispecific antibody of any one of embodiments 45 to 60, wherein the
first
and the second antigen binding domain are fused to each other, optionally via
a peptide
linker.
62. The bispecific antibody of embodiment 61, wherein the first and the
second
antigen binding domain are each a Fab molecule and either (i) the second
antigen binding
domain is fused at the C-terminus of the Fab heavy chain to the N-terminus of
the Fab
heavy chain of the first antigen binding domain, or (ii) the first antigen
binding domain is
fused at the C-terminus of the Fab heavy chain to the N-terminus of the Fab
heavy chain of
the second antigen binding domain.
108
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
63. The bispecific antibody of any one of embodiments 45 to 62, wherein the
bispecific antibody provides monovalent binding to CD3.
64. The bispecific antibody of embodiment 63, which comprises two antigen
binding
domains that specifically bind to glyco-MUC1.
65. The bispecific antibody of embodiment 64, wherein the two antigen
binding
domains that specifically bind to glyco-MUC1 comprise the same amino acid
sequences.
66. The bispecific antibody of any one of embodiments 45 to 65, wherein the
bispecific antibody further comprises an Fc domain composed of a first and a
second
subunit.
67. The bispecific antibody of embodiment 66, wherein the Fc domain is an
IgG Fc
domain.
68. The bispecific antibody of embodiment 67, wherein the Fc domain is an
IgGi Fc
domain.
69. The bispecific antibody of embodiment 67, wherein the Fc domain is an
IgG4 Fc
domain.
70. The bispecific antibody of embodiment 69, wherein the IgG4 Fc domain
comprises an amino acid substitution at position S228 (Kabat EU index
numbering),
preferably the amino acid substitution S228P.
71. The bispecific antibody of embodiment 66, wherein the Fc domain is a
human Fc
domain.
72. The bispecific antibody of embodiment 71, wherein the Fc domain is a
human
IgGi Fc domain, which optionally comprises SEQ ID NO:42.
73. The bispecific antibody of any one of embodiments 66 to 72, wherein the
first,
the second and, where present, the third antigen binding domain are each a Fab
molecule,
and (a) either (i) the second antigen binding domain is fused at the C-
terminus of the Fab
heavy chain to the N-terminus of the Fab heavy chain of the first antigen
binding domain
and the first antigen binding domain is fused at the C-terminus of the Fab
heavy chain to the
N-terminus of the first subunit of the Fc domain, or (ii) the first antigen
binding domain is
fused at the C-terminus of the Fab heavy chain to the N-terminus of the Fab
heavy chain of
the second antigen binding domain and the second antigen binding domain is
fused at the
C-terminus of the Fab heavy chain to the N-terminus of the first subunit of
the Fc domain;
109
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
and (b) the third antigen binding domain, where present, is fused at the C-
terminus of the
Fab heavy chain to the N-terminus of the second subunit of the Fc domain.
74. The bispecific antibody of any one of embodiments 66 to 73, wherein the
Fc
domain comprises a modification promoting the association of the first and the
second
subunit of the Fc domain.
75. The bispecific antibody of any one of embodiments 66 to 74, wherein the
Fc
domain comprises one or more amino acid substitutions that reduces binding to
an Fc
receptor and/or effector function.
76. The bispecific antibody of embodiment 45, comprising
a. a first antigen binding domain that specifically binds to CD3, wherein the
first
antigen binding domain is a crossover Fab molecule wherein either the variable
or the constant regions, preferably the variable regions, of the Fab light
chain
and the Fab heavy chain are exchanged;
b. a second and a third antigen binding domain that specifically bind to glyco-
MUC1, comprising a heavy chain variable region comprising the heavy chain
CDR-H1 of SEQ ID NO: 5, the CDR-H2 of SEQ ID NO: 6, and the CDR-H3 of
SEQ ID NO: 7; and a light chain variable region comprising the light chain CDR-
L1 of SEQ ID NO: 8, the CDR-L2 of SEQ ID NO: 9 and the CDR-L3 of SEQ ID
NO:10, wherein the second and third antigen binding domain are each a Fab
molecule;
c. an Fc domain composed of a first and a second subunit capable of stable
association,
wherein the second antigen binding domain is fused at the C-terminus of the
Fab heavy
chain to the N-terminus of the Fab heavy chain of the first antigen binding
domain, and
the first antigen binding domain is fused at the C-terminus of the Fab heavy
chain to the
N-terminus of the first subunit of the Fc domain, and wherein the third
antigen binding
domain is fused at the C-terminus of the Fab heavy chain to the N-terminus of
the
second subunit of the Fc domain.
77. The bispecific antibody of embodiment 77, wherein the first antigen
binding
domain comprises a heavy chain variable region comprising the heavy chain CDR-
H1 of
SEQ ID NO:34, the CDR-H2 of SEQ ID NO:35, and the CDR-H3 of SEQ ID NO:36; and
a
light chain variable region comprising the light chain CDR-L1 of SEQ ID NO:37,
the CDR-L2
of SEQ ID NO:38 and the CDR-L3 of SEQ ID NO:39.
110
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
78. The bispecific antibody of embodiment 77, wherein the first antigen
binding
domain comprises a heavy chain variable region sequence that is at least about
95%, 96%,
97%, 98%, 99% or 100% identical to the amino acid sequence of SEQ ID NO:40 and
a light
chain variable region sequence that is at least about 95%, 96%, 97%, 98%, 99%
or 100%
identical to the amino acid sequence of SEQ ID NO:41.
79. The bispecific antibody of embodiment 78, wherein the first antigen
binding
domain comprises the heavy chain variable region sequence of SEQ ID NO:40 and
the light
chain variable region sequence of SEQ ID NO:41.
80. The bispecific antibody of any one of embodiments 76 to 79, wherein the
second
and third antigen binding domain comprise a heavy chain variable region
sequence that is
at least about 95%, 96%, 97%, 98%, 99% or 100% identical to the amino acid
sequence of
SEQ ID NO:3 and a light chain variable region sequence that is at least about
95%, 96%,
97%, 98%, 99% or 100% identical to the amino acid sequence of SEQ ID NO:4.
81. The bispecific antibody of embodiment 80, wherein the second and third
antigen
binding domains comprise the heavy chain variable region of SEQ ID NO:3 and
the light
chain variable region of SEQ ID NO:4.
82. The bispecific antibody of any one of embodiments 76 to 81, wherein the
Fc
domain incorporates, singly or in combination, all of the features described
in Sections 5.1
and 5.2 in relation to Fc domains.
83. The bispecific antibody of any one of embodiments 76 to 82, wherein the
antigen
binding domains and the Fc region are fused to each other by peptide linkers.
84. The bispecific antibody of embodiment 83, wherein the peptide linkers
comprise
the peptide linkers as in SEQ ID NO:45 and/or SEQ ID NO:46.
85. The bispecific antibody of any one of embodiments 76 to 84, wherein in
the
constant domain CL of the second and the third Fab molecule, the amino acid at
position
124 is substituted by lysine (K) (numbering according to Kabat) and the amino
acid at
position 123 is substituted by lysine (K) or arginine (R), preferably by
arginine (R)
(numbering according to Kabat), and in the constant domain CH1 of the second
and the
third Fab molecule under (ii) the amino acid at position 147 is substituted by
glutamic acid
(E) (numbering according to Kabat EU index) and the amino acid at position 213
is
substituted by glutamic acid (E) (numbering according to Kabat EU index).
86. The bispecific antibody of any one of embodiments 76 to 85, which
comprises a
polypeptide comprising a sequence that is at least 80%, 85%, 90%, 95%, 96%,
97%, 98%,
111
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
or 99% identical to the sequence of SEQ ID NO:43, a polypeptide comprising a
sequence
that is at least 80%, 85%, 90%, 95%, 96%, 97%, 98%, or 99% identical to the
sequence of
SEQ ID NO:44, a polypeptide comprising a sequence that is at least 80%, 85%,
90%, 95%,
96%, 97%, 98%, or 99% identical to the sequence of SEQ ID NO:45, and a
polypeptide
comprising a sequence that is at least 80%, 85%, 90%, 95%, 96%, 97%, 98%, or
99%
identical to the sequence of SEQ ID NO:46.
87. The bispecific antibody of embodiment 86, wherein the bispecific
antibody
comprises a polypeptide comprising the sequence of SEQ ID NO:43, a polypeptide
comprising the sequence of SEQ ID NO:44, a polypeptide comprising the sequence
of SEQ
ID NO:45, and a polypeptide comprising the sequence of SEQ ID NO:46.
88. The bispecific antibody of embodiment 87, which comprises two
polypeptides
comprising the sequence of SEQ ID NO:43.
89. The bispecific antibody of any one of embodiments 45 to 88 which is
conjugated
to a detectable moiety.
90. The bispecific antibody of embodiment 89 in which the detectable marker
is an
enzyme, a radioisotope, or a fluorescent label.
91. A fusion protein comprising the amino acid sequence of the anti-glyco-M
UC1
antibody or antigen-binding fragment of any of embodiments 1 to 44 or the
bispecific
antibody of any one of embodiments 45 to 90 operably linked to at least a
second amino
acid sequence.
92. The fusion protein of embodiment 91, wherein the second amino acid
sequence
is that of 4-1BB, CD3-zeta, or a fragment thereof.
93. The fusion protein of embodiment 91, wherein the second amino acid
sequence
is that of a fusion peptide.
94. The fusion protein of embodiment 93, wherein the fusion peptide is a
0D28-
CD3-zeta or 4-IBB (0D137)-CD3-zeta fusion peptide.
95. The fusion protein of embodiment 91, wherein the second amino acid
sequence
is that of a modulator of T cell activation or a fragment thereof.
96. The fusion protein of embodiment 95, wherein the modulator of T cell
activation
is IL-15 or IL-15Ra.
112
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
97. A chimeric antigen receptor (CAR) comprising the scFv of any one of
embodiments 24 to 26.
98. The CAR of embodiment 97, comprising in amino- to carboxy-terminal
order: a
human CD8 leader peptide, the scFv, a human CD8 hinge domain, a human CD8
transmembrane domain, and a CD3-zeta signaling domain.
99. An antibody-drug conjugate comprising the anti-glyco-MUC1 antibody or
antigen-binding fragment of any of embodiments 1 to 44 or the bispecific
antibody of any
one of embodiments 45 to 90 or the fusion protein of any one of embodiments 91
to 96
conjugated to a cytotoxic agent.
100. The antibody-drug conjugate of embodiment 99, wherein the cytotoxic agent
is
an auristatin, a DNA minor groove binding agent, an alkylating agent, an
enediyne, a
lexitropsin, a duocarmycin, a taxane, a dolastatin, a maytansinoid, or a vinca
alkaloid.
101. The antibody-drug conjugate of embodiment 100, wherein the anti-glyco-
MUC1
antibody or antigen-binding fragment or bispecific antibody is conjugated to
the cytotoxic
agent via a linker.
102. The antibody-drug conjugate of embodiment 101, wherein the linker is
cleavable
under intracellular conditions.
103. The antibody-drug conjugate of embodiment 102, wherein the cleavable
linker is
cleavable by an intracellular protease.
104. The antibody-drug conjugate of embodiment 103, wherein the linker
comprises a
dipeptide.
105. The antibody-drug conjugate of embodiment 104, wherein the dipeptide is
val-cit
or phe-lys.
106. The antibody-drug conjugate of embodiment 102, wherein the cleavable
linker is
hydrolyzable at a pH of less than 5.5.
107. The antibody-drug conjugate of embodiment 106, wherein the hydrolyzable
linker is a hydrazone linker.
108. The antibody-drug conjugate of embodiment 102, wherein the cleavable
linker is
a disulfide linker.
109. A nucleic acid comprising a coding region for an anti-glyco-MUC1 antibody
or
antigen-binding fragment of any of embodiments 1 to 44 or the bispecific
antibody of any
113
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
one of embodiments 45 to 90, the fusion protein of any one of embodiments 91
to 96, or
the CAR of embodiment 97 or embodiment 98.
110. The nucleic acid of embodiment 109 in which the coding region is codon-
optimized for expression in a human cell.
111. A vector comprising the nucleic acid of embodiment 109 or embodiment 110.
112. The vector of embodiment 111 which is a viral vector.
113. The vector of embodiment 112 wherein the viral vector is a lentiviral
vector.
114. A host cell engineered to express the nucleic acid of embodiment 109 or
embodiment 110.
115. The host cell of embodiment 114, which is a human T-cell engineered to
express
the CAR of embodiment 97 or embodiment 98.
116. A host cell comprising the vector of any one of embodiments 111 to 113.
117. The host cell of embodiment 116 which is a T-cell and wherein the vector
encodes the CAR of embodiment 97 or embodiment 98.
118. A pharmaceutical composition comprising (a) the anti-glyco-MUC1 antibody
or
antigen binding fragment of any of embodiments 1 to 44, the bispecific
antibody of any one
of embodiments 45 to 90, the fusion protein of any one of embodiments 91 to
96, the CAR
of embodiment 97 or embodiment 98, the antibody-drug conjugate of any one of
embodiments 99 to 108, the nucleic acid of embodiment 109 or embodiment 110,
the vector
of any one of embodiments 111 to 113, or the host cell of embodiment any one
of
embodiments 114 to 117, and (b) a physiologically suitable buffer, adjuvant or
diluent.
119. A method treating cancer comprising administering to a subject in need
thereof
an effective amount of the anti-glyco-M UC1 antibody or antigen binding
fragment of any of
embodiments 1 to 44, the bispecific antibody of any one of embodiments 45 to
90, the
fusion protein of any one of embodiments 91 to 96, the CAR of embodiment 97 or
embodiment 98, the antibody-drug conjugate of any one of embodiments 99 to
108, the
nucleic acid of embodiment 109 or embodiment 110, the vector of any one of
embodiments
111 to 113, the host cell of embodiment any one of embodiments 114 to 117, or
the
pharmaceutical composition of embodiment 118.
114
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
120. The method of embodiment 119, wherein the subject is suffering from
breast
cancer, non-small cell lung cancer, prostate cancer, pancreatic cancer,
esophageal cancer,
or colorectal cancer.
121. A method of detecting cancer in a biological sample, comprising
contacting a
sample with an anti-glyco-MUC1 antibody or antigen-binding fragment according
to any one
of embodiments 1 to 44 and detecting binding of the anti-glyco-MUC1 antibody
or antigen-
binding fragment.
122. The method of embodiment 121, further comprising quantitating the binding
of
the anti-glyco-MUC1 antibody or antigen-binding fragment.
123. The method of embodiment 121 or embodiment 122, wherein the binding is
compared to a normal tissue control as a negative/baseline control and/or to a
cancerous
tissue control as a positive control.
[0363] All publications, patents, patent applications and other documents
cited in this
application are hereby incorporated by reference in their entireties for all
purposes to the same
extent as if each individual publication, patent, patent application or other
document were
individually indicated to be incorporated by reference for all purposes. In
the event that there is
an inconsistency between the teachings of one or more of the references
incorporated herein
and the present disclosure, the teachings of the present specification are
intended.
8. REFERENCES
1. Bennett, E.P., Hassan, H., Mandel, U., Mirgorodskaya, E., Roepstorff, P.,
Burchell, J.,
Taylor-Papadimitriou, J., Hollingsworth, M.A., Merkx, G., van Kessel, A.G.,
and others.
(1998) Cloning of a human UDP-N- acetyl-alpha-D-galactosamine:polypeptide N-
acetylgalactosaminyl- transferase that complements other GaINAc-transferases
in
complete 0-glycosylation of the MUC1 tandem repeat. J. Biol. Chem., 273, 30472-
30481.
2. Fontenot, J.D., Finn, 0.J., Dales, N., Andrews, P.C., and Montelaro, R.C.
(1993)
Synthesis of large multideterminant peptide immunogens using a poly-proline
beta-turn
helix motif. Pept. Res., 6, 330-336.
3. Mandel, U., Petersen, 0.W., Sorensen, H., Vedtofte, P., Hakomori, S.I.,
Clausen, H.,
and Dabelsteen, E. (1991) Simple mucin-type carbohy- drates in oral stratified
squamous and salivary-gland epithelia. J. Invest. Dermatol., 97, 713-721.
4. Miles, D.W., Linehan, J., Smith, P., and Filipe, I. (1995) Expression of
sialyl-Tn in gastric
cancer: correlation with known prognostic factors. Br. J. Cancer., 71, 1074-
1076.
115
CA 03078812 2020-04-08
WO 2019/083506 PCT/US2017/058036
5. Schwientek, T., Bennett, E.P., Flores, C., Thacker, J., Hol!mann, M., Reis,
C.A.,
Behrens, J., Mandel, U., Keck, B., Schafer, M.A., and others. (2002)
Functional
conservation of subfamilies of putative UDP-N- acetylgalactosamine:polypeptide
N-
acetylgalactosaminyltransferases in Drosophila, Caenorhabditis elegans, and
mammals.
One subfamily composed of1(2)35Aa is essential in Drosophila. J. Biol. Chem.,
277,
22623-22638.
6. Soares, R., Marinho, A., and Schmitt, F. (1996) Expression of Sialyl-Tn in
breast cancer.
Correlation with prognostic parameters. Pathol. Res. Pract., 192,1181-1186.
7. Springer, G.F. (1984) T and Tn, general carcinoma auto-antigens. Science,
224,1198-
1206.
8. Werther, J.L., Tatematsu, M., Klein, R., Kurihara, M., Kumagai, K.,
Llorens, P., Guidugli,
N.J., Bodian, C., Pertsemlidis, D., Yamachika, T., and others. (1996) Sialosyl-
Tn antigen
as a marker of gastric cancer progression: an international study. Int. J.
Cancer., 69,
193-199.
9. Taylor-Papadimitriou, J., Burchell, J., Miles, D.W., and Dalziel, M. (1999)
MUC1 and
cancer. Biochim. Biophys. Acta, 1455,301-313.
116