Note: Descriptions are shown in the official language in which they were submitted.
CA 03100492 2020-11-16
WO 2019/221769 PCT/US2018/054188
SEQUENCE SPECIFIC METHYLATION ENRICHMENT AND DETECTION
Cross Reference to Related Applications
This application is a continuation-in-part of U.S. Non-Provisional Application
No.
16/018,926, filed June 26, 2018, which claims priority to and the benefit of
U.S. Provisional
Application No. 62/526,091, filed June 28, 2017 and U.S. Provisional
Application No.
62/672,217, filed May 16, 2018, the contents of each of which are incorporated
by reference.
Technical Field
The invention relates to molecular genetics.
Background
When testing for diseases, such as cancer, physicians often rely on liquid and
tissue
biopsy. Conventional biopsy sample analysis methods typically require
expensive and time-
consuming sample preparation procedures, kits, and reagents.
For example, in a liquid biopsy, a blood sample is obtained from a patient and
may be
centrifuged to remove whole blood cells, leaving plasma or serum that includes
cell-free DNA
(cfDNA). Typically, the sample must be subject to a sample preparation
protocol before any
genetic analysis is performed. For example, laboratory technicians use a
commercially-available
kit to aliquot the serum through a series of steps that use proteinase
solutions to digest away
proteins, lysis buffers to dissociate vesicles and other lipid fragments, and
cleaning and
suspension buffers. In some protocols, the resultant mixture is washed through
a membrane
within a column under vacuum after which the cfDNA is eluted from the column
with a specialty
wash buffer. The entire process can require hours or more and the use of
expensive kits.
Epigenetic changes, such as DNA methylation, are common in the genome and can
be
indicative of disease. Methylation is the most studied epigenetic change and
has been linked to
cancer and certain metabolic disorders. Methylation is a chemical modification
of DNA in which
a methyl group is added to certain cytosines in DNA to yield 5-methylcytosine,
Methylation
appears to influence gene expression by affecting the interactions of both
chromatin proteins and
specific transcription factors with DNA. In cancer cells, hypermethylation or
hypomethylation
1
CA 03100492 2020-11-16
WO 2019/221769 PCT/US2018/054188
of a specific region of DNA is associated with cancer. The detection of, for
example,
hypermethylation in regions associated with cancer provides a useful approach
to early detection.
Conventional protocols for detecting methylation utilize bisulfite treatment
to convert
unmethylated cytosine to uracil in the DNA. Any methylated cytosine will not
be converted in
response to bisulfite treatment. Then, various sequencing techniques such as
bisulfite sequencing
or methylation-specific PCR, are used to detect methylated cytosines. These
methods are not
only time consuming and require expensive kits, but damage to the DNA and
incomplete
conversion during bisulfite treatment, render them unsuitable for reliable and
targeted sequence-
specific analysis.
Methylation is not the only epigenetic change that has been associated with
disease.
Histone modification, chromatin rearrangement, and RNA silencing are examples
of other
epigenetic modifications that can have an impact on health and disease. Since
epigenetic changes
may be good diagnostic markers for disease, there remains a need for rapid and
simple targeted
detection.
Summary
The present invention provides methods for detecting epigenetic changes,
including but
not limited to methylation changes, in DNA directly from biological samples
without the need
for significant sample preparation steps or kits.
Methods of the invention use Cas endonuclease to bind target regions of DNA
suspected
to be affected by epigenetic changes. According to the invention, Cas
endonuclease is
complexed with one or more guide RNAs that bind to protospacer adjacent motif
(PAM)
sequences near a region of interest, such as a methylation locus known to be
associated with
cancer, when hypermethylated. The Cas endonuclease binds to and protects
target regions of
DNA even when a target DNA is only present as a small fraction of the sample.
Thus, methods
of the invention are useful when analyzing DNA present in low abundance in a
sample such as
blood or other bodily fluids.
In a preferred embodiment, Cas proteins, along with their sequence-specific
guide RNAs
(gRNA), are complexed and introduced directly into the biological sample. The
Cas/guide RNA
complex may be introduced as part of sample collection, or added into
collection tubes
containing the sample. The gRNA mediates binding of the Cas proteins to a
target region of
DNA of interest, such as a tumor DNA fragment suspected to be hypermethylated.
2
CA 03100492 2020-11-16
WO 2019/221769 PCT/US2018/054188
The target region is enriched relative to other materials in the sample by any
suitable
enrichment methods, such as by elution of bound Cas proteins. The target
region may be
enriched by elimination of non-target DNA acid using, for example,
exonucleases. Enrichment
methods may be used alone or in combination with other enrichment methods. As
a non-limiting
example, the sample is enriched by isolating complex-bound target regions by
amplification, size
fractionation, or hybrid capture. In another embodiment, exonuclease digestion
may be used
alone, or may be used before or after elution of bound Cas proteins.
Epigenetic modifications are detected using available methods. For example,
for
methylation, one can use antibody binding, immunoassay, amplification, or gel
electrophoresis,
or other techniques known in the art. Detection of DNA methylation in the
target region is
indicative of increased risk, or even presence, of cancer.
Methods and related kits described herein are useful to detect the presence of
methylation
in a sample. Due to the nature by which a protein, such as a Cas complex,
binds nucleic acid,
methods may be used even where the target is present only in very small
quantities, e.g., even as
low as 0.01% frequency of mutant fragments among normal fragments in a sample
(i.e., where
about 50 copies of a circulating tumor DNA fragment harboring hypermethylation
sites are
present among about 500,000 unrelated fragments of similar size). Thus,
methods of the
invention may have particular applicability in discovering very rare, yet
clinically important,
information, such as site-specific methylation of tumor related genes and may
be used to detect
abnormal methylation within cell-free DNA, such as target region-specific
hypermethylation in
circulating tumor DNA.
In a preferred method, CRISPR/Cas systems and associated guide RNAs are
introduced
to a biological sample. When used according to methods of the invention, Cas
endonuclease¨
whether catalytically active or inactive¨will bind to a target consistently
via a guide RNA and
will protect that target (i.e., stay bound), thereby allowing the target to be
pulled out of the
sample by, for example, elution of the captured sequence or elimination of non-
target DNA. In
certain aspects, the invention provides methods for detecting hypermethylation
of a target region
of DNA. Methods include obtaining a biological sample from a subject,
introducing Cas proteins
and guide RNA into the sample, and binding the Cas proteins to ends of a
target region of DNA.
The Cas protein may be a Cas endonuclease or a catalytically deficient homolog
thereof. The
target region is then enriched by isolating the target region from the sample.
3
CA 03100492 2020-11-16
WO 2019/221769 PCT/US2018/054188
The target region may be any naturally-occurring or artificial DNA. In other
embodiments, the nucleic acid may be any naturally-occurring or artificial
nucleic acid. The
nucleic acid may be DNA, RNA, hybrid DNA/RNA, peptide nucleic acid (PNA),
morpholino
and locked nucleic acid (LNA), glycol nucleic acid (GNA), threose nucleic acid
(TNA), or Xeno
nucleic acid. The RNA may be a subpopulation of RNA, such as mRNA, tRNA, rRNA,
miRNA,
or siRNA. Preferably the nucleic acid is DNA.
The target region or feature of interest may be any site of epigenetic
modification that is
indicative of disease, such as a nucleic acid modification, a histone
modification, or chromatin
remodeling. An example of a nucleic acid modification may be a modification to
methylation
status of DNA. As such, a feature of interest may be DNA hypermethylation or
hypomethylation.
In certain embodiments, the methods of the invention involve the detection of
DNA
hypermethylation of the target region.
The target may be from a sub-population of nucleic acid within the nucleic
acid sample.
For example, the target may contain cell-free DNA, such as cell-free fetal DNA
or circulating
tumor DNA. In some embodiments, the sample includes plasma from the subject
and the target is
cell-free DNA (cfDNA). The plasma may be maternal plasma and the target may be
of fetal
DNA. In certain embodiments, the sample includes plasma from the subject and
the target is
circulating tumor DNA (ctDNA). In some embodiments, the sample includes at
least one
circulating tumor cell from a tumor and the target is tumor DNA from the tumor
cell. In some
embodiments, the target is complementary DNA (cDNA), which is made by reverse
transcribing
RNA. In some embodiments, detecting cDNA is a way to detecting target RNA.
The target may be from any source of DNA. In preferred embodiments, the target
is DNA
from a biological sample from a human or other animal. In preferred
embodiments, the
biological sample is a body fluid sample, such as bile, blood, plasma, serum,
sweat, saliva, urine,
feces, phlegm, mucus, sputum, tears, cerebrospinal fluid, synovial fluid,
pericardial fluid,
lymphatic fluid, semen, vaginal secretion, products of lactation or
menstruation, amniotic fluid,
pleural fluid, rheum, vomit, or the like. In preferred embodiments, the
biological sample is a
blood sample, serum sample, plasma sample, urine sample, saliva sample, semen
sample, feces
sample, phlegm sample, or liquid biopsy. The biological sample may be a tissue
sample from an
animal, such as skin, conjunctiva, gastrointestinal tract, respiratory tract,
vagina, placenta, uterus,
oral cavity or nasal cavity. The biological sample may be a liquid biopsy or a
tissue biopsy.
4
CA 03100492 2020-11-16
WO 2019/221769 PCT/US2018/054188
In some embodiments, obtaining the sample includes obtaining a biological
sample from
a subject in a collection tube. In a non-limiting example, the biological
sample is blood and the
collection tube is centrifuged to isolate serum or plasma from blood cells.
The Cas endonuclease
or catalytically deficient homolog thereof is introduced into the serum or
plasma. In a preferred
embodiment, the Cas endonuclease, or the catalytically deficient homolog
thereof, is complexed
with the guide RNA and introduced into the serum or plasma. In an embodiment,
the Cas
endonuclease, or the catalytically deficient homolog thereof, is introduced
into the serum or
plasma as a ribonucleoprotein (RNP) in which the endonuclease is complexed
with the guide
RNA. Preferably, the guide RNA includes at least two single guide RNA
molecules that each
complex with a Cas endonuclease, where the complexes target PAM sequences that
are adjacent
a target region and the guide RNA guides Cas endonuclease to hybridize to one
of the targets,
wherein the target region includes a methylation site associated with cancer
when
hypermethylated.
The method may include enriching the sample by isolating the complex-bound
target
region from some or all of the unbound DNA. For example, the method may
include binding the
complex-bound target region to a particle. The particle may include magnetic
or paramagnetic
material. The method may include applying a magnetic field to the sample. The
particle may
include an agent that binds to a protein bound to an end of the target DNA.
The agent may an
antibody or fragment thereof. The method may include chromatography, applying
the sample to
a column, or gel electrophoresis. The method may include separating the
complex-bound target
from some or all of the unbound nucleic by size exclusion, ion exchange, or
adsorption.
Each of the proteins may independently be any protein that binds a nucleic
acid in a
sequence-specific manner. The protein may be a programmable nuclease. For
example, the
protein may be a CRISPR-associated (Cas) endonuclease, zinc-finger nuclease
(ZFN),
transcription activator-like effector nuclease (TALEN), or RNA-guided
engineered nuclease
(RGEN). The protein may be a catalytically inactive form of a nuclease, such
as a programmable
nuclease described above. The protein may be a transcription activator-like
effector (TALE). The
protein may be complexed with a nucleic acid that guides the protein to an end
of the nucleic
acid. For example, the protein may be a Cas endonuclease in a complex with one
or more guide
RNAs. Preferably, the protein is a Cas endonuclease or a catalytically
deficient homolog thereof.
In a preferred embodiment, the Cas endonuclease is in a complex with one or
more guide RNAs.
CA 03100492 2020-11-16
WO 2019/221769 PCT/US2018/054188
Once the sample is enriched, the target region may be detected by any means
known in
the art. For example and without limitation, the target region may be detected
by ligand binding
assay, DNA staining, spectrophotometry, sequencing, fluorescent probe
hybridization,
fluorescence resonance energy transfer, optical microscopy, or electron
microscopy. Methods of
the invention may include detecting and quantifying the DNA methylation of the
target region.
Detecting and quantifying the DNA methylation of the target region may include
ligand binding
assays, immunosorbent assays, such as enzyme-linked immunosorbent assay
(ELISA), or other
signal amplification techniques. In certain embodiments, the methods include
quantifying
relative amounts of hypermethylation of the target region.
In some embodiments, the target is DNA and includes hypermethylation specific
to a
tumor. In some other embodiments, the target is nucleic acid and includes a
mutation specific to
a tumor. The target may be present at no more than about 0.01% of cell-free
DNA in the plasma
or serum. By methods herein, the target is enriched by isolating the target
from the serum or
plasma.
Furthermore, methods of the invention may include negative enrichment. As an
example,
Cas endonuclease may be provided with one or more guide RNAs that targets and
binds to one
or more target regions of DNA, such as a region or locus suspected to be
hypermethylated. In
other embodiments, the Cas endonuclease/guide RNA complex targets and binds to
a target
nucleic acid and flanks a loci of interest, such as a locus of a known cancer-
associated mutation
or a specific genetic allele of clinical interest. The Cas endonuclease binds
to, and protects, DNA
containing hypermethylated sites even when the mutation is only present as a
small fraction of
the sample. In other embodiments, the Cas endonuclease binds to, and protects
mutation-
containing nucleic acid even when the mutation is only present as a small
fraction of the sample.
The bound Cas proteins prevent exonuclease from digesting the target and,
after incubation with
exonuclease, the only nucleic acid substantially present in the sample is that
of the target. The
target is thus isolated or enriched in a sequence-specific manner. The target
may then be subject
to any suitable detection or analysis assay such as antibody signal or
immunosorbent assays.
In a preferred method, CRISPR/Cas systems using guide RNAs specific for a
methylation
site is introduced to the sample under conditions such that DNA containing the
methylation site
is protected from exonuclease digestion while non-target DNA is digested by an
exonuclease.
When used according to methods of the invention, Cas endonuclease¨whether
catalytically
6
CA 03100492 2020-11-16
WO 2019/221769 PCT/US2018/054188
active or inactive¨will bind to a target consistently via a guide RNA and will
protect that target
(i.e., stay bound) for at least long enough that a promiscuous exonuclease can
be reliably used to
digest unbound, non-target DNA. By protection of the target with digestion of
the non-target, a
sample is effectively enriched for the target, and those remaining target
fragments are captured,
stored, isolated, preserved, detected, sequenced, or otherwise assayed with
success that would be
unobtainable without methods of the invention.
In other embodiments of the invention, CRISPR/Cas systems using guide RNAs
specific
for a mutation is introduced to the sample under conditions such that nucleic
acid containing the
mutation is protected from exonuclease digestion while non-target nucleic acid
is digested by an
exonuclease. When used according to methods of the invention, Cas
endonuclease¨whether
catalytically active or inactive¨will bind to a target consistently via a
guide RNA and will
protect that target (i.e., stay bound) for at least long enough that a
promiscuous exonuclease can
be reliably used to digest unbound, non-target nucleic acid. By protection of
the target with
digestion of the non-target, a sample is effectively enriched for the target,
and those remaining
target fragments are captured, stored, isolated, preserved, detected,
sequenced, or otherwise
assayed with success that would be unobtainable without methods of the
invention. In certain
aspects, the invention provides a method for detecting a target nucleic acid.
The method includes
obtaining a serum or plasma sample from a subject, introducing Cas proteins
and guide RNA
into the serum or plasma, and binding the Cas proteins to ends of a target
nucleic acid. The Cas
protein may be a Cas endonuclease or a catalytically deficient homolog
thereof. Unbound nucleic
acid is digested from the sample by introducing exonuclease while the Cas
proteins prevent the
exonuclease from digesting the target nucleic acid, thereby enriching the
sample for the target
nucleic acid. The target nucleic acid may then be isolated from the enriched
sample by
amplification, size fractionation, or hybrid capture. Methods may include
inactivating the
exonuclease (e.g., by heating) prior to the isolating step. Preferably, two
Cas proteins bind to
ends of the target nucleic acid and prevent the exonuclease from digesting the
target nucleic acid.
In a preferred embodiment, the invention provides a method for detecting
methylation in
DNA. The method includes obtaining a biological sample, exposing the
biological sample to a
Cas /guide RNA complex. The Cas protein may be a Cas endonuclease or a
catalytically
deficient homolog thereof. The complex targets and binds to one or more target
regions of DNA
suspected to be hypermethylated in the sample. Preferably, the complexes are
targeted to PAM
7
CA 03100492 2020-11-16
WO 2019/221769 PCT/US2018/054188
sequences that are adjacent a target region of DNA suspected to be
hypermethylated in the
sample. The sample may be enriched by isolating the target DNA by
amplification, size
fractionation, and hybrid capture.
In other embodiments, the sample may be enriched by digesting the unbound DNA
by
introducing exonuclease while the Cas proteins prevent the exonuclease from
digesting the target
region, thereby enriching the sample for the target region DNA. As such,
methods may include
inactivating the exonuclease (e.g., by heating) prior to isolating the target
DNA.
Brief Description of the Drawings
FIG. 1 shows a table of the inputs and the dilation amounts used in the
Example
described herein. Dilution 11 is at 3x concentration from previous experiments
because the
experiment uses 3x as much input DNA volume in the reaction. The copies per ul
of stock,
copies per ul in 50 ul reaction, amount of previous dilution (ul), plasma, and
total volume (ul) are
indicated.
FIG. 2 shows a table of the dilutions used in the Example. For the percent of
plasma in
the final reaction, the percent of plasma in 2x sample, plasma dilution (ul),
and tris dilution (ul)
are shown in the table.
FIG. 3 shows a graph of the qPCR results after amplification from the post-
cutting
dilutions described in the Example.
FIG. 4 shows the tabulated qPCR results from the Example. Percent plasma, use
of a
Streck tube, amount of no Cas9 present, amount of Cas9 present, and percent
cutting are
indicated.
FIG. 5 shows a chart of the binding efficiency from the Example, particularly
showing
the relationship between percent cleavage and percent plasma. In particular,
the percent cleavage
is shown as a function of the amount or percent of plasma in the cutting
reaction. Results are
shown for samples with no tube and samples using a Streck tube.
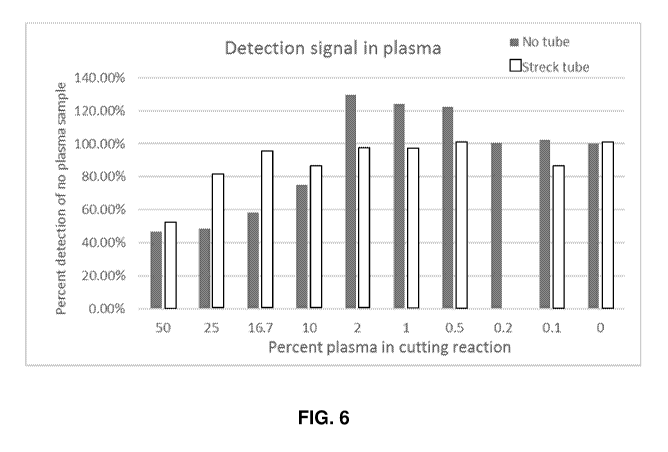
FIG. 6 shows a chart of the detection signal in plasma from the Example,
particularly
showing the relationship between qPCR signal and percent plasma. In
particular, the percent
detection of no plasma in the sample is shown as a function of the percent
plasma in the cutting
reaction. Results are shown for samples with no tube and sample using a Streck
tube.
8
CA 03100492 2020-11-16
WO 2019/221769 PCT/US2018/054188
Detailed Description
Methods of the invention provide for the enrichment of a target nucleic acid,
in a
sequence-specific manner, directly from bodily fluid samples without the need
for complex
sample preparation. Preferred embodiments include obtaining a bodily fluid
sample from a
subject. Certain embodiments of the invention provide a method for detecting a
target nucleic
acid in the bodily fluid sample. Certain other embodiments of the invention
also provide for the
detection of methylation in DNA, in a sequence-specific manner, directly from
biological
samples without the need for complex sample preparation or sequencing. The
sample is
positively enriched for the target region and methylation is detected. In
preferred embodiments
of the invention, the target region is suspected to be hypermethylated.
Methods of the invention include introducing the Cas endonuclease,
catalytically inactive
Cas endonuclease, or homolog thereof and guide RNA into the bodily fluid
sample. In a
preferred embodiment, the binding proteins are provided by Cas
endonuclease/guide RNA
complexes. Embodiments of the invention use Cas endonuclease proteins that are
originally
encoded by genes that are associated with clustered regularly interspaced
short palindromic
repeats (CRISPR) in bacterial genomes. A CRISPR-associated (Cas) endonuclease
may be
introduced directly into the bodily fluid sample.
The Cas proteins bind to ends of a target nucleic acid. The target nucleic
acid is thus
isolated or enriched in a sequence-specific manner. The enriched target
nucleic acid may then be
subject to any suitable detection or analysis assay such as amplification or
sequencing. The
enriched target nucleic acid may be further enriched by digesting other,
unbound nucleic acids
present in the sample with exonuclease. The bound Cas proteins prevent the
exonuclease from
digesting the target nucleic acid, thereby leaving the only the target nucleic
acid substantially
present in the sample. The target nucleic acid is thus isolated or enriched in
a sequence-specific
manner. The target nucleic acid may then be subject to any suitable detection
or analysis assay
such as amplification or sequencing. The target may be subjected to a signal
amplification assay,
for example, an immunoassay.
Preferably, the Cas endonuclease is complexed with a guide RNA that targets
the Cas
endonuclease to a specific sequence. In a preferred embodiment, the Cas
endonuclease/guide
RNA complex targets and binds to one or more target regions of DNA suspected
to be
hypermethylated. Any suitable Cas endonuclease or homolog thereof may be used.
A Cas
9
CA 03100492 2020-11-16
WO 2019/221769 PCT/US2018/054188
endonuclease (catalytically active or deactivated) may be Cas9 (e.g., spCas9),
catalytically
inactive Cas (dCas such as dCas9), Cpfl (aka Cas12a), C2c2, Cas13, Cas13a,
Cas13b, e.g.,
PsmCas13b, LbaCas13a, LwaCas13a, AsCas12a, others, modified variants thereof,
and similar
proteins or macromolecular complexes. The Cas13 proteins may be preferred
where the target
includes RNA. A Cas endonuclease/guide RNA complex includes a first Cas
endonuclease and a
first guide RNA. In the depicted embodiment, the complex comprises the Cas
endonuclease or
the catalytically deficient homolog thereof being introduced into the serum or
plasma as a
ribonucleoprotein (RNP) in which the Cas endonuclease or catalytically
deficient homolog
thereof is complexed with the guide RNA. The Cas endonuclease will bind to the
target. The
target may then be isolated or enriched, allowing for detection of the target.
The proteins that bind to ends of the target nucleic acid may be any proteins
that bind to a
nucleic acid in a sequence-specific manner. The protein may be a programmable
nuclease. For
example, the protein may be a CRISPR-associated (Cas) endonuclease, zinc-
finger nuclease
(ZFN), transcription activator-like effector nuclease (TALEN), or RNA-guided
engineered
nuclease (RGEN). Programmable nucleases and their uses are described in, for
example, Zhang,
2014, "CRISPR/Cas9 for genome editing: progress, implications and challenges",
Hum Mol
Genet 23 (R1):R40-6; Ledford, 2016. CRISPR: gene editing is just the
beginning, Nature. 531
(7593): 156-9; Hsu, 2014, Development and applications of CRISPR-Cas9 for
genome
engineering, Cell 157(6):1262-78; Boch, 2011, TALEs of genome targeting, Nat
Biotech
29(2):135-6; Wood, 2011, Targeted genome editing across species using ZFNs and
TALENs,
Science 333(6040):307; Carroll, 2011, Genome engineering with zinc-finger
nucleases, Genetics
Soc Amer 188(4):773-782; and Urnov, 2010, Genome Editing with Engineered Zinc
Finger
Nucleases, Nat Rev Genet 11(9):636-646, each incorporated by reference.
The protein may be a catalytically inactive form of a nuclease, such as a
programmable
nuclease described above. The protein may be a transcription activator-like
effector (TALE). The
protein may be complexed with a nucleic acid that guides the protein to an end
of the target
nucleic acid. For example, the protein may be a Cas endonuclease in a complex
with one or more
guide RNAs. In preferred embodiments, the protein is a Cas endonuclease,
catalytically inactive
Cas endonuclease, or homologs thereof.
In certain embodiments, the sample includes cfDNA from a subject. The sample
is
exposed to a first Cas endonuclease/guide RNA complex that binds to a target
nucleic acid (e.g.,
CA 03100492 2020-11-16
WO 2019/221769 PCT/US2018/054188
a mutation of interest) in a sequence-specific fashion. In some embodiments,
the complex binds
to a mutation in a sequence-specific manner. A segment of the nucleic acid,
i.e., the target
nucleic acid, is protected by introducing the first Cas endonuclease/guide RNA
complex and a
second Cas endonuclease/guide RNA complex that also binds to the nucleic acid.
In preferred
embodiments of the method, the guide RNA comprises at least two guide RNA
molecules that
each complex with a Cas endonuclease and guide the Cas endonuclease to
hybridize to one target
nucleic acid, wherein the target nucleic acid includes a loci know to harbor a
cancer-associated
mutation.
In certain embodiments, the sample includes cfDNA from a subject. The sample
is
exposed to a first Cas endonuclease/guide RNA complex that binds to a target
region of DNA
(e.g., a region suspected to be hypermethylated) in a sequence-specific
fashion. In some
embodiments, the complex targets and binds to a one or more PAM sequences in a
sequence
specific manner that are adjacent the target region suspected to be
hypermethylated. A segment
of the DNA, i.e., the target region, is protected by introducing the first Cas
endonuclease/guide
RNA complex and a second Cas endonuclease/guide RNA complex that also binds to
the DNA.
In preferred embodiments of the method, the guide RNA comprises at least two
guide RNA
molecules that each complex with a Cas endonuclease and guide the Cas
endonuclease to
hybridize to a target region of DNA, wherein the target region includes a loci
know to be
hypermethylated.
Optionally, unprotected nucleic acid is digested. For example, one or more
exonucleases
may be introduced that promiscuously digest unbound, unprotected nucleic acid.
Any suitable
exonuclease may be used. Suitable exonucleases include, for example, Lambda
exonuclease,
Reaf, Exonuclease III, Exonuclease I, Exonuclease T, Exonuclease V,
Exonuclease VII, T5
Exonuclease, and T7 Exonuclease, most of which are available from New England
Biolabs
(Ipswich, MA). While the exonucleases act, the target nucleic acid is
protected by the bound
complexes and survives the digestion step intact.
The described steps including the digestion by the exonuclease leave a
reaction product
that includes principally only the mutant segment of nucleic acid, as well as
any spent reagents,
Cas endonuclease complexes, exonuclease, nucleotide monophosphates, and
pyrophosphate as
may be present.
11
CA 03100492 2020-11-16
WO 2019/221769 PCT/US2018/054188
In certain embodiments, the exonuclease is deactivated. For example,
exonuclease may
be deactivated by following the manufacturer's instructions e.g., by heating
to 90 degrees for a
few minutes. After digestion, a positive selection step may be performed which
may include, for
example, amplification of the target nucleic acid by known methods or
selection by an affinity
assays.
The nucleic acid may be any naturally-occurring or artificial nucleic acid.
The nucleic
acid may be DNA, RNA, hybrid DNA/RNA, peptide nucleic acid (PNA), morpholino
and locked
nucleic acid (LNA), glycol nucleic acid (GNA), threose nucleic acid (TNA), or
Xeno nucleic
acid. The RNA may be a subpopulation of RNA, such as mRNA, tRNA, rRNA, miRNA,
or
siRNA. Preferably the nucleic acid is DNA.
The target or feature of interest may be any feature of a nucleic acid. The
feature may be
a mutation. For example and without limitation, the feature may be an
insertion, deletion,
substitution, inversion, amplification, duplication, translocation, or
polymorphism. The feature
may be a nucleic acid from an infectious agent or pathogen. For example, the
nucleic acid
sample may be obtained from an organism, and the feature may contain a
sequence foreign to the
genome of that organism. In other embodiments, the target region or feature of
interest may be
any site of epigenetic modification that is indicative of a disease, such as a
nucleic acid
modification, a histone modification, or chromatin remodeling. An example of a
nucleic acid
modification may be a modification to methylation status of DNA. As such, a
feature of interest
may be DNA hypermethylation or hypomethylation. In certain embodiments, the
methods of the
invention involve the detection of DNA hypermethylation. Histone modifications
can also
constitute an epigenetic modification indicative of disease. Examples of
histone modifications
include but are not limited to methylation, acetylation, deacetylation, ADP-
ribosylation,
ubiquitination or phosphorylation. Other examples of epigenetic elements
indicative of disease
include RNA interference (e.g., short interfering RNA, also known as siRNA)
and prions.
The target nucleic acid may be from a sub-population of nucleic acid within
the nucleic
acid sample. For example, the target nucleic acid may contain cell-free DNA,
such as cell-free
fetal DNA or circulating tumor DNA. In some embodiments, the sample includes
plasma from
the subject and the target nucleic acid is cell-free DNA (cfDNA). The plasma
may be maternal
plasma and the target may be of fetal DNA. In certain embodiments, the sample
includes plasma
from the subject and the target is circulating tumor DNA (ctDNA). In some
embodiments, the
12
CA 03100492 2020-11-16
WO 2019/221769 PCT/US2018/054188
sample includes at least one circulating tumor cell from a tumor and the
target is tumor DNA
from the tumor cell. In some embodiments, the target nucleic acid is
complementary DNA
(cDNA), which is made by reverse transcribing RNA. In some embodiments,
detecting cDNA is
a way to detecting target RNA.
The target nucleic acid may be from any source of nucleic acid. In preferred
embodiments, the target nucleic acid is from a bodily fluid sample from a
human. In preferred
embodiments, the bodily fluid sample is a liquid or bodily fluid from a
subject, such as bile,
blood, plasma, serum, sweat, saliva, urine, feces, phlegm, mucus, sputum,
tears, cerebrospinal
fluid, synovial fluid, pericardial fluid, lymphatic fluid, semen, vaginal
secretion, products of
lactation or menstruation, amniotic fluid, pleural fluid, rheum, vomit, or the
like. In preferred
embodiments, the bodily fluid sample is a blood sample, serum sample, plasma
sample, urine
sample, saliva sample, semen sample, feces sample, phlegm sample, or liquid
biopsy. The
sample may be a tissue sample from an animal, such as skin, conjunctiva,
gastrointestinal tract,
respiratory tract, vagina, placenta, uterus, oral cavity or nasal cavity. The
sample may be a liquid
biopsy or a tissue biopsy.
The method optionally includes detecting the target nucleic acid (which may
harbor the
mutation). Any suitable technique may be used to detect the target nucleic
acid. For example,
detection may be performed using DNA staining, spectrophotometry, sequencing,
fluorescent
probe hybridization, fluorescence resonance energy transfer, optical
microscopy, electron
microscopy, others, or combinations thereof. Detecting the target nucleic acid
may indicate the
presence of the mutation in the subject (i.e., a patient), and a report may be
provided describing
the mutation in the patient.
In an embodiment of the invention, detecting methylation in DNA of the target
region is
performed. Any suitable technique may be used to detect methylation in the
target region of
DNA. For example, detection may be performed using known hybridization assays,
such as those
that utilize antibodies, or other signal amplification techniques, such as an
immunoassay, like an
ELISA. The described methodology may be used to detect hypermethylation in the
target region.
In such cases, site-specific hypermethylation detection may be used to detect
the presence of
cancer.
In an embodiment of the invention, a sample may contain a mutant fragment of
DNA, a
wild-type fragment of DNA, or both. A locus of interest is identified where a
mutation may be
13
CA 03100492 2020-11-16
WO 2019/221769 PCT/US2018/054188
present proximal to, or within, a protospacer adjacent motif (PAM). When the
wild-type
fragment is present, it may contain a wild-type allele at a homologous
location in the fragment,
also proximal to, or within, a PAM. A guide RNA is introduced to the sample
that has a targeting
portion complementary to the portion of the mutant fragment that includes the
mutation. When a
Cas endonuclease is introduced, it will form a complex with the guide RNA and
bind to the
mutant fragment but not to the wild-type fragment. The first Cas
endonuclease/guide RNA
complex includes a guide RNA with a targeting region that binds to the
mutation but that does
not bind to other variants at a loci of the mutation. The described
methodology may be used to
target a mutation that is proximal to a PAM, or it may be used to target and
detect a mutation in a
PAM, e.g., a loss-of-PAM or gain-of-PAM mutation.
The described methodology may be used to target a mutation that is proximal to
a PAM,
or it may be used to target and detect a mutation in a PAM, e.g., a loss-of-
PAM or gain-of-PAM
mutation. The PAM is typically specific to, or defined by, the Cas
endonuclease being used. For
example, for Streptococcus pyogenes Cas9, the PAM includes NGG, and the
targeted portion
includes the 20 bases immediately 5' to the PAM. As such, the targetable
portion of the DNA
includes any twenty-three consecutive bases that terminate in GG or that are
mutated to
terminate in GG. Such a pattern may be found to be distributed over ctDNA at
such frequency
that the potentially detectable mutations are abundant enough as to be
representative of
mutations over the tumor DNA at large. In such cases, mutation-specific
enrichment may be used
to detect mutations from a tumor. Moreover, methods may be used to determine a
number of
mutations over the representative, targetable portion of tumor DNA. Since the
targetable portion
of the genome is representative of the tumor DNA overall, the number of
mutations may be used
to infer a mutational burden for the tumor.
In another embodiment of the invention, a sample may contain a mutant fragment
of
DNA, a wild-type fragment of DNA, or both. A locus of interest is identified
where
hypermethylation may be present adjacent to a protospacer adjacent motif
(PAM). A guide RNA
is introduced to the sample that has a targeting portion complementary to PAM
sequences that
are adjacent a target region suspected to be hypermethylated. When a Cas
endonuclease is
introduced, it will form a complex with the guide RNA and the complex will
target and bind to
the PAM sequences. The Cas endonuclease/guide RNA complex includes a guide RNA
with a
targeting region that binds to the PAM but that does not bind to other regions
at a loci of the
14
CA 03100492 2020-11-16
WO 2019/221769 PCT/US2018/054188
target region. The described methodology may be used to target and detect DNA
methylation
adjacent to a PAM. The described methodology may be used to target DNA
methylation adjacent
to a PAM, or it may be used to target a region and detect DNA hypermethylation
adjacent a
PAM. As such, the targetable portion of the DNA includes any twenty-three
consecutive bases
that terminate in GG. In such cases, enrichment may be used to detect
methylated target regions
of DNA known to be associated with cancer. Moreover, methods may be used to
quantify DNA
methylation levels of the target region of DNA. Since the targetable portion
of the genome is
representative of the tumor DNA overall, the hypermethylation levels may be
used to infer a
mutational burden for the tumor.
A feature of the method is that a specific mutation may be detected by a
technique that
includes detecting only the presence or absence of a fragment of DNA, and it
need not be
necessary to sequence DNA from a subject to describe mutations. Methods of the
invention use
protection at one or both ends of DNA segments. The gRNA selects for a known
mutation on
one end. A positive selection may be performed to positively select out the
bound, target nucleic
acid. If the gRNA does not find the mutation, no protection is provided and
the molecule may be
digested, e.g. in negative enrichment, and the remaining molecules are either
counted or
sequenced. Methods are well suited for the analysis of samples in which the
target of interest is
extremely rare, and particularly for the analysis of maternal plasma or serum
(e.g., for fetal
DNA) or a liquid biopsy (e.g., for ctDNA). Such methods of the invention are
useful for
detection of hypermethylation of target region of DNA known to be associated
with cancer. The
Cas endonuclease/gRNA complex targets and binds to PAM adjacent to the target
region
suspected to be hypermethylated.
Methods are useful for the isolation of intact DNA fragments of any arbitrary
length and
may preferably be used in some embodiments to isolate (or enrich for)
arbitrarily long fragments
of DNA, e.g., tens, hundreds, thousands, or tens of thousands of bases in
length or longer. Long,
isolated, intact fragments of DNA may be analyzed by any suitable method such
as simple
detection (e.g., via staining with ethidium bromide) or by single-molecule
sequencing. It is noted
that the Cas9/gRNA complexes may be subsequently or previously labeled using
standard
procedures. The complexes may be fluorescently labeled, e.g., with distinct
fluorescent labels
such that detecting involves detecting both labels together (e.g., after a
dilution into fluid
partitions). Preferred embodiments of the detection do not require PCR
amplification and
CA 03100492 2020-11-16
WO 2019/221769 PCT/US2018/054188
therefore significantly reduces cost and sequence bias associated with PCR
amplification.
Sample analysis can also be performed by a number of approaches, such as next
generation
sequencing (NGS), etc. However, many analytical platforms may require PCR
amplification
prior to analysis. Therefore, preferred embodiments of analysis of the
reaction products include
single molecule analysis that avoids the requirement of amplification.
Kits and methods of the invention are useful with methods disclosed in U.S.
Provisional
Patent Application 62/526,091, filed June 28, 2017, for POLYNUCLEIC ACID
MOLECULE
ENRICHMENT METHODOLOGIES and U.S. Provisional Patent Application 62/519,051,
filed
June 13, 2017, for POLYNUCLEIC ACID MOLECULE ENRICHMENT METHODOLOGIES,
both incorporated by reference.
The target nucleic acid may be detected, sequenced, or counted. Where a
plurality of
fragments are present or expected, the fragment may be quantified, e.g., by
qPCR.
The target nucleic acid may further be isolated or detected by any suitable
method in
order to separate the target segment from other nucleic acids in the sample.
For example, the
isolation or detection method may include separating the protein-bound target
nucleic acid from
some or all of the unbound nucleic acid. The isolation or detection method may
include binding
the protein-bound target nucleic acid to a particle. The particle may include
magnetic or
paramagnetic material. The isolation or detection method may include applying
a magnetic field
to the sample. The particle may include an agent that binds to a protein bound
to an end of the
target nucleic acid. The agent may an antibody or fragment thereof. The
isolation or detection
method may include chromatography. The isolation or detection method may
include applying
the sample to a column. The isolation or detection method may include
separating the protein-
bound target nucleic acid from some or all of the unbound nucleic acid by size
exclusion, ion
exchange, or adsorption. The isolation or detection method may include gel
electrophoresis.
The target DNA may further be enriched or isolated by any suitable method in
order to
separate the target segment from other components in the sample. For example,
the isolation or
detection method may include separating the complex-bound target region DNA
from some or
all of the unbound DNA. In a preferred embodiment, the isolation method may
include binding
the complex-bound target region to a particle. The particle may include
magnetic or
paramagnetic material. The isolation or detection method may include applying
a magnetic field
to the sample. The particle may include an agent that binds to a protein bound
to an end of the
16
CA 03100492 2020-11-16
WO 2019/221769 PCT/US2018/054188
target DNA. The agent may an antibody or fragment thereof. The isolation or
detection method
may include chromatography. The isolation or detection method may include
applying the
sample to a column. The isolation or detection method may include separating
the complex-
bound target DNA from some or all of the unbound DNA by size exclusion, ion
exchange, or
adsorption. The enrichment or isolation method may include gel
electrophoresis.
Embodiments of the invention may include detecting the target nucleic acid and
optionally providing a report describing a mutation as present in the patient.
In some
embodiments, the target nucleic acid is a target region of DNA suspected to be
hypermethylated.
The mutation-containing fragments may be detected by a suitable assay, such as
sequencing, gel
electrophoresis, a probe-based assay. The detection of the isolated segment of
the target nucleic
acid may be done by sequencing. The detection of the methylation of the
isolated target r may be
done by any known signal amplification technique in the art. The digestion or
the isolation
provides a reaction product that includes principally only the target nucleic
acid, as well as any
spent reagents, Cas endonuclease complexes, exonuclease (e.g. when negative
enrichment is
performed), nucleotide monophosphates, or pyrophosphate as may be present. The
reaction
product may be provided as an aliquot (e.g., in a micro centrifuge tube such
as that sold under
the trademark EPPENDORF by Eppendorf North America (Hauppauge, NY) or glass
cuvette).
The reaction product aliquot may be disposed on a substrate. For example, the
reaction product
may be pipetted onto a glass slide and subsequently combed or dried to extend
the fragment
across the glass slide. The reaction product may optionally be amplified.
Optionally, adaptors are
ligated to ends of the reaction product, which adaptors may contain primer
sites or sequencing
adaptors. The presence of the segment in the reaction product aliquot may then
be detected using
an instrument.
The target nucleic acid may be detected by any means known in the art. For
example and
without limitation, the target nucleic acid may be detected by DNA staining,
spectrophotometry,
sequencing, fluorescent probe hybridization, fluorescence resonance energy
transfer, optical
microscopy, or electron microscopy. Detecting the nucleic acid may include
identifying a
mutation in the nucleic acid. In other embodiments, detecting the nucleic acid
may include
detecting methylation at the Identifying the mutation may include sequencing
the nucleic acid
(e.g., on a next-generation sequencing instrument), allele-specific
amplification, and hybridizing
a probe to the nucleic acid. Methods of DNA sequencing are known in the art
and described in,
17
CA 03100492 2020-11-16
WO 2019/221769 PCT/US2018/054188
for example, Peterson, 2009, Generations of sequencing technologies, Genomics
93(2):105-11;
Goodwin, 2016, Coming of age: ten years of next-generation sequencing
technologies, Nat Rev
Genet 17(6):333-51; and Morey, 2013, A glimpse into past, present, and future
DNA
sequencing, Mol Genet Metab 110(1-2):3-24, each incorporated by reference.
Other methods of
DNA detection are known in the art and described in, for example, Xu, 2014,
Label-Free DNA
Sequence Detection through FRET from a Fluorescent Polymer with Pyrene Excimer
to SG,
ACS Macro Lett 3(9):845-848, incorporated by reference.
One method for detection of protein-bound nucleic acids is immunomagnetic
separation.
Magnetic or paramagnetic particles are coated with an antibody that binds the
protein bound to
the segment, and a magnetic field is applied to separate particle-bound
segment from other
nucleic acids. Methods of immunomagnetic purification of biological materials
such as cells and
macromolecules are known in the art and described in, for example, U.S. Patent
No. 8,318,445;
Safarik and Safarikova, Magnetic techniques for the isolation and purification
of proteins and
peptides, Biomagn Res Technol. 2004; 2:7, doi: 10.1186/1477-044X-2-7, the
contents of each of
which are incorporated herein by reference. The antibody may be a full-length
antibody, a
fragment of an antibody, a naturally occurring antibody, a synthetic antibody,
an engineered
antibody, or a fragment of the aforementioned antibodies. Alternatively or
additionally, the
particles may be coated with another protein-binding moiety, such as an
aptamer, peptide,
receptor, ligand, or the like. Preferably, the particles are coated with
biotin-binding protein, such
as Streptavidin. In other embodiments, the magnetic separation of the complex-
bound regions
provides for enrichment and isolation of the target region. The target region
may be suspected to
be hypermethylated.
Chromatographic methods may be used for detection. In such methods, the bodily
fluid
sample is applied to a column, and the target nucleic acid is separated from
other nucleic acids
based on a difference in the properties of the target nucleic acid and the
other nucleic acids. Size
exclusion chromatography is useful for separating molecules based on
differences in size and
thus is useful when the segment is larger than other nucleic acids, for
example the residual
nucleic acids left from a digestion step. Methods of size exclusion
chromatography are known in
the art and described in, for example, Ballou, David P.; Benore, Marilee;
Ninfa, Alexander J.
(2008). Fundamental laboratory approaches for biochemistry and biotechnology
(2nd ed.).
Hoboken, N.J.: Wiley. p. 129. ISBN 9780470087664; Striegel, A. M.; and
Kirkland, J. J.; Yau,
18
CA 03100492 2020-11-16
WO 2019/221769 PCT/US2018/054188
W. W.; Bly, D. D.; Modern Size Exclusion Chromatography, Practice of Gel
Permeation and Gel
Filtration Chromatography, 2nd ed.; Wiley: NY, 2009, the contents of each of
which are
incorporated herein by reference.
Ion exchange chromatography uses an ion exchange mechanism to separate
analytes
based on their respective charges. Thus, ion exchange chromatography can be
used with the
proteins bound to the target nucleic acid impart a differential charge as
compared to other nucleic
acids. Methods of ion exchange chromatography are known in the art and
described in, for
example, Small, Hamish (1989). Ion chromatography. New York: Plenum Press.
ISBN 0-306-
43290-0; Tatjana Weiss, and Joachim Weiss (2005). Handbook of Ion
Chromatography.
Weinheim: Wiley-VCH. ISBN 3-527-28701-9; Gjerde, Douglas T.; Fritz, James S.
(2000). Ion
Chromatography. Weinheim: Wiley-VCH. ISBN 3-527-29914-9; and Jackson, Peter;
Haddad,
Paul R. (1990). Ion chromatography: principles and applications. Amsterdam:
Elsevier. ISBN 0-
444-88232-4, the contents of each of which are incorporated herein by
reference.
Adsorption chromatography relies on difference in the ability of molecule to
adsorb to a
solid phase material. Larger nucleic acid molecules are more adsorbent on
stationary phase
surfaces than smaller nucleic acid molecules, so adsorption chromatography is
useful when the
target nucleic acid is larger than other nucleic acids, for example the
residual nucleic acids left
from a digestion step. Methods of adsorption chromatography are known in the
art and described
in, for example, Cady, 2003, Nucleic acid purification using microfabricated
silicon structures.
Biosensors and Bioelectronics, 19:59-66; Melzak, 1996, Driving Forces for DNA
Adsorption to
Silica in Perchlorate Solutions, J Colloid Interface Sci 181:635-644; Tian,
2000, Evaluation of
Silica Resins for Direct and Efficient Extraction of DNA from Complex
Biological Matrices in a
Miniaturized Format, Anal Biochem 283:175-191; and Wolfe, 2002, Toward a
microchip-based
solid-phase extraction method for isolation of nucleic acids, Electrophoresis
23:727-733, each
incorporated by reference.
Another method for detection is gel electrophoresis. Gel electrophoresis
allows
separation of molecules based on differences in their sizes and is thus useful
when the target
nucleic acid is larger than other nucleic acids, for example the residual
nucleic acids left from a
digestion step. Methods of gel electrophoresis are known in the art and
described in, for example,
Tom Maniatis; E. F. Fritsch; Joseph Sambrook. "Chapter 5, protocol 1".
Molecular Cloning - A
Laboratory Manual. 1 (3rd ed.). p. 5.2-5.3. ISBN 978-0879691363; and Ninfa,
Alexander J.;
19
CA 03100492 2020-11-16
WO 2019/221769 PCT/US2018/054188
Ballou, David P.; Benore, Marilee (2009). fundamental laboratory approaches
for biochemistry
and biotechnology. Hoboken, NJ: Wiley. p. 161. ISBN 0470087668, the contents
of which are
incorporated herein by reference.
Methods of the invention involve signal amplification techniques, such as
antibody-based
detection and analysis. In order to detect methylation of a target region of
DNA the use of signal
amplification techniques are employed. For example, antibody-based detection
methods are
generally based on the transformation of a specific biomolecular interaction
between antigen and
antibody into a macroscopically detectable signal or change in the physical
properties of the
media. See e.g., Sveshnikov, Peter; "The Potential of Different Biotechnology
Methods in BTW
Agent Detection: Antibody Based Methods" The Role of Biotechnology in
Countering BTW
Agents; Vol. 34 of the series NATO Science Series, pp. 69-77 (2001),
incorporated herein by
reference.
Particular antibody detection methods are known in the art. Proteins can be
detected and
quantified through epitopes recognized by polyclonal and/or monoclonal
antibodies used in
methods such as enzyme-linked immunoabsorbent assay (ELISA), immunoblot
assays, flow
cytometric assays, radioimmuno assays, immunocytochemical assays, Western blot
assays, an
immunofluorescent assays, chemiluminescent assays, flow cytometry and
fluorescence-activated
cell sorting (FACS), immunoprecipitation, enzyme linked immunospot (ELISPOT),
and other
polypeptide detection strategies. Proteins may also be detected by mass
spectrometry assays
(potentially coupled to immunoaffinity assays) including matrix-assisted laser
desorption/ionization time-of-flight (MALDI-TOF) mass mapping and liquid
chromatography/quadrupole time-of-flight electro spray ionization tandem mass
spectrometry
(LC/Q-TOF-ESI-MS/MS). Additionally, methods of the disclosed invention may
include tagging
of proteins separated by two-dimensional polyacrylamide gel electrophoresis
(2D-PAGE),
(Kiernan et al, Anal Biochem 301, 49-56 (2002); Poutanen et al, Mass Spectrom
15, 1685-1692
(2001) the content of each of which is incorporated by reference herein in its
entirety) or any
other method of detecting protein. Methods for making monoclonal antibodies
are well known
(see, e.g., Harlow and Lane, 1988, ANTIBODIES: A LABORATORY MANUAL, Cold
Spring
Harbor, N.Y., which is incorporated in its entirety for all purposes). In some
embodiments,
immunohistochemistry methods may be used for detecting the presence of DNA
hypermethylation of a target region. In these methods, antibodies (monoclonal
or polyclonal)
CA 03100492 2020-11-16
WO 2019/221769 PCT/US2018/054188
specific for each target region are used to detect hypermethylation.
Immunohistochemistry
protocols and kits are well known in the art and are commercially available.
Certain preferred embodiments include obtaining a blood, plasma, or serum
sample from
a patient. The blood, plasma, or serum may include cfDNA and thus also include
ctDNA among
the cfDNA. Specific sequences of the ctDNA are isolated or enriched and
analyzed or detected to
detect or report genetic information from the subject, such as a presence or
count of certain
tumor mutations. Methods of the invention include introduce Cas endonucleases
(or catalytically
inactive homologs thereof such as dCas9) directly into serum or plasma. The
Cas endonucleases
are complexed with guide RNAs that include targeting portions specific for a
target nucleic acid.
In the plasma or serum, the complexes bind to ends of the target and protect
it. Exonuclease may
be introduced to digest unbound nucleic acid into monomers and fragments too
small for further
meaningful detection, sequencing, or amplification.
Embodiments of the invention provide for treatment of a sample. For example, a
blood
sample may be obtained from a patient. The sample may be collected in any
suitable blood
collection tube such as the collection tube sold under the trademark
VACUTAINER by BD
(Franklin Lakes, NJ). In certain embodiments, the collection tube comprises an
EDTA collection
tube, and Na-EDTA collection tube or the collection tube sold under the
trademark CELL-FREE
DNA BCT by Streck, Inc. (La Vista, NE), sometimes referred to in the art as a
Streck tube. Use
of a Streck tube stabilizes nucleated blood cells and prevents the release of
genomic DNA into
the sample. This facilitates the collection of sample that includes cell-free
DNA.
The sample may be centrifuged to generate a sample that includes a pellet of
blood cells
and a supernatant, which contains serum or plasma. Serum is the liquid
supernatant of whole
blood that is collected after the blood is allowed to clot and centrifuged.
Plasma is produced
when the process includes an anticoagulant. To collect serum, blood is
collected in tubes. After
collection, the blood is allowed to clot by leaving it undisturbed at room
temperature (about 15-
30 minutes). The clot is removed by centrifuging, e.g., at 1,000-2,000 x g for
10 minutes in a
refrigerated centrifuge. The resulting supernatant is designated serum and may
be transferred to a
clean polypropylene tube using a Pasteur pipette. For plasma, blood is
collected into
commercially available anticoagulant-treated tubes e.g., EDTA-treated
(lavender tops), citrate-
treated (light blue tops), or heparinized tubes (green tops), followed by
centrifugation to collect
the supernatant. The supernatant is preferably transferred to a fresh tube,
away from the pellet,
21
CA 03100492 2020-11-16
WO 2019/221769 PCT/US2018/054188
which may be discarded. Particularly where the collection tube included an
anticoagulant, the
transfer should give a good separation of the plasma from the whole blood
cells. After transfer,
the sample includes plasma or serum, which includes cfDNA.
In an exemplary embodiment, serum or plasma is transferred from a centrifuge
tube to a
new tube, complexes comprising Cas9 and guide RNA are added, and the mixture
is incubated.
For example, amplification or an affinity assay may be performed to positively
select out the
bound, target nucleic acid. In another embodiment, exonuclease may be
introduced to digest
unbound, non-target DNA, and then the exonuclease may be deactivated (e.g., by
heat). A
positive selection may then follow (e.g., amplification or an affinity assay)
to positively select
out the bound, target nucleic acid.
In another exemplary embodiment, plasma or serum is removed from the
centrifuge tube
(the supernatant) and transferred into a new tube. Appropriate
buffers/reagents are added to
modify a chemical environment to promote binding of Cas endonuclease to the
target nucleic
acid. For example, pH can be adjusted, as may temperature, salinity, or co-
factors present. The
Cas complexes are added and allowed to incubate. For example, amplification or
an affinity may
be performed to positively select out the bound, target nucleic acid. An
exonuclease may
optionally be added, which ablates all free, non-target nucleic acid. The
target may be positively
selected such as by amplification or an affinity assay after exonuclease
digestion of the non-
target nucleic acid.
Methods may include detection or isolation of circulating tumor cells (CTCs)
from a
blood sample. Cytometric approaches use immunostaining profiles to identify
CTCs. CTC
methods may employ an enrichment step to optimize the probability of rare cell
detection,
achievable through immune-magnetic separation, centrifugation, or filtration.
Cytometric CTC
technology includes the CTC analysis platform sold under the trademark
CELLSEARCH by
Veridex LLC (Huntingdon Valley, PA). Such systems provide semi-automation and
proven
reproducibility, reliability, sensitivity, linearity and accuracy. See Krebs,
2010, Circulating tumor
cells, Ther Adv Med Oncol 2(6):351-365 and Miller, 2010, Significance of
circulating tumor
cells detected by the CellSearch system in patients with metastatic breast
colorectal and prostate
cancer, J Oncol 2010:617421-617421, both incorporated by reference.
Certain embodiments of the invention may provide a kit. The kit preferably
includes
reagents and materials useful for performing methods of the invention. For
example, the kit may
22
CA 03100492 2020-11-16
WO 2019/221769 PCT/US2018/054188
include one or more guide RNA that, taken in pairs, are designed to flank
cancer-associated
mutations. The kit may include one or more guide RNAs that are mutation
specific and only
hybridize to target that includes a mutation. The kit may include a Cas
endonuclease or a nucleic
acid encoding a Cas endonuclease such as a plasmid. The kit may optionally
include
exonuclease. The kit may include reagents for adjusting conditions such as pH,
salinity, co-
factors, etc., to promote binding or activity of Cas endonuclease (including
to promote binding of
catalytically inactive Cas endonuclease, which may be included as the Cas
endonuclease) in the
bodily fluid sample, such as plasma or serum. The kit may further include
instructional materials
for performing methods of the invention, and components of the kit may be
packaged in a box
suitable for shipping or storage. Preferably, the kit contains one or more
collection tubes, such as
a blood collection tube. In other embodiments, the kit may include one or more
guide RNA that,
taken in pairs, are designed to flank cancer-associated methylation sites. The
kit may include one
or more guide RNAs that are target specific and only hybridize to PAM
sequences adjacent to
the target region that includes a methylation site suspected to be
hypermethylated.
The Cas endonuclease/guide RNA complexes can be designed to bind to mutations
of
clinical significance, such as a mutation specific to a tumor. In other
embodiments, the
complexes can be designed to target and bind to one or more target regions of
DNA suspected to
be hypermethylated, where such hypermethylation is specific to a tumor. When a
mutation is
thus detected, a report may be provided to, for example, describe the mutation
in a patient or a
subject. Thus, certain embodiments may comprise providing a report. The report
preferably
includes a description of the mutation in the subject (e.g., a patient). The
method for detecting
rare nucleic acid may be used in conjunction with a method of describing
mutations (e.g., as
described herein). Either or both detection processes may be performed over
any number of loci
in a patient's genome or preferably in a patient's tumor DNA. As such, the
report may include a
description of a plurality of structural alterations, mutations, or both in
the patient's genome or
tumor DNA. As such, the report may give a description of a mutational
landscape of a tumor. In
other embodiments, the report may provide a description of the DNA methylation
in the target
region of the subject.
Knowledge of a mutational landscape of a tumor may be used to inform treatment
decisions, monitor therapy, detect remissions, or combinations thereof. For
example, where the
report includes a description of a plurality of mutations, the report may also
include an estimate
23
CA 03100492 2020-11-16
WO 2019/221769 PCT/US2018/054188
of a tumor mutation burden (TMB) for a tumor. It may be found that TMB is
predictive of
success of immunotherapy in treating a tumor, and thus methods described
herein may be used
for treating a tumor. Furthermore, knowledge of DNA methylation of a target
region may also be
used similarly.
Methods of the invention thus may be used to detect and report clinically
actionable
information about a patient or a tumor in a patient. For example, the method
may be used to
provide a report describing the presence of the genomic alteration in a genome
of a subject. In
other embodiments, the method may be used to provide a report describing the
DNA methylation
of a target region known to be associated with cancer of a subject.
Additionally, protecting a
segment of DNA, and optionally digesting unprotected DNA, provides a method
for isolation or
enrichment of DNA fragments, i.e., the protected segment. It may be found that
the described
enrichment techniques are well-suited to the isolation/enrichment of
arbitrarily long DNA
fragments, e.g., thousands to tens of thousands of bases in length or longer.
Long DNA fragment targeted enrichment, or negative enrichment, creates the
opportunity
of applying long read platforms in clinical diagnostics. Negative enrichment
may be used to
enrich "representative" genomic regions that can allow an investigator to
identify "off rate"
when performing CRISPR Cas9 experimentation, as well as enrich for genomic
regions that
would be used to determine TMB for immuno-oncology associated therapeutic
treatments. In
such applications, the negative enrichment technology is utilized to enrich
large regions (>50
kb) within the genome of interest.
The methods described herein provide the ability to assay for methylation and
other
epigenetic changes that are indicative of disease. Methods of the invention
are conducive to
high-throughput testing, and may be performed, for example, in droplets on a
microfluidic
device, to rapidly assay a large number of aliquots from a sample for one or
any number of
genomic structural alterations. Furthermore, using the methods described
herein, a biological
sample can be assayed for hypermethylation or other epigenetic changes in
target regions using a
technique that does not require bisulfite treatment of DNA to detect
methylation.
Example
The cutting efficiency of amplicons by Cas9 in plasma is shown by experiment.
Results
from the experiment indicated that Cas proteins bind to expected cognate
targets under guide
24
CA 03100492 2020-11-16
WO 2019/221769 PCT/US2018/054188
RNA guidance in plasma or serum. In particular, Cas9 was tested for cutting
activity in plasma in
an experimental protocol.
Plasma samples were placed in Streck tubes and in standard tubes. The
experiments used
an 800 bp amplicon from the cystic fibrosis transmembrane receptor gene.
Dilutions were made
of CFTR F2 800 bp into plasma with 5 million copies per reaction total (Figure
1). The percent
plasma in reaction after dilution was 50%, 25%, 16.7%, 10%, 2%, 1%, 0.5%,
0.2%, 0.1%,
and 0% (Figure 2).
Cas9 with guide RNA was added and allowed to cut. qPCR was then used to probe
across
the cut site. For qPCR, samples were diluted 1/100, and then 5 ul were used
per 20 ul reaction.
The qPCR results were analyzed from amplifying, post-cutting, from dilutions
(Figures 3 and 4).
The qPCR results indicated cleavage as a function of plasma amount (Figure 5).
For example,
every replicate in a Streck tube demonstrated greater than 60% cutting
efficiency by Cas9 in the
CFTR amplicon. Cas9 exhibited detectable cutting, even in standard, non-Streck
tubes.
The results also indicated a relationship between the qPCR signal and percent
plasma
(Figure 6). For example, the data show Cas9 exhibits detectable cutting in Na-
EDTA plasma. For
the reactions performed in straight plasma, cutting efficiency in 2% plasma or
lower resembled
no plasma cutting efficiency (82.82% for in plasma compared to 79.97% in no
plasma). For the
reactions performed in plasma incubated in a Streck tube, the cutting
efficiency in 25% plasma
or lower resembled no-plasma cutting efficiency (83.14% compared to 78.90%).
Further, there
was 60-67% cutting for the 50% plasma samples. In 50% plasma, CRISPR/Cas9
complexes
retained 75% activity. Results of the data show that Cas endonuclease and
homologs thereof bind
to target DNA under guidance of guide RNA in plasma.
In another example, sequence specific Cas9/gRNA complexes were added directly
to a
biological sample, a plasma sample. Here, a complex of Cas9-guide RNA and a
particle, in this
example, a magnetic bead coated with streptavidin, is directly inserted into
the plasma sample.
That is, the magnetic bead bound Cas proteins bind to expected targets under
guide RNA
guidance in plasma (or serum) and cut and protect the target region to achieve
enrichment. In this
example the target region is suspected to be hypermethylated and the complexes
were
complementary to both sides the target region of DNA. The complexes therefore
targeted and
bound to PAM sequences that were adjacent to the target region suspected to be
hypermethylated.
CA 03100492 2020-11-16
WO 2019/221769 PCT/US2018/054188
The particle-bound complexes were then isolated from the remaining sample
using a
separator, in this case, a magnetic field was used to separate out the
particle-bound complexes.
Once target specific positive enrichment was completed, antibodies that bind
only to methylated
DNA, and not un-methylated DNA were added to the enriched sample. An ELISA was
then
performed to detect the signal produced by the binding of the methylation
specific antibodies to
the methylated target region of DNA. Results from the experiment indicated
that Cas proteins
bind to expected target regions that are suspected to be hypermethylated under
guide RNA
guidance in a liquid biopsy, such as plasma or serum, allowing for the use of
binding assays,
such as an ELISA to detect the presence of methylation at the specific target
region. Such an
ability to detect the methylation status of known cancer-associated regions,
allows for the early
detection of cancer and tumor growth.
Incorporation by Reference
References and citations to other documents, such as patents, patent
applications, patent
publications, journals, books, papers, web contents, have been made throughout
this disclosure.
All such documents are hereby incorporated herein by reference in their
entirety for all purposes.
Equivalents
The invention may be embodied in other specific forms without departing from
the spirit
or essential characteristics thereof. The foregoing embodiments are therefore
to be considered in
all respects illustrative rather than limiting on the invention described
herein. Scope of the
invention is thus indicated by the appended claims rather than by the
foregoing description, and
all changes which come within the meaning and range of equivalency of the
claims are therefore
intended to be embraced therein.
26