Note: Descriptions are shown in the official language in which they were submitted.
CA 03105726 2021-01-05
WO 2020/010304 PCT/US2019/040683
1
BC1-2 Antibodies and Immunoassay for Diagnosis of Cancer
Priority and Incorporation by Reference
[001] This application claims priority to United States Provisional
Application 62/694,142
filed on July 5, 2018. All references cited herein are expressly incorporated
by
reference.
Background
[002] The use of BCL-2 as a cancer marker is described in U.S. patent
8,034,549 and
published patent applications US 2011-0318763 Al and US 2016-0258960 Al. While
these references teach antibodies to BCL-2, an antibody meeting regulatory
manufacturing requirements and having improved specificity was required.
[003] There are several anti-BCL-2 monoclonal antibodies described in the
literature,
however, a need exists for new anti-BCL-2 antibodies having unique genetic and
amino
acid structures, including unique binding and functional characteristics.
Brief Description of the Invention
[004] The present invention is directed to antibodies and fragments thereof
that bind to human
BCL-2. The antibodies may be labeled with one or more labels selected from the
group
consisting of a biotin label, a fluorescent label, an enzyme label, a coenzyme
label, a
chemiluminescent label, colloidal particles, and a radioactive isotope label.
[005] The invention is also directed to a hybridoma cell line that produces
the antibody, and
to methods of treating cancer and purifying exosomes using the antibody or
antigen-
binding fragments thereof.
[006] The present invention is also directed to a method for diagnosing
cancer, wherein the
method comprises: reacting an anti- BCL-2 antibody with a sample collected
from the
subject; detecting an BCL-2 protein in the sample; and diagnosing cancer when
the
level of BCL-2 protein is higher in the sample than in a normal sample,
wherein the
sample collected from the subject is at least one clinical sample selected
from the group
consisting of a urine sample, saliva sample, tissue sample, a blood sample, a
serum
sample, a plasma sample, tears, or a mucous sample.
[007] The antibodies of the present invention are preferably isolated
monoclonal antibodies
having specific binding properties against a human BCL-2 protein, more
preferably
against human BCL-2 in its native form.
CA 03105726 2021-01-05
WO 2020/010304 PCT/US2019/040683
2
[008] The invention is also directed to anti- BCL-2 antibody based
immunoassays for
detecting ovarian cancer.
Description of the Figures
[009] Figure 1 Human Bc1-2 Sequence & secondary structure prediction
[010] Figure 2: Homology of BCL-2 and BCL-x:
[011] Figure 3: Homology of Human and Mouse Bc1-2.
[012] Figure 4 shows the alignment of epitopes to Bc1-2.
[013] Figure 5A-C: Structural predictions of Bc1-2 binding sites.
[014] Figure 6 is an electropherogram of a Bc1-2 antibody of the present
invention.
[015] Figure 7 is an electropherogram of a Bc1-2 antibody of the present
invention.
[016] Figure 8 is an image of an electrophoresis gel of a Bc1-2 antibody of
the present
invention.
[017] Figure 9 is a chromatograph of a Bc1-2 antibody of the present
invention.
[018] Figure 10 is an isoelectric gel of a Bc1-2 antibody of the present
invention.
[019] Figure 11 is an image of an electrophoresis gel of a Bc1-2 antibody of
the present
invention.
[020] Figure 12 is a chromatograph of a Bc1-2 antibody of the present
invention.
[021] Figure 13 is an isoelectric gel of a Bc1-2 antibody of the present
invention.
[022] Figure 14 is an image of an electrophoresis gel of a Bc1-2 antibody of
the present
invention.
[023] Figure 15 is an isoelectric gel of a Bc1-2 antibody of the present
invention.
[024] Figure 16 is a chromatograph of a Bc1-2 antibody of the present
invention.
[025] Figure 17 is an image of an electrophoresis gel of a Bc1-2 antibody of
the present
invention.
[026] Figure 18 is a chromatograph of a Bc1-2 antibody of the present
invention.
[027] Figure 19 is an isoelectric gel of a Bc1-2 antibody of the present
invention.
[028] Figure 20 is an image of an electrophoresis gel of a Bc1-2 antibody of
the present
invention.
[029] Figure 21 is an isoelectric gel of a Bc1-2 antibody of the present
invention.
[030] Figure 22 is a chromatograph of a Bc1-2 antibody of the present
invention.
[031] Figure 23 is a graph showing absorbance of antibodies of the present
invention.
[032] Figure 24 is a DNA amino acid alignment for Clone 10Al2 Heavy Chain
[033] Figure 25 DNA is a DNA amino acid alignment for Clone 10Al2 Light Chain
CA 03105726 2021-01-05
WO 2020/010304 PCT/US2019/040683
3
[034] Figure 26 DNA is a DNA amino acid alignment for Clone 14A09 Heavy Chain
[035] Figure 27 DNA is a DNA amino acid alignment Clone 14A09 Light Chain
[036] Figure 28 is a photograph of a lateral flow assay.
[037] Figure 29 is a photograph of a lateral flow assay.
[038] Figure 30 is a photograph of a lateral flow assay.
[039] Figure 31 is a photograph of a lateral flow assay.
[040] Figure 32 is a photograph of a lateral flow assay.
[041] Figure 33 is a photograph of a lateral flow assay.
[042] Figure 34 is a photograph of a lateral flow assay.
[043] Figure 35 is a photograph of a lateral flow assay.
[044] Figure 36 is a photograph of a lateral flow assay.
[045] Figure 37 is a photograph of the lateral flow assay in a cassette.
Detailed Description of the Invention
[046] The present invention is directed to antibodies and fragments thereof
that bind to human
BCL-2. The antibodies may be labeled with one or more labels selected from the
group
consisting of a biotin label, a fluorescent label, an enzyme label, a coenzyme
label, a
chemiluminescent label, colloidal particles, and a radioactive isotope label.
[047] The invention is also directed to a hybridoma cell line that produces
the antibody, and
to methods of treating cancer and purifying exosomes using the antibody or
antigen-
binding fragments thereof.
[048] The present invention is also directed to a method for diagnosing
cancer, wherein the
method comprises: reacting an anti- BCL-2 antibody with a sample collected
from the
subject; detecting an BCL-2 protein in the sample; and diagnosing cancer when
the
level of BCL-2 protein is higher in the sample than in a normal sample,
wherein the
sample collected from the subject is at least one sample selected from the
group
consisting of a tissue sample, a urine sample, a saliva sample, a blood
sample, a serum
sample, and a plasma sample.
[049] The antibodies of the present invention are preferably isolated
monoclonal antibodies
having specific binding properties against a human BCL-2 protein, more
preferably
against human BCL-2 in its native form.
[050] In one form the invention comprises an ELISA in which a first antibody
binds to Bc1-2
anchoring it to a substrate and a second antibody is labelled for
identification. The label
is typically selected from a biotin label, a fluorescent label, an enzyme
label, a
CA 03105726 2021-01-05
WO 2020/010304 PCT/US2019/040683
4
coenzyme label, a chemiluminescent label, colloidal particles, and a
radioactive isotope
label.
[051] The term "antibody" as used herein refers to any naturally occurring
antibody or
antigen-binding protein, the production of which is induced by an immune
system
(immunoglobulins or IgGs). "Conventional" antibodies comprise two heavy chains
linked together by disulfide bonds and two light chains, one light chain being
linked to
each of the heavy chains by disulfide bonds. Each heavy chain has at one end a
variable
domain (VH) followed by a number of constant domains (three or four constant
domains, CHL CH2, CH3 and CH4, depending on the antibody class). Each light
chain
has a variable domain (VL) at one end and a constant domain (CL) at its other
end; the
constant domains of the light chains each align with the first constant
domains of the
heavy chains, and the light chain variable domains each align with the
variable domains
of the heavy chains. This type of antibodies exist in camels, dromedaries and
llamas
along with an "unconventional" naturally occurring type of antibodies
consisting of
only two heavy chains, and thus being devoid of light chains. Other
"unconventional"
naturally occurring antibodies exist in in the serum of nurse sharks
(Ginglymostomatidae) and wobbegong sharks (Orectolobidae). These latter
antibodies
are called Ig new antigen receptors (IgNARs). They are disulfide-bonded
homodimers
consisting of five constant domains (CNAR) and one variable domain (VNAR).
There
is no light chain, and the individual variable domains are independent in
solution and
do not appear to associate across a hydrophobic interface (Greenberg et al.
1995, Nature
374, 168-173; Nuttall et al. 2001, Mol Immunol 38, 313-326; Diaz et al. 2002,
Immunogenetics 54, 501-512; Nuttall et al. 2003, Eur J Biochem 270, 3543-
3554). Due
to the heavy chain dimer structure characteristic of camelid and shark
antibodies, these
are sometimes termed "Heavy-Chain Mini-Antibodies" (mnHCAbs) or simply "Mini-
Antibodies" (mnAbs) (Holliger & Hudson 2005, Nature Biotechnol 23, 1126-1136).
The complementary determining region 3 (CDR3) of camel antibodies and shark
antibodies is usually longer (comprising about 16-21 amino acids, and about 16-
27
amino acids, respectively) than the CDR3 of mouse VH region (comprising about
9
amino acids) (Muyldermans et al. 1994, Prot Eng 7, 1129-1135; Dooley & Flajnik
2005,
Eur J Immunol 35, 936-945). Without the light chain, these heavy-chain
antibodies bind
to their antigens by one single domain, the variable antigen binding domain of
the
heavy-chain immunoglobulin, referred to as Vab (camelid antibodies) or V-NAR
(shark
antibodies). These smallest intact and independently functional antigen-
binding
CA 03105726 2021-01-05
WO 2020/010304 PCT/US2019/040683
fragment Vab is referred to as nano-antibody or nanobody (Muyldermans 2001, J
Biotechnol 74, 277-302). Multivalent (etc. divalent, trivalent, tetravalent
and
pentavalent) Vab and/or V-NAR domains may be preferred in some instances due
to
their potentially higher cellular intake and retention and may be made by
recombinant
technology or by chemical means, such as described in WO 2010/033913. The
variable
domains of the light and/or heavy chains are involved directly in binding the
antibody
to the antigen. The variable domains of naturally occurring light and heavy
chains have
the same general structure: four framework regions (FRs) connected by three
complementarity determining regions (CDRs) (see e.g. Kabat et al. 1991,
Sequences of
Proteins of Immunological Interest, 5th Ed. Public Health Service,
National
Institutes of Health, Bethesda, Md.). The CDRs in a light or heavy chain are
held in
close proximity by the FRs and contribute to the formation of the antigen
binding site.
The term "antibody fragment" refers to any molecule comprising one or more
fragments
(usually one or more CDRs) of an antibody (the parent antibody) such that it
binds to
the same antigen to which the parent antibody binds. Antibody fragments
include Fv,
Fab, Fab', Fab'-SH, single-chain antibody molecules (such as scFv),
F(ab')2, single
variable VH domains, and single variable VL domains (Holliger & Hudson 2005,
Nature Biotechnol 23, 1126-1136). The term further includes microantibodies,
i.e. the
minimum recognition unit of a parent antibody usually comprising just one CDR
(Heap
et al. 2005, J Gen Virol 86, 1791-1800). Any of the fragments can be
incorporated in a
multivalent and/or multispecific larger molecule, e.g. mono- or bispecific
Fab2,
mono- or tri-specific Fab3, bis-scFv (mono- or bispecific), diabodies
(mono- or
bispecific), triabodies (e.g. trivalent monospecific), tetrabodies (e.g.
tetravalent
monospecific), minibodies and the like (Holliger & Hudson 2005, Nature
Biotechnol
23, 1126-1136). Any of the fragments can further be incorporated in e.g. V-NAR
domains of shark antibodies or VhH domains of camelid antibodies (nanobodies).
All
these are included in the term "antibody fragment".
[052] The term "monoclonal antibody" refers to a population of substantially
homogeneous
antibodies. In contrast to polyclonal antibody preparations that typically
include
different antibodies directed against different determinants (epitopes), each
monoclonal
antibody is directed against a single determinant on the antigen. The modifier
"monoclonal" is not to be construed as requiring production of the antibody by
any
particular method. For example, the monoclonal antibodies to be used in
accordance
with the present invention may be made by the hybridoma method first described
by
CA 03105726 2021-01-05
WO 2020/010304 PCT/US2019/040683
6
Kohler & Milstein 1975, Nature 256, 495-497), or may be made by recombinant
DNA
methods (e.g. U.S. Pat. No. 4,816,567). Monoclonal antibodies may also be
isolated
from phage antibody libraries using the techniques described in, e.g.,
Clackson et al.
1991, Nature 352, 624-628 or Marks et al. 1991, J Mol Biol 222, 581-597, or by
yet
other techniques or technologies.
[053] In the hybridoma method, a mouse or other appropriate host animal, such
as a hamster
or macaque monkey, is immunized to elicit lymphocytes that produce or are
capable of
producing antibodies that will specifically bind to the protein used for
immunization
(Harlow & Lane; Antibodies: A Laboratory Manual, Cold Spring Harbor Laboratory
Press: Cold Spring Harbor, N.Y. (1988).
[054] The present invention also encompasses nucleic acid molecules encoding
antibodies of
the invention. In some embodiments, different nucleic acid molecules encode a
heavy
chain variable region and a light chain variable region of an antigen-specific
antibody.
In other embodiments, the same nucleic acid molecule encodes a heavy chain and
a
light chain variable regions of an antigen-specific antibody.
[055] DNA encoding a monoclonal antibody of the invention may be isolated and
sequenced
from a hybridoma cell secreting the antibody using conventional procedures
(e.g., by
using oligonucleotide probes that are capable of binding specifically to genes
encoding
the heavy and light chains of the monoclonal antibodies). Sequence
determination will
generally require isolation of at least a portion of the gene or cDNA of
interest. Usually
this requires cloning the DNA or, preferably, mRNA (i.e., cDNA) encoding the
monoclonal antibodies. Cloning is carried out using standard techniques (see,
e.g.,
Sambrook et al. (1989) Molecular Cloning: A Laboratory Guide, Vols 1-3, Cold
Spring
Harbor Press, which is incorporated herein by reference). For example, a cDNA
library
may be constructed by reverse transcription of polyA+ mRNA, preferably
membrane-
associated mRNA, and the library screened using probes specific for human
immunoglobulin polypeptide gene sequences. Nucleotide probe reactions and
other
nucleotide hybridization reactions are carried out at conditions enabling the
identification of polynucleotides which hybridize to each other under
specified
conditions.
[056] One exemplary set of conditions is as follows: stringent hybridization
at 42 C. in 50%
formamide, 5 times saline sodium citrate "5 SC", 20 mM Na.PO4, pH 6.8; and
washing
in 1xSSC at 55 C for 30 minutes. Formula for calculating equivalent
hybridization
conditions and/or selecting other conditions to achieve a desired level of
stringency are
CA 03105726 2021-01-05
WO 2020/010304 PCT/US2019/040683
7
well known. It is understood in the art that conditions of equivalent
stringency can be
achieved through variation of temperature and buffer, or salt concentration as
described
Ausubel, et al. (Eds.), Protocols in Molecular Biology, John Wiley & Sons
(1994), pp.
6Ø3 to 6.4.10. Modifications in hybridization conditions can be empirically
determined or precisely calculated based on the length and the percentage of
guanosine/cytosine (GC) base pairing of the probe. The hybridization
conditions can
be calculated as described in Sambrook, et al., (Eds.), Molecular Cloning: A
Laboratory
Manual, Cold Spring Harbor Laboratory Press: Cold Spring Harbor, N.Y. (1989),
pp.
9.47 to 9.51
The immunoassay of the present invention can be used with any suitable
clinical
specimen. Preferred specimens include urine, saliva, blood, serum, and plasma.
The
specimens are collected and processed in conventional ways before tested using
the
immunoassay of the present invention.
Example 1: Human BCL-2 Antigenicity Determination
[057] The challenge was to produce a matched pair of monoclonal antibodies to
the human
BCL-2 protein that will work in a sandwich ELISA format. To accomplish this,
peptides were be designed as antigens to regions with a high likelihood of
generating specificity to the BCL-2 protein, but not cross react with BCL-x.
[058] Bioinformatic tools were used to analyze the protein as well as try to
predict the best
options for choosing peptides to be developed as antigens for the hybridoma
projects.
It should be noted that this analysis is a computational prediction and is
valuable in
assisting in the decision process, however, these predictions are in no way
considered
guarantees for success of each individual project or the overall stated goal.
Human BCL-2 Sequence & secondary structure prediction:
[059] The secondary structure is presented graphically in Figure 1. Alpha
helical
predictions are shown by pink cylinders, beta strands by yellow arrows, and
turns by
blue arrows. Unstructured or loop regions are shown by grey squiggly lines.
CA 03105726 2021-01-05
WO 2020/010304 PCT/US2019/040683
8
Homology of BCL-2 & BCL-x:
[060] Referring to Figure 2, overall the homology of BCL-2 with BCL-x is 41%.
However,
there are several structural domains that are conserved between the two
proteins and
the structural homology may be higher than is predicted by the sequence
homology.
Of note are two regions that are quite dissimilar between the two: region 30-
100 (BCL-
2 sequence) & 205-241. The region between these two areas are quite conserved
at
both the sequence and structural level, therefore these two non-homologous
regions
would likely be the most appropriate regions to develop antibodies that target
BCL- 2
and are not cross reactive with BCL-x.
Homology of Human and Mouse BCL-2:
[061] Since the monoclonal antibodies are going to be generated in mouse, the
similarity
of the human and mouse BCL-2 protein needs to be considered. Referring to
Figure
3, the overall homology is 90% between the two species, however there is a
region
between residues 35 and 81 that shows the highest level of divergence. With
the
exception of aa 99, the remainder of the protein is 100% identical. Therefore
the most
likely region for antibody generation would be in this area.
Linear Epitope Prediction:
[062] To predict areas that might be antigenic and therefore be good targets
for antibody
generation, analysis of potential epitopes was done using two methods that
predict
linear epitopes based on the sequence of the protein. This can be useful in
identifying
specific peptides to use for immunization. The results are shown in the Table
1 below
and those that fall within the region of non-homology (between mouse & human)
are
highlighted in yellow.
[063] Table 1
KT Analysis
No Start End Sequence
Length
1 15 26 VMKYIHYKL SQR(SEQ ID NO 1)
12
2 35 52 VGAAPPGAAPAPGIFSSQ (SEQ ID NO 2)
18
3 54
98 GHTPHPAASRDPVARTSPLQTPAAPGAAAGPALSPVPP 45
VVHLTLR (SEQ ID NO 3)
4 116 124 SSQLHLTPF (SEQ ID NO 4)
9
CA 03105726 2021-01-05
WO 2020/010304 PCT/US2019/040683
9
130 137 FATVVEEL (SEQ ID NO 5) 8
6 145 151 GRIVAFF (SEQ ID NO 6) 7
7 155 162 GVMCVESV (SEQ ID NO 7) 8
8 167 173 SPLVDNI (SEQ ID NO 8) 7
196 204 DAFVELYGP (SEQ ID NO 9) 9
BiPred
Analysis
1 3 11 HAGRTGYDN (SEQ ID NO 10) 9
2 27 90 GYEWDAGDVGAAPPGAAPAPGIFSSQPGHTPHPAASR 64
DPVARTSPLQTPAAPGAAAGPALSPVP (SEQ ID NO 11)
3 100 106 AGDDF SR (SEQ ID NO 12) 7
4 190 196 QDNGGWD (SEQ ID NO 13) 7
Example 2: Hybridoma Development
[064] Generation of Antigens
[065] A C terminus 10X His BCL-2 fragment HuBc1-2 10x His pET303 was generated
in
BL21 (DE3) E.coli and the resultant protein purified on Immobilized Metal
Affinity
Chromatography = IMAC (RL/P0003). 20mM Sodium Phosphate, 500mM NaC1
pH 7.4 w/ 40mM Imidazole, 20% Glycerol & 2mM DTT was used for binding and
20mM Sodium Phosphate, 500mM NaCl, pH 7.4, 500mM Imidazole, 20% Glycerol &
2mM DTT was used for elution. 5.2 gm of protein were recovered and
reconstituted in
20mM Tris pH 7.5,100mM NaCl, 10% Glycerol, & 5mM DTT, 2mM EDTA at a final
concentration: 0.7 mg/mL in a final volume of 7.5 mL. Referring to Figure 6,
purity
was determined to be greater than 95% as determined by Experion Pro260
Automated
Electrophoresis. Molecular weight was approximately 26 KD. The process was
repeated a second time and yielded a purity of 99%. See Figure 7.
Hybridoma 1
[066] Antibodies were generated using C57BL mice and hybridomas were created.
MBS
Accession: 15-0264RB2 for Cell Line: Bc1-2-41 Pep-KLH-BA-1.3-17D07-02D12
was conjugated with biotin and molar ratio quantified with (4'-
hydroxyazobenzene-
2-carboxylic acid) ("HABN") avidin assay which determined that 3.5 molecules
of
biotin per conjugated antibody from this hybridoma. Antibodies were purified
using
protein A affinity chromatography with a binding buffer of 0.5M NaCl in 0.1M
Citrate/Phosphate buffer, pH=9.0 and an elution buffer of 0.1M
citrate/phosphate ,
0.15M NaCl, pH=3Ø The antibody eluted along a linear gradient with a pH
range of
elution from 4.7-3.3. See Figure 8. 28.3mg antibodies were produced.
CA 03105726 2021-01-05
WO 2020/010304 PCT/US2019/040683
[067] Referring to Figure 9 purity was determined to be 100 percent using size
exclusion
chromatography. Referring to Figure 10, the isoelectric point was determined
to be
around 7.8 using 20 1 per well with a focusing conditions of 100v for 1 hour,
250v for
1 hour and 500v for 30 minutes.
Hybridorna 2
[068] Antibodies were generated using C57BL mice and hy bridomas were created.
MBS
Accession: 15-0264RE31 for Cell :Line: Bc1-2-41 Pep-KLH-BA-1.3-10Al2-02F02
was conjugated with biotin and molar ratio quantified with HABN avidin assay
which determined that 6.2 molecules of biotin per conjugated antibody from
this
hybridoma. Antibodies were purified using protein A affinity chromatography
with
a binding buffer of 0.5M NaC1 in 0.1M Citrate/Phosphate buffer, pH=9.0 and an
elution buffer of 0.1M citrate/phosphate, 0.15M NaC1, pH=3Ø The antibody
eluted along a linear gradient with a p1-1 range of elution from 6.2-3.9. See
Figure
11. 33.0 mg of antibody were recovered.
[069] Referring to Figure 12 purity was determined to be 100 percent using
size exclusion
chromatography. Referring to Figure 13, the isoelectric point was determined
to
be around 6.7 using 20g1 per well with a focusing conditions of 100v for 1
hour,
250v for] hour and 500v for 30 minutes.
Hybridoma 3
[070] Antibodies were generated using C57RL mice and hybridomas were created.
MBS
Accession: 15-0271 RBI for Cell Line: Bc1-2-61 Pep-KLH-BA-1.1-1 8E10-02G02
was conjugated with biotin and molar ratio quantified with HABN avidin assay
which determined that 6.2 molecules of biotin per conjugated antibody from
this
hybridoma. Anti bodies were purified using protein A affinity chromatography
with
a binding buffer of 0.5M NaC1 in 0.1M Citrate/Phosphate buffer, pH=9.0 and an
elution buffer of 0.1M citrate/phosphate, 0.15M: NaC1, pH=3Ø The antibody
eluted along a linear gradient with a pH range of elution from 6.7-4.6. See
Figure
14. 33.0 mg of antibody were recovered.
[071] Referring to Figure 15 purity was determined to be 100 percent using
size exclusion
chromatography. Referring to Figure 16, the isoelectric point was determined
to
be around 6.7 using 20111 per well with a focusing conditions of 100v for 1
hour,
250v for 1 hour and 500v for 30 minutes.
CA 03105726 2021-01-05
WO 2020/010304 PCT/US2019/040683
11
Hybridoma 4
[072] Antibodies were generated using C57131_, mice and hybridomas were
created. MBS
Accession: 15-0271RB2 for Cell Line: Bc1-2-61 Pep-KLH-BA-1. 1-02C10-02D02-
02B07 was conjugated with biotin and molar ratio quantified with HABN avidin
assay which determined that 10.5 molecules of biotin per conjugated antibody
from
this hybridoina. Antibodies were purified using protein A affinity
chromatography
with a binding buffer of 0.5M NaCl in 0.1M Citrate/Phosphate buffer, pH=9.0
and
an elution buffer of 0.1M citrate/phosphate, 0.15M NaC1, pH=3Ø The antibody
eluted along a linear gradient with a pH range of elution from 6.4-3.9. See
Figure
17. 21.8 mg of antibody were recovered.
[073] Referring to Figure 18 purity was determined to be 100 percent using
size exclusion
chromatography. Referrinp.. to Figure 19, the isoelectric point was determined
to
be around 6.5 using 20pil per well with a focusing conditions of 100v for 1
hour,
250v for 1 hour and 500v for 30 minutes.
Hybridoma 5
[074] Antibodies were generated using C57BL mice and hybridomas were created.
MBS
Accession: 15-0271RB3 for Cell Line: Bc1-2-61 Pep-KLH-BA-1.1-14A09-02C08-
02F06 was conjugated with biotin and molar ratio quantified with HABN avidin
assay
which determined that 7.0 molecules of biotin per conjugated antibody from
this
hybridoma. Antibodies were purified using protein A affinity chromatography
with a
binding buffer of 0.5M NaC1 in 0.1M Citrate/Phosphate buffer, pH=9.0 and an
elution
buffer of 0.1M citrate/phosphate, 0.15M NaC1, pH=3Ø The antibody eluted
along a
linear gradient with a pH range of elution from 4.7-3.3. See Figure 20. 23.5
mg of
antibody were recovered.
[075] Referring to Figure 21 purity was determined to be 100 percent using
size exclusion
chromatography. Referring to Figure 22, the isoelectric point was determined
to be
around 6.7 using 20 1 per well with a focusing conditions of 100v for 1 hour,
250v for
1 hour and 500v for 30 minutes.
CA 03105726 2021-01-05
WO 2020/010304 PCT/US2019/040683
12
Example 3: ELISA
[076] Research was conducted to identify monoclonal antibody pairs capable of
specific
detection of Bc1-2 in human urine sample matrix using a sandwich ELISA format
at a
clinically-relevant 1-20ng/mL sensitivity range.
Sandwich ELISA pre-work with Commercial Reagents:
[077] The commercial Bc1-2 reagents used for the initial assays are shown in
Table 2.
[078] Table 2
Reagent Source
mAb Bc1-2/100 Identified as an assay component in the Bender/eBioscience
kit). Received two vials of 100 ug. One was biotinylated.
mAb Identified as an assay component in the
4D7 Bender/eBioscience kit, received additional 100 ug
and performed biotinylation.
rBc1-2 R&D Systems product, Ala2-Asp211, with a C-terminal
10X His tag.
Normal For use in matrix testing. Received sample pool from
human urine three donors; Lee BioSolutions.
SKOV-3 cell Cancer cell line, over-expressing human Bc1-2, for
lysate evaluating detection of native, clinical samples.
Received
one vial of 100 ug protein; Origene.
Bc1-2 HEK293T Origene; received two 20 ug vials, along with HEK293
overexpression negative control lysate.
lysate:
rBc1-2 MBS-produced recombinant Bc1-2, Metl-Asp211, with
a C-terminal 10X His tag.
[079] A sandwich ELISA was developed reproducing the standard curve
performance of the
Bender Platinum Bc1-2 kit, using rBc1-2 proteins above from R&D Systems and an
inhouse manufactured protein. Free and biotinylated forms of mAb Bc1-2/100 and
mAb 4D7 were crossed in a sandwich ELISA with the R&D Systems protein.
[080] The most sensitive results were obtained with Bc1-2/100 capture antibody
and 4D7
biotin conjugate. However, the sensitivity limited by the high negative sample
optical
density's (OD) observed with this antibody pairing.
[081] Side by side comparisons of the R&D Systems protein and the inhouse
protein
showed them to be functionally equivalent. OD values of pertinent
concentrations of
CA 03105726 2021-01-05
WO 2020/010304 PCT/US2019/040683
13
protein were directly comparable to the average ODs published in the Bender
Platinum ELISA product insert.
[082] However, during semi-optimization of assay format, found lack of day to
day
reproducibility with the protein calibrators. Storage was at minus 80 C, and
activity
appeared to decline with freeze-thaw cycles.
[083] This led to investigations with multiple storage buffers and assay
buffers. Selected a
PBS-BSA system of blocking, sample diluent and conjugate diluent to minimize
background while maximizing signal. Theorized that slow re- solubilization of
the
protein stock upon thawing is part of the consistency problem. Also believe
that the
problem is with the immune-stability of the protein, which may be alleviated
with
different antibody pairs.
[084] The final assay with these reagents included the SKOV-3 and HEK293
lysates. Each
was run at a 1:5 dilution in sample diluent. The SKOV-3 sample had a blank-
subtracted OD of 0.1, confounded by the negative HEK293 lysate at 0.09. The 10
ng/mL protein sample ran at 0.67, which puts it in line with the Platinum
ELISA kit.
The Bc1-2-expressing lysate ran at 0.24 OD, but used an entire vial of the
product for
just the one assay.
[085] With the goal of measuring Bc1-2 in urine at concentrations less than 2
ng/mL in
normal subjects to an average of only 4 ng/mL in cancer patients', a more
sensitive
ELISA is needed. Urine sample matrices are known to interfere with
immunoassays2'3, often ameliorated by sample dilution. The more sensitive the
assay,
the higher the dilution factor can be while maintaining the target sensitivity
range
[086] With this goal in mind, the generated anti-Bc1-2 hybridoma products will
be screened
for sandwich detection of rBc1-2 alongside the commercial antibody pairing.
Matched Pair Assay Characterization of Hybridoma Fusion Products:
[087] Materials:
[088] Hybridoma project BCL2-41 Pep-KLH-1.3 yielded 19 purified (MultiPure)
fusion
products; samples of each were conjugated to biotin. Four of the 19 yielded
<0.2 mg
antibody and were excluded from further testing.
[089] Hybridoma project BCL2-61 Pep-KLH-BA-1.1 yielded 20 purified (MultiPure)
fusion products; samples of each were conjugated to biotin. Three of the 20
yielded
<0.2 mg antibody and were excluded from further testing.
[090] Commercial mAb Bc1-2/100 capture antibody Commercial mAb 4D7-biotin
conjugate
CA 03105726 2021-01-05
WO 2020/010304 PCT/US2019/040683
14
[091] rBc1-2 protein ¨ MBS lot 161071BP.1P
[092] Matched pair screenings were performed in two orientations: 15 Pep-41
captures x 17
Pep-61-biotin conjugates, and 17 Pep-61 captures x 15 Pep-41 biotin
conjugates. All
coatings were at 2 ug/mL and all biotins were at 1 ug/mL. Samples: Negative
(sample
diluent) and 25 ng/mL rBc1-2 in sample diluent. The commercial mAb pairing was
repeated throughout the plates for comparison.
[093] Referring to Table 3 and Table 4, Initial heat maps showed clear
superiority of 10Al2
as a capture with 14A09 and 18E10 as the tracer antibodies. Table 5 is a
condensed
heat map
[094]
Table 3
0
Bc1-2 Pep-41 Captures with Pep-61 Biotins
6'
Absorbance Difference: 25 ng/mL Positive Well minus Negative well. OD <0.2
-a,
Fusion Positive-Biotin Tracers
W
0
o o 0 0 0 0 0 0 0
FA '' N '-' 1-' NJ 4,
. N NJ W U.3 Do --.1 OD OD NJ A 4.
Do FA I--, Co ,D 0 O Avg.
Coated > co n -n CO > CO > = > > 2 > rn 0
. 1--, FA FA c, 0 0 0 , 0 0 0 0
F., 0 0 ,A Capture
NJ NJ 0 0 ,-A W OD ln CO OD to J J
0 A CO .
OD
Fusion
Positives
...:.........:.,:......4i:r.
01G11 003 EVAM.OE -0.01 0.11 0.02 0.00 0.01 0.05
-0.03 _AM. 0.01 -0 ... .02 ..:... ... . 6 ....: 010
0.3.
02807 ' 0.01 :::i.= ===== .-....9. -0.01 0.10
0.02 0.00 0.01 0.05 -0.01 iii 2.0,Ei 0.02 -0.01
mitigni::iiiiiginiff mic 0.16 0.4
............. ? :::,,: =:::.:::.
02D04 0.02 : ::',;':0'2,..,:. ,:r.,,:xo.''''::: 0.00 0.05 0.01
0.00 0.00 0.02 -0.01 0.16 0.01 0.00 0.13 0.13
0.07 0.02 0.1
. = = = = `:i'?X::
I
03F04 0.01 : A.i., A.*,,1:. 0.00 0.09 0.01 0.01 0.00
0.04 0.00 ;:.? 0.01 -0.01 0.85 0.94 0,53 0.11 0.3
06E06 . 0.00 : 016 .::t..i:: 0.00 0.04 0.02 0.00 0.00
0.03 0.00 ,.*:i't 0.01 0.00 0.35 all 009 0.02 0.1
.......,,:::::::
P
06F06 0.01 :Wi:t:' v.$:.L aoo 0.04 0.01 0.01
0.00 0.04 0.00 *i*i:0::::%i*i: 001 aoo (L.?.
.;..1...:5.1.9:. .9.,?.. 0.10 0.3
........ .........
06H05 0.00 0.04 017 -0.13 0.02 0.01 0.00
0.00 0.01 0.00 mg.c .... ro.p. 9 7ØT...
................ 31 002 0.02 0.0 o
w
06H08 -0.01 0.05 0.07 -0.01 0.01 0.01 -0.01 aoo
002 -am. :J....A .õ aol -0.02 144
1I c. :3,...; ....!?..1 0.2 i--µ
ul
10Al2 AU PiNiigninglik 0.00 *4:0 g 0.10 0.07 44,A
0.00 "13: 0.17 0.01 t1 14
1.4
I,
iv
Gil
cn
15C07 0.03 . ;=-= =;:- .4i7 0.00 0.13 0.03 0.02
0.00 0.07 0.00 iiiil.,:.,iniiiiii...: 0.03 0.00
Iiii*.4.iimiii.i3:] .?.,..;-: 0.18 0.4
-::':'..." ' = *,:',: ,. : !*: : : : : .......'
rO'
, 0.02 m....,;.:7'.,.i....:: :.:=:.:*:*1*.i..:0*.õ:......::':::'
0.00 0.13 0.02 0.02 0.00 0.08 0.00
:i.a4is20g 0.02 0.01 Q9 1, AiOk 0.16 0.4 Iv
16E07
'
o
17D07 0.01 õ. *0 ww 0.00 0.08 0.02 0.01 0.00 0.05
0.00 .:.:t......W..:...: 0.01 0.00 ;µ,=,;::...=:M.......
õ.9....i.g..... W4t 0.10 0.3 '7
ul
18A07 0Ø0 . 4a: 0.00 0.04 0.02 0.01 0.00 0.04
0.00 ;:fmi 0.01 0.00 *gi vg gogi 0.07 0.2
19C04 0.01 ::::::::::::.. .....,....:.: -0.01 0.04
0.02 0.00 0.00 0.04 -0.01 :::,;1....:L. 001 aoa n...
:.t.,. 0.10 0.2
.,:.'.,.($:::
....'*i
20H06 0.01 :...i., ,,,,,......Am,,... 0.00 0.05
0.02 0.00 0.00 0.06 0.00 ..M.A.:.iii 0.02 0.00
ii*ii*i.i'i.iI.:.S:.1`.#:.:.A.:.:::.:::.:::.:::.:::.:::.:::.:::::,.:Sõ.::.::.::
.:.: r;,.e......IA.:.:::.:. 0.18 0.3
No Ab Ctl. -0.01 i 0.00 0.00 0.00 0.00 0.00 0.00
0.01 0.00 0.00 0.00 0.00 0.0
100/ 4D7 Bt in4N :::::::::::a *.3%: :::::::::14::::::::
.............................
:i*:.*ia'..;g7..:i:i
::::::::::13::::::: 1.3
Avg Conj OD 0.02 0.62 0.86 -0.01 0.12 0.03 0.01 0.01
0.08 0.00 1.00 0.02 0.00 0.99 1.03 0.63 0.22
. .
.0
n
,-i
cp
t..)
-a,
4,.
c.,
oe
,.,.,
[095] Table 4
BcI-2 Pep-61 Captures with Pep-41 Biotins
0
..
Absorbance Difference: 25 ng/mL Positive Well minus Negative well. OD <0.2
0
:: = === = === '. -------
----- :::::::::::::::;;;;;;;;..f.:.:.:.:.::::::::::::::::::
::::::::::::.:.:.:.:.:.:::::..:::::::::.:.::::::::::::::::::
k..)
0
1-,
Antibody-Biotin Tracers
Avg. Capture
. 8 8 8 A A tal
1
1 sT1 A
OD
:
01Al2 0.01 : 0.12 0.07 -0.01 0.00 0.00 0.00 -0.01
0.01 0.00 : 0.03 0.06 0.07 0.00 0.01 0.0
02B12 Wigi 194 000 o.00 000 000
aoo 0.01 mt aoo a'.,i'i fiii i4i:;.g6!::4 aoo 0.04 0.5
=:Y:7i:i:::::========:=============:========.,:',::i:i:fff:
:==:=:=g ;;-= :=:..i:i:i:i:i:i:i:i:i:i:i:i:i:i:i
i:i:i:i:i:i:i:i:i:i:i:i:i:i:i:
02C10 0.06 Ap... m..::. g 0.00 0.00 0.00 0.00 0.00 1118 000 a?,..m a1 114 OLO
0.03 0.3
03F10 0.01 0.06 0.03 0.00 0.00 0.00 0.00 -0.01
0.01 0.00 0.01 0.05 0.05 0.00 0.00 0.0
03G01 0.00 0.05 0.04 0.00 0.00 0.00 0.00 -0.02
0.01 0.00 0.02 0.07 0.13 0.00 0.02 0.0
05A05 0.03 0..N MC 0.00 0.00 0.01
0.00 -0.01 0.08 0.00 0.09 ining iMEN OLO 0.03 0.1
07609 0.02 0.09 0.06 0.00 0.00 0.00 0.00 0.00
0.02 0.00 0.02 0.04 0.02 0.00 0.01 0.0
P
09A05 0.01 0.07 0.05 -0.01 0.00 0.00 0.00 0.00
0.01 0.00 0.02 0.03 0.03 0.00 0.02
0.0 o
L.
r
09 H 08 0.02 Ai:Mii 0.14 0.00 0.00 0.00 0.00 0.00
0.02 0.00 0.02 0.07 0.06 0.00 0.01
0.0 o
ol
12A09 0.05 õP..:::3 :6,1:1L.; 0.00 0.00 0.01 0.00
0.01 um. 000 0.10 M5.33.N azo 0.00
0.02 0.1 ....1
I,
IV
7.,_.''''''.: ::i:K:K.,:v::::::::::::::::::::::::::::::::::.::=:::::::::::
...:::., . Mi :i:]:;:::::K:::::::K*K
14A09 g.ft iiiiiiiiigia&i:,i:,i:,i:,i:,i:,i:,i:,igai
0.00 0.00 0.01 0.01 0.01 gog. 0.01
ma...,.m2.....,:....,:magi 0.00 0.04 0.6
o
Iv
14 H 07 0.02 0.14 0.10 0.00 0.00 0.01 0.00 0.00
0.01 0.00 0.01 0.04 0.02 0.00 0.01
0.0 r
o1
15A07 0.01 0.02 0.01 0.00 0.00 0.00 0.00 0.01
0.00 0.00 0.01 0.01 0.01 -0.01
0.00 0.0 r
o1
M 0.00 0.00 0.00 001 001 Mi
aoo at'ii ::::)::::::.:SiEla4.Yi.1:i 0.00 0.04 0.7 ol
_...: \ """"""======= """"
===============================
18 E 10 I.A., ,,,,:z.. \\.. .*
.: ::....:t.... ':?.: mis
iiiiiiT.....7.:5iiiiiiiii iiiiiiiIi.:48i*i*:
19 D 04 iM:,:''.iiiMi1...i9U aoo 0.00 aoo aoo
001 (1- nn 0 00 0 03 0 6 :
iiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiii] lak:
iiiiAiiniiiiiiiii iiiiiiii]a8i:i:i: = ' '
20A08 0.08 1.30 0.,.:T 0.00 0.00
0.00 0.00 0.01 0.19 ' 0.01
iiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiiai 0.00 0.04 0.3
20G11 0.06 ..#..et.: t.:.'..,.... 0.00 0.00 0.00 0.01
0.00 0.06 0.00 0.07 ....il: '31 c'....?.&. 0.00 0.02
0.1
No Ab Ctl. 0.00 0.01 0.00 0.00 0.00 0.00 0.01 0.00
0.00 0.00 0.00 0.01 0.00 0.00 0.00 0.0
100/ 4D-Bt
:i:i:i:i:IA&:i:i:i:ii iiiiM 1.5
Avg. Conj OD 0.10 0.92 0.70 0.00 0.00 0.00 0.00
0.00 0.14 0.00 0.13 0.61 0.54
0.00 0.02 .0
n
,-i
cp
,...)
,..,
---,-7.5
cT
Cie
tin)
[096] Table 5
Absorbance Difference: 25 ng/mL Positive Well minus Negative well.
0
BcI-2 Pep-41 BcI-2 Pep-61
n.)
o
Coated
Antibody-Biotin Tracers Coated
Antibody-Biotin Tracers t,.)
o
'a
o o o o NJ NJ w 4.. 00 ks) 0
G')" iv
ca iv
0 00 0
ca o G) m o Fusion
Fusion 1- 1- 0 8 . 0 8
N, , . . 0
4,.
Positives =Positives
............................*:*::Nom::::.::.::::
02612 1.94 1.60 :t:.g ,.;:.o.: 01G11 11 15 0.11 (..87
a Ã.4 0.67 0.33 0.10 .
02607 0 . ==== 0.9:1:.: 0.10 :i*,..:,... 1 10 1-22
11.1.:....i.....afF 016 02C10 0.79 C. 66 tat 114 !i!
- - ..
õ, õ __
..............:::::::::::::::::::::::::::......................................
..:::::
03F04 0.3M
..:,..,..:õ.:,.: .. -, ,,; c: qn 0.E5,...:, 0.11 I
*!...-:i?X: :: 0.09 u "24 t.....c, ....
14/AuJ .:-...i...i.......2-..:91-......................................-2-
.49:!:!:::::.....i:-. -:-.........-:-... -.....-:-.... -:-:
____________________________________________________________________________
=.i....:...Ogg.g.:::::::-:' ''''': .:;;:;44.: :..,.---:i4.-,;;;i:
06F06 .-'.:Ift M;==1:.' 0.04 .;.:::::µ,0pi c.,..,...,...
1.00 0.52 0.10 I 18E10 ::::::1:16::::=21.44 ii,i.!w r,,..:.y..
- ........................::::,
.;,.....,,..,......,,......,...........,..,..,..,..,..,..,.. 1 w
06H08 0.05 0.07 1 0.01 .... :,.'', '-'
'''s
:==*...r..."$:"$:"..i"......"....:"...:"...:"...:"...::tr..........."..ii"i"...
.!..........Ø.....8....?.....................Ø...:1.....1 19D04
::::::iiiiiiI9i3U: - ,..,...:...: .......:
.. =:":'.?...:::::=..*========.::::::::::::::::=?..*?...:-.,i--
..,iiiiiii.ii.,:mr--ifff
10Al2 iiiiiiiiiiiiiiiiiiiiiiiiiiiiiiii --
iigriNn';:::aii...ii.ii.i=iii....i.iiiiiiiiii1R......i.õ1.:.:..i!....Ø..õ....
......õ......,r1.i.,õi:i:i 20A08 1.30 :.) =.--:' i= 1.23 . .
1.16
, gi.i.........i.........iiiiiiiiiit
n 1 q ]*.........ii.iiiiiiiiiiigimiw.
0.62 0.18 P
15C07 '1'..1:5 piiii.?!..e.iii ---
::::,..........M=ii=iiiiiiiiiiiii........::.ff............... . ..c 0
,,,...=-=:1,.*=:,:,= .::::::::E05,::;:::::: n i z
iiiapiiiiiiiiiiiiii:iiiiilMiiiiiiiiiiiiiiii:440.iiiiiiiiiii 0 E:,: 0.16
r
0
.......:..4.. ==========:***********':'*':::: --- :.**-
ii...iii...*::::-...iii...i...i..:-....iiiiiiiii...*-....i.:-...i.:-...?:-...:-
...:-...:-...:-...:-...:-...:-...:-...:-...:-...:-...:-
....?.......C..............?.?.?.?.?.?..:-.
16E07,::,..,::::............................i.:x:
,
::":-:::::............:.::::::::::::
..............................................
17D07 c_:=:*:' :;::::!.4,;,?.11.i::i ....
0.08 u al C...79 0.92 0.4 '_-':
0.10 1 ND
,
= = = = = = = = = =
=============================.:=::::::ii*iiiiiiiiiii , _, ND
. 1 0.05 q.....-in,..46......iiiiiii.......in,...:-..gnniiI5?-i=-?-i..i..i
0.õ, 0.18 I
20E106 (}..* i).34
i.i.........."........"..............i.i.i.i.i.i.i.i."..?.?....................
........i*i*?.?.?.?...........................................................
i
o
i--
i
o
u,
1.44 1.13
1-d
n
,-i
cp
t..)
=
'a
4,.
=
c,
oe
,...,
CA 03105726 2021-01-05
WO 2020/010304
PCT/US2019/040683
18
[097] Note that negative sample ODs for the pertinent pairings were all 0.08
or lower.
[098] Referring to Figure 23, in a subsequent ELISA, each of the 16 candidate
capture
antibodies were tested in pairs with 4 different biotin conjugates. Samples
ranged
from 0 to 20 ng/mL rBc1-2, and included 2 ug/mL rBc1- XL. None of the pairs
showed any cross-reactivity to the 2 ug/mL rBc1-XL sample. The capture and
conjugate antibody concentrations were not changed from the initial screen.
The data
below illustrates samples of the resulting standard curves.
[099] Table 6 summarizes characteristics of all of the fusion positives.
CA 03105726 2021-01-05
WO 2020/010304
PCT/US2019/040683
19
[100]
[101] Table 6
TM-
Pure yield Capture Conjugate
mg/mL IEF Estimated pl senstivity sensitivity Comments
MultiPures eliminated due to low antibody yield:
01F03 0.13 n/a
08H01 0.11 n/a
12806 0.02 n/a
12C12 0.11 n/a
06Al2 0.07 n/a
11F06 0.12 n/a
14E08 0.1 n/a
MultiPures eliminated due to poor sandwich performance:
06E06 0.75 7.6- 7.8 +1- -
06H05 0.54 8.2 -/-F -
19C04 0.16 7.9 + -
01Al2 0.18 too faint 6.8-7? - -
03F10 0.18 faint 8.2? - -
05A05 1.32 6.8- 7 +1- -1+
07609 1.01 6.7 - 7.1 - -
09A05 0.28 faint 6.7 - 7.1 - -
09H08 0.51 6.8 - 7.6 -1+ +1-
12A09 0.25 7.5 - 8.1 +/- -
14H07 0.73 6.9 - 7.2 - -
15A07 0.16 faint 6.6 - 7.1 - -
Functional MultiPures for further investigation:
01G11 0.7 7.1 - 7.8 ++ +1-
02607 1.3 6.8 - 7.1 ++ +++
02D04 0.5 8.1 + +++
03F04 0.2 7.9 + -
06F06 0.6 7.9 - 8 ++ -
06H08 0.4 8.2 ++ -
10Al2 0.6 6.4- 6.6 +++ +1-
15C07 0.3 7.9- 8 ++ -
16E07 1.1 8.2 ++ +1-
17D07 1.1 7.6 - 7.9 + ++ Sister to 18A07
18A07 0.8 7.6- 7.9 + ++ Sister to 17D07 - eliminate
20H06 1.1 6.9 - 7.1 ++ -
02612 0.3 6.1- 6.7 & 6.8- 7 ++ +++ Eliminate -
polyclonal
02C10 1.1 6.75 - 7 ++ +++
03G01 0.9 6.8- 7.2 - +
14A09 0.8 6.75 - 7.1 +++ +++
18E10 0.7 6.4 - 6.6 +++ +++
19D04 1.0 6.6- 6.8 & 7.5- 7.8 +++ +++ Eliminate -
polyclonal
20A08 1.1 6.7 - 7.2 ++ +++
20G11 1.0 6.6 - 7.1 + ++
[102] Shading indicates possible sister clones by IEF profile.
[103] The pairings that exhibited good sensitivity were semi-optimized for
coating
conditions and conjugate concentrations. An assay format similar to that of
the Bc1-2
CA 03105726 2021-01-05
WO 2020/010304
PCT/US2019/040683
Platinum ELISA Kit was used.
[104] These antibody pairs were tested for matrix effect by fortifying rBc1-2
protein into
varied dilutions of normal human urine (Lee Biosolutions lot 991-03-P). Note
that this
urine sample did not exhibit detectable levels of Bc1-2 in any of the
pairings.
[105] Table 7 below illustrates the relative sensitivity of these pairings
(absorbance values
for rBc1-2 calibrators) as well as the recovery of the rBc1-2 fortifications
(at 4 or 5
ng/mL).
[106] Table 7
_____________________________________________________________ normal Hu
urine; urine
rBCI-2 (ng/mL)
dilution:
1 2.5 5 10 20 30 40 1:2 1:5
1:8
CAPTURE CONJUGATE Absorbance 450 nm minus 650 nm % Recovery
10Al2 18E10 0.3 0.9 1.6 2.7 76% 96%
108%
10Al2 14A09 0.2 0.7 1.4 2.5 74% 94%
106%
10Al2 02C10 0.1 0.3 0.7 1.6 72% 92%
94%
18E10 17D07 0.08 0.2 0.6 1.4 64% 76%
88%
18E10 02607 0.08 0.2 0.5 1.3 72% 88%
100%
16E07 14A09 0.2 0.4 0.9 1.8 2.3 2.7 68%
95%
20A08 17D07 0.1 0.3 0.6 1.2 1.7 2.0 85%
85%
20A08 02607 0.1 0.3 0.6 1.1 1.5 1.9 63%
68%
14A09 17007 0.15 0.2 0.5 1.2 1.7 60% 78%
90%
20H06 14A09 0.1 0.2 0.5 1.1 1.5 1.8
72% 92% 110%
02C10 17D07 0.1 0.2 0.3 0.5 0.8 1.0 78%
85%
[107] Peptide 41 Ab is in bold. Peptide 61 Ab is in normal font.
[108] The top two pairings of 10Al2/18E10 and 10Al2/14A09 were standouts for
sensitivity. There was a trend for better recovery of fortified sample in
urine with
captures from the Peptide 41 hybridoma project. However, recoveries in the
less
sensitive pairings are likely affected by the 4 -5 ng/mL sample running at
very low
absorbance values.
Example 4 Sequencing
[109] The antibodies created in the examples above were sequenced. Two
cryovials
containing approximately 2 X 106 cells for clone 10Al2 and 14A09 were sent to
Lake
Pharma for sequencing. Lake Pharma isolated RNA from each sample and cloned
the
Heavy Chain Variable region (Vii) and Light Chain Variable region (VL) by RACE
PCR followed by ligation into a suitable vector. Multiple isolates of each
clone were
sequenced for each VH and VL.
CA 03105726 2021-01-05
WO 2020/010304 PCT/US2019/040683
21
[110] The nucleotide sequences obtained from Lake Pharma for each clone was
submitted
to IgBlast to determine the Complementarity-determining regions ("CDR") and
Framework Regions ("FR") regions as well as determine the Vh, Dh, and Jh heavy
chain (HC) gene usage and the V1 and Jl light chain (LC) gene usage for each
clone.
[111] Clone 10Al2
[112] Referring to Figure 24, the protein sequence of the heavy chain is:
[113] QVQLQQSGPQLVRPGASVKISCKASGYSFTSYWMEIWVKQRPGQGLEWIGMI
DPSDSETRLNQKFKDKATLTVDKSSSTAYMQLSRPTFEDSAVYYCERGDYYY
GSSYFAYWGQGTLVTVSA (SEQ ID NO 14)
[114] Wherein CDR1 is GYSFTSYW (SEQ ID NO 15 and CDR2 is IDPSDSET (SEQ ID
NO16 ) and CDR3 is ERGDYYYGSSYFAY (SEQ ID NO 17)
[115] The amino acid sequence is coded by the DNA sequence of SEQ ID NO 18:
[116] CAGGT GCAACT GCAGCAGT CT GGGCCTCAGCT GGTTAGGCCT GGGGCTTCAGT GAAGATAT C
CT G
CAAGGC T T CT GGT TAT T CAT T CACCAGCTACT GGAT GCACT GGGT GAAGCAGAGGCCT
GGACAAG
GT CT T GAGT GGATT GGCAT GAT T GAT CCTT CC GATAGT GAAACTAGGTTAAATCAGAAGTT CAAG
GACAAGGCCACATT GACT GTAGACAAAT CC T CCAGCACAGCC TACAT GCAACTCAGCAGACC GAC
AT TT GAGGAC T CT GC GGT CTAT TACT GT GAAAGAGGGGAT TAT TAC TAC GGTAGTAGC TACT
TT G
CT TAT T GGGGCCAGGGGACT CT GGTCACT GT CT C T GCA (SEQ ID NO 18)
[117] Referring to Figure 25, the protein sequence of the light chain is:
[118] QAVVTQESALTTSPGETITLTCRSSTGAVTTSNYATWVQEKPDHLFTGLMGG
TTYRAPGVPARF SGSLIGDKAALTITGAQTEDEAMYFCALWF SNHFWVFGGG
TKLTVL (SEQ ID NO 19)
[119] Wherein CDR1 is TGAVTTSNY (SEQ ID NO 20) and CDR2 is GTT (SEQ ID NO
21) and CDR3 is ALWFSNHFWV (SEQ ID NO 22)
[120] The amino acid sequence is coded by the DNA sequence of SEQ ID NO 23:
[121] CAGGCT GT T GT GACT CAGGAAT CT GCACT CACCACAT CACCT GGT GAAACAAT CACACT
CACT T GT CGC
T CAAGTACT GGGGCT GT TACAAC TAGTAAC TAT GCCACCT GGGT CCAAGAAAAAC CAGAT CAT T
TAT T C
ACT GGT CT GAT GGGT GGTACCACCTACCGAGCT CCAGGT GT T CCT GCCAGAT T CT CAGGCT
CCCT GATT
GGAGACAAGGCT GCCCT CACCAT CACAGGGGCACAGACT GAGGAT GAGGCAAT GTAT T T CT GT GCT
CT T
T GGT T CAGCAACCAT T T CT GGGT GT T CGGT GGAGGAACCAAACT GACT GT CCTAG (SEQ ID
NO 23)
[122] By combining a sequence having a high homology with the above-
described
heavy chain amino acid sequence with a sequence having a high homology with
the
above-described light chain amino acid sequence, it is possible to select an
antibody
having improved specificity for BCL-2. The homology is generally a homology of
CA 03105726 2021-01-05
WO 2020/010304 PCT/US2019/040683
22
80% or more, preferably a homology of 90% or more, more preferably a homology
of
95% or 96% or 97%, 98% or more, most preferably a homology of 99% or more.
Further, by combining an amino acid sequence including a substitution,
deletion, or
addition of one to several amino acid residues in the heavy chain or light
chain amino
acid sequence, it is also possible to select an antibody having a cytotoxic
activity
equivalent to that of each of the above-described antibodies. The number of
amino
acid residues to be substituted, deleted, or added is generally 10 or fewer,
preferably 5
to 6 or fewer, more preferably 2 to 3 or fewer, most preferably 1.
[123] Clone 14A09
[124] Referring to Figure 26, the protein sequence of the heavy chain is:
[125] EVQLQQSGPELVNPGASVKMSCKASGYTFTDYYLDWVKQSHGESFEWIGRA
NPYNGVTNSNQKFKGKATLTVDMSSSTAFMELNSLTFEDSAVYYCARSSFDV
WGAGTTVTVSS (SEQ ID NO 24)
[126] Wherein CDR1 is GYTFTDYY (SEQ ID NO 25) and CDR2 is ANPYNGVT (SEQ
ID NO 26) and CDR3 is ARSSFDV (SEQ ID NO 27)
[127] The amino acid sequence is coded by the DNA sequence of SEQ ID NO 28:
[128] GAGGTCCAGCTGCAACAGTCTGGACCTGAGCTGGTGAACCCTGGGGCTTC
AGTGAAGATGTCCTGTAAGGCTTCTGGATACACATTCACTGACTACTACCT
GGACTGGGTGAAGCAGAGCCATGGAGAAAGCTTTGAGTGGATTGGACGT
GCCAATCCTTACAATGGTGTTACTAACTCCAACCAGAAGTTCAAGGGCAA
GGCCACATTGACTGTTGACATGTCCTCCAGCACAGCCTTCATGGAGCTCA
ACAGCCTGACATTTGAGGACTCTGCGGTCTATTATTGTGCAAGATCAAGCT
TCGATGTCTGGGGCGCAGGGACCACGGTCACCGTCTCCTCA (SEQ ID NO
28)
[129] Referring to Figure 27, the protein sequence of the light chain is:
[130] DIVITQDELSHPVTSGESVSISCRSSKSLLYKDGKTYLNWFLQRPGQSPQLLIY
LVSTRASGVSDRFSGSGSGTDFTLEISRVKAEDVGVYYCQQPVEYPFTFGSGT
KLEIK (SEQ ID NO 29)
[131] Wherein CDR1 is KSLLYKDGKTY (SEQ ID NO 30) and CDR2 is LVS (SEQ ID
NO 31) and CDR3 is QQPVEYPFT (SEQ ID NO 32)
CA 03105726 2021-01-05
WO 2020/010304 PCT/US2019/040683
23
[132] The amino acid sequence is coded by the DNA sequence of SEQ ID NO 33:
[133] GATATTGTGATAACCCAGGATGAACTCTCCCATCCTGTCACTTCTGGAGAATCAGTTTCCATCTCCTGC
AGGTCTAGTAAGAGTCTCCTATATAAGGATGGGAAGACATACTTGAATTGGTTTCTGCAGAGACCAGGA
CAATCTCCTCAGCTCCTGATCTATTTGGTGTCCACCCGTGCATCAGGAGTCTCAGACCGGTTTAGTGGC
AGT GGGT CAGGAACAGATTT CACCCT GGAAAT CAGTAGAGT GAAGGCT GAGGAT GT GGGT GT
GTATTAC
T GT CAACAACCT GTAGAGTAT CCATT CACGTT CGGCT CGGGGACAAAGTT GGAAATAAAA (SEQ ID
NO 33)
[134] By combining a sequence having a high homology with the above-described
heavy
chain amino acid sequence with a sequence having a high homology with the
above-
described light chain amino acid sequence, it is possible to select an
antibody having
improved specificity for BCL-2. The homology is generally a homology of 80% or
more, preferably a homology of 90% or more, more preferably a homology of 95%
or
96% or 97%, 98% or more, most preferably a homology of 99% or more. Further,
by
combining an amino acid sequence including a substitution, deletion, or
addition of
one to several amino acid residues in the heavy chain or light chain amino
acid
sequence, it is also possible to select an antibody having a cytotoxic
activity
equivalent to that of each of the above-described antibodies. The number of
amino
acid residues to be substituted, deleted, or added is generally 10 or fewer,
preferably 5
to 6 or fewer, more preferably 2 to 3 or fewer, most preferably 1.
Example 5: Lateral flow Assay
[135] The antibodies described in Example 2 were tested for use in a lateral
flow assay.
[136] For the capture, each of the 5 clones was spotted at 0.5mg/mL on 4mm
wide strips
using moderate flow nitrocellulose membrane, (10 strips per clone so that
negative vs.
+100ng/mL positive sample can be analyzed for each of the 5 clones), with a
basic
striping buffer, & a control spot (rPA). The sample pads and nitrocellulose
were
untreated.
[137] For the detector: each of the 5 clones were conjugated to 40nm gold
under a standard
conjugation protocol: middle pH, with medium-strong loading of the antibody
onto
the gold, a standard gold blocker, and then combined the gold conjugates 1:1
with a
basic conjugate diluent containing sugar, buffer, protein & surfactant. We
spot dried
the gold + CD mixture onto each test strip, with each of the 5 conjugate + CD
mixtures 2 at a time for each of the strip sets from above (5 conjugates x2
strips each
= 10 strips, for each of the 5 clone sets = 50 strips).
CA 03105726 2021-01-05
WO 2020/010304 PCT/US2019/040683
24
[138] We used a simple "synthetic" running buffer and for positive sample
spiked @
10Ong/mL BCL-2.
A= 10Al2
B = 14A09
C = 18E10
D = 02C10
E= 17D07
[139] We looked for the best signal-noise (s/n) between negative vs.
positive signals,
with lowest false positive possible. Zero false positives was not a
requirement.
[140] Figure 28 shows Set "A" (A=Capture). The results were recorded at 20
mins
and the best results are boxed in Figure 28. The best detector from Set-A = B.
C and
D looked ok, but C/D/E conjugates crashed out of solution, leaving only B-
conjugate
Detector as a viable pair w/ A-Capture)
[141] Referring to Figure 29 set B, there was no Best Detector from Set-B.
Referring
to Figure 30 Set C, the best Detector from Set C was A (boxed). Referring to
Figures
31 and 32 sets D and E were not as good as A for detector. This may be because
the
C/D/E gold conjugates all crashed out of solution over time showing the
difficulty of
this assay. Also, E did not release from the pad well. The best Detector from
Set-D =
A. There was no best detector from set E. The E detector showed the strongest
overall signals, however E did not show any specificity for the target vs.
sample
without target, hence E was not a good Capture.
[142] Referring to Figures 33-35, for each of the 3 "best" pairings noted
above, we
striped the 3 best Capture antibodies from A/C/D, at 0.5mg/mL with std.
striping
buffer, goat anti-mouse control line, and for each of the 3 Capture
antibodies, their
best pair:
[143] A-capture with B-Detector (Figure 33)
[144] C-Capture with A-Detector (Figure 34)
[145] D-Capture with A-Detector (Figure 35): best
[146] In each picture we ran 3 sets. Within each set we ran 4 strips: 0,
+1, +10,
+100ng/mL BCL-2. Each of the 3 sets were different conjugate conditions of
loading
& pH comparison.
[147] After 4 more rounds of optimization of the conjugate & particularly
the
conjugate diluent, we selected the best condition in "dip strip" (standing
upright)
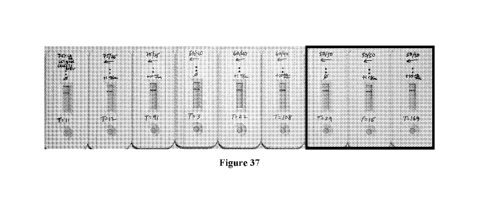
Lateral Flow Assay format (See Figure 36and then tried it in Cassette format
(Figure
CA 03105726 2021-01-05
WO 2020/010304 PCT/US2019/040683
37), whereby we could also obtain digital readings with our reader designed
for the
cassettes. In Figure 37 we selected the best condition from top & then ran
ratios of
CD/CG. Dip strip vs. cassette requires further optimization.
[148] The best pairs were 1) 02C10 Capture ("D") with 10Al2 Detector ("A"),
slightly better than set #2; 2) 18E10 Capture ("C") with 10Al2 Detector ("A"),
definitively better than set #3; and 3) 10Al2 Capture ("A") with 14A09
Detector
("B").
Example 6: Retesting of Clones in EIA
[149] We performed additional testing of ETA testing of clones for BCL-2.
Well
capture plates were coated in 200 ng/well in 1xPBS, lhr, blocked with 1% BSA,
rinsed 4x (Phospahte buffered saline with tween) , stabilized with Stabilguard
immunoassay stabilizer.
[150] We tested each biotinylated monoclonal at 1:1K dilution with 1:1K SA-
HRP
(Streptavadin conjugated with horseradish peroxidase enzume) with and without
2ng/mL rBCL-2, 30 min incubation shaking at RT, 4x PBST wash. The plates were
incubated with 3,3,5,5' 3,3',5,5'-Tetramethylbenzidine -Tetramethylbenzidine
("TMB") substrate for 7 min, stopped with an appropriate stop solution and
read at
405nm. The results are shown in Table 8.
[151] Table 8
plate DO? Al2 Al2 EH) C10 A09 AV-Al2
Abs=>
_ + _ + _ + _ + _ + _ +
DO? 1.23 1.23 0.19 0.24 0.18 0.46 0.21 0.53 0.4 1.07 0.10 0.18
1:1K 1 8 5 2 5 4 2 1 9 5
Al2 0.05 0.04 0.01 0.03 0.02 0.18 0.02 0.13 0.05 0.11 0.03 0.12
1:1K 1 7 8 2 8 6 1 1 6 7 6 3
HO 1:1K 0.05 0.06 0.01 0.03 0.06 0.11 0.04 0.05 0.15
0.07 0.03 0.03
3 6 4 2 1 4 9 7 1 9 3
C10 0.04 0.04 0.01 0.03 0.05 0.18 0.04 0.02 0.13 0.03 0.00 0.00
1:1K 2 3 1 5 8 1 4 6 3
A09 0.19 0.58 0.05 0.20 0.14 0.19 0.08 0.05 0.13 0.09 0.01 0.00
1:1K 4 5 4 7 9 2 5 2
AV-Al2 0.03 0.02 0.01 0.05 0.12 0.12 0.00 0.03 0.02 0.08 0.02 0.00
1:1K 8 8 7 1 5 8 9 5 2 8 3
no 0.03 0.03 0.01 0.01 0.04 0.08 0.01 0.01 0.01 0.00 0.04 0
CA 03105726 2021-01-05
WO 2020/010304 PCT/US2019/040683
26
secondar 8 3 4 7 3 1 3 4
no 0.03 0.01 0.01 0.07 0.04 0.00 0.04 0.00 0.01 0.00 0.02 0
secondar 2 6 1 4 2 2 1 3 5 6 2
(+)=2ng/mL; 30 min incubation, 1:1K detector + 1:1K SA-HRP; 4xPBST wash,
Xtreme
TMB 7' w/stop; read at 405nm (200ng/well capture in 1xPBS for lhr; BSA block,
w/stabilcoat on Greiner plates)
[152] The signal to noise ratio is shown in Table 9 below.
[153] Table 9 Signal to Noise
plate Abs=> D07 Al2 E10 C10 A09 AV-Al2
D07 1:1K 1.01 1.29 2.55 2.49 2.68 1.70
Al2 1:1K 0.92 1.78 6.64 6.24 2.09 3.42
E10 1:1K 1.25 2.29 1.87 1.16 0.52 1.10
Clo 1:1K 1.05 2.38 3.27 0.70 0.26 0.50
A09 1:1K 3.07 4.10 1.39 0.71 0.66 0.13
AV-Al2 1:1K 0.74 3.00 1.02 3.33 3.28 0.11
no secondary 0.87 1.21 1.86 0.91 0.30 0.00
no secondary 0.50 6.73 0.05 0.07 0.40 0.00
[154] The best pairs are shown in bold. The best pairs for LFS are the Al2
detector
with Eli) or C10 capture. The secondary set was A09 detector and Al2 capture
[155] The best pairs for ETA are Al2 detector with Eli) or C10 capture. The
secondary set was A09 detector and Al2 capture
References
1 Anderson N, Bermudez Y, Badgwell D, Chen R, Nicosia S, Bast Jr. R, Kruk P.
Urinary
levels of Cc1-2 are elevated in ovarian cancer patients. Gynecologic Oncology
112 (2009)
60-67.
2Chatziharalambous D, Lygirou V, Latosinska A, et al. Analytical Performance
of ELISA
Assays in Urine: One More Bottleneck towards Biomarker Validation and Clinical
Implementation. PLoS ONE 11(2):e0149471. Koi:10.137/journal.pone 0149471.
2016.
3Taylor T, Janech, M, Slate E, et. al. Overcoming the Effects of Matrix
Interference in the
Measurement of Urine Protein Analytes. Biomarker Insights 7 (2012) 1-8.