Note: Descriptions are shown in the official language in which they were submitted.
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
DATA BASED CANCER RESEARCH AND TREATMENT
SYSTEMS AND METHODS
CROSS-REFERENCE TO RELATED APPLICATIONS
[0001] This application claims priority to US provisional patent
application No.
62/746,997 which was filed on October 17, 201, titled "Data Based Cancer
Research
and Treatment Systems and Methods", which if incorporated herein in its
entirety by
reference.
STATEMENT REGARDING FEDERALLY SPONSORED RESEARCH OR
DEVELOPMENT
[0002] Not applicable.
BACKGROUND OF THE DISCLOSURE
[0003] The present invention relates to systems and methods for obtaining
and employing data related to physical and genomic patient characteristics as
well
as diagnosis, treatments and treatment efficacy to provide a suite of tools to
healthcare providers, researchers and other interested parties enabling those
entities
to develop new cancer state-treatment-results insights and/or improve overall
patient
healthcare and treatment plans for specific patients.
[0004] The present disclosure is described in the context of a system
related
to cancer research, diagnosis, treatment and results analysis. Nevertheless,
it
should be appreciated that the present disclosure is intended to teach
concepts,
features and aspects that will be useful in many different health related
contexts and
therefore the specification should not be considered limited to a cancer
related
systems unless specifically indicated for some system aspect.
[0005] Hereafter, unless indicated otherwise, the following terms and
phrases
will be used in this disclosure as described. The term "provider" will be used
to refer
to an entity that operates the overall system disclosed herein and, in most
cases, will
include a company or other entity that runs servers and maintains databases
and
that employs people with many different skill sets required to construct,
maintain and
adapt the disclosed system to accommodate new data types, new medical and
treatment insights, and other needs. Exemplary provider employees may include
-1-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
researchers, data abstractors, physicians, pathologists, radiologists, data
scientists,
and many other persons with specialized skill sets.
[0006] The term "physician" will be used to refer generally to any health
care
provider including but not limited to a primary care physician, a medical
specialist, a
physician, a nurse, a medical assistant, etc.,
[0007] The term "researcher" will be used to refer generally to any person
that
performs research including but not limited to a pathologist, a radiologist, a
physician, a data scientist, or some other health care provider. One person
may
operate both a physician and a researcher while others may simply operate in
one of
those capacities.
[0008] The phrase "system specialist" will be used generally to refer to
any
provider employee that operates within the disclosed systems to collect,
develop,
analyze or otherwise process system data, tissue samples or other information
types
(e.g., medical images) to generate any intermediate system work product or
final
work product where intermediate work product includes any data set,
conclusions,
tissue or other samples, grown tissues or samples, or other information for
consumption by one or more other system specialists and where final work
product
includes data, conclusions or other information that is placed in a final or
conclusory
report for a system client or that operates within the system to perform
research, to
adapt the system to changing needs, data types or client requirements. For
instance, the phrase "abstractor specialist" will be used to refer to a person
that
consumes data available in clinical records provided by a physician to
generate
normalized and structured data for use by other system specialists, the phrase
"programming specialist" will be used to refer to a person that generates or
modifies
application program code to accommodate new data types and or clinical
insights,
etc.
[0009] The phrase "system user" will be used generally to refer to any
person
that uses the disclosed system to access or manipulate system data for any
purpose
and therefore will generally include physicians and researchers that work for
the
provider or that partner with the provider to perform services for patients or
for other
partner research institutions as well as system specialists that work for the
provider.
[0010] The phrase "cancer state" will be used to refer to a cancer
patient's
overall condition including diagnosed cancer, location of cancer, cancer
stage, other
cancer characteristics (e.g., tumor characteristics), other user conditions
(e.g., age,
gender, weight, race, habits (e.g., smoking, drinking, diet)), other pertinent
medical
conditions (e.g., high blood pressure, dry skin, other diseases, etc.),
medications,
-2-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
allergies, other pertinent medical history, current side effects of cancer
treatments
and other medications, etc..
[0011] The term "consume" will be used to refer to any type of
consideration,
use, modification, or other activity related to any type of system data,
tissue samples,
etc., whether or not that consumption is exhaustive (e.g., used only once, as
in the
case of a tissue sample that cannot be reproduced) or inexhaustible so that
the data,
sample, etc., persists for consumption by multiple entities (e.g., used
multiple times
as in the case of a simple data value).
[0012] The term "consumer" will be used to refer to any system entity that
consumes any system data, samples, or other information in any way including
each
of specialists, physicians, researchers, clients that consume any system work
product, and software application programs or operational code that
automatically
consume data, samples, information or other system work product independent of
any initiating human activity.
[0013] The phrase "treatment planning process" will be used to refer to an
overall process that includes one or more sub-processes that process clinical
and
other patient data and samples (e.g., tumor tissue) to generate intermediate
data
deliverables and eventually final work product in the form of one or more
final reports
provided to system clients. These processes typically include varying levels
of
exploration of treatment options for a patient's specific cancer state but are
typically
related to treatment of a specific patient as opposed to more general
exploration for
the purpose of more general research activities. Thus, treatment planning may
include data generation and processes used to generate that data,
consideration of
different treatment options and effects of those options on patient illness,
etc.,
resulting in ultimate prescriptive plans for addressing specific patient
ailments.
[0014] Medical treatment prescriptions or plans are typically based on an
understanding of how treatments affect illness (e.g., treatment results)
including how
well specific treatments eradicate illness, duration of specific treatments,
duration of
healing processes associated with specific treatments and typical treatment
specific
side effects. Ideally treatments result in complete elimination of an illness
in a short
period with minimal or no adverse side effects. In some cases cost is also a
consideration when selecting specific medical treatments for specific
ailments.
[0015] Knowledge about treatment results is often based on analysis of
empirical data developed over decades or even longer time periods during which
physicians and/or researchers have recorded treatment results for many
different
patients and reviewed those results to identify generally successful ailment
specific
-3-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
treatments. Researchers and physicians give medicine to patients or treat an
ailment in some other fashion, observe results and, if the results are good,
the
researchers and physicians use the treatments again to treat similar ailments.
If
treatment results are bad, a researcher foregoes prescribing the associated
treatment for a next encountered similar ailment and instead tries some other
treatment, hopefully based on prior treatment efficacy data. Treatment results
are
sometimes published in medical journals and/or periodicals so that many
physicians
can benefit from a treating physician's insights and treatment results.
[0016] In many cases treatment results for specific illnesses vary for
different
patients. In particular, in the case of cancer treatments and results,
different patients
often respond differently to identical or similar treatments. Recognizing that
different
patients experience different results given effectively the same treatments in
some
cases, researchers and physicians often develop additional guidelines around
how to
optimize ailment treatments based on specific patient cancer state. For
instance,
while a first treatment may be best for a young relatively healthy woman
suffering
colon cancer, a second treatment associated with fewer adverse side effects
may be
optimal for an older relatively frail man with a similar colon same cancer
diagnosis.
In many cases patient conditions related to cancer state may be gleaned from
clinical medical records, via a medical examination and/or via a patient
interview,
and may be used to develop a personalized treatment plan for a patient's
specific
cancer state. The idea here is to collect data on as many factors as possible
that
have any cause-effect relationship with treatment results and use those
factors to
design optimal personalized treatment plans.
[0017] In treatment of at least some cancer states, treatment and results
data
is simply inconclusive. To this end, in treatment of some cancer states,
seemingly
indistinguishable patients with similar conditions often react differently to
similar
treatment plans so that there is no cause and effect between patient
conditions and
disparate treatment results. For instance, two women may be the same age,
indistinguishably physically fit and diagnosed with the same exact cancer
state (e.g.,
cancer type, stage, tumor characteristics, etc.). Here, the first woman may
respond
to a cancer treatment plan well and may recover from her disease completely in
8
months with minimal side effects while the second woman, administered the same
treatment plan, may suffer several severe adverse side effects and may never
fully
recover from her diagnosed cancer. Disparate treatment results for seemingly
similar cancer states exacerbate efforts to develop treatment and results data
sets
and prescriptive activities. In these cases, unfortunately, there are cancer
state
-4-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
factors that have cause and effect relationships to specific treatment results
that are
simply currently unknown and therefore those factors cannot be used to
optimize
specific patient treatments at this time.
[0018] Genomic sequencing has been explored to some extent as another
cancer state factor (e.g., another patient condition) that can affect cancer
treatment
efficacy. To this end, at least some studies have shown that genetic features
(e.g.,
DNA related patient factors (e.g., DNA and DNA alterations) and/or DNA related
cancerous material factors (e.g., DNA of a tumor)) as well as RNA and other
genetic
sequencing data can have cause and effect relationships with at least some
cancer
treatment results for at least some patients. For instance, in one
chemotherapy
study using SULT1A1, a gene known to have many polymorphisms that contribute
to
a reduction of enzyme activity in the metabolic pathways that process drugs to
fight
breast cancer, patients with a SULT1A1 mutation did not respond optimally to
tamoxifen, a widely used treatment for breast cancer. In some cases these
patients
were simply resistant to the drug and in others a wrong dosage was likely
lethal.
Side effects ranged in severity depending on varying abilities to metabolize
tamoxifen. Raftogianis R, Zalatoris J. Walther S. The role of pharmacogenetics
in
cancer therapy, prevention and risk. Medical Science Division. 1999: 243-247.
Other cases where genetic features of a patient and/or a tumor affect
treatment
efficacy are well known.
[0019] While corollaries between genomic features and treatment efficacy
have been shown in a small number of cases, it is believed that there are
likely many
more genomic features and treatment results cause and effect relationships
that
have yet to be discovered. Despite this belief, genetic testing in cancer
cases is the
rare exception, not the norm, for several reasons. One problem with genetic
testing
is that testing is expensive and has been cost prohibitive in many cases.
[0020] Another problem with genetic testing for treatment planning is that,
as
indicated above, cause and effect relationships have only been shown in a
small
number of cases and therefore, in most cancer cases, if genetic testing is
performed,
there is no linkage between resulting genetic factors and treatment efficacy.
In other
words, in most cases how genetic test results can be used to prescribe better
treatment plans for patients is unknown so the extra expense associated with
genetic
testing in specific cases cannot be justified. Thus, while promising, genetic
testing
as part of first-line cancer treatment planning has been minimal or sporadic
at best.
[0021] While the lack of genetic and treatment efficacy data makes it
difficult
to justify genetic testing for most cancer patients, perhaps the greater
problem is that
-5-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
the dearth of genomic data in most cancer cases impedes processes required to
develop cause and effect insights between genetics and treatment efficacy in
the first
place. Thus, without massive amounts of genetic data, there is no way to
correlate
genetic factors with treatment efficacy to develop justification for the
expense
associated with genetic testing in future cancer cases.
[0022] Yet one other problem posed by lack of genomic data is that if a
researcher develops a genomic based treatment efficacy hypothesis based on a
small genomic data set in a lab, the data needed to evaluate and clinically
assess
the hypothesis simply does not exist and it often takes months or even years
to
generate the data needed to properly evaluate the hypothesis. Here, if the
hypothesis is wrong, the researcher may develop a different hypothesis which,
again, may not be properly evaluated without developing a whole new set of
genomic data for multiple patients over another several year period.
[0023] For some cancer states treatments and associated results are fully
developed and understood and are generally consistent and acceptable (e.g.,
high
cure rate, no long term effects, minimal or at least understood side effects,
etc.). In
other cases, however, treatment results cause and effect data associated with
other
cancer states is underdeveloped and/or inaccessible for several reasons.
First,
there are more than 250 known cancer types and each type may be in one of
first
through four stages where, in each stage, the cancer may have many different
characteristics so that the number of possible "cancer varieties" is
relatively large
which makes the sheer volume of knowledge required to fully comprehend all
treatment results unwieldy and effectively inaccessible.
[0024] Second, there are many factors that affect treatment efficacy
including
many different types of patient conditions where different conditions render
some
treatments more efficacious for one patient than other treatments or for one
patient
as opposed to other patients. Clearly capturing specific patient conditions or
cancer
state factors that do or may have a cause and effect relationship to treatment
results
is not easy and some causal conditions may not be appreciated and memorialized
at
all.
[0025] Third, for most cancer states, there are several different treatment
options where each general option can be customized for a specific cancer
state and
patient condition set. The plethora of treatment and customization options in
many
cases makes it difficult to accurately capture treatment and results data in a
normalized fashion as there are no clear standardized guidelines for how to
capture
that type of information.
-6-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
[0026] Fourth, in most cases patient treatments and results are not
published
for general consumption and therefore are simply not accessible to be combined
with
other treatment and results data to provide a more fulsome overall data set.
In this
regard, many physicians see treatment results that are within an expected
range of
efficacy and conclude that those results cannot add to the overall cancer
treatment
knowledge base and therefore those results are never published. The problem
here
is that the expected range of efficacy can be large (e.g., 20% of patients
fully heal
and recover, 40% live for an extended duration, 40% live for an intermediate
duration
and 20% do not appreciably respond to a treatment plan) so that all treatment
results
are within an "expected" efficacy range and treatment result nuances are
simply lost.
[0027] Fifth, currently there is no easy way to build on and supplement
many
existing illness-treatment-results databases so that as more data is
generated, the
new data and associated results cannot be added to existing databases as
evidence
of treatment efficacy or to challenge efficacy. Thus, for example, if a
researcher
publishes a study in a medical journal, there is no easy way for other
physicians or
researchers to supplement the data captured in the study. Without data
supplementation over time, treatment and results corollaries cannot be tested
and
confirmed or challenged.
[0028] Sixth, the knowledge base around cancer treatments is always
growing
with different clinical trials in different stages around the world so that if
a physician's
knowledge is current today, her knowledge will be dated within months if not
weeks.
Thousands of oncological articles are published each year and many are verbose
and/or intellectually arduous to consume (e.g., the articles are difficult to
read and
internalize), especially by extremely busy physicians that have limited time
to absorb
new materials and information. Distilling publications down to those that are
pertinent to a specific physician's practice takes time and is an inexact
endeavor in
many cases.
[0029] Seventh, in most cases there is no clear incentive for physicians
to
memorialize a complete set of treatment and results data and, in fact, the
time
required to memorialize such data can operate as an impediment to collecting
that
data in a useful and complete form. To this end, prescribing and treating
physicians
are busy diagnosing and treating patients based on what they currently
understand
and painstakingly capturing a complete set of cancer state, treatment and
results
data without instantaneously reaping some benefit for patients being treated
in return
(e.g. a new insight, a better prescriptive treatment tool, etc.) is often
perceived as a
"waste" of time. In addition, because time is often of the essence in cancer
-7-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
treatment planning and plan implementation (e.g., starting treatment as soon
as
possible can increase efficacy in many cases), most physicians opt to take
more
time attending to their patients instead of generating perfect and fulsome
treatments
and results data sets.
[0030] Eighth, the field of next generation sequencing ("NGS") for cancer
genomics is new and NGS faces significant challenges in managing related
sequencing, bioinformatics, variant calling, analysis, and reporting data.
Next
generation sequencing involves using specialized equipment such as a next
generation gene sequencer, which is an automated instrument that determines
the
order of nucleotides in DNA and RNA. The instrument reports the sequences as a
string of letters, called a read, which the analyst compares to one or more
reference
genomes of the same genes, which is like a library of normal and variant gene
sequences associated with certain conditions. With no settled NGS standards,
different NGS providers have different approaches for sequencing cancer
patient
genomics and, based on their sequencing approaches, generate different types
and
quantities of genomics data to share with physicians, researchers, and
patients.
Different genomic datasets exacerbate the task of discerning and, in some
cases,
render it impossible to discern, meaningful genetics-treatment efficacy
insights as
required data is not in a normalized form, was never captured or simply was
never
generated.
[0031] In addition to problems associated with collecting and
memorializing
treatment and results data sets, there are problems with digesting or
consuming
recorded data to generate useful conclusions. For instance, recorded cancer
state,
treatment and results data is often incomplete. In most cases physicians are
not
researchers and they do not follow clearly defined research techniques that
enforce
tracking of all aspects of cancer states, treatments and results and therefore
data
that is recorded is often missing key information such as, for instance,
specific
patient conditions that may be of current or future interest, reasons why a
specific
treatment was selected and other treatments were rejected, specific results,
etc. In
many cases where cause and effect relationships exist between cancer state
factors
and treatment results, if a physician fails to identify and record a causal
factor, the
results cannot be tied to existing cause and effect data sets and therefore
simply
cannot be consumed and added the overall cancer knowledge data set in a
meaningful way.
[0032] Another impediment to digesting collected data is that physicians
often
capture cancer state, treatment and results data in forms that make it
difficult if not
-8-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
impossible to process the collected information so that the data can be
normalized
and used with other data from similar patient treatments to identify more
nuanced
insights and to draw more robust conclusions. For instance, many physicians
prefer
to use pen and paper to track patient care and/or use personal shorthand or
abbreviations for different cancer state descriptions, patient conditions,
treatments,
results and even conclusions. Using software to glean accurate information
from
hand written notes is difficult at best and the task is exacerbated when hand
written
records include personal abbreviations and shorthand representations of
information
that software simply cannot identify with the physician's intended meaning.
[0033] One positive development in the area of cancer treatment planning
has
been establishment of cancer committees or boards at cancer treating
institutions
where committee members routinely consider treatment planning for specific
patient
cancer states as a committee. To this end, it has been recognized that the
task of
prescribing optimized treatment plans for diagnosed cancer states is
exacerbated by
the fact that many physicians do not specialize in more than one or a small
handful
of cancer treatment options (e.g., radiation therapy, chemotherapy, surgery,
etc.).
For this reason, many physicians are not aware of many treatment options for
specific ailment-patient condition combinations, related treatment efficacy
and/or
how to implement those treatment options. In the case of cancer boards, the
idea is
that different board members bring different treatment experiences, expertise
and
perspectives to bear so that each patient can benefit from the combined
knowledge
of all board members and so that each board member's awareness of treatment
options continually expands.
[0034] While treatment boards are useful and facilitate at least some
sharing
of experiences among physicians and other healthcare providers, unfortunately
treatment committees only consider small snapshots of treatment options and
associated results based on personal knowledge of board members. In many cases
boards are forced to extrapolate from "most similar" cancer states they are
aware of
to craft patient treatment plans instead of relying on a more fulsome
collection of
cancer state-treatment-results data, insights and conclusions. In many cases
the
combined knowledge of board members may not include one or several important
perspectives or represent important experience bases so that a final treatment
plan
simply cannot be optimized.
[0035] To be useful cancer state, treatment and efficacy data and
conclusions
based thereon have to be rendered accessible to physicians, researchers and
other
interested parties. In the case of cancer treatments where cancer states,
treatments,
-9-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
results and conclusions are extremely complicated and nuanced, physician and
researcher interfaces have to present massive amounts of information and show
many data corollaries and relationships. When massive amounts of information
are
presented via an interface, interfaces often become extremely complex and
intimidating which can result in misunderstanding and underutilization. What
is
needed are well designed interfaces that make complex data sets simple to
understand and digest. For instance, in the case of cancer states, treatments
and
results, it would be useful to provide interfaces that enable physicians to
consider de-
identified patient data for many patients where the data is specifically
arranged to
trigger important treatment and results insights. It would also be useful if
interfaces
had interactive aspects so that the physicians could use filters to access
different
treatment and results data sets, again, to trigger different insights, to
explore
anomalies in data sets, and to better think out treatment plans for their own
specific
patients.
[0036] In some cases specific cancers are extremely uncommon so that when
they do occur, there is little if any data related to treatments previously
administered
and associated results. With no proven best or even somewhat efficacious
treatment option to choose from, in many of these cases physicians turn to
clinical
trials.
[0037] Cancer research is progressing all the time at many hospitals and
research institutions where clinical trials are always being performed to test
new
medications and treatment plans, each trial associated with one or a small
subset of
specific cancer states (e.g., cancer type, state, tumor location and tumor
characteristics). A cancer patient without other effective treatment options
can opt to
participate in a clinical trial if the patient's cancer state meets trial
requirements and if
the trial is not yet fully subscribed (e.g., there is often a limit to the
number of patients
that can participate in a trial).
[0038] At any time there are several thousand clinical trials progressing
around the world and identifying trial options for specific patients can be a
daunting
endeavor. Matching patient cancer state to a subset of ongoing trials is
complicated
and time consuming. Pairing down matching trials to a best match given
location,
patient and physician requirements and other factors exacerbates the task of
considering trial participation. In addition, considering whether or not to
recommend
a clinical trial to a specific patient given the possibility of trial
treatment efficacy
where the treatments are by their very nature experimental, especially in
light of
specific patient conditions, is a daunting activity that most physicians do
not take
-10-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
lightly. It would be advantageous to have a tool that could help physicians
identify
clinical trial options for specific patients with specific cancer states and
to access
information associated with trial options.
[0039] As described above, optimized cancer treatment deliberation and
planning involves consideration of many different cancer state factors,
treatment
options and treatment results as well as activities performed by many
different types
of service providers including, for instance, physicians, radiologists,
pathologists, lab
technicians, etc. One cancer treatment consideration most physicians agree
affects
treatment efficacy is treatment timing where earlier treatment is almost
always better.
For this reason, there is always a tension between treatment planning speed
and
thoroughness where one or the other of speed and thoroughness suffers.
[0040] One other problem with current cancer treatment planning processes
is
that it is difficult to integrate new pertinent treatment factors, treatment
efficacy data
and insights into existing planning databases. In this regard, known treatment
planning databases and application programs have been developed based on a
predefined set of factors and insights and changing those databases and
applications often requires a substantial effort on the part of a software
engineer to
accommodate and integrate the new factors or insights in a meaningful way
where
those factors and insights are properly considered along with other known
factors
and insights. In some cases the substantial effort required to integrate new
factors
and insights simply means that the new factors or insights will not be
captured in the
database or used to affect planning. In other cases the effort means that the
new
factors or insights are only added to the system at some delayed time after a
software engineer has applied the required and substantial reprogramming
effort. In
still other cases, the required effort means that physicians that want to
apply new
insights and factors may attempt to do so based on their own experiences and
understandings instead of in a more scripted and rules based manner.
Unfortunately, rendering a new insight actionable in the case of cancer
treatment is a
literal matter of life and death and therefore any delay or inaccurate
application can
have the worst effect on current patient prognosis.
[0041] One other problem with existing cancer treatment efficacy databases
and systems is that they are simply incapable of optimally supporting
different types
of system users. To this end, data access, views and interfaces needed for
optimal
use are often dependent upon what a system user is using the system for. For
instance, physicians often want treatment options, results and efficacy data
distilled
down to simple correlations while a cancer researcher often requires much more
-11-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
detailed data access required to develop new hypothesis related to cancer
state,
treatment and efficacy relationships. In known systems, data access, views and
interfaces are often developed with one consuming client in mind such as, for
instance, physicians, pathologists, radiologists, a cancer treatment
researcher, etc.,
and are therefore optimized for that specific system user type which means
that the
system is not optimized for other user types and cannot be easily changed to
accommodate needs of those other user types.
[0042] With the advent of NGS it has become possible to accurately detect
genetic alterations in relevant cancer genes in a single comprehensive assay
with
high sensitivity and specificity. However, the routine use of NGS testing in a
clinical
context faces several challenges. First, many tissue samples include minimal
high
quality DNA and RNA required for meaningful testing. In this regard, nearly
all
clinical specimens comprise formalin fixed paraffin embedded tissue (FFPET),
which, in many cases, has been shown to include degraded DNA and RNA.
Exacerbating matters, many samples available for testing contain limited
amounts of
tissue, which in turn limits the amount of nucleic acid attainable from the
tissue. For
this reason, accurate profiling in clinical specimens requires an extremely
sensitive
assay capable of detecting gene alterations in specimens with a low tumor
percentage. Second, millions of bases within the tumor genome are assayed. For
this reason, rigorous statistical and analytical approaches for validation are
required
in order to demonstrate the accuracy of NGS technology for use in clinical
settings
and in developing cause and effect efficacy insights.
[0043] Thus, what is needed is a system that is capable of efficiently
capturing
all treatment relevant data including cancer state factors, treatment
decisions,
treatment efficacy and exploratory factors (e.g., factors that may have a
causal
relationship to treatment efficacy) and structuring that data to optimally
drive different
system activities including memorialization of data and treatment decisions,
database analytics and user applications and interfaces. In addition, the
system
should be highly and rapidly adaptable so that it can be modified to absorb
new data
types and new treatment and research insights as well as to enable development
of
new user applications and interfaces optimized to specific user activities.
BRIEF SUMMARY OF THE DISCLOSURE
[0044] It has been recognized that an architecture where system processes
are compartmentalized into loosely coupled and distinct micro-services that
consume
-12-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
defined subsets of system data to generate new data products for consumption
by
other micro-services as well as other system resources enables maximum system
adaptability so that new data types as well as treatment and research insights
can be
rapidly accommodated. To this end, because micro-services operate
independently
of other system resources to perform defined processes where the only
development
constraints are related to system data consumed and data products generated,
small
autonomous teams of scientists and software engineers can develop new micro-
services with minimal system constraints thereby enabling expedited service
development.
[0045] The system enables rapid changes to existing micro-services as well
as development of new micro-services to meet any data handling and analytical
needs. For instance, in a case where a new record type is to be ingested into
an
existing system, a new record ingestion micro-service can be rapidly developed
for
new record intake purposes resulting in addition of the new record in a raw
data form
to a system database as well as a system alert notifying other system
resources that
the new record is available for consumption. Here, the intra-micro-service
process is
independent of all other system processes and therefore can be developed as
efficiently and rapidly as possible to achieve the service specific goal. As
an
alternative, an existing record ingestion micro-service may be modified
independent
of other system processes to accommodate some aspect of the new record type.
The micro-service architecture enables many service development teams to work
independently to simultaneously develop many different micro-services so that
many
aspects of the overall system can be rapidly adapted and improved at the same
time.
[0046] According to another aspect of the present disclosure, in at least
some
disclosed embodiments system data may be represented in several differently
structured databases that are optimally designed for different purposes. To
this end,
it has been recognized that system data is used for many different purposes
such as
memorialization of original records or documents, for data progression
memorialization and auditing, for internal system resource consumption to
generate
interim data products, for driving research and analytics, and for supporting
user
application programs and related interfaces, among others. It has also been
recognized that a data structure that is optimal for one purpose often is sub-
optimal
for other purposes. For instance, data structured to optimize for database
searching
by a data scientist may have a completely different structure than data
optimized to
drive a physician's application program and associated user interface. As
another
instance, data optimized for database searching by a data scientist usually
has a
-13-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
different structure than raw data represented in an original clinical medical
record
that is stored to memorialize the original record.
[0047] By storing system data in purpose specific data structures, a
diverse
array of system functionality is optimally enabled. Advantages include simpler
and
more rapid application and micro-service development, faster analytics and
other
system processes and more rapid user application program operations.
[0048] Particularly useful systems disclosed herein include three separate
databases including a "data lake" database, a "data vault" database and a
"data
marts" database. The data lake database includes, among other data, original
raw
data as well as interim micro-service data products and is used primarily to
memorialize original raw data and data progression for auditing purposes and
to
enable data recreation that is tied to prior points in time. The data vault
database
includes data structured optimally to support database access and manipulation
and
typically includes routinely accessed original data as well as derived data.
The data
marts database includes data structured to support specific user application
programs and user interfaces including original as well as derived data.
[0049] In some cases the disclosed inventions include a method for
conducting genomic sequencing, the method comprising the steps of storing a
set of
user application programs wherein each of the programs requires an application
specific subset of data to perform application processes and generate user
output,
for each of a plurality of patients that have cancerous cells and that receive
cancer
treatment, (a) obtaining clinical records data in original forms where the
clinical
records data includes cancer state information, treatment types and treatment
efficacy information; (b) storing the clinical records data in a semi-
structured first
database, (c) for each patient, using a next generation genomic sequencer to
generate genomic sequencing data for the patient's cancerous cells and normal
cells, d) storing the sequencing data in the first database, (e) shaping at
least a
subset of the first database data to generate system structured data including
clinical
record data and sequencing data wherein the system structured data is
optimized for
searching, (f) storing the system structured data in a second database, (g)
for each
user application program, (i) selecting the application specific subset of
data from the
second database and (ii) storing the application specific subset of data in a
structure
optimized for application program interfacing in a third database.
[0050] In at least some cases the method includes the step of storing a
plurality of micro-service programs where each micro-service program includes
a
data consume definition, a data product to generate definition and a data
shaping
-14-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
process that converts consumed data to a data product, the step of shaping
including running a sequence of micro-service programs on data in the first
database
to retrieve data, shape the retrieved data into data products and publish the
data
products back to the second database as structured data.
[0051] In at least some cases the method includes storing a new data alert
in
an alert list in response to a new clinical record or a new micro-service data
product
being stored in the second database. In at least some cases the method
includes
each micro-service program monitoring the alert list and determining if stored
data is
to be consumed by that micro-service program independent of all other micro-
service
programs. In at least some embodiments at least a subset of the micro-service
programs operate sequentially to condition data.
[0052] In at least some embodiments at least a subset of the micro-service
programs specify the same data to consume definition. In at least some
embodiments the step of shaping includes at least one manual step to be
performed
by a system user and wherein the system adds a data shaping activity to a
user's
work queue in response to at least one of the alerts being added to the alert
list. In
at least some embodiments the first database includes both unstructured
original
clinical data records and semi-structured data generated by the micro-service
programs.
[0053] In at least some embodiments each micro-service program operates
automatically and independently when data that meets the data to consume
definition is stored to the first database. In at least some embodiments the
application programs include operational programs and wherein at least a
subset of
the operational programs comprise a physician suite of programs useable to
consider cancer state treatment options. In at least some embodiments at least
a
subset of the operational programs comprise a suite of data shaping programs
usable by a system user to shape data stored in the first database. In at
least some
embodiments the data shaping programs are for use by a radiologist.
[0054] In at least some embodiments the data shaping programs are for use
by a pathologist. In at least some cases the method includes a set of
visualization
tools and associated interfaces useable by a system user to analyze the second
database data. In at least some embodiments the third database includes a
subset
of the second database data. In at least some embodiments the third database
includes data derived from the second database data. In at least some cases
the
method includes the steps of presenting a user interface to a system user that
-15-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
includes data that indicates how genomic sequencing data affects different
treatment
efficacies.
[0055] In at least some embodiments each cancer state includes a plurality
of
factors, the method further including the steps of using a processor to
automatically
perform the steps of analyzing patient genomic sequencing data that is
associated
with patients having at least a common subset of cancer state factors to
identify
treatments of genomically similar patients that experience treatment
efficacies above
a threshold level. In at least some embodiments each cancer state includes a
plurality of factors, the method further including the steps of using a
processor to
automatically identify, for specific cancer types, highly efficacious cancer
treatments
and, for each highly efficacious cancer treatment, identify at least one
genomic
sequencing data subset that is different for patients that experienced
treatment
efficacy above a first threshold level when compared to patients that
experienced
treatment efficacy below a second threshold level.
[0056] In other embodiments the invention includes a method for conducting
genomic sequencing, the method comprising the steps of, for each of a
plurality of
patients that have cancerous cells and that receive cancer treatment, (a)
obtaining
clinical records data in original forms where the clinical records data
includes cancer
state information, treatment types and treatment efficacy information, (b)
storing the
clinical records data in a semi-structured first database, (c) obtaining a
tumor
specimen from the patient, (d) growing the tumor specimen into a plurality of
tissue
organoids, (e) treating each tissue organoids with an organoid specific
treatment, (f)
collecting and storing organoid treatment efficacy information in the first
database,
(g) using a processor to examining the first database data including organoid
treatment efficacy and clinical record data to identify at least one optimal
treatment
for a specific cancer patient.
[0057] In at least some cases the method includes the steps of storing a
set of
user application programs wherein each of the programs requires an application
specific subset of data to perform application processes and generate user
output,
shaping at least a subset of the first database data to generate system
structured
data including clinical record data and organoid treatment efficacy data
wherein the
system structured data is optimized for searching, storing the system
structured data
in a second database, for each user application program, selecting the
application
specific subset of data from at least one of the first and second databases
and
storing the application specific subset of data in a structure optimized for
application
program interfacing in a third database. In at least some cases the method
includes
-16-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
the steps of using a genomic sequencer to generate genomic sequencing data for
each of the patients and the patient's cancerous cells and storing the
sequencing
data in the first database, the step of examining the first database data
including
examining each of the organoid treatment efficacy data, the genomic sequencing
data and the clinical record data to identify at least one optimal treatment
for a
specific cancer patient.
[0058] In at least some embodiments the sequencing data includes DNA
sequencing data. In at least some embodiments the sequencing data include RNA
sequencing data. In at least some embodiments the sequencing data includes
only
DNA sequencing data. In at least some embodiments the sequencing data includes
only RNA sequencing data. In at least some embodiments the sequencing is
conducted using the xT gene panel. In at least some embodiments the sequencing
is conducted using a plurality of genes from the xT gene panel. In at least
some
embodiments the sequencing is conducted using at least one gene from the xF
gene
panel. In at least some embodiments the sequencing is conducted using the xE
gene panel. In at least some embodiments the sequencing is conducted using at
least one gene from the xE gene panel.
[0059] In at least some embodiments sequencing is done on the KRAS gene.
In at least some embodiments sequencing is done on the PIK3CA gene. In at
least
some embodiments sequencing is done on the CDKN2A gene. In at least some
embodiments sequencing is done on the PTEN gene. In at least some embodiments
sequencing is done on the ARID1A gene. In at least some embodiments
sequencing is done on the APO gene. In at least some embodiments sequencing is
done on the ERBB2 gene. In at least some embodiments sequencing is done on the
EGFR gene. In at least some embodiments sequencing is done on the IDH1 gene.
In at least some embodiments sequencing is done on the CDKN2B gene. In at
least
some embodiments the sequencing includes MAP kinase cascade. In at least some
embodiments the sequencing includes EGFR. In at least some embodiments the
sequencing includes BRA. In at least some embodiments the sequencing includes
NRAS.
[0060] In at least some embodiments the sequencing is performed on a
particular cancer type. In at least some embodiments at least one of the micro-
services is a variant annotation service. In at least some embodiments the
application programs include operational programs and wherein at least one of
the
operational programs is a variant annotation program. In at least some
embodiments the application programs include operational programs and wherein
at
-17-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
least one of the operational programs is a clinical data structuring
application for
converting unstructured raw clinical medical records into structured records.
In at
least some embodiments the data vault database includes a database of
molecular
sequencing data. In at least some embodiments the molecular sequencing data
includes DNA data.
[0061] In at least some embodiments the molecular sequencing data includes
RNA data. In at least some embodiments the molecular sequencing data includes
normalized RNA data. In at least some embodiments the molecular sequencing
data
includes tumor-normal sequencing data. In at least some embodiments the
molecular sequencing data includes variant calls. In at least some embodiments
the
molecular sequencing data includes variants of unknown significance. In at
least
some embodiments the molecular sequencing data includes germline variants. In
at
least some embodiments the molecular sequencing data includes MSI information.
[0062] In at least some embodiments the molecular sequencing data includes
TMB information. In at least some cases the method includes the step of
determining an MSI value for the cancerous cells. In at least some cases the
method includes determining a TMB value for the cancerous cells. In at least
some
cases the method includes identifying a TMB value greater than 9 mutations/Mb.
In
at least some cases the method includes detecting a genomic alteration that
results
in a chimeric protein product. In at least some cases the method includes
detecting
a genomic alteration that drives EML4-ALK. In at least some cases the method
includes the step of determining neoantigen load. In at least some cases the
method
includes the step of identifying a cytolytic index. In at least some cases the
method
includes distinguishing a population of immune cells (dependent: TMG-high /
TMB-
low).
[0063] In at least some cases the method includes the step of determining
0D274 expression. In at least some cases the method includes reporting an
overexpression of MYC. In at least some cases the method includes detecting a
fusion event. In at least some embodiments the fusion event is a TMPRSS-ERG
fusion. In at least some cases the method includes the step of detecting a PD-
L1 in
a lung cancer patient. In at least some cases the method includes indicating a
PARP inhibitor. In at least some embodiments the PARP inhibitor is for BRCA1.
In
at least some embodiments the PARP inhibitor is for BRCA2. In at least some
cases
the method includes the steps of recommending an immunotherapy. In at least
some embodiments the recommended immunotherapy is one of CAR-T therapy,
-18-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
antibody therapy, cytokine therapy, adoptive t-cell therapy, anti-0D47
therapy, anti-
GD2 therapy, immune checkpoint inhibitor and neoantigen therapy.
[0064] In at least some embodiments the cancer cells are from a tumor
tissue
and the non-cancer cells are blood cells. In at least some embodiments the
cancerous cells are cell free DNA from blood. In at least some embodiments the
cancer cells are from fresh tissue. In at least some embodiments the cancer
cells
are from a FFPE slide. In at least some embodiments the cancer cells are from
frozen tissue. In at least some embodiments the cancer cells are from biopsied
tissue. In at least some embodiments sequencing is done on the TP53 gene.
[0065] To the accomplishment of the foregoing and related ends, the
invention, then, comprises the features hereinafter fully described. The
following
description and the annexed drawings set forth in detail certain illustrative
aspects of
the invention. However, these aspects are indicative of but a few of the
various ways
in which the principles of the invention can be employed. Other aspects,
advantages
and novel features of the invention will become apparent from the following
detailed
description of the invention when considered in conjunction with the drawings.
[0066]
BRIEF DESCRIPTION OF THE SEVERAL VIEWS OF THE DRAWINGS
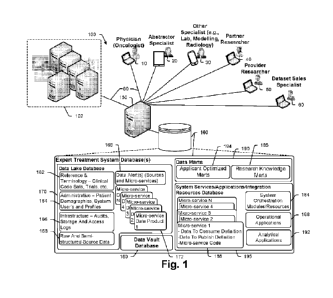
[0067] Fig. 1 is a schematic diagram illustrating a computer and
communication system that is consistent with at least some aspects of the
present
disclosure:
[0068] Fig. 2 is a schematic diagram illustrating another view of the Fig.
1
system where functional components that are implemented by the Fig. 1
components are shown in some detail;
[0069] Fig. 3 is a schematic diagram illustrating yet another view of the
Fig. 1
system where additional system components are illustrated;
[0070] Fig. 3a is a schematic diagram showing a data platform that is
consistent with at least some aspects of the present disclosure;
[0071] Fig. 4 is a data handling flow chart that is consistent with at
least some
aspects of the present disclosure;
[0072] Fig. 5 is a flow chart that shows a process for ingesting raw data
into
the system and alerting other system components that the raw data is available
for
consumption;
[0073] Fig. 6 is a flow chart that shows a micro-service based process for
retrieving data from a database, consuming that data to generate new data
products
-19-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
and publishing the new data products back to a database while publishing an
alert
that the new data products are available for consumption;
[0074] Fig. 7 is a flow chart illustrating a process similar to the Fig. 6
process,
albeit where the micro-service is an OCR service;
[0075] Fig. 8 is a is a flow chart illustrating a process similar to the
Fig. 6
process, albeit where the micro-service is a data structuring service; and
[0076] Fig. 9 is a schematic view of an abstractor's display screen used
to
generate a structured data record from data in an unstructured or semi-
structured
record;
[0077] Fig. 10 is a schematic illustrating a multi-micro-service process
for
ingesting a clinical medical record into the system of Fig. 1;
[0078] Fig. 11 is a schematic illustrating a multi-micro-service process
for
generating genomic sequencing and related data that is consistent with at
least
some aspects of the present disclosure;
[0079] Flg. lla is a flow chart illustrating an exemplary variant calling
process
that is consistent with at least some aspects of the present disclosure;
[0080] Fig. llb is a schematic illustrating an exemplary bioinformatics
pipeline
process that is consistent with at least some embodiments of the present
disclosure;
[0081] Flg. 11c is a schematic illustrating various system features
including a
therapy matching engine;
[0082] Fig. 12 is a schematic illustrating a multi-micro-service process
for
generating organoid modelling data that is consistent with at least some
aspects of
the present disclosure;
[0083] Fig. 13 is a schematic illustrating a multi-micro-service process
for
generating a 3D model of a patient's tumor as well as identifying a large
number of
tumor features and characteristics that is consistent with at least some
aspects of the
present disclosure;
[0084] Fig, 14 is a screenshot illustrating a patient list view that may
be
accessed by a physician using the disclosed system to consider treatment
options
for a patient;
[0085] Fig. 15 is a screenshot illustrating an overview view that may be
accessed by a physician using the disclosed system to review prior treatment
or
case activities related to the patient.
[0086] Fig. 16 is a screenshot illustrating screenshot illustrating a
reports view
that may be used to access patient reports generated by the system 100;
-20-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
[0087] Fig. 17 is a screenshot illustrating a second reports view that
shows
one report in a larger format;
[0088] Fig. 17a shows an initial view of an RNA sequence reporting
screenshot that is consistent with at least some aspects of the present
disclosure;
[0089] Fig. 18 is a screenshot illustrating an alterations view accessible
by a
physician to consider molecular tumor alterations;
[0090] Fig. 18a is an exemplary top portion of a screenshot of a user
interface
for reporting and exploring approved therapies while Fig. 18b shows the lower
portion of the Fig. 18a screenshot,
[0091] Fig. 19 is a screenshot illustrating a trials view in which a
physician
views information related to clinical trials on conjunction with considering
treatment
options for a patient;
[0092] Fig. 20 is a screenshot illustrating an immunotherapy screenshot
accessible to a physician for considering immunotherapy efficacy options for
treating
a patient's cancer state;
[0093] Fig. 21 is a screenshot illustrating an efficacy exploration view
where
molecular differences between a patient's tumor and other tumors of the same
general type are used a primary factor in generating the illustrated graph;
[0094] Figs. 22a through 22j include an exemplary 1711 gene panel listing
that may be interrogated during genomic sequencing in at least some
embodiments
of the present disclosure;
[0095] Fig. 23 includes a clinically actionable 130 gene panel listing
that may
be interrogated during genomic sequencing in at least some embodiments of the
present disclosure;
[0096] Fig. 24 includes a clinically actionable 41 RNA based gene
rearrangements listing that may be interrogated during genomic sequencing in
at
least some embodiments of the present disclosure;
[0097] Fig. 25 includes a table that lists exemplary variant data that is
consistent with at least some aspects of the present disclosure;
[0098] Fig. 26 includes exemplary OVA data that is consistent with at
least
some implementations and aspects of the present disclosure;
[0099] Figs. 27a through 27d includes additional gene panel tables that
may
be interrogated in at least some embodiments of the present disclosure;
[00100] Figs. 28a and 28b include yet one other gene panel table that may
be
interrogated;
-21-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
[00101] Fig. 29 is a bar chart illustrating data for a 500 patient group
that
clusters mutation similarities for gene, mutation type, and cancer type
derived for an
exemplary xT panel using techniques that are consistent with aspects of the
present
disclosure;
[00102] Fig. 30 is a bar chart comparing study results generated for the
exemplary xT panel using at least some processes described in this
specification
with previously published pan-cancer analysis using an IMPACT panel;
[00103] Fig. 31 is a graph illustrating expression profiles for tumor types
related
to the exemplary xT panel described in the present disclosure;
[00104] Fig. 32 is a graph illustrating clustering of samples by TCGA
cancer
group in a t-SNE plot for the exemplary xT panel;
[00105] Fig. 33 is a plot of genomic rearrangements using DNA and RNA
assays for the exemplary xT panel;
[00106] Fig. 34 is a schematic illustrating data related to one
rearrangement
detected via RNA sequencing related to the exemplary xT panel;
[00107] Fig. 35 is a schematic illustrating data related to a second
rearrangement detected via RNA sequencing related to the exemplary xT panel;
[00108] Fig. 36 includes a chart that illustrates the distribution of TMB
varied by
cancer type identified using techniques that are consistent with at least some
aspects of the present disclosure related to the exemplary xT panel;
[00109] Fig. 37 includes data represented on a two dimensional plot showing
TMB on one axis and predicted antigenic mutations with RNA support on the
other
axis that was generated using techniques that are consistent with at least
some
aspects of the present disclosure related to the exemplary xT panel;
[00110] Fig. 38 includes additional data related to TMB generated using
techniques that are consistent with at least some aspects of the present
disclosure
related to the exemplary xT panel;
[00111] Fig. 39 includes two schematics illustrating two gene expression
scores
for low and high TMB and MSI populations generated using techniques that are
consistent with at least some aspects of the present disclosure related to the
exemplary xT panel;
[00112] Fig. 40 includes three schematics illustrating data related to
propensity
of different types inflammatory immune and non-inflammatory immune cells in
low
and high TMB samples generated for the related xT panel;
[00113] Fig. 41 includes a schematic illustrating data related to
prevalence of
CD274 expression in low and high TMB samples generated using techniques
-22-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
consistent with at least some aspects of the present disclosure generated for
the
related xT panel;
[00114] Fig. 42 includes two schematics illustrating correlations between
0D274 expression and other cell types generated using techniques consistent
with
at least some aspects of the present disclosure generated for the related xT
panel;
[00115] Fig. 43 is a schematic illustrating data generated via a 28 gene
interferon gamma-related signature that is consistent with at least some
aspects of
the present disclosure;
[00116] Fig. 44 includes data shown as a graph illustrating levels of
interferon
gamma-related genes versus TMB-high, MSI-high and PDL1 IHC positive tumors
generated using techniques consistent with at least some aspects of the
present
disclosure;
[00117] Fig. 45 includes a bar graph illustrating data related to
therapeutic
evidence as it varies among different cancer types generated using techniques
consistent with at least some aspects of the present disclosure;
[00118] Fig. 46 includes a bar graph illustrating data related to specific
therapeutic evidence matches based on copy number variants generating using
techniques consistent with at least some aspects of the present disclosure;
[00119] Fig. 47 includes a bar graph illustrating data related to specific
therapeutic evidence matches based on single nucleotide variants and indels
generating using techniques consistent with at least some aspects of the
present
disclosure;
[00120] Fig. 48 includes a plot illustrating data related to single
nucleotide
variants and indels or CNVs by cancer type generating using techniques
consistent
with at least some aspects of the present disclosure;
[00121] Fig. 49 includes a bar graph illustrating data that shows percent
of
patients with gene calls and evidence for association between gene expression
and
drug response where the data was generated using techniques consistent with at
least some aspects of the present disclosure;
[00122] Fig. 50 includes a bar graph illustrating response to therapeutic
options
based on evidence tiers and broken down by cancer type;
[00123] Fig. 51 includes a bar graph showing data related to patients that
are
potential candidates for immunotherapy broken down by cancer type where the
data
is based on techniques consistent with the present disclosure;
[00124] Fig. 52 is a bar graph presenting data related to relevant
molecular
insights for a patent group based on CNVs, indels, CNVs, gene expression calls
and
-23-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
immunotherapy biomarker assays where the data was generated using techniques
that are consistent with various aspects of the present disclosure;
[00125] Fig. 53 includes a bar graph illustrating disease-based trial
matches
and biomarker based match percentages based that reflect results of techniques
that
are consistent with at least some aspects of the present disclosure;
[00126] Fig. 54 includes a bar graph including data that shows exemplary
distribution of expression calls by sample that was generated using techniques
that
are consistent with at least some aspects of the present disclosure;
[00127] Fig. 55 includes a bar graph including data that shows exemplary
distribution of expression calls by gene that was generated using techniques
that are
consistent with at least some aspects of the present disclosure;
[00128] Fig. 56 includes a graph illustrating response evidence to
therapies
across all cancer types in an exemplary study using techniques consistent with
at
least some aspects of the present disclosure;
[00129] Fig. 57 includes a graph illustrating evidence of resistance to
therapies
across all cancer types in an exemplary study using techniques consistent with
at
least some aspects of the present disclosure; and
[00130] Fig. 58 includes a graph illustrating therapeutic evidence tiers
for all
cancer types in an exemplary study using techniques consistent with at least
some
aspects of the present disclosure.
[00131] While the invention is susceptible to various modifications and
alternative forms, specific embodiments thereof have been shown by way of
example in the drawings and are herein described in detail. It should be
understood,
however, that the description herein of specific embodiments is not intended
to limit
the invention to the particular forms disclosed, but on the contrary, the
intention is to
cover all modifications, equivalents, and alternatives falling within the
spirit and
scope of the invention as defined by the appended claims.
DETAILED DESCRIPTION OF THE DISCLOSURE
[00132] The various aspects of the subject invention are now described with
reference to the annexed drawings, wherein like reference numerals correspond
to
similar elements throughout the several views. It should be understood,
however,
that the drawings and detailed description hereafter relating thereto are not
intended
to limit the claimed subject matter to the particular form disclosed. Rather,
the
-24-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
intention is to cover all modifications, equivalents, and alternatives falling
within the
spirit and scope of the claimed subject matter.
[00133] As used herein, the terms "component," "system" and the like are
intended to refer to a computer-related entity, either hardware, a combination
of
hardware and software, software, or software in execution. For example, a
component may be, but is not limited to being, a process running on a
processor, a
processor, an object, an executable, a thread of execution, a program, and/or
a
computer. By way of illustration, both an application running on a computer
and the
computer can be a component. One or more components may reside within a
process and/or thread of execution and a component may be localized on one
computer and/or distributed between two or more computers or processors.
[00134] The word "exemplary" is used herein to mean serving as an example,
instance, or illustration. Any aspect or design described herein as
"exemplary" is not
necessarily to be construed as preferred or advantageous over other aspects or
designs.
[00135] The phrase "Allelic Fraction" or "AF" will be used to refer to the
percentage of reads supporting a candidate variant divided by a total number
of
reads covering a candidate locus.
[00136] The phrase "base pair" or "bp" will be used to refer to a unit
consisting
of two nucleobases bound to each other by hydrogen bonds. The size of an
organism's genome is measured in base pairs because DNA is typically double
stranded.
[00137] The phrase "Single Nucleotide Polymorphism" or "SNP" will be used
to
refer to a variation within a DNA sequence with respect to a known reference
at a
level of a single base pair of DNA.
[00138] The phrase "insertions and deletions" or "indels" will be used to
refer to
a variant resulting from the gain or loss of DNA base pairs within an analyzed
region.
[00139] The phrase "Multiple Nucleotide Polymorphism" or "MNP" will be used
to refer to a variation within a DNA sequence with respect to a known
reference at a
level of two or more base pairs of DNA, but not varying with respect to total
count of
base pairs. For example an AA to CC would be an MNP, but an AA to C would be a
different form of variation (e.g., an indel).
[00140] The phrase "Copy Number Variation" or "CNV" will be used to refer
to
the process by which large structural changes in a genome associated with
tumor
aneuploidy and other dysregulated repair systems are detected. These processes
are used to detect large scale insertions or deletions of entire genomic
regions. CNV
-25-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
is defined as structural insertions or deletions greater than a certain base
pair ("bp")
in size, such as 500 bp.
[00141] The phrase "Germ line Variants" will be used to refer to genetic
variants
inherited from maternal and paternal DNA. Germline variants may be determined
through a matched tumor-normal calling pipeline.
[00142] The phrase "Somatic Variants" will be used to refer to variants
arising
as a result of dysregulated cellular processes associated with neoplastic
cells.
Somatic variants may be detected via subtraction from a matched normal sample.
[00143] The phrase "Gene Fusion" will be used to refer to the product of
large
scale chromosomal aberrations resulting in the creation of a chimeric protein.
These
expressed products can be non-functional, or they can be highly over or under
active. This can cause deleterious effects in cancer such as hyper-
proliferative or
anti-apoptotic phenotypes.
[00144] The phrase "RNA Fusion Assay" will be used to refer to a fusion
assay
which uses RNA as the analytical substrate. These assays may analyze for
expressed RNA transcripts with junctional breakpoints that do not map to
canonical
regions within a reference range.
[00145] The term "Microsatellites" refers to short, repeated sequences of
DNA.
[00146] The phrase "Microsatellite instability" or "MSI" refers to a change
that
occurs in the DNA of certain cells (such as tumor cells) in which the number
of
repeats of microsatellites is different than the number of repeats that was in
the DNA
when it was inherited. The cause of microsatellite instability may be a defect
in the
ability to repair mistakes made when DNA is copied in the cell.
[00147] "Microsatellite Instability-High" or "MSI-H" tumors are those
tumors
where the number of repeats of microsatellites in the cancer cell is
significantly
different than the number of repeats that are in the DNA of a benign cell.
This
phenotype may result from defective DNA mismatch repair. In MSI FOR testing,
tumors where 2 or more of the 5 microsatellite markers on the Bethesda panel
are
unstable are considered MSI-H.
[00148] "Microsatellite Stable" or "MSS" tumors are tumors that have no
functional defects in DNA mismatch repair and have no significant differences
in
microsatellite regions between tumor and normal tissue.
[00149] "Microsatellite Equivocal" or "MSE" tumors are tumors with an
intermediate phenotype that cannot be clearly classified as MSI-H or MSS based
on
the statistical cutoffs used to define those two categories.
-26-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
[00150] The phrase "Limit of Detection" or "LOD" refers to the minimal
quantity
of variant present that an assay can reliably detect. All measures of
precision and
recall are with respect to the assay LOD.
[00151] The phrase "BAM File" means a (B)inary file containing (A)lignment
(M)aps that include genomic data aligned to a reference genome.
[00152] The phrase "Sensitivity of called variants" refers to a number of
correctly called variants divided by a total number of loci that are positive
for
variation within a sample.
[00153] The phrase "specificity of called variants" refers to a number of
true
negative sites called as negative by an assay divided by a total number of
true
negative sites within a sample. Specificity can be expressed as (True
negatives) /
(True negatives + false positives).
[00154] The phrase "Positive Predictive Value" or "PPV" means the
likelihood
that a variant is properly called given that a variant has been called by an
assay.
PPV can be expressed as (number of true positives) / (number of false
positives +
number of true positives).
[00155] The disclosed subject matter may be implemented as a system,
method, apparatus, or article of manufacture using standard programming and/or
engineering techniques to produce software, firmware, hardware, or any
combination
thereof to control a computer or processor based device to implement aspects
detailed herein. The term "article of manufacture" (or alternatively,
"computer
program product") as used herein is intended to encompass a computer program
accessible from any computer-readable device, carrier, or media. For example,
computer readable media can include but are not limited to magnetic storage
devices (e.g., hard disk, floppy disk, magnetic strips. . . ), optical disks
(e.g.,
compact disk (CD), digital versatile disk (DVD) . . . ), smart cards, and
flash memory
devices (e.g., card, stick). Additionally it should be appreciated that a
carrier wave
can be employed to carry computer-readable electronic data such as those used
in
transmitting and receiving electronic mail or in accessing a network such as
the
Internet or a local area network (LAN). Of course, those skilled in the art
will
recognize many modifications may be made to this configuration without
departing
from the scope or spirit of the claimed subject matter.
[00156] Unless indicated otherwise, while the disclosed system is used for
many different purposes (e.g., data collection, data analysis, treatment,
research,
etc.), in the interest of simplicity and consistency, the overall disclosed
system will be
-27-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
referred to hereinafter as "the disclosed system".
I. System Overview
[00157] Referring now to the figures that accompany this written
description
and more specifically referring to Fig. 1, the present disclosure will be
described in
the context of an exemplary system 100 where data is received at a system
server
150 from many different data sources 102, is stored in a database 160, is
manipulated in many different ways by internal system micro-service programs
to
condition or "shape" the data to generate new interim data or to structure
data in
different structured formats for consumption by user application programs and
to
then drive the user application programs to provide user interfaces via any of
several
different types of user interface devices. While a single server 150 and a
single
database 160 are shown in Fig. 1 in the interest of simplifying this
explanation, it
should be appreciated that in most cases, the system 100 will include a
plurality of
distributed servers and databases that are linked via local and/or wide area
networks
and/or the Internet or some other type of communication infrastructure. An
exemplary simplified communication network is labelled 80 in Fig. 1. Network
connections can be any type including hard wired, wireless, etc., and may
operate
pursuant to any suitable communication protocols.
[00158] The disclosed system 10 enables many different system clients to
securely link to server 150 using various types of computing devices to access
system application program interfaces optimized to facilitate specific
activities
performed by those clients. For instance, in Fig. 1 a physician 10 is shown
using a
laptop computer (not labelled) to link to server 150, an abstractor specialist
20 is
shown using a tablet type computing device to link, another specialist 30 is
shown
using a smartphone device to link to server 150, etc. Other types of personal
computing devices are contemplated including virtual and augmented reality
headsets, projectors, wearable devices (e.g., a smart watch, etc.). Fig. 1
shows
other exemplary system users linked to server 150 including a partner
researcher 40,
a provider researcher 50 and a data sales specialist 60, all of which are
shown using
laptop computers.
[00159] In at least some embodiments when a physician uses system 100, a
physician's user interface(s) is optimally designed to support typical
physician
activities that the system supports including activities geared toward patient
treatment planning. Similarly, when a researcher like a pathologist or a
radiologist
-28-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
uses system 100, interfaces optimally designed to support activities performed
by
those system clients are provided.
[00160] System specialists (e.g. employees of the provider that
controls/maintains overall system 100) also use interface computing devices to
link
to server 150 to perform various processes and functions. In Fig. 1 exemplary
system specialists include abstractor 20, the dataset sales specialist 60 and
a
"general" specialist 30 referred to as a "lab, modeling, radiology" specialist
to
indicate that the system accommodates many different additional specialist
types.
Different specialists will use system 100 to perform many different functions
where
each specialist requires specific skill sets needed to perform those
functions. For
instance, abstractor specialists are trained to ingest clinical records from
sources
102 and convert that data to normalized and system optimized structured data
sets.
A lab specialist is trained to acquire and process non-tumorous patient and/or
tumor
tissue samples, grow organoids, generate one or both of DNA and RNA genomic
data for one or each of non-tumorous and tumorous tissue, treat organoids and
generate results. Other specialists are trained to assess treatment efficacy,
perform
data research to identify new insights of various types and/or to modify the
existing
system to adapt to new insights, new data types, etc. The system interfaces
and tool
sets available to provider specialists are optimized for specific needs and
tasks
performed by those specialists.
[00161] Referring yet again to Fig. 1, system database 160 includes several
different sub-databases including, in at least some embodiments, a data lake
database 170 (hereinafter "the lake database"), a data vault database 180, a
data
marts database 190 and a system services/applications and integration resource
database 195. While database 195 is shown to includes several different types
of
information as well as system programs, in other cases one or each of the sets
of
information or programs in database 195 may be stored in a different one of
the
databases 170, 180 or 190. In general, data lake database 170 is used to store
several different data types including system reference data 162, system
administration data 164, infrastructure data 166, raw source data 168 and
micro-
service data products 172 (e.g., data generated by micro-services).
[00162] Reference data 162 includes references and terminology used within
data received from source devices 102 when available such as, for instance,
clinical
code sets, specialized terms and phrases, etc. In addition, reference data 162
includes reference information related to clinical trials including detailed
trial
-29-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
descriptions, qualifications, requirements, caveats, current phases, interim
results,
conclusions, insights, hypothesis, etc.
[00163] In at least some cases reference data 162 includes gene
descriptions,
variant descriptions, etc. Variant descriptions may be incorporated in whole
or in
part from known sources, such as the Catalogue of Somatic Mutations in Cancer
(COSMIC) (Wellcome Sanger Institute, operated by Genome Research Limited,
London, England, available at https://cancer.sanger.ac.uk/cosmic). In some
cases,
reference data 162 may structure and format data to support clinical
workflows, for
instance in the areas of variant assessment and therapies selection. The
reference
data 162 may also provide a set of assertions about genes in cancer and
evidence-
based precision therapy options. Inputs to reference data 162 may include
NCCN,
FDA, PubMed, conference abstracts, journal articles, etc. Information in the
reference data 162 may be annotated by gene; mutation type (somatic, germline,
copy number variant, fusion, expression, epigenetic, somatic genome wide,
etc.);
disease; evidence type (therapeutic, prognostic, diagnostic, associated,
etc.); and
other notes.
[00164] Referring still to Fig. 1, reference data 162 may further comprise
gene
curation information. A sequencing panel often has a predetermined number of
gene profiles that are sequenced as part of the panel. For instance, one type
of
sequencing panel in the market (i.e., xT, Tempus Labs, Inc, Chicago, Illinois)
makes
use of 595 gene profiles (see tables in Fig. 27 series of figures) while
another makes
use of 1711 gene profiles (see tables in Fig. 22 series of figures). Reference
data
162 may store a centralized gene knowledge base and comprise variant
prioritization
and filtering information that may be utilized for Gain Of Function (GOF),
Loss Of
Function (LOF), CNV, and fusions. For purposes of precision care, evidence may
be
annotated based on mutation type and disease; therapeutic evidence may include
drug(s) and effect (response, resistance, etc.); prognostic effect may include
outcome (favorable, unfavorable, etc.). Therapeutic evidence and prognostic
evidence may include evidence source level (preclinical, case study, clinical
research, guidelines, etc.). Preclinical information may be from mouse models,
PDX,
cell lines, etc. Case study information may be from groups of one or more
patients.
Clinical research may be information from a larger study or results from
clinical trials.
Guideline information may come from NCCN, WHO, etc.
[00165] The administrative data 164 includes patient demographic data as
well
as system user information including user identifications, user verification
information
(e.g., usernames, passwords, etc.), constraints on system features usable by
-30-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
specific system users, constraints on data access by users including
limitations to
specific patient data, data types, data uses, time and other data access
limits, etc.
[00166] In at least some cases system 100 is designed to memorialize entire
life cycles of every dataset or element collected or generated by system 100
so that
a system user can recreate any dataset corresponding to any point in time by
replicating system processes up to that point in time. Here, the idea is that
a
researcher or other system user can use this data re-creation capability to
verify data
and conclusions based thereon, to manipulate interim data products as part of
an
exploration process designed to test other hypothesis based on system data,
etc. To
this end, infrastructure data 166 includes complete data storage, access,
audit and
manipulation logs that can be used to recreate any system data previously
generated. In addition, infrastructure data 166 is usable to trace user access
and
storage for access auditing purposes.
[00167] Referring still to Fig. 1, lake database 170 also includes raw
unmodified
data 168 from sources 102. For instance, original clinical medical records
from
physicians are stored in their original format as are any medical images and
radiology reports, pathology reports, organoid documentation, and any other
data
type related to patient treatment, treatment efficacy, etc. In addition the
raw original
data, metadata related thereto is also identified and stored at 168. Exemplary
metadata includes source identity, data type, date and time data received, any
data
formatting information available, etc. The metadata listed here is not
exhaustive and
other metadata types may also be obtained and stored. Raw sequencing data,
such
as BAM files, may be stored in lake database 170. Unless indicated otherwise
hereafter, the data stored in lake database 170 will be referred to generally
as "lake
data".
[00168] It has been recognized that a fulsome database suitable for cancer
research and treatment planning must account for a massive number of complex
factors. It has also been recognized that the unstructured or semi-structured
lake
data is unsuitable for performing many data search processes, analytics and
other
calculations and data manipulations that are required to support the overall
system.
In this regard, searching or otherwise manipulating a massive database data
set that
includes data having many disparate data formats or structures can slow down
or
even halt system applications. For this reason the disclosed system converts
much
of the lake data to a system data structure optimized for database
manipulation (e.g.,
for searching, analyzing, calculating, etc.). For example, genomic data may be
-31-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
converted to JSON or Apache Parquet format, however, others are contemplated.
The optimized structured data is referred to herein as the "data vault
database" 180.
[00169] Thus, in Fig. 1, data vault database 180 includes data that has
been
normalized and optimally structured for storage and database manipulation. For
instance, raw original clinical medical records stored at 168 in lake database
170
may be processed to normalize data formats and placed in specific structured
data
fields optimized for data searching and other data manipulation processes. For
instance, raw original clinical medical records, such as progress notes,
pathology
reports, etc. may be processed into specific structured data fields.
Structured data
fields may be focused in certain clinical areas, such as demographics,
diagnosis,
treatment and outcomes, and genetic testing / labs. For instance, structured
diagnosis information may include primary diagnosis; tissue of origin; date of
diagnosis; date of recurrence; date of biochemical recurrence; date of CRPC,
alternative grade; gleason score; gleason score primary; gleason score
secondary;
gleason score overall; lymphovascular invasion; perineural invasion; venous
invasion. Structured diagnosis information may also include tumor
characterization,
which may be described with a set of structured data, including the type of
characterization; date of characterization; diagnosis; standard grade; AJCC
values
such as AJCC status, AJCC status T, AJCC status N, AJCC Status M, AJCC status
stage, and FIGO status stage. Structured diagnosis information may also
include
tumor size, which may be described with a set of structured size data,
including
tumor size (greatest dimension), tumor size measure, and tumor size units.
Structured diagnosis information may also include structured metastases
information. Each metastasis may be described with a set of structured data,
including location, date of identification, tumor size, diagnosis, grade, and
AJCC
values. Structured diagnosis information may also include additional
diagnoses.
Additional diagnoses may be described with a set of structured data, including
tissue
of origin, date of diagnosis, date of recurrence, date of biochemical
recurrence, date
of CRPC, tumor characterizations, and metastases.
[00170] As another instance, 2 dimensional slice type images through a
patient's tumor may be used to generate a normalized 3 dimensional
radiological
tumor model having specific attributes of interest and those attributes may be
gleaned and stored along with the 3D tumor model in the structured data vault
for
access by other system resources. In Fig. 2, the data vault database 180 is
shown
including a structured clinical database 181 for storage of structured
clinical data, a
molecular sequencing database 183 for storage of molecular sequencing data, a
-32-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
structure imaging database 185 for storage of imaging data, and a predictive
modeling database 187 for storage of organoid and other modeling data.
Additional
databases for specific lines of data may also be added to the data vault
database
180. RNA sequencing data in the molecular sequencing data may be normalized,
for instance using the methods disclosed in U.S. Provisional Patent App. No.
62/735,349, METHODS OF NORMALIZING AND CORRECTING RNA
EXPRESSION DATA, incorporated by reference herein in its entirety. Unless
indicated otherwise hereafter, the phrase "canonical data" will be used to
refer to the
data vault data in its system optimized structured form.
[00171] It has further been recognized that certain data manipulations,
calculations, aggregates, etc., are routinely consumed by application programs
and
other system consumers on a recurring albeit often random basis. By shaping at
least subsets of normalized system data, smaller sub-databases including
application and research specific data sets can be generated and published for
consumption by many different applications and research entities which
ultimately
speeds up the data access and manipulation processes.
[00172] Thus, in Fig. 1, data marts database 190 includes data that is
specifically structured to support user application programs 194 and/or
specific
research activities 196. Here, it is contemplated that different user
application
programs may require different data models (e.g., different data structures)
and
therefore data marts 190 will typically include many different application or
research
specific structured data sets. For instance, a first data mart data set may
include
data arranged consistent with a first data structure model optimized to
support a
physician's user interfaces, a second data mart data set may include data
arranged
consistent with a second data structure model optimized to support a
radiologist
specialist, a third data mart data set may include data arranged consistent
with a
third data structure model optimized to support a partner researcher, and so
on. A
single user type may have multiple data mart data sets structured to support
different
workflows on the same or different raw data.
[00173] Similarly, in the case of specific research activities, specific
data sets
and formats are optimal for specific research activities and the data marts
provide a
vehicle by which optimized data sets are optimally structured to ensure speedy
access and manipulation during research activities. Unless indicated otherwise
hereafter, the phrase "mart data" will be used to generally refer to data
stored in the
data marts 190.
-33-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
[00174] In most cases mart data is mined out of the data vault 180 and is
restructured pursuant to application and research data models to generate the
mart
data for application and research support. In some embodiments system
orchestration modules or software programs that are described hereafter will
be
provided for orchestrating data mining in the system databases as well as
restructuring data per different system models when required.
[00175] Referring still to Fig. 1, the system
services/applications/integration
resources database 195 includes various programs and services run by system
server 150 to perform and/or guide system functions. To this end, exemplary
database 195 includes system orchestration modules/resources 184, a set of
first
through N micro-services collectively identified by numeral 186, operational
user
application programs 188 and analytical user application programs 192.
[00176] Orchestration modules/resources 184 include overall scheduling
programs that define workflows and overall system flow. For instance, one
orchestration program may specify that once a new unstructured or semi-
structured
clinical medical record is stored in lake database 170, several additional
processes
occur, some in series and some in parallel, to shape and structure new data
and
data derived from the new data to instantiate new sets of canonical data and
mart
data in databases 180 and 190. Here, the orchestration program would manage
all
sub-processes and data handoffs required to orchestrate the overall system
processes. One type of orchestration program that could be utilized is a
programmatic workflow application, which uses programming to author, schedule
and monitor "workflows". A "workflow" is a series of tasks automatically
executed in
whole or in part by one or more micro-services. In one embodiment, the
workflow
may be implemented as a series of directed acyclic graphs (DAGs) of tasks or
micro-
services.
[00177] Micro-services 186 are system services that generate interim system
data products to be consumed by other system consumers (e.g., applications,
other
micro-services, etc.). In Fig. 1, first through Nth micro-service data
products
corresponding to micro-services 186 are shown stored in lake database 170 at
172.
When a micro-service data product is published to lake database 170, a data
alert or
event is added to a data alerts list 169 to announce availability of the newly
published data for consumption by other micro-services, application programs,
etc.
Micro-services are independent and autonomous in that, once a service obtains
data
required to initiate the service, the service operates independent of other
system
resources to generate output data products.
-34-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
[00178] In many cases micro-services are completely automated software
programs that consume system data and generate interim data products without
requiring any user input. For instance, an exemplary fully automated micro-
service
may include an optical character recognition (OCR) program that accesses an
original clinical record in the raw source data 168 and performs an OCR
process on
that data to generate an OCR tagged clinical record which is stored in lake
database
170 as a data product 172. As another instance, another fully automated micro-
service may glean data subsets from an OCR tagged clinical record and populate
structured record fields automatically with the gleaned data as a first
attempt to
convert unstructured or semi-structured raw data to a system optimized
structure.
[00179] In other cases a micro-service requires at least some system user
activities including, for instance, data abstraction and structuring services
or lab
activities, to generate interim data products 172. For instance, in the case
of clinical
medical record ingestion, in many cases an original clinical record will be
unstructured or semi-structured and structuring will require an abstractor
specialist
20 (see again Fig. 1) to at least verify data in structured data record fields
and in
many cases to manually add data to those fields to generate a completely
instantiated instance of the structured record as a data product 172. As
another
instance, in the case of genetic sequencing, a lab technician is required to
obtain
and load sample tumor or other tissue into a sequencing machine as part of a
sequencing process. In cases where a service requires at least some user
activities,
the service will typically be divided into separate micro-services where a
user
application operates on a micro-service data product to queue user activities
in a
user work queue or the like and a separate micro-service responds to the user
activity being completed to continue an overall process. While this disclosure
describes a small set of micro-services, a working system 100 will typically
employ a
massive number (e.g., hundreds or even many thousands) of micro-services to
drive
all of the system capabilities contemplated. It is possible that in the life
cycle of
analysis for a patient that hundreds or thousands of executions of micro-
services will
be performed.
[00180] In an embodiment, a micro-service creates a data product that may
be
accessed by an application, where the application provides a worklist and user
interface that allows a user to act upon the data product. One example set of
micro-
services is the set of micro-services for genomic variant characterization and
classification. An exemplary micro-service set for genomic variant
characterization
includes but is not limited to the following set: (1) Variant characterization
(a data
-35-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
package containing characterized variant calls for a case, which may include
overall
classification, reference criteria and other singles used to determine
classification,
exclusion rules, other flags, etc.); (2) Therapy match (including therapies
matched to
a variant characterization's list of SNV, indel, CNV, etc. variants via
therapy
templates); (3) Report (a machine-readable version of the data delivered to a
physician for a case); (4) Variants reference sets (a set of unique variants
analyzed
across all cases); (5) Unique indel regions reference sets (gene-specific
regions
where pathogenic inframe indels and/or frameshift variants are known to
occur); (6)
DNA reports; (7) RNA reports; (8) Tumor Mutation Burden (TMB) calculations,
etc.
Once genomic variant characterization and classification has been completed,
other
applications and micro-services provide tools for variant scientists or other
clinicians
or even other micro-services to act upon the data results.
[00181] Referring still to Fig. 1, each micro-service includes a service
specification including definitions of data that the specified service is to
consume,
micro-service code defining the service to be performed by the specific micro-
service
and a definition of the data that is to be published to the lake as an interim
data
product 172. In each case, the service to be performed includes monitoring the
data
alerts list 169 or published data on the system communication network for data
to be
consumed (e.g., monitor for data that fits subscriptions associated with the
microservice) by the service and, once the service generates a data product,
publishing that data product to the data lake and placing an alert in alerts
list 169 or
publishing that data. In operation, when a micro-service is to consume a
published
data product, the service obtains the data product, consumes the product as
part of
performing the service, publishes new data product(s) to lake database 170 and
then
places a new data alert in list 169 to announce to other system consumers that
the
new data is ready for consumption.
[00182] Another system for asynchronous communication between micro-
services is a publish-subscribe message passing ("pub/sub") system which uses
the
alerts list 169. In this system type, alerts list 169 may be implemented in
the form of
a message bus. One example of a message bus that may be utilized is Amazon
Simple Notifications Service (SNS). In this system type, micro-services
publish
messages about their activities on message bus topics that they define. Other
micro-
services subscribe to these messages as needed to take action in response to
activities that occur in other micro-services.
[00183] In at least some embodiments, micro-services are not required to
directly subscribe to SNS topics. Rather, they set up message queues via a
queue
-36-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
service, and subscribe their queues to the SNS Topics that they are interested
in.
The micro-services then pull messages from their queues at any time for
processing,
without worrying about missing messages. One example of a queue service is the
Amazon Simple Queue Service (SQS) although others are contemplated.
[00184] Granularity of SNS topics may be defined on a message subject basis
(for instance, 1 topic per message subject), on a domain object basis (for
instance,
one topic per domain object basis), and/or on a per micro-service basis (for
instance,
one topic per micro-service basis). Message content may include only essential
information for the message in order to prioritize small message size. In at
least
some cases message content is architectured to avoid inclusion of patient
health
information or other information for which authorization is required to
access.
[00185] Different alerts may be employed throughout the system. For
instance,
alerts may be utilized in connection with the registration of a patient. One
example
of an alert is "services-patients.created", which is triggered by creation of
a new
patient in the system. Alerts may be utilized in connection with the analysis
of
variant call files. One example is "variant-analysis_staging", which is
triggered upon
the completion of a new variant calling result. Another example is "variant-
analysis_staging.ready", which is triggered upon completed ingestion of all
input files
for a variant calling result. Another example is "case_staging.ready", which
is
triggered when information in the system is ready for manual user review. Many
other alerts are contemplated.
[00186] Both orchestration workflows and micro-service alerts may be
employed in the system, either alone or in combination. In an example, an
event-
based micro-service architecture may be utilized to implement a complex
workflow
orchestration. Orchestrations may be integrated into the system so that they
are
tailored for specific needs of users. For instance, a provider or another
partner who
requires the ability to provide structured data into the lake may utilize a
partner-
specific orchestration to land structured data in the lake, pre-process files,
map data,
and load data into the data fault. As another example, a provider or other
partner
who requires the ability to provide unstructured data into the lake may
utilize a
partner-specific orchestration for pre-processing and providing unstructured
data to
the data lake. As another example, an orchestration may, upon publishing of
data
that is qualified for a particular use case (such as for research, or third-
party
delivery), transform the data and load it into a columnar data store
technology. As
another example, a "data vault to clinical mart" orchestration may take stable
points
in time of the data published to data vault by other orchestrations; transform
the data
-37-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
into a mart model, and transform the mart data through a de-identification
pipeline.
As another example, a "commercial partner egress file gateway" may utilize a
cohort
of patients whose data is defined for delivery, sourcing the data from de-
identified
data marts and the data lake (including molecular sequencing data) and publish
the
same to a third-party partner.
[00187] Referring still to Fig. 1, operational and analytical applications
188 and
192, respectively, are application programs that provide functionality to
various
system user types as well as interfaces optimized for use by those system
users.
Operational applications 188 include application programs that are primarily
required
to enable cancer state treatment planning processes for specific patients. For
instance, operational applications include application programs used by a
cancer
treating physician to assess treatment options and efficacy for a specific
patient. As
another instance, operational applications also include application programs
used by
an abstractor specialist to convert unstructured raw clinical medical records
or semi-
structured records to system optimized structured records. As another
instance,
operational applications may also include application programs used by
bioinformatics scientists or molecular pathologists to annotate variants. As
another
instance, operational applications also include application programs used by
clinicians to determine whether a patient is a good match for a clinical
trial. As yet
one other instance, operational applications may include application programs
used
by physicians to finalize patient reports.
[00188] Analytical applications 192, in contrast, include application
programs
that are provided primarily for research purposes and use by either provider
client
researchers or provider specialist researchers. For instance, analytical
applications
192 include programs that enable a researcher to generate and analyze data
sets or
derived data sets corresponding to a researcher specified subset of de-
identified
(e.g., not associated with a specific patient) cancer state characteristics.
Here,
analysis may include various data views and manipulation tools which are
optimized
for the types of data presented. Some applications may have features of both
analytical applications 192 and operational applications 188.
[00189] II. System Database Architecture And General Data Flow
[00190] Referring now to Fig. 2, a second representation of disclosed
system
100 shows many of the components shown in Fig. 1 in an operational
arrangement.
The Fig. 2 system includes system data sources 102 and operational system
components including an integration layer 220 in addition to the lake database
170,
-38-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
data vault database 180, operational applications 188 and analytical
applications 192
that are described above. Exemplary data sources 102 include physician
clinical
records systems 200, radiology imaging systems 202, provider genomic
sequencers
204, organoid modeling labs 206, partner genomic sequencers 208 and research
partner records systems 210. The source data types are only exemplary and are
not
intended to be limiting. In fact, it is contemplated that many other data
source types
generating other clinically relevant data types will be added to the system
over time
as other sources and data types of interest are identified and integrated into
the
overall system.
[00191] Referring again to Fig. 2, integration layer 220 includes
integration
gateways 312/314, a data lake catalog 226 and the data marts database 190
described above with respect to Fig. 1. The integration gateways receive data
files
and messages from sources 102, glean metadata from those files and messages
and route those files and messages on to other system components including
data
lake database 170 and catalog 226 as well as various system applications. New
files are stored in lake database 170 and metadata useful for searching and
otherwise accessing the lake data is stored in catalog 226. Again, non-
structured
and semi-structured raw and micro-service data is stored in lake database 170
and
system optimized structured data is stored in vault database 180 while
application
optimized structured data is stored in data marts database 190.
[00192] Referring again to Fig. 2, system users 10, 20, 3040, 50 and 60
access system data and functionality via the operational and/or analytical
applications 188 and 192, respectively. In some instances, in order to protect
patient
confidentiality, the system user cannot have access to patient medical records
that
are tied to specific and identified patients. For this reason, integration
layer 220 may
include a de-identification module which accesses system data, scrubs that
data to
remove any specific patient identification information and then serves up the
de-
identified data to the application platform. In other examples, the data vault
database may have its structure duplicated, such that a de-identified copy of
the data
in the data vault database 180 is retained separately from the non de-
identified copy
of the data in the data vault database. Data in the de-identified copy may be
stripped of its identifiers, including patient names; geographic subdivisions
smaller
than a state, including street address, city, county, precinct, ZIP code, and
their
equivalent geocodes, except for the initial three digits of the ZIP code if,
according to
the current publicly available data from the Bureau of the Census: (1) The
geographic unit formed by combining all ZIP codes with the same three initial
digits
-39-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
contains more than 20,000 people; and (2) The initial three digits of a ZIP
code for all
such geographic units containing 20,000 or fewer people is changed to 000;
elements of dates (except year) for dates that are directly related to an
individual,
including birth date, admission date, discharge date, death date, and all ages
over
89 and all elements of dates (including year) indicative of such age, except
that such
ages and elements may be aggregated into a single category of age 90 or older;
Telephone numbers; Vehicle identifiers and serial numbers, including license
plate
numbers; Fax numbers; Device identifiers and serial numbers; Email addresses;
Web Universal Resource Locators (URLs), Social security numbers; Internet
Protocol (IP) addresses; Medical record numbers; Biometric identifiers,
including
finger and voice prints; Health plan beneficiary numbers; Full-face
photographs and
any comparable images; Account numbers and other unique identifying numbers,
characteristics, or codes; and Certificate/license numbers. Because data in
the data
vault database 180 is structured, much of the information not permitted for
inclusion
in the de-identified copy is absent by virtue of the fact that a structured
location does
not exist for inclusion of such information. For instance, the structure of
the data
vault database for storing the de-identified copy may not include a field for
storing a
social security number. As another example, data in the data vault database
may be
segregated by customer. For example, if one physician 10 wishes for his or her
patients to have their data segregated from other data in the data lake
database 170,
their data may be segregated in a single tenant data vault, such as the single
tenant
data vault arrangement shown in Fig. 3a.
[00193] Many users employing the operational applications 188 do have
physician-patient relationships, or otherwise are permitted to access records
in
furtherance of treatment, and so have authority to access patent identified
medical,
healthcare and other personal records. Other users employing the operational
applications have authority to access such records as business associates of a
health care provider that is a covered entity. Therefore, in at least some
cases,
operational applications will link directly into the integration layer of the
system
without passing through de-identification module 224, or will provide access
to the
non de-identified data in the database 160. Thus, for instance, a physician
treating a
specific patient clearly requires access to patient specific information and
therefore
would use an operational application that presents, among other information,
patient
identifying information.
[00194] In some cases, users employing operational applications will want
access to at least some de-identified analytical applications and
functionality. For
-40-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
instance, in some cases an operational application may enable a physician to
compare a specific patient's cancer state to multiple other patient's cancer
states,
treatments and treatment efficacies. Here, while the physician clearly needs
access
to her patient's identifying information and state factors, there is no need
and no right
for the physician to have access to information specifically identifying the
other
patients that are associated with the data to be compared. Thus, in some cases
one
operational application will access a set of patient identified data and other
sets of
patient de-identified data and may consume all of those data sets.
[00195] Referring now to Fig. 3, a system representation 100 akin to the
one in
Fig. 2 is shown, albeit where the Fig. 3 representation is more detailed. In
Fig. 3
integration layer 220 includes separate message and file gateways 312 and 314,
respectively, an event reporting bus 316, system micro-services 186, various
data
lake APIs 332, 334 and 336, an ETL module 338, data lake query and analytics
modules 346 and 348, respectively, an ETL platform 360 as well as data marts
database 190.
[00196] Referring to Fig. 3, sources 102 are linked via the internet or
some
other communication network to system 100 via message gateway 312 and file
gateway 314. Messages received from data sources 102 at gateway 312 are
forwarded on to event bus 326 which routes those messages to other system
modules as shown. Messages from other system modules can be routed to the data
sources via message gateway 312.
[00197] File gateway 314 receives source files and controls the process of
adding those files to lake database 170. To this end, the file gateway runs
system
access security software to glean metadata from any received file and to then
determine if the file should be added to the lake database 170 or rejected as,
for
instance, from an unauthorized source. Once a file is to be added to the lake
database, gateway 314 transfers the file to lake database 170 for storage,
uses the
metadata gleaned from the file to catalog the new file in the lake catalog 226
and
posts an alert in the data alert list 169 (see again Fig. 1) announcing that
the new
data has been published to the lake for consumption.
[00198] Referring still to Fig. 3, a subset of micro-services monitoring
alert list
169 for data of the type published to lake database 170 access the new data or
consume that data when published to the network, perform their data
consumption
processes, publish new data products to lake database 170 and post new data
alerts
in list 169 or publish the new data on the network per the publication-
subscription
architecture described above. In cases where system user activities are
required as
-41-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
part of a micro-service, the service schedules those activities to be
completed by
provider specialists when needed and ingests data generated thereby,
eventually
publishing new data products to the lake database 170.
[00199] The orchestration modules and resources monitor the entire data
process and determine when data lake data is to be replicated within the data
vault
and/or within the data marts in different system or application optimized
model
formats. Whenever lake data is to be restructured and placed in the data vault
or the
data marts, ETL platform 360 extracts the data to restructure, transforms the
data to
the system or application specific data structure required and then loads that
data
into the respective database 180 or 190. In some cases it is contemplated that
ETL
platform may only be capable of transforming data from the data lake structure
to the
data vault structure and from the data vault structure to the application
specific data
models required in data marts 190.
[00200] Referring still to Fig. 3, analytical applications 192 are shown to
include, among other applications, "self-service" applications. Here, the
phrase "self-
service" is used to refer to applications that enable a system user to, in
effect, use
query tools and data visualization tools, to access and manipulate data sets
that are
not optimally supported by other user applications. Here, the idea is that,
especially
in the context of research, system users should not be constrained to specific
data
sets and analysis and instead should be able to explore different data sets
associated with different cancer state factors, different treatments and
different
treatment efficacies. The self-service tools are designed to allow an
authorized
system user to develop different data visualizations, unique SQL or other
database
queries and/or to prepare data in whatever format desired. Hereinafter, unless
indicated otherwise, the term "explore" will be used to refer to any self-
service
activities performed within the disclosed system.
[00201] Referring still to Fig. 3, self-service applications 356 enable a
system
user to explore all system databases in at least some embodiments including
the
data marts 190, the lake database 170 and the data vault database 180. In
other
embodiments, because lake database 170 data is either unstructured or only
semi-
structured, self-service applications may be limited to exploring only the
data mart
database 190 or the data vault database 180.
[00202] III. Data Ingestion, Normalization And Publication
[00203] Referring to Fig. 4, a high level data distribution process 400 is
illustrated that is consistent with at least some aspects of the present
disclosure. At
process block 402, data is collected from various data sources 102 (see again
Figs.
-42-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
1 through 3) and at block 404, assuming that data is to be ingested into the
system
100, the data is stored in lake database 170. Here, data collection is
continual over
time as more and more data for increasing the system knowledge base is
generated
regularly by physicians, provider and partner researchers and provider
specialists.
Specific steps in at least some exemplary data collection processes are
described
hereafter. The collected original data is stored in the lake database 170 as
raw
original data (e.g., documents, images, records, files, etc.).
[00204] At process block 406, at least a subset of the collected data is
"shaped"
or otherwise processed to generate structured data that is optimal for
database
access, searching, processing and manipulation. Here, the data shaping process
may take many forms and may include a plurality of data processing steps that
ultimately result in optimal system structured data sets. At step 408 the
database
optimized shaped data is added to similarly structured data already maintained
in
data vault database 180.
[00205] Continuing, at block 410, at least a subset of the data vault data
or the
lake data is "shaped" or otherwise processed to generate structured data that
is
optimal to support specific user application programs 188 and 192 (see again
Fig. 2).
Here, again, the data shaping process may take many forms and may include a
plurality of data processing steps that ultimately result in optimal
application
supporting structured data sets. At step 412 the optimized application
structured
data is added to similarly structured data already maintained in data marts
database
190.
[00206] Referring again to Fig. 4, at block 414, system users employ
various
application programs to access and manipulate system data including the data
in
any of the lake database 170, data vault database 180 and data marts 190. At
block
212, as users use the system, data related to system use is collected after
which
control passes backup to block 206 where the collected use data is shaped and
eventually stored for driving additional applications.
[00207] Fig. 5 includes a flow chart illustrating a process 500 that is
consistent
with at least some aspects of the present disclosure for ingesting initial raw
data into
the disclosed system. At process block 502 new raw data is received at the
file
gateway 314 (see Fig. 2) which, at block 504, determines whether or not the
data
should be rejected or ingested based on the data source, data format or other
transport data used to transmit the received data to the gateway. If the data
is to be
ingested, gateway 312 gleans metadata from the received data at block 506
which is
stored in the data lake catalog 226 (see Fig. 2) while the received data set
is stored
-43-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
in data lake 170 at 508. At block 510, an alert is added to the alert list 169
indicting
the new data is available to be consumed along with a data type so that other
data
consumers can recognize when to consume the newly stored data. Control passes
back up to block 502 where the process described above continues.
[00208] Fig. 6 is a flow chart illustrating a general process 600 by which
system
micro-services consume lake data and generate micro-service data products that
are
published back to the lake database for further consumption by other micro-
services.
At process block 602 a micro-service process is specified that includes data
consumption and data product definitions as well as micro-service code for
carrying
out process steps. At block 604 the micro-service monitors the data lake 170
for
alerts specifying new data that meets the data consumption definition for the
specific
micro-service. At block 606, where new lake data alerts do not specify data
that
meets the data consumption definition, control passes back up to block 604
where
steps 604 and 606 continue to cycle.
[00209] Referring still to Fig. 6, once an alert indicates new data that
meets the
micro-service data consumption definition, control passes to block 608 where
the
micro-service accesses the lake data to be consumed and that data is consumed
at
block 610 which generates a new data product. Continuing, at block 612, the
new
data product is published to data lake database 170 and at 614 another alert
is
added to the data alert list 169.
[00210] Referring still to Fig. 6, process 600 is associated with a single
system
micro-service. It should be understood that hundreds and in some cases even
thousands of micro-services will be performed simultaneously and that two or
more
micro-services may be performed on the same raw data or using prior generated
micro-service data product(s) at the same time. In many cases a micro-service
will
require two or more data sets at the same time and, in those cases, a micro-
service
will be programmed to monitor for all required data in the data lake and may
only be
initiated once all required data is indicated in the alerts list 169.
[00211] As described above, some micro-services will be completely
automated, so that no user activities are required, while other micro-services
will
require at least some user activities to perform some service steps. Fig. 7
illustrates
a simple fully automated micro-service 700 while Fig. 8 illustrates a micro-
service
800 where a user has to perform some activities. In Fig. 7, at process block
702, an
OCR micro-service is specified that requires consumption of raw clinical
medical
records to generate semi-structured clinical medical records with OCR tags
appended to document characters. At block 704 the OCR micro-service monitors
-44-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
the system alert list 169 for alerts indicating that new raw clinical records
data is
stored in the data lake.
[00212] At block 706, where there is no new clinical record to be ingested
into
the system, control passes back up to block 704 and the process 700 cycles
through
blocks 704 and 706. Once a new clinical record is saved to lake database 170
and
an alert related thereto is detected by the OCR micro-service, the micro-
service
accesses the new raw clinical record from the data lake at 708 and that record
is
consumed at block 710 to generate a new OCR tagged record. The new OCR
tagged record is published back to the lake at 712 and an alert related
thereto is
added to the data alert list 169 at 714. Once the OCR tagged record is stored
in lake
database 170, it can be consumed by other micro-services or other system
modules
or components as required.
[00213] The Fig. 8 process 800 is associated with a micro-service for
generating a system optimized structured clinical record assuming that an
unstructured clinical medical record that has already been tagged with medical
terms, phrases and contextual meaning has been generated as a micro-service
data
product by a prior micro-service. At process block 802, the record structuring
micro-
service process is defined and includes a data consumption definition that
requires
OCR, NLP records to be consumed and a data production definition where the
system optimized data structure is generated as a micro-service data product.
At
block 804 the structuring micro-service listens for alerts that new records to
consume
have been stored in lake database 170. At block 806, where new data to consume
has not been stored in the lake database 170, control cycles back through
blocks
804 and 806 continually. Once new data to consume has been stored in lake
database 170, control passes to block 808 where the micro-service places an
alert in
an abstractor specialist's work queue identifying the record to consume as
requiring
specialist activities to complete the micro-service.
[00214] Referring still to Fig. 8, at block 810, the system monitors for
specialist
selection of the queued record for consumption and the system cycles between
blocks 808 and 810 until the record is selected. Once the record is selected
by the
abstractor specialist at 810, control passes to block 812 where the record to
be
consumed is accessed in database 170. At block 814, the micro-service accesses
a
structured clinical record file which includes data fields to be populated
with data
from the accessed clinical record. The micro-service attempts to identify data
in the
clinical record to populate each field in the structured record at 814 and
populates
fields with data whenever possible to generate a structured clinical record
draft.
-45-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
[00215] Continuing, at block 816 a micro-service presents an abstractor
application interface to the abstractor specialist that can be used to verify
draft field
entries, modify entries or to aid the abstractor specialist in identifying
data to
populate unfilled structured record fields. To this end, see Fig. 9 that shows
an
exemplary abstractor interface screenshot 914 that may be viewed by an
abstractor
specialist which includes an original record in an original record field 900
on the right
hand side of the shot and a structured record area 902 on the left hand side
of the
screenshot. The structured record in area 902 includes a set of fields to be
populated with information from the original record or in some other fashion
to
prepare the structured record for use by system applications. The structured
record
shown in area 902 only shows a portion of the structured record that fits
within area
902 and in most cases the structured record will have hundreds or even
thousands
of record fields that need to be populated with data. Exemplary structured
record
fields shown include a site field 904, year fields 905 and a histology field
906.
[00216] Referring still to Fig. 9, the original record shown in field 900
has
already been subjected to OCR and NLP so that words and phrases have been
recognized by a system processor and the text in the document is associated
with
specific medical words and phrases or other meaning (e.g., dates are
recognized as
dates, a "Patient's Name" label on an original record is recognized as the
phrase"
patient's name" and an adjacent field is recognized as a field that likely
includes a
patient's name, etc.). Again, the processor examines the original record for
data that
can be used to populate the structured record fields in order to create at
least a
partially complete draft of the structured record for consideration and
completion by
the abstractor specialist.
[00217] Data in the original record used to populate any field in the
structured
record is highlighted (see 910, 912) or somehow visually distinguished within
the
original record to aid the abstractor specialist in located that data in the
original
record when reviewing data in the structured record fields. The specialist
moves
through the structured record reviewing data in each field, checking that data
against
the original record and confirming a match (e.g., via selection of a
confirmation icon
or the like) or modifying the structured record field data if the
automatically populated
data is inaccurate (see block 818 in Fig. 8).
[00218] In cases where the processor cannot automatically identify data to
populate one or more fields in the structured record, the specialist reviews
the
original record manually to attempt to locate the data required for the field
and then
enters data if appropriate data is located. Where the micro-service fills in
fields that
-46-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
are then to be checked by the specialist, in at least some cases original
record data
used to populate a next structured record field to be considered by the
specialist may
be especially highlighted as a further aid to locating the data in the
original record. In
some cases the micro-service will be able to recognize data in several
different
formats to be used to fill in a structured record field and will be able to
reformat that
data to fill in the structured record field with a required form.
[00219] Referring again to Fig. 8, at block 820, once the structured
clinical
record has been completed, the complete system optimized structured clinical
record
is stored in lake database 170 and then a new data alert is added to alert
list 169 at
822 to alert other micro-services and orchestration resources that the
complete
record is available to be consumed.
[00220] In some cases a system micro-service will "learn" from specialist
decisions regarding data appropriate for populating different structured data
sets.
For instance, if a specialist routinely converts an abbreviation in clinical
records to a
specific medical phrase, in at least some cases the system will automatically
learn a
new rule related to that persistent conversion and may, in future structured
draft
records, automatically convert the abbreviation to its expanded form. Many
other
system learning techniques are contemplated.
[00221] In cases where a system micro-service can confirm structured record
field information with high confidence, the micro-service may reduce the
confirmation
burden on the specialist by not highlighting the accurate information in the
structured
record. For instance, where a patient's date of birth is known, the micro-
service may
not highlight a patient DOB field in the structured record for confirmation.
[00222] Referring now to Fig. 10, an exemplary multi-micro-service process
1000 for ingesting a clinical medical record and structuring the record
optimally for
database activities is illustrated. At step 1001, a medical record is acquired
in digital
form. Here, where an original record is in paper form, acquiring a digital
record may
include scanning that record into the system via a scanner 1012 to generate a
PDF
or other digital representation which is then provided to a system server 150
for
storage in database 160. In other cases where the record is already in digital
form
(e.g., an EMR), the digital record can simply be stored by server 150 in
database
160.
[00223] A data normalization and shaping process is performed at 1002 that
includes accessing an original clinical record from database 160 and
presenting that
record to a system specialist 40 as shown in Fig. 9. As the original record is
accessed or at some other prior time, an OCR micro-service 700 (see again Fig.
7)
-47-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
is used to tag letters in the record. The tagged record is stored in the data
lake and
an alert is added to the alert list 169. Next, an NLP micro-service 1008
accesses the
OCR tagged record and performs an NLP process on the text in that record to
generate an NLP processed record which is again stored in the data lake and
another alert is added to the alert list 169.
[00224] At 800 (see Fig. 8), a draft structured clinical medical record is
generated for the patient and is presented to an abstractor specialist via an
interface
as in Fig. 9 so that the specialist can correct errors.
[00225] Referring again to Fig. 10, once the structured record has been
filled in
to the extent possible based on an original medical record, at block 1020 the
specialist may perform some task to attempt to complete record fields that
have not
been filled. For instance, in a case where a specific structured record field
cannot be
filled based on information from the original record, the specialist may
attempt to
track down information related to the field from some other source. For
example, in
a simple case the specialist may call 1024 a physician that generated the
original
record to track down missing information. As another example, the specialist
may
access some other patient record (e.g., an insurance record, a pharmacy
record,
etc.) that may include additional information useable to populate an empty
field.
Once the structured record is as complete as possible, that record is stored
at 1022
back to the system database 160.
[00226] Referring now to Fig. 11, an exemplary process 1100 for generating
genomic patient and tumor data is illustrated. Robust nucleic acid extraction
protocols and sequencing library construction protocols may be applied, and
appropriately deep coverage across all targeted regions and appropriately
designed
analysis algorithms may be utilized. Prior to process 1100, a genomic
sequencing
order may be received at file gateway 314 and, once ingested, may be stored in
lake
database 170 for subsequent consumption. Here, when a tumor sample
corresponding to the sequencing order is received 1114, the sample is
associated
with the order and process 1100 continues with the order being assigned to a
lab
technician's work queue to commence specimen sequencing 1116. At 1116 the
specimens are subjected to a genetic sequencing process using sequencing
machine 1132 to generate genomic data for both the patient and the tumor
specimens. At 1118 alterations from raw molecular data are called and at block
1120 pathogenicity of the variants is classified. At 1122 genomic phenotypes
may
be calculated. At 1123 an MSI assay may be performed. At 1124 at least a
subset of
-48-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
the genomic data and/or an analysis of at least the subset of the genomic data
is
stored in system database 160.
[00227] Referring still to Fig. 11, different approaches may be utilized to
implement the genetic sequencing process at 1116. In one example, an oncology
assay may be implemented that interrogates all or a subset of cancer-related
genes
in matched tumor and normal tissue. As used herein, "tumor" tissue or specimen
refers to a tumor biopsy or other biospecimen from which the DNA and/or RNA of
a
cancer tumor may be determined. As used herein, "normal" tissue or specimen
refers to a non-tumor biopsy or other biospecimen from which DNA and/or RNA
may
be determined. As used herein, "matched" refers to the tumor tissue and the
normal
tissue being correlated at the same position in a DNA and/or RNA sequence,
such
as a reference sequence. The assay may further provide whole transcriptome RNA
sequencing for gene rearrangement detection. The assay may combine tumor and
normal DNA sequencing panels with tumor RNA sequencing to detect somatic and
germline variants, as well as fusion mRNAs created from chromosomal
rearrangements.
[00228] The assay may be capable of detecting somatic and germline single
nucleotide polymorphisms (SNPs), indels, copy number variants, and gene
rearrangements causing chimeric mRNA transcript expression. The assay may
identify actionable oncologic variants in a wide array of solid tumor types.
The assay
may make use of FFPE tumor samples and matched normal blood or saliva
samples. The subtraction of variants detected in the normal sample from
variants
detected in the tumor sample in at least some embodiments provides greater
somatic variant calling accuracy. Base substitutions, insertions and deletions
(indels), focal gene amplifications and homozygous gene deletions of tumor and
germline may be assayed through DNA hybrid capture sequencing. Gene
rearrangement events may be assayed through RNA sequencing.
[00229] In one example, the assay interrogates one or more of the 1711
cancer-related genes listed in the tables shown in Fig. 22a-22j (referred to
herein as
the "xE" assay). This targeted gene panel may be divided into a clinically
actionable
tier, wherein 130 tier 1 genes (see table in Fig. 23) that can influence
treatment
decisions are assayed with an assigned detection cutoff of 5% variant allele
fraction
(VAF) i.e. the limit of detection is 5% VAF or lower, and a secondary tier,
wherein an
additional 1,581 genes (e.g., the difference between the gene set in Figs. 22a-
22j
and Fig. 23) are assayed for analytical purposes with an assigned detection
cutoff of
10% VAF (limit of detection 10% VAF or lower). The RNA based gene
-49-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
rearrangement detection may also be divided into a primary clinically-
actionable tier
containing 41 rearrangements (See table in Fig. 24), and a secondary tier that
may
contain some or all known fusions within the wider literature or novel fusions
of
putative clinical importance detected by the assay. "Tier 1" genes are genes
linked
with response or resistance to targeted therapies, resistance to standard of
care, or
toxicities associated with treatment. The VAF cutoff percentages described
herein
are exemplary and other cutoff values may be utilized. Reads may be mapped to
a
human reference genome, such as hg16, hg17, hg18, hg19, etc. (available from
the
Genome Reference Consortium, at httosliwww.ncth nim nih.govicirc). In another
example, the assay may interrogate other gene panels, such as the panels
listed in
the tables shown in Figs. 27a, 27b1, 27b2, 27c1 and 27c2 and 27d (herein "the
xT
panel") or the panel listed in the table shown in Figs. 28a and 28b.
[00230] Referring still to Fig. 11, the alterations called in sub-process
1118 may
be called through a clinical variant calling process. An exemplary variant
calling
process is shown in Fig. 11a. At 1134 acceptance criteria are applied to the
raw
molecular data for clinical variant calling. There may be one or more
acceptance
criteria, and multiple acceptance criteria may be applied.
[00231] One type of acceptance criteria is that a certain percentage of
loci
assay must exceed a certain coverage. For instance, a first percentage of loci
must
exceed a certain first coverage and a second percentage of loci must exceed a
second coverage. The first percentage of loci may be 60%, 65%, 70%, 75%, 80%,
85%, etc. and the first coverage level may be 150x, 200x, 250x, 300x, etc. The
second percentage of loci may be 60%, 65%, 70%, 75%, 80%, 85%, etc. and the
second coverage level may be 150x, 200x, 250x, 300x, etc. The first percentage
of
loci assayed may be lower than the second percentage of loci assayed while the
first
coverage level may be deeper than the second coverage level.
[00232] Another type of acceptance criteria may be that the mean coverage
in
the tumor sample meets or exceeds a certain coverage threshold, such as 300x,
400x, 500x, 600x, 700x, etc.
[00233] Another type of acceptance criteria may be that the total number of
reads exceeds a predefined first threshold for the tumor sample and a
predefined
second threshold for the normal sample. For instance, the total number of
reads for
the tumor sample must exceed 5 million, 10 million, 15 million, 20 million, 25
million,
30 million, 35 million, 40 million, etc. reads and the total number of reads
for the
normal sample must exceed 5 million, 10 million, 15 million, 20 million, 25
million, 30
million, 35 million, 40 million, etc. reads. In one example, the threshold for
the total
-50-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
number of the reads for the tumor sample may be greater than the total number
of
reads for the normal sample. For instance, the threshold for the total number
of the
reads for the tumor sample may be greater than the total number of reads for
the
normal sample by 5 million, 10 million, 5 million, 10 million, 15 million, 20
million, 25
million, 30 million, 35 million, 40 million, etc. reads.
[00234] Another type of acceptance criteria is that reads must maintain an
average quality score. The quality score may be an average PHRED quality
score,
which is a measure of the quality of the identification of the nucleobases
generated
by automated DNA sequencing. The quality score may be applied to a portion of
the
raw molecular data. For instance, the quality score may be applied to the
forward
read. Another type of acceptance criteria is that the percentage of reads that
map to
the human reference genome. For instance, at least 60%, 65%, 70%, 75%, 80%,
85%, 80%, 95%, etc. of reads must map to the human reference genome.
[00235] Still at 1134, RNA acceptance criteria may additionally be
reviewed.
One type of RNA acceptance criteria is that a threshold level of read pairs
will be
generated by the sequencer and pass quality trimming in order to continue with
fusion analysis. For instance, the threshold level may be 5 million, 10
million, 15
million, 20 million, 25 million, 30 million, 35 million, 40 million, etc.
Another type of
acceptance criteria is that reads must maintain an average quality score. The
quality
score may be an average RNA PHRED quality score, which is a measure of the
quality of the identification of the nucleobases generated by automated RNA
sequencing. The quality score may be applied to a portion of the raw molecular
data. For instance, the quality score may be applied to the forward read.
[00236] Yet another type of acceptance criteria is that the percentage of
reads
that map to the human reference genome. For instance, at least 60%, 65%, 70%,
75%, 80%, 85%, 80%, 95%, etc. of reads must map to the human reference
genome.
[00237] If RNA analysis fails pre or post-analytic quality control, DNA
analysis
may still be reported. Due to the difficulties of RNA-seq from FFPE, a higher
than
normal failure rate is expected. Because of this, it may be standard to report
the
DNA variant calling and copy number analysis section of the assay, no matter
the
outcome of RNA analysis.
[00238] At 1138, the step of variant quality filtering may be performed.
Variant
quality filtering may be performed for somatic and germline variations. For
somatic
variant filtering, the variant may have at least a minimum number of reads
supporting
the variant allele in regions of average genomic complexity. For instance, the
-51-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
minimum number of reads may be 1, 2, 3, 4, 5, 6, 7, etc. A region of the
genome
may be determined free of variation at a percentage of LLOD (for instance, 5%
of
LLOD) if it is sequenced to at least a certain read depth. For instance, the
read
depth may be 100x, 150x, 200x, 250x, 300x, 350x, etc.
[00239] The somatic variant may have a minimum threshold for SNPs. For
instance, it may have at least 20x, 25x, 30x, 35x, 40x, 45x, 50x, etc.
coverage for
SNPs. The somatic variant may have a minimum threshold for indels. For
instance,
at least 50x, 55x, 60x, 65x, 70x, 75x, 80x, 85x, 90x, 95x, 100x, etc. coverage
for
indels may be required. The variant allele may have at least a certain variant
allele
fraction for SNPs. For instance, it may have at least 1%, 3%, 5%, 7%, 9%, etc.
variant allele fraction for SNPs. The variant allele may have at least a
certain variant
allele fraction for indels. For instance, it may have a 6%, 8%, 10%, 12%, 14%,
etc.
variant allele fraction for indels.
[00240] The variant allele may have at least a certain read depth coverage
of
the variant fraction in the tumor compared to the variant fraction in the
normal
sample. For instance, the variant allele may have 4x, 6x, 8x, 10x etc. the
variant
fraction in the tumor compared to the variant fraction in the normal sample.
Another
type of filtering criteria may be that the bases contributing to the variant
must have
mapping quality greater than a threshold value. For instance, the threshold
value
may be 20, 25, 30, 35, 40, 45, 50, etc.
[00241] Another type of filtering criteria may be that alignments
contributing to
the variant must have a base quality score greater than a threshold value. For
instance, the threshold value may be 10, 15, 20, 25, 30, 35, etc. Variants
around
homopolymer and multimer regions known to generate artifacts may be filtered
in
various manners. For instance, strand specific filtering may occur in the
direction of
the read in order to minimize stranded artifacts. If variants do not exceed
the
stranded minimum deviation for a specific locus within known artifact
generating
regions, they may be filtered as artifacts.
[00242] Variants may be required to exceed a standard deviation multiple
above the median base fraction observed in greater than a predetermined
percentage of samples from a process matched germline group in order to ensure
the variants are not caused by observed artifact generating processes. For
instance,
the standard deviation multiple may be 3x, 4x, 5x, 6x, 7x, etc. For instance,
the
predetermined percentage of samples may be 15%, 20%, 25%, 30%, 35%, etc.
[00243] Still at 1138, for germline variant filtering, the germline variant
may
have a minimum threshold for SNPs. For instance, it may have at least 20x,
25x,
-52-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
30x, 35x, 40x, 45x, 50x, etc. coverage for SNPs. The germline variant may have
a
minimum threshold for indels. For instance, at least 50x, 55x, 60x, 65x, 70x,
75x,
80x, 85x, 90x, 95x, 100x, etc. coverage for indels may be required. The
germline
variant calling may require at least a certain variant allele fraction. For
instance, it
may require at least 15%, 20%, 25%, 30%, 35%, 40%, 45% etc. variant allelic
fraction.
[00244] Another type of filtering criteria may be that the bases
contributing to
the variant must have mapping quality greater than a threshold value. For
instance,
the threshold value may be 20, 25, 30, 35, 40, 45, 50, etc. Another type of
filtering
criteria may be that alignments contributing to the variant must have a base
quality
score greater than a threshold value. For instance, the threshold value may be
10,
15, 20, 25, 30, 35, etc.
[00245] At 1142, copy number analysis may be performed. Copy number
alteration may be reported if more than a certain number of copies are
detected by
the assay, such as 3, 4, 5, 6, 7, 8, 9, 10, etc. Copy number losses may be
reported
if the ratio of the segments is below a certain threshold. For instance, copy
number
losses may be reported if the 10g2 ratio of the segment is less than -1Ø
[00246] At 1146, RNA fusion calling analysis may be conducted. RNA fusions
may be compared to information in a gene-drug knowledge database 1148, such as
a database described in "Prospective: Database of Genomic Biomarkers for
Cancer
Drugs and Clinical Targetability in Solid Tumors." Cancer Discovery 5, no. 2
(February 2015): 118-23. doi:10.1158/2159-8290.CD-14-1118. If the RNA fusion
is
not present within the gene-drug knowledge database 1148, the RNA fusion may
not
be presented. RNA fusions may not be called if they display fewer than a
threshold
of breakpoint spanning reads, such as fewer than 2, 3, 4, 5, 6, 7, 8, 9, 10,
etc.
breakpoint spanning reads. If an RNA fusion breakpoint is not within the body
of two
genes (including promotor regions), the fusion may not be called.
[00247] At 1150, DNA fusion calling analysis may be performed. At 1154,
joint
tumor normal variant calling data may be prepared for further downstream
processing and analysis. Germline and somatic variant data are loaded to the
pipeline database for storage and reporting. For example, for both somatic and
germline variations, the data may include information on chromosome, position,
reference, alt, sample type, variant caller, variant type, coverage, base
fraction,
mutation effect, gene, mutation name, and filtering. Fig. 25 shows an
exemplary
data set in table form that is consistent with at least some embodiments of
the above
disclosure.
-53-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
[00248] Copy Number Variant (CNV) data may also be loaded to the pipeline
database for downstream analysis. For example, the data may include
information
on chromosome, start position, end position, gene, amplification, copy number,
and
10g2 ratios. Fig. 26 includes exemplary CNV data.
[00249] Following analysis, a workflow processing system may extract and
upload the variant data to the bioinformatics database. In one example, the
variant
data from a normal sample may be compared to the variant data from a tumor
sample. If the variant is found in the normal and in the tumor, then it may be
determined that the variant is not a cause of the patient's cancer. As a
result, the
related information for that variant as a cancer-causing variant may not
appear on a
patient report. Similarly, that variant may not be included in the expert
treatment
system database 160 with respect to the particular patient. Variant data may
include
translation information, CNV region findings, single nucleotide variants,
single
nucleotide variant findings, indel variants, indel variant findings, variant
gene
findings. Files, such as BAM, FASTQ, and VCF files, may be stored in the
expert
treatment system database 160.
[00250] Referring again to Fig. 11, at 1123, an MSI assay may be performed
as
a next generation sequencing based test for microsatellite instability. The
MSI assay
may comprise a panel of microsatellites that are frequently unstable in tumors
with
mismatch repair deficiencies to determine the frequency of DNA slippage
events.
Using the assay methods, tumors may be classified into different categories,
such as
microsatellite instability high (MSI-H), microsatellite stable (MSS), or
microsatellite
equivocal (MSE). The assay may require FFPE tumor samples with matched normal
saliva or blood to determine the MSI status of a tumor. MSI status can provide
doctors with clinical insight into therapeutic and clinical trial options for
patient care,
as well as the need for further genetic testing for conditions such as Lynch
Syndrome. The MSI algorithm may be initiated after the raw sequencing data is
processed through the bioinformatics pipeline. Upon completion of the MSI
algorithm, results may be stored in the expert treatment system database 160.
U.S.
Prov. Pat. App. No. 62/745,946, filed October 15, 2018, incorporated by
reference in
its entirety, describes exemplary systems and methods for MSI algorithms.
[00251] Referring still to Fig. 11, sub-processes 1116 through 1123 may be
substantially or, in some cases even completely automated so that there is
little if
any lab technician activity required to complete those processes. In other
cases
each of the sub-processes 1116 through 1123 may include one or more lab
technician activities and one or more automated micro-service steps or
calculations.
-54-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
Again, in cases where a lab technician performs service steps, the micro-
service
may present instructions or other interface tools to help guide the technician
through
the manual service steps. At the end of each manual step some indication that
the
step has been completed is received by the micro-service. For instance, in
some
cases a system machine (e.g., the sequencing computer 1132) may provide one or
more data products to the micro-service that indicate completion of the step.
As
another instance, a technician may be queried for specific data related to the
stage
of the service. As yet one other instance, a technician may simply enter some
status
indication like, step completed, to indicate that process 1100 should
continue.
[00252] One exemplary workflow 1153 with respect to the bioinformatics
pipeline is shown in Fig. 11b. Referring also to Fig. 11c, a client, such as
an entity
that generates a bioinformatics pipeline, can register new samples 1157 and
upload
variant call text files 1159 for processing to a cloud service 1161. The cloud
service
1161 may initiate an alert by adding a message 1163 to a queue service 1165
(e.g.,
to an alert list) for each uploaded file. Input micro-services 1167 (1167 in
Fig. 11c)
receive messages 1169 about each incoming file and process each of those files
one at a time (see 1171) as they are received to process and validate each
file. The
input micro-services 1167 may run as separate node processes and, in at least
some cases, generate SQL insertion statements 1173 to add each validated file
to
the expert treatment system database 160.
[00253] Referring still to Figs. 11b and 11c, the input micro-services 1167
may
also run a variant classification engine 1360 on the variant files utilizing a
knowledge
database of variant information 1175 to calculate many different types of
variant
criteria, further classification and addition database insertion. The variant
micro-
service 1167 may publish an alert 1183 when a key event occurs, to which other
services 1179 can subscribe in order to react. After a variant call text file
is parsed,
the variant micro-service may insert variant analysis data into the expert
treatment
system database 160 including criteria, classifications, variants, findings,
and sample
information.
[00254] Other micro-services 1179 can query 1181 samples, findings,
variants,
classifications, etc. via an interface 1177 and SQL queries 1187. Authorized
users
may also be permitted to register samples and post classifications via the
other
micro-services.
[00255] Referring to Fig. 12, an organoid modelling process 1200 is
illustrated
that is consistent with at least some aspects of the present disclosure. At
1201 a
tumor specimen 1230 is obtained which is divided into multiple specimens and
each
-55-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
specimen is then grown 1202 as a 3D organoid 1232 in a special growth media
designed to promote organoid development. At 1204 different cancer treatments
are
applied to each of the organoids to elicit responses. At 1206 a provider
specialist
observes the treatment results and at 1208 the results are characterized to
assess
efficacy of each treatment. At 1210 the results are stored in the system
database
160 as part of the unified structured data set for the patient.
[00256] Referring to Fig. 13, a process 1300 for ingesting radiological
images
into the disclosed system and for identifying treatment relevant tumor
features is
illustrated. At 1302 a set of 2D medical images including a tumor and
surrounding
tissue are either generated or acquired from some other source and are stored
in
system database 160 (e.g., as unaltered images in the lake database). In many
cases the 2D images will be in a digital format suitable for processing by a
system
processor. In other cases the 2D images will be in a format that has to be
converted
to a data set suitable for system analysis. For instance, in some cases the
original
images may be on film and may need to be scanned into a digital format prior
to
creating a 3D tumor model. In some cases original images may not be useable to
generate a 3D tumor model and in those cases additional imaging may be
required
to generate the model.
[00257] At 1304 tumor tissue is detected and segmented within each of the
2D
images so that tumor tissue and different tissue types are clearly
distinguished from
surrounding tissues and substances and so that different tumor tissue types
are
distinguishable within each image. At 1306 the tissue segments within the 2D
images are used as a guide for contouring the tissue segments to generate a 3D
model of the tumor tissue. At 908 a system processor runs various algorithms
to
examine the 3D model and identify a set of radiomic (e.g., quantitative
features
based on data characterization algorithms that are unable to be appreciated
via the
naked eye) features of the segmented tumor tissue that are clinically and/or
biologically meaningful and that can be used to diagnose tumors, assess cancer
state, be used in treatment planning and/or for research activities. At 1310
the 3D
model and identified features are stored in the system database 160.
[00258] While not shown, in some cases a normalization process is performed
on the medical images before the 3D model is generated, for example, to ensure
a
normalization of image intensity distribution, image color, and voxel size for
the 3D
model. In other cases the normalization process may be performed on a 3D model
generated by the disclosed system. In at least some cases the system will
support
many different segmentation and normalization processes so that 3D models can
be
-56-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
generated from many different types of original 2D medical images and from
many
different imaging modalities (e.g., X-ray, MRI, CT, etc.). US provisional
patent
application No. 62/693,371 which is titled "3D Radiomic Platform For Managing
Biomarker Development" and which was filed on July 2, 2018 teaches a system
for
ingesting radiological images into the disclosed system and that reference is
incorporated herein in its entirety by reference.
[00259] Referring again to Fig. 11c, a therapy matching engine 1358 may
match therapies based on the information stored in database 160. In one
example,
the therapy matching engine 1358 matches therapies at the gene level and uses
variant-level information to rank the therapies within a case. For each
variant in a
case, the therapy matching engine 1358 retrieves therapies matching a variant
gene
from an actionability database 1350. The actionability database 1350 may store
a
variety of information for different kinds of variants, such as somatic
functional,
somatic positional, germline functional, germline positional, along with
therapies
associated with SNVs and indels.
[00260] Therapy matching engine 1358 may rank therapies for each gene
based on one or more factors. For instance, the therapy matching engine may
rank
the therapies based on whether the patient disease (such as pancreatic cancer)
matches the disease type associated with the therapy evidence, whether the
patient
variant matches the evidence, and the evidence level for the therapy. For
CNVs, the
therapy matching engine may automatically determine that the patient variant
matches the evidence. For SNVs or indels, the therapy matching engine may
evaluate whether the therapy data came from a functional input or a positional
input.
For positional SNV/indels, if a variant value falls within the range of the
variant locus
start and variant locus end associated with the evidence, the therapy matching
engine may determine that the patient variant matches the evidence. The
variant
locus start and variant locus end may reflect those locations of the variant
in the
protein product (an amino acid sequence position).
[00261] For functional SNV/indels, if a variant mechanism matches the
mechanism associate with the evidence, the therapy matching engine may
determine that the patient variant matches the evidence. Therapies may then be
ranked by evidence level. The first level may be "consensus" evidence
determined
by the medical community, such as medical practice guidelines. The next level
may
be "clinical research" evidence, such as evidence from a clinical trial or
other human
subject research that a therapy is effective. The next level may be "case
study"
evidence, such as evidence from a case study published in a medical journal.
The
-57-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
next level may be "preclinical" evidence, such as evidence from animal studies
or in
vitro studies. Ultimately, pdf or other format reports 1368 are generated for
consumption.
[00262] While a set of data sources and types are described above, it
should
be appreciated that many other data sets that may be meaningful from a
research or
treatment planning perspective are contemplated and may be accommodated in the
present system to further enhance research and treatment planning
capabilities.
[00263] Referring now to Fig. 3a, a schematic is shown that represents an
exemplary data platform 364 that is consistent with at least some aspects of
the
present disclosure. The exemplary platform shows data, information and samples
as
they exist throughout a system where different system processes and functions
are
controlled by different entities including an overall system provider that
operates both
single tenant and multi-tenant cloud service platforms 368 and 372,
respectively,
partners 366 that provide clinical files as well as tissue samples and related
test
requisition orders as well as other partners 374 that access processed data
and
information stored on the service platforms 368 and 372. Partners 366 provide
secure clinical files 375 via a file transfer to the single tenant cloud
platform 368 and
are stored as unstructured and identified files in the lake database. Those
files are
abstracted and shaped as described above to generate normalized structured
clinical data that is stored in a single tenant data vault as well as in a
multi-tenant
data vault 388. The data from the vault is then de-identified and stored in a
de-
identified clinical data database which is accessible to authorized partners
374 via
system interfaces 383 and applications 381 as described herein.
[00264] Referring still to Fig. 3a, partners 366 also provide tissue
samples and
test requisition orders that drive next generation sequencing lab activity at
385 to
generate the bioinformatics pipeline 386 which is stored in both a molecular
data
lake database 389 and the multi-tenant data vault 388. The data in vault 388
is de-
identified and stored in an aggregate de-identified clinical data database 390
where it
is accessible to authorized partners via system interfaces 393 and
applications 382
as described herein. In addition, the molecular lake data 389 and the de-
identified
single tenant files 380 are accessible to other authorized partners via other
interfaces 384.
[00265] IV. User Interfaces
[00266] Referring again to Fig. 3, the disclosed system 100 is accessible
by
many different types of system users that have many different needs and goals
-58-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
including clinical physicians 10 as well as provider specialists like data
abstractors
20, lab, modeling and radiology specialists 30, partner researchers 40,
provider
researchers 50 and dataset sales specialists 60, among others. Because each
user
type performs different activities aimed at achieving different goals, the
application
suites 188, 192 and associated user interfaces employed by each user type will
typically be at least somewhat if not very different. For instance, a
physician's
application suite may include 9 separate application programs that are
designed to
optimally support many oncological treatment consideration and planning
processes
while an abstractor specialist's application suite may include 5 application
programs
that are completely separate from the 9 programs in the physician's suite and
that
are designed to optimally facilitate record abstraction and data structuring
processes.
[00267] In some cases a system user's program suite will be internally
facing
meaning that the user is typically a provider employee and that the suite
generates
data or other information deliverables that are to be consumed within the
system 100
itself. For instance, an abstractor application program for structuring data
from a raw
data set to be consumed by micro-services and other system resources is an
example of an internally facing application program. Other system user
programs or
suites will be externally facing meaning that the user is typically a provider
customer
and that the suite generates data or other information deliverables that are
primarily
for use outside the system. For instance, a physician's application program
suite
that facilitates treatment planning is an example of an externally facing
program
suite.
[00268] Referring now to Figs. 14 through 21, screenshots of an exemplary
physician's user interface that include a series of hyperlinked user interface
views
that are consistent with at least some aspects of the present disclosure are
shown.
The screenshots show one natural progression of information consideration
wherein
each interface is associated with one of the physician's program suite
applications
188. While some of the illustrated screenshots are complete, others are only
partial
and additional screen data would be accessible via either scrolling downward
as well
known in the graphical arts or by selection of a hyperlink within the
presented view
that accesses additional information related to the screenshot that includes
the
selected hyperlink.
[00269] Referring to Fig. 14, once a physician logs onto system 10 via
entry of
a username and password or via some other security protocol, the physician is
either
presented with a patient list screen 1400 or can navigate to that screen. The
patient
list screen 1400 includes a first navigation bar or ribbon that extends along
an upper
-59-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
edge of the view as well as a patient list area 1405 that includes a separate
cell or
field (two labelled 1402 and 1404) for each of the physician's patients for
which the
system 100 stores data. Each patient cell (e.g., 1404) includes basic patient
information including the patient's name, an identification number and a
cancer type
and operates as a hyperlink phrase for accessing applications where the system
loads data for the patient indicated in the cell. The screen 1400 also
includes a
"New Patient" icon 1406 that is selectable to add a new patient to the
physician's
view. The screen 1400 may display all patients of the physician's who have
received
genomic testing. Each patient cell can represent one or more reports created
based
on tissue samples. Physicians can also see in-progress patients along with a
status
indicating an order's progress, such as if the sample has been received. Some
physicians may be provided with an additional section displaying reference
patients.
In these cases, the physician signed into the system 10 is not the patient's
ordering
physician, but has some other reason to access the patient information, such
as
because the the ordering physician indicated he or she should receive a copy
of the
report and be permitted other appropriate access. Certain users of the system
10,
such as administrators, may have access to browse all patients within their
institution.
[00270] Referring again to Fig. 14, upon selecting cell 1404 associated
with a
patient named Dwayne Holder, the system presents the screenshot 1500 shown in
Fig. 15 that includes a second level navigation bar 1502 near the top of the
screen
1500 and a workspace 1504 below bar 1502. Navigation bar 1502 persistently
identifies the patient 1506 associated with the data currently being viewed by
the
physician throughout the screenshots illustrated and also includes a separate
hyperlink text term for each of several system data views or application
programs
that can be selected by the physician. In Fig. 15 the view and applications
options
include an "Overview" option 1508, a "Reports" option 1510, an "Alterations"
option
1512, a "Trials" option 1514, an "Immunotherapy" option 1516, a "Cohort"
option
1518, a "Board" option 1520 and a "Modelling" option 1522. Many other options
will
be added to bar 1502 over time as they are developed. A view or application
currently accessed by the physician is underlined or otherwise visually
distinguished
in bar 1502. For instance, in Fig. 15 the overview icon 1508 is shown
highlighted to
indicate that the information presented in workspace 1504 is associated with
the
overview data view.
[00271] Referring still to Fig. 15, the exemplary overview view includes a
patient care timeline 1509 along a left edge of workspace 1504, high level
patient
-60-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
cancer state information 1550 in a central portion of workspace 1504 and view
selection icons 1540 along a right edge of workspace 1504. Timeline 1509
includes
a set of patient care cells 1570, 1580, etc., each of which corresponds to a
meaningful care related event associated with treatment of the patient's
cancer state.
The cells are vertically stacked with earliest cells in time near the bottom
of the stack
and more recent cells near the top of the stack. Each cell is typically
restricted to
activities or information associated with a specific date and, in addition to
the
associated date, may include any subset of several different information types
including hospital or clinic admission and release dates, medical imaging
descriptors,
procedure descriptors, medication start and end dates, treatment procedure
start
and end descriptors, test descriptors, test or procedure results descriptors
and other
descriptors. This list is exemplary and not intended to be exhaustive. For
instance,
cell 1532 that is dated 12/29/2017 indicates that a lung biopsy occurred as
well as a
brain CT imaging session and an MRI of the patient's abdomen. Information in
the
timeline 1509 may be loaded from the structured data that results from using
the
systems and methods described herein, such as those with reference to Fig. 10.
Information in the timeline 1509 may also include references to genomic
sequencing
tests ordered for a patient.
[00272] Referring still to Fig. 15, in addition to including the patient
care cell
stack, the care timeline 1509 includes a vertical activity icon progression
1534 that
extends along the left edge of the cell stack. The activity icons in
progression 1534
are horizontally aligned with associated textual descriptions of care events
in the cell
stack. Each activity icon is designed to glanceably indicate an activity type
so that a
physician can quickly identify activities of specific types within the stacked
cells by
simply viewing the icons and associated stack event descriptors. For instance,
exemplary activity icons include a gene panel publication icon 1552, a
medication
start/stop icon 1554, a facility admit/release icon 1556 and an imaging
session icon
1558. Other icons corresponding to surgery, detected patient medical
conditions,
and other procedures or important medical events are contemplated.
[00273] Referring still to Fig. 15, in at least some cases detailed data
related to
a care event will be further accessible by selecting one of the activity icons
along the
left of the cells or events in a cell to hyperlink to the additional
information. For
instance, the "CT:Brain" text at 1662 may be selectable to link to a CT image
viewer
to view CT images of the patient's brain that correspond to the event. Other
links are
contemplated.
-61-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
[00274] Referring again to Fig. 15, general cancer state and patient
information
at 1550 includes diagnosis, stage, patient date of birth and gender
information 1530
as well as an anatomical image that shows a representation of a tumor within a
body
that is generally consistent with the patient's cancer state. In some cases
the tumor
representation is just representative of the patient's condition as opposed to
directly
tied to actual tumor images while in other cases the tumor representation is
derived
from actual medical images of the patient's tumor.
[00275] Referring again to Fig. 15, the patient body image 1550 may be
overlaid with structured contours 1560 from the patient's radiology imaging.
Represented structures may include primary or metastatic lesions, organs,
edema,
etc. A physician may click each structured contour to obtain an additional
level of
detail of information. Clicking the structured contour may isolate it visually
for the
physician. In the case of a tumor contour, the additional level of detail may
include
supporting information such as tumor volume, longest 3D diameter, or other
features. Certain radiomic features that may be presented to the physician are
described in further detail in, for instance, U.S. Provisional Patent
Application No.
62/693,371, titled 3D Radiomic Platform for Imaging Biomarker Development,
which
has been incorporated herein by reference in its entirety.
[00276] From this detailed view, the physician may further drill down to an
additional, microscopic level of detail. Here, a patient's histopathology
results may
be displayed. Clinical interpretations are shown, where available from an
issued
report. The microscopic detail may also display thumbnail images of microscope
slides of a patient's specimens.
[00277] View selection icons 1540 include a set of icons that allow the
physician to select different views of the patient's cancer condition and are
progressively more granular. To this end, the exemplary view icons include a
body
view icon 1572 corresponding to the body view shown in Fig. 15, a medical
imaging
view icon 1574 for accessing medical X-ray, CT, MRI and other images, a
cellular
view icon 1576 that shows cellular level images and genomic sequencing data
icon
1578 for accessing genomic data views.
[00278] Referring again to Fig. 15, to access specific issued reports
associated
with the patient the physician selects reports icon 1510 to access a reports
screen
1600 shown in Fig. 16. Reports screen 1600 shows the reports icon 1510
highlighted
to help orient the physician and includes a report list indicating all reports
stored in
the system that are associated with the patient. In the exemplary reports
view, each
report is represented in the list by a reduced size image of the first page of
the report
-62-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
and with a general report description field near the bottom of the image. For
exemplary report images are shown at 1602 and 1604 and a general report
description of the report associated with image 1602 is provided at 1606
indicating
report type, date and other characterizing information.
[00279] The physician can select one of the report images to access the
full
report. For instance, if the physician selects image icon 1602, the screenshot
1700
shown in Fig. 17 is presented that splits the display screen into a report
list section
1702 along the left edge of the screen and an enlarged report section 1704
that
covers about the right two thirds of the screen where the selected report is
presented
in a larger format for viewing. The report presents clinically significant
information
and may take many different forms. Each report is listed again in section 1702
as a
reduced size hyper linkable image as shown at 1602 and 1604 where the
currently
selected report 1602 is highlighted or otherwise visually distinguished. The
physician can select a PDF icon 1708 to download a copy of the report to the
physician's computer.
[00280] A patient may have multiple reports for each specimen or specimen
set
sequenced. Reports may include DNA sequencing reports, IHC staining reports,
RNA expression level reports, organoid growth reports, imaging and/or
radiology
reports, etc. Each report may contain results of sequencing of the patient's
tumor
tissue and, where available the normal tissue as well. Normal tissue can be
used to
identify which alterations, if any, are inherited versus those that the tumor
uniquely
acquired. Such differentiation often has therapeutic implications.
[00281] Fig. 17a shows an exemplary first page of a report screenshot
indicating the results of one RNA sequencing process. Profiling of whole RNA
transcriptome provides molecular information that is complementary to DNA
sequencing and can be clinically important to physicians. For example, RNA
sequencing can assist in clinically validated unbiased translocation
detection.
Overexpression and underexpression of certain genes may be presented to the
physician as a result of RNA sequencing. Likewise, treatment implications may
be
provided to the physician which the physician may take into consideration when
determining the best type of treatment for a patient. The physician may decide
to
verify results, for instance, through an orthogonal assay methodology, before
using
the results in clinical decision making.
[00282] To examine information related to a patient's genomic tumor
alterations
and possible treatment options, the physician selects alterations icon 1512 to
access
screen 1800 shown in Fig. 18. Screen 1800 includes an approved therapies list
-63-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
1802 and a pertinent genes list 1804. The therapies list 1802 includes a list
of genes
for which variants have been identified and for each gene in the list, the
associated
variant, how the variant is indicated and other information including details
regarding
considerations corresponding to the associated therapy option. Other screens
for
considering alterations are contemplated to enable a physician to consider
many
aspects of treatment efficacy. Additional details may be provided to add
context to
alterations, such as gene descriptions, explanation of mutation effect, and
variant
allelic fraction. Alterations may be reported by category, ranging from highly
relevant
genes to variants of unknown significance.
[00283] Selecting an alteration may take the physician to an additional
view,
shown at Figs. 18a and 18b (showing different scrolled sections of one view in
the
two figures), where the physician can delve deeper into the alteration's
effect, with
supporting data visualizations. Germline alterations associated with diseases
may
be reported as incidental findings. In Fig. 18a, approved therapies are listed
with
relevant related information including a gene and variant indicator along with
hyperlinks to evidence associated with the therapy and details about each of
the
therapies.
[00284] The physician application suite also provides tools to help the
physician
identify and consider clinical trials that may be related to treatment options
for his
patient. To access the trials tools, the physician selects trials icon 1514 to
access
the screen (not shown) that lists all clinical trials that may be of any
interest to the
physician given patent cancer state characteristics. For instance, for a
patient
suffering from pancreatic cancer, the list may indicate 12 different trials
occurring
within the United States. In some cases the trials may be arranged according
to
likely most relevant given detailed cancer state factors for the specific
patient. The
physician can select one of the clinical trials from the list to access a
screen 1900
like the one shown in Fig. 19. Screen 1900 includes a map 1904 with markers
(three
labelled 1906, 1908 and 1910) at map locations corresponding to institutions
are
participating in the selected trial as well as a general description 1920 of
the trial.
Screen 1900 also provides a set of filtering tools 1930 in the form of pull
down
menus the physician can use to narrow down trial options by different factors
including distance from the patient's location, trial phase (e.g., not yet
initiated,
progressing, wrapping up, etc.), and other factors. Here, the idea is that the
physician can explore trial options for specific patient cancer states quickly
by
focusing consideration on the most relevant and convenient trial options for
specific
patients.
-64-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
[00285] The physician application suite provides tools for the physician to
consider different immunotherapies that are accessible by selecting
immunotherapy
icon 1516 from the navigation bar. When icon 1516 is selected, an exemplary
immunotherapy screenshot 2000 shown in Fig. 20 is presented. Screenshot 2000
includes a menu of immunotherapy interface options 2002 extending vertically
along
a left area of the screen and a detailed information area 2004 to the right of
the
options 2002. In at least some cases the immunotherapy options 2002 will
include a
summary option, a tumor mutation burden option, a microsatellite instability
status
option, an immune resistance risk option and an immune infiltration option
where
each option is selectable to access specific immunotherapy data related to the
patient's case. lmmunotherapy options 2002 may provide the physician with an
indication that an immunotherapy, such as an FDA approved immunotherapy, may
be appropriate to prescribe the patient. Examples may include dendritic cell
therapies, CAR-T cell therapies, antibody therapies, cytokine therapies,
combination
immunotherapies, adoptive t-cell therapies, anti-0D47 therapies, anti-GD2
therapies,
immune checkpoint inhibitors, oncolytic viruses, polysaccharides, or
neoantigens,
among others. Area 2004 shows summary information presented when the
summary option is selected from the option list 2002. When other list options
are
selected, related information is used to populate area 2004 with additional
related
information.
[00286] Referring to Fig. 21, the cohort option 1518 can be selected to
access
an analytical tool that enables the physician to explore prior treatment
responses of
patients that have the same type of cancer as the patient that the physician
is
planning treatment for in light of similarities in molecular data between the
patients.
To this end, once genomic sequencing has been completed for each patient in a
set
of patients, molecular similarities can be identified between any patients and
used as
a distance plotting factor on a chart 2110. In Fig. 21, the screen 2100
includes a
graph at 2110, filter options at 2120, some view options 2140, graph
information at
2150 and additional treatment efficacy bar graphs at 2160.
[00287] Referring still to Fig. 21, the illustrated graph presents a tumor
associated with the patient for which planning is progressing at a center
location as a
star and other patient tumors of a similar type (e.g., pancreatic) at
different radial
distances from the central tumor where molecular similarity is based on
distance
from the central location so that tumors more similar to the central tumor are
near the
center and tumors other than the central tumor are located in proximity to one
another based on their respective similarity. Angular displacements between
the
-65-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
other tumors represented indicate dissimilarity or similarity between any two
tumors
where a greater angular distance between two tumors indicates greater
dissimilarity.
Except for the central tumor (e.g., indicated via the star), each of the other
tumors is
color coded to indicate treatment efficacy. For instance, a green dot may
represent
a tumor that completely responded to treatment, a yellow dot may indicate a
tumor
that responded minimally while a red dot indicates a tumor that did not
respond. An
efficacy legend at 2130 is provided that associates tumor colors with
efficacies "e.g.,
"Complete Response", "Partial Response", etc.). the physician can select
different
options to show in the graph including response, adverse reaction, or both
using
icons at 2140.
[00288] Referring still to Fig. 21, an initial view 2110 may include all
patient
tumors that are of the same general type as the central tumor presented on the
graph 2110, regardless of other cancer state factors. In Fig. 21, a number "n"
is
equal to 975 indicating that 975 tumors and associated patients are
represented on
graph 2110. Filters at 2120 can be used by the physician to select different
cancer
state filter factors to reduce the n count to include patients that have other
factors in
common with the patient associated with the central tumor. For instance,
patient sex
or age or tumor mutations or any factor combination supported by the system
may
be used to filter n down to a smaller number where multiple factors are common
among associated patients.
[00289] Referring again to Fig. 21, the efficacy bar graphs 2160 present
efficacy data for different treatment types. To this end, screen area 2160
presents a
list of medications or combinations thereof that have been used in the past to
treat
the tumors represented in graph 2110. A separate bar graph is provided for
each of
the treatment medications or combinations where each bar graph includes
different
length color coded sub-sections that show efficacy percentages. For instance,
for
Germcitabine, the bar graph 2170 may include a green section that extends 11%
of
the length of the total bar graph and a blue section that extends 5% of the
length of
the total bar graph to indicate that 11% of patients treated with Germcitabine
experienced a complete response while 5% experienced only a partial response.
Other color coded sections of bar 2170 would indicate other efficacies. The
illustrated list only includes two treatment regimens but in most cases the
list would
be much longer and each list regimen would include its own efficacy bar graph.
[00290] IV. Automated Cancer State-Treatment-Efficacy Insights
Across Patient Populations
-66-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
[00291] Referring again to Fig. 21, the cohort tool shown allows a
physician to
select different cancer state filters 2120 to be applied to the system
database
thereby changing the set of patients for which the system presents treatment
efficacy
data to help the physician explore effects of different factors on efficacy
which is
intended to lead to new treatment insights like factor-treatment-efficacy
relationships.
While powerful, this physician driven system is only as good as the physician
that
operates it and in many cases cancer state-treatment-efficacy relationships
simply
will not even be considered by a physician if clinically relevant state
factors are not
selected via the filter tools. While a physician could try every filter
combination
possible, time restraints would prohibit such an effort. In addition, while a
large
number of filter options could be added to the filter tools 2120 in Fig. 21,
it would be
impractical to support all state factors as filter options so that some filter
combinations simply could not be considered.
[00292] To further the pursuit of new cancer state-treatment-efficacy
exploration and research, in at least some embodiments it is contemplated that
system processors may be programmed to continually and automatically perform
efficacy studies on data sets in an attempt to identify statistically
meaningful state
factor-treatment-efficacy insights. These insights can be confirmed by
researchers
or physicians and used thereafter to suggest treatments to physicians for
specific
cancer states.
[00293] V. Exemplary System Techniques And Results
[00294] The systems and methods described above may be used with a variety
of sequencing panels. One exemplary panel, the 595 gene xT panel referred to
above (See again the Flg. 27 series of figures), is focused on actionable
mutations.
Hereafter we present a description of various techniques and associated
results that
are consistent with aspects of the present disclosure in the context of an
exemplary
xT panel.
[00295] Techniques and results include the following. SNVs (single
nucleotide
variants), indels, and CNVs (copy number variants) were detected in all 595
genes.
Genomic rearrangements were detected on a 21 gene subset by next generation
DNA sequencing, with other genomic rearrangements detected by next generation
RNA sequencing (RNA Seq). The panel also indicated MSI (microsatellite
instability
status) and TMB (tumor mutational burden). DNA tumor coverage was provided at
500x read sequencing depth. Full transcriptome was also provided by RNA
-67-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
sequencing, with unbiased gene rearrangement detection from fusion transcripts
and
expression changes, sequenced at 50 million reads.
[00296] In addition to reporting on somatic variants, when a normal sample
is
provided, the assay permits reporting of germline incidental findings on a
limited set
of variants within genes selected based on recommendations from the American
College of Medical Genetics (ACMG) and published literature on inherited
cancer
syndromes.
[00297] Mutation Spectrum Analysis For Exemplary 500 Patient xT Group
[00298] Subsequent to selection, patients were binned by pre-specified
cancer
type and filtered for only those variants being classified as therapeutically
relevant.
The gene set was then filtered for only those genes having greater than 5
variants
across the entire group so as to select for recurrently mutated genes. Having
collated this set, patients were clustered by mutational similarity across
SNPs,
indels, amplifications, and homozygous deletions. Subsequently, mutation
prevalence data for the MSKCC IMPACT data were extracted from MSKCC
Cbioportal (http://www.cbioportal.ord/study?id=msk impact 2017#summary) in
order
to compare the xT assay variant calls against publicly available variant data
for solid
tumors. After selecting for only those genes on both panels, variants with a
minimum
of 2.5% prevalence within their respective group were plotted.
[00299] Detection Of Gene Rearrangements From DNA By The xT Assay
[00300] Gene rearrangements were detected and analyzed via separate
parallel workflows optimized for the detection of structural alterations
developed in
the JANE workflow language. Following de-multiplexing, tumor FASTQ files were
aligned against the human reference genome using BWA (Li et al., 2009). Reads
were sorted and duplicates were marked with SAMBlaster (Faust et al., 2014).
Utilizing this process, discordant and split reads are further identified and
separated.
These data were then read into LUMPY (Layer et al., 2014) for structural
variant
detection. A VCF was generated and then parsed by a fusion VCF parser and the
data was pushed to a Bioinformatics database. Structural alterations were then
grouped by type, recurrence, and presence within the database and displayed
through a quality control application. Known and previously known fusions were
highlighted by the application and selected by a variant science team for
loading into
-68-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
a patient report.
[00301] Detection Of Gene Rearrangements From RNA By The xT Assay
[00302] Gene rearrangements in RNA were analyzed via a separate workflow
that quantitated gene level expression as well as chimeric transcripts via non-
canonical exon-exon junctions mapped via split or discordant read pairs. In
brief,
RNA-sequencing data was aligned to GRCh38 using STAR (Dobin et al., 2009) and
expression quantitation per gene was computed via FeatureCounts (Liao et al.,
2014). Subsequent to expression quantitation, reads were mapped across exon-
exon boundaries to un-annotated splice junctions and evidence was computed for
potential chimeric gene products. If sufficient evidence was present for the
chimeric
transcript, a rearrangement was called as detected.
[00303] Gene Expression Data Collection
[00304] RNA sequencing data was generated from FFPE tumor samples using
an exome-capture based RNA seq protocol. Raw RNA seq reads were aligned
using CRISP and gene expression was quantified via the RNA bioinformatics
pipeline. One RNA bioinformatics pipeline is now described. Tissues with
highest
tumor content for each patient may be disrupted by 5mm beads on a Tissuelyser
II
(Qiagen). Tumor genomic DNA and total RNA may be purified from the same sample
using the AllPrep DNA/RNA/miRNA kit (Qiagen). Matched normal genomic DNA
from blood, buccal swab or saliva may be isolated using the DNeasy Blood &
Tissue
Kit (Qiagen). RNA integrity may be measured on an Agilent 2100 Bioanalyzer
using
RNA Nano reagents (Agilent Technologies). RNA sequencing may be performed
either by poly(A)+ transcriptome or exome-capture transcriptome platform. Both
poly(A)+ and capture transcriptome libraries may be prepared using 1-2 ug of
total
RNA. Poly(A)+ RNA may be isolated using Sera-Mag oligo(dT) beads (Thermo
Scientific) and fragmented with the Ambion Fragmentation Reagents kit (Ambion,
Austin, TX). cDNA synthesis, end-repair, A-base addition, and ligation of the
Illumine
index adapters may be performed according to IIlumina's TruSeq RNA protocol
(Illumine). Libraries may be size-selected on 3% agarose gel. Recovered
fragments
may be enriched by FOR using Phusion DNA polymerase (New England Biolabs)
and purified using AMPure XP beads (Beckman Coulter). Capture transcriptomes
may be prepared as above without the up-front mRNA selection and captured by
-69-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
Agilent SureSelect Human all exon v4 probes following the manufacturer's
protocol.
Library quality may be measured on an Agilent 2100 Bioanalyzer for product
size
and concentration. Paired-end libraries may be sequenced by the Illumine HiSeq
2000 or HiSeq 2500 (2x100 nucleotide read length), with sequence coverage to
40-75M paired reads. Reads that passed the chastity filter of Illumine
BaseCall
software may be used for subsequent analysis. Further details of the pipeline
raw
read counts may be normalized to correct for GC content and gene length using
full
quantile normalization and adjusted for sequencing depth via the size factor
method
(see Robinson, D. R. et al. Integrative clinical genomics of metastatic
cancer. Nature
548, 297-303 (2017)). Normalized gene expression data was log, base 10,
transformed and used for all subsequent analyses.
[00305] Reference Database
[00306] Gene expression data generated (as previously described) was
combined with publicly available gene expression data for cancer samples and
normal tissue samples to create a Reference Database. For this analysis, we
specifically include data from The Cancer Genome Atlas (TOGA) Project and
Genotype-Tissue Expression (GTEx) project. Raw data from these publically
available datasets were downloaded via the GDC or SRA and processed via an
RNAseq pipeline (described above). In total 4,865 TOGA samples and 6,541 GTEx
samples were processed and included as part of the larger Reference Database
for
this analysis. After processing, these datasets were corrected to account for
batch
effect differences between sequencing protocols across institutions (i.e. TOGA
& and
the Reference Database). For example, TOGA and GTEx both sequenced fresh,
frozen tissue using a standard polyA capture based protocol.
[00307] Gene Expression Calling
[00308] For each patient, the expression of key genes was compared to the
Reference Database to determine overexpression or underexpression. 42 genes
for
over- or under-expression based on the specific cancer type of the sample were
evaluated. The list of genes evaluated can vary based on expression calls,
cancer
type, and time of sample collection. In order to make an expression call, the
percentile of expression of the new patient was calculated relative to all
cancer
samples in the database, all normal samples in the database, matched cancer
-70-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
samples, and matched normal samples. For example, a breast cancer patient's
tumor expression was compared to all cancer samples, all normal samples, all
breast cancer samples, and all breast normal tissue samples within the
Reference
Database. Based on these percentiles criteria specific to each gene and cancer
type
to determine overexpression was identified.
[00309] t-Distributed Stochastic Neighbor Embedding (t-SNE) RNA
analysis
[00310] The t-SNE plot was generated using the Rtsne package in R [R
version
3.4.4 and Rtsne version 0.13] based on principal components analysis of all
samples
(N = 482) across all genes (N = 17,869). A perplexity parameter of 30 and
theta
parameter of 0.3 was used for this analysis.
[00311] Cancer Type Prediction
[00312] A random forest model was used to generate cancer type predictions.
The model was trained on 804 samples and 4,526 TOGA samples across cancer
types from the Reference Database. For the purposes of this analysis,
hematological malignancies were excluded. Both datasets were sampled equally
during the construction of the model to account for differences in the size of
the
training data. The random forest model was calculated using the Ranger package
in
R [R version 3.4.4 and ranger 0.9.0]. Model accuracy was calculated within the
training dataset using a leave-one-out approach. Based on this data, the
overall
classification accuracy was 81%.
[00313] Tumor Mutational Burden (TMB)
[00314] TMB was calculated by determining the dividend of the number of non-
synonymous mutations divided by the megabase size of the panel (2.4 MB). All
non-
silent somatic coding mutations, including missense, indel, and stop loss
variants,
with coverage greater than 100X and an allelic fraction greater than 5% were
included in the number of non-synonymous mutations.
[00315] Human Leukocyte Antigen (H LA) Class I Typing
[00316] HLA class I typing for each patient was performed using Optitype on
DNA sequencing (Szolek 2014). Normal samples were used as the default
reference
-71-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
for matched tumor-normal samples. Tumor sample-determined HLA type was used
in cases where the normal sample did not meet internal HLA coverage thresholds
or
the sample was run as tumor-only.
[00317] Neoantigen Prediction
[00318] Neoantigen prediction was performed on all non-silent mutations
identified by the xT pipeline. For each mutation, the binding affinities for
all possible
8-11aa peptides containing that mutation were predicted using MHCflurry
(Rubinsteyn 2016). For alleles where there was insufficient training data to
generate
an allele-specific MHCflurry model, binding affinities were predicted for the
nearest
neighbor HLA allele as assessed by amino acid homology. A mutation was
determined to be antigenic if any resulting peptide was predicted to bind to
any of the
patient's HLA alleles using a 500nM affinity threshold. RNA support was
calculated
for each variant using varlens (https://dithub.com/openvax/varlens). Predicted
neoantigens were determined to have RNA support if at least one read
supporting
the variant allele could be detected in the RNA-seq data.
[00319] Microsatellite Instability (MSI) Status
[00320] The exemplary xT panel includes probes for 43 microsatellites that
are
frequently unstable in tumors with mismatch repair deficiencies. The MSI
classification algorithm uses reads mapping to those regions to classify
tumors into
three categories: microsatellite instability-high (MSI-H), microsatellite
stable (MSS),
or microsatellite equivocal (MSE). This assay can be performed with paired
tumor-
normal samples or tumor-only samples.
[00321] MSI testing in paired mode begins with identifying accurately
mapped
reads to the microsatellite loci. To be a microsatellite locus mapping read,
the read
must be mapped to the microsatellite locus during the alignment step of the
exemplary xT bioinformatics pipeline and also contain the 5 base pairs in both
the
front and rear flank of the microsatellite, with any number of expected
repeating units
in between. All the loci with sufficient coverage are tested for instability,
as
measured by changes in the distribution of the number of repeat units in the
tumor
reads compared to the normal reads using the Kolmogorov-Smimov test. If p <=
0.05, the locus is considered unstable. The proportion of unstable loci is fed
into a
logistic regression classifier trained on samples from the TOGA colorectal and
-72-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
endometrial groups that have clinically determined MSI statuses.
[00322] MSI testing in unpaired mode also begins with identifying
accurately
mapped reads to the microsatellite loci, using the same requirements as
described
above. The mean number of repeat units and the variance of the number of
repeat
units is calculated for each microsatellite locus. A vector containing the
mean and
variance data for each microsatellite locus is put into a support vector
machine
classification algorithm trained on samples from the TOGA colorectal and
endometrial groups that have clinically determined MSI statuses.
[00323] Both algorithms return the probability of the patient being MSI-H,
which
is then translated into a MSI status of MSS, MSE, or MSI-H.
[00324] Cytolytic Index (CYT)
[00325] CYT was calculated as the geometric mean of the normalized RNA
counts of granzyme A (GZMA) and perforin (PRF1) (Rooney, M. S., Shukla, S. A.,
Wu, C. J., Getz, G. & Hacohen, N. Molecular and Genetic Properties of Tumors
Associated with Local Immune Cytolytic Activity. Cell 160, 48-61 (2015)).
[00326] Interferon Gamma Gene Signature Score
[00327] Twenty-eight interferon gamma (IFNG) pathway-related genes (Ayers
M., J Olin Invest 2017) were used as the basis for an IFNG gene. Hierarchical
clustering was performed based on Euclidean distance using the R package
ComplexHeatmap (version 1.17.1) and the heatmap was annotated with PD-L1
positive IHC staining, TMB-high, or MSI-high status. IFNG score was calculated
using the arithmetic mean of the 28 genes.
[00328] Knowledge Database (KDB)
[00329] In order to determine therapeutic actionability for sequenced
patients, a
KDB with structured data regarding drug/gene interactions and precision
medicine
assertions is maintained. The KDB of therapeutic and prognostic evidence is
compiled from a combination of external sources (including but not exclusive
to
NOON, CIViC{28138153}, and DGIdb{28356508}) and from constant annotation by
provider experts. Clinical actionability entries in the KDB are structured by
both the
disease in which the evidence applies, and by the level of evidence.
Therapeutic
actionability entries are binned into Tiers of somatic evidence by patient
disease
matches as laid out by the ASCO/AMP/CAP working group {27993330}. Briefly,
Tier
-73-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
I Level A (IA) evidence are biomarkers that follow consensus guidelines and
match
disease type. Tier I Level B (IB) evidence are biomarkers that follow clinical
research
and match disease type. Tier II Level C (IIC) evidence biomarkers follow the
off-label
use of consensus guidelines and Tier II Level D (IID) evidence biomarkers
follow
clinical research or case reports. Tier III evidence are variants with no
therapies.
Patients are then matched to actionability entries by gene, specific variant,
patient
disease, and level of evidence.
[00330] Alteration Classification, Prioritization, And Reporting
[00331] Somatic alterations are interpreted based on a collection of
internally
weighted criteria that are composed of knowledge of known evolutionary models,
functional data, clinical data, hotspot regions within genes, internal and
external
somatic databases, primary literature, and other features of somatic drivers
{24768039}{29218886}. The criteria are features of a derived heuristic
algorithm that
buckets them into one of four categories (Pathogenic/ VUS/ Benign/
Reportable).
Pathogenic variants are typically defined as driver events or tumor prognostic
signals. Benign variants are defined as those alterations that have evidence
indicating a neutral state in the population and are removed from reporting.
VUS
variants are variants of unknown significance and are seen as passenger
events.
Reportable variants are those that could be seen as diagnostic, offer
therapeutic
guidance or are associated with disease but are not key driver events. Gene
amplifications, deletions and translocations were reported based on the
features of
known gene fusions, relevant breakpoints, biological relevance and therapeutic
actionability.
[00332] For the tumor-only analysis germline variants were computationally
identified and removed using by an internal algorithm that takes copy number,
tumor
purity, and sequencing depth into account. There was further filtering on
observed
frequency in a population database (positions with AF>1% ExAC non-TOGA group).
The algorithm was purposely tuned to be conservative when calling germline
variants in therapeutic genes minimizing removal of true somatic pathogenic
alterations that occur within the general population. Alterations observed in
an
internal pool of 50 unmatched normal samples were also removed. The remaining
variants were analyzed as somatic at a VAF>=5 /0 and Coverage>=90. Using
normal tissue, true germline variants were able to be flagged and somatic
analysis
-74-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
contamination was evaluated. The Tumor/Normal variants were also set at the
Tumor-only VAF/Coverage thresholds for analysis.
[00333] Clinical trial matching occurs through a process of associating a
patient's actionable variants and clinical data to a curated database of
clinical trials.
Clinical trials are verified as open and recruiting patients before report
generation.
[00334] Germline Pathogenic And Variants of Unknown Significance (VUS)
[00335] Alterations identified in the Tumor/Normal match samples are
reported
as secondary findings for consenting patients. These are a subset of genes
recommended by the ACMG (Richards, S. et al. Standards and guidelines for the
interpretation of sequence variants: a joint consensus recommendation of the
American
College of Medical Genetics and Genomics and the Association for Molecular
Pathology.
Genet. Med. 17, 405-24 (2015)) and genes associated with cancer predisposition
or
drug resistance.
[00336] In an example patient group analysis, a group of 500 cancer
patients
was selected where each patient had undergone clinical tumor and germline
matched sequencing using the panel of genes at Figs. 27a, 27b, 27c1, 27c2, and
27d (known herein as the "xT" assay). In order to be eligible for inclusion in
the
group, each case was required to have complete data elements for tumor-normal
matched DNA sequencing, RNA sequencing, clinical data, and therapeutic data.
Subsequent to filtering for eligibility, a set of patients was randomly
sampled via a
pseudo-random number generator. Patients were divided among seven broad
cancer categories including tumors from brain (50 patients), breast (50
patients),
colorectal (51 patients), lung (49 patients), ovarian and endometrial (99
patients),
pancreas (50 patients), and prostate (52 patients). Additionally, 48 tumors
from a
combined set of rare malignancies and 51 tumors of unknown origin were
included
for analyses for a total of nine broad cancer categories. These patients were
collated together as a single group and used for subsequent group analyses.
[00337] The mutational spectra for the studied group was compared with
broad
patterns of genomic alterations observed in large-scale studies across major
cancer
types. First, data from all 500 patients was plotted by gene, mutation type,
and
cancer type, and then clustered by mutational similarity (Fig. 29). The most
commonly mutated genes included well-known driver mutations, including
mutations
in more than 5% of all cases in the group for TP53, KRAS, PIK3CA, CDKN2A,
PTEN, ARID IA, APC, ERBB2, EGFR, IDH1, and CDKN2B. These genes are known
hallmarks of cancer and commonly found in solid tumors. Of these genes,
-75-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
CDKN2A, CDKN2B, and PTEN were most commonly found to be homozygously
deleted, indicating loss-of-function mutations likely coinciding with loss of
heterozygosity. These data demonstrate expected molecular signatures commonly
seen in clinical solid tumor samples.
[00338] Previous pan-cancer mutation analyses have established mutational
spectra within and across tumor types, and provide context to which the study
group
sequencing data may be compared. In Fig. 30, the study group results were
compared to a previously published pan-cancer analysis using the Memorial
Sloan
Kettering Cancer Center (MSKCC) IMPACT panel (Zehir, A. et al. Mutational
landscape of metastatic cancer revealed from prospective clinical sequencing
of
10,000 patients. Nat. Med. 23, 703-713 (2017)). In both datasets, we observed
the
same commonly mutated genes, including TP53, KRAS, APC and PIK3CA. These
genes were observed at similar relative frequencies compared to the MSKCC
group.
These results indicate the mutation spectra within the study group is
representative
of the broader population of tumors that have been sequenced in large-scale
studies.
[00339] Because both tumor and germline samples were sequenced in the
group, the effect of germ line sequencing on the accuracy of somatic mutation
identification could be examined. Fiftyone cases were randomly selected from
the
study group with a range of tumor mutational burden profiles. Their variants
were re-
evaluated using a tumor-only analytical pipeline. After filtering the dataset
using a
population database and focusing on coding variants from the 51 samples, 2,544
variants were identified that had a false positive rate of 12.5%. By further
filtering
with an internally developed list of technical artifacts (e.g., artifacts from
DNA
sequencing process), an internal pool of matched normal samples, and
classification
criteria, 74% of the false somatic variants (false positive rate of 2.3%) were
removed
while still retaining all true somatic alterations.
[00340] To further characterize the tumors in the study group, RNA
expression
profiles for patients in the group were examined. Similar tumor types tend to
have
similar expression profiles (Fig. 31). On average, samples within a cancer
type as
determined by pathologic diagnosis showed a higher pairwise correlation within
the
corresponding TCGA cancer group compared to between TCGA cancer groups (p-
values=10-6-10-16). This clustering of samples by TCGA cancer group is
observed in
the t-SNE plot shown in Fig. 32. For some tumor types, such as prostate
cancer,
metastatic samples cluster very closely to non-metastatic tumor samples.
However
other cancer types, most notably pancreatic cancer and colorectal cancer, form
a
-76-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
distinct metastatic tumor cluster that also contains breast tumors and tumors
of
unknown origin. This effect is likely due to the effect of the background
tissue on the
expression profile of the tumor sample. For example, metastatic samples from
the
liver frequently, but not always, cluster together. This effect can also
depend on the
level of tumor purity within the sample.
[00341] Given the high-dimensionality of the data, we sought to determine
whether we could predict cancer types using gene expression data. We developed
a
random forest cancer type predictor using a combination of publically
available
TOGA expression data and expression data generated at Tempus Labs. TOGA
cancer type predictions compared to the xT group samples are shown in Fig. 32.
For example, 100% of breast cancer samples were correctly classified.
Interestingly,
using this method we are able to accurately classify these tumors even when
the
samples are biopsied from metastatic sites.
[00342] Additionally, it is notable that some of the "misclassified"
samples may
actually represent biologically and pathologically relevant classifications.
For
example, of the 50 brain tumors in our dataset, 48 (96%) were classified as
gliomas,
while 2 were classified as sarcomas.
[00343] One of these tumors carries a histopathologic diagnosis of
"solitary
fibrous tumor, hemangiopericytoma type, WHO grade III", which is indeed a
sarcoma. The other was diagnosed as "glioblastoma, WHO grade IV (gliosarcoma),
with smooth muscle and epithelial differentiation". The immunohistochemical
profile
is GFAP negative with desmin and SMA focally positive, supporting the
diagnosis of
gliosarcoma. It can be argued that the algorithm classified this tumor
correctly by
grouping it with sarcomas, and in fact, gliosarcomas carry a worse prognosis
and
have the ability to metastasize, differentiating them clinically from
traditional
glioblastoma.
[00344] Similarly, a case with a histopathologic diagnosis favoring
carcinosarcoma was identified by the model as SARC in a patient with a history
of
prostate cancer presenting with a pelvic mass five years after surgery. The
immunohistochemical profile of the tumor showed it was negative for the
prostate
markers prostatic acid phosphatase (PSAP) and prostatic specific antigen (PSA)
and
positive for SMA, consistent with sarcoma, which was thought to be secondary
to
-77-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
prostate fossa radiation treatment. However, gene rearrangement analysis
identified
a TMPRSS2-ERG, suggesting that the tumor was in fact recurrent prostate cancer
with sarcomatoid features.
[00345] The constellation of gene rearrangements and fusions in the study
group were also examined. These types of genomic alterations can result in
proteins
that drive malignancies, such as EML4-ALK, which results in constitutive
activation
of ALK through removal of the transmembrane domain.
[00346] In order to assess assay decision support for clinically relevant
genomic rearrangements, alterations detected using DNA or RNA sequencing
assays were compared across assay type and for evidence matching them to
therapeutic interventions. Overall, 28 total genomic rearrangements resulting
in
chimeric protein products were detected in the study group. 22 rearrangements
were concordantly detected between assay type, four were detected via DNA-only
assay, and two were detected via RNA-only assay (Fig. 33). Of the three
rearrangements detected via RNA sequencing, two of the three were not targets
on
the DNA sequencing assay and thus not expected to be detected via DNA
sequencing. The functionality of these fusions were further analyzed via their
predicted structures (Figs. 34 and 35). In all cases, algorithms predicted
fully intact
tyrosine kinase domains for RET and NTRK3 exemplar rearrangements, which may
be potential therapeutic targets for tyrosine kinase inhibitors. This analysis
indicates
the utility of genomic rearrangement analyses as a source of clinically
relevant
information for therapeutic interventions.
[00347] To characterize the mutational landscape in all patients, the
distribution
of the mutational load across cancer types was analyzed. The median TMB across
the study group was 2.09 mutations per megabase (Mb) of DNA with a range of 0-
54.2 mutations/Mb.
[00348] The distribution of TMB varied by cancer type. For example, cancers
that are associated with higher levels of mutagenesis, like lung cancer, had a
higher
median TMB (Fig. 36). We found that there is a population of hypermutated
tumors
with significantly higher TMB than the overall distribution of TMB for solid
tumors.
These hypermutators are found in all cancer types, including cancers typically
associated with low TMB, like glioblastoma (Fig. 36). These hypermutated
tumors
are referred to as TMB-high, which are defined as tumors with a TMB greater
than 9
mutations/Mb. This threshold was established by testing for the enrichment of
-78-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
tumors with orthogonally defined hypermutation (MSI-H) in a larger clinical
database
using the hypergeometric test. In this group, all MSI-H samples are in the TMB-
high
population (Figs. 37 and 38). The high mutational burdens from the remaining
TMB-
high samples were primarily explained by mutational signatures associated with
smoking, UV exposure, and APOBEC mediated mutagenesis.
[00349] While TMB is a measure of the number of mutations in a tumor, the
neoantigen load is a more qualitative estimate of the number of somatic
mutations
that are actually presented to the immune system. We calculated neoantigen
load
as the number of mutations that have a predicted binding affinity of 500nM or
less to
any of a patient's HLA class I alleles as well as at least one read supporting
the
variant allele in RNA sequencing data. TMB was found to be highly correlated
with
neoantigen load (R=0.933, p=2.42x10-211) (Fig. 37). This suggests that a
higher
tumor mutational burden likely results in a greater number of potential
neoantigens.
[00350] The association of high TMB and MSI-H status with response to
immunotherapy has been attributed to the greater immunogenicity of these
highly
mutated tumors. We used whole transcriptome sequencing to measure whether
greater immunogenicity results in higher levels of immune infiltration and
activation.
[00351] To test this, we assessed the relative levels of cytotoxic immune
activity using a gene expression score, cytolytic index (CYT) (Rooney, M. S.,
Shukla,
S. A., Wu, C. J., Getz, G. & Hacohen, N. Molecular and Genetic Properties of
Tumors Associated with Local Immune Cytolytic Activity. Cell 160, 48-61
(2015)).
We found that this two gene expression score is significantly higher in our
TMB-high
and MSI-high populations (p=4.3x10-5 and p=0.015, respectively) (Fig. 39).
This
result demonstrates that even in patients with heavily pre-treated and
advanced
stage disease, a hypermutator status is strongly associated with greater
cytotoxic
immune activity.
[00352] Next, whether specific immune cell populations were differentially
represented in the immune cell composition of TMB-high tumors compared to TMB-
low was analyzed. We implemented a support vector regression-based
deconvolution model to computationally estimate the relative proportion of 22
immune cell types in each tumor (Newman, A. M. et al. Robust enumeration of
cell
subsets from tissue expression profiles. Nat. Methods 12, 453-7 (2015)). In
accordance to our cytolytic index analysis, we also found that inflammatory
immune
cells, like CD8 T cells and M1 polarized macrophages, were significantly
higher in
TMB-high samples, while non-inflammatory immune cells, like monocytes, were
-79-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
significantly lower in TMB-low samples (p=0.0001, p=2.8x10-7, p=0.0008) (see
Fig.
40).
[00353] Increased immune pressure, like infiltration of more inflammatory
immune cells, can lead tumors to express higher levels of immune checkpoint
molecules like PD-L1 (CD274). These immune checkpoints function as a brake on
the immune system, turning activated immune cells into quiescent ones.
Accordingly, whole transcriptome analysis determined CD274 expression is
significantly higher in the more immune-infiltrated TMB-high tumors (p=0.0002)
(Fig.
41). CD274 expression is also highly correlated with the expression of its
binding
partner on immune cells, PDCD1 (PD-1), as well as other T cell lineage-
specific
markers like CD3E (Fig. 42). Furthermore, samples that stained positive for PD-
L1
protein via clinically-validated IHC tests cluster with higher CD274 RNA
expression
levels (Fig. 42), suggesting the expression of CD274 may be used as a proxy
for
protein levels of PD-L1.
[00354] Transcriptomic markers were utilized to further determine whether
patients that lack classically defined immunotherapy biomarkers still
exhibited
immunologically similar tumors. Using a 28 gene interferon gamma-related
signature, it was found that tumor samples could be broadly categorized as
either
immunologically active "hot" tumors or immunologically silent "cold" tumors
based on
gene expression (Fig. 43). The 28-gene set encompassed genes related to
cytolytic
activity (e.g., granzyme NB/K, PRF1), cytokines/chemokines for initiation of
inflammation (CXCR6, CXCL9, CCL5, and CCR5), T cell markers (CD3D, CD3E,
CD2, IL2RG [encoding IL-2RA), NK cell activity (NKG7, HLA-E), antigen
presentation (Cl/TA, HLA-DRA), and additional immunomodulatory factors (LAG3,
ID01, SLAMF6). Results support this stratification, with the immunologically
"hot"
population enriched for samples that were TMB-high, MSI-high or PDL1 IHC
positive. Furthermore, TMB-high, MSI-high, or PD-L1 IHC positive tumors
expressed higher levels of interferon gamma-related genes versus tumors
without
any of those biomarkers (p=2.2x10-5) (Fig. 44). Hence, patients within this
immunologically active cluster that lack traditional immunotherapy biomarkers
represent an interesting patient population that may potentially benefit from
immunotherapy.
[00355] The ultimate goal of the broad molecular profiling done in the xT
assay
is to match patients to therapies as effectively as possible, with targeted or
immunotherapy options being the most desirable. We evaluated whether patients
in
the xT group matched to response and resistance therapeutic evidence based on
-80-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
consensus clinical guidelines by cancer type (see KDB in Methods). Across all
cancer types, 90.6% matched to therapeutic evidence based on response to
therapy
(Fig. 56), and 22.6% matched to evidence based on resistance to therapy (Fig.
57).
[00356] For both response and resistance therapeutic evidence,
approximately
24% of the group could be matched to a precision medicine option with at least
a tier
IB level. In particular, tier IA therapeutic evidence, as defined by joint
AMP, ASCO,
and CAP guidelines, was returned for 15.8% of patients (Fig. 58). The maximum
tier
of therapeutic evidence per patient varied significantly by cancer type (Fig.
45). For
example, 58.0% of colorectal patients could be matched to tier IA evidence,
the
majority of which were for resistance to therapy based on detected KRAS
mutations;
while no pancreatic cancer patients could be matched to tier IA evidence. This
is
expected, as there are several molecularly based consensus guidelines in
colorectal
cancer, but fewer or none for other cancer types. Additionally, specific
therapeutic
evidence matches were made based on copy number variants (CNVs) (Fig. 46) and
single nucleotide variants (SNVs) and indels (Fig. 47) for each cancer
category.
[00357] Therapies were also matched to single gene alterations, either SNVs
and indels or CNVs, and plotted by cancer type (Fig. 48). Unfortunately, the
two
most commonly mutated genes in cancer are TP53 and KRAS, with TP53 only
having Tier IIC evidence and drugs in clinical trials, and KRAS having Tier 1A
evidence, but as resistance to therapies targeting other proteins (36
patients).
However, many less commonly mutated genes have Tier 1A evidence for targeted
therapies across a variety of cancer types. Notable in this category are the
PARP
inhibitors for BRCA1 and BRCA2 mutated breast and ovarian cancer (16
patients),
which are currently also in clinical trials or being used off-label in other
disease types
harboring BRCA mutations, such as prostate and pancreatic cancer. The majority
of
the remaining targetable mutations with Tier 1A evidence are from the
druggable
portions of the MAP kinase cascade (MAPK/ERK pathway), including EGFR, BRAF
and NRAS across colorectal and lung cancer (18 patients).
[00358] Therapeutic options were further matched based on RNA sequencing
data. We focused on the expression of 42 clinically relevant genes selected
based
on their relevance to disease diagnosis, prognosis, and/or possible
therapeutic
intervention. Over or underexpression of these genes may be reported to
physicians.
-81-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
[00359] Expression calls were made by comparison of the patient tumor
expression to the tumor and normal tissue expression in the data vault
database 180
based on overall comparisons as well as tissue-specific comparisons. For
example,
each breast cancer case was compared to all cancer samples, all normal
samples,
all breast cancer samples, and all normal breast samples. At least one gene in
76%
of patients with gene expression data was reported. The distribution of
expression
calls is shown by sample (Fig. 54) and by gene (Fig. 55). It was found that
metastatic cases are equally as likely to have at least one reportable
expression call
compared to non-metastatic tumors (79% vs 75%, p-value=0.288). The most
commonly reported gene is overexpression of MYC, which was seen in 80 (17%)
patient tumors across the group. Next, the percent of patients with gene
expression
calls was determined and evidence for the association between gene expression
and
drug response (Fig. 49) was identified. Among the cases with reported
expression
calls, 25% of cases across cancer types included evidence based on clinical
studies,
case studies, and preclinical studies reported in the literature.
[00360] Fusion proteins are proteins made from RNA that has been generated
by a DNA chromosomal rearrangement, also known as a "fusion event." Fusion
proteins can be oncogenic drivers that are among the most druggable targets in
cancer. Of the 28 chromosomal rearrangements detected in the study group, 26
were associated with evidence of response to various therapeutic options based
on
evidence tiers and cancer type (Fig. 50). The majority of fusion events were
TMPRSS-ERG fusions within prostate cancer patients in the group. TMPRSS-ERG
fusions in prostate cancer were given a IID evidence level due to the early
evidence
around therapeutic response. Of the seven non-prostate cancer fusions, one was
rated as evidence level IA, one was rated as IIC and five were rated evidence
level
IID. These detected fusions are clear drivers of cancer, part of consensus
therapeutic guidelines and shown to be present with high sensitivity by the xT
assay
referred to herein.
[00361] Based on the immunotherapy biomarkers identified by the xT assays,
we investigated what percentage of the group would be eligible for
immunotherapy.
We discovered 10.1% of the xT group would be considered potential candidates
for
immunotherapy based on TMB, MSI status, and PD-L1 IHC results alone (Fig. 51).
The number of MSI-high and TMB-high cases were distributed among cancer types.
This represents the most common immunotherapy biomarkers measured in the
group with 4% of patients positive for both TMB-high and MSI-high status. PD-
L1
-82-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
positive IHC alone were measured in 3% of the eligibility group, and was found
to be
the highest among lung cancer patients. TMB-high status alone was measured in
2.6% of the eligibility group, primarily in lung and breast cancer cases. PD-
L1
positive IHC and TMB-high status was the minority of cases and measured in
only
0.4% of the eligibility group.
[00362] Overall, clinically relevant molecular insights were uncovered for
over
90% of the group based on SNVs, indels, CNVs, gene expression calls, and
immunotherapy biomarker assays (Fig. 52). The majority of therapeutic matches
to
patients were based on clinically relevant xT findings reported on SNVs and
indels.
This was followed by matches based on CNVs, gene expression calls, fusion
detection, and immunotherapy biomarkers. In addition to therapeutic matching,
we
determined clinical-trial matching for the group based on molecular insights
from the
xT assay.
[00363] In total, 1952 clinical trials were reported for the xT 500 patient
group.
The majority of patients, 91.6%, were matched to at least one clinical trial,
with
73.6% matched with at least one biomarker-based clinical trial for a gene
variant on
their final report. The frequency of biomarker-based clinical trial matches
varied by
diagnosis and outnumbered disease-based clinical trial matches (Fig. 53). For
example, gynecological and pancreatic cancers were typically matched to a
biomarker-based clinical trial; while rare cancers had the least number of
biomarker-
based clinical trial matches and an almost equal ratio of biomarker-based to
disease-
based trial matching. The differences between biomarker versus disease-based
trial
matching appears to be due to the frequency of targetable alterations and
heterogeneity of those cancer types.
[00364] The particular embodiments disclosed above are illustrative only,
as
the invention may be modified and practiced in different but equivalent
manners
apparent to those skilled in the art having the benefit of the teachings
herein.
Furthermore, no limitations are intended to the details of construction or
design
herein shown, other than as described in the claims below. It is therefore
evident
that the particular embodiments disclosed above may be altered or modified and
all
such variations are considered within the scope and spirit of the invention.
Accordingly, the protection sought herein is as set forth in the claims below.
[00365] Thus, the invention is to cover all modifications, equivalents, and
alternatives falling within the spirit and scope of the invention as defined
by the
following appended claims.
-83-
CA 03116712 2021-04-15
WO 2020/081795
PCT/US2019/056713
[00366] To
apprise the public of the scope of this invention, the following claims
are made:
-84-