Note: Descriptions are shown in the official language in which they were submitted.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
1
Nucleic acids for inhibiting expression of ALDH2 in a cell
The present invention relates to products and compositions and their uses. In
particular
the invention relates to nucleic acid products that interfere with or inhibit
the aldehyde
dehydrogenase-2 (ALDH2) gene expression and therapeutic uses of such
repression
such as for the treatment of alcohol use disorder and associated pathologies
or
syndromes.
Background
Double-stranded RNA (dsRNA) able to complementarily bind expressed mRNA has
been
shown to be able to block gene expression (Fire et al., 1998, Nature. 1998 Feb
19;391(6669):806-11 and Elbashir et al., 2001, Nature. 2001 May
24;411(6836):494-8) by
a mechanism that has been termed RNA interference (RNAi). Short dsRNAs direct
gene-specific, post-transcriptional silencing in many organisms, including
vertebrates, and
have become a useful tool for studying gene function. RNAi is mediated by the
RNA-induced silencing complex (RISC), a sequence-specific, multi-component
nuclease
that degrades messenger RNAs homologous to the silencing trigger loaded into
the RISC
complex. Interfering RNA (termed herein iRNA) such as siRNAs, antisense RNA,
and
micro RNA are oligonucleotides that prevent the formation of proteins by gene-
silencing
i.e. inhibiting gene translation of the protein through degradation of mRNA
molecules.
Gene-silencing agents are becoming increasingly important for therapeutic
applications in
medicine.
According to Watts and Corey in the Journal of Pathology (2012; Vol 226, p 365-
379)
there are algorithms that can be used to design nucleic acid silencing
triggers, but all of
these have severe limitations. It may take various experimental methods to
identify potent
iRNAs, as algorithms do not take into account factors such as tertiary
structure of the
target mRNA or the involvement of RNA binding proteins. Therefore, the
discovery of a
potent nucleic acid silencing trigger with minimal off-target effects is a
complex process.
For the pharmaceutical development of these highly charged molecules it is
necessary
that they can be synthesised economically, distributed to target tissues,
enter cells and
function within acceptable limits of toxicity.
Compulsive, excessive alcohol consumption beyond social drinking (aka.
alcoholism,
alcohol abuse, alcohol dependence) is a worldwide increasing health problem
with severe
personal and socioeconomic impact. Current medications that aid alcohol
dishabituation
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
2
are either not very effective or they have considerable undesired side
effects. As a
therapeutic approach, reduction of activity of a rate-limiting key enzyme of
the hepatic
alcohol metabolism pathway, the aldehyde dehydrogenase-2 (ALDH2), leads to
accumulation of the toxic metabolite acetaldehyde. This accumulation results
in
unpleasant physiological effects in alcohol-consuming individuals. The
association of
unpleasant to severe feelings affects the patient's alcohol seeking behaviour,
reduces
alcohol consumption and, thereby, assists dishabituation. This therapeutic
avenue has
been exploited successfully for decades using small molecule ALDH-inhibiting
compounds
that act systemically (Kragh et al., 2008, Bull. Hist. Chem., Vol. 33, 2). The
safety profile of
those systemically acting inhibitors is worsened by the unwanted side effects
even in
individuals that do not consume alcohol or are produced by involuntary
exposure to
environmental aldehydes present in e.g. food, perfumes or other forms harmless
to the
organism in normal circumstances.
The current antidipsotropic medications that are aimed at keeping alcohol
dependent
patients away from drinking have low molecular and tissue specificity and
show, therefore,
a bad safety profile with considerable effects even in non-alcohol consuming
patients.
Treatment failure of those therapies is high due to high abandonment and
cheating (not
taking the medicine in order to be able to resume the consumption of alcohol).
So there is
a medical need to improve such therapeutic approach.
A first aspect of the invention relates to a nucleic acid for inhibiting
expression of ALDH2
in a cell, comprising at least one duplex region that comprises at least a
portion of a first
strand and at least a portion of a second strand that is at least partially
complementary to
the first strand, wherein said first strand is at least partially
complementary to at least a
portion of a RNA transcribed from the ALDH2 gene, wherein said first strand
comprises a
nucleotide sequence selected from the following sequences: SEQ ID NOs:1, 3, 5,
7, 9, 11,
13, 15, 17, 19, 21, 23, 25, 27 or 55.
In one embodiment, the nucleic acid comprises in the first strand a sequence
of at least
15, preferably at least 16, more preferably at least 17, yet more preferably
at least 18 and
most preferably at least 19 nucleotides of any one of the reference sequences
SEQ ID
NO: 11, 17,3, 1, 5, 7, 9, 13, 15, 19, 21, 23, 25 27 or 55.
In one embodiment, the number of single nucleotide mismatches in the first
strand
sequence relative to the portion of the reference sequence that is comprised
in the first
strand sequence is at most three, preferably at most two, more preferably at
most one and
most preferably zero
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
3
The second strand may comprise a nucleotide sequence of SEQ ID NO:2, 4, 6, 8,
10, 12,
14, 16, 18, 20, 22, 24, 26 or 28.
The first strand may comprise the nucleotide sequence of SEQ ID NO:3, SEQ ID
NO:11
or SEQ ID NO:17 and/or the second strand may comprise the nucleotide sequence
of
SEQ ID NO:4, SEQ ID NO:12 or SEQ ID NO:18.
The first strand and/or the second strand may each be from 17-35 nucleotides
in length
and at least one duplex region may be from 10-25 nucleotides in length. The
duplex may
comprise two separate strands or it may comprise a single strand which
comprises the
first strand and the second strand.
The nucleic acid may: a) be blunt ended at both ends; b) have an overhang at
one end
and a blunt end at the other; or c) have an overhang at both ends.
One or more nucleotides on the first and/or second strand may be modified, to
form
modified nucleotides. One or more of the odd numbered nucleotides of the first
strand
may be modified. One or more of the even numbered nucleotides of the first
strand may
be modified by at least a second modification, wherein the at least second
modification is
different from the modification on the one or more odd nucleotides. At least
one of the one
or more modified even numbered nucleotides may be adjacent to at least one of
the one
or more modified odd numbered nucleotides.
A plurality of odd numbered nucleotides in the first strand may be modified in
the nucleic
acid of the invention. A plurality of even numbered nucleotides in the first
strand may be
modified by a second modification. The first strand may comprise adjacent
nucleotides
that are modified by a common modification. The first strand may also comprise
adjacent
nucleotides that are modified by a second different modification.
One or more of the odd numbered nucleotides of the second strand may be
modified by a
modification that is different to the modification of the odd numbered
nucleotides on the
first strand and/or one or more of the even numbered nucleotides of the second
strand
may be modified by the same modification of the odd numbered nucleotides of
the first
strand. At least one of the one or more modified even numbered nucleotides of
the
second strand may be adjacent to the one or more modified odd numbered
nucleotides. A
plurality of odd numbered nucleotides of the second strand may be modified by
a common
modification and/or a plurality of even numbered nucleotides may be modified
by the
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
4
same modification that is present on the first strand odd numbered
nucleotides. A plurality
of odd numbered nucleotides on the second strand may be modified by a second
modification, wherein the second modification is different from the
modification of the first
strand odd numbered nucleotides.
The second strand may comprise adjacent nucleotides that are modified by a
common
modification, which may be a second modification that is different from the
modification of
the odd numbered nucleotides of the first strand.
In the nucleic acid of the invention, each of the odd numbered nucleotides in
the first
strand and each of the even numbered nucleotides in the second strand may be
modified
with a common modification and, each of the even numbered nucleotides may be
modified in the first strand with a second modification and each of the odd
numbered
nucleotides may be modified in the second strand with a second different
modification.
The nucleic acid of the invention may have the modified nucleotides of the
first strand
shifted by at least one nucleotide relative to the unmodified or differently
modified
nucleotides of the second strand.
The modification and / or modifications may each and individually be selected
from the
group consisting of 3'-terminal deoxy-thymine, 2'-0-methyl, a 2'-deoxy-
modification, a
2'-amino-modification, a 2'-alkyl-modification, a morpholino modification, a
phosphoramidate modification, 5'-phosphorothioate group modification, a 5'
phosphate or
5' phosphate mimic modification and a cholesteryl derivative or a dodecanoic
acid
bisdecylamide group modification and/or the modified nucleotide may be any one
of a
locked nucleotide, an abasic nucleotide or a non-natural base comprising
nucleotide.
At least one modification may be 2'-0-methyl and/or at least one modification
may be 2'-F.
The invention further provides, as a second aspect, a nucleic acid for
inhibiting expression
of ALDH2 in a cell, comprising at least one duplex region that comprises at
least a portion
of a first strand and at least a portion of a second strand that is at least
partially
complementary to the first strand, wherein said first strand is at least
partially
complementary to at least a portion of an RNA transcribed from the ALDH2 gene,
wherein
said first strand comprises a nucleotide sequence selected from the following
sequences:
SEQ ID NOs:1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25 or 27, wherein the
nucleotides of
first strand are modified by a first modification on the odd numbered
nucleotides, and
modified by a second modification on the even numbered nucleotides, and
nucleotides of
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
the second strand are modified by a third modification on the even numbered
nucleotides
and modified by a fourth modification on the odd numbered nucleotides, wherein
at least
the first modification is different to the second modification and the third
modification is
different to the fourth modification. The second strand may comprise a
nucleotide
5 sequence of SEQ ID NO: 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26 or
28. The third
modification and the first modification may be the same and/or the second
modification
and the fourth modification may be the same.
The first modification may be 2'0Me and the second modification may be 2'F.
1.13
In the nucleic acid of the second aspect, the first strand may comprise the
nucleotide
sequence of SEQ ID NO:3, SEQ ID NO:11 or SEQ ID NO:17 and the second strand
may
comprise the nucleotide sequence of SEQ ID NO:4, SEQ ID NO:12 or SEQ ID NO:18.
The sequence and modifications may be as shown in the Table below; which shows
preferred sequences based on an extract of Table 1 as provided herein:
Sequence
siRNA ID sequence modifications
ID No
3 5'-AAUGUUUUCCUGCUGACGG-3' 6254515173547182748
ALDH2-hcm-2
4 5'-CCGUCAGCAGGAAAACAUU-3' 3745364728462627251
11 5'-UCUUCUUAAACUGAGUUUC-3' 5351715262718281517
ALDH2-hcm-6
12 5'-GAAACUCAGUUUAAGAAGA-3' 4626353645152646282
17 5'-AUGUAGCCGAGGAUCUUCU-3' 6181647382846171535
ALDH2-hcm-9
18 5'-AGAAGAUCCUCGGCUACAU-3' 2826461735384716361
wherein the specific modifications are depicted by the following numbers
1=2"F-dU,
2=2`F-dA,
3=2"F-dC,
4=2"F-dG,
5=2'-0Me-rU;
6=2'-0Me-rA;
7=2'-0Me-rC;
8=2'-0Me-rG.
A nucleic acid of the invention may comprise a phosphorothioate linkage
between the
terminal one, two or three 3' nucleotides and/or one, two or three 5'
nucleotides of the first
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
6
and/or the second strand. It may comprise two phosphorothioate linkages
between each
of the three terminal 3' and between each of the three terminal 5' nucleotides
on the first
strand, and two phosphorothioate linkages between the three terminal
nucleotides of the
3' end of the second strand.
Such a nucleic acid may be conjugated to a ligand.
The invention further provides, as a third aspect, a nucleic acid for
inhibiting expression of
ALDH2 in a cell, comprising at least one duplex region that comprises at least
a portion of
a first strand and at least a portion of a second strand that is at least
partially
complementary to the first strand, wherein said first strand is at least
partially
complementary to at least a portion of a RNA transcribed from the ALDH2 gene,
wherein
said first strand comprises a nucleotide sequence selected from the following
sequences:
SEQ ID NOs:1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25 or 27, and wherein
the nucleic
acid is conjugated to a ligand.
The ligand may comprise (i) one or more N-acetyl galactosamine (GaINAc)
moieties and
derivatives thereof, and (ii) a linker, wherein the linker conjugates the
GaINAc moieties to
a sequence as defined in any preceding aspects. The linker may be a bivalent
or trivalent
or tetravalent branched structure. The nucleotides may be modified as defined
herein.
The ligand may comprise the formula I:
[S-X1-P-X93-A-X3- (I)
wherein:
S represents a saccharide, wherein the saccharide is N-acetyl galactosamine;
X1 represents C3-C6 alkylene or (-CH2-CH2-0)m(-CH2)2- wherein m is 1, 2, or 3;
P is a phosphate or modified phosphate (preferably a thiophosphate);
X2 is alkylene or an alkylene ether of the formula (-CH2)n-O-CH2- where n = 1-
6;
A is a branching unit;
X3 represents a bridging unit;
wherein a nucleic acid according to the present invention is conjugated to
X3via a
phosphate or modified phosphate (preferably a thiophosphate).
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
7
The present invention therefore additionally provides a conjugated nucleic
acid having one
of the following structures
OH
OH
N -00H
AcH A:FITCH
0
0
0
H:)0
0
S=L0-
I OH
(D
s.0
/7c-0H
PcH
11
0 OH
/ ___________________ 0 0
H
S ______________
Z-0---IFLO/
0
Jr s
011_0--
b-
CA 03118327 2021-04-30
WO 2019/092282
PCT/EP2018/081105
8
OH
1-1 OH
OH OH
0
HO ,./.1...,... AcHN
0 0
NHAc
Ll*sl
0
1 e
0 =P ¨S
&l 0
-) 1 e
0 =P¨S
1
0 OH
67
0 --/¨j AcHN OH
i
1-0 OH
/ I /
0
0
If
Z ¨0¨ P ¨0
le
S L.1 0
o-A-cr
le
S I
1
OH
HO\,_,.. 0H
OH OH
0
HOND AcHN
0 0
NHAc
L1*1
0
I 0
0 =P ¨S
1
01 0
I e
0 =P ¨S
i
0 OH
OH
0 AcHN
_______________________________________ / 0/' OH
/
0 0
II /
Z-0 ¨P-0 -__
I0
S
? /
0¨P¨O
to
S
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
9
OH
HO\,...,011
OH OH
0
AcHN
0 0
NHAc
0
Li)
i 0
0=P¨S
i
0.1 0
i 0
0=P¨S
0
/I OH
0
/ AcHN C
0¨ OH
'N
_______________________________________ / r().-R'OH
_Ex J-0/ 0
-.,
0 0
II
I¨O¨P-0
LI,µ 1? f
,.
S 0¨P-0
so
S,
OH
H0µ,_ OH
OH OH
0
AcHN
0 0
NHAc
1')
0
I 0
0 =P ¨S
i
01 0
I 0
0 = P ¨S
0
/ I OH
0,, o j OH
/
AcHN
, r i c ( 1 -FON
_x_rxi¨ 0 _____________________ õ, 0
0 .--
0
II ,
z-0¨P¨O 0¨p-0
i 0 10
S S
CA 03118327 2021-04-30
WO 2019/092282
PCT/EP2018/081105
OH
HO k,___01.1
OH OH 0
AcHN
0
NHAc
0
04¨Se \
i
01 0
04¨Se
i
0
AcHN OH
OH
0 ,,, 0 --/¨/ (12/)(OH
0/ L /
0
OLL /0
II
Z ¨0 ¨P ¨0 ¨rj---
le
s o
0
0¨p-0
le
s
5
OH
HO OH
OH OH
AcH 0N
0
NHAc
0
i 0
0=P ¨S
\
1
01 0
I 0
0 =P ¨ S
I
0 AcHN OH
OH
0 0 ¨r0 OH
/
0
/ I
0
0
0
2 ¨0 ¨11-0 ¨T.-1j¨ LI,. 0 /
10 0
S 0 ¨P ¨0
S
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
11
DH
HL
,,.---OH
OH OH b
t f
AcHN
0
r H ; \,....-\.....\.....\0
\
1 0
0 =P -S
i
0%1 0
0P-
1....1 t S0
=
t
0
AcHiv H
i.......\ OH
0 0 OH
_____________________________________________ / 0
0/ co
0
11 li /
2 -0 -P -0 LI,,, 0 -P -0
t 0 t 0
S s
wherein Z represents a nucleic acid as defined herein before.
Alternatively, a nucleic acid according to the present invention may be
conjugated to a
ligand of the following structure
OH
0H H FlA
H
H o
1004
c 1N
NNc 01 OH
O-P=0 H
0
4
H
H
11 ICIY
0
HAc
Hae7.0/ANNN
H
H
The present invention also relates to a conjugate for inhibiting expression of
a ALDH2 gene
in a cell, said conjugate comprising a nucleic acid portion, comprising the
nucleic acid of
any aspect of the invention, and ligand portions, said nucleic acid portion
comprising at least
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
12
one duplex region that comprises at least a portion of a first RNA strand and
at least a
portion of a second RNA strand that is at least partially complementary to the
first strand,
wherein said first strand is at least partially complementary to at least a
portion of RNA
transcribed from said ALDH2 gene, said ligand portions comprising a linker
moiety,
preferably a serinol-derived linker moiety, and a targeting ligand for in vivo
targeting of cells
and being conjugated exclusively to the 3' and/or 5' ends of one or both RNA
strands,
wherein the 5' end of the first RNA strand is not conjugated, wherein:
(i) the second RNA strand is conjugated at the 5' end to the targeting
ligand, and
wherein (a) the second RNA strand is also conjugated at the 3' end to the
targeting ligand and the 3' end of the first RNA strand is not conjugated; or
(b)
the first RNA strand is conjugated at the 3' end to the targeting ligand and
the 3'
end of the second RNA strand is not conjugated; or (c) both the second RNA
strand and the first RNA strand are also conjugated at the 3' ends to the
targeting
ligand; or
(ii) both the second RNA strand and the first RNA strand are conjugated at
the 3'
ends to the targeting ligand and the 5' end of the second RNA strand is not
conjugated, or
the present invention relates to a conjugate for inhibiting expression of a
ALDH2 gene in a
cell, said conjugate comprising a nucleic acid portion and ligand portions,
said nucleic acid
portion comprising at least one duplex region that comprises at least a
portion of a first RNA
strand and at least a portion of a second RNA strand that is at least
partially complementary
to the first strand, wherein said first strand is at least partially
complementary to at least a
portion of RNA transcribed from said ALDH2 gene, said ligand portions
comprising a linker
moiety and a targeting ligand for in vivo targeting of cells and being
conjugated exclusively
to the 3' and/or 5' ends of one or both RNA strands, wherein the 5' end of the
first RNA
strand is not conjugated, wherein:
(i)
the second RNA strand is conjugated at the 5' end to the targeting ligand, and
wherein (a) the second RNA strand is also conjugated at the 3' end to the
targeting ligand and the 3' end of the first RNA strand is not conjugated; or
(b)
the first RNA strand is conjugated at the 3' end to the targeting ligand and
the 3'
end of the second RNA strand is not conjugated; or (c) both the second RNA
strand and the first RNA strand are also conjugated at the 3' ends to the
targeting
ligand; or
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
13
(ii) both the second RNA strand and the first RNA strand are
conjugated at the 3'
ends to the targeting ligand and the 5' end of the second RNA strand is not
conjugated.
The linker moiety may for example be a serinol-derived linker moiety or one of
the other
linker types described herein.
The invention also provides a composition comprising the nucleic acid or
conjugated
nucleic acid of any aspect of the invention, and a physiologically acceptable
excipient.
Also provided is a nucleic acid or conjugated nucleic acid according to any
aspect of the
invention for use in the treatment of a disease, disorder or syndrome and/or
in the
manufacture of a medicament for treating a disease, disorder, or syndrome.
The invention provides a method of treating or preventing a disease, disorder
or syndrome
comprising administration of a composition comprising a nucleic acid or
conjugated
nucleic acid according to any aspect of the invention to an individual in need
of treatment.
The nucleic acid or conjugated nucleic acid may be administered to the subject
subcutaneously, intravenously or using any other application routes such as
oral, rectal or
intraperitoneal.
After subcutaneous application, the invention may be delivered in a tissue
specific manner
to liver (hepatocytes) and target specifically ALDH2, in order to reduce
unwanted side
effects and achieve a lower therapeutic dose necessary to achieve the desired
effect.
The disease, disorder or syndrome may be alcohol use disorder which includes
acute
alcohol sensitivity (hangover), alcoholic neuropathy (alcohol-related
polyneuropathy)
alcohol dependence, alcohol abuse (alcoholism) or foetal alcohol syndrome
(alcohol-
related neurodevelopmental disorder) or any other pathology associated to
acute or
prolonged excessive alcohol consumption.
A method of making the nucleic acid or conjugated nucleic acid according to
the invention
is also included.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
14
Detailed Description of Invention
The present invention relates to a nucleic acid which is double stranded and
directed to an
expressed RNA transcript of ALDH2 and compositions thereof. These nucleic
acids or
conjugated nucleic acids can be used in the treatment and prevention of a
variety of
diseases, disorders and syndromes where reduced expression of ALDH2 gene
product is
desirable.
A first aspect of the invention relates to a nucleic acid for inhibiting
expression of ALDH2
in a cell, comprising at least one duplex region that comprises at least a
portion of a first
strand and at least a portion of a second strand that is at least partially
complementary to
the first strand, wherein said first strand is at least partially
complementary to at least a
portion of a RNA transcribed from the ALDH2 gene, wherein said first strand
comprises a
nucleotide sequence selected from the following sequences: SEQ ID NOs:1, 3, 5,
7, 9, 11,
13, 15, 17, 19, 21, 23, 25 or 27.
A related aspect of the invention is a nucleic acid for inhibiting expression
of ALDH2 in a
cell, wherein the nucleic acid comprises at least one duplex region that
comprises a first
strand and a second strand, wherein said second strand is at least partially
complementary
to the first strand,wherein said first strand comprises a sequence of at least
15, preferably
at least 16, more preferably at least 17, yet more preferably at least 18 and
most preferably
at least 19 nucleotides of any one of the reference sequences SEQ ID NO: 11,
17, 3, 1, 5,
7, 9, 13, 15, 19, 21, 23, 25, 27 or 55, and wherein the number of single
nucleotide
mismatches and/or deletions and/or insertions in the first strand sequence
relative to the
portion of the reference sequence that is comprised in the first strand
sequence is at most
three, preferably at most two, more preferably at most one and most preferably
zero.
In one aspect, the first strand of the nucleic acid comprises a sequence of at
least 18
nucleotides of any one of the reference sequences, preferably of any one of
the reference
sequences SEQ ID NO: 11, 17 and 3, and wherein the number of single-nucleotide
mismatches and/or deletions and/or insertions in the first strand sequence
relative to the
portion of the reference sequence that is comprised in the first strand
sequence is at most
one, and preferably zero.
In one aspect, the first strand of the nucleic acid comprises a sequence of at
least 19
nucleotides of any of the reference sequences SEQ ID NO: 11, 17 and 3.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
A certain number of mismatches, deletions or insertions between the first
(antisense)
strand and the target sequence, or between the first strand and the second
(sense) strand
can be tolerated in the context of siRNA and even have the potential in
certain cases to
increase activity.
5
By nucleic acid it is meant a nucleic acid comprising two strands comprising
nucleotides,
that is able to interfere with gene expression. Inhibition may be complete or
partial and
results in down regulation of gene expression in a targeted manner. The
nucleic acid
comprises two separate polynucleotide strands; the first strand, which may
also be a
10 guide strand; and a second strand, which may also be a passenger strand.
The first
strand and the second strand may be part of the same polynucleotide molecule
that is
self-complementary which 'folds' back to form a double stranded molecule. The
nucleic
acid may be an siRNA molecule.
15 The nucleic acid may comprise ribonucleotides, modified ribonucleotides,
deoxynucleotides, deoxyribonucleotides, or nucleotide analogues non-
nucleotides that are
able to mimic nucleotides such that they may 'pair' with the corresponding
base on the
target sequence or complementary strand. The nucleic acid may further comprise
a
double-stranded nucleic acid portion or duplex region formed by all or a
portion of the first
strand (also known in the art as a guide strand) and all or a portion of the
second strand
(also known in the art as a passenger strand). The duplex region is defined as
beginning
with the first base pair formed between the first strand and the second strand
and ending
with the last base pair formed between the first strand and the second strand,
inclusive.
By duplex region it is meant the region in two complementary or substantially
complementary oligonucleotides that form base pairs with one another, either
by Watson-
Crick base pairing or any other manner that allows for a duplex between
oligonucleotide
strands that are complementary or substantially complementary. For example, an
oligonucleotide strand having 21 nucleotide units can base pair with another
oligonucleotide of 21 nucleotide units, yet only 19 nucleotides on each strand
are
complementary or substantially complementary, such that the "duplex region"
consists of
19 base pairs. The remaining base pairs may exist as 5' and 3' overhangs, or
as single
stranded regions. Further, within the duplex region, 100% complementarity is
not required;
substantial complementarity is allowable within a duplex region. Substantial
complementarity refers to complementarity between the strands such that they
are
capable of annealing under biological conditions. Techniques to empirically
determine if
two strands are capable of annealing under biological conditions are well
known in the art.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
16
Alternatively, two strands can be synthesised and added together under
biological
conditions to determine if they anneal to one another.
The portion of the first strand and second strand that form at least one
duplex region may
be fully complementary and is at least partially complementary to each other.
Depending on the length of a nucleic acid, a perfect match in terms of base
complementarity between the first strand and the second strand is not
necessarily
required. However, the first and second strands must be able to hybridise
under
physiological conditions.
The complementarity between the first strand and second strand in the at least
one duplex
region may be perfect in that there are no nucleotide mismatches or
additional/deleted
nucleotides in either strand. Alternatively, the complementarity may not be
perfect. The
complementarity may be from about 70% to about 100%. More specifically, the
complementarity may be at least 70%, 75%, 80%, 85%, 90% or 95% and
intermediate
values.
In the context of this invention, "a portion of" as for example in "one duplex
region that
comprises at least a portion of a first strand" should be understood to mean
that the
duplex region comprises at least 10, preferably at least 12, more preferably
at least 14,
yet more preferably at least 16, even more preferably at least 18 and most
preferably all of
the nucleotides of a given reference strand sequence. The portion of the
reference
sequence in the dublex region is at least 70%, preferably at least 80%, more
preferably at
least 90%, yet more preferably at least 95% and most preferably 100% identical
to the
corresponding portion of the reference sequence. Alternatively, the number of
single
nucleotide mismatches relative to the portion of the reference sequence is at
most three,
preferably at most two, more preferably at most one and most preferably zero.
The first strand and the second strand may each comprise a region of
complementarity
which comprises at least 15 contiguous nucleotides differing by no more than 3
nucleotides from any one of the sequences listed in Table 1.
The nucleic acid may comprise a second sequence comprising a nucleotide
sequence of
SEQ ID NO:2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26 or 28.
The nucleic acid may comprise a first strand that comprises a nucleotide
sequence of
SEQ ID NO: 11, and optionally wherein the second strand comprises a nucleotide
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
17
sequence of SEQ ID NO: 12; or a first strand that comprises a nucleotide
sequence of
SEQ ID NO: 17, and optionally wherein the second strand comprises a nucleotide
sequence of SEQ ID NO: 18; or a first strand that comprises a nucleotide
sequence of
SEQ ID NO: 3, and optionally wherein the second strand comprises a nucleotide
sequence of SEQ ID NO: 4; or a first strand that comprises a nucleotide
sequence of SEQ
ID NO:1, and optionally wherein the second strand comprises a nucleotide
sequence of
SEQ ID NO: 2; or a first strand that comprises a nucleotide sequence of SEQ ID
NO:5,
and optionally wherein the second strand comprises a nucleotide sequence of
SEQ ID
NO: 6; or a first strand that comprises a nucleotide sequence of SEQ ID NO: 7,
and
optionally wherein the second strand comprises a nucleotide sequence of SEQ ID
NO: 8;
or a first strand that comprises a nucleotide sequence of SEQ ID NO: 9, and
optionally
wherein the second strand comprises a nucleotide sequence of SEQ ID NO: 10; or
a first
strand that comprises a nucleotide sequence of SEQ ID NO: 13, and optionally
wherein
the second strand comprises a nucleotide sequence of SEQ ID NO: 14; or a first
strand
that comprises a nucleotide sequence of SEQ ID NO: 15, and optionally wherein
the
second strand comprises a nucleotide sequence of SEQ ID NO: 16; or a first
strand that
comprises a nucleotide sequence of SEQ ID NO: 19, and optionally wherein the
second
strand comprises a nucleotide sequence of SEQ ID NO: 20; or a first strand
that
comprises a nucleotide sequence of SEQ ID NO: 21, and optionally wherein the
second
strand comprises a nucleotide sequence of SEQ ID NO: 22; or a first strand
that
comprises a nucleotide sequence of SEQ ID NO: 23, and optionally wherein the
second
strand comprises a nucleotide sequence of SEQ ID NO: 24; or a first strand
that
comprises a nucleotide sequence of SEQ ID NO:25, and optionally wherein the
second
strand comprises a nucleotide sequence of SEQ ID NO: 26; or a first strand
that
comprises a nucleotide sequence of SEQ ID NO: 27, and optionally wherein the
second
strand comprises a nucleotide sequence of SEQ ID NO: 28.
Use of a nucleic acid according to the present invention involves the
formation of a duplex
region between all or a portion of the first strand and a portion of a target
nucleic acid. The
portion of the target nucleic acid that forms a duplex region with the first
strand, defined as
beginning with the first base pair formed between the first strand and the
target sequence
and ending with the last base pair formed between the first strand and the
target
sequence, inclusive, is the target nucleic acid sequence or simply, target
sequence. The
duplex region formed between the first strand and the second strand need not
be the
same as the duplex region formed between the first strand and the target
sequence. That
is, the second strand may have a sequence different from the target sequence;
however,
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
18
the first strand must be able to form a duplex structure with both the second
strand and
the target sequence, at least under physiological conditions.
The complementarity between the first strand and the target sequence may be
perfect (no
nucleotide mismatches or additional/deleted nucleotides in either nucleic
acid).
The complementarity between the first strand and the target sequence may not
be perfect.
The complementarity may be from about 70% to about 100%. More specifically,
the
complementarity may be at least 70%, 80%, 85%, 90% or 95% and intermediate
values.
The identity between the first strand and the complementary sequence of the
target
sequence may range from about 75% to about 100%. More specifically, the
complementarity may be at least 75%, 80%, 85%, 90% or 95% and intermediate
values,
provided a nucleic acid is capable of reducing or inhibiting the expression of
ALDH2.
A nucleic acid having less than 100% complementarity between the first strand
and the
target sequence may be able to reduce the expression of ALDH2 to the same
level as a
nucleic acid having perfect complementarity between the first strand and
target sequence.
Alternatively, it may be able to reduce expression of ALDH2 to a level that is
15% - 100%
of the level of reduction achieved by the nucleic acid with perfect
complementarity.
The nucleic acid may comprise a first strand and a second strand that are each
from
19-25 nucleotides in length. The first strand and the second strand may be of
different
lengths.
The nucleic acid may be 15-25 nucleotide pairs in length. The nucleic acid may
be 17-23
nucleotide pairs in length. The nucleic acid may be 17-25 nucleotide pairs in
length. The
nucleic acid may be 23-24 nucleotide pairs in length. The nucleic acid may be
19-21
nucleotide pairs in length. The nucleic acid may be 21-23 nucleotide pairs in
length.
The nucleic acid may comprise a duplex region that consists of 19-25
nucleotide base
pairs. The duplex region may consist of 17, 18, 19, 20, 21, 22, 23, 24 or 25
base pairs,
which may be contiguous.
The nucleic acid may comprise a first strand sequence of SEQ ID NO:3, SEQ ID
NO:11 or
SEQ ID NO:17. The nucleic acid may comprise a second strand sequence of SEQ ID
NO:4, SEQ ID NO:12 or SEQ ID NO:18.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
19
Preferably, the nucleic acid mediates RNA interference.
In one embodiment, the nucleic acid for inhibiting expression of ALDH2 in a
cell,
comprises at least one duplex region that comprises a first strand and a
second strand
that is at least partially complementary to the first strand, wherein said
first strand
comprises a sequence of at least 15, preferably at least 16, more preferably
at least 17,
yet more preferably at least 18 and most preferably at least 19 nucleotides
with a
sequence identity of at least 70%, preferably at least 80%, more preferably at
least 90%,
yet more preferably at least 95% and most preferably 100% of any of sequences
SEQ ID
NOs: 11, 17,3, 1, 5, 7, 9, 13, 15, 19, 21, 23, 25, 27 or 55.
SEQ ID NO: 55 is a 32-nucleotide sequence that corresponds to the RNA reverse
complement of nucleotides 1186 to 1217 of the ALDH2 reference sequence
NM_000690.3 (SEQ ID NO: 55 ¨ AUGUAGCCGAGGAUCUUCUUAAACUGAGUUUC).
This locus is particularly well suited as a target for RNA interference
because siRNAs with
sequences SEQ ID NO: 11 and SEQ ID NO: 17, which both allow efficiently
reducing the
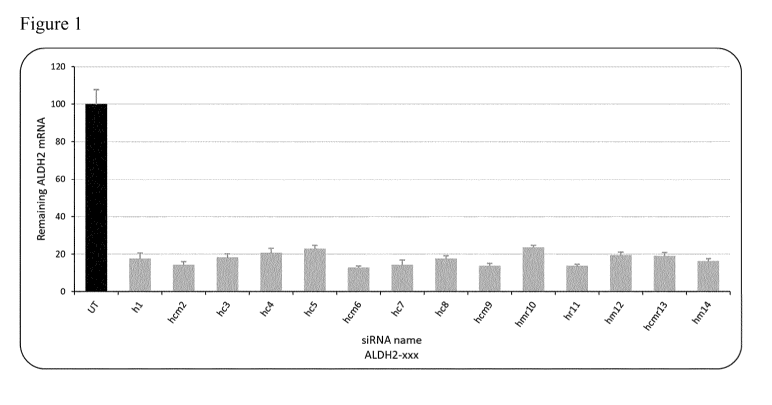
ALDH2 mRNA level, both bind to parts of this short locus.
In a further aspect the nucleic acid or conjugated nucleic acid as described
may reduce
the expression of ALDH2 by at least 15% compared to the expression observed in
the
absence of the nucleic acid or conjugated nucleic acid. All preferred features
of any of the
previous aspects also apply to this aspect. In particular, the expression of
ALDH2 may be
reduced to at least the following given % or less than 90%, 80%, 70%, 60%,
50%, 40%,
30%, 20%, 15% or less, and intermediate values, than that observed in the
absence of the
nucleic acid or conjugated nucleic acid or in the presence of a non-silencing
control.
The nucleic acid may be blunt ended at both ends; have an overhang at one end
and a
blunt end at the other end; or have an overhang at both ends.
An "overhang" as used herein has its normal and customary meaning in the art,
i.e. a
single stranded portion of a nucleic acid that extends beyond the terminal
nucleotide of a
complementary strand in a double strand nucleic acid. The term "blunt end"
includes
double stranded nucleic acid whereby both strands terminate at the same
position,
regardless of whether the terminal nucleotide(s) are base-paired. The terminal
nucleotide
of a first strand and a second strand at a blunt end may be base paired. The
terminal
nucleotide of a first strand and a second strand at a blunt end may not be
paired. The
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
terminal two nucleotides of a first strand and a second strand at a blunt end
may be base-
paired. The terminal two nucleotides of a first strand and a second strand at
a blunt end
may not be paired.
5 The nucleic acid may have an overhang at one end and a blunt end at the
other. The
nucleic acid may have an overhang at both ends. The nucleic acid may be blunt
ended at
both ends. The nucleic acid may be blunt ended at the end with the 5'-end of
the first
strand and the 3'-end of the second strand or at the 3'-end of the first
strand and the
5'-end of the second strand.
The nucleic acid may comprise an overhang at a 3'- or 5'-end. The nucleic acid
may have
a 3'-overhang on the first strand. The nucleic acid may have a 3'-overhang on
the second
strand. The nucleic acid may have a 5'-overhang on the first strand. The
nucleic acid may
have a 5'-overhang on the second strand. The nucleic acid may have an overhang
at both
the 5'-end and 3'-end of the first strand. The nucleic acid may have an
overhang at both
the 5'-end and 3'-end of the second strand. The nucleic acid may have a 5'
overhang on
the first strand and a 3' overhang on the second strand. The nucleic acid may
have a 3'
overhang on the first strand and a 5' overhang on the second strand. The
nucleic acid
may have a 3' overhang on the first strand and a 3' overhang on the second
strand. The
nucleic acid may have a 5' overhang on the first strand and a 5' overhang on
the second
strand.
An overhang at the 3'-end or 5' end of the second strand or the first strand
may be
selected from consisting of 1,2, 3,4 and 5 nucleotides in length. Optionally,
an overhang
may consist of 1 or 2 nucleotides, which may or may not be modified.
Unmodified polynucleotides, particularly ribonucleotides, may be prone to
degradation by
cellular nucleases, and, as such, modifications/ modified nucleotides may be
included in
the nucleic acid of the invention. Such modifications may help to stabilise
the nucleic acid
by making them more resistant against nucleases. This improved resistance
allows
nucleic acids to be active in mediating RNA interference for longer time
periods and is
especially desirable when the nucleic acids are to be used for treatment.
One or more nucleotides on the second and/or first strand of the nucleic acid
of the
invention may be modified.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
21
Modifications of the nucleic acid of the present invention generally provide a
powerful tool
in overcoming potential limitations including, but not limited to, in vitro
and in vivo stability
and bioavailability inherent to native RNA molecules. The nucleic acid
according to the
invention may be modified by chemical modifications. Modified nucleic acids
can also
minimise the possibility of inducing interferon activity in humans.
Modifications can further
enhance the functional delivery of a nucleic acid to a target cell. The
modified nucleic acid
of the present invention may comprise one or more chemically modified
ribonucleotides of
either or both of the first strand or the second strand. A ribonucleotide may
comprise a
chemical modification of the base, sugar or phosphate moieties. The
ribonucleic acid may
be modified by substitution or insertion with analogues of nucleic acids or
bases.
One or more nucleotides of a nucleic acid of the present invention may be
modified. The
nucleic acid may comprise at least one modified nucleotide. The modified
nucleotide may
be in the first strand. The modified nucleotide may be in the second strand.
The modified
nucleotide may be in the duplex region. The modified nucleotide may be outside
the
duplex region, i.e., in a single stranded region. The modified nucleotide may
be on the first
strand and may be outside the duplex region. The modified nucleotide may be on
the
second strand and may be outside the duplex region. The 3'-terminal nucleotide
of the
first strand may be a modified nucleotide. The 3'-terminal nucleotide of the
second strand
may be a modified nucleotide. The 5'-terminal nucleotide of the first strand
may be a
modified nucleotide. The 5'-terminal nucleotide of the second strand may be a
modified
nucleotide.
A nucleic acid of the invention may have 1 modified nucleotide or a nucleic
acid of the
invention may have about 2-4 modified nucleotides, or a nucleic acid may have
about 4-6
modified nucleotides, about 6-8 modified nucleotides, about 8-10 modified
nucleotides,
about 10-12 modified nucleotides, about 12-14 modified nucleotides, about 14-
16
modified nucleotides about 16-18 modified nucleotides, about 18-20 modified
nucleotides,
about 20-22 modified nucleotides, about 22-24 modified nucleotides, 24-26
modified
nucleotides or about 26-28 modified nucleotides. In each case the nucleic acid
comprising
said modified nucleotides retains at least 50% of its activity as compared to
the same
nucleic acid but without said modified nucleotides or vice versa. The nucleic
acid may
retain 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 95% or 100% and intermediate
values of its activity as compared to the same nucleic acid but without said
modified
nucleotides, or may have more than 100% of the activity of the same nucleic
acid without
said modified nucleotides.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
22
The modified nucleotide may be a purine or a pyrimidine. At least half of the
purines may
be modified. At least half of the pyrimidines may be modified. All of the
purines may be
modified. All of the pyrimidines may be modified. The modified nucleotides may
be
selected from the group consisting of a 3'-terminal deoxy-thymine (dT)
nucleotide, a
2'-0-methyl modified nucleotide, a 2' modified nucleotide, a 2'-deoxy-modified
nucleotide,
a locked nucleotide, an abasic nucleotide, a 2'-amino-modified nucleotide, a
2'-alkyl-modified nucleotide, a morpholino nucleotide, a phosphoramidate, a
non-natural
base comprising nucleotide, a nucleotide comprising a 5'-phosphorothioate
group, a
nucleotide comprising a 5' phosphate or 5' phosphate mimic and a terminal
nucleotide
linked to a cholesteryl derivative or a dodecanoic acid bisdecylamide group.
The nucleic acid may comprise a nucleotide comprising a modified nucleotide,
wherein
the base is selected from 2-aminoadenosine, 2,6-diaminopurine,inosine, pyridin-
4-one,
pyridin-2-one, phenyl, pseudouracil, 2, 4, 6-trimethoxy benzene, 3-methyl
uracil,
dihydrouridine, naphthyl, aminophenyl, 5-alkylcytidine (e.g., 5-
methylcytidine), 5-
alkyluridine (e.g., ribothymidine), 5-halouridine (e.g., 5-bromouridine), 6-
azapyrimidine, 6-
alkylpyrimidine (e.g. 6-methyluridine), propyne, quesosine, 2-thiouridine, 4-
thiouridine,
wybutosine, wybutoxosine, 4-acetylcytidine, 5-(carboxyhydroxymethyl)uridine,
5'-
carboxymethylaminomethy1-2-thiouridine, 5-carboxymethylaminomethyluridine,
beta-D-
galactosylqueosine, 1-methyladenosine, 1-methylinosine, 2,2-dimethylguanosine,
3-
methylcytidine, 2-methyladenosine, 2-methylguanosine, N6-methyladenosine, 7-
methylguanosine, 5-methoxyaminomethy1-2-thiouridine, 5-
methylaminomethyluridine, 5-
methylcarbonylmethyluridine, 5-methyloxyuridine, 5-methyl-2-thiouridine, 2-
methylthio-N6-
isopentenyladenosine, beta-D-mannosylqueosine, uridine-5-oxyacetic acid and 2-
thiocytidine.
Nucleic acids discussed herein include unmodified RNA as well as RNA which has
been
modified, e.g., to improve efficacy, and polymers of nucleoside surrogates.
Unmodified
RNA refers to a molecule in which the components of the nucleic acid, namely
sugars,
bases, and phosphate moieties, are the same or essentially the same as that
which occur
in nature, for example as occur naturally in the human body. Modified
nucleotide as used
herein refers to a nucleotide in which one or more of the components of the
nucleotides,
namely sugars, bases, and phosphate moieties, are different from those which
occur in
nature. While they are referred to as modified nucleotides they will of
course, because of
the modification, the term also includes molecules which are not nucleotides,
for example
a polynucleotide molecule in which the ribophosphate backbone is replaced with
a non-
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
23
ribophosphate construct that allows hybridisation between strands i.e. the
modified
nucleotides mimic the ribophosphate backbone.
Many of the modifications described below that occur within a nucleic acid
will be
repeated within a polynucleotide molecule, such as a modification of a base,
or a
phosphate moiety, or a non-linking 0 of a phosphate moiety. In some cases the
modification will occur at all of the possible positions/nucleotides in the
polynucleotide but
in many cases it will not. A modification may only occur at a 3' or 5'
terminal position, may
only occur in a terminal regions, such as at a position on a terminal
nucleotide or in the
last 2, 3, 4, 5, or 10 nucleotides of a strand. A modification may occur in a
double strand
region, a single strand region, or in both. A modification may occur only in
the double
strand region of a nucleic acid of the invention or may only occur in a single
strand region
of a nucleic acid of the invention. A phosphorothioate modification at a non-
linking 0
position may only occur at one or both termini, may only occur in a terminal
region, e.g., at
a position on a terminal nucleotide or in the last 2, 3, 4 or 5 nucleotides of
a strand, or may
occur in duplex and/or in single strand regions, particularly at termini. The
5' end or 3'
ends may be phosphorylated.
Stability of a nucleic acid of the invention may be increased by including
particular bases
in overhangs, or to include modified nucleotides, in single strand overhangs,
e.g., in a 5' or
3' overhang, or in both. Purine nucleotides may be included in overhangs. All
or some of
the bases in a 3' or 5' overhang may be modified. Modifications can include
the use of
modifications at the 2' OH group of the ribose sugar, the use of
deoxyribonucleotides,
instead of ribonucleotides, and modifications in the phosphate group, such as
phosphorothioate modifications. Overhangs need not be homologous with the
target
sequence.
Nucleases can hydrolyse nucleic acid phosphodiester bonds. However, chemical
modifications to nucleic acids can confer improved properties, and, can render
oligoribonucleotides more stable to nucleases.
Modified nucleic acids, as used herein, can include one or more of:
(i) alteration, e.g., replacement, of one or both of the non-linking phosphate
oxygens
and/or of one or more of the linking phosphate oxygens (referred to as linking
even if at
the 5' and 3' terminus of the nucleic acid of the invention);
(ii) alteration, e.g., replacement, of a constituent of the ribose sugar,
e.g., of the 2'
hydroxyl on the ribose sugar;
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
24
(iii) replacement of the phosphate moiety with "dephospho" linkers;
(iv) modification or replacement of a naturally occurring base;
(v) replacement or modification of the ribose-phosphate backbone;
(vi) modification of the 3' end or 5' end of the RNA, e.g., removal,
modification or
replacement of a terminal phosphate group or conjugation of a moiety, e.g., a
fluorescently labelled moiety, to either the 3' or 5' end of RNA.
The terms replacement, modification, alteration, indicate a difference from a
naturally
occurring molecule.
Specific modifications are discussed in more detail below.
Examples of modified phosphate groups include phosphorothioate,
phosphoroselenates,
borano phosphates, borano phosphate esters, hydrogen phosphonates,
phosphoroamidates, alkyl or aryl phosphonates and phosphotriesters.
Phosphorodithioates have both non-linking oxygens replaced by sulphur. One,
each or
both non-linking oxygens in the phosphate group can be independently any one
of S, Se,
B, C, H, N, or OR (R is alkyl or aryl).
The phosphate linker can also be modified by replacement of a linking oxygen
with
nitrogen (bridged phosphoroamidates), sulfur (bridged phosphorothioates) and
carbon
(bridged methylenephosphonates). The replacement can occur at a terminal
oxygen.
Replacement of the non-linking oxygens with nitrogen is possible.
A modified nucleotide can include modification of the sugar groups. The 2'
hydroxyl group
(OH) can be modified or replaced with a number of different "oxy" or "deoxy"
substituents.
Examples of "oxy"-2' hydroxyl group modifications include alkoxy or aryloxy
(OR, e.g.,
R=H, alkyl, cycloalkyl, aryl, aralkyl, heteroaryl or sugar);
polyethyleneglycols (PEG),
0(CH2CH20)nCH2CH2OR; "locked" nucleic acids (LNA) in which the 2' hydroxyl is
connected, e.g., by a methylene bridge, to the 4' carbon of the same ribose
sugar; 0-
AMINE (AMINE=NH2; alkylamino, dialkylamino, heterocyclyl, arylamino, diaryl
amino,
heteroaryl amino, or diheteroaryl amino, ethylene diamine, polyamino) and
aminoalkoxy,
0(CH2)nAMINE, (e.g., AMINE=NH2; alkylamino, dialkylamino, heterocyclyl,
arylamino,
diaryl amino, heteroaryl amino, or diheteroaryl amino, ethylene diamine,
polyamino).
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
"Deoxy" modifications include hydrogen, halogen, amino (e.g., NH2; alkylamino,
dialkylamino, heterocyclyl, arylamino, diaryl amino, heteroaryl amino,
diheteroaryl amino,
or amino acid); NH(CH2CH2NH)nCH2CH2-AMINE (AMINE=NH2; alkylamino,
dialkylamino, heterocyclyl, arylamino, diaryl amino, heteroaryl amino, or
diheteroaryl
5 amino), ¨NHC(0)R (R=alkyl, cycloalkyl, aryl, aralkyl, heteroaryl or
sugar), cyano;
mercapto; alkyl-thio-alkyl; thioalkoxy; and alkyl, cycloalkyl, aryl, alkenyl
and alkynyl, which
may be optionally substituted with e.g., an amino functionality. Other
substitutents of
certain embodiments include 2'-methoxyethyl, 2'-OCH3, 2'-0-allyl, 2'-C-allyl,
and 2'-fluoro.
10 The sugar group can also contain one or more carbons that possess the
opposite
stereochemical configuration than that of the corresponding carbon in ribose.
Thus, a
modified nucleotides may contain a sugar such as arabinose.
Modified nucleotides can also include "abasic" sugars, which lack a nucleobase
at C-1'.
15 These abasic sugars can further contain modifications at one or more of
the constituent
sugar atoms.
The 2' modifications may be used in combination with one or more phosphate
linker
modifications (e.g., phosphorothioate).
The phosphate groups can individually be replaced by non-phosphorus containing
connectors.
Examples of moieties which can replace the phosphate group include siloxane,
carbonate,
carboxymethyl, carbamate, amide, thioether, ethylene oxide linker, sulfonate,
sulfonamide, thioformacetal, formacetal, oxime, methyleneimino,
methylenemethylimino,
methylenehydrazo, methylenedimethylhydrazo and methyleneoxymethylimino. In
certain
embodiments, replacements may include the methylenecarbonylamino and
methylenemethylimino groups.
The phosphate linker and ribose sugar may be replaced by nuclease resistant
nucleotides.
Examples include the morpholino, cyclobutyl, pyrrolidine and peptide nucleic
acid (PNA)
nucleoside surrogates. In certain embodiments, PNA surrogates may be used.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
26
The 3' and 5' ends of an oligonucleotide can be modified. Such modifications
can be at
the 3' end or the 5' end or both ends of the molecule. They can include
modification or
replacement of an entire terminal phosphate or of one or more of the atoms of
the
phosphate group. For example, the 3' and 5' ends of an oligonucleotide can be
conjugated
to other functional molecular entities such as labelling moieties, e.g.,
fluorophores (e.g.,
pyrene, TAMRA, fluorescein, Cy3 or Cy5 dyes) or protecting groups (based e.g.,
on sulfur,
silicon, boron or ester). The functional molecular entities can be attached to
the sugar
through a phosphate group and/or a linker. The terminal atom of the linker can
connect to
or replace the linking atom of the phosphate group or the C-3' or C-5' 0, N, S
or C group
of the sugar. Alternatively, the linker can connect to or replace the terminal
atom of a
nucleotide surrogate (e.g., PNAs). These spacers or linkers can include e.g.,
¨(CH2)n¨,
¨(CH2)nN¨, ¨(CH2)n0¨, ¨(CH2)nS¨, ¨ (CH2CH20)nCH2CH20¨ (e.g., n=3 or 6),
abasic sugars, amide, carboxy, amine, oxyamine, oxyimine, thioether,
disulfide, thiourea,
sulfonamide, or morpholino, or biotin and fluorescein reagents. The 3' end can
be an -
OH group.
Other examples of terminal modifications include dyes, intercalating agents
(e.g.,
acridines), cross-linkers (e.g., psoralene, mitomycin C), porphyrins (TPPC4,
texaphyrin,
Sapphyrin), polycyclic aromatic hydrocarbons (e.g., phenazine,
dihydrophenazine),
artificial endonucleases, EDTA, lipophilic carriers (e.g., cholesterol, cholic
acid,
adamantane acetic acid, 1-pyrene butyric acid, dihydrotestosterone, 1,3-Bis-
0(hexadecyl)glycerol, geranyloxyhexyl group, hexadecylglycerol, borneol,
menthol, 1,3-
propanediol, heptadecyl group, palmitic acid, myristic acid, 03-
(oleoyl)lithocholic acid, 03-
(oleoyl)cholenic acid, dimethoxytrityl, or phenoxazine) and peptide conjugates
(e.g.,
antennapedia peptide, Tat peptide), alkylating agents, phosphate, amino,
mercapto, PEG
(e.g., PEG-40K), MPEG, [MPEG]2, polyamino, alkyl, substituted alkyl,
radiolabeled
markers, enzymes, haptens (e.g., biotin), transport/absorption facilitators
(e.g., aspirin,
vitamin E, folic acid), synthetic ribonucleases (e.g., imidazole,
bisimidazole, histamine,
imidazole clusters, acridine-imidazole conjugates, Eu3+ complexes of
tetraazamacrocycles).
Terminal modifications can be added for a number of reasons, including to
modulate
activity or to modulate resistance to degradation. Terminal modifications
useful for
modulating activity include modification of the 5' end with phosphate or
phosphate
analogues. Nucleic acids of the invention, on the first or second strand, may
be 5'
phosphorylated or include a phosphoryl analogue at the 5' prime terminus. 5'-
phosphate
modifications include those which are compatible with RISC mediated gene
silencing.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
27
Suitable modifications include: 5'-monophosphate ((H0)2(0)P-0-5'); 5'-
diphosphate
((H0)2(0)P¨O¨P(H0)(0)-0-5'); 5'-triphosphate ((H0)2(0)P-0¨(H0)(0)P¨O¨
P(H0)(0)-0-5'); 5'-guanosine cap (7-methylated or non-methylated) (7m-G-0-5'-
(H0)(0)P-0¨(H0)(0)P¨O¨P(H0)(0)-0-5'); 5'-adenosine cap (Appp), and any
modified or unmodified nucleotide cap structure (N-0-5'-(H0)(0)P-0¨(H0)(0)P¨O¨
P(H0)(0)-0-5'); 5'-monothiophosphate (phosphorothioate; (H0)2(S)P-0-5'); 5'-
monodithiophosphate (phosphorodithioate; (H0)(HS)(S)P-0-5'), 5'-
phosphorothiolate
((H0)2(0)P¨S-5'); any additional combination of oxygen/sulfur replaced
monophosphate,
diphosphate and triphosphates (e.g., 5'-alpha-thiotriphosphate, 5'-gamma-
thiotriphosphate, etc.), 5'-phosphoramidates ((H0)2(0)P¨NH-5', (H0)(NH2)(0)P-0-
5'),
5'-alkylphosphonates (R=alkyl=methyl, ethyl, isopropyl, propyl, etc., e.g.,
RP(OH)(0)-0-
5'-, (OH)2(0)P-5'-CH2-), 51vinylphosphonate, 5'-alkyletherphosphonates
(R=alkylether=methoxymethyl (MeOCH2-), ethoxymethyl, etc., e.g., RP(OH)(0)-0-
5'-).
The nucleic acid of the present invention may include one or more
phosphorothioate
modifications on one or more of the terminal ends of the first and/or the
second strand.
Optionally, each or either end of the first strand may comprise one or two or
three
phosphorothioate modified nucleotides. Optionally, each or either end of the
second
strand may comprise one or two or three phosphorothioate modified nucleotides.
Terminal modifications can also be useful for monitoring distribution, and in
such cases
the groups to be added may include fluorophores, e.g., fluorescein or an Alexa
dye.
Terminal modifications can also be useful for enhancing uptake, useful
modifications for
this include cholesterol. Terminal modifications can also be useful for cross-
linking an
RNA agent to another moiety.
Adenine, guanine, cytosine and uracil are the most common bases found in RNA.
These
bases can be modified or replaced to provide RNAs having improved properties.
E.g.,
nuclease resistant oligoribonucleotides can be prepared with these bases or
with synthetic
and natural nucleobases (e.g., inosine, thymine, xanthine, hypoxanthine,
nubularine,
isoguanisine, or tubercidine) and any one of the above modifications.
Alternatively,
substituted or modified analogues of any of the above bases and "universal
bases" can be
employed. Examples include 2-aminoadenine, 6-methyl and other alkyl
derivatives of
adenine and guanine, 2-propyl and other alkyl derivatives of adenine and
guanine, 5-
halouracil and cytosine, 5-propynyl uracil and cytosine, 6-azo uracil,
cytosine and thymine,
5-uracil (pseudouracil), 4-thiouracil, 5-halouracil, 5-(2-aminopropyl)uracil,
5-amino ally!
uracil, 8-halo, amino, thiol, thioalkyl, hydroxyl and other 8-substituted
adenines and
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
28
guanines, 5-trifluoromethyl and other 5-substituted uracils and cytosines, 7-
methylguanine, 5-substituted pyrimidines, 6-azapyrimidines and N-2, N-6 and 0-
6
substituted purines, including 2-aminopropyladenine, 5-propynyluracil and 5-
propynylcytosine, dihydrouracil, 3-deaza-5-azacytosine, 2-aminopurine, 5-
alkyluracil, 7-
alkylguanine, 5-alkyl cytosine, 7-deazaadenine, N6,N6-dimethyladenine, 2,6-
diaminopurine, 5-amino-allyl-uracil, N3-methyluracil, substituted 1,2,4-
triazoles, 2-
pyridinone, 5-nitroindole, 3-nitropyrrole, 5-methoxyuracil, uracil-5-oxyacetic
acid, 5-
methoxycarbonylmethyluracil, 5-methyl-2-thiouracil, 5-methoxycarbonylmethy1-2-
thiouracil, 5-methylaminomethy1-2-thiouracil, 3-(3-amino-3-
carboxypropyl)uracil, 3-
methylcytosine, 5-methylcytosine, N<4>-acetyl cytosine, 2-thiocytosine, N6-
methyladenine, N6-isopentyladenine, 2-methylthio-N6-isopentenyladenine, N-
methylguanines, or 0-alkylated bases.
As used herein, the terms "non-pairing nucleotide analogue" means a nucleotide
analogue
which includes a non-base pairing moiety including but not limited to: 6 des
amino
adenosine (Nebularine), 4-Me-indole, 3-nitropyrrole, 5-nitroindole, Ds, Pa, N3-
Me ribo U,
N3-Me riboT, N3-Me dC, N3-Me-dT, N1-Me-dG, N1-Me-dA, N3-ethyl-dC, N3-Me dC. In
some embodiments the non-base pairing nucleotide analogue is a ribonucleotide.
In other
embodiments it is a deoxyribonucleotide.
As used herein, the term, "terminal functional group" includes without
limitation a halogen,
alcohol, amine, carboxylic, ester, amide, aldehyde, ketone, ether groups.
Certain moieties may be linked to the 5' terminus of the first strand or the
second strand.
These include abasic ribose moiety, abasic deoxyribose moiety, modifications
abasic
ribose and abasic deoxyribose moieties including 2' 0 alkyl modifications;
inverted abasic
ribose and abasic deoxyribose moieties and modifications thereof, C6-imino-Pi;
a mirror
nucleotide including L-DNA and L-RNA; 5'0Me nucleotide; and nucleotide
analogues
including 4',5'-methylene nucleotide; 1-(6-D-erythrofuranosyl)nucleotide; 4'-
thio nucleotide,
carbocyclic nucleotide; 5'-amino-alkyl phosphate; 1,3-diamino-2-propyl
phosphate, 3-
aminopropyl phosphate; 6-aminohexyl phosphate; 12-aminododecyl phosphate;
hydroxypropyl phosphate; 1,5-anhydrohexitol nucleotide; alpha-nucleotide;
threo-
pentofuranosyl nucleotide; acyclic 3',4'-seco nucleotide; 3,4-dihydroxybutyl
nucleotide;
3,5-dihydroxypentyl nucleotide, 5'-5'-inverted abasic moiety; 1,4-butanediol
phosphate; 5'-
amino; and bridging or non-bridging methylphosphonate and 5'-mercapto
moieties.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
29
The nucleic acids of the invention may include one or more inverted
nucleotides, for
example inverted thymidine or inverted adenine (for example see Takei, et al.,
2002. JBC
277 (26):23800-06).
As used herein, the term "inhibit", "down-regulate", or "reduce" with respect
to gene
expression means the expression of the gene, or level of RNA molecules or
equivalent
RNA molecules encoding one or more proteins or protein subunits (e.g., mRNA),
or
activity of one or more proteins or protein subunits, is reduced below that
observed in the
absence of the nucleic acid or conjugated nucleic acid of the invention or in
reference to
an siRNA molecule with no known homology to human transcripts (herein termed
non-
silencing control). Such control may be conjugated and modified in an
analogous manner
to the molecule of the invention and delivered into the target cell by the
same route; for
example the expression may be reduced to 90%, 80%, 70%, 60%, 50%, 40%, 30%,
20%,
15%, or to intermediate values, or less than that observed in the absence of
the nucleic
acid or conjugated nucleic acid or in the presence of a non-silencing control.
The nucleic acid of the present invention may comprise an abasic nucleotide.
The term
"abasic" as used herein, refers to moieties lacking a base or having other
chemical groups
in place of a base at the 1' position, for example a 3',3'-linked or 5',5'-
linked deoxyabasic
ribose derivative.
The nucleic acid may comprise one or more nucleotides on the second and/or
first strands
that are modified. Alternating nucleotides may be modified, to form modified
nucleotides.
Alternating as described herein means to occur one after another in a regular
way. In
other words, alternating means to occur in turn repeatedly. For example if one
nucleotide
is modified, the next contiguous nucleotide is not modified and the following
contiguous
nucleotide is modified and so on. One nucleotide may be modified with a first
modification,
the next contiguous nucleotide may be modified with a second modification and
the
following contiguous nucleotide is modified with the first modification and so
on, where the
first and second modifications are different.
One or more of the odd numbered nucleotides of the first strand of the nucleic
acid of the
invention may be modified wherein the first strand is numbered 5' to 3', the
5'-most
nucleotide being nucleotide number 1 of the first strand. The term "odd
numbered" as
described herein means a number not divisible by two. Examples of odd numbers
are 1,
3, 5, 7, 9, 11 and so on. One or more of the even numbered nucleotides of the
first strand
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
of the nucleic acid of the invention may be modified, wherein the first strand
is numbered
5' to 3'. The term "even numbered" as described herein means a number which is
evenly
divisible by two. Examples of even numbers are 2, 4, 6, 8, 10, 12, 14 and so
on. One or
more of the odd numbered nucleotides of the second strand of the nucleic acid
of the
5 invention may be modified wherein the second strand is numbered 3' to 5',
the 3'-most
nucleotide being nucleotide number 1 of the second strand. One or more of the
even
numbered nucleotides of the second strand of the nucleic acid of the invention
may be
modified, wherein the second strand is numbered 3' to 5'.
10 One or more nucleotides on the first and/or second strand may be
modified, to form
modified nucleotides. One or more of the odd numbered nucleotides of the first
strand
may be modified. One or more of the even numbered nucleotides of the first
strand may
be modified by at least a second modification, wherein the at least second
modification is
different from the modification on the one or more odd nucleotides. At least
one of the one
15 or more modified even numbered nucleotides may be adjacent to at least
one of the one
or more modified odd numbered nucleotides.
A plurality of odd numbered nucleotides in the first strand may be modified in
the nucleic
acid of the invention. A plurality of even numbered nucleotides in the first
strand may be
20 modified by a second modification. The first strand may comprise
adjacent nucleotides
that are modified by a common modification. The first strand may also comprise
adjacent
nucleotides that are modified by a second different modification.
One or more of the odd numbered nucleotides of the second strand may be
modified by a
25 modification that is different to the modification of the odd numbered
nucleotides on the
first strand and/or one or more of the even numbered nucleotides of the second
strand
may be modified by the same modification of the odd numbered nucleotides of
the first
strand. At least one of the one or more modified even numbered nucleotides of
the
second strand may be adjacent to the one or more modified odd numbered
nucleotides. A
30 plurality of odd numbered nucleotides of the second strand may be
modified by a common
modification and/or a plurality of even numbered nucleotides may be modified
by the
same modification that is present on the first stand odd numbered nucleotides.
A plurality
of odd numbered nucleotides on the second strand may be modified by a second
modification, wherein the second modification is different from the
modification of the first
strand odd numbered nucleotides.
The second strand may comprise adjacent nucleotides that are modified by a
common
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
31
modification, which may be a second modification that is different from the
modification of
the odd numbered nucleotides of the first strand.
In the nucleic acid of the invention, each of the odd numbered nucleotides in
the first
strand and each of the even numbered nucleotides in the second strand may be
modified
with a common modification and, each of the even numbered nucleotides may be
modified in the first strand with a second modification and each of the odd
numbered
nucleotides may be modified in the second strand with the second modification.
The nucleic acid of the invention may have the modified nucleotides of the
first strand
shifted by at least one nucleotide relative to the unmodified or differently
modified
nucleotides of the second strand.
One or more or each of the odd numbered nucleotides may be modified in the
first strand
and one or more or each of the even numbered nucleotides may be modified in
the
second strand. One or more or each of the alternating nucleotides on either or
both
strands may be modified by a second modification. One or more or each of the
even
numbered nucleotides may be modified in the first strand and one or more or
each of the
even numbered nucleotides may be modified in the second strand. One or more or
each
of the alternating nucleotides on either or both strands may be modified by a
second
modification. One or more or each of the odd numbered nucleotides may be
modified in
the first strand and one or more of the odd numbered nucleotides may be
modified in the
second strand by a common modification. One or more or each of the alternating
nucleotides on either or both strands may be modified by a second
modification. One or
more or each of the even numbered nucleotides may be modified in the first
strand and
one or more or each of the odd numbered nucleotides may be modified in the
second
strand by a common modification. One or more or each of the alternating
nucleotides on
either or both strands may be modified by a second modification.
The nucleic acid of the invention may comprise single or double stranded
constructs that
comprise at least two regions of alternating modifications in one or both of
the strands.
These alternating regions can comprise up to about 12 nucleotides but
preferably
comprise from about 3 to about 10 nucleotides. The regions of alternating
nucleotides
may be located at the termini of one or both strands of the nucleic acid of
the invention.
The nucleic acid may comprise from 4 to about 10 nucleotides of alternating
nucleotides
at each termini (3' and 5') and these regions may be separated by from about 5
to about
12 contiguous unmodified or differently or commonly modified nucleotides.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
32
The odd numbered nucleotides of the first strand may be modified and the even
numbered nucleotides may be modified with a second modification. The second
strand
may comprise adjacent nucleotides that are modified with a common
modification, which
may be the same as the modification of the odd numbered nucleotides of the
first strand.
One or more nucleotides of second strand may also be modified with the second
modification. One or more nucleotides with the second modification may be
adjacent to
each other and to nucleotides having a modification that is the same as the
modification of
the odd numbered nucleotides of the first strand. The first strand may also
comprise
phosphorothioate linkages between the two nucleotides at the 3' end and at the
5' end.
The second strand may comprise a phosphorothioate linkage between the two
nucleotides at 5' end. The second strand may also be conjugated to a ligand at
the 5' end.
The nucleic acid of the invention may comprise a first strand comprising
adjacent
nucleotides that are modified with a common modification. One or more of such
nucleotides may be adjacent to one or more nucleotides which may be modified
with a
second modification. One or more nucleotides with the second modification may
be
adjacent. The second strand may comprise adjacent nucleotides that are
modified with a
common modification, which may be the same as one of the modifications of one
or more
nucleotides of the first strand. One or more nucleotides of second strand may
also be
modified with the second modification. One or more nucleotides with the second
modification may be adjacent. The first strand may also comprise
phosphorothioate
linkages between the two nucleotides at the 5' end and at the 3' end. The
second strand
may comprise a phosphorothioate linkage between the two nucleotides at the 3'
end. The
second strand may also be conjugated to a ligand at the 5' end.
The nucleotides numbered from 5' to 3' on the first strand and 3' to 5' on the
second
strand, 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23 and 25 may be modified by a
modification
on the first strand. The nucleotides numbered 2, 4, 6, 8, 10, 12, 14, 16, 18,
20, 22 and 24
may be modified by a second modification on the first strand. The nucleotides
numbered
1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23 may be modified by a modification on
the second
strand. The nucleotides numbered 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22 and 24
may be
modified by a second modification on the second strand. Nucleotides are
numbered for
the sake of the nucleic acid of the present invention from 5' to 3' on the
first strand and 3'
to 5' on the second strand
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
33
The nucleotides numbered 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22 and 24 may be
modified
by a modification on the first strand. The nucleotides numbered 1, 3, 5, 7, 9,
11, 13, 15,
17, 19, 21,23 may be modified by a second modification on the first strand.
The
nucleotides numbered 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23 may be modified
by a
modification on the second strand. The nucleotides numbered 2, 4, 6, 8, 10,
12, 14, 16,
18, 20, 22 and 24 may be modified by a second modification on the second
strand.
Clearly, if the first and/or the second strand are shorter than 25 nucleotides
in length, such
as 19 nucleotides in length, there are no nucleotides numbered 20, 21, 22, 23,
24 and 25
to be modified. The skilled person understands the description above to apply
to shorter
strands, accordingly.
One or more modified nucleotides on the first strand may be paired with
modified
nucleotides on the second strand having a common modification. One or more
modified
nucleotides on the first strand may be paired with modified nucleotides on the
second
strand having a different modification. One or more modified nucleotides on
the first strand
may be paired with unmodified nucleotides on the second strand. One or more
modified
nucleotides on the second strand may be paired with unmodified nucleotides on
the first
strand. In other words, the alternating nucleotides can be aligned on the two
strands such
as, for example, all the modifications in the alternating regions of the
second strand are
paired with identical modifications in the first strand or alternatively the
modifications can
be offset by one nucleotide with the common modifications in the alternating
regions of
one strand pairing with dissimilar modifications (i.e. a second or further
modification) in the
other strand. Another option is to have dissimilar modifications in each of
the strands.
The modifications on the first strand may be shifted by one nucleotide
relative to the
modified nucleotides on the second strand, such that common modified
nucleotides are
not paired with each other.
The modification and/or modifications may each and individually be selected
from the
group consisting of 3'-terminal deoxy-thymine, 2'-0-methyl, a 2'-deoxy-
modification, a
2'-amino-modification, a 2'-alkyl-modification, a morpholino modification, a
phosphoramidate modification, 5'-phosphorothioate group modification, a 5'
phosphate or
5' phosphate mimic modification and a cholesteryl derivative or a dodecanoic
acid
bisdecylamide group modification and/or the modified nucleotide may be any one
of a
locked nucleotide, an abasic nucleotide or a non-natural base comprising
nucleotide.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
34
At least one modification may be 2'-0-methyl and/or at least one modification
may be 2'-F.
Further modifications as described herein may be present on the first and/or
second
strand.
The nucleic acid of the invention may comprise an inverted RNA nucleotide at
one or
several of the strand ends. Such inverted nucleotides provide stability to the
nucleic acid.
Preferably, the nucleic acid comprises at least an inverted nucleotide at one
or several of
the 3' end of at least one of the strands and/or at the 5' end of the of the
second strand.
More preferably, the nucleic acid comprises an inverted nucleotde at the 3'
end of the
second strand. Most preferably, the nucleic acid comprises an inverted RNA
nucleotide at
the 3' end of the second strand and this nucleotide is preferably an inverted
A. The
inverted nucleotide is preferably present at an end of a strand not as an
overhang but
opposite a corresponding nucleotide in the other strand. A nucleic acid with
such a
modification is stable and easy to synthesise.
Throughout the description of the invention, "same or common modification"
means the
same modification to any nucleotide, be that A, G, C or U modified with a
group such as a
methyl group or a fluoro group. Is it not taken to mean the same addition on
the same
nucleotide. For example, 2"F-dU, 2"F-dA, 2"F-dC, 2"F-dG are all considered to
be the
same or common modification, as are 2'-0Me-rU, 2'-0Me-rA; 2'-0Me-rC; 2'-0Me-
rG. A
2'F modification is a different modification to a 2'0Me modification.
Some representative modified nucleic acid sequences of the present invention
are shown
in the examples. These examples are meant to be representative and not
limiting.
Preferably, the nucleic acid may comprise a modification and a second or
further
modification which are each and individually selected from the group
comprising 2'-0-
methyl modification and 2'-F modification. The nucleic acid may comprise a
modification
that is 2'-0-methyl (2'0Me) that may be a first modification, and a second
modification that
is 2'-F. The nucleic acid of the invention may also include a phosphorothioate
modification
and/or a deoxy modification which may be present in or between the terminal 1,
2 or 3
nucleotides of each or any end of each or both strands.
The invention provides as a further aspect, a nucleic acid for inhibiting
expression of
ALDH2 in a cell, comprising a nucleotide sequence of SEQ ID NO:1, 3, 5, 7, 9,
11, 13, 15,
17, 19, 21, 23, 25 or 27, wherein the nucleotides of first strand are modified
by a first
modification on the odd numbered nucleotides, and modified by a second
modification on
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
the even numbered nucleotides, and nucleotides of the second strand are
modified by a
third modification on the even numbered nucleotides and modified by a fourth
modification
the odd numbered nucleotides, wherein at least the first modification is
different to the
second modification and the third modification is different to the fourth
modification. The
5 third and first modifications may be the same or different, the second
and fourth
modifications may be the same or different. The first and second modifications
may be
different to each other and the third and fourth modifications may be
different to each
other.
10 The second strand may comprise a nucleotide sequence of SEQ ID NO:2, 4,
6, 8, 10, 12,
14, 16, 18, 20, 22, 24, 26 or 28. The nucleotides of the first strand may be
modified by a
first modification on the odd numbered nucleotides, and modified with a second
modification on the even numbered nucleotides, and the second strand may be
modified
on the odd numbered nucleotides with the second modification and modified with
the first
15 modification on the even numbered nucleotides. The first modification
may be 2'0Me and
the second modification may be 2' F. The first strand may comprise the
nucleotide
sequence of SEQ ID NO:3, SEQ ID NO:11 or SEQ ID NO:17 and/or the second strand
may comprise the nucleotide sequence of SEQ ID NO:4, SEQ ID NO:12 or SEQ ID
NO:18. The modifications may be those as set out in Table 1.
The nucleic acid of the invention may be conjugated to a ligand. Efficient
delivery of
oligonucleotides, in particular double stranded nucleic acids of the
invention, to cells in
vivo is important and requires specific targeting and substantial protection
from the
extracellular environment, particularly serum proteins. One method of
achieving specific
targeting is to conjugate a ligand to the nucleic acid. The ligand helps in
targeting the
nucleic acid to the required target site. There is a need to conjugate
appropriate ligands
for the desired receptor molecules in order for the conjugated molecules to be
taken up by
the target cells by mechanisms such as different receptor-mediated endocytosis
pathways
or functionally analogous processes.
One example is the asialoglycoprotein receptor complex (ASGP-R) composed by
varying
ratios of multimers of membrane ASGR1 and ASGR2 receptors, which is highly
abundant
on hepatocytes and has high affinity to the here described GaINAc moiety. One
of the first
disclosures of the use of triantennary cluster glycosides as conjugated
ligands was in US
patent number US 5,885,968. Conjugates having three GaINAc ligands and
comprising
phosphate groups are known and are described in Dubber et al. (Bioconjug.
Chem. 2003
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
36
Jan-Feb;14(1):239-46.). The ASGP-R complex shows a 50-fold higher affinity for
N-
Acetyl-D-Galactosylamine (GaINAc) than D-Gal.
The asialoglycoprotein receptor complex (ASGP-R), which recognizes
specifically terminal
8-galactosyl subunits of glycosylated proteins or other oligosaccharides
(Weigel, P.H. et.
al., Biochim. Biophys. Acta. 2002 Sep 19;1572(2-3):341-63) can be used for
delivering a
drug to the liver's hepatocytes expressing the receptor complex by covalent
coupling of
galactose or galactosamine to the drug substance (Ishibashi,S.; et. al., J
Biol. Chem. 1994
Nov 11;269(45):27803-6). Furthermore the binding affinity can be significantly
increased
by the multi-valency effect, which is achieved by the repetition of the
targeting moiety
(Biessen EA, et al., J Med Chem. 1995 Apr 28;38(9):1538-46).
The ASGP-R complex is a mediator for an active uptake of terminal 8-galactosyl
containing glycoproteins to the cell's endosomes. Thus, the ASGPR is highly
suitable for
targeted delivery of drug candidates conjugated to such ligands like, e.g.,
nucleic acids
into receptor-expressing cells (Akinc et al., Mol Ther. 2010 Jul;18(7):1357-
64).
More generally the ligand can comprise a saccharide that is selected to have
an affinity for
at least one type of receptor on a target cell. In particular, the receptor is
on the surface of
a mammalian liver cell, for example, the hepatic asialoglycoprotein receptor
complex
described before (ASGP-R).
The saccharide may be selected from N-acetyl galactosamine, mannose,
galactose,
glucose, glucosamine and fucose. The saccharide may be N-acetyl galactosamine
(GaINAc).
A ligand for use in the present invention may therefore comprise (i) one or
more N-acetyl
galactosamine (GaINAc) moieties and derivatives thereof, and (ii) a linker,
wherein the
linker conjugates the GaINAc moieties to a sequence as defined in any
preceding
aspects. The linker may be a bivalent or trivalent or tetravalent branched
structure. The
nucleotides may be modified as defined herein.
"GaINAc" refers to 2-(Acetylamino)-2-deoxy-D- galactopyranose, commonly
referred to in
the literature as N-acetyl galactosamine. Reference to "GaINAc" or "N-acetyl
galactosamine" includes both the 13- form: 2-(Acetylamino)-2-deoxy-8 -D-
galactopyranose
and the a-form: 2-(Acetylamino)-2-deoxy-a-D- galactopyranose. Both the 13-
form: 2-
(Acetylarnino)-2-deoxy-8-D-galactopyranose and a-form: 2-(Acetylamino)-2-deoxy-
a-D-
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
37
galactopyranose may be used interchangeably. Preferably, the compounds of the
invention comprise the 13-form, 2-(Acetylarnino)-2-deoxy-13-D-galactopyranose.
The ligand may therefore comprise GaINAc.
The ligand may comprise a compound of formula I:
[S-X1-P-X93-A-X3- (I)
wherein:
S represents a saccharide, wherein the saccharide is N-acetyl galactosamine;
X1 represents C3-C6 alkylene or (-CH2-CH2-0)m(-CH2)2- wherein m is 1, 2, or 3;
P is a phosphate or modified phosphate (preferably a thiophosphate);
X2 is alkylene or an alkylene ether of the formula (-CH2)n-O-CH2- where n = 1-
6;
A is a branching unit;
X3 represents a bridging unit;
wherein a nucleic acid according to the present invention is conjugated to X3
via a
phosphate or modified phosphate (preferably a thiophosphate).
In formula I, branching unit "A" branches into three in order to accommodate
the three
saccharide ligands. The branching unit is covalently attached to the remaining
tethered
portions of the ligand and the nucleic acid. The branching unit may comprise a
branched
aliphatic group comprising groups selected from alkyl, amide, disulphide,
polyethylene
glycol, ether, thioether and hydroxyamino groups. The branching unit may
comprise groups
selected from alkyl and ether groups.
The branching unit A may have a structure selected from:
I I
Al.._ -4,. At
4in All
A1,..,....¨Al/
Ai 1
/ and I
¨....,õ
wherein each A1 independently represents 0, S, C=0 or NH; and
each n independently represents an integer from 1 to 20.
The branching unit may have a structure selected from:
CA 03118327 2021-04-30
WO 2019/092282
PCT/EP2018/081105
38
Ad
1-4 A1
41 1:in FAJ! )nf , (iA=A
n
¨ A and
)0
wherein each A1 independently represents 0, S, C=0 or NH; and
each n independently represents an integer from 1 to 20.
The branching unit may have a structure selected from:
rris siss
)n 111 1ln 1%.
111.1: n n and n n
wherein A1 is 0, S, C=0 or NH; and
each n independently represents an integer from 1 to 20.
The branching unit may have the structure:
0, -7,
The branching unit may have the structure:
0
An".
=
The branching unit may have the structure:
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
39
/
\
_______________ /1\
\ss
s'
Optionally, the branching unit consists of only a carbon atom.
The "X3" portion is a bridging unit. The bridging unit is linear and is
covalently bound to the
branching unit and the nucleic acid.
X3 may be selected from -C1-C20 alkylene-, -C2-C20 alkenylene-, an alkylene
ether of formula
-(C1-C20 alkylene)-0¨(Ci-C20 alkylene)-, -C(0)-C1-C20 alkylene-, -Co-Ca
alkylene(Cy)Co-Ca
alkylene- wherein Cy represents a substituted or unsubstituted 5 or 6 membered
cycloalkylene, arylene, heterocyclylene or heteroarylene ring, -Ci-Ca alkylene-
NHC(0)-Ci-
C4 alkylene-, -Ci-Ca alkylene-C(0)NH-C1-C4 alkylene-, -Ci-Ca alkylene-SC(0)-Ci-
C4
alkylene-, -Ci-Ca alkylene-C(0)S-Ci-C4 alkylene-, -Ci-Ca alkylene-OC(0)-Ci-C4
alkylene-,
-Ci-Ca alkylene-C(0)0-Ci-C4 alkylene-, and -C1-C6 alkylene-S-S-Ci-Co alkylene-
.
X3 may be an alkylene ether of formula -(C1-C20 alkylene)-0¨(Ci-C20 alkylene)-
. X3 may be
an alkylene ether of formula -(C1-C20 alkylene)-0¨(C4-C20 alkylene)-, wherein
said (C4-C20
alkylene) is linked to Z. X3 may be selected from the group consisting of -CH2-
0-C3H6-, -
CH2-0-C4F-18-, -CH2-0-C6F-112- and -CH2-0-C8H16-, especially -CH2-0-C4F-18-, -
CH2-0-C6H12-
and -CH2-0-C81-116-, wherein in each case the -CH2- group is linked to A.
The ligand may comprise a compound of formula (II):
[S-X1-P-X2]3-A-X3- (I I )
wherein:
S represents a saccharide;
X1 represents C3-C6 alkylene or (-CH2-CH2-0)m(-CH2)2- wherein m is 1, 2, or 3;
P is a phosphate or modified phosphate (preferably a thiophosphate);
X2 is Ci-Co alkylene;
A is a branching unit selected from:
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
K
Al n )
n Al AlyA¨Al
n n
Al Al Al A2-1
Al = 0, NH Al = 0, NH A2 = NH, CH2, 0
n = 1 to 4 n = 1 to 4
X3 is a bridging unit;
wherein a nucleic acid according to the present invention is conjugated to
X3via a
phosphate or modified phosphate (preferably a thiophosphate)
5
Branching unit A may have the structure:
AoxoN
0 0
Branching unit A may have the structure:
-40¨v-0N
0 __ /IIN A
X , wherein X3 is attached to the nitrogen atom.
X3 may be C1-C20 alkylene. Preferably, X3 is selected from the group
consisting of -C3H6-, -
C4H8-,-C6F-112- and -C81-116-, especially -C4H8-, -C6F-112- and -C81-116-=
The ligand may comprise a compound of formula (Ill):
[S-X1-P-X93-A-X3- (Ill)
wherein:
S represents a saccharide;
X1 represents C3-C6 alkylene or (-CH2-CH2-0)m(-CH2)2- wherein m is 1, 2, or 3;
P is a phosphate or modified phosphate (preferably a thiophosphate);
X2 is an alkylene ether of formula -C3H6-0-CH2-;
A is a branching unit;
X3 is an alkylene ether of formula selected from the group consisting of -CH2-
0-CH2-, -
CH2-0-C2H4-, -CH2-0-C3H6-, -CH2-0-C4H8-, -CH2-0-C6H10-, -CH2-0-C6F-112-, -CH2-
0-
C7I-114-, and -CH2-0-C81-116-, wherein in each case the -CH2- group is linked
to A,
and wherein X3 is conjugated to a nucleic acid according to the present
invention by a
phosphate or modified phosphate (preferably a thiophosphate).
The branching unit may comprise carbon. Preferably, the branching unit is
carbon.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
41
X3 may be selected from the group consisting of -CH2-0-C4H8-, -CH2-0-05H10-, -
CH2-0-
C6H12-, -CH2-0-C7H14-, and -CH2-0-C8H16-. Preferably, X3 is selected from the
group
consisting of -CH2-0-C4H8-, -CH2-0-C6H12- and -CH2-0-C8H16.
For any of the above aspects, when P represents a modified phosphate group, P
can be
represented by:
1
Y
1
E0_P- 01
19
Y-
wherein Y1 and Y2 each independently represent =0, =S, -0-, -OH, -SH, -BH3, -
OCH2CO2,
-OCH2CO2Rx, -OCH2C(S)0Rx, and ¨0Rx, wherein Rx represents Ci-C6 alkyl and
wherein
¨I indicates attachment to the remainder of the compound.
By modified phosphate it is meant a phosphate group wherein one or more of the
non-
linking oxygens is replaced. Examples of modified phosphate groups include
phosphorothioate, phosphoroselenates, borano phosphates, borano phosphate
esters,
hydrogen phosphonates, phosphoroamidates, alkyl or aryl phosphonates and
phosphotriesters. Phosphorodithioates have both non-linking oxygens replaced
by
sulphur. One, each or both non-linking oxygens in the phosphate group can be
independently any one of S, Se, B, C, H, N, or OR (R is alkyl or aryl).
The phosphate can also be modified by replacement of a linking oxygen with
nitrogen
(bridged phosphoroamidates), sulfur (bridged phosphorothioates) and carbon
(bridged
methylenephosphonates). The replacement can occur at a terminal oxygen.
Replacement
of the non-linking oxygens with nitrogen is possible.
For example, Y1 may represent -OH and Y2 may represent =0 or =S; or
Y1 may represent -0- and Y2 may represent =0 or =S;
Y1 may represent =0 and Y2 may represent ¨CH3, -SH, -0Rx, or ¨BH3
Y1 may represent =S and Y2 may represent ¨CH3, ORx or ¨SH.
It will be understood by the skilled person that in certain instances there
will be
delocalisation between Y1 and Y2.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
42
Preferably, the modified phosphate group is a thiophosphate group.
Thiophosphate
groups include bithiophosphate (i.e. where Y1 represents =S and Y2 represents
¨S-) and
monothiophosphate (i.e. where Y1 represents -0- and Y2 represents =S, or where
Y1
represents =0 and Y2 represents ¨S-). Preferably, P is a monothiophosphate.
The
inventors have found that conjugates having thiophosphate groups in
replacement of
phosphate groups have improved potency and duration of action in vivo.
P may also be an ethylphosphate (i.e. where Y1 represents =0 and Y2 represents
OCH2CH3).
The saccharide may be selected to have an affinity for at least one type of
receptor on a
target cell. In particular, the receptor is on the surface of a mammalian
liver cell, for
example, the hepatic asialoglycoprotein receptor complex (ASGP-R).
For any of the above aspects, the saccharide may be selected from N-acetyl
with one or
more of galactosamine, mannose, galactose, glucose, glucosamine and fructose.
Typically a ligand to be used in the present invention may include N-acetyl
galactosamine
(GaINAc). Preferably the compounds of the invention may have 3 ligands, which
will each
preferably include N-acetyl galactosamine.
"GaINAc" refers to 2-(Acetylamino)-2-deoxy-D- galactopyranose, commonly
referred to in
the literature as N-acetyl galactosamine. Reference to "GaINAc" or "N-acetyl
galactosamine" includes both the 13- form: 2-(Acetylamino)-2-deoxyl3 -D-
galactopyranose
and the a-form: 2-(Acetylamino)-2-deoxy-a-D- galactopyranose. In certain
embodiments,
both the 13-form: 2-(Acetylarnino)-2-deoxy13-D-galactopyranose and a-form: 2-
(Acetylamino)-2-deoxy-a-D-galactopyranose may be used interchangeably.
Preferably,
the compounds of the invention comprise the 13-form, 2-(Acetylarnino)-2-
deoxy13-D-
galactopyranose.
.-:
FF'44.41%...e," ''=-,.-FtOH
1 1
1-1C= . r~-, 1 1
0 H
2-(Acetylamino)-2-deoxy-D-galactopyranose
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
43
OH
HO
',j [ i Ac
2-(Acetylamino)-2-deoxy-P-D-galactopyranose
OH
HO
.....,.....\____.-0
HO \\6 \\,....
I !A:
.......\---__
......'s/
2-(Acetylamino)-2-deoxy-a-D-galactopyranose
For any of the above compounds of formula (III), X1 may be (-CH2-CH2-0)(-CH2)2-
. X' may
be (-CH2-CH2-0)2(-CH2)2-. X1 may be (-CH2-CH2-0)3(-CH2)2-. Preferably, X1 is (-
CH2-CH2-
0)2(-CH2)2-. Alternatively, X' represents C3-C6 alkylene. X1 may be propylene.
X1 may be
butylene. X' may be pentylene. X1 may be hexylene. Preferably the alkyl is a
linear
alkylene. In particular, X1 may be butylene.
For compounds of formula (III), X2 represents an alkylene ether of formula -
C3H6-0-CH2-
i.e. C3 alkoxy methylene, or ¨CH2CH2CH2OCH2-.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
44
The invention provides a conjugated nucleic acid having one of the following
structures:
OH
OH
OH H7:)
AcH OH
N.01-1
0
AcH
0
0
0
.0
0
(
S-11-0-
01 OH
0
it AcHN7-
c-00:
0
(:)
0¨/¨/
/
S /-0
0
Z-011-0/
0
(!)- s
1-
CA 03118327 2021-04-30
WO 2019/092282
PCT/EP2018/081105
OH
HOµ,_4Dii
OH OH
0
HO,:L._.... AcHN
0 0
NHAc
LI)
0
1 e
01-S
O 0
0 =P-S
1
NNN1 0 OH
0 0_f AcHNF
OH
/
criOH
/
0 ..,,,
0 0
II
le
S
N, 9 /
0 -P-0
10 1
S 1
OH
H0µ,. OH
OH OH
0
HON.14.1....õ AcHN
0 0
NHAc
ININ1
0
1 e
0 =P -S
i
O 0
--,, I 0
O=P-S
Nsi i
0 OH
0 ___/¨1
0 AcHN OH
_______________________________________ /
r(i)OH
/
O _j__ j-0 =,,o /0
0
Z -0 -P-0 L,
I0
S 0
.., II
0-P-0
I
S
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
46
OH
HO\,...,011
OH OH
0
AcHN
0 0
NHAc
0
Li)
i 0
0=P¨S
i
0.1 0
i 0
0=P¨S
0
/I OH
0
/ AcHN C
0¨ OH
'N
_______________________________________ / r().-R'OH
_Ex J-0/ 0
-.,
0 0
II
I¨O¨P-0
LI,µ 1? f
,.
S 0¨P-0
so
S,
OH
H0µ,_ OH
OH OH
0
AcHN
0 0
NHAc
1')
0
I 0
0 =P ¨S
i
01 0
I 0
0 = P ¨S
0
/ I OH
0,, o j OH
/
AcHN
, r i c ( 1 -FON
_x_rxi¨ 0 _____________________ õ, 0
0 .--
0
II ,
z-0¨P¨O 0¨p-0
i 0 10
S S
CA 03118327 2021-04-30
WO 2019/092282
PCT/EP2018/081105
47
OH
HO k,,,_01.1
OH OH
HO 0
AcHN
0
0
NHAc
0
0 = -Se \
i
01 0
o4-s
i
0
AcHN OH
OH
0
1 0 ¨OH(2)
0
0/ L /
0
OLL /0
II
Z-0 -P -0 ¨rj---
le
s o
0
0 -P -0
le
s
OH
HO OH
OH OH 0
AcHN
0
\
NHAc
0
I 0
0 =I) -S
i
01 0
1/4-) 0 -4 -Se
I
0
AcHN OH
/..-6 )0H
0 0 --/--/ 0 H
I /
/
0
0 T 0 /o
1
I0 it
S 0 -1, -0
t 0
S
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
48
DH
H` <,..i.---OH
OH OH b
t f
AcHti
0
\
I 0
0.1,-s
1
0,1 0
oP-
1...1 I S0
=
I
0
AcH i iv H
.......\ OH
0 0 OH
_____________________________________________ / 0
j. j..../...... Jr- 0/ c)
0
II ?I /
2 ¨0 ¨P ¨0 CL,,, 0 ¨P ¨0
t 0 t 0
S s
wherein Z is a nucleic acid as defined herein before.
A ligand of formula (I), (II) or (III) can be attached at the 3'-end of the
first (antisense)
strand and/or at any of the 3'- and/or 5'-end of the second (sense) strand.
The nucleic
acid can comprise more than one ligand of formula (I), (II), or (III).
However, a single
ligand of formula (I), (II) or (III) is preferred because a single such ligand
is sufficient for
efficient targeting of the nucleic acid to the target cells.
Preferably, the 5'-end of the first (antisense) strand is not attached to a
ligand of formula
(I), (II) or (III), since a ligand in this position can potentially interfere
with the biological
activity of the nucleic acid.
A nucleic acid with a single ligand of formula (I), (II) or (III) at the 5'-
end of a strand is
easier and therefore cheaper to synthesis than the same nucleic acid with the
same ligand
at the 3'-end. Preferably therefore, a single ligand of any of formulae (I),
(II) or (III) is
covalently attached to (conjugated with) the 5'-end of the second strand of
the nucleic
acid.
In one embodiment, the nucleic acid is conjugated to a ligand that comprises a
lipid, and
more preferably a ligand that comprises a cholesterol.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
49
A conjugate of the invention can comprise any nucleic acid as disclosed herein
conjugated to any ligand or ligands as disclosed herein.
The present invention also relates to a conjugate for inhibiting expression of
a ALDH2 gene
in a cell, said conjugate comprising a nucleic acid portion, comprising the
nucleic acid of
any aspect of the invention, and at least one ligand portion, said nucleic
acid portion
comprising at least one duplex region that comprises at least a portion of a
first RNA strand
and at least a portion of a second RNA strand that is at least partially
complementary to the
first strand, wherein said first strand is at least partially complementary to
at least a portion
of RNA transcribed from said ALDH2 gene, said at least one ligand portion
comprising a
linker moiety, preferably a serinol-derived linker moiety, and a targeting
ligand for in vivo
targeting of cells and being conjugated exclusively to the 3' and/or 5' ends
of one or both
RNA strands, wherein the 5' end of the first RNA strand is not conjugated,
wherein:
(I) the
second RNA strand is conjugated at the 5' end to the targeting ligand, and
wherein (a) the second RNA strand is also conjugated at the 3' end to the
targeting ligand and the 3' end of the first RNA strand is not conjugated; or
(b)
the first RNA strand is conjugated at the 3' end to the targeting ligand and
the 3'
end of the second RNA strand is not conjugated; or (c) both the second RNA
strand and the first RNA strand are also conjugated at the 3' ends to the
targeting
ligand; or
(ii) both the second RNA strand and the first RNA strand are conjugated
at the 3'
ends to the targeting ligand and the 5' end of the second RNA strand is not
conjugated.
In an embodiment of the present invention, the second RNA strand (i.e. the
sense strand)
is conjugated at the 5' end to a targeting ligand, the first RNA strand (i.e.
the antisense
strand) is conjugated at the 3' end to the targeting ligand and the 3' end of
the second RNA
strand (i.e. the sense strand) is not conjugated, such that a conjugate with
the schematic
structure as shown in Figure 59A is formed.
In an embodiment of the present invention, the second RNA strand (i.e. the
sense strand)
is conjugated at the 5' end to the targeting ligand, the second RNA strand
(i.e. the sense
strand) is also conjugated at the 3' end to the targeting ligand and the 3'
end of the first RNA
strand (i.e. the antisense strand) is not conjugated, such that a conjugate
with the schematic
structure as shown in Figure 59B is formed.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
In an embodiment of the present invention, both the second RNA strand (i.e.
the sense
strand) and the first RNA strand (i.e. the antisense strand) are conjugated at
the 3' ends to
the targeting ligand and the 5' end of the second RNA strand (i.e. the sense
strand) is not
conjugated , such that a conjugate with the schematic structure as shown in
Figure 59C is
5 formed.
In an embodiment of the present invention, the second RNA strand (i.e. the
sense strand)
is conjugated at the 5' end to the targeting ligand and both the second RNA
strand (i.e. the
sense strand) and the first RNA strand (i.e. the antisense strand) are also
conjugated at the
10 3' ends to the targeting ligand, such that a conjugate with the
schematic structure as shown
in Figure 59D is formed.
In any one of the above embodiments, ."-",-,-r indicates the linker which
conjugates the
ligand to the ends of the nucleic acid portion; the ligand may be a GaINAc
moiety such as
15 GaINAc; and the schematic structure as shown in Figure 59E represents
the nucleic acid
portion.
These schematic diagrams are not intended to limit the number of nucleotides
in the first
or second strand, nor do the diagrams represent any kind of limitation on
complementarity
20 of the bases or any other limitation.
The ligands may be monomeric or multimeric (e.g. dimeric, trimeric, etc.).
Suitably, the ligands are monomeric, thus containing a single targeting ligand
moiety, e.g.
25 a single GaINAc moiety.
Alternatively, the ligands may be dimeric ligands wherein the ligand portions
comprise two
linker moieties, such as serinol-derived linker moieties or non-serinol linker
moieties, each
linked to a single targeting ligand moiety.
The ligands may be trimeric ligands wherein the ligand portions comprise three
linker
moieties, such as serinol-derived linker moieties or non-serinol linker
moieties, each linked
to a single targeting ligand moiety.
The two or three serinol-derived linker moieties may be linked in series e.g.
as shown
below:
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
51
,
HN, OH HN
wherein n is 1 or 2 and Y is S or 0.
Preferably, the ligands are monomeric.
Suitably, the conjugated RNA strands are conjugated to a targeting ligand via
a linker moiety
including a further linker wherein the further linker is or comprises a
saturated, unbranched
or branched C1_15 alkyl chain, wherein optionally one or more carbons (for
example 1, 2 or
3 carbons, suitably 1 or 2, in particular 1) is/are replaced by a heteroatom
selected from 0,
N, S(0)p, wherein p is 0, 1 or 2 (for example a CH2 group is replaced with 0,
or with NH, or
with S, or with SO2 or a ¨CH3 group at the terminus of the chain or on a
branch is replaced
with OH or with NH2) wherein said chain is optionally substituted by one or
more oxo groups
(for example 1 to 3, such as 1 group).
Suitably, the linker moiety is a serinol-derived linker moiety.
More suitably, the further linker comprises a saturated, unbranched C1_15
alkyl chain wherein
one or more carbons (for example 1, 2 or 3 carbons, suitably 1 or 2, in
particular 1) is/are
replaced by an oxygen atom.
More suitably, the further linker comprises a PEG-chain.
More suitably, the further linker comprises a saturated, unbranched C1_15
alkyl chain.
More suitably, the further linker comprises a saturated, unbranched C1_6 alkyl
chain.
More suitably, the further linker comprises a saturated, unbranched C4 or C6
alkyl chain,
e.g. a C4 alkyl chain.
In an embodiment, ,n-rtrtf is a linking moiety of formula (V):
CA 03118327 2021-04-30
WO 2019/092282
PCT/EP2018/081105
52
7 \II \ Y
ll .
-i¨i¨Li¨O¨P-0--Li¨O¨P-0¨:¨
I I
\ OH i
/ OH
(V)
wherein n, Y and L1 are defined below and the 0 of the phosphoro-group is
attached to
the terminal oligonucleoside of the RNA strands.
Thus in an embodiment, the targeting ligand portion is a linking moiety of
formula (VI):
7 11 \ Y
11
\
GaINAc ______ L1 0 Pi 0 L1 0 11 0 OH 1 OH
/
(VI)
wherein n, Y and L1 are defined below and the 0 of the phosphoro-group is
attached to
the terminal oligonucleoside of the RNA strands.
Suitably, sr%"-"f is a linking moiety of formula (XIV):
L L
/ /
HN HN
/ Y Y
_______________ \ II \ ___ \ II .
HO _____________ 0 P 0 0¨P-0-1,¨
I I
\ Ri OH /n R1 OH
(XIV)
wherein n, Y, R1 and L are defined below, L is connected to the targeting
ligand e.g.
GaINAc and the 0 of the phosphoro-group is attached to the terminal
oligonucleoside of
the RNA strands.
Suitably, the targeting ligand portion is a linking moiety of formula (IV):
GaINAc
GaINAc
L/ L/
/ /
25 HN
Y
\ II Y
__________________________________ \ II .
HO ________________________ 0 P 0 I ) 0¨P-0¨
. I
\ R1 OH R1 OH
n (IV)
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
53
wherein n, Y, R1 and L are defined below and the 0 of the phosphoro-group is
attached to
the terminal oligonucleoside of the RNA strands.
Suitably, sr%"-"f is a linking moiety of formula (VII):
-( Y \ Y
II II
I
OH /
in OH
(VII)
wherein n, Y and L2 are defined below and the 0 of the phosphoro-group is
attached to
the terminal oligonucleoside of the RNA strands.
Suitably, the targeting ligand portion is a linking moiety of formula (VIII):
7
1(1 Y
ll _ .
GaINAc _______ L2 0 P 0 ___ L2 0 1:,) u-
I I
\ OH / OH
VIII
wherein n, Y and L2 are defined below and the 0 of the phosphoro-group is
attached to
the terminal oligonucleoside of the RNA strands.
Suitably, -ruN-ry is a linking moiety of formula (IX):
Y
Il ' I
,
-,'-L 0¨P¨O-H
= 1¨F/ 1- I
OH (IX)
wherein F, Y and L are defined below and the 0 of the phosphoro-group is
attached to the
terminal oligonucleoside of the RNA strands.
Suitably, the targeting ligand portion is a linking moiety of formula (IXa):
Y
II .
GaINAc¨L 0¨P¨O-H
1¨F/ 1- I
OH (IXa)
wherein F, Y and L are defined below and the 0 of the phosphoro-group is
attached to the
terminal oligonucleoside of the RNA strands.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
54
Suitably, L is:
0
, .
In any of the above structures, suitably the ligands are selected from GaINAc
and galactose
moieties, especially GaINAc moieties. Alternatively, GalNac may be replaced by
another
targeting ligand, e.g. a saccharide.
In an embodiment of the invention, the first RNA strand is a compound of
formula (X):
_
¨
Y Y \
II II
5'Z1-3' O¨P-0 L1 ________________________ 0 P 0 L1--O _______ H
I I
OH OH
/
¨ ¨ b (x)
wherein b is 0 or 1; and
the second RNA strand is a compound of formula (XI):
-
Y \ Y
II Y
5' 3' II Y
-( \
H-0 Li-0-111-0-'-1_1-0¨P-0-4-0¨P-0¨Li _________________ 0-111-0¨L1-LO¨H
I I I I
OH / OH OH OH
/
- -c - - d (Xi);
wherein:
c and d are independently 0 or 1;
Z1 and Z2 are the RNA portions of the first and second RNA strands
respectively;
Y is 0 or S;
n is 0, 1, 2 or 3; and
L1 is a linker to which a ligand is attached;
and wherein b + c + d is 2 or 3.
Suitably, the first RNA strand is a compound of formula (XV):
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
¨ _
GaINAc GaINAc
\ \
L\ L
\
NH NH
/ _____________________________________ / II /
Z1-0¨P-0 0 P 0 0¨H
I I
OH R1 \ OH Ri /
n
b¨ ¨ (XV)
wherein b is 0 or 1; and
the second RNA strand is a compound of formula (XVI):
_ _ _ _
GaINAc ,GaINAc GaINAc GaINAc
/µ µ
L L L L
HN HN NH NH
\
Yil \ Y Y / Yil
H-0-/R OPO OPO Z20P0 OP d R1 -)-
0¨H
I I I I I
\ Ri i OH Ri OH OH R1 \ OH
n n
c _ ¨ d ¨ ¨
5 (XVI);
wherein c and d are independently 0 or 1;
wherein:
Z1 and Z2 are the RNA portions of the first and second RNA strands
respectively;
Y is 0 or S;
10 R1 is H or methyl;
n is 0, 1,2 or 3; and
L is the same or different in formulae (XV) and (XVI) and is selected from the
group
consisting of:
-(CH2)r-C(0)-, wherein r = 2-12;
15 -(CH2-CH2-0)s-CH2-C(0)-,
wherein s = 1-5;
-(CH2)1-CO-NH-(CH2)1-NH-C(0)-, wherein t is independently is 1-5;
-(CH2).-CO-NH-(CH2).-C(0)-, wherein u is independently is 1-5; and
-(CH2),-NH-C(0)-, wherein v is 2-12; and
wherein the terminal C(0) (if present) is attached to the NH group;
20 and wherein b + c + d is 2 or 3.
Suitably, the first RNA strand is a compound of formula (XII):
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
56
Y Y
II II
5,Z1-3, 0¨P-0 L2 ________________________ 0 P¨O¨L2 O¨H
I I
OH OH
_ n
¨ b (XII)
wherein b is 0 or 1; and
the second RNA strand is a compound of formula (XIII):
¨
_
Y \ Y Y Y
5' 3' II Il
H-0 L2-0JI-0¨H_2-0-11:1-0¨Z2-0¨P¨O¨L2 0¨PI-0¨L2 0 H
I I
OH / OH OH OH
n
¨ / _ c ¨
n _d (MO;
wherein:
c and d are independently 0 or 1;
Z1 and Z2 are the RNA portions of the first and second RNA strands
respectively;
Y is 0 or S;
n is 0, 1,2 or 3; and
L2 is the same or different in formulae (XII) and (XIII) and is the same or
different in
moieties bracketed by b, c and d, and is selected from the group consisting
of:
0 y F. ,L, il GaINAc
H
H NI, GaINAc QN
FLGaINAc F L
(.)g, õ...k........L......),µ,.
.....I.....:3õ,
= . ; and 1 ; or
,
,
n is 0 and L2 is:
, H
-1 ,NõGaINIAc
iF 1_-
and the terminal OH group is absent such that the following moiety is formed:
Y
II .
GaINAc¨L 0¨P-0-1
I-1\1¨F/ I
= OH ,
wherein
F is a saturated branched or unbranched (such as unbranched) Ci_salkyl (e.g.
C1-
6alkyl) chain wherein one of the carbon atoms is optionally replaced with an
oxygen atom
provided that said oxygen atom is separated from another heteroatom (e.g. an 0
or N atom)
by at least 2 carbon atoms;
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
57
L is the same or different in formulae (XII) and (XIII) and is selected from
the group
consisting of:
-(CH2)r-C(0)-, wherein r = 2-12;
-(CH2-CH2-0)s-CH2-C(0)-, wherein s = 1-5;
-(CH2)1-CO-NH-(CH2)1-NH-C(0)-, wherein t is independently is 1-5;
-(CH2).-CO-NH-(CH2).-C(0)-, wherein u is independently is 1-5; and
-(CH2)v-NH-C(0)-, wherein v is 2-12; and
wherein the terminal C(0) (if present) is attached to the NH group;
and wherein b + c + d is 2 or 3.
In any one of the above formulae where GaINAc is present, the GaINAc may be
substituted
for any other targeting ligand, such as those mentioned herein.
Suitably, b is 0, c is 1 and d is 1; b is 1, c is 0 and d is 1; b is 1, c is 1
and d is 0; or b is 1, c
is 1 and d is 1.
More suitably, b is 0, c is 1 and d is 1; b is 1, c is 0 and d is 1; or b is
1, c is 1 and d is 1.
Most suitably, b is 0, c is 1 and d is 1.
In one embodiment, Y is 0. In another embodiment, Y is S.
In one embodiment, R1 is H or methyl. In one embodiment, R1 is H. In another
embodiment,
R1 is methyl.
In one embodiment, n is 0, 1, 2 or 3. Suitably, n is 0.
In one embodiment, L is selected from the group consisting of:
-(CH2)r-C(0)-, wherein r = 2-12;
-(CH2-CH2-0)s-CH2-C(0)-, wherein s = 1-5;
-(CH2)1-CO-NH-(CH2)1-NH-C(0)-, wherein t is independently is 1-5;
-(CH2).-CO-NH-(CH2).-C(0)-, wherein u is independently is 1-5; and
-(CH2)v-NH-C(0)-, wherein v is 2-12;
wherein the terminal C(0) is attached to the NH group.
Suitably, L is -(CH2)r-C(0)-, wherein r = 2-12. Suitably, r = 2-6. More
suitably, r = 4 or 6 e.g.
4.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
58
Suitably, L is:
0
%,..",..,............/.7....N......A./...
, .
Example F moieties include (CH2)1_6 e.g. (CH2)1-4 e.g. CH2, (CH2)4, (CH2)5 or
(CH2)6, or
CH20(CH2)2_3, e.g. CH20(CH2)CH3.
Suitably, L2 is:
H
N, GaINAc
L
Suitably, L2 is:
H
aON I_GaINAc
. ,
Suitably, L2 is:
N'L GaINAc
H
i .
. .
Suitably, L2 is:
L 0,),......--W .õ ..,..
N GaINAc
H
N
1
.
Suitably, n is 0 and L2 is:
H
c..N, LGaINAc
,
,
and the terminal OH group is absent such that the following moiety is formed:
GaINAc
\
L¨NH
\ Y
' 20 OH ,
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
59
wherein Y is as defined elsewhere herein.
Within the moiety bracketed by b, c and d, L2 is typically the same. Between
moieties
bracketed by b, c and d, L2 may be the same or different. In an embodiment, L2
in the
moiety bracketed by c is the same as the L2 in the moiety bracketed by d. In
an
embodiment, L2 in the moiety bracketed by c is not the same as L2 in the
moiety bracketed
by d. In an embodiment, the L2 in the moieties bracketed by b, c and d is the
same, for
example when the linker moiety is a serinol-derived linker moiety.
Serinol derived linker moieties may be based on serinol in any stereochemistry
i.e.
derived from L-serine isomer, D-serine isomer, a racemic serine or other
combination of
isomers. In a preferred aspect of the invention, the serinol-GaINAc moiety
(SerGN) has
the following stereochemistry:
yiPr2
DMTo oPoCN
0 \ NH
ss,
HOOH *
NH2 H .\
DMT0 oN =L-Serine 0
Serinol derived linker moieties
2\
(S)-Serinol building blocks
.
i.e. is based on an (S)-serinol-amidite or (S)-serinol succinate solid
supported building
block derived from L-serine isomer.
In one embodiment, the targeted cells are hepatocytes.
General synthesis schemes
Example compounds can be synthesised according to methods described below and
known
to the person skilled in the art. Whilst the schemes illustrate the synthesis
of particular
conjugates, it will be understood that other claimed conjugates may be
prepared by
analogous methods. Assembly of the oligonucleotide chain and linker building
blocks may,
for example, be performed by solid phase synthesis applying phosphoramidite
methodology. Solid phase synthesis may start from a base or modified building
block loaded
lcaa CPG. Phosphoramidite synthesis coupling cycle consists of 1) DMT-removal,
2) chain
elongation using the required DMT-masked phosphoramidite and an activator,
which may
be benzylthiotetrazole (BTT), 3) capping of non-elongated oligonucleotide
chains, followed
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
by oxidation of the P(III) to P(V) either by Iodine (if phosphodiester linkage
is desired) or
EDITH (if phosphorothioate linkage is desired) and again capping (Cap/Ox/Cap
or
Cap/Thio/Cap). GaINAc conjugation may be achieved by peptide bond formation of
a
GaINAc-carboxylic acid building block to the prior assembled and purified
oligonucleotide
5 having the necessary number of amino modified linker building blocks
attached. The
necessary building blocks are either commercially available or synthesis is
described below.
All final single stranded products were analysed by AEX-H PLC to prove their
purity. Purity
is given in %FLP (% full length product) which is the percentage of the UV-
area under the
assigned product signal in the UV-trace of the AEX-HPLC analysis of the final
product.
10 Identity of the respective single stranded products was proved by LC-MS
analysis.
Synthesis of Synthons
Scheme 1: Synthesis of DMT-serinol(TFA) linker synthons
NH3C1 HN 0 ii HN 0 iii NH2
HO - C) HO - ¨1" DMTOyO DMTOOH
0 0 0
1 2 3 4
i
0
0
HNACF3 0
HNACF3
DMTO0y)LOH
DMTOOH
0
DMT-Serinol(TFA)-succinate 6 5
vil iv
0
0
HNACF3 0
II HNACF3
DMTOO
11
N DMTOOõOCN
0
NiPr2
DMT-Serinol(TFA)-CEP 7
i) ethyl trifluoroacetate, NEt3, Me0H, 0 C, 16h, 2: 86% 5: 90%, ii) DMTCI,
pyridine, 0 C,
16h, 74%, iii) LiBH4, Et0H/THF (1/1, v/v), 0 C, 1h, 76%, iv) 2-cyanoethyl-N,N-
diisopropylchloro phosphoramidite, EtNPr2, CH2Cl2, 56%, v) succinic anhydride,
DMAP,
pyridine, RT, 16h, 38%, vi) HBTU, DIEA, amino-lcaa CPG (500A), RT, 18h, 29%
(26 umol/g loading).
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
61
(S)-DMT-Serinol(TFA)-phosphoramidite 7 can be synthesised from (L)-serine
methyl ester
derivative 1 according to literature published methods (Hoevelmann et al.
Chem. Sci.,
2016,7, 128-135).
(S)-DMT-Serinol(TFA)-succinate 6 can be made by conversion of intermediate 5
with
succinic anhydride in presence of a catalyst such as DMAP.
Loading of 6 to a solid support such as a controlled pore glass (CPG) support
may be
achieved by peptide bond formation to a solid support such as an amino
modified native
CPG support (500A) using a coupling reagent such as HBTU. The (S)-DMT-
Serinol(TFA)-
succinate 6 and a coupling reagent such as HBTU is dissolved in a solvent such
as CH3CN.
A base, such as diisopropylethylamine, is added to the solution, and the
reaction mixture is
stirred for 2 min. A solid support such as a native amino-lcaa-CPG support
(500 A, 3 g,
amine content: 136 umol/g) is added to the reaction mixture and a suspension
forms. The
suspension is gently shaken at room temperature on a wrist-action shaker for
16h then
filtered, and washed with solvent such as DCM and Et0H. The support is dried
under
vacuum for 2 h. The unreacted amines on the support can be capped by stirring
with acetic
anhydride/lutidine/N-methylimidazole at room temperature. Washing of the
support may be
repeated as above. The solid support is dried under vacuum to yield solid
support 10.
Scheme 2: Synthesis of GaINAc synthon 9
OAc OAc OAc OAc
CL vii, viii
1&.....T2..\___
Ac01&"\'`\--0Ac ¨"- Ac0 OrOH
NHAc NHAc
0
8 GaIN(Ac4)-04-acid 9
(vii) TMSOTf, DCM, hexenol, viii) RuC13, Nalat, DCM, CH3CN, H20, 46% over two
steps.
Synthesis of the GaINAc synthon 9 can be prepared according to methods as
described in
Nair et al. (2014), starting from commercially available per-acetylated
galactose amine 8.
Synthesis of single stranded serinol-derived GalNAc conjugates
Scheme 3: General procedure of oligonucleotide synthesis for serinol-derived
linkers
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
62
0
HN Steps
).LCF3 0 Ny12 ' H2N \
(see Figure 25)
_
P
DMTOOL ND ¨I-- HO - 0õ0 5oligonucleotide strand3'
H
0 0 SH 0 SH
im
n
11
n=0,m=1:11 =A0264
n = 1, m = 0: 11 =A0220
n=1,m=1:11 =A0329
Oligonucleotide synthesis of 3' mono-GaINAc conjugated oligonucleotides (such
as
compound A0264) is outlined in Figure 25 and summarised in Scheme 3. Synthesis
is
commenced using (S)-DMT-Serinol(TFA) ¨succinate-lcaa-CPG 10 as in example
5 compound A0264. In case additional serinol building blocks are needed the
(S)-DMT-
serinol(TFA) amidite (7) is used in the appropriate solid phase synthesis
cycle. For example,
to make compound A0329, the chain assembly is finished with an additional
serinol amidite
coupling after the base sequence is fully assembled. Further, oligonucleotide
synthesis of
5' mono-GaINAc conjugated oligonucleotides may be commenced from a solid
support
10 loaded with the appropriate nucleoside of its respected sequence. In
example compound
A0220 this may be 2'fA. The oligonucleotide chain is assembled according to
its sequence
and as appropriate, the building block (S)-DMT-serinol(TFA)-amidite (7) is
used. Upon
completion of chain elongation, the protective DMT group of the last coupled
amidite
building block is removed, as in step 1) of the phosphoramidite synthesis
cycle.
Upon completion of the last synthesizer step, the single strands can be
cleaved off the solid
support by treatment with an amine such as 40% aq. methylamine treatment. Any
remaining
protecting groups are also removed in this step and methylamine treatment also
liberates
the serinol amino function. The crude products were then purified each by AEX-
H PLC and
SEC to yield the precursor oligonucleotide for further GaINAc conjugation.
Scheme 4: GaINAc conjugation synthesis of serinol-derived precursor
oligonucleotides
CA 03118327 2021-04-30
WO 2019/092282
PCT/EP2018/081105
63
(Ac OAc
Ac0
NHAc
OH _I-1 OH OH OH
9 0
HO
HO
NHAc
NHAc
1. HBTU, DIPEA, DMSO, 2min
2. 11 in DMSO/H20, 30 min
3. 40% MeNH aq., 15min /I-IN 0
5' 3' H2N
\ 0
HOO,D,0 oligonucleotide strand 0,p,0 OH
ir\
XH 0 XH
im
12
n = 0, m = 1:12 =A0268
n = 1, m = 0:12 =A0241
n = 1, m = 1:12 =A0330
Post solid phase synthesis GaINAc-conjugation was achieved by pre-activation
of the
GaIN(Ac4)-C4-acid (9) by a peptide coupling reagent such as HBTU. The pre-
activated acid
9 was then reacted with the amino-groups in 11 (e.g. A0264) to form the
intermediate
GaIN(Ac4)-conjugates. The acetyl groups protecting the hydroxyl groups in the
GaINAc-
moieties were cleaved off by methylamine treatment to yield the desired
example
compounds 12 (e.g. A0268), which were further purified by AEX-HPLC and SEC.
Synthesis of single stranded non-serinol-derived GalNAc conjugates
Amino modified building blocks other than serinol are commercially available
from various
suppliers and can be used instead of serinol to provide reactive amino-groups
that allow for
GaINAc conjugation. For example the commercially available building blocks
shown in
Table 6 below can be used to provide non-serinol-derived amino modified
precursor
oligonucleotides 14 (Scheme 5A) by using amino-modifier loaded CPG such as 10-
1 to 10-
3 followed by sequence assembly as described above and finally coupling of
amino-modifier
phosophoramidites such as 13-1, 13-2 or 13-4.
For example, to make 14 (A0653) GlyC3Am-CPG (10-2) was used in combination
with
GlyC3Am-Amidite 13-2. Structurally differing modifiers can be used to make 14,
for example
for A0651 C7Am-CPG was used in combination with C6Am-Amidite as second amino
modification. In a similar fashion commercially available amino-modifier
loaded CPG 10-5
and amino-modified phosphoramidite 13-5 can be used to synthesise amino-
modified
precursor molecules 14 (A0655).
Table 6: Commercially available building blocks
C3Am-CPG (10-1) is: GlyC3Am-CPG (10-2) is:
CA 03118327 2021-04-30
WO 2019/092282
PCT/EP2018/081105
64
NHFnnoc 0.NHTFA
.0 CPG .0 ,CPG
DMTr0 0 DMTr0 0
C7Am-CPG (10-3) is: PipAm-CPG (10-5) is:
01,......======,..../..,.....====,.
NHTFA
0C.0NHFmoc r
DMTr CPG 0.0
TMTr CPG
C3Am-Phos (13-1) is: GlyC3Am-Phos (13-2) is:
NHFmoc 0.-.NHFmoc
.0 .0 CEP
DMTr0 0CEP DMTr0 0
C6Am-Phos (13-4) is: PipAm-Phos (13-5) is:
NHTFA 0.).,õ,-w
NHTFA
CEP N
0
c<0
TMTrO CEP
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
Scheme 5: General procedure for oligonucleotide synthesis
A)
Ga
Ga
HN, I
HN,
Oa 0 Steps L.,a 0
5' 3'
DMTOL5a L3 N HO-oligonucleotide strand-0Põ0L5a, p).r)-
LNO
' L3
H (see Figure 25)
S 0
o 'cN 0
10-1, 10-2, 10-3 1 1. 13, BIT,
Acetonitrile
10-1, L5a = CH2, L3a absent, Lsa = CH2, G = Fmoc 2. further steps
10-2, L5a = CH2, L3a absent, Lsa = CH20(CH2)3, G = TFA
10-3, L5a = CH2, L3a = CH2, Oa = (CH2)4, G = Fmoc H2N,Lsb
H2N.Lsa
5' 3'
HO, -1 p L5b ,0-
oligonucleotide strand-0õ L5a L3 0, pH
L3 'F) P
0 SH 0 SH
14
1 Sb = ru nif-u \
A0653: L5a, L5b = CH2, L3a, L3b absent, Lsa , , L,n2,-,ks-
,..2i3
A0563: L5a, L5b = CH2, L3a, L3b absent, Lsa , Lsb = CH2
or
H2N,Lsb H2N.Lsa
5' 3'
3 p ,0-
oligonuc1eotide strand-0õ0, L3 pH
L 'P P L5a
I/ \
0 SH 0 SH
14
A0561: Oa = CH2, L3a, L3b absent, Lsa = CH20(CH2)3, Lsb = (CH2)5
A0651: L5a, Ca = CH2, L3b absent, Lsa = (CH2)4, Lsb =(CH2)5
Gb Gb
I I
HN, HN,
Lsb Lsb
DMT0-.L513-LL3P--P-- -.....--"-cN L- q Pip ,C1
' CN
I I
N
13-1, 13-2 rr
13-1, L5b = CH2, L3b absent, Lscb = CH2, G = Fmoc 13-4, L3b = absent, Lscb
= (CH2)5, G = TEA
13-2, L5b = CH2, L3b absent, Lscb = CH20(CH2)3, G = TEA
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
66
0yl_aNH2
B)
Ga
N
41sa
, Steps 0
L 0 3'
HO-Oligonucleotide strand-0Põ0 3, p
TMT13'L5ajL3PIN L5a L )-HN 0
H (see Figure 25)
0 S OCN 0
10-5 1. 13,
BTT, Acetonitrile
10-5, Oa = CH2, L3a = CH2, Lsa = (CH2)5, G = TEA 2. further steps
0 Ls,a
y Ga
0yl_bNH2 0yl_aNH2
N
--- :-...
N N
..-- --.
TMTO X p OCN 5' 3'
I HO, X põ0-oligonucleotide strand-0,p-
O,LXL3pH
-.TNT- L b L3 P
6 \SH /, \
0 SH
14
13-5, Lsb = CH2, L3b = CH2, Lscb = (CH2)5, G = TFA A0655:
L5a, L5b = CH2, L38, L3b = CH2, LSa , LSb = (CH2)5
The resulting precursor oligonucleotides 14 can then be conjugated with
GaIN(Ac4)-C4-
acid (9) to yield the desired example compounds 15 (Scheme 6).
CA 03118327 2021-04-30
WO 2019/092282
PCT/EP2018/081105
67
Scheme 6: GaINAc conjugation synthesis of precursor oligonucleotides
OH _OH OH _OH
0 0
HO ===\--0 HO
====\o
NHAc
NHAc
HNSID, HN,
L
3'
HO,L5b-LL3,p,p,CA' ligonucleotide strand¨O..p,0,0gLopH
6 \SH 6 \SH
OAc OAc
O CHH2
A0654: Oa, L5b = CH2, Oa, L3b absent, Lsa i , _Sb = _ - , - 0(CH
2,3
Ac0 OrOH A0564: Oa, L5b = CH2, Oa, L3b
absent, Lsa , L Sb = c
NHAc
9 0 OL _H OH OH
or
0 0
1. HBTU, DIPEA, DMSO, 2min HO HO
NHAc
2. 14 in DMSO/H20, 30 min
NHAc
3. 40% MeNH aq., 15min
IN- '15
HN, HN,
LSb
5' 3'
Lop,p_O-oligonucleotide strand-0,p,0,0a-LopH
OSH 6 \SH
A0562: L5a = CH2, L3a, L3b absent, Lsa = CH20(CH2)3, Lsb = (CH2)5
A0652: L5a, L3a = CH2, L3b absent, Lsa = (CH2)4, Lsb =(CH2)5
or
OH _OH OH /DH
0 0
HO HO
NHAc0 LT' NHAc0
y N y N 0
5' 3'
HO, 51,cZ 3b0õ0-oligonucleotide strand-0õ0, 3pH
L L P P L L
6 \SH OSH
A0656: L5a, L5b = CH2, L3a, L3b = CH2, Lsa , _sb I = (CH 2)5 Synthesis of the
single stranded tri-antennary GaINAc conjugates in reference conjugates
3-4
Oligonucleotides synthesis of tri-antennary GaINAc-cluster conjugated siRNA is
outlined in
Figure 26. Oligonucleotide chain assembly is commenced using base loaded
support e.g.
5'DMT-2'FdA(bz)-succinate-lcaa-CPG as in example compound A0006.
Phosphoramidite
synthesis coupling cycle consisting of 1) DMT-removal, 2) chain elongation
using the
required DMT-masked phosphoramidite, 3) capping of non-elongated
oligonucleotide
chains, followed by oxidation of the P(III) to P(V) either by Iodine or EDITH
(if
phosphorothioate linkage is desired) and again capping (Cap/Ox/Cap or
Cap/Thio/Cap) is
repeated until full length of the product is reached. For the on column
conjugation of a
trivalent tri-antennary GaINAc cluster the same synthesis cycle was applied
with using the
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
68
necessary trivalent branching amidite C4XLT-phos followed by another round of
the
synthesis cycle using the GaINAc amidite ST23-phos. Upon completion of this
last
synthesizer step, the oligonucleotide was cleaved from the solid support and
additional
protecting groups may be removed by methylamine treatment. The crude products
were
then purified each by AEX-H PLC and SEC.
General procedure of double strand formation
In order to obtain the double stranded conjugates, individual single strands
are dissolved in
a concentration of 60 OD/mL in H20. Both individual oligonucleotide solutions
can be added
together to a reaction vessel. For reaction monitoring a titration can be
performed. The first
strand is added in 25% excess over the second strand as determined by UV-
absorption at
260nm. The reaction mixture is heated e.g. to 80 C for 5min and then slowly
cooled to RT.
Double strand formation may be monitored by ion pairing reverse phase HPLC.
From the
UV-area of the residual single strand the needed amount of the second strand
can be
calculated and added to the reaction mixture. The reaction is heated e.g. to
80 C again and
slowly cooled to RT. This procedure can be repeated until less than 10% of
residual single
strand is detected.
The above process (including Schemes 1-6 and Figures 25 and 26) may be easily
adapted
to replace GalNac with another targeting ligand e.g. a saccharide.
In any of the above aspects, instead of post solid phase synthesis conjugation
it is possible
to make a preformed Serinol(GN)-phosphoramidite and use this for on-column
conjugation.
General synthesis schemes: 2
Example compounds can be synthesised according to methods described below and
known
to the person skilled in the art. Assembly of the oligonucleotide chain and
linker building
blocks may, for example, be performed by solid phase synthesis applying
phosphoramidite
methodology. GaINAc conjugation may be achieved by peptide bond formation of a
GaINAc-carboxylic acid building block to the prior assembled and purified
oligonucleotide
having the necessary number of amino modified linker building blocks attached.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
69
Scheme 1: Synthesis of DMT-serinol(TFA) linker synthons
..(4?
NH3ci HN 0 ii HN 0 iii NH2
HO - C) HO - ¨1" DMTOyO DMTOOH
0 0 0
1 2 3 4
i
0
0
HNACF3 0
A
DMTO0y)LOH HN CF3
DMTOOH
0
DMT-Serinol(TFA)-succinate 6 5
vil iv
0
0
HNACF3 0
HNACF3
DMT00
y N 4111 DMTO- 0,p,
CN
0
NiPr2
DMT-Serinol(TFA)-CEP 7
i) ethyl trifluoroacetate, NEt3, Me0H, 0 C, 16h, 2: 86% 5: 90%, ii) DMTCI,
pyridine, 0 C,
16h, 74%, iii) LiBH4, Et0H/THF (1/1, v/v), 0 C, 1h, 76%, iv) 2-cyanoethyl-N,N-
diisopropylchloro phosphoramidite, EtNPr2, CH2Cl2, 56%, v) succinic anhydride,
DMAP,
pyridine, RT, 16h, 38%, vi) HBTU, DIEA, amino-lcaa CPG (500A), RT, 18h, 29%.
DMT-Serinol(TFA)-phosphoramidite 7 can be synthesised from serinol derivative
1
according to literature published methods (Hoevelmann et al. Chem. Sci.,
2016,7, 128-135).
DMT-Serinol(TFA)-succinate 6 can be made by conversion of intermediate 5 with
succinic
5 anhydride in presence of a catalyst such as DMAP.
Loading of 6 to a solid support such as a CPG support may be achieved by
peptide bond
formation to a solid support such as an amino modified native CPG support
(500A) using a
coupling reagent such as HBTU. The DMT-Serinol(TFA)-succinate 6 and a coupling
reagent
such as HBTU is dissolved in a solvent such as CH3CN. A base, such as
10 diisopropylethylamine, is added to the solution, and the reaction
mixture is stirred for 2 min.
A solid support such as a native amino-lcaa-CPG support (500 A, 3 g, amine
content: 136
micromol/g) is added to the reaction mixture and a suspension forms. The
suspension is
gently shaken at room temperature on a wrist-action shaker for 16h then
filtered and washed
with solvent such as DCM and Et0H. The support is dried under vacuum for 2 h.
The
unreacted amines on the support can be capped by stirring with acetic
anhydride/lutidine/N-
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
methylimidazole at room temperature. Washing of the support may be repeated as
above.
The solid support is dried under vacuum to yield solid support 10.
Scheme 2: Synthesis of GaINAc synthon 9
_Ac OAc OAc OAc
vii, viii
Ac0 OAc Ac0 OH
NHAc NHAc
8 GaIN(Ac4)-04-acid 9
5 (vii) TMSOTf, DCM, hexenol, viii) RuC13, Nalat, DCM, CH3CN, H20,46% over
two steps.
Synthesis of the GaINAc synthon 9 can be prepared according to methods as
described in
Nair et al. J. Am. Chem. Soc., 2014,136(49), pp 16958-16961, starting from
commercially
available per-acetylated galactose amine 8.
Scheme 3: General procedure of oligonucleotide synthesis
HNACF3 o Steps NH2
DMTOONX-)
HOO
oligonucleotide strand
0 (see Figure 29)
11
10 10
All Oligonucleotides can be synthesized on an AKTA oligopilot 10 synthesizer
using
standard phosphoramidite chemistry which is described in detail below.
Oligonucleotide synthesis of 3' and 5' GaINAc conjugated oligonucleotides
precursors (such
as compound X0385B-prec) is outlined in Figure 29 and summarised in Scheme 3.
15 Synthesis is commenced using DMT-Serinol(TFA)¨succinate-lcaa-CPG 10. A
phosphoramidite synthesis cycle is applied until full length of the product
was reached.
Upon completion of chain elongation, the protective DMT group of the last
coupled amidite
building block can be removed in the same manner as in every individual
synthesis cycle.
To complete synthesis of example compound X0385B-prec (which has a serinol-
derived
20 linker moiety at the 3' and 5' ends of the second strand), the chain
assembly was finished
with an additional serinol amidite coupling after the base sequence was fully
assembled.
Upon completion of the last synthesizer step, the single strands can be
cleaved off the solid
support by treatment with an amine such as 40% aq. methylamine. Any remaining
protecting groups are also removed in this step and methylamine treatment also
liberates
25 the serinol amino function. The resulting crude oligonucleotide can be
purified by ion
exchange chromatography (Resource Q, 6mL, GE Healthcare) on a AKTA Pure HPLC
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
71
System using a gradient such as a sodium chloride gradient. Excess salt from
IEX
purification can be removed by SEC to yield the amino modified precursor
oligonucleotide
11. Product containing fractions are pooled, desalted on a size exclusion
column (Zetadex,
EMP Biotech) and lyophilised.
Scheme 4: General procedure for GaINAc conjugation
OAc OAc OH (OH
NH2 (I)
Ac0"\=2-\--0 OH + HOO,oligo
NHAc NHAc
9 11 12
(i) 9, HBTU, DIPEA, DMSO; 11, H20, DMSO, DIPEA; then activated 9, 11; then 40%
MeNH2, H20
Conjugation of the GaINAc synthon 9, as described above, can be achieved by
coupling 9
to the serinol-amino function of the respective oligonucleotide strand 11
using standard
peptide coupling conditions known to the skilled person. For example, the
respective amino-
modified precursor molecule 11 is dissolved in H20 and a polar solvent such as
DMSO (e.g.
DMSO/H20, 2/1, v/v) is added, followed by a base such as DIPEA (e.g. 2.5% of
total
volume). In a separate reaction vessel pre-activation of the GaINAc synthon 9
can be
performed by reacting 2 eq. (per amino function in the amino-modified
precursor
oligonucleotide) of the carboxylic acid component with 2 eq. of a coupling
reagent such as
HBTU in presence of 8 eq. of a base, such as DIPEA, in a polar solvent such as
DMSO.
After 2 min the activated compound 9 is added to the solution of the
respective amino-
modified precursor molecule 11. The reaction progress can be monitored by LCMS
or AEX-
HPLC. Upon completion of the conjugation reaction (e.g. 30 minutes) the crude
product can
be precipitated by addition of 10x PrOH 0.1x 2M NaCI and harvested by
centrifugation
decantation. The acetyl hydroxy-protecting groups are removed under basic
conditions,
such as 40% MeNH2 (1mL per 500 OD). After 15 min at RT H20 (1:10 v/v) is added
and
compound 12 (such as X0385B shown in Figure 7) are isolated, purified again by
anion
exchange and size exclusion chromatography and then lyophilised.
General procedure of double strand formation
Individual single strands are dissolved in a concentration of 60 OD/mL in H20.
Both
individual oligonucleotide solutions can be added together to a reaction
vessel. For reaction
monitoring a titration can be performed. The first strand is added in 25%
excess over the
second strand as determined by UV-absorption at 260nm. The reaction mixture is
heated
e.g. to 80 C for 5min and then slowly cooled to RT. Double strand formation
may be
monitored by ion pairing reverse phase HPLC. From the UV-area of the residual
single
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
72
strand the needed amount of the second strand can be calculated and added to
the reaction
mixture. The reaction is heated e.g. to 80 C again and slowly cooled to RT.
This procedure
can be repeated until less than 10% of residual single strand is detected.
The above process (including Schemes 1-4 and Figure 29) may be easily adapted
to
replace GalNac with another targeting ligand e.g. a saccharide.
The present invention relates to a conjugate for inhibiting expression of a
ALDH2 gene in a
cell, said conjugate comprising a nucleic acid portion and ligand portions,
said nucleic acid
portion comprising at least one duplex region that comprises at least a
portion of a first RNA
strand and at least a portion of a second RNA strand that is at least
partially complementary
to the first strand, wherein said first strand is at least partially
complementary to at least a
portion of RNA transcribed from said ALDH2 gene, said ligand portions
comprising a linker
moiety and a targeting ligand for in vivo targeting of cells and being
conjugated exclusively
to the 3' and/or 5' ends of one or both RNA strands, wherein the 5' end of the
first RNA
strand is not conjugated, wherein:
(I) the second RNA strand is conjugated at the 5' end to the targeting
ligand, and
wherein (a) the second RNA strand is also conjugated at the 3' end to the
targeting ligand and the 3' end of the first RNA strand is not conjugated; or
(b)
the first RNA strand is conjugated at the 3' end to the targeting ligand and
the 3'
end of the second RNA strand is not conjugated; or (c) both the second RNA
strand and the first RNA strand are also conjugated at the 3' ends; or
(ii) both the second RNA strand and the first RNA strand are
conjugated at the 3'
ends and the 5' end of the second RNA strand is not conjugated.
The linker may be a serinol-derived linker moiety.
The invention provides, as another aspect, a nucleic acid for inhibiting
expression of
ALDH2 in a cell, comprising at least one duplex region that comprises at least
a portion of
a first strand and at least a portion of a second strand that is at least
partially
complementary to the first strand, wherein said first strand is at least
partially
complementary to at least a portion of a RNA transcribed from the ALDH2 gene,
wherein
said first strand comprises a nucleotide sequence selected from the following
sequences:
SEQ ID NOs:1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25 or 27, wherein the
nucleic acid is
conjugated to a ligand. The second strand may comprise a nucleotide sequence
of SEQ
ID NO:2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26 or 28. The nucleotides of
the first and/or
second strand may be modified, as herein described.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
73
Preferably, the nucleic acid comprises SEQ ID NO:3, SEQ ID NO:11 or SEQ ID
NO:17
and SEQ ID NO:4, SEQ ID NO:12 or SEQ ID NO:18 conjugated to a ligand of
formula I
(as set out above), wherein the ligand is conjugated to the nucleic acid as
described and
wherein the first strand is modified with a 2'0Me modification on the odd
numbered
nucleotides, and modified with a 2'F on the even numbered nucleotides, and the
second
strand is modified with a 2'0Me on the even numbered nucleotides and modified
with a
2'F on the odd numbered nucleotides.
More preferably, the nucleic acid comprises SEQ ID NO:3, SEQ ID NO:11 or SEQ
ID
NO:17 and SEQ ID NO:4, SEQ ID NO:12 or SEQ ID NO:18, wherein the nucleic acid
is
conjugated to a ligand of formula I (as set out above), and furthermore
wherein the nucleic
acid has a modification pattern as shown below which is an extract of Table 1
as herein
provided.
Sequence
siRNA ID sequence modifications
ID No
3 5'-AAUGUUUUCCUGCUGACGG-3' 6254515173547182748
ALDH2-hcm-2
4 5'-CCGUCAGCAGGAAAACAUU-3' 3745364728462627251
11 5'-UCUUCUUAAACUGAGUUUC-3' 5351715262718281517
ALDH2-hcm-6
12 5'-GAAACUCAGUUUAAGAAGA-3' 4626353645152646282
17 5'-AUGUAGCCGAGGAUCUUCU-3' 6181647382846171535
ALDH2-hcm-9
18 5'-AGAAGAUCCUCGGCUACAU-3' 2826461735384716361
wherein the specific modifications are depicted by numbers
1=2"F-dU,
2=2`-F-dA,
3=2"F-dC,
4=2"F-dG,
5=2'-0Me-rU;
6=2'-0Me-rA;
7=2'-0Me-rC;
8=2'-0Me-rG.
The ligand may comprise GaINAc and Figure 10A or Figure 10B further illustrate
examples of the present invention.
The present invention also provides pharmaceutical compositions comprising the
nucleic
acid or conjugated nucleic acid of the invention. The pharmaceutical
compositions may be
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
74
used as medicaments or as diagnostic agents, alone or in combination with
other agents.
For example, one or more nucleic acid conjugates of the invention can be
combined with
a delivery vehicle (e.g., liposomes) and/or excipients, such as carriers,
diluents. Other
agents such as preservatives and stabilizers can also be added. Methods for
the delivery
of nucleic acids are known in the art and within the knowledge of the person
skilled in the
art.
The nucleic acid or conjugated nucleic acid of the present invention can also
be
administered in combination with other therapeutic compounds, either
administrated
separately or simultaneously, e.g., as a combined unit dose. The invention
also includes a
pharmaceutical composition comprising one or more nucleic acids or conjugated
nucleic
acids according to the present invention in a physiologically/pharmaceutically
acceptable
excipient, such as a stabilizer, preservative, diluent, buffer, and the like.
Dosage levels for the medicament and pharmaceutical compositions of the
invention can
be determined by those skilled in the art by routine experimentation. In one
embodiment,
a unit dose may contain between about 0.01 mg/kg and about 100 mg/kg body
weight of
nucleic acid or conjugated nucleic acid. Alternatively, the dose can be from
10 mg/kg to 25
mg/kg body weight, or 1 mg/kg to 10 mg/kg body weight, or 0.05 mg/kg to 5
mg/kg body
weight, or 0.1 mg/kg to 5 mg/kg body weight, or 0.1 mg/kg to1 mg/kg body
weight, or 0.1
mg/kg to 0.5 mg/kg body weight, or 0.5 mg/kg to 1 mg/kg body weight. Dosage
levels may
also be calculated via other parameters such as, e.g., body surface area.
The pharmaceutical composition may be a sterile injectable aqueous suspension
or
solution, or in a lyophilized form.
The pharmaceutical compositions and medicaments of the present invention may
be
administered to a mammalian subject in a pharmaceutically effective dose. The
mammal
may be selected from a human, a non-human primate, a simian or prosimian, a
dog, a cat,
a horse, cattle, a pig, a goat, a sheep, a mouse, a rat, a hamster, a hedgehog
and a
guinea pig, or other species of relevance. On this basis, the wording "ALDH2"
or "ALDH2"
as used herein denotes nucleic acid or protein in any of the above mentioned
species, if
expressed therein naturally or artificially, but preferably this wording
denotes human
nucleic acids or proteins.
A further aspect of the invention relates to a nucleic acid or conjugated
nucleic acid of the
invention or the pharmaceutical composition comprising the nucleic acid or
conjugated
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
nucleic acid of the invention for use in the treatment of a disease, disorder
or syndrome.
The disease, disorder or syndrome may be alcohol use disorder which includes
acute
alcohol sensitivity (hangover), alcoholic neuropathy (alcohol-related
polyneuropathy),
alcohol dependence, alcohol abuse (alcoholism), or foetal alcohol syndrome
(alcohol-
5 related neurodevelopmental disorder) or any other pathology associated to
acute or
prolonged excessive alcohol consumption. The invention includes a
pharmaceutical
composition comprising one or more nucleic acids or conjugated nucleic acids
according
to the present invention in a physiologically/ pharmaceutically acceptable
excipient, such
as a stabiliser, preservative, diluent, buffer and the like.
The pharmaceutical composition may be a sterile injectable aqueous suspension
or
solution, or in a lyophilised form or adhered, absorbed or included to or into
any other
suitable galenic carrier substance such as pellets, tablets, capsules,
nanoparticles, gels,
tablets, beads or similar structures.
The nucleic acid described herein may be capable of inhibiting the expression
of ALDH2.
The nucleic acid described herein may be capable of partially inhibiting the
expression of
ALDH2. Inhibition may be complete, i.e. 0% compared of the expression level of
ALDH2 in
the absence of the nucleic acid of the invention. Inhibition of ALDH2
expression may be
partial, i.e. it may be 15%, 20%, 30%, 40%, 50%, 60%, 70%, 75%, 80%, 85%, 90%,
95%
or intermediate values of ALDH2 expression in the absence of a nucleic acid of
the
invention. Inhibition may last 2 weeks, 3 weeks, 4 weeks, 5 weeks, 6 weeks, 7
weeks, 8
weeks, 10 weeks, 11 weeks, 12 weeks, 13 weeks, 14 weeks or up to 3 months,
when
used in a subject, such as a human patient. A nucleic acid or conjugated
nucleic acid of
the invention, or compositions including the same, may be for use in a regimen
comprising
treatments once or twice weekly, every week, every two weeks, every three
weeks, every
four weeks, every five weeks, every six weeks, every seven weeks, or every
eight weeks,
or in regimens with varying dosing frequency such as combinations of the
before-
mentioned intervals. The nucleic acid may be for use subcutaneously,
intravenously or
using any other application routes such as oral, rectal or intraperitoneal.
In cells and/or subjects treated with or receiving the nucleic acid or
conjugated nucleic
acid of the present invention, the ALDH2 expression may be inhibited compared
to
untreated cells and/or subjects by a range from 15% up to 100% but at least
about 30%,
35%, 40%, 45%, 50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 95%, 98%, or 100%
or intermediate values. The level of inhibition may allow treatment of a
disease associated
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
76
with ALDH2 expression or overexpression, or may serve to further investigate
the
functions and physiological roles of the ALDH2 gene product.
A further aspect of the invention relates to a nucleic acid or conjugated
nucleic acid of the
invention in the manufacture of a medicament for treating a disease, disorder
or
syndromes, such as those as listed above or additional pathologies associated
with acute
or prolonged excessive alcohol consumption, or additional therapeutic
approaches where
inhibition of ALDH2 expression is desired.
Also included in the invention is a method of treating or preventing a
disease, disorder or
syndrome, such as those listed above, comprising administration of a
pharmaceutical
composition comprising a nucleic acid or conjugated nucleic acid as described
herein, to
an individual in need of treatment (to improve such pathologies). The nucleic
acid
composition may be administered in a regimen comprising treatments twice every
week,
once every week, every two weeks, every three weeks, every four weeks, every
five
weeks, every six weeks, every seven weeks, or every eight weeks or in regimens
with
varying dosing frequency such as combinations of the before-mentioned
intervals. The
nucleic acid or conjugated nucleic acid may be for use subcutaneously or
intravenously or
other application routes such as oral, rectal or intraperitoneal.
The nucleic acid or conjugated nucleic acid of the present invention can also
be
administered for use in combination with other therapeutic compounds, either
administered separately or simultaneously, e.g., as a combined unit dose. A
molecular
conjugation to other biologically active molecular entities such as peptides,
cellular or
artificial ligands or small and large molecules is also possible.
The nucleic acid or conjugated nucleic acid of the present invention can be
produced
using routine methods in the art including chemical synthesis or expressing
the nucleic
acid either in vitro (e.g., run off transcription) or in vivo. For example,
using solid phase
chemical synthesis or using a nucleic acid-based expression vector including
viral
derivates or partially or completely synthetic expression systems. In one
embodiment, the
expression vector can be used to produce the nucleic acid of the invention in
vitro, within
an intermediate host organism or cell type, within an intermediate or the
final organism or
within the desired target cell. Methods for the production (synthesis or
enzymatic
transcription) of the nucleic acid described herein are known to persons
skilled in the art.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
77
The invention consists of chemical molecular entities that mediate ALDH2 mRNA
degradation by binding to the ALDH2 gene transcripts through cellular RNA
interference
mechanisms. The molecular compounds invented may be used as conjugates with,
but
are not limited to an N-acetylgalactosamin (GaINAc) sugar moiety that ensures
hepatocyte-specific cellular uptake, though specific binding to the
asialoglycoprotein
receptor complex (ASGPR). The invention may be linked to other different
chemical
structures conferring different properties as referred to in the following.
The use of a
chemical modification pattern of the nucleic acids confers nuclease stability
in serum and
makes for example subcutaneous application route feasible.
Low substrate specificity of known drugs inhibiting up to 20 different enzymes
in addition
to ALDH2 leads to severe undesired physiological effects in several tissues
such as heart
muscle and the central and peripheral nervous system even in non-drinking
patients. The
present invention aims at conferring assistance in alcohol dishabituation
inducing alcohol
aversion with a better safety profile than the currently available treatments.
The invention
is characterized by high specificity at the molecular and tissue-directed
delivery level.
The invention is characterized by high specificity at the molecular and tissue-
directed
delivery level, potentially conferring a better safety profile than the
currently available
treatments.
The invention also provides a nucleic acid according to any aspect of the
invention
described herein, wherein the first RNA strand has a terminal 5' (E)-
vinylphosphonate
nucleotide, and the terminal 5' (E)-vinylphosphonate nucleotide is linked to
the second
nucleotide in the first strand by a phosphodiester linkage.
In one embodiment, the first strand may include more than 1 phosphodiester
linkage.
In one embodiment, the first strand may comprise phosphodiester linkages
between at
least the terminal three 5' nucleotides.
In one embodiment, the first strand may comprise phosphodiester linkages
between at
least the terminal four 5' nucleotides.
In one embodiment, the first strand may comprise formula (XVII):
(Vp)-N(po)[N(po)]n-
(XVII)
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
78
where '(vp)-' is the 5' (E)-vinylphosphonate, 'N' is a nucleotide, 'po' is a
phosphodiester
linkage, and n is from 1 to (the total number of nucleotides in the first
strand - 2),
preferably wherein n is from 1 to (the total number of nucleotides in the
first strand -3),
more preferably wherein n is from 1 to (the total number of nucleotides in the
first strand -
4).
In one embodiment, the first strand may include at least one phosphorothioate
(ps)
linkage.
In one embodiment, the first strand may further comprise a phosphorothioate
linkage
between the terminal two 3' nucleotides or phosphorothioate linkages between
the
terminal three 3' nucleotides.
In one embodiment, the linkages between the other nucleotides in the first
strand are
phosphodiester linkages.
In one embodiment, the first strand may include more than 1 phosphorothioate
linkage.
In a further embodiment, the second strand may comprise a phosphorothioate
linkage
between the terminal two 3' nucleotides or phosphorothioate linkages between
the
terminal three 3' nucleotides.
In another further embodiment, the second strand may comprise a
phosphorothioate
linkage between the terminal two 5' nucleotides or phosphorothioate linkages
between the
terminal three 5' nucleotides.
In an embodiment, the terminal 5' (E)-vinylphosphonate nucleotide is an RNA
nucleotide.
A terminal 5' (E)-vinylphosphonate nucleotide is a nucleotide wherein the
natural phosphate
group at the 5'-end has been replaced with a E-vinylphosphonate, in which the
bridging 5'-
oxygen atom of the terminal nucleotide of the 5' phosphorylated strand is
replaced with a
methynyl (-CH=) group:
0
Nucleotides with a natural phosphate at the 5'-end
-0'
"~1 Rase
6 OMe
C=P-0
6
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
79
Nucleotide with a E-vinylphosphonate at the 5'-end
Base
0
"8-
0=P-C
0
5(E) vinylphosphonate is a 5' phosphate mimic. A biological mimic is a
molecule that is
capable of carrying out the same function as and is structurally very similar
to the original
molecule that is being mimicked. In the context of the present invention, 5'
(E)
vinylphosphonate mimics the function of a normal 5' phosphate, e.g. enabling
efficient RISC
loading. In addition, because of its slightly altered structure, 5(E)
vinylphosphonate is
capable of stabilizing the 5'-end nucleotide by protecting it from
dephosphorylation by
enzymes such as phosphatases.
One aspect of the invention is a nucleic acid as disclosed herein for
inhibiting expression
of a target gene in a cell, comprising at least one duplex region that
comprises at least a
portion of a first strand and at least a portion of a second strand that is at
least partially
complementary to the first strand, wherein said first strand is at least
partially
complementary to at least a portion of RNA transcribed from said target gene,
wherein
said first strand includes modified nucleotides or unmodified nucleotides at a
plurality of
positions in order to facilitate processing of the nucleic acid by RISC.
In one aspect "facilitate processing by RISC" means that the nucleic acid can
be
processed by RISC, for example any modification present will permit the
nucleic acid to be
processed by RISC, suitably such that siRNA activity can take place.
A nucleic acid as disclosed herein, wherein the nucleotides at positions 2 and
14 from the
5' end of the first strand are not modified with a 2' 0-methyl modification,
and the
nucleotide on the second strand which corresponds to position 13 of the first
strand is not
modified with a 2' 0-methyl modification.
A nucleotide on the second strand that "corresponds to" a position on the
first strand is
suitably the nucleotide that base pairs with that nucleotide on the first
strand.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
In one aspect the nucleotide on the second strand which corresponds to
position 13 of the
first strand is the nucleotide that forms a base pair with position 13 of the
first strand.
In one aspect the nucleotide on the second strand which corresponds to
position 11 of the
5 first strand is the nucleotide that forms a base pair with position 11 of
the first strand.
In one aspect the nucleotide on the second strand which corresponds to
position 12 of the
first strand is the nucleotide that forms a base pair with position 12 of the
first strand.
10 This nomenclature may be applied to other positions of the second
strand. For example,
in a 19-mer nucleic acid which is double stranded and blunt ended, position 13
of the first
strand would pair with position 7 of the second strand. Position 11 of the
first strand would
pair with position 9 of the second strand. This nomenclature may be applied to
other
positions of the second strand.
The nucleotide that corresponds to position 13 of the first strand is suitably
position 13 of
the second strand, counting from the 3' of the second strand, starting from
the first
nucleotide of the double stranded region. Likewise position 11 of the second
strand is
suitably the 11th nucleotide from the 3' of the second strand, starting from
the first
nucleotide of the double stranded region. This nomenclature may be applied to
other
positions of the second strand.
In one aspect, in the case of a partially complementary first and second
strand, the
nucleotide on the second strand that "corresponds to" a position on the first
strand may
not necessarily form a base pair if that position is the position in which
there is a
mismatch, but the principle of the nomenclature still applies.
Preferred is a first and second strand that are fully complementary over the
duplex region
(ignoring any overhang regions) and there are no mismatches within the double
stranded
region of the nucleic acid.
Also preferred are:
A nucleic acid as disclosed herein, wherein the nucleotides at positions 2 and
14 from the
5' end of the first strand are not modified with a 2' 0-methyl modification,
and the
nucleotide on the second strand which corresponds to position 11 of the first
strand is not
modified with a 2' 0-methyl modification.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
81
A nucleic acid as disclosed herein, wherein the nucleotides at positions 2 and
14 from the
5' end of the first strand are not modified with a 2' 0-methyl modification,
and the
nucleotides on the second strand which corresponds to position 11 and 13 of
the first
strand are not modified with a 2' 0-methyl modification.
In one aspect the nucleotide on the second strand which corresponds to
position 12 of the
first strand is not modified with a 2' 0-methyl modification. This limitation
on the nucleic
acid may be seen with any other limitation described herein.
Therefore another aspect of the invention is a nucleic acid as disclosed
herein, wherein
the nucleotides at positions 2 and 14 from the Send of the first strand are
not modified
with a 2' 0-methyl modification, and the nucleotides on the second strand
which
corresponds to position 11-13 of the first strand are not modified with a 2' 0-
methyl
modification.
A nucleic acid as disclosed herein, wherein the nucleotides at positions 2 and
14 from the
5' end of the first strand are not modified with a 2' 0-methyl modification,
and the
nucleotides on the second strand which correspond to position 11, or 13, or 11
and 13, or
11-13 of the first strand are modified with a 2' fluoro modification.
A nucleic acid as disclosed herein, wherein the nucleotides at positions 2 and
14 from the
5' end of the first strand are modified with a 2' fluoro modification, and the
nucleotides on
the second strand which correspond to position 11, or 13, or 11 and 13, or 11-
13 of the
first strand are not modified with a 2' 0-methyl modification.
A nucleic acid as disclosed herein, wherein the nucleotides at positions 2 and
14 from the
5' end of the first strand are modified with a 2' fluoro modification, and the
nucleotides on
the second strand which correspond to position 11, or 13, or 11 and 13, or 11-
13 of the
first strand are modified with a 2' fluoro modification.
A nucleic acid as disclosed herein wherein greater than 50% of the nucleotides
of the first
and/or second strand comprise a 2' 0-methyl modification, such as greater than
55%,
60%, 65%, 70%, 75%, 80%, or 85%, or more, of the first and/or second strand
comprise a
2' 0-methyl modification, preferably measured as a percentage of the total
nucleotides of
both the first and second strands.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
82
A nucleic acid as disclosed herein wherein greater than 50% of the nucleotides
of the first
and/or second strand comprise a naturally occurring RNA modification, such as
wherein
greater than 55%, 60%, 65%, 70%, 75%, 80%, or 85% or more of the first and/or
second
strands comprise such a modification, preferably measured as a percentage of
the total
nucleotides of both the first and second strands. Suitable naturally occurring
modifications
include, as well as 2 0' methyl, other 2' sugar modifications, in particular a
2' H
modification resulting in a DNA nucleotide.
A nucleic acid as disclosed herein comprising no more than 20%, such as no
more than
15% such as more than 10%, of nucleotides which have 2' modifications that are
not 2' 0
methyl modifications on the first and/or second strand, preferably as a
percentage of the
total nucleotides of both the first and second strands.
A nucleic acid as disclosed herein comprising no more than 20%, (such as no
more than
15% or no more than 10%) of 2' fluoro modifications on the first and/or second
strand,
preferably as a percentage of the total nucleotides of both strands.
A nucleic acid as disclosed herein, wherein all nucleotides are modified with
a 2' 0-methyl
modification except positions 2 and 14 from the 5' end of the first strand and
the
nucleotides on the second strand which correspond to position 11, or 13, or 11
and 13, or
11-13 of the first strand. Preferably the nucleotides that are not modified
with 2' 0-methyl
are modified with fluoro at the 2' position.
Preferred is a nucleic acid as disclosed herein wherein all nucleotides of the
nucleic acid
are modified at the 2' position of the sugar. Preferably these nucleotides are
modified with
a 2'- fluoro modification where the modification is not a 2' 0-Methyl
modification.
Nucleic acids of the invention may comprise one or more nucleotides modified
at the 2'
position with a 2' H, and therefore having a DNA nucleotide within the nucleic
acid.
Nucleic acids of the invention may comprise DNA nucleotides at positions 2
and/or 14 of
the first strand counting from the 5' end of the first strand. Nucleic acids
may comprise
DNA nucleotides on the second strand which correspond to position 11, or 13,
or 11 and
13, or 11-13 of the first strand.
In one aspect there is no more than one DNA per nucleic acid of the invention.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
83
Nucleic acids of the invention may comprise one or more LNA nucleotides.
Nucleic acids
of the invention may comprise LNA nucleotides at positions 2 and/or 14 of the
first strand
counting from the 5' end of the first strand. Nucleic acids may comprise LNA
on the
second strand which correspond to position 11, or 13, or 11 and 13, or 11-13
of the first
strand.
In one aspect the nucleic acid is modified on the first strand with
alternating 2-0 methyl
modifications and 2 fluoro modifications, and positions 2 and 14 (starting
from the 5' end)
are modified with 2' fluoro. Preferably the second strand is modified with 2'
fluoro
modifications at nucleotides on the second strand which correspond to position
11, or 13,
or 11 and 13, or 11-13 of the first strand. Preferably the second strand is
modified with 2'
fluoro modifications at positions 11-13 counting from the 3' end starting at
the first position
of the complementary (double stranded) region, and the remaining modifications
are
naturally occurring modifications, preferably 2' 0-methyl.
In one aspect the nucleic acids of the invention comprise one or more inverted
ribonucleotides, preferably an inverted adenine, using a 5'-5' linkage or a 3'-
3' linkage,
preferably a 3'-3' linkage at the 3' end of the second strand.
In one aspect the nucleic acid comprises one or more phosphorodithioate
linkages, such
as 1, 2, 3 or 4 phosphorodithioate linkages. Preferably there are up to 4
phosphorodithioate linkages, one each at the 5' and 3' ends of the first and
second
strands.
All the features of the nucleic acids can be combined with all other aspects
of the
invention disclosed herein.
In particular, preferred are nucleic acids which are siRNA molecules wherein
the
nucleotides at positions 2 and 14 from the 5' end of the first strand are not
modified with a
2' 0-methyl modification, and the nucleic acid comprises one or more or all
of:
(i) an inverted nucleotide, preferably a 3'-3' linkage at the 3' end of the
second strand;
(ii) one or more phosphorodithioate linkages;
(iii) the second strand nucleotide corresponding to position 11 or 13 of the
first strand is
not modified with a 2' 0-methyl modification, preferably wherein one or both
of these
positions comprise a 2' fluoro modification;
(iv) the nucleic acid comprises at least 80% of all nucleotides having a 2'-0-
methly
modification;
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
84
(v) the nucleic acid comprises no more than 20% of nucleotides which have 2'
fluoro
modifications.
Also provided by the present invention is a nucleic acid as disclosed herein,
wherein the
nucleotides at positions 2 and 14 from the 5' end of the first strand and the
nucleotides at
positions 7 and/or 9, or 7 - 9 from the 5' end of the second strand are
modified with a 2'
fluoro modification, and at least 90% of the remaining nucleotides are 2'-0
methyl
modified or comprise another naturally occurring 2' modification.
Specific preferred examples, for a blunt double stranded 19 base nucleic acid,
with no
overhang, are:
A nucleic acid as disclosed herein, wherein the nucleotides at positions 2 and
14 from the
5' end of the first strand are not modified with a 2' 0-methyl modification,
and the
nucleotide at position 7 from the 5' end of the second strand is not modified
with a 2' 0-
methyl modification.
A nucleic acid as disclosed herein, wherein the nucleotides at positions 2 and
14 from the
5' end of the first strand are not modified with a 2' 0-methyl modification,
and the
nucleotide at position 9 from the 5' end of the second strand is not modified
with a 2' 0-
methyl modification
A nucleic acid as disclosed herein, wherein the nucleotides at positions 2 and
14 from the
5' end of the first strand are not modified with a 2' 0-methyl modification,
and the
nucleotides at position 7 and 9 from the 5' end of the second strand are not
modified with
a 2' 0-methyl modification.
A nucleic acid as disclosed herein, wherein the nucleotides at positions 2 and
14 from the
5' end of the first strand are not modified with a 2' 0-methyl modification,
and the
nucleotides at positions 7 - 9 from the 5' end of the second strand are not
modified with a
2' 0-methyl modification.
A nucleic acid as disclosed herein, wherein the nucleotides at positions 2 and
14 from the
5' end of the first strand are not modified with a 2' 0-methyl modification,
and the
nucleotides at positions 7 and/or 9, or 7-9 from the 5' end of the second
strand are
modified with a 2' fluoro modification.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
A nucleic acid as disclosed herein, wherein the nucleotides at positions 2 and
14 from the
5' end of the first strand are modified with a 2' fluoro modification, and the
nucleotides at
positions 7 and/or 9, or 7 - 9 from the 5' end of the second strand are not
modified with a
2' 0-methyl modification.
5
A nucleic acid as disclosed herein, wherein the nucleotides at positions 2 and
14 from the
5' end of the first strand are modified with a 2' fluoro modification, and the
nucleotides at
positions 7 and/or 9, or 7 - 9 from the 5' end of the second strand are
modified with a 2'
fluoro modification.
A nucleic acid as disclosed herein wherein greater than 50% of the nucleotides
of the first
and/or second strand comprise a 2' 0-methyl modification, such as greater than
55%,
60%, 65%, 70%, 75%, 80%, or 85%, or more, of the first and/or second strand
comprise a
2' 0-methyl modification, preferably measured as a percentage of the total
nucleotides of
both the first and second strands.
A nucleic acid as disclosed herein wherein greater than 50% of the nucleotides
of the first
and/or second strand comprise a naturally occurring RNA modification, such as
wherein
greater than 55%, 60%, 65%, 70%, 75%, 80%, or 85% or more of the first and/or
second
strands comprise such a modification, preferably measured as a percentage of
the total
nucleotides of both the first and second strands. Suitable naturally occurring
modifications
include, as well as 2 0' methyl, other 2' sugar modifications, in particular a
2' H
modification resulting in a DNA nucleotide.
A nucleic acid as disclosed herein comprising no more than 20%, such as no
more than
15% such as more than 10%, of nucleotides which have 2' modifications that are
not 2' 0
methyl modifications on the first and/or second strand, preferably as a
percentage of the
total nucleotides of both the first and second strands.
A nucleic acid as disclosed herein comprising no more than 20%, (such as no
more than
15% or no more than 10%) of 2' fluoro modifications on the first and/or second
strand,
preferably as a percentage of the total nucleotides of both strands.
A nucleic acid as disclosed herein, wherein all nucleotides are modified with
a 2' 0-methyl
modification except positions 2 and 14 from the 5' end of the first strand and
the
nucleotides at positions 7 and/or 9 from the 5' end of the second strand.
Preferably the
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
86
nucleotides that are not modified with 2' 0-methyl are modified with fluoro at
the 2'
position.
A nucleic acid as disclosed herein, wherein all nucleotides are modified with
a 2' 0-methyl
modification except positions 2 and 14 from the 5' end of the first strand and
the
nucleotides at positions 7 - 9 from the 5' end of the second strand.
Preferably the
nucleotides that are not modified with 2' 0-methyl are modified with fluoro at
the 2'
position.
For a nucleic acid comprising a 20 base pair duplex region, the second strand
preferably
does not have a 2' 0-methyl group at nucleotides 8 or 9 or 10 counting from
the 5' end of
the duplex corresponding to positions 13, 12, and 11 of the first strand
respectively.
For a nucleic acid comprising a 21 base pair duplex region, the second strand
preferably
does not have a 2' 0-methyl group at nucleotides 9 or 10 or 11 counting from
the 5' end of
the duplex corresponding to positions 13, 12, and 11 of the first strand
respectively.
The present invention also relates to the unmodified sequences of all modified
sequences
disclosed herein.
The invention will now be described with reference to the following non-
limiting Figures
and Examples.
Figures
Figure 1 shows the results of a unconjugated RNAi molecule screen for
inhibition of
ALDH2 expression in human Hep3B cells.
Figure 2 shows the dose response of ALDH2 siRNA molecules in human HepG2
cells.
Figure 3 shows mRNA and protein reduction by ALDH2-hcm-2 siRNA in human HepG2
cells as well as its effect on ALDH enzymatic activity in HepG2 total cell
extracts.
Figure 4 shows the dose-response for GaINAc-coupled siRNAs targeting Aldh2
mRNA in
primary mouse hepatocytes.
Figure 5 shows the dose response for Gal NAc-coupled siRNAs against ALDH2 mRNA
in
primary human hepatocytes.
Figure 6 shows the reduction of ALDH2 mRNA by 15nM of GaINAc-coupled ALDH2-hcm-
2 siRNA in 10 human, cynomolgus and mouse hepatocytes.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
87
Figure 7 shows the comparison in dose-response on ALDH2 mRNA knockdown by
GaINAc-conjugated ALDH2 siRNAs hcm2, hcm6, hcm9 in conjugation to L1 linker
vs.
conjugation to the L6 GaINAc-linker in 1 human hepatocytes.
Figure 8a show Aldh2 mRNA knockdown in mice in vivo 10 days post treatment, 8b
the
resulting reduction of ALDH2 protein and 8c the resulting hepatic acetaldehyde
metabolite
levels in the same mice.
Figure 9 shows the dose-dependent duration of the Aldh2 mRNA knockdown up to
41
days upon a single dose application in mice using as an example GaINAc-ALDH2-
hcm6
and hcm9 conjugated to the L6 linked GaINAc moiety.
Figures 10a and 10b show the structure of the GaINAc L1 and L6 ligands,
respectively, to
which the example oligonucleotides were conjugated.
Figure 11 shows different siRNA duplexes containing inverted RNA nucleotides
at both T-
ends tested for serum stability.
Figure 12 shows the influence on ALDH2 mRNA knockdown of inverted A, U, C and
G
RNA nucleotides at 3`-overhang positions using derivatives of the siRNA ALDH2-
hcm2
(see Table 3).
Figure 13 shows the influence on ALDH2 mRNA knockdown of inverted A, U, C and
G
RNA nucleotides at terminal 3' positions using derivatives of the siRNA ALDH2-
hcm2 (see
Table 3).
Figure 14 shows different GaINAc-L6-linker conjugates of ALDH2-hcm2 containing
inverted RNA nucleotides (see Table 4) tested for serum stability.
Figure 15 shows the dose response of inverted RNA nucleotides at terminal 3'
positions
on the efficacy of ALDH2 mRNA knockdown using GaINAc-L6-conjugated derivatives
of
the siRNA ALDH2-hcm2 (see Table 4) by ASGPR receptor complex-mediated uptake
in
mouse primary hepatocytes.
Figure 16 shows the results of an in vivo study in mice testing the influence
of ivA at the
first strand's 3`-end in GaINAc-L6 derivatives of ALDH2-hcm6 and ALDH2-hcm9
siRNA
conjugates on the efficacy of ALDH2 mRNA knockdown.
Figure 17 shows the effect of different modified derivatives of the GaINAc-
siRNA
conjugates of ALDH2-hcm6 and ALDH2-hcm9 (Table 5) on knockdown efficacy on
Aldh2
mRNA expression levels in vivo in mice.
Figure 18 depicts Conjugate 1.
Figure 19 depicts Conjugate 2.
Figure 20 depicts Conjugate 3.
Figure 21 depicts Reference Conjugate 1.
Figure 22 depicts Reference Conjugate 2.
Figure 23 depicts Reference Conjugate 3.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
88
Figure 24 depicts Reference Conjugate 4.
In each of Figures 18-24 and 37-49, the top strand is the antisense strand and
the bottom
strand is the sense strand. In addition, to show more clearly the connection
between the
nucleic acid and ligand portions, the nucleotide at the end of the respective
conjugated
strands is drawn in full.
Figure 25 shows the synthesis of A0268, which is a 3' mono-GaINAc conjugated
single
stranded oligonucleotide and is the starting material in the synthesis of
Conjugate 1 and
Conjugate 3. (ps) denotes phosphorothioate linkage.
Figure 26 shows the synthesis of A0006, which is a 5' tri-antennary GaINAc
conjugated
single stranded oligonucleotide used for the synthesis of Reference Conjugate
4. (ps)
denotes phosphorothioate linkage.
Figure 27 illustrates the in vitro determination of TTR knockdown. In
particular, Figure 27A
shows the in vitro determination of TTR knockdown by Reference Conjugates (RC)
1 and
3 as well as the untreated control "UT"; Figure 27B shows the in vitro
determination of TTR
knockdown by Reference Conjugates (RC) 2 and 3, as well as the untreated
control "UT";
and Figure 27C shows the in vitro determination of TTR knockdown by Conjugates
1, 2 and
3, as well as by RC3 and untreated control "UT". Reference Conjugates 1 and 2
represent
comparator conjugates. Reference Conjugate 3 represents a non-targeting GaINAc
siRNA
and "untreated" ("UT") represents untreated cells. Both RC3 and UT are
negative controls.
mRNA levels were normalised against Ptenll.
Figure 28 shows a time course of serum TTR in c57BL/6 mice cohorts of n=4 at
7, 14, and
27 days post s.c. treatment with lmg/kg - Conjugates 1-3, Reference Conjugates
(RC) 1, 2
and 4 and mock treated (PBS) individuals.
Figure 29 shows oligonucleotide synthesis of 3' and 5' GaINAc conjugated
oligonucleotides
precursors (such as compound X0385B-prec).
Figure 30 discloses GaINAc conjugates with each one serinol-linked GaINAc
moiety at both
termini of the second strand and with 2'-F at positions 7-9 of the second
strand reducing
ALDH2 mRNA levels in vitro.
Figure 31 discloses GaINAc siRNA conjugates with vinylphosphonate at the 5'
end of the
first strand and phosphodiester internucleotide linkages at the 5' end of the
first strand effect
reduction of ALDH2 target mRNA levels in vitro.
Figure 32 discloses GaINAc siRNA conjugates with vinylphosphonate at the 5'
end of the
first strand and phosphodiester internucleotide linkages at the 5' end of the
first strand effect
improved reduction of ALDH2 target mRNA levels in vitro.
Figure 33 shows that GaINAc siRNA conjugates with vinylphosphonate at the 5'
end of the
first strand and phosphodiester internucleotide linkages at the 5' end of the
first strand are
stable in acidic tritosome lysate.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
89
Figure 34 shows that GaINAc siRNA conjugates with vinylphosphonate at the 5'
end of the
first strand and phosphodiester internucleotide linkages at the 5' end of the
first strand are
stable in acidic tritosome lysate.
Figure 35 discloses GaINAc siRNA conjugates with vinylphosphonate at the 5'
end of the
first strand and phosphodiester internucleotide linkages at the 5' end of the
first strand effect
reduction of ALDH2 target mRNA levels in vitro.
Figure 36 shows improved duration of Aldh2 mRNA knockdown in mice following
s.c.
administration of 1mg/kg ALDH2-hcm9 siRNA conjugate 16 with 5' & 3' serinol-
GaINAc
second strand conjugation as compared to conjugate 14 with 5' second strand
triantennary GaINAc-conjugation.
Figure 37 depicts Conjugate 4. The last three nucleotides at the 5' and 3'
ends of the
antisense and sense strands are connected by a phosphorothioate linker between
each
nucleotide. The serinol-GaINAc-linkers are conjugated via a phosphodiester
bond to the 3'
end and the 5' end of the sense strand.
Figure 38 depicts Conjugate 5. The last three nucleotides at the 5' and 3'
ends of the
antisense strand are connected by a phosphorothioate linker between each
nucleotide. The
serinol-GaINAc-linkers are conjugated via a phosphorothioate bond to the 3'
end and the 5'
end of the sense strand.
Figure 39 depicts Conjugate 6. The last three nucleotides at the 5' and 3'
ends of the
antisense strand are connected by a phosphorothioate linker between each
nucleotide. The
serinol-GaINAc-linkers are conjugated via a phosphodiester bond to the 3' end
and the 5'
end of the sense strand.
Figure 40 depicts Conjugate 7. The last three nucleotides at the 5' and 3'
ends of the
antisense and sense strands are connected by a phosphorothioate linker between
each
nucleotide. The serinol-GaINAc-linkers are conjugated via a phosphorothioate
bond to the
3' end and the 5' end of the sense strand. The serinol-GaINAc-linkers are
connected to
each other via a phosphorothioate bond.
Figure 41 depicts Conjugate 8. The last three nucleotides at the 5' and 3'
ends of the
antisense and sense strands are connected by a phosphorothioate linker between
each
nucleotide. A GaINAc-C6-amino-modifier linker is conjugated at the 5' end of
the sense
strand and a GaINAc-C7-amino-modifier linker is conjugated at the 3' end of
the sense
strand.
Figure 42 depicts Conjugate 9. The last three nucleotides at the 5' and 3'
ends of the
antisense and sense strands are connected by a phosphorothioate linker between
each
nucleotide. A GaINAc-GlyC3-amino-modifier linker is conjugated at the 5' and
3' ends of the
sense strand.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
Figure 43 depicts Conjugate 10. The last three nucleotides at the 5' and 3'
ends of the
antisense and sense strands are connected by a phosphorothioate linker between
each
nucleotide. and sense strands are connected by a phosphorothioate linker
between each
nucleotide. A GaINAc-piperidyl-amino-modifier linker is conjugated at the 5'
and 3' ends of
5 the sense strand.
Figure 44 depicts Conjugate 11. The last three nucleotides at the 5' and 3'
ends of the
antisense and sense strands are connected by a phosphorothioate linker between
each
nucleotide. A GaINAc-C3-amino-modifier linker is conjugated at the 5' and 3'
ends of the
sense strand.
10 Figure 45 depicts Conjugate 12. The last three nucleotides at the 5' and
3' ends of the
antisense and sense strands are connected by a phosphorothioate linker between
each
nucleotide. A GaINAc-C6-amino-modifier linker is conjugated at the 5' end of
the sense
strand and a GaINAc-GlyC3-amino-modifier linker is conjugated at the 3' end of
the sense
strand.
15 Figure 46 depicts Conjugates 15, 16, 18 and 19 which differ only by
their RNA sequences.
The last three nucleotides at the 5' and 3' ends of the antisense and sense
strands are
connected by a phosphorothioate linker between each nucleotide in each
conjugate. The
serinol-GaINAc-linkers are conjugated via a phosphorothioate bond to the 3'
end and the 5'
end of the sense strand.
20 Figure 47 depicts Reference Conjugate 5 and Reference Conjugate 9 which
differ only by
their RNA sequences. The last three nucleotides at the 5' and 3' ends of the
antisense
strand and 3' end of the sense strand are connected by a phosphorothioate
linker between
each nucleotide in both conjugates. The trimeric GaINAc-linker is conjugated
via a
phosphorothioate bond to the 5' end of the sense strand in both conjugates.
25 Figure 48 depicts Reference Conjugate 6 and Reference Conjugate 7 which
differ only by
their RNA sequences. The last three nucleotides at the 5' and 3' ends of the
antisense
strand and 3' end of the sense strand are connected by a phosphorothioate
linker between
each nucleotide in both conjugates. The trimeric GaINAc-linker is conjugated
via a
phosphorothioate bond to the 5' end of the sense strand in both conjugates.
30 Figure 49 depicts Reference Conjugate 8. The last three nucleotides at
the 5' and 3' ends
of the antisense strand and 3' end of the sense strand are connected by a
phosphorothioate
linker between each nucleotide. The trimeric GaINAc-linker is conjugated via a
phosphorothioate bond to the 5' end of the sense strand.
Figure 50 illustrates the in vitro determination of TTR knockdown. In
particular, Figure 50A
35 shows the in vitro determination of TTR knockdown by Conjugates 4, 5, 6
and 2 compared
to "Luc" (Reference Conjugate 3) as well as the untreated control "UT"; Figure
50B shows
the in vitro determination of TTR knockdown by Conjugates 7 and 2, compared to
"Luc"
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
91
(Reference Conjugate 3) as well as the untreated control "UT". Luc or
Reference Conjugate
3 (RC3) represents a non-targeting GaINAc siRNA and "untreated" ("UT")
represents
untreated cells. Both RC3 and UT are negative controls. mRNA level were
normalised
against Ptenll.
Figure 51 illustrates the in vitro determination of TTR knockdown. In
particular, Figure 51A
shows the in vitro determination of TTR knockdown by Conjugates 8, 9, 10, 11
and 2
compared to "Luc" (Reference Conjugate 3) as well as the untreated control
"UT"; Figure
51B shows the in vitro determination of TTR knockdown by Conjugates 12 and 2,
compared
to "Luc" (Reference Conjugate 3) as well as the untreated control "UT". Luc or
Reference
Conjugate 3 represents a non-targeting GaINAc siRNA and "untreated" ("UT")
represents
untreated cells. Both RC3 and UT are negative controls. mRNA levels were
normalised
against Ptenll.
Figure 52 illustrates the in vitro determination of LPA mRNA knockdown by
Conjugate 19
compared to controls. Ctr represents a non-targeting GaINAc siRNA and
"untreated" ("UT")
represents untreated cells. Both Ctr and UT are negative controls. mRNA level
were
normalised against ACTB.
Figure 53 shows a time course of liver ALDH2 mRNA in c57BL/6 mice cohorts of
n=6 at 14,
28 and 42 days post s.c. treatment with lmg/kg - Conjugate 15, Reference
Conjugate (RC)
6 and mock treated (PBS) individuals. mRNA levels were normalised against
Pten.
Figure 54 shows a time course of liver ALDH2 mRNA in c57BL/6 mice cohorts of
n=6 at 14,
28 and 42 days post s.c. treatment with lmg/kg - Conjugate 16, Reference
Conjugate (RC)
7 and mock treated (PBS) individuals. mRNA level were normalised against Pten.
Figure 55 shows a time course of liver TMPRSS6 mRNA in c57BL/6 mice cohorts of
n=6 at
14, 28 and 42 days post s.c. treatment with 1mg/kg - Conjugate 18, Reference
Conjugate
(RC) 8 and mock treated (PBS) individuals. mRNA level were normalised against
Pten.
Figure 56 shows serum stability of Conjugates 4, 5, 6, 7 and 2, and untreated
control (UT)
at 37 C over 3 days.
Figure 57 shows serum stability of Conjugates 8, 9, 10, 11, 12 and 2, and
untreated control
(UT) at 37 C over 3 days.
Figure 58 shows extended reduction of Aldh2 mRNA levels in mice by an ALDH2-
targeting
GaINAc conjugates with an optimised 2'-0Me/2'-F modification pattern in the
second strand
and with end-stabilising inverted RNA at the 3' end of the second strand.
Figure 59 shows schematic representations of varous embodiments of nucleic
acids
conjugated with ligands via linkers.
CA 03118327 2021-04-30
WO 2019/092282
PCT/EP2018/081105
92
Examples
The numbering referred to in each example is specific for said example.
Example 1
Nucleic acids in accordance with the invention were synthesised, using the
oligonucleotides as set out in the tables below.
The method of synthesis was as follows, using one of the sequences of the
invention as
an example:
ALDH2-hcm-2
GaINAc-ALDH2-hcm-2-L1:
First strand (SEQ ID NO: 84, based on SEQ ID NO: 3)
5'0MeA(ps)- FA-(ps)-0MeU-FG-0MeU-FU-OMeU-FU-OMeC-FC-OMeU-FG-OMeC-FU-
OMeG-FA-OMeC-(ps)-FG-(ps)-0MeG 3'
Second strand (SEQ ID NO: 85, based on SEQ ID NO: 4)
5'[5-123 (ps)]3 long trebler (ps) FC-0MeC-FG-0MeU-FC-OMeA-FG-OMeC-FA-OMeG-
FG-OMeA-FA-OMeA-FA-OMeC-FA-(ps)-0MeU-(ps)-FU 3'
GaINAc-ALDH2-hcm-2-L6:
First strand (SEQ ID NO: 86, based on SEQ ID NO: 3)
5'0MeA(ps)- FA-(ps)-0MeU-FG-0MeU-FU-OMeU-FU-OMeC-FC-OMeU-FG-OMeC-FU-
OMeG-FA-OMeC-(ps)-FG-(ps)-0MeG 3'
Second strand (SEQ ID NO: 87, based on SEQ ID NO: 4)
5'[5-123 (ps)]3 5T43 (ps) FC-0MeC-FG-0MeU-FC-OMeA-FG-OMeC-FA-OMeG-FG-
OMeA-FA-0MeA-FA-0MeC-FA-(ps)-0MeU-(ps)-FU 3'
FN (N=A, C, G, U) denotes 2'Fluoro, 2' DeoxyNucleosides
OMeN (N=A, C, G, U) denotes 2'0 Methyl Nucleosides
(ps) indicates a phosphorothioate linkage
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
93
ST23 and ST43 are as below.
A further example is ALDH2-hcm-6
GaINAc-ALDH2-hcm6-L1
First strand (SEQ ID NO: 88, based on SEQ ID NO: 11)
5' OMeU-(ps)-FC-(ps)-0MeU-FU-OMeC-FU-OMeU-FA-OMeA-FA-OMeC-FU-OMeG-FA-
OMeG-FU-OMeU-(ps)-FU-(ps)-0MeC
Second strand (SEQ ID NO: 89, based on SEQ ID NO: 12)
5'[5-123 (ps)]3 long trebler (ps) FG-0MeA-FA-0MeA-FC-OMeU-FC-OMeA-FG-OMeU-
FU-OMeU-FA-0MeA-FG-OMeA-FA-(ps)-0MeG-(ps)-FA 3'
GaINAc-ALDH2-hcm6-L6
First strand (SEQ ID NO: 90, based on SEQ ID NO: 11)
5' OMeU-(ps)-FC-(ps)-0MeU-FU-OMeC-FU-OMeU-FA-OMeA-FA-OMeC-FU-OMeG-FA-
OMeG-FU-OMeU-(ps)-FU-(ps)-0MeC
Second strand (SEQ ID NO: 91, based on SEQ ID NO: 12)
5'[5-123 (ps)]3 5T43 (ps) FG-0MeA-FA-0MeA-FC-OMeU-FC-OMeA-FG-OMeU-FU-
OMeU-FA-OMeA-FG-OMeA-FA-(ps)-0MeG-(ps)-FA 3'
FN (N=A, C, G, U) denotes 2'Fluoro, 2' DeoxyNucleosides
OMeN (N=A, C, G, U) denotes 2'0 Methyl Nucleosides
(ps) indicates a phosphorothioate linkage
5T23 and 5T43 are as below.
A further example is ALDH2-hcm-9
GaINAc-ALDH2-hcm9-L1
First strand (SEQ ID NO: 92, based on SEQ ID NO: 17)
5'0MeA-(ps)-FU-(ps)-0MeG-FU-OMeA-FG-0MeC-FC-OMeG-FA-OMeG-FG-OMeA-FU-
OMeC-FU-OMeU-(ps)-FC-(ps)-0MeU 3'
CA 03118327 2021-04-30
WO 2019/092282
PCT/EP2018/081105
94
Second strand (SEQ ID NO: 93, based on SEQ ID NO:18)
5.[ST23 (ps)]3 long trebler (ps) FA-0MeG-FA-0MeA-FG-OMeA-FU-OMeC-FC-OMeU-
FC-OMeG-FG-OMeC-FU-OMeA-FC-(ps)-0MeA-(ps)-FU 3'
GaINAc-ALDH2-hcm9-L6
First strand (SEQ ID NO: 94, based on SEQ ID NO: 17)
5'0MeA-(ps)-FU-(ps)-0MeG-FU-OMeA-FG-0MeC-FC-OMeG-FA-OMeG-FG-OMeA-FU-
OMeC-FU-OMeU-(ps)-FC-(ps)-0MeU 3'
Second strand (SEQ ID NO: 95, SEQ ID NO:18)
5'[5-123 (ps)]3 5T43 (ps) FA-0MeG-FA-0MeA-FG-OMeA-FU-OMeC-FC-OMeU-FC-
OMeG-FG-0MeC-FU-OMeA-FC-(ps)-0MeA-(ps)-FU 3'
GaINAc-ALDH2-hcm9-L4
First strand (SEQ ID NO: 96, based on SEQ ID NO: 17)
5'0MeA-(ps)-FU-(ps)-0MeG-FU-OMeA-FG-0MeC-FC-OMeG-FA-OMeG-FG-OMeA-FU-
OMeC-FU-OMeU-(ps)-FC-(ps)-0MeU 3'
Second strand (SEQ ID NO: 97, based on SEQ ID NO:18)
5'[5-123 (ps)]3 5T41 (ps) FA-0MeG-FA-0MeA-FG-OMeA-FU-OMeC-FC-OMeU-FC-
OMeG-FG-OMeC-FU-OMeA-FC-(ps)-0MeA-(ps)-FU 3'
FN (N=A, C, G, U) denotes 2'Fluoro, 2' DeoxyNucleosides
OMeN (N=A, C, G, U) denotes 2'0 Methyl Nucleosides
(ps) indicates a phosphorothioate linkage
5T23, ST41and 5T43 are as below.
5T23-phos is a GalNac C4 phosphoramidite (structure components as below)
OAc
N
Ac0
0 I
Ac0 (:)/\
NHAc -
5T23-phos
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
ODMT
r/DOCOODMT
NCC'
1
NOP02 \ __ \
\ _________________________________________________________ ODMT
long trebler (STKS)
ST41-phos is as follows:
DMTr
0 0 )LN'L
DMIr
0 0 Oc:115Ø,CN
DMTr.
0 0
ST41
5 ST43-phos is as follows and as described in W02017/174657:
DMTr.
DMTr,
0 0 0015,10,,CN
DMTr,
0 0
ST43-phos
10 All oligonucleotides were either obtained from a commercial
oligonucleotide manufacturer
(Biospring, Frankfurt, Germany) or synthesized on an AKTA oligopilot
synthesizer (in
house) using standard phosphoramidite chemistry. Commercially available solid
support
and 2"0-Methyl RNA phosphoramidites, 2"Fluoro DNA phosphoramidites (all
standard
protection) and commercially available long trebler phosphoramidite (Glen
research) were
15 used. Synthesis was performed using 0.1 M solutions of the
phosphoramidite in dry
acetonitrile and benzylthiotetrazole (BTT) was used as activator (0.3M in
acetonitrile). All
other reagents were commercially available standard reagents.
Conjugation of the respective GalNac synthon (e.g., 5T23, 5T41 or 5T43) was
achieved
20 by coupling of the respective phosphoramidite to the 5"end of the
oligochain under
standard phosphoramidite coupling conditions. Phosphorothioates were
introduced using
standard commercially available thiolation reagents (EDITH, Link
technologies).
The single strands were cleaved off the CPG by using methylamine (40% aqueous)
and
25 the resulting crude oligonucleotide was purified by Ion exchange
chromatography
(Resource Q, 6mL, GE Healthcare) on a AKTA Pure HPLC System using a Sodium
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
96
chloride gradient. Product containing fractions were pooled, desalted on a
size exclusion
column (Zetadex, EMP Biotech) and lyophilised.
For annealing, equimolar amounts of the respective single strands were
dissolved in water
and heated to 80 C for 5min. After gradual cooling to RT the resulting duplex
was
lyophilised.
The sequences of the resulting nucleic acids (siRNAs) are set out in Table 1
below.
Table 1: nucleic acid sequences tested for inhibition of ALDH2 expression.
Nucleic acids
were synthesized by Biospring, Frankfurt, Germany.
Sequence
siRNA ID sequence modifications
ID No
1 5'-
UUGUUUAUGAAAAUCUGGU-3' 5181516182626171845
ALDH2-h-1
2 5'-
ACCAGAUUUUCAUAAACAA-3' 2736461515361626362
3 5'-
AAUGUUUUCCUGCUGACGG-3' 6254515173547182748
ALDH2-hcm-2
4 5'-
CCGUCAGCAGGAAAACAUU-3' 3745364728462627251
5 5'-
UUGAACUUCAGGAUCUGCA-3' 5182635172846171836
ALDH2-hc-3
6 5'-
UGCAGAUCCUGAAGUUCAA-3' 1836461735462815362
7 5'-
UUUCACUUCAGUGUAUGCC-3' 5153635172818161837
ALDH2-hc-4
8 5'-
GGCAUACACUGAAGUGAAA-3' 4836163635462818262
9 5'-
UUCUUAUGAGUUCUUCUGA-3' 5171525464517153546
ALDH2-hc-5
10 5'-
UCAGAAGAACUCAUAAGAA-3' 1728264627172526462
11 5'-
UCUUCUUAAACUGAGUUUC-3' 5351715262718281517
ALDH2-hcm-6
12 5'-
GAAACUCAGUUUAAGAAGA-3' 4626353645152646282
13 5'-
UCCUUGAUCAGGUUGGCCA-3' 5371546172845184736
ALDH2-hc-7
14 5'-
UGGCCAACCUGAUCAAGGA-3' 1847362735461726482
5'-UUGAAGAACAGGGCGAAGU-3' 5182646272848382645
ALDH2-hc-8
16 5'-
ACUUCGCCCUGUUCUUCAA-3' 2715383735451715362
17 5'-
AUGUAGCCGAGGAUCUUCU-3' 6181647382846171535
ALDH2-hcm-9
18 5'-
AGAAGAUCCUCGGCUACAU-3' 2826461735384716361
19 5'-
AUCUGGUUGCAGAAGACCU-3' 6171845183646282735
ALDH2-hmr-10
5'-AGGUCUUCUGCAACCAGAU-3' 2845351718362736461
21 5'-
AAAUCCACCAGGUAGGAGA-3' 6261736372845284646
ALDH2-hr-11
22 5'-
UCUCCUACCUGGUGGAUUU-3' 1717352735481846151
23 5'-
ACUCUCUUGAGGUUGCUGC-3' 6353535182845183547
ALDH2-hm-12
24 5'-
GCAGCAACCUCAAGAGAGU-3' 4728362735362828281
5'-AUGUCCAAAUCCACCAGGU-3' 6181736261736372845
ALDH2-hcmr-13
26 5'-
ACCUGGUGGAUUUGGACAU-3' 2735481846151846361
27
ALDH2-hm-14 5'-CCAUGCUUGCAUCAGGAGC-3' 7361835183617284647
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
97
1 28 15'-GCUCCUGAUGCAAGCAUGG-3' 14717354618362836184 I
Table 1
Nucleotides modifications are depicted by the following numbers (column 4),
1=2"F-dU,
2=2'F-dA, 3=2"F-dC, 4=2"F-dG, 5=2'-0Me-rU; 6=2'-0Me-rA; 7=2'-0Me-rC; 8=2'-0Me-
rG.
Example 2
Screening for inhibition ALDH2 mRNA expression in human Hep3B cells was
carried out
as follows.
Cells were plated at a density of 150,000 cells per well in 6-well format. 24
hours post
seeding, cells were transfected with 20 nM of indicated non-conjugated siRNAs
using 1
pg/ml of AtuFECT as a liposomal transfection reagent. 48 hours after
transfection, RNA
was isolated using the Spin Cell Mini Kit 250 (Stratec). ALDH2 mRNA levels
were
determined by gRT-PCR relative to PTEN mRNA expression as housekeeper. Values
were normalized to the amount of ALDH2 mRNA in untreated cells (UT). Means and
SD
of normalized triplicate measurements are shown. Results are shown in Figure
1.
Table 2: Sequences of ALDH2, ApoB, beta-Actin and PTEN qPCR amplicon sets that
were used to measure mRNA levels are shown below.
SEQ ID
NO:: Gene Species Sequences
29 ALDH2 (upper) 5' GGCAAGCCCTATGTCATCTCCT 3'
human,
ALDH2 (lower) cynomolgus
30 5' GGATGGTTTTCCCGTGGTACTT 3'
ALDH2 (probe)
31 5' TGGTCCTCAAATGTCTCCGGTATTATGCC 3'
ALDH2 (upper)
32 5' GGCAAGCCTTATGTCATCTCGT 3'
ALDH2 (lower)
33 mouse 5' GGAATGGTTTTCCCATGGTACTT 3'
ALDH2 (probe)
34 5' TGAAATGTCTCCGCTATTACGCTGGCTG 3'
35 ApoB (upper) 5' TCATTCCTTCCCCAAAGAGACC 3'
36 ApoB (lower) human 5' CACCTCCGTTTTGGTGGTAGAG 3'
37 ApoB (probe) 5' CAAGCTGCTCAGTGGAGGCAACACATTA 3'
38 ApoB (upper) 5' TCATTCCTTCCCCAAAGAAACC 3'
39 ApoB (lower) cynomolgus 5' CACCTCCGTTTTGGTGGTAGAG 3'
40 ApoB (probe) 5' TCAAGCTGTTAAGTGGCAGCAACACGTT 3'
41 beta-Actin (upper) 5' GCATGGGTCAGAAGGATTCCTAT 3'
human
42 beta-Actin (lower) 5' TGTAGAAGGTGTGGTGCCAGATT 3'
CA 03118327 2021-04-30
WO 2019/092282
PCT/EP2018/081105
98
43 beta-Actin (probe) 5' TCGAGCACGGCATCGTCACCAA 3'
44 beta-Actin (upper) 5' AAGGCCAACCGCGAGAAG 3'
45 beta-Actin (lower) cynomolgus 5' AGAGGCGTACAGGGACAGCA 3'
46 beta-Actin (probe) 5' TGAGACCTTCAACACCCCAGCCATGTAC 3'
- _______________________________________
47 beta-Actin (upper) 5' CCTAAGGCCAACCGTGAAAAG 3'
48 beta-Actin (lower) mouse 5' AGGCATACAGGGACAGCACAG 3'
49 beta-Actin (probe) 5' TGAGACCTTCAACACCCCAGCCATGTAC 3'
50 PTEN (upper) 5' CACCGCCAAATTTAACTGCAGA 3'
51 PTEN (lower) human 5' AAGGGTTTGATAAGTTCTAGCTGT 3'
52 PTEN (probe) 5' TGCACAGTATCCTTTTGAAGACCATAACCCA
3'
Table 2
Table 3: Non-conjugated derivatives of ALDH2-hcm2 with the indicated
modification
pattern are shown:
Duplex sequence and chemistry SEQ ID
ID top: first strand, bottom: second strand, both 5'-3 NO:
ALD01 mA(ps)fA(ps)mUfGmUfUmUfUmCfCmUfGmCfUmGfAmC(ps)fG(ps)mG 98
fC(ps)mC(ps)fGmUfCmAfGmCfAmGfGmAfAmAfAmCfA(ps)mU(ps)fU 99
ALD02 mA(ps)fA(ps)mUfGmUfUmUfUmCfCmUfGmCfUmGfAmCfGmG ivA 100
fC(ps)mC(ps)fGmUfCmAfGmCfAmGfGmAfAmAfAmCfAmUfU ivA 101
ALD03 mA(ps)fA(ps)mUfGmUfUmUfUmCfCmUfGmCfUmGfAmCfGmG ivU 102
fC(ps)mC(ps)fGmUfCmAfGmCfAmGfGmAfAmAfAmCfAmUfU ivU 103
ALD04 mA(ps)fA(ps)mUfGmUfUmUfUmCfCmUfGmCfUmGfAmCfGmG ivC 104
fC(ps)mC(ps)fGmUfCmAfGmCfAmGfGmAfAmAfAmCfAmUfU ivC 105
ALD05 mA(ps)fA(ps)mUfGmUfUmUfUmCfCmUfGmCfUmGfAmCfGmG ivG 106
fC(ps)mC(ps)fGmUfCmAfGmCfAmGfGmAfAmAfAmCfAmUfU ivG 107
ALD06 mA(ps)fA(ps)mUfGmUfUmUfUmCfCmUfGmCfUmGfAmCfG ivA 108
fC(ps)mC(ps)fGmUfCmAfGmCfAmGfGmAfAmAfAmCfAmU ivA 109
ALD07 mA(ps)fA(ps)mUfGmUfUmUfUmCfCmUfGmCfUmGfAmCfG ivU 110
fC(ps)mC(ps)fGmUfCmAfGmCfAmGfGmAfAmAfAmCfAmU ivU 111
ALD08 mA(ps)fA(ps)mUfGmUfUmUfUmCfCmUfGmCfUmGfAmCfG ivC 112
fC(ps)mC(ps)fGmUfCmAfGmCfAmGfGmAfAmAfAmCfAmU ivC 113
ALD09 mA(ps)fA(ps)mUfGmUfUmUfUmCfCmUfGmCfUmGfAmCfG ivG 114
fC(ps)mC(ps)fGmUfCmAfGmCfAmGfGmAfAmAfAmCfAmU ivG 115
mA, mU, mC, mG ¨ 2`-0Me RNA
fA, fU, fC, fG ¨ 2`-F RNA
ivA, ivU, ivC, ivG - inverted RNA (3'-3')
(ps) ¨ phosphorothioate
Table 3
CA 03118327 2021-04-30
WO 2019/092282
PCT/EP2018/081105
99
Table 4: L6-GaINAc linker conjugated derivatives of ALDH2-hcm2 with the
indicated
modification pattern are shown
SEQ
Duplex sequence and chemistry ID
ID top: first strand, bottom: second strand, both 5'-3
NO:
STS22002L6
mA(ps)fA(ps)mUfGmUfUmUfUmCfCmUfGmCfUmGfAmC(ps)fG(ps)mG 116
ST23(ps)3 ST
117
43(ps)fCmCfGmUfCmAfGmCfAmGfGmAfAmAfAmCfA(ps)mU(ps)fU
STS22002V1L6 mA(ps)fA(ps)mUfGmUfUmUfUmCfCmUfGmCfUmGfAmC(ps)fG(ps)mG 118
5T23(ps)3 ST
119
43(ps)fCmCfGmUfCmAfGmCfAmGfGmAfAmAfAmCfAmUivA
5T522002V2L6 mA(ps)fA(ps)mUfGmUfUmUfUmCfCmUfGmCfUmGfAmC(ps)fG(ps)mG 120
5T23(ps)3 ST 121
43(ps)fCmCfGmUfCmAfGmCfAmGfGmAfAmAfAmCfAmUivG
5T522002V3L6 mA(ps)fA(ps)mUfGmUfUmUfUmCfCmUfGmCfUmGfAmCfGmGivA
122
5T23(ps)3 ST
123
43(ps)fCmCfGmUfCmAfGmCfAmGfGmAfAmAfAmCfA(ps)mU(ps)fU
5T522002V4L6 mA(ps)fA(ps)mUfGmUfUmUfUmCfCmUfGmCfUmGfAmCfGmGivG
124
5T23(ps)3 ST
125
43(ps)fCmCfGmUfCmAfGmCfAmGfGmAfAmAfAmCfA(ps)mU(ps)fU
mA, mU, mC, mG ¨ 2`-0Me RNA
fA, fU, fC, fG ¨ 2`-F RNA
ivA - inverted RNA (3'-3')
(ps) ¨ phosphorothioate
Table 4
Table 5: L6-GalNac linker conjugated derivatives of ALDH2-hcm6 and ALDH2-hcm9
with
the indicated modification pattern tested in vivo in mice are shown
SEQ
Duplex sequence and chemistry ID
ID top: first strand, bottom: second strand, both 5%3'
NO:
S1S22006L6 mU(ps)fC(ps)mUfUmCfUmUfAmAfAmCfUmGfAmGfUmU(ps)fU(ps)mC
126
S123(ps)3 ST 43(ps)fGmAfAmAfCmUfCmAfGmUfUmUfAmAfGmAfAmGfA
127
STS22006V1L6 mU(ps)fC(ps)mUfUmCfUmUfAmAfAmCfUmGfAmGfUmU(ps)fU(ps)mC
128
S123(ps)3 ST 43(ps)mGmAmAmAmCmUfCfAfGmUmUmUmAmAmGmAmAmGivA 129
S1S22009L6 mA(ps)fU(ps)mGfUmAfGmCfCmGfAmGfGmAfUmCfUmU(ps)fC(ps)mU
130
S123(ps)3 ST 43(ps)fAmGfAmAfGmAfUmCfCmUfCmGfGmCfUmAfCmAfU
131
STS22009V1L6 mA(ps)fU(ps)mGfUmAfGmCfCmGfAmGfGmAfUmCfUmU(ps)fC(ps)mU
132
S123(ps)3 ST 43(ps)mAmGmAmAmGmAfUfCfCmUmCmGmGmCmUmAmCmAivA 133
S1S22009V2L6 mA(ps)fU(ps)mGmUmAmGmCmCmGmAmGmGmAfUmCmUmU(ps)mC(ps)mU
134
S123(ps)3 ST 43(ps)mAmGmAmAmGmAfUfCfCmUmCmGmGmCmUmAmCmAivA 135
mA, mU, mC, mG ¨ 2'-0Me RNA
fA, fU, fC, fG ¨ 2`-F RNA
ivA - inverted RNA (3'-3')
(ps) ¨ phosphorothioate
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
100
Table 5
Example 3
Dose responses of ALDH2-siRNA molecules in human hepatocarinoma HepG2 cells
were
measured as follows.
Cells were plated at a density of 150,000 cells per well in 6-well format. 24
hours post
seeding, cells were transfected with indicated amounts of non-conjugated
siRNAs using 1
pg/ml of AtuFECT as liposomal transfection reagent. 48 hours after
transfection, RNA was
isolated using the Spin Cell Mini Kit 250 (Stratec). ALDH2 mRNA levels were
determined
by qRT-PCR relative to the geometric mean of PTEN and ACTIN mRNA expression as
housekeepers. Values were normalized to the amount of ALDH2 mRNA in untreated
cells.
Means and SD of normalized triplicate measurements are shown. IC50 values, 95%
confidence intervals and coefficients of determination are given. Results are
shown in
Figure 2.
Example 4
ALDH2 mRNA and protein levels and reduction of ALDH enzyme activity by ALDH2-
hcm-
2 siRNA in human HepG2-cells were investigated as follows.
Cells were seeded at 500,000 cells per well in 6-cm dishes. 24 hours post
seeding, cells
were transfected in triplicate with 1nM of ALDH2-hcm-2 siRNA or an siRNA
against firefly
luciferase as a non-targeting control (Fluc) using 1pg/m1 of Atufect as
liposomal
transfection reagent. Three days post transfection, cells were lysed using
InviTrap Spin
Cell RNA Mini Kit (Stratec). ALDH2 mRNA levels were determined by qRT-PCR
relative to
actin expression as housekeeper. Mean and SD of normalized values are shown.
For Western Blotting, cells were lysed in 500 pl lysis buffer (Tris/HCI, 15%
Glycerin, 1%
Triton). Protein amount was determined using the Pierce BCA Protein Assay Kit
(Thermo
Scientific), and 30 pg per sample were used for electrophoresis in Laemmli
buffer.
Membranes were probed with antibodies against ALDH2 (Abcam #194587, 1:1200)
and
alpha-ACTININ (NEB #3134, 1:1000). Quantification was performed Using a Stella
Imaging System (Raytest). Aldehyde-Dehydrogenase activity assay was performed
with
total protein (12.2 micro g) extract using the Mitochondria! Aldehyde
Dehydrogenase
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
101
Activity Assay Kit (Abcam) according to manufacturer's protocol. Results are
shown in
Figure 3.
Example 5
Dose-response for GaINAc- L1-linker-coupled siRNAs against Aldh2 mRNA in
primary
mouse hepatocytes was measured as follows.
Mouse hepatocytes were plated at a density of 20.000 cells per well on
collagen-coated
96-well plates (Life Technologies). 3 hours after plating, cells were rinsed
once with PBS
and L1-GaINAc-conjugated siRNAs were added at the indicated concentrations in
triplicates. 24 hours post treatment, cells were lysed with InviTrap Spin Cell
RNA Mini Kit
(Stratec). Aldh2 mRNA levels were determined by gRT-PCR relative to Actin mRNA
expression as housekeeper. Values were normalized to the amount of Aldh2 in
untreated
cells. Means and SD of normalized triplicate values are shown. Results are
shown in
Figure 4.
Example 6
Dose-response for GaINAc- L1-linker-coupled siRNAs against ALDH2 in primary
human
hepatocytes was measured as follows.
Human hepatocytes (Thermo Fisher) were plated at a density of 30.000 cells per
well on
collagen-coated 96-well plates (Life Technologies). GaINAc-L1 linker
conjugated siRNAs
were added at the indicated concentrations in triplicates immediately after
plating. 24
hours post treatment, cells were lysed with InviTrap Spin Cell RNA Mini Kit
(Stratec).
ALDH2 mRNA levels were determined by gRT-PCR relative to ACTIN mRNA expression
as housekeeper. Values were normalized to the amount of ALDH2 mRNA in
untreated
cells. Means and SD of normalized triplicate values are shown. Results are
shown in
Figure 5.
Example 7
Reduction of ALDH2-mRNA by 15 nM of GaINAc-L1 linker-coupled ALDH2-hcm-2 siRNA
in primary hepatocytes of different species indicated 6 days post treatment
was measured
as follows.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
102
Hepatocytes (Thermo Fisher) were plated at a density of 250.000 (human and
cynomolgus) or 150.000 (mouse) cells per well on collagen-coated 24-well
plates (Life
Technologies). L1-GaINAc conjugated ALDH2-hcm-2 or non-silencing control siRNA
were
added at 15 nM concentration in triplicates immediately after plating. 6 days
post
treatment, cells were lysed with InviTrap Spin Cell RNA Mini Kit (Stratec).
ALDH2 mRNA
levels of the different species were determined by qRT-PCR relative to
geometric means
of ACTIN and APOB expression (human and cynomolgus) or to Actin alone (mouse)
as
housekeeper. Means and SD of normalized triplicate values are shown. Results
are
shown in Figure 6.
Example 8
Comparison of ALDH2 mRNA knockdown by GaINAc-conjugated ALDH2 siRNAs with
linkers L1 and L6 in human primary hepatocytes was measured as follows.
Human hepatocytes (Thermo Fisher) were seeded at 30.000 cells per well in
collagen-
coated 96-well plates (Life Technologies). Immediately after seeding, GaINAc-
conjugated
siRNAs were added as indicated in graphs. 24 hours post treatment, cells were
lysed with
with InviTrap Spin Cell RNA Mini Kit (Stratec). ALDH2 mRNA levels were
determined by
qRT-PCR relative to geometric means of ACTIN and APOB expression as
housekeepers.
Values were normalized to the amount of ALDH2 mRNA in untreated cells. Means
and SD
of normalized triplicate values are shown. IC50 values and 90% confidence
intervals were
calculated. Results are shown in Figure 7.
Example 9
Activity of GaINAc-siRNAs against ALDH2 in vivo was measured as follows.
Male mice (C5761/6J) were treated subcutaneously with 3 or 10 mg/kg of the
indicated
compounds (6 animals per group). At necropsy 10 days post treatment, liver was
harvested. Tissue was homogenized in a Tissue Lyser II machine (Qiagen).
A -RNA was extracted from 10 mg of liver tissue using an InviTrap Spin Tissue
RNA Mini
Kit (Stratec). Aldh2 mRNA levels were determined by qRT-PCR relative to the
geometric
means of Actin and ApoB (mouse) as housekeepers. Values were normalized to the
amount of Aldh2 mRNA in mice treated with the negative control siRNA (GN-
Dmcn).
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
103
Means and SD of normalized values are shown as well as values for individual
animals.
Measurements of untreated control mice (PBS) is shown. Results are shown in
Figure 8A.
B - Protein was extracted from 10 mg of liver tissue in RIPA buffer (Cell
Signaling) from
three animals treated with 10 mg/kg of the indicated compounds per group.
Protein
amount was determined using the Pierce BCA Protein Assay Kit (Thermo
Scientific), and
30 pg per sample were used for electrophoresis in Laemmli buffer. Membranes
were
probed with antibodies against ALDH2 (Abcam #194587, 1:1200) and alpha-ACTININ
(NEB #3134, 1:1000) and developed with HRP secondary antbody. Visualizations
of the
membranes are shown in the lower panel. Quantification (upper left panel) was
performed
Using a Stella Imaging System (Raytest). Aldehyde-Dehydrogenase activity assay
(upper
right panel) was performed using liver protein extracts of three mice treated
with the
indicated siRNAs or PBS using the Mitochondria! Aldehyde Dehydrogenase
Activity Assay
Kit (Abcam) according to manufacturer's protocol. Results are shown in Figure
8B.
C - Livers from mice untreated (PBS) or treated with the indicated GaINAc-
siRNAs (n=6)
at the indicated doses for 10 days were analysed for its amount of hepatic
acetaldehyde.
Acetaldehyde content was measured in whole liver tissue homogenates (100 mg)
in
aqueous solution using an Agilent gas chromatography (GC) 6890 and 7890 series
system coupled with a 5975 C MS detector (Agilent), MSD Chemstation (Agilent)
and
Maestro software for data analyses. Internal standard was 2-propanol and the
following
settings were applied: Incubation: 70 C for 30 min; Syringe temp.: 80 C;
Injector temp.:
200 C; Injection mode: SHS, Split 1/5; Injection volume: 1 ml (gas syringe);
Column: DB-
WAX MS, 30m x 0.25 mm; 1pm; Helium Flow: 1 ml/min; Oven : 37 C for 5 min then
30 C/min to 120 C for 1 min; MS Transfer Line: 240 C; Incubation solvent:
Purified Water;
MS detection: El - Scan from 10 to 300 uma. Individual measurements as well as
mean
+/-SD are depicted. Results are shown in Figure 8C.
Example 10
Activity of GaINAc-siRNAs against Aldh2 in vivo ¨ duration and dose-response.
Male mice (C5761/6J) were treated once subcutaneously with 1, 3 or 10 mg/kg of
the
indicated compounds (5 animals per group). At necropsy at 10, 20, 30, and 41
days post
treatment, liver was harvested. Tissue was homogenized in a Tissue Lyser II
machine
(Qiagen). Total RNA was extracted from 10 mg of liver tissue using an InviTrap
Spin
Tissue RNA Mini Kit (Stratec). Aldh2 mRNA levels were determined by gRT-PCR
relative
to the geometric means of Actin and ApoB expression (mouse) as housekeepers.
Values
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
104
were normalized to the amount of Aldh2 mRNA in mice treated with the non-
silencing
control siRNA. Means and SD of normalized values are shown as well as values
for
individual animals. Results are shown in Figure 9.
Example 11
Different siRNA duplexes containing inverted RNA nucleotides at both 3`-ends
were tested
for serum stability. ALD02-ALD05 (Table 3) contain inverted RNA in addition to
the last
nucleotide in the first strand and second strand. ALD06-ALD09 contain inverted
RNA
instead of the last nucleotide in first strand and second strand. All inverted
RNA
nucleotides substitute for terminally used phosphorothioates. In both designs,
ivA and ivG
confer higher stability to the tested sequence than ivU and ivC. "UT"
indicates untreated
samples. "FBS" indicates siRNA duplexes which were incubated at 37 C at 5 pM
concentration with 50 % FBS for 3 days, before being phenol/chloroform-
extracted and
precipitated with Ethanol. Samples were analyzed on 20% TBE polyacrylamide
gels in
native gel electrophoresis. UV pictures of the ethidium bromide-stained gels
are shown in
Figure 11.
Example 12:
The influence of inverted A, U, C and G RNA nucleotides at 3'-overhang
positions on the
efficacy of ALDH2 mRNA knockdown was analyzed using derivatives of the siRNA
ALDH2-hcm2 (see Table 3). ALD01 contains phosphorothioates at all termini,
whereas
ALD02-ALD05 contain ivA (ALD01), ivU (ALD03), ivC (ALD04) and ivG (ALD05) at
the 3'-
end of the first strand and at the 3`-end of the second strand. Both inverted
nucleotides
are present in addition to the terminal nucleotide of the respective strands
and substitute
for terminal phosphorothioates. A non-related siRNA (PTEN) and a control siRNA
targeting firefly luciferase (Luci) were included. All tested variants show
comparable
activity under the tested conditions. The experiment was conducted in the
human
hepatocarcinoma cell line Hep3B. Cells were seeded at a density of 150,000
cells per 6-
well, transfected with 0.1 nM and 1 nM siRNA, respectively, using 1 pg/ml
Atufect as a
liposomal transfection reagent after 24 h. Cells were lysed after 48 h. Total
RNA was
extracted and ALDH2 and ACTIN mRNA levels were determined by Taqman gRT-PCR.
Each bar represents mean SD of three technical replicates normalized to the
house
keeping gene expression (ACTIN). Results are shown in Figure 12.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
105
Example 13
The influence of inverted A, U, C and G RNA nucleotides at terminal 3'
positions on the
efficacy of ALDH2 mRNA knockdown was analyzed using derivatives of the siRNA
ALDH2-hcm2 (see Table 3). ALD01 contains phosphorothioates at all termini,
whereas
ALD06-ALD09 contain ivA (ALD06), ivU (ALD07), ivC (ALD08) and ivG (ALD09) at
the 3`-
end of the first strand and at the 3`-end of the second strand. Both inverted
nucleotides
replace the terminal nucleotide of the respective strands and substitute for
terminal
phosphorothioates used in other derivatives for stabilization. A non-related
siRNA (PTEN)
and a control siRNA targeting firefly luciferase (Luci) were included. All
tested variants
show comparable activity under the tested conditions. The experiment was
conducted in
the human hepatocarcinoma cell line Hep3B. Cells were seeded at a density of
150,000
cells per 6-well, transfected with 0.1 nM and 1 nM siRNA, respectively,using
and 1 pg/ml
Atufect as a liposomal transfection reagent after 24 h. Cells were lysed after
48 h. Total
RNA was extracted and ALDH2 and ACTIN mRNA levels were determined by Taqman
gRT-PCR. Each bar represents mean SD of three technical replicates
normalized to the
house keeping gene expression (ACTIN). Results are shown in Figure 13.
Example 14
Different GaINAc-L6-linker conjugates of ALDH2-hcm2 containing inverted RNA
nucleotides (see Table 4) were tested for serum stability. 5T522002L6 contains
phosphorothioates at all non-conjugated ends, whereas 5T522002V1-L6 and
5T522002V2-L6 contain inverted RNA nucleotides at the second strand's 3`-end,
where
the respective inverted nucleotide is present instead of the last nucleotide.
5T522002V3-
L6 and 5T522002V4-L6 contain inverted RNA nucleotides at the first strand's 3`-
end,
whereby the respective inverted nucleotide is present in addition to the last
nucleotide. ivA
was used in 5T522002V1-L6 and 5T522002V3-L6, whereas ivG was used in
5T522002V2-L6 and 5T522002V4-L6. All inverted RNA nucleotides substitute for
terminal phosphorothioates used in other derivatives for stabilization.
5T522002V1-L6
and 5T522002V2-L6 are slightly more stable than the other variants tested
here. "UT"
indicates untreated samples. JUK04 represents a control compound not serum
stabilized
as a positive control. "FBS" indicates GaINAc-siRNA conjugates which were
incubated at
37 C at 5 pM concentration with 50% FBS for 3 days before being
phenol/chloroform-
extracted and precipitated with Ethanol. Samples were analyzed on 20% TBE
polyacrylamide gels in native gel electrophoresis. UV pictures of the ethidium
bromide-
stained gels are shown in Figure 14.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
106
Example 15
The dose response of inverted RNA nucleotides at terminal 3' positions on the
efficacy of
ALDH2 mRNA knockdown was analyzed using GalNac-L6-conjugated derivatives of
the
siRNA ALDH2-hcm2 (see Table 4) by ASGPR receptor complex-mediated uptake in
mouse primary hepatocytes. STS22002L6 contains phosphorothioates at all non-
conjugated ends, whereas STS22002V1-L6 and STS22002V2-L6 contain inverted RNA
nucleotides at the second strand's 3`-end, where the respective inverted
nucleotide is
present instead of the last nucleotide. STS22002V3-L6 and STS22002V4-L6
contain
inverted RNA nucleotides at the first strand's 3`-end, whereby the nucleotide
is present in
addition to the last nucleotide. ivA was used in STS22002V1-L6 and STS22002V3-
L6,
whereas ivG was used in STS22002V2-L6 and STS22002V4-L6. All inverted RNA
nucleotides substitute for terminal phosphorothioates used in other
derivatives for
stabilization. All tested variants show comparable activity. The experiment
was conducted
in mouse primary hepatocytes. Cells were seeded at a density of 20,000 cells
per 96-well,
treated with the indicated doses from 125 nM to 0.04 nM of the respective
indicated
siRNA conjugates directly after plating. Non-treated cells and a control siRNA
targeting
firefly luciferase at 10 nM (Luc) were included. Cells were lysed after 24 h.
Total RNA was
extracted and Aldh2 and Actin mRNA levels were determined by Taqman gRT-PCR.
Each
bar represents mean SD of three biological replicates normalized to the
house keeping
gene expression (Actin). Results are shown in Figure 15.
Example 16
The influence of ivA at the first strand's 3`-end on the efficacy of ALDH2
mRNA
knockdown was analyzed in vivo. Therefore, GaINAc-L6 derivatives of ALDH2-hcm6
and
ALDH2-hcm9 siRNA conjugates targeting Aldh2 were used in an in vivo efficacy
study.
5T522002L6 contains phosphorothioates at all non-conjugated termini, whereas
5T522002V3-L6 contains an ivA at the first strand's 3`-end in addition to the
last
nucleotide. C57BL/6 male mice (n=6) were treated subcutaneously with vehicle
(PBS), 10
mg/kg and 3 mg/kg of the indicated GaINAc-L6 conjugate, respectively. Seven
days after
treatment, at necropsy liver tissue was harvested and total RNA was extracted.
Aldh2 and
ApoB mRNA levels were analyzed by Taqman gRT-PCR. Each bar represents mean
normalized (ApoB) expression SD of six animals. Individual values normalized
to the
house keeping gene expression (ApoB) are plotted, additionally. Results are
shown in
Figure 16.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
107
Example 17
Different modified derivatives of the GaINAc-siRNA conjugates of ALDH2-hcm6
and
ALDH2-hcm9 (Table 5) were analyzed for knockdown efficacy on ALDH2 mRNA
expression levels in vivo in mice. "V1" variants contain a different 2`-0Me/2`-
F modification
pattern in the second strand and an ivA nucleotide at the 3`-end of the second
strand,
substituting for the last nucleotide and for terminal phosphorothioates at
this end. "V2"
variants additionally contain a different 2`-0Me/2`-F modification pattern in
the first strand
as specified in Table 5. C57BL/6 male mice (n=6) were subcutaneously treated
with
vehicle (PBS) or 3 mg/kg and 1 mg/kg GaINAc-L6 conjugate derivatives,
respectively.
Nine days after treatment, at necropsy liver tissue was harvested and total
RNA was
extracted. Aldh2 and ApoB mRNA levels were analyzed by Taqman gRT-PCR. Each
bar
represents mean normalized (ApoB) expression SD of six animals. Individual
values
normalized to the house keeping gene expression (ApoB) are plotted,
additionally.
Statistical analysis is based on Kruskal-Wallis test with Dunn's multiple
comparisons test
respective to the control group (PBS). Results are shown in Figure 17.
Example 18 - Synthesis of conjugates 1
Example compounds were synthesised according to methods described below and
methods known to the person skilled in the art. Assembly of the
oligonucleotide chain and
linker building blocks was performed by solid phase synthesis applying
phosphoramidite
methodology. GaINAc conjugation was achieved by peptide bond formation of a
GaINAc-
carboxylic acid building block to the prior assembled and purified
oligonucleotide having the
necessary number of amino modified linker building blocks attached.
Oligonucleotide synthesis, deprotection and purification followed standard
procedures that
are known in the art.
All Oligonucleotides were synthesized on an AKTA oligopilot synthesizer using
standard
phosphoramidite chemistry. Commercially available solid support and 2"0-Methyl
RNA
phosphoramidites, 2"Fluoro, 2"Deoxy RNA phosphoramidites (all standard
protection,
ChemGenes, LinkTech) and commercially available 3'-Amino Modifier TFA Amino C-
6 lcaa
CPG 500A (Chemgenes) were used. Per-acetylated galactose amine 8 is
commercially
available.
Ancillary reagents were purchased from EMP Biotech. Synthesis was performed
using a
0.1 M solution of the phosphoramidite in dry acetonitrile and
benzylthiotetrazole (BTT) was
used as activator (0.3M in acetonitrile). Coupling time was 15 min. A
Cap/OX/Cap or
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
108
Cap/Thio/Cap cycle was applied (Cap: Ac20/NMI/Lutidine/Acetonitrile, Oxidizer:
0.1M 12 in
pyridine/H20). Phosphorothioates were introduced using standard commercially
available
thiolation reagent (EDITH, Link technologies). DMT cleavage was achieved by
treatment
with 3% dichloroacetic acid in toluene. Upon completion of the programmed
synthesis
cycles a diethylamine (DEA) wash was performed. All oligonucleotides were
synthesized in
DMT-off mode.
Attachment of the serinol-derived linker moiety was achieved by use of either
base-loaded
(S)-DMT-Serinol(TFA)-succinate-lcaa-CPG 10 or a (S)-DMT-Serinol(TFA)
phosphoramidite
7 (synthesis was performed as described in Hoevelmann etal. (Chem. Sci., 2016,
7, 128-
135)). Tri-antennary GaINAc clusters (ST23/C4XLT) were introduced by
successive
coupling of the respective trebler amidite derivatives (C4XLT-phos) followed
by the GaINAc
amidite (ST23-phos).
The single strands were cleaved off the CPG by 40% aq. methylamine treatment.
The
resulting crude oligonucleotide was purified by ion exchange chromatography
(Resource Q,
6mL, GE Healthcare) on a AKTA Pure HPLC System using a sodium chloride
gradient.
Product containing fractions were pooled, desalted on a size exclusion column
(Zetadex,
EMP Biotech) and lyophilised.
Individual single strands were dissolved in a concentration of 60 OD/mL in
H20. Both
individual oligonucleotide solutions were added together in a reaction vessel.
For easier
reaction monitoring a titration was performed. The first strand was added in
25% excess
over the second strand as determined by UV-absorption at 260nm. The reaction
mixture
was heated to 80 C for 5min and then slowly cooled to RT. Double strand
formation was
monitored by ion pairing reverse phase HPLC. From the UV-area of the residual
single
strand the needed amount of the second strand was calculated and added to the
reaction
mixture. The reaction was heated to 80 C again and slowly cooled to RT. This
procedure
was repeated until less than 10% of residual single strand was detected.
Synthesis of compounds 2-10
Compounds 2 to 5 and (S)-DMT-Serinol(TFA)-phosphoramidite 7 were synthesised
according to literature published methods (Hoevelmann et al. Chem. Sci.,
2016,7, 128-135).
(S)-4-(3-(bis(4-methoxyphenyl)(phenyl)methoxy)-2-(2,2,2-
trifluoroacetamido)propoxy)-4-oxobutanoic acid (6).
To a solution of 5 in pyridine was added succinic anhydride, followed by DMAP.
The
resulting mixture was stirred at room temperature overnight. All starting
material was
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
109
consumed, as judged by TLC. The reaction was concentrated. The crude material
was
chromatographed in silica gel using a gradient 0% to 5% methanol in DCM (+ 1%
triethylamine) to afford 1.33 g of 6 (yield = 38%). m/z (ESI-): 588.2 (100%),
(calcd. for
C30H29F3N08- [M-H] 588.6). 1 H-NMR: (400 MHz, CDCI3) 6 [ppm] = 7.94 (d, 1H,
NH),
7.39 - 7.36 (m, 2H, CHary1), 7.29 - 7.25 (m, 7H, CHary1), 6.82-6.79 (m, 4H,
CHary1), 4.51 ¨
4.47 (m, 1H), 4.31 ¨4.24 (m, 2H), 3.77 (s, 6H, 2xDMTr-OMe), 3.66 - 3.60 (m,
16H, HNEt3+),
3.26 - 3.25 (m, 2H), 2.97 ¨ 2.81 (m, 20H, NEt3), 2.50-2.41 (4H, m), 1.48¨ 1.45
(m, 26H,
HNEt3+), 1.24 - 1.18 (m, 29H, NEt3).
(S)-DMT-Serinol(TFA)-succinate-lcaa-CPG (10)
The (S)-DMT-Serinol(TFA)-succinate (159 mg, 270 umol) and HBTU (113 mg, 299
umol)
were dissolved in CH3CN (10 mL). Diisopropylethylamine (DIPEA, 94 pL, 540
umol) was
added to the solution, and the mixture was swirled for 2 min followed by
addition native
amino-lcaa-CPG (500 A, 3 g, amine content: 136 umol/g). The suspension was
gently
shaken at room temperature on a wrist-action shaker for 16h then filtered, and
washed with
DCM and Et0H. The solid support was dried under vacuum for 2 h. The unreacted
amines
on the support were capped by stirring with acetic anhydride/lutidine/N-
methylimidazole at
room temperature. The washing of the support was repeated as above. The solid
was dried
under vacuum to yield solid support 10 (3 g, 26 umol/g loading).
GaINAc Synthon (9)
Synthesis of the GaINAc synthon 9 was performed as described in Nair et al. J.
Am. Chem.
Soc., 2014, 136 (49), pp 16958-16961, in 46% yield over two steps.
The characterising data matched the published data.
Synthesis of Oligonucleotides
All single stranded oligonucleotides were synthesised according to the
reaction conditions
described above and in Figure 25 and 26.
All final single stranded products were analysed by AEX-H PLC to prove their
purity. Purity
is given in %FLP (% full length product) which is the percentage of the UV-
area under the
assigned product signal in the UV-trace of the AEX-HPLC analysis of the final
product.
Identity of the respective single stranded products (non-modified, amino-
modified
precursors or GaINAc conjugated oligonucleotides) was proved by LC-MS
analysis.
Table 7: Single stranded un-conjugated oligonucleotides
Product Name MW MW (ESI-) YoFLP
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
110
(11) calc. found (AEX-
HPLC)
A0002 STS16001A 6943.3 Da 6943.0 Da 86.6%
A0006 STS16001BL4 8387.5 Da 8387.5 Da 94.1%
A0130 STS18001A 6259.9 Da 6259.8 Da 76.5%
A0131 STS18001BL4 7813.2 Da 7813.1 Da 74.3%
A0220 STS16001B-5'1xNH2 6982.2 Da 6982.1 Da 95.7%
A0237 STS16001A 6943.3 Da 6943.3 Da 95.6%
A0244 STS16001BV1 6845.2 Da 6844.9 Da 98.2%
A0264 STS16001AV4-3'1xN H2 7112.4 Da 7112.2 Da 95.4%
A0329 STS16001BV6-3'5'1xNH2 7183.3 Da 7183.2 Da 88.8%
51 x NH2 means refers to the position (5' end) and number (1 x NH2) of free
serinol derived
amino groups which are available for conjugation. For example, 1x3'NH2 on
A0264 means
there is free amino group which can be reacted with GaINAc synthon 9 at the 3'
end of the
strand A0264. 3'5'1xNH2 means there is one serinol-derived free amino group
which can
be reacted with GaINAc linker 9 at the 3' end and the 5' end of the strand.
Synthesis of conjugates 1-3 and reference conjugates 1-2
Conjugated singles strands for conjugates 1-2 and reference conjugates 1-2
Conjugation of the GalNac synthon (9) was achieved by coupling to the serinol-
amino
function of the respective oligonucleotide strand 11 using a peptide coupling
reagent.
Therefore, the respective amino-modified precursor molecule 11 was dissolved
in H20 (500
OD/mL) and DMSO (DMSO/H20, 2/1, v/v) was added, followed by DIPEA (2.5% of
total
volume). In a separate reaction vessel pre-activation of the GaIN(Ac4)-C4-acid
(9) was
performed by reacting 2 eq. (per amino function in the amino-modified
precursor
oligonucleotide 11) of the carboxylic acid component with 2 eq. of HBTU in
presence of 8
eq. DIPEA in DMSO. After 2 min the pre-activated compound 9 was added to the
solution
of the respective amino-modified precursor molecule. After 30 min the reaction
progress
was monitored by LCMS or AEX-HPLC. Upon completion of the conjugation reaction
the
crude product was precipitated by addition of 10x PrOH and 0.1x 2M NaCI and
harvested
by centrifugation and decantation. To set free the acetylated hydroxyl groups
in the GaINAc
moieties the resulting pellet was dissolved in 40% MeNH2 (1mL per 500 OD) and
after 15
min at RT diluted in H20 (1:10) and finally purified again by anion exchange
and size
exclusion chromatography and lyophilised to yield the final product 12.
Table 8: Single stranded GaINAc-conjuqated oliqonucleotides
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
111
Product Starting Name MW MW (ESI-) YoFLP
(12) Material calc. found (AEX-
HPLC)
A0241 A0220 STS16001BL20 7285.5 Da 7285.3
Da 91.8%
A0268 A0264 STS16001AV4L33 7415.7 Da 7415.4 Da 96.9%
A0330 A0329 STS16001BV6L42 7789.8 Da 7789.8 Da 95.5%
Double strand formation
Double strand formation was performed according to the methods described
above.
The double strand purity is given in % double strand which is the percentage
of the UV-area
under the assigned product signal in the UV-trace of the IP-RP-HPLC analysis.
Table 9: Nucleic acid conjugates
Product Starting Materials Name % double strand
First Strand Second Strand
Ref. Conj. 1 A0237 A0241 STS16001L20 97.7%
Ref. Conj. 2 A0268 A0244 STS16001L33 97.8%
Ref. Conj. 3 A0130 A0131 STS18001L4 96.8%
Ref. Conj. 4 A0002 A0006 STS16001L4 90.1%
Conjugate 1 A0268 A0241 STS16001L24 96.0%
Conjugate 2 A0237 A0330 STS16001V1L42 98.5%
Conjugate 3 A0268 A0330 STS16001V1L43 98.2%
Sequences
Modifications key for the following sequences:
f denotes 2"Fluoro 2"deoxyribonucleotide or 2'-fluoro ribonucleotide (the
terms are
interchangeable)
m denotes 2'0 Methyl ribonucleotide
(ps) denotes phosphorothioate linkage
Ser(GN) is a GaINAc-C4 building block attached to serinol derived linker
moiety:
OH
HO ONH
NHAc
wherein the 0--- is the linkage between the oxygen atom and e.g. H,
phosphordiester
linkage or phosphorothioate linkage.
C4XLT is:
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
112
s'soo
ST23 is:
OAc OAc
AcO
NHAc
Synthesis of the phosphoramidite derivatives of C4XLT (C4XLT-phos) as well as
5T23
(5T23-phos) can be performed as described in W02017/174657.
C4XLT-phos:
DMTroo JN1
CN
0 0
ST23-phos:
OAc OAc
0
NHAc
Conjugate 1
Antisense strand - 5T516001AL33 (SEQ ID NO: 136)
5' mU (ps) fU (ps) mA fU mA fG mA fG mC fA mA fG mA fA mC fA mC fU mG (ps) fU
(ps)
mU (ps) Ser(GN) 3'
Sense strand - STS16001BL20 (SEQ ID NO: 137)
5' Ser(GN) (ps) fA mA fC mA fG mU fG mU fU mC fU mU fG mC fU mC fU mA fU (ps)
mA
(ps) fA 3'
Conjugate 2
Antisense strand - STS16001A (SEQ ID NO: 138)
mU (ps) fU (ps) mA fU mA fG mA fG mC fA mA fG mA fA mC fA mC fU mG (ps) fU
(ps) mU
Sense strand - STS16001BV1L42 (SEQ ID NO: 139)
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
113
Ser(GN) (ps) fA (ps) mA (ps) fC mA fG mU fG mU fU mC fU mU fG mC fU mC fU mA
fU
(ps) mA (ps) fA (ps) Ser(GN)
Conjugate 3
Antisense strand - STS16001AL33 (SEQ ID NO: 136)
5' mU (ps) fU (ps) mA fU mA fG mA fG mC fA mA fG mA fA mC fA mC fU mG (ps) fU
(ps)
mU (ps) Ser(GN) 3'
Sense strand - STS16001BV1L42 (SEQ ID NO: 139)
5' Ser(GN) (ps) fA (ps) mA (ps) fC mA fG mU fG mU fU mC fU mU fG mC fU mC fU
mA fU
(ps) mA (ps) fA (ps) Ser(GN) 3'
Reference conjugate 1
Antisense strand - STS16001A (SEQ ID NO: 138)
mU (ps) fU (ps) mA fU mA fG mA fG mC fA mA fG mA fA mC fA mC fU mG (ps) fU
(ps) mU
Sense strand ¨ STS16001BL20 (SEQ ID NO: 137)
Ser(GN) (ps) fA mA fC mA fG mU fG mU fU mC fU mU fG mC fU mC fU mA fU (ps) mA
(ps) fA
Reference conjugate 2
Antisense strand - 5T516001AL33 (SEQ ID NO: 136)
mU (ps) fU (ps) mA fU mA fG mA fG mC fA mA fG mA fA mC fA mC fU mG (ps) fU
(ps) mU
(ps) Ser(GN)
Sense strand - STS16001V1B (SEQ ID NO: 140)
fA (ps) mA (ps) fC mA fG mU fG mU fU mC fU mU fG mC fU mC fU mA fU (ps) mA
(ps) fA
Reference Conjugate 3
Antisense strand - STS18001A (A0130; SEQ ID NO: 141)
mU (ps) fC (ps) mG fA mA fG mU fA mU fU mC fC mG fC mG fU mA (ps) fC (ps) mG
Sense strand - STS18001BL4 (A0131; SEQ ID NO: 142)
[(5T23) (ps)13 C4XLT (ps) fC mG fU mA fC mG fC mG fG mA fA mU fA mC fU mU fC
(ps)
mG (ps) fA
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
114
Reference Conjugate 4
Antisense strand - STS16001AL33 (SEQ ID NO: 136)
mU (ps) fU (ps) mA fU mA fG mA fG mC fA mA fG mA fA mC fA mC fU mG (ps) fU
(ps) mU
Sense strand - STS16001BL4 (SEQ ID NO: 143)
5"[(5T23) (ps)13 C4XLT(ps) fA (ps) mA (ps) fC mA fG mU fG mU fU mC fU mU fG mC
fU
mC fU mA fU (ps) mA (ps) fA
Example 19 ¨ In vitro determination of TTR knockdown of various TTR siRNA
GaINAc
conjugates
Murine primary hepatocytes were seeded into collagen pre-coated 96 well plates
(Thermo
Fisher Scientific, #A1142803) at a cell density of 30,000 cells per well and
treated with
siRNA-conjugates at concentrations ranging from 10nM to 0.0001nM. 24h post
treatment
cells were lysed and RNA extracted with InviTrap RNA Cell HTS 96 Kit / C24 x
96 preps
(Stratec #7061300400) according to the manufactures protocol. Transcripts
levels of TTR
and housekeeping mRNA (Ptenl I) were quantified by TaqMan analysis.
Target gene expression in primary murine hepatocytes 24h following treatment
with the
conjugates of the invention, Conjugates 1-3, showed that target gene
expression decreases
as the dose of the conjugate increased compared to the negative controls (see
"UT" column
and Reference Conjugate 3), as shown in Figure 27. This indicates that the
first strand is
binding to the target gene, thus lowering gene expression. Figure 27 also
shows the target
gene expression levels of Reference Conjugates 1 and 2 which act as comparator
conjugates. As can be seen from a comparison between the data presented in
Figures 27A
and 27C, and 27B and 27C, the conjugates of the invention (Conjugates 1-3)
decrease the
target gene expression compared to Reference Conjugates 1 and 2. The most
effective
conjugate at 0.01 nM appears to be Conjugate 2. The most effective conjugate
at 0.1 nM,
0.5 nM, 1 nM and 10 nM appears to be Conjugate 3.
Example 20 ¨ In vivo time course of serum TTR in mice
C57BL/6 mice were treated s.c. with 1mg/kg siRNA-conjugates at day 0. Serum
samples
were taken at day 7, 14, and 27 by orbital sinus bleeding and stored at -20 C
until analysis.
Serum TTR quantification was performed with a Mouse Prealbumin ELISA (ALPCO,
41-
PALMS/lot 22, 2008003B) according to the manufacturers protocol (sample
dilution 1:8000
or 1:800).
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
115
The results of the time course of serum TTR in c57BL/6 mice cohorts of n=4 at
7, 14, and
27 days post s.c. treatment with 1 mg/kg Conjugates 1-3, Reference Conjugates
1, 2 and
4, and mock treated (PBS) individuals is shown in Figure 28. As indicated by
the data in
Figure 28, the conjugates of the invention are particularly effective at
reducing target gene
expression compared to the negative control (PBS) and Reference Conjugates 1,
2, and in
particular to Reference Conjugate 4. Conjugates 2 and 3 are also more
effective than
Reference Conjugates 1, 2 and 4. The most effective conjugate is Conjugate 2.
Thus, it may
be expected that the dosing level of Conjugate 3 would be about three times
lower to
achieve the same initial knock down and would also result in longer duration
of knock down
as compared to Reference Conjugate 4.
Example 21 - Synthesis of conjugates 2
Example compounds were synthesised according to methods described below and
methods known to the person skilled in the art. Assembly of the
oligonucleotide chain and
linker building blocks was performed by solid phase synthesis applying
phosphoramidite
methodology. GaINAc conjugation was achieved by peptide bond formation of a
GaINAc-
carboxylic acid building block to the prior assembled and purified
oligonucleotide having the
necessary number of amino modified linker building blocks attached.
Oligonucleotide synthesis, deprotection and purification followed standard
procedures that
are known in the art.
All Oligonucleotides were synthesized on an AKTA oligopilot synthesizer using
standard
phosphoramidite chemistry. Commercially available solid support and 2"0-Methyl
RNA
phosphoramidites, 2"Fluoro, 2"Deoxy RNA phosphoramidites (all standard
protection,
ChemGenes, LinkTech) and commercially available 3'-Amino Modifier TFA Amino C-
6 lcaa
CPG 500A (Chemgenes) were used. Per-acetylated galactose amine 8 is
commercially
available.
Ancillary reagents were purchased from EMP Biotech. Synthesis was performed
using a
0.1 M solution of the phosphoramidite in dry acetonitrile and
benzylthiotetrazole (BTT) was
used as activator (0.3M in acetonitrile). Coupling time was 15 min. A
Cap/OX/Cap or
Cap/Thio/Cap cycle was applied (Cap: Ac20/NMI/Lutidine/Acetonitrile, Oxidizer:
0.1M 12 in
pyridine/H20). Phosphorothioates were introduced using standard commercially
available
thiolation reagent (EDITH, Link technologies). DMT cleavage was achieved by
treatment
with 3% dichloroacetic acid in toluene. Upon completion of the programmed
synthesis
cycles a diethylamine (DEA) wash was performed. All oligonucleotides were
synthesized in
DMT-off mode.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
116
Attachment of the serinol-derived linker moiety was achieved by use of either
base-loaded
(S)-DMT-Serinol(TFA)-succinate-lcaa-CPG 10 or a (S)-DMT-Serinol(TFA)
phosphoramidite
7 (synthesis was performed as described in literature Hoevelmann etal. Chem.
Sci., 2016,7,
128-135). Tri-antennary GaINAc clusters (ST23/C4XLT or ST23/C6XLT) were
introduced
by successive coupling of the respective trebler amid ite derivatives (C4XLT-
phos or C6XLT-
phos) followed by the GaINAc amidite (ST23-phos).
The single strands were cleaved off the CPG by 40% aq. methylamine treatment.
The
resulting crude oligonucleotide was purified by ion exchange chromatography
(Resource Q,
6mL, GE Healthcare) on a AKTA Pure HPLC System using a sodium chloride
gradient.
Product containing fractions were pooled, desalted on a size exclusion column
(Zetadex,
EMP Biotech) and lyophilised.
Individual single strands were dissolved in a concentration of 60 OD/mL in
H20. Both
individual oligonucleotide solutions were added together in a reaction vessel.
For easier
reaction monitoring a titration was performed. The first strand was added in
25% excess
over the second strand as determined by UV-absorption at 260nm. The reaction
mixture
was heated to 80 C for 5min and then slowly cooled to RT. Double strand
formation was
monitored by ion pairing reverse phase HPLC. From the UV-area of the residual
single
strand the needed amount of the second strand was calculated and added to the
reaction
mixture. The reaction was heated to 80 C again and slowly cooled to RT. This
procedure
was repeated until less than 10% of residual single strand was detected.
Synthesis of compounds 2-10
Compounds 2 to 5 and (S)-DMT-Serinol(TFA)-phosphoramidite 7 were synthesised
according to literature published methods (Hoevelmann et al. Chem. Sci.,
2016,7, 128-135).
(S)-4-(3-(bis(4-methoxyphenyl)(phenyl)methoxy)-2-(2,2,2-
trifluoroacetamido)propoxy)-4-oxobutanoic acid (6).
To a solution of 5 in pyridine was added succinic anhydride, followed by DMAP.
The
resulting mixture was stirred at room temperature overnight. All starting
material was
consumed, as judged by TLC. The reaction was concentrated. The crude material
was
chromatographed in silica gel using a gradient 0% to 5% methanol in DCM (+ 1%
triethylamine) to afford 1.33 g of 6 (yield = 38%). m/z (ESI-): 588.2 (100%),
(calcd. for
C30H29F3N08- [M-H] 588.6). 1 H-NMR: (400 MHz, CDCI3) 6 [ppm] = 7.94 (d, 1H,
NH),
7.39 - 7.36 (m, 2H, CHary1), 7.29 - 7.25 (m, 7H, CHary1), 6.82-6.79 (m, 4H,
CHary1), 4.51 ¨
4.47 (m, 1H), 4.31 ¨4.24 (m, 2H), 3.77 (s, 6H, 2xDMTr-OMe), 3.66 - 3.60 (m,
16H, HNEt3+),
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
117
3.26 - 3.25 (m, 2H), 2.97 ¨ 2.81 (m, 20H, NEt3), 2.50-2.41 (4H, m), 1.48¨ 1.45
(m, 26H,
HNEt3+), 1.24 - 1.18 (m, 29H, NEt3).
(S)-DMT-Serinol(TFA)-succinate-lcaa-CPG (10)
The (S)-DMT-Serinol(TFA)-succinate (159 mg, 270 umol) and HBTU (113 mg, 299
umol)
were dissolved in CH3CN (10 mL). Diisopropylethylamine (DIPEA, 94 pL, 540
umol) was
added to the solution, and the mixture was swirled for 2 min followed by
addition native
amino-lcaa-CPG (500 A, 3 g, amine content: 136 umol/g). The suspension was
gently
shaken at room temperature on a wrist-action shaker for 16h then filtered and
washed with
DCM and Et0H. The solid support was dried under vacuum for 2 h. The unreacted
amines
on the support were capped by stirring with acetic anhydride/lutidine/N-
methylimidazole at
room temperature. The washing of the support was repeated as above. The solid
was dried
under vacuum to yield solid support 10 (3 g, 26 umol/g loading).
GaINAc Synthon (9)
Synthesis of the GaINAc synthon 9 was performed as described in Nair et al. J.
Am. Chem.
Soc., 2014, 136 (49), pp 16958-16961, in 46% yield over two steps.
The characterising data matched the published data.
Synthesis of Oligonucleotides
All single stranded oligonucleotides were synthesised according to the
reaction conditions
described above and in Figure 25 and 26.
All final single stranded products were analysed by AEX-H PLC to prove their
purity. Purity
is given in %FLP (% full length product) which is the percentage of the UV-
area under the
assigned product signal in the UV-trace of the AEX-HPLC analysis of the final
product.
Identity of the respective single stranded products (non-modified, amino-
modified
precursors, C4XLT/5T23 or C6XLT/5T23 GaINAc conjugated oligonucleotides) was
proved
by LC-MS analysis.
Table 10: Single stranded un-conjugated and on-column conjugated
oligonucleotides
Product MW MW (ESI-) YoFLP
(11) calc. Found (AEX-
HPLC)
X0385A 6315.0 Da 6314.6 Da 91.0%
X0385B-prec 6593.1 Da 6593.1 Da 87.5%
X038BA 6315.0 Da 6314.6 Da 91.0%
X0386B-prec 6547.1 Da 6546.9 Da 87.5%
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
118
X0383A 6315.0 Da 6314.5 Da 91.9%
X0383B-prec 6508.8 Da 6508.6 Da 84.6%
X0371A 6416.1 Da 6416.1 Da 88.4%
X0371B-prec 6522.0 Da 6521.8 Da 91.9%
X0320A 6143.8 Da 6143.7 Da 94.6%
X0320B-prec 6665.0 Da 6664.8 Da 87.0%
X0477A 6143.8 Da 6143.4 Da 85.6%
X0477B-prec 6749.3 Da 6749.2 Da 83.1%
X0027A 6416.1 Da 6415.8 Da 92.8%
X0027B 7642.0 Da 7641.8 Da 88.2%
Synthesis of conjugates 1-3 and reference conjugates 1-2
Conjugated single strands for conjugates 1-2 and reference conjugates 1-2
Conjugation of the GalNac synthon (9) was achieved by coupling to the serinol-
amino
function of the respective oligonucleotide strand 11 using a peptide coupling
reagent.
Therefore, the respective amino-modified precursor molecule 11 was dissolved
in H20 (500
OD/mL) and DMSO (DMSO/H20, 2/1, v/v) was added, followed by DIPEA (2.5% of
total
volume). In a separate reaction vessel pre-activation of the GaIN(Ac4)-C4-acid
(9) was
performed by reacting 2 eq. (per amino function in the amino-modified
precursor
oligonucleotide 11) of the carboxylic acid component with 2 eq. of HBTU in
presence of 8
eq. DIPEA in DMSO. After 2 min the pre-activated compound 9 was added to the
solution
of the respective amino-modified precursor molecule. After 30 min the reaction
progress
was monitored by LCMS or AEX-HPLC. Upon completion of the conjugation reaction
the
crude product was precipitated by addition of 10x PrOH and 0.1x 2M NaCI and
harvested
by centrifugation and decantation. To set free the acetylated hydroxyl groups
in the GaINAc
moieties the resulting pellet was dissolved in 40% MeNH2 (1mL per 500 OD) and
after 15
min at RT diluted in H20 (1:10) and finally purified again by anion exchange
and size
exclusion chromatography and lyophilised to yield the final product 12.
Table 11: Single stranded GaINAc-conjugated oligonucleotides
Product Starting MW MW (ESI-) YoFLP
(12) Material (11) calc. found (AEX-
HPLC)
X0385B X0385B-prec 7199.8 Da 7199.3 Da 93.2%
X0386B X0386B-prec 7153.8 da 7153.0 Da 86.2%
X0383B X0383B-prec 7115.5 Da 7115.4 Da 93.7%
X0320B X0320B-prec 7271.7 Da 7271.7 Da 90.0%
X0371B X0371B-prec 7128.8 Da 7128.3 Da 95.0%
X0477B X0477B-prec 7356.0 Da 7355.7 Da 91.4%
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
119
Double strand formation
Double strand formation was performed according to the methods described
above.
The double strand purity is given in % double strand which is the percentage
of the UV-area
under the assigned product signal in the UV-trace of the IP-RP-HPLC analysis.
Table 12: Nucleic acid conjugates
Product Starting Materials cyo
First Second double
Strand Strand
strand
X0385 X0385A X0385B 97.5%
X0386 X0386A X0386B 96.9%
X0383 X0383A X0383B 91.9%
X0371 X0371A X0371B 97.7%
X0027 X0027A X0027B 93.4%
X0320 X0320A X0320B 98.6%
X0477 X0477A X0477B 96.0%
Sequences
Modifications key for the following sequences:
f denotes 2"Fluoro 2"deoxyribonucleotide or 2'-fluoro ribonucleotide (the
terms are
interchangeable)
m denotes 2'0 Methyl ribonucleotide
(ps) denotes phosphorothioate linkage
Ser(GN) is a GaINAc-C4 building block attached to serinol derived linker
moiety:
OH
HO ONH
NHAc
wherein the 0--- is the linkage between the oxygen atom and e.g. H,
phosphordiester
linkage or phosphorothioate linkage.
C4XLT is:
C6XLT is:
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
120
---oo
ST23 is:
OAc OAc
11(f Ac0
NHAc
Synthesis of the phosphoramidite derivatives of C4XLT (C4XLT-phos), C6XLT
(C6XLT-
phos) as well as 5T23 (5T23-phos) can be performed as described in
W02017/174657.
C4XLT-phos:
DMTroc) )1\lj
0 0
C6XLT-phos:
)1\1
DMIN,0,-...,...õ--,0,-,......õ 1
0-,....."..õ.õ-- ,P,.. ..-".......-CN
0 0
DMTr-,0,--........õ..--..õ0õ--
5T23-phos:
OAc OAc ,...\,..C..)...\___ i
Ac0 0(:),PCN
NHAc
Example 22
GaINAc conjugates with each one serinol-linked GaINAc moiety at both termini
of the
second strand and with 2'-F at positions 7-9 of the second strand reduce ALDH2
mRNA
levels in vitro.
Different siRNA conjugates targeting human ALDH2 were tested for in vitro
activity in
mouse primary hepatocytes. Both conjugates X0320 and X0477 contain a similar
first
strand, which is modified with alternating 2'-0Me/2'-F. In both conjugates
X0320 and
X0477, serinol-linked GaINAc moieties are conjugated to both ends of the
second strand.
In X0320, the second strand is modified with alternating 2'-F/2'-0Me. In
X0477, the
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
121
second strand is modified with 2'-0Me at positions 1-6 and 10-19 and with 2'-F
at
positions 7-9. Both conjugates mediate dose-dependent reduction of Aldh2
target gene
levels. "UT" indicates an untreated sample all other samples were normalized
to. "Luc"
indicates a GaINAc-conjugated siRNA (X0028) targeting Luciferase, which was
used as
non-targeting control and does not reduce target mRNA levels.
The experiment was conducted in mouse primary hepatocytes. The cells were
seeded at
a density of 20,000 cells per 96-well and treated with 1 nM, 10 nM, and 100 nM
GaINAc
conjugates directly after plating. Transfections with 10 nM GaINAc-siRNA and 1
pg/ml
Atufect liposomal transfection reagent served as control. Cells were lysed
after 24 h, total
RNA was extracted and ALDH2 and ACTB mRNA levels were determined by Taqman
qRT-PCR. Each bar represents mean SD from three technical replicates.
Data are shown in Figure 30.
Sequence Table
Single Sequence (A, first strand; B, second strand, both SEQ ID
Duplex strands 5'-3`) NO:
X0028 X0028A mU (ps) fC (ps) mG fA mA fG mU fA mU fU mC fC 144
mG fC mG fU mA (ps) fC (ps) mG
X0028B [5T23(ps)]3 5T41(ps)fC mG fU mA fC mG fC mG fG 145
mA fA mU fA mC fU mU fC (ps) mG (ps) fA
X0320 X0320A mU (ps) fC (ps) mU fU mC fU mU fA mA fA mC fU 146
mG fA mG fU mU (ps) fU (ps) mC
X0320B Ser(GN) (ps) fG (ps) mA (ps) fA mA fC mU fC mA fG 147
mU fU mU fA mA fG mA fA (ps) mG (ps) fA (ps)
Ser(GN)
X0477 X0477A mU (ps) fC (ps) mU fU mC fU mU fA mA fA mC fU 148
mG fA mG fU mU (ps) fU (ps) mC
X0477B Ser(GN) (ps) mG (ps) mA (ps) mA mA mC mU fC fA 149
fG mU mU mU mA mA mG mA mA (ps) mG (ps) mA
(ps) Ser(GN)
mA, mU, mC, mG 2`-0-Methyl RNA
fA, fU, fC, fG 2'-deoxy-2'-fluoro RNA
irA, irC, irU, irG inverted RNA (3'-3' or 5'-5')
(ps) phosphorothioate
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
122
Example 23
GaINAc siRNA conjugates with vinylphosphonate at the 5' end of the first
strand and
phosphodiester internucleotide linkages at the 5' end of the first strand
effect reduction of
Aldh2 target mRNA levels in vitro.
All tested conjugates contain each one Serinol-linked GaINAc moiety at the 5'
end and at
the 3' end of the second strand. The siRNAs are modified with alternating 2'-
0Me/2'-F and
contain each two phosphorothioate internucleotide linkages at their 5' and 3'
termini, if not
stated differently. X0319 contains two phosphorothioate internucleotide
linkages at the 5'
end of the first strand. X0362 contains a vinylphosphonate modification at the
first
nucleotide and no phosphorothioate internucleotide linkages at the 5' end of
the first
strand. Both siRNA conjugates reduce Aldh2 target gene levels in vitro. "ut"
indicates an
untreated sample, which the other samples were normalised to. "Luc" indicates
an siRNA
targeting Luciferase (X0028), which was used as non-targeting control and does
not
reduce target mRNA levels.
The experiment was conducted in mouse primary hepatocates. 25,000 cells were
seeded
per 96-well and treated with 0.1 ¨ 100 nM GaINAc-conjugated siRNA directly
after plating.
Cells were lysed after 24 h, total RNA was extracted and Aldh2 and ApoB mRNA
levels
were determined by Taqman gRT-PCR. Each bar represents mean SD from three
technical replicates.
Data are shown in Figure 31.
Example 24
GaINAc siRNA conjugates with vinylphosphonate at the 5' end of the first
strand and
phosphodiester internucleotide linkages at the 5' end of the first strand
effect improved
reduction of Aldh2 target mRNA levels in vitro.
All tested conjugates contain each one Serinol-linked GaINAc moiety at the 5'
end and at
the 3' end of the second strand. The siRNAs are modified with alternating 2'-
0Me/2'-F and
contain each two phosphorothioate internucleotide linkages at their 5' and 3'
termini, if not
stated differently. X0320 contains two phosphorothioate internucleotide
linkages at the 5'
end of the first strand. X0363 contains a vinylphosphonate modification at the
first
nucleotide and no phosphorothioate internucleotide linkages at the 5' end of
the first
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
123
strand. Compared to X0320, X0363 shows improved reduction of Aldh2 target gene
levels
in vitro. "ut" indicates an untreated sample, which the other samples were
normalised to.
"Luc" indicates an siRNA targeting Luciferase (X0028), which was used as non-
targeting
control and does not reduce target mRNA levels.
The experiment was conducted in mouse primary hepatocates. 25,000 cells were
seeded
per 96-well and treated with 0.1 ¨ 100 nM GaINAc-conjugated siRNA directly
after plating.
Cells were lysed after 24 h, total RNA was extracted and Aldh2 and ApoB mRNA
levels
were determined by Taqman gRT-PCR. Each bar represents mean SD from three
technical replicates.
Data are shown in Figure 32.
Example 25
GaINAc siRNA conjugates with vinylphosphonate at the 5' end of the first
strand and
phosphodiester internucleotide linkages at the 5' end of the first strand are
stable in acidic
tritosome lysate.
Both tested siRNA conjugates contain each one Serinol-linked GaINAc moiety at
the 5'
end and at the 3' end of the second strand. The siRNAs are modified with
alternating 2'-
OMe/2'-F and contain each two phosphorothioate internucleotide linkages at
their 5' and
3' termini, if not stated differently. X0319 contains two phosphorothioate
internucleotide
linkages at the 5' end of the first strand. X0362 contains a vinylphosphonate
modification
at the first nucleotide and no phosphorothioate internucleotide linkages at
the 5' end of the
first strand. Both GaINAc siRNA conjugates are stable for at least 72 hours.
To assess stability, 5 pM siRNA conjugate was incubated with acidic rat
tritosome extract
(pH 5) at 37 C for 0, 4, and 72 hours. After incubation, RNA was purified,
separated on
20% TBE polyacrylamide gels and visualised by ethidium bromide staining.
Data are shown in Figure 33.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
124
Example 26
GaINAc siRNA conjugates with vinylphosphonate at the 5' end of the first
strand and
phosphodiester internucleotide linkages at the 5' end of the first strand are
stable in acidic
tritosome lysate.
Both tested siRNA conjugates contain each one Serinol-linked GaINAc moiety at
the 5'
end and at the 3' end of the second strand. The siRNAs are modified with
alternating 2'-
OMe/2'-F and contain each two phosphorothioate internucleotide linkages at
their 5' and
3' termini, if not stated differently. X0320 contains two phosphorothioate
internucleotide
linkages at the 5' end of the first strand. X0363 contains a vinylphosphonate
modification
at the first nucleotide and no phosphorothioate internucleotide linkages at
the 5' end of the
first strand. Both GaINAc siRNA conjugates are stable for at least 72 hours.
To assess stability, 5 pM siRNA conjugate was incubated with acidic rat
tritosome extract
(pH 5) at 37 C for 0, 4, and 72 hours. After incubation, RNA was purified,
separated on
20% TBE polyacrylamide gels and visualised by ethidium bromide staining.
Data are shown in Figure 34.
Example 27
GaINAc siRNA conjugates with vinylphosphonate at the 5' end of the first
strand and
phosphodiester internucleotide linkages at the 5' end of the first strand
effect reduction of
Aldh2 target mRNA levels in vitro.
All tested conjugates contain each one Serinol-linked GaINAc moiety at the 5'
end and at
the 3' end of the second strand. The siRNAs contain each two phosphorothioate
internucleotide linkages at their 5' and 3' termini, if not stated
differently. X0320 and X363
are modified with alternating 2'-0Me/2'-F. X0477 and X0478 are modified with
alternating
2'-0Me/2'-F in the first strand and with 2'-0Me at positions 1-6 and 10-19 of
the second
strand and with 2'-F at positions 7-9 of the second strand. X0320 and X0477
contain two
phosphorothioate internucleotide linkages at the 5' end of their first
strands. X0363 and
X0478 contains a vinylphosphonate modification at the first nucleotide and no
phosphorothioate internucleotide linkages at the 5' end of the first strand.
Compared to
X0320, X0363 reduced Aldh2 mRNA levels more. Compared to X0477, X0478 reduced
Aldh2 mRNA levels more. "ut" indicates an untreated sample, which the other
samples
CA 03118327 2021-04-30
WO 2019/092282
PCT/EP2018/081105
125
were normalised to. "Luc" indicates an siRNA targeting Luciferase (X0028),
which was
used as non-targeting control and does not reduce target mRNA levels.
The experiment was conducted in mouse primary hepatocates. 20,000 cells were
seeded
per 96-well and treated with 1 ¨ 100 nM GaINAc-conjugated siRNA directly after
plating.
Cells were lysed after 24 h, total RNA was extracted and Aldh2 and Actb mRNA
levels
were determined by Taqman gRT-PCR. Each bar represents mean SD from three
technical replicates.
Data are shown in Figure 35.
Material & Methods:
Primer:
SEQ ID NO:
fw GGCAAGCCTTATGTCATCTCGT 32
ALDH2 rev GGAATGGTTTTCCCATGGTACTT 33
probe BHQ1-TGAAATGTCTCCGCTATTACGCTGGCTG-FAM 150
fw AAAGAGGCCAGTCAAGCTGTTC 53
ApoB rev GGTGGGATCACTTCTGTTTTGG 54
probe BHQ1-CAGCAACACACTGCATCTGGTCTCTACCA-VIC 151
fw CACCGCCAAATTTAACTGCAGA 50
PTEN rev AAGGGTTTGATAAGTTCTAGCTGT 51
BHQ1-TGCACAGTATCCTTTTGAAGACCATAACCCA- 52
probe VIC
Sequences
SEQ ID
duplex Strand Sequence (A first stand; B, second stand, both 6-3`)
NO:
X0319A mA(ps)fA(ps)mUfG mUfU mUfU mCfC mUfG mCfU mGfAmC 152
(ps)fG(ps)mG
X0319
Ser(GN)(ps)fC (ps) mC (ps)fG mU fC mAfG mCfA mGfG mAfA mA 153
X0319B
fA mCfA (ps)mU (ps)fU (ps)Ser(GN)
(vp)-mUfAmUfG mUfU mUfU mCfCmUfG mCfU mGfAmC(ps) 154
X0362A
fG(ps)mG
X0362
Ser(GN)(ps)fC (ps) mC (ps)fG mU fC mAfG mCfA mGfG mAfA mA 155
X0362B
fA mCfA (ps)mU (ps)fA (ps)Ser(GN)
X0320A mU (ps)fC (ps)mUit1 mCU mUfAmAfAmCfU mGfAmGU mU 156
(ps)1U (ps)mC
X0320
Ser(GN)(ps)fG (ps)mA (ps)fA mAfC mU C mAfG mUIU mU fA mA 147
X0320B
fG mAfA (ps) mG (ps)fA (ps)Ser(GN)
(vp)-mUfCmUit1 mCU mUfAmAfAmCU mGfAmGIU mU (ps)1U 157
X0363A
X0363 (ps)mC
Ser(GN)(ps)fG (ps)mA (ps)fA mAfC mU C mAfG mUIU mU fA mA 147
X0363B
fG mAfA (ps) mG (ps)fA (ps)Ser(GN)
X0028 X0028A mU (ps)fC (ps) mGfA mAfG mUfAmUit1 mCfCmGfCmGU mA 141
(ps)fC(ps)mG
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
126
X0028B [ST23(ps)]3 ST41(ps)fC mGfU mAfC mGfC mGfG mAfA mU fA mC 142
11_1 mUfC (ps)mG (ps)fA
X0477A mU (ps)fC (ps) mU 11_1 mCU mUfAmAfAmCfU mGfAmGU mU 153
X0477 (ps)1U (ps)mC
X0477B Ser(GN)(ps)mG (ps)mA (ps)mA mA mC mUfCfAfG mU mU mU 158
mAmAmG mAmA(ps)mG (ps)mA(ps)Ser(GN)
(vp)-mUfCmUit1 mCU mUfAmAfAmCU mGfAmGIU mU (ps)1U X0478A 157
X0478 (ps)mC
X0478B Ser(GN)(ps)mG (ps)mA (ps)mA mA mC mUfCfAfG mU mU mU 158
mAmAmG mAmA(ps)mG (ps)mA(ps)Ser(GN)
Legend
mA, mU, mC, mG 2`-0-Methyl RNA
fA, fU, fC, fG 2'-deoxy-2'-fluoro RNA
(ps) phosphorothioate
(vp)-mU (E)-vinylphosphonate mU
Example 28
Improved duration of Aldh2 mRNA knockdown in mice following s.c.
administration of
1mg/kg ALDH2-hcm9 siRNA conjugate 16 with 5' & 3' serinol-GaINAc second strand
conjugation as compared to conjugate 14 with 5' second strand triantennary
GaINAc-
conjugation.
The siRNAs are modified with alternating 2'-0Me/2'-F and contain each two
phosphorothioate (PS) internucleotide linkages at their 5' and 3' terminal two
internucleotide linkages. In conjugate 16 two serinol-GaINAc units are
attached via a PS-
bonds to the Sand 3' of the second strand. In conjugate 14 the two terminal 5'
internucleotide linkages of the second are phosphodiesters and a triantennary
GaINAc
linker is attached via a PS bond to this 5' end.
In vivo activity was assessed by s.c. application of 1mg/kg siRNA to c57BL/6
mice. The
vehicle control was PBS. At indicated time points following treatment animals
were
sacrificed, RNA isolated from liver tissue and transcript levels quantified by
MultiPLEX RT-
qPCR. The Aldh2 transcript ct-value for each treatment group was normalized to
the Pten
transcript ct value (Act) and to the PBS cohort (8, A ct).
Data are shown in Figure 36.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
127
Material & Methods:
siRNAs
SEQ
name batch strand sequence
ID NO:
X0025A mA (ps) fU (ps) mG fU mA fG mC fC mG fA 159
Reference mG fG mA fU mC fU mU (ps) fC (ps) mU
Conjugate X0025 [ST23 (ps)]3 ltrb (ps) fA mG fA mA fG mA fU
160
7 X0025B mC fC mU fC mG fG mC fU mA fC (ps) mA
(ps) fU
X0321A mA (ps) fU (ps) mG fU mA fG mC fC mG fA 161
mG fG mA fU mC fU mU (ps) fC (ps) mU
Conjugate
X0321 Ser(GN) (ps) fA (ps) mG (ps) fA mA fG mA 162
16
X0321B fU mC fC mU fC mG fG mC fU mA fC (ps)
mA (ps) fU (ps) Ser(GN)
Legend
mA, mU, mC, mG 2`-0-Methyl RNA
fA, fU, fC, fG 2'-deoxy-2'-fluoro RNA
(ps) phosphorothioate
(po) phosphodiester
Primer:
SEQ ID NO:
fw GGCAAGCCTTATGTCATCTCGT 32
ALDH2 rev GGAATGGTTTTCCCATGGTACTT 33
probe BHQ1-TGAAATGTCTCCGCTATTACGCTGGCTG-FAM 150
General Methods
In vitro experiments
Primary murine hepatocytes (Thermo Scientific: GIBCO Lot: #MC798) were thawed
and
cryo-preservation medium exchanged for Williams E medium supplemented with 5%
FBS,
1 pM dexamethasone, 2 mM GlutaMax, 1% PenStrep, 4mg/m1 human recombinant
insulin,
15mM Hepes. Cell density was adjusted to 250000 cells per lml. 100p1 per well
of this cell
suspension were seeded into collagen pre-coated 96 well plates. The test
article was
prediluted in the same medium (5 times concentrated) for each concentration
and 25p1 of
this prediluted siRNA or medium only were added to the cells. Cells were
cultured at 37 C
and 5% CO2. 24 h post treatment the supernatant was discarded, and cells were
washed
in cold PBS and 250 pl RNA- Lysis Buffer S (Stratec) was added. Following 15
min
incubation at room temperature plates were storage at -80 C until RNA
isolation according
to the manufacturers protocol.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
128
TaqMan analysis
For mTTR & PTEN MultiPlex TaqMan analysis 10p1 isolated RNA for each treatment
group
were mixed with 10 pl PCR mastermix (TAKYON low Rox) containing 600 nM mTTR-
primer,
400 nM ApoB-primer and 200nM of each probe as well as 0.5 units Euroscript 11
RT
polymerase with 0.2 units RNAse inhibitor. TaqMan analysis was performed in
384-well
plate with a 10 min RT step at 48 C, 3 min initial denaturation at 95 C and 40
cycles of 95 C
for 10 sec and 60 C for 1 min. The primers contain two of BHQ1, FAM and YY,
one at each
end of the sequence.
For TMPRSS6 & ApoB MultiPlex TaqMan analysis 10 pl isolated RNA for each
treatment
group were mixed with 10 pl PCR mastermix (TAKYON low Rox) containing 800 nM
TMPRSS6 primer, 100 nM ApoB primer and 200 nM of either probe as well as 0.5
units
Euroscriptll RT polymerase with 0.2 units RNAse inhibitor. TaqMan analysis was
performed
in 384-well plate with a 10min RT step at 48 C, 3min initial denaturation at
95 C and 40
cycles of 95 C for 10 sec and 60 C for 1 min.
In vivo experiments
To compare in vivo potency of different siRNA conjugates 1 mg/kg siRNA
dissolved in PBS
was administered sub cutaneous in the scapular region of c57BL/6 mice. Cohorts
of of n=6
for were treated with siRNA targeting Aldh2 or Tmprss6 at day 1 and sacrificed
at selected
times points post treatment. Liver samples were snap frozen in liquid nitrogen
and stored
at -80 C until extraction RNA with InviTrap Spin Tissue RNA Mini Kit (stratec)
according to
the manufacturers manual. Following, transcript level of Aldh2, Tmprss6 and
Pten were
quantified as described above.
Tritosome stability assay
To probe for RNAase stability in the endosomal / lysosomal compartment of
hepatic cells in
vitro siRNA was incubated for 0 h, 4 h, 24 h or 72 h in Sprague Dawley Rat
Liver Tritosomes
(Tebu- Bio, CatN.: R0610.LT, lot: 1610405, pH: 7.4, 2.827 Units/m1). To mimic
the acidified
environment the Tritosomes were mixed 1:10 with low pH buffer (1.5M acetic
acid, 1.5M
sodium acetate pH 4.75). 30 pl of this acidified Tritosomes. Following 10p1
siRNA (20pM)
were mixed with and incubated for the indicated times at 37 C. Following
incubation RNA
was isolated with the Clarity OTX Starter Kit-Cartriges (Phenomenex CatNo: KSO-
8494)
according to the manufactures protocol for biological fluids. Lyophilized RNA
was
reconstituted in 30 pl H20, mixed with 4xloading buffer and 5 pl were loaded
to a 20% TBE-
polyacrylamide gel electrophoresis (PAGE) for separation qualitative semi-
quantitative
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
129
analysis. PAGE was run at 120 V for 2 h and RNA visualized by Ethidum-bromide
staining
with subsequent digital imaging with a Biorad Imaging system.
Example 29 - Synthesis of conjugates 3
Example compounds were synthesised according to methods described below and
methods known to the person skilled in the art. Assembly of the
oligonucleotide chain and
linker building blocks was performed by solid phase synthesis applying
phosphoramidite
methodology. GaINAc conjugation was achieved by peptide bond formation of a
GaINAc-
carboxylic acid building block to the prior assembled and purified
oligonucleotide having the
necessary number of amino modified linker building blocks attached.
Oligonucleotide synthesis, deprotection and purification followed standard
procedures that
are known in the art.
All Oligonucleotides were synthesized on an AKTA oligopilot synthesizer using
standard
phosphoramidite chemistry. Commercially available solid support and 2"0-Methyl
RNA
phosphoramidites, 2"Fluoro, 2"Deoxy RNA phosphoramidites (all standard
protection,
ChemGenes, LinkTech) and commercially available 3'-Amino Modifier TFA Amino C-
6 lcaa
CPG 500A (Chemgenes), Fmoc-Amino-DMT C-7 CE phosphoramidite (GlyC3Am), 3'-
Amino Modifier C-3 lcaa CPG 500A (C3Am), Fmoc-Amino-DMT C-3 CED
phosphoramidite
(C3Am) and TFA-Amino C-6 CED phosphoramidite (C6Am) (Chemgenes), 3'-Amino-
Modifier C7 CPG (C7Am) (Glen Research), Non-nucleosidic TFA amino
Phosphoramidite
(Pip), Non-nucleosidic TFA amino Solid Support (PipAm) (AM Chemicals) were
used. Per-
acetylated galactose amine 8 is commercially available.
Ancillary reagents were purchased from EMP Biotech. Synthesis was performed
using a
0.1 M solution of the phosphoramidite in dry acetonitrile and
benzylthiotetrazole (BTT) was
used as activator (0.3M in acetonitrile). Coupling time was 15 min. A
Cap/OX/Cap or
Cap/Thio/Cap cycle was applied (Cap: Ac20/NMI/Lutidine/Acetonitrile, Oxidizer:
0.1M 12 in
pyridine/H20). Phosphorothioates were introduced using standard commercially
available
thiolation reagent (EDITH, Link technologies). DMT cleavage was achieved by
treatment
with 3% dichloroacetic acid in toluene. Upon completion of the programmed
synthesis
cycles a diethylamine (DEA) wash was performed. All oligonucleotides were
synthesized in
DMT-off mode.
Attachment of the serinol-derived linker moiety was achieved by use of either
base-loaded
(S)-DMT-Serinol(TFA)-succinate-lcaa-CPG 10 or a (S)-DMT-Serinol(TFA)
phosphoramidite
7 (synthesis was performed as described in Hoevelmann et al. (2016)). Tri-
antennary
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
130
GaINAc clusters (ST23/C4XLT) were introduced by successive coupling of the
respective
trebler amid ite derivatives (C4XLT-phos) followed by the GaINAc amid ite
(ST23-phos).
Attachment of amino modified moieties (non-serinol-derived linkers) was
achieved by use
of either the respective commercially available amino modified building block
CPG or
amidite.
The single strands were cleaved off the CPG by 40% aq. methylamine treatment.
The
resulting crude oligonucleotide was purified by ion exchange chromatography
(Resource Q,
6mL, GE Healthcare) on a AKTA Pure HPLC System using a sodium chloride
gradient.
Product containing fractions were pooled, desalted on a size exclusion column
(Zetadex,
EMP Biotech) and lyophilised.
Individual single strands were dissolved in a concentration of 60 OD/mL in
H20. Both
individual oligonucleotide solutions were added together in a reaction vessel.
For easier
reaction monitoring a titration was performed. The first strand was added in
25% excess
over the second strand as determined by UV-absorption at 260nm. The reaction
mixture
was heated to 80 C for 5min and then slowly cooled to RT. Double strand
formation was
monitored by ion pairing reverse phase HPLC. From the UV-area of the residual
single
strand the needed amount of the second strand was calculated and added to the
reaction
mixture. The reaction was heated to 80 C again and slowly cooled to RT. This
procedure
was repeated until less than 10% of residual single strand was detected.
Synthesis of compounds 2-10
Compounds 2 to 5 and (S)-DMT-Serinol(TFA)-phosphoramidite 7 were synthesised
according to literature published methods (Hoevelmann et al. Chem. Sci.,
2016,7, 128-135).
(S)-4-(3-(bis(4-methoxyphenyl)(phenyl)methoxy)-2-(2,2,2-
trifluoroacetamido)propoxy)-4-oxobutanoic acid (6).
To a solution of 5 in pyridine was added succinic anhydride, followed by DMAP.
The
resulting mixture was stirred at room temperature overnight. All starting
material was
consumed, as judged by TLC. The reaction was concentrated. The crude material
was
chromatographed in silica gel using a gradient 0% to 5% methanol in DCM (+ 1%
triethylamine) to afford 1.33 g of 6 (yield = 38%). m/z (ESI-): 588.2 (100%),
(calcd. for
C30H29F3N08- [M-H] 588.6). 1 H-NMR: (400 MHz, CDCI3) 6 [ppm] = 7.94 (d, 1H,
NH),
7.39 - 7.36 (m, 2H, CHary1), 7.29 - 7.25 (m, 7H, CHary1), 6.82-6.79 (m, 4H,
CHary1), 4.51 ¨
4.47 (m, 1H), 4.31 ¨4.24 (m, 2H), 3.77 (s, 6H, 2xDMTr-OMe), 3.66 - 3.60 (m,
16H, HNEt3+),
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
131
3.26 - 3.25 (m, 2H), 2.97 ¨ 2.81 (m, 20H, NEt3), 2.50-2.41 (4H, m), 1.48¨ 1.45
(m, 26H,
HNEt3+), 1.24 - 1.18 (m, 29H, NEt3).
(S)-DMT-Serinol(TFA)-succinate-lcaa-CPG (10)
The (S)-DMT-Serinol(TFA)-succinate (159 mg, 270 umol) and HBTU (113 mg, 299
umol)
were dissolved in CH3CN (10 mL). Diisopropylethylamine (DIPEA, 94 pL, 540
umol) was
added to the solution, and the mixture was swirled for 2 min followed by
addition native
amino-lcaa-CPG (500 A, 3 g, amine content: 136 umol/g). The suspension was
gently
shaken at room temperature on a wrist-action shaker for 16h then filtered, and
washed with
DCM and Et0H. The solid support was dried under vacuum for 2 h. The unreacted
amines
on the support were capped by stirring with acetic anhydride/lutidine/N-
methylimidazole at
room temperature. The washing of the support was repeated as above. The solid
was dried
under vacuum to yield solid support 10 (3 g, 26 umol/g loading).
GaINAc Synthon (9)
Synthesis of the GaINAc synthon 9 was performed as described in Nair et al. J.
Am. Chem.
Soc., 2014, 136 (49), pp 16958-16961, in 46% yield over two steps.
The characterising data matched the published data.
Synthesis of Oligonucleotides
All single stranded oligonucleotides were synthesised according to the
reaction conditions
described above and in Figure 25 and 26, and are outlined in Tables 13 and 14.
All final single stranded products were analysed by AEX-H PLC to prove their
purity. Purity
is given in %FLP (% full length product) which is the percentage of the UV-
area under the
assigned product signal in the UV-trace of the AEX-HPLC analysis of the final
product.
Identity of the respective single stranded products (non-modified, amino-
modified
precursors or GaINAc conjugated oligonucleotides) was proved by LC-MS
analysis.
Table 13: Single stranded un-conjugated oligonucleotides
Product Name MW MW (ESI-) YoFLP
(11) calc. found (AEX-
HPLC)
A0002 STS16001A 6943.3 Da 6943.0 Da 86.6%
A0006 STS16001BL4 8387.5 Da 8387.5 Da 94.1%
A0114 STS22006A 6143.8 Da 6143.7 Da 94.3%
A0115 STS22006BL1 7855.1 Da 7855.1 Da 92.8%
A0122 STS22009A 6260.9 Da 6260.6 Da 92.8%
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
132
A0123 STS22009BL1 7783.0 Da 7782.9 Da 87.1%
A0130 STS18001A 6259.9 Da 6259.8 Da 76.5%
A0131 STS18001BL4 7813.2 Da 7813.1 Da 74.3%
A0220 STS16001B-5'1xNH2 6982.2 Da 6982.1 Da 95.7%
A0237 STS16001A 6943.3 Da 6943.3 Da 95.6%
A0244 STS16001BV1 6845.2 Da 6844.9 Da 98.2%
A0264 STS16001AV4-3'1xN H2 7112.4 Da 7112.2 Da
95.4%
A0329 STS16001BV6-3'5'1xNH2 7183.3 Da 7183.2 Da
88.8%
A0560 STS16001A 6943.3 Da 6943.3 Da 96.7%
A0541 STS16001BV1-3'5'NH2 7151,3 Da 7151,0 Da 85,6%
A0547 STS16001BV16-3'5'NH2 7119,3 Da 7119,1 Da 89,9%
A0617 STS16001BV20-3'5'NH2 7087,3Da 7086,7 Da 90,1%
A0619 STS16001BV1-3'5'2xNH2 7521,3 Da 7521,3 Da 93.4%
A0680 STS16001A 6943.3 Da 6942.9 Da 91.2%
A0514 STS22006A 6143.8 Da 6143.7 Da 94.6%
A0516 STS22009BV11-3'5'NH2 6665.0 Da 6664.8 Da 87.0%
A0517 STS22009BV11-3'5'NH2 6593.0 Da 6593.0 Da 86.0%
A0521 STS12009BV1-3'5'NH2 6437,7 Da 6437.8 Da 91.1%
A0303 STS12209BL4 7665.0 Da 7664.9 Da 90.4%
A0304 STS12209A 6393.1 Da 6392.9 Da 77.6%
A0319 STS22009A 6260,9 Da 6260.5 Da 86.9%
A0353 STS12009A 6416.1 Da 6416.1 Da 94.1%
A0216 STS17001A 6178.8 Da 6178.7 Da 87.2%
A0217 STS17001BL6 7937.2 Da 7937.2 Da 78.3%
51 x NH2 means refers to the position (5' end) and number (1 x NH2) of free
serinol derived
amino groups which are available for conjugation. For example, 1x3'NH2 on
A0264 means
there is free amino group which can be reacted with GaINAc synthon 9 at the 3'
end of the
strand A0264. 3'5'1 xNH2 means there is one serinol-derived free amino group
which can
be reacted with GaINAc linker 9 at the 3' end and the 5' end of the strand.
Table 14: Single stranded oligonucleotides with 5' and 3' modifications
Product Name 5'mod 3'mod MW MW (ESI-) YoFLP
calc. found
(AEX-
HPLC)
A0561 STS16001BV1-3'5'1xNH2 C6Am GlyC3Am 7267.5 Da 7267.5 Da
66.7%
A0563 STS16001BV1-3'5'1xNH2 C3Am C3Am 7183.4 Da 7183.1 Da
75.1%
A0651 STS16001BV1-3'5'1xNH2 C6Am C7Am 7265.6 Da 7265.2 Da
99.8%
A0653 STS16001BV1-3'5'1xNH2 GlyC3Am GlyC3Am 7299.5 Da 7299.3 Da
88.1%
A0655 STS16001BV1-3'5'1xNH2 pipAm PipAm 7517.7 Da 7517.5 Da
89.8%
Similarly, 351 x NH2 refers to the position (3' and 5' end) and number (1 x
NH2 each) of
free amino groups which are available for conjugation. For example, 3'5'1xNH2
on A0561
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
133
means there are 2 free amino group (1 at the 3' AND 1 at the 5' end) which can
be reacted
with GaINAc synthon 9 at the 3' end of the strand A0561.
Synthesis of certain conjugates of the invention and reference conjugates 1-2
Conjugation of the GalNac synthon (9) was achieved by coupling to the serinol-
amino
function of the respective oligonucleotide strand 11 using a peptide coupling
reagent.
Therefore, the respective amino-modified precursor molecule 11 was dissolved
in H20 (500
OD/mL) and DMSO (DMSO/H20, 2/1, v/v) was added, followed by DIPEA (2.5% of
total
volume). In a separate reaction vessel pre-activation of the GaIN(Ac4)-C4-acid
(9) was
performed by reacting 2 eq. (per amino function in the amino-modified
precursor
oligonucleotide 11) of the carboxylic acid component with 2 eq. of HBTU in
presence of 8
eq. DIPEA in DMSO. After 2 min the pre-activated compound 9 was added to the
solution
of the respective amino-modified precursor molecule. After 30 min the reaction
progress
was monitored by LCMS or AEX-HPLC. Upon completion of the conjugation reaction
the
crude product was precipitated by addition of 10x PrOH and 0.1x 2M NaCI and
harvested
by centrifugation and decantation. To set free the acetylated hydroxyl groups
in the GaINAc
moieties the resulting pellet was dissolved in 40% MeNH2 (1mL per 500 OD) and
after 15
min at RT diluted in H20 (1:10) and finally purified again by anion exchange
and size
exclusion chromatography and lyophilised to yield the final product 12 (Table
15).
Table 15: Single stranded GaINAc-conjugated oligonucleotides
Product Starting Name MW MW (ESI-) YoFLP
(12) Material calc. found (AEX-
HPLC)
A0241 A0220 STS16001BL20 7285.5 Da 7285.3 Da 91.8%
A0268 A0264 STS16001AV4L33 7415.7 Da 7415.4 Da 96.9%
A0330 A0329 STS16001BV6L42 7789.8 Da 7789.8 Da 95.5%
A0544 A0541 STS16001BV1L75 7757,9 Da 7757,7 Da 93.3%
A0550 A0547 STS16001BV16L42 7725,9 Da 7725.7 Da 88.5%
A0620 A0617 STS16001BV20L75 7693,91 Da 7693,2 Da 90.9%
A0622 A0619 STS16001BV1L94 8734,3 Da 8734,6 Da 82.9%
A0519 A0516 STS22006BV11L42 7271.7 Da 7271.7 Da 90.0%
A0520 A0517 STS22009BV11L42 7199.6 Da 7199.7 Da 92.9%
A0522 A0521 STS12009BV1L42 7044.4 Da 7044.4 Da 96.0%
A0603 A0602 STS20041BV1L42 7280.7 Da 7280.4 Da 93.4%
Synthesis of certain conjugates of the invention
Conjugation of the GalNac synthon (9) was achieved by coupling to the amino
function of
the respective oligonucleotide strand 14 using a peptide coupling reagent.
Therefore, the
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
134
respective amino-modified precursor molecule 14 was dissolved in H20 (500
OD/mL) and
DMSO (DMSO/H20, 2/1, v/v) was added, followed by DIPEA (2.5% of total volume).
In a
separate reaction vessel pre-activation of the GaIN(Ac4)-C4-acid (9) was
performed by
reacting 2 eq. (per amino function in the amino-modified precursor
oligonucleotide 14) of
the carboxylic acid component with 2 eq. of HBTU in presence of 8 eq. DIPEA in
DMSO.
After 2 min the pre-activated compound 9 was added to the solution of the
respective amino-
modified precursor molecule. After 30 min the reaction progress was monitored
by LCMS
or AEX-HPLC. Upon completion of the conjugation reaction the crude product was
precipitated by addition of 10x PrOH and 0.1x 2M NaCI and harvested by
centrifugation
and decantation. To set free the acetylated hydroxyl groups in the GaINAc
moieties the
resulting pellet was dissolved in 40% MeNH2 (1mL per 500 OD) and after 15 min
at RT
diluted in H20 (1:10) and finally purified again by anion exchange and size
exclusion
chromatography and lyophilised to yield the final product 15 (Table 16).
Table 16: Single stranded GaINAc-conjuqated olicionucleotides
Product Starting Name MW MW (ESI-) YoFLP
(15) Material calc. found (AEX-
HPLC)
A0562 A0561 STS16001BV1L87 7874.2 Da 7874.0 Da 82.7%
A0564 A0563 STS16001BV1L88 7790.0 Da 7789.4 Da 90.4%
A0652 A0651 STS16001BV1L96 7872.2 Da 7871.8 Da 94.6%
A0654 A0653 STS16001BV1L97 7906.2 Da 7905.6 Da 89.9%
A0656 A0655 STS16001BV1L98 8124.3 Da 8124.0 Da 93.6%
Double strand formation
Double strand formation was performed according to the methods described
above.
The double strand purity is given in % double strand which is the percentage
of the UV-area
under the assigned product signal in the UV-trace of the IP-RP-HPLC analysis
(Table 17).
Table 17: Nucleic acid conjugates
Product Starting Materials Name % double strand
First Strand Second Strand
Ref. Conj. 1 A0237 A0241 STS16001L20 97.7%
Ref. Conj. 2 A0268 A0244 STS16001L33 97.8%
Ref. Conj. 3 A0130 A0131 STS18001L4 96.8%
Ref. Conj. 4 A0002 A0006 STS16001L4 90.1%
Ref. Conj. 5 A0216 A0217 STS17001L6 88.4%
Conjugate 1 A0268 A0241 STS16001L24 96.0%
Conjugate 2 A0237 A0330 STS16001V1L42 98.5%
Conjugate 3 A0268 A0330 STS16001V1L43 98.2%
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
135
Conjugate 4 A0560 A0544 STS16001V1L75
92.5%
Conjugate 5 A0560 A0550
STS16001V16L42 95.3%
Conjugate 6 A0237 A0620
STS16001V20L75 97.8%
Conjugate 7 A0237 A0622 STS16001V1L94
93.7%
Conjugate 8 A0680 A0652 STS16001V1L96
98.4%
Conjugate 9 A0680 A0654 STS16001V1L97
95.8%
Conjugate 10 A0680 A0656 STS16001V1L98
97.6%
Conjugate 11 A0560 A0564 STS16001V1L88
95.0%
Conjugate 12 A0237 A0562 STS16001V1L87
96.8%
Conjugate 13 A0114 A0115 STS22006L1 85.6%
Conjugate 14 A0122 A0123 STS22009L1 96.4%
Conjugate 15 A0514 A0519
STS22006V11L42 98.6%
Conjugate 16 A0319 A0520
STS22009V11L42 97.9%
Conjugate 17 A0304 A0303 STS12209L4 93.0%
Conjugate 18 A0353 A0522 STS12009V1L42
98.9%
Conjugate 19 A0601 A0603 STS20041BL42 97.6%
Sequences
Modifications key for the following sequences:
f denotes 2"Fluoro 2"deoxyribonucleotide or 2'-fluoro ribonucleotide (the
terms are
interchangeable)
m denotes 2'0 Methyl ribonucleotide
(ps) denotes phosphorothioate linkage
FAM = 6-Carboxyfluorescein
BHQ = Black Hole Quencher 1
YY = Yakima Yellow
Definitions
Ser(GN) is a GaINAc-C4 building block attached to serinol derived linker
moiety:
OH
HO.
ONH
HO"\I"====¨.\/
NHAc
wherein the 0--- is the linkage between the oxygen atom and e.g. H,
phosphordiester
linkage or phosphorothioate linkage.
GN is:
OH
HO
NHAc
C4XLT (also known as ST41) is:
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
136
---00
---000 ..........õ,¨....õ..¨..õ.0,õ-
---00
ST23 is:
OAc OAc
Ac0&1111L-00õ-
NHAc
Synthesis of the phosphoramidite derivatives of C4XLT (C4XLT-phos) as well as
5T23
(5T23-phos) can be performed as described in W02017/174657.
C4XLT-phos:
DMTroo LI\I
DMTr(:)0 1
0õ,õ0,--.õ....
0 0
DMTroe
5T23-phos:
OAc OAc
Ac0,. __...\,..C.)...\_ I
0(:),PCN
NHAc
C3Am is: Itrb is: GlyC3Am is:
--'00 H
H
C6Am is: Pip Am is: C7Am is:
H 01,w,
NG N'G
.7..N,G
H H
0
,00.
wherein G = H (pre conjugation) or G = GN (post conjugation).
Conjugate 1
Antisense strand - 5T516001AL33 (SEQ ID NO: 136)
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
137
5' mU (ps) fU (ps) mA fU mA fG mA fG mC fA mA fG mA fA mC fA mC fU mG (ps) fU
(ps)
mU (ps) Ser(GN) 3'
Sense strand - STS16001BL20 (SEQ ID NO: 137)
5' Ser(GN) (ps) fA mA fC mA fG mU fG mU fU mC fU mU fG mC fU mC fU mA fU (ps)
mA
(ps) fA 3'
Conjugate 2
Antisense strand - STS16001A (SEQ ID NO: 138)
mU (ps) fU (ps) mA fU mA fG mA fG mC fA mA fG mA fA mC fA mC fU mG (ps) fU
(ps) mU
Sense strand - STS16001BV1L42 (SEQ ID NO: 139)
Ser(GN) (ps) fA (ps) mA (ps) fC mA fG mU fG mU fU mC fU mU fG mC fU mC fU mA
fU
(ps) mA (ps) fA (ps) Ser(GN)
Conjugate 3
Antisense strand - 5T516001AL33 (SEQ ID NO: 136)
5' mU (ps) fU (ps) mA fU mA fG mA fG mC fA mA fG mA fA mC fA mC fU mG (ps) fU
(ps)
mU (ps) Ser(GN) 3'
Sense strand - STS16001BV1L42 (SEQ ID NO: 139)
5' Ser(GN) (ps) fA (ps) mA (ps) fC mA fG mU fG mU fU mC fU mU fG mC fU mC fU
mA fU
(ps) mA (ps) fA (ps) Ser(GN) 3'
Conjugate 4
Antisense strand - STS16001A (SEQ ID NO: 138)
mU (ps) fU (ps) mA fU mA fG mA fG mC fA mA fG mA fA mC fA mC fU mG (ps) fU
(ps) mU
Sense strand - STS16001BV1L75 (SEQ ID NO: 163)
5' Ser(GN) fA (ps) mA (ps) fC mA fG mU fG mU fU mC fU mU fG mC fU mC fU mA fU
(ps)
mA (ps) fA Ser(GN) 3'
Conjugate 5
Antisense strand - STS16001A (SEQ ID NO: 138)
mU (ps) fU (ps) mA fU mA fG mA fG mC fA mA fG mA fA mC fA mC fU mG (ps) fU
(ps) mU
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
138
Sense strand - 5T516001BV16L42 (SEQ ID NO: 164)
5' Ser(GN) (ps) fA mA fC mA fG mU fG mU fU mC fU mU fG mC fU mC fU mA fU mA fA
(ps) Ser(GN) 3'
Conjugate 6
Antisense strand - STS16001A (SEQ ID NO: 138)
mU (ps) fU (ps) mA fU mA fG mA fG mC fA mA fG mA fA mC fA mC fU mG (ps) fU
(ps) mU
Sense strand - 5T516001BV20L75 (SEQ ID NO: 165)
5' Ser(GN)fA mA fC mA fG mU fG mU fU mC fU mU fG mC fU mC fU mA fU mA fA
Ser(GN)
3'
Conjugate 7
Antisense strand - STS16001A (SEQ ID NO: 138)
mU (ps) fU (ps) mA fU mA fG mA fG mC fA mA fG mA fA mC fA mC fU mG (ps) fU
(ps) mU
Sense strand - STS16001BV1L94 (SEQ ID NO: 166)
5' Ser(GN) (ps) Ser(GN) (ps) fA (ps) mA (ps) fC mA fG mU fG mU fU mC fU mU fG
mC fU
mC fU mA fU (ps) mA (ps) fA (ps) Ser(GN) (ps) Ser(GN) 3'
Conjugate 8
Antisense strand - STS16001A (SEQ ID NO: 138)
5' mU (ps) fU (ps) mA fU mA fG mA fG mC fA mA fG mA fA mC fA mC fU mG (ps) fU
(ps)
mU 3'
Sense strand - STS16001V1BL96 (SEQ ID NO: 167)
5' C6Am(GN) (ps) fA (ps) mA (ps) fC mA fG mU fG mU fU mC fU mU fG mC fU mC fU
mA
fU (ps) mA (ps) fA (ps) C7Am(GN) 3'
Conjugate 9
Antisense strand - STS16001A (SEQ ID NO: 138)
5' mU (ps) fU (ps) mA fU mA fG mA fG mC fA mA fG mA fA mC fA mC fU mG (ps) fU
(ps)
mU 3'
Sense strand - STS16001V1BL97 (SEQ ID NO: 168)
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
139
5' GlyC3Am(GN) (ps) fA (ps) mA (ps) fC mA fG mU fG mU fU mC fU mU fG mC fU mC
fU
mA fU (ps) mA (ps) fA (ps) GlyC3Am(GN) 3'
Conjugate 10
Antisense strand - STS16001A (SEQ ID NO: 138)
5' mU (ps) fU (ps) mA fU mA fG mA fG mC fA mA fG mA fA mC fA mC fU mG (ps) fU
(ps)
mU 3'
Sense strand (SEQ ID NO: 169)
5' PipAm(GN) (ps) fA (ps) mA (ps) fC mA fG mU fG mU fU mC fU mU fG mC fU mC fU
mA
fU (ps) mA (ps) fA (ps) PipAm(GN) 3'
Conjugate 11
Antisense strand - STS16001A (SEQ ID NO: 138)
5' mU (ps) fU (ps) mA fU mA fG mA fG mC fA mA fG mA fA mC fA mC fU mG (ps) fU
(ps)
mU 3'
Sense strand - STS16001V1BL88 (SEQ ID NO: 170)
5' C3Am(GN) (ps) fA (ps) mA (ps) fC mA fG mU fG mU fU mC fU mU fG mC fU mC fU
mA
fU (ps) mA (ps) fA (ps) C3Am(GN) 3'
Conjugate 12
Antisense strand - STS16001A (SEQ ID NO: 138)
5' mU (ps) fU (ps) mA fU mA fG mA fG mC fA mA fG mA fA mC fA mC fU mG (ps) fU
(ps)
mU 3'
Sense strand - STS16001V1BL87 (SEQ ID NO: 171)
5' C6Am(GN) (ps) fA (ps) mA (ps) fC mA fG mU fG mU fU mC fU mU fG mC fU mC fU
mA
fU (ps) mA (ps) fA (ps) GlyC3Am(GN) 3'
Conjugate 15
Antisense strand (SEQ ID NO: 146)
mU (ps) fC (ps) mU fU mC fU mU fA mA fA mC fU mG fA mG fU mU (ps) fU (ps) mC
Sense strand (SEQ ID NO: 147)
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
140
Ser(GN) (ps) fG (ps) mA (ps) fA mA fC mU fC mA fG mU fU mU fA mA fG mA fA (ps)
mG
(ps) fA (ps) Ser(GN)
Conjugate 16
Antisense strand (SEQ ID NO: 159)
mA (ps) fU (ps) mG fU mA fG mC fC mG fA mG fG mA fU mC fU mU (ps) fC (ps) mU
Sense strand (SEQ ID NO: 162)
Ser(GN) (ps) fA (ps) mG (ps) fA mA fG mA fU mC fC mU fC mG fG mC fU mA fC (ps)
mA
(ps) fU (ps) Ser(GN)
Conjugate 18
Antisense strand (SEQ ID NO: 172)
mA (ps) fA (ps) mC fC mA fG mA fA mG fA mA fG mC fA mG fG mU (ps) fG (ps) mA
Sense strand (SEQ ID NO: 173)
Ser(GN) (ps) fU (ps) mC (ps) fA mC fC mU fG mC fU mU fC mU fU mC fU mG fG (ps)
mU
(ps) fU (ps) Ser(GN)
Conjugate 19
Antisense strand (SEQ ID NO: 174)
mA (ps) fU (ps) mA fA mC fU mC fU mG fU mC fC mA fU mU fA mC (ps) fC (ps) mG
Sense strand (SEQ ID NO: 175)
Ser(GN) (ps) fC (ps) mG (ps) fG mU fA mA fU mG fG mA fC mA fG mA fG mU fU (ps)
mA
(ps) fU (ps) Ser(GN)
Reference conjugate 1
Antisense strand - STS16001A (SEQ ID NO: 138)
mU (ps) fU (ps) mA fU mA fG mA fG mC fA mA fG mA fA mC fA mC fU mG (ps) fU
(ps) mU
Sense strand ¨ STS16001BL20 (SEQ ID NO: 137)
Ser(GN) (ps) fA mA fC mA fG mU fG mU fU mC fU mU fG mC fU mC fU mA fU (ps) mA
(ps) fA
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
141
Reference conjugate 2
Antisense strand - STS16001AL33 (SEQ ID NO: 136)
mU (ps) fU (ps) mA fU mA fG mA fG mC fA mA fG mA fA mC fA mC fU mG (ps) fU
(ps) mU
(ps) Ser(GN)
Sense strand - STS16001BV1 (SEQ ID NO: 140)
fA (ps) mA (ps) fC mA fG mU fG mU fU mC fU mU fG mC fU mC fU mA fU (ps) mA
(ps) fA
Reference Conjugate 3 ¨ "Luc"
Antisense strand - STS18001A (A0130; SEQ ID NO: 141)
mU (ps) fC (ps) mG fA mA fG mU fA mU fU mC fC mG fC mG fU mA (ps) fC (ps) mG
Sense strand - STS18001BL4 (A0131; SEQ ID NO: 142)
[(5T23) (p5)13 C4XLT (ps) fC mG fU mA fC mG fC mG fG mA fA mU fA mC fU mU fC
(ps)
mG (ps) fA
Reference Conjugate 4
Antisense strand - 5T516001AL33 (SEQ ID NO: 136)
mU (ps) fU (ps) mA fU mA fG mA fG mC fA mA fG mA fA mC fA mC fU mG (ps) fU
(ps) mU
Sense strand - STS16001BL4 (SEQ ID NO: 143)
5"[(5T23) (p5)13 C4XLT(ps) fA (ps) mA (ps) fC mA fG mU fG mU fU mC fU mU fG mC
fU
mC fU mA fU (ps) mA (ps) fA
Reference Conjugate 5 ¨ "Ctr"
Antisense strand (SEQ ID NO: 176)
mC (ps) fU (ps) mU fA mC fU mC fU mC fG mC fC mC fA mA fG mC (ps) fG (ps) mA
Sense strand (SEQ ID NO: 177)
[(5T23) (ps)]3 (C6XLT) (ps) fU mC fG mC fU mU fG mG fG mC fG mA fG mA fG mU fA
(ps)
mA (ps) fG
Reference Conjugate 6
Antisense strand (SEQ ID NO: 146)
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
142
mU (ps) fC (ps) mU fU mC fU mU fA mA fA mC fU mG fA mG fU mU (ps) fU (ps) mC
Sense strand (SEQ ID NO: 178)
[5T23 (ps)]3 Itrb (ps) fG mA fA mA fC mU fC mA fG mU fU mU fA mA fG mA fA (ps)
mG
(ps) fA
Reference Conjugate 7
Antisense strand (SEQ ID NO: 159)
mA (ps) fU (ps) mG fU mA fG mC fC mG fA mG fG mA fU mC fU mU (ps) fC (ps) mU
Sense strand (SEQ ID NO: 160)
[5T23 (ps)]3 Itrb (ps) fA mG fA mA fG mA fU mC fC mU fC mG fG mC fU mA fC (ps)
mA
(ps) fU
Reference Conjugate 8
Antisense strand (SEQ ID NO: 179)
mU (ps) fA (ps) mC fC mA fG mA fA mG fA mA fG mC fA mG fG mU (ps) fG (ps) mA
Sense strand
[5T23 (ps)]3 5T41 (ps)fU mC fA mC fC mU fG mC fU mU fC mU fU mC fU mG fG (ps)
mU
(ps) fA
Reference Conjugate 9
Antisense strand (SEQ ID NO: 174)
mA (ps) fU (ps) mA fA mC fU mC fU mG fU mC fC mA fU mU fA mC (ps) fC (ps) mG
Sense strand (SEQ ID NO: 181)
[5T23 (ps)]3 C6XLT (ps) fC mG fG mU fA mA fU mG fG mA fC mA fG mA fG mU fU
(ps)
mA (ps) fU
Example 30 ¨ In vitro determination of TTR knockdown of various TTR siRNA
GaINAc
conjugates
Conjugates 1 to 3
Murine primary hepatocytes were seeded into collagen pre-coated 96 well plates
(Thermo
Fisher Scientific, #A1142803) at a cell density of 30,000 cells per well and
treated with
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
143
siRNA-conjugates at concentrations ranging from 10nM to 0.0001nM. 24h post
treatment
cells were lysed and RNA extracted with InviTrap RNA Cell HTS 96 Kit / C24 x
96 preps
(Stratec #7061300400) according to the manufactures protocol. Transcripts
levels of TTR
and housekeeping mRNA (Ptenl I) were quantified by TaqMan analysis.
Target gene expression in primary murine hepatocytes 24h following treatment
at 0.01 nM,
0.1 nM, 0.5 nM, 1nM and 10nM with the conjugates of the invention, Conjugates
1-3,
showed that target gene expression decreases as the dose of the conjugate
increased
compared to the negative controls (see "UT" column and Reference Conjugate 3),
as shown
in Figure 27. This indicates that the first strand is binding to the target
gene, thus lowering
gene expression. Figure 27 also shows the target gene expression levels of
Reference
Conjugates 1 and 2 which act as comparator conjugates. As can be seen from a
comparison
between the data presented in Figures 27A and 27C, and 27B and 27C, the
conjugates of
the invention (Conjugates 1-3) decrease the target gene expression compared to
Reference
Conjugates 1 and 2. The most effective conjugate at 0.01 nM appears to be
Conjugate 2.
The most effective conjugate at 0.1 nM, 0.5 nM, 1 nM and 10 nM appears to be
Conjugate
3.
Conjugates 4 to 7
The method described above under "In vitro experiments" in the General Method
section
was followed.
Target gene expression in primary murine hepatocytes 24h following treatment
at 0.01 nM,
0.1 nM, 0.5 nM, 1nM and 10nM with the conjugates of the invention, Conjugates
4-7,
showed that target gene expression decreases as the dose of the conjugate
increased
compared to the negative controls (see "UT" column and Luc [Reference
Conjugate 3]), as
shown in Figure 50. This indicates that the first strand is binding to the
target gene, thus
lowering gene expression.
The in vitro data show that in the context of one or two serinol-derived
linker moieties being
provided at 5' and 3' ends of the sense strand in Conjugates 4-7, the number
of
phosphorothioate (PS) bonds between the terminal nucleotide and the linker,
and/or
between the terminal three nucleotides in the sense strand, can be varied
whilst maintaining
efficacy for decreasing target gene expression.
Conjugates 8 to 12 and 19
The method described above under "In vitro experiments" in the General Method
section
was followed.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
144
Target gene expression in primary murine hepatocytes 24h following treatment
at 0.01 nM,
0.1 nM, 0.5 nM, 1nM and 10nM with the conjugates of the invention, Conjugates
8-12,
showed that target gene expression decreases as the dose of the conjugate
increased
compared to the negative controls (see "UT" column and Luc [Reference
Conjugate 3]), as
shown in Figure 51. This indicates that the first strand is binding to the
target gene, thus
lowering gene expression. In particular, Conjugates 8, 9, 10 and 11 appear to
be
comparable to or better than Conjugate 2 which was previously shown to be the
most
effective conjugate at 0.01 nM.
Conjugate 19 was also shown to decrease target gene expression compared to the
negative
controls (see "UT" column and Ctr which is a non-targeting siRNA and also
referred to as
Reference Conjugate 5), as shown in Figure 52. This indicates that the first
strand is binding
to the target gene, thus lowering gene expression.
The in vitro data for Conjugates 8-12 and 19 show that a number of linkers
which are
structurally diverse and which are conjugated at both termini of the sense
strand are
effective at decreasing target gene expression. Conjugates 8-12 and 19
decrease target
gene expression more effectively than "Luc" which is Reference Conjugate 3
(for
Conjugates 8-12), "Ctr" which is Reference Conjugate 5 (for Conjugate 19) and
untreated
control.
Example 31 ¨ In vivo time course of serum TTR, ALDH2 and TMPRSS6 in mice
Conjugates 1 to 3
C57BL/6 mice were treated s.c. with 1mg/kg siRNA-conjugates at day 0. Serum
samples
were taken at day 7, 14, and 27 by orbital sinus bleeding and stored at -20 C
until analysis.
Serum TTR quantification was performed with a Mouse Prealbumin ELISA (ALPCO,
41-
PALMS/lot 22, 2008003B) according to the manufacturers protocol (sample
dilution 1:8000
or 1:800).
The results of the time course of serum TTR in c57BL/6 mice cohorts of n=4 at
7, 14, and
27 days post s.c. treatment with lmg/kg Conjugates 1-3, Reference Conjugates
1, 2 and 4,
and mock treated (PBS) individuals is shown in Figure 28. As indicated by the
data in Figure
28, the conjugates of the invention are particularly effective at reducing
target gene
expression compared to the negative control (PBS) and Reference Conjugates 1,
2, and in
particular to Reference Conjugate 4. Conjugates 2 and 3 are also more
effective than
Reference Conjugates 1, 2 and 4. The most effective conjugate is Conjugate 2.
Thus, it
may be expected that the dosing level of Conjugate 2 would be about three
times lower to
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
145
achieve the same initial knock down and would also result in longer duration
of knock down
as compared to Reference Conjugate 4.
More specifically, Conjugate 2 resulted in 3-fold lower target protein level
in serum at day
seven and 4-fold lower target protein level in serum at day 27 compared to
Reference
Conjugate 4 at equimolar dose in wild type mice. Furthermore, Conjugate 2
resulted in 85%
reduction of target serum protein level at day 27 after single injection,
compared to 36%
reduction by equimolar amount of Reference Conjugate 4.
Conjugates 15 to 18
The method described above under "In vivo experiments" in the General Method
section
was followed.
The results of the time course of serum Aldh2 in c57BL/6 mice cohorts of n=6
at 14,28 and
42 days post s.c. treatment with 1 mg/kg Conjugates 15 and 16, Reference
Conjugates 6
and 7, and mock treated (PBS) individuals is shown in Figures 53 and 54. As
indicated by
the data in Figures 53 and 54, the conjugates of the invention are
particularly effective at
reducing target gene expression compared to the negative control (PBS) and
Reference
Conjugates 6 and 7 respectively.
The results of the time course of serum Tmprss6 in c57BL/6 mice cohorts of n=6
at 14, 28
and 42 days post s.c. treatment with 1 mg/kg Conjugate 18, Reference Conjugate
8, and
mock treated (PBS) individuals is shown in Figure 55. As indicated by the data
in Figure 55,
the conjugates of the invention are particularly effective at reducing target
gene expression
compared to the negative control (PBS) and Reference Conjugate 8.
Overall, the in vivo data show that a variety of example linkers which are
conjugated at both
termini of the second strand are effective at decreasing target gene
expression in vivo. The
positioning of the linker improves in vivo potency conjugates, as compared to
a triantenary
GaINAc-linker control at the 5' terminus of the second strand (Reference
Conjugates 6, 7
and 8).
Example 32 ¨ Serum Stability Studies
The method described above under "Tritosome stability assay" in the General
Method
section was followed.
Figure 56 shows the results from the serum stability studies in respect of
Conjugates 2, 4,
5, 6 and 7. Figure 57 shows the serum stability of Conjugates 2, 8, 9, 10, 11
and 12.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
146
All conjugates of the invention that were tested are more stable in serum
compared to
control.
All tested conjugates contain each one GaINAc linker unit at the 5' end and
another at the
3' end of the second strand. The siRNAs are modified with alternating 2'-
0Me/2'-F and
contain each two phosphorothioate (PS) internucleotide linkages at their 5'
and 3' terminal
two internucleotide linkages, unless stated differently.
In Conjugate 4 the serinol-GaINAc units are attached via a phosphodiester
bond. In
Conjugate 5 the serinol-GaINAc units are conjugated via PS, whereas all
internucleotide
linkage in the second strand are phosphodiesters. In Conjugate 6 the second
strand
contains no PS. In Conjugate 7 two serinol-GaINAc units are attached to each
second
strand terminus and to each other via a PS-bonds at the respective ends. In
Conjugate 8 a
C6-amino-modifier at 5' and a C7-amino-modifier at the 3' end of the second
strand were
applied for ligand attachment. In Conjugate 9 Gly-C3-amino-modifiers, in
Conjugate 10
piperidyl-amino-modifiers, in Conjugate 11 C3-amino-modifiers and in Conjugate
2 serinol-
GaINAc units were used as linkers for conjugation to both ends of the second
strand. In
Conjugate 2 both terminal internucleotides as well as the nucleotide-serinol
bonds are PS.
In Conjugate 12 a C6-amino-modifier at the 5' and a GlyC3-amino-modifier at
the 3' end of
second strand were applied for ligand attachment. "ut" indicates an untreated
sample which
the other samples were normalised to. "Luc" indicates an siRNA targeting
Luciferase
(Reference Conjugate 3), which was used as non-targeting control and does not
reduce
target mRNA levels.
The data show that in context of a serinol-derived linker moiety being
provided at 5' and 3'
ends of the sense strand, the number of phosphorothioate (PS) bonds between
the terminal
nucleotide and the linker, and/or between the terminal three nucleotides in
the sense strand,
can be varied whilst maintaining stability in serum.
Example 33
ALDH2-targeting GaINAc conjugates with an optimized 2'-0Me/2'-F modification
pattern in
the second strand and with end-stabilising inverted RNA at the 3' end of the
second strand
show extended reduction of ALDH2 mRNA levels in mice. Modified variants of the
GaINAc-
siRNA conjugates 5T522006 and 5T522009 were analyzed for knockdown activity in
vivo.
5T522006L6 and 5T522009L6 contain alternating 2'-0Me/2'-F modifications in
both first
and second strand. 5T522006V1L6 and 5T522009V1L6 contain alternating 2'-0Me/2'-
F
only in the first strand. In the second strand, these conjugates contain 2'-
0Me at positions
1-6 and 10-18 and 2'-F at positions 7-9. An inverted RNA nucleotide is present
at the 3' end
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
147
of the second strand. There, it substitutes for the last nucleotide of the
second strand. The
3' end of the second strand does not contain any phosphorothioate linkages,
whereas all
other, non-conjugated ends are stabilized by two terminal phosphorothioates.
All
conjugates reduce ALDH2 target gene levels. Among the timepoints analysed
here,
strongest reduction is achieved 10 days after treatment. STS22006V1L6 has
improved
initial knockdown activity and duration of action compared to STS22006L6.
STS22009L6
and STS22009V1L6 show equal levels of activity.
C57BL/6 male mice were subcutaneously treated with 3 mg/kg GaINAc conjugate.
Liver
sections were prepared 10, 20, and 30 days after treatment (labeled "d11",
"d21" and "d31"),
total RNA was extracted from the tissue and ALDH2 and ApoB mRNA levels were
analyzed
by Taqman qRT-PCR. Each bar represents mean SD of six animals.
Results are shown in Figure 58.
Statements of Invention
1. A nucleic acid for inhibiting expression of ALDH2 in a cell, comprising
at least one
duplex region that comprises at least a portion of a first strand and at least
a portion of a
second strand that is at least partially complementary to the first strand,
wherein said first
strand is at least partially complementary to at least a portion of RNA
transcribed from the
ALDH2 gene, wherein said first strand comprises a nucleotide sequence selected
from the
following sequences: SEQ ID NOs: 11, 1, 3, 5, 7, 9, 13, 15, 17, 19, 21, 23, 25
or 27 or any
sequence disclosed herein.
2. A nucleic acid of statement 1, wherein the second strand comprises a
nucleotide
sequence of SEQ ID NO: 12, 2, 4, 6, 8, 10, 14, 16, 18, 20, 22, 24, 26 or 28,
or any
sequence disclosed herein.
3. A nucleic acid of statement 1 or statement 2, wherein said first strand
comprises a
nucleotide sequence of SEQ ID NO:11, SEQ ID NO:3 or SEQ ID NO:17.
4. A nucleic acid of any one of statements 1 to 3, wherein said second
strand
comprises the nucleotide sequence of SEQ ID NO:12, SEQ ID NO:4 or SEQ ID
NO:18.
5. A nucleic acid according to any one of statements 1 to 4, wherein said
first strand
and/or said second strand are each from 17-35 nucleotides in length.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
148
6. A nucleic acid of any one of statements 1 to 5, wherein the at
least one duplex
region consists of 19-25 consecutive nucleotide base pairs.
7. A nucleic acid of any preceding statement, which
a) is blunt ended at both ends; or
b) has an overhang at one end and a blunt end at the other; or
c) has an overhang at both ends.
8. A nucleic acid according to any preceding statement, wherein one
or more
nucleotides on the first and / or second strand are modified, to form modified
nucleotides.
9. A nucleic acid of statement 8, wherein one or more of the odd
numbered
nucleotides of the first strand are modified.
10. A nucleic acid according to statement 9, wherein one or more of the
even
numbered nucleotides of the first strand are modified by at least a second
modification,
wherein the at least second modification is different from the modification of
statement 9.
11. A nucleic acid of statement 10, wherein at least one of the one or more
modified
even numbered nucleotides is adjacent to at least one of the one or more
modified odd
numbered nucleotides.
12. A nucleic acid of any one of statements 9 to 11, wherein a plurality of
odd
numbered nucleotides are modified.
13. A nucleic acid of statement 10 to 12, wherein a plurality of even
numbered
nucleotides are modified by a second modification.
14. A nucleic acid of any of statements 8 to 13, wherein the first strand
comprises
adjacent nucleotides that are modified by a common modification.
15. A nucleic acid of any of statements 9 to 14, wherein the first strand
comprises
adjacent nucleotides that are modified by a second modification that is
different to the
modification of statement 9.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
149
16. A nucleic acid of any of statements 9 to 15, wherein one or more
of the odd
numbered nucleotides of the second strand are modified by a modification that
is different
to the modification of statement 9.
17. A nucleic acid according to any of statements 9 to 15, wherein one or
more of the
even numbered nucleotides of the second strand are modified by the
modification of
statement 9.
18. A nucleic acid of statement 16 or 17, wherein at least one of the one
or more
modified even numbered nucleotides of the second strand is adjacent to the one
or more
modified odd numbered nucleotides of the second strand.
19. A nucleic acid of any of statements 16 to 18, wherein a plurality of
odd numbered
nucleotides of the second strand are modified by a common modification.
20. A nucleic acid of any of statements 16 to 19, wherein a plurality of
even numbered
nucleotides are modified by a modification according to statement 9.
21. A nucleic acid of any of statements 16 to 20, wherein a plurality of
odd numbered
nucleotides on the second strand are modified by a second modification,
wherein the
second modification is different from the modification of statement 9.
22. A nucleic acid of any of statements 16 to 21, wherein the second strand
comprises
adjacent nucleotides that are modified by a common modification.
23. A nucleic acid of any of statements 16 to 22, wherein the second strand
comprises
adjacent nucleotides that are modified by a second modification that is
different from the
modification of statement 9.
24. A nucleic acid according to any one of statements 8 to 23, wherein each
of the odd
numbered nucleotides in the first strand and each of the even numbered
nucleotides in
the second strand are modified with a common modification.
25. A nucleic acid of statement 24, wherein each of the even numbered
nucleotides
are modified in the first strand with a second modification and each of the
odd numbered
nucleotides are modified in the second strand with a second modification.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
150
26. A nucleic acid according to any one of statements 8 to 25, wherein
the modified
nucleotides of the first strand are shifted by at least one nucleotide
relative to the
unmodified or differently modified nucleotides of the second strand.
27. A nucleic acid according to any one of statements 8 to 26, wherein the
modification and/or modifications are each and individually selected from the
group
consisting of 3'-terminal deoxy-thymine, 2'-0-methyl, a 2'-deoxy-modification,
a
2'-amino-modification, a 2'-alkyl-modification, a morpholino modification, a
phosphoramidate modification, 5'-phosphorothioate group modification, a 5'
phosphate or
5' phosphate mimic modification and a cholesteryl derivative or a dodecanoic
acid
bisdecylamide group modification.
28. A nucleic acid according to any one of statements 8 to 27, wherein the
modification is any one of a locked nucleotide, an abasic nucleotide or a non-
natural base
comprising nucleotide.
29. A nucleic acid according to any one of statements 8 to 28, wherein at
least one
modification is 2'-0-methyl.
30. A nucleic acid according to any one of statements 8 to 29, wherein at
least one
modification is 2'-F.
31. A nucleic acid for inhibiting expression of ALDH2 in a cell, comprising
at least one
duplex region that comprises at least a portion of a first strand and at least
a portion of a
second strand that is at least partially complementary to the first strand,
wherein said first
strand is at least partially complementary to at least a portion of a RNA
transcribed from
the ALDH2 gene, wherein said first strand comprises a nucleotide sequence
selected from
the following sequences: SEQ ID NOs: 11, 1, 3, 5, 7, 9, 13, 15, 17, 19, 21,
23, 25 or 27,
wherein the nucleotides of first strand are modified by first modification on
the odd
numbered nucleotides, and modified by a second modification on the even
numbered
nucleotides, and nucleotides of the second strand are modified by a third
modification on
the even numbered nucleotides and modified by a fourth modification on the odd
numbered nucleotides, wherein at least the first modification is different to
the second
modification and the third modification is different to the fourth
modification.
32. A nucleic acid of statement 31, wherein second sequence comprises a
nucleotide
sequence of SEQ ID NO: 12, 2, 4, 6, 8, 10, 14, 16, 18, 20, 22, 24, 26 or 28.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
151
33. A nucleic acid of statement 31 or 32, wherein the fourth
modification and the
second modification are the same.
34. A nucleic acid of any one of statements 31 to 33, wherein the first
modification and
the third modification are the same.
35. A nucleic acid of any one of statements 31 to 34, wherein the first
modification is
2'0-Me and the second modification is 2'F.
36. A nucleic acid of any one of statements 31 to 35, wherein the first
strand
comprises the nucleotide sequence of SEQ ID NO:11, SEQ ID NO:3 or SEQ ID NO:17
and the second strand comprises the nucleotide sequence of SEQ ID NO:12, SEQ
ID
NO:4 or SEQ ID NO:18.
37. A nucleic acid of any one of statements 31 to 36, comprising a sequence
and
modifications as shown below:
Sequence
siRNA ID sequence modifications
ID No
3 5'-AAUGUUUUCCUGCUGACGG-3' 6254515173547182748
ALDH2-hcm-2
4 5'-CCGUCAGCAGGAAAACAUU-3' 3745364728462627251
11 5'-UCUUCUUAAACUGAGUUUC-3' 5351715262718281517
ALDH2-hcm-6
12 5'-GAAACUCAGUUUAAGAAGA-3' 4626353645152646282
17 5'-AUGUAGCCGAGGAUCUUCU-3' 6181647382846171535
ALDH2-hcm-9
18 5'-AGAAGAUCCUCGGCUACAU-3' 2826461735384716361
wherein, the specific modifications are depicted by numbers
1=2"F-dU,
2=2`F-dA,
3=2"F-dC,
4=2"F-dG,
5=2'-0Me-rU;
6=2'-0Me-rA;
7=2'-0Me-rC;
8=2'-0Me-rG.
38. A nucleic acid according to any one of statements 1 to 37, conjugated
to a ligand.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
152
39. A nucleic acid according to any one of statements 1 to 38, comprising a
phosphorothioate linkage between the terminal one, two or three 3' nucleotides
and/or 5'
nucleotides of the first and/or the second strand.
40. A nucleic acid according to any one of statements 1 to 39 comprising
two
phosphorothioate linkage between each of the three terminal 3' and between
each of the
three terminal 5' nucleotides on the first strand, and two phosphorothioate
linkages
between the three terminal nucleotides of the 3' end of the second strand.
41. A nucleic acid for inhibiting expression of ALDH2 in a cell, comprising
at least one
duplex region that comprises at least a portion of a first strand and at least
a portion of a
second strand that is at least partially complementary to the first strand,
wherein said first
strand is at least partially complementary to at least a portion of a RNA
transcribed from
the ALDH2 gene, wherein said first strand comprises a nucleotide sequence
selected from
the following sequences: SEQ ID NOs: 11, 1, 3, 5, 7, 9, 13, 15, 17, 19, 21,
23, 25 or 27,
and wherein the nucleic acid is conjugated to a ligand.
42. A nucleic acid according to statement 41, wherein the ligand comprises
(i) one or
more N-acetyl galactosamine (GalNac) moieties and derivatives thereof, and
(ii) a linker,
wherein the linker conjugates the GalNac moieties to a nucleic acid as defined
in
statement 41.
43. A nucleic acid according to any of statements 41 or 42, wherein linker
may be a
bivalent or trivalent or tetravalent branched structure.
44. A nucleic acid of any of statements 41 to 43, wherein the nucleotides
are modified
as defined in any preceding statements.
45. A nucleic acid of any preceding statement, which is conjugated to a
ligand
comprising the formula I:
[S-X1-P-X2]3-A-X3- (I)
wherein:
S represents a saccharide, wherein the saccharide is N-acetyl galactosamine;
X1 represents C3-C6 alkylene or (-CH2-CH2-0)m(-CH2)2- wherein m is 1, 2, or 3;
P is a phosphate or modified phosphate (preferably a thiophosphate);
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
153
X2 is alkylene or an alkylene ether of the formula (-CH2)n-O-CH2- where n = 1-
6;
A is a branching unit;
X3 represents a bridging unit;
wherein a nucleic acid as defined in any of statements 1 to 40 is conjugated
to X3 via a
phosphate or modified phosphate (preferably a thiophosphate).
46. A conjugated nucleic acid having one of the following structures
OH
OH
OH
PcH Ft¨OH
\OH
0
0
L
0
S¨LO-
t OH
OH
0- Pd-1
0 OH
0
0¨/¨/
CA 03118327 2021-04-30
WO 2019/092282
PCT/EP2018/081105
154
OH
HOI,&_.0H
OH OH
HO õ1....) ...... AcHNks
0 0
NHAc
1'1')
0
1 9
0=P ¨S
$
01 0
..$) $ 9
0 =P¨S
(!) OH
0 o.1¨/ OH
AcHN
_______________________________________ /
Cr1(10H
/
0
0 /......"--0 ..,
0
II
Z-0¨P-0
S
L 1 9 if
0 ¨P-0
$0
S :
OH
HO\,_ 0H
OH OH
0
HO,,,L....) .... AcHN
0 0
NHAc
L.I1
0
I 9
0 =P ¨ S
i
0 0
0 =P ¨S
0
/ I OH
0,,,. o¨/ OH
AcHN
_______________________________________ /
$CrIOH
/
_ j_ j--0 0
'...ct
0
$1
Z ¨0¨P-0
S
? /
0¨P-0
I0
S
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
155
OH
HO\,...,011
OH OH
0
AcHN
0 0
NHAc
0
Li)
i 0
0=P¨S
i
0.1 0
i 0
0=P¨S
0
/I OH
0
/ AcHN C
0¨ OH
'N
_______________________________________ / r().-R'OH
_Ex J-0/ 0
-.,
0 0
II
I¨O¨P-0
LI,µ 1? f
,.
S 0¨P-0
so
S,
OH
H0µ,_ OH
OH OH
0
AcHN
0 0
NHAc
1')
0
I 0
0 =P ¨S
i
01 0
I 0
0 = P ¨S
0
/ I OH
0,, o j OH
/
AcHN
, r i c ( 1 -FON
_x_rxi¨ 0 _____________________ õ, 0
0 .--
0
II ,
z-0¨P¨O 0¨p-0
i 0 10
S S
CA 03118327 2021-04-30
WO 2019/092282
PCT/EP2018/081105
156
OH
HO k,,,_01.1
OH OH 0
AcHN
0
NHAc
0
04¨Se \
i
01 0
04¨Se
i
0
AcHN OH
OH
0 ,,, 0 --/¨/ (12/)(OH
0/ L /
0
OLL /0
0
Z ¨0 ¨P ¨0 ¨rj---
le
s o
0
0¨p-0
le
s
OH
HO OH
OH OH
AcH 0N
0
NHAc
0
i 0
0=P ¨S
\
1
01 0
I 0
0 =P ¨ S
I
0 AcHN OH
OH
0 0 ¨r0 OH
/
0
/ I
0
0
0
2 ¨0 ¨11-0 ¨T.-1j¨ LI,. 0 /
0
S 0 ¨P ¨0
S
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
157
OH
H\,,_ 0 li
OH OH , 0
AcHN
HO..õ...&.,,) 0
0
r1HAc
0
\
i 0 =P -Se
1
0,1 0
0
OH
0 =P -S
of
AcHN;;OH
0 0 OH
0
0
r-17C.. ? /
II
0 -P -0
1 < 10
S s
wherein Z is a nucleic acid according to any of statements 1 to 40.
47. A nucleic acid according to any of statements 1 to 40, which is
conjugated to a
ligand of the following structure
HO
c04. H (:)
H FiAc 1
r\INc 01 OH
0-P=0 n(
0
(---*V
µAcH orliN 0
H
H
ir.1 k 0
0
HAc
H H Ce 0 /AN==vNN
H
48. A nucleic acid or conjugated nucleic acid of any preceding statement,
wherein the
duplex comprises separate strands.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
158
49. A nucleic acid or conjugated nucleic acid of any preceding statement,
wherein the
duplex comprises a single strand comprising a first strand and a second
strand.
50. A composition comprising a nucleic acid or conjugated nucleic acid of
any
preceding statement and a physiologically acceptable excipient.
51. A nucleic acid or conjugated nucleic acid according to any preceding
statement for
use in the prevention or treatment of a disease, disorder or syndrome.
52. Use of a nucleic acid or conjugated nucleic acid according to any
preceding
statement in the manufacture of a medicament for preventing or treating a
disease,
disorder or syndrome.
53. A method of preventing or treating a disease, disorder or syndrome
comprising
administration of a composition comprising a nucleic acid or conjugated
nucleic acid
according to any preceding statement to an individual in need of treatment.
54. The method of statement 53, wherein the nucleic acid or conjugated
nucleic acid is
administered to the subject subcutaneously, intravenously or using any other
application
routes such as oral, rectal or intraperitoneal.
55. A use or method according to statements 51 to 54, wherein said disease
or
disorder is alcohol use disorder.
56. A use or method according to statement 55, wherein the alcohol use
disorder is
acute alcohol sensitivity, alcoholic neuropathy, alcohol abuse or fetal
alcohol disorder or
other pathologies associated to acute or prolonged excessive alcohol
consumption.
57. A process for making a nucleic acid or conjugated nucleic acid of any
one of
statements 1 to 49.
58. A nucleic acid for inhibiting expression of ALDH2, comprising at least
one duplex
region that comprises at least a portion of a first strand and at least a
portion of a second
strand that is at least partially complementary to the first strand, wherein
said first strand is
at least partially complementary to at least a portion of RNA transcribed from
the ALDH2
gene, wherein the expression of ALDH2 is reduced to levels which are at least
15% lower
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
159
than expression levels observed in same test conditions but in the absence of
the nucleic
acid or conjugated nucleic acid or in the presence of a non-silencing control.
59. A nucleic acid for inhibiting expression of ALDH2, comprising at
least one duplex
region that comprises at least a portion of a first strand and at least a
portion of a second
strand that is at least partially complementary to the first strand, wherein
said first strand is
at least partially complementary to at least a portion of a RNA transcribed
from the ALDH2
gene, wherein said first strand comprises a nucleotide sequence selected from
the
following sequences: SEQ ID NOs: 11, 1, 3, 5, 7, 9, 13, 15, 17, 19, 21, 23, 25
or 27,
wherein the expression of ALDH2 is reduced to levels which are at least 15%
lower than
expression levels observed in same test conditions but in the absence of the
nucleic acid
or conjugated nucleic acid or in the presence of a non-silencing control.
Other clauses of the invention include:
1. A conjugate for inhibiting expression of a ALDH2 gene in a cell,
said conjugate
comprising a nucleic acid portion and ligand portions, said nucleic acid
portion comprising
at least one duplex region that comprises at least a portion of a first RNA
strand and at least
a portion of a second RNA strand that is at least partially complementary to
the first strand,
wherein said first strand is at least partially complementary to at least a
portion of RNA
transcribed from said ALDH2 gene, said ligand portions comprising a serinol-
derived linker
moiety and a targeting ligand for in vivo targeting of cells and being
conjugated exclusively
to the 3' and/or 5' ends of one or both RNA strands, wherein the 5' end of the
first RNA
strand is not conjugated, wherein:
(i) the second RNA strand is conjugated at the 5' end to the targeting
ligand, and
wherein (a) the second RNA strand is also conjugated at the 3' end to the
targeting ligand and the 3' end of the first RNA strand is not conjugated; or
(b)
the first RNA strand is conjugated at the 3' end to the targeting ligand and
the 3'
end of the second RNA strand is not conjugated; or (c) both the second RNA
strand and the first RNA strand are also conjugated at the 3' ends to the
targeting
ligand; or
(ii) both the second RNA strand and the first RNA strand are conjugated at
the 3'
ends to the targeting ligand and the 5' end of the second RNA strand is not
conjugated
or
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
160
a conjugate for inhibiting expression of a ALDH2 gene in a cell, said
conjugate comprising
a nucleic acid portion and ligand portions, said nucleic acid portion
comprising at least one
duplex region that comprises at least a portion of a first RNA strand and at
least a portion
of a second RNA strand that is at least partially complementary to the first
strand, wherein
said first strand is at least partially complementary to at least a portion of
RNA transcribed
from said ALDH2 gene, said ligand portions comprising a linker moiety and a
targeting
ligand for in vivo targeting of cells and being conjugated exclusively to the
3' and/or 5' ends
of one or both RNA strands, wherein the 5' end of the first RNA strand is not
conjugated,
wherein:
(i) the second RNA strand is conjugated at the 5' end to the targeting
ligand, and
wherein (a) the second RNA strand is also conjugated at the 3' end to the
targeting ligand and the 3' end of the first RNA strand is not conjugated; or
(b)
the first RNA strand is conjugated at the 3' end to the targeting ligand and
the 3'
end of the second RNA strand is not conjugated; or (c) both the second RNA
strand and the first RNA strand are also conjugated at the 3' ends to the
targeting
ligand; or
(ii) both the second RNA strand and the first RNA strand are conjugated at
the 3'
ends to the targeting ligand and the 5' end of the second RNA strand is not
conjugated, optionally
wherein the linker moiety is a serinol-derived linker moiety or one of the
other linker types
described herein.
2. The conjugate according to clause 1 wherein the second RNA strand is
conjugated
at the 5' end to the targeting ligand, the second RNA strand is also
conjugated at the 3' end
to the targeting ligand and the 3' end of the first RNA strand is not
conjugated.
3. The conjugate according to clause 1 wherein the second RNA strand is
conjugated
at the 5' end to the targeting ligand, the first RNA strand is conjugated at
the 3' end to the
targeting ligand and the 3' end of the second RNA strand is not conjugated.
4. The conjugate according to clause 1 wherein the second RNA strand is
conjugated
at the 5' end to the targeting ligand and both the second RNA strand and the
first RNA
strand are also conjugated at the 3' ends to the targeting ligand.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
161
5. The conjugate according to clause 1 wherein both the second RNA
strand and the
first RNA strand are conjugated at the 3' ends to the targeting ligand and the
5' end of the
second RNA strand is not conjugated.
6. The conjugate according to any one of clauses 1 to 5 wherein the ligands
are
monomeric ligands.
7. The conjugate according to any one of clauses 1 to 6 wherein the ligands
are
selected from GaINAc and galactose moieties, especially GaINAc moieties.
8. The conjugate according to any one of clauses 1 to 7 wherein the
conjugated RNA
strands are conjugated to a targeting ligand via a serinol-derived linker
moiety including a
further linker wherein the further linker is or comprises a saturated,
unbranched or branched
C1-16 alkyl chain, wherein optionally one or more carbons (for example 1, 2 or
3 carbons,
suitably 1 or 2, in particular 1) is/are replaced by a heteroatom selected
from 0, N, S(0)p
wherein p is 0, 1 or 2, (for example a CH2 group is replaced with 0, or with
NH, or with S,
or with SO2 or a ¨CH3 group at the terminus of the chain or on a branch is
replaced with OH
or with NH2) wherein said chain is optionally substituted by one or more oxo
groups (for
example 1 to 3, such as 1 group).
9. The conjugate according to clause 8 wherein the further linker comprises
a
saturated, unbranched C1_15 alkyl chain wherein one or more carbons (for
example 1, 2 or
3 carbons, suitably 1 or 2, in particular 1) is/are replaced by an oxygen
atom.
10. The conjugate according to clause 9 wherein the further linker
comprises a PEG-
chain.
11. The conjugate according to clause 8 wherein the further linker
comprises a
saturated, unbranched Ci_16 alkyl chain.
12. The conjugate according to clause 11 wherein the further linker
comprises a
saturated, unbranched C1_6 alkyl chain.
13. The conjugate according to clause 12 wherein the further linker
comprises a
saturated, unbranched C4 or C6 alkyl chain, e.g. a C4 alkyl chain.
CA 03118327 2021-04-30
WO 2019/092282
PCT/EP2018/081105
162
14. The conjugate according to clause 1 wherein the first RNA strand
is a compound of
formula (XV):
_
GaINAc GaINAc
µ \
L L
\ \
NH NH
Y
Zi¨O¨P-0 0 P 0 _______ 0 __ H
I I
OH R1 \ OH Ri /
n
b¨ ¨ (XV)
wherein b is 0 or 1; and
the second RNA strand is a compound of formula (XVI):
GaINAc GaINAc GaINAc GaINAc
/
/ \ \
L L L L
/ / \ \
HN HN NH NH
0
H 0 0
0 P 0 Z2 0 P 0 0 __
H0 P 0
I I I
n n
\ Ri OH i R1 OH OH R1 \ OH R1 i
c _ ¨ d ¨ ¨
(XVI);
wherein c and d are independently 0 or 1;
wherein:
Z1 and Z2 are the RNA portions of the first and second RNA strands
respectively;
Y is 0 or S;
R1 is H or methyl;
n is 0, 1,2 or 3; and
L is the same or different in formulae (XV) and (XVI) and is selected from the
group
consisting of:
-(CH2)q, wherein q = 2-12;
-(CH2),-C(0)-, wherein r = 2-12;
-(CH2-CH2-0)s-CH2-C(0)-, wherein s = 1-5;
-(CH2)1-CO-NH-(CH2)1-NH-C(0)-, wherein t is independently is 1-5;
-(CH2).-CO-NH-(CH2).-C(0)-, wherein u is independently is 1-5; and
-(CH2),-NH-C(0)-, wherein v is 2-12; and
wherein the terminal C(0) (if present) is attached to the NH group;
and wherein b + c + d is 2 or 3.
15. The conjugate according to clause 14 wherein b is 0, c is 1 and d is 1.
16. The conjugate according to clause 14 wherein b is 1, c is 0 and d
is 1.
CA 03118327 2021-04-30
WO 2019/092282 PCT/EP2018/081105
163
17. The conjugate according to clause 14 wherein b is 1, c is 1 and d is O.
18. The conjugate according to clause 14 wherein b is 1, c is 1 and d is 1.
19. The conjugate according to any one of clauses 14-18 wherein Y is 0.
20. The conjugate according to any one of clauses 14-18 wherein Y is S.
21. The conjugate according to any one of clauses 14-20 wherein R1 is H.
22. The conjugate according to any one of clauses 14-20 wherein R1 is
methyl.
23. The conjugate according to any one of clauses 14-22 wherein n is 0.
1.5 24. The conjugate according to any one of clauses 14-23 wherein L is
-(CH2)r-C(0)-,
wherein r = 2-12.
25. The conjugate according to clause 24 wherein r = 2-6.
26. The conjugate according to clause 25 wherein r = 4 or 6 e.g. 4.
27. The conjugate according to any preceding clause, with any feature or
combination
of features disclosed herein.
Summary SEQUENCE TABLE
0
t..)
o
SEQ ID Sequence (5'-3')
Unmodified Sequence counterpart (5'-3')
Name
O-
NO
t..)
t..)
oe
ALDH2-h-1 1st
t..)
1 strand UUGUUUAUGAAAAUCUGGU
UUGUUUAUGAAAAUCUGGU
ALDH2-h-1 2nd
2 ACCAGAUUUUCAUAAACAA
ACCAGAUUUUCAUAAACAA
strand
ALDH2-hcm-2
3 1st strand AAUGUUUUCCUGCUGACGG
AAUGUUUUCCUGCUGACGG
ALDH2-hcm-2
4 2nd strand CCGUCAGCAGGAAAACAUU
CCGUCAGCAGGAAAACAUU
ALDH2-hc-3 1st
P
UUGAACUUCAGGAUCUGCA UUGAACUUCAGGAUCUGCA .
strand
,
,
ALDH2-hc-3 2nd
6 UGCAGAUCCUGAAGUUCAA
UGCAGAUCCUGAAGUUCAA o ,"
strand
4. "
0
ALDH2-hc-4 1st
N),
7 UUUCACUUCAGUGUAUGCC
UUUCACUUCAGUGUAUGCC , c,
strand
.
,
ALDH2-hc-4 2nd
.
8 GGCAUACACUGAAGUGAAA
GGCAUACACUGAAGUGAAA
strand
ALDH2-hc-5 1st
9 UUCUUAUGAGUUCUUCUGA
UUCUUAUGAGUUCUUCUGA
strand
ALDH2-hc-5 2nd
UCAGAAGAACUCAUAAGAA UCAGAAGAACUCAUAAGAA
strand
ALDH2-hcm-6
11 1st strand UCUUCUUAAACUGAGUUUC
UCUUCUUAAACUGAGUUUC od
n
ALDH2-hcm-6
12 GAAACUCAGUUUAAGAAGA
GAAACUCAGUUUAAGAAGA t=1
2nd strand
od
t..)
ALDH2-hc-7 1st
13 UCCUUGAUCAGGUUGGCCA
UCCUUGAUCAGGUUGGCCA
oe
strand
O-
oe
ALDH2-hc-7 2nd
14 UGGCCAACCUGAUCAAGGA
UGGCCAACCUGAUCAAGGA
strand
o
u,
ALDH2-hc-8 1st
0
15 UUGAAGAACAGGGCGAAGU
UUGAAGAACAGGGCGAAGU t..)
strand
o
ALDH2-hc-8 2nd
,z
16 ACUUCGCCCUGUUCUUCAA
ACUUCGCCCUGUUCUUCAA O-
strand
,z
t..)
t..)
ALDH2-hcm-9
oe
17 AUGUAGCCGAGGAUCUUCU
AUGUAGCCGAGGAUCUUCU t..)
1st strand
ALDH2-hcm-9
18 2nd strand AGAAGAUCCUCGGCUACAU
AGAAGAUCCUCGGCUACAU
ALDH2-hmr-10
19 AUCUGGUUGCAGAAGACCU
AUCUGGUUGCAGAAGACCU
1st strand
ALDH2-hmr-10
20 AGGUCUUCUGCAACCAGAU
AGGUCUUCUGCAACCAGAU
2nd strand
ALDH2-hr-11 1st
21 AAAUCCACCAGGUAGGAGA
AAAUCCACCAGGUAGGAGA P
strand
-
,
ALDH2-hr-11 2nd
,
.3
22 UCUCCUACCUGGUGGAUUU
UCUCCUACCUGGUGGAUUU
strand
"
u,
ALDH2-hm-12
N)-
23 ACUCUCUUGAGGUUGCUGC
ACUCUCUUGAGGUUGCUGC
1st strand
,I,
ALDH2-hm-12
24 GCAGCAACCUCAAGAGAGU
GCAGCAACCUCAAGAGAGU 0
2nd strand
ALDH2-hcmr-13
25 AUGUCCAAAUCCACCAGGU
AUGUCCAAAUCCACCAGGU
1st strand
ALDH2-hcmr-13
26 ACCUGGUGGAUUUGGACAU
ACCUGGUGGAUUUGGACAU
2nd strand
ALDH2-hm-14
27 CCAUGCUUGCAUCAGGAGC
CCAUGCUUGCAUCAGGAGC
1st strand
od
ALDH2-hm-14
n
28 GCUCCUGAUGCAAGCAUGG
GCUCCUGAUGCAAGCAUGG
2nd strand
m
od
ALDH2 (upper)
t..)
o
29 Human, GGCAAGCCCTATGTCATCTCCT
GGCAAGCCCTATGTCATCTCCT
cio
O-
Cynomolgus
oe
o
vi
ALDH2 (lower)
0
t..)
30 Human, GGATGGTTTTCCCGTGGTACTT
GGATGGTTTTCCCGTGGTACTT o
Cynomolgus
O-
ALDH2 (probe)
t..)
t..)
31 Human, TGGTCCTCAAATGTCTCCGGTATTATGCC
TGGTCCTCAAATGTCTCCGGTATTATGCC cio
t..)
Cynomolgus
32 ALDH2 (upper)
GGCAAGCCTTATGTCATCTCGT
GGCAAGCCTTATGTCATCTCGT
Mouse
33 ALDH2 (lower)
GGAATGGTTTTCCCATGGTACTT
GGAATGGTTTTCCCATGGTACTT
Mouse
34 ALDH2 (probe)
TGAAATGTCTCCGCTATTACGCTGGCTG
TGAAATGTCTCCGCTATTACGCTGGCTG
Mouse
ApoB (upper)
35 T
P TCATTCCTTCCCCAAAGAGACC CATTCCTTCCCCAAAGAGACC
Human
,
,
36
ApoB (lower) CACCTCCGTTTTGGTGGTAGAG
CACCTCCGTTTTGGTGGTAGAG .3
"
Human
o, ,,
.
ApoB (probe)
CAAGCTGCTCAGTGGAGGCAACACATTA " 37
CAAGCTGCTCAGTGGAGGCAACACATTA ,
, Human
.
'
38
ApoB (upper) TCATTCCTTCCCCAAAGAAACC
TCATTCCTTCCCCAAAGAAACC
0
Cynomolgus
ApoB (lower)
39 CACCTCCGTTTTGGTGGTAGAGCACCTCCGTTTTGGTGGTAGAG
Cynomolgus
ApoB (probe) TCAAGCTGTTAAGTGGCAGCAACACGTT
TCAAGCTGTTAAGTGGCAGCAACACGTT
Cynomolgus
41
beta-Actin GCATGGGTCAGAAGGATTCCTAT
GCATGGGTCAGAAGGATTCCTAT
(upper) Human
oo
n
42 beta-Actin (lower)
TGTAGAAGGTGTGGTGCCAGATT
TGTAGAAGGTGTGGTGCCAGATT
t=1 Human
oo
t..)
43
beta-Actin TCGAGCACGGCATCGTCACCAA
TCGAGCACGGCATCGTCACCAA cio (probe) Human
O-
oe
o
u,
beta-Actin
0
44 (upper) AAGGCCAACCGCGAGAAG
AAGGCCAACCGCGAGAAG t..)
o
Cynomolgus
O-
45 beta-Actin (lower)
AGAGGCGTACAGGGACAGCA
AGAGGCGTACAGGGACAGCA t..)
t..)
Cynomolgus
oe
t..)
beta-Actin
46 (probe) TGAGACCTTCAACACCCCAGCCATGTAC
TGAGACCTTCAACACCCCAGCCATGTAC
Cynomolgus
beta-Actin
47 CCTAAGGCCAACCGTGAAAAG
CCTAAGGCCAACCGTGAAAAG
(upper) Mouse
48 beta-Actin (lower)
AGGCATACAGGGACAGCACAG
AGGCATACAGGGACAGCACAG
Mouse
beta-Actin P 49
TGAGACCTTCAACACCCCAGCCATGTAC TGAGACCTTCAACACCCCAGCCATGTAC
(probe) Mouse
.
,
,
50 PTEN (upper)
CACCGCCAAATTTAACTGCAGA
CACCGCCAAATTTAACTGCAGA
Human
o, ,
-4
,,
PTEN (lower)
.
51 AAGGGTTTGATAAGTTCTAGCTGT
AAGGGTTTGATAAGTTCTAGCTGT
N) ,
, Human
0
' TGCACAGTATCCTTTTGAAGACCATAACCC
52 PTEN (probe) TGCACAGTATCCTTTTGAAGACCATAACCCA
A
53 ApoB fw Mouse AAAGAGGCCAGTCAAGCTGTTC
AAAGAGGCCAGTCAAGCTGTTC
54 ApoB rev Mouse GGTGGGATCACTTCTGTTTTGG
GGTGGGATCACTTCTGTTTTGG
ALDH2 siRNA
AUGUAGCCGAGGAUCUUCUUAAACUGAG
55 AUGUAGCCGAGGAUCUUCUUAAACUGAGUUUC
binding locus
UUUC
Modified SEQ ID
56 NO: 1 5181516182626171845
UUGUUUAUGAAAAUCUGGU oo
n
Modified SEQ ID
57 NO: 2 2736461515361626362
ACCAGAUUUUCAUAAACAA m
oo
t..)
Modified SEQ ID
58 NO: 3 6254515173547182748
AAUGUUUUCCUGCUGACGG oe
O-
oe
Modified SEQ ID
59 NO: 4 3745364728462627251
CCGUCAGCAGGAAAACAUU
u,
Modified SEQ ID
0
60 NO: 5 5182635172846171836
UUGAACUUCAGGAUCUGCA t..)
o
,-,
Modified SEQ ID
61 NO: 6 1836461735462815362
UGCAGAUCCUGAAGUUCAA O-
t..)
t..)
Modified SEQ ID
oe
62 NO: 7 5153635172818161837
UUUCACUUCAGUGUAUGCC t..)
Modified SEQ ID
63 NO: 8 4836163635462818262
GGCAUACACUGAAGUGAAA
Modified SEQ ID
64 NO: 9 5171525464517153546
UUCUUAUGAGUUCUUCUGA
Modified SEQ ID
65 NO: 10 1728264627172526462
UCAGAAGAACUCAUAAGAA
Modified SEQ ID
66 5351715262718281517
UCUUCUUAAACUGAGUUUC P
NO: 11
0
,
Modified SEQ ID
,
.3
67 NO: 12 4626353645152646282
GAAACUCAGUUUAAGAAGA ,-,
oe
Modified SEQ ID
"
-
68 5371546172845184736
UCCUUGAUCAGGUUGGCCA
NO: 13
,I,
Modified SEQ ID
69 NO: 14 1847362735461726482
UGGCCAACCUGAUCAAGGA
Modified SEQ ID
70 NO: 15 5182646272848382645
UUGAAGAACAGGGCGAAGU
Modified SEQ ID
71 NO: 16 2715383735451715362
ACUUCGCCCUGUUCUUCAA
Modified SEQ ID
72 NO: 17 6181647382846171535
AUGUAGCCGAGGAUCUUCU
od
Modified SEQ ID
n
73 NO: 18 2826461735384716361
AGAAGAUCCUCGGCUACAU
m
od
Modified SEQ ID
t..)
74 NO: 19 6171845183646282735
AUCUGGUUGCAGAAGACCU o
,-,
cio
Modified
O-
SEQ ID
oe
75 NO: 20 2845351718362736461
AGGUCUUCUGCAACCAGAU
,-,
o
u,
Modified SEQ ID
0
76 6261736372845284646
AAAUCCACCAGGUAGGAGA t..)
NO: 21
o
,-,
Modified SEQ ID
77 1717352735481846151
UCUCCUACCUGGUGGAUUU NO: 22
t..)
t..)
Modified SEQ ID
cio
78 6353535182845183547
ACUCUCUUGAGGUUGCUGC t..)
NO: 23
Modified SEQ ID
79 4728362735362828281
GCAGCAACCUCAAGAGAGU
NO: 24
Modified SEQ ID
80 6181736261736372845
AUGUCCAAAUCCACCAGGU
NO: 25
Modified SEQ ID
81 2735481846151846361
ACCUGGUGGAUUUGGACAU
NO: 26
Modified SEQ ID
82 7361835183617284647
CCAUGCUUGCAUCAGGAGC P
NO: 27
o
,
Modified SEQ ID
,
.3
83 4717354618362836184
GCUCCUGAUGCAAGCAUGG ,-,
" NO: 28
o, ,
,,
GaINAc-ALDH2- OMeA(ps)-FA-(ps)-0MeU-FG-0MeU-FU-OMeU-FU-OMeC-
-
,,
,
' 84 hcm-2-L1 1st FC-0MeU-FG-0MeC-FU-OMeG-FA-OMeC-(ps)-FG-(ps)-
AAUGUUUUCCUGCUGACGG .
' strand
OMeG
.
GaINAc-ALDH2- [5T23 (ps)]3 long trebler (ps) FC-0MeC-FG-0MeU-FC-OMeA-
85 hcm-2-L1 2nd FG-0MeC-FA-OMeG-FG-OMeA-FA-OMeA-FA-OMeC-FA-(ps)-
CCGUCAGCAGGAAAACAUU
strand OMeU-(ps)-FU
GaINAc-ALDH2- OMeA(ps)- FA-(ps)-0MeU-FG-0MeU-FU-OMeU-FU-OMeC-
86 hcm-2-L6 1st FC-0MeU-FG-0MeC-FU-OMeG-FA-OMeC-(ps)-FG-(ps)-
AAUGUUUUCCUGCUGACGG
strand OMeG
GaINAc-ALDH2- [5T23 (ps)]3 5T43 (ps) FC-0MeC-FG-0MeU-FC-OMeA-FG-
oo
87 hcm-2-L6 2nd OMeC-FA-OMeG-FG-0MeA-FA-0MeA-FA-0MeC-FA-(ps)-
CCGUCAGCAGGAAAACAUU n
1-i
strand OMeU-(ps)-FU
m
oo
GaINAc-ALDH2- OMeU-(ps)-FC-(ps)-0MeU-FU-OMeC-FU-OMeU-FA-OMeA-
t..)
o
88 hcm6-L1 1st FA-0MeC-FU-OMeG-FA-OMeG-FU-OMeU-(ps)-FU-(ps)-
UCUUCUUAAACUGAGUUUC
oe
strand OMeC
O-
oo
,-,
,-,
o
u,
GaINAc-ALDH2- [ST23 (ps)]3 long trebler (ps) FG-0MeA-FA-0MeA-FC-OMeU-
0
89 hcm6-L1 2nd FC-0MeA-FG-OMeU-FU-OMeU-FA-OMeA-FG-OMeA-FA-(ps)-
GAAACUCAGUUUAAGAAGA t..)
o
,-,
strand OMeG-(ps)-FA
O-
GaINAc-ALDH2- OMeU-(ps)-FC-(ps)-0MeU-FU-OMeC-FU-OMeU-FA-OMeA-
t..)
90 hcm6-L6 1st FA-0MeC-FU-OMeG-FA-OMeG-FU-OMeU-(ps)-FU-(ps)-
UCUUCUUAAACUGAGUUUC t..)
oe
t..)
strand OMeC
GaINAc-ALDH2- [ST23 (ps)]3 ST43 (ps) FG-0MeA-FA-0MeA-FC-OMeU-FC-
91 hcm6-L6 2nd OMeA-FG-OMeU-FU-OMeU-FA-0MeA-FG-0MeA-FA-(ps)-
GAAACUCAGUUUAAGAAGA
strand OMeG-(ps)-FA
GaINAc-ALDH2- OMeA-(ps)-FU-(ps)-0MeG-FU-OMeA-FG-OMeC-FC-OMeG-
92 hcm9-L1 1st FA-0MeG-FG-0MeA-FU-OMeC-FU-OMeU-(ps)-FC-(ps)-
AUGUAGCCGAGGAUCUUCU
strand OMeU
GaINAc-ALDH2- [ST23 (ps)]3 long trebler (ps) FA-0MeG-FA-0MeA-FG-OMeA-
P
93 hcm9-L1 2nd FU-OMeC-FC-OMeU-FC-OMeG-FG-OMeC-FU-OMeA-FC-
AGAAGAUCCUCGGCUACAU .
strand (ps)-0MeA-(ps)-FU
,
,
.3
GaINAc-ALDH2- OMeA-(ps)-FU-(ps)-0MeG-FU-OMeA-FG-OMeC-FC-OMeG-
o
94 hcm9-L6 1st FA-0MeG-FG-0MeA-FU-OMeC-FU-OMeU-(ps)-FC-(ps)-
AUGUAGCCGAGGAUCUUCU 0"
IV
strand OMeU
,
,
GaINAc-ALDH2- [ST23 (ps)]3 ST43 (ps) FA-0MeG-FA-0MeA-FG-OMeA-FU- ' 95
hcm9-L6 2nd OMeC-FC-OMeU-FC-OMeG-FG-OMeC-FU-OMeA-FC-(ps)-
AGAAGAUCCUCGGCUACAU
strand OMeA-(ps)-FU
GaINAc-ALDH2- OMeA-(ps)-FU-(ps)-0MeG-FU-OMeA-FG-OMeC-FC-OMeG-
96 hcm9-L4 1st FA-0MeG-FG-0MeA-FU-OMeC-FU-OMeU-(ps)-FC-(ps)-
AUGUAGCCGAGGAUCUUCU
strand OMeU
GaINAc-ALDH2- [ST23 (ps)]3 ST41 (ps) FA-0MeG-FA-0MeA-FG-OMeA-FU-
97 hcm9-L4 2nd OMeC-FC-OMeU-FC-OMeG-FG-OMeC-FU-OMeA-FC-(ps)-
AGAAGAUCCUCGGCUACAU oo
strand OMeA-(ps)-FU
n
1-i
mA(ps)fA(ps)mUfGmUfUmUfUmCfCmUfGmCfUmGfAmC(ps)f
m
98 ALD01 1st strand
AAUGUUUUCCUGCUGACGG oo
G(ps)mG
t..)
o
ALD01 2nd fC(ps)mC(ps)fGmUfCmAfGmCfAmGfGmAfAmAfAmCfA(ps)mU
,-,
99
CCGUCAGCAGGAAAACAUU O-
strand (ps)fU
.
,-,
,-,
o
u,
mA(ps)fA(ps)mUfGmUfUmUfUmCfCmUfGmCfUmGfAmCfGmG
0
100 ALD02 1st strand
AAUGUUUUCCUGCUGACGGA t..)
ivA
o
,-,
ALD02 2nd fC(ps)mC(ps)fGmUfCmAfGmCfAmGfGmAfAmAfAmCfAmUfU
101 CCGUCAGCAGGAAAACAUUA strand
ivA
ivA
t..)
t..)
mA(ps)fA(ps)mUfGmUfUmUfUmCfCmUfGmCfUmGfAmCfGmG
oe
102 ALD03 1st strand
AAUGUUUUCCUGCUGACGGU t..)
ivU
ALD03 2nd fC(ps)mC(ps)fGmUfCmAfGmCfAmGfGmAfAmAfAmCfAmUfU
103
CCGUCAGCAGGAAAACAUUU
strand ivU
mA(ps)fA(ps)mUfGmUfUmUfUmCfCmUfGmCfUmGfAmCfGmG
104 ALD04 1st strand
AAUGUUUUCCUGCUGACGGC
ivC
ALD04 2nd fC(ps)mC(ps)fGmUfCmAfGmCfAmGfGmAfAmAfAmCfAmUfU
105
CCGUCAGCAGGAAAACAUUC
strand ivC
mA(ps)fA(ps)mUfGmUfUmUfUmCfCmUfGmCfUmGfAmCfGmG
106 ALD05 1st strand
AAUGUUUUCCUGCUGACGGG P
ivG
.
,
ALD05 2nd fC(ps)mC(ps)fGmUfCmAfGmCfAmGfGmAfAmAfAmCfAmUfU
,
0
107
CCGUCAGCAGGAAAACAUUG ,-,
strand ivG
,-,
mA(ps)fA(ps)mUfGmUfUmUfUmCfCmUfGmCfUmGfAmCfG
IV
0
108 ALD06 1st strand
AAUGUUUUCCUGCUGACGA
ivA
0'
ALD06 2nd
109 fC(ps)mC(ps)fGmUfCmAfGmCfAmGfGmAfAmAfAmCfAmU ivA
CCGUCAGCAGGAAAACAUA 0
strand
mA(ps)fA(ps)mUfGmUfUmUfUmCfCmUfGmCfUmGfAmCfG
110 ALD07 1st strand
AAUGUUUUCCUGCUGACGU
ivU
ALD07 2nd
111 fC(ps)mC(ps)fGmUfCmAfGmCfAmGfGmAfAmAfAmCfAmU ivU
CCGUCAGCAGGAAAACAUU
strand
mA(ps)fA(ps)mUfGmUfUmUfUmCfCmUfGmCfUmGfAmCfG
112 ALD08 1st strand
AAUGUUUUCCUGCUGACGC
ivC
oo
ALD08 2nd
n
1-i
113 fC(ps)mC(ps)fGmUfCmAfGmCfAmGfGmAfAmAfAmCfAmU ivC
CCGUCAGCAGGAAAACAUC
strand
m
oo
mA(ps)fA(ps)mUfGmUfUmUfUmCfCmUfGmCfUmGfAmCfG
t..)
114 ALD09 1st strand
AAUGUUUUCCUGCUGACGG o
,-,
ivG
oe
O-
ALD09 2nd
oe
115 fC(ps)mC(ps)fGmUfCmAfGmCfAmGfGmAfAmAfAmCfAmU ivG
CCGUCAGCAGGAAAACAUG
strand
o
u,
0
116
STS22002L6 1st mA(ps)fA(ps)mUfGmUfUmUfUmCfCmUfGmCfUmGfAmC(ps)f
AAUGUUUUCCUGCUGACGG
t..)
o strand
G(ps)mG
,o
ST23(ps)3 ST
O-
5T522002L6 2nd
,o
117 43(ps)fCmCfGmUfCmAfGmCfAmGfGmAfAmAfAmCfA(ps)mU(
CCGUCAGCAGGAAAACAUU t..)
t..)
strand
oo
ps)fU
t..)
118 STS22002V1L6 mA(ps)fA(ps)mUfGmUfUmUfUmCfCmUfGmCfUmGfAmC(ps)f
AAUGUUUUCCUGCUGACGG
1st strand G(ps)mG
119 STS22002V1L6 5T23(ps)3 ST
CCGUCAGCAGGAAAACAUA
2nd strand 43(ps)fCmCfGmUfCmAfGmCfAmGfGmAfAmAfAmCfAmUivA
120 5T522002V2L6 mA(ps)fA(ps)mUfGmUfUmUfUmCfCmUfGmCfUmGfAmC(ps)f
AAUGUUUUCCUGCUGACGG
1st strand G(ps)mG
121 5T522002V2L6 5T23(ps)3 ST
CCGUCAGCAGGAAAACAUG
2nd strand 43(ps)fCmCfGmUfCmAfGmCfAmGfGmAfAmAfAmCfAmUivG
P
5T522002V3L6 mA(ps)fA(ps)mUfGmUfUmUfUmCfCmUfGmCfUmGfAmCfGmG
AAUGUUUUCCUGCUGACGGA
.
122
,
,
1st strand ivA
a'
,-,
" 5T23(ps)3 ST
-4 ,
t..)
STS22002V3L6
,,
123 43(ps)fCmCfGmUfCmAfGmCfAmGfGmAfAmAfAmCfA(ps)mU(
CCGUCAGCAGGAAAACAUU
" , 2nd strand
,
ps)fU
.
'
124 5T522002V4L6 mA(ps)fA(ps)mUfGmUfUmUfUmCfCmUfGmCfUmGfAmCfGmG
AAUGUUUUCCUGCUGACGGG
',;
1st strand ivG
S
5T522002V4L6 T23(ps)3 ST
125 2nd strand 43(ps)fCmCfGmUfCmAfGmCfAmGfGmAfAmAfAmCfA(ps)mU(
CCGUCAGCAGGAAAACAUU
ps)fU
AA 126
5T522006L6 1st mA(ps)fA(ps)mUfGmUfUmUfUmCfCmUfGmCfUmGfAmC(ps)f
UGUUUUCCUGCUGACGG
strand G(ps)mG
5T23(ps)3 ST
oo
5T522006 L6 2nd
n
127 43(ps)fCmCfGmUfCmAfGmCfAmGfGmAfAmAfAmCfA(ps)mU(
CCGUCAGCAGGAAAACAUU
strand
m ps)fU
oo
t..)
128
STS22006V1L6 mA(ps)fA(ps)mUfGmUfUmUfUmCfCmUfGmCfUmGfAmC(ps)f
AAUGUUUUCCUGCUGACGG
,-,
oe 1st strand
G(ps)mG
129
O-
STS22006V1L6 5T23(ps)3 ST
CCGUCAGCAGGAAAACAUA
oe
,-,
,-,
2nd strand 43(ps)fCmCfGmUfCmAfGmCfAmGfGmAfAmAfAmCfAmUivA
o
u,
130 STS22009L6 1st mA(ps)fA(ps)mUfGmUfUmUfUmCfCmUfGmCfUmGfAmC(ps)f
AAUGUUUUCCUGCUGACGG
o strand G(ps)mG
131 STS22009L6 2nd ST23(ps)3 ST
CCGUCAGCAGGAAAACAUG
O-
strand 43(ps)fCmCfGmUfCmAfGmCfAmGfGmAfAmAfAmCfAmUivG
t..)
t..)
oe
132 STS22009V1L6 mA(ps)fA(ps)mUfGmUfUmUfUmCfCmUfGmCfUmGfAmCfGmG
AAUGUUUUCCUGCUGACGGA
t..)
1st strand ivA
5T23(ps)3 ST
133 5T522009V1L6
43(ps)fCmCfGmUfCmAfGmCfAmGfGmAfAmAfAmCfA(ps)mU( CCGUCAGCAGGAAAACAUU
2nd strand
ps)fU
AA 134 5T522009V2L6 mA(ps)fA(ps)mUfGmUfUmUfUmCfCmUfGmCfUmGfAmCfGmG
UGUUUUCCUGCUGACGGG
1st strand ivG
5T23(ps)3 ST
135 5T522009V2L6
43(ps)fCmCfGmUfCmAfGmCfAmGfGmAfAmAfAmCfA(ps)mU( CCGUCAGCAGGAAAACAUU
p
2nd strand
ps)fU
-
,
,
136 5T516001AL33 mU (ps) fU (ps) mA fU mA fG mA fG mC fA mA fG mA fA mC
,-, UUAUAGAGCAAGAACACUGUU
.3
fA mC fU mG (ps) fU (ps) mU (ps) Ser(GN)
"
(...)
,,
.
137 STS16001BL20 Ser(GN) (ps) fA mA fC mA fG mU fG mU fU mC fU mU fG mC
AACAGUGUUCUUGCUCUAUAA
N),
,
fU mC fU mA fU (ps) mA (ps) fA
.
' mU (ps) fU (ps) mA fU mA fG mA fG mC fA mA fG mA fA mC
UUAUAGAGCAAGAACACUGUU
0
138 STS16001A
fA mC fU mG (ps) fU (ps) mU
139 STS16001BV1L4 Ser(GN) (ps) fA (ps) mA (ps) fC mA fG mU fG mU fU mC fU
AACAGUGUUCUUGCUCUAUAA
2 mU fG mC fU mC fU mA fU (ps) mA (ps) fA (ps) Ser(GN)
fA (ps) mA (ps) fC mA fG mU fG mU fU mC fU mU fG mC fU
AACAGUGUUCUUGCUCUAUAA
140 STS16001V1B
mC fU mA fU (ps) mA (ps) fA
141 STS18001A mU (ps) fC (ps) mG fA mA fG mU fA mU fU mC fC mG fC mG
UCGAAGUAUUCCGCGUACG
fU mA (ps) fC (ps) mG
oo
n
[(5T23) (ps)]3 C4XLT (ps) fC mG fU mA fC mG fC mG fG mA
CGUACGCGGAAUACUUCGA
142 STS18001BL4
fA mU fA mC fU mU fC (ps) mG (ps) fA
m
oo
t..)
143 STS16001BL4 [(5T23) (ps)]3 C4XLT(ps) fA (ps) mA (ps) fC mA fG mU
fG mU
AACAGUGUUCUUGCUCUAUAA
'
,-,
fU mC fU mU fG mC fU mC fU mA fU (ps) mA (ps) fA
cio
O-
cio mU (ps) fC (ps) mG fA mA fG mU fA mU fU mC fC mG fC mG
UCGAAGUAUUCCGCGUACG
144 X0028A
fU mA (ps) fC (ps) mG
o
u,
[ST23(ps)]3 ST41(ps) fC mG fU mA fC mG fC mG fG mA fA
0
145 X0028B
CGUACGCGGAAUACUUCGA
mU fA mC fU mU fC (ps) mG (ps) fA
t..)
o
,-,
mU (ps) fC (ps) mU fU mC fU mU fA mA fA mC fU mG fA mG
146 X0320A
UCUUCUUAAACUGAGUUUC fU mU mU (ps) fU (ps) mC
t..)
Ser(GN) (ps) fG (ps) mA (ps) fA mA fC mU fC mA fG mU fU
t..)
cio
147 X0320B
GAAACUCAGUUUAAGAAGA t..)
mU fA mA fG mA fA (ps) mG (ps) fA (ps) Ser(GN)
mU (ps) fC (ps) mU fU mC fU mU fA mA fA mC fU mG fA mG
148 X0477A
UCUUCUUAAACUGAGUUUC
fU mU (ps) fU (ps) mC
149 X0477B Ser(GN) (ps) mG (ps) mA (ps) mA mA mC mU fC fA fG mU
mU
GAAACUCAGUUUAAGAAGA
mU mA mA mG mA mA (ps) mG (ps) mA (ps) Ser(GN)
ALDH2 probe
150 based on SEQ ID BHQ1-TGAAATGTCTCCGCTATTACGCTGGCTG-FAM
TGAAATGTCTCCGCTATTACGCTGGCTGA
NO: 34
P
ApoB probe
.
151 BHQ1-CAGCAACACACTGCATCTGGTCTCTACCA-VIC
CAGCAACACACTGCATCTGGTCTCTACCA
,
Mouse
,
.3
mA (ps) fA (ps) mU fG mU fU mU fU mC fC mU fG mC fU mG
,-,
"
152 X0319A
AAUGUUUUCCUGCUGACGG
fA mC (ps) fG (ps) mG
"
,,
Ser(GN) (ps) fC (ps) mC (ps) fG mU fC mA fG mC fA mG fG
,
'
153 X0319B
CCGUCAGCAGGAAAACAUU .
' mA fA mA fA mC fA (ps) mU (ps) fU (ps) Ser(GN)
.
(vp)-mU fA mU fG mU fU mU fU mC fC mU fG mC fU mG fA
154 X0362A
UAUGUUUUCCUGCUGACGG
mC (ps) fG (ps) mG
Ser(GN) (ps) fC (ps) mC (ps) fG mU fC mA fG mC fA mG fG
155 X0362B
CCGUCAGCAGGAAAACAUA
mA fA mA fA mC fA (ps) mU (ps) fA (ps) Ser(GN)
mU (ps) fC (ps) mU fU mC fU mU fA mA fA mC fU mG fA mG
156 X0320A
UCUUCUUAAACUGAGUUUC
fU mU (ps) fU (ps) mC
(vp)-mU fC mU fU mC fU mU fA mA fA mC fU mG fA mG fU
oo
157 X0363A
UCUUCUUAAACUGAGUUUC
mU (ps) fU (ps) mC
n
1-i
Ser(GN) (ps) mG (ps) mA (ps) mA mA mC mU fC fA fG mU mU
t=1
158 X0477B
GAAACUCAGUUUAAGAAGA oo
mU mA mA mG mA mA (ps) mG (ps) mA (ps) Ser(GN)
t..)
o
mA (ps) fU (ps) mG fU mA fG mC fC mG fA mG fG mA fU mC
,-,
oe
159 X0025A
AUGUAGCCGAGGAUCUUCU
fU mU (ps) fC (ps) mU
O-
oe
,-,
,-,
o
u,
[ST23 (ps)]3 ltrb (ps) fA mG fA mA fG mA fU mC fC mU fC mG
0
160 X0025B
AGAAGAUCCUCGGCUACAU t..)
fG mC fU mA fC (ps) mA (ps) fU
o
,-.
mA (ps) fU (ps) mG fU mA fG mC fC mG fA mG fG mA fU mC
161 X0321A
AUGUAGCCGAGGAUCUUCU fU mU mU (ps) fC (ps) mU
t..)
t..)
Ser(GN) (ps) fA (ps) mG (ps) fA mA fG mA fU mC fC mU fC
tc!.
162 X0321B
AGAAGAUCCUCGGCUACAU
mG fG mC fU mA fC (ps) mA (ps) fU (ps) Ser(GN)
STS16001BV1L7 Ser(GN) fA (ps) mA (ps) fC mA fG mU fG mU fU mC fU mU fG
163
AACAGUGUUCUUGCUCUAUAA
mC fU mC fU mA fU (ps) mA (ps) fA Ser(GN)
STS16001BV16L Ser(GN) (ps) fA mA fC mA fG mU fG mU fU mC fU mU fG mC
164
AACAGUGUUCUUGCUCUAUAA
42 fU mC fU mA fU mA fA (ps) Ser(GN)
STS16001BV2OL Ser(GN) fA mA fC mA fG mU fG mU fU mC fU mU fG mC fU
165
AACAGUGUUCUUGCUCUAUAA
75 mC fU mA fU mA fA Ser(GN)
Ser(GN) (ps) Ser(GN) (ps) fA (ps) mA (ps) fC mA fG mU fG mU
STS16001BV1L9
P
166 fU mC fU mU fG mC fU mC fU mA fU (ps) mA (ps) fA (ps)
AACAGUGUUCUUGCUCUAUAA 2
4
,
Ser(GN) (ps) Ser(GN)
,
.3
,-.
STS16001V1BL9 C6Am(GN) (ps) fA (ps) mA (ps) fC mA fG mU fG mU fU mC fU
167
AACAGUGUUCUUGCUCUAUAA
6 mU fG mC fU mC fU mA fU (ps) mA (ps) fA (ps) C7Am(GN)
.
,,
,
GlyC3Am(GN) (ps) fA (ps) mA (ps) fC mA fG mU fG mU fU mC
,
0
STS16001V1BL9
.
168 fU mU fG mC fU mC fU mA fU (ps) mA (ps) fA (ps)
AACAGUGUUCUUGCUCUAUAA
7 .
GlyC3Am(GN)
Conjugate 10 PipAm(GN) (ps) fA (ps) mA (ps) fC mA fG mU fG mU fU mC
fU
169
AACAGUGUUCUUGCUCUAUAA
sense strand mU fG mC fU mC fU mA fU (ps) mA (ps) fA (ps) PipAm(GN)
STS16001V1BL8 C3Am(GN) (ps) fA (ps) mA (ps) fC mA fG mU fG mU fU mC fU
170
AACAGUGUUCUUGCUCUAUAA
8 mU fG mC fU mC fU mA fU (ps) mA (ps) fA (ps) C3Am(GN)
STS16001V1BL8 C6Am(GN) (ps) fA (ps) mA (ps) fC mA fG mU fG mU fU mC fU
171
AACAGUGUUCUUGCUCUAUAA
7 mU fG mC fU mC fU mA fU (ps) mA (ps) fA (ps)
GlyC3Am(GN) oo
Conjugate 18 mA (ps) fA (ps) mC fC mA fG mA fA mG fA mA fG mC fA mG
n
172
AACCAGAAGAAGCAGGUGA
antisense strand fG mU (ps) fG (ps) mA
m
oo
Conjugate 18 Ser(GN) (ps) fU (ps) mC (ps) fA mC fC mU fG mC fU mU
fC t..)
173
UCACCUGCUUCUUCUGGUU '
sense strand mU fU mC fU mG fG (ps) mU (ps) fU (ps) Ser(GN)
re
-a
Conjugate 19 mA (ps) fU (ps) mA fA mC fU mC fU mG fU mC fC mA fU mU
oe
174
AUAACUCUGUCCAUUACCG
antisense strand fA mC (ps) fC (ps) mG
o
u,
Conjugate 19 Ser(GN) (ps) fC (ps) mG (ps) fG mU fA mA fU mG fG mA
fC 0
175
CGGUAAUGGACAGAGUUAU t..)
sense strand mA fG mA fG mU fU (ps) mA (ps) fU (ps) Ser(GN)
o
,-,
Reference
mC (ps) fU (ps) mU fA mC fU mC fU mC fG mC fC mC fA mA
O-
176 Conjugate 5
CUUACUCUCGCCCAAGCGA
fG mC (ps) fG (ps) mA
t..)
t..)
antisense
oe
t..)
Reference
[(ST23) (ps)]3 (C6XLT) (ps) fU mC fG mC fU mU fG mG fG mC
177 Conjugate 5
UCGCUUGGGCGAGAGUAAG
fG mA fG mA fG mU fA (ps) mA (ps) fG
sense
Reference
[ST23 (ps)]3 ltrb (ps) fG mA fA mA fC mU fC mA fG mU fU mU
178 Conjugate 6
GAAACUCAGU U UAAGAAGA
fA mA fG mA fA (ps) mG (ps) fA
sense
Reference
mU (ps) fA (ps) mC fC mA fG mA fA mG fA mA fG mC fA mG
179 Conjugate 8
UACCAGAAGAAGCAGGUGA
fG mU (ps) fG (ps) mA
P
antisense
.
,
Reference
,
[ST23 (ps)]3 ST41 (ps)fU mC fA mC fC mU fG mC fU mU fC
,-,
180 Conjugate 8
UCACCUGCUUCUUCUGGUA
mU fU mC fU mG fG (ps) mU (ps) fA
o, IV
sense
.
IV
F'
Reference
,
[ST23 (ps)]3 C6XLT (ps) fC mG fG mU fA mA fU mG fG mA fC
,?.
181 Conjugate 9
CGGUAAUGGACAGAGUUAU ' mA fG mA fG mU fU (ps) mA (ps)
fU -
sense
Key
1 = 2'F-dU
2 = 2'F-dA
oo
3 = 2'F-dC
n
1-i
4 = 2'F-dG
m
oo
t..)
o
= 2'0Me-rU
oe
O-
6 = 2'0Me-rA
oe
,-,
,-,
o
u,
7 = 2'0Me-rC
0
t..)
o
8 = 2'0Me-rG
,¨
,o
O-
,o
t..)
t..)
mA, mU, mC, mG, OMeA, OMeU, OMeC, OMeG ¨ 2`-0Me RNA
oe
t..)
fA, fU, fC, fG, FA, FU, FG, FC ¨ 2'deoxy-2'-F RNA
(ps) ¨ phosphorothioate
(vp) - Vinyl-(E)-phosphonate
ivA, ivC, ivU, ivG - inverted RNA (3'-3')
P
.
w
,
,
A single sequence may have more than one name. In those cases, one of those
names is given in the summary sequence table. .3
I,
L'
IV
,j
IV
0
IV
F'
I
Where specific linkers and or modified linkages are taught within an RNA
sequence, such as PS and [ST23 (ps)]3 ST41 (ps) etc, these are optional
,
parts of the sequence, but are a preferred embodiment of that sequence.
The following abbreviations may be used:
,
_
FAM 6-Carboxyfluorescein
BHQ Black Hole Quencher 1
Iv
n
ST23 oi._ ( _Ac OAc
1-3
M
IV
n.)
Ac0 ---400,.-
o
NHAc
oe
oe
1-.
1-.
o
vi
ST41/C4XLT ''-oo
0
t..)
..........õ..¨..._õ..¨...._o-,--
o
---.00
-a-,
t..,
t..)
ST43-phos/C6XLT DMTr
oo
0 0
N
DMTrN
0 0 001:1.0,CN
DMTrN
0 0
Long trebler/Itrb/STKS (phosphoramidite)
ODMT
IDCDOCOODMT
NC()
I
N(/P02 \
P
\-0DMT
w
00
I,
L'
Ser(GN) (phosphoramidite) OH
--1
,
oo
HO.\...o.
,,
.
,,
,
HO NH
,
0
,
NHAc
w
0 .
C3Am(GN) 0 0.,y,"....õ.õ-
-...õ,. -,
GaINAc
NH
õ.0j --
0-
od
n
GlyC3Am(GN) 0
0.y....õ..."...,..../.. ..,
GaINAc
m
od
t..)
ONH
o
oo
-a-,
,oj
--
o'
.
=
u,
CA 03118327 2021-04-30
WO 2019/092282
PCT/EP2018/081105
179
C-)
<
Z
TO
0 0
< \
Z 0
c73
<ct) 0
\
Z 0
To
0
/
0
0
Z 2
=
0 :
, /
=
\O
R R
\ ,
,
2 2 2
O 0 0
E E E
< < <
CD N=-= 0-
O 0 il